code
stringlengths 2.5k
6.36M
| kind
stringclasses 2
values | parsed_code
stringlengths 0
404k
| quality_prob
float64 0
0.98
| learning_prob
float64 0.03
1
|
|---|---|---|---|---|
```
pip install tweepy
pip install yweather
pip install google-cloud
pip install google
pip install protobuf
pip install google-cloud-bigquery
pip install google-cloud-language
import json
import csv
import tweepy
import re
import yweather
import pandas as pd
#Twitter API credentials
consumer_key = ""
consumer_secret = ""
access_key = ""
access_secret = ""
#authorize twitter, initialize tweepy
auth = tweepy.OAuthHandler(consumer_key, consumer_secret)
auth.set_access_token(access_key, access_secret)
api = tweepy.API(auth)
import os
from google.cloud import language_v1
credential_path = "Z:\BU\2021Fall\EC601\Project2\credential.json"
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = credential_path
# Instantiates a client
client = language_v1.LanguageServiceClient()
def sentimentScore(text):
document = language_v1.Document(content=text, type_=language_v1.Document.Type.PLAIN_TEXT)
# Detects the sentiment of the text
sentiment = client.analyze_sentiment(request={'document': document}).document_sentiment
return sentiment.score
def getCorodinate(place):
from geopy.geocoders import Nominatim
geolocator = Nominatim(user_agent="myapp")
location = geolocator.geocode(place)
return location.latitude, location.longitude
def getWOEID(place):
try:
trends = api.trends_available()
for val in trends:
if (val['name'].lower() == place.lower()):
return(val['woeid'])
print('Location Not Found')
except Exception as e:
print('Exception:',e)
return(0)
def get_trends_by_location(loc_id,count=50):
try:
trends = api.trends_place(loc_id)
df = pd.DataFrame([trending['name'], trending['tweet_volume'], trending['url']] for trending in trends[0]['trends'])
df.columns = ['Trends','Volume','url']
# df = df.sort_values('Volume', ascending = False)
# print(df[:count])
return(df['Trends'][:count])
except Exception as e:
print("An exception occurred",e)
print(get_trends_by_location(getWOEID('boston'),10))
def search_for_phrase(phrase,place,amount):
try:
df = pd.DataFrame( columns = ["text",'sentiment score'])
latitude = getCorodinate(place)[0]
longitude = getCorodinate(place)[1]
for tweet in tweepy.Cursor(api.search, q=phrase.encode('utf-8') +' -filter:retweets'.encode('utf-8'),geocode=str(latitude)+","+str(longitude)+",100km",lang='en',result_type='recent',tweet_mode='extended').items(amount):
txt = tweet.full_text.replace('\n',' ').encode('utf-8')
df=df.append({"text": txt,'sentiment score': sentimentScore(txt)},ignore_index=True)
# print (df)
return phrase, df['sentiment score'].mean(), df['sentiment score'].var()
except Exception as e:
print("An exception occurred",e)
search_for_phrase('pizza','boston',10)
def getResult(place):
data=[]
trends = get_trends_by_location(getWOEID(place),10)
for phrase in trends:
data.append(search_for_phrase(phrase,place,10))
df = pd.DataFrame(data,columns=['trends','mean of sentiment-score','variance of sentiment-score'])
print (df)
if __name__ == '__main__':
getResult("boston")
```
|
github_jupyter
|
pip install tweepy
pip install yweather
pip install google-cloud
pip install google
pip install protobuf
pip install google-cloud-bigquery
pip install google-cloud-language
import json
import csv
import tweepy
import re
import yweather
import pandas as pd
#Twitter API credentials
consumer_key = ""
consumer_secret = ""
access_key = ""
access_secret = ""
#authorize twitter, initialize tweepy
auth = tweepy.OAuthHandler(consumer_key, consumer_secret)
auth.set_access_token(access_key, access_secret)
api = tweepy.API(auth)
import os
from google.cloud import language_v1
credential_path = "Z:\BU\2021Fall\EC601\Project2\credential.json"
os.environ['GOOGLE_APPLICATION_CREDENTIALS'] = credential_path
# Instantiates a client
client = language_v1.LanguageServiceClient()
def sentimentScore(text):
document = language_v1.Document(content=text, type_=language_v1.Document.Type.PLAIN_TEXT)
# Detects the sentiment of the text
sentiment = client.analyze_sentiment(request={'document': document}).document_sentiment
return sentiment.score
def getCorodinate(place):
from geopy.geocoders import Nominatim
geolocator = Nominatim(user_agent="myapp")
location = geolocator.geocode(place)
return location.latitude, location.longitude
def getWOEID(place):
try:
trends = api.trends_available()
for val in trends:
if (val['name'].lower() == place.lower()):
return(val['woeid'])
print('Location Not Found')
except Exception as e:
print('Exception:',e)
return(0)
def get_trends_by_location(loc_id,count=50):
try:
trends = api.trends_place(loc_id)
df = pd.DataFrame([trending['name'], trending['tweet_volume'], trending['url']] for trending in trends[0]['trends'])
df.columns = ['Trends','Volume','url']
# df = df.sort_values('Volume', ascending = False)
# print(df[:count])
return(df['Trends'][:count])
except Exception as e:
print("An exception occurred",e)
print(get_trends_by_location(getWOEID('boston'),10))
def search_for_phrase(phrase,place,amount):
try:
df = pd.DataFrame( columns = ["text",'sentiment score'])
latitude = getCorodinate(place)[0]
longitude = getCorodinate(place)[1]
for tweet in tweepy.Cursor(api.search, q=phrase.encode('utf-8') +' -filter:retweets'.encode('utf-8'),geocode=str(latitude)+","+str(longitude)+",100km",lang='en',result_type='recent',tweet_mode='extended').items(amount):
txt = tweet.full_text.replace('\n',' ').encode('utf-8')
df=df.append({"text": txt,'sentiment score': sentimentScore(txt)},ignore_index=True)
# print (df)
return phrase, df['sentiment score'].mean(), df['sentiment score'].var()
except Exception as e:
print("An exception occurred",e)
search_for_phrase('pizza','boston',10)
def getResult(place):
data=[]
trends = get_trends_by_location(getWOEID(place),10)
for phrase in trends:
data.append(search_for_phrase(phrase,place,10))
df = pd.DataFrame(data,columns=['trends','mean of sentiment-score','variance of sentiment-score'])
print (df)
if __name__ == '__main__':
getResult("boston")
| 0.272799 | 0.132543 |
## LA's COVID-19 Reopening Indicators
This report contains information about how LA County and the City of LA performed yesterday, on a number of key COVID-19 indicators related to the speed at which opening up can occur. Taken together, performing well against the benchmarks provide confidence in moving through each phase of reopening.
**LA is certainly an epicenter.**
* [NYT analysis of LA County neighborhoods](https://www.nytimes.com/interactive/2021/01/29/us/los-angeles-county-covid-rates.html)
* By mid Jan 2021, it is estimated that [1 in 3](https://www.latimes.com/california/story/2021-01-14/one-in-three-la-county-residents-infected-coronavirus) already have had COVID in LA County, triple the confirmed/reported cases.
* By early Jan 2021, it is estimated that [1 in 17 people currently have COVID in LA County](https://www.sfgate.com/bayarea/article/Los-Angeles-County-COVID-data-1-in-17-hospitals-15848658.php)
* By late Dec, southern CA and central CA have [run out of ICU beds](https://www.nytimes.com/2020/12/26/world/central-and-southern-california-icu-capacity.html)
* In mid Dec, LA County Department of Public Health [estimated that 1 in 80 people were infectious](http://file.lacounty.gov/SDSInter/dhs/1082410_COVID-19ProjectionPublicUpdateLewis12.16.20English.pdf)
* LA reaches [5,000 deaths](https://www.latimes.com/california/story/2020-08-11/13000-more-coronavirus-cases-california-test-results-backlog) by mid-August, and is ranked as 3rd highest US county in terms of deaths, after Queens (NY) and Kings (NY) Counties.
* LA County is [projected](https://www.planetizen.com/news/2020/07/109756-californias-coronavirus-infections-and-hospitalizations-surge) to [run out](https://www.latimes.com/california/story/2020-06-29/l-a-county-issues-dire-warning-amid-alarming-increases-in-coronavirus) of ICU beds by early to mid July, with acute care beds [reaching capacity](https://www.nytimes.com/2020/07/03/health/coronavirus-mortality-testing.html) soon after.
* In late June, LA County Department of Public Health (DPH) [estimated that 1 in 400 people were infectious](https://www.latimes.com/california/story/2020-06-29/o-c-reports-highest-weekly-covid-19-death-toll-as-california-sees-spike-in-cases), asymptomatic, and not isolated. One week later, as of June 29, LA DPH estimated the risk increased threefold, up to [1 in 140](https://www.nytimes.com/2020/06/29/us/california-coronavirus-reopening.html).
As long as LA consistently tests large portions of its population with fairly low positive COVID-19 results, sustains decreases in cases and deaths, has stable or decreasing COVID-related hospitalizations, and stocks ample available hospital equipment for a potential surge, we are positioned to continue loosening restrictions. When any one indicator fails to meet the benchmark, we should slow down to consider why that is happening. When multiple indicators fail to meet the benchmark, we should pause our reopening plans and even enact more stringent physical and social distancing protocols by moving back a phase.
* [Federal Gating Criteria](https://www.whitehouse.gov/wp-content/uploads/2020/04/Guidelines-for-Opening-Up-America-Again.pdf)
* [State Gating Criteria](https://covid19.ca.gov/roadmap-counties/)
* CA Department of Public Health's [Blueprint](https://covid19.ca.gov/safer-economy/#reopening-data) toward Reopening, [rules for tier assignments](https://www.cdph.ca.gov/Programs/CID/DCDC/Pages/COVID-19/COVID19CountyMonitoringOverview.aspx) and [Dimmer Switch Framework](https://www.cdph.ca.gov/Programs/CID/DCDC/CDPH%20Document%20Library/COVID-19/Dimmer-Framework-September_2020.pdf)
* [CA Reopening, Take 2](https://www.nytimes.com/2020/08/31/us/california-coronavirus-reopening.html)
* [WHO Testing and Positivity Rate Guidelines](https://coronavirus.jhu.edu/testing/testing-positivity)
Below, you will see how LA performed yesterday on the following indicators. The data does have a one day lag. Whenever City of LA (subset of LA County) data is available, it is also reported.
#### Symptoms
* Downward trajectory of influenza-like illnesses (ILI) reported within a 14-day period **and**
* Downward trajectory of COVID-like syndromic cases reported within a 14-day period
#### Cases
* Downward trajectory of documented cases within a 14-day period **or**
* Downward trajectory of positive tests as a percent of total tests within a 14-day period (flat or increasing volume of tests)
#### Hospitals
* Treat all patients without crisis care **and**
* Robust testing program in place for at-risk healthcare workers, including emerging antibody testing
### References
* [Reopening Indicators](https://github.com/CityOfLosAngeles/covid19-indicators/blob/master/Reopening_Indicators_Comparison.xlsx) from [New York State](https://www.nytimes.com/2020/05/04/nyregion/coronavirus-reopen-cuomo-ny.html) and [Chicago](https://www.chicagotribune.com/coronavirus/ct-coronavirus-chicago-reopening-lightfoot-20200508-ztpnouwexrcvfdfcr2yccbc53a-story.html)
* [Collection of articles](https://github.com/CityOfLosAngeles/covid19-indicators/blob/master/reopening-sources.md) related to what experts say about reopening and the known unknowns ahead
* [LA and Chicago](https://www.nytimes.com/2020/05/09/us/coronavirus-chicago.html), after NYC, have the most persistent virus caseloads
* [LA, DC, and Chicago](https://www.latimes.com/california/story/2020-05-22/white-house-concerned-with-coronavirus-spread-in-l-a-area-asks-cdc-to-investigate) remain hotspots within the US
**Related daily reports:**
1. **[US counties report on cases and deaths for select major cities](https://cityoflosangeles.github.io/covid19-indicators/us-county-trends.html)**
1. **[CA counties report on cases, deaths, and hospitalizations](https://cityoflosangeles.github.io/covid19-indicators/ca-county-trends.html)**
1. **[Los Angeles County neighborhoods report on cases and deaths](https://cityoflosangeles.github.io/covid19-indicators/la-neighborhoods-trends.html)**
```
import numpy as np
import pandas as pd
import utils
import default_parameters
import make_charts
import meet_indicators
import ca_reopening_tiers
from IPython.display import display_html, Markdown, HTML
# Default parameters
county_state_name = default_parameters.county_state_name
state_name = default_parameters.state_name
msa_name = default_parameters.msa_name
time_zone = default_parameters.time_zone
fulldate_format = default_parameters.fulldate_format
monthdate_format = default_parameters.monthdate_format
start_date = default_parameters.start_date
yesterday_date = default_parameters.yesterday_date
today_date = default_parameters.today_date
one_week_ago = default_parameters.one_week_ago
two_weeks_ago = default_parameters.two_weeks_ago
three_weeks_ago = default_parameters.three_weeks_ago
two_days_ago = default_parameters.two_days_ago
eight_days_ago = default_parameters.eight_days_ago
# Daily testing upper and lower bound
county_test_lower_bound = 15_000
county_test_upper_bound = 16_667
positive_lower_bound = 0.04
positive_upper_bound = 0.08
positive_2weeks_bound = 0.05
hospital_bound = 0.30
ca_hospitalization_bound = 0.05
# Set cut-offs for CA reopening
ca_case_minimal_bound = 1
ca_case_moderate_bound = 4
ca_case_substantial_bound = 7
ca_test_minimal_bound = 0.020
ca_test_moderate_bound = 0.050
ca_test_substantial_bound = 0.080
def check_report_readiness(county_state_name, state_name, msa_name, start_date, yesterday_date):
"""
Check if each dataframe has yesterday's date's info.
If all datasets are complete, report can be run.
"""
df = utils.prep_county(county_state_name, start_date)
if df.date.max() < yesterday_date:
raise Exception("Data incomplete")
df = utils.prep_lacity_cases(start_date)
if df.date.max() < yesterday_date:
raise Exception("Data incomplete")
df = utils.prep_testing(start_date)
if (df.date.max() < yesterday_date) or (
(df.date.max() == today_date) and (df.County_Performed == 0) ):
raise Exception("Data incomplete")
df = utils.prep_hospital_surge(county_state_name, start_date)
if df.date.max() < two_days_ago:
raise Exception("Data incomplete")
check_report_readiness(county_state_name, state_name, msa_name, start_date, yesterday_date)
# Check cases according to some criterion
def check_cases(row):
status = ["failed" if x < 14 else "met" if x >= 14 else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_deaths(row):
status = ["failed" if x < 14 else "met" if x >= 14 else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_tests(lower_bound, upper_bound, row):
status = ["failed" if x < lower_bound
else "met" if ((x >= lower_bound) and (x < upper_bound))
else "exceeded" if x >= upper_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_positives(row):
status = ["failed" if x > positive_upper_bound
else "met" if ((x >= positive_lower_bound) and (x <= positive_upper_bound))
else "exceeded" if x < positive_lower_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_positives_two_weeks(row):
status = ["met" if x <= positive_2weeks_bound
else "failed" if x>= positive_2weeks_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_hospitalizations(row):
status = ["met" if x < ca_hospitalization_bound
else "failed" if x >= ca_hospitalization_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
county_fnmap = {
"Cases": check_cases,
"Deaths": check_deaths,
"Daily Testing": lambda row: check_tests(county_test_lower_bound, county_test_upper_bound, row),
"Positive Tests": check_positives,
"Positive Tests (WHO)": check_positives_two_weeks,
"COVID Hospitalizations": check_hospitalizations,
"COVID ICU Hospitalizations": check_hospitalizations,
}
city_fnmap = {
"Cases": check_cases,
"Deaths": check_deaths,
"Daily Testing": lambda row: check_tests(county_test_lower_bound, county_test_upper_bound, row),
"Positive Tests": check_positives,
"Positive Tests (WHO)": check_positives_two_weeks,
"COVID Hospitalizations": check_hospitalizations,
"COVID ICU Hospitalizations": check_hospitalizations,
}
red = make_charts.maroon
green = make_charts.green
blue = make_charts.blue
stylemap = {
"failed": f"background-color: {red}; color: white; font-weight: bold; opacity: 0.7" ,
"met": f"background-color: {green}; color: white; font-weight: bold; opacity: 0.7",
"exceeded": f"background-color: {blue}; color: white; font-weight: bold; opacity: 0.7",
"": "background-color: white; color: black; font-weight: bold; opacity: 0.7",
}
# Add CA Blueprint for Safer Economy (reopening, take 2) criterion to get colors
def check_ca_case(row):
status = ["minimal" if x < ca_case_minimal_bound
else "moderate" if ((x >= ca_case_minimal_bound) and (x < ca_case_moderate_bound))
else "substantial" if ((x >= ca_case_moderate_bound) and (x <= ca_case_substantial_bound))
else "widespread" if x > ca_case_substantial_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_ca_test(row):
status = ["minimal" if x < ca_test_minimal_bound
else "moderate" if (x >= ca_test_minimal_bound) and (x < ca_test_moderate_bound)
else "substantial" if (x >= ca_test_moderate_bound) and (x <= ca_test_substantial_bound)
else "widespread" if x > ca_test_substantial_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_ca_overall_tier(row):
status = ["minimal" if x==1
else "moderate" if x==2
else "substantial" if x==3
else "widespread" if x==4
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
ca_tier_fnmap = {
"Overall Tier": check_ca_overall_tier,
"Case Rate per 100k": check_ca_case,
"Test Positivity Rate": check_ca_test,
}
purple = make_charts.purple
red = make_charts.maroon
orange = make_charts.orange
yellow = make_charts.yellow
stylemap_ca = {
"widespread": f"background-color: {purple}; color: white; font-weight: bold; opacity: 0.7" ,
"substantial": f"background-color: {red}; color: white; font-weight: bold; opacity: 0.7",
"moderate": f"background-color: {orange}; color: white; font-weight: bold; opacity: 0.7",
"minimal": f"background-color: {yellow}; color: white; font-weight: bold; opacity: 0.7",
"": "background-color: white; color: black; font-weight: bold; opacity: 0.7",
}
# These are functions used to clean the CA county indicators df
def put_in_tiers(row, indicator, minimal_bound, moderate_bound, substantial_bound):
if row[indicator] < minimal_bound:
return 1
elif (row[indicator] >= minimal_bound) and (row[indicator] < moderate_bound):
return 2
elif (row[indicator] >= moderate_bound) and (row[indicator] <= substantial_bound):
return 3
elif row[indicator] > substantial_bound:
return 4
else:
return np.nan
def add_tiers(df):
df = df.assign(
case_tier = df.apply(lambda x:
put_in_tiers(x, "case",
ca_case_minimal_bound, ca_case_moderate_bound,
ca_case_substantial_bound), axis=1).astype("Int64"),
test_tier = df.apply(lambda x:
put_in_tiers(x, "test",
ca_test_minimal_bound, ca_test_moderate_bound,
ca_test_substantial_bound), axis=1).astype("Int64")
)
# If 2 indicators belong in different tiers, most restrictive (max) is assigned overall
df = (df.assign(
overall_tier = df[["case_tier", "test_tier"]].max(axis=1).astype(int)
)[["overall_tier", "case", "test"]]
.rename(columns = {"overall_tier": "Overall Tier",
"case": "Case Rate per 100k",
"test": "Test Positivity Rate"
})
)
# Transpose
df = (df.T
.rename(columns = {0: "Current Week",
1: "One Week Ago",
2: "Two Weeks Ago",})
)
return df
def summary_of_ca_reopening_indicators():
# Grab indicators
ca_case_today_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "today")
ca_case_last_week_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "one_week_ago")
ca_case_two_week_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "two_weeks_ago")
ca_test_last_week_indicator = ca_reopening_tiers.positive_rate(start_date, "one_week_ago")
ca_test_two_week_indicator = ca_reopening_tiers.positive_rate(start_date, "two_weeks_ago")
# Create a df for the county's indicators based off of CA Blueprint to Safer Economy
ca_county = pd.DataFrame(
{"case": [ca_case_today_indicator, ca_case_last_week_indicator, ca_case_two_week_indicator],
"test": [np.nan, ca_test_last_week_indicator, ca_test_two_week_indicator]
}
)
# Clean up df
df = add_tiers(ca_county)
# Style the table
ca_county_html = (df.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: ca_tier_fnmap[row.name](row).map(stylemap_ca), axis=1)
.render()
)
display(Markdown(f"<strong>CA Reopening Metrics: {county_state_name.split(',')[0]} County</strong>"))
display(HTML(ca_county_html))
def summary_of_indicators():
# Grab indicators
county_case_indicator = meet_indicators.meet_case("county", county_state_name, start_date)
county_death_indicator = meet_indicators.meet_death("county", county_state_name, start_date)
county_test_indicator = meet_indicators.meet_daily_testing(yesterday_date, "county",
county_test_lower_bound, county_test_upper_bound)
county_positive_indicator = meet_indicators.meet_positive_share(yesterday_date, "county",
positive_lower_bound, positive_upper_bound)
county_positive_2wks_indicator = meet_indicators.meet_positive_share_for_two_weeks(yesterday_date, "county")
# 8/11: Ignore LA County HavBed data, since it seems to be reported much more infrequently
# Use CA data portal's hospitalizations
hospitalization_indicator = meet_indicators.meet_all_hospitalization(county_state_name, yesterday_date)
icu_hospitalization_indicator = meet_indicators.meet_icu_hospitalization(county_state_name, yesterday_date)
city_case_indicator = meet_indicators.meet_case("lacity", "City of LA", start_date)
city_death_indicator = meet_indicators.meet_death("lacity", "City of LA", start_date)
indicator_names = ["Cases", "Deaths",
"Daily Testing", "Positive Tests", "Positive Tests (WHO)",
"COVID Hospitalizations", "COVID ICU Hospitalizations"]
# Create separate df for county and city
county = pd.DataFrame(
{"LA County": [county_case_indicator, county_death_indicator,
county_test_indicator, county_positive_indicator, county_positive_2wks_indicator,
hospitalization_indicator, icu_hospitalization_indicator]},
index=indicator_names
)
city = pd.DataFrame(
{"City of LA": [city_case_indicator, city_death_indicator,
np.nan, np.nan, np.nan,
np.nan, np.nan]},
index=indicator_names
)
# Style the table
def display_side_by_side(*args):
html_str=''
for df in args:
html_str+=df
display_html(html_str.replace('table','table style="display:inline"'),raw=True)
county_html = (county.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: county_fnmap[row.name](row).map(stylemap), axis=1)
.render()
)
city_html = (city.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: city_fnmap[row.name](row).map(stylemap), axis=1)
.hide_index()
.render()
)
display(Markdown("<strong>Indicators for LA County and City of LA</strong>"))
display_side_by_side(county_html, city_html)
display(Markdown(f"### Summary of Indicators as of {yesterday_date.strftime(fulldate_format)}:"))
```
#### Indicators Based on CA State Guidelines
CA's [Blueprint for a Safer Economy](https://www.cdph.ca.gov/Programs/CID/DCDC/Pages/COVID-19/COVID19CountyMonitoringOverview.aspx) assigns each county to a tier based on case rate and test positivity rate. If counties fall into 2 different tiers on the two metrics, they are assigned to the more restrictive tier. Tiers, from most severe to least severe, categorizes coronavirus spread as <strong><span style='color:#6B1F84'>widespread; </span></strong>
<strong><span style='color:#F3324C'>substantial; </span></strong><strong><span style='color:#F7AE1D'>moderate; </span></strong><strong><span style = 'color:#D0E700'>or minimal.</span></strong>
**Counties must stay in the current tier for 3 consecutive weeks and metrics from the last 2 consecutive weeks must fall into less restrictive tier before moving into a less restrictive tier.**
* **Case Rate per 100k**: the unadjusted case rate per 100k. <span style = 'color:#6B1F84'>Any value above 7 is widespread; </span><span style = 'color:#F3324C'>a value of 4-7 is substantial. </span> CA does adjust the case rate based on testing volume, but that is not done here.
* **Test Positivity Rate**: percent of tests that are COVID-positive. <span style = 'color:#6B1F84'>Any value above 8% is widespread; </span><span style = 'color:#F3324C'>a value of 5-8% is substantial. </span>
#### Indicators Based on Federal Guidelines
These indicators can <strong><span style='color:#F3324C'>fail to meet the lower benchmark; </span></strong><strong><span style='color:#10DE7A'>meet the lower benchmark; </span></strong>
<strong><span style = 'color:#1696D2'>or exceed the higher benchmark.</span></strong>
* **Cases and deaths**: the number of days with declining values from the prior day over the past 14 days. Guidelines state both should sustain a 14-day downward trajectory. <span style = 'color:red'>Any value less than 14 means we failed to meet this benchmark.</span>
* **Daily Testing**: number of daily tests conducted for the county 2 days ago (accounting for a time lag in results reported). LA County's goal is to test 15,000 daily (45 tests per 1,000 residents) *(lower bound)*. Chicago's goal was 50 tests per 1,000 residents, translating to 16,667 tests daily *(upper bound)*.
<span style = 'color:red'>Below 15,000 (county) means we failed this benchmark.</span>
* **Positive Tests**: proportion of positive tests last week, values fall between 0 and 1. CA's positivity requirement is 8% or below *(upper bound)*, but experts say that [less than 4%](https://www.nytimes.com/2020/05/25/health/coronavirus-testing-trump.html) is necessary to halt the spread of the virus *(lower bound)*.
<span style = 'color:red'>More than 8% positive for the past week means we failed to meet this benchmark.</span>
* **Positive Tests (WHO)**: proportion of positive tests in the past 2 weeks, values fall between 0 and 1. The weeks are weighted by the number of tests conducted. The WHO recommends that tests return less than 5% positive for 14 days prior to reopening. JHU has a [state-by-state analysis](https://coronavirus.jhu.edu/testing/testing-positivity) of this.
<span style = 'color:red'>More than 5% positive over two weeks means we failed to meet this benchmark.</span>
* **Hospitalizations**: the 7-day averaged daily percent change in all COVID hospitalizations and COVID ICU hospitalizations; values fall between 0 and 1. CA guidelines ask for stable or downward trends, not exceeding a 5% daily change.
<span style = 'color:red'>Above 5% means we failed to meet this benchmark.</span>
```
summary_of_ca_reopening_indicators()
summary_of_indicators()
display(Markdown("## Caseload Charts"))
display(Markdown(
f"These are the trends in cases and deaths for the county and the city since {start_date.strftime(fulldate_format)}, using a 7-day rolling average. "
)
)
```
The **cases and deaths requirement is that both have been decreasing for the past 14 days.** The past 14 days are shaded in gray.
```
la_county = utils.county_case_charts(county_state_name, start_date)
la_city = utils.lacity_case_charts(start_date)
display(Markdown("## Testing Charts"))
display(Markdown(
f"These charts show the amount of daily testing conducted and the percent of tests that came back positive for COVID-19 by week since {start_date.strftime(fulldate_format)}. "
)
)
```
#### Daily Testing
LA County's goal is to conduct an average of 15,000 tests a day, a rate of 45 tests per 1,000 residents *(lower
bound)*. Chicago, another region faced with a severe outbreak, set the precedent for regional benchmarks being more stringent than statewide requirements if a particular region underwent a more severe outbreak. Chicago's goal is 50 tests per 1,000 residents, or 16,667 tests per day *(upper bound)*.
**The daily testing requirement is that we are conducting at least 15,000 tests daily until a vaccine is ready.** We need to **consistently record testing levels at or above** the lower dashed line.
```
county_tests = utils.lacounty_testing_charts(start_date, county_test_lower_bound, county_test_upper_bound)
```
#### Share of Positive COVID-19 Test Results by Week
LA County's data, though subject to a time lag, does report the number of positive tests per testing batch. We aggregate the results by week. Only weeks with all 7 days of data available is used for the chart, which means the current week is excluded.
The chart compares the percent of positive test results, the number of positive cases, and the number of tests conducted. The percent of positive test results is the indicator of interest, but it is extremely dependent on the number of tests conducted. A higher percentage of positive tests can be due to more confirmed cases or fewer tests conducted. Therefore, the next chart shows the number of tests conducted each week (blue) and the number of positive tests (gray). It also shows the testing upper and lower bounds, which is simply the daily testing upper and lower bounds multiplied by 7.
**How to Interpret Results**
1. If the number of positive tests and the percent of positive tests increase while daily testing is conducted at a similar level, there is increased transmission of the virus.
1. If we keep up our daily testing levels yet see a corresponding drop in the share of positive tests and the number of positive cases, we are curbing the asymptomatic transmission of the virus.
1. If daily testing drops and we see a corresponding drop in positive test results, the decrease in positive results is due to a lack of testing, not because there is less hidden, community transmission of the virus.
1. If daily testing is stable or increasing, the share of positive tests is stable or decreasing, yet the number of positive cases is growing, then our tests are finding the new cases.
**CA's weekly COVID-19 positive share requirement is that tests coming back positive is 8% or below** *(upper bound)*, but experts say that [less than 4% positive](https://www.nytimes.com/2020/05/25/health/coronavirus-testing-trump.html) is necessary to halt the spread of the virus *(lower bound)*.
*Caveat 1:* Testing data for city only counts tests done at city sites. Oral swabs are used by county/city sites, which have an approximate 10% false negative rate. On average, 10% of negative tests are falsely identified to be negative when they are actually positive. At the county level, we do have information on total number of tests, which include county/city sites and private healthcare providers (anyone who provides electronic data reporting), such as Kaiser Permanente. We use the [Tests by Date](http://dashboard.publichealth.lacounty.gov/covid19_surveillance_dashboard/) table. There is a time lag in results reported, and we are not sure if this lag is 3, 5, or 7 days for the results from an entire testing batch to come back.
*Caveat 2:* The situation on-the-ground is important for contextualizing the data's interpretation. When testing capacity is stretched and [testing is rationed](https://calmatters.org/health/coronavirus/2020/07/california-new-coronavirus-testing-guidelines/), those who are able to obtain tests are more likely to test positive. This distorts the share of positive results, and we expect the positivity rate to increase when we target the riskiest subgroups. We are excluding a subset of cases that would test positive, if testing supplies were available. Factoring in the high false negative rate from oral swabs used at county/city sites, we are **undercounting the actual number of cases, but are observing trends from the riskiest or most vulnerable subgroups**.
```
county_positive_tests = utils.lacounty_positive_test_charts(start_date, positive_lower_bound, positive_upper_bound,
county_test_lower_bound, county_test_upper_bound)
```
## Hospitalizations Charts
Data on all COVID-related hospitalizations and ICU hospitalizations comes from the CA open data portal made available 6/25/20; this data **covers the entire county**. These charts show the number of hospitalizations from all COVID cases and also ICU hospitalizations for severe COVID cases.
CA guidelines state that hospitalizations should be stable or downtrending on 7-day average of daily percent change of less than 5%. LA County's all COVID-related hospitalizations and COVID-related ICU hospitalizations (subset of all hospitalizations) are shown.
```
hospitalizations = utils.county_covid_hospital_charts(county_state_name, start_date)
```
If you have any questions, please email [email protected].
|
github_jupyter
|
import numpy as np
import pandas as pd
import utils
import default_parameters
import make_charts
import meet_indicators
import ca_reopening_tiers
from IPython.display import display_html, Markdown, HTML
# Default parameters
county_state_name = default_parameters.county_state_name
state_name = default_parameters.state_name
msa_name = default_parameters.msa_name
time_zone = default_parameters.time_zone
fulldate_format = default_parameters.fulldate_format
monthdate_format = default_parameters.monthdate_format
start_date = default_parameters.start_date
yesterday_date = default_parameters.yesterday_date
today_date = default_parameters.today_date
one_week_ago = default_parameters.one_week_ago
two_weeks_ago = default_parameters.two_weeks_ago
three_weeks_ago = default_parameters.three_weeks_ago
two_days_ago = default_parameters.two_days_ago
eight_days_ago = default_parameters.eight_days_ago
# Daily testing upper and lower bound
county_test_lower_bound = 15_000
county_test_upper_bound = 16_667
positive_lower_bound = 0.04
positive_upper_bound = 0.08
positive_2weeks_bound = 0.05
hospital_bound = 0.30
ca_hospitalization_bound = 0.05
# Set cut-offs for CA reopening
ca_case_minimal_bound = 1
ca_case_moderate_bound = 4
ca_case_substantial_bound = 7
ca_test_minimal_bound = 0.020
ca_test_moderate_bound = 0.050
ca_test_substantial_bound = 0.080
def check_report_readiness(county_state_name, state_name, msa_name, start_date, yesterday_date):
"""
Check if each dataframe has yesterday's date's info.
If all datasets are complete, report can be run.
"""
df = utils.prep_county(county_state_name, start_date)
if df.date.max() < yesterday_date:
raise Exception("Data incomplete")
df = utils.prep_lacity_cases(start_date)
if df.date.max() < yesterday_date:
raise Exception("Data incomplete")
df = utils.prep_testing(start_date)
if (df.date.max() < yesterday_date) or (
(df.date.max() == today_date) and (df.County_Performed == 0) ):
raise Exception("Data incomplete")
df = utils.prep_hospital_surge(county_state_name, start_date)
if df.date.max() < two_days_ago:
raise Exception("Data incomplete")
check_report_readiness(county_state_name, state_name, msa_name, start_date, yesterday_date)
# Check cases according to some criterion
def check_cases(row):
status = ["failed" if x < 14 else "met" if x >= 14 else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_deaths(row):
status = ["failed" if x < 14 else "met" if x >= 14 else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_tests(lower_bound, upper_bound, row):
status = ["failed" if x < lower_bound
else "met" if ((x >= lower_bound) and (x < upper_bound))
else "exceeded" if x >= upper_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_positives(row):
status = ["failed" if x > positive_upper_bound
else "met" if ((x >= positive_lower_bound) and (x <= positive_upper_bound))
else "exceeded" if x < positive_lower_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_positives_two_weeks(row):
status = ["met" if x <= positive_2weeks_bound
else "failed" if x>= positive_2weeks_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_hospitalizations(row):
status = ["met" if x < ca_hospitalization_bound
else "failed" if x >= ca_hospitalization_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
county_fnmap = {
"Cases": check_cases,
"Deaths": check_deaths,
"Daily Testing": lambda row: check_tests(county_test_lower_bound, county_test_upper_bound, row),
"Positive Tests": check_positives,
"Positive Tests (WHO)": check_positives_two_weeks,
"COVID Hospitalizations": check_hospitalizations,
"COVID ICU Hospitalizations": check_hospitalizations,
}
city_fnmap = {
"Cases": check_cases,
"Deaths": check_deaths,
"Daily Testing": lambda row: check_tests(county_test_lower_bound, county_test_upper_bound, row),
"Positive Tests": check_positives,
"Positive Tests (WHO)": check_positives_two_weeks,
"COVID Hospitalizations": check_hospitalizations,
"COVID ICU Hospitalizations": check_hospitalizations,
}
red = make_charts.maroon
green = make_charts.green
blue = make_charts.blue
stylemap = {
"failed": f"background-color: {red}; color: white; font-weight: bold; opacity: 0.7" ,
"met": f"background-color: {green}; color: white; font-weight: bold; opacity: 0.7",
"exceeded": f"background-color: {blue}; color: white; font-weight: bold; opacity: 0.7",
"": "background-color: white; color: black; font-weight: bold; opacity: 0.7",
}
# Add CA Blueprint for Safer Economy (reopening, take 2) criterion to get colors
def check_ca_case(row):
status = ["minimal" if x < ca_case_minimal_bound
else "moderate" if ((x >= ca_case_minimal_bound) and (x < ca_case_moderate_bound))
else "substantial" if ((x >= ca_case_moderate_bound) and (x <= ca_case_substantial_bound))
else "widespread" if x > ca_case_substantial_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_ca_test(row):
status = ["minimal" if x < ca_test_minimal_bound
else "moderate" if (x >= ca_test_minimal_bound) and (x < ca_test_moderate_bound)
else "substantial" if (x >= ca_test_moderate_bound) and (x <= ca_test_substantial_bound)
else "widespread" if x > ca_test_substantial_bound
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
def check_ca_overall_tier(row):
status = ["minimal" if x==1
else "moderate" if x==2
else "substantial" if x==3
else "widespread" if x==4
else "" for x in row]
return pd.Series(status, index=row.index, dtype="category")
ca_tier_fnmap = {
"Overall Tier": check_ca_overall_tier,
"Case Rate per 100k": check_ca_case,
"Test Positivity Rate": check_ca_test,
}
purple = make_charts.purple
red = make_charts.maroon
orange = make_charts.orange
yellow = make_charts.yellow
stylemap_ca = {
"widespread": f"background-color: {purple}; color: white; font-weight: bold; opacity: 0.7" ,
"substantial": f"background-color: {red}; color: white; font-weight: bold; opacity: 0.7",
"moderate": f"background-color: {orange}; color: white; font-weight: bold; opacity: 0.7",
"minimal": f"background-color: {yellow}; color: white; font-weight: bold; opacity: 0.7",
"": "background-color: white; color: black; font-weight: bold; opacity: 0.7",
}
# These are functions used to clean the CA county indicators df
def put_in_tiers(row, indicator, minimal_bound, moderate_bound, substantial_bound):
if row[indicator] < minimal_bound:
return 1
elif (row[indicator] >= minimal_bound) and (row[indicator] < moderate_bound):
return 2
elif (row[indicator] >= moderate_bound) and (row[indicator] <= substantial_bound):
return 3
elif row[indicator] > substantial_bound:
return 4
else:
return np.nan
def add_tiers(df):
df = df.assign(
case_tier = df.apply(lambda x:
put_in_tiers(x, "case",
ca_case_minimal_bound, ca_case_moderate_bound,
ca_case_substantial_bound), axis=1).astype("Int64"),
test_tier = df.apply(lambda x:
put_in_tiers(x, "test",
ca_test_minimal_bound, ca_test_moderate_bound,
ca_test_substantial_bound), axis=1).astype("Int64")
)
# If 2 indicators belong in different tiers, most restrictive (max) is assigned overall
df = (df.assign(
overall_tier = df[["case_tier", "test_tier"]].max(axis=1).astype(int)
)[["overall_tier", "case", "test"]]
.rename(columns = {"overall_tier": "Overall Tier",
"case": "Case Rate per 100k",
"test": "Test Positivity Rate"
})
)
# Transpose
df = (df.T
.rename(columns = {0: "Current Week",
1: "One Week Ago",
2: "Two Weeks Ago",})
)
return df
def summary_of_ca_reopening_indicators():
# Grab indicators
ca_case_today_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "today")
ca_case_last_week_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "one_week_ago")
ca_case_two_week_indicator = ca_reopening_tiers.case_rate(county_state_name, start_date, "two_weeks_ago")
ca_test_last_week_indicator = ca_reopening_tiers.positive_rate(start_date, "one_week_ago")
ca_test_two_week_indicator = ca_reopening_tiers.positive_rate(start_date, "two_weeks_ago")
# Create a df for the county's indicators based off of CA Blueprint to Safer Economy
ca_county = pd.DataFrame(
{"case": [ca_case_today_indicator, ca_case_last_week_indicator, ca_case_two_week_indicator],
"test": [np.nan, ca_test_last_week_indicator, ca_test_two_week_indicator]
}
)
# Clean up df
df = add_tiers(ca_county)
# Style the table
ca_county_html = (df.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: ca_tier_fnmap[row.name](row).map(stylemap_ca), axis=1)
.render()
)
display(Markdown(f"<strong>CA Reopening Metrics: {county_state_name.split(',')[0]} County</strong>"))
display(HTML(ca_county_html))
def summary_of_indicators():
# Grab indicators
county_case_indicator = meet_indicators.meet_case("county", county_state_name, start_date)
county_death_indicator = meet_indicators.meet_death("county", county_state_name, start_date)
county_test_indicator = meet_indicators.meet_daily_testing(yesterday_date, "county",
county_test_lower_bound, county_test_upper_bound)
county_positive_indicator = meet_indicators.meet_positive_share(yesterday_date, "county",
positive_lower_bound, positive_upper_bound)
county_positive_2wks_indicator = meet_indicators.meet_positive_share_for_two_weeks(yesterday_date, "county")
# 8/11: Ignore LA County HavBed data, since it seems to be reported much more infrequently
# Use CA data portal's hospitalizations
hospitalization_indicator = meet_indicators.meet_all_hospitalization(county_state_name, yesterday_date)
icu_hospitalization_indicator = meet_indicators.meet_icu_hospitalization(county_state_name, yesterday_date)
city_case_indicator = meet_indicators.meet_case("lacity", "City of LA", start_date)
city_death_indicator = meet_indicators.meet_death("lacity", "City of LA", start_date)
indicator_names = ["Cases", "Deaths",
"Daily Testing", "Positive Tests", "Positive Tests (WHO)",
"COVID Hospitalizations", "COVID ICU Hospitalizations"]
# Create separate df for county and city
county = pd.DataFrame(
{"LA County": [county_case_indicator, county_death_indicator,
county_test_indicator, county_positive_indicator, county_positive_2wks_indicator,
hospitalization_indicator, icu_hospitalization_indicator]},
index=indicator_names
)
city = pd.DataFrame(
{"City of LA": [city_case_indicator, city_death_indicator,
np.nan, np.nan, np.nan,
np.nan, np.nan]},
index=indicator_names
)
# Style the table
def display_side_by_side(*args):
html_str=''
for df in args:
html_str+=df
display_html(html_str.replace('table','table style="display:inline"'),raw=True)
county_html = (county.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: county_fnmap[row.name](row).map(stylemap), axis=1)
.render()
)
city_html = (city.style
.format(lambda s: f"{s:,g}", na_rep="-")
.apply(lambda row: city_fnmap[row.name](row).map(stylemap), axis=1)
.hide_index()
.render()
)
display(Markdown("<strong>Indicators for LA County and City of LA</strong>"))
display_side_by_side(county_html, city_html)
display(Markdown(f"### Summary of Indicators as of {yesterday_date.strftime(fulldate_format)}:"))
summary_of_ca_reopening_indicators()
summary_of_indicators()
display(Markdown("## Caseload Charts"))
display(Markdown(
f"These are the trends in cases and deaths for the county and the city since {start_date.strftime(fulldate_format)}, using a 7-day rolling average. "
)
)
la_county = utils.county_case_charts(county_state_name, start_date)
la_city = utils.lacity_case_charts(start_date)
display(Markdown("## Testing Charts"))
display(Markdown(
f"These charts show the amount of daily testing conducted and the percent of tests that came back positive for COVID-19 by week since {start_date.strftime(fulldate_format)}. "
)
)
county_tests = utils.lacounty_testing_charts(start_date, county_test_lower_bound, county_test_upper_bound)
county_positive_tests = utils.lacounty_positive_test_charts(start_date, positive_lower_bound, positive_upper_bound,
county_test_lower_bound, county_test_upper_bound)
hospitalizations = utils.county_covid_hospital_charts(county_state_name, start_date)
| 0.289975 | 0.910942 |
## Project 3 Codebook
### Topic: Spatial Analysis of Exposure to Respiratory Hazards and School Performance in Riverside, California.
This codebook contains a step by step guide to download, retrieve, subset and visualize data pooled from different sources (EPA EJscreen, NCES, SEDA, ACS).
