code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
import matplotlib.pyplot as plt
from matplotlib.ticker import MultipleLocator
def extractMetrics(number, result):
# 对各个引擎的统计结果画图
Submit = {"Comfort": result['COMFORT_info']['Submitted'], "DIE":result['DIE_info']['Submitted'],
"Fuzzilli":result['Fuzzilli_info']['Submitted'], "Montage": result['Montage_info']['Submitted'],
"Deepsmith": result['DeepSmith_info']['Submitted'], "CodeAlchemist": result['CodeAlchemist_info']['Submitted']}
Submitted = Submit[number]
Confirm = {"Comfort": result['COMFORT_info']['Confirmed'], "DIE":result['DIE_info']['Confirmed'],
"Fuzzilli":result['Fuzzilli_info']['Confirmed'], "Montage": result['Montage_info']['Confirmed'],
"Deepsmith": result['DeepSmith_info']['Confirmed'], "CodeAlchemist": result['CodeAlchemist_info']['Confirmed']}
Confirmed = Confirm[number]
Fix = {"Comfort": result['COMFORT_info']['Fixed'], "DIE":result['DIE_info']['Fixed'],
"Fuzzilli":result['Fuzzilli_info']['Fixed'], "Montage": result['Montage_info']['Fixed'],
"Deepsmith": result['DeepSmith_info']['Fixed'], "CodeAlchemist": result['CodeAlchemist_info']['Fixed']}
Fixed = Fix[number]
return [Submitted, Confirmed, Fixed]
def drawBars(result):
arguments = ["Submitted", "Confirmed", "Fixed"]
comfort = extractMetrics("Comfort", result)
die = extractMetrics("DIE", result)
fuzzilli = extractMetrics("Fuzzilli", result)
montage = extractMetrics("Montage", result)
deepsmith = extractMetrics("Deepsmith", result)
codeAlchemist = extractMetrics("CodeAlchemist", result)
fuzzers = [comfort, die, fuzzilli, montage, deepsmith, codeAlchemist]
fuzzer_names = ["Comfort", "DIE", "Fuzzilli", "Montage", "Deepsmith", "CodeAlchemist"]
fc = ['k', 'dimgray', 'grey', 'darkgray', 'lightgray', 'gainsboro']
x = list(range(len(comfort)))
total_width, n = 0.8, 6
width = total_width / n
# 设置主次刻度间隔
ymajorLocator = MultipleLocator(20)
yminorLocator = MultipleLocator(10)
# 设置y轴刻度值
plt.yticks([0, 20, 40, 60, 80])
plt.ylim(0, 80)
# 设置主次刻度线
plt.grid(which="major", axis="y", linestyle="-")
plt.grid(which="minor", axis="y", linestyle="--")
# 显示主次刻度
plt.gca().yaxis.set_major_locator(ymajorLocator)
plt.gca().yaxis.set_minor_locator(yminorLocator)
plt.xlabel("Bug State")
plt.ylabel("Numbers of Bugs")
# 显示柱状图
for i in range(len(fuzzers)):
if i == len(fuzzers) - 3:
# zorder越大,表示柱子越靠后,不会被虚线覆盖
plt.bar(x, fuzzers[i], width=width, label=fuzzer_names[i], tick_label=arguments, fc=fc[i], zorder=2)
else:
plt.bar(x, fuzzers[i], width=width, label=fuzzer_names[i], fc=fc[i], zorder=2)
for j in range(len(x)):
x[j] = x[j] + width
plt.legend(loc='upper center', fontsize=10, ncol=3)
plt.show()
plt.style.use('ggplot')
if __name__ == "__main__":
result = {'COMFORT_info': {'Submitted': 60, 'Confirmed': 50, 'Fixed': 48}, 'DIE_info': {'Submitted': 30, 'Confirmed': 19, 'Fixed': 9}, 'Fuzzilli_info': {'Submitted': 16, 'Confirmed': 12, 'Fixed': 9}, 'Montage_info': {'Submitted': 15, 'Confirmed': 7, 'Fixed': 5}, 'DeepSmith_info': {'Submitted': 6, 'Confirmed': 6, 'Fixed': 4}, 'CodeAlchemist_info': {'Submitted': 11, 'Confirmed': 8, 'Fixed': 5}}
drawBars(result)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
from matplotlib.ticker import MultipleLocator
def extractMetrics(number, result):
# 对各个引擎的统计结果画图
Submit = {"Comfort": result['COMFORT_info']['Submitted'], "DIE":result['DIE_info']['Submitted'],
"Fuzzilli":result['Fuzzilli_info']['Submitted'], "Montage": result['Montage_info']['Submitted'],
"Deepsmith": result['DeepSmith_info']['Submitted'], "CodeAlchemist": result['CodeAlchemist_info']['Submitted']}
Submitted = Submit[number]
Confirm = {"Comfort": result['COMFORT_info']['Confirmed'], "DIE":result['DIE_info']['Confirmed'],
"Fuzzilli":result['Fuzzilli_info']['Confirmed'], "Montage": result['Montage_info']['Confirmed'],
"Deepsmith": result['DeepSmith_info']['Confirmed'], "CodeAlchemist": result['CodeAlchemist_info']['Confirmed']}
Confirmed = Confirm[number]
Fix = {"Comfort": result['COMFORT_info']['Fixed'], "DIE":result['DIE_info']['Fixed'],
"Fuzzilli":result['Fuzzilli_info']['Fixed'], "Montage": result['Montage_info']['Fixed'],
"Deepsmith": result['DeepSmith_info']['Fixed'], "CodeAlchemist": result['CodeAlchemist_info']['Fixed']}
Fixed = Fix[number]
return [Submitted, Confirmed, Fixed]
def drawBars(result):
arguments = ["Submitted", "Confirmed", "Fixed"]
comfort = extractMetrics("Comfort", result)
die = extractMetrics("DIE", result)
fuzzilli = extractMetrics("Fuzzilli", result)
montage = extractMetrics("Montage", result)
deepsmith = extractMetrics("Deepsmith", result)
codeAlchemist = extractMetrics("CodeAlchemist", result)
fuzzers = [comfort, die, fuzzilli, montage, deepsmith, codeAlchemist]
fuzzer_names = ["Comfort", "DIE", "Fuzzilli", "Montage", "Deepsmith", "CodeAlchemist"]
fc = ['k', 'dimgray', 'grey', 'darkgray', 'lightgray', 'gainsboro']
x = list(range(len(comfort)))
total_width, n = 0.8, 6
width = total_width / n
# 设置主次刻度间隔
ymajorLocator = MultipleLocator(20)
yminorLocator = MultipleLocator(10)
# 设置y轴刻度值
plt.yticks([0, 20, 40, 60, 80])
plt.ylim(0, 80)
# 设置主次刻度线
plt.grid(which="major", axis="y", linestyle="-")
plt.grid(which="minor", axis="y", linestyle="--")
# 显示主次刻度
plt.gca().yaxis.set_major_locator(ymajorLocator)
plt.gca().yaxis.set_minor_locator(yminorLocator)
plt.xlabel("Bug State")
plt.ylabel("Numbers of Bugs")
# 显示柱状图
for i in range(len(fuzzers)):
if i == len(fuzzers) - 3:
# zorder越大,表示柱子越靠后,不会被虚线覆盖
plt.bar(x, fuzzers[i], width=width, label=fuzzer_names[i], tick_label=arguments, fc=fc[i], zorder=2)
else:
plt.bar(x, fuzzers[i], width=width, label=fuzzer_names[i], fc=fc[i], zorder=2)
for j in range(len(x)):
x[j] = x[j] + width
plt.legend(loc='upper center', fontsize=10, ncol=3)
plt.show()
plt.style.use('ggplot')
if __name__ == "__main__":
result = {'COMFORT_info': {'Submitted': 60, 'Confirmed': 50, 'Fixed': 48}, 'DIE_info': {'Submitted': 30, 'Confirmed': 19, 'Fixed': 9}, 'Fuzzilli_info': {'Submitted': 16, 'Confirmed': 12, 'Fixed': 9}, 'Montage_info': {'Submitted': 15, 'Confirmed': 7, 'Fixed': 5}, 'DeepSmith_info': {'Submitted': 6, 'Confirmed': 6, 'Fixed': 4}, 'CodeAlchemist_info': {'Submitted': 11, 'Confirmed': 8, 'Fixed': 5}}
drawBars(result)
| 0.194253 | 0.648821 |
# Creating a Filter, Edge Detection
### Import resources and display image
```
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import cv2
import numpy as np
%matplotlib inline
# Read in the image
image = mpimg.imread('data/curved_lane.jpg')
plt.imshow(image)
```
### Convert the image to grayscale
```
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
plt.imshow(gray, cmap='gray')
```
### TODO: Create a custom kernel
Below, you've been given one common type of edge detection filter: a Sobel operator.
The Sobel filter is very commonly used in edge detection and in finding patterns in intensity in an image. Applying a Sobel filter to an image is a way of **taking (an approximation) of the derivative of the image** in the x or y direction, separately. The operators look as follows.
<img src="images/sobel_ops.png" width=200 height=200>
**It's up to you to create a Sobel x operator and apply it to the given image.**
For a challenge, see if you can put the image through a series of filters: first one that blurs the image (takes an average of pixels), and then one that detects the edges.
```
# Create a custom kernel
# 3x3 array for edge detection
sobel_y = np.array([[ -1, -2, -1],
[ 0, 0, 0],
[ 1, 2, 1]])
## TODO: Create and apply a Sobel x operator
sobel_x = np.array([[-1, 0, 1],
[-2, 0, 2],
[-1, 0, 1]])
# Filter the image using filter2D, which has inputs: (grayscale image, bit-depth, kernel)
filtered_image = cv2.filter2D(gray, -1, sobel_y)
filtered_image = cv2.filter2D(gray, -1, sobel_x)
plt.imshow(filtered_image, cmap='gray')
```
### Test out other filters!
You're encouraged to create other kinds of filters and apply them to see what happens! As an **optional exercise**, try the following:
* Create a filter with decimal value weights.
* Create a 5x5 filter
* Apply your filters to the other images in the `images` directory.
```
image = mpimg.imread('data/udacity_sdc.png')
plt.imshow(image)
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
plt.imshow(gray, cmap='gray')
# Filters
custom_filter_1 = np.array([[0, -10.8, 0],
[-10.8, 0, 10.8],
[0, 10.8, 0]])
custom_filter_2 = np.array([[-4, -2, 0, 2, 4],
[-6, -3, 0, 3, 6],
[-4, -2, 0, 2, 4]])
# Apply filter 1
filtered_image = cv2.filter2D(gray, -1, custom_filter_1)
plt.imshow(filtered_image, cmap='gray')
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
filtered_image = cv2.filter2D(gray, -1, custom_filter_2)
plt.imshow(filtered_image, cmap='gray')
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import cv2
import numpy as np
%matplotlib inline
# Read in the image
image = mpimg.imread('data/curved_lane.jpg')
plt.imshow(image)
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
plt.imshow(gray, cmap='gray')
# Create a custom kernel
# 3x3 array for edge detection
sobel_y = np.array([[ -1, -2, -1],
[ 0, 0, 0],
[ 1, 2, 1]])
## TODO: Create and apply a Sobel x operator
sobel_x = np.array([[-1, 0, 1],
[-2, 0, 2],
[-1, 0, 1]])
# Filter the image using filter2D, which has inputs: (grayscale image, bit-depth, kernel)
filtered_image = cv2.filter2D(gray, -1, sobel_y)
filtered_image = cv2.filter2D(gray, -1, sobel_x)
plt.imshow(filtered_image, cmap='gray')
image = mpimg.imread('data/udacity_sdc.png')
plt.imshow(image)
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
plt.imshow(gray, cmap='gray')
# Filters
custom_filter_1 = np.array([[0, -10.8, 0],
[-10.8, 0, 10.8],
[0, 10.8, 0]])
custom_filter_2 = np.array([[-4, -2, 0, 2, 4],
[-6, -3, 0, 3, 6],
[-4, -2, 0, 2, 4]])
# Apply filter 1
filtered_image = cv2.filter2D(gray, -1, custom_filter_1)
plt.imshow(filtered_image, cmap='gray')
# Convert to grayscale for filtering
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
filtered_image = cv2.filter2D(gray, -1, custom_filter_2)
plt.imshow(filtered_image, cmap='gray')
| 0.328206 | 0.989161 |
Installing pre-requisites
```
!pip install pytorch-metric-learning
!pip install faiss-gpu
from google.colab import drive
drive.mount('/content/drive')
```
The notebook is divided into 3 parts:
- In the first part we import the images and create the dataframe
- In the second part we define our embedding network and train it on the given images
- In the last part we use the embedding network to create a classifier and use it to classify the given images
## Part 1: Data import and dataframe creation
Importing important libraries for the section
```
import numpy as np
import pandas as pd
import os
from PIL import Image
import torch
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from torch.utils.data import Dataset
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
```
Just like the previous section, we will repeat the same steps for dataset creation
Replace the path below with the path to the folder containing the 0-9 folders from Task 1
```
pathToDataset = '/content/drive/MyDrive/MIDAS/TASK 2/train'
imagePath = []
labels = []
for folder in os.listdir(pathToDataset):
for images in os.listdir(os.path.join(pathToDataset,folder)):
image = os.path.join(pathToDataset,folder,images)
imagePath.append(image)
labels.append(folder)
data = {'Images':imagePath, 'Labels':labels}
data = pd.DataFrame(data)
data.head()
labelEncoder = LabelEncoder()
data['Encoded Labels'] = labelEncoder.fit_transform(data['Labels'])
data.head()
batchSize = 128
validationSplit = 0.15
shuffleDataset = True
randomSeed = 17
datasetSize = len(data)
indices = list(range(datasetSize))
split = int(np.floor(validationSplit*datasetSize))
if shuffleDataset:
np.random.seed(randomSeed)
np.random.shuffle(indices)
trainIndices, validationIndices = indices[split:], indices[:split]
trainSampler = SubsetRandomSampler(trainIndices)
validationSampler = SubsetRandomSampler(validationIndices)
class CustomDataset(Dataset):
def __init__(self, imageData, imagePath, transform=None):
self.imagePath = imagePath
self.imageData = imageData
self.transform = transform
def __len__(self):
return len(self.imageData)
def __getitem__(self, index):
imageName = os.path.join(self.imagePath, self.imageData.loc[index, 'Labels'],self.imageData.loc[index,'Images'])
image = Image.open(imageName).convert('L')
image = image.resize((32,32))
label = torch.tensor(self.imageData.loc[index, 'Encoded Labels'])
if self.transform is not None:
image = self.transform(image)
return image,label
transform = transforms.Compose(
[transforms.Resize(220),
transforms.ToTensor(),
transforms.Normalize(mean=[0.5], std=[0.5])])
dataset = CustomDataset(data,pathToDataset,transform)
trainLoader = torch.utils.data.DataLoader(dataset, batch_size = batchSize, sampler = trainSampler)
validationLoader = torch.utils.data.DataLoader(dataset, batch_size = batchSize, sampler = validationSampler)
def displayImage(image):
image = image/2 + 0.5
image = image.numpy()
image = image.reshape(220,220)
return image
dataIterator = iter(trainLoader)
images, labels = dataIterator.next()
figure, axis = plt.subplots(3,5, figsize=(16,16))
for i, ax in enumerate(axis.flat):
with torch.no_grad():
image, label = images[i], labels[i]
ax.imshow(displayImage(image))
ax.set(title=f"{label.item()}")
```
## Part 2: Classification (Pre-training before MNIST)
In this part we first pre-train our model on the provided dataset and then fine-tune it on the MNIST dataset.
```
#the pytorch metric learning library comes with inbuilt methods for triplet mining and computing triplet losses between anchor, positive class and negative class
from pytorch_metric_learning import losses, miners
from pytorch_metric_learning.distances import CosineSimilarity
from pytorch_metric_learning.reducers import ThresholdReducer
from pytorch_metric_learning.regularizers import LpRegularizer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import accuracy_score
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision.models as models
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
print("Training the model on: ",device, " Available GPU: ", torch.cuda.get_device_name(0))
```
This time we shall be using a much simpler neural network as opposed to the previous task, as the features to be extracted for Numerical pictures are much less.
```
class EmbeddingNetwork(nn.Module):
def __init__(self):
super(EmbeddingNetwork, self).__init__()
self.conv1 = nn.Sequential(
nn.Conv2d(1, 64, (7,7), stride=(2,2), padding=(3,3)),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3, 3), 2, padding=(1,1))
)
self.conv2 = nn.Sequential(
nn.Conv2d(64,64,(1,1), stride=(1,1)),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.001),
nn.Conv2d(64,192, (3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(192),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3),2, padding=(1,1))
)
self.conv3 = nn.Sequential(
nn.Conv2d(192,192,(1,1), stride=(1,1)),
nn.BatchNorm2d(192),
nn.LeakyReLU(0.001),
nn.Conv2d(192,384,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(384),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3), 2, padding=(1,1))
)
self.conv4 = nn.Sequential(
nn.Conv2d(384,384,(1,1), stride=(1,1)),
nn.BatchNorm2d(384),
nn.LeakyReLU(0.001),
nn.Conv2d(384,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001)
)
self.conv5 = nn.Sequential(
nn.Conv2d(256,256,(1,1), stride=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.Conv2d(256,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001)
)
self.conv6 = nn.Sequential(
nn.Conv2d(256,256,(1,1), stride=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.Conv2d(256,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3),2, padding=(1,1)),
nn.Flatten()
)
self.fullyConnected = nn.Sequential(
nn.Linear(7*7*256,32*128),
nn.BatchNorm1d(32*128),
nn.LeakyReLU(0.001),
nn.Linear(32*128,128)
)
def forward(self,x):
x = self.conv1(x)
x = self.conv2(x)
x = self.conv3(x)
x = self.conv4(x)
x = self.conv5(x)
x = self.conv6(x)
x = self.fullyConnected(x)
return torch.nn.functional.normalize(x, p=2, dim=-1)
embeddingNetwork = EmbeddingNetwork().to(device)
print(embeddingNetwork)
```
Trainer function similar to SUB TASK 1
```
def train(model, lossFunction, miningFunction, device, trainLoader, optimizer, epoch):
print("Training started for Epoch: ",epoch)
model.train()
for batchIndex, (data, labels) in enumerate(trainLoader):
data, labels = data.to(device), labels.to(device)
optimizer.zero_grad()
embeddings = model(data)
hardPairs = miningFunction(embeddings, labels)
loss = lossFunction(embeddings, labels, hardPairs)
loss.backward()
optimizer.step()
if batchIndex%3==0:
print("Training Stats for Epoch {} Iteration {}: Loss= {}, Number of mined triplets {}".format(epoch, batchIndex, loss, miningFunction.num_triplets))
```
Defining loss function and other parameters
```
#distance this tells the model how to calculate the distance between the generated embeddings
distance = CosineSimilarity()
reducer = ThresholdReducer(low=0.0)
lossFunction = losses.TripletMarginLoss(margin = 0.2, distance = distance, reducer = reducer)
miningFunction = miners.TripletMarginMiner(margin = 0.2, distance = distance, type_of_triplets = "semi-hard")
optimizer = torch.optim.Adam(embeddingNetwork.parameters(), lr=0.05)
```
Standard test function using a KNN Classifier to test the embedding model
```
def tester(maxValidationAccuracy):
trainEmbeddings = []
trainLabels = []
validationEmbeddings = []
validationLabels = []
with torch.no_grad():
embeddingNetwork.eval()
for (dataTr, labelTr) in (trainLoader):
dataTr, labelTr = dataTr.to(device), labelTr.to(device)
embeddingTr = embeddingNetwork(dataTr)
trainEmbeddings.append(embeddingTr.cpu().detach().numpy())
trainLabels.append(labelTr.cpu().detach().numpy())
for (dataTe, labelTe) in (validationLoader):
dataTe, labelTe = dataTe.to(device), labelTe.to(device)
embeddingsTe = embeddingNetwork(dataTe)
validationEmbeddings.append(embeddingsTe.cpu().detach().numpy())
validationLabels.append(labelTe.cpu().detach().numpy())
trainEmbeddings1 = []
trainLabels1 = []
validationEmbeddings1 = []
validationLabels1 = []
for bat in trainEmbeddings:
for exm in bat:
trainEmbeddings1.append(exm)
for bat in trainLabels:
for exm in bat:
trainLabels1.append(exm)
for bat in validationEmbeddings:
for exm in bat:
validationEmbeddings1.append(exm)
for bat in validationLabels:
for exm in bat:
validationLabels1.append(exm)
neigh = KNeighborsClassifier(n_neighbors=13)
neigh.fit(trainEmbeddings1, trainLabels1)
prediction = neigh.predict(validationEmbeddings1)
currentAccuracy = accuracy_score(validationLabels1,prediction)
print("Accuracy: ",currentAccuracy)
if currentAccuracy > maxValidationAccuracy:
maxValidationAccuracy = currentAccuracy
print("New highest validation accuracy, saving the embedding model")
torch.save(embeddingNetwork.state_dict(), "embeddingNetworkTask2.pt")
return maxValidationAccuracy
```
Now that our training and testing functions are ready, we shall pre-train our model on the given dataset. We also save the model having highest validation accuracy, these saved weights will be used to fine-tune the MNIST dataset and make further inferences. This is done below:
```
maxValidationAccuracy = 0
for epoch in range(1, 81):
train(embeddingNetwork, lossFunction, miningFunction, device, trainLoader, optimizer, epoch)
print("Training completed for the Epoch:", epoch)
maxValidationAccuracy = tester(maxValidationAccuracy)
print("Highest Validation Accuracy acheived during training: ", maxValidationAccuracy)
```
Loading the standard MNIST training and validation sets
```
from torchvision import datasets
mnistTrain = datasets.MNIST('.', train=True, download=True, transform=transform)
mnistTest = datasets.MNIST('.', train=False, transform=transform)
mnistTrainLoader = torch.utils.data.DataLoader(mnistTrain, batch_size=128, shuffle=True)
mnistTestLoader = torch.utils.data.DataLoader(mnistTest, batch_size=128)
```
Visualizing MNIST Train Images
```
dataIterator = iter(mnistTrainLoader)
images, labels = dataIterator.next()
figure, axis = plt.subplots(3,5, figsize=(16,16))
for i, ax in enumerate(axis.flat):
with torch.no_grad():
image, label = images[i], labels[i]
ax.imshow(displayImage(image))
ax.set(title=f"{label.item()}")
```
Now that our MNIST datasets are ready we shall fine-tune and test the model for MNIST dataset. Before we create the classifier, we shall fine tune the embedding network on the MNIST data too.
```
#loading best validated weights into the classifier
embeddingNetwork = EmbeddingNetwork().to(device)
embeddingNetwork.load_state_dict(torch.load('embeddingNetworkTask2.pt'))
def trainMNIST(model, lossFunction, miningFunction, device, trainLoader, optimizer, epoch):
print("Training started for Epoch: ",epoch)
model.train()
for batchIndex, (data, labels) in enumerate(trainLoader):
data, labels = data.to(device), labels.to(device)
optimizer.zero_grad()
embeddings = model(data)
hardPairs = miningFunction(embeddings, labels)
loss = lossFunction(embeddings, labels, hardPairs)
loss.backward()
optimizer.step()
if batchIndex%300==0:
print("Training Stats for Epoch {} Iteration {}: Loss= {}, Number of mined triplets {}".format(epoch, batchIndex, loss, miningFunction.num_triplets))
#distance this tells the model how to calculate the distance between the generated embeddings
distance = CosineSimilarity()
reducer = ThresholdReducer(low=0.0)
lossFunction = losses.TripletMarginLoss(margin = 0.2, distance = distance, reducer = reducer)
miningFunction = miners.TripletMarginMiner(margin = 0.2, distance = distance, type_of_triplets = "semi-hard")
optimizer = torch.optim.Adam(embeddingNetwork.parameters(), lr=0.05)
maxValidationAccuracyMNIST = 0.0
def testerMNIST(maxValidationAccuracyMNIST):
trainEmbeddings = []
trainLabels = []
validationEmbeddings = []
validationLabels = []
with torch.no_grad():
embeddingNetwork.eval()
for (dataTr, labelTr) in (mnistTrainLoader):
dataTr, labelTr = dataTr.to(device), labelTr.to(device)
embeddingTr = embeddingNetwork(dataTr)
trainEmbeddings.append(embeddingTr.cpu().detach().numpy())
trainLabels.append(labelTr.cpu().detach().numpy())
for (dataTe, labelTe) in (mnistTestLoader):
dataTe, labelTe = dataTe.to(device), labelTe.to(device)
embeddingsTe = embeddingNetwork(dataTe)
validationEmbeddings.append(embeddingsTe.cpu().detach().numpy())
validationLabels.append(labelTe.cpu().detach().numpy())
trainEmbeddings1 = []
trainLabels1 = []
validationEmbeddings1 = []
validationLabels1 = []
for bat in trainEmbeddings:
for exm in bat:
trainEmbeddings1.append(exm)
for bat in trainLabels:
for exm in bat:
trainLabels1.append(exm)
for bat in validationEmbeddings:
for exm in bat:
validationEmbeddings1.append(exm)
for bat in validationLabels:
for exm in bat:
validationLabels1.append(exm)
neigh = KNeighborsClassifier(n_neighbors=13)
neigh.fit(trainEmbeddings1, trainLabels1)
prediction = neigh.predict(validationEmbeddings1)
currentAccuracy = accuracy_score(validationLabels1,prediction)
print("Accuracy: ",currentAccuracy)
if currentAccuracy > maxValidationAccuracyMNIST:
maxValidationAccuracyMNIST = currentAccuracy
print("New highest validation accuracy, saving the embedding model")
torch.save(embeddingNetwork.state_dict(), "embeddingNetworkMNIST.pt")
return maxValidationAccuracyMNIST
maxValidationAccuracyMNIST = 0
for epoch in range(1, 5):
trainMNIST(embeddingNetwork, lossFunction, miningFunction, device, mnistTrainLoader, optimizer, epoch)
print("Training completed for the Epoch:", epoch)
maxValidationAccuracyMNIST = testerMNIST(maxValidationAccuracyMNIST)
```
Classifying the MNIST Dataset using pre-trained model
```
class classifierNet(nn.Module):
def __init__(self, EmbeddingNet):
super(classifierNet, self).__init__()
self.embeddingLayer = EmbeddingNet
self.linearLayer = nn.Sequential(nn.Linear(128, 64), nn.ReLU())
self.classifierLayer = nn.Linear(64,10)
self.dropout = nn.Dropout(0.5)
def forward(self, x):
x = self.dropout(self.embeddingLayer(x))
x = self.dropout(self.linearLayer(x))
x = self.classifierLayer(x)
return F.log_softmax(x, dim=1)
bestEmbeddingNetwork = EmbeddingNetwork().to(device)
bestEmbeddingNetwork.load_state_dict(torch.load('embeddingNetworkMNIST.pt'))
classifier = classifierNet(embeddingNetwork).to(device)
print(classifier)
for param in classifier.embeddingLayer.parameters():
param.requires_grad = False
```
Defining the NNL Loss function for classification as we are using log softmax layer in the end.
```
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(classifier.parameters(), lr=0.01)
def accuracy(output, labels):
_, predictions = torch.max(output, dim=1)
return torch.sum(predictions==labels).item()
numberOfEpochs = 20
validAccuracyMaxTransfer = 0.0
validationLossTransfer = []
validationAccuracyTransfer = []
trainingLossTransfer = []
trainingAccuracyTransfer = []
totalSteps = len(mnistTrainLoader)
for epoch in range(1, numberOfEpochs):
classifier.train()
runningLoss = 0.0
correct = 0
total = 0
print("Training started for Epoch: ",epoch)
for batchIndex, (data, target) in enumerate(mnistTrainLoader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
outputs = classifier(data)
loss = criterion(outputs,target)
loss.backward()
optimizer.step()
runningLoss += loss.item()
_, pred = torch.max(outputs, dim=1)
correct += torch.sum(pred==target).item()
total += target.size(0)
if (batchIndex)%100 ==0:
print("Epoch [{}/{}] Step [{}/{}] Loss: {:.4f}".format(epoch,numberOfEpochs,batchIndex,totalSteps,loss.item()))
trainingAccuracyTransfer.append(100*correct/total)
trainingLossTransfer.append(runningLoss/totalSteps)
print("Training Accuracy: ",(100*correct/total))
batchLoss = 0
totalV = 0
correctV = 0
with torch.no_grad():
classifier.eval()
for dataV, targetV in (mnistTestLoader):
dataV, targetV = dataV.to(device), targetV.to(device)
outputV = classifier(dataV)
lossV = criterion(outputV,targetV)
batchLoss += lossV.item()
_, predV = torch.max(outputV, dim=1)
correctV += torch.sum(predV==targetV).item()
totalV += targetV.size(0)
validationAccuracyTransfer.append(100*correctV/totalV)
validationLossTransfer.append(batchLoss/len(mnistTestLoader))
print("Validation Accuracy: ",(100*correctV/totalV))
if (100*correctV/totalV)>validAccuracyMaxTransfer:
validAccuracyMaxTransfer = 100*correctV/totalV
print("Validation accuracy improved, network improvement detected, saving network")
torch.save(classifier.state_dict(), "classifierNetworkTransferLearningTask2.pt")
classifier.train()
fig = plt.figure(figsize=(20,10))
plt.title("Transfer Learning Model Train-Validation Loss Plot")
plt.plot(trainingLossTransfer, label='training loss')
plt.plot(validationLossTransfer, label='validation loss')
plt.xlabel('Number of epochs')
plt.ylabel('Loss')
plt.legend(loc='best')
fig = plt.figure(figsize=(20,10))
plt.title("Transfer Learning Train-Validation Accuracy Plot")
plt.plot(trainingAccuracyTransfer, label='training accuracy')
plt.plot(validationAccuracyTransfer, label='validation accuracy')
plt.xlabel('Number of epochs')
plt.ylabel('Accuracy')
plt.legend(loc='best')
```
|
github_jupyter
|
!pip install pytorch-metric-learning
!pip install faiss-gpu
from google.colab import drive
drive.mount('/content/drive')
import numpy as np
import pandas as pd
import os
from PIL import Image
import torch
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from torch.utils.data import Dataset
import torchvision.transforms as transforms
from torch.utils.data.sampler import SubsetRandomSampler
pathToDataset = '/content/drive/MyDrive/MIDAS/TASK 2/train'
imagePath = []
labels = []
for folder in os.listdir(pathToDataset):
for images in os.listdir(os.path.join(pathToDataset,folder)):
image = os.path.join(pathToDataset,folder,images)
imagePath.append(image)
labels.append(folder)
data = {'Images':imagePath, 'Labels':labels}
data = pd.DataFrame(data)
data.head()
labelEncoder = LabelEncoder()
data['Encoded Labels'] = labelEncoder.fit_transform(data['Labels'])
data.head()
batchSize = 128
validationSplit = 0.15
shuffleDataset = True
randomSeed = 17
datasetSize = len(data)
indices = list(range(datasetSize))
split = int(np.floor(validationSplit*datasetSize))
if shuffleDataset:
np.random.seed(randomSeed)
np.random.shuffle(indices)
trainIndices, validationIndices = indices[split:], indices[:split]
trainSampler = SubsetRandomSampler(trainIndices)
validationSampler = SubsetRandomSampler(validationIndices)
class CustomDataset(Dataset):
def __init__(self, imageData, imagePath, transform=None):
self.imagePath = imagePath
self.imageData = imageData
self.transform = transform
def __len__(self):
return len(self.imageData)
def __getitem__(self, index):
imageName = os.path.join(self.imagePath, self.imageData.loc[index, 'Labels'],self.imageData.loc[index,'Images'])
image = Image.open(imageName).convert('L')
image = image.resize((32,32))
label = torch.tensor(self.imageData.loc[index, 'Encoded Labels'])
if self.transform is not None:
image = self.transform(image)
return image,label
transform = transforms.Compose(
[transforms.Resize(220),
transforms.ToTensor(),
transforms.Normalize(mean=[0.5], std=[0.5])])
dataset = CustomDataset(data,pathToDataset,transform)
trainLoader = torch.utils.data.DataLoader(dataset, batch_size = batchSize, sampler = trainSampler)
validationLoader = torch.utils.data.DataLoader(dataset, batch_size = batchSize, sampler = validationSampler)
def displayImage(image):
image = image/2 + 0.5
image = image.numpy()
image = image.reshape(220,220)
return image
dataIterator = iter(trainLoader)
images, labels = dataIterator.next()
figure, axis = plt.subplots(3,5, figsize=(16,16))
for i, ax in enumerate(axis.flat):
with torch.no_grad():
image, label = images[i], labels[i]
ax.imshow(displayImage(image))
ax.set(title=f"{label.item()}")
#the pytorch metric learning library comes with inbuilt methods for triplet mining and computing triplet losses between anchor, positive class and negative class
from pytorch_metric_learning import losses, miners
from pytorch_metric_learning.distances import CosineSimilarity
from pytorch_metric_learning.reducers import ThresholdReducer
from pytorch_metric_learning.regularizers import LpRegularizer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import accuracy_score
import torch
import torch.nn as nn
import torch.nn.functional as F
import torchvision.models as models
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
print("Training the model on: ",device, " Available GPU: ", torch.cuda.get_device_name(0))
class EmbeddingNetwork(nn.Module):
def __init__(self):
super(EmbeddingNetwork, self).__init__()
self.conv1 = nn.Sequential(
nn.Conv2d(1, 64, (7,7), stride=(2,2), padding=(3,3)),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3, 3), 2, padding=(1,1))
)
self.conv2 = nn.Sequential(
nn.Conv2d(64,64,(1,1), stride=(1,1)),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.001),
nn.Conv2d(64,192, (3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(192),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3),2, padding=(1,1))
)
self.conv3 = nn.Sequential(
nn.Conv2d(192,192,(1,1), stride=(1,1)),
nn.BatchNorm2d(192),
nn.LeakyReLU(0.001),
nn.Conv2d(192,384,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(384),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3), 2, padding=(1,1))
)
self.conv4 = nn.Sequential(
nn.Conv2d(384,384,(1,1), stride=(1,1)),
nn.BatchNorm2d(384),
nn.LeakyReLU(0.001),
nn.Conv2d(384,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001)
)
self.conv5 = nn.Sequential(
nn.Conv2d(256,256,(1,1), stride=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.Conv2d(256,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001)
)
self.conv6 = nn.Sequential(
nn.Conv2d(256,256,(1,1), stride=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.Conv2d(256,256,(3,3), stride=(1,1), padding=(1,1)),
nn.BatchNorm2d(256),
nn.LeakyReLU(0.001),
nn.MaxPool2d((3,3),2, padding=(1,1)),
nn.Flatten()
)
self.fullyConnected = nn.Sequential(
nn.Linear(7*7*256,32*128),
nn.BatchNorm1d(32*128),
nn.LeakyReLU(0.001),
nn.Linear(32*128,128)
)
def forward(self,x):
x = self.conv1(x)
x = self.conv2(x)
x = self.conv3(x)
x = self.conv4(x)
x = self.conv5(x)
x = self.conv6(x)
x = self.fullyConnected(x)
return torch.nn.functional.normalize(x, p=2, dim=-1)
embeddingNetwork = EmbeddingNetwork().to(device)
print(embeddingNetwork)
def train(model, lossFunction, miningFunction, device, trainLoader, optimizer, epoch):
print("Training started for Epoch: ",epoch)
model.train()
for batchIndex, (data, labels) in enumerate(trainLoader):
data, labels = data.to(device), labels.to(device)
optimizer.zero_grad()
embeddings = model(data)
hardPairs = miningFunction(embeddings, labels)
loss = lossFunction(embeddings, labels, hardPairs)
loss.backward()
optimizer.step()
if batchIndex%3==0:
print("Training Stats for Epoch {} Iteration {}: Loss= {}, Number of mined triplets {}".format(epoch, batchIndex, loss, miningFunction.num_triplets))
#distance this tells the model how to calculate the distance between the generated embeddings
distance = CosineSimilarity()
reducer = ThresholdReducer(low=0.0)
lossFunction = losses.TripletMarginLoss(margin = 0.2, distance = distance, reducer = reducer)
miningFunction = miners.TripletMarginMiner(margin = 0.2, distance = distance, type_of_triplets = "semi-hard")
optimizer = torch.optim.Adam(embeddingNetwork.parameters(), lr=0.05)
def tester(maxValidationAccuracy):
trainEmbeddings = []
trainLabels = []
validationEmbeddings = []
validationLabels = []
with torch.no_grad():
embeddingNetwork.eval()
for (dataTr, labelTr) in (trainLoader):
dataTr, labelTr = dataTr.to(device), labelTr.to(device)
embeddingTr = embeddingNetwork(dataTr)
trainEmbeddings.append(embeddingTr.cpu().detach().numpy())
trainLabels.append(labelTr.cpu().detach().numpy())
for (dataTe, labelTe) in (validationLoader):
dataTe, labelTe = dataTe.to(device), labelTe.to(device)
embeddingsTe = embeddingNetwork(dataTe)
validationEmbeddings.append(embeddingsTe.cpu().detach().numpy())
validationLabels.append(labelTe.cpu().detach().numpy())
trainEmbeddings1 = []
trainLabels1 = []
validationEmbeddings1 = []
validationLabels1 = []
for bat in trainEmbeddings:
for exm in bat:
trainEmbeddings1.append(exm)
for bat in trainLabels:
for exm in bat:
trainLabels1.append(exm)
for bat in validationEmbeddings:
for exm in bat:
validationEmbeddings1.append(exm)
for bat in validationLabels:
for exm in bat:
validationLabels1.append(exm)
neigh = KNeighborsClassifier(n_neighbors=13)
neigh.fit(trainEmbeddings1, trainLabels1)
prediction = neigh.predict(validationEmbeddings1)
currentAccuracy = accuracy_score(validationLabels1,prediction)
print("Accuracy: ",currentAccuracy)
if currentAccuracy > maxValidationAccuracy:
maxValidationAccuracy = currentAccuracy
print("New highest validation accuracy, saving the embedding model")
torch.save(embeddingNetwork.state_dict(), "embeddingNetworkTask2.pt")
return maxValidationAccuracy
maxValidationAccuracy = 0
for epoch in range(1, 81):
train(embeddingNetwork, lossFunction, miningFunction, device, trainLoader, optimizer, epoch)
print("Training completed for the Epoch:", epoch)
maxValidationAccuracy = tester(maxValidationAccuracy)
print("Highest Validation Accuracy acheived during training: ", maxValidationAccuracy)
from torchvision import datasets
mnistTrain = datasets.MNIST('.', train=True, download=True, transform=transform)
mnistTest = datasets.MNIST('.', train=False, transform=transform)
mnistTrainLoader = torch.utils.data.DataLoader(mnistTrain, batch_size=128, shuffle=True)
mnistTestLoader = torch.utils.data.DataLoader(mnistTest, batch_size=128)
dataIterator = iter(mnistTrainLoader)
images, labels = dataIterator.next()
figure, axis = plt.subplots(3,5, figsize=(16,16))
for i, ax in enumerate(axis.flat):
with torch.no_grad():
image, label = images[i], labels[i]
ax.imshow(displayImage(image))
ax.set(title=f"{label.item()}")
#loading best validated weights into the classifier
embeddingNetwork = EmbeddingNetwork().to(device)
embeddingNetwork.load_state_dict(torch.load('embeddingNetworkTask2.pt'))
def trainMNIST(model, lossFunction, miningFunction, device, trainLoader, optimizer, epoch):
print("Training started for Epoch: ",epoch)
model.train()
for batchIndex, (data, labels) in enumerate(trainLoader):
data, labels = data.to(device), labels.to(device)
optimizer.zero_grad()
embeddings = model(data)
hardPairs = miningFunction(embeddings, labels)
loss = lossFunction(embeddings, labels, hardPairs)
loss.backward()
optimizer.step()
if batchIndex%300==0:
print("Training Stats for Epoch {} Iteration {}: Loss= {}, Number of mined triplets {}".format(epoch, batchIndex, loss, miningFunction.num_triplets))
#distance this tells the model how to calculate the distance between the generated embeddings
distance = CosineSimilarity()
reducer = ThresholdReducer(low=0.0)
lossFunction = losses.TripletMarginLoss(margin = 0.2, distance = distance, reducer = reducer)
miningFunction = miners.TripletMarginMiner(margin = 0.2, distance = distance, type_of_triplets = "semi-hard")
optimizer = torch.optim.Adam(embeddingNetwork.parameters(), lr=0.05)
maxValidationAccuracyMNIST = 0.0
def testerMNIST(maxValidationAccuracyMNIST):
trainEmbeddings = []
trainLabels = []
validationEmbeddings = []
validationLabels = []
with torch.no_grad():
embeddingNetwork.eval()
for (dataTr, labelTr) in (mnistTrainLoader):
dataTr, labelTr = dataTr.to(device), labelTr.to(device)
embeddingTr = embeddingNetwork(dataTr)
trainEmbeddings.append(embeddingTr.cpu().detach().numpy())
trainLabels.append(labelTr.cpu().detach().numpy())
for (dataTe, labelTe) in (mnistTestLoader):
dataTe, labelTe = dataTe.to(device), labelTe.to(device)
embeddingsTe = embeddingNetwork(dataTe)
validationEmbeddings.append(embeddingsTe.cpu().detach().numpy())
validationLabels.append(labelTe.cpu().detach().numpy())
trainEmbeddings1 = []
trainLabels1 = []
validationEmbeddings1 = []
validationLabels1 = []
for bat in trainEmbeddings:
for exm in bat:
trainEmbeddings1.append(exm)
for bat in trainLabels:
for exm in bat:
trainLabels1.append(exm)
for bat in validationEmbeddings:
for exm in bat:
validationEmbeddings1.append(exm)
for bat in validationLabels:
for exm in bat:
validationLabels1.append(exm)
neigh = KNeighborsClassifier(n_neighbors=13)
neigh.fit(trainEmbeddings1, trainLabels1)
prediction = neigh.predict(validationEmbeddings1)
currentAccuracy = accuracy_score(validationLabels1,prediction)
print("Accuracy: ",currentAccuracy)
if currentAccuracy > maxValidationAccuracyMNIST:
maxValidationAccuracyMNIST = currentAccuracy
print("New highest validation accuracy, saving the embedding model")
torch.save(embeddingNetwork.state_dict(), "embeddingNetworkMNIST.pt")
return maxValidationAccuracyMNIST
maxValidationAccuracyMNIST = 0
for epoch in range(1, 5):
trainMNIST(embeddingNetwork, lossFunction, miningFunction, device, mnistTrainLoader, optimizer, epoch)
print("Training completed for the Epoch:", epoch)
maxValidationAccuracyMNIST = testerMNIST(maxValidationAccuracyMNIST)
class classifierNet(nn.Module):
def __init__(self, EmbeddingNet):
super(classifierNet, self).__init__()
self.embeddingLayer = EmbeddingNet
self.linearLayer = nn.Sequential(nn.Linear(128, 64), nn.ReLU())
self.classifierLayer = nn.Linear(64,10)
self.dropout = nn.Dropout(0.5)
def forward(self, x):
x = self.dropout(self.embeddingLayer(x))
x = self.dropout(self.linearLayer(x))
x = self.classifierLayer(x)
return F.log_softmax(x, dim=1)
bestEmbeddingNetwork = EmbeddingNetwork().to(device)
bestEmbeddingNetwork.load_state_dict(torch.load('embeddingNetworkMNIST.pt'))
classifier = classifierNet(embeddingNetwork).to(device)
print(classifier)
for param in classifier.embeddingLayer.parameters():
param.requires_grad = False
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(classifier.parameters(), lr=0.01)
def accuracy(output, labels):
_, predictions = torch.max(output, dim=1)
return torch.sum(predictions==labels).item()
numberOfEpochs = 20
validAccuracyMaxTransfer = 0.0
validationLossTransfer = []
validationAccuracyTransfer = []
trainingLossTransfer = []
trainingAccuracyTransfer = []
totalSteps = len(mnistTrainLoader)
for epoch in range(1, numberOfEpochs):
classifier.train()
runningLoss = 0.0
correct = 0
total = 0
print("Training started for Epoch: ",epoch)
for batchIndex, (data, target) in enumerate(mnistTrainLoader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
outputs = classifier(data)
loss = criterion(outputs,target)
loss.backward()
optimizer.step()
runningLoss += loss.item()
_, pred = torch.max(outputs, dim=1)
correct += torch.sum(pred==target).item()
total += target.size(0)
if (batchIndex)%100 ==0:
print("Epoch [{}/{}] Step [{}/{}] Loss: {:.4f}".format(epoch,numberOfEpochs,batchIndex,totalSteps,loss.item()))
trainingAccuracyTransfer.append(100*correct/total)
trainingLossTransfer.append(runningLoss/totalSteps)
print("Training Accuracy: ",(100*correct/total))
batchLoss = 0
totalV = 0
correctV = 0
with torch.no_grad():
classifier.eval()
for dataV, targetV in (mnistTestLoader):
dataV, targetV = dataV.to(device), targetV.to(device)
outputV = classifier(dataV)
lossV = criterion(outputV,targetV)
batchLoss += lossV.item()
_, predV = torch.max(outputV, dim=1)
correctV += torch.sum(predV==targetV).item()
totalV += targetV.size(0)
validationAccuracyTransfer.append(100*correctV/totalV)
validationLossTransfer.append(batchLoss/len(mnistTestLoader))
print("Validation Accuracy: ",(100*correctV/totalV))
if (100*correctV/totalV)>validAccuracyMaxTransfer:
validAccuracyMaxTransfer = 100*correctV/totalV
print("Validation accuracy improved, network improvement detected, saving network")
torch.save(classifier.state_dict(), "classifierNetworkTransferLearningTask2.pt")
classifier.train()
fig = plt.figure(figsize=(20,10))
plt.title("Transfer Learning Model Train-Validation Loss Plot")
plt.plot(trainingLossTransfer, label='training loss')
plt.plot(validationLossTransfer, label='validation loss')
plt.xlabel('Number of epochs')
plt.ylabel('Loss')
plt.legend(loc='best')
fig = plt.figure(figsize=(20,10))
plt.title("Transfer Learning Train-Validation Accuracy Plot")
plt.plot(trainingAccuracyTransfer, label='training accuracy')
plt.plot(validationAccuracyTransfer, label='validation accuracy')
plt.xlabel('Number of epochs')
plt.ylabel('Accuracy')
plt.legend(loc='best')
| 0.808332 | 0.898678 |
```
# Importing dependencies
import pandas as pd
import datetime
# Seclecting only data from 2009 due to the size of the dataset
nine_df = pd.read_csv('Resources/CSV/2009.csv')
# nine_df.head()
nine_df.columns
# Get a list of all of the carriers in the dataset
carriers_list = []
for carrier in nine_df['OP_CARRIER']:
if carrier not in carriers_list:
carriers_list.append(carrier)
print(carriers_list)
# Dropping columns that do not provide required or useful information
# Columns:
# CRS_DEP_TIME, DEP_TIME, DEP_DELAY, TAXI_OUT, WHEELS_OFF, TAXI_IN, CRS_ARR_TIME, ARR_TIME,
# CANCELLED, CANCELLATION_CODE, DIVERTED, CRS_ELAPSED_TIME, ACTUAL_ELAPSED_TIME, AIR_TIME, DISTANCE, CARRIER_DELAY,
# WEATHER_DELAY, NAS_DELAY, SECURITY_DELAY, LATE_AIRCRAFT_DELAY
nine_dropped_df = nine_df[['FL_DATE',
'OP_CARRIER',
'OP_CARRIER_FL_NUM',
'ORIGIN',
'DEST',
'ARR_DELAY']]
# Only keeping OO, SkyWest Airlines, due to the size of the data
oo_df = nine_dropped_df.loc[nine_dropped_df['OP_CARRIER'] == 'OO']
oo_df.head()
# Checking data for NaNs
oo_df.isna().sum()
# Dropping all flights that have a NaN
oo_df.dropna(inplace=True)
# Converting flight date to day of the week in order to convert to the weekday name
# monday:0, tuesday:1, wednesday:2, thursday:3, friday:4, saturday:5, sunday:6
oo_df['WEEKDAY'] = oo_df['FL_DATE'].apply(lambda x: datetime.datetime.strptime(x, "%d-%m-%y").weekday())
oo_df.head()
# Encoding flight status
# delayed:0, on_time:1, early:2
oo_df.loc[oo_df['ARR_DELAY'] > 0, 'flight_status'] = 0
oo_df.loc[oo_df['ARR_DELAY'] == 0, 'flight_status'] = 1
oo_df.loc[oo_df['ARR_DELAY'] < 0, 'flight_status'] = 2
#Replacing original ARR_Delay column with encoded flight_status column
oo_df.drop(['ARR_DELAY'], axis=1, inplace=True)
oo_df.head()
# Replacing FL_Date with WEEKDAY and dropping OP_Carrier because it is no longer needed
cleaned_oo_df = oo_df.drop(['FL_DATE'], axis=1, inplace=False)
cleaned_oo_df = cleaned_oo_df.drop(['OP_CARRIER'], axis=1, inplace=False)
cleaned_oo_df.head()
# Renaming data columns to match database table
cleaned_oo_df = cleaned_oo_df.rename(columns={"OP_CARRIER_FL_NUM":
"fl_num",
"ORIGIN":
"origin",
"DEST":
"dest",
"WEEKDAY":
"weekday"})
cleaned_oo_df.head()
# Creating a list of unique airport callsigns with enumerated airport id
unique_airport_list = list(set(cleaned_oo_df['origin'].values).union(set(cleaned_oo_df['dest'].values)))
airport_update = {e: i for i, e in enumerate(unique_airport_list)}
# Adding updated airport list and id to cleaned_oo_df and checking data types and NaNs
flight_data = cleaned_oo_df.replace(airport_update)
flight_data = flight_data.dropna()
# Reseting index to match postgres table and dropping original index
flight_data.reset_index(inplace=True)
flight_data['flight_id'] = flight_data['index']
flight_data = flight_data.drop('index', axis=1)
# Creating the airports dataframe and reseting the index
airports = pd.DataFrame.from_dict(airport_update, orient='index').reset_index()
airports = airports.rename(columns={"index": "airport", 0: "airport_id"})
airports.head()
# Saving the cleaned dataframe to a zipped CSV file
cleaned_oo_df.to_csv('C:/Users/maxke/Documents/programming/data_analysis/UCB_Data_analytics/Pandas-Project/Resources/CSV/cleaned_oo_data.zip',
index=False,
compression='zip')
# Creating flight status dataframe with id for flight_status Postgres table
flight_status = pd.DataFrame({'flight_status': ['early',
'delayed',
'on_time'],
'flight_status_id': [2, 0, 1]})
flight_status
# Creating flight status dataframe with id for days_of_the_week Postgres table
days_of_the_week = pd.DataFrame({'weekday': ['monday',
'tuesday',
'wednesday',
'thursday',
'friday',
'saturday',
'sunday'],
'weekday_id': range(7)})
days_of_the_week
# Inputing settings for sqlalchemy connection with postgres
protocol = "postgres"
user = "postgres"
password = "0UyJ3HQUDBTs*1^4FnqX"
location = "localhost"
port = "5432"
database = "flight_delays"
connection_string = f"{protocol}://{user}:{password}@{location}:{port}/{database}"
print(connection_string)
from sqlalchemy import create_engine
engine = create_engine(connection_string)
# Writing dataframes to postgres tables
days_of_the_week.to_sql('days_of_the_week', engine, if_exists = 'append', index=False)
flight_status.to_sql('flight_status', engine, if_exists = 'append', index=False)
airports.to_sql('airports', engine, if_exists = 'append', index=False)
flight_data.to_sql('flight_data', engine, if_exists = 'append', index=False)
```
|
github_jupyter
|
# Importing dependencies
import pandas as pd
import datetime
# Seclecting only data from 2009 due to the size of the dataset
nine_df = pd.read_csv('Resources/CSV/2009.csv')
# nine_df.head()
nine_df.columns
# Get a list of all of the carriers in the dataset
carriers_list = []
for carrier in nine_df['OP_CARRIER']:
if carrier not in carriers_list:
carriers_list.append(carrier)
print(carriers_list)
# Dropping columns that do not provide required or useful information
# Columns:
# CRS_DEP_TIME, DEP_TIME, DEP_DELAY, TAXI_OUT, WHEELS_OFF, TAXI_IN, CRS_ARR_TIME, ARR_TIME,
# CANCELLED, CANCELLATION_CODE, DIVERTED, CRS_ELAPSED_TIME, ACTUAL_ELAPSED_TIME, AIR_TIME, DISTANCE, CARRIER_DELAY,
# WEATHER_DELAY, NAS_DELAY, SECURITY_DELAY, LATE_AIRCRAFT_DELAY
nine_dropped_df = nine_df[['FL_DATE',
'OP_CARRIER',
'OP_CARRIER_FL_NUM',
'ORIGIN',
'DEST',
'ARR_DELAY']]
# Only keeping OO, SkyWest Airlines, due to the size of the data
oo_df = nine_dropped_df.loc[nine_dropped_df['OP_CARRIER'] == 'OO']
oo_df.head()
# Checking data for NaNs
oo_df.isna().sum()
# Dropping all flights that have a NaN
oo_df.dropna(inplace=True)
# Converting flight date to day of the week in order to convert to the weekday name
# monday:0, tuesday:1, wednesday:2, thursday:3, friday:4, saturday:5, sunday:6
oo_df['WEEKDAY'] = oo_df['FL_DATE'].apply(lambda x: datetime.datetime.strptime(x, "%d-%m-%y").weekday())
oo_df.head()
# Encoding flight status
# delayed:0, on_time:1, early:2
oo_df.loc[oo_df['ARR_DELAY'] > 0, 'flight_status'] = 0
oo_df.loc[oo_df['ARR_DELAY'] == 0, 'flight_status'] = 1
oo_df.loc[oo_df['ARR_DELAY'] < 0, 'flight_status'] = 2
#Replacing original ARR_Delay column with encoded flight_status column
oo_df.drop(['ARR_DELAY'], axis=1, inplace=True)
oo_df.head()
# Replacing FL_Date with WEEKDAY and dropping OP_Carrier because it is no longer needed
cleaned_oo_df = oo_df.drop(['FL_DATE'], axis=1, inplace=False)
cleaned_oo_df = cleaned_oo_df.drop(['OP_CARRIER'], axis=1, inplace=False)
cleaned_oo_df.head()
# Renaming data columns to match database table
cleaned_oo_df = cleaned_oo_df.rename(columns={"OP_CARRIER_FL_NUM":
"fl_num",
"ORIGIN":
"origin",
"DEST":
"dest",
"WEEKDAY":
"weekday"})
cleaned_oo_df.head()
# Creating a list of unique airport callsigns with enumerated airport id
unique_airport_list = list(set(cleaned_oo_df['origin'].values).union(set(cleaned_oo_df['dest'].values)))
airport_update = {e: i for i, e in enumerate(unique_airport_list)}
# Adding updated airport list and id to cleaned_oo_df and checking data types and NaNs
flight_data = cleaned_oo_df.replace(airport_update)
flight_data = flight_data.dropna()
# Reseting index to match postgres table and dropping original index
flight_data.reset_index(inplace=True)
flight_data['flight_id'] = flight_data['index']
flight_data = flight_data.drop('index', axis=1)
# Creating the airports dataframe and reseting the index
airports = pd.DataFrame.from_dict(airport_update, orient='index').reset_index()
airports = airports.rename(columns={"index": "airport", 0: "airport_id"})
airports.head()
# Saving the cleaned dataframe to a zipped CSV file
cleaned_oo_df.to_csv('C:/Users/maxke/Documents/programming/data_analysis/UCB_Data_analytics/Pandas-Project/Resources/CSV/cleaned_oo_data.zip',
index=False,
compression='zip')
# Creating flight status dataframe with id for flight_status Postgres table
flight_status = pd.DataFrame({'flight_status': ['early',
'delayed',
'on_time'],
'flight_status_id': [2, 0, 1]})
flight_status
# Creating flight status dataframe with id for days_of_the_week Postgres table
days_of_the_week = pd.DataFrame({'weekday': ['monday',
'tuesday',
'wednesday',
'thursday',
'friday',
'saturday',
'sunday'],
'weekday_id': range(7)})
days_of_the_week
# Inputing settings for sqlalchemy connection with postgres
protocol = "postgres"
user = "postgres"
password = "0UyJ3HQUDBTs*1^4FnqX"
location = "localhost"
port = "5432"
database = "flight_delays"
connection_string = f"{protocol}://{user}:{password}@{location}:{port}/{database}"
print(connection_string)
from sqlalchemy import create_engine
engine = create_engine(connection_string)
# Writing dataframes to postgres tables
days_of_the_week.to_sql('days_of_the_week', engine, if_exists = 'append', index=False)
flight_status.to_sql('flight_status', engine, if_exists = 'append', index=False)
airports.to_sql('airports', engine, if_exists = 'append', index=False)
flight_data.to_sql('flight_data', engine, if_exists = 'append', index=False)
| 0.378229 | 0.317645 |
# Joint Coordinate System
> Marcos Duarte
> Laboratory of Biomechanics and Motor Control ([http://demotu.org/](http://demotu.org/))
> Federal University of ABC, Brazil
<div class='center-align'><figure><img src='./../images/JCSpelvisV.png' width=420 alt='Pelvic anatomical frame'/> <figcaption><center><i>Pelvic anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSthighV.png' width=280 alt='Femur anatomical frame'/> <figcaption><center><i>Femur anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSlegV.png' width=320 alt='Tibial/Fibula anatomical frame'/> <figcaption><center><i>Tibial/Fibula anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSfootV.png' width=350 alt='Foot anatomical frame'/> <figcaption><center><i>Foot anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
## References
- Cappozzo A, Catani F, Croce UD, Leardini AC (1995) [Position and orientation in space of bones during movement: anatomical frame definition and determination](http://www.ncbi.nlm.nih.gov/pubmed/11415549). Clinical Biomechanics, 10(4):171-178.
- Grood ES, Suntay WJ (1983) [A Joint Coordinate System for the Clinical Description of Three-Dimensional Motions: Application to the Knee](http://www.ncbi.nlm.nih.gov/pubmed/6865355). Journal of Biomechanical Engineering, 105, 136–144.
- MacWilliams BA, Davis RB (2013) [Addressing some misperceptions of the joint coordinate system](http://www.ncbi.nlm.nih.gov/pubmed/24231967). Journal of Biomechanical Engineering, 135:54506. doi: 10.1115/1.4024142.
- Virtual Animation of the Kinematics of the Human for Industrial, Educational and Research Purposes (VAKHUM). [D3.2. Technical Report on Data Collection Procedure - ANNEX I](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf).
- Wu G, Cavanagh PR (1995) [ISB recommendations for standardization in the reporting data](http://www.ncbi.nlm.nih.gov/pubmed/8550644). Journal of Biomechanics, 28, 1257-1261.
- Zatsiorsky VM (1997) [Kinematics of Human Motion](http://books.google.com.br/books/about/Kinematics_of_Human_Motion.html?id=Pql_xXdbrMcC&redir_esc=y). Champaign, Human Kinetics.
|
github_jupyter
|
# Joint Coordinate System
> Marcos Duarte
> Laboratory of Biomechanics and Motor Control ([http://demotu.org/](http://demotu.org/))
> Federal University of ABC, Brazil
<div class='center-align'><figure><img src='./../images/JCSpelvisV.png' width=420 alt='Pelvic anatomical frame'/> <figcaption><center><i>Pelvic anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSthighV.png' width=280 alt='Femur anatomical frame'/> <figcaption><center><i>Femur anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSlegV.png' width=320 alt='Tibial/Fibula anatomical frame'/> <figcaption><center><i>Tibial/Fibula anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
<div class='center-align'><figure><img src='./../images/JCSfootV.png' width=350 alt='Foot anatomical frame'/> <figcaption><center><i>Foot anatomical frame (figure from the [VAKHUM project](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf)).</i></center></figcaption> </figure></div>
## References
- Cappozzo A, Catani F, Croce UD, Leardini AC (1995) [Position and orientation in space of bones during movement: anatomical frame definition and determination](http://www.ncbi.nlm.nih.gov/pubmed/11415549). Clinical Biomechanics, 10(4):171-178.
- Grood ES, Suntay WJ (1983) [A Joint Coordinate System for the Clinical Description of Three-Dimensional Motions: Application to the Knee](http://www.ncbi.nlm.nih.gov/pubmed/6865355). Journal of Biomechanical Engineering, 105, 136–144.
- MacWilliams BA, Davis RB (2013) [Addressing some misperceptions of the joint coordinate system](http://www.ncbi.nlm.nih.gov/pubmed/24231967). Journal of Biomechanical Engineering, 135:54506. doi: 10.1115/1.4024142.
- Virtual Animation of the Kinematics of the Human for Industrial, Educational and Research Purposes (VAKHUM). [D3.2. Technical Report on Data Collection Procedure - ANNEX I](http://www.ulb.ac.be/project/vakhum/public_dataset/Doc/VAKHUM-3-Frame_Convention.pdf).
- Wu G, Cavanagh PR (1995) [ISB recommendations for standardization in the reporting data](http://www.ncbi.nlm.nih.gov/pubmed/8550644). Journal of Biomechanics, 28, 1257-1261.
- Zatsiorsky VM (1997) [Kinematics of Human Motion](http://books.google.com.br/books/about/Kinematics_of_Human_Motion.html?id=Pql_xXdbrMcC&redir_esc=y). Champaign, Human Kinetics.
| 0.682574 | 0.685542 |
(parallel)=
# Parallelization
```
%config InlineBackend.figure_format = "retina"
from matplotlib import rcParams
rcParams["savefig.dpi"] = 100
rcParams["figure.dpi"] = 100
rcParams["font.size"] = 20
import multiprocessing
multiprocessing.set_start_method("fork")
```
:::{note}
Some builds of NumPy (including the version included with Anaconda) will automatically parallelize some operations using something like the MKL linear algebra. This can cause problems when used with the parallelization methods described here so it can be good to turn that off (by setting the environment variable `OMP_NUM_THREADS=1`, for example).
:::
```
import os
os.environ["OMP_NUM_THREADS"] = "1"
```
With emcee, it's easy to make use of multiple CPUs to speed up slow sampling.
There will always be some computational overhead introduced by parallelization so it will only be beneficial in the case where the model is expensive, but this is often true for real research problems.
All parallelization techniques are accessed using the `pool` keyword argument in the :class:`EnsembleSampler` class but, depending on your system and your model, there are a few pool options that you can choose from.
In general, a `pool` is any Python object with a `map` method that can be used to apply a function to a list of numpy arrays.
Below, we will discuss a few options.
In all of the following examples, we'll test the code with the following convoluted model:
```
import time
import numpy as np
def log_prob(theta):
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
```
This probability function will randomly sleep for a fraction of a second every time it is called.
This is meant to emulate a more realistic situation where the model is computationally expensive to compute.
To start, let's sample the usual (serial) way:
```
import emcee
np.random.seed(42)
initial = np.random.randn(32, 5)
nwalkers, ndim = initial.shape
nsteps = 100
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
serial_time = end - start
print("Serial took {0:.1f} seconds".format(serial_time))
```
## Multiprocessing
The simplest method of parallelizing emcee is to use the [multiprocessing module from the standard library](https://docs.python.org/3/library/multiprocessing.html).
To parallelize the above sampling, you could update the code as follows:
```
from multiprocessing import Pool
with Pool() as pool:
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_time))
print("{0:.1f} times faster than serial".format(serial_time / multi_time))
```
I have 4 cores on the machine where this is being tested:
```
from multiprocessing import cpu_count
ncpu = cpu_count()
print("{0} CPUs".format(ncpu))
```
We don't quite get the factor of 4 runtime decrease that you might expect because there is some overhead in the parallelization, but we're getting pretty close with this example and this will get even closer for more expensive models.
## MPI
Multiprocessing can only be used for distributing calculations across processors on one machine.
If you want to take advantage of a bigger cluster, you'll need to use MPI.
In that case, you need to execute the code using the `mpiexec` executable, so this demo is slightly more convoluted.
For this example, we'll write the code to a file called `script.py` and then execute it using MPI, but when you really use the MPI pool, you'll probably just want to edit the script directly.
To run this example, you'll first need to install [the schwimmbad library](https://github.com/adrn/schwimmbad) because emcee no longer includes its own `MPIPool`.
```
with open("script.py", "w") as f:
f.write("""
import sys
import time
import emcee
import numpy as np
from schwimmbad import MPIPool
def log_prob(theta):
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5*np.sum(theta**2)
with MPIPool() as pool:
if not pool.is_master():
pool.wait()
sys.exit(0)
np.random.seed(42)
initial = np.random.randn(32, 5)
nwalkers, ndim = initial.shape
nsteps = 100
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps)
end = time.time()
print(end - start)
""")
mpi_time = !mpiexec -n {ncpu} python script.py
mpi_time = float(mpi_time[0])
print("MPI took {0:.1f} seconds".format(mpi_time))
print("{0:.1f} times faster than serial".format(serial_time / mpi_time))
```
There is often more overhead introduced by MPI than multiprocessing so we get less of a gain this time.
That being said, MPI is much more flexible and it can be used to scale to huge systems.
## Pickling, data transfer & arguments
All parallel Python implementations work by spinning up multiple `python` processes with identical environments then and passing information between the processes using `pickle`.
This means that the probability function [must be picklable](https://docs.python.org/3/library/pickle.html#pickle-picklable).
Some users might hit issues when they use `args` to pass data to their model.
These args must be pickled and passed every time the model is called.
This can be a problem if you have a large dataset, as you can see here:
```
def log_prob_data(theta, data):
a = data[0] # Use the data somehow...
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
data = np.random.randn(5000, 200)
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob_data, args=(data,))
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
serial_data_time = end - start
print("Serial took {0:.1f} seconds".format(serial_data_time))
```
We basically get no change in performance when we include the `data` argument here.
Now let's try including this naively using multiprocessing:
```
with Pool() as pool:
sampler = emcee.EnsembleSampler(
nwalkers, ndim, log_prob_data, pool=pool, args=(data,)
)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_data_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_data_time))
print(
"{0:.1f} times faster(?) than serial".format(serial_data_time / multi_data_time)
)
```
Brutal.
We can do better than that though.
It's a bit ugly, but if we just make `data` a global variable and use that variable within the model calculation, then we take no hit at all.
```
def log_prob_data_global(theta):
a = data[0] # Use the data somehow...
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
with Pool() as pool:
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob_data_global, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_data_global_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_data_global_time))
print(
"{0:.1f} times faster than serial".format(
serial_data_time / multi_data_global_time
)
)
```
That's better!
This works because, in the global variable case, the dataset is only pickled and passed between processes once (when the pool is created) instead of once for every model evaluation.
|
github_jupyter
|
%config InlineBackend.figure_format = "retina"
from matplotlib import rcParams
rcParams["savefig.dpi"] = 100
rcParams["figure.dpi"] = 100
rcParams["font.size"] = 20
import multiprocessing
multiprocessing.set_start_method("fork")
import os
os.environ["OMP_NUM_THREADS"] = "1"
import time
import numpy as np
def log_prob(theta):
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
import emcee
np.random.seed(42)
initial = np.random.randn(32, 5)
nwalkers, ndim = initial.shape
nsteps = 100
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
serial_time = end - start
print("Serial took {0:.1f} seconds".format(serial_time))
from multiprocessing import Pool
with Pool() as pool:
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_time))
print("{0:.1f} times faster than serial".format(serial_time / multi_time))
from multiprocessing import cpu_count
ncpu = cpu_count()
print("{0} CPUs".format(ncpu))
with open("script.py", "w") as f:
f.write("""
import sys
import time
import emcee
import numpy as np
from schwimmbad import MPIPool
def log_prob(theta):
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5*np.sum(theta**2)
with MPIPool() as pool:
if not pool.is_master():
pool.wait()
sys.exit(0)
np.random.seed(42)
initial = np.random.randn(32, 5)
nwalkers, ndim = initial.shape
nsteps = 100
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps)
end = time.time()
print(end - start)
""")
mpi_time = !mpiexec -n {ncpu} python script.py
mpi_time = float(mpi_time[0])
print("MPI took {0:.1f} seconds".format(mpi_time))
print("{0:.1f} times faster than serial".format(serial_time / mpi_time))
def log_prob_data(theta, data):
a = data[0] # Use the data somehow...
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
data = np.random.randn(5000, 200)
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob_data, args=(data,))
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
serial_data_time = end - start
print("Serial took {0:.1f} seconds".format(serial_data_time))
with Pool() as pool:
sampler = emcee.EnsembleSampler(
nwalkers, ndim, log_prob_data, pool=pool, args=(data,)
)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_data_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_data_time))
print(
"{0:.1f} times faster(?) than serial".format(serial_data_time / multi_data_time)
)
def log_prob_data_global(theta):
a = data[0] # Use the data somehow...
t = time.time() + np.random.uniform(0.005, 0.008)
while True:
if time.time() >= t:
break
return -0.5 * np.sum(theta ** 2)
with Pool() as pool:
sampler = emcee.EnsembleSampler(nwalkers, ndim, log_prob_data_global, pool=pool)
start = time.time()
sampler.run_mcmc(initial, nsteps, progress=True)
end = time.time()
multi_data_global_time = end - start
print("Multiprocessing took {0:.1f} seconds".format(multi_data_global_time))
print(
"{0:.1f} times faster than serial".format(
serial_data_time / multi_data_global_time
)
)
| 0.4436 | 0.842863 |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter GoogleAds Segmentology Parameters
GoogleAds funnel analysis using Census data.
1. Wait for <b>BigQuery->->->Census_Join</b> to be created.
1. Join the <a hre='https://groups.google.com/d/forum/starthinker-assets' target='_blank'>StarThinker Assets Group</a> to access the following assets
1. Copy <a href='https://datastudio.google.com/c/u/0/reporting/3673497b-f36f-4448-8fb9-3e05ea51842f/' target='_blank'>GoogleAds Segmentology Sample</a>. Leave the Data Source as is, you will change it in the next step.
1. Click Edit Connection, and change to <b>BigQuery->->->Census_Join</b>.
1. Or give these instructions to the client.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'auth_read': 'user', # Credentials used for reading data.
'customer_id': '', # Google Ads customer.
'developer_token': '', # Google Ads developer token.
'login_id': '', # Google Ads login.
'recipe_project': '', # Project ID hosting dataset.
'auth_write': 'service', # Authorization used for writing data.
'recipe_slug': '', # Name of Google BigQuery dataset to create.
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute GoogleAds Segmentology
This does NOT need to be modified unless you are changing the recipe, click play.
```
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import commandline_parser
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'description': 'Create a dataset for bigquery tables.',
'hour': [
4
],
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}}
}
},
{
'bigquery': {
'auth': 'user',
'function': 'Pearson Significance Test',
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'googleads',
'version': 'v5',
'function': 'customers.googleAds.search',
'kwargs': {
'customerId': {'field': {'name': 'customer_id','kind': 'string','description': 'Google Ads customer.','default': ''}},
'body': {
'query': 'SELECT campaign.name, ad_group.name, segments.geo_target_postal_code, metrics.impressions, metrics.clicks, metrics.conversions, metrics.interactions FROM user_location_view '
}
},
'headers': {
'developer-token': {'field': {'name': 'developer_token','kind': 'string','description': 'Google Ads developer token.','default': ''}},
'login-customer-id': {'field': {'name': 'login_id','kind': 'string','description': 'Google Ads login.','default': ''}}
},
'iterate': True,
'results': {
'bigquery': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'GoogleAds_KPI',
'schema': [
{
'name': 'userLocationView',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'segments',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'geoTargetPostalCode',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'metrics',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'interactions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'impressions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'conversions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'clicks',
'type': 'INTEGER',
'mode': 'NULLABLE'
}
]
},
{
'name': 'adGroup',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'name',
'type': 'STRING',
'mode': 'NULLABLE'
},
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'campaign',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'name',
'type': 'STRING',
'mode': 'NULLABLE'
},
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
}
]
}
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': 'SELECT campaign.name AS Campaign, adGRoup.name AS Ad_Group, segments.geoTargetPostalCode AS Postal_Code, SAFE_DIVIDE(metrics.impressions, SUM(metrics.impressions) OVER()) AS Impression, SAFE_DIVIDE(metrics.clicks, metrics.impressions) AS Click, SAFE_DIVIDE(metrics.conversions, metrics.impressions) AS Conversion, SAFE_DIVIDE(metrics.interactions, metrics.impressions) AS Interaction, metrics.impressions AS Impressions FROM `{project}.{dataset}.GoogleAds_KPI`; ',
'parameters': {
'project': {'field': {'name': 'recipe_project','kind': 'string','description': 'Project ID hosting dataset.'}},
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be written in BigQuery.'}},
'view': 'GoogleAds_KPI_Normalized'
}
}
},
{
'census': {
'auth': 'user',
'normalize': {
'census_geography': 'zip_codes',
'census_year': '2018',
'census_span': '5yr'
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'type': 'view'
}
}
},
{
'census': {
'auth': 'user',
'correlate': {
'join': 'Postal_Code',
'pass': [
'Campaign',
'Ad_Group'
],
'sum': [
'Impressions'
],
'correlate': [
'Impression',
'Click',
'Conversion',
'Interaction'
],
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'GoogleAds_KPI_Normalized',
'significance': 80
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'type': 'view'
}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
```
|
github_jupyter
|
!pip install git+https://github.com/google/starthinker
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
CLIENT_CREDENTIALS = 'PASTE CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
FIELDS = {
'auth_read': 'user', # Credentials used for reading data.
'customer_id': '', # Google Ads customer.
'developer_token': '', # Google Ads developer token.
'login_id': '', # Google Ads login.
'recipe_project': '', # Project ID hosting dataset.
'auth_write': 'service', # Authorization used for writing data.
'recipe_slug': '', # Name of Google BigQuery dataset to create.
}
print("Parameters Set To: %s" % FIELDS)
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import commandline_parser
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'description': 'Create a dataset for bigquery tables.',
'hour': [
4
],
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}}
}
},
{
'bigquery': {
'auth': 'user',
'function': 'Pearson Significance Test',
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'googleads',
'version': 'v5',
'function': 'customers.googleAds.search',
'kwargs': {
'customerId': {'field': {'name': 'customer_id','kind': 'string','description': 'Google Ads customer.','default': ''}},
'body': {
'query': 'SELECT campaign.name, ad_group.name, segments.geo_target_postal_code, metrics.impressions, metrics.clicks, metrics.conversions, metrics.interactions FROM user_location_view '
}
},
'headers': {
'developer-token': {'field': {'name': 'developer_token','kind': 'string','description': 'Google Ads developer token.','default': ''}},
'login-customer-id': {'field': {'name': 'login_id','kind': 'string','description': 'Google Ads login.','default': ''}}
},
'iterate': True,
'results': {
'bigquery': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'GoogleAds_KPI',
'schema': [
{
'name': 'userLocationView',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'segments',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'geoTargetPostalCode',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'metrics',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'interactions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'impressions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'conversions',
'type': 'INTEGER',
'mode': 'NULLABLE'
},
{
'name': 'clicks',
'type': 'INTEGER',
'mode': 'NULLABLE'
}
]
},
{
'name': 'adGroup',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'name',
'type': 'STRING',
'mode': 'NULLABLE'
},
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
},
{
'name': 'campaign',
'type': 'RECORD',
'mode': 'NULLABLE',
'fields': [
{
'name': 'name',
'type': 'STRING',
'mode': 'NULLABLE'
},
{
'name': 'resourceName',
'type': 'STRING',
'mode': 'NULLABLE'
}
]
}
]
}
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': 'SELECT campaign.name AS Campaign, adGRoup.name AS Ad_Group, segments.geoTargetPostalCode AS Postal_Code, SAFE_DIVIDE(metrics.impressions, SUM(metrics.impressions) OVER()) AS Impression, SAFE_DIVIDE(metrics.clicks, metrics.impressions) AS Click, SAFE_DIVIDE(metrics.conversions, metrics.impressions) AS Conversion, SAFE_DIVIDE(metrics.interactions, metrics.impressions) AS Interaction, metrics.impressions AS Impressions FROM `{project}.{dataset}.GoogleAds_KPI`; ',
'parameters': {
'project': {'field': {'name': 'recipe_project','kind': 'string','description': 'Project ID hosting dataset.'}},
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be created in BigQuery.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','description': 'Place where tables will be written in BigQuery.'}},
'view': 'GoogleAds_KPI_Normalized'
}
}
},
{
'census': {
'auth': 'user',
'normalize': {
'census_geography': 'zip_codes',
'census_year': '2018',
'census_span': '5yr'
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'type': 'view'
}
}
},
{
'census': {
'auth': 'user',
'correlate': {
'join': 'Postal_Code',
'pass': [
'Campaign',
'Ad_Group'
],
'sum': [
'Impressions'
],
'correlate': [
'Impression',
'Click',
'Conversion',
'Interaction'
],
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'GoogleAds_KPI_Normalized',
'significance': 80
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'type': 'view'
}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
| 0.329607 | 0.761694 |
An alternate way to handle this problem is to train two separate resnet models for the two image tasks. We can then use Octopod to combine them into an ensemble model with the text model that was trained on both tasks.
This notebook trains a gender model, Step6 trains a season model, but they could be run in parallel.
This notebook was run on an AWS p3.2xlarge
# Octopod Image Model Training Pipeline
```
%load_ext autoreload
%autoreload 2
import sys
sys.path.append('../../')
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torch.utils.data import Dataset, DataLoader
```
Note: for images, we use the MultiInputMultiTaskLearner since we will send in the full image and a center crop of the image.
```
from octopod import MultiInputMultiTaskLearner, MultiDatasetLoader
from octopod.vision.dataset import OctopodImageDataset
from octopod.vision.models import ResnetForMultiTaskClassification
```
## Load in train and validation datasets
First we load in the csv's we created in Step 1.
Remember to change the path if you stored your data somewhere other than the default.
```
TRAIN_GENDER_DF = pd.read_csv('/home/ec2-user/fashion_dataset/gender_train.csv')
VALID_GENDER_DF = pd.read_csv('/home/ec2-user/fashion_dataset/gender_valid.csv')
#TRAIN_SEASON_DF = pd.read_csv('/home/ubuntu/fashion_dataset/season_train.csv')
#VALID_SEASON_DF = pd.read_csv('/home/ubuntu/fashion_dataset/season_valid.csv')
```
You will most likely have to alter this to however big your batches can be on your machine
```
batch_size = 64
```
We use the `OctopodImageDataSet` class to create train and valid datasets for each task.
Check out the documentation for infomation about the transformations.
```
gender_train_dataset = OctopodImageDataset(
x=TRAIN_GENDER_DF['image_urls'],
y=TRAIN_GENDER_DF['gender_cat'],
transform='train',
crop_transform='train'
)
gender_valid_dataset = OctopodImageDataset(
x=VALID_GENDER_DF['image_urls'],
y=VALID_GENDER_DF['gender_cat'],
transform='val',
crop_transform='val'
)
# season_train_dataset = OctopodImageDataset(
# x=TRAIN_SEASON_DF['image_urls'],
# y=TRAIN_SEASON_DF['season_cat'],
# transform='train',
# crop_transform='train'
# )
# season_valid_dataset = OctopodImageDataset(
# x=VALID_SEASON_DF['image_urls'],
# y=VALID_SEASON_DF['season_cat'],
# transform='val',
# crop_transform='val'
# )
```
We then put the datasets into a dictionary of dataloaders.
Each task is a key.
```
train_dataloaders_dict = {
'gender': DataLoader(gender_train_dataset, batch_size=batch_size, shuffle=True, num_workers=4),
#'season': DataLoader(season_train_dataset, batch_size=batch_size, shuffle=True, num_workers=4),
}
valid_dataloaders_dict = {
'gender': DataLoader(gender_valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4),
#'season': DataLoader(season_valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4),
}
```
The dictionary of dataloaders is then put into an instance of the Octopod `MultiDatasetLoader` class.
```
TrainLoader = MultiDatasetLoader(loader_dict=train_dataloaders_dict)
len(TrainLoader)
ValidLoader = MultiDatasetLoader(loader_dict=valid_dataloaders_dict, shuffle=False)
len(ValidLoader)
```
We need to create a dictionary of the tasks and the number of unique values so that we can create our model.
```
new_task_dict = {
'gender': TRAIN_GENDER_DF['gender_cat'].nunique(),
#'season': TRAIN_SEASON_DF['season_cat'].nunique(),
}
new_task_dict
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
print(device)
```
Create Model and Learner
===
These are completely new tasks so we use `new_task_dict`. If we had already trained a model on some tasks, we would use `pretrained_task_dict`.
And since these are new tasks, we set `load_pretrained_renset=True` to use the weights from Torch.
```
model = ResnetForMultiTaskClassification(
new_task_dict=new_task_dict,
load_pretrained_resnet=True
)
```
You will likely need to explore different values in this section to find some that work
for your particular model.
```
lr_last = 1e-2
lr_main = 1e-4
optimizer = optim.Adam([
{'params': model.resnet.parameters(), 'lr': lr_main},
{'params': model.dense_layers.parameters(), 'lr': lr_last},
{'params': model.new_classifiers.parameters(), 'lr': lr_last},
])
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size= 4, gamma= 0.1)
loss_function_dict = {'gender': 'categorical_cross_entropy', 'season': 'categorical_cross_entropy'}
metric_function_dict = {'gender': 'multi_class_acc', 'season': 'multi_class_acc'}
learn = MultiInputMultiTaskLearner(model, TrainLoader, ValidLoader, new_task_dict, loss_function_dict, metric_function_dict)
```
Train model
===
As your model trains, you can see some output of how the model is performing overall and how it is doing on each individual task.
```
learn.fit(
num_epochs=10,
scheduler=exp_lr_scheduler,
step_scheduler_on_batch=False,
optimizer=optimizer,
device=device,
best_model=True
)
```
Validate model
===
We provide a method on the learner called `get_val_preds`, which makes predictions on the validation data. You can then use this to analyze your model's performance in more detail.
```
pred_dict = learn.get_val_preds(device)
pred_dict
```
Save/Export Model
===
Once we are happy with our training we can save (or export) our model, using the `save` method (or `export`).
See the docs for the difference between `save` and `export`.
We will need the saved model later to use in the ensemble model
```
model.save(folder='/home/ec2-user/fashion_dataset/models/', model_id='GENDER_IMAGE_MODEL1')
model.export(folder='/home/ec2-user/fashion_dataset/models/', model_id='GENDER_IMAGE_MODEL1')
```
Now that we have a gender image model, we can move to `Step6_train_season_image_model`.
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import sys
sys.path.append('../../')
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torch.utils.data import Dataset, DataLoader
from octopod import MultiInputMultiTaskLearner, MultiDatasetLoader
from octopod.vision.dataset import OctopodImageDataset
from octopod.vision.models import ResnetForMultiTaskClassification
TRAIN_GENDER_DF = pd.read_csv('/home/ec2-user/fashion_dataset/gender_train.csv')
VALID_GENDER_DF = pd.read_csv('/home/ec2-user/fashion_dataset/gender_valid.csv')
#TRAIN_SEASON_DF = pd.read_csv('/home/ubuntu/fashion_dataset/season_train.csv')
#VALID_SEASON_DF = pd.read_csv('/home/ubuntu/fashion_dataset/season_valid.csv')
batch_size = 64
gender_train_dataset = OctopodImageDataset(
x=TRAIN_GENDER_DF['image_urls'],
y=TRAIN_GENDER_DF['gender_cat'],
transform='train',
crop_transform='train'
)
gender_valid_dataset = OctopodImageDataset(
x=VALID_GENDER_DF['image_urls'],
y=VALID_GENDER_DF['gender_cat'],
transform='val',
crop_transform='val'
)
# season_train_dataset = OctopodImageDataset(
# x=TRAIN_SEASON_DF['image_urls'],
# y=TRAIN_SEASON_DF['season_cat'],
# transform='train',
# crop_transform='train'
# )
# season_valid_dataset = OctopodImageDataset(
# x=VALID_SEASON_DF['image_urls'],
# y=VALID_SEASON_DF['season_cat'],
# transform='val',
# crop_transform='val'
# )
train_dataloaders_dict = {
'gender': DataLoader(gender_train_dataset, batch_size=batch_size, shuffle=True, num_workers=4),
#'season': DataLoader(season_train_dataset, batch_size=batch_size, shuffle=True, num_workers=4),
}
valid_dataloaders_dict = {
'gender': DataLoader(gender_valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4),
#'season': DataLoader(season_valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4),
}
TrainLoader = MultiDatasetLoader(loader_dict=train_dataloaders_dict)
len(TrainLoader)
ValidLoader = MultiDatasetLoader(loader_dict=valid_dataloaders_dict, shuffle=False)
len(ValidLoader)
new_task_dict = {
'gender': TRAIN_GENDER_DF['gender_cat'].nunique(),
#'season': TRAIN_SEASON_DF['season_cat'].nunique(),
}
new_task_dict
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
print(device)
model = ResnetForMultiTaskClassification(
new_task_dict=new_task_dict,
load_pretrained_resnet=True
)
lr_last = 1e-2
lr_main = 1e-4
optimizer = optim.Adam([
{'params': model.resnet.parameters(), 'lr': lr_main},
{'params': model.dense_layers.parameters(), 'lr': lr_last},
{'params': model.new_classifiers.parameters(), 'lr': lr_last},
])
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size= 4, gamma= 0.1)
loss_function_dict = {'gender': 'categorical_cross_entropy', 'season': 'categorical_cross_entropy'}
metric_function_dict = {'gender': 'multi_class_acc', 'season': 'multi_class_acc'}
learn = MultiInputMultiTaskLearner(model, TrainLoader, ValidLoader, new_task_dict, loss_function_dict, metric_function_dict)
learn.fit(
num_epochs=10,
scheduler=exp_lr_scheduler,
step_scheduler_on_batch=False,
optimizer=optimizer,
device=device,
best_model=True
)
pred_dict = learn.get_val_preds(device)
pred_dict
model.save(folder='/home/ec2-user/fashion_dataset/models/', model_id='GENDER_IMAGE_MODEL1')
model.export(folder='/home/ec2-user/fashion_dataset/models/', model_id='GENDER_IMAGE_MODEL1')
| 0.493164 | 0.867766 |
## 使用卷积神经网络对CIFAR-10数据集进行分类
```
import os
import pickle as p
import tarfile
import urllib.request
from time import time
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn.preprocessing import OneHotEncoder
```
## 准备数据集
```
url = "https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz"
filepath = "Data/CIFAR-10/cifar-10-python.tar.gz"
# 下载数据集
if not os.path.isfile(filepath):
result = urllib.request.urlretrieve(url, filepath)
print("downloaded: ", result)
else:
print("Data file already exists")
# 解压数据集
if not os.path.exists("Data/CIFAR-10/DataSets"):
tfile = tarfile.open("Data/CIFAR-10/cifar-10-python.tar.gz", "r:gz")
result = tfile.extractall("Data/CIFAR-10/DataSets")
print("Extract to Data/CIFAR-10/DataSets")
else:
print("Data file already exists")
```
## 导入数据集
```
# 按批次导入
def load_CIFAR_batch(filename):
with open(filename, "rb") as f:
data_dict = p.load(f, encoding="bytes")
images = data_dict[b"data"]
labels = data_dict[b"labels"]
# 将原始数据调整为BCWH
images = images.reshape(10000, 3, 32, 32)
# tensorflow处理图象数据的结构:BWHC
# 把通道数数据C移动到最后一个维度
images = images.transpose(0, 2, 3, 1)
labels = np.array(labels)
return images, labels
# 导入数据集
def load_CIFAR_data(data_dir):
images_train = []
labels_train = []
for i in range(5):
f = os.path.join(data_dir, "data_batch_%d" % (i + 1))
print("loading", f)
# 调用load_CIFAR_batch()获得批量的图像机器对应的标签
image_batch, label_batch = load_CIFAR_batch(f)
images_train.append(image_batch)
labels_train.append(label_batch)
Xtrain = np.concatenate(images_train)
Ytrain = np.concatenate(labels_train)
del image_batch, label_batch
Xtest, Ytest = load_CIFAR_batch(os.path.join(data_dir, "test_batch"))
print("Finished loading CIFAR-10 data")
# 返回训练集的体香和标签,测试集的图像和标签
return Xtrain, Ytrain, Xtest, Ytest
data_dir = "Data/CIFAR-10/DataSets/cifar-10-batches-py"
Xtrain, Ytrain, Xtest, Ytest = load_CIFAR_data(data_dir)
```
## 查看数据集
```
print("Training data shape: ", Xtrain.shape)
print("Training labels shape: ", Ytrain.shape)
print("Test im shape: ", Xtest.shape)
print("Test labels shape: ", Ytest.shape)
# 查看单项数据
plt.imshow(Xtrain[6])
print(Ytrain[6])
```
## 定义多项images与label
```
# 定义标签字典,每一个数字所代表的图像类别的名称
label_dict = {0: "airplane", 1: "automobile", 2: "bird", 3: "cat", 4: "deer", 5: "dog", 6: "frog", 7: "horse",
8: "ship", 9: "truck"}
# 定义显示图像数据及其对应标签的函数
def plot_images_labels_prediction(images, labels, prediction, idx, num=10):
fig = plt.gcf()
fig.set_size_inches(12, 6)
if num > 10:
num = 10
for i in range(0, num):
ax = plt.subplot(2, 5, 1 + i)
ax.imshow(images[idx], cmap="binary")
title = str(i) + ", " + label_dict[labels[idx]]
if len(prediction) > 0:
title += "=>" + label_dict[prediction[idx]]
ax.set_title(title, fontsize=10)
idx += 1
plt.show()
plot_images_labels_prediction(Xtest, Ytest, [], 1, 10)
```
## 数据预处理
```
# 查看图像数据信息
# 显示第一个图的第一个像素点
Xtrain[0][0][0]
# 将图像进行数字标准化
Xtrain_normalize = Xtrain.astype("float32") / 255.0
Xtest_normalize = Xtest.astype("float32") / 255.0
# 查看预处理后的图像数据信息
Xtrain_normalize[0][0][0]
```
## 标签数据预处理 - 独热编码
```
# 查看标签数据
Ytrain[:10]
encoder = OneHotEncoder(sparse=False)
yy = [[0], [1], [2], [3], [4], [5], [6], [7], [8], [9]]
encoder.fit(yy)
Ytrain_reshape = Ytrain.reshape(-1, 1)
Ytrain_onehot = encoder.transform(Ytrain_reshape)
Ytest_reshape = Ytest.reshape(-1, 1)
Ytest_onehot = encoder.transform(Ytest_reshape)
Ytrain_onehot.shape
Ytrain_onehot[:5]
```
## 定义共享函数
```
# 定义权值
def weight(shape):
# 在构建模型时,需要使用tfVariable来创建一个变量
# 在训练时,这个变量不断更新
# 使用函数tf.truncated.normal(截断的正态分布)生成标准差为0.1的随机数来初始化权值
return tf.Variable(tf.truncated_normal(shape, stddev=0.1), name="W")
# 定义偏置
# 初始化0.1
def bias(shape):
return tf.Variable(tf.constant(0.1, shape=shape), name="b")
# 定义卷积操作
# 步长为1,padding为"SAME"
def conv2d(x, W):
# tf.nn.conv2d(input, filter, strides, padding, use_cudnn_oon_gpu=None, name=None)
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding="SAME")
# 定义池化操作
# 步长为2,即原尺寸的长和宽各除以2
def max_pool_2x2(x):
# tf.nn.max_pool(value, ksize, strides, padding, name=None)
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding="SAME")
```
## 定义网络结构
+ 图像的特征提取:通过卷积层1,降采样层1,卷积层2以及降采样层的处理,提取图像的特征
+ 全连接神经网路:全连接层、输出层所组成的网络结构
| 输入层 | 卷积层1 | 降采样层1 | 卷积层2 | 降采样层2 | 全连接层 | 输出层 |
|--------------------|-----------------------------------------|--------------------------------------------|------------------------------------------|------------------------------------------|--------------------------------------|--------------------------|
| 32*32图像,通道为3(RGB) | 第1次卷积:输入通道:3,输出通道:32,卷积后图像尺寸不变,依然是32*32 | 第一次降采样:将32*32图像缩小为16*16;池化不改变通道数量,因此依然是32个 | 第2次卷积:输入通道:32,输出通道:64,卷积后图像尺寸不变,依然是16*16 | 第二次降采样:将16*16图像缩小为8*8;池化不改变通道数量,因此依然是64个 | 将64个8*8的图像转换为长度为4096的一维向量,该层有128个神经元 | 输出层共有10个神经元,对应到0-9这10个类别 |
```
# 输入层
# 32*32图像,通道为3(RGB)
with tf.name_scope("input_layer"):
x = tf.placeholder("float", shape=[None, 32, 32, 3], name="X")
# 第1个卷积层
# 输入通道:3,输出通道:32,卷积后图像尺寸不变,依然是32*32
with tf.name_scope("conv_1"):
# [k_width, k_height, input_chn, output_chn]
W1 = weight([3, 3, 3, 32])
b1 = bias([32])
conv_1 = conv2d(x, W1) + b1
conv_1 = tf.nn.relu(conv_1)
# 第1个池化层
# 将32*32图像缩小为16*16,池化不改变通道数量,因此依然是32个
with tf.name_scope("pool_1"):
pool_1 = max_pool_2x2(conv_1)
# 第2个卷积层
# 输入通道:32,输出通道:64,卷积后图像尺寸不变,依然是16*16
with tf.name_scope("conv_2"):
W2 = weight([3, 3, 32, 64])
b2 = bias([64])
conv_2 = conv2d(pool_1, W2) + b2
conv_2 = tf.nn.relu(conv_2)
# 第2个池化层
# 将16*16图像缩小为8*8,池化不改变通道数量,因此以此是64个
with tf.name_scope("pool_2"):
pool_2 = max_pool_2x2(conv_2)
# 全连接层
# 将第2个池化层的64个8*8的图像转换为一维的向量,长度是64*8*8=4096
with tf.name_scope("fc"):
W3 = weight([4096, 128])
b3 = bias([128])
flat = tf.reshape(pool_2, [-1, 4096])
h = tf.nn.relu(tf.matmul(flat, W3) + b3)
h_dropout = tf.nn.dropout(h, keep_prob=0.8)
# 输出层
# 输出层共有10个神经元,对应到0~9这10个类别
with tf.name_scope("output_layer"):
W4 = weight([128, 10])
b4 = bias([10])
pred = tf.nn.softmax(tf.matmul(h_dropout, W4) + b4)
```
## 构建模型
```
with tf.name_scope("optimizer"):
# 定义占位符
y = tf.placeholder("float", shape=[None, 10], name="label")
# 定义损失函数
loss_function = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
# 选择优化器
optimizer = tf.train.AdamOptimizer(learning_rate=0.0001).minimize(loss_function)
```
## 定义准确率
```
with tf.name_scope("evaluation"):
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, "float"))
```
## 定义模型参数
```
train_epochs = 25
batch_size = 50
total_batch = int(len(Xtrain) / batch_size)
```
## 启动会话
```
epoch_list = []
accuracy_list = []
loss_list = []
epoch = tf.Variable(0, name="epoch", trainable=False)
startTime = time()
session = tf.Session()
init = tf.global_variables_initializer()
session.run(init)
```
## 断点续训
```
# 设置检查点存储目录
ckpt_dir = "Model_ckpt/8-CIFAR/"
if not os.path.exists(ckpt_dir):
os.makedirs(ckpt_dir)
# 生成saver
saver = tf.train.Saver(max_to_keep=1)
# 如果有检查点文件,读取最新的检查点文件,恢复各种变量值
ckpt = tf.train.latest_checkpoint(ckpt_dir)
if ckpt != None:
# 恢复所有参数
saver.restore(session, ckpt)
else:
# 从恢复点使用模型预测,或接着训练
print("Training from scratch")
# 获取续训参数
start = session.run(epoch)
print("Training starts from {} epoch".format(start + 1))
```
## 迭代训练
改进方式(根据个人可利用的计算资源)
+ 增加网络层数
+ 增加迭代次数
+ 增加全连接层数
+ 增加全连接层的神经元个数
+ 数据扩增
+ ...
```
def get_train_batch(number, batch_size):
return Xtrain_normalize[number *
batch_size:(number + 1) * batch_size], Ytrain_onehot[number *
batch_size:(number + 1) * batch_size]
for ep in range(start, train_epochs):
for i in range(total_batch):
batch_x, batch_y = get_train_batch(i, batch_size)
session.run(optimizer, feed_dict={x: batch_x, y: batch_y})
if i % 100 == 0:
print("Step: {}".format(i), "Finish")
loss, acc = session.run([loss_function, accuracy], feed_dict={x: batch_x, y: batch_y})
epoch_list.append(ep + 1)
loss_list.append(loss)
accuracy_list.append(acc)
print("Train epoch:", "%02d" % (session.run(epoch) + 1),
"Loss=", "{:.6f}".format(loss),
"Accuracy=", acc)
# 保存检查点
saver.save(session, ckpt_dir + "CIFAR10_cnn_model.ckpt", global_step=ep + 1)
session.run(epoch.assign(ep + 1))
duration = time() - startTime
print("Train finished takes: ", duration)
```
## 可视化损失值
```
flg = plt.gcf()
# flg.set_size_inches(4,2)
plt.plot(epoch_list, loss_list, label="loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend(["loss"], loc="upper right")
```
## 可视化准确率
```
plt.plot(epoch_list, accuracy_list, label="accuracy")
fig = plt.gcf()
plt.ylim(0.1, 1)
plt.xlabel("epoch")
plt.ylabel("accuracy")
plt.legend()
plt.show()
```
## 评估模型
```
test_total_batch = int(len(Xtest_normalize) / batch_size)
test_acc_sum = 0.0
for i in range(test_total_batch):
test_image_batch = Xtest_normalize[i * batch_size:(i + 1) * batch_size]
test_label_batch = Ytest_onehot[i * batch_size:(i + 1) * batch_size]
test_batch_acc = session.run(accuracy, feed_dict={x: test_image_batch, y: test_label_batch})
test_acc_sum += test_batch_acc
test_acc = float(test_acc_sum / test_total_batch)
print("Test accuracy: {:.6f}".format(test_acc))
```
## 利用模型进行预测
```
test_pred = session.run(pred, feed_dict={x: Xtest_normalize[:10]})
prediction_result = session.run(tf.argmax(test_pred, 1))
```
## 可视化预测结果
```
plot_images_labels_prediction(Xtest, Ytest, prediction_result, 0, 10)
```
|
github_jupyter
|
import os
import pickle as p
import tarfile
import urllib.request
from time import time
import matplotlib.pyplot as plt
import numpy as np
import tensorflow as tf
from sklearn.preprocessing import OneHotEncoder
url = "https://www.cs.toronto.edu/~kriz/cifar-10-python.tar.gz"
filepath = "Data/CIFAR-10/cifar-10-python.tar.gz"
# 下载数据集
if not os.path.isfile(filepath):
result = urllib.request.urlretrieve(url, filepath)
print("downloaded: ", result)
else:
print("Data file already exists")
# 解压数据集
if not os.path.exists("Data/CIFAR-10/DataSets"):
tfile = tarfile.open("Data/CIFAR-10/cifar-10-python.tar.gz", "r:gz")
result = tfile.extractall("Data/CIFAR-10/DataSets")
print("Extract to Data/CIFAR-10/DataSets")
else:
print("Data file already exists")
# 按批次导入
def load_CIFAR_batch(filename):
with open(filename, "rb") as f:
data_dict = p.load(f, encoding="bytes")
images = data_dict[b"data"]
labels = data_dict[b"labels"]
# 将原始数据调整为BCWH
images = images.reshape(10000, 3, 32, 32)
# tensorflow处理图象数据的结构:BWHC
# 把通道数数据C移动到最后一个维度
images = images.transpose(0, 2, 3, 1)
labels = np.array(labels)
return images, labels
# 导入数据集
def load_CIFAR_data(data_dir):
images_train = []
labels_train = []
for i in range(5):
f = os.path.join(data_dir, "data_batch_%d" % (i + 1))
print("loading", f)
# 调用load_CIFAR_batch()获得批量的图像机器对应的标签
image_batch, label_batch = load_CIFAR_batch(f)
images_train.append(image_batch)
labels_train.append(label_batch)
Xtrain = np.concatenate(images_train)
Ytrain = np.concatenate(labels_train)
del image_batch, label_batch
Xtest, Ytest = load_CIFAR_batch(os.path.join(data_dir, "test_batch"))
print("Finished loading CIFAR-10 data")
# 返回训练集的体香和标签,测试集的图像和标签
return Xtrain, Ytrain, Xtest, Ytest
data_dir = "Data/CIFAR-10/DataSets/cifar-10-batches-py"
Xtrain, Ytrain, Xtest, Ytest = load_CIFAR_data(data_dir)
print("Training data shape: ", Xtrain.shape)
print("Training labels shape: ", Ytrain.shape)
print("Test im shape: ", Xtest.shape)
print("Test labels shape: ", Ytest.shape)
# 查看单项数据
plt.imshow(Xtrain[6])
print(Ytrain[6])
# 定义标签字典,每一个数字所代表的图像类别的名称
label_dict = {0: "airplane", 1: "automobile", 2: "bird", 3: "cat", 4: "deer", 5: "dog", 6: "frog", 7: "horse",
8: "ship", 9: "truck"}
# 定义显示图像数据及其对应标签的函数
def plot_images_labels_prediction(images, labels, prediction, idx, num=10):
fig = plt.gcf()
fig.set_size_inches(12, 6)
if num > 10:
num = 10
for i in range(0, num):
ax = plt.subplot(2, 5, 1 + i)
ax.imshow(images[idx], cmap="binary")
title = str(i) + ", " + label_dict[labels[idx]]
if len(prediction) > 0:
title += "=>" + label_dict[prediction[idx]]
ax.set_title(title, fontsize=10)
idx += 1
plt.show()
plot_images_labels_prediction(Xtest, Ytest, [], 1, 10)
# 查看图像数据信息
# 显示第一个图的第一个像素点
Xtrain[0][0][0]
# 将图像进行数字标准化
Xtrain_normalize = Xtrain.astype("float32") / 255.0
Xtest_normalize = Xtest.astype("float32") / 255.0
# 查看预处理后的图像数据信息
Xtrain_normalize[0][0][0]
# 查看标签数据
Ytrain[:10]
encoder = OneHotEncoder(sparse=False)
yy = [[0], [1], [2], [3], [4], [5], [6], [7], [8], [9]]
encoder.fit(yy)
Ytrain_reshape = Ytrain.reshape(-1, 1)
Ytrain_onehot = encoder.transform(Ytrain_reshape)
Ytest_reshape = Ytest.reshape(-1, 1)
Ytest_onehot = encoder.transform(Ytest_reshape)
Ytrain_onehot.shape
Ytrain_onehot[:5]
# 定义权值
def weight(shape):
# 在构建模型时,需要使用tfVariable来创建一个变量
# 在训练时,这个变量不断更新
# 使用函数tf.truncated.normal(截断的正态分布)生成标准差为0.1的随机数来初始化权值
return tf.Variable(tf.truncated_normal(shape, stddev=0.1), name="W")
# 定义偏置
# 初始化0.1
def bias(shape):
return tf.Variable(tf.constant(0.1, shape=shape), name="b")
# 定义卷积操作
# 步长为1,padding为"SAME"
def conv2d(x, W):
# tf.nn.conv2d(input, filter, strides, padding, use_cudnn_oon_gpu=None, name=None)
return tf.nn.conv2d(x, W, strides=[1, 1, 1, 1], padding="SAME")
# 定义池化操作
# 步长为2,即原尺寸的长和宽各除以2
def max_pool_2x2(x):
# tf.nn.max_pool(value, ksize, strides, padding, name=None)
return tf.nn.max_pool(x, ksize=[1, 2, 2, 1], strides=[1, 2, 2, 1], padding="SAME")
# 输入层
# 32*32图像,通道为3(RGB)
with tf.name_scope("input_layer"):
x = tf.placeholder("float", shape=[None, 32, 32, 3], name="X")
# 第1个卷积层
# 输入通道:3,输出通道:32,卷积后图像尺寸不变,依然是32*32
with tf.name_scope("conv_1"):
# [k_width, k_height, input_chn, output_chn]
W1 = weight([3, 3, 3, 32])
b1 = bias([32])
conv_1 = conv2d(x, W1) + b1
conv_1 = tf.nn.relu(conv_1)
# 第1个池化层
# 将32*32图像缩小为16*16,池化不改变通道数量,因此依然是32个
with tf.name_scope("pool_1"):
pool_1 = max_pool_2x2(conv_1)
# 第2个卷积层
# 输入通道:32,输出通道:64,卷积后图像尺寸不变,依然是16*16
with tf.name_scope("conv_2"):
W2 = weight([3, 3, 32, 64])
b2 = bias([64])
conv_2 = conv2d(pool_1, W2) + b2
conv_2 = tf.nn.relu(conv_2)
# 第2个池化层
# 将16*16图像缩小为8*8,池化不改变通道数量,因此以此是64个
with tf.name_scope("pool_2"):
pool_2 = max_pool_2x2(conv_2)
# 全连接层
# 将第2个池化层的64个8*8的图像转换为一维的向量,长度是64*8*8=4096
with tf.name_scope("fc"):
W3 = weight([4096, 128])
b3 = bias([128])
flat = tf.reshape(pool_2, [-1, 4096])
h = tf.nn.relu(tf.matmul(flat, W3) + b3)
h_dropout = tf.nn.dropout(h, keep_prob=0.8)
# 输出层
# 输出层共有10个神经元,对应到0~9这10个类别
with tf.name_scope("output_layer"):
W4 = weight([128, 10])
b4 = bias([10])
pred = tf.nn.softmax(tf.matmul(h_dropout, W4) + b4)
with tf.name_scope("optimizer"):
# 定义占位符
y = tf.placeholder("float", shape=[None, 10], name="label")
# 定义损失函数
loss_function = tf.reduce_mean(
tf.nn.softmax_cross_entropy_with_logits(logits=pred, labels=y))
# 选择优化器
optimizer = tf.train.AdamOptimizer(learning_rate=0.0001).minimize(loss_function)
with tf.name_scope("evaluation"):
correct_prediction = tf.equal(tf.argmax(pred, 1), tf.argmax(y, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, "float"))
train_epochs = 25
batch_size = 50
total_batch = int(len(Xtrain) / batch_size)
epoch_list = []
accuracy_list = []
loss_list = []
epoch = tf.Variable(0, name="epoch", trainable=False)
startTime = time()
session = tf.Session()
init = tf.global_variables_initializer()
session.run(init)
# 设置检查点存储目录
ckpt_dir = "Model_ckpt/8-CIFAR/"
if not os.path.exists(ckpt_dir):
os.makedirs(ckpt_dir)
# 生成saver
saver = tf.train.Saver(max_to_keep=1)
# 如果有检查点文件,读取最新的检查点文件,恢复各种变量值
ckpt = tf.train.latest_checkpoint(ckpt_dir)
if ckpt != None:
# 恢复所有参数
saver.restore(session, ckpt)
else:
# 从恢复点使用模型预测,或接着训练
print("Training from scratch")
# 获取续训参数
start = session.run(epoch)
print("Training starts from {} epoch".format(start + 1))
def get_train_batch(number, batch_size):
return Xtrain_normalize[number *
batch_size:(number + 1) * batch_size], Ytrain_onehot[number *
batch_size:(number + 1) * batch_size]
for ep in range(start, train_epochs):
for i in range(total_batch):
batch_x, batch_y = get_train_batch(i, batch_size)
session.run(optimizer, feed_dict={x: batch_x, y: batch_y})
if i % 100 == 0:
print("Step: {}".format(i), "Finish")
loss, acc = session.run([loss_function, accuracy], feed_dict={x: batch_x, y: batch_y})
epoch_list.append(ep + 1)
loss_list.append(loss)
accuracy_list.append(acc)
print("Train epoch:", "%02d" % (session.run(epoch) + 1),
"Loss=", "{:.6f}".format(loss),
"Accuracy=", acc)
# 保存检查点
saver.save(session, ckpt_dir + "CIFAR10_cnn_model.ckpt", global_step=ep + 1)
session.run(epoch.assign(ep + 1))
duration = time() - startTime
print("Train finished takes: ", duration)
flg = plt.gcf()
# flg.set_size_inches(4,2)
plt.plot(epoch_list, loss_list, label="loss")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend(["loss"], loc="upper right")
plt.plot(epoch_list, accuracy_list, label="accuracy")
fig = plt.gcf()
plt.ylim(0.1, 1)
plt.xlabel("epoch")
plt.ylabel("accuracy")
plt.legend()
plt.show()
test_total_batch = int(len(Xtest_normalize) / batch_size)
test_acc_sum = 0.0
for i in range(test_total_batch):
test_image_batch = Xtest_normalize[i * batch_size:(i + 1) * batch_size]
test_label_batch = Ytest_onehot[i * batch_size:(i + 1) * batch_size]
test_batch_acc = session.run(accuracy, feed_dict={x: test_image_batch, y: test_label_batch})
test_acc_sum += test_batch_acc
test_acc = float(test_acc_sum / test_total_batch)
print("Test accuracy: {:.6f}".format(test_acc))
test_pred = session.run(pred, feed_dict={x: Xtest_normalize[:10]})
prediction_result = session.run(tf.argmax(test_pred, 1))
plot_images_labels_prediction(Xtest, Ytest, prediction_result, 0, 10)
| 0.305905 | 0.825414 |
```
import pandas as pd
import numpy as np
import matplotlib as plt
import seaborn as sns
from sklearn.svm import SVC
```
## Question 5) String kernels
*Note: data taken from https://cseweb.ucsd.edu/classes/wi17/cse151-a/hw5.pdf along with inspiration for this problem.*
As we've seen, kernels have wide applications. We've seen kernels be a time efficient way to lift a feature matrix in many different contexts like PCA, Perceptrons, SVM's and Ridge Regression. One great property of kernels is the ability to supply any valid kernel function to one of our kernelized model it will work within that space.
For this problem, we will be working with **string kernels**, which apply kernel functions to text data so that models like SVM can work with them. Remember kernel functions are essentially similarity functions. Therefore, string kernel functions generally tell us how similar two strings are.
### Question 5a)
First, let's take a look at our data.
```
train_data = pd.read_csv('string_train.csv')
train_data.head()
```
The sequence is a sequence of amino acids, and the classification is whether or not the sequence belongs to a particular protein family or not.
Visualize the number of positive and negative sequences we have using a barplot.
```
# START TODO
# END TODO
```
Answer the following questions.
* **Brainstorm some ways to deal with this text data. How can we compute similarity between two strings?**
* **Does the imbalance in data matter? Why or why not?**
### Question 5b)
For this problem, we are going to be using the **Spectrum Kernel**, one of the most simple string kernels. The basic idea behind the kernel is that two strings with more common substrings will be more similar. What is a substring? A substring is any contiguous sequence of characters within a string. The kernel function the **p-spectrum kernel** expresses this idea. $k(s_1, s_2)$ counts all size $p$ substrings that are present in both the string $s_1$ and the string $s_2$.
Fill out the code below to finish the implementation of the kernel.
*Note 1: There are many other valid string kernels, many of which are much better and domain specific than this one. For more string kernels, check here https://people.eecs.berkeley.edu/~jordan/kernels/0521813972c11_p344-396.pdf. In fact, Michael I Jordan has a whole book on kernels for pattern recognition that might be fun to look at.*
*Note 2: Section 1.4.6.2.2 https://scikit-learn.org/stable/modules/svm.html#kernel-functions might be useful.*
```
def p_spectrum(s, t, p):
num_in_common = ...
# START TODO
num_in_common = 0
for i in range(len(s) - p + 1):
curr = s[i:i+p]
if curr in t:
num_in_common += 1
# END TODO
return num_in_common
```
Now that we have our kernel function defined, let's actually use it to classify our dataset. We will be using an SVM classifier. Unfortunately, scikit learn doesn't support string kernel functions for SVM's, but there are a few work arounds. The first work around is precomputing the **Gram matrix** $K$ for our data, and then running an SVM on it. Remember, all we need is the Gram matrix to do classification.
*Note: it will take a few seconds to a minute to run this part of the code.*
```
X_train = np.array(train_data["Sequence"])
y_train = np.array(train_data["Classification"])
def compute_gram_matrix(X_one, X_two, y, p=3):
# START COMPUTE GRAM MATRIX
K = np.zeros((X_one.shape[0], X_two.shape[0]))
for s_index, s in enumerate(X_one):
for t_index, t in enumerate(X_two):
K[s_index][t_index] = p_spectrum(s, t, p)
# END COMPUTE GRAM MATRIX
return K
K_train = ...
# START TODO
K_train = compute_gram_matrix(X_train, X_train, y_train)
# END TODO
print (K_train.shape)
K_train
```
### Question 5c)
Now let's try some classifiers out. Let's start with the SVM classifier. Complete the below code to fit an SVM model with our precomputed Gram matrix and print out the accuracy on the train set.
```
clf = SVC(kernel='precomputed')
clf.fit(K_train, y_train)
score = clf.score(K_train, y_train)
print ("Accuracy on the training data: " + str(score))
```
Now calculate the accuracy on the testing data.
```
test_data = pd.read_csv('string_test.csv')
X_test = np.array(test_data["Sequence"])
y_test = np.array(test_data["Classification"])
# START TODO
K_test = compute_gram_matrix(X_test, X_train, y_test)
y_pred = clf.predict(K_test)
# END TODO
score = clf.score(K_test, y_test)
print ("Accuracy on the testing data: " + str(score))
```
Answer the following questions.
* **What was your accuracy on the test dataset? Was it different from the training dataset?**
* **Can you think of any improvements to our kernel function?**
Congrats! You just used string kernels to classify a real dataset of Amino Acid sequences! Hopefully you can see the power of kernels from this example. Further applications include Graph kernels, Tree kernels, kernels for images... the possibilities are endless!
If you want to learn more, read some of Michael I Jordan's stuff on this subject here: https://people.eecs.berkeley.edu/~jordan/kernels/0521813972pre_pi-xiv.pdf
If you're interested in string kernels, here are some good papers to read that build upon the simple p-spectrum kernel we used in this example!
* Mismatch kernels - https://papers.nips.cc/paper/2179-mismatch-string-kernels-for-svm-protein-classification.pdf
* Gappy kernels - https://www.semanticscholar.org/paper/A-fast-%2C-large-scale-learning-method-for-protein-Kuksa-Huang/bd5a49164b7d0a9179ef5cb39148279825877a7f
* Motif kernels - https://almob.biomedcentral.com/articles/10.1186/1748-7188-1-21
* More spectrum kernels - https://pubmed.ncbi.nlm.nih.gov/11928508/
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib as plt
import seaborn as sns
from sklearn.svm import SVC
train_data = pd.read_csv('string_train.csv')
train_data.head()
# START TODO
# END TODO
def p_spectrum(s, t, p):
num_in_common = ...
# START TODO
num_in_common = 0
for i in range(len(s) - p + 1):
curr = s[i:i+p]
if curr in t:
num_in_common += 1
# END TODO
return num_in_common
X_train = np.array(train_data["Sequence"])
y_train = np.array(train_data["Classification"])
def compute_gram_matrix(X_one, X_two, y, p=3):
# START COMPUTE GRAM MATRIX
K = np.zeros((X_one.shape[0], X_two.shape[0]))
for s_index, s in enumerate(X_one):
for t_index, t in enumerate(X_two):
K[s_index][t_index] = p_spectrum(s, t, p)
# END COMPUTE GRAM MATRIX
return K
K_train = ...
# START TODO
K_train = compute_gram_matrix(X_train, X_train, y_train)
# END TODO
print (K_train.shape)
K_train
clf = SVC(kernel='precomputed')
clf.fit(K_train, y_train)
score = clf.score(K_train, y_train)
print ("Accuracy on the training data: " + str(score))
test_data = pd.read_csv('string_test.csv')
X_test = np.array(test_data["Sequence"])
y_test = np.array(test_data["Classification"])
# START TODO
K_test = compute_gram_matrix(X_test, X_train, y_test)
y_pred = clf.predict(K_test)
# END TODO
score = clf.score(K_test, y_test)
print ("Accuracy on the testing data: " + str(score))
| 0.160299 | 0.95096 |
```
from SeismicReduction import *
import pickle
```
## Modify training and VaeModel to return loss value from training:
```
def train(epoch, model, optimizer, train_loader, beta=1, recon_loss_method='mse'):
"""
Trains a single epoch of the vae model.
Parameters
----------
epoch : int
epoch number being trained
model : torch.nn.module
model being trained, here a vae
optimizer : torch.optim
optmizer used to train model
train_loader : torch.utils.data.DataLoader
data loader used for training
beta : float
beta parameter for the beta-vae
recon_loss_method : str
specifies the reconstruction loss technique
Returns
-------
trains the model and returns training loss for the epoch
"""
model.train()
train_loss = 0
for batch_idx, (data, _) in enumerate(train_loader):
data = Variable(data)
optimizer.zero_grad()
recon_batch, mu, logvar, _ = model(data)
loss = loss_function(recon_batch,
data,
mu,
logvar,
window_size=data.shape[-1],
beta=beta,
recon_loss_method=recon_loss_method)
# print('batch:', batch_idx, 'loss:', loss.item())
loss.backward()
# 'loss' is the SUM of all vector to vector losses in batch
train_loss += loss.item() # * data.size(0) # originally
# print('batch:', batch_idx, 'to add to total:', loss.item())
optimizer.step()
train_loss /= len(train_loader.dataset)
# print('====> Epoch: {} Average loss: {:.4f}'.format(epoch, train_loss), len(train_loader.dataset))
return train_loss
class VaeModel(ModelAgent):
"""
Runs the VAE model to reduce the seismic data to an arbitrary sized dimension, visualised in 2 via UMAP.
"""
def __init__(self, data):
super().__init__(data)
self.name = 'VAE'
def create_dataloader(self, batch_size=32):
"""
Create pytorch data loaders for use in vae training, testing and running.
Parameters
----------
batch_size : int
Size of data loader batches.
Returns
-------
Modifies object data loader attributes.
"""
# create torch tensor
assert self.input.shape[1] == 2, 'Expected a three dimensional input with 2 channels'
X = torch.from_numpy(self.input).float()
# Create a stacked representation and a zero tensor so we can use the standard Pytorch TensorDataset
y = torch.from_numpy(np.zeros((X.shape[0], 1))).float()
split = ShuffleSplit(n_splits=1, test_size=0.5)
for train_index, test_index in split.split(X):
X_train, y_train = X[train_index], y[train_index]
X_test, y_test = X[test_index], y[test_index]
train_dset = TensorDataset(X_train, y_train)
test_dset = TensorDataset(X_test, y_test)
all_dset = TensorDataset(X, y)
kwargs = {'num_workers': 1, 'pin_memory': True}
self.train_loader = torch.utils.data.DataLoader(train_dset,
batch_size=batch_size,
shuffle=True,
**kwargs)
self.test_loader = torch.utils.data.DataLoader(test_dset,
batch_size=batch_size,
shuffle=False,
**kwargs)
self.all_loader = torch.utils.data.DataLoader(all_dset,
batch_size=batch_size,
shuffle=False,
**kwargs)
def train_vae(self, epochs=5, hidden_size=8, lr=1e-2, recon_loss_method='mse'):
"""
Handles the training of the vae model.
Parameters
----------
epochs : int
Number of complete passes over the whole training set.
hidden_size : int
Size of the latent space of the vae.
lr : float.
Learning rate for the vae model training.
recon_loss_method : str
Method for reconstruction loss calculation
Returns
-------
None
"""
set_seed(42) # Set the random seed
self.model = VAE(hidden_size,
self.input.shape) # Inititalize the model
optimizer = optim.Adam(self.model.parameters(),
lr=lr,
betas=(0.9, 0.999))
if self.plot_loss:
liveloss = PlotLosses()
liveloss.skip_first = 0
liveloss.figsize = (16, 10)
# Start training loop
for epoch in range(1, epochs + 1):
tl = train(epoch,
self.model,
optimizer,
self.train_loader,
recon_loss_method=recon_loss_method) # Train model on train dataset
testl = test(epoch, self.model, self.test_loader, recon_loss_method=recon_loss_method)
if self.plot_loss: # log train and test losses for dynamic plot
logs = {}
logs['' + 'ELBO'] = tl
logs['val_' + 'ELBO'] = testl
liveloss.update(logs)
liveloss.draw()
return testl
def run_vae(self):
"""
Run the full data set through the trained vae model.
Returns
-------
Modifies the zs attribute, an array of shape (number_traces, latent_space)
"""
_, zs = forward_all(self.model, self.all_loader)
return zs.numpy()
def reduce(self, epochs=5, hidden_size=8, lr=1e-2, recon_loss_method='mse', plot_loss=True):
"""
Controller function for the vae model.
Parameters
----------
epochs : int
Number of epochs to run vae model.
hidden_size : int
Size of the vae model latent space representation.
lr : float
Learning rate for vae model training.
recon_loss_method : str
Method for reconstruction loss calculation
plot_loss : bool
Control on whether to plot the loss on vae training.
Returns
-------
Modifies embedding attribute via generation of the low dimensional representation.
"""
if hidden_size < 2:
raise Exception('Please use hidden size > 1')
self.plot_loss = plot_loss # define whether to plot training losses or not
self.create_dataloader()
if not self.loaded_model:
loss = self.train_vae(epochs=epochs, hidden_size=hidden_size, lr=lr, recon_loss_method=recon_loss_method)
self.embedding = self.run_vae() # arbitrary dimension output from VAE
return loss
# load data
file_pi2 = open('../pickled/data.pickle', 'rb')
dataholder = pickle.load(file_pi2)
file_pi2.close()
### Processor
processor = Processor(dataholder)
input1 = processor(flatten=[True, 12, 52], normalise=True)
```
# Latent dimension testing:
```
vae = VaeModel(input1)
losses = []
embeddings = []
latent_dims = [i for i in range(2,64,4)]
print('dimensions tested:', latent_dims)
for i in latent_dims:
loss = vae.reduce(epochs=100, hidden_size=i, lr=0.0005, plot_loss=False)
print('dim', i, 'loss', loss)
embeddings.append(vae.embedding)
losses.append(loss)
print(losses)
fig, ax = plt.subplots(figsize=(8, 5))
ax.plot(latent_dims, losses, marker='o', color='black')
ax.set_title('ELBO performance latent dimension')
ax.set_ylabel('ELBO')
ax.set_xlabel('Latent dimension')
fig.tight_layout()
```
|
github_jupyter
|
from SeismicReduction import *
import pickle
def train(epoch, model, optimizer, train_loader, beta=1, recon_loss_method='mse'):
"""
Trains a single epoch of the vae model.
Parameters
----------
epoch : int
epoch number being trained
model : torch.nn.module
model being trained, here a vae
optimizer : torch.optim
optmizer used to train model
train_loader : torch.utils.data.DataLoader
data loader used for training
beta : float
beta parameter for the beta-vae
recon_loss_method : str
specifies the reconstruction loss technique
Returns
-------
trains the model and returns training loss for the epoch
"""
model.train()
train_loss = 0
for batch_idx, (data, _) in enumerate(train_loader):
data = Variable(data)
optimizer.zero_grad()
recon_batch, mu, logvar, _ = model(data)
loss = loss_function(recon_batch,
data,
mu,
logvar,
window_size=data.shape[-1],
beta=beta,
recon_loss_method=recon_loss_method)
# print('batch:', batch_idx, 'loss:', loss.item())
loss.backward()
# 'loss' is the SUM of all vector to vector losses in batch
train_loss += loss.item() # * data.size(0) # originally
# print('batch:', batch_idx, 'to add to total:', loss.item())
optimizer.step()
train_loss /= len(train_loader.dataset)
# print('====> Epoch: {} Average loss: {:.4f}'.format(epoch, train_loss), len(train_loader.dataset))
return train_loss
class VaeModel(ModelAgent):
"""
Runs the VAE model to reduce the seismic data to an arbitrary sized dimension, visualised in 2 via UMAP.
"""
def __init__(self, data):
super().__init__(data)
self.name = 'VAE'
def create_dataloader(self, batch_size=32):
"""
Create pytorch data loaders for use in vae training, testing and running.
Parameters
----------
batch_size : int
Size of data loader batches.
Returns
-------
Modifies object data loader attributes.
"""
# create torch tensor
assert self.input.shape[1] == 2, 'Expected a three dimensional input with 2 channels'
X = torch.from_numpy(self.input).float()
# Create a stacked representation and a zero tensor so we can use the standard Pytorch TensorDataset
y = torch.from_numpy(np.zeros((X.shape[0], 1))).float()
split = ShuffleSplit(n_splits=1, test_size=0.5)
for train_index, test_index in split.split(X):
X_train, y_train = X[train_index], y[train_index]
X_test, y_test = X[test_index], y[test_index]
train_dset = TensorDataset(X_train, y_train)
test_dset = TensorDataset(X_test, y_test)
all_dset = TensorDataset(X, y)
kwargs = {'num_workers': 1, 'pin_memory': True}
self.train_loader = torch.utils.data.DataLoader(train_dset,
batch_size=batch_size,
shuffle=True,
**kwargs)
self.test_loader = torch.utils.data.DataLoader(test_dset,
batch_size=batch_size,
shuffle=False,
**kwargs)
self.all_loader = torch.utils.data.DataLoader(all_dset,
batch_size=batch_size,
shuffle=False,
**kwargs)
def train_vae(self, epochs=5, hidden_size=8, lr=1e-2, recon_loss_method='mse'):
"""
Handles the training of the vae model.
Parameters
----------
epochs : int
Number of complete passes over the whole training set.
hidden_size : int
Size of the latent space of the vae.
lr : float.
Learning rate for the vae model training.
recon_loss_method : str
Method for reconstruction loss calculation
Returns
-------
None
"""
set_seed(42) # Set the random seed
self.model = VAE(hidden_size,
self.input.shape) # Inititalize the model
optimizer = optim.Adam(self.model.parameters(),
lr=lr,
betas=(0.9, 0.999))
if self.plot_loss:
liveloss = PlotLosses()
liveloss.skip_first = 0
liveloss.figsize = (16, 10)
# Start training loop
for epoch in range(1, epochs + 1):
tl = train(epoch,
self.model,
optimizer,
self.train_loader,
recon_loss_method=recon_loss_method) # Train model on train dataset
testl = test(epoch, self.model, self.test_loader, recon_loss_method=recon_loss_method)
if self.plot_loss: # log train and test losses for dynamic plot
logs = {}
logs['' + 'ELBO'] = tl
logs['val_' + 'ELBO'] = testl
liveloss.update(logs)
liveloss.draw()
return testl
def run_vae(self):
"""
Run the full data set through the trained vae model.
Returns
-------
Modifies the zs attribute, an array of shape (number_traces, latent_space)
"""
_, zs = forward_all(self.model, self.all_loader)
return zs.numpy()
def reduce(self, epochs=5, hidden_size=8, lr=1e-2, recon_loss_method='mse', plot_loss=True):
"""
Controller function for the vae model.
Parameters
----------
epochs : int
Number of epochs to run vae model.
hidden_size : int
Size of the vae model latent space representation.
lr : float
Learning rate for vae model training.
recon_loss_method : str
Method for reconstruction loss calculation
plot_loss : bool
Control on whether to plot the loss on vae training.
Returns
-------
Modifies embedding attribute via generation of the low dimensional representation.
"""
if hidden_size < 2:
raise Exception('Please use hidden size > 1')
self.plot_loss = plot_loss # define whether to plot training losses or not
self.create_dataloader()
if not self.loaded_model:
loss = self.train_vae(epochs=epochs, hidden_size=hidden_size, lr=lr, recon_loss_method=recon_loss_method)
self.embedding = self.run_vae() # arbitrary dimension output from VAE
return loss
# load data
file_pi2 = open('../pickled/data.pickle', 'rb')
dataholder = pickle.load(file_pi2)
file_pi2.close()
### Processor
processor = Processor(dataholder)
input1 = processor(flatten=[True, 12, 52], normalise=True)
vae = VaeModel(input1)
losses = []
embeddings = []
latent_dims = [i for i in range(2,64,4)]
print('dimensions tested:', latent_dims)
for i in latent_dims:
loss = vae.reduce(epochs=100, hidden_size=i, lr=0.0005, plot_loss=False)
print('dim', i, 'loss', loss)
embeddings.append(vae.embedding)
losses.append(loss)
print(losses)
fig, ax = plt.subplots(figsize=(8, 5))
ax.plot(latent_dims, losses, marker='o', color='black')
ax.set_title('ELBO performance latent dimension')
ax.set_ylabel('ELBO')
ax.set_xlabel('Latent dimension')
fig.tight_layout()
| 0.933741 | 0.842086 |
Based on https://github.com/Savvysherpa/slda .
Modified to the prediction of multivariate normal (diagonal covariance) responses.
```
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
V = 25 # number of vocabulary
K = 10 # number of topics
N = 100 # number of words in each document
D = 1000 # total number of documents
```
## Generate topics
```
topics = []
topic_base = np.concatenate((np.ones((1, 5)) * 0.2, np.zeros((4, 5))), axis=0).ravel()
for i in range(5):
topics.append(np.roll(topic_base, i * 5))
topic_base = np.concatenate((np.ones((5, 1)) * 0.2, np.zeros((5, 4))), axis=1).ravel()
for i in range(5):
topics.append(np.roll(topic_base, i))
topics = np.array(topics)
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(topics[k, :].reshape((5, 5)))
plt.show()
```
## Generate documents from topics
```
alpha = np.ones(K)
thetas = np.random.dirichlet(alpha, size=D)
topic_assignments = np.array([np.random.choice(range(K), size=N, p=theta)
for theta in thetas])
word_assignments = np.array([[np.random.choice(range(V), size=1, p=topics[topic_assignments[d, n]])[0]
for n in range(N)] for d in range(D)])
doc_term_matrix = np.array([np.histogram(word_assignments[d], bins=V, range=(0, V - 1))[0] for d in range(D)])
fig, ax = plt.subplots(figsize=(4,3))
ax.imshow(doc_term_matrix, aspect='auto')
plt.show()
```
## Generate responses
```
nu2 = 10
sigma2 = 1
eta1 = np.random.normal(scale=nu2, size=K)
eta2 = np.random.normal(scale=nu2, size=K)
fig, axes = plt.subplots(figsize=(9,3), ncols=2)
axes[0].plot(range(K), eta1, color='b')
axes[1].plot(range(K), eta2, color='r')
axes[0].set_title('eta1'); axes[1].set_title('eta2')
plt.show()
y1 = [np.dot(eta1, thetas[i]) for i in range(D)] + np.random.normal(scale=sigma2, size=D)
y2 = [np.dot(eta2, thetas[i]) for i in range(D)] + np.random.normal(scale=sigma2, size=D)
y = np.hstack((y1[:, np.newaxis], y2[:, np.newaxis]))
fig, ax = plt.subplots()
ax.hist(y[:, 0], bins=20, alpha=.5, label='y1')
ax.hist(y[:, 1], bins=20, alpha=.5, label='y2')
ax.legend()
plt.show()
np.savetxt('y.txt', y)
with open('train.dat', 'w') as ofp:
for i in range(D):
ofp.write(' '.join([str(ID)+':'+str(Cnt) for ID, Cnt in zip(range(1,V+1), doc_term_matrix[i,:])])+'\n')
```
## Estimate parameters
```
!../src/mvslda -I 300 -K 10 -Y 2 ./train.dat ./y.txt ./model
# likelihood
lik = np.loadtxt('./model.lik')
fig, ax = plt.subplots()
ax.plot(range(len(lik)), lik)
plt.show()
# phi
phi = np.loadtxt('./model.phi')
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(phi[:, k].reshape((5, 5)))
plt.show()
# topic reordering
from scipy.spatial.distance import cdist
topic_reorder = np.argsort([np.argmin(cdist([phi[:, k]], topics)[0]) for k in range(K)])
phi = phi[:, topic_reorder]
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(phi[:, k].reshape((5, 5)))
plt.show()
# eta
eta = np.loadtxt('./model.eta')
eta = eta[topic_reorder, :]
fig, axes = plt.subplots(figsize=(9,3), ncols=2)
axes[0].plot(range(K), eta1, color='b', label='truth')
axes[0].plot(range(K), eta[:, 0], color='g', linestyle=':', label='predicted')
axes[1].plot(range(K), eta2, color='r', label='truth')
axes[1].plot(range(K), eta[:, 1], color='g', linestyle=':', label='predicted')
axes[0].set_title('eta1'); axes[1].set_title('eta2')
axes[0].legend(); axes[1].legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
V = 25 # number of vocabulary
K = 10 # number of topics
N = 100 # number of words in each document
D = 1000 # total number of documents
topics = []
topic_base = np.concatenate((np.ones((1, 5)) * 0.2, np.zeros((4, 5))), axis=0).ravel()
for i in range(5):
topics.append(np.roll(topic_base, i * 5))
topic_base = np.concatenate((np.ones((5, 1)) * 0.2, np.zeros((5, 4))), axis=1).ravel()
for i in range(5):
topics.append(np.roll(topic_base, i))
topics = np.array(topics)
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(topics[k, :].reshape((5, 5)))
plt.show()
alpha = np.ones(K)
thetas = np.random.dirichlet(alpha, size=D)
topic_assignments = np.array([np.random.choice(range(K), size=N, p=theta)
for theta in thetas])
word_assignments = np.array([[np.random.choice(range(V), size=1, p=topics[topic_assignments[d, n]])[0]
for n in range(N)] for d in range(D)])
doc_term_matrix = np.array([np.histogram(word_assignments[d], bins=V, range=(0, V - 1))[0] for d in range(D)])
fig, ax = plt.subplots(figsize=(4,3))
ax.imshow(doc_term_matrix, aspect='auto')
plt.show()
nu2 = 10
sigma2 = 1
eta1 = np.random.normal(scale=nu2, size=K)
eta2 = np.random.normal(scale=nu2, size=K)
fig, axes = plt.subplots(figsize=(9,3), ncols=2)
axes[0].plot(range(K), eta1, color='b')
axes[1].plot(range(K), eta2, color='r')
axes[0].set_title('eta1'); axes[1].set_title('eta2')
plt.show()
y1 = [np.dot(eta1, thetas[i]) for i in range(D)] + np.random.normal(scale=sigma2, size=D)
y2 = [np.dot(eta2, thetas[i]) for i in range(D)] + np.random.normal(scale=sigma2, size=D)
y = np.hstack((y1[:, np.newaxis], y2[:, np.newaxis]))
fig, ax = plt.subplots()
ax.hist(y[:, 0], bins=20, alpha=.5, label='y1')
ax.hist(y[:, 1], bins=20, alpha=.5, label='y2')
ax.legend()
plt.show()
np.savetxt('y.txt', y)
with open('train.dat', 'w') as ofp:
for i in range(D):
ofp.write(' '.join([str(ID)+':'+str(Cnt) for ID, Cnt in zip(range(1,V+1), doc_term_matrix[i,:])])+'\n')
!../src/mvslda -I 300 -K 10 -Y 2 ./train.dat ./y.txt ./model
# likelihood
lik = np.loadtxt('./model.lik')
fig, ax = plt.subplots()
ax.plot(range(len(lik)), lik)
plt.show()
# phi
phi = np.loadtxt('./model.phi')
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(phi[:, k].reshape((5, 5)))
plt.show()
# topic reordering
from scipy.spatial.distance import cdist
topic_reorder = np.argsort([np.argmin(cdist([phi[:, k]], topics)[0]) for k in range(K)])
phi = phi[:, topic_reorder]
fig, axes = plt.subplots(figsize=(12,6), nrows=2, ncols=5)
for k in range(K):
row_ind = int(k / 5)
col_ind = k % 5
axes[row_ind, col_ind].imshow(phi[:, k].reshape((5, 5)))
plt.show()
# eta
eta = np.loadtxt('./model.eta')
eta = eta[topic_reorder, :]
fig, axes = plt.subplots(figsize=(9,3), ncols=2)
axes[0].plot(range(K), eta1, color='b', label='truth')
axes[0].plot(range(K), eta[:, 0], color='g', linestyle=':', label='predicted')
axes[1].plot(range(K), eta2, color='r', label='truth')
axes[1].plot(range(K), eta[:, 1], color='g', linestyle=':', label='predicted')
axes[0].set_title('eta1'); axes[1].set_title('eta2')
axes[0].legend(); axes[1].legend()
plt.show()
| 0.402627 | 0.923316 |
# [**Broutonlab**](https://broutonlab.com/) face recognition with masks pipeline
## [**github repo**](https://github.com/broutonlab/face-id-with-medical-masks) with solution
```
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Check GPU resources</font></b>
!nvidia-smi
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Import requirements</font></b>
import os
import sys
import cv2
from matplotlib import pyplot as plt
import sys
import numpy as np
import torch
from torch import nn
from tqdm.notebook import tqdm
from torch.utils.data import DataLoader
%matplotlib inline
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Clone and build face-alignment git repo</font></b>
!git clone https://github.com/1adrianb/face-alignment
%cd face-alignment
!pip install -r requirements.txt
!python setup.py install
import face_alignment
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Download and import face mask SDK</font></b>
!git clone https://github.com/broutonlab/face-id-with-medical-masks.git
%cd face-id-with-medical-masks
from masked_face_sdk.mask_generation_utils import end2end_mask_generation
from masked_face_sdk.pipeline_dataset_loader import PipelineFacesDatasetGenerator
from masked_face_sdk.pipeline_dataset_loader \
import PipelineFacesDatasetGenerator
from masked_face_sdk.neural_network_modules \
import Backbone, ArcFaceLayer, FaceRecognitionModel, resnet18
from masked_face_sdk.training_utils import default_acc_function, test_embedding_net
!gdown --id 1b64prOr4_E8gcD1Q_cVZkFnSzNVfGwU_
!unzip face_recognition_with_masks_dataset.zip
!ls
# Pathes to datasets for face recognition in Keras-like format
root_train_dataset_path = 'test_large/'
root_test_dataset_path = 'test_small/'
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Generate masks database</font></b>
# Generate masks database
!python3 generate_masks_database.py \
--masks-folder=data/masked_faces/ \
--database-file=data/masks_base.json \
--verbose --skip-warnings
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Prepare training dataset</font></b>
# Prepare training dataset
!python3 apply_masks_to_face_recognition_dataset.py \
--face-dataset-folder={root_train_dataset_path} \
--masks-database=data/masks_base.json \
--verbose \
--use-cuda
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Prepare test dataset</font></b>
# Prepare test dataset
!python3 apply_masks_to_face_recognition_dataset.py \
--face-dataset-folder={root_test_dataset_path} \
--masks-database=data/masks_base.json \
--verbose \
--use-cuda
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Initialize constants</font></b>
# Init constants
batch_size = 100
n_jobs = 4
epochs = 3000
image_shape = (112, 112)
embedding_size = 256
device = 'cuda:0'
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Initialize base variables for training</font></b>
# Init base variables for training
generator_train_dataset = PipelineFacesDatasetGenerator(
root_train_dataset_path,
image_shape
)
train_loader = DataLoader(
generator_train_dataset,
batch_size=batch_size,
num_workers=n_jobs,
shuffle=True,
drop_last=True
)
model = FaceRecognitionModel(
backbone=Backbone(
backbone=resnet18(pretrained=True),
embedding_size=embedding_size,
input_shape=(3, image_shape[0], image_shape[1])
),
head=ArcFaceLayer(
embedding_size=embedding_size,
num_classes=generator_train_dataset.num_classes
)
)
model = model.to(device)
loss_function = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(params=model.parameters(), lr=0.00001)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Run test for embedding net</font></b>
print(
'Start accuracy rate = {:.5f}'.format(
test_embedding_net(root_test_dataset_path, image_shape, model, device)
)
)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Perform training process</font></b>
# Training process
epoch_loss = []
epoch_test_acc = []
for epoch in range(1, epochs + 1):
model.train()
batches_count = len(train_loader)
avg_epoch_loss = 0
avg_epoch_acc = 0
with tqdm(total=batches_count) as pbar:
for i, (_img, _y_true) in enumerate(train_loader):
img = _img.to(device)
y_true = _y_true.to(device)
optimizer.zero_grad()
y_pred = model(img, y_true)
loss = loss_function(
y_pred,
y_true
)
loss.backward()
optimizer.step()
acc = default_acc_function(
y_pred,
torch.nn.functional.one_hot(
y_true,
num_classes=y_pred.size(-1)
).to(y_pred.dtype).to(device)
).numpy()
pbar.postfix = \
'Epoch: {}/{}, loss: {:.8f}, ' \
'avg acc: {:.8f}'.format(
epoch,
epochs,
loss.item(),
acc
)
avg_epoch_loss += \
loss.item() / y_true.size(0) / batches_count
avg_epoch_acc += acc / batches_count
pbar.update(1)
test_acc = test_embedding_net(root_test_dataset_path, image_shape, model, device)
print('Test accuracy rate: {:.5f}'.format(test_acc))
epoch_loss.append(avg_epoch_loss)
epoch_test_acc.append(test_acc)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Plot the results</font></b>
plt.figure(figsize=(8, 8))
plt.title('Train loss per epoch')
plt.xlabel('Epoch number')
plt.ylabel('Binary crossentropy value')
plt.plot(list(range(1, len(epoch_loss) + 1)), epoch_loss)
plt.figure(figsize=(8, 8))
plt.title('Test accuracy rate per epoch')
plt.xlabel('Epoch number')
plt.ylabel('Accuracy rate')
plt.plot(list(range(1, len(epoch_test_acc) + 1)), epoch_test_acc, color='orange')
plt.show()
```
|
github_jupyter
|
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Check GPU resources</font></b>
!nvidia-smi
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Import requirements</font></b>
import os
import sys
import cv2
from matplotlib import pyplot as plt
import sys
import numpy as np
import torch
from torch import nn
from tqdm.notebook import tqdm
from torch.utils.data import DataLoader
%matplotlib inline
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Clone and build face-alignment git repo</font></b>
!git clone https://github.com/1adrianb/face-alignment
%cd face-alignment
!pip install -r requirements.txt
!python setup.py install
import face_alignment
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Download and import face mask SDK</font></b>
!git clone https://github.com/broutonlab/face-id-with-medical-masks.git
%cd face-id-with-medical-masks
from masked_face_sdk.mask_generation_utils import end2end_mask_generation
from masked_face_sdk.pipeline_dataset_loader import PipelineFacesDatasetGenerator
from masked_face_sdk.pipeline_dataset_loader \
import PipelineFacesDatasetGenerator
from masked_face_sdk.neural_network_modules \
import Backbone, ArcFaceLayer, FaceRecognitionModel, resnet18
from masked_face_sdk.training_utils import default_acc_function, test_embedding_net
!gdown --id 1b64prOr4_E8gcD1Q_cVZkFnSzNVfGwU_
!unzip face_recognition_with_masks_dataset.zip
!ls
# Pathes to datasets for face recognition in Keras-like format
root_train_dataset_path = 'test_large/'
root_test_dataset_path = 'test_small/'
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Generate masks database</font></b>
# Generate masks database
!python3 generate_masks_database.py \
--masks-folder=data/masked_faces/ \
--database-file=data/masks_base.json \
--verbose --skip-warnings
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Prepare training dataset</font></b>
# Prepare training dataset
!python3 apply_masks_to_face_recognition_dataset.py \
--face-dataset-folder={root_train_dataset_path} \
--masks-database=data/masks_base.json \
--verbose \
--use-cuda
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Prepare test dataset</font></b>
# Prepare test dataset
!python3 apply_masks_to_face_recognition_dataset.py \
--face-dataset-folder={root_test_dataset_path} \
--masks-database=data/masks_base.json \
--verbose \
--use-cuda
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Initialize constants</font></b>
# Init constants
batch_size = 100
n_jobs = 4
epochs = 3000
image_shape = (112, 112)
embedding_size = 256
device = 'cuda:0'
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Initialize base variables for training</font></b>
# Init base variables for training
generator_train_dataset = PipelineFacesDatasetGenerator(
root_train_dataset_path,
image_shape
)
train_loader = DataLoader(
generator_train_dataset,
batch_size=batch_size,
num_workers=n_jobs,
shuffle=True,
drop_last=True
)
model = FaceRecognitionModel(
backbone=Backbone(
backbone=resnet18(pretrained=True),
embedding_size=embedding_size,
input_shape=(3, image_shape[0], image_shape[1])
),
head=ArcFaceLayer(
embedding_size=embedding_size,
num_classes=generator_train_dataset.num_classes
)
)
model = model.to(device)
loss_function = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(params=model.parameters(), lr=0.00001)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Run test for embedding net</font></b>
print(
'Start accuracy rate = {:.5f}'.format(
test_embedding_net(root_test_dataset_path, image_shape, model, device)
)
)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Perform training process</font></b>
# Training process
epoch_loss = []
epoch_test_acc = []
for epoch in range(1, epochs + 1):
model.train()
batches_count = len(train_loader)
avg_epoch_loss = 0
avg_epoch_acc = 0
with tqdm(total=batches_count) as pbar:
for i, (_img, _y_true) in enumerate(train_loader):
img = _img.to(device)
y_true = _y_true.to(device)
optimizer.zero_grad()
y_pred = model(img, y_true)
loss = loss_function(
y_pred,
y_true
)
loss.backward()
optimizer.step()
acc = default_acc_function(
y_pred,
torch.nn.functional.one_hot(
y_true,
num_classes=y_pred.size(-1)
).to(y_pred.dtype).to(device)
).numpy()
pbar.postfix = \
'Epoch: {}/{}, loss: {:.8f}, ' \
'avg acc: {:.8f}'.format(
epoch,
epochs,
loss.item(),
acc
)
avg_epoch_loss += \
loss.item() / y_true.size(0) / batches_count
avg_epoch_acc += acc / batches_count
pbar.update(1)
test_acc = test_embedding_net(root_test_dataset_path, image_shape, model, device)
print('Test accuracy rate: {:.5f}'.format(test_acc))
epoch_loss.append(avg_epoch_loss)
epoch_test_acc.append(test_acc)
#@title <b><font color="red" size="+3">←</font><font color="black" size="+3"> Plot the results</font></b>
plt.figure(figsize=(8, 8))
plt.title('Train loss per epoch')
plt.xlabel('Epoch number')
plt.ylabel('Binary crossentropy value')
plt.plot(list(range(1, len(epoch_loss) + 1)), epoch_loss)
plt.figure(figsize=(8, 8))
plt.title('Test accuracy rate per epoch')
plt.xlabel('Epoch number')
plt.ylabel('Accuracy rate')
plt.plot(list(range(1, len(epoch_test_acc) + 1)), epoch_test_acc, color='orange')
plt.show()
| 0.524882 | 0.776411 |
# Position-specific feature importance analysis for LbCpf1
# Calculation of frequency of nucleotides at each location
```
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset_features_clean2.csv", encoding="cp1252")
df = pd.DataFrame(OT_data)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT), len(NOT))
l1=['A_OTSeqPosition1', 'A_OTSeqPosition2', 'A_OTSeqPosition3', 'A_OTSeqPosition4', 'A_OTSeqPosition5', 'A_OTSeqPosition6', 'A_OTSeqPosition7', 'A_OTSeqPosition8', 'A_OTSeqPosition9', 'A_OTSeqPosition10', 'A_OTSeqPosition11', 'A_OTSeqPosition12', 'A_OTSeqPosition13', 'A_OTSeqPosition14', 'A_OTSeqPosition15', 'A_OTSeqPosition16', 'A_OTSeqPosition17', 'A_OTSeqPosition18', 'A_OTSeqPosition19', 'A_OTSeqPosition20', 'A_OTSeqPosition21', 'A_OTSeqPosition22', 'A_OTSeqPosition23', 'A_OTSeqPosition24', 'A_OTSeqPosition25', 'A_OTSeqPosition26', 'A_OTSeqPosition27']
l2=['T_OTSeqPosition1', 'T_OTSeqPosition2', 'T_OTSeqPosition3', 'T_OTSeqPosition4', 'T_OTSeqPosition5', 'T_OTSeqPosition6', 'T_OTSeqPosition7', 'T_OTSeqPosition8', 'T_OTSeqPosition9', 'T_OTSeqPosition10', 'T_OTSeqPosition11', 'T_OTSeqPosition12', 'T_OTSeqPosition13', 'T_OTSeqPosition14', 'T_OTSeqPosition15', 'T_OTSeqPosition16', 'T_OTSeqPosition17', 'T_OTSeqPosition18', 'T_OTSeqPosition19', 'T_OTSeqPosition20', 'T_OTSeqPosition21', 'T_OTSeqPosition22', 'T_OTSeqPosition23', 'T_OTSeqPosition24', 'T_OTSeqPosition25', 'T_OTSeqPosition26', 'T_OTSeqPosition27']
l3=['G_OTSeqPosition1', 'G_OTSeqPosition2', 'G_OTSeqPosition3', 'G_OTSeqPosition4', 'G_OTSeqPosition5', 'G_OTSeqPosition6', 'G_OTSeqPosition7', 'G_OTSeqPosition8', 'G_OTSeqPosition9', 'G_OTSeqPosition10', 'G_OTSeqPosition11', 'G_OTSeqPosition12', 'G_OTSeqPosition13', 'G_OTSeqPosition14', 'G_OTSeqPosition15', 'G_OTSeqPosition16', 'G_OTSeqPosition17', 'G_OTSeqPosition18', 'G_OTSeqPosition19', 'G_OTSeqPosition20', 'G_OTSeqPosition21', 'G_OTSeqPosition22', 'G_OTSeqPosition23', 'G_OTSeqPosition24', 'G_OTSeqPosition25', 'G_OTSeqPosition26', 'G_OTSeqPosition27']
l4=['C_OTSeqPosition1', 'C_OTSeqPosition2', 'C_OTSeqPosition3', 'C_OTSeqPosition4', 'C_OTSeqPosition5', 'C_OTSeqPosition6', 'C_OTSeqPosition7', 'C_OTSeqPosition8', 'C_OTSeqPosition9', 'C_OTSeqPosition10', 'C_OTSeqPosition11', 'C_OTSeqPosition12', 'C_OTSeqPosition13', 'C_OTSeqPosition14', 'C_OTSeqPosition15', 'C_OTSeqPosition16', 'C_OTSeqPosition17', 'C_OTSeqPosition18', 'C_OTSeqPosition19', 'C_OTSeqPosition20', 'C_OTSeqPosition21', 'C_OTSeqPosition22', 'C_OTSeqPosition23', 'C_OTSeqPosition24', 'C_OTSeqPosition25', 'C_OTSeqPosition26', 'C_OTSeqPosition27']
print("positive off-targets")
POT_l1=[]
POT_l2=[]
POT_l3=[]
POT_l4=[]
for i, j, k, l in zip(l1, l2, l3, l4):
t0=POT[i].sum()
t1=POT[j].sum()
t2=POT[k].sum()
t3=POT[l].sum()
POT_A=t0/481
POT_T=t1/481
POT_G=t2/481
POT_C=t3/481
POT_l1.append(POT_A)
POT_l2.append(POT_T)
POT_l3.append(POT_G)
POT_l4.append(POT_C)
print("Position-wise frequency of A in positive off-targets \n", POT_l1, "\n")
print("Position-wise frequency of T in positive off-targets \n", POT_l2, "\n")
print("Position-wise frequency of G in positive off-targets \n", POT_l3, "\n")
print("Position-wise frequency of C in positive off-targets \n", POT_l4, "\n")
print("negative off-targets")
NOT_l1=[]
NOT_l2=[]
NOT_l3=[]
NOT_l4=[]
for i, j, k, l in zip(l1, l2, l3, l4):
t0=NOT[i].sum()
t1=NOT[j].sum()
t2=NOT[k].sum()
t3=NOT[l].sum()
NOT_A=t0/58474
NOT_T=t1/58474
NOT_G=t2/58474
NOT_C=t3/58474
NOT_l1.append(NOT_A)
NOT_l2.append(NOT_T)
NOT_l3.append(NOT_G)
NOT_l4.append(NOT_C)
print("Position-wise frequency of A in negative off-targets \n", POT_l1, "\n")
print("Position-wise frequency of T in negative off-targets \n", POT_l2, "\n")
print("Position-wise frequency of G in negative off-targets \n", POT_l3, "\n")
print("Position-wise frequency of C in negative off-targets \n", POT_l4, "\n")
```
# Enrichment analysis to study the position-specific favour and disfavour of Nucleotides
```
import pandas as pd
l1=['C_OTSeqPosition1','G_OTSeqPosition1', 'T_OTSeqPosition1', 'A_OTSeqPosition1', 'C_OTSeqPosition2', 'G_OTSeqPosition2', 'T_OTSeqPosition2', 'A_OTSeqPosition2', 'C_OTSeqPosition3', 'G_OTSeqPosition3', 'T_OTSeqPosition3', 'A_OTSeqPosition3', 'C_OTSeqPosition4', 'G_OTSeqPosition4', 'T_OTSeqPosition4', 'A_OTSeqPosition4', 'C_OTSeqPosition5', 'G_OTSeqPosition5', 'T_OTSeqPosition5', 'A_OTSeqPosition5', 'C_OTSeqPosition6', 'G_OTSeqPosition6', 'T_OTSeqPosition6', 'A_OTSeqPosition6', 'C_OTSeqPosition7', 'G_OTSeqPosition7', 'T_OTSeqPosition7', 'A_OTSeqPosition7', 'C_OTSeqPosition8', 'G_OTSeqPosition8', 'T_OTSeqPosition8', 'A_OTSeqPosition8', 'C_OTSeqPosition9', 'G_OTSeqPosition9', 'T_OTSeqPosition9', 'A_OTSeqPosition9', 'C_OTSeqPosition10', 'G_OTSeqPosition10', 'T_OTSeqPosition10', 'A_OTSeqPosition10', 'C_OTSeqPosition11', 'G_OTSeqPosition11', 'T_OTSeqPosition11', 'A_OTSeqPosition11', 'C_OTSeqPosition12', 'G_OTSeqPosition12', 'T_OTSeqPosition12', 'A_OTSeqPosition12', 'C_OTSeqPosition13', 'G_OTSeqPosition13', 'T_OTSeqPosition13', 'A_OTSeqPosition13', 'C_OTSeqPosition14', 'G_OTSeqPosition14', 'T_OTSeqPosition14', 'A_OTSeqPosition14', 'C_OTSeqPosition15', 'G_OTSeqPosition15', 'T_OTSeqPosition15', 'A_OTSeqPosition15', 'C_OTSeqPosition16', 'G_OTSeqPosition16', 'T_OTSeqPosition16', 'A_OTSeqPosition16', 'C_OTSeqPosition17', 'G_OTSeqPosition17', 'T_OTSeqPosition17', 'A_OTSeqPosition17', 'C_OTSeqPosition18', 'G_OTSeqPosition18', 'T_OTSeqPosition18', 'A_OTSeqPosition18', 'C_OTSeqPosition19', 'G_OTSeqPosition19', 'T_OTSeqPosition19', 'A_OTSeqPosition19', 'C_OTSeqPosition20', 'G_OTSeqPosition20', 'T_OTSeqPosition20', 'A_OTSeqPosition20', 'C_OTSeqPosition21', 'G_OTSeqPosition21', 'T_OTSeqPosition21', 'A_OTSeqPosition21', 'C_OTSeqPosition22', 'G_OTSeqPosition22', 'T_OTSeqPosition22', 'A_OTSeqPosition22', 'C_OTSeqPosition23', 'G_OTSeqPosition23', 'T_OTSeqPosition23', 'A_OTSeqPosition23', 'C_OTSeqPosition24', 'G_OTSeqPosition24', 'T_OTSeqPosition24', 'A_OTSeqPosition24', 'C_OTSeqPosition25', 'G_OTSeqPosition25', 'T_OTSeqPosition25', 'A_OTSeqPosition25', 'C_OTSeqPosition26', 'G_OTSeqPosition26', 'T_OTSeqPosition26', 'A_OTSeqPosition26', 'C_OTSeqPosition27', 'G_OTSeqPosition27', 'T_OTSeqPosition27', 'A_OTSeqPosition27']
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv")
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", encoding="cp1252")
df = pd.DataFrame(OT_data)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(df))
l1=['C_OTSeqPosition1','G_OTSeqPosition1', 'T_OTSeqPosition1', 'A_OTSeqPosition1', 'C_OTSeqPosition2', 'G_OTSeqPosition2', 'T_OTSeqPosition2', 'A_OTSeqPosition2', 'C_OTSeqPosition3', 'G_OTSeqPosition3', 'T_OTSeqPosition3', 'A_OTSeqPosition3', 'C_OTSeqPosition4', 'G_OTSeqPosition4', 'T_OTSeqPosition4', 'A_OTSeqPosition4', 'C_OTSeqPosition5', 'G_OTSeqPosition5', 'T_OTSeqPosition5', 'A_OTSeqPosition5', 'C_OTSeqPosition6', 'G_OTSeqPosition6', 'T_OTSeqPosition6', 'A_OTSeqPosition6', 'C_OTSeqPosition7', 'G_OTSeqPosition7', 'T_OTSeqPosition7', 'A_OTSeqPosition7', 'C_OTSeqPosition8', 'G_OTSeqPosition8', 'T_OTSeqPosition8', 'A_OTSeqPosition8', 'C_OTSeqPosition9', 'G_OTSeqPosition9', 'T_OTSeqPosition9', 'A_OTSeqPosition9', 'C_OTSeqPosition10', 'G_OTSeqPosition10', 'T_OTSeqPosition10', 'A_OTSeqPosition10', 'C_OTSeqPosition11', 'G_OTSeqPosition11', 'T_OTSeqPosition11', 'A_OTSeqPosition11', 'C_OTSeqPosition12', 'G_OTSeqPosition12', 'T_OTSeqPosition12', 'A_OTSeqPosition12', 'C_OTSeqPosition13', 'G_OTSeqPosition13', 'T_OTSeqPosition13', 'A_OTSeqPosition13', 'C_OTSeqPosition14', 'G_OTSeqPosition14', 'T_OTSeqPosition14', 'A_OTSeqPosition14', 'C_OTSeqPosition15', 'G_OTSeqPosition15', 'T_OTSeqPosition15', 'A_OTSeqPosition15', 'C_OTSeqPosition16', 'G_OTSeqPosition16', 'T_OTSeqPosition16', 'A_OTSeqPosition16', 'C_OTSeqPosition17', 'G_OTSeqPosition17', 'T_OTSeqPosition17', 'A_OTSeqPosition17', 'C_OTSeqPosition18', 'G_OTSeqPosition18', 'T_OTSeqPosition18', 'A_OTSeqPosition18', 'C_OTSeqPosition19', 'G_OTSeqPosition19', 'T_OTSeqPosition19', 'A_OTSeqPosition19', 'C_OTSeqPosition20', 'G_OTSeqPosition20', 'T_OTSeqPosition20', 'A_OTSeqPosition20', 'C_OTSeqPosition21', 'G_OTSeqPosition21', 'T_OTSeqPosition21', 'A_OTSeqPosition21', 'C_OTSeqPosition22', 'G_OTSeqPosition22', 'T_OTSeqPosition22', 'A_OTSeqPosition22', 'C_OTSeqPosition23', 'G_OTSeqPosition23', 'T_OTSeqPosition23', 'A_OTSeqPosition23', 'C_OTSeqPosition24', 'G_OTSeqPosition24', 'T_OTSeqPosition24', 'A_OTSeqPosition24', 'C_OTSeqPosition25', 'G_OTSeqPosition25', 'T_OTSeqPosition25', 'A_OTSeqPosition25', 'C_OTSeqPosition26', 'G_OTSeqPosition26', 'T_OTSeqPosition26', 'A_OTSeqPosition26', 'C_OTSeqPosition27', 'G_OTSeqPosition27', 'T_OTSeqPosition27', 'A_OTSeqPosition27']
print("positive off-targets")
POT_l1=[]
for i in l1:
total = POT[i].sum()
POT_ratio=total/524
POT_l1.append(POT_ratio)
print(POT_l1)
print("negative off-targets")
NOT_l1=[]
for i in l1:
total = NOT[i].sum()
NOT_ratio=total/525
NOT_l1.append(NOT_ratio)
print(NOT_l1)
enrichment_ratio=[]
for i, j in zip(POT_l1, NOT_l1):
enrichment_ratio1=i/j
enrichment_ratio.append(enrichment_ratio1)
print(enrichment_ratio)
```
# mismatch distribution analysis LbCpf1
```
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset_features_POT.csv", index_col=[0], encoding="cp1252")
df = pd.DataFrame(OT_data)
print(len(df))
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
for i in l1:
total = df[i].sum()
print(total/524)
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
#print(len(df))
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
for i in l1:
total = POT[i].sum()
l2.append(total/481)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/481)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv")
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
# Position specific mismatch type analysis LbCpf1
# mismatch at position 4
```
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS4', 'MM_type_A–C_POS4', 'MM_type_A–G_POS4', 'MM_type_T–C_POS4', 'MM_type_T–G_POS4', 'MM_type_T–A_POS4', 'MM_type_G–A_POS4', 'MM_type_G–T_POS4', 'MM_type_G–C_POS4', 'MM_type_C–A_POS4', 'MM_type_C–T_POS4', 'MM_type_C–G_POS4', 'MM_type_other_POS4']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS4', 'MM_type_A–C_POS4', 'MM_type_A–G_POS4', 'MM_type_T–C_POS4', 'MM_type_T–G_POS4', 'MM_type_T–A_POS4', 'MM_type_G–A_POS4', 'MM_type_G–T_POS4', 'MM_type_G–C_POS4', 'MM_type_C–A_POS4', 'MM_type_C–T_POS4', 'MM_type_C–G_POS4', 'MM_type_other_POS4']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
# mismatch at position 16
```
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS16', 'MM_type_A–C_POS16', 'MM_type_A–G_POS16', 'MM_type_T–C_POS16', 'MM_type_T–G_POS16', 'MM_type_T–A_POS16', 'MM_type_G–A_POS16', 'MM_type_G–T_POS16', 'MM_type_G–C_POS16', 'MM_type_C–A_POS16', 'MM_type_C–T_POS16', 'MM_type_C–G_POS16', 'MM_type_other_POS16']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS16', 'MM_type_A–C_POS16', 'MM_type_A–G_POS16', 'MM_type_T–C_POS16', 'MM_type_T–G_POS16', 'MM_type_T–A_POS16', 'MM_type_G–A_POS16', 'MM_type_G–T_POS16', 'MM_type_G–C_POS16', 'MM_type_C–A_POS16', 'MM_type_C–T_POS16', 'MM_type_C–G_POS16', 'MM_type_other_POS16']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
# mismatch at position 17
```
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS17', 'MM_type_A–C_POS17', 'MM_type_A–G_POS17', 'MM_type_T–C_POS17', 'MM_type_T–G_POS17', 'MM_type_T–A_POS17', 'MM_type_G–A_POS17', 'MM_type_G–T_POS17', 'MM_type_G–C_POS17', 'MM_type_C–A_POS17', 'MM_type_C–T_POS17', 'MM_type_C–G_POS17', 'MM_type_other_POS17']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS17', 'MM_type_A–C_POS17', 'MM_type_A–G_POS17', 'MM_type_T–C_POS17', 'MM_type_T–G_POS17', 'MM_type_T–A_POS17', 'MM_type_G–A_POS17', 'MM_type_G–T_POS17', 'MM_type_G–C_POS17', 'MM_type_C–A_POS17', 'MM_type_C–T_POS17', 'MM_type_C–G_POS17', 'MM_type_other_POS17']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
# mismatch at position 18
```
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS18', 'MM_type_A–C_POS18', 'MM_type_A–G_POS18', 'MM_type_T–C_POS18', 'MM_type_T–G_POS18', 'MM_type_T–A_POS18', 'MM_type_G–A_POS18', 'MM_type_G–T_POS18', 'MM_type_G–C_POS18', 'MM_type_C–A_POS18', 'MM_type_C–T_POS18', 'MM_type_C–G_POS18', 'MM_type_other_POS18']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS18', 'MM_type_A–C_POS18', 'MM_type_A–G_POS18', 'MM_type_T–C_POS18', 'MM_type_T–G_POS18', 'MM_type_T–A_POS18', 'MM_type_G–A_POS18', 'MM_type_G–T_POS18', 'MM_type_G–C_POS18', 'MM_type_C–A_POS18', 'MM_type_C–T_POS18', 'MM_type_C–G_POS18', 'MM_type_other_POS18']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
# mismatch at position 23
```
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS23', 'MM_type_A–C_POS23', 'MM_type_A–G_POS23', 'MM_type_T–C_POS23', 'MM_type_T–G_POS23', 'MM_type_T–A_POS23', 'MM_type_G–A_POS23', 'MM_type_G–T_POS23', 'MM_type_G–C_POS23', 'MM_type_C–A_POS23', 'MM_type_C–T_POS23', 'MM_type_C–G_POS23', 'MM_type_other_POS23']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS23', 'MM_type_A–C_POS23', 'MM_type_A–G_POS23', 'MM_type_T–C_POS23', 'MM_type_T–G_POS23', 'MM_type_T–A_POS23', 'MM_type_G–A_POS23', 'MM_type_G–T_POS23', 'MM_type_G–C_POS23', 'MM_type_C–A_POS23', 'MM_type_C–T_POS23', 'MM_type_C–G_POS23', 'MM_type_other_POS23']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
```
|
github_jupyter
|
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset_features_clean2.csv", encoding="cp1252")
df = pd.DataFrame(OT_data)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT), len(NOT))
l1=['A_OTSeqPosition1', 'A_OTSeqPosition2', 'A_OTSeqPosition3', 'A_OTSeqPosition4', 'A_OTSeqPosition5', 'A_OTSeqPosition6', 'A_OTSeqPosition7', 'A_OTSeqPosition8', 'A_OTSeqPosition9', 'A_OTSeqPosition10', 'A_OTSeqPosition11', 'A_OTSeqPosition12', 'A_OTSeqPosition13', 'A_OTSeqPosition14', 'A_OTSeqPosition15', 'A_OTSeqPosition16', 'A_OTSeqPosition17', 'A_OTSeqPosition18', 'A_OTSeqPosition19', 'A_OTSeqPosition20', 'A_OTSeqPosition21', 'A_OTSeqPosition22', 'A_OTSeqPosition23', 'A_OTSeqPosition24', 'A_OTSeqPosition25', 'A_OTSeqPosition26', 'A_OTSeqPosition27']
l2=['T_OTSeqPosition1', 'T_OTSeqPosition2', 'T_OTSeqPosition3', 'T_OTSeqPosition4', 'T_OTSeqPosition5', 'T_OTSeqPosition6', 'T_OTSeqPosition7', 'T_OTSeqPosition8', 'T_OTSeqPosition9', 'T_OTSeqPosition10', 'T_OTSeqPosition11', 'T_OTSeqPosition12', 'T_OTSeqPosition13', 'T_OTSeqPosition14', 'T_OTSeqPosition15', 'T_OTSeqPosition16', 'T_OTSeqPosition17', 'T_OTSeqPosition18', 'T_OTSeqPosition19', 'T_OTSeqPosition20', 'T_OTSeqPosition21', 'T_OTSeqPosition22', 'T_OTSeqPosition23', 'T_OTSeqPosition24', 'T_OTSeqPosition25', 'T_OTSeqPosition26', 'T_OTSeqPosition27']
l3=['G_OTSeqPosition1', 'G_OTSeqPosition2', 'G_OTSeqPosition3', 'G_OTSeqPosition4', 'G_OTSeqPosition5', 'G_OTSeqPosition6', 'G_OTSeqPosition7', 'G_OTSeqPosition8', 'G_OTSeqPosition9', 'G_OTSeqPosition10', 'G_OTSeqPosition11', 'G_OTSeqPosition12', 'G_OTSeqPosition13', 'G_OTSeqPosition14', 'G_OTSeqPosition15', 'G_OTSeqPosition16', 'G_OTSeqPosition17', 'G_OTSeqPosition18', 'G_OTSeqPosition19', 'G_OTSeqPosition20', 'G_OTSeqPosition21', 'G_OTSeqPosition22', 'G_OTSeqPosition23', 'G_OTSeqPosition24', 'G_OTSeqPosition25', 'G_OTSeqPosition26', 'G_OTSeqPosition27']
l4=['C_OTSeqPosition1', 'C_OTSeqPosition2', 'C_OTSeqPosition3', 'C_OTSeqPosition4', 'C_OTSeqPosition5', 'C_OTSeqPosition6', 'C_OTSeqPosition7', 'C_OTSeqPosition8', 'C_OTSeqPosition9', 'C_OTSeqPosition10', 'C_OTSeqPosition11', 'C_OTSeqPosition12', 'C_OTSeqPosition13', 'C_OTSeqPosition14', 'C_OTSeqPosition15', 'C_OTSeqPosition16', 'C_OTSeqPosition17', 'C_OTSeqPosition18', 'C_OTSeqPosition19', 'C_OTSeqPosition20', 'C_OTSeqPosition21', 'C_OTSeqPosition22', 'C_OTSeqPosition23', 'C_OTSeqPosition24', 'C_OTSeqPosition25', 'C_OTSeqPosition26', 'C_OTSeqPosition27']
print("positive off-targets")
POT_l1=[]
POT_l2=[]
POT_l3=[]
POT_l4=[]
for i, j, k, l in zip(l1, l2, l3, l4):
t0=POT[i].sum()
t1=POT[j].sum()
t2=POT[k].sum()
t3=POT[l].sum()
POT_A=t0/481
POT_T=t1/481
POT_G=t2/481
POT_C=t3/481
POT_l1.append(POT_A)
POT_l2.append(POT_T)
POT_l3.append(POT_G)
POT_l4.append(POT_C)
print("Position-wise frequency of A in positive off-targets \n", POT_l1, "\n")
print("Position-wise frequency of T in positive off-targets \n", POT_l2, "\n")
print("Position-wise frequency of G in positive off-targets \n", POT_l3, "\n")
print("Position-wise frequency of C in positive off-targets \n", POT_l4, "\n")
print("negative off-targets")
NOT_l1=[]
NOT_l2=[]
NOT_l3=[]
NOT_l4=[]
for i, j, k, l in zip(l1, l2, l3, l4):
t0=NOT[i].sum()
t1=NOT[j].sum()
t2=NOT[k].sum()
t3=NOT[l].sum()
NOT_A=t0/58474
NOT_T=t1/58474
NOT_G=t2/58474
NOT_C=t3/58474
NOT_l1.append(NOT_A)
NOT_l2.append(NOT_T)
NOT_l3.append(NOT_G)
NOT_l4.append(NOT_C)
print("Position-wise frequency of A in negative off-targets \n", POT_l1, "\n")
print("Position-wise frequency of T in negative off-targets \n", POT_l2, "\n")
print("Position-wise frequency of G in negative off-targets \n", POT_l3, "\n")
print("Position-wise frequency of C in negative off-targets \n", POT_l4, "\n")
import pandas as pd
l1=['C_OTSeqPosition1','G_OTSeqPosition1', 'T_OTSeqPosition1', 'A_OTSeqPosition1', 'C_OTSeqPosition2', 'G_OTSeqPosition2', 'T_OTSeqPosition2', 'A_OTSeqPosition2', 'C_OTSeqPosition3', 'G_OTSeqPosition3', 'T_OTSeqPosition3', 'A_OTSeqPosition3', 'C_OTSeqPosition4', 'G_OTSeqPosition4', 'T_OTSeqPosition4', 'A_OTSeqPosition4', 'C_OTSeqPosition5', 'G_OTSeqPosition5', 'T_OTSeqPosition5', 'A_OTSeqPosition5', 'C_OTSeqPosition6', 'G_OTSeqPosition6', 'T_OTSeqPosition6', 'A_OTSeqPosition6', 'C_OTSeqPosition7', 'G_OTSeqPosition7', 'T_OTSeqPosition7', 'A_OTSeqPosition7', 'C_OTSeqPosition8', 'G_OTSeqPosition8', 'T_OTSeqPosition8', 'A_OTSeqPosition8', 'C_OTSeqPosition9', 'G_OTSeqPosition9', 'T_OTSeqPosition9', 'A_OTSeqPosition9', 'C_OTSeqPosition10', 'G_OTSeqPosition10', 'T_OTSeqPosition10', 'A_OTSeqPosition10', 'C_OTSeqPosition11', 'G_OTSeqPosition11', 'T_OTSeqPosition11', 'A_OTSeqPosition11', 'C_OTSeqPosition12', 'G_OTSeqPosition12', 'T_OTSeqPosition12', 'A_OTSeqPosition12', 'C_OTSeqPosition13', 'G_OTSeqPosition13', 'T_OTSeqPosition13', 'A_OTSeqPosition13', 'C_OTSeqPosition14', 'G_OTSeqPosition14', 'T_OTSeqPosition14', 'A_OTSeqPosition14', 'C_OTSeqPosition15', 'G_OTSeqPosition15', 'T_OTSeqPosition15', 'A_OTSeqPosition15', 'C_OTSeqPosition16', 'G_OTSeqPosition16', 'T_OTSeqPosition16', 'A_OTSeqPosition16', 'C_OTSeqPosition17', 'G_OTSeqPosition17', 'T_OTSeqPosition17', 'A_OTSeqPosition17', 'C_OTSeqPosition18', 'G_OTSeqPosition18', 'T_OTSeqPosition18', 'A_OTSeqPosition18', 'C_OTSeqPosition19', 'G_OTSeqPosition19', 'T_OTSeqPosition19', 'A_OTSeqPosition19', 'C_OTSeqPosition20', 'G_OTSeqPosition20', 'T_OTSeqPosition20', 'A_OTSeqPosition20', 'C_OTSeqPosition21', 'G_OTSeqPosition21', 'T_OTSeqPosition21', 'A_OTSeqPosition21', 'C_OTSeqPosition22', 'G_OTSeqPosition22', 'T_OTSeqPosition22', 'A_OTSeqPosition22', 'C_OTSeqPosition23', 'G_OTSeqPosition23', 'T_OTSeqPosition23', 'A_OTSeqPosition23', 'C_OTSeqPosition24', 'G_OTSeqPosition24', 'T_OTSeqPosition24', 'A_OTSeqPosition24', 'C_OTSeqPosition25', 'G_OTSeqPosition25', 'T_OTSeqPosition25', 'A_OTSeqPosition25', 'C_OTSeqPosition26', 'G_OTSeqPosition26', 'T_OTSeqPosition26', 'A_OTSeqPosition26', 'C_OTSeqPosition27', 'G_OTSeqPosition27', 'T_OTSeqPosition27', 'A_OTSeqPosition27']
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv")
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", encoding="cp1252")
df = pd.DataFrame(OT_data)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(df))
l1=['C_OTSeqPosition1','G_OTSeqPosition1', 'T_OTSeqPosition1', 'A_OTSeqPosition1', 'C_OTSeqPosition2', 'G_OTSeqPosition2', 'T_OTSeqPosition2', 'A_OTSeqPosition2', 'C_OTSeqPosition3', 'G_OTSeqPosition3', 'T_OTSeqPosition3', 'A_OTSeqPosition3', 'C_OTSeqPosition4', 'G_OTSeqPosition4', 'T_OTSeqPosition4', 'A_OTSeqPosition4', 'C_OTSeqPosition5', 'G_OTSeqPosition5', 'T_OTSeqPosition5', 'A_OTSeqPosition5', 'C_OTSeqPosition6', 'G_OTSeqPosition6', 'T_OTSeqPosition6', 'A_OTSeqPosition6', 'C_OTSeqPosition7', 'G_OTSeqPosition7', 'T_OTSeqPosition7', 'A_OTSeqPosition7', 'C_OTSeqPosition8', 'G_OTSeqPosition8', 'T_OTSeqPosition8', 'A_OTSeqPosition8', 'C_OTSeqPosition9', 'G_OTSeqPosition9', 'T_OTSeqPosition9', 'A_OTSeqPosition9', 'C_OTSeqPosition10', 'G_OTSeqPosition10', 'T_OTSeqPosition10', 'A_OTSeqPosition10', 'C_OTSeqPosition11', 'G_OTSeqPosition11', 'T_OTSeqPosition11', 'A_OTSeqPosition11', 'C_OTSeqPosition12', 'G_OTSeqPosition12', 'T_OTSeqPosition12', 'A_OTSeqPosition12', 'C_OTSeqPosition13', 'G_OTSeqPosition13', 'T_OTSeqPosition13', 'A_OTSeqPosition13', 'C_OTSeqPosition14', 'G_OTSeqPosition14', 'T_OTSeqPosition14', 'A_OTSeqPosition14', 'C_OTSeqPosition15', 'G_OTSeqPosition15', 'T_OTSeqPosition15', 'A_OTSeqPosition15', 'C_OTSeqPosition16', 'G_OTSeqPosition16', 'T_OTSeqPosition16', 'A_OTSeqPosition16', 'C_OTSeqPosition17', 'G_OTSeqPosition17', 'T_OTSeqPosition17', 'A_OTSeqPosition17', 'C_OTSeqPosition18', 'G_OTSeqPosition18', 'T_OTSeqPosition18', 'A_OTSeqPosition18', 'C_OTSeqPosition19', 'G_OTSeqPosition19', 'T_OTSeqPosition19', 'A_OTSeqPosition19', 'C_OTSeqPosition20', 'G_OTSeqPosition20', 'T_OTSeqPosition20', 'A_OTSeqPosition20', 'C_OTSeqPosition21', 'G_OTSeqPosition21', 'T_OTSeqPosition21', 'A_OTSeqPosition21', 'C_OTSeqPosition22', 'G_OTSeqPosition22', 'T_OTSeqPosition22', 'A_OTSeqPosition22', 'C_OTSeqPosition23', 'G_OTSeqPosition23', 'T_OTSeqPosition23', 'A_OTSeqPosition23', 'C_OTSeqPosition24', 'G_OTSeqPosition24', 'T_OTSeqPosition24', 'A_OTSeqPosition24', 'C_OTSeqPosition25', 'G_OTSeqPosition25', 'T_OTSeqPosition25', 'A_OTSeqPosition25', 'C_OTSeqPosition26', 'G_OTSeqPosition26', 'T_OTSeqPosition26', 'A_OTSeqPosition26', 'C_OTSeqPosition27', 'G_OTSeqPosition27', 'T_OTSeqPosition27', 'A_OTSeqPosition27']
print("positive off-targets")
POT_l1=[]
for i in l1:
total = POT[i].sum()
POT_ratio=total/524
POT_l1.append(POT_ratio)
print(POT_l1)
print("negative off-targets")
NOT_l1=[]
for i in l1:
total = NOT[i].sum()
NOT_ratio=total/525
NOT_l1.append(NOT_ratio)
print(NOT_l1)
enrichment_ratio=[]
for i, j in zip(POT_l1, NOT_l1):
enrichment_ratio1=i/j
enrichment_ratio.append(enrichment_ratio1)
print(enrichment_ratio)
import pandas as pd
OT_data=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset_features_POT.csv", index_col=[0], encoding="cp1252")
df = pd.DataFrame(OT_data)
print(len(df))
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
for i in l1:
total = df[i].sum()
print(total/524)
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
#print(len(df))
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
for i in l1:
total = POT[i].sum()
l2.append(total/481)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/481)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
l1=['mismatch_POS1', 'mismatch_POS2', 'mismatch_POS3', 'mismatch_POS4', 'mismatch_POS5', 'mismatch_POS6', 'mismatch_POS7', 'mismatch_POS8', 'mismatch_POS9', 'mismatch_POS10', 'mismatch_POS11', 'mismatch_POS12', 'mismatch_POS13', 'mismatch_POS14', 'mismatch_POS15', 'mismatch_POS16', 'mismatch_POS17', 'mismatch_POS18', 'mismatch_POS19', 'mismatch_POS20', 'mismatch_POS21', 'mismatch_POS22', 'mismatch_POS23', 'mismatch_POS24', 'mismatch_POS25', 'mismatch_POS26', 'mismatch_POS27']
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv")
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS4', 'MM_type_A–C_POS4', 'MM_type_A–G_POS4', 'MM_type_T–C_POS4', 'MM_type_T–G_POS4', 'MM_type_T–A_POS4', 'MM_type_G–A_POS4', 'MM_type_G–T_POS4', 'MM_type_G–C_POS4', 'MM_type_C–A_POS4', 'MM_type_C–T_POS4', 'MM_type_C–G_POS4', 'MM_type_other_POS4']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS4', 'MM_type_A–C_POS4', 'MM_type_A–G_POS4', 'MM_type_T–C_POS4', 'MM_type_T–G_POS4', 'MM_type_T–A_POS4', 'MM_type_G–A_POS4', 'MM_type_G–T_POS4', 'MM_type_G–C_POS4', 'MM_type_C–A_POS4', 'MM_type_C–T_POS4', 'MM_type_C–G_POS4', 'MM_type_other_POS4']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS16', 'MM_type_A–C_POS16', 'MM_type_A–G_POS16', 'MM_type_T–C_POS16', 'MM_type_T–G_POS16', 'MM_type_T–A_POS16', 'MM_type_G–A_POS16', 'MM_type_G–T_POS16', 'MM_type_G–C_POS16', 'MM_type_C–A_POS16', 'MM_type_C–T_POS16', 'MM_type_C–G_POS16', 'MM_type_other_POS16']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS16', 'MM_type_A–C_POS16', 'MM_type_A–G_POS16', 'MM_type_T–C_POS16', 'MM_type_T–G_POS16', 'MM_type_T–A_POS16', 'MM_type_G–A_POS16', 'MM_type_G–T_POS16', 'MM_type_G–C_POS16', 'MM_type_C–A_POS16', 'MM_type_C–T_POS16', 'MM_type_C–G_POS16', 'MM_type_other_POS16']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS17', 'MM_type_A–C_POS17', 'MM_type_A–G_POS17', 'MM_type_T–C_POS17', 'MM_type_T–G_POS17', 'MM_type_T–A_POS17', 'MM_type_G–A_POS17', 'MM_type_G–T_POS17', 'MM_type_G–C_POS17', 'MM_type_C–A_POS17', 'MM_type_C–T_POS17', 'MM_type_C–G_POS17', 'MM_type_other_POS17']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS17', 'MM_type_A–C_POS17', 'MM_type_A–G_POS17', 'MM_type_T–C_POS17', 'MM_type_T–G_POS17', 'MM_type_T–A_POS17', 'MM_type_G–A_POS17', 'MM_type_G–T_POS17', 'MM_type_G–C_POS17', 'MM_type_C–A_POS17', 'MM_type_C–T_POS17', 'MM_type_C–G_POS17', 'MM_type_other_POS17']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS18', 'MM_type_A–C_POS18', 'MM_type_A–G_POS18', 'MM_type_T–C_POS18', 'MM_type_T–G_POS18', 'MM_type_T–A_POS18', 'MM_type_G–A_POS18', 'MM_type_G–T_POS18', 'MM_type_G–C_POS18', 'MM_type_C–A_POS18', 'MM_type_C–T_POS18', 'MM_type_C–G_POS18', 'MM_type_other_POS18']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS18', 'MM_type_A–C_POS18', 'MM_type_A–G_POS18', 'MM_type_T–C_POS18', 'MM_type_T–G_POS18', 'MM_type_T–A_POS18', 'MM_type_G–A_POS18', 'MM_type_G–T_POS18', 'MM_type_G–C_POS18', 'MM_type_C–A_POS18', 'MM_type_C–T_POS18', 'MM_type_C–G_POS18', 'MM_type_other_POS18']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
import pandas as pd
from scipy import stats
from scipy.stats import sem
OT_data1=pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l2=[]
l3=[]
df = pd.DataFrame(OT_data1)
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
print(len(POT))
print(len(NOT))
#print(len(df))
l1=['MM_type_A–T_POS23', 'MM_type_A–C_POS23', 'MM_type_A–G_POS23', 'MM_type_T–C_POS23', 'MM_type_T–G_POS23', 'MM_type_T–A_POS23', 'MM_type_G–A_POS23', 'MM_type_G–T_POS23', 'MM_type_G–C_POS23', 'MM_type_C–A_POS23', 'MM_type_C–T_POS23', 'MM_type_C–G_POS23', 'MM_type_other_POS23']
for i in l1:
total = POT[i].sum()
l2.append(total/524)
l3.append(sem(POT[i]))
print("\n Positive off-target \n")
print(l2)
print(l3)
l2=[]
l3=[]
for i in l1:
total = NOT[i].sum()
l2.append(total/525)
l3.append(sem(NOT[i]))
print("\n Negative off-targets \n")
print(l2)
print(l3)
import pandas as pd
df= pd.read_csv("D://PhD_related/manuscript_obj1/seq_encoding_map/LbCpf1/LbCpf1_dataset-sampled-525PN.csv", index_col=[0], encoding="cp1252")
l1=['MM_type_A–T_POS23', 'MM_type_A–C_POS23', 'MM_type_A–G_POS23', 'MM_type_T–C_POS23', 'MM_type_T–G_POS23', 'MM_type_T–A_POS23', 'MM_type_G–A_POS23', 'MM_type_G–T_POS23', 'MM_type_G–C_POS23', 'MM_type_C–A_POS23', 'MM_type_C–T_POS23', 'MM_type_C–G_POS23', 'MM_type_other_POS23']
for i in l1:
#print(df.groupby("Y")[i].describe())
POT = df[(df['Y'] == 1)]
NOT = df[(df['Y'] == 0)]
from scipy import stats
print(i)
print(stats.shapiro(POT[i]))
print(stats.shapiro(NOT[i]))
print(stats.ttest_ind(POT[i], NOT[i], equal_var = False))
print("\n")
| 0.232833 | 0.887156 |
# Use examples of [premise](https://github.com/romainsacchi/premise)
Author: [romainsacchi](https://github.com/romainsacchi)
This notebook shows examples on how to use `premise` to adapt the life cycle inventory database [ecoinvent](https://www.ecoinvent.org/) for prospective environmental impact assessment.
This library extract useful information from IAM model output files (such as those of REMIND or IMAGE) and aligns inventories in the ecoinvent database accordingly.
With version 0.3.7, the following transformation are available:
* `update_electricity()`: create regional electricity markets and adjust efficiency of power plants
* `update_cement()`: creates regional markets for clinker production and adjust clinker production efficiency
* `update_steel()`: creates regional markets for steel and adjust steel production efficiency and the supply of secondary steel
* `update_cars()`: produces fleet average cars and relinks to activities consuming pasenger car trnasport
* `update_trucks()`: produces fleet average trucks and relinks to activities consuming lorry trnasport
* `update_solar_PV()`: updates efficiency of solar PV modules
Additional documentation on the methodology is available [here](https://premise.readthedocs.io/en/latest/introduction.html).
There's also a **pre-print publication** about `premise` [here](https://www.psi.ch/en/ta/preprint).
## Requirements
* **Pyhton 3.9 is highly recommended**
* a user license for ecoinvent v.3
* a **decryption key**, to be asked from [Romain Sacchi](mailto:[email protected])
# Use case with [brightway2](https://brightway.dev/)
`brightway2` is an open source LCA framework for Python.
To use `premise` from `brightway2`, it requires that you have an opened `brightway2` project with a biosphere database as well as an ecoinvent v.3 cut-off database registered in that project.
```
from premise import *
import brightway2 as bw
bw.projects
bw.projects.set_current("my_bw_project")
list(bw.databases)
```
### List of available scenarios
Some scenarios come installed with the library.
They are stored in `data/iam_ouput_files` from the root directory.
They are all within the same Shared Socio-Economic Pathway (SSP): SSP2 (nicknamed "middle of the road"), which descrobes a future world (in terms of GDP and demographics development, education, intergovernmental collaboration) very much in line with what has been observed historically..
But they are proposed in combination with different climate mitigation targets, called Representative Concentration Pathways (RCP).