Time Frame for Analysis
1. EJScreen data - 2020
2. NCES/Districts - 2018/2019
3. ACS- 2018
4. SEDA- Standardized test scores administered in 3rd through 8 th grade in mathematics and Reading Language Arts (RLA) over the 2008-09 through 2017-18 school years.
**Note:Some cells have # infront of code(remove hashtag to run the code in the cells)**
### 1: Download software
```
# Import software
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as ctx
import numpy as np
import quilt3
from geopandas_view import view
import libpysal as lps
import seaborn as sns
import tobler as tob
plt.rcParams['figure.figsize'] = [20, 10]
```
### 2: Retrieve and Adjust Data
#### 2.1 Environmental Justice Screen Data From EPA
```
# Retrieve EPA EJ Screen data from UCR CGS Quilt Bucket
b = quilt3.Bucket("s3://spatial-ucr")
b.fetch("epa/ejscreen/ejscreen_2020.parquet", "./ejscreen_2020.parquet"), # Might be a good idea to get 2018
ej = pd.read_parquet('ejscreen_2020.parquet')
ej.head()
# Rename EJ column to state GEOID
ej.rename(columns = {'ID' : 'GEOID'}, inplace = True)
# ej.head()
ej.columns
# Download USA Census block groups from the 2018 ACS via Quilt
geoms = gpd.read_parquet('s3://spatial-ucr/census/acs/acs_2018_bg.parquet')
geoms.head()
# Merge EJ and ACS data
ej = geoms.merge(ej, on='GEOID')
# ej.head()
# Filter EJ Screen data so it only displays CA
ca_ej = ej[ej.GEOID.str.startswith('06')]
ca_ej.head()
# Filter out EJ Index for Air toxics respiratory hazard index
D_RESP_2 = ca_ej.D_RESP_2
ca_ej.D_RESP_2 = D_RESP_2.replace(to_replace= "None", value=np.nan).astype(float)
# Create variable for Riverside, Orange and LA County using the fips code
riv_ej = ca_ej[ca_ej.GEOID.str.startswith("06065")] # Riverside County EJSCREEN
# Checking Data for Orange County
#riv_ej.head()
riv_ej.shape
# New dataframe for respiratory hazards index, minority percent , low income percent
new_riverside = riv_ej[['D_RESP_2','MINORPCT','LOWINCPCT', 'RESP','geometry']]
new_riverside.head()
#new_riverside.groupby(by ='MINORPCT').sum()
#Statistics
new_riverside.describe()
```
### 3: Merge District Data w/ EJ Data
```
# Upload school districts data
districts = gpd.read_parquet('s3://spatial-ucr/nces/districts/school_districts_1819.parquet')
# Subset of the california schools from the US data
CA_dist = districts[districts.STATEFP == "06"]
CA_dist.head()
# Locate Riverside Unified School District (RUSD)- better to make sure its in California
rusd_CA = districts[districts['STATEFP'].str.lower().str.contains('06')]
rusd = rusd_CA[rusd_CA['NAME'].str.lower().str.contains('riverside unified')]
rusd.head()
# Now, let's overlay the EPA EJ data (that was combined with ACS data) on top of the RUSD shape
rusd_ej = gpd.overlay(riv_ej,rusd, how='intersection')
```
### 4: Combine School Locations with School District/EJ Map
```
# Download NCES school location data
schools = gpd.read_parquet("s3://spatial-ucr/nces/schools/schools_1819.parquet")
#schools.head()
# Download SEDA learning outcomes data
seda = pd.read_csv("https://stacks.stanford.edu/file/druid:db586ns4974/seda_school_pool_gcs_4.0.csv",converters={"sedasch":str})
seda.sedasch=seda.sedasch.str.rjust(12, "0")
#seda.head()
# Convert NCES data into a GeoDataFrame
school_geoms = schools[['NCESSCH','CNTY','NMCNTY', 'geometry']]
school_geoms.head()
# Merge SEDA and NCES data
seda_merge = seda.merge(school_geoms, left_on="sedasch", right_on= "NCESSCH")
#seda_merge.head()
# Convert merged NCES/SEDA data into a GeoDataFrame and plot it
seda_merge= gpd.GeoDataFrame(seda_merge)
#seda_merge.plot()
# Subset data to only locate schools in Riverside County
riv_schools = seda_merge[seda_merge['CNTY']=='06065']
#riv_schools.plot()
# Subset school data to find schools in RUSD
rusd_schools = gpd.overlay(riv_schools, rusd, how='intersection')
```
### 5: Voronoi Polygons
```
# Subset of rusd schools with only mean scores
rusd_pts= rusd_schools[['gcs_mn_avg_ol','geometry']]
rusd_pts.head()
rusd_pts.plot(column='gcs_mn_avg_ol', legend=True)
# Subset for EJ screen for RUSD only containing Respiratory index and geometry
EJ_RUSD= rusd_ej[['D_RESP_2','geometry']]
EJ_RUSD.plot(column='D_RESP_2', cmap='Greens', scheme='Quantiles', k=3,edgecolor='grey',
legend=True)
```
### 5.1 Overlays
#### Approach 1 Spatial Join
```
# 1 for 1 mapping
base = EJ_RUSD.plot(column='D_RESP_2', legend=True)
rusd_pts.plot(color='red', ax=base)
rusd_sch= gpd.sjoin(rusd_pts, EJ_RUSD, how='left', op='within')
rusd_sch.head()
rusd_pts['D_RESP_2'] = rusd_sch.D_RESP_2
rusd_pts.head()
rusd_pts.crs
```
#### Approach 2 Areal Interpolation
```
x = rusd_pts.geometry.x
y = rusd_pts.geometry.y
cents = np.array([x,y]).T
cents
schools_vd, school_cents = lps.cg.voronoi_frames(cents)
base = schools_vd.plot()
rusd_pts.plot(ax=base, color='red')
EJ_RUSD.crs
base = EJ_RUSD.geometry.boundary.plot(edgecolor='green')
schools_vd.plot(ax=base)
rusd_pts.plot(ax=base, color='red')
schools_vd, school_cents = lps.cg.voronoi_frames(cents, clip = EJ_RUSD.unary_union)
base = EJ_RUSD.geometry.boundary.plot(edgecolor='green')
schools_vd.plot(ax=base)
rusd_pts.plot(ax=base, color='red')
base = EJ_RUSD.plot(column='D_RESP_2')
schools_vd.geometry.boundary.plot(ax=base, edgecolor='red')
rusd_pts.plot(ax=base, color='red')
```
Estimate the escore for a school using areal interpolation
```
RESP = tob.area_weighted.area_interpolate(source_df=EJ_RUSD,
target_df=schools_vd,
intensive_variables=['D_RESP_2'])
RESP
base = RESP.plot(column='D_RESP_2')
rusd_pts.plot(ax=base, color='red')
x = EJ_RUSD.to_crs(epsg=4269)
gdf_proj = x.to_crs(EJ_RUSD.crs)
gdf_proj.head()
boundary_shape = cascaded_union(x.geometry)
coords = points_to_coords(gdf_proj.geometry)
# Calculate Voronoi Regions
poly_shapes, pts, poly_to_pt_assignments = voronoi_regions_from_coords(coords, boundary_shape)
```
|
github_jupyter
|
# Import software
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as ctx
import numpy as np
import quilt3
from geopandas_view import view
import libpysal as lps
import seaborn as sns
import tobler as tob
plt.rcParams['figure.figsize'] = [20, 10]
# Retrieve EPA EJ Screen data from UCR CGS Quilt Bucket
b = quilt3.Bucket("s3://spatial-ucr")
b.fetch("epa/ejscreen/ejscreen_2020.parquet", "./ejscreen_2020.parquet"), # Might be a good idea to get 2018
ej = pd.read_parquet('ejscreen_2020.parquet')
ej.head()
# Rename EJ column to state GEOID
ej.rename(columns = {'ID' : 'GEOID'}, inplace = True)
# ej.head()
ej.columns
# Download USA Census block groups from the 2018 ACS via Quilt
geoms = gpd.read_parquet('s3://spatial-ucr/census/acs/acs_2018_bg.parquet')
geoms.head()
# Merge EJ and ACS data
ej = geoms.merge(ej, on='GEOID')
# ej.head()
# Filter EJ Screen data so it only displays CA
ca_ej = ej[ej.GEOID.str.startswith('06')]
ca_ej.head()
# Filter out EJ Index for Air toxics respiratory hazard index
D_RESP_2 = ca_ej.D_RESP_2
ca_ej.D_RESP_2 = D_RESP_2.replace(to_replace= "None", value=np.nan).astype(float)
# Create variable for Riverside, Orange and LA County using the fips code
riv_ej = ca_ej[ca_ej.GEOID.str.startswith("06065")] # Riverside County EJSCREEN
# Checking Data for Orange County
#riv_ej.head()
riv_ej.shape
# New dataframe for respiratory hazards index, minority percent , low income percent
new_riverside = riv_ej[['D_RESP_2','MINORPCT','LOWINCPCT', 'RESP','geometry']]
new_riverside.head()
#new_riverside.groupby(by ='MINORPCT').sum()
#Statistics
new_riverside.describe()
# Upload school districts data
districts = gpd.read_parquet('s3://spatial-ucr/nces/districts/school_districts_1819.parquet')
# Subset of the california schools from the US data
CA_dist = districts[districts.STATEFP == "06"]
CA_dist.head()
# Locate Riverside Unified School District (RUSD)- better to make sure its in California
rusd_CA = districts[districts['STATEFP'].str.lower().str.contains('06')]
rusd = rusd_CA[rusd_CA['NAME'].str.lower().str.contains('riverside unified')]
rusd.head()
# Now, let's overlay the EPA EJ data (that was combined with ACS data) on top of the RUSD shape
rusd_ej = gpd.overlay(riv_ej,rusd, how='intersection')
# Download NCES school location data
schools = gpd.read_parquet("s3://spatial-ucr/nces/schools/schools_1819.parquet")
#schools.head()
# Download SEDA learning outcomes data
seda = pd.read_csv("https://stacks.stanford.edu/file/druid:db586ns4974/seda_school_pool_gcs_4.0.csv",converters={"sedasch":str})
seda.sedasch=seda.sedasch.str.rjust(12, "0")
#seda.head()
# Convert NCES data into a GeoDataFrame
school_geoms = schools[['NCESSCH','CNTY','NMCNTY', 'geometry']]
school_geoms.head()
# Merge SEDA and NCES data
seda_merge = seda.merge(school_geoms, left_on="sedasch", right_on= "NCESSCH")
#seda_merge.head()
# Convert merged NCES/SEDA data into a GeoDataFrame and plot it
seda_merge= gpd.GeoDataFrame(seda_merge)
#seda_merge.plot()
# Subset data to only locate schools in Riverside County
riv_schools = seda_merge[seda_merge['CNTY']=='06065']
#riv_schools.plot()
# Subset school data to find schools in RUSD
rusd_schools = gpd.overlay(riv_schools, rusd, how='intersection')
# Subset of rusd schools with only mean scores
rusd_pts= rusd_schools[['gcs_mn_avg_ol','geometry']]
rusd_pts.head()
rusd_pts.plot(column='gcs_mn_avg_ol', legend=True)
# Subset for EJ screen for RUSD only containing Respiratory index and geometry
EJ_RUSD= rusd_ej[['D_RESP_2','geometry']]
EJ_RUSD.plot(column='D_RESP_2', cmap='Greens', scheme='Quantiles', k=3,edgecolor='grey',
legend=True)
# 1 for 1 mapping
base = EJ_RUSD.plot(column='D_RESP_2', legend=True)
rusd_pts.plot(color='red', ax=base)
rusd_sch= gpd.sjoin(rusd_pts, EJ_RUSD, how='left', op='within')
rusd_sch.head()
rusd_pts['D_RESP_2'] = rusd_sch.D_RESP_2
rusd_pts.head()
rusd_pts.crs
x = rusd_pts.geometry.x
y = rusd_pts.geometry.y
cents = np.array([x,y]).T
cents
schools_vd, school_cents = lps.cg.voronoi_frames(cents)
base = schools_vd.plot()
rusd_pts.plot(ax=base, color='red')
EJ_RUSD.crs
base = EJ_RUSD.geometry.boundary.plot(edgecolor='green')
schools_vd.plot(ax=base)
rusd_pts.plot(ax=base, color='red')
schools_vd, school_cents = lps.cg.voronoi_frames(cents, clip = EJ_RUSD.unary_union)
base = EJ_RUSD.geometry.boundary.plot(edgecolor='green')
schools_vd.plot(ax=base)
rusd_pts.plot(ax=base, color='red')
base = EJ_RUSD.plot(column='D_RESP_2')
schools_vd.geometry.boundary.plot(ax=base, edgecolor='red')
rusd_pts.plot(ax=base, color='red')
RESP = tob.area_weighted.area_interpolate(source_df=EJ_RUSD,
target_df=schools_vd,
intensive_variables=['D_RESP_2'])
RESP
base = RESP.plot(column='D_RESP_2')
rusd_pts.plot(ax=base, color='red')
x = EJ_RUSD.to_crs(epsg=4269)
gdf_proj = x.to_crs(EJ_RUSD.crs)
gdf_proj.head()
boundary_shape = cascaded_union(x.geometry)
coords = points_to_coords(gdf_proj.geometry)
# Calculate Voronoi Regions
poly_shapes, pts, poly_to_pt_assignments = voronoi_regions_from_coords(coords, boundary_shape)
| 0.520253 | 0.916633 |
# Collaboration and Competition
---
### 1. Start the Environment
We begin by importing the necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).
```
from unityagents import UnityEnvironment
import numpy as np
```
Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.
- **Mac**: `"path/to/Tennis.app"`
- **Windows** (x86_64): `"path/to/Tennis_Windows_x86_64/Tennis.exe"`
For instance, if you are using a Mac, then you downloaded `Tennis.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:
```
env = UnityEnvironment(file_name="Tennis.app")
```
```
# Imports
import random
import torch
import os
import numpy as np
from collections import deque
import time
import matplotlib.pyplot as plt
import sys
sys.path.append("scripts/")
# Set plotting options
%matplotlib inline
plt.style.use('ggplot')
np.set_printoptions(precision=3, linewidth=120)
# Hide Matplotlib deprecate warnings
import warnings
warnings.filterwarnings("ignore")
# High resolution plot outputs for retina display
%config InlineBackend.figure_format = 'retina'
# Path to save the mdoels and collect the Tensorboard logs
model_dir= os.getcwd()+"/model_dir"
os.makedirs(model_dir, exist_ok=True)
model_dir
env = UnityEnvironment(file_name="Tennis_Windows_x86_64/Tennis.exe")
```
Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.
```
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
```
### 2. Examine the State and Action Spaces
In this environment, two agents control rackets to bounce a ball over a net. If an agent hits the ball over the net, it receives a reward of +0.1. If an agent lets a ball hit the ground or hits the ball out of bounds, it receives a reward of -0.01. Thus, the goal of each agent is to keep the ball in play.
The observation space consists of 8 variables corresponding to the position and velocity of the ball and racket. Two continuous actions are available, corresponding to movement toward (or away from) the net, and jumping.
Run the code cell below to print some information about the environment.
```
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
# size of each action
ENV_ACTION_SIZE = brain.vector_action_space_size
# size of the state space
ENV_STATE_SIZE = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
```
### 3. Take Random Actions in the Environment
In the next code cell, you will learn how to use the Python API to control the agents and receive feedback from the environment.
Once this cell is executed, you will watch the agents' performance, if they select actions at random with each time step. A window should pop up that allows you to observe the agents.
Of course, as part of the project, you'll have to change the code so that the agents are able to use their experiences to gradually choose better actions when interacting with the environment!
```
for i in range(1, 6): # play game for 5 episodes
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Score (max over agents) from episode {}: {}'.format(i, np.max(scores)))
# Helper function to plot the scores
def plot_training(scores):
# Plot the Score evolution during the training
fig = plt.figure()
ax = fig.add_subplot(111)
ax.tick_params(axis='x', colors='deepskyblue')
ax.tick_params(axis='y', colors='deepskyblue')
plt.plot(np.arange(1, len(scores)+1), scores, color='deepskyblue')
plt.ylabel('Score', color='deepskyblue')
plt.xlabel('Episode #', color='deepskyblue')
plt.show()
from scripts.maddpg_agents import Maddpg
from scripts.hyperparameters import *
def train():
# Seeding
np.random.seed(SEED)
torch.manual_seed(SEED)
# Instantiate the MADDPG agents
maddpg = Maddpg(ENV_STATE_SIZE, ENV_ACTION_SIZE, num_agents, SEED)
# Monitor the score
scores_deque = deque(maxlen=100)
all_scores = []
all_avg_score = []
# Intialize amplitude OU noise (will decay during training)
noise = NOISE
all_steps =0 # Monitor total number of steps performed
# Training Loop
for i_episode in range(NB_EPISODES+1):
env_info = env.reset(train_mode=True)[brain_name] # reset the environment
maddpg.reset() # reset the agents
states = env_info.vector_observations # get the current state for each agent
scores = np.zeros(num_agents) # initialize the score (for each agent)
for steps in range(NB_STEPS):
all_steps+=1
actions = maddpg.act(states, noise) # retrieve actions to performe for each agents
noise *= NOISE_REDUCTION # Decrease action noise
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state for each agent
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
# Save experience in replay memory, and use random sample from buffer to learn
maddpg.step(states, actions, rewards, next_states, dones, i_episode)
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
#print(" ** Debug: episode= {} steps={} rewards={} dones={}".format(i_episode, steps,rewards,dones))
break
# Save scores and compute average score over last 100 episodes
episode_score = np.max(scores) # Consider the maximum score amongs all Agents
all_scores.append(episode_score)
scores_deque.append(episode_score)
avg_score = np.mean(scores_deque)
# Display statistics
print('\rEpisode {}\tAverage Score: {:.2f}\tEpisode score (max over agents): {:.2f}'.format(i_episode, avg_score, episode_score), end="")
if i_episode>0 and i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f} (nb of total steps={} noise={:.4f})'.format(i_episode, avg_score, all_steps, noise))
maddpg.checkpoints()
all_avg_score.append(avg_score)
# Early stop
if (i_episode > 99) and (avg_score >=0.5):
print('\rEnvironment solved in {} episodes with an Average Score of {:.2f}'.format(i_episode, avg_score))
maddpg.checkpoints()
return all_scores
return all_scores
scores = train()
plot_training(scores)
maddpg = Maddpg(ENV_STATE_SIZE, ENV_ACTION_SIZE, num_agents, SEED)
actor_local_filename = 'model_dir/checkpoint_actor_local_0.pth'
critic_local_filename = 'model_dir/checkpoint_critic_local_0.pth'
actor_target_filename = 'model_dir/checkpoint_actor_target_0.pth'
critic_target_filename = 'model_dir/checkpoint_critic_target_0.pth'
actor_local_filename1 = 'model_dir/checkpoint_actor_local_1.pth'
critic_local_filename1 = 'model_dir/checkpoint_critic_local_1.pth'
actor_target_filename1 = 'model_dir/checkpoint_actor_target_1.pth'
critic_target_filename1 = 'model_dir/checkpoint_critic_target_1.pth'
maddpg.agents[0].actor_local.load_state_dict(torch.load(actor_local_filename))
maddpg.agents[1].actor_local.load_state_dict(torch.load(actor_local_filename1))
maddpg.agents[0].actor_target.load_state_dict(torch.load(actor_target_filename))
maddpg.agents[1].actor_target.load_state_dict(torch.load(actor_target_filename1))
maddpg.agents[0].critic_local.load_state_dict(torch.load(critic_local_filename))
maddpg.agents[1].critic_local.load_state_dict(torch.load(critic_local_filename1))
maddpg.agents[0].critic_target.load_state_dict(torch.load(critic_target_filename))
maddpg.agents[1].critic_target.load_state_dict(torch.load(critic_target_filename1))
num_episodes = 3
noise = NOISE
scores = []
for i_episode in range(1,num_episodes+1):
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state for each agent
scores = np.zeros(num_agents) # initialize the score
while True:
actions = maddpg.act(states, noise) # retrieve actions to performe for each agents
noise *= NOISE_REDUCTION # Decrease action noise
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state for each agent
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
# Save experience in replay memory, and use random sample from buffer to learn
maddpg.step(states, actions, rewards, next_states, dones, i_episode)
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
#print(" ** Debug: episode= {} steps={} rewards={} dones={}".format(i_episode, steps,rewards,dones))
break
```
When finished, you can close the environment.
```
env.close()
```
### 4. It's Your Turn!
Now it's your turn to train your own agent to solve the environment! When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:
```python
env_info = env.reset(train_mode=True)[brain_name]
```
|
github_jupyter
|
from unityagents import UnityEnvironment
import numpy as np
env = UnityEnvironment(file_name="Tennis.app")
# Imports
import random
import torch
import os
import numpy as np
from collections import deque
import time
import matplotlib.pyplot as plt
import sys
sys.path.append("scripts/")
# Set plotting options
%matplotlib inline
plt.style.use('ggplot')
np.set_printoptions(precision=3, linewidth=120)
# Hide Matplotlib deprecate warnings
import warnings
warnings.filterwarnings("ignore")
# High resolution plot outputs for retina display
%config InlineBackend.figure_format = 'retina'
# Path to save the mdoels and collect the Tensorboard logs
model_dir= os.getcwd()+"/model_dir"
os.makedirs(model_dir, exist_ok=True)
model_dir
env = UnityEnvironment(file_name="Tennis_Windows_x86_64/Tennis.exe")
# get the default brain
brain_name = env.brain_names[0]
brain = env.brains[brain_name]
# reset the environment
env_info = env.reset(train_mode=True)[brain_name]
# number of agents
num_agents = len(env_info.agents)
print('Number of agents:', num_agents)
# size of each action
action_size = brain.vector_action_space_size
print('Size of each action:', action_size)
# examine the state space
states = env_info.vector_observations
state_size = states.shape[1]
# size of each action
ENV_ACTION_SIZE = brain.vector_action_space_size
# size of the state space
ENV_STATE_SIZE = states.shape[1]
print('There are {} agents. Each observes a state with length: {}'.format(states.shape[0], state_size))
print('The state for the first agent looks like:', states[0])
for i in range(1, 6): # play game for 5 episodes
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state (for each agent)
scores = np.zeros(num_agents) # initialize the score (for each agent)
while True:
actions = np.random.randn(num_agents, action_size) # select an action (for each agent)
actions = np.clip(actions, -1, 1) # all actions between -1 and 1
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state (for each agent)
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
break
print('Score (max over agents) from episode {}: {}'.format(i, np.max(scores)))
# Helper function to plot the scores
def plot_training(scores):
# Plot the Score evolution during the training
fig = plt.figure()
ax = fig.add_subplot(111)
ax.tick_params(axis='x', colors='deepskyblue')
ax.tick_params(axis='y', colors='deepskyblue')
plt.plot(np.arange(1, len(scores)+1), scores, color='deepskyblue')
plt.ylabel('Score', color='deepskyblue')
plt.xlabel('Episode #', color='deepskyblue')
plt.show()
from scripts.maddpg_agents import Maddpg
from scripts.hyperparameters import *
def train():
# Seeding
np.random.seed(SEED)
torch.manual_seed(SEED)
# Instantiate the MADDPG agents
maddpg = Maddpg(ENV_STATE_SIZE, ENV_ACTION_SIZE, num_agents, SEED)
# Monitor the score
scores_deque = deque(maxlen=100)
all_scores = []
all_avg_score = []
# Intialize amplitude OU noise (will decay during training)
noise = NOISE
all_steps =0 # Monitor total number of steps performed
# Training Loop
for i_episode in range(NB_EPISODES+1):
env_info = env.reset(train_mode=True)[brain_name] # reset the environment
maddpg.reset() # reset the agents
states = env_info.vector_observations # get the current state for each agent
scores = np.zeros(num_agents) # initialize the score (for each agent)
for steps in range(NB_STEPS):
all_steps+=1
actions = maddpg.act(states, noise) # retrieve actions to performe for each agents
noise *= NOISE_REDUCTION # Decrease action noise
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state for each agent
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
# Save experience in replay memory, and use random sample from buffer to learn
maddpg.step(states, actions, rewards, next_states, dones, i_episode)
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
#print(" ** Debug: episode= {} steps={} rewards={} dones={}".format(i_episode, steps,rewards,dones))
break
# Save scores and compute average score over last 100 episodes
episode_score = np.max(scores) # Consider the maximum score amongs all Agents
all_scores.append(episode_score)
scores_deque.append(episode_score)
avg_score = np.mean(scores_deque)
# Display statistics
print('\rEpisode {}\tAverage Score: {:.2f}\tEpisode score (max over agents): {:.2f}'.format(i_episode, avg_score, episode_score), end="")
if i_episode>0 and i_episode % 100 == 0:
print('\rEpisode {}\tAverage Score: {:.2f} (nb of total steps={} noise={:.4f})'.format(i_episode, avg_score, all_steps, noise))
maddpg.checkpoints()
all_avg_score.append(avg_score)
# Early stop
if (i_episode > 99) and (avg_score >=0.5):
print('\rEnvironment solved in {} episodes with an Average Score of {:.2f}'.format(i_episode, avg_score))
maddpg.checkpoints()
return all_scores
return all_scores
scores = train()
plot_training(scores)
maddpg = Maddpg(ENV_STATE_SIZE, ENV_ACTION_SIZE, num_agents, SEED)
actor_local_filename = 'model_dir/checkpoint_actor_local_0.pth'
critic_local_filename = 'model_dir/checkpoint_critic_local_0.pth'
actor_target_filename = 'model_dir/checkpoint_actor_target_0.pth'
critic_target_filename = 'model_dir/checkpoint_critic_target_0.pth'
actor_local_filename1 = 'model_dir/checkpoint_actor_local_1.pth'
critic_local_filename1 = 'model_dir/checkpoint_critic_local_1.pth'
actor_target_filename1 = 'model_dir/checkpoint_actor_target_1.pth'
critic_target_filename1 = 'model_dir/checkpoint_critic_target_1.pth'
maddpg.agents[0].actor_local.load_state_dict(torch.load(actor_local_filename))
maddpg.agents[1].actor_local.load_state_dict(torch.load(actor_local_filename1))
maddpg.agents[0].actor_target.load_state_dict(torch.load(actor_target_filename))
maddpg.agents[1].actor_target.load_state_dict(torch.load(actor_target_filename1))
maddpg.agents[0].critic_local.load_state_dict(torch.load(critic_local_filename))
maddpg.agents[1].critic_local.load_state_dict(torch.load(critic_local_filename1))
maddpg.agents[0].critic_target.load_state_dict(torch.load(critic_target_filename))
maddpg.agents[1].critic_target.load_state_dict(torch.load(critic_target_filename1))
num_episodes = 3
noise = NOISE
scores = []
for i_episode in range(1,num_episodes+1):
env_info = env.reset(train_mode=False)[brain_name] # reset the environment
states = env_info.vector_observations # get the current state for each agent
scores = np.zeros(num_agents) # initialize the score
while True:
actions = maddpg.act(states, noise) # retrieve actions to performe for each agents
noise *= NOISE_REDUCTION # Decrease action noise
env_info = env.step(actions)[brain_name] # send all actions to tne environment
next_states = env_info.vector_observations # get next state for each agent
rewards = env_info.rewards # get reward (for each agent)
dones = env_info.local_done # see if episode finished
# Save experience in replay memory, and use random sample from buffer to learn
maddpg.step(states, actions, rewards, next_states, dones, i_episode)
scores += env_info.rewards # update the score (for each agent)
states = next_states # roll over states to next time step
if np.any(dones): # exit loop if episode finished
#print(" ** Debug: episode= {} steps={} rewards={} dones={}".format(i_episode, steps,rewards,dones))
break
env.close()
env_info = env.reset(train_mode=True)[brain_name]
| 0.467332 | 0.979413 |
[View in Colaboratory](https://colab.research.google.com/github/Kremer80/LEOgit/blob/master/zalando-model01.ipynb)
```
# TensorFlow and tf.keras
import tensorflow as tf
from tensorflow import keras
# Helper libraries
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
fashion_mnist = keras.datasets.fashion_mnist
(train_images, train_labels), (test_images, test_labels) = fashion_mnist.load_data()
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
train_images.shape
len(train_labels)
train_labels
test_images.shape
len(test_labels)
plt.figure()
plt.imshow(train_images[0])
plt.colorbar()
plt.gca().grid(True)
train_images = train_images / 255.0
test_images = test_images / 255.0
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=(10,10))
for i in range(36):
plt.subplot(6,6,i+1)
plt.xticks([])
plt.yticks([])
plt.grid('off')
plt.imshow(train_images[i], cmap=plt.cm.binary)
plt.xlabel(class_names[train_labels[i]])
model = keras.Sequential([
keras.layers.Flatten(input_shape=(28, 28)),
keras.layers.Dense(128, activation=tf.nn.relu),
keras.layers.Dense(10, activation=tf.nn.softmax)
])
print(model)
model.compile(optimizer=tf.train.AdamOptimizer(),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
model.fit(train_images, train_labels, epochs=5)
test_loss, test_acc = model.evaluate(test_images, test_labels)
print('Test accuracy:', test_acc)
predictions = model.predict(test_images)
predictions[2]
np.argmax(predictions[2])
# Plot the first 25 test images, their predicted label, and the true label
# Color correct predictions in green, incorrect predictions in red
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid('off')
plt.imshow(test_images[i], cmap=plt.cm.binary)
predicted_label = np.argmax(predictions[i])
true_label = test_labels[i]
if predicted_label == true_label:
color = 'green'
else:
color = 'red'
plt.xlabel("{} ({})".format(class_names[predicted_label],
class_names[true_label]),
color=color)
# Grab an image from the test dataset
img = test_images[0]
print(img.shape)
# Add the image to a batch where it's the only member.
img = (np.expand_dims(img,0))
print(img.shape)
predictions = model.predict(img)
print(predictions)
prediction = predictions[0]
np.argmax(prediction)
```
|
github_jupyter
|
# TensorFlow and tf.keras
import tensorflow as tf
from tensorflow import keras
# Helper libraries
import numpy as np
import matplotlib.pyplot as plt
print(tf.__version__)
fashion_mnist = keras.datasets.fashion_mnist
(train_images, train_labels), (test_images, test_labels) = fashion_mnist.load_data()
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
train_images.shape
len(train_labels)
train_labels
test_images.shape
len(test_labels)
plt.figure()
plt.imshow(train_images[0])
plt.colorbar()
plt.gca().grid(True)
train_images = train_images / 255.0
test_images = test_images / 255.0
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=(10,10))
for i in range(36):
plt.subplot(6,6,i+1)
plt.xticks([])
plt.yticks([])
plt.grid('off')
plt.imshow(train_images[i], cmap=plt.cm.binary)
plt.xlabel(class_names[train_labels[i]])
model = keras.Sequential([
keras.layers.Flatten(input_shape=(28, 28)),
keras.layers.Dense(128, activation=tf.nn.relu),
keras.layers.Dense(10, activation=tf.nn.softmax)
])
print(model)
model.compile(optimizer=tf.train.AdamOptimizer(),
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
model.fit(train_images, train_labels, epochs=5)
test_loss, test_acc = model.evaluate(test_images, test_labels)
print('Test accuracy:', test_acc)
predictions = model.predict(test_images)
predictions[2]
np.argmax(predictions[2])
# Plot the first 25 test images, their predicted label, and the true label
# Color correct predictions in green, incorrect predictions in red
plt.figure(figsize=(10,10))
for i in range(25):
plt.subplot(5,5,i+1)
plt.xticks([])
plt.yticks([])
plt.grid('off')
plt.imshow(test_images[i], cmap=plt.cm.binary)
predicted_label = np.argmax(predictions[i])
true_label = test_labels[i]
if predicted_label == true_label:
color = 'green'
else:
color = 'red'
plt.xlabel("{} ({})".format(class_names[predicted_label],
class_names[true_label]),
color=color)
# Grab an image from the test dataset
img = test_images[0]
print(img.shape)
# Add the image to a batch where it's the only member.
img = (np.expand_dims(img,0))
print(img.shape)
predictions = model.predict(img)
print(predictions)
prediction = predictions[0]
np.argmax(prediction)
| 0.883713 | 0.944791 |
#1. Install Dependencies
First install the libraries needed to execute recipes, this only needs to be done once, then click play.
```
!pip install git+https://github.com/google/starthinker
```
#2. Get Cloud Project ID
To run this recipe [requires a Google Cloud Project](https://github.com/google/starthinker/blob/master/tutorials/cloud_project.md), this only needs to be done once, then click play.
```
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
```
#3. Get Client Credentials
To read and write to various endpoints requires [downloading client credentials](https://github.com/google/starthinker/blob/master/tutorials/cloud_client_installed.md), this only needs to be done once, then click play.
```
CLIENT_CREDENTIALS = 'PASTE CLIENT CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
```
#4. Enter DV360 User Audit Parameters
Gives DV clients ability to see which users have access to which parts of an account. Loads DV user profile mappings using the API into BigQuery and connects to a DataStudio dashboard.
1. DV360 only permits SERVICE accounts to access the user list API endpoint, be sure to provide and permission one.
1. Wait for <b>BigQuery->->->DV_*</b> to be created.
1. Wait for <b>BigQuery->->->Barnacle_*</b> to be created, then copy and connect the following data sources.
1. Join the <a href='https://groups.google.com/d/forum/starthinker-assets' target='_blank'>StarThinker Assets Group</a> to access the following assets
1. Copy <a href='https://datastudio.google.com/c/u/0/reporting/9f6b9e62-43ec-4027-849a-287e9c1911bd' target='_blank'>Barnacle DV Report</a>.
1. Click Edit->Resource->Manage added data sources, then edit each connection to connect to your new tables above.
1. Or give these intructions to the client.
Modify the values below for your use case, can be done multiple times, then click play.
```
FIELDS = {
'auth_read': 'user', # Credentials used for writing data.
'auth_write': 'service', # Credentials used for writing data.
'partner': '', # Partner ID to run user audit on.
'recipe_slug': '', # Name of Google BigQuery dataset to create.
}
print("Parameters Set To: %s" % FIELDS)
```
#5. Execute DV360 User Audit
This does NOT need to be modified unless you are changing the recipe, click play.
```
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
}
},
{
'google_api': {
'auth': 'user',
'api': 'doubleclickbidmanager',
'version': 'v1.1',
'function': 'queries.listqueries',
'alias': 'list',
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Reports'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'partners.list',
'kwargs': {
'fields': 'partners.displayName,partners.partnerId,nextPageToken'
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Partners'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.list',
'kwargs': {
'partnerId': {'field': {'name': 'partner','kind': 'integer','order': 2,'default': '','description': 'Partner ID to run user audit on.'}},
'fields': 'advertisers.displayName,advertisers.advertiserId,nextPageToken'
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Advertisers'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'users.list',
'kwargs': {
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Users'
}
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': "SELECT U.userId, U.name, U.email, U.displayName, REGEXP_EXTRACT(U.email, r'@(.+)') AS Domain, IF (ENDS_WITH(U.email, '.gserviceaccount.com'), 'Service', 'User') AS Authentication, IF((Select COUNT(advertiserId) from UNNEST(U.assignedUserRoles)) = 0, 'Partner', 'Advertiser') AS Scope, STRUCT( AUR.partnerId, P.displayName AS partnerName, AUR.userRole, AUR.advertiserId, A.displayName AS advertiserName, AUR.assignedUserRoleId ) AS assignedUserRoles, FROM `{dataset}.DV_Users` AS U, UNNEST(assignedUserRoles) AS AUR LEFT JOIN `{dataset}.DV_Partners` AS P ON AUR.partnerId=P.partnerId LEFT JOIN `{dataset}.DV_Advertisers` AS A ON AUR.advertiserId=A.advertiserId ",
'parameters': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'view': 'Barnacle_User_Roles'
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': "SELECT R.*, P.displayName AS partnerName, A.displayName AS advertiserName, FROM ( SELECT queryId, (SELECT CAST(value AS INT64) FROM UNNEST(R.params.filters) WHERE type = 'FILTER_PARTNER' LIMIT 1) AS partnerId, (SELECT CAST(value AS INT64) FROM UNNEST(R.params.filters) WHERE type = 'FILTER_ADVERTISER' LIMIT 1) AS advertiserId, R.schedule.frequency, R.params.metrics, R.params.type, R.metadata.dataRange, R.metadata.sendNotification, DATE(TIMESTAMP_MILLIS(R.metadata.latestReportRunTimeMS)) AS latestReportRunTime, FROM `{dataset}.DV_Reports` AS R) AS R LEFT JOIN `{dataset}.DV_Partners` AS P ON R.partnerId=P.partnerId LEFT JOIN `{dataset}.DV_Advertisers` AS A ON R.advertiserId=A.advertiserId ",
'parameters': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'view': 'Barnacle_Reports'
}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
```
|
github_jupyter
|
!pip install git+https://github.com/google/starthinker
CLOUD_PROJECT = 'PASTE PROJECT ID HERE'
print("Cloud Project Set To: %s" % CLOUD_PROJECT)
CLIENT_CREDENTIALS = 'PASTE CLIENT CREDENTIALS HERE'
print("Client Credentials Set To: %s" % CLIENT_CREDENTIALS)
FIELDS = {
'auth_read': 'user', # Credentials used for writing data.
'auth_write': 'service', # Credentials used for writing data.
'partner': '', # Partner ID to run user audit on.
'recipe_slug': '', # Name of Google BigQuery dataset to create.