Read more about SSPs and RCPs, [here](https://www.carbonbrief.org/explainer-how-shared-socioeconomic-pathways-explore-future-climate-change).
With REMIND, we have the following SSP/RCP scenarios:
* "SSP2-Base"
* "SSP2-NPi"
* "SSP2-NDC"
* "SSP2-PkBudg1300"
* "SSP2-PkBudg1100"
* "SSP2-PkBudg900"
With IMAGE, we have the following SSP/RCP scenarios:
* "SSP2-Base"
* "SSP2-RCP26"
* "SSP2-RCP19"
For reference:
* the "Base" scenario in REMIND and IMAGE corresponds to RCP 6 (W/m^2), with a temperature increase by 2100 superior to **3.5 C**.
* a RCP of **2.6** (W/m^2) corresponds to the soft target of the Paris Agreement (just **below 2 C** of atmospheric temperature increase by 2100)
* a RCP of **1.9** (W/m^2) corresponds to the ambitious target of the Paris Agreement (**1.5 C** of atmospheric temperature increase by 2100)
* On REMIND's side, "SSP2-Base", "SSP2-PkBudg1300" and "SSP2-PkBudg900" are roughly equivalent in terms of climate mitigation target to "SSP2-Base", "SSP2-RCP26" and "SSP2-RCP19" on IMAGE's side.
### Database creation from default scenarios
To create a scenario using REMIND's SSP2 Base pathway, from ecoinvent 3.6 for the year 2028, one would execute the following cell. This leads to the extraction of the database, some cleanup as well as importing a few additional inventories.
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2029}
],
source_db="ecoinvent 3.5 cutoff", # <-- name of the database in the BW2 project. Must be a string.
source_version="3.5", # <-- version of ecoinvent. Can be "3.5", "3.6", "3.7" or "3.7.1". Must be a string.
key='xxxxxxxxxxxxxxxxxxxxxxxxxxxxxx' # <-- decryption key
# to be requested from the library maintainers if you want ot use default scenarios included in `premise`
)
```
If you do not want to integrate the IAM projections in the database, but only wish to have the additional inventories, you can stop here and export the database back to Brightway or other destinations, by using the `write_db_to` methods, like so:
```
ndb.write_db_to_brightway()
```
Howver, if you wish first to proceed with the IAM integration, you need to use the `updated_` methods, like so:
```
ndb.update_all()
ndb.write_db_to_brightway()
```
or here with ecoinvent 3.7.1
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2028}
],
source_db="ecoinvent 3.7 cutoff", # <-- this is NEW.
source_version="3.7.1", # <-- this is NEW
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
)
```
If you want to create multiple databases at once, just populate the `scenarios` list.
You will notice the key `exclude` for which we can list the transformations we do not wish to perform.
In this case, we do not wish to update the electricity sector.
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2020, "exclude": ["update_electricity"]},
{"model":"remind", "pathway":"SSP2-Base", "year":2030},
{"model":"remind", "pathway":"SSP2-Base", "year":2040},
{"model":"remind", "pathway":"SSP2-Base", "year":2050},
],
source_db="ecoinvent 3.7 cutoff", # <-- name of the database. Must be a string.
source_version="3.7.1", # <-- version of ecoinvent. Can be "3.5", "3.6", "3.7" or "3.7.1"
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
)
ndb.update_all()
```
When the database is loaded and the additional inventories imported, you can apply a transformation function.
For example here, we adjust the efficiency of the solar PVs to the two scenarios we have loaded.
We go more in details later.
```
ndb.update_solar_PV()
```
And then, we register these two databases back into brightway2.
```
ndb.write_db_to_brightway()
```
### Database creation from non-default scenarios
If you have some specific IAM scenarios (one that is not included in `premise`) you would like to build a database from, you can specify the directory to those.
**Important remark**: your scenario file must begin with "remind_" or "image_". When using a non-default scenario that you provide yourself, you do not have to provide a decryption key.
```
ndb = NewDatabase(
scenarios = [{"model":"remind", "pathway":"my_special_scenario", "year":2028, "filepath":r"C:\Users\sacchi_r\Downloads\REMIND"}],
source_db="ecoinvent 3.6 cutoff", # <-- name of the database
source_version=3.6, # <-- version of ecoinvent
)
```
### Adding inventories
Upon the database extraction, you can import some of your Brightway2-compatible inventories like so:
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030},
],
source_db="ecoinvent 3.7 cutoff",
source_version="3.7.1",
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
additional_inventories= [ # <-- this is NEW
{"filepath": r"filepath\to\excel_file.xlsx", "ecoinvent version": "3.7"}, # <-- this is NEW
{"filepath": r"filepath\to\another_excel_file.xlsx", "ecoinvent version": "3.7"}, # <-- this is NEW
] # <-- this is NEW
)
```
# Use case with ecospold2
The source database does not have to be from a brightway2 project.
It can be directly extracted from the bunch of ecospold2 files one gets when downloaded from the [ecoinvent website](https://ecoinvent.org).
For this, one needs to specify the argument `source_db = "ecospold"` as well as `source_file_path`, which is the directory leading to the ecospold files.
For example, here we combine the use of a specific (non-default) IAM scenario file with the use of ecospold2 files as data source (ecoinvent 3.5 in this case).
```
ndb = NewDatabase(
scenarios = [
{"model":"remind", "pathway":"my_special_scenario", "year":2028, "filepath":r"C:\Users\sacchi_r\Downloads\REMIND"}
],
source_type="ecospold", # <--- this is NEW
source_file_path=r"C:\Users\sacchi_r\Dropbox\Public\ecoinvent 3.5_cutoff_ecoSpold02\datasets", # <-- this is NEW
source_version="3.5",
)
```
# Transformation functions
These functions modify the extracted database:
* **update_electricity()**: alignment of regional electricity production mixes as well as efficiencies for a number of
electricity production technologies, including Carbon Capture and Storage technologies.
* **update_cement()**: adjustment of technologies for cement production (dry, semi-dry, wet, with pre-heater or not),
fuel efficiency of kilns, fuel mix of kilns (including biomass and waste fuels) and clinker-to-cement ratio.
* **update_steel()**: adjustment of process efficiency, fuel mix and share of secondary steel in steel markets.
* **update_solar_PV()**: adjustment of solar PV panels efficiency to the year considered.
* **update_cars()**: creates updated inventories for fleet average passenger cars and links back to activities that consume
transport.
* **update_trucks()**: creates updated inventories for fleet average lorry trucks and links back to activities that consume
transport. Such inventories are generated by [carculator](https://github.com/romainsacchi/carculator) for passenger cars and [carculator_truck](https://github.com/romainsacchi/carculator_truck) for medium and heavy-duty trucks.
They can be applied *separately*, *consecutively* or *altogether* (using instead **.update_all()**).
They will apply to all the scenario-specific databases listed in `scenarios`.
```
ndb.update_all()
from premise import *
import brightway2 as bw
bw.projects.set_current("article_carculator")
ndb = NewDatabase(
scenarios=[
{'model':'remind','pathway':'SSP2-Base','year':'2020'},
{"model":"image", "pathway":"SSP2-Base", "year":2034},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7",
)
ndb.update_all()
ndb.write_db_to_brightway()
```
You can also give your datababases a custom name.
```
ndb.write_db_to_brightway(name=["my_custom_name_1", "my_custom_name_2"])
```
### Fleet files
A last word about passenger cars and trucks: it is possible to pass a custom fleet composition file to generate fleet average inventories and limit those to specific regions (here, the European region).
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030,
"passenger cars": {"regions":["EUR", "NEU"],
"fleet file":r"filepath/to/fleet_file.csv.csv"},
"trucks": {"regions":["EUR"]}
},
{"model":"image", "pathway":"SSP2-Base", "year":2030,},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7"
)
```
### Exclude specific functions
Finally, we can exclude some transformation functions when executing `update_all()` like so:
```
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030,
"exclude": ["update_steel"], # <-- do not execute update_seel()
"passenger cars": {"regions":["EUR"]},"trucks": {"regions":["EUR"]}
},
{"model":"remind", "pathway":"SSP2-Base", "year":2030,},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7",
)
```
# Export
### As a Brightway2 database
Export the modified database to brightway2
```
ndb.write_db_to_brightway()
```
### As a sparse matrix representation
Or export it as a sparse matrix representation.
This will export four files:
* "A_matrix.csv": matrix coordinates and values of shape (index of activity; index of product; value) for the technosphere
* "A_matrix_index.csv": labels for indices for A matrix of shape (name of activity, reference product, unit, location, index)
* "B_matrix.csv": matrix coordinates and values of shape (index of activity; index of biosphere flow; value) for the biosphere
* "B_matrix_index.csv": labels for indices for B matrix of shape (name of biosphere flow, main compartment, sub-compartmnet, unit, index)
As a convenience, you can specifiy a directory where to store the exported matrices.
If the directory does not exist, it will be created.
If you leave it unspecified, they will be stored in **data/matrices** in the root folder of the library.
```
ndb.write_db_to_matrices(filepath=r"C:/Users/sacchi_r/Downloads/exported_matrices")
```
### As a SimaPro CSV file
```
ndb.write_db_to_simapro(filepath=r"C:/Users/sacchi_r/Downloads/exported_simapro_file")
```
### As a Superstructure database
A superstructure database is a database that can accomodate several scenarios, as described [here](https://github.com/dgdekoning/brightway-superstructure), to be then used in [Activity-Browser](https://github.com/LCA-ActivityBrowser/activity-browser).
This function will export the superstructure database as well as produce a "scenario difference file". Hence, even though you create multiple scenarios, **you only need to write to disk one database**.
```
ndb.write_superstructure_db_to_brightway()
```
|
github_jupyter
|
from premise import *
import brightway2 as bw
bw.projects
bw.projects.set_current("my_bw_project")
list(bw.databases)
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2029}
],
source_db="ecoinvent 3.5 cutoff", # <-- name of the database in the BW2 project. Must be a string.
source_version="3.5", # <-- version of ecoinvent. Can be "3.5", "3.6", "3.7" or "3.7.1". Must be a string.
key='xxxxxxxxxxxxxxxxxxxxxxxxxxxxxx' # <-- decryption key
# to be requested from the library maintainers if you want ot use default scenarios included in `premise`
)
ndb.write_db_to_brightway()
ndb.update_all()
ndb.write_db_to_brightway()
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2028}
],
source_db="ecoinvent 3.7 cutoff", # <-- this is NEW.
source_version="3.7.1", # <-- this is NEW
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
)
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2020, "exclude": ["update_electricity"]},
{"model":"remind", "pathway":"SSP2-Base", "year":2030},
{"model":"remind", "pathway":"SSP2-Base", "year":2040},
{"model":"remind", "pathway":"SSP2-Base", "year":2050},
],
source_db="ecoinvent 3.7 cutoff", # <-- name of the database. Must be a string.
source_version="3.7.1", # <-- version of ecoinvent. Can be "3.5", "3.6", "3.7" or "3.7.1"
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
)
ndb.update_all()
ndb.update_solar_PV()
ndb.write_db_to_brightway()
ndb = NewDatabase(
scenarios = [{"model":"remind", "pathway":"my_special_scenario", "year":2028, "filepath":r"C:\Users\sacchi_r\Downloads\REMIND"}],
source_db="ecoinvent 3.6 cutoff", # <-- name of the database
source_version=3.6, # <-- version of ecoinvent
)
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030},
],
source_db="ecoinvent 3.7 cutoff",
source_version="3.7.1",
key='xxxxxxxxxxxxxxxxxxxxxxxxx'
additional_inventories= [ # <-- this is NEW
{"filepath": r"filepath\to\excel_file.xlsx", "ecoinvent version": "3.7"}, # <-- this is NEW
{"filepath": r"filepath\to\another_excel_file.xlsx", "ecoinvent version": "3.7"}, # <-- this is NEW
] # <-- this is NEW
)
ndb = NewDatabase(
scenarios = [
{"model":"remind", "pathway":"my_special_scenario", "year":2028, "filepath":r"C:\Users\sacchi_r\Downloads\REMIND"}
],
source_type="ecospold", # <--- this is NEW
source_file_path=r"C:\Users\sacchi_r\Dropbox\Public\ecoinvent 3.5_cutoff_ecoSpold02\datasets", # <-- this is NEW
source_version="3.5",
)
ndb.update_all()
from premise import *
import brightway2 as bw
bw.projects.set_current("article_carculator")
ndb = NewDatabase(
scenarios=[
{'model':'remind','pathway':'SSP2-Base','year':'2020'},
{"model":"image", "pathway":"SSP2-Base", "year":2034},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7",
)
ndb.update_all()
ndb.write_db_to_brightway()
ndb.write_db_to_brightway(name=["my_custom_name_1", "my_custom_name_2"])
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030,
"passenger cars": {"regions":["EUR", "NEU"],
"fleet file":r"filepath/to/fleet_file.csv.csv"},
"trucks": {"regions":["EUR"]}
},
{"model":"image", "pathway":"SSP2-Base", "year":2030,},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7"
)
ndb = NewDatabase(
scenarios=[
{"model":"remind", "pathway":"SSP2-Base", "year":2030,
"exclude": ["update_steel"], # <-- do not execute update_seel()
"passenger cars": {"regions":["EUR"]},"trucks": {"regions":["EUR"]}
},
{"model":"remind", "pathway":"SSP2-Base", "year":2030,},
],
key='xxxxxxxxxxxxxxxxxxxxxxxxx',
source_db="ecoinvent 3.7 cutoff",
source_version="3.7",
)
ndb.write_db_to_brightway()
ndb.write_db_to_matrices(filepath=r"C:/Users/sacchi_r/Downloads/exported_matrices")
ndb.write_db_to_simapro(filepath=r"C:/Users/sacchi_r/Downloads/exported_simapro_file")
ndb.write_superstructure_db_to_brightway()
| 0.356447 | 0.96738 |
# Data Mining Process Overview
The process we will be following throughout this course is known as the **Cross-Industry Standard Process for Data Mining (CRISP-DM),** one of the most widely used methods for data mining.
|  |
|:--:|
| <b>Fig. 1 - This is the CRISP-DM Process.</b>|
The CRISP-DM process is made up of several parts:
1. **Data**
2. **Business Understanding and Data Understanding** (iterative)
3. **Data Preparation and Data Modeling** (iterative/dependent)
4. **Evaluation**
5. **Deployment**
Data mining is not a clear-cut process and sometimes may require you to go back to part one even after getting to your evaluation.
The first part of the process, **Business Understanding,** requires you to understand what it is you are trying to achieve from a business perspective. This will involve measuring the criteria which will determine whether or not the project was successful.
When developing a business understanding, it should be known there are 3 potential outcomes when it comes to data mining:
1. **Descriptive** - using data to develop insights into what already happened
2. **Predictive** - using data to forecast potential future outcomes
3. **Prescriptive** - using data to guide decision making
Let's look at Zillow, an online real estate marketplace, as an example of how someone may pose questions differently depending on these 3 potential outcomes:
1. Descriptive - What kind of houses sell fast?
2. Predictive - How much will this house sell for?
3. Prescriptive - What listings should be featured?
The next step in our process, which goes hand-in-hand with **Business Understanding,** is **Data Understanding.** Understanding your data will require you to process what data you will need and how to get that data. You may collect it yourself, use publicly available data, or purchase data.
Once there is a clear understanding of what you are trying to achieve and how you plan to achieve it, you will move on to the next step, **Data Preparation,** which will allow you to perform data mining techniques on that data. Preparing your data can involve many different steps, including but not limited to: dealing with missing variables or outliers, rescaling your data, and creating dummy variables for qualitative values.
At the **Modeling** stage, you will need to determine what type of data mining process to apply. This will require you to determine if this is a problem where there is an intended problem or outcome that can be a variable. If so, it will have you using a **supervised model.** If there is not then you will be using an **unsupervised model.** We will discuss these models in more detail later on in the chapter.
We see that the **Data Preparation** and **Modeling** stages are closely related; this is because our data modeling is dependent on the way we structure our data. Once we have decided on a model for our data, it is time for the **Evaluation** stage. For predictive models, we see how well the model works based on the use of an entirely _new_ dataset. The comparison of this new data with the original model will gauge predictive accuracy.
Depending on the results of the evaluation, we now may be ready for the final stage or we may need to revert back to the original stage. The final stage of the process is the **Deployment** stage. This is where the model is ready to be released and shared for implementation. It is important to note that though this is the final stage, the data mining cycle is continuous and ongoing. As such, it is very likely this model will be revised in the future.
**(Concluding Paragraph on DM Process)**
## Data Sets
**(Introductory paragraph for data set info)**
Now we are going to see what a data set could look like. We use a table to represent the dataset where each row is a record/observation and each column is variable. A record/observation could be information about a specific customer or a specific property if you are a company like Zillow. The variables reflect different pieces of information about each record.
Suppose we want to collect information about Gies Business students.
| | YEAR | MAJOR | INTERNSHIP TYPE |
|-------------------|-----------|-------|-----------------|
| Student 1 | Freshman | ACCY | Non-profit |
| Student 2 | Junior | FIN | Banking |
| Student 3 | Sophomore | OM | Tech |
| Student 4 | Senior | ACCY | Banking |
| Student 5 | Senior | IS | Tech |
| Student 6 | Freshman | SCM | Retail |
| Student 7 | Sophomore | FIN | Banking |
| Student 8 | Sophomore | OM | Retail |
| (Table continues) | | | |
Each row will correspond to a single student and the columns represent information about each student, such as their year, major and what type of internship they most recently completed. It is likely that you will be dealing with large databases when working for a company. In this case, it is more practical to work with a sample of the dataset in order to build and test your model.
The process of taking a sample from the dataset and using it to create your final model with which you will make predictions for records in the larger database is called **scoring**. In this case, sample refers to a collection of observations, but in the machine learning community, sample refers to a single record.
It is important to note data points in your sample will have an impact on your model. Particularly, an imbalanced sample which doesn’t reflect the broader population will lead to algorithmic bias.
An example of this is Amazon using AI when trying to screen job applications. Going through thousands of applications seemed like a monotonous job, so Amazon figured they could use AI to automate the process and they did just that in 2014. One issue came with how Amazon built their model: by scanning through past applicant resumes. The majority of past applications were from male applicants, which led the AI algorithm to penalize women applicants for going to woman schools and even having the word “woman” in their resume. Amazon ultimately discarded this project in 2018 as its algorithm was shown to be discriminatory. Algorithms are meant to alleviate human biases, not to exacerbate them.
## Types of Data
There are two main types of variables: numerical and categorical.
**Numerical variables** can either be continuous, meaning they can be any real number, or an integer. These variables usually do not require special processing.
**Categorical variables** are used to represent categories or classes. They can be coded as either text or integers. Categorical variables can be either nominal or ordinal, nominal being unordered and ordinal being ordered. An example of a **nominal variable** would be college majors, as there is no inherent ranking between different fields of study. **Ordinal variables**, on the other hand, do have an inherent characteristic order with labels such as low, medium and high. In order for a machine learning algorithm to process categorical data that is text, it needs to be converted to numbers.
Nominal variables would be converted to dummy variables, usually taking a value of either 0 or 1, whereas ordinal variables would be converted to integers which represent their inherent ranking. Let’s look at an example of this using student data.
|  |
|:--:|
| <b>Fig. 2 - Nominal variables do not have an inherent order, whereas ordinal variables do.</b>|
As mentioned earlier, majors are an example of nominal variables. In order to turn these variables into numbers, we need to create dummy variables.
We would create 2 new columns: Major_Accy and Major_OM. In the Major_Accy column, we would put a 1 for that row if the student is an accounting major and a 0 otherwise. We would follow the same rule for the Major_OM column. The other column would be considered an ordinal value as order does matter, so we would use values such as “First” through “Fourth” year, and convert those to numbers 1-4 to place in the appropriate row based on the student’s year in school.
```
random_df = pd.get_dummies(data_df, prefix_sep='_', drop_first=True)
random_df.columns
```
Converting variables is a very common data pre-processing step. Other common steps in the data pre-processing stage include detecting and possibly removing outliers, handling missing data, and normalizing data.
As you may have noticed, many of our variable names include underscores as opposed to spaces. Stripping leading and trailing spaces is an important part of cleaning up your data and reformatting it so that it is Python-friendly.
```
new_names = [s.strip().replace(' ', '_') for s in random_df.columns]
random_df.columns = new_names
random_df.columns
```
Additionally, in order to begin analyzing our data, we will need to split the predictor and outcome variables. We can accomplish this by dropping the outcome_variable from the initial dataframe(random_df) and assigning the outcome_variable to ‘y’:
```
X = random_df.drop('outcome_variable', axis=1) # all variables EXCEPT outcome variable
y = random_df.outcome_variable # just the outcome variable
```
**Outliers**
An **outlier** is an extreme observation, meaning that it is something that is further away from the other data points. These outliers can end up significantly affecting the outcome of the model.

In the figure, we see the independent variable speed on the horizontal axis and the dependent variable distance on the vertical axis. We’ve plotted two graphs: one with outliers (left) and one without (right). We see that the majority of our data points are on the bottom left of the graph, with only a few points at the upper right hand corner (graph on the left). A simple regression line is fit, and we observe the slope of this graph is pretty steep.
When we remove the outliers and fit the model again (graph on the right), the line has a significantly less steep slope. In the second model, the prediction of distance for larger speed values is significantly smaller than that in the first model.
When outliers are identified, removing them is not always necessary. However, outliers must be detected and then carefully examined. You must use your domain knowledge to determine whether or not they are appropriate to keep in the dataset.
**Missing Values**
Another issue you will face is **missing values** in your dataset. You should expect that when you get a dataset, some records will not have data for every single variable.
There are different ways to deal with this. The first solution to this problem is **omission**.
| | Variable 1 | Variable 2 | Variable 3 |
|----------|------------|------------|------------|
| Record 1 | x | x | x |
| Record 2 | x | x | x |
| Record 3 | | | x |
| Record 4 | x | x | x |
| Record 5 | x | x | x |
| Record 6 | x | | |
| Record 7 | x | x | x |
| Record 8 | x | x | x |
Your missing data could be concentrated in a small number of records. In the example shown above, missing data appears only in Records 3 and 6, so we could simply choose to omit those records.
If your data is concentrated in a small number of variables, you can remove that variable from your dataset. Since we see missing data only occurs in variable 2, we would omit that column from our data frame. However, you may not want to omit entire records or variables because you would lose the information that they do contain.
Another option would be to replace the missing values with a substitute such as the mean or median of all the records. This process is called **imputation**.
| | Variable 1 | Variable 2 | Variable 3 | Variable 4 |
|----------|------------|------------|------------|------------|
| Record 1 | 1 | 4 | 5 | 2 |
| Record 2 | | 3 | | 3 |
| Record 3 | 5 | 2 | 9 | |
| Record 4 | 3 | 8 | 2 | 3 |
Let’s use imputation to handle missing values in the dataset above and replace all missing values with the **mean** of the remaining records.
The first missing value is Variable 1 for Record 2:
Sum of the variable values for the other three records: $ 1 + 5 + 3 = 9 $
Mean: $ 9 /3 = 3 $
The second missing value is Variable 3 for Record 2:
Sum of the variable values for the other three records: $ 5 + 9 + 2 = 16 $
Mean: $ 16 / 3 \approx 5.34 $
The final missing value is Variable 4 for Record 3:
Sum of the variable values for the other three records: $ 2 + 3 + 3 = 8 $
Mean: $ 8/3 \approx 2.67 $
**Rescaling Data**
The fourth and final pre-processing step we’ll discuss is **rescaling data**.
Sometimes you may come across a dataset that has vastly different ranges of values. Take for example a data frame storing values for prices of homes, which would be in the range of thousands, and other variables such as number of rooms, which would likely not be over the teen values.
For some machine learning algorithms, this difference in scale will negatively affect performance, so we rescale the data.
There are two ways we can rescale:
First, we can **standardize** the data for a given variable. To do this we would need to calculate the mean and standard deviation for that variable. Then, for each observation for that variable, we subtract the average and divide by the standard deviation.
The formula to standardize data is: $$ x_{new} = \frac{x - \mu}{\sigma} $$
A second option is to **normalize** the data. In this case, we want to rescale all of the values for a given variable to be between 0 and 1. We do this by subtracting the minimum variable value from each possible value and dividing it by the range of values.
The formula to normalize data is: $$ x_{new} = \frac{x - x_{min}}{x_{max} - x_{min}} $$
For the following example where we have 6 records and we want to normalize the variable # of rooms, how would we do this?
| | # of Rooms |
|----------|------------|
| Record 1 | 3 |
| Record 2 | 2 |
| Record 3 | 1 |
| Record 4 | 6 |
| Record 5 | 7 |
| Record 6 | 5 |
We must subtract the minimum value and divide by the range.
We will do this step by step.
First, we see the minimum value of this variable, # of rooms, is 1 and the range (max-min) is 6. Let's subtract 1 from each record, which yields variable values of 2, 1, 0, 5, 6, and 4. Now we will divide by our range, 6, giving us ⅓, ⅙ , 0, ⅚ ,1, ⅔ We have now normalized the data for this variable.
**$ (x - x_{min}) $ yields:**
| |$(Record - Min)$|
|----------|----------------|
| Record 1 | $2$ |
| Record 2 | $1$ |
| Record 3 | $0$ |
| Record 4 | $5$ |
| Record 5 | $6$ |
| Record 6 | $4$ |
**$ \frac{x - x_{min}}{x_{max} - x_{min}} $ yields:**
| |$ x_{new} $ |
|-------------|--------------|
| Record 1 |$\frac {1}{3}$|
| Record 2 |$\frac {1}{6}$|
| Record 3 |$0 $ |
| Record 4 |$\frac {5}{6}$|
| Record 5 |$ 1$ |
| Record 6 |$\frac {2}{3}$|
To recap the steps of data preparation:
* First, you begin by sampling data from the larger database.
* Next, you convert some variables, usually converting categorical variables from text to integers. Nominal variables are converted into a series of dummy variables, and ordinal variables are converted to integers which preserve order.
* Then, you inspect the data for outliers and use your domain knowledge to determine whether or not to remove them.
* You also must decide how to handle missing data, either through omission of variables/ records or using imputation to fill them in with the median or mean values.
* Finally, you might need to rescale your data to account for extremely different ranges. You can do this either by standardizing or normalization data.
We will talk about different data mining algorithms we can use and how to evaluate and select the best model.
## References:
**(relevant links + additional readings)**
## Glossary:
**Business Understanding:**
**Descriptive Data Mining:**
**Predictive Data Mining:**
**Prescriptive Data Mining:**
**Data Understanding:**
**Data Preparation:**
**Modeling:**
**Supervised Model:**
**Unsupervised Model:**
**Evaluation:**
**Deployment:**
**Sample:**
**Scoring:**
**Numerical Variables:**
**Categorical Variables:**
**Nominal Variables:**
**Ordinal Variables:**
**Dummy Variables:**
**Outliers:**
**Missing Values:**
**Rescaling Data:**
**Standardization:**
**Normalization:**
|
github_jupyter
|
random_df = pd.get_dummies(data_df, prefix_sep='_', drop_first=True)
random_df.columns
new_names = [s.strip().replace(' ', '_') for s in random_df.columns]
random_df.columns = new_names
random_df.columns
X = random_df.drop('outcome_variable', axis=1) # all variables EXCEPT outcome variable
y = random_df.outcome_variable # just the outcome variable
| 0.159119 | 0.990857 |
```
%matplotlib inline
import adaptive
import matplotlib.pyplot as plt
import pycqed as pq
import numpy as np
from pycqed.measurement import measurement_control
import pycqed.measurement.detector_functions as det
from qcodes import station
station = station.Station()
```
## Setting up the mock device
Measurements are controlled through the `MeasurementControl` usually instantiated as `MC`
```
from pycqed.instrument_drivers.virtual_instruments.mock_device import Mock_Device
MC = measurement_control.MeasurementControl('MC',live_plot_enabled=True, verbose=True)
MC.station = station
station.add_component(MC)
mock_device = Mock_Device('mock_device')
mock_device.mw_pow(-20)
mock_device.res_freq(7.400023457e9)
mock_device.cw_noise_level(.0005)
mock_device.acq_delay(.05)
```
## Measuring a resonator using the conventional method
Points are chosen on a linspace of 100 points. This is enough to identify the location of the resonator.
```
freqs = np.linspace(7.39e9, 7.41e9, 100)
d = det.Function_Detector(mock_device.S21,value_names=['Magn', 'Phase'],
value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_sweep_points(freqs)
MC.set_detector_function(d)
dat=MC.run('test')
```
## Using 1D adaptive sampler from the MC
This can also be done using an adaptive `Leaner1D` object, chosing 100 points optimally in the interval.
```
mock_device.acq_delay(.05)
d = det.Function_Detector(mock_device.S21, value_names=['Magn', 'Phase'], value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_detector_function(d)
MC.set_adaptive_function_parameters({'adaptive_function': adaptive.Learner1D,
'goal':lambda l: l.npoints>100,
'bounds':(7.39e9, 7.41e9)})
dat = MC.run(mode='adaptive')
from pycqed.analysis import measurement_analysis as ma
# ma.Homodyne_Analysis(close_fig=False, label='M')
```
## Two D learner
The learner can also be used to adaptively sample a 2D /heatmap type experiment.
However, currently we do not have easy plotting function for that and we still need to rely on the adaptive Learner plotting methods.
It would be great to have this working with a realtime pyqtgraph based plotting window so that we can use this without the notebooks.
```
d = det.Function_Detector(mock_device.S21, value_names=['Magn', 'Phase'], value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_sweep_function_2D(mock_device.mw_pow)
MC.set_detector_function(d)
MC.set_adaptive_function_parameters({'adaptive_function': adaptive.Learner2D,
'goal':lambda l: l.npoints>20*20,
'bounds':((7.398e9, 7.402e9),
(-20, -10))})
dat = MC.run(mode='adaptive')
# Required to be able to use the fancy interpolating plot
adaptive.notebook_extension()
MC.learner.plot(tri_alpha=.1)
```
|
github_jupyter
|
%matplotlib inline
import adaptive
import matplotlib.pyplot as plt
import pycqed as pq
import numpy as np
from pycqed.measurement import measurement_control
import pycqed.measurement.detector_functions as det
from qcodes import station
station = station.Station()
from pycqed.instrument_drivers.virtual_instruments.mock_device import Mock_Device
MC = measurement_control.MeasurementControl('MC',live_plot_enabled=True, verbose=True)
MC.station = station
station.add_component(MC)
mock_device = Mock_Device('mock_device')
mock_device.mw_pow(-20)
mock_device.res_freq(7.400023457e9)
mock_device.cw_noise_level(.0005)
mock_device.acq_delay(.05)
freqs = np.linspace(7.39e9, 7.41e9, 100)
d = det.Function_Detector(mock_device.S21,value_names=['Magn', 'Phase'],
value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_sweep_points(freqs)
MC.set_detector_function(d)
dat=MC.run('test')
mock_device.acq_delay(.05)
d = det.Function_Detector(mock_device.S21, value_names=['Magn', 'Phase'], value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_detector_function(d)
MC.set_adaptive_function_parameters({'adaptive_function': adaptive.Learner1D,
'goal':lambda l: l.npoints>100,
'bounds':(7.39e9, 7.41e9)})
dat = MC.run(mode='adaptive')
from pycqed.analysis import measurement_analysis as ma
# ma.Homodyne_Analysis(close_fig=False, label='M')
d = det.Function_Detector(mock_device.S21, value_names=['Magn', 'Phase'], value_units=['V', 'deg'])
MC.set_sweep_function(mock_device.mw_freq)
MC.set_sweep_function_2D(mock_device.mw_pow)
MC.set_detector_function(d)
MC.set_adaptive_function_parameters({'adaptive_function': adaptive.Learner2D,
'goal':lambda l: l.npoints>20*20,
'bounds':((7.398e9, 7.402e9),
(-20, -10))})
dat = MC.run(mode='adaptive')
# Required to be able to use the fancy interpolating plot
adaptive.notebook_extension()
MC.learner.plot(tri_alpha=.1)
| 0.567098 | 0.880026 |
# 3D Image Classification from CT Scans
**Author:** [Hasib Zunair](https://twitter.com/hasibzunair)<br>
**Date created:** 2020/09/23<br>
**Last modified:** 2020/09/23<br>
**Description:** Train a 3D convolutional neural network to predict presence of pneumonia.
## Introduction
This example will show the steps needed to build a 3D convolutional neural network (CNN)
to predict the presence of viral pneumonia in computer tomography (CT) scans. 2D CNNs are
commonly used to process RGB images (3 channels). A 3D CNN is simply the 3D
equivalent: it takes as input a 3D volume or a sequence of 2D frames (e.g. slices in a CT scan),
3D CNNs are a powerful model for learning representations for volumetric data.
## References
- [A survey on Deep Learning Advances on Different 3D DataRepresentations](https://arxiv.org/pdf/1808.01462.pdf)
- [VoxNet: A 3D Convolutional Neural Network for Real-Time Object Recognition](https://www.ri.cmu.edu/pub_files/2015/9/voxnet_maturana_scherer_iros15.pdf)
- [FusionNet: 3D Object Classification Using MultipleData Representations](http://3ddl.cs.princeton.edu/2016/papers/Hegde_Zadeh.pdf)
- [Uniformizing Techniques to Process CT scans with 3D CNNs for Tuberculosis Prediction](https://arxiv.org/abs/2007.13224)
## Setup
```
import os
import zipfile
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
```
## Downloading the MosMedData: Chest CT Scans with COVID-19 Related Findings
In this example, we use a subset of the
[MosMedData: Chest CT Scans with COVID-19 Related Findings](https://www.medrxiv.org/content/10.1101/2020.05.20.20100362v1).
This dataset consists of lung CT scans with COVID-19 related findings, as well as without such findings.
We will be using the associated radiological findings of the CT scans as labels to build
a classifier to predict presence of viral pneumonia.
Hence, the task is a binary classification problem.
```
# Download url of normal CT scans.
url = "https://github.com/hasibzunair/3D-image-classification-tutorial/releases/download/v0.2/CT-0.zip"
filename = os.path.join(os.getcwd(), "CT-0.zip")
keras.utils.get_file(filename, url)
# Download url of abnormal CT scans.
url = "https://github.com/hasibzunair/3D-image-classification-tutorial/releases/download/v0.2/CT-23.zip"
filename = os.path.join(os.getcwd(), "CT-23.zip")
keras.utils.get_file(filename, url)
# Make a directory to store the data.
os.makedirs("MosMedData")
# Unzip data in the newly created directory.
with zipfile.ZipFile("CT-0.zip", "r") as z_fp:
z_fp.extractall("./MosMedData/")
with zipfile.ZipFile("CT-23.zip", "r") as z_fp:
z_fp.extractall("./MosMedData/")
```
## Loading data and preprocessing
The files are provided in Nifti format with the extension .nii. To read the
scans, we use the `nibabel` package.
You can install the package via `pip install nibabel`. CT scans store raw voxel
intensity in Hounsfield units (HU). They range from -1024 to above 2000 in this dataset.
Above 400 are bones with different radiointensity, so this is used as a higher bound. A threshold
between -1000 and 400 is commonly used to normalize CT scans.
To process the data, we do the following:
* We first rotate the volumes by 90 degrees, so the orientation is fixed
* We scale the HU values to be between 0 and 1.
* We resize width, height and depth.
Here we define several helper functions to process the data. These functions
will be used when building training and validation datasets.
```
import nibabel as nib
from scipy import ndimage
def read_nifti_file(filepath):
"""Read and load volume"""
# Read file
scan = nib.load(filepath)
# Get raw data
scan = scan.get_fdata()
return scan
def normalize(volume):
"""Normalize the volume"""
min = -1000
max = 400
volume[volume < min] = min
volume[volume > max] = max
volume = (volume - min) / (max - min)
volume = volume.astype("float32")
return volume
def resize_volume(img):
"""Resize across z-axis"""
# Set the desired depth
desired_depth = 64
desired_width = 128
desired_height = 128
# Get current depth
current_depth = img.shape[-1]
current_width = img.shape[0]
current_height = img.shape[1]
# Compute depth factor
depth = current_depth / desired_depth
width = current_width / desired_width
height = current_height / desired_height
depth_factor = 1 / depth
width_factor = 1 / width
height_factor = 1 / height
# Rotate
img = ndimage.rotate(img, 90, reshape=False)
# Resize across z-axis
img = ndimage.zoom(img, (width_factor, height_factor, depth_factor), order=1)
return img
def process_scan(path):
"""Read and resize volume"""
# Read scan
volume = read_nifti_file(path)
# Normalize
volume = normalize(volume)
# Resize width, height and depth
volume = resize_volume(volume)
return volume
```
Let's read the paths of the CT scans from the class directories.
```
# Folder "CT-0" consist of CT scans having normal lung tissue,
# no CT-signs of viral pneumonia.
normal_scan_paths = [
os.path.join(os.getcwd(), "MosMedData/CT-0", x)
for x in os.listdir("MosMedData/CT-0")
]
# Folder "CT-23" consist of CT scans having several ground-glass opacifications,
# involvement of lung parenchyma.
abnormal_scan_paths = [
os.path.join(os.getcwd(), "MosMedData/CT-23", x)
for x in os.listdir("MosMedData/CT-23")
]
print("CT scans with normal lung tissue: " + str(len(normal_scan_paths)))
print("CT scans with abnormal lung tissue: " + str(len(abnormal_scan_paths)))
```
## Build train and validation datasets
Read the scans from the class directories and assign labels. Downsample the scans to have
shape of 128x128x64. Rescale the raw HU values to the range 0 to 1.
Lastly, split the dataset into train and validation subsets.
```
# Read and process the scans.
# Each scan is resized across height, width, and depth and rescaled.
abnormal_scans = np.array([process_scan(path) for path in abnormal_scan_paths])
normal_scans = np.array([process_scan(path) for path in normal_scan_paths])
# For the CT scans having presence of viral pneumonia
# assign 1, for the normal ones assign 0.
abnormal_labels = np.array([1 for _ in range(len(abnormal_scans))])
normal_labels = np.array([0 for _ in range(len(normal_scans))])
# Split data in the ratio 70-30 for training and validation.
x_train = np.concatenate((abnormal_scans[:70], normal_scans[:70]), axis=0)
y_train = np.concatenate((abnormal_labels[:70], normal_labels[:70]), axis=0)
x_val = np.concatenate((abnormal_scans[70:], normal_scans[70:]), axis=0)
y_val = np.concatenate((abnormal_labels[70:], normal_labels[70:]), axis=0)
print(
"Number of samples in train and validation are %d and %d."
% (x_train.shape[0], x_val.shape[0])
)
```
## Data augmentation
The CT scans also augmented by rotating at random angles during training. Since
the data is stored in rank-3 tensors of shape `(samples, height, width, depth)`,
we add a dimension of size 1 at axis 4 to be able to perform 3D convolutions on
the data. The new shape is thus `(samples, height, width, depth, 1)`. There are
different kinds of preprocessing and augmentation techniques out there,
this example shows a few simple ones to get started.
```
import random
from scipy import ndimage
@tf.function
def rotate(volume):
"""Rotate the volume by a few degrees"""
def scipy_rotate(volume):
# define some rotation angles
angles = [-20, -10, -5, 5, 10, 20]
# pick angles at random
angle = random.choice(angles)
# rotate volume
volume = ndimage.rotate(volume, angle, reshape=False)
volume[volume < 0] = 0
volume[volume > 1] = 1
return volume
augmented_volume = tf.numpy_function(scipy_rotate, [volume], tf.float32)
return augmented_volume
def train_preprocessing(volume, label):
"""Process training data by rotating and adding a channel."""
# Rotate volume
volume = rotate(volume)
volume = tf.expand_dims(volume, axis=3)
return volume, label
def validation_preprocessing(volume, label):
"""Process validation data by only adding a channel."""
volume = tf.expand_dims(volume, axis=3)
return volume, label
```
While defining the train and validation data loader, the training data is passed through
and augmentation function which randomly rotates volume at different angles. Note that both
training and validation data are already rescaled to have values between 0 and 1.
```
# Define data loaders.
train_loader = tf.data.Dataset.from_tensor_slices((x_train, y_train))
validation_loader = tf.data.Dataset.from_tensor_slices((x_val, y_val))
batch_size = 2
# Augment the on the fly during training.
train_dataset = (
train_loader.shuffle(len(x_train))
.map(train_preprocessing)
.batch(batch_size)
.prefetch(2)
)
# Only rescale.
validation_dataset = (
validation_loader.shuffle(len(x_val))
.map(validation_preprocessing)
.batch(batch_size)
.prefetch(2)
)
```
Visualize an augmented CT scan.
```
import matplotlib.pyplot as plt
data = train_dataset.take(1)
images, labels = list(data)[0]
images = images.numpy()
image = images[0]
print("Dimension of the CT scan is:", image.shape)
plt.imshow(np.squeeze(image[:, :, 30]), cmap="gray")
```
Since a CT scan has many slices, let's visualize a montage of the slices.
```
def plot_slices(num_rows, num_columns, width, height, data):
"""Plot a montage of 20 CT slices"""
data = np.rot90(np.array(data))
data = np.transpose(data)
data = np.reshape(data, (num_rows, num_columns, width, height))
rows_data, columns_data = data.shape[0], data.shape[1]
heights = [slc[0].shape[0] for slc in data]
widths = [slc.shape[1] for slc in data[0]]
fig_width = 12.0
fig_height = fig_width * sum(heights) / sum(widths)
f, axarr = plt.subplots(
rows_data,
columns_data,
figsize=(fig_width, fig_height),
gridspec_kw={"height_ratios": heights},
)
for i in range(rows_data):
for j in range(columns_data):
axarr[i, j].imshow(data[i][j], cmap="gray")
axarr[i, j].axis("off")
plt.subplots_adjust(wspace=0, hspace=0, left=0, right=1, bottom=0, top=1)
plt.show()
# Visualize montage of slices.
# 4 rows and 10 columns for 100 slices of the CT scan.
plot_slices(4, 10, 128, 128, image[:, :, :40])
```
## Define a 3D convolutional neural network
To make the model easier to understand, we structure it into blocks.
The architecture of the 3D CNN used in this example
is based on [this paper](https://arxiv.org/abs/2007.13224).
```
def get_model(width=128, height=128, depth=64):
"""Build a 3D convolutional neural network model."""
inputs = keras.Input((width, height, depth, 1))
x = layers.Conv3D(filters=64, kernel_size=3, activation="relu")(inputs)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=64, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=128, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=256, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.GlobalAveragePooling3D()(x)
x = layers.Dense(units=512, activation="relu")(x)
x = layers.Dropout(0.3)(x)
outputs = layers.Dense(units=1, activation="sigmoid")(x)
# Define the model.
model = keras.Model(inputs, outputs, name="3dcnn")
return model
# Build model.
model = get_model(width=128, height=128, depth=64)
model.summary()
```
## Train model
```
# Compile model.
initial_learning_rate = 0.0001
lr_schedule = keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate, decay_steps=100000, decay_rate=0.96, staircase=True
)
model.compile(
loss="binary_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=lr_schedule),
metrics=["acc"],
)
# Define callbacks.
checkpoint_cb = keras.callbacks.ModelCheckpoint(
"3d_image_classification.h5", save_best_only=True
)
early_stopping_cb = keras.callbacks.EarlyStopping(monitor="val_acc", patience=15)
# Train the model, doing validation at the end of each epoch
epochs = 100
model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=epochs,
shuffle=True,
verbose=2,
callbacks=[checkpoint_cb, early_stopping_cb],
)
```
It is important to note that the number of samples is very small (only 200) and we don't
specify a random seed. As such, you can expect significant variance in the results. The full dataset
which consists of over 1000 CT scans can be found [here](https://www.medrxiv.org/content/10.1101/2020.05.20.20100362v1). Using the full
dataset, an accuracy of 83% was achieved. A variability of 6-7% in the classification
performance is observed in both cases.
## Visualizing model performance
Here the model accuracy and loss for the training and the validation sets are plotted.
Since the validation set is class-balanced, accuracy provides an unbiased representation
of the model's performance.
```
fig, ax = plt.subplots(1, 2, figsize=(20, 3))
ax = ax.ravel()
for i, metric in enumerate(["acc", "loss"]):
ax[i].plot(model.history.history[metric])
ax[i].plot(model.history.history["val_" + metric])
ax[i].set_title("Model {}".format(metric))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(metric)
ax[i].legend(["train", "val"])
```
## Make predictions on a single CT scan
```
# Load best weights.
model.load_weights("3d_image_classification.h5")
prediction = model.predict(np.expand_dims(x_val[0], axis=0))[0]
scores = [1 - prediction[0], prediction[0]]
class_names = ["normal", "abnormal"]
for score, name in zip(scores, class_names):
print(
"This model is %.2f percent confident that CT scan is %s"
% ((100 * score), name)
)
```
|
github_jupyter
|
import os
import zipfile
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
# Download url of normal CT scans.
url = "https://github.com/hasibzunair/3D-image-classification-tutorial/releases/download/v0.2/CT-0.zip"
filename = os.path.join(os.getcwd(), "CT-0.zip")
keras.utils.get_file(filename, url)
# Download url of abnormal CT scans.
url = "https://github.com/hasibzunair/3D-image-classification-tutorial/releases/download/v0.2/CT-23.zip"
filename = os.path.join(os.getcwd(), "CT-23.zip")
keras.utils.get_file(filename, url)
# Make a directory to store the data.
os.makedirs("MosMedData")
# Unzip data in the newly created directory.
with zipfile.ZipFile("CT-0.zip", "r") as z_fp:
z_fp.extractall("./MosMedData/")
with zipfile.ZipFile("CT-23.zip", "r") as z_fp:
z_fp.extractall("./MosMedData/")
import nibabel as nib
from scipy import ndimage
def read_nifti_file(filepath):
"""Read and load volume"""
# Read file
scan = nib.load(filepath)
# Get raw data
scan = scan.get_fdata()
return scan
def normalize(volume):
"""Normalize the volume"""
min = -1000
max = 400
volume[volume < min] = min
volume[volume > max] = max
volume = (volume - min) / (max - min)
volume = volume.astype("float32")
return volume
def resize_volume(img):
"""Resize across z-axis"""
# Set the desired depth
desired_depth = 64
desired_width = 128
desired_height = 128
# Get current depth
current_depth = img.shape[-1]
current_width = img.shape[0]
current_height = img.shape[1]
# Compute depth factor
depth = current_depth / desired_depth
width = current_width / desired_width
height = current_height / desired_height
depth_factor = 1 / depth
width_factor = 1 / width
height_factor = 1 / height
# Rotate
img = ndimage.rotate(img, 90, reshape=False)
# Resize across z-axis
img = ndimage.zoom(img, (width_factor, height_factor, depth_factor), order=1)
return img
def process_scan(path):
"""Read and resize volume"""
# Read scan
volume = read_nifti_file(path)
# Normalize
volume = normalize(volume)
# Resize width, height and depth
volume = resize_volume(volume)
return volume
# Folder "CT-0" consist of CT scans having normal lung tissue,
# no CT-signs of viral pneumonia.
normal_scan_paths = [
os.path.join(os.getcwd(), "MosMedData/CT-0", x)
for x in os.listdir("MosMedData/CT-0")
]
# Folder "CT-23" consist of CT scans having several ground-glass opacifications,
# involvement of lung parenchyma.
abnormal_scan_paths = [
os.path.join(os.getcwd(), "MosMedData/CT-23", x)
for x in os.listdir("MosMedData/CT-23")
]
print("CT scans with normal lung tissue: " + str(len(normal_scan_paths)))
print("CT scans with abnormal lung tissue: " + str(len(abnormal_scan_paths)))
# Read and process the scans.
# Each scan is resized across height, width, and depth and rescaled.
abnormal_scans = np.array([process_scan(path) for path in abnormal_scan_paths])
normal_scans = np.array([process_scan(path) for path in normal_scan_paths])
# For the CT scans having presence of viral pneumonia
# assign 1, for the normal ones assign 0.
abnormal_labels = np.array([1 for _ in range(len(abnormal_scans))])
normal_labels = np.array([0 for _ in range(len(normal_scans))])
# Split data in the ratio 70-30 for training and validation.
x_train = np.concatenate((abnormal_scans[:70], normal_scans[:70]), axis=0)
y_train = np.concatenate((abnormal_labels[:70], normal_labels[:70]), axis=0)
x_val = np.concatenate((abnormal_scans[70:], normal_scans[70:]), axis=0)
y_val = np.concatenate((abnormal_labels[70:], normal_labels[70:]), axis=0)
print(
"Number of samples in train and validation are %d and %d."
% (x_train.shape[0], x_val.shape[0])
)
import random
from scipy import ndimage
@tf.function
def rotate(volume):
"""Rotate the volume by a few degrees"""
def scipy_rotate(volume):
# define some rotation angles
angles = [-20, -10, -5, 5, 10, 20]
# pick angles at random
angle = random.choice(angles)
# rotate volume
volume = ndimage.rotate(volume, angle, reshape=False)
volume[volume < 0] = 0
volume[volume > 1] = 1
return volume
augmented_volume = tf.numpy_function(scipy_rotate, [volume], tf.float32)
return augmented_volume
def train_preprocessing(volume, label):
"""Process training data by rotating and adding a channel."""
# Rotate volume
volume = rotate(volume)
volume = tf.expand_dims(volume, axis=3)
return volume, label
def validation_preprocessing(volume, label):
"""Process validation data by only adding a channel."""
volume = tf.expand_dims(volume, axis=3)
return volume, label
# Define data loaders.
train_loader = tf.data.Dataset.from_tensor_slices((x_train, y_train))
validation_loader = tf.data.Dataset.from_tensor_slices((x_val, y_val))
batch_size = 2
# Augment the on the fly during training.
train_dataset = (
train_loader.shuffle(len(x_train))
.map(train_preprocessing)
.batch(batch_size)
.prefetch(2)
)
# Only rescale.
validation_dataset = (
validation_loader.shuffle(len(x_val))
.map(validation_preprocessing)
.batch(batch_size)
.prefetch(2)
)
import matplotlib.pyplot as plt
data = train_dataset.take(1)
images, labels = list(data)[0]
images = images.numpy()
image = images[0]
print("Dimension of the CT scan is:", image.shape)
plt.imshow(np.squeeze(image[:, :, 30]), cmap="gray")
def plot_slices(num_rows, num_columns, width, height, data):
"""Plot a montage of 20 CT slices"""
data = np.rot90(np.array(data))
data = np.transpose(data)
data = np.reshape(data, (num_rows, num_columns, width, height))
rows_data, columns_data = data.shape[0], data.shape[1]
heights = [slc[0].shape[0] for slc in data]
widths = [slc.shape[1] for slc in data[0]]
fig_width = 12.0
fig_height = fig_width * sum(heights) / sum(widths)
f, axarr = plt.subplots(
rows_data,
columns_data,
figsize=(fig_width, fig_height),
gridspec_kw={"height_ratios": heights},
)
for i in range(rows_data):
for j in range(columns_data):
axarr[i, j].imshow(data[i][j], cmap="gray")
axarr[i, j].axis("off")
plt.subplots_adjust(wspace=0, hspace=0, left=0, right=1, bottom=0, top=1)
plt.show()
# Visualize montage of slices.
# 4 rows and 10 columns for 100 slices of the CT scan.
plot_slices(4, 10, 128, 128, image[:, :, :40])
def get_model(width=128, height=128, depth=64):
"""Build a 3D convolutional neural network model."""
inputs = keras.Input((width, height, depth, 1))
x = layers.Conv3D(filters=64, kernel_size=3, activation="relu")(inputs)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=64, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=128, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.Conv3D(filters=256, kernel_size=3, activation="relu")(x)
x = layers.MaxPool3D(pool_size=2)(x)
x = layers.BatchNormalization()(x)
x = layers.GlobalAveragePooling3D()(x)
x = layers.Dense(units=512, activation="relu")(x)
x = layers.Dropout(0.3)(x)
outputs = layers.Dense(units=1, activation="sigmoid")(x)
# Define the model.
model = keras.Model(inputs, outputs, name="3dcnn")
return model
# Build model.
model = get_model(width=128, height=128, depth=64)
model.summary()
# Compile model.
initial_learning_rate = 0.0001
lr_schedule = keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate, decay_steps=100000, decay_rate=0.96, staircase=True
)
model.compile(
loss="binary_crossentropy",
optimizer=keras.optimizers.Adam(learning_rate=lr_schedule),
metrics=["acc"],
)
# Define callbacks.
checkpoint_cb = keras.callbacks.ModelCheckpoint(
"3d_image_classification.h5", save_best_only=True
)
early_stopping_cb = keras.callbacks.EarlyStopping(monitor="val_acc", patience=15)
# Train the model, doing validation at the end of each epoch
epochs = 100
model.fit(
train_dataset,
validation_data=validation_dataset,
epochs=epochs,
shuffle=True,
verbose=2,
callbacks=[checkpoint_cb, early_stopping_cb],
)
fig, ax = plt.subplots(1, 2, figsize=(20, 3))
ax = ax.ravel()
for i, metric in enumerate(["acc", "loss"]):
ax[i].plot(model.history.history[metric])
ax[i].plot(model.history.history["val_" + metric])
ax[i].set_title("Model {}".format(metric))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(metric)
ax[i].legend(["train", "val"])
# Load best weights.
model.load_weights("3d_image_classification.h5")
prediction = model.predict(np.expand_dims(x_val[0], axis=0))[0]
scores = [1 - prediction[0], prediction[0]]
class_names = ["normal", "abnormal"]
for score, name in zip(scores, class_names):
print(
"This model is %.2f percent confident that CT scan is %s"
% ((100 * score), name)
)
| 0.738198 | 0.986165 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
#export
from exp.nb_05b import *
torch.set_num_threads(2)
x_train,y_train,x_valid,y_valid = get_data()
#export
def normalize_to(train, valid):
m,s = train.mean(),train.std()
return normalize(train, m, s), normalize(valid, m, s)
x_train, x_valid = normalize_to(x_train, x_valid)
train_ds, valid_ds = Dataset(x_train, y_train), Dataset(x_valid, y_valid)
x_train.mean(),x_train.std()
nh = 50
bs = 512
c = y_train.max().item() + 1
loss_func = F.cross_entropy
data = DataBunch(*get_dls(train_ds, valid_ds, bs), c)
#export
class Lambda(nn.Module):
def __init__(self, func):
super().__init__()
self.func = func
def forward(self, x):
return self.func(x)
def __repr__(self):
return f"Lambda({self.func})"
def flatten(x):
return x.view(x.shape[0], -1)
def mnist_resize(x):
return x.view(-1, 1, 28, 28)
def get_cnn_model(data):
return nn.Sequential(
Lambda(mnist_resize),
nn.Conv2d( 1, 8, 5, padding=2,stride=2), nn.ReLU(), #14
# nn.Conv2d( 1, 8, 3, padding=1,stride=2), nn.ReLU(), #14
nn.Conv2d( 8,16, 3, padding=1,stride=2), nn.ReLU(), # 7
nn.Conv2d(16,32, 3, padding=1,stride=2), nn.ReLU(), # 4
nn.Conv2d(32,32, 3, padding=1,stride=2), nn.ReLU(), # 2
nn.AdaptiveAvgPool2d(1),
Lambda(flatten),
nn.Linear(32,data.c)
)
all_entries = [i for i in iter(train_ds)]
img1 = all_entries[0][0]
model = get_cnn_model(data)
model
mods = [mod for mod in model.modules()]
x1 = mods[1](all_entries[0][0])
for i in range(len(mods) - 2):
x1 = mods[i + 2](x1)
print(i, mods[i + 2], x1.shape)
cbfs = [Recorder, partial(AvgStatsCallback, accuracy)]
def create_runner(model, cbfs):
opt = optim.SGD(model.parameters(), lr=0.4)
learn = Learner(model, opt, loss_func, data)
run = Runner(cb_funcs=cbfs)
return run, learn
run, learn = create_runner(model, cbfs)
%%time
run.fit(1, learn)
device = torch.device('cuda', 0)
class CudaCallback(Callback):
def __init__(self):
self.device = device
def begin_fit(self):
self.model.to(self.device)
def begin_batch(self):
self.run.xb = self.run.xb.to(device)
self.run.yb = self.run.yb.to(device)
torch.cuda.set_device(device)
#export
class CudaCallback(Callback):
def begin_fit(self):
self.model.cuda()
def begin_batch(self):
self.run.xb = self.run.xb.cuda()
self.run.yb = self.run.yb.cuda()
cbfs.append(CudaCallback)
model = get_cnn_model(data)
run, learn = create_runner(model, cbfs)
%%time
run.fit(3, learn)
```
## Refactor model
```
def conv2d(ni, nf, ks=3, stride=2):
return nn.Sequential(
nn.Conv2d(ni, nf, kernel_size=ks, padding=ks // 2, stride=stride), nn.ReLU()
)
#export
class BatchTransformXCallback(Callback):
_order=2
def __init__(self, tfm):
self.tfm = tfm
def begin_batch(self):
self.run.xb = self.tfm(self.xb)
def view_tfm(*size):
def _inner(x):
return x.view(*((-1,) + size))
return _inner
mnist_view = view_tfm(1, 28, 28)
cbfs.append(partial(BatchTransformXCallback, mnist_view))
nfs = [8, 16, 32, 32]
def get_cnn_layers(data, nfs):
nfs = [1] + nfs
return [
conv2d(nfs[i], nfs[i + 1], 5 if i == 0 else 3) for i in range(len(nfs) - 1)
] + [nn.AdaptiveAvgPool2d(1), Lambda(flatten), nn.Linear(nfs[-1], data.c)]
def get_cnn_model(data, nfs):
return nn.Sequential(*get_cnn_layers(data, nfs))
#export
def get_runner(model, data, lr=.6, cbs=None, opt_func=None, loss_func = F.cross_entropy):
if opt_func is None:
opt_func = optim.SGD
opt = opt_func(model.parameters(), lr=lr)
learn = Learner(model, opt, loss_func, data)
return learn, Runner(cb_funcs=listify(cbs))
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.4, cbs=cbfs)
model
run.fit(3, learn)
```
## Hooks
### Manual Insertion
```
class SequentialModel(nn.Module):
def __init__(self, *layers):
super().__init__()
self.layers = nn.ModuleList(layers)
self.act_means = [[] for _ in layers]
self.act_stds = [[] for _ in layers]
def __call__(self, x):
for i, l in enumerate(self.layers):
x = l(x)
self.act_means[i].append(x.data.mean())
self.act_stds[i].append(x.data.std())
return x
def __iter__(self):
return iter(self.layers)
model = SequentialModel(*get_cnn_layers(data, nfs))
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
run.fit(2, learn)
for l in model.act_means:
plt.plot(l)
plt.legend(range(6))
for l in model.act_stds:
plt.plot(l)
plt.legend(range(6))
for l in model.act_means:
plt.plot(l[:10])
plt.legend(range(6))
for l in model.act_stds:
plt.plot(l[:10])
plt.legend(range(6))
```
## Pytorch Hooks
```
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.5, cbs = cbfs)
act_means = [[] for _ in model]
act_stds = [[] for _ in model]
def append_stats(i, mod, inp, out):
act_means[i].append(out.data.mean())
act_stds[i].append(out.data.std())
for i,m in enumerate(model): m.register_forward_hook(partial(append_stats, i))
run.fit(1, learn)
for o in act_means:
plt.plot(o)
plt.legend(range(5));
for o in act_stds:
plt.plot(o)
plt.legend(range(5));
for l in act_means:
plt.plot(l[:40])
plt.legend(range(6))
```
## Hook Class
```
#export
def children(m):
return list(m.children())
class Hook():
def __init__(self, model, fun):
self.hook = model.register_forward_hook(partial(fun, self))
def remove(self):
self.hook.remove()
def __del__(self):
self.remove()
def append_stats(hook, mod, inp, out):
if not hasattr(hook, 'stats'):
hook.stats = ([], [])
means, stds = hook.stats
means.append(out.data.mean())
stds.append(out.data.std())
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.5, cbs = cbfs)
hooks = [Hook(l, append_stats) for l in children(model[:4])]
run.fit(1, learn)
for h in hooks:
plt.plot(h.stats[0])
h.remove()
plt.legend(range(4))
for h in hooks:
plt.plot(h.stats[1])
h.remove()
plt.legend(range(4))
```
### A Hooks class
```
#export
class ListContainer():
def __init__(self, items):
self.items = listify(items)
def __getitem__(self, idx):
if isinstance(idx, (int, slice)):
return self.items[idx]
if isinstance(idx[0], bool):
assert len(idx) == len(self.items)
return [o for m,o in zip(idx,self.items) if m]
return [self.items[i] for i in idx]
def __len__(self):
return len(self.items)
def __iter__(self):
return iter(self.items)
def __setitem__(self, i, o):
self.items[i] = o
def __delitem__(self, i):
del(self.items[i])
def __repr__(self):
res = f'{self.__class__.__name__} ({len(self)} items)\n{self.items[:10]}'
if len(self)>10: res = res[:-1]+ '...]'
return res
lc = ListContainer([1,2,3])
print(len(lc))
print(lc[:2])
print(lc[[True, False, True]])
lc[1] = 10
del lc[2]
for i in lc:
print(':', i)
lc
ListContainer(range(10))
#export
from torch.nn import init
class Hooks(ListContainer):
def __init__(self, layers, fun):
super().__init__([Hook(l, fun) for l in layers])
def __enter__(self, *args):
return self
def __exit__(self, *args):
self.remove()
def __del__(self, *args):
self.remove()
def __delitem__(self, i):
self[i].remove()
super().__delitem__(i)
def remove(self):
for h in self: h.remove()
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
hooks = Hooks(model, append_stats)
hooks
run.fit(1, learn)
for h in hooks:
plt.plot(h.stats[0])
h.remove()
plt.legend(range(4))
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
hooks = Hooks(model, append_stats)
hooks
model.cuda()
x,y = next(iter(data.train_dl))
x = mnist_resize(x).cuda()
x.mean(), x.std()
p = model[0](x)
p.mean(),p.std()
for l in model:
if isinstance(l, nn.Sequential):
init.kaiming_normal_(l[0].weight)
l[0].bias.data.zero_()
p = model[0](x)
p.mean(),p.std()
def plot_ms_ss(hooks, slice_):
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss = h.stats
ax1.plot(ms[slice_])
ax2.plot(ss[slice_])
plt.legend(range(6))
with Hooks(model, append_stats) as hooks:
run.fit(2, learn)
plot_ms_ss(hooks, slice(0, 10))
plot_ms_ss(hooks, slice(0, len(h.stats[0])))
```
### Other statistics
```
def append_stats_main(hook, mod, inp, outp, histc1, histc2, histc3):
if not hasattr(hook,'stats'):
hook.stats = ([],[],[])
means,stds,hists = hook.stats
means.append(outp.data.mean().cpu())
stds .append(outp.data.std().cpu())
hists.append(outp.data.cpu().histc(histc1,histc2,histc3)) #histc isn't implemented on the GPU
def append_stats(hook, mod, inp, outp):
append_stats_main(hook, mod, inp, outp, 40,0,10)
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
for l in model:
if isinstance(l, nn.Sequential):
init.kaiming_normal_(l[0].weight)
l[0].bias.data.zero_()
with Hooks(model, append_stats) as hooks:
run.fit(1, learn)
# Thanks to @ste for initial version of histgram plotting code
def get_hist(h):
return torch.stack(h.stats[2]).t().float().log1p()
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.imshow(get_hist(h), origin='lower')
ax.axis('off')
plt.tight_layout()
def get_min(h):
h1 = torch.stack(h.stats[2]).t().float()
return h1[:2].sum(0) / h1.sum(0)
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.plot(get_min(h))
ax.set_ylim(0,1)
plt.tight_layout()
```
## Generalized ReLU
Now let's use our model with a generalized ReLU that can be shifted and with maximum value.
```
#export
def get_cnn_layers(data, nfs, layer, **kwargs):
nfs = [1] + nfs
return [layer(nfs[i], nfs[i + 1], 5 if i == 0 else 3, **kwargs) for i in range(len(nfs)-1)] + [nn.AdaptiveAvgPool2d(1), Lambda(flatten), nn.Linear(nfs[-1], data.c)]
#export
def conv_layer(ni, nf, ks=3, stride=2, **kwargs):
return nn.Sequential(nn.Conv2d(ni, nf, ks, padding=ks//2, stride=stride), GeneralRelu(**kwargs))
#export
class GeneralRelu(nn.Module):
def __init__(self, leak=None, sub=None, maxv=None):
super().__init__()
self.leak,self.sub,self.maxv = leak,sub,maxv
def forward(self, x):
x = F.leaky_relu(x, self.leak) if self.leak is not None else F.relu(x)
if self.sub is not None:
x.sub_(self.sub)
if self.maxv is not None:
x.clamp_max_(self.maxv)
return x
#export
def init_cnn(m, uniform=False):
f = init.kaiming_uniform_ if uniform else init.kaiming_normal_
for l in m:
if isinstance(l, nn.Sequential):
f(l[0].weight, a=.1)
l[0].bias.data.zero_()
#export
def get_cnn_model(data, nfs, layer, **kwargs):
return nn.Sequential(*get_cnn_layers(data, nfs, layer, **kwargs))
def append_stats(hook, mod, inp, outp):
append_stats_main(hook, mod, inp, outp, 40,-7,7)
model = get_cnn_model(data, nfs, conv_layer, leak=0.1, sub=0.4, maxv=6.)
init_cnn(model)
learn,run = get_runner(model, data, lr=0.9, cbs=cbfs)
with Hooks(model, append_stats) as hooks:
run.fit(1, learn)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss, hi = h.stats
ax1.plot(ms[:10])
ax2.plot(ss[:10])
h.remove()
plt.legend(range(5))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss, hi = h.stats
ax1.plot(ms[:])
ax2.plot(ss[:])
h.remove()
plt.legend(range(5))
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.imshow(get_hist(h), origin='lower')
ax.axis('off')
plt.tight_layout()
def get_min(h):
h1 = torch.stack(h.stats[2]).t().float()
return h1[19:22].sum(0)/h1.sum(0)
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.plot(get_min(h))
ax.set_ylim(0,1)
plt.tight_layout()
#export
def get_learn_run(nfs, data, lr, layer, cbs=None, opt_func=None, uniform=False, **kwargs):
model = get_cnn_model(data, nfs, layer, **kwargs)
init_cnn(model, uniform=uniform)
return get_runner(model, data, lr=lr, cbs=cbs, opt_func=opt_func)
sched = combine_scheds([0.5, 0.5], [sched_cos(0.2, 1.), sched_cos(1., 0.1)])
learn,run = get_learn_run(nfs, data, 1., conv_layer, cbs=cbfs + [partial(ParamScheduler, 'lr', sched)])
run.fit(8, learn)
learn,run = get_learn_run(nfs, data, 1., conv_layer, uniform=True,
cbs=cbfs+[partial(ParamScheduler,'lr', sched)])
run.fit(8, learn)
#export
from IPython.display import display, Javascript
def nb_auto_export():
display(Javascript("""{
const ip = IPython.notebook
if (ip) {
ip.save_notebook()
console.log('a')
const s = `!python notebook2script.py ${ip.notebook_name}`
if (ip.kernel) { ip.kernel.execute(s) }
}
}"""))
nb_auto_export()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
#export
from exp.nb_05b import *
torch.set_num_threads(2)
x_train,y_train,x_valid,y_valid = get_data()
#export
def normalize_to(train, valid):
m,s = train.mean(),train.std()
return normalize(train, m, s), normalize(valid, m, s)
x_train, x_valid = normalize_to(x_train, x_valid)
train_ds, valid_ds = Dataset(x_train, y_train), Dataset(x_valid, y_valid)
x_train.mean(),x_train.std()
nh = 50
bs = 512
c = y_train.max().item() + 1
loss_func = F.cross_entropy
data = DataBunch(*get_dls(train_ds, valid_ds, bs), c)
#export
class Lambda(nn.Module):
def __init__(self, func):
super().__init__()
self.func = func
def forward(self, x):
return self.func(x)
def __repr__(self):
return f"Lambda({self.func})"
def flatten(x):
return x.view(x.shape[0], -1)
def mnist_resize(x):
return x.view(-1, 1, 28, 28)
def get_cnn_model(data):
return nn.Sequential(
Lambda(mnist_resize),
nn.Conv2d( 1, 8, 5, padding=2,stride=2), nn.ReLU(), #14
# nn.Conv2d( 1, 8, 3, padding=1,stride=2), nn.ReLU(), #14
nn.Conv2d( 8,16, 3, padding=1,stride=2), nn.ReLU(), # 7
nn.Conv2d(16,32, 3, padding=1,stride=2), nn.ReLU(), # 4
nn.Conv2d(32,32, 3, padding=1,stride=2), nn.ReLU(), # 2
nn.AdaptiveAvgPool2d(1),
Lambda(flatten),
nn.Linear(32,data.c)
)
all_entries = [i for i in iter(train_ds)]
img1 = all_entries[0][0]
model = get_cnn_model(data)
model
mods = [mod for mod in model.modules()]
x1 = mods[1](all_entries[0][0])
for i in range(len(mods) - 2):
x1 = mods[i + 2](x1)
print(i, mods[i + 2], x1.shape)
cbfs = [Recorder, partial(AvgStatsCallback, accuracy)]
def create_runner(model, cbfs):
opt = optim.SGD(model.parameters(), lr=0.4)
learn = Learner(model, opt, loss_func, data)
run = Runner(cb_funcs=cbfs)
return run, learn
run, learn = create_runner(model, cbfs)
%%time
run.fit(1, learn)
device = torch.device('cuda', 0)
class CudaCallback(Callback):
def __init__(self):
self.device = device
def begin_fit(self):
self.model.to(self.device)
def begin_batch(self):
self.run.xb = self.run.xb.to(device)
self.run.yb = self.run.yb.to(device)
torch.cuda.set_device(device)
#export
class CudaCallback(Callback):
def begin_fit(self):
self.model.cuda()
def begin_batch(self):
self.run.xb = self.run.xb.cuda()
self.run.yb = self.run.yb.cuda()
cbfs.append(CudaCallback)
model = get_cnn_model(data)
run, learn = create_runner(model, cbfs)
%%time
run.fit(3, learn)
def conv2d(ni, nf, ks=3, stride=2):
return nn.Sequential(
nn.Conv2d(ni, nf, kernel_size=ks, padding=ks // 2, stride=stride), nn.ReLU()
)
#export
class BatchTransformXCallback(Callback):
_order=2
def __init__(self, tfm):
self.tfm = tfm
def begin_batch(self):
self.run.xb = self.tfm(self.xb)
def view_tfm(*size):
def _inner(x):
return x.view(*((-1,) + size))
return _inner
mnist_view = view_tfm(1, 28, 28)
cbfs.append(partial(BatchTransformXCallback, mnist_view))
nfs = [8, 16, 32, 32]
def get_cnn_layers(data, nfs):
nfs = [1] + nfs
return [
conv2d(nfs[i], nfs[i + 1], 5 if i == 0 else 3) for i in range(len(nfs) - 1)
] + [nn.AdaptiveAvgPool2d(1), Lambda(flatten), nn.Linear(nfs[-1], data.c)]
def get_cnn_model(data, nfs):
return nn.Sequential(*get_cnn_layers(data, nfs))
#export
def get_runner(model, data, lr=.6, cbs=None, opt_func=None, loss_func = F.cross_entropy):
if opt_func is None:
opt_func = optim.SGD
opt = opt_func(model.parameters(), lr=lr)
learn = Learner(model, opt, loss_func, data)
return learn, Runner(cb_funcs=listify(cbs))
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.4, cbs=cbfs)
model
run.fit(3, learn)
class SequentialModel(nn.Module):
def __init__(self, *layers):
super().__init__()
self.layers = nn.ModuleList(layers)
self.act_means = [[] for _ in layers]
self.act_stds = [[] for _ in layers]
def __call__(self, x):
for i, l in enumerate(self.layers):
x = l(x)
self.act_means[i].append(x.data.mean())
self.act_stds[i].append(x.data.std())
return x
def __iter__(self):
return iter(self.layers)
model = SequentialModel(*get_cnn_layers(data, nfs))
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
run.fit(2, learn)
for l in model.act_means:
plt.plot(l)
plt.legend(range(6))
for l in model.act_stds:
plt.plot(l)
plt.legend(range(6))
for l in model.act_means:
plt.plot(l[:10])
plt.legend(range(6))
for l in model.act_stds:
plt.plot(l[:10])
plt.legend(range(6))
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.5, cbs = cbfs)
act_means = [[] for _ in model]
act_stds = [[] for _ in model]
def append_stats(i, mod, inp, out):
act_means[i].append(out.data.mean())
act_stds[i].append(out.data.std())
for i,m in enumerate(model): m.register_forward_hook(partial(append_stats, i))
run.fit(1, learn)
for o in act_means:
plt.plot(o)
plt.legend(range(5));
for o in act_stds:
plt.plot(o)
plt.legend(range(5));
for l in act_means:
plt.plot(l[:40])
plt.legend(range(6))
#export
def children(m):
return list(m.children())
class Hook():
def __init__(self, model, fun):
self.hook = model.register_forward_hook(partial(fun, self))
def remove(self):
self.hook.remove()
def __del__(self):
self.remove()
def append_stats(hook, mod, inp, out):
if not hasattr(hook, 'stats'):
hook.stats = ([], [])
means, stds = hook.stats
means.append(out.data.mean())
stds.append(out.data.std())
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.5, cbs = cbfs)
hooks = [Hook(l, append_stats) for l in children(model[:4])]
run.fit(1, learn)
for h in hooks:
plt.plot(h.stats[0])
h.remove()
plt.legend(range(4))
for h in hooks:
plt.plot(h.stats[1])
h.remove()
plt.legend(range(4))
#export
class ListContainer():
def __init__(self, items):
self.items = listify(items)
def __getitem__(self, idx):
if isinstance(idx, (int, slice)):
return self.items[idx]
if isinstance(idx[0], bool):
assert len(idx) == len(self.items)
return [o for m,o in zip(idx,self.items) if m]
return [self.items[i] for i in idx]
def __len__(self):
return len(self.items)
def __iter__(self):
return iter(self.items)
def __setitem__(self, i, o):
self.items[i] = o
def __delitem__(self, i):
del(self.items[i])
def __repr__(self):
res = f'{self.__class__.__name__} ({len(self)} items)\n{self.items[:10]}'
if len(self)>10: res = res[:-1]+ '...]'
return res
lc = ListContainer([1,2,3])
print(len(lc))
print(lc[:2])
print(lc[[True, False, True]])
lc[1] = 10
del lc[2]
for i in lc:
print(':', i)
lc
ListContainer(range(10))
#export
from torch.nn import init
class Hooks(ListContainer):
def __init__(self, layers, fun):
super().__init__([Hook(l, fun) for l in layers])
def __enter__(self, *args):
return self
def __exit__(self, *args):
self.remove()
def __del__(self, *args):
self.remove()
def __delitem__(self, i):
self[i].remove()
super().__delitem__(i)
def remove(self):
for h in self: h.remove()
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
hooks = Hooks(model, append_stats)
hooks
run.fit(1, learn)
for h in hooks:
plt.plot(h.stats[0])
h.remove()
plt.legend(range(4))
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
hooks = Hooks(model, append_stats)
hooks
model.cuda()
x,y = next(iter(data.train_dl))
x = mnist_resize(x).cuda()
x.mean(), x.std()
p = model[0](x)
p.mean(),p.std()
for l in model:
if isinstance(l, nn.Sequential):
init.kaiming_normal_(l[0].weight)
l[0].bias.data.zero_()
p = model[0](x)
p.mean(),p.std()
def plot_ms_ss(hooks, slice_):
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss = h.stats
ax1.plot(ms[slice_])
ax2.plot(ss[slice_])
plt.legend(range(6))
with Hooks(model, append_stats) as hooks:
run.fit(2, learn)
plot_ms_ss(hooks, slice(0, 10))
plot_ms_ss(hooks, slice(0, len(h.stats[0])))
def append_stats_main(hook, mod, inp, outp, histc1, histc2, histc3):
if not hasattr(hook,'stats'):
hook.stats = ([],[],[])
means,stds,hists = hook.stats
means.append(outp.data.mean().cpu())
stds .append(outp.data.std().cpu())
hists.append(outp.data.cpu().histc(histc1,histc2,histc3)) #histc isn't implemented on the GPU
def append_stats(hook, mod, inp, outp):
append_stats_main(hook, mod, inp, outp, 40,0,10)
model = get_cnn_model(data, nfs)
learn, run = get_runner(model, data, lr=.9, cbs = cbfs)
for l in model:
if isinstance(l, nn.Sequential):
init.kaiming_normal_(l[0].weight)
l[0].bias.data.zero_()
with Hooks(model, append_stats) as hooks:
run.fit(1, learn)
# Thanks to @ste for initial version of histgram plotting code
def get_hist(h):
return torch.stack(h.stats[2]).t().float().log1p()
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.imshow(get_hist(h), origin='lower')
ax.axis('off')
plt.tight_layout()
def get_min(h):
h1 = torch.stack(h.stats[2]).t().float()
return h1[:2].sum(0) / h1.sum(0)
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.plot(get_min(h))
ax.set_ylim(0,1)
plt.tight_layout()
#export
def get_cnn_layers(data, nfs, layer, **kwargs):
nfs = [1] + nfs
return [layer(nfs[i], nfs[i + 1], 5 if i == 0 else 3, **kwargs) for i in range(len(nfs)-1)] + [nn.AdaptiveAvgPool2d(1), Lambda(flatten), nn.Linear(nfs[-1], data.c)]
#export
def conv_layer(ni, nf, ks=3, stride=2, **kwargs):
return nn.Sequential(nn.Conv2d(ni, nf, ks, padding=ks//2, stride=stride), GeneralRelu(**kwargs))
#export
class GeneralRelu(nn.Module):
def __init__(self, leak=None, sub=None, maxv=None):
super().__init__()
self.leak,self.sub,self.maxv = leak,sub,maxv
def forward(self, x):
x = F.leaky_relu(x, self.leak) if self.leak is not None else F.relu(x)
if self.sub is not None:
x.sub_(self.sub)
if self.maxv is not None:
x.clamp_max_(self.maxv)
return x
#export
def init_cnn(m, uniform=False):
f = init.kaiming_uniform_ if uniform else init.kaiming_normal_
for l in m:
if isinstance(l, nn.Sequential):
f(l[0].weight, a=.1)
l[0].bias.data.zero_()
#export
def get_cnn_model(data, nfs, layer, **kwargs):
return nn.Sequential(*get_cnn_layers(data, nfs, layer, **kwargs))
def append_stats(hook, mod, inp, outp):
append_stats_main(hook, mod, inp, outp, 40,-7,7)
model = get_cnn_model(data, nfs, conv_layer, leak=0.1, sub=0.4, maxv=6.)
init_cnn(model)
learn,run = get_runner(model, data, lr=0.9, cbs=cbfs)
with Hooks(model, append_stats) as hooks:
run.fit(1, learn)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss, hi = h.stats
ax1.plot(ms[:10])
ax2.plot(ss[:10])
h.remove()
plt.legend(range(5))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
for h in hooks:
ms, ss, hi = h.stats
ax1.plot(ms[:])
ax2.plot(ss[:])
h.remove()
plt.legend(range(5))
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.imshow(get_hist(h), origin='lower')
ax.axis('off')
plt.tight_layout()
def get_min(h):
h1 = torch.stack(h.stats[2]).t().float()
return h1[19:22].sum(0)/h1.sum(0)
fig,axes = plt.subplots(2,2, figsize=(15,6))
for ax,h in zip(axes.flatten(), hooks[:4]):
ax.plot(get_min(h))
ax.set_ylim(0,1)
plt.tight_layout()
#export
def get_learn_run(nfs, data, lr, layer, cbs=None, opt_func=None, uniform=False, **kwargs):
model = get_cnn_model(data, nfs, layer, **kwargs)
init_cnn(model, uniform=uniform)
return get_runner(model, data, lr=lr, cbs=cbs, opt_func=opt_func)
sched = combine_scheds([0.5, 0.5], [sched_cos(0.2, 1.), sched_cos(1., 0.1)])
learn,run = get_learn_run(nfs, data, 1., conv_layer, cbs=cbfs + [partial(ParamScheduler, 'lr', sched)])
run.fit(8, learn)
learn,run = get_learn_run(nfs, data, 1., conv_layer, uniform=True,
cbs=cbfs+[partial(ParamScheduler,'lr', sched)])
run.fit(8, learn)
#export
from IPython.display import display, Javascript
def nb_auto_export():
display(Javascript("""{
const ip = IPython.notebook
if (ip) {
ip.save_notebook()
console.log('a')
const s = `!python notebook2script.py ${ip.notebook_name}`
if (ip.kernel) { ip.kernel.execute(s) }
}
}"""))
nb_auto_export()
| 0.802013 | 0.749958 |
# Introduction to the Azure ML SDK
Azure Machine Learning (*Azure ML*) is a cloud-based service for creating and managing machine learning solutions. It's designed to help data scientists leverage their existing data processing and model development skills and frameworks, and help them scale their workloads to the cloud. The Azure ML SDK for Python provides classes you can use to work with Azure ML in your Azure subscription.
## Check the Azure ML SDK Version
Let's start by importing the **azureml-core** package and checking the version of the SDK that is installed.
```
import azureml.core
print("Ready to use Azure ML", azureml.core.VERSION)
```
## Connect to Your Workspace
All experiments and associated resources are managed within you Azure ML workspace. You can connect to an existing workspace, or create a new one using the Azure ML SDK.
In most cases, you should store the workspace configuration in a JSON configuration file. This makes it easier to reconnect without needing to remember details like your Azure subscription ID. You can download the JSON configuration file from the blade for your workspace in the Azure portal, but if you're using a Compute Instance within your wokspace, the configuration file has already been downloaded to the root folder.
The code below uses the configuration file to connect to your workspace. The first time you run it in a notebook session, you'll be prompted to sign into Azure by clicking the https://microsoft.com/devicelogin link, entering an automatically generated code, and signing into Azure. After you have successfully signed in, you can close the browser tab that was opened and return to this notebook.
```
from azureml.core import Workspace
ws = Workspace.from_config()
print(ws.name, "loaded")
```
## View Azure ML Resources
Now that you have a connection to your workspace, you can view the resources it contains.
```
from azureml.core import ComputeTarget, Datastore, Dataset
print("Compute Targets:")
for compute_name in ws.compute_targets:
compute = ws.compute_targets[compute_name]
print("\t", compute.name, ':', compute.type)
print("Datastores:")
for datastore_name in ws.datastores:
datastore = Datastore.get(ws, datastore_name)
print("\t", datastore.name, ':', datastore.datastore_type)
print("Datasets:")
for dataset_name in list(ws.datasets.keys()):
dataset = Dataset.get_by_name(ws, dataset_name)
print("\t", dataset.name)
```
Now you've seen how to use the Azure ML SDK to view the resources in your workspace. The SDK provides a great way to script the creation and configuration of the resources you need to operate machine learning workloads using Azure ML. For more details, see the [Azure ML SDK documentation](https://docs.microsoft.com/python/api/overview/azure/ml/intro?view=azure-ml-py).
|
github_jupyter
|
import azureml.core
print("Ready to use Azure ML", azureml.core.VERSION)
from azureml.core import Workspace
ws = Workspace.from_config()
print(ws.name, "loaded")
from azureml.core import ComputeTarget, Datastore, Dataset
print("Compute Targets:")
for compute_name in ws.compute_targets:
compute = ws.compute_targets[compute_name]
print("\t", compute.name, ':', compute.type)
print("Datastores:")
for datastore_name in ws.datastores:
datastore = Datastore.get(ws, datastore_name)
print("\t", datastore.name, ':', datastore.datastore_type)
print("Datasets:")
for dataset_name in list(ws.datasets.keys()):
dataset = Dataset.get_by_name(ws, dataset_name)
print("\t", dataset.name)
| 0.313525 | 0.986376 |
```
# This notebook demonstrates fitting a PS by bspline
%pylab inline
import jax_cosmo as jc
import jax.numpy as np
import jax
# Here is a function that produces a power spectrum for provided
# params
k = np.logspace(-2.5,0.5,512)
@jax.jit
@jax.vmap
def get_ps(params):
# Retrieve cosmology
cosmo = jc.Cosmology(sigma8=params[0], Omega_c=params[1], Omega_b=params[2],
h=params[3], n_s=params[4], w0=params[5],
Omega_k=0., wa=0.)
k2 = k / cosmo.h
return k2*jc.power.linear_matter_power(cosmo,k2)
fid_params = np.array([0.801, 0.2545, 0.0485, 0.682, 0.971, -1])
pk = get_ps(fid_params.reshape((1,-1)))
plot(pk[0])
scale_pk = pk.max()
from functools import partial
class Bspline():
"""Numpy implementation of Cox - de Boor algorithm in 1D.
From here: https://github.com/johntfoster/bspline/blob/master/bspline/bspline.py
"""
def __init__(self, knot_vector, order):
"""Create a Bspline object.
Parameters:
knot_vector: Python list or rank-1 Numpy array containing knot vector
entries
order: Order of interpolation, e.g. 0 -> piecewise constant between
knots, 1 -> piecewise linear between knots, etc.
Returns:
Bspline object, callable to evaluate basis functions at given
values of `x` inside the knot span.
"""
kv = np.atleast_1d(knot_vector)
if kv.ndim > 1:
raise ValueError("knot_vector must be Python list or rank-1 array, but got rank = %d" % (kv.ndim))
self.knot_vector = kv
order = int(order)
if order < 0:
raise ValueError("order must be integer >= 0, but got %d" % (order))
self.p = order
def __basis0(self, xi):
"""Order zero basis (for internal use)."""
return np.where(np.all([self.knot_vector[:-1] <= xi,
xi < self.knot_vector[1:]],axis=0), 1.0, 0.0)
def __basis(self, xi, p, compute_derivatives=False):
"""Recursive Cox - de Boor function (for internal use).
Compute basis functions and optionally their first derivatives.
"""
if p == 0:
return self.__basis0(xi)
else:
basis_p_minus_1 = self.__basis(xi, p - 1)
first_term_numerator = xi - self.knot_vector[:-p]
first_term_denominator = self.knot_vector[p:] - self.knot_vector[:-p]
second_term_numerator = self.knot_vector[(p + 1):] - xi
second_term_denominator = (self.knot_vector[(p + 1):] -
self.knot_vector[1:-p])
#Change numerator in last recursion if derivatives are desired
if compute_derivatives and p == self.p:
first_term_numerator = p
second_term_numerator = -p
#Disable divide by zero error because we check for it
first_term = np.where(first_term_denominator != 0.0,
(first_term_numerator /
first_term_denominator), 0.0)
second_term = np.where(second_term_denominator != 0.0,
(second_term_numerator /
second_term_denominator), 0.0)
return (first_term[:-1] * basis_p_minus_1[:-1] +
second_term * basis_p_minus_1[1:])
def __call__(self, xi):
"""Convenience function to make the object callable. Also 'memoized' for speed."""
@jax.vmap
def fn(x):
return self.__basis(x, self.p, compute_derivatives=False)
return fn(xi)
def d(self, xi):
"""Convenience function to compute first derivative of basis functions. 'Memoized' for speed."""
@jax.vmap
def fn(x):
return self.__basis(x, self.p, compute_derivatives=True)
return fn(xi)
import flax
from flax import nn, optim
class emulator(nn.Module):
def apply(self, p, x):
net = nn.leaky_relu(nn.Dense(p, 256))
net = nn.leaky_relu(nn.Dense(net, 256))
net = nn.leaky_relu(nn.Dense(net, 256))
w = nn.Dense(net, 60)
k = nn.Dense(net, 64)
# make sure the knots sum to 1 and are in the interval 0,1
k = np.cumsum(nn.activation.softmax(k,axis=1), axis=1)
@jax.vmap
def eval_spline(a):
return Bspline(a, order=3)(x)
toto = eval_spline(k)
return np.einsum('bij,bj->bi', eval_spline(k), w)
x = np.linspace(0,1,512)
_, initial_params = emulator.init_by_shape(jax.random.PRNGKey(0),
[((1, 6,), np.float32), ((512,), np.float32)])
model = flax.nn.Model(emulator, initial_params)
model(fid_params.reshape((1,-1)), x).shape
plot(x, model(fid_params.reshape((1,-1)), x)[0])
# function that generates batches of params and pk from prior
import numpy as onp
batch_size=128
def get_batch():
om = onp.random.uniform(0.1,0.9, batch_size)
s8 = onp.random.uniform(0.4,1.0, batch_size)
ob = onp.random.uniform(0.03,0.07, batch_size)
h = onp.random.uniform(0.55,0.9, batch_size)
ns = onp.random.uniform(0.87,1.97, batch_size)
w0 = onp.random.uniform(-2.0,-0.33, batch_size)
p = np.stack([om,s8,ob,h,ns,w0],axis=1)
pk = get_ps(p)/scale_pk
return {'x':p, 'y':pk}
batch = get_batch()
batch['x'].shape, batch['y'].shape
model(batch['x'], x).shape
plot(x, model(batch['x'], x)[0])
plot(x, model(batch['x'], x)[1])
plot(x, model(batch['x'], x)[3])
plot(batch['y'][8])
plot(batch['y'][0])
plot(batch['y'][1])
plot(batch['y'][2])
plot(batch['y'][3])
plot(batch['y'][4])
@jax.jit
def train_step(optimizer, batch):
def loss_fn(model):
pred_y = model(batch['x'], x)
loss = np.sum(((pred_y - batch['y'])**2), axis=1).mean()
return loss
l, grad = jax.value_and_grad(loss_fn)(optimizer.target)
optimizer = optimizer.apply_gradient(grad)
return optimizer,l
# We also need an optimizer
optimizer = flax.optim.Momentum(learning_rate=0.001, beta=0.9).create(model)
#optimizer = flax.optim.Momentum(learning_rate=0.00002, beta=0.9).create(optimizer.target)
losses = []
for i in range(10000):
batch = get_batch()
optimizer, l = train_step(optimizer, batch)
losses.append(l)
if i%100 ==0:
print(l)
loglog(losses)
plot(x, optimizer.target(batch['x'], x)[0], label='b spline fit')
plot(x,batch['y'][0],'--', label='data')
legend()
xlim(0,1)
plot(x, optimizer.target(batch['x'], x)[1], label='b spline fit')
plot(x,batch['y'][1],'--', label='data')
plot(x, optimizer.target(batch['x'], x)[2], label='b spline fit')
plot(x,batch['y'][2],'--', label='data')
plot(batch['y'][0])
plot(batch['y'][8])
plot(x[100:-100], ((y-optimizer.target(x))/y)[100:-100])
xlim(0,1)
loglog(k,pk)
loglog(k,(optimizer.target(x)*y0.max() + y0.min()))
#xlim(0,0.6)
```
|
github_jupyter
|
# This notebook demonstrates fitting a PS by bspline
%pylab inline
import jax_cosmo as jc
import jax.numpy as np
import jax
# Here is a function that produces a power spectrum for provided
# params
k = np.logspace(-2.5,0.5,512)
@jax.jit
@jax.vmap
def get_ps(params):
# Retrieve cosmology
cosmo = jc.Cosmology(sigma8=params[0], Omega_c=params[1], Omega_b=params[2],
h=params[3], n_s=params[4], w0=params[5],
Omega_k=0., wa=0.)
k2 = k / cosmo.h
return k2*jc.power.linear_matter_power(cosmo,k2)
fid_params = np.array([0.801, 0.2545, 0.0485, 0.682, 0.971, -1])
pk = get_ps(fid_params.reshape((1,-1)))
plot(pk[0])
scale_pk = pk.max()
from functools import partial
class Bspline():
"""Numpy implementation of Cox - de Boor algorithm in 1D.
From here: https://github.com/johntfoster/bspline/blob/master/bspline/bspline.py
"""
def __init__(self, knot_vector, order):
"""Create a Bspline object.
Parameters:
knot_vector: Python list or rank-1 Numpy array containing knot vector
entries
order: Order of interpolation, e.g. 0 -> piecewise constant between
knots, 1 -> piecewise linear between knots, etc.
Returns:
Bspline object, callable to evaluate basis functions at given
values of `x` inside the knot span.
"""
kv = np.atleast_1d(knot_vector)
if kv.ndim > 1:
raise ValueError("knot_vector must be Python list or rank-1 array, but got rank = %d" % (kv.ndim))
self.knot_vector = kv
order = int(order)
if order < 0:
raise ValueError("order must be integer >= 0, but got %d" % (order))
self.p = order
def __basis0(self, xi):
"""Order zero basis (for internal use)."""
return np.where(np.all([self.knot_vector[:-1] <= xi,
xi < self.knot_vector[1:]],axis=0), 1.0, 0.0)
def __basis(self, xi, p, compute_derivatives=False):
"""Recursive Cox - de Boor function (for internal use).
Compute basis functions and optionally their first derivatives.
"""
if p == 0:
return self.__basis0(xi)
else:
basis_p_minus_1 = self.__basis(xi, p - 1)
first_term_numerator = xi - self.knot_vector[:-p]
first_term_denominator = self.knot_vector[p:] - self.knot_vector[:-p]
second_term_numerator = self.knot_vector[(p + 1):] - xi
second_term_denominator = (self.knot_vector[(p + 1):] -
self.knot_vector[1:-p])
#Change numerator in last recursion if derivatives are desired
if compute_derivatives and p == self.p:
first_term_numerator = p
second_term_numerator = -p
#Disable divide by zero error because we check for it
first_term = np.where(first_term_denominator != 0.0,
(first_term_numerator /
first_term_denominator), 0.0)
second_term = np.where(second_term_denominator != 0.0,
(second_term_numerator /
second_term_denominator), 0.0)
return (first_term[:-1] * basis_p_minus_1[:-1] +
second_term * basis_p_minus_1[1:])
def __call__(self, xi):
"""Convenience function to make the object callable. Also 'memoized' for speed."""
@jax.vmap
def fn(x):
return self.__basis(x, self.p, compute_derivatives=False)
return fn(xi)
def d(self, xi):
"""Convenience function to compute first derivative of basis functions. 'Memoized' for speed."""
@jax.vmap
def fn(x):
return self.__basis(x, self.p, compute_derivatives=True)
return fn(xi)
import flax
from flax import nn, optim
class emulator(nn.Module):
def apply(self, p, x):
net = nn.leaky_relu(nn.Dense(p, 256))
net = nn.leaky_relu(nn.Dense(net, 256))
net = nn.leaky_relu(nn.Dense(net, 256))
w = nn.Dense(net, 60)
k = nn.Dense(net, 64)
# make sure the knots sum to 1 and are in the interval 0,1
k = np.cumsum(nn.activation.softmax(k,axis=1), axis=1)
@jax.vmap
def eval_spline(a):
return Bspline(a, order=3)(x)
toto = eval_spline(k)
return np.einsum('bij,bj->bi', eval_spline(k), w)
x = np.linspace(0,1,512)
_, initial_params = emulator.init_by_shape(jax.random.PRNGKey(0),
[((1, 6,), np.float32), ((512,), np.float32)])
model = flax.nn.Model(emulator, initial_params)
model(fid_params.reshape((1,-1)), x).shape
plot(x, model(fid_params.reshape((1,-1)), x)[0])
# function that generates batches of params and pk from prior
import numpy as onp
batch_size=128
def get_batch():
om = onp.random.uniform(0.1,0.9, batch_size)
s8 = onp.random.uniform(0.4,1.0, batch_size)
ob = onp.random.uniform(0.03,0.07, batch_size)
h = onp.random.uniform(0.55,0.9, batch_size)
ns = onp.random.uniform(0.87,1.97, batch_size)
w0 = onp.random.uniform(-2.0,-0.33, batch_size)
p = np.stack([om,s8,ob,h,ns,w0],axis=1)
pk = get_ps(p)/scale_pk
return {'x':p, 'y':pk}
batch = get_batch()
batch['x'].shape, batch['y'].shape
model(batch['x'], x).shape
plot(x, model(batch['x'], x)[0])
plot(x, model(batch['x'], x)[1])
plot(x, model(batch['x'], x)[3])
plot(batch['y'][8])
plot(batch['y'][0])
plot(batch['y'][1])
plot(batch['y'][2])
plot(batch['y'][3])
plot(batch['y'][4])
@jax.jit
def train_step(optimizer, batch):
def loss_fn(model):
pred_y = model(batch['x'], x)
loss = np.sum(((pred_y - batch['y'])**2), axis=1).mean()
return loss
l, grad = jax.value_and_grad(loss_fn)(optimizer.target)
optimizer = optimizer.apply_gradient(grad)
return optimizer,l
# We also need an optimizer
optimizer = flax.optim.Momentum(learning_rate=0.001, beta=0.9).create(model)
#optimizer = flax.optim.Momentum(learning_rate=0.00002, beta=0.9).create(optimizer.target)
losses = []
for i in range(10000):
batch = get_batch()
optimizer, l = train_step(optimizer, batch)
losses.append(l)
if i%100 ==0:
print(l)
loglog(losses)
plot(x, optimizer.target(batch['x'], x)[0], label='b spline fit')
plot(x,batch['y'][0],'--', label='data')
legend()
xlim(0,1)
plot(x, optimizer.target(batch['x'], x)[1], label='b spline fit')
plot(x,batch['y'][1],'--', label='data')
plot(x, optimizer.target(batch['x'], x)[2], label='b spline fit')
plot(x,batch['y'][2],'--', label='data')
plot(batch['y'][0])
plot(batch['y'][8])
plot(x[100:-100], ((y-optimizer.target(x))/y)[100:-100])
xlim(0,1)
loglog(k,pk)
loglog(k,(optimizer.target(x)*y0.max() + y0.min()))
#xlim(0,0.6)
| 0.908496 | 0.677141 |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CLIENT CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter BigQuery Dataset Parameters
Create and permission a dataset in BigQuery.
1. Specify the name of the dataset.
1. If dataset exists, it is inchanged.
1. Add emails and / or groups to add read permission.
1. CAUTION: Removing permissions in StarThinker has no effect.
1. CAUTION: To remove permissions you have to edit the dataset.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'auth_write': 'service', # Credentials used for writing data.
'dataset_dataset': '', # Name of Google BigQuery dataset to create.
'dataset_emails': [], # Comma separated emails.
'dataset_groups': [], # Comma separated groups.
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute BigQuery Dataset
This does NOT need to be modified unless you are changing the recipe, click play.
```
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'auth': 'user',
'dataset': {'field': {'name': 'dataset_dataset','kind': 'string','order': 1,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'emails': {'field': {'name': 'dataset_emails','kind': 'string_list','order': 2,'default': [],'description': 'Comma separated emails.'}},
'groups': {'field': {'name': 'dataset_groups','kind': 'string_list','order': 3,'default': [],'description': 'Comma separated groups.'}}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
```
|
github_jupyter
|
!pip install git+https://github.com/google/starthinker
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
CLIENT_CREDENTIALS = 'PASTE CLIENT CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
FIELDS = {
'auth_write': 'service', # Credentials used for writing data.
'dataset_dataset': '', # Name of Google BigQuery dataset to create.
'dataset_emails': [], # Comma separated emails.
'dataset_groups': [], # Comma separated groups.
}
print("Parameters Set To: %s" % FIELDS)
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'auth': 'user',
'dataset': {'field': {'name': 'dataset_dataset','kind': 'string','order': 1,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'emails': {'field': {'name': 'dataset_emails','kind': 'string_list','order': 2,'default': [],'description': 'Comma separated emails.'}},
'groups': {'field': {'name': 'dataset_groups','kind': 'string_list','order': 3,'default': [],'description': 'Comma separated groups.'}}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
| 0.325628 | 0.735071 |
```
!pip install -q -r requirements.txt
```
## Import Libraries
```
import gc
import logging
import warnings
import itertools
import multiprocessing
import numpy as np
import pandas as pd
from sklearn.model_selection import GroupKFold
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
import pytorch_lightning as pl
from pytorch_lightning.utilities.seed import seed_everything
from transformers import (
BertForTokenClassification,
BertJapaneseTokenizer,
get_linear_schedule_with_warmup
)
warnings.filterwarnings('ignore')
logging.getLogger('pytorch_lightning').setLevel(logging.ERROR)
logging.getLogger('transformers').setLevel(logging.ERROR)
class Cfg:
debug = False
seed = 42
epochs = 5
lr = 1e-5
weight_decay = 1e-2
max_len = 193
n_folds = 5
num_entities = 8
train_batch_size = 32
val_batch_size = 256
group_col = 'curid'
label_col = 'label'
model_name = 'cl-tohoku/bert-base-japanese-whole-word-masking'
n_gpus = torch.cuda.device_count()
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
def read_data(config):
data = pd.read_csv(
'data/preprocessed_data.csv',
dtype={
'curid': object,
'text_body': object,
'text': object,
'start': np.int32,
'end': np.int32,
'label': np.int32
})
if config.debug:
return data.sample(1000, random_state=config.seed)
return data
def init_logger(file_path):
logger = logging.getLogger(__name__)
logger.setLevel(logging.INFO)
stream_handler = logging.StreamHandler()
stream_handler.setFormatter(logging.Formatter('%(message)s'))
file_handler = logging.FileHandler(filename=file_path)
file_handler.setFormatter(logging.Formatter('%(message)s'))
logger.addHandler(stream_handler)
logger.addHandler(file_handler)
return logger
```
## Create Dataset
```
class NERTokenizer(BertJapaneseTokenizer):
def bio_tagger(self, text, label, num_entities):
'''
IO法でラベリングされているテキストに、
トークナイズと合わせて、BIO法を適用する
'''
tokens = self.tokenize(text)
if label > 0:
labels = [label + num_entities] * len(tokens)
labels[0] = label
else:
labels = [0] * len(tokens)
return tokens, labels
def encode_plus_tagged(self, text, label, max_length, num_entities):
'''
トークナイズ結果に合わせてラベル付けをし、
エンコーディング
'''
token_arr, label_arr = [], []
tokens, labels = self.bio_tagger(text, label, num_entities)
token_arr.extend(tokens)
label_arr.extend(labels)
input_ids = self.convert_tokens_to_ids(token_arr)
encoded = self.prepare_for_model(input_ids,
max_length=max_length,
padding='max_length',
truncation=True)
# [CLS], [SEP], [PAD]のラベルを0として追加
label_arr = [0] + label_arr[:max_length-2] + [0]
encoded['labels'] = label_arr + [0] * (max_length - len(label_arr))
return encoded
def encode_plus_untagged(self, text_body, text, max_length):
'''
トークナイズとスパン取得を行い、
エンコーディング
'''
tokens, tokens_for_spans = [], []
words = self.word_tokenizer.tokenize(text)
for word in words:
subwords = self.subword_tokenizer.tokenize(word)
tokens.extend(subwords)
if subwords[0] == '[UNK]':
tokens_for_spans.append(word)
else:
tokens_for_spans.extend([subword.replace('##','') for subword in subwords])
pos = 0
spans = []
for token in tokens_for_spans:
token_len = len(token)
while True:
if token != text_body[pos:pos+token_len]:
pos += 1
else:
spans.append([pos, pos+token_len])
pos += token_len
break
input_ids = self.convert_tokens_to_ids(tokens)
encoded = self.prepare_for_model(input_ids,
max_length=max_length,
padding='max_length',
truncation=True)
# [CLS], [SEP], [PAD]に対応するスパン追加
n_seq = len(encoded['input_ids'])
spans = [[-1, -1]] + spans[:n_seq-2]
spans = spans + [[-1, -1]] * (n_seq - len(spans))
encoded = {k: torch.tensor([v]) for k, v in encoded.items()}
return encoded, spans
@staticmethod
def viterbi_optimizer(preds, num_entities, penalty=10000):
'''
BIO法のルールに従わない予測ラベル列に、
ペナルティを与えて、予測値を最適化する
'''
m = 2 * num_entities + 1
penalty_matrix = np.zeros([m,m])
for i in range(m):
for j in range(num_entities+1, m):
if not ((i == j) or (num_entities+i == j)):
penalty_matrix[i,j] = penalty
path = [[i] for i in range(m)]
preds_path = preds[0] - penalty_matrix[0,:]
preds = preds[1:]
for pred in preds:
assert len(pred) == 2 * num_entities + 1
pred_matrix = np.array(preds_path).reshape(-1,1) + np.array(pred).reshape(1,-1)
pred_matrix -= penalty_matrix
preds_path = pred_matrix.max(axis=0)
pred_argmax = pred_matrix.argmax(axis=0)
path = [path[idx]+[i] for i, idx in enumerate(pred_argmax)]
optimized_preds = path[np.argmax(preds_path)]
return optimized_preds
def convert_bert_output_to_entities(self, text_body, preds, spans, num_entities):
'''
同じラベルが連続するトークンをまとめて、
固有表現として抽出する
'''
assert len(spans) == len(preds)
# [CLS], [SEP], [PAD]に対応する箇所を削除
preds = [pred for pred, span in zip(preds, spans) if span[0] != -1]
spans = [span for span in spans if span[0] != -1]
preds = self.viterbi_optimizer(preds, num_entities)
entities = []
for pred, group in itertools.groupby(enumerate(preds), key=lambda x: x[1]):
group = list(group)
start = spans[group[0][0]][0]
end = spans[group[-1][0]][1]
if pred != 0:
# Bならば
if 1 <= pred <= num_entities:
entity = {
'name': text_body[start:end],
'span': [start, end],
'type_id': pred
}
entities.append(entity)
# Iならば
else:
entity['span'][1] = end
entity['name'] = text_body[entity['span'][0]:entity['span'][1]]
return entities
class NERDataset(Dataset):
def __init__(self, data, tokenizer, config):
super().__init__()
self.data = data
self.tokenizer = tokenizer
self.config = config
def __len__(self):
return len(self.data)
def __getitem__(self, index):
data_row = self.data.iloc[index]
text = data_row['text']
label = data_row['label']
encoded = self.tokenizer.encode_plus_tagged(
text, label, self.config.max_len, self.config.num_entities
)
encoded = {k: torch.tensor(v) for k, v in encoded.items()}
return {
'input_ids': encoded['input_ids'].flatten(),
'token_type_ids': encoded['token_type_ids'].flatten(),
'attention_mask': encoded['attention_mask'].flatten(),
'labels': encoded['labels'].flatten()
}
class NERDataModule(pl.LightningDataModule):
def __init__(self, train_data, val_data, tokenizer, config):
super().__init__()
self.train_data = train_data
self.val_data = val_data
self.tokenizer = tokenizer
self.config = config
def create_dataset(self, mode):
return (
NERDataset(self.train_data, self.tokenizer, self.config)
if mode == 'train'
else NERDataset(self.val_data, self.tokenizer, self.config)
)
def train_dataloader(self):
train_ds = self.create_dataset(mode='train')
train_loader = DataLoader(train_ds,
batch_size=self.config.train_batch_size,
num_workers=multiprocessing.cpu_count(),
pin_memory=True,
drop_last=True,
shuffle=True)
return train_loader
def val_dataloader(self):
val_ds = self.create_dataset(mode='val')
val_loader = DataLoader(val_ds,
batch_size=self.config.val_batch_size,
num_workers=multiprocessing.cpu_count(),
pin_memory=True,
drop_last=False,
shuffle=False)
return val_loader
```
## Create Model
```
class NERModel(pl.LightningModule):
def __init__(self, config, num_training_steps):
super().__init__()
self.config = config
self.num_training_steps = num_training_steps
self.bert = BertForTokenClassification.from_pretrained(
self.config.model_name,
num_labels=2 * self.config.num_entities + 1
)
def forward(self, input_ids, token_type_ids, attention_mask):
preds = self.bert(
input_ids,
token_type_ids=token_type_ids,
attention_mask=attention_mask
)
return preds
def training_step(self, batch, batch_idx):
output = self.bert(**batch)
loss = output.loss
self.log('train_loss', loss)
return loss
def validation_step(self, batch, batch_idx):
output = self.bert(**batch)
val_loss = output.loss
self.log('val_loss', val_loss)
def configure_optimizers(self):
no_decay = ['bias', 'LayerNorm.weight']
optimizer_grouped_parameters = [
{
'params': [p for n, p in self.bert.named_parameters()
if not any(nd in n for nd in no_decay)],
'weight_decay': self.config.weight_decay
},
{
'params': [p for n, p in self.bert.named_parameters()
if any(nd in n for nd in no_decay)],
'weight_decay': 0.0
}
]
optimizer = torch.optim.AdamW(optimizer_grouped_parameters, lr=self.config.lr)
scheduler = get_linear_schedule_with_warmup(
optimizer,
num_warmup_steps=0,
num_training_steps=self.num_training_steps
)
return [optimizer], [scheduler]
```
## FineTuning -> Inference -> Evaluation
```
def ner_inference(model, val_data, tokenizer, config):
all_entities, all_targets = [], []
n_val = len(val_data)
for i in range(n_val):
encoded, spans = tokenizer.encode_plus_untagged(
val_data.iloc[i]['text_body'],
val_data.iloc[i]['text'],
config.max_len
)
encoded = {k: v.to(config.device) for k, v in encoded.items()}
model.to(config.device)
model.eval()
with torch.no_grad():
output = model(**encoded)
preds = output.logits[0].cpu().detach().numpy().tolist()
entities = tokenizer.convert_bert_output_to_entities(
val_data.iloc[i]['text_body'],
preds,
spans,
config.num_entities
)
all_entities.append(entities)
# モデル評価の為に、ターゲットのエンティティを作成
target_name = val_data.iloc[i]['text']
target_span = [val_data.iloc[i]['start'], val_data.iloc[i]['end']]
target_typeId = val_data.iloc[i]['label']
if target_typeId == 0:
targets = []
else:
targets = [{'name': target_name, 'span': target_span, 'type_id': target_typeId}]
all_targets.append(targets)
return all_entities, all_targets
def ner_evaluation(entities_arr, targets_arr):
n_entities, n_targets, n_correct = 0, 0, 0
for entities, targets in zip(entities_arr, targets_arr):
get_span_type = lambda x: (x['span'][0], x['span'][1], x['type_id'])
set_entities = set(get_span_type(entity) for entity in entities)
set_targets = set(get_span_type(target) for target in targets)
n_targets += len(targets)
n_entities += len(entities)
n_correct += len(set_entities & set_targets)
precision = n_correct / n_entities
recall = n_correct / n_targets
f1 = 2 * precision * recall / (precision + recall)
return {'precision': precision, 'recall': recall, 'f1': f1}
def run_train(fold, tokenizer, data, tr_idx, val_idx, logger, config):
logger.info(f'\t----- Fold: {fold} -----')
train, val = data.iloc[tr_idx], data.iloc[val_idx]
checkpoint = pl.callbacks.ModelCheckpoint(monitor='val_loss',
mode='min',
save_top_k=1,
save_weights_only=True,
dirpath=f'model_folds_seed_{config.seed}/model_fold{fold}/')
es_callback = pl.callbacks.EarlyStopping(monitor='val_loss',
patience=1)
tb_logger = pl.loggers.TensorBoardLogger(f'model_folds_seed_{config.seed}/model_fold{fold}_logs/')
trainer = pl.Trainer(max_epochs=config.epochs,
gpus=config.n_gpus,
logger=tb_logger,
callbacks=[checkpoint,es_callback],
progress_bar_refresh_rate=0)
num_training_steps = ((len(train)) // (config.train_batch_size)) * float(config.epochs)
model = NERModel(config, num_training_steps)
datamodule = NERDataModule(train, val, tokenizer, config)
trainer.fit(model, datamodule=datamodule)
model.load_state_dict(torch.load(checkpoint.best_model_path)['state_dict'])
entities_arr, targets_arr = ner_inference(model, val, tokenizer, config)
del datamodule, model
gc.collect()
torch.cuda.empty_cache()
return entities_arr, targets_arr
```
## Metrics
#### Precision, Recall, F1
## CV
#### GroupKFold
`curid`をGroup IDとして、 バリデーション分割をする
```
def run_all(config):
seed_everything(config.seed, workers=True)
data = read_data(config)
gkf = GroupKFold(n_splits=config.n_folds)
tokenizer = NERTokenizer.from_pretrained(config.model_name)
logger = init_logger('cv_results/cv.log')
precision_score = 0.0
recall_score = 0.0
f1_score = 0.0
for i, (tr_idx, val_idx) in enumerate(gkf.split(data, data[config.label_col], data[config.group_col])):
entities_arr, targets_arr = run_train(i, tokenizer, data, tr_idx, val_idx, logger, config)
eval_result = ner_evaluation(entities_arr, targets_arr)
precision_score += eval_result['precision']
recall_score += eval_result['recall']
f1_score += eval_result['f1']
logger.info(f'FOLD{i} PRECISION SCORE: {eval_result["precision"]:.5f}')
logger.info(f'FOLD{i} RECALL SCORE: {eval_result["recall"]:.5f}')
logger.info(f'FOLD{i} F1 SCORE: {eval_result["f1"]:.5f}')
logger.info(f'{config.n_folds}FOLDS PRECISION CV SCORE: {precision_score/config.n_folds:.5f}')
logger.info(f'{config.n_folds}FOLDS RECALL CV SCORE: {recall_score/config.n_folds:.5f}')
logger.info(f'{config.n_folds}FOLDS F1 CV SCORE: {f1_score/config.n_folds:.5f}')
run_all(Cfg)
```
|
github_jupyter
|
!pip install -q -r requirements.txt
import gc
import logging
import warnings
import itertools
import multiprocessing
import numpy as np
import pandas as pd
from sklearn.model_selection import GroupKFold
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
import pytorch_lightning as pl
from pytorch_lightning.utilities.seed import seed_everything
from transformers import (
BertForTokenClassification,
BertJapaneseTokenizer,
get_linear_schedule_with_warmup
)
warnings.filterwarnings('ignore')
logging.getLogger('pytorch_lightning').setLevel(logging.ERROR)
logging.getLogger('transformers').setLevel(logging.ERROR)
class Cfg:
debug = False
seed = 42
epochs = 5
lr = 1e-5
weight_decay = 1e-2
max_len = 193
n_folds = 5
num_entities = 8
train_batch_size = 32
val_batch_size = 256
group_col = 'curid'
label_col = 'label'
model_name = 'cl-tohoku/bert-base-japanese-whole-word-masking'
n_gpus = torch.cuda.device_count()
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
def read_data(config):
data = pd.read_csv(
'data/preprocessed_data.csv',
dtype={
'curid': object,
'text_body': object,
'text': object,
'start': np.int32,
'end': np.int32,
'label': np.int32
})
if config.debug:
return data.sample(1000, random_state=config.seed)
return data
def init_logger(file_path):
logger = logging.getLogger(__name__)
logger.setLevel(logging.INFO)
stream_handler = logging.StreamHandler()
stream_handler.setFormatter(logging.Formatter('%(message)s'))
file_handler = logging.FileHandler(filename=file_path)
file_handler.setFormatter(logging.Formatter('%(message)s'))
logger.addHandler(stream_handler)
logger.addHandler(file_handler)
return logger
class NERTokenizer(BertJapaneseTokenizer):
def bio_tagger(self, text, label, num_entities):
'''
IO法でラベリングされているテキストに、
トークナイズと合わせて、BIO法を適用する
'''
tokens = self.tokenize(text)
if label > 0:
labels = [label + num_entities] * len(tokens)
labels[0] = label
else:
labels = [0] * len(tokens)
return tokens, labels
def encode_plus_tagged(self, text, label, max_length, num_entities):
'''
トークナイズ結果に合わせてラベル付けをし、
エンコーディング
'''
token_arr, label_arr = [], []
tokens, labels = self.bio_tagger(text, label, num_entities)
token_arr.extend(tokens)
label_arr.extend(labels)
input_ids = self.convert_tokens_to_ids(token_arr)
encoded = self.prepare_for_model(input_ids,
max_length=max_length,
padding='max_length',
truncation=True)
# [CLS], [SEP], [PAD]のラベルを0として追加
label_arr = [0] + label_arr[:max_length-2] + [0]
encoded['labels'] = label_arr + [0] * (max_length - len(label_arr))
return encoded
def encode_plus_untagged(self, text_body, text, max_length):
'''
トークナイズとスパン取得を行い、
エンコーディング
'''
tokens, tokens_for_spans = [], []
words = self.word_tokenizer.tokenize(text)
for word in words:
subwords = self.subword_tokenizer.tokenize(word)
tokens.extend(subwords)
if subwords[0] == '[UNK]':
tokens_for_spans.append(word)
else:
tokens_for_spans.extend([subword.replace('##','') for subword in subwords])
pos = 0
spans = []
for token in tokens_for_spans:
token_len = len(token)
while True:
if token != text_body[pos:pos+token_len]:
pos += 1
else:
spans.append([pos, pos+token_len])
pos += token_len
break
input_ids = self.convert_tokens_to_ids(tokens)
encoded = self.prepare_for_model(input_ids,
max_length=max_length,
padding='max_length',
truncation=True)
# [CLS], [SEP], [PAD]に対応するスパン追加
n_seq = len(encoded['input_ids'])
spans = [[-1, -1]] + spans[:n_seq-2]
spans = spans + [[-1, -1]] * (n_seq - len(spans))
encoded = {k: torch.tensor([v]) for k, v in encoded.items()}
return encoded, spans
@staticmethod
def viterbi_optimizer(preds, num_entities, penalty=10000):
'''
BIO法のルールに従わない予測ラベル列に、
ペナルティを与えて、予測値を最適化する
'''
m = 2 * num_entities + 1
penalty_matrix = np.zeros([m,m])
for i in range(m):
for j in range(num_entities+1, m):
if not ((i == j) or (num_entities+i == j)):
penalty_matrix[i,j] = penalty
path = [[i] for i in range(m)]
preds_path = preds[0] - penalty_matrix[0,:]
preds = preds[1:]
for pred in preds:
assert len(pred) == 2 * num_entities + 1
pred_matrix = np.array(preds_path).reshape(-1,1) + np.array(pred).reshape(1,-1)
pred_matrix -= penalty_matrix
preds_path = pred_matrix.max(axis=0)
pred_argmax = pred_matrix.argmax(axis=0)
path = [path[idx]+[i] for i, idx in enumerate(pred_argmax)]
optimized_preds = path[np.argmax(preds_path)]
return optimized_preds
def convert_bert_output_to_entities(self, text_body, preds, spans, num_entities):
'''
同じラベルが連続するトークンをまとめて、
固有表現として抽出する
'''
assert len(spans) == len(preds)
# [CLS], [SEP], [PAD]に対応する箇所を削除
preds = [pred for pred, span in zip(preds, spans) if span[0] != -1]
spans = [span for span in spans if span[0] != -1]
preds = self.viterbi_optimizer(preds, num_entities)
entities = []
for pred, group in itertools.groupby(enumerate(preds), key=lambda x: x[1]):
group = list(group)
start = spans[group[0][0]][0]
end = spans[group[-1][0]][1]
if pred != 0:
# Bならば
if 1 <= pred <= num_entities:
entity = {
'name': text_body[start:end],
'span': [start, end],
'type_id': pred
}
entities.append(entity)
# Iならば
else:
entity['span'][1] = end
entity['name'] = text_body[entity['span'][0]:entity['span'][1]]
return entities
class NERDataset(Dataset):
def __init__(self, data, tokenizer, config):
super().__init__()
self.data = data
self.tokenizer = tokenizer
self.config = config
def __len__(self):
return len(self.data)
def __getitem__(self, index):
data_row = self.data.iloc[index]
text = data_row['text']
label = data_row['label']
encoded = self.tokenizer.encode_plus_tagged(
text, label, self.config.max_len, self.config.num_entities
)
encoded = {k: torch.tensor(v) for k, v in encoded.items()}
return {
'input_ids': encoded['input_ids'].flatten(),
'token_type_ids': encoded['token_type_ids'].flatten(),
'attention_mask': encoded['attention_mask'].flatten(),
'labels': encoded['labels'].flatten()
}
class NERDataModule(pl.LightningDataModule):
def __init__(self, train_data, val_data, tokenizer, config):
super().__init__()
self.train_data = train_data
self.val_data = val_data
self.tokenizer = tokenizer
self.config = config
def create_dataset(self, mode):
return (
NERDataset(self.train_data, self.tokenizer, self.config)
if mode == 'train'
else NERDataset(self.val_data, self.tokenizer, self.config)
)
def train_dataloader(self):
train_ds = self.create_dataset(mode='train')
train_loader = DataLoader(train_ds,
batch_size=self.config.train_batch_size,
num_workers=multiprocessing.cpu_count(),
pin_memory=True,
drop_last=True,
shuffle=True)
return train_loader
def val_dataloader(self):
val_ds = self.create_dataset(mode='val')
val_loader = DataLoader(val_ds,
batch_size=self.config.val_batch_size,
num_workers=multiprocessing.cpu_count(),
pin_memory=True,
drop_last=False,
shuffle=False)
return val_loader
class NERModel(pl.LightningModule):
def __init__(self, config, num_training_steps):
super().__init__()
self.config = config
self.num_training_steps = num_training_steps
self.bert = BertForTokenClassification.from_pretrained(
self.config.model_name,
num_labels=2 * self.config.num_entities + 1
)
def forward(self, input_ids, token_type_ids, attention_mask):
preds = self.bert(
input_ids,
token_type_ids=token_type_ids,
attention_mask=attention_mask
)
return preds
def training_step(self, batch, batch_idx):
output = self.bert(**batch)
loss = output.loss
self.log('train_loss', loss)
return loss
def validation_step(self, batch, batch_idx):
output = self.bert(**batch)
val_loss = output.loss
self.log('val_loss', val_loss)
def configure_optimizers(self):
no_decay = ['bias', 'LayerNorm.weight']
optimizer_grouped_parameters = [
{
'params': [p for n, p in self.bert.named_parameters()
if not any(nd in n for nd in no_decay)],
'weight_decay': self.config.weight_decay
},
{
'params': [p for n, p in self.bert.named_parameters()
if any(nd in n for nd in no_decay)],
'weight_decay': 0.0
}
]
optimizer = torch.optim.AdamW(optimizer_grouped_parameters, lr=self.config.lr)
scheduler = get_linear_schedule_with_warmup(
optimizer,
num_warmup_steps=0,
num_training_steps=self.num_training_steps
)
return [optimizer], [scheduler]
def ner_inference(model, val_data, tokenizer, config):
all_entities, all_targets = [], []
n_val = len(val_data)
for i in range(n_val):
encoded, spans = tokenizer.encode_plus_untagged(
val_data.iloc[i]['text_body'],
val_data.iloc[i]['text'],
config.max_len
)
encoded = {k: v.to(config.device) for k, v in encoded.items()}
model.to(config.device)
model.eval()
with torch.no_grad():
output = model(**encoded)
preds = output.logits[0].cpu().detach().numpy().tolist()
entities = tokenizer.convert_bert_output_to_entities(
val_data.iloc[i]['text_body'],
preds,
spans,
config.num_entities
)
all_entities.append(entities)
# モデル評価の為に、ターゲットのエンティティを作成
target_name = val_data.iloc[i]['text']
target_span = [val_data.iloc[i]['start'], val_data.iloc[i]['end']]
target_typeId = val_data.iloc[i]['label']
if target_typeId == 0:
targets = []
else:
targets = [{'name': target_name, 'span': target_span, 'type_id': target_typeId}]
all_targets.append(targets)
return all_entities, all_targets
def ner_evaluation(entities_arr, targets_arr):
n_entities, n_targets, n_correct = 0, 0, 0
for entities, targets in zip(entities_arr, targets_arr):
get_span_type = lambda x: (x['span'][0], x['span'][1], x['type_id'])
set_entities = set(get_span_type(entity) for entity in entities)
set_targets = set(get_span_type(target) for target in targets)
n_targets += len(targets)
n_entities += len(entities)
n_correct += len(set_entities & set_targets)
precision = n_correct / n_entities
recall = n_correct / n_targets
f1 = 2 * precision * recall / (precision + recall)
return {'precision': precision, 'recall': recall, 'f1': f1}
def run_train(fold, tokenizer, data, tr_idx, val_idx, logger, config):
logger.info(f'\t----- Fold: {fold} -----')
train, val = data.iloc[tr_idx], data.iloc[val_idx]
checkpoint = pl.callbacks.ModelCheckpoint(monitor='val_loss',
mode='min',
save_top_k=1,
save_weights_only=True,
dirpath=f'model_folds_seed_{config.seed}/model_fold{fold}/')
es_callback = pl.callbacks.EarlyStopping(monitor='val_loss',
patience=1)
tb_logger = pl.loggers.TensorBoardLogger(f'model_folds_seed_{config.seed}/model_fold{fold}_logs/')
trainer = pl.Trainer(max_epochs=config.epochs,
gpus=config.n_gpus,
logger=tb_logger,
callbacks=[checkpoint,es_callback],
progress_bar_refresh_rate=0)
num_training_steps = ((len(train)) // (config.train_batch_size)) * float(config.epochs)
model = NERModel(config, num_training_steps)
datamodule = NERDataModule(train, val, tokenizer, config)
trainer.fit(model, datamodule=datamodule)
model.load_state_dict(torch.load(checkpoint.best_model_path)['state_dict'])
entities_arr, targets_arr = ner_inference(model, val, tokenizer, config)
del datamodule, model
gc.collect()
torch.cuda.empty_cache()
return entities_arr, targets_arr
def run_all(config):
seed_everything(config.seed, workers=True)
data = read_data(config)
gkf = GroupKFold(n_splits=config.n_folds)
tokenizer = NERTokenizer.from_pretrained(config.model_name)
logger = init_logger('cv_results/cv.log')
precision_score = 0.0
recall_score = 0.0
f1_score = 0.0
for i, (tr_idx, val_idx) in enumerate(gkf.split(data, data[config.label_col], data[config.group_col])):
entities_arr, targets_arr = run_train(i, tokenizer, data, tr_idx, val_idx, logger, config)
eval_result = ner_evaluation(entities_arr, targets_arr)
precision_score += eval_result['precision']
recall_score += eval_result['recall']
f1_score += eval_result['f1']
logger.info(f'FOLD{i} PRECISION SCORE: {eval_result["precision"]:.5f}')
logger.info(f'FOLD{i} RECALL SCORE: {eval_result["recall"]:.5f}')
logger.info(f'FOLD{i} F1 SCORE: {eval_result["f1"]:.5f}')
logger.info(f'{config.n_folds}FOLDS PRECISION CV SCORE: {precision_score/config.n_folds:.5f}')
logger.info(f'{config.n_folds}FOLDS RECALL CV SCORE: {recall_score/config.n_folds:.5f}')
logger.info(f'{config.n_folds}FOLDS F1 CV SCORE: {f1_score/config.n_folds:.5f}')
run_all(Cfg)
| 0.541166 | 0.583144 |
```
# Задание на повторение материала предыдущего семестра
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet, LogisticRegression
from sklearn.neighbors import KNeighborsRegressor, KNeighborsClassifier
from sklearn.naive_bayes import MultinomialNB
from sklearn.cluster import KMeans
from sklearn.metrics import mean_squared_error, f1_score, silhouette_score
# Генерируем уникальный seed
my_code = "Johnson"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Данные загружены отсюда: https://www.kaggle.com/dwdkills/russian-demography
# Читаем данные из файла
example_data = pd.read_csv("datasets/russian_demography.csv")
# "year" - год (1990-2017)
# "region" - название региона
# "npg" - естественный прирост населения на 1000 человек
# "birth_rate" - количество рождений на 1000 человек
# "death_rate" - количество смертей на 1000 человек
# "gdw" - коэффициент демографической нагрузки на 100 человек (Отношение числа нетрудоспособных к числу трудоспособных).
# "urbanization" - процент городского населения
example_data.head()
# Так как список регионов меняется от года к году, в данных есть строки без значений. Удалим их
example_data.dropna(inplace=True)
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(example_data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(example_data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
columns_to_scale = ['year', 'npg', 'birth_rate', 'death_rate', 'gdw', 'urbanization']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), columns_to_scale)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_test = pd.DataFrame(ct.transform(test))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = columns_to_scale + ['region']
sc_train.columns = column_names
sc_test.columns = column_names
sc_val.columns = column_names
sc_train
# Вспоминаем алгоритмы решения задачи регрессии: линейную регрессию и метод k ближайших соседей
r_models = []
# Линейная регрессия
# Для использования регуляризации, вместо LinearRegression используем Lasso, Ridge или ElasticNet
# Параметр alpha - коэффициент регуляризации для Lasso и Ridge, по умолчанию равен 1
# Для ElasticNet, если регуляризация иммет вид a*L1+b*L2, то
# параметр alpha = a + b, по умолчанию равен 1
# параметр l1_ratio = a / (a + b), по умолчанию равен 0.5
r_models.append(LinearRegression())
r_models.append(Lasso(alpha=1.0))
r_models.append(Ridge(alpha=1.0))
r_models.append(ElasticNet(alpha=1.0, l1_ratio=0.5))
# K ближайших соседей
# Параметр n_neighbors - количество соседей, по умолчания равен 5
r_models.append(KNeighborsRegressor(n_neighbors=5))
r_models.append(KNeighborsRegressor(n_neighbors=10))
r_models.append(KNeighborsRegressor(n_neighbors=15))
# Выделим предикторы и зависимую переменную
x_labels = column_names[0:-2]
y_labels = ['urbanization']
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_labels]
y_test = sc_test[y_labels]
y_val = sc_val[y_labels]
# Обучаем модели
for model in r_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
mses = []
for model in r_models:
val_pred = model.predict(x_val)
mse = mean_squared_error(y_val, val_pred)
mses.append(mse)
print(mse)
# Выбираем лучшую модель
i_min = mses.index(min(mses))
best_r_model = r_models[i_min]
best_r_model
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_r_model.predict(x_test)
mse = mean_squared_error(y_test, test_pred)
print(mse)
# Вспоминаем алгоритмы решения задачи классификации:
# логистическую регрессию, наивный байесовский классификатор и (снова) метод k ближайших соседей
c_models = []
# Логистическая регрессия
# Параметр penalty - тип регуляризации: 'l1', 'l2', 'elasticnet', 'none'}, по умолчанию 'l2'
# Для некоторых типов регуляризации доступны не все алгоритмы (параметр solver)
# Для elasticnet регуляризации необходимо уазывать параметр l1_ratio (0 - l2, 1 - l1)
c_models.append(LogisticRegression(penalty='none', solver='saga'))
c_models.append(LogisticRegression(penalty='l1', solver='saga'))
c_models.append(LogisticRegression(penalty='l2', solver='saga'))
c_models.append(LogisticRegression(penalty='elasticnet', l1_ratio=0.5, solver='saga'))
c_models.append(LogisticRegression())
# Наивный байесовский классификатор
# Параметр alpha - параметр сглаживания, по умолчанию равен 1 (сглаживание Лапласа)
c_models.append(MultinomialNB(alpha=0.0))
c_models.append(MultinomialNB(alpha=0.5))
c_models.append(MultinomialNB(alpha=1.0))
# K ближайших соседей
# Параметр n_neighbors - количество соседей, по умолчания равен 5
c_models.append(KNeighborsClassifier(n_neighbors=5))
c_models.append(KNeighborsClassifier(n_neighbors=10))
c_models.append(KNeighborsClassifier(n_neighbors=15))
# Выделим предикторы и метки классов
x_labels = column_names[0:-1]
y_labels = ['region']
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = np.ravel(sc_train[y_labels])
y_test = np.ravel(sc_test[y_labels])
y_val = np.ravel(sc_val[y_labels])
# Обучаем модели
for model in c_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
f1s = []
for model in c_models:
val_pred = model.predict(x_val)
f1 = f1_score(y_val, val_pred, average='weighted')
f1s.append(f1)
print(f1)
# Выбираем лучшую модель
i_min = f1s.index(min(f1s))
best_c_model = c_models[i_min]
best_c_model
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_c_model.predict(x_test)
f1 = f1_score(y_test, test_pred, average='weighted')
print(f1)
# Вспоминаем алгоритм решения задачи кластеризации - метод k-средних
# Параметр n_clusters - количество кластеров, по умолчанию равен 8
k_models = []
k_models.append(KMeans(n_clusters=5))
k_models.append(KMeans(n_clusters=8))
k_models.append(KMeans(n_clusters=20))
k_models.append(KMeans(n_clusters=50))
# Выделим используемые параметры
x_labels = column_names[0:-1]
x = pd.concat([sc_train[x_labels], sc_val[x_labels], sc_test[x_labels]])
x
# Произведем кластеризацию
for model in k_models:
model.fit(x)
# Оценим качество результата
sils = []
for model in k_models:
cluster_labels = model.predict(x)
s = silhouette_score(x, cluster_labels)
sils.append(s)
print(s)
# Выбираем лучшую модель
i_min = sils.index(min(sils))
best_k_model = k_models[i_min]
print(best_k_model)
print(sils[i_min])
# Задание №1 - анализ моделей для задачи регрессии
# Общий список моделей
r_models = [
LinearRegression(),
Lasso(alpha=1.0),
Lasso(alpha=0.5),
Ridge(alpha=1.0),
Ridge(alpha=0.5),
ElasticNet(alpha=1.0, l1_ratio=0.5),
ElasticNet(alpha=1.0, l1_ratio=0.25),
ElasticNet(alpha=1.0, l1_ratio=0.75),
ElasticNet(alpha=0.5, l1_ratio=0.5),
ElasticNet(alpha=0.5, l1_ratio=0.25),
ElasticNet(alpha=0.5, l1_ratio=0.75),
KNeighborsRegressor(n_neighbors=5),
KNeighborsRegressor(n_neighbors=10),
KNeighborsRegressor(n_neighbors=15),
KNeighborsRegressor(n_neighbors=20),
KNeighborsRegressor(n_neighbors=25)
]
# Выбор моделей для задания
n = 4
random.seed(my_seed)
my_models1 = random.sample(r_models, n)
print(my_models1)
# Загрузим данные для задачи регрессии
data = pd.read_csv("datasets/weather.csv")
data
# Зависимая переменная для всех одна и та же. Предикторы выбираем случайнм образом.
columns = list(data.columns)
n_x = 5
y_label = 'water_level'
x_labels = random.sample(columns[1:], n_x)
print(x_labels)
# Преобразуйте значения всех необходимых параметров к отрезку [0,1].
# Решите получившуюся задачу регрессии с помощью выбранных моделей и сравните их эффективность.
# Укажите, какая модель решает задачу лучше других.
# Задание №2 - анализ моделей для задачи классификации
# Общий список моделей
c_models = [
LogisticRegression(penalty='none', solver='saga'),
LogisticRegression(penalty='l1', solver='saga'),
LogisticRegression(penalty='l2', solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.25, solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.5, solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.75, solver='saga'),
LogisticRegression(),
MultinomialNB(alpha=0.0),
MultinomialNB(alpha=0.25),
MultinomialNB(alpha=0.5),
MultinomialNB(alpha=0.75),
MultinomialNB(alpha=1.0),
KNeighborsClassifier(n_neighbors=5),
KNeighborsClassifier(n_neighbors=10),
KNeighborsClassifier(n_neighbors=15),
KNeighborsClassifier(n_neighbors=20),
KNeighborsClassifier(n_neighbors=25)
]
# Выбор моделей для задания
n = 5
my_models2 = random.sample(c_models, n)
print(my_models2)
# Загрузим данные для задачи классификации
data = pd.read_csv("datasets/zoo2.csv")
data
# Метка класса для всех одна и та же. Параметры выбираем случайнм образом.
columns = list(data.columns)
n_x = 8
y_label = 'class_type'
x_labels = random.sample(columns[:-1], n_x)
print(x_labels)
# Преобразуйте значения всех необходимых параметров к отрезку [0,1].
# Решите получившуюся задачу классификации с помощью выбранных моделей и сравните их эффективность.
# Укажите, какая модель решает задачу лучше других.
```
|
github_jupyter
|
# Задание на повторение материала предыдущего семестра
# Зависимости
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.compose import ColumnTransformer
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet, LogisticRegression
from sklearn.neighbors import KNeighborsRegressor, KNeighborsClassifier
from sklearn.naive_bayes import MultinomialNB
from sklearn.cluster import KMeans
from sklearn.metrics import mean_squared_error, f1_score, silhouette_score
# Генерируем уникальный seed
my_code = "Johnson"
seed_limit = 2 ** 32
my_seed = int.from_bytes(my_code.encode(), "little") % seed_limit
# Данные загружены отсюда: https://www.kaggle.com/dwdkills/russian-demography
# Читаем данные из файла
example_data = pd.read_csv("datasets/russian_demography.csv")
# "year" - год (1990-2017)
# "region" - название региона
# "npg" - естественный прирост населения на 1000 человек
# "birth_rate" - количество рождений на 1000 человек
# "death_rate" - количество смертей на 1000 человек
# "gdw" - коэффициент демографической нагрузки на 100 человек (Отношение числа нетрудоспособных к числу трудоспособных).
# "urbanization" - процент городского населения
example_data.head()
# Так как список регионов меняется от года к году, в данных есть строки без значений. Удалим их
example_data.dropna(inplace=True)
# Определим размер валидационной и тестовой выборок
val_test_size = round(0.2*len(example_data))
print(val_test_size)
# Создадим обучающую, валидационную и тестовую выборки
random_state = my_seed
train_val, test = train_test_split(example_data, test_size=val_test_size, random_state=random_state)
train, val = train_test_split(train_val, test_size=val_test_size, random_state=random_state)
print(len(train), len(val), len(test))
# Значения в числовых столбцах преобразуем к отрезку [0,1].
# Для настройки скалировщика используем только обучающую выборку.
columns_to_scale = ['year', 'npg', 'birth_rate', 'death_rate', 'gdw', 'urbanization']
ct = ColumnTransformer(transformers=[('numerical', MinMaxScaler(), columns_to_scale)], remainder='passthrough')
ct.fit(train)
# Преобразуем значения, тип данных приводим к DataFrame
sc_train = pd.DataFrame(ct.transform(train))
sc_test = pd.DataFrame(ct.transform(test))
sc_val = pd.DataFrame(ct.transform(val))
# Устанавливаем названия столбцов
column_names = columns_to_scale + ['region']
sc_train.columns = column_names
sc_test.columns = column_names
sc_val.columns = column_names
sc_train
# Вспоминаем алгоритмы решения задачи регрессии: линейную регрессию и метод k ближайших соседей
r_models = []
# Линейная регрессия
# Для использования регуляризации, вместо LinearRegression используем Lasso, Ridge или ElasticNet
# Параметр alpha - коэффициент регуляризации для Lasso и Ridge, по умолчанию равен 1
# Для ElasticNet, если регуляризация иммет вид a*L1+b*L2, то
# параметр alpha = a + b, по умолчанию равен 1
# параметр l1_ratio = a / (a + b), по умолчанию равен 0.5
r_models.append(LinearRegression())
r_models.append(Lasso(alpha=1.0))
r_models.append(Ridge(alpha=1.0))
r_models.append(ElasticNet(alpha=1.0, l1_ratio=0.5))
# K ближайших соседей
# Параметр n_neighbors - количество соседей, по умолчания равен 5
r_models.append(KNeighborsRegressor(n_neighbors=5))
r_models.append(KNeighborsRegressor(n_neighbors=10))
r_models.append(KNeighborsRegressor(n_neighbors=15))
# Выделим предикторы и зависимую переменную
x_labels = column_names[0:-2]
y_labels = ['urbanization']
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = sc_train[y_labels]
y_test = sc_test[y_labels]
y_val = sc_val[y_labels]
# Обучаем модели
for model in r_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
mses = []
for model in r_models:
val_pred = model.predict(x_val)
mse = mean_squared_error(y_val, val_pred)
mses.append(mse)
print(mse)
# Выбираем лучшую модель
i_min = mses.index(min(mses))
best_r_model = r_models[i_min]
best_r_model
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_r_model.predict(x_test)
mse = mean_squared_error(y_test, test_pred)
print(mse)
# Вспоминаем алгоритмы решения задачи классификации:
# логистическую регрессию, наивный байесовский классификатор и (снова) метод k ближайших соседей
c_models = []
# Логистическая регрессия
# Параметр penalty - тип регуляризации: 'l1', 'l2', 'elasticnet', 'none'}, по умолчанию 'l2'
# Для некоторых типов регуляризации доступны не все алгоритмы (параметр solver)
# Для elasticnet регуляризации необходимо уазывать параметр l1_ratio (0 - l2, 1 - l1)
c_models.append(LogisticRegression(penalty='none', solver='saga'))
c_models.append(LogisticRegression(penalty='l1', solver='saga'))
c_models.append(LogisticRegression(penalty='l2', solver='saga'))
c_models.append(LogisticRegression(penalty='elasticnet', l1_ratio=0.5, solver='saga'))
c_models.append(LogisticRegression())
# Наивный байесовский классификатор
# Параметр alpha - параметр сглаживания, по умолчанию равен 1 (сглаживание Лапласа)
c_models.append(MultinomialNB(alpha=0.0))
c_models.append(MultinomialNB(alpha=0.5))
c_models.append(MultinomialNB(alpha=1.0))
# K ближайших соседей
# Параметр n_neighbors - количество соседей, по умолчания равен 5
c_models.append(KNeighborsClassifier(n_neighbors=5))
c_models.append(KNeighborsClassifier(n_neighbors=10))
c_models.append(KNeighborsClassifier(n_neighbors=15))
# Выделим предикторы и метки классов
x_labels = column_names[0:-1]
y_labels = ['region']
x_train = sc_train[x_labels]
x_test = sc_test[x_labels]
x_val = sc_val[x_labels]
y_train = np.ravel(sc_train[y_labels])
y_test = np.ravel(sc_test[y_labels])
y_val = np.ravel(sc_val[y_labels])
# Обучаем модели
for model in c_models:
model.fit(x_train, y_train)
# Оценииваем качество работы моделей на валидационной выборке.
f1s = []
for model in c_models:
val_pred = model.predict(x_val)
f1 = f1_score(y_val, val_pred, average='weighted')
f1s.append(f1)
print(f1)
# Выбираем лучшую модель
i_min = f1s.index(min(f1s))
best_c_model = c_models[i_min]
best_c_model
# Вычислим ошибку лучшей модели на тестовой выборке.
test_pred = best_c_model.predict(x_test)
f1 = f1_score(y_test, test_pred, average='weighted')
print(f1)
# Вспоминаем алгоритм решения задачи кластеризации - метод k-средних
# Параметр n_clusters - количество кластеров, по умолчанию равен 8
k_models = []
k_models.append(KMeans(n_clusters=5))
k_models.append(KMeans(n_clusters=8))
k_models.append(KMeans(n_clusters=20))
k_models.append(KMeans(n_clusters=50))
# Выделим используемые параметры
x_labels = column_names[0:-1]
x = pd.concat([sc_train[x_labels], sc_val[x_labels], sc_test[x_labels]])
x
# Произведем кластеризацию
for model in k_models:
model.fit(x)
# Оценим качество результата
sils = []
for model in k_models:
cluster_labels = model.predict(x)
s = silhouette_score(x, cluster_labels)
sils.append(s)
print(s)
# Выбираем лучшую модель
i_min = sils.index(min(sils))
best_k_model = k_models[i_min]
print(best_k_model)
print(sils[i_min])
# Задание №1 - анализ моделей для задачи регрессии
# Общий список моделей
r_models = [
LinearRegression(),
Lasso(alpha=1.0),
Lasso(alpha=0.5),
Ridge(alpha=1.0),
Ridge(alpha=0.5),
ElasticNet(alpha=1.0, l1_ratio=0.5),
ElasticNet(alpha=1.0, l1_ratio=0.25),
ElasticNet(alpha=1.0, l1_ratio=0.75),
ElasticNet(alpha=0.5, l1_ratio=0.5),
ElasticNet(alpha=0.5, l1_ratio=0.25),
ElasticNet(alpha=0.5, l1_ratio=0.75),
KNeighborsRegressor(n_neighbors=5),
KNeighborsRegressor(n_neighbors=10),
KNeighborsRegressor(n_neighbors=15),
KNeighborsRegressor(n_neighbors=20),
KNeighborsRegressor(n_neighbors=25)
]
# Выбор моделей для задания
n = 4
random.seed(my_seed)
my_models1 = random.sample(r_models, n)
print(my_models1)
# Загрузим данные для задачи регрессии
data = pd.read_csv("datasets/weather.csv")
data
# Зависимая переменная для всех одна и та же. Предикторы выбираем случайнм образом.
columns = list(data.columns)
n_x = 5
y_label = 'water_level'
x_labels = random.sample(columns[1:], n_x)
print(x_labels)
# Преобразуйте значения всех необходимых параметров к отрезку [0,1].
# Решите получившуюся задачу регрессии с помощью выбранных моделей и сравните их эффективность.
# Укажите, какая модель решает задачу лучше других.
# Задание №2 - анализ моделей для задачи классификации
# Общий список моделей
c_models = [
LogisticRegression(penalty='none', solver='saga'),
LogisticRegression(penalty='l1', solver='saga'),
LogisticRegression(penalty='l2', solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.25, solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.5, solver='saga'),
LogisticRegression(penalty='elasticnet', l1_ratio=0.75, solver='saga'),
LogisticRegression(),
MultinomialNB(alpha=0.0),
MultinomialNB(alpha=0.25),
MultinomialNB(alpha=0.5),
MultinomialNB(alpha=0.75),
MultinomialNB(alpha=1.0),
KNeighborsClassifier(n_neighbors=5),
KNeighborsClassifier(n_neighbors=10),
KNeighborsClassifier(n_neighbors=15),
KNeighborsClassifier(n_neighbors=20),
KNeighborsClassifier(n_neighbors=25)
]
# Выбор моделей для задания
n = 5
my_models2 = random.sample(c_models, n)
print(my_models2)
# Загрузим данные для задачи классификации
data = pd.read_csv("datasets/zoo2.csv")
data
# Метка класса для всех одна и та же. Параметры выбираем случайнм образом.
columns = list(data.columns)
n_x = 8
y_label = 'class_type'
x_labels = random.sample(columns[:-1], n_x)
print(x_labels)
# Преобразуйте значения всех необходимых параметров к отрезку [0,1].
# Решите получившуюся задачу классификации с помощью выбранных моделей и сравните их эффективность.
# Укажите, какая модель решает задачу лучше других.
| 0.306527 | 0.746982 |
<div>
<a href="https://www.audiolabs-erlangen.de/fau/professor/mueller"><img src="data_layout/PCP_Teaser.png" width=100% style="float: right;" alt="PCP Teaser"></a>
</div>
# Overview
The PCP notebooks serve two purposes. First, they introduce some basic material on Python programming as required for more advanced lab courses offered in FAU study programs such as <a href="https://www.cme.studium.fau.de/">Communications and Multimedia Engineering (CME)</a> or <a href="https://www.asc.studium.fau.de/">Advanced Signal Processing and Communications Engineering (ASC)</a>. Second, the PCP notebooks may be used as a gentle introduction to programming as needed in the more advanced <a href="https://www.audiolabs-erlangen.de/FMP">FMP Notebooks on Fundamentals of Music Processing</a>. While the first half of the PCP notebooks covers general Python concepts, the second half introduces and requires fundamental concepts in signal processing. The PCP notebooks are not intended to give a comprehensive overview of Python programming, nor are the notebooks self-contained. For a systematic introduction to Python programming, we refer to online sources such as <a href="https://docs.python.org/3/tutorial/index.html">The Python Tutorial</a> or the <a href="https://scipy-lectures.org/">Scipy Lecture Notes</a>. The PCP notebooks have been inspired and borrow material from the <a href="https://www.audiolabs-erlangen.de/FMP">FMP Notebooks on Fundamentals of Music Processing</a>.
<div class="alert alert-block alert-warning">
<strong>Note:</strong> The code, text, and figures of the PCP notebooks are licensed under the
<a href="https://opensource.org/licenses/MIT">MIT License</a>. The latest version of the PCP notebooks is hosted on <a href="https://github.com/meinardmueller/PCP">GitHub</a>. Alternatively, you can download a <a href="https://www.audiolabs-erlangen.de/resources/MIR/PCP/PCP_1.0.0.zip">zip-compressed archive</a> containing the PCP notebooks and all data. We work continuously on the PCP notebooks and provide updates on a regular basis (current version: 1.0.0). For suggestions and feedback, please contact <a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Meinard Müller</a>.
<br>
</div>
## Get Started
If a static view of the PCP notebooks is enough for you, the exported HTML versions can be used right away without any installation. All material including the explanations, the figures, and the audio examples can be accessed by just following the **HTML links**. If you want to **execute** the Python code cells, you have to download the notebooks (along with the data), create an environment, and start a Jupyter server. You then need to follow the **IPYNB links** within the Jupyter session. The necessary steps are explained in detail in the [PCP notebook on how to get started](PCP_GetStarted.html).
## Content
The collection of PCP notebooks is organized in ten units. Each unit, corresponding to an individual notebook, introduces some Python concepts, which are then applied and explored in exercises. The following table gives an overview of these units and provides links. In the first unit, we provide basic information on how to set up the Python and Jupyter framework, and discuss some tools used throughout the PCP notebooks.
<table class="table table-hover" style="border:none; font-size: 90%; width:100%; text-align:left">
<colgroup>
<col style="width:10%; text-align:left">
<col style="width:25%; text-align:left">
<col style="width:55%; text-align:left">
<col style="width:10%; text-align:left">
<col style="width:10%; text-align:left">
</colgroup>
<tr text-align="left" style="border:1px solid #C8C8C8; background-color:#F0F0F0" >
<td style="border:none; text-align:left"><b>Unit</b></td>
<td style="border:none; text-align:left"><b>Title</b></td>
<td style="border:none; text-align:left"><b>Notions, Techniques & Algorithms</b></td>
<td style="border:none; text-align:left"><b>HTML</b></td>
<td style="border:none; text-align:left"><b>IPYNB</b></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>1</strong></td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.html">Get Started</a></td>
<td style="border:none; text-align:left">Download; Conda; Python environment; Jupyter</td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>2</strong></td>
<td style="border:none; text-align:left"><a href="PCP_python.html">Python Basics</a></td>
<td style="border:none; text-align:left">Help; variables; basic operators; list; tuple; boolean values; set; dictionary; type conversion; shallow and deep copy</td>
<td style="border:none; text-align:left"><a href="PCP_python.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_python.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>3</strong></td>
<td style="border:none; text-align:left"><a href="PCP_numpy.html">NumPy Basics</a></td>
<td style="border:none; text-align:left">Array; reshape; array operations; type conversion; constants; matrix</td>
<td style="border:none; text-align:left"><a href="PCP_numpy.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_numpy.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>4</strong></td>
<td style="border:none; text-align:left"><a href="PCP_control.html">Control Structures and Functions</a></td>
<td style="border:none; text-align:left">Loop; for; while; break; continue; Python function; efficiency; runtime</td>
<td style="border:none; text-align:left"><a href="PCP_control.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_control.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>5</strong></td>
<td style="border:none; text-align:left"><a href="PCP_vis.html">Visualization Using Matplotlib</a></td>
<td style="border:none; text-align:left">Plot (1D); figure; imshow (2D); surface (3D); logarithmic axis</td>
<td style="border:none; text-align:left"><a href="PCP_vis.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_vis.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>6</strong></td>
<td style="border:none; text-align:left"><a href="PCP_complex.html">Complex Numbers</a></td>
<td style="border:none; text-align:left">Real part; imaginary part; absolute value; angle; polar representation; complex operations; conjugate; polar coordinate plot; roots; Mandelbrot </td>
<td style="border:none; text-align:left"><a href="PCP_complex.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_complex.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>7</strong></td>
<td style="border:none; text-align:left"><a href="PCP_exp.html">Exponential Function</a></td>
<td style="border:none; text-align:left">Power series; exponentiation identity; Euler's formula; differential equation; roots of unity; Gaussian function; spiral</td>
<td style="border:none; text-align:left"><a href="PCP_exp.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_exp.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>8</strong></td>
<td style="border:none; text-align:left"><a href="PCP_signal.html">Signals and Sampling</a></td>
<td style="border:none; text-align:left">Continuous-time signal; periodic; frequency; Hertz; amplitude; phase; discrete-time signal; sampling; aliasing; interference; beating; </td>
<td style="border:none; text-align:left"><a href="PCP_signal.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_signal.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>9</strong></td>
<td style="border:none; text-align:left"><a href="PCP_dft.html">Discrete Fourier Transform (DFT)</a></td>
<td style="border:none; text-align:left">Inner product; DFT; phase; optimality; DFT matrix; fast Fourier transform (FFT); FFT algorithm; runtime; time localization; chirp signal; inverse DFT</td>
<td style="border:none; text-align:left"><a href="PCP_dft.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_dft.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>10</strong></td>
<td style="border:none; text-align:left"><a href="PCP_module.html">Python Modules and Packages</a></td>
<td style="border:none; text-align:left">Python modules; Python packages; LibPCP; documentation; docstring</td>
<td style="border:none; text-align:left"><a href="PCP_module.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_module.ipynb">[ipynb]</a></td>
</tr>
</table>
## Contact
<p>
<a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Prof. Dr. Meinard Müller</a> <br>
Friedrich-Alexander Universität Erlangen-Nürnberg <br>
International Audio Laboratories Erlangen <br>
Lehrstuhl Semantic Audio Processing <br>
Am Wolfsmantel 33, 91058 Erlangen <br>
Email: [email protected]
</p>
## Acknowledgment
We want to thank the various people who have contributed to the design, implementation, and code examples of the notebooks. We mention the main contributors in alphabetical order:
<ul>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/krause">Michael Krause</a></li>
<li><a href="https://www.lms.tf.fau.eu/person/loellmann-heinrich/">Heinrich Löllmann</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Meinard Müller</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/rosenzweig">Sebastian Rosenzweig</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/zalkow">Frank Zalkow</a></li>
</ul>
The [International Audio Laboratories Erlangen](https://www.audiolabs-erlangen.de/) are a joint institution of the [Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU)](https://www.fau.eu/) and [Fraunhofer Institute for Integrated Circuits IIS](https://www.iis.fraunhofer.de/en.html).
<div>
<a href="https://opensource.org/licenses/MIT"><img src="data_layout/PCP_License.png" width=100% style="float: right;" alt="PCP License"></a>
</div>
|
github_jupyter
|
<div>
<a href="https://www.audiolabs-erlangen.de/fau/professor/mueller"><img src="data_layout/PCP_Teaser.png" width=100% style="float: right;" alt="PCP Teaser"></a>
</div>
# Overview
The PCP notebooks serve two purposes. First, they introduce some basic material on Python programming as required for more advanced lab courses offered in FAU study programs such as <a href="https://www.cme.studium.fau.de/">Communications and Multimedia Engineering (CME)</a> or <a href="https://www.asc.studium.fau.de/">Advanced Signal Processing and Communications Engineering (ASC)</a>. Second, the PCP notebooks may be used as a gentle introduction to programming as needed in the more advanced <a href="https://www.audiolabs-erlangen.de/FMP">FMP Notebooks on Fundamentals of Music Processing</a>. While the first half of the PCP notebooks covers general Python concepts, the second half introduces and requires fundamental concepts in signal processing. The PCP notebooks are not intended to give a comprehensive overview of Python programming, nor are the notebooks self-contained. For a systematic introduction to Python programming, we refer to online sources such as <a href="https://docs.python.org/3/tutorial/index.html">The Python Tutorial</a> or the <a href="https://scipy-lectures.org/">Scipy Lecture Notes</a>. The PCP notebooks have been inspired and borrow material from the <a href="https://www.audiolabs-erlangen.de/FMP">FMP Notebooks on Fundamentals of Music Processing</a>.
<div class="alert alert-block alert-warning">
<strong>Note:</strong> The code, text, and figures of the PCP notebooks are licensed under the
<a href="https://opensource.org/licenses/MIT">MIT License</a>. The latest version of the PCP notebooks is hosted on <a href="https://github.com/meinardmueller/PCP">GitHub</a>. Alternatively, you can download a <a href="https://www.audiolabs-erlangen.de/resources/MIR/PCP/PCP_1.0.0.zip">zip-compressed archive</a> containing the PCP notebooks and all data. We work continuously on the PCP notebooks and provide updates on a regular basis (current version: 1.0.0). For suggestions and feedback, please contact <a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Meinard Müller</a>.
<br>
</div>
## Get Started
If a static view of the PCP notebooks is enough for you, the exported HTML versions can be used right away without any installation. All material including the explanations, the figures, and the audio examples can be accessed by just following the **HTML links**. If you want to **execute** the Python code cells, you have to download the notebooks (along with the data), create an environment, and start a Jupyter server. You then need to follow the **IPYNB links** within the Jupyter session. The necessary steps are explained in detail in the [PCP notebook on how to get started](PCP_GetStarted.html).
## Content
The collection of PCP notebooks is organized in ten units. Each unit, corresponding to an individual notebook, introduces some Python concepts, which are then applied and explored in exercises. The following table gives an overview of these units and provides links. In the first unit, we provide basic information on how to set up the Python and Jupyter framework, and discuss some tools used throughout the PCP notebooks.
<table class="table table-hover" style="border:none; font-size: 90%; width:100%; text-align:left">
<colgroup>
<col style="width:10%; text-align:left">
<col style="width:25%; text-align:left">
<col style="width:55%; text-align:left">
<col style="width:10%; text-align:left">
<col style="width:10%; text-align:left">
</colgroup>
<tr text-align="left" style="border:1px solid #C8C8C8; background-color:#F0F0F0" >
<td style="border:none; text-align:left"><b>Unit</b></td>
<td style="border:none; text-align:left"><b>Title</b></td>
<td style="border:none; text-align:left"><b>Notions, Techniques & Algorithms</b></td>
<td style="border:none; text-align:left"><b>HTML</b></td>
<td style="border:none; text-align:left"><b>IPYNB</b></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>1</strong></td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.html">Get Started</a></td>
<td style="border:none; text-align:left">Download; Conda; Python environment; Jupyter</td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_getstarted.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>2</strong></td>
<td style="border:none; text-align:left"><a href="PCP_python.html">Python Basics</a></td>
<td style="border:none; text-align:left">Help; variables; basic operators; list; tuple; boolean values; set; dictionary; type conversion; shallow and deep copy</td>
<td style="border:none; text-align:left"><a href="PCP_python.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_python.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>3</strong></td>
<td style="border:none; text-align:left"><a href="PCP_numpy.html">NumPy Basics</a></td>
<td style="border:none; text-align:left">Array; reshape; array operations; type conversion; constants; matrix</td>
<td style="border:none; text-align:left"><a href="PCP_numpy.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_numpy.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>4</strong></td>
<td style="border:none; text-align:left"><a href="PCP_control.html">Control Structures and Functions</a></td>
<td style="border:none; text-align:left">Loop; for; while; break; continue; Python function; efficiency; runtime</td>
<td style="border:none; text-align:left"><a href="PCP_control.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_control.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>5</strong></td>
<td style="border:none; text-align:left"><a href="PCP_vis.html">Visualization Using Matplotlib</a></td>
<td style="border:none; text-align:left">Plot (1D); figure; imshow (2D); surface (3D); logarithmic axis</td>
<td style="border:none; text-align:left"><a href="PCP_vis.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_vis.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>6</strong></td>
<td style="border:none; text-align:left"><a href="PCP_complex.html">Complex Numbers</a></td>
<td style="border:none; text-align:left">Real part; imaginary part; absolute value; angle; polar representation; complex operations; conjugate; polar coordinate plot; roots; Mandelbrot </td>
<td style="border:none; text-align:left"><a href="PCP_complex.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_complex.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>7</strong></td>
<td style="border:none; text-align:left"><a href="PCP_exp.html">Exponential Function</a></td>
<td style="border:none; text-align:left">Power series; exponentiation identity; Euler's formula; differential equation; roots of unity; Gaussian function; spiral</td>
<td style="border:none; text-align:left"><a href="PCP_exp.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_exp.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>8</strong></td>
<td style="border:none; text-align:left"><a href="PCP_signal.html">Signals and Sampling</a></td>
<td style="border:none; text-align:left">Continuous-time signal; periodic; frequency; Hertz; amplitude; phase; discrete-time signal; sampling; aliasing; interference; beating; </td>
<td style="border:none; text-align:left"><a href="PCP_signal.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_signal.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>9</strong></td>
<td style="border:none; text-align:left"><a href="PCP_dft.html">Discrete Fourier Transform (DFT)</a></td>
<td style="border:none; text-align:left">Inner product; DFT; phase; optimality; DFT matrix; fast Fourier transform (FFT); FFT algorithm; runtime; time localization; chirp signal; inverse DFT</td>
<td style="border:none; text-align:left"><a href="PCP_dft.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_dft.ipynb">[ipynb]</a></td>
</tr>
<tr text-align="left" style="border:1px solid #C8C8C8">
<td style="border:none; text-align:left"><strong>10</strong></td>
<td style="border:none; text-align:left"><a href="PCP_module.html">Python Modules and Packages</a></td>
<td style="border:none; text-align:left">Python modules; Python packages; LibPCP; documentation; docstring</td>
<td style="border:none; text-align:left"><a href="PCP_module.html">[html]</a></td>
<td style="border:none; text-align:left"><a href="PCP_module.ipynb">[ipynb]</a></td>
</tr>
</table>
## Contact
<p>
<a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Prof. Dr. Meinard Müller</a> <br>
Friedrich-Alexander Universität Erlangen-Nürnberg <br>
International Audio Laboratories Erlangen <br>
Lehrstuhl Semantic Audio Processing <br>
Am Wolfsmantel 33, 91058 Erlangen <br>
Email: [email protected]
</p>
## Acknowledgment
We want to thank the various people who have contributed to the design, implementation, and code examples of the notebooks. We mention the main contributors in alphabetical order:
<ul>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/krause">Michael Krause</a></li>
<li><a href="https://www.lms.tf.fau.eu/person/loellmann-heinrich/">Heinrich Löllmann</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/professor/mueller">Meinard Müller</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/rosenzweig">Sebastian Rosenzweig</a></li>
<li><a href="https://www.audiolabs-erlangen.de/fau/assistant/zalkow">Frank Zalkow</a></li>
</ul>
The [International Audio Laboratories Erlangen](https://www.audiolabs-erlangen.de/) are a joint institution of the [Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU)](https://www.fau.eu/) and [Fraunhofer Institute for Integrated Circuits IIS](https://www.iis.fraunhofer.de/en.html).
<div>
<a href="https://opensource.org/licenses/MIT"><img src="data_layout/PCP_License.png" width=100% style="float: right;" alt="PCP License"></a>
</div>
| 0.766905 | 0.929376 |
```
import numpy as np
import pandas as pd
import xarray as xr
from tqdm import tqdm
import gc
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import matplotlib.ticker as mticker
def open_chi(path):
ds=(xr.open_dataset(path)*100)
ds=ds.assign_coords(lon=(((ds.lon + 180) % 360) - 180))
ds=ds.reindex(lon=sorted(ds.lon))
return ds
method_ls = ["ML","MAM4","diff","diff_abs"]
chi_ls = ["chi_b","chi_c","chi_h"]
file_path = {}
file_path["MAM4"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_chi/"
file_path["ML"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/ml_chi/"
file_path["diff"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_minus_ml_chi/"
file_path["diff_abs"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_minus_ml_chi/"
mask_path = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mask/"
name_ls = {}
# name_ls["chi_abd"]=r'$\overline{\chi_{\mathrm{a}}}$'+" (%)"
name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}}$'+" (%)"
name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}}$'+" (%)"
name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}}$'+" (%)"
mam4_name_ls = {}
mam4_name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}^{\mathrm{MAM4}}}$ (%)'
mam4_name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}^{\mathrm{MAM4}}}$ (%)'
mam4_name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}^{\mathrm{MAM4}}}$ (%)'
ml_name_ls = {}
ml_name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}^{\mathrm{ML}}}$ (%)'
ml_name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}^{\mathrm{ML}}}$ (%)'
ml_name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}^{\mathrm{ML}}}$ (%)'
diff_name_ls = {}
# diff_name_ls["chi_abd"]=r'$\overline{\Delta\chi_{\mathrm{a}}}$'+" (%)"
diff_name_ls["chi_h"]=r'$\overline{\Delta\chi_{\mathrm{h}}}$'+" (%)"
diff_name_ls["chi_b"]=r'$\overline{\Delta\chi_{\mathrm{o}}}$'+" (%)"
diff_name_ls["chi_c"]=r'$\overline{\Delta\chi_{\mathrm{c}}}$'+" (%)"
diff_abs_name_ls = {}
# diff_abs_name_ls["chi_abd"]=r'$\overline{|\Delta\chi_{\mathrm{a}}|}$'+" (%)"
diff_abs_name_ls["chi_h"]=r'$\overline{|\Delta\chi_{\mathrm{h}}|}$'+" (%)"
diff_abs_name_ls["chi_b"]=r'$\overline{|\Delta\chi_{\mathrm{o}}|}$'+" (%)"
diff_abs_name_ls["chi_c"]=r'$\overline{|\Delta\chi_{\mathrm{c}}|}$'+" (%)"
char_ls="abcdefghijklmnop"
```
# Hatch for all figure
```
%%time
def ax_mesh_hatch(da,mask,nrows,ncols,idx,ax_title,ctext,vmin=0,vmax=100,cmap='RdYlBu_r'):
ax = plt.subplot(nrows,ncols,idx,projection=ccrs.EqualEarth())
# plot area with mask
p = da.where(mask).plot(vmin=vmin,vmax=vmax,
cmap=cmap,
ax=ax,transform=ccrs.PlateCarree(),
add_colorbar=False,rasterized=True)
# plot inverse mask (using hatch)
mask_inverse = mask.where(mask==0)
ax.pcolor(mask_inverse.lon, mask_inverse.lat,
mask_inverse,hatch="///",alpha=0,
transform=ccrs.PlateCarree(),
rasterized=True)
ax.set_title(ax_title)
ax.set_global()
ax.coastlines()
cbar = plt.colorbar(p, ax=ax,
orientation="horizontal",
fraction=0.1,
shrink=0.75,
pad=0.05,
extend="neither")
cbar.ax.set_xlabel(ctext)
g = ax.gridlines(color='grey', linestyle='--', draw_labels=False)
g.xlocator = mticker.FixedLocator([-90, 0, 90])
#################################
year = "2011"
rc={'axes.labelsize':12, 'font.size':12, 'legend.fontsize':12, 'axes.titlesize':12}
plt.rcParams.update(**rc)
i=1
fig = plt.figure(figsize=(12,8))
for chi in tqdm(chi_ls):
for method in tqdm(method_ls):
if method=="diff_abs":
da = open_chi(file_path[method]+str(year)+"_"+chi+"_mean_abs.nc")[chi]
mask = open_chi(mask_path+str(year)+"_"+chi+".nc")["mask"]
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
diff_abs_name_ls[chi],vmin=0,vmax=100)
else:
da = open_chi(file_path[method]+str(year)+"_"+chi+"_mean.nc")[chi]
mask = open_chi(mask_path+str(year)+"_"+chi+".nc")["mask"]
if method=="diff":
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
diff_name_ls[chi],vmin=-100,vmax=100,cmap="bwr")
elif method=="ML":
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
ml_name_ls[chi])
else:
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
mam4_name_ls[chi])
i+=1
del da, mask
gc.collect()
plt.tight_layout()
fig.savefig("../figures/diff_map.pdf",dpi=288)
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import xarray as xr
from tqdm import tqdm
import gc
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import matplotlib.ticker as mticker
def open_chi(path):
ds=(xr.open_dataset(path)*100)
ds=ds.assign_coords(lon=(((ds.lon + 180) % 360) - 180))
ds=ds.reindex(lon=sorted(ds.lon))
return ds
method_ls = ["ML","MAM4","diff","diff_abs"]
chi_ls = ["chi_b","chi_c","chi_h"]
file_path = {}
file_path["MAM4"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_chi/"
file_path["ML"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/ml_chi/"
file_path["diff"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_minus_ml_chi/"
file_path["diff_abs"] = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mam4_minus_ml_chi/"
mask_path = "/data/keeling/a/zzheng25/d/mam4_paper_data/chi_only/mask/"
name_ls = {}
# name_ls["chi_abd"]=r'$\overline{\chi_{\mathrm{a}}}$'+" (%)"
name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}}$'+" (%)"
name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}}$'+" (%)"
name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}}$'+" (%)"
mam4_name_ls = {}
mam4_name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}^{\mathrm{MAM4}}}$ (%)'
mam4_name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}^{\mathrm{MAM4}}}$ (%)'
mam4_name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}^{\mathrm{MAM4}}}$ (%)'
ml_name_ls = {}
ml_name_ls["chi_b"]=r'$\overline{\chi_{\mathrm{o}}^{\mathrm{ML}}}$ (%)'
ml_name_ls["chi_c"]=r'$\overline{\chi_{\mathrm{c}}^{\mathrm{ML}}}$ (%)'
ml_name_ls["chi_h"]=r'$\overline{\chi_{\mathrm{h}}^{\mathrm{ML}}}$ (%)'
diff_name_ls = {}
# diff_name_ls["chi_abd"]=r'$\overline{\Delta\chi_{\mathrm{a}}}$'+" (%)"
diff_name_ls["chi_h"]=r'$\overline{\Delta\chi_{\mathrm{h}}}$'+" (%)"
diff_name_ls["chi_b"]=r'$\overline{\Delta\chi_{\mathrm{o}}}$'+" (%)"
diff_name_ls["chi_c"]=r'$\overline{\Delta\chi_{\mathrm{c}}}$'+" (%)"
diff_abs_name_ls = {}
# diff_abs_name_ls["chi_abd"]=r'$\overline{|\Delta\chi_{\mathrm{a}}|}$'+" (%)"
diff_abs_name_ls["chi_h"]=r'$\overline{|\Delta\chi_{\mathrm{h}}|}$'+" (%)"
diff_abs_name_ls["chi_b"]=r'$\overline{|\Delta\chi_{\mathrm{o}}|}$'+" (%)"
diff_abs_name_ls["chi_c"]=r'$\overline{|\Delta\chi_{\mathrm{c}}|}$'+" (%)"
char_ls="abcdefghijklmnop"
%%time
def ax_mesh_hatch(da,mask,nrows,ncols,idx,ax_title,ctext,vmin=0,vmax=100,cmap='RdYlBu_r'):
ax = plt.subplot(nrows,ncols,idx,projection=ccrs.EqualEarth())
# plot area with mask
p = da.where(mask).plot(vmin=vmin,vmax=vmax,
cmap=cmap,
ax=ax,transform=ccrs.PlateCarree(),
add_colorbar=False,rasterized=True)
# plot inverse mask (using hatch)
mask_inverse = mask.where(mask==0)
ax.pcolor(mask_inverse.lon, mask_inverse.lat,
mask_inverse,hatch="///",alpha=0,
transform=ccrs.PlateCarree(),
rasterized=True)
ax.set_title(ax_title)
ax.set_global()
ax.coastlines()
cbar = plt.colorbar(p, ax=ax,
orientation="horizontal",
fraction=0.1,
shrink=0.75,
pad=0.05,
extend="neither")
cbar.ax.set_xlabel(ctext)
g = ax.gridlines(color='grey', linestyle='--', draw_labels=False)
g.xlocator = mticker.FixedLocator([-90, 0, 90])
#################################
year = "2011"
rc={'axes.labelsize':12, 'font.size':12, 'legend.fontsize':12, 'axes.titlesize':12}
plt.rcParams.update(**rc)
i=1
fig = plt.figure(figsize=(12,8))
for chi in tqdm(chi_ls):
for method in tqdm(method_ls):
if method=="diff_abs":
da = open_chi(file_path[method]+str(year)+"_"+chi+"_mean_abs.nc")[chi]
mask = open_chi(mask_path+str(year)+"_"+chi+".nc")["mask"]
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
diff_abs_name_ls[chi],vmin=0,vmax=100)
else:
da = open_chi(file_path[method]+str(year)+"_"+chi+"_mean.nc")[chi]
mask = open_chi(mask_path+str(year)+"_"+chi+".nc")["mask"]
if method=="diff":
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
diff_name_ls[chi],vmin=-100,vmax=100,cmap="bwr")
elif method=="ML":
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
ml_name_ls[chi])
else:
ax_mesh_hatch(da,mask,3,4,i,"("+char_ls[i-1]+")",
mam4_name_ls[chi])
i+=1
del da, mask
gc.collect()
plt.tight_layout()
fig.savefig("../figures/diff_map.pdf",dpi=288)
plt.show()
| 0.210848 | 0.560553 |
```
import re
import requests
import mercantile
from rasterio.io import MemoryFile
from rasterio.features import sieve
from rasterio import features
from shapely.geometry import shape, mapping, Polygon
from shapely.ops import cascaded_union
from shapely.ops import transform
import numpy as np
from affine import Affine
from pyproj import Transformer
import json
import matplotlib.pyplot as plt
from descartes.patch import PolygonPatch
from scipy.ndimage.filters import gaussian_filter
from ipywidgets import interact, interactive, FloatSlider, interact_manual, HTML
import warnings
warnings.simplefilter("ignore")
style = """
<style>
.output_scroll {
height: unset !important;
border-radius: unset !important;
-webkit-box-shadow: unset !important;
box-shadow: unset !important;
}
</style>
"""
display(HTML(style))
%matplotlib inline
print("Enter URL: E.g. https://mapproxy.osm.ch/tiles/AGIS2019/EPSG900913/{zoom}/{x}/{y}.png?origin=nw")
print("")
url = input()
print("")
print("Enter Zoom level: E.g. 12")
print("")
zoom = int(input())
print("")
print("Enter extent: (Format: min_lon,min_lat,max_lon,max_lat) E.g. 7.66646,47.08713,8.45118,47.63340")
print("")
bbox_str = input()
min_lon, min_lat, max_lon, max_lat = map(float, bbox_str.split(','))
print("Extent:")
print("West: {}".format(min_lon))
print("East: {}".format(max_lon))
print("South: {}".format(min_lat))
print("North: {}".format(max_lat))
parameters = {}
# {z} instead of {zoom}
if '{z}' in url:
print('{z} found instead of {zoom} in tile url')
exit()
if '{apikey}' in url:
print("Not possible to check URL, apikey is required.")
exit
if "{switch:" in url:
match = re.search(r'switch:?([^}]*)', url)
switches = match.group(1).split(',')
tms_url = url.replace(match.group(0), 'switch')
parameters['switch'] = switches[0]
def process_tile(tile):
query_url = url
if '{-y}' in url:
y = 2 ** tile.z - 1 - tile.y
query_url = query_url.replace('{-y}', str(y))
elif '{!y}' in url:
y = 2 ** (tile.z - 1) - 1 - tile.y
query_url = query_url.replace('{!y}', str(y))
else:
query_url = query_url.replace('{y}', str(tile.y))
parameters['x'] = tile.x
parameters['zoom'] = tile.z
query_url = query_url.format(**parameters)
print("Request tile url:", query_url)
data = None
for i in range(3):
try:
r = requests.get(query_url)
data = r.content
except Exception as e:
print(str(e))
break
if data is None:
return []
bounds = mercantile.bounds(*tile)
try:
with MemoryFile(data) as memfile:
with memfile.open() as dataset:
pixel_x = (bounds.east - bounds.west) / dataset.width
pixel_y = (bounds.north - bounds.south) / dataset.height
geotransform = (bounds.west, pixel_x, 0, bounds.north, 0, -pixel_y)
data = np.zeros(shape=(dataset.height, dataset.width))
for band in range(dataset.count):
data += dataset.read(band + 1)
# Convert extrema to 0
data[data < 2] = 0
data[data > 254] = 0
# Blur data to avoid holes
data = gaussian_filter(data, sigma=3)
# Convert other pixels to 1
data[data > 0] = 1
# Filter small areas
data = sieve(data.astype(np.uint8),
size=int(dataset.height * dataset.width * 0.001),
connectivity=8)
# Create shapes for all areas with value = 1
shapes = list(features.shapes(data.astype(np.uint8), transform=Affine.from_gdal(*geotransform)))
buffer_dist = max(pixel_x, pixel_y) * 5
geoms = [shape(s[0]).buffer(buffer_dist).buffer(-buffer_dist)
for s in shapes if int(s[1]) == 1]
return geoms
except Exception as e:
print(str(e))
return []
def simplify(geom, distance=200):
""" Simplify geometry in epsg:3857"""
transformer = Transformer.from_crs("epsg:4326", "epsg:3857")
transformer_back = Transformer.from_crs("epsg:3857", "epsg:4326")
geom_3857 = transform(transformer.transform, geom)
geom_3857_simplified = geom_3857.simplify(distance, preserve_topology=False)
geom_simplified = transform(transformer_back.transform, geom_3857_simplified)
return geom_simplified
geoms = []
tiles = list(mercantile.tiles(west=min_lon, south=min_lat, east=max_lon, north=max_lat, zooms=zoom))
for tile in tiles:
tile_geoms = process_tile(tile)
if len(tile_geoms) > 0:
geoms.extend(tile_geoms)
geom = cascaded_union(geoms)
geom = simplify(geom)
def plot_geometry(geom):
bounds = geom.bounds
fig, ax = plt.subplots()
if isinstance(geom, Polygon):
geom = [geom]
for g in geom:
patch = PolygonPatch(g, facecolor='#6699cc', edgecolor='#6699cc',
alpha=0.5, zorder=2)
ax.add_patch(patch)
ax.set_xlim(bounds[0], bounds[2])
ax.set_ylim(bounds[1], bounds[3])
plt.show()
plot_geometry(geom)
print(json.dumps(mapping(geom), indent=4))
```
|
github_jupyter
|
import re
import requests
import mercantile
from rasterio.io import MemoryFile
from rasterio.features import sieve
from rasterio import features
from shapely.geometry import shape, mapping, Polygon
from shapely.ops import cascaded_union
from shapely.ops import transform
import numpy as np
from affine import Affine
from pyproj import Transformer
import json
import matplotlib.pyplot as plt
from descartes.patch import PolygonPatch
from scipy.ndimage.filters import gaussian_filter
from ipywidgets import interact, interactive, FloatSlider, interact_manual, HTML
import warnings
warnings.simplefilter("ignore")
style = """
<style>
.output_scroll {
height: unset !important;
border-radius: unset !important;
-webkit-box-shadow: unset !important;
box-shadow: unset !important;
}
</style>
"""
display(HTML(style))
%matplotlib inline
print("Enter URL: E.g. https://mapproxy.osm.ch/tiles/AGIS2019/EPSG900913/{zoom}/{x}/{y}.png?origin=nw")
print("")
url = input()
print("")
print("Enter Zoom level: E.g. 12")
print("")
zoom = int(input())
print("")
print("Enter extent: (Format: min_lon,min_lat,max_lon,max_lat) E.g. 7.66646,47.08713,8.45118,47.63340")
print("")
bbox_str = input()
min_lon, min_lat, max_lon, max_lat = map(float, bbox_str.split(','))
print("Extent:")
print("West: {}".format(min_lon))
print("East: {}".format(max_lon))
print("South: {}".format(min_lat))
print("North: {}".format(max_lat))
parameters = {}
# {z} instead of {zoom}
if '{z}' in url:
print('{z} found instead of {zoom} in tile url')
exit()
if '{apikey}' in url:
print("Not possible to check URL, apikey is required.")
exit
if "{switch:" in url:
match = re.search(r'switch:?([^}]*)', url)
switches = match.group(1).split(',')
tms_url = url.replace(match.group(0), 'switch')
parameters['switch'] = switches[0]
def process_tile(tile):
query_url = url
if '{-y}' in url:
y = 2 ** tile.z - 1 - tile.y
query_url = query_url.replace('{-y}', str(y))
elif '{!y}' in url:
y = 2 ** (tile.z - 1) - 1 - tile.y
query_url = query_url.replace('{!y}', str(y))
else:
query_url = query_url.replace('{y}', str(tile.y))
parameters['x'] = tile.x
parameters['zoom'] = tile.z
query_url = query_url.format(**parameters)
print("Request tile url:", query_url)
data = None
for i in range(3):
try:
r = requests.get(query_url)
data = r.content
except Exception as e:
print(str(e))
break
if data is None:
return []
bounds = mercantile.bounds(*tile)
try:
with MemoryFile(data) as memfile:
with memfile.open() as dataset:
pixel_x = (bounds.east - bounds.west) / dataset.width
pixel_y = (bounds.north - bounds.south) / dataset.height
geotransform = (bounds.west, pixel_x, 0, bounds.north, 0, -pixel_y)
data = np.zeros(shape=(dataset.height, dataset.width))
for band in range(dataset.count):
data += dataset.read(band + 1)
# Convert extrema to 0
data[data < 2] = 0
data[data > 254] = 0
# Blur data to avoid holes
data = gaussian_filter(data, sigma=3)
# Convert other pixels to 1
data[data > 0] = 1
# Filter small areas
data = sieve(data.astype(np.uint8),
size=int(dataset.height * dataset.width * 0.001),
connectivity=8)
# Create shapes for all areas with value = 1
shapes = list(features.shapes(data.astype(np.uint8), transform=Affine.from_gdal(*geotransform)))
buffer_dist = max(pixel_x, pixel_y) * 5
geoms = [shape(s[0]).buffer(buffer_dist).buffer(-buffer_dist)
for s in shapes if int(s[1]) == 1]
return geoms
except Exception as e:
print(str(e))
return []
def simplify(geom, distance=200):
""" Simplify geometry in epsg:3857"""
transformer = Transformer.from_crs("epsg:4326", "epsg:3857")
transformer_back = Transformer.from_crs("epsg:3857", "epsg:4326")
geom_3857 = transform(transformer.transform, geom)
geom_3857_simplified = geom_3857.simplify(distance, preserve_topology=False)
geom_simplified = transform(transformer_back.transform, geom_3857_simplified)
return geom_simplified
geoms = []
tiles = list(mercantile.tiles(west=min_lon, south=min_lat, east=max_lon, north=max_lat, zooms=zoom))
for tile in tiles:
tile_geoms = process_tile(tile)
if len(tile_geoms) > 0:
geoms.extend(tile_geoms)
geom = cascaded_union(geoms)
geom = simplify(geom)
def plot_geometry(geom):
bounds = geom.bounds
fig, ax = plt.subplots()
if isinstance(geom, Polygon):
geom = [geom]
for g in geom:
patch = PolygonPatch(g, facecolor='#6699cc', edgecolor='#6699cc',
alpha=0.5, zorder=2)
ax.add_patch(patch)
ax.set_xlim(bounds[0], bounds[2])
ax.set_ylim(bounds[1], bounds[3])
plt.show()
plot_geometry(geom)
print(json.dumps(mapping(geom), indent=4))
| 0.438545 | 0.304614 |

[](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/streamlit_notebooks/KEYPHRASE_EXTRACTION.ipynb)
# **Extract keyphrases from documents**
You can look at the example outputs stored at the bottom of the notebook to see what the model can do, or enter your own inputs to transform in the "Inputs" section. Find more about this keyphrase extraction model in another notebook [here](https://colab.research.google.com/github/JohnSnowLabs/spark-nlp-workshop/blob/master/tutorials/Certification_Trainings/Healthcare/9.Keyword_Extraction_YAKE.ipynb).
## 1. Colab setup
Install dependencies
```
# Install Java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
! pip install --ignore-installed -q spark-nlp
```
Import dependencies
```
import json
import os
import pandas as pd
import numpy as np
os.environ['JAVA_HOME'] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ['PATH'] = os.environ['JAVA_HOME'] + "/bin:" + os.environ['PATH']
# Import pyspark
from pyspark.sql import SparkSession
from pyspark.ml import PipelineModel
from pyspark.sql import functions as F
# Import SparkNLP
import sparknlp
from sparknlp.annotator import *
from sparknlp.base import *
# Start Spark session
spark = sparknlp.start()
```
## 2. Inputs
Enter inputs as strings in this list. Later cells of the notebook will extract keyphrases from whatever inputs are entered here.
```
input_list = [
"""Extracting keywords from texts has become a challenge for individuals and organizations as the information grows in complexity and size. The need to automate this task so that text can be processed in a timely and adequate manner has led to the emergence of automatic keyword extraction tools. Yake is a novel feature-based system for multi-lingual keyword extraction, which supports texts of different sizes, domain or languages. Unlike other approaches, Yake does not rely on dictionaries nor thesauri, neither is trained against any corpora. Instead, it follows an unsupervised approach which builds upon features extracted from the text, making it thus applicable to documents written in different languages without the need for further knowledge. This can be beneficial for a large number of tasks and a plethora of situations where access to training corpora is either limited or restricted.""",
"""Iodine deficiency is a lack of the trace element iodine, an essential nutrient in the diet. It may result in metabolic problems such as goiter, sometimes as an endemic goiter as well as cretinism due to untreated congenital hypothyroidism, which results in developmental delays and other health problems. Iodine deficiency is an important global health issue, especially for fertile and pregnant women. It is also a preventable cause of intellectual disability.
Iodine is an essential dietary mineral for neurodevelopment among offsprings and toddlers. The thyroid hormones thyroxine and triiodothyronine contain iodine. In areas where there is little iodine in the diet, typically remote inland areas where no marine foods are eaten, iodine deficiency is common. It is also common in mountainous regions of the world where food is grown in iodine-poor soil.
Prevention includes adding small amounts of iodine to table salt, a product known as iodized salt. Iodine compounds have also been added to other foodstuffs, such as flour, water and milk, in areas of deficiency. Seafood is also a well known source of iodine.""",
"""The Prague Quadrennial of Performance Design and Space was established in 1967 to bring the best of design for performance, scenography, and theatre architecture to the front line of cultural activities to be experienced by professional and emerging artists as well as the general public. The quadrennial exhibitions, festivals, and educational programs act as a global catalyst of creative progress by encouraging experimentation, networking, innovation, and future collaborations. PQ aims to honor, empower and celebrate the work of designers, artists and architects while inspiring and educating audiences, who are the most essential element of any live performance. The Prague Quadrennial strives to present performance design as an art form concerned with creation of active performance environments, that are far beyond merely decorative or beautiful, but emotionally charged, where design can become a quest, a question, an argument, a threat, a resolution, an agent of change, or a provocation. Performance design is a collaborative field where designers mix, fuse and blur the lines between multiple artistic disciplines to search for new approaches and new visions.
The Prague Quadrennial organizes an expansive program of international projects and activities between the main quadrennial events – performances, exhibitions, symposia, workshops, residencies, and educational initiatives serve as an international platform for exploring the practice, theory and education of contemporary performance design in the most encompassing terms.""",
"""Author Nathan Wiseman-Trowse explained that the "approach to the sheer physicality of sound" integral to dream pop was "arguably pioneered in popular music by figures such as Phil Spector and Brian Wilson". The music of the Velvet Underground in the 1960s and 1970s, which experimented with repetition, tone, and texture over conventional song structure, was also an important touchstone in the genre's development George Harrison's 1970 album All Things Must Pass, with its Spector-produced Wall of Sound and fluid arrangements, led music journalist John Bergstrom to credit it as a progenitor of the genre.
Reynolds described dream pop bands as "a wave of hazy neo-psychedelic groups", noting the influence of the "ethereal soundscapes" of bands such as Cocteau Twins. Rolling Stone's Kory Grow described "modern dream pop" as originating with the early 1980s work of Cocteau Twins and their contemporaries, while PopMatters' AJ Ramirez noted an evolutionary line from gothic rock to dream pop. Grow considered Julee Cruise's 1989 album Floating into the Night, written and produced by David Lynch and Angelo Badalamenti, as a significant development of the dream pop sound which "gave the genre its synthy sheen." The influence of Cocteau Twins extended to the expansion of the genre's influence into Cantopop and Mandopop through the music of Faye Wong, who covered multiple Cocteau Twins songs, including tracks featured in Chungking Express, in which she also acted. Cocteau Twins would go on to collaborate with Wong on original songs of hers, and Wong contributed vocals to a limited release of a late Cocteau Twins single.