}
print("Parameters Set To: %s" % FIELDS)
from starthinker.util.configuration import Configuration
from starthinker.util.configuration import execute
from starthinker.util.recipe import json_set_fields
USER_CREDENTIALS = '/content/user.json'
TASKS = [
{
'dataset': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
}
},
{
'google_api': {
'auth': 'user',
'api': 'doubleclickbidmanager',
'version': 'v1.1',
'function': 'queries.listqueries',
'alias': 'list',
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Reports'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'partners.list',
'kwargs': {
'fields': 'partners.displayName,partners.partnerId,nextPageToken'
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Partners'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'advertisers.list',
'kwargs': {
'partnerId': {'field': {'name': 'partner','kind': 'integer','order': 2,'default': '','description': 'Partner ID to run user audit on.'}},
'fields': 'advertisers.displayName,advertisers.advertiserId,nextPageToken'
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Advertisers'
}
}
}
},
{
'google_api': {
'auth': 'user',
'api': 'displayvideo',
'version': 'v1',
'function': 'users.list',
'kwargs': {
},
'results': {
'bigquery': {
'auth': 'user',
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'table': 'DV_Users'
}
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': "SELECT U.userId, U.name, U.email, U.displayName, REGEXP_EXTRACT(U.email, r'@(.+)') AS Domain, IF (ENDS_WITH(U.email, '.gserviceaccount.com'), 'Service', 'User') AS Authentication, IF((Select COUNT(advertiserId) from UNNEST(U.assignedUserRoles)) = 0, 'Partner', 'Advertiser') AS Scope, STRUCT( AUR.partnerId, P.displayName AS partnerName, AUR.userRole, AUR.advertiserId, A.displayName AS advertiserName, AUR.assignedUserRoleId ) AS assignedUserRoles, FROM `{dataset}.DV_Users` AS U, UNNEST(assignedUserRoles) AS AUR LEFT JOIN `{dataset}.DV_Partners` AS P ON AUR.partnerId=P.partnerId LEFT JOIN `{dataset}.DV_Advertisers` AS A ON AUR.advertiserId=A.advertiserId ",
'parameters': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'view': 'Barnacle_User_Roles'
}
}
},
{
'bigquery': {
'auth': 'user',
'from': {
'query': "SELECT R.*, P.displayName AS partnerName, A.displayName AS advertiserName, FROM ( SELECT queryId, (SELECT CAST(value AS INT64) FROM UNNEST(R.params.filters) WHERE type = 'FILTER_PARTNER' LIMIT 1) AS partnerId, (SELECT CAST(value AS INT64) FROM UNNEST(R.params.filters) WHERE type = 'FILTER_ADVERTISER' LIMIT 1) AS advertiserId, R.schedule.frequency, R.params.metrics, R.params.type, R.metadata.dataRange, R.metadata.sendNotification, DATE(TIMESTAMP_MILLIS(R.metadata.latestReportRunTimeMS)) AS latestReportRunTime, FROM `{dataset}.DV_Reports` AS R) AS R LEFT JOIN `{dataset}.DV_Partners` AS P ON R.partnerId=P.partnerId LEFT JOIN `{dataset}.DV_Advertisers` AS A ON R.advertiserId=A.advertiserId ",
'parameters': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}}
},
'legacy': False
},
'to': {
'dataset': {'field': {'name': 'recipe_slug','kind': 'string','order': 4,'default': '','description': 'Name of Google BigQuery dataset to create.'}},
'view': 'Barnacle_Reports'
}
}
}
]
json_set_fields(TASKS, FIELDS)
execute(Configuration(project=CLOUD_PROJECT, client=CLIENT_CREDENTIALS, user=USER_CREDENTIALS, verbose=True), TASKS, force=True)
| 0.31944 | 0.819533 |
# Bert Evaluation
## BERT - Eval.ipynb
```
import torch
import numpy as np
import pickle
from sklearn.metrics import matthews_corrcoef, confusion_matrix
from torch.utils.data import (DataLoader, RandomSampler, SequentialSampler,
TensorDataset)
from torch.utils.data.distributed import DistributedSampler
from torch.nn import CrossEntropyLoss, MSELoss
from tools import *
from multiprocessing import Pool, cpu_count
import convert_examples_to_features
from tqdm import tqdm_notebook, trange
import os
from pytorch_pretrained_bert import BertTokenizer, BertModel, BertForMaskedLM, BertForSequenceClassification
from pytorch_pretrained_bert.optimization import BertAdam, WarmupLinearSchedule
# OPTIONAL: if you want to have more information on what's happening, activate the logger as follows
import logging
logging.basicConfig(level=logging.INFO)
#device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device = torch.device("cpu")
# The input data dir. Should contain the .tsv files (or other data files) for the task.
DATA_DIR = "Bert_Dev/"
# Bert pre-trained model selected in the list: bert-base-uncased,
# bert-large-uncased, bert-base-cased, bert-large-cased, bert-base-multilingual-uncased,
# bert-base-multilingual-cased, bert-base-chinese.
BERT_MODEL = 'tmu.tar.gz'
# The name of the task to train.I'm going to name this 'yelp'.
TASK_NAME = 'TMU'
# The output directory where the fine-tuned model and checkpoints will be written.
OUTPUT_DIR = f'outputs/{TASK_NAME}/'
# The directory where the evaluation reports will be written to.
REPORTS_DIR = f'reports/{TASK_NAME}_evaluation_reports/'
# This is where BERT will look for pre-trained models to load parameters from.
CACHE_DIR = 'cache/'
# The maximum total input sequence length after WordPiece tokenization.
# Sequences longer than this will be truncated, and sequences shorter than this will be padded.
MAX_SEQ_LENGTH = 128
TRAIN_BATCH_SIZE = 24
EVAL_BATCH_SIZE = 8
LEARNING_RATE = 2e-5
NUM_TRAIN_EPOCHS = 1
RANDOM_SEED = 42
GRADIENT_ACCUMULATION_STEPS = 1
WARMUP_PROPORTION = 0.1
OUTPUT_MODE = 'classification'
CONFIG_NAME = "config.json"
WEIGHTS_NAME = "pytorch_model.bin"
if os.path.exists(REPORTS_DIR) and os.listdir(REPORTS_DIR):
REPORTS_DIR += f'/report_{len(os.listdir(REPORTS_DIR))}'
os.makedirs(REPORTS_DIR)
if not os.path.exists(REPORTS_DIR):
os.makedirs(REPORTS_DIR)
REPORTS_DIR += f'/report_{len(os.listdir(REPORTS_DIR))}'
os.makedirs(REPORTS_DIR)
def get_eval_report(task_name, labels, preds):
mcc = matthews_corrcoef(labels, preds)
tn, fp, fn, tp = confusion_matrix(labels, preds).ravel()
return {
"task": task_name,
"mcc": mcc,
"tp": tp,
"tn": tn,
"fp": fp,
"fn": fn
}
def compute_metrics(task_name, labels, preds):
assert len(preds) == len(labels)
return get_eval_report(task_name, labels, preds)
# Load pre-trained model tokenizer (vocabulary)
tokenizer = BertTokenizer.from_pretrained(OUTPUT_DIR + 'vocab.txt', do_lower_case=False)
processor = BinaryClassificationProcessor()
eval_examples = processor.get_dev_examples(DATA_DIR)
label_list = processor.get_labels() # [0, 1] for binary classification
num_labels = len(label_list)
eval_examples_len = len(eval_examples)
label_map = {label: i for i, label in enumerate(label_list)}
eval_examples_for_processing = [(example, label_map, MAX_SEQ_LENGTH, tokenizer, OUTPUT_MODE) for example in eval_examples]
process_count = cpu_count() - 1
if __name__ == '__main__':
print(f'Preparing to convert {eval_examples_len} examples..')
print(f'Spawning {process_count} processes..')
with Pool(process_count) as p:
eval_features = list(tqdm_notebook(p.imap(convert_examples_to_features.convert_example_to_feature, eval_examples_for_processing), total=eval_examples_len))
all_input_ids = torch.tensor([f.input_ids for f in eval_features], dtype=torch.long)
all_input_mask = torch.tensor([f.input_mask for f in eval_features], dtype=torch.long)
all_segment_ids = torch.tensor([f.segment_ids for f in eval_features], dtype=torch.long)
if OUTPUT_MODE == "classification":
all_label_ids = torch.tensor([f.label_id for f in eval_features], dtype=torch.long)
elif OUTPUT_MODE == "regression":
all_label_ids = torch.tensor([f.label_id for f in eval_features], dtype=torch.float)
eval_data = TensorDataset(all_input_ids, all_input_mask, all_segment_ids, all_label_ids)
# Run prediction for full data
eval_sampler = SequentialSampler(eval_data)
eval_dataloader = DataLoader(eval_data, sampler=eval_sampler, batch_size=EVAL_BATCH_SIZE)
# Load pre-trained model (weights)
model = BertForSequenceClassification.from_pretrained(CACHE_DIR + BERT_MODEL, cache_dir=CACHE_DIR, num_labels=len(label_list))
model.to(device)
model.eval()
eval_loss = 0
nb_eval_steps = 0
preds = []
for input_ids, input_mask, segment_ids, label_ids in tqdm_notebook(eval_dataloader, desc="Evaluating"):
input_ids = input_ids.to(device)
input_mask = input_mask.to(device)
segment_ids = segment_ids.to(device)
label_ids = label_ids.to(device)
with torch.no_grad():
logits = model(input_ids, segment_ids, input_mask, labels=None)
# create eval loss and other metric required by the task
if OUTPUT_MODE == "classification":
loss_fct = CrossEntropyLoss()
tmp_eval_loss = loss_fct(logits.view(-1, num_labels), label_ids.view(-1))
elif OUTPUT_MODE == "regression":
loss_fct = MSELoss()
tmp_eval_loss = loss_fct(logits.view(-1), label_ids.view(-1))
eval_loss += tmp_eval_loss.mean().item()
nb_eval_steps += 1
if len(preds) == 0:
preds.append(logits.detach().cpu().numpy())
else:
preds[0] = np.append(
preds[0], logits.detach().cpu().numpy(), axis=0)
eval_loss = eval_loss / nb_eval_steps
preds = preds[0]
if OUTPUT_MODE == "classification":
preds = np.argmax(preds, axis=1)
elif OUTPUT_MODE == "regression":
preds = np.squeeze(preds)
result = compute_metrics(TASK_NAME, all_label_ids.numpy(), preds)
result['eval_loss'] = eval_loss
output_eval_file = os.path.join(REPORTS_DIR, "eval_results.txt")
with open(output_eval_file, "w") as writer:
logger.info("***** Eval results *****")
for key in (result.keys()):
logger.info(" %s = %s", key, str(result[key]))
writer.write("%s = %s\n" % (key, str(result[key])))
```
|
github_jupyter
|
import torch
import numpy as np
import pickle
from sklearn.metrics import matthews_corrcoef, confusion_matrix
from torch.utils.data import (DataLoader, RandomSampler, SequentialSampler,
TensorDataset)
from torch.utils.data.distributed import DistributedSampler
from torch.nn import CrossEntropyLoss, MSELoss
from tools import *
from multiprocessing import Pool, cpu_count
import convert_examples_to_features
from tqdm import tqdm_notebook, trange
import os
from pytorch_pretrained_bert import BertTokenizer, BertModel, BertForMaskedLM, BertForSequenceClassification
from pytorch_pretrained_bert.optimization import BertAdam, WarmupLinearSchedule
# OPTIONAL: if you want to have more information on what's happening, activate the logger as follows
import logging
logging.basicConfig(level=logging.INFO)
#device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device = torch.device("cpu")
# The input data dir. Should contain the .tsv files (or other data files) for the task.
DATA_DIR = "Bert_Dev/"
# Bert pre-trained model selected in the list: bert-base-uncased,
# bert-large-uncased, bert-base-cased, bert-large-cased, bert-base-multilingual-uncased,
# bert-base-multilingual-cased, bert-base-chinese.
BERT_MODEL = 'tmu.tar.gz'
# The name of the task to train.I'm going to name this 'yelp'.
TASK_NAME = 'TMU'
# The output directory where the fine-tuned model and checkpoints will be written.
OUTPUT_DIR = f'outputs/{TASK_NAME}/'
# The directory where the evaluation reports will be written to.
REPORTS_DIR = f'reports/{TASK_NAME}_evaluation_reports/'
# This is where BERT will look for pre-trained models to load parameters from.
CACHE_DIR = 'cache/'
# The maximum total input sequence length after WordPiece tokenization.
# Sequences longer than this will be truncated, and sequences shorter than this will be padded.
MAX_SEQ_LENGTH = 128
TRAIN_BATCH_SIZE = 24
EVAL_BATCH_SIZE = 8
LEARNING_RATE = 2e-5
NUM_TRAIN_EPOCHS = 1
RANDOM_SEED = 42
GRADIENT_ACCUMULATION_STEPS = 1
WARMUP_PROPORTION = 0.1
OUTPUT_MODE = 'classification'
CONFIG_NAME = "config.json"
WEIGHTS_NAME = "pytorch_model.bin"
if os.path.exists(REPORTS_DIR) and os.listdir(REPORTS_DIR):
REPORTS_DIR += f'/report_{len(os.listdir(REPORTS_DIR))}'
os.makedirs(REPORTS_DIR)
if not os.path.exists(REPORTS_DIR):
os.makedirs(REPORTS_DIR)
REPORTS_DIR += f'/report_{len(os.listdir(REPORTS_DIR))}'
os.makedirs(REPORTS_DIR)
def get_eval_report(task_name, labels, preds):
mcc = matthews_corrcoef(labels, preds)
tn, fp, fn, tp = confusion_matrix(labels, preds).ravel()
return {
"task": task_name,
"mcc": mcc,
"tp": tp,
"tn": tn,
"fp": fp,
"fn": fn
}
def compute_metrics(task_name, labels, preds):
assert len(preds) == len(labels)
return get_eval_report(task_name, labels, preds)
# Load pre-trained model tokenizer (vocabulary)
tokenizer = BertTokenizer.from_pretrained(OUTPUT_DIR + 'vocab.txt', do_lower_case=False)
processor = BinaryClassificationProcessor()
eval_examples = processor.get_dev_examples(DATA_DIR)
label_list = processor.get_labels() # [0, 1] for binary classification
num_labels = len(label_list)
eval_examples_len = len(eval_examples)
label_map = {label: i for i, label in enumerate(label_list)}
eval_examples_for_processing = [(example, label_map, MAX_SEQ_LENGTH, tokenizer, OUTPUT_MODE) for example in eval_examples]
process_count = cpu_count() - 1
if __name__ == '__main__':
print(f'Preparing to convert {eval_examples_len} examples..')
print(f'Spawning {process_count} processes..')
with Pool(process_count) as p:
eval_features = list(tqdm_notebook(p.imap(convert_examples_to_features.convert_example_to_feature, eval_examples_for_processing), total=eval_examples_len))
all_input_ids = torch.tensor([f.input_ids for f in eval_features], dtype=torch.long)
all_input_mask = torch.tensor([f.input_mask for f in eval_features], dtype=torch.long)
all_segment_ids = torch.tensor([f.segment_ids for f in eval_features], dtype=torch.long)
if OUTPUT_MODE == "classification":
all_label_ids = torch.tensor([f.label_id for f in eval_features], dtype=torch.long)
elif OUTPUT_MODE == "regression":
all_label_ids = torch.tensor([f.label_id for f in eval_features], dtype=torch.float)
eval_data = TensorDataset(all_input_ids, all_input_mask, all_segment_ids, all_label_ids)
# Run prediction for full data
eval_sampler = SequentialSampler(eval_data)
eval_dataloader = DataLoader(eval_data, sampler=eval_sampler, batch_size=EVAL_BATCH_SIZE)
# Load pre-trained model (weights)
model = BertForSequenceClassification.from_pretrained(CACHE_DIR + BERT_MODEL, cache_dir=CACHE_DIR, num_labels=len(label_list))
model.to(device)
model.eval()
eval_loss = 0
nb_eval_steps = 0
preds = []
for input_ids, input_mask, segment_ids, label_ids in tqdm_notebook(eval_dataloader, desc="Evaluating"):
input_ids = input_ids.to(device)
input_mask = input_mask.to(device)
segment_ids = segment_ids.to(device)
label_ids = label_ids.to(device)
with torch.no_grad():
logits = model(input_ids, segment_ids, input_mask, labels=None)
# create eval loss and other metric required by the task
if OUTPUT_MODE == "classification":
loss_fct = CrossEntropyLoss()
tmp_eval_loss = loss_fct(logits.view(-1, num_labels), label_ids.view(-1))
elif OUTPUT_MODE == "regression":
loss_fct = MSELoss()
tmp_eval_loss = loss_fct(logits.view(-1), label_ids.view(-1))
eval_loss += tmp_eval_loss.mean().item()
nb_eval_steps += 1
if len(preds) == 0:
preds.append(logits.detach().cpu().numpy())
else:
preds[0] = np.append(
preds[0], logits.detach().cpu().numpy(), axis=0)
eval_loss = eval_loss / nb_eval_steps
preds = preds[0]
if OUTPUT_MODE == "classification":
preds = np.argmax(preds, axis=1)
elif OUTPUT_MODE == "regression":
preds = np.squeeze(preds)
result = compute_metrics(TASK_NAME, all_label_ids.numpy(), preds)
result['eval_loss'] = eval_loss
output_eval_file = os.path.join(REPORTS_DIR, "eval_results.txt")
with open(output_eval_file, "w") as writer:
logger.info("***** Eval results *****")
for key in (result.keys()):
logger.info(" %s = %s", key, str(result[key]))
writer.write("%s = %s\n" % (key, str(result[key])))
| 0.769167 | 0.667717 |
```
from vpython import sphere, canvas, box, vec, color, rate
import math
math.tau = np.tau = 2*math.pi
def cart2pol(vec):
theta = np.arctan2(vec[:, 1], vec[:, 0])
rho = np.hypot(vec[:, 0], vec[:, 1])
return theta, rho
def pol2cart(theta, rho):
x = rho * np.cos(theta)
y = rho * np.sin(theta)
return x, y
def uniform_circle_sample(theta, rho):
x = np.sqrt(rho) * np.cos(theta)
y = np.sqrt(rho) * np.sin(theta)
return x, y
class FanSimulator(object):
g_Forces={'mars':np.array([0, 0, -3.80]),
'earth':np.array([0, 0, -9.81])}
radius = 0.1
start = vec(0, 0, radius)
win = 600
L = 30.
gray = vec(0.7, 0.7, 0.7)
up = vec(0, 0, 1)
def __init__(self, N, vent_radius=0.5, vmax=50, dt=1e-2, location='mars'):
np.random.seed(42)
self.N = N
self.dt = dt
self.vent_radius = vent_radius
self.vmax = vmax
self.particles = []
self.t = None # set to 0 in init_positions
self.g = self.g_Forces[location]
def init_positions(self, vent_radius=None, N=None):
if vent_radius is None:
vent_radius = self.vent_radius
if N is None:
N = self.N
radii = np.random.uniform(0, vent_radius, N)
thetas = np.random.uniform(0, math.tau, N)
X, Y = uniform_circle_sample(thetas, radii)
self.positions = np.stack([X, Y, np.full_like(X, self.radius/2)], axis=1)
self.radii = radii
self.init_pos = self.positions.copy()
self.t = 0
def init_velocities(self, vmax=None):
if vmax is None:
vmax = self.vmax
# using Hagen-Poiseulle flow's parabolic velocity distribution
vz = vmax * (1 - self.radii**2/(self.vent_radius*1.05)**2)
velocities = np.zeros((self.N, 3))
# setting z-column to vz
velocities[:, -1] = vz
self.velocities = velocities
def incline_and_vary_jet(self, incline=1, jitter=0.1):
self.incline = incline
self.velocities[:, 0] = incline
self.jitter = jitter
radii = np.random.uniform(0, jitter, self.N)
thetas = np.random.uniform(0, math.tau, self.N)
vx, vy = uniform_circle_sample(thetas, radii)
self.velocities[:, 0] += vx
self.velocities[:, 1] += vy
def update(self):
to_update = self.positions[:, -1] > 0
self.positions[to_update] += self.velocities[to_update]*self.dt
self.velocities[to_update] += self.g*self.dt
self.t += self.dt
@property
def something_in_the_air(self):
return any(self.positions[:, -1] > 0)
def loop(self):
while self.something_in_the_air:
self.update()
if self.particles:
rate(200)
for p,pos in zip(sim.particles, sim.positions):
if p.update:
p.pos = vec(*pos)
if p.pos.z < start.z:
p.update = False
def plot(self, save=False, equal=True):
fig, axes = plt.subplots(ncols=1, squeeze=False)
axes = axes.ravel()
axes[0].scatter(self.positions[:,0], self.positions[:,1], 5)
for ax in axes:
if equal:
ax.set_aspect('equal')
ax.set_xlabel('Distance [m]')
ax.set_ylabel('Spread [m]')
ax.set_title("{0} particles, v0_z={1}, v0_x= {2}, jitter={3} [m/s]\n"
"dt={4}"
.format(self.N, self.vmax, self.incline, self.jitter, self.dt))
if save:
root = "/Users/klay6683/Dropbox/SSW_2015_cryo_venting/figures/"
fig.savefig(root+'fan_vmax{}_incline{}_vent_radius{}.png'
.format(self.vmax, self.incline, self.vent_radius),
dpi=150)
def init_vpython(self):
scene = canvas(title="Fans", width=self.win, height=self.win, x=0, y=0,
center=vec(0, 0, 0), forward=vec(1,0,-1),
up=self.up)
scene.autoscale = False
scene.range = 25
h = 0.1
mybox = box(pos=vec(0, 0, -h/2), length=self.L, height=h, width=L, up=self.up,
color=color.white)
# create dust particles
for pos in self.positions:
p = sphere(pos=vec(*pos), radius=self.radius, color=color.red)
p.update = True # to determine if needs position update
self.particles.append(p)
#%matplotlib nbagg
import seaborn as sns
sns.set_context('notebook')
sim = FanSimulator(5000, vent_radius=0.1, dt=0.01)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(jitter=0.2, incline=10.0)
sim.loop()
sim.plot(save=True, equal=False)
sim = FanSimulator(5000, vent_radius=0.1, dt=0.001)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(jitter=0.2, incline=10.0)
sim.loop()
sim.plot(save=True, equal=False)
from pypet import Environment, cartesian_product
def add_parameters(traj, dt=1e-2):
traj.f_add_parameter('N', 5000, comment='number of particles')
traj.f_add_parameter('vent_radius', 0.5, comment='radius of particle emitting vent')
traj.f_add_parameter('vmax', 50, comment='vmax in center of vent')
traj.f_add_parameter('dt', dt, comment='dt of simulation')
traj.f_add_parameter('incline', 10.0, comment='inclining vx value')
traj.f_add_parameter('jitter', 0.1, comment='random x,y jitter for velocities')
traj.f_add_parameter('location', 'mars', comment='location determining g-force')
def run_simulation(traj):
sim = FanSimulator(traj.N, vent_radius=traj.vent_radius, vmax=traj.vmax,
dt=traj.dt, location=traj.location)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(incline=traj.incline, jitter=traj.jitter)
sim.loop()
sim.plot(save=True, equal=False)
traj.f_add_result('positions', sim.positions, comment='End positions of particles')
traj.f_add_result('t', sim.t, comment='duration of flight')
env = Environment(trajectory='FanSimulation', filename='./pypet/',
large_overview_tables=True,
add_time=True,
multiproc=False,
ncores=6,
log_config='DEFAULT')
traj = env.v_trajectory
add_parameters(traj, dt=1e-2)
explore_dict = {'vent_radius':[0.1, 0.5, 1.0],
'vmax':[10, 50, 100],
'incline':[0.1, 1.0, 5.0]}
to_explore = cartesian_product(explore_dict)
traj.f_explore(to_explore)
env.f_run(run_simulation)
env.f_disable_logging()
```
|
github_jupyter
|
from vpython import sphere, canvas, box, vec, color, rate
import math
math.tau = np.tau = 2*math.pi
def cart2pol(vec):
theta = np.arctan2(vec[:, 1], vec[:, 0])
rho = np.hypot(vec[:, 0], vec[:, 1])
return theta, rho
def pol2cart(theta, rho):
x = rho * np.cos(theta)
y = rho * np.sin(theta)
return x, y
def uniform_circle_sample(theta, rho):
x = np.sqrt(rho) * np.cos(theta)
y = np.sqrt(rho) * np.sin(theta)
return x, y
class FanSimulator(object):
g_Forces={'mars':np.array([0, 0, -3.80]),
'earth':np.array([0, 0, -9.81])}
radius = 0.1
start = vec(0, 0, radius)
win = 600
L = 30.
gray = vec(0.7, 0.7, 0.7)
up = vec(0, 0, 1)
def __init__(self, N, vent_radius=0.5, vmax=50, dt=1e-2, location='mars'):
np.random.seed(42)
self.N = N
self.dt = dt
self.vent_radius = vent_radius
self.vmax = vmax
self.particles = []
self.t = None # set to 0 in init_positions
self.g = self.g_Forces[location]
def init_positions(self, vent_radius=None, N=None):
if vent_radius is None:
vent_radius = self.vent_radius
if N is None:
N = self.N
radii = np.random.uniform(0, vent_radius, N)
thetas = np.random.uniform(0, math.tau, N)
X, Y = uniform_circle_sample(thetas, radii)
self.positions = np.stack([X, Y, np.full_like(X, self.radius/2)], axis=1)
self.radii = radii
self.init_pos = self.positions.copy()
self.t = 0
def init_velocities(self, vmax=None):
if vmax is None:
vmax = self.vmax
# using Hagen-Poiseulle flow's parabolic velocity distribution
vz = vmax * (1 - self.radii**2/(self.vent_radius*1.05)**2)
velocities = np.zeros((self.N, 3))
# setting z-column to vz
velocities[:, -1] = vz
self.velocities = velocities
def incline_and_vary_jet(self, incline=1, jitter=0.1):
self.incline = incline
self.velocities[:, 0] = incline
self.jitter = jitter
radii = np.random.uniform(0, jitter, self.N)
thetas = np.random.uniform(0, math.tau, self.N)
vx, vy = uniform_circle_sample(thetas, radii)
self.velocities[:, 0] += vx
self.velocities[:, 1] += vy
def update(self):
to_update = self.positions[:, -1] > 0
self.positions[to_update] += self.velocities[to_update]*self.dt
self.velocities[to_update] += self.g*self.dt
self.t += self.dt
@property
def something_in_the_air(self):
return any(self.positions[:, -1] > 0)
def loop(self):
while self.something_in_the_air:
self.update()
if self.particles:
rate(200)
for p,pos in zip(sim.particles, sim.positions):
if p.update:
p.pos = vec(*pos)
if p.pos.z < start.z:
p.update = False
def plot(self, save=False, equal=True):
fig, axes = plt.subplots(ncols=1, squeeze=False)
axes = axes.ravel()
axes[0].scatter(self.positions[:,0], self.positions[:,1], 5)
for ax in axes:
if equal:
ax.set_aspect('equal')
ax.set_xlabel('Distance [m]')
ax.set_ylabel('Spread [m]')
ax.set_title("{0} particles, v0_z={1}, v0_x= {2}, jitter={3} [m/s]\n"
"dt={4}"
.format(self.N, self.vmax, self.incline, self.jitter, self.dt))
if save:
root = "/Users/klay6683/Dropbox/SSW_2015_cryo_venting/figures/"
fig.savefig(root+'fan_vmax{}_incline{}_vent_radius{}.png'
.format(self.vmax, self.incline, self.vent_radius),
dpi=150)
def init_vpython(self):
scene = canvas(title="Fans", width=self.win, height=self.win, x=0, y=0,
center=vec(0, 0, 0), forward=vec(1,0,-1),
up=self.up)
scene.autoscale = False
scene.range = 25
h = 0.1
mybox = box(pos=vec(0, 0, -h/2), length=self.L, height=h, width=L, up=self.up,
color=color.white)
# create dust particles
for pos in self.positions:
p = sphere(pos=vec(*pos), radius=self.radius, color=color.red)
p.update = True # to determine if needs position update
self.particles.append(p)
#%matplotlib nbagg
import seaborn as sns
sns.set_context('notebook')
sim = FanSimulator(5000, vent_radius=0.1, dt=0.01)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(jitter=0.2, incline=10.0)
sim.loop()
sim.plot(save=True, equal=False)
sim = FanSimulator(5000, vent_radius=0.1, dt=0.001)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(jitter=0.2, incline=10.0)
sim.loop()
sim.plot(save=True, equal=False)
from pypet import Environment, cartesian_product
def add_parameters(traj, dt=1e-2):
traj.f_add_parameter('N', 5000, comment='number of particles')
traj.f_add_parameter('vent_radius', 0.5, comment='radius of particle emitting vent')
traj.f_add_parameter('vmax', 50, comment='vmax in center of vent')
traj.f_add_parameter('dt', dt, comment='dt of simulation')
traj.f_add_parameter('incline', 10.0, comment='inclining vx value')
traj.f_add_parameter('jitter', 0.1, comment='random x,y jitter for velocities')
traj.f_add_parameter('location', 'mars', comment='location determining g-force')
def run_simulation(traj):
sim = FanSimulator(traj.N, vent_radius=traj.vent_radius, vmax=traj.vmax,
dt=traj.dt, location=traj.location)
sim.init_positions()
sim.init_velocities()
sim.incline_and_vary_jet(incline=traj.incline, jitter=traj.jitter)
sim.loop()
sim.plot(save=True, equal=False)
traj.f_add_result('positions', sim.positions, comment='End positions of particles')
traj.f_add_result('t', sim.t, comment='duration of flight')
env = Environment(trajectory='FanSimulation', filename='./pypet/',
large_overview_tables=True,
add_time=True,
multiproc=False,
ncores=6,
log_config='DEFAULT')
traj = env.v_trajectory
add_parameters(traj, dt=1e-2)
explore_dict = {'vent_radius':[0.1, 0.5, 1.0],
'vmax':[10, 50, 100],
'incline':[0.1, 1.0, 5.0]}
to_explore = cartesian_product(explore_dict)
traj.f_explore(to_explore)
env.f_run(run_simulation)
env.f_disable_logging()
| 0.781581 | 0.587914 |
# Sequence classification
In this exercise, you will get familiar with how to build RNNs in Keras. You will build a recurrent model to classify moview reviews as either positive or negative.
```
%matplotlib inline
import numpy as np
from keras.preprocessing import sequence
from keras.utils import np_utils
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Embedding
from keras.layers import LSTM, SimpleRNN, GRU
from keras.datasets import imdb
```
## IMDB Sentiment Dataset
The large movie review dataset is a collection of 25k positive and 25k negative movie reviews from [IMDB](http://www.imdb.com). Here are some excerpts from the dataset, both easy and hard, to get a sense of why this dataset is challenging:
> Ah, I loved this movie.
> Quite honestly, The Omega Code is the worst movie I have seen in a very long time.
> The wit and pace and three show stopping Busby Berkley numbers put this ahead of the over-rated 42nd Street.
> There simply was no suspense, precious little excitement and too many dull spots, most of them trying to show why "Nellie" (Monroe) was so messed up.
The dataset can be found at http://ai.stanford.edu/~amaas/data/sentiment/. Since this is a common dataset for RNNs, Keras has a preprocessed version built-in.
```
# We will limit to the most frequent 20k words defined by max_features, our vocabulary size
max_features = 20000
(X_train, y_train), (X_test, y_test) = imdb.load_data(num_words=max_features)
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
```
The data is preprocessed by replacing words with indexes - review [Keras's docs](http://keras.io/datasets/#imdb-movie-reviews-sentiment-classification). Here's the first review in the training set.
```
review = X_train[0]
review
```
We can convince ourselves that these are movies reviews, using the vocabulary provided by keras:
```
word_index = imdb.get_word_index()
```
First we create a dictionary from index to word, notice that words are indexed starting from the number 3, while the first three entries are for special characters:
```
index_word = {i+3: w for w, i in word_index.items()}
index_word[0]=''
index_word[1]='start_char'
index_word[2]='oov'
```
Then we can covert the first review to text:
```
' '.join([index_word[i] for i in review])
```
#### Exercise 1 - prepare the data
The reviews are different lengths but we need to fit them into a matrix to feed to Keras. We will do this by picking a maximum word length and cutting off words from the examples that are over that limit and padding the examples with 0 if they are under the limit.
Refer to the [Keras docs](http://keras.io/preprocessing/sequence/#pad_sequences) for the `pad_sequences` function. Use `pad_sequences` to prepare both `X_train` and `X_test` to be `maxlen` long at the most.
```
maxlen = 80
# Pad and clip the example sequences
X_train = sequence.pad_sequences(X_train, maxlen=maxlen)
X_test = sequence.pad_sequences(X_test, maxlen=maxlen)
print('X_train shape:', X_train.shape)
print('X_test shape:', X_test.shape)
```
#### Exercise 2 - build an RNN for classifying reviews as positive or negative
Build a single-layer RNN model and train it. You will need to include these parts:
* An `Embedding` layer for efficiently one-hot encoding the inputs - [docs](http://keras.io/layers/embeddings/)
* A recurrent layer. Keras has a [few variants](http://keras.io/layers/recurrent/) you could use. LSTM layers are by far the most popular for RNNs.
* A `Dense` layer for the hidden to output connection.
* A softmax to produce the final prediction.
You will need to decide how large your hidden state will be. You may also consider using some dropout on your recurrent or embedding layers - refers to docs for how to do this.
Training for longer will be much better overall, but since RNNs are expensive to train, you can use 1 epoch to test. You should be able to get > 70% accuracy with 1 epoch. How high can you get?
```
# Design an recurrent model
model = Sequential()
model.add(Embedding(max_features, 128, input_length=maxlen))
model.add(LSTM(128, dropout=0.2, recurrent_dropout=0.2))
model.add(Dense(2))
model.add(Activation('softmax'))
# The Adam optimizer can automatically adjust learning rates for you
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy'])
model.summary()
model.fit(X_train, y_train, batch_size=32, epochs=1, validation_data=(X_test, y_test))
loss, acc = model.evaluate(X_test, y_test, batch_size=32)
print('Test loss:', loss)
print('Test accuracy:', acc)
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
from keras.preprocessing import sequence
from keras.utils import np_utils
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Embedding
from keras.layers import LSTM, SimpleRNN, GRU
from keras.datasets import imdb
# We will limit to the most frequent 20k words defined by max_features, our vocabulary size
max_features = 20000
(X_train, y_train), (X_test, y_test) = imdb.load_data(num_words=max_features)
y_train = np_utils.to_categorical(y_train)
y_test = np_utils.to_categorical(y_test)
review = X_train[0]
review
word_index = imdb.get_word_index()
index_word = {i+3: w for w, i in word_index.items()}
index_word[0]=''
index_word[1]='start_char'
index_word[2]='oov'
' '.join([index_word[i] for i in review])
maxlen = 80
# Pad and clip the example sequences
X_train = sequence.pad_sequences(X_train, maxlen=maxlen)
X_test = sequence.pad_sequences(X_test, maxlen=maxlen)
print('X_train shape:', X_train.shape)
print('X_test shape:', X_test.shape)
# Design an recurrent model
model = Sequential()
model.add(Embedding(max_features, 128, input_length=maxlen))
model.add(LSTM(128, dropout=0.2, recurrent_dropout=0.2))
model.add(Dense(2))
model.add(Activation('softmax'))
# The Adam optimizer can automatically adjust learning rates for you
model.compile(loss='categorical_crossentropy',
optimizer='adam',
metrics=['accuracy'])
model.summary()
model.fit(X_train, y_train, batch_size=32, epochs=1, validation_data=(X_test, y_test))
loss, acc = model.evaluate(X_test, y_test, batch_size=32)
print('Test loss:', loss)
print('Test accuracy:', acc)
| 0.803482 | 0.989055 |
For images, packages such as Pillow, OpenCV are useful
For audio, packages such as scipy and librosa
For text, either raw Python or Cython based loading, or NLTK and SpaCy are useful
# Training an image Classifier
1. Load and normalize the CIFAR10 training and test datasets using torchvision
2. Define CNN.
3. Define a loss function
4. Train the network on training data
5. Test the network on test data
```
import torch
import torchvision
import torchvision.transforms as transforms
```
The output of torchvision datasets are PILImage images of range [0, 1]. We transform them to Tensors of normalized range [-1, 1].
```
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5,0.5,0.5),(0.5,0.5,0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data' ,train=True,download=True,transform=transform)
trainloader = torch.utils.data.DataLoader(trainset,batch_size=4, shuffle=True,num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data',train=False,download=True,transform=transform)
testloader = torch.utils.data.DataLoader(testset,batch_size=4,shuffle=False,num_workers=2)
classes = ('plane','car','bird','cat','deer','dog','frog','horse','ship','truck')
import matplotlib.pyplot as plt
import numpy as np
#fucntion to show images
def imshow(img):
img=img/4 +0.5 #unnormalize
npimg=img.numpy()
plt.imshow(np.transpose(npimg,(1,2,0)))
plt.show()
# get some random training images
dataiter = iter(trainloader)
images,labels = dataiter.next()
#show images
imshow(torchvision.utils.make_grid(images))
#print labels
print(' '.join('%5s'% classes[labels[j]] for j in range(4)))
```
# Define a Convolutional Neural Network
```
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
#3 input image , 6 output channels and 5x5 squares
self.conv1 = nn.Conv2d(3,6,5)
self.pool = nn.MaxPool2d(2,2)
self.conv2 = nn.Conv2d(6,16,5)
self.fc1 = nn.Linear(16*5*5,120)
self.fc2 = nn.Linear(120,84)
self.fc3 = nn.Linear(84,10)
def forward(self,x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(-1,16*5*5)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
#Define a loss function
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(),lr=0.001,momentum = 0.9)
# Train the network
for epoch in range(2):
running_loss = 0.0
for i,data in enumerate(trainloader,0):
#get the inputs
inputs, labels = data
#zero the parameters gradients
optimizer.zero_grad()
#forward + backward+ optimize
outputs = net(inputs)
loss = criterion(outputs,labels)
loss.backward()
optimizer.step()
#print the statistics
running_loss += loss.item()
if i %2000 == 1999: #print every 2000 mini-batches
print('[%d,%5d] loss: %.3f'% (epoch +1,i+1,running_loss/2000))
running_loss = 0.0
print('Finished Training')
dataiter = iter(testloader)
images,labels = dataiter.next()
imshow(torchvision.utils.make_grid(images))
print('GroundTruth:', ' '.join('%5s' %classes[labels[j]] for j in range(4)))
output = net(images)
_, predicted = torch.max(outputs, 1)
print('Predicted: ', ' '.join('%5s' % classes[predicted[j]]
for j in range(4)))
correct =0
total =0
with torch.no_grad():
for data in testloader:
images , labels = data
outputs = net(images)
_,predicted = torch.max(outputs.data,1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of network on the 10000 test images: %d %%'%(100*correct/total))
class_correct = list(0. for i in range(10))
class_total = list(0. for i in range(10))
with torch.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = torch.max(outputs, 1)
c = (predicted == labels).squeeze()
for i in range(4):
label = labels[i]
class_correct[label] += c[i].item()
class_total[label] += 1
for i in range(10):
print('Accuracy of %5s : %2d %%' % (
classes[i], 100 * class_correct[i] / class_total[i]))
#Testing the cuda and training the data on GPU
device = torch.device('cuda:0' if torch.cuda.is_available() else "cpu")
print(device)
```
|
github_jupyter
|
import torch
import torchvision
import torchvision.transforms as transforms
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5,0.5,0.5),(0.5,0.5,0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data' ,train=True,download=True,transform=transform)
trainloader = torch.utils.data.DataLoader(trainset,batch_size=4, shuffle=True,num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data',train=False,download=True,transform=transform)
testloader = torch.utils.data.DataLoader(testset,batch_size=4,shuffle=False,num_workers=2)
classes = ('plane','car','bird','cat','deer','dog','frog','horse','ship','truck')
import matplotlib.pyplot as plt
import numpy as np
#fucntion to show images
def imshow(img):
img=img/4 +0.5 #unnormalize
npimg=img.numpy()
plt.imshow(np.transpose(npimg,(1,2,0)))
plt.show()
# get some random training images
dataiter = iter(trainloader)
images,labels = dataiter.next()
#show images
imshow(torchvision.utils.make_grid(images))
#print labels
print(' '.join('%5s'% classes[labels[j]] for j in range(4)))
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net,self).__init__()
#3 input image , 6 output channels and 5x5 squares
self.conv1 = nn.Conv2d(3,6,5)
self.pool = nn.MaxPool2d(2,2)
self.conv2 = nn.Conv2d(6,16,5)
self.fc1 = nn.Linear(16*5*5,120)
self.fc2 = nn.Linear(120,84)
self.fc3 = nn.Linear(84,10)
def forward(self,x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(-1,16*5*5)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
#Define a loss function
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(),lr=0.001,momentum = 0.9)
# Train the network
for epoch in range(2):
running_loss = 0.0
for i,data in enumerate(trainloader,0):
#get the inputs
inputs, labels = data
#zero the parameters gradients
optimizer.zero_grad()
#forward + backward+ optimize
outputs = net(inputs)
loss = criterion(outputs,labels)
loss.backward()
optimizer.step()
#print the statistics
running_loss += loss.item()
if i %2000 == 1999: #print every 2000 mini-batches
print('[%d,%5d] loss: %.3f'% (epoch +1,i+1,running_loss/2000))
running_loss = 0.0
print('Finished Training')
dataiter = iter(testloader)
images,labels = dataiter.next()
imshow(torchvision.utils.make_grid(images))
print('GroundTruth:', ' '.join('%5s' %classes[labels[j]] for j in range(4)))
output = net(images)
_, predicted = torch.max(outputs, 1)
print('Predicted: ', ' '.join('%5s' % classes[predicted[j]]
for j in range(4)))
correct =0
total =0
with torch.no_grad():
for data in testloader:
images , labels = data
outputs = net(images)
_,predicted = torch.max(outputs.data,1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of network on the 10000 test images: %d %%'%(100*correct/total))
class_correct = list(0. for i in range(10))
class_total = list(0. for i in range(10))
with torch.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predicted = torch.max(outputs, 1)
c = (predicted == labels).squeeze()
for i in range(4):
label = labels[i]
class_correct[label] += c[i].item()
class_total[label] += 1
for i in range(10):
print('Accuracy of %5s : %2d %%' % (
classes[i], 100 * class_correct[i] / class_total[i]))
#Testing the cuda and training the data on GPU
device = torch.device('cuda:0' if torch.cuda.is_available() else "cpu")
print(device)
| 0.843799 | 0.973164 |
```
import datetime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import datetime as dt
import re
import nltk
from nltk.sentiment.vader import SentimentIntensityAnalyzer
import snscrape.modules.twitter as sntwitter
from textblob import TextBlob
nltk.download('vader_lexicon')
```
# SNSCRAPE to scrape through Twitter
```
#Creating list to append tweet data
tweets_list = []
#keyword
keyword = 'bitcoin'
# No of tweets
noOfTweet = 50000
#Loop through the usernames:
#user_names = open('News_Station.txt','r')
user_names = open('Influential_People.txt','r')
for user in user_names:
print (user)
# Using TwitterSearchScraper to scrape data and append tweets to list
for i,tweet in enumerate(sntwitter.TwitterSearchScraper("from:"+ user +" "+keyword ).get_items()):
if i > int(noOfTweet):
break
tweets_list.append([tweet.date, tweet.id, tweet.content, tweet.username])
```
# Creating and cleaning the dataframe from the tweets list above
```
# Creating a dataframe from the tweets list above
df = pd.DataFrame(tweets_list, columns=['Datetime', 'Tweet Id', 'Text', 'Username'])
df['Datetime'] = pd.to_datetime(df['Datetime'],unit='ms').dt.tz_convert('Asia/Singapore')
df['Datetime'] = df['Datetime'].apply(lambda a: datetime.datetime.strftime(a,"%d-%m-%Y %H:%M:%S"))
df['Datetime'] = pd.to_datetime(df['Datetime'])
df['Tweet Id'] = ('"'+ df['Tweet Id'].astype(str) + '"')
# Create a function to clean the tweets
def cleanTxt(text):
text = re.sub('@[A-Za-z0–9]+', '', text) #Removing @mentions
text = re.sub('#', '', text) # Removing '#' hash tag
text = re.sub('RT[\s]+', '', text) # Removing RT
text = re.sub('https?:\/\/\S+', '', text) # Removing hyperlink
return text
df["Text"] = df["Text"].apply(cleanTxt)
```
# Sentiment Analysis
NLTK
```
#Sentiment Analysis
def percentage(part,whole):
return 100 * float(part)/float(whole)
#Iterating over the tweets in the dataframe
def apply_analysis(tweet):
return SentimentIntensityAnalyzer().polarity_scores(tweet)
df[['neg','neu','pos','compound']] = df['Text'].apply(apply_analysis).apply(pd.Series)
def sentimental_analysis(df):
if df['neg'] > df['pos']:
return 'Negative'
elif df['pos'] > df['neg']:
return 'Positive'
elif df['pos'] == df['neg']:
return 'Neutral'
df['Sentiment_NLTK'] = df.apply(sentimental_analysis, axis = 1)
```
Textblob
```
def getSubjectivity(twt):
return TextBlob(twt).sentiment.subjectivity
def getPolarity(twt):
return TextBlob(twt).sentiment.polarity
def getSentiment(score):
if score<0:
return 'Negative'
elif score==0:
return 'Neutral'
else:
return 'Positive'
df['Subjectivity']=df['Text'].apply(getSubjectivity)
df['Polarity']=df['Text'].apply(getPolarity)
df['Sentiment_TB']=df['Polarity'].apply(getSentiment)
```
# Generating csv
```
#df.to_csv('News_Station.csv', encoding='utf-8-sig')
df.to_csv('Influential_People.csv', encoding='utf-8-sig' ,index= False)
print('Done')
```
|
github_jupyter
|
import datetime
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import datetime as dt
import re
import nltk
from nltk.sentiment.vader import SentimentIntensityAnalyzer
import snscrape.modules.twitter as sntwitter
from textblob import TextBlob
nltk.download('vader_lexicon')
#Creating list to append tweet data
tweets_list = []
#keyword
keyword = 'bitcoin'
# No of tweets
noOfTweet = 50000
#Loop through the usernames:
#user_names = open('News_Station.txt','r')
user_names = open('Influential_People.txt','r')
for user in user_names:
print (user)
# Using TwitterSearchScraper to scrape data and append tweets to list
for i,tweet in enumerate(sntwitter.TwitterSearchScraper("from:"+ user +" "+keyword ).get_items()):
if i > int(noOfTweet):
break
tweets_list.append([tweet.date, tweet.id, tweet.content, tweet.username])
# Creating a dataframe from the tweets list above
df = pd.DataFrame(tweets_list, columns=['Datetime', 'Tweet Id', 'Text', 'Username'])
df['Datetime'] = pd.to_datetime(df['Datetime'],unit='ms').dt.tz_convert('Asia/Singapore')
df['Datetime'] = df['Datetime'].apply(lambda a: datetime.datetime.strftime(a,"%d-%m-%Y %H:%M:%S"))
df['Datetime'] = pd.to_datetime(df['Datetime'])
df['Tweet Id'] = ('"'+ df['Tweet Id'].astype(str) + '"')
# Create a function to clean the tweets
def cleanTxt(text):
text = re.sub('@[A-Za-z0–9]+', '', text) #Removing @mentions
text = re.sub('#', '', text) # Removing '#' hash tag
text = re.sub('RT[\s]+', '', text) # Removing RT
text = re.sub('https?:\/\/\S+', '', text) # Removing hyperlink
return text
df["Text"] = df["Text"].apply(cleanTxt)
#Sentiment Analysis
def percentage(part,whole):
return 100 * float(part)/float(whole)
#Iterating over the tweets in the dataframe
def apply_analysis(tweet):
return SentimentIntensityAnalyzer().polarity_scores(tweet)
df[['neg','neu','pos','compound']] = df['Text'].apply(apply_analysis).apply(pd.Series)
def sentimental_analysis(df):
if df['neg'] > df['pos']:
return 'Negative'
elif df['pos'] > df['neg']:
return 'Positive'
elif df['pos'] == df['neg']:
return 'Neutral'
df['Sentiment_NLTK'] = df.apply(sentimental_analysis, axis = 1)
def getSubjectivity(twt):
return TextBlob(twt).sentiment.subjectivity
def getPolarity(twt):
return TextBlob(twt).sentiment.polarity
def getSentiment(score):
if score<0:
return 'Negative'
elif score==0:
return 'Neutral'
else:
return 'Positive'
df['Subjectivity']=df['Text'].apply(getSubjectivity)
df['Polarity']=df['Text'].apply(getPolarity)
df['Sentiment_TB']=df['Polarity'].apply(getSentiment)
#df.to_csv('News_Station.csv', encoding='utf-8-sig')
df.to_csv('Influential_People.csv', encoding='utf-8-sig' ,index= False)
print('Done')
| 0.319015 | 0.485539 |
# Logging data
```
from planout.ops.random import *
from planout.experiment import SimpleExperiment
import pandas as pd
import json
```
### Log data
Here we explain what all the fields are in the log data. Run this:
```
class LoggedExperiment(SimpleExperiment):
def assign(self, params, userid):
params.x = UniformChoice(choices=["What's on your mind?", "Say something."], unit=userid)
params.y = BernoulliTrial(p=0.5, unit=userid)
print LoggedExperiment(userid=5).get('x')
```
Then open your terminal, navigate to the directory this notebook is in, and type:
```
> tail -f LoggedExperiment.log
```
You can now see how data is logged to your experiment as its run.
#### Exposure logs
Whenever you request a parameter, an exposure is automatically logged. In a production environment, one would use caching (e.g., memcache) so that we only exposure log once per unit. `SimpleExperiment` exposure logs once per instance.
```
e = LoggedExperiment(userid=4)
print e.get('x')
print e.get('y')
```
#### Manual exposure logging
Calling `log_exposure()` will force PlanOut to log an exposure event. You can optionally pass in additional data.
```
e.log_exposure()
e.log_exposure({'endpoint': 'home.py'})
```
#### Event logging
You can also log arbitrary events. The first argument to `log_event()` is a required parameter that specifies the event type.
```
e.log_event('post_status_update')
e.log_event('post_status_update', {'type': 'photo'})
```
## Putting it all together
We simulate the components of a PlanOut-driven website and show how data analysis would work in conjunction with the data generated from the simulation.
This hypothetical experiment looks at the effect of sorting a music album's songs by popularity (instead of say track number) on a Web-based music store.
Our website simulation consists of four main parts:
* Code to render the web page (which uses PlanOut to decide how to display items)
* Code to handle item purchases (this logs the "conversion" event)
* Code to simulate the process of users' purchase decision-making
* A loop that simulates many users viewing many albums
```
class MusicExperiment(SimpleExperiment):
def assign(self, params, userid, albumid):
params.sort_by_rating = BernoulliTrial(p=0.2, unit=[userid, albumid])
import random
def get_price(albumid):
"look up the price of an album"
# this would realistically hook into a database
return 11.99
```
#### Rendering the web page
```
def render_webpage(userid, albumid):
'simulated web page rendering function'
# get experiment for the given user / album pair.
e = MusicExperiment(userid=userid, albumid=albumid)
# use log_exposure() so that we can also record the price
e.log_exposure({'price': get_price(albumid)})
# use a default value with get() in production settings, in case
# your experimentation system goes down
if e.get('sort_by_rating', False):
songs = "some sorted songs" # this would sort the songs by rating
else:
songs = "some non-sorted songs"
html = "some HTML code involving %s" % songs # most valid html ever.
# render html
```
#### Logging outcomes
```
def handle_purchase(userid, albumid):
'handles purchase of an album'
e = MusicExperiment(userid=userid, albumid=albumid)
e.log_event('purchase', {'price': get_price(albumid)})
# start album download
```
#### Generative model of user decision making
```
def simulate_user_decision(userid, albumid):
'simulate user experience'
# This function should be thought of as simulating a users' decision-making
# process for the given stimulus - and so we don't actually want to do any
# logging here.
e = MusicExperiment(userid=userid, albumid=albumid)
e.set_auto_exposure_logging(False) # turn off auto-logging
# users with sorted songs have a higher purchase rate
if e.get('sort_by_rating'):
prob_purchase = 0.15
else:
prob_purchase = 0.10
# make purchase with probability prob_purchase
return random.random() < prob_purchase
```
#### Running the simulation
```
# We then simulate 500 users' visitation to 20 albums, and their decision to purchase
random.seed(0)
for u in xrange(500):
for a in xrange(20):
render_webpage(u, a)
if simulate_user_decision(u, a):
handle_purchase(u, a)
```
### Loading data into Python for analysis
Data is logged to `MusicExperiment.log`. Each line is JSON-encoded dictionary that contains information about the event types, inputs, and parameter assignments.
```
raw_log_data = [json.loads(i) for i in open('MusicExperiment.log')]
raw_log_data[:2]
```
It's preferable to deal with the data as a flat set of columns. We use this handy-dandy function Eytan found on stackoverflow to flatten dictionaries.
```
# stolen from http://stackoverflow.com/questions/23019119/converting-multilevel-nested-dictionaries-to-pandas-dataframe
from collections import OrderedDict
def flatten(d):
"Flatten an OrderedDict object"
result = OrderedDict()
for k, v in d.items():
if isinstance(v, dict):
result.update(flatten(v))
else:
result[k] = v
return result
```
Here is what the flattened dataframe looks like:
```
log_data = pd.DataFrame.from_dict([flatten(i) for i in raw_log_data])
log_data[:5]
```
### Joining exposure data with event data
We first extract all user-album pairs that were exposed to an experiemntal treatment, and their parameter assignments.
```
all_exposures = log_data[log_data.event=='exposure']
unique_exposures = all_exposures[['userid','albumid','sort_by_rating']].drop_duplicates()
```
Tabulating the users' assignments, we find that the assignment probabilities correspond to the design at the beginning of this notebook.
```
unique_exposures[['userid','sort_by_rating']].groupby('sort_by_rating').agg(len)
```
Now we can merge with the conversion data.
```
conversions = log_data[log_data.event=='purchase'][['userid', 'albumid','price']]
df = pd.merge(unique_exposures, conversions, on=['userid', 'albumid'], how='left')
df['purchased'] = df.price.notnull()
df['revenue'] = df.purchased * df.price.fillna(0)
```
Here is a sample of the merged rows. Most rows contain missing values for price, because the user didn't purchase the item.
```
df[:5]
```
Restricted to those who bought something...
```
df[df.price > 0][:5]
```
### Analyzing the experimental results
```
df.groupby('sort_by_rating')[['purchased', 'price', 'revenue']].agg(mean)
```
If you were actually analyzing the experiment you would want to compute confidence intervals.
|
github_jupyter
|
from planout.ops.random import *
from planout.experiment import SimpleExperiment
import pandas as pd
import json
class LoggedExperiment(SimpleExperiment):
def assign(self, params, userid):
params.x = UniformChoice(choices=["What's on your mind?", "Say something."], unit=userid)
params.y = BernoulliTrial(p=0.5, unit=userid)
print LoggedExperiment(userid=5).get('x')
> tail -f LoggedExperiment.log
e = LoggedExperiment(userid=4)
print e.get('x')
print e.get('y')
e.log_exposure()
e.log_exposure({'endpoint': 'home.py'})
e.log_event('post_status_update')
e.log_event('post_status_update', {'type': 'photo'})
class MusicExperiment(SimpleExperiment):
def assign(self, params, userid, albumid):
params.sort_by_rating = BernoulliTrial(p=0.2, unit=[userid, albumid])
import random
def get_price(albumid):
"look up the price of an album"
# this would realistically hook into a database
return 11.99
def render_webpage(userid, albumid):
'simulated web page rendering function'
# get experiment for the given user / album pair.
e = MusicExperiment(userid=userid, albumid=albumid)
# use log_exposure() so that we can also record the price
e.log_exposure({'price': get_price(albumid)})
# use a default value with get() in production settings, in case
# your experimentation system goes down
if e.get('sort_by_rating', False):
songs = "some sorted songs" # this would sort the songs by rating
else:
songs = "some non-sorted songs"
html = "some HTML code involving %s" % songs # most valid html ever.
# render html
def handle_purchase(userid, albumid):
'handles purchase of an album'
e = MusicExperiment(userid=userid, albumid=albumid)
e.log_event('purchase', {'price': get_price(albumid)})
# start album download
def simulate_user_decision(userid, albumid):
'simulate user experience'
# This function should be thought of as simulating a users' decision-making
# process for the given stimulus - and so we don't actually want to do any
# logging here.
e = MusicExperiment(userid=userid, albumid=albumid)
e.set_auto_exposure_logging(False) # turn off auto-logging
# users with sorted songs have a higher purchase rate
if e.get('sort_by_rating'):
prob_purchase = 0.15
else:
prob_purchase = 0.10
# make purchase with probability prob_purchase
return random.random() < prob_purchase
# We then simulate 500 users' visitation to 20 albums, and their decision to purchase
random.seed(0)
for u in xrange(500):
for a in xrange(20):
render_webpage(u, a)
if simulate_user_decision(u, a):
handle_purchase(u, a)
raw_log_data = [json.loads(i) for i in open('MusicExperiment.log')]
raw_log_data[:2]
# stolen from http://stackoverflow.com/questions/23019119/converting-multilevel-nested-dictionaries-to-pandas-dataframe
from collections import OrderedDict
def flatten(d):
"Flatten an OrderedDict object"
result = OrderedDict()
for k, v in d.items():
if isinstance(v, dict):
result.update(flatten(v))
else:
result[k] = v
return result
log_data = pd.DataFrame.from_dict([flatten(i) for i in raw_log_data])
log_data[:5]
all_exposures = log_data[log_data.event=='exposure']
unique_exposures = all_exposures[['userid','albumid','sort_by_rating']].drop_duplicates()
unique_exposures[['userid','sort_by_rating']].groupby('sort_by_rating').agg(len)
conversions = log_data[log_data.event=='purchase'][['userid', 'albumid','price']]
df = pd.merge(unique_exposures, conversions, on=['userid', 'albumid'], how='left')
df['purchased'] = df.price.notnull()
df['revenue'] = df.purchased * df.price.fillna(0)
df[:5]
df[df.price > 0][:5]
df.groupby('sort_by_rating')[['purchased', 'price', 'revenue']].agg(mean)
| 0.405096 | 0.8308 |
```
#!/usr/bin/env python
import os
import unittest
from unittest import TestCase
from cogent import LoadTable
def bonferroni_correction(table, alpha):
"""apply bonferroni correction to summary.txt file, the p-adjusted value is calculated as 0.05/n, where n is
number of total tests"""
# this should be the sequential Bonferroni method -- CHECK!!
sig_results = []
num_rows = table.Shape[0]
prob_index = table.Header.index('prob')
table = table.sorted(columns='prob')
#compare the smallest probability p_1 to p_adjusted (alpha / n). If p is greater than p_adjusted, declare
#all tests not significant; if p is samller than p_adjusted, declare this test is significant,
#and compare the second smallest p_2...
for row in table.getRawData():
p_adjusted = alpha / (num_rows - len(sig_results))
if row[prob_index] > p_adjusted:
break
sig_results.append(row)
if not sig_results:
return None
sub_table = LoadTable(header=table.Header, rows=sig_results)
return sub_table
class TestBonferroni(TestCase):
table_data = LoadTable('test_bonferroni.txt', sep='\t')
alpha = 0.05
def test_bonferroni(self):
"""an example table is constructed according to BIOMETRY 3rd edition, pp241, with known
probs and significant results; raise the AssertionError when significant results
are not correctly produces after Bonferroni correction"""
bon_adj_tbl = bonferroni_correction(self.table_data, self.alpha)
sig_results = bon_adj_tbl.getRawData('prob')
expect_results = [1e-06, 1e-06, 0.000002, 0.004765]
self.assertEqual(sig_results, expect_results)
unittest.TextTestRunner(verbosity=2).run(TestBonferroni('test_bonferroni'))
seq_classes = ['ENU_variants/autosomes', 'ENU_variants/sex_chroms', 'ENU_variants/long_flanks/autosomes', 'ENU_variants/long_flanks/sex_chroms',
'ENU_vs_germline/autosomes', 'ENU_vs_germline/sex_chroms',
'germline_variants/autosomes', 'germline_variants/sex_chroms', 'germline_variants/long_flanks/autosomes', 'germline_variants/long_flanks/sex_chroms',
'ENU_A_vs_X', 'germline_A_vs_X']
summary_paths = []
for seq_class in seq_classes:
seq_paths = !find ../results/$seq_class/* -name "summary.txt"
summary_paths.append(seq_paths)
for paths in summary_paths:
output_dir = os.path.commonprefix(paths)
output_filename = output_dir + "bonferroni.txt"
header = ['directions', 'positions', 'probs']
rows = []
for path in paths:
direction = path.split('/')[-2]
table = LoadTable(path, sep='\t')
alpha = 0.05
adjusted_tbl = bonferroni_correction(table, alpha)
if adjusted_tbl is None:
print "all tests in %s are not significant" % path
continue
adjusted_tbl_ncol = adjusted_tbl.withNewColumn('direction', lambda x: direction, columns=adjusted_tbl.Header[0])
for row in adjusted_tbl_ncol.getRawData(['direction', 'Position', 'prob']):
rows.append(row)
summary_tbl = LoadTable(header=header, rows=rows, sep='\t')
summary_tbl.writeToFile(output_filename, sep='\t')
print "bonferroni corrected table %s is saved" % output_filename
```
A sequential Bonferroni correction involves dividing the significance level (0.05) by the total number of hypothesis tests. If the most "significant" test passes that criteria, we remove it and repeat the process.
We want another cogent Table that has the following columns:
* direction
* Position (see the summary.txt tables)
* prob
From these we will produce synopses of broad patterns, e.g. significant interactions are always between adjacent positions in sequence.
```
from cogent import LoadTable
##constructed a testing sample tbale with known probs and seq bonf sigc
header = ['Contrasts', 'prob']
rows = [['Sugars vs. control (C)(G,F,F + F,S)', 1e-6],
['Mixed vs. pure sugars (G + F)(G,F,S)', 0.004765],
['Among pure sugars (G)(F)(S)', 0.000002],
['Mixed ns. average of G and F (G+F)(G,F)', 0.418749],
['Monosaccharides vs. disaccharides (G,F)(S)', 1e-6]]
table = LoadTable(header = header, rows = rows)
print(table)
table.writeToFile('test_bonferroni.txt', sep='\t')
a <= 1e-6
print (a)
```
|
github_jupyter
|
#!/usr/bin/env python
import os
import unittest
from unittest import TestCase
from cogent import LoadTable
def bonferroni_correction(table, alpha):
"""apply bonferroni correction to summary.txt file, the p-adjusted value is calculated as 0.05/n, where n is
number of total tests"""
# this should be the sequential Bonferroni method -- CHECK!!
sig_results = []
num_rows = table.Shape[0]
prob_index = table.Header.index('prob')
table = table.sorted(columns='prob')
#compare the smallest probability p_1 to p_adjusted (alpha / n). If p is greater than p_adjusted, declare
#all tests not significant; if p is samller than p_adjusted, declare this test is significant,
#and compare the second smallest p_2...
for row in table.getRawData():
p_adjusted = alpha / (num_rows - len(sig_results))
if row[prob_index] > p_adjusted:
break
sig_results.append(row)
if not sig_results:
return None
sub_table = LoadTable(header=table.Header, rows=sig_results)
return sub_table
class TestBonferroni(TestCase):
table_data = LoadTable('test_bonferroni.txt', sep='\t')
alpha = 0.05
def test_bonferroni(self):
"""an example table is constructed according to BIOMETRY 3rd edition, pp241, with known
probs and significant results; raise the AssertionError when significant results
are not correctly produces after Bonferroni correction"""
bon_adj_tbl = bonferroni_correction(self.table_data, self.alpha)
sig_results = bon_adj_tbl.getRawData('prob')
expect_results = [1e-06, 1e-06, 0.000002, 0.004765]
self.assertEqual(sig_results, expect_results)
unittest.TextTestRunner(verbosity=2).run(TestBonferroni('test_bonferroni'))
seq_classes = ['ENU_variants/autosomes', 'ENU_variants/sex_chroms', 'ENU_variants/long_flanks/autosomes', 'ENU_variants/long_flanks/sex_chroms',
'ENU_vs_germline/autosomes', 'ENU_vs_germline/sex_chroms',
'germline_variants/autosomes', 'germline_variants/sex_chroms', 'germline_variants/long_flanks/autosomes', 'germline_variants/long_flanks/sex_chroms',
'ENU_A_vs_X', 'germline_A_vs_X']
summary_paths = []
for seq_class in seq_classes:
seq_paths = !find ../results/$seq_class/* -name "summary.txt"
summary_paths.append(seq_paths)
for paths in summary_paths:
output_dir = os.path.commonprefix(paths)
output_filename = output_dir + "bonferroni.txt"
header = ['directions', 'positions', 'probs']
rows = []
for path in paths:
direction = path.split('/')[-2]
table = LoadTable(path, sep='\t')
alpha = 0.05
adjusted_tbl = bonferroni_correction(table, alpha)
if adjusted_tbl is None:
print "all tests in %s are not significant" % path
continue
adjusted_tbl_ncol = adjusted_tbl.withNewColumn('direction', lambda x: direction, columns=adjusted_tbl.Header[0])
for row in adjusted_tbl_ncol.getRawData(['direction', 'Position', 'prob']):
rows.append(row)
summary_tbl = LoadTable(header=header, rows=rows, sep='\t')
summary_tbl.writeToFile(output_filename, sep='\t')
print "bonferroni corrected table %s is saved" % output_filename
from cogent import LoadTable
##constructed a testing sample tbale with known probs and seq bonf sigc
header = ['Contrasts', 'prob']
rows = [['Sugars vs. control (C)(G,F,F + F,S)', 1e-6],
['Mixed vs. pure sugars (G + F)(G,F,S)', 0.004765],
['Among pure sugars (G)(F)(S)', 0.000002],
['Mixed ns. average of G and F (G+F)(G,F)', 0.418749],
['Monosaccharides vs. disaccharides (G,F)(S)', 1e-6]]
table = LoadTable(header = header, rows = rows)
print(table)
table.writeToFile('test_bonferroni.txt', sep='\t')
a <= 1e-6
print (a)
| 0.401101 | 0.599339 |
# Removing fragments from simulation data
## Outline
1. [Starting point](#starting_point)
2. [Volume-of-fluid data](#vof_data)
3. [Parameterization](#parameterization)
4. [Simple function approximation](#function_approximation)
5. [Direct approximation of the radius](#direct_approximation)
6. [Using prior/domain knowledge](#prior_knowledge)
1. [Re-scaling the data](#rescaling)
2. [Adding artificial data](#artificial_data)
3. [Creating ensemble models](#ensemble_models)
7. [Final notes](#final_notes)
## Starting point<a id="starting_point"></a>
- detect and remove fragments
- show image of bubble and fragments
```
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import sklearn as sk
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
import urllib
import cloudpickle as cp
else:
import pickle
matplotlib.rcParams['figure.dpi'] = 80
print("Pandas version: {}".format(pd.__version__))
print("Numpy version: {}".format(np.__version__))
print("Sklearn version: {}".format(sk.__version__))
print("Running notebook {}".format("in colab." if IN_COLAB else "locally."))
if not IN_COLAB:
data_file = "../data/7mm_water_air_plic.pkl"
with open(data_file, 'rb') as file:
data = pickle.load(file).drop(["element"], axis=1)
else:
data_file = "https://github.com/AndreWeiner/machine-learning-applied-to-cfd/blob/master/data/7mm_water_air_plic.pkl?raw=true"
response = urllib.request.urlopen(data_file)
data = cp.load(response).drop(["element"], axis=1)
print("The data set contains {} points.".format(data.shape[0]))
data.sample(5)
%matplotlib inline
every = 100
fontsize = 14
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 10), sharey=True)
ax1.scatter(data.px.values[::every], data.py.values[::every],
marker='x', color='C0', s=30, linewidth=0.5)
ax2.scatter(data.pz.values[::every], data.py.values[::every],
marker='x', color='C0', s=30, linewidth=0.5)
ax1.set_xlabel(r"$x$", fontsize=fontsize)
ax1.set_ylabel(r"$y$", fontsize=fontsize)
ax2.set_xlabel(r"$z$", fontsize=fontsize)
plt.show()
from sklearn.ensemble import IsolationForest
def fit_isolation_forest(contamination):
clf = IsolationForest(n_estimators=100, contamination=contamination)
return clf.fit(data[['px', 'py', 'pz']].values).predict(data[['px', 'py', 'pz']].values)
fig, axarr = plt.subplots(1, 6, figsize=(15, 10), sharey=True)
colors = np.array(['C0', 'C1'])
for i, cont in enumerate(np.arange(0.08, 0.135, 0.01)):
pred = fit_isolation_forest(cont)
axarr[i].scatter(data.px.values[::every], data.py.values[::every],
marker='x', color=colors[(pred[::every] + 1) // 2], s=30, linewidth=0.5)
axarr[i].set_title("Contamination: {:2.2f}".format(cont))
axarr[i].set_xlabel(r"$x$", fontsize=fontsize)
axarr[0].set_ylabel(r"$y$", fontsize=fontsize)
plt.show()
clf = IsolationForest(n_estimators=200, contamination=0.11)
pred = clf.fit(data[['px', 'py', 'pz']].values).predict(data[['px', 'py', 'pz']].values)
points_clean = data[['px', 'py', 'pz']].values[pred > 0]
print("Removed {} points from data set.".format(data.shape[0] - points_clean.shape[0]))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 8), sharey=True)
ax1.scatter(points_clean[:,0], points_clean[:,1],
marker='x', color='C1', s=2, linewidth=0.5)
ax2.scatter(points_clean[:,2], points_clean[:,1],
marker='x', color='C1', s=2, linewidth=0.5)
ax1.set_xlabel(r"$x$", fontsize=fontsize)
ax1.set_ylabel(r"$y$", fontsize=fontsize)
ax2.set_xlabel(r"$z$", fontsize=fontsize)
plt.show()
if IN_COLAB:
%matplotlib inline
else:
%matplotlib notebook
every = 50
fig = plt.figure(figsize=(12, 8))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(points_clean[:,0][::every], points_clean[:,2][::every], points_clean[:,1][::every],
marker='x', color='C1', s=10, linewidth=0.5)
ax.set_xlabel(r"$x$", fontsize=fontsize)
ax.set_ylabel(r"$z$", fontsize=fontsize)
ax.set_zlabel(r"$y$", fontsize=fontsize)
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import sklearn as sk
import sys
IN_COLAB = 'google.colab' in sys.modules
if IN_COLAB:
import urllib
import cloudpickle as cp
else:
import pickle
matplotlib.rcParams['figure.dpi'] = 80
print("Pandas version: {}".format(pd.__version__))
print("Numpy version: {}".format(np.__version__))
print("Sklearn version: {}".format(sk.__version__))
print("Running notebook {}".format("in colab." if IN_COLAB else "locally."))
if not IN_COLAB:
data_file = "../data/7mm_water_air_plic.pkl"
with open(data_file, 'rb') as file:
data = pickle.load(file).drop(["element"], axis=1)
else:
data_file = "https://github.com/AndreWeiner/machine-learning-applied-to-cfd/blob/master/data/7mm_water_air_plic.pkl?raw=true"
response = urllib.request.urlopen(data_file)
data = cp.load(response).drop(["element"], axis=1)
print("The data set contains {} points.".format(data.shape[0]))
data.sample(5)
%matplotlib inline
every = 100
fontsize = 14
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 10), sharey=True)
ax1.scatter(data.px.values[::every], data.py.values[::every],
marker='x', color='C0', s=30, linewidth=0.5)
ax2.scatter(data.pz.values[::every], data.py.values[::every],
marker='x', color='C0', s=30, linewidth=0.5)
ax1.set_xlabel(r"$x$", fontsize=fontsize)
ax1.set_ylabel(r"$y$", fontsize=fontsize)
ax2.set_xlabel(r"$z$", fontsize=fontsize)
plt.show()
from sklearn.ensemble import IsolationForest
def fit_isolation_forest(contamination):
clf = IsolationForest(n_estimators=100, contamination=contamination)
return clf.fit(data[['px', 'py', 'pz']].values).predict(data[['px', 'py', 'pz']].values)
fig, axarr = plt.subplots(1, 6, figsize=(15, 10), sharey=True)
colors = np.array(['C0', 'C1'])
for i, cont in enumerate(np.arange(0.08, 0.135, 0.01)):
pred = fit_isolation_forest(cont)
axarr[i].scatter(data.px.values[::every], data.py.values[::every],
marker='x', color=colors[(pred[::every] + 1) // 2], s=30, linewidth=0.5)
axarr[i].set_title("Contamination: {:2.2f}".format(cont))
axarr[i].set_xlabel(r"$x$", fontsize=fontsize)
axarr[0].set_ylabel(r"$y$", fontsize=fontsize)
plt.show()
clf = IsolationForest(n_estimators=200, contamination=0.11)
pred = clf.fit(data[['px', 'py', 'pz']].values).predict(data[['px', 'py', 'pz']].values)
points_clean = data[['px', 'py', 'pz']].values[pred > 0]
print("Removed {} points from data set.".format(data.shape[0] - points_clean.shape[0]))
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 8), sharey=True)
ax1.scatter(points_clean[:,0], points_clean[:,1],
marker='x', color='C1', s=2, linewidth=0.5)
ax2.scatter(points_clean[:,2], points_clean[:,1],
marker='x', color='C1', s=2, linewidth=0.5)
ax1.set_xlabel(r"$x$", fontsize=fontsize)
ax1.set_ylabel(r"$y$", fontsize=fontsize)
ax2.set_xlabel(r"$z$", fontsize=fontsize)
plt.show()
if IN_COLAB:
%matplotlib inline
else:
%matplotlib notebook
every = 50
fig = plt.figure(figsize=(12, 8))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(points_clean[:,0][::every], points_clean[:,2][::every], points_clean[:,1][::every],
marker='x', color='C1', s=10, linewidth=0.5)
ax.set_xlabel(r"$x$", fontsize=fontsize)
ax.set_ylabel(r"$z$", fontsize=fontsize)
ax.set_zlabel(r"$y$", fontsize=fontsize)
plt.show()
| 0.426083 | 0.865281 |
# Transfer Learning
Most of the time you won't want to train a whole convolutional network yourself. Modern ConvNets training on huge datasets like ImageNet take weeks on multiple GPUs. Instead, most people use a pretrained network either as a fixed feature extractor, or as an initial network to fine tune. In this notebook, you'll be using [VGGNet](https://arxiv.org/pdf/1409.1556.pdf) trained on the [ImageNet dataset](http://www.image-net.org/) as a feature extractor. Below is a diagram of the VGGNet architecture.
<img src="assets/cnnarchitecture.jpg" width=700px>
VGGNet is great because it's simple and has great performance, coming in second in the ImageNet competition. The idea here is that we keep all the convolutional layers, but replace the final fully connected layers with our own classifier. This way we can use VGGNet as a feature extractor for our images then easily train a simple classifier on top of that. What we'll do is take the first fully connected layer with 4096 units, including thresholding with ReLUs. We can use those values as a code for each image, then build a classifier on top of those codes.
You can read more about transfer learning from [the CS231n course notes](http://cs231n.github.io/transfer-learning/#tf).
## Pretrained VGGNet
We'll be using a pretrained network from https://github.com/machrisaa/tensorflow-vgg.
This is a really nice implementation of VGGNet, quite easy to work with. The network has already been trained and the parameters are available from this link.
```
from urllib.request import urlretrieve
from os.path import isfile, isdir
from tqdm import tqdm
vgg_dir = 'tensorflow_vgg/'
# Make sure vgg exists
if not isdir(vgg_dir):
raise Exception("VGG directory doesn't exist!")
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile(vgg_dir + "vgg16.npy"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='VGG16 Parameters') as pbar:
urlretrieve(
'https://s3.amazonaws.com/content.udacity-data.com/nd101/vgg16.npy',
vgg_dir + 'vgg16.npy',
pbar.hook)
else:
print("Parameter file already exists!")
```
## Flower power
Here we'll be using VGGNet to classify images of flowers. To get the flower dataset, run the cell below. This dataset comes from the [TensorFlow inception tutorial](https://www.tensorflow.org/tutorials/image_retraining).
```
import tarfile
dataset_folder_path = 'flower_photos'
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile('flower_photos.tar.gz'):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='Flowers Dataset') as pbar:
urlretrieve(
'http://download.tensorflow.org/example_images/flower_photos.tgz',
'flower_photos.tar.gz',
pbar.hook)
if not isdir(dataset_folder_path):
with tarfile.open('flower_photos.tar.gz') as tar:
tar.extractall()
tar.close()
```
## ConvNet Codes
Below, we'll run through all the images in our dataset and get codes for each of them. That is, we'll run the images through the VGGNet convolutional layers and record the values of the first fully connected layer. We can then write these to a file for later when we build our own classifier.
Here we're using the `vgg16` module from `tensorflow_vgg`. The network takes images of size $244 \times 224 \times 3$ as input. Then it has 5 sets of convolutional layers. The network implemented here has this structure (copied from [the source code](https://github.com/machrisaa/tensorflow-vgg/blob/master/vgg16.py):
```
self.conv1_1 = self.conv_layer(bgr, "conv1_1")
self.conv1_2 = self.conv_layer(self.conv1_1, "conv1_2")
self.pool1 = self.max_pool(self.conv1_2, 'pool1')
self.conv2_1 = self.conv_layer(self.pool1, "conv2_1")
self.conv2_2 = self.conv_layer(self.conv2_1, "conv2_2")
self.pool2 = self.max_pool(self.conv2_2, 'pool2')
self.conv3_1 = self.conv_layer(self.pool2, "conv3_1")
self.conv3_2 = self.conv_layer(self.conv3_1, "conv3_2")
self.conv3_3 = self.conv_layer(self.conv3_2, "conv3_3")
self.pool3 = self.max_pool(self.conv3_3, 'pool3')
self.conv4_1 = self.conv_layer(self.pool3, "conv4_1")
self.conv4_2 = self.conv_layer(self.conv4_1, "conv4_2")
self.conv4_3 = self.conv_layer(self.conv4_2, "conv4_3")
self.pool4 = self.max_pool(self.conv4_3, 'pool4')
self.conv5_1 = self.conv_layer(self.pool4, "conv5_1")
self.conv5_2 = self.conv_layer(self.conv5_1, "conv5_2")
self.conv5_3 = self.conv_layer(self.conv5_2, "conv5_3")
self.pool5 = self.max_pool(self.conv5_3, 'pool5')
self.fc6 = self.fc_layer(self.pool5, "fc6")
self.relu6 = tf.nn.relu(self.fc6)
```
So what we want are the values of the first fully connected layer, after being ReLUd (`self.relu6`). To build the network, we use
```
with tf.Session() as sess:
vgg = vgg16.Vgg16()
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
with tf.name_scope("content_vgg"):
vgg.build(input_)
```
This creates the `vgg` object, then builds the graph with `vgg.build(input_)`. Then to get the values from the layer,
```
feed_dict = {input_: images}
codes = sess.run(vgg.relu6, feed_dict=feed_dict)
```
```
import os
import numpy as np
import tensorflow as tf
from tensorflow_vgg import vgg16
from tensorflow_vgg import utils
data_dir = 'flower_photos/'
contents = os.listdir(data_dir)
classes = [each for each in contents if os.path.isdir(data_dir + each)]
```
Below I'm running images through the VGG network in batches.
```
# Set the batch size higher if you can fit in in your GPU memory
batch_size = 10
codes_list = []
labels = []
batch = []
codes = None
with tf.Session() as sess:
vgg = vgg16.Vgg16()
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
with tf.name_scope("content_vgg"):
vgg.build(input_)
for each in classes:
print("Starting {} images".format(each))
class_path = data_dir + each
files = os.listdir(class_path)
for ii, file in enumerate(files, 1):
# Add images to the current batch
# utils.load_image crops the input images for us, from the center
img = utils.load_image(os.path.join(class_path, file))
batch.append(img.reshape((1, 224, 224, 3)))
labels.append(each)
# Running the batch through the network to get the codes
if ii % batch_size == 0 or ii == len(files):
images = np.concatenate(batch)
feed_dict = {input_: images}
codes_batch = sess.run(vgg.relu6, feed_dict=feed_dict)
# Here I'm building an array of the codes
if codes is None:
codes = codes_batch
else:
codes = np.concatenate((codes, codes_batch))
# Reset to start building the next batch
batch = []
print('{} images processed'.format(ii))
# write codes to file
with open('codes', 'w') as f:
codes.tofile(f)
# write labels to file
import csv
with open('labels', 'w') as f:
writer = csv.writer(f, delimiter='\n')
writer.writerow(labels)
```
## Building the Classifier
Now that we have codes for all the images, we can build a simple classifier on top of them. The codes behave just like normal input into a simple neural network. Below I'm going to have you do most of the work.
```
# read codes and labels from file
import csv
with open('labels') as f:
reader = csv.reader(f, delimiter='\n')
labels = np.array([each for each in reader if len(each) > 0]).squeeze()
with open('codes') as f:
codes = np.fromfile(f, dtype=np.float32)
codes = codes.reshape((len(labels), -1))
```
### Data prep
As usual, now we need to one-hot encode our labels and create validation/test sets. First up, creating our labels!
> **Exercise:** From scikit-learn, use [LabelBinarizer](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.LabelBinarizer.html) to create one-hot encoded vectors from the labels.
```
from sklearn.preprocessing import LabelBinarizer
lb = LabelBinarizer()
lb.fit(labels)
labels_vecs = lb.transform(labels)
```
Now you'll want to create your training, validation, and test sets. An important thing to note here is that our labels and data aren't randomized yet. We'll want to shuffle our data so the validation and test sets contain data from all classes. Otherwise, you could end up with testing sets that are all one class. Typically, you'll also want to make sure that each smaller set has the same the distribution of classes as it is for the whole data set. The easiest way to accomplish both these goals is to use [`StratifiedShuffleSplit`](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.StratifiedShuffleSplit.html) from scikit-learn.
You can create the splitter like so:
```
ss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
```
Then split the data with
```
splitter = ss.split(x, y)
```
`ss.split` returns a generator of indices. You can pass the indices into the arrays to get the split sets. The fact that it's a generator means you either need to iterate over it, or use `next(splitter)` to get the indices. Be sure to read the [documentation](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.StratifiedShuffleSplit.html) and the [user guide](http://scikit-learn.org/stable/modules/cross_validation.html#random-permutations-cross-validation-a-k-a-shuffle-split).
> **Exercise:** Use StratifiedShuffleSplit to split the codes and labels into training, validation, and test sets.
```
def gen():
i = 5
while i<10:
i+=1
yield i
for idx, i in enumerate(gen()):
print(idx, i)
from sklearn.model_selection import StratifiedShuffleSplit
ss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
train_idx, val_idx = next(ss.split(codes, labels_vecs))
half_val_len = int(len(val_idx)/2)
val_idx, test_idx = val_idx[:half_val_len], val_idx[half_val_len:]
train_x, train_y = codes[train_idx], labels_vecs[train_idx]
val_x, val_y = codes[val_idx], labels_vecs[val_idx]
test_x, test_y = codes[test_idx], labels_vecs[test_idx]
print("Train shapes (x, y):", train_x.shape, train_y.shape)
print("Validation shapes (x, y):", val_x.shape, val_y.shape)
print("Test shapes (x, y):", test_x.shape, test_y.shape)
```
If you did it right, you should see these sizes for the training sets:
```
Train shapes (x, y): (2936, 4096) (2936, 5)
Validation shapes (x, y): (367, 4096) (367, 5)
Test shapes (x, y): (367, 4096) (367, 5)
```
### Classifier layers
Once you have the convolutional codes, you just need to build a classfier from some fully connected layers. You use the codes as the inputs and the image labels as targets. Otherwise the classifier is a typical neural network.