In the early 1990s, some dream pop acts influenced by My Bloody Valentine, such as Seefeel, were drawn to techno and began utilizing elements such as samples and sequenced rhythms. Ambient pop music was described by AllMusic as "essentially an extension of the dream pop that emerged in the wake of the shoegazer movement", distinct for its incorporation of electronic textures.
Much of the music associated with the 2009-coined term "chillwave" could be considered dream pop. In the opinion of Grantland's David Schilling, when "chillwave" was popularized, the discussion that followed among music journalists and bloggers revealed that labels such as "shoegaze" and "dream pop" were ultimately "arbitrary and meaningless".""",
"""North Ingria was located in the Karelian Isthmus, between Finland and Soviet Russia. It was established 23 January 1919. The republic was first served by a post office at the Rautu railway station on the Finnish side of the border. As the access across the border was mainly restricted, the North Ingrian postal service was finally launched in the early 1920. The man behind the idea was the lieutenant colonel Georg Elfvengren, head of the governing council of North Ingria. He was also known as an enthusiastic stamp collector. The post office was opened at the capital village of Kirjasalo.
The first series of North Ingrian stamps were issued in 21 March 1920. They were based on the 1917 Finnish "Model Saarinen" series, a stamp designed by the Finnish architect Eliel Saarinen. The first series were soon sold to collectors, as the postage stamps became the major financial source of the North Ingrian government. The second series was designed for the North Ingrian postal service and issued 2 August 1920. The value of both series was in Finnish marks and similar to the postal fees of Finland. The number of letters sent from North Ingria was about 50 per day, most of them were carried to Finland. They were mainly sent by the personnel of the Finnish occupying forces. Large number of letters were also sent in pure philatelic purposes.
With the Treaty of Tartu, the area was re-integrated into Soviet Russia and the use of the North Ingrian postage stamps ended in 4 December 1920. Stamps were still sold in Finland in 1921 with an overprinting "Inkerin hyväksi" (For the Ingria), but they were no longer valid. Funds of the sale went for the North Ingrian refugees."""
]
# Change these to wherever you want your inputs and outputs to go
INPUT_FILE_PATH = "inputs"
OUTPUT_FILE_PATH = "outputs"
```
Write the example inputs to the input folder.
```
! mkdir -p $INPUT_FILE_PATH
for i, text in enumerate(input_list):
open(f'{INPUT_FILE_PATH}/Example{i + 1}.txt', 'w') \
.write(text[:min(len(text) - 10, 100)] + '... \n' + text)
```
## 3. Pipeline creation
Create the NLP pipeline.
```
# Transforms the raw text into a document readable by the later stages of the
# pipeline
document_assembler = DocumentAssembler() \
.setInputCol('text') \
.setOutputCol('document')
# Separates the document into sentences
sentence_detector = SentenceDetector() \
.setInputCols(['document']) \
.setOutputCol('sentences')# \
#.setDetectLists(True)
# Separates sentences into individial tokens (words)
tokenizer = Tokenizer() \
.setInputCols(['sentences']) \
.setOutputCol('tokens') \
.setContextChars(['(', ')', '?', '!', '.', ','])
# The keyphrase extraction model. Change MinNGrams and MaxNGrams to set the
# minimum and maximum length of possible keyphrases, and change NKeywords to
# set the amount of potential keyphrases identified per document.
keywords = YakeModel() \
.setInputCols('tokens') \
.setOutputCol('keywords') \
.setMinNGrams(2) \
.setMaxNGrams(5) \
.setNKeywords(100) \
.setStopWords(StopWordsCleaner().getStopWords())
# Assemble all of these stages into a pipeline, then fit the pipeline on an
# empty data frame so it can be used to transform new inputs.
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
keywords
])
empty_df = spark.createDataFrame([[""]]).toDF('text')
pipeline_model = pipeline.fit(empty_df)
# LightPipeline is faster than Pipeline for small datasets
light_pipeline = LightPipeline(pipeline_model)
```
## 4. Output creation
Utility functions to create more useful sets of keyphrases from the raw data frame produced by the model.
```
def adjusted_score(row, pow=2.5):
"""This function adjusts the scores of potential key phrases to give better
scores to phrases with more words (which will naturally have worse scores
due to the nature of the model). You can change the exponent to reward
longer phrases more or less. Higher exponents reward longer phrases."""
return ((row.result.count(' ') + 1) ** pow /
(float(row.metadata['score']) + 0.1))
def get_top_ranges(phrases, input_text):
"""Combine phrases that overlap."""
starts = sorted([row['begin'] for row in phrases])
ends = sorted([row['end'] for row in phrases])
ranges = [[starts[0], None]]
for i in range(len(starts) - 1):
if ends[i] < starts[i + 1]:
ranges[-1][1] = ends[i]
ranges.append([starts[i + 1], None])
ranges[-1][1] = ends[-1]
return [{
'begin': range[0],
'end': range[1],
'phrase': input_text[range[0]:range[1] + 1]
} for range in ranges]
def remove_duplicates(phrases):
"""Remove phrases that appear multiple times."""
i = 0
while i < len(phrases):
j = i + 1
while j < len(phrases):
if phrases[i]['phrase'] == phrases[j]['phrase']:
phrases.remove(phrases[j])
j += 1
i += 1
return phrases
def get_output_lists(df_row):
"""Returns a tuple with two lists of five phrases each. The first combines
key phrases that overlap to create longer kep phrases, which is best for
highlighting key phrases in text, and the seocnd is simply the keyphrases
with the highest scores, which is best for summarizing a document."""
keyphrases = []
for row in df_row.keywords:
keyphrases.append({
'begin': row.begin,
'end': row.end,
'phrase': row.result,
'score': adjusted_score(row)
})
keyphrases = sorted(keyphrases, key=lambda x: x['score'], reverse=True)
return (
get_top_ranges(keyphrases[:20], df_row.text)[:5],
remove_duplicates(keyphrases[:10])[:5]
)
```
Transform the example inputs to create a data frame storing the identified keyphrases.
```
df = spark.createDataFrame(pd.DataFrame({'text': input_list}))
result = light_pipeline.transform(df).toPandas()
```
For each example, create two JSON files containing selections of the best keyphrases for the document. See the docstring of `get_output_lists` two cells above to learn more about the two JSON files produced. These JSON files are used directly in the public demo app for this model.
```
! mkdir -p $OUTPUT_FILE_PATH
for i in range(len(result)):
top_ranges, top_summaries = get_output_lists(result.iloc[i])
with open(f'{OUTPUT_FILE_PATH}/Example{i + 1}.json', 'w') as ranges_file:
json.dump(top_ranges, ranges_file)
with open(f'{OUTPUT_FILE_PATH}/Example{i + 1}_summaries.json', 'w') \
as summaries_file:
json.dump(top_summaries, summaries_file)
```
## 5. Visualize outputs
The raw pandas data frame containing the outputs
```
result
```
The list of the top keyphrases (with overlapping keyphrases merged) for the last example
```
top_ranges
```
The list of the best summary kephrases (with duplicates removed) for the last example
```
top_summaries
```
|
github_jupyter
|
# Install Java
! apt-get update -qq
! apt-get install -y openjdk-8-jdk-headless -qq > /dev/null
! java -version
# Install pyspark
! pip install --ignore-installed -q pyspark==2.4.4
! pip install --ignore-installed -q spark-nlp
import json
import os
import pandas as pd
import numpy as np
os.environ['JAVA_HOME'] = "/usr/lib/jvm/java-8-openjdk-amd64"
os.environ['PATH'] = os.environ['JAVA_HOME'] + "/bin:" + os.environ['PATH']
# Import pyspark
from pyspark.sql import SparkSession
from pyspark.ml import PipelineModel
from pyspark.sql import functions as F
# Import SparkNLP
import sparknlp
from sparknlp.annotator import *
from sparknlp.base import *
# Start Spark session
spark = sparknlp.start()
input_list = [
"""Extracting keywords from texts has become a challenge for individuals and organizations as the information grows in complexity and size. The need to automate this task so that text can be processed in a timely and adequate manner has led to the emergence of automatic keyword extraction tools. Yake is a novel feature-based system for multi-lingual keyword extraction, which supports texts of different sizes, domain or languages. Unlike other approaches, Yake does not rely on dictionaries nor thesauri, neither is trained against any corpora. Instead, it follows an unsupervised approach which builds upon features extracted from the text, making it thus applicable to documents written in different languages without the need for further knowledge. This can be beneficial for a large number of tasks and a plethora of situations where access to training corpora is either limited or restricted.""",
"""Iodine deficiency is a lack of the trace element iodine, an essential nutrient in the diet. It may result in metabolic problems such as goiter, sometimes as an endemic goiter as well as cretinism due to untreated congenital hypothyroidism, which results in developmental delays and other health problems. Iodine deficiency is an important global health issue, especially for fertile and pregnant women. It is also a preventable cause of intellectual disability.
Iodine is an essential dietary mineral for neurodevelopment among offsprings and toddlers. The thyroid hormones thyroxine and triiodothyronine contain iodine. In areas where there is little iodine in the diet, typically remote inland areas where no marine foods are eaten, iodine deficiency is common. It is also common in mountainous regions of the world where food is grown in iodine-poor soil.
Prevention includes adding small amounts of iodine to table salt, a product known as iodized salt. Iodine compounds have also been added to other foodstuffs, such as flour, water and milk, in areas of deficiency. Seafood is also a well known source of iodine.""",
"""The Prague Quadrennial of Performance Design and Space was established in 1967 to bring the best of design for performance, scenography, and theatre architecture to the front line of cultural activities to be experienced by professional and emerging artists as well as the general public. The quadrennial exhibitions, festivals, and educational programs act as a global catalyst of creative progress by encouraging experimentation, networking, innovation, and future collaborations. PQ aims to honor, empower and celebrate the work of designers, artists and architects while inspiring and educating audiences, who are the most essential element of any live performance. The Prague Quadrennial strives to present performance design as an art form concerned with creation of active performance environments, that are far beyond merely decorative or beautiful, but emotionally charged, where design can become a quest, a question, an argument, a threat, a resolution, an agent of change, or a provocation. Performance design is a collaborative field where designers mix, fuse and blur the lines between multiple artistic disciplines to search for new approaches and new visions.
The Prague Quadrennial organizes an expansive program of international projects and activities between the main quadrennial events – performances, exhibitions, symposia, workshops, residencies, and educational initiatives serve as an international platform for exploring the practice, theory and education of contemporary performance design in the most encompassing terms.""",
"""Author Nathan Wiseman-Trowse explained that the "approach to the sheer physicality of sound" integral to dream pop was "arguably pioneered in popular music by figures such as Phil Spector and Brian Wilson". The music of the Velvet Underground in the 1960s and 1970s, which experimented with repetition, tone, and texture over conventional song structure, was also an important touchstone in the genre's development George Harrison's 1970 album All Things Must Pass, with its Spector-produced Wall of Sound and fluid arrangements, led music journalist John Bergstrom to credit it as a progenitor of the genre.
Reynolds described dream pop bands as "a wave of hazy neo-psychedelic groups", noting the influence of the "ethereal soundscapes" of bands such as Cocteau Twins. Rolling Stone's Kory Grow described "modern dream pop" as originating with the early 1980s work of Cocteau Twins and their contemporaries, while PopMatters' AJ Ramirez noted an evolutionary line from gothic rock to dream pop. Grow considered Julee Cruise's 1989 album Floating into the Night, written and produced by David Lynch and Angelo Badalamenti, as a significant development of the dream pop sound which "gave the genre its synthy sheen." The influence of Cocteau Twins extended to the expansion of the genre's influence into Cantopop and Mandopop through the music of Faye Wong, who covered multiple Cocteau Twins songs, including tracks featured in Chungking Express, in which she also acted. Cocteau Twins would go on to collaborate with Wong on original songs of hers, and Wong contributed vocals to a limited release of a late Cocteau Twins single.
In the early 1990s, some dream pop acts influenced by My Bloody Valentine, such as Seefeel, were drawn to techno and began utilizing elements such as samples and sequenced rhythms. Ambient pop music was described by AllMusic as "essentially an extension of the dream pop that emerged in the wake of the shoegazer movement", distinct for its incorporation of electronic textures.
Much of the music associated with the 2009-coined term "chillwave" could be considered dream pop. In the opinion of Grantland's David Schilling, when "chillwave" was popularized, the discussion that followed among music journalists and bloggers revealed that labels such as "shoegaze" and "dream pop" were ultimately "arbitrary and meaningless".""",
"""North Ingria was located in the Karelian Isthmus, between Finland and Soviet Russia. It was established 23 January 1919. The republic was first served by a post office at the Rautu railway station on the Finnish side of the border. As the access across the border was mainly restricted, the North Ingrian postal service was finally launched in the early 1920. The man behind the idea was the lieutenant colonel Georg Elfvengren, head of the governing council of North Ingria. He was also known as an enthusiastic stamp collector. The post office was opened at the capital village of Kirjasalo.
The first series of North Ingrian stamps were issued in 21 March 1920. They were based on the 1917 Finnish "Model Saarinen" series, a stamp designed by the Finnish architect Eliel Saarinen. The first series were soon sold to collectors, as the postage stamps became the major financial source of the North Ingrian government. The second series was designed for the North Ingrian postal service and issued 2 August 1920. The value of both series was in Finnish marks and similar to the postal fees of Finland. The number of letters sent from North Ingria was about 50 per day, most of them were carried to Finland. They were mainly sent by the personnel of the Finnish occupying forces. Large number of letters were also sent in pure philatelic purposes.
With the Treaty of Tartu, the area was re-integrated into Soviet Russia and the use of the North Ingrian postage stamps ended in 4 December 1920. Stamps were still sold in Finland in 1921 with an overprinting "Inkerin hyväksi" (For the Ingria), but they were no longer valid. Funds of the sale went for the North Ingrian refugees."""
]
# Change these to wherever you want your inputs and outputs to go
INPUT_FILE_PATH = "inputs"
OUTPUT_FILE_PATH = "outputs"
! mkdir -p $INPUT_FILE_PATH
for i, text in enumerate(input_list):
open(f'{INPUT_FILE_PATH}/Example{i + 1}.txt', 'w') \
.write(text[:min(len(text) - 10, 100)] + '... \n' + text)
# Transforms the raw text into a document readable by the later stages of the
# pipeline
document_assembler = DocumentAssembler() \
.setInputCol('text') \
.setOutputCol('document')
# Separates the document into sentences
sentence_detector = SentenceDetector() \
.setInputCols(['document']) \
.setOutputCol('sentences')# \
#.setDetectLists(True)
# Separates sentences into individial tokens (words)
tokenizer = Tokenizer() \
.setInputCols(['sentences']) \
.setOutputCol('tokens') \
.setContextChars(['(', ')', '?', '!', '.', ','])
# The keyphrase extraction model. Change MinNGrams and MaxNGrams to set the
# minimum and maximum length of possible keyphrases, and change NKeywords to
# set the amount of potential keyphrases identified per document.
keywords = YakeModel() \
.setInputCols('tokens') \
.setOutputCol('keywords') \
.setMinNGrams(2) \
.setMaxNGrams(5) \
.setNKeywords(100) \
.setStopWords(StopWordsCleaner().getStopWords())
# Assemble all of these stages into a pipeline, then fit the pipeline on an
# empty data frame so it can be used to transform new inputs.
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
keywords
])
empty_df = spark.createDataFrame([[""]]).toDF('text')
pipeline_model = pipeline.fit(empty_df)
# LightPipeline is faster than Pipeline for small datasets
light_pipeline = LightPipeline(pipeline_model)
def adjusted_score(row, pow=2.5):
"""This function adjusts the scores of potential key phrases to give better
scores to phrases with more words (which will naturally have worse scores
due to the nature of the model). You can change the exponent to reward
longer phrases more or less. Higher exponents reward longer phrases."""
return ((row.result.count(' ') + 1) ** pow /
(float(row.metadata['score']) + 0.1))
def get_top_ranges(phrases, input_text):
"""Combine phrases that overlap."""
starts = sorted([row['begin'] for row in phrases])
ends = sorted([row['end'] for row in phrases])
ranges = [[starts[0], None]]
for i in range(len(starts) - 1):
if ends[i] < starts[i + 1]:
ranges[-1][1] = ends[i]
ranges.append([starts[i + 1], None])
ranges[-1][1] = ends[-1]
return [{
'begin': range[0],
'end': range[1],
'phrase': input_text[range[0]:range[1] + 1]
} for range in ranges]
def remove_duplicates(phrases):
"""Remove phrases that appear multiple times."""
i = 0
while i < len(phrases):
j = i + 1
while j < len(phrases):
if phrases[i]['phrase'] == phrases[j]['phrase']:
phrases.remove(phrases[j])
j += 1
i += 1
return phrases
def get_output_lists(df_row):
"""Returns a tuple with two lists of five phrases each. The first combines
key phrases that overlap to create longer kep phrases, which is best for
highlighting key phrases in text, and the seocnd is simply the keyphrases
with the highest scores, which is best for summarizing a document."""
keyphrases = []
for row in df_row.keywords:
keyphrases.append({
'begin': row.begin,
'end': row.end,
'phrase': row.result,
'score': adjusted_score(row)
})
keyphrases = sorted(keyphrases, key=lambda x: x['score'], reverse=True)
return (
get_top_ranges(keyphrases[:20], df_row.text)[:5],
remove_duplicates(keyphrases[:10])[:5]
)
df = spark.createDataFrame(pd.DataFrame({'text': input_list}))
result = light_pipeline.transform(df).toPandas()
! mkdir -p $OUTPUT_FILE_PATH
for i in range(len(result)):
top_ranges, top_summaries = get_output_lists(result.iloc[i])
with open(f'{OUTPUT_FILE_PATH}/Example{i + 1}.json', 'w') as ranges_file:
json.dump(top_ranges, ranges_file)
with open(f'{OUTPUT_FILE_PATH}/Example{i + 1}_summaries.json', 'w') \
as summaries_file:
json.dump(top_summaries, summaries_file)
result
top_ranges
top_summaries
| 0.524151 | 0.978405 |
```
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
pd.options.display.max_rows = 8
```
# Reshaping data with `stack` and `unstack`
## Pivoting
Data is often stored in CSV files or databases in so-called “stacked” or “record” format:
```
df = pd.DataFrame({'subject':['A', 'A', 'B', 'B'],
'treatment':['CH', 'DT', 'CH', 'DT'],
'concentration':range(4)},
columns=['subject', 'treatment', 'concentration'])
df
```
A better representation might be one where the different subjects are in rows, the applied treatments are in columns and outcomes are in the data frame values.
<img src="img/stack.png" width=70%>
You can achieve this by `pivot` function:
```
pivoted = df.pivot(index='subject', columns='treatment', values='concentration')
pivoted
```
If there is more that one record for each pair of "subject" and "treatment" (for example, the subject was tested twice with the same treatment at different times) you can use `pivot_table`. It works just like `pivot` but it allows to specify additionally an aggregation function (`'mean'` by default).
To take another example, we will use some data from expeditions to the [Pole of Inaccessibility](https://www.google.com/maps/place/82%C2%B053'14.0%22S+55%C2%B004'30.0%22E/@-82.887222,55.075,577m/data=!3m1!1e3!4m2!3m1!1s0x0:0x0?hl=en). We will read the data from SQL database.
```
from sqlalchemy import create_engine
engine = create_engine('sqlite:///data/survey.db')
visited = pd.read_sql('Visited', engine, index_col='ident', parse_dates=['dated'])
visited
readings = pd.read_sql('Survey', engine).dropna()
readings = readings.drop_duplicates()
readings
```
<div class="alert alert-success">
<b>EXERCISE</b>: Join the `readings` and `visited` tables.
</div>
<div class="alert alert-success">
<b>EXERCISE</b>: Pivot the table such that we have sites in rows and different quantities in columns.
</div>
## Hierarchical index
Hierarchical index of pandas is a way of introducing another dimension to a (two-dimensional) data frame. This is implemented by having multiple levels of the index. Let's look at an example.
```
multi = df.set_index(['subject', 'treatment'])
multi
```
Note how the two indexes are nested: 2nd level index ('treatment') is grouped under the first level index ('subject'). To access the two levels you can use labels from the first level or both levels using a tuple.
```
multi.loc['A'] # first level only
```
Note that it creates a standard data frame with "flat" index.
```
multi.loc[('A', 'CH')] # two level
```
Indexing on the second index only may be slightly involved:
```
multi.loc[(slice(None), 'CH'), :]
```
Consult the [documentation](http://pandas.pydata.org/pandas-docs/stable/advanced.html#advanced-indexing-with-hierarchical-index) for other methods.
To return to orginal format with columns insted of indexes use `reset_index`:
```
multi.reset_index()
```
<div class="alert alert-success">
<b>EXERCISE</b>: Group the survey data by sites, date of measurement on each site and the quantity measured. List all readings for `site` DR-1; all readings of radiation using the hierchical index.
</div>
## `stack/unstack`
`stack` — shifts last level of hierarchical rows to columns
`unstack` — does the opposite, i.e. shifts last level of hierarchical columns to rows
```
result = multi['concentration'].unstack()
result
```
`unstack` reverses the operation:
```
result.stack()
```
We can "stack" it even further:
```
df = multi.stack()
df
```
<div class="alert alert-success">
<b>EXERCISE</b>: Rearange the data frame from last exercise, such that rows contain sites and dates (hierchical index) and columns different quantities. List all readings of radiation.
</div>
# Formatting data — Case study
Going further with the time series case study [test](05 - Time series data.ipynb) on the AirBase (The European Air quality dataBase) data.
One of the actual downloaded raw data files of AirBase is included in the repo:
```
!head -1 ./data/BETR8010000800100hour.1-1-1990.31-12-2012
```
Just reading the tab-delimited data:
```
data = pd.read_csv("data/BETR8010000800100hour.1-1-1990.31-12-2012", sep='\t')#, header=None)
data.head()
```
The above data is clearly not ready to be used! Each row contains the 24 measurements for each hour of the day, and also contains a flag (0/1) indicating the quality of the data.
Lets replace the negative numbers by missing values and give columns proper names.
```
hours = map(str, range(24))
flags = ['flag'] * 24
col_names = ['date'] + list(sum(zip(hours, flags), ()))
col_names[:5]
data = pd.read_csv("data/BETR8010000800100hour.1-1-1990.31-12-2012", sep='\t',
na_values=['-999', '-9999'],
names=col_names,
index_col='date')#, header=None)
```
For now, we disregard the 'flag' columns
```
data = data.drop('flag', axis=1)
data.head()
```
Now, we want to reshape it: our goal is to have the different hours as row indices, merged with the date into a datetime-index.
## `stack` at work
We can now use `stack` and some other functions to create a timeseries from the original dataframe:
<div class="alert alert-success">
<b>EXERCISE</b>: Reshape the dataframe to a timeseries
</div>
The end result should look like:
<div>
<table border="1" class="dataframe">
<thead>
<tr style="text-align: right;">
<th></th>
<th>BETR801</th>
</tr>
</thead>
<tbody>
<tr>
<th>1990-01-02 09:00:00</th>
<td>48.0</td>
</tr>
<tr>
<th>1990-01-02 12:00:00</th>
<td>48.0</td>
</tr>
<tr>
<th>1990-01-02 13:00:00</th>
<td>50.0</td>
</tr>
<tr>
<th>1990-01-02 14:00:00</th>
<td>55.0</td>
</tr>
<tr>
<th>...</th>
<td>...</td>
</tr>
<tr>
<th>2012-12-31 20:00:00</th>
<td>16.5</td>
</tr>
<tr>
<th>2012-12-31 21:00:00</th>
<td>14.5</td>
</tr>
<tr>
<th>2012-12-31 22:00:00</th>
<td>16.5</td>
</tr>
<tr>
<th>2012-12-31 23:00:00</th>
<td>15.0</td>
</tr>
</tbody>
</table>
<p>170794 rows × 1 columns</p>
</div>
First, reshape the dataframe so that each row consists of one observation for one date + hour combination:
Now, combine the date and hour colums into a datetime (tip: string columns can be summed to concatenate the strings):
## Acknowledgement
> *© 2015, Stijn Van Hoey and Joris Van den Bossche (<mailto:[email protected]>, <mailto:[email protected]>)*.
> *© 2015, modified by Bartosz Teleńczuk (original sources available from https://github.com/jorisvandenbossche/2015-EuroScipy-pandas-tutorial)*
> *Licensed under [CC BY 4.0 Creative Commons](http://creativecommons.org/licenses/by/4.0/)*
---
|
github_jupyter
|
%matplotlib inline
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
pd.options.display.max_rows = 8
df = pd.DataFrame({'subject':['A', 'A', 'B', 'B'],
'treatment':['CH', 'DT', 'CH', 'DT'],
'concentration':range(4)},
columns=['subject', 'treatment', 'concentration'])
df
pivoted = df.pivot(index='subject', columns='treatment', values='concentration')
pivoted
from sqlalchemy import create_engine
engine = create_engine('sqlite:///data/survey.db')
visited = pd.read_sql('Visited', engine, index_col='ident', parse_dates=['dated'])
visited
readings = pd.read_sql('Survey', engine).dropna()
readings = readings.drop_duplicates()
readings
multi = df.set_index(['subject', 'treatment'])
multi
multi.loc['A'] # first level only
multi.loc[('A', 'CH')] # two level
multi.loc[(slice(None), 'CH'), :]
multi.reset_index()
result = multi['concentration'].unstack()
result
result.stack()
df = multi.stack()
df
!head -1 ./data/BETR8010000800100hour.1-1-1990.31-12-2012
data = pd.read_csv("data/BETR8010000800100hour.1-1-1990.31-12-2012", sep='\t')#, header=None)
data.head()
hours = map(str, range(24))
flags = ['flag'] * 24
col_names = ['date'] + list(sum(zip(hours, flags), ()))
col_names[:5]
data = pd.read_csv("data/BETR8010000800100hour.1-1-1990.31-12-2012", sep='\t',
na_values=['-999', '-9999'],
names=col_names,
index_col='date')#, header=None)
data = data.drop('flag', axis=1)
data.head()
| 0.284278 | 0.956957 |
# Pytorch
Como dijimos anteriormente, PyTorch es un paquete de Python diseñado para realizar cálculos numéricos haciendo uso de la programación de tensores. Además permite su ejecución en GPU para acelerar los cálculos.
En la práctica es un sustituto bastante potente de Numpy, una librería casi estándar para trabajar con arrays en python.
## ¿Cómo funciona pytorch?
Vamos a ver un tutorial rápido del tipo de datos de pytorch y cómo trabaja internamente esta librería. Para esto tendrás que haber seguido correctamente todos los pasos anteriores. Para esto necesitas la **versión interactiva del notebook**.
Para esta sección:
* **Abre Jupyter** (consultar arriba)
* Navega hasta el notebook `00 Práctica Deep Learning - Introducción.ipynb` y ábrelo.
* Baja hasta esta sección.
Pero antes de nada os cuento algunas diferencias entre matlab y python:
* Python es un **lenguaje de propósito general** mientras que matlab es un lenguaje **específico para ciencia e ingeniería**. Esto no es ni bueno ni malo; matlab es más fácil de utilizar para ingeniería sin preparación, pero python es más versátil.
* Debido a ello, **Matlab carga automáticamente todas las funciones** mientras que en Python, **hay que cargar las librerías que vamos a utilizar**. Esto hace que usar funciones en matlab sea más sencillo (dos letras menos que escribir), pero a costa de que es más difícil gestionar la memoria, y los nombres de funciones se puden superponer. Supon que `A` es una matriz. Para hacer la pseudoinversa, en matlab hacemos:
```matlab
pinv(A)
```
* en python tenemos que cargar la librería:
```python
import scipy as sp
sp.pinv(A)
```
* Esto genera una cosa llamada **espacio de nombres**, en el que las funciones de cada librería van precedidas por su abreviatura (si importamos con `import x as y`) o el propio nombre si usamos `import torch`, `torch.tensor()`, mientras que en matlab basta con llamar a la función. Por ejemplo, cuando en matlab escribimos:
- `vector = [1, 2, 3]`
* en python+pytorch necesitamos especificar que es un tensor (un array multidimensional):
- `vector = torch.tensor([1,2,3])`
Vamos a cargar la librería con `import torch` y ver que podemos, por ejemplo, construir una matriz de 5x3 aleatoria. Para ejecutar una celda, basta con seleccionarla (bien con las flechas del teclado, bien con el ratón) y pulsando `Ctrl+Enter` (o bien pulsando "Run" en la barra superior).
```
import torch
x = torch.rand(5, 3)
print(x)
```
O una matriz de ceros:
```
x = torch.zeros(5, 3, dtype=torch.long)
print(x)
```
O a partir de unos datos dados, y podemos mostrarla con `print`, pero también acceder a sus características, como el tamaño de la matriz:
```
x = torch.tensor([[5.5, 3, 3],[2,1, 5], [3,4,2],[7,6,5],[2,1,2]])
print(x)
print(x.shape)
```
Con tensores se puede operar de forma normal:
```
y = torch.rand(5, 3)
print(x + y)
```
Pero OJO CUIDAO, tienen que ser del mismo tamaño, si no, va a dar error:
```
y = torch.rand(2,3)
print(x+y)
```
Se puede hacer *slicing* como en numpy o Matlab. Por ejemplo, para extraer la primera columna:
```
print(x[:, 1])
```
Otra característica que nos será de mucha utilidad es cambiar la forma de la matriz, que en otros lenguajes se conoce como `reshape`, y aquí es un método del objeto tensor llamado `view()`:
```
x = torch.randn(4, 4)
y = x.view(16)
z = x.view(-1, 8) # the size -1 is inferred from other dimensions
print(x.size(), y.size(), z.size())
```
Podemos operar con tensores y valores escalares:
```
y = x + 2
print(y)
```
Y también podemos definir funciones que realicen estas operaciones que apliquemos a los diferentes tensores:
```
def modulo(x,y):
aux = x**2 + y**2
salida = torch.sqrt(aux)
return salida
print(modulo(x,y))
```
Y, una parte fundamental es que pytorch conserva memoria de las operaciones realizadas en un vector:
```
x = torch.ones(2, 2, requires_grad=True)
y = x + 2
print(y)
```
La propiedad `grad_fn` será fundamental en el entrenamiento de redes neuronales, ya que guarda el gradiente de la operación o función que se haya aplicado a los datos. Esto se conserva a traves de todas las operaciones:
```
z = y * y * 3
out = z.mean()
print(z, out)
```
O incluso llevan cuenta de las operaciones realizadas con funciones:
```
print(modulo(x,y))
```
Para calcular el gradiente a lo largo de estas operaciones se utiliza la función `.backward()`, que realiza la propagación del gradiente hacia atrás. Podemos mostrar el gradiente $\frac{\partial out}{\partial x}$ con la propiedad `x.grad`, así que lo vemos:
```
out.backward()
print(x.grad)
```
Habrá aquí una matriz de 2x2 con valores 4.5. Si llamamos el tensor de salida $o$, tenemos que:
$$
o = \frac{1}{4} \sum_iz_i, \quad z_i = 3(x_i + 2)^2
$$
Así que $z_i|_{x_i=1} = 27$. Entonces, la $\frac{\partial o}{\partial x_i} = \frac{3}{2}(x_i+2)$ y $\frac{\partial o}{\partial x_i} |_{x_i=1} = \frac{9}{2} = 4.5$
Gracias a esto, y a las matemáticas del algoritmo de propagación hacia atrás (*backpropagation*, ver video de introducción a la práctica), se pueden actualizar los pesos en función de una función de pérdida en las redes neuronales. Se puede activar y desactivar el cálculo del gradiente con la expresión `torch.no_grad()`.
```
print(x.requires_grad)
print((x ** 2).requires_grad)
with torch.no_grad():
print((x ** 2).requires_grad)
```
En la próxima sección, `01 Práctica Deep Learning - Perceptrón Multicapa.ipynb`, veremos como se construye y se entrena nuestra primera red neuronal utilizando estas características de pytorch.
|
github_jupyter
|
pinv(A)
import scipy as sp
sp.pinv(A)
import torch
x = torch.rand(5, 3)
print(x)
x = torch.zeros(5, 3, dtype=torch.long)
print(x)
x = torch.tensor([[5.5, 3, 3],[2,1, 5], [3,4,2],[7,6,5],[2,1,2]])
print(x)
print(x.shape)
y = torch.rand(5, 3)
print(x + y)
y = torch.rand(2,3)
print(x+y)
print(x[:, 1])
x = torch.randn(4, 4)
y = x.view(16)
z = x.view(-1, 8) # the size -1 is inferred from other dimensions
print(x.size(), y.size(), z.size())
y = x + 2
print(y)
def modulo(x,y):
aux = x**2 + y**2
salida = torch.sqrt(aux)
return salida
print(modulo(x,y))
x = torch.ones(2, 2, requires_grad=True)
y = x + 2
print(y)
z = y * y * 3
out = z.mean()
print(z, out)
print(modulo(x,y))
out.backward()
print(x.grad)
print(x.requires_grad)
print((x ** 2).requires_grad)
with torch.no_grad():
print((x ** 2).requires_grad)
| 0.447943 | 0.988668 |
<a href="https://colab.research.google.com/github/skywalker00001/Conterfactual-Reasoning-Project/blob/main/data_cleaning_4_2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# how to find out the different or changed blocks in sentence b compared to the sentence a (base)
```
# Importing stock libraries
import numpy as np
import pandas as pd
import difflib
import nltk
import copy
import regex as re
nltk.download("punkt")
# a = "I paid the cashier and patiently waited for my drink."
# b = "I paid the cashier and patiently waited at the counter for my drink."
a = "I want to acchieve something to school."
b = "I do something to go for it to school."
a_list = nltk.word_tokenize(a)
b_list = nltk.word_tokenize(b)
a_list = "qabxcd"
b_list = "abycdf"
print(a_list)
print(b_list)
s = difflib.SequenceMatcher(None, a_list, b_list)
for block in s.get_matching_blocks():
print(block)
print(a_list[2])
print(a_list[0:0+8])
print(a_list[8:8+3])
print(a_list[11:11+0])
matches = []
matches.append([0, 0, 0])
for block in s.get_matching_blocks():
#matches.append([block[0], block[1], block[2]])
matches.append([i for i in block])
#matches.append(block)
print(matches)
# explanation: matches[i][0] are the a index, matches[i][1] are the b index, matches[i] [2] are the lengths of same (matched) words.
changes = []
for i in range(len(matches) - 1):
#print(matches[i])
if ((matches[i][0]+ matches[i][2] < matches[i+1][0]) & (matches[i][1]+ matches[i][2] < matches[i+1][1])): # replace
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"replacing_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
elif ((matches[i][0]+ matches[i][2] < matches[i+1][0]) & (matches[i][1]+ matches[i][2] == matches[i+1][1])): # delete
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"deleting_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
elif ((matches[i][0]+ matches[i][2] == matches[i+1][0]) & (matches[i][1]+ matches[i][2] < matches[i+1][1])): # insert
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"inserting_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
print(changes)
c = copy.deepcopy(a_list)
c[7:7] = nltk.word_tokenize("at the counter")
print(a_list)
```
```
print(c)
```
# another way
```
# a = "qabxcd"
# b = "abycdf"
cgs = []
s = difflib.SequenceMatcher(None, a_list, b_list)
for tag, i1, i2, j1, j2 in s.get_opcodes():
print('{:7} a[{}:{}] --> b[{}:{}] {!r:>8} --> {!r}'.format(tag, i1, i2, j1, j2, a_list[i1:i2], b_list[j1:j2]))
#print('{:7} a[{}:{}] --> b[{}:{}] {!r} --> {!r}'.format(tag, i1, i2, j1, j2, a[i1:i2], b[j1:j2]))
if (tag != 'equal'):
#cgs.append(f'{tag}_{i1}-{i2}:\"{(" ").join(b_list[j1:j2])}\".')
cgs.append(f{i1}-{i2}:\"{(" ").join(b_list[j1:j2])}\".')
print(cgs)
t=(" ".join(cgs))
print(t)
print(t.split(' '))
```
function
```
def origin2edited(origin, edited):
origin_list = nltk.word_tokenize(origin)
editeed_list = nltk.word_tokenize(edited)
s = difflib.SequenceMatcher(None, origin_list, editeed_list)
cgs = []
for tag, i1, i2, j1, j2 in s.get_opcodes():
if (tag != 'equal'):
cgs.append(f'{i1}-{i2}:\"{(" ").join(editeed_list[j1:j2])}\".')
conclusion = "".join(cgs)
return conclusion
print(origin2edited(a, b))
cgs = sen.split('.')[0:-1]
print(len(cgs))
def restore(cg, origin):
cgs = cg.split('.')[0:-1]
origin_list = nltk.word_tokenize(origin)
#blocks = copy.deepcopy(origin)
for j in list(reversed(cgs)):
#print("unchanged: ", origin_list)
#print(j)
pattern = re.compile(r'^(\d+)-(\d+):\"(.*?)\"') # matches 1-2:"do something". (1), (2), (do something)
results = re.search(pattern, j) # i3 means the content between ""
i1, i2, i3 = results.group(1), results.group(2), results.group(3)
origin_list[int(i1): int(i2)] = nltk.word_tokenize(i3)
#print("changed: ", origin_list)
return (" ").join(origin_list[0:-1])+'.' #because we don't want any space before '.', which is the last element in the origin_list
print(a)
print(sen)
b = "I do something to go for it to school."
print(restore(origin2edited(a, b), a))
i1 = re.compile(r'^(\d+)-(\d+):\"(.*?)\"')
#i1 = re.compile(r'(\d+)')
sen = "1-2:\"do something\".3-5:\"go for it\"."
i3 = re.search(i1, sen).group(1)
print(i3)
pattern = re.compile(r'^(\d+)-(\d+):\"(.*?)\"') # matches 1-2:"do something". (1), (2), (do something)
results = re.search(pattern, sen) # i3 means the content between ""
i1, i2, i3 = results.group(1), results.group(2), results.group(3)
print(i1, i2, i3)
print(restore(orgin2edited(a, b), a))
```
|
github_jupyter
|
# Importing stock libraries
import numpy as np
import pandas as pd
import difflib
import nltk
import copy
import regex as re
nltk.download("punkt")
# a = "I paid the cashier and patiently waited for my drink."
# b = "I paid the cashier and patiently waited at the counter for my drink."
a = "I want to acchieve something to school."
b = "I do something to go for it to school."
a_list = nltk.word_tokenize(a)
b_list = nltk.word_tokenize(b)
a_list = "qabxcd"
b_list = "abycdf"
print(a_list)
print(b_list)
s = difflib.SequenceMatcher(None, a_list, b_list)
for block in s.get_matching_blocks():
print(block)
print(a_list[2])
print(a_list[0:0+8])
print(a_list[8:8+3])
print(a_list[11:11+0])
matches = []
matches.append([0, 0, 0])
for block in s.get_matching_blocks():
#matches.append([block[0], block[1], block[2]])
matches.append([i for i in block])
#matches.append(block)
print(matches)
# explanation: matches[i][0] are the a index, matches[i][1] are the b index, matches[i] [2] are the lengths of same (matched) words.
changes = []
for i in range(len(matches) - 1):
#print(matches[i])
if ((matches[i][0]+ matches[i][2] < matches[i+1][0]) & (matches[i][1]+ matches[i][2] < matches[i+1][1])): # replace
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"replacing_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
elif ((matches[i][0]+ matches[i][2] < matches[i+1][0]) & (matches[i][1]+ matches[i][2] == matches[i+1][1])): # delete
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"deleting_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
elif ((matches[i][0]+ matches[i][2] == matches[i+1][0]) & (matches[i][1]+ matches[i][2] < matches[i+1][1])): # insert
string = (" ").join(b_list[(matches[i][1]+matches[i][2]) : matches[i+1][1]])
changes.append(f"inserting_{matches[i][0]+matches[i][2]}-{matches[i+1][0]}:\"{string}\".")
print(changes)
c = copy.deepcopy(a_list)
c[7:7] = nltk.word_tokenize("at the counter")
print(a_list)
print(c)
# a = "qabxcd"
# b = "abycdf"
cgs = []
s = difflib.SequenceMatcher(None, a_list, b_list)
for tag, i1, i2, j1, j2 in s.get_opcodes():
print('{:7} a[{}:{}] --> b[{}:{}] {!r:>8} --> {!r}'.format(tag, i1, i2, j1, j2, a_list[i1:i2], b_list[j1:j2]))
#print('{:7} a[{}:{}] --> b[{}:{}] {!r} --> {!r}'.format(tag, i1, i2, j1, j2, a[i1:i2], b[j1:j2]))
if (tag != 'equal'):
#cgs.append(f'{tag}_{i1}-{i2}:\"{(" ").join(b_list[j1:j2])}\".')
cgs.append(f{i1}-{i2}:\"{(" ").join(b_list[j1:j2])}\".')
print(cgs)
t=(" ".join(cgs))
print(t)
print(t.split(' '))
def origin2edited(origin, edited):
origin_list = nltk.word_tokenize(origin)
editeed_list = nltk.word_tokenize(edited)
s = difflib.SequenceMatcher(None, origin_list, editeed_list)
cgs = []
for tag, i1, i2, j1, j2 in s.get_opcodes():
if (tag != 'equal'):
cgs.append(f'{i1}-{i2}:\"{(" ").join(editeed_list[j1:j2])}\".')
conclusion = "".join(cgs)
return conclusion
print(origin2edited(a, b))
cgs = sen.split('.')[0:-1]
print(len(cgs))
def restore(cg, origin):
cgs = cg.split('.')[0:-1]
origin_list = nltk.word_tokenize(origin)
#blocks = copy.deepcopy(origin)
for j in list(reversed(cgs)):
#print("unchanged: ", origin_list)
#print(j)
pattern = re.compile(r'^(\d+)-(\d+):\"(.*?)\"') # matches 1-2:"do something". (1), (2), (do something)
results = re.search(pattern, j) # i3 means the content between ""
i1, i2, i3 = results.group(1), results.group(2), results.group(3)
origin_list[int(i1): int(i2)] = nltk.word_tokenize(i3)
#print("changed: ", origin_list)
return (" ").join(origin_list[0:-1])+'.' #because we don't want any space before '.', which is the last element in the origin_list
print(a)
print(sen)
b = "I do something to go for it to school."
print(restore(origin2edited(a, b), a))
i1 = re.compile(r'^(\d+)-(\d+):\"(.*?)\"')
#i1 = re.compile(r'(\d+)')
sen = "1-2:\"do something\".3-5:\"go for it\"."
i3 = re.search(i1, sen).group(1)
print(i3)
pattern = re.compile(r'^(\d+)-(\d+):\"(.*?)\"') # matches 1-2:"do something". (1), (2), (do something)
results = re.search(pattern, sen) # i3 means the content between ""
i1, i2, i3 = results.group(1), results.group(2), results.group(3)
print(i1, i2, i3)
print(restore(orgin2edited(a, b), a))
| 0.088694 | 0.760139 |
**This notebook is an exercise in the [Data Cleaning](https://www.kaggle.com/learn/data-cleaning) course. You can reference the tutorial at [this link](https://www.kaggle.com/alexisbcook/character-encodings).**
---
In this exercise, you'll apply what you learned in the **Character encodings** tutorial.
# Setup
The questions below will give you feedback on your work. Run the following cell to set up the feedback system.
```
from learntools.core import binder
binder.bind(globals())
from learntools.data_cleaning.ex4 import *
print("Setup Complete")
```
# Get our environment set up
The first thing we'll need to do is load in the libraries we'll be using.
```
# modules we'll use
import pandas as pd
import numpy as np
# helpful character encoding module
import chardet
# set seed for reproducibility
np.random.seed(0)
```
# 1) What are encodings?
You're working with a dataset composed of bytes. Run the code cell below to print a sample entry.
```
sample_entry = b'\xa7A\xa6n'
print(sample_entry)
print('data type:', type(sample_entry))
```
You notice that it doesn't use the standard UTF-8 encoding.
Use the next code cell to create a variable `new_entry` that changes the encoding from `"big5-tw"` to `"utf-8"`. `new_entry` should have the bytes datatype.
```
sample_entry.decode("big5-tw")
sample_entry.decode("big5-tw").encode("utf-8")
new_entry = sample_entry.decode("big5-tw").encode("utf-8")
# Check your answer
q1.check()
# Lines below will give you a hint or solution code
q1.hint()
#q1.solution()
```
# 2) Reading in files with encoding problems
Use the code cell below to read in this file at path `"../input/fatal-police-shootings-in-the-us/PoliceKillingsUS.csv"`.
Figure out what the correct encoding should be and read in the file to a DataFrame `police_killings`.
```
with open("../input/fatal-police-shootings-in-the-us/PoliceKillingsUS.csv", 'rb') as rawdata:
result = chardet.detect(rawdata.read(100000))
# check what the character encoding might be
print(result)
# TODO: Load in the DataFrame correctly.
police_killings = pd.read_csv("../input/fatal-police-shootings-in-the-us/PoliceKillingsUS.csv", encoding='Windows-1252')
# Check your answer
q2.check()
```
Feel free to use any additional code cells for supplemental work. To get credit for finishing this question, you'll need to run `q2.check()` and get a result of **Correct**.
```
# (Optional) Use this code cell for any additional work.
# Lines below will give you a hint or solution code
q2.hint()
#q2.solution()
```
# 3) Saving your files with UTF-8 encoding
Save a version of the police killings dataset to CSV with UTF-8 encoding. Your answer will be marked correct after saving this file.
Note: When using the `to_csv()` method, supply only the name of the file (e.g., `"my_file.csv"`). This saves the file at the filepath `"/kaggle/working/my_file.csv"`.
```
# TODO: Save the police killings dataset to CSV
police_killings.to_csv('PoliceKillingsUS.csv')
# Check your answer
q3.check()
# Lines below will give you a hint or solution code
#q3.hint()
#q3.solution()
```
# (Optional) More practice
Check out [this dataset of files in different character encodings](https://www.kaggle.com/rtatman/character-encoding-examples). Can you read in all the files with their original encodings and them save them out as UTF-8 files?
If you have a file that's in UTF-8 but has just a couple of weird-looking characters in it, you can try out the [ftfy module](https://ftfy.readthedocs.io/en/latest/#) and see if it helps.
# Keep going
In the final lesson, learn how to [**clean up inconsistent text entries**](https://www.kaggle.com/alexisbcook/inconsistent-data-entry) in your dataset.
---
*Have questions or comments? Visit the [Learn Discussion forum](https://www.kaggle.com/learn-forum/172650) to chat with other Learners.*
|
github_jupyter
|
from learntools.core import binder
binder.bind(globals())
from learntools.data_cleaning.ex4 import *
print("Setup Complete")
# modules we'll use
import pandas as pd
import numpy as np
# helpful character encoding module
import chardet
# set seed for reproducibility
np.random.seed(0)
sample_entry = b'\xa7A\xa6n'
print(sample_entry)
print('data type:', type(sample_entry))
sample_entry.decode("big5-tw")
sample_entry.decode("big5-tw").encode("utf-8")
new_entry = sample_entry.decode("big5-tw").encode("utf-8")
# Check your answer
q1.check()
# Lines below will give you a hint or solution code
q1.hint()
#q1.solution()
with open("../input/fatal-police-shootings-in-the-us/PoliceKillingsUS.csv", 'rb') as rawdata:
result = chardet.detect(rawdata.read(100000))
# check what the character encoding might be
print(result)
# TODO: Load in the DataFrame correctly.
police_killings = pd.read_csv("../input/fatal-police-shootings-in-the-us/PoliceKillingsUS.csv", encoding='Windows-1252')
# Check your answer
q2.check()
# (Optional) Use this code cell for any additional work.
# Lines below will give you a hint or solution code
q2.hint()
#q2.solution()
# TODO: Save the police killings dataset to CSV
police_killings.to_csv('PoliceKillingsUS.csv')
# Check your answer
q3.check()
# Lines below will give you a hint or solution code
#q3.hint()
#q3.solution()
| 0.291687 | 0.9255 |
# Ensemble Optimization for Robust Pulses
```
# NBVAL_IGNORE_OUTPUT
%load_ext watermark
import sys
import os
import qutip
import numpy as np
import scipy
import matplotlib
import matplotlib.pylab as plt
import krotov
from qutip import Qobj
import pickle
%watermark -v --iversions
```
$\newcommand{tr}[0]{\operatorname{tr}}
\newcommand{diag}[0]{\operatorname{diag}}
\newcommand{abs}[0]{\operatorname{abs}}
\newcommand{pop}[0]{\operatorname{pop}}
\newcommand{aux}[0]{\text{aux}}
\newcommand{opt}[0]{\text{opt}}
\newcommand{tgt}[0]{\text{tgt}}
\newcommand{init}[0]{\text{init}}
\newcommand{lab}[0]{\text{lab}}
\newcommand{rwa}[0]{\text{rwa}}
\newcommand{bra}[1]{\langle#1\vert}
\newcommand{ket}[1]{\vert#1\rangle}
\newcommand{Bra}[1]{\left\langle#1\right\vert}
\newcommand{Ket}[1]{\left\vert#1\right\rangle}
\newcommand{Braket}[2]{\left\langle #1\vphantom{#2}\mid{#2}\vphantom{#1}\right\rangle}
\newcommand{ketbra}[2]{\vert#1\rangle\!\langle#2\vert}
\newcommand{op}[1]{\hat{#1}}
\newcommand{Op}[1]{\hat{#1}}
\newcommand{dd}[0]{\,\text{d}}
\newcommand{Liouville}[0]{\mathcal{L}}
\newcommand{DynMap}[0]{\mathcal{E}}
\newcommand{identity}[0]{\mathbf{1}}
\newcommand{Norm}[1]{\lVert#1\rVert}
\newcommand{Abs}[1]{\left\vert#1\right\vert}
\newcommand{avg}[1]{\langle#1\rangle}
\newcommand{Avg}[1]{\left\langle#1\right\rangle}
\newcommand{AbsSq}[1]{\left\vert#1\right\vert^2}
\newcommand{Re}[0]{\operatorname{Re}}
\newcommand{Im}[0]{\operatorname{Im}}
\newcommand{toP}[0]{\omega_{12}}
\newcommand{toS}[0]{\omega_{23}}$
This example revisits the [Optimization of a State-to-State Transfer in a
Lambda System in the RWA](02_example_lambda_system_rwa_complex_pulse.ipynb),
attempting to make the control pulses robustness with respect to variations in
the pulse amplitude, through "ensemble optimization".
**Note**: This notebook uses some parallelization features (`parallel_map`/`multiprocessing`). Unfortunately, on Windows (and macOS with Python >= 3.8), `multiprocessing` does not work correctly for functions defined in a Jupyter notebook (due to the [spawn method](https://docs.python.org/3/library/multiprocessing.html#contexts-and-start-methods) being used on Windows, instead of Unix-`fork`, see also https://stackoverflow.com/questions/45719956). We can use the third-party [loky](https://loky.readthedocs.io/) library to fix this, but this significantly increases the overhead of multi-process parallelization. The use of parallelization here is for illustration only and makes no guarantee of actually improving the runtime of the optimization.
```
krotov.parallelization.set_parallelization(use_loky=True)
from krotov.parallelization import parallel_map
```
## Control objectives for population transfer in the Lambda system
As in the original example, we define the Hamiltonian for a Lambda system in
the rotating wave approximation, like this:

We set up the control fields and the Hamiltonian exactly as before:
```
def Omega_P1(t, args):
"""Guess for the real part of the pump pulse"""
Ω0 = 5.0
return Ω0 * krotov.shapes.blackman(t, t_start=2.0, t_stop=5.0)
def Omega_P2(t, args):
"""Guess for the imaginary part of the pump pulse"""
return 0.0
def Omega_S1(t, args):
"""Guess for the real part of the Stokes pulse"""
Ω0 = 5.0
return Ω0 * krotov.shapes.blackman(t, t_start=0.0, t_stop=3.0)
def Omega_S2(t, args):
"""Guess for the imaginary part of the Stokes pulse"""
return 0.0
tlist = np.linspace(0, 5, 500)
def hamiltonian(E1=0.0, E2=10.0, E3=5.0, omega_P=9.5, omega_S=4.5):
"""Lambda-system Hamiltonian in the RWA"""
# detunings
ΔP = E1 + omega_P - E2
ΔS = E3 + omega_S - E2
H0 = Qobj([[ΔP, 0.0, 0.0], [0.0, 0.0, 0.0], [0.0, 0.0, ΔS]])
HP_re = -0.5 * Qobj([[0.0, 1.0, 0.0], [1.0, 0.0, 0.0], [0.0, 0.0, 0.0]])
HP_im = -0.5 * Qobj([[0.0, 1.0j, 0.0], [-1.0j, 0.0, 0.0], [0.0, 0.0, 0.0]])
HS_re = -0.5 * Qobj([[0.0, 0.0, 0.0], [0.0, 0.0, 1.0], [0.0, 1.0, 0.0]])
HS_im = -0.5 * Qobj([[0.0, 0.0, 0.0], [0.0, 0.0, 1.0j], [0.0, -1.0j, 0.0]])
return [
H0,
[HP_re, Omega_P1],
[HP_im, Omega_P2],
[HS_re, Omega_S1],
[HS_im, Omega_S2],
]
H = hamiltonian()
```
The control objective is the realization of a phase sensitive $\ket{1}
\rightarrow \ket{3}$ transition in the lab frame. Thus, in the rotating frame,
we must take into account an additional phase factor.
```
ket1 = qutip.Qobj(np.array([1.0, 0.0, 0.0]))
ket2 = qutip.Qobj(np.array([0.0, 1.0, 0.0]))
ket3 = qutip.Qobj(np.array([0.0, 0.0, 1.0]))
def rwa_target_state(ket3, E2=10.0, omega_S=4.5, T=5):
return np.exp(1j * (E2 - omega_S) * T) * ket3
psi_target = rwa_target_state(ket3)
objective = krotov.Objective(initial_state=ket1, target=psi_target, H=H)
objectives = [objective]
objectives
```
## Robustness to amplitude fluctuations
A potential source of error is fluctuations in the pulse amplitude between
different runs of the experiment. To account for this, the `hamiltonian`
function above include a parameter `mu` that scales the pulse amplitudes by the
given factor.
We can analyze the result of the [Optimization of a State-to-State Transfer in
a Lambda System in the RWA](02_example_lambda_system_rwa_complex_pulse.ipynb)
with respect to such fluctuations. We load the earlier optimization result from
disk, and verify that the optimized controls produce the $\ket{1} \rightarrow
\ket{3}$ transition as desired.
```
opt_result_unperturbed = krotov.result.Result.load(
'lambda_rwa_opt_result.dump', objectives=[objective]
)
proj1 = qutip.ket2dm(ket1)
proj2 = qutip.ket2dm(ket2)
proj3 = qutip.ket2dm(ket3)
opt_unperturbed_dynamics = (
opt_result_unperturbed
.optimized_objectives[0]
.mesolve(tlist, e_ops=[proj1, proj2, proj3])
)
def plot_population(result):
fig, ax = plt.subplots()
ax.plot(result.times, result.expect[0], label='1')
ax.plot(result.times, result.expect[1], label='2')
ax.plot(result.times, result.expect[2], label='3')
ax.legend()
ax.set_xlabel('time')
ax.set_ylabel('population')
plt.show(fig)
plot_population(opt_unperturbed_dynamics)
```
Now we can analyze how robust this control is for variations of ±20% of the
pulse amplitude. Numerically, this is achieved by scaling the control
Hamiltonians with a pre-factor $\mu$.
```
def scale_control(H, *, mu):
"""Scale all control Hamiltonians by `mu`."""
H_scaled = []
for spec in H:
if isinstance(spec, list):
H_scaled.append([mu * spec[0], spec[1]])
else:
H_scaled.append(spec)
return H_scaled
```
For the analysis, we take the following sample of $\mu$ values:
```
mu_vals = np.linspace(0.75, 1.25, 33)
```
We measure the success of the transfer via the "population error", i.e., the
deviation from 1.0 of the population in state $\ket{3}$ at final time $T$.
```
def pop_error(obj, mu):
res = obj.mesolve(tlist, H=scale_control(obj.H, mu=mu), e_ops=[proj3])
return 1 - res.expect[0][-1]
def _f(mu):
# parallel_map needs a global function
return pop_error(opt_result_unperturbed.optimized_objectives[0], mu=mu)
pop_errors_norobust = parallel_map(_f, mu_vals)
def plot_robustness(mu_vals, pop_errors, pop_errors0=None):
fig, ax = plt.subplots()
ax.plot(mu_vals, pop_errors, label='1')
if pop_errors0 is not None:
ax.set_prop_cycle(None) # reset colors
if isinstance(pop_errors0, list):
for (i, pop_errors_prev) in enumerate(pop_errors0):
ax.plot(
mu_vals, pop_errors_prev, ls='dotted', label=("%d" % (-i))
)
else:
ax.plot(mu_vals, pop_errors0, ls='dotted', label='0')
ax.set_xlabel("relative coupling strength")
ax.set_ylabel(r"$1 - \vert \langle \Psi \vert 3 \rangle \vert^2$")
ax.axvspan(0.9, 1.1, alpha=0.25, color='red')
ax.set_yscale('log')
if pop_errors0 is not None:
ax.legend()
plt.show(fig)
plot_robustness(mu_vals, pop_errors_norobust)
```
The plot shows that as the pulse amplitude deviates from the optimal value, the
error rises quickly: our previous optimization result is not robust.
The highlighted region of ±10% is our "region of interest" within
which we would like the control to be robust by applying optimal control.
## Setting the ensemble objectives
They central idea of optimizing for robustness is to take multiple copies of
the Hamiltonian, sampling over the space of variations to which would
like to be robust, and optimize over the average of this ensemble.
Here, we sample 5 values of $\mu$ (including the unperturbed $\mu=1$) in the
region of interest, $\mu \in [0.9, 1.1]$.
```
ensemble_mu = [0.9, 0.95, 1.0, 1.05, 1.1]
```
The corresponding Hamiltonians are
```
ham_ensemble = [scale_control(objective.H, mu=mu) for mu in ensemble_mu]
```
The `krotov.objectives.ensemble_objectives` extends the original objective of a
single unperturbed state-to-state transition with one additional objective for
each ensemble Hamiltonian for $\mu \neq 1$:
```
ensemble_objectives = krotov.objectives.ensemble_objectives(
objectives, ham_ensemble, keep_original_objectives=False,
)
ensemble_objectives
```
It is important that all five objectives reference the same four control
pulses, as is the case here.
## Optimize
We use the same update shape $S(t)$ and $\lambda_a$ value as in the original optimization:
```
def S(t):
"""Scales the Krotov methods update of the pulse value at the time t"""
return krotov.shapes.flattop(t, 0.0, 5, 0.3, func='sinsq')
λ = 0.5
pulse_options = {
H[1][1]: dict(lambda_a=λ, update_shape=S),
H[2][1]: dict(lambda_a=λ, update_shape=S),
H[3][1]: dict(lambda_a=λ, update_shape=S),
H[4][1]: dict(lambda_a=λ, update_shape=S),
}
```
It will be interesting to see how the optimization progresses for each
individual element of the ensemble. Thus, we write an `info_hook` routine that
prints out a tabular overview of $1 - \Re\Braket{\Psi(T)}{3}_{\Op{H}_i}$ for
all $\Op{H}_i$ in the ensemble, as well as their average (the total functional
$J_T$ that is being minimized)
```
def print_J_T_per_target(**kwargs):
iteration = kwargs['iteration']
N = len(ensemble_mu)
if iteration == 0:
print(
"iteration "
+ "%11s " % "J_T(avg)"
+ " ".join([("J_T(μ=%.2f)" % μ) for μ in ensemble_mu])
)
J_T_vals = 1 - kwargs['tau_vals'].real
J_T = np.sum(J_T_vals) / N
print(
("%9d " % iteration)
+ ("%11.2e " % J_T)
+ " ".join([("%11.2e" % v) for v in J_T_vals])
)
```
We'll also want to look at the output of ``krotov.info_hooks.print_table``, but
in order to keep the output orderly, we will write that information to a file
`ensemble_opt.log`.
```
log_fh = open("ensemble_opt.log", "w", encoding="utf-8")
```
To speed up the optimization slightly, we parallelize across the five
objectives with appropriate `parallel_map` functions.
The optimization starts for the same guess pulses as the original [Optimization
of a State-to-State Transfer in a Lambda System in the
RWA](02_example_lambda_system_rwa_complex_pulse.ipynb). Generally, for a
robustness ensemble optimization, this will yield better results than trying to
take the optimized pulses for the unperturbed system as a guess.
```
opt_result = krotov.optimize_pulses(
ensemble_objectives,
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.chain(
print_J_T_per_target,
krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re, out=log_fh
),
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1e-3, name='J_T'),
krotov.convergence.check_monotonic_error,
),
parallel_map=(
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map_fw_prop_step,
),
iter_stop=12,
)
```
After twelve iterations (which were sufficient to produce an error $<10^{-3}$
in the original optimization), we find the average error over the ensemble to
be still above $>10^{-2}$. However, the error for $\mu = 1$ is only *slightly*
larger than in the original optimization; the lack of success is entirely due
to the large error for the other elements of the ensemble for $\mu \neq 1$.
Achieving robustness is hard!
We continue the optimization until the *average* error falls below $10^{-3}$:
```
dumpfile = "./ensemble_opt_result.dump"
if os.path.isfile(dumpfile):
opt_result = krotov.result.Result.load(dumpfile, objectives)
print_J_T_per_target(iteration=0, tau_vals=opt_result.tau_vals[12])
print(" ...")
n_iters = len(opt_result.tau_vals)
for i in range(n_iters - 10, n_iters):
print_J_T_per_target(iteration=i, tau_vals=opt_result.tau_vals[i])
else:
opt_result = krotov.optimize_pulses(
ensemble_objectives,
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.chain(
print_J_T_per_target,
krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re, out=log_fh
),
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1e-3, name='J_T'),
krotov.convergence.check_monotonic_error,
),
parallel_map=(
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map_fw_prop_step,
),
iter_stop=1000,
continue_from=opt_result,
)
opt_result.dump(dumpfile)
opt_result
log_fh.close()
```
Even now, the ideal Hamiltonian ($\mu = 1$) has the lowest error in the
ensemble by a significant margin. However, notice that the error in the $J_T$
for $\mu = 1$ is actually rising, while the errors for values of $\mu \neq 1$
are falling by a much larger value! This is a good thing: we sacrifice a little
bit of fidelity in the unperturbed dynamics to an increase in robustness.
The optimized "robust" pulse looks as follows:
```
def plot_pulse_amplitude_and_phase(pulse_real, pulse_imaginary, tlist):
ax1 = plt.subplot(211)
ax2 = plt.subplot(212)
amplitudes = [
np.sqrt(x * x + y * y) for x, y in zip(pulse_real, pulse_imaginary)
]
phases = [
np.arctan2(y, x) / np.pi for x, y in zip(pulse_real, pulse_imaginary)
]
ax1.plot(tlist, amplitudes)
ax1.set_xlabel('time')
ax1.set_ylabel('pulse amplitude')
ax2.plot(tlist, phases)
ax2.set_xlabel('time')
ax2.set_ylabel('pulse phase (π)')
plt.show()
print("pump pulse amplitude and phase:")
plot_pulse_amplitude_and_phase(
opt_result.optimized_controls[0], opt_result.optimized_controls[1], tlist
)
print("Stokes pulse amplitude and phase:")
plot_pulse_amplitude_and_phase(
opt_result.optimized_controls[2], opt_result.optimized_controls[3], tlist
)
```
and produces the dynamics (in the unperturbed system) shown below:
```
opt_robust_dynamics = opt_result.optimized_objectives[0].mesolve(
tlist, e_ops=[proj1, proj2, proj3]
)
plot_population(opt_robust_dynamics)
```
### Robustness analysis
When comparing the robustness of the "robust" optimized pulse to that obtained
from the original optimization for the unperturbed Hamiltonian, we should make
sure that we have converged to a comparable error: We would like to avoid the
suspicion that the ensemble error is below our threshold only because the error
for $\mu = 1$ is so much lower. Therefore, we continue the original unperturbed
optimization for a few more iterations, until we reach the same error $\approx
1.13 \times 10^{-4}$ that we found as the result of the ensemble optimization,
looking at $\mu=1$ only:
```
print("J_T(μ=1) = %.2e" % (1 - opt_result.tau_vals[-1][0].real))
opt_result_unperturbed_cont = krotov.optimize_pulses(
[objective],
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re,
show_g_a_int_per_pulse=True,
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1.13e-4, name='J_T'),
krotov.convergence.check_monotonic_error,
),
iter_stop=50,
continue_from=opt_result_unperturbed,
)
```
Now, we can compare the robustness of the optimized pulses from the original
unperturbed optimization (label "-1"), the continued unperturbed optimization
(label "0"), and the ensemble optimization (label "1"):
```
def _f(mu):
return pop_error(
opt_result_unperturbed_cont.optimized_objectives[0], mu=mu
)
pop_errors_norobust_cont = parallel_map(_f, mu_vals)
def _f(mu):
return pop_error(opt_result.optimized_objectives[0], mu=mu)
pop_errors_robust = parallel_map(_f, mu_vals)
plot_robustness(
mu_vals,
pop_errors_robust,
pop_errors0=[pop_errors_norobust_cont, pop_errors_norobust],
)
```
We see that without the ensemble optimization, we only lower the error for
exactly $\mu = 1$: the more we converge, the less robust the result. In
contrast, the ensemble optimization results in considerably lower errors (order
of magnitude!) throughout the highlighted "region of interest" and beyond.
|
github_jupyter
|
# NBVAL_IGNORE_OUTPUT
%load_ext watermark
import sys
import os
import qutip
import numpy as np
import scipy
import matplotlib
import matplotlib.pylab as plt
import krotov
from qutip import Qobj
import pickle
%watermark -v --iversions
krotov.parallelization.set_parallelization(use_loky=True)
from krotov.parallelization import parallel_map
def Omega_P1(t, args):
"""Guess for the real part of the pump pulse"""
Ω0 = 5.0
return Ω0 * krotov.shapes.blackman(t, t_start=2.0, t_stop=5.0)
def Omega_P2(t, args):
"""Guess for the imaginary part of the pump pulse"""
return 0.0
def Omega_S1(t, args):
"""Guess for the real part of the Stokes pulse"""
Ω0 = 5.0
return Ω0 * krotov.shapes.blackman(t, t_start=0.0, t_stop=3.0)
def Omega_S2(t, args):
"""Guess for the imaginary part of the Stokes pulse"""
return 0.0
tlist = np.linspace(0, 5, 500)
def hamiltonian(E1=0.0, E2=10.0, E3=5.0, omega_P=9.5, omega_S=4.5):
"""Lambda-system Hamiltonian in the RWA"""
# detunings
ΔP = E1 + omega_P - E2
ΔS = E3 + omega_S - E2
H0 = Qobj([[ΔP, 0.0, 0.0], [0.0, 0.0, 0.0], [0.0, 0.0, ΔS]])
HP_re = -0.5 * Qobj([[0.0, 1.0, 0.0], [1.0, 0.0, 0.0], [0.0, 0.0, 0.0]])
HP_im = -0.5 * Qobj([[0.0, 1.0j, 0.0], [-1.0j, 0.0, 0.0], [0.0, 0.0, 0.0]])
HS_re = -0.5 * Qobj([[0.0, 0.0, 0.0], [0.0, 0.0, 1.0], [0.0, 1.0, 0.0]])
HS_im = -0.5 * Qobj([[0.0, 0.0, 0.0], [0.0, 0.0, 1.0j], [0.0, -1.0j, 0.0]])
return [
H0,
[HP_re, Omega_P1],
[HP_im, Omega_P2],
[HS_re, Omega_S1],
[HS_im, Omega_S2],
]
H = hamiltonian()
ket1 = qutip.Qobj(np.array([1.0, 0.0, 0.0]))
ket2 = qutip.Qobj(np.array([0.0, 1.0, 0.0]))
ket3 = qutip.Qobj(np.array([0.0, 0.0, 1.0]))
def rwa_target_state(ket3, E2=10.0, omega_S=4.5, T=5):
return np.exp(1j * (E2 - omega_S) * T) * ket3
psi_target = rwa_target_state(ket3)
objective = krotov.Objective(initial_state=ket1, target=psi_target, H=H)
objectives = [objective]
objectives
opt_result_unperturbed = krotov.result.Result.load(
'lambda_rwa_opt_result.dump', objectives=[objective]
)
proj1 = qutip.ket2dm(ket1)
proj2 = qutip.ket2dm(ket2)
proj3 = qutip.ket2dm(ket3)
opt_unperturbed_dynamics = (
opt_result_unperturbed
.optimized_objectives[0]
.mesolve(tlist, e_ops=[proj1, proj2, proj3])
)
def plot_population(result):
fig, ax = plt.subplots()
ax.plot(result.times, result.expect[0], label='1')
ax.plot(result.times, result.expect[1], label='2')
ax.plot(result.times, result.expect[2], label='3')
ax.legend()
ax.set_xlabel('time')
ax.set_ylabel('population')
plt.show(fig)
plot_population(opt_unperturbed_dynamics)
def scale_control(H, *, mu):
"""Scale all control Hamiltonians by `mu`."""
H_scaled = []
for spec in H:
if isinstance(spec, list):
H_scaled.append([mu * spec[0], spec[1]])
else:
H_scaled.append(spec)
return H_scaled
mu_vals = np.linspace(0.75, 1.25, 33)
def pop_error(obj, mu):
res = obj.mesolve(tlist, H=scale_control(obj.H, mu=mu), e_ops=[proj3])
return 1 - res.expect[0][-1]
def _f(mu):
# parallel_map needs a global function
return pop_error(opt_result_unperturbed.optimized_objectives[0], mu=mu)
pop_errors_norobust = parallel_map(_f, mu_vals)
def plot_robustness(mu_vals, pop_errors, pop_errors0=None):
fig, ax = plt.subplots()
ax.plot(mu_vals, pop_errors, label='1')
if pop_errors0 is not None:
ax.set_prop_cycle(None) # reset colors
if isinstance(pop_errors0, list):
for (i, pop_errors_prev) in enumerate(pop_errors0):
ax.plot(
mu_vals, pop_errors_prev, ls='dotted', label=("%d" % (-i))
)
else:
ax.plot(mu_vals, pop_errors0, ls='dotted', label='0')
ax.set_xlabel("relative coupling strength")
ax.set_ylabel(r"$1 - \vert \langle \Psi \vert 3 \rangle \vert^2$")
ax.axvspan(0.9, 1.1, alpha=0.25, color='red')
ax.set_yscale('log')
if pop_errors0 is not None:
ax.legend()
plt.show(fig)
plot_robustness(mu_vals, pop_errors_norobust)
ensemble_mu = [0.9, 0.95, 1.0, 1.05, 1.1]
ham_ensemble = [scale_control(objective.H, mu=mu) for mu in ensemble_mu]
ensemble_objectives = krotov.objectives.ensemble_objectives(
objectives, ham_ensemble, keep_original_objectives=False,
)
ensemble_objectives
def S(t):
"""Scales the Krotov methods update of the pulse value at the time t"""
return krotov.shapes.flattop(t, 0.0, 5, 0.3, func='sinsq')
λ = 0.5
pulse_options = {
H[1][1]: dict(lambda_a=λ, update_shape=S),
H[2][1]: dict(lambda_a=λ, update_shape=S),
H[3][1]: dict(lambda_a=λ, update_shape=S),
H[4][1]: dict(lambda_a=λ, update_shape=S),
}
def print_J_T_per_target(**kwargs):
iteration = kwargs['iteration']
N = len(ensemble_mu)
if iteration == 0:
print(
"iteration "
+ "%11s " % "J_T(avg)"
+ " ".join([("J_T(μ=%.2f)" % μ) for μ in ensemble_mu])
)
J_T_vals = 1 - kwargs['tau_vals'].real
J_T = np.sum(J_T_vals) / N
print(
("%9d " % iteration)
+ ("%11.2e " % J_T)
+ " ".join([("%11.2e" % v) for v in J_T_vals])
)
log_fh = open("ensemble_opt.log", "w", encoding="utf-8")
opt_result = krotov.optimize_pulses(
ensemble_objectives,
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.chain(
print_J_T_per_target,
krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re, out=log_fh
),
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1e-3, name='J_T'),
krotov.convergence.check_monotonic_error,
),
parallel_map=(
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map_fw_prop_step,
),
iter_stop=12,
)
dumpfile = "./ensemble_opt_result.dump"
if os.path.isfile(dumpfile):
opt_result = krotov.result.Result.load(dumpfile, objectives)
print_J_T_per_target(iteration=0, tau_vals=opt_result.tau_vals[12])
print(" ...")
n_iters = len(opt_result.tau_vals)
for i in range(n_iters - 10, n_iters):
print_J_T_per_target(iteration=i, tau_vals=opt_result.tau_vals[i])
else:
opt_result = krotov.optimize_pulses(
ensemble_objectives,
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.chain(
print_J_T_per_target,
krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re, out=log_fh
),
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1e-3, name='J_T'),
krotov.convergence.check_monotonic_error,
),
parallel_map=(
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map,
krotov.parallelization.parallel_map_fw_prop_step,
),
iter_stop=1000,
continue_from=opt_result,
)
opt_result.dump(dumpfile)
opt_result
log_fh.close()
def plot_pulse_amplitude_and_phase(pulse_real, pulse_imaginary, tlist):
ax1 = plt.subplot(211)
ax2 = plt.subplot(212)
amplitudes = [
np.sqrt(x * x + y * y) for x, y in zip(pulse_real, pulse_imaginary)
]
phases = [
np.arctan2(y, x) / np.pi for x, y in zip(pulse_real, pulse_imaginary)
]
ax1.plot(tlist, amplitudes)
ax1.set_xlabel('time')
ax1.set_ylabel('pulse amplitude')
ax2.plot(tlist, phases)
ax2.set_xlabel('time')
ax2.set_ylabel('pulse phase (π)')
plt.show()
print("pump pulse amplitude and phase:")
plot_pulse_amplitude_and_phase(
opt_result.optimized_controls[0], opt_result.optimized_controls[1], tlist
)
print("Stokes pulse amplitude and phase:")
plot_pulse_amplitude_and_phase(
opt_result.optimized_controls[2], opt_result.optimized_controls[3], tlist
)
opt_robust_dynamics = opt_result.optimized_objectives[0].mesolve(
tlist, e_ops=[proj1, proj2, proj3]
)
plot_population(opt_robust_dynamics)
print("J_T(μ=1) = %.2e" % (1 - opt_result.tau_vals[-1][0].real))
opt_result_unperturbed_cont = krotov.optimize_pulses(
[objective],
pulse_options,
tlist,
propagator=krotov.propagators.expm,
chi_constructor=krotov.functionals.chis_re,
info_hook=krotov.info_hooks.print_table(
J_T=krotov.functionals.J_T_re,
show_g_a_int_per_pulse=True,
),
check_convergence=krotov.convergence.Or(
krotov.convergence.value_below(1.13e-4, name='J_T'),
krotov.convergence.check_monotonic_error,
),
iter_stop=50,
continue_from=opt_result_unperturbed,
)
def _f(mu):
return pop_error(
opt_result_unperturbed_cont.optimized_objectives[0], mu=mu
)
pop_errors_norobust_cont = parallel_map(_f, mu_vals)
def _f(mu):
return pop_error(opt_result.optimized_objectives[0], mu=mu)
pop_errors_robust = parallel_map(_f, mu_vals)
plot_robustness(
mu_vals,
pop_errors_robust,
pop_errors0=[pop_errors_norobust_cont, pop_errors_norobust],
)
| 0.549399 | 0.91939 |
# Digital House - Data Science a Distancia
## Trabajo Práctico 2
Prepara el dataset original con las características que se presentan en el [valuador de Properati](https://www.properati.com.ar/tools/valuador-propiedades)
### Autores: Daniel Borrino, Ivan Mongi, Jessica Polakoff, Julio Tentor
<p style="text-align:right;">Mayo 2022</p>
#### Aspectos técnicos
La notebook se ejecuta correctamente en una instalación estándar de Anaconda versión 4.11.0 build 3.21.6, Python 3.9.7
#### Librerías necesarias
```
import pandas as pd
import numpy as np
import seaborn as sns
import re
import matplotlib.pyplot as plt
data_url = "../Data/properatti.csv"
data = pd.read_csv(data_url, encoding="utf-8")
```
---
### Generación del dataset final
#### Eliminamos los valores nulos de la variable Target
```
#Limpiamos los NaN en el precio
data = data.dropna(axis=0, how='any', subset=['price_aprox_usd']).copy()
# Corregir incoherencias
mask = data['price_aprox_usd'] < 4000000
data = data[mask].copy()
mask = data['surface_covered_in_m2'] > 10
mask = np.logical_and(mask, data['surface_covered_in_m2'] < 1000)
data = data[mask].copy()
```
#### Seleccionamos solo Capital Federal y Bs.As. zonas Norte, Sur y Oeste
```
#Seleccionamos solo Capital Federal y Bs.As. zonas Norte, Sur y Oeste
iterar_state = ['Capital Federal',
'Bs.As. G.B.A. Zona Norte',
'Bs.As. G.B.A. Zona Sur',
'Bs.As. G.B.A. Zona Oeste']
data['state_name'] = [x if x in iterar_state else np.NaN for x in data['state_name']]
data = data.dropna(axis=0, how='any', subset=['state_name']).copy()
```
#### Seleccionamos solo Departamento, Casa y PH
```
#Seleccionamos solo Departamento, Casa y PH
iterar_tipo = data['property_type'].value_counts().head(3)
iterar_tipo = iterar_tipo.index
data['property_type'] = [x if x in iterar_tipo else np.NaN for x in data['property_type']]
data = data.dropna(axis=0, how='any', subset=['property_type']).copy()
data['place_name'].value_counts().head(100)
```
#### Seleccionamos solo Lugares con muchas observaciones
```
#Seleccionamos solo Lugares con muchas observaciones
iterar_place = data['place_name'].value_counts()[:100]
iterar_place = iterar_place.index
data['place_name'] = [x if x in iterar_place else np.NaN for x in data['place_name']]
data = data.dropna(axis=0, how='any', subset=['place_name']).copy()
```
#### Eliminamos Outliers para las variables que vamos a correlacionar:
---
#### Creacion de nuevas variables con valor predictivo:
##### Analisis para Cantidad de ambientes
```
def regex_to_values(col, reg, not_match=0) :
u"""Returns a serie with the result of apply the regular expresion to the column
the serie have a float value only when regular expression search() method found a match
col : column where to apply regular expresion
reg : regular expresion compiled
"""
serie = col.apply(lambda x : x if x is np.NaN else reg.search(x))
serie = serie.apply(lambda x : not_match if x is np.NaN or x is None else float(x.group(1)))
return serie
#Buscamos cantidad de ambientes
_pattern = '([1-2][0-9]?)(?= amb)'
_express = re.compile(_pattern, flags = re.IGNORECASE)
work = regex_to_values(data['description'], _express, 1)
data['ambientes'] = work
#realizamos la imputacion
mask = data['rooms'].notnull()
data.loc[mask, 'ambientes'] = data.loc[mask, 'rooms']
```
##### Analisis para Cantidad de baños
```
_pattern = '([1-2][0-9]?)(?= baño)'
_express = re.compile(_pattern, flags = re.IGNORECASE)
work = regex_to_values(data['description'], _express, 1)
data['baños'] = work
```
---
##### Nos proponemos encontrar amenities
```
def regex_to_tags(col, reg, match, not_match = np.NaN) :
u"""Returns a series with 'match' values result of apply the regular expresion to the column
the 'match' value will be when the regular expression search() method found a match
the 'not_match' value will be when the regular expression serach() method did not found a match
col : column where to apply regular expresion
reg : regular expresion compiled
"""
serie = col.apply(lambda x : x if x is np.NaN else reg.search(x))
serie = serie.apply(lambda x : match if x is not None else not_match)
return serie
#Buscamos Balcón
_pattern = 'balcon|balcón'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['balcón'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Cocheras
_pattern = 'cochera|garage|auto'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['cochera'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Parrillas
_pattern = 'parrilla'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['parrilla'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Piletas
_pattern = 'piscina|pileta'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['pileta'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Amoblado
_pattern = 'amoblado'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['amoblado'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Lavadero
_pattern = 'lavadero'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['lavadero'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Patio
_pattern = 'patio'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['patio'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Terraza
_pattern = 'terraza'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['terraza'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Jardin
_pattern = 'jardin'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['jardin'] = regex_to_tags(data['description'], _express, 1, 0)
data.reset_index(inplace=True)
data = data.drop(columns=['index', 'Unnamed: 0','operation', 'place_with_parent_names',
'country_name','geonames_id', 'lat-lon', 'lat', 'lon', 'price', 'currency',
'price_aprox_local_currency','floor', 'surface_total_in_m2', 'price_usd_per_m2',
'price_per_m2', 'rooms', 'expenses', 'properati_url', 'description', 'title', 'image_thumbnail']).copy()
data
data.shape
```
---
#### Sanity checks
```
data.isnull().sum()
data_final_url = "../Data/properatti_final7.csv"
data.to_csv(data_final_url)
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import re
import matplotlib.pyplot as plt
data_url = "../Data/properatti.csv"
data = pd.read_csv(data_url, encoding="utf-8")
#Limpiamos los NaN en el precio
data = data.dropna(axis=0, how='any', subset=['price_aprox_usd']).copy()
# Corregir incoherencias
mask = data['price_aprox_usd'] < 4000000
data = data[mask].copy()
mask = data['surface_covered_in_m2'] > 10
mask = np.logical_and(mask, data['surface_covered_in_m2'] < 1000)
data = data[mask].copy()
#Seleccionamos solo Capital Federal y Bs.As. zonas Norte, Sur y Oeste
iterar_state = ['Capital Federal',
'Bs.As. G.B.A. Zona Norte',
'Bs.As. G.B.A. Zona Sur',
'Bs.As. G.B.A. Zona Oeste']
data['state_name'] = [x if x in iterar_state else np.NaN for x in data['state_name']]
data = data.dropna(axis=0, how='any', subset=['state_name']).copy()
#Seleccionamos solo Departamento, Casa y PH
iterar_tipo = data['property_type'].value_counts().head(3)
iterar_tipo = iterar_tipo.index
data['property_type'] = [x if x in iterar_tipo else np.NaN for x in data['property_type']]
data = data.dropna(axis=0, how='any', subset=['property_type']).copy()
data['place_name'].value_counts().head(100)
#Seleccionamos solo Lugares con muchas observaciones
iterar_place = data['place_name'].value_counts()[:100]
iterar_place = iterar_place.index
data['place_name'] = [x if x in iterar_place else np.NaN for x in data['place_name']]
data = data.dropna(axis=0, how='any', subset=['place_name']).copy()
def regex_to_values(col, reg, not_match=0) :
u"""Returns a serie with the result of apply the regular expresion to the column
the serie have a float value only when regular expression search() method found a match
col : column where to apply regular expresion
reg : regular expresion compiled
"""
serie = col.apply(lambda x : x if x is np.NaN else reg.search(x))
serie = serie.apply(lambda x : not_match if x is np.NaN or x is None else float(x.group(1)))
return serie
#Buscamos cantidad de ambientes
_pattern = '([1-2][0-9]?)(?= amb)'
_express = re.compile(_pattern, flags = re.IGNORECASE)
work = regex_to_values(data['description'], _express, 1)
data['ambientes'] = work
#realizamos la imputacion
mask = data['rooms'].notnull()
data.loc[mask, 'ambientes'] = data.loc[mask, 'rooms']
_pattern = '([1-2][0-9]?)(?= baño)'
_express = re.compile(_pattern, flags = re.IGNORECASE)
work = regex_to_values(data['description'], _express, 1)
data['baños'] = work
def regex_to_tags(col, reg, match, not_match = np.NaN) :
u"""Returns a series with 'match' values result of apply the regular expresion to the column
the 'match' value will be when the regular expression search() method found a match
the 'not_match' value will be when the regular expression serach() method did not found a match
col : column where to apply regular expresion
reg : regular expresion compiled
"""
serie = col.apply(lambda x : x if x is np.NaN else reg.search(x))
serie = serie.apply(lambda x : match if x is not None else not_match)
return serie
#Buscamos Balcón
_pattern = 'balcon|balcón'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['balcón'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Cocheras
_pattern = 'cochera|garage|auto'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['cochera'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Parrillas
_pattern = 'parrilla'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['parrilla'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Piletas
_pattern = 'piscina|pileta'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['pileta'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Amoblado
_pattern = 'amoblado'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['amoblado'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Lavadero
_pattern = 'lavadero'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['lavadero'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Patio
_pattern = 'patio'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['patio'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Terraza
_pattern = 'terraza'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['terraza'] = regex_to_tags(data['description'], _express, 1, 0)
#Buscamos Jardin
_pattern = 'jardin'
_express = re.compile(_pattern, flags = re.IGNORECASE)
data['jardin'] = regex_to_tags(data['description'], _express, 1, 0)
data.reset_index(inplace=True)
data = data.drop(columns=['index', 'Unnamed: 0','operation', 'place_with_parent_names',
'country_name','geonames_id', 'lat-lon', 'lat', 'lon', 'price', 'currency',
'price_aprox_local_currency','floor', 'surface_total_in_m2', 'price_usd_per_m2',
'price_per_m2', 'rooms', 'expenses', 'properati_url', 'description', 'title', 'image_thumbnail']).copy()
data
data.shape
data.isnull().sum()
data_final_url = "../Data/properatti_final7.csv"
data.to_csv(data_final_url)
| 0.410047 | 0.939443 |
### Determine the optimal K number
* This notebook determines the optimal number of K based on K-means algorithm using Elbow method
* Three data processing approaches:
* Approach I. 514 features were selected out of the 1063 features. See the details in separate feature selection files. All variables have been transformed into numerical values using low rank representation method, the reduced dimension matrix is used here to determine optimal K.
* Approach II. Drop all the categorical values and impute the numerical values with means, the method will be applied on three parallel ramdonly generated subsamples (1.25%) of the original size data.
* Approach III. Drop all the categorical values and impute the numerical values with means, use PCA to reduce the original 1063 dimensions. the method will be applied on three parallel ramdonly generated subsamples (1.25%) of the original size data.
* Approach IV 514 features were selected out of the 1063 features. Then use LASSO and PCA to contidue reduce dimension to 44 variables (all of them turned out to be numerical variables). Impute NA with means.
Reference: : https://github.com/sarguido/k-means-clustering/blob/master/k-means-clustering.ipynb
```
from sklearn.cluster import KMeans
import pandas as pd
import numpy as np
from scipy.spatial.distance import cdist, pdist
from matplotlib import pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import silhouette_score, silhouette_samples
import matplotlib.cm as cm
from sklearn.decomposition import PCA
%matplotlib inline
```
#### Approach I. All variables have been transformed into numerical values using low rank representation method, the reduced dimension matrix is used here to determine optimal K. --This results was not completed due to memory error.
```
df = pd.read_csv('/mnt/UW/outputDataset/lowrank_rep.csv.gz')
df.shape
df.head()
# records number is too large, use a sample to perform analysis
import random
random.seed(123)
df_sample = df.sample(frac=0.0125, replace=False)
df_sample = df_sample.fillna(df_sample.mean())
X = StandardScaler().fit_transform(df_sample)
import time
start = time.time()
# Determine your k range
k_range = range(3,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
#### Approach II. Drop all the categorical values and impute the numerical values with means, the method will be applied on three parallel ramdonly generated subsamples (1.25%) of the original size data
```
# Use a smaller data set to save time
df1 = pd.read_csv('PHBsample14_sss.csv', low_memory=False)
df2 = pd.read_csv('PHBsample15_sss.csv', low_memory=False)
df3 = pd.read_csv('PHBsample16_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df1.drop('Unnamed: 0', axis=1, inplace=True)
df2.drop('Unnamed: 0', axis=1, inplace=True)
df3.drop('Unnamed: 0', axis=1, inplace=True)
selected_variable = pd.read_csv('selectedVariables.csv')
selected_variable.drop('Unnamed: 0', axis=1, inplace=True)
df1_1 = df1[df1.columns.intersection(selected_variable.columns)]
df2_1 = df2[df2.columns.intersection(selected_variable.columns)]
df3_1 = df3[df3.columns.intersection(selected_variable.columns)]
# Drop all the categoricald data for now.
df1_2 = df1_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df1_3 = df1_2.fillna(df1_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df1_3)
```
##### Subsample 1
```
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
##### Subsample 2
```
# Drop all the categoricald data for now.
df2_2 = df2_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df2_3 = df2_2.fillna(df2_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df2_3)
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
##### Subsample 3
```
# Drop all the categoricald data for now.
df3_2 = df3_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df3_3 = df3_2.fillna(df3_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df3_3)
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
fig.savefig('OptimalK_elbum.png', dpi=100)
```
#### Approach III. Use PCA to reduce the original 1063 dimensions. Drop all the categorical values and impute the numerical values with means, the method will be applied on a ramdonly generated subsamples (1.25%) of the original size data.
##### subsample 1
```
df = pd.read_csv('PHBsample14_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
##### subsample 2
```
df = pd.read_csv('PHBsample15_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
##### subsample 3
```
df = pd.read_csv('PHBsample16_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
#### Approach IV 514 features were selected out of the 1063 features. Then use LASSO and PCA to contidue reduce dimension to 44 variables (all of them turned out to be numerical variables). Impute NA with means.
```
df = pd.read_csv('/mnt/UW/outputDataset/pca_reduced_LASSO.csv')
# records number is too large, use a sample to perform analysis
import random
random.seed(123)
df_sample = df.sample(frac=0.0125, replace=False)
df_sample.shape
df_sample = df_sample.fillna(df_sample.mean())
X = StandardScaler().fit_transform(df_sample)
import time
start = time.time()
# Determine your k range
k_range = range(3,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares also known as inertia
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
```
|
github_jupyter
|
from sklearn.cluster import KMeans
import pandas as pd
import numpy as np
from scipy.spatial.distance import cdist, pdist
from matplotlib import pyplot as plt
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import silhouette_score, silhouette_samples
import matplotlib.cm as cm
from sklearn.decomposition import PCA
%matplotlib inline
df = pd.read_csv('/mnt/UW/outputDataset/lowrank_rep.csv.gz')
df.shape
df.head()
# records number is too large, use a sample to perform analysis
import random
random.seed(123)
df_sample = df.sample(frac=0.0125, replace=False)
df_sample = df_sample.fillna(df_sample.mean())
X = StandardScaler().fit_transform(df_sample)
import time
start = time.time()
# Determine your k range
k_range = range(3,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
# Use a smaller data set to save time
df1 = pd.read_csv('PHBsample14_sss.csv', low_memory=False)
df2 = pd.read_csv('PHBsample15_sss.csv', low_memory=False)
df3 = pd.read_csv('PHBsample16_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df1.drop('Unnamed: 0', axis=1, inplace=True)
df2.drop('Unnamed: 0', axis=1, inplace=True)
df3.drop('Unnamed: 0', axis=1, inplace=True)
selected_variable = pd.read_csv('selectedVariables.csv')
selected_variable.drop('Unnamed: 0', axis=1, inplace=True)
df1_1 = df1[df1.columns.intersection(selected_variable.columns)]
df2_1 = df2[df2.columns.intersection(selected_variable.columns)]
df3_1 = df3[df3.columns.intersection(selected_variable.columns)]
# Drop all the categoricald data for now.
df1_2 = df1_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df1_3 = df1_2.fillna(df1_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df1_3)
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
# Drop all the categoricald data for now.
df2_2 = df2_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df2_3 = df2_2.fillna(df2_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df2_3)
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
# Drop all the categoricald data for now.
df3_2 = df3_1.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df3_3 = df3_2.fillna(df3_2.mean())
# Standarize the data points
X = StandardScaler().fit_transform(df3_3)
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
fig.savefig('OptimalK_elbum.png', dpi=100)
df = pd.read_csv('PHBsample14_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
df = pd.read_csv('PHBsample15_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
df = pd.read_csv('PHBsample16_sss.csv', low_memory=False)
# drop the column resulted from sampling of the original data set
df.drop('Unnamed: 0', axis=1, inplace=True)
# In order to run K-means, drop all the categoricald data for now.
df = df.select_dtypes(include=['float64', 'int64'])
# Impute missing values with means
df = df.fillna(df.mean())
pca = PCA(2, svd_solver='randomized')
pca.fit(df)
df_reduced = pca.fit_transform(df)
df_reduced = StandardScaler().fit_transform(df_reduced)
X = df_reduced
import time
start = time.time()
# Determine your k range
k_range = range(1,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
df = pd.read_csv('/mnt/UW/outputDataset/pca_reduced_LASSO.csv')
# records number is too large, use a sample to perform analysis
import random
random.seed(123)
df_sample = df.sample(frac=0.0125, replace=False)
df_sample.shape
df_sample = df_sample.fillna(df_sample.mean())
X = StandardScaler().fit_transform(df_sample)
import time
start = time.time()
# Determine your k range
k_range = range(3,14)
# Fit the kmeans model for each n_clusters = k
k_means_var = [KMeans(n_clusters=k).fit(X) for k in k_range]
# Pull out the cluster centers for each model
centroids = [X.cluster_centers_ for X in k_means_var]
# Calculate the Euclidean distance from
# each point to each cluster center
k_euclid = [cdist(X, cent, 'euclidean') for cent in centroids]
dist = [np.min(ke,axis=1) for ke in k_euclid]
# Total within-cluster sum of squares also known as inertia
wcss = [sum(d**2) for d in dist]
# The total sum of squares
tss = sum(pdist(X)**2)/X.shape[0]
# The between-cluster sum of squares
bss = tss - wcss
end = time.time()
hours, rem = divmod(end-start, 3600)
minutes, seconds = divmod(rem, 60)
print("{:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds))
# elbow curve
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot(k_range, bss/tss*100, 'b*-')
ax.set_ylim((0,100))
plt.grid(True)
plt.xlabel('n_clusters')
plt.ylabel('Percentage of variance explained')
plt.title('Variance Explained vs. k')
| 0.865281 | 0.972389 |
# Logic
This Jupyter notebook acts as supporting material for topics covered in __Chapter 6 Logical Agents__, __Chapter 7 First-Order Logic__ and __Chapter 8 Inference in First-Order Logic__ of the book *[Artificial Intelligence: A Modern Approach](http://aima.cs.berkeley.edu)*. We make use the implementations in the [logic.py](https://github.com/aimacode/aima-python/blob/master/logic.py) module. See the [intro notebook](https://github.com/aimacode/aima-python/blob/master/intro.ipynb) for instructions.
Let's first import everything from the `logic` module.
```
from utils import *
from logic import *
from notebook import psource
```
## CONTENTS
- Logical sentences
- Expr
- PropKB
- Knowledge-based agents
- Inference in propositional knowledge base
- Truth table enumeration
- Proof by resolution
- Forward and backward chaining
- DPLL
- WalkSAT
- SATPlan
- FolKB
- Inference in first order knowledge base
- Unification
- Forward chaining algorithm
- Backward chaining algorithm
## Logical Sentences
The `Expr` class is designed to represent any kind of mathematical expression. The simplest type of `Expr` is a symbol, which can be defined with the function `Symbol`:
```
Symbol('x')
```
Or we can define multiple symbols at the same time with the function `symbols`:
```
(x, y, P, Q, f) = symbols('x, y, P, Q, f')
```
We can combine `Expr`s with the regular Python infix and prefix operators. Here's how we would form the logical sentence "P and not Q":
```
P & ~Q
```
This works because the `Expr` class overloads the `&` operator with this definition:
```python
def __and__(self, other): return Expr('&', self, other)```
and does similar overloads for the other operators. An `Expr` has two fields: `op` for the operator, which is always a string, and `args` for the arguments, which is a tuple of 0 or more expressions. By "expression," I mean either an instance of `Expr`, or a number. Let's take a look at the fields for some `Expr` examples:
```
sentence = P & ~Q
sentence.op
sentence.args
P.op
P.args
Pxy = P(x, y)
Pxy.op
Pxy.args
```
It is important to note that the `Expr` class does not define the *logic* of Propositional Logic sentences; it just gives you a way to *represent* expressions. Think of an `Expr` as an [abstract syntax tree](https://en.wikipedia.org/wiki/Abstract_syntax_tree). Each of the `args` in an `Expr` can be either a symbol, a number, or a nested `Expr`. We can nest these trees to any depth. Here is a deply nested `Expr`:
```
3 * f(x, y) + P(y) / 2 + 1
```
## Operators for Constructing Logical Sentences
Here is a table of the operators that can be used to form sentences. Note that we have a problem: we want to use Python operators to make sentences, so that our programs (and our interactive sessions like the one here) will show simple code. But Python does not allow implication arrows as operators, so for now we have to use a more verbose notation that Python does allow: `|'==>'|` instead of just `==>`. Alternately, you can always use the more verbose `Expr` constructor forms:
| Operation | Book | Python Infix Input | Python Output | Python `Expr` Input
|--------------------------|----------------------|-------------------------|---|---|
| Negation | ¬ P | `~P` | `~P` | `Expr('~', P)`
| And | P ∧ Q | `P & Q` | `P & Q` | `Expr('&', P, Q)`
| Or | P ∨ Q | `P`<tt> | </tt>`Q`| `P`<tt> | </tt>`Q` | `Expr('`|`', P, Q)`
| Inequality (Xor) | P ≠ Q | `P ^ Q` | `P ^ Q` | `Expr('^', P, Q)`
| Implication | P → Q | `P` <tt>|</tt>`'==>'`<tt>|</tt> `Q` | `P ==> Q` | `Expr('==>', P, Q)`
| Reverse Implication | Q ← P | `Q` <tt>|</tt>`'<=='`<tt>|</tt> `P` |`Q <== P` | `Expr('<==', Q, P)`
| Equivalence | P ↔ Q | `P` <tt>|</tt>`'<=>'`<tt>|</tt> `Q` |`P <=> Q` | `Expr('<=>', P, Q)`
Here's an example of defining a sentence with an implication arrow:
```
~(P & Q) |'==>'| (~P | ~Q)
```
## `expr`: a Shortcut for Constructing Sentences
If the `|'==>'|` notation looks ugly to you, you can use the function `expr` instead:
```
expr('~(P & Q) ==> (~P | ~Q)')
```
`expr` takes a string as input, and parses it into an `Expr`. The string can contain arrow operators: `==>`, `<==`, or `<=>`, which are handled as if they were regular Python infix operators. And `expr` automatically defines any symbols, so you don't need to pre-define them:
```
expr('sqrt(b ** 2 - 4 * a * c)')
```
For now that's all you need to know about `expr`. If you are interested, we explain the messy details of how `expr` is implemented and how `|'==>'|` is handled in the appendix.
## Propositional Knowledge Bases: `PropKB`
The class `PropKB` can be used to represent a knowledge base of propositional logic sentences.
We see that the class `KB` has four methods, apart from `__init__`. A point to note here: the `ask` method simply calls the `ask_generator` method. Thus, this one has already been implemented, and what you'll have to actually implement when you create your own knowledge base class (though you'll probably never need to, considering the ones we've created for you) will be the `ask_generator` function and not the `ask` function itself.
The class `PropKB` now.
* `__init__(self, sentence=None)` : The constructor `__init__` creates a single field `clauses` which will be a list of all the sentences of the knowledge base. Note that each one of these sentences will be a 'clause' i.e. a sentence which is made up of only literals and `or`s.
* `tell(self, sentence)` : When you want to add a sentence to the KB, you use the `tell` method. This method takes a sentence, converts it to its CNF, extracts all the clauses, and adds all these clauses to the `clauses` field. So, you need not worry about `tell`ing only clauses to the knowledge base. You can `tell` the knowledge base a sentence in any form that you wish; converting it to CNF and adding the resulting clauses will be handled by the `tell` method.
* `ask_generator(self, query)` : The `ask_generator` function is used by the `ask` function. It calls the `tt_entails` function, which in turn returns `True` if the knowledge base entails query and `False` otherwise. The `ask_generator` itself returns an empty dict `{}` if the knowledge base entails query and `None` otherwise. This might seem a little bit weird to you. After all, it makes more sense just to return a `True` or a `False` instead of the `{}` or `None` But this is done to maintain consistency with the way things are in First-Order Logic, where an `ask_generator` function is supposed to return all the substitutions that make the query true. Hence the dict, to return all these substitutions. I will be mostly be using the `ask` function which returns a `{}` or a `False`, but if you don't like this, you can always use the `ask_if_true` function which returns a `True` or a `False`.
* `retract(self, sentence)` : This function removes all the clauses of the sentence given, from the knowledge base. Like the `tell` function, you don't have to pass clauses to remove them from the knowledge base; any sentence will do fine. The function will take care of converting that sentence to clauses and then remove those.
## Wumpus World KB
Let us create a `PropKB` for the wumpus world with the sentences mentioned in `section 7.4.3`.
```
wumpus_kb = PropKB()
```
We define the symbols we use in our clauses.<br/>
$P_{x, y}$ is true if there is a pit in `[x, y]`.<br/>
$B_{x, y}$ is true if the agent senses breeze in `[x, y]`.<br/>
```
P11, P12, P21, P22, P31, B11, B21 = expr('P11, P12, P21, P22, P31, B11, B21')
```
Now we tell sentences based on `section 7.4.3`.<br/>
There is no pit in `[1,1]`.
```
wumpus_kb.tell(~P11)
```
A square is breezy if and only if there is a pit in a neighboring square. This has to be stated for each square but for now, we include just the relevant squares.
```
wumpus_kb.tell(B11 | '<=>' | ((P12 | P21)))
wumpus_kb.tell(B21 | '<=>' | ((P11 | P22 | P31)))
```
Now we include the breeze percepts for the first two squares leading up to the situation in `Figure 7.3(b)`
```
wumpus_kb.tell(~B11)
wumpus_kb.tell(B21)
```
We can check the clauses stored in a `KB` by accessing its `clauses` variable
```
wumpus_kb.clauses
```
We see that the equivalence $B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was automatically converted to two implications which were inturn converted to CNF which is stored in the `KB`.<br/>
$B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was split into $B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ and $B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$.<br/>
$B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ was converted to $P_{1, 2} \lor P_{2, 1} \lor \neg B_{1, 1}$.<br/>
$B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$ was converted to $\neg (P_{1, 2} \lor P_{2, 1}) \lor B_{1, 1}$ which becomes $(\neg P_{1, 2} \lor B_{1, 1}) \land (\neg P_{2, 1} \lor B_{1, 1})$ after applying De Morgan's laws and distributing the disjunction.<br/>
$B_{2, 1} \iff (P_{1, 1} \lor P_{2, 2} \lor P_{3, 2})$ is converted in similar manner.
## Knowledge based agents
A knowledge-based agent is a simple generic agent that maintains and handles a knowledge base.
The knowledge base may initially contain some background knowledge.
<br>
The purpose of a KB agent is to provide a level of abstraction over knowledge-base manipulation and is to be used as a base class for agents that work on a knowledge base.
<br>
Given a percept, the KB agent adds the percept to its knowledge base, asks the knowledge base for the best action, and tells the knowledge base that it has infact taken that action.
<br>
Our implementation of `KB-Agent` is encapsulated in a class `KB_AgentProgram` which inherits from the `KB` class.
<br>
Let's have a look.
```
psource(KB_AgentProgram)
```
The helper functions `make_percept_sentence`, `make_action_query` and `make_action_sentence` are all aptly named and as expected,
`make_percept_sentence` makes first-order logic sentences about percepts we want our agent to receive,
`make_action_query` asks the underlying `KB` about the action that should be taken and
`make_action_sentence` tells the underlying `KB` about the action it has just taken.
## Inference in Propositional Knowledge Base
In this section we will look at two algorithms to check if a sentence is entailed by the `KB`. Our goal is to decide whether $\text{KB} \vDash \alpha$ for some sentence $\alpha$.
### Truth Table Enumeration
It is a model-checking approach which, as the name suggests, enumerates all possible models in which the `KB` is true and checks if $\alpha$ is also true in these models. We list the $n$ symbols in the `KB` and enumerate the $2^{n}$ models in a depth-first manner and check the truth of `KB` and $\alpha$.
```
psource(tt_check_all)
```
The algorithm basically computes every line of the truth table $KB\implies \alpha$ and checks if it is true everywhere.
<br>
If symbols are defined, the routine recursively constructs every combination of truth values for the symbols and then,
it checks whether `model` is consistent with `kb`.
The given models correspond to the lines in the truth table,
which have a `true` in the KB column,
and for these lines it checks whether the query evaluates to true
<br>
`result = pl_true(alpha, model)`.
<br>
<br>
In short, `tt_check_all` evaluates this logical expression for each `model`
<br>
`pl_true(kb, model) => pl_true(alpha, model)`
<br>
which is logically equivalent to
<br>
`pl_true(kb, model) & ~pl_true(alpha, model)`
<br>
that is, the knowledge base and the negation of the query are logically inconsistent.
<br>
<br>
`tt_entails()` just extracts the symbols from the query and calls `tt_check_all()` with the proper parameters.
```
psource(tt_entails)
```
Keep in mind that for two symbols P and Q, P => Q is false only when P is `True` and Q is `False`.
Example usage of `tt_entails()`:
```
tt_entails(P & Q, Q)
```
P & Q is True only when both P and Q are True. Hence, (P & Q) => Q is True
```
tt_entails(P | Q, Q)
tt_entails(P | Q, P)
```
If we know that P | Q is true, we cannot infer the truth values of P and Q.
Hence (P | Q) => Q is False and so is (P | Q) => P.
```
(A, B, C, D, E, F, G) = symbols('A, B, C, D, E, F, G')
tt_entails(A & (B | C) & D & E & ~(F | G), A & D & E & ~F & ~G)
```
We can see that for the KB to be true, A, D, E have to be True and F and G have to be False.
Nothing can be said about B or C.
Coming back to our problem, note that `tt_entails()` takes an `Expr` which is a conjunction of clauses as the input instead of the `KB` itself.
You can use the `ask_if_true()` method of `PropKB` which does all the required conversions.
Let's check what `wumpus_kb` tells us about $P_{1, 1}$.
```
wumpus_kb.ask_if_true(~P11), wumpus_kb.ask_if_true(P11)
```
Looking at Figure 7.9 we see that in all models in which the knowledge base is `True`, $P_{1, 1}$ is `False`. It makes sense that `ask_if_true()` returns `True` for $\alpha = \neg P_{1, 1}$ and `False` for $\alpha = P_{1, 1}$. This begs the question, what if $\alpha$ is `True` in only a portion of all models. Do we return `True` or `False`? This doesn't rule out the possibility of $\alpha$ being `True` but it is not entailed by the `KB` so we return `False` in such cases. We can see this is the case for $P_{2, 2}$ and $P_{3, 1}$.
```
wumpus_kb.ask_if_true(~P22), wumpus_kb.ask_if_true(P22)
```
### Proof by Resolution
Recall that our goal is to check whether $\text{KB} \vDash \alpha$ i.e. is $\text{KB} \implies \alpha$ true in every model. Suppose we wanted to check if $P \implies Q$ is valid. We check the satisfiability of $\neg (P \implies Q)$, which can be rewritten as $P \land \neg Q$. If $P \land \neg Q$ is unsatisfiable, then $P \implies Q$ must be true in all models. This gives us the result "$\text{KB} \vDash \alpha$ <em>if and only if</em> $\text{KB} \land \neg \alpha$ is unsatisfiable".<br/>
This technique corresponds to <em>proof by <strong>contradiction</strong></em>, a standard mathematical proof technique. We assume $\alpha$ to be false and show that this leads to a contradiction with known axioms in $\text{KB}$. We obtain a contradiction by making valid inferences using inference rules. In this proof we use a single inference rule, <strong>resolution</strong> which states $(l_1 \lor \dots \lor l_k) \land (m_1 \lor \dots \lor m_n) \land (l_i \iff \neg m_j) \implies l_1 \lor \dots \lor l_{i - 1} \lor l_{i + 1} \lor \dots \lor l_k \lor m_1 \lor \dots \lor m_{j - 1} \lor m_{j + 1} \lor \dots \lor m_n$. Applying the resolution yeilds us a clause which we add to the KB. We keep doing this until:
* There are no new clauses that can be added, in which case $\text{KB} \nvDash \alpha$.
* Two clauses resolve to yield the <em>empty clause</em>, in which case $\text{KB} \vDash \alpha$.
The <em>empty clause</em> is equivalent to <em>False</em> because it arises only from resolving two complementary
unit clauses such as $P$ and $\neg P$ which is a contradiction as both $P$ and $\neg P$ can't be <em>True</em> at the same time.
There is one catch however, the algorithm that implements proof by resolution cannot handle complex sentences.
Implications and bi-implications have to be simplified into simpler clauses.
We already know that *every sentence of a propositional logic is logically equivalent to a conjunction of clauses*.
We will use this fact to our advantage and simplify the input sentence into the **conjunctive normal form** (CNF) which is a conjunction of disjunctions of literals.
For eg:
<br>
$$(A\lor B)\land (\neg B\lor C\lor\neg D)\land (D\lor\neg E)$$
This is equivalent to the POS (Product of sums) form in digital electronics.
<br>
Here's an outline of how the conversion is done:
1. Convert bi-implications to implications
<br>
$\alpha\iff\beta$ can be written as $(\alpha\implies\beta)\land(\beta\implies\alpha)$
<br>
This also applies to compound sentences
<br>
$\alpha\iff(\beta\lor\gamma)$ can be written as $(\alpha\implies(\beta\lor\gamma))\land((\beta\lor\gamma)\implies\alpha)$
<br>
2. Convert implications to their logical equivalents
<br>
$\alpha\implies\beta$ can be written as $\neg\alpha\lor\beta$
<br>
3. Move negation inwards
<br>
CNF requires atomic literals. Hence, negation cannot appear on a compound statement.
De Morgan's laws will be helpful here.
<br>
$\neg(\alpha\land\beta)\equiv(\neg\alpha\lor\neg\beta)$
<br>
$\neg(\alpha\lor\beta)\equiv(\neg\alpha\land\neg\beta)$
<br>
4. Distribute disjunction over conjunction
<br>
Disjunction and conjunction are distributive over each other.
Now that we only have conjunctions, disjunctions and negations in our expression,
we will distribute disjunctions over conjunctions wherever possible as this will give us a sentence which is a conjunction of simpler clauses,
which is what we wanted in the first place.
<br>
We need a term of the form
<br>
$(\alpha_{1}\lor\alpha_{2}\lor\alpha_{3}...)\land(\beta_{1}\lor\beta_{2}\lor\beta_{3}...)\land(\gamma_{1}\lor\gamma_{2}\lor\gamma_{3}...)\land...$
<br>
<br>
The `to_cnf` function executes this conversion using helper subroutines.
```
psource(to_cnf)
```
`to_cnf` calls three subroutines.
<br>
`eliminate_implications` converts bi-implications and implications to their logical equivalents.
<br>
`move_not_inwards` removes negations from compound statements and moves them inwards using De Morgan's laws.
<br>
`distribute_and_over_or` distributes disjunctions over conjunctions.
<br>
Run the cell below for implementation details.
```
psource(eliminate_implications)
psource(move_not_inwards)
psource(distribute_and_over_or)
```
Let's convert some sentences to see how it works
```
A, B, C, D = expr('A, B, C, D')
to_cnf(A |'<=>'| B)
to_cnf(A |'<=>'| (B & C))
to_cnf(A & (B | (C & D)))
to_cnf((A |'<=>'| ~B) |'==>'| (C | ~D))
```
Coming back to our resolution problem, we can see how the `to_cnf` function is utilized here
```
psource(pl_resolution)
pl_resolution(wumpus_kb, ~P11), pl_resolution(wumpus_kb, P11)
pl_resolution(wumpus_kb, ~P22), pl_resolution(wumpus_kb, P22)
```
### Forward and backward chaining
Previously, we said we will look at two algorithms to check if a sentence is entailed by the `KB`,
but here's a third one.
The difference here is that our goal now is to determine if a knowledge base of definite clauses entails a single proposition symbol *q* - the query.
There is a catch however, the knowledge base can only contain **Horn clauses**.
<br>
#### Horn Clauses
Horn clauses can be defined as a *disjunction* of *literals* with **at most** one positive literal.
<br>
A Horn clause with exactly one positive literal is called a *definite clause*.
<br>
A Horn clause might look like
<br>
$\neg a\lor\neg b\lor\neg c\lor\neg d... \lor z$
<br>
This, coincidentally, is also a definite clause.
<br>
Using De Morgan's laws, the example above can be simplified to
<br>
$a\land b\land c\land d ... \implies z$
<br>
This seems like a logical representation of how humans process known data and facts.
Assuming percepts `a`, `b`, `c`, `d` ... to be true simultaneously, we can infer `z` to also be true at that point in time.
There are some interesting aspects of Horn clauses that make algorithmic inference or *resolution* easier.
- Definite clauses can be written as implications:
<br>
The most important simplification a definite clause provides is that it can be written as an implication.
The premise (or the knowledge that leads to the implication) is a conjunction of positive literals.
The conclusion (the implied statement) is also a positive literal.
The sentence thus becomes easier to understand.
The premise and the conclusion are conventionally called the *body* and the *head* respectively.
A single positive literal is called a *fact*.
- Forward chaining and backward chaining can be used for inference from Horn clauses:
<br>
Forward chaining is semantically identical to `AND-OR-Graph-Search` from the chapter on search algorithms.
Implementational details will be explained shortly.
- Deciding entailment with Horn clauses is linear in size of the knowledge base:
<br>
Surprisingly, the forward and backward chaining algorithms traverse each element of the knowledge base at most once, greatly simplifying the problem.
<br>
<br>
The function `pl_fc_entails` implements forward chaining to see if a knowledge base `KB` entails a symbol `q`.
<br>
Before we proceed further, note that `pl_fc_entails` doesn't use an ordinary `KB` instance.
The knowledge base here is an instance of the `PropDefiniteKB` class, derived from the `PropKB` class,
but modified to store definite clauses.
<br>
The main point of difference arises in the inclusion of a helper method to `PropDefiniteKB` that returns a list of clauses in KB that have a given symbol `p` in their premise.
```
psource(PropDefiniteKB.clauses_with_premise)
```
Let's now have a look at the `pl_fc_entails` algorithm.
```
psource(pl_fc_entails)
```
The function accepts a knowledge base `KB` (an instance of `PropDefiniteKB`) and a query `q` as inputs.
<br>
<br>
`count` initially stores the number of symbols in the premise of each sentence in the knowledge base.
<br>
The `conjuncts` helper function separates a given sentence at conjunctions.
<br>
`inferred` is initialized as a *boolean* defaultdict.
This will be used later to check if we have inferred all premises of each clause of the agenda.
<br>
`agenda` initially stores a list of clauses that the knowledge base knows to be true.
The `is_prop_symbol` helper function checks if the given symbol is a valid propositional logic symbol.
<br>
<br>
We now iterate through `agenda`, popping a symbol `p` on each iteration.
If the query `q` is the same as `p`, we know that entailment holds.
<br>
The agenda is processed, reducing `count` by one for each implication with a premise `p`.
A conclusion is added to the agenda when `count` reaches zero. This means we know all the premises of that particular implication to be true.
<br>
`clauses_with_premise` is a helpful method of the `PropKB` class.
It returns a list of clauses in the knowledge base that have `p` in their premise.
<br>
<br>
Now that we have an idea of how this function works, let's see a few examples of its usage, but we first need to define our knowledge base. We assume we know the following clauses to be true.
```
clauses = ['(B & F)==>E',
'(A & E & F)==>G',
'(B & C)==>F',
'(A & B)==>D',
'(E & F)==>H',
'(H & I)==>J',
'A',
'B',
'C']
```
We will now `tell` this information to our knowledge base.
```
definite_clauses_KB = PropDefiniteKB()
for clause in clauses:
definite_clauses_KB.tell(expr(clause))
```
We can now check if our knowledge base entails the following queries.
```
pl_fc_entails(definite_clauses_KB, expr('G'))
pl_fc_entails(definite_clauses_KB, expr('H'))
pl_fc_entails(definite_clauses_KB, expr('I'))
pl_fc_entails(definite_clauses_KB, expr('J'))
```
### Effective Propositional Model Checking
The previous segments elucidate the algorithmic procedure for model checking.
In this segment, we look at ways of making them computationally efficient.
<br>
The problem we are trying to solve is conventionally called the _propositional satisfiability problem_, abbreviated as the _SAT_ problem.
In layman terms, if there exists a model that satisfies a given Boolean formula, the formula is called satisfiable.
<br>
The SAT problem was the first problem to be proven _NP-complete_.
The main characteristics of an NP-complete problem are:
- Given a solution to such a problem, it is easy to verify if the solution solves the problem.
- The time required to actually solve the problem using any known algorithm increases exponentially with respect to the size of the problem.
<br>
<br>
Due to these properties, heuristic and approximational methods are often applied to find solutions to these problems.
<br>
It is extremely important to be able to solve large scale SAT problems efficiently because
many combinatorial problems in computer science can be conveniently reduced to checking the satisfiability of a propositional sentence under some constraints.
<br>
We will introduce two new algorithms that perform propositional model checking in a computationally effective way.
<br>
### 1. DPLL (Davis-Putnam-Logeman-Loveland) algorithm
This algorithm is very similar to Backtracking-Search.
It recursively enumerates possible models in a depth-first fashion with the following improvements over algorithms like `tt_entails`:
1. Early termination:
<br>
In certain cases, the algorithm can detect the truth value of a statement using just a partially completed model.
For example, $(P\lor Q)\land(P\lor R)$ is true if P is true, regardless of other variables.
This reduces the search space significantly.
2. Pure symbol heuristic:
<br>
A symbol that has the same sign (positive or negative) in all clauses is called a _pure symbol_.
It isn't difficult to see that any satisfiable model will have the pure symbols assigned such that its parent clause becomes _true_.
For example, $(P\lor\neg Q)\land(\neg Q\lor\neg R)\land(R\lor P)$ has P and Q as pure symbols
and for the sentence to be true, P _has_ to be true and Q _has_ to be false.
The pure symbol heuristic thus simplifies the problem a bit.
3. Unit clause heuristic:
<br>
In the context of DPLL, clauses with just one literal and clauses with all but one _false_ literals are called unit clauses.
If a clause is a unit clause, it can only be satisfied by assigning the necessary value to make the last literal true.
We have no other choice.
<br>
Assigning one unit clause can create another unit clause.
For example, when P is false, $(P\lor Q)$ becomes a unit clause, causing _true_ to be assigned to Q.
A series of forced assignments derived from previous unit clauses is called _unit propagation_.
In this way, this heuristic simplifies the problem further.
<br>
The algorithm often employs other tricks to scale up to large problems.
However, these tricks are currently out of the scope of this notebook. Refer to section 7.6 of the book for more details.
<br>
<br>
Let's have a look at the algorithm.
```
psource(dpll)
```
The algorithm uses the ideas described above to check satisfiability of a sentence in propositional logic.
It recursively calls itself, simplifying the problem at each step. It also uses helper functions `find_pure_symbol` and `find_unit_clause` to carry out steps 2 and 3 above.
<br>
The `dpll_satisfiable` helper function converts the input clauses to _conjunctive normal form_ and calls the `dpll` function with the correct parameters.
```
psource(dpll_satisfiable)
```
Let's see a few examples of usage.
```
A, B, C, D = expr('A, B, C, D')
dpll_satisfiable(A & B & ~C & D)
```
This is a simple case to highlight that the algorithm actually works.
```
dpll_satisfiable((A & B) | (C & ~A) | (B & ~D))
```
If a particular symbol isn't present in the solution,
it means that the solution is independent of the value of that symbol.
In this case, the solution is independent of A.
```
dpll_satisfiable(A |'<=>'| B)
dpll_satisfiable((A |'<=>'| B) |'==>'| (C & ~A))
dpll_satisfiable((A | (B & C)) |'<=>'| ((A | B) & (A | C)))
```
### 2. WalkSAT algorithm
This algorithm is very similar to Hill climbing.
On every iteration, the algorithm picks an unsatisfied clause and flips a symbol in the clause.
This is similar to finding a neighboring state in the `hill_climbing` algorithm.
<br>
The symbol to be flipped is decided by an evaluation function that counts the number of unsatisfied clauses.
Sometimes, symbols are also flipped randomly, to avoid local optima. A subtle balance between greediness and randomness is required. Alternatively, some versions of the algorithm restart with a completely new random assignment if no solution has been found for too long, as a way of getting out of local minima of numbers of unsatisfied clauses.
<br>
<br>
Let's have a look at the algorithm.
```
psource(WalkSAT)
```
The function takes three arguments:
<br>
1. The `clauses` we want to satisfy.
<br>
2. The probability `p` of randomly changing a symbol.
<br>
3. The maximum number of flips (`max_flips`) the algorithm will run for. If the clauses are still unsatisfied, the algorithm returns `None` to denote failure.
<br>
The algorithm is identical in concept to Hill climbing and the code isn't difficult to understand.
<br>
<br>
Let's see a few examples of usage.
```
A, B, C, D = expr('A, B, C, D')
WalkSAT([A, B, ~C, D], 0.5, 100)
```
This is a simple case to show that the algorithm converges.
```
WalkSAT([A & B, A & C], 0.5, 100)
WalkSAT([A & B, C & D, C & B], 0.5, 100)
WalkSAT([A & B, C | D, ~(D | B)], 0.5, 1000)
```
This one doesn't give any output because WalkSAT did not find any model where these clauses hold. We can solve these clauses to see that they together form a contradiction and hence, it isn't supposed to have a solution.
One point of difference between this algorithm and the `dpll_satisfiable` algorithms is that both these algorithms take inputs differently.
For WalkSAT to take complete sentences as input,
we can write a helper function that converts the input sentence into conjunctive normal form and then calls WalkSAT with the list of conjuncts of the CNF form of the sentence.
```
def WalkSAT_CNF(sentence, p=0.5, max_flips=10000):
return WalkSAT(conjuncts(to_cnf(sentence)), 0, max_flips)
```
Now we can call `WalkSAT_CNF` and `DPLL_Satisfiable` with the same arguments.
```
WalkSAT_CNF((A & B) | (C & ~A) | (B & ~D), 0.5, 1000)
```
It works!
<br>
Notice that the solution generated by WalkSAT doesn't omit variables that the sentence doesn't depend upon.
If the sentence is independent of a particular variable, the solution contains a random value for that variable because of the stochastic nature of the algorithm.
<br>
<br>
Let's compare the runtime of WalkSAT and DPLL for a few cases. We will use the `%%timeit` magic to do this.
```
sentence_1 = A |'<=>'| B
sentence_2 = (A & B) | (C & ~A) | (B & ~D)
sentence_3 = (A | (B & C)) |'<=>'| ((A | B) & (A | C))
%%timeit
dpll_satisfiable(sentence_1)
dpll_satisfiable(sentence_2)
dpll_satisfiable(sentence_3)
%%timeit
WalkSAT_CNF(sentence_1)
WalkSAT_CNF(sentence_2)
WalkSAT_CNF(sentence_3)
```
On an average, for solvable cases, `WalkSAT` is quite faster than `dpll` because, for a small number of variables,
`WalkSAT` can reduce the search space significantly.
Results can be different for sentences with more symbols though.
Feel free to play around with this to understand the trade-offs of these algorithms better.
### SATPlan
In this section we show how to make plans by logical inference. The basic idea is very simple. It includes the following three steps:
1. Constuct a sentence that includes:
1. A colection of assertions about the initial state.
2. The successor-state axioms for all the possible actions at each time up to some maximum time t.
3. The assertion that the goal is achieved at time t.
2. Present the whole sentence to a SAT solver.
3. Assuming a model is found, extract from the model those variables that represent actions and are assigned true. Together they represent a plan to achieve the goals.
Lets have a look at the algorithm
```
psource(SAT_plan)
```
Let's see few examples of its usage. First we define a transition and then call `SAT_plan`.
```
transition = {'A': {'Left': 'A', 'Right': 'B'},
'B': {'Left': 'A', 'Right': 'C'},
'C': {'Left': 'B', 'Right': 'C'}}
print(SAT_plan('A', transition, 'C', 2))
print(SAT_plan('A', transition, 'B', 3))
print(SAT_plan('C', transition, 'A', 3))
```
Let us do the same for another transition.
```
transition = {(0, 0): {'Right': (0, 1), 'Down': (1, 0)},
(0, 1): {'Left': (1, 0), 'Down': (1, 1)},
(1, 0): {'Right': (1, 0), 'Up': (1, 0), 'Left': (1, 0), 'Down': (1, 0)},
(1, 1): {'Left': (1, 0), 'Up': (0, 1)}}
print(SAT_plan((0, 0), transition, (1, 1), 4))
```
## First-Order Logic Knowledge Bases: `FolKB`
The class `FolKB` can be used to represent a knowledge base of First-order logic sentences. You would initialize and use it the same way as you would for `PropKB` except that the clauses are first-order definite clauses. We will see how to write such clauses to create a database and query them in the following sections.
## Criminal KB
In this section we create a `FolKB` based on the following paragraph.<br/>
<em>The law says that it is a crime for an American to sell weapons to hostile nations. The country Nono, an enemy of America, has some missiles, and all of its missiles were sold to it by Colonel West, who is American.</em><br/>
The first step is to extract the facts and convert them into first-order definite clauses. Extracting the facts from data alone is a challenging task. Fortunately, we have a small paragraph and can do extraction and conversion manually. We'll store the clauses in list aptly named `clauses`.
```
clauses = []
```
<em>“... it is a crime for an American to sell weapons to hostile nations”</em><br/>
The keywords to look for here are 'crime', 'American', 'sell', 'weapon' and 'hostile'. We use predicate symbols to make meaning of them.
* `Criminal(x)`: `x` is a criminal
* `American(x)`: `x` is an American
* `Sells(x ,y, z)`: `x` sells `y` to `z`
* `Weapon(x)`: `x` is a weapon
* `Hostile(x)`: `x` is a hostile nation
Let us now combine them with appropriate variable naming to depict the meaning of the sentence. The criminal `x` is also the American `x` who sells weapon `y` to `z`, which is a hostile nation.
$\text{American}(x) \land \text{Weapon}(y) \land \text{Sells}(x, y, z) \land \text{Hostile}(z) \implies \text{Criminal} (x)$
```
clauses.append(expr("(American(x) & Weapon(y) & Sells(x, y, z) & Hostile(z)) ==> Criminal(x)"))
```
<em>"The country Nono, an enemy of America"</em><br/>
We now know that Nono is an enemy of America. We represent these nations using the constant symbols `Nono` and `America`. the enemy relation is show using the predicate symbol `Enemy`.
$\text{Enemy}(\text{Nono}, \text{America})$
```
clauses.append(expr("Enemy(Nono, America)"))
```
<em>"Nono ... has some missiles"</em><br/>
This states the existence of some missile which is owned by Nono. $\exists x \text{Owns}(\text{Nono}, x) \land \text{Missile}(x)$. We invoke existential instantiation to introduce a new constant `M1` which is the missile owned by Nono.
$\text{Owns}(\text{Nono}, \text{M1}), \text{Missile}(\text{M1})$
```
clauses.append(expr("Owns(Nono, M1)"))
clauses.append(expr("Missile(M1)"))
```
<em>"All of its missiles were sold to it by Colonel West"</em><br/>
If Nono owns something and it classifies as a missile, then it was sold to Nono by West.
$\text{Missile}(x) \land \text{Owns}(\text{Nono}, x) \implies \text{Sells}(\text{West}, x, \text{Nono})$
```
clauses.append(expr("(Missile(x) & Owns(Nono, x)) ==> Sells(West, x, Nono)"))
```
<em>"West, who is American"</em><br/>
West is an American.
$\text{American}(\text{West})$
```
clauses.append(expr("American(West)"))
```
We also know, from our understanding of language, that missiles are weapons and that an enemy of America counts as “hostile”.
$\text{Missile}(x) \implies \text{Weapon}(x), \text{Enemy}(x, \text{America}) \implies \text{Hostile}(x)$
```
clauses.append(expr("Missile(x) ==> Weapon(x)"))
clauses.append(expr("Enemy(x, America) ==> Hostile(x)"))
```
Now that we have converted the information into first-order definite clauses we can create our first-order logic knowledge base.
```
crime_kb = FolKB(clauses)
```
The `subst` helper function substitutes variables with given values in first-order logic statements.
This will be useful in later algorithms.
It's implementation is quite simple and self-explanatory.
```
psource(subst)
```
Here's an example of how `subst` can be used.
```
subst({x: expr('Nono'), y: expr('M1')}, expr('Owns(x, y)'))
```
## Inference in First-Order Logic
In this section we look at a forward chaining and a backward chaining algorithm for `FolKB`. Both aforementioned algorithms rely on a process called <strong>unification</strong>, a key component of all first-order inference algorithms.
### Unification
We sometimes require finding substitutions that make different logical expressions look identical. This process, called unification, is done by the `unify` algorithm. It takes as input two sentences and returns a <em>unifier</em> for them if one exists. A unifier is a dictionary which stores the substitutions required to make the two sentences identical. It does so by recursively unifying the components of a sentence, where the unification of a variable symbol `var` with a constant symbol `Const` is the mapping `{var: Const}`. Let's look at a few examples.
```
unify(expr('x'), 3)
unify(expr('A(x)'), expr('A(B)'))
unify(expr('Cat(x) & Dog(Dobby)'), expr('Cat(Bella) & Dog(y)'))
```
In cases where there is no possible substitution that unifies the two sentences the function return `None`.
```
print(unify(expr('Cat(x)'), expr('Dog(Dobby)')))
```
We also need to take care we do not unintentionally use the same variable name. Unify treats them as a single variable which prevents it from taking multiple value.
```
print(unify(expr('Cat(x) & Dog(Dobby)'), expr('Cat(Bella) & Dog(x)')))
```
### Forward Chaining Algorithm
We consider the simple forward-chaining algorithm presented in <em>Figure 9.3</em>. We look at each rule in the knoweldge base and see if the premises can be satisfied. This is done by finding a substitution which unifies each of the premise with a clause in the `KB`. If we are able to unify the premises, the conclusion (with the corresponding substitution) is added to the `KB`. This inferencing process is repeated until either the query can be answered or till no new sentences can be added. We test if the newly added clause unifies with the query in which case the substitution yielded by `unify` is an answer to the query. If we run out of sentences to infer, this means the query was a failure.
The function `fol_fc_ask` is a generator which yields all substitutions which validate the query.
```
psource(fol_fc_ask)
```
Let's find out all the hostile nations. Note that we only told the `KB` that Nono was an enemy of America, not that it was hostile.
```
answer = fol_fc_ask(crime_kb, expr('Hostile(x)'))
print(list(answer))
```
The generator returned a single substitution which says that Nono is a hostile nation. See how after adding another enemy nation the generator returns two substitutions.
```
crime_kb.tell(expr('Enemy(JaJa, America)'))
answer = fol_fc_ask(crime_kb, expr('Hostile(x)'))
print(list(answer))
```
<strong><em>Note</em>:</strong> `fol_fc_ask` makes changes to the `KB` by adding sentences to it.
### Backward Chaining Algorithm
This algorithm works backward from the goal, chaining through rules to find known facts that support the proof. Suppose `goal` is the query we want to find the substitution for. We find rules of the form $\text{lhs} \implies \text{goal}$ in the `KB` and try to prove `lhs`. There may be multiple clauses in the `KB` which give multiple `lhs`. It is sufficient to prove only one of these. But to prove a `lhs` all the conjuncts in the `lhs` of the clause must be proved. This makes it similar to <em>And/Or</em> search.
#### OR
The <em>OR</em> part of the algorithm comes from our choice to select any clause of the form $\text{lhs} \implies \text{goal}$. Looking at all rules's `lhs` whose `rhs` unify with the `goal`, we yield a substitution which proves all the conjuncts in the `lhs`. We use `parse_definite_clause` to attain `lhs` and `rhs` from a clause of the form $\text{lhs} \implies \text{rhs}$. For atomic facts the `lhs` is an empty list.
```
psource(fol_bc_or)
```
#### AND
The <em>AND</em> corresponds to proving all the conjuncts in the `lhs`. We need to find a substitution which proves each <em>and</em> every clause in the list of conjuncts.
```
psource(fol_bc_and)
```
Now the main function `fl_bc_ask` calls `fol_bc_or` with substitution initialized as empty. The `ask` method of `FolKB` uses `fol_bc_ask` and fetches the first substitution returned by the generator to answer query. Let's query the knowledge base we created from `clauses` to find hostile nations.
```
# Rebuild KB because running fol_fc_ask would add new facts to the KB
crime_kb = FolKB(clauses)
crime_kb.ask(expr('Hostile(x)'))
```
You may notice some new variables in the substitution. They are introduced to standardize the variable names to prevent naming problems as discussed in the [Unification section](#Unification)
## Appendix: The Implementation of `|'==>'|`
Consider the `Expr` formed by this syntax:
```
P |'==>'| ~Q
```
What is the funny `|'==>'|` syntax? The trick is that "`|`" is just the regular Python or-operator, and so is exactly equivalent to this:
```
(P | '==>') | ~Q
```
In other words, there are two applications of or-operators. Here's the first one:
```
P | '==>'
```
What is going on here is that the `__or__` method of `Expr` serves a dual purpose. If the right-hand-side is another `Expr` (or a number), then the result is an `Expr`, as in `(P | Q)`. But if the right-hand-side is a string, then the string is taken to be an operator, and we create a node in the abstract syntax tree corresponding to a partially-filled `Expr`, one where we know the left-hand-side is `P` and the operator is `==>`, but we don't yet know the right-hand-side.
The `PartialExpr` class has an `__or__` method that says to create an `Expr` node with the right-hand-side filled in. Here we can see the combination of the `PartialExpr` with `Q` to create a complete `Expr`:
```
partial = PartialExpr('==>', P)
partial | ~Q
```
This [trick](http://code.activestate.com/recipes/384122-infix-operators/) is due to [Ferdinand Jamitzky](http://code.activestate.com/recipes/users/98863/), with a modification by [C. G. Vedant](https://github.com/Chipe1),
who suggested using a string inside the or-bars.
## Appendix: The Implementation of `expr`
How does `expr` parse a string into an `Expr`? It turns out there are two tricks (besides the Jamitzky/Vedant trick):
1. We do a string substitution, replacing "`==>`" with "`|'==>'|`" (and likewise for other operators).
2. We `eval` the resulting string in an environment in which every identifier
is bound to a symbol with that identifier as the `op`.
In other words,
```
expr('~(P & Q) ==> (~P | ~Q)')
```
is equivalent to doing:
```
P, Q = symbols('P, Q')
~(P & Q) |'==>'| (~P | ~Q)
```
One thing to beware of: this puts `==>` at the same precedence level as `"|"`, which is not quite right. For example, we get this:
```
P & Q |'==>'| P | Q
```
which is probably not what we meant; when in doubt, put in extra parens:
```
(P & Q) |'==>'| (P | Q)
```
## Examples
```
from notebook import Canvas_fol_bc_ask
canvas_bc_ask = Canvas_fol_bc_ask('canvas_bc_ask', crime_kb, expr('Criminal(x)'))
```
# Authors
This notebook by [Chirag Vartak](https://github.com/chiragvartak) and [Peter Norvig](https://github.com/norvig).
|
github_jupyter
|
from utils import *
from logic import *
from notebook import psource
Symbol('x')
(x, y, P, Q, f) = symbols('x, y, P, Q, f')
P & ~Q
def __and__(self, other): return Expr('&', self, other)```
and does similar overloads for the other operators. An `Expr` has two fields: `op` for the operator, which is always a string, and `args` for the arguments, which is a tuple of 0 or more expressions. By "expression," I mean either an instance of `Expr`, or a number. Let's take a look at the fields for some `Expr` examples:
It is important to note that the `Expr` class does not define the *logic* of Propositional Logic sentences; it just gives you a way to *represent* expressions. Think of an `Expr` as an [abstract syntax tree](https://en.wikipedia.org/wiki/Abstract_syntax_tree). Each of the `args` in an `Expr` can be either a symbol, a number, or a nested `Expr`. We can nest these trees to any depth. Here is a deply nested `Expr`:
## Operators for Constructing Logical Sentences
Here is a table of the operators that can be used to form sentences. Note that we have a problem: we want to use Python operators to make sentences, so that our programs (and our interactive sessions like the one here) will show simple code. But Python does not allow implication arrows as operators, so for now we have to use a more verbose notation that Python does allow: `|'==>'|` instead of just `==>`. Alternately, you can always use the more verbose `Expr` constructor forms:
| Operation | Book | Python Infix Input | Python Output | Python `Expr` Input
|--------------------------|----------------------|-------------------------|---|---|
| Negation | ¬ P | `~P` | `~P` | `Expr('~', P)`
| And | P ∧ Q | `P & Q` | `P & Q` | `Expr('&', P, Q)`
| Or | P ∨ Q | `P`<tt> | </tt>`Q`| `P`<tt> | </tt>`Q` | `Expr('`|`', P, Q)`
| Inequality (Xor) | P ≠ Q | `P ^ Q` | `P ^ Q` | `Expr('^', P, Q)`
| Implication | P → Q | `P` <tt>|</tt>`'==>'`<tt>|</tt> `Q` | `P ==> Q` | `Expr('==>', P, Q)`
| Reverse Implication | Q ← P | `Q` <tt>|</tt>`'<=='`<tt>|</tt> `P` |`Q <== P` | `Expr('<==', Q, P)`
| Equivalence | P ↔ Q | `P` <tt>|</tt>`'<=>'`<tt>|</tt> `Q` |`P <=> Q` | `Expr('<=>', P, Q)`
Here's an example of defining a sentence with an implication arrow:
## `expr`: a Shortcut for Constructing Sentences
If the `|'==>'|` notation looks ugly to you, you can use the function `expr` instead:
`expr` takes a string as input, and parses it into an `Expr`. The string can contain arrow operators: `==>`, `<==`, or `<=>`, which are handled as if they were regular Python infix operators. And `expr` automatically defines any symbols, so you don't need to pre-define them:
For now that's all you need to know about `expr`. If you are interested, we explain the messy details of how `expr` is implemented and how `|'==>'|` is handled in the appendix.
## Propositional Knowledge Bases: `PropKB`
The class `PropKB` can be used to represent a knowledge base of propositional logic sentences.
We see that the class `KB` has four methods, apart from `__init__`. A point to note here: the `ask` method simply calls the `ask_generator` method. Thus, this one has already been implemented, and what you'll have to actually implement when you create your own knowledge base class (though you'll probably never need to, considering the ones we've created for you) will be the `ask_generator` function and not the `ask` function itself.
The class `PropKB` now.
* `__init__(self, sentence=None)` : The constructor `__init__` creates a single field `clauses` which will be a list of all the sentences of the knowledge base. Note that each one of these sentences will be a 'clause' i.e. a sentence which is made up of only literals and `or`s.
* `tell(self, sentence)` : When you want to add a sentence to the KB, you use the `tell` method. This method takes a sentence, converts it to its CNF, extracts all the clauses, and adds all these clauses to the `clauses` field. So, you need not worry about `tell`ing only clauses to the knowledge base. You can `tell` the knowledge base a sentence in any form that you wish; converting it to CNF and adding the resulting clauses will be handled by the `tell` method.
* `ask_generator(self, query)` : The `ask_generator` function is used by the `ask` function. It calls the `tt_entails` function, which in turn returns `True` if the knowledge base entails query and `False` otherwise. The `ask_generator` itself returns an empty dict `{}` if the knowledge base entails query and `None` otherwise. This might seem a little bit weird to you. After all, it makes more sense just to return a `True` or a `False` instead of the `{}` or `None` But this is done to maintain consistency with the way things are in First-Order Logic, where an `ask_generator` function is supposed to return all the substitutions that make the query true. Hence the dict, to return all these substitutions. I will be mostly be using the `ask` function which returns a `{}` or a `False`, but if you don't like this, you can always use the `ask_if_true` function which returns a `True` or a `False`.
* `retract(self, sentence)` : This function removes all the clauses of the sentence given, from the knowledge base. Like the `tell` function, you don't have to pass clauses to remove them from the knowledge base; any sentence will do fine. The function will take care of converting that sentence to clauses and then remove those.
## Wumpus World KB
Let us create a `PropKB` for the wumpus world with the sentences mentioned in `section 7.4.3`.
We define the symbols we use in our clauses.<br/>
$P_{x, y}$ is true if there is a pit in `[x, y]`.<br/>
$B_{x, y}$ is true if the agent senses breeze in `[x, y]`.<br/>
Now we tell sentences based on `section 7.4.3`.<br/>
There is no pit in `[1,1]`.
A square is breezy if and only if there is a pit in a neighboring square. This has to be stated for each square but for now, we include just the relevant squares.
Now we include the breeze percepts for the first two squares leading up to the situation in `Figure 7.3(b)`
We can check the clauses stored in a `KB` by accessing its `clauses` variable
We see that the equivalence $B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was automatically converted to two implications which were inturn converted to CNF which is stored in the `KB`.<br/>
$B_{1, 1} \iff (P_{1, 2} \lor P_{2, 1})$ was split into $B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ and $B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$.<br/>
$B_{1, 1} \implies (P_{1, 2} \lor P_{2, 1})$ was converted to $P_{1, 2} \lor P_{2, 1} \lor \neg B_{1, 1}$.<br/>
$B_{1, 1} \Longleftarrow (P_{1, 2} \lor P_{2, 1})$ was converted to $\neg (P_{1, 2} \lor P_{2, 1}) \lor B_{1, 1}$ which becomes $(\neg P_{1, 2} \lor B_{1, 1}) \land (\neg P_{2, 1} \lor B_{1, 1})$ after applying De Morgan's laws and distributing the disjunction.<br/>
$B_{2, 1} \iff (P_{1, 1} \lor P_{2, 2} \lor P_{3, 2})$ is converted in similar manner.
## Knowledge based agents
A knowledge-based agent is a simple generic agent that maintains and handles a knowledge base.
The knowledge base may initially contain some background knowledge.
<br>
The purpose of a KB agent is to provide a level of abstraction over knowledge-base manipulation and is to be used as a base class for agents that work on a knowledge base.
<br>
Given a percept, the KB agent adds the percept to its knowledge base, asks the knowledge base for the best action, and tells the knowledge base that it has infact taken that action.
<br>
Our implementation of `KB-Agent` is encapsulated in a class `KB_AgentProgram` which inherits from the `KB` class.
<br>
Let's have a look.
The helper functions `make_percept_sentence`, `make_action_query` and `make_action_sentence` are all aptly named and as expected,
`make_percept_sentence` makes first-order logic sentences about percepts we want our agent to receive,
`make_action_query` asks the underlying `KB` about the action that should be taken and
`make_action_sentence` tells the underlying `KB` about the action it has just taken.
## Inference in Propositional Knowledge Base
In this section we will look at two algorithms to check if a sentence is entailed by the `KB`. Our goal is to decide whether $\text{KB} \vDash \alpha$ for some sentence $\alpha$.
### Truth Table Enumeration
It is a model-checking approach which, as the name suggests, enumerates all possible models in which the `KB` is true and checks if $\alpha$ is also true in these models. We list the $n$ symbols in the `KB` and enumerate the $2^{n}$ models in a depth-first manner and check the truth of `KB` and $\alpha$.
The algorithm basically computes every line of the truth table $KB\implies \alpha$ and checks if it is true everywhere.
<br>
If symbols are defined, the routine recursively constructs every combination of truth values for the symbols and then,
it checks whether `model` is consistent with `kb`.
The given models correspond to the lines in the truth table,
which have a `true` in the KB column,
and for these lines it checks whether the query evaluates to true
<br>
`result = pl_true(alpha, model)`.
<br>
<br>
In short, `tt_check_all` evaluates this logical expression for each `model`
<br>
`pl_true(kb, model) => pl_true(alpha, model)`
<br>
which is logically equivalent to
<br>
`pl_true(kb, model) & ~pl_true(alpha, model)`
<br>
that is, the knowledge base and the negation of the query are logically inconsistent.
<br>
<br>
`tt_entails()` just extracts the symbols from the query and calls `tt_check_all()` with the proper parameters.
Keep in mind that for two symbols P and Q, P => Q is false only when P is `True` and Q is `False`.
Example usage of `tt_entails()`:
P & Q is True only when both P and Q are True. Hence, (P & Q) => Q is True
If we know that P | Q is true, we cannot infer the truth values of P and Q.
Hence (P | Q) => Q is False and so is (P | Q) => P.
We can see that for the KB to be true, A, D, E have to be True and F and G have to be False.
Nothing can be said about B or C.
Coming back to our problem, note that `tt_entails()` takes an `Expr` which is a conjunction of clauses as the input instead of the `KB` itself.
You can use the `ask_if_true()` method of `PropKB` which does all the required conversions.
Let's check what `wumpus_kb` tells us about $P_{1, 1}$.
Looking at Figure 7.9 we see that in all models in which the knowledge base is `True`, $P_{1, 1}$ is `False`. It makes sense that `ask_if_true()` returns `True` for $\alpha = \neg P_{1, 1}$ and `False` for $\alpha = P_{1, 1}$. This begs the question, what if $\alpha$ is `True` in only a portion of all models. Do we return `True` or `False`? This doesn't rule out the possibility of $\alpha$ being `True` but it is not entailed by the `KB` so we return `False` in such cases. We can see this is the case for $P_{2, 2}$ and $P_{3, 1}$.
### Proof by Resolution
Recall that our goal is to check whether $\text{KB} \vDash \alpha$ i.e. is $\text{KB} \implies \alpha$ true in every model. Suppose we wanted to check if $P \implies Q$ is valid. We check the satisfiability of $\neg (P \implies Q)$, which can be rewritten as $P \land \neg Q$. If $P \land \neg Q$ is unsatisfiable, then $P \implies Q$ must be true in all models. This gives us the result "$\text{KB} \vDash \alpha$ <em>if and only if</em> $\text{KB} \land \neg \alpha$ is unsatisfiable".<br/>
This technique corresponds to <em>proof by <strong>contradiction</strong></em>, a standard mathematical proof technique. We assume $\alpha$ to be false and show that this leads to a contradiction with known axioms in $\text{KB}$. We obtain a contradiction by making valid inferences using inference rules. In this proof we use a single inference rule, <strong>resolution</strong> which states $(l_1 \lor \dots \lor l_k) \land (m_1 \lor \dots \lor m_n) \land (l_i \iff \neg m_j) \implies l_1 \lor \dots \lor l_{i - 1} \lor l_{i + 1} \lor \dots \lor l_k \lor m_1 \lor \dots \lor m_{j - 1} \lor m_{j + 1} \lor \dots \lor m_n$. Applying the resolution yeilds us a clause which we add to the KB. We keep doing this until:
* There are no new clauses that can be added, in which case $\text{KB} \nvDash \alpha$.
* Two clauses resolve to yield the <em>empty clause</em>, in which case $\text{KB} \vDash \alpha$.
The <em>empty clause</em> is equivalent to <em>False</em> because it arises only from resolving two complementary
unit clauses such as $P$ and $\neg P$ which is a contradiction as both $P$ and $\neg P$ can't be <em>True</em> at the same time.
There is one catch however, the algorithm that implements proof by resolution cannot handle complex sentences.
Implications and bi-implications have to be simplified into simpler clauses.
We already know that *every sentence of a propositional logic is logically equivalent to a conjunction of clauses*.
We will use this fact to our advantage and simplify the input sentence into the **conjunctive normal form** (CNF) which is a conjunction of disjunctions of literals.
For eg:
<br>
$$(A\lor B)\land (\neg B\lor C\lor\neg D)\land (D\lor\neg E)$$
This is equivalent to the POS (Product of sums) form in digital electronics.
<br>
Here's an outline of how the conversion is done:
1. Convert bi-implications to implications
<br>
$\alpha\iff\beta$ can be written as $(\alpha\implies\beta)\land(\beta\implies\alpha)$
<br>
This also applies to compound sentences
<br>
$\alpha\iff(\beta\lor\gamma)$ can be written as $(\alpha\implies(\beta\lor\gamma))\land((\beta\lor\gamma)\implies\alpha)$
<br>
2. Convert implications to their logical equivalents
<br>
$\alpha\implies\beta$ can be written as $\neg\alpha\lor\beta$
<br>
3. Move negation inwards
<br>
CNF requires atomic literals. Hence, negation cannot appear on a compound statement.
De Morgan's laws will be helpful here.
<br>
$\neg(\alpha\land\beta)\equiv(\neg\alpha\lor\neg\beta)$
<br>
$\neg(\alpha\lor\beta)\equiv(\neg\alpha\land\neg\beta)$
<br>
4. Distribute disjunction over conjunction
<br>
Disjunction and conjunction are distributive over each other.
Now that we only have conjunctions, disjunctions and negations in our expression,
we will distribute disjunctions over conjunctions wherever possible as this will give us a sentence which is a conjunction of simpler clauses,
which is what we wanted in the first place.
<br>
We need a term of the form
<br>
$(\alpha_{1}\lor\alpha_{2}\lor\alpha_{3}...)\land(\beta_{1}\lor\beta_{2}\lor\beta_{3}...)\land(\gamma_{1}\lor\gamma_{2}\lor\gamma_{3}...)\land...$
<br>
<br>
The `to_cnf` function executes this conversion using helper subroutines.
`to_cnf` calls three subroutines.
<br>
`eliminate_implications` converts bi-implications and implications to their logical equivalents.
<br>
`move_not_inwards` removes negations from compound statements and moves them inwards using De Morgan's laws.
<br>
`distribute_and_over_or` distributes disjunctions over conjunctions.
<br>
Run the cell below for implementation details.
Let's convert some sentences to see how it works
Coming back to our resolution problem, we can see how the `to_cnf` function is utilized here
### Forward and backward chaining
Previously, we said we will look at two algorithms to check if a sentence is entailed by the `KB`,
but here's a third one.
The difference here is that our goal now is to determine if a knowledge base of definite clauses entails a single proposition symbol *q* - the query.
There is a catch however, the knowledge base can only contain **Horn clauses**.
<br>
#### Horn Clauses
Horn clauses can be defined as a *disjunction* of *literals* with **at most** one positive literal.
<br>
A Horn clause with exactly one positive literal is called a *definite clause*.
<br>
A Horn clause might look like
<br>
$\neg a\lor\neg b\lor\neg c\lor\neg d... \lor z$
<br>
This, coincidentally, is also a definite clause.
<br>
Using De Morgan's laws, the example above can be simplified to
<br>
$a\land b\land c\land d ... \implies z$
<br>
This seems like a logical representation of how humans process known data and facts.
Assuming percepts `a`, `b`, `c`, `d` ... to be true simultaneously, we can infer `z` to also be true at that point in time.
There are some interesting aspects of Horn clauses that make algorithmic inference or *resolution* easier.
- Definite clauses can be written as implications:
<br>
The most important simplification a definite clause provides is that it can be written as an implication.
The premise (or the knowledge that leads to the implication) is a conjunction of positive literals.
The conclusion (the implied statement) is also a positive literal.
The sentence thus becomes easier to understand.
The premise and the conclusion are conventionally called the *body* and the *head* respectively.
A single positive literal is called a *fact*.
- Forward chaining and backward chaining can be used for inference from Horn clauses:
<br>
Forward chaining is semantically identical to `AND-OR-Graph-Search` from the chapter on search algorithms.
Implementational details will be explained shortly.
- Deciding entailment with Horn clauses is linear in size of the knowledge base:
<br>
Surprisingly, the forward and backward chaining algorithms traverse each element of the knowledge base at most once, greatly simplifying the problem.
<br>
<br>
The function `pl_fc_entails` implements forward chaining to see if a knowledge base `KB` entails a symbol `q`.
<br>
Before we proceed further, note that `pl_fc_entails` doesn't use an ordinary `KB` instance.
The knowledge base here is an instance of the `PropDefiniteKB` class, derived from the `PropKB` class,
but modified to store definite clauses.
<br>
The main point of difference arises in the inclusion of a helper method to `PropDefiniteKB` that returns a list of clauses in KB that have a given symbol `p` in their premise.
Let's now have a look at the `pl_fc_entails` algorithm.
The function accepts a knowledge base `KB` (an instance of `PropDefiniteKB`) and a query `q` as inputs.
<br>
<br>
`count` initially stores the number of symbols in the premise of each sentence in the knowledge base.
<br>
The `conjuncts` helper function separates a given sentence at conjunctions.
<br>
`inferred` is initialized as a *boolean* defaultdict.
This will be used later to check if we have inferred all premises of each clause of the agenda.
<br>
`agenda` initially stores a list of clauses that the knowledge base knows to be true.
The `is_prop_symbol` helper function checks if the given symbol is a valid propositional logic symbol.
<br>
<br>
We now iterate through `agenda`, popping a symbol `p` on each iteration.
If the query `q` is the same as `p`, we know that entailment holds.
<br>
The agenda is processed, reducing `count` by one for each implication with a premise `p`.
A conclusion is added to the agenda when `count` reaches zero. This means we know all the premises of that particular implication to be true.
<br>
`clauses_with_premise` is a helpful method of the `PropKB` class.
It returns a list of clauses in the knowledge base that have `p` in their premise.
<br>
<br>
Now that we have an idea of how this function works, let's see a few examples of its usage, but we first need to define our knowledge base. We assume we know the following clauses to be true.
We will now `tell` this information to our knowledge base.
We can now check if our knowledge base entails the following queries.
### Effective Propositional Model Checking
The previous segments elucidate the algorithmic procedure for model checking.
In this segment, we look at ways of making them computationally efficient.
<br>
The problem we are trying to solve is conventionally called the _propositional satisfiability problem_, abbreviated as the _SAT_ problem.
In layman terms, if there exists a model that satisfies a given Boolean formula, the formula is called satisfiable.
<br>
The SAT problem was the first problem to be proven _NP-complete_.
The main characteristics of an NP-complete problem are:
- Given a solution to such a problem, it is easy to verify if the solution solves the problem.
- The time required to actually solve the problem using any known algorithm increases exponentially with respect to the size of the problem.
<br>
<br>
Due to these properties, heuristic and approximational methods are often applied to find solutions to these problems.
<br>
It is extremely important to be able to solve large scale SAT problems efficiently because
many combinatorial problems in computer science can be conveniently reduced to checking the satisfiability of a propositional sentence under some constraints.
<br>
We will introduce two new algorithms that perform propositional model checking in a computationally effective way.
<br>
### 1. DPLL (Davis-Putnam-Logeman-Loveland) algorithm
This algorithm is very similar to Backtracking-Search.
It recursively enumerates possible models in a depth-first fashion with the following improvements over algorithms like `tt_entails`:
1. Early termination:
<br>
In certain cases, the algorithm can detect the truth value of a statement using just a partially completed model.
For example, $(P\lor Q)\land(P\lor R)$ is true if P is true, regardless of other variables.
This reduces the search space significantly.
2. Pure symbol heuristic:
<br>
A symbol that has the same sign (positive or negative) in all clauses is called a _pure symbol_.
It isn't difficult to see that any satisfiable model will have the pure symbols assigned such that its parent clause becomes _true_.
For example, $(P\lor\neg Q)\land(\neg Q\lor\neg R)\land(R\lor P)$ has P and Q as pure symbols
and for the sentence to be true, P _has_ to be true and Q _has_ to be false.
The pure symbol heuristic thus simplifies the problem a bit.
3. Unit clause heuristic:
<br>
In the context of DPLL, clauses with just one literal and clauses with all but one _false_ literals are called unit clauses.
If a clause is a unit clause, it can only be satisfied by assigning the necessary value to make the last literal true.
We have no other choice.
<br>
Assigning one unit clause can create another unit clause.
For example, when P is false, $(P\lor Q)$ becomes a unit clause, causing _true_ to be assigned to Q.
A series of forced assignments derived from previous unit clauses is called _unit propagation_.
In this way, this heuristic simplifies the problem further.
<br>
The algorithm often employs other tricks to scale up to large problems.
However, these tricks are currently out of the scope of this notebook. Refer to section 7.6 of the book for more details.
<br>
<br>
Let's have a look at the algorithm.
The algorithm uses the ideas described above to check satisfiability of a sentence in propositional logic.
It recursively calls itself, simplifying the problem at each step. It also uses helper functions `find_pure_symbol` and `find_unit_clause` to carry out steps 2 and 3 above.
<br>
The `dpll_satisfiable` helper function converts the input clauses to _conjunctive normal form_ and calls the `dpll` function with the correct parameters.
Let's see a few examples of usage.
This is a simple case to highlight that the algorithm actually works.
If a particular symbol isn't present in the solution,
it means that the solution is independent of the value of that symbol.
In this case, the solution is independent of A.
### 2. WalkSAT algorithm
This algorithm is very similar to Hill climbing.
On every iteration, the algorithm picks an unsatisfied clause and flips a symbol in the clause.
This is similar to finding a neighboring state in the `hill_climbing` algorithm.
<br>
The symbol to be flipped is decided by an evaluation function that counts the number of unsatisfied clauses.
Sometimes, symbols are also flipped randomly, to avoid local optima. A subtle balance between greediness and randomness is required. Alternatively, some versions of the algorithm restart with a completely new random assignment if no solution has been found for too long, as a way of getting out of local minima of numbers of unsatisfied clauses.
<br>
<br>
Let's have a look at the algorithm.
The function takes three arguments:
<br>
1. The `clauses` we want to satisfy.
<br>
2. The probability `p` of randomly changing a symbol.
<br>
3. The maximum number of flips (`max_flips`) the algorithm will run for. If the clauses are still unsatisfied, the algorithm returns `None` to denote failure.
<br>
The algorithm is identical in concept to Hill climbing and the code isn't difficult to understand.
<br>
<br>
Let's see a few examples of usage.
This is a simple case to show that the algorithm converges.
This one doesn't give any output because WalkSAT did not find any model where these clauses hold. We can solve these clauses to see that they together form a contradiction and hence, it isn't supposed to have a solution.
One point of difference between this algorithm and the `dpll_satisfiable` algorithms is that both these algorithms take inputs differently.
For WalkSAT to take complete sentences as input,
we can write a helper function that converts the input sentence into conjunctive normal form and then calls WalkSAT with the list of conjuncts of the CNF form of the sentence.
Now we can call `WalkSAT_CNF` and `DPLL_Satisfiable` with the same arguments.
It works!
<br>
Notice that the solution generated by WalkSAT doesn't omit variables that the sentence doesn't depend upon.
If the sentence is independent of a particular variable, the solution contains a random value for that variable because of the stochastic nature of the algorithm.
<br>
<br>
Let's compare the runtime of WalkSAT and DPLL for a few cases. We will use the `%%timeit` magic to do this.
On an average, for solvable cases, `WalkSAT` is quite faster than `dpll` because, for a small number of variables,
`WalkSAT` can reduce the search space significantly.
Results can be different for sentences with more symbols though.
Feel free to play around with this to understand the trade-offs of these algorithms better.
### SATPlan
In this section we show how to make plans by logical inference. The basic idea is very simple. It includes the following three steps:
1. Constuct a sentence that includes:
1. A colection of assertions about the initial state.
2. The successor-state axioms for all the possible actions at each time up to some maximum time t.
3. The assertion that the goal is achieved at time t.
2. Present the whole sentence to a SAT solver.
3. Assuming a model is found, extract from the model those variables that represent actions and are assigned true. Together they represent a plan to achieve the goals.
Lets have a look at the algorithm
Let's see few examples of its usage. First we define a transition and then call `SAT_plan`.
Let us do the same for another transition.
## First-Order Logic Knowledge Bases: `FolKB`
The class `FolKB` can be used to represent a knowledge base of First-order logic sentences. You would initialize and use it the same way as you would for `PropKB` except that the clauses are first-order definite clauses. We will see how to write such clauses to create a database and query them in the following sections.
## Criminal KB
In this section we create a `FolKB` based on the following paragraph.<br/>
<em>The law says that it is a crime for an American to sell weapons to hostile nations. The country Nono, an enemy of America, has some missiles, and all of its missiles were sold to it by Colonel West, who is American.</em><br/>
The first step is to extract the facts and convert them into first-order definite clauses. Extracting the facts from data alone is a challenging task. Fortunately, we have a small paragraph and can do extraction and conversion manually. We'll store the clauses in list aptly named `clauses`.
<em>“... it is a crime for an American to sell weapons to hostile nations”</em><br/>
The keywords to look for here are 'crime', 'American', 'sell', 'weapon' and 'hostile'. We use predicate symbols to make meaning of them.
* `Criminal(x)`: `x` is a criminal
* `American(x)`: `x` is an American
* `Sells(x ,y, z)`: `x` sells `y` to `z`
* `Weapon(x)`: `x` is a weapon
* `Hostile(x)`: `x` is a hostile nation
Let us now combine them with appropriate variable naming to depict the meaning of the sentence. The criminal `x` is also the American `x` who sells weapon `y` to `z`, which is a hostile nation.
$\text{American}(x) \land \text{Weapon}(y) \land \text{Sells}(x, y, z) \land \text{Hostile}(z) \implies \text{Criminal} (x)$
<em>"The country Nono, an enemy of America"</em><br/>
We now know that Nono is an enemy of America. We represent these nations using the constant symbols `Nono` and `America`. the enemy relation is show using the predicate symbol `Enemy`.
$\text{Enemy}(\text{Nono}, \text{America})$
<em>"Nono ... has some missiles"</em><br/>
This states the existence of some missile which is owned by Nono. $\exists x \text{Owns}(\text{Nono}, x) \land \text{Missile}(x)$. We invoke existential instantiation to introduce a new constant `M1` which is the missile owned by Nono.
$\text{Owns}(\text{Nono}, \text{M1}), \text{Missile}(\text{M1})$
<em>"All of its missiles were sold to it by Colonel West"</em><br/>
If Nono owns something and it classifies as a missile, then it was sold to Nono by West.
$\text{Missile}(x) \land \text{Owns}(\text{Nono}, x) \implies \text{Sells}(\text{West}, x, \text{Nono})$
<em>"West, who is American"</em><br/>
West is an American.
$\text{American}(\text{West})$
We also know, from our understanding of language, that missiles are weapons and that an enemy of America counts as “hostile”.
$\text{Missile}(x) \implies \text{Weapon}(x), \text{Enemy}(x, \text{America}) \implies \text{Hostile}(x)$
Now that we have converted the information into first-order definite clauses we can create our first-order logic knowledge base.
The `subst` helper function substitutes variables with given values in first-order logic statements.
This will be useful in later algorithms.
It's implementation is quite simple and self-explanatory.
Here's an example of how `subst` can be used.
## Inference in First-Order Logic
In this section we look at a forward chaining and a backward chaining algorithm for `FolKB`. Both aforementioned algorithms rely on a process called <strong>unification</strong>, a key component of all first-order inference algorithms.
### Unification
We sometimes require finding substitutions that make different logical expressions look identical. This process, called unification, is done by the `unify` algorithm. It takes as input two sentences and returns a <em>unifier</em> for them if one exists. A unifier is a dictionary which stores the substitutions required to make the two sentences identical. It does so by recursively unifying the components of a sentence, where the unification of a variable symbol `var` with a constant symbol `Const` is the mapping `{var: Const}`. Let's look at a few examples.
In cases where there is no possible substitution that unifies the two sentences the function return `None`.
We also need to take care we do not unintentionally use the same variable name. Unify treats them as a single variable which prevents it from taking multiple value.
### Forward Chaining Algorithm
We consider the simple forward-chaining algorithm presented in <em>Figure 9.3</em>. We look at each rule in the knoweldge base and see if the premises can be satisfied. This is done by finding a substitution which unifies each of the premise with a clause in the `KB`. If we are able to unify the premises, the conclusion (with the corresponding substitution) is added to the `KB`. This inferencing process is repeated until either the query can be answered or till no new sentences can be added. We test if the newly added clause unifies with the query in which case the substitution yielded by `unify` is an answer to the query. If we run out of sentences to infer, this means the query was a failure.
The function `fol_fc_ask` is a generator which yields all substitutions which validate the query.
Let's find out all the hostile nations. Note that we only told the `KB` that Nono was an enemy of America, not that it was hostile.
The generator returned a single substitution which says that Nono is a hostile nation. See how after adding another enemy nation the generator returns two substitutions.
<strong><em>Note</em>:</strong> `fol_fc_ask` makes changes to the `KB` by adding sentences to it.
### Backward Chaining Algorithm
This algorithm works backward from the goal, chaining through rules to find known facts that support the proof. Suppose `goal` is the query we want to find the substitution for. We find rules of the form $\text{lhs} \implies \text{goal}$ in the `KB` and try to prove `lhs`. There may be multiple clauses in the `KB` which give multiple `lhs`. It is sufficient to prove only one of these. But to prove a `lhs` all the conjuncts in the `lhs` of the clause must be proved. This makes it similar to <em>And/Or</em> search.
#### OR
The <em>OR</em> part of the algorithm comes from our choice to select any clause of the form $\text{lhs} \implies \text{goal}$. Looking at all rules's `lhs` whose `rhs` unify with the `goal`, we yield a substitution which proves all the conjuncts in the `lhs`. We use `parse_definite_clause` to attain `lhs` and `rhs` from a clause of the form $\text{lhs} \implies \text{rhs}$. For atomic facts the `lhs` is an empty list.
#### AND
The <em>AND</em> corresponds to proving all the conjuncts in the `lhs`. We need to find a substitution which proves each <em>and</em> every clause in the list of conjuncts.
Now the main function `fl_bc_ask` calls `fol_bc_or` with substitution initialized as empty. The `ask` method of `FolKB` uses `fol_bc_ask` and fetches the first substitution returned by the generator to answer query. Let's query the knowledge base we created from `clauses` to find hostile nations.
You may notice some new variables in the substitution. They are introduced to standardize the variable names to prevent naming problems as discussed in the [Unification section](#Unification)
## Appendix: The Implementation of `|'==>'|`
Consider the `Expr` formed by this syntax:
What is the funny `|'==>'|` syntax? The trick is that "`|`" is just the regular Python or-operator, and so is exactly equivalent to this:
In other words, there are two applications of or-operators. Here's the first one:
What is going on here is that the `__or__` method of `Expr` serves a dual purpose. If the right-hand-side is another `Expr` (or a number), then the result is an `Expr`, as in `(P | Q)`. But if the right-hand-side is a string, then the string is taken to be an operator, and we create a node in the abstract syntax tree corresponding to a partially-filled `Expr`, one where we know the left-hand-side is `P` and the operator is `==>`, but we don't yet know the right-hand-side.
The `PartialExpr` class has an `__or__` method that says to create an `Expr` node with the right-hand-side filled in. Here we can see the combination of the `PartialExpr` with `Q` to create a complete `Expr`:
This [trick](http://code.activestate.com/recipes/384122-infix-operators/) is due to [Ferdinand Jamitzky](http://code.activestate.com/recipes/users/98863/), with a modification by [C. G. Vedant](https://github.com/Chipe1),
who suggested using a string inside the or-bars.
## Appendix: The Implementation of `expr`
How does `expr` parse a string into an `Expr`? It turns out there are two tricks (besides the Jamitzky/Vedant trick):
1. We do a string substitution, replacing "`==>`" with "`|'==>'|`" (and likewise for other operators).
2. We `eval` the resulting string in an environment in which every identifier
is bound to a symbol with that identifier as the `op`.
In other words,
is equivalent to doing:
One thing to beware of: this puts `==>` at the same precedence level as `"|"`, which is not quite right. For example, we get this:
which is probably not what we meant; when in doubt, put in extra parens:
## Examples
| 0.870707 | 0.987911 |
# HHL を利用して線形方程式系を解きQiskit で実装する
このチュートリアルでは、HHL アルゴリズムを紹介したあと量子回路を導出し、Qiskit を利用して実装します。HHL をどのようにシミュレーターと5量子ビットデバイスで実行するかも示します。
## 内容
1. [はじめに](#introduction)
2. [HHLアルゴリズム](#hhlalg)
1. [いくつかの数学的背景](#mathbackground)
2. [HHL アルゴリムの説明](#hhldescription)
3. [HHLでの量子位相推定(QPE) について](#qpe)
4. [正確でない QPE の場合](#qpe2)
3. [例: 4量子ビット HHL](#example1)
4. [Qiskit実装](#implementation)
1. [HHL をシミュレーターで実行する: 一般的な方法](#implementationsim)
2. [HHL を実量子デバイスで実行する:最適化例](#implementationdev)
5. [演習](#problems)
6. [参考文献](#references)
## 1. はじめに <a id='introduction'></a>
線形方程式系は様々な分野で、多くの実世界アプリケーションとして自然に現れます。例えば、偏微分方程式の解、金融モデルの校正(キャリブレーション)、流体シミュレーション、あるいは数値場計算などです。問題は次のように定義できます:行列
$A\in\mathbb{C}^{N\times N}$ とベクトル $\vec{b}\in\mathbb{C}^{N}$ が与えられている時、 $A\vec{x}=\vec{b} $ を満足する $\vec{x}\in\mathbb{C}^{N} $ を求める。
$N=2$ の場合に以下の例を見てみましょう。
$$A = \begin{pmatrix}1 & -1/3\\-1/3 & 1 \end{pmatrix},\quad \vec{x}=\begin{pmatrix} x_{1}\\ x_{2}\end{pmatrix}\quad , \quad \vec{b}=\begin{pmatrix}1 \\ 0\end{pmatrix}.$$
この問題は以下のように $x_{1}, x_{2}\in\mathbb{C}$ を見つけるというものに書き換えることもできます。
$$\begin{cases}x_{1} - \frac{x_{2}}{3} = 1 \\ -\frac{x_{1}}{3} + x_{2} = 0\end{cases}. $$
線形方程式系は$A$の行もしくは列に高々$s$個の非ゼロ成分を持つ時に、$s$-sparse (スパース、疎行列)と呼ばれます。$N$ サイズの $s$-sparse系を古典コンピューターで解くには、共役勾配法(conjugate gradient method) を用いても $\mathcal{ O }(Ns\kappa\log(1/\epsilon))$ の実行時間が必要です <sup>[1](#conjgrad)</sup>。ここで、$\kappa$ はシステムの条件数、$\epsilon$ は近似の正確度です。
HHLは、データのローディングを実施する効果的なオラクル(Oracle)が存在し、ハミルトニアン シミュレーションと解の関数の計算が可能であるという仮定のもとで、$A$ がエルミート行列である時、複雑な式 $\mathcal{ O }(\log(N)s^{2}\kappa^{2}/\epsilon)$<sup>[2](#hhl)</sup> に比例した時間で解の関数を推測するという量子アルゴリズムです。これはシステムのサイズに対して指数関数的スピードアップです。ただし、非常に重要な注意点があり、古典アルゴリズムが完全解を返すのに対して、HHL は解となるベクトルを与える関数を近似するだけです。
## 2. HHLアルゴリズム<a id='hhlalg'></a>
### A. いくつかの数学的背景<a id='mathbackground'></a>
量子コンピューターを利用して線形方程式系を解く最初のステップは、問題を量子の言葉に落とし込むことです。系を再スケールすることで、$\vec{b}$ と $\vec{x}$ は正規化できると仮定でき、それぞれ量子状態 $|b\rangle$ と $|x\rangle$ にマップできます。通常、利用されるマッピングは次のようなものです。すなわち、$\vec{b}$ (resp. $\vec{x}$) の $i$ 番目の成分は、量子状態 $|b\rangle$ (resp. $|x\rangle$) の $i$番目の基底状態の振幅に対応するというものです。ここからは、再スケールされた問題にフォーカスします。
$$ A|x\rangle=|b\rangle.$$
$A$ はエルミートなので、スペクトル分解を持ちます。
$$
A=\sum_{j=0}^{N-1}\lambda_{j}|u_{j}\rangle\langle u_{j}|,\quad \lambda_{j}\in\mathbb{ R },
$$
ここで、$|u_{j}\rangle$ は $A$ の $j$ 番目の固有ベクトルで、固有値 はそれぞれ $\lambda_{j}$ です。次に、逆行列です。
$$
A^{-1}=\sum_{j=0}^{N-1}\lambda_{j}^{-1}|u_{j}\rangle\langle u_{j}|,
$$
系の右側は $A$ の固有基底を用いて次のように書けます。
$$
|b\rangle=\sum_{j=0}^{N-1}b_{j}|u_{j}\rangle,\quad b_{j}\in\mathbb{ C }.
$$
HHLの目的は、次の状態でレジスターを読み込むことでアルゴリズムを終了させることを銘記してください。
$$
|x\rangle=A^{-1}|b\rangle=\sum_{j=0}^{N-1}\lambda_{j}^{-1}b_{j}|u_{j}\rangle.
$$
量子状態について話しているので、既に暗黙的な規格化定数が入っていることに着目してください。
### B. HHL アルゴリムの説明 <a id='hhldescription'></a>
アルゴリズムは3つの量子レジスターを利用し、アルゴリズムの開始時点ではすべて $|0\rangle $ にセットされます。一つのレジスター(ここでは $n_{l}$ とサブインデックスで示します)は $A$ の固有値のバイナリー表現の保管に利用されます。2つ目のレジスター、$n_{b}$はベクトル解が保管されます。ここからは $N=2^{n_{b}}$ とします。補助量子ビットとして追加のレジスターがあります。追加レジスターは、個々の計算の中間段階で利用されますが、各計算の最初に $|0\rangle $ にセットされ、個々の操作の最後には、$|0\rangle $ 状態に戻されるため、以下の説明では省略しています。
以下の手順で対応する回路のハイレベルな図形で HHL アルゴリズムのアウトラインを説明します。簡単のため、引き続く説明ではすべての計算が正確であると仮定し、正確でない場合の詳細なセクション [2.D.](#qpe2) で説明します。
<img src="images/hhlcircuit.png" width = "75%" height = "75%">
1. データ $|b\rangle\in\mathbb{ C }^{N}$ のロード。すなわち、以下の変換を実行します。
$$ |0\rangle _{n_{b}} \mapsto |b\rangle _{n_{b}}. $$
2. 量子位相推定(Quantum Phase Estimation - QPE) を適用します。
$$
U = e ^ { i A t } := \sum _{j=0}^{N-1}e ^ { i \lambda _ { j } t } |u_{j}\rangle\langle u_{j}|.
$$
$A$ の固有基底で表現されるレジスターの量子状態は次のようになります。
$$
\sum_{j=0}^{N-1} b _ { j } |\lambda _ {j }\rangle_{n_{l}} |u_{j}\rangle_{n_{b}},
$$
ここで、$|\lambda _ {j }\rangle_{n_{l}}$ は $\lambda _ {j }$ の $n_{l}$ ビットバイナリー表現です。
3. 補助量子ビットを追加し、$|\lambda_{ j }\rangle$ の条件に応じて回転を適用します。
$$
\sum_{j=0}^{N-1} b _ { j } |\lambda _ { j }\rangle_{n_{l}}|u_{j}\rangle_{n_{b}} \left( \sqrt { 1 - \frac { C^{2} } { \lambda _ { j } ^ { 2 } } } |0\rangle + \frac { C } { \lambda _ { j } } |1\rangle \right),
$$ $C$ は規格化定数です。
4. QPE$^{\dagger}$ を適用します。QPE で発生しうるエラーを無視すると、次の結果になります。
$$
\sum_{j=0}^{N-1} b _ { j } |0\rangle_{n_{l}}|u_{j}\rangle_{n_{b}} \left( \sqrt { 1 - \frac {C^{2} } { \lambda _ { j } ^ { 2 } } } |0\rangle + \frac { C } { \lambda _ { j } } |1\rangle \right).
$$
5. 計算基底にて補助量子ビットを測定します。結果が $1$ の場合、測定後のレジスターの状態は次のようになります。
$$
\left( \sqrt { \frac { 1 } { \sum_{j=0}^{N-1} \left| b _ { j } \right| ^ { 2 } / \left| \lambda _ { j } \right| ^ { 2 } } } \right) \sum _{j=0}^{N-1} \frac{b _ { j }}{\lambda _ { j }} |0\rangle_{n_{l}}|u_{j}\rangle_{n_{b}},
$$
規格化定数を除き、解になっています。
6. オブザーバブル $M$ を適用し、$F(x):=\langle x|M|x\rangle$ を計算します。
### C. HHLでの量子位相推定(QPE) について <a id='qpe'></a>
量子位相推定については第3章でより詳しく説明がされていますが、HHLアルゴリズムではこの量子処理が要になっているので、ここで定義を再確認したいと思います。大雑把にいうと、固有ベクトリ $|\psi\rangle_{m}$ と 固有値 $e^{2\pi i\theta}$ をもつユニタリー $U$ が与えられた場合に $\theta$ を求めるという量子アルゴリズムです。次のように正確に定義できます。
**定義:** $U\in\mathbb{ C }^{2^{m}\times 2^{m}}$ がユニタリーであり、$|\psi\rangle_{m}\in\mathbb{ C }^{2^{m}}$ がそれぞれ固有値 $e^{2\pi i\theta}$ を持つ固有ベクトルである時、**量子位相推定(Quantum Phase Estimation)** (省略して **QPE**) アルゴリズムは、$U$ に対応するユニタリーゲートと状態 $|0\rangle_{n}|\psi\rangle_{m}$ を入力として、状態 $|\tilde{\theta}\rangle_{n}|\psi\rangle_{m}$ を返すアルゴリズムです。
$\tilde{\theta}$ は $2^{n}\theta$ に対するバイナリー近似を示しており、$n$ の添え文字は $n$ ディジットに切り捨てられていることを示しています。
$$
\operatorname { QPE } ( U , |0\rangle_{n}|\psi\rangle_{m} ) = |\tilde{\theta}\rangle_{n}|\psi\rangle_{m}.
$$
HHLでは QPE を $U = e ^ { i A t }$ に対して適用しています。ここで $A$ は行列で解きたい系に関係しています。この場合、
$$
e ^ { i A t } = \sum_{j=0}^{N-1}e^{i\lambda_{j}t}|u_{j}\rangle\langle u_{j}|.
$$
です。さらに、固有値 $e ^ { i \lambda _ { j } t }$ をもつ固有ベクトル $|u_{j}\rangle_{n_{b}}$ に対しては QPE は $|\tilde{\lambda }_ { j }\rangle_{n_{l}}|u_{j}\rangle_{n_{b}}$ を出力します。$\tilde{\lambda }_ { j }$ は、$2^{n_l}\frac{\lambda_ { j }t}{2\pi}$ に対する $n_{l}$-bit バイナリー近似です。従って、仮にそれぞれの $\lambda_{j}$ が $n_{l}$ビットで正確に記述できる場合には、
$$
\operatorname { QPE } ( e ^ { i A 2\pi } , \sum_{j=0}^{N-1}b_{j}|0\rangle_{n_{l}}|u_{j}\rangle_{n_{b}} ) = \sum_{j=0}^{N-1}b_{j}|\lambda_{j}\rangle_{n_{l}}|u_{j}\rangle_{n_{b}}.
$$
となります。
### D. 正確でない QPE の場合<a id='qpe2'></a>
現実には、QPE を初期状態に適用したあとのレジスターの量子状態は、
$$
\sum _ { j=0 }^{N-1} b _ { j } \left( \sum _ { l = 0 } ^ { 2 ^ { n_{l} } - 1 } \alpha _ { l | j } |l\rangle_{n_{l}} \right)|u_{j}\rangle_{n_{b}},
$$
です。ここで、
$$
\alpha _ { l | j } = \frac { 1 } { 2 ^ { n_{l} } } \sum _ { k = 0 } ^ { 2^{n_{l}}- 1 } \left( e ^ { 2 \pi i \left( \frac { \lambda _ { j } t } { 2 \pi } - \frac { l } { 2 ^ { n_{l} } } \right) } \right) ^ { k }.
$$
$\tilde{\lambda_{j}}$ で表すのは、 $\lambda_{j}$, $1\leq j\leq N$ に対する ベストな $n_{l}$ビット近似です。次に、$n_{l}$-レジスターを再ラベルし、$\alpha _ { l | j }$ が $|l + \tilde { \lambda } _ { j } \rangle_{n_{l}}$ の振幅を表すようにします。そうすると、以下のように変形できます。
$$
\alpha _ { l | j } : = \frac { 1 } { 2 ^ { n_{l}} } \sum _ { k = 0 } ^ { 2 ^ { n_{l} } - 1 } \left( e ^ { 2 \pi i \left( \frac { \lambda _ { j } t } { 2 \pi } - \frac { l + \tilde { \lambda } _ { j } } { 2 ^ { n_{l} } } \right) } \right) ^ { k }.
$$
各 $\frac { \lambda _ { j } t } { 2 \pi }$ が $n_{l}$ バイナリービットで正確に表現できる場合には、$\frac { \lambda _ { j } t } { 2 \pi }=\frac { \tilde { \lambda } _ { j } } { 2 ^ { n_{l} } }$ $\forall j$ となります。従って、$\forall j$, $1\leq j \leq N$ の場合は、 $\alpha _ { 0 | j } = 1$ と $\alpha _ { l | j } = 0 \quad \forall l \neq 0$ を保持します。この場合のみ、QPE を適用した後のレジスターの状態を次のように書けます。
$$
\sum_{j=0}^{N-1} b _ { j } |\lambda _ {j }\rangle_{n_{l}} |u_{j}\rangle_{n_{b}}.
$$
それ以外の場合は、 $|\alpha _ { l | j }|$ は $\frac { \lambda _ { j } t } { 2 \pi } \approx \frac { l + \tilde { \lambda } _ { j } } { 2 ^ { n_{l} } }$ の場合のみ大きく、レジスターの状態は次のようになります。
$$
\sum _ { j=0 }^{N-1} \sum _ { l = 0 } ^ { 2 ^ { n_{l} } - 1 } \alpha _ { l | j } b _ { j }|l\rangle_{n_{l}} |u_{j}\rangle_{n_{b}}.
$$
## 3. 例: 4量子ビット HHL<a id='example1'></a>
はじめにのセクションで紹介した小さな例をとってアルゴリズムを説明しましょう。こうでした。
$$A = \begin{pmatrix}1 & -1/3\\-1/3 & 1 \end{pmatrix}\quad , \quad |b\rangle=\begin{pmatrix}1 \\ 0\end{pmatrix}.$$
$|b\rangle$ には $n_{b}=1$ 量子ビットを利用し、解 $|x\rangle$ には、$n_{l}=2$ 量子ビットを固有値のバイナリー表現を利用し、$1$ 補助量子ビットを制御回転に利用します。このようにするとアルゴリズムが成功します。
アルゴリズムの例示のため、少しごまかして $A$ の固有値を計算して、$n_{l}$-レジスターの再スケールされた固有値の正確なバイナリー表現を得るために、 $t$ を選択できるようにします。なお、HHL アルゴリズムの実装には固有値の事前知識は必要ないことは心に留めてください。とは言ったものの、簡単に計算すると、
$$\lambda_{1} = 2/3\quad , \quad\lambda_{2}=4/3.$$
が得られます。
前のセクションを思い出すと、 QPE は $\frac{\lambda_ { j }t}{2\pi}$ に対する $n_{l}$ビット (この例では $2$ビット) バイナリー近似を出力するものでした。従って、
$$t=2\pi\cdot \frac{3}{8},$$
を設定すると、QPE は
$$\frac{\lambda_ { 1 }t}{2\pi} = 1/4\quad , \quad\frac{\lambda_ { 2 }t}{2\pi}=1/2,$$
への$2$ビットバイナリー近似を与えます。これらはそれぞれ、
$$|01\rangle_{n_{l}}\quad , \quad|10\rangle_{n_{l}}.$$
に対応します。
固有ベクトルはそれぞれ、
$$|u_{1}\rangle=\begin{pmatrix}1 \\ -1\end{pmatrix}\quad , \quad|u_{2}\rangle=\begin{pmatrix}1 \\ 1\end{pmatrix}.$$
再度、HHL の実装には固有ベクトルの計算は必要ないことを心に銘記してください。事実、$N$ 次元の一般的なエルミート行列 $A$ は $N$ 個の異なる固有値を持つことがあり、従って計算には $\mathcal{O}(N)$ 時間かかるので、量子アドバンテージが失われてしまいます。
次に、$A$ の固有基底で $|b\rangle$ が次のように書けます。
$$|b\rangle _{n_{b}}=\sum_{j=0}^{N-1}\frac{1}{\sqrt{2}}|u_{j}\rangle _{n_{b}}.$$
ここで、HHL アルゴリズムを適用する準備が整いましたので、ひとつひとつ見ていきます。
1. この例における初期状態準備は簡単です。 $|b\rangle=|0\rangle$。
2. QPE を適用します。
$$
\frac{1}{\sqrt{2}}|01\rangle|u_{1}\rangle + \frac{1}{\sqrt{2}}|10\rangle|u_{2}\rangle.
$$
3. 固有値を再スケールした結果を補うため $C=3/8$ で制御回転させます。
$$\frac{1}{\sqrt{2}}|01\rangle|u_{1}\rangle\left( \sqrt { 1 - \frac { (3/8)^{2} } {(1/4)^{2} } } |0\rangle + \frac { 3/8 } { 1/4 } |1\rangle \right) + \frac{1}{\sqrt{2}}|10\rangle|u_{2}\rangle\left( \sqrt { 1 - \frac { (3/8)^{2} } {(1/2)^{2} } } |0\rangle + \frac { 3/8 } { 1/2 } |1\rangle \right)
$$
$$
=\frac{1}{\sqrt{2}}|01\rangle|u_{1}\rangle\left( \sqrt { 1 - \frac { 9 } {4 } } |0\rangle + \frac { 3 } { 2 } |1\rangle \right) + \frac{1}{\sqrt{2}}|10\rangle|u_{2}\rangle\left( \sqrt { 1 - \frac { 9 } {16 } } |0\rangle + \frac { 3 } { 4 } |1\rangle \right).
$$
4. QPE$^{\dagger}$ を量子コンピューターで適用した後は、次の状態になります。
$$
\frac{1}{\sqrt{2}}|00\rangle|u_{1}\rangle\left( \sqrt { 1 - \frac { 9 } {4 } } |0\rangle + \frac { 3 } { 2 } |1\rangle \right) + \frac{1}{\sqrt{2}}|00\rangle|u_{2}\rangle\left( \sqrt { 1 - \frac { 9 } {16 } } |0\rangle + \frac { 3 } { 4 } |1\rangle \right).
$$
5. 補助ビットを計測し、$1$ が出た場合には状態は次のようになります。
$$
\frac{\frac{1}{\sqrt{2}}|00\rangle|u_{1}\rangle\frac { 3 } { 2 } |1\rangle + \frac{1}{\sqrt{2}}|00\rangle|u_{2}\rangle\frac { 3 } { 4 } |1\rangle}{\sqrt{45/32}}.
$$
簡単に計算すると:
$$
\frac{\frac{3}{2\sqrt{2}}|u_{1}\rangle+ \frac{3}{4\sqrt{2}}|u_{2}\rangle}{\sqrt{45/32}} = \frac{|x\rangle}{||x||}.
$$
6. 追加のゲートを利用しなくても、$|x\rangle$ のノルムを計算できます、それは前のステップで補助ビットを $1$ で測定する確率になります。
$$
P[|1\rangle] = \left(\frac{3}{2\sqrt{2}}\right)^{2} + \left(\frac{3}{4\sqrt{2}}\right)^{2} = \frac{45}{32} = |||x\rangle||^{2}.
$$
## 4. Qiskit 実装<a id='implementation'></a>
例を利用して問題を分析的に解きましたが、HHL を量子シミュレーターと実ハードウェアで実行することを示したいと思います。量子シミュレーターの場合は、Qiskit Aqua に行列$A$ と $|b\rangle$ を基本入力として要求する HHL アルゴリズム実装が既に提供されています。主なアドバンテージは一般的なエルミート行列と任意の初期状態を入力することができる点です。これが意味することは、このアルゴリズムは一般の目的のために設計されており、特定の問題のための回路の最適化は実施しないということです。そのため、既存の実ハードウェアで実行することが目的の場合には問題が生じます。この章を記述している時点で、既存の量子コンピューターはノイズがあり、小さな回路しか実行できません。そのため、セクション[4.B.](#implementationdev)では、この例が帰属する問題のクラスに利用できる最適化された回路を見ることにし、ノイズのある量子コンピューターに対する既存の処理方法について述べます。
## A. HHL をシミュレーターで実行する: 一般的な方法<a id='implementationsim'></a>
Qiskit Aqua が提供する HHL アルゴリズムを実行するには、適切なモジュールをインポートし以下のようにパラメータをセットするだけです。いままで見てきた例では、ハミルトニアンシミュレーションの時間を $t=2\pi\cdot \frac{3}{8}$ にセットしましたが、固有値の知識が必要ないことを示すためこのパラメータをセットしないでシミュレーションを実行してみます。それにも関わらず、行列がある構造がある場合には、固有値情報を得て適切な $t$ を選択し HHLが返す解の精度を向上させることができるかも知れません。8の演習で、$t=2\pi\cdot \frac{3}{8}$ をセットし正常に実行されると解のフィデリティーは $1$ になることを確認します。
```
from qiskit import Aer
from qiskit.circuit.library import QFT
from qiskit.aqua import QuantumInstance, aqua_globals
from qiskit.quantum_info import state_fidelity
from qiskit.aqua.algorithms import HHL, NumPyLSsolver
from qiskit.aqua.components.eigs import EigsQPE
from qiskit.aqua.components.reciprocals import LookupRotation
from qiskit.aqua.operators import MatrixOperator
from qiskit.aqua.components.initial_states import Custom
import numpy as np
def create_eigs(matrix, num_ancillae, num_time_slices, negative_evals):
ne_qfts = [None, None]
if negative_evals:
num_ancillae += 1
ne_qfts = [QFT(num_ancillae - 1), QFT(num_ancillae - 1).inverse()]
return EigsQPE(MatrixOperator(matrix=matrix),
QFT(num_ancillae).inverse(),
num_time_slices=num_time_slices,
num_ancillae=num_ancillae,
expansion_mode='suzuki',
expansion_order=2,
evo_time=None, # This is t, can set to: np.pi*3/4
negative_evals=negative_evals,
ne_qfts=ne_qfts)
```
次の関数を使って HHL アルゴリズムが返した解のフィデリティーを計算します。
```
def fidelity(hhl, ref):
solution_hhl_normed = hhl / np.linalg.norm(hhl)
solution_ref_normed = ref / np.linalg.norm(ref)
fidelity = state_fidelity(solution_hhl_normed, solution_ref_normed)
print("Fidelity:\t\t %f" % fidelity)
matrix = [[1, -1/3], [-1/3, 1]]
vector = [1, 0]
orig_size = len(vector)
matrix, vector, truncate_powerdim, truncate_hermitian = HHL.matrix_resize(matrix, vector)
# Initialize eigenvalue finding module
eigs = create_eigs(matrix, 3, 50, False)
num_q, num_a = eigs.get_register_sizes()
# Initialize initial state module
init_state = Custom(num_q, state_vector=vector)
# Initialize reciprocal rotation module
reci = LookupRotation(negative_evals=eigs._negative_evals, evo_time=eigs._evo_time)
algo = HHL(matrix, vector, truncate_powerdim, truncate_hermitian, eigs,
init_state, reci, num_q, num_a, orig_size)
```
$t=2\pi\cdot \frac{3}{8}$ を選択した理由は固有値を再スケールさせるためでした。今はこの場合に当てはまらないので、表現は近似になり、QPE は非正確で結果の解も近似になります。
```
result = algo.run(QuantumInstance(Aer.get_backend('statevector_simulator')))
print("Solution:\t\t", np.round(result['solution'], 5))
result_ref = NumPyLSsolver(matrix, vector).run()
print("Classical Solution:\t", np.round(result_ref['solution'], 5))
print("Probability:\t\t %f" % result['probability_result'])
fidelity(result['solution'], result_ref['solution'])
```
アルゴリズムは利用したリソースを出力します。depth は単一量子ビットに適用されたゲート数の最大値で、width は利用した量子ビットの数として定義されます。また、CNOT の数も出力してみます。この数をもちいて、実行している回路が現在の実ハードウェアで実行することができるかを判断することができます。
```
print("circuit_width:\t", result['circuit_info']['width'])
print("circuit_depth:\t", result['circuit_info']['depth'])
print("CNOT gates:\t", result['circuit_info']['operations']['cx'])
```
## B. HHL を実量子デバイスで実行する:最適化例<a id='implementationdev'></a>
前のセクションでは Qiskit が提供する標準のアルゴリズムを実行し、$7$ 量子ビットを利用し、depth が $102$ ゲートで、かつ $54$ CNOTゲートを利用しているのを見ました。この数は現在利用可能なハードウェアで実行するには不適切で、量を減らす必要があります。特にゴールは単一量子ゲートに比べフィデリティーが悪化する CNOTの数を $10$ のファクターで減らすことです。さらに量子ビットの数を $4$ にします。これはもともとの問題での数です。Qiskit の方法は一般的な問題のために書かれているため、余分な $3$ 補助量子ビットが追加されました。
ところが、単純にゲートや量子ビットの数を減らすだけでは実ハードウェア上の解としてよい近似が出てきません。二つのエラーソースが原因です:実行時に発生するものと読み取りエラーです。
Qiskit には、個別に全ての基底状態を準備測定することで読み取りエラーを回避するモジュールが提供されています。このトピックの詳細は Dewes et al.<sup>[3](#readouterr)</sup> を参照してください。回路の実行時に発生するエラーを取り扱うために、回路を3回実行し、それぞれ CNOTゲートを $1$, $3$, $5$ CNOT で置き換えるという Richardson 外挿を利用します<sup>[4](#richardson)</sup>。
理論的には3つの回路は同じ結果をもたらしますが、実ハードウェアでは CNOT を追加するということはエラーを増幅することになるというアイデアに基づきます。得られた結果がエラーが増幅したことによるということを知るので、それぞれの場合でどの程度エラーが増幅したのかを推定できます。これらの量を組み合わせることで、前の詳細な値よりも解析解により近い近似値をだすあたらしい結果を得ることができます。
以下はどのようなフォームの問題でも利用できる最適化された回路です。
$$A = \begin{pmatrix}a & b\\b & a \end{pmatrix}\quad , \quad |b\rangle=\begin{pmatrix}\cos(\theta) \\ \sin(\theta)\end{pmatrix},\quad a,b,\theta\in\mathbb{R}.$$
以下の最適化は HHL for tridiagonal symmetric matrices<sup>[5](#tridi)</sup> での結果から引用したもので、特に回路は UniversalQCompiler software<sup>[6](#qcompiler)</sup> の助けを借りて導出しました。
```
from qiskit import QuantumRegister, QuantumCircuit
import numpy as np
t = 2 # This is not optimal; As an exercise, set this to the
# value that will get the best results. See section 8 for solution.
nqubits = 4 # Total number of qubits
nb = 1 # Number of qubits representing the solution
nl = 2 # Number of qubits representing the eigenvalues
theta = 0 # Angle defining |b>
a = 1 # Matrix diagonal
b = -1/3 # Matrix off-diagonal
# Initialise the quantum and classical registers
qr = QuantumRegister(nqubits)
# Create a Quantum Circuit
qc = QuantumCircuit(qr)
qrb = qr[0:nb]
qrl = qr[nb:nb+nl]
qra = qr[nb+nl:nb+nl+1]
# State preparation.
qc.ry(2*theta, qrb[0])
# QPE with e^{iAt}
for qu in qrl:
qc.h(qu)
qc.u1(a*t, qrl[0])
qc.u1(a*t*2, qrl[1])
qc.u3(b*t, -np.pi/2, np.pi/2, qrb[0])
# Controlled e^{iAt} on \lambda_{1}:
params=b*t
qc.u1(np.pi/2,qrb[0])
qc.cx(qrl[0],qrb[0])
qc.ry(params,qrb[0])
qc.cx(qrl[0],qrb[0])
qc.ry(-params,qrb[0])
qc.u1(3*np.pi/2,qrb[0])
# Controlled e^{2iAt} on \lambda_{2}:
params = b*t*2
qc.u1(np.pi/2,qrb[0])
qc.cx(qrl[1],qrb[0])
qc.ry(params,qrb[0])
qc.cx(qrl[1],qrb[0])
qc.ry(-params,qrb[0])
qc.u1(3*np.pi/2,qrb[0])
# Inverse QFT
qc.h(qrl[1])
qc.rz(-np.pi/4,qrl[1])
qc.cx(qrl[0],qrl[1])
qc.rz(np.pi/4,qrl[1])
qc.cx(qrl[0],qrl[1])
qc.rz(-np.pi/4,qrl[0])
qc.h(qrl[0])
# Eigenvalue rotation
t1=(-np.pi +np.pi/3 - 2*np.arcsin(1/3))/4
t2=(-np.pi -np.pi/3 + 2*np.arcsin(1/3))/4
t3=(np.pi -np.pi/3 - 2*np.arcsin(1/3))/4
t4=(np.pi +np.pi/3 + 2*np.arcsin(1/3))/4
qc.cx(qrl[1],qra[0])
qc.ry(t1,qra[0])
qc.cx(qrl[0],qra[0])
qc.ry(t2,qra[0])
qc.cx(qrl[1],qra[0])
qc.ry(t3,qra[0])
qc.cx(qrl[0],qra[0])
qc.ry(t4,qra[0])
qc.measure_all()
print("Depth: %i" % qc.depth())
print("CNOTS: %i" % qc.count_ops()['cx'])
qc.draw(fold=100)
```
以下のコードでは回路、実ハードウェアのバックエンド、及び利用したい量子ビットのセットを入力とし、特定のデバイスで実行した結果とインスタンス情報を出力します。
実デバイスは定期的にキャリブレーションが必要なため、時間によっては特定の量子ビットもしくはゲートのフィデリティーが異なります。さらには、異なるチップは異なる接続性を持ちます。特定のデバイスで接続されていない2つの量子ビットゲートを実行する時には、トランスパイラーは SWAP を追加します。ですので、実行する前に、特定の時間で正しい接続性をもつかエラー率が低いかを IBM Q Experience Webページ<sup>[7](#qexperience)</sup> で確認することを推奨します。
```
from qiskit import execute, BasicAer, ClassicalRegister, IBMQ
from qiskit.compiler import transpile
from qiskit.ignis.mitigation.measurement import (complete_meas_cal, # Measurement error mitigation functions
CompleteMeasFitter,
MeasurementFilter)
provider = IBMQ.load_account()
backend = provider.get_backend('ibmqx2') # calibrate using real hardware
layout = [2,3,0,4]
chip_qubits = 5
# Transpiled circuit for the real hardware
qc_qa_cx = transpile(qc, backend=backend, initial_layout=layout)
```
次のステップでは読み取りエラーを回避するための回路を追加しています<sup>[3](#readouterr)</sup>。
```
meas_cals, state_labels = complete_meas_cal(qubit_list=layout, qr=QuantumRegister(chip_qubits))
qcs = meas_cals + [qc_qa_cx]
shots = 10
job = execute(qcs, backend=backend, shots=shots, optimization_level=0)
```
次のプロットは <sup>[5](#tridi)</sup> 上記の回路を $10$ の異なる初期状態から実行して得られた結果を示しています。$x$軸は各々の場合の初期状態で定義される $\theta$ 角です。結果は読み取りエラーを回避、$1$, $3$, $5$ の回路結果から得られたエラーを外挿した後の結果を表示しています。
<img src="images/norm_public.png">
以下はエラー回避や CNOT からの外挿がない場合の比較です <sup>[5](#tridi)</sup>。
<img src="images/noerrmit_public.png">
## 8. 演習<a id='problems'></a>
1. 'evo_time': $2\pi(3/8)$ でシミュレーションを実行する。フィデリティーが $1$ となることを確認せよ。
##### 実ハードウェア:
2. 最適化された例で時間のパラメータをセットする。
<details>
<summary> 解 (Click to expand)</summary>
t = 2.344915690192344
ベストな結果はこの値をセットします。逆数は解に対して大きな貢献をするので、最小の固有値が表現できるようになります。
</details>
2. 与えられた回路で $3$ と $5$ CNOTs を実行する。回路を作成する時には引き続く CNOT ゲートが transpile() メソッドでキャンセルされないようにバリアを追加する必要がある。
3. 実ハードウェアで回路を実行し、結果に対して2次フィットを適用し外挿の値を求めよ。
## 9. 参考文献<a id='references'></a>
1. J. R. Shewchuk. An Introduction to the Conjugate Gradient Method Without the Agonizing Pain. Technical Report CMU-CS-94-125, School of Computer Science, Carnegie Mellon University, Pittsburgh, Pennsylvania, March 1994.<a id='conjgrad'></a>
2. A. W. Harrow, A. Hassidim, and S. Lloyd, “Quantum algorithm for linear systems of equations,” Phys. Rev. Lett. 103.15 (2009), p. 150502.<a id='hhl'></a>
3. A. Dewes, F. R. Ong, V. Schmitt, R. Lauro, N. Boulant, P. Bertet, D. Vion, and D. Esteve, “Characterization of a two-transmon processor with individual single-shot qubit readout,” Phys. Rev. Lett. 108, 057002 (2012). <a id='readouterr'></a>
4. N. Stamatopoulos, D. J. Egger, Y. Sun, C. Zoufal, R. Iten, N. Shen, and S. Woerner, “Option Pricing using Quantum Computers,” arXiv:1905.02666 . <a id='richardson'></a>
5. A. Carrera Vazquez, A. Frisch, D. Steenken, H. S. Barowski, R. Hiptmair, and S. Woerner, “Enhancing Quantum Linear System Algorithm by Richardson Extrapolation,” (to be included).<a id='tridi'></a>
6. R. Iten, O. Reardon-Smith, L. Mondada, E. Redmond, R. Singh Kohli, R. Colbeck, “Introduction to UniversalQCompiler,” arXiv:1904.01072 .<a id='qcompiler'></a>
7. https://quantum-computing.ibm.com/ .<a id='qexperience'></a>
8. D. Bucher, J. Mueggenburg, G. Kus, I. Haide, S. Deutschle, H. Barowski, D. Steenken, A. Frisch, "Qiskit Aqua: Solving linear systems of equations with the HHL algorithm" https://github.com/Qiskit/qiskit-tutorials/blob/master/legacy_tutorials/aqua/linear_systems_of_equations.ipynb
|
github_jupyter
|
from qiskit import Aer
from qiskit.circuit.library import QFT
from qiskit.aqua import QuantumInstance, aqua_globals
from qiskit.quantum_info import state_fidelity
from qiskit.aqua.algorithms import HHL, NumPyLSsolver
from qiskit.aqua.components.eigs import EigsQPE
from qiskit.aqua.components.reciprocals import LookupRotation
from qiskit.aqua.operators import MatrixOperator
from qiskit.aqua.components.initial_states import Custom
import numpy as np
def create_eigs(matrix, num_ancillae, num_time_slices, negative_evals):
ne_qfts = [None, None]
if negative_evals:
num_ancillae += 1
ne_qfts = [QFT(num_ancillae - 1), QFT(num_ancillae - 1).inverse()]
return EigsQPE(MatrixOperator(matrix=matrix),
QFT(num_ancillae).inverse(),
num_time_slices=num_time_slices,
num_ancillae=num_ancillae,
expansion_mode='suzuki',
expansion_order=2,
evo_time=None, # This is t, can set to: np.pi*3/4
negative_evals=negative_evals,
ne_qfts=ne_qfts)
def fidelity(hhl, ref):
solution_hhl_normed = hhl / np.linalg.norm(hhl)
solution_ref_normed = ref / np.linalg.norm(ref)
fidelity = state_fidelity(solution_hhl_normed, solution_ref_normed)
print("Fidelity:\t\t %f" % fidelity)
matrix = [[1, -1/3], [-1/3, 1]]
vector = [1, 0]
orig_size = len(vector)
matrix, vector, truncate_powerdim, truncate_hermitian = HHL.matrix_resize(matrix, vector)
# Initialize eigenvalue finding module
eigs = create_eigs(matrix, 3, 50, False)
num_q, num_a = eigs.get_register_sizes()
# Initialize initial state module
init_state = Custom(num_q, state_vector=vector)
# Initialize reciprocal rotation module
reci = LookupRotation(negative_evals=eigs._negative_evals, evo_time=eigs._evo_time)
algo = HHL(matrix, vector, truncate_powerdim, truncate_hermitian, eigs,
init_state, reci, num_q, num_a, orig_size)
result = algo.run(QuantumInstance(Aer.get_backend('statevector_simulator')))
print("Solution:\t\t", np.round(result['solution'], 5))
result_ref = NumPyLSsolver(matrix, vector).run()
print("Classical Solution:\t", np.round(result_ref['solution'], 5))
print("Probability:\t\t %f" % result['probability_result'])
fidelity(result['solution'], result_ref['solution'])
print("circuit_width:\t", result['circuit_info']['width'])
print("circuit_depth:\t", result['circuit_info']['depth'])
print("CNOT gates:\t", result['circuit_info']['operations']['cx'])
from qiskit import QuantumRegister, QuantumCircuit
import numpy as np
t = 2 # This is not optimal; As an exercise, set this to the
# value that will get the best results. See section 8 for solution.
nqubits = 4 # Total number of qubits
nb = 1 # Number of qubits representing the solution
nl = 2 # Number of qubits representing the eigenvalues
theta = 0 # Angle defining |b>
a = 1 # Matrix diagonal
b = -1/3 # Matrix off-diagonal
# Initialise the quantum and classical registers
qr = QuantumRegister(nqubits)
# Create a Quantum Circuit
qc = QuantumCircuit(qr)
qrb = qr[0:nb]
qrl = qr[nb:nb+nl]
qra = qr[nb+nl:nb+nl+1]
# State preparation.
qc.ry(2*theta, qrb[0])
# QPE with e^{iAt}
for qu in qrl:
qc.h(qu)
qc.u1(a*t, qrl[0])
qc.u1(a*t*2, qrl[1])
qc.u3(b*t, -np.pi/2, np.pi/2, qrb[0])
# Controlled e^{iAt} on \lambda_{1}:
params=b*t
qc.u1(np.pi/2,qrb[0])
qc.cx(qrl[0],qrb[0])
qc.ry(params,qrb[0])
qc.cx(qrl[0],qrb[0])
qc.ry(-params,qrb[0])
qc.u1(3*np.pi/2,qrb[0])
# Controlled e^{2iAt} on \lambda_{2}:
params = b*t*2
qc.u1(np.pi/2,qrb[0])
qc.cx(qrl[1],qrb[0])
qc.ry(params,qrb[0])
qc.cx(qrl[1],qrb[0])
qc.ry(-params,qrb[0])
qc.u1(3*np.pi/2,qrb[0])
# Inverse QFT
qc.h(qrl[1])
qc.rz(-np.pi/4,qrl[1])
qc.cx(qrl[0],qrl[1])
qc.rz(np.pi/4,qrl[1])
qc.cx(qrl[0],qrl[1])
qc.rz(-np.pi/4,qrl[0])
qc.h(qrl[0])
# Eigenvalue rotation
t1=(-np.pi +np.pi/3 - 2*np.arcsin(1/3))/4
t2=(-np.pi -np.pi/3 + 2*np.arcsin(1/3))/4
t3=(np.pi -np.pi/3 - 2*np.arcsin(1/3))/4
t4=(np.pi +np.pi/3 + 2*np.arcsin(1/3))/4
qc.cx(qrl[1],qra[0])
qc.ry(t1,qra[0])
qc.cx(qrl[0],qra[0])
qc.ry(t2,qra[0])
qc.cx(qrl[1],qra[0])
qc.ry(t3,qra[0])
qc.cx(qrl[0],qra[0])
qc.ry(t4,qra[0])
qc.measure_all()
print("Depth: %i" % qc.depth())
print("CNOTS: %i" % qc.count_ops()['cx'])
qc.draw(fold=100)
from qiskit import execute, BasicAer, ClassicalRegister, IBMQ
from qiskit.compiler import transpile
from qiskit.ignis.mitigation.measurement import (complete_meas_cal, # Measurement error mitigation functions
CompleteMeasFitter,
MeasurementFilter)
provider = IBMQ.load_account()
backend = provider.get_backend('ibmqx2') # calibrate using real hardware
layout = [2,3,0,4]
chip_qubits = 5
# Transpiled circuit for the real hardware
qc_qa_cx = transpile(qc, backend=backend, initial_layout=layout)
meas_cals, state_labels = complete_meas_cal(qubit_list=layout, qr=QuantumRegister(chip_qubits))
qcs = meas_cals + [qc_qa_cx]
shots = 10
job = execute(qcs, backend=backend, shots=shots, optimization_level=0)
| 0.712032 | 0.984124 |
# ONNX side by side
The notebook compares two runtimes for the same ONNX and looks into differences at each step of the graph.