> **Exercise:** With the codes and labels loaded, build the classifier. Consider the codes as your inputs, each of them are 4096D vectors. You'll want to use a hidden layer and an output layer as your classifier. Remember that the output layer needs to have one unit for each class and a softmax activation function. Use the cross entropy to calculate the cost.
```
inputs_ = tf.placeholder(tf.float32, shape=[None, codes.shape[1]])
labels_ = tf.placeholder(tf.int64, shape=[None, labels_vecs.shape[1]])
fc = tf.contrib.layers.fully_connected(inputs_, 256)
logits = tf.contrib.layers.fully_connected(fc, labels_vecs.shape[1], activation_fn=None)
cross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=labels_, logits=logits)
cost = tf.reduce_mean(cross_entropy)
optimizer = tf.train.AdamOptimizer().minimize(cost)
predicted = tf.nn.softmax(logits)
correct_pred = tf.equal(tf.argmax(predicted, 1), tf.argmax(labels_, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
```
### Batches!
Here is just a simple way to do batches. I've written it so that it includes all the data. Sometimes you'll throw out some data at the end to make sure you have full batches. Here I just extend the last batch to include the remaining data.
```
def get_batches(x, y, n_batches=10):
""" Return a generator that yields batches from arrays x and y. """
batch_size = len(x)//n_batches
for ii in range(0, n_batches*batch_size, batch_size):
# If we're not on the last batch, grab data with size batch_size
if ii != (n_batches-1)*batch_size:
X, Y = x[ii: ii+batch_size], y[ii: ii+batch_size]
# On the last batch, grab the rest of the data
else:
X, Y = x[ii:], y[ii:]
# I love generators
yield X, Y
```
### Training
Here, we'll train the network.
> **Exercise:** So far we've been providing the training code for you. Here, I'm going to give you a bit more of a challenge and have you write the code to train the network. Of course, you'll be able to see my solution if you need help.
```
epochs = 10
iteration = 0
saver = tf.train.Saver()
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for x, y in get_batches(train_x, train_y):
feed = {inputs_: x,
labels_: y}
loss, _ = sess.run([cost, optimizer], feed_dict=feed)
print("Epoch: {}/{}".format(e+1, epochs),
"Iteration: {}".format(iteration),
"Training loss: {:.5f}".format(loss))
iteration += 1
if iteration % 5 == 0:
feed = {inputs_: val_x,
labels_: val_y}
val_acc = sess.run(accuracy, feed_dict=feed)
print("Epoch: {}/{}".format(e, epochs),
"Iteration: {}".format(iteration),
"Validation Acc: {:.4f}".format(val_acc))
saver.save(sess, "checkpoints/flowers.ckpt")
```
### Testing
Below you see the test accuracy. You can also see the predictions returned for images.
```
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
feed = {inputs_: test_x,
labels_: test_y}
test_acc = sess.run(accuracy, feed_dict=feed)
print("Test accuracy: {:.4f}".format(test_acc))
%matplotlib inline
import matplotlib.pyplot as plt
from scipy.ndimage import imread
```
Below, feel free to choose images and see how the trained classifier predicts the flowers in them.
```
test_img_path = 'flower_photos/roses/10894627425_ec76bbc757_n.jpg'
test_img = imread(test_img_path)
plt.imshow(test_img)
# Run this cell if you don't have a vgg graph built
with tf.Session() as sess:
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
vgg = vgg16.Vgg16()
vgg.build(input_)
with tf.Session() as sess:
img = utils.load_image(test_img_path)
img = img.reshape((1, 224, 224, 3))
feed_dict = {input_: img}
code = sess.run(vgg.relu6, feed_dict=feed_dict)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
feed = {inputs_: code}
prediction = sess.run(predicted, feed_dict=feed).squeeze()
plt.imshow(test_img)
plt.barh(np.arange(5), prediction)
_ = plt.yticks(np.arange(5), lb.classes_)
```
|
github_jupyter
|
from urllib.request import urlretrieve
from os.path import isfile, isdir
from tqdm import tqdm
vgg_dir = 'tensorflow_vgg/'
# Make sure vgg exists
if not isdir(vgg_dir):
raise Exception("VGG directory doesn't exist!")
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile(vgg_dir + "vgg16.npy"):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='VGG16 Parameters') as pbar:
urlretrieve(
'https://s3.amazonaws.com/content.udacity-data.com/nd101/vgg16.npy',
vgg_dir + 'vgg16.npy',
pbar.hook)
else:
print("Parameter file already exists!")
import tarfile
dataset_folder_path = 'flower_photos'
class DLProgress(tqdm):
last_block = 0
def hook(self, block_num=1, block_size=1, total_size=None):
self.total = total_size
self.update((block_num - self.last_block) * block_size)
self.last_block = block_num
if not isfile('flower_photos.tar.gz'):
with DLProgress(unit='B', unit_scale=True, miniters=1, desc='Flowers Dataset') as pbar:
urlretrieve(
'http://download.tensorflow.org/example_images/flower_photos.tgz',
'flower_photos.tar.gz',
pbar.hook)
if not isdir(dataset_folder_path):
with tarfile.open('flower_photos.tar.gz') as tar:
tar.extractall()
tar.close()
self.conv1_1 = self.conv_layer(bgr, "conv1_1")
self.conv1_2 = self.conv_layer(self.conv1_1, "conv1_2")
self.pool1 = self.max_pool(self.conv1_2, 'pool1')
self.conv2_1 = self.conv_layer(self.pool1, "conv2_1")
self.conv2_2 = self.conv_layer(self.conv2_1, "conv2_2")
self.pool2 = self.max_pool(self.conv2_2, 'pool2')
self.conv3_1 = self.conv_layer(self.pool2, "conv3_1")
self.conv3_2 = self.conv_layer(self.conv3_1, "conv3_2")
self.conv3_3 = self.conv_layer(self.conv3_2, "conv3_3")
self.pool3 = self.max_pool(self.conv3_3, 'pool3')
self.conv4_1 = self.conv_layer(self.pool3, "conv4_1")
self.conv4_2 = self.conv_layer(self.conv4_1, "conv4_2")
self.conv4_3 = self.conv_layer(self.conv4_2, "conv4_3")
self.pool4 = self.max_pool(self.conv4_3, 'pool4')
self.conv5_1 = self.conv_layer(self.pool4, "conv5_1")
self.conv5_2 = self.conv_layer(self.conv5_1, "conv5_2")
self.conv5_3 = self.conv_layer(self.conv5_2, "conv5_3")
self.pool5 = self.max_pool(self.conv5_3, 'pool5')
self.fc6 = self.fc_layer(self.pool5, "fc6")
self.relu6 = tf.nn.relu(self.fc6)
with tf.Session() as sess:
vgg = vgg16.Vgg16()
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
with tf.name_scope("content_vgg"):
vgg.build(input_)
feed_dict = {input_: images}
codes = sess.run(vgg.relu6, feed_dict=feed_dict)
import os
import numpy as np
import tensorflow as tf
from tensorflow_vgg import vgg16
from tensorflow_vgg import utils
data_dir = 'flower_photos/'
contents = os.listdir(data_dir)
classes = [each for each in contents if os.path.isdir(data_dir + each)]
# Set the batch size higher if you can fit in in your GPU memory
batch_size = 10
codes_list = []
labels = []
batch = []
codes = None
with tf.Session() as sess:
vgg = vgg16.Vgg16()
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
with tf.name_scope("content_vgg"):
vgg.build(input_)
for each in classes:
print("Starting {} images".format(each))
class_path = data_dir + each
files = os.listdir(class_path)
for ii, file in enumerate(files, 1):
# Add images to the current batch
# utils.load_image crops the input images for us, from the center
img = utils.load_image(os.path.join(class_path, file))
batch.append(img.reshape((1, 224, 224, 3)))
labels.append(each)
# Running the batch through the network to get the codes
if ii % batch_size == 0 or ii == len(files):
images = np.concatenate(batch)
feed_dict = {input_: images}
codes_batch = sess.run(vgg.relu6, feed_dict=feed_dict)
# Here I'm building an array of the codes
if codes is None:
codes = codes_batch
else:
codes = np.concatenate((codes, codes_batch))
# Reset to start building the next batch
batch = []
print('{} images processed'.format(ii))
# write codes to file
with open('codes', 'w') as f:
codes.tofile(f)
# write labels to file
import csv
with open('labels', 'w') as f:
writer = csv.writer(f, delimiter='\n')
writer.writerow(labels)
# read codes and labels from file
import csv
with open('labels') as f:
reader = csv.reader(f, delimiter='\n')
labels = np.array([each for each in reader if len(each) > 0]).squeeze()
with open('codes') as f:
codes = np.fromfile(f, dtype=np.float32)
codes = codes.reshape((len(labels), -1))
from sklearn.preprocessing import LabelBinarizer
lb = LabelBinarizer()
lb.fit(labels)
labels_vecs = lb.transform(labels)
ss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
splitter = ss.split(x, y)
def gen():
i = 5
while i<10:
i+=1
yield i
for idx, i in enumerate(gen()):
print(idx, i)
from sklearn.model_selection import StratifiedShuffleSplit
ss = StratifiedShuffleSplit(n_splits=1, test_size=0.2)
train_idx, val_idx = next(ss.split(codes, labels_vecs))
half_val_len = int(len(val_idx)/2)
val_idx, test_idx = val_idx[:half_val_len], val_idx[half_val_len:]
train_x, train_y = codes[train_idx], labels_vecs[train_idx]
val_x, val_y = codes[val_idx], labels_vecs[val_idx]
test_x, test_y = codes[test_idx], labels_vecs[test_idx]
print("Train shapes (x, y):", train_x.shape, train_y.shape)
print("Validation shapes (x, y):", val_x.shape, val_y.shape)
print("Test shapes (x, y):", test_x.shape, test_y.shape)
Train shapes (x, y): (2936, 4096) (2936, 5)
Validation shapes (x, y): (367, 4096) (367, 5)
Test shapes (x, y): (367, 4096) (367, 5)
inputs_ = tf.placeholder(tf.float32, shape=[None, codes.shape[1]])
labels_ = tf.placeholder(tf.int64, shape=[None, labels_vecs.shape[1]])
fc = tf.contrib.layers.fully_connected(inputs_, 256)
logits = tf.contrib.layers.fully_connected(fc, labels_vecs.shape[1], activation_fn=None)
cross_entropy = tf.nn.softmax_cross_entropy_with_logits(labels=labels_, logits=logits)
cost = tf.reduce_mean(cross_entropy)
optimizer = tf.train.AdamOptimizer().minimize(cost)
predicted = tf.nn.softmax(logits)
correct_pred = tf.equal(tf.argmax(predicted, 1), tf.argmax(labels_, 1))
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
def get_batches(x, y, n_batches=10):
""" Return a generator that yields batches from arrays x and y. """
batch_size = len(x)//n_batches
for ii in range(0, n_batches*batch_size, batch_size):
# If we're not on the last batch, grab data with size batch_size
if ii != (n_batches-1)*batch_size:
X, Y = x[ii: ii+batch_size], y[ii: ii+batch_size]
# On the last batch, grab the rest of the data
else:
X, Y = x[ii:], y[ii:]
# I love generators
yield X, Y
epochs = 10
iteration = 0
saver = tf.train.Saver()
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for x, y in get_batches(train_x, train_y):
feed = {inputs_: x,
labels_: y}
loss, _ = sess.run([cost, optimizer], feed_dict=feed)
print("Epoch: {}/{}".format(e+1, epochs),
"Iteration: {}".format(iteration),
"Training loss: {:.5f}".format(loss))
iteration += 1
if iteration % 5 == 0:
feed = {inputs_: val_x,
labels_: val_y}
val_acc = sess.run(accuracy, feed_dict=feed)
print("Epoch: {}/{}".format(e, epochs),
"Iteration: {}".format(iteration),
"Validation Acc: {:.4f}".format(val_acc))
saver.save(sess, "checkpoints/flowers.ckpt")
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
feed = {inputs_: test_x,
labels_: test_y}
test_acc = sess.run(accuracy, feed_dict=feed)
print("Test accuracy: {:.4f}".format(test_acc))
%matplotlib inline
import matplotlib.pyplot as plt
from scipy.ndimage import imread
test_img_path = 'flower_photos/roses/10894627425_ec76bbc757_n.jpg'
test_img = imread(test_img_path)
plt.imshow(test_img)
# Run this cell if you don't have a vgg graph built
with tf.Session() as sess:
input_ = tf.placeholder(tf.float32, [None, 224, 224, 3])
vgg = vgg16.Vgg16()
vgg.build(input_)
with tf.Session() as sess:
img = utils.load_image(test_img_path)
img = img.reshape((1, 224, 224, 3))
feed_dict = {input_: img}
code = sess.run(vgg.relu6, feed_dict=feed_dict)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
feed = {inputs_: code}
prediction = sess.run(predicted, feed_dict=feed).squeeze()
plt.imshow(test_img)
plt.barh(np.arange(5), prediction)
_ = plt.yticks(np.arange(5), lb.classes_)
| 0.446012 | 0.962918 |
```
import numpy as np
import ipyvuetify as v
import ipywidgets as widgets
from bqplot import pyplot as plt
import bqplot
```
# Deploying voilà
## First histogram plot: bqplot
```
n = 200
x = np.linspace(0.0, 10.0, n)
y = np.cumsum(np.random.randn(n)*10).astype(int)
fig = plt.figure( title='Histogram')
np.random.seed(0)
hist = plt.hist(y, bins=25)
fig
slider = v.Slider(thumb_label='always', class_="px-4", v_model=30)
widgets.link((slider, 'v_model'), (hist, 'bins'))
slider
```
# Line chart
```
fig2 = plt.figure( title='Line Chart')
np.random.seed(0)
p = plt.plot(x, y)
fig2
selector = bqplot.interacts.BrushIntervalSelector(scale=p.scales['x'])
def update_range(*args):
if selector.selected is not None and selector.selected.shape == (2,):
mask = (x > selector.selected[0]) & (x < selector.selected[1])
hist.sample = y[mask]
selector.observe(update_range, 'selected')
fig2.interaction = selector
```
# Set up voila vuetify layout
The voila vuetify template does not render output from the notebook, it only shows widget with the mount_id metadata.
```
fig.layout.width = 'auto'
fig.layout.height = 'auto'
fig.layout.min_height = '300px' # so it still shows nicely in the notebook
fig2.layout.width = 'auto'
fig2.layout.height = 'auto'
fig2.layout.min_height = '300px' # so it still shows nicely in the notebook
content_main = v.Tabs(_metadata={'mount_id': 'content-main'}, children=[
v.Tab(children=['Tab1']),
v.Tab(children=['Tab2']),
v.TabItem(children=[
v.Layout(row=True, wrap=True, align_center=True, children=[
v.Flex(xs12=True, lg6=True, xl4=True, children=[
fig, slider
]),
v.Flex(xs12=True, lg6=True, xl4=True, children=[
fig2
]),
])
]),
v.TabItem(children=[
v.Container(children=['Lorum ipsum'])
])
])
# no need to display content_main for the voila-vuetify template
# but might be useful for debugging
# content_main
```
# Running locally with voila
```
$ voila --template voila-vuetify --enable_nbextensions=True ./notebooks/voila-vuetify.ipynb
```
# Run it on mybinder
* Put it on a github repo, e.g. https://github.com/maartenbreddels/voila-demo
* Add a noteook, e.g. voila-vuetify.ipynb
* Make sure the kernelspec name is vanilla "python3" (modify this in the classical notebook under Edit->Edit Notebook Metadata)
* put in a requirements.txt, with at least voila and voila-vuetify
* Create a mybinder on https://ovh.mybinder.org/
* GitHub URL: e.g. https://github.com/maartenbreddels/voila-demo/
* Change 'File' to 'URL'
* URL to open: e.g. `voila/render/voila-vuetify.ipynb`
|
github_jupyter
|
import numpy as np
import ipyvuetify as v
import ipywidgets as widgets
from bqplot import pyplot as plt
import bqplot
n = 200
x = np.linspace(0.0, 10.0, n)
y = np.cumsum(np.random.randn(n)*10).astype(int)
fig = plt.figure( title='Histogram')
np.random.seed(0)
hist = plt.hist(y, bins=25)
fig
slider = v.Slider(thumb_label='always', class_="px-4", v_model=30)
widgets.link((slider, 'v_model'), (hist, 'bins'))
slider
fig2 = plt.figure( title='Line Chart')
np.random.seed(0)
p = plt.plot(x, y)
fig2
selector = bqplot.interacts.BrushIntervalSelector(scale=p.scales['x'])
def update_range(*args):
if selector.selected is not None and selector.selected.shape == (2,):
mask = (x > selector.selected[0]) & (x < selector.selected[1])
hist.sample = y[mask]
selector.observe(update_range, 'selected')
fig2.interaction = selector
fig.layout.width = 'auto'
fig.layout.height = 'auto'
fig.layout.min_height = '300px' # so it still shows nicely in the notebook
fig2.layout.width = 'auto'
fig2.layout.height = 'auto'
fig2.layout.min_height = '300px' # so it still shows nicely in the notebook
content_main = v.Tabs(_metadata={'mount_id': 'content-main'}, children=[
v.Tab(children=['Tab1']),
v.Tab(children=['Tab2']),
v.TabItem(children=[
v.Layout(row=True, wrap=True, align_center=True, children=[
v.Flex(xs12=True, lg6=True, xl4=True, children=[
fig, slider
]),
v.Flex(xs12=True, lg6=True, xl4=True, children=[
fig2
]),
])
]),
v.TabItem(children=[
v.Container(children=['Lorum ipsum'])
])
])
# no need to display content_main for the voila-vuetify template
# but might be useful for debugging
# content_main
$ voila --template voila-vuetify --enable_nbextensions=True ./notebooks/voila-vuetify.ipynb
| 0.307982 | 0.723456 |
# Artist Plays
> Tracking artist plays over time.
- toc: true
- badges: true
- comments: false
- categories: [asot, artists]
- image: images/artist-plays.png
```
#hide
import os
import yaml
import spotipy
import json
import altair as alt
import numpy as np
import pandas as pd
from spotipy.oauth2 import SpotifyClientCredentials
with open('spotipy_credentials.yaml', 'r') as spotipy_credentials_file:
credentials = yaml.safe_load(spotipy_credentials_file)
os.environ["SPOTIPY_CLIENT_ID"] = credentials['spotipy_credentials']['spotipy_client_id']
os.environ["SPOTIPY_CLIENT_SECRET"] = credentials['spotipy_credentials']['spotipi_client_seret']
sp = spotipy.Spotify(client_credentials_manager=SpotifyClientCredentials())
asot_radio_id = '25mFVpuABa9GkGcj9eOPce'
albums = []
results = sp.artist_albums(asot_radio_id, album_type='album')
albums.extend(results['items'])
while results['next']:
results = sp.next(results)
albums.extend(results['items'])
seen = set() # to avoid dups
for album in albums:
name = album['name']
if name not in seen:
seen.add(name)
singles = []
results = sp.artist_albums(asot_radio_id, album_type='single')
singles.extend(results['items'])
while results['next']:
results = sp.next(results)
singles.extend(results['items'])
seen = set() # to avoid dups
for single in singles:
name = single['name']
if name not in seen:
seen.add(name)
episodes = singles + albums
episodes.sort(key=lambda x: x['release_date']) # Sort by release date
```
## Introduction
How many unique artists have been played on A State of Trance over the years? Who's been played the most? And the least?
In this post, we'll examine artist plays over time.
## Getting Started
First, some baseline analysis. Let's first figure out how many tracks played on A State of Trance are available on Spotify:
```
tracks_counted = 0
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
tracks_counted += 1
except:
pass
print(tracks_counted)
```
Wow - 17,000+ total _(not unique)_ tracks have been played! Remember, as we learned in the ["Methodology" post](https://scottbrenner.github.io/asot-jupyter/asot/bpm/2020/04/27/methodology.html) some episodes - especially early ones - are incomplete.
How many unique artists?
```
unique_artists = set()
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
for artist in track['artists']:
unique_artists.add(artist['name'])
except:
pass
print(len(unique_artists))
```
As always, this is a "best guess" - an approximation.
For the sake of tallying unique artists, we are treating collaborators as individuals. A track produced by "Artist A & Artist B" is recorded here as a production by Artist A and Artist B invididually.
## Calculating
Let's crunch some numbers.
Which artists have the most plays?
```
from collections import defaultdict
artist_counter = defaultdict(int)
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
for artist in track['artists']:
artist_counter[artist['name']] += 1
except:
pass
top_artists = sorted(artist_counter.items(), key=lambda k_v: k_v[1], reverse=True)
```
Alright, let's see the top 25 in a graph..
```
source = pd.DataFrame.from_dict(top_artists[:25])
bars = alt.Chart(source).mark_bar().encode(
x=alt.X('1:Q', title='Plays'),
y=alt.Y('0:N', sort='-x', title='Artist')
).properties(
title="A State of Trance - Most-played artists",
width=600
)
text = bars.mark_text(
align='left',
baseline='middle',
dx=3 # Nudges text to right so it doesn't appear on top of the bar
).encode(
text='1:Q'
)
bars + text
```
First place goes to the big man himself of course. Keep in mind we're counting a remix of an artist's track as a play for that artist.
From our numbers, a track that credits Armin van Buuren has been played 1210 times across 960 episodes. From this, we can say the average episode of A State of Trance has
```
top_artists[0][1] / len(episodes)
```
tracks produced by Armin van Buuren in some form or another, which is totally useless to know.
Earlier we found 17,000+ total tracks played from 4000+ unique artists based on what's currently available on Spotify. How many artists have been played exactly once?
```
artists_played_once = 0
one_hit_wonders = []
for artist, plays in artist_counter.items():
if plays == 1:
one_hit_wonders.append(artist)
artists_played_once += 1
print(artists_played_once)
```
Seems like quite a bit, what percentage is that?
```
print(100 * artists_played_once / len(unique_artists))
```
From the data available on Spotify, we can say 43% of artists played on A State of Trance were played exactly once. A full list of artists played once appears below:
```
print(sorted(one_hit_wonders))
```
Highlights include Steve Aoki and What So Not, among others. Again, this is based on how artists are reported in Spotify and as such is not entirely accurate. In the above we see 'tAudrey Gallagher' and "Thomas Datt's", both typos, report as separate from their namesake.
## Summary
In conclusion, across the current (as of writing) 960 episodes of A State of Trance...
* 17,000+ total tracks have been played
* 4,000+ unique artists have been featured
* 1,200+ tracks produced by Armin van Buuren in some form or another have been played
* 1,700+ artists (43%) have been played exactly once
And there we have it! There's plenty more to investigate, stay tuned..
|
github_jupyter
|
#hide
import os
import yaml
import spotipy
import json
import altair as alt
import numpy as np
import pandas as pd
from spotipy.oauth2 import SpotifyClientCredentials
with open('spotipy_credentials.yaml', 'r') as spotipy_credentials_file:
credentials = yaml.safe_load(spotipy_credentials_file)
os.environ["SPOTIPY_CLIENT_ID"] = credentials['spotipy_credentials']['spotipy_client_id']
os.environ["SPOTIPY_CLIENT_SECRET"] = credentials['spotipy_credentials']['spotipi_client_seret']
sp = spotipy.Spotify(client_credentials_manager=SpotifyClientCredentials())
asot_radio_id = '25mFVpuABa9GkGcj9eOPce'
albums = []
results = sp.artist_albums(asot_radio_id, album_type='album')
albums.extend(results['items'])
while results['next']:
results = sp.next(results)
albums.extend(results['items'])
seen = set() # to avoid dups
for album in albums:
name = album['name']
if name not in seen:
seen.add(name)
singles = []
results = sp.artist_albums(asot_radio_id, album_type='single')
singles.extend(results['items'])
while results['next']:
results = sp.next(results)
singles.extend(results['items'])
seen = set() # to avoid dups
for single in singles:
name = single['name']
if name not in seen:
seen.add(name)
episodes = singles + albums
episodes.sort(key=lambda x: x['release_date']) # Sort by release date
tracks_counted = 0
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
tracks_counted += 1
except:
pass
print(tracks_counted)
unique_artists = set()
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
for artist in track['artists']:
unique_artists.add(artist['name'])
except:
pass
print(len(unique_artists))
from collections import defaultdict
artist_counter = defaultdict(int)
for episode in episodes:
try:
for track in sp.album_tracks(episode['uri'])['items']:
if "a state of trance" in track['name'].lower() or "- interview" in track['name'].lower():
continue
else:
for artist in track['artists']:
artist_counter[artist['name']] += 1
except:
pass
top_artists = sorted(artist_counter.items(), key=lambda k_v: k_v[1], reverse=True)
source = pd.DataFrame.from_dict(top_artists[:25])
bars = alt.Chart(source).mark_bar().encode(
x=alt.X('1:Q', title='Plays'),
y=alt.Y('0:N', sort='-x', title='Artist')
).properties(
title="A State of Trance - Most-played artists",
width=600
)
text = bars.mark_text(
align='left',
baseline='middle',
dx=3 # Nudges text to right so it doesn't appear on top of the bar
).encode(
text='1:Q'
)
bars + text
top_artists[0][1] / len(episodes)
artists_played_once = 0
one_hit_wonders = []
for artist, plays in artist_counter.items():
if plays == 1:
one_hit_wonders.append(artist)
artists_played_once += 1
print(artists_played_once)
print(100 * artists_played_once / len(unique_artists))
print(sorted(one_hit_wonders))
| 0.136637 | 0.511412 |
# Lecture 7: Bias-Variance Tradeoff, Regularization
## 4/1/19
### Hosted by and maintained by the [Statistics Undergraduate Students Association (SUSA)](https://susa.berkeley.edu). Authored by [Ajay Raj](mailto:[email protected]), [Nichole Sun](mailto:[email protected]), [Rosa Choe](mailto:[email protected]), [Calvin Chen](mailto:[email protected]), and [Roland Chin](mailto:[email protected]).
### Table Of Contents
* [Recap](#recap)
* [Bias-Variance Tradeoff](#bv-tradeoff)
* [Bias](#bias)
* [Variance](#variance)
* [The Tradeoff](#the-tradeoff)
* [Polynomial Regression](#polynomial-regression)
* [Regularization](#regularization)
* [Ridge](#ridge)
* [LASSO](#lasso)
* [Visualizing Ridge and Lasso](#visualizing-ridge-and-lasso)
* [Regularization and Bias Variance](#regularization-and-bias-variance)
* [Lambda](#lambda)
* [Validation on Lambda](#validation-on-lambda)
* [Exercises](#exercises)
```
import matplotlib.pyplot as plt
import random
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from plotting import overfittingDemo, plot_multiple_linear_regression, ridgeRegularizationDemo, lassoRegularizationDemo
from scipy.optimize import curve_fit
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
```
<a id='recap'></a>
# Recap

High bias corresponds to underfitting. If we look at the first model, the points seem to follow some sort of curve, but our predictor is linear and therefore, unable to capture all the points. In this case, we have chosen a model which is not complex enough to accurately capture all the information from our data set.
If we look at the last model, the predictor is now overly complex because it adjusts based on every point in order to get as close to every data point as possible. In this case, the model changes too much based on small fluctuations caused by insignificant details in the data. This model is fitting more to noise than signal.
<a id='bv-tradeoff'></a>
# Bias-Variance Tradeoff
Today we'll perform **model evaluation**, where we'll judge how our linear regression models actually perform. Last week, we talked about **loss functions**, which describes a numerical value for how far your model is from the true values.
$$\text{Mean Squared Error: } \frac{1}{n}\sum_{i=1}^n \left(y_i - f(x_i)\right)^2$$
In this loss function, $y_i$ is a scalar, and $x_i$ is a $p$ dimensional vector, because there are $p$ features. This loss is called **mean squared error**, or **MSE**.
Now, we'll talk about other ways to evaluate a model.
**First, let's define some terms.**
We can say that everything in the universe can be described with the following equation:
$$y = h(x) + \epsilon$$
- $y$ is the quantity you are trying to model or approximate
- $x$ are the features (independent variables)
- $h$ is the **true model** for $y$ in terms of $x$
- $\epsilon$ represents **noise**, a random variable which has mean zero
Let $f$ be your model for $y$ in terms of $x$.
<a id='bias'></a>
## Bias
When evaluating a model, the most intuitive first step is to look at how well the model performs. For classification, this may be the percentage of data points correctly classified, or for regression it may be how close the predicted values are to actual. The **bias** of a model is *a measure of how close our prediction is to the actual value on average from an average model*.
Note that bias is **not** a measure of a single model, it encapuslates the scenario in which we collect many datasets, create models for each dataset, and average the error over all of models. Bias is not a measure of error for a single model, but a more abstract concept describing the average error over all errors. A low value for the bias of a model describes that on average, our predictions are similar to the actual values.
<a id='variance'></a>
## Variance
The **variance** of a model relates to the variance of the distribution of all models. In the previous section about bias, we envisoned the scenario of collecting many datasets, creating models for each dataset, and averaging the error overall the datasets. Instead, the variance of a model describes the variance in prediction. While we might be able to predict a value very well on average, if the variance of predictions is very high this may not be very helpful, as when we train a model we only have one such instance, and a high model variance tells us little about the true nature of the predictions. A low variance describes that our model will not predict very different values for different datasets.
**We can take a look at how bias and variance differ and can be explained in a dataset with the following diagram:**

Image from http://scott.fortmann-roe.com/docs/BiasVariance.html
The image describes what bias and variance are in a more simplified example. Consider that we would like to create a model that selects a point close to the center. The models on the top row have low bias, meaning the center of the cluster is close to the red dot on the target. The models on the left column have low variance, the clusters are quite tight, meaning our predictions are close together.
**Question: What do the blue dots represent? What about the bullseye?**
**Question: What is the order of best scenarios?**
<a id='the-tradeoff'></a>
## The Tradeoff
We are trying to minimize **expected error**, or the average **MSE** over all datasets. It turns out (with some advanced probability gymnastics), that:
$$\text{Mean Squared Error} = \text{Noise Variance} + \text{Bias}^2 + \text{Variance}$$
Note that $\text{Noise Variance}$ is constant: we assume there is some noise, and $\text{moise variance}$ is simply a value that describes how noisy your dataset will be on average. This is often also called "irreducible noise", as it is literally irreducible, we cannot avoid this.
Furthermore, notice that the equation above is the sum of (squared) bias and variance. Thus there is a literal tradeoff between these two, since decreasing one increases the other. This defines what is known as the **bias variance tradeoff**.

Image from http://scott.fortmann-roe.com/docs/BiasVariance.html
**Why does this happen?**
At some point, as we decrease **bias**, instead of getting closer to the **true model** $h$, we go past and try to fit to the $\epsilon$ (noise) that is part of our current dataset. This is equivalent to making our model more noisy, or **overfit** on our dataset, which means that over all datasets, it has more **variance**.
**Questions for understanding**:
> 1. Where does underfitting and overfitting lie in the graph above? How do they relate to bias and variance?
> 2. Why can't we usually just make a bunch of models with low bias and high variance and average them?
> 3. Why is low variance important in models?
<a id='polynonmial-regression'></a>
## Polynomial Regression
Let's revisit the polynomial problem that we have discussed.
In this case, if our model has degree $d$, we have $d + 1$ features: $x = [x^0, x^1, ..., x^d]$. Now, we have a linear model with $d + 1$ features:
$$\hat{f}(x) = \sum_{i=0}^{d} a_i x^i$$
Model complexity in this case is the degree of the polynomial. As we saw last week, as $d$ increases, model complexity increases. The model gets better, but then gets erratic. This directly corresponds to the bias-variance graph above.
```
overfittingDemo()
```
As we saw from last time, the best model was a degree 3 model.
<a id='regularization'></a>
# Regularization
We talked about **validation** as a means of combating overfitting. However, this is not the only method to combat overfitting. Another method to do so is to add *regularization* terms to our loss function. **Regularization** basically penalizes complexity in our models. This allows us to add explanatory variables to our model without worrying as much about overfitting. Here's what our ordinary least squares model looks like with a regularization term:
$$\hat{\boldsymbol{\theta}} = \arg\!\min_\theta \sum_{i=1}^n (y_i - f_\boldsymbol{\theta}(x_i))^2 + \lambda R(\boldsymbol{\theta})$$
We've written the model a little differently here, but the first term is the same as the ordinary least squares regression model you learned last week. This time it's just generalized to any function of $x$ where $\theta$ is a list of parameters, or weights on our explanatory variables, such as coefficients to a polynomial. We're minimizing a loss function to find the best coefficients for our model.
The second term is the **regularization** term. The $\lambda$ parameter in front of it dictates how much we care about our regularization term – the higher $\lambda$ is, the more we penalize large weights, and the more the regularization makes our weights deviate from OLS.
**Question**: What happens when $\lambda = 0$?
So, what is $R(\theta)$ in the equation? There are a variety of different regularization functions that could take its place, but today, we'll just talk about the two most common types of functions: **ridge regression** and **LASSO regression**.
$$\begin{align}\text{ Ridge (L2 Norm)}: &\ R(\boldsymbol{\theta}) = \|\theta\|_2^2 = \sum_{i=1}^p \theta_i^2\\
\text{ LASSO (L1 Norm)}: &\ R(\boldsymbol{\theta}) = \|\theta\|_1=\sum_{i=1}^p \lvert \theta_i\rvert\end{align}$$
<a id='ridge'></a>
## Ridge
$$\hat{\boldsymbol{\theta}} = \arg\!\min_\theta \sum_{i=1}^n (y_i - f_\boldsymbol{\theta}(x_i))^2 + \lambda \|\theta\|_2^2$$
In **ridge** regression, the regularization function is the sum of squared weights. One nice thing about ridge regression is that there is always a unique, mathematical solution to minimizing the loss of that term. The solution involves some linear algebra, which we won't get into in this notebook, but the existence of this formula makes this minimization computationally easy to solve!
$$\hat{\boldsymbol{\theta}} = \left(\boldsymbol{X}^T \boldsymbol{X} + \lambda\boldsymbol{I}\right)^{-1}\boldsymbol{X}^T\boldsymbol{Y}$$
If you recall, the solution to linear regression was of the form:
$$\hat{\boldsymbol{\theta}} = \left(\boldsymbol{X}^T \boldsymbol{X}\right)^{-1}\boldsymbol{X}^T\boldsymbol{Y}$$
And we said that the $\boldsymbol{X}^T\boldsymbol{X}$ isn't always invertible. **What about $\boldsymbol{X}^T \boldsymbol{X} + \lambda\boldsymbol{I}$?**
Turns out, this is always invertible! If you are familiar with linear algebra, this is equivalent to adding $\lambda$ to all the eigenvalues of $X^TX$.
**To see this in practice**, we'll first create a regular linear regression model, and compare how it does against models using regularization on the `mpg` dataset we used from last week! We'll be constructing models of `displacement` vs. `mpg`, and seeing the difference from there!
First, let's construct the `mpg_train` dataset!
```
mpg = pd.read_csv("mpg.csv", index_col='name')# load mpg dataset
mpg = mpg.loc[mpg["horsepower"] != '?'].astype(float) # remove columns with missing horsepower values
mpg_train, mpg_test = train_test_split(mpg, test_size = .2, random_state = 0) # split into training set and test set
mpg_train, mpg_validation = train_test_split(mpg_train, test_size = .5, random_state = 0)
mpg_train.head()
```
**Exercise:** Now, let's create a regular linear regression model using the same process we've learned before (fitting, predicting, finding the loss)!
```
from sklearn.linear_model import LinearRegression
x_train = np.vander(mpg_train["displacement"], 13)
y_train = mpg_train[["mpg"]]
x_validation = np.vander(mpg_validation["displacement"], 13)
y_validation = mpg_validation[["mpg"]]
# instantiate your model
linear_model = LinearRegression()
# fit the model
linear_model.fit(x_train, y_train)
# make predictions on validation set
linear_prediction = linear_model.predict(x_validation)
# find error
linear_loss = mean_squared_error(linear_prediction, y_validation)
print("Root Mean Squared Error of linear model: {:.2f}".format(linear_loss))
```
**Exercise:** Using what you did above as reference, do the same using a Ridge regression model!
```
from sklearn.linear_model import Ridge
ridge_model = Ridge()
ridge_model.fit(x_train, y_train)
ridge_prediction = ridge_model.predict(x_validation)
ridge_loss = mean_squared_error(ridge_prediction, y_validation) # mean squared error of ridge model
print("Root Mean Squared Error of Linear Model: {:.2f}".format(linear_loss))
print("Root Mean Squared Error of Ridge Model: {:.2f}".format(ridge_loss))
```
<a id='lasso'></a>
## LASSO
$$\hat{\boldsymbol{\theta}} = \arg\!\min_\theta \sum_{i=1}^n (y_i - f_\boldsymbol{\theta}(x_i))^2 + \lambda \|\theta\|_1$$
In **LASSO** regression, the regularization function is **the sum of absolute values of the weights**. One key thing to note about **LASSO** is that it is **sparsity inducing**, meaning it forces weights to be zero values rather than really small values (which can happen in **Ridge Regression**), leaving you with fewer explanatory variables in the resulting model! Unlike Ridge Regression, LASSO doesn't necessarily have a unique solution that can be solved for with linear algebra, so there's no formula that determines what the optimal weights should be.
```
from sklearn.linear_model import Lasso
lasso_model = Lasso()
lasso_model.fit(x_train, y_train)
lasso_prediction = lasso_model.predict(x_validation)
lasso_loss = mean_squared_error(lasso_prediction, y_validation) # mean squared error of lasso model
print("Root Mean Squared Error of Linear Model: {:.2f}".format(linear_loss))
print("Root Mean Squared Error of LASSO Model: {:.2f}".format(lasso_loss))
```
As we can see, both **Ridge Regression and LASSO Regression** minimized the loss of our linear regression models, so maybe penalizing some features allowed us to prevent overfitting on our dataset!
<a id='visualizing-ridge-and-lasso'></a>
## Visualizing Ridge and LASSO
We went through a lot about **ridge** and **LASSO**, but we didn't really get into what they look like for understanding! And so, here are some visualizations that might help build the intution behind some of the characteristics of these two regularization methods.
Another way to describe the modified minimization function above is that it's the same loss function as before, with the *additional constraint* that $R(\boldsymbol{\theta}) \leq t$. Now, $t$ is related to $\lambda$ but the exact relationship between the two parameters depends on your data. Regardless, let's take a look at what this means in the two-dimensional case. For ridge,
$$\theta_0^2 + \theta_1^2 = t$$
Lasso is of the form $$\left|\theta_0\right| + \left|\theta_1\right| =t$$
<a id='norm-balls'></a>
### Norm Balls
Let's take at another visualization that may help build some intuition behind how both of these regularization methods work!
<img src='https://upload.wikimedia.org/wikipedia/commons/f/f8/L1_and_L2_balls.svg' width=400/>
Image from https://upload.wikimedia.org/wikipedia/commons/f/f8/L1_and_L2_balls.svg.
<img src='norm_balls.png' width=400/>
Image from https://towardsdatascience.com/regression-analysis-lasso-ridge-and-elastic-net-9e65dc61d6d3.
The rhombus and circle as a visualization of the regularization term, while the blue circles are the topological curves/level sets representing the loss function based on the weights. You want to minimize the sum of these, which means you want to minimize each of those. The point that minimizes the sum is the minimum point at which they intersect.
**Question**: Based on these visualizations, could you explain why LASSO is sparsity-inducing?
Turns out that the $L2-norm$ is always some sort of smooth surface, from a circle in 2D to a sphere in 3D. On the other hand, LASSO always has sharp corners. In higher dimensions, it forms an octahedron. This is exactly the feature that makes it sparsiy-inducing. As you might imagine, just as humans are more likely to bump into sharp corners than smooth surfaces, the loss term is also most likely to intersect the $L2-norm$ at one of the corners.
<a id='regularization-and-bias-variance'></a>
## Regularization and Bias Variance
As we mentioned earlier, **bias** is the average linear squares loss term across multiple models of the same family (e.g. same degree polynomial) trained on separate datasets. **Variance** is the average variance of the weight vectors (coefficients) on your features.
Without the regularization term, we’re just minimizing bias; the regularization term means we won’t get the lowest possible bias, but we’re exchanging that for some lower variance so that our model does better at generalizing to data points outside of our training data.
<a id='lambda'></a>
## Lambda
We said that $\lambda$ is how much we care about the regularization term, but what does that look like? Let's return to the polynomial example from last week, and see what the resulting models look like with different values of $\lambda$ given a degree 8 polynomial.
```
ridgeRegularizationDemo([0, 0.5, 1.0, 5.0, 10.0], 8)
```
From the diagram above, it's difficult to determine which lambda value help fit our model the closest to the true data distribution. So, **how do we know what to use for $\lambda$ (or `alpha` in the `sklearn.linear_model` constructors)?**
That's right, let's use the process of **validation** here! In this case, we'd be finding the value for lambda that **minimizes the loss for ridge regression, and then the one that minimizes the loss for LASSO regression**!
<a id='validation-on-lambda'></a>
## Validation on Lambda
Let's try to find the best $\lambda$ for the degree 20 polynomial on `displacement` from above.