```
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%load_ext mlprodict
%matplotlib inline
```
## The ONNX model
We convert kernel function used in [GaussianProcessRegressor](https://scikit-learn.org/stable/modules/generated/sklearn.gaussian_process.GaussianProcessRegressor.html). First some values to use for testing.
```
import numpy
import pandas
from io import StringIO
Xtest = pandas.read_csv(StringIO("""
1.000000000000000000e+02,1.061277971307766705e+02,1.472195004809226493e+00,2.307125069497626552e-02,4.539948095743629591e-02,2.855191098141335870e-01
1.000000000000000000e+02,9.417031896832908444e+01,1.249743892709246573e+00,2.370416174339620707e-02,2.613847280316268853e-02,5.097165413593484073e-01
1.000000000000000000e+02,9.305231488674536422e+01,1.795726729335217264e+00,2.473274733802270642e-02,1.349765645107412620e-02,9.410288840541443378e-02
1.000000000000000000e+02,7.411264142156210255e+01,1.747723020195752319e+00,1.559695663417645997e-02,4.230394035515055301e-02,2.225492746314280956e-01
1.000000000000000000e+02,9.326006195761877393e+01,1.738860294343326229e+00,2.280160135767652502e-02,4.883335335161764074e-02,2.806808409247734115e-01
1.000000000000000000e+02,8.341529291866362428e+01,5.119682123742423929e-01,2.488795768635816003e-02,4.887573336092913834e-02,1.673462179673477768e-01
1.000000000000000000e+02,1.182436477919874562e+02,1.733516391831658954e+00,1.533520930349476820e-02,3.131213519485807895e-02,1.955345358785769427e-01
1.000000000000000000e+02,1.228982583299257101e+02,1.115599996405831629e+00,1.929354155079938959e-02,3.056996308544096715e-03,1.197052763998271013e-01
1.000000000000000000e+02,1.160303269386108838e+02,1.018627021014927303e+00,2.248784981616459844e-02,2.688111547114307651e-02,3.326105131778724355e-01
1.000000000000000000e+02,1.163414374640396005e+02,6.644299545804077667e-01,1.508088417713602906e-02,4.451836657613789106e-02,3.245643044204808425e-01
""".strip("\n\r ")), header=None).values
```
Then the kernel.
```
from sklearn.gaussian_process.kernels import RBF, ConstantKernel as CK, Sum
ker = Sum(
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=10,
length_scale_bounds=(1e-3, 1e3)),
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=1,
length_scale_bounds=(1e-3, 1e3))
)
ker
ker(Xtest)
```
## Conversion to ONNX
The function is not an operator, the function to use is specific to this usage.
```
from skl2onnx.operator_converters.gaussian_process import convert_kernel
from skl2onnx.common.data_types import FloatTensorType, DoubleTensorType
from skl2onnx.algebra.onnx_ops import OnnxIdentity
onnx_op = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float32, op_version=12)
onnx_op = OnnxIdentity(onnx_op, output_names=['Y'], op_version=12)
model_onnx = model_onnx = onnx_op.to_onnx(
inputs=[('X', FloatTensorType([None, None]))],
target_opset=12)
with open("model_onnx.onnx", "wb") as f:
f.write(model_onnx.SerializeToString())
```
``[('X', FloatTensorType([None, None]))]`` means the function applies on every tensor whatever its dimension is.
```
%onnxview model_onnx
from mlprodict.onnxrt import OnnxInference
from mlprodict.tools.asv_options_helper import get_ir_version_from_onnx
# line needed when onnx is more recent than onnxruntime
model_onnx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(model_onnx, inplace=False)
rtrun = OnnxInference(model_onnx, runtime="onnxruntime1")
pyres = pyrun.run({'X': Xtest.astype(numpy.float32)})
pyres
rtres = rtrun.run({'X': Xtest.astype(numpy.float32)})
rtres
from mlprodict.onnxrt.validate.validate_difference import measure_relative_difference
measure_relative_difference(pyres['Y'], rtres['Y'])
```
The last runtime uses the same runtime but with double instead of floats.
```
onnx_op_64 = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float64, op_version=12)
onnx_op_64 = OnnxIdentity(onnx_op_64, output_names=['Y'], op_version=12)
model_onnx_64 = onnx_op_64.to_onnx(
inputs=[('X', DoubleTensorType([None, None]))],
target_opset=12)
pyrun64 = OnnxInference(model_onnx_64, runtime="python", inplace=False)
pyres64 = pyrun64.run({'X': Xtest.astype(numpy.float64)})
measure_relative_difference(pyres['Y'], pyres64['Y'])
```
## Side by side
We run every node independently and we compare the output at each step.
```
%matplotlib inline
from mlprodict.onnxrt.validate.side_by_side import side_by_side_by_values
from pandas import DataFrame
def run_sbs(r1, r2, r3, x):
sbs = side_by_side_by_values([r1, r2, r3],
inputs=[
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float64)},
])
df = DataFrame(sbs)
dfd = df.drop(['value[0]', 'value[1]', 'value[2]'], axis=1).copy()
dfd.loc[dfd.cmp == 'ERROR->=inf', 'v[1]'] = 10
return dfd, sbs
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, Xtest)
dfd
ax = dfd[['name', 'v[2]']].iloc[1:].set_index('name').plot(kind='bar', figsize=(14,4), logy=True)
ax.set_title("relative difference for each output between python and onnxruntime");
```
Let's try for other inputs.
```
import warnings
from matplotlib.cbook.deprecation import MatplotlibDeprecationWarning
import matplotlib.pyplot as plt
with warnings.catch_warnings():
warnings.simplefilter("ignore", MatplotlibDeprecationWarning)
values = [4, 6, 8, 12]
fig, ax = plt.subplots(len(values), 2, figsize=(14, len(values) * 4))
for i, d in enumerate(values):
for j, dim in enumerate([3, 8]):
mat = numpy.random.rand(d, dim)
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, mat)
dfd[['name', 'v[1]']].iloc[1:].set_index('name').plot(
kind='bar', figsize=(14,4), logy=True, ax=ax[i, j])
ax[i, j].set_title("abs diff input shape {}".format(mat.shape))
if i < len(values) - 1:
for xlabel_i in ax[i, j].get_xticklabels():
xlabel_i.set_visible(False)
```
## Further analysis
If there is one issue, we can create a simple graph to test. We consider ``Y = A + B`` where *A* and *B* have the following name in the *ONNX* graph:
```
node = pyrun.sequence_[-2].onnx_node
final_inputs = list(node.input)
final_inputs
_, sbs = run_sbs(pyrun, rtrun, pyrun64, Xtest)
names = final_inputs + ['Y']
values = {}
for row in sbs:
if row.get('name', '#') not in names:
continue
name = row['name']
values[name] = [row["value[%d]" % i] for i in range(3)]
list(values.keys())
```
Let's check.
```
for name in names:
if name not in values:
raise Exception("Unable to find '{}' in\n{}".format(
name, [_.get('name', "?") for _ in sbs]))
a, b, c = names
for i in [0, 1, 2]:
A = values[a][i]
B = values[b][i]
Y = values[c][i]
diff = Y - (A + B)
dabs = numpy.max(numpy.abs(diff))
print(i, diff.dtype, dabs)
```
If the second runtime has issue, we can create a single node to check something.
```
from skl2onnx.algebra.onnx_ops import OnnxAdd
onnx_add = OnnxAdd('X1', 'X2', output_names=['Y'], op_version=12)
add_onnx = onnx_add.to_onnx({'X1': A, 'X2': B}, target_opset=12)
add_onnx.ir_version = get_ir_version_from_onnx()
pyrun_add = OnnxInference(add_onnx, inplace=False)
rtrun_add = OnnxInference(add_onnx, runtime="onnxruntime1")
res1 = pyrun_add.run({'X1': A, 'X2': B})
res2 = rtrun_add.run({'X1': A, 'X2': B})
measure_relative_difference(res1['Y'], res2['Y'])
```
No mistake here.
## onnxruntime
```
from onnxruntime import InferenceSession, RunOptions, SessionOptions
opt = SessionOptions()
opt.enable_mem_pattern = True
opt.enable_cpu_mem_arena = True
sess = InferenceSession(model_onnx.SerializeToString(), opt)
sess
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
```
## Side by side for MLPRegressor
```
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.neural_network import MLPRegressor
iris = load_iris()
X, y = iris.data, iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
clr = MLPRegressor()
clr.fit(X_train, y_train)
from mlprodict.onnx_conv import to_onnx
onx = to_onnx(clr, X_train.astype(numpy.float32), target_opset=12)
onx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(onx, runtime="python", inplace=False)
rtrun = OnnxInference(onx, runtime="onnxruntime1")
rt_partial_run = OnnxInference(onx, runtime="onnxruntime2")
dfd, _ = run_sbs(rtrun, rt_partial_run, pyrun, X_test)
dfd
%onnxview onx
```
|
github_jupyter
|
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%load_ext mlprodict
%matplotlib inline
import numpy
import pandas
from io import StringIO
Xtest = pandas.read_csv(StringIO("""
1.000000000000000000e+02,1.061277971307766705e+02,1.472195004809226493e+00,2.307125069497626552e-02,4.539948095743629591e-02,2.855191098141335870e-01
1.000000000000000000e+02,9.417031896832908444e+01,1.249743892709246573e+00,2.370416174339620707e-02,2.613847280316268853e-02,5.097165413593484073e-01
1.000000000000000000e+02,9.305231488674536422e+01,1.795726729335217264e+00,2.473274733802270642e-02,1.349765645107412620e-02,9.410288840541443378e-02
1.000000000000000000e+02,7.411264142156210255e+01,1.747723020195752319e+00,1.559695663417645997e-02,4.230394035515055301e-02,2.225492746314280956e-01
1.000000000000000000e+02,9.326006195761877393e+01,1.738860294343326229e+00,2.280160135767652502e-02,4.883335335161764074e-02,2.806808409247734115e-01
1.000000000000000000e+02,8.341529291866362428e+01,5.119682123742423929e-01,2.488795768635816003e-02,4.887573336092913834e-02,1.673462179673477768e-01
1.000000000000000000e+02,1.182436477919874562e+02,1.733516391831658954e+00,1.533520930349476820e-02,3.131213519485807895e-02,1.955345358785769427e-01
1.000000000000000000e+02,1.228982583299257101e+02,1.115599996405831629e+00,1.929354155079938959e-02,3.056996308544096715e-03,1.197052763998271013e-01
1.000000000000000000e+02,1.160303269386108838e+02,1.018627021014927303e+00,2.248784981616459844e-02,2.688111547114307651e-02,3.326105131778724355e-01
1.000000000000000000e+02,1.163414374640396005e+02,6.644299545804077667e-01,1.508088417713602906e-02,4.451836657613789106e-02,3.245643044204808425e-01
""".strip("\n\r ")), header=None).values
from sklearn.gaussian_process.kernels import RBF, ConstantKernel as CK, Sum
ker = Sum(
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=10,
length_scale_bounds=(1e-3, 1e3)),
CK(0.1, (1e-3, 1e3)) * RBF(length_scale=1,
length_scale_bounds=(1e-3, 1e3))
)
ker
ker(Xtest)
from skl2onnx.operator_converters.gaussian_process import convert_kernel
from skl2onnx.common.data_types import FloatTensorType, DoubleTensorType
from skl2onnx.algebra.onnx_ops import OnnxIdentity
onnx_op = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float32, op_version=12)
onnx_op = OnnxIdentity(onnx_op, output_names=['Y'], op_version=12)
model_onnx = model_onnx = onnx_op.to_onnx(
inputs=[('X', FloatTensorType([None, None]))],
target_opset=12)
with open("model_onnx.onnx", "wb") as f:
f.write(model_onnx.SerializeToString())
%onnxview model_onnx
from mlprodict.onnxrt import OnnxInference
from mlprodict.tools.asv_options_helper import get_ir_version_from_onnx
# line needed when onnx is more recent than onnxruntime
model_onnx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(model_onnx, inplace=False)
rtrun = OnnxInference(model_onnx, runtime="onnxruntime1")
pyres = pyrun.run({'X': Xtest.astype(numpy.float32)})
pyres
rtres = rtrun.run({'X': Xtest.astype(numpy.float32)})
rtres
from mlprodict.onnxrt.validate.validate_difference import measure_relative_difference
measure_relative_difference(pyres['Y'], rtres['Y'])
onnx_op_64 = convert_kernel(ker, 'X', output_names=['final_after_op_Add'],
dtype=numpy.float64, op_version=12)
onnx_op_64 = OnnxIdentity(onnx_op_64, output_names=['Y'], op_version=12)
model_onnx_64 = onnx_op_64.to_onnx(
inputs=[('X', DoubleTensorType([None, None]))],
target_opset=12)
pyrun64 = OnnxInference(model_onnx_64, runtime="python", inplace=False)
pyres64 = pyrun64.run({'X': Xtest.astype(numpy.float64)})
measure_relative_difference(pyres['Y'], pyres64['Y'])
%matplotlib inline
from mlprodict.onnxrt.validate.side_by_side import side_by_side_by_values
from pandas import DataFrame
def run_sbs(r1, r2, r3, x):
sbs = side_by_side_by_values([r1, r2, r3],
inputs=[
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float32)},
{'X': x.astype(numpy.float64)},
])
df = DataFrame(sbs)
dfd = df.drop(['value[0]', 'value[1]', 'value[2]'], axis=1).copy()
dfd.loc[dfd.cmp == 'ERROR->=inf', 'v[1]'] = 10
return dfd, sbs
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, Xtest)
dfd
ax = dfd[['name', 'v[2]']].iloc[1:].set_index('name').plot(kind='bar', figsize=(14,4), logy=True)
ax.set_title("relative difference for each output between python and onnxruntime");
import warnings
from matplotlib.cbook.deprecation import MatplotlibDeprecationWarning
import matplotlib.pyplot as plt
with warnings.catch_warnings():
warnings.simplefilter("ignore", MatplotlibDeprecationWarning)
values = [4, 6, 8, 12]
fig, ax = plt.subplots(len(values), 2, figsize=(14, len(values) * 4))
for i, d in enumerate(values):
for j, dim in enumerate([3, 8]):
mat = numpy.random.rand(d, dim)
dfd, _ = run_sbs(pyrun, rtrun, pyrun64, mat)
dfd[['name', 'v[1]']].iloc[1:].set_index('name').plot(
kind='bar', figsize=(14,4), logy=True, ax=ax[i, j])
ax[i, j].set_title("abs diff input shape {}".format(mat.shape))
if i < len(values) - 1:
for xlabel_i in ax[i, j].get_xticklabels():
xlabel_i.set_visible(False)
node = pyrun.sequence_[-2].onnx_node
final_inputs = list(node.input)
final_inputs
_, sbs = run_sbs(pyrun, rtrun, pyrun64, Xtest)
names = final_inputs + ['Y']
values = {}
for row in sbs:
if row.get('name', '#') not in names:
continue
name = row['name']
values[name] = [row["value[%d]" % i] for i in range(3)]
list(values.keys())
for name in names:
if name not in values:
raise Exception("Unable to find '{}' in\n{}".format(
name, [_.get('name', "?") for _ in sbs]))
a, b, c = names
for i in [0, 1, 2]:
A = values[a][i]
B = values[b][i]
Y = values[c][i]
diff = Y - (A + B)
dabs = numpy.max(numpy.abs(diff))
print(i, diff.dtype, dabs)
from skl2onnx.algebra.onnx_ops import OnnxAdd
onnx_add = OnnxAdd('X1', 'X2', output_names=['Y'], op_version=12)
add_onnx = onnx_add.to_onnx({'X1': A, 'X2': B}, target_opset=12)
add_onnx.ir_version = get_ir_version_from_onnx()
pyrun_add = OnnxInference(add_onnx, inplace=False)
rtrun_add = OnnxInference(add_onnx, runtime="onnxruntime1")
res1 = pyrun_add.run({'X1': A, 'X2': B})
res2 = rtrun_add.run({'X1': A, 'X2': B})
measure_relative_difference(res1['Y'], res2['Y'])
from onnxruntime import InferenceSession, RunOptions, SessionOptions
opt = SessionOptions()
opt.enable_mem_pattern = True
opt.enable_cpu_mem_arena = True
sess = InferenceSession(model_onnx.SerializeToString(), opt)
sess
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
res = sess.run(None, {'X': Xtest.astype(numpy.float32)})[0]
measure_relative_difference(pyres['Y'], res)
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.neural_network import MLPRegressor
iris = load_iris()
X, y = iris.data, iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y)
clr = MLPRegressor()
clr.fit(X_train, y_train)
from mlprodict.onnx_conv import to_onnx
onx = to_onnx(clr, X_train.astype(numpy.float32), target_opset=12)
onx.ir_version = get_ir_version_from_onnx()
pyrun = OnnxInference(onx, runtime="python", inplace=False)
rtrun = OnnxInference(onx, runtime="onnxruntime1")
rt_partial_run = OnnxInference(onx, runtime="onnxruntime2")
dfd, _ = run_sbs(rtrun, rt_partial_run, pyrun, X_test)
dfd
%onnxview onx
| 0.423935 | 0.892469 |
```
# default_exp data.preprocessing
```
# Data preprocessing
> Functions used to preprocess time series (both X and y).
```
#export
from tsai.imports import *
from tsai.utils import *
from tsai.data.external import *
from tsai.data.core import *
dsid = 'NATOPS'
X, y, splits = get_UCR_data(dsid, return_split=False)
tfms = [None, Categorize()]
dsets = TSDatasets(X, y, tfms=tfms, splits=splits)
#export
class ToNumpyCategory(Transform):
"Categorize a numpy batch"
order = 90
def __init__(self, **kwargs):
super().__init__(**kwargs)
def encodes(self, o: np.ndarray):
self.type = type(o)
self.cat = Categorize()
self.cat.setup(o)
self.vocab = self.cat.vocab
return np.asarray(stack([self.cat(oi) for oi in o]))
def decodes(self, o: (np.ndarray, torch.Tensor)):
return stack([self.cat.decode(oi) for oi in o])
t = ToNumpyCategory()
y_cat = t(y)
y_cat[:10]
test_eq(t.decode(tensor(y_cat)), y)
test_eq(t.decode(np.array(y_cat)), y)
#export
class OneHot(Transform):
"One-hot encode/ decode a batch"
order = 90
def __init__(self, n_classes=None, **kwargs):
self.n_classes = n_classes
super().__init__(**kwargs)
def encodes(self, o: torch.Tensor):
if not self.n_classes: self.n_classes = len(np.unique(o))
return torch.eye(self.n_classes)[o]
def encodes(self, o: np.ndarray):
o = ToNumpyCategory()(o)
if not self.n_classes: self.n_classes = len(np.unique(o))
return np.eye(self.n_classes)[o]
def decodes(self, o: torch.Tensor): return torch.argmax(o, dim=-1)
def decodes(self, o: np.ndarray): return np.argmax(o, axis=-1)
oh_encoder = OneHot()
y_cat = ToNumpyCategory()(y)
oht = oh_encoder(y_cat)
oht[:10]
n_classes = 10
n_samples = 100
t = torch.randint(0, n_classes, (n_samples,))
oh_encoder = OneHot()
oht = oh_encoder(t)
test_eq(oht.shape, (n_samples, n_classes))
test_eq(torch.argmax(oht, dim=-1), t)
test_eq(oh_encoder.decode(oht), t)
n_classes = 10
n_samples = 100
a = np.random.randint(0, n_classes, (n_samples,))
oh_encoder = OneHot()
oha = oh_encoder(a)
test_eq(oha.shape, (n_samples, n_classes))
test_eq(np.argmax(oha, axis=-1), a)
test_eq(oh_encoder.decode(oha), a)
#export
class Nan2Value(Transform):
"Replaces any nan values by a predefined value or median"
order = 90
def __init__(self, value=0, median=False, by_sample_and_var=True):
store_attr()
def encodes(self, o:TSTensor):
mask = torch.isnan(o)
if mask.any():
if self.median:
if self.by_sample_and_var:
median = torch.nanmedian(o, dim=2, keepdim=True)[0].repeat(1, 1, o.shape[-1])
o[mask] = median[mask]
else:
# o = torch.nan_to_num(o, torch.nanmedian(o)) # Only available in Pytorch 1.8
o = torch_nan_to_num(o, torch.nanmedian(o))
# o = torch.nan_to_num(o, self.value) # Only available in Pytorch 1.8
o = torch_nan_to_num(o, self.value)
return o
o = TSTensor(torch.randn(16, 10, 100))
o[0,0] = float('nan')
o[o > .9] = float('nan')
o[[0,1,5,8,14,15], :, -20:] = float('nan')
nan_vals1 = torch.isnan(o).sum()
o2 = Pipeline(Nan2Value(), split_idx=0)(o.clone())
o3 = Pipeline(Nan2Value(median=True, by_sample_and_var=True), split_idx=0)(o.clone())
o4 = Pipeline(Nan2Value(median=True, by_sample_and_var=False), split_idx=0)(o.clone())
nan_vals2 = torch.isnan(o2).sum()
nan_vals3 = torch.isnan(o3).sum()
nan_vals4 = torch.isnan(o4).sum()
test_ne(nan_vals1, 0)
test_eq(nan_vals2, 0)
test_eq(nan_vals3, 0)
test_eq(nan_vals4, 0)
# export
class TSStandardize(Transform):
"""Standardizes batch of type `TSTensor`
Args:
- mean: you can pass a precalculated mean value as a torch tensor which is the one that will be used, or leave as None, in which case
it will be estimated using a batch.
- std: you can pass a precalculated std value as a torch tensor which is the one that will be used, or leave as None, in which case
it will be estimated using a batch. If both mean and std values are passed when instantiating TSStandardize, the rest of arguments won't be used.
- by_sample: if True, it will calculate mean and std for each individual sample. Otherwise based on the entire batch.
- by_var:
* False: mean and std will be the same for all variables.
* True: a mean and std will be be different for each variable.
* a list of ints: (like [0,1,3]) a different mean and std will be set for each variable on the list. Variables not included in the list
won't be standardized.
* a list that contains a list/lists: (like[0, [1,3]]) a different mean and std will be set for each element of the list. If multiple elements are
included in a list, the same mean and std will be set for those variable in the sublist/s. (in the example a mean and std is determined for
variable 0, and another one for variables 1 & 3 - the same one). Variables not included in the list won't be standardized.
- by_step: if False, it will standardize values for each time step.
- eps: it avoids dividing by 0
- use_single_batch: if True a single training batch will be used to calculate mean & std. Else the entire training set will be used.
"""
parameters, order = L('mean', 'std'), 90
_setup = True # indicates it requires set up
def __init__(self, mean=None, std=None, by_sample=False, by_var=False, by_step=False, eps=1e-8, use_single_batch=True, verbose=False):
self.mean = tensor(mean) if mean is not None else None
self.std = tensor(std) if std is not None else None
self._setup = (mean is None or std is None) and not by_sample
self.eps = eps
self.by_sample, self.by_var, self.by_step = by_sample, by_var, by_step
drop_axes = []
if by_sample: drop_axes.append(0)
if by_var: drop_axes.append(1)
if by_step: drop_axes.append(2)
self.axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes])
if by_var and is_listy(by_var):
self.list_axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes]) + (1,)
self.use_single_batch = use_single_batch
self.verbose = verbose
if self.mean is not None or self.std is not None:
pv(f'{self.__class__.__name__} mean={self.mean}, std={self.std}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n', self.verbose)
@classmethod
def from_stats(cls, mean, std): return cls(mean, std)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
mean = torch.zeros(*shape, device=o.device)
std = torch.ones(*shape, device=o.device)
for v in self.by_var:
if not is_listy(v): v = [v]
mean[:, v] = torch_nanmean(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True)
std[:, v] = torch.clamp_min(torch_nanstd(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True), self.eps)
else:
mean = torch_nanmean(o, dim=self.axes, keepdim=self.axes!=())
std = torch.clamp_min(torch_nanstd(o, dim=self.axes, keepdim=self.axes!=()), self.eps)
self.mean, self.std = mean, std
if len(self.mean.shape) == 0:
pv(f'{self.__class__.__name__} mean={self.mean}, std={self.std}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
else:
pv(f'{self.__class__.__name__} mean shape={self.mean.shape}, std shape={self.std.shape}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
self._setup = False
elif self.by_sample: self.mean, self.std = torch.zeros(1), torch.ones(1)
def encodes(self, o:TSTensor):
if self.by_sample:
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
mean = torch.zeros(*shape, device=o.device)
std = torch.ones(*shape, device=o.device)
for v in self.by_var:
if not is_listy(v): v = [v]
mean[:, v] = torch_nanmean(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True)
std[:, v] = torch.clamp_min(torch_nanstd(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True), self.eps)
else:
mean = torch_nanmean(o, dim=self.axes, keepdim=self.axes!=())
std = torch.clamp_min(torch_nanstd(o, dim=self.axes, keepdim=self.axes!=()), self.eps)
self.mean, self.std = mean, std
return (o - self.mean) / self.std
def decodes(self, o:TSTensor):
if self.mean is None or self.std is None: return o
return o * self.std + self.mean
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step})'
batch_tfms=[TSStandardize(by_sample=True, by_var=False, verbose=True)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, batch_tfms=batch_tfms)
xb, yb = next(iter(dls.train))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
from tsai.data.validation import TimeSplitter
X_nan = np.random.rand(100, 5, 10)
idxs = np.random.choice(len(X_nan), int(len(X_nan)*.5), False)
X_nan[idxs, 0] = float('nan')
idxs = np.random.choice(len(X_nan), int(len(X_nan)*.5), False)
X_nan[idxs, 1, -10:] = float('nan')
batch_tfms = TSStandardize(by_var=True)
dls = get_ts_dls(X_nan, batch_tfms=batch_tfms, splits=TimeSplitter(show_plot=False)(range_of(X_nan)))
test_eq(torch.isnan(dls.after_batch[0].mean).sum(), 0)
test_eq(torch.isnan(dls.after_batch[0].std).sum(), 0)
xb = first(dls.train)[0]
test_ne(torch.isnan(xb).sum(), 0)
test_ne(torch.isnan(xb).sum(), torch.isnan(xb).numel())
batch_tfms = [TSStandardize(by_var=True), Nan2Value()]
dls = get_ts_dls(X_nan, batch_tfms=batch_tfms, splits=TimeSplitter(show_plot=False)(range_of(X_nan)))
xb = first(dls.train)[0]
test_eq(torch.isnan(xb).sum(), 0)
batch_tfms=[TSStandardize(by_sample=True, by_var=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = next(iter(dls.valid))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
tfms = [None, TSClassification()]
batch_tfms = TSStandardize(by_sample=True)
dls = get_ts_dls(X, y, splits=splits, tfms=tfms, batch_tfms=batch_tfms, bs=[64, 128], inplace=True)
xb, yb = dls.train.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = dls.valid.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
tfms = [None, TSClassification()]
batch_tfms = TSStandardize(by_sample=True, by_var=False, verbose=False)
dls = get_ts_dls(X, y, splits=splits, tfms=tfms, batch_tfms=batch_tfms, bs=[64, 128], inplace=False)
xb, yb = dls.train.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = dls.valid.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
#export
@patch
def mul_min(x:(torch.Tensor, TSTensor, NumpyTensor), axes=(), keepdim=False):
if axes == (): return retain_type(x.min(), x)
axes = reversed(sorted(axes if is_listy(axes) else [axes]))
min_x = x
for ax in axes: min_x, _ = min_x.min(ax, keepdim)
return retain_type(min_x, x)
@patch
def mul_max(x:(torch.Tensor, TSTensor, NumpyTensor), axes=(), keepdim=False):
if axes == (): return retain_type(x.max(), x)
axes = reversed(sorted(axes if is_listy(axes) else [axes]))
max_x = x
for ax in axes: max_x, _ = max_x.max(ax, keepdim)
return retain_type(max_x, x)
class TSNormalize(Transform):
"Normalizes batch of type `TSTensor`"
parameters, order = L('min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, min=None, max=None, range=(-1, 1), by_sample=False, by_var=False, by_step=False, clip_values=True,
use_single_batch=True, verbose=False):
self.min = tensor(min) if min is not None else None
self.max = tensor(max) if max is not None else None
self._setup = (self.min is None and self.max is None) and not by_sample
self.range_min, self.range_max = range
self.by_sample, self.by_var, self.by_step = by_sample, by_var, by_step
drop_axes = []
if by_sample: drop_axes.append(0)
if by_var: drop_axes.append(1)
if by_step: drop_axes.append(2)
self.axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes])
if by_var and is_listy(by_var):
self.list_axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes]) + (1,)
self.clip_values = clip_values
self.use_single_batch = use_single_batch
self.verbose = verbose
if self.min is not None or self.max is not None:
pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n', self.verbose)
@classmethod
def from_stats(cls, min, max, range_min=0, range_max=1): return cls(min, max, self.range_min, self.range_max)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
_min = torch.zeros(*shape, device=o.device) + self.range_min
_max = torch.zeros(*shape, device=o.device) + self.range_max
for v in self.by_var:
if not is_listy(v): v = [v]
_min[:, v] = o[:, v].mul_min(self.axes if len(v) == 1 else self.list_axes, keepdim=self.axes!=())
_max[:, v] = o[:, v].mul_max(self.axes if len(v) == 1 else self.list_axes, keepdim=self.axes!=())
else:
_min, _max = o.mul_min(self.axes, keepdim=self.axes!=()), o.mul_max(self.axes, keepdim=self.axes!=())
self.min, self.max = _min, _max
if len(self.min.shape) == 0:
pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
else:
pv(f'{self.__class__.__name__} min shape={self.min.shape}, max shape={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
self._setup = False
elif self.by_sample: self.min, self.max = -torch.ones(1), torch.ones(1)
def encodes(self, o:TSTensor):
if self.by_sample:
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
_min = torch.zeros(*shape, device=o.device) + self.range_min
_max = torch.ones(*shape, device=o.device) + self.range_max
for v in self.by_var:
if not is_listy(v): v = [v]
_min[:, v] = o[:, v].mul_min(self.axes, keepdim=self.axes!=())
_max[:, v] = o[:, v].mul_max(self.axes, keepdim=self.axes!=())
else:
_min, _max = o.mul_min(self.axes, keepdim=self.axes!=()), o.mul_max(self.axes, keepdim=self.axes!=())
self.min, self.max = _min, _max
output = ((o - self.min) / (self.max - self.min)) * (self.range_max - self.range_min) + self.range_min
if self.clip_values:
if self.by_var and is_listy(self.by_var):
for v in self.by_var:
if not is_listy(v): v = [v]
output[:, v] = torch.clamp(output[:, v], self.range_min, self.range_max)
else:
output = torch.clamp(output, self.range_min, self.range_max)
return output
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step})'
batch_tfms = [TSNormalize()]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
batch_tfms=[TSNormalize(by_sample=True, by_var=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
batch_tfms = [TSNormalize(by_var=[0, [1, 2]], use_single_batch=False, clip_values=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb[:, [0, 1, 2]].max() <= 1
assert xb[:, [0, 1, 2]].min() >= -1
#export
class TSClipOutliers(Transform):
"Clip outliers batch of type `TSTensor` based on the IQR"
parameters, order = L('min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, min=None, max=None, by_sample=False, by_var=False, use_single_batch=False, verbose=False):
self.min = tensor(min) if min is not None else tensor(-np.inf)
self.max = tensor(max) if max is not None else tensor(np.inf)
self.by_sample, self.by_var = by_sample, by_var
self._setup = (min is None or max is None) and not by_sample
if by_sample and by_var: self.axis = (2)
elif by_sample: self.axis = (1, 2)
elif by_var: self.axis = (0, 2)
else: self.axis = None
self.use_single_batch = use_single_batch
self.verbose = verbose
if min is not None or max is not None:
pv(f'{self.__class__.__name__} min={min}, max={max}\n', self.verbose)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
min, max = get_outliers_IQR(o, self.axis)
self.min, self.max = tensor(min), tensor(max)
if self.axis is None: pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
else: pv(f'{self.__class__.__name__} min={self.min.shape}, max={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
self._setup = False
def encodes(self, o:TSTensor):
if self.axis is None: return torch.clamp(o, self.min, self.max)
elif self.by_sample:
min, max = get_outliers_IQR(o, axis=self.axis)
self.min, self.max = o.new(min), o.new(max)
return torch_clamp(o, self.min, self.max)
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var})'
batch_tfms=[TSClipOutliers(-1, 1, verbose=True)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
test_close(xb.min(), -1, eps=1e-1)
test_close(xb.max(), 1, eps=1e-1)
xb, yb = next(iter(dls.valid))
test_close(xb.min(), -1, eps=1e-1)
test_close(xb.max(), 1, eps=1e-1)
# export
class TSClip(Transform):
"Clip batch of type `TSTensor`"
parameters, order = L('min', 'max'), 90
def __init__(self, min=-6, max=6):
self.min = torch.tensor(min)
self.max = torch.tensor(max)
def encodes(self, o:TSTensor):
return torch.clamp(o, self.min, self.max)
def __repr__(self): return f'{self.__class__.__name__}(min={self.min}, max={self.max})'
t = TSTensor(torch.randn(10, 20, 100)*10)
test_le(TSClip()(t).max().item(), 6)
test_ge(TSClip()(t).min().item(), -6)
#export
class TSRobustScale(Transform):
r"""This Scaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range)"""
parameters, order = L('median', 'min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, median=None, min=None, max=None, by_sample=False, by_var=False, quantile_range=(25.0, 75.0), use_single_batch=True, verbose=False):
self.median = tensor(median) if median is not None else tensor(0)
self.min = tensor(min) if min is not None else tensor(-np.inf)
self.max = tensor(max) if max is not None else tensor(np.inf)
self._setup = (median is None or min is None or max is None) and not by_sample
self.by_sample, self.by_var = by_sample, by_var
if by_sample and by_var: self.axis = (2)
elif by_sample: self.axis = (1, 2)
elif by_var: self.axis = (0, 2)
else: self.axis = None
self.use_single_batch = use_single_batch
self.verbose = verbose
self.quantile_range = quantile_range
if median is not None or min is not None or max is not None:
pv(f'{self.__class__.__name__} median={median} min={min}, max={max}\n', self.verbose)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
median = get_percentile(o, 50, self.axis)
min, max = get_outliers_IQR(o, self.axis, quantile_range=self.quantile_range)
self.median, self.min, self.max = tensor(median), tensor(min), tensor(max)
if self.axis is None: pv(f'{self.__class__.__name__} median={self.median} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
else: pv(f'{self.__class__.__name__} median={self.median.shape} min={self.min.shape}, max={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
self._setup = False
def encodes(self, o:TSTensor):
if self.by_sample:
median = get_percentile(o, 50, self.axis)
min, max = get_outliers_IQR(o, axis=self.axis, quantile_range=self.quantile_range)
self.median, self.min, self.max = o.new(median), o.new(min), o.new(max)
return (o - self.median) / (self.max - self.min)
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var})'
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, num_workers=0)
xb, yb = next(iter(dls.train))
clipped_xb = TSRobustScale(by_sample=true)(xb)
test_ne(clipped_xb, xb)
clipped_xb.min(), clipped_xb.max(), xb.min(), xb.max()
#export
class TSDiff(Transform):
"Differences batch of type `TSTensor`"
order = 90
def __init__(self, lag=1, pad=True):
self.lag, self.pad = lag, pad
def encodes(self, o:TSTensor):
return torch_diff(o, lag=self.lag, pad=self.pad)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor(torch.arange(24).reshape(2,3,4))
test_eq(TSDiff()(t)[..., 1:].float().mean(), 1)
test_eq(TSDiff(lag=2, pad=False)(t).float().mean(), 2)
#export
class TSLog(Transform):
"Log transforms batch of type `TSTensor` + 1. Accepts positive and negative numbers"
order = 90
def __init__(self, ex=None, **kwargs):
self.ex = ex
super().__init__(**kwargs)
def encodes(self, o:TSTensor):
output = torch.zeros_like(o)
output[o > 0] = torch.log1p(o[o > 0])
output[o < 0] = -torch.log1p(torch.abs(o[o < 0]))
if self.ex is not None: output[...,self.ex,:] = o[...,self.ex,:]
return output
def decodes(self, o:TSTensor):
output = torch.zeros_like(o)
output[o > 0] = torch.exp(o[o > 0]) - 1
output[o < 0] = -torch.exp(torch.abs(o[o < 0])) + 1
if self.ex is not None: output[...,self.ex,:] = o[...,self.ex,:]
return output
def __repr__(self): return f'{self.__class__.__name__}()'
t = TSTensor(torch.rand(2,3,4)) * 2 - 1
tfm = TSLog()
enc_t = tfm(t)
test_ne(enc_t, t)
test_close(tfm.decodes(enc_t).data, t.data)
#export
class TSCyclicalPosition(Transform):
"""Concatenates the position along the sequence as 2 additional variables (sine and cosine)
Args:
magnitude: added for compatibility. It's not used.
"""
order = 90
def __init__(self, magnitude=None, **kwargs):
super().__init__(**kwargs)
def encodes(self, o: TSTensor):
bs,_,seq_len = o.shape
sin, cos = sincos_encoding(seq_len, device=o.device)
output = torch.cat([o, sin.reshape(1,1,-1).repeat(bs,1,1), cos.reshape(1,1,-1).repeat(bs,1,1)], 1)
return output
bs, c_in, seq_len = 1,3,100
t = TSTensor(torch.rand(bs, c_in, seq_len))
enc_t = TSCyclicalPosition()(t)
test_ne(enc_t, t)
assert t.shape[1] == enc_t.shape[1] - 2
plt.plot(enc_t[0, -2:].cpu().numpy().T)
plt.show()
#export
class TSLinearPosition(Transform):
"""Concatenates the position along the sequence as 1 additional variable
Args:
magnitude: added for compatibility. It's not used.
"""
order = 90
def __init__(self, magnitude=None, lin_range=(-1,1), **kwargs):
self.lin_range = lin_range
super().__init__(**kwargs)
def encodes(self, o: TSTensor):
bs,_,seq_len = o.shape
lin = linear_encoding(seq_len, device=o.device, lin_range=self.lin_range)
output = torch.cat([o, lin.reshape(1,1,-1).repeat(bs,1,1)], 1)
return output
bs, c_in, seq_len = 1,3,100
t = TSTensor(torch.rand(bs, c_in, seq_len))
enc_t = TSLinearPosition()(t)
test_ne(enc_t, t)
assert t.shape[1] == enc_t.shape[1] - 1
plt.plot(enc_t[0, -1].cpu().numpy().T)
plt.show()
#export
class TSLogReturn(Transform):
"Calculates log-return of batch of type `TSTensor`. For positive values only"
order = 90
def __init__(self, lag=1, pad=True):
self.lag, self.pad = lag, pad
def encodes(self, o:TSTensor):
return torch_diff(torch.log(o), lag=self.lag, pad=self.pad)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor([1,2,4,8,16,32,64,128,256]).float()
test_eq(TSLogReturn(pad=False)(t).std(), 0)
#export
class TSAdd(Transform):
"Add a defined amount to each batch of type `TSTensor`."
order = 90
def __init__(self, add):
self.add = add
def encodes(self, o:TSTensor):
return torch.add(o, self.add)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor([1,2,3]).float()
test_eq(TSAdd(1)(t), TSTensor([2,3,4]).float())
```
# sklearn API transforms
```
#export
from sklearn.base import BaseEstimator, TransformerMixin
from fastai.data.transforms import CategoryMap
from joblib import dump, load
class TSShrinkDataFrame(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, skip=[], obj2cat=True, int2uint=False, verbose=True):
self.columns, self.skip, self.obj2cat, self.int2uint, self.verbose = listify(columns), skip, obj2cat, int2uint, verbose
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
self.old_dtypes = X.dtypes
if not self.columns: self.columns = X.columns
self.dt = df_shrink_dtypes(X[self.columns], self.skip, obj2cat=self.obj2cat, int2uint=self.int2uint)
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
if self.verbose:
start_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe is {start_memory} MB")
X[self.columns] = X[self.columns].astype(self.dt)
if self.verbose:
end_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe after reduction {end_memory} MB")
print(f"Reduced by {100 * (start_memory - end_memory) / start_memory} % ")
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
if self.verbose:
start_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe is {start_memory} MB")
X = X.astype(self.old_dtypes)
if self.verbose:
end_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe after reduction {end_memory} MB")
print(f"Reduced by {100 * (start_memory - end_memory) / start_memory} % ")
return X
df = pd.DataFrame()
df["ints64"] = np.random.randint(0,3,10)
df['floats64'] = np.random.rand(10)
tfm = TSShrinkDataFrame()
tfm.fit(df)
df = tfm.transform(df)
test_eq(df["ints64"].dtype, "int8")
test_eq(df["floats64"].dtype, "float32")
#export
class TSOneHotEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, drop=True, add_na=True, dtype=np.int64):
self.columns = listify(columns)
self.drop, self.add_na, self.dtype = drop, add_na, dtype
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
handle_unknown = "ignore" if self.add_na else "error"
self.ohe_tfm = sklearn.preprocessing.OneHotEncoder(handle_unknown=handle_unknown)
if len(self.columns) == 1:
self.ohe_tfm.fit(X[self.columns].to_numpy().reshape(-1, 1))
else:
self.ohe_tfm.fit(X[self.columns])
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
if len(self.columns) == 1:
output = self.ohe_tfm.transform(X[self.columns].to_numpy().reshape(-1, 1)).toarray().astype(self.dtype)
else:
output = self.ohe_tfm.transform(X[self.columns]).toarray().astype(self.dtype)
new_cols = []
for i,col in enumerate(self.columns):
for cats in self.ohe_tfm.categories_[i]:
new_cols.append(f"{str(col)}_{str(cats)}")
X[new_cols] = output
if self.drop: X = X.drop(self.columns, axis=1)
return X
df = pd.DataFrame()
df["a"] = np.random.randint(0,2,10)
df["b"] = np.random.randint(0,3,10)
unique_cols = len(df["a"].unique()) + len(df["b"].unique())
tfm = TSOneHotEncoder()
tfm.fit(df)
df = tfm.transform(df)
test_eq(df.shape[1], unique_cols)
#export
class TSCategoricalEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, add_na=True):
self.columns = listify(columns)
self.add_na = add_na
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
self.cat_tfms = []
for column in self.columns:
self.cat_tfms.append(CategoryMap(X[column], add_na=self.add_na))
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
for cat_tfm, column in zip(self.cat_tfms, self.columns):
X[column] = cat_tfm.map_objs(X[column])
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
for cat_tfm, column in zip(self.cat_tfms, self.columns):
X[column] = cat_tfm.map_ids(X[column])
return X
```
Stateful transforms like TSCategoricalEncoder can easily be serialized.
```
import joblib
df = pd.DataFrame()
df["a"] = alphabet[np.random.randint(0,2,100)]
df["b"] = ALPHABET[np.random.randint(0,3,100)]
a_unique = len(df["a"].unique())
b_unique = len(df["b"].unique())
tfm = TSCategoricalEncoder()
tfm.fit(df)
joblib.dump(tfm, "TSCategoricalEncoder.joblib")
tfm = joblib.load("TSCategoricalEncoder.joblib")
df = tfm.transform(df)
test_eq(df['a'].max(), a_unique)
test_eq(df['b'].max(), b_unique)
#export
default_date_attr = ['Year', 'Month', 'Week', 'Day', 'Dayofweek', 'Dayofyear', 'Is_month_end', 'Is_month_start',
'Is_quarter_end', 'Is_quarter_start', 'Is_year_end', 'Is_year_start']
class TSDateTimeEncoder(BaseEstimator, TransformerMixin):
def __init__(self, datetime_columns=None, prefix=None, drop=True, time=False, attr=default_date_attr):
self.datetime_columns = listify(datetime_columns)
self.prefix, self.drop, self.time, self.attr = prefix, drop, time ,attr
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if self.time: self.attr = self.attr + ['Hour', 'Minute', 'Second']
if not self.datetime_columns:
self.datetime_columns = X.columns
self.prefixes = []
for dt_column in self.datetime_columns:
self.prefixes.append(re.sub('[Dd]ate$', '', dt_column) if self.prefix is None else self.prefix)
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
for dt_column,prefix in zip(self.datetime_columns,self.prefixes):
make_date(X, dt_column)
field = X[dt_column]
# Pandas removed `dt.week` in v1.1.10
week = field.dt.isocalendar().week.astype(field.dt.day.dtype) if hasattr(field.dt, 'isocalendar') else field.dt.week
for n in self.attr: X[prefix + "_" + n] = getattr(field.dt, n.lower()) if n != 'Week' else week
if self.drop: X = X.drop(self.datetime_columns, axis=1)
return X
import datetime
df = pd.DataFrame()
df.loc[0, "date"] = datetime.datetime.now()
df.loc[1, "date"] = datetime.datetime.now() + pd.Timedelta(1, unit="D")
tfm = TSDateTimeEncoder()
joblib.dump(tfm, "TSDateTimeEncoder.joblib")
tfm = joblib.load("TSDateTimeEncoder.joblib")
tfm.fit_transform(df)
#export
class TSMissingnessEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None):
self.columns = listify(columns)
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
self.missing_columns = [f"{cn}_missing" for cn in self.columns]
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
X[self.missing_columns] = X[self.columns].isnull().astype(int)
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
X.drop(self.missing_columns, axis=1, inplace=True)
return X
data = np.random.rand(10,3)
data[data > .8] = np.nan
df = pd.DataFrame(data, columns=["a", "b", "c"])
tfm = TSMissingnessEncoder()
tfm.fit(df)
joblib.dump(tfm, "TSMissingnessEncoder.joblib")
tfm = joblib.load("TSMissingnessEncoder.joblib")
df = tfm.transform(df)
df
```
## y transforms
```
# export
class Preprocessor():
def __init__(self, preprocessor, **kwargs):
self.preprocessor = preprocessor(**kwargs)
def fit(self, o):
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
self.fit_preprocessor = self.preprocessor.fit(o)
return self.fit_preprocessor
def transform(self, o, copy=True):
if type(o) in [float, int]: o = array([o]).reshape(-1,1)
o_shape = o.shape
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
output = self.fit_preprocessor.transform(o).reshape(*o_shape)
if isinstance(o, torch.Tensor): return o.new(output)
return output
def inverse_transform(self, o, copy=True):
o_shape = o.shape
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
output = self.fit_preprocessor.inverse_transform(o).reshape(*o_shape)
if isinstance(o, torch.Tensor): return o.new(output)
return output
StandardScaler = partial(sklearn.preprocessing.StandardScaler)
setattr(StandardScaler, '__name__', 'StandardScaler')
RobustScaler = partial(sklearn.preprocessing.RobustScaler)
setattr(RobustScaler, '__name__', 'RobustScaler')
Normalizer = partial(sklearn.preprocessing.MinMaxScaler, feature_range=(-1, 1))
setattr(Normalizer, '__name__', 'Normalizer')
BoxCox = partial(sklearn.preprocessing.PowerTransformer, method='box-cox')
setattr(BoxCox, '__name__', 'BoxCox')
YeoJohnshon = partial(sklearn.preprocessing.PowerTransformer, method='yeo-johnson')
setattr(YeoJohnshon, '__name__', 'YeoJohnshon')
Quantile = partial(sklearn.preprocessing.QuantileTransformer, n_quantiles=1_000, output_distribution='normal', random_state=0)
setattr(Quantile, '__name__', 'Quantile')
# Standardize
from tsai.data.validation import TimeSplitter
y = random_shuffle(np.random.randn(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(StandardScaler)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# RobustScaler
y = random_shuffle(np.random.randn(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(RobustScaler)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# Normalize
y = random_shuffle(np.random.rand(1000) * 3 + .5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(Normalizer)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# BoxCox
y = random_shuffle(np.random.rand(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(BoxCox)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# YeoJohnshon
y = random_shuffle(np.random.randn(1000) * 10 + 5)
y = np.random.beta(.5, .5, size=1000)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(YeoJohnshon)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# QuantileTransformer
y = - np.random.beta(1, .5, 10000) * 10
splits = TimeSplitter()(y)
preprocessor = Preprocessor(Quantile)
preprocessor.fit(y[splits[0]])
plt.hist(y, 50, label='ori',)
y_tfm = preprocessor.transform(y)
plt.legend(loc='best')
plt.show()
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
test_close(preprocessor.inverse_transform(y_tfm), y, 1e-1)
#export
def ReLabeler(cm):
r"""Changes the labels in a dataset based on a dictionary (class mapping)
Args:
cm = class mapping dictionary
"""
def _relabel(y):
obj = len(set([len(listify(v)) for v in cm.values()])) > 1
keys = cm.keys()
if obj:
new_cm = {k:v for k,v in zip(keys, [listify(v) for v in cm.values()])}
return np.array([new_cm[yi] if yi in keys else listify(yi) for yi in y], dtype=object).reshape(*y.shape)
else:
new_cm = {k:v for k,v in zip(keys, [listify(v) for v in cm.values()])}
return np.array([new_cm[yi] if yi in keys else listify(yi) for yi in y]).reshape(*y.shape)
return _relabel
vals = {0:'a', 1:'b', 2:'c', 3:'d', 4:'e'}
y = np.array([vals[i] for i in np.random.randint(0, 5, 20)])
labeler = ReLabeler(dict(a='x', b='x', c='y', d='z', e='z'))
y_new = labeler(y)
test_eq(y.shape, y_new.shape)
y, y_new
#hide
from tsai.imports import create_scripts
from tsai.export import get_nb_name
nb_name = get_nb_name()
create_scripts(nb_name);
```
|
github_jupyter
|
# default_exp data.preprocessing
#export
from tsai.imports import *
from tsai.utils import *
from tsai.data.external import *
from tsai.data.core import *
dsid = 'NATOPS'
X, y, splits = get_UCR_data(dsid, return_split=False)
tfms = [None, Categorize()]
dsets = TSDatasets(X, y, tfms=tfms, splits=splits)
#export
class ToNumpyCategory(Transform):
"Categorize a numpy batch"
order = 90
def __init__(self, **kwargs):
super().__init__(**kwargs)
def encodes(self, o: np.ndarray):
self.type = type(o)
self.cat = Categorize()
self.cat.setup(o)
self.vocab = self.cat.vocab
return np.asarray(stack([self.cat(oi) for oi in o]))
def decodes(self, o: (np.ndarray, torch.Tensor)):
return stack([self.cat.decode(oi) for oi in o])
t = ToNumpyCategory()
y_cat = t(y)
y_cat[:10]
test_eq(t.decode(tensor(y_cat)), y)
test_eq(t.decode(np.array(y_cat)), y)
#export
class OneHot(Transform):
"One-hot encode/ decode a batch"
order = 90
def __init__(self, n_classes=None, **kwargs):
self.n_classes = n_classes
super().__init__(**kwargs)
def encodes(self, o: torch.Tensor):
if not self.n_classes: self.n_classes = len(np.unique(o))
return torch.eye(self.n_classes)[o]
def encodes(self, o: np.ndarray):
o = ToNumpyCategory()(o)
if not self.n_classes: self.n_classes = len(np.unique(o))
return np.eye(self.n_classes)[o]
def decodes(self, o: torch.Tensor): return torch.argmax(o, dim=-1)
def decodes(self, o: np.ndarray): return np.argmax(o, axis=-1)
oh_encoder = OneHot()
y_cat = ToNumpyCategory()(y)
oht = oh_encoder(y_cat)
oht[:10]
n_classes = 10
n_samples = 100
t = torch.randint(0, n_classes, (n_samples,))
oh_encoder = OneHot()
oht = oh_encoder(t)
test_eq(oht.shape, (n_samples, n_classes))
test_eq(torch.argmax(oht, dim=-1), t)
test_eq(oh_encoder.decode(oht), t)
n_classes = 10
n_samples = 100
a = np.random.randint(0, n_classes, (n_samples,))
oh_encoder = OneHot()
oha = oh_encoder(a)
test_eq(oha.shape, (n_samples, n_classes))
test_eq(np.argmax(oha, axis=-1), a)
test_eq(oh_encoder.decode(oha), a)
#export
class Nan2Value(Transform):
"Replaces any nan values by a predefined value or median"
order = 90
def __init__(self, value=0, median=False, by_sample_and_var=True):
store_attr()
def encodes(self, o:TSTensor):
mask = torch.isnan(o)
if mask.any():
if self.median:
if self.by_sample_and_var:
median = torch.nanmedian(o, dim=2, keepdim=True)[0].repeat(1, 1, o.shape[-1])
o[mask] = median[mask]
else:
# o = torch.nan_to_num(o, torch.nanmedian(o)) # Only available in Pytorch 1.8
o = torch_nan_to_num(o, torch.nanmedian(o))
# o = torch.nan_to_num(o, self.value) # Only available in Pytorch 1.8
o = torch_nan_to_num(o, self.value)
return o
o = TSTensor(torch.randn(16, 10, 100))
o[0,0] = float('nan')
o[o > .9] = float('nan')
o[[0,1,5,8,14,15], :, -20:] = float('nan')
nan_vals1 = torch.isnan(o).sum()
o2 = Pipeline(Nan2Value(), split_idx=0)(o.clone())
o3 = Pipeline(Nan2Value(median=True, by_sample_and_var=True), split_idx=0)(o.clone())
o4 = Pipeline(Nan2Value(median=True, by_sample_and_var=False), split_idx=0)(o.clone())
nan_vals2 = torch.isnan(o2).sum()
nan_vals3 = torch.isnan(o3).sum()
nan_vals4 = torch.isnan(o4).sum()
test_ne(nan_vals1, 0)
test_eq(nan_vals2, 0)
test_eq(nan_vals3, 0)
test_eq(nan_vals4, 0)
# export
class TSStandardize(Transform):
"""Standardizes batch of type `TSTensor`
Args:
- mean: you can pass a precalculated mean value as a torch tensor which is the one that will be used, or leave as None, in which case
it will be estimated using a batch.
- std: you can pass a precalculated std value as a torch tensor which is the one that will be used, or leave as None, in which case
it will be estimated using a batch. If both mean and std values are passed when instantiating TSStandardize, the rest of arguments won't be used.
- by_sample: if True, it will calculate mean and std for each individual sample. Otherwise based on the entire batch.
- by_var:
* False: mean and std will be the same for all variables.
* True: a mean and std will be be different for each variable.
* a list of ints: (like [0,1,3]) a different mean and std will be set for each variable on the list. Variables not included in the list
won't be standardized.
* a list that contains a list/lists: (like[0, [1,3]]) a different mean and std will be set for each element of the list. If multiple elements are
included in a list, the same mean and std will be set for those variable in the sublist/s. (in the example a mean and std is determined for
variable 0, and another one for variables 1 & 3 - the same one). Variables not included in the list won't be standardized.
- by_step: if False, it will standardize values for each time step.
- eps: it avoids dividing by 0
- use_single_batch: if True a single training batch will be used to calculate mean & std. Else the entire training set will be used.
"""
parameters, order = L('mean', 'std'), 90
_setup = True # indicates it requires set up
def __init__(self, mean=None, std=None, by_sample=False, by_var=False, by_step=False, eps=1e-8, use_single_batch=True, verbose=False):
self.mean = tensor(mean) if mean is not None else None
self.std = tensor(std) if std is not None else None
self._setup = (mean is None or std is None) and not by_sample
self.eps = eps
self.by_sample, self.by_var, self.by_step = by_sample, by_var, by_step
drop_axes = []
if by_sample: drop_axes.append(0)
if by_var: drop_axes.append(1)
if by_step: drop_axes.append(2)
self.axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes])
if by_var and is_listy(by_var):
self.list_axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes]) + (1,)
self.use_single_batch = use_single_batch
self.verbose = verbose
if self.mean is not None or self.std is not None:
pv(f'{self.__class__.__name__} mean={self.mean}, std={self.std}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n', self.verbose)
@classmethod
def from_stats(cls, mean, std): return cls(mean, std)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
mean = torch.zeros(*shape, device=o.device)
std = torch.ones(*shape, device=o.device)
for v in self.by_var:
if not is_listy(v): v = [v]
mean[:, v] = torch_nanmean(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True)
std[:, v] = torch.clamp_min(torch_nanstd(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True), self.eps)
else:
mean = torch_nanmean(o, dim=self.axes, keepdim=self.axes!=())
std = torch.clamp_min(torch_nanstd(o, dim=self.axes, keepdim=self.axes!=()), self.eps)
self.mean, self.std = mean, std
if len(self.mean.shape) == 0:
pv(f'{self.__class__.__name__} mean={self.mean}, std={self.std}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
else:
pv(f'{self.__class__.__name__} mean shape={self.mean.shape}, std shape={self.std.shape}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
self._setup = False
elif self.by_sample: self.mean, self.std = torch.zeros(1), torch.ones(1)
def encodes(self, o:TSTensor):
if self.by_sample:
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
mean = torch.zeros(*shape, device=o.device)
std = torch.ones(*shape, device=o.device)
for v in self.by_var:
if not is_listy(v): v = [v]
mean[:, v] = torch_nanmean(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True)
std[:, v] = torch.clamp_min(torch_nanstd(o[:, v], dim=self.axes if len(v) == 1 else self.list_axes, keepdim=True), self.eps)
else:
mean = torch_nanmean(o, dim=self.axes, keepdim=self.axes!=())
std = torch.clamp_min(torch_nanstd(o, dim=self.axes, keepdim=self.axes!=()), self.eps)
self.mean, self.std = mean, std
return (o - self.mean) / self.std
def decodes(self, o:TSTensor):
if self.mean is None or self.std is None: return o
return o * self.std + self.mean
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step})'
batch_tfms=[TSStandardize(by_sample=True, by_var=False, verbose=True)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, batch_tfms=batch_tfms)
xb, yb = next(iter(dls.train))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
from tsai.data.validation import TimeSplitter
X_nan = np.random.rand(100, 5, 10)
idxs = np.random.choice(len(X_nan), int(len(X_nan)*.5), False)
X_nan[idxs, 0] = float('nan')
idxs = np.random.choice(len(X_nan), int(len(X_nan)*.5), False)
X_nan[idxs, 1, -10:] = float('nan')
batch_tfms = TSStandardize(by_var=True)
dls = get_ts_dls(X_nan, batch_tfms=batch_tfms, splits=TimeSplitter(show_plot=False)(range_of(X_nan)))
test_eq(torch.isnan(dls.after_batch[0].mean).sum(), 0)
test_eq(torch.isnan(dls.after_batch[0].std).sum(), 0)
xb = first(dls.train)[0]
test_ne(torch.isnan(xb).sum(), 0)
test_ne(torch.isnan(xb).sum(), torch.isnan(xb).numel())
batch_tfms = [TSStandardize(by_var=True), Nan2Value()]
dls = get_ts_dls(X_nan, batch_tfms=batch_tfms, splits=TimeSplitter(show_plot=False)(range_of(X_nan)))
xb = first(dls.train)[0]
test_eq(torch.isnan(xb).sum(), 0)
batch_tfms=[TSStandardize(by_sample=True, by_var=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = next(iter(dls.valid))
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
tfms = [None, TSClassification()]
batch_tfms = TSStandardize(by_sample=True)
dls = get_ts_dls(X, y, splits=splits, tfms=tfms, batch_tfms=batch_tfms, bs=[64, 128], inplace=True)
xb, yb = dls.train.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = dls.valid.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
tfms = [None, TSClassification()]
batch_tfms = TSStandardize(by_sample=True, by_var=False, verbose=False)
dls = get_ts_dls(X, y, splits=splits, tfms=tfms, batch_tfms=batch_tfms, bs=[64, 128], inplace=False)
xb, yb = dls.train.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
xb, yb = dls.valid.one_batch()
test_close(xb.mean(), 0, eps=1e-1)
test_close(xb.std(), 1, eps=1e-1)
#export
@patch
def mul_min(x:(torch.Tensor, TSTensor, NumpyTensor), axes=(), keepdim=False):
if axes == (): return retain_type(x.min(), x)
axes = reversed(sorted(axes if is_listy(axes) else [axes]))
min_x = x
for ax in axes: min_x, _ = min_x.min(ax, keepdim)
return retain_type(min_x, x)
@patch
def mul_max(x:(torch.Tensor, TSTensor, NumpyTensor), axes=(), keepdim=False):
if axes == (): return retain_type(x.max(), x)
axes = reversed(sorted(axes if is_listy(axes) else [axes]))
max_x = x
for ax in axes: max_x, _ = max_x.max(ax, keepdim)
return retain_type(max_x, x)
class TSNormalize(Transform):
"Normalizes batch of type `TSTensor`"
parameters, order = L('min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, min=None, max=None, range=(-1, 1), by_sample=False, by_var=False, by_step=False, clip_values=True,
use_single_batch=True, verbose=False):
self.min = tensor(min) if min is not None else None
self.max = tensor(max) if max is not None else None
self._setup = (self.min is None and self.max is None) and not by_sample
self.range_min, self.range_max = range
self.by_sample, self.by_var, self.by_step = by_sample, by_var, by_step
drop_axes = []
if by_sample: drop_axes.append(0)
if by_var: drop_axes.append(1)
if by_step: drop_axes.append(2)
self.axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes])
if by_var and is_listy(by_var):
self.list_axes = tuple([ax for ax in (0, 1, 2) if ax not in drop_axes]) + (1,)
self.clip_values = clip_values
self.use_single_batch = use_single_batch
self.verbose = verbose
if self.min is not None or self.max is not None:
pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n', self.verbose)
@classmethod
def from_stats(cls, min, max, range_min=0, range_max=1): return cls(min, max, self.range_min, self.range_max)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
_min = torch.zeros(*shape, device=o.device) + self.range_min
_max = torch.zeros(*shape, device=o.device) + self.range_max
for v in self.by_var:
if not is_listy(v): v = [v]
_min[:, v] = o[:, v].mul_min(self.axes if len(v) == 1 else self.list_axes, keepdim=self.axes!=())
_max[:, v] = o[:, v].mul_max(self.axes if len(v) == 1 else self.list_axes, keepdim=self.axes!=())
else:
_min, _max = o.mul_min(self.axes, keepdim=self.axes!=()), o.mul_max(self.axes, keepdim=self.axes!=())
self.min, self.max = _min, _max
if len(self.min.shape) == 0:
pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
else:
pv(f'{self.__class__.__name__} min shape={self.min.shape}, max shape={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step}\n',
self.verbose)
self._setup = False
elif self.by_sample: self.min, self.max = -torch.ones(1), torch.ones(1)
def encodes(self, o:TSTensor):
if self.by_sample:
if self.by_var and is_listy(self.by_var):
shape = torch.mean(o, dim=self.axes, keepdim=self.axes!=()).shape
_min = torch.zeros(*shape, device=o.device) + self.range_min
_max = torch.ones(*shape, device=o.device) + self.range_max
for v in self.by_var:
if not is_listy(v): v = [v]
_min[:, v] = o[:, v].mul_min(self.axes, keepdim=self.axes!=())
_max[:, v] = o[:, v].mul_max(self.axes, keepdim=self.axes!=())
else:
_min, _max = o.mul_min(self.axes, keepdim=self.axes!=()), o.mul_max(self.axes, keepdim=self.axes!=())
self.min, self.max = _min, _max
output = ((o - self.min) / (self.max - self.min)) * (self.range_max - self.range_min) + self.range_min
if self.clip_values:
if self.by_var and is_listy(self.by_var):
for v in self.by_var:
if not is_listy(v): v = [v]
output[:, v] = torch.clamp(output[:, v], self.range_min, self.range_max)
else:
output = torch.clamp(output, self.range_min, self.range_max)
return output
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var}, by_step={self.by_step})'
batch_tfms = [TSNormalize()]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
batch_tfms=[TSNormalize(by_sample=True, by_var=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
batch_tfms = [TSNormalize(by_var=[0, [1, 2]], use_single_batch=False, clip_values=False, verbose=False)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb[:, [0, 1, 2]].max() <= 1
assert xb[:, [0, 1, 2]].min() >= -1
#export
class TSClipOutliers(Transform):
"Clip outliers batch of type `TSTensor` based on the IQR"
parameters, order = L('min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, min=None, max=None, by_sample=False, by_var=False, use_single_batch=False, verbose=False):
self.min = tensor(min) if min is not None else tensor(-np.inf)
self.max = tensor(max) if max is not None else tensor(np.inf)
self.by_sample, self.by_var = by_sample, by_var
self._setup = (min is None or max is None) and not by_sample
if by_sample and by_var: self.axis = (2)
elif by_sample: self.axis = (1, 2)
elif by_var: self.axis = (0, 2)
else: self.axis = None
self.use_single_batch = use_single_batch
self.verbose = verbose
if min is not None or max is not None:
pv(f'{self.__class__.__name__} min={min}, max={max}\n', self.verbose)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
min, max = get_outliers_IQR(o, self.axis)
self.min, self.max = tensor(min), tensor(max)
if self.axis is None: pv(f'{self.__class__.__name__} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
else: pv(f'{self.__class__.__name__} min={self.min.shape}, max={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
self._setup = False
def encodes(self, o:TSTensor):
if self.axis is None: return torch.clamp(o, self.min, self.max)
elif self.by_sample:
min, max = get_outliers_IQR(o, axis=self.axis)
self.min, self.max = o.new(min), o.new(max)
return torch_clamp(o, self.min, self.max)
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var})'
batch_tfms=[TSClipOutliers(-1, 1, verbose=True)]
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, bs=128, num_workers=0, after_batch=batch_tfms)
xb, yb = next(iter(dls.train))
assert xb.max() <= 1
assert xb.min() >= -1
test_close(xb.min(), -1, eps=1e-1)
test_close(xb.max(), 1, eps=1e-1)
xb, yb = next(iter(dls.valid))
test_close(xb.min(), -1, eps=1e-1)
test_close(xb.max(), 1, eps=1e-1)
# export
class TSClip(Transform):
"Clip batch of type `TSTensor`"
parameters, order = L('min', 'max'), 90
def __init__(self, min=-6, max=6):
self.min = torch.tensor(min)
self.max = torch.tensor(max)
def encodes(self, o:TSTensor):
return torch.clamp(o, self.min, self.max)
def __repr__(self): return f'{self.__class__.__name__}(min={self.min}, max={self.max})'
t = TSTensor(torch.randn(10, 20, 100)*10)
test_le(TSClip()(t).max().item(), 6)
test_ge(TSClip()(t).min().item(), -6)
#export
class TSRobustScale(Transform):
r"""This Scaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range)"""
parameters, order = L('median', 'min', 'max'), 90
_setup = True # indicates it requires set up
def __init__(self, median=None, min=None, max=None, by_sample=False, by_var=False, quantile_range=(25.0, 75.0), use_single_batch=True, verbose=False):
self.median = tensor(median) if median is not None else tensor(0)
self.min = tensor(min) if min is not None else tensor(-np.inf)
self.max = tensor(max) if max is not None else tensor(np.inf)
self._setup = (median is None or min is None or max is None) and not by_sample
self.by_sample, self.by_var = by_sample, by_var
if by_sample and by_var: self.axis = (2)
elif by_sample: self.axis = (1, 2)
elif by_var: self.axis = (0, 2)
else: self.axis = None
self.use_single_batch = use_single_batch
self.verbose = verbose
self.quantile_range = quantile_range
if median is not None or min is not None or max is not None:
pv(f'{self.__class__.__name__} median={median} min={min}, max={max}\n', self.verbose)
def setups(self, dl: DataLoader):
if self._setup:
if not self.use_single_batch:
o = dl.dataset.__getitem__([slice(None)])[0]
else:
o, *_ = dl.one_batch()
median = get_percentile(o, 50, self.axis)
min, max = get_outliers_IQR(o, self.axis, quantile_range=self.quantile_range)
self.median, self.min, self.max = tensor(median), tensor(min), tensor(max)
if self.axis is None: pv(f'{self.__class__.__name__} median={self.median} min={self.min}, max={self.max}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
else: pv(f'{self.__class__.__name__} median={self.median.shape} min={self.min.shape}, max={self.max.shape}, by_sample={self.by_sample}, by_var={self.by_var}\n',
self.verbose)
self._setup = False
def encodes(self, o:TSTensor):
if self.by_sample:
median = get_percentile(o, 50, self.axis)
min, max = get_outliers_IQR(o, axis=self.axis, quantile_range=self.quantile_range)
self.median, self.min, self.max = o.new(median), o.new(min), o.new(max)
return (o - self.median) / (self.max - self.min)
def __repr__(self): return f'{self.__class__.__name__}(by_sample={self.by_sample}, by_var={self.by_var})'
dls = TSDataLoaders.from_dsets(dsets.train, dsets.valid, num_workers=0)
xb, yb = next(iter(dls.train))
clipped_xb = TSRobustScale(by_sample=true)(xb)
test_ne(clipped_xb, xb)
clipped_xb.min(), clipped_xb.max(), xb.min(), xb.max()
#export
class TSDiff(Transform):
"Differences batch of type `TSTensor`"
order = 90
def __init__(self, lag=1, pad=True):
self.lag, self.pad = lag, pad
def encodes(self, o:TSTensor):
return torch_diff(o, lag=self.lag, pad=self.pad)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor(torch.arange(24).reshape(2,3,4))
test_eq(TSDiff()(t)[..., 1:].float().mean(), 1)
test_eq(TSDiff(lag=2, pad=False)(t).float().mean(), 2)
#export
class TSLog(Transform):
"Log transforms batch of type `TSTensor` + 1. Accepts positive and negative numbers"
order = 90
def __init__(self, ex=None, **kwargs):
self.ex = ex
super().__init__(**kwargs)
def encodes(self, o:TSTensor):
output = torch.zeros_like(o)
output[o > 0] = torch.log1p(o[o > 0])
output[o < 0] = -torch.log1p(torch.abs(o[o < 0]))
if self.ex is not None: output[...,self.ex,:] = o[...,self.ex,:]
return output
def decodes(self, o:TSTensor):
output = torch.zeros_like(o)
output[o > 0] = torch.exp(o[o > 0]) - 1
output[o < 0] = -torch.exp(torch.abs(o[o < 0])) + 1
if self.ex is not None: output[...,self.ex,:] = o[...,self.ex,:]
return output
def __repr__(self): return f'{self.__class__.__name__}()'
t = TSTensor(torch.rand(2,3,4)) * 2 - 1
tfm = TSLog()
enc_t = tfm(t)
test_ne(enc_t, t)
test_close(tfm.decodes(enc_t).data, t.data)
#export
class TSCyclicalPosition(Transform):
"""Concatenates the position along the sequence as 2 additional variables (sine and cosine)
Args:
magnitude: added for compatibility. It's not used.
"""
order = 90
def __init__(self, magnitude=None, **kwargs):
super().__init__(**kwargs)
def encodes(self, o: TSTensor):
bs,_,seq_len = o.shape
sin, cos = sincos_encoding(seq_len, device=o.device)
output = torch.cat([o, sin.reshape(1,1,-1).repeat(bs,1,1), cos.reshape(1,1,-1).repeat(bs,1,1)], 1)
return output
bs, c_in, seq_len = 1,3,100
t = TSTensor(torch.rand(bs, c_in, seq_len))
enc_t = TSCyclicalPosition()(t)
test_ne(enc_t, t)
assert t.shape[1] == enc_t.shape[1] - 2
plt.plot(enc_t[0, -2:].cpu().numpy().T)
plt.show()
#export
class TSLinearPosition(Transform):
"""Concatenates the position along the sequence as 1 additional variable
Args:
magnitude: added for compatibility. It's not used.
"""
order = 90
def __init__(self, magnitude=None, lin_range=(-1,1), **kwargs):
self.lin_range = lin_range
super().__init__(**kwargs)
def encodes(self, o: TSTensor):
bs,_,seq_len = o.shape
lin = linear_encoding(seq_len, device=o.device, lin_range=self.lin_range)
output = torch.cat([o, lin.reshape(1,1,-1).repeat(bs,1,1)], 1)
return output
bs, c_in, seq_len = 1,3,100
t = TSTensor(torch.rand(bs, c_in, seq_len))
enc_t = TSLinearPosition()(t)
test_ne(enc_t, t)
assert t.shape[1] == enc_t.shape[1] - 1
plt.plot(enc_t[0, -1].cpu().numpy().T)
plt.show()
#export
class TSLogReturn(Transform):
"Calculates log-return of batch of type `TSTensor`. For positive values only"
order = 90
def __init__(self, lag=1, pad=True):
self.lag, self.pad = lag, pad
def encodes(self, o:TSTensor):
return torch_diff(torch.log(o), lag=self.lag, pad=self.pad)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor([1,2,4,8,16,32,64,128,256]).float()
test_eq(TSLogReturn(pad=False)(t).std(), 0)
#export
class TSAdd(Transform):
"Add a defined amount to each batch of type `TSTensor`."
order = 90
def __init__(self, add):
self.add = add
def encodes(self, o:TSTensor):
return torch.add(o, self.add)
def __repr__(self): return f'{self.__class__.__name__}(lag={self.lag}, pad={self.pad})'
t = TSTensor([1,2,3]).float()
test_eq(TSAdd(1)(t), TSTensor([2,3,4]).float())
#export
from sklearn.base import BaseEstimator, TransformerMixin
from fastai.data.transforms import CategoryMap
from joblib import dump, load
class TSShrinkDataFrame(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, skip=[], obj2cat=True, int2uint=False, verbose=True):
self.columns, self.skip, self.obj2cat, self.int2uint, self.verbose = listify(columns), skip, obj2cat, int2uint, verbose
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
self.old_dtypes = X.dtypes
if not self.columns: self.columns = X.columns
self.dt = df_shrink_dtypes(X[self.columns], self.skip, obj2cat=self.obj2cat, int2uint=self.int2uint)
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
if self.verbose:
start_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe is {start_memory} MB")
X[self.columns] = X[self.columns].astype(self.dt)
if self.verbose:
end_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe after reduction {end_memory} MB")
print(f"Reduced by {100 * (start_memory - end_memory) / start_memory} % ")
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
if self.verbose:
start_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe is {start_memory} MB")
X = X.astype(self.old_dtypes)
if self.verbose:
end_memory = X.memory_usage().sum() / 1024**2
print(f"Memory usage of dataframe after reduction {end_memory} MB")
print(f"Reduced by {100 * (start_memory - end_memory) / start_memory} % ")
return X
df = pd.DataFrame()
df["ints64"] = np.random.randint(0,3,10)
df['floats64'] = np.random.rand(10)
tfm = TSShrinkDataFrame()
tfm.fit(df)
df = tfm.transform(df)
test_eq(df["ints64"].dtype, "int8")
test_eq(df["floats64"].dtype, "float32")
#export
class TSOneHotEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, drop=True, add_na=True, dtype=np.int64):
self.columns = listify(columns)
self.drop, self.add_na, self.dtype = drop, add_na, dtype
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
handle_unknown = "ignore" if self.add_na else "error"
self.ohe_tfm = sklearn.preprocessing.OneHotEncoder(handle_unknown=handle_unknown)
if len(self.columns) == 1:
self.ohe_tfm.fit(X[self.columns].to_numpy().reshape(-1, 1))
else:
self.ohe_tfm.fit(X[self.columns])
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
if len(self.columns) == 1:
output = self.ohe_tfm.transform(X[self.columns].to_numpy().reshape(-1, 1)).toarray().astype(self.dtype)
else:
output = self.ohe_tfm.transform(X[self.columns]).toarray().astype(self.dtype)
new_cols = []
for i,col in enumerate(self.columns):
for cats in self.ohe_tfm.categories_[i]:
new_cols.append(f"{str(col)}_{str(cats)}")
X[new_cols] = output
if self.drop: X = X.drop(self.columns, axis=1)
return X
df = pd.DataFrame()
df["a"] = np.random.randint(0,2,10)
df["b"] = np.random.randint(0,3,10)
unique_cols = len(df["a"].unique()) + len(df["b"].unique())
tfm = TSOneHotEncoder()
tfm.fit(df)
df = tfm.transform(df)
test_eq(df.shape[1], unique_cols)
#export
class TSCategoricalEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None, add_na=True):
self.columns = listify(columns)
self.add_na = add_na
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
self.cat_tfms = []
for column in self.columns:
self.cat_tfms.append(CategoryMap(X[column], add_na=self.add_na))
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
for cat_tfm, column in zip(self.cat_tfms, self.columns):
X[column] = cat_tfm.map_objs(X[column])
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
for cat_tfm, column in zip(self.cat_tfms, self.columns):
X[column] = cat_tfm.map_ids(X[column])
return X
import joblib
df = pd.DataFrame()
df["a"] = alphabet[np.random.randint(0,2,100)]
df["b"] = ALPHABET[np.random.randint(0,3,100)]
a_unique = len(df["a"].unique())
b_unique = len(df["b"].unique())
tfm = TSCategoricalEncoder()
tfm.fit(df)
joblib.dump(tfm, "TSCategoricalEncoder.joblib")
tfm = joblib.load("TSCategoricalEncoder.joblib")
df = tfm.transform(df)
test_eq(df['a'].max(), a_unique)
test_eq(df['b'].max(), b_unique)
#export
default_date_attr = ['Year', 'Month', 'Week', 'Day', 'Dayofweek', 'Dayofyear', 'Is_month_end', 'Is_month_start',
'Is_quarter_end', 'Is_quarter_start', 'Is_year_end', 'Is_year_start']
class TSDateTimeEncoder(BaseEstimator, TransformerMixin):
def __init__(self, datetime_columns=None, prefix=None, drop=True, time=False, attr=default_date_attr):
self.datetime_columns = listify(datetime_columns)
self.prefix, self.drop, self.time, self.attr = prefix, drop, time ,attr
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if self.time: self.attr = self.attr + ['Hour', 'Minute', 'Second']
if not self.datetime_columns:
self.datetime_columns = X.columns
self.prefixes = []
for dt_column in self.datetime_columns:
self.prefixes.append(re.sub('[Dd]ate$', '', dt_column) if self.prefix is None else self.prefix)
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
for dt_column,prefix in zip(self.datetime_columns,self.prefixes):
make_date(X, dt_column)
field = X[dt_column]
# Pandas removed `dt.week` in v1.1.10
week = field.dt.isocalendar().week.astype(field.dt.day.dtype) if hasattr(field.dt, 'isocalendar') else field.dt.week
for n in self.attr: X[prefix + "_" + n] = getattr(field.dt, n.lower()) if n != 'Week' else week
if self.drop: X = X.drop(self.datetime_columns, axis=1)
return X
import datetime
df = pd.DataFrame()
df.loc[0, "date"] = datetime.datetime.now()
df.loc[1, "date"] = datetime.datetime.now() + pd.Timedelta(1, unit="D")
tfm = TSDateTimeEncoder()
joblib.dump(tfm, "TSDateTimeEncoder.joblib")
tfm = joblib.load("TSDateTimeEncoder.joblib")
tfm.fit_transform(df)
#export
class TSMissingnessEncoder(BaseEstimator, TransformerMixin):
def __init__(self, columns=None):
self.columns = listify(columns)
def fit(self, X:pd.DataFrame, y=None, **fit_params):
assert isinstance(X, pd.DataFrame)
if not self.columns: self.columns = X.columns
self.missing_columns = [f"{cn}_missing" for cn in self.columns]
return self
def transform(self, X:pd.DataFrame, y=None, **transform_params):
assert isinstance(X, pd.DataFrame)
X[self.missing_columns] = X[self.columns].isnull().astype(int)
return X
def inverse_transform(self, X):
assert isinstance(X, pd.DataFrame)
X.drop(self.missing_columns, axis=1, inplace=True)
return X
data = np.random.rand(10,3)
data[data > .8] = np.nan
df = pd.DataFrame(data, columns=["a", "b", "c"])
tfm = TSMissingnessEncoder()
tfm.fit(df)
joblib.dump(tfm, "TSMissingnessEncoder.joblib")
tfm = joblib.load("TSMissingnessEncoder.joblib")
df = tfm.transform(df)
df
# export
class Preprocessor():
def __init__(self, preprocessor, **kwargs):
self.preprocessor = preprocessor(**kwargs)
def fit(self, o):
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
self.fit_preprocessor = self.preprocessor.fit(o)
return self.fit_preprocessor
def transform(self, o, copy=True):
if type(o) in [float, int]: o = array([o]).reshape(-1,1)
o_shape = o.shape
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
output = self.fit_preprocessor.transform(o).reshape(*o_shape)
if isinstance(o, torch.Tensor): return o.new(output)
return output
def inverse_transform(self, o, copy=True):
o_shape = o.shape
if isinstance(o, pd.Series): o = o.values.reshape(-1,1)
else: o = o.reshape(-1,1)
output = self.fit_preprocessor.inverse_transform(o).reshape(*o_shape)
if isinstance(o, torch.Tensor): return o.new(output)
return output
StandardScaler = partial(sklearn.preprocessing.StandardScaler)
setattr(StandardScaler, '__name__', 'StandardScaler')
RobustScaler = partial(sklearn.preprocessing.RobustScaler)
setattr(RobustScaler, '__name__', 'RobustScaler')
Normalizer = partial(sklearn.preprocessing.MinMaxScaler, feature_range=(-1, 1))
setattr(Normalizer, '__name__', 'Normalizer')
BoxCox = partial(sklearn.preprocessing.PowerTransformer, method='box-cox')
setattr(BoxCox, '__name__', 'BoxCox')
YeoJohnshon = partial(sklearn.preprocessing.PowerTransformer, method='yeo-johnson')
setattr(YeoJohnshon, '__name__', 'YeoJohnshon')
Quantile = partial(sklearn.preprocessing.QuantileTransformer, n_quantiles=1_000, output_distribution='normal', random_state=0)
setattr(Quantile, '__name__', 'Quantile')
# Standardize
from tsai.data.validation import TimeSplitter
y = random_shuffle(np.random.randn(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(StandardScaler)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# RobustScaler
y = random_shuffle(np.random.randn(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(RobustScaler)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# Normalize
y = random_shuffle(np.random.rand(1000) * 3 + .5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(Normalizer)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# BoxCox
y = random_shuffle(np.random.rand(1000) * 10 + 5)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(BoxCox)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# YeoJohnshon
y = random_shuffle(np.random.randn(1000) * 10 + 5)
y = np.random.beta(.5, .5, size=1000)
splits = TimeSplitter()(y)
preprocessor = Preprocessor(YeoJohnshon)
preprocessor.fit(y[splits[0]])
y_tfm = preprocessor.transform(y)
test_close(preprocessor.inverse_transform(y_tfm), y)
plt.hist(y, 50, label='ori',)
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
# QuantileTransformer
y = - np.random.beta(1, .5, 10000) * 10
splits = TimeSplitter()(y)
preprocessor = Preprocessor(Quantile)
preprocessor.fit(y[splits[0]])
plt.hist(y, 50, label='ori',)
y_tfm = preprocessor.transform(y)
plt.legend(loc='best')
plt.show()
plt.hist(y_tfm, 50, label='tfm')
plt.legend(loc='best')
plt.show()
test_close(preprocessor.inverse_transform(y_tfm), y, 1e-1)
#export
def ReLabeler(cm):
r"""Changes the labels in a dataset based on a dictionary (class mapping)
Args:
cm = class mapping dictionary
"""
def _relabel(y):
obj = len(set([len(listify(v)) for v in cm.values()])) > 1
keys = cm.keys()
if obj:
new_cm = {k:v for k,v in zip(keys, [listify(v) for v in cm.values()])}
return np.array([new_cm[yi] if yi in keys else listify(yi) for yi in y], dtype=object).reshape(*y.shape)
else:
new_cm = {k:v for k,v in zip(keys, [listify(v) for v in cm.values()])}
return np.array([new_cm[yi] if yi in keys else listify(yi) for yi in y]).reshape(*y.shape)
return _relabel
vals = {0:'a', 1:'b', 2:'c', 3:'d', 4:'e'}
y = np.array([vals[i] for i in np.random.randint(0, 5, 20)])
labeler = ReLabeler(dict(a='x', b='x', c='y', d='z', e='z'))
y_new = labeler(y)
test_eq(y.shape, y_new.shape)
y, y_new
#hide
from tsai.imports import create_scripts
from tsai.export import get_nb_name
nb_name = get_nb_name()
create_scripts(nb_name);
| 0.722233 | 0.883034 |
# NoSQL (HBase) (sesión 5)

Esta hoja muestra cómo acceder a bases de datos HBase y también a conectar la salida con Jupyter.
Se puede utilizar el *shell* propio de HBase en el contenedor.
Con HBase vamos a simular un _clúster_ de varias máquinas con varios contenedores conectados. En el directorio `hbase` del repositorio git hay un script para ejecutar la instalación con `docker-compose`.
Para conectarse al _clúster_ con un _shell_ de hbase, hay que ejecutar, desde una terminal el siguiente comando de docker:
```bash
$ docker exec -ti hbase-regionserver hbase shell
Base Shell; enter 'help<RETURN>' for list of supported commands.
Type "exit<RETURN>" to leave the HBase Shell
Version 1.2.7, rac57c51f7ad25e312b4275665d62b34a5945422f, Fri Sep 7 16:11:05 CDT 2018
hbase(main):001:0>
```
```
from pprint import pprint as pp
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
%matplotlib inline
matplotlib.style.use('ggplot')
```
Usaremos la librería `happybase` para python. La cargamos a continuación y hacemos la conexión.
```
import os
import os.path as path
from urllib.request import urlretrieve
def download_file_upper_dir(baseurl, filename):
file = path.abspath(path.join(os.getcwd(),os.pardir,filename))
if not os.path.isfile(file):
urlretrieve(baseurl + '/' + filename, file)
baseurl = 'http://neuromancer.inf.um.es:8080/es.stackoverflow/'
download_file_upper_dir(baseurl, 'Posts.csv')
download_file_upper_dir(baseurl, 'Users.csv')
download_file_upper_dir(baseurl, 'Tags.csv')
download_file_upper_dir(baseurl, 'Comments.csv')
download_file_upper_dir(baseurl, 'Votes.csv')
!pip install happybase
import happybase
host = 'hbase-thriftserver'
pool = happybase.ConnectionPool(size=5, host=host)
with pool.connection() as connection:
print(connection.tables())
```
Para la carga inicial, vamos a crear todas las tablas con una única familia de columnas, `rawdata`, donde meteremos toda la información _raw_ comprimida. Después podremos hacer reorganizaciones de los datos para hacer el acceso más eficiente. Es una de las muchas ventajas de no tener un esquema.
```
# Create tables
tables = ['posts', 'votes', 'users', 'tags', 'comments']
for t in tables:
try:
with pool.connection() as connection:
connection.create_table(
t,
{
'rawdata': dict(max_versions=1,compression='GZ')
})
except Exception as e:
print("Database already exists: {0}. {1}".format(t, e))
pass
with pool.connection() as connection:
print(connection.tables())
```
El código de importación es siempre el mismo, ya que se coge la primera fila del CSV que contiene el nombre de las columnas y se utiliza para generar nombres de columnas dentro de la familia de columnas dada como parámetro. La función `csv_to_hbase()` acepta un fichero CSV a abrir, un nombre de tabla y una familia de columnas donde agregar las columnas del fichero CSV. En nuestro caso siempre va a ser `rawdata`.
```
import csv
def csv_to_hbase(file, tablename, cf):
with pool.connection() as connection, open(file) as f:
table = connection.table(tablename)
# La llamada csv.reader() crea un iterador sobre un fichero CSV
reader = csv.reader(f, dialect='excel')
# Se leen las columnas. Sus nombres se usarán para crear las diferentes columnas en la familia
columns = next(reader)
columns = [cf + ':' + c for c in columns]
with table.batch(batch_size=500) as b:
for row in reader:
# La primera columna se usará como Row Key
b.put(row[0], dict(zip(columns[1:], row[1:])))
for t in tables:
print("Importando tabla {0}...".format(t))
%time csv_to_hbase('../'+t.capitalize() + '.csv', t, 'rawdata')
```
### Consultas sencillas desde Python
A continuación veremos algunas consultas sencillas desde python usando el API de `happybase`.
```
with pool.connection() as connection:
posts = connection.table('posts')
```
Obtener el Post con `Id` 5. La orden más sencilla e inmediata de HBase es obtener una fila, opcionalmente limitando las columnas a mostrar:
```
posts.row(b'5',columns=[b'rawdata:Body'])
```
El siguiente código permite mostrar de forma amigable las tablas extraídas de la base de datos en forma de diccionario:
```
# http://stackoverflow.com/a/30525061/62365
class DictTable(dict):
# Overridden dict class which takes a dict in the form {'a': 2, 'b': 3},
# and renders an HTML Table in IPython Notebook.
def _repr_html_(self):
htmltext = ["<table width=100%>"]
for key, value in self.items():
htmltext.append("<tr>")
htmltext.append("<td>{0}</td>".format(key.decode('utf-8')))
htmltext.append("<td>{0}</td>".format(value.decode('utf-8')))
htmltext.append("</tr>")
htmltext.append("</table>")
return ''.join(htmltext)
# Muestra cómo queda la fila del Id del Post 9997
DictTable(posts.row(b'5'))
DictTable(posts.row(b'5',columns=[b'rawdata:AnswerCount',b'rawdata:AcceptedAnswerId']))
```
Y también se puede recorrer como un diccionario normal (el `decode` se utiliza para convertir los valores binarios de la base de datos a una codificación UTF-8):
```
row = posts.row(b'5')
for key, value in row.items():
print("Key = '%s', Value = '%s'" % (key, value.decode('utf-8')[:40]))
```
Finalmente, también se puede recorrer toda la tabla estableciendo filtros, que se estudiarán después. Se utiliza la función `scan`. Se puede iterar con los parámetros `key` y `data`. Por ejemplo, calcular el tamaño máximo de la longitud del texto de los posts:
**(OJO, es un ejemplo, no se debería hacer así)**
```
max_len = 0
for key, data in posts.scan():
cur_len = len(data[b'rawdata:Body'].decode('utf-8'))
if cur_len > max_len:
max_len = cur_len
print("Máxima longitud: %s caracteres." % (max_len))
```
### Construcción de estructuras anidadas
Al igual que pasaba con MongoDB, las bases de datos NoSQL como en este caso HBase permiten almacenar estructuras de datos complejas. En nuestro caso vamos a agregar los comentarios de cada pregunta o respuesta (post) en columnas del mismo. Para ello, creamos una nueva familia de columnas `comments`.
HBase es bueno para añadir columnas sencillas, por ejemplo que contengan un valor. Sin embargo, si queremos añadir objetos complejos, tenemos que jugar con la codificación de la familia de columnas y columna.
Usaremos el shell porque `happybase` no permite alterar tablas ya creadas. Para acceder al shell de HBase, tenemos que contactar al contenedor `hbase-regionserver`, de esta forma:
```bash
$ docker exec -ti hbase-regionserver hbase shell
```
En el `shell` de HBase pondremos lo siguiente:
```
disable 'posts'
alter 'posts', {NAME => 'comments', VERSIONS => 1}
enable 'posts'
```
Cada comentario que añadimos contiene, al menos:
- un id único
- un texto
- un autor
- etc.
¿Cómo se consigue meterlo en una única familia de columnas?
Hay varias formas. La que usaremos aquí, añadiremos el **id** de cada comentario como parte del nombre de la columna. Por ejemplo, el comentario con Id 2000, generará las columnas:
- `Id_2000` (valor 2000)
- `UserId_2000`
- `PostId_2000`
- `Text_2000`
con sus correspondientes valores. Así, todos los datos relativos al comentario con Id original 2000, estarán almacenados en todas las columnas que terminen en "`_2000`". La base de datos permite implementar filtros que nos permiten buscar esto de forma muy sencilla. Los veremos después.
```
with pool.connection() as connection:
comments = connection.table('comments')
posts = connection.table('posts')
with posts.batch(batch_size=500) as bp:
# Hacer un scan de la tabla
for key, data in comments.scan():
comment = {'comments:' +
d.decode('utf-8').split(':')[1] + "_" +
key.decode('utf-8') :
data[d].decode('utf-8') for d in data.keys()}
bp.put(data[b'rawdata:PostId'], comment)
DictTable(posts.row(b'7251'))
%timeit q = posts.row(b'7251')
from functools import reduce
def doit():
q = posts.row(b'7251')
(s,n) = reduce(lambda res, e:
(res[0]+len(e[1].decode('utf-8')), res[1]+1) if e[0].decode('utf-8').startswith('comments:Text') else res
, q.items(), (0,0))
return (s/n)
%timeit doit()
# MySQL -> 1.12 ms
# HBase -> 1.47 ms
```
## EJERCICIO: ¿Cómo sería el código para saber qué usuarios han comentado un post en particular?
## Wikipedia
Como otro ejemplo de carga de datos y de organización en HBase, veremos de manera simplificada el ejemplo de la wikipedia visto en teoría.
A continuación se descarga una pequeña parte del fichero de la wikipedia en XML:
```
download_file_upper_dir('http://neuromancer.inf.um.es:8080/wikipedia/','eswiki.xml.gz')
```
Se crea la tabla para albergar la `wikipedia`. Igual que la vista en teoría, pero aquí se usa `wikipedia` en vez de `wiki` para que no colisionen la versión completa con la reducida.
De nuevo en el `shell` de HBase:
```
create 'wikipedia' , 'text', 'revision'
disable 'wikipedia' # Para evitar su uso temporal
alter 'wikipedia' , { NAME => 'text', VERSIONS => org.apache.hadoop.hbase.HConstants::ALL_VERSIONS }
alter 'wikipedia' , { NAME => 'revision', VERSIONS => org.apache.hadoop.hbase.HConstants::ALL_VERSIONS }
alter 'wikipedia' , { NAME => 'text', COMPRESSION => 'GZ', BLOOMFILTER => 'ROW'}
enable 'wikipedia'
```
Este código, visto en teoría, recorre el árbol XML construyendo documentos y llamando a la función `callback` con cada uno. Los documentos son diccionarios con las claves encontradas dentro de los tags `<page>...</page>`.
```
import xml.sax
import re
class WikiHandler(xml.sax.handler.ContentHandler):
def __init__(self):
self._charBuffer = ''
self.document = {}
def _getCharacterData(self):
data = self._charBuffer
self._charBuffer = ''
return data
def parse(self, f, callback):
self.callback = callback
xml.sax.parse(f, self)
def characters(self, data):
self._charBuffer = self._charBuffer + data
def startElement(self, name, attrs):
if name == 'page':
# print 'Start of page'
self.document = {}
if re.match(r'title|timestamp|username|comment|text', name):
self._charBuffer = ''
def endElement(self, name):
if re.match(r'title|timestamp|username|comment|text', name):
self.document[name] = self._getCharacterData()
# print(name, ': ', self.document[name][:20])
if 'revision' == name:
self.callback(self.document)
```
El codigo a continuación, cada vez que el código anterior llama a la función `processdoc()` se añade un documento a la base de datos.
```
import time
import os
import gzip
class FillWikiTable():
"""Llena la tabla Wiki"""
def __init__(self,connection):
# Conectar a la base de datos a través de Thrift
self.table = connection.table('wikipedia')
def run(_s):
def processdoc(d):
print("Callback called with {0}".format(d['title']))
tuple_time = time.strptime(d['timestamp'], "%Y-%m-%dT%H:%M:%SZ")
timestamp = int(time.mktime(tuple_time))
_s.table.put(d['title'],
{'text:': d.get('text',''),
'revision:author': d.get('username',''),
'revision:comment': d.get('comment','')},
timestamp=timestamp)
with gzip.open(os.path.join(os.pardir,'eswiki.xml.gz'),'r') as f:
start = time.time()
WikiHandler().parse(f, processdoc)
end = time.time()
print ("End adding documents. Time: %.5f" % (end - start))
with pool.connection() as connection:
FillWikiTable(connection).run()
```
El código a continuación permite ver las diferentes versiones de una revisión. Como la versión reducida es muy pequeña no da lugar a que haya ninguna revisión, pero con este código se vería. Hace uso del _shell_ de HBase:
```
get 'wikipedia', 'Commodore Amiga', {COLUMN => 'revision',VERSIONS=>10}
```
### Enlazado de documentos en la wikipedia
Los artículos de la wikipedia llevan enlaces entre sí, incluyendo referencias del tipo `[[artículo referenciado]]`. Se pueden extraer estos enlaces y se puede construir un grafo de conexiones. Para cada artículo, se anotarán qué enlaces hay que salen de él y hacia qué otros artículos enlazan y también qué enlaces llegan a él. Esto se hará con dos familias de columnas, `from` y `to`.
En cada momento, se añadirá una columna `from:artículo` cuando un artículo nos apunte, y otras columnas `to:articulo` con los artículos que nosotros enlazamos.
```
import sys
class BuildLinks():
"""Llena la tabla de Links"""
def __init__(self,connection):
# Create table
try:
connection.create_table(
"wikilinks",
{
'from': dict(bloom_filter_type='ROW',max_versions=1),
'to' : dict(bloom_filter_type='ROW',max_versions=1)
})
except:
print ("Database wikilinks already exists.")
pass
self.table = connection.table('wikilinks')
self.wikitable = connection.table('wikipedia')
def run(self):
print("run")
linkpattern = r'\[\[([^\[\]\|\:\#][^\[\]\|:]*)(?:\|([^\[\]\|]+))?\]\]'
# target, label
with self.table.batch(batch_size=500) as b:
for key, data in self.wikitable.scan():
to_dict = {}
doc = key.strip().decode('utf-8')
print("\n{0}:".format(doc))
for mo in re.finditer(linkpattern, data[b'text:'].decode('utf-8')):
(target, label) = mo.groups()
target = target.strip()
if target == '':
continue
label = '' if not label else label
label = label.strip()
to_dict['to:' + target] = label
sys.stdout.write(".")
b.put(target, {'from:' + doc : label})
if bool(to_dict):
b.put(doc, to_dict)
with pool.connection() as connection:
BuildLinks(connection).run()
```
En la siguiente sesión veremos técnicas más sofisticadas de filtrado, pero por ahora se puede jugar con estas construcciones. Se puede seleccionar qué columnas se quiere mostrar e incluso filtros. En el _shell_:
```
scan 'wikilinks', {COLUMNS=>'to', FILTER => "ColumnPrefixFilter('A')", LIMIT => 300}
```
El proceso de `scan` recorre toda la tabla mostrando sólo las filas seleccionadas. HBase ofrece ciertas optimizaciones para que el escaneo sea eficiente, que veremos en la siguiente sesión.
Una introducción a los filtros y parámetros disponibles se puede ver [aquí](http://www.hadooptpoint.com/filters-in-hbase-shell/). En el _shell_:
```
scan 'wikipedia', {COLUMNS=>['revision'] , STARTROW => 'A', ENDROW=>'B'}
```
## EJERCICIO: Encontrar páginas que estén enlazadas y que ambas estén en la tabla `wikipedia`
(Ojo, no estarán todas porque es una versión reducida de la wikipedia)
## EJERCICIO: Probar diversas búsquedas sobre las tablas `wikipedia` y `wikilinks`
## EJERCICIO: Modificar la tabla `posts` para añadir una familia de columnas que guarde el histórico de ediciones guardado en `PostHistory.csv`. Usar como ejemplo la función `csv_to_hbase`
|
github_jupyter
|
$ docker exec -ti hbase-regionserver hbase shell
Base Shell; enter 'help<RETURN>' for list of supported commands.
Type "exit<RETURN>" to leave the HBase Shell
Version 1.2.7, rac57c51f7ad25e312b4275665d62b34a5945422f, Fri Sep 7 16:11:05 CDT 2018
hbase(main):001:0>
from pprint import pprint as pp
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
%matplotlib inline
matplotlib.style.use('ggplot')
import os
import os.path as path
from urllib.request import urlretrieve
def download_file_upper_dir(baseurl, filename):
file = path.abspath(path.join(os.getcwd(),os.pardir,filename))
if not os.path.isfile(file):
urlretrieve(baseurl + '/' + filename, file)
baseurl = 'http://neuromancer.inf.um.es:8080/es.stackoverflow/'
download_file_upper_dir(baseurl, 'Posts.csv')
download_file_upper_dir(baseurl, 'Users.csv')
download_file_upper_dir(baseurl, 'Tags.csv')
download_file_upper_dir(baseurl, 'Comments.csv')
download_file_upper_dir(baseurl, 'Votes.csv')
!pip install happybase
import happybase
host = 'hbase-thriftserver'
pool = happybase.ConnectionPool(size=5, host=host)
with pool.connection() as connection:
print(connection.tables())
# Create tables
tables = ['posts', 'votes', 'users', 'tags', 'comments']
for t in tables:
try:
with pool.connection() as connection:
connection.create_table(
t,
{
'rawdata': dict(max_versions=1,compression='GZ')
})
except Exception as e:
print("Database already exists: {0}. {1}".format(t, e))
pass
with pool.connection() as connection:
print(connection.tables())
import csv
def csv_to_hbase(file, tablename, cf):
with pool.connection() as connection, open(file) as f:
table = connection.table(tablename)
# La llamada csv.reader() crea un iterador sobre un fichero CSV
reader = csv.reader(f, dialect='excel')
# Se leen las columnas. Sus nombres se usarán para crear las diferentes columnas en la familia
columns = next(reader)
columns = [cf + ':' + c for c in columns]
with table.batch(batch_size=500) as b:
for row in reader:
# La primera columna se usará como Row Key
b.put(row[0], dict(zip(columns[1:], row[1:])))
for t in tables:
print("Importando tabla {0}...".format(t))
%time csv_to_hbase('../'+t.capitalize() + '.csv', t, 'rawdata')
with pool.connection() as connection:
posts = connection.table('posts')
posts.row(b'5',columns=[b'rawdata:Body'])
# http://stackoverflow.com/a/30525061/62365
class DictTable(dict):
# Overridden dict class which takes a dict in the form {'a': 2, 'b': 3},
# and renders an HTML Table in IPython Notebook.
def _repr_html_(self):
htmltext = ["<table width=100%>"]
for key, value in self.items():
htmltext.append("<tr>")
htmltext.append("<td>{0}</td>".format(key.decode('utf-8')))
htmltext.append("<td>{0}</td>".format(value.decode('utf-8')))
htmltext.append("</tr>")
htmltext.append("</table>")
return ''.join(htmltext)
# Muestra cómo queda la fila del Id del Post 9997
DictTable(posts.row(b'5'))
DictTable(posts.row(b'5',columns=[b'rawdata:AnswerCount',b'rawdata:AcceptedAnswerId']))
row = posts.row(b'5')
for key, value in row.items():
print("Key = '%s', Value = '%s'" % (key, value.decode('utf-8')[:40]))
max_len = 0
for key, data in posts.scan():
cur_len = len(data[b'rawdata:Body'].decode('utf-8'))
if cur_len > max_len:
max_len = cur_len
print("Máxima longitud: %s caracteres." % (max_len))
$ docker exec -ti hbase-regionserver hbase shell
disable 'posts'
alter 'posts', {NAME => 'comments', VERSIONS => 1}
enable 'posts'
with pool.connection() as connection:
comments = connection.table('comments')
posts = connection.table('posts')
with posts.batch(batch_size=500) as bp:
# Hacer un scan de la tabla
for key, data in comments.scan():
comment = {'comments:' +
d.decode('utf-8').split(':')[1] + "_" +
key.decode('utf-8') :
data[d].decode('utf-8') for d in data.keys()}
bp.put(data[b'rawdata:PostId'], comment)
DictTable(posts.row(b'7251'))
%timeit q = posts.row(b'7251')
from functools import reduce
def doit():
q = posts.row(b'7251')
(s,n) = reduce(lambda res, e:
(res[0]+len(e[1].decode('utf-8')), res[1]+1) if e[0].decode('utf-8').startswith('comments:Text') else res
, q.items(), (0,0))
return (s/n)
%timeit doit()
# MySQL -> 1.12 ms
# HBase -> 1.47 ms
download_file_upper_dir('http://neuromancer.inf.um.es:8080/wikipedia/','eswiki.xml.gz')
create 'wikipedia' , 'text', 'revision'
disable 'wikipedia' # Para evitar su uso temporal
alter 'wikipedia' , { NAME => 'text', VERSIONS => org.apache.hadoop.hbase.HConstants::ALL_VERSIONS }
alter 'wikipedia' , { NAME => 'revision', VERSIONS => org.apache.hadoop.hbase.HConstants::ALL_VERSIONS }
alter 'wikipedia' , { NAME => 'text', COMPRESSION => 'GZ', BLOOMFILTER => 'ROW'}
enable 'wikipedia'
import xml.sax
import re
class WikiHandler(xml.sax.handler.ContentHandler):
def __init__(self):
self._charBuffer = ''
self.document = {}
def _getCharacterData(self):
data = self._charBuffer
self._charBuffer = ''
return data
def parse(self, f, callback):
self.callback = callback
xml.sax.parse(f, self)
def characters(self, data):
self._charBuffer = self._charBuffer + data
def startElement(self, name, attrs):
if name == 'page':
# print 'Start of page'
self.document = {}
if re.match(r'title|timestamp|username|comment|text', name):
self._charBuffer = ''
def endElement(self, name):
if re.match(r'title|timestamp|username|comment|text', name):
self.document[name] = self._getCharacterData()
# print(name, ': ', self.document[name][:20])
if 'revision' == name:
self.callback(self.document)
import time
import os
import gzip
class FillWikiTable():
"""Llena la tabla Wiki"""
def __init__(self,connection):
# Conectar a la base de datos a través de Thrift
self.table = connection.table('wikipedia')
def run(_s):
def processdoc(d):
print("Callback called with {0}".format(d['title']))
tuple_time = time.strptime(d['timestamp'], "%Y-%m-%dT%H:%M:%SZ")
timestamp = int(time.mktime(tuple_time))
_s.table.put(d['title'],
{'text:': d.get('text',''),
'revision:author': d.get('username',''),
'revision:comment': d.get('comment','')},
timestamp=timestamp)
with gzip.open(os.path.join(os.pardir,'eswiki.xml.gz'),'r') as f:
start = time.time()
WikiHandler().parse(f, processdoc)
end = time.time()
print ("End adding documents. Time: %.5f" % (end - start))
with pool.connection() as connection:
FillWikiTable(connection).run()
get 'wikipedia', 'Commodore Amiga', {COLUMN => 'revision',VERSIONS=>10}
import sys
class BuildLinks():
"""Llena la tabla de Links"""
def __init__(self,connection):
# Create table
try:
connection.create_table(
"wikilinks",
{
'from': dict(bloom_filter_type='ROW',max_versions=1),
'to' : dict(bloom_filter_type='ROW',max_versions=1)
})
except:
print ("Database wikilinks already exists.")
pass
self.table = connection.table('wikilinks')
self.wikitable = connection.table('wikipedia')
def run(self):
print("run")
linkpattern = r'\[\[([^\[\]\|\:\#][^\[\]\|:]*)(?:\|([^\[\]\|]+))?\]\]'
# target, label
with self.table.batch(batch_size=500) as b:
for key, data in self.wikitable.scan():
to_dict = {}
doc = key.strip().decode('utf-8')
print("\n{0}:".format(doc))
for mo in re.finditer(linkpattern, data[b'text:'].decode('utf-8')):
(target, label) = mo.groups()
target = target.strip()
if target == '':
continue
label = '' if not label else label
label = label.strip()
to_dict['to:' + target] = label
sys.stdout.write(".")
b.put(target, {'from:' + doc : label})
if bool(to_dict):
b.put(doc, to_dict)
with pool.connection() as connection:
BuildLinks(connection).run()
scan 'wikilinks', {COLUMNS=>'to', FILTER => "ColumnPrefixFilter('A')", LIMIT => 300}
scan 'wikipedia', {COLUMNS=>['revision'] , STARTROW => 'A', ENDROW=>'B'}
| 0.234056 | 0.788461 |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings(action="ignore")
pd.set_option('display.max_columns', 100)
pd.set_option('display.max_rows',300)
path_processed = "../data/processed/pjud"
df_causas = pd.read_feather(f"{path_processed}/Consolidado_FULL_feather")
print(f"{len(df_causas)} registros en el dataset")
df_causas.head()
causas_oral = df_causas[df_causas.tipo_juzgado=='ORAL']
df_delitos_oral = causas_oral.materia.value_counts().rename_axis('delitos').to_frame('total').query("total>3000")
df_delitos_oral
fig, axes = plt.subplots(1, 1, figsize=(29, 18))
sns.barplot(y=df_delitos_oral.index, x="total", data=df_delitos_oral, orient="h")
plt.xticks(rotation= 90 );
df_audiencias_oral = causas_oral.tribunal.value_counts().rename_axis('tribunal').to_frame('total')
df_audiencias_oral
causas_oral.tipologia_materia.unique()
causas_oral.tipo_audiencia.unique()
causas_oral[causas_oral.tipo_audiencia=='AUDIENCIA DE JUICIO ORAL TOP']
causas_ley20000 = causas_oral[(causas_oral.tipologia_materia.str.contains('LEY 20.000')) & (causas_oral.tipo_audiencia=='AUDIENCIA DE JUICIO ORAL TOP')]
print(f"Existen {len(causas_ley20000)} audiencias de Juicio Oral relacionada a Ley 20000 desde 2015 a 2019")
```
## Por Tribunal y por delitos
```
causas_ley20000.groupby(by=['materia','tribunal'])
data = []
for (materia,tribunal), sub_df in causas_ley20000.groupby(by=['materia','tribunal']):
total = len(sub_df.tribunal_rit.unique())
poblacion = sub_df.iloc[0].poblacion
urbano = sub_df.iloc[0].urbano
rural = sub_df.iloc[0].rural
dotacion = sub_df.iloc[0].dotacion_jueces
proporcion = total / poblacion
territorial = rural / urbano
causas_jueces = total / dotacion
row = [tribunal, materia, total, poblacion, proporcion, territorial, causas_jueces]
data.append(row)
ley20000 = pd.DataFrame(data, columns=['tribunal','materia','total_audiencias','poblacion','proporcion', 'territorial','causas_jueces']).sort_values(["proporcion","materia"], ascending=False)[:50]
ley20000
sns.set_style(style="ticks")
sns.pairplot(ley20000, hue="tribunal", diag_kws={'bw': 0.2})
f, ax = plt.subplots(figsize=(7, 5))
sns.despine(f)
sns.histplot(
ley20000,
x="MATERIA", hue="TRIBUNAL",
multiple="stack",
palette="light:m_r",
edgecolor=".3",
linewidth=.5,
log_scale=True,
)
ax.xaxis.set_major_formatter(mpl.ticker.ScalarFormatter())
ax.set_xticks([500, 1000, 2000, 5000, 10000])
nodrogas = causas_oral.query("`TIPOLOGIA MATERIA` != 'LEY 20.000 TRAFICO ILICITO DE ESTUPEFACIENTES Y SUSTANCIAS SICOTROPICAS'")
data = []
for (materia,tribunal), sub_df in nodrogas.groupby(by=['MATERIA_x','TRIBUNAL']):
total = len(sub_df.RIT.unique())
poblacion = sub_df.iloc[0].POBLACION
proporcion = total / poblacion
row = [materia, tribunal, total, poblacion, proporcion]
data.append(row)
pd.DataFrame(data, columns=['MATERIA','TRIBUNAL','TOTAL','POBLACION','PROPORCION']).sort_values("PROPORCION", ascending=False)[:50]
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings(action="ignore")
pd.set_option('display.max_columns', 100)
pd.set_option('display.max_rows',300)
path_processed = "../data/processed/pjud"
df_causas = pd.read_feather(f"{path_processed}/Consolidado_FULL_feather")
print(f"{len(df_causas)} registros en el dataset")
df_causas.head()
causas_oral = df_causas[df_causas.tipo_juzgado=='ORAL']
df_delitos_oral = causas_oral.materia.value_counts().rename_axis('delitos').to_frame('total').query("total>3000")
df_delitos_oral
fig, axes = plt.subplots(1, 1, figsize=(29, 18))
sns.barplot(y=df_delitos_oral.index, x="total", data=df_delitos_oral, orient="h")
plt.xticks(rotation= 90 );
df_audiencias_oral = causas_oral.tribunal.value_counts().rename_axis('tribunal').to_frame('total')
df_audiencias_oral
causas_oral.tipologia_materia.unique()
causas_oral.tipo_audiencia.unique()
causas_oral[causas_oral.tipo_audiencia=='AUDIENCIA DE JUICIO ORAL TOP']
causas_ley20000 = causas_oral[(causas_oral.tipologia_materia.str.contains('LEY 20.000')) & (causas_oral.tipo_audiencia=='AUDIENCIA DE JUICIO ORAL TOP')]
print(f"Existen {len(causas_ley20000)} audiencias de Juicio Oral relacionada a Ley 20000 desde 2015 a 2019")
causas_ley20000.groupby(by=['materia','tribunal'])
data = []
for (materia,tribunal), sub_df in causas_ley20000.groupby(by=['materia','tribunal']):
total = len(sub_df.tribunal_rit.unique())
poblacion = sub_df.iloc[0].poblacion
urbano = sub_df.iloc[0].urbano
rural = sub_df.iloc[0].rural
dotacion = sub_df.iloc[0].dotacion_jueces
proporcion = total / poblacion
territorial = rural / urbano
causas_jueces = total / dotacion
row = [tribunal, materia, total, poblacion, proporcion, territorial, causas_jueces]
data.append(row)
ley20000 = pd.DataFrame(data, columns=['tribunal','materia','total_audiencias','poblacion','proporcion', 'territorial','causas_jueces']).sort_values(["proporcion","materia"], ascending=False)[:50]
ley20000
sns.set_style(style="ticks")
sns.pairplot(ley20000, hue="tribunal", diag_kws={'bw': 0.2})
f, ax = plt.subplots(figsize=(7, 5))
sns.despine(f)
sns.histplot(
ley20000,
x="MATERIA", hue="TRIBUNAL",
multiple="stack",
palette="light:m_r",
edgecolor=".3",
linewidth=.5,
log_scale=True,
)
ax.xaxis.set_major_formatter(mpl.ticker.ScalarFormatter())
ax.set_xticks([500, 1000, 2000, 5000, 10000])
nodrogas = causas_oral.query("`TIPOLOGIA MATERIA` != 'LEY 20.000 TRAFICO ILICITO DE ESTUPEFACIENTES Y SUSTANCIAS SICOTROPICAS'")
data = []
for (materia,tribunal), sub_df in nodrogas.groupby(by=['MATERIA_x','TRIBUNAL']):
total = len(sub_df.RIT.unique())
poblacion = sub_df.iloc[0].POBLACION
proporcion = total / poblacion
row = [materia, tribunal, total, poblacion, proporcion]
data.append(row)
pd.DataFrame(data, columns=['MATERIA','TRIBUNAL','TOTAL','POBLACION','PROPORCION']).sort_values("PROPORCION", ascending=False)[:50]
| 0.19853 | 0.427994 |
<a href="https://colab.research.google.com/github/jmend01/DS-Unit-1-Sprint-1-Dealing-With-Data/blob/master/Jonathan%20Mendoza%20DS_Unit_1_Sprint_Challenge_1.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Data Science Unit 1 Sprint Challenge 1
## Loading, cleaning, visualizing, and analyzing data
In this sprint challenge you will look at a dataset of the survival of patients who underwent surgery for breast cancer.
http://archive.ics.uci.edu/ml/datasets/Haberman%27s+Survival
Data Set Information:
The dataset contains cases from a study that was conducted between 1958 and 1970 at the University of Chicago's Billings Hospital on the survival of patients who had undergone surgery for breast cancer.
Attribute Information:
1. Age of patient at time of operation (numerical)
2. Patient's year of operation (year - 1900, numerical)
3. Number of positive axillary nodes detected (numerical)
4. Survival status (class attribute)
-- 1 = the patient survived 5 years or longer
-- 2 = the patient died within 5 year
Sprint challenges are evaluated based on satisfactory completion of each part. It is suggested you work through it in order, getting each aspect reasonably working, before trying to deeply explore, iterate, or refine any given step. Once you get to the end, if you want to go back and improve things, go for it!
## Part 1 - Load and validate the data
- Load the data as a `pandas` data frame.
- Validate that it has the appropriate number of observations (you can check the raw file, and also read the dataset description from UCI).
- Validate that you have no missing values.
- Add informative names to the features.
- The survival variable is encoded as 1 for surviving >5 years and 2 for not - change this to be 0 for not surviving and 1 for surviving >5 years (0/1 is a more traditional encoding of binary variables)
At the end, print the first five rows of the dataset to demonstrate the above.
### Import libraries, load & validate dataframe, update header names
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
!pip install pandas==0.23.4 #downgrade pandas for functinality on crosstab
col_names = ['operation_age','operation_year','axillary_nodes','survival_status']
df = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data', header = None, names = col_names )
df.head()
print(df.shape)
print(df.survival_status.value_counts())
df.isna().sum()
```
verified no NaN values present, checked data values for next operation
```
df['survival_status'] = np.where(df['survival_status']==1, 1, 0)
print(df.survival_status.value_counts())
df.head()
```
changed values under survival_status to conform with a 0/1 standard
## Part 2 - Examine the distribution and relationships of the features
Explore the data - create at least *2* tables (can be summary statistics or crosstabulations) and *2* plots illustrating the nature of the data.
This is open-ended, so to remind - first *complete* this task as a baseline, then go on to the remaining sections, and *then* as time allows revisit and explore further.
Hint - you may need to bin some variables depending on your chosen tables/plots.
### Figure 1
```
age_bins = pd.cut(df['operation_age'], [29,39,49,59,69,85])
print(age_bins.value_counts())
pd.crosstab(age_bins, df['survival_status'], normalize = 'index').plot.bar(figsize = (12,6))
plt.xlabel('Age at Time of Operation')
plt.ylabel('%')
plt.title('Age at Time of Operation vs Survival Status')
plt.show()
pd.crosstab(age_bins, df['survival_status'],normalize = 'index')
```
if you get the operation before age 40, you are more likely to survive past year 5 from the operation date.
### Figure 2
```
ax_bins = pd.cut(df['axillary_nodes'], [-1,0,5,10,15,20,30,52])
pd.crosstab(ax_bins,df['survival_status'], normalize = 'index').plot.bar(figsize = (9,6))
plt.xlabel('# of Axillary Nodes')
plt.ylabel('%')
plt.title('# of Axillary Nodes Found vs Survival Status')
plt.show()
pd.crosstab(ax_bins,df['survival_status'], normalize = 'index')
```
if no axillary nodes are found then survival beyond year 5 is more likely. There is an inverse relationship between the number of nodes and suvival status
### Figure 3
```
pd.crosstab(age_bins, ax_bins,normalize = 'index').plot.bar(figsize = (12,8), stacked = True)
plt.xlabel('Age at Time of Operation')
plt.ylabel('%')
plt.title('Age and # of Axillary Nodes Found')
plt.show()
pd.crosstab(age_bins, ax_bins,normalize = 'index')
```
positive correlation between age and number of axillary nodes found
## Part 3 - Analysis and Interpretation
Now that you've looked at the data, answer the following questions:
- What is at least one feature that looks to have a positive relationship with survival?
- What is at least one feature that looks to have a negative relationship with survival?
- How are those two features related with each other, and what might that mean?
Answer with text, but feel free to intersperse example code/results or refer to it from earlier.
Figure 2 shows a positive correlation between # of axillary nodes detected and survival of less than 5 years.
Figure 1 shows there is a negative relationship between age and survival of more than 5 years.
Figure 3 shows that with increased age there were more axillary nodes detected and those people were less likely to survive past year 5
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
!pip install pandas==0.23.4 #downgrade pandas for functinality on crosstab
col_names = ['operation_age','operation_year','axillary_nodes','survival_status']
df = pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/haberman/haberman.data', header = None, names = col_names )
df.head()
print(df.shape)
print(df.survival_status.value_counts())
df.isna().sum()
df['survival_status'] = np.where(df['survival_status']==1, 1, 0)
print(df.survival_status.value_counts())
df.head()
age_bins = pd.cut(df['operation_age'], [29,39,49,59,69,85])
print(age_bins.value_counts())
pd.crosstab(age_bins, df['survival_status'], normalize = 'index').plot.bar(figsize = (12,6))
plt.xlabel('Age at Time of Operation')
plt.ylabel('%')
plt.title('Age at Time of Operation vs Survival Status')
plt.show()
pd.crosstab(age_bins, df['survival_status'],normalize = 'index')
ax_bins = pd.cut(df['axillary_nodes'], [-1,0,5,10,15,20,30,52])
pd.crosstab(ax_bins,df['survival_status'], normalize = 'index').plot.bar(figsize = (9,6))
plt.xlabel('# of Axillary Nodes')
plt.ylabel('%')
plt.title('# of Axillary Nodes Found vs Survival Status')
plt.show()
pd.crosstab(ax_bins,df['survival_status'], normalize = 'index')
pd.crosstab(age_bins, ax_bins,normalize = 'index').plot.bar(figsize = (12,8), stacked = True)
plt.xlabel('Age at Time of Operation')
plt.ylabel('%')
plt.title('Age and # of Axillary Nodes Found')
plt.show()
pd.crosstab(age_bins, ax_bins,normalize = 'index')
| 0.394084 | 0.989327 |
# Lesson 7 Class Exercises: Matplotlib
With these class exercises we learn a few new things. When new knowledge is introduced you'll see the icon shown on the right:
<span style="float:right; margin-left:10px; clear:both;"></span>
## Get Started
Import the Numpy, Pandas, Matplotlib packages and the Jupyter notebook Matplotlib magic
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
```
## Exercise 1. Load and clean the data for plotting
Import the Real Minimum Wages dataset from https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv
```
minwages = pd.read_csv('https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv')
print(minwages.shape)
minwages.head()
```
Clean the data by performing the following:
1. Add a new column containing just the year
2. Drop rows with missing values
3. Keep only rows in the series "In 2015 constant prices at 2015 USD PPPs"
4. Keep only rows where the pay period is 'Annual'
5. Drop unwanted columns: 'Unnamed: 0', 'Time' and 'Series'
6. Rename the 'value' column as 'Salary'
7. Reset the indexes
```
minwages['Year'] = pd.to_datetime(minwages['Time']).dt.year
minwages.dropna(inplace=True)
minwages = minwages[minwages['Series'] == "In 2015 constant prices at 2015 USD PPPs"]
minwages = minwages[minwages['Pay period'] == "Annual"]
print(minwages.shape)
minwages.drop(['Unnamed: 0', 'Time', 'Series'], inplace=True, axis=1)
minwages.rename({'value' : 'Minimum_Salary'}, inplace=True, axis=1)
minwages.reset_index(drop=True, inplace=True)
minwages.head()
```
## Exercise 2. Add a quartile group column
Find the quartiles for the minimal annual salary. Add a new column to the dataframe named `Group` that contains the values QG1, QG2, QG3 and QG4 representeding the quartile gropu (QG) to which the row belongs. Rows with a value between 0 and the first quartile get the value QG1, rows between the 1st and 2nd quartile get the value QG2, etc.
```
q1 = minwages['Minimum_Salary'].quantile(q=0.25)
q2 = minwages['Minimum_Salary'].quantile(q=0.5)
q3 = minwages['Minimum_Salary'].quantile(q=0.75)
q4 = minwages['Minimum_Salary'].quantile(q=1)
print(q1, q2, q3, q4)
group = pd.Series(np.zeros(minwages.shape[0]))
group[(minwages['Minimum_Salary'] > 0) & (minwages['Minimum_Salary'] <= q1)] = 'QG1'
group[(minwages['Minimum_Salary'] > q1) & (minwages['Minimum_Salary'] <= q2)] = 'QG2'
group[(minwages['Minimum_Salary'] > q2) & (minwages['Minimum_Salary'] <= q3)] = 'QG3'
group[(minwages['Minimum_Salary'] > q3) & (minwages['Minimum_Salary'] <= q4)] = 'QG4'
group.unique()
minwages['Group'] = group
minwages.head()
```
## Exercise 3. Create a boxplot
Create a graph using a single axis that shows the boxplots of the four groups. This will allow us to see if we properly separated rows by quartiles. It will also allow us to see the spread of the data in each quartile. Be sure to lable the x-axis tick marks with the proper quantile group name.
```
group1 = minwages[(minwages['Group'] == "QG1")]
group2 = minwages[(minwages['Group'] == "QG2")]
group3 = minwages[(minwages['Group'] == "QG3")]
group4 = minwages[(minwages['Group'] == "QG4")]
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
plot = ax.boxplot([group1['Minimum_Salary'], group2['Minimum_Salary'],
group3['Minimum_Salary'], group4['Minimum_Salary']],
labels=['QG1', 'QG2','QG3', 'QG4'])
```
## Exercise 4. Create a Scatterplot
Create a single scatterplot to explore if the salaries in quartile group 1 and quartile group 4 are correlated. Hint: to compare two categories we must have an observational unit that is common between them. Be sure to add the x and y axis labels.
```
qgroup_minwages = minwages.pivot_table(values = 'Minimum_Salary', columns='Group', index='Year', aggfunc='mean')
qgroup_minwages
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.axes.set_xlabel('Lowest Quartile Salaries')
ax.axes.set_ylabel('Highest Quartile Salaries')
ax.scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'])
plt.show()
qgroup_minwages['QG1']
```
Recreate the plot above, but set a different color per year and size the points to be larger for later years and smaller for earlier years.
```
colors = np.arange(10, 120, 10)
sizes = np.arange(10, 120, 10)
colors
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.axes.set_xlabel('Lowest Quartile Salaries')
ax.axes.set_ylabel('Highest Quartile Salaries')
ax.scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.50)
plt.show()
```
## Exercise 5. Create a grid of scatterplots
Now, let's see the pairwise scatterplot of each quartile group with every other group. Create a 4x4 grid of subplots. The rows and columns of the subplot represent one of the 4 groups and each plot represents the scatterplot of those groups. You can skip the plots in the diagonal as these will always the same quartile group.
<span style="float:right; margin-left:10px; clear:both;"></span>
Use the following code to ensure that the plot is large enough to see detail:
```python
plt.rcParams["figure.figsize"] = (12, 12)
```
The code above sets the size of the image in "inches" (i.e. 12 x 12 inches). Also, because the x-axis and y-axis labels will be repeated, we only need to set them on the first column and last rows. You can set the y-axis labels on the first column by using the `set` function and providing the `ylabel` argument. For example.
```python
axes[0, 0].set(ylabel="QG1")
```
You can do the same for the x-axis on the bottom row using the same style:
```python
axes[3, 0].set(xlabel="QG1")
```
```
plt.rcParams["figure.figsize"] = (12, 12)
fig = plt.figure()
axes = fig.subplots(4, 4)
#axes[0, 0].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[0, 1].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[0, 2].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[0, 3].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[1, 0].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
#axes[1, 1].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[1, 2].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[1, 3].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[2, 0].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[2, 1].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
#axes[2, 2].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[2, 3].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[3, 0].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[3, 1].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[3, 2].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
#axes[3, 3].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
#axes[0, 0].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[0, 2].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[0, 3].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[1, 0].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
#axes[1, 1].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[1, 2].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[1, 3].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[2, 0].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[2, 1].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
#axes[2, 2].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[2, 3].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[3, 0].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[3, 1].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[3, 2].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
#axes[3, 3].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[0, 0].set(ylabel="QG1")
axes[1, 0].set(ylabel="QG2")
axes[2, 0].set(ylabel="QG3")
axes[3, 0].set(ylabel="QG4")
axes[3, 0].set(xlabel="QG1")
axes[3, 1].set(xlabel="QG2")
axes[3, 2].set(xlabel="QG3")
axes[3, 3].set(xlabel="QG4")
plt.show()
```
Do you see any correlation between any of the groups? If so, why do you suspect this is?
## Exercise 6. Create a grid of line plots
Now, let's create a line graph of changes over time for each quartile group. Let's use a 2x2 subplot grid with each grid showing a different group.
```
plt.rcParams["figure.figsize"] = (8, 8)
fig = plt.figure()
axes = fig.subplots(2, 2)
axes[0, 0].plot(qgroup_minwages['QG1'])
axes[0, 1].plot(qgroup_minwages['QG2'])
axes[1, 0].plot(qgroup_minwages['QG3'])
axes[1, 1].plot(qgroup_minwages['QG4'])
axes[0, 0].set(xlabel = 'QG1')
axes[0, 1].set(xlabel = 'QG2')
axes[1, 0].set(xlabel = 'QG3')
axes[1, 1].set(xlabel = 'QG4')
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
minwages = pd.read_csv('https://raw.githubusercontent.com/QuantEcon/lecture-source-py/master/source/_static/lecture_specific/pandas_panel/realwage.csv')
print(minwages.shape)
minwages.head()
minwages['Year'] = pd.to_datetime(minwages['Time']).dt.year
minwages.dropna(inplace=True)
minwages = minwages[minwages['Series'] == "In 2015 constant prices at 2015 USD PPPs"]
minwages = minwages[minwages['Pay period'] == "Annual"]
print(minwages.shape)
minwages.drop(['Unnamed: 0', 'Time', 'Series'], inplace=True, axis=1)
minwages.rename({'value' : 'Minimum_Salary'}, inplace=True, axis=1)
minwages.reset_index(drop=True, inplace=True)
minwages.head()
q1 = minwages['Minimum_Salary'].quantile(q=0.25)
q2 = minwages['Minimum_Salary'].quantile(q=0.5)
q3 = minwages['Minimum_Salary'].quantile(q=0.75)
q4 = minwages['Minimum_Salary'].quantile(q=1)
print(q1, q2, q3, q4)
group = pd.Series(np.zeros(minwages.shape[0]))
group[(minwages['Minimum_Salary'] > 0) & (minwages['Minimum_Salary'] <= q1)] = 'QG1'
group[(minwages['Minimum_Salary'] > q1) & (minwages['Minimum_Salary'] <= q2)] = 'QG2'
group[(minwages['Minimum_Salary'] > q2) & (minwages['Minimum_Salary'] <= q3)] = 'QG3'
group[(minwages['Minimum_Salary'] > q3) & (minwages['Minimum_Salary'] <= q4)] = 'QG4'
group.unique()
minwages['Group'] = group
minwages.head()
group1 = minwages[(minwages['Group'] == "QG1")]
group2 = minwages[(minwages['Group'] == "QG2")]
group3 = minwages[(minwages['Group'] == "QG3")]
group4 = minwages[(minwages['Group'] == "QG4")]
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
plot = ax.boxplot([group1['Minimum_Salary'], group2['Minimum_Salary'],
group3['Minimum_Salary'], group4['Minimum_Salary']],
labels=['QG1', 'QG2','QG3', 'QG4'])
qgroup_minwages = minwages.pivot_table(values = 'Minimum_Salary', columns='Group', index='Year', aggfunc='mean')
qgroup_minwages
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.axes.set_xlabel('Lowest Quartile Salaries')
ax.axes.set_ylabel('Highest Quartile Salaries')
ax.scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'])
plt.show()
qgroup_minwages['QG1']
colors = np.arange(10, 120, 10)
sizes = np.arange(10, 120, 10)
colors
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.axes.set_xlabel('Lowest Quartile Salaries')
ax.axes.set_ylabel('Highest Quartile Salaries')
ax.scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.50)
plt.show()
plt.rcParams["figure.figsize"] = (12, 12)
axes[0, 0].set(ylabel="QG1")
axes[3, 0].set(xlabel="QG1")
plt.rcParams["figure.figsize"] = (12, 12)
fig = plt.figure()
axes = fig.subplots(4, 4)
#axes[0, 0].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[0, 1].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[0, 2].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[0, 3].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[1, 0].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
#axes[1, 1].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[1, 2].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[1, 3].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[2, 0].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[2, 1].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
#axes[2, 2].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[2, 3].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[3, 0].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[3, 1].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[3, 2].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
#axes[3, 3].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
#axes[0, 0].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[0, 2].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[0, 3].scatter(qgroup_minwages['QG1'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[1, 0].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
#axes[1, 1].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[1, 2].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[1, 3].scatter(qgroup_minwages['QG2'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[2, 0].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[2, 1].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
#axes[2, 2].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
axes[2, 3].scatter(qgroup_minwages['QG3'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[3, 0].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG1'], c=colors, s=sizes, alpha=0.75)
axes[3, 1].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG2'], c=colors, s=sizes, alpha=0.75)
axes[3, 2].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG3'], c=colors, s=sizes, alpha=0.75)
#axes[3, 3].scatter(qgroup_minwages['QG4'], qgroup_minwages['QG4'], c=colors, s=sizes, alpha=0.75)
axes[0, 0].set(ylabel="QG1")
axes[1, 0].set(ylabel="QG2")
axes[2, 0].set(ylabel="QG3")
axes[3, 0].set(ylabel="QG4")
axes[3, 0].set(xlabel="QG1")
axes[3, 1].set(xlabel="QG2")
axes[3, 2].set(xlabel="QG3")
axes[3, 3].set(xlabel="QG4")
plt.show()
plt.rcParams["figure.figsize"] = (8, 8)
fig = plt.figure()
axes = fig.subplots(2, 2)
axes[0, 0].plot(qgroup_minwages['QG1'])
axes[0, 1].plot(qgroup_minwages['QG2'])
axes[1, 0].plot(qgroup_minwages['QG3'])
axes[1, 1].plot(qgroup_minwages['QG4'])
axes[0, 0].set(xlabel = 'QG1')
axes[0, 1].set(xlabel = 'QG2')
axes[1, 0].set(xlabel = 'QG3')
axes[1, 1].set(xlabel = 'QG4')
plt.show()
| 0.465145 | 0.98631 |
# Information Sampling Task Analysis (Comparison to Hunt)
Here, we explain how the IST is analyzed and compare the output from our own script to Hunt's.
```
import itertools
import pandas as pd
import numpy as np
# Calculating which row wins in each possible game
cards = list(range(1,11))
games = list(itertools.product(cards, repeat=4))
games = pd.DataFrame(games, columns = [1,2,3,4])
# In each of the games row A either wins or loses
games['row_a'] = games[[1,2]].sum(axis = 1)
games['row_b'] = games[[3,4]].sum(axis = 1)
games['row_a_wins'] = games.row_a > games.row_b # Note that ties are allowed
games['row_b_wins'] = games.row_a < games.row_b
# A dataframe with all possible states
states_0 = [()] # No card revealed at stage 0
states_1 = list(itertools.product(cards, repeat=1))
states_2 = list(itertools.product(cards, repeat=2))
states_3 = list(itertools.product(cards, repeat=3))
states_4 = list(itertools.product(cards, repeat=4))
states = states_0 + states_1 + states_2 + states_3 + states_4
states = pd.DataFrame({'state':states})
states['stage'] = states.state.apply(lambda x: len(x))
states['total_turn_cost'] = states.stage.apply(lambda x: np.sum([0,10,15,20][:x]))
# Calculating p_win for each state
def get_p_win(state):
state = {location+1:value for location,value in enumerate(list(state))}
possible_games = games.loc[(games[list(state)] == pd.Series(state)).all(axis=1)]
p_a_win = possible_games.row_a_wins.mean()
p_b_win = possible_games.row_b_wins.mean()
choice = 'row_a_wins' if p_a_win > p_b_win else 'row_b_wins'
p_win = possible_games[choice].mean()
return p_win
states['p_win'] = states.state.apply(get_p_win)
states['q_s_guess'] = states.p_win*60-(1-states.p_win)*50-states.total_turn_cost
df = states
df = states
def choose_best_action(df):
# This will fail if q_s_guess equals q_s_seek
df['best_action'] = (df['q_s_guess'] > df['q_s_seek']).replace({True:"guess",False:"seek"})
df.loc[df.q_s_guess.isnull() | df.q_s_seek.isnull(), 'best_action'] = np.nan
df['expected_best_value'] = df[['q_s_guess','q_s_seek']].max(axis=1)
df.loc[df.q_s_guess.isnull() | df.q_s_seek.isnull(), 'expected_best_value'] = np.nan
# We do not allow seeking in stage 4
states.loc[states.stage==4,'best_action'] = 'guess'
return df
def get_q_s_seek(state):
if len(state) == 4:
return -np.inf # We do not allow seeking in stage 4
expected_best_values = []
for i in range(1,11):
next_state = state + (i,)
expected_best_values.append(df[df.state==next_state].expected_best_value)
return np.mean(expected_best_values)
#df.loc[df.stage==4, 'q_s_guess'] = 15 # This is incorrect (but implied in Hunt's paper)
for stage in range(4,-1,-1): # Looping through stage 4 to 0
df.loc[df.stage==stage, 'q_s_seek'] = df.loc[df.stage==stage].state.apply(get_q_s_seek)
df = choose_best_action(df)
display(df.groupby('stage').tail(3))
test = df.query('stage==1')
test['first_card'] = test.state.apply(lambda x: list(x)[0])
test['rel_val'] = test.q_s_guess - test.q_s_seek
display(test)
test.plot(x='first_card',y='rel_val')
```
<img src="images/dynamic_programming_stage_1.png" alt="Drawing" style="width: 600px; border-right:10px solid white"/>
Expected: 1: 6, 2: 2.42, 3:-2.52, 4: -8.6
This almost matches for the first-card cases. In the next step, I could compare data created from my model (e.g. optimum turns) against that of Hunt, by importing a few of his trials and associated outputs.
# Comparing to Hunt
Getting optimal turns from Hunt (see SST Hunt Script Notebook):
```
with open('opt_string.txt','r') as f: # This file is produced based on infotask_dyprog_AA_trials.m from Hunt
optimal_turns = f.read()
# Getting optimal turns from Hunt's model
ot = optimal_turns.split('\n')
ot = [x.split(':') for x in ot][:-1]
ot = {eval(x[0]):int(x[1]) for x in ot}
```
Calculating based on my model:
```
turn_dict[(5,)]
ot
turn_dict = dict(zip(states.state, states.best_action=='guess'))
ot2 = {}
for i in range(1, 11):
for j in range(1, 11):
for k in range(1, 11):
for l in range(1, 11):
state = (i,j,k,l)
if turn_dict[(i,)]:
ot2[state] = 1
elif turn_dict[(i,j)]:
ot2[state] = 2
elif turn_dict[(i,j,k)]:
ot2[state] = 3
else:
ot2[state] = 4
```
Hunt and I predict the same number of optimal turns.
```
ot
print(ot == ot2)
```
To run trial-by-trial, first calculate diff score between optimal and actual and the use as dependent.
|
github_jupyter
|
import itertools
import pandas as pd
import numpy as np
# Calculating which row wins in each possible game
cards = list(range(1,11))
games = list(itertools.product(cards, repeat=4))
games = pd.DataFrame(games, columns = [1,2,3,4])
# In each of the games row A either wins or loses
games['row_a'] = games[[1,2]].sum(axis = 1)
games['row_b'] = games[[3,4]].sum(axis = 1)
games['row_a_wins'] = games.row_a > games.row_b # Note that ties are allowed
games['row_b_wins'] = games.row_a < games.row_b
# A dataframe with all possible states
states_0 = [()] # No card revealed at stage 0
states_1 = list(itertools.product(cards, repeat=1))
states_2 = list(itertools.product(cards, repeat=2))
states_3 = list(itertools.product(cards, repeat=3))
states_4 = list(itertools.product(cards, repeat=4))
states = states_0 + states_1 + states_2 + states_3 + states_4
states = pd.DataFrame({'state':states})
states['stage'] = states.state.apply(lambda x: len(x))
states['total_turn_cost'] = states.stage.apply(lambda x: np.sum([0,10,15,20][:x]))
# Calculating p_win for each state
def get_p_win(state):
state = {location+1:value for location,value in enumerate(list(state))}
possible_games = games.loc[(games[list(state)] == pd.Series(state)).all(axis=1)]
p_a_win = possible_games.row_a_wins.mean()
p_b_win = possible_games.row_b_wins.mean()
choice = 'row_a_wins' if p_a_win > p_b_win else 'row_b_wins'
p_win = possible_games[choice].mean()
return p_win
states['p_win'] = states.state.apply(get_p_win)
states['q_s_guess'] = states.p_win*60-(1-states.p_win)*50-states.total_turn_cost
df = states
df = states
def choose_best_action(df):
# This will fail if q_s_guess equals q_s_seek
df['best_action'] = (df['q_s_guess'] > df['q_s_seek']).replace({True:"guess",False:"seek"})
df.loc[df.q_s_guess.isnull() | df.q_s_seek.isnull(), 'best_action'] = np.nan
df['expected_best_value'] = df[['q_s_guess','q_s_seek']].max(axis=1)
df.loc[df.q_s_guess.isnull() | df.q_s_seek.isnull(), 'expected_best_value'] = np.nan
# We do not allow seeking in stage 4
states.loc[states.stage==4,'best_action'] = 'guess'
return df
def get_q_s_seek(state):
if len(state) == 4:
return -np.inf # We do not allow seeking in stage 4
expected_best_values = []
for i in range(1,11):
next_state = state + (i,)
expected_best_values.append(df[df.state==next_state].expected_best_value)
return np.mean(expected_best_values)
#df.loc[df.stage==4, 'q_s_guess'] = 15 # This is incorrect (but implied in Hunt's paper)
for stage in range(4,-1,-1): # Looping through stage 4 to 0
df.loc[df.stage==stage, 'q_s_seek'] = df.loc[df.stage==stage].state.apply(get_q_s_seek)
df = choose_best_action(df)
display(df.groupby('stage').tail(3))
test = df.query('stage==1')
test['first_card'] = test.state.apply(lambda x: list(x)[0])
test['rel_val'] = test.q_s_guess - test.q_s_seek
display(test)
test.plot(x='first_card',y='rel_val')
with open('opt_string.txt','r') as f: # This file is produced based on infotask_dyprog_AA_trials.m from Hunt
optimal_turns = f.read()
# Getting optimal turns from Hunt's model
ot = optimal_turns.split('\n')
ot = [x.split(':') for x in ot][:-1]
ot = {eval(x[0]):int(x[1]) for x in ot}
turn_dict[(5,)]
ot
turn_dict = dict(zip(states.state, states.best_action=='guess'))
ot2 = {}
for i in range(1, 11):
for j in range(1, 11):
for k in range(1, 11):
for l in range(1, 11):
state = (i,j,k,l)
if turn_dict[(i,)]:
ot2[state] = 1
elif turn_dict[(i,j)]:
ot2[state] = 2
elif turn_dict[(i,j,k)]:
ot2[state] = 3
else:
ot2[state] = 4
ot
print(ot == ot2)
| 0.20462 | 0.900223 |
# Example of recipe computation, model fit, predict* and conversion to raster using API of antares3 and [kale](https://github.com/kubeflow-kale/kale) functionality
*Prediction is pixel wise.
**Will use an already ingested and processed Landsat8 data via antares3**
## Some imports
```
import sys
import os
import json
from datetime import datetime
import matplotlib
from matplotlib.patches import Patch
from matplotlib import pyplot as plt
import numpy as np
import xarray as xr
from shapely.geometry import Point
import rasterio
import dill
import geopandas as gpd
import fiona
from affine import Affine
from rasterio.features import rasterize
import datacube
from datacube.api import GridWorkflow
from datacube.storage import masking
from datacube.drivers.netcdf import write_dataset_to_netcdf
from madmex.util.db import get_cmap_from_scheme
from madmex.models import Tag
from madmex.overlay.extractions import zonal_stats_xarray
from madmex.io.vector_db import VectorDb
from madmex.wrappers import gwf_query
from madmex.modeling.supervised.xgb import Model
from madmex.models import Tag
from madmex.overlay.extractions import zonal_stats_xarray
from madmex.util import randomword, mid_date, join_dicts
from madmex.util.xarray import to_float, to_int
from django.contrib.gis.geos.geometry import GEOSGeometry
from madmex.models import PredictObject
```
## Recipe computation
Following [landsat_madmex_003.py](https://github.com/CONABIO/antares3/blob/develop/madmex/recipes/landsat_madmex_003.py)
Also could be helpful:
[1c_clusterization_for_agriculture_inecol](https://github.com/CONABIO/antares3-sandbox/blob/master/notebooks/agriculture_madmex_app/1c_clusterization_for_agriculture_inecol.ipynb)
[1d_clusterization_for_agriculture_inecol](https://github.com/CONABIO/antares3-sandbox/blob/master/notebooks/agriculture_madmex_app/1d_clusterization_for_agriculture_inecol.ipynb)
[2_clusterization_for_agriculture_inecol_intersect_with_area_of_interest.](https://github.com/CONABIO/antares3-sandbox/blob/master/notebooks/agriculture_madmex_app/2_clusterization_for_agriculture_inecol_intersect_with_area_of_interest.ipynb)
```
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
region = 'Chiapas'
products = ['ls8_espa_mexico']
begin = '2017-01-01'
end = '2017-12-31'
gwf_kwargs = {'region': region,
'begin': begin,
'end':end}
#query
dict_list = []
for prod in products:
gwf_kwargs.update(product = prod)
try:
dict_list.append(gwf_query(**gwf_kwargs, view=False))
# Exception is in case one of the product hasn't been registered in the datacube
except Exception as e:
pass
iterable = join_dicts(*dict_list, join='full').items()
list_iter = list(iterable)
list_iter_sorted = sorted(list_iter, key = lambda x: (x[0][0], x[0][1]))
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
# Select datacube tile index: (54, -38)
tile = [index for index in list_iter_sorted if index[0] == (54, -38)][0]
center_dt = mid_date(datetime.strptime(begin, '%Y-%m-%d'),
datetime.strptime(end, '%Y-%m-%d'))
crs = tile[1][0].geobox.crs
center_dt = center_dt.strftime("%Y-%m-%d")
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
# Load via Grid Workflow API
ds = xr.combine_by_coords([GridWorkflow.load(x, dask_chunks={'x': 1200, 'y': 1200})
for x in tile[1]], data_vars='minimal', coords='minimal')
ds.attrs['geobox'] = tile[1][0].geobox
# Mask clouds, shadow, water, ice,... and drop qa layer
clear = masking.make_mask(ds.pixel_qa, cloud=False, cloud_shadow=False,
snow=False)
ds_1 = ds.where(clear)
ds_1 = ds_1.drop('pixel_qa')
ds_1 = ds_1.apply(func=to_float, keep_attrs=True)
# Compute vegetation indices
ds_1['ndvi'] = ((ds_1.nir - ds_1.red) / (ds_1.nir + ds_1.red)) * 10000
ds_1['ndvi'].attrs['nodata'] = -9999
ds_1['ndmi'] = ((ds_1.nir - ds_1.swir1) / (ds_1.nir + ds_1.swir1)) * 10000
ds_1['ndmi'].attrs['nodata'] = -9999
# Run temporal reductions and rename DataArrays
ds_mean = ds_1.mean('time', keep_attrs=True, skipna=True)
ds_mean = ds_mean.rename({'blue': 'blue_mean',
'green': 'green_mean',
'red': 'red_mean',
'nir': 'nir_mean',
'swir1': 'swir1_mean',
'swir2': 'swir2_mean',
'ndmi': 'ndmi_mean',
'ndvi': 'ndvi_mean'})
# Compute min/max/std only for vegetation indices
ndvi_max = ds_1.ndvi.max('time', keep_attrs=True, skipna=True)
ndvi_max = ndvi_max.rename('ndvi_max')
ndvi_max.attrs['nodata'] = -9999
ndvi_min = ds_1.ndvi.min('time', keep_attrs=True, skipna=True)
ndvi_min = ndvi_min.rename('ndvi_min')
ndvi_min.attrs['nodata'] = -9999
# ndmi
ndmi_max = ds_1.ndmi.max('time', keep_attrs=True, skipna=True)
ndmi_max = ndmi_max.rename('ndmi_max')
ndmi_max.attrs['nodata'] = -9999
ndmi_min = ds_1.ndmi.min('time', keep_attrs=True, skipna=True)
ndmi_min = ndmi_min.rename('ndmi_min')
ndmi_min.attrs['nodata'] = -9999
# Load terrain metrics using same spatial parameters than sr
dc = datacube.Datacube(app = 'landsat_madmex_003_%s' % randomword(5))
terrain = dc.load(product='srtm_cgiar_mexico', like=ds,
time=(datetime(1970, 1, 1), datetime(2018, 1, 1)),
dask_chunks={'x': 1200, 'y': 1200})
dc.close()
# Merge dataarrays
combined = xr.merge([ds_mean.apply(to_int),
to_int(ndvi_max),
to_int(ndvi_min),
to_int(ndmi_max),
to_int(ndmi_min),
terrain])
combined.attrs['crs'] = crs
combined.attrs['affine'] = Affine(*list(ds.affine)[0:6])
#write_dataset_to_netcdf(combined.compute(scheduler='threads'), nc_filename)
plt.imshow(combined.ndvi_mean[:,:])
plt.show()
```
## Model Fit
```
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
training_data = "train_chiapas_dummy"
loader = VectorDb()
fc_train_0 = loader.load_training_from_dataset(dataset=combined,
training_set=training_data,
sample=1)
fc_train_0 = list(fc_train_0)
#Assign code level to this training data according to next scheme...
scheme = "madmex"
qs = Tag.objects.filter(scheme=scheme)
tag_mapping = {x.id:x.numeric_code for x in qs}
tag_id_list = [x['properties']['class'] for x in fc_train_0]
fc_train = [{'geometry': x[0]['geometry'],
'properties': {'code': tag_mapping[x[1]]},
'type': 'feature'} for x in zip(fc_train_0, tag_id_list)]
```
### Extract some zonal statistics using dataset and feature collection of training data...
```
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
X_train, y_train = zonal_stats_xarray(combined, fc_train, 'code')
```
### Model fit
```
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
path_result = "/shared_volume/land_cover_results"
if not os.path.exists(path_result):
os.makedirs(path_result)
xgb_model = Model()
xgb_model.fit(X_train, y_train)
```
## Predict and write raster to FS
```
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
arr_3d = combined.to_array().squeeze().values #squeeze to remove time dimension
#because has length 1
arr_3d = np.moveaxis(arr_3d, 0, 2)
shape_2d = (arr_3d.shape[0] * arr_3d.shape[1], arr_3d.shape[2])
arr_2d = arr_3d.reshape(shape_2d)
predicted_array = xgb_model.predict(arr_2d)
#write to FS
predicted_array = predicted_array.reshape((arr_3d.shape[0], arr_3d.shape[1]))
predicted_array = predicted_array.astype('uint8')
rasterio_meta = {'width': predicted_array.shape[1],
'height': predicted_array.shape[0],
'transform': combined.affine,
'crs': combined.crs.crs_str,
'count': 1,
'dtype': 'uint8',
'compress': 'lzw',
'driver': 'GTiff',
'nodata': 0}
filename_raster = 'raster_landsat8_chiapas_madmex_31_clases_pixel_wise_via_kale' + '_%d_%d' %(tile[0][0],tile[0][1]) + '.tif'
filename_raster = os.path.join(path_result, filename_raster)
with rasterio.open(filename_raster, 'w', **rasterio_meta) as dst:
dst.write(predicted_array, indexes = 1)
```
# Is next necessary?
**Next will use:**
[ingest_recipe_products](https://github.com/CONABIO/antares3-sandbox/blob/master/notebooks/ingest_recipe_products/ingest_recipe_products.ipynb)
**Need to create `/shared_volume/.config/madmex/indexing/`:**
```
mkdir -p /shared_volume/.config/madmex/indexing/
cp ~/.config/madmex/indexing/landsat_madmex_003.yaml /shared_volume/.config/madmex/indexing/
```
```
from madmex.indexing import add_product_from_yaml, add_dataset, metadict_from_netcdf
from madmex.util import yaml_to_dict
yaml_file = '/shared_volume/.config/madmex/indexing/landsat_madmex_003.yaml'
recipe = 'landsat_madmex_003'
product_description = yaml_to_dict(yaml_file)
args = {'description': product_description,
'center_dt': datetime.strptime(center_dt, '%Y-%m-%d'),
'from_dt': datetime.strptime(begin, '%Y-%m-%d'),
'to_dt': datetime.strptime(end, '%Y-%m-%d'),
'algorithm': recipe}
pr, dt = add_product_from_yaml(yaml_file, name_of_recipe_product)
result = metadict_from_netcdf(nc_filename, **args)
print("Adding %s to datacube database" % result[0])
r_add_dataset = add_dataset(pr=pr, dt=dt, metadict=result[1], file=result[0])
print(r_add_dataset)
```
|
github_jupyter
|
import sys
import os
import json
from datetime import datetime
import matplotlib
from matplotlib.patches import Patch
from matplotlib import pyplot as plt
import numpy as np
import xarray as xr
from shapely.geometry import Point
import rasterio
import dill
import geopandas as gpd
import fiona
from affine import Affine
from rasterio.features import rasterize
import datacube
from datacube.api import GridWorkflow
from datacube.storage import masking
from datacube.drivers.netcdf import write_dataset_to_netcdf
from madmex.util.db import get_cmap_from_scheme
from madmex.models import Tag
from madmex.overlay.extractions import zonal_stats_xarray
from madmex.io.vector_db import VectorDb
from madmex.wrappers import gwf_query
from madmex.modeling.supervised.xgb import Model
from madmex.models import Tag
from madmex.overlay.extractions import zonal_stats_xarray
from madmex.util import randomword, mid_date, join_dicts
from madmex.util.xarray import to_float, to_int
from django.contrib.gis.geos.geometry import GEOSGeometry
from madmex.models import PredictObject
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
region = 'Chiapas'
products = ['ls8_espa_mexico']
begin = '2017-01-01'
end = '2017-12-31'
gwf_kwargs = {'region': region,
'begin': begin,
'end':end}
#query
dict_list = []
for prod in products:
gwf_kwargs.update(product = prod)
try:
dict_list.append(gwf_query(**gwf_kwargs, view=False))
# Exception is in case one of the product hasn't been registered in the datacube
except Exception as e:
pass
iterable = join_dicts(*dict_list, join='full').items()
list_iter = list(iterable)
list_iter_sorted = sorted(list_iter, key = lambda x: (x[0][0], x[0][1]))
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
# Select datacube tile index: (54, -38)
tile = [index for index in list_iter_sorted if index[0] == (54, -38)][0]
center_dt = mid_date(datetime.strptime(begin, '%Y-%m-%d'),
datetime.strptime(end, '%Y-%m-%d'))
crs = tile[1][0].geobox.crs
center_dt = center_dt.strftime("%Y-%m-%d")
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
# Load via Grid Workflow API
ds = xr.combine_by_coords([GridWorkflow.load(x, dask_chunks={'x': 1200, 'y': 1200})
for x in tile[1]], data_vars='minimal', coords='minimal')
ds.attrs['geobox'] = tile[1][0].geobox
# Mask clouds, shadow, water, ice,... and drop qa layer
clear = masking.make_mask(ds.pixel_qa, cloud=False, cloud_shadow=False,
snow=False)
ds_1 = ds.where(clear)
ds_1 = ds_1.drop('pixel_qa')
ds_1 = ds_1.apply(func=to_float, keep_attrs=True)
# Compute vegetation indices
ds_1['ndvi'] = ((ds_1.nir - ds_1.red) / (ds_1.nir + ds_1.red)) * 10000
ds_1['ndvi'].attrs['nodata'] = -9999
ds_1['ndmi'] = ((ds_1.nir - ds_1.swir1) / (ds_1.nir + ds_1.swir1)) * 10000
ds_1['ndmi'].attrs['nodata'] = -9999
# Run temporal reductions and rename DataArrays
ds_mean = ds_1.mean('time', keep_attrs=True, skipna=True)
ds_mean = ds_mean.rename({'blue': 'blue_mean',
'green': 'green_mean',
'red': 'red_mean',
'nir': 'nir_mean',
'swir1': 'swir1_mean',
'swir2': 'swir2_mean',
'ndmi': 'ndmi_mean',
'ndvi': 'ndvi_mean'})
# Compute min/max/std only for vegetation indices
ndvi_max = ds_1.ndvi.max('time', keep_attrs=True, skipna=True)
ndvi_max = ndvi_max.rename('ndvi_max')
ndvi_max.attrs['nodata'] = -9999
ndvi_min = ds_1.ndvi.min('time', keep_attrs=True, skipna=True)
ndvi_min = ndvi_min.rename('ndvi_min')
ndvi_min.attrs['nodata'] = -9999
# ndmi
ndmi_max = ds_1.ndmi.max('time', keep_attrs=True, skipna=True)
ndmi_max = ndmi_max.rename('ndmi_max')
ndmi_max.attrs['nodata'] = -9999
ndmi_min = ds_1.ndmi.min('time', keep_attrs=True, skipna=True)
ndmi_min = ndmi_min.rename('ndmi_min')
ndmi_min.attrs['nodata'] = -9999
# Load terrain metrics using same spatial parameters than sr
dc = datacube.Datacube(app = 'landsat_madmex_003_%s' % randomword(5))
terrain = dc.load(product='srtm_cgiar_mexico', like=ds,
time=(datetime(1970, 1, 1), datetime(2018, 1, 1)),
dask_chunks={'x': 1200, 'y': 1200})
dc.close()
# Merge dataarrays
combined = xr.merge([ds_mean.apply(to_int),
to_int(ndvi_max),
to_int(ndvi_min),
to_int(ndmi_max),
to_int(ndmi_min),
terrain])
combined.attrs['crs'] = crs
combined.attrs['affine'] = Affine(*list(ds.affine)[0:6])
#write_dataset_to_netcdf(combined.compute(scheduler='threads'), nc_filename)
plt.imshow(combined.ndvi_mean[:,:])
plt.show()
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
training_data = "train_chiapas_dummy"
loader = VectorDb()
fc_train_0 = loader.load_training_from_dataset(dataset=combined,
training_set=training_data,
sample=1)
fc_train_0 = list(fc_train_0)
#Assign code level to this training data according to next scheme...
scheme = "madmex"
qs = Tag.objects.filter(scheme=scheme)
tag_mapping = {x.id:x.numeric_code for x in qs}
tag_id_list = [x['properties']['class'] for x in fc_train_0]
fc_train = [{'geometry': x[0]['geometry'],
'properties': {'code': tag_mapping[x[1]]},
'type': 'feature'} for x in zip(fc_train_0, tag_id_list)]
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
X_train, y_train = zonal_stats_xarray(combined, fc_train, 'code')
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
path_result = "/shared_volume/land_cover_results"
if not os.path.exists(path_result):
os.makedirs(path_result)
xgb_model = Model()
xgb_model.fit(X_train, y_train)
os.environ.setdefault("DJANGO_ALLOW_ASYNC_UNSAFE", "true")
arr_3d = combined.to_array().squeeze().values #squeeze to remove time dimension
#because has length 1
arr_3d = np.moveaxis(arr_3d, 0, 2)
shape_2d = (arr_3d.shape[0] * arr_3d.shape[1], arr_3d.shape[2])
arr_2d = arr_3d.reshape(shape_2d)
predicted_array = xgb_model.predict(arr_2d)
#write to FS
predicted_array = predicted_array.reshape((arr_3d.shape[0], arr_3d.shape[1]))
predicted_array = predicted_array.astype('uint8')
rasterio_meta = {'width': predicted_array.shape[1],
'height': predicted_array.shape[0],
'transform': combined.affine,
'crs': combined.crs.crs_str,
'count': 1,
'dtype': 'uint8',
'compress': 'lzw',
'driver': 'GTiff',
'nodata': 0}
filename_raster = 'raster_landsat8_chiapas_madmex_31_clases_pixel_wise_via_kale' + '_%d_%d' %(tile[0][0],tile[0][1]) + '.tif'
filename_raster = os.path.join(path_result, filename_raster)
with rasterio.open(filename_raster, 'w', **rasterio_meta) as dst:
dst.write(predicted_array, indexes = 1)
mkdir -p /shared_volume/.config/madmex/indexing/
cp ~/.config/madmex/indexing/landsat_madmex_003.yaml /shared_volume/.config/madmex/indexing/
from madmex.indexing import add_product_from_yaml, add_dataset, metadict_from_netcdf
from madmex.util import yaml_to_dict
yaml_file = '/shared_volume/.config/madmex/indexing/landsat_madmex_003.yaml'
recipe = 'landsat_madmex_003'
product_description = yaml_to_dict(yaml_file)
args = {'description': product_description,
'center_dt': datetime.strptime(center_dt, '%Y-%m-%d'),
'from_dt': datetime.strptime(begin, '%Y-%m-%d'),
'to_dt': datetime.strptime(end, '%Y-%m-%d'),
'algorithm': recipe}
pr, dt = add_product_from_yaml(yaml_file, name_of_recipe_product)
result = metadict_from_netcdf(nc_filename, **args)
print("Adding %s to datacube database" % result[0])
r_add_dataset = add_dataset(pr=pr, dt=dt, metadict=result[1], file=result[0])
print(r_add_dataset)
| 0.520984 | 0.754486 |
# SMILES String Autoencoder
Goal: Use an autoencoder into a continuous vector to represent SMILES strings. We want the network to understand SMILES well-enough to provide valid outputs when the encoding is manipulated.
General Strategy
1. Download a batch of SMILES from pubchem
2. Encode SMILES into a matrix representation that captures characters correctly (Cl, Br, etc.)
3. Train a simple autoencoder archiecture
4. Measure the rate of valid SMILES strings produced
## Gather Data
```
#!conda install -c mcs07 pubchempy
import pandas as pd
import numpy as np
import sklearn
from sklearn.preprocessing import OneHotEncoder
import pubchempy as pcp
import torch
from torch.utils.data import TensorDataset, random_split
from torch import nn
from fastai.basics import *
data = pd.read_csv('../data/raw/csv/CID-malaria-1-stats.csv')
data.head()
cid_list = list(data.cid)
smiles = [x['CanonicalSMILES'] for
x in pcp.get_properties('CanonicalSMILES', cid_list)]
smiles[:30]
```
## Pre-processing
We don't want the model to have to figure out that 'Cl' and 'Br' are actually just one atom - so let's replace these with single characters. We'll also make all the compounds the same length - if they're shorter than 150 characters, we'll pad it with spaces.
```
sub_dict = {
'Cl' : 'R',
'Br' : 'M',
'Ca' : 'A',
'Be' : 'E',
'Na' : 'X',
'Li' : 'L'
}
smiles_sub = []
length = 120
for atom in sub_dict:
for s in smiles:
if len(s) > 120: continue
smile = s.replace(atom, sub_dict[atom])
while len(smile) < length:
smile += ' '
smiles_sub.append(smile)
smiles = smiles_sub
# What are all the possible characters?
bank = []
for s in smiles:
for char in set(s):
if char not in bank:
bank.append(char)
bank = sorted(bank)
print(bank, len(bank))
smiles[:10]
def smiles_vectorizer(s, bank):
vector = [[0 if symbol != char else 1 for symbol in bank]
for char in s]
return vector
def smiles_decoder(t, bank):
'''
Inputs a vector with one-hot encoding of SMILES string
Returns a SMILES string
'''
smiles = ''
for vector in t:
npv = vector.numpy()
idx = np.where(npv == 1)[0][0]
smiles += bank[idx]
return smiles
smiles_decoder(smiles_tensor[0], bank)
smiles_tensor = []
for s in smiles:
smiles_tensor.append(smiles_vectorizer(s, bank))
smiles_tensor = torch.tensor(smiles_tensor, dtype=torch.long) #need long for cross entropy loss
smiles_tensor
torch.save(smiles_tensor, 'smiles_tensor.pkl')
```
## Simple Autoencoder with PyTorch
```
smiles_tensor = torch.load('smiles_tensor.pkl')
smiles_tensor.type()
train = smiles_tensor[:239382, :, :]
valid = smiles_tensor[239382:, :, :]
print(train.shape, valid.shape)
bs = 64
dataloader = DataLoader(train, batch_size=bs, shuffle=True)
class autoencoder(nn.Module):
def __init__(self):
super(autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Linear(120 * 37, 128),
nn.ReLU(True),
nn.Linear(128, 64),
nn.ReLU(True),
nn.Linear(64, 32))
self.decoder = nn.Sequential(
nn.Linear(32, 64),
nn.ReLU(True),
nn.Linear(64, 128),
nn.ReLU(True),
nn.Linear(128, 120 * 37),
nn.Softmax())
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = autoencoder().cuda()
lr=2e-2
loss_func = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(
model.parameters(), lr=lr, weight_decay=1e-5)
'''
num_epochs = 1
def train(dataloader)
for epoch in range(num_epochs):
for data in dataloader:
data
img, _ = data
img = img.view(img.size(0), -1)
img = Variable(img).cuda()
# ===================forward=====================
output = model(img)
loss = criterion(output, img)
# ===================backward====================
optimizer.zero_grad()
loss.backward()
optimizer.step()
# ===================log========================
print('epoch [{}/{}], loss:{:.4f}'
.format(epoch + 1, num_epochs, loss.data[0]))
if epoch % 10 == 0:
pic = to_img(output.cpu().data)
save_image(pic, './mlp_img/image_{}.png'.format(epoch))
'''
def accuracy_1(input:Tensor, targs:Tensor)->Rank0Tensor:
'Compute accuracy with targs when input is bs * n_classes.'
targs = targs.view(-1).long()
n = targs.shape[0]
input = input.argmax(dim=-1).view(n,-1)
targs = targs.view(n,-1)
return (input==targs).float().mean()
class MyCrossEntropy(nn.CrossEntropyLoss):
def forward(self, input, target):
target = target.long() # ADDED
return F.cross_entropy(input, target, weight=self.weight, ignore_index=self.ignore_index, reduction=self.reduction)
# FASTAI Make a databunch
bs=64
train_ds = TensorDataset(train, train)
valid_ds = TensorDataset(valid, valid)
db = DataBunch.create(train_ds, valid_ds, bs=bs)
# LEARNER
learn = Learner(db, autoencoder(), loss_func=nn.CrossEntropyLoss, metrics=accuracy_1)
learn.fit_one_cycle(4)
```
## Simple Autoencoder with Keras
|
github_jupyter
|
#!conda install -c mcs07 pubchempy
import pandas as pd
import numpy as np
import sklearn
from sklearn.preprocessing import OneHotEncoder
import pubchempy as pcp
import torch
from torch.utils.data import TensorDataset, random_split
from torch import nn
from fastai.basics import *
data = pd.read_csv('../data/raw/csv/CID-malaria-1-stats.csv')
data.head()
cid_list = list(data.cid)
smiles = [x['CanonicalSMILES'] for
x in pcp.get_properties('CanonicalSMILES', cid_list)]
smiles[:30]
sub_dict = {
'Cl' : 'R',
'Br' : 'M',
'Ca' : 'A',
'Be' : 'E',
'Na' : 'X',
'Li' : 'L'
}
smiles_sub = []
length = 120
for atom in sub_dict:
for s in smiles:
if len(s) > 120: continue
smile = s.replace(atom, sub_dict[atom])
while len(smile) < length:
smile += ' '
smiles_sub.append(smile)
smiles = smiles_sub
# What are all the possible characters?
bank = []
for s in smiles:
for char in set(s):
if char not in bank:
bank.append(char)
bank = sorted(bank)
print(bank, len(bank))
smiles[:10]
def smiles_vectorizer(s, bank):
vector = [[0 if symbol != char else 1 for symbol in bank]
for char in s]
return vector
def smiles_decoder(t, bank):
'''
Inputs a vector with one-hot encoding of SMILES string
Returns a SMILES string
'''
smiles = ''
for vector in t:
npv = vector.numpy()
idx = np.where(npv == 1)[0][0]
smiles += bank[idx]
return smiles
smiles_decoder(smiles_tensor[0], bank)
smiles_tensor = []
for s in smiles:
smiles_tensor.append(smiles_vectorizer(s, bank))
smiles_tensor = torch.tensor(smiles_tensor, dtype=torch.long) #need long for cross entropy loss
smiles_tensor
torch.save(smiles_tensor, 'smiles_tensor.pkl')
smiles_tensor = torch.load('smiles_tensor.pkl')
smiles_tensor.type()
train = smiles_tensor[:239382, :, :]
valid = smiles_tensor[239382:, :, :]
print(train.shape, valid.shape)
bs = 64
dataloader = DataLoader(train, batch_size=bs, shuffle=True)
class autoencoder(nn.Module):
def __init__(self):
super(autoencoder, self).__init__()
self.encoder = nn.Sequential(
nn.Linear(120 * 37, 128),
nn.ReLU(True),
nn.Linear(128, 64),
nn.ReLU(True),
nn.Linear(64, 32))
self.decoder = nn.Sequential(
nn.Linear(32, 64),
nn.ReLU(True),
nn.Linear(64, 128),
nn.ReLU(True),
nn.Linear(128, 120 * 37),
nn.Softmax())
def forward(self, x):
x = self.encoder(x)
x = self.decoder(x)
return x
model = autoencoder().cuda()
lr=2e-2
loss_func = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(
model.parameters(), lr=lr, weight_decay=1e-5)
'''
num_epochs = 1
def train(dataloader)
for epoch in range(num_epochs):
for data in dataloader:
data
img, _ = data
img = img.view(img.size(0), -1)
img = Variable(img).cuda()
# ===================forward=====================
output = model(img)
loss = criterion(output, img)
# ===================backward====================
optimizer.zero_grad()
loss.backward()
optimizer.step()
# ===================log========================
print('epoch [{}/{}], loss:{:.4f}'
.format(epoch + 1, num_epochs, loss.data[0]))
if epoch % 10 == 0:
pic = to_img(output.cpu().data)
save_image(pic, './mlp_img/image_{}.png'.format(epoch))
'''
def accuracy_1(input:Tensor, targs:Tensor)->Rank0Tensor:
'Compute accuracy with targs when input is bs * n_classes.'
targs = targs.view(-1).long()
n = targs.shape[0]
input = input.argmax(dim=-1).view(n,-1)
targs = targs.view(n,-1)
return (input==targs).float().mean()
class MyCrossEntropy(nn.CrossEntropyLoss):
def forward(self, input, target):
target = target.long() # ADDED
return F.cross_entropy(input, target, weight=self.weight, ignore_index=self.ignore_index, reduction=self.reduction)
# FASTAI Make a databunch
bs=64
train_ds = TensorDataset(train, train)
valid_ds = TensorDataset(valid, valid)
db = DataBunch.create(train_ds, valid_ds, bs=bs)
# LEARNER
learn = Learner(db, autoencoder(), loss_func=nn.CrossEntropyLoss, metrics=accuracy_1)
learn.fit_one_cycle(4)
| 0.802052 | 0.886273 |
# Appendix 1: Using spin symmetry to optimize the calculations
Author: Gediminas Kiršanskas
In this Appendix we show how numerical calculations can be optimized by using spin symmetry in the system (if it is present).
```
# Prerequisites
from __future__ import division, print_function
import numpy as np
import qmeq
```
## Convention
In order to be able to use spin symmetry in the problem the single particle Hamiltonian, tunneling amplitudes, and the lead parameters need to be set up in a particular way. Let us say we have $n=2m$ quantum dot single particle states counting with spin. Then we use the convention that the states with spin up have the labels $0\ldots m-1$ and the states with spin down have the labels $m\ldots n-1$. Also there should be no coupling between the up and down states to get correct results.
Additionally, when the spin degeneracy is present and we want to have a number $m_{\alpha}$ of physical leads, then we need to specify parameters for $n_{\alpha}=2m_{\alpha}$ number of leads. For example, if there are source and drain leads with chemical potentials $\mu_{L}=V/2$ and $\mu_{R}=-V/2$, then we need to specify the following list for chemical potentials:
```
vbias = 1.0
#L,up R,up L,down R,down
mulst = [vbias/2, -vbias/2, vbias/2, -vbias/2]
```
## Example system
Random single particle Hamiltonian with SU(2) spin symmetry:
```
# Number of single particle states counting with spin
nsingle = 4
vgate = 30.3
# Spin polarized Hamiltonian
hsingle0 = np.random.rand(nsingle//2, nsingle//2)
hsingle0 = hsingle0+hsingle0.T+vgate*np.eye(nsingle//2)
# Introduce spin
hsingle = np.kron(np.eye(2), hsingle0)
```
Random inter and intra dot coulomb matrix elements:
```
uinter = 10
uintra = 30
coulomb = np.zeros((nsingle*(nsingle-1)//2, 5), dtype=int)
ind = 0
for j1 in range(nsingle):
for j2 in range(j1+1, nsingle):
if j2 == nsingle//2+j1:
coulomb[ind] = [j1, j2, j2, j1, uintra]
else:
coulomb[ind] = [j1, j2, j2, j1, uinter]
ind += 1
print(coulomb)
```
Lead parameters and random tunneling amplitudes:
```
nleads = 4
tleads0 = 0.1*np.random.rand(2, nsingle//2)
tleads = np.kron(np.eye(2), tleads0)
vbias = 0.5
temp = 5.0
dband = 100.0
mulst = [vbias/2, -vbias/2, vbias/2, -vbias/2]
tlst = [temp, temp, temp, temp]
```
Now we set up three systems where no symmetries are used (*indexing='charge'*), just spin projection symmetry $S_{z}$ (*indexing='sz'*) is used, and a full spin symmetry is used (*indexing='ssq'*):
```
sys_charge = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='charge', kerntype='1vN')
sys_sz = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='sz', kerntype='1vN')
sys_ssq = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='ssq', kerntype='1vN')
```
In all three cases we obtain the same current:
```
sys_charge.solve()
sys_sz.solve()
sys_ssq.solve()
print(sys_charge.current)
print(sys_sz.current)
print(sys_ssq.current)
```
However, the matrix size of Liouvillian (master equation kernel) is reduced for 'sz' and 'ssq' indexing compared to 'charge' indexing:
```
print(sys_charge.kern.shape)
print(sys_sz.kern.shape)
print(sys_ssq.kern.shape)
```
This can be achieved, because a lot of reduced density matrix elements are zero or are related by symmetry:
```
print(sys_charge.phi0.shape)
print(sys_sz.phi0.shape)
print(sys_ssq.phi0.shape)
```
|
github_jupyter
|
# Prerequisites
from __future__ import division, print_function
import numpy as np
import qmeq
vbias = 1.0
#L,up R,up L,down R,down
mulst = [vbias/2, -vbias/2, vbias/2, -vbias/2]
# Number of single particle states counting with spin
nsingle = 4
vgate = 30.3
# Spin polarized Hamiltonian
hsingle0 = np.random.rand(nsingle//2, nsingle//2)
hsingle0 = hsingle0+hsingle0.T+vgate*np.eye(nsingle//2)
# Introduce spin
hsingle = np.kron(np.eye(2), hsingle0)
uinter = 10
uintra = 30
coulomb = np.zeros((nsingle*(nsingle-1)//2, 5), dtype=int)
ind = 0
for j1 in range(nsingle):
for j2 in range(j1+1, nsingle):
if j2 == nsingle//2+j1:
coulomb[ind] = [j1, j2, j2, j1, uintra]
else:
coulomb[ind] = [j1, j2, j2, j1, uinter]
ind += 1
print(coulomb)
nleads = 4
tleads0 = 0.1*np.random.rand(2, nsingle//2)
tleads = np.kron(np.eye(2), tleads0)
vbias = 0.5
temp = 5.0
dband = 100.0
mulst = [vbias/2, -vbias/2, vbias/2, -vbias/2]
tlst = [temp, temp, temp, temp]
sys_charge = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='charge', kerntype='1vN')
sys_sz = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='sz', kerntype='1vN')
sys_ssq = qmeq.Builder(nsingle, hsingle, coulomb, nleads, tleads, mulst, tlst, dband, indexing='ssq', kerntype='1vN')
sys_charge.solve()
sys_sz.solve()
sys_ssq.solve()
print(sys_charge.current)
print(sys_sz.current)
print(sys_ssq.current)
print(sys_charge.kern.shape)
print(sys_sz.kern.shape)
print(sys_ssq.kern.shape)
print(sys_charge.phi0.shape)
print(sys_sz.phi0.shape)
print(sys_ssq.phi0.shape)
| 0.41324 | 0.983053 |
Copyright (c) 2020 Martin Holle. Alle Rechte vorbehalten. Lizensiert unter der MIT-Lizenz.
# Covid-19 Statistics Aachen: Datenabfrage
Abfrage der Daten von der Website der Städteregion Aachen und Speichern in einer Excel-Datei für die Datenübergabe an den nächsten Schritt, in dem die Daten aufbereitet werden.
## Vorbereitungen
- Benötigte Imports
- Konfiguration aus zentraler `.ini`-Datei einlesen
- Konfiguration und Instanzierung des Loggers
- Globale Variablen definieren
```
import pandas as pd
import numpy as np
import re
from datetime import date
from datetime import timedelta
from datetime import date
from datetime import time
from datetime import datetime
import requests
from bs4 import BeautifulSoup
import logging
import configparser
# Konfiguration einlesen
config = configparser.ConfigParser()
config.read('config.ini')
# Konfiguration des Loggings
# - Die Logging-Ausgaben werden in der lokalen Datei covid-19-datenabfrage.log geschrieben
# - Für die Ausgabe wird eine bestimmte Formatierung konfiguriert
fhandler = logging.FileHandler(filename=config['Logging']['LogFileName'], mode='a')
# TODO: Formatierung finalisieren (Tausendstel-Sekunden, Tag des Monats, 1. Zeichen des Levels)
formatter = logging.Formatter('%(asctime)s %(levelname)-1.1s %(name)-20.20s - %(message)s')
fhandler.setFormatter(formatter)
# Instanzierung und Konfigurierung des Loggers
log = logging.getLogger("datenabfrage")
log.addHandler(fhandler)
log.setLevel(logging.DEBUG)
# Für die Zwischenspeicherung des eingelesenen HTML
# - Wenn die Website in der aktuellen Session schon einmal abgefragt wurde, wird das Ergebnis
# der Abfrage in dieser Variablen gesichert
# - Dies erleichtert die Entwicklung der nachfolgenden Verabeitungsschritte und führt nicht immer wieder zu
# neuen überflüssigen Abfragen der Website
html_payload = None
```
## Einlesen der existierenden Excel-Datei
- Datei und Seite der Excel-Datei: Siehe `config.ini`
- Einzulesende Spalten:
- **A**: Datum im Format 'DD.MM.'
- **B**: Akkumulierte Anzahl der Infektionen für gesamte Städteregion (inkl. Aachen) als Integerzahl
- **C**: Akkumulierte Anzahl der Infektionen für die Stadt Aachen als Integerzahl
- **D**: Anzahl neuer Todesfälle durch Covid-19 für gesamte Städteregion (inkl. Aachen) als Integerzahl
- **E**: Akkumulierte Anzahl der Todesfälle durch Covid-19 für gesamte Städteregion (inkl. Aachen) als Integerzahl
- **F**: Akkumulierte Anzahl der Genesenen für gesamte Städteregion (inkl. Aachen) als Integerzahl
- Spalte A als Datum interpretieren
- Die erste Zeile (Header) überspringen
- Label der Spalten explizit setzen
```
col_names = ['Uhrzeit', 'Summe', 'Summe Aachen', 'Summe Todesfälle', 'Summe genesen', 'Akute Fälle' ]
try:
c19_cases = pd.read_excel(config['Rohdaten']['FileName'],
sheet_name=config['Rohdaten']['SheetName'],
index_col=0,
parse_dates=[0],
skiprows=[],
names=col_names)
except FileNotFoundError as err:
log.warning('Error during pd.read_excel(): {0}'.format(err))
# Leere DataFrame für den Start erzeugen
c19_cases = pd.DataFrame(columns=col_names, index=pd.DatetimeIndex([], name='Datum'))
c19_cases.tail(14)
```
## Datenabfrage
### Funktionen für die Datenabfrage und Extraktion der Rohtexte
- `robot_access_allowed()` - Via robots.txt prüfen, ob die Website durch ein Skript abgefragt und verabeitet werden darf
- `gather_html()` - Website abfragen und HTML zurückliefern
- `gather_text()` - Relevante Texte aus dem von der Website geliefertem HTML extrahieren und zurückliefern
*<u>Anmerkungen:</u>
Die Website der Städteregion Aachen verwendet keine `robots.txt`, damit wäre die Funktion `robot_access_allowed()` eigentlich überflüssig. Hier ist sie nur der Vollständigkeit halber definiert und nicht ausimplementiert.*
*Eine leicht andere Struktur bei der Meldung am 12.08.2020 erforderte auch eine Änderung an den CSS-Selektor in `gather_html()`.*
```
default_user_agent = config['Rohdaten']['UserAgent']
def robot_access_allowed(url: str, user_agent: str=default_user_agent) -> bool:
"""
Return True if scraping is allowed according to robots.txt.
Request robots.txt. If exist, parse robots.txt and return True if scraping by this script is allowed.
Parameters
----------
url: str
URL of the website, which is to be scraped
user_agent: str
User agent string, which will be used when requesting the website
Returns
-------
bool
True, if web scraping is allowed by robots.txt or if robots.txt does not exist.
False, if web scraping is not allowed by robots.txt
"""
log.debug("access_allowed(" + url + ", " + user_agent + ")")
headers = { 'user-agent': user_agent }
# Extract root of the website's URL
# Request robots.txt from root of the website
# If robots.txt exist, parse content of the file
return True
def gather_html(url: str, user_agent: str=default_user_agent) -> str:
"""
Request website from <url> and return the HTML delivered by the website as text.
Parameters
----------
url: str
URL of the website, which is to be scraped
user_agent: str
User agent string, which will be used when requesting the website
Returns
-------
str
The HTML which was gathered from the website
"""
log.debug("gather_html(" + url + ", " + user_agent + ")")
# Website abfragen
headers = { 'user-agent': user_agent }
page = requests.get(url)
log.debug("gather_html/page.status_code: %d", page.status_code)
for key, val in page.headers.items():
log.debug("gather_html/page.header(%s): %s", key, val)
if page.status_code == requests.codes.ALL_OK: # Alles ok
return page.text
return None
def gather_text(html_text: str) -> []:
"""
Extract relevant text content from HTML and return extracted text for news entry parts.
Extract relevant text content, i.e. the distinctive news entries, from <html_text>.
Return array with news entries, each entry consisting of a dictionary with extracted text parts
for 'Header', 'Abstract', 'Main'.
Parameters
----------
html_str: str
The HTML code, which was gathered from the website.
Returns
-------
[]
Array with entries, each consisting of a dictionary with extracted text for 'Header', 'Abstract', 'Main'
part of the news entries.
"""
log.debug(f"gather_text(%d chars)", len(html_text))
records = []
# Parser instanzieren
soup = BeautifulSoup(html_text, 'html.parser')
# Relevante Objekte aus geliefertem HTML extrahieren
# - Header
# - Abstract (existiert nur für die aktuellen Meldungen, nicht im Meldungsarchiv)
# - Main
divs = soup.select('div.mid-col article > div.textcontent > div')
for div in divs:
# Header erkennen: Nun wenn dieser gefunden wird, macht es Sinn, nachfolgend nach
# Abstract und Haupttext zu suchen
header = div.select('h2')
if header:
# Header: Text extrahieren
header_text = next(header[0].stripped_strings)
# TODO: Ab hier überarbeiten, sodass der gesamte Body-Text eines Eintrags eingelesen wird
body_text = ''
body_elems = div.select('div > div.ce-bodytext > p, div > div.ce-bodytext > ul')
for be in body_elems:
# Body: Text extrahieren
body_text += next(be.stripped_strings)
print(body_text)
# Neuen Eintrag mit den extahierten Texten hinzufügen
records.append({'Header': header_text, 'Body': body_text })
log.debug("gather_text: %d records extracted from HTML", len(records))
return records
```
### Funktionen für die Extraktion der Falldaten aus den Rohtexten
In der ersten Version erfolgt die Extraktion der Daten aus den Texten mit Hilfe regulärer Ausdrücke:
- Datum/Uhrzeit der Meldung wird aus dem `Header` extrahiert.
- Die eigentlichen Daten werden zweimal extrahiert: Einmal aus dem `Abstract` und zum zweiten Mal aus dem `Main`-Text.
- Falls sowohl `Abstract` als auch `Main` eingelesen wurden, werden anschließend die extrahierten Daten miteinander verglichen. Nur wenn sie übereinstimmen, wird der Datensatz übernommen.
Die für das Parsen der eingelesenen Texte verwendeten regulären Ausdrücke versuchen einerseits, auf Nummer sicher zu gehen, um die richtigen Textstellen für das Einlesen der Zahlen zu treffen, und andererseits einige Freiheitsgrade zuzulassen:
- Leerzeichen und andere "White Spaces" werden von Menschen hin und wieder vergesssen oder mehrfach eingegeben
- Textvariationen kommen vor, in denen einzelne Wörter nicht erscheinen oder hinzugefügt werden
- Manchmal werden einzelne Buchstaben weggelassen, ohne dass dies ein Schreibfehler wäre
- Und natürlich kommen auch Schreibfehler vor
Die regulären Ausdrücke sind dadurch relativ unübersichtlich geworden und berücksichtigen dennoch nicht alle Situationen. Beispielsweise wurde bis zum 10.08.2020 im `Abstract` die folgende Formulierung verwendet:
> Aktuell 2157 bestätigte Coronafälle in der StädteRegion Aachen (davon 1060 in der Stadt Aachen).
Der `Main`-Text sah ähnlich, aber ein wenig anders aus:
> Es gibt insgesamt in der StädteRegion nunmehr 2157 positive Fälle, davon 1060 in der Stadt Aachen.
Am 10.08.2020 wurde dann die Formulierung für den `Main`-Text minimal variert (Klammerausdruck statt Nebensatz):
> Es gibt insgesamt in der StädteRegion nunmehr 2170 positive Fälle (davon 1068 in der Stadt Aachen).
Damit passte der reguläre Ausdruck nicht mehr, den ich dann anpassen musste. Obwohl reguläre Ausdrücke ein sehr mächtiges Mittel darstellen, um Texte zu verarbeiten, bleiben sie zwangsläufig auf der Ebene eines reinen Mustervergleichs stehen, die fehlerresistente Verarbeitung natürlicher Sprache stößt mit regulären Ausdrücken immer wieder an eine Grenze.
Ab dem 12.08.2020 wurden die Zahlen für die Stadt Aachen nicht mehr wie oben angegeben. Stattdessen, und nur im `Main`-Text der Meldung, wird nun eine Liste aller Kommunen in der Städteregion mit den jeweils auf sie entfallenden Fallzahlen mitgeteilt. Die Extraktion und Verarbeitung der Zahlen für die Stadt Aachen wurde daher zunächst deaktiviert und dann durch ein neues Verfahren ersetzt. Ebenfalls wurden das Verfahren und die regulären Ausdrücke für die Ermittlung der Todesfälle angepasst.
```
class NewsMeta():
"""Value class for meta data of news entry."""
pass
class CaseFigures():
"""Value class for case figures, extracted from the news entries."""
pass
def parse_header(header: str) -> NewsMeta:
"""
Analyse <header> of the news and return date and time of news or None.
The <header> is expected to have the following format:
'Aktuelle Lage Stadt und StädteRegion Aachen zum Corona-Virus; Montag, 22. Juni, 10:00 Uhr'
Variables weekday name, date, and time will be extracted from header. Year assumed to be 2020.
Parameters
----------
header : str
Extracted raw text of header
Returns
-------
None
If no matching header found
NewsMeta(weekday_name, day_of_month, month_name, hour, minutes, date, time, datetime)
Object with meta data of news entry
"""
monthnames = [ "Januar", "Februar", "März", "April", "Mai", "Juni", "Juli", "August", "September",
"Oktober", "November", "Dezember" ]
log.debug("parse_header(" + header[:40] + "...)")
pattern = (r"^Aktuelle Lage Stadt und StädteRegion Aachen zum Corona-Virus;\s*"
r"(Montag|Dienstag|Mittwoch|Donnerstag|Freitag|Samstag|Sonntag),\s*"
r"([0-9]{1,2})\.\s*(Januar|Februar|März|April|Mai|Juni|Juli|August|"
r"September|Oktober|November|Dezember),\s*"
r"([0-9]{1,2}):([0-9]{2})\s*Uhr.*$")
match = re.search(pattern, header)
if match:
meta = NewsMeta()
try:
meta.weekday_name = match.group(1)
meta.day_of_month = int(match.group(2))
meta.month_name = match.group(3)
meta.hour = int(match.group(4))
meta.minutes = int(match.group(5))
meta.date = date(2020, monthnames.index(meta.month_name) + 1, meta.day_of_month)
meta.time = time(meta.hour, meta.minutes)
meta.datetime = datetime.combine(meta.date, meta.time)
log.debug(f"parse_header/meta.weekday: %s", meta.weekday_name)
log.debug(f"parse_header/meta.day_of_month: %d", meta.day_of_month)
log.debug(f"parse_header/meta.month_name: %s", meta.month_name)
log.debug(f"parse_header/meta.hour: %d", meta.hour)
log.debug(f"parse_header/meta.minutes: %d", meta.minutes)
log.debug(f"parse_header/meta.date: %s", str(meta.date))
log.debug(f"parse_header/meta.time: %s", str(meta.time))
log.debug(f"parse_header/meta.datetime: %s", str(meta.datetime))
return meta
except:
log.warning("parse_header() failed parsing news header [1]")
return None
else:
log.warning("parse_header() failed parsing news header [2]")
return None
def parse_cases(kind: str, meta: NewsMeta, text: str, pattern_total: str, patterns_total_AC: [],
pattern_recovered: str, patterns_deaths: [], pattern_active: str) -> CaseFigures:
"""
Analyse <abstract> or <main> text of the news and return Covid-19 case numbers included therein.
Parameters
----------
kind : str
"Main" - Parse main part of news entry
"Abstract" - Parse abstract of news entry
meta : NewsMeta
Meta data of news entry
text : str
Text to be parsed
pattern_total : str
Regular expression for parsing total number of cases in Städteregion Aachen
patterns_total_AC : str
Regular expression for parsing total number of cases in Aachen
pattern_recovered : str
Regular expression for parsing total number of recovered cases in Städteregion Aachen
patterns_deaths : [ str ]
Array with regular expressions for parsing total number of deaths in Städteregion Aachen
pattern_active : str
Regular expression for parsing number of active cases in Städteregion Aachen
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug(f"parse_cases(%s, %s)", kind, text[:80])
if (not text.strip()):
# Der übergebene Text ist leer
return None
def log_debug(kind: str, meta: NewsMeta, par_name: str, par_value: int):
log.debug(f"parse_cases(%s,%s)/figures.%s: %d", kind, str(meta.datetime), par_name, par_value)
def log_warn(kind: str, meta: NewsMeta, reason: int):
log.warning(f"parse_cases(%s,%s) failed [%d]", kind, str(meta.datetime), reason)
figures = CaseFigures()
# Summe der Corona-Fälle in der Städteregion extrahieren
match = re.search(pattern_total, text)
if match:
try:
figures.total = int(match.group(1))
log_debug(kind, meta, "total", figures.total)
except:
log_warn(kind, meta, 1)
return None
else:
log_warn(kind, meta, 2)
return None
# Summe der Corona-Fälle in der Stadt Aachen extrahieren
figures.total_AC = None
for ptac in patterns_total_AC:
match = re.search(ptac, text)
if match:
try:
figures.total_AC = int(match.group(1))
log_debug(kind, meta, "total_AC", figures.total_AC)
break
except:
log_warn(kind, meta, 3)
return None
else:
log.debug(f"parse_cases(%s)/total_AC: No match for pattern %s", kind, ptac)
if figures.total_AC is None:
log_warn(kind, meta, 4)
# Summe der wieder Genesenen extrahieren
match = re.search(pattern_recovered, text)
if match:
try:
figures.recovered = int(match.group(1))
log_debug(kind, meta, "recovered", figures.recovered)
except:
log_warn(kind, meta, 5)
return None
else:
log_warn(kind, meta, 6)
return None
# Summe der Corona-Toten extrahieren
figures.deaths = None
for pd in patterns_deaths:
match = re.search(pd, text)
if match:
try:
figures.deaths = int(match.group(1))
log_debug(kind, meta, "deaths", figures.deaths)
break
except:
log_warn(kind, meta, 7)
return None
else:
log.debug(f"parse_cases(%s)/deaths: No match for pattern %s", kind, pd)
if figures.deaths is None:
log_warn(kind, meta, 8)
return None
# Anzahl der aktiven Infektionen extrahieren
match = re.search(pattern_active, text)
if match:
try:
figures.active = int(match.group(1))
log_debug(kind, meta, "active", figures.active)
except:
log_warn(kind, meta, 9)
return None
else:
log_warn(kind, meta, 10)
return None
return figures
def parse_abstract(abstract: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <abstract> of the news and return Covid-19 case numbers included in abstract.
The relevant area of the <abstract> is expected to have the following format:
'Aktuell 1999 bestätigte Coronafälle in der StädteRegion Aachen (davon 994 in der Stadt Aachen). '
'1880 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund.'
'Bislang 98 Todesfälle.' | 'neuer Todesfall, somit insgesamt 98.' | 'neue Todesfälle, somit insgesamt 98.'
'Damit aktuell 21 nachgewiesen(e) Infizierte.'
Values will be extracted from abstract text.
Parameters
----------
meta : NewsMeta
Meta data of news entry
abstract : str
Abstract text to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug("parse_abstract(" + abstract[:40] + "...)")
pattern_total = r"Aktuell\s*([0-9]{1,6})\s*bestätigte\s*Coronafälle\s*in\s*der\s*StädteRegion\s*"
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund.*")
pattern_deaths = [
r"(?:Insgesamt|Bislang)\s*([0-9]{1,6})\s*Todesfälle\.",
r"(?:neuer\s*Todesfall|neue\s*Todesfälle),\s*somit\s*insgesamt\s*([0-9]{1,6})\."
]
pattern_active = r"Damit\s*aktuell\s*([0-9]{1,6})\s*nachgewiesene?\s*Infizie.*"
return parse_cases("Abstract", meta, abstract,
pattern_total, patterns_total_AC,
pattern_recovered, pattern_deaths, pattern_active)
def parse_main(main: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <main> text of the news and return Covid-19 case numbers included in main text.
The relevant area of the <abstract> is expected to have the following format:
'Es gibt insgesamt in der StädteRegion [nunmehr] 1997 positive Fälle, davon 992 in der Stadt Aachen'
'1876 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund'
'Die Zahl der gemeldeten Todesfälle liegt [nach wie vor] bei 98'
'Damit sind aktuell 23 Menschen in der StädteRegion nachgewiesen infiziert'
Parameters
----------
meta : NewsMeta
Meta data of news entry
main : str
Main text to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug("parse_main(" + main[:40] + "...)")
pattern_total = (r"Es\s*gibt\s*insgesamt\s*in\s*der\s*StädteRegion\s*(?:nunmehr)?\s*([0-9]{1,6})\s*"
r"positive\s*Fälle")
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen",
r"Aachen\s*(?:[0-9]{1,6})\s*\(([0-9]{1,6})\)"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund")
patterns_deaths = [
r"Die\s*Zahl\s*der\s*gemeldeten\s*Todesfälle\s*liegt\s*(?:nach\s*wie\s*vor)?\s*bei\s*([0-9]{1,6})"
]
pattern_active = (r"Damit\s*sind\s*aktuell\s*([0-9]{1,6})\s*Menschen\s*in\s*der\s*StädteRegion\s*"
r"(?:Aachen)?\s*nachgewiesen\s*infiziert")
return parse_cases("Main", meta, main,
pattern_total, patterns_total_AC,
pattern_recovered, patterns_deaths, pattern_active)
def parse_body(body: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <body> text of the news and return Covid-19 case numbers included in main text.
The relevant area of the <body> is expected to have the following format:
'Es gibt insgesamt in der StädteRegion [nunmehr] 1997 positive Fälle, davon 992 in der Stadt Aachen'
'1876 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund'
'Die Zahl der gemeldeten Todesfälle liegt [nach wie vor] bei 98'
'Damit sind aktuell 23 Menschen in der StädteRegion nachgewiesen infiziert'
Parameters
----------
meta : NewsMeta
Meta data of news entry
Body : str
Text in news body to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures
"""
log.debug("parse_body(" + body[:40] + "...)")
pattern_total = (r"Es\s*gibt\s*insgesamt\s*in\s*der\s*StädteRegion\s*(?:nunmehr)?\s*([0-9]{1,6})\s*"
r"positive\s*Fälle")
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen",
r"Aachen\s*(?:[0-9]{1,6})\s*\(([0-9]{1,6})\)"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund")
patterns_deaths = [
r"Die\s*Zahl\s*der\s*gemeldeten\s*Todesfälle\s*liegt\s*(?:nach\s*wie\s*vor)?\s*bei\s*([0-9]{1,6})"
]
pattern_active = (r"Damit\s*sind\s*aktuell\s*([0-9]{1,6})\s*Menschen\s*in\s*der\s*StädteRegion\s*"
r"(?:Aachen)?\s*nachgewiesen\s*infiziert")
return parse_cases("Body", meta, body,
pattern_total, patterns_total_AC,
pattern_recovered, patterns_deaths, pattern_active)
def checked_data1(meta: NewsMeta, fig_abstract: CaseFigures, fig_main: CaseFigures) -> []:
"""
Check extracted data for consistency and return valid figures or None.
Parameters
----------
meta : NewMeta
Meta data of news entry
fig_abstract: CaseFigures
Object with figures extracted from abstract of the news entry
fig_main: CaseFigures
Object with figures extracted from main text of the news entry
Returns
-------
None
If consistency check fails
[ Date, Time, Total, Total_AC, Deaths, Recovered, Active ]
Array with extracted Covid-19 numbers
"""
if meta != None:
# Der Header konnte erfolgreich analysiert werden
a, m = fig_abstract, fig_main
if (a != None) and (m != None):
# Sowohl Abstract als auch Main konnten erfolgreich analysiert werden
if (m.total == a.total) and (m.deaths == a.deaths) and (m.recovered == a.recovered) and \
(m.active == a.active):
# Die eingelesenen Zahlen aus Abstract und Main stimmmen überein
if m.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
m.active = m.total - m.deaths - m.recovered
if m.total - m.deaths - m.recovered == m.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [ meta.date, meta.time, m.total, m.total_AC, m.deaths, m.recovered, m.active ]
else:
log.warning('Data consistency error: (1) Main and Abstract found. Inconsistent figures read.')
return None
elif m != None:
# Mindestens Main konnte erfolgreich analysiert werden
if m.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
m.active = m.total - m.deaths - m.recovered
if m.total - m.deaths - m.recovered == m.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [ meta.date, meta.time, m.total, m.total_AC, m.deaths, m.recovered, m.active ]
else:
log.warning('Data onsistency error: (2) Only Main found. Inconsistent figures read. ')
return None
else:
log.warning('Data consistency error: (3) Header not found.')
return None
def checked_data2(meta: NewsMeta, fig_body: CaseFigures) -> []:
"""
Check extracted data for consistency and return valid figures or None.
Parameters
----------
meta : NewMeta
Meta data of news entry
fig_body: CaseFigures
Object with figures extracted from body of the news entry
Returns
-------
None
If consistency check fails
[ Date, Time, Total, Total_AC, Deaths, Recovered, Active ]
Array with extracted Covid-19 numbers
"""
if meta != None:
# Der Header konnte erfolgreich analysiert werden
if fig_body != None:
if fig_body.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
fig_body.active = fig_body.total - fig_body.deaths - fig_body.recovered
if fig_body.total - fig_body.deaths - fig_body.recovered == fig_body.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [
meta.date, meta.time,
fig_body.total, fig_body.total_AC, fig_body.deaths, fig_body.recovered, fig_body.active
]
else:
log.warning('Data onsistency error: Inconsistent figures read. ')
return None
else:
log.warning('Data consistency error: Header not found.')
return None
def fill_dataframe_from_text(text_records: []) -> pd.DataFrame:
"""
Analyse <text_records> and return Pandas dataframe with parsed Covid-19 case numbers.
Parameters
----------
text_records : []
Array with extracted text
Returns
-------
pd.DataFrame
Pandas dataframe with extracted data
"""
log.debug(f"fill_dataframe_from_text(%d records)", len(text_records))
figures = [] # Zwischenspeicherung der eingelesenen Zahlen
dates = [] # Zwischenspeicherung der Datumsangaben
for tr in text_records:
meta = parse_header(tr['Header'])
if tr['Body']:
fig_body = parse_body(tr['Body'], meta)
rec = checked_data2(meta, fig_body)
else:
if tr['Abstract']:
fig_abstract = parse_abstract(tr['Abstract'], meta)
if tr['Main']:
fig_main = parse_main(tr['Main'], meta)
rec = checked_data1(meta, fig_abstract, fig_main)
if rec:
dates.append(rec[0])
figures.append(rec[1:])
if figures.count:
cols = ['Uhrzeit', 'Summe', 'Summe Aachen', 'Summe Todesfälle', 'Summe genesen', 'Akute Fälle' ]
index = pd.DatetimeIndex(dates, name='Datum')
df = pd.DataFrame(np.array(figures), columns=cols, index=index).sort_index()
return df
return pd.DataFrame() # Leer
```
## Datenabfrage durchführen
1. Datum des letzten Datensatzes in der Excel-Datei ermitteln und mit aktuellem Datum vergleichen.
2. Wenn mindestens 1 Tag seit der letzten Datenabfrage vergangen ist, neue Abfrage durchführen.
3. Von der Website gelieferte Rohtexte auswerten und Fallzahlen extrahieren
4. Extrahierte Fallzahlen auf Konsistenz prüfen
5. Geprüfte Fallzahlen zu neuem DataFrame hinzufügen
6. Den neuen mit dem existierendem DataFrame zusammenführen
7. Zusammengeführte Daten speichern
```
new_request = (c19_cases.size == 0) or \
(date.today() >= (date.fromtimestamp(c19_cases.index.max().timestamp()) + timedelta(days=1)))
if new_request:
log.info("New request initiated")
if not html_payload:
url = config['Rohdaten']['SourceURLDefault']
if robot_access_allowed(url):
html_payload = gather_html(url)
if html_payload:
records = gather_text(html_payload)
log.info("Processing {0} text records".format(len(records)))
if len(records):
new_cases = fill_dataframe_from_text(records)
if not new_cases.empty:
# Nur Zeilen mit neuerem Datum hinzufügen
merged_cases = pd.concat([c19_cases, new_cases[new_cases.index > c19_cases.index[-1]]], join='outer')
log.info("{0} new case records appended".format(len(merged_cases.index) - len(c19_cases.index)))
else:
log.info("No new case figures extracted")
merged_cases.to_excel(config['Rohdaten']['FileName'],
sheet_name=config['Rohdaten']['SheetName'], index_label='Datum')
merged_cases.tail(10)
else:
log.info("No new request required")
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import re
from datetime import date
from datetime import timedelta
from datetime import date
from datetime import time
from datetime import datetime
import requests
from bs4 import BeautifulSoup
import logging
import configparser
# Konfiguration einlesen
config = configparser.ConfigParser()
config.read('config.ini')
# Konfiguration des Loggings
# - Die Logging-Ausgaben werden in der lokalen Datei covid-19-datenabfrage.log geschrieben
# - Für die Ausgabe wird eine bestimmte Formatierung konfiguriert
fhandler = logging.FileHandler(filename=config['Logging']['LogFileName'], mode='a')
# TODO: Formatierung finalisieren (Tausendstel-Sekunden, Tag des Monats, 1. Zeichen des Levels)
formatter = logging.Formatter('%(asctime)s %(levelname)-1.1s %(name)-20.20s - %(message)s')
fhandler.setFormatter(formatter)
# Instanzierung und Konfigurierung des Loggers
log = logging.getLogger("datenabfrage")
log.addHandler(fhandler)
log.setLevel(logging.DEBUG)
# Für die Zwischenspeicherung des eingelesenen HTML
# - Wenn die Website in der aktuellen Session schon einmal abgefragt wurde, wird das Ergebnis
# der Abfrage in dieser Variablen gesichert
# - Dies erleichtert die Entwicklung der nachfolgenden Verabeitungsschritte und führt nicht immer wieder zu
# neuen überflüssigen Abfragen der Website
html_payload = None
col_names = ['Uhrzeit', 'Summe', 'Summe Aachen', 'Summe Todesfälle', 'Summe genesen', 'Akute Fälle' ]
try:
c19_cases = pd.read_excel(config['Rohdaten']['FileName'],
sheet_name=config['Rohdaten']['SheetName'],
index_col=0,
parse_dates=[0],
skiprows=[],
names=col_names)
except FileNotFoundError as err:
log.warning('Error during pd.read_excel(): {0}'.format(err))
# Leere DataFrame für den Start erzeugen
c19_cases = pd.DataFrame(columns=col_names, index=pd.DatetimeIndex([], name='Datum'))
c19_cases.tail(14)
default_user_agent = config['Rohdaten']['UserAgent']
def robot_access_allowed(url: str, user_agent: str=default_user_agent) -> bool:
"""
Return True if scraping is allowed according to robots.txt.
Request robots.txt. If exist, parse robots.txt and return True if scraping by this script is allowed.
Parameters
----------
url: str
URL of the website, which is to be scraped
user_agent: str
User agent string, which will be used when requesting the website
Returns
-------
bool
True, if web scraping is allowed by robots.txt or if robots.txt does not exist.
False, if web scraping is not allowed by robots.txt
"""
log.debug("access_allowed(" + url + ", " + user_agent + ")")
headers = { 'user-agent': user_agent }
# Extract root of the website's URL
# Request robots.txt from root of the website
# If robots.txt exist, parse content of the file
return True
def gather_html(url: str, user_agent: str=default_user_agent) -> str:
"""
Request website from <url> and return the HTML delivered by the website as text.
Parameters
----------
url: str
URL of the website, which is to be scraped
user_agent: str
User agent string, which will be used when requesting the website
Returns
-------
str
The HTML which was gathered from the website
"""
log.debug("gather_html(" + url + ", " + user_agent + ")")
# Website abfragen
headers = { 'user-agent': user_agent }
page = requests.get(url)
log.debug("gather_html/page.status_code: %d", page.status_code)
for key, val in page.headers.items():
log.debug("gather_html/page.header(%s): %s", key, val)
if page.status_code == requests.codes.ALL_OK: # Alles ok
return page.text
return None
def gather_text(html_text: str) -> []:
"""
Extract relevant text content from HTML and return extracted text for news entry parts.
Extract relevant text content, i.e. the distinctive news entries, from <html_text>.
Return array with news entries, each entry consisting of a dictionary with extracted text parts
for 'Header', 'Abstract', 'Main'.
Parameters
----------
html_str: str
The HTML code, which was gathered from the website.
Returns
-------
[]
Array with entries, each consisting of a dictionary with extracted text for 'Header', 'Abstract', 'Main'
part of the news entries.
"""
log.debug(f"gather_text(%d chars)", len(html_text))
records = []
# Parser instanzieren
soup = BeautifulSoup(html_text, 'html.parser')
# Relevante Objekte aus geliefertem HTML extrahieren
# - Header
# - Abstract (existiert nur für die aktuellen Meldungen, nicht im Meldungsarchiv)
# - Main
divs = soup.select('div.mid-col article > div.textcontent > div')
for div in divs:
# Header erkennen: Nun wenn dieser gefunden wird, macht es Sinn, nachfolgend nach
# Abstract und Haupttext zu suchen
header = div.select('h2')
if header:
# Header: Text extrahieren
header_text = next(header[0].stripped_strings)
# TODO: Ab hier überarbeiten, sodass der gesamte Body-Text eines Eintrags eingelesen wird
body_text = ''
body_elems = div.select('div > div.ce-bodytext > p, div > div.ce-bodytext > ul')
for be in body_elems:
# Body: Text extrahieren
body_text += next(be.stripped_strings)
print(body_text)
# Neuen Eintrag mit den extahierten Texten hinzufügen
records.append({'Header': header_text, 'Body': body_text })
log.debug("gather_text: %d records extracted from HTML", len(records))
return records
class NewsMeta():
"""Value class for meta data of news entry."""
pass
class CaseFigures():
"""Value class for case figures, extracted from the news entries."""
pass
def parse_header(header: str) -> NewsMeta:
"""
Analyse <header> of the news and return date and time of news or None.
The <header> is expected to have the following format:
'Aktuelle Lage Stadt und StädteRegion Aachen zum Corona-Virus; Montag, 22. Juni, 10:00 Uhr'
Variables weekday name, date, and time will be extracted from header. Year assumed to be 2020.
Parameters
----------
header : str
Extracted raw text of header
Returns
-------
None
If no matching header found
NewsMeta(weekday_name, day_of_month, month_name, hour, minutes, date, time, datetime)
Object with meta data of news entry
"""
monthnames = [ "Januar", "Februar", "März", "April", "Mai", "Juni", "Juli", "August", "September",
"Oktober", "November", "Dezember" ]
log.debug("parse_header(" + header[:40] + "...)")
pattern = (r"^Aktuelle Lage Stadt und StädteRegion Aachen zum Corona-Virus;\s*"
r"(Montag|Dienstag|Mittwoch|Donnerstag|Freitag|Samstag|Sonntag),\s*"
r"([0-9]{1,2})\.\s*(Januar|Februar|März|April|Mai|Juni|Juli|August|"
r"September|Oktober|November|Dezember),\s*"
r"([0-9]{1,2}):([0-9]{2})\s*Uhr.*$")
match = re.search(pattern, header)
if match:
meta = NewsMeta()
try:
meta.weekday_name = match.group(1)
meta.day_of_month = int(match.group(2))
meta.month_name = match.group(3)
meta.hour = int(match.group(4))
meta.minutes = int(match.group(5))
meta.date = date(2020, monthnames.index(meta.month_name) + 1, meta.day_of_month)
meta.time = time(meta.hour, meta.minutes)
meta.datetime = datetime.combine(meta.date, meta.time)
log.debug(f"parse_header/meta.weekday: %s", meta.weekday_name)
log.debug(f"parse_header/meta.day_of_month: %d", meta.day_of_month)
log.debug(f"parse_header/meta.month_name: %s", meta.month_name)
log.debug(f"parse_header/meta.hour: %d", meta.hour)
log.debug(f"parse_header/meta.minutes: %d", meta.minutes)
log.debug(f"parse_header/meta.date: %s", str(meta.date))
log.debug(f"parse_header/meta.time: %s", str(meta.time))
log.debug(f"parse_header/meta.datetime: %s", str(meta.datetime))
return meta
except:
log.warning("parse_header() failed parsing news header [1]")
return None
else:
log.warning("parse_header() failed parsing news header [2]")
return None
def parse_cases(kind: str, meta: NewsMeta, text: str, pattern_total: str, patterns_total_AC: [],
pattern_recovered: str, patterns_deaths: [], pattern_active: str) -> CaseFigures:
"""
Analyse <abstract> or <main> text of the news and return Covid-19 case numbers included therein.
Parameters
----------
kind : str
"Main" - Parse main part of news entry
"Abstract" - Parse abstract of news entry
meta : NewsMeta
Meta data of news entry
text : str
Text to be parsed
pattern_total : str
Regular expression for parsing total number of cases in Städteregion Aachen
patterns_total_AC : str
Regular expression for parsing total number of cases in Aachen
pattern_recovered : str
Regular expression for parsing total number of recovered cases in Städteregion Aachen
patterns_deaths : [ str ]
Array with regular expressions for parsing total number of deaths in Städteregion Aachen
pattern_active : str
Regular expression for parsing number of active cases in Städteregion Aachen
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug(f"parse_cases(%s, %s)", kind, text[:80])
if (not text.strip()):
# Der übergebene Text ist leer
return None
def log_debug(kind: str, meta: NewsMeta, par_name: str, par_value: int):
log.debug(f"parse_cases(%s,%s)/figures.%s: %d", kind, str(meta.datetime), par_name, par_value)
def log_warn(kind: str, meta: NewsMeta, reason: int):
log.warning(f"parse_cases(%s,%s) failed [%d]", kind, str(meta.datetime), reason)
figures = CaseFigures()
# Summe der Corona-Fälle in der Städteregion extrahieren
match = re.search(pattern_total, text)
if match:
try:
figures.total = int(match.group(1))
log_debug(kind, meta, "total", figures.total)
except:
log_warn(kind, meta, 1)
return None
else:
log_warn(kind, meta, 2)
return None
# Summe der Corona-Fälle in der Stadt Aachen extrahieren
figures.total_AC = None
for ptac in patterns_total_AC:
match = re.search(ptac, text)
if match:
try:
figures.total_AC = int(match.group(1))
log_debug(kind, meta, "total_AC", figures.total_AC)
break
except:
log_warn(kind, meta, 3)
return None
else:
log.debug(f"parse_cases(%s)/total_AC: No match for pattern %s", kind, ptac)
if figures.total_AC is None:
log_warn(kind, meta, 4)
# Summe der wieder Genesenen extrahieren
match = re.search(pattern_recovered, text)
if match:
try:
figures.recovered = int(match.group(1))
log_debug(kind, meta, "recovered", figures.recovered)
except:
log_warn(kind, meta, 5)
return None
else:
log_warn(kind, meta, 6)
return None
# Summe der Corona-Toten extrahieren
figures.deaths = None
for pd in patterns_deaths:
match = re.search(pd, text)
if match:
try:
figures.deaths = int(match.group(1))
log_debug(kind, meta, "deaths", figures.deaths)
break
except:
log_warn(kind, meta, 7)
return None
else:
log.debug(f"parse_cases(%s)/deaths: No match for pattern %s", kind, pd)
if figures.deaths is None:
log_warn(kind, meta, 8)
return None
# Anzahl der aktiven Infektionen extrahieren
match = re.search(pattern_active, text)
if match:
try:
figures.active = int(match.group(1))
log_debug(kind, meta, "active", figures.active)
except:
log_warn(kind, meta, 9)
return None
else:
log_warn(kind, meta, 10)
return None
return figures
def parse_abstract(abstract: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <abstract> of the news and return Covid-19 case numbers included in abstract.
The relevant area of the <abstract> is expected to have the following format:
'Aktuell 1999 bestätigte Coronafälle in der StädteRegion Aachen (davon 994 in der Stadt Aachen). '
'1880 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund.'
'Bislang 98 Todesfälle.' | 'neuer Todesfall, somit insgesamt 98.' | 'neue Todesfälle, somit insgesamt 98.'
'Damit aktuell 21 nachgewiesen(e) Infizierte.'
Values will be extracted from abstract text.
Parameters
----------
meta : NewsMeta
Meta data of news entry
abstract : str
Abstract text to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug("parse_abstract(" + abstract[:40] + "...)")
pattern_total = r"Aktuell\s*([0-9]{1,6})\s*bestätigte\s*Coronafälle\s*in\s*der\s*StädteRegion\s*"
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund.*")
pattern_deaths = [
r"(?:Insgesamt|Bislang)\s*([0-9]{1,6})\s*Todesfälle\.",
r"(?:neuer\s*Todesfall|neue\s*Todesfälle),\s*somit\s*insgesamt\s*([0-9]{1,6})\."
]
pattern_active = r"Damit\s*aktuell\s*([0-9]{1,6})\s*nachgewiesene?\s*Infizie.*"
return parse_cases("Abstract", meta, abstract,
pattern_total, patterns_total_AC,
pattern_recovered, pattern_deaths, pattern_active)
def parse_main(main: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <main> text of the news and return Covid-19 case numbers included in main text.
The relevant area of the <abstract> is expected to have the following format:
'Es gibt insgesamt in der StädteRegion [nunmehr] 1997 positive Fälle, davon 992 in der Stadt Aachen'
'1876 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund'
'Die Zahl der gemeldeten Todesfälle liegt [nach wie vor] bei 98'
'Damit sind aktuell 23 Menschen in der StädteRegion nachgewiesen infiziert'
Parameters
----------
meta : NewsMeta
Meta data of news entry
main : str
Main text to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures.
"""
log.debug("parse_main(" + main[:40] + "...)")
pattern_total = (r"Es\s*gibt\s*insgesamt\s*in\s*der\s*StädteRegion\s*(?:nunmehr)?\s*([0-9]{1,6})\s*"
r"positive\s*Fälle")
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen",
r"Aachen\s*(?:[0-9]{1,6})\s*\(([0-9]{1,6})\)"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund")
patterns_deaths = [
r"Die\s*Zahl\s*der\s*gemeldeten\s*Todesfälle\s*liegt\s*(?:nach\s*wie\s*vor)?\s*bei\s*([0-9]{1,6})"
]
pattern_active = (r"Damit\s*sind\s*aktuell\s*([0-9]{1,6})\s*Menschen\s*in\s*der\s*StädteRegion\s*"
r"(?:Aachen)?\s*nachgewiesen\s*infiziert")
return parse_cases("Main", meta, main,
pattern_total, patterns_total_AC,
pattern_recovered, patterns_deaths, pattern_active)
def parse_body(body: str, meta: NewsMeta) -> CaseFigures:
"""
Analyse <body> text of the news and return Covid-19 case numbers included in main text.
The relevant area of the <body> is expected to have the following format:
'Es gibt insgesamt in der StädteRegion [nunmehr] 1997 positive Fälle, davon 992 in der Stadt Aachen'
'1876 ehemals positiv auf das Corona-Virus getestete Personen sind inzwischen wieder gesund'
'Die Zahl der gemeldeten Todesfälle liegt [nach wie vor] bei 98'
'Damit sind aktuell 23 Menschen in der StädteRegion nachgewiesen infiziert'
Parameters
----------
meta : NewsMeta
Meta data of news entry
Body : str
Text in news body to be parsed
Returns
-------
None
If text could not be parsed successfully
CaseFigures(total, total_AC, recovered, deaths, active)
Object with extracted case figures
"""
log.debug("parse_body(" + body[:40] + "...)")
pattern_total = (r"Es\s*gibt\s*insgesamt\s*in\s*der\s*StädteRegion\s*(?:nunmehr)?\s*([0-9]{1,6})\s*"
r"positive\s*Fälle")
patterns_total_AC = [
r"davon\s*([0-9]{1,6})\s*in\s*der\s*Stadt\s*Aachen",
r"Aachen\s*(?:[0-9]{1,6})\s*\(([0-9]{1,6})\)"
]
pattern_recovered = (r"([0-9]{1,6})\s*ehemals\s*positiv\s*auf\s*das\s*Corona-Virus\s*getestete\s*"
r"Personen\s*sind\s*inzwischen\s*wieder\s*gesund")
patterns_deaths = [
r"Die\s*Zahl\s*der\s*gemeldeten\s*Todesfälle\s*liegt\s*(?:nach\s*wie\s*vor)?\s*bei\s*([0-9]{1,6})"
]
pattern_active = (r"Damit\s*sind\s*aktuell\s*([0-9]{1,6})\s*Menschen\s*in\s*der\s*StädteRegion\s*"
r"(?:Aachen)?\s*nachgewiesen\s*infiziert")
return parse_cases("Body", meta, body,
pattern_total, patterns_total_AC,
pattern_recovered, patterns_deaths, pattern_active)
def checked_data1(meta: NewsMeta, fig_abstract: CaseFigures, fig_main: CaseFigures) -> []:
"""
Check extracted data for consistency and return valid figures or None.
Parameters
----------
meta : NewMeta
Meta data of news entry
fig_abstract: CaseFigures
Object with figures extracted from abstract of the news entry
fig_main: CaseFigures
Object with figures extracted from main text of the news entry
Returns
-------
None
If consistency check fails
[ Date, Time, Total, Total_AC, Deaths, Recovered, Active ]
Array with extracted Covid-19 numbers
"""
if meta != None:
# Der Header konnte erfolgreich analysiert werden
a, m = fig_abstract, fig_main
if (a != None) and (m != None):
# Sowohl Abstract als auch Main konnten erfolgreich analysiert werden
if (m.total == a.total) and (m.deaths == a.deaths) and (m.recovered == a.recovered) and \
(m.active == a.active):
# Die eingelesenen Zahlen aus Abstract und Main stimmmen überein
if m.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
m.active = m.total - m.deaths - m.recovered
if m.total - m.deaths - m.recovered == m.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [ meta.date, meta.time, m.total, m.total_AC, m.deaths, m.recovered, m.active ]
else:
log.warning('Data consistency error: (1) Main and Abstract found. Inconsistent figures read.')
return None
elif m != None:
# Mindestens Main konnte erfolgreich analysiert werden
if m.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
m.active = m.total - m.deaths - m.recovered
if m.total - m.deaths - m.recovered == m.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [ meta.date, meta.time, m.total, m.total_AC, m.deaths, m.recovered, m.active ]
else:
log.warning('Data onsistency error: (2) Only Main found. Inconsistent figures read. ')
return None
else:
log.warning('Data consistency error: (3) Header not found.')
return None
def checked_data2(meta: NewsMeta, fig_body: CaseFigures) -> []:
"""
Check extracted data for consistency and return valid figures or None.
Parameters
----------
meta : NewMeta
Meta data of news entry
fig_body: CaseFigures
Object with figures extracted from body of the news entry
Returns
-------
None
If consistency check fails
[ Date, Time, Total, Total_AC, Deaths, Recovered, Active ]
Array with extracted Covid-19 numbers
"""
if meta != None:
# Der Header konnte erfolgreich analysiert werden
if fig_body != None:
if fig_body.active == 0:
# Die Anzahl aktiver Fälle war nicht explizit angegeben und wird stattdessen berechnet
fig_body.active = fig_body.total - fig_body.deaths - fig_body.recovered
if fig_body.total - fig_body.deaths - fig_body.recovered == fig_body.active:
# Die eingelesenen Daten haben die Konsistenzprüfung bestanden
return [
meta.date, meta.time,
fig_body.total, fig_body.total_AC, fig_body.deaths, fig_body.recovered, fig_body.active
]
else:
log.warning('Data onsistency error: Inconsistent figures read. ')
return None
else:
log.warning('Data consistency error: Header not found.')
return None
def fill_dataframe_from_text(text_records: []) -> pd.DataFrame:
"""
Analyse <text_records> and return Pandas dataframe with parsed Covid-19 case numbers.
Parameters
----------
text_records : []
Array with extracted text
Returns
-------
pd.DataFrame
Pandas dataframe with extracted data
"""
log.debug(f"fill_dataframe_from_text(%d records)", len(text_records))
figures = [] # Zwischenspeicherung der eingelesenen Zahlen
dates = [] # Zwischenspeicherung der Datumsangaben
for tr in text_records:
meta = parse_header(tr['Header'])
if tr['Body']:
fig_body = parse_body(tr['Body'], meta)
rec = checked_data2(meta, fig_body)
else:
if tr['Abstract']:
fig_abstract = parse_abstract(tr['Abstract'], meta)
if tr['Main']:
fig_main = parse_main(tr['Main'], meta)
rec = checked_data1(meta, fig_abstract, fig_main)
if rec:
dates.append(rec[0])
figures.append(rec[1:])
if figures.count:
cols = ['Uhrzeit', 'Summe', 'Summe Aachen', 'Summe Todesfälle', 'Summe genesen', 'Akute Fälle' ]
index = pd.DatetimeIndex(dates, name='Datum')
df = pd.DataFrame(np.array(figures), columns=cols, index=index).sort_index()
return df
return pd.DataFrame() # Leer
new_request = (c19_cases.size == 0) or \
(date.today() >= (date.fromtimestamp(c19_cases.index.max().timestamp()) + timedelta(days=1)))
if new_request:
log.info("New request initiated")
if not html_payload:
url = config['Rohdaten']['SourceURLDefault']
if robot_access_allowed(url):
html_payload = gather_html(url)
if html_payload:
records = gather_text(html_payload)
log.info("Processing {0} text records".format(len(records)))
if len(records):
new_cases = fill_dataframe_from_text(records)
if not new_cases.empty:
# Nur Zeilen mit neuerem Datum hinzufügen
merged_cases = pd.concat([c19_cases, new_cases[new_cases.index > c19_cases.index[-1]]], join='outer')
log.info("{0} new case records appended".format(len(merged_cases.index) - len(c19_cases.index)))
else:
log.info("No new case figures extracted")
merged_cases.to_excel(config['Rohdaten']['FileName'],
sheet_name=config['Rohdaten']['SheetName'], index_label='Datum')
merged_cases.tail(10)
else:
log.info("No new request required")
| 0.419886 | 0.632559 |
## Dependencies
```
import json, warnings, shutil
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_text import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras.models import Model
from tensorflow.keras import optimizers, metrics, losses, layers
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
SEED = 0
seed_everything(SEED)
warnings.filterwarnings("ignore")
class RectifiedAdam(tf.keras.optimizers.Optimizer):
"""Variant of the Adam optimizer whose adaptive learning rate is rectified
so as to have a consistent variance.
It implements the Rectified Adam (a.k.a. RAdam) proposed by
Liyuan Liu et al. in [On The Variance Of The Adaptive Learning Rate
And Beyond](https://arxiv.org/pdf/1908.03265v1.pdf).
Example of usage:
```python
opt = tfa.optimizers.RectifiedAdam(lr=1e-3)
```
Note: `amsgrad` is not described in the original paper. Use it with
caution.
RAdam is not a placement of the heuristic warmup, the settings should be
kept if warmup has already been employed and tuned in the baseline method.
You can enable warmup by setting `total_steps` and `warmup_proportion`:
```python
opt = tfa.optimizers.RectifiedAdam(
lr=1e-3,
total_steps=10000,
warmup_proportion=0.1,
min_lr=1e-5,
)
```
In the above example, the learning rate will increase linearly
from 0 to `lr` in 1000 steps, then decrease linearly from `lr` to `min_lr`
in 9000 steps.
Lookahead, proposed by Michael R. Zhang et.al in the paper
[Lookahead Optimizer: k steps forward, 1 step back]
(https://arxiv.org/abs/1907.08610v1), can be integrated with RAdam,
which is announced by Less Wright and the new combined optimizer can also
be called "Ranger". The mechanism can be enabled by using the lookahead
wrapper. For example:
```python
radam = tfa.optimizers.RectifiedAdam()
ranger = tfa.optimizers.Lookahead(radam, sync_period=6, slow_step_size=0.5)
```
"""
def __init__(self,
learning_rate=0.001,
beta_1=0.9,
beta_2=0.999,
epsilon=1e-7,
weight_decay=0.,
amsgrad=False,
sma_threshold=5.0,
total_steps=0,
warmup_proportion=0.1,
min_lr=0.,
name='RectifiedAdam',
**kwargs):
r"""Construct a new RAdam optimizer.
Args:
learning_rate: A `Tensor` or a floating point value. or a schedule
that is a `tf.keras.optimizers.schedules.LearningRateSchedule`
The learning rate.
beta_1: A float value or a constant float tensor.
The exponential decay rate for the 1st moment estimates.
beta_2: A float value or a constant float tensor.
The exponential decay rate for the 2nd moment estimates.
epsilon: A small constant for numerical stability.
weight_decay: A floating point value. Weight decay for each param.
amsgrad: boolean. Whether to apply AMSGrad variant of this
algorithm from the paper "On the Convergence of Adam and
beyond".
sma_threshold. A float value.
The threshold for simple mean average.
total_steps: An integer. Total number of training steps.
Enable warmup by setting a positive value.
warmup_proportion: A floating point value.
The proportion of increasing steps.
min_lr: A floating point value. Minimum learning rate after warmup.
name: Optional name for the operations created when applying
gradients. Defaults to "RectifiedAdam".
**kwargs: keyword arguments. Allowed to be {`clipnorm`,
`clipvalue`, `lr`, `decay`}. `clipnorm` is clip gradients
by norm; `clipvalue` is clip gradients by value, `decay` is
included for backward compatibility to allow time inverse
decay of learning rate. `lr` is included for backward
compatibility, recommended to use `learning_rate` instead.
"""
super(RectifiedAdam, self).__init__(name, **kwargs)
self._set_hyper('learning_rate', kwargs.get('lr', learning_rate))
self._set_hyper('beta_1', beta_1)
self._set_hyper('beta_2', beta_2)
self._set_hyper('decay', self._initial_decay)
self._set_hyper('weight_decay', weight_decay)
self._set_hyper('sma_threshold', sma_threshold)
self._set_hyper('total_steps', float(total_steps))
self._set_hyper('warmup_proportion', warmup_proportion)
self._set_hyper('min_lr', min_lr)
self.epsilon = epsilon or tf.keras.backend.epsilon()
self.amsgrad = amsgrad
self._initial_weight_decay = weight_decay
self._initial_total_steps = total_steps
def _create_slots(self, var_list):
for var in var_list:
self.add_slot(var, 'm')
for var in var_list:
self.add_slot(var, 'v')
if self.amsgrad:
for var in var_list:
self.add_slot(var, 'vhat')
def set_weights(self, weights):
params = self.weights
num_vars = int((len(params) - 1) / 2)
if len(weights) == 3 * num_vars + 1:
weights = weights[:len(params)]
super(RectifiedAdam, self).set_weights(weights)
def _resource_apply_dense(self, grad, var):
var_dtype = var.dtype.base_dtype
lr_t = self._decayed_lr(var_dtype)
m = self.get_slot(var, 'm')
v = self.get_slot(var, 'v')
beta_1_t = self._get_hyper('beta_1', var_dtype)
beta_2_t = self._get_hyper('beta_2', var_dtype)
epsilon_t = tf.convert_to_tensor(self.epsilon, var_dtype)
local_step = tf.cast(self.iterations + 1, var_dtype)
beta_1_power = tf.pow(beta_1_t, local_step)
beta_2_power = tf.pow(beta_2_t, local_step)
if self._initial_total_steps > 0:
total_steps = self._get_hyper('total_steps', var_dtype)
warmup_steps = total_steps *\
self._get_hyper('warmup_proportion', var_dtype)
min_lr = self._get_hyper('min_lr', var_dtype)
decay_steps = tf.maximum(total_steps - warmup_steps, 1)
decay_rate = (min_lr - lr_t) / decay_steps
lr_t = tf.where(
local_step <= warmup_steps,
lr_t * (local_step / warmup_steps),
lr_t + decay_rate * tf.minimum(local_step - warmup_steps,
decay_steps),
)
sma_inf = 2.0 / (1.0 - beta_2_t) - 1.0
sma_t = sma_inf - 2.0 * local_step * beta_2_power / (
1.0 - beta_2_power)
m_t = m.assign(
beta_1_t * m + (1.0 - beta_1_t) * grad,
use_locking=self._use_locking)
m_corr_t = m_t / (1.0 - beta_1_power)
v_t = v.assign(
beta_2_t * v + (1.0 - beta_2_t) * tf.square(grad),
use_locking=self._use_locking)
if self.amsgrad:
vhat = self.get_slot(var, 'vhat')
vhat_t = vhat.assign(
tf.maximum(vhat, v_t), use_locking=self._use_locking)
v_corr_t = tf.sqrt(vhat_t / (1.0 - beta_2_power))
else:
vhat_t = None
v_corr_t = tf.sqrt(v_t / (1.0 - beta_2_power))
r_t = tf.sqrt((sma_t - 4.0) / (sma_inf - 4.0) * (sma_t - 2.0) /
(sma_inf - 2.0) * sma_inf / sma_t)
sma_threshold = self._get_hyper('sma_threshold', var_dtype)
var_t = tf.where(sma_t >= sma_threshold,
r_t * m_corr_t / (v_corr_t + epsilon_t), m_corr_t)
if self._initial_weight_decay > 0.0:
var_t += self._get_hyper('weight_decay', var_dtype) * var
var_update = var.assign_sub(
lr_t * var_t, use_locking=self._use_locking)
updates = [var_update, m_t, v_t]
if self.amsgrad:
updates.append(vhat_t)
return tf.group(*updates)
def _resource_apply_sparse(self, grad, var, indices):
var_dtype = var.dtype.base_dtype
lr_t = self._decayed_lr(var_dtype)
beta_1_t = self._get_hyper('beta_1', var_dtype)
beta_2_t = self._get_hyper('beta_2', var_dtype)
epsilon_t = tf.convert_to_tensor(self.epsilon, var_dtype)
local_step = tf.cast(self.iterations + 1, var_dtype)
beta_1_power = tf.pow(beta_1_t, local_step)
beta_2_power = tf.pow(beta_2_t, local_step)
if self._initial_total_steps > 0:
total_steps = self._get_hyper('total_steps', var_dtype)
warmup_steps = total_steps *\
self._get_hyper('warmup_proportion', var_dtype)
min_lr = self._get_hyper('min_lr', var_dtype)
decay_steps = tf.maximum(total_steps - warmup_steps, 1)
decay_rate = (min_lr - lr_t) / decay_steps
lr_t = tf.where(
local_step <= warmup_steps,
lr_t * (local_step / warmup_steps),
lr_t + decay_rate * tf.minimum(local_step - warmup_steps,
decay_steps),
)
sma_inf = 2.0 / (1.0 - beta_2_t) - 1.0
sma_t = sma_inf - 2.0 * local_step * beta_2_power / (
1.0 - beta_2_power)
m = self.get_slot(var, 'm')
m_scaled_g_values = grad * (1 - beta_1_t)
m_t = m.assign(m * beta_1_t, use_locking=self._use_locking)
with tf.control_dependencies([m_t]):
m_t = self._resource_scatter_add(m, indices, m_scaled_g_values)
m_corr_t = m_t / (1.0 - beta_1_power)
v = self.get_slot(var, 'v')
v_scaled_g_values = (grad * grad) * (1 - beta_2_t)
v_t = v.assign(v * beta_2_t, use_locking=self._use_locking)
with tf.control_dependencies([v_t]):
v_t = self._resource_scatter_add(v, indices, v_scaled_g_values)
if self.amsgrad:
vhat = self.get_slot(var, 'vhat')
vhat_t = vhat.assign(
tf.maximum(vhat, v_t), use_locking=self._use_locking)
v_corr_t = tf.sqrt(vhat_t / (1.0 - beta_2_power))
else:
vhat_t = None
v_corr_t = tf.sqrt(v_t / (1.0 - beta_2_power))
r_t = tf.sqrt((sma_t - 4.0) / (sma_inf - 4.0) * (sma_t - 2.0) /
(sma_inf - 2.0) * sma_inf / sma_t)
sma_threshold = self._get_hyper('sma_threshold', var_dtype)
var_t = tf.where(sma_t >= sma_threshold,
r_t * m_corr_t / (v_corr_t + epsilon_t), m_corr_t)
if self._initial_weight_decay > 0.0:
var_t += self._get_hyper('weight_decay', var_dtype) * var
with tf.control_dependencies([var_t]):
var_update = self._resource_scatter_add(
var, indices, tf.gather(-lr_t * var_t, indices))
updates = [var_update, m_t, v_t]
if self.amsgrad:
updates.append(vhat_t)
return tf.group(*updates)
def get_config(self):
config = super(RectifiedAdam, self).get_config()
config.update({
'learning_rate':
self._serialize_hyperparameter('learning_rate'),
'beta_1':
self._serialize_hyperparameter('beta_1'),
'beta_2':
self._serialize_hyperparameter('beta_2'),
'decay':
self._serialize_hyperparameter('decay'),
'weight_decay':
self._serialize_hyperparameter('weight_decay'),
'sma_threshold':
self._serialize_hyperparameter('sma_threshold'),
'epsilon':
self.epsilon,
'amsgrad':
self.amsgrad,
'total_steps':
self._serialize_hyperparameter('total_steps'),
'warmup_proportion':
self._serialize_hyperparameter('warmup_proportion'),
'min_lr':
self._serialize_hyperparameter('min_lr'),
})
return config
```
# Load data
```
database_base_path = '/kaggle/input/tweet-dataset-split-roberta-base-96-text/'
k_fold = pd.read_csv(database_base_path + '5-fold.csv')
display(k_fold.head())
# Unzip files
!tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_1.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_2.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_3.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_4.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_5.tar.gz
```
# Model parameters
```
vocab_path = database_base_path + 'vocab.json'
merges_path = database_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
config = {
"MAX_LEN": 96,
"BATCH_SIZE": 32,
"EPOCHS": 6,
"LEARNING_RATE": 3e-5,
"ES_PATIENCE": 1,
"N_FOLDS": 1,
"base_model_path": base_path + 'roberta-base-tf_model.h5',
"config_path": base_path + 'roberta-base-config.json'
}
with open('config.json', 'w') as json_file:
json.dump(json.loads(json.dumps(config)), json_file)
```
# Model
```
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
x_start = layers.Dropout(.1)(last_hidden_state)
x_start = layers.Conv1D(1, 1)(x_start)
x_start = layers.Flatten()(x_start)
x_end = layers.Dropout(.1)(last_hidden_state)
x_end = layers.Conv1D(1, 1)(x_end)
x_end = layers.Flatten()(x_end)
y_start = layers.Subtract()([x_start, x_end])
y_start = layers.Activation('softmax', name='y_start')(x_start)
y_end = layers.Subtract()([x_end, x_start])
y_end = layers.Activation('softmax', name='y_end')(x_end)
x_sentiment = layers.GlobalAveragePooling1D()(last_hidden_state)
x_sentiment = layers.Dropout(.1)(x_sentiment)
y_sentiment = layers.Dense(3, activation='softmax', name='y_sentiment')(x_sentiment)
model = Model(inputs=[input_ids, attention_mask], outputs=[y_start, y_end, y_sentiment])
optimizer = RectifiedAdam(lr=config['LEARNING_RATE'],
total_steps=(len(k_fold[k_fold['fold_1'] == 'train']) // config['BATCH_SIZE']) * config['EPOCHS'],
warmup_proportion=0.1,
min_lr=1e-7)
model.compile(optimizer, loss={'y_start': losses.CategoricalCrossentropy(),
'y_end': losses.CategoricalCrossentropy(),
'y_sentiment': losses.SparseCategoricalCrossentropy()},
loss_weights=[1, 1, .5],
metrics={'y_start': metrics.CategoricalAccuracy(),
'y_end': metrics.CategoricalAccuracy(),
'y_sentiment': metrics.SparseCategoricalCrossentropy()})
return model
```
# Tokenizer
```
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
tokenizer.save('./')
```
# Train
```
history_list = []
for n_fold in range(config['N_FOLDS']):
n_fold +=1
print('\nFOLD: %d' % (n_fold))
# Load data
base_data_path = 'fold_%d/' % (n_fold)
x_train = np.load(base_data_path + 'x_train.npy')
y_train = np.load(base_data_path + 'y_train.npy')
x_valid = np.load(base_data_path + 'x_valid.npy')
y_valid = np.load(base_data_path + 'y_valid.npy')
y_train_aux = np.load(base_data_path + 'y_train_aux.npy')[0].astype(int)
y_valid_aux = np.load(base_data_path + 'y_valid_aux.npy')[0].astype(int)
### Delete data dir
shutil.rmtree(base_data_path)
# Train model
model_path = 'model_fold_%d.h5' % (n_fold)
model = model_fn(config['MAX_LEN'])
es = EarlyStopping(monitor='val_loss', mode='min', patience=config['ES_PATIENCE'],
restore_best_weights=True, verbose=1)
checkpoint = ModelCheckpoint(model_path, monitor='val_loss', mode='min',
save_best_only=True, save_weights_only=True)
history = model.fit(list(x_train), list([y_train[0], y_train[1], y_train_aux]),
validation_data=(list(x_valid), list([y_valid[0], y_valid[1], y_valid_aux])),
batch_size=config['BATCH_SIZE'],
callbacks=[checkpoint, es],
epochs=config['EPOCHS'],
verbose=2).history
history_list.append(history)
# Make predictions
train_preds = model.predict(list(x_train))
valid_preds = model.predict(list(x_valid))
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'start_fold_%d' % (n_fold)] = train_preds[0].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'end_fold_%d' % (n_fold)] = train_preds[1].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'start_fold_%d' % (n_fold)] = valid_preds[0].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'end_fold_%d' % (n_fold)] = valid_preds[1].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'sentiment_fold_%d' % (n_fold)] = train_preds[2].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'sentiment_fold_%d' % (n_fold)] = valid_preds[2].argmax(axis=-1)
k_fold['end_fold_%d' % (n_fold)] = k_fold['end_fold_%d' % (n_fold)].astype(int)
k_fold['start_fold_%d' % (n_fold)] = k_fold['start_fold_%d' % (n_fold)].astype(int)
k_fold['end_fold_%d' % (n_fold)].clip(0, k_fold['text_len'], inplace=True)
k_fold['start_fold_%d' % (n_fold)].clip(0, k_fold['end_fold_%d' % (n_fold)], inplace=True)
k_fold['prediction_fold_%d' % (n_fold)] = k_fold.apply(lambda x: decode(x['start_fold_%d' % (n_fold)], x['end_fold_%d' % (n_fold)], x['text'], tokenizer), axis=1)
k_fold['prediction_fold_%d' % (n_fold)].fillna(k_fold["text"], inplace=True)
k_fold['jaccard_fold_%d' % (n_fold)] = k_fold.apply(lambda x: jaccard(x['selected_text'], x['prediction_fold_%d' % (n_fold)]), axis=1)
```
# Model loss graph
```
sns.set(style="whitegrid")
for n_fold in range(config['N_FOLDS']):
print('Fold: %d' % (n_fold+1))
plot_metrics(history_list[n_fold])
```
# Model evaluation
```
display(evaluate_model_kfold(k_fold, config['N_FOLDS']).style.applymap(color_map))
```
# Visualize predictions
```
display(k_fold[[c for c in k_fold.columns if not (c.startswith('textID') or
c.startswith('text_len') or
c.startswith('selected_text_len') or
c.startswith('text_wordCnt') or
c.startswith('selected_text_wordCnt') or
c.startswith('fold_') or
c.startswith('start_fold_') or
c.startswith('end_fold_'))]].head(15))
```
|
github_jupyter
|
import json, warnings, shutil
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_text import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras.models import Model
from tensorflow.keras import optimizers, metrics, losses, layers
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
SEED = 0
seed_everything(SEED)
warnings.filterwarnings("ignore")
class RectifiedAdam(tf.keras.optimizers.Optimizer):
"""Variant of the Adam optimizer whose adaptive learning rate is rectified
so as to have a consistent variance.
It implements the Rectified Adam (a.k.a. RAdam) proposed by
Liyuan Liu et al. in [On The Variance Of The Adaptive Learning Rate
And Beyond](https://arxiv.org/pdf/1908.03265v1.pdf).
Example of usage:
```python
opt = tfa.optimizers.RectifiedAdam(lr=1e-3)
```
Note: `amsgrad` is not described in the original paper. Use it with
caution.
RAdam is not a placement of the heuristic warmup, the settings should be
kept if warmup has already been employed and tuned in the baseline method.
You can enable warmup by setting `total_steps` and `warmup_proportion`:
```python
opt = tfa.optimizers.RectifiedAdam(
lr=1e-3,
total_steps=10000,
warmup_proportion=0.1,
min_lr=1e-5,
)
```
In the above example, the learning rate will increase linearly
from 0 to `lr` in 1000 steps, then decrease linearly from `lr` to `min_lr`
in 9000 steps.
Lookahead, proposed by Michael R. Zhang et.al in the paper
[Lookahead Optimizer: k steps forward, 1 step back]
(https://arxiv.org/abs/1907.08610v1), can be integrated with RAdam,
which is announced by Less Wright and the new combined optimizer can also
be called "Ranger". The mechanism can be enabled by using the lookahead
wrapper. For example:
```python
radam = tfa.optimizers.RectifiedAdam()
ranger = tfa.optimizers.Lookahead(radam, sync_period=6, slow_step_size=0.5)
```
"""
def __init__(self,
learning_rate=0.001,
beta_1=0.9,
beta_2=0.999,
epsilon=1e-7,
weight_decay=0.,
amsgrad=False,
sma_threshold=5.0,
total_steps=0,
warmup_proportion=0.1,
min_lr=0.,
name='RectifiedAdam',
**kwargs):
r"""Construct a new RAdam optimizer.
Args:
learning_rate: A `Tensor` or a floating point value. or a schedule
that is a `tf.keras.optimizers.schedules.LearningRateSchedule`
The learning rate.
beta_1: A float value or a constant float tensor.
The exponential decay rate for the 1st moment estimates.
beta_2: A float value or a constant float tensor.
The exponential decay rate for the 2nd moment estimates.
epsilon: A small constant for numerical stability.
weight_decay: A floating point value. Weight decay for each param.
amsgrad: boolean. Whether to apply AMSGrad variant of this
algorithm from the paper "On the Convergence of Adam and
beyond".
sma_threshold. A float value.
The threshold for simple mean average.
total_steps: An integer. Total number of training steps.
Enable warmup by setting a positive value.
warmup_proportion: A floating point value.
The proportion of increasing steps.
min_lr: A floating point value. Minimum learning rate after warmup.
name: Optional name for the operations created when applying
gradients. Defaults to "RectifiedAdam".
**kwargs: keyword arguments. Allowed to be {`clipnorm`,
`clipvalue`, `lr`, `decay`}. `clipnorm` is clip gradients
by norm; `clipvalue` is clip gradients by value, `decay` is
included for backward compatibility to allow time inverse
decay of learning rate. `lr` is included for backward
compatibility, recommended to use `learning_rate` instead.
"""
super(RectifiedAdam, self).__init__(name, **kwargs)
self._set_hyper('learning_rate', kwargs.get('lr', learning_rate))
self._set_hyper('beta_1', beta_1)
self._set_hyper('beta_2', beta_2)
self._set_hyper('decay', self._initial_decay)
self._set_hyper('weight_decay', weight_decay)
self._set_hyper('sma_threshold', sma_threshold)
self._set_hyper('total_steps', float(total_steps))
self._set_hyper('warmup_proportion', warmup_proportion)
self._set_hyper('min_lr', min_lr)
self.epsilon = epsilon or tf.keras.backend.epsilon()
self.amsgrad = amsgrad
self._initial_weight_decay = weight_decay
self._initial_total_steps = total_steps
def _create_slots(self, var_list):
for var in var_list:
self.add_slot(var, 'm')
for var in var_list:
self.add_slot(var, 'v')
if self.amsgrad:
for var in var_list:
self.add_slot(var, 'vhat')
def set_weights(self, weights):
params = self.weights
num_vars = int((len(params) - 1) / 2)
if len(weights) == 3 * num_vars + 1:
weights = weights[:len(params)]
super(RectifiedAdam, self).set_weights(weights)
def _resource_apply_dense(self, grad, var):
var_dtype = var.dtype.base_dtype
lr_t = self._decayed_lr(var_dtype)
m = self.get_slot(var, 'm')
v = self.get_slot(var, 'v')
beta_1_t = self._get_hyper('beta_1', var_dtype)
beta_2_t = self._get_hyper('beta_2', var_dtype)
epsilon_t = tf.convert_to_tensor(self.epsilon, var_dtype)
local_step = tf.cast(self.iterations + 1, var_dtype)
beta_1_power = tf.pow(beta_1_t, local_step)
beta_2_power = tf.pow(beta_2_t, local_step)
if self._initial_total_steps > 0:
total_steps = self._get_hyper('total_steps', var_dtype)
warmup_steps = total_steps *\
self._get_hyper('warmup_proportion', var_dtype)
min_lr = self._get_hyper('min_lr', var_dtype)
decay_steps = tf.maximum(total_steps - warmup_steps, 1)
decay_rate = (min_lr - lr_t) / decay_steps
lr_t = tf.where(
local_step <= warmup_steps,
lr_t * (local_step / warmup_steps),
lr_t + decay_rate * tf.minimum(local_step - warmup_steps,
decay_steps),
)
sma_inf = 2.0 / (1.0 - beta_2_t) - 1.0
sma_t = sma_inf - 2.0 * local_step * beta_2_power / (
1.0 - beta_2_power)
m_t = m.assign(
beta_1_t * m + (1.0 - beta_1_t) * grad,
use_locking=self._use_locking)
m_corr_t = m_t / (1.0 - beta_1_power)
v_t = v.assign(
beta_2_t * v + (1.0 - beta_2_t) * tf.square(grad),
use_locking=self._use_locking)
if self.amsgrad:
vhat = self.get_slot(var, 'vhat')
vhat_t = vhat.assign(
tf.maximum(vhat, v_t), use_locking=self._use_locking)
v_corr_t = tf.sqrt(vhat_t / (1.0 - beta_2_power))
else:
vhat_t = None
v_corr_t = tf.sqrt(v_t / (1.0 - beta_2_power))
r_t = tf.sqrt((sma_t - 4.0) / (sma_inf - 4.0) * (sma_t - 2.0) /
(sma_inf - 2.0) * sma_inf / sma_t)
sma_threshold = self._get_hyper('sma_threshold', var_dtype)
var_t = tf.where(sma_t >= sma_threshold,
r_t * m_corr_t / (v_corr_t + epsilon_t), m_corr_t)
if self._initial_weight_decay > 0.0:
var_t += self._get_hyper('weight_decay', var_dtype) * var
var_update = var.assign_sub(
lr_t * var_t, use_locking=self._use_locking)
updates = [var_update, m_t, v_t]
if self.amsgrad:
updates.append(vhat_t)
return tf.group(*updates)
def _resource_apply_sparse(self, grad, var, indices):
var_dtype = var.dtype.base_dtype
lr_t = self._decayed_lr(var_dtype)
beta_1_t = self._get_hyper('beta_1', var_dtype)
beta_2_t = self._get_hyper('beta_2', var_dtype)
epsilon_t = tf.convert_to_tensor(self.epsilon, var_dtype)
local_step = tf.cast(self.iterations + 1, var_dtype)
beta_1_power = tf.pow(beta_1_t, local_step)
beta_2_power = tf.pow(beta_2_t, local_step)
if self._initial_total_steps > 0:
total_steps = self._get_hyper('total_steps', var_dtype)
warmup_steps = total_steps *\
self._get_hyper('warmup_proportion', var_dtype)
min_lr = self._get_hyper('min_lr', var_dtype)
decay_steps = tf.maximum(total_steps - warmup_steps, 1)
decay_rate = (min_lr - lr_t) / decay_steps
lr_t = tf.where(
local_step <= warmup_steps,
lr_t * (local_step / warmup_steps),
lr_t + decay_rate * tf.minimum(local_step - warmup_steps,
decay_steps),
)
sma_inf = 2.0 / (1.0 - beta_2_t) - 1.0
sma_t = sma_inf - 2.0 * local_step * beta_2_power / (
1.0 - beta_2_power)
m = self.get_slot(var, 'm')
m_scaled_g_values = grad * (1 - beta_1_t)
m_t = m.assign(m * beta_1_t, use_locking=self._use_locking)
with tf.control_dependencies([m_t]):
m_t = self._resource_scatter_add(m, indices, m_scaled_g_values)
m_corr_t = m_t / (1.0 - beta_1_power)
v = self.get_slot(var, 'v')
v_scaled_g_values = (grad * grad) * (1 - beta_2_t)
v_t = v.assign(v * beta_2_t, use_locking=self._use_locking)
with tf.control_dependencies([v_t]):
v_t = self._resource_scatter_add(v, indices, v_scaled_g_values)
if self.amsgrad:
vhat = self.get_slot(var, 'vhat')
vhat_t = vhat.assign(
tf.maximum(vhat, v_t), use_locking=self._use_locking)
v_corr_t = tf.sqrt(vhat_t / (1.0 - beta_2_power))
else:
vhat_t = None
v_corr_t = tf.sqrt(v_t / (1.0 - beta_2_power))
r_t = tf.sqrt((sma_t - 4.0) / (sma_inf - 4.0) * (sma_t - 2.0) /
(sma_inf - 2.0) * sma_inf / sma_t)
sma_threshold = self._get_hyper('sma_threshold', var_dtype)
var_t = tf.where(sma_t >= sma_threshold,
r_t * m_corr_t / (v_corr_t + epsilon_t), m_corr_t)
if self._initial_weight_decay > 0.0:
var_t += self._get_hyper('weight_decay', var_dtype) * var
with tf.control_dependencies([var_t]):
var_update = self._resource_scatter_add(
var, indices, tf.gather(-lr_t * var_t, indices))
updates = [var_update, m_t, v_t]
if self.amsgrad:
updates.append(vhat_t)
return tf.group(*updates)
def get_config(self):
config = super(RectifiedAdam, self).get_config()
config.update({
'learning_rate':
self._serialize_hyperparameter('learning_rate'),
'beta_1':
self._serialize_hyperparameter('beta_1'),
'beta_2':
self._serialize_hyperparameter('beta_2'),
'decay':
self._serialize_hyperparameter('decay'),
'weight_decay':
self._serialize_hyperparameter('weight_decay'),
'sma_threshold':
self._serialize_hyperparameter('sma_threshold'),
'epsilon':
self.epsilon,
'amsgrad':
self.amsgrad,
'total_steps':
self._serialize_hyperparameter('total_steps'),
'warmup_proportion':
self._serialize_hyperparameter('warmup_proportion'),
'min_lr':
self._serialize_hyperparameter('min_lr'),
})
return config
database_base_path = '/kaggle/input/tweet-dataset-split-roberta-base-96-text/'
k_fold = pd.read_csv(database_base_path + '5-fold.csv')
display(k_fold.head())
# Unzip files
!tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_1.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_2.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_3.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_4.tar.gz
# !tar -xvf /kaggle/input/tweet-dataset-split-roberta-base-96-text/fold_5.tar.gz
vocab_path = database_base_path + 'vocab.json'
merges_path = database_base_path + 'merges.txt'
base_path = '/kaggle/input/qa-transformers/roberta/'
config = {
"MAX_LEN": 96,
"BATCH_SIZE": 32,
"EPOCHS": 6,
"LEARNING_RATE": 3e-5,
"ES_PATIENCE": 1,
"N_FOLDS": 1,
"base_model_path": base_path + 'roberta-base-tf_model.h5',
"config_path": base_path + 'roberta-base-config.json'
}
with open('config.json', 'w') as json_file:
json.dump(json.loads(json.dumps(config)), json_file)
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
x_start = layers.Dropout(.1)(last_hidden_state)
x_start = layers.Conv1D(1, 1)(x_start)
x_start = layers.Flatten()(x_start)
x_end = layers.Dropout(.1)(last_hidden_state)
x_end = layers.Conv1D(1, 1)(x_end)
x_end = layers.Flatten()(x_end)
y_start = layers.Subtract()([x_start, x_end])
y_start = layers.Activation('softmax', name='y_start')(x_start)
y_end = layers.Subtract()([x_end, x_start])
y_end = layers.Activation('softmax', name='y_end')(x_end)
x_sentiment = layers.GlobalAveragePooling1D()(last_hidden_state)
x_sentiment = layers.Dropout(.1)(x_sentiment)
y_sentiment = layers.Dense(3, activation='softmax', name='y_sentiment')(x_sentiment)
model = Model(inputs=[input_ids, attention_mask], outputs=[y_start, y_end, y_sentiment])
optimizer = RectifiedAdam(lr=config['LEARNING_RATE'],
total_steps=(len(k_fold[k_fold['fold_1'] == 'train']) // config['BATCH_SIZE']) * config['EPOCHS'],
warmup_proportion=0.1,
min_lr=1e-7)
model.compile(optimizer, loss={'y_start': losses.CategoricalCrossentropy(),
'y_end': losses.CategoricalCrossentropy(),
'y_sentiment': losses.SparseCategoricalCrossentropy()},
loss_weights=[1, 1, .5],
metrics={'y_start': metrics.CategoricalAccuracy(),
'y_end': metrics.CategoricalAccuracy(),
'y_sentiment': metrics.SparseCategoricalCrossentropy()})
return model
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
tokenizer.save('./')
history_list = []
for n_fold in range(config['N_FOLDS']):
n_fold +=1
print('\nFOLD: %d' % (n_fold))
# Load data
base_data_path = 'fold_%d/' % (n_fold)
x_train = np.load(base_data_path + 'x_train.npy')
y_train = np.load(base_data_path + 'y_train.npy')
x_valid = np.load(base_data_path + 'x_valid.npy')
y_valid = np.load(base_data_path + 'y_valid.npy')
y_train_aux = np.load(base_data_path + 'y_train_aux.npy')[0].astype(int)
y_valid_aux = np.load(base_data_path + 'y_valid_aux.npy')[0].astype(int)
### Delete data dir
shutil.rmtree(base_data_path)
# Train model
model_path = 'model_fold_%d.h5' % (n_fold)
model = model_fn(config['MAX_LEN'])
es = EarlyStopping(monitor='val_loss', mode='min', patience=config['ES_PATIENCE'],
restore_best_weights=True, verbose=1)
checkpoint = ModelCheckpoint(model_path, monitor='val_loss', mode='min',
save_best_only=True, save_weights_only=True)
history = model.fit(list(x_train), list([y_train[0], y_train[1], y_train_aux]),
validation_data=(list(x_valid), list([y_valid[0], y_valid[1], y_valid_aux])),
batch_size=config['BATCH_SIZE'],
callbacks=[checkpoint, es],
epochs=config['EPOCHS'],
verbose=2).history
history_list.append(history)
# Make predictions
train_preds = model.predict(list(x_train))
valid_preds = model.predict(list(x_valid))
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'start_fold_%d' % (n_fold)] = train_preds[0].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'end_fold_%d' % (n_fold)] = train_preds[1].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'start_fold_%d' % (n_fold)] = valid_preds[0].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'end_fold_%d' % (n_fold)] = valid_preds[1].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'train', 'sentiment_fold_%d' % (n_fold)] = train_preds[2].argmax(axis=-1)
k_fold.loc[k_fold['fold_%d' % (n_fold)] == 'validation', 'sentiment_fold_%d' % (n_fold)] = valid_preds[2].argmax(axis=-1)
k_fold['end_fold_%d' % (n_fold)] = k_fold['end_fold_%d' % (n_fold)].astype(int)
k_fold['start_fold_%d' % (n_fold)] = k_fold['start_fold_%d' % (n_fold)].astype(int)
k_fold['end_fold_%d' % (n_fold)].clip(0, k_fold['text_len'], inplace=True)
k_fold['start_fold_%d' % (n_fold)].clip(0, k_fold['end_fold_%d' % (n_fold)], inplace=True)
k_fold['prediction_fold_%d' % (n_fold)] = k_fold.apply(lambda x: decode(x['start_fold_%d' % (n_fold)], x['end_fold_%d' % (n_fold)], x['text'], tokenizer), axis=1)
k_fold['prediction_fold_%d' % (n_fold)].fillna(k_fold["text"], inplace=True)
k_fold['jaccard_fold_%d' % (n_fold)] = k_fold.apply(lambda x: jaccard(x['selected_text'], x['prediction_fold_%d' % (n_fold)]), axis=1)
sns.set(style="whitegrid")
for n_fold in range(config['N_FOLDS']):
print('Fold: %d' % (n_fold+1))
plot_metrics(history_list[n_fold])
display(evaluate_model_kfold(k_fold, config['N_FOLDS']).style.applymap(color_map))
display(k_fold[[c for c in k_fold.columns if not (c.startswith('textID') or
c.startswith('text_len') or
c.startswith('selected_text_len') or
c.startswith('text_wordCnt') or
c.startswith('selected_text_wordCnt') or
c.startswith('fold_') or
c.startswith('start_fold_') or
c.startswith('end_fold_'))]].head(15))
| 0.922961 | 0.800653 |
# Forecasting II: state space models
This tutorial covers state space modeling with the [pyro.contrib.forecast](http://docs.pyro.ai/en/latest/contrib.forecast.html) module. This tutorial assumes the reader is already familiar with [SVI](http://pyro.ai/examples/svi_part_ii.html), [tensor shapes](http://pyro.ai/examples/tensor_shapes.html), and [univariate forecasting](http://pyro.ai/examples/forecasting_i.html).
See also:
- [Forecasting I: univariate, heavy tailed](http://pyro.ai/examples/forecasting_i.html)
- [Forecasting III: hierarchical models](http://pyro.ai/examples/forecasting_iii.html)
#### Summary
- Pyro's [ForecastingModel](http://docs.pyro.ai/en/latest/contrib.forecast.html#pyro.contrib.forecast.forecaster.ForecastingModel) can combine regression, variational inference, and exact inference.
- To model a linear-Gaussian dynamical system, use a [GaussianHMM](http://docs.pyro.ai/en/latest/distributions.html#gaussianhmm) `noise_dist`.
- To model a heavy-tailed linear dynamical system, use [LinearHMM](http://docs.pyro.ai/en/latest/distributions.html#linearhmm) with heavy-tailed distributions.
- To enable inference with [LinearHMM](http://docs.pyro.ai/en/latest/distributions.html#linearhmm), use a [LinearHMMReparam](http://docs.pyro.ai/en/latest/infer.reparam.html#pyro.infer.reparam.hmm.LinearHMMReparam) reparameterizer.
```
import math
import torch
import pyro
import pyro.distributions as dist
import pyro.poutine as poutine
from pyro.contrib.examples.bart import load_bart_od
from pyro.contrib.forecast import ForecastingModel, Forecaster, eval_crps
from pyro.infer.reparam import LinearHMMReparam, StableReparam, SymmetricStableReparam
from pyro.ops.tensor_utils import periodic_repeat
from pyro.ops.stats import quantile
import matplotlib.pyplot as plt
%matplotlib inline
assert pyro.__version__.startswith('1.3.1')
pyro.enable_validation(True)
pyro.set_rng_seed(20200305)
```
## Intro to state space models
In the [univariate tutorial](http://pyro.ai/examples/forecasting_i.html) we saw how to model time series as regression plus a local level model, using variational inference. This tutorial covers a different way to model time series: state space models and exact inference. Pyro's forecasting module allows these two paradigms to be combined, for example modeling seasonality with regression, including a slow global trend, and using a state-space model for short-term local trend.
Pyro implements a few state space models, but the most important are the [GaussianHMM](http://docs.pyro.ai/en/latest/distributions.html#gaussianhmm) distribution and its heavy-tailed generalization the [LinearHMM](http://docs.pyro.ai/en/latest/distributions.html#linearhmm) distribution. Both of these model a linear dynamical system with hidden state; both are multivariate, and both allow learning of all process parameters. On top of these the [pyro.contrib.timeseries](http://docs.pyro.ai/en/latest/contrib.timeseries.html) module implements a variety of multivariate Gaussian Process models that compile down to `GaussianHMM`s.
Pyro's inference for `GaussianHMM` uses parallel-scan Kalman filtering, allowing fast analysis of very long time series. Similarly, Pyro's inference for `LinearHMM` uses entirely parallel auxiliary variable methods to reduce to a `GaussianHMM`, which then permits parallel-scan inference. Thus both methods allow parallelization of long time series analysis, even for a single univariate time series.
Let's again look at the [BART train](https://www.bart.gov/about/reports/ridership) ridership dataset:
```
dataset = load_bart_od()
print(dataset.keys())
print(dataset["counts"].shape)
print(" ".join(dataset["stations"]))
data = dataset["counts"].sum([-1, -2]).unsqueeze(-1).log1p()
print(data.shape)
plt.figure(figsize=(9, 3))
plt.plot(data, 'b.', alpha=0.1, markeredgewidth=0)
plt.title("Total hourly ridership over nine years")
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(0, len(data));
plt.figure(figsize=(9, 3))
plt.plot(data)
plt.title("Total hourly ridership over one month")
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(len(data) - 24 * 30, len(data));
```
## GaussianHMM
Let's start by modeling hourly seasonality together with a local linear trend, where we model seasonality via regression and local linear trend via a [GaussianHMM](http://docs.pyro.ai/en/latest/distributions.html#gaussianhmm). This noise model includes a mean-reverting hidden state (an [Ornstein-Uhlenbeck process](https://en.wikipedia.org/wiki/Ornstein%E2%80%93Uhlenbeck_process)) plus Gaussian observation noise.
```
T0 = 0 # beginning
T2 = data.size(-2) # end
T1 = T2 - 24 * 7 * 2 # train/test split
means = data[:T1 // (24 * 7) * 24 * 7].reshape(-1, 24 * 7).mean(0)
class Model1(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
# We'll hard-code the periodic part of this model, learning only the local model.
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
# On top of this mean prediction, we'll learn a linear dynamical system.
# This requires specifying five pieces of data, on which we will put structured priors.
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
# Note timescale is a scalar but we need a 1x1 transition matrix (hidden_dim=1),
# thus we unsqueeze twice using [..., None, None].
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
trans_dist = dist.Normal(0, trans_scale.unsqueeze(-1)).to_event(1)
# Note the obs_matrix has shape hidden_dim x obs_dim = 1 x 1.
obs_matrix = torch.tensor([[1.]])
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_dist = dist.Normal(0, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.GaussianHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
self.predict(noise_dist, prediction)
```
We can then train the model on many years of data. Note that because we are being variational about only time-global variables, and exactly integrating out time-local variables (via `GaussianHMM`), stochastic gradients are very low variance; this allows us to use a large learning rate and few steps.
```
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model1(), data[:T1], covariates[:T1], learning_rate=0.1, num_steps=400)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
```
Plotting forecasts of the next two weeks of data, we see mostly reasonable forecasts, but an anomaly on Christmas when rides were overpredicted. This is to be expected, as we have not modeled yearly seasonality or holidays.
```
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
print(samples.shape, p10.shape)
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
```
Next let's change the model to use heteroskedastic observation noise, depending on the hour of week.
```
class Model2(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
trans_dist = dist.Normal(0, trans_scale.unsqueeze(-1)).to_event(1)
obs_matrix = torch.tensor([[1.]])
# To model heteroskedastic observation noise, we'll sample obs_scale inside a plate,
# then repeat to full duration. This is the only change from Model1.
with pyro.plate("hour_of_week", 24 * 7, dim=-1):
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_scale = periodic_repeat(obs_scale, duration, dim=-1)
obs_dist = dist.Normal(0, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.GaussianHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
self.predict(noise_dist, prediction)
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model2(), data[:T1], covariates[:T1], learning_rate=0.1, num_steps=400)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
```
Note this gives us a much longer timescale and thereby more accurate short-term predictions:
```
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 2, T1 + 24 * 4)
plt.legend(loc="best");
```
## Heavy-tailed modeling with LinearHMM
Next let's change our model to a linear-[Stable](http://docs.pyro.ai/en/latest/distributions.html#pyro.distributions.Stable) dynamical system, exhibiting learnable heavy tailed behavior in both the process noise and observation noise. As we've already seen in the [univariate tutorial](http://pyro.ai/examples/forecasting_i.html), this will require special handling of stable distributions by [poutine.reparam()](http://docs.pyro.ai/en/latest/poutine.html#pyro.poutine.handlers.reparam). For state space models, we combine [LinearHMMReparam](http://docs.pyro.ai/en/latest/infer.reparam.html#pyro.infer.reparam.hmm.LinearHMMReparam) with other reparameterizers like [StableReparam](http://docs.pyro.ai/en/latest/infer.reparam.html#pyro.infer.reparam.stable.StableReparam) and [SymmetricStableReparam](http://docs.pyro.ai/en/latest/infer.reparam.html#pyro.infer.reparam.stable.SymmetricStableReparam). All reparameterizers preserve behavior of the generative model, and only serve to enable inference via auxiliary variable methods.
```
class Model3(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
# First sample the Gaussian-like parameters as in previous models.
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
obs_matrix = torch.tensor([[1.]])
with pyro.plate("hour_of_week", 24 * 7, dim=-1):
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_scale = periodic_repeat(obs_scale, duration, dim=-1)
# In addition to the Gaussian parameters, we will learn a global stability
# parameter to determine tail weights, and an observation skew parameter.
stability = pyro.sample("stability", dist.Uniform(1, 2).expand([1]).to_event(1))
skew = pyro.sample("skew", dist.Uniform(-1, 1).expand([1]).to_event(1))
# Next we construct stable distributions and a linear-stable HMM distribution.
trans_dist = dist.Stable(stability, 0, trans_scale.unsqueeze(-1)).to_event(1)
obs_dist = dist.Stable(stability, skew, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.LinearHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
# Finally we use a reparameterizer to enable inference.
rep = LinearHMMReparam(None, # init_dist is already Gaussian.
SymmetricStableReparam(), # trans_dist is symmetric.
StableReparam()) # obs_dist is asymmetric.
with poutine.reparam(config={"residual": rep}):
self.predict(noise_dist, prediction)
```
Note that since this model introduces auxiliary variables that are learned by variational inference, gradients are higher variance and we need to train for longer.
```
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model3(), data[:T1], covariates[:T1], learning_rate=0.1)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 2, T1 + 24 * 4)
plt.legend(loc="best");
```
|
github_jupyter
|
import math
import torch
import pyro
import pyro.distributions as dist
import pyro.poutine as poutine
from pyro.contrib.examples.bart import load_bart_od
from pyro.contrib.forecast import ForecastingModel, Forecaster, eval_crps
from pyro.infer.reparam import LinearHMMReparam, StableReparam, SymmetricStableReparam
from pyro.ops.tensor_utils import periodic_repeat
from pyro.ops.stats import quantile
import matplotlib.pyplot as plt
%matplotlib inline
assert pyro.__version__.startswith('1.3.1')
pyro.enable_validation(True)
pyro.set_rng_seed(20200305)
dataset = load_bart_od()
print(dataset.keys())
print(dataset["counts"].shape)
print(" ".join(dataset["stations"]))
data = dataset["counts"].sum([-1, -2]).unsqueeze(-1).log1p()
print(data.shape)
plt.figure(figsize=(9, 3))
plt.plot(data, 'b.', alpha=0.1, markeredgewidth=0)
plt.title("Total hourly ridership over nine years")
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(0, len(data));
plt.figure(figsize=(9, 3))
plt.plot(data)
plt.title("Total hourly ridership over one month")
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(len(data) - 24 * 30, len(data));
T0 = 0 # beginning
T2 = data.size(-2) # end
T1 = T2 - 24 * 7 * 2 # train/test split
means = data[:T1 // (24 * 7) * 24 * 7].reshape(-1, 24 * 7).mean(0)
class Model1(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
# We'll hard-code the periodic part of this model, learning only the local model.
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
# On top of this mean prediction, we'll learn a linear dynamical system.
# This requires specifying five pieces of data, on which we will put structured priors.
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
# Note timescale is a scalar but we need a 1x1 transition matrix (hidden_dim=1),
# thus we unsqueeze twice using [..., None, None].
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
trans_dist = dist.Normal(0, trans_scale.unsqueeze(-1)).to_event(1)
# Note the obs_matrix has shape hidden_dim x obs_dim = 1 x 1.
obs_matrix = torch.tensor([[1.]])
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_dist = dist.Normal(0, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.GaussianHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
self.predict(noise_dist, prediction)
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model1(), data[:T1], covariates[:T1], learning_rate=0.1, num_steps=400)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
print(samples.shape, p10.shape)
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
class Model2(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
trans_dist = dist.Normal(0, trans_scale.unsqueeze(-1)).to_event(1)
obs_matrix = torch.tensor([[1.]])
# To model heteroskedastic observation noise, we'll sample obs_scale inside a plate,
# then repeat to full duration. This is the only change from Model1.
with pyro.plate("hour_of_week", 24 * 7, dim=-1):
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_scale = periodic_repeat(obs_scale, duration, dim=-1)
obs_dist = dist.Normal(0, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.GaussianHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
self.predict(noise_dist, prediction)
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model2(), data[:T1], covariates[:T1], learning_rate=0.1, num_steps=400)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 2, T1 + 24 * 4)
plt.legend(loc="best");
class Model3(ForecastingModel):
def model(self, zero_data, covariates):
duration = zero_data.size(-2)
prediction = periodic_repeat(means, duration, dim=-1).unsqueeze(-1)
# First sample the Gaussian-like parameters as in previous models.
init_dist = dist.Normal(0, 10).expand([1]).to_event(1)
timescale = pyro.sample("timescale", dist.LogNormal(math.log(24), 1))
trans_matrix = torch.exp(-1 / timescale)[..., None, None]
trans_scale = pyro.sample("trans_scale", dist.LogNormal(-0.5 * math.log(24), 1))
obs_matrix = torch.tensor([[1.]])
with pyro.plate("hour_of_week", 24 * 7, dim=-1):
obs_scale = pyro.sample("obs_scale", dist.LogNormal(-2, 1))
obs_scale = periodic_repeat(obs_scale, duration, dim=-1)
# In addition to the Gaussian parameters, we will learn a global stability
# parameter to determine tail weights, and an observation skew parameter.
stability = pyro.sample("stability", dist.Uniform(1, 2).expand([1]).to_event(1))
skew = pyro.sample("skew", dist.Uniform(-1, 1).expand([1]).to_event(1))
# Next we construct stable distributions and a linear-stable HMM distribution.
trans_dist = dist.Stable(stability, 0, trans_scale.unsqueeze(-1)).to_event(1)
obs_dist = dist.Stable(stability, skew, obs_scale.unsqueeze(-1)).to_event(1)
noise_dist = dist.LinearHMM(
init_dist, trans_matrix, trans_dist, obs_matrix, obs_dist, duration=duration)
# Finally we use a reparameterizer to enable inference.
rep = LinearHMMReparam(None, # init_dist is already Gaussian.
SymmetricStableReparam(), # trans_dist is symmetric.
StableReparam()) # obs_dist is asymmetric.
with poutine.reparam(config={"residual": rep}):
self.predict(noise_dist, prediction)
%%time
pyro.set_rng_seed(1)
pyro.clear_param_store()
covariates = torch.zeros(len(data), 0) # empty
forecaster = Forecaster(Model3(), data[:T1], covariates[:T1], learning_rate=0.1)
for name, value in forecaster.guide.median().items():
if value.numel() == 1:
print("{} = {:0.4g}".format(name, value.item()))
samples = forecaster(data[:T1], covariates, num_samples=100)
samples.clamp_(min=0) # apply domain knowledge: the samples must be positive
p10, p50, p90 = quantile(samples, (0.1, 0.5, 0.9)).squeeze(-1)
crps = eval_crps(samples, data[T1:])
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 7, T2)
plt.text(78732, 3.5, "Christmas", rotation=90, color="green")
plt.legend(loc="best");
plt.figure(figsize=(9, 3))
plt.fill_between(torch.arange(T1, T2), p10, p90, color="red", alpha=0.3)
plt.plot(torch.arange(T1, T2), p50, 'r-', label='forecast')
plt.plot(torch.arange(T1 - 24 * 7, T2),
data[T1 - 24 * 7: T2], 'k-', label='truth')
plt.title("Total hourly ridership (CRPS = {:0.3g})".format(crps))
plt.ylabel("log(# rides)")
plt.xlabel("Hour after 2011-01-01")
plt.xlim(T1 - 24 * 2, T1 + 24 * 4)
plt.legend(loc="best");
| 0.821259 | 0.994336 |
# Chapter 1 - The Python Data Model
> "Notes from Luciano Ramalho's book Fluent Python"
- badges: true
- categories: [Fluent Python Notes]
- Data model as a description of Python as a framework. It formalizes the interfaces of the building blocks of the language itself, such as sequences, iterators, functions, classes, context managers and so on.
- The special method names allows objects to implement, support and interact with basic language constructs such as: iteration, collections, attribute access, operator overloading, function and method invocation, string representation and formating, manage contexts.
**Two advantages of using special methods**
- the users of user defined class don't have to memorize method names for standard operations("How to get the number of items?, etc")
- It's easier to benefit from the rich Python standard library and avoid reinventing the wheel like the random.choice function.
**Note**
- Just by implementing the \_\_getitem\_\_ special method, the class also becomes iterable.(why?)
**Why len is not a method?**
- len is not called as a method because it gets special treatment as part of the Python Data Model, just like abs. But thanks to the special method \_\_len\_\_ you can also make len work with your own custom objects
- Think of len and abs as unary operators.
### namedtuple — Tuple Subclass with Named Fields
Each kind of namedtuple is represented by its own class, which is created by using the namedtuple() factory function.
Named tuples assign meaning to each position in a tuple and allow for more readable, self-documenting code.
```
import collections
Person = collections.namedtuple('Person','name age')
```
- Returns a new tuple subclass named Person
- Any valid Python identifier may be used for a fieldname except for names starting with an underscore. Valid identifiers consist of letters, digits, and underscores but do not start with a digit or underscore and cannot be a keyword such as class, for, return, global, pass, or raise
```
Person.__doc__
bob = Person(name = 'bob', age = 30)
print("Representation:", bob)
```
it is possible to access the fields of the namedtuple by name using dotted notation (obj.attr) as well as by using the positional indexes of standard tuples.
```
# accessing the values using obj.attr
print("name: ",bob.name)
print("age: ",bob.age)
# accessing the values using index
print("name: ",bob[0])
print("age: ",bob[1])
```
Just like a regular tuple, a namedtuple is immutable.
```
bob.name = "Borat"
```
**Some special methods defined in namedtuple**
```
bob._fields
bob._asdict()
rob = bob._replace(name='Robert')
rob is bob # bob and rob are two objects
```
Class method that makes a new instance from an existing sequence or iterable. _make method can be thought of as an alternative constructor
```
Person._make(['ME',200])
```
**Various methods and behaviour of namedtuples**
```
for i in dir(Person):
print(i,end=" ")
```
### Return a random element from a list
```
import random
mylist = ["apple","mango","banana"]
random.choice(mylist)
x = "OPEN_SESAME"
random.choice(x)
```
|
github_jupyter
|
import collections
Person = collections.namedtuple('Person','name age')
Person.__doc__
bob = Person(name = 'bob', age = 30)
print("Representation:", bob)
# accessing the values using obj.attr
print("name: ",bob.name)
print("age: ",bob.age)
# accessing the values using index
print("name: ",bob[0])
print("age: ",bob[1])
bob.name = "Borat"
bob._fields
bob._asdict()
rob = bob._replace(name='Robert')
rob is bob # bob and rob are two objects
Person._make(['ME',200])
for i in dir(Person):
print(i,end=" ")
import random
mylist = ["apple","mango","banana"]
random.choice(mylist)
x = "OPEN_SESAME"
random.choice(x)
| 0.14817 | 0.987946 |
```
import pandas as pd
items_fh = "./files/items.csv"
```
## Base dataframe
```
df = pd.read_csv(items_fh)
df.head()
```
### $0 values
```
df["0"].value_counts()
```
### $1 values
```
df["1"].value_counts()
```
### $2 values
```
df["2"].value_counts()
```
### $4 values
```
df["4"].value_counts()
df["4"].value_counts().to_csv("./files/sub_$4.csv", header=True)
```
### $5 values
```
df["5"].value_counts()
df["5"].value_counts().to_csv("./files/sub_$5.csv", header=True)
```
### $7 values
```
df["7"].value_counts().to_csv("./files/sub_$7.csv", header=True)
```
### $A values
```
df["A"].value_counts()
```
### $a values
```
df["a"].value_counts()
df["a"].value_counts().to_csv("./files/sub_$a.csv", header=True)
```
### $B values
```
df["B"].value_counts()
df["B"].value_counts().to_csv("./files/sub_$B.csv", header=True)
```
### $b values
```
df["b"].value_counts()
df["b"].value_counts().to_csv("./files/sub_$b_.csv", header=True)
```
### $F values
```
df["F"].value_counts()
df["F"].value_counts().to_csv("./files/sub_$F.csv", header=True)
```
### $f values
```
df["f"].value_counts()
df["f"].value_counts().to_csv("./files/sub_$f_.csv", header=True)
```
### $H value
```
df["H"].value_counts()
```
### $h values
```
df["h"].value_counts()
df["h"].value_counts().to_csv("./files/sub_$h.csv", header=True)
```
### $I values
```
df["I"].value_counts()
df["I"].value_counts().to_csv("./files/sub_$I.csv", header=True)
```
### $i values
```
df["i"].value_counts()
df["i"].value_counts().to_csv("./files/sub_$i_.csv", header=True)
```
### $J values
```
df["J"].value_counts()
df["J"].value_counts().to_csv("./files/sub_$J.csv", header=True)
```
### $j values
```
df["j"].value_counts()
df["j"].value_counts().to_csv("./files/sub_j_.csv", header=True)
```
### $k values
```
df["k"].value_counts()
df["k"].value_counts().to_csv("./files/sub_$k.csv", header=True)
```
### $m1 values
```
df["m1"].value_counts()
df["m1"].value_counts().to_csv("./files/sub_$m1.csv", header=True)
```
### $m2 values
```
df["m2"].value_counts()
df["m2"].value_counts().to_csv("./files/sub_$m2.csv", header=True)
```
### $P values
```
df["P"].value_counts()
df["P"].value_counts().to_csv("./files/sub_$P.csv", header=True)
```
### $p values
```
df["p"].value_counts()
df["p"].value_counts().to_csv("./files/sub_$p_.csv", header=True)
```
### $Q values
```
df["Q"].value_counts()
df["Q"].value_counts().to_csv("./files/sub_$Q.csv", header=True)
```
### $R values
```
df["R"].value_counts()
df["R"].value_counts().to_csv("./files/sub_$R.csv", header=True)
```
### $S values
```
df["S"].value_counts()
df["S"].value_counts().to_csv("./files/sub_$S.csv", header=True)
```
### $w values
```
df["w"].value_counts()
df["w"].value_counts().to_csv("./files/sub_$w.csv", header=True)
```
|
github_jupyter
|
import pandas as pd
items_fh = "./files/items.csv"
df = pd.read_csv(items_fh)
df.head()
df["0"].value_counts()
df["1"].value_counts()
df["2"].value_counts()
df["4"].value_counts()
df["4"].value_counts().to_csv("./files/sub_$4.csv", header=True)
df["5"].value_counts()
df["5"].value_counts().to_csv("./files/sub_$5.csv", header=True)
df["7"].value_counts().to_csv("./files/sub_$7.csv", header=True)
df["A"].value_counts()
df["a"].value_counts()
df["a"].value_counts().to_csv("./files/sub_$a.csv", header=True)
df["B"].value_counts()
df["B"].value_counts().to_csv("./files/sub_$B.csv", header=True)
df["b"].value_counts()
df["b"].value_counts().to_csv("./files/sub_$b_.csv", header=True)
df["F"].value_counts()
df["F"].value_counts().to_csv("./files/sub_$F.csv", header=True)
df["f"].value_counts()
df["f"].value_counts().to_csv("./files/sub_$f_.csv", header=True)
df["H"].value_counts()
df["h"].value_counts()
df["h"].value_counts().to_csv("./files/sub_$h.csv", header=True)
df["I"].value_counts()
df["I"].value_counts().to_csv("./files/sub_$I.csv", header=True)
df["i"].value_counts()
df["i"].value_counts().to_csv("./files/sub_$i_.csv", header=True)
df["J"].value_counts()
df["J"].value_counts().to_csv("./files/sub_$J.csv", header=True)
df["j"].value_counts()
df["j"].value_counts().to_csv("./files/sub_j_.csv", header=True)
df["k"].value_counts()
df["k"].value_counts().to_csv("./files/sub_$k.csv", header=True)
df["m1"].value_counts()
df["m1"].value_counts().to_csv("./files/sub_$m1.csv", header=True)
df["m2"].value_counts()
df["m2"].value_counts().to_csv("./files/sub_$m2.csv", header=True)
df["P"].value_counts()
df["P"].value_counts().to_csv("./files/sub_$P.csv", header=True)
df["p"].value_counts()
df["p"].value_counts().to_csv("./files/sub_$p_.csv", header=True)
df["Q"].value_counts()
df["Q"].value_counts().to_csv("./files/sub_$Q.csv", header=True)
df["R"].value_counts()
df["R"].value_counts().to_csv("./files/sub_$R.csv", header=True)
df["S"].value_counts()
df["S"].value_counts().to_csv("./files/sub_$S.csv", header=True)
df["w"].value_counts()
df["w"].value_counts().to_csv("./files/sub_$w.csv", header=True)
| 0.211743 | 0.792785 |
```
%matplotlib inline
from __future__ import print_function, division
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
import numpy as np
import torchvision
from torchvision import datasets, models, transforms
import matplotlib.pyplot as plt
import time
import os
import copy
import PIL
from torch.utils.data.sampler import SubsetRandomSampler
plt.ion() # interactive mode
# path to folder with data
path_to_data = '/content/drive/My Drive/Colab Notebooks/data'
# means and st.dev of data
means, stds =[0.43,0.44,0.47], [0.20,0.20,0.20]
data_train = datasets.SVHN(path_to_data,
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=means,
std=stds)
])
)
data_test = datasets.SVHN(path_to_data,
split='test',
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=means,
std=stds
)
]))
batch_size = 64
data_size = data_train.data.shape[0]
validation_split = .2
split = int(np.floor(validation_split * data_size))
indices = list(range(data_size))
np.random.shuffle(indices)
train_indices, val_indices = indices[split:], indices[:split]
train_sampler = SubsetRandomSampler(train_indices)
val_sampler = SubsetRandomSampler(val_indices)
train_loader = torch.utils.data.DataLoader(data_train, batch_size=batch_size,
sampler=train_sampler)
val_loader = torch.utils.data.DataLoader(data_train, batch_size=batch_size,
sampler=val_sampler)
dataset_sizes = {'train': data_size-split, 'val': split}
lables_names = list(range(10))
```
Visualize a few images
^^^^^^^^^^^^^^^^^^^^^^
Let's visualize a few training images so as to understand the data augmentations.
```
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array(means)
std = np.array(stds)
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, classes = next(iter(train_loader))
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[lables_names[x] for x in classes])
```
Training the model
------------------
Now, let's write a general function to train a model. Here, we will
illustrate:
- Scheduling the learning rate
- Saving the best model
In the following, parameter ``scheduler`` is an LR scheduler object from
``torch.optim.lr_scheduler``.
```
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
data = train_loader
scheduler.step()
model.train() # Set model to training mode
else:
data = val_loader
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
# Iterate over data.
for inputs, labels in data:
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
```
Visualizing the model predictions
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
Generic function to display predictions for a few images
```
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(val_loader):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(lables_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
```
Finetuning the convnet
----------------------
Load a pretrained model and reset final fully connected layer.
```
device = torch.device("cuda:0")
model_ft = models.resnet18(pretrained=True)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Linear(num_ftrs, 10)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
# Observe that all parameters are being optimized
optimizer_ft = optim.SGD(model_ft.parameters(), lr=0.001, momentum=0.9)
# Decay LR by a factor of 0.1 every 7 epochs
exp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)
```
Train and evaluate
^^^^^^^^^^^^^^^^^^
```
model_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler,
num_epochs=25)
visualize_model(model_ft)
```
|
github_jupyter
|
%matplotlib inline
from __future__ import print_function, division
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
import numpy as np
import torchvision
from torchvision import datasets, models, transforms
import matplotlib.pyplot as plt
import time
import os
import copy
import PIL
from torch.utils.data.sampler import SubsetRandomSampler
plt.ion() # interactive mode
# path to folder with data
path_to_data = '/content/drive/My Drive/Colab Notebooks/data'
# means and st.dev of data
means, stds =[0.43,0.44,0.47], [0.20,0.20,0.20]
data_train = datasets.SVHN(path_to_data,
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=means,
std=stds)
])
)
data_test = datasets.SVHN(path_to_data,
split='test',
transform=transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=means,
std=stds
)
]))
batch_size = 64
data_size = data_train.data.shape[0]
validation_split = .2
split = int(np.floor(validation_split * data_size))
indices = list(range(data_size))
np.random.shuffle(indices)
train_indices, val_indices = indices[split:], indices[:split]
train_sampler = SubsetRandomSampler(train_indices)
val_sampler = SubsetRandomSampler(val_indices)
train_loader = torch.utils.data.DataLoader(data_train, batch_size=batch_size,
sampler=train_sampler)
val_loader = torch.utils.data.DataLoader(data_train, batch_size=batch_size,
sampler=val_sampler)
dataset_sizes = {'train': data_size-split, 'val': split}
lables_names = list(range(10))
def imshow(inp, title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array(means)
std = np.array(stds)
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.imshow(inp)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, classes = next(iter(train_loader))
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[lables_names[x] for x in classes])
def train_model(model, criterion, optimizer, scheduler, num_epochs=25):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(num_epochs):
print('Epoch {}/{}'.format(epoch, num_epochs - 1))
print('-' * 10)
# Each epoch has a training and validation phase
for phase in ['train', 'val']:
if phase == 'train':
data = train_loader
scheduler.step()
model.train() # Set model to training mode
else:
data = val_loader
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
# Iterate over data.
for inputs, labels in data:
inputs = inputs.to(device)
labels = labels.to(device)
# zero the parameter gradients
optimizer.zero_grad()
# forward
# track history if only in train
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == 'train':
loss.backward()
optimizer.step()
# statistics
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
# deep copy the model
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best val Acc: {:4f}'.format(best_acc))
# load best model weights
model.load_state_dict(best_model_wts)
return model
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_so_far = 0
fig = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(val_loader):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_so_far += 1
ax = plt.subplot(num_images//2, 2, images_so_far)
ax.axis('off')
ax.set_title('predicted: {}'.format(lables_names[preds[j]]))
imshow(inputs.cpu().data[j])
if images_so_far == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
device = torch.device("cuda:0")
model_ft = models.resnet18(pretrained=True)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Linear(num_ftrs, 10)
model_ft = model_ft.to(device)
criterion = nn.CrossEntropyLoss()
# Observe that all parameters are being optimized
optimizer_ft = optim.SGD(model_ft.parameters(), lr=0.001, momentum=0.9)
# Decay LR by a factor of 0.1 every 7 epochs
exp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)
model_ft = train_model(model_ft, criterion, optimizer_ft, exp_lr_scheduler,
num_epochs=25)
visualize_model(model_ft)
| 0.83772 | 0.807878 |
<div class="alert alert-block alert-info">
<b><h1>ENGR 1330 Computational Thinking with Data Science </h1></b>
</div>
Copyright © 2021 Theodore G. Cleveland and Farhang Forghanparast
Last GitHub Commit Date:
# 22: Testing Hypothesis - Comparing Collections
- Comparing two (or more) collections
- Parametric and Non-Parametric Tests
- Type 1 & Type 2 errors
## Background
In engineering, when we wish to start asking questions about the data and interpret the results, we use statistical methods that provide a confidence or likelihood about the answers. In general, this class of methods is called statistical hypothesis testing, or significance tests. The material for today's lecture is inspired by and gathered from several resources including:
- *Hypothesis testing in Machine learning using Python* by *Yogesh Agrawal* available at https://towardsdatascience.com/hypothesis-testing-in-machine-learning-using-python-a0dc89e169ce
- *Demystifying hypothesis testing with simple Python examples* by *Tirthajyoti Sarkar* available at https://towardsdatascience.com/demystifying-hypothesis-testing-with-simple-python-examples-4997ad3c5294
- *A Gentle Introduction to Statistical Hypothesis Testing* by *Jason Brownlee* available at https://machinelearningmastery.com/statistical-hypothesis-tests/
Let's go over a few important concepts first.
### <font color=crimson>What is hypothesis testing ?</font><br>
Hypothesis testing is a statistical method that is used in making statistical decisions (about population) using experimental data (samples). Hypothesis Testing is basically an assumption that we make about the population parameter.<br>
Ex : you say on average, students in the class are taller than 5 ft and 4 inches or an average boy is taller than girls or a specific treatment is effective in treating COVID-19 patients. <br>
We need some mathematical conclusion that whatever we are assuming is true. We will validate our hypotheses, basing our conclusion on random samples and empirical distributions._
### <font color=crimson>Why do we use it ?</font><br>
Hypothesis testing is an essential procedure in statistics. A hypothesis test evaluates two mutually exclusive statements about a population to determine which statement is best supported by the sample data. When we say that a finding is statistically significant, it’s thanks to a hypothesis test._
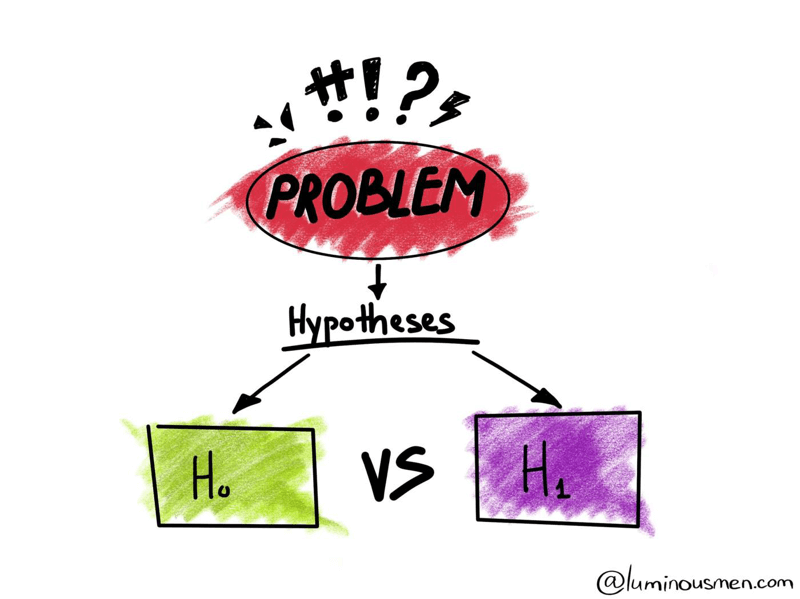
### <font color=crimson>Which are important elements of hypothesis testing ?</font><br>
**Null hypothesis:**<br>
The assertion of a statistical test is called the null hypothesis, or hypothesis 0 (H0 for short). It is often called the default assumption, or the assumption that nothing has changed. In inferential statistics, the null hypothesis is a general statement or default position that there is no relationship between two measured phenomena, or no association among groups. In other words it is a basic assertion made based on domain or problem knowledge.
Example : a company's gadget production is = 50 unit/per day.
**Alternative hypothesis:**<br>
A violation of the test’s assertion is often called the first hypothesis, hypothesis 1 or H1 for short. H1 is really a short hand for “some other hypothesis,” as all we know is that the evidence suggests that the H0 can be rejected. The alternative hypothesis is the hypothesis used in hypothesis testing that is contrary to the null hypothesis. It is usually taken to be that the observations are the result of a real effect (with some amount of chance variation superposed).
Example : a company's production is !=50 unit/per day.
### <font color=crimson>What are basic of hypothesis ?</font><br>
A fundamental stipulation is normalisation and standard normalisation. all our assertions and alternatives revolve around these 2 terms.<br>
 <br>
 <br>
in the 1st image, you can see there are different normal curves. Those normal curves have different means and variances. In the 2nd image if you notice the graph is properly distributed with a mean =0 and variance =1. Concept of z-score comes in picture when we use standardized normal data.
#### Normal Distribution:
A variable is said to be normally distributed or have a normal distribution if its distribution has the shape of a normal curve — a special bell-shaped curve. The graph of a normal distribution is called the normal curve, for which the mean, median, and mode are equal. (The 1st Image)
#### Standardised Normal Distribution:
A standard normal distribution is a normal distribution with mean 0 and standard deviation 1 (The 2nd Image)
#### <font color=crimson>Z score:</font><br>
It is a method of expressing data in relation to the group mean. To obtain the Z-score of a particular data, we calculate its deviation from the mean and then divide it by the SD.<br>
 <br>
The Z score is one way of standardizing a score so that it can be referred to a standard normal distribution curve.<br>
 <br>
_Read more on Z-Score @_
- __*Z-Score: Definition, Formula and Calculation* available at https://www.statisticshowto.com/probability-and-statistics/z-score/__
- __*Z-Score: Definition, Calculation and Interpretation* by *Saul McLeod* available at https://www.simplypsychology.org/z-score.html__
#### <font color=crimson>Tailing of Hypothesis:</font><br>
Depending on the research question hypothesis can be of 2 types. In the Nondirectional (two-tailed) test the Research Question is like: Is there a (statistically) significant difference between scores of Group-A and Group-B in a certain competition? In Directional (one-tailed) test the Research Question is like: Do Group-A score significantly higher than Group-B in a certain competition?<br>
 <br>
_Read more on Tailing @_
- *One- and two-tailed tests* available at https://en.wikipedia.org/wiki/One-_and_two-tailed_tests
- *Z-Score: Definition, Calculation and Interpretation* by *Saul McLeod* available at https://www.simplypsychology.org/z-score.html
#### <font color=crimson>Level of significance:</font><br>
<br>
Refers to the degree of significance in which we accept or reject the null-hypothesis. 100% accuracy is not possible for accepting or rejecting a hypothesis, so we therefore select a level of significance.
This significance level is usually denoted with alpha $\alpha$ and often it is set to 0.05 or 5% , which means your output should be 95% confident to give similar kind of result in each sample. A smaller alpha value suggests a more robust interpretation of the null hypothesis, such as 1% or 0.1%.
#### <font color=crimson>P-value :</font><br>
The P value, or attained (calculated) probability, is the probability (p-value) of the collected data, given that the null hypothesis was true. The p-value reflects the strength of evidence against the null hypothesis. Accordingly, we’ll encounter two situations: the strength is strong enough or not strong enough to reject the null hypothesis.
The p-value is compared to the pre-chosen alpha value. A result is statistically significant when the p-value is less than alpha. If your P value is less than the chosen significance level then you reject the null hypothesis i.e. accept that your sample gives reasonable evidence to support the alternative hypothesis.
```{note}
If p-value > alpha: Do not reject the null hypothesis (i.e. not significant result).<br>
If p-value <= alpha: Reject the null hypothesis (i.e. significant result).<br>
```
<br>
For example, if we were performing a test of whether a data sample was normally distributed and we calculated a p-value of .07, we could state something like:
"The test found that the data sample was normal, failing to reject the null hypothesis at a 5% significance level.""
The significance level can be inverted by subtracting it from 1 to give a **confidence level** of the hypothesis given the observed sample data.
Therefore, statements such as the following can also be made:
"The test found that the data was normal, failing to reject the null hypothesis at a 95% confidence level.""
### Example :<br>
You have a coin and you don’t know whether that is fair or tricky so let’s decide null and alternate hypothes is<br>
1. H0 : a coin is a fair coin.<br>
2. H1 : a coin is a tricky coin. <br>
3. alpha = 5% or 0.05<br>
Now let’s toss the coin and calculate p-value (probability value).<br>
Toss a coin 1st time (sample size =1) and result is tail
- P-value = 50% (as head and tail have equal probability)<br>
Toss a coin 2nd time (sample size =2) and result is tail, now
- P-value = 50/2 = 25%<br>
and similarly suppose we Toss 6 consecutive times (sample size =6) and got result as P-value = 1.5%
```
print("probability of 6 tails in 6 tosses if coin is fair",round((0.5)**6,3))
```
but we set our significance level as 5%. Here we see we are beyond that level i.e. our null- hypothesis does not hold good so we need to reject and propose that this coin is not fair. It does not necessarily mean that the coin is tricky, but 6 tails in a row is quite unlikely with a fair coin, and a good "bet" would be to reject the coin as unfair, and 95% of the time you would be correct.
Alternatively, one could phrase the result as a fair coin would produce a result **other** than 6 tails in a row 98.5% of the time.
Read more on p-value @<br>
- *P-values Explained By Data Scientist For Data Scientists* by *Admond Lee* available at https://towardsdatascience.com/p-values-explained-by-data-scientist-f40a746cfc8<br>
- *What a p-Value Tells You about Statistical Data* by *Deborah J. Rumsey* available at https://www.dummies.com/education/math/statistics/what-a-p-value-tells-you-about-statistical-data/<br>
- *Key to statistical result interpretation: P-value in plain English* by *Tran Quang Hung* available at https://s4be.cochrane.org/blog/2016/03/21/p-value-in-plain-english-2/<br>
Watch more on p-value @<br>
- *StatQuest: P Values, clearly explained* available at https://www.youtube.com/watch?v=5Z9OIYA8He8<br>
- *Understanding the p-value - Statistics Help* available at https://www.youtube.com/watch?v=eyknGvncKLw<br>
- *What Is A P-Value? - Clearly Explained* available at https://www.youtube.com/watch?v=ukcFrzt6cHk<br>
#### <font color=crimson>“Reject” vs “Failure to Reject”</font><br>
The p-value is probabilistic. This means that when we interpret the result of a statistical test, we do not know what is true or false, only what is likely. Rejecting the null hypothesis means that there is sufficient statistical evidence (from the samples) that the null hypothesis does not look likely (for the population). Otherwise, it means that there is not sufficient statistical evidence to reject the null hypothesis.<br>
We may think about the statistical test in terms of the dichotomy of rejecting and accepting the null hypothesis. The danger is that if we say that we “accept” the null hypothesis, the "language" implies that the null hypothesis is true. Instead, it is more preferred to say that we “fail to reject” the null hypothesis, as in, there is insufficient statistical evidence to reject it.<br>
#### <font color=crimson>Errors in Statistical Tests</font><br>
The interpretation of a statistical hypothesis test is probabilistic. That means that the evidence of the test may suggest an outcome and be mistaken. For example, if alpha was 5%, it suggests that (at most) 1 time in 20 that the null hypothesis would be mistakenly rejected or failed to be rejected (e.g., because of the statistical noise in the data sample).<br>
Having a small p-value (rejecting the null hypothesis) either means that the null hypothesis is false (we got it right) or it is true and some rare and unlikely event has been observed (we made a mistake). If this type of error is made, it is called a false positive. We falsely rejected of the null hypothesis.
Alternately, given a large p-value (failing to reject the null hypothesis), it may mean that the null hypothesis is true (we got it right) or that the null hypothesis is false and some unlikely event occurred (we made a mistake). If this type of error is made, it is called a false negative. We falsely believe the null hypothesis or assumption of the statistical test.<br>
<br>
Each of these two types of error has a specific name:<br>
- Type I Error: The incorrect rejection of a true null hypothesis or a false positive.<br>
- Type II Error: The incorrect failure of rejection of a false null hypothesis or a false negative.<br>
<br>
All statistical hypothesis tests have a risk of making either of these types of errors. False findings are more than just possible; they are probable!<br>
Ideally, we want to choose a significance level that minimizes the likelihood of one of these errors. E.g. a very small significance level. Although significance levels such as 0.05 and 0.01 are common in many fields of science, harder sciences (as defined by Dr. Sheldon Cooper), such as physics, are more aggressive.
Read more on Type I and Type II Errors @<br>
- *Type I and type II errors* available at https://en.wikipedia.org/wiki/Type_I_and_type_II_errors#:~:text=In%20statistical%20hypothesis%20testing%2C%20a,false%20negative%22%20finding%20or%20conclusion<br>
- *To Err is Human: What are Type I and II Errors?* available at https://www.statisticssolutions.com/to-err-is-human-what-are-type-i-and-ii-errors/<br>
- *Statistics: What are Type 1 and Type 2 Errors?* available at https://www.abtasty.com/blog/type-1-and-type-2-errors/<br>
#### <font color=crimson>Some Important Statistical Hypothesis Tests</font><br>
__Variable Distribution Type Tests (Gaussian)__
- Shapiro-Wilk Test
- D’Agostino’s K^2 Test
- Anderson-Darling Test
__Compare Sample Means (parametric)__
- Student’s t-test
- Paired Student’s t-test
- Analysis of Variance Test (ANOVA)
- Repeated Measures ANOVA Test
__Compare Sample Means (nonparametric)__
- Mann-Whitney U Test
- Wilcoxon Signed-Rank Test
- Kruskal-Wallis H Test
- Friedman Test
Check these excellent links to read more on different Statistical Hypothesis Tests:_<br>
- *17 Statistical Hypothesis Tests in Python (Cheat Sheet)* by *Jason Brownlee * available at https://machinelearningmastery.com/statistical-hypothesis-tests-in-python-cheat-sheet/<br>
- *Statistical Tests — When to use Which ?* by *vibhor nigam* available at https://towardsdatascience.com/statistical-tests-when-to-use-which-704557554740<br>
- *Comparing Hypothesis Tests for Continuous, Binary, and Count Data* by *Jim Frost* available at https://statisticsbyjim.com/hypothesis-testing/comparing-hypothesis-tests-data-types/<br>
*****
#### <font color=crimson>Normality Tests: Shapiro-Wilk Test</font><br>
Tests whether a data sample has a Gaussian distribution.<br>
Assumptions:<br>
Observations in each sample are independent and identically distributed (iid).<br>
Interpretation:<br>
- H0: the sample has a Gaussian distribution.
- H1: the sample does not have a Gaussian distribution.
```
# Example of the Shapiro-Wilk Normality Test
from scipy.stats import shapiro
data = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
stat, p = shapiro(data)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably Gaussian')
else:
print('Probably not Gaussian')
```
#### <font color=crimson>Normality Tests: D’Agostino’s K^2 Test</font><br>
Tests whether a data sample has a Gaussian distribution.<br>
Assumptions:<br>
Observations in each sample are independent and identically distributed (iid).<br>
Interpretation:<br>
- H0: the sample has a Gaussian distribution.
- H1: the sample does not have a Gaussian distribution.
```
# Example of the D'Agostino's K^2 Normality Test
from scipy.stats import normaltest
data = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
stat, p = normaltest(data)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably Gaussian')
else:
print('Probably not Gaussian')
```
_Read more on Normality Tests @_<br>
- __*A Gentle Introduction to Normality Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/a-gentle-introduction-to-normality-tests-in-python/__<br>
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Student’s t-test</font><br>
Tests whether the means of two independent samples are significantly different.
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
Interpretation:
- H0: the means of the samples are equal.<br>
- H1: the means of the samples are unequal.<br>
```
# Example of the Student's t-test
from scipy.stats import ttest_ind
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
stat, p = ttest_ind(data1, data2)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Paired Student’s t-test</font><br>
Tests whether the means of two paired samples are significantly different.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
- Observations across each sample are paired.<br>
Interpretation:<br>
- H0: the means of the samples are equal.<br>
- H1: the means of the samples are unequal.<br>
```
# Example of the Paired Student's t-test
from scipy.stats import ttest_rel
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
stat, p = ttest_rel(data1, data2)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Analysis of Variance Test (ANOVA)</font><br>
Tests whether the means of two or more independent samples are significantly different.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
Interpretation:<br>
- H0: the means of the samples are equal.<br>
- H1: one or more of the means of the samples are unequal.<br>
```
# Example of the Analysis of Variance Test
from scipy.stats import f_oneway
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
data3 = [-0.208, 0.696, 0.928, -1.148, -0.213, 0.229, 0.137, 0.269, -0.870, -1.204]
stat, p = f_oneway(data1, data2, data3)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
_Read more on Parametric Statistical Hypothesis Tests @_<br>
- __*How to Calculate Parametric Statistical Hypothesis Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/parametric-statistical-significance-tests-in-python/__<br>
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Mann-Whitney U Test</font><br>
Tests whether the distributions of two independent samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample can be ranked.<br>
Interpretation:<br>
- H0: the distributions of both samples are equal.<br>
- H1: the distributions of both samples are not equal.<br>
```
# Example of the Mann-Whitney U Test
from scipy.stats import mannwhitneyu
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
stat, p = mannwhitneyu(data1, data2)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Wilcoxon Signed-Rank Test</font><br>
Tests whether the distributions of two paired samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).:<br>
- Observations in each sample can be ranked.<br>
- Observations across each sample are paired.<br>
Interpretation:<br>
- H0: the distributions of both samples are equal.<br>
- H1: the distributions of both samples are not equal.<br>
```
# Example of the Wilcoxon Signed-Rank Test
from scipy.stats import wilcoxon
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
stat, p = wilcoxon(data1, data2)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Kruskal-Wallis H Test</font><br>
Tests whether the distributions of two or more independent samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample can be ranked.<br>
Interpretation:<br>
- H0: the distributions of all samples are equal.<br>
- H1: the distributions of one or more samples are not equal.<br>
```
# Example of the Kruskal-Wallis H Test
from scipy.stats import kruskal
data1 = [0.873, 2.817, 0.121, -0.945, -0.055, -1.436, 0.360, -1.478, -1.637, -1.869]
data2 = [1.142, -0.432, -0.938, -0.729, -0.846, -0.157, 0.500, 1.183, -1.075, -0.169]
stat, p = kruskal(data1, data2)
print('stat=%.3f, p=%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
_Read more on Nonparametric Statistical Hypothesis Tests @_<br>
- __*How to Calculate Nonparametric Statistical Hypothesis Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/nonparametric-statistical-significance-tests-in-python/__<br>
</hr>
#### <font color=crimson>Example with REAL data: Do construction activities impact stormwater solids metrics?</font><br>
### Background
The Clean Water Act (CWA) prohibits storm water discharge from construction sites
that disturb 5 or more acres, unless authorized by a National Pollutant Discharge
Elimination System (NPDES) permit. Permittees must provide a site description,
identify sources of contaminants that will affect storm water, identify appropriate
measures to reduce pollutants in stormwater discharges, and implement these measures.
The appropriate measures are further divided into four classes: erosion and
sediment control, stabilization practices, structural practices, and storm water management.
Collectively the site description and accompanying measures are known as
the facility’s Storm Water Pollution Prevention Plan (SW3P).
The permit contains no specific performance measures for construction activities,
but states that ”EPA anticipates that storm water management will be able to
provide for the removal of at least 80% of the total suspended solids (TSS).” The
rules also note ”TSS can be used as an indicator parameter to characterize the
control of other pollutants, including heavy metals, oxygen demanding pollutants,
and nutrients commonly found in stormwater discharges”; therefore, solids control is
critical to the success of any SW3P.
Although the NPDES permit requires SW3Ps to be in-place, it does not require
any performance measures as to the effectiveness of the controls with respect to
construction activities. The reason for the exclusion was to reduce costs associated
with monitoring storm water discharges, but unfortunately the exclusion also makes
it difficult for a permittee to assess the effectiveness of the controls implemented at
their site. Assessing the effectiveness of controls will aid the permittee concerned
with selecting the most cost effective SW3P.<br>
### Problem Statement <br>
The files precon.CSV and durcon.CSV contain observations of cumulative
rainfall, total solids, and total suspended solids collected from a construction
site on Nasa Road 1 in Harris County. <br>
The data in the file precon.CSV was collected `before` construction began. The data in the file durcon.CSV were collected `during` the construction activity.<br>
The first column is the date that the observation was made, the second column the total solids (by standard methods), the third column is is the total suspended solids (also by standard methods), and the last column is the cumulative rainfall for that storm.<br>
```{note}
Script to get the files automatically is listed below this note:
```
```
import requests # Module to process http/https requests
remote_url="http://54.243.252.9/engr-1330-webroot/9-MyJupyterNotebooks/41A-HypothesisTests/precon.csv" # set the url
rget = requests.get(remote_url, allow_redirects=True) # get the remote resource, follow imbedded links
open('precon.csv','wb').write(rget.content) # extract from the remote the contents, assign to a local file same name
remote_url="http://54.243.252.9/engr-1330-webroot/9-MyJupyterNotebooks/41A-HypothesisTests/durcon.csv" # set the url
rget = requests.get(remote_url, allow_redirects=True) # get the remote resource, follow imbedded links
open('durcon.csv','wb').write(rget.content) # extract from the remote the contents, assign to a local file same name
```
These data are not time series (there was sufficient time between site visits that you can safely assume each storm was independent.
__Our task is to analyze these two data sets and decide if construction activities impact stormwater quality in terms of solids measures.__
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
```
Read and examine the files, see if we can understand their structure
```
precon = pd.read_csv("precon.csv")
durcon = pd.read_csv("durcon.csv")
precon
durcon
precon.describe()
durcon.describe()
precon.plot.box()
durcon.plot.box()
```
Here we see that the scales of the two data sets are quite different. Let's see if the two construction phases represent approximately the same rainfall conditions?
```
precon['RAIN.PRE'].describe()
durcon['RAIN.DUR'].describe()
```
If we look at the summary statistics, we might conclude there is more rainfall during construction, which could bias our interpretation, a box plot of just rainfall might be useful, as would hypothesis tests.
```
precon['RAIN.PRE'].plot.box()
durcon['RAIN.DUR'].plot.box()
```
Hard to tell from the plots, they look a little different, but are they? Lets apply some hypothesis tests
```
from scipy.stats import mannwhitneyu # import a useful non-parametric test
stat, p = mannwhitneyu(precon['RAIN.PRE'],durcon['RAIN.DUR'])
print('statistic=%.3f, p-value at rejection =%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
from scipy import stats
results = stats.ttest_ind(precon['RAIN.PRE'], durcon['RAIN.DUR'])
print('statistic=%.3f, p-value at rejection =%.3f ' % (results[0], results[1]))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
From these two tests (the data are NOT paired) we conclude that the two sets of data originate from the same distribution. Thus the question "Do the two construction phases represent approximately the same rainfall conditions?" can be safely answered in the affirmative.
Continuing, lets ask the same about total solids, first plots:
```
precon['TS.PRE'].plot.box()
durcon['TS.DUR'].plot.box()
```
Look at the difference in scales, the during construction phase, is about 5 to 10 times greater.
But lets apply some tests to formalize our interpretation.
```
stat, p = mannwhitneyu(precon['TS.PRE'],durcon['TS.DUR'])
print('statistic=%.3f, p-value at rejection =%.3f' % (stat, p))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
results = stats.ttest_ind(precon['TS.PRE'], durcon['TS.DUR'])
print('statistic=%.3f, p-value at rejection =%.3f ' % (results[0], results[1]))
if p > 0.05:
print('Probably the same distribution')
else:
print('Probably different distributions')
```
Both these tests indicate that the data derive from distirbutions with different measures of central tendency (means). Lets now ask the question about normality, we will apply a test called normaltest. This function tests a null hypothesis that a sample comes from a normal distribution. It is based on D’Agostino and Pearson’s test that combines skew and kurtosis to produce an omnibus test of normality. We will likely get a warning because our sample size is pretty small.
```
stat, p = stats.normaltest(precon['TS.PRE'])
print('statistic=%.3f, p-value at rejection =%.3f' % (stat, p))
if p > 0.05:
print('Probably normal distributed')
else:
print('Probably Not-normal distributed')
stat, p = stats.normaltest(durcon['TS.DUR'])
print('statistic=%.3f, p-value at rejection =%.3f' % (stat, p))
if p > 0.05:
print('Probably normal distributed')
else:
print('Probably Not-normal distributed')
```
#### References
D’Agostino, R. B. (1971), “An omnibus test of normality for moderate and large sample size”, Biometrika, 58, 341-348
D’Agostino, R. and Pearson, E. S. (1973), “Tests for departure from normality”, Biometrika, 60, 613-622
## References
<hr>
## Laboratory 22
**Examine** (click) Laboratory 22 as a webpage at [Laboratory 22.html](http://54.243.252.9/engr-1330-webroot/8-Labs/Lab22/Lab22.html)
**Download** (right-click, save target as ...) Laboratory 22 as a jupyterlab notebook from [Laboratory 22.ipynb](http://54.243.252.9/engr-1330-webroot/8-Labs/Lab22/Lab22.ipynb)
<hr><hr>
## Exercise Set 22
**Examine** (click) Exercise Set 22 as a webpage at [Exercise 22.html](http://54.243.252.9/engr-1330-webroot/8-Labs/Lab22/Lab22-TH.html)
**Download** (right-click, save target as ...) Exercise Set 21 as a jupyterlab notebook at [Exercise Set 22.ipynb](http://54.243.252.9/engr-1330-webroot/8-Labs/Lab22/Lab22-TH.ipynb)
|
github_jupyter
|
<br>
For example, if we were performing a test of whether a data sample was normally distributed and we calculated a p-value of .07, we could state something like:
"The test found that the data sample was normal, failing to reject the null hypothesis at a 5% significance level.""
The significance level can be inverted by subtracting it from 1 to give a **confidence level** of the hypothesis given the observed sample data.
Therefore, statements such as the following can also be made:
"The test found that the data was normal, failing to reject the null hypothesis at a 95% confidence level.""
### Example :<br>
You have a coin and you don’t know whether that is fair or tricky so let’s decide null and alternate hypothes is<br>
1. H0 : a coin is a fair coin.<br>
2. H1 : a coin is a tricky coin. <br>
3. alpha = 5% or 0.05<br>
Now let’s toss the coin and calculate p-value (probability value).<br>
Toss a coin 1st time (sample size =1) and result is tail
- P-value = 50% (as head and tail have equal probability)<br>
Toss a coin 2nd time (sample size =2) and result is tail, now
- P-value = 50/2 = 25%<br>
and similarly suppose we Toss 6 consecutive times (sample size =6) and got result as P-value = 1.5%
but we set our significance level as 5%. Here we see we are beyond that level i.e. our null- hypothesis does not hold good so we need to reject and propose that this coin is not fair. It does not necessarily mean that the coin is tricky, but 6 tails in a row is quite unlikely with a fair coin, and a good "bet" would be to reject the coin as unfair, and 95% of the time you would be correct.
Alternatively, one could phrase the result as a fair coin would produce a result **other** than 6 tails in a row 98.5% of the time.
Read more on p-value @<br>
- *P-values Explained By Data Scientist For Data Scientists* by *Admond Lee* available at https://towardsdatascience.com/p-values-explained-by-data-scientist-f40a746cfc8<br>
- *What a p-Value Tells You about Statistical Data* by *Deborah J. Rumsey* available at https://www.dummies.com/education/math/statistics/what-a-p-value-tells-you-about-statistical-data/<br>
- *Key to statistical result interpretation: P-value in plain English* by *Tran Quang Hung* available at https://s4be.cochrane.org/blog/2016/03/21/p-value-in-plain-english-2/<br>
Watch more on p-value @<br>
- *StatQuest: P Values, clearly explained* available at https://www.youtube.com/watch?v=5Z9OIYA8He8<br>
- *Understanding the p-value - Statistics Help* available at https://www.youtube.com/watch?v=eyknGvncKLw<br>
- *What Is A P-Value? - Clearly Explained* available at https://www.youtube.com/watch?v=ukcFrzt6cHk<br>
#### <font color=crimson>“Reject” vs “Failure to Reject”</font><br>
The p-value is probabilistic. This means that when we interpret the result of a statistical test, we do not know what is true or false, only what is likely. Rejecting the null hypothesis means that there is sufficient statistical evidence (from the samples) that the null hypothesis does not look likely (for the population). Otherwise, it means that there is not sufficient statistical evidence to reject the null hypothesis.<br>
We may think about the statistical test in terms of the dichotomy of rejecting and accepting the null hypothesis. The danger is that if we say that we “accept” the null hypothesis, the "language" implies that the null hypothesis is true. Instead, it is more preferred to say that we “fail to reject” the null hypothesis, as in, there is insufficient statistical evidence to reject it.<br>
#### <font color=crimson>Errors in Statistical Tests</font><br>
The interpretation of a statistical hypothesis test is probabilistic. That means that the evidence of the test may suggest an outcome and be mistaken. For example, if alpha was 5%, it suggests that (at most) 1 time in 20 that the null hypothesis would be mistakenly rejected or failed to be rejected (e.g., because of the statistical noise in the data sample).<br>
Having a small p-value (rejecting the null hypothesis) either means that the null hypothesis is false (we got it right) or it is true and some rare and unlikely event has been observed (we made a mistake). If this type of error is made, it is called a false positive. We falsely rejected of the null hypothesis.
Alternately, given a large p-value (failing to reject the null hypothesis), it may mean that the null hypothesis is true (we got it right) or that the null hypothesis is false and some unlikely event occurred (we made a mistake). If this type of error is made, it is called a false negative. We falsely believe the null hypothesis or assumption of the statistical test.<br>
<br>
Each of these two types of error has a specific name:<br>
- Type I Error: The incorrect rejection of a true null hypothesis or a false positive.<br>
- Type II Error: The incorrect failure of rejection of a false null hypothesis or a false negative.<br>
<br>
All statistical hypothesis tests have a risk of making either of these types of errors. False findings are more than just possible; they are probable!<br>
Ideally, we want to choose a significance level that minimizes the likelihood of one of these errors. E.g. a very small significance level. Although significance levels such as 0.05 and 0.01 are common in many fields of science, harder sciences (as defined by Dr. Sheldon Cooper), such as physics, are more aggressive.
Read more on Type I and Type II Errors @<br>
- *Type I and type II errors* available at https://en.wikipedia.org/wiki/Type_I_and_type_II_errors#:~:text=In%20statistical%20hypothesis%20testing%2C%20a,false%20negative%22%20finding%20or%20conclusion<br>
- *To Err is Human: What are Type I and II Errors?* available at https://www.statisticssolutions.com/to-err-is-human-what-are-type-i-and-ii-errors/<br>
- *Statistics: What are Type 1 and Type 2 Errors?* available at https://www.abtasty.com/blog/type-1-and-type-2-errors/<br>
#### <font color=crimson>Some Important Statistical Hypothesis Tests</font><br>
__Variable Distribution Type Tests (Gaussian)__
- Shapiro-Wilk Test
- D’Agostino’s K^2 Test
- Anderson-Darling Test
__Compare Sample Means (parametric)__
- Student’s t-test
- Paired Student’s t-test
- Analysis of Variance Test (ANOVA)
- Repeated Measures ANOVA Test
__Compare Sample Means (nonparametric)__
- Mann-Whitney U Test
- Wilcoxon Signed-Rank Test
- Kruskal-Wallis H Test
- Friedman Test
Check these excellent links to read more on different Statistical Hypothesis Tests:_<br>
- *17 Statistical Hypothesis Tests in Python (Cheat Sheet)* by *Jason Brownlee * available at https://machinelearningmastery.com/statistical-hypothesis-tests-in-python-cheat-sheet/<br>
- *Statistical Tests — When to use Which ?* by *vibhor nigam* available at https://towardsdatascience.com/statistical-tests-when-to-use-which-704557554740<br>
- *Comparing Hypothesis Tests for Continuous, Binary, and Count Data* by *Jim Frost* available at https://statisticsbyjim.com/hypothesis-testing/comparing-hypothesis-tests-data-types/<br>
*****
#### <font color=crimson>Normality Tests: Shapiro-Wilk Test</font><br>
Tests whether a data sample has a Gaussian distribution.<br>
Assumptions:<br>
Observations in each sample are independent and identically distributed (iid).<br>
Interpretation:<br>
- H0: the sample has a Gaussian distribution.
- H1: the sample does not have a Gaussian distribution.
#### <font color=crimson>Normality Tests: D’Agostino’s K^2 Test</font><br>
Tests whether a data sample has a Gaussian distribution.<br>
Assumptions:<br>
Observations in each sample are independent and identically distributed (iid).<br>
Interpretation:<br>
- H0: the sample has a Gaussian distribution.
- H1: the sample does not have a Gaussian distribution.
_Read more on Normality Tests @_<br>
- __*A Gentle Introduction to Normality Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/a-gentle-introduction-to-normality-tests-in-python/__<br>
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Student’s t-test</font><br>
Tests whether the means of two independent samples are significantly different.
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
Interpretation:
- H0: the means of the samples are equal.<br>
- H1: the means of the samples are unequal.<br>
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Paired Student’s t-test</font><br>
Tests whether the means of two paired samples are significantly different.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
- Observations across each sample are paired.<br>
Interpretation:<br>
- H0: the means of the samples are equal.<br>
- H1: the means of the samples are unequal.<br>
#### <font color=crimson>Parametric Statistical Hypothesis Tests: Analysis of Variance Test (ANOVA)</font><br>
Tests whether the means of two or more independent samples are significantly different.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample are normally distributed.<br>
- Observations in each sample have the same variance.<br>
Interpretation:<br>
- H0: the means of the samples are equal.<br>
- H1: one or more of the means of the samples are unequal.<br>
_Read more on Parametric Statistical Hypothesis Tests @_<br>
- __*How to Calculate Parametric Statistical Hypothesis Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/parametric-statistical-significance-tests-in-python/__<br>
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Mann-Whitney U Test</font><br>
Tests whether the distributions of two independent samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample can be ranked.<br>
Interpretation:<br>
- H0: the distributions of both samples are equal.<br>
- H1: the distributions of both samples are not equal.<br>
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Wilcoxon Signed-Rank Test</font><br>
Tests whether the distributions of two paired samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).:<br>
- Observations in each sample can be ranked.<br>
- Observations across each sample are paired.<br>
Interpretation:<br>
- H0: the distributions of both samples are equal.<br>
- H1: the distributions of both samples are not equal.<br>
#### <font color=crimson>Nonparametric Statistical Hypothesis Tests: Kruskal-Wallis H Test</font><br>
Tests whether the distributions of two or more independent samples are equal or not.<br>
Assumptions:<br>
- Observations in each sample are independent and identically distributed (iid).<br>
- Observations in each sample can be ranked.<br>
Interpretation:<br>
- H0: the distributions of all samples are equal.<br>
- H1: the distributions of one or more samples are not equal.<br>
_Read more on Nonparametric Statistical Hypothesis Tests @_<br>
- __*How to Calculate Nonparametric Statistical Hypothesis Tests in Python* by *Jason Brownlee* available at https://machinelearningmastery.com/nonparametric-statistical-significance-tests-in-python/__<br>
</hr>
#### <font color=crimson>Example with REAL data: Do construction activities impact stormwater solids metrics?</font><br>
### Background
The Clean Water Act (CWA) prohibits storm water discharge from construction sites
that disturb 5 or more acres, unless authorized by a National Pollutant Discharge
Elimination System (NPDES) permit. Permittees must provide a site description,
identify sources of contaminants that will affect storm water, identify appropriate
measures to reduce pollutants in stormwater discharges, and implement these measures.
The appropriate measures are further divided into four classes: erosion and
sediment control, stabilization practices, structural practices, and storm water management.
Collectively the site description and accompanying measures are known as
the facility’s Storm Water Pollution Prevention Plan (SW3P).
The permit contains no specific performance measures for construction activities,
but states that ”EPA anticipates that storm water management will be able to
provide for the removal of at least 80% of the total suspended solids (TSS).” The
rules also note ”TSS can be used as an indicator parameter to characterize the
control of other pollutants, including heavy metals, oxygen demanding pollutants,
and nutrients commonly found in stormwater discharges”; therefore, solids control is
critical to the success of any SW3P.
Although the NPDES permit requires SW3Ps to be in-place, it does not require
any performance measures as to the effectiveness of the controls with respect to
construction activities. The reason for the exclusion was to reduce costs associated
with monitoring storm water discharges, but unfortunately the exclusion also makes
it difficult for a permittee to assess the effectiveness of the controls implemented at
their site. Assessing the effectiveness of controls will aid the permittee concerned
with selecting the most cost effective SW3P.<br>
### Problem Statement <br>
The files precon.CSV and durcon.CSV contain observations of cumulative
rainfall, total solids, and total suspended solids collected from a construction
site on Nasa Road 1 in Harris County. <br>
The data in the file precon.CSV was collected `before` construction began. The data in the file durcon.CSV were collected `during` the construction activity.<br>
The first column is the date that the observation was made, the second column the total solids (by standard methods), the third column is is the total suspended solids (also by standard methods), and the last column is the cumulative rainfall for that storm.<br>
These data are not time series (there was sufficient time between site visits that you can safely assume each storm was independent.
__Our task is to analyze these two data sets and decide if construction activities impact stormwater quality in terms of solids measures.__
Read and examine the files, see if we can understand their structure
Here we see that the scales of the two data sets are quite different. Let's see if the two construction phases represent approximately the same rainfall conditions?
If we look at the summary statistics, we might conclude there is more rainfall during construction, which could bias our interpretation, a box plot of just rainfall might be useful, as would hypothesis tests.
Hard to tell from the plots, they look a little different, but are they? Lets apply some hypothesis tests
From these two tests (the data are NOT paired) we conclude that the two sets of data originate from the same distribution. Thus the question "Do the two construction phases represent approximately the same rainfall conditions?" can be safely answered in the affirmative.
Continuing, lets ask the same about total solids, first plots:
Look at the difference in scales, the during construction phase, is about 5 to 10 times greater.
But lets apply some tests to formalize our interpretation.
Both these tests indicate that the data derive from distirbutions with different measures of central tendency (means). Lets now ask the question about normality, we will apply a test called normaltest. This function tests a null hypothesis that a sample comes from a normal distribution. It is based on D’Agostino and Pearson’s test that combines skew and kurtosis to produce an omnibus test of normality. We will likely get a warning because our sample size is pretty small.
| 0.885848 | 0.993759 |
Tutorial 5: Linking Phases
==========================
So, we've learnt that if our parameter space is too complex, our `NonLinearSearch` might fail to find the global
maximum solution. However, we also learnt how to ensure this doesn`t happen, by:
1) Tuning our priors to the strong lens we're fitting.
2) Making our lens model less complex.
3) Searching non-linear parameter space for longer.
However, each of the above approaches has disadvantages. The more we tune our priors, the less we can generalize our
analysis to a different strong lens. The less complex we make our model, the less realistic it is. And if we rely too
much on searching parameter space for longer, we could end up with phase`s that take days, weeks or months to run.
In this exercise, we're going to combine these 3 approaches so that we can fit complex and realistic lens models in a
way that that can be generalized to many different strong lenses. To do this, we'll run 2 phases, and link the lens
model inferred in the first phase to the priors of the second phase`s lens model.
Our first phase will make the same light-traces-mass assumption we made in the previous tutorial. We saw that this
gives a reasonable lens model. However, we'll make a couple of extra simplifying assumptions, to really try and bring
our lens model complexity down and get the `NonLinearSearch` running fast.
The model we infer above will therefore be a lot less realistic. But it doesn`t matter, because in the second phase
we're going to relax these assumptions and get back our more realistic lens model. The beauty is that, by running the
first phase, we can use its results to tune the priors of our second phase. For example:
1) The first phase should give us a pretty good idea of the lens `Galaxy`'s light and mass profiles, for example its
intensity, effective radius and einstein radius.
2) It should also give us a pretty good fit to the lensed source galaxy. This means we'll already know where in
source-plane its is located and what its intensity and effective are.
```
%matplotlib inline
from pyprojroot import here
workspace_path = str(here())
%cd $workspace_path
print(f"Working Directory has been set to `{workspace_path}`")
import numpy as np
from os import path
import autolens as al
import autolens.plot as aplt
import autofit as af
```
we'll use the same strong lensing data as the previous tutorial, where:
- The lens `Galaxy`'s `LightProfile` is an `EllipticalSersic`.
- The lens `Galaxy`'s total mass distribution is an `EllipticalIsothermal`.
- The source `Galaxy`'s `LightProfile` is an `EllipticalExponential`.
```
dataset_name = "light_sersic__mass_sie__source_exp"
dataset_path = path.join("dataset", "howtolens", "chapter_2", dataset_name)
imaging = al.Imaging.from_fits(
image_path=path.join(dataset_path, "image.fits"),
noise_map_path=path.join(dataset_path, "noise_map.fits"),
psf_path=path.join(dataset_path, "psf.fits"),
pixel_scales=0.1,
)
```
we'll create and use a smaller 2.0" `Mask2D` again.
```
mask = al.Mask2D.circular(
shape_2d=imaging.shape_2d, pixel_scales=imaging.pixel_scales, radius=2.0
)
```
When plotted, the lens light`s is clearly visible in the centre of the image.
```
aplt.Imaging.subplot_imaging(imaging=imaging, mask=mask)
```
Like in the previous tutorial, we use a `SettingsPhaseImaging` object to specify our model-fitting procedure uses a
regular `Grid`.
```
settings_masked_imaging = al.SettingsMaskedImaging(grid_class=al.Grid, sub_size=2)
settings = al.SettingsPhaseImaging(settings_masked_imaging=settings_masked_imaging)
```
As we've eluded to before, one can look at an image and immediately identify the centre of the lens galaxy. It's
that bright blob of light in the middle! Given that we know we're going to make the lens model more complex in the
next phase, lets take a more liberal approach than before and fix the lens centre to $(y,x)$ = (0.0", 0.0").
```
lens = al.GalaxyModel(
redshift=0.5, bulge=al.lp.EllipticalSersic, mass=al.mp.EllipticalIsothermal
)
source = al.GalaxyModel(redshift=1.0, bulge=al.lp.EllipticalExponential)
```
You haven`t actually seen a line like this one before. By setting a parameter to a number (and not a prior) it is be
removed from non-linear parameter space and always fixed to that value. Pretty neat, huh?
```
lens.bulge.centre_0 = 0.0
lens.bulge.centre_1 = 0.0
lens.mass.centre_0 = 0.0
lens.mass.centre_1 = 0.0
```
Lets use the same approach of making the ellipticity of the mass trace that of the sersic.
"""
lens.mass.elliptical_comps = lens.bulge.elliptical_comps
Now, you might be thinking, doesn`t this prevent our phase from generalizing to other strong lenses? What if the
centre of their lens galaxy isn't at (0.0", 0.0")?
Well, this is true if our dataset reduction centres the lens galaxy somewhere else. But we get to choose where we
centre it when we make the image. Therefore, I`d recommend you always centre the lens galaxy at the same location,
and (0.0", 0.0") seems the best choice!
We also discussed that the Sersic index of most lens galaxies is around 4. Lets fix it to 4 this time.
```
lens.bulge.sersic_index = 4.0
```
Now lets create the phase.
```
phase1 = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_1", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(lens=lens, source=source),
)
```
Lets run the phase, noting that our liberal approach to reducing the lens model complexity has reduced it to just
11 parameters. (The results are still preloaded for you, but feel free to run it yourself, its fairly quick).
```
print(
"Dynesty has begun running - checkout the workspace/output/5_linking_phases"
" folder for live output of the results, images and lens model."
" This Jupyter notebook cell with progress once Dynesty has completed - this could take some time!"
)
phase1_result = phase1.run(dataset=imaging, mask=mask)
print("Dynesty has finished run - you may now continue the notebook.")
```
And indeed, we get a reasonably good model and fit to the data - in a much shorter space of time!
```
aplt.FitImaging.subplot_fit_imaging(fit=phase1_result.max_log_likelihood_fit)
```
Now all we need to do is look at the results of phase 1 and tune our priors in phase 2 to those result. Lets setup
a custom phase that does exactly that.
GaussianPriors are a nice way to do this. They tell the `NonLinearSearch` where to look, but leave open the
possibility that there might be a better solution nearby. In contrast, UniformPriors put hard limits on what values a
parameter can or can`t take. It makes it more likely we'll accidently cut-out the global maxima solution.
```
lens = al.GalaxyModel(
redshift=0.5, bulge=al.lp.EllipticalSersic, mass=al.mp.EllipticalIsothermal
)
source = al.GalaxyModel(redshift=1.0, bulge=al.lp.EllipticalExponential)
```
What I've done below is looked at the results of phase 1 and manually specified a prior for every parameter. If a
parameter was fixed in the previous phase, its prior is based around the previous value. Don't worry about the sigma
values for now, I've chosen values that I know will ensure reasonable sampling, but we'll cover this later.
```
"""LENS LIGHT PRIORS"""
lens.bulge.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.bulge.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.bulge.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.33333, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
lens.bulge.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=0.0, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
lens.bulge.intensity = af.GaussianPrior(
mean=0.02, sigma=0.01, lower_limit=0.0, upper_limit=np.inf
)
lens.bulge.effective_radius = af.GaussianPrior(
mean=0.62, sigma=0.2, lower_limit=0.0, upper_limit=np.inf
)
lens.bulge.sersic_index = af.GaussianPrior(
mean=4.0, sigma=2.0, lower_limit=0.0, upper_limit=np.inf
)
"""LENS MASS PRIORS"""
lens.mass.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.mass.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.mass.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.33333, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
lens.mass.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=0.0, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
lens.mass.einstein_radius = af.GaussianPrior(
mean=0.8, sigma=0.1, lower_limit=0.0, upper_limit=np.inf
)
"""SOURCE LIGHT PRIORS"""
source.bulge.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
source.bulge.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
source.bulge.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.0, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
source.bulge.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=-0.33333, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
source.bulge.intensity = af.GaussianPrior(
mean=0.14, sigma=0.05, lower_limit=0.0, upper_limit=np.inf
)
source.bulge.effective_radius = af.GaussianPrior(
mean=0.27, sigma=0.2, lower_limit=0.0, upper_limit=np.inf
)
```
Lets setup and run the phase. As expected, it gives us the correct lens model. However, it does so significantly faster
than we're used to - I didn`t have to edit the config files to get this phase to run fast!
```
phase2 = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_2", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(lens=lens, source=source),
)
print(
"Dynesty has begun running - checkout the workspace/output/5_linking_phases"
" folder for live output of the results, images and lens model."
" This Jupyter notebook cell with progress once Dynesty has completed - this could take some time!"
)
phase2_result = phase2.run(dataset=imaging, mask=mask)
print("Dynesty has finished run - you may now continue the notebook.")
```
Look at that, the right lens model, again!
```
aplt.FitImaging.subplot_fit_imaging(fit=phase2_result.max_log_likelihood_fit)
```
Our choice to link two phases together was a huge success. We managed to fit a complex and realistic model, but were
able to begin by making simplifying assumptions that eased our search of non-linear parameter space. We could apply
phase 1 to pretty much any strong lens and therefore get ourselves a decent lens model with which to tune phase 2`s
priors.
You`re probably thinking though that there is one huge, giant, glaring flaw in all of this that I've not mentioned.
Phase 2 can`t be generalized to another lens - it`s priors are tuned to the image we fitted. If we had a lot of lenses,
we`d have to write a new phase2 for every single one. This isn't ideal, is it?
Fortunately, we can pass priors in **PyAutoLens** without specifying the specific values, using what we call promises. The
code below sets up phase2 with priors fully linked, but without specifying each individual prior!
```
phase2_pass = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_2_pass", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(
lens=phase1_result.model.galaxies.lens,
source=phase1_result.model.galaxies.source,
),
)
# phase2_pass.run(dataset=imaging, mask=mask)
```
By using the following API to link the result to the next model:
lens = phase1_result.model.galaxies.lens
source = phase1_result.model.galaxies.source
Once the above phase is running, you should checkout its `model.info` file. The parameters do not use the default
priors we saw in phase 1 (which are typically broad UniformPriors). Instead, it uses GaussianPrior`s where:
- The mean values are the median PDF results of every parameter in phase 1.
- Many sigma values are the errors computed at 3.0 sigma confidence of every parameter in phase 1.
- Other sigma values are higher than the errors computed at 3.0 sigma confidence. These instead use the value
specified in the `width_modifier` field of the `Profile`'s entry in the `json_config` files (we will discuss
why this is used in a moment).
Thus, much like the manual GaussianPriors I specified above, we have set up the phase with GaussianPriors centred on
the high likelihood regions of parameter space!
The priors passed above retained the model parameterization of phase 1, including the fixed values of (0.0, 0.0) for
the centres of the light and mass profiles and the alignment between their elliptical components. However, we often
want to pass priors *and* change the model parameterization.
To do this, we have to use the ``.riorModel__ object in AutoFit, which allows us to turn light and mass profiles into
`model components` whose parameters have priors that can be manipulated in an analogous fashion to to ``.alaxyModel__.
In fact, the individual components of the ``.alaxyModel__ class have been ``.riorModel__`s all along!
```
print(lens.bulge)
print(lens.mass)
print(source.bulge)
```
We can thus set up the ``.alaxyModel__ we desire, by first creating the individual ``.riorModel__`s of each
component and then passing the priors of each individual parameter.
LENS LIGHT PRIORS"""
sersic = af.PriorModel(al.lp.EllipticalSersic)
sersic.elliptical_comps.elliptical_comps = (
phase1_result.model.galaxies.lens.bulge.elliptical_comps
)
sersic.intensity = phase1_result.model.galaxies.lens.bulge.intensity
sersic.effective_radius = phase1_result.model.galaxies.lens.bulge.effective_radius
"""LENS MASS PRIORS"""
mass = af.PriorModel(al.mp.EllipticalIsothermal)
lens.mass.elliptical_comps.elliptical_comps = (
phase1_result.model.galaxies.lens.mass.elliptical_comps
)
lens.mass.einstein_radius = phase1_result.model.galaxies.lens.mass.einstein_radius
lens = al.GalaxyModel(redshift=0.5, bulge=sersic, mass=mass)
We now create and run the phase, using the lens ``.alaxyModel__ we created above.
```
phase2_pass = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens",
name="phase_t5_linking_phases_2_pass_individual",
n_live_points=40,
),
settings=settings,
galaxies=af.CollectionPriorModel(
lens=lens, source=phase1_result.model.galaxies.source
),
)
# phase2_pass.run(dataset=imaging, mask=mask)
```
Don't worry too much about whether you fully understand the prior passing API yet, as this will be a key subject in
chapter 3 when we consider pipelines. Furthermore, in the `autolens_workspace/pipelines` directly you'll find
numerous example pipelines that give examples of how to perform prior passing for many common lens models.
To end, lets consider how we passed priors using the `model` attribute of the phase 1 results above, as its not clear
how priors are passed. Do they use a UniformPrior or GaussianPrior? What are the limits / mean / width of
these priors?
Lets say I link two parameters as follows:
mass.einstein_radius = phase1_result.model.galaxies.lens.mass.einstein_radius
By invoking the `model` attribute, the prioris passed following 3 rules:
1) The new parameter, in this case the einstein radius, uses a GaussianPrior. A GaussianPrior is ideal, as the 1D
pdf results we compute at the end of a phase are easily summarized as a Gaussian.
2) The mean of the GaussianPrior is the median PDF value of the parameter estimated in phase 1.
This ensures that the initial sampling of the new phase`s non-linear starts by searching the region of non-linear
parameter space that correspond to highest log likelihood solutions in the previous phase. Thus, we're setting
our priors to look in the `correct` regions of parameter space.
3) The sigma of the Gaussian will use the maximum of two values:
(i) the 1D error of the parameter computed at an input sigma value (default sigma=3.0).
(ii) The value specified for the profile in the `config/priors/*.json` config file`s `width_modifer`
field (check these files out now).
The idea here is simple. We want a value of sigma that gives a GaussianPrior wide enough to search a broad
region of parameter space, so that the lens model can change if a better solution is nearby. However, we want it
to be narrow enough that we don't search too much of parameter space, as this will be slow or risk leading us
into an incorrect solution! A natural choice is the errors of the parameter from the previous phase.
Unfortunately, this doesn`t always work. Lens modeling is prone to an effect called `over-fitting` where we
underestimate the errors on our lens model parameters. This is especially true when we take the shortcuts in
early phases - fast `NonLinearSearch` settings, simplified lens models, etc.
Therefore, the `width_modifier` in the json config files are our fallback. If the error on a parameter is
suspiciously small, we instead use the value specified in the widths file. These values are chosen based on
our experience as being a good balance broadly sampling parameter space but not being so narrow important solutions
are missed.
There are two ways a value is specified using the priors/width file:
1) Absolute: In this case, the error assumed on the parameter is the value given in the config file.
For example, if for the width on centre_0 of a `LightProfile`, the width modifier reads "Absolute" with a value
0.05. This means if the error on the parameter centre_0 was less than 0.05 in the previous phase, the sigma of
its GaussianPrior in this phase will be 0.05.
2) Relative: In this case, the error assumed on the parameter is the % of the value of the
estimate value given in the config file. For example, if the intensity estimated in the previous phase was 2.0,
and the relative error in the config file reads "Relative" with a value 0.5, then the sigma of the GaussianPrior
will be 50% of this value, i.e. sigma = 0.5 * 2.0 = 1.0.
We use absolute and relative values for different parameters, depending on their properties. For example, using the
relative value of a parameter like the `Profile` centre makes no sense. If our lens galaxy is centred at (0.0, 0.0),
the relative error will always be tiny and thus poorly defined. Therefore, the default configs in **PyAutoLens** use
absolute errors on the centre.
However, there are parameters where using an absolute value does not make sense. Intensity is a good example of this.
The intensity of an image depends on its unit_label, S/N, galaxy brightness, etc. There is no single absolute value
that one can use to generically link the intensity of any two proflies. Thus, it makes more sense to link them using
the relative value from a previous phase.
We can customize how priors are passed from the results of a phase and `NonLinearSearch` by inputting to the search
a PriorPasser object:
"""
search = af.DynestyStatic(
prior_passer=af.PriorPasser(sigma=2.0, use_widths=False, use_errors=True)
)
The PriorPasser allows us to customize at what sigma the error values the model results are computed at to compute
the passed sigma values and customizes whether the widths in the config file, these computed errors, or both,
are used to set the sigma values of the passed priors.
The default values of the PriorPasser are found in the config file of every non-linear search, in the [prior_passer]
section. All non-linear searches by default use a sigma value of 3.0, use_width=True and use_errors=True. We anticipate
you should not need to change these values to get lens modeling to work proficiently!
__EXAMPLE__
Lets go through an example using a real parameter. Lets say in phase 1 we fit the lens `Galaxy`'s light with an
elliptical Sersic profile, and we estimate that its sersic index is equal to 4.0 +- 2.0 where the error value of 2.0
was computed at 3.0 sigma confidence. To pass this as a prior to phase 2, we would write:
lens.bulge.sersic_index = phase1.result.model.lens.bulge.sersic_index
The prior on the lens `Galaxy`'s sersic `LightProfile` in phase 2 would thus be a GaussianPrior, with mean=4.0 and
sigma=2.0. If we had used a sigma value of 1.0 to compute the error, which reduced the estimate from 4.0 +- 2.0 to
4.0 +- 1.0, the sigma of the Gaussian prior would instead be 1.0.
If the error on the Sersic index in phase 1 had been really small, lets say, 0.01, we would instead use the value of the
Sersic index width in the priors config file to set sigma instead. In this case, the prior config file specifies
that we use an "Absolute" value of 0.8 to link this prior. Thus, the GaussianPrior in phase 2 would have a mean=4.0 and
sigma=0.8.
If the prior config file had specified that we use an relative value of 0.8, the GaussianPrior in phase 2 would have a
mean=4.0 and sigma=3.2.
And with that, we're done. Linking priors is a bit of an art form, but one that tends to work really well. Its true to
say that things can go wrong - maybe we `trim` out the solution we're looking for, or underestimate our errors a bit
due to making our priors too narrow. However, in general, things are okay, and the example pipelines in
`autolens_workspace/pipelines` have been thoroughly tested to ensure prior linking works effectively.
|
github_jupyter
|
%matplotlib inline
from pyprojroot import here
workspace_path = str(here())
%cd $workspace_path
print(f"Working Directory has been set to `{workspace_path}`")
import numpy as np
from os import path
import autolens as al
import autolens.plot as aplt
import autofit as af
dataset_name = "light_sersic__mass_sie__source_exp"
dataset_path = path.join("dataset", "howtolens", "chapter_2", dataset_name)
imaging = al.Imaging.from_fits(
image_path=path.join(dataset_path, "image.fits"),
noise_map_path=path.join(dataset_path, "noise_map.fits"),
psf_path=path.join(dataset_path, "psf.fits"),
pixel_scales=0.1,
)
mask = al.Mask2D.circular(
shape_2d=imaging.shape_2d, pixel_scales=imaging.pixel_scales, radius=2.0
)
aplt.Imaging.subplot_imaging(imaging=imaging, mask=mask)
settings_masked_imaging = al.SettingsMaskedImaging(grid_class=al.Grid, sub_size=2)
settings = al.SettingsPhaseImaging(settings_masked_imaging=settings_masked_imaging)
lens = al.GalaxyModel(
redshift=0.5, bulge=al.lp.EllipticalSersic, mass=al.mp.EllipticalIsothermal
)
source = al.GalaxyModel(redshift=1.0, bulge=al.lp.EllipticalExponential)
lens.bulge.centre_0 = 0.0
lens.bulge.centre_1 = 0.0
lens.mass.centre_0 = 0.0
lens.mass.centre_1 = 0.0
lens.bulge.sersic_index = 4.0
phase1 = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_1", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(lens=lens, source=source),
)
print(
"Dynesty has begun running - checkout the workspace/output/5_linking_phases"
" folder for live output of the results, images and lens model."
" This Jupyter notebook cell with progress once Dynesty has completed - this could take some time!"
)
phase1_result = phase1.run(dataset=imaging, mask=mask)
print("Dynesty has finished run - you may now continue the notebook.")
aplt.FitImaging.subplot_fit_imaging(fit=phase1_result.max_log_likelihood_fit)
lens = al.GalaxyModel(
redshift=0.5, bulge=al.lp.EllipticalSersic, mass=al.mp.EllipticalIsothermal
)
source = al.GalaxyModel(redshift=1.0, bulge=al.lp.EllipticalExponential)
"""LENS LIGHT PRIORS"""
lens.bulge.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.bulge.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.bulge.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.33333, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
lens.bulge.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=0.0, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
lens.bulge.intensity = af.GaussianPrior(
mean=0.02, sigma=0.01, lower_limit=0.0, upper_limit=np.inf
)
lens.bulge.effective_radius = af.GaussianPrior(
mean=0.62, sigma=0.2, lower_limit=0.0, upper_limit=np.inf
)
lens.bulge.sersic_index = af.GaussianPrior(
mean=4.0, sigma=2.0, lower_limit=0.0, upper_limit=np.inf
)
"""LENS MASS PRIORS"""
lens.mass.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.mass.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
lens.mass.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.33333, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
lens.mass.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=0.0, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
lens.mass.einstein_radius = af.GaussianPrior(
mean=0.8, sigma=0.1, lower_limit=0.0, upper_limit=np.inf
)
"""SOURCE LIGHT PRIORS"""
source.bulge.centre.centre_0 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
source.bulge.centre.centre_1 = af.GaussianPrior(
mean=0.0, sigma=0.1, lower_limit=-np.inf, upper_limit=np.inf
)
source.bulge.elliptical_comps.elliptical_comps_0 = af.GaussianPrior(
mean=0.0, sigma=0.15, lower_limit=-1.0, upper_limit=1.0
)
source.bulge.elliptical_comps.elliptical_comps_1 = af.GaussianPrior(
mean=-0.33333, sigma=0.2, lower_limit=-1.0, upper_limit=1.0
)
source.bulge.intensity = af.GaussianPrior(
mean=0.14, sigma=0.05, lower_limit=0.0, upper_limit=np.inf
)
source.bulge.effective_radius = af.GaussianPrior(
mean=0.27, sigma=0.2, lower_limit=0.0, upper_limit=np.inf
)
phase2 = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_2", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(lens=lens, source=source),
)
print(
"Dynesty has begun running - checkout the workspace/output/5_linking_phases"
" folder for live output of the results, images and lens model."
" This Jupyter notebook cell with progress once Dynesty has completed - this could take some time!"
)
phase2_result = phase2.run(dataset=imaging, mask=mask)
print("Dynesty has finished run - you may now continue the notebook.")
aplt.FitImaging.subplot_fit_imaging(fit=phase2_result.max_log_likelihood_fit)
phase2_pass = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens", name="phase_t5_linking_phases_2_pass", n_live_points=40
),
settings=settings,
galaxies=af.CollectionPriorModel(
lens=phase1_result.model.galaxies.lens,
source=phase1_result.model.galaxies.source,
),
)
# phase2_pass.run(dataset=imaging, mask=mask)
print(lens.bulge)
print(lens.mass)
print(source.bulge)
phase2_pass = al.PhaseImaging(
search=af.DynestyStatic(
path_prefix="howtolens",
name="phase_t5_linking_phases_2_pass_individual",
n_live_points=40,
),
settings=settings,
galaxies=af.CollectionPriorModel(
lens=lens, source=phase1_result.model.galaxies.source
),
)
# phase2_pass.run(dataset=imaging, mask=mask)
| 0.674694 | 0.983502 |
# Machine Learning artifacts management
This notebook contains steps and code to demonstrate how to manage and clean up Watson Machine Learning instance. This notebook contains steps and code to work with [ibm-watson-machine-learning](https://pypi.python.org/pypi/ibm-watson-machine-learning) library available in PyPI repository. This notebook introduces commands for listing artifacts, getting artifacts details and deleting them.
Some familiarity with Python is helpful. This notebook uses Python 3.8.
## Learning goals
The learning goals of this notebook are:
- List Watson Machine Learning artifacts.
- Get artifacts details.
- Delete artifacts.
## Contents
This notebook contains the following parts:
1. [Setup](#setup)
2. [Manage pipelines](#pipelines)
3. [Manage model definitions](#model_definitions)
4. [Manage models](#models)
5. [Manage functions](#functions)
6. [Manage experiments](#experiments)
7. [Manage trainings](#trainings)
8. [Manage deployments](#deployments)
9. [Summary and next steps](#summary)
<a id="setup"></a>
## 1. Set up the environment
Before you use the sample code in this notebook, you must perform the following setup tasks:
- Contact with your Cloud Pack for Data administrator and ask him for your account credentials
### Connection to WML
Authenticate the Watson Machine Learning service on IBM Cloud Pack for Data. You need to provide platform `url`, your `username` and `api_key`.
```
username = 'PASTE YOUR USERNAME HERE'
api_key = 'PASTE YOUR API_KEY HERE'
url = 'PASTE THE PLATFORM URL HERE'
wml_credentials = {
"username": username,
"apikey": api_key,
"url": url,
"instance_id": 'openshift',
"version": '4.0'
}
```
Alternatively you can use `username` and `password` to authenticate WML services.
```
wml_credentials = {
"username": ***,
"password": ***,
"url": ***,
"instance_id": 'openshift',
"version": '4.0'
}
```
### Install and import the `ibm-watson-machine-learning` package
**Note:** `ibm-watson-machine-learning` documentation can be found <a href="http://ibm-wml-api-pyclient.mybluemix.net/" target="_blank" rel="noopener no referrer">here</a>.
```
!pip install -U ibm-watson-machine-learning
from ibm_watson_machine_learning import APIClient
client = APIClient(wml_credentials)
```
### Working with spaces
First of all, you need to create a space that will be used for your work. If you do not have space already created, you can use `{PLATFORM_URL}/ml-runtime/spaces?context=icp4data` to create one.
- Click New Deployment Space
- Create an empty space
- Go to space `Settings` tab
- Copy `space_id` and paste it below
**Tip**: You can also use SDK to prepare the space for your work. More information can be found [here](https://github.com/IBM/watson-machine-learning-samples/blob/master/cpd4.0/notebooks/python_sdk/instance-management/Space%20management.ipynb).
**Action**: Assign space ID below
```
space_id = 'PASTE YOUR SPACE ID HERE'
```
You can use `list` method to print all existing spaces.
```
client.spaces.list(limit=10)
```
To be able to interact with all resources available in Watson Machine Learning, you need to set **space** which you will be using.
```
client.set.default_space(space_id)
```
<a id="pipelines"></a>
## 2. Manage pipelines
List existing pipelines. If you want to list only part of pipelines use `client.pipelines.list(limit=n_pipelines)`.
```
client.pipelines.list(limit=10)
```
Get pipelines details. If you want to get only part of pipelines details use `client.pipelines.get_details(limit=n_pipelines)`.
You can get each pipeline details by calling `client.pipelines.get_details()` and providing pipeline id from listed pipelines.
```
pipelines_details = client.pipelines.get_details(limit=10)
print(pipelines_details)
```
Delete all pipelines. You can delete one pipeline by calling `client.pipelines.delete()` and providing pipeline id from listed pipelines.
```
for pipeline in pipelines_details['resources']:
client.pipelines.delete(pipeline['metadata']['id'])
```
<a id="model_definitions"></a>
## 3. Manage model definitions
List existing model definitions. If you want to list only part of model definitions use `client.model_definitions.list(limit=n_model_definitions)`.
```
client.model_definitions.list(limit=10)
```
Get model definiton details by copying model definition uid from above cell and running `client.model_definitions.get_details(model_definition_guid)`.
```
model_definition_guid = "PUT_YOUR_MODEL_DEFINITION_GUID"
model_definitions_details = client.model_definitions.get_details(model_definition_guid)
print(model_definitions_details)
```
Delete model definitions by calling `client.model_definitions.delete(model_definition_guid)`.
```
client.model_definitions.delete(model_definition_guid)
```
<a id="models"></a>
## 4. Manage models
List existing models. If you want to list only part of models use `client.repository.list_models(limit=n_models)`.
```
client.repository.list_models(limit=10)
```
Get model details by copying model uid from above cell and running `client.repository.get_details(model_guid)`.