```
lambdas = np.arange(0, 200) # create a list of potential lambda values
# create a list containing the corresponding mean_squared_error for each lambda usinb both ridge and lasso regression
ridge_errors = []
lasso_errors = []
for l in lambdas:
ridge_model = Ridge(l)
ridge_model.fit(x_train, y_train)
ridge_predictions = ridge_model.predict(x_validation)
ridge_errors.append(mean_squared_error(ridge_predictions, y_validation))
lasso_model = Lasso(l)
lasso_model.fit(x_train, y_train)
lasso_predictions = lasso_model.predict(x_validation)
lasso_errors.append(mean_squared_error(lasso_predictions, y_validation))
answer = ridge_errors.index(min(ridge_errors)), lasso_errors.index(min(lasso_errors))
answer
```
As we can see from above, we've been able to determine which lambdas minimizes our ridge regression model and our LASSO regression model through validation by iterating through potential lambda values and finding the ones that minimize our loss for each model!
<a id='conclusion'></a>
# Conclusion
Through the course of the notebook, we introduced two main concepts, **bias** and **variance**, and how the two relate with one another when it comes to finding the best model for our dataset! We also went into different methods we can use to minimize overfitting our model, and in turn lower variance, by taking look at a process called **regularization**. We saw that the two main regression models, **ridge regression** and **LASSO regression**, and saw the difference between the two (ridge -> penalize large weights, LASSO -> make weights sparse). We also took a look at different visualizations between the two to build up some more intuition behind how they work, through **graphs** and **norm balls**. Finally, we went through a familiar process (**validation**) to determine what the best values of lambda were for our models, officially ending our journey of **bias** and **variance**, and how we can minimze both in our models!
# Congratulations! You are now a Bias + Variance master!
<a id='exercises'></a>
## Exercises
1. What happens as $\lambda$ increases?
1. bias increases, variance increases
2. bias increases, variance decreases
3. bias decreases, variance increases
4. bias decreases, variance decreases
**Insert answer here**:
2. **True** or **False**? Bias is how much error your model makes.
**Insert answer here:**
3. What is **sparsity**?
**Insert answer here:**
4. For each of the following, choose **ridge**, **lasso**, **both**, or **neither**:
1. L1-norm
2. L2-norm
3. Induces sparsity
4. Has analytic (mathematical) solution
5. Increases bias
6. Increases variance
**Insert answer here:**
5. Which one is better to use: Ridge Regression or LASSO Regression?
**Insert answer here:**
### Congrats! You've finished our few conceptual questions, now you can help out the rest of your peers and use the rest of the time to work on the intermediate project with your project group!
|
github_jupyter
|
import matplotlib.pyplot as plt
import random
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from plotting import overfittingDemo, plot_multiple_linear_regression, ridgeRegularizationDemo, lassoRegularizationDemo
from scipy.optimize import curve_fit
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
overfittingDemo()
mpg = pd.read_csv("mpg.csv", index_col='name')# load mpg dataset
mpg = mpg.loc[mpg["horsepower"] != '?'].astype(float) # remove columns with missing horsepower values
mpg_train, mpg_test = train_test_split(mpg, test_size = .2, random_state = 0) # split into training set and test set
mpg_train, mpg_validation = train_test_split(mpg_train, test_size = .5, random_state = 0)
mpg_train.head()
from sklearn.linear_model import LinearRegression
x_train = np.vander(mpg_train["displacement"], 13)
y_train = mpg_train[["mpg"]]
x_validation = np.vander(mpg_validation["displacement"], 13)
y_validation = mpg_validation[["mpg"]]
# instantiate your model
linear_model = LinearRegression()
# fit the model
linear_model.fit(x_train, y_train)
# make predictions on validation set
linear_prediction = linear_model.predict(x_validation)
# find error
linear_loss = mean_squared_error(linear_prediction, y_validation)
print("Root Mean Squared Error of linear model: {:.2f}".format(linear_loss))
from sklearn.linear_model import Ridge
ridge_model = Ridge()
ridge_model.fit(x_train, y_train)
ridge_prediction = ridge_model.predict(x_validation)
ridge_loss = mean_squared_error(ridge_prediction, y_validation) # mean squared error of ridge model
print("Root Mean Squared Error of Linear Model: {:.2f}".format(linear_loss))
print("Root Mean Squared Error of Ridge Model: {:.2f}".format(ridge_loss))
from sklearn.linear_model import Lasso
lasso_model = Lasso()
lasso_model.fit(x_train, y_train)
lasso_prediction = lasso_model.predict(x_validation)
lasso_loss = mean_squared_error(lasso_prediction, y_validation) # mean squared error of lasso model
print("Root Mean Squared Error of Linear Model: {:.2f}".format(linear_loss))
print("Root Mean Squared Error of LASSO Model: {:.2f}".format(lasso_loss))
ridgeRegularizationDemo([0, 0.5, 1.0, 5.0, 10.0], 8)
lambdas = np.arange(0, 200) # create a list of potential lambda values
# create a list containing the corresponding mean_squared_error for each lambda usinb both ridge and lasso regression
ridge_errors = []
lasso_errors = []
for l in lambdas:
ridge_model = Ridge(l)
ridge_model.fit(x_train, y_train)
ridge_predictions = ridge_model.predict(x_validation)
ridge_errors.append(mean_squared_error(ridge_predictions, y_validation))
lasso_model = Lasso(l)
lasso_model.fit(x_train, y_train)
lasso_predictions = lasso_model.predict(x_validation)
lasso_errors.append(mean_squared_error(lasso_predictions, y_validation))
answer = ridge_errors.index(min(ridge_errors)), lasso_errors.index(min(lasso_errors))
answer
| 0.842378 | 0.992961 |
# Bonsai log analysis
This notebook shows you how to access your bonsai brain logs from a jupyter notebook, and carry analysis/vizualization in the notebook. Episode level and iteration level logs during training and assessment are stored on [Log Anlytics](https://docs.microsoft.com/en-us/azure/azure-monitor/platform/data-platform-logs). Logs on Azure Monitor can be probed using [KQL language](https://docs.microsoft.com/en-us/sharepoint/dev/general-development/keyword-query-language-kql-syntax-reference). The notebook uses **[Kqlmagic](https://github.com/Microsoft/jupyter-Kqlmagic)** an python library and extension for jupyter notebook [more info + more complete functionality on Kqlmagic](https://docs.microsoft.com/en-us/azure/data-explorer/kqlmagic).
## Description
Logging for bonsai service currently works for un-managed sims only (will soon be enabled for managed sims). Logging is user-enabled per specific sim's session-id. The logs are sent to a Log Analytics workspace and can be queried via KQL language. A sample to query and vizualize data in a jupyter notebook is provided here.
## Prerequisites
1. Install requirements: [enviroment.yml](https://gist.github.com/akzaidi/ed687b492b0f9e77682b0a0a83397659/)
1. Temporary manual step: If your azure subscription has not yet been registered to allow Log Analytics workspace resource provider, it needs to be registered.
1. Determine if registering is required. <SUBCRIPTION_ID> can be found on preview.bons.ai by clicking on id/Workspace info.
```
az provider show --namespace 'Microsoft.OperationalInsights' -o table --subscription <SUBCRIPTION_ID>
```
2. If the registrationState is `Registered`, you can skip this step. If not registered, we will need to register it. This is a one-time step per subscription and user will need owner-level permission. If you don't have the appropriate permission, work with your IT admin to execute that step.
```
az login
az provider register --namespace 'Microsoft.OperationalInsights' --subscription <SUBCRIPTION_ID>
```
## Usage
1. Start an unmanaged sim and brain training as you normally would:
1. register a sim by launching your sim. For example `python main.py` (or through our partner sims AnyLogic or Simulink)
1. start brain training `bonsai brain version start-training --name <BRAIN_NAME>`
1. connect your registered sim to a brain `bonsai simulator connect --simulator-name <SIM_NAME> --brain-name <BRAIN_NAME> --version <VERSION_#> --action Train --concept-name <CONCEPT_NAME>`
3. Find the session-id of un-managed sim using Bonsai CLI: `bonsai simulator unmanaged list`
4. When you're ready to start logging:
`bonsai brain version start-logging -n <BRAIN_NAME> --session-id <SESSION_ID>`
1.Note: A Log Analytics workspace will get created on Azure if it does not already exist
6. Temporary: You can find the Log Analytics workspace id on portal.azure.com. It will be created under your provisioned resource group `bonsai-rg-<BONSAI WORKSPACE NAME >-<WORKSPACE-ID>`
1. Note: It might take ~5 minutes to show up if it is the first time using logging feature as the log analytics workspace gets created.
1. Logs will start populating the Log Analytics workspace 3-5 minutes after starting logging
1. Note: if this is the first time you're using logging you may need to wait for the first episode to finish so that episode-level (EpisodeLog_CL table) logs gets created and filled with at least 1 row of data.
1. 1. Optional: Navigate to https://ms.portal.azure.com/#blade/Microsoft_Azure_Monitoring_Logs/LogsBlade to query logs in the webUI. Sample KQL query, take 10 samples of IterationLog_CL table for the corresponding sim's session-id:
```KQL
IterationLog_CL
| where SessionId_s == <SESSION_ID>
| take 1
```
## Prerequisite: Install KQL Magic (skip if already installed)
You can use the following conda [environment.yml](https://gist.github.com/akzaidi/ed687b492b0f9e77682b0a0a83397659/).
Load Kqlmagic commands
```
import sys
print(sys.path)
%reload_ext Kqlmagic
%config Kqlmagic.display_limit = 5 #limiting the number of rows displayed (full rows will still be stored)
```
## Login to the log analytics workspace
The `LOG_ANALYTICS_WORKSPACE_ID` is the `workspace-id` of the log analytics workspace, not your bonsai workspace
Please see **Usage** above on how to find your `LOG_ANALYTICS_WORKSPACE_ID`
```
#Login to workspace
LOG_ANALYTICS_WORKSPACE_ID = '0c26bd37-834f-43ec-80f5-21e8ba6b86bc' #this is the workdpace-id
ALIAS = '' #optional
%kql loganalytics://code;workspace=LOG_ANALYTICS_WORKSPACE_ID;alias=ALIAS
```
Locate the simulator you have run locally and you'd like to log. You'll need the simulator's session id.
```
!bonsai simulator unmanaged list
```
## Iteration and Episode Level Logs
Let's extract both iteration (IterationLog_CL table) and episode-level (EpisodeLog_CL table) logs and join them together via a KQL query. We then export the query results in a dataframe.
**Note**: if this is the first time you're using logging you may need to wait for the first episode to finish so that episode-level (EpisodeLog_CL table) logs gets created and filled with at least 1 row of data.
```
session_id = "661870409_10.244.39.115" #define sim's session_id to pull logs from
number_of_rows = 1000 # define number of rows to pull
%%kql
let _session_id = session_id;
let _number_of_rows = number_of_rows;
EpisodeLog_CL
| where SessionId_s == _session_id
| join kind=inner (
IterationLog_CL
| where SessionId_s == _session_id
| sort by Timestamp_t desc
| take _number_of_rows
) on EpisodeId_g
| project
Timestamp = Timestamp_t,
EpisodeIndex = EpisodeIndex_d,
IterationIndex = IterationIndex_d,
BrainName = BrainName_s,
BrainVersion = BrainVersion_d,
SimState = parse_json(SimState_s),
SimAction = parse_json(SimAction_s),
Reward = Reward_d,
CumulativeReward = CumulativeReward_d,
Terminal = Terminal_b,
LessonIndex = LessonIndex_d,
SimConfig = parse_json(SimConfig_s),
GoalMetrics = parse_json(GoalMetrics_s),
EpisodeType = EpisodeType_s
| order by EpisodeIndex, IterationIndex
# convert query results in a dataframe
iter_df = _kql_raw_result_.to_dataframe()
iter_df.head(5)
```
## Converting Nested Array Into New Columns
Notice that the array-data as stored in `SimState`, `SimAction` and `SimConfig` are dictionaries. You can cast them into new columns using the operations below:
```
import pandas as pd
def format_kql_logs(df: pd.DataFrame) -> pd.DataFrame:
''' Function to format a dataframe obtained from KQL query.
Output format: keeps only selected columns, and flatten nested columns [SimAction, SimState, SimConfig]
Parameters
----------
df : DataFrame
dataframe obtained from running KQL query then exporting `_kql_raw_result_.to_dataframe()`
'''
selected_columns = ["Timestamp","EpisodeIndex", "IterationIndex", "Reward", "Terminal", "SimState", "SimAction", "SimConfig"]
nested_columns = [ "SimState", "SimAction", "SimConfig"]
df_selected_columns = df[selected_columns]
series_lst = []
ordered_columns = ["Timestamp","EpisodeIndex", "IterationIndex", "Reward", "Terminal", "IterationSpeed_s"]
for i in nested_columns:
new_series = df_selected_columns[i].apply(pd.Series)
column_names = new_series.columns.values.tolist()
series_lst.append(new_series)
if len(column_names) > 0:
ordered_columns.extend(column_names)
del(df_selected_columns[i])
series_lst.append(df_selected_columns)
formated_df = pd.concat(series_lst, axis=1)
formated_df = formated_df.sort_values(by='Timestamp',ascending=True) # reorder df based on Timestamp
formated_df.index = range(len(formated_df)) # re-index
formated_df['Timestamp']=pd.to_datetime(formated_df['Timestamp']) # convert Timestamp to datetime
formated_df['IterationSpeed_s']=formated_df['Timestamp'].diff().dt.total_seconds() # convert Timestamp to datetime
formated_df = formated_df[ordered_columns]
return formated_df
df =format_kql_logs(iter_df)
df.head(5)
```
## Example Vizualizations
```
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import plotly
import plotly.tools as tls
import cufflinks as cf
import pylab
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
plotly.offline.init_notebook_mode(connected=True)
cf.go_offline()
sns.set()#setting seaborn theme for matplotlib
sns.set_context("talk")# to plot figures in the notebook
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
pylab.rcParams['figure.figsize'] = (17, 8)
pylab.rcParams['axes.titlesize'] = 20
pylab.rcParams['axes.labelsize'] = 20
pylab.rcParams['xtick.labelsize'] = 20
pylab.rcParams['ytick.labelsize'] = 20
#pylab.rcParams['legend.fontsize'] = 20
```
## Interactive plot of all columns vs index
```
df.iplot(
subplots=True,
shared_xaxes=True,
#title = 'Title'
)
```
## Distributions of values
```
plot_df = df.pivot_table(
index=['IterationIndex'],
values= ['angle_position','angle_velocity', 'x_position','x_velocity']
)
plot_df.plot(kind='hist',alpha=0.5)
```
## Iteration speed
```
plotdf = df
plotdf.index= df['Timestamp']
plotdf.iplot(
subplots=True,
shared_xaxes=True,
title = 'vs Timestamp'
)
plotdf['IterationSpeed_s'].iplot(
title = 'IterationSpeed distribution',
kind='hist',
xTitle='IterationSpeed_s',
yTitle='Count'
)
```
## State trajectory for multiple episodes
TODO: Consider plotting considering vs SimConfig
```
plot_df = df.pivot(
index=['IterationIndex'],
columns = ['EpisodeIndex'],
values = ['x_position', 'angle_position','x_velocity','angle_velocity'])
plot_df.head(10)
```
|
github_jupyter
|
az provider show --namespace 'Microsoft.OperationalInsights' -o table --subscription <SUBCRIPTION_ID>
```
2. If the registrationState is `Registered`, you can skip this step. If not registered, we will need to register it. This is a one-time step per subscription and user will need owner-level permission. If you don't have the appropriate permission, work with your IT admin to execute that step.
```
az login
az provider register --namespace 'Microsoft.OperationalInsights' --subscription <SUBCRIPTION_ID>
```
## Usage
1. Start an unmanaged sim and brain training as you normally would:
1. register a sim by launching your sim. For example `python main.py` (or through our partner sims AnyLogic or Simulink)
1. start brain training `bonsai brain version start-training --name <BRAIN_NAME>`
1. connect your registered sim to a brain `bonsai simulator connect --simulator-name <SIM_NAME> --brain-name <BRAIN_NAME> --version <VERSION_#> --action Train --concept-name <CONCEPT_NAME>`
3. Find the session-id of un-managed sim using Bonsai CLI: `bonsai simulator unmanaged list`
4. When you're ready to start logging:
`bonsai brain version start-logging -n <BRAIN_NAME> --session-id <SESSION_ID>`
1.Note: A Log Analytics workspace will get created on Azure if it does not already exist
6. Temporary: You can find the Log Analytics workspace id on portal.azure.com. It will be created under your provisioned resource group `bonsai-rg-<BONSAI WORKSPACE NAME >-<WORKSPACE-ID>`
1. Note: It might take ~5 minutes to show up if it is the first time using logging feature as the log analytics workspace gets created.
1. Logs will start populating the Log Analytics workspace 3-5 minutes after starting logging
1. Note: if this is the first time you're using logging you may need to wait for the first episode to finish so that episode-level (EpisodeLog_CL table) logs gets created and filled with at least 1 row of data.
1. 1. Optional: Navigate to https://ms.portal.azure.com/#blade/Microsoft_Azure_Monitoring_Logs/LogsBlade to query logs in the webUI. Sample KQL query, take 10 samples of IterationLog_CL table for the corresponding sim's session-id:
```KQL
IterationLog_CL
| where SessionId_s == <SESSION_ID>
| take 1
```
## Prerequisite: Install KQL Magic (skip if already installed)
You can use the following conda [environment.yml](https://gist.github.com/akzaidi/ed687b492b0f9e77682b0a0a83397659/).
Load Kqlmagic commands
## Login to the log analytics workspace
The `LOG_ANALYTICS_WORKSPACE_ID` is the `workspace-id` of the log analytics workspace, not your bonsai workspace
Please see **Usage** above on how to find your `LOG_ANALYTICS_WORKSPACE_ID`
Locate the simulator you have run locally and you'd like to log. You'll need the simulator's session id.
## Iteration and Episode Level Logs
Let's extract both iteration (IterationLog_CL table) and episode-level (EpisodeLog_CL table) logs and join them together via a KQL query. We then export the query results in a dataframe.
**Note**: if this is the first time you're using logging you may need to wait for the first episode to finish so that episode-level (EpisodeLog_CL table) logs gets created and filled with at least 1 row of data.
## Converting Nested Array Into New Columns
Notice that the array-data as stored in `SimState`, `SimAction` and `SimConfig` are dictionaries. You can cast them into new columns using the operations below:
## Example Vizualizations
## Interactive plot of all columns vs index
## Distributions of values
## Iteration speed
## State trajectory for multiple episodes
TODO: Consider plotting considering vs SimConfig
| 0.788176 | 0.945248 |
# 2A.ml - Réduction d'une forêt aléatoire - correction
Le modèle Lasso permet de sélectionner des variables, une forêt aléatoire produit une prédiction comme étant la moyenne d'arbres de régression. Et si on mélangeait les deux ?
```
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%matplotlib inline
```
## Datasets
Comme il faut toujours des données, on prend ce jeu [Boston](https://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_boston.html).
```
from sklearn.datasets import load_boston
data = load_boston()
X, y = data.data, data.target
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
```
## Une forêt aléatoire
```
from sklearn.ensemble import RandomForestRegressor as model_class
clr = model_class()
clr.fit(X_train, y_train)
```
Le nombre d'arbres est...
```
len(clr.estimators_)
from sklearn.metrics import r2_score
r2_score(y_test, clr.predict(X_test))
```
## Random Forest = moyenne des prédictions
On recommence en faisant la moyenne soi-même.
```
import numpy
dest = numpy.zeros((X_test.shape[0], len(clr.estimators_)))
estimators = numpy.array(clr.estimators_).ravel()
for i, est in enumerate(estimators):
pred = est.predict(X_test)
dest[:, i] = pred
average = numpy.mean(dest, axis=1)
r2_score(y_test, average)
```
A priori, c'est la même chose.
## Pondérer les arbres à l'aide d'une régression linéaire
La forêt aléatoire est une façon de créer de nouvelles features, 100 exactement qu'on utilise pour caler une régression linéaire.
```
from sklearn.linear_model import LinearRegression
def new_features(forest, X):
dest = numpy.zeros((X.shape[0], len(forest.estimators_)))
estimators = numpy.array(forest.estimators_).ravel()
for i, est in enumerate(estimators):
pred = est.predict(X)
dest[:, i] = pred
return dest
X_train_2 = new_features(clr, X_train)
lr = LinearRegression()
lr.fit(X_train_2, y_train)
X_test_2 = new_features(clr, X_test)
r2_score(y_test, lr.predict(X_test_2))
```
Un peu moins bien, un peu mieux, le risque d'overfitting est un peu plus grand avec ces nombreuses features car la base d'apprentissage ne contient que 379 observations (regardez ``X_train.shape`` pour vérifier).
```
lr.coef_
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.bar(numpy.arange(0, len(lr.coef_)), lr.coef_)
ax.set_title("Coefficients pour chaque arbre calculés avec une régression linéaire");
```
Le score est avec une régression linéaire sur les variables initiales est nettement moins élevé.
```
lr_raw = LinearRegression()
lr_raw.fit(X_train, y_train)
r2_score(y_test, lr_raw.predict(X_test))
```
## Sélection d'arbres
L'idée est d'utiliser un algorithme de sélection de variables type [Lasso](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.Lasso.html) pour réduire la forêt aléatoire sans perdre en performance. C'est presque le même code.
```
from sklearn.linear_model import Lasso
lrs = Lasso(max_iter=10000)
lrs.fit(X_train_2, y_train)
lrs.coef_
```
Pas mal de zéros donc pas mal d'arbres non utilisés.
```
r2_score(y_test, lrs.predict(X_test_2))
```
Pas trop de perte... Ca donne envie d'essayer plusieurs valeur de `alpha`.
```
from tqdm import tqdm
alphas = [0.01 * i for i in range(100)] +[1 + 0.1 * i for i in range(100)]
obs = []
for i in tqdm(range(0, len(alphas))):
alpha = alphas[i]
lrs = Lasso(max_iter=20000, alpha=alpha)
lrs.fit(X_train_2, y_train)
obs.append(dict(
alpha=alpha,
null=len(lrs.coef_[lrs.coef_!=0]),
r2=r2_score(y_test, lrs.predict(X_test_2))
))
from pandas import DataFrame
df = DataFrame(obs)
df.tail()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
df[["alpha", "null"]].set_index("alpha").plot(ax=ax[0], logx=True)
ax[0].set_title("Nombre de coefficients non nulls")
df[["alpha", "r2"]].set_index("alpha").plot(ax=ax[1], logx=True)
ax[1].set_title("r2");
```
Dans ce cas, supprimer des arbres augmentent la performance, comme évoqué ci-dessus, cela réduit l'overfitting. Le nombre d'arbre peut être réduit des deux tiers avec ce modèle.
|
github_jupyter
|
from jyquickhelper import add_notebook_menu
add_notebook_menu()
%matplotlib inline
from sklearn.datasets import load_boston
data = load_boston()
X, y = data.data, data.target
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
from sklearn.ensemble import RandomForestRegressor as model_class
clr = model_class()
clr.fit(X_train, y_train)
len(clr.estimators_)
from sklearn.metrics import r2_score
r2_score(y_test, clr.predict(X_test))
import numpy
dest = numpy.zeros((X_test.shape[0], len(clr.estimators_)))
estimators = numpy.array(clr.estimators_).ravel()
for i, est in enumerate(estimators):
pred = est.predict(X_test)
dest[:, i] = pred
average = numpy.mean(dest, axis=1)
r2_score(y_test, average)
from sklearn.linear_model import LinearRegression
def new_features(forest, X):
dest = numpy.zeros((X.shape[0], len(forest.estimators_)))
estimators = numpy.array(forest.estimators_).ravel()
for i, est in enumerate(estimators):
pred = est.predict(X)
dest[:, i] = pred
return dest
X_train_2 = new_features(clr, X_train)
lr = LinearRegression()
lr.fit(X_train_2, y_train)
X_test_2 = new_features(clr, X_test)
r2_score(y_test, lr.predict(X_test_2))
lr.coef_
import matplotlib.pyplot as plt
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.bar(numpy.arange(0, len(lr.coef_)), lr.coef_)
ax.set_title("Coefficients pour chaque arbre calculés avec une régression linéaire");
lr_raw = LinearRegression()
lr_raw.fit(X_train, y_train)
r2_score(y_test, lr_raw.predict(X_test))
from sklearn.linear_model import Lasso
lrs = Lasso(max_iter=10000)
lrs.fit(X_train_2, y_train)
lrs.coef_
r2_score(y_test, lrs.predict(X_test_2))
from tqdm import tqdm
alphas = [0.01 * i for i in range(100)] +[1 + 0.1 * i for i in range(100)]
obs = []
for i in tqdm(range(0, len(alphas))):
alpha = alphas[i]
lrs = Lasso(max_iter=20000, alpha=alpha)
lrs.fit(X_train_2, y_train)
obs.append(dict(
alpha=alpha,
null=len(lrs.coef_[lrs.coef_!=0]),
r2=r2_score(y_test, lrs.predict(X_test_2))
))
from pandas import DataFrame
df = DataFrame(obs)
df.tail()
fig, ax = plt.subplots(1, 2, figsize=(12, 4))
df[["alpha", "null"]].set_index("alpha").plot(ax=ax[0], logx=True)
ax[0].set_title("Nombre de coefficients non nulls")
df[["alpha", "r2"]].set_index("alpha").plot(ax=ax[1], logx=True)
ax[1].set_title("r2");
| 0.545528 | 0.977735 |
# A user-guide for `yelpgoogletool`
This package aim to help its users find their ideal dinner places. It offers assistance such as:
* searching for specific restaurants in their neighborhood;
* reporting use information like Yelp ratings and customer reviews to help users decide their dinner places;
* ranking a list of restaurants and reporting the best ones by various criteria;
* generating a navigation sheet to guide the users to the restaurant they choose.
In this vignette, I will demonstrate how these functionalities can be realized.
## 1. Installation
```
# Install from github
!pip install git+http://github.com/Simon-YG/yelpgoogletool.git#egg=yelpgoogletool
```
## 2. Get your tokens
In order to make full use of this package, the users should get their own tokens for [Yelp Fusion API](https://www.yelp.com/developers/documentation/v3) and [Google Direction API](https://developers.google.com/maps/documentation/directions). It is recommended to save the keys in environment vairables by running the following commands.
```
export POETRY_YELP_KEY=<Your key for Yelp Fusion API>
export POETRY_GOOGLE_KEY=<Your key for Google Direction API>
```
This will save you the energy to manually input the keys upon importing the package.
## 3. Import the package
To import the package, simply run the following codes. If the keys are not found in the environment the users will be asked to input their keys manually.
```
# Import the package
from yelpgoogletool import yelpgoogletool
```
## 4. Functions in `yelpgoogletool`
### 4.1. `Where2Eat()`
This is the **main function of the whole package**, which works interactively to assist the user to decide where to have dinner. The function wraps up almost all the functionalities of the package. Users can simply call the function without any parameter specifications and follow the on-screen instructions to decide their dinner places. The following example illustrate how the function works.
```
# call Where2Eat() function to get an advise on dinner place interactively
yelpgoogletool.Where2Eat()
```
### 4.2. `ExactRestaurantID()`
A supporting function that returns the unique Yelp ID of a restaurant by its name and location interactively. Upon calling the function, it will print a list of five restaurants and ask if the target restaurant is in the list. If yes, it will ask the user to indicate which one in the list is the target one. Otherwise, another five restaurants will be shown. The process will repeat until the target resataurant is found.
* **Parameters:**
- **`restaurant_name`** (*str*) – Required. The name of the restaurant. It does not need to be exact.
- **`location`** (*str*) – Required. A string describe the address of the location around which the search is conducted.
* **Return:** A string that serves as the unique identifier of the restaurant of interest.
* **Return type:** *str*
**Example:**
```
# Find the unique Yelp ID of a steakhouse named "Peter Luger" in Brooklyn, New York.
yelpgoogletool.ExactRestaurantID("Peter Luger","NYC")
```
### 4.3. `FindBestRestaurant()`
A function that sorts a list of restaurants by various criteria and return the top choices.
* **Parameters:**
- **`list_of_restaurants`** (*pandas.core.frame.DataFrame*) – Required. A dataframe of restaurants from which the best ones are looked for. A typical choice is the output from `SearchingRestaurant()` function.conducted.
- **`by`** (*str*) – Optional. A string represent the criterion of sorting. The default choice is “rating and review counts”. The details are as follows:
- ”rating and review counts”: sort by the Yelp rating. In the case that ratings are tied, use the number of reviews as the second criterion;
- “rating and distance”: sort by the Yelp rating. In the case that ratings are tied, use the distance to current location as the second criterion;
- “review count”: sort by the number of reviews;
- “distance”: sort by the distance to the current location.
- **`result_len`** (*int*) – Optional. The number of the top-ranked restaurants to return. The default value is 5.
* **Return:** A sub-dataframe of the original dataframe`list_of_restaurant`, which consists the top restaurants from the searching results.
* **Return type:** *pandas.core.frame.DataFrame*
**Example:**
```
# List five restaurants on Yelp which are closest to Columbia University
list_of_restaurant = yelpgoogletool.SearchRestaurant(location = "Columbia University, NYC",list_len=40)
yelpgoogletool.FindBestRestaurants(list_of_restaurant,"distance")
```
### 4.4. `GetDirection()`
A function that returns the navigation from the current location a specific restaurant.
- **Parameters:**
- **`restaurant_id`** (*str*) – Required. The unique identifier of the restaurant.
- **`verbose`** (*bool*) – Optional. If "True", generate a detailed navigation; if "False", generate a concise one. The default value is "True".
- **`mode`** (*str*) – Optional. The mode of transportation. Should be one of “driving” (default), “walking” and “transit”. The default value is "driving".
- **`start_location`** (*str*) – Optional. A string describe the address of the location as the origin.
- **`start_latitude`** (*str*) – Required if start_location is not specified. The latitude of the origin.
- **`start_longitude`** (*str*) – Required if start_location is not specified. The longitude of the origin.
- **Returns:** A string that stores the detailed instruction to get to the restaurant.
- **Return type:** str
**Example:**
```
# Print out how to get to Peter Luger from Columbia University by public transportation
str_direction = yelpgoogletool.GetDirection(restaurant_id = '4yPqqJDJOQX69gC66YUDkA', # restaurant_id is provided by `ExactRestaurantID()`
verbose = True,
mode = "transit",
start_location = "Columbia University, New York")
print(str_direction)
```
### 4.5. `GetReviews()`
Get three most recent review for a specific restaurant and store them in a Dataframe. It is recommended to pass the result further to `review_report() function` for a more readable output.
* **Parameters:**
- **`restaurant_id`** (*str*) – Required. Required. The unique identifier of the restaurant, of which the reviews are desired.
* **Returns:** A pandas dataframe that stores basic information about reviews on the restaurant.
* **Return type:** pandas.core.frame.DataFrame
**Example: (See 4.8 for a more reader friendly version)**
```
# Get reviews for "Peter Luger"
yelpgoogletool.GetReviews('4yPqqJDJOQX69gC66YUDkA')
```
### 4.6. `ParsingAddress()`
A supporting function that parses the raw location info from Yelp Fusion API to make it more readable.
- **Parameters:**
- **`raw_location_list`** (*pandas.core.frame.DataFrame*) – Required. A pd.Series of dictionaries containing address information in the JSON output from Fusion API.
- **Returns:** A list that stores more readable result. A typical element from the output list is a string of format: "\<street address>, \<City\>, \<State\> \<ZIP code\>”. E.g. “509 Amsterdam Ave, New York, NY 10024”.
- **Return type:** pandas.core.series.Series
### 4.7. `SearchRestaurant()`
A function that searches restaurant on Yelp.
- **Parameters:**
- **`searching_keywords`** (*str*) – Optional. The keywords for Yelp searching. If not specified, the general term “restaurant” is searched.
- **`location`** (*str*) – Optional. A string describe the address of the location around which the search is conducted.
- **`longitude`** (*float*) – Required if location is not specified. The longitude of the current location.
- **`latitude`** (*float*) – Required if location is not specified. The latitude of the current location.
- **`distance_max`** (*int*) – Optional. A suggested search radius in meters.
- **`list_len`** (*int*) – Optional. The number of restaurants to show in the resulting dataframe.
- **`price`** (*str*) – Optional. Pricing levels to filter the search result with: 1 = \\$ 2 = \\$\\$, 3 = \\$\\$\\$, 4 = \\$\\$\\$\\$. The price filter can be a list of comma delimited pricing levels. For example, “1, 2, 3” will filter the results to show the ones that are \\$, \\$\\$, or \\$\\$\\$.
- **Returns:** A pandas dataframe that include essential information about the restaurants in the resarching result.
- **Return type:** pandas.core.frame.Dataframe
**Example:**
```
yelpgoogletool.SearchRestaurant(location = "Columbia University, NYC",
list_len = 10)
```
### 4.8. `review_report()`
- **Parameters:**
- **`df_reviews`** (*pandas.core.frame.DataFrame*) – Required. A pandas dataframe stores basic information about reviews on the restaurant. It is typically the output from `GetReviews()` function.
- **Returns:** None
- **Return type:** NoneType
**Example:**
```
# Get reviews for "Peter Luger"
df_review = yelpgoogletool.GetReviews('4yPqqJDJOQX69gC66YUDkA')
yelpgoogletool.review_report(df_review)
```
|
github_jupyter
|
# Install from github
!pip install git+http://github.com/Simon-YG/yelpgoogletool.git#egg=yelpgoogletool
export POETRY_YELP_KEY=<Your key for Yelp Fusion API>
export POETRY_GOOGLE_KEY=<Your key for Google Direction API>
# Import the package
from yelpgoogletool import yelpgoogletool
# call Where2Eat() function to get an advise on dinner place interactively
yelpgoogletool.Where2Eat()
# Find the unique Yelp ID of a steakhouse named "Peter Luger" in Brooklyn, New York.
yelpgoogletool.ExactRestaurantID("Peter Luger","NYC")
# List five restaurants on Yelp which are closest to Columbia University
list_of_restaurant = yelpgoogletool.SearchRestaurant(location = "Columbia University, NYC",list_len=40)
yelpgoogletool.FindBestRestaurants(list_of_restaurant,"distance")
# Print out how to get to Peter Luger from Columbia University by public transportation
str_direction = yelpgoogletool.GetDirection(restaurant_id = '4yPqqJDJOQX69gC66YUDkA', # restaurant_id is provided by `ExactRestaurantID()`
verbose = True,
mode = "transit",
start_location = "Columbia University, New York")
print(str_direction)
# Get reviews for "Peter Luger"
yelpgoogletool.GetReviews('4yPqqJDJOQX69gC66YUDkA')
yelpgoogletool.SearchRestaurant(location = "Columbia University, NYC",
list_len = 10)
# Get reviews for "Peter Luger"
df_review = yelpgoogletool.GetReviews('4yPqqJDJOQX69gC66YUDkA')
yelpgoogletool.review_report(df_review)
| 0.494629 | 0.923661 |
# Analysis of MAESTRO results
### MAESTRO
* An Open-source Infrastructure for Modeling Dataflows within Deep Learning Accelerators ([https://github.com/maestro-project/maestro](https://github.com/maestro-project/maestro))
* The throughput and energy efficiency of a dataflow changes dramatically depending on both the DNN topology (i.e., layer shapes and sizes), and accelerator hardware resources (buffer size, and network-on-chip (NoC) bandwidth). This demonstrates the importance of dataflow as a first-order consideration for deep learning accelerator ASICs, both at design-time when hardware resources (buffers and interconnects) are being allocated on-chip, and compile-time when different layers need to be optimally mapped for high utilization and energy-efficiency.
### MAESTRO Result Analysis
* This script is written to analyze the detailed output of MAESTRO per layer using various graphing techniques to showcase interesting results and filter out important results from the gigantic output file that the tool dumps emphasising the importance of Hardware Requirements and Dataflow used per layer for a DNN Model.
## 1 - Packages
Let's first import all the packages that you will need during this assignment.
- [numpy](www.numpy.org) is the fundamental package for scientific computing with Python.
- [matplotlib](http://matplotlib.org) is a library to plot graphs in Python.
- [pandas](https://pandas.pydata.org) is a fast, powerful and easy to use open source data analysis and manipulation tool
```
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from graph_util import *
```
## 2 - Reading the output file
Please provide the output file for your Maestro run to be read by the script with the path relative to the script.
pd.read_csv("name of your csv file")
As an example we are reading file in tools/jupyter_notebook/data folder including **data/Resnet50_kcp_ws.csv**
```
resnet50_kcp_ws_pe256_data = pd.read_csv('data/Resnet50_kcp_ws_pe256.csv')
resnet50_rs_pe256_data = pd.read_csv('data/Resnet50_rs_pe256.csv')
mobilenetv2_kcp_ws_pe256_data = pd.read_csv('data/MobileNetV2_kcp_ws_pe256.csv')
resnet50_kcp_ws_pe1024_data = pd.read_csv('data/Resnet50_kcp_ws_pe1024.csv')
```
## 2 - Creating Graphs
Graphs can be generated by assigning values to the variables provided below in the code.
The graphs are created by draw_graph function which operates on the variables provided below.
## 3- Understanding the Variables
* x = "x axis of your graph"
- One can simply assign the column name from the csv to this graph as shown below in the code. Also, an alternate way is to select the column number from dataframe.columns. For example: for this case it will be new_data.columns[1] that corresponds to 'Layer Number'.
* y = "y axis of your graph"
- One can simply assign the column name from the csv to this graph as shown below in the code. Also, an alternate way is to select the column number from dataframe.columns. For example: for this case it will be new_data.columns[5] that corresponds to 'Throughput (MACs/Cycle)'.
- User can compare multiple bar graphs by providing a list to y.
* color = "color of the graph"
* figsize = "Controls the size of the graph"
* Legend =" Do you want a legend or not" -> values (True/False)
* title = "title of your graph"
* xlabel = "xlabel name on the graph"
* ylabel = "ylabel name on the graph"
* start_layer = "start layer that you want to analyse"
- a subset of layers to be plotted which are of interest. This variable allows you to do that.
* end_layer ="last layer that you want to analyse"
- a subset of layers to be plotted which are of interest. This variable allows you to do that. Please provide "all" if you want all the layers to be seen in the graph
```
# Compare two different dataflow on the same DNN and HW
x = 'Layer Number'
y = 'Avg number of utilized PEs'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'PE Utilization on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Avg number of utilized PEs'
color = 'red'
figsize = (20,7)
legend = 'true'
title = 'PE Utilization on resnet50_rs_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_rs_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
#draw_two_graph(resnet50_kcp_ws_data, resnet50_rs_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different dataflow on same DNN and HW
x = 'Layer Number'
y = 'Activity count-based Energy (nJ)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Activity Count-Based Energy on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Activity count-based Energy (nJ)'
color = 'red'
figsize = (20,7)
legend = 'true'
title = 'Activity Count-Based Energy on resnet50_rs_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_rs_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different DNNs with same dataflow and HW
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'green'
figsize = (20,7)
legend = 'true'
title = 'Runtime on mobilenetv2_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(mobilenetv2_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different HWs with same dataflow and DNN
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'brown'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe1024'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe1024_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
```
|
github_jupyter
|
%matplotlib inline
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from graph_util import *
resnet50_kcp_ws_pe256_data = pd.read_csv('data/Resnet50_kcp_ws_pe256.csv')
resnet50_rs_pe256_data = pd.read_csv('data/Resnet50_rs_pe256.csv')
mobilenetv2_kcp_ws_pe256_data = pd.read_csv('data/MobileNetV2_kcp_ws_pe256.csv')
resnet50_kcp_ws_pe1024_data = pd.read_csv('data/Resnet50_kcp_ws_pe1024.csv')
# Compare two different dataflow on the same DNN and HW
x = 'Layer Number'
y = 'Avg number of utilized PEs'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'PE Utilization on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Avg number of utilized PEs'
color = 'red'
figsize = (20,7)
legend = 'true'
title = 'PE Utilization on resnet50_rs_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_rs_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
#draw_two_graph(resnet50_kcp_ws_data, resnet50_rs_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different dataflow on same DNN and HW
x = 'Layer Number'
y = 'Activity count-based Energy (nJ)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Activity Count-Based Energy on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Activity count-based Energy (nJ)'
color = 'red'
figsize = (20,7)
legend = 'true'
title = 'Activity Count-Based Energy on resnet50_rs_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_rs_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different DNNs with same dataflow and HW
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'green'
figsize = (20,7)
legend = 'true'
title = 'Runtime on mobilenetv2_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(mobilenetv2_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
# Compare two different HWs with same dataflow and DNN
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'blue'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe256'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe256_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
x = 'Layer Number'
y = 'Runtime (Cycles)'
color = 'brown'
figsize = (20,7)
legend = 'true'
title = 'Runtime on resnet50_kcp_ws_pe1024'
xlabel = x
ylabel = y
start_layer = 0
end_layer = 'all'
draw_graph(resnet50_kcp_ws_pe1024_data, y, x, color, figsize, legend, title, xlabel, ylabel, start_layer, end_layer)
| 0.363082 | 0.982014 |
<a href="https://colab.research.google.com/github/Sylwiaes/numpy_pandas_cwiczenia/blob/main/02_pandas_cwiczenia/141_150_exercises.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Pandas
### Spis treści:
* [Import biblioteki](#0)
* [Ćwiczenie 141](#1)
* [Ćwiczenie 142](#2)
* [Ćwiczenie 143](#3)
* [Ćwiczenie 144](#4)
* [Ćwiczenie 145](#5)
* [Ćwiczenie 146](#6)
* [Ćwiczenie 147](#7)
* [Ćwiczenie 148](#8)
* [Ćwiczenie 149](#9)
* [Ćwiczenie 150](#10)
### <a name='0'></a>Import biblioteki
```
import numpy as np
import pandas as pd
np.random.seed(42)
np.__version__
```
### <a name='1'></a> Ćwiczenie 141
Podane są dwa obiekty typu _Series_:
```
s1 = pd.Series(np.random.rand(20))
s2 = pd.Series(np.random.randn(20))
```
Połącz te dwa obiekty w jeden obiekt typu _DataFrame_ (jako dwie kolumny) i przypisz do zmiennej _df_. Nadaj nazwy kolumn odpowiednio _col1_ oraz _col2_.