```
model_guid = "PUT_YOUR_MODEL_GUID"
model_details = client.repository.get_details(model_guid)
print(model_details)
```
To download selected model from repository use:
```
client.repository.download(model_guid, <path_to_model>)
# To obtain serialized model first decompress it
!tar xzvf <path_to_model>
```
```
client.repository.download(model_guid)
```
Instead of downloading model can be also loaded directly to runtime using:
```
model = client.repository.load(model_guid)
# Loaded model can be used to perform prediction locally
# If loaded model was a scikit-learn pipeline we can use 'predict' method
model.predict(<test_data>)
```
```
client.repository.load(model_guid)
```
Delete model from repository by calling `client.repository.delete(model_guid)`.
```
client.repository.delete(model_guid)
```
<a id="functions"></a>
## 5. Manage functions
List existing functions. If you want to list only part of functions use `client.repository.list_functions(limit=n_functions)`.
```
client.repository.list_functions(limit=10)
```
Get function details by copying function uid from above cell and running `client.repository.get_details(function_guid)`.
```
function_guid = "PUT_YOUR_FUNCTION_GUID"
function_details = client.repository.get_details(function_guid)
print(function_details)
```
Delete function from repository by calling `client.repository.delete(function_guid)`.
```
client.repository.delete(function_guid)
```
<a id="experiments"></a>
## 6. Manage experiments
List existing experiments. If you want to list only part of experiments use `client.pipelines.list(limit=n_experiments)`.
```
client.experiments.list(limit=10)
```
Get experiments details. If you want to get only part of experiments details use `client.experiments.get_details(limit=n_experiments)`.
You can get each experiment details by calling `client.experiments.get_details()` and providing experiment id from listed experiments.
```
experiments_details = client.experiments.get_details()
print(experiments_details)
```
Delete all experiments. You can delete one experiment by calling `client.experiments.delete()` and providing experiment id from listed experiments.
```
for experiment in experiments_details['resources']:
client.experiments.delete(experiment['metadata']['id'])
```
<a id="trainings"></a>
## 7. Manage trainings
List existing trainings. If you want to list only part of trainings use `client.training.list(limit=n_trainings)`.
```
client.training.list(limit=10)
```
Get trainings details. If you want to get only part of trainings details use `client.training.get_details(limit=n_trainings)`.
You can get each training details by calling `client.training.get_details()` and providing training id from listed trainings.
```
trainings_details = client.training.get_details(limit=10)
print(trainings_details)
```
Delete all trainings. You can delete one training by calling `client.training.cancel()` and providing training id from listed trainings.
**Note** The `client.training.cancel()` method has `hard_delete` parameter. Please change it to:
- True - to delete the completed or canceled training runs.
- False - to cancel the currently running training run.
Default value is `False`.
```
for training in trainings_details['resources']:
client.training.cancel(training['metadata']['id'])
```
<a id="deployments"></a>
## 8. Manage deployments
List existing deployments. If you want to list only part of deployments use `client.deployments.list(limit=n_deployments)`.
```
client.deployments.list(limit=10)
```
Get deployments details. If you want to get only part of deployments details use `client.deployments.get_details(limit=n_deployments)`.
You can get each deployment details by calling `client.deployments.get_details()` and providing deployment id from listed deployments.
```
deployments_details = client.deployments.get_details()
print(deployments_details)
```
Delete all deployments. You can delete one deployment by calling `client.deployments.delete()` and providing deployment id from listed deployments.
```
for deployment in deployments_details['resources']:
client.deployments.delete(deployment['metadata']['id'])
```
<a id="summary"></a>
## 9. Summary and next steps
You successfully completed this notebook! You learned how to use ibm-watson-machine-learning client for Watson Machine Learning instance management and clean up.
Check out our <a href="https://dataplatform.cloud.ibm.com/docs/content/analyze-data/wml-setup.html" target="_blank" rel="noopener noreferrer">Online Documentation</a> for more samples, tutorials, documentation, how-tos, and blog posts.
### Authors
**Szymon Kucharczyk**, Software Engineer at IBM.
Copyright © 2020, 2021, 2022 IBM. This notebook and its source code are released under the terms of the MIT License.
|
github_jupyter
|
username = 'PASTE YOUR USERNAME HERE'
api_key = 'PASTE YOUR API_KEY HERE'
url = 'PASTE THE PLATFORM URL HERE'
wml_credentials = {
"username": username,
"apikey": api_key,
"url": url,
"instance_id": 'openshift',
"version": '4.0'
}
wml_credentials = {
"username": ***,
"password": ***,
"url": ***,
"instance_id": 'openshift',
"version": '4.0'
}
!pip install -U ibm-watson-machine-learning
from ibm_watson_machine_learning import APIClient
client = APIClient(wml_credentials)
space_id = 'PASTE YOUR SPACE ID HERE'
client.spaces.list(limit=10)
client.set.default_space(space_id)
client.pipelines.list(limit=10)
pipelines_details = client.pipelines.get_details(limit=10)
print(pipelines_details)
for pipeline in pipelines_details['resources']:
client.pipelines.delete(pipeline['metadata']['id'])
client.model_definitions.list(limit=10)
model_definition_guid = "PUT_YOUR_MODEL_DEFINITION_GUID"
model_definitions_details = client.model_definitions.get_details(model_definition_guid)
print(model_definitions_details)
client.model_definitions.delete(model_definition_guid)
client.repository.list_models(limit=10)
model_guid = "PUT_YOUR_MODEL_GUID"
model_details = client.repository.get_details(model_guid)
print(model_details)
client.repository.download(model_guid, <path_to_model>)
# To obtain serialized model first decompress it
!tar xzvf <path_to_model>
client.repository.download(model_guid)
model = client.repository.load(model_guid)
# Loaded model can be used to perform prediction locally
# If loaded model was a scikit-learn pipeline we can use 'predict' method
model.predict(<test_data>)
client.repository.load(model_guid)
client.repository.delete(model_guid)
client.repository.list_functions(limit=10)
function_guid = "PUT_YOUR_FUNCTION_GUID"
function_details = client.repository.get_details(function_guid)
print(function_details)
client.repository.delete(function_guid)
client.experiments.list(limit=10)
experiments_details = client.experiments.get_details()
print(experiments_details)
for experiment in experiments_details['resources']:
client.experiments.delete(experiment['metadata']['id'])
client.training.list(limit=10)
trainings_details = client.training.get_details(limit=10)
print(trainings_details)
for training in trainings_details['resources']:
client.training.cancel(training['metadata']['id'])
client.deployments.list(limit=10)
deployments_details = client.deployments.get_details()
print(deployments_details)
for deployment in deployments_details['resources']:
client.deployments.delete(deployment['metadata']['id'])
| 0.495361 | 0.933854 |
<a href="https://colab.research.google.com/github/gulgis/boot-igti-python/blob/main/magic_functions.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Magic methods
```
!pip install flake8
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
def __str__(self):
return f"{self.title} by {self.author}, costs $ {self.price}"
def __repr__(self):
return f"title={self.title},author={self.author},price={self.price}"
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
print(str(b1))
print(repr(b2))
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
# the __eq__ method checks for equality between two objects
def __eq__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return (
self.title == value.title and
self.author == value.author and
self.price == value.price
)
# the __ge__ establishes >= relationship with another obj
def __ge__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return self.price >= value.price
# the __lt__ establishes < relationship with another obj
def __lt__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return self.price < value.price
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
b3 = Book("War and Peace", "Leo Tolstoy", 39.95)
b4 = Book("To Kill a Mockingbird", "Harper Lee", 24.95)
# Check for equality
# print(b1 == b3)
# print(b1 == b4)
# Check for greater and lesser value
# print(b2 >= b1)
# print(b2 < b1)
# sort
books = [b1, b3, b2, b4]
books.sort()
print([book.title for book in books])
```
**the __getattr__ version of __getattribute__:**
this version of the function only gets called if the __getattribute__ version
* either doesn't exist, or
* if it throws an exception, or
* if the attribute doesn't actually exist.
```
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
self._discount = 0.1
def __str__(self):
return f"{self.title} by {self.author}, costs ${self.price}"
def __getattribute__(self, name):
if name == "price":
without_discount = super().__getattribute__("price")
discount = super().__getattribute__("_discount")
return without_discount - (without_discount * discount)
return super().__getattribute__(name)
def __setattr__(self, name, value):
if name == "price":
if type(value) is not float:
raise ValueError("The price must be a float")
return super().__setattr__(name, value)
def __getattr__(self, name):
return name + " is not here :O"
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
# b1.price = float(45)
# print(b1)
# print(b1.isbn)
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
def __str__(self):
return f"{self.title} by {self.author}, costs ${self.price}"
# method can be used to call the object like a function
def __call__(self, title, author, price):
self.title = title
self.author = author
self.price = price
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
# call the object as if it were a function
print(b1)
b1("How Democracies Die", "Steven Levitsky", 27.88)
print(b1)
```
|
github_jupyter
|
!pip install flake8
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
def __str__(self):
return f"{self.title} by {self.author}, costs $ {self.price}"
def __repr__(self):
return f"title={self.title},author={self.author},price={self.price}"
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
print(str(b1))
print(repr(b2))
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
# the __eq__ method checks for equality between two objects
def __eq__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return (
self.title == value.title and
self.author == value.author and
self.price == value.price
)
# the __ge__ establishes >= relationship with another obj
def __ge__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return self.price >= value.price
# the __lt__ establishes < relationship with another obj
def __lt__(self, value):
if not isinstance(value, Book):
raise ValueError("Can't compare two different objects!")
return self.price < value.price
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
b3 = Book("War and Peace", "Leo Tolstoy", 39.95)
b4 = Book("To Kill a Mockingbird", "Harper Lee", 24.95)
# Check for equality
# print(b1 == b3)
# print(b1 == b4)
# Check for greater and lesser value
# print(b2 >= b1)
# print(b2 < b1)
# sort
books = [b1, b3, b2, b4]
books.sort()
print([book.title for book in books])
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
self._discount = 0.1
def __str__(self):
return f"{self.title} by {self.author}, costs ${self.price}"
def __getattribute__(self, name):
if name == "price":
without_discount = super().__getattribute__("price")
discount = super().__getattribute__("_discount")
return without_discount - (without_discount * discount)
return super().__getattribute__(name)
def __setattr__(self, name, value):
if name == "price":
if type(value) is not float:
raise ValueError("The price must be a float")
return super().__setattr__(name, value)
def __getattr__(self, name):
return name + " is not here :O"
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
# b1.price = float(45)
# print(b1)
# print(b1.isbn)
class Book:
def __init__(self, title, author, price):
super().__init__()
self.title = title
self.author = author
self.price = price
def __str__(self):
return f"{self.title} by {self.author}, costs ${self.price}"
# method can be used to call the object like a function
def __call__(self, title, author, price):
self.title = title
self.author = author
self.price = price
b1 = Book("War and Peace", "Leo Tolstoy", 39.95)
b2 = Book("The Catcher in the Rye", "JD Salinger", 29.95)
# call the object as if it were a function
print(b1)
b1("How Democracies Die", "Steven Levitsky", 27.88)
print(b1)
| 0.607547 | 0.808408 |
# House Data exercise
https://openclassrooms.com/fr/courses/4011851-initiez-vous-au-machine-learning/6785036-entrainez-vous-a-entrainer-un-algorithme-de-machine-learning
```
# Import libraries
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import linear_model
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import mean_absolute_error, median_absolute_error, r2_score
import seaborn as sns
from pylab import *
# open datas
df = pd.read_csv("house_data.csv", sep=',', header=0)
df.head()
```
## Basic exploration of data
```
df.describe()
df.info()
df = df.dropna()
df["arrondissement"]= df["arrondissement"].astype(int)
df.info()
t = pd.DataFrame(df["arrondissement"])
j = t["arrondissement"].value_counts().sort_index()
y_pos = np.arange(j.shape[0])
data = list(j)
plt.bar(y_pos, data, align='center', alpha=0.5)
plt.xticks(y_pos, list(j.index))
plt.ylabel('arrondissement')
plt.xlabel('arrondissement number')
plt.title('Number of instance per arrondissement')
rcParams['figure.figsize'] = (12, 8)
plt.show()
```
Probably less efficient on the second arrondissement, else correctly distributed.
We now want to see if there are outliers and drop them.
```
ax = sns.scatterplot(x=df.index, y="price", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.ylabel('price in €')
plt.xlabel('index')
plt.title('Price repartition over arrondissement')
plt.show()
ax = sns.scatterplot(x=df.index, y="surface", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.ylabel('surface in €')
plt.xlabel('index')
plt.title('surface repartition over arrondissement')
plt.show()
ax = sns.scatterplot(x="surface", y="price", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.show()
price = df["price"]
removed_price = price.between(price.quantile(0), price.quantile(0.95))
index_kept = df[removed_price].index
df2 = df.loc[index_kept]
df2.info()
ax = sns.scatterplot(x="surface", y="price", hue="arrondissement", data=df2,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.show()
```
## Data preprocessing
In order to use categorical data in a regression, we will convert them to bag of words (One hot encoder)
```
oneHot = pd.get_dummies(df2["arrondissement"])
arr = pd.DataFrame(df2["arrondissement"])
df3 = df2.drop(labels="arrondissement", axis=1)
df4 = df3.merge(oneHot, left_index=True, right_index=True)
df4.head()
```
# Training regression
```
Y = pd.DataFrame(df4["price"])
X = df4.drop(labels="price", axis=1)
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2)
print("Training set count X:{}/Y:{}, Test set count X:{}/Y:{}".format(
int(X_train.shape[0]), int(Y_train.shape[0]), int(X_test.shape[0]), int(Y_test.shape[0])))
LinearRegression = linear_model.LinearRegression()
LinearRegression.fit(X_train, Y_train)
prediction = LinearRegression.predict(X_test)
print('Coefficients: \n', LinearRegression.coef_)
def evaluation_linearRegression(Y_test,prediction):
# The mean squared error
print("Mean absolute error: %.2f"% mean_absolute_error(Y_test, prediction))
print("Median absolute error: %.2f"% median_absolute_error(Y_test, prediction))
print('Variance score: %.2f' % r2_score(Y_test, prediction))
evaluation_linearRegression(Y_test,prediction)
LinearRegression = linear_model.LinearRegression()
scores = cross_val_score(LinearRegression, X, Y, cv=3)
print("Variance : {}".format(scores.mean()))
```
Our initial score is about 3/4
## Optimizing the model
We will now try to improve our model by separating different categories
```
cat = []
for i in df2["arrondissement"].value_counts().sort_index().index:
cat.append(df2[df2["arrondissement"] == i].drop(labels="arrondissement", axis=1))
scores_cat = []
for c in cat:
Y = pd.DataFrame(c["price"])
X = c.drop(labels="price", axis=1)
LinearRegression = linear_model.LinearRegression()
scores = cross_val_score(LinearRegression, X, Y, cv=3)
scores_cat.append(scores.mean())
j = list(df2["arrondissement"].value_counts().sort_index().index)
y_pos = np.arange(len(j))
data = scores_cat
plt.bar(y_pos, data, align='center', alpha=0.5)
plt.xticks(y_pos, list(j))
plt.ylabel('score R2')
plt.xlabel('arrondissement')
plt.title('Linear Regression scores for each arrondissement')
rcParams['figure.figsize'] = (12, 8)
plt.show()
print(np.array(scores_cat).mean())
```
Our optimization didn't improve the model, we can conclude that due to the poor amount of data, the initial linear regression seems to be better than the categorical linear regression approach
|
github_jupyter
|
# Import libraries
import math
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import linear_model
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import mean_absolute_error, median_absolute_error, r2_score
import seaborn as sns
from pylab import *
# open datas
df = pd.read_csv("house_data.csv", sep=',', header=0)
df.head()
df.describe()
df.info()
df = df.dropna()
df["arrondissement"]= df["arrondissement"].astype(int)
df.info()
t = pd.DataFrame(df["arrondissement"])
j = t["arrondissement"].value_counts().sort_index()
y_pos = np.arange(j.shape[0])
data = list(j)
plt.bar(y_pos, data, align='center', alpha=0.5)
plt.xticks(y_pos, list(j.index))
plt.ylabel('arrondissement')
plt.xlabel('arrondissement number')
plt.title('Number of instance per arrondissement')
rcParams['figure.figsize'] = (12, 8)
plt.show()
ax = sns.scatterplot(x=df.index, y="price", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.ylabel('price in €')
plt.xlabel('index')
plt.title('Price repartition over arrondissement')
plt.show()
ax = sns.scatterplot(x=df.index, y="surface", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.ylabel('surface in €')
plt.xlabel('index')
plt.title('surface repartition over arrondissement')
plt.show()
ax = sns.scatterplot(x="surface", y="price", hue="arrondissement", data=df,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.show()
price = df["price"]
removed_price = price.between(price.quantile(0), price.quantile(0.95))
index_kept = df[removed_price].index
df2 = df.loc[index_kept]
df2.info()
ax = sns.scatterplot(x="surface", y="price", hue="arrondissement", data=df2,
palette=['green','orange','brown','dodgerblue','red'], legend='full')
plt.show()
oneHot = pd.get_dummies(df2["arrondissement"])
arr = pd.DataFrame(df2["arrondissement"])
df3 = df2.drop(labels="arrondissement", axis=1)
df4 = df3.merge(oneHot, left_index=True, right_index=True)
df4.head()
Y = pd.DataFrame(df4["price"])
X = df4.drop(labels="price", axis=1)
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2)
print("Training set count X:{}/Y:{}, Test set count X:{}/Y:{}".format(
int(X_train.shape[0]), int(Y_train.shape[0]), int(X_test.shape[0]), int(Y_test.shape[0])))
LinearRegression = linear_model.LinearRegression()
LinearRegression.fit(X_train, Y_train)
prediction = LinearRegression.predict(X_test)
print('Coefficients: \n', LinearRegression.coef_)
def evaluation_linearRegression(Y_test,prediction):
# The mean squared error
print("Mean absolute error: %.2f"% mean_absolute_error(Y_test, prediction))
print("Median absolute error: %.2f"% median_absolute_error(Y_test, prediction))
print('Variance score: %.2f' % r2_score(Y_test, prediction))
evaluation_linearRegression(Y_test,prediction)
LinearRegression = linear_model.LinearRegression()
scores = cross_val_score(LinearRegression, X, Y, cv=3)
print("Variance : {}".format(scores.mean()))
cat = []
for i in df2["arrondissement"].value_counts().sort_index().index:
cat.append(df2[df2["arrondissement"] == i].drop(labels="arrondissement", axis=1))
scores_cat = []
for c in cat:
Y = pd.DataFrame(c["price"])
X = c.drop(labels="price", axis=1)
LinearRegression = linear_model.LinearRegression()
scores = cross_val_score(LinearRegression, X, Y, cv=3)
scores_cat.append(scores.mean())
j = list(df2["arrondissement"].value_counts().sort_index().index)
y_pos = np.arange(len(j))
data = scores_cat
plt.bar(y_pos, data, align='center', alpha=0.5)
plt.xticks(y_pos, list(j))
plt.ylabel('score R2')
plt.xlabel('arrondissement')
plt.title('Linear Regression scores for each arrondissement')
rcParams['figure.figsize'] = (12, 8)
plt.show()
print(np.array(scores_cat).mean())
| 0.437824 | 0.917303 |
# Working with JAX numpy and calculating perplexity: Ungraded Lecture Notebook
Normally you would import `numpy` and rename it as `np`.
However in this week's assignment you will notice that this convention has been changed.
Now standard `numpy` is not renamed and `trax.fastmath.numpy` is renamed as `np`.
The rationale behind this change is that you will be using Trax's numpy (which is compatible with JAX) far more often. Trax's numpy supports most of the same functions as the regular numpy so the change won't be noticeable in most cases.
```
import numpy
import trax
import trax.fastmath.numpy as np
# Setting random seeds
trax.supervised.trainer_lib.init_random_number_generators(32)
numpy.random.seed(32)
```
One important change to take into consideration is that the types of the resulting objects will be different depending on the version of numpy. With regular numpy you get `numpy.ndarray` but with Trax's numpy you will get `jax.interpreters.xla.DeviceArray`. These two types map to each other. So if you find some error logs mentioning DeviceArray type, don't worry about it, treat it like you would treat an ndarray and march ahead.
You can get a randomized numpy array by using the `numpy.random.random()` function.
This is one of the functionalities that Trax's numpy does not currently support in the same way as the regular numpy.
```
numpy_array = numpy.random.random((5,10))
print(f"The regular numpy array looks like this:\n\n {numpy_array}\n")
print(f"It is of type: {type(numpy_array)}")
```
You can easily cast regular numpy arrays or lists into trax numpy arrays using the `trax.fastmath.numpy.array()` function:
```
trax_numpy_array = np.array(numpy_array)
print(f"The trax numpy array looks like this:\n\n {trax_numpy_array}\n")
print(f"It is of type: {type(trax_numpy_array)}")
```
Hope you now understand the differences (and similarities) between these two versions and numpy. **Great!**
The previous section was a quick look at Trax's numpy. However this notebook also aims to teach you how you can calculate the perplexity of a trained model.
## Calculating Perplexity
The perplexity is a metric that measures how well a probability model predicts a sample and it is commonly used to evaluate language models. It is defined as:
$$P(W) = \sqrt[N]{\prod_{i=1}^{N} \frac{1}{P(w_i| w_1,...,w_{n-1})}}$$
As an implementation hack, you would usually take the log of that formula (to enable us to use the log probabilities we get as output of our `RNN`, convert exponents to products, and products into sums which makes computations less complicated and computationally more efficient). You should also take care of the padding, since you do not want to include the padding when calculating the perplexity (because we do not want to have a perplexity measure artificially good). The algebra behind this process is explained next:
$$log P(W) = {log\big(\sqrt[N]{\prod_{i=1}^{N} \frac{1}{P(w_i| w_1,...,w_{n-1})}}\big)}$$
$$ = {log\big({\prod_{i=1}^{N} \frac{1}{P(w_i| w_1,...,w_{n-1})}}\big)^{\frac{1}{N}}}$$
$$ = {log\big({\prod_{i=1}^{N}{P(w_i| w_1,...,w_{n-1})}}\big)^{-\frac{1}{N}}} $$
$$ = -\frac{1}{N}{log\big({\prod_{i=1}^{N}{P(w_i| w_1,...,w_{n-1})}}\big)} $$
$$ = -\frac{1}{N}{\big({\sum_{i=1}^{N}{logP(w_i| w_1,...,w_{n-1})}}\big)} $$
You will be working with a real example from this week's assignment. The example is made up of:
- `predictions` : batch of tensors corresponding to lines of text predicted by the model.
- `targets` : batch of actual tensors corresponding to lines of text.
```
from trax import layers as tl
# Load from .npy files
predictions = numpy.load('predictions.npy')
targets = numpy.load('targets.npy')
# Cast to jax.interpreters.xla.DeviceArray
predictions = np.array(predictions)
targets = np.array(targets)
# Print shapes
print(f'predictions has shape: {predictions.shape}')
print(f'targets has shape: {targets.shape}')
```
Notice that the predictions have an extra dimension with the same length as the size of the vocabulary used.
Because of this you will need a way of reshaping `targets` to match this shape. For this you can use `trax.layers.one_hot()`.
Notice that `predictions.shape[-1]` will return the size of the last dimension of `predictions`.
```
reshaped_targets = tl.one_hot(targets, predictions.shape[-1]) #trax's one_hot function takes the input as one_hot(x, n_categories, dtype=optional)
print(f'reshaped_targets has shape: {reshaped_targets.shape}')
```
By calculating the product of the predictions and the reshaped targets and summing across the last dimension, the total log perplexity can be computed:
```
total_log_ppx = np.sum(predictions * reshaped_targets, axis= -1)
```
Now you will need to account for the padding so this metric is not artificially deflated (since a lower perplexity means a better model). For identifying which elements are padding and which are not, you can use `np.equal()` and get a tensor with `1s` in the positions of actual values and `0s` where there are paddings.
```
non_pad = 1.0 - np.equal(targets, 0)
print(f'non_pad has shape: {non_pad.shape}\n')
print(f'non_pad looks like this: \n\n {non_pad}')
```
By computing the product of the total log perplexity and the non_pad tensor we remove the effect of padding on the metric:
```
real_log_ppx = total_log_ppx * non_pad
print(f'real perplexity still has shape: {real_log_ppx.shape}')
```
You can check the effect of filtering out the padding by looking at the two log perplexity tensors:
```
print(f'log perplexity tensor before filtering padding: \n\n {total_log_ppx}\n')
print(f'log perplexity tensor after filtering padding: \n\n {real_log_ppx}')
```
To get a single average log perplexity across all the elements in the batch you can sum across both dimensions and divide by the number of elements. Notice that the result will be the negative of the real log perplexity of the model:
```
log_ppx = np.sum(real_log_ppx) / np.sum(non_pad)
log_ppx = -log_ppx
print(f'The log perplexity and perplexity of the model are respectively: {log_ppx} and {np.exp(log_ppx)}')
```
**Congratulations on finishing this lecture notebook!** Now you should have a clear understanding of how to work with Trax's numpy and how to compute the perplexity to evaluate your language models. **Keep it up!**
|
github_jupyter
|
import numpy
import trax
import trax.fastmath.numpy as np
# Setting random seeds
trax.supervised.trainer_lib.init_random_number_generators(32)
numpy.random.seed(32)
numpy_array = numpy.random.random((5,10))
print(f"The regular numpy array looks like this:\n\n {numpy_array}\n")
print(f"It is of type: {type(numpy_array)}")
trax_numpy_array = np.array(numpy_array)
print(f"The trax numpy array looks like this:\n\n {trax_numpy_array}\n")
print(f"It is of type: {type(trax_numpy_array)}")
from trax import layers as tl
# Load from .npy files
predictions = numpy.load('predictions.npy')
targets = numpy.load('targets.npy')
# Cast to jax.interpreters.xla.DeviceArray
predictions = np.array(predictions)
targets = np.array(targets)
# Print shapes
print(f'predictions has shape: {predictions.shape}')
print(f'targets has shape: {targets.shape}')
reshaped_targets = tl.one_hot(targets, predictions.shape[-1]) #trax's one_hot function takes the input as one_hot(x, n_categories, dtype=optional)
print(f'reshaped_targets has shape: {reshaped_targets.shape}')
total_log_ppx = np.sum(predictions * reshaped_targets, axis= -1)
non_pad = 1.0 - np.equal(targets, 0)
print(f'non_pad has shape: {non_pad.shape}\n')
print(f'non_pad looks like this: \n\n {non_pad}')
real_log_ppx = total_log_ppx * non_pad
print(f'real perplexity still has shape: {real_log_ppx.shape}')
print(f'log perplexity tensor before filtering padding: \n\n {total_log_ppx}\n')
print(f'log perplexity tensor after filtering padding: \n\n {real_log_ppx}')
log_ppx = np.sum(real_log_ppx) / np.sum(non_pad)
log_ppx = -log_ppx
print(f'The log perplexity and perplexity of the model are respectively: {log_ppx} and {np.exp(log_ppx)}')
| 0.67694 | 0.985773 |
# Overview of `tobler`s Interpolation Methods
```
import geopandas as gpd
import matplotlib.pyplot as plt
from tobler.dasymetric import masked_area_interpolate
from tobler.model import glm
from tobler.area_weighted import area_interpolate
from libpysal.examples import load_example
```
Let's say we want to represent the poverty rate and male employment using zip code geographies, but the only data available is at the census-tract level. The `tobler` package provides several different ways to estimate data collected at one geography using the boundaries of a different geography.
## Load Data
First we'll grab two geodataframes representing chaleston, sc--one with census tracts and the other with zctas
```
c1 = load_example('Charleston1')
c2 = load_example('Charleston2')
```
Since areal interpolation uses spatial ovelays, we should make sure to use a reasonable projection. If a `tobler` detects that a user is performing an analysis on an unprojected geodataframe, it will do its best to reproject the data into the appropriate UTM zone to ensure accuracy. All the same, its best to set the CRS explicitly
```
crs = 6569 # https://epsg.io/6569
tracts = gpd.read_file(c1.get_path('sc_final_census2.shp')).to_crs(crs)
zip_codes = gpd.read_file(c2.get_path('CharlestonMSA2.shp')).to_crs(crs)
fig, ax = plt.subplots(1,2, figsize=(14,7))
tracts.plot(ax=ax[0])
zip_codes.plot(ax=ax[1])
for ax in ax:
ax.axis('off')
tracts.columns
tracts['pct_poverty'] = tracts.POV_POP/tracts.POV_TOT
```
## Areal Interpolation
The simplest technique available in `tobler` is simple areal interpolation in which variables from the source data are weighted according to the overlap between source and target polygons, then reaggregated to fit the target polygon geometries
```
results = area_interpolate(source_df=tracts, target_df=zip_codes, intensive_variables=['pct_poverty'], extensive_variables=['EMP_MALE'])
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
```
---
## Dasymetric Interpolation
To help improve accuracy in interpolation we can use axiliary information to mask out areas we know aren't inhabited. For example we can use raster data like <https://www.mrlc.gov/national-land-cover-database-nlcd-2016> to mask out uninhabited land uses. To do so, we need to provide a path to the raster and a list of pixel values that are considered developed. Default values are taken from NLCD
`tobler` can accept any kind of raster data that can be read by rasterio, so you can provide your own, or download directly from NLCD linked above. Alternatively, you can download a compressed version of the NLCD we host in the spatialucr quilt bucket using three short lines of code:
```
from quilt3 import Package
p = Package.browse("rasters/nlcd", "s3://spatial-ucr")
p["nlcd_2011.tif"].fetch()
results = masked_area_interpolate(raster="nlcd_2011.tif",
source_df=tracts,
target_df=zip_codes,
intensive_variables=['pct_poverty'],
extensive_variables=['EMP_MALE'])
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
```
---
## Model-Based Interpolation
Rather than assume that extensive and intensive variables are distributed uniformly throughout developed land, tobler can also use a model-based approach to assign weights to different cell types, e.g. if we assume that "high intensity development" raster cells are likely to contain more people than "low intensity development", even though we definitely want to allocate population to both types. In these cases, `tobler` consumes a raster layer as additional input and a patsy-style model formula string. A default formula string is provided but results can be improved by fitting a model more appropriate for local settings
Unlike area weighted and dasymetric techniques, model-based interpolation estimates only a single variable at a time
```
emp_results = glm(raster="nlcd_2011.tif",source_df=tracts, target_df=zip_codes, variable='EMP_MALE', )
fig, ax = plt.subplots(1,2, figsize=(14,7))
emp_results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
pov_results = glm(raster="nlcd_2011.tif",source_df=tracts, target_df=zip_codes, variable='pct_poverty', )
fig, ax = plt.subplots(1,2, figsize=(14,7))
pov_results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
```
conda remove --name myenv --allFrom the maps, it looks like the model-based approach is better at estimating the extensive variable (male employment) than the intensive variable (poverty rate), at least using the default regression equation. That's not a surprising result, though, because its much easier to intuit the relationship between land-use types and total employment levels than land-use type and poverty rate
|
github_jupyter
|
import geopandas as gpd
import matplotlib.pyplot as plt
from tobler.dasymetric import masked_area_interpolate
from tobler.model import glm
from tobler.area_weighted import area_interpolate
from libpysal.examples import load_example
c1 = load_example('Charleston1')
c2 = load_example('Charleston2')
crs = 6569 # https://epsg.io/6569
tracts = gpd.read_file(c1.get_path('sc_final_census2.shp')).to_crs(crs)
zip_codes = gpd.read_file(c2.get_path('CharlestonMSA2.shp')).to_crs(crs)
fig, ax = plt.subplots(1,2, figsize=(14,7))
tracts.plot(ax=ax[0])
zip_codes.plot(ax=ax[1])
for ax in ax:
ax.axis('off')
tracts.columns
tracts['pct_poverty'] = tracts.POV_POP/tracts.POV_TOT
results = area_interpolate(source_df=tracts, target_df=zip_codes, intensive_variables=['pct_poverty'], extensive_variables=['EMP_MALE'])
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
from quilt3 import Package
p = Package.browse("rasters/nlcd", "s3://spatial-ucr")
p["nlcd_2011.tif"].fetch()
results = masked_area_interpolate(raster="nlcd_2011.tif",
source_df=tracts,
target_df=zip_codes,
intensive_variables=['pct_poverty'],
extensive_variables=['EMP_MALE'])
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
fig, ax = plt.subplots(1,2, figsize=(14,7))
results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
emp_results = glm(raster="nlcd_2011.tif",source_df=tracts, target_df=zip_codes, variable='EMP_MALE', )
fig, ax = plt.subplots(1,2, figsize=(14,7))
emp_results.plot('EMP_MALE', scheme='quantiles', ax=ax[0])
tracts.plot('EMP_MALE', scheme='quantiles', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Male Employment (extensive)')
pov_results = glm(raster="nlcd_2011.tif",source_df=tracts, target_df=zip_codes, variable='pct_poverty', )
fig, ax = plt.subplots(1,2, figsize=(14,7))
pov_results.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[0])
tracts.plot('pct_poverty', scheme='quantiles', cmap='magma', ax=ax[1])
ax[0].set_title('interpolated')
ax[1].set_title('original')
for ax in ax:
ax.axis('off')
fig.suptitle('Poverty Rate (intensive)')
| 0.631822 | 0.971238 |
# First BigQuery ML models for Taxifare Prediction
In this notebook, we will use BigQuery ML to build our first models for taxifare prediction.BigQuery ML provides a fast way to build ML models on large structured and semi-structured datasets.
## Learning Objectives
1. Choose the correct BigQuery ML model type and specify options
2. Evaluate the performance of your ML model
3. Improve model performance through data quality cleanup
4. Create a Deep Neural Network (DNN) using SQL
Each learning objective will correspond to a __#TODO__ in the [student lab notebook](../labs/first_model.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
We'll start by creating a dataset to hold all the models we create in BigQuery
```
%%bash
export PROJECT=$(gcloud config list project --format "value(core.project)")
echo "Your current GCP Project Name is: "$PROJECT
%%bash
!pip install tensorflow==2.1.0 --user
```
Let's make sure we install the necessary version of tensorflow. After doing the pip install above, click __Restart the kernel__ on the notebook so that the Python environment picks up the new packages.
```
import os
PROJECT = "qwiklabs-gcp-bdc77450c97b4bf6" # REPLACE WITH YOUR PROJECT NAME
REGION = "us-central1" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1
import tensorflow as tf
print("TensorFlow version: ",tf.version.VERSION)
# Do not change these
os.environ["PROJECT"] = PROJECT
os.environ["REGION"] = REGION
os.environ["BUCKET"] = PROJECT # DEFAULT BUCKET WILL BE PROJECT ID
if PROJECT == "your-gcp-project-here":
print("Don't forget to update your PROJECT name! Currently:", PROJECT)
```
## Create a BigQuery Dataset and Google Cloud Storage Bucket
A BigQuery dataset is a container for tables, views, and models built with BigQuery ML. Let's create one called __serverlessml__ if we have not already done so in an earlier lab. We'll do the same for a GCS bucket for our project too.
```
%%bash
## Create a BigQuery dataset for serverlessml if it doesn't exist
datasetexists=$(bq ls -d | grep -w serverlessml)
if [ -n "$datasetexists" ]; then
echo -e "BigQuery dataset already exists, let's not recreate it."
else
echo "Creating BigQuery dataset titled: serverlessml"
bq --location=US mk --dataset \
--description 'Taxi Fare' \
$PROJECT:serverlessml
echo "\nHere are your current datasets:"
bq ls
fi
## Create GCS bucket if it doesn't exist already...
exists=$(gsutil ls -d | grep -w gs://${PROJECT}/)
if [ -n "$exists" ]; then
echo -e "Bucket exists, let's not recreate it."
else
echo "Creating a new GCS bucket."
gsutil mb -l ${REGION} gs://${PROJECT}
echo "\nHere are your current buckets:"
gsutil ls
fi
```
## Model 1: Raw data
Let's build a model using just the raw data. It's not going to be very good, but sometimes it is good to actually experience this.
The model will take a minute or so to train. When it comes to ML, this is blazing fast.
```
%%bigquery
CREATE OR REPLACE MODEL serverlessml.model1_rawdata
OPTIONS(input_label_cols=['fare_amount'], model_type='linear_reg') AS
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 1
```
Once the training is done, visit the [BigQuery Cloud Console](https://console.cloud.google.com/bigquery) and look at the model that has been trained. Then, come back to this notebook.
Note that BigQuery automatically split the data we gave it, and trained on only a part of the data and used the rest for evaluation. We can look at eval statistics on that held-out data:
```
%%bigquery
SELECT * FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata)
```
Let's report just the error we care about, the Root Mean Squared Error (RMSE)
```
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata)
```
We told you it was not going to be good! Recall that our heuristic got 8.13, and our target is $6.
Note that the error is going to depend on the dataset that we evaluate it on.
We can also evaluate the model on our own held-out benchmark/test dataset, but we shouldn't make a habit of this (we want to keep our benchmark dataset as the final evaluation, not make decisions using it all along the way. If we do that, our test dataset won't be truly independent).
```
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata,
(
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 2
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
))
```
## Model 2: Apply data cleanup
Recall that we did some data cleanup in the previous lab. Let's do those before training.
This is a dataset that we will need quite frequently in this notebook, so let's extract it first.
```
%%bigquery
CREATE OR REPLACE TABLE serverlessml.cleaned_training_data AS
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 1
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
%%bigquery
-- LIMIT 0 is a free query; this allows us to check that the table exists.
SELECT * FROM serverlessml.cleaned_training_data
LIMIT 0
%%bigquery
CREATE OR REPLACE MODEL serverlessml.model2_cleanup
OPTIONS(input_label_cols=['fare_amount'], model_type='linear_reg') AS
SELECT
*
FROM
serverlessml.cleaned_training_data
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model2_cleanup)
```
## Model 3: More sophisticated models
What if we try a more sophisticated model? Let's try Deep Neural Networks (DNNs) in BigQuery:
### DNN
To create a DNN, simply specify __dnn_regressor__ for the model_type and add your hidden layers.
```
%%bigquery
-- This model type is in alpha, so it may not work for you yet. This training takes on the order of 15 minutes.
CREATE OR REPLACE MODEL serverlessml.model3b_dnn
OPTIONS(input_label_cols=['fare_amount'], model_type='dnn_regressor', hidden_units=[32, 8]) AS
SELECT
*
FROM
serverlessml.cleaned_training_data
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model3b_dnn)
```
Nice!
## Evaluate DNN on benchmark dataset
Let's use the same validation dataset to evaluate -- remember that evaluation metrics depend on the dataset. You can not compare two models unless you have run them on the same withheld data.
```
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse
FROM ML.EVALUATE(MODEL serverlessml.model3b_dnn,
(
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_datetime,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers,
'unused' AS key
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 10000)) = 2
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
))
```
Wow! Later in this sequence of notebooks, we will get to below $4, but this is quite good, for very little work.
In this notebook, we showed you how to use BigQuery ML to quickly build ML models. We will come back to BigQuery ML when we want to experiment with different types of feature engineering. The speed of BigQuery ML is very attractive for development.
Copyright 2019 Google Inc.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
|
github_jupyter
|
%%bash
export PROJECT=$(gcloud config list project --format "value(core.project)")
echo "Your current GCP Project Name is: "$PROJECT
%%bash
!pip install tensorflow==2.1.0 --user
import os
PROJECT = "qwiklabs-gcp-bdc77450c97b4bf6" # REPLACE WITH YOUR PROJECT NAME
REGION = "us-central1" # REPLACE WITH YOUR BUCKET REGION e.g. us-central1
import tensorflow as tf
print("TensorFlow version: ",tf.version.VERSION)
# Do not change these
os.environ["PROJECT"] = PROJECT
os.environ["REGION"] = REGION
os.environ["BUCKET"] = PROJECT # DEFAULT BUCKET WILL BE PROJECT ID
if PROJECT == "your-gcp-project-here":
print("Don't forget to update your PROJECT name! Currently:", PROJECT)
%%bash
## Create a BigQuery dataset for serverlessml if it doesn't exist
datasetexists=$(bq ls -d | grep -w serverlessml)
if [ -n "$datasetexists" ]; then
echo -e "BigQuery dataset already exists, let's not recreate it."
else
echo "Creating BigQuery dataset titled: serverlessml"
bq --location=US mk --dataset \
--description 'Taxi Fare' \
$PROJECT:serverlessml
echo "\nHere are your current datasets:"
bq ls
fi
## Create GCS bucket if it doesn't exist already...
exists=$(gsutil ls -d | grep -w gs://${PROJECT}/)
if [ -n "$exists" ]; then
echo -e "Bucket exists, let's not recreate it."
else
echo "Creating a new GCS bucket."
gsutil mb -l ${REGION} gs://${PROJECT}
echo "\nHere are your current buckets:"
gsutil ls
fi
%%bigquery
CREATE OR REPLACE MODEL serverlessml.model1_rawdata
OPTIONS(input_label_cols=['fare_amount'], model_type='linear_reg') AS
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 1
%%bigquery
SELECT * FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata)
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata)
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model1_rawdata,
(
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 2
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
))
%%bigquery
CREATE OR REPLACE TABLE serverlessml.cleaned_training_data AS
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 100000)) = 1
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
%%bigquery
-- LIMIT 0 is a free query; this allows us to check that the table exists.
SELECT * FROM serverlessml.cleaned_training_data
LIMIT 0
%%bigquery
CREATE OR REPLACE MODEL serverlessml.model2_cleanup
OPTIONS(input_label_cols=['fare_amount'], model_type='linear_reg') AS
SELECT
*
FROM
serverlessml.cleaned_training_data
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model2_cleanup)
%%bigquery
-- This model type is in alpha, so it may not work for you yet. This training takes on the order of 15 minutes.
CREATE OR REPLACE MODEL serverlessml.model3b_dnn
OPTIONS(input_label_cols=['fare_amount'], model_type='dnn_regressor', hidden_units=[32, 8]) AS
SELECT
*
FROM
serverlessml.cleaned_training_data
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse FROM ML.EVALUATE(MODEL serverlessml.model3b_dnn)
%%bigquery
SELECT SQRT(mean_squared_error) AS rmse
FROM ML.EVALUATE(MODEL serverlessml.model3b_dnn,
(
SELECT
(tolls_amount + fare_amount) AS fare_amount,
pickup_datetime,
pickup_longitude AS pickuplon,
pickup_latitude AS pickuplat,
dropoff_longitude AS dropofflon,
dropoff_latitude AS dropofflat,
passenger_count*1.0 AS passengers,
'unused' AS key
FROM `nyc-tlc.yellow.trips`
WHERE ABS(MOD(FARM_FINGERPRINT(CAST(pickup_datetime AS STRING)), 10000)) = 2
AND
trip_distance > 0
AND fare_amount >= 2.5
AND pickup_longitude > -78
AND pickup_longitude < -70
AND dropoff_longitude > -78
AND dropoff_longitude < -70
AND pickup_latitude > 37
AND pickup_latitude < 45
AND dropoff_latitude > 37
AND dropoff_latitude < 45
AND passenger_count > 0
))
| 0.343232 | 0.949809 |
# Documentation
## Reading images and videos
```
import cv2 as cv
import numpy as np
img = cv.imread("Files/tatsuya.jpg") #takes path or image
cv.imshow("Tatsuya Shiba",img)
cv.waitKey(0) #makes the shell wait for an infinite amt of time till a key is pressed.
capture = cv.VideoCapture("C:/Users/admin/Videos/anime/inari kon kon/EP.1.480p.mp4")
while True:
isTrue, frame = capture.read()
cv.imshow("Video",frame)
if cv.waitKey(20) & 0xFF==ord("d"):
break
capture.release()
cv.destroyAllWindows()
```
## Resizing & Rescaling
```
#image video and live video
def rescaleFrame(frame,scale=0.7):
width = int(frame.shape[1]*scale)
height = int(frame.shape[0]*scale)
dimensions = (width,height)
return cv.resize(frame,dimensions,interpolation=cv.INTER_AREA)
#live video
def changeRes(width,height):
capture.set(3,width)
capture.set(4,height)
img_resized = rescaleFrame(img,scale=0.7)
cv.imshow("tatsuya_img",img_resized)
cv.waitKey(0)
capture = cv.VideoCapture("C:/Users/admin/Videos/anime/inari kon kon/EP.1.480p.mp4")
while True:
isTrue, frame = capture.read()
frame_resize = rescaleFrame(frame,scale=0.7)
cv.imshow("Video",frame_resize)
if cv.waitKey(20) & 0xFF==ord("d"):
break
capture.release()
cv.destroyAllWindows()
```
## Draw shapes and write text
```
import numpy as np
blank_1 = np.zeros((500,500,3),dtype = 'uint8')
blank_2 = np.zeros((500,500,3),dtype = 'uint8')
blank_3 = np.zeros((500,500,3),dtype = 'uint8')
blank_4 = np.zeros((500,500,3),dtype = 'uint8')
cv.imshow("blank",blank_1)
#painting green
blank_1[:]=0,255,0
cv.imshow("green",blank_1)
#painting red
blank_2[:]=0,0,255
cv.imshow("red",blank_2)
#painting blue
blank_3[:]=255,0,0
cv.imshow("blue",blank_3)
#painting a portion only
blank_4[200:300, 300:400] = 0,0,255
cv.imshow("portion",blank_4)
cv.waitKey(0)
#rectangle
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(250,250),(0,255,0),thickness=-1)
cv.imshow("Rectangle",blank)
cv.waitKey(0)
#circle
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(blank.shape[1]//2,blank.shape[0]//2),(0,255,0),thickness=-1)
cv.imshow("Circle",blank)
cv.waitKey(0)
#draw a line
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(blank.shape[1]//2,blank.shape[0]//2),(0,255,0))
cv.imshow("Line",blank)
cv.waitKey(0)
# writeing
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.putText(blank,"Hello",(225,225),cv.FONT_HERSHEY_TRIPLEX,1.0,(0,255,0),2)
cv.imshow("Line",blank)
cv.waitKey(0)
```
## 5 essential functions
```
img = cv.imread("C:/Users/admin/Pictures/anime cool stuff/tatsuya.jpg") #takes path
cv.imshow("Tatsuya Shiba",img)
cv.waitKey(0) #makes the shell wait for an infinite amt of time till a key is pressed.
# Converting an image to GrayScale
gray = cv.cvtColor(img,cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba but in gray",gray)
cv.waitKey(0)
# Blur
blur = cv.GaussianBlur(img,(3,3),cv.BODER_DEFAULT)
cv.imshow("Tatsuya Shiba but in gray",blur)
cv.waitKey(0)
# Edge cascade
canny = cv.Canny(img,125,175)
cv.imshow("Tatsuya Shiba but only edges",canny)
cv.waitKey(0)
#dilation
dilated = cv.dilate(canny,(7,7),iterations=5)
cv.imshow("Tatsuya Shiba with dilated edges",dilated)
cv.waitKey(0)
# eroding
eroded = cv.erode(dilated,(3,3),iteration=2)
cv.imshow("Tatsuya Shiba with dilated edges",eroded)
cv.waitKey(0)
#resize
resized = cv.resize(img,(500,500), interpolation=cv.INTER_CUBIC)
cv.imshow("Tatsuya Shiba resized",resized)
cv.waitKey(0)
#Cropping
cropped = img[50:200,200:400]
cv.imshow("Cropped",cropped)
```
## Image Transformation in openCV
```
# Translation (shifting x or y axxis or both)
def translate(img,x,y):
transMat = np.float32([[1,0,x],[0,1,y]])
dimensions = (img.shape[1],img.shape[0])
return cv.warpAffine(img,transMat,dimensions)
#a -x--> left
#a +x--> right
#a +y--> down
#a -y--> Up
translated = translate(img,100,100)
cv.imshow("translated",translated)
cv.waitKey(0)
#Rotation
def rotate(imf, angle, rotPoint = None):
(height,width)=img.shape[:2]
if rotPoint is None:
rotPoint = (width//2,height//2)
rotMat = cv.getRotationMatrix2D(rotPoint, angle, 1.0)
dimensions = (width,height)
return cv.warpAffine(img, rotMat, dimensions)
rotated = rotate(img,45)
cv.imshow("rotated",rotated)
cv.waitKey(0)
#Resizing
resized =cv.resize(img, (500,500), interpolation=cv.INTER_CUBIC)
cv.imshow("Resized", resized)
#Flipping
flip = cv.flip(img, 0)
# 0 ->vertically via x-axis
# 1 -> horiziontally via y-axis
# -1 -> both
cv.imshow("flip",flip)
cv.waitKey(0)
#Cropping
cropped = img[200:400,300:400]
cv.imshow("crop",cropped)
cv.waitKey(0)
```
## Contour Detection
### Method 1 -
```
gray = cv.cvtColor(img, cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba",gray)
#blur = cv.GaussianBlur(gray, (5,5), cv.BODER_DEFAULT)
#cv.imshow("BLUR",blur)
#blur the image in case there are too many contours
canny = cv.Canny(img,125,175)
cv.imshow("Tatsuya Shiba",canny)
contours, heiarchies = cv.findContours(canny, cv.RETR_LIST, cv.CHAIN_APPROX_NONE)
#RETR_LIST = all
#RETR_EXTERNAL = boundaries
#RETR_TREE = contours in a heiarchy
#CHAIN_APPROX_NONE = returns all
#CHAIN_APPROX_SIMPLE = compreses the contours
print(f'There are {len(contours)} contours')
cv.waitKey(0)
```
### Method 2 -
```
gray = cv.cvtColor(img, cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba",gray)
ret, thresh = cv.threshold(gray,125,255, cv.THRESH_BINARY)
cv.imshow("Tatsuya Shiba",thresh)
contours, heiarchies = cv.findContours(thresh, cv.RETR_LIST, cv.CHAIN_APPROX_NONE)
print(f'There are {len(contours)} contours')
cv.waitKey(0)
```
### Drawing the contours
```
cv.drawContours(blank, contours,-1,(0,0,255),1)
cv.imshow("Contours", blank)
cv.waitKey(0)
```
|
github_jupyter
|
import cv2 as cv
import numpy as np
img = cv.imread("Files/tatsuya.jpg") #takes path or image
cv.imshow("Tatsuya Shiba",img)
cv.waitKey(0) #makes the shell wait for an infinite amt of time till a key is pressed.
capture = cv.VideoCapture("C:/Users/admin/Videos/anime/inari kon kon/EP.1.480p.mp4")
while True:
isTrue, frame = capture.read()
cv.imshow("Video",frame)
if cv.waitKey(20) & 0xFF==ord("d"):
break
capture.release()
cv.destroyAllWindows()
#image video and live video
def rescaleFrame(frame,scale=0.7):
width = int(frame.shape[1]*scale)
height = int(frame.shape[0]*scale)
dimensions = (width,height)
return cv.resize(frame,dimensions,interpolation=cv.INTER_AREA)
#live video
def changeRes(width,height):
capture.set(3,width)
capture.set(4,height)
img_resized = rescaleFrame(img,scale=0.7)
cv.imshow("tatsuya_img",img_resized)
cv.waitKey(0)
capture = cv.VideoCapture("C:/Users/admin/Videos/anime/inari kon kon/EP.1.480p.mp4")
while True:
isTrue, frame = capture.read()
frame_resize = rescaleFrame(frame,scale=0.7)
cv.imshow("Video",frame_resize)
if cv.waitKey(20) & 0xFF==ord("d"):
break
capture.release()
cv.destroyAllWindows()
import numpy as np
blank_1 = np.zeros((500,500,3),dtype = 'uint8')
blank_2 = np.zeros((500,500,3),dtype = 'uint8')
blank_3 = np.zeros((500,500,3),dtype = 'uint8')
blank_4 = np.zeros((500,500,3),dtype = 'uint8')
cv.imshow("blank",blank_1)
#painting green
blank_1[:]=0,255,0
cv.imshow("green",blank_1)
#painting red
blank_2[:]=0,0,255
cv.imshow("red",blank_2)
#painting blue
blank_3[:]=255,0,0
cv.imshow("blue",blank_3)
#painting a portion only
blank_4[200:300, 300:400] = 0,0,255
cv.imshow("portion",blank_4)
cv.waitKey(0)
#rectangle
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(250,250),(0,255,0),thickness=-1)
cv.imshow("Rectangle",blank)
cv.waitKey(0)
#circle
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(blank.shape[1]//2,blank.shape[0]//2),(0,255,0),thickness=-1)
cv.imshow("Circle",blank)
cv.waitKey(0)
#draw a line
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.rectangle(blank,(0,0),(blank.shape[1]//2,blank.shape[0]//2),(0,255,0))
cv.imshow("Line",blank)
cv.waitKey(0)
# writeing
blank = np.zeros((500,500,3),dtype = 'uint8')
cv.putText(blank,"Hello",(225,225),cv.FONT_HERSHEY_TRIPLEX,1.0,(0,255,0),2)
cv.imshow("Line",blank)
cv.waitKey(0)
img = cv.imread("C:/Users/admin/Pictures/anime cool stuff/tatsuya.jpg") #takes path
cv.imshow("Tatsuya Shiba",img)
cv.waitKey(0) #makes the shell wait for an infinite amt of time till a key is pressed.
# Converting an image to GrayScale
gray = cv.cvtColor(img,cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba but in gray",gray)
cv.waitKey(0)
# Blur
blur = cv.GaussianBlur(img,(3,3),cv.BODER_DEFAULT)
cv.imshow("Tatsuya Shiba but in gray",blur)
cv.waitKey(0)
# Edge cascade
canny = cv.Canny(img,125,175)
cv.imshow("Tatsuya Shiba but only edges",canny)
cv.waitKey(0)
#dilation
dilated = cv.dilate(canny,(7,7),iterations=5)
cv.imshow("Tatsuya Shiba with dilated edges",dilated)
cv.waitKey(0)
# eroding
eroded = cv.erode(dilated,(3,3),iteration=2)
cv.imshow("Tatsuya Shiba with dilated edges",eroded)
cv.waitKey(0)
#resize
resized = cv.resize(img,(500,500), interpolation=cv.INTER_CUBIC)
cv.imshow("Tatsuya Shiba resized",resized)
cv.waitKey(0)
#Cropping
cropped = img[50:200,200:400]
cv.imshow("Cropped",cropped)
# Translation (shifting x or y axxis or both)
def translate(img,x,y):
transMat = np.float32([[1,0,x],[0,1,y]])
dimensions = (img.shape[1],img.shape[0])
return cv.warpAffine(img,transMat,dimensions)
#a -x--> left
#a +x--> right
#a +y--> down
#a -y--> Up
translated = translate(img,100,100)
cv.imshow("translated",translated)
cv.waitKey(0)
#Rotation
def rotate(imf, angle, rotPoint = None):
(height,width)=img.shape[:2]
if rotPoint is None:
rotPoint = (width//2,height//2)
rotMat = cv.getRotationMatrix2D(rotPoint, angle, 1.0)
dimensions = (width,height)
return cv.warpAffine(img, rotMat, dimensions)
rotated = rotate(img,45)
cv.imshow("rotated",rotated)
cv.waitKey(0)
#Resizing
resized =cv.resize(img, (500,500), interpolation=cv.INTER_CUBIC)
cv.imshow("Resized", resized)
#Flipping
flip = cv.flip(img, 0)
# 0 ->vertically via x-axis
# 1 -> horiziontally via y-axis
# -1 -> both
cv.imshow("flip",flip)
cv.waitKey(0)
#Cropping
cropped = img[200:400,300:400]
cv.imshow("crop",cropped)
cv.waitKey(0)
gray = cv.cvtColor(img, cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba",gray)
#blur = cv.GaussianBlur(gray, (5,5), cv.BODER_DEFAULT)
#cv.imshow("BLUR",blur)
#blur the image in case there are too many contours
canny = cv.Canny(img,125,175)
cv.imshow("Tatsuya Shiba",canny)
contours, heiarchies = cv.findContours(canny, cv.RETR_LIST, cv.CHAIN_APPROX_NONE)
#RETR_LIST = all
#RETR_EXTERNAL = boundaries
#RETR_TREE = contours in a heiarchy
#CHAIN_APPROX_NONE = returns all
#CHAIN_APPROX_SIMPLE = compreses the contours
print(f'There are {len(contours)} contours')
cv.waitKey(0)
gray = cv.cvtColor(img, cv.COLOR_BGR2GRAY)
cv.imshow("Tatsuya Shiba",gray)
ret, thresh = cv.threshold(gray,125,255, cv.THRESH_BINARY)
cv.imshow("Tatsuya Shiba",thresh)
contours, heiarchies = cv.findContours(thresh, cv.RETR_LIST, cv.CHAIN_APPROX_NONE)
print(f'There are {len(contours)} contours')
cv.waitKey(0)
cv.drawContours(blank, contours,-1,(0,0,255),1)
cv.imshow("Contours", blank)
cv.waitKey(0)
| 0.249722 | 0.862004 |
# Clustering
##### Identifying different types of passes
---
```
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
```
By now, you should be familiar with all of these libraries
---
```
base_url = "https://raw.githubusercontent.com/statsbomb/open-data/master/data/"
comp_url = base_url + "matches/{}/{}.json"
match_url = base_url + "events/{}.json"
def parse_data(competition_id, season_id):
matches = requests.get(url=comp_url.format(competition_id, season_id)).json()
match_ids = [m['match_id'] for m in matches]
all_events = []
for match_id in tqdm(match_ids):
events = requests.get(url=match_url.format(match_id)).json()
passes = [x for x in events if x['type']['name'] == "Pass"]
for p in passes:
attributes = {
"x": p['location'][0],
"y": p['location'][1],
"end_x": p['pass']['end_location'][0],
"end_y": p['pass']['end_location'][1],
"outcome": 0 if 'outcome' in p['pass'].keys() else 1,
}
all_events.append(attributes)
return pd.DataFrame(all_events)
```
The `parse_data` function has been adjusted such that only passes are collected, and some new attributes, including:
- `x` - the x-coordinate of the origin of the pass
- ranges from `0 to 120`
- `y` - the y-coordinate of the origin of the pass
- ranges from `0 to 80`
- `end_x` - the x-coordinate of the end of the pass
- ranges from `0 to 120`
- `end_y` - the y-coordinate of the end of the pass
- ranges from `0 to 80`
- `outcome` - did the pass complete successfully
---
```
competition_id = 43
season_id = 3
df = parse_data(competition_id, season_id)
from sklearn.cluster import KMeans
model = KMeans(n_clusters=50)
```
This time, we import `KMeans` from `sklearn.cluster` and create an object of it.
---
```
features = df[['x', 'y', 'end_x', 'end_y']]
fit = model.fit(features)
```
We create a new DataFrame represents the model features (the attributes we think are important).
Notice, this time we don't have a `labels` variable. This is because we don't have a prior understanding of what those labels should be. This makes this an **unsupervised** modeling approach.
We fit the model using .fit(), just like the other `sklearn` models.
---
```
df['cluster'] = model.predict(features)
df.head(10)
```
We create a new column on the `pandas` DataFrame that represents which cluster each pass was classified into.
Again, we're benefitting from broadcasting so we don't need to loop through the data structure.
---
```
for i, (x, y, end_x, end_y) in enumerate(fit.cluster_centers_):
plt.arrow(x, y, end_x-x, end_y-y,
head_width=1,
head_length=1,
color='red',
alpha=0.5,
length_includes_head=True)
plt.text((x+end_x)/2, (y+end_y)/2, str(i+1))
plt.xlim(0,120)
plt.ylim(0,80)
plt.show()
```
This loop plots (using `matplotlib`) each cluster `centroid` (or simply, `center` but that sounds less cool).
Notice the use of `enumeration` here. It's a nice python trick for iterating over a list while keeping an index.
This gives us a visually striking result, showing clear (but not perfect) symmetry. You can play with the `n_clusters` variable in the model declaration to adjust the model to the correct level of coarseness for your personal purposes.
Pass clusters are great to use as starting points for pass difficulty models. The typical components you would put into a pass difficulty model (e.g. `pass_distance`, `pass_angle`) can have very different effects on different pass types.
---
Devin Pleuler 2020
|
github_jupyter
|
import requests
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from tqdm import tqdm
base_url = "https://raw.githubusercontent.com/statsbomb/open-data/master/data/"
comp_url = base_url + "matches/{}/{}.json"
match_url = base_url + "events/{}.json"
def parse_data(competition_id, season_id):
matches = requests.get(url=comp_url.format(competition_id, season_id)).json()
match_ids = [m['match_id'] for m in matches]
all_events = []
for match_id in tqdm(match_ids):
events = requests.get(url=match_url.format(match_id)).json()
passes = [x for x in events if x['type']['name'] == "Pass"]
for p in passes:
attributes = {
"x": p['location'][0],
"y": p['location'][1],
"end_x": p['pass']['end_location'][0],
"end_y": p['pass']['end_location'][1],
"outcome": 0 if 'outcome' in p['pass'].keys() else 1,
}
all_events.append(attributes)
return pd.DataFrame(all_events)
competition_id = 43
season_id = 3
df = parse_data(competition_id, season_id)
from sklearn.cluster import KMeans
model = KMeans(n_clusters=50)
features = df[['x', 'y', 'end_x', 'end_y']]
fit = model.fit(features)
df['cluster'] = model.predict(features)
df.head(10)
for i, (x, y, end_x, end_y) in enumerate(fit.cluster_centers_):
plt.arrow(x, y, end_x-x, end_y-y,
head_width=1,
head_length=1,
color='red',
alpha=0.5,
length_includes_head=True)
plt.text((x+end_x)/2, (y+end_y)/2, str(i+1))
plt.xlim(0,120)
plt.ylim(0,80)
plt.show()
| 0.317215 | 0.888372 |
```
%matplotlib inline
```
# Post processing of displacement on distributed processes {#ref_distributed_total_disp}
To help understand this example the following diagram is provided. It
shows the operator chain used to compute the final result.
{.align-center width="400px"}
Import dpf module and its examples files
```
from ansys.dpf import core as dpf
from ansys.dpf.core import examples
from ansys.dpf.core import operators as ops
```
# Configure the servers
Make a list of ip addresses and port numbers on which dpf servers are
started. Operator instances will be created on each of those servers to
address each a different result file. In this example, we will post
process an analysis distributed in 2 files, we will consequently require
2 remote processes. To make this example easier, we will start local
servers here, but we could get connected to any existing servers on the
network.
```
remote_servers = [dpf.start_local_server(as_global=False), dpf.start_local_server(as_global=False)]
ips = [remote_server.ip for remote_server in remote_servers]
ports = [remote_server.port for remote_server in remote_servers]
```
Print the ips and ports
```
print("ips:", ips)
print("ports:", ports)
```
Here we show how we could send files in temporary directory if we were
not in shared memory
```
files = examples.download_distributed_files()
server_file_paths = [dpf.upload_file_in_tmp_folder(files[0], server=remote_servers[0]),
dpf.upload_file_in_tmp_folder(files[1], server=remote_servers[1])]
```
Create the operators on the servers
\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~ On each server we
create two new operators for \'displacement\' and \'norm\' computations
and define their data sources. The displacement operator receives data
from the data file in its respective server. And the norm operator,
being chained to the displacement operator, receives input from the
output of this one.
```
remote_operators = []
for i, server in enumerate(remote_servers):
displacement = ops.result.displacement(server=server)
norm = ops.math.norm_fc(displacement, server=server)
remote_operators.append(norm)
ds = dpf.DataSources(server_file_paths[i], server=server)
displacement.inputs.data_sources(ds)
```
Create a merge_fields_containers operator able to merge the results
\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~\~
```
merge = ops.utility.merge_fields_containers()
```
# Connect the operators together and get the output
```
for i, server in enumerate(remote_servers):
merge.connect(i, remote_operators[i], 0)
fc = merge.get_output(0, dpf.types.fields_container)
print(fc)
print(fc[0].min().data)
print(fc[0].max().data)
```
|
github_jupyter
|
%matplotlib inline
from ansys.dpf import core as dpf
from ansys.dpf.core import examples
from ansys.dpf.core import operators as ops
remote_servers = [dpf.start_local_server(as_global=False), dpf.start_local_server(as_global=False)]
ips = [remote_server.ip for remote_server in remote_servers]
ports = [remote_server.port for remote_server in remote_servers]
print("ips:", ips)
print("ports:", ports)
files = examples.download_distributed_files()
server_file_paths = [dpf.upload_file_in_tmp_folder(files[0], server=remote_servers[0]),
dpf.upload_file_in_tmp_folder(files[1], server=remote_servers[1])]
remote_operators = []
for i, server in enumerate(remote_servers):
displacement = ops.result.displacement(server=server)
norm = ops.math.norm_fc(displacement, server=server)
remote_operators.append(norm)
ds = dpf.DataSources(server_file_paths[i], server=server)
displacement.inputs.data_sources(ds)
merge = ops.utility.merge_fields_containers()
for i, server in enumerate(remote_servers):
merge.connect(i, remote_operators[i], 0)
fc = merge.get_output(0, dpf.types.fields_container)
print(fc)
print(fc[0].min().data)
print(fc[0].max().data)
| 0.238196 | 0.883889 |
# Initialisation steps
## dependancies install
Process daily rainfall hosted on Google Earth Engine.
## Authentification to GEE
```
import ee
# Trigger the authentication flow.
ee.Authenticate()
# Initialize the library.
ee.Initialize()
```
# CHIRPS Processing
[CHIRPS](https://www.chc.ucsb.edu/data/chirps) data is hosted (not uniquely) on Google Earth Engine.
[chirps_gee](https://developers.google.com/earth-engine/datasets/catalog/UCSB-CHG_CHIRPS_DAILY)
As always with large data analysis we want to bring the compute to the data which is exactly one of the features of GEE.
So most of the analysis will be performed on GEE and result downloaded.
## Parameters Definition
```
gee_collection_id = "UCSB-CHG/CHIRPS/DAILY"
band_name = "precipitation"
aoi_url = 'https://github.com/matthieu-bernard/gee-processing/raw/main/gee_processing/data/aoi.geojson'
```
## Metadata collection
Google Actually publish SpatioTemporal Catalog Assets for the catalogs they host in GEE.
For now it seems like processing of the data with a Open Source platform (like Pangeo) is not there yet due to the 'proprietary' storage format they use. But lets at least salute the effort of publishing metadata to the STAC standard.
[CHIRPS_stac_metadata](https://gee.stac.cloud/3cRrouu75vEzeP4uufqKgwzeubDQYizmMUQm?t=bands)
```
import intake
url = 'https://earthengine-stac.storage.googleapis.com/catalog/UCSB-CHG_CHIRPS_DAILY.json'
cat = intake.open_stac_catalog(url)
cat.metadata
```
## Region of Interest
The CHIRPS dataset cover the all globe from -50 to +50. We are interested in a spatial subset.
```
# Read region of interest vector file
import geopandas as gpd
gpd_aoi = gpd.GeoDataFrame.from_file(aoi_url)
gjson = gpd_aoi.to_crs(epsg='4326').to_json()
# Really simple plot of the region of interest.
# Just for the sake of control.
import folium
import shapely
from typing import Tuple
def centroid(geom: shapely.geometry) -> Tuple[float, float]:
centroid = geom.centroid
return [centroid.y, centroid.x]
aoi_polygon = gpd_aoi['geometry'][0]
env = aoi_polygon.envelope
mapa = folium.Map(centroid(aoi_polygon),
zoom_start=5,
tiles='cartodbpositron')
aoi = folium.features.GeoJson(gjson)
bbox = folium.features.GeoJson(env)
mapa.add_children(aoi)
folium.features.GeoJson(env).add_to(mapa)
mapa
print(env)
```
## Area of interets to GEE server
The area of interest need to be communicated to the GEE server for server-side computation.
There seems to be 2 options here:
- Pushing the shapefile to GEE assets
- Manual upload from the GUI tool: https://developers.google.com/earth-engine/guides/importing
- Use the cli to upload from GCS (cli is written in python so no need for a external call though https://github.com/google/earthengine-api/blob/7a6f605e1be30002c4a3ec516669703dea9e3f71/python/ee/cli/commands.py).
- Just construct an ee.Geometry from the Polygon (or a boundingBox if the shapefile is too big (or reduce it first...))
Pushind files to GEE assets, either need a Google Cloud account or manual intervention in the pipeline.
Creating a ee.Geomtry object is more flexible although it definitely have a the obvious drawback that we would send the geometry definiton over the network each time we run the pipeline.
```
import os
import json
def geojson_to_ee(geo_json: str, geodesic=True) -> ee.Geometry:
"""Converts a geojson to ee.Geometry()
Args:
geo_json: geojson definition as str.
Returns:
ee_object: An ee.Geometry object
"""
geo_json = json.loads(geo_json)
features = ee.FeatureCollection(geo_json['features'])
return features
aoi = geojson_to_ee(gjson)
#aoi_shp = ee.FeatureCollection('users/matthieu_bernard/aoi').geometry()
```
## GEE (Server side) processing pipeline
```
import datetime
import dateutil.parser
# Here we get the extent of the datastet to get all of it expect the data for the current
# since a partial month could pollute or monthly statistics
start = dateutil.parser.isoparse(cat.metadata['extent']['temporal']['interval'][0][0])
stop = datetime.date.today().replace(day=1)
# Select dataset of interest
precip = ee.ImageCollection(gee_collection_id).select(band_name)
precip = (ee.ImageCollection(gee_collection_id)
.select(band_name)
.filter(ee.Filter.date(start.isoformat(), stop.isoformat()))
)
# Convert from daily precipitation values bin (rainy/not-rainy).
precip_event = precip.map(lambda img: img.gt(0).copyProperties(img, img.propertyNames()))
# get sum of rainy days per month (one image per month per year)
months = ee.List.sequence(1, 12)
years = ee.List.sequence(start.year, stop.year)
by_month_year = ee.ImageCollection.fromImages(
years.map(lambda y: months.map(lambda m:
(precip_event
.filter(ee.Filter.calendarRange(y, y, 'year'))
.filter(ee.Filter.calendarRange(m, m, 'month'))
.sum()
.set('month', m).set('year', y))
)
).flatten())
# Get the monthly average over all years
by_month = ee.ImageCollection.fromImages(
months.map(lambda m:
(by_month_year
.filterMetadata('month', 'equals', m)
.mean()
.clip(aoi)
.round()
.set('month', m))
.set('description', ('number of rainy days mean the number of days that have non null rainfall amount over'
'the aggregation period'))
.set('aggregation_period', 'calendar_month')
.set('long_name', 'number_of_rainy_days')
.set('original_dataset', {k: cat.metadata[k] for k in ['title', 'description', 'version']})
.set('provider', '[email protected]')
))
images = by_month.toBands()
def coord_list(geom):
coords = list(geom.exterior.coords)
return (coords)
tasks = []
for i in range(12):
task = ee.batch.Export.image(images.select(f'{i}_precipitation'), f'monthly_average_rainy_days_{i+1:02d}',
{
'scale': 5000,
'maxPixels': 1.0E13,
'region': coord_list(gpd_aoi.geometry[0]),
'fileFormat': 'GeoTIFF',
'formatOptions': {
'cloudOptimized': True
},
'folder': 'chirps'
})
task.start()
tasks.append(task)
```
## Visualisation
Visualisation of the exported dataset
Will also set metadata a bit better than what is done on the GEE side.
### One file per month. Average number of rainy day (climato from 1981 to 2020)
Here I plot from local files already downloaded for the demonstration.
```
import intake
local_cat = intake.cat()
import matplotlib.pyplot as plt
import xarray as xr
%matplotlib inline
f, axs = plt.subplots(4, 3, figsize=(12, 16))
for m in range(1,13):
da=local_cat.gee.datasets.rainy_days_chirps_int(month=m).read()
da.plot(ax=axs.flatten()[m-1])
```
### One band per month. Average number of rainy day (climato from 1981 to 2020) as float
```
da = local_cat.gee.datasets.rainy_days_chirps().read()
da
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import xarray as xr
%matplotlib inline
f, axs = plt.subplots(4, 3, figsize=(12, 16))
for band in range(12):
da.isel(band=band).plot(ax=axs.flatten()[band])
```
### Metadata filling
```
!pip install rioxarray
da.attrs.update({
'description': ('number of rainy days mean the number of days that have non null rainfall amount over'
'the :ggregation period'),
'aggregation_period': 'calendar_month',
'long_name': 'number_of_rainy_days',
'original_dataset': {k: cat.metadata[k] for k in ['title', 'description', 'version']},
'provider': '[email protected]',
})
# Here we should save to the raster nut for the sake of showing that attributes are not exported by GEE
import rioxarray
da.rio.to_raster('/tmp/test.tif')
da
import xarray as xr
rio = xr.open_rasterio("/tmp/test.tif")
rio
```
|
github_jupyter
|
import ee
# Trigger the authentication flow.
ee.Authenticate()
# Initialize the library.
ee.Initialize()
gee_collection_id = "UCSB-CHG/CHIRPS/DAILY"
band_name = "precipitation"
aoi_url = 'https://github.com/matthieu-bernard/gee-processing/raw/main/gee_processing/data/aoi.geojson'
import intake
url = 'https://earthengine-stac.storage.googleapis.com/catalog/UCSB-CHG_CHIRPS_DAILY.json'
cat = intake.open_stac_catalog(url)
cat.metadata
# Read region of interest vector file
import geopandas as gpd
gpd_aoi = gpd.GeoDataFrame.from_file(aoi_url)
gjson = gpd_aoi.to_crs(epsg='4326').to_json()
# Really simple plot of the region of interest.
# Just for the sake of control.
import folium
import shapely
from typing import Tuple
def centroid(geom: shapely.geometry) -> Tuple[float, float]:
centroid = geom.centroid
return [centroid.y, centroid.x]
aoi_polygon = gpd_aoi['geometry'][0]
env = aoi_polygon.envelope
mapa = folium.Map(centroid(aoi_polygon),
zoom_start=5,
tiles='cartodbpositron')
aoi = folium.features.GeoJson(gjson)
bbox = folium.features.GeoJson(env)
mapa.add_children(aoi)
folium.features.GeoJson(env).add_to(mapa)
mapa
print(env)
import os
import json
def geojson_to_ee(geo_json: str, geodesic=True) -> ee.Geometry:
"""Converts a geojson to ee.Geometry()
Args:
geo_json: geojson definition as str.
Returns:
ee_object: An ee.Geometry object
"""
geo_json = json.loads(geo_json)
features = ee.FeatureCollection(geo_json['features'])
return features
aoi = geojson_to_ee(gjson)
#aoi_shp = ee.FeatureCollection('users/matthieu_bernard/aoi').geometry()
import datetime
import dateutil.parser
# Here we get the extent of the datastet to get all of it expect the data for the current
# since a partial month could pollute or monthly statistics
start = dateutil.parser.isoparse(cat.metadata['extent']['temporal']['interval'][0][0])
stop = datetime.date.today().replace(day=1)
# Select dataset of interest
precip = ee.ImageCollection(gee_collection_id).select(band_name)
precip = (ee.ImageCollection(gee_collection_id)
.select(band_name)
.filter(ee.Filter.date(start.isoformat(), stop.isoformat()))
)
# Convert from daily precipitation values bin (rainy/not-rainy).
precip_event = precip.map(lambda img: img.gt(0).copyProperties(img, img.propertyNames()))
# get sum of rainy days per month (one image per month per year)
months = ee.List.sequence(1, 12)
years = ee.List.sequence(start.year, stop.year)
by_month_year = ee.ImageCollection.fromImages(
years.map(lambda y: months.map(lambda m:
(precip_event
.filter(ee.Filter.calendarRange(y, y, 'year'))
.filter(ee.Filter.calendarRange(m, m, 'month'))
.sum()
.set('month', m).set('year', y))
)
).flatten())
# Get the monthly average over all years
by_month = ee.ImageCollection.fromImages(
months.map(lambda m:
(by_month_year
.filterMetadata('month', 'equals', m)
.mean()
.clip(aoi)
.round()
.set('month', m))
.set('description', ('number of rainy days mean the number of days that have non null rainfall amount over'
'the aggregation period'))
.set('aggregation_period', 'calendar_month')
.set('long_name', 'number_of_rainy_days')
.set('original_dataset', {k: cat.metadata[k] for k in ['title', 'description', 'version']})
.set('provider', '[email protected]')
))
images = by_month.toBands()
def coord_list(geom):
coords = list(geom.exterior.coords)
return (coords)
tasks = []
for i in range(12):
task = ee.batch.Export.image(images.select(f'{i}_precipitation'), f'monthly_average_rainy_days_{i+1:02d}',
{
'scale': 5000,
'maxPixels': 1.0E13,
'region': coord_list(gpd_aoi.geometry[0]),
'fileFormat': 'GeoTIFF',
'formatOptions': {
'cloudOptimized': True
},
'folder': 'chirps'
})
task.start()
tasks.append(task)
import intake
local_cat = intake.cat()
import matplotlib.pyplot as plt
import xarray as xr
%matplotlib inline
f, axs = plt.subplots(4, 3, figsize=(12, 16))
for m in range(1,13):
da=local_cat.gee.datasets.rainy_days_chirps_int(month=m).read()
da.plot(ax=axs.flatten()[m-1])
da = local_cat.gee.datasets.rainy_days_chirps().read()
da
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
import xarray as xr
%matplotlib inline
f, axs = plt.subplots(4, 3, figsize=(12, 16))
for band in range(12):
da.isel(band=band).plot(ax=axs.flatten()[band])
!pip install rioxarray
da.attrs.update({
'description': ('number of rainy days mean the number of days that have non null rainfall amount over'
'the :ggregation period'),
'aggregation_period': 'calendar_month',
'long_name': 'number_of_rainy_days',
'original_dataset': {k: cat.metadata[k] for k in ['title', 'description', 'version']},
'provider': '[email protected]',
})
# Here we should save to the raster nut for the sake of showing that attributes are not exported by GEE
import rioxarray
da.rio.to_raster('/tmp/test.tif')
da
import xarray as xr
rio = xr.open_rasterio("/tmp/test.tif")
rio
| 0.677901 | 0.927724 |
<a href="https://colab.research.google.com/github/danidavid/Tensorflow-Keras-Repo/blob/master/Housing_Classification_Project_3.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Project 3: Housing Classification
# Step 1: Importing dataset
```
import pandas as pd #This will be used for online dataset import
import io
import requests
url='https://raw.githubusercontent.com/jbrownlee/Datasets/master/housing.data'
s=requests.get(url).content
c=pd.read_csv(io.StringIO(s.decode('UTF-8')),delim_whitespace=True, header=None)
#c
```
## Importing packages
```
import numpy as np
import numpy
import pandas
import keras
from keras import layers
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasRegressor
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
```
## Importing X and Y from dataset
```
dataset = c.values
X = dataset[:,0:13]
Y = dataset[:,13]
```
## Seed
```
# fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
```
# Step 2: Making model
```
#baseline model
def baseline_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=baseline_model, epochs=100, batch_size=5, verbose=0)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(estimator, X, Y, cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
# Step 3: Modeling The Standardized Dataset
```
# evaluate model with standardized dataset
numpy.random.seed(seed)
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=baseline_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Standardized: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
## Extension of Step 3: Modeling The Standardized Dataset
```
#baseline model
def baseline_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
#model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
# evaluate model with standardized dataset
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=baseline_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Standardized: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
# Step 4. Tune The Neural Network Topology
## Step 4.1. Evaluate a Deeper Network Topology
```
#larger model
def larger_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
numpy.random.seed(seed)
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=larger_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Larger: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
## Step 4.2. Evaluate a Wider Network Topology
```
#wider model
def wider_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(20,activation='relu',input_shape=(13,)))
#model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=wider_model, epochs=100, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Wider: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
# Step 5. Developing a model that overfits
```
#overfitting model
def create_overfit():
#create model
model = keras.Sequential()
#adding more layers & making them bigger
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(5,activation='relu'))
model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(5,activation='relu'))
model.add(layers.Dense(4,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#training for more epochs
model.fit(X,Y,epochs=20,batch_size=32,verbose=0)
return model
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=create_overfit, epochs=100, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Overfit: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
# Step 6. Tuning the Model
```
#baseline model
def tuned_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(9,activation='relu'))
model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=tuned_model, epochs=600, batch_size=5, verbose=0)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(estimator, X, Y, cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
# Step 7. Rewriting the code using Keras functional API
## Creating baseline model
```
#creating baseline model
def kf_baseline():
#create model
input = keras.Input(shape=(13,))
a = layers.Dense(13,activation='relu')(input)
a = layers.Dense(3,activation='relu')(a)
outputs = layers.Dense(1)(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=kf_baseline,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
```
## Now with smaller network
```
#smaller model
def create_kfsmaller():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfsmaller,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Smaller: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
```
## Now with larger network
```
#larger model
def create_kflarger():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfsmaller,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Larger: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
```
## Developing a model that overfits
```
#overfit model
def create_kfoverfit():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(4,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adamax',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfoverfit, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=5,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Overfit: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
```
## Tuning the model
```
#tuning model
def create_kftune():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(4,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adamax',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=200,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kftune, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=5,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Tuned: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
estimator=[]
estimator = KerasClassifier(build_fn=create_kftune, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=9,shuffle=True,random_state=seed) # n_splits cahnged to 9 (just experimenting)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Tuned: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
```
# Step 8. Rewriting the code by doing Model Subclassing
```
input = keras.Input(shape=(13,))
#making a class
class MyModel(keras.Model):
def __init__(self):
super(MyModel, self).__init__()
self.dense1 = layers.Dense(13,activation='relu')
self.dense2 = layers.Dense(3,activation='relu')
self.dense3 = layers.Dense(1)
def call(self,inputs):
x = self.dense1(inputs)
x = self.dense2(x)
return self.dense3(x)
model = MyModel()
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
```
# Step 9. Rewriting the code without using scikit-learn
```
def create_baseline():
#create model
input = keras.Input(shape=(13,))
a = layers.Dense(13,activation='relu')(input)
a = layers.Dense(3,activation='relu')(a)
outputs = layers.Dense(1)(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32,verbose=0)
return model
k = 9
num_val_samples = len(X) // k
num_epochs = 900
all_scores = []
for i in range(k):
print('processing fold #', i)
val_data = X[i * num_val_samples: (i + 1) * num_val_samples]
val_targets = Y[i * num_val_samples: (i + 1) * num_val_samples]
partial_train_data = np.concatenate([X[:i * num_val_samples],X[(i + 1) * num_val_samples:]],axis=0)
partial_train_targets = np.concatenate([Y[:i * num_val_samples],Y[(i + 1) * num_val_samples:]],axis=0)
model = create_baseline()
model.fit(partial_train_data, partial_train_targets,epochs=num_epochs, batch_size=32, verbose=0)
val_mse, val_mae = model.evaluate(val_data, val_targets, verbose=0)
all_scores.append(val_mae)
score = np.average(all_scores)
print("Result: %.2f%% (%.2f%%)" % (score.mean()*100,score.std()*100))
```
|
github_jupyter
|
import pandas as pd #This will be used for online dataset import
import io
import requests
url='https://raw.githubusercontent.com/jbrownlee/Datasets/master/housing.data'
s=requests.get(url).content
c=pd.read_csv(io.StringIO(s.decode('UTF-8')),delim_whitespace=True, header=None)
#c
import numpy as np
import numpy
import pandas
import keras
from keras import layers
from keras.models import Sequential
from keras.layers import Dense
from keras.wrappers.scikit_learn import KerasRegressor
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
dataset = c.values
X = dataset[:,0:13]
Y = dataset[:,13]
# fix random seed for reproducibility
seed = 7
numpy.random.seed(seed)
#baseline model
def baseline_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=baseline_model, epochs=100, batch_size=5, verbose=0)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(estimator, X, Y, cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
# evaluate model with standardized dataset
numpy.random.seed(seed)
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=baseline_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Standardized: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#baseline model
def baseline_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
#model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
# evaluate model with standardized dataset
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=baseline_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Standardized: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#larger model
def larger_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
numpy.random.seed(seed)
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=larger_model, epochs=50, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Larger: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#wider model
def wider_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(20,activation='relu',input_shape=(13,)))
#model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=wider_model, epochs=100, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Wider: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#overfitting model
def create_overfit():
#create model
model = keras.Sequential()
#adding more layers & making them bigger
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(5,activation='relu'))
model.add(layers.Dense(6,activation='relu'))
model.add(layers.Dense(5,activation='relu'))
model.add(layers.Dense(4,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#training for more epochs
model.fit(X,Y,epochs=20,batch_size=32,verbose=0)
return model
estimators = []
estimators.append(('standardize', StandardScaler()))
estimators.append(('mlp', KerasRegressor(build_fn=create_overfit, epochs=100, batch_size=5, verbose=0)))
pipeline = Pipeline(estimators)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(pipeline, X, Y, cv=kfold)
print("Overfit: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#baseline model
def tuned_model():
#created model
model = keras.Sequential()
model.add(layers.Dense(13,activation='relu',input_shape=(13,)))
model.add(layers.Dense(9,activation='relu'))
model.add(layers.Dense(3,activation='relu'))
model.add(layers.Dense(1))
#compiled model
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
#model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=tuned_model, epochs=600, batch_size=5, verbose=0)
kfold = KFold(n_splits=10, random_state=seed)
results = cross_val_score(estimator, X, Y, cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#creating baseline model
def kf_baseline():
#create model
input = keras.Input(shape=(13,))
a = layers.Dense(13,activation='relu')(input)
a = layers.Dense(3,activation='relu')(a)
outputs = layers.Dense(1)(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
#evaluate model with standardized dataset
estimator=[]
estimator = KerasRegressor(build_fn=kf_baseline,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Results: %.2f (%.2f) MSE" % (results.mean(), results.std()))
#smaller model
def create_kfsmaller():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfsmaller,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Smaller: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
#larger model
def create_kflarger():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfsmaller,epochs=100,batch_size=5,verbose=0)
kfold = KFold(n_splits=10,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Larger: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
#overfit model
def create_kfoverfit():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(4,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adamax',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kfoverfit, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=5,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Overfit: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
#tuning model
def create_kftune():
#create model
input = keras.Input(shape=(4,))
a = layers.Dense(8,activation='relu')(input)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(6,activation='relu')(a)
a = layers.Dense(5,activation='relu')(a)
a = layers.Dense(4,activation='relu')(a)
outputs = layers.Dense(3,activation='softmax')(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adamax',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=200,batch_size=32)
return model
estimator=[]
estimator = KerasClassifier(build_fn=create_kftune, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=5,shuffle=True,random_state=seed)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Tuned: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
estimator=[]
estimator = KerasClassifier(build_fn=create_kftune, epochs=200, batch_size=5, verbose=0)
kfold = KFold(n_splits=9,shuffle=True,random_state=seed) # n_splits cahnged to 9 (just experimenting)
results = cross_val_score(estimator,X,Y,cv=kfold)
print("Tuned: %.2f%% (%.2f%%)" % (results.mean()*100,results.std()*100))
input = keras.Input(shape=(13,))
#making a class
class MyModel(keras.Model):
def __init__(self):
super(MyModel, self).__init__()
self.dense1 = layers.Dense(13,activation='relu')
self.dense2 = layers.Dense(3,activation='relu')
self.dense3 = layers.Dense(1)
def call(self,inputs):
x = self.dense1(inputs)
x = self.dense2(x)
return self.dense3(x)
model = MyModel()
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32)
def create_baseline():
#create model
input = keras.Input(shape=(13,))
a = layers.Dense(13,activation='relu')(input)
a = layers.Dense(3,activation='relu')(a)
outputs = layers.Dense(1)(a)
#compile model
model = keras.Model(input, outputs)
model.compile(optimizer='Adam',loss='mse',metrics=['accuracy'])
model.fit(X,Y,epochs=20,batch_size=32,verbose=0)
return model
k = 9
num_val_samples = len(X) // k
num_epochs = 900
all_scores = []
for i in range(k):
print('processing fold #', i)
val_data = X[i * num_val_samples: (i + 1) * num_val_samples]
val_targets = Y[i * num_val_samples: (i + 1) * num_val_samples]
partial_train_data = np.concatenate([X[:i * num_val_samples],X[(i + 1) * num_val_samples:]],axis=0)
partial_train_targets = np.concatenate([Y[:i * num_val_samples],Y[(i + 1) * num_val_samples:]],axis=0)
model = create_baseline()
model.fit(partial_train_data, partial_train_targets,epochs=num_epochs, batch_size=32, verbose=0)
val_mse, val_mae = model.evaluate(val_data, val_targets, verbose=0)
all_scores.append(val_mae)
score = np.average(all_scores)
print("Result: %.2f%% (%.2f%%)" % (score.mean()*100,score.std()*100))
| 0.689724 | 0.913677 |
# Python für Geisteswissenschaftler/innen
(Teil 2)
Melanie Andresen
[[email protected]](mailto:[email protected])
## Agenda
### Im letzten Teil:
* Python im interaktiven Modus
* Variablen und Datentypen
* Pythonskripte speichern und ausführen
* Reguläre Ausdrücke
### In diesem Teil:
* Listen
* Boolsche Ausdrücke
* Bedingungen
* Schleifen
* Dateien einlesen und schreiben
## Listen
Listen werden in Python mit eckigen Klammern dargestellt. Die Elemente der Liste werden mit Kommata getrennt:
```
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
```
Die Elemente in der Liste können alle möglichen Datentypen haben.
```
lottozahlen = [3, 6, 18, 31, 39, 40]
listen = [['Äpfel', 'Schokolade', 'Klopapier'], [3, 6, 18, 31, 39, 40]]
```
Die String-Methode `split()`erzeugt eine Liste, vgl. letzte Woche:
```
satz = 'Käsebrot ist ein gutes Brot.'
satz.split()
```
## Listen erweitern
```
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
```
Ein Element ergänzen:
```
einkaufsliste.append('Brot')
print(einkaufsliste)
einkaufsliste = einkaufsliste + ['Käse']
print(einkaufsliste)
```
Achtung: Die Methode `append` verändert die Liste, ohne dass der modifizierte Inhalt der Variablen neu zugewiesen werden müsste! Die Addition leistet das nicht.
Liste um zweite Liste erweitern:
```
einkaufsliste_2 = ['Joghurt', 'Milch', 'Käse']
einkaufsliste.extend(einkaufsliste_2)
print(einkaufsliste)
```
## Listen als Sequenzen
Listen und Strings sind Sequenzen. Sie bieten deshalb teilweise ähnliche Möglichkeiten:
Slicing:
```
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
einkaufsliste[1]
```
Länge feststellen:
```
len(einkaufsliste)
len('Käsebrot')
```
Wir können prüfen, ob ein Element in einer Sequenz enthalten ist:
```
'Schokolade' in einkaufsliste
'Ente' in einkaufsliste
'miau' in 'Krimiautorin'
'wau' in 'Krimiautorin'
```
## Listen
Listen sortieren:
```
einkaufsliste.sort()
print(einkaufsliste)
```
Reihenfolge der Elemente umkehren:
```
einkaufsliste.reverse()
print(einkaufsliste)
```
Vorsicht beim Kopieren von Listen:
```
liste = [1,2,3]
neue_liste = liste
neue_liste
neue_liste.append(4)
neue_liste
```
Welche Belegung der Variablen `liste` erwarten wir?
```
liste
```
Zum Kopieren von Listen die Methode `copy()` verwenden:
```
liste = [1,2,3]
neue_liste = liste.copy()
neue_liste
neue_liste.append(4)
neue_liste
liste
```
## Boolsche Ausdrücke
```
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
'Schokolade' in einkaufsliste
```
Diese Abfrage gibt einen neuen Datentyp zurück: einen sog. Boolschen Ausdruck.
Eine boolsche Variable hat nur zwei mögliche Werte: `True` oder `False`. Die Zuweisung erfolgt ohne Anführungsstriche und mit Großschreibung:
```
mittagspause = False
```
Boolsche Operatoren liefern Informationen über Zustände.
```
alter_karin = 32
alter_theo = 60
```
Wir können Werte prüfen auf z. B.
```
# Gleichheit:
alter_theo == alter_karin
# Größenverhältnisse (<, >, <=, >=):
alter_theo > alter_karin
# Ungleichheit:
alter_theo != alter_karin
```
## Boolsche Operatoren
Komplexere Zustände ausdrücken:
```
not alter_theo == alter_karin
alter_theo > alter_karin and alter_theo > 60
alter_theo > 60 or alter_karin < 20
```
## Bedingungen
Oft soll ein Programm Dinge nur unter bestimmten Bedingungen machen:
```
if 'Schokolade' in einkaufsliste:
print('Sehr gut!')
```
In der `if`-Zeile wird ein Test ausgeführt. Ist das Testergebnis positiv, wird das Statement darunter ebenfalls ausgeführt.
Python benutzt Doppelpunkte und Whitespace, um Hierarchien anzuzeigen. Eine Einrückung entspricht dabei vier Leerzeichen oder einem Tabstopp.
Wir können eine `else`-Zeile ergänzen, die festlegt, was das Programm machen soll, wenn das Testergebnis negativ ist:
```
if 'Schokolade' in einkaufsliste:
print('Sehr gut!')
else:
einkaufsliste.append('Schokolade')
```
Bei mehr als zwei möglichen Zuständen:
```
temperatur = 21.5
if temperatur > 30:
print('Was für eine Hitze!')
elif temperatur > 20:
print('So lässt es sich aushalten!')
else:
print('Frisch heute!')
```
Als Test lassen sich sehr gut Funktionen aus dem re-Modul nutzen.
Beispiel - Nach großgeschriebenen Wörter filtern:
```
import re
words = ['Dinosaurier', 'nachmittags', 'Eis', 'essen']
for word in words:
if re.match('[A-ZÖÄÜ]', word):
print(word)
```
## Schleifen
Viele Sachen soll ein Programm mehrmals machen, z. B. für jedes Element einer Sequenz (hier: einer Liste):
```
for item in einkaufsliste:
print('Wir brauchen noch ' + item + '!')
```
`item` ist hier ein frei vergebener Variablenname, der in jedem Durchlauf der Schleife für ein anderes Element der Liste steht.
Ein anderer Schleifentyp:
```
i = 0
while i < 10:
i = i + 1
print(i)
print('Fertig!')
```
Solange die Bedingung nach `while` gegeben ist, werden alle Befehle darunter immer wieder ausgeführt. Danach geht es ggf. mit dem folgenden Code weiter.
### Abbruchkriterium festlegen!
A programmer goes out to get some dry cleaning. His wife told him, "While you're out, buy some milk." He never came home.
`while out = True:
buy_milk()`
→ Bei `while`-Schleifen immer darauf achten, dass es ein Abbruchkriterium gibt!
## Dateien lesen und schreiben
## Dateien lesen
Inhalt einer Datei einlesen:
```
with open('Daten/cathaskueche_04.txt', 'r', encoding='utf8') as input_file:
text = input_file.read()
```
(Der Text in der Datei stammt von [hier](https://cathaskueche.wordpress.com/2013/05/18/babaganoush/).)
Dateipfad: Name der Datei und ggf. der dazugehörige Pfad, von der aktuellen Position im Terminal aus gesehen.

## Dateien schreiben
Inhalt in eine Datei schreiben:
```
text = 'Was ich schon immer sagen wollte'
with open('Daten/my_file.txt', 'w') as output_file:
output_file.write(text)
```
Inhalt über eine Schleife in eine Datei schreiben:
```
with open('Daten/my_file.txt', 'w') as output:
for item in einkaufsliste:
output.write(item + '\n')
```
## Im nächsten Teil:
* Debugging: Wo ist der Fehler?
* Dictionaries
* Module einbinden und nutzen
* os-Modul
* NumPy
|
github_jupyter
|
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
lottozahlen = [3, 6, 18, 31, 39, 40]
listen = [['Äpfel', 'Schokolade', 'Klopapier'], [3, 6, 18, 31, 39, 40]]
satz = 'Käsebrot ist ein gutes Brot.'
satz.split()
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
einkaufsliste.append('Brot')
print(einkaufsliste)
einkaufsliste = einkaufsliste + ['Käse']
print(einkaufsliste)
einkaufsliste_2 = ['Joghurt', 'Milch', 'Käse']
einkaufsliste.extend(einkaufsliste_2)
print(einkaufsliste)
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
einkaufsliste[1]
len(einkaufsliste)
len('Käsebrot')
'Schokolade' in einkaufsliste
'Ente' in einkaufsliste
'miau' in 'Krimiautorin'
'wau' in 'Krimiautorin'
einkaufsliste.sort()
print(einkaufsliste)
einkaufsliste.reverse()
print(einkaufsliste)
liste = [1,2,3]
neue_liste = liste
neue_liste
neue_liste.append(4)
neue_liste
liste
liste = [1,2,3]
neue_liste = liste.copy()
neue_liste
neue_liste.append(4)
neue_liste
liste
einkaufsliste = ['Äpfel', 'Schokolade', 'Klopapier']
'Schokolade' in einkaufsliste
mittagspause = False
alter_karin = 32
alter_theo = 60
# Gleichheit:
alter_theo == alter_karin
# Größenverhältnisse (<, >, <=, >=):
alter_theo > alter_karin
# Ungleichheit:
alter_theo != alter_karin
not alter_theo == alter_karin
alter_theo > alter_karin and alter_theo > 60
alter_theo > 60 or alter_karin < 20
if 'Schokolade' in einkaufsliste:
print('Sehr gut!')
if 'Schokolade' in einkaufsliste:
print('Sehr gut!')
else:
einkaufsliste.append('Schokolade')
temperatur = 21.5
if temperatur > 30:
print('Was für eine Hitze!')
elif temperatur > 20:
print('So lässt es sich aushalten!')
else:
print('Frisch heute!')
import re
words = ['Dinosaurier', 'nachmittags', 'Eis', 'essen']
for word in words:
if re.match('[A-ZÖÄÜ]', word):
print(word)
for item in einkaufsliste:
print('Wir brauchen noch ' + item + '!')
i = 0
while i < 10:
i = i + 1
print(i)
print('Fertig!')
with open('Daten/cathaskueche_04.txt', 'r', encoding='utf8') as input_file:
text = input_file.read()
text = 'Was ich schon immer sagen wollte'
with open('Daten/my_file.txt', 'w') as output_file:
output_file.write(text)
with open('Daten/my_file.txt', 'w') as output:
for item in einkaufsliste:
output.write(item + '\n')
| 0.067263 | 0.92944 |
```
import json
import math
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
non_perturbed_deck = 'standard'
decks = [
'batman_joker',
'captain_america',
'adversarial_standard',
'adversarial_batman_joker',
'adversarial_captain_america'
]
noise_pcts = [10,20,30,40,50,60,70,80,90]
FONT_SIZE=18
TICK_FONT_SIZE=12
mpl.rcParams['xtick.labelsize'] = TICK_FONT_SIZE
mpl.rcParams['ytick.labelsize'] = TICK_FONT_SIZE
def get_nsl_results(net_type, deck):
struc_unstruc = 'structured_test_data'
acc_metric = 'accuracy'
# need to build array of results for noise pcts
acc = []
acc_stds = []
# get standard
np_0 = json.loads(open('../nsl/'+struc_unstruc+'/'+net_type+'/standard.json').read())
acc.append(np_0['noise_pct_0'][acc_metric]['mean'])
acc_stds.append(np_0['noise_pct_0'][acc_metric]['std_err'])
# other noise pcts
np_res = json.loads(open('../nsl/'+struc_unstruc+'/'+net_type+'/'+deck+'.json').read())
for n in noise_pcts:
acc.append(np_res['noise_pct_'+str(n)][acc_metric]['mean'])
acc_stds.append(np_res['noise_pct_'+str(n)][acc_metric]['std_err'])
return acc, acc_stds
fig2 = plt.figure(constrained_layout=True, figsize=(16,10))
spec2 = gridspec.GridSpec(ncols=3, nrows=2, figure=fig2)
f2_ax1 = fig2.add_subplot(spec2[0, 0])
f2_ax2 = fig2.add_subplot(spec2[0, 1])
f2_ax3 = fig2.add_subplot(spec2[0, 2])
f2_ax4 = fig2.add_subplot(spec2[1, 0])
f2_ax5 = fig2.add_subplot(spec2[1, 1])
f2_ax6 = fig2.add_subplot(spec2[1, 2])
axes = [f2_ax1, f2_ax2, f2_ax3, f2_ax4, f2_ax5]
nps_x = [0]+noise_pcts
for i in range(5):
# Softmax
softmax, softmax_err = get_nsl_results('softmax', decks[i])
axes[i].plot(nps_x, softmax, label = "FF-NSL Softmax 104 examples", color="b", linestyle='-.')
axes[i].errorbar(nps_x, softmax, yerr=softmax_err, color="b", capsize=7,linestyle='-.')
# EDL-GEN
edl_gen, edl_gen_err = get_nsl_results('edl_gen', decks[i])
axes[i].plot(nps_x, edl_gen, label = "FF-NSL EDL-GEN 104 examples", color="k", linestyle='-.')
axes[i].errorbar(nps_x, edl_gen, yerr=edl_gen_err, color="k", capsize=7,linestyle='-.')
# Constant
constant_softmax, constant_err = get_nsl_results('constant_softmax', decks[i])
axes[i].plot(nps_x, constant_softmax, label = "FF-NSL Softmax (constant penalty) 104 examples", color="r", linestyle=':')
axes[i].errorbar(nps_x, constant_softmax, yerr=constant_err, color="r", capsize=5,linestyle=':')
constant_edl_gen, constant_err = get_nsl_results('constant_edl_gen', decks[i])
axes[i].plot(nps_x, constant_edl_gen, label = "FF-NSL EDL-GEN (constant penalty) 104 examples", color="g", linestyle=':')
axes[i].errorbar(nps_x, constant_edl_gen, yerr=constant_err, color="g", capsize=5,linestyle=':')
# Without rank higher
without_rh, without_rh_err = get_nsl_results('without_suit', decks[i])
axes[i].plot(nps_x, without_rh, label = "FF-NSL EDL-GEN (without suit) 104 examples", color="tab:orange", linestyle=':')
axes[i].errorbar(nps_x, without_rh, yerr=without_rh_err, color="tab:orange", capsize=5,linestyle=':')
# Without rank higher
without_rh, without_rh_err = get_nsl_results('without_rank_higher', decks[i])
axes[i].plot(nps_x, without_rh, label = "FF-NSL EDL-GEN (without rank_higher) 104 examples", color="tab:purple", linestyle=':')
axes[i].errorbar(nps_x, without_rh, yerr=without_rh_err, color="tab:purple", capsize=5,linestyle=':')
axes[i].set_xticks(nps_x)
#axes[i].set_yticks(np.arange(0.45,1.01,0.05))
axes[i].set_xlabel('Training ex. subject to distributional shift (%)', fontsize=FONT_SIZE)
axes[i].set_ylabel('Structured test set accuracy', fontsize=FONT_SIZE)
axes[i].grid(True)
axes[i].set_title(decks[i])
# Legend
f2_ax6.legend(*axes[0].get_legend_handles_labels(), loc='center')
f2_ax6.get_xaxis().set_visible(False)
f2_ax6.get_yaxis().set_visible(False)
f2_ax6.set_title('Legend')
plt.show()
```
|
github_jupyter
|
import json
import math
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib as mpl
non_perturbed_deck = 'standard'
decks = [
'batman_joker',
'captain_america',
'adversarial_standard',
'adversarial_batman_joker',
'adversarial_captain_america'
]
noise_pcts = [10,20,30,40,50,60,70,80,90]
FONT_SIZE=18
TICK_FONT_SIZE=12
mpl.rcParams['xtick.labelsize'] = TICK_FONT_SIZE
mpl.rcParams['ytick.labelsize'] = TICK_FONT_SIZE
def get_nsl_results(net_type, deck):
struc_unstruc = 'structured_test_data'
acc_metric = 'accuracy'
# need to build array of results for noise pcts
acc = []
acc_stds = []
# get standard
np_0 = json.loads(open('../nsl/'+struc_unstruc+'/'+net_type+'/standard.json').read())
acc.append(np_0['noise_pct_0'][acc_metric]['mean'])
acc_stds.append(np_0['noise_pct_0'][acc_metric]['std_err'])
# other noise pcts
np_res = json.loads(open('../nsl/'+struc_unstruc+'/'+net_type+'/'+deck+'.json').read())
for n in noise_pcts:
acc.append(np_res['noise_pct_'+str(n)][acc_metric]['mean'])
acc_stds.append(np_res['noise_pct_'+str(n)][acc_metric]['std_err'])
return acc, acc_stds
fig2 = plt.figure(constrained_layout=True, figsize=(16,10))
spec2 = gridspec.GridSpec(ncols=3, nrows=2, figure=fig2)
f2_ax1 = fig2.add_subplot(spec2[0, 0])
f2_ax2 = fig2.add_subplot(spec2[0, 1])
f2_ax3 = fig2.add_subplot(spec2[0, 2])
f2_ax4 = fig2.add_subplot(spec2[1, 0])
f2_ax5 = fig2.add_subplot(spec2[1, 1])
f2_ax6 = fig2.add_subplot(spec2[1, 2])
axes = [f2_ax1, f2_ax2, f2_ax3, f2_ax4, f2_ax5]
nps_x = [0]+noise_pcts
for i in range(5):
# Softmax
softmax, softmax_err = get_nsl_results('softmax', decks[i])
axes[i].plot(nps_x, softmax, label = "FF-NSL Softmax 104 examples", color="b", linestyle='-.')
axes[i].errorbar(nps_x, softmax, yerr=softmax_err, color="b", capsize=7,linestyle='-.')
# EDL-GEN
edl_gen, edl_gen_err = get_nsl_results('edl_gen', decks[i])
axes[i].plot(nps_x, edl_gen, label = "FF-NSL EDL-GEN 104 examples", color="k", linestyle='-.')
axes[i].errorbar(nps_x, edl_gen, yerr=edl_gen_err, color="k", capsize=7,linestyle='-.')
# Constant
constant_softmax, constant_err = get_nsl_results('constant_softmax', decks[i])
axes[i].plot(nps_x, constant_softmax, label = "FF-NSL Softmax (constant penalty) 104 examples", color="r", linestyle=':')
axes[i].errorbar(nps_x, constant_softmax, yerr=constant_err, color="r", capsize=5,linestyle=':')
constant_edl_gen, constant_err = get_nsl_results('constant_edl_gen', decks[i])
axes[i].plot(nps_x, constant_edl_gen, label = "FF-NSL EDL-GEN (constant penalty) 104 examples", color="g", linestyle=':')
axes[i].errorbar(nps_x, constant_edl_gen, yerr=constant_err, color="g", capsize=5,linestyle=':')
# Without rank higher
without_rh, without_rh_err = get_nsl_results('without_suit', decks[i])
axes[i].plot(nps_x, without_rh, label = "FF-NSL EDL-GEN (without suit) 104 examples", color="tab:orange", linestyle=':')
axes[i].errorbar(nps_x, without_rh, yerr=without_rh_err, color="tab:orange", capsize=5,linestyle=':')
# Without rank higher
without_rh, without_rh_err = get_nsl_results('without_rank_higher', decks[i])
axes[i].plot(nps_x, without_rh, label = "FF-NSL EDL-GEN (without rank_higher) 104 examples", color="tab:purple", linestyle=':')
axes[i].errorbar(nps_x, without_rh, yerr=without_rh_err, color="tab:purple", capsize=5,linestyle=':')
axes[i].set_xticks(nps_x)
#axes[i].set_yticks(np.arange(0.45,1.01,0.05))
axes[i].set_xlabel('Training ex. subject to distributional shift (%)', fontsize=FONT_SIZE)
axes[i].set_ylabel('Structured test set accuracy', fontsize=FONT_SIZE)
axes[i].grid(True)
axes[i].set_title(decks[i])
# Legend
f2_ax6.legend(*axes[0].get_legend_handles_labels(), loc='center')
f2_ax6.get_xaxis().set_visible(False)
f2_ax6.get_yaxis().set_visible(False)
f2_ax6.set_title('Legend')
plt.show()
| 0.318803 | 0.355859 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sea
from kneed import KneeLocator
from sklearn.datasets import make_blobs
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from mpl_toolkits.mplot3d import Axes3D
```
Data is obtained from the following site:
https://www.kaggle.com/kaushiksuresh147/customer-segmentation
### Read Data and Make Sense of It
```
auto1 = pd.read_csv("/Users/owner/Desktop/DS_AI_projects/Customer segmentation project/Train.csv")
auto2 = pd.read_csv("/Users/owner/Desktop/DS_AI_projects/Customer segmentation project/Test.csv")
automobile = pd.concat([auto1, auto2])
automobile.head()
automobile.describe()
```
### Imputation, Filling Missing Values, One Hot Encoding
```
automobile.columns = ['id', 'gender', 'married', 'age', 'graduated', 'profession',
'work_experience', 'spending_score', 'family_size', 'var_1',
'segmentation']
automobile = automobile.drop(columns=["id", "segmentation"])
automobile.head()
# treating numerical features of null values with mean values
automobile.fillna(automobile[["age", "work_experience", "family_size"]].mean(), inplace=True)
for c in automobile.columns:
a = automobile[c].value_counts()
print(a)
automobile.isnull().sum()
# binary encoding for categorical features with only 2 possible values.
# dict = {# some kind of dictionary with categorical keys mapping to numerical values}
# df['feature'] = df['feature'].map(dict)
# one-hot encoding for categorical features with 2 or more possible values.
automobiles = pd.get_dummies(automobile, prefix = ['profession'], columns = ['profession'])
automobiles = pd.get_dummies(automobiles, prefix = ['gender'], columns = ['gender'])
automobiles = pd.get_dummies(automobiles, prefix = ['married'], columns = ['married'])
automobiles = pd.get_dummies(automobiles, prefix = ['graduated'], columns = ['graduated'])
automobiles = pd.get_dummies(automobiles, prefix = ['var_1'], columns = ['var_1'])
automobiles.head(10)
automobiles.drop(columns=["gender_Male"], inplace=True)
# Ordincal feature encoding for features that have ordered values
from sklearn.preprocessing import LabelEncoder
#label encoder can't handle missing values
automobiles['spending_score'] = automobiles['spending_score'].fillna('None')
# Label encode ord_1 feature
label_encoder = LabelEncoder()
automobiles['spending_score'] = label_encoder.fit_transform(automobiles['spending_score'])
# Print sample of dataset
automobiles.head(10)
# feature standardization
scaler = StandardScaler().fit(automobiles)
scaled_features = scaler.transform(automobiles)
scaled_features = pd.DataFrame(scaled_features, columns = automobiles.columns)
scaled_features.head()
```
### Make a Plot on the "Elbow Method"
```
# store the squared sum of distances from the mean of each cluster to the data points to that mean
sse = []
for cluster in range(1,10):
kmeans = KMeans(n_clusters = cluster, init='k-means++')
kmeans.fit(automobiles)
sse.append(kmeans.inertia_)
# converting the results into a dataframe and plotting them
clustering = pd.DataFrame({'Cluster':range(1,10), 'SSE':sse})
#plt.figure(figsize=(12,6))
plt.plot(clustering['Cluster'], clustering['SSE'], marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Inertia')
# based on the elbow method, the best number of clusters to set would be 4,
# since after K=4, the inertia decreases in a linear manner.
# Build a model with 4 clusters
kmeans = KMeans(n_clusters=4, init='k-means++')
kmeans.fit(automobiles)
# Print the silhouette score for the model above
print(silhouette_score(automobiles, kmeans.labels_, metric='euclidean'))
```
### Try Improve the Quality of Clusters by Selecting Useful Features
```
# attempt to improve the model using PCA to reduce dimensionality.
pca = PCA(n_components=5)
pca_fit = pca.fit_transform(automobiles)
n_comp = range(pca.n_components_)
plt.bar(n_comp, pca.explained_variance_ratio_, color='blue')
plt.xlabel('PCA features')
plt.ylabel('variance %')
pca_components = pd.DataFrame(pca_fit)
pca_components.head()
# building a new model after PCA.
inertia = []
for cluster in range(1,10):
kmeans = KMeans(n_clusters = cluster)
kmeans.fit(pca_components.iloc[:,:2])
inertia.append(kmeans.inertia_)
# converting the results into a dataframe and plotting them
clusters = pd.DataFrame({'Cluster':range(1,10), 'Inertia':inertia})
# plt.figure(figsize=(12,6))
plt.plot(clusters['Cluster'], clusters['Inertia'], marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Inertia')
# still, K=4 seems like the best approach.
kmeans_new = KMeans(n_clusters=4)
kmeans_new.fit(pca_components.iloc[:,:2])
# silhouette score
print(silhouette_score(pca_components.iloc[:,:2], kmeans_new.labels_, metric='euclidean'))
```
We see roughly 4.5% increase in the Silhouette score!
```
model = KMeans(n_clusters=4)
clusters = model.fit_predict(pca_components.iloc[:,:2])
automobiles["label"] = clusters
fig = plt.figure(figsize=(21,10))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(automobiles.age[automobiles.label == 0], automobiles["work_experience"][automobiles.label == 0], automobiles["spending_score"][automobiles.label == 0], c='blue', s=60)
ax.scatter(automobiles.age[automobiles.label == 1], automobiles["work_experience"][automobiles.label == 1], automobiles["spending_score"][automobiles.label == 1], c='red', s=60)
ax.scatter(automobiles.age[automobiles.label == 2], automobiles["work_experience"][automobiles.label == 2], automobiles["spending_score"][automobiles.label == 2], c='green', s=60)
ax.scatter(automobiles.age[automobiles.label == 3], automobiles["work_experience"][automobiles.label == 3], automobiles["spending_score"][automobiles.label == 3], c='orange', s=60)
ax.view_init(30, 40)
plt.show()
automobiles.columns
```
As you can see, the clusters look like they can be separated quite easily, except a few spots where different colors overlap a bit. As far as the results go, I'm satisfied with the clusters!
### Take a Look at the Demographics
```
sns.barplot(x='label', y='age', data=automobiles).set(title='Cluster vs Age')
sns.barplot(x='label', y='family_size', data=automobiles).set(title='Cluster vs Family Size')
sns.barplot(x='label', y='spending_score', data=automobiles).set(title='Cluster vs Spending Score')
sns.barplot(x='label', y='work_experience', data=automobiles).set(title='Cluster vs Work Experience')
```
### Now take a look at the count of each cluster
```
automobile = automobile.fillna('Unknown')
automobile["cluster"] = clusters
automobile.head()
sns.catplot(x="cluster", kind="count", data=automobile)
```
The above barplots show the following insights:
1. Cluster 1: The averge or typical customer in this cluster is middle-aged (around 40) with an average family size of 2.5 and a moderate ~ moderately high spending score. This customer group has the longest years of work_experience on average. This is the largest customer group. My recommendation to this company is to focus on utility —— advertise versatile models that cater to a variety of driving consitions and preferences to maximize the reach to the largest pool of potential buyers.
2. Cluster 2: The typical customer in this cluster is a senior (over 70) with an average family size of 2 and a moderate ~ moderately high spending score. This is the smallest group and its members should be mostly retired.
3. Cluster 3: The typical customer is a slightly older middle-aged person with an average family size of 2.8 and the lowest spending score out of all groups with a less than 1.0 average. This group won't be able to afford expensive vehicles, probably because it has more family members to spend money on or this group as a whole do not spend beyond their needs. The company's strategy should be to advertise less expensive, mid-to-low tier vehicles.
4. Cluster 4: The typical customer is a young adult aged around 25 who has the highest spending score of all groups and the largest average family size! This is surprising because we normally assume older customers to have more spending power and larger family size, but for this automobile company, this is simply not the case. This cluster is likely a group of young working professionals with high pay and/or heritage. The company should promote the most top-of-the-line car models to this group as their primary target consumer base because this group is most likely able to afford more expensive cars.
```
automobiles.columns
automobiles.groupby("label")["graduated_Yes"].mean()
plt.figure(figsize=(20,5))
sns.catplot(x="profession", kind="count", hue='cluster', data=automobile, palette="ch:.25",
height=5, aspect=3)
```
We can see the following trends:
1. Artists, entertainment workers, and engineers are most represented in the 1st cluster.
2. Lawyers, artists, and executives are most represented in the 2nd cluster.
3. Artists, entertainment workers, and executives are most represented in the 3rd cluster.
4. Healthcare professionals, doctors, and artists are most represented in the 4th cluster.
```
sns.catplot(x="married", y="age", kind="bar", hue='cluster', data=automobile, palette="ch:.25",
height=4, aspect=2)
sns.catplot(x="married", kind="count", hue='cluster', data=automobile, palette="ch:.25",
height=4, aspect=2)
```
For all clusters except cluster 3 (actually the cluster 4 because python starts counting from 0), there are more married people than unmarried. The last cluster (represented by black) is the young professional group. It's understandable that most of them are unmarried.
```
sns.catplot(x="married", y="age", col="gender", kind="bar", hue='cluster', data=automobile, palette="ch:.25")
```
The charts show that the age for each cluster for unmarried and married people is roughly equivalent. Age for female customers in each cluster is lower than the male counterparts.
```
automobile["gender"].value_counts()
print(f"Male to Female Ratio is: {5841/4854}")
```
### This is the end of the analysis!
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sea
from kneed import KneeLocator
from sklearn.datasets import make_blobs
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from mpl_toolkits.mplot3d import Axes3D
auto1 = pd.read_csv("/Users/owner/Desktop/DS_AI_projects/Customer segmentation project/Train.csv")
auto2 = pd.read_csv("/Users/owner/Desktop/DS_AI_projects/Customer segmentation project/Test.csv")
automobile = pd.concat([auto1, auto2])
automobile.head()
automobile.describe()
automobile.columns = ['id', 'gender', 'married', 'age', 'graduated', 'profession',
'work_experience', 'spending_score', 'family_size', 'var_1',
'segmentation']
automobile = automobile.drop(columns=["id", "segmentation"])
automobile.head()
# treating numerical features of null values with mean values
automobile.fillna(automobile[["age", "work_experience", "family_size"]].mean(), inplace=True)
for c in automobile.columns:
a = automobile[c].value_counts()
print(a)
automobile.isnull().sum()
# binary encoding for categorical features with only 2 possible values.
# dict = {# some kind of dictionary with categorical keys mapping to numerical values}
# df['feature'] = df['feature'].map(dict)
# one-hot encoding for categorical features with 2 or more possible values.
automobiles = pd.get_dummies(automobile, prefix = ['profession'], columns = ['profession'])
automobiles = pd.get_dummies(automobiles, prefix = ['gender'], columns = ['gender'])
automobiles = pd.get_dummies(automobiles, prefix = ['married'], columns = ['married'])
automobiles = pd.get_dummies(automobiles, prefix = ['graduated'], columns = ['graduated'])
automobiles = pd.get_dummies(automobiles, prefix = ['var_1'], columns = ['var_1'])
automobiles.head(10)
automobiles.drop(columns=["gender_Male"], inplace=True)
# Ordincal feature encoding for features that have ordered values
from sklearn.preprocessing import LabelEncoder
#label encoder can't handle missing values
automobiles['spending_score'] = automobiles['spending_score'].fillna('None')
# Label encode ord_1 feature
label_encoder = LabelEncoder()
automobiles['spending_score'] = label_encoder.fit_transform(automobiles['spending_score'])
# Print sample of dataset
automobiles.head(10)
# feature standardization
scaler = StandardScaler().fit(automobiles)
scaled_features = scaler.transform(automobiles)
scaled_features = pd.DataFrame(scaled_features, columns = automobiles.columns)
scaled_features.head()
# store the squared sum of distances from the mean of each cluster to the data points to that mean
sse = []
for cluster in range(1,10):
kmeans = KMeans(n_clusters = cluster, init='k-means++')
kmeans.fit(automobiles)
sse.append(kmeans.inertia_)
# converting the results into a dataframe and plotting them
clustering = pd.DataFrame({'Cluster':range(1,10), 'SSE':sse})
#plt.figure(figsize=(12,6))
plt.plot(clustering['Cluster'], clustering['SSE'], marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Inertia')
# based on the elbow method, the best number of clusters to set would be 4,
# since after K=4, the inertia decreases in a linear manner.
# Build a model with 4 clusters
kmeans = KMeans(n_clusters=4, init='k-means++')
kmeans.fit(automobiles)
# Print the silhouette score for the model above
print(silhouette_score(automobiles, kmeans.labels_, metric='euclidean'))
# attempt to improve the model using PCA to reduce dimensionality.
pca = PCA(n_components=5)
pca_fit = pca.fit_transform(automobiles)
n_comp = range(pca.n_components_)
plt.bar(n_comp, pca.explained_variance_ratio_, color='blue')
plt.xlabel('PCA features')
plt.ylabel('variance %')
pca_components = pd.DataFrame(pca_fit)
pca_components.head()
# building a new model after PCA.
inertia = []
for cluster in range(1,10):
kmeans = KMeans(n_clusters = cluster)
kmeans.fit(pca_components.iloc[:,:2])
inertia.append(kmeans.inertia_)
# converting the results into a dataframe and plotting them
clusters = pd.DataFrame({'Cluster':range(1,10), 'Inertia':inertia})
# plt.figure(figsize=(12,6))
plt.plot(clusters['Cluster'], clusters['Inertia'], marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Inertia')
# still, K=4 seems like the best approach.
kmeans_new = KMeans(n_clusters=4)
kmeans_new.fit(pca_components.iloc[:,:2])
# silhouette score
print(silhouette_score(pca_components.iloc[:,:2], kmeans_new.labels_, metric='euclidean'))
model = KMeans(n_clusters=4)
clusters = model.fit_predict(pca_components.iloc[:,:2])
automobiles["label"] = clusters
fig = plt.figure(figsize=(21,10))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(automobiles.age[automobiles.label == 0], automobiles["work_experience"][automobiles.label == 0], automobiles["spending_score"][automobiles.label == 0], c='blue', s=60)
ax.scatter(automobiles.age[automobiles.label == 1], automobiles["work_experience"][automobiles.label == 1], automobiles["spending_score"][automobiles.label == 1], c='red', s=60)
ax.scatter(automobiles.age[automobiles.label == 2], automobiles["work_experience"][automobiles.label == 2], automobiles["spending_score"][automobiles.label == 2], c='green', s=60)
ax.scatter(automobiles.age[automobiles.label == 3], automobiles["work_experience"][automobiles.label == 3], automobiles["spending_score"][automobiles.label == 3], c='orange', s=60)
ax.view_init(30, 40)
plt.show()
automobiles.columns
sns.barplot(x='label', y='age', data=automobiles).set(title='Cluster vs Age')
sns.barplot(x='label', y='family_size', data=automobiles).set(title='Cluster vs Family Size')
sns.barplot(x='label', y='spending_score', data=automobiles).set(title='Cluster vs Spending Score')
sns.barplot(x='label', y='work_experience', data=automobiles).set(title='Cluster vs Work Experience')
automobile = automobile.fillna('Unknown')
automobile["cluster"] = clusters
automobile.head()
sns.catplot(x="cluster", kind="count", data=automobile)
automobiles.columns
automobiles.groupby("label")["graduated_Yes"].mean()
plt.figure(figsize=(20,5))
sns.catplot(x="profession", kind="count", hue='cluster', data=automobile, palette="ch:.25",
height=5, aspect=3)
sns.catplot(x="married", y="age", kind="bar", hue='cluster', data=automobile, palette="ch:.25",
height=4, aspect=2)
sns.catplot(x="married", kind="count", hue='cluster', data=automobile, palette="ch:.25",
height=4, aspect=2)
sns.catplot(x="married", y="age", col="gender", kind="bar", hue='cluster', data=automobile, palette="ch:.25")
automobile["gender"].value_counts()
print(f"Male to Female Ratio is: {5841/4854}")
| 0.521227 | 0.879716 |
```
# Liberally borrowing from various notebooks to get this all in one place
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import astropy.units as u
from astropy.io import fits
from astroduet.background import background_pixel_rate
from astroduet.config import Telescope
from astroduet.models import load_model_fluence, load_model_ABmag
from astroduet.lightcurve import get_lightcurve, lightcurve_through_image
from astroduet.utils import suppress_stdout, duet_fluence_to_abmag
from astroduet.image_utils import construct_image, estimate_background
from astroduet.diff_image import py_zogy
# Set up
duet = Telescope()
exposure = 300 * u.s
distance = 100 * u.Mpc
dist_mod = 5. * np.log10(distance.to_value(u.pc)) - 5.
[bgd_band1, bgd_band2] = background_pixel_rate(duet, low_zodi = True)
# Load EMGW lightcurves
shock_5e10_time, shock_5e10_fluence1, shock_5e10_fluence2 = load_model_fluence('shock_5e10.dat',distance)
shock_5e10_rate1, shock_5e10_rate2 = duet.fluence_to_rate(shock_5e10_fluence1), duet.fluence_to_rate(shock_5e10_fluence2)
_, shock_5e10_ABmag1, shock_5e10_ABmag2 = load_model_ABmag('shock_5e10.dat',distance)
blukn_04_time, blukn_04_fluence1, blukn_04_fluence2 = load_model_fluence('kilonova_0.04.dat',distance)
blukn_04_rate1, blukn_04_rate2 = duet.fluence_to_rate(blukn_04_fluence1), duet.fluence_to_rate(blukn_04_fluence2)
_, blukn_04_ABmag1, blukn_04_ABmag2 = load_model_ABmag('kilonova_0.04.dat',distance)
# Plot smooth model inputs
# Plot lightcurve for shock and for blue kilonova, in the two bands, at 100 Mpc
font = {'size': 22}
matplotlib.rc('font', **font)
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1, color='lightcoral', linestyle='-', linewidth=2, label='Shock model (5e10), DUET 1')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag2, color='lightcoral', linestyle='--', linewidth=2, label='Shock model (5e10), DUET 2')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1, color='deepskyblue', linestyle='-', linewidth=2, label='Blue Kilonova (0.04), DUET 1')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag2, color='deepskyblue', linestyle='--', linewidth=2, label='Blue Kilonova (0.04), DUET 2')
plt.axhline(y=22,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(24,17)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel(r'DUET ABmag')
plt.title('Shock and blue kilonova models @ 100 Mpc')
plt.show()
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel(r'DUET1 - DUET2')
plt.title('Color evolution @ 100 Mpc')
plt.show()
# Get lightcurves through images
shock_lightcurve_init = get_lightcurve("shock_5e10.dat", distance=distance)
shock_lightcurve = lightcurve_through_image(shock_lightcurve_init, exposure=exposure)
shock_lightcurve_rebin = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, final_resolution=2400*u.s)
blukn_lightcurve_init = get_lightcurve("kilonova_0.04.dat", distance=distance)
blukn_lightcurve = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure)
blukn_lightcurve_rebin = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, final_resolution=2400*u.s)
# And with a spiral galaxy in place
shock_lightcurve_gal = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, gal_type='spiral')
blukn_lightcurve_gal = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, gal_type='spiral')
shock_lightcurve_gal_rebin = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, gal_type='spiral',
final_resolution=2400*u.s)
blukn_lightcurve_gal_rebin = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, gal_type='spiral',
final_resolution=2400*u.s)
# Note: surface brightness is independent of distance, galaxy angular size is not
def get_plottables(lc):
good1 = (lc['fluence_D1_fit'] > 0) & (lc['fluence_D1_fiterr'] < lc['fluence_D1_fit'])
good2 = (lc['fluence_D2_fit'] > 0) & (lc['fluence_D2_fiterr'] < lc['fluence_D2_fit'])
good = good1 & good2
rate_meas1, rate_meas2 = duet.fluence_to_rate(lc['fluence_D1_fit']), duet.fluence_to_rate(lc['fluence_D2_fit'])
rate_meas_err1, rate_meas_err2 = duet.fluence_to_rate(lc['fluence_D1_fiterr']), duet.fluence_to_rate(lc['fluence_D2_fiterr'])
mag_meas1, mag_meas2 = duet_fluence_to_abmag(lc['fluence_D1_fit'], band=duet.bandpass1), duet_fluence_to_abmag(lc['fluence_D2_fit'], band=duet.bandpass2)
color = mag_meas1 - mag_meas2
return rate_meas1, rate_meas2, rate_meas_err1, rate_meas_err2, mag_meas1, mag_meas2, color, good
# Measured stuff
shock_rate_meas1, shock_rate_meas2, shock_rate_meas_err1, shock_rate_meas_err2, shock_mag_meas1, shock_mag_meas2, shock_color, sgood = get_plottables(shock_lightcurve)
blukn_rate_meas1, blukn_rate_meas2, blukn_rate_meas_err1, blukn_rate_meas_err2, blukn_mag_meas1, blukn_mag_meas2, blukn_color, bgood = get_plottables(blukn_lightcurve)
shock_rate_rebin_meas1, shock_rate_rebin_meas2, shock_rate_rebin_meas_err1, shock_rate_rebin_meas_err2, shock_mag_rebin_meas1, shock_mag_rebin_meas2, shock_rebin_color, srgood = get_plottables(shock_lightcurve_rebin)
blukn_rate_rebin_meas1, blukn_rate_rebin_meas2, blukn_rate_rebin_meas_err1, blukn_rate_rebin_meas_err2, blukn_mag_rebin_meas1, blukn_mag_rebin_meas2, blukn_rebin_color, brgood = get_plottables(blukn_lightcurve_rebin)
shock_rate_gal_meas1, shock_rate_gal_meas2, shock_rate_gal_meas_err1, shock_rate_gal_meas_err2, shock_mag_gal_meas1, shock_mag_gal_meas2, shock_color_gal, sggood = get_plottables(shock_lightcurve_gal)
blukn_rate_gal_meas1, blukn_rate_gal_meas2, blukn_rate_gal_meas_err1, blukn_rate_gal_meas_err2, blukn_mag_gal_meas1, blukn_mag_gal_meas2, blukn_color_gal, bggood = get_plottables(blukn_lightcurve_gal)
shock_rate_gal_rebin_meas1, shock_rate_gal_rebin_meas2, shock_rate_gal_rebin_meas_err1, shock_rate_gal_rebin_meas_err2, shock_mag_gal_rebin_meas1, shock_mag_gal_rebin_meas2, shock_gal_rebin_color, sgrgood = get_plottables(shock_lightcurve_gal_rebin)
blukn_rate_gal_rebin_meas1, blukn_rate_gal_rebin_meas2, blukn_rate_gal_rebin_meas_err1, blukn_rate_gal_rebin_meas_err2, blukn_mag_gal_rebin_meas1, blukn_mag_gal_rebin_meas2, blukn_gal_rebin_color, bgrgood = get_plottables(blukn_lightcurve_gal_rebin)
font = {'size': 22}
matplotlib.rc('font', **font)
plt.figure(figsize=(12, 8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate1, color='coral', linestyle='-')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate2, color='lightcoral', linestyle='--')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate1, color='deepskyblue', linestyle='-')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate2, color='skyblue', linestyle='--')
plt.errorbar(shock_lightcurve['time'].to(u.d).value[sgood], shock_rate_meas1.value[sgood], fmt='o',
markersize=5, yerr=shock_rate_meas_err1.value[sgood], color='coral', label='Shock model (5e10), DUET 1')
plt.errorbar(shock_lightcurve['time'].to(u.d).value[sgood], shock_rate_meas2.value[sgood], fmt='o',
markersize=5, yerr=shock_rate_meas_err2.value[sgood], color='lightcoral', label='Shock model (5e10), DUET 2')
plt.errorbar(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rate_rebin_meas1.value[srgood], fmt='o',
markersize=5, yerr=shock_rate_rebin_meas_err1.value[srgood], color='red', label='Shock model rebinned')
plt.errorbar(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rate_rebin_meas2.value[srgood], fmt='o',
markersize=5, yerr=shock_rate_rebin_meas_err2.value[srgood], color='red')
plt.errorbar(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_rate_meas1.value[bgood], fmt='o',
markersize=5, yerr=blukn_rate_meas_err1.value[bgood], color='deepskyblue', label='Blue Kilonova (0.04), DUET 1')
plt.errorbar(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_rate_meas2.value[bgood], fmt='o',
markersize=5, yerr=blukn_rate_meas_err2.value[bgood], color='skyblue', label='Blue Kilonova (0.04), DUET 2')
plt.errorbar(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rate_rebin_meas1.value[brgood], fmt='o',
markersize=5, yerr=blukn_rate_rebin_meas_err1.value[brgood], color='blue', label='Blue kilonova rebinned')
plt.errorbar(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rate_rebin_meas2.value[brgood], fmt='o',
markersize=5, yerr=blukn_rate_rebin_meas_err2.value[brgood], color='blue')
plt.legend()
plt.ylabel("Count rate (ph/s)")
plt.xlabel("Time after merger (d)")
plt.ylim(0,9)
plt.xlim(-0.1,1)
plt.show()
'''
plt.figure(figsize=(12, 8))
plt.scatter(shock_lightcurve['time'].to(u.d).value, shock_lightcurve['snr_D1'], s=5, color='coral', label='Shock model (2.5e10)')
plt.scatter(shock_lightcurve['time'].to(u.d).value, shock_lightcurve['snr_D2'], s=5, color='lightcoral', label='_Shock model (2.5e10)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value, blukn_lightcurve['snr_D1'], s=5, color='deepskyblue', label='Blue Kilonova (0.01)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value, blukn_lightcurve['snr_D2'], s=5, color='skyblue', label='_Blue Kilonova (0.01)')
plt.ylabel("S/N")
plt.xlabel("Time after merger (d)")
plt.xlim(-0.1,1)
plt.legend()
plt.show()
'''
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='_Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='_Blue Kilonova (0.04)')
plt.scatter(shock_lightcurve['time'].to(u.d).value[sgood], shock_color.value[sgood], s=5,
color='coral', linewidth=2, label='Shock model (5e10)')
plt.scatter(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rebin_color.value[srgood], s=5,
color='red', linewidth=2, label='_Shock model (5e10)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_color.value[bgood], s=5,
color='deepskyblue', linewidth=2, label='Blue Kilonova (0.04)')
plt.scatter(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rebin_color.value[brgood], s=5,
color='blue', linewidth=2, label='_Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel('DUET1 - DUET2')
plt.title('Color evolution @ 100 Mpc')
plt.show()
# This time with added galaxy
plt.figure(figsize=(12, 8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate1, color='coral', linestyle='-')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate2, color='lightcoral', linestyle='--')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate1, color='deepskyblue', linestyle='-')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate2, color='skyblue', linestyle='--')
plt.errorbar(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_rate_gal_meas1.value[sggood], fmt='o',
markersize=5, yerr=shock_rate_gal_meas_err1.value[sggood], color='coral', label='Shock model (5e10), DUET 1')
plt.errorbar(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_rate_gal_meas2.value[sggood], fmt='o',
markersize=5, yerr=shock_rate_gal_meas_err2.value[sggood], color='lightcoral', label='Shock model (5e10), DUET 2')
plt.errorbar(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_rate_gal_meas1.value[bggood], fmt='o',
markersize=5, yerr=blukn_rate_gal_meas_err1.value[bggood], color='deepskyblue', label='Blue Kilonova (0.04), DUET 1')
plt.errorbar(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_rate_gal_meas2.value[bggood], fmt='o',
markersize=5, yerr=blukn_rate_gal_meas_err2.value[bggood], color='skyblue', label='Blue Kilonova (0.04), DUET 2')
#plt.errorbar(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_rate_gal_rebin_meas1.value[sgrgood], fmt='o',
# markersize=5, yerr=shock_rate_gal_rebin_meas_err1.value[sgrgood], color='darkred', label='_Shock model (5e10)')
#plt.errorbar(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_rate_gal_rebin_meas2.value[sgrgood], fmt='o',
# markersize=5, yerr=shock_rate_gal_rebin_meas_err2.value[sgrgood], color='red', label='_Shock model (5e10)')
#plt.errorbar(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_rate_gal_rebin_meas1.value[bgrgood], fmt='o',
# markersize=5, yerr=blukn_rate_gal_rebin_meas_err1.value[bgrgood], color='darkblue', label='_Blue Kilonova (0.04)')
#plt.errorbar(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_rate_gal_rebin_meas2.value[bgrgood], fmt='o',
# markersize=5, yerr=blukn_rate_gal_rebin_meas_err2.value[bgrgood], color='blue', label='_Blue Kilonova (0.04)')
plt.legend()
plt.title('Shock and blue kilonova, spiral galaxy @ 100 Mpc')
plt.ylabel("Count rate (ph/s)")
plt.xlabel("Time after merger (d)")
plt.ylim(0,9)
plt.xlim(-0.1,1)
plt.show()
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='_Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='_Blue Kilonova (0.04)')
plt.scatter(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_color_gal.value[sggood], s=5,
color='coral', linewidth=2, label='Shock model (5e10)')
plt.scatter(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_color_gal.value[bggood], s=5,
color='deepskyblue', linewidth=2, label='Blue Kilonova (0.04)')
#plt.scatter(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_gal_rebin_color.value[sgrgood], s=5,
# color='red', linewidth=2, label='_Shock model (5e10)')
#plt.scatter(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_gal_rebin_color.value[bgrgood], s=5,
# color='blue', linewidth=2, label='_Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel("DUET1 - DUET2")
plt.title('Color evolution @ 100 Mpc')
plt.show()
# Show example images
frame = np.array([30, 30])
nexp = 5
ref_image1 = construct_image(frame, exposure, duet=duet, band=duet.bandpass1,
gal_type='spiral', sky_rate=bgd_band1, n_exp=nexp)
ref_image_rate1 = ref_image1 / (exposure * nexp)
ref_bkg1, ref_bkg_rms_median1 = estimate_background(ref_image_rate1, method='1D', sigma=2)
ref_rate_bkgsub1 = ref_image_rate1 - ref_bkg1
psf_array = duet.psf_model(x_size=5,y_size=5).array
# Example image at peak of shock light curve
src1 = shock_rate_gal_meas1[7]
image1 = construct_image(frame, exposure, duet=duet, band=duet.bandpass1, source=src1,
gal_type='spiral', sky_rate=bgd_band1)
image_rate1 = image1 / exposure
plt.imshow(image_rate1.value, cmap='viridis', aspect=1, origin='lower')
plt.colorbar()
plt.show()
# Difference image
image_bkg, image_bkg_rms_median = estimate_background(image_rate1, method='1D', sigma=2)
ref_bkg, ref_bkg_rms_median = estimate_background(ref_image_rate1, method='1D', sigma=2)
image_rate_bkgsub, ref_rate_bkgsub = image_rate1 - image_bkg, ref_image_rate1 - ref_bkg
s_n, s_r = np.sqrt(image_rate1), np.sqrt(ref_image_rate1) # 2D uncertainty (sigma) - that is, noise on the background
sn, sr = np.mean(s_n), np.mean(s_r) # Average uncertainty (sigma)
dx, dy = 0.1, 0.01 # Astrometric uncertainty (sigma)
diff_image, d_psf, s_corr = py_zogy(image_rate_bkgsub.value,
ref_rate_bkgsub.value,
psf_array,psf_array,
s_n.value,s_r.value,
sn.value,sr.value,dx,dy)
diff_image *= image_rate_bkgsub.unit
plt.imshow(diff_image.value, cmap='viridis', aspect=1, origin='lower')
plt.colorbar()
plt.show()
# To what volume are we going to get 'quality lightcurves to distinguish models' ?
# - 'quality light curve' = sufficient to distinguish between the two models to x sigma
# To what volume will we get a detection/localisation?
# If we stack x images, we can get a detection to y sigma out to blah Mpc
# Two-pointing strategy vs. six-pointing strategy. Only two-inteferometer events.
# What is the advanced LIGO sensitivity
# With probabilities of getting there in time from Kristin
# and binning from Matteo
# For shock and for kilonova:
# As above, only go through a number of distances.
# For each distance, first a detection test: get difference image in both bands, sum, locate source. (Summing all exposures in an orbit, in a worst-case where we miss the peak.)
# Second, a lightcurve test: can we rule out the alternative model with this lightcurve?
# These probabilities above - multiply them with the probability of getting there in time from Kristin
# How many stacked exposures required for a detection as a function of distance? That informs Kristin's operations simulations.
# For different start times: 30 mins, 50 mins, 70 mins
# For different distances
# For different positions in the galaxy: sources at r_eff/2, r_eff, r_eff*2
# Function to get detection significance at 1 exposure, 2 exposures etc... until 5-sigma detection is reached
```
|
github_jupyter
|
# Liberally borrowing from various notebooks to get this all in one place
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import astropy.units as u
from astropy.io import fits
from astroduet.background import background_pixel_rate
from astroduet.config import Telescope
from astroduet.models import load_model_fluence, load_model_ABmag
from astroduet.lightcurve import get_lightcurve, lightcurve_through_image
from astroduet.utils import suppress_stdout, duet_fluence_to_abmag
from astroduet.image_utils import construct_image, estimate_background
from astroduet.diff_image import py_zogy
# Set up
duet = Telescope()
exposure = 300 * u.s
distance = 100 * u.Mpc
dist_mod = 5. * np.log10(distance.to_value(u.pc)) - 5.
[bgd_band1, bgd_band2] = background_pixel_rate(duet, low_zodi = True)
# Load EMGW lightcurves
shock_5e10_time, shock_5e10_fluence1, shock_5e10_fluence2 = load_model_fluence('shock_5e10.dat',distance)
shock_5e10_rate1, shock_5e10_rate2 = duet.fluence_to_rate(shock_5e10_fluence1), duet.fluence_to_rate(shock_5e10_fluence2)
_, shock_5e10_ABmag1, shock_5e10_ABmag2 = load_model_ABmag('shock_5e10.dat',distance)
blukn_04_time, blukn_04_fluence1, blukn_04_fluence2 = load_model_fluence('kilonova_0.04.dat',distance)
blukn_04_rate1, blukn_04_rate2 = duet.fluence_to_rate(blukn_04_fluence1), duet.fluence_to_rate(blukn_04_fluence2)
_, blukn_04_ABmag1, blukn_04_ABmag2 = load_model_ABmag('kilonova_0.04.dat',distance)
# Plot smooth model inputs
# Plot lightcurve for shock and for blue kilonova, in the two bands, at 100 Mpc
font = {'size': 22}
matplotlib.rc('font', **font)
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1, color='lightcoral', linestyle='-', linewidth=2, label='Shock model (5e10), DUET 1')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag2, color='lightcoral', linestyle='--', linewidth=2, label='Shock model (5e10), DUET 2')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1, color='deepskyblue', linestyle='-', linewidth=2, label='Blue Kilonova (0.04), DUET 1')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag2, color='deepskyblue', linestyle='--', linewidth=2, label='Blue Kilonova (0.04), DUET 2')
plt.axhline(y=22,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(24,17)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel(r'DUET ABmag')
plt.title('Shock and blue kilonova models @ 100 Mpc')
plt.show()
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel(r'DUET1 - DUET2')
plt.title('Color evolution @ 100 Mpc')
plt.show()
# Get lightcurves through images
shock_lightcurve_init = get_lightcurve("shock_5e10.dat", distance=distance)
shock_lightcurve = lightcurve_through_image(shock_lightcurve_init, exposure=exposure)
shock_lightcurve_rebin = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, final_resolution=2400*u.s)
blukn_lightcurve_init = get_lightcurve("kilonova_0.04.dat", distance=distance)
blukn_lightcurve = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure)
blukn_lightcurve_rebin = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, final_resolution=2400*u.s)
# And with a spiral galaxy in place
shock_lightcurve_gal = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, gal_type='spiral')
blukn_lightcurve_gal = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, gal_type='spiral')
shock_lightcurve_gal_rebin = lightcurve_through_image(shock_lightcurve_init, exposure=exposure, gal_type='spiral',
final_resolution=2400*u.s)
blukn_lightcurve_gal_rebin = lightcurve_through_image(blukn_lightcurve_init, exposure=exposure, gal_type='spiral',
final_resolution=2400*u.s)
# Note: surface brightness is independent of distance, galaxy angular size is not
def get_plottables(lc):
good1 = (lc['fluence_D1_fit'] > 0) & (lc['fluence_D1_fiterr'] < lc['fluence_D1_fit'])
good2 = (lc['fluence_D2_fit'] > 0) & (lc['fluence_D2_fiterr'] < lc['fluence_D2_fit'])
good = good1 & good2
rate_meas1, rate_meas2 = duet.fluence_to_rate(lc['fluence_D1_fit']), duet.fluence_to_rate(lc['fluence_D2_fit'])
rate_meas_err1, rate_meas_err2 = duet.fluence_to_rate(lc['fluence_D1_fiterr']), duet.fluence_to_rate(lc['fluence_D2_fiterr'])
mag_meas1, mag_meas2 = duet_fluence_to_abmag(lc['fluence_D1_fit'], band=duet.bandpass1), duet_fluence_to_abmag(lc['fluence_D2_fit'], band=duet.bandpass2)
color = mag_meas1 - mag_meas2
return rate_meas1, rate_meas2, rate_meas_err1, rate_meas_err2, mag_meas1, mag_meas2, color, good
# Measured stuff
shock_rate_meas1, shock_rate_meas2, shock_rate_meas_err1, shock_rate_meas_err2, shock_mag_meas1, shock_mag_meas2, shock_color, sgood = get_plottables(shock_lightcurve)
blukn_rate_meas1, blukn_rate_meas2, blukn_rate_meas_err1, blukn_rate_meas_err2, blukn_mag_meas1, blukn_mag_meas2, blukn_color, bgood = get_plottables(blukn_lightcurve)
shock_rate_rebin_meas1, shock_rate_rebin_meas2, shock_rate_rebin_meas_err1, shock_rate_rebin_meas_err2, shock_mag_rebin_meas1, shock_mag_rebin_meas2, shock_rebin_color, srgood = get_plottables(shock_lightcurve_rebin)
blukn_rate_rebin_meas1, blukn_rate_rebin_meas2, blukn_rate_rebin_meas_err1, blukn_rate_rebin_meas_err2, blukn_mag_rebin_meas1, blukn_mag_rebin_meas2, blukn_rebin_color, brgood = get_plottables(blukn_lightcurve_rebin)
shock_rate_gal_meas1, shock_rate_gal_meas2, shock_rate_gal_meas_err1, shock_rate_gal_meas_err2, shock_mag_gal_meas1, shock_mag_gal_meas2, shock_color_gal, sggood = get_plottables(shock_lightcurve_gal)
blukn_rate_gal_meas1, blukn_rate_gal_meas2, blukn_rate_gal_meas_err1, blukn_rate_gal_meas_err2, blukn_mag_gal_meas1, blukn_mag_gal_meas2, blukn_color_gal, bggood = get_plottables(blukn_lightcurve_gal)
shock_rate_gal_rebin_meas1, shock_rate_gal_rebin_meas2, shock_rate_gal_rebin_meas_err1, shock_rate_gal_rebin_meas_err2, shock_mag_gal_rebin_meas1, shock_mag_gal_rebin_meas2, shock_gal_rebin_color, sgrgood = get_plottables(shock_lightcurve_gal_rebin)
blukn_rate_gal_rebin_meas1, blukn_rate_gal_rebin_meas2, blukn_rate_gal_rebin_meas_err1, blukn_rate_gal_rebin_meas_err2, blukn_mag_gal_rebin_meas1, blukn_mag_gal_rebin_meas2, blukn_gal_rebin_color, bgrgood = get_plottables(blukn_lightcurve_gal_rebin)
font = {'size': 22}
matplotlib.rc('font', **font)
plt.figure(figsize=(12, 8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate1, color='coral', linestyle='-')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate2, color='lightcoral', linestyle='--')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate1, color='deepskyblue', linestyle='-')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate2, color='skyblue', linestyle='--')
plt.errorbar(shock_lightcurve['time'].to(u.d).value[sgood], shock_rate_meas1.value[sgood], fmt='o',
markersize=5, yerr=shock_rate_meas_err1.value[sgood], color='coral', label='Shock model (5e10), DUET 1')
plt.errorbar(shock_lightcurve['time'].to(u.d).value[sgood], shock_rate_meas2.value[sgood], fmt='o',
markersize=5, yerr=shock_rate_meas_err2.value[sgood], color='lightcoral', label='Shock model (5e10), DUET 2')
plt.errorbar(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rate_rebin_meas1.value[srgood], fmt='o',
markersize=5, yerr=shock_rate_rebin_meas_err1.value[srgood], color='red', label='Shock model rebinned')
plt.errorbar(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rate_rebin_meas2.value[srgood], fmt='o',
markersize=5, yerr=shock_rate_rebin_meas_err2.value[srgood], color='red')
plt.errorbar(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_rate_meas1.value[bgood], fmt='o',
markersize=5, yerr=blukn_rate_meas_err1.value[bgood], color='deepskyblue', label='Blue Kilonova (0.04), DUET 1')
plt.errorbar(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_rate_meas2.value[bgood], fmt='o',
markersize=5, yerr=blukn_rate_meas_err2.value[bgood], color='skyblue', label='Blue Kilonova (0.04), DUET 2')
plt.errorbar(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rate_rebin_meas1.value[brgood], fmt='o',
markersize=5, yerr=blukn_rate_rebin_meas_err1.value[brgood], color='blue', label='Blue kilonova rebinned')
plt.errorbar(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rate_rebin_meas2.value[brgood], fmt='o',
markersize=5, yerr=blukn_rate_rebin_meas_err2.value[brgood], color='blue')
plt.legend()
plt.ylabel("Count rate (ph/s)")
plt.xlabel("Time after merger (d)")
plt.ylim(0,9)
plt.xlim(-0.1,1)
plt.show()
'''
plt.figure(figsize=(12, 8))
plt.scatter(shock_lightcurve['time'].to(u.d).value, shock_lightcurve['snr_D1'], s=5, color='coral', label='Shock model (2.5e10)')
plt.scatter(shock_lightcurve['time'].to(u.d).value, shock_lightcurve['snr_D2'], s=5, color='lightcoral', label='_Shock model (2.5e10)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value, blukn_lightcurve['snr_D1'], s=5, color='deepskyblue', label='Blue Kilonova (0.01)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value, blukn_lightcurve['snr_D2'], s=5, color='skyblue', label='_Blue Kilonova (0.01)')
plt.ylabel("S/N")
plt.xlabel("Time after merger (d)")
plt.xlim(-0.1,1)
plt.legend()
plt.show()
'''
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='_Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='_Blue Kilonova (0.04)')
plt.scatter(shock_lightcurve['time'].to(u.d).value[sgood], shock_color.value[sgood], s=5,
color='coral', linewidth=2, label='Shock model (5e10)')
plt.scatter(shock_lightcurve_rebin['time'].to(u.d).value[srgood], shock_rebin_color.value[srgood], s=5,
color='red', linewidth=2, label='_Shock model (5e10)')
plt.scatter(blukn_lightcurve['time'].to(u.d).value[bgood], blukn_color.value[bgood], s=5,
color='deepskyblue', linewidth=2, label='Blue Kilonova (0.04)')
plt.scatter(blukn_lightcurve_rebin['time'].to(u.d).value[brgood], blukn_rebin_color.value[brgood], s=5,
color='blue', linewidth=2, label='_Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel('DUET1 - DUET2')
plt.title('Color evolution @ 100 Mpc')
plt.show()
# This time with added galaxy
plt.figure(figsize=(12, 8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate1, color='coral', linestyle='-')
plt.plot((shock_5e10_time).to(u.d), shock_5e10_rate2, color='lightcoral', linestyle='--')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate1, color='deepskyblue', linestyle='-')
plt.plot((blukn_04_time).to(u.d), blukn_04_rate2, color='skyblue', linestyle='--')
plt.errorbar(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_rate_gal_meas1.value[sggood], fmt='o',
markersize=5, yerr=shock_rate_gal_meas_err1.value[sggood], color='coral', label='Shock model (5e10), DUET 1')
plt.errorbar(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_rate_gal_meas2.value[sggood], fmt='o',
markersize=5, yerr=shock_rate_gal_meas_err2.value[sggood], color='lightcoral', label='Shock model (5e10), DUET 2')
plt.errorbar(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_rate_gal_meas1.value[bggood], fmt='o',
markersize=5, yerr=blukn_rate_gal_meas_err1.value[bggood], color='deepskyblue', label='Blue Kilonova (0.04), DUET 1')
plt.errorbar(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_rate_gal_meas2.value[bggood], fmt='o',
markersize=5, yerr=blukn_rate_gal_meas_err2.value[bggood], color='skyblue', label='Blue Kilonova (0.04), DUET 2')
#plt.errorbar(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_rate_gal_rebin_meas1.value[sgrgood], fmt='o',
# markersize=5, yerr=shock_rate_gal_rebin_meas_err1.value[sgrgood], color='darkred', label='_Shock model (5e10)')
#plt.errorbar(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_rate_gal_rebin_meas2.value[sgrgood], fmt='o',
# markersize=5, yerr=shock_rate_gal_rebin_meas_err2.value[sgrgood], color='red', label='_Shock model (5e10)')
#plt.errorbar(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_rate_gal_rebin_meas1.value[bgrgood], fmt='o',
# markersize=5, yerr=blukn_rate_gal_rebin_meas_err1.value[bgrgood], color='darkblue', label='_Blue Kilonova (0.04)')
#plt.errorbar(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_rate_gal_rebin_meas2.value[bgrgood], fmt='o',
# markersize=5, yerr=blukn_rate_gal_rebin_meas_err2.value[bgrgood], color='blue', label='_Blue Kilonova (0.04)')
plt.legend()
plt.title('Shock and blue kilonova, spiral galaxy @ 100 Mpc')
plt.ylabel("Count rate (ph/s)")
plt.xlabel("Time after merger (d)")
plt.ylim(0,9)
plt.xlim(-0.1,1)
plt.show()
plt.figure(figsize=(12,8))
plt.plot((shock_5e10_time).to(u.d), shock_5e10_ABmag1-shock_5e10_ABmag2, color='lightcoral', linestyle='-', linewidth=2, label='_Shock model (5e10)')
plt.plot((blukn_04_time).to(u.d), blukn_04_ABmag1-blukn_04_ABmag2, color='deepskyblue', linestyle='-', linewidth=2, label='_Blue Kilonova (0.04)')
plt.scatter(shock_lightcurve_gal['time'].to(u.d).value[sggood], shock_color_gal.value[sggood], s=5,
color='coral', linewidth=2, label='Shock model (5e10)')
plt.scatter(blukn_lightcurve_gal['time'].to(u.d).value[bggood], blukn_color_gal.value[bggood], s=5,
color='deepskyblue', linewidth=2, label='Blue Kilonova (0.04)')
#plt.scatter(shock_lightcurve_gal_rebin['time'].to(u.d).value[sgrgood], shock_gal_rebin_color.value[sgrgood], s=5,
# color='red', linewidth=2, label='_Shock model (5e10)')
#plt.scatter(blukn_lightcurve_gal_rebin['time'].to(u.d).value[bgrgood], blukn_gal_rebin_color.value[bgrgood], s=5,
# color='blue', linewidth=2, label='_Blue Kilonova (0.04)')
plt.axhline(y=0,xmin=0,xmax=1,color='black',linestyle=':')
plt.ylim(-1,3)
plt.xlim(-0.1,1)
plt.legend()
plt.xlabel('Time after merger (d)')
plt.ylabel("DUET1 - DUET2")
plt.title('Color evolution @ 100 Mpc')
plt.show()
# Show example images
frame = np.array([30, 30])
nexp = 5
ref_image1 = construct_image(frame, exposure, duet=duet, band=duet.bandpass1,
gal_type='spiral', sky_rate=bgd_band1, n_exp=nexp)
ref_image_rate1 = ref_image1 / (exposure * nexp)
ref_bkg1, ref_bkg_rms_median1 = estimate_background(ref_image_rate1, method='1D', sigma=2)
ref_rate_bkgsub1 = ref_image_rate1 - ref_bkg1
psf_array = duet.psf_model(x_size=5,y_size=5).array
# Example image at peak of shock light curve
src1 = shock_rate_gal_meas1[7]
image1 = construct_image(frame, exposure, duet=duet, band=duet.bandpass1, source=src1,
gal_type='spiral', sky_rate=bgd_band1)
image_rate1 = image1 / exposure
plt.imshow(image_rate1.value, cmap='viridis', aspect=1, origin='lower')
plt.colorbar()
plt.show()
# Difference image
image_bkg, image_bkg_rms_median = estimate_background(image_rate1, method='1D', sigma=2)
ref_bkg, ref_bkg_rms_median = estimate_background(ref_image_rate1, method='1D', sigma=2)
image_rate_bkgsub, ref_rate_bkgsub = image_rate1 - image_bkg, ref_image_rate1 - ref_bkg
s_n, s_r = np.sqrt(image_rate1), np.sqrt(ref_image_rate1) # 2D uncertainty (sigma) - that is, noise on the background
sn, sr = np.mean(s_n), np.mean(s_r) # Average uncertainty (sigma)
dx, dy = 0.1, 0.01 # Astrometric uncertainty (sigma)
diff_image, d_psf, s_corr = py_zogy(image_rate_bkgsub.value,
ref_rate_bkgsub.value,
psf_array,psf_array,
s_n.value,s_r.value,
sn.value,sr.value,dx,dy)
diff_image *= image_rate_bkgsub.unit
plt.imshow(diff_image.value, cmap='viridis', aspect=1, origin='lower')
plt.colorbar()
plt.show()
# To what volume are we going to get 'quality lightcurves to distinguish models' ?
# - 'quality light curve' = sufficient to distinguish between the two models to x sigma
# To what volume will we get a detection/localisation?
# If we stack x images, we can get a detection to y sigma out to blah Mpc
# Two-pointing strategy vs. six-pointing strategy. Only two-inteferometer events.
# What is the advanced LIGO sensitivity
# With probabilities of getting there in time from Kristin
# and binning from Matteo
# For shock and for kilonova:
# As above, only go through a number of distances.
# For each distance, first a detection test: get difference image in both bands, sum, locate source. (Summing all exposures in an orbit, in a worst-case where we miss the peak.)
# Second, a lightcurve test: can we rule out the alternative model with this lightcurve?
# These probabilities above - multiply them with the probability of getting there in time from Kristin
# How many stacked exposures required for a detection as a function of distance? That informs Kristin's operations simulations.
# For different start times: 30 mins, 50 mins, 70 mins
# For different distances
# For different positions in the galaxy: sources at r_eff/2, r_eff, r_eff*2
# Function to get detection significance at 1 exposure, 2 exposures etc... until 5-sigma detection is reached
| 0.854339 | 0.583648 |
# Week 3 Lab Task
## More Jupyter Tips
Hopefully by this week, you are growing more comfortable with starting Jupyter Notebooks and adding/editing cells. Remember that the keyboard shortcuts are invaluable: running a cell with `Ctrl+Enter`, or adding a new cell below with `B` (in command mode).
Two tricks to try this week: autocompletion and retrieving documentation.
**Autocomplete**
If you start typing a known object or function into Jupyter, you can press `TAB` to finish it. This is especially useful for seeing what functions are available.
```
test = "this is a string"
```
Above, I've set a string to `test`. If I type `te` then press tab, it will complete the word. This is especially useful for long variable names that you don't want to keep typing. Note that it only completed because there no other options: in that case, there's a scrollable list of candidates for what you might be looking for.
The `test` variable is a string. Last week, we saw a two functions that can be performed on strings: `split()` and `join()`. If you would like to see what other options there are for strings, try typing `test.` then press TAB. Magic!

** Documentation reference **
If you want to look up information about a function, you can precede the code running that function with a `?`. For example, if I want to learn how I would use `split()` on `test`, I can type:
```
?test.split()
```
This will open a panel that looks like this in Jupyter:

The documentation is only as good as what the library is documented, so some libraries might be more or less detailed in this feature.
*Questions*
- 1) What does `test.isalpha()` do? Copy the documentation string.
- 2) Strings have access to a function (whose name starts with a `ce`) that will let you change "HEADING" to "====HEADING====" (that is, padding with `=` to make the string 15 characters wide). What's the code to do that? (tip: this is an auto-fill question!)
## Intro to the NLTK
This week we'll start using the Natural Language toolkit. For the remaining questions, follow along with:
- [Getting NLTK for Text Processing](https://github.com/sgsinclair/alta/blob/2acb6ed09f298f631e4025d33f062f980758a1ce/ipynb/GettingNltk.ipynb), Art of Literary Text Analysis
Two notes. First, the tutorial suggests downloading "all" packages. However, install the packages from 'book' should be sufficient for now.
Also, skip the text processing section, which deals with automatically downloading and cleaning a book. Instead, download this [already-cleaned version of Mary Shelley's Frankenstein](https://raw.githubusercontent.com/organisciak/Text-Mining-Course/master/data/frankenstein.txt), put it into the same folder as your notebook, and load it as follows:
```
with open("../data/frankenstein.txt") as f:
frankensteinString = f.read()
```
Here's a quick way of viewing part of our string: the first 250 characters. Notice that you can select subsets of strings like you select subsets of lists.
```
frankensteinString[0:250]
```
> Side-note for the Python novice: you don't actually need the zero in [0:250]. If left blank, like '[:250]`, Python will assume "from the very start", which is the same as using a 0. If you leave the second part blank, Python will assume "until the very end".
For the rest of the ALTA chapter, follow along using `frankensteinString` string instead of `goldBugString`.
__Questions__
- 3) Use the `word_tokenize` function on Frankenstein, as shown in ALTA. What are tokens 39:67? Hint: this is a full sentence. Include your code.
- 4) Create a sample of only the tokens where the first character is an alphabetical character. In this sample, what are tokens 1215:1221? Again, this will be a sentence, but won't include punctuation as tokens. Include your code.
_For the next questions use the list of tokens that start with an alphabetical character._
- 5) What are the ten most frequent words in this book? Create a frequency distribution of the words from question 4, then tabulate the top 10 words. Include your code.
- 6) After case-folding, what are the ten most frequent words in this book? Include your code.
- 7) Rewrite this list comprehension as a `for` loop (what ALTA called technique 1): `[word for word in listOfWords if word.find('-') >= 0]`. No output necessary, just the code, but feel free to test it out.
- 8) We're going to use a customized stoplist. First, load the NLTK stoplist, and add the words 'could', 'would', 'upon', and 'yet' to the stoplist. What are the top ten case-folded words when stopping against the stoplist. Include your code and paste the tabulated output.
Using the autocomplete in Jupyter, you may notice that a list of tokens converted to a `FreqDist` object has more methods than just `tabulate()`. One really cool one is `plot()`.
`plot` gives you a visualization of the top frequency words. However, you may notice that if you try to run it, the visualization doesn't show up.
It _is_ created, but Jupyter just doesn't know that you want the visualization shown _within_ the notebook. To turn that option on, run the following line of code:
```
%matplotlib inline
```
This is only necessary once: it tells Jupyter to show plots 'inline' (ie. inside the notebook).
**Questions**
- 9) Write the code to plot the top forty stoplisted, lowercase words (from question 8). And again, remember the docs! The output will look similar to this:

- 10) Enter the first 5 concordances for the word "monster" in the original token list - the list straight from word_tokenize that included punctuation and numbers - narrowing the search to a 49-characters window. Include the code. Tip: See the docs for the concordance tool in Jupyter.
|
github_jupyter
|
test = "this is a string"
?test.split()
with open("../data/frankenstein.txt") as f:
frankensteinString = f.read()
frankensteinString[0:250]
%matplotlib inline
| 0.18628 | 0.979413 |
# Test Model Saving
This notebook is designed to make sure saving and loading models works correctly
```
import argparse
import logging
import numpy as np
import pickle
import random
import sys
import torch
# Add whistl modules to the path
sys.path.append('../whistl')
import classifier
import dataset
import model
import plot_util
import util
# Tell pytorch to use the gpu
device = torch.device('cuda')
# Set up logging
logging.basicConfig(level=logging.ERROR)
logger = logging.getLogger(__name__)
# Ensure the models train deterministically
seed = 42
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
if torch.backends.cudnn.enabled:
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# Select a classifier architecture
label_to_encoding = {'tb': 1, 'healthy': 0}
net = model.ThreeLayerNet
# Split train and test data
train_dirs, tune_dirs = util.train_tune_split('../data/', 2)
# Initialize arguments to use in training the models
map_file = '../data/sample_classifications.pkl'
gene_file = '../data/intersection_genes.csv'
num_epochs = 1500
loss_scaling_factor = 1
```
## Train a three layer neural network with IRM
```
irm_results = classifier.train_with_irm(net, map_file, train_dirs, tune_dirs, gene_file, num_epochs,
loss_scaling_factor, label_to_encoding, device, logger, '../logs/irm.pkl', 5)
```
## Load the model and ensure the weights saved properly
```
trained_net = torch.load('../logs/irm.pkl')
sample_to_label = util.parse_map_file(map_file)
tune_dataset = dataset.ExpressionDataset(tune_dirs, sample_to_label, label_to_encoding, gene_file)
tune_loader = torch.utils.data.DataLoader(tune_dataset, batch_size=16, num_workers=4, pin_memory=True)
```
### Test trained network
```
tune_loss = 0
tune_correct = 0
for tune_batch in tune_loader:
expression, labels, ids = tune_batch
tune_expression = expression.to(device)
tune_labels = labels.to(device).double()
loss_function = torch.nn.BCEWithLogitsLoss()
tune_preds = trained_net(tune_expression)
loss = loss_function(tune_preds, tune_labels)
tune_loss += float(loss)
tune_correct += util.count_correct(tune_preds, tune_labels)
avg_loss = tune_loss / len(tune_dataset)
tune_acc = tune_correct / len(tune_dataset)
print('Trained network tune accuracy: {}'.format(tune_acc))
print('Trained network tune loss: {}'.format(avg_loss))
```
### Test untrained network
```
input_size = tune_dataset[0][0].shape[0]
untrained_net = model.ThreeLayerNet(input_size).double().to(device)
tune_loss = 0
tune_correct = 0
for tune_batch in tune_loader:
expression, labels, ids = tune_batch
tune_expression = expression.to(device)
tune_labels = labels.to(device).double()
loss_function = torch.nn.BCEWithLogitsLoss()
tune_preds = untrained_net(tune_expression)
loss = loss_function(tune_preds, tune_labels)
tune_loss += float(loss)
tune_correct += util.count_correct(tune_preds, tune_labels)
avg_loss = tune_loss / len(tune_dataset)
tune_acc = tune_correct / len(tune_dataset)
print('Untrained network tune accuracy: {}'.format(tune_acc))
print('Untrained network tune loss: {}'.format(avg_loss))
```
## Save results to a file to keep track of genes and samples used
```
with open('../logs/model_saving_test_results.pkl', 'wb') as out_file:
pickle.dump(irm_results, out_file)
```
## Conclusion
The model saving functions in `classifier.py` work, and the trained network outperforms an untrained one.
|
github_jupyter
|
import argparse
import logging
import numpy as np
import pickle
import random
import sys
import torch
# Add whistl modules to the path
sys.path.append('../whistl')
import classifier
import dataset
import model
import plot_util
import util
# Tell pytorch to use the gpu
device = torch.device('cuda')
# Set up logging
logging.basicConfig(level=logging.ERROR)
logger = logging.getLogger(__name__)
# Ensure the models train deterministically
seed = 42
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
if torch.backends.cudnn.enabled:
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# Select a classifier architecture
label_to_encoding = {'tb': 1, 'healthy': 0}
net = model.ThreeLayerNet
# Split train and test data
train_dirs, tune_dirs = util.train_tune_split('../data/', 2)
# Initialize arguments to use in training the models
map_file = '../data/sample_classifications.pkl'
gene_file = '../data/intersection_genes.csv'
num_epochs = 1500
loss_scaling_factor = 1
irm_results = classifier.train_with_irm(net, map_file, train_dirs, tune_dirs, gene_file, num_epochs,
loss_scaling_factor, label_to_encoding, device, logger, '../logs/irm.pkl', 5)
trained_net = torch.load('../logs/irm.pkl')
sample_to_label = util.parse_map_file(map_file)
tune_dataset = dataset.ExpressionDataset(tune_dirs, sample_to_label, label_to_encoding, gene_file)
tune_loader = torch.utils.data.DataLoader(tune_dataset, batch_size=16, num_workers=4, pin_memory=True)
tune_loss = 0
tune_correct = 0
for tune_batch in tune_loader:
expression, labels, ids = tune_batch
tune_expression = expression.to(device)
tune_labels = labels.to(device).double()
loss_function = torch.nn.BCEWithLogitsLoss()
tune_preds = trained_net(tune_expression)
loss = loss_function(tune_preds, tune_labels)
tune_loss += float(loss)
tune_correct += util.count_correct(tune_preds, tune_labels)
avg_loss = tune_loss / len(tune_dataset)
tune_acc = tune_correct / len(tune_dataset)
print('Trained network tune accuracy: {}'.format(tune_acc))
print('Trained network tune loss: {}'.format(avg_loss))
input_size = tune_dataset[0][0].shape[0]
untrained_net = model.ThreeLayerNet(input_size).double().to(device)
tune_loss = 0
tune_correct = 0
for tune_batch in tune_loader:
expression, labels, ids = tune_batch
tune_expression = expression.to(device)
tune_labels = labels.to(device).double()
loss_function = torch.nn.BCEWithLogitsLoss()
tune_preds = untrained_net(tune_expression)
loss = loss_function(tune_preds, tune_labels)
tune_loss += float(loss)
tune_correct += util.count_correct(tune_preds, tune_labels)
avg_loss = tune_loss / len(tune_dataset)
tune_acc = tune_correct / len(tune_dataset)
print('Untrained network tune accuracy: {}'.format(tune_acc))
print('Untrained network tune loss: {}'.format(avg_loss))
with open('../logs/model_saving_test_results.pkl', 'wb') as out_file:
pickle.dump(irm_results, out_file)
| 0.471223 | 0.807612 |
```
import sys
from markdown import markdown
sys.path.insert(0, "..")
from versions import (
JUPYTER_LSP_VERSION,
JUPYTERLAB_LSP_VERSION,
JUPYTERLAB_NEXT_MAJOR_VERSION,
JUPYTERLAB_VERSION,
REQUIRED_JUPYTER_SERVER,
REQUIRED_JUPYTERLAB,
REQUIRED_PYTHON,
)
```
## Installation
```
%%markdown
### Please Read This First
Delivering LSP features to your JupyterLab **requires** three pieces:
#### `jupyter-lsp`
- runs in your `jupyter_server` web application on your server to handle requests from
the browser to _language servers_
- to run, you need:
- `python {REQUIRED_PYTHON}`
- `jupyter_server {REQUIRED_JUPYTER_SERVER}`
#### `jupyterlab-lsp`
- runs in your browser, as an extension to JupyterLab
- to install it, you need:
- `jupyterlab {REQUIRED_JUPYTERLAB}`
#### Language Servers
- run on your server
- probably in another language runtime than python
- some can be automatically [detected](./Language%20Servers.ipynb) if installed
- others also need to be [configured](./Configuring.ipynb#language_servers)
```
### Fast Paths
Here are two approaches based on Jupyter documentation. If these do not meet
your needs, try [The Harder Way](#The-Harder-Way).
```
%%markdown
#### conda (minimal python)
```bash
conda create -c conda-forge -n lsp 'python {REQUIRED_PYTHON}' 'jupyterlab={JUPYTERLAB_VERSION}' 'jupyterlab-lsp={JUPYTERLAB_LSP_VERSION}' 'jupyter-lsp-python={JUPYTER_LSP_VERSION}'
# jupyter-lsp-python includes both the server extension (jupyter-lsp) and pyls third-party server (python-language-server)
# if you swap it with another pre-made bundle, jupyter-lsp-r, you will get the server extension and r-languageserver;
# alternatively, manually install a language server of your choice (see the table below).
conda activate lsp
```
Then run
```bash
jupyter lab
```
Your browser should open to your local server.
```
#### docker (data science)
This approach is based roughly on the
[Jupyter docker-stacks documentation](https://github.com/jupyter/docker-stacks/tree/master/examples/docker-compose/notebook),
which should be consulted for more about connecting volumes, passwords, and
other advanced features:
> Note: docker instructions were **not** updated for JupyterLab 3.0 and
> extension 3.0. Please consider submitting a PR to fix it.
```
%%markdown
##### `Dockerfile`
```dockerfile
# This already contains the python, r, julia, latex, and nodejs runtimes
FROM jupyter/datascience-notebook@sha256:73a577b006b496e1a1c02f5be432f4aab969c456881c4789e0df77c89a0a60c2
RUN conda install --quiet --yes --freeze-installed -c conda-forge \
'python-language-server' \
'jupyterlab={JUPYTERLAB_VERSION}' \
'r-languageserver' \
'texlab' \
'chktex' \
'jupyter-lsp={JUPYTER_LSP_VERSION}' \
&& jupyter labextension install --no-build \
'@krassowski/jupyterlab-lsp@{JUPYTERLAB_LSP_VERSION}' \
&& jupyter lab build --dev-build=False --minimize=True \
&& conda clean --all -f -y \
&& rm -rf \
$CONDA_DIR/share/jupyter/lab/staging \
/home/$NB_USER/.cache/yarn \
&& fix-permissions $CONDA_DIR \
&& fix-permissions /home/$NB_USER
```
```
##### `docker-compose.yml`
```yaml
version: '2'
services:
lsp-lab:
build: .
ports:
- '18888:8888'
```
##### Build and Start
```bash
docker-compose up
```
You should now be able to access `http://localhost:18888/lab`, using the `token`
provided in the log.
### The Harder Way
#### Get A Working JupyterLab environment
Refer to the official
[JupyterLab Installation Documentation](https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html)
for your installation approach.
| pip | conda | pipenv | poetry | `*` |
| -------------- | ---------------- | ------ | ------ | --- |
| [lab][lab-pip] | [lab][lab-conda] | `*` | `*` | `*` |
> `*` PRs welcome!
Verify your lab works:
```bash
jupyter lab --version
jupyter lab
```
[lab-conda]:
https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html#conda
[lab-pip]:
https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html#pip
```
%%markdown
#### Install Jupyter[Lab] LSP
##### conda
```bash
conda install jupyterlab-lsp={JUPYTERLAB_LSP_VERSION}
```
##### pip
```bash
pip install jupyterlab-lsp={JUPYTERLAB_LSP_VERSION}
```
```
### Next Step: Language Servers
Now that you have `jupyterlab-lsp`, `jupyter-lsp` and all of their dependencies,
you'll need some language servers. See:
- [Language Servers](./Language%20Servers.ipynb) that will be found
automatically once installed
- [configuring](./Configuring.ipynb) `jupyter-lsp` for more control over which
servers to load
|
github_jupyter
|
import sys
from markdown import markdown
sys.path.insert(0, "..")
from versions import (
JUPYTER_LSP_VERSION,
JUPYTERLAB_LSP_VERSION,
JUPYTERLAB_NEXT_MAJOR_VERSION,
JUPYTERLAB_VERSION,
REQUIRED_JUPYTER_SERVER,
REQUIRED_JUPYTERLAB,
REQUIRED_PYTHON,
)
%%markdown
### Please Read This First
Delivering LSP features to your JupyterLab **requires** three pieces:
#### `jupyter-lsp`
- runs in your `jupyter_server` web application on your server to handle requests from
the browser to _language servers_
- to run, you need:
- `python {REQUIRED_PYTHON}`
- `jupyter_server {REQUIRED_JUPYTER_SERVER}`
#### `jupyterlab-lsp`
- runs in your browser, as an extension to JupyterLab
- to install it, you need:
- `jupyterlab {REQUIRED_JUPYTERLAB}`
#### Language Servers
- run on your server
- probably in another language runtime than python
- some can be automatically [detected](./Language%20Servers.ipynb) if installed
- others also need to be [configured](./Configuring.ipynb#language_servers)
%%markdown
#### conda (minimal python)
Then run
Your browser should open to your local server.
%%markdown
##### `Dockerfile`
```
##### `docker-compose.yml`
##### Build and Start
You should now be able to access `http://localhost:18888/lab`, using the `token`
provided in the log.
### The Harder Way
#### Get A Working JupyterLab environment
Refer to the official
[JupyterLab Installation Documentation](https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html)
for your installation approach.
| pip | conda | pipenv | poetry | `*` |
| -------------- | ---------------- | ------ | ------ | --- |
| [lab][lab-pip] | [lab][lab-conda] | `*` | `*` | `*` |
> `*` PRs welcome!
Verify your lab works:
[lab-conda]:
https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html#conda
[lab-pip]:
https://jupyterlab.readthedocs.io/en/stable/getting_started/installation.html#pip
conda install jupyterlab-lsp={JUPYTERLAB_LSP_VERSION}
pip install jupyterlab-lsp={JUPYTERLAB_LSP_VERSION}
| 0.433502 | 0.471284 |
# Scikit-Criteria: SIMUS implementations
## Abstract
En este documento se presenta la sintaxis y opciones de la implementacion del método SIMUS, utilizando el stack provisto por Scikit-Criteria.
## Problema
Se utilizara el problema enunciado en el trabajo
> Munier, N., Carignano, C., & Alberto, C. UN MÉTODO DE PROGRAMACIÓN MULTIOBJETIVO. Revista de la Escuela de Perfeccionamiento en Investigación Operativa, 24(39).
### Enunciado
A fin de mostrar una aplicación del método, se utiliza un ejemplo
48
SECCION APLICACIONESINVESTIGACION OPERATIVA - AÑO XXIV - No 39 - PAGINAS 44 a 54 - MAYO 2016
sobre planificación urbana, propuesto en Lliso (2014) 2 .
Desde hace algunas décadas y debido a los cambios en la industria
del transporte ferroviario y marítimo en casi todas las ciudades hay espacios
vacíos, por lo general céntricos, los cuales eran ocupados por las estaciones
de ferrocarril, patios de ferrocarril y muelles. Estas parcelas abandonadas son
el blanco de los municipios para ser usadas en la construcción de parques,
edificios de viviendas, oficinas gubernamentales, centros comerciales, etc.
Suelen ser grandes parcelas que pueden ser adecuadas para varios usos
diferentes al mismo tiempo, razón por la cual el gobierno municipal se
encuentra ante un dilema ya que es deseable seleccionar la alternativa de uso
que mejor sirva a la ciudad.
#### CASO: REHABILITACIÓN DE TIERRAS
Una importante ciudad portuaria se ha visto afectada por el cambio en
la modalidad de transporte marítimo, cuando comenzó el transporte de
contenedores a mediados del siglo 20. La ciudad se quedó con 39 hectáreas
de muelles vacíos, así como los almacenes y una terminal ferroviaria.
El municipio tiene que decidir qué hacer con esta tierra y desarrolló un
Plan Maestro basado en tres proyectos diferentes:
- Proyecto 1: Torres Corporativas - Hoteles - Marina - Pequeño Parque.
- Proyecto 2: Torres de Viviendas - Zona Comercial en la antigua
estación de ferrocarril.
- Proyecto 3: Centro de Convenciones - Gran Parque y área recreativa.
Los criterios que considera para el análisis de las propuestas se
refieren a:
- Criterio 1: Generación de Trabajo
- Criterio 2: Espacio Verde Recuperado
- Criterio 3: Factibilidad Financiera
- Criterio 4: Impacto Ambiental
Para el criterio 2 se especifica un límite máximo, mientras que los
restantes criterios no tienen restricción definida para el lado derecho.
El Decisor considera a los cuatro criterios como objetivos, por lo que
se deberán resolver cuatro programas lineales con tres restricciones cada
uno.
Los datos se detallan en la tabla siguiente.
| |Proyecto 1 | Proyecto 2 | Proyecto 3 | VLD | Acción |
| ------------- |-----------|------------|------------|-----|-----------|
| Criterio 1 | 250 | 130 | 350 | - | Maximizar |
| Criterio 2 | 120 | 200 | 340 | 500 | Maximizar |
| Criterio 3 | 20 | 40 | 15 | - | Minimizar |
| Criterio 4 | 800 | 1000 | 600 | - | Maximizar |
## El modelo
```python
from skcriteria.madm import simus
dm = simus.SIMUS()
```
- El metodo SIMUS se configura con la llamada `simus.SIMUS`.
- El unico parametro opcional es **solver**. Este parámetro se utiliza
para determinar que resolutor de programación lineal se ha de utilizar.
- Entre Los resolutores disponibles se encuentran
[coin-mp](https://projects.coin-or.org/CoinMP) (este es el por defecto),
[cplex](https://en.wikipedia.org/wiki/CPLEX),
[gurobi](https://en.wikipedia.org/wiki/Gurobi) y
[glpk](https://en.wikipedia.org/wiki/GNU_Linear_Programming_Kit)
- Para utilizar cplex, gurobi y glpk es necesario instalarlos y configurarlos
dentro de scikit-criteria (se explicara cuando se implemente)
Ejemplo de creación de un modelo con los solvers gplk y cplex:
```python
# utilizando glpk
dm = simus.SIMUS(solver="glpk")
# utilizando cplex
dm = simus.SIMUS(solver="cplex")
```
## Los Datos
El metodo implementado dentro scikit-criteria recibe los datos en el mismo formato que todos los demas métodos:
- Una matriz de alternativa, donde cada fila es una alaternativas y cada columna es un criterio. Internamente SIMUS transpone esta matris para su operación)
- Una vector de criterios que indica si el criterio es maximizaro o minimizar.
Opcionales:
- Vector de pesos (ignorado por SIMUS)
- Nombre de las alternativas
- Nombre de los criterios
**Ejemplo del paper:**
```
from skcriteria import Data
data = Data(
mtx=[[250, 120, 20, 800],
[130, 200, 40, 1000],
[350, 340, 15, 600]],
criteria=[max, max, min, max],
anames=["Proyecto 1", "Proyecto 2", "Proyecto 3"],
cnames=["Criterio 1", "Criterio 2", "Criterio 3", "Criterio 4"])
data
```
### Resolviendo el problema
Anteriormente habiamos creado el modelo llamado **`dm`**. **`dm`** tiene una funcón llamada solve que recibe el objeto **`data`** que creamos y opcionalmente un parametro llamado **`b`** que representa el vector del lado
derecho y que tiene que tener la misma cantidad de valores que criterios tenga los datos.