```
# tutaj wpisz rozwiązanie
s1 = pd.Series(np.random.rand(20))
s2 = pd.Series(np.random.randn(20))
df = pd.concat([s1, s2], axis=1)
df.columns = ['col1', 'col2']
df
```
### <a name='2'></a> Ćwiczenie 142
Wytnij z obiektu _df_ wiersze, dla których wartość w kolumnie _col2_ jest pomiędzy 0.0 a 1.0 (włącznie).
```
# tutaj wpisz rozwiązanie
df[df['col2'].between(0.0, 1.0)]
df[(df['col2'] >= 0.0) & (df['col2'] <= 1.0)]
```
### <a name='3'></a> Ćwiczenie 143
Przypisz nową kolumnę o nazwie _col3_, która przyjmie wartość 1 w momencie gdy w kolumnie _col2_ jest wartość nieujemna i przeciwnie -1.
```
# tutaj wpisz rozwiązanie
df['col3'] = df['col2'].map(lambda x: 1 if x >= 0 else -1)
df
```
### <a name='4'></a> Ćwiczenie 144
Przypisz nową kolumnę o nazwie _col4_, która przytnie wartości z kolumny _col2_ do przedziału $[-1.0, 1.0]$.
Innymi słowy, dla wartości poniżej -1.0 należy wstawić wartość -1.0, dla wartości powyżej 1.0 wartość 1.0.
```
# tutaj wpisz rozwiązanie
df['col4'] = df['col2'].clip(-1.0, 1.0)
df
```
### <a name='5'></a> Ćwiczenie 145
Znajdź 5 największych wartości dla kolumny _col2_.
```
# tutaj wpisz rozwiązanie
df['col2'].nlargest(5)
```
Znajdź 5 najmniejszych wartości dla kolumny _col2_.
```
# tutaj wpisz rozwiązanie
df['col2'].nsmallest(5)
```
### <a name='6'></a> Ćwiczenie 146
Wyznacz skumulowaną sumę dla każdej kolumny.
__Wskazówka:__ Użyj metody _pd.DataFrame.cumsum()_.
```
# tutaj wpisz rozwiązanie
df.cumsum()
```
### <a name='7'></a> Ćwiczenie 147
Wyznacz medianę dla zmiennej _col2_ (inaczej kwantyl rzędu 0.5).
```
# tutaj wpisz rozwiązanie
df['col2'].median()
```
### <a name='8'></a> Ćwiczenie 148
Wytnij wiersze, dla których zmienna _col2_ przyjmuje wartości większe od 0.0.
```
# tutaj wpisz rozwiązanie
df.query("col2 > 0") #I sposób
df[df['col2'] > 0] #II sposób
```
### <a name='9'></a> Ćwiczenie 149
Wytnij 5 pierwszych wierszy obiektu _df_ i przekształć je do słownika.
```
# tutaj wpisz rozwiązanie
df.head().to_dict()
```
### <a name='10'></a> Ćwiczenie 150
Wytnij 5 pierwszych wierszy obiektu _df_, przekształć je do formatu Markdown i przypisz do zmiennej _df_markdown_.
```
# tutaj wpisz rozwiązanie
df_markdown = df.head().to_markdown()
df_markdown
```
Wydrukuj do konsoli zawartość zmiennej _df_markdown_.
```
# tutaj wpisz rozwiązanie
print(df_markdown)
```
Skopiuj wynik uruchomienia powyższej komórki i wklej poniżej:
| | col1 | col2 | col3 | col4 |
|---:|---------:|-----------:|-------:|-----------:|
| 0 | 0.760785 | 0.915402 | 1 | 0.915402 |
| 1 | 0.561277 | 0.328751 | 1 | 0.328751 |
| 2 | 0.770967 | -0.52976 | -1 | -0.52976 |
| 3 | 0.493796 | 0.513267 | 1 | 0.513267 |
| 4 | 0.522733 | 0.0970775 | 1 | 0.0970775 |
|
github_jupyter
|
import numpy as np
import pandas as pd
np.random.seed(42)
np.__version__
s1 = pd.Series(np.random.rand(20))
s2 = pd.Series(np.random.randn(20))
# tutaj wpisz rozwiązanie
s1 = pd.Series(np.random.rand(20))
s2 = pd.Series(np.random.randn(20))
df = pd.concat([s1, s2], axis=1)
df.columns = ['col1', 'col2']
df
# tutaj wpisz rozwiązanie
df[df['col2'].between(0.0, 1.0)]
df[(df['col2'] >= 0.0) & (df['col2'] <= 1.0)]
# tutaj wpisz rozwiązanie
df['col3'] = df['col2'].map(lambda x: 1 if x >= 0 else -1)
df
# tutaj wpisz rozwiązanie
df['col4'] = df['col2'].clip(-1.0, 1.0)
df
# tutaj wpisz rozwiązanie
df['col2'].nlargest(5)
# tutaj wpisz rozwiązanie
df['col2'].nsmallest(5)
# tutaj wpisz rozwiązanie
df.cumsum()
# tutaj wpisz rozwiązanie
df['col2'].median()
# tutaj wpisz rozwiązanie
df.query("col2 > 0") #I sposób
df[df['col2'] > 0] #II sposób
# tutaj wpisz rozwiązanie
df.head().to_dict()
# tutaj wpisz rozwiązanie
df_markdown = df.head().to_markdown()
df_markdown
# tutaj wpisz rozwiązanie
print(df_markdown)
| 0.128922 | 0.920718 |
Import Libraries
```
import csv
import numpy as np
import tensorflow as tf
```
```
attack_set_1_raw = np.genfromtxt('set2_attack_udp_zodiac.csv',
delimiter=',',
skip_header=2)
attack_set_2_raw = np.genfromtxt('set2_attack2_udp_zodiac.csv',
delimiter=',',
skip_header=2)
no_attack_set_1_raw = np.genfromtxt('set2_no_attack_udp_none_zodiac.csv',
delimiter=',',
skip_header=2)
no_attack_set_2_raw = np.genfromtxt('set2_no_attack2_udp_zodiac.csv',
delimiter=',',
skip_header=2)
```
```
attack_set_1 = np.delete(attack_set_1_raw, 2, axis=1)
attack_set_2 = np.delete(attack_set_2_raw, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1_raw, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2_raw, 2, axis=1)
'''
attack_set_1 = np.delete(attack_set_1, 2, axis=1)
attack_set_2 = np.delete(attack_set_2, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2, 2, axis=1)
attack_set_1 = np.delete(attack_set_1, 2, axis=1)
attack_set_2 = np.delete(attack_set_2, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2, 2, axis=1)
'''
```
```
zeroes_set = np.zeros((len(attack_set_1),1))
ones_set = np.ones((len(no_attack_set_1),1))
```
```
training_set = np.concatenate((attack_set_1,no_attack_set_1,no_attack_set_2), axis=0)
training_targets = np.concatenate((ones_set, zeroes_set, zeroes_set), axis=0)
training_set
```
```
training_set_standardized = tf.keras.utils.normalize(
training_set,
axis = -1,
order = 2
)
```
```
input_size = 4
output_size = 1
model = tf.keras.Sequential([
tf.keras.layers.Dense(100, activation='relu', input_shape=(input_size,)),
tf.keras.layers.Dense(1, activation='sigmoid'),
])
model.compile(optimizer=tf.keras.optimizers.Adam(lr=0.00001),
loss='binary_crossentropy',
metrics=['accuracy'])
model.fit(training_set, training_targets, epochs=300, verbose=1)
```
```
model.layers[0].get_weights()[1]
```
```
attack_set_2_standardized = tf.keras.utils.normalize(
attack_set_2,
axis = -1,
order = 2
)
model.predict(attack_set_1)
model.predict(attack_set_2)
no_attack_set_2_standardized = tf.keras.utils.normalize(
no_attack_set_2,
axis = -1,
order = 2
)
model.predict(no_attack_set_1)
model.predict(no_attack_set_2)
```
```
model.save('my_model.h5')
model = tf.keras.models.load_model('my_model.h5')
model.predict([[1,1,1,1]])
```
|
github_jupyter
|
import csv
import numpy as np
import tensorflow as tf
attack_set_1_raw = np.genfromtxt('set2_attack_udp_zodiac.csv',
delimiter=',',
skip_header=2)
attack_set_2_raw = np.genfromtxt('set2_attack2_udp_zodiac.csv',
delimiter=',',
skip_header=2)
no_attack_set_1_raw = np.genfromtxt('set2_no_attack_udp_none_zodiac.csv',
delimiter=',',
skip_header=2)
no_attack_set_2_raw = np.genfromtxt('set2_no_attack2_udp_zodiac.csv',
delimiter=',',
skip_header=2)
attack_set_1 = np.delete(attack_set_1_raw, 2, axis=1)
attack_set_2 = np.delete(attack_set_2_raw, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1_raw, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2_raw, 2, axis=1)
'''
attack_set_1 = np.delete(attack_set_1, 2, axis=1)
attack_set_2 = np.delete(attack_set_2, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2, 2, axis=1)
attack_set_1 = np.delete(attack_set_1, 2, axis=1)
attack_set_2 = np.delete(attack_set_2, 2, axis=1)
no_attack_set_1 = np.delete(no_attack_set_1, 2, axis=1)
no_attack_set_2 = np.delete(no_attack_set_2, 2, axis=1)
'''
zeroes_set = np.zeros((len(attack_set_1),1))
ones_set = np.ones((len(no_attack_set_1),1))
training_set = np.concatenate((attack_set_1,no_attack_set_1,no_attack_set_2), axis=0)
training_targets = np.concatenate((ones_set, zeroes_set, zeroes_set), axis=0)
training_set
training_set_standardized = tf.keras.utils.normalize(
training_set,
axis = -1,
order = 2
)
input_size = 4
output_size = 1
model = tf.keras.Sequential([
tf.keras.layers.Dense(100, activation='relu', input_shape=(input_size,)),
tf.keras.layers.Dense(1, activation='sigmoid'),
])
model.compile(optimizer=tf.keras.optimizers.Adam(lr=0.00001),
loss='binary_crossentropy',
metrics=['accuracy'])
model.fit(training_set, training_targets, epochs=300, verbose=1)
model.layers[0].get_weights()[1]
attack_set_2_standardized = tf.keras.utils.normalize(
attack_set_2,
axis = -1,
order = 2
)
model.predict(attack_set_1)
model.predict(attack_set_2)
no_attack_set_2_standardized = tf.keras.utils.normalize(
no_attack_set_2,
axis = -1,
order = 2
)
model.predict(no_attack_set_1)
model.predict(no_attack_set_2)
model.save('my_model.h5')
model = tf.keras.models.load_model('my_model.h5')
model.predict([[1,1,1,1]])
| 0.562898 | 0.652961 |
[@LorenaABarba](https://twitter.com/LorenaABarba)
12 steps to Navier-Stokes
======
***
You should have completed Steps [1](./01_Step_1.ipynb) and [2](./02_Step_2.ipynb) before continuing. This IPython notebook continues the presentation of the **12 steps to Navier-Stokes**, the practical module taught in the interactive CFD class of [Prof. Lorena Barba](http://lorenabarba.com).
Step 3: Diffusion Equation in 1-D
-----
***
The one-dimensional diffusion equation is:
$$\frac{\partial u}{\partial t}= \nu \frac{\partial^2 u}{\partial x^2}$$
The first thing you should notice is that —unlike the previous two simple equations we have studied— this equation has a second-order derivative. We first need to learn what to do with it!
### Discretizing $\frac{\partial ^2 u}{\partial x^2}$
The second-order derivative can be represented geometrically as the line tangent to the curve given by the first derivative. We will discretize the second-order derivative with a Central Difference scheme: a combination of Forward Difference and Backward Difference of the first derivative. Consider the Taylor expansion of $u_{i+1}$ and $u_{i-1}$ around $u_i$:
$u_{i+1} = u_i + \Delta x \frac{\partial u}{\partial x}\bigg|_i + \frac{\Delta x^2}{2} \frac{\partial ^2 u}{\partial x^2}\bigg|_i + \frac{\Delta x^3}{3!} \frac{\partial ^3 u}{\partial x^3}\bigg|_i + O(\Delta x^4)$
$u_{i-1} = u_i - \Delta x \frac{\partial u}{\partial x}\bigg|_i + \frac{\Delta x^2}{2} \frac{\partial ^2 u}{\partial x^2}\bigg|_i - \frac{\Delta x^3}{3!} \frac{\partial ^3 u}{\partial x^3}\bigg|_i + O(\Delta x^4)$
If we add these two expansions, you can see that the odd-numbered derivative terms will cancel each other out. If we neglect any terms of $O(\Delta x^4)$ or higher (and really, those are very small), then we can rearrange the sum of these two expansions to solve for our second-derivative.
$u_{i+1} + u_{i-1} = 2u_i+\Delta x^2 \frac{\partial ^2 u}{\partial x^2}\bigg|_i + O(\Delta x^4)$
Then rearrange to solve for $\frac{\partial ^2 u}{\partial x^2}\bigg|_i$ and the result is:
$$\frac{\partial ^2 u}{\partial x^2}=\frac{u_{i+1}-2u_{i}+u_{i-1}}{\Delta x^2} + O(\Delta x^2)$$
### Back to Step 3
We can now write the discretized version of the diffusion equation in 1D:
$$\frac{u_{i}^{n+1}-u_{i}^{n}}{\Delta t}=\nu\frac{u_{i+1}^{n}-2u_{i}^{n}+u_{i-1}^{n}}{\Delta x^2}$$
As before, we notice that once we have an initial condition, the only unknown is $u_{i}^{n+1}$, so we re-arrange the equation solving for our unknown:
$$u_{i}^{n+1}=u_{i}^{n}+\frac{\nu\Delta t}{\Delta x^2}(u_{i+1}^{n}-2u_{i}^{n}+u_{i-1}^{n})$$
The above discrete equation allows us to write a program to advance a solution in time. But we need an initial condition. Let's continue using our favorite: the hat function. So, at $t=0$, $u=2$ in the interval $0.5\le x\le 1$ and $u=1$ everywhere else. We are ready to number-crunch!
```
import numpy #loading our favorite library
from matplotlib import pyplot #and the useful plotting library
%matplotlib inline
nx = 41
dx = 2 / (nx - 1)
nt = 20 #the number of timesteps we want to calculate
nu = 0.3 #the value of viscosity
sigma = .2 #sigma is a parameter, we'll learn more about it later
dt = sigma * dx**2 / nu #dt is defined using sigma ... more later!
u = numpy.ones(nx) #a numpy array with nx elements all equal to 1.
u[int(.5 / dx):int(1 / dx + 1)] = 2 #setting u = 2 between 0.5 and 1 as per our I.C.s
un = numpy.ones(nx) #our placeholder array, un, to advance the solution in time
for n in range(nt): #iterate through time
un = u.copy() ##copy the existing values of u into un
for i in range(1, nx - 1):
u[i] = un[i] + nu * dt / dx**2 * (un[i+1] - 2 * un[i] + un[i-1])
pyplot.plot(numpy.linspace(0, 2, nx), u);
```
## Learn More
For a careful walk-through of the discretization of the diffusion equation with finite differences (and all steps from 1 to 4), watch **Video Lesson 4** by Prof. Barba on YouTube.
```
from IPython.display import YouTubeVideo
YouTubeVideo('y2WaK7_iMRI')
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
```
> (The cell above executes the style for this notebook.)
|
github_jupyter
|
import numpy #loading our favorite library
from matplotlib import pyplot #and the useful plotting library
%matplotlib inline
nx = 41
dx = 2 / (nx - 1)
nt = 20 #the number of timesteps we want to calculate
nu = 0.3 #the value of viscosity
sigma = .2 #sigma is a parameter, we'll learn more about it later
dt = sigma * dx**2 / nu #dt is defined using sigma ... more later!
u = numpy.ones(nx) #a numpy array with nx elements all equal to 1.
u[int(.5 / dx):int(1 / dx + 1)] = 2 #setting u = 2 between 0.5 and 1 as per our I.C.s
un = numpy.ones(nx) #our placeholder array, un, to advance the solution in time
for n in range(nt): #iterate through time
un = u.copy() ##copy the existing values of u into un
for i in range(1, nx - 1):
u[i] = un[i] + nu * dt / dx**2 * (un[i+1] - 2 * un[i] + un[i-1])
pyplot.plot(numpy.linspace(0, 2, nx), u);
from IPython.display import YouTubeVideo
YouTubeVideo('y2WaK7_iMRI')
from IPython.core.display import HTML
def css_styling():
styles = open("../styles/custom.css", "r").read()
return HTML(styles)
css_styling()
| 0.353986 | 0.993203 |
# Overview
This week we are going to learn a bit about __Data Visualization__, which is an important aspect in Computational Social Science. Why is it so important to make nice plots if we can use stats and modelling? I hope I will convince that it is _very_ important to make meaningful visualizations. Then, we will try to produce some beautiful figures using the data we downloaded last week.
Here is the plan:
* __Part 1__: Some talking from me on __why do we even care about visualizing data__.
* __Part 2__: Here is where you convince yourself that data visualization is useful by doing a __little visualization exercise__.
* __Part 3__: We will look at the relation between the attention to GME on Reddit and the evolution of the GME market indicators.
* __Part 4__: We will visualize the activity of Redditors posting about GME.
## Part 1: Intro to visualization
Start by watching this short introduction video to Data Visualization.
> * _Video Lecture_: Intro to Data Visualization
```
from IPython.display import YouTubeVideo
YouTubeVideo("oLSdlg3PUO0",width=800, height=450)
```
There are many types of data visualizations, serving different purposes. Today we will look at some of those types for visualizing single variable data: _line graphs_ and _histograms_. We will also use _scatter plots_ two visualize two variables against each other.
Before starting, read the following sections of the data visualization book.
> * _Reading_ [Sections 2,3.2 and 5 of the data visualization book](https://clauswilke.com/dataviz/aesthetic-mapping.html)
## Part 2: A little visualization exercise
Ok, but is data visualization really so necessary? Let's see if I can convince you of that with this little visualization exercise.
> *Exercise 1: Visualization vs stats*
>
> Start by downloading these four datasets: [Data 1](https://raw.githubusercontent.com/suneman/socialdataanalysis2020/master/files/data1.tsv), [Data 2](https://raw.githubusercontent.com/suneman/socialdataanalysis2020/master/files/data2.tsv), [Data 3](https://raw.githubusercontent.com/suneman/socialdataanalysis2020/master/files/data3.tsv), and [Data 4](https://raw.githubusercontent.com/suneman/socialdataanalysis2020/master/files/data4.tsv). The format is `.tsv`, which stands for _tab separated values_.
> Each file has two columns (separated using the tab character). The first column is $x$-values, and the second column is $y$-values.
>
> * Using the `numpy` function `mean`, calculate the mean of both $x$-values and $y$-values for each dataset.
> * Use python string formatting to print precisely two decimal places of these results to the output cell. Check out [this _stackoverflow_ page](http://stackoverflow.com/questions/8885663/how-to-format-a-floating-number-to-fixed-width-in-python) for help with the string formatting.
> * Now calculate the variance for all of the various sets of $x$- and $y$-values (to three decimal places).
> * Use [`scipy.stats.pearsonr`](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.pearsonr.html) to calculate the [Pearson correlation](https://en.wikipedia.org/wiki/Pearson_product-moment_correlation_coefficient) between $x$- and $y$-values for all four data sets (also to three decimal places).
> * The next step is use _linear regression_ to fit a straight line $f(x) = a x + b$ through each dataset and report $a$ and $b$ (to two decimal places). An easy way to fit a straight line in Python is using `scipy`'s `linregress`. It works like this
> ```
> from scipy import stats
> slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
>```
> * Finally, it's time to plot the four datasets using `matplotlib.pyplot`. Use a two-by-two [`subplot`](http://matplotlib.org/examples/pylab_examples/subplot_demo.html) to put all of the plots nicely in a grid and use the same $x$ and $y$ range for all four plots. And include the linear fit in all four plots. (To get a sense of what I think the plot should look like, you can take a look at my version [here](https://raw.githubusercontent.com/suneman/socialdataanalysis2017/master/files/anscombe.png).)
> * Explain - in your own words - what you think my point with this exercise is.
Get more insight in the ideas behind this exercise by reading [here](https://en.wikipedia.org/wiki/Anscombe%27s_quartet).
And the video below generalizes in the coolest way imaginable. It's a treat, but don't watch it until **after** you've done the exercises.
```
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
data1 = pd.read_csv("data1.tsv",header=None,sep = "\t")
data2 = pd.read_csv("data2.tsv",header=None,sep = "\t")
data3 = pd.read_csv("data3.tsv",header=None,sep = "\t")
data4 = pd.read_csv("data4.tsv",header=None,sep = "\t")
data = [data1,data2,data3,data4]
for i in range(4):
dat = data[i]
mean1, mean2 = np.mean(dat[0]),np.mean(dat[1])
var1, var2 = np.var(dat[0]),np.var(dat[1])
print("X: {:}, Y: {:}".format(mean1, mean2))
print(f'Corr is {stats.pearsonr(dat[0], dat[1])[0]} and p-val {stats.pearsonr(dat[0], dat[1])[1]}')
slope, intercept, r_value, p_value, std_err = stats.linregress(dat[0],dat[1])
x = np.linspace(min(dat[0]),max(dat[0]),100)
y = slope*x+intercept
plt.plot(dat[0],dat[1],'o')
plt.plot(x, y, '-r')
plt.xlabel('x', color='#1C2833')
plt.ylabel('y', color='#1C2833')
plt.legend(loc='upper left')
plt.grid()
plt.show()
print(mean1,mean2)
data.describe()
from IPython.display import YouTubeVideo
YouTubeVideo("DbJyPELmhJc",width=800, height=450)
```
## Prelude to Part 3: Some tips to make nicer figures.
Before even starting visualizing some cool data, I just want to give a few tips for making nice plots in matplotlib. Unless you are already a pro-visualizer, those should be pretty useful to make your plots look much nicer.
Paying attention to details can make an incredible difference when we present our work to others.
```
from IPython.display import YouTubeVideo
YouTubeVideo("sdszHGaP_ag",width=800, height=450)
```
## Part 3: Time series of Reddit activity and market indicators.
```
import matplotlib as mpl
import matplotlib.dates as mdates
def setup_mpl():
mpl.rcParams['font.family'] = 'Helvetica Neue'
mpl.rcParams['lines.linewidth'] = 1
setup_mpl()
GME_data = pd.read_csv('GME.csv',parse_dates = ['Date']).set_index('Date')
rolled_series = GME_data['Volume'].rolling('7D').mean()
myFmt = mdates.DateFormatter('%b %Y')
fig , ax = plt.subplots(figsize=(10,2.5),dpi=400)
ax.plot(GME_data.index, GME_data.Volume, ls = '--', alpha=0.5, label = 'daily value')
ax.plot(rolled_series.index, rolled_series.values, color = 'k',label = '7 days rolling average')
ax.set_ylabel('Volume (USD)')
ax.set_yscale('log')
ax.legend()
ax.xaxis.set_major_formatter(myFmt)
```
It's really time to put into practice what we learnt by plotting some data! We will start by looking at the time series describing the number of comments about GME in wallstreetbets over time. We will try to see how that relates to the volume and price of GME over time, through some exploratory data visualization.
We will use two datasets today:
* the _GME market data_, that you can download from [here](https://finance.yahoo.com/quote/GME/history/).
* the dataset you downloaded in Week1, Exercise 3. We will refer to this as the _comments dataset_.
> _Exercise 2 : Plotting prices and comments using line-graphs._
> 1. Plot the daily volume of the GME stock over time using the _GME market data_. On top of the daily data, plot the rolling average, using a 7 days window (you can use the function [``pd.rolling``](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.rolling.html)). Use a [log-scale on the y-axis](https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.yscale.html) to appreciate changes across orders of magnitude.
> 2. Now make a second plot where you plot the total number of comments on Reddit per day. Follow the same steps you followed in step 1.
> 3. Now take a minute to __look at these two figures__. Then write in a couple of lines: What are the three most important observations you can draw by looking at the figures?
```
import matplotlib as mpl
import matplotlib.dates as mdates
def setup_mpl():
mpl.rcParams['font.family'] = 'Helvetica Neue'
mpl.rcParams['lines.linewidth'] = 1
setup_mpl()
GME_data = pd.read_csv('gmecomments.csv', index_col= 0, parse_dates = ["created"]).set_index('created')
#rolled_series = GME_data['Volume'].rolling('7D').mean()
unq_dat = GME_data.resample("1D")["id"].nunique()
coms_counts = GME_data.resample("1D")["id"].count()
rolled_series = coms_counts.rolling('7D').mean()
myFmt = mdates.DateFormatter('%b %Y')
fig , ax = plt.subplots(figsize=(10,2.5),dpi=400)
ax.plot(unq_dat.index, coms_counts, ls = '--', alpha=0.5, label = 'Daily Number')
ax.plot(rolled_series.index, rolled_series.values, color = 'k',label = '7 days rolling average')
ax.set_ylabel('# of Comments')
ax.set_yscale('log')
#axes.set_ylim([-10000,max(coms_counts)+20])
ax.legend()
ax.xaxis.set_major_formatter(myFmt)
plt.show()
```
> _Exercise 3 : Returns vs comments using scatter-plots_.
> In this exercise, we will look at the association between GME market indicators and the attention on Reddit. First, we will create the time-series of daily [returns](https://en.wikipedia.org/wiki/Price_return). Returns measure the change in price given two given points in time (in our case two consecutive days). They really constitute the quantity of interest when it comes to stock time-series, because they tell us how much _money_ one would make if he/she bought the stock on a given day and sold it at a later time. For consistency, we will also compute returns (corresponding to daily changes) for the number of Reddit comments over time.
> 1. Compute the daily log-returns as ``np.log(Close_price(t)/Close_price(t-1))``, where ``Close_price(t)`` is the Close Price of GME on day t. You can use the function [pd.Series.shift](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.shift.html). Working with log-returns instead of regular returns is a standard thing to do in economics, if you are interested in why, check out [this blog post](https://quantivity.wordpress.com/2011/02/21/why-log-returns/).
> 2. Compute the daily log-change in number of new submissions as ``np.log(submissions(t)/submissions(t-1))`` where ``submissions(t)`` is the number of submissions on day t.
> 3. Compute the [Pearson correlation](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.pearsonr.html) between the series computed in step 1 and step 2 (note that you need to first remove days without any comments from the time-series). Is the correlation statistically significant?
> 4. Make a [scatter plot](https://matplotlib.org/api/_as_gen/matplotlib.pyplot.scatter.html) of the daily log-return on investment for the GME stock against the daily log-change in number of submission. Color the markers for 2020 and 2021 in different colors, and make the marker size proportional to the price.
> 5. Now take a minute to __look at the figure you just prepared__. Then write in a couple of lines: What are the three most salient observations you can draw by looking at it?
## Part 4 : The activity of Redditors
It is time to start looking at redditors activity. The [r/wallstreetbets]() subreddit has definitely become really popular in recent weeks. But probably many users only jumped on board recently, while only a few were discussing about investing on GME [for a long time](https://www.reddit.com/user/DeepFuckingValue/). Now, we wil look at the activity of redditors over time? How different are authors?
> _Video Lecture_: Start by watching the short video lecture below about plotting histograms in matplotlib.
> _Reading_: [Section 7 of the Data Visualization book](https://clauswilke.com/dataviz/histograms-density-plots.html)
```
YouTubeVideo("UpwEsguMtY4",width=800, height=450)
```
> _Exercise 4: Authors overall activity_
> 1. Compute the total number of comments per author using the _comments dataset_. Then, make a histogram of the number of comments per author, using the function [``numpy.histogram``](https://numpy.org/doc/stable/reference/generated/numpy.histogram.html), using logarithmic binning. Here are some important points on histograms (they should be already quite clear if you have watched the video above):
> * __Binning__: By default numpy makes 10 equally spaced bins, but you always have to customize the binning. The number and size of bins you choose for your histograms can completely change the visualization. If you use too few bins, the histogram doesn't portray well the data. If you have too many, you get a broken comb look. Unfortunately is no "best" number of bins, because different bin sizes can reveal different features of the data. Play a bit with the binning to find a suitable number of bins. Define a vector $\nu$ including the desired bins and then feed it as a parameter of numpy.histogram, by specifying _bins=\nu_ as an argument of the function. You always have at least two options:
> * _Linear binning_: Use linear binning, when the data is not heavy tailed, by using ``np.linspace`` to define bins.
> * _Logarithmic binning_: Use logarithmic binning, when the data is [heavy tailed](https://en.wikipedia.org/wiki/Fat-tailed_distribution), by using ``np.logspace`` to define your bins.
> * __Normalization__: To plot [probability densities](https://en.wikipedia.org/wiki/Probability_density_function), you can set the argument _density=True_ of the ``numpy.histogram`` function.
>
> 3. Compute the mean and the median value of the number of comments per author and plot them as vertical lines on top of your histogram. What do you observe? Which value do you think is more meaningful?
> _Exercise 5: Authors lifespan_
>
> 1. For each author, find the time of publication of their first comment, _minTime_, and the time of publication of their last comment, _maxTime_, in [unix timestamp](https://www.unixtimestamp.com/).
> 2. Compute the "lifespan" of authors as the difference between _maxTime_ and _minTime_. Note that timestamps are measured in seconds, but it is appropriate here to compute the lifespan in days. Make a histogram showing the distribution of lifespans, choosing appropriate binning. What do you observe?
> 3. Now, we will look at how many authors joined and abandoned the discussion on GME over time. First, use the numpy function [numpy.histogram2d](https://numpy.org/doc/stable/reference/generated/numpy.histogram2d.html) to create a 2-dimensional histogram for the two variables _minTime_ and _maxTime_. A 2D histogram, is nothing but a histogram where bins have two dimensions, as we look simultaneously at two variables. You need to specify two arrays of bins, one for the values along the x-axis (_minTime_) and the other for the values along the y-axis (_maxTime_). Choose bins with length 1 week.
> 4. Now, use the matplotlib function [``plt.imshow``](https://matplotlib.org/api/_as_gen/matplotlib.pyplot.imshow.html) to visualize the 2d histogram. You can follow [this example](https://stackoverflow.com/questions/2369492/generate-a-heatmap-in-matplotlib-using-a-scatter-data-set) on StackOverflow. To show dates instead of unix timestamps in the x and y axes, use [``mdates.date2num``](https://matplotlib.org/api/dates_api.html#matplotlib.dates.date2num). More details in this [StackOverflow example](https://stackoverflow.com/questions/23139595/dates-in-the-xaxis-for-a-matplotlib-plot-with-imshow), see accepted answer.
> 5. Make sure that the colormap allows to well interpret the data, by passing ``norm=mpl.colors.LogNorm()`` as an argument to imshow. This will ensure that your colormap is log-scaled. Then, add a [colorbar](https://matplotlib.org/3.1.0/gallery/color/colorbar_basics.html) on the side of the figure, with the appropriate [colorbar label](https://matplotlib.org/3.1.1/api/colorbar_api.html#matplotlib.colorbar.ColorbarBase.set_label).
> 6. As usual :) Look at the figure, and write down three key observations.
|
github_jupyter
|
from IPython.display import YouTubeVideo
YouTubeVideo("oLSdlg3PUO0",width=800, height=450)
> from scipy import stats
> slope, intercept, r_value, p_value, std_err = stats.linregress(x,y)
>```
> * Finally, it's time to plot the four datasets using `matplotlib.pyplot`. Use a two-by-two [`subplot`](http://matplotlib.org/examples/pylab_examples/subplot_demo.html) to put all of the plots nicely in a grid and use the same $x$ and $y$ range for all four plots. And include the linear fit in all four plots. (To get a sense of what I think the plot should look like, you can take a look at my version [here](https://raw.githubusercontent.com/suneman/socialdataanalysis2017/master/files/anscombe.png).)
> * Explain - in your own words - what you think my point with this exercise is.
Get more insight in the ideas behind this exercise by reading [here](https://en.wikipedia.org/wiki/Anscombe%27s_quartet).
And the video below generalizes in the coolest way imaginable. It's a treat, but don't watch it until **after** you've done the exercises.
## Prelude to Part 3: Some tips to make nicer figures.
Before even starting visualizing some cool data, I just want to give a few tips for making nice plots in matplotlib. Unless you are already a pro-visualizer, those should be pretty useful to make your plots look much nicer.
Paying attention to details can make an incredible difference when we present our work to others.
## Part 3: Time series of Reddit activity and market indicators.
It's really time to put into practice what we learnt by plotting some data! We will start by looking at the time series describing the number of comments about GME in wallstreetbets over time. We will try to see how that relates to the volume and price of GME over time, through some exploratory data visualization.
We will use two datasets today:
* the _GME market data_, that you can download from [here](https://finance.yahoo.com/quote/GME/history/).
* the dataset you downloaded in Week1, Exercise 3. We will refer to this as the _comments dataset_.
> _Exercise 2 : Plotting prices and comments using line-graphs._
> 1. Plot the daily volume of the GME stock over time using the _GME market data_. On top of the daily data, plot the rolling average, using a 7 days window (you can use the function [``pd.rolling``](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.rolling.html)). Use a [log-scale on the y-axis](https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.yscale.html) to appreciate changes across orders of magnitude.
> 2. Now make a second plot where you plot the total number of comments on Reddit per day. Follow the same steps you followed in step 1.
> 3. Now take a minute to __look at these two figures__. Then write in a couple of lines: What are the three most important observations you can draw by looking at the figures?
> _Exercise 3 : Returns vs comments using scatter-plots_.
> In this exercise, we will look at the association between GME market indicators and the attention on Reddit. First, we will create the time-series of daily [returns](https://en.wikipedia.org/wiki/Price_return). Returns measure the change in price given two given points in time (in our case two consecutive days). They really constitute the quantity of interest when it comes to stock time-series, because they tell us how much _money_ one would make if he/she bought the stock on a given day and sold it at a later time. For consistency, we will also compute returns (corresponding to daily changes) for the number of Reddit comments over time.
> 1. Compute the daily log-returns as ``np.log(Close_price(t)/Close_price(t-1))``, where ``Close_price(t)`` is the Close Price of GME on day t. You can use the function [pd.Series.shift](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.Series.shift.html). Working with log-returns instead of regular returns is a standard thing to do in economics, if you are interested in why, check out [this blog post](https://quantivity.wordpress.com/2011/02/21/why-log-returns/).
> 2. Compute the daily log-change in number of new submissions as ``np.log(submissions(t)/submissions(t-1))`` where ``submissions(t)`` is the number of submissions on day t.
> 3. Compute the [Pearson correlation](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.pearsonr.html) between the series computed in step 1 and step 2 (note that you need to first remove days without any comments from the time-series). Is the correlation statistically significant?
> 4. Make a [scatter plot](https://matplotlib.org/api/_as_gen/matplotlib.pyplot.scatter.html) of the daily log-return on investment for the GME stock against the daily log-change in number of submission. Color the markers for 2020 and 2021 in different colors, and make the marker size proportional to the price.
> 5. Now take a minute to __look at the figure you just prepared__. Then write in a couple of lines: What are the three most salient observations you can draw by looking at it?
## Part 4 : The activity of Redditors
It is time to start looking at redditors activity. The [r/wallstreetbets]() subreddit has definitely become really popular in recent weeks. But probably many users only jumped on board recently, while only a few were discussing about investing on GME [for a long time](https://www.reddit.com/user/DeepFuckingValue/). Now, we wil look at the activity of redditors over time? How different are authors?
> _Video Lecture_: Start by watching the short video lecture below about plotting histograms in matplotlib.
> _Reading_: [Section 7 of the Data Visualization book](https://clauswilke.com/dataviz/histograms-density-plots.html)
| 0.911952 | 0.984002 |
```
BRANCH = 'main'
"""
You can run either this notebook locally (if you have all the dependencies and a GPU) or on Google Colab.
Instructions for setting up Colab are as follows:
1. Open a new Python 3 notebook.
2. Import this notebook from GitHub (File -> Upload Notebook -> "GITHUB" tab -> copy/paste GitHub URL)
3. Connect to an instance with a GPU (Runtime -> Change runtime type -> select "GPU" for hardware accelerator)
4. Run this cell to set up dependencies.
"""
# If you're using Google Colab and not running locally, run this cell.
# install NeMo
!python -m pip install git+https://github.com/NVIDIA/NeMo.git@$BRANCH#egg=nemo_toolkit[all]
import json
import os
import wget
from IPython.display import Audio
import numpy as np
import scipy.io.wavfile as wav
! pip install pandas
# optional
! pip install plotly
from plotly import graph_objects as go
```
# Introduction
End-to-end Automatic Speech Recognition (ASR) systems surpassed traditional systems in performance but require large amounts of labeled data for training.
This tutorial will show how to use a pre-trained with Connectionist Temporal Classification (CTC) ASR model, such as [QuartzNet Model](https://arxiv.org/abs/1910.10261) to split long audio files and the corresponding transcripts into shorter fragments that are suitable for an ASR model training.
We're going to use [ctc-segmentation](https://github.com/lumaku/ctc-segmentation) Python package based on the algorithm described in [CTC-Segmentation of Large Corpora for German End-to-end Speech Recognition](https://arxiv.org/pdf/2007.09127.pdf).
```
! pip install ctc_segmentation==1.1.0
! pip install num2words
! apt-get install -y ffmpeg
# If you're running the notebook locally, update the TOOLS_DIR path below
# In Colab, a few required scripts will be downloaded from NeMo github
TOOLS_DIR = '<UPDATE_PATH_TO_NeMo_root>/tools/ctc_segmentation/scripts'
if 'google.colab' in str(get_ipython()):
TOOLS_DIR = 'scripts/'
os.makedirs(TOOLS_DIR, exist_ok=True)
required_files = ['prepare_data.py',
'normalization_helpers.py',
'run_ctc_segmentation.py',
'verify_segments.py',
'cut_audio.py',
'process_manifests.py',
'utils.py']
for file in required_files:
if not os.path.exists(os.path.join(TOOLS_DIR, file)):
file_path = 'https://raw.githubusercontent.com/NVIDIA/NeMo/' + BRANCH + '/tools/ctc_segmentation/' + TOOLS_DIR + file
print(file_path)
wget.download(file_path, TOOLS_DIR)
elif not os.path.exists(TOOLS_DIR):
raise ValueError(f'update path to NeMo root directory')
```
`TOOLS_DIR` should now contain scripts that we are going to need in the next steps, all necessary scripts could be found [here](https://github.com/NVIDIA/NeMo/tree/main/tools/ctc_segmentation/scripts).
```
print(TOOLS_DIR)
! ls -l $TOOLS_DIR
```
# Data Download
First, let's download an audio file from [https://librivox.org/](https://librivox.org/).
```
## create data directory and download an audio file
WORK_DIR = 'WORK_DIR'
DATA_DIR = WORK_DIR + '/DATA'
os.makedirs(DATA_DIR, exist_ok=True)
audio_file = 'childrensshortworks019_06acarriersdog_am_128kb.mp3'
if not os.path.exists(os.path.join(DATA_DIR, audio_file)):
print('Downloading audio file')
wget.download('http://archive.org/download/childrens_short_works_vol_019_1310_librivox/' + audio_file, DATA_DIR)
```
Next, we need to get the corresponding transcript.
Note, the text file and the audio file should have the same base name, for example, an audio file `example.wav` or `example.mp3` should have corresponding text data stored under `example.txt` file.
```
# text source: http://www.gutenberg.org/cache/epub/24263/pg24263.txt
text = """
A carrier on his way to a market town had occasion to stop at some houses
by the road side, in the way of his business, leaving his cart and horse
upon the public road, under the protection of a passenger and a trusty
dog. Upon his return he missed a led horse, belonging to a gentleman in
the neighbourhood, which he had tied to the end of the cart, and likewise
one of the female passengers. On inquiry he was informed that during his
absence the female, who had been anxious to try the mettle of the pony,
had mounted it, and that the animal had set off at full speed. The carrier
expressed much anxiety for the safety of the young woman, casting at the
same time an expressive look at his dog. Oscar observed his master's eye,
and aware of its meaning, instantly set off in pursuit of the pony, which
coming up with soon after, he made a sudden spring, seized the bridle, and
held the animal fast. Several people having observed the circumstance, and
the perilous situation of the girl, came to relieve her. Oscar, however,
notwithstanding their repeated endeavours, would not quit his hold, and
the pony was actually led into the stable with the dog, till such time as
the carrier should arrive. Upon the carrier entering the stable, Oscar
wagged his tail in token of satisfaction, and immediately relinquished the
bridle to his master.
"""
with open(os.path.join(DATA_DIR, audio_file.replace('mp3', 'txt')), 'w') as f:
f.write(text)
```
The `DATA_DIR` should now contain both audio and text files:
```
!ls -l $DATA_DIR
```
Listen to the audio:
```
Audio(os.path.join(DATA_DIR, audio_file))
```
As one probably noticed, the audio file contains a prologue and an epilogue that are missing in the corresponding text. The segmentation algorithm could handle extra audio fragments at the end and the beginning of the audio, but prolonged untranscribed audio segments in the middle of the file could deteriorate segmentation results. That's why to improve the segmentation quality, it is recommended to normalize text, so that transcript contains spoken equivalents of abbreviations and numbers.
# Prepare Text and Audio
We're going to use `prepare_data.py` script to prepare both text and audio data for segmentation.
Text preprocessing:
* the text will be roughly split into sentences and stored under '$OUTPUT_DIR/processed/*.txt' where each sentence is going to start with a new line (we're going to find alignments for these sentences in the next steps)
* to change the lengths of the final sentences/fragments, use `min_length` and `max_length` arguments, that specify min/max number of chars of the text segment for alignment.
* to specify additional punctuation marks to split the text into fragments, use `--additional_split_symbols` argument. If segments produced after splitting the original text based on the end of sentence punctuation marks is longer than `--max_length`, `--additional_split_symbols` are going to be used to shorten the segments. Use `|` as a separator between symbols, for example: `--additional_split_symbols=;|:`
* out-of-vocabulary words will be removed based on pre-trained ASR model vocabulary, (optionally) text will be changed to lowercase
* sentences for alignment with the original punctuation and capitalization will be stored under `$OUTPUT_DIR/processed/*_with_punct.txt`
* numbers will be converted from written to their spoken form with `num2words` package. To use NeMo normalization tool use `--use_nemo_normalization` argument (not supported if running this segmentation tutorial in Colab, see the text normalization tutorial: [`tutorials/text_processing/Text_Normalization.ipynb`](https://colab.research.google.com/github/NVIDIA/NeMo/blob/stable/tutorials/text_processing/Text_Normalization.ipynb) for more details). Such normalization is usually enough for proper segmentation. However, it does not take audio into account. NeMo supports audio-based normalization for English and Russian languages that can be applied to the segmented data as a post-processing step. Audio-based normalization produces multiple normalization options. For example, `901` could be normalized as `nine zero one` or `nine hundred and one`. The audio-based normalization chooses the best match among the possible normalization options and the transcript based on the character error rate. Note, the audio-based normalization of long audio samples is not supported due to many possible normalization options. See [https://github.com/NVIDIA/NeMo/blob/main/nemo_text_processing/text_normalization/normalize_with_audio.py](https://github.com/NVIDIA/NeMo/blob/main/nemo_text_processing/text_normalization/normalize_with_audio.py) for more details.
Audio preprocessing:
* `.mp3` files will be converted to `.wav` files
* audio files will be resampled to use the same sampling rate as was used to pre-train the ASR model we're using for alignment
* stereo tracks will be converted to mono
* since librivox.org audio contains relatively long prologues, we're also cutting a few seconds from the beginning of the audio files (optional step, see `--cut_prefix` argument). In some cases, if an audio contains a very long untranscribed prologue, increasing `--cut_prefix` value might help improve segmentation quality.
The `prepare_data.py` will preprocess all `.txt` files found in the `--in_text=$DATA_DIR` and all `.mp3` files located at `--audio_dir=$DATA_DIR`.
```
MODEL = 'QuartzNet15x5Base-En'
OUTPUT_DIR = WORK_DIR + '/output'
! python $TOOLS_DIR/prepare_data.py \
--in_text=$DATA_DIR \
--output_dir=$OUTPUT_DIR/processed/ \
--language='eng' \
--cut_prefix=3 \
--model=$MODEL \
--audio_dir=$DATA_DIR
```
The following four files should be generated and stored at the `$OUTPUT_DIR/processed` folder:
* childrensshortworks019_06acarriersdog_am_128kb.txt
* childrensshortworks019_06acarriersdog_am_128kb.wav
* childrensshortworks019_06acarriersdog_am_128kb_with_punct.txt
* childrensshortworks019_06acarriersdog_am_128kb_with_punct_normalized.txt
```
! ls -l $OUTPUT_DIR/processed
```
The `.txt` file without punctuation contains preprocessed text phrases that we're going to align within the audio file. Here, we split the text into sentences. Each line should contain a text snippet for alignment.
```
with open(os.path.join(OUTPUT_DIR, 'processed', audio_file.replace('.mp3', '.txt')), 'r') as f:
for line in f:
print (line)
```
# Run CTC-Segmentation
In this step, we're going to use the [`ctc-segmentation`](https://github.com/lumaku/ctc-segmentation) to find the start and end time stamps for the segments we created during the previous step.
As described in the [CTC-Segmentation of Large Corpora for German End-to-end Speech Recognition](https://arxiv.org/pdf/2007.09127.pdf), the algorithm is relying on a CTC-based ASR model to extract utterance segments with exact time-wise alignments. For this tutorial, we're using a pre-trained 'QuartzNet15x5Base-En' model.
```
WINDOW = 8000
! python $TOOLS_DIR/run_ctc_segmentation.py \
--output_dir=$OUTPUT_DIR \
--data=$OUTPUT_DIR/processed \
--model=$MODEL \
--window_len=$WINDOW \
--no_parallel
```
`WINDOW` parameter might need to be adjusted depending on the length of the utterance one wants to align, the default value should work in most cases.
Let's take a look at the generated alignments.
The expected output for our audio sample with 'QuartzNet15x5Base-En' model looks like this:
```
<PATH_TO>/processed/childrensshortworks019_06acarriersdog_am_128kb.wav
16.03 32.39 -4.5911999284929115 | a carrier on ... a trusty dog. | ...
33.31 45.01 -0.22886803973405373 | upon his ... passengers. | ...
46.17 58.57 -0.3523662826061572 | on inquiry ... at full speed. | ...
59.75 69.43 -0.04128918756038118 | the carrier ... dog. | ...
69.93 85.31 -0.3595261826390344 | oscar observed ... animal fast. | ...
85.95 93.43 -0.04447770533708611 | several people ... relieve her. | ...
93.61 105.95 -0.07326174931639003 | oscar however ... arrive. | ...
106.65 116.91 -0.14680841514778062 | upon the carrier ... his master. | ...
```
Details of the file content:
- the first line of the file contains the path to the original audio file
- all subsequent lines contain:
* the first number is the start of the segment (in seconds)
* the second one is the end of the segment (in seconds)
* the third value - alignment confidence score (in log space)
* text fragments corresponding to the timestamps
* original text without pre-processing
* normalized text
```
alignment_file = str(WINDOW) + '_' + audio_file.replace('.mp3', '_segments.txt')
! cat $OUTPUT_DIR/segments/$alignment_file
```
Finally, we're going to split the original audio file into segments based on the found alignments. We're going to create three subsets and three corresponding manifests:
* high scored clips (segments with the segmentation score above the threshold value, default threshold value = -5)
* low scored clips (segments with the segmentation score below the threshold)
* deleted segments (segments that were excluded during the alignment. For example, in our sample audio file, the prologue and epilogue that don't have the corresponding transcript were excluded. Oftentimes, deleted files also contain such things as clapping, music, or hard breathing.
The alignment score values depend on the pre-trained model quality and the dataset, the `THRESHOLD` parameter might be worth adjusting based on the analysis of the low/high scored clips.
Also note, that the `OFFSET` parameter is something one might want to experiment with since timestamps have a delay (offset) depending on the model.
```
OFFSET = 0
THRESHOLD = -5
! python $TOOLS_DIR/cut_audio.py \
--output_dir=$OUTPUT_DIR \
--model=$MODEL \
--alignment=$OUTPUT_DIR/segments/ \
--threshold=$THRESHOLD \
--offset=$OFFSET
```
`manifests` folder should be created under `OUTPUT_DIR`, and it should contain
corresponding manifests for the three groups of clips described above:
```
! ls -l $OUTPUT_DIR/manifests
def plot_signal(signal, sample_rate):
""" Plot the signal in time domain """
fig_signal = go.Figure(
go.Scatter(x=np.arange(signal.shape[0])/sample_rate,
y=signal, line={'color': 'green'},
name='Waveform',
hovertemplate='Time: %{x:.2f} s<br>Amplitude: %{y:.2f}<br><extra></extra>'),
layout={
'height': 200,
'xaxis': {'title': 'Time, s'},
'yaxis': {'title': 'Amplitude'},
'title': 'Audio Signal',
'margin': dict(l=0, r=0, t=40, b=0, pad=0),
}
)
fig_signal.show()
def display_samples(manifest):
""" Display audio and reference text."""
with open(manifest, 'r') as f:
for line in f:
sample = json.loads(line)
sample_rate, signal = wav.read(sample['audio_filepath'])
plot_signal(signal, sample_rate)
display(Audio(sample['audio_filepath']))
display('Reference text: ' + sample['text_no_preprocessing'])
display('ASR transcript: ' + sample['pred_text'])
print('\n' + '-' * 110)
```
Let's examine the high scored segments we obtained.
The `Reference text` in the next cell represents the original text without pre-processing, while `ASR transcript` is an ASR model prediction with greedy decoding. Also notice, that `ASR transcript` in some cases contains errors that could decrease the alignment score, but usually it doesn’t hurt the quality of the aligned segments.
```
high_score_manifest = str(WINDOW) + '_' + audio_file.replace('.mp3', '_high_score_manifest.json')
display_samples(os.path.join(OUTPUT_DIR, 'manifests', high_score_manifest))
! cat $OUTPUT_DIR/manifests/$high_score_manifest
```
# Multiple files alignment
Up until now, we were processing only one file at a time, but to create a large dataset processing of multiple files simultaneously could help speed up things considerably.
Let's download another audio file and corresponding text.
```
# https://librivox.org/frost-to-night-by-edith-m-thomas/
audio_file_2 = 'frosttonight_thomas_bk_128kb.mp3'
if not os.path.exists(os.path.join(DATA_DIR, audio_file_2)):
print('Downloading audio file')
wget.download('http://www.archive.org/download/frost_to-night_1710.poem_librivox/frosttonight_thomas_bk_128kb.mp3', DATA_DIR)
# text source: text source: https://www.bartleby.com/267/151.html
text = """
APPLE-GREEN west and an orange bar,
And the crystal eye of a lone, one star …
And, “Child, take the shears and cut what you will,
Frost to-night—so clear and dead-still.”
Then, I sally forth, half sad, half proud,
And I come to the velvet, imperial crowd,
The wine-red, the gold, the crimson, the pied,—
The dahlias that reign by the garden-side.
The dahlias I might not touch till to-night!
A gleam of the shears in the fading light,
And I gathered them all,—the splendid throng,
And in one great sheaf I bore them along.
. . . . . .
In my garden of Life with its all-late flowers
I heed a Voice in the shrinking hours:
“Frost to-night—so clear and dead-still” …
Half sad, half proud, my arms I fill.
"""
with open(os.path.join(DATA_DIR, audio_file_2.replace('mp3', 'txt')), 'w') as f:
f.write(text)
```
`DATA_DIR` should now contain two .mp3 files and two .txt files:
```
! ls -l $DATA_DIR
Audio(os.path.join(DATA_DIR, audio_file_2))
```
Finally, we need to download a script to perform all the above steps starting from the text and audio preprocessing to segmentation and manifest creation in a single step.
```
if 'google.colab' in str(get_ipython()) and not os.path.exists('run_sample.sh'):
wget.download('https://raw.githubusercontent.com/NVIDIA/NeMo/' + BRANCH + '/tools/ctc_segmentation/run_sample.sh', '.')
```
`run_sample.sh` script takes `DATA_DIR` argument and assumes that it contains folders `text` and `audio`.
An example of the `DATA_DIR` folder structure:
--DATA_DIR
|----audio
|---1.mp3
|---2.mp3
|-----text
|---1.txt
|---2.txt
Let's move our files to subfolders to follow the above structure.
```
! mkdir $DATA_DIR/text && mkdir $DATA_DIR/audio
! mv $DATA_DIR/*txt $DATA_DIR/text/. && mv $DATA_DIR/*mp3 $DATA_DIR/audio/.
! ls -l $DATA_DIR
```
Next, we're going to execute `run_sample.sh` script to find alignment for two audio/text samples. By default, if the alignment is not found for an initial WINDOW size, the initial window size will be doubled a few times to re-attempt alignment.
`run_sample.sh` applies two initial WINDOW sizes, 8000 and 12000, and then adds segments that were similarly aligned with two window sizes to `verified_segments` folder. This could be useful to reduce the amount of manual work while checking the alignment quality.
```
if 'google.colab' in str(get_ipython()):
OUTPUT_DIR_2 = f'/content/{WORK_DIR}/output_multiple_files'
else:
OUTPUT_DIR_2 = os.path.join(WORK_DIR, 'output_multiple_files')
! bash $TOOLS_DIR/../run_sample.sh \
--MODEL_NAME_OR_PATH=$MODEL \
--DATA_DIR=$DATA_DIR \
--OUTPUT_DIR=$OUTPUT_DIR_2 \
--SCRIPTS_DIR=$TOOLS_DIR \
--CUT_PREFIX=3 \
--MIN_SCORE=$THRESHOLD \
--USE_NEMO_NORMALIZATION=False
```
High scored manifests for the data samples were aggregated to the `all_manifest.json` under `OUTPUT_DIR_2`.
```
display_samples(os.path.join(OUTPUT_DIR_2, 'all_manifest.json'))
```
# Next Steps
Check out [NeMo Speech Data Explorer tool](https://github.com/NVIDIA/NeMo/tree/main/tools/speech_data_explorer#speech-data-explorer) to interactively evaluate the aligned segments.
# References
Kürzinger, Ludwig, et al. ["CTC-Segmentation of Large Corpora for German End-to-End Speech Recognition."](https://arxiv.org/abs/2007.09127) International Conference on Speech and Computer. Springer, Cham, 2020.
|
github_jupyter
|
BRANCH = 'main'
"""
You can run either this notebook locally (if you have all the dependencies and a GPU) or on Google Colab.
Instructions for setting up Colab are as follows:
1. Open a new Python 3 notebook.
2. Import this notebook from GitHub (File -> Upload Notebook -> "GITHUB" tab -> copy/paste GitHub URL)
3. Connect to an instance with a GPU (Runtime -> Change runtime type -> select "GPU" for hardware accelerator)
4. Run this cell to set up dependencies.
"""
# If you're using Google Colab and not running locally, run this cell.
# install NeMo
!python -m pip install git+https://github.com/NVIDIA/NeMo.git@$BRANCH#egg=nemo_toolkit[all]
import json
import os
import wget
from IPython.display import Audio
import numpy as np
import scipy.io.wavfile as wav
! pip install pandas
# optional
! pip install plotly
from plotly import graph_objects as go
! pip install ctc_segmentation==1.1.0
! pip install num2words
! apt-get install -y ffmpeg
# If you're running the notebook locally, update the TOOLS_DIR path below
# In Colab, a few required scripts will be downloaded from NeMo github
TOOLS_DIR = '<UPDATE_PATH_TO_NeMo_root>/tools/ctc_segmentation/scripts'
if 'google.colab' in str(get_ipython()):
TOOLS_DIR = 'scripts/'
os.makedirs(TOOLS_DIR, exist_ok=True)
required_files = ['prepare_data.py',
'normalization_helpers.py',
'run_ctc_segmentation.py',
'verify_segments.py',
'cut_audio.py',
'process_manifests.py',
'utils.py']
for file in required_files:
if not os.path.exists(os.path.join(TOOLS_DIR, file)):
file_path = 'https://raw.githubusercontent.com/NVIDIA/NeMo/' + BRANCH + '/tools/ctc_segmentation/' + TOOLS_DIR + file
print(file_path)
wget.download(file_path, TOOLS_DIR)
elif not os.path.exists(TOOLS_DIR):
raise ValueError(f'update path to NeMo root directory')
print(TOOLS_DIR)
! ls -l $TOOLS_DIR
## create data directory and download an audio file
WORK_DIR = 'WORK_DIR'
DATA_DIR = WORK_DIR + '/DATA'
os.makedirs(DATA_DIR, exist_ok=True)
audio_file = 'childrensshortworks019_06acarriersdog_am_128kb.mp3'
if not os.path.exists(os.path.join(DATA_DIR, audio_file)):
print('Downloading audio file')
wget.download('http://archive.org/download/childrens_short_works_vol_019_1310_librivox/' + audio_file, DATA_DIR)
# text source: http://www.gutenberg.org/cache/epub/24263/pg24263.txt
text = """
A carrier on his way to a market town had occasion to stop at some houses
by the road side, in the way of his business, leaving his cart and horse
upon the public road, under the protection of a passenger and a trusty
dog. Upon his return he missed a led horse, belonging to a gentleman in
the neighbourhood, which he had tied to the end of the cart, and likewise
one of the female passengers. On inquiry he was informed that during his
absence the female, who had been anxious to try the mettle of the pony,
had mounted it, and that the animal had set off at full speed. The carrier
expressed much anxiety for the safety of the young woman, casting at the
same time an expressive look at his dog. Oscar observed his master's eye,
and aware of its meaning, instantly set off in pursuit of the pony, which
coming up with soon after, he made a sudden spring, seized the bridle, and
held the animal fast. Several people having observed the circumstance, and
the perilous situation of the girl, came to relieve her. Oscar, however,
notwithstanding their repeated endeavours, would not quit his hold, and
the pony was actually led into the stable with the dog, till such time as
the carrier should arrive. Upon the carrier entering the stable, Oscar
wagged his tail in token of satisfaction, and immediately relinquished the
bridle to his master.
"""
with open(os.path.join(DATA_DIR, audio_file.replace('mp3', 'txt')), 'w') as f:
f.write(text)
!ls -l $DATA_DIR
Audio(os.path.join(DATA_DIR, audio_file))
MODEL = 'QuartzNet15x5Base-En'
OUTPUT_DIR = WORK_DIR + '/output'
! python $TOOLS_DIR/prepare_data.py \
--in_text=$DATA_DIR \
--output_dir=$OUTPUT_DIR/processed/ \
--language='eng' \
--cut_prefix=3 \
--model=$MODEL \
--audio_dir=$DATA_DIR
! ls -l $OUTPUT_DIR/processed
with open(os.path.join(OUTPUT_DIR, 'processed', audio_file.replace('.mp3', '.txt')), 'r') as f:
for line in f:
print (line)
WINDOW = 8000
! python $TOOLS_DIR/run_ctc_segmentation.py \
--output_dir=$OUTPUT_DIR \
--data=$OUTPUT_DIR/processed \
--model=$MODEL \
--window_len=$WINDOW \
--no_parallel
<PATH_TO>/processed/childrensshortworks019_06acarriersdog_am_128kb.wav
16.03 32.39 -4.5911999284929115 | a carrier on ... a trusty dog. | ...
33.31 45.01 -0.22886803973405373 | upon his ... passengers. | ...
46.17 58.57 -0.3523662826061572 | on inquiry ... at full speed. | ...
59.75 69.43 -0.04128918756038118 | the carrier ... dog. | ...
69.93 85.31 -0.3595261826390344 | oscar observed ... animal fast. | ...
85.95 93.43 -0.04447770533708611 | several people ... relieve her. | ...
93.61 105.95 -0.07326174931639003 | oscar however ... arrive. | ...
106.65 116.91 -0.14680841514778062 | upon the carrier ... his master. | ...
alignment_file = str(WINDOW) + '_' + audio_file.replace('.mp3', '_segments.txt')
! cat $OUTPUT_DIR/segments/$alignment_file
OFFSET = 0
THRESHOLD = -5
! python $TOOLS_DIR/cut_audio.py \
--output_dir=$OUTPUT_DIR \
--model=$MODEL \
--alignment=$OUTPUT_DIR/segments/ \
--threshold=$THRESHOLD \
--offset=$OFFSET
! ls -l $OUTPUT_DIR/manifests
def plot_signal(signal, sample_rate):
""" Plot the signal in time domain """
fig_signal = go.Figure(
go.Scatter(x=np.arange(signal.shape[0])/sample_rate,
y=signal, line={'color': 'green'},
name='Waveform',
hovertemplate='Time: %{x:.2f} s<br>Amplitude: %{y:.2f}<br><extra></extra>'),
layout={
'height': 200,
'xaxis': {'title': 'Time, s'},
'yaxis': {'title': 'Amplitude'},
'title': 'Audio Signal',
'margin': dict(l=0, r=0, t=40, b=0, pad=0),
}
)
fig_signal.show()
def display_samples(manifest):
""" Display audio and reference text."""
with open(manifest, 'r') as f:
for line in f:
sample = json.loads(line)
sample_rate, signal = wav.read(sample['audio_filepath'])
plot_signal(signal, sample_rate)
display(Audio(sample['audio_filepath']))
display('Reference text: ' + sample['text_no_preprocessing'])
display('ASR transcript: ' + sample['pred_text'])
print('\n' + '-' * 110)
high_score_manifest = str(WINDOW) + '_' + audio_file.replace('.mp3', '_high_score_manifest.json')
display_samples(os.path.join(OUTPUT_DIR, 'manifests', high_score_manifest))
! cat $OUTPUT_DIR/manifests/$high_score_manifest
# https://librivox.org/frost-to-night-by-edith-m-thomas/
audio_file_2 = 'frosttonight_thomas_bk_128kb.mp3'
if not os.path.exists(os.path.join(DATA_DIR, audio_file_2)):
print('Downloading audio file')
wget.download('http://www.archive.org/download/frost_to-night_1710.poem_librivox/frosttonight_thomas_bk_128kb.mp3', DATA_DIR)
# text source: text source: https://www.bartleby.com/267/151.html
text = """
APPLE-GREEN west and an orange bar,
And the crystal eye of a lone, one star …
And, “Child, take the shears and cut what you will,
Frost to-night—so clear and dead-still.”
Then, I sally forth, half sad, half proud,
And I come to the velvet, imperial crowd,
The wine-red, the gold, the crimson, the pied,—
The dahlias that reign by the garden-side.
The dahlias I might not touch till to-night!
A gleam of the shears in the fading light,
And I gathered them all,—the splendid throng,
And in one great sheaf I bore them along.
. . . . . .
In my garden of Life with its all-late flowers
I heed a Voice in the shrinking hours:
“Frost to-night—so clear and dead-still” …
Half sad, half proud, my arms I fill.
"""
with open(os.path.join(DATA_DIR, audio_file_2.replace('mp3', 'txt')), 'w') as f:
f.write(text)
! ls -l $DATA_DIR
Audio(os.path.join(DATA_DIR, audio_file_2))
if 'google.colab' in str(get_ipython()) and not os.path.exists('run_sample.sh'):
wget.download('https://raw.githubusercontent.com/NVIDIA/NeMo/' + BRANCH + '/tools/ctc_segmentation/run_sample.sh', '.')
! mkdir $DATA_DIR/text && mkdir $DATA_DIR/audio
! mv $DATA_DIR/*txt $DATA_DIR/text/. && mv $DATA_DIR/*mp3 $DATA_DIR/audio/.
! ls -l $DATA_DIR
if 'google.colab' in str(get_ipython()):
OUTPUT_DIR_2 = f'/content/{WORK_DIR}/output_multiple_files'
else:
OUTPUT_DIR_2 = os.path.join(WORK_DIR, 'output_multiple_files')
! bash $TOOLS_DIR/../run_sample.sh \
--MODEL_NAME_OR_PATH=$MODEL \
--DATA_DIR=$DATA_DIR \
--OUTPUT_DIR=$OUTPUT_DIR_2 \
--SCRIPTS_DIR=$TOOLS_DIR \
--CUT_PREFIX=3 \
--MIN_SCORE=$THRESHOLD \
--USE_NEMO_NORMALIZATION=False
display_samples(os.path.join(OUTPUT_DIR_2, 'all_manifest.json'))
| 0.590543 | 0.792143 |
```
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
class MySequence :
def __init__(self) :
self.dummy = 1
keras.utils.Sequence = MySequence
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from deepexplain.tensorflow import DeepExplain
#Define dataset/experiment name
dataset_name = "apa_doubledope"
#Load cached dataframe
cached_dict = pickle.load(open('apa_doubledope_cached_set.pickle', 'rb'))
data_df = cached_dict['data_df']
print("len(data_df) = " + str(len(data_df)) + " (loaded)")
#Make generators
valid_set_size = 0.05
test_set_size = 0.05
batch_size = 32
#Generate training and test set indexes
data_index = np.arange(len(data_df), dtype=np.int)
train_index = data_index[:-int(len(data_df) * (valid_set_size + test_set_size))]
valid_index = data_index[train_index.shape[0]:-int(len(data_df) * test_set_size)]
test_index = data_index[train_index.shape[0] + valid_index.shape[0]:]
print('Training set size = ' + str(train_index.shape[0]))
print('Validation set size = ' + str(valid_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
data_gens = {
gen_id : isol.DataGenerator(
idx,
{'df' : data_df},
batch_size=batch_size,
inputs = [
{
'id' : 'seq',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : isol.SequenceExtractor('padded_seq', start_pos=180, end_pos=180 + 205),
'encoder' : isol.OneHotEncoder(seq_length=205),
'dim' : (1, 205, 4),
'sparsify' : False
}
],
outputs = [
{
'id' : 'hairpin',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['proximal_usage'],
'transformer' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [],
shuffle = True if gen_id == 'train' else False
) for gen_id, idx in [('all', data_index), ('train', train_index), ('valid', valid_index), ('test', test_index)]
}
#Load data matrices
x_train = np.concatenate([data_gens['train'][i][0][0] for i in range(len(data_gens['train']))], axis=0)
x_test = np.concatenate([data_gens['test'][i][0][0] for i in range(len(data_gens['test']))], axis=0)
y_train = np.concatenate([data_gens['train'][i][1][0] for i in range(len(data_gens['train']))], axis=0)
y_test = np.concatenate([data_gens['test'][i][1][0] for i in range(len(data_gens['test']))], axis=0)
print("x_train.shape = " + str(x_train.shape))
print("x_test.shape = " + str(x_test.shape))
print("y_train.shape = " + str(y_train.shape))
print("y_test.shape = " + str(y_test.shape))
#Define sequence template (APA Doubledope sublibrary)
sequence_template = 'CTTCCGATCTNNNNNNNNNNNNNNNNNNNNCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTAC'
sequence_mask = np.array([1 if sequence_template[j] == 'N' else 0 for j in range(len(sequence_template))])
#Visualize background sequence distribution
pseudo_count = 1.0
x_mean = (np.sum(x_train, axis=(0, 1)) + pseudo_count) / (x_train.shape[0] + 4. * pseudo_count)
x_mean_logits = np.log(x_mean / (1. - x_mean))
'''
#APARENT parameters
seq_input_shape = (1, 205, 4)
lib_input_shape = (13,)
distal_pas_shape = (1,)
num_outputs_iso = 1
num_outputs_cut = 206
#Shared model definition
layer_1 = Conv2D(96, (8, 4), padding='valid', activation='relu')
layer_1_pool = MaxPooling2D(pool_size=(2, 1))
layer_2 = Conv2D(128, (6, 1), padding='valid', activation='relu')
layer_dense = Dense(256, activation='relu')
layer_drop = Dropout(0.2)
def shared_model(seq_input, distal_pas_input) :
return layer_drop(
layer_dense(
Concatenate()([
Flatten()(
layer_2(
layer_1_pool(
layer_1(
seq_input
)
)
)
),
distal_pas_input
])
)
)
#Inputs
seq_input = Input(name="seq_input", shape=seq_input_shape)
lib_input = Input(name="lib_input", shape=lib_input_shape)
distal_pas_input = Input(name="distal_pas_input", shape=distal_pas_shape)
permute_layer = Lambda(lambda x: K.permute_dimensions(x, (0, 2, 3, 1)))
plasmid_out_shared = Concatenate()([shared_model(permute_layer(seq_input), distal_pas_input), lib_input])
plasmid_out_cut = Dense(num_outputs_cut, activation='softmax', kernel_initializer='zeros')(plasmid_out_shared)
plasmid_out_iso = Dense(num_outputs_iso, activation='linear', kernel_initializer='zeros', name="apa_logodds")(plasmid_out_shared)
predictor_temp = Model(
inputs=[
seq_input,
lib_input,
distal_pas_input
],
outputs=[
plasmid_out_iso,
plasmid_out_cut
]
)
predictor_temp.load_weights('../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5')
predictor = Model(
inputs=predictor_temp.inputs,
outputs=[
predictor_temp.outputs[0]
]
)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
'''
#APARENT parameters
seq_input_shape = (1, 205, 4)
lib_input_shape = (13,)
distal_pas_shape = (1,)
num_outputs_iso = 1
num_outputs_cut = 206
#Shared model definition
layer_1 = Conv2D(96, (8, 4), padding='valid', activation='relu')
layer_1_pool = MaxPooling2D(pool_size=(2, 1))
layer_2 = Conv2D(128, (6, 1), padding='valid', activation='relu')
layer_dense = Dense(256, activation='relu')
layer_drop = Dropout(0.2)
def shared_model(seq_input, distal_pas_input) :
return layer_drop(
layer_dense(
Concatenate()([
Flatten()(
layer_2(
layer_1_pool(
layer_1(
seq_input
)
)
)
),
distal_pas_input
])
)
)
#Inputs
seq_input = Input(name="seq_input", shape=seq_input_shape)
permute_layer = Lambda(lambda x: K.permute_dimensions(x, (0, 2, 3, 1)))
lib_input = Lambda(lambda x: K.tile(K.expand_dims(K.constant(np.array([0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0.])), axis=0), (K.shape(x)[0], 1)))(seq_input)
distal_pas_input = Lambda(lambda x: K.tile(K.expand_dims(K.constant(np.array([1.])), axis=0), (K.shape(x)[0], 1)))(seq_input)
plasmid_out_shared = Concatenate()([shared_model(permute_layer(seq_input), distal_pas_input), lib_input])
plasmid_out_cut = Dense(num_outputs_cut, activation='softmax', kernel_initializer='zeros')(plasmid_out_shared)
plasmid_out_iso = Dense(num_outputs_iso, activation='linear', kernel_initializer='zeros', name="apa_logodds")(plasmid_out_shared)
predictor_temp = Model(
inputs=[
seq_input
],
outputs=[
plasmid_out_iso,
plasmid_out_cut
]
)
predictor_temp.load_weights('../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5')
predictor = Model(
inputs=predictor_temp.inputs,
outputs=[
predictor_temp.outputs[0]
]
)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
predictor.summary()
#Tile the ref background to same shape as test set
x_mean_baseline = np.expand_dims(x_mean, axis=0)#np.expand_dims(x_mean, axis=-1)
x_mean_baseline.shape
aparent_l_test = np.zeros((x_test.shape[0], 13))
aparent_l_test[:, 4] = 1.
aparent_d_test = np.ones((x_test.shape[0], 1))
def _deep_explain(method_name, predictor=predictor, x_test=x_test, baseline=x_mean_baseline, aparent_l_test=aparent_l_test, aparent_d_test=aparent_d_test) :
attributions = None
with DeepExplain(session=K.get_session()) as de :
input_tensor = predictor.get_layer("seq_input").input
fModel = Model(inputs=input_tensor, outputs = predictor.get_layer("apa_logodds").output)
target_tensor = fModel(input_tensor)
if method_name == 'deeplift' :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test, baseline=baseline)
elif method_name == 'intgrad' :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test, baseline=baseline, steps=10)
else :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test)
return attributions
#Specify deeplift attribution models
attribution_suffixes = [
'gradient',
'rescale',
'integrated_gradients',
#'eta_lrp'
]
attribution_method_names = [
'grad*input',
'deeplift',
'intgrad',
#'elrp'
]
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
dna_letter_at(ref_seq[i], i + 0.5, 0, mutability_score, ax)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
plt.tight_layout()
plt.show()
#Run attribution methods
encoder = isol.OneHotEncoder(205)
score_clip = 1.5
for attr_suffix, attr_method_name in zip(attribution_suffixes, attribution_method_names) :
print("Attribution method = '" + attr_suffix + "'")
importance_scores_test = _deep_explain(attr_method_name)#[0]
importance_scores_test_signed = np.copy(importance_scores_test)
importance_scores_test = np.abs(importance_scores_test)
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(14, 0.65), plot_start=0, plot_end=205)
plot_importance_scores(importance_scores_test[plot_i, 0, :, :].T, encoder.decode(x_test[plot_i, 0, :, :]), figsize=(14, 0.65), score_clip=score_clip, sequence_template=sequence_template, plot_start=0, plot_end=205)
#Save predicted importance scores
model_name = "deepexplain_" + dataset_name + "_method_" + attr_suffix
np.save(model_name + "_importance_scores_test", importance_scores_test)
np.save(model_name + "_importance_scores_test_signed", importance_scores_test_signed)
```
|
github_jupyter
|
import keras
import keras.backend as K
from keras.datasets import mnist
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Conv2DTranspose, Embedding, Multiply, Activation
from functools import partial
from collections import defaultdict
import os
import pickle
import numpy as np
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
class MySequence :
def __init__(self) :
self.dummy = 1
keras.utils.Sequence = MySequence
import isolearn.io as isoio
import isolearn.keras as isol
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from deepexplain.tensorflow import DeepExplain
#Define dataset/experiment name
dataset_name = "apa_doubledope"
#Load cached dataframe
cached_dict = pickle.load(open('apa_doubledope_cached_set.pickle', 'rb'))
data_df = cached_dict['data_df']
print("len(data_df) = " + str(len(data_df)) + " (loaded)")
#Make generators
valid_set_size = 0.05
test_set_size = 0.05
batch_size = 32
#Generate training and test set indexes
data_index = np.arange(len(data_df), dtype=np.int)
train_index = data_index[:-int(len(data_df) * (valid_set_size + test_set_size))]
valid_index = data_index[train_index.shape[0]:-int(len(data_df) * test_set_size)]
test_index = data_index[train_index.shape[0] + valid_index.shape[0]:]
print('Training set size = ' + str(train_index.shape[0]))
print('Validation set size = ' + str(valid_index.shape[0]))
print('Test set size = ' + str(test_index.shape[0]))
data_gens = {
gen_id : isol.DataGenerator(
idx,
{'df' : data_df},
batch_size=batch_size,
inputs = [
{
'id' : 'seq',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : isol.SequenceExtractor('padded_seq', start_pos=180, end_pos=180 + 205),
'encoder' : isol.OneHotEncoder(seq_length=205),
'dim' : (1, 205, 4),
'sparsify' : False
}
],
outputs = [
{
'id' : 'hairpin',
'source_type' : 'dataframe',
'source' : 'df',
'extractor' : lambda row, index: row['proximal_usage'],
'transformer' : lambda t: t,
'dim' : (1,),
'sparsify' : False
}
],
randomizers = [],
shuffle = True if gen_id == 'train' else False
) for gen_id, idx in [('all', data_index), ('train', train_index), ('valid', valid_index), ('test', test_index)]
}
#Load data matrices
x_train = np.concatenate([data_gens['train'][i][0][0] for i in range(len(data_gens['train']))], axis=0)
x_test = np.concatenate([data_gens['test'][i][0][0] for i in range(len(data_gens['test']))], axis=0)
y_train = np.concatenate([data_gens['train'][i][1][0] for i in range(len(data_gens['train']))], axis=0)
y_test = np.concatenate([data_gens['test'][i][1][0] for i in range(len(data_gens['test']))], axis=0)
print("x_train.shape = " + str(x_train.shape))
print("x_test.shape = " + str(x_test.shape))
print("y_train.shape = " + str(y_train.shape))
print("y_test.shape = " + str(y_test.shape))
#Define sequence template (APA Doubledope sublibrary)
sequence_template = 'CTTCCGATCTNNNNNNNNNNNNNNNNNNNNCATTACTCGCATCCANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCAGCCAATTAAGCCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCTAC'
sequence_mask = np.array([1 if sequence_template[j] == 'N' else 0 for j in range(len(sequence_template))])
#Visualize background sequence distribution
pseudo_count = 1.0
x_mean = (np.sum(x_train, axis=(0, 1)) + pseudo_count) / (x_train.shape[0] + 4. * pseudo_count)
x_mean_logits = np.log(x_mean / (1. - x_mean))
'''
#APARENT parameters
seq_input_shape = (1, 205, 4)
lib_input_shape = (13,)
distal_pas_shape = (1,)
num_outputs_iso = 1
num_outputs_cut = 206
#Shared model definition
layer_1 = Conv2D(96, (8, 4), padding='valid', activation='relu')
layer_1_pool = MaxPooling2D(pool_size=(2, 1))
layer_2 = Conv2D(128, (6, 1), padding='valid', activation='relu')
layer_dense = Dense(256, activation='relu')
layer_drop = Dropout(0.2)
def shared_model(seq_input, distal_pas_input) :
return layer_drop(
layer_dense(
Concatenate()([
Flatten()(
layer_2(
layer_1_pool(
layer_1(
seq_input
)
)
)
),
distal_pas_input
])
)
)
#Inputs
seq_input = Input(name="seq_input", shape=seq_input_shape)
lib_input = Input(name="lib_input", shape=lib_input_shape)
distal_pas_input = Input(name="distal_pas_input", shape=distal_pas_shape)
permute_layer = Lambda(lambda x: K.permute_dimensions(x, (0, 2, 3, 1)))
plasmid_out_shared = Concatenate()([shared_model(permute_layer(seq_input), distal_pas_input), lib_input])
plasmid_out_cut = Dense(num_outputs_cut, activation='softmax', kernel_initializer='zeros')(plasmid_out_shared)
plasmid_out_iso = Dense(num_outputs_iso, activation='linear', kernel_initializer='zeros', name="apa_logodds")(plasmid_out_shared)
predictor_temp = Model(
inputs=[
seq_input,
lib_input,
distal_pas_input
],
outputs=[
plasmid_out_iso,
plasmid_out_cut
]
)
predictor_temp.load_weights('../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5')
predictor = Model(
inputs=predictor_temp.inputs,
outputs=[
predictor_temp.outputs[0]
]
)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
'''
#APARENT parameters
seq_input_shape = (1, 205, 4)
lib_input_shape = (13,)
distal_pas_shape = (1,)
num_outputs_iso = 1
num_outputs_cut = 206
#Shared model definition
layer_1 = Conv2D(96, (8, 4), padding='valid', activation='relu')
layer_1_pool = MaxPooling2D(pool_size=(2, 1))
layer_2 = Conv2D(128, (6, 1), padding='valid', activation='relu')
layer_dense = Dense(256, activation='relu')
layer_drop = Dropout(0.2)
def shared_model(seq_input, distal_pas_input) :
return layer_drop(
layer_dense(
Concatenate()([
Flatten()(
layer_2(
layer_1_pool(
layer_1(
seq_input
)
)
)
),
distal_pas_input
])
)
)
#Inputs
seq_input = Input(name="seq_input", shape=seq_input_shape)
permute_layer = Lambda(lambda x: K.permute_dimensions(x, (0, 2, 3, 1)))
lib_input = Lambda(lambda x: K.tile(K.expand_dims(K.constant(np.array([0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0.])), axis=0), (K.shape(x)[0], 1)))(seq_input)
distal_pas_input = Lambda(lambda x: K.tile(K.expand_dims(K.constant(np.array([1.])), axis=0), (K.shape(x)[0], 1)))(seq_input)
plasmid_out_shared = Concatenate()([shared_model(permute_layer(seq_input), distal_pas_input), lib_input])
plasmid_out_cut = Dense(num_outputs_cut, activation='softmax', kernel_initializer='zeros')(plasmid_out_shared)
plasmid_out_iso = Dense(num_outputs_iso, activation='linear', kernel_initializer='zeros', name="apa_logodds")(plasmid_out_shared)
predictor_temp = Model(
inputs=[
seq_input
],
outputs=[
plasmid_out_iso,
plasmid_out_cut
]
)
predictor_temp.load_weights('../../../aparent/saved_models/aparent_plasmid_iso_cut_distalpas_all_libs_no_sampleweights_sgd.h5')
predictor = Model(
inputs=predictor_temp.inputs,
outputs=[
predictor_temp.outputs[0]
]
)
predictor.trainable = False
predictor.compile(
optimizer=keras.optimizers.SGD(lr=0.1),
loss='mean_squared_error'
)
predictor.summary()
#Tile the ref background to same shape as test set
x_mean_baseline = np.expand_dims(x_mean, axis=0)#np.expand_dims(x_mean, axis=-1)
x_mean_baseline.shape
aparent_l_test = np.zeros((x_test.shape[0], 13))
aparent_l_test[:, 4] = 1.
aparent_d_test = np.ones((x_test.shape[0], 1))
def _deep_explain(method_name, predictor=predictor, x_test=x_test, baseline=x_mean_baseline, aparent_l_test=aparent_l_test, aparent_d_test=aparent_d_test) :
attributions = None
with DeepExplain(session=K.get_session()) as de :
input_tensor = predictor.get_layer("seq_input").input
fModel = Model(inputs=input_tensor, outputs = predictor.get_layer("apa_logodds").output)
target_tensor = fModel(input_tensor)
if method_name == 'deeplift