En el caso del ejemplo
```python
decision = dm.decide(data, b=[None, 500, None, None]
decision
```

Todos los calculos intermedios como: vectores de subordinacion, puntajes por ambos procedimientos, los resultados de los algoritmos de programacion lineal y otros; estan
guardados dentro de la variable de **`dec.e_`** (esta variable de llama "e" por extra, ya que depende el metodo a utilizar guarda diferentes cosas (en topsis guarda el vector de cercanias por ejemplo))


#### Por que data tiene esta forma?
Por que con los mismos datos podemos alimentar y comparar diferentes metodos. Por ejemplo
**TOPSIS**
```
from skcriteria.madm import closeness
closeness.TOPSIS().decide(data)
```
**ELECTRE 1**
```
from skcriteria.madm import electre
electre.ELECTRE1().decide(data)
```
**Suma Ponderada**
```
from skcriteria.madm import simple
simple.WeightedSum().decide(data)
```
Ademas que el objeto data tiene capacidades graficas como:
```
data.plot();
data.plot.box();
```
|
github_jupyter
|
from skcriteria.madm import simus
dm = simus.SIMUS()
# utilizando glpk
dm = simus.SIMUS(solver="glpk")
# utilizando cplex
dm = simus.SIMUS(solver="cplex")
from skcriteria import Data
data = Data(
mtx=[[250, 120, 20, 800],
[130, 200, 40, 1000],
[350, 340, 15, 600]],
criteria=[max, max, min, max],
anames=["Proyecto 1", "Proyecto 2", "Proyecto 3"],
cnames=["Criterio 1", "Criterio 2", "Criterio 3", "Criterio 4"])
data
decision = dm.decide(data, b=[None, 500, None, None]
decision
from skcriteria.madm import closeness
closeness.TOPSIS().decide(data)
from skcriteria.madm import electre
electre.ELECTRE1().decide(data)
from skcriteria.madm import simple
simple.WeightedSum().decide(data)
data.plot();
data.plot.box();
| 0.390825 | 0.910784 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import datetime
aaxis_df=pd.read_csv('TECHM.NS.csv')
aaxis_df[['Open', 'High','Low', 'Close','Adj Close','Volume']] = aaxis_df[['Open', 'High','Low', 'Close','Adj Close','Volume']].apply(pd.to_numeric, errors='coerce')
aaxis_df["Open"].fillna(value=aaxis_df["Open"].mean(), inplace=True)
aaxis_df["High"].fillna(value=aaxis_df["High"].mean(), inplace=True)
aaxis_df["Low"].fillna(value=aaxis_df["Low"].mean(), inplace=True)
aaxis_df["Close"].fillna(value=aaxis_df["Close"].mean(), inplace=True)
aaxis_df["Adj Close"].fillna(value=aaxis_df["Adj Close"].mean(), inplace=True)
aaxis_df["Volume"].fillna(value=aaxis_df["Volume"].mean(), inplace=True)
date_column=aaxis_df["Date"]
from sklearn import preprocessing, cross_validation, svm
from sklearn.linear_model import LinearRegression
#Visualization
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
#aaxis_df.set_index("Date", inplace = True)
aaxis_df[['Adj Close','Open','High','Low','Close']].plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
plt.ylabel('Price')
from sklearn import preprocessing, cross_validation, svm
from sklearn.linear_model import LinearRegression
#Visualization
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
#Moving Average
def MA(df, n):
name = 'SMA_' + str(n)
#MA = pd.Series(pd.rolling_mean(df['Close'], n), name = 'SMA_' + str(n))
#df = df.join(MA)
df[name]=pd.rolling_mean(df['Adj Close'],n)
return df
#Exponential Moving Average
def EMA(df, n):
name = 'EMA_' + str(n)
#MA = pd.Series(pd.rolling_mean(df['Close'], n), name = 'SMA_' + str(n))
#df = df.join(MA)
df[name]=pd.ewma(df['Adj Close'], span = n, min_periods = n - 1)
return df
for i in [30,40,50]:
MA(aaxis_df,i)
for i in [30,40,50]:
EMA(aaxis_df,i)
aaxis_df[['Adj Close', 'Close']].plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
dates = np.array(aaxis_df["Date"])
#print(dates)
dates_check = dates[-30:]
dates = dates[:-30]
# define a new feature, HL_PCT
aaxis_df['HL_PCT'] = (aaxis_df['High'] - aaxis_df['Low'])/(aaxis_df['Low']*100)
# define a new feature percentage change
aaxis_df['PCT_CHNG'] = (aaxis_df['Close'] - aaxis_df['Open'])/(aaxis_df['Open']*100)
columns_main=['Adj Close', 'HL_PCT', 'PCT_CHNG', 'Volume' ,'SMA_30', 'SMA_40', 'SMA_50', 'EMA_30', 'EMA_40', 'EMA_50']
aaxis_df = aaxis_df[columns_main]
aaxis_df.fillna( value=0, inplace=True)
aaxis_df.isnull().sum()
# pick a forecast column
forecast_col = 'Adj Close'
# Chosing 30 days as number of forecast days
forecast_out = int(30)
print('length =',len(aaxis_df), "and forecast_out =", forecast_out)
# Creating label by shifting 'Adj. Close' according to 'forecast_out'
aaxis_df['label'] = aaxis_df[forecast_col].shift(-forecast_out)
print(aaxis_df.head(2))
print('\n')
# If we look at the tail, it consists of n(=forecast_out) rows with NAN in Label column
print(aaxis_df.tail(2))
# Define features Matrix X by excluding the label column which we just created
X = np.array(aaxis_df.drop(['label'], 1))
# Using a feature in sklearn, preposessing to scale features
X = preprocessing.scale(X)
print(X[1,:])
X_forecast_out = X[-forecast_out:]
X = X[:-forecast_out]
print ("Length of X_forecast_out:", len(X_forecast_out), "& Length of X :", len(X))
# A good test is to make sure length of X and y are identical
y = np.array(aaxis_df['label'])
y = y[:-forecast_out]
print('Length of y: ',len(y))
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size = 0.2)
print('length of X_train and x_test: ', len(X_train), len(X_test))
# Train
from sklearn.ensemble import RandomForestRegressor
clf = RandomForestRegressor()
clf.fit(X_train,y_train)
# Test
accuracy = clf.score(X_test, y_test)
print("Accuracy of Linear Regression: ", accuracy)
forecast_prediction = clf.predict(X_forecast_out)
print(forecast_prediction)
#Make the final DataFrame containing Dates, ClosePrices, and Forecast values
actual = pd.DataFrame(dates, columns = ["Date"])
actual["ClosePrice"] = aaxis_df["Adj Close"]
actual["Forecast"] = np.nan
actual.set_index("Date", inplace = True)
forecast = pd.DataFrame(dates_check, columns=["Date"])
forecast["Forecast"] = forecast_prediction
forecast["ClosePrice"] = np.nan
forecast.set_index("Date", inplace = True)
var = [actual, forecast]
result = pd.concat(var) #This is the final DataFrame
result.info()
#Plot the results
result.plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
plt.xlabel('Date')
plt.ylabel('Price')
a=result['ClosePrice'].iloc[-31]
b=result['Forecast'].iloc[-1]
ret=((b-a)/a)*100
ret
sub=pd.read_csv('submission.csv')
sub
sub=pd.read_csv('submission.csv')
sub.iloc[2,2]
for i in range(12):
if sub.loc[i]['Symbol']=='TECHM.NS':
sub.iloc[i,2]="{0:.2f}".format(ret)
sub.to_csv('submission.csv',index=False)
sub
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import datetime
aaxis_df=pd.read_csv('TECHM.NS.csv')
aaxis_df[['Open', 'High','Low', 'Close','Adj Close','Volume']] = aaxis_df[['Open', 'High','Low', 'Close','Adj Close','Volume']].apply(pd.to_numeric, errors='coerce')
aaxis_df["Open"].fillna(value=aaxis_df["Open"].mean(), inplace=True)
aaxis_df["High"].fillna(value=aaxis_df["High"].mean(), inplace=True)
aaxis_df["Low"].fillna(value=aaxis_df["Low"].mean(), inplace=True)
aaxis_df["Close"].fillna(value=aaxis_df["Close"].mean(), inplace=True)
aaxis_df["Adj Close"].fillna(value=aaxis_df["Adj Close"].mean(), inplace=True)
aaxis_df["Volume"].fillna(value=aaxis_df["Volume"].mean(), inplace=True)
date_column=aaxis_df["Date"]
from sklearn import preprocessing, cross_validation, svm
from sklearn.linear_model import LinearRegression
#Visualization
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
#aaxis_df.set_index("Date", inplace = True)
aaxis_df[['Adj Close','Open','High','Low','Close']].plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
plt.ylabel('Price')
from sklearn import preprocessing, cross_validation, svm
from sklearn.linear_model import LinearRegression
#Visualization
import matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
#Moving Average
def MA(df, n):
name = 'SMA_' + str(n)
#MA = pd.Series(pd.rolling_mean(df['Close'], n), name = 'SMA_' + str(n))
#df = df.join(MA)
df[name]=pd.rolling_mean(df['Adj Close'],n)
return df
#Exponential Moving Average
def EMA(df, n):
name = 'EMA_' + str(n)
#MA = pd.Series(pd.rolling_mean(df['Close'], n), name = 'SMA_' + str(n))
#df = df.join(MA)
df[name]=pd.ewma(df['Adj Close'], span = n, min_periods = n - 1)
return df
for i in [30,40,50]:
MA(aaxis_df,i)
for i in [30,40,50]:
EMA(aaxis_df,i)
aaxis_df[['Adj Close', 'Close']].plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
dates = np.array(aaxis_df["Date"])
#print(dates)
dates_check = dates[-30:]
dates = dates[:-30]
# define a new feature, HL_PCT
aaxis_df['HL_PCT'] = (aaxis_df['High'] - aaxis_df['Low'])/(aaxis_df['Low']*100)
# define a new feature percentage change
aaxis_df['PCT_CHNG'] = (aaxis_df['Close'] - aaxis_df['Open'])/(aaxis_df['Open']*100)
columns_main=['Adj Close', 'HL_PCT', 'PCT_CHNG', 'Volume' ,'SMA_30', 'SMA_40', 'SMA_50', 'EMA_30', 'EMA_40', 'EMA_50']
aaxis_df = aaxis_df[columns_main]
aaxis_df.fillna( value=0, inplace=True)
aaxis_df.isnull().sum()
# pick a forecast column
forecast_col = 'Adj Close'
# Chosing 30 days as number of forecast days
forecast_out = int(30)
print('length =',len(aaxis_df), "and forecast_out =", forecast_out)
# Creating label by shifting 'Adj. Close' according to 'forecast_out'
aaxis_df['label'] = aaxis_df[forecast_col].shift(-forecast_out)
print(aaxis_df.head(2))
print('\n')
# If we look at the tail, it consists of n(=forecast_out) rows with NAN in Label column
print(aaxis_df.tail(2))
# Define features Matrix X by excluding the label column which we just created
X = np.array(aaxis_df.drop(['label'], 1))
# Using a feature in sklearn, preposessing to scale features
X = preprocessing.scale(X)
print(X[1,:])
X_forecast_out = X[-forecast_out:]
X = X[:-forecast_out]
print ("Length of X_forecast_out:", len(X_forecast_out), "& Length of X :", len(X))
# A good test is to make sure length of X and y are identical
y = np.array(aaxis_df['label'])
y = y[:-forecast_out]
print('Length of y: ',len(y))
X_train, X_test, y_train, y_test = cross_validation.train_test_split(X, y, test_size = 0.2)
print('length of X_train and x_test: ', len(X_train), len(X_test))
# Train
from sklearn.ensemble import RandomForestRegressor
clf = RandomForestRegressor()
clf.fit(X_train,y_train)
# Test
accuracy = clf.score(X_test, y_test)
print("Accuracy of Linear Regression: ", accuracy)
forecast_prediction = clf.predict(X_forecast_out)
print(forecast_prediction)
#Make the final DataFrame containing Dates, ClosePrices, and Forecast values
actual = pd.DataFrame(dates, columns = ["Date"])
actual["ClosePrice"] = aaxis_df["Adj Close"]
actual["Forecast"] = np.nan
actual.set_index("Date", inplace = True)
forecast = pd.DataFrame(dates_check, columns=["Date"])
forecast["Forecast"] = forecast_prediction
forecast["ClosePrice"] = np.nan
forecast.set_index("Date", inplace = True)
var = [actual, forecast]
result = pd.concat(var) #This is the final DataFrame
result.info()
#Plot the results
result.plot(figsize=(20,10), linewidth=1.5)
plt.legend(loc=2, prop={'size':20})
plt.xlabel('Date')
plt.ylabel('Price')
a=result['ClosePrice'].iloc[-31]
b=result['Forecast'].iloc[-1]
ret=((b-a)/a)*100
ret
sub=pd.read_csv('submission.csv')
sub
sub=pd.read_csv('submission.csv')
sub.iloc[2,2]
for i in range(12):
if sub.loc[i]['Symbol']=='TECHM.NS':
sub.iloc[i,2]="{0:.2f}".format(ret)
sub.to_csv('submission.csv',index=False)
sub
| 0.408749 | 0.41653 |
What if your data is actually more complex than a simple straight line? Surprisingly,
you can actually use a linear model to fit nonlinear data. A simple way to do this is to
add powers of each feature as new features, then train a linear model on this extended
set of features. This technique is called Polynomial Regression.
```
import numpy as np
import numpy.random as rnd
import matplotlib.pyplot as plt
np.random.seed(42)
m = 100
X = 6 * np.random.rand(m, 1) - 3
y = 0.5 * X**2 + X + 2 + np.random.randn(m, 1)
plt.plot(X, y, "b.")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.axis([-3, 3, 0, 10])
plt.show()
from sklearn.preprocessing import PolynomialFeatures
poly_features = PolynomialFeatures(degree=2, include_bias=False)
X_poly = poly_features.fit_transform(X)
X[0]
X_poly[0]
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(X_poly, y)
lin_reg.intercept_, lin_reg.coef_
X_new=np.linspace(-3, 3, 100).reshape(100, 1)
X_new_poly = poly_features.transform(X_new)
y_new = lin_reg.predict(X_new_poly)
plt.plot(X, y, "b.")
plt.plot(X_new, y_new, "r-", linewidth=2, label="Predictions")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.legend(loc="upper left", fontsize=14)
plt.axis([-3, 3, 0, 10])
plt.show()
```
# high-degree Polynomial Regression model
```
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
for style, width, degree in (("g-", 1, 300), ("b--", 2, 2), ("r-+", 2, 1)):
polybig_features = PolynomialFeatures(degree=degree, include_bias=False)
std_scaler = StandardScaler()
lin_reg = LinearRegression()
polynomial_regression = Pipeline([
("poly_features", polybig_features),
("std_scaler", std_scaler),
("lin_reg", lin_reg),
])
polynomial_regression.fit(X, y)
y_newbig = polynomial_regression.predict(X_new)
plt.plot(X_new, y_newbig, style, label=str(degree), linewidth=width)
plt.plot(X, y, "b.", linewidth=3)
plt.legend(loc="upper left")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.axis([-3, 3, 0, 10])
plt.show()
```
you used cross-validation to get an estimate of a model’s generalization
performance. If a model performs well on the training data but generalizes poorly
according to the cross-validation metrics, then your model is overfitting. If it per‐
forms poorly on both, then it is underfitting. This is one way to tell when a model is
too simple or too complex.
Another method is Learning curves to know how can you decide how complex your model should be? How can you tell
that your model is overfitting or underfitting the data?
```
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
def plot_learning_curves(model, X, y):
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.2, random_state=10)
train_errors, val_errors = [], []
for m in range(1, len(X_train)):
model.fit(X_train[:m], y_train[:m])
y_train_predict = model.predict(X_train[:m])
y_val_predict = model.predict(X_val)
train_errors.append(mean_squared_error(y_train[:m], y_train_predict))
val_errors.append(mean_squared_error(y_val, y_val_predict))
plt.plot(np.sqrt(train_errors), "r-+", linewidth=2, label="train")
plt.plot(np.sqrt(val_errors), "b-", linewidth=3, label="val")
plt.legend(loc="upper right", fontsize=14)
plt.xlabel("Trainig set Size", fontsize=14)
plt.ylabel("RMSE", fontsize=14)
lin_reg = LinearRegression()
plot_learning_curves(lin_reg, X, y)
plt.axis([0, 80, 0, 3])
plt.show()
```
This deserves a bit of explanation. First, let’s look at the performance on the training
data: when there are just one or two instances in the training set, the model can fit
them perfectly, which is why the curve starts at zero. But as new instances are added
to the training set, it becomes impossible for the model to fit the training data per‐
fectly, both because the data is noisy and because it is not linear at all. So the error on
the training data goes up until it reaches a plateau, at which point adding new instan‐
ces to the training set doesn’t make the average error much better or worse. Now let’s
look at the performance of the model on the validation data. When the model is
trained on very few training instances, it is incapable of generalizing properly, which
is why the validation error is initially quite big. Then as the model is shown more
training examples, it learns and thus the validation error slowly goes down. However,
once again a straight line cannot do a good job modeling the data, so the error ends
up at a plateau, very close to the other curve.
These learning curves are typical of an underfitting model. Both curves have reached
a plateau; they are close and fairly high.
If your model is underfitting the training data, adding more train‐
ing examples will not help. You need to use a more complex model
or come up with better features.
```
from sklearn.pipeline import Pipeline
polynomial_regression = Pipeline([
("poly_features", PolynomialFeatures(degree=10, include_bias=False)),
("lin_reg", LinearRegression()),
])
plot_learning_curves(polynomial_regression, X, y)
plt.axis([0, 80, 0, 3])
plt.show()
```
These learning curves look a bit like the previous ones, but there are two very impor‐
tant differences:
• The error on the training data is much lower than with the Linear Regression
model.
• There is a gap between the curves. This means that the model performs signifi‐
cantly better on the training data than on the validation data, which is the hall‐
mark of an overfitting model. However, if you used a much larger training set,
the two curves would continue to get closer.
# One way to improve an overfitting model is to feed it more training
data until the validation error reaches the training error.
# The bias variance Trade OFF
An important theoretical result of statistics and Machine Learning is the fact that a
model’s generalization error can be expressed as the sum of three very different
errors:
Bias
This part of the generalization error is due to wrong assumptions, such as assum‐
ing that the data is linear when it is actually quadratic. A high-bias model is most
likely to underfit the training data. 10
Variance
This part is due to the model’s excessive sensitivity to small variations in the
training data. A model with many degrees of freedom (such as a high-degree pol‐
ynomial model) is likely to have high variance, and thus to overfit the training
data.
Irreducible error
This part is due to the noisiness of the data itself. The only way to reduce this
part of the error is to clean up the data (e.g., fix the data sources, such as broken
sensors, or detect and remove outliers).
Increasing a model’s complexity will typically increase its variance and reduce its bias.
Conversely, reducing a model’s complexity increases its bias and reduces its variance.
This is why it is called a tradeoff.
# Regularized Linear Model
a good way to reduce overfitting is to regularize the
model (i.e., to constrain it): the fewer degrees of freedom it has, the harder it will be
for it to overfit the data. For example, a simple way to regularize a polynomial model
is to reduce the number of polynomial degrees.
For a linear model, regularization is typically achieved by constraining the weights of
the model. We will now look at Ridge Regression, Lasso Regression, and Elastic Net,
which implement three different ways to constrain the weights.
# Ridge Regression
Ridge Regression (also called Tikhonov regularization) is a regularized version of Lin‐
ear Regression: a regularization term equal to α∑ n i = 1 θ i 2 is added to the cost function.
This forces the learning algorithm to not only fit the data but also keep the model
weights as small as possible. Note that the regularization term should only be added
to the cost function during training. Once the model is trained, you want to evaluate
the model’s performance using the unregularized performance measure.
The difference between the ridge and the linear regression is just the value of alpha ... if the alpha is 0 then the ridge regression is just the linear regression.If α is very large, then all weights end
up very close to zero and the result is a flat line going through the data’s mean.
```
np.random.seed(42)
m = 20
X = 3 * np.random.rand(m,1)
y = 1 + 0.5 * X + np.random.randn(m,1) / 1.5
X_new = np.linspace(0,3,100).reshape(100,1)
from sklearn.linear_model import Ridge
ridge_reg = Ridge(alpha=1 , solver="cholesky", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]])
ridge_reg = Ridge(alpha=1, solver="sag", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]])
from sklearn.linear_model import Ridge
def plot_model(model_class, polynomial, alphas, **model_kargs):
for alpha, style in zip(alphas, ("b-", "g--", "r:")):
model = model_class(alpha, **model_kargs) if alpha > 0 else LinearRegression()
if polynomial:
model = Pipeline([
("poly_features", PolynomialFeatures(degree=10, include_bias=False)),
("std_scaler", StandardScaler()),
("regul_reg", model),
])
model.fit(X, y)
y_new_regul = model.predict(X_new)
lw = 2 if alpha > 0 else 1
plt.plot(X_new, y_new_regul, style, linewidth=lw, label=r"$\alpha = {}$".format(alpha))
plt.plot(X, y, "b.", linewidth=3)
plt.legend(loc="upper left", fontsize=15)
plt.xlabel("$x_1$", fontsize=18)
plt.axis([0, 3, 0, 4])
plt.figure(figsize=(8,4))
plt.subplot(121)
plot_model(Ridge, polynomial=False, alphas=(0, 10, 100), random_state=42)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(122)
plot_model(Ridge, polynomial=True, alphas=(0, 10**-5, 1), random_state=42)
from sklearn.linear_model import SGDRegressor
sgd_reg = SGDRegressor(penalty="l2", max_iter=1000, tol=1e-3, random_state=42)
sgd_reg.fit(X, y.ravel())
sgd_reg.predict([[1.5]])
```
# Lasso Regression
Least Absolute Shrinkage and Selection Operator Regression (simply called Lasso
Regression) is another regularized version of Linear Regression: just like Ridge
Regression, it adds a regularization term to the cost function, but it uses the l 1 norm
of the weight vector instead of half the square of the l 2 norm.
```
from sklearn.linear_model import Lasso
plt.figure(figsize=(8,4))
plt.subplot(121)
plot_model(Lasso, polynomial=False, alphas=(0, 0.1, 1), random_state=42)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(122)
plot_model(Lasso, polynomial=True, alphas=(0, 10**-7, 1), random_state=42)
plt.show()
from sklearn.linear_model import Lasso
lasso_reg = Lasso(alpha=0.1)
lasso_reg.fit(X, y)
lasso_reg.predict([[1.5]])
```
# Elastic Net
Elastic Net is a middle ground between Ridge Regression and Lasso Regression. The
regularization term is a simple mix of both Ridge and Lasso’s regularization terms,
and you can control the mix ratio r. When r = 0, Elastic Net is equivalent to Ridge
Regression, and when r = 1, it is equivalent to Lasso Regression.
```
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5, random_state=42)
elastic_net.fit(X, y)
elastic_net.predict([[1.5]])
np.random.seed(42)
m = 100
X = 6 * np.random.rand(m, 1) - 3
y = 2 + X + 0.5 * X**2 + np.random.randn(m, 1)
X_train, X_val, y_train, y_val = train_test_split(X[:50], y[:50].ravel(), test_size=0.5, random_state=10)
```
# Early Stopping
```
from copy import deepcopy
poly_scaler = Pipeline([
("poly_features", PolynomialFeatures(degree=90, include_bias=False)),
("std_scaler", StandardScaler())
])
X_train_poly_scaled = poly_scaler.fit_transform(X_train)
X_val_poly_scaled = poly_scaler.transform(X_val)
sgd_reg = SGDRegressor(max_iter=1, tol=-np.infty, warm_start=True,
penalty=None, learning_rate="constant", eta0=0.0005, random_state=42)
minimum_val_error = float("inf")
best_epoch = None
best_model = None
for epoch in range(1000):
sgd_reg.fit(X_train_poly_scaled, y_train) # continues where it left off
y_val_predict = sgd_reg.predict(X_val_poly_scaled)
val_error = mean_squared_error(y_val, y_val_predict)
if val_error < minimum_val_error:
minimum_val_error = val_error
best_epoch = epoch
best_model = deepcopy(sgd_reg)
sgd_reg = SGDRegressor(max_iter=1, tol=-np.infty, warm_start=True,
penalty=None, learning_rate="constant", eta0=0.0005, random_state=42)
n_epochs = 500
train_errors, val_errors = [], []
for epoch in range(n_epochs):
sgd_reg.fit(X_train_poly_scaled, y_train)
y_train_predict = sgd_reg.predict(X_train_poly_scaled)
y_val_predict = sgd_reg.predict(X_val_poly_scaled)
train_errors.append(mean_squared_error(y_train, y_train_predict))
val_errors.append(mean_squared_error(y_val, y_val_predict))
best_epoch = np.argmin(val_errors)
best_val_rmse = np.sqrt(val_errors[best_epoch])
plt.annotate('Best model',
xy=(best_epoch, best_val_rmse),
xytext=(best_epoch, best_val_rmse + 1),
ha="center",
arrowprops=dict(facecolor='black', shrink=0.05),
fontsize=16,
)
best_val_rmse -= 0.03 # just to make the graph look better
plt.plot([0, n_epochs], [best_val_rmse, best_val_rmse], "k:", linewidth=2)
plt.plot(np.sqrt(val_errors), "b-", linewidth=3, label="Validation set")
plt.plot(np.sqrt(train_errors), "r--", linewidth=2, label="Training set")
plt.legend(loc="upper right", fontsize=14)
plt.xlabel("Epoch", fontsize=14)
plt.ylabel("RMSE", fontsize=14)
plt.show()
best_epoch, best_model
```
# lasso vs Ridge
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
t1a, t1b, t2a, t2b = -1, 3, -1.5, 1.5
t1s = np.linspace(t1a, t1b, 500)
t2s = np.linspace(t2a, t2b, 500)
t1, t2 = np.meshgrid(t1s, t2s)
T = np.c_[t1.ravel(), t2.ravel()]
Xr = np.array([[1, 1], [1, -1], [1, 0.5]])
yr = 2 * Xr[:, :1] + 0.5 * Xr[:, 1:]
J = (1/len(Xr) * np.sum((T.dot(Xr.T) - yr.T)**2, axis=1)).reshape(t1.shape)
N1 = np.linalg.norm(T, ord=1, axis=1).reshape(t1.shape)
N2 = np.linalg.norm(T, ord=2, axis=1).reshape(t1.shape)
t_min_idx = np.unravel_index(np.argmin(J), J.shape)
t1_min, t2_min = t1[t_min_idx], t2[t_min_idx]
t_init = np.array([[0.25], [-1]])
def bgd_path(theta, X, y, l1, l2, core = 1, eta = 0.05, n_iterations = 200):
path = [theta]
for iteration in range(n_iterations):
gradients = core * 2/len(X) * X.T.dot(X.dot(theta) - y) + l1 * np.sign(theta) + l2 * theta
theta = theta - eta * gradients
path.append(theta)
return np.array(path)
fig, axes = plt.subplots(2, 2, sharex=True, sharey=True, figsize=(10.1, 8))
for i, N, l1, l2, title in ((0, N1, 2., 0, "Lasso"), (1, N2, 0, 2., "Ridge")):
JR = J + l1 * N1 + l2 * 0.5 * N2**2
tr_min_idx = np.unravel_index(np.argmin(JR), JR.shape)
t1r_min, t2r_min = t1[tr_min_idx], t2[tr_min_idx]
levelsJ=(np.exp(np.linspace(0, 1, 20)) - 1) * (np.max(J) - np.min(J)) + np.min(J)
levelsJR=(np.exp(np.linspace(0, 1, 20)) - 1) * (np.max(JR) - np.min(JR)) + np.min(JR)
levelsN=np.linspace(0, np.max(N), 10)
path_J = bgd_path(t_init, Xr, yr, l1=0, l2=0)
path_JR = bgd_path(t_init, Xr, yr, l1, l2)
path_N = bgd_path(np.array([[2.0], [0.5]]), Xr, yr, np.sign(l1)/3, np.sign(l2), core=0)
ax = axes[i, 0]
ax.grid(True)
ax.axhline(y=0, color='k')
ax.axvline(x=0, color='k')
ax.contourf(t1, t2, N / 2., levels=levelsN)
ax.plot(path_N[:, 0], path_N[:, 1], "y--")
ax.plot(0, 0, "ys")
ax.plot(t1_min, t2_min, "ys")
ax.set_title(r"$\ell_{}$ penalty".format(i + 1), fontsize=16)
ax.axis([t1a, t1b, t2a, t2b])
if i == 1:
ax.set_xlabel(r"$\theta_1$", fontsize=16)
ax.set_ylabel(r"$\theta_2$", fontsize=16, rotation=0)
ax = axes[i, 1]
ax.grid(True)
ax.axhline(y=0, color='k')
ax.axvline(x=0, color='k')
ax.contourf(t1, t2, JR, levels=levelsJR, alpha=0.9)
ax.plot(path_JR[:, 0], path_JR[:, 1], "w-o")
ax.plot(path_N[:, 0], path_N[:, 1], "y--")
ax.plot(0, 0, "ys")
ax.plot(t1_min, t2_min, "ys")
ax.plot(t1r_min, t2r_min, "rs")
ax.set_title(title, fontsize=16)
ax.axis([t1a, t1b, t2a, t2b])
if i == 1:
ax.set_xlabel(r"$\theta_1$", fontsize=16)
plt.show()
```
|
github_jupyter
|
import numpy as np
import numpy.random as rnd
import matplotlib.pyplot as plt
np.random.seed(42)
m = 100
X = 6 * np.random.rand(m, 1) - 3
y = 0.5 * X**2 + X + 2 + np.random.randn(m, 1)
plt.plot(X, y, "b.")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.axis([-3, 3, 0, 10])
plt.show()
from sklearn.preprocessing import PolynomialFeatures
poly_features = PolynomialFeatures(degree=2, include_bias=False)
X_poly = poly_features.fit_transform(X)
X[0]
X_poly[0]
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(X_poly, y)
lin_reg.intercept_, lin_reg.coef_
X_new=np.linspace(-3, 3, 100).reshape(100, 1)
X_new_poly = poly_features.transform(X_new)
y_new = lin_reg.predict(X_new_poly)
plt.plot(X, y, "b.")
plt.plot(X_new, y_new, "r-", linewidth=2, label="Predictions")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.legend(loc="upper left", fontsize=14)
plt.axis([-3, 3, 0, 10])
plt.show()
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
for style, width, degree in (("g-", 1, 300), ("b--", 2, 2), ("r-+", 2, 1)):
polybig_features = PolynomialFeatures(degree=degree, include_bias=False)
std_scaler = StandardScaler()
lin_reg = LinearRegression()
polynomial_regression = Pipeline([
("poly_features", polybig_features),
("std_scaler", std_scaler),
("lin_reg", lin_reg),
])
polynomial_regression.fit(X, y)
y_newbig = polynomial_regression.predict(X_new)
plt.plot(X_new, y_newbig, style, label=str(degree), linewidth=width)
plt.plot(X, y, "b.", linewidth=3)
plt.legend(loc="upper left")
plt.xlabel("$x_1$", fontsize=18)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.axis([-3, 3, 0, 10])
plt.show()
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
def plot_learning_curves(model, X, y):
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.2, random_state=10)
train_errors, val_errors = [], []
for m in range(1, len(X_train)):
model.fit(X_train[:m], y_train[:m])
y_train_predict = model.predict(X_train[:m])
y_val_predict = model.predict(X_val)
train_errors.append(mean_squared_error(y_train[:m], y_train_predict))
val_errors.append(mean_squared_error(y_val, y_val_predict))
plt.plot(np.sqrt(train_errors), "r-+", linewidth=2, label="train")
plt.plot(np.sqrt(val_errors), "b-", linewidth=3, label="val")
plt.legend(loc="upper right", fontsize=14)
plt.xlabel("Trainig set Size", fontsize=14)
plt.ylabel("RMSE", fontsize=14)
lin_reg = LinearRegression()
plot_learning_curves(lin_reg, X, y)
plt.axis([0, 80, 0, 3])
plt.show()
from sklearn.pipeline import Pipeline
polynomial_regression = Pipeline([
("poly_features", PolynomialFeatures(degree=10, include_bias=False)),
("lin_reg", LinearRegression()),
])
plot_learning_curves(polynomial_regression, X, y)
plt.axis([0, 80, 0, 3])
plt.show()
np.random.seed(42)
m = 20
X = 3 * np.random.rand(m,1)
y = 1 + 0.5 * X + np.random.randn(m,1) / 1.5
X_new = np.linspace(0,3,100).reshape(100,1)
from sklearn.linear_model import Ridge
ridge_reg = Ridge(alpha=1 , solver="cholesky", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]])
ridge_reg = Ridge(alpha=1, solver="sag", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]])
from sklearn.linear_model import Ridge
def plot_model(model_class, polynomial, alphas, **model_kargs):
for alpha, style in zip(alphas, ("b-", "g--", "r:")):
model = model_class(alpha, **model_kargs) if alpha > 0 else LinearRegression()
if polynomial:
model = Pipeline([
("poly_features", PolynomialFeatures(degree=10, include_bias=False)),
("std_scaler", StandardScaler()),
("regul_reg", model),
])
model.fit(X, y)
y_new_regul = model.predict(X_new)
lw = 2 if alpha > 0 else 1
plt.plot(X_new, y_new_regul, style, linewidth=lw, label=r"$\alpha = {}$".format(alpha))
plt.plot(X, y, "b.", linewidth=3)
plt.legend(loc="upper left", fontsize=15)
plt.xlabel("$x_1$", fontsize=18)
plt.axis([0, 3, 0, 4])
plt.figure(figsize=(8,4))
plt.subplot(121)
plot_model(Ridge, polynomial=False, alphas=(0, 10, 100), random_state=42)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(122)
plot_model(Ridge, polynomial=True, alphas=(0, 10**-5, 1), random_state=42)
from sklearn.linear_model import SGDRegressor
sgd_reg = SGDRegressor(penalty="l2", max_iter=1000, tol=1e-3, random_state=42)
sgd_reg.fit(X, y.ravel())
sgd_reg.predict([[1.5]])
from sklearn.linear_model import Lasso
plt.figure(figsize=(8,4))
plt.subplot(121)
plot_model(Lasso, polynomial=False, alphas=(0, 0.1, 1), random_state=42)
plt.ylabel("$y$", rotation=0, fontsize=18)
plt.subplot(122)
plot_model(Lasso, polynomial=True, alphas=(0, 10**-7, 1), random_state=42)
plt.show()
from sklearn.linear_model import Lasso
lasso_reg = Lasso(alpha=0.1)
lasso_reg.fit(X, y)
lasso_reg.predict([[1.5]])
from sklearn.linear_model import ElasticNet
elastic_net = ElasticNet(alpha=0.1, l1_ratio=0.5, random_state=42)
elastic_net.fit(X, y)
elastic_net.predict([[1.5]])
np.random.seed(42)
m = 100
X = 6 * np.random.rand(m, 1) - 3
y = 2 + X + 0.5 * X**2 + np.random.randn(m, 1)
X_train, X_val, y_train, y_val = train_test_split(X[:50], y[:50].ravel(), test_size=0.5, random_state=10)
from copy import deepcopy
poly_scaler = Pipeline([
("poly_features", PolynomialFeatures(degree=90, include_bias=False)),
("std_scaler", StandardScaler())
])
X_train_poly_scaled = poly_scaler.fit_transform(X_train)
X_val_poly_scaled = poly_scaler.transform(X_val)
sgd_reg = SGDRegressor(max_iter=1, tol=-np.infty, warm_start=True,
penalty=None, learning_rate="constant", eta0=0.0005, random_state=42)
minimum_val_error = float("inf")
best_epoch = None
best_model = None
for epoch in range(1000):
sgd_reg.fit(X_train_poly_scaled, y_train) # continues where it left off
y_val_predict = sgd_reg.predict(X_val_poly_scaled)
val_error = mean_squared_error(y_val, y_val_predict)
if val_error < minimum_val_error:
minimum_val_error = val_error
best_epoch = epoch
best_model = deepcopy(sgd_reg)
sgd_reg = SGDRegressor(max_iter=1, tol=-np.infty, warm_start=True,
penalty=None, learning_rate="constant", eta0=0.0005, random_state=42)
n_epochs = 500
train_errors, val_errors = [], []
for epoch in range(n_epochs):
sgd_reg.fit(X_train_poly_scaled, y_train)
y_train_predict = sgd_reg.predict(X_train_poly_scaled)
y_val_predict = sgd_reg.predict(X_val_poly_scaled)
train_errors.append(mean_squared_error(y_train, y_train_predict))
val_errors.append(mean_squared_error(y_val, y_val_predict))
best_epoch = np.argmin(val_errors)
best_val_rmse = np.sqrt(val_errors[best_epoch])
plt.annotate('Best model',
xy=(best_epoch, best_val_rmse),
xytext=(best_epoch, best_val_rmse + 1),
ha="center",
arrowprops=dict(facecolor='black', shrink=0.05),
fontsize=16,
)
best_val_rmse -= 0.03 # just to make the graph look better
plt.plot([0, n_epochs], [best_val_rmse, best_val_rmse], "k:", linewidth=2)
plt.plot(np.sqrt(val_errors), "b-", linewidth=3, label="Validation set")
plt.plot(np.sqrt(train_errors), "r--", linewidth=2, label="Training set")
plt.legend(loc="upper right", fontsize=14)
plt.xlabel("Epoch", fontsize=14)
plt.ylabel("RMSE", fontsize=14)
plt.show()
best_epoch, best_model
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
t1a, t1b, t2a, t2b = -1, 3, -1.5, 1.5
t1s = np.linspace(t1a, t1b, 500)
t2s = np.linspace(t2a, t2b, 500)
t1, t2 = np.meshgrid(t1s, t2s)
T = np.c_[t1.ravel(), t2.ravel()]
Xr = np.array([[1, 1], [1, -1], [1, 0.5]])
yr = 2 * Xr[:, :1] + 0.5 * Xr[:, 1:]
J = (1/len(Xr) * np.sum((T.dot(Xr.T) - yr.T)**2, axis=1)).reshape(t1.shape)
N1 = np.linalg.norm(T, ord=1, axis=1).reshape(t1.shape)
N2 = np.linalg.norm(T, ord=2, axis=1).reshape(t1.shape)
t_min_idx = np.unravel_index(np.argmin(J), J.shape)
t1_min, t2_min = t1[t_min_idx], t2[t_min_idx]
t_init = np.array([[0.25], [-1]])
def bgd_path(theta, X, y, l1, l2, core = 1, eta = 0.05, n_iterations = 200):
path = [theta]
for iteration in range(n_iterations):
gradients = core * 2/len(X) * X.T.dot(X.dot(theta) - y) + l1 * np.sign(theta) + l2 * theta
theta = theta - eta * gradients
path.append(theta)
return np.array(path)
fig, axes = plt.subplots(2, 2, sharex=True, sharey=True, figsize=(10.1, 8))
for i, N, l1, l2, title in ((0, N1, 2., 0, "Lasso"), (1, N2, 0, 2., "Ridge")):
JR = J + l1 * N1 + l2 * 0.5 * N2**2
tr_min_idx = np.unravel_index(np.argmin(JR), JR.shape)
t1r_min, t2r_min = t1[tr_min_idx], t2[tr_min_idx]
levelsJ=(np.exp(np.linspace(0, 1, 20)) - 1) * (np.max(J) - np.min(J)) + np.min(J)
levelsJR=(np.exp(np.linspace(0, 1, 20)) - 1) * (np.max(JR) - np.min(JR)) + np.min(JR)
levelsN=np.linspace(0, np.max(N), 10)
path_J = bgd_path(t_init, Xr, yr, l1=0, l2=0)
path_JR = bgd_path(t_init, Xr, yr, l1, l2)
path_N = bgd_path(np.array([[2.0], [0.5]]), Xr, yr, np.sign(l1)/3, np.sign(l2), core=0)
ax = axes[i, 0]
ax.grid(True)
ax.axhline(y=0, color='k')
ax.axvline(x=0, color='k')
ax.contourf(t1, t2, N / 2., levels=levelsN)
ax.plot(path_N[:, 0], path_N[:, 1], "y--")
ax.plot(0, 0, "ys")
ax.plot(t1_min, t2_min, "ys")
ax.set_title(r"$\ell_{}$ penalty".format(i + 1), fontsize=16)
ax.axis([t1a, t1b, t2a, t2b])
if i == 1:
ax.set_xlabel(r"$\theta_1$", fontsize=16)
ax.set_ylabel(r"$\theta_2$", fontsize=16, rotation=0)
ax = axes[i, 1]
ax.grid(True)
ax.axhline(y=0, color='k')
ax.axvline(x=0, color='k')
ax.contourf(t1, t2, JR, levels=levelsJR, alpha=0.9)
ax.plot(path_JR[:, 0], path_JR[:, 1], "w-o")
ax.plot(path_N[:, 0], path_N[:, 1], "y--")
ax.plot(0, 0, "ys")
ax.plot(t1_min, t2_min, "ys")
ax.plot(t1r_min, t2r_min, "rs")
ax.set_title(title, fontsize=16)
ax.axis([t1a, t1b, t2a, t2b])
if i == 1:
ax.set_xlabel(r"$\theta_1$", fontsize=16)
plt.show()
| 0.712432 | 0.988279 |
RMinimum : Phase 3 - Jupyter
```
# Imports
import random
import math
```
Testfall : $X = [0, \cdots, n/2 - 1, n/2 + n/k, \cdots, n+n/k]$, $|X|=n$, $minele = [n/2, \cdots, n/2 + n/k - 1]$, $|minele| = n/k$, $k$
```
# User input
n = 2**8
k = 2**4
# Automatic
# W = [0, ..., 3/4 * n - 1, 3/4 * n + n/k, ..., n + n/k ]
# M = [3/4 * n, ..., 3/4 * n + n/k - 1]
W = [i for i in range(int(n + math.ceil(n / k)))]
M = [i for i in range(int(3 / 4 * n), int(3 / 4 * n + math.ceil(n / k)))]
for m in M:
if m in W:
W.remove(m)
cnt = [0 for _ in range(int(n + math.ceil(n / k)))]
# Show Testcase
print(' Testcase: ')
print('=============================')
print('|W| :', len(W))
print(' n :', n)
print('-----------------------------')
print('|M| :', len(M))
print(' n/k :', int(n/k))
print('=============================')
```
Algorithmus : Phase 3
```
def phase3(W, k, M, cnt):
random.shuffle(W)
subsets = [W[i * k:(i + 1) * k] for i in range((len(W) + k - 1) // k)]
subsets_filtered = [0 for _ in range(len(subsets))]
for i in range(len(subsets_filtered)):
subsets_filtered[i] = [elem for elem in subsets[i] if elem < M[i]]
cnt[M[i]] += len(subsets[i])
for elem in subsets[i]:
cnt[elem] += 1
Wnew = [item for sublist in subsets_filtered for item in sublist]
return subsets, subsets_filtered, Wnew, cnt
# ==================================================
# Testfall
subsets, subsets_filtered, Wnew, cnt = phase3(W, k, M, cnt)
```
Resultat :
```
def test(W, Wnew, k, M, subsets, subsets_filtered, cnt):
# Test: Filter
filt = True
# No larger elements left
for w in Wnew:
if w > max(M):
filt = False
# No larger elements in subsets according to the respective m_i
for subset in subsets_filtered:
for i in range(len(subsets_filtered)):
if max(subsets_filtered[i]) > M[i]:
filt = False
print('')
print('Testfall n / k:', len(W), '/', k)
print('====================================')
print('Size of subsets:')
print('----------------')
print("|W'| / '|W|' :", len(Wnew), '/', len(W))
print('# W_i :', len(subsets))
print('|W_i| :', len(subsets[0]))
print('|M| :', len(M))
print('====================================')
print('Fragile Complexity:')
print('-------------------')
print('min(f_w(n)) :', min([cnt[w] for w in Wnew]))
print('max(f_w(n)) :', max([cnt[w] for w in Wnew]))
print('min(f_m(n)) :', min([cnt[m] for m in M]))
print('max(f_m(n)) :', max([cnt[m] for m in M]))
print('====================================')
print('Process:')
print('--------')
print('# Ele filtered :', len(W) - len(Wnew))
print('Correct Filter :', filt)
print('------------------------------------')
print('min(f_m(n)) = max(f_m(n)) :', min([cnt[m] for m in M]) == max([cnt[m] for m in M]))
print(' = # W_i :', max([cnt[m] for m in M]) == len(subsets))
print(' = |M| :', len(subsets) == len(M))
print(' = n / k :', len(M) == math.ceil(len(W) / k))
print('====================================')
return
# ==================================================
# Testfall
test(W, Wnew, k, M, subsets, subsets_filtered, cnt)
```
|
github_jupyter
|
# Imports
import random
import math
# User input
n = 2**8
k = 2**4
# Automatic
# W = [0, ..., 3/4 * n - 1, 3/4 * n + n/k, ..., n + n/k ]
# M = [3/4 * n, ..., 3/4 * n + n/k - 1]
W = [i for i in range(int(n + math.ceil(n / k)))]
M = [i for i in range(int(3 / 4 * n), int(3 / 4 * n + math.ceil(n / k)))]
for m in M:
if m in W:
W.remove(m)
cnt = [0 for _ in range(int(n + math.ceil(n / k)))]
# Show Testcase
print(' Testcase: ')
print('=============================')
print('|W| :', len(W))
print(' n :', n)
print('-----------------------------')
print('|M| :', len(M))
print(' n/k :', int(n/k))
print('=============================')
def phase3(W, k, M, cnt):
random.shuffle(W)
subsets = [W[i * k:(i + 1) * k] for i in range((len(W) + k - 1) // k)]
subsets_filtered = [0 for _ in range(len(subsets))]
for i in range(len(subsets_filtered)):
subsets_filtered[i] = [elem for elem in subsets[i] if elem < M[i]]
cnt[M[i]] += len(subsets[i])
for elem in subsets[i]:
cnt[elem] += 1
Wnew = [item for sublist in subsets_filtered for item in sublist]
return subsets, subsets_filtered, Wnew, cnt
# ==================================================
# Testfall
subsets, subsets_filtered, Wnew, cnt = phase3(W, k, M, cnt)
def test(W, Wnew, k, M, subsets, subsets_filtered, cnt):
# Test: Filter
filt = True
# No larger elements left
for w in Wnew:
if w > max(M):
filt = False
# No larger elements in subsets according to the respective m_i
for subset in subsets_filtered:
for i in range(len(subsets_filtered)):
if max(subsets_filtered[i]) > M[i]:
filt = False
print('')
print('Testfall n / k:', len(W), '/', k)
print('====================================')
print('Size of subsets:')
print('----------------')
print("|W'| / '|W|' :", len(Wnew), '/', len(W))
print('# W_i :', len(subsets))
print('|W_i| :', len(subsets[0]))
print('|M| :', len(M))
print('====================================')
print('Fragile Complexity:')
print('-------------------')
print('min(f_w(n)) :', min([cnt[w] for w in Wnew]))
print('max(f_w(n)) :', max([cnt[w] for w in Wnew]))
print('min(f_m(n)) :', min([cnt[m] for m in M]))
print('max(f_m(n)) :', max([cnt[m] for m in M]))
print('====================================')
print('Process:')
print('--------')
print('# Ele filtered :', len(W) - len(Wnew))
print('Correct Filter :', filt)
print('------------------------------------')
print('min(f_m(n)) = max(f_m(n)) :', min([cnt[m] for m in M]) == max([cnt[m] for m in M]))
print(' = # W_i :', max([cnt[m] for m in M]) == len(subsets))
print(' = |M| :', len(subsets) == len(M))
print(' = n / k :', len(M) == math.ceil(len(W) / k))
print('====================================')
return
# ==================================================
# Testfall
test(W, Wnew, k, M, subsets, subsets_filtered, cnt)
| 0.208179 | 0.813275 |
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.decomposition import PCA
from imblearn.over_sampling import SMOTE
pd.set_option('max_columns', 200)
market = pd.read_csv('data/estaticos_market.csv', index_col='Unnamed: 0')
market.head()
portfolio1 = pd.read_csv('data/estaticos_portfolio1.csv', index_col='Unnamed: 0')
portfolio2 = pd.read_csv('data/estaticos_portfolio2.csv', index_col='Unnamed: 0')
portfolio3 = pd.read_csv('data/estaticos_portfolio3.csv', index_col='Unnamed: 0')
print('Market shape: {}'.format(market.shape))
print('Portfolio 1 shape: {}'.format(portfolio1.shape))
print('Portfolio 2 shape: {}'.format(portfolio2.shape))
print('Portfolio 3 shape: {}'.format(portfolio3.shape))
market.info()
market_infos = pd.DataFrame({'column': market.columns,
'dtype': market.dtypes,
'missing_percent': market.isna().sum()/market.shape[0],
'n_unique': market.nunique()}).reset_index().drop('index', axis=1)
market_infos.head()
booleans = ['fl_rm', 'fl_spa', 'fl_antt', 'fl_veiculo', 'fl_optante_simples', 'fl_optante_simei',
'fl_simples_irregular', 'fl_passivel_iss']
market[booleans] = market[booleans].astype('bool')
market_infos.sort_values('missing_percent', ascending=False).head(20)
```
Seems like the market dataframe has a lot of columns with a big quantity of missing values. Let's check this quantity for values bigger than 70%, 80%, 90% and 95% of missing data.
```
print('More than 70% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .7].shape[0]))
print('More than 80% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .8].shape[0]))
print('More than 90% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .9].shape[0]))
print('More than 95% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .95].shape[0]))
```
As market dataframe has 182 columns and 462298 rows, it's possible to maintain all columns that have less than 90% and remove columns with more than that.
```
column_remove = list(market_infos[market_infos['missing_percent'] > .9]['column'])
try:
market.drop(column_remove, axis=1, inplace=True)
except KeyError:
pass
market_infos = market_infos[~market_infos['column'].isin(column_remove)]
market_infos
market_infos[market_infos['n_unique'] == 1].shape
```
Let's remove columns with only 1 unique value, as it gives no relevant information to the training set
```
column_remove2 = list(market_infos[market_infos['n_unique'] == 1]['column'])
try:
market.drop(column_remove2, axis=1, inplace=True)
except KeyError:
pass
market_infos = market_infos[~market_infos['column'].isin(column_remove2)]
market_infos.reset_index(drop=True, inplace=True)
market_infos
column_others = ['dt_situacao']
try:
market.drop(column_others, axis=1, inplace=True)
except:
pass
market_infos = market_infos[~market_infos['column'].isin(column_others)]
```
## EDA
```
market.describe()
columns_num = market.select_dtypes(include=['int64', 'float64'])
for column in columns_num:
sns.distplot(market[column][~np.isnan(market[column])], kde=False)
plt.show()
```
## Filling missing values
```
def fill_missing(df: pd.DataFrame) -> pd.DataFrame:
"""
Parameters
----------
df: pd.DataFrame
Dataframe that will have it's missing values filled
Returns
-------
pd.DataFrame
Dataframe with missing values filled
"""
fill_median = ['idade_empresa_anos', 'nu_meses_rescencia', 'empsetorcensitariofaixarendapopulacao',
'percent_func_genero_masc', 'percent_func_genero_fem',
'grau_instrucao_macro_escolaridade_media', 'total', 'qt_funcionarios',
'qt_funcionarios_12meses', 'qt_funcionarios_24meses', 'tx_crescimento_12meses',
'tx_crescimento_24meses', 'tx_rotatividade']
fill_0 = ['vl_total_veiculos_pesados_grupo', 'vl_total_veiculos_leves_grupo', 'qt_socios',
'qt_socios_pf', 'qt_socios_pj', 'idade_media_socios', 'idade_maxima_socios',
'idade_minima_socios', 'qt_socios_st_regular', 'qt_socios_masculino',
'qt_socios_feminino', 'qt_coligados', 'qt_socios_coligados', 'qt_coligados_matriz',
'qt_coligados_ativo', 'qt_coligados_baixada', 'qt_coligados_inapta', 'qt_coligados_suspensa',
'idade_media_coligadas', 'idade_maxima_coligadas', 'idade_minima_coligadas',
'coligada_mais_nova_ativa', 'coligada_mais_antiga_ativa', 'idade_media_coligadas_ativas',
'qt_coligados_sa', 'qt_coligados_me', 'qt_coligados_mei', 'qt_coligados_ltda',
'qt_coligados_epp', 'qt_coligados_norte', 'qt_coligados_sul', 'qt_coligados_nordeste',
'qt_coligados_centro', 'qt_coligados_sudeste', 'qt_coligados_exterior', 'qt_ufs_coligados',
'qt_regioes_coligados', 'qt_ramos_coligados', 'qt_coligados_industria', 'qt_coligados_agropecuaria',
'qt_coligados_comercio', 'qt_coligados_serviço', 'qt_coligados_ccivil', 'faturamento_est_coligados',
'media_faturamento_est_coligados', 'max_faturamento_est_coligados', 'min_faturamento_est_coligados',
'faturamento_est_coligados_gp', 'media_faturamento_est_coligados_gp',
'max_faturamento_est_coligados_gp', 'min_faturamento_est_coligados_gp', 'qt_coligadas',
'sum_faturamento_estimado_coligadas', 'vl_faturamento_estimado_aux',
'vl_faturamento_estimado_grupo_aux', 'qt_ex_funcionarios', 'qt_funcionarios_grupo',
'meses_ultima_contratacaco', 'qt_admitidos_12meses', 'qt_desligados_12meses',
'qt_desligados', 'qt_admitidos', 'media_meses_servicos_all', 'max_meses_servicos_all',
'min_meses_servicos_all', 'media_meses_servicos', 'max_meses_servicos',
'min_meses_servicos', 'qt_filiais']
fill_mode = list(market_infos[market_infos['dtype'] == 'object']['column'])
fill_mode.remove('id')
df[fill_0] = df[fill_0].fillna(0)
imp_median = SimpleImputer(missing_values=np.nan, strategy='median')
imp_median.fit(df[fill_median])
df[fill_median] = imp_median.transform(df[fill_median])
for column in fill_mode:
market[column] = market[column].fillna(market[column].mode()[0])
return df
%%time
train_data = fill_missing(df=market)
```
## Standardization
```
%%time
numerical_columns = train_data.select_dtypes(include=['float64', 'int64']).columns
scaler = StandardScaler()
scaler.fit(train_data[numerical_columns])
train_data[numerical_columns] = scaler.transform(train_data[numerical_columns])
```
## Encoding
```
bool_columns = train_data.select_dtypes(include='bool').columns
train_data[bool_columns] = train_data[bool_columns].replace({False: 0, True: 1})
object_columns = list(market.select_dtypes(include=['object']).columns)
object_columns.remove('id')
train_data = pd.get_dummies(market, sparse=True, columns=object_columns)
train_data.shape
```
## Selecting Train and Test Data
```
y_portfolio1 = train_data['id'].isin(portfolio1['id']).replace({False: 0, True: 1})
y_portfolio2 = train_data['id'].isin(portfolio2['id']).replace({False: 0, True: 1})
y_portfolio3 = train_data['id'].isin(portfolio3['id']).replace({False: 0, True: 1})
X = train_data.drop('id', axis=1)
X_train_p1, X_test_p1, y_train_p1, y_test_p1 = train_test_split(X, y_portfolio1,
random_state=42,
test_size=.25)
X_train_p2, X_test_p2, y_train_p2, y_test_p2 = train_test_split(X, y_portfolio2,
random_state=42,
test_size=.25)
X_train_p3, X_test_p3, y_train_p3, y_test_p3 = train_test_split(X, y_portfolio3,
random_state=42,
test_size=.25)
```
## PCA
```
pca1 = PCA(n_components=2)
imb_pca1 = pca1.fit_transform(X_train_p1)
pca2 = PCA(n_components=2)
imb_pca2 = pca2.fit_transform(X_train_p2)
pca3 = PCA(n_components=2)
imb_pca3 = pca3.fit_transform(X_train_p3)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca1[:, 0], y=imb_pca1[:, 1], hue=y_train_p1)
plt.show()
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca2[:, 0], y=imb_pca2[:, 1], hue=y_train_p2)
plt.show()
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca3[:, 0], y=imb_pca3[:, 1], hue=y_train_p3)
plt.show()
print(y_train_p1.sum()/y_train_p1.shape[0])
print(y_train_p2.sum()/y_train_p2.shape[0])
print(y_train_p3.sum()/y_train_p3.shape[0])
```
## SMOTE
```
smote1 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p1, y_smote_p1 = smote1.fit_resample(X_train_p1, y_train_p1)
smote2 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p2, y_smote_p2 = smote2.fit_resample(X_train_p2, y_train_p2)
smote3 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p3, y_smote_p3 = smote3.fit_resample(X_train_p3, y_train_p3)
imb_pca_smote1 = pca1.transform(X_smote_p1)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote1[:, 0], y=imb_pca_smote1[:, 1], hue=y_smote_p1)
plt.show()
imb_pca_smote2 = pca2.transform(X_smote_p2)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote2[:, 0], y=imb_pca_smote2[:, 1], hue=y_smote_p2)
plt.show()
imb_pca_smote3 = pca3.transform(X_smote_p3)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote3[:, 0], y=imb_pca_smote3[:, 1], hue=y_smote_p3)
plt.show()
print(y_smote_p1.sum()/y_smote_p1.shape[0])
print(y_smote_p2.sum()/y_smote_p2.shape[0])
print(y_smote_p3.sum()/y_smote_p3.shape[0])
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.decomposition import PCA
from imblearn.over_sampling import SMOTE
pd.set_option('max_columns', 200)
market = pd.read_csv('data/estaticos_market.csv', index_col='Unnamed: 0')
market.head()
portfolio1 = pd.read_csv('data/estaticos_portfolio1.csv', index_col='Unnamed: 0')
portfolio2 = pd.read_csv('data/estaticos_portfolio2.csv', index_col='Unnamed: 0')
portfolio3 = pd.read_csv('data/estaticos_portfolio3.csv', index_col='Unnamed: 0')
print('Market shape: {}'.format(market.shape))
print('Portfolio 1 shape: {}'.format(portfolio1.shape))
print('Portfolio 2 shape: {}'.format(portfolio2.shape))
print('Portfolio 3 shape: {}'.format(portfolio3.shape))
market.info()
market_infos = pd.DataFrame({'column': market.columns,
'dtype': market.dtypes,
'missing_percent': market.isna().sum()/market.shape[0],
'n_unique': market.nunique()}).reset_index().drop('index', axis=1)
market_infos.head()
booleans = ['fl_rm', 'fl_spa', 'fl_antt', 'fl_veiculo', 'fl_optante_simples', 'fl_optante_simei',
'fl_simples_irregular', 'fl_passivel_iss']
market[booleans] = market[booleans].astype('bool')
market_infos.sort_values('missing_percent', ascending=False).head(20)
print('More than 70% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .7].shape[0]))
print('More than 80% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .8].shape[0]))
print('More than 90% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .9].shape[0]))
print('More than 95% of missing data: {}'.format(market_infos[market_infos['missing_percent'] > .95].shape[0]))
column_remove = list(market_infos[market_infos['missing_percent'] > .9]['column'])
try:
market.drop(column_remove, axis=1, inplace=True)
except KeyError:
pass
market_infos = market_infos[~market_infos['column'].isin(column_remove)]
market_infos
market_infos[market_infos['n_unique'] == 1].shape
column_remove2 = list(market_infos[market_infos['n_unique'] == 1]['column'])
try:
market.drop(column_remove2, axis=1, inplace=True)
except KeyError:
pass
market_infos = market_infos[~market_infos['column'].isin(column_remove2)]
market_infos.reset_index(drop=True, inplace=True)
market_infos
column_others = ['dt_situacao']
try:
market.drop(column_others, axis=1, inplace=True)
except:
pass
market_infos = market_infos[~market_infos['column'].isin(column_others)]
market.describe()
columns_num = market.select_dtypes(include=['int64', 'float64'])
for column in columns_num:
sns.distplot(market[column][~np.isnan(market[column])], kde=False)
plt.show()
def fill_missing(df: pd.DataFrame) -> pd.DataFrame:
"""
Parameters
----------
df: pd.DataFrame
Dataframe that will have it's missing values filled
Returns
-------
pd.DataFrame
Dataframe with missing values filled
"""
fill_median = ['idade_empresa_anos', 'nu_meses_rescencia', 'empsetorcensitariofaixarendapopulacao',
'percent_func_genero_masc', 'percent_func_genero_fem',
'grau_instrucao_macro_escolaridade_media', 'total', 'qt_funcionarios',
'qt_funcionarios_12meses', 'qt_funcionarios_24meses', 'tx_crescimento_12meses',
'tx_crescimento_24meses', 'tx_rotatividade']
fill_0 = ['vl_total_veiculos_pesados_grupo', 'vl_total_veiculos_leves_grupo', 'qt_socios',
'qt_socios_pf', 'qt_socios_pj', 'idade_media_socios', 'idade_maxima_socios',
'idade_minima_socios', 'qt_socios_st_regular', 'qt_socios_masculino',
'qt_socios_feminino', 'qt_coligados', 'qt_socios_coligados', 'qt_coligados_matriz',
'qt_coligados_ativo', 'qt_coligados_baixada', 'qt_coligados_inapta', 'qt_coligados_suspensa',
'idade_media_coligadas', 'idade_maxima_coligadas', 'idade_minima_coligadas',
'coligada_mais_nova_ativa', 'coligada_mais_antiga_ativa', 'idade_media_coligadas_ativas',
'qt_coligados_sa', 'qt_coligados_me', 'qt_coligados_mei', 'qt_coligados_ltda',
'qt_coligados_epp', 'qt_coligados_norte', 'qt_coligados_sul', 'qt_coligados_nordeste',
'qt_coligados_centro', 'qt_coligados_sudeste', 'qt_coligados_exterior', 'qt_ufs_coligados',
'qt_regioes_coligados', 'qt_ramos_coligados', 'qt_coligados_industria', 'qt_coligados_agropecuaria',
'qt_coligados_comercio', 'qt_coligados_serviço', 'qt_coligados_ccivil', 'faturamento_est_coligados',
'media_faturamento_est_coligados', 'max_faturamento_est_coligados', 'min_faturamento_est_coligados',
'faturamento_est_coligados_gp', 'media_faturamento_est_coligados_gp',
'max_faturamento_est_coligados_gp', 'min_faturamento_est_coligados_gp', 'qt_coligadas',
'sum_faturamento_estimado_coligadas', 'vl_faturamento_estimado_aux',
'vl_faturamento_estimado_grupo_aux', 'qt_ex_funcionarios', 'qt_funcionarios_grupo',
'meses_ultima_contratacaco', 'qt_admitidos_12meses', 'qt_desligados_12meses',
'qt_desligados', 'qt_admitidos', 'media_meses_servicos_all', 'max_meses_servicos_all',
'min_meses_servicos_all', 'media_meses_servicos', 'max_meses_servicos',
'min_meses_servicos', 'qt_filiais']
fill_mode = list(market_infos[market_infos['dtype'] == 'object']['column'])
fill_mode.remove('id')
df[fill_0] = df[fill_0].fillna(0)
imp_median = SimpleImputer(missing_values=np.nan, strategy='median')
imp_median.fit(df[fill_median])
df[fill_median] = imp_median.transform(df[fill_median])
for column in fill_mode:
market[column] = market[column].fillna(market[column].mode()[0])
return df
%%time
train_data = fill_missing(df=market)
%%time
numerical_columns = train_data.select_dtypes(include=['float64', 'int64']).columns
scaler = StandardScaler()
scaler.fit(train_data[numerical_columns])
train_data[numerical_columns] = scaler.transform(train_data[numerical_columns])
bool_columns = train_data.select_dtypes(include='bool').columns
train_data[bool_columns] = train_data[bool_columns].replace({False: 0, True: 1})
object_columns = list(market.select_dtypes(include=['object']).columns)
object_columns.remove('id')
train_data = pd.get_dummies(market, sparse=True, columns=object_columns)
train_data.shape
y_portfolio1 = train_data['id'].isin(portfolio1['id']).replace({False: 0, True: 1})
y_portfolio2 = train_data['id'].isin(portfolio2['id']).replace({False: 0, True: 1})
y_portfolio3 = train_data['id'].isin(portfolio3['id']).replace({False: 0, True: 1})
X = train_data.drop('id', axis=1)
X_train_p1, X_test_p1, y_train_p1, y_test_p1 = train_test_split(X, y_portfolio1,
random_state=42,
test_size=.25)
X_train_p2, X_test_p2, y_train_p2, y_test_p2 = train_test_split(X, y_portfolio2,
random_state=42,
test_size=.25)
X_train_p3, X_test_p3, y_train_p3, y_test_p3 = train_test_split(X, y_portfolio3,
random_state=42,
test_size=.25)
pca1 = PCA(n_components=2)
imb_pca1 = pca1.fit_transform(X_train_p1)
pca2 = PCA(n_components=2)
imb_pca2 = pca2.fit_transform(X_train_p2)
pca3 = PCA(n_components=2)
imb_pca3 = pca3.fit_transform(X_train_p3)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca1[:, 0], y=imb_pca1[:, 1], hue=y_train_p1)
plt.show()
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca2[:, 0], y=imb_pca2[:, 1], hue=y_train_p2)
plt.show()
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca3[:, 0], y=imb_pca3[:, 1], hue=y_train_p3)
plt.show()
print(y_train_p1.sum()/y_train_p1.shape[0])
print(y_train_p2.sum()/y_train_p2.shape[0])
print(y_train_p3.sum()/y_train_p3.shape[0])
smote1 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p1, y_smote_p1 = smote1.fit_resample(X_train_p1, y_train_p1)
smote2 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p2, y_smote_p2 = smote2.fit_resample(X_train_p2, y_train_p2)
smote3 = SMOTE(sampling_strategy='minority', random_state=42)
X_smote_p3, y_smote_p3 = smote3.fit_resample(X_train_p3, y_train_p3)
imb_pca_smote1 = pca1.transform(X_smote_p1)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote1[:, 0], y=imb_pca_smote1[:, 1], hue=y_smote_p1)
plt.show()
imb_pca_smote2 = pca2.transform(X_smote_p2)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote2[:, 0], y=imb_pca_smote2[:, 1], hue=y_smote_p2)
plt.show()
imb_pca_smote3 = pca3.transform(X_smote_p3)
plt.figure(figsize=(8, 8))
sns.scatterplot(x=imb_pca_smote3[:, 0], y=imb_pca_smote3[:, 1], hue=y_smote_p3)
plt.show()
print(y_smote_p1.sum()/y_smote_p1.shape[0])
print(y_smote_p2.sum()/y_smote_p2.shape[0])
print(y_smote_p3.sum()/y_smote_p3.shape[0])
| 0.436862 | 0.720884 |
# History and Resources
<p>
<img src = "assets/1.png">
</p>
- 1943: If you connect very simple binary neurons, you can do logical operations or reasoning. They had an hypothesis that brain has a very large network (actually a fact) of binary neurons which can turn on/off.
- 1947: Donald Hebb, a psychologist came up with the proposal that neurons in the brain can change their function by sort of changing the strength of the connection between the neurons. He proposed that if two neurons are active at the same time then the synapse that connects them strengthens and if they're not active then it gets depressed. This is called **Hebbian Learning**.
- 1948: Founded a discipline called cybernetics, now called as system theory or stuff like that; out of that came the idea of self-organization - that if you connect lots of very simple elements with a very simple rule to make them compute, then they might be able to self-organize themselves into having an emergent property.
- 1957: **Frank Rosenblatt** introduced the idea of Perceptron, which has now become the basis of Supervised Learning. This idea introduced the concept of weights and how they are updated with the corresponding loss.
- 1961, 62: Adaline was introduced, much similar to a basic linear classifier. Around that time, neuroscientists discovered some basic properties about the visual cortex i.e., the neurons in the visual cortex basically look at kind of a small area of the visual field and many neurons at different places of individual cortex basically perform similar operation.
- 1969: Marvin Minsky publihsed the limits of perceptrons and asked people to work on something else so the field died; people started working on logical reasoning and stuff.
**The main reasons for the field dying off in 1960 are:**
- The researchers used neurons that were binary. However, the way to get backpropagation to work is to use activation functions that are continuous. At that time, researchers didn’t have the idea of using continuous neurons and they didn’t think they can train with gradients because binary neurons are not differential.
- With continuous neurons, one would have to multiply the activation of a neuron by a weight to get a contribution to the weighted sum. However, before 1980, the multiplication of two numbers, especially floating-point numbers, were extremely slow. This resulted in another incentive to avoid using continuous neurons.
<p>
<img src = "assets/2.png">
</p>
<p>
<img src = "assets/3.png">
</p>
- 1970s: People started working on Statistical Pattern Recognition or basically changed the name of what they were working on
- 1979: He came up with the idea of Neocognitron getting inspired by Hubel and Weisel and tried to simulate the visual cortex
- 1982: Physicists started getting interested in NN; Hopfield came up with the theory for a type of recurrent neural network that he showed could be used as associative memories.
- 1983: Introduced Boltzmann machines; answered the cons of previous models like perceptrons, which were single layer models. But these had multiple layers between them, called the hidden layers.
- 1985,86: With backprop, a major conceptual change was introduced to switch from binary neurons to continuous neurons because they were differential.
- 1989: Yann LeCun introduced CNN's.
- 2003: Yoshua bengio came up with neural language model; GPT-3 uses this idea.
<p>
<img src = "assets/4.png">
</p>
Hot Future things to work on acc. to Yann:
- Self-supervised Learning
- Transformers in CV
- Machine reasoning
- Autonomous intelligent machines (AGI he mean)
<p>
<img src = "assets/5.png">
</p>
<p>
<img src = "assets/6.png">
</p>
<p>
<img src = "assets/7.png">
</p>
<p>
<img src = "assets/8.png">
</p>
### Hierarchical representation of the Visual Cortex
Experiments by Fukushima gave us an understanding of how our brain interprets the input to our eyes. In summary, it was discovered that neurons in front of our retina compress the input (known as contrast normalization) and the signal travels from our eyes to our brain. After this, the image gets processed in stages and certain neurons get activated for certain categories. Hence, the visual cortex does pattern recognition in a hierarchical manner.
Experiments in which researchers poked electrodes in specific areas of the visual cortex, specifically the V1 area made researchers realize that certain neurons react to motifs that appear in a very small area in a visual field and similarly with neighbouring neurons and neighbouring areas in the visual field. Additionally, neurons that react to the same visual field, react to different types of edges in an organized manner (e.g. vertical or horizontal edges). It is also important to note that there’s also the idea that the visual process is essentially a feed forward process. Hence, somehow fast recognition can be done without some recurrent connections.
<p>
<img src = "assets/9.png">
</p>
<p>
<img src = "assets/10.png">
</p>
<p>
<img src = "assets/11.png">
</p>
**Multilayer networks are successful because they exploit the compositional structure of natural data.** In compositional hierarchy, combinations of objects at one layer in the hierarchy form the objects at the next layer. If we mimic this hierarchy as multiple layers and let the network learn the appropriate combination of features, we get what is called Deep Learning architecture. Thus, Deep Learning networks are hierarchical in nature.
<p>
<img src = "assets/12.png">
</p>
<p>
<img src = "assets/13.png">
</p>
Feature extraction consists of expanding the representational dimension such that the expanded features are more likely to be linearly separable; data points in higher dimensional space are more likely to be linearly separable due to the increase in the number of possible separating planes.
Earlier machine learning practitioners relied on high quality, hand crafted, and task specific features to build artificial intelligence models, but with the advent of Deep Learning, the models are able to extract the generic features automatically. Some common approaches used in feature extraction algorithms are highlighted below:
Space tiling
Random Projections
Polynomial Classifier (feature cross-products)
Radial basis functions
Kernel Machines
Because of the compositional nature of data, learned features have a hierarchy of representations with increasing level of abstractions. For example:
Images - At the most granular level, images can be thought of as pixels. Combination of pixels constitute edges which when combined forms textons (multi-edge shapes). Textons form motifs and motifs form parts of the image. By combining these parts together we get the final image.
Text - Similarly, there is an inherent hierarchy in textual data. Characters form words, when we combine words together we get word-groups, then clauses, then by combining clauses we get sentences. Sentences finally tell us what story is being conveyed.
Speech - In speech, samples compose bands, which compose sounds, which compose phones, then phonemes, then whole words, then sentences, thus showing a clear hierarchy in representation.
<p>
<img src = "assets/14.png">
</p>
<p>
<img src = "assets/15.png">
</p>
There are those who dismiss Deep Learning: if we can approximate any function with 2 layers, why have more?
For example: SVMs find a separating hyperplane “in the span of the data”, meaning predictions are based on comparisons to training examples. SVMs are essentially a very simplistic 2 layer neural net, where the first layer defines “templates” and the second layer is a linear classifier. The problem with 2 layer fallacy is that the complexity and size of the middle layer is exponential in NN (to do well with a difficult task, need LOTS of templates). But if you expand the number of layers to \log(N)log(N), the layers become linear in NN. There is a trade-off between time and space.
An analogy is designing a circuit to compute a boolean function with no more than two layers of gates – we can compute any boolean function this way! But, the complexity and resources of the first layer (number of gates) quickly becomes infeasible for complex functions.
What is “deep”?
An SVM isn’t deep because it only has two layers
A classification tree isn’t deep because every layer analyses the same (raw) features
A deep network has several layers and uses them to build a hierarchy of features of increasing complexity
How can models learn representations (good features)?
Manifold hypothesis: natural data lives in a low-dimensional manifold. Set of possible images is essentially infinite, set of “natural” images is a tiny subset. For example: for an image of a person, the set of possible images is on the order of magnitude of the number of face muscles they can move (degrees of freedom) ~ 50. An ideal (and unrealistic) feature extractor represents all the factors of variation (each of the muscles, lighting, etc.).
|
github_jupyter
|
# History and Resources
<p>
<img src = "assets/1.png">
</p>
- 1943: If you connect very simple binary neurons, you can do logical operations or reasoning. They had an hypothesis that brain has a very large network (actually a fact) of binary neurons which can turn on/off.
- 1947: Donald Hebb, a psychologist came up with the proposal that neurons in the brain can change their function by sort of changing the strength of the connection between the neurons. He proposed that if two neurons are active at the same time then the synapse that connects them strengthens and if they're not active then it gets depressed. This is called **Hebbian Learning**.
- 1948: Founded a discipline called cybernetics, now called as system theory or stuff like that; out of that came the idea of self-organization - that if you connect lots of very simple elements with a very simple rule to make them compute, then they might be able to self-organize themselves into having an emergent property.
- 1957: **Frank Rosenblatt** introduced the idea of Perceptron, which has now become the basis of Supervised Learning. This idea introduced the concept of weights and how they are updated with the corresponding loss.
- 1961, 62: Adaline was introduced, much similar to a basic linear classifier. Around that time, neuroscientists discovered some basic properties about the visual cortex i.e., the neurons in the visual cortex basically look at kind of a small area of the visual field and many neurons at different places of individual cortex basically perform similar operation.
- 1969: Marvin Minsky publihsed the limits of perceptrons and asked people to work on something else so the field died; people started working on logical reasoning and stuff.
**The main reasons for the field dying off in 1960 are:**
- The researchers used neurons that were binary. However, the way to get backpropagation to work is to use activation functions that are continuous. At that time, researchers didn’t have the idea of using continuous neurons and they didn’t think they can train with gradients because binary neurons are not differential.
- With continuous neurons, one would have to multiply the activation of a neuron by a weight to get a contribution to the weighted sum. However, before 1980, the multiplication of two numbers, especially floating-point numbers, were extremely slow. This resulted in another incentive to avoid using continuous neurons.
<p>
<img src = "assets/2.png">
</p>
<p>
<img src = "assets/3.png">
</p>
- 1970s: People started working on Statistical Pattern Recognition or basically changed the name of what they were working on
- 1979: He came up with the idea of Neocognitron getting inspired by Hubel and Weisel and tried to simulate the visual cortex
- 1982: Physicists started getting interested in NN; Hopfield came up with the theory for a type of recurrent neural network that he showed could be used as associative memories.
- 1983: Introduced Boltzmann machines; answered the cons of previous models like perceptrons, which were single layer models. But these had multiple layers between them, called the hidden layers.
- 1985,86: With backprop, a major conceptual change was introduced to switch from binary neurons to continuous neurons because they were differential.
- 1989: Yann LeCun introduced CNN's.
- 2003: Yoshua bengio came up with neural language model; GPT-3 uses this idea.
<p>
<img src = "assets/4.png">
</p>
Hot Future things to work on acc. to Yann:
- Self-supervised Learning
- Transformers in CV
- Machine reasoning
- Autonomous intelligent machines (AGI he mean)
<p>
<img src = "assets/5.png">
</p>
<p>
<img src = "assets/6.png">
</p>
<p>
<img src = "assets/7.png">
</p>
<p>
<img src = "assets/8.png">
</p>
### Hierarchical representation of the Visual Cortex
Experiments by Fukushima gave us an understanding of how our brain interprets the input to our eyes. In summary, it was discovered that neurons in front of our retina compress the input (known as contrast normalization) and the signal travels from our eyes to our brain. After this, the image gets processed in stages and certain neurons get activated for certain categories. Hence, the visual cortex does pattern recognition in a hierarchical manner.
Experiments in which researchers poked electrodes in specific areas of the visual cortex, specifically the V1 area made researchers realize that certain neurons react to motifs that appear in a very small area in a visual field and similarly with neighbouring neurons and neighbouring areas in the visual field. Additionally, neurons that react to the same visual field, react to different types of edges in an organized manner (e.g. vertical or horizontal edges). It is also important to note that there’s also the idea that the visual process is essentially a feed forward process. Hence, somehow fast recognition can be done without some recurrent connections.
<p>
<img src = "assets/9.png">
</p>
<p>
<img src = "assets/10.png">
</p>
<p>
<img src = "assets/11.png">
</p>
**Multilayer networks are successful because they exploit the compositional structure of natural data.** In compositional hierarchy, combinations of objects at one layer in the hierarchy form the objects at the next layer. If we mimic this hierarchy as multiple layers and let the network learn the appropriate combination of features, we get what is called Deep Learning architecture. Thus, Deep Learning networks are hierarchical in nature.
<p>
<img src = "assets/12.png">
</p>
<p>
<img src = "assets/13.png">
</p>
Feature extraction consists of expanding the representational dimension such that the expanded features are more likely to be linearly separable; data points in higher dimensional space are more likely to be linearly separable due to the increase in the number of possible separating planes.
Earlier machine learning practitioners relied on high quality, hand crafted, and task specific features to build artificial intelligence models, but with the advent of Deep Learning, the models are able to extract the generic features automatically. Some common approaches used in feature extraction algorithms are highlighted below:
Space tiling
Random Projections
Polynomial Classifier (feature cross-products)
Radial basis functions
Kernel Machines
Because of the compositional nature of data, learned features have a hierarchy of representations with increasing level of abstractions. For example:
Images - At the most granular level, images can be thought of as pixels. Combination of pixels constitute edges which when combined forms textons (multi-edge shapes). Textons form motifs and motifs form parts of the image. By combining these parts together we get the final image.
Text - Similarly, there is an inherent hierarchy in textual data. Characters form words, when we combine words together we get word-groups, then clauses, then by combining clauses we get sentences. Sentences finally tell us what story is being conveyed.
Speech - In speech, samples compose bands, which compose sounds, which compose phones, then phonemes, then whole words, then sentences, thus showing a clear hierarchy in representation.
<p>
<img src = "assets/14.png">
</p>
<p>
<img src = "assets/15.png">
</p>
There are those who dismiss Deep Learning: if we can approximate any function with 2 layers, why have more?
For example: SVMs find a separating hyperplane “in the span of the data”, meaning predictions are based on comparisons to training examples. SVMs are essentially a very simplistic 2 layer neural net, where the first layer defines “templates” and the second layer is a linear classifier. The problem with 2 layer fallacy is that the complexity and size of the middle layer is exponential in NN (to do well with a difficult task, need LOTS of templates). But if you expand the number of layers to \log(N)log(N), the layers become linear in NN. There is a trade-off between time and space.
An analogy is designing a circuit to compute a boolean function with no more than two layers of gates – we can compute any boolean function this way! But, the complexity and resources of the first layer (number of gates) quickly becomes infeasible for complex functions.
What is “deep”?
An SVM isn’t deep because it only has two layers
A classification tree isn’t deep because every layer analyses the same (raw) features
A deep network has several layers and uses them to build a hierarchy of features of increasing complexity
How can models learn representations (good features)?
Manifold hypothesis: natural data lives in a low-dimensional manifold. Set of possible images is essentially infinite, set of “natural” images is a tiny subset. For example: for an image of a person, the set of possible images is on the order of magnitude of the number of face muscles they can move (degrees of freedom) ~ 50. An ideal (and unrealistic) feature extractor represents all the factors of variation (each of the muscles, lighting, etc.).
| 0.723505 | 0.984649 |
# Francy Package
Francy is responsible for creating a representational structure that can be rendered using pretty much any UI framework
in any language for any OS.
Francy JS renders this model in Jupyter, and allows interactivity with GAP.
# Draw
Draw is the main function of Francy. It renders a canvas and all the child objects in Jupyter environment or any other environment which connects to GAP somehow, e.g. a webssh console with websockets, etc.
### DrawSplash
DrawSplash uses Draw to generate the data and creates a Static HTML Page that can be embedded or viewed in any browser, in "offline" mode.
**NOTE: When using this, there will be no interaction back to gap as there is no kernel orchestrating the communication between francy and gap! This might change in the future, but no plans to implement such functionality using websockets at the moment.**
# Load Package
```
LoadPackage("francy");
```
# Canvas
Canvas are the base where graphics are produced. A Canvas is constituted by a Main Menu and an area where the graphics are produced.
## How to create a Canvas?
It is possible to set some default configurations for the canvas:
```gap
gap> defaults := CanvasDefaults;
gap> Sanitize(defaults);
rec( height := 600, width := 800, zoomToFit := true )
gap> SetWidth(defaults, 830);
gap> SetHeight(defaults, 630);
gap> SetZoomToFit(defaults, false);
gap> SetTexTypesetting(defaults, true);
gap> canvas := Canvas("Example Canvas", defaults);
```
Or it can be done after, by:
```gap
gap> canvas := Canvas("Example Canvas");
gap> SetWidth(canvas, 850);
gap> SetHeight(canvas, 650);
gap> SetZoomToFit(canvas, true);
```
```
canvas := Canvas("Callbacks in action");;
SetTexTypesetting(canvas, true);;
SetHeight(canvas, 100);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE, "$x^2$");; # will go throughout shapes later
Add(graph, shape);;
Add(canvas, graph);;
HelloWorld := function(name)
Add(canvas, FrancyMessage(Concatenation("Hello, ", name))); # will go throughout messages later
return Draw(canvas);
end;;
callback1 := Callback(HelloWorld);;
arg1 := RequiredArg(ArgType.STRING, "Your Name?");;
Add(callback1, arg1);;
menu := Menu("Example Menu Holder");;
menu1 := Menu( "Hello Menu Action", callback1 );;
Add(menu, menu1);;
Add(canvas, menu);;
Add(canvas, menu1);;
Add(shape, menu1);;
Draw(canvas);
```
# Menus
Menus can be added to the Canvas, where they will be added to the Main Menu on the Top, or to Shapes, where they will appear as Context Menu - Mouse right click.
The Main Menu has by default a Menu entry called Francy with 3 Sub Menus: Zoom to Fit, Save to PNG and About. When a Graph is produce the Main Menu will also contain a Menu entry called Graph Options with 3 Sub Menus: Enable/Disable Drag, Enable/Disable Neighbours, Clear Selected Nodes.
# Callbacks
A Callback is a function that is triggered in GAP and can be added to Menus and/or Shapes.
## How to create a Callback?
Callbacks can be created in many different ways, and it will depend on what you want to do.
Callbacks are triggered with mouse events. Available TriggerTypes are:
* TriggerType.DOUBLE_CLICK
* TriggerType.RIGHT_CLICK
* TriggerType.CLICK
*NOTE: No matter what you choose for TriggerType on a callback that is used on a Menu will always default to TriggerType.CLICK!*
Calling a Simple function that doesn't require any argument is the simplest form:
```gap
gap> MyFunction := function()
> # Must return allways! This is because GAP CallFuncList is used and requires it
> return;
> end;
gap> callback := Callback(MyFunction); # defaults to CLICK event
gap> callback := Callback(TriggerType.DOUBLE_CLICK, MyFunction);
```
Calling a Function with a "known" argument is also simple:
```gap
gap> canvas := Canvas("Callbacks in Action!");
gap> MyFunction := function(someKnownArg)
> # Do some crazy computation
> # Redraw
> return Draw(canvas);
> end;
gap> something := NumericalSemigroup(10,11,19);
gap> callback := Callback(MyFunction, [something]);
```
What if we want the user to give some input? Well, this is the case you have "required" arguments:
```gap
gap> canvas := Canvas("Callbacks in Action!");
gap> MyFunction := function(someKnownArg, someUserInputArg)
> # Do Some Crazy computation
> # Redraw
> return Draw(canvas);
> end;
gap> something := NumericalSemigroup(10,11,19);
gap> callback := Callback(MyFunction, [something]);
gap> arg := RequiredArg(ArgType.NUMBER, "Give me a Prime?");
gap> Add(callback, arg);
```
It is possible to add a confirmation message that gets displayed before executing the callback:
```gap
...
gap> SetConfirmMessage(callback, "This is a confirmation message! Click OK to proceed...");
```
Required Arguments type defines the data type. Available ArgTypes are:
* ArgType.SELECT
* ArgType.BOOLEAN
* ArgType.STRING
* ArgType.NUMBER
## How to create a Menu?
Menus can include a Callback or not. Menus without callback are useful for holding Submenus.
```gap
gap> callback := Callback(MyCallbackFunction);
gap> menu := Menu("Example Holder Menu");
gap> submenu := Menu("I'm a Submenu!", callback);
gap> Add(menu, submenu);
gap> Add(canvas, menu);
```
Or as a top Menu:
```gap
gap> callback := Callback(MyCallbackFunction);
gap> menu := Menu("Menu", callback);
gap> Add(canvas, menu);
```
The same menu objects can be used in Shapes:
* NOTE: Submenus are flatenned in context menus!*
```gap
gap> shape := Shape(SpaheType.CIRCLE); # will go throughout shapes and graphs later
gap> Add(shape, menu);
```
```
canvas2 := Canvas("Callbacks in action");;
SetHeight(canvas2, 100);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE);; # will go throughout shapes later
SetColor(shape, "#2E8B57");;
Add(graph, shape);;
Add(canvas2, graph);;
HelloWorld := function(name, node)
Add(canvas2, FrancyMessage(Concatenation("Hello, ", name, String(node)))); # will go throughout messages later
return Draw(canvas2);
end;;
callback1 := Callback(HelloWorld);;
SetConfirmMessage(callback1, "This is a confirmation message...");;
arg1 := RequiredArg(ArgType.STRING, "Your Name?");;
arg2 := RequiredArg(ArgType.SELECT, "Selected Nodes");;
Add(callback1, arg1);;
Add(callback1, arg2);;
Add(shape, callback1);;
menu := Menu("Example Menu Holder");;
menu1 := Menu( "Hello Menu Action", callback1 );;
Add(menu, menu1);;
Add(canvas2, menu);;
Add(canvas2, menu1);;
Add(shape, menu1);;
Draw(canvas2);
canvas3 := Canvas("Example Callbacks with Known Arguments");;
SetHeight(canvas3, 100);;
graph := Graph(GraphType.DIRECTED);;
shape := Shape(ShapeType.CIRCLE, "Click Me 1");;
shape1 := Shape(ShapeType.CIRCLE, "Click Me 2");;
Add(graph, shape);;
Add(graph, shape1);;
WhichNode := function(node)
Add(canvas3, FrancyMessage(node!.title));
return Draw(canvas3);
end;;
Add(shape, Callback(WhichNode, [shape]));; # similar to Add(shape, Callback(TriggerEvent.CLICK, WhichNode, [shape]));
Add(shape1, Callback(WhichNode, [shape1]));; # similar to Add(shape1, Callback(TriggerEvent.CLICK, WhichNode, [shape1]));
link := Link(shape, shape1);;
SetWeight(link, 2);;
SetColor(link, "red");;
SetTitle(link, "or");;
Add(graph, link);;
Add(canvas3, graph);;
Draw(canvas3);
```
# Messages
Messages are usefull for providing information to the user. Messages can be added to the Canvas and/or to Shapes.
Messages added to a Canvas are displayed as messages using colors to differentiate types, they appear on the top left corner and can be dismissed by clicking on them.
Messages added to a Shape are displayed as tooltips and their types are not taken in account, they appear when the user moves the mouse hover the Shape.
## How to create Messages?
Once again, creating messages is fairly simple and depends on the purpose of the message.
Messages can be of the following types:
* FrancyMessageType.INFO
* FrancyMessageType.ERROR
* FrancyMessageType.SUCCESS
* FrancyMessageType.WARNING
* FrancyMessageType.DEFAULT
The simplest Message with the default type would be:
```gap
gap> FrancyMessage("Hello", "World"); # title and text
gap> FrancyMessage("Hello"); # without title
```
Messages with a custom type:
```gap
gap> FrancyMessage(FrancyMessageType.INFO, "Hello", "World"); # title and text
gap> FrancyMessage(FrancyMessageType.INFO, "Hello World"); # without title
```
```
canvas4 := Canvas("Example Callbacks with Known Arguments");;
SetHeight(canvas4, 100);;
graph := Graph(GraphType.DIRECTED);;
Add(canvas4, graph);;
shape := Shape(ShapeType.CIRCLE, "Click Me");;
shape1 := Shape(ShapeType.CIRCLE, "Click Me");;
Add(graph, shape);;
Add(graph, shape1);;
WhichNode := function(c, node)
Add(c, FrancyMessage(String(node!.title)));
return Draw(c);
end;;
Add(shape, Callback(WhichNode, [canvas4, shape]));; # similar to Add(shape, Callback(TriggerEvent.CLICK, WhichNode, [shape]));
Add(shape1, Callback(WhichNode, [canvas4, shape1]));; # similar to Add(shape1, Callback(TriggerEvent.CLICK, WhichNode, [shape1]));
Draw(canvas4);
canvas5 := Canvas("Example Canvas / Shape with Messages");;
SetTexTypesetting(canvas5, true);;
SetHeight(canvas5, 250);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE);; # will go throughout shapes later
Add(graph, shape);;
Add(canvas5, graph);;
Add(canvas5, FrancyMessage(FrancyMessageType.INFO, "Hello $x^2$"));;
Add(shape, FrancyMessage(FrancyMessageType.INFO, "Hello $x^2$"));;
Add(canvas5, FrancyMessage(FrancyMessageType.ERROR, "Oops", "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.ERROR, "Oops", "Hello"));;
Add(canvas5, FrancyMessage(FrancyMessageType.WARNING, "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.WARNING, "Hello"));;
Add(canvas5, FrancyMessage(FrancyMessageType.SUCCESS, "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.SUCCESS, "Hello"));;
Add(canvas5, FrancyMessage("Hello", "World"));;
Add(shape, FrancyMessage("Hello", "World"));;
Draw(canvas5);
```
# Graphs
Graphs, according to wikipedia: *a graph is a structure amounting to a set of objects in which some pairs of the objects are in some sense "related"*
In Francy, Graphs can be created using Shapes (nodes) and Links (edges). Francy, in this case the D3 library, will try its best to shape the graph according to a set of "forces". If the Shapes provide x and y coordinates, these will be used instead and the graph will be fixed to those.
Supported GraphTypes are:
* GraphType.UNDIRECTED
* GraphType.DIRECTED
* GraphType.TREE
By default, Graphs are created with default options set to:
* GraphDefaults.simulation [true] - does not work on type **tree**. applies d3 forces to the diagram and arranges the nodes without fixed positions
* GraphDefaults.collapsed [true] - only works on type **tree**! whether the graph will be collapsed or not by default.
Supported ShapeTypes are (Node Shapes):
* ShapeType.TRIANGLE
* ShapeType.DIAMOND
* ShapeType.CIRCLE
* ShapeType.SQUARE
* ShapeType.CROSS
* ShapeType.STAR
* ShapeType.WYE
By default, Shapes are created with default options set to:
* ShapeDefaults.layer [0] - used to create hasse diagrams to set indexes
* ShapeDefaults.size [10]
* ShapeDefaults.x [0] - x position in canvas
* ShapeDefaults.y [0] - y position in canvas
*NOTE: Please note that Francy is not a Graph Library and thus no graph operations, in mathematical terms, are available*
## How to create Graphs?
Let's see how to create a graph of each type, starting with the Hasse. The Hasse diagram requires the layer to be set, in order to fix y positions to this layer option:
```gap
gap> graph := Graph(GraphType.UNDIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> SetLayer(shape, 1);
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> SetLayer(shape1, 2);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
```
*NOTE: This might change in order to ease the creation of HASSE diagrams without having to specify the layer. Suggestions are welcome!*
A Directed graph instead is simpler:
```gap
gap> graph := Graph(GraphType.DIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
```
Undirected graphs are as simple as Directed ones, but the arrows are not present:
```gap
gap> graph := Graph(GraphType.UNDIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
```
Tree graphs are as simple as the previous ones, but you need to specify the parent node:
```gap
gap> graph := Graph(GraphType.TREE);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> SetParentShape(shape1, shape);
```
**NOTE: Blue nodes are clickable on trees and allows to expand and collapse.**
```
canvas6 := Canvas("Example Hasse Graph");;
SetHeight(canvas6, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas6, graph);;
Draw(canvas6);
canvas7 := Canvas("Example Directed Graph");;
SetHeight(canvas7, 100);;
graph := Graph(GraphType.DIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas7, graph);;
Draw(canvas7);
canvas8 := Canvas("Example Undirected Graph");;
SetHeight(canvas8, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas8, graph);;
Draw(canvas8);
canvas9 := Canvas("Example Multiple Shapes Graph");;
SetHeight(canvas9, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shapeG := Shape(ShapeType.DIAMOND, "G");;
Add(graph, shapeG);;
shape1 := Shape(ShapeType.WYE, "1");;
Add(graph, shape1);;
shapeSG1 := Shape(ShapeType.SQUARE, "SG1");;
Add(graph, shapeSG1);;
Add(graph, Link(shapeG, shapeSG1));;
shapeSG2 := Shape(ShapeType.TRIANGLE, "SG2");;
Add(graph, shapeSG2);;
Add(graph, Link(shapeSG1, shapeSG2));;
Add(graph, Link(shapeSG2, shape1));;
shapeSG3 := Shape(ShapeType.CROSS, "SG3");;
Add(graph, shapeSG3);;
Add(graph, Link(shapeSG1, shapeSG3));;
Add(graph, Link(shapeSG3, shape1));;
shapeSG4 := Shape(ShapeType.STAR, "SG4");;
Add(graph, shapeSG4);;
Add(graph, Link(shapeSG1, shapeSG4));;
Add(graph, Link(shapeSG4, shape1));;
Add(canvas9, graph);;
Draw(canvas9);
canvas10 := Canvas("Example Tree Graph");;
SetHeight(canvas10, 100);;
graph := Graph(GraphType.TREE);;
SetCollapsed(graph, false);;
shapeG := Shape(ShapeType.CIRCLE, "G");;
Add(graph, shapeG);;
shape1 := Shape(ShapeType.SQUARE, "1");;
Add(graph, shape1);;
shapeSG1 := Shape(ShapeType.CIRCLE, "SG1");;
Add(graph, shapeSG1);;
shapeSG2 := Shape(ShapeType.CIRCLE, "SG2");;
Add(graph, shapeSG2);;
SetParentShape(shapeG, shape1);;
SetParentShape(shapeSG1, shapeG);;
SetParentShape(shapeSG2, shapeG);;
Add(canvas10, graph);;
Draw(canvas10);
```
# Charts
Charts are another graphical way to represent data.
Supported ChartTypes:
* ChartType.LINE
* ChartType.BAR
* ChartType.SCATTER
By default, Chart are created with default options set to:
* ChartDefaults.labels [true]
* ChartDefaults.legend [true]
## How to create Charts?
Let's see how to create a Chart of each type, starting with the LINE. LINE Charts don't support providing a custom domain, this needs more work!
```gap
gap> chart := Chart(ChartType.LINE);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
```
The same data in a Bar Chart:
```gap
gap> chart := Chart(ChartType.BAR);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisXDomain(chart, ["domain1", "domain2", "domain3", "domain4", "domain5"]);
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
```
Same data in a SCATTER Chart:
```gap
gap> chart := Chart(ChartType.SCATTER);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
```
**NOTE: Charts need more work in general**
```
canvas11 := Canvas("Example Line Chart");;
SetHeight(canvas11, 200);;
chart := Chart(ChartType.LINE);;
SetAxisXTitle(chart, "X Axis");;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas11, chart);;
Draw(canvas11);
canvas12 := Canvas("Example Bar Chart");;
SetHeight(canvas12, 200);;
chart := Chart(ChartType.BAR);;
SetAxisXTitle(chart, "X Axis");;
SetAxisXDomain(chart, ["domain1", "domain2", "domain3", "domain4", "domain5"]);;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas12, chart);;
Draw(canvas12);
canvas13 := Canvas("Example Scatter Chart");;
SetHeight(canvas13, 200);;
chart := Chart(ChartType.SCATTER);;
SetAxisXTitle(chart, "X Axis");;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas13, chart);;
Draw(canvas13);
```
|
github_jupyter
|
LoadPackage("francy");
gap> defaults := CanvasDefaults;
gap> Sanitize(defaults);
rec( height := 600, width := 800, zoomToFit := true )
gap> SetWidth(defaults, 830);
gap> SetHeight(defaults, 630);
gap> SetZoomToFit(defaults, false);
gap> SetTexTypesetting(defaults, true);
gap> canvas := Canvas("Example Canvas", defaults);
gap> canvas := Canvas("Example Canvas");
gap> SetWidth(canvas, 850);
gap> SetHeight(canvas, 650);
gap> SetZoomToFit(canvas, true);
canvas := Canvas("Callbacks in action");;
SetTexTypesetting(canvas, true);;
SetHeight(canvas, 100);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE, "$x^2$");; # will go throughout shapes later
Add(graph, shape);;
Add(canvas, graph);;
HelloWorld := function(name)
Add(canvas, FrancyMessage(Concatenation("Hello, ", name))); # will go throughout messages later
return Draw(canvas);
end;;
callback1 := Callback(HelloWorld);;
arg1 := RequiredArg(ArgType.STRING, "Your Name?");;
Add(callback1, arg1);;
menu := Menu("Example Menu Holder");;
menu1 := Menu( "Hello Menu Action", callback1 );;
Add(menu, menu1);;
Add(canvas, menu);;
Add(canvas, menu1);;
Add(shape, menu1);;
Draw(canvas);
gap> MyFunction := function()
> # Must return allways! This is because GAP CallFuncList is used and requires it
> return;
> end;
gap> callback := Callback(MyFunction); # defaults to CLICK event
gap> callback := Callback(TriggerType.DOUBLE_CLICK, MyFunction);
gap> canvas := Canvas("Callbacks in Action!");
gap> MyFunction := function(someKnownArg)
> # Do some crazy computation
> # Redraw
> return Draw(canvas);
> end;
gap> something := NumericalSemigroup(10,11,19);
gap> callback := Callback(MyFunction, [something]);
gap> canvas := Canvas("Callbacks in Action!");
gap> MyFunction := function(someKnownArg, someUserInputArg)
> # Do Some Crazy computation
> # Redraw
> return Draw(canvas);
> end;
gap> something := NumericalSemigroup(10,11,19);
gap> callback := Callback(MyFunction, [something]);
gap> arg := RequiredArg(ArgType.NUMBER, "Give me a Prime?");
gap> Add(callback, arg);
...
gap> SetConfirmMessage(callback, "This is a confirmation message! Click OK to proceed...");
gap> callback := Callback(MyCallbackFunction);
gap> menu := Menu("Example Holder Menu");
gap> submenu := Menu("I'm a Submenu!", callback);
gap> Add(menu, submenu);
gap> Add(canvas, menu);
gap> callback := Callback(MyCallbackFunction);
gap> menu := Menu("Menu", callback);
gap> Add(canvas, menu);
gap> shape := Shape(SpaheType.CIRCLE); # will go throughout shapes and graphs later
gap> Add(shape, menu);
canvas2 := Canvas("Callbacks in action");;
SetHeight(canvas2, 100);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE);; # will go throughout shapes later
SetColor(shape, "#2E8B57");;
Add(graph, shape);;
Add(canvas2, graph);;
HelloWorld := function(name, node)
Add(canvas2, FrancyMessage(Concatenation("Hello, ", name, String(node)))); # will go throughout messages later
return Draw(canvas2);
end;;
callback1 := Callback(HelloWorld);;
SetConfirmMessage(callback1, "This is a confirmation message...");;
arg1 := RequiredArg(ArgType.STRING, "Your Name?");;
arg2 := RequiredArg(ArgType.SELECT, "Selected Nodes");;
Add(callback1, arg1);;
Add(callback1, arg2);;
Add(shape, callback1);;
menu := Menu("Example Menu Holder");;
menu1 := Menu( "Hello Menu Action", callback1 );;
Add(menu, menu1);;
Add(canvas2, menu);;
Add(canvas2, menu1);;
Add(shape, menu1);;
Draw(canvas2);
canvas3 := Canvas("Example Callbacks with Known Arguments");;
SetHeight(canvas3, 100);;
graph := Graph(GraphType.DIRECTED);;
shape := Shape(ShapeType.CIRCLE, "Click Me 1");;
shape1 := Shape(ShapeType.CIRCLE, "Click Me 2");;
Add(graph, shape);;
Add(graph, shape1);;
WhichNode := function(node)
Add(canvas3, FrancyMessage(node!.title));
return Draw(canvas3);
end;;
Add(shape, Callback(WhichNode, [shape]));; # similar to Add(shape, Callback(TriggerEvent.CLICK, WhichNode, [shape]));
Add(shape1, Callback(WhichNode, [shape1]));; # similar to Add(shape1, Callback(TriggerEvent.CLICK, WhichNode, [shape1]));
link := Link(shape, shape1);;
SetWeight(link, 2);;
SetColor(link, "red");;
SetTitle(link, "or");;
Add(graph, link);;
Add(canvas3, graph);;
Draw(canvas3);
gap> FrancyMessage("Hello", "World"); # title and text
gap> FrancyMessage("Hello"); # without title
gap> FrancyMessage(FrancyMessageType.INFO, "Hello", "World"); # title and text
gap> FrancyMessage(FrancyMessageType.INFO, "Hello World"); # without title
canvas4 := Canvas("Example Callbacks with Known Arguments");;
SetHeight(canvas4, 100);;
graph := Graph(GraphType.DIRECTED);;
Add(canvas4, graph);;
shape := Shape(ShapeType.CIRCLE, "Click Me");;
shape1 := Shape(ShapeType.CIRCLE, "Click Me");;
Add(graph, shape);;
Add(graph, shape1);;
WhichNode := function(c, node)
Add(c, FrancyMessage(String(node!.title)));
return Draw(c);
end;;
Add(shape, Callback(WhichNode, [canvas4, shape]));; # similar to Add(shape, Callback(TriggerEvent.CLICK, WhichNode, [shape]));
Add(shape1, Callback(WhichNode, [canvas4, shape1]));; # similar to Add(shape1, Callback(TriggerEvent.CLICK, WhichNode, [shape1]));
Draw(canvas4);
canvas5 := Canvas("Example Canvas / Shape with Messages");;
SetTexTypesetting(canvas5, true);;
SetHeight(canvas5, 250);;
graph := Graph(GraphType.UNDIRECTED);; # will go throughout graphs later
shape := Shape(ShapeType.CIRCLE);; # will go throughout shapes later
Add(graph, shape);;
Add(canvas5, graph);;
Add(canvas5, FrancyMessage(FrancyMessageType.INFO, "Hello $x^2$"));;
Add(shape, FrancyMessage(FrancyMessageType.INFO, "Hello $x^2$"));;
Add(canvas5, FrancyMessage(FrancyMessageType.ERROR, "Oops", "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.ERROR, "Oops", "Hello"));;
Add(canvas5, FrancyMessage(FrancyMessageType.WARNING, "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.WARNING, "Hello"));;
Add(canvas5, FrancyMessage(FrancyMessageType.SUCCESS, "Hello"));;
Add(shape, FrancyMessage(FrancyMessageType.SUCCESS, "Hello"));;
Add(canvas5, FrancyMessage("Hello", "World"));;
Add(shape, FrancyMessage("Hello", "World"));;
Draw(canvas5);
gap> graph := Graph(GraphType.UNDIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> SetLayer(shape, 1);
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> SetLayer(shape1, 2);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
gap> graph := Graph(GraphType.DIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
gap> graph := Graph(GraphType.UNDIRECTED);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> link := Link(shape, shape1);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> Add(graph, link);
gap> graph := Graph(GraphType.TREE);
gap> shape := Shape(ShapeType.CIRCLE, "Title");
gap> shape1 := Shape(ShapeType.CIRCLE);
gap> Add(graph, shape);
gap> Add(graph, shape1);
gap> SetParentShape(shape1, shape);
canvas6 := Canvas("Example Hasse Graph");;
SetHeight(canvas6, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas6, graph);;
Draw(canvas6);
canvas7 := Canvas("Example Directed Graph");;
SetHeight(canvas7, 100);;
graph := Graph(GraphType.DIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas7, graph);;
Draw(canvas7);
canvas8 := Canvas("Example Undirected Graph");;
SetHeight(canvas8, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shape := Shape(ShapeType.CIRCLE, "G");;
SetLayer(shape, 1);;
shape1 := Shape(ShapeType.CIRCLE, "1");;
SetLayer(shape1, 2);;
Add(graph, shape);;
Add(graph, shape1);;
link := Link(shape, shape1);;
Add(graph, link);;
Add(canvas8, graph);;
Draw(canvas8);
canvas9 := Canvas("Example Multiple Shapes Graph");;
SetHeight(canvas9, 100);;
graph := Graph(GraphType.UNDIRECTED);;
shapeG := Shape(ShapeType.DIAMOND, "G");;
Add(graph, shapeG);;
shape1 := Shape(ShapeType.WYE, "1");;
Add(graph, shape1);;
shapeSG1 := Shape(ShapeType.SQUARE, "SG1");;
Add(graph, shapeSG1);;
Add(graph, Link(shapeG, shapeSG1));;
shapeSG2 := Shape(ShapeType.TRIANGLE, "SG2");;
Add(graph, shapeSG2);;
Add(graph, Link(shapeSG1, shapeSG2));;
Add(graph, Link(shapeSG2, shape1));;
shapeSG3 := Shape(ShapeType.CROSS, "SG3");;
Add(graph, shapeSG3);;
Add(graph, Link(shapeSG1, shapeSG3));;
Add(graph, Link(shapeSG3, shape1));;
shapeSG4 := Shape(ShapeType.STAR, "SG4");;
Add(graph, shapeSG4);;
Add(graph, Link(shapeSG1, shapeSG4));;
Add(graph, Link(shapeSG4, shape1));;
Add(canvas9, graph);;
Draw(canvas9);
canvas10 := Canvas("Example Tree Graph");;
SetHeight(canvas10, 100);;
graph := Graph(GraphType.TREE);;
SetCollapsed(graph, false);;
shapeG := Shape(ShapeType.CIRCLE, "G");;
Add(graph, shapeG);;
shape1 := Shape(ShapeType.SQUARE, "1");;
Add(graph, shape1);;
shapeSG1 := Shape(ShapeType.CIRCLE, "SG1");;
Add(graph, shapeSG1);;
shapeSG2 := Shape(ShapeType.CIRCLE, "SG2");;
Add(graph, shapeSG2);;
SetParentShape(shapeG, shape1);;
SetParentShape(shapeSG1, shapeG);;
SetParentShape(shapeSG2, shapeG);;
Add(canvas10, graph);;
Draw(canvas10);
gap> chart := Chart(ChartType.LINE);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
gap> chart := Chart(ChartType.BAR);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisXDomain(chart, ["domain1", "domain2", "domain3", "domain4", "domain5"]);
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
gap> chart := Chart(ChartType.SCATTER);
gap> SetAxisXTitle(chart, "X Axis");
gap> SetAxisYTitle(chart, "Y Axis");
gap> data1 := Dataset("data1", [100,20,30,47,90]);
gap> data2 := Dataset("data2", [51,60,72,38,97]);
gap> data3 := Dataset("data3", [50,60,70,80,90]);
gap> Add(chart, [data1, data2, data3]);
canvas11 := Canvas("Example Line Chart");;
SetHeight(canvas11, 200);;
chart := Chart(ChartType.LINE);;
SetAxisXTitle(chart, "X Axis");;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas11, chart);;
Draw(canvas11);
canvas12 := Canvas("Example Bar Chart");;
SetHeight(canvas12, 200);;
chart := Chart(ChartType.BAR);;
SetAxisXTitle(chart, "X Axis");;
SetAxisXDomain(chart, ["domain1", "domain2", "domain3", "domain4", "domain5"]);;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas12, chart);;
Draw(canvas12);
canvas13 := Canvas("Example Scatter Chart");;
SetHeight(canvas13, 200);;
chart := Chart(ChartType.SCATTER);;
SetAxisXTitle(chart, "X Axis");;
SetAxisYTitle(chart, "Y Axis");;
data1 := Dataset("data1", [100,20,30,47,90]);;
data2 := Dataset("data2", [51,60,72,38,97]);;
data3 := Dataset("data3", [50,60,70,80,90]);;
Add(chart, [data1, data2, data3]);;
Add(canvas13, chart);;
Draw(canvas13);
| 0.59843 | 0.858303 |
# Video Dataset Transformation
Transforming our dataset can help us make the most out of a limited dataset. Especially for video data that is often difficult to get in the first place, using transformations can really help reduce the total amount of video footage that is needed.
In this notebook, we show examples of what different transformations look like, to help decide what transformations to use during training.
### Initialization
Import all the functions we need.
```
import sys
sys.path.append("../../")
import os
import time
import decord
import matplotlib.pyplot as plt
import numpy as np
import warnings
import shutil
from sklearn.metrics import accuracy_score
import torch
import torch.cuda as cuda
import torch.nn as nn
import torchvision
import urllib.request
from utils_cv.action_recognition.dataset import VideoDataset, DEFAULT_MEAN, DEFAULT_STD
from utils_cv.action_recognition.references.functional_video import denormalize
from utils_cv.action_recognition.references.transforms_video import (
CenterCropVideo,
NormalizeVideo,
RandomCropVideo,
RandomHorizontalFlipVideo,
RandomResizedCropVideo,
ResizeVideo,
ToTensorVideo,
)
from utils_cv.common.gpu import system_info
from utils_cv.common.data import data_path
system_info()
warnings.filterwarnings('ignore')
%load_ext autoreload
%autoreload 2
```
Set parameters for this notebook:
```
VIDEO_PATH = os.path.join(str(data_path()), "drinking.mp4")
```
In this notebook, we'll be showing various transformations, so here's a simple helper function to easily display clips from a video.
```
def show_clip(clip, size_factor=600):
""" Show frames in a clip """
if isinstance(clip, torch.Tensor):
# Convert [C, T, H, W] tensor to [T, H, W, C] numpy array
clip = np.moveaxis(clip.numpy(), 0, -1)
figsize = np.array([clip[0].shape[1] * len(clip), clip[0].shape[0]]) / size_factor
fig, axs = plt.subplots(1, len(clip), figsize=figsize)
for i, f in enumerate(clip):
axs[i].axis("off")
axs[i].imshow(f)
```
# Prepare a Sample Video
For this notebook, we'll use a sample video that we've stored in Azure blob to demonstrate the various kinds of transformations. The video is saved to this location:
```
url = (
"https://cvbp-secondary.z19.web.core.windows.net/datasets/action_recognition/drinking.mp4"
)
```
Download the file from `url` and save it locally under VIDEO_PATH:
```
with urllib.request.urlopen(url) as response, open(VIDEO_PATH, "wb") as out_file:
shutil.copyfileobj(response, out_file)
```
We'll use `decord` to see how many frames the video has.
```
video_reader = decord.VideoReader(VIDEO_PATH)
video_length = len(video_reader)
print("Video length = {} frames".format(video_length))
```
For the rest of the notebook, we'll use the first, middle, and last frames to visualize what the video transformations look like.
```
clip = [
video_reader[0].asnumpy(),
video_reader[video_length//2].asnumpy(),
video_reader[video_length-1].asnumpy(),
]
```
Here's what the first, middle and last frame looks like without transformations:
```
show_clip(clip)
```
Finally, we'll create a `t_clip` tensor that contains each of these three frames so that we can apply our transformations to all of them at once.
```
# [T, H, W, C] numpy array to [C, T, H, W] tensor
t_clip = ToTensorVideo()(torch.from_numpy(np.array(clip)))
t_clip.shape
```
# Video Transformations
This section of the notebook shows a variety of different transformations you can apply when training your own model.
1. Resizing with the original ratio
```
show_clip(ResizeVideo(size=800)(t_clip))
```
2. Resizing
```
show_clip(ResizeVideo(size=800, keep_ratio=False)(t_clip))
```
3. Center cropping
```
show_clip(CenterCropVideo(size=800)(t_clip))
```
4. Random cropping
```
random_crop = RandomCropVideo(size=800)
show_clip(random_crop(t_clip))
show_clip(random_crop(t_clip))
```
5. Randomly resized cropping
```
random_resized_crop = RandomResizedCropVideo(size=800)
show_clip(random_resized_crop(t_clip))
show_clip(random_resized_crop(t_clip))
```
6. Normalizing (and denormalizing to verify)
```
norm_t_clip = NormalizeVideo(mean=DEFAULT_MEAN, std=DEFAULT_STD)(t_clip)
show_clip(norm_t_clip)
show_clip(denormalize(norm_t_clip, mean=DEFAULT_MEAN, std=DEFAULT_STD))
```
7. Horizontal flipping
```
show_clip(RandomHorizontalFlipVideo(p=.5)(t_clip))
```
|
github_jupyter
|
import sys
sys.path.append("../../")
import os
import time
import decord
import matplotlib.pyplot as plt
import numpy as np
import warnings
import shutil
from sklearn.metrics import accuracy_score
import torch
import torch.cuda as cuda
import torch.nn as nn
import torchvision
import urllib.request
from utils_cv.action_recognition.dataset import VideoDataset, DEFAULT_MEAN, DEFAULT_STD
from utils_cv.action_recognition.references.functional_video import denormalize
from utils_cv.action_recognition.references.transforms_video import (
CenterCropVideo,
NormalizeVideo,
RandomCropVideo,
RandomHorizontalFlipVideo,
RandomResizedCropVideo,
ResizeVideo,
ToTensorVideo,
)
from utils_cv.common.gpu import system_info
from utils_cv.common.data import data_path
system_info()
warnings.filterwarnings('ignore')
%load_ext autoreload
%autoreload 2
VIDEO_PATH = os.path.join(str(data_path()), "drinking.mp4")
def show_clip(clip, size_factor=600):
""" Show frames in a clip """
if isinstance(clip, torch.Tensor):
# Convert [C, T, H, W] tensor to [T, H, W, C] numpy array
clip = np.moveaxis(clip.numpy(), 0, -1)
figsize = np.array([clip[0].shape[1] * len(clip), clip[0].shape[0]]) / size_factor
fig, axs = plt.subplots(1, len(clip), figsize=figsize)
for i, f in enumerate(clip):
axs[i].axis("off")
axs[i].imshow(f)
url = (
"https://cvbp-secondary.z19.web.core.windows.net/datasets/action_recognition/drinking.mp4"
)
with urllib.request.urlopen(url) as response, open(VIDEO_PATH, "wb") as out_file:
shutil.copyfileobj(response, out_file)
video_reader = decord.VideoReader(VIDEO_PATH)
video_length = len(video_reader)
print("Video length = {} frames".format(video_length))
clip = [
video_reader[0].asnumpy(),
video_reader[video_length//2].asnumpy(),
video_reader[video_length-1].asnumpy(),
]
show_clip(clip)
# [T, H, W, C] numpy array to [C, T, H, W] tensor
t_clip = ToTensorVideo()(torch.from_numpy(np.array(clip)))
t_clip.shape
show_clip(ResizeVideo(size=800)(t_clip))
show_clip(ResizeVideo(size=800, keep_ratio=False)(t_clip))
show_clip(CenterCropVideo(size=800)(t_clip))
random_crop = RandomCropVideo(size=800)
show_clip(random_crop(t_clip))
show_clip(random_crop(t_clip))
random_resized_crop = RandomResizedCropVideo(size=800)
show_clip(random_resized_crop(t_clip))
show_clip(random_resized_crop(t_clip))
norm_t_clip = NormalizeVideo(mean=DEFAULT_MEAN, std=DEFAULT_STD)(t_clip)
show_clip(norm_t_clip)
show_clip(denormalize(norm_t_clip, mean=DEFAULT_MEAN, std=DEFAULT_STD))
show_clip(RandomHorizontalFlipVideo(p=.5)(t_clip))
| 0.521959 | 0.948058 |
```
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
```
se tienen las desigualdades
$$50x_1 + 24x_2\leq 2400 $$
$$30x_1 + 33x_2\leq 2100 $$
Para hacer el plot pasamos a igualdades y depejamos para una variable
$$50x_1 + 24x_2 = 2400 $$
$$30x_1 + 33x_2 = 2100 $$
El despeje queda entonces como
$$x_1 = (2400 - 24 x_1)/24$$
$$x_1 = (2100 - 30 x_1)/33$$
```
def x2_v1(x1):
return (2400 - 50*x1)/24
def x2_v2(x1):
return (2100 - 30*x1)/33
x1 = np.linspace(0, 100)
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)) # desigualdad 2
plt.plot([45, 45], [0, 25], 'k')
plt.scatter([45, 45], [0, 25], color = 'red', s = 200)
plt.plot([43, 47], [5, 5], 'r')
plt.scatter([43, 47], [5, 5])
plt.xlim(xmin = 25, xmax = 50)
plt.ylim(ymin = -5, ymax = 45)
#plt.scatter([45], [6.25], color = 'red')
#plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
# 5*np.ones(2), alpha = .2, color = 'orange')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)) # desigualdad 2
plt.plot([45, 45], [0, 25], 'k')
plt.scatter([45, 45], [0, 25], color = 'red', s = 200)
plt.plot([43, 47], [5, 5], 'r')
plt.scatter([43, 47], [5, 5])
plt.xlim(xmin = 44, xmax = 46)
plt.ylim(ymin = 4, ymax = 8)
#plt.scatter([45], [6.25], color = 'red')
plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
5*np.ones(2), alpha = .2, color = 'blue')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
```
Ya tenemos un piunto posible para la solucion, $(45, 5)$. Los otros son (46.5, 5) y (45, 6.25)
### TAREA
Actividad.
Mónica hace aretes y cadenitas de joyería. Es tan buena, que todo lo que hace lo vende.
Le toma 30 minutos hacer un par de aretes y una hora hacer una cadenita, y como Mónica también es estudihambre, solo dispone de 10 horas a la semana para hacer las joyas. Por otra parte, el material que compra solo le alcanza para hacer 15 unidades (el par de aretes cuenta como unidad) de joyas por semana.
La utilidad que le deja la venta de las joyas es $15 en cada par de aretes y $20 en cada cadenita.
¿Cuántos pares de aretes y cuántas cadenitas debería hacer Mónica para maximizar su utilidad?
```
import matplotlib.pyplot as plt
import numpy as np
import scipy.optimize as opt
```
Se tienen las desigualdades
$$ .5x_1 + 1x_2\leq 10 $$
$$ x_1 + x_2\leq 15 $$
Despejamos
$$ x_2 = 10 - .5 x_1 $$
$$ x_2 = 15 - x_1 $$
```
def x2_v1(x1):
return (10 - .5*x1 )# ec 1
def x2_v2(x1):
return (15 - 1*x1) # de la ec 2
x1 = np.linspace(0, 100)
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)), 'blue' # desigualdad 2
#plt.plot([45, 45], [0, 25], 'k')
plt.scatter([10], [5], s = 200, color= "red")
#plt.plot([43, 47], [5, 5], 'r')
plt.xlim(xmin = 9, xmax = 11)
plt.ylim(ymin = 4, ymax = 6)
#plt.scatter([45, 45, 45.6], [6.25, 5, 5], color = 'green')
#plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
# 5*np.ones(2), alpha = .2, color = 'orange')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
c = np.array([-15, -20])
A = np.array([[0.5, 1], [1, 1]])
b = np.array([10, 15])
x1_cota = (0, None)
x2_cota = (0, None)
res_monica = opt.linprog(c=c, A_ub=A, b_ub=b, bounds = (x1_cota,x2_cota))
res_monica
```
|
github_jupyter
|
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
def x2_v1(x1):
return (2400 - 50*x1)/24
def x2_v2(x1):
return (2100 - 30*x1)/33
x1 = np.linspace(0, 100)
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)) # desigualdad 2
plt.plot([45, 45], [0, 25], 'k')
plt.scatter([45, 45], [0, 25], color = 'red', s = 200)
plt.plot([43, 47], [5, 5], 'r')
plt.scatter([43, 47], [5, 5])
plt.xlim(xmin = 25, xmax = 50)
plt.ylim(ymin = -5, ymax = 45)
#plt.scatter([45], [6.25], color = 'red')
#plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
# 5*np.ones(2), alpha = .2, color = 'orange')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)) # desigualdad 2
plt.plot([45, 45], [0, 25], 'k')
plt.scatter([45, 45], [0, 25], color = 'red', s = 200)
plt.plot([43, 47], [5, 5], 'r')
plt.scatter([43, 47], [5, 5])
plt.xlim(xmin = 44, xmax = 46)
plt.ylim(ymin = 4, ymax = 8)
#plt.scatter([45], [6.25], color = 'red')
plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
5*np.ones(2), alpha = .2, color = 'blue')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
import matplotlib.pyplot as plt
import numpy as np
import scipy.optimize as opt
def x2_v1(x1):
return (10 - .5*x1 )# ec 1
def x2_v2(x1):
return (15 - 1*x1) # de la ec 2
x1 = np.linspace(0, 100)
plt.plot(x1, x2_v1(x1), 'magenta') #desigualdad 1
plt.plot(x1, x2_v2(x1)), 'blue' # desigualdad 2
#plt.plot([45, 45], [0, 25], 'k')
plt.scatter([10], [5], s = 200, color= "red")
#plt.plot([43, 47], [5, 5], 'r')
plt.xlim(xmin = 9, xmax = 11)
plt.ylim(ymin = 4, ymax = 6)
#plt.scatter([45, 45, 45.6], [6.25, 5, 5], color = 'green')
#plt.fill_between(np.array([45, 45.6]), x2_v1(np.array([45, 45.6])),
# 5*np.ones(2), alpha = .2, color = 'orange')
plt.xlabel(r"$x_1$", fontsize = 18)
plt.ylabel(r"$x_2$", fontsize = 18)
plt.show()
c = np.array([-15, -20])
A = np.array([[0.5, 1], [1, 1]])
b = np.array([10, 15])
x1_cota = (0, None)
x2_cota = (0, None)
res_monica = opt.linprog(c=c, A_ub=A, b_ub=b, bounds = (x1_cota,x2_cota))
res_monica
| 0.394551 | 0.975012 |
# Spectrum simulation
## Prerequisites
- Knowledge of spectral extraction and datasets used in gammapy, see for instance the [spectral analysis tutorial](spectrum_analysis.ipynb)
## Context
To simulate a specific observation, it is not always necessary to simulate the full photon list. For many uses cases, simulating directly a reduced binned dataset is enough: the IRFs reduced in the correct geometry are combined with a source model to predict an actual number of counts per bin. The latter is then used to simulate a reduced dataset using Poisson probability distribution.
This can be done to check the feasibility of a measurement, to test whether fitted parameters really provide a good fit to the data etc.
Here we will see how to perform a 1D spectral simulation of a CTA observation, in particular, we will generate OFF observations following the template background stored in the CTA IRFs.
**Objective: simulate a number of spectral ON-OFF observations of a source with a power-law spectral model with CTA using the CTA 1DC response, fit them with the assumed spectral model and check that the distribution of fitted parameters is consistent with the input values.**
## Proposed approach:
We will use the following classes:
* `~gammapy.datasets.SpectrumDatasetOnOff`
* `~gammapy.datasets.SpectrumDataset`
* `~gammapy.irf.load_cta_irfs`
* `~gammapy.modeling.models.PowerLawSpectralModel`
## Setup
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord, Angle
from regions import CircleSkyRegion
from gammapy.datasets import SpectrumDatasetOnOff, SpectrumDataset, Datasets
from gammapy.makers import SpectrumDatasetMaker
from gammapy.modeling import Fit
from gammapy.modeling.models import (
PowerLawSpectralModel,
SkyModel,
)
from gammapy.irf import load_cta_irfs
from gammapy.data import Observation
from gammapy.maps import MapAxis
```
## Simulation of a single spectrum
To do a simulation, we need to define the observational parameters like the livetime, the offset, the assumed integration radius, the energy range to perform the simulation for and the choice of spectral model. We then use an in-memory observation which is convolved with the IRFs to get the predicted number of counts. This is Poission fluctuated using the `fake()` to get the simulated counts for each observation.
```
# Define simulation parameters parameters
livetime = 1 * u.h
pointing = SkyCoord(0, 0, unit="deg", frame="galactic")
offset = 0.5 * u.deg
# Reconstructed and true energy axis
energy_axis = MapAxis.from_edges(
np.logspace(-0.5, 1.0, 10), unit="TeV", name="energy", interp="log"
)
energy_axis_true = MapAxis.from_edges(
np.logspace(-1.2, 2.0, 31), unit="TeV", name="energy_true", interp="log"
)
on_region_radius = Angle("0.11 deg")
center = pointing.directional_offset_by(
position_angle=0 * u.deg, separation=offset
)
on_region = CircleSkyRegion(center=center, radius=on_region_radius)
# Define spectral model - a simple Power Law in this case
model_simu = PowerLawSpectralModel(
index=3.0,
amplitude=2.5e-12 * u.Unit("cm-2 s-1 TeV-1"),
reference=1 * u.TeV,
)
print(model_simu)
# we set the sky model used in the dataset
model = SkyModel(spectral_model=model_simu, name="source")
# Load the IRFs
# In this simulation, we use the CTA-1DC irfs shipped with gammapy.
irfs = load_cta_irfs(
"$GAMMAPY_DATA/cta-1dc/caldb/data/cta/1dc/bcf/South_z20_50h/irf_file.fits"
)
obs = Observation.create(pointing=pointing, livetime=livetime, irfs=irfs)
print(obs)
# Make the SpectrumDataset
dataset_empty = SpectrumDataset.create(
e_reco=energy_axis, e_true=energy_axis_true, region=on_region, name="obs-0"
)
maker = SpectrumDatasetMaker(selection=["exposure", "edisp", "background"])
dataset = maker.run(dataset_empty, obs)
# Set the model on the dataset, and fake
dataset.models = model
dataset.fake(random_state=42)
print(dataset)
```
You can see that backgound counts are now simulated
### On-Off analysis
To do an on off spectral analysis, which is the usual science case, the standard would be to use `SpectrumDatasetOnOff`, which uses the acceptance to fake off-counts
```
dataset_on_off = SpectrumDatasetOnOff.from_spectrum_dataset(
dataset=dataset, acceptance=1, acceptance_off=5
)
dataset_on_off.fake(npred_background=dataset.npred_background())
print(dataset_on_off)
```
You can see that off counts are now simulated as well. We now simulate several spectra using the same set of observation conditions.
```
%%time
n_obs = 100
datasets = Datasets()
for idx in range(n_obs):
dataset_on_off.fake(
random_state=idx, npred_background=dataset.npred_background()
)
dataset_fake = dataset_on_off.copy(name=f"obs-{idx}")
dataset_fake.meta_table["OBS_ID"] = [idx]
datasets.append(dataset_fake)
table = datasets.info_table()
table
```
Before moving on to the fit let's have a look at the simulated observations.
```
fix, axes = plt.subplots(1, 3, figsize=(12, 4))
axes[0].hist(table["counts"])
axes[0].set_xlabel("Counts")
axes[1].hist(table["counts_off"])
axes[1].set_xlabel("Counts Off")
axes[2].hist(table["excess"])
axes[2].set_xlabel("excess");
```
Now, we fit each simulated spectrum individually
```
%%time
results = []
for dataset in datasets:
dataset.models = model.copy()
fit = Fit([dataset])
result = fit.optimize()
results.append(
{
"index": result.parameters["index"].value,
"amplitude": result.parameters["amplitude"].value,
}
)
```
We take a look at the distribution of the fitted indices. This matches very well with the spectrum that we initially injected.
```
index = np.array([_["index"] for _ in results])
plt.hist(index, bins=10, alpha=0.5)
plt.axvline(x=model_simu.parameters["index"].value, color="red")
print(f"index: {index.mean()} += {index.std()}")
```
## Exercises
* Change the observation time to something longer or shorter. Does the observation and spectrum results change as you expected?
* Change the spectral model, e.g. add a cutoff at 5 TeV, or put a steep-spectrum source with spectral index of 4.0
* Simulate spectra with the spectral model we just defined. How much observation duration do you need to get back the injected parameters?
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import astropy.units as u
from astropy.coordinates import SkyCoord, Angle
from regions import CircleSkyRegion
from gammapy.datasets import SpectrumDatasetOnOff, SpectrumDataset, Datasets
from gammapy.makers import SpectrumDatasetMaker
from gammapy.modeling import Fit
from gammapy.modeling.models import (
PowerLawSpectralModel,
SkyModel,
)
from gammapy.irf import load_cta_irfs
from gammapy.data import Observation
from gammapy.maps import MapAxis
# Define simulation parameters parameters
livetime = 1 * u.h
pointing = SkyCoord(0, 0, unit="deg", frame="galactic")
offset = 0.5 * u.deg
# Reconstructed and true energy axis
energy_axis = MapAxis.from_edges(
np.logspace(-0.5, 1.0, 10), unit="TeV", name="energy", interp="log"
)
energy_axis_true = MapAxis.from_edges(
np.logspace(-1.2, 2.0, 31), unit="TeV", name="energy_true", interp="log"
)
on_region_radius = Angle("0.11 deg")
center = pointing.directional_offset_by(
position_angle=0 * u.deg, separation=offset
)
on_region = CircleSkyRegion(center=center, radius=on_region_radius)
# Define spectral model - a simple Power Law in this case
model_simu = PowerLawSpectralModel(
index=3.0,
amplitude=2.5e-12 * u.Unit("cm-2 s-1 TeV-1"),
reference=1 * u.TeV,
)
print(model_simu)
# we set the sky model used in the dataset
model = SkyModel(spectral_model=model_simu, name="source")
# Load the IRFs
# In this simulation, we use the CTA-1DC irfs shipped with gammapy.
irfs = load_cta_irfs(
"$GAMMAPY_DATA/cta-1dc/caldb/data/cta/1dc/bcf/South_z20_50h/irf_file.fits"
)
obs = Observation.create(pointing=pointing, livetime=livetime, irfs=irfs)
print(obs)
# Make the SpectrumDataset
dataset_empty = SpectrumDataset.create(
e_reco=energy_axis, e_true=energy_axis_true, region=on_region, name="obs-0"
)
maker = SpectrumDatasetMaker(selection=["exposure", "edisp", "background"])
dataset = maker.run(dataset_empty, obs)
# Set the model on the dataset, and fake
dataset.models = model
dataset.fake(random_state=42)
print(dataset)
dataset_on_off = SpectrumDatasetOnOff.from_spectrum_dataset(
dataset=dataset, acceptance=1, acceptance_off=5
)
dataset_on_off.fake(npred_background=dataset.npred_background())
print(dataset_on_off)
%%time
n_obs = 100
datasets = Datasets()
for idx in range(n_obs):
dataset_on_off.fake(
random_state=idx, npred_background=dataset.npred_background()
)
dataset_fake = dataset_on_off.copy(name=f"obs-{idx}")
dataset_fake.meta_table["OBS_ID"] = [idx]
datasets.append(dataset_fake)
table = datasets.info_table()
table
fix, axes = plt.subplots(1, 3, figsize=(12, 4))
axes[0].hist(table["counts"])
axes[0].set_xlabel("Counts")
axes[1].hist(table["counts_off"])
axes[1].set_xlabel("Counts Off")
axes[2].hist(table["excess"])
axes[2].set_xlabel("excess");
%%time
results = []
for dataset in datasets:
dataset.models = model.copy()
fit = Fit([dataset])
result = fit.optimize()
results.append(
{
"index": result.parameters["index"].value,
"amplitude": result.parameters["amplitude"].value,
}
)
index = np.array([_["index"] for _ in results])
plt.hist(index, bins=10, alpha=0.5)
plt.axvline(x=model_simu.parameters["index"].value, color="red")
print(f"index: {index.mean()} += {index.std()}")
| 0.750004 | 0.97959 |
# Supervised learning models to predicting football matches outcomes
### Notebook by [Martim Pinto da Silva](https://github.com/motapinto), [Luis Ramos](https://github.com/luispramos), [Francisco Gonçalves](https://github.com/kiko-g)
#### Supported by [Luis Paulo Reis](https://web.fe.up.pt/~lpreis/)
#### [Faculdade de Engenharia da Universidade do Porto](https://sigarra.up.pt/feup/en/web_page.inicial)
#### It is recommended to [view this notebook in nbviewer](https://nbviewer.ipython.org/github.com/motapinto/football-classification-predications/blob/master/src/Supervised%20Learning%20Models.ipynb) for the best overall experience
#### You can also execute the code on this notebook using [Jupyter Notebook](https://jupyter.org/) or [Binder](https://mybinder.org/)(no local installation required)
## Table of contents
1. * [Introduction](#Introduction)
2. * [Required libraries and models](#Required-libraries-and-models)
- [Libraries](#Libraries)
- [Models](#Models)
3. * [The problem domain](#The-problem-domain)
4. * [Step 1: Data analysis](#Step-1:-Data-analysis)
- [Extracting data from the database](#Extracting-data-from-the-database)
- [Matches](#Matches)
- [Team Stats - Team Attributes](#Team-Stats---Team-Attributes)
- [Team Stats - Shots](#Team-Stats---Shots)
- [Team Stats - Possession](#Team-Stats---Possession)
- [Team Stats - Crosses](#Team-Stats---Crosses)
- [FIFA data](#FIFA-data)
- [Joining all features](#Joining-all-features)
5. * [Step 2: Classification & Results Interpretation](#Step-2:-Classification-&-Results-Interpretation)
- [Training and Evaluating Models](#Training-and-Evaluating-Models)
- [The basis](#The-basis)
- [KNN](#KNN)
- [Decision Tree](#Decision-Tree)
- [SVC](#SVC)
- [Naive Bayes](#Naive-Bayes)
- [Gradient Boosting](#Gradient-Boosting)
- [Neural Network](#Neural-Network)
- [Deep Neural Network](#Deep-Neural-Network)
6. * [Conclusion](#Conclusion)
- [What did we learn](#What-did-we-learn)
- [Choosing best model](#Choosing-best-model)
7. * [Resources](#Resources)
## Introduction
[go back to the top](#Table-of-contents)
In the most recent years there's been a major influx of data. In response to this situation, Machine Learning alongside the field of Data Science have come to the forefront, representing the desire of humans to better understand and make sense of the current abundance of data in the world we live in.
In this notebook, we look forward to use Supervised Learning models to harness a dataset of around 25k football matches in order to be able to predict the outcome of other matchups according to a set of classes (win, draw, loss, etc.)
## Required libraries and models
[go back to the top](#Table-of-contents)
### Libraries
If you don't have Python on your computer, you can use the [Anaconda Python distribution](http://continuum.io/downloads) to install most of the Python packages you need. Anaconda provides a simple double-click installer for your convenience.
This notebook uses several Python packages that come standard with the Anaconda Python distribution. The primary libraries that we'll be using are:
**NumPy**: Provides a fast numerical array structure and helper functions.
**pandas**: Provides a DataFrame structure to store data in memory and work with it easily and efficiently.
**scikit-learn**: The essential Machine Learning package for a variaty of supervised learning models, in Python.
**tensorflow**: The essential Machine Learning package for deep learning, in Python.
**matplotlib**: Basic plotting library in Python; most other Python plotting libraries are built on top of it.
### Models
Regarding the supervised learning models, we are using:
* [Gaussian Naive Bayes](https://scikit-learn.org/stable/modules/generated/sklearn.naive_bayes.GaussianNB.html)
* [Nearest Neighbors](https://scikit-learn.org/stable/modules/neighbors.html)
* [DecisionTree](https://scikit-learn.org/stable/modules/generated/sklearn.tree.DecisionTreeClassifier.html)
* [Support Vector Machines](https://scikit-learn.org/stable/modules/svm.html)
* [XGBoost](https://xgboost.readthedocs.io/en/latest/)
* [Neural Networks](https://keras.io/guides/sequential_model/)
* [Deep Neural Networks](https://keras.io/guides/sequential_model/)
```
# Primary libraries
from time import time
import numpy as np
import pandas as pd
import sqlite3
import matplotlib.pyplot as plt
# Models
from sklearn.naive_bayes import GaussianNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from xgboost import XGBClassifier
# Neural Networks
from tensorflow import keras
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Flatten
from keras.layers import Input
from keras.models import Model
from keras.utils import np_utils
# Measures
from sklearn.preprocessing import Normalizer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import f1_score, accuracy_score, recall_score, confusion_matrix, precision_score, ConfusionMatrixDisplay, classification_report
from sklearn.model_selection import KFold
from sklearn.preprocessing import LabelEncoder
```
## The problem domain
[go back to the top](#Table-of-contents)
The first step to any data analysis project is to define the question or problem we're looking to solve, and to define a measure (or set of measures) for our success at solving that task. The data analysis checklist has us answer a handful of questions to accomplish that, so let's work through those questions.
#### Did you specify the type of data analytic question (e.g. exploration, association causality) before touching the data?
> We are trying to design a predictive model capable of accurately predicting if the home team will either win, lose or draw, i.e., predict the outcome of football matche based on a set of measurements, including player ratings, team ratings, team average stats(possession, corners, shoots), team style(pressing, possession, defending, counter attacking, speed of play, ..) and team match history(previous games).
#### Did you define the metric for success before beginning?
> Let's do that now. Since we're performing classification, we can use [accuracy](https://en.wikipedia.org/wiki/Accuracy_and_precision) — the fraction of correctly classified matches — to quantify how well our model is performing. Knowing that most bookkeepers predict matches with an accuracy of 50%, we will try to match or beat that value. We will also use a confusion matrix, and analyse the precision, recall and f1-score.
#### Did you consider whether the question could be answered with the available data?",
> The data provided has information about more than 25k matches across multiple leagues. Even though the usability isn't great, after some processing and cleansing of the data, we will be able to predict matches with great confidence. To answer the question, yes, we have more than enough data to analyse football matches..
## Step 1: Data analysis
[go back to the top](#Table-of-contents)
The first step we have, is to look at the data, and after extracting, analyse it. We know that most datasets can contain minor issues, so we have to search for possible null or not defined values, and if so how do we proceed? Do we remove an entire row of a Dataframe? Maybe we just need to purify and substitute it's value? This analysis is done below.
Before analysing the data, we need to first extract it. For that we use multiple methods to have a cleaner code
### Extracting data from the database
```
!pip install kaggle --quiet
!mkdir /root/.kaggle
!echo '{"username":"YOUR USERNAME","key":"YOUR API KEY"}' > /root/.kaggle/kaggle.json
!kaggle datasets download -d hugomathien/soccer -p dataset/
import zipfile
import os
os.chdir('dataset')
for file in os.listdir():
if file.endswith('.zip'):
zip_ref = zipfile.ZipFile(file, 'r')
zip_ref.extractall()
zip_ref.close()
with sqlite3.connect("database.sqlite") as con:
matches = pd.read_sql_query("SELECT * from Match", con)
team_attributes = pd.read_sql_query("SELECT distinct * from Team_Attributes",con)
player = pd.read_sql_query("SELECT * from Player",con)
player_attributes = pd.read_sql_query("SELECT * from Player_Attributes",con)
```
### Matches
We start by cleaning the match data and defining some methods for the data extraction and the labels
```
''' Derives a label for a given match. '''
def get_match_outcome(match):
#Define variables
home_goals = match['home_team_goal']
away_goals = match['away_team_goal']
outcome = pd.DataFrame()
outcome.loc[0,'match_api_id'] = match['match_api_id']
#Identify match outcome
if home_goals > away_goals:
outcome.loc[0,'outcome'] = "Win"
if home_goals == away_goals:
outcome.loc[0,'outcome'] = "Draw"
if home_goals < away_goals:
outcome.loc[0,'outcome'] = "Defeat"
#Return outcome
return outcome.loc[0]
''' Get the last x matches of a given team. '''
def get_last_matches(matches, date, team, x = 10):
#Filter team matches from matches
team_matches = matches[(matches['home_team_api_id'] == team) | (matches['away_team_api_id'] == team)]
#Filter x last matches from team matches
last_matches = team_matches[team_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:x,:]
#Return last matches
return last_matches
''' Get the last team stats of a given team. '''
def get_last_team_stats(team_id, date, team_stats):
#Filter team stats
all_team_stats = teams_stats[teams_stats['team_api_id'] == team_id]
#Filter last stats from team
last_team_stats = all_team_stats[all_team_stats.date < date].sort_values(by='date', ascending=False)
if last_team_stats.empty:
last_team_stats = all_team_stats[all_team_stats.date > date].sort_values(by='date', ascending=True)
#Return last matches
return last_team_stats.iloc[0:1,:]
''' Get the last x matches of two given teams. '''
def get_last_matches_against_eachother(matches, date, home_team, away_team, x = 10):
#Find matches of both teams
home_matches = matches[(matches['home_team_api_id'] == home_team) & (matches['away_team_api_id'] == away_team)]
away_matches = matches[(matches['home_team_api_id'] == away_team) & (matches['away_team_api_id'] == home_team)]
total_matches = pd.concat([home_matches, away_matches])
#Get last x matches
try:
last_matches = total_matches[total_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:x,:]
except:
last_matches = total_matches[total_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:total_matches.shape[0],:]
#Check for error in data
if(last_matches.shape[0] > x):
print("Error in obtaining matches")
#Return data
return last_matches
''' Get the goals[home & away] of a specfic team from a set of matches. '''
def get_goals(matches, team):
home_goals = int(matches.home_team_goal[matches.home_team_api_id == team].sum())
away_goals = int(matches.away_team_goal[matches.away_team_api_id == team].sum())
total_goals = home_goals + away_goals
return total_goals
''' Get the goals[home & away] conceided of a specfic team from a set of matches. '''
def get_goals_conceided(matches, team):
home_goals = int(matches.home_team_goal[matches.away_team_api_id == team].sum())
away_goals = int(matches.away_team_goal[matches.home_team_api_id == team].sum())
total_goals = home_goals + away_goals
return total_goals
''' Get the number of wins of a specfic team from a set of matches. '''
def get_wins(matches, team):
#Find home and away wins
home_wins = int(matches.home_team_goal[(matches.home_team_api_id == team) & (matches.home_team_goal > matches.away_team_goal)].count())
away_wins = int(matches.away_team_goal[(matches.away_team_api_id == team) & (matches.away_team_goal > matches.home_team_goal)].count())
total_wins = home_wins + away_wins
return total_wins
''' Create match specific features for a given match. '''
def get_match_features(match, matches, teams_stats, x = 10):
#Define variables
date = match.date
home_team = match.home_team_api_id
away_team = match.away_team_api_id
# Gets home and away team_stats
home_team_stats = get_last_team_stats(home_team, date, teams_stats);
away_team_stats = get_last_team_stats(away_team, date, teams_stats);
#Get last x matches of home and away team
matches_home_team = get_last_matches(matches, date, home_team, x = 5)
matches_away_team = get_last_matches(matches, date, away_team, x = 5)
#Get last x matches of both teams against each other
last_matches_against = get_last_matches_against_eachother(matches, date, home_team, away_team, x = 3)
#Create goal variables
home_goals = get_goals(matches_home_team, home_team)
away_goals = get_goals(matches_away_team, away_team)
home_goals_conceided = get_goals_conceided(matches_home_team, home_team)
away_goals_conceided = get_goals_conceided(matches_away_team, away_team)
#Define result data frame
result = pd.DataFrame()
#Define ID features
result.loc[0, 'match_api_id'] = match.match_api_id
result.loc[0, 'league_id'] = match.league_id
#Create match features and team stats
if(not home_team_stats.empty):
result.loc[0, 'home_team_buildUpPlaySpeed'] = home_team_stats['buildUpPlaySpeed'].values[0]
result.loc[0, 'home_team_buildUpPlayPassing'] = home_team_stats['buildUpPlayPassing'].values[0]
result.loc[0, 'home_team_chanceCreationPassing'] = home_team_stats['chanceCreationPassing'].values[0]
result.loc[0, 'home_team_chanceCreationCrossing'] = home_team_stats['chanceCreationCrossing'].values[0]
result.loc[0, 'home_team_chanceCreationShooting'] = home_team_stats['chanceCreationShooting'].values[0]
result.loc[0, 'home_team_defencePressure'] = home_team_stats['defencePressure'].values[0]
result.loc[0, 'home_team_defenceAggression'] = home_team_stats['defenceAggression'].values[0]
result.loc[0, 'home_team_defenceTeamWidth'] = home_team_stats['defenceTeamWidth'].values[0]
result.loc[0, 'home_team_avg_shots'] = home_team_stats['avg_shots'].values[0]
result.loc[0, 'home_team_avg_corners'] = home_team_stats['avg_corners'].values[0]
result.loc[0, 'home_team_avg_crosses'] = away_team_stats['avg_crosses'].values[0]
if(not away_team_stats.empty):
result.loc[0, 'away_team_buildUpPlaySpeed'] = away_team_stats['buildUpPlaySpeed'].values[0]
result.loc[0, 'away_team_buildUpPlayPassing'] = away_team_stats['buildUpPlayPassing'].values[0]
result.loc[0, 'away_team_chanceCreationPassing'] = away_team_stats['chanceCreationPassing'].values[0]
result.loc[0, 'away_team_chanceCreationCrossing'] = away_team_stats['chanceCreationCrossing'].values[0]
result.loc[0, 'away_team_chanceCreationShooting'] = away_team_stats['chanceCreationShooting'].values[0]
result.loc[0, 'away_team_defencePressure'] = away_team_stats['defencePressure'].values[0]
result.loc[0, 'away_team_defenceAggression'] = away_team_stats['defenceAggression'].values[0]
result.loc[0, 'away_team_defenceTeamWidth'] = away_team_stats['defenceTeamWidth'].values[0]
result.loc[0, 'away_team_avg_shots'] = away_team_stats['avg_shots'].values[0]
result.loc[0, 'away_team_avg_corners'] = away_team_stats['avg_corners'].values[0]
result.loc[0, 'away_team_avg_crosses'] = away_team_stats['avg_crosses'].values[0]
result.loc[0, 'home_team_goals_difference'] = home_goals - home_goals_conceided
result.loc[0, 'away_team_goals_difference'] = away_goals - away_goals_conceided
result.loc[0, 'games_won_home_team'] = get_wins(matches_home_team, home_team)
result.loc[0, 'games_won_away_team'] = get_wins(matches_away_team, away_team)
result.loc[0, 'games_against_won'] = get_wins(last_matches_against, home_team)
result.loc[0, 'games_against_lost'] = get_wins(last_matches_against, away_team)
result.loc[0, 'B365H'] = match.B365H
result.loc[0, 'B365D'] = match.B365D
result.loc[0, 'B365A'] = match.B365A
#Return match features
return result.loc[0]
''' Create and aggregate features and labels for all matches. '''
def get_features(matches, teams_stats, fifa, x = 10, get_overall = False):
#Get fifa stats features
fifa_stats = get_overall_fifa_rankings(fifa, get_overall)
#Get match features for all matches
match_stats = matches.apply(lambda i: get_match_features(i, matches, teams_stats, x = 10), axis = 1)
#Create dummies for league ID feature
dummies = pd.get_dummies(match_stats['league_id']).rename(columns = lambda x: 'League_' + str(x))
match_stats = pd.concat([match_stats, dummies], axis = 1)
match_stats.drop(['league_id'], inplace = True, axis = 1)
#Create match outcomes
outcomes = matches.apply(get_match_outcome, axis = 1)
#Merges features and outcomes into one frame
features = pd.merge(match_stats, fifa_stats, on = 'match_api_id', how = 'left')
features = pd.merge(features, outcomes, on = 'match_api_id', how = 'left')
#Drop NA values
features.dropna(inplace = True)
#Return preprocessed data
return features
def get_overall_fifa_rankings(fifa, get_overall = False):
''' Get overall fifa rankings from fifa data. '''
temp_data = fifa
#Check if only overall player stats are desired
if get_overall == True:
#Get overall stats
data = temp_data.loc[:,(fifa.columns.str.contains('overall_rating'))]
data.loc[:,'match_api_id'] = temp_data.loc[:,'match_api_id']
else:
#Get all stats except for stat date
cols = fifa.loc[:,(fifa.columns.str.contains('date_stat'))]
temp_data = fifa.drop(cols.columns, axis = 1)
data = temp_data
#Return data
return data
viable_matches = matches
viable_matches.describe()
```
Looking at the match data we can see that most columns have 25979 values. This means we are analysing this number of matches from the database. We can start by looking at the bookkeeper data. We can see that the number of bookkepper match data is different for each bookkeper. We start by selecting the bookeeper with the most predictions data available.
```
viable_matches = matches.sample(n=5000)
b365 = viable_matches.dropna(subset=['B365H', 'B365D', 'B365A'],inplace=False)
b365.drop(['BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace = True, axis = 1)
bw = viable_matches.dropna(subset=['BWH', 'BWD', 'BWA'],inplace=False)
bw.drop(['B365H', 'B365D', 'B365A',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
iw = viable_matches.dropna(subset=['IWH', 'IWD', 'IWA'],inplace=False)
iw.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
lb = viable_matches.dropna(subset=['LBH', 'LBD', 'LBA'],inplace=False)
lb.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
ps = viable_matches.dropna(subset=['PSH', 'PSD', 'PSA'],inplace=False)
ps.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
wh = viable_matches.dropna(subset=['WHH', 'WHD', 'WHA'],inplace=False)
wh.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
sj = viable_matches.dropna(subset=['SJH', 'SJD', 'SJA'],inplace=False)
sj.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
vc = viable_matches.dropna(subset=['VCH', 'VCD', 'VCA'],inplace=False)
vc.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
gb = viable_matches.dropna(subset=['GBH', 'GBD', 'GBA'],inplace=False)
gb.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
bs = viable_matches.dropna(subset=['BSH', 'BSD', 'BSA'],inplace=False)
bs.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA'], inplace=True, axis = 1)
lis = [b365, bw, iw, lb, ps, wh, sj, vc, gb, bs]
viable_matches = max(lis, key = lambda datframe: datframe.shape[0])
viable_matches.describe()
```
Analysing the description of the dataframe, we can see that the bookkeper regarding Bet 365 has the most available information and, has such, we will decide to selected it as a feature input for our models.
We also need to consider that some of these matches may not be on the team attributes that we will clean after this. In that case, we need to remove any matches that does not contain any team stats information, since **mean imputation** would't work in these case.
We also need to remove some rows that do not contain any information about the position of the players for some matches.
```
teams_stats = team_attributes
viable_matches = viable_matches.dropna(inplace=False)
home_teams = viable_matches['home_team_api_id'].isin(teams_stats['team_api_id'].tolist())
away_teams = viable_matches['away_team_api_id'].isin(teams_stats['team_api_id'].tolist())
viable_matches = viable_matches[home_teams & away_teams]
viable_matches.describe()
```
### Team Stats - Team Attributes
```
teams_stats.describe()
```
Looking at the description of team attributes we can see that there are a lot of values missing from the column buildUpPlayDribbling, and all the other values seem to have the right amout of rows. This means that there are a lot of values with 'Nan' on this column.
It's not ideal that we just drop those rows. Seems like the missing data on the column is systematic - all of the missing values are in the same column - this error could potentially bias our analysis.
One way to deal with missing values is **mean imputation**. If we know that the values for a measurement fall in a certain range, we can fill in empty values with the average of that measure.
```
teams_stats['buildUpPlayDribbling'].hist();
```
We can see that most buildUpPlayDribbling values fall within the 45 - 55 range, so let's fill in these entries with the average measured buildUpPlaySpeed
```
build_up_play_drib_avg = teams_stats['buildUpPlayDribbling'].mean()
# mean imputation
teams_stats.loc[(teams_stats['buildUpPlayDribbling'].isnull()), 'buildUpPlayDribbling'] = build_up_play_drib_avg
# showing new values
teams_stats.loc[teams_stats['buildUpPlayDribbling'] == build_up_play_drib_avg].head()
teams_stats.loc[(teams_stats['buildUpPlayDribbling'].isnull())]
```
Having done the **mean imputation** for team_attributes we can see that there are no longer missing values for the buildUpPlayDribbling. After that, we decided to select only continuous data, i.e, select only columns that "store" numerical values that we will provide to the input of the supervised learning models.
```
teams_stats.drop(['buildUpPlaySpeedClass', 'buildUpPlayDribblingClass', 'buildUpPlayPassingClass',
'buildUpPlayPositioningClass', 'chanceCreationPassingClass', 'chanceCreationCrossingClass',
'chanceCreationShootingClass','chanceCreationPositioningClass','defencePressureClass', 'defenceAggressionClass',
'defenceTeamWidthClass','defenceDefenderLineClass'], inplace = True, axis = 1)
teams_stats.describe()
```
### Team Stats - Shots
After cleaning the team attributes data we need to consider adding some more stats to each match. We will start by adding the average of the number of shots per team. The number of shots consists on the sum of the shots on target and the shots of target. After merging all the information to teams_stats we have to analyse the data again.
```
shots_off = pd.read_csv("shotoff_detail.csv")
shots_on = pd.read_csv("shoton_detail.csv")
shots = pd.concat([shots_off[['match_id', 'team']], shots_on[['match_id', 'team']]])
total_shots = shots["team"].value_counts()
total_matches = shots.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_shots in total_shots.iteritems():
n_matches = total_matches[index]
avg_shots = n_shots / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_shots'] = avg_shots
teams_stats.describe()
```
As we can see, there are a lot of Nan values on the avg_shots column. This represents teams that did not have shots data on this dataset. Instead of removing thoose rows, and give less input to our models we need again to do **mean imputation** and deal with these values.
```
teams_stats['avg_shots'].hist();
```
We can see that most avg_shots values fall within the 7 - 14 range, so let's fill in these entries with the average measured avg_shots
```
shots_avg_team_avg = teams_stats['avg_shots'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_shots'].isnull()), 'avg_shots'] = shots_avg_team_avg
# showing new values
teams_stats.describe()
teams_stats.loc[(teams_stats['avg_shots'].isnull())]
```
Having done the **mean imputation** for team_attributes we can see that there are no longer missing values for the avg_shots.
### Team Stats - Possession
We will now add another stat, the ball possession. One of the more important statistics to predict the match results. If we look closely the most dominating teams usually control the ball possession very easily. We can see first that the csv have repeated values for the ball possession, for each match, based on the elapsed time of the match. We need to remove all the values that do not refer to the 90 minutes mark of the elapsed time.
```
# possessions read, cleanup and merge
possessions_data = pd.read_csv("../dataset/possession_detail.csv")
last_possessions = possessions_data.sort_values(['elapsed'], ascending=False).drop_duplicates(subset=['match_id'])
last_possessions = last_possessions[['match_id', 'homepos', 'awaypos']]
```
After reading it, we need to see if the number of possession data we have available is enough to be considered
```
# get the ids of the home_team and away_team to be able to join with teams later
possessions = pd.DataFrame(columns=['team', 'possession', 'match'])
for index, row in last_possessions.iterrows():
match = matches.loc[matches['id'] == row['match_id'], ['home_team_api_id', 'away_team_api_id']]
if match.empty:
continue
hometeam = match['home_team_api_id'].values[0]
awayteam = match['away_team_api_id'].values[0]
possessions = possessions.append({'team': hometeam, 'possession': row['homepos'], 'match': row['match_id']}, ignore_index=True)
possessions = possessions.append({'team': awayteam, 'possession': row['awaypos'], 'match': row['match_id']}, ignore_index=True)
total_possessions = possessions.groupby(by=['team'])['possession'].sum()
total_matches = possessions.drop_duplicates(['team', 'match'])["team"].value_counts()
total_possessions.to_frame().describe()
```
Since the number of average possession regarding the number of viable matches is very low it doesn't make any sense to do **mean imputation** in this instance. After carefully consideration, we decided to scrap this attribute, even though this attribute as a very important meaning. This reflects the poor usability of this dataset.
### Team Stats - Corners
We will try to add yet another feature. This is time the corners, also an important measurement of domination in a football match. After merging all corners data that were given to us we can see that there are some values missing.
```
corners_data = pd.read_csv("../dataset/corner_detail.csv")
corners = corners_data[['match_id', 'team']]
total_corners = corners["team"].value_counts()
total_matches = corners.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_corners in total_shots.iteritems():
n_matches = total_matches[index]
avg_corners = n_corners / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_corners'] = avg_corners
teams_stats.describe()
```
As we can see, there are a lot of Nan values on the avg_corners column. This represents teams that did not have corners data on this dataset. Instead of removing thoose rows, and give less input to our models we need again to do **mean imputation** and deal with these values.
```
teams_stats['avg_corners'].hist();
```
We can see that most avg_corners values fall within the 8.5 - 12 range, so let's fill in these entries with the average measured avg_corners
```
corners_avg_team_avg = teams_stats['avg_corners'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_corners'].isnull()), 'avg_corners'] = corners_avg_team_avg
# showing new values
teams_stats.describe()
```
Having done the **mean imputation** for team_attributes we can see that there are no longer missing values for the avg_corners.
```
teams_stats.loc[(teams_stats['avg_corners'].isnull())]
```
### Team Stats - Crosses
The final feature to be added is the crosses data. Normally the more dominant team has more crosses because it creates more opportunity of goals during a match. After merging all the data we need to watch for missing rows on the new added column.
```
crosses_data = pd.read_csv("../dataset/cross_detail.csv")
crosses = crosses_data[['match_id', 'team']]
total_crosses = crosses["team"].value_counts()
total_matches = crosses.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_crosses in total_crosses.iteritems():
n_matches = total_matches[index]
avg_crosses = n_crosses / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_crosses'] = avg_crosses
teams_stats.describe()
```
As we can see, there are a lot of Nan values on the avg_crosses column. This represents teams that did not have crosses data on this dataset. Instead of removing thoose rows, and give less input to our models we need again to do **mean imputation** and deal with these values
```
teams_stats['avg_crosses'].hist();
```
We can see that most avg_crosses values fall within the 12.5 - 17.5 range, so let's fill in these entries with the average measured avg_corners
```
crosses_avg_team_avg = teams_stats['avg_crosses'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_crosses'].isnull()), 'avg_crosses'] = crosses_avg_team_avg
# showing new values
teams_stats.describe()
```
Having done the **mean imputation** for team_attributes we can see that there are no longer missing values for the avg_crosses.
```
teams_stats.loc[(teams_stats['avg_crosses'].isnull())]
```
### FIFA data
We will now gather the fifa data regarding the overrall rating of the teams. This will create some columns that will include the overall ratings of the players that belong to a team. This way we can more easily compare the value of each team.
```
def get_fifa_stats(match, player_stats):
''' Aggregates fifa stats for a given match. '''
#Define variables
match_id = match.match_api_id
date = match['date']
players = ['home_player_1', 'home_player_2', 'home_player_3', "home_player_4", "home_player_5",
"home_player_6", "home_player_7", "home_player_8", "home_player_9", "home_player_10",
"home_player_11", "away_player_1", "away_player_2", "away_player_3", "away_player_4",
"away_player_5", "away_player_6", "away_player_7", "away_player_8", "away_player_9",
"away_player_10", "away_player_11"]
player_stats_new = pd.DataFrame()
names = []
#Loop through all players
for player in players:
#Get player ID
player_id = match[player]
#Get player stats
stats = player_stats[player_stats.player_api_id == player_id]
#Identify current stats
current_stats = stats[stats.date < date].sort_values(by = 'date', ascending = False)[:1]
if np.isnan(player_id) == True:
overall_rating = pd.Series(0)
else:
current_stats.reset_index(inplace = True, drop = True)
overall_rating = pd.Series(current_stats.loc[0, "overall_rating"])
#Rename stat
name = "{}_overall_rating".format(player)
names.append(name)
#Aggregate stats
player_stats_new = pd.concat([player_stats_new, overall_rating], axis = 1)
player_stats_new.columns = names
player_stats_new['match_api_id'] = match_id
player_stats_new.reset_index(inplace = True, drop = True)
#Return player stats
return player_stats_new.iloc[0]
def get_fifa_data(matches, player_stats, path = None, data_exists = False):
''' Gets fifa data for all matches. '''
#Check if fifa data already exists
if data_exists == True:
fifa_data = pd.read_pickle(path)
else:
print("Collecting fifa data for each match...")
start = time()
#Apply get_fifa_stats for each match
fifa_data = matches.apply(lambda x :get_fifa_stats(x, player_stats), axis = 1)
end = time()
print("Fifa data collected in {:.1f} minutes".format((end - start)/60))
#Return fifa_data
return fifa_data
fifa_data = get_fifa_data(viable_matches, player_attributes, None, data_exists = False)
fifa_data.describe()
```
### Joining all features
In this instance we need to join all features, select the input we will pass to our models and drop the column regarding the outcome label.
To improve the overall measures of the supervised learning models we need to normalize our features before training our models.
```
# Creates features and labels based on the provided data
viables = get_features(viable_matches, teams_stats, fifa_data, 10, False)
inputs = viables.drop('match_api_id', axis=1)
outcomes = inputs.loc[:, 'outcome']
# all features except outcomes
features = inputs.drop('outcome', axis=1)
features.iloc[:,:] = Normalizer(norm='l1').fit_transform(features)
features.head()
```
## Step 2: Classification & Results Interpretation
[go back to the top](#Table-of-contents)
### Training and Evaluating Models
We used **K-Fold Cross validation**. The idea is that we split the dataset, after processing it, in k bins of equal size to better estimate the skill of a model on unseen data. That is, to use a limited sample in order to estimate how the model is expected to perform in general when used to make predictions on data not used during the training of the model. It results in a less biased estimate of the model skill in comparision to the typical train_test_split.
```
from sklearn import metrics
def train_predict(clf, data, outcomes):
y_predict = train_model(clf, data, outcomes)
predict_metrics(outcomes, y_predict)
def train_predict_nn(clf, data, outcomes):
le = LabelEncoder()
y_outcomes = le.fit_transform(outcomes)
y_outcomes = np_utils.to_categorical(y_outcomes)
y_predict = train_model_nn(clf, data, y_outcomes)
y_predict_reverse = [np.argmax(y, axis=None, out=None) for y in y_predict]
y_predict_decoded = le.inverse_transform(y_predict_reverse)
predict_metrics(outcomes, y_predict_decoded)
def train_model(clf, data, labels):
kf = KFold(n_splits=5)
predictions = []
for train, test in kf.split(data):
X_train, X_test = data[data.index.isin(train)], data[data.index.isin(test)]
y_train, y_test = labels[data.index.isin(train)], labels[data.index.isin(test)]
clf.fit(X_train, y_train)
predictions.append(clf.predict(X_test))
y_predict = predictions[0]
y_predict = np.append(y_predict, predictions[1], axis=0)
y_predict = np.append(y_predict, predictions[2], axis=0)
y_predict = np.append(y_predict, predictions[3], axis=0)
y_predict = np.append(y_predict, predictions[4], axis=0)
return y_predict
def train_model_nn(clf, data, labels):
kf = KFold(n_splits=5, shuffle=False)
predictions = []
for train, test in kf.split(data):
X_train, X_test = data[data.index.isin(train)], data[data.index.isin(test)]
y_train, y_test = labels[data.index.isin(train)], labels[data.index.isin(test)]
clf.fit(X_train, y_train, epochs=20, verbose=0)
predictions.append(clf.predict(X_test))
y_predict = predictions[0]
y_predict = np.append(y_predict, predictions[1], axis=0)
y_predict = np.append(y_predict, predictions[2], axis=0)
y_predict = np.append(y_predict, predictions[3], axis=0)
y_predict = np.append(y_predict, predictions[4], axis=0)
return y_predict
def predict_metrics(y_test, y_predict):
ls = ['Win', 'Draw', 'Defeat']
cm = confusion_matrix(y_test, y_predict, ls, normalize='true')
cmap = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=ls)
fig, ax = plt.subplots(figsize=(8, 8))
cmap.plot(ax=ax, cmap='YlOrBr')
plt.show()
print(classification_report(y_test, y_predict, target_names=ls))
print("\n\nAccuracy: ", accuracy_score(y_test, y_predict))
print("Recall: ", recall_score(y_test, y_predict, average='macro'))
print("Precision: ", precision_score(y_test, y_predict, average='macro'))
print("F1 Score: ", f1_score(y_test, y_predict, average='macro'))
```
### The basis
* The **accuracy** measure is great for balanced classes, but because this is not the case we can't do much with it.
* **Precision-Recall** is a useful measure of success of prediction when the classes are very imbalanced.
* **Precision** is a measure of the ability of a classification model to identify only the relevant data points, ie, answers the question: what portion of **predicted Positives** is truly Positive?
* **Recall** is a measure of the ability of a model to find all the relevant cases within a dataset, ie, answers the question what portion of **actual Positives** is correctly classified?
* **F1-Score** is a combination of both recall and precision. Because precision and recall are often in tension, i.e, improving one will lead to a reduction in the other, this way the f1-score is a great measure also.
### KNN
```
clf = KNeighborsClassifier(n_neighbors=100)
train_predict(clf, features, outcomes)
```
### Decision Tree
```
clf = DecisionTreeClassifier(random_state=0, criterion='entropy', splitter='random', max_depth=5)
train_predict(clf, features, outcomes)
```
### SVC
```
clf = SVC(coef0=5, kernel='poly')
train_predict(clf, features, outcomes)
```
### Naive Bayes
```
clf = GaussianNB(var_smoothing=1.1)
train_predict(clf, features, outcomes)
```
### Gradient Boosting
```
clf = XGBClassifier(max_depth=20)
train_predict(clf, features, outcomes)
```
### Neural Network
```
visible = Input(shape=(features.shape[1],))
hidden = Dense(500, activation='relu')(visible)
output = Dense(3, activation='softmax')(hidden)
clf = Model(inputs=visible, outputs=output)
print(clf.summary())
from keras import metrics
from keras import losses
from keras import optimizers
clf.compile(optimizer=optimizers.Adam(),
loss=losses.CategoricalCrossentropy(),
metrics=[metrics.Precision(), metrics.Recall()])
train_predict_nn(clf, features, outcomes)
```
### Deep Neural Network
```
visible = Input(shape=(features.shape[1],))
hidden1 = Dense(500, activation='relu')(visible)
hidden2 = Dense(100, activation='relu')(hidden1)
hidden3 = Dense(50, activation='relu')(hidden2)
hidden4 = Dense(20, activation='relu')(hidden3)
output = Dense(3, activation='softmax')(hidden4)
clf = Model(inputs=visible, outputs=output)
print(clf.summary())
from keras import metrics
from keras import losses
from keras import optimizers
clf.compile(optimizer=optimizers.Adam(),
loss=losses.CategoricalCrossentropy(),
metrics=[metrics.Precision(), metrics.Recall()])
train_predict_nn(clf, features, outcomes)
```
## Conclusion
[go back to the top](#Table-of-contents)
### What did we learn
Regarding data analysis we learn so many things we did not knew before. Regarding data analysys and processment of data, we learned the importance of usability of the dataset and in case it is not perfect we bust fixed it.
After looking at our dataset we noticed some inconsistences and fixes by either removing some rows and **mean imputation** to allow the models to have multiple features.
Before fitting the models, we noticed also, the importance of splitting in a good manner the data or train and test and used **K-Fold Cross validation**.
Finally, the analysis of the models is very important... We cannot look only for the accuracy measure because many times it's misleading and to counter that, we must focus on other metrics to evaluate our models.
### Choosing best model
To evaluate the different models we need to choose a metric for comparison. The **accuracy** is only useful when we are dealing with a balanced dataset, which is not the case. Because of that, we need to consider only the other 3 possible metric, **recall**, **precison** or **f-measure**.
Since a predicton is associated with a cost in our case (given that to predict a match we have to spend money to bet on it), the most valuable measure is the precision. Then, our objective is to try to get the maximum percentage of **true positives** that are correctly classified by our models.
In other cases, such as for medical applications, the objective of a model is to maximize the recall. The f1-score is also an important measure to evaluate both the recall and the precision, when the 2 have the same importance.
Considering this, we think that both Naives Bayes and KNN were the most effective. Both of them got the most precision alongside with Gradient Boosting and SVC. The test were done thoroughly and exhaustive, with shuffling the data and measuring more than once obviously. This way, by running once **the results may vary** but not considerably.
### All in all
All in all, it was a great experience and very valuable. We noticed the interest of the professors to shift the lecture to do a more pratical work. In that way we can learn much more by ourselves, learn with our mistakes, become very interested in this topic and pursue it in future years.
## References
[go back to the top](#Table-of-contents)
* [Europen Soccer Database](https://www.kaggle.com/hugomathien/soccer)
* [Football Data Analysis](https://www.kaggle.com/pavanraj159/european-football-data-analysis#The-ultimate-Soccer-database-for-data-analysis-and-machine-learning)
* [Data Analysis and Machine Learning Projects](https://github.com/rhiever/Data-Analysis-and-Machine-Learning-Projects/blob/master/example-data-science-notebook/Example%20Machine%20Learning%20Notebook.ipynb)
* [Match Outcome Prediction in Football](https://www.kaggle.com/airback/match-outcome-prediction-in-football)
* [European Soccer Database Supplementary (XML Events to CSV)](https://www.kaggle.com/jiezi2004/soccer)
* [A deep learning framework for football match prediction](https://link.springer.com/article/10.1007/s42452-019-1821-5)
* [Predicting Football Match Outcome using Machine Learning: Football Match prediction using machine learning algorithms in jupyter notebook](https://github.com/prathameshtari/Predicting-Football-Match-Outcome-using-Machine-Learning)
* [(PDF) Football Result Prediction by Deep Learning Algorithms](https://www.researchgate.net/publication/334415630_Football_Result_Prediction_by_Deep_Learning_Algorithms)
* [Predicting Football Results Using Machine Learning Techniques](https://www.imperial.ac.uk/media/imperial-college/faculty-of-engineering/computing/public/1718-ug-projects/Corentin-Herbinet-Using-Machine-Learning-techniques-to-predict-the-outcome-of-profressional-football-matches.pdf)
* [A machine learning framework for sport result prediction](https://www.sciencedirect.com/science/article/pii/S2210832717301485)
|
github_jupyter
|
# Primary libraries
from time import time
import numpy as np
import pandas as pd
import sqlite3
import matplotlib.pyplot as plt
# Models
from sklearn.naive_bayes import GaussianNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from xgboost import XGBClassifier
# Neural Networks
from tensorflow import keras
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Flatten
from keras.layers import Input
from keras.models import Model
from keras.utils import np_utils
# Measures
from sklearn.preprocessing import Normalizer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.metrics import f1_score, accuracy_score, recall_score, confusion_matrix, precision_score, ConfusionMatrixDisplay, classification_report
from sklearn.model_selection import KFold
from sklearn.preprocessing import LabelEncoder
!pip install kaggle --quiet
!mkdir /root/.kaggle
!echo '{"username":"YOUR USERNAME","key":"YOUR API KEY"}' > /root/.kaggle/kaggle.json
!kaggle datasets download -d hugomathien/soccer -p dataset/
import zipfile
import os
os.chdir('dataset')
for file in os.listdir():
if file.endswith('.zip'):
zip_ref = zipfile.ZipFile(file, 'r')
zip_ref.extractall()
zip_ref.close()
with sqlite3.connect("database.sqlite") as con:
matches = pd.read_sql_query("SELECT * from Match", con)
team_attributes = pd.read_sql_query("SELECT distinct * from Team_Attributes",con)
player = pd.read_sql_query("SELECT * from Player",con)
player_attributes = pd.read_sql_query("SELECT * from Player_Attributes",con)
''' Derives a label for a given match. '''
def get_match_outcome(match):
#Define variables
home_goals = match['home_team_goal']
away_goals = match['away_team_goal']
outcome = pd.DataFrame()
outcome.loc[0,'match_api_id'] = match['match_api_id']
#Identify match outcome
if home_goals > away_goals:
outcome.loc[0,'outcome'] = "Win"
if home_goals == away_goals:
outcome.loc[0,'outcome'] = "Draw"
if home_goals < away_goals:
outcome.loc[0,'outcome'] = "Defeat"
#Return outcome
return outcome.loc[0]
''' Get the last x matches of a given team. '''
def get_last_matches(matches, date, team, x = 10):
#Filter team matches from matches
team_matches = matches[(matches['home_team_api_id'] == team) | (matches['away_team_api_id'] == team)]
#Filter x last matches from team matches
last_matches = team_matches[team_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:x,:]
#Return last matches
return last_matches
''' Get the last team stats of a given team. '''
def get_last_team_stats(team_id, date, team_stats):
#Filter team stats
all_team_stats = teams_stats[teams_stats['team_api_id'] == team_id]
#Filter last stats from team
last_team_stats = all_team_stats[all_team_stats.date < date].sort_values(by='date', ascending=False)
if last_team_stats.empty:
last_team_stats = all_team_stats[all_team_stats.date > date].sort_values(by='date', ascending=True)
#Return last matches
return last_team_stats.iloc[0:1,:]
''' Get the last x matches of two given teams. '''
def get_last_matches_against_eachother(matches, date, home_team, away_team, x = 10):
#Find matches of both teams
home_matches = matches[(matches['home_team_api_id'] == home_team) & (matches['away_team_api_id'] == away_team)]
away_matches = matches[(matches['home_team_api_id'] == away_team) & (matches['away_team_api_id'] == home_team)]
total_matches = pd.concat([home_matches, away_matches])
#Get last x matches
try:
last_matches = total_matches[total_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:x,:]
except:
last_matches = total_matches[total_matches.date < date].sort_values(by = 'date', ascending = False).iloc[0:total_matches.shape[0],:]
#Check for error in data
if(last_matches.shape[0] > x):
print("Error in obtaining matches")
#Return data
return last_matches
''' Get the goals[home & away] of a specfic team from a set of matches. '''
def get_goals(matches, team):
home_goals = int(matches.home_team_goal[matches.home_team_api_id == team].sum())
away_goals = int(matches.away_team_goal[matches.away_team_api_id == team].sum())
total_goals = home_goals + away_goals
return total_goals
''' Get the goals[home & away] conceided of a specfic team from a set of matches. '''
def get_goals_conceided(matches, team):
home_goals = int(matches.home_team_goal[matches.away_team_api_id == team].sum())
away_goals = int(matches.away_team_goal[matches.home_team_api_id == team].sum())
total_goals = home_goals + away_goals
return total_goals
''' Get the number of wins of a specfic team from a set of matches. '''
def get_wins(matches, team):
#Find home and away wins
home_wins = int(matches.home_team_goal[(matches.home_team_api_id == team) & (matches.home_team_goal > matches.away_team_goal)].count())
away_wins = int(matches.away_team_goal[(matches.away_team_api_id == team) & (matches.away_team_goal > matches.home_team_goal)].count())
total_wins = home_wins + away_wins
return total_wins
''' Create match specific features for a given match. '''
def get_match_features(match, matches, teams_stats, x = 10):
#Define variables
date = match.date
home_team = match.home_team_api_id
away_team = match.away_team_api_id
# Gets home and away team_stats
home_team_stats = get_last_team_stats(home_team, date, teams_stats);
away_team_stats = get_last_team_stats(away_team, date, teams_stats);
#Get last x matches of home and away team
matches_home_team = get_last_matches(matches, date, home_team, x = 5)
matches_away_team = get_last_matches(matches, date, away_team, x = 5)
#Get last x matches of both teams against each other
last_matches_against = get_last_matches_against_eachother(matches, date, home_team, away_team, x = 3)
#Create goal variables
home_goals = get_goals(matches_home_team, home_team)
away_goals = get_goals(matches_away_team, away_team)
home_goals_conceided = get_goals_conceided(matches_home_team, home_team)
away_goals_conceided = get_goals_conceided(matches_away_team, away_team)
#Define result data frame
result = pd.DataFrame()
#Define ID features
result.loc[0, 'match_api_id'] = match.match_api_id
result.loc[0, 'league_id'] = match.league_id
#Create match features and team stats
if(not home_team_stats.empty):
result.loc[0, 'home_team_buildUpPlaySpeed'] = home_team_stats['buildUpPlaySpeed'].values[0]
result.loc[0, 'home_team_buildUpPlayPassing'] = home_team_stats['buildUpPlayPassing'].values[0]
result.loc[0, 'home_team_chanceCreationPassing'] = home_team_stats['chanceCreationPassing'].values[0]
result.loc[0, 'home_team_chanceCreationCrossing'] = home_team_stats['chanceCreationCrossing'].values[0]
result.loc[0, 'home_team_chanceCreationShooting'] = home_team_stats['chanceCreationShooting'].values[0]
result.loc[0, 'home_team_defencePressure'] = home_team_stats['defencePressure'].values[0]
result.loc[0, 'home_team_defenceAggression'] = home_team_stats['defenceAggression'].values[0]
result.loc[0, 'home_team_defenceTeamWidth'] = home_team_stats['defenceTeamWidth'].values[0]
result.loc[0, 'home_team_avg_shots'] = home_team_stats['avg_shots'].values[0]
result.loc[0, 'home_team_avg_corners'] = home_team_stats['avg_corners'].values[0]
result.loc[0, 'home_team_avg_crosses'] = away_team_stats['avg_crosses'].values[0]
if(not away_team_stats.empty):
result.loc[0, 'away_team_buildUpPlaySpeed'] = away_team_stats['buildUpPlaySpeed'].values[0]
result.loc[0, 'away_team_buildUpPlayPassing'] = away_team_stats['buildUpPlayPassing'].values[0]
result.loc[0, 'away_team_chanceCreationPassing'] = away_team_stats['chanceCreationPassing'].values[0]
result.loc[0, 'away_team_chanceCreationCrossing'] = away_team_stats['chanceCreationCrossing'].values[0]
result.loc[0, 'away_team_chanceCreationShooting'] = away_team_stats['chanceCreationShooting'].values[0]
result.loc[0, 'away_team_defencePressure'] = away_team_stats['defencePressure'].values[0]
result.loc[0, 'away_team_defenceAggression'] = away_team_stats['defenceAggression'].values[0]
result.loc[0, 'away_team_defenceTeamWidth'] = away_team_stats['defenceTeamWidth'].values[0]
result.loc[0, 'away_team_avg_shots'] = away_team_stats['avg_shots'].values[0]
result.loc[0, 'away_team_avg_corners'] = away_team_stats['avg_corners'].values[0]
result.loc[0, 'away_team_avg_crosses'] = away_team_stats['avg_crosses'].values[0]
result.loc[0, 'home_team_goals_difference'] = home_goals - home_goals_conceided
result.loc[0, 'away_team_goals_difference'] = away_goals - away_goals_conceided
result.loc[0, 'games_won_home_team'] = get_wins(matches_home_team, home_team)
result.loc[0, 'games_won_away_team'] = get_wins(matches_away_team, away_team)
result.loc[0, 'games_against_won'] = get_wins(last_matches_against, home_team)
result.loc[0, 'games_against_lost'] = get_wins(last_matches_against, away_team)
result.loc[0, 'B365H'] = match.B365H
result.loc[0, 'B365D'] = match.B365D
result.loc[0, 'B365A'] = match.B365A
#Return match features
return result.loc[0]
''' Create and aggregate features and labels for all matches. '''
def get_features(matches, teams_stats, fifa, x = 10, get_overall = False):
#Get fifa stats features
fifa_stats = get_overall_fifa_rankings(fifa, get_overall)
#Get match features for all matches
match_stats = matches.apply(lambda i: get_match_features(i, matches, teams_stats, x = 10), axis = 1)
#Create dummies for league ID feature
dummies = pd.get_dummies(match_stats['league_id']).rename(columns = lambda x: 'League_' + str(x))
match_stats = pd.concat([match_stats, dummies], axis = 1)
match_stats.drop(['league_id'], inplace = True, axis = 1)
#Create match outcomes
outcomes = matches.apply(get_match_outcome, axis = 1)
#Merges features and outcomes into one frame
features = pd.merge(match_stats, fifa_stats, on = 'match_api_id', how = 'left')
features = pd.merge(features, outcomes, on = 'match_api_id', how = 'left')
#Drop NA values
features.dropna(inplace = True)
#Return preprocessed data
return features
def get_overall_fifa_rankings(fifa, get_overall = False):
''' Get overall fifa rankings from fifa data. '''
temp_data = fifa
#Check if only overall player stats are desired
if get_overall == True:
#Get overall stats
data = temp_data.loc[:,(fifa.columns.str.contains('overall_rating'))]
data.loc[:,'match_api_id'] = temp_data.loc[:,'match_api_id']
else:
#Get all stats except for stat date
cols = fifa.loc[:,(fifa.columns.str.contains('date_stat'))]
temp_data = fifa.drop(cols.columns, axis = 1)
data = temp_data
#Return data
return data
viable_matches = matches
viable_matches.describe()
viable_matches = matches.sample(n=5000)
b365 = viable_matches.dropna(subset=['B365H', 'B365D', 'B365A'],inplace=False)
b365.drop(['BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace = True, axis = 1)
bw = viable_matches.dropna(subset=['BWH', 'BWD', 'BWA'],inplace=False)
bw.drop(['B365H', 'B365D', 'B365A',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
iw = viable_matches.dropna(subset=['IWH', 'IWD', 'IWA'],inplace=False)
iw.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
lb = viable_matches.dropna(subset=['LBH', 'LBD', 'LBA'],inplace=False)
lb.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
ps = viable_matches.dropna(subset=['PSH', 'PSD', 'PSA'],inplace=False)
ps.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
wh = viable_matches.dropna(subset=['WHH', 'WHD', 'WHA'],inplace=False)
wh.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
sj = viable_matches.dropna(subset=['SJH', 'SJD', 'SJA'],inplace=False)
sj.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
vc = viable_matches.dropna(subset=['VCH', 'VCD', 'VCA'],inplace=False)
vc.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'GBH', 'GBD', 'GBA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
gb = viable_matches.dropna(subset=['GBH', 'GBD', 'GBA'],inplace=False)
gb.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'BSH', 'BSD', 'BSA'], inplace=True, axis = 1)
bs = viable_matches.dropna(subset=['BSH', 'BSD', 'BSA'],inplace=False)
bs.drop(['B365H', 'B365D', 'B365A',
'BWH', 'BWD', 'BWA',
'IWH', 'IWD', 'IWA',
'LBH', 'LBD', 'LBA',
'PSH', 'PSD', 'PSA',
'WHH', 'WHD', 'WHA',
'SJH', 'SJD', 'SJA',
'VCH', 'VCD', 'VCA',
'GBH', 'GBD', 'GBA'], inplace=True, axis = 1)
lis = [b365, bw, iw, lb, ps, wh, sj, vc, gb, bs]
viable_matches = max(lis, key = lambda datframe: datframe.shape[0])
viable_matches.describe()
teams_stats = team_attributes
viable_matches = viable_matches.dropna(inplace=False)
home_teams = viable_matches['home_team_api_id'].isin(teams_stats['team_api_id'].tolist())
away_teams = viable_matches['away_team_api_id'].isin(teams_stats['team_api_id'].tolist())
viable_matches = viable_matches[home_teams & away_teams]
viable_matches.describe()
teams_stats.describe()
teams_stats['buildUpPlayDribbling'].hist();
build_up_play_drib_avg = teams_stats['buildUpPlayDribbling'].mean()
# mean imputation
teams_stats.loc[(teams_stats['buildUpPlayDribbling'].isnull()), 'buildUpPlayDribbling'] = build_up_play_drib_avg
# showing new values
teams_stats.loc[teams_stats['buildUpPlayDribbling'] == build_up_play_drib_avg].head()
teams_stats.loc[(teams_stats['buildUpPlayDribbling'].isnull())]
teams_stats.drop(['buildUpPlaySpeedClass', 'buildUpPlayDribblingClass', 'buildUpPlayPassingClass',
'buildUpPlayPositioningClass', 'chanceCreationPassingClass', 'chanceCreationCrossingClass',
'chanceCreationShootingClass','chanceCreationPositioningClass','defencePressureClass', 'defenceAggressionClass',
'defenceTeamWidthClass','defenceDefenderLineClass'], inplace = True, axis = 1)
teams_stats.describe()
shots_off = pd.read_csv("shotoff_detail.csv")
shots_on = pd.read_csv("shoton_detail.csv")
shots = pd.concat([shots_off[['match_id', 'team']], shots_on[['match_id', 'team']]])
total_shots = shots["team"].value_counts()
total_matches = shots.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_shots in total_shots.iteritems():
n_matches = total_matches[index]
avg_shots = n_shots / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_shots'] = avg_shots
teams_stats.describe()
teams_stats['avg_shots'].hist();
shots_avg_team_avg = teams_stats['avg_shots'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_shots'].isnull()), 'avg_shots'] = shots_avg_team_avg
# showing new values
teams_stats.describe()
teams_stats.loc[(teams_stats['avg_shots'].isnull())]
# possessions read, cleanup and merge
possessions_data = pd.read_csv("../dataset/possession_detail.csv")
last_possessions = possessions_data.sort_values(['elapsed'], ascending=False).drop_duplicates(subset=['match_id'])
last_possessions = last_possessions[['match_id', 'homepos', 'awaypos']]
# get the ids of the home_team and away_team to be able to join with teams later
possessions = pd.DataFrame(columns=['team', 'possession', 'match'])
for index, row in last_possessions.iterrows():
match = matches.loc[matches['id'] == row['match_id'], ['home_team_api_id', 'away_team_api_id']]
if match.empty:
continue
hometeam = match['home_team_api_id'].values[0]
awayteam = match['away_team_api_id'].values[0]
possessions = possessions.append({'team': hometeam, 'possession': row['homepos'], 'match': row['match_id']}, ignore_index=True)
possessions = possessions.append({'team': awayteam, 'possession': row['awaypos'], 'match': row['match_id']}, ignore_index=True)
total_possessions = possessions.groupby(by=['team'])['possession'].sum()
total_matches = possessions.drop_duplicates(['team', 'match'])["team"].value_counts()
total_possessions.to_frame().describe()
corners_data = pd.read_csv("../dataset/corner_detail.csv")
corners = corners_data[['match_id', 'team']]
total_corners = corners["team"].value_counts()
total_matches = corners.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_corners in total_shots.iteritems():
n_matches = total_matches[index]
avg_corners = n_corners / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_corners'] = avg_corners
teams_stats.describe()
teams_stats['avg_corners'].hist();
corners_avg_team_avg = teams_stats['avg_corners'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_corners'].isnull()), 'avg_corners'] = corners_avg_team_avg
# showing new values
teams_stats.describe()
teams_stats.loc[(teams_stats['avg_corners'].isnull())]
crosses_data = pd.read_csv("../dataset/cross_detail.csv")
crosses = crosses_data[['match_id', 'team']]
total_crosses = crosses["team"].value_counts()
total_matches = crosses.drop_duplicates(['match_id', 'team'])["team"].value_counts()
for index, n_crosses in total_crosses.iteritems():
n_matches = total_matches[index]
avg_crosses = n_crosses / n_matches
teams_stats.loc[teams_stats['team_api_id'] == index, 'avg_crosses'] = avg_crosses
teams_stats.describe()
teams_stats['avg_crosses'].hist();
crosses_avg_team_avg = teams_stats['avg_crosses'].mean()
# mean imputation
teams_stats.loc[(teams_stats['avg_crosses'].isnull()), 'avg_crosses'] = crosses_avg_team_avg
# showing new values
teams_stats.describe()
teams_stats.loc[(teams_stats['avg_crosses'].isnull())]
def get_fifa_stats(match, player_stats):
''' Aggregates fifa stats for a given match. '''
#Define variables
match_id = match.match_api_id
date = match['date']
players = ['home_player_1', 'home_player_2', 'home_player_3', "home_player_4", "home_player_5",
"home_player_6", "home_player_7", "home_player_8", "home_player_9", "home_player_10",
"home_player_11", "away_player_1", "away_player_2", "away_player_3", "away_player_4",
"away_player_5", "away_player_6", "away_player_7", "away_player_8", "away_player_9",
"away_player_10", "away_player_11"]
player_stats_new = pd.DataFrame()
names = []
#Loop through all players
for player in players:
#Get player ID
player_id = match[player]
#Get player stats
stats = player_stats[player_stats.player_api_id == player_id]
#Identify current stats
current_stats = stats[stats.date < date].sort_values(by = 'date', ascending = False)[:1]
if np.isnan(player_id) == True:
overall_rating = pd.Series(0)
else:
current_stats.reset_index(inplace = True, drop = True)
overall_rating = pd.Series(current_stats.loc[0, "overall_rating"])
#Rename stat
name = "{}_overall_rating".format(player)
names.append(name)
#Aggregate stats
player_stats_new = pd.concat([player_stats_new, overall_rating], axis = 1)
player_stats_new.columns = names
player_stats_new['match_api_id'] = match_id
player_stats_new.reset_index(inplace = True, drop = True)
#Return player stats
return player_stats_new.iloc[0]
def get_fifa_data(matches, player_stats, path = None, data_exists = False):
''' Gets fifa data for all matches. '''
#Check if fifa data already exists
if data_exists == True:
fifa_data = pd.read_pickle(path)
else:
print("Collecting fifa data for each match...")
start = time()
#Apply get_fifa_stats for each match
fifa_data = matches.apply(lambda x :get_fifa_stats(x, player_stats), axis = 1)
end = time()
print("Fifa data collected in {:.1f} minutes".format((end - start)/60))
#Return fifa_data
return fifa_data
fifa_data = get_fifa_data(viable_matches, player_attributes, None, data_exists = False)
fifa_data.describe()
# Creates features and labels based on the provided data
viables = get_features(viable_matches, teams_stats, fifa_data, 10, False)
inputs = viables.drop('match_api_id', axis=1)
outcomes = inputs.loc[:, 'outcome']
# all features except outcomes
features = inputs.drop('outcome', axis=1)
features.iloc[:,:] = Normalizer(norm='l1').fit_transform(features)
features.head()
from sklearn import metrics
def train_predict(clf, data, outcomes):
y_predict = train_model(clf, data, outcomes)
predict_metrics(outcomes, y_predict)
def train_predict_nn(clf, data, outcomes):
le = LabelEncoder()
y_outcomes = le.fit_transform(outcomes)
y_outcomes = np_utils.to_categorical(y_outcomes)
y_predict = train_model_nn(clf, data, y_outcomes)
y_predict_reverse = [np.argmax(y, axis=None, out=None) for y in y_predict]
y_predict_decoded = le.inverse_transform(y_predict_reverse)
predict_metrics(outcomes, y_predict_decoded)
def train_model(clf, data, labels):
kf = KFold(n_splits=5)
predictions = []
for train, test in kf.split(data):
X_train, X_test = data[data.index.isin(train)], data[data.index.isin(test)]
y_train, y_test = labels[data.index.isin(train)], labels[data.index.isin(test)]
clf.fit(X_train, y_train)
predictions.append(clf.predict(X_test))
y_predict = predictions[0]
y_predict = np.append(y_predict, predictions[1], axis=0)
y_predict = np.append(y_predict, predictions[2], axis=0)
y_predict = np.append(y_predict, predictions[3], axis=0)
y_predict = np.append(y_predict, predictions[4], axis=0)
return y_predict
def train_model_nn(clf, data, labels):
kf = KFold(n_splits=5, shuffle=False)
predictions = []
for train, test in kf.split(data):
X_train, X_test = data[data.index.isin(train)], data[data.index.isin(test)]
y_train, y_test = labels[data.index.isin(train)], labels[data.index.isin(test)]
clf.fit(X_train, y_train, epochs=20, verbose=0)
predictions.append(clf.predict(X_test))
y_predict = predictions[0]
y_predict = np.append(y_predict, predictions[1], axis=0)
y_predict = np.append(y_predict, predictions[2], axis=0)
y_predict = np.append(y_predict, predictions[3], axis=0)
y_predict = np.append(y_predict, predictions[4], axis=0)
return y_predict
def predict_metrics(y_test, y_predict):
ls = ['Win', 'Draw', 'Defeat']
cm = confusion_matrix(y_test, y_predict, ls, normalize='true')
cmap = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=ls)
fig, ax = plt.subplots(figsize=(8, 8))
cmap.plot(ax=ax, cmap='YlOrBr')
plt.show()
print(classification_report(y_test, y_predict, target_names=ls))
print("\n\nAccuracy: ", accuracy_score(y_test, y_predict))
print("Recall: ", recall_score(y_test, y_predict, average='macro'))
print("Precision: ", precision_score(y_test, y_predict, average='macro'))
print("F1 Score: ", f1_score(y_test, y_predict, average='macro'))
clf = KNeighborsClassifier(n_neighbors=100)
train_predict(clf, features, outcomes)
clf = DecisionTreeClassifier(random_state=0, criterion='entropy', splitter='random', max_depth=5)
train_predict(clf, features, outcomes)
clf = SVC(coef0=5, kernel='poly')
train_predict(clf, features, outcomes)
clf = GaussianNB(var_smoothing=1.1)
train_predict(clf, features, outcomes)
clf = XGBClassifier(max_depth=20)
train_predict(clf, features, outcomes)
visible = Input(shape=(features.shape[1],))
hidden = Dense(500, activation='relu')(visible)
output = Dense(3, activation='softmax')(hidden)
clf = Model(inputs=visible, outputs=output)
print(clf.summary())
from keras import metrics
from keras import losses
from keras import optimizers
clf.compile(optimizer=optimizers.Adam(),
loss=losses.CategoricalCrossentropy(),
metrics=[metrics.Precision(), metrics.Recall()])
train_predict_nn(clf, features, outcomes)
visible = Input(shape=(features.shape[1],))
hidden1 = Dense(500, activation='relu')(visible)
hidden2 = Dense(100, activation='relu')(hidden1)
hidden3 = Dense(50, activation='relu')(hidden2)
hidden4 = Dense(20, activation='relu')(hidden3)
output = Dense(3, activation='softmax')(hidden4)
clf = Model(inputs=visible, outputs=output)
print(clf.summary())
from keras import metrics
from keras import losses
from keras import optimizers
clf.compile(optimizer=optimizers.Adam(),
loss=losses.CategoricalCrossentropy(),
metrics=[metrics.Precision(), metrics.Recall()])
train_predict_nn(clf, features, outcomes)
| 0.577257 | 0.990301 |
Copyright 2022 Google LLC
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
# Visualize models training/validation loss
## Prepare colab
```
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1" # no need to use gpu
# bazel build -c opt --copt=-mavx2 //kws_streaming/colab:colab_notebook.par
# ./bazel-bin/kws_streaming/colab/colab_notebook.par
```
## Imports
```
import tensorflow as tf
import os
import numpy as np
import math
import os.path
import sys
import matplotlib.pyplot as plt
tf = tf.compat.v1
```
## Utils
```
def read_log(fname, tag="accuracy"):
eval_acc = []
eval_loss = []
try:
for event in tf.train.summary_iterator(fname):
for value in event.summary.value:
if value.tag == tag:
eval_acc.append(value.simple_value)
except:
print("stop parsing " + fname)
return eval_acc, eval_loss
def scan_log(log_path):
acc = []
found = False
if os.path.isdir(log_path):
for file in os.listdir(log_path):
if file.endswith(".gtc"):
validation_file = os.path.join(log_path, file)
acc = []
if os.path.isfile(validation_file):
if found:
raise ValueError("found duplicate log")
acc, _ = read_log(validation_file)
found = True
return acc
def train_eval_log(model_path):
log_path_train = os.path.join(model_path, "logs", "train")
acc_train = scan_log(log_path_train)
log_path_val = os.path.join(model_path, "logs", "validation")
acc_val = scan_log(log_path_val)
return acc_train, acc_val
```
## Read logs
```
# set path to trained models
model_path1 = "/tmp/kws_streaming/models2/att_mh_rnn/"
model_path2 = "/tmp/kws_streaming/models2/tc_resnet/"
acc_train1, acc_val1 = train_eval_log(model_path1)
acc_train2, acc_val2 = train_eval_log(model_path2)
```
## Plot logs and compare models training/validation loss
```
# smoothing filter
window = np.hanning(70)
window = window/window.sum()
plt.figure(figsize=(20, 10))
data_val1 = np.convolve(window, acc_val1, mode='valid')
data_tr1 = np.convolve(window, acc_train1, mode='valid')
data_val2 = np.convolve(window, acc_val2, mode='valid')
data_tr2 = np.convolve(window, acc_train2, mode='valid')
ratio1 = (float)(len(data_tr1)) / len(data_val1)
ratio2 = (float)(len(data_tr2)) / len(data_val2)
xx1 = ratio1 * np.arange(len(data_val1))
plt_val1, = plt.plot(xx1, data_val1, label='val1')
plt_train1, = plt.plot(data_tr1, label='train1')
xx2 = ratio2 * np.arange(len(data_val2))
plt_val2, = plt.plot(xx2, data_val2, label='val2')
plt_train2, = plt.plot(data_tr2, label='train2')
plt.ylim(0.6, 1.0)
plt.legend(handles=[plt_val1, plt_train1, plt_val2, plt_train2])
plt.figure(figsize=(20, 10))
plt_val1, = plt.plot(xx1, data_val1, label='val1')
plt_val2, = plt.plot(xx2, data_val2, label='val2')
plt.ylim(0.90, 1.0)
plt.legend(handles=[plt_val1, plt_val2])
plt.show()
```
|
github_jupyter
|
import os
os.environ["CUDA_VISIBLE_DEVICES"] = "-1" # no need to use gpu
# bazel build -c opt --copt=-mavx2 //kws_streaming/colab:colab_notebook.par
# ./bazel-bin/kws_streaming/colab/colab_notebook.par
import tensorflow as tf
import os
import numpy as np
import math
import os.path
import sys
import matplotlib.pyplot as plt
tf = tf.compat.v1
def read_log(fname, tag="accuracy"):
eval_acc = []
eval_loss = []
try:
for event in tf.train.summary_iterator(fname):
for value in event.summary.value:
if value.tag == tag:
eval_acc.append(value.simple_value)
except:
print("stop parsing " + fname)
return eval_acc, eval_loss
def scan_log(log_path):
acc = []
found = False
if os.path.isdir(log_path):
for file in os.listdir(log_path):
if file.endswith(".gtc"):
validation_file = os.path.join(log_path, file)
acc = []
if os.path.isfile(validation_file):
if found:
raise ValueError("found duplicate log")
acc, _ = read_log(validation_file)
found = True
return acc
def train_eval_log(model_path):
log_path_train = os.path.join(model_path, "logs", "train")
acc_train = scan_log(log_path_train)
log_path_val = os.path.join(model_path, "logs", "validation")
acc_val = scan_log(log_path_val)
return acc_train, acc_val
# set path to trained models
model_path1 = "/tmp/kws_streaming/models2/att_mh_rnn/"
model_path2 = "/tmp/kws_streaming/models2/tc_resnet/"
acc_train1, acc_val1 = train_eval_log(model_path1)
acc_train2, acc_val2 = train_eval_log(model_path2)
# smoothing filter
window = np.hanning(70)
window = window/window.sum()
plt.figure(figsize=(20, 10))
data_val1 = np.convolve(window, acc_val1, mode='valid')
data_tr1 = np.convolve(window, acc_train1, mode='valid')
data_val2 = np.convolve(window, acc_val2, mode='valid')
data_tr2 = np.convolve(window, acc_train2, mode='valid')
ratio1 = (float)(len(data_tr1)) / len(data_val1)
ratio2 = (float)(len(data_tr2)) / len(data_val2)
xx1 = ratio1 * np.arange(len(data_val1))
plt_val1, = plt.plot(xx1, data_val1, label='val1')
plt_train1, = plt.plot(data_tr1, label='train1')
xx2 = ratio2 * np.arange(len(data_val2))
plt_val2, = plt.plot(xx2, data_val2, label='val2')
plt_train2, = plt.plot(data_tr2, label='train2')
plt.ylim(0.6, 1.0)
plt.legend(handles=[plt_val1, plt_train1, plt_val2, plt_train2])
plt.figure(figsize=(20, 10))
plt_val1, = plt.plot(xx1, data_val1, label='val1')
plt_val2, = plt.plot(xx2, data_val2, label='val2')
plt.ylim(0.90, 1.0)
plt.legend(handles=[plt_val1, plt_val2])
plt.show()
| 0.338405 | 0.700639 |
```
import os
import random
import numpy as np
import scipy.stats
import seaborn as sbn
import torch
from sklearn.datasets import load_boston
from prior_elicitation.models.folded_logistic_glm import FoldedLogisticGLM
from prior_elicitation.models.linear_glm import LinearGLM
#parameters
MODEL: str = "folded-logistic"
COLUMN: str = "intercept"
PERCENTILE: int = 25
PRIOR_INFO: dict = {
'AGE': {'dist': 'norm', 'loc': 0, 'scale': 1},
'B': {'dist': 'norm', 'loc': 0, 'scale': 1},
'CHAS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'CRIM': {'dist': 'norm', 'loc': 0, 'scale': 1},
'DIS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'INDUS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'LSTAT': {'dist': 'norm', 'loc': 0, 'scale': 1},
'NOX': {'dist': 'norm', 'loc': 0, 'scale': 1},
'PTRATIO': {'dist': 'norm', 'loc': 0, 'scale': 1},
'RAD': {'dist': 'norm', 'loc': 0, 'scale': 1},
'RM': {'dist': 'norm', 'loc': 0, 'scale': 1},
'TAX': {'dist': 'norm', 'loc': 0, 'scale': 1},
'ZN': {'dist': 'norm', 'loc': 0, 'scale': 1},
'intercept': {'dist': 'norm', 'loc': 0, 'scale': 1},
'scale': {'c': 0, 'dist': 'foldnorm', 'loc': 10, 'scale': 1}}
NUM_PRIOR_SIM: int = 100 # Establish a number of simulations from the prior
SEED: int = 29 # What's the random seed for reproducibility
PLOT_PATH: str = (
# "../reports/figures/_001/25th_percentile_intercept_times_outcome.png"
"../reports/figures/test_plot.png"
)
SAVE: bool = True
# Parameters
MODEL = "folded-logistic"
COLUMN = "intercept"
PERCENTILE = 25
PRIOR_INFO = {
"AGE": {"dist": "norm", "loc": 0, "scale": 1},
"B": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"CHAS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"CRIM": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"DIS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"INDUS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"LSTAT": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"NOX": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"PTRATIO": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"RAD": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"RM": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"TAX": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"ZN": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"intercept": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"scale": {"dist": "foldnorm", "loc": 1.0, "scale": 0.01, "c": 0},
"Age": {"dist": "norm", "loc": 0.0, "scale": 5.0},
}
NUM_PRIOR_SIM = 100
SEED = 129
PLOT_PATH = (
"/Users/timothyb0912/Documents/prior_elicitation/reports/figures/test_plot.png"
)
SAVE = True
```
# Load and transform data
```
data_container = load_boston()
raw_design = data_container["data"]
training_num_obs = raw_design.shape[0]
training_design_np = np.concatenate(
(np.ones((training_num_obs, 1)), raw_design), axis=1
)
training_outcomes_np = data_container["target"].ravel()
training_design_column_names = (
["intercept"] + [col for col in data_container["feature_names"]]
)
parameter_names = training_design_column_names + ["scale"]
```
# Load models
```
training_design_torch = torch.from_numpy(training_design_np).double()
training_outcomes_torch = torch.from_numpy(training_outcomes_np).double()
linear_model = LinearGLM.from_input(training_design_torch)
folded_logistic_model = FoldedLogisticGLM.from_input(training_design_torch)
with torch.no_grad():
linear_preds = linear_model(training_design_torch)
folded_preds = folded_logistic_model(training_design_torch)
```
# Sample parameters from the prior distribution
```
# Choose the model to simulate from
model_dict = {"folded-logistic": folded_logistic_model, "linear": linear_model}
current_model = model_dict[MODEL]
# Set a random seed for reproducility
np.random.seed(SEED)
torch.manual_seed(SEED)
# Simulate parameters from the prior
prior_sim_parameters = np.empty((len(PRIOR_INFO), NUM_PRIOR_SIM), dtype=float)
for pos, key in enumerate(parameter_names):
if PRIOR_INFO[key]["dist"] == "norm":
prior_sim_parameters[pos, :] = scipy.stats.norm.rvs(
loc = PRIOR_INFO[key]["loc"],
scale = PRIOR_INFO[key]["scale"],
size = NUM_PRIOR_SIM,
)
elif PRIOR_INFO[key]["dist"] == "foldnorm":
prior_sim_parameters[pos, :] = scipy.stats.foldnorm.rvs(
c = PRIOR_INFO[key]["c"],
loc = PRIOR_INFO[key]["loc"],
scale = PRIOR_INFO[key]["scale"],
size=NUM_PRIOR_SIM,
)
print(prior_sim_parameters.shape)
```
# Sample outcomes from the prior distribution
```
# Draw from the prior predictive distribution
prior_sim_outcomes = np.empty((training_design_np.shape[0], NUM_PRIOR_SIM), dtype=float)
with torch.no_grad():
for i in range(NUM_PRIOR_SIM):
current_params = prior_sim_parameters[:, i]
current_model.set_params_numpy(current_params)
prior_sim_outcomes[:, i] = current_model.simulate(
training_design_torch, num_sim=1,
).ravel()
```
# Create desired plots
```
def percentile_closure(percentile):
def _calc_25th_percentile(array_like):
return np.percentile(array_like, percentile, axis=0)
return _calc_25th_percentile
def make_percentile_plot(
calc_percentile_func, design, obs_outcomes, sim_outcomes, col_names, save
):
column_idx = col_names.index(COLUMN)
sim_product = sim_outcomes * design[:, column_idx][:, None]
obs_product = obs_outcomes * design[:, column_idx]
percentiles_of_sim_product = calc_percentile_func(sim_product)
percentiles_of_obs_product = calc_percentile_func(obs_product)
p_value = (percentiles_of_sim_product > percentiles_of_obs_product).mean()
sim_label = "Simulated"
obs_label = "Observed = {:.1f}\nP-value = {:.0%}".format(
percentiles_of_obs_product, p_value
)
plot = sbn.distplot(
percentiles_of_sim_product, kde=False, hist=True, label=sim_label
)
ymin, ymax = plot.get_ylim()
plot.vlines(
percentiles_of_obs_product,
ymin,
ymax,
label=obs_label,
linestyle="dashed",
color="black",
)
max_obs_product = obs_product.max()
min_obs_product = obs_product.min()
plot.vlines(
obs_product.min(),
ymin,
ymax,
label=f"Minimum Outcome * {COLUMN} = {min_obs_product}",
linestyle="dotted",
color="black",
)
plot.vlines(
obs_product.max(),
ymin,
ymax,
label=f"Maximum Outcome * {COLUMN} = {max_obs_product}",
linestyle="dashdot",
color="black",
)
plot.legend(loc="best")
xlabel = (
"{}-Percentile of [Median Home Value * {}]".format(PERCENTILE, COLUMN)
)
plot.set_xlabel(xlabel)
plot.set_ylabel("Count", rotation=0, labelpad=40)
plot.set_title(MODEL)
plot.figure.set_size_inches(10, 6)
sbn.despine()
plot.figure.tight_layout()
if save:
plot.figure.savefig(PLOT_PATH, dpi=500, bbox_inches="tight")
return plot
calc_percentile = percentile_closure(PERCENTILE)
percentile_plot = make_percentile_plot(
calc_percentile,
training_design_np,
training_outcomes_np,
prior_sim_outcomes,
training_design_column_names,
SAVE,
)
```
# To-Do:
Add complete compendium of desired prior predictive plots.
```
# Make desired prior predictive plots
```
|
github_jupyter
|
import os
import random
import numpy as np
import scipy.stats
import seaborn as sbn
import torch
from sklearn.datasets import load_boston
from prior_elicitation.models.folded_logistic_glm import FoldedLogisticGLM
from prior_elicitation.models.linear_glm import LinearGLM
#parameters
MODEL: str = "folded-logistic"
COLUMN: str = "intercept"
PERCENTILE: int = 25
PRIOR_INFO: dict = {
'AGE': {'dist': 'norm', 'loc': 0, 'scale': 1},
'B': {'dist': 'norm', 'loc': 0, 'scale': 1},
'CHAS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'CRIM': {'dist': 'norm', 'loc': 0, 'scale': 1},
'DIS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'INDUS': {'dist': 'norm', 'loc': 0, 'scale': 1},
'LSTAT': {'dist': 'norm', 'loc': 0, 'scale': 1},
'NOX': {'dist': 'norm', 'loc': 0, 'scale': 1},
'PTRATIO': {'dist': 'norm', 'loc': 0, 'scale': 1},
'RAD': {'dist': 'norm', 'loc': 0, 'scale': 1},
'RM': {'dist': 'norm', 'loc': 0, 'scale': 1},
'TAX': {'dist': 'norm', 'loc': 0, 'scale': 1},
'ZN': {'dist': 'norm', 'loc': 0, 'scale': 1},
'intercept': {'dist': 'norm', 'loc': 0, 'scale': 1},
'scale': {'c': 0, 'dist': 'foldnorm', 'loc': 10, 'scale': 1}}
NUM_PRIOR_SIM: int = 100 # Establish a number of simulations from the prior
SEED: int = 29 # What's the random seed for reproducibility
PLOT_PATH: str = (
# "../reports/figures/_001/25th_percentile_intercept_times_outcome.png"
"../reports/figures/test_plot.png"
)
SAVE: bool = True
# Parameters
MODEL = "folded-logistic"
COLUMN = "intercept"
PERCENTILE = 25
PRIOR_INFO = {
"AGE": {"dist": "norm", "loc": 0, "scale": 1},
"B": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"CHAS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"CRIM": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"DIS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"INDUS": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"LSTAT": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"NOX": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"PTRATIO": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"RAD": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"RM": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"TAX": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"ZN": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"intercept": {"dist": "norm", "loc": 0.0, "scale": 5.0},
"scale": {"dist": "foldnorm", "loc": 1.0, "scale": 0.01, "c": 0},
"Age": {"dist": "norm", "loc": 0.0, "scale": 5.0},
}
NUM_PRIOR_SIM = 100
SEED = 129
PLOT_PATH = (
"/Users/timothyb0912/Documents/prior_elicitation/reports/figures/test_plot.png"
)
SAVE = True
data_container = load_boston()
raw_design = data_container["data"]
training_num_obs = raw_design.shape[0]
training_design_np = np.concatenate(
(np.ones((training_num_obs, 1)), raw_design), axis=1
)
training_outcomes_np = data_container["target"].ravel()
training_design_column_names = (
["intercept"] + [col for col in data_container["feature_names"]]
)
parameter_names = training_design_column_names + ["scale"]
training_design_torch = torch.from_numpy(training_design_np).double()
training_outcomes_torch = torch.from_numpy(training_outcomes_np).double()
linear_model = LinearGLM.from_input(training_design_torch)
folded_logistic_model = FoldedLogisticGLM.from_input(training_design_torch)
with torch.no_grad():
linear_preds = linear_model(training_design_torch)
folded_preds = folded_logistic_model(training_design_torch)
# Choose the model to simulate from
model_dict = {"folded-logistic": folded_logistic_model, "linear": linear_model}
current_model = model_dict[MODEL]
# Set a random seed for reproducility
np.random.seed(SEED)
torch.manual_seed(SEED)
# Simulate parameters from the prior
prior_sim_parameters = np.empty((len(PRIOR_INFO), NUM_PRIOR_SIM), dtype=float)
for pos, key in enumerate(parameter_names):
if PRIOR_INFO[key]["dist"] == "norm":
prior_sim_parameters[pos, :] = scipy.stats.norm.rvs(
loc = PRIOR_INFO[key]["loc"],
scale = PRIOR_INFO[key]["scale"],
size = NUM_PRIOR_SIM,
)
elif PRIOR_INFO[key]["dist"] == "foldnorm":
prior_sim_parameters[pos, :] = scipy.stats.foldnorm.rvs(
c = PRIOR_INFO[key]["c"],
loc = PRIOR_INFO[key]["loc"],
scale = PRIOR_INFO[key]["scale"],
size=NUM_PRIOR_SIM,
)
print(prior_sim_parameters.shape)
# Draw from the prior predictive distribution
prior_sim_outcomes = np.empty((training_design_np.shape[0], NUM_PRIOR_SIM), dtype=float)
with torch.no_grad():
for i in range(NUM_PRIOR_SIM):
current_params = prior_sim_parameters[:, i]
current_model.set_params_numpy(current_params)
prior_sim_outcomes[:, i] = current_model.simulate(
training_design_torch, num_sim=1,
).ravel()
def percentile_closure(percentile):
def _calc_25th_percentile(array_like):
return np.percentile(array_like, percentile, axis=0)
return _calc_25th_percentile
def make_percentile_plot(
calc_percentile_func, design, obs_outcomes, sim_outcomes, col_names, save
):
column_idx = col_names.index(COLUMN)
sim_product = sim_outcomes * design[:, column_idx][:, None]
obs_product = obs_outcomes * design[:, column_idx]
percentiles_of_sim_product = calc_percentile_func(sim_product)
percentiles_of_obs_product = calc_percentile_func(obs_product)
p_value = (percentiles_of_sim_product > percentiles_of_obs_product).mean()
sim_label = "Simulated"
obs_label = "Observed = {:.1f}\nP-value = {:.0%}".format(
percentiles_of_obs_product, p_value
)
plot = sbn.distplot(
percentiles_of_sim_product, kde=False, hist=True, label=sim_label
)
ymin, ymax = plot.get_ylim()
plot.vlines(
percentiles_of_obs_product,
ymin,
ymax,
label=obs_label,
linestyle="dashed",
color="black",
)
max_obs_product = obs_product.max()
min_obs_product = obs_product.min()
plot.vlines(
obs_product.min(),
ymin,
ymax,
label=f"Minimum Outcome * {COLUMN} = {min_obs_product}",
linestyle="dotted",
color="black",
)
plot.vlines(
obs_product.max(),
ymin,
ymax,
label=f"Maximum Outcome * {COLUMN} = {max_obs_product}",
linestyle="dashdot",
color="black",
)
plot.legend(loc="best")
xlabel = (
"{}-Percentile of [Median Home Value * {}]".format(PERCENTILE, COLUMN)
)
plot.set_xlabel(xlabel)
plot.set_ylabel("Count", rotation=0, labelpad=40)
plot.set_title(MODEL)
plot.figure.set_size_inches(10, 6)
sbn.despine()
plot.figure.tight_layout()
if save:
plot.figure.savefig(PLOT_PATH, dpi=500, bbox_inches="tight")
return plot
calc_percentile = percentile_closure(PERCENTILE)
percentile_plot = make_percentile_plot(
calc_percentile,
training_design_np,
training_outcomes_np,
prior_sim_outcomes,
training_design_column_names,
SAVE,
)
# Make desired prior predictive plots
| 0.613931 | 0.637708 |
# 2A.eco - Web Scrapping et API avec Pocket
Le notebook revient sur le webscrapping et l'utilisation d'API avec [pocket](https://getpocket.com/).
```
from jyquickhelper import add_notebook_menu
add_notebook_menu()
```
## Objectifs des prochaines séances
Connaissez-vous l'application [Pocket](https://getpocket.com/) ? C'est une application qui simplifie le bookmarking. Elle prend la forme d'une extension Chrome / Firefox. Quand on tombe sur un site intéressant, on peut le bookmaker, et ajouter, ou non, des tags pour "qualifier" le contenu.
Cette application répond au besoin de conserver le contenu web pertinent et de le classer.
Au cours des prochaines séances, nous allons construire un outil de machine learning qui :
- se connecte à un compte pocket
- récupère les sites bookmarqués et les tags éventuels
- à partir des articles taggés, prédit les meilleurs tags des sites non-taggés
- tag les articles non taggés
Bref, nous allons concevoir un programme de classification automatique des articles !
## Objectif de la séance
- Créer un compte Pocket
- S'authentifier auprès de l'API
- Populer le compte avec des données via l'API
- Récupérer les données via l'API
- Scraper les sites bookmarqués pour enrichir les données
## Mais c'est quoi une API ?
On vous explique tout ici :
- [Définition](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_eco_les_API.html#definition)
- [Les API qui existent](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_eco_les_API.html#les-api-qui-existent)
- [Comment parler à une API ?](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_eco_les_API.html#comment-parler-a-une-api)
Donc un API est une interface permettant de _communiquer_ avec une application. En général, on veut récupérer des données. Donc la communication consiste à envoyer une requete HTTP (le plus souvent GET ou POST) et à récupérer des données, souvent au format json. Ici nous voulons récupérer des données d'un compte utilisateur pocket.
Pour savoir comment on communique précisément avec l'API de pocket, il n'y a pas de secret : il faut lire la doc de ceux qui l'ont codée. On vous a simplifié un peu les étapes ci-après.
## Création d'un compte Pocket
Créer un compte sur https://getpocket.com/signup?ep=4. Il n'y a pas de vérification d'email, donc vous pouvez mettre un faux mail.
Aller sur la _console developer_ de pocket: https://getpocket.com/developer/apps/index.php
<img src="./images/console_developer_pocket.png" width="450"/>
Cliquer sur CREATE AN APPLICATION
Compléter le formulaire comme suit (vous pouvez changer le nom de l'application et la description)
<img src="./images/screen_consumer_key.png" width="450"/>
Cliquer sur CREATE APPLICATION
```
# keyring pour récupérer un mot de passe
import keyring
import os
CONSUMER_KEY = keyring.get_password("web", os.environ["COMPUTERNAME"] + "pocket")
# input pour le demander
# CONSUMER_KEY = input("Insérer ici le CONSUMER KEY de la plateform WEB ")
```
<img src="./images/screen_consumer_key2.png" width="450"/>
Vous en aurez besoin pour vous connecter à l'API de pocket.
## Authentification
D'abord il faut s'[authentifier](https://getpocket.com/developer/docs/authentication)
Protocole utilisé ici : [OAUTH2](https://tools.ietf.org/html/rfc6749) (très classique).
<img src="./images/screen_oauth2.png"/>
6 étapes donc avant d'avoir le droit de récupérer les données. De temps en temps, il existe une librairie python. C'est notre cas : https://github.com/tapanpandita/pocket. On va s'en servir pour s'authentifier. Mais pas pour récupérer les données (elle n'est plus à jour pour faire ça).
### Etape 1 : Obtenir un code d'authorisation => get_request_token
```
import pocket
from pocket import Pocket
REDIRECT_URI = "http://localhost:8888/notebooks/API%20Pocket.ipynb"
# c'est l'url à laquelle vous allez rediriger l'utilisateur (ici, vous) après que pocket a authentifié l'utilisateur (vous)
REQUEST_TOKEN = Pocket.get_request_token(consumer_key=CONSUMER_KEY, redirect_uri=REDIRECT_URI)
# print(REQUEST_TOKEN)
```
### Etape 2: Authoriser l'accès
Il faut le faire à chaque exécution du notebook.
```
# Enlever les guillemets autour de REQUEST_TOKEN
url = "https://getpocket.com/auth/authorize?request_token={0}&redirect_uri={1}".format("REQUEST_TOKEN", REDIRECT_URI)
print("Aller à l'url : \n" + url)
```
<img src="./images/screen_authorization_pocket.png" width="450"/>
Cliquer sur Autoriser.
### Etape 3: Récupérer le token d'accès
```
try:
USER_CREDENTIALS = Pocket.get_credentials(consumer_key=CONSUMER_KEY, code=REQUEST_TOKEN)
except Exception as e:
print(e)
# print(USER_CREDENTIALS)
ACCESS_TOKEN = USER_CREDENTIALS['access_token']
# print(ACCESS_TOKEN)
```
## Chargement de données sur le nouveau compte
Comme vous venez de créer un compte, vous n'avez pas encore d'articles sauvegardés. On vous a préparé un peu moins de 500 articles (format json). Un tiers de ces articles sont taggés (catégorisés). Un article peut comprendre un ou plusieurs tags.
On vous rappelle que l'objectif à termes sera de prédire les meilleurs tags pour les articles non taggés, étant donnés les mots qui caractérisent ces articles (titre, résumé, et ensemble des mots présents dans le html de la page).
### Chargement du fichier json en mémoire
```
import json
with open('./images/data_pocket.json') as fp:
data = json.load(fp)
```
On affiche le premier élément.
```
from pprint import pprint
keys = list(data.keys())
pprint(data[keys[0]])
```
### Exercice 1
Chargement des données du json dans le compte nouvellement créé.
Pour communiquer avec une API, il faut envoyer des requêtes HTTP. Ici on veut ajouter les données du fichier json ("given_url" et "tags") dans le compte Pocket.
Que nous dit la doc ? Consulter https://getpocket.com/developer/docs/v3/add.
Par exemple, pour l'ajout d'un seul item, la doc nous donne l'url à laquelle il faut envoyer une requête (https://getpocket.com/v3/add), et la méthode qu'il faut employer. Ici, il s'agit d'une méthode [POST](https://en.wikipedia.org/wiki/POST_(HTTP)).
Pour envoyer une requête en python, il y a plusieurs solutions. Un des plus simples consiste à utiliser la librairie [requests](http://docs.python-requests.org/en/master/user/quickstart/#make-a-request).
A vous de jouer ! Commencez par un seul (par exemple http://docs.python-requests.org/en/master/user/quickstart/ avec les tags "python, requests"), puis si cela a fonctionné, vous pouvez passer 100 items (pas plus, le maximum de requêtes autorisées par l'API est de 500 par heure et par utilisateur).
Pour voir si cela a marché, il suffit d'aller sur votre compte Pocket :)
## Récupération des données disponibles dans l'API
### Exercice 2
Récupérer les urls et les tags des items qui contiennent le tag "python".
C'est par ici : https://getpocket.com/developer/docs/v3/retrieve. A vous de jouer !
### Exercice 3
Récupérer les urls et les titres des items qui contiennent le mot "python" dans le titre ou l'url
Dans le fichier qui servira à la catégorisation automatique, nous allons avoir besoin : de l'url, du titre, de l'extrait, des tags et du contenu.
L'api ne permet pas d'accéder au contenu des sites épinglés. Mais nous pouvons le récupérer grâce à l'url.
### Exercice 4
Constituer d'abord un DataFrame avec les champs resolved_url, resolved_title, excerpt, et les tags des items comprenant le terme python.
## Compléter les données avec du webscraping
### Mais c'est quoi le webscraping ?
On vous explique tout ici :
- [Définition](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_Eco_Web_Scraping.html#a-eco-web-scraping)
- [Un détour par le Web : comment fonctionne un site ?](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_Eco_Web_Scraping.html#un-detour-par-le-web-comment-fonctionne-un-site)
- [Scrapper avec python](http://www.xavierdupre.fr/app/ensae_teaching_cs/helpsphinx/notebooks/TD2A_Eco_Web_Scraping.html#scrapper-avec-python)
### Beautiful Soup
Reprenons notre liste de sites sur python et analysons le contenu HTML.
```
from bs4 import BeautifulSoup
from pprint import pprint
#première url de la liste
if 'df' in locals():
url = df['url'].iloc[0]
print(url)
else:
url = "http://www.xavierdupre.fr"
# récupération du contenu (même librairie que pour les requetes api =>
#requests est une librairie qui permet de faire des requetes http, que cela soit pour des api ou du webscraping)
import requests
r = requests.get(url)
#pprint : librairie pour "pretty print" (essayer sans : on voit pas grand chose)
text = r.text if len(r.text) < 1000 else r.text[:1000] + "\n..."
pprint(text)
# on stock le contenu html dans la variable html
html = r.text
# on "parse" le html grâce à la librairie beautiful soup
soup = BeautifulSoup(html, "html5lib")
# c'est plus "joli" encore que pprint et surtout,
# il y a plein de méthodes pour extraire les informations que l'on souhaite
soup
type(soup)
```
_soup_ est une instance de la classe BeautifulSoup (que l'on a importé de la librairie bs4). C'est une représentation du code html avec des méthodes pratiques, telles que __find__. __find__ permet de trouver la première occurence d'une balise html qu'on lui passe. Par exemple le 1er lien de la page :
```
soup.find("a")
```
Si on veut tous les liens, il faut utiliser __findAll__. Cela renvoie une objet qui se comporte comme une liste. On peut itérer dessus, ou facilement le transformer en objet "list" (en faisant list())
```
print(type(soup.findAll("a")))
soup.findAll("a")
```
On peut sélectionner des tags qui ont certains attributs css. Par exemple ici, on peut vouloir seulement les liens internes du site. Ils ont la classe "internal". Les autres "external".
```
list(soup.findAll("a", {"class": "internal"}))
```
Ce n'est pas toujours comme ça, la plupart du temps, il faudra la forme de la valeur de l'attribut href. les liens externes pourront etre identifiés parce qu'ils sont absolus (on indique l'ensemble du lien comme href="https://docs.python.org/3/reference/expressions.html etc. et non pas un lien relatif comme href="../c_exception/exception.html ('..' signifie "le répertoire parent).
On peut lister toutes les balises h1 et h2 de cette page, en une ligne.
```
list(soup.findAll({"h1", "h2"}))
```
Naviguer vers les enfants
```
h1 = soup.find("h1")
child = h1.a if h1 else None
child if child else "vide"
```
Retrouver le parent
```
child.parent
```
Récupérer tous les enfants
```
children = soup.find("h1").findAll("a")
children
```
Récupérer les attributs des éléments html
```
[c.attrs for c in children]
```
Accéder à la valeur d'un attribut en particulier
```
[c.attrs['href'] for c in children]
```
Jusqu'ici, on passe systématiquement par une balise html. Comment faire si on veut récupérer les élements qui ont une class css, quelle que soit la balise html ?
```
internals = soup.findAll("", {"class": "internal"})
internals
```
On a presque fini de passer en revue la librairie. Il reste la notion de "sibling", pratique pour parcourir un tableau. nextSibling : permet de récupérer l'élément html suivant et "de même niveau" dans l'arbre (le prochain frère), previousSibling, le précédent. findChildren permet de trouver tous les enfants.
Enfin la méthode "get_text()" pour récupérer le contenu des balises html. A appliquer en dernier ! En effet, une fois appliquer, on perd toute trace à l'arbre html. SI vous voulez aller plus loin, la doc est bien faite : https://www.crummy.com/software/BeautifulSoup/bs4/doc/
### Exercice 5
Récupérer le contenu des code snippets de la page : http://www.xavierdupre.fr/app/teachpyx/helpsphinx/c_lang/types.html
### Exercice 6
Récupérer la 3ème colonne (intitulée "exemples") du tableau des opérateurs : http://www.xavierdupre.fr/app/teachpyx/helpsphinx/c_lang/types.html
Astuce : explorer les CSS selectors : https://www.crummy.com/software/BeautifulSoup/bs4/doc/.
### Exercice 7
Récupérer le contenu des titres (h1, h2, h3, etc.) et des balise p de l'ensemble des liens de notre liste et compter le nombre d'occurences du mot python, et la porportion que cela représente dans l'ensemble des mots
|
github_jupyter
|
from jyquickhelper import add_notebook_menu
add_notebook_menu()
# keyring pour récupérer un mot de passe
import keyring
import os
CONSUMER_KEY = keyring.get_password("web", os.environ["COMPUTERNAME"] + "pocket")
# input pour le demander
# CONSUMER_KEY = input("Insérer ici le CONSUMER KEY de la plateform WEB ")
import pocket
from pocket import Pocket
REDIRECT_URI = "http://localhost:8888/notebooks/API%20Pocket.ipynb"
# c'est l'url à laquelle vous allez rediriger l'utilisateur (ici, vous) après que pocket a authentifié l'utilisateur (vous)
REQUEST_TOKEN = Pocket.get_request_token(consumer_key=CONSUMER_KEY, redirect_uri=REDIRECT_URI)
# print(REQUEST_TOKEN)
# Enlever les guillemets autour de REQUEST_TOKEN
url = "https://getpocket.com/auth/authorize?request_token={0}&redirect_uri={1}".format("REQUEST_TOKEN", REDIRECT_URI)
print("Aller à l'url : \n" + url)
try:
USER_CREDENTIALS = Pocket.get_credentials(consumer_key=CONSUMER_KEY, code=REQUEST_TOKEN)
except Exception as e:
print(e)
# print(USER_CREDENTIALS)
ACCESS_TOKEN = USER_CREDENTIALS['access_token']
# print(ACCESS_TOKEN)
import json
with open('./images/data_pocket.json') as fp:
data = json.load(fp)
from pprint import pprint
keys = list(data.keys())
pprint(data[keys[0]])
from bs4 import BeautifulSoup
from pprint import pprint
#première url de la liste
if 'df' in locals():
url = df['url'].iloc[0]
print(url)
else:
url = "http://www.xavierdupre.fr"
# récupération du contenu (même librairie que pour les requetes api =>
#requests est une librairie qui permet de faire des requetes http, que cela soit pour des api ou du webscraping)
import requests
r = requests.get(url)
#pprint : librairie pour "pretty print" (essayer sans : on voit pas grand chose)
text = r.text if len(r.text) < 1000 else r.text[:1000] + "\n..."
pprint(text)
# on stock le contenu html dans la variable html
html = r.text
# on "parse" le html grâce à la librairie beautiful soup
soup = BeautifulSoup(html, "html5lib")
# c'est plus "joli" encore que pprint et surtout,
# il y a plein de méthodes pour extraire les informations que l'on souhaite
soup
type(soup)
soup.find("a")
print(type(soup.findAll("a")))
soup.findAll("a")
list(soup.findAll("a", {"class": "internal"}))
list(soup.findAll({"h1", "h2"}))
h1 = soup.find("h1")
child = h1.a if h1 else None
child if child else "vide"
child.parent
children = soup.find("h1").findAll("a")
children
[c.attrs for c in children]
[c.attrs['href'] for c in children]
internals = soup.findAll("", {"class": "internal"})
internals
| 0.085792 | 0.886273 |
```
from keras.datasets import mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
import numpy
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Dropout
from keras.utils import np_utils
# flatten 28*28 images to a 784 vector for each image
num_pixels = X_train.shape[1] * X_train.shape[2]
X_train = X_train.reshape(X_train.shape[0], num_pixels).astype('float32')
X_test = X_test.reshape(X_test.shape[0], num_pixels).astype('float32')
# one hot encode outputs
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
num_classes = y_test.shape[1]
from keras.callbacks import TensorBoard
from time import time
model = Sequential()
model.add(Dense(1000, input_dim=784, activation='sigmoid'))
model.add(Dense(10, activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy'])
tensorboard=TensorBoard(log_dir='logs/tensor9')
model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=10, batch_size=1024, verbose=2,callbacks=[tensorboard])
from google.datalab.ml import TensorBoard as tb
tb.start('logs/tensor9')
from keras import backend as K
K.clear_session()
from keras.callbacks import TensorBoard
from time import time
model = Sequential()
model.add(Dense(num_pixels, input_dim=784, kernel_initializer='normal',activation='sigmoid',name='input_layer1'))
model.add(Dense(1000, activation='relu',name='hidden_layer1'))
model.add(Dense(10, activation='softmax',name='output_layer1'))
model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy'])
tensorboard=TensorBoard(log_dir='logs/tensor10',histogram_freq=1,batch_size=10000)
model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=5, batch_size=1024, verbose=2,callbacks=[tensorboard])
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
%matplotlib inline
from tensorflow.examples.tutorials.mnist import input_data
data = input_data.read_data_sets('data/MNIST/', one_hot=True)
data.test.cls = np.argmax(data.test.labels, axis=1)
data.train.cls = np.argmax(data.train.labels, axis=1)
train_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x2": np.array(data.train.images)},
y=np.array(data.train.cls),
num_epochs=None,
batch_size=1024,
shuffle=True)
test_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x2": np.array(data.test.images)},
y=np.array(data.test.cls),
num_epochs=1,
shuffle=False)
feature_x = tf.feature_column.numeric_column("x2", shape=(784))
feature_columns = [feature_x]
feature_columns
def model_fn(features, labels, mode, params):
x = features["x2"]
net = tf.layers.dense(inputs=x, name='h1',units=512, activation=tf.nn.relu)
net2 = tf.layers.dense(inputs=net, name='h2',units=256, activation=tf.nn.relu)
net3 = tf.layers.dense(inputs=net2, name='h3',units=128, activation=tf.nn.relu)
net4 = tf.layers.dense(inputs=net3, name='softmax',units=10,activation=tf.nn.softmax)
y_pred_cls = tf.argmax(net4, axis=1)
if mode == tf.estimator.ModeKeys.PREDICT:
spec = tf.estimator.EstimatorSpec(mode=mode,predictions=y_pred_cls)
else:
cross_entropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=labels,logits = net4)
loss = tf.reduce_mean(cross_entropy)
optimizer = tf.train.ProximalAdagradOptimizer(learning_rate=params["learning_rate"],l1_regularization_strength=0.001)
train_op = optimizer.minimize(loss=loss, global_step=tf.train.get_global_step())
accuracy = tf.metrics.accuracy(labels, y_pred_cls)
metrics = {'accuracy': accuracy}
tf.summary.scalar('train_accuracy', accuracy[1])
tf.summary.histogram("hidden1",net)
tf.summary.histogram("hidden2",net2)
tf.summary.histogram("hidden3",net3)
spec = tf.estimator.EstimatorSpec(mode=mode,loss=loss,train_op=train_op,eval_metric_ops=metrics)
return spec
params = {"learning_rate": 0.1}
model = tf.estimator.Estimator(model_fn=model_fn,
params=params,model_dir='/content/datalab/docs/log11/')
model.train(input_fn=train_input_fn, steps=1000)
from google.datalab.ml import TensorBoard as tb
tb.start('/content/datalab/docs/log11/')
num_hidden_units = [512, 256, 128]
model = tf.estimator.DNNClassifier(feature_columns=feature_columns,
hidden_units=num_hidden_units,
activation_fn=tf.nn.relu,
n_classes=num_classes,
model_dir='/content/datalab/docs/log4')
model.train(input_fn=train_input_fn, steps=2000)
from google.datalab.ml import TensorBoard as tb
tb.start('/content/datalab/docs/log4/')
```
|
github_jupyter
|
from keras.datasets import mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
import numpy
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Dropout
from keras.utils import np_utils
# flatten 28*28 images to a 784 vector for each image
num_pixels = X_train.shape[1] * X_train.shape[2]
X_train = X_train.reshape(X_train.shape[0], num_pixels).astype('float32')
X_test = X_test.reshape(X_test.shape[0], num_pixels).astype('float32')
# one hot encode outputs
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
num_classes = y_test.shape[1]
from keras.callbacks import TensorBoard
from time import time
model = Sequential()
model.add(Dense(1000, input_dim=784, activation='sigmoid'))
model.add(Dense(10, activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy'])
tensorboard=TensorBoard(log_dir='logs/tensor9')
model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=10, batch_size=1024, verbose=2,callbacks=[tensorboard])
from google.datalab.ml import TensorBoard as tb
tb.start('logs/tensor9')
from keras import backend as K
K.clear_session()
from keras.callbacks import TensorBoard
from time import time
model = Sequential()
model.add(Dense(num_pixels, input_dim=784, kernel_initializer='normal',activation='sigmoid',name='input_layer1'))
model.add(Dense(1000, activation='relu',name='hidden_layer1'))
model.add(Dense(10, activation='softmax',name='output_layer1'))
model.compile(loss='categorical_crossentropy', optimizer='sgd', metrics=['accuracy'])
tensorboard=TensorBoard(log_dir='logs/tensor10',histogram_freq=1,batch_size=10000)
model.fit(X_train, y_train, validation_data=(X_test, y_test), epochs=5, batch_size=1024, verbose=2,callbacks=[tensorboard])
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
%matplotlib inline
from tensorflow.examples.tutorials.mnist import input_data
data = input_data.read_data_sets('data/MNIST/', one_hot=True)
data.test.cls = np.argmax(data.test.labels, axis=1)
data.train.cls = np.argmax(data.train.labels, axis=1)
train_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x2": np.array(data.train.images)},
y=np.array(data.train.cls),
num_epochs=None,
batch_size=1024,
shuffle=True)
test_input_fn = tf.estimator.inputs.numpy_input_fn(
x={"x2": np.array(data.test.images)},
y=np.array(data.test.cls),
num_epochs=1,
shuffle=False)
feature_x = tf.feature_column.numeric_column("x2", shape=(784))
feature_columns = [feature_x]
feature_columns
def model_fn(features, labels, mode, params):
x = features["x2"]
net = tf.layers.dense(inputs=x, name='h1',units=512, activation=tf.nn.relu)
net2 = tf.layers.dense(inputs=net, name='h2',units=256, activation=tf.nn.relu)
net3 = tf.layers.dense(inputs=net2, name='h3',units=128, activation=tf.nn.relu)
net4 = tf.layers.dense(inputs=net3, name='softmax',units=10,activation=tf.nn.softmax)
y_pred_cls = tf.argmax(net4, axis=1)
if mode == tf.estimator.ModeKeys.PREDICT:
spec = tf.estimator.EstimatorSpec(mode=mode,predictions=y_pred_cls)
else:
cross_entropy = tf.nn.sparse_softmax_cross_entropy_with_logits(labels=labels,logits = net4)
loss = tf.reduce_mean(cross_entropy)
optimizer = tf.train.ProximalAdagradOptimizer(learning_rate=params["learning_rate"],l1_regularization_strength=0.001)
train_op = optimizer.minimize(loss=loss, global_step=tf.train.get_global_step())
accuracy = tf.metrics.accuracy(labels, y_pred_cls)
metrics = {'accuracy': accuracy}
tf.summary.scalar('train_accuracy', accuracy[1])
tf.summary.histogram("hidden1",net)
tf.summary.histogram("hidden2",net2)
tf.summary.histogram("hidden3",net3)
spec = tf.estimator.EstimatorSpec(mode=mode,loss=loss,train_op=train_op,eval_metric_ops=metrics)
return spec
params = {"learning_rate": 0.1}
model = tf.estimator.Estimator(model_fn=model_fn,
params=params,model_dir='/content/datalab/docs/log11/')
model.train(input_fn=train_input_fn, steps=1000)
from google.datalab.ml import TensorBoard as tb
tb.start('/content/datalab/docs/log11/')
num_hidden_units = [512, 256, 128]
model = tf.estimator.DNNClassifier(feature_columns=feature_columns,
hidden_units=num_hidden_units,
activation_fn=tf.nn.relu,
n_classes=num_classes,
model_dir='/content/datalab/docs/log4')
model.train(input_fn=train_input_fn, steps=2000)
from google.datalab.ml import TensorBoard as tb
tb.start('/content/datalab/docs/log4/')
| 0.89187 | 0.64848 |
# Introduction to Pandas
- prepared by [Rita Colaço](https://www.cpr.ku.dk/staff/?id=621366&vis=medarbejder)
Pandas is a library for data analysis and its powertool is the **DataFrame**.
Pandas is well suited for many different kinds of data:
- Tabular data with heterogeneously-typed columns, as in an Excel spreadsheet
- Ordered and unordered time series data.
- Any other form of observational / statistical data sets. The data actually need not be labeled at all to be placed into a pandas data structure
## Learning goals
- Concept of
- vectors (1d-arrays) as series
- tables as data frames
- Organization of a table: index, columns
- Filtering and slicing
- Data types a Pandas dataframe can handle
- Applying statistics and grouping
- Modifying a table
## What is a DataFrame?
A DataFrame is basically, a **Table** of data (or a tabular data structure) with labeled rows and columns. The rows are labeled by a special data structure called an Index, taht permits fast look-up and powerful relational operations.
For example:
| (index) | Name | Age | Height | LikesIceCream |
| :---: | :--: | :--: | :--: | :--: |
| 0 | "Nick" | 22 | 3.4 | True |
| 1 | "Jenn" | 55 | 1.2 | True |
| 2 | "Joe" | 25 | 2.2 | True |
## Importing Pandas library
```
import pandas as pd
```
## Create a DataFrame directly
### From a List of Lists
```
data = [
[2.23, 1, "test"],
[3.45, 2, "train"],
[4.5, 3, "test"],
[6.0, 4, "train"]
]
df = pd.DataFrame(data, columns=['A', 'B', 'C'])
df
```
### From a List of Dicts
```
data = [
{'A':2.23, 'B':1, 'C':"test"},
{'A':3.45, 'B':2, 'C':"train"},
{'A':4.5, 'B':3, 'C':"test"},
{'A':6.0, 'B':4, 'C':"train"}
]
df = pd.DataFrame(data)
df
```
### From a Dict of Lists
```
df = pd.DataFrame({
'A': [2.23, 3.45, 4.5, 6.0],
'B': [1, 2, 3, 4],
'C': ["test", "train", "test", "train"]
})
df
```
### From an empty DataFrame
```
df = pd.DataFrame()
df['A'] = [2.23, 3.45, 4.5, 6.0]
df['B'] = [1, 2, 3, 4]
df['C'] = ["test", "train", "test", "train"]
df
```
### Exercise 1
Please recreate the table below as a Dataframe using one of the approaches detailed above:
| Year | Product | Cost |
| :--: | :----: | :--: |
| 2015 | Apples | 0.35 |
| 2016 | Apples | 0.45 |
| 2015 | Bananas | 0.75 |
| 2016 | Bananas | 1.10 |
Which approach did you prefer? Why?
## Making DataFrames from a Data File
Pandas has functions that can make DataFrames from a wide variety of file types. To do this, use one of the functions in Pandas that start with "read_". Here is a non-exclusive list of examples:
| File Type | Function Name |
| :----: | :---: |
| Excel | `pd.read_excel()` |
| CSV, TSV | `pd.read_csv()` |
| H5, HDF, HDF5 | `pd.read_hdf()` |
| JSON | `pd.read_json()` |
| SQL | `pd.read_sql_table()` |
### Loading the Data
```
# url = 'https://raw.githubusercontent.com/mwaskom/seaborn-data/master/titanic.csv'
# df = pd.read_csv(url)
url_ecdc_daily_cases = "https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv"
df = pd.read_csv(url_ecdc_daily_cases, parse_dates=True, infer_datetime_format=True)
df.head()
```
## Examining the Dataset
Sometimes, we might just want to quickly inspect the DataFrame:
### Attributes
```python
df.shape # Shape of the object (2D)
df.dtypes # Data types in each column
df.index # Index range
df.columns # Column names
```
### Methods
```python
df.describe() # Descriptive statistics of columns
df.info() # DataFrame information
```
### Shape
```
df.shape
```
### Data types
```
df.dtypes
```
### Index and Columns
```
df.index
df.columns
```
## Selecting Data
Pandas has a lot of flexibility in the number of syntaxes it supports. For example, to select columns in a DataFrame:
```python
df['Column1']
df.Column1 # no whitespaces possible!
```
Multiple Columns can also be selected by providing a list:
```python
df[['Column1', 'Column2']]
```
Rows are selected with the **iloc** and **loc** attributes:
```python
df.iloc[5] # Used to get the "integer" index of the row.
df.loc['Row5'] # Used if rows are named.
```
However, with large DataFrames, we often just want to see the first or last rows, or even just a sample of the rows.
| Method | Description |
| --- | --- |
| `df.head(5)` | the first 5 rows |
| `df.tail(5)` | the last 5 rows |
| `df.sample(5)` | a random 5 rows |
### Exercise 2
Please open the file `titanic.csv` (using `pd.read_csv`) and use it to answer the following questions about the rdataset. If you reach the end of the exercises, explore the dataset and DataFrames more and see what you can find about it!
Display the first 5 lines of the dataset.
Show the last 3 lines of the "alive" column.
Check 3 random lines of the dataset
Make a new dataframe containing just the "survived", "sex", and "age" columns
Make a new dataframe containing just the 10th, 15th and 16th lines of the dataset
## Query/Filtering Data
To get rows based on their value, Pandas supports both Numpy's logical indexing:
```python
select_rows = df[df['Column1'] > 0]
```
and an SQL-like query string:
```python
df.query('Colummn1 > 0')
```
One can also filter based on multiple conditions, using the element-wise ("bit-wise") logical operators **&** data intersection, or **|** for the data union.
```python
select_rows = df[(df['Column1'] > 0) & (df['Column2'] > 2)]
```
```python
select_rows = df[(df['Column1'] > 0) | (df['Column2'] > 2)]
```
### Exercise 3
Using the Titanic dataset, let's do some data querying exercises.
What is ticket fare for the 1st class? The 2nd? The 3rd?
```
pd.Series([True, True]) & pd.Series([False, False])
```
Did the oldest passenger on the Titanic survive?
Was the youngest passenger on the Titanic alone?
How many passengers on the Titanic embarked from Cherbourg?
How much money did the Titanic make from passengers from Southampton? From Cherbourg? From Queenstown?
Considering only those passengers older than 22, were there more Males travelling alone from Southampton or Females in Third class from Cherbourg?
## Summarizing/Statistics in DataFrames
Pandas' Series and DataFrames are iterables, and can be given to any function that expects a list or Numpy Array, which allows them to be useful to many different libraries' functions. For example, to compute basic statistics:
```python
df['Column1'].count()
df['Column1'].max()
df['Column2'][df['Column1'] == 'string'].sum()
```
You can also use the "pipe" method to call a function on the rows or columns of a DataFrame:
```python
df['Column1'].pipe(np.mean)
```
### Exercise 4
What is the mean ticket fare that the passengers paid on the titanic? And the median?
How many passengers does this dataset contain?
What class ticket did the 10th (index = 9) passenger in this dataset buy?
What proportion of the passengers were alone on the titanic?
How many different classes were on the titanic?
How many men and women are in this dataset? (value_counts())
How many passengers are sitting in each class?
## Transforming/Modifying Data
Any transformation function can be performed on each element of a column or on the entire DataFrame. For example:
```python
df['Column1'] * 5
np.sqrt(df['Column1'])
df['Column1'].str.upper()
del df['B']
df['Column1'] = [3, 9. 27, 81] # Replace the entire column with other values (length must match)
```
### Exercise 5
Get everyone's age if they were still alive today (hint: Titanic sunk in 1912)
Make the class name title-cased (the first letter capitalized)
Make a columns called "not_survived", the opposite of the "survived" column
## GroupBy Operations
In most of our tasks, getting single metrics from a dataset is not enough, and we often actually want to compare metrics between groups or conditions.
The **groupby()** method essentially splits the data into different groups depending on a variable of your choice, and allows you to apply summary functions on each group. For example, if you wanted to calculate the mean temperature by month from a given data frame:
```python
df.groupby('month').temperature.mean()
```
where "month" and "temperature" are column names from the data frame.
You can also group by multiple columns, by providing a list of column names:
```python
df.groupby(['year', 'month']).temperature.mean()
```
The **groupby()** function returns a GroupBy object, where the **.groups** variable is a dictionary whose keys are the computed unique groups.
Groupby objects are **lazy**, meaning they don't start calculating anything until they know the full pipeline. This approach is called the **"Split-Apply-Combine"** workflow. You can get more info on it [here](https://pandas.pydata.org/pandas-docs/stable/user_guide/groupby.html).
### Exercise 6
Let's try this out on the Titanic dataset.
What was the median ticket fare for each class?
What was the survival rate for each class?
What was the survival rate for each sex?
What was the survival rate, broken down by both sex and class?
Which class tended to travel alone more often? Did it matter where they were embarking from?
What was the ticket fare for each embarking city?
What was the median age of the survivors vs non-survivors, when sex is considered as a factor?
# For Track 2 (or not, what do you think?)
## GroupBy Operations: Multiple Statistics per Group
Another piece of syntax we are going to look at, is the **agg()** function for Pandas. The aggregation functionality provided by this function allows multiple statistics to be calculated per group in one calculation.
The instructions to the function **agg()** are provided in the form of a dictionary, where the keys specify the columns upon which to apply the operations, and the value specify the function to run:
```python
df.groupby(['year', 'month']).agg({'duration':sum,
'network_type':'count',
'date':'first'})
```
You can also apply multiple functions to one column in groups:
```python
df.groupby(['year', 'month']).agg({'duration':[min, max, sum],
'network_type':'count',
'date':[min, 'first', 'nunique']})
```
### Exercise 7
Now, let's try to apply it to our Titanic dataset, and answer the following questions.
How many man, women and childern survived, and what was their average age?
```
# df.groupby('who').agg({'survived':sum,
# 'age':"mean"})
```
How many males and females, embarking on different towns, were alive? And how many of those were alone?
```
# df.groupby(['sex', 'class', 'embark_town']).agg({'alive':"count",
# "alone":sum})
```
## Handling Missing Values
Missing values are often a concern in data science, for example in proteomics, and can be indicated with a **`None`** or **`NaN`** (np.nan in Numpy). Pandas DataFrames have several methods for detecting, removing and replacing these values:
| method | description
| ---: | :---- |
**`isna()`** | Returns True for each NaN |
**`notna()`** | Returns False for each NaN |
**`dropna()`** | Returns just the rows without any NaNs |
### Exercise 8
What proportion of the "deck" column is missing data?
How many rows don't contain any missing data at all?
Make a dataframe with only the rows containing no missing data.
## Imputation
Imputation means replacing the missing values with real values.
| method | description |
| ----: | :---- |
| **`fillna()`** | Replaces the NaNs with values (provides lots of options) |
| **`ffill()`** | Replaces the Nans with the previous non-NaN value (equivalent to df.fillna(method='ffill') |
| **`bfill()`** | Replaces the Nans with the following non-NaN value (equivalent to df.fillna(method='bfill') |
| **`interpolate()`** | interpolates nans with previous and following values |
### Exercise 9
Using the following DataFrame, solve the exercises below.
```
data = pd.DataFrame({'time': [0.5, 1., 1.5, None, 2.5, 3., 3.5, None], 'value': [6, 4, 5, 8, None, 10, 11, None]})
data
```
Replace all the missing "value" rows with zeros.
Replace the missing "time" rows with the previous value.
Replace all of the missing values with the data from the next row. What do you notice when you do this with this dataset?
Linearly interpolate the missing data. What is the result for this dataset?
|
github_jupyter
|
import pandas as pd
data = [
[2.23, 1, "test"],
[3.45, 2, "train"],
[4.5, 3, "test"],
[6.0, 4, "train"]
]
df = pd.DataFrame(data, columns=['A', 'B', 'C'])
df
data = [
{'A':2.23, 'B':1, 'C':"test"},
{'A':3.45, 'B':2, 'C':"train"},
{'A':4.5, 'B':3, 'C':"test"},
{'A':6.0, 'B':4, 'C':"train"}
]
df = pd.DataFrame(data)
df
df = pd.DataFrame({
'A': [2.23, 3.45, 4.5, 6.0],
'B': [1, 2, 3, 4],
'C': ["test", "train", "test", "train"]
})
df
df = pd.DataFrame()
df['A'] = [2.23, 3.45, 4.5, 6.0]
df['B'] = [1, 2, 3, 4]
df['C'] = ["test", "train", "test", "train"]
df
# url = 'https://raw.githubusercontent.com/mwaskom/seaborn-data/master/titanic.csv'
# df = pd.read_csv(url)
url_ecdc_daily_cases = "https://opendata.ecdc.europa.eu/covid19/casedistribution/csv/data.csv"
df = pd.read_csv(url_ecdc_daily_cases, parse_dates=True, infer_datetime_format=True)
df.head()
df.shape # Shape of the object (2D)
df.dtypes # Data types in each column
df.index # Index range
df.columns # Column names
df.describe() # Descriptive statistics of columns
df.info() # DataFrame information
df.shape
df.dtypes
df.index
df.columns
df['Column1']
df.Column1 # no whitespaces possible!
df[['Column1', 'Column2']]
df.iloc[5] # Used to get the "integer" index of the row.
df.loc['Row5'] # Used if rows are named.
select_rows = df[df['Column1'] > 0]
df.query('Colummn1 > 0')
select_rows = df[(df['Column1'] > 0) & (df['Column2'] > 2)]
select_rows = df[(df['Column1'] > 0) | (df['Column2'] > 2)]
pd.Series([True, True]) & pd.Series([False, False])
df['Column1'].count()
df['Column1'].max()
df['Column2'][df['Column1'] == 'string'].sum()
df['Column1'].pipe(np.mean)
df['Column1'] * 5
np.sqrt(df['Column1'])
df['Column1'].str.upper()
del df['B']
df['Column1'] = [3, 9. 27, 81] # Replace the entire column with other values (length must match)
df.groupby('month').temperature.mean()
df.groupby(['year', 'month']).temperature.mean()
df.groupby(['year', 'month']).agg({'duration':sum,
'network_type':'count',
'date':'first'})
df.groupby(['year', 'month']).agg({'duration':[min, max, sum],
'network_type':'count',
'date':[min, 'first', 'nunique']})
# df.groupby('who').agg({'survived':sum,
# 'age':"mean"})
# df.groupby(['sex', 'class', 'embark_town']).agg({'alive':"count",
# "alone":sum})
data = pd.DataFrame({'time': [0.5, 1., 1.5, None, 2.5, 3., 3.5, None], 'value': [6, 4, 5, 8, None, 10, 11, None]})
data
| 0.387574 | 0.986258 |
```
# Load Data
import glob
import pandas as pd
def Carga_All_Files( ):
regexp='../data/covi*'
df = pd.DataFrame()
# Iterate trough LIST DIR and
for my_file in glob.glob(regexp):
this_df = pd.read_csv(my_file)
this_df['Fecha'] = my_file
df = pd.concat([df,this_df])
return df
df = Carga_All_Files( )
df.tail()
def Get_Comunidades_List( ):
return Carga_All_Files( )['CCAA'].unique()
#Get_Comunidades_List()
def Preprocesado():
df = Carga_All_Files( )
# Formateamos la fecha
df['Fecha'].replace({
'../data/covi': '2020-',
'.csv' : ''}, inplace=True, regex=True)
df['Fecha'] = pd.to_datetime(df['Fecha'], format='%Y-%d%m')
#
return df.sort_values(by='Fecha')
df = Preprocesado()
import numpy as np
def Enrich_Columns(comunidad):
del comunidad['ID']
del comunidad['IA']
del comunidad['Nuevos']
if 'Fecha' in comunidad.columns :
comunidad.set_index('Fecha', inplace=True)
# Datos de fallecimientos diarios, en totales y tanto por uno.
comunidad['Fallecidos hoy absoluto'] = comunidad['Fallecidos'] - comunidad['Fallecidos'].shift(1)
comunidad['Fallecidos hoy porcentaje'] = comunidad['Fallecidos hoy absoluto'] / comunidad['Fallecidos']
comunidad['Fallecidos hoy variacion respecto ayer'] = comunidad['Fallecidos hoy absoluto'] - comunidad['Fallecidos hoy absoluto'].shift(1)
# Datos de Casos diarios, en totales y tanto por uno.
comunidad['Casos hoy absoluto'] = comunidad['Casos'] - comunidad['Casos'].shift(1)
comunidad['Casos hoy porcentaje'] = comunidad['Casos hoy absoluto'] / comunidad['Casos']
comunidad['Casos hoy variacion respecto ayer'] = comunidad['Casos hoy absoluto'] - comunidad['Casos hoy absoluto'].shift(1)
# Convertimos a entero, para quitar decimales
CONVERT_INT_COLUMNS = ['Fallecidos hoy absoluto',
'Fallecidos hoy variacion respecto ayer',
'Casos hoy variacion respecto ayer',
'Casos hoy absoluto',
'Hospitalizados',
'Curados']
for column in CONVERT_INT_COLUMNS :
comunidad[column] = comunidad[column].fillna(0)
comunidad[column] = comunidad[column].astype(np.int64)
comunidad['Curados hoy absoluto'] = comunidad['Curados'] - comunidad['Curados'].shift(1)
try :
comunidad['Proporcion Curados hoy absoluto / Casos hoy absoluto'] = comunidad['Curados hoy absoluto'] / comunidad['Casos hoy absoluto']
except:
pass
comunidad['Casos excluidos curados'] = comunidad['Casos'] - comunidad['Curados']
comunidad['Tasa Mortalidad'] = comunidad['Fallecidos'] / comunidad['Casos']
# ordenamos las filas y columnas
columnsTitles = ['CCAA',
'Casos' , 'Casos hoy absoluto' , 'Casos hoy variacion respecto ayer', 'Casos hoy porcentaje' ,
'Fallecidos', 'Fallecidos hoy absoluto', 'Fallecidos hoy variacion respecto ayer', 'Fallecidos hoy porcentaje' ,
'Tasa Mortalidad',
'Curados', 'Curados hoy absoluto', 'Casos excluidos curados', 'Proporcion Curados hoy absoluto / Casos hoy absoluto',
'UCI',
'Hospitalizados']
comunidad = comunidad.reindex(columns=columnsTitles)
comunidad = comunidad.sort_values(by=['Fecha'], ascending=False)
comunidad = comunidad.rename(columns = {'CCAA':'Lugar'})
return comunidad
def Get_Comunidad(nombre_comunidad):
# Trabajamos solo con una comunidad
df = Preprocesado()
df = df[(df['CCAA'] == nombre_comunidad)].sort_values(by='Fecha')
df = Enrich_Columns(df)
return df
def Get_Nacion():
df = Preprocesado()
df = df.sort_values(by='Fecha')
df = df.groupby(['Fecha']).sum()
df['CCAA'] = 'España'
df = Enrich_Columns(df)
return df
# Just for debug purposes
def Debug_Get_Comunidad():
comunidad = Get_Comunidad('Madrid')
return comunidad
Debug_Get_Comunidad()
# Just for debug purposes
def Debug_Get_Nacion():
return Get_Nacion()
Debug_Get_Nacion()
```
|
github_jupyter
|
# Load Data
import glob
import pandas as pd
def Carga_All_Files( ):
regexp='../data/covi*'
df = pd.DataFrame()
# Iterate trough LIST DIR and
for my_file in glob.glob(regexp):
this_df = pd.read_csv(my_file)
this_df['Fecha'] = my_file
df = pd.concat([df,this_df])
return df
df = Carga_All_Files( )
df.tail()
def Get_Comunidades_List( ):
return Carga_All_Files( )['CCAA'].unique()
#Get_Comunidades_List()
def Preprocesado():
df = Carga_All_Files( )
# Formateamos la fecha
df['Fecha'].replace({
'../data/covi': '2020-',
'.csv' : ''}, inplace=True, regex=True)
df['Fecha'] = pd.to_datetime(df['Fecha'], format='%Y-%d%m')
#
return df.sort_values(by='Fecha')
df = Preprocesado()
import numpy as np
def Enrich_Columns(comunidad):
del comunidad['ID']
del comunidad['IA']
del comunidad['Nuevos']
if 'Fecha' in comunidad.columns :
comunidad.set_index('Fecha', inplace=True)
# Datos de fallecimientos diarios, en totales y tanto por uno.
comunidad['Fallecidos hoy absoluto'] = comunidad['Fallecidos'] - comunidad['Fallecidos'].shift(1)
comunidad['Fallecidos hoy porcentaje'] = comunidad['Fallecidos hoy absoluto'] / comunidad['Fallecidos']
comunidad['Fallecidos hoy variacion respecto ayer'] = comunidad['Fallecidos hoy absoluto'] - comunidad['Fallecidos hoy absoluto'].shift(1)
# Datos de Casos diarios, en totales y tanto por uno.
comunidad['Casos hoy absoluto'] = comunidad['Casos'] - comunidad['Casos'].shift(1)
comunidad['Casos hoy porcentaje'] = comunidad['Casos hoy absoluto'] / comunidad['Casos']
comunidad['Casos hoy variacion respecto ayer'] = comunidad['Casos hoy absoluto'] - comunidad['Casos hoy absoluto'].shift(1)
# Convertimos a entero, para quitar decimales
CONVERT_INT_COLUMNS = ['Fallecidos hoy absoluto',
'Fallecidos hoy variacion respecto ayer',
'Casos hoy variacion respecto ayer',
'Casos hoy absoluto',
'Hospitalizados',
'Curados']
for column in CONVERT_INT_COLUMNS :
comunidad[column] = comunidad[column].fillna(0)
comunidad[column] = comunidad[column].astype(np.int64)
comunidad['Curados hoy absoluto'] = comunidad['Curados'] - comunidad['Curados'].shift(1)
try :
comunidad['Proporcion Curados hoy absoluto / Casos hoy absoluto'] = comunidad['Curados hoy absoluto'] / comunidad['Casos hoy absoluto']
except:
pass
comunidad['Casos excluidos curados'] = comunidad['Casos'] - comunidad['Curados']
comunidad['Tasa Mortalidad'] = comunidad['Fallecidos'] / comunidad['Casos']
# ordenamos las filas y columnas
columnsTitles = ['CCAA',
'Casos' , 'Casos hoy absoluto' , 'Casos hoy variacion respecto ayer', 'Casos hoy porcentaje' ,
'Fallecidos', 'Fallecidos hoy absoluto', 'Fallecidos hoy variacion respecto ayer', 'Fallecidos hoy porcentaje' ,
'Tasa Mortalidad',
'Curados', 'Curados hoy absoluto', 'Casos excluidos curados', 'Proporcion Curados hoy absoluto / Casos hoy absoluto',
'UCI',
'Hospitalizados']
comunidad = comunidad.reindex(columns=columnsTitles)
comunidad = comunidad.sort_values(by=['Fecha'], ascending=False)
comunidad = comunidad.rename(columns = {'CCAA':'Lugar'})
return comunidad
def Get_Comunidad(nombre_comunidad):
# Trabajamos solo con una comunidad
df = Preprocesado()
df = df[(df['CCAA'] == nombre_comunidad)].sort_values(by='Fecha')
df = Enrich_Columns(df)
return df
def Get_Nacion():
df = Preprocesado()
df = df.sort_values(by='Fecha')
df = df.groupby(['Fecha']).sum()
df['CCAA'] = 'España'
df = Enrich_Columns(df)
return df
# Just for debug purposes
def Debug_Get_Comunidad():
comunidad = Get_Comunidad('Madrid')
return comunidad
Debug_Get_Comunidad()
# Just for debug purposes
def Debug_Get_Nacion():
return Get_Nacion()
Debug_Get_Nacion()
| 0.335024 | 0.333612 |
## Ensemble Methods
### Agenda
<hr>
1. Introduction to Ensemble Methods
2. RandomForest
3. AdaBoost
4. GradientBoostingTree
5. VotingClassifier
<hr>
### 1. Introduction to Ensemble Method
* Objective of ensemble methods is to combine the predictions of serveral base estimators ( Linear Regression, Decisison Tree, etc. ) to create a combined effect or more genralized model.
* Two types of Ensemble Method
- Averaging Method : Build several estimators independently & average their predictions. Examples are RandomForest etc.
- Boosting Method : Base estimators are built sequentially using weighted version of data .i.e fitting models with data that were mis-classified. Examples are AdaBoost
<img src="https://cdn-images-1.medium.com/max/1000/1*PaXJ8HCYE9r2MgiZ32TQ2A.png">
### 2. RandomForest
* Recap - Limitations of decison tree is that it overfits & shows high variance.
* RandomForest is an averaging ensemble method whose prediction is function of prediction of 'n' decision trees.
<img src="https://www.researchgate.net/profile/Stavros_Dimitriadis/publication/324517994/figure/fig1/AS:615965951799303@1523869135381/Classification-process-based-on-the-Random-Forest-algorithm-2.png">
##### Algorithm
* Data consist of R rows & M features.
* Sample of training data is taken.
* Random set of features are selected.
* As many as configured number of trees are created using above two steps.
* Final prediction in case of classification is majority prediction.
* Final prediction in case of regression is mean/median of individual tree prediction
##### Comparing Decision Tree & Random Forest for MNIST data
```
from sklearn.datasets import load_digits
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
digits = load_digits()
X = digits.data
y = digits.target
trainX, testX, trainY, testY = train_test_split(X,y)
dt = DecisionTreeClassifier()
dt.fit(trainX,trainY)
dt.score(testX,testY)
rf = RandomForestClassifier()
rf.fit(trainX,trainY)
rf.score(testX,testY)
```
##### Important Hyper-parameters
* n_estimators : number of trees to be configured, larger is better but compute cost.
* max_features : maximum number of features to be considered for splitting the node. For classification this equals to sqrt(n_features). And, for regression max_features = n_features.
* n_jobs : Configure as -1 so that we can make use of all cores.
#### Advantages
* Minimal data cleaning or dealing with missing values required.
* Works well with high dimensional datasets
* Minimizes variance even for low variance models
* RandomForest can tell importance of features. We can find important features & use them in model training
```
rf.feature_importances_
```
### 3. AdaBoost
* Boosting in general is about building a model from the training data, then creating a second model that attempts to correct the errors from the first model. Models are added until the training set is predicted perfectly or a maximum number of models are added.
* AdaBoost was first boosting algorithm.
* AdaBoost can be used for both classification & regression
##### Algorithm
* Core concept of adaboost is to fit weak learners ( like decision tree ) sequantially on repeatedly modifying data.
* Initially, each data is assigned equal weights.
* A base estimator is fitted with this data.
* Weights of misclassified data are increased & weights of correctly classified data is decreased.
* Repeat the above two steps till all data are correctly classified or max number of iterations configured.
* Making Prediction : The predictions from all of them are then combined through a weighted majority vote (or sum) to produce the final prediction.
```
from sklearn.ensemble import AdaBoostClassifier
ab = AdaBoostClassifier(base_estimator=DecisionTreeClassifier(max_depth=8),n_estimators=600)
ab.fit(trainX,trainY)
ab.score(testX,testY)
ab = AdaBoostClassifier(base_estimator=RandomForestClassifier(n_estimators=20),n_estimators=600)
ab.fit(trainX,trainY)
ab.score(testX,testY)
```
### 4. GradientBoostingTree
* A machine learning technique for regression and classification problems, which produces a prediction model in the form of an ensemble of weak prediction models, typically decision trees.
* One of the very basic assumption of linear regression is that it's sum of residuals is 0.
* These residuals as mistakes committed by our predictor model.
* Although, tree based models are not based on any of such assumptions, but if sum of residuals is not 0, then most probably there is some pattern in the residuals of our model which can be leveraged to make our model better.
* So, the intuition behind gradient boosting algorithm is to leverage the pattern in residuals and strenghten a weak prediction model, until our residuals don't show any pattern.
* Algorithmically, we are minimizing our loss function, such that test loss reach it’s minima.
##### Problem : House Price Prediction using GradientBoostingTree
```
from sklearn.datasets import load_boston
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
house_data = load_boston()
X = house_data.data
y = house_data.target
from sklearn.ensemble import GradientBoostingRegressor
gbt = GradientBoostingRegressor()
gbt
from sklearn.model_selection import train_test_split
trainX, testX, trainY, testY = train_test_split(X,y)
gbt.fit(trainX,trainY)
test_score = np.zeros(100, dtype=np.float64)
for i, y_pred in enumerate(gbt.staged_predict(testX)):
test_score[i] = gbt.loss_(testY, y_pred)
plt.plot(test_score)
plt.xlabel('Iterations')
plt.ylabel('Least squares Loss')
```
### 5. VotingClassifier
* Core concept of VotingClassifier is to combine conceptually different machine learning classifiers and use a majority vote or weighted vote to predict the class labels.
* Voting classifier is quite effective with good estimators & handles individual's limitations, ensemble methods can also participate.
* Types of Voting Classifier
- Soft Voting Classifier, different weights configured to different estimator
- Hard Voting Classifier, all estimators have equal weighage
##### Problem : DIGIT identification using VotingClassifier
```
from sklearn.ensemble import VotingClassifier,RandomForestClassifier,AdaBoostClassifier
from sklearn.svm import SVC
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
estimators = [
('rf',RandomForestClassifier(n_estimators=20)),
('svc',SVC(kernel='rbf', probability=True)),
('knc',KNeighborsClassifier()),
('abc',AdaBoostClassifier(base_estimator=DecisionTreeClassifier() ,n_estimators=20)),
('lr',LogisticRegression())
]
vc = VotingClassifier(estimators=estimators, voting='hard')
digits = load_digits()
X,y = digits.data, digits.target
trainX, testX, trainY, testY = train_test_split(X,y)
vc.fit(trainX,trainY)
vc.score(testX,testY)
for est,name in zip(vc.estimators_,vc.estimators):
print (name[0], est.score(testX,testY))
vc = VotingClassifier(estimators=estimators, voting='soft', weights=[2,.1,3,2,2])
vc.fit(trainX,trainY)
vc.score(testX,testY)
```
|
github_jupyter
|
from sklearn.datasets import load_digits
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
digits = load_digits()
X = digits.data
y = digits.target
trainX, testX, trainY, testY = train_test_split(X,y)
dt = DecisionTreeClassifier()
dt.fit(trainX,trainY)
dt.score(testX,testY)
rf = RandomForestClassifier()
rf.fit(trainX,trainY)
rf.score(testX,testY)
rf.feature_importances_
from sklearn.ensemble import AdaBoostClassifier
ab = AdaBoostClassifier(base_estimator=DecisionTreeClassifier(max_depth=8),n_estimators=600)
ab.fit(trainX,trainY)
ab.score(testX,testY)
ab = AdaBoostClassifier(base_estimator=RandomForestClassifier(n_estimators=20),n_estimators=600)
ab.fit(trainX,trainY)
ab.score(testX,testY)
from sklearn.datasets import load_boston
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
house_data = load_boston()
X = house_data.data
y = house_data.target
from sklearn.ensemble import GradientBoostingRegressor
gbt = GradientBoostingRegressor()
gbt
from sklearn.model_selection import train_test_split
trainX, testX, trainY, testY = train_test_split(X,y)
gbt.fit(trainX,trainY)
test_score = np.zeros(100, dtype=np.float64)
for i, y_pred in enumerate(gbt.staged_predict(testX)):
test_score[i] = gbt.loss_(testY, y_pred)
plt.plot(test_score)
plt.xlabel('Iterations')
plt.ylabel('Least squares Loss')
from sklearn.ensemble import VotingClassifier,RandomForestClassifier,AdaBoostClassifier
from sklearn.svm import SVC
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.datasets import load_digits
from sklearn.model_selection import train_test_split
estimators = [
('rf',RandomForestClassifier(n_estimators=20)),
('svc',SVC(kernel='rbf', probability=True)),
('knc',KNeighborsClassifier()),
('abc',AdaBoostClassifier(base_estimator=DecisionTreeClassifier() ,n_estimators=20)),
('lr',LogisticRegression())
]
vc = VotingClassifier(estimators=estimators, voting='hard')
digits = load_digits()
X,y = digits.data, digits.target
trainX, testX, trainY, testY = train_test_split(X,y)
vc.fit(trainX,trainY)
vc.score(testX,testY)
for est,name in zip(vc.estimators_,vc.estimators):
print (name[0], est.score(testX,testY))
vc = VotingClassifier(estimators=estimators, voting='soft', weights=[2,.1,3,2,2])
vc.fit(trainX,trainY)
vc.score(testX,testY)
| 0.717705 | 0.990714 |
# Custom Dataset
<div style="text-align: right"> by <a href="https://scholar.google.com/citations?user=f-4YHeMAAAAJ&hl=en">Lorenzo Bertoni</a> 13/04/2021 </div>
## Overview
In this section of the guide, we will see how to train and evaluate OpenPifPaf on a custom dataset. OpenPifPaf is based on the concept of __[Plugin architecture pattern](https://cs.uwaterloo.ca/~m2nagapp/courses/CS446/1195/Arch_Design_Activity/PlugIn.pdf)__, and the overall system is composed of a core component and auxiliary plug-in modules. To train a model on a custom dataset, you don't need to change the core system, only to create a small plugin for it.
This tutorial will go through the steps required to create a new plugin for a custom dataset.
Let's go through the steps of implementing a 2D pose estimator for vehicles, as a case study. If you are interested in how this specific plugin works, please check its {doc}`guide section <plugins_apollocar3d>`. We suggest to create and debug your own plugin copying a pre-existing plugin, changing its name, and adapting its files to your needs.
Below, a description of the structure of the plugin to give you some intuition of what you will need to change.
## Plugin structure
### 1. Data Module
This module handles the interface of your custom dataset with the core system and it is the main component of the plugin. For the [ApolloCar3D Dataset](http://apolloscape.auto/car_instance.html), we created a module called _apollo_kp.py_ containing the class [ApolloKp](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_kp.py) which inherits from the [DataModule](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/datasets/module.py) class.
The base class to inherit from has the following structure:
```{eval-rst}
.. autoclass:: openpifpaf.datasets.DataModule
:members: cli, configure, train_loader, val_loader, eval_loader, metrics
:noindex:
```
Now that you have a general view of the structure of a data module, we suggest you to refer to the implementation of the [ApolloKp](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_kp.py) class. You can get started by copying and modifying this class according to your needs.
### 2. Plugin Registration
For the core system to recognize the new plugin you need to create a [\_\_init\_\_.py](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/__init__.py) file specifying:
1. __Name Convention__: include all the plugins file into a folder named __openpifpaf\_\<plugin_name\>__. Only folders, which names start with openpifpaf\_ are recognized
2. __Registration__: Inside the folder, create an _init.py_ file and add to the list of existing plugins the datamodule that we have just created (__ApolloKp__). In this case, the name __apollo__ represents the name of the dataset.
```
def register():
openpifpaf.DATAMODULES['apollo'] = ApolloKp
```
### 3. Constants
Create a module _constants.py_ containing all the constants needed to define the 2D keypoints of vehicles. The most important are:
- __Names__ of the keypoints, as a list of strings. In our plugin this is called CAR_KEYPOINTS
- __Skeleton__: the connections between the keypooints, as a list of lists of two elements indicating the indeces of the starting and ending connections. In our plugin this is called CAR_SKELETON
- __Sigmas__: the size of the area to compute the object keypoint similarity (OKS), if you wish to use average precision (AP) as a metric.
- __Score weights__:the weights to compute the overall score of an object (e.g. car or person). When computing the overall score the highest weights will be assigned to the most confident joints.
- __Categories__ of the keypoints. In this case, the only category is car.
- __Standard pose__ of the keypoints, to visualize the connections between the keypoints and as an argument for the head network.
- __Horizontal flip__ equivalents, if you use horizontal flipping as augmentation technique, you will need to define the corresponding left and right and keypoints as a dictionary. E.g. left_ear --> right_ear. In our plugin this is called HFLIP
In addition to the constants, the module contains two functions to draw the skeleton and save it as an image. The functions are only for debugging and can usually be used as they are, only changing the arguments with the new constants. For additional information, refer to the file [constants.py](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/constants.py).
### 4. Data Loading
If you are using COCO-style annotations, there is no need to create a datalader to load images and annotations. A default [CocoLoader](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/coco/dataset.py) is already available to be called inside the data module [ApolloKp](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_kp.py)
If you wish to load different annotations, you can either write your own dataloader, or you can transform your annotations to COCO style .json files. In this plugin, we first convert ApolloCar3D annotations into COCO style .json files and then load them as standard annotations.
### 5. Annotations format
This step transforms custom annotations, in this case from ApolloCar3D, into COCO-style annotations. Below we describe how to populate a json file using COCO-style format. For the full working example, check the module [apollo_to_coco.py](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_to_coco.py) inside the plugin.
```
def initiate_json(self):
"""
Initiate json file: one for training phase and another one for validation.
"""
self.json_file["info"] = dict(url="https://github.com/openpifpaf/openpifpaf",
date_created=time.strftime("%a, %d %b %Y %H:%M:%S +0000", time.localtime()),
description="Conversion of ApolloCar3D dataset into MS-COCO format")
self.json_file["categories"] = [dict(name='', # Category name
id=1, # Id of category
skeleton=[], # Skeleton connections (check constants.py)
supercategory='', # Same as category if no supercategory
keypoints=[])] # Keypoint names
self.json_file["images"] = [] # Empty for initialization
self.json_file["annotations"] = [] # Empty for initialization
def process_image(json_file):
"""
Update image field in json file
"""
# ------------------
# Add here your code
# -------------------
json_file["images"].append({
'coco_url': "unknown",
'file_name': '', # Image name
'id': 0, # Image id
'license': 1, # License type
'date_captured': "unknown",
'width': 0, # Image width (pixels)
'height': 0}) # Image height (pixels)
def process_annotation(json_file):
"""
Process and include in the json file a single annotation (instance) from a given image
"""
# ------------------
# Add here your code
# -------------------
json_file["annotations"].append({
'image_id': 0, # Image id
'category_id': 1, # Id of the category (like car or person)
'iscrowd': 0, # 1 to mask crowd regions, 0 if the annotation is not a crowd annotation
'id': 0, # Id of the annotations
'area': 0, # Bounding box area of the annotation (width*height)
'bbox': [], # Bounding box coordinates (x0, y0, width, heigth), where x0, y0 are the left corner
'num_keypoints': 0, # number of keypoints
'keypoints': [], # Flattened list of keypoints [x, y, visibility, x, y, visibility, .. ]
'segmentation': []}) # To add a segmentation of the annotation, empty otherwise
```
## Training
We have seen all the elements needed to create your own plugin on a custom dataset. To train the dataset, all OpenPifPaf commands are still valid. There are only two differences:
1. Specify the dataset name in the training command. In this case, we have called our dataset _apollo_ during the registration phase, therefore we will have `--dataset=apollo`.
2. Include the commands we have created in the data module for this specific dataset, for example `--apollo-square-edge` to define the size of the training crops.
A training command may look like this:
```sh
python3 -m openpifpaf.train --dataset apollo \
--apollo-square-edge=769 \
--basenet=shufflenetv2k16 --lr=0.00002 --momentum=0.95 --b-scale=5.0 \
--epochs=300 --lr-decay 160 260 --lr-decay-epochs=10 --weight-decay=1e-5 \
--weight-decay=1e-5 --val-interval 10 --loader-workers 16 --apollo-upsample 2 \
--apollo-bmin 2 --batch-size 8
```
## Evaluation
Evaluation on the COCO metric is supported by pifpaf and a simple evaluation command may look like this:
```sh
python3 -m openpifpaf.eval --dataset=apollo --checkpoint <path of the model>
```
To evaluate on custom metrics, we would need to define a new metric and add it in the list of metrics, inside the data module. In our case, we have a a DataModule class called __ApolloKp__, and its function _**metrics**_ returns the list of metrics to run. Each metric is defined as a class that inherits from openpifpaf.metric.base.Base
For more information, please check how we implemented a simple metric for the ApolloCar3D dataset called MeanPixelError, that calculate mean pixel error and detection rate for a given image.
## Prediction
To run your model trained on a different dataset, you simply need to run the standard OpenPifPaf command specifying your model. A prediction command looks like this:
```sh
python3 -m openpifpaf.predict --checkpoint <model path>
```
All the command line options are still valid, check them with:
```sh
python3 -m openpifpaf.predict --help
```
## Final remarks
We hope you'll find this guide useful to create your own plugin. For more information check the guide section for the {doc}`ApolloCar3D plugin <plugins_apollocar3d>`.
Please keep us posted on issues you encounter (using the issue section on GitHub) and especially on your successes! We will be more than happy to add your plugin to the list of OpenPifPaf [related projects](https://openpifpaf.github.io/openpifpaf/dev/intro.html#related-projects).
Finally, if you find OpenPifPaf useful for your research, we would be happy if you cite us!
```
@article{kreiss2021openpifpaf,
title = {{OpenPifPaf: Composite Fields for Semantic Keypoint Detection and Spatio-Temporal Association}},
author = {Sven Kreiss and Lorenzo Bertoni and Alexandre Alahi},
journal = {arXiv preprint arXiv:2103.02440},
month = {March},
year = {2021}
}
@InProceedings{kreiss2019pifpaf,
author = {Kreiss, Sven and Bertoni, Lorenzo and Alahi, Alexandre},
title = {{PifPaf: Composite Fields for Human Pose Estimation}},
booktitle = {The IEEE Conference on Computer Vision and Pattern Recognition (CVPR)},
month = {June},
year = {2019}
}
```
|
github_jupyter
|
Now that you have a general view of the structure of a data module, we suggest you to refer to the implementation of the [ApolloKp](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_kp.py) class. You can get started by copying and modifying this class according to your needs.
### 2. Plugin Registration
For the core system to recognize the new plugin you need to create a [\_\_init\_\_.py](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/__init__.py) file specifying:
1. __Name Convention__: include all the plugins file into a folder named __openpifpaf\_\<plugin_name\>__. Only folders, which names start with openpifpaf\_ are recognized
2. __Registration__: Inside the folder, create an _init.py_ file and add to the list of existing plugins the datamodule that we have just created (__ApolloKp__). In this case, the name __apollo__ represents the name of the dataset.
### 3. Constants
Create a module _constants.py_ containing all the constants needed to define the 2D keypoints of vehicles. The most important are:
- __Names__ of the keypoints, as a list of strings. In our plugin this is called CAR_KEYPOINTS
- __Skeleton__: the connections between the keypooints, as a list of lists of two elements indicating the indeces of the starting and ending connections. In our plugin this is called CAR_SKELETON
- __Sigmas__: the size of the area to compute the object keypoint similarity (OKS), if you wish to use average precision (AP) as a metric.
- __Score weights__:the weights to compute the overall score of an object (e.g. car or person). When computing the overall score the highest weights will be assigned to the most confident joints.
- __Categories__ of the keypoints. In this case, the only category is car.
- __Standard pose__ of the keypoints, to visualize the connections between the keypoints and as an argument for the head network.
- __Horizontal flip__ equivalents, if you use horizontal flipping as augmentation technique, you will need to define the corresponding left and right and keypoints as a dictionary. E.g. left_ear --> right_ear. In our plugin this is called HFLIP
In addition to the constants, the module contains two functions to draw the skeleton and save it as an image. The functions are only for debugging and can usually be used as they are, only changing the arguments with the new constants. For additional information, refer to the file [constants.py](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/constants.py).
### 4. Data Loading
If you are using COCO-style annotations, there is no need to create a datalader to load images and annotations. A default [CocoLoader](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/coco/dataset.py) is already available to be called inside the data module [ApolloKp](https://github.com/openpifpaf/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_kp.py)
If you wish to load different annotations, you can either write your own dataloader, or you can transform your annotations to COCO style .json files. In this plugin, we first convert ApolloCar3D annotations into COCO style .json files and then load them as standard annotations.
### 5. Annotations format
This step transforms custom annotations, in this case from ApolloCar3D, into COCO-style annotations. Below we describe how to populate a json file using COCO-style format. For the full working example, check the module [apollo_to_coco.py](https://github.com/vita-epfl/openpifpaf/blob/main/openpifpaf/plugins/apollocar3d/apollocar3d/apollo_to_coco.py) inside the plugin.
## Training
We have seen all the elements needed to create your own plugin on a custom dataset. To train the dataset, all OpenPifPaf commands are still valid. There are only two differences:
1. Specify the dataset name in the training command. In this case, we have called our dataset _apollo_ during the registration phase, therefore we will have `--dataset=apollo`.
2. Include the commands we have created in the data module for this specific dataset, for example `--apollo-square-edge` to define the size of the training crops.
A training command may look like this:
## Evaluation
Evaluation on the COCO metric is supported by pifpaf and a simple evaluation command may look like this:
To evaluate on custom metrics, we would need to define a new metric and add it in the list of metrics, inside the data module. In our case, we have a a DataModule class called __ApolloKp__, and its function _**metrics**_ returns the list of metrics to run. Each metric is defined as a class that inherits from openpifpaf.metric.base.Base
For more information, please check how we implemented a simple metric for the ApolloCar3D dataset called MeanPixelError, that calculate mean pixel error and detection rate for a given image.
## Prediction
To run your model trained on a different dataset, you simply need to run the standard OpenPifPaf command specifying your model. A prediction command looks like this:
All the command line options are still valid, check them with:
## Final remarks
We hope you'll find this guide useful to create your own plugin. For more information check the guide section for the {doc}`ApolloCar3D plugin <plugins_apollocar3d>`.
Please keep us posted on issues you encounter (using the issue section on GitHub) and especially on your successes! We will be more than happy to add your plugin to the list of OpenPifPaf [related projects](https://openpifpaf.github.io/openpifpaf/dev/intro.html#related-projects).
Finally, if you find OpenPifPaf useful for your research, we would be happy if you cite us!
| 0.839043 | 0.915469 |
# Searching PDB for binders to desired protein
Idea:
- complete partial complexes
- start with single chain/ sequence
- find homologs which are in a complex
- check if complex could also be formed with query instead of the homolog
- use as template for homology modelling
Dan Farrell & Stefen Bienert
```
from pyspark.sql import SparkSession
from mmtfPyspark.io import mmtfReader
from mmtfPyspark.webfilters import SequenceSimilarity
from mmtfPyspark.mappers import StructureToPolymerChains
```
__Configure Spark__
```
spark = SparkSession.builder.master("local[4]").appName("Search").getOrCreate()
```
### Prepare an example query
Download a structure and extract its sequence for the query.
```
query_id = '4LGR.A'
query_tf = mmtfReader.download_reduced_mmtf_files([query_id[:4]])
primary = query_tf.flatMap(StructureToPolymerChains())
query_seq = primary.lookup(query_id)[0].entity_list[0]['sequence']
```
### Run BLAST against PDB
Load the PDB and use the query sequence on it with BLAST.
```
path = "../resources/mmtf_reduced_sample"
pdb = mmtfReader.read_sequence_file(path).cache()
pdb = pdb.flatMap(StructureToPolymerChains())\
.filter(SequenceSimilarity(sequence=query_seq, searchTool=SequenceSimilarity.BLAST,
eValueCutoff=0.1, sequenceIdentityCutoff=10,
maskLowComplexity=True)).collect()
```
Print out hits:
```
for pdbId, structure in pdb:
print(f"{pdbId} : {structure.entity_list[0]['sequence']}")
```
### TODO: Find homologs which are oligomers
### TODO: Extract cahins bound to homologs
## Part 2:
##### General idea:
1. Align your possible binder to the query protein's ideal binder.
* Do sequence alignment between the two
* Do structural alignment based on the sequence alignment
2. Combine your protein of interest with a possible binder, then combine them into one structure
3. Generate a matrix around the interface for **both** (anchor on the constant protein) and fill the matrix with vdw spheres that represent the CAlphas.
* **both**= input structure + ideal binder && input structure + possible binder
4. Then compare the two matricies and give it a score based on the cross correlation
5. Sort by score and see if you have found any unexpected binders!
```
import os
import Bio.PDB
from Bio.PDB import PDBIO
from Bio.Align.Applications import ClustalOmegaCommandline
from Bio.PDB.mmtf import MMTFParser
from Bio.PDB.Polypeptide import is_aa, three_to_one
from Bio import AlignIO
import py3Dmol
def get_sequence_from_biopython_and_chain_name(bp_structure, chain_name):
"""Get the AA sequence of a chain from biopython
Args:
bp_structure: A biopython structure
chain_name: A chain in that biopython structure to get the AA sequence of
Returns:
sequence
Notes:
Some horrible pdbs only have one atom for whole residues (IE just N, instead of NCC)
so you MUST check for CA since that's what we're aligning on
"""
sequence = ""
for chain in bp_structure:
if chain.id != chain_name:
continue
for res in chain:
if is_aa(res):
for atom in res:
if atom.name == "CA":
sequence += three_to_one(res.get_resname())
return sequence
def get_aln_atoms(bp_structure, residues, chain_name):
"""Get CA atoms from aligned residues
Alignments are residue based, but we want to do our alignment based on CA atoms only
essentially convert residue to atom['CA']
Args:
bp_structure: a biopython structure
residues: the index of aligned residues that you're interested in
chain_name: The chain name of interest, (tells us what chains to iterate over)
Retruns:
biopython CA atoms that we want to use in the alignment
"""
ret_atoms = []
residue_count = 0
for chain in bp_structure:
if chain.id != chain_name:
continue
for res in chain:
if is_aa(res):
for atom in res:
if atom.name == "CA" and residue_count in residues:
ret_atoms.append(atom)
residue_count += 1
return ret_atoms
def make_combo_pose(input_chainID, target_chain, ref_model, target_model):
"""Combine the best parts of our target/reference
Args:
input_chainID: The original quereied seuqence's chainID
target_chain: The chainID of the target
ref_model: The biopython model of the reference(input)
target_model: the biopython model of the target pdb
Returns:
a biopython structure containing the reference_model(input_chainID)
"""
base_structure = Bio.PDB.Structure.Structure("combo_pose")
base_model = Bio.PDB.Model.Model(0)
base_structure.add(base_model)
ref_chains_to_mv = []
for i, chain in enumerate(ref_model):
if chain.id != input_chainID:
continue
ref_chains_to_mv.append(i)
ref_chains = [x for x in ref_model]
for x in reversed(ref_chains_to_mv):
ref_chains[x].detach_parent()
ref_chains[x].id = 'A'
base_structure[0].add(ref_chains[x])
target_chains_to_mv = []
for i, chain in enumerate(target_model):
if chain.id != target_chain:
continue
target_chains_to_mv.append(i)
target_chains = [x for x in target_model]
for x in reversed(target_chains_to_mv):
target_chains[x].detach_parent()
target_chains[x].id = 'B'
base_structure[0].add(target_chains[x])
io = Bio.PDB.PDBIO()
io.set_structure(base_structure)
io.save("test_align.pdb")
return base_structure
def align_two_structures(input_pdbID_chainID, reference, target):
mmtf_parser = MMTFParser()
# Only get first model
ref_structure = mmtf_parser.get_structure_from_url(reference.split('_')[0])
ref_model = ref_structure[0]
ref_sequence = get_sequence_from_biopython_and_chain_name(ref_model, reference.split('_')[1])
target_structure = mmtf_parser.get_structure_from_url(target.split('_')[0])
target_model = target_structure[0]
target_sequence = get_sequence_from_biopython_and_chain_name(target_model, target.split('_')[1])
if os.path.isfile("out.clust.aln"):
os.remove("out.clust.aln")
with open("ref_targ.fasta", 'w') as f:
f.write(">reference\n{}\n".format(ref_sequence))
f.write(">target\n{}\n".format(target_sequence))
clustalomega_cline = ClustalOmegaCommandline(infile="ref_targ.fasta", outfile="out.clust.aln",
verbose=True, auto=True)
clustalomega_cline()
align = AlignIO.read("out.clust.aln", "fasta")
# We look for aligned residues in the clustalO alignment, and mark them to be used in super imposition
ref_atoms = []
ref_count = 0
target_atoms = []
target_count = 0
for i, _ in enumerate(align[0].seq):
use = True
# Reference on top
if align[0].seq[i] == '-':
use = False
else:
ref_count += 1
if align[1].seq[i] == '-':
use = False
else:
target_count += 1
if use:
ref_atoms.append(ref_count-1)
target_atoms.append(target_count-1)
ref_atoms_bp = get_aln_atoms(ref_model, ref_atoms, reference.split('_')[1])
target_atoms_bp = get_aln_atoms(target_model, target_atoms, target.split('_')[1])
super_imposer = Bio.PDB.Superimposer()
super_imposer.set_atoms(ref_atoms_bp, target_atoms_bp)
super_imposer.apply(target_model.get_atoms())
combo_pose = make_combo_pose(input_pdbID_chainID.split('_')[-1], target.split('_')[-1], ref_model, target_model)[0]
# A = reference protein, B = target protein
def part_2_main(inputs):
"""Run alignment and scoring
Args:
inputs: a list of tuples, each tuple has the format
(input pdbID_chainID of base protein of interest,
found pdbID_chainID of protein that interacts with tuple[0],
protein that is homologous, or structurally similar to tuple[1])
Notes:
General Idea:
combine your protein of interest with a possible binder, then combine them into one,
generate a matrix around the interface for both (anchor on the constant protein) and fill the matrix
with vdw spheres that represent the CAlphas. then compare that matrix to the matrix of the ideal
binder, and give it a score based on the cross correlation
TODO:
SCORING based on vdw spheres cross correlation (overlap)
of result and input 'answer'
"""
for pair in inputs:
align_two_structures(pair[0], pair[1], pair[2])
inputs = [("2BJN_A", "3KXC_A", "3KXC_C")]
# part_2_main(inputs)
viewer = py3Dmol.view(query='pdb:2BJN')
# setting styles will be covered in the next tutorial
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
viewer = py3Dmol.view(query='pdb:3KXC')
# setting styles will be covered in the next tutorial
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
structure = open('test_align.pdb','r').read()
viewer = py3Dmol.view()
viewer.addModel(structure,'pdb')
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
spark.stop()
```
|
github_jupyter
|
from pyspark.sql import SparkSession
from mmtfPyspark.io import mmtfReader
from mmtfPyspark.webfilters import SequenceSimilarity
from mmtfPyspark.mappers import StructureToPolymerChains
spark = SparkSession.builder.master("local[4]").appName("Search").getOrCreate()
query_id = '4LGR.A'
query_tf = mmtfReader.download_reduced_mmtf_files([query_id[:4]])
primary = query_tf.flatMap(StructureToPolymerChains())
query_seq = primary.lookup(query_id)[0].entity_list[0]['sequence']
path = "../resources/mmtf_reduced_sample"
pdb = mmtfReader.read_sequence_file(path).cache()
pdb = pdb.flatMap(StructureToPolymerChains())\
.filter(SequenceSimilarity(sequence=query_seq, searchTool=SequenceSimilarity.BLAST,
eValueCutoff=0.1, sequenceIdentityCutoff=10,
maskLowComplexity=True)).collect()
for pdbId, structure in pdb:
print(f"{pdbId} : {structure.entity_list[0]['sequence']}")
import os
import Bio.PDB
from Bio.PDB import PDBIO
from Bio.Align.Applications import ClustalOmegaCommandline
from Bio.PDB.mmtf import MMTFParser
from Bio.PDB.Polypeptide import is_aa, three_to_one
from Bio import AlignIO
import py3Dmol
def get_sequence_from_biopython_and_chain_name(bp_structure, chain_name):
"""Get the AA sequence of a chain from biopython
Args:
bp_structure: A biopython structure
chain_name: A chain in that biopython structure to get the AA sequence of
Returns:
sequence
Notes:
Some horrible pdbs only have one atom for whole residues (IE just N, instead of NCC)
so you MUST check for CA since that's what we're aligning on
"""
sequence = ""
for chain in bp_structure:
if chain.id != chain_name:
continue
for res in chain:
if is_aa(res):
for atom in res:
if atom.name == "CA":
sequence += three_to_one(res.get_resname())
return sequence
def get_aln_atoms(bp_structure, residues, chain_name):
"""Get CA atoms from aligned residues
Alignments are residue based, but we want to do our alignment based on CA atoms only
essentially convert residue to atom['CA']
Args:
bp_structure: a biopython structure
residues: the index of aligned residues that you're interested in
chain_name: The chain name of interest, (tells us what chains to iterate over)
Retruns:
biopython CA atoms that we want to use in the alignment
"""
ret_atoms = []
residue_count = 0
for chain in bp_structure:
if chain.id != chain_name:
continue
for res in chain:
if is_aa(res):
for atom in res:
if atom.name == "CA" and residue_count in residues:
ret_atoms.append(atom)
residue_count += 1
return ret_atoms
def make_combo_pose(input_chainID, target_chain, ref_model, target_model):
"""Combine the best parts of our target/reference
Args:
input_chainID: The original quereied seuqence's chainID
target_chain: The chainID of the target
ref_model: The biopython model of the reference(input)
target_model: the biopython model of the target pdb
Returns:
a biopython structure containing the reference_model(input_chainID)
"""
base_structure = Bio.PDB.Structure.Structure("combo_pose")
base_model = Bio.PDB.Model.Model(0)
base_structure.add(base_model)
ref_chains_to_mv = []
for i, chain in enumerate(ref_model):
if chain.id != input_chainID:
continue
ref_chains_to_mv.append(i)
ref_chains = [x for x in ref_model]
for x in reversed(ref_chains_to_mv):
ref_chains[x].detach_parent()
ref_chains[x].id = 'A'
base_structure[0].add(ref_chains[x])
target_chains_to_mv = []
for i, chain in enumerate(target_model):
if chain.id != target_chain:
continue
target_chains_to_mv.append(i)
target_chains = [x for x in target_model]
for x in reversed(target_chains_to_mv):
target_chains[x].detach_parent()
target_chains[x].id = 'B'
base_structure[0].add(target_chains[x])
io = Bio.PDB.PDBIO()
io.set_structure(base_structure)
io.save("test_align.pdb")
return base_structure
def align_two_structures(input_pdbID_chainID, reference, target):
mmtf_parser = MMTFParser()
# Only get first model
ref_structure = mmtf_parser.get_structure_from_url(reference.split('_')[0])
ref_model = ref_structure[0]
ref_sequence = get_sequence_from_biopython_and_chain_name(ref_model, reference.split('_')[1])
target_structure = mmtf_parser.get_structure_from_url(target.split('_')[0])
target_model = target_structure[0]
target_sequence = get_sequence_from_biopython_and_chain_name(target_model, target.split('_')[1])
if os.path.isfile("out.clust.aln"):
os.remove("out.clust.aln")
with open("ref_targ.fasta", 'w') as f:
f.write(">reference\n{}\n".format(ref_sequence))
f.write(">target\n{}\n".format(target_sequence))
clustalomega_cline = ClustalOmegaCommandline(infile="ref_targ.fasta", outfile="out.clust.aln",
verbose=True, auto=True)
clustalomega_cline()
align = AlignIO.read("out.clust.aln", "fasta")
# We look for aligned residues in the clustalO alignment, and mark them to be used in super imposition
ref_atoms = []
ref_count = 0
target_atoms = []
target_count = 0
for i, _ in enumerate(align[0].seq):
use = True
# Reference on top
if align[0].seq[i] == '-':
use = False
else:
ref_count += 1
if align[1].seq[i] == '-':
use = False
else:
target_count += 1
if use:
ref_atoms.append(ref_count-1)
target_atoms.append(target_count-1)
ref_atoms_bp = get_aln_atoms(ref_model, ref_atoms, reference.split('_')[1])
target_atoms_bp = get_aln_atoms(target_model, target_atoms, target.split('_')[1])
super_imposer = Bio.PDB.Superimposer()
super_imposer.set_atoms(ref_atoms_bp, target_atoms_bp)
super_imposer.apply(target_model.get_atoms())
combo_pose = make_combo_pose(input_pdbID_chainID.split('_')[-1], target.split('_')[-1], ref_model, target_model)[0]
# A = reference protein, B = target protein
def part_2_main(inputs):
"""Run alignment and scoring
Args:
inputs: a list of tuples, each tuple has the format
(input pdbID_chainID of base protein of interest,
found pdbID_chainID of protein that interacts with tuple[0],
protein that is homologous, or structurally similar to tuple[1])
Notes:
General Idea:
combine your protein of interest with a possible binder, then combine them into one,
generate a matrix around the interface for both (anchor on the constant protein) and fill the matrix
with vdw spheres that represent the CAlphas. then compare that matrix to the matrix of the ideal
binder, and give it a score based on the cross correlation
TODO:
SCORING based on vdw spheres cross correlation (overlap)
of result and input 'answer'
"""
for pair in inputs:
align_two_structures(pair[0], pair[1], pair[2])
inputs = [("2BJN_A", "3KXC_A", "3KXC_C")]
# part_2_main(inputs)
viewer = py3Dmol.view(query='pdb:2BJN')
# setting styles will be covered in the next tutorial
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
viewer = py3Dmol.view(query='pdb:3KXC')
# setting styles will be covered in the next tutorial
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
structure = open('test_align.pdb','r').read()
viewer = py3Dmol.view()
viewer.addModel(structure,'pdb')
viewer.setStyle({'cartoon': {'color': 'spectrum'}})
viewer.setStyle({'hetflag': True}, {'stick':{'radius': 0.3, 'singleBond': False}})
viewer.zoomTo()
viewer.show()
spark.stop()
| 0.777215 | 0.900486 |
# Exercise 2.2
> A sawtooth signal has a waveform that ramps up linearly from -1 to 1, then drops to -1 and repeats. See http://en.wikipedia.org/wiki/Sawtooth_wave
> Write a class called SawtoothSignal that extends `Signal` and provides `evaluate` to evaluate a sawtooth signal.
> Compute the spectrum of a sawtooth wave. How does the harmonic structure compare to triangle and square waves?
```
from thinkdsp import Signal, Sinusoid, normalize, unbias, PI2
import numpy as np
class MySawtooth(Sinusoid):
def evaluate(self, ts):
cycles = self.freq * ts + self.offset / PI2
frac, _ = np.modf(cycles)
# ys = np.abs(frac - 0.5)
# ys = normalize(unbias(ys), self.amp)
# ys = self.amp * np.sign(unbias(frac))
ys = self.amp * frac
return ys
from thinkdsp import decorate
test_saw = MySawtooth(200)
test_wave = test_saw.make_wave(test_saw.period*3, framerate=10000)
test_wave.plot()
decorate(xlabel='Time (s)')
test_wave = test_saw.make_wave(duration=0.5, framerate=10000)
test_wave.apodize()
test_wave.make_audio()
test_spectrum = test_wave.make_spectrum()
test_spectrum.plot()
decorate(xlabel='Frequency (Hz)')
```
## Excercise 2.3
> Make a square signal at 1100 Hz and make a wave that samplesit at 10000 frames per second. If you plot the spectrum, you can see that most of the harmonics are aliased. When you listen to the wave, can you hear the aliased harmonics?
```
from thinkdsp import SquareSignal
signal = SquareSignal(1100)
# signal.plot()
wave = signal.make_wave(duration=0.5, framerate=10_000)
# wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
wave.apodize()
wave.make_audio()
wave2 = signal.make_wave(duration=0.5, framerate=signal.freq*10)
spectrum2 = wave2.make_spectrum()
spectrum2.plot()
wave2.apodize()
wave2.make_audio()
```
## Exercise 2.3
> If you have a spectrum object, `spectrum`, and print the first few values of `spectrum.fs`, you’ll see that they start at zero. So `spectrum.hs[0]` is the magnitude of the component with frequency 0. But what does that mean?
> Try this experiment:
> 1. Make a triangle signal with frequency 440 and make a Wave with duration 0.01 seconds. Plot the waveform.
> 1. Make a Spectrum object and print `spectrum.hs[0]`. What is the amplitude and phase of this component?
> 1. Set `spectrum.hs[0] = 100`. Make a Wave from the modified Spectrum and plot it. What effect does this operation have on the waveform?
```
from thinkdsp import TriangleSignal
signal = TriangleSignal(440)
wave = signal.make_wave(duration=0.01, framerate=11025)
wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
spectrum.hs[0]
spectrum.hs[0] = 0
wave2 = spectrum.make_wave()
wave2.plot()
```
## Exercise 2.5
> Write a function that takes a Spectrum as a parameter and modifies it by dividing each element of `hs` by the corresponding frequency from `fs`. Hint: since division by zero is undefined, you might want to set `spectrum.hs[0] = 0`. Test your function using a square, triangle, or sawtooth wave.
> 1. Compute the Spectrum and plot it.
> 1. Modify the Spectrum using your function and plot it again.
> 1. Make a Wave from the modified Spectrum and listen to it. What effect does this operation have on the signal?
```
def modify(spectrum):
for it in range(1, len(spectrum.hs)):
spectrum.hs[it] /= spectrum.fs[it]
spectrum.hs[0] = 0
from thinkdsp import SawtoothSignal
signal = SawtoothSignal(440)
wave = signal.make_wave(duration=0.5, framerate=440 * 100)
spectrum = wave.make_spectrum()
spectrum.plot()
wave.make_audio()
modify(spectrum)
spectrum.plot(high=100)
wave2 = spectrum.make_wave()
wave2.make_audio()
```
## Exercise 2.6
> Triangle and square waves have odd harmonics only; the sawtooth wave has both even and odd harmonics. The harmonics of the square and sawtooth waves drop off in proportion to 1/f; the harmonics of the triangle wave drop off like 1/f2. Can you find a waveform that has even and odd harmonics that drop off like 1/f2? Hint: There are two ways you could approach this: you could construct the signal you want by adding up sinusoids, or you could start with a signal that is similar to what you want and modify it.
```
signal = SawtoothSignal(100)
wave = signal.make_wave(duration=signal.period*10, framerate=10_000)
wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
modify(spectrum)
spectrum.plot()
wave2 = spectrum.make_wave()
wave2.plot()
```
It looks like sinusoids, but it can't be just a single sinusoid. Othersize we'd see a single frequency component. To be honest, I couldn't come to this idea myself, so I consulted the solution, and realized it's `ParabolicSignal`.
```
from thinkdsp import ParabolicSignal
signal4 = ParabolicSignal(100)
wave4 = signal4.make_wave(duration=signal4.period*4, framerate=10_000)
wave4.plot()
```
|
github_jupyter
|
from thinkdsp import Signal, Sinusoid, normalize, unbias, PI2
import numpy as np
class MySawtooth(Sinusoid):
def evaluate(self, ts):
cycles = self.freq * ts + self.offset / PI2
frac, _ = np.modf(cycles)
# ys = np.abs(frac - 0.5)
# ys = normalize(unbias(ys), self.amp)
# ys = self.amp * np.sign(unbias(frac))
ys = self.amp * frac
return ys
from thinkdsp import decorate
test_saw = MySawtooth(200)
test_wave = test_saw.make_wave(test_saw.period*3, framerate=10000)
test_wave.plot()
decorate(xlabel='Time (s)')
test_wave = test_saw.make_wave(duration=0.5, framerate=10000)
test_wave.apodize()
test_wave.make_audio()
test_spectrum = test_wave.make_spectrum()
test_spectrum.plot()
decorate(xlabel='Frequency (Hz)')
from thinkdsp import SquareSignal
signal = SquareSignal(1100)
# signal.plot()
wave = signal.make_wave(duration=0.5, framerate=10_000)
# wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
wave.apodize()
wave.make_audio()
wave2 = signal.make_wave(duration=0.5, framerate=signal.freq*10)
spectrum2 = wave2.make_spectrum()
spectrum2.plot()
wave2.apodize()
wave2.make_audio()
from thinkdsp import TriangleSignal
signal = TriangleSignal(440)
wave = signal.make_wave(duration=0.01, framerate=11025)
wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
spectrum.hs[0]
spectrum.hs[0] = 0
wave2 = spectrum.make_wave()
wave2.plot()
def modify(spectrum):
for it in range(1, len(spectrum.hs)):
spectrum.hs[it] /= spectrum.fs[it]
spectrum.hs[0] = 0
from thinkdsp import SawtoothSignal
signal = SawtoothSignal(440)
wave = signal.make_wave(duration=0.5, framerate=440 * 100)
spectrum = wave.make_spectrum()
spectrum.plot()
wave.make_audio()
modify(spectrum)
spectrum.plot(high=100)
wave2 = spectrum.make_wave()
wave2.make_audio()
signal = SawtoothSignal(100)
wave = signal.make_wave(duration=signal.period*10, framerate=10_000)
wave.plot()
spectrum = wave.make_spectrum()
spectrum.plot()
modify(spectrum)
spectrum.plot()
wave2 = spectrum.make_wave()
wave2.plot()
from thinkdsp import ParabolicSignal
signal4 = ParabolicSignal(100)
wave4 = signal4.make_wave(duration=signal4.period*4, framerate=10_000)
wave4.plot()
| 0.429669 | 0.981542 |
### Data frames
In addition to the `Series`, Pandas also provides a `DataFrame` which has rows and columns, like a table or a spreadsheet. They're similar to (and based on) data frames in the statistics programming language R.
We can build a data frame from a dictionary where the _columns_ are entries in a dictionary. Each dictionary _key_ is a column header, and the associated _value_ is a list. The `pd.DataFrame()` function creates a data frame.
```
nucls = pd.DataFrame({'letter': [ 'A', 'C', 'G', 'T' ],
'name': ['adenine', 'cytosine', 'guanine', 'thymine'],
'ring': ['purine', 'pyrimidine', 'purine', 'pyrimidine']})
```
We can extract one column of a `DataFrame` as a `Series` using square brackets to index it by the name of the column:
```
nucls['name']
```
We can then index by row into the `Series` with a second set of square brackets
```
nucls['letter'][2]
```
Here is some Python code to create a data frame with observed nucleotide counts from 389 TATA boxes taken from eukaryotic promoters (Bucher, J Mol Biol (1990) 212, 563-578).
```
tata_counts = pd.DataFrame({'A': [ 16, 352, 3, 354, 268, 360, 222, 155],
'C': [ 46, 0, 10, 0, 0, 3, 2, 44],
'G': [ 18, 2, 2, 5, 0, 20, 44, 157],
'T': [ 309, 35, 374, 30, 121, 6, 121, 33]})
```
Each row is a position in the TATA motif, and each column is a nucleotide. It's possible to read off the consensus sequence of TATA(A/T)A(A/T)(A/G), sometimes written TATAWAWR, just from looking at the counts in the table.
Data frames have many useful methods. For instance, we can use the .sum() method to take the sums across rows or columns. The argument `0` will calculate column sums and the argument `1` will calculate row sums.
We can then turn these counts into probabilities by dividing each nucleotide count by the total number of sequences counted. That is if 35 out of 389 TATA-box sequences have a `T` at the second position, then the probability of a `T` at position 1 in a random TATA-box sequence is 35/389, just under 10%.
```
tata_counts / 389
```
will make a new data frame dividing each individual entry in our data frame by 389. We'll use this to make a new `tata_probs` data frame with the _probabilities_ of each nucleotide.
We can now look up, e.g., the probability of a `T` at the second position, which is position 1 in Python counting
```
tata_probs['T'][1]
```
We're most of the way to a probabilistic model of a TATA box. We will assume that each of the nucleotides in the TATA box is independent, so we can multiply these probabilities together
$$P(\;\mathtt{TATAAAG}\;|\;\mathrm{TATA-box}\;) =
P(\;\mathtt{T}\mathrm{\,at\,0\;}) \times
P(\;\mathtt{A}\mathrm{\,at\,1\;}) \times
P(\;\mathtt{T}\mathrm{\,at\,2\;}) \times
P(\;\mathtt{A}\mathrm{\,at\,3\;}) \times
P(\;\mathtt{A}\mathrm{\,at\,4\;}) \times
P(\;\mathtt{A}\mathrm{\,at\,5\;}) \times
P(\;\mathtt{G}\mathrm{\,at\,6\;})
$$
We need to keep track of which position is which, because $P(\;\mathtt{T}\mathrm{\,at\,0\;}) \neq P(\;\mathtt{T}\mathrm{\,at\,1\;})$. The `enumerate()` function lets us keep track of a position when we're iterating over a sequence.
```
for position, nt in enumerate(sequ):
print('position = ' + str(position) + ', nt = ' + str(nt))
```
Now, we'll write a `for` loop to iterate over the positions in a sequence and compute a running probability.
We'll start with probability 1
```
prob = 1
```
and then multiply the probability for each independent position
```
for position, nt in enumerate(sequ):
p = tata_probs[nt][position]
prob = prob * p
print(position, nt, p, prob)
```
We can use this to compute the probability of a "very good" TATA-box like `TATATATA`. We can also try the worst possible TATA box, `ACGCGCCT`.
Our final probability is 0! While $P(\;\mathtt{ACGCGCCT}\;|\;\textrm{TATA-box}\;)$ is definitely very small, it's probably not 0. We see zero `C` nucleotides at position 1 out of 389 TATA-boxes, but what if we counted 389,000? Would we find 100, 10, or 1?
We often handle these situations by adding a _pseudocount_ to our data. We add a fake count for each nucleotide, at each position, in order to eliminate zeros. The impact of this pseudocount depends on the number of real counts. If we add a pseudocount with 9 real observations, it represents 10% of our overall counts, but if we add a pseudocount with 999 real observations, it's only 0.1%.
We can just add 1 to every entry and use this table with pseudocounts to make our new data.
```
tata_counts_pseudo = tata_counts + 1
```
Now we can use the new tata_probs to compute the probability of the best TATA-box, which is pretty similar. We can also compute the worst TATA-box, which is very low but not zero.
It's getting tedious to write the same for loop every time we want to try a different sequence.
We can write our own function, `likelihood_tata()`, that will compute the likelihood of a sequence under our TATA-box probability model. We define a function with def followed by the function name. The arguments to the function are named in parentheses, and inside the function, these become variables that take on a different value each time we use the function. The `return` keyword gives the computed "value" for the function.
```
def likelihood_tata(sequ):
prob = 1
for position, nt in enumerate(sequ):
p = tata_probs[nt][position]
prob = prob * p
print(position, nt, p, prob)
return(prob)
```
Now we can easily use our function to compute the likelihood of some other possible TATA-box sequences. For example, the three sequences below are "very good" TATA-boxes that differ from the "best" TATA box at one of the three "degenerate" positions in the motif. Notice that the overall probability of getting one of these three imperfect motifs is substantially higher than the probability of the perfect TATA-box. In fact, although the TATA-box is a strong motif, fewer than 10% of the sequences generated according to our model will actually match the "best" sequence.
```
TATATAAG
TATAAATG
TATAAAAA
```
If we want to use our Bayesian framework to think about TATA-boxes, we need some additional information. What is $P(\;\mathtt{TATAAAAG}\;|\;\textit{not}\,\textrm{TATA-box}\;)$? We need a model for all the other sequences in the genome, often called a "background" model.
The easy background model is independent nucleotides, with probabilities determined by the overall composition of the genome. We just counted the overall number of `A`s etc in the yeast genome. A rough estimate is
```
background = pd.Series({'A': 0.31, 'C': 0.19, 'G': 0.19, 'T': 0.31})
```
_Exercise_ Use the `background` defined above to write a `likelihood_background()` function that calculates the likelihood of generating a given sequence under the model of random yeast genome.
Since the "worst" TATA-box is GC-rich and the "best" TATA-box is AT-rich, the odds of getting the "best" TATA-box by chance in random sequence is somewhat higher. Of course, the chance of getting the "best" sequence under our TATA-box probabilistic model is dramatically higher than the chance of getting the "worst" sequence. We can use the _ratio of the likelihoods_ as a measure of how well two different models fit a given sequence.
Below, we compute the likelihood ratios for the "best" sequence TATAAAAG, the "worst" sequence ACGCGCCT, and getting any one of the three very-good sequences TATAAATG and TATAAAAA.
```
print(likelihood_tata('TATAAAAG') / likelihood_background('TATAAAAG'))
print(likelihood_tata('ACGCGCCT') / likelihood_background('ACGCGCCT'))
print( (likelihood_tata('TATATAAG') + likelihood_tata('TATAAATG') + likelihood_tata('TATAAAAA'))
/ (likelihood_background('TATATAAG') + likelihood_background('TATAAATG') + likelihood_background('TATAAAAA')) )
```
We can go one step further and turn this likelihood ratio into a function
```
def likelihood_ratio(sequ):
return(likelihood_tata(sequ) / likelihood_background(sequ))
```
We might want to scan a whole promoter to find a TATA-box. Here is the promoter region for the yeast _CDC19_ gene.
```
cdc19_prm = 'TATGATGCTAGGTACCTTTAGTGTCTTCCTAAAAAAAAAAAAAGGCTCGCCATCAAAACGATATTCGTTGGCTTTTTTTTCTGAATTATAAATACTCTTTGGTAACTTTTCATTTCCAAGAACCTCTTTTTTCCAGTTATATCATG'
```
We need to extract 8-nucleotide chunks out of the promoter. Square brackets can extract a _range_ of values from a string or a list. To do this, we do `[start:end]` where the start is _included_ and the end is _excluded_.
```
alphabet = 'abcdefghijklmnopqrstuvwxyz'
alphabet[2:6]
```
This code goes from index 2 (the 3rd entry, `c`) to index 5 (`f`) and does not include index 6 (`g`).
We can use this to run
```
likelihood_ratio(cdc19_prm[0:8])
likelihood_ratio(cdc19_prm[1:9])
```
Now we can loop over each starting position in `cdc19_prm` and compute its likelihood.
We start at position 0 and we run until the _end_ of our 8-position window is at the end of the promoter. This happens when `start+8 = len(cdc19_prm)` or equivalently `start = len(cdc19_prm) - 8`.
The `range(start, end)` function creates a series of numbers.
To start, we can write the loop
```
for start in range(0, len(cdc19_prm) - 8):
print(str(start) + ' ' + cdc19_prm[start:start+8])
```
and if all of that looks good we can add in a `likelihood_ratio()`.
Then we can build a _list_ of these likelihoods and covert it into a Pandas `Series`.
|
github_jupyter
|
nucls = pd.DataFrame({'letter': [ 'A', 'C', 'G', 'T' ],
'name': ['adenine', 'cytosine', 'guanine', 'thymine'],
'ring': ['purine', 'pyrimidine', 'purine', 'pyrimidine']})
nucls['name']
nucls['letter'][2]
tata_counts = pd.DataFrame({'A': [ 16, 352, 3, 354, 268, 360, 222, 155],
'C': [ 46, 0, 10, 0, 0, 3, 2, 44],
'G': [ 18, 2, 2, 5, 0, 20, 44, 157],
'T': [ 309, 35, 374, 30, 121, 6, 121, 33]})
tata_counts / 389
tata_probs['T'][1]
for position, nt in enumerate(sequ):
print('position = ' + str(position) + ', nt = ' + str(nt))
prob = 1
for position, nt in enumerate(sequ):
p = tata_probs[nt][position]
prob = prob * p
print(position, nt, p, prob)
tata_counts_pseudo = tata_counts + 1
def likelihood_tata(sequ):
prob = 1
for position, nt in enumerate(sequ):
p = tata_probs[nt][position]
prob = prob * p
print(position, nt, p, prob)
return(prob)
TATATAAG
TATAAATG
TATAAAAA
background = pd.Series({'A': 0.31, 'C': 0.19, 'G': 0.19, 'T': 0.31})
print(likelihood_tata('TATAAAAG') / likelihood_background('TATAAAAG'))
print(likelihood_tata('ACGCGCCT') / likelihood_background('ACGCGCCT'))
print( (likelihood_tata('TATATAAG') + likelihood_tata('TATAAATG') + likelihood_tata('TATAAAAA'))
/ (likelihood_background('TATATAAG') + likelihood_background('TATAAATG') + likelihood_background('TATAAAAA')) )
def likelihood_ratio(sequ):
return(likelihood_tata(sequ) / likelihood_background(sequ))
cdc19_prm = 'TATGATGCTAGGTACCTTTAGTGTCTTCCTAAAAAAAAAAAAAGGCTCGCCATCAAAACGATATTCGTTGGCTTTTTTTTCTGAATTATAAATACTCTTTGGTAACTTTTCATTTCCAAGAACCTCTTTTTTCCAGTTATATCATG'
alphabet = 'abcdefghijklmnopqrstuvwxyz'
alphabet[2:6]
likelihood_ratio(cdc19_prm[0:8])
likelihood_ratio(cdc19_prm[1:9])
for start in range(0, len(cdc19_prm) - 8):
print(str(start) + ' ' + cdc19_prm[start:start+8])
| 0.410284 | 0.994402 |
# "[Kaggle] Otto Group"
> "Feature가 명확하지 않음"
- toc: true
- branch: master
- badges: true
- comments: true
- author: Rauk
- categories: [SSUDA, Machine Learning]
## Data fields
- id - an anonymous id unique to a product
- feat_1, feat_2, ..., feat_93 - the various features of a product
- target - the class of a product
```
from google.colab import drive
drive.mount('/content/drive')
from google.colab import files
files.upload()
!mkdir -p ~/.kaggle
!cp kaggle.json ~/.kaggle/
!chmod 600 /root/.kaggle/kaggle.json
!kaggle competitions download -c otto-group-product-classification-challenge
!unzip sampleSubmission.csv.zip
!unzip test.csv.zip
!unzip train.csv.zip
```
## CODING
```
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import pandas as pd
train = pd.read_csv('train.csv')
train.head(7)
test = pd.read_csv('test.csv')
test.head(7)
sns.countplot(x = train.target)
class_to_order = dict()
order_to_class = dict()
for idx, col in enumerate(train.target.unique()):
order_to_class[idx] = col
class_to_order[col] = idx
train["target_ord"] = train["target"].map(class_to_order).astype("int16")
feature_columns = [col for col in train.columns if col.startswith("feat_")]
target_column = ["target_ord"]
order_to_class
class_to_order
```
## Skewness
```
skew = []
for i in train[feature_columns].columns:
skew.append(train[str(i)].skew())
skew_df = pd.DataFrame({'Feature': train[feature_columns].columns, 'Skewness': skew})
skew_df.plot(kind='bar',figsize=(18,10))
```
## Quantile Transformer
- We are now going to apply the QuantileTransformer from scikit-learn. I first used StandardScaler but found that there was no change in the skew value of the features.
- QuantileTransformer는 각 형상의 확률 밀도 함수가 균등 또는 가우스 분포에 매핑되도록 비선형 변환을 적용한다. 이 경우, 특이치를 포함한 모든 데이터는 [0,1] 범위의 균등 분포에 매핑되어 특이치와 inlier를 구분할 수 없게 됩니다.
- 기본적으로 1000개 분위를 사용하여 데이터를 '균등분포' 시킵니다.Robust처럼 이상치에 민감X, 0~1사이로 압축합니다. -> 이후 output_distribution='normal'을 적용하면 정규분포형태의 값으로 나타난다.
- RobustScaler와 QuantileTransformer는 훈련 세트에서 특이치를 추가하거나 제거하면 거의 동일한 변환을 얻을 수 있다는 점에서 특이치에 강하다. 그러나 RobustScaler와 반대로 QuantileTransformer는 또한 이상치를 사전 정의된 범위 경계(0 및 1)로 설정하여 자동으로 축소한다. 이로 인해 극단값의 포화 아티팩트가 발생할 수 있습니다.
```
from sklearn.preprocessing import QuantileTransformer
train[feature_columns] = QuantileTransformer(copy=False, output_distribution='normal').fit_transform(train[feature_columns])
test[feature_columns] = QuantileTransformer(copy=False, output_distribution='normal').fit_transform(test[feature_columns])
# 조정 후 Skewness
skew = []
for i in train[feature_columns].columns:
skew.append(train[str(i)].skew())
skew_df = pd.DataFrame({'Feature': train[feature_columns].columns, 'Skewness': skew})
skew_df.plot(kind='bar',figsize=(18,10))
# check features for skew
skew_feats = train[feature_columns].skew().sort_values(ascending=False)
skewness = pd.DataFrame({'Skew': skew_feats})
skewness = skewness[abs(skewness) > 3.75].dropna()
skewed_features = skewness.index.values.tolist()
skewed_features
train_new = train.drop(skewed_features, axis = 1)
train_new
```
## Train_test 분리
```
from sklearn.model_selection import train_test_split
from sklearn.metrics import log_loss
X_train, X_valid, y_train, y_valid = train_test_split(
train_new.drop(['id', 'target', 'target_ord'], axis = 1),
train_new[target_column],
test_size = 0.275,
random_state = 7,
stratify = train_new[target_column]
)
```
## KNN
```
from sklearn.neighbors import KNeighborsClassifier
knc = KNeighborsClassifier(n_neighbors = 25, weights = 'distance')
knc.fit(X_train, y_train)
yhat = knc.predict(X_valid)
from sklearn.metrics import classification_report, confusion_matrix
result = confusion_matrix(y_valid, yhat)
print("Confusion Matrix:")
print(result)
result1 = classification_report(y_valid, yhat)
print("Classification Report:")
print(result1)
yhat_KNN = knc.predict_proba(X_valid)
logloss_KNN = log_loss(y_valid, yhat_KNN)
print('Log loss using KNN classifier:', logloss_KNN)
```
## Support Vector Machine
```
from sklearn import svm
svm = svm.SVC(kernel = 'rbf', probability = True, random_state = 7)
svm.fit(X_train, y_train)
yhat = svm.predict(X_valid)
yhat_svm = svm.predict_proba(X_valid)
logloss_svm = log_loss(y_valid, yhat_svm)
print('Logloss using Support Vector Machines:', logloss_svm)
cnf_matrix = confusion_matrix(y_valid, yhat, labels = train_new.target_ord.unique().tolist())
plt.figure()
plot_confusion_matrix(cnf_matrix, classes = train_new.target.unique().tolist())
print('Classification Report:')
print(classification_report(y_valid, yhat))
```
## Catboost
```
pip install catboost
from catboost import CatBoostClassifier
CBC_params = {
'iterations': 5000,
'od_wait': 250,
'use_best_model': True,
'loss_function': 'MultiClass',
'eval_metric': 'MultiClass',
'leaf_estimation_method': 'Newton',
'bootstrap_type': 'Bernoulli',
'subsample': 0.5,
'learning_rate': 0.08,
'l2_leaf_reg': 0.4, #L2 Regularization
'random_strength': 10, #amount of randomness to use for scoring splits when tree structure is selected
'depth': 7, #Tree depth
'min_data_in_leaf': 3, #minimum number of training samples in a leaf
'leaf_estimation_iterations': 4, #Earlier = 7
'task_type': 'GPU',
'border_count': 128, #Number of splits for numerical features
}
cbc = CatBoostClassifier(**CBC_params)
cbc.fit(X_train, y_train,
eval_set = [(X_valid, y_valid)],
early_stopping_rounds = 100,
)
yhat_CBC = cbc.predict_proba(X_valid)
logloss_CBC = log_loss(y_valid, yhat_CBC)
print('Log loss using CatBoost Classifier:', logloss_CBC)
```
## Submission
```
submission = pd.read_csv("sampleSubmission.csv")
test.drop(skewed_features, axis = 1, inplace = True)
test.drop("id", axis = 1)
pred = cbc.predict_proba(test.drop("id", axis = 1))
res = pd.concat([pd.DataFrame(test.id.copy()), pd.DataFrame(pred)], axis = 1)
res.columns = submission.columns
res
res.to_csv("submission.csv", index = False)
!kaggle competitions submit -c otto-group-product-classification-challenge -f submission.csv -m "Message"
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive')
from google.colab import files
files.upload()
!mkdir -p ~/.kaggle
!cp kaggle.json ~/.kaggle/
!chmod 600 /root/.kaggle/kaggle.json
!kaggle competitions download -c otto-group-product-classification-challenge
!unzip sampleSubmission.csv.zip
!unzip test.csv.zip
!unzip train.csv.zip
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import pandas as pd
train = pd.read_csv('train.csv')
train.head(7)
test = pd.read_csv('test.csv')
test.head(7)
sns.countplot(x = train.target)
class_to_order = dict()
order_to_class = dict()
for idx, col in enumerate(train.target.unique()):
order_to_class[idx] = col
class_to_order[col] = idx
train["target_ord"] = train["target"].map(class_to_order).astype("int16")
feature_columns = [col for col in train.columns if col.startswith("feat_")]
target_column = ["target_ord"]
order_to_class
class_to_order
skew = []
for i in train[feature_columns].columns:
skew.append(train[str(i)].skew())
skew_df = pd.DataFrame({'Feature': train[feature_columns].columns, 'Skewness': skew})
skew_df.plot(kind='bar',figsize=(18,10))
from sklearn.preprocessing import QuantileTransformer
train[feature_columns] = QuantileTransformer(copy=False, output_distribution='normal').fit_transform(train[feature_columns])
test[feature_columns] = QuantileTransformer(copy=False, output_distribution='normal').fit_transform(test[feature_columns])
# 조정 후 Skewness
skew = []
for i in train[feature_columns].columns:
skew.append(train[str(i)].skew())
skew_df = pd.DataFrame({'Feature': train[feature_columns].columns, 'Skewness': skew})
skew_df.plot(kind='bar',figsize=(18,10))
# check features for skew
skew_feats = train[feature_columns].skew().sort_values(ascending=False)
skewness = pd.DataFrame({'Skew': skew_feats})
skewness = skewness[abs(skewness) > 3.75].dropna()
skewed_features = skewness.index.values.tolist()
skewed_features
train_new = train.drop(skewed_features, axis = 1)
train_new
from sklearn.model_selection import train_test_split
from sklearn.metrics import log_loss
X_train, X_valid, y_train, y_valid = train_test_split(
train_new.drop(['id', 'target', 'target_ord'], axis = 1),
train_new[target_column],
test_size = 0.275,
random_state = 7,
stratify = train_new[target_column]
)
from sklearn.neighbors import KNeighborsClassifier
knc = KNeighborsClassifier(n_neighbors = 25, weights = 'distance')
knc.fit(X_train, y_train)
yhat = knc.predict(X_valid)
from sklearn.metrics import classification_report, confusion_matrix
result = confusion_matrix(y_valid, yhat)
print("Confusion Matrix:")
print(result)
result1 = classification_report(y_valid, yhat)
print("Classification Report:")
print(result1)
yhat_KNN = knc.predict_proba(X_valid)
logloss_KNN = log_loss(y_valid, yhat_KNN)
print('Log loss using KNN classifier:', logloss_KNN)
from sklearn import svm
svm = svm.SVC(kernel = 'rbf', probability = True, random_state = 7)
svm.fit(X_train, y_train)
yhat = svm.predict(X_valid)
yhat_svm = svm.predict_proba(X_valid)
logloss_svm = log_loss(y_valid, yhat_svm)
print('Logloss using Support Vector Machines:', logloss_svm)
cnf_matrix = confusion_matrix(y_valid, yhat, labels = train_new.target_ord.unique().tolist())
plt.figure()
plot_confusion_matrix(cnf_matrix, classes = train_new.target.unique().tolist())
print('Classification Report:')
print(classification_report(y_valid, yhat))
pip install catboost
from catboost import CatBoostClassifier
CBC_params = {
'iterations': 5000,
'od_wait': 250,
'use_best_model': True,
'loss_function': 'MultiClass',
'eval_metric': 'MultiClass',
'leaf_estimation_method': 'Newton',
'bootstrap_type': 'Bernoulli',
'subsample': 0.5,
'learning_rate': 0.08,
'l2_leaf_reg': 0.4, #L2 Regularization
'random_strength': 10, #amount of randomness to use for scoring splits when tree structure is selected
'depth': 7, #Tree depth
'min_data_in_leaf': 3, #minimum number of training samples in a leaf
'leaf_estimation_iterations': 4, #Earlier = 7
'task_type': 'GPU',
'border_count': 128, #Number of splits for numerical features
}
cbc = CatBoostClassifier(**CBC_params)
cbc.fit(X_train, y_train,
eval_set = [(X_valid, y_valid)],
early_stopping_rounds = 100,
)
yhat_CBC = cbc.predict_proba(X_valid)
logloss_CBC = log_loss(y_valid, yhat_CBC)
print('Log loss using CatBoost Classifier:', logloss_CBC)
submission = pd.read_csv("sampleSubmission.csv")
test.drop(skewed_features, axis = 1, inplace = True)
test.drop("id", axis = 1)
pred = cbc.predict_proba(test.drop("id", axis = 1))
res = pd.concat([pd.DataFrame(test.id.copy()), pd.DataFrame(pred)], axis = 1)
res.columns = submission.columns
res
res.to_csv("submission.csv", index = False)
!kaggle competitions submit -c otto-group-product-classification-challenge -f submission.csv -m "Message"
| 0.504883 | 0.789964 |
# How do I make my custom QRenderer
This notebook demonstrates how to create a user-defined QRenderer. These steps are needed when you intend to configure Qiskit Metal to interface with your favorite (and presently not supported) external tool/simulator.
To execute all the steps in this notebook, you will be modifying the core code. Therefore, we assume that you have installed qiskit-metal from the Github repository, using the README instructions, which will install qiskit-metal in 'editable' mode.
### Preparations
To get started, enable [automatic reloading of modules] (https://ipython.readthedocs.io/en/stable/config/extensions/autoreload.html?highlight=autoreload). This will allow you to modify the source code and immediately observe the effects of the changes in the notebook, without the need for reinitiating the kernel or reinstalling the package.
```
%load_ext autoreload
%autoreload 2
```
Pre-load all the Qiskit Metal libraries that are needed for the rest of this notebook.
```
import qiskit_metal as metal
from qiskit_metal import designs, draw
from qiskit_metal import MetalGUI, Dict, Headings
from qiskit_metal.qlibrary.qubits.transmon_pocket import TransmonPocket
from qiskit_metal.qlibrary.qubits.transmon_cross import TransmonCross
from qiskit_metal.renderers.renderer_gds.gds_renderer import QGDSRenderer
```
## Integrating the user-defined renderer with the rest of Qiskit Metal
### Architectural insights
This section will give you the architectural overview of how Qiskit Metal manages renderers, and how you can add your own.
We will refer to your custom renderer as the `skeleton` renderer, since we will not code tool-specific methods/classes, but only worry about how to bootstrap one without functionality.
Note that all renderers (existing `gds`, `hfss` and `q3d` as well as the newly created `skeleton`) have to be identified in the config.py file. Therefore, you will be required to modify the `qiskit_metal/config.py` file.
The following image describes how the QRenderer (superclass of all renderers) interacts with the rest of Qiskit Metal. The key take-away is that creating a QDesign class object initiates all the QRenderer subclass objects as well. Specifically, the `QDesign.__init__()` method reads the `renderers_to_load` dictionary from the config.py file, which enumerates which QRenderers subclasses need to be instantiated. After instantiating the renderer objects, the `QDesign.__init__()` registers them in the `QDesign._renderers` dictionary for later reference.

### QRenderer inheritance and subclass management
Presently, the config.py file references three QRenderers subclasses, which handle the `gds`, `hfss` and `q3d` interfaces. Explicitly, QGDSRenderer is a subclass of QRenderer. Both QHFSSRenderer and QQ3DRenderer subclass from QAnsysRenderer. The class QAnsysRenderer is a subclass of QRenderer.
The `renderers_to_load` dictionary in the config.py file needs to be updated to inform Qiskit Metal about the new renderer `skeleton` you are going to create. `renderers_to_load` stores the explicit path and class name so that Qiskit Metal will load to memory by default only those specified renderers. This happens during the `QDesign.__init__()`.
For this notebook, we created a sample class named QSkeletonRender in `tutorials/resources/skeleton_renderer`. This class is your skeleton to develop a new QRenderer subclass. Feel free to edit the class content at will. If you change the path to the file, please reflect that in the remainder of this notebook. Presently, you can find the production QRenderers subclasses in the package directory `qiskit_metal.renderers`.
### TODO: Let's tell Qiskit Metal where to find your new custom renderer
As the first step, please locate and open the file config.py in the qiskit-metal package and edit the `renderers_to_load` dictionary to add the new renderer `skeleton`, like so:
`renderers_to_load = Dict(
hfss=Dict(path_name='qiskit_metal.renderers.renderer_ansys.hfss_renderer',
class_name='QHFSSRenderer'),
q3d=Dict(path_name='qiskit_metal.renderers.renderer_ansys.q3d_renderer',
class_name='QQ3DRenderer'),
gds=Dict(path_name='qiskit_metal.renderers.renderer_gds.gds_renderer',
class_name='QGDSRenderer'),
skeleton=Dict(path_name='tutorials.resources.skeleton_renderer',
class_name='QSkeletonRenderer'),
)`
### TODO: Add the following functions to make your renderer compatible with the default structure
These lines may be added immediately after the init function.
def _initiate_renderer(self):
"""Not used by the skeleton renderer at this time. only returns True.
"""
return True
def _close_renderer(self):
"""Not used by the skeleton renderer at this time. only returns True.
"""
return True
def render_design(self):
"""Export the design to Skeleton."""
self.write_qgeometry_table_names_to_file(
file_name=self.options.file_geometry_tables,
highlight_qcomponents=[])
### Confirm QDesign is able to load your renderer
Create a QDesign instance.
```
design = designs.DesignPlanar()
```
If you modified the config.py correctly, the previous command should have instantiated and registered the `skeleton` renderer. Verify that by inspecting the renderers dictionary property of the QDesign instance.
If executing the next cell does not show the `skeleton` renderer in the list, please make sure you correctly updated the `config.py` file, next you could try resetting the jupyter notebook kernel, or restarting jupyter notebook.
```
design.renderers.keys()
```
For convenience, let's create a short-handle alias to refer to the renderer during the remainder of this notebook.
```
a_skeleton = design.renderers.skeleton
```
## Interact with your new user-custom renderer
### Verify and modify the options of your renderer
In the QSkeletonRenderer class some sample `default_options` class parameter has been defined. <br>
`default_options = Dict(
number_of_bones='206')
`
The instance `a_skeleton` will contain a dictionary `options` that is initiated using the `default_options`. (This works similarly to `options` for QComponents, which has been introduced in the jupyter notebooks found in the folder: `tutorials/2 Front End User`.)
You can access and modify the options in the QSkeletonRenderer class instance as follows. For example, let's update the skeleton from that of a human to that of a dog (319 bones).
```
a_skeleton.options.number_of_bones = '319'
a_skeleton.options
```
Original values will continue being accessible like so:
```
a_skeleton.get_template_options(design)
```
### Populate a sample QDesign to demonstrate interaction with the renderer
This portion is described in notebooks within directory `tutorials/2 Front End User`. Some of the options have been made distinctly different to show what can be done, i.e., fillet value, fillet='25um', varies for each cpw. However, that may not be what user will implement for their design.
```
gui = MetalGUI(design)
design.overwrite_enabled = True
from qiskit_metal.qlibrary.qubits.transmon_pocket import TransmonPocket
from qiskit_metal.qlibrary.tlines.meandered import RouteMeander
## Custom options for all the transmons
options = dict(
pad_width = '425 um',
pad_gap = '80 um',
pocket_height = '650um',
# Adding 4 connectors (see below for defaults)
connection_pads=dict(
a = dict(loc_W=+1,loc_H=+1),
b = dict(loc_W=-1,loc_H=+1, pad_height='30um'),
c = dict(loc_W=+1,loc_H=-1, pad_width='200um'),
d = dict(loc_W=-1,loc_H=-1, pad_height='50um')
)
)
## Create 4 TransmonPockets
q1 = TransmonPocket(design, 'Q1', options = dict(
pos_x='+2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_02', **options))
q2 = TransmonPocket(design, 'Q2', options = dict(
pos_x='+0.0mm', pos_y='-0.9mm', orientation = '90', gds_cell_name='FakeJunction_02', **options))
q3 = TransmonPocket(design, 'Q3', options = dict(
pos_x='-2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_01',**options))
q4 = TransmonPocket(design, 'Q4', options = dict(
pos_x='+0.0mm', pos_y='+0.9mm', orientation = '90', gds_cell_name='my_other_junction', **options))
options = Dict(
meander=Dict(
lead_start='0.1mm',
lead_end='0.1mm',
asymmetry='0 um')
)
def connect(component_name: str, component1: str, pin1: str, component2: str, pin2: str,
length: str, asymmetry='0 um', flip=False, fillet='50um'):
"""Connect two pins with a CPW."""
myoptions = Dict(
fillet=fillet,
pin_inputs=Dict(
start_pin=Dict(
component=component1,
pin=pin1),
end_pin=Dict(
component=component2,
pin=pin2)),
lead=Dict(
start_straight='0.13mm',
end_straight='0.13mm'
),
total_length=length)
myoptions.update(options)
myoptions.meander.asymmetry = asymmetry
myoptions.meander.lead_direction_inverted = 'true' if flip else 'false'
return RouteMeander(design, component_name, myoptions)
asym = 90
cpw1 = connect('cpw1', 'Q1', 'd', 'Q2', 'c', '5.7 mm', f'+{asym}um', fillet='25um')
cpw2 = connect('cpw2', 'Q3', 'c', 'Q2', 'a', '5.6 mm', f'-{asym}um', flip=True, fillet='100um')
cpw3 = connect('cpw3', 'Q3', 'a', 'Q4', 'b', '5.5 mm', f'+{asym}um', fillet='75um')
cpw4 = connect('cpw4', 'Q1', 'b', 'Q4', 'd', '5.8 mm', f'-{asym}um', flip=True)
gui.rebuild()
gui.autoscale()
```
### Export list of the design QGeometries to file using your custom QSkeletonRenderer
The QSkeletonRenderer class contains several sample methods. Let's use one intended to print out the name of the QGeometry tables to a text file (Remember: QGeometry contains the list of the raw layout shapes that compose the design, which we have created in the previous cell).
```
a_skeleton.write_qgeometry_table_names_to_file('./simple_output.txt')
```
Here another example where we sub select a single QComponent instance (`cpw1`) of type RouteMeander. This will only export the name of tables containing shapes related to that instance, which in this case is only paths, and not junctions or poly.
```
a_skeleton.write_qgeometry_table_names_to_file('./simple_output_cpw1.txt',highlight_qcomponents=['cpw1'])
```
## What if my new tool requires additional parameters that Qiskit Metal does not natively support?
### QRenderers can request special tool parameters from the user
External tools, such as Ansys, might require special parameters to be able to render (interpret) correctly the QGeometries that Qiskit Metal wants to pass (render) to them. Every tool might need a different set of special parameters; thus we architected a solution that allows individual QRenderers to communicate to qiskit-metal what additional parameters their associated tool requires.
The implementation consists of enabling the QRenderers to add new columns (parameters) and tables (geometry types) to the QGeometry table collection. The QRenderer should also specify what is the default value to use to populate those columns/tables. The user can then update them to a value different than default by editing them at run-time, which can happen through the QComponent options (or directly, for advanced users). Note that older QComponents remain valid also for newer QRenderers, thanks to the defaults provided by the QRenderer.
Our QSkeletonRenderer class for example is designed to add a `a_column_name` column to the `junction` table, with default value `a_default_value`. This is implemented by creating the following class parameter:
<br>`element_table_data = dict(junction=dict(a_column_name='a_default_value'))`
Note that the final column name will be `skeleton_a_column_name` because the provided column name is prefixed with the renderer name (`QSkeletonRenderer.name`).
The method that executes the magic described above is `QRenderer.load()`, which is called from the `QSkeletonRenderer.__init__()`.
### Let's observe and update the additional properties that our QSkeletonRenderer needs
First, make sure that the registration of the QRenderer added the additional parameter as expected. Search for the column `skeleton_a_column_name` in the qgeometry table `junction`
```
design.qgeometry.tables['junction']
```
If you cannot locate the new column (might need to scroll to the far right), then something must be amiss, so please start over this notebook and execute all of the cells.
Once you can locate the new column, and observe the set default value, let's now try to update the value in the column by modifying the design of the correspondent QComponent. All we need to do is pass a different set of options to the component, like so:
```
q1.options.skeleton_a_column_name = 'q1 skeleton'
q2.options.skeleton_a_column_name = 'q2 skeleton'
q3.options.skeleton_a_column_name = 'q3 skeleton'
q4.options.skeleton_a_column_name = 'q4 skeleton'
gui.rebuild()
gui.autoscale()
design.qgeometry.tables['junction']
```
You can also create the components by directly passing the options you know the renderer will require, like so:
```
q1.delete()
q2.delete()
q3.delete()
q4.delete()
q1 = TransmonPocket(design, 'Q1', options = dict(
pos_x='+2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_02', skeleton_a_column_name='q1 skeleton 2', **options))
q2 = TransmonPocket(design, 'Q2', options = dict(
pos_x='+0.0mm', pos_y='-0.9mm', orientation = '90', gds_cell_name='FakeJunction_02', skeleton_a_column_name='q2 skeleton 2', **options))
q3 = TransmonPocket(design, 'Q3', options = dict(
pos_x='-2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_01', skeleton_a_column_name='q3 skeleton 2', **options))
q4 = TransmonPocket(design, 'Q4', options = dict(
pos_x='+0.0mm', pos_y='+0.9mm', orientation = '90', gds_cell_name='my_other_junction', skeleton_a_column_name='q4 skeleton 2', **options))
design.qgeometry.tables['junction']
```
## Can my user-defined renderer change/interact with the design?
### Accessing information and methods
It is possible that the result of a rendering action, or analysis requires a design update back to qiskit-metal. This can be achieved without the user intervention by simply controlling the QDesign instance from within the QRenderer.
Just as an example, the next three cells inspect the current design QComponent, QGeometry table, and QRenderer names.
```
a_skeleton.design.components.keys()
a_skeleton.design.qgeometry.tables.keys()
a_skeleton.design.renderers.keys()
```
The base QRenderer class comes with useful methods to more easily access some of the information. You will find more method described in the QRenderer documentation. The example below for example returns the QComponent's IDs.
```
a_skeleton.get_unique_component_ids(highlight_qcomponents = ['Q1', 'Q1', 'Q4', 'cpw1', 'cpw2', 'cpw3', 'cpw4'])
```
The following instead shows three ways to access the same QGeometry table.
```
a_skeleton.design.components['Q1'].qgeometry_table('junction') # via QComonent name
a_skeleton.design._components[9].qgeometry_table('junction') # via QComponent ID
q1.qgeometry_table('junction') # via the QComponent instance
```
The method `QSkeletonRenderer.get_qgeometry_tables_for_skeleton()` exemplifies how to iterate through chips and tables.
```
from tutorials.resources.skeleton_renderer import QSkeletonRenderer
?QSkeletonRenderer.get_qgeometry_tables_for_skeleton
```
### Communicate state
We can also interact with any other method of the QDesign instance, for example we can generate a warning into the logger as shown in the next cell. This is particularly useful to document problems with the user defined QRenderer execution
```
# Purposefully generates a warning message.
a_skeleton.logger.warning('Show a warning message for plugin developer.')
```
## Qiskit Metal Version
```
metal.about();
# This command below is if the user wants to close the Metal GUI.
gui.main_window.close()
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import qiskit_metal as metal
from qiskit_metal import designs, draw
from qiskit_metal import MetalGUI, Dict, Headings
from qiskit_metal.qlibrary.qubits.transmon_pocket import TransmonPocket
from qiskit_metal.qlibrary.qubits.transmon_cross import TransmonCross
from qiskit_metal.renderers.renderer_gds.gds_renderer import QGDSRenderer
design = designs.DesignPlanar()
design.renderers.keys()
a_skeleton = design.renderers.skeleton
a_skeleton.options.number_of_bones = '319'
a_skeleton.options
a_skeleton.get_template_options(design)
gui = MetalGUI(design)
design.overwrite_enabled = True
from qiskit_metal.qlibrary.qubits.transmon_pocket import TransmonPocket
from qiskit_metal.qlibrary.tlines.meandered import RouteMeander
## Custom options for all the transmons
options = dict(
pad_width = '425 um',
pad_gap = '80 um',
pocket_height = '650um',
# Adding 4 connectors (see below for defaults)
connection_pads=dict(
a = dict(loc_W=+1,loc_H=+1),
b = dict(loc_W=-1,loc_H=+1, pad_height='30um'),
c = dict(loc_W=+1,loc_H=-1, pad_width='200um'),
d = dict(loc_W=-1,loc_H=-1, pad_height='50um')
)
)
## Create 4 TransmonPockets
q1 = TransmonPocket(design, 'Q1', options = dict(
pos_x='+2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_02', **options))
q2 = TransmonPocket(design, 'Q2', options = dict(
pos_x='+0.0mm', pos_y='-0.9mm', orientation = '90', gds_cell_name='FakeJunction_02', **options))
q3 = TransmonPocket(design, 'Q3', options = dict(
pos_x='-2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_01',**options))
q4 = TransmonPocket(design, 'Q4', options = dict(
pos_x='+0.0mm', pos_y='+0.9mm', orientation = '90', gds_cell_name='my_other_junction', **options))
options = Dict(
meander=Dict(
lead_start='0.1mm',
lead_end='0.1mm',
asymmetry='0 um')
)
def connect(component_name: str, component1: str, pin1: str, component2: str, pin2: str,
length: str, asymmetry='0 um', flip=False, fillet='50um'):
"""Connect two pins with a CPW."""
myoptions = Dict(
fillet=fillet,
pin_inputs=Dict(
start_pin=Dict(
component=component1,
pin=pin1),
end_pin=Dict(
component=component2,
pin=pin2)),
lead=Dict(
start_straight='0.13mm',
end_straight='0.13mm'
),
total_length=length)
myoptions.update(options)
myoptions.meander.asymmetry = asymmetry
myoptions.meander.lead_direction_inverted = 'true' if flip else 'false'
return RouteMeander(design, component_name, myoptions)
asym = 90
cpw1 = connect('cpw1', 'Q1', 'd', 'Q2', 'c', '5.7 mm', f'+{asym}um', fillet='25um')
cpw2 = connect('cpw2', 'Q3', 'c', 'Q2', 'a', '5.6 mm', f'-{asym}um', flip=True, fillet='100um')
cpw3 = connect('cpw3', 'Q3', 'a', 'Q4', 'b', '5.5 mm', f'+{asym}um', fillet='75um')
cpw4 = connect('cpw4', 'Q1', 'b', 'Q4', 'd', '5.8 mm', f'-{asym}um', flip=True)
gui.rebuild()
gui.autoscale()
a_skeleton.write_qgeometry_table_names_to_file('./simple_output.txt')
a_skeleton.write_qgeometry_table_names_to_file('./simple_output_cpw1.txt',highlight_qcomponents=['cpw1'])
design.qgeometry.tables['junction']
q1.options.skeleton_a_column_name = 'q1 skeleton'
q2.options.skeleton_a_column_name = 'q2 skeleton'
q3.options.skeleton_a_column_name = 'q3 skeleton'
q4.options.skeleton_a_column_name = 'q4 skeleton'
gui.rebuild()
gui.autoscale()
design.qgeometry.tables['junction']
q1.delete()
q2.delete()
q3.delete()
q4.delete()
q1 = TransmonPocket(design, 'Q1', options = dict(
pos_x='+2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_02', skeleton_a_column_name='q1 skeleton 2', **options))
q2 = TransmonPocket(design, 'Q2', options = dict(
pos_x='+0.0mm', pos_y='-0.9mm', orientation = '90', gds_cell_name='FakeJunction_02', skeleton_a_column_name='q2 skeleton 2', **options))
q3 = TransmonPocket(design, 'Q3', options = dict(
pos_x='-2.55mm', pos_y='+0.0mm', gds_cell_name='FakeJunction_01', skeleton_a_column_name='q3 skeleton 2', **options))
q4 = TransmonPocket(design, 'Q4', options = dict(
pos_x='+0.0mm', pos_y='+0.9mm', orientation = '90', gds_cell_name='my_other_junction', skeleton_a_column_name='q4 skeleton 2', **options))
design.qgeometry.tables['junction']
a_skeleton.design.components.keys()
a_skeleton.design.qgeometry.tables.keys()
a_skeleton.design.renderers.keys()
a_skeleton.get_unique_component_ids(highlight_qcomponents = ['Q1', 'Q1', 'Q4', 'cpw1', 'cpw2', 'cpw3', 'cpw4'])
a_skeleton.design.components['Q1'].qgeometry_table('junction') # via QComonent name
a_skeleton.design._components[9].qgeometry_table('junction') # via QComponent ID
q1.qgeometry_table('junction') # via the QComponent instance
from tutorials.resources.skeleton_renderer import QSkeletonRenderer
?QSkeletonRenderer.get_qgeometry_tables_for_skeleton
# Purposefully generates a warning message.
a_skeleton.logger.warning('Show a warning message for plugin developer.')
metal.about();
# This command below is if the user wants to close the Metal GUI.
gui.main_window.close()
| 0.574992 | 0.959154 |
```
import os
os.environ['CUDA_VISIBLE_DEVICES'] = '5'
from pycocotools.coco import COCO
site = 'train'
img_dir = f'./data/yunnan_512/{site}/JPEGImages'
json_path = f'./data/yunnan_512/{site}/{site}.json'
coco = COCO(json_path)
img_ids = set(_['image_id'] for _ in coco.anns.values())
from mmdet.apis import init_detector, inference_detector, show_result_pyplot
import mmcv
import glob
work_dir = './checkpoints/yunnan'
config_file = glob.glob(os.path.join(work_dir, '*.py'))[0]
checkpoint_file = glob.glob(os.path.join(work_dir, '*.pth'))[0]
cfg_options = {}
cfg_options = {'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.kernel_size': 3,
'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.type': 'gaussian'}
model = init_detector(config_file, checkpoint_file, device='cuda:0', cfg_options=cfg_options)
import numpy as np
import mmcv
import cv2
import torch
import numpy as np
import mmcv
import cv2
import torch
import tifffile as tiff
def show_mask_result(img, result, score_thr):
# 基于掩码,提取轮廓显示
bbox_result, segm_result = result[:2]
bboxes = np.vstack(bbox_result)
labels = [
np.full(bbox.shape[0], i, dtype=np.int32)
for i, bbox in enumerate(bbox_result)
]
labels = np.concatenate(labels)
# draw segmentation masks
segms = None
if segm_result is not None and len(labels) > 0: # non empty
segms = mmcv.concat_list(segm_result)
if isinstance(segms[0], torch.Tensor):
segms = torch.stack(segms, dim=0).detach().cpu().numpy()
else:
segms = np.stack(segms, axis=0)
scores = bboxes[:, -1]
inds = scores > score_thr
bboxes = bboxes[inds, :]
labels = labels[inds]
if segms is not None:
segms = segms[inds, ...]
points = []
for i in range(len(bboxes)):
mask = segms[i].astype(np.uint8)
contours, _ = cv2.findContours(mask[..., None], cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
cv2.drawContours(img, contours, -1, (0,0,255), 2)
for contour in contours:
points.append(len(contour))
return img, sum(points) / (len(points) + 1e-5)
def show_polygon_result(img, result, score_thr):
# 根据点结果,显示轮廓
bbox_result, _, polygon_result = result
bboxes = np.vstack(bbox_result)
scores = bboxes[:, -1]
inds = np.nonzero(scores > score_thr)[0]
points = []
for i in inds:
poly = np.array(polygon_result[0][i]).reshape(-1, 2).astype(np.int32)
points.append(len(poly))
cv2.polylines(img, [poly], True, (0, 0, 255), 2)
return img, sum(points) / (len(points) + 1e-5)
def show_det_result(img, result, score_thr):
bbox_result = result
bboxes = np.vstack(bbox_result)
scores = bboxes[:, -1]
inds = scores > score_thr
bboxes = bboxes[inds, :]
for box in bboxes:
x1, y1, x2, y2 = box.astype(np.int32).tolist()[:4]
cv2.rectangle(img, (x1, y1), (x2, y2), (0, 0, 255), 2)
return img, 4
def show_result(img_path, result, score_thr, ret_det=False):
if os.path.splitext(img_path)[1].lower() == '.tif':
img = tiff.imread(img_path)
img = img[:, :, :3][:, :, ::-1]
img = img.copy()
else:
img = cv2.imread(img_path)
if ret_det:
if isinstance(result, tuple):
result = result[0]
return show_det_result(img, result, score_thr=score_thr)
if len(result) == 3:
return show_polygon_result(img, result, score_thr=score_thr)
else:
return show_mask_result(img, result, score_thr=score_thr)
print(len(img_ids))
# output_dir = './visual_yunnan_val_poly'
# os.makedirs(output_dir, exist_ok=True)
import matplotlib.pyplot as plt
from tqdm import tqdm
img_ids = list(img_ids)
for bi, img_id in enumerate(tqdm(img_ids)):
fn = coco.load_imgs([img_id])[0]['file_name']
img_path = os.path.join(img_dir, fn)
result = inference_detector(model, img_path)
# show_result_pyplot(model, img_path, result, score_thr=0.2)
img, _ = show_result(img_path, result, score_thr=0.6)
img = img[:, :, ::-1]
plt.figure(figsize=(15, 10))
plt.axis('off')
plt.imshow(img)
plt.show()
# out_path = os.path.join(output_dir, fn)
# cv2.imwrite(out_path, img)
if bi > 10:
break
!ls data/yunnan_512_1/val/JPEGImages | wc -l
```
### RUN POLYRNN
```
import os
os.environ['CUDA_VISIBLE_DEVICES'] = '1'
from subprocess import Popen
import glob
test_work_dir = './venus_last_tf/15'
cfg_options = ''
cfg_options += 'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.kernel_size=3 '
cfg_options += 'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.type=gaussian'
log = os.path.join(test_work_dir, 'result_out.log')
# config = './configs/polygon/polyrnn_r50_fpn_1x_building_0329_3.py'
config = glob.glob(os.path.join(test_work_dir, '*.py'))[0]
checkpoint = os.path.join(test_work_dir, 'latest.pth')
if cfg_options:
execmd = f'python tools/test.py {config} {checkpoint} --cfg-options {cfg_options} --out {test_work_dir}/result.pkl --eval bbox segm > {log}'
else:
execmd = f'python tools/test.py {config} {checkpoint} --out {test_work_dir}/result.pkl --eval bbox segm > {log}'
execmd
p = Popen(execmd, shell=True)
p.wait()
```
|
github_jupyter
|
import os
os.environ['CUDA_VISIBLE_DEVICES'] = '5'
from pycocotools.coco import COCO
site = 'train'
img_dir = f'./data/yunnan_512/{site}/JPEGImages'
json_path = f'./data/yunnan_512/{site}/{site}.json'
coco = COCO(json_path)
img_ids = set(_['image_id'] for _ in coco.anns.values())
from mmdet.apis import init_detector, inference_detector, show_result_pyplot
import mmcv
import glob
work_dir = './checkpoints/yunnan'
config_file = glob.glob(os.path.join(work_dir, '*.py'))[0]
checkpoint_file = glob.glob(os.path.join(work_dir, '*.pth'))[0]
cfg_options = {}
cfg_options = {'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.kernel_size': 3,
'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.type': 'gaussian'}
model = init_detector(config_file, checkpoint_file, device='cuda:0', cfg_options=cfg_options)
import numpy as np
import mmcv
import cv2
import torch
import numpy as np
import mmcv
import cv2
import torch
import tifffile as tiff
def show_mask_result(img, result, score_thr):
# 基于掩码,提取轮廓显示
bbox_result, segm_result = result[:2]
bboxes = np.vstack(bbox_result)
labels = [
np.full(bbox.shape[0], i, dtype=np.int32)
for i, bbox in enumerate(bbox_result)
]
labels = np.concatenate(labels)
# draw segmentation masks
segms = None
if segm_result is not None and len(labels) > 0: # non empty
segms = mmcv.concat_list(segm_result)
if isinstance(segms[0], torch.Tensor):
segms = torch.stack(segms, dim=0).detach().cpu().numpy()
else:
segms = np.stack(segms, axis=0)
scores = bboxes[:, -1]
inds = scores > score_thr
bboxes = bboxes[inds, :]
labels = labels[inds]
if segms is not None:
segms = segms[inds, ...]
points = []
for i in range(len(bboxes)):
mask = segms[i].astype(np.uint8)
contours, _ = cv2.findContours(mask[..., None], cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
cv2.drawContours(img, contours, -1, (0,0,255), 2)
for contour in contours:
points.append(len(contour))
return img, sum(points) / (len(points) + 1e-5)
def show_polygon_result(img, result, score_thr):
# 根据点结果,显示轮廓
bbox_result, _, polygon_result = result
bboxes = np.vstack(bbox_result)
scores = bboxes[:, -1]
inds = np.nonzero(scores > score_thr)[0]
points = []
for i in inds:
poly = np.array(polygon_result[0][i]).reshape(-1, 2).astype(np.int32)
points.append(len(poly))
cv2.polylines(img, [poly], True, (0, 0, 255), 2)
return img, sum(points) / (len(points) + 1e-5)
def show_det_result(img, result, score_thr):
bbox_result = result
bboxes = np.vstack(bbox_result)
scores = bboxes[:, -1]
inds = scores > score_thr
bboxes = bboxes[inds, :]
for box in bboxes:
x1, y1, x2, y2 = box.astype(np.int32).tolist()[:4]
cv2.rectangle(img, (x1, y1), (x2, y2), (0, 0, 255), 2)
return img, 4
def show_result(img_path, result, score_thr, ret_det=False):
if os.path.splitext(img_path)[1].lower() == '.tif':
img = tiff.imread(img_path)
img = img[:, :, :3][:, :, ::-1]
img = img.copy()
else:
img = cv2.imread(img_path)
if ret_det:
if isinstance(result, tuple):
result = result[0]
return show_det_result(img, result, score_thr=score_thr)
if len(result) == 3:
return show_polygon_result(img, result, score_thr=score_thr)
else:
return show_mask_result(img, result, score_thr=score_thr)
print(len(img_ids))
# output_dir = './visual_yunnan_val_poly'
# os.makedirs(output_dir, exist_ok=True)
import matplotlib.pyplot as plt
from tqdm import tqdm
img_ids = list(img_ids)
for bi, img_id in enumerate(tqdm(img_ids)):
fn = coco.load_imgs([img_id])[0]['file_name']
img_path = os.path.join(img_dir, fn)
result = inference_detector(model, img_path)
# show_result_pyplot(model, img_path, result, score_thr=0.2)
img, _ = show_result(img_path, result, score_thr=0.6)
img = img[:, :, ::-1]
plt.figure(figsize=(15, 10))
plt.axis('off')
plt.imshow(img)
plt.show()
# out_path = os.path.join(output_dir, fn)
# cv2.imwrite(out_path, img)
if bi > 10:
break
!ls data/yunnan_512_1/val/JPEGImages | wc -l
import os
os.environ['CUDA_VISIBLE_DEVICES'] = '1'
from subprocess import Popen
import glob
test_work_dir = './venus_last_tf/15'
cfg_options = ''
cfg_options += 'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.kernel_size=3 '
cfg_options += 'model.roi_head.polygon_head.polyrnn_head.weight_kernel_params.type=gaussian'
log = os.path.join(test_work_dir, 'result_out.log')
# config = './configs/polygon/polyrnn_r50_fpn_1x_building_0329_3.py'
config = glob.glob(os.path.join(test_work_dir, '*.py'))[0]
checkpoint = os.path.join(test_work_dir, 'latest.pth')
if cfg_options:
execmd = f'python tools/test.py {config} {checkpoint} --cfg-options {cfg_options} --out {test_work_dir}/result.pkl --eval bbox segm > {log}'
else:
execmd = f'python tools/test.py {config} {checkpoint} --out {test_work_dir}/result.pkl --eval bbox segm > {log}'
execmd
p = Popen(execmd, shell=True)
p.wait()
| 0.251464 | 0.247419 |
```
from google.colab import drive
drive.mount('/content/drive/')
! pip3 install transformers
! pip3 install jsonlines
%cd /content/drive/My Drive/Sentence_pair_modeling/Models/BERTs/
```
## **LCQMC**
## LCQMC train-validate-test
```
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
lcqmc_path = "/content/drive/My Drive/Sentence_pair_modeling/LCQMC/"
train_df = pd.read_csv(os.path.join(lcqmc_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(lcqmc_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(lcqmc_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
target_dir = os.path.join(lcqmc_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
```
## LCQMC infer by other pretrained models
```
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric
import os
train_df = pd.read_csv(os.path.join(lcqmc_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(lcqmc_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(lcqmc_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(bq_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(bq_path, "output/Infer_LCQMC") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
```
# **XiAn**
## XiAn train-validate-test
```
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
xian_path = "/content/drive/My Drive/Sentence_pair_modeling/XiAn_STS/"
train_df = pd.read_csv(os.path.join(xian_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(xian_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(xian_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
target_dir = os.path.join(xian_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
```
## XiAn infer by other pretrained models
```
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric
import os
train_df = pd.read_csv(os.path.join(xian_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(xian_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(xian_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(lcqmc_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(lcqmc_path, "output/Infer_XiAn") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
```
# **BQ Corpus**
## BQ train-validate-test
```
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric, json2df
import os
bq_path = "/content/drive/My Drive/Sentence_pair_modeling/BQ Corpus/"
train_df = json2df(os.path.join(bq_path, "data/train.json"))
dev_df = json2df(os.path.join(bq_path, "data/dev.json"))
test_df = json2df(os.path.join(bq_path, "data/test.json"))
target_dir = os.path.join(bq_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
```
## BQ infer by other pretrained models
```
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric, json2df
import os
bq_path = "/content/drive/My Drive/Sentence_pair_modeling/BQ Corpus/"
train_df = json2df(os.path.join(bq_path, "data/train.json"))
dev_df = json2df(os.path.join(bq_path, "data/dev.json"))
test_df = json2df(os.path.join(bq_path, "data/test.json"))
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(lcqmc_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(lcqmc_path, "output/Infer_BQ") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
```
# OCNLI
```
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
ocnli_path = "/content/drive/My Drive/Sentence_pair_modeling/OCNLI/"
train_df = pd.read_csv(os.path.join(ocnli_path, "data/train.csv"),header=None, names=['s1','s2','label','genre'])
dev_df = pd.read_csv(os.path.join(ocnli_path, "data/dev.csv"),header=None, names=['s1','s2','label','genre'])
test_df = pd.read_csv(os.path.join(ocnli_path, "data/test.csv"),header=None, names=['s1','s2','label','genre'])
target_dir = os.path.join(ocnli_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=3,
epochs=6,
batch_size=32,
lr=1e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
# torch.cuda.empty_cache() #释放cuda的显存
```
# CMNLI
```
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
cmnli_path = "/content/drive/My Drive/Sentence_pair_modeling/CMNLI/"
train_df = pd.read_csv(os.path.join(cmnli_path, "data/train1.csv"),header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(cmnli_path, "data/dev.csv"),header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(cmnli_path, "data/test.csv"),header=None, names=['s1','s2','label'])
target_dir = os.path.join(cmnli_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=3,
epochs=3,
batch_size=64,
lr=3e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
```
|
github_jupyter
|
from google.colab import drive
drive.mount('/content/drive/')
! pip3 install transformers
! pip3 install jsonlines
%cd /content/drive/My Drive/Sentence_pair_modeling/Models/BERTs/
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
lcqmc_path = "/content/drive/My Drive/Sentence_pair_modeling/LCQMC/"
train_df = pd.read_csv(os.path.join(lcqmc_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(lcqmc_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(lcqmc_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
target_dir = os.path.join(lcqmc_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric
import os
train_df = pd.read_csv(os.path.join(lcqmc_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(lcqmc_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(lcqmc_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(bq_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(bq_path, "output/Infer_LCQMC") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
xian_path = "/content/drive/My Drive/Sentence_pair_modeling/XiAn_STS/"
train_df = pd.read_csv(os.path.join(xian_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(xian_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(xian_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
target_dir = os.path.join(xian_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric
import os
train_df = pd.read_csv(os.path.join(xian_path, "data/train.tsv"),sep='\t',header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(xian_path, "data/dev.tsv"),sep='\t',header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(xian_path, "data/test.tsv"),sep='\t',header=None, names=['s1','s2','label'])
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(lcqmc_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(lcqmc_path, "output/Infer_XiAn") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric, json2df
import os
bq_path = "/content/drive/My Drive/Sentence_pair_modeling/BQ Corpus/"
train_df = json2df(os.path.join(bq_path, "data/train.json"))
dev_df = json2df(os.path.join(bq_path, "data/dev.json"))
test_df = json2df(os.path.join(bq_path, "data/test.json"))
target_dir = os.path.join(bq_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=2,
epochs=10,
batch_size=32,
lr=2e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
from run_Roberta_model import model_load_test
import pandas as pd
from utils import Metric, json2df
import os
bq_path = "/content/drive/My Drive/Sentence_pair_modeling/BQ Corpus/"
train_df = json2df(os.path.join(bq_path, "data/train.json"))
dev_df = json2df(os.path.join(bq_path, "data/dev.json"))
test_df = json2df(os.path.join(bq_path, "data/test.json"))
data = pd.concat([train_df,dev_df,test_df]).reset_index(drop=True)
target_dir = os.path.join(lcqmc_path, "output/Roberta") # load pretrained model
test_prediction_dir = os.path.join(lcqmc_path, "output/Infer_BQ") # where to save the infer result
test_prediction_name = 'Roberta_test_prediction.csv' # the infer result name
model_load_test(test_df = data,
target_dir = target_dir,
test_prediction_dir = test_prediction_dir,
test_prediction_name = test_prediction_name)
test_result = pd.read_csv(os.path.join(test_prediction_dir, test_prediction_name))
Metric(data.label, test_result.prediction)
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
ocnli_path = "/content/drive/My Drive/Sentence_pair_modeling/OCNLI/"
train_df = pd.read_csv(os.path.join(ocnli_path, "data/train.csv"),header=None, names=['s1','s2','label','genre'])
dev_df = pd.read_csv(os.path.join(ocnli_path, "data/dev.csv"),header=None, names=['s1','s2','label','genre'])
test_df = pd.read_csv(os.path.join(ocnli_path, "data/test.csv"),header=None, names=['s1','s2','label','genre'])
target_dir = os.path.join(ocnli_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=3,
epochs=6,
batch_size=32,
lr=1e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
# torch.cuda.empty_cache() #释放cuda的显存
from run_Roberta_model import model_train_validate_test
import pandas as pd
from utils import Metric
import os
cmnli_path = "/content/drive/My Drive/Sentence_pair_modeling/CMNLI/"
train_df = pd.read_csv(os.path.join(cmnli_path, "data/train1.csv"),header=None, names=['s1','s2','label'])
dev_df = pd.read_csv(os.path.join(cmnli_path, "data/dev.csv"),header=None, names=['s1','s2','label'])
test_df = pd.read_csv(os.path.join(cmnli_path, "data/test.csv"),header=None, names=['s1','s2','label'])
target_dir = os.path.join(cmnli_path, "output/Roberta/")
model_train_validate_test(train_df, dev_df, test_df, target_dir,
max_seq_len=64,
num_labels=3,
epochs=3,
batch_size=64,
lr=3e-05,
patience=1,
max_grad_norm=10.0,
if_save_model=True,
checkpoint=None)
test_result = pd.read_csv(os.path.join(target_dir, 'test_prediction.csv'))
Metric(test_df.label, test_result.prediction)
| 0.311636 | 0.395193 |
# Обработка пропусков в данных, кодирование категориальных признаков, масштабирование данных.
### Задание:
1. Выбрать набор данных (датасет), содержащий категориальные признаки и пропуски в данных. Для выполнения следующих пунктов можно использовать несколько различных наборов данных (один для обработки пропусков, другой для категориальных признаков и т.д.)
2. Для выбранного датасета (датасетов) на основе материалов лекции решить следующие задачи:
<br>2.1. обработку пропусков в данных;
<br>2.2. кодирование категориальных признаков;
<br>2.3. масштабирование данных.
```
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.impute import SimpleImputer
from sklearn.impute import MissingIndicator
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.preprocessing import MinMaxScaler, StandardScaler, Normalizer
%matplotlib inline
sns.set(style="ticks")
data = pd.read_csv('train.csv', sep=",")
```
##### Размер датасета
```
data.shape
```
##### Типы данных колонок
```
data.dtypes
```
##### Проверим были ли пропущены <br/>значения в каких-нибудь колонках
```
data.isnull().sum()
```
##### Первые пять строк датасета
```
data.head()
total_count = data.shape[0]
print(f'Всего строк: {total_count}')
```
## 1. Обработка пропущенных данных
### 1.1. Простые стратегии - удаление или заполнение нулями
#### 1.1.1. Удаление колонок содержащих пустые значения
```
data_new_1 = data.dropna(axis=1, how='any')
(data.shape, data_new_1.shape)
```
#### 1.1.2. Удаление строк содержащих пустые значения
```
data_new_2 = data.dropna(axis=0, how='any')
(data.shape, data_new_2.shape)
```
#### 1.1.2. Заполнение всех пропущенных значений нулями, что некорректно для категориальных знаичений
```
data_new_3 = data.fillna(0)
data_new_3.head()
```
### "Внедрение значений" - импьютация
#### 1.2.1. Обработка пропусков в числовых данных
<br>Выберем числовые колонки с пропущенными значениями
```
num_cols = [col for col in data.columns if (data[data[col].isnull()].shape[0] > 0 \
and (data[col].dtype=='float64' or data[col].dtype=='int64'))]
for col in num_cols:
print(f"Колонка {col}, количество пропусков {data[col].isnull().sum()} - \
{round((data[col].isnull().sum()/total_count)*100,2)}%")
```
##### Фильтр по колонкам с пропущенными значениями
```
data_num = data[num_cols]
data_num
for col in data_num:
plt.hist(data[col], 75)
plt.xlabel(col)
plt.show()
data[data['Age'].isnull()]
missedValues = data[data['Age'].isnull()].index
missedValues
data[data.index.isin(missedValues)]
data_num[data_num.index.isin(missedValues)]['Age']
data_num_age=data_num[['Age']]
data_num_age.head()
```
<br>Фильтр для проверки заполнения пустых значений
```
indicator = MissingIndicator()
mask_missing_values_only = indicator.fit_transform(data_num_age)
mask_missing_values_only
strategies=['mean', 'median','most_frequent']
```
т.е. нашими стратегиями будут:
* Среднее значение
* Медиана
* Наиболее часто встречающаяся величина
<br> Определим функцию для импьютации в которую <br/>будет отправляться название стратегии как аргумент
```
def test_num_impute(strat):
# Определяем сратегию
imp_num = SimpleImputer(strategy=strat)
data_num_imp = imp_num.fit_transform(data_num_age)
return data_num_imp[mask_missing_values_only]
strategies[0], test_num_impute(strategies[0])
strategies[1], test_num_impute(strategies[1])
strategies[2], test_num_impute(strategies[2])
```
###### 1.2.2. Обработка пропусков в категориальных данных
Выберем категорильные колонки с пропущенными значениями
```
cat_cols = [col for col in data.columns if (data[data[col].isnull()].shape[0] > 0 \
and data[col].dtype=='object')]
for col in cat_cols:
print(f"Колонка {col}, количество пропусков {data[col].isnull().sum()} - \
{round((data[col].isnull().sum()/total_count)*100,2)}%")
cat_temp_data = data[['Cabin']]
cat_temp_data.head()
```
<br> Получим уникальныые значения для колонки
```
cat_temp_data['Cabin'].unique()
cat_temp_data[cat_temp_data['Cabin'].isnull()].shape
```
Импьютация наиболее частыми выражениями
```
imp2 = SimpleImputer(missing_values=np.nan, strategy='most_frequent')
data_imp2 = imp2.fit_transform(cat_temp_data)
data_imp2
```
Проверим, что пустые значения отсутствуют
```
np.unique(data_imp2)
```
Импьютация константой
```
imp3 = SimpleImputer(missing_values=np.nan, strategy='constant', fill_value='MyConst')
data_imp3 = imp3.fit_transform(cat_temp_data)
data_imp3
np.unique(data_imp3)
data_imp3[data_imp3=='MyConst'].size
```
<br>
### 2. Форматирование категориальных признаков в числовые
Рассмотрим набор, в котором мы заменили пропущенные значения на саме частое значение
```
cat_enc = pd.DataFrame({'Cabin':data_imp2.T[0]})
cat_enc
```
#### 2.1 Label encoding - кодирование целыми значениями
```
le = LabelEncoder()
cat_enc_le = le.fit_transform(cat_enc['Cabin'])
cat_enc['Cabin'].unique()
np.unique(cat_enc_le)
le.inverse_transform([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38,
39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51,
52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142,
143, 144, 145, 146])
```
#### 2.2. One-hot encoding - Кодирование наборами бинарных значений
```
ohe = OneHotEncoder()
cat_enc_ohe = ohe.fit_transform(cat_enc[['Cabin']])
cat_enc.shape
cat_enc_ohe.shape
cat_enc_ohe
cat_enc_ohe.todense()[0:10]
cat_enc.head(10)
```
#### Pandas get_dummies - быстрый вариант one-hot кодирования
```
pd.get_dummies(cat_enc).head()
```
dummy_na - параметр, который создает отдельый столбец для NaNов, если False, то игнорирует NaNы
```
pd.get_dummies(cat_temp_data, dummy_na=True).head()
### 3. Масштабированние данных
```
### 3. Заполним пропуски и закодируем пропуски в нашей выборке
```
data[data['Age'].isnull()]
imp_num = SimpleImputer(strategy='mean')
data['Age'] = imp_num.fit_transform(data_num_age)
data['Age'].head(10)
data[data['Age'].isnull()]
```
### 4. Масштабирование
#### 4.1. MinMax - масштабирование
```
sc1 = MinMaxScaler()
sc1_data = sc1.fit_transform(data[['Age']])
plt.hist(data['Age'], 50)
plt.show()
plt.hist(sc1_data, 50)
plt.show()
```
#### 4.2. Z-оценка - StandartScaling
```
sc2 = StandardScaler()
sc2_data = sc2.fit_transform(data[['Age']])
plt.hist(data['Age'],50)
plt.show()
plt.hist(sc2_data,50)
plt.show()
```
### 5. Нормализация данных
```
sc3 = Normalizer()
sc3_data = sc3.fit_transform(data[['Age']])
plt.hist(data['Age'],50)
plt.show()
plt.hist(sc3_data,50)
plt.show()
```
|
github_jupyter
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.impute import SimpleImputer
from sklearn.impute import MissingIndicator
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.preprocessing import MinMaxScaler, StandardScaler, Normalizer
%matplotlib inline
sns.set(style="ticks")
data = pd.read_csv('train.csv', sep=",")
data.shape
data.dtypes
data.isnull().sum()
data.head()
total_count = data.shape[0]
print(f'Всего строк: {total_count}')
data_new_1 = data.dropna(axis=1, how='any')
(data.shape, data_new_1.shape)
data_new_2 = data.dropna(axis=0, how='any')
(data.shape, data_new_2.shape)
data_new_3 = data.fillna(0)
data_new_3.head()
num_cols = [col for col in data.columns if (data[data[col].isnull()].shape[0] > 0 \
and (data[col].dtype=='float64' or data[col].dtype=='int64'))]
for col in num_cols:
print(f"Колонка {col}, количество пропусков {data[col].isnull().sum()} - \
{round((data[col].isnull().sum()/total_count)*100,2)}%")
data_num = data[num_cols]
data_num
for col in data_num:
plt.hist(data[col], 75)
plt.xlabel(col)
plt.show()
data[data['Age'].isnull()]
missedValues = data[data['Age'].isnull()].index
missedValues
data[data.index.isin(missedValues)]
data_num[data_num.index.isin(missedValues)]['Age']
data_num_age=data_num[['Age']]
data_num_age.head()
indicator = MissingIndicator()
mask_missing_values_only = indicator.fit_transform(data_num_age)
mask_missing_values_only
strategies=['mean', 'median','most_frequent']
def test_num_impute(strat):
# Определяем сратегию
imp_num = SimpleImputer(strategy=strat)
data_num_imp = imp_num.fit_transform(data_num_age)
return data_num_imp[mask_missing_values_only]
strategies[0], test_num_impute(strategies[0])
strategies[1], test_num_impute(strategies[1])
strategies[2], test_num_impute(strategies[2])
cat_cols = [col for col in data.columns if (data[data[col].isnull()].shape[0] > 0 \
and data[col].dtype=='object')]
for col in cat_cols:
print(f"Колонка {col}, количество пропусков {data[col].isnull().sum()} - \
{round((data[col].isnull().sum()/total_count)*100,2)}%")
cat_temp_data = data[['Cabin']]
cat_temp_data.head()
cat_temp_data['Cabin'].unique()
cat_temp_data[cat_temp_data['Cabin'].isnull()].shape
imp2 = SimpleImputer(missing_values=np.nan, strategy='most_frequent')
data_imp2 = imp2.fit_transform(cat_temp_data)
data_imp2
np.unique(data_imp2)
imp3 = SimpleImputer(missing_values=np.nan, strategy='constant', fill_value='MyConst')
data_imp3 = imp3.fit_transform(cat_temp_data)
data_imp3
np.unique(data_imp3)
data_imp3[data_imp3=='MyConst'].size
cat_enc = pd.DataFrame({'Cabin':data_imp2.T[0]})
cat_enc
le = LabelEncoder()
cat_enc_le = le.fit_transform(cat_enc['Cabin'])
cat_enc['Cabin'].unique()
np.unique(cat_enc_le)
le.inverse_transform([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38,
39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51,
52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 141, 142,
143, 144, 145, 146])
ohe = OneHotEncoder()
cat_enc_ohe = ohe.fit_transform(cat_enc[['Cabin']])
cat_enc.shape
cat_enc_ohe.shape
cat_enc_ohe
cat_enc_ohe.todense()[0:10]
cat_enc.head(10)
pd.get_dummies(cat_enc).head()
pd.get_dummies(cat_temp_data, dummy_na=True).head()
### 3. Масштабированние данных
data[data['Age'].isnull()]
imp_num = SimpleImputer(strategy='mean')
data['Age'] = imp_num.fit_transform(data_num_age)
data['Age'].head(10)
data[data['Age'].isnull()]
sc1 = MinMaxScaler()
sc1_data = sc1.fit_transform(data[['Age']])
plt.hist(data['Age'], 50)
plt.show()
plt.hist(sc1_data, 50)
plt.show()
sc2 = StandardScaler()
sc2_data = sc2.fit_transform(data[['Age']])
plt.hist(data['Age'],50)
plt.show()
plt.hist(sc2_data,50)
plt.show()
sc3 = Normalizer()
sc3_data = sc3.fit_transform(data[['Age']])
plt.hist(data['Age'],50)
plt.show()
plt.hist(sc3_data,50)
plt.show()
| 0.241847 | 0.980839 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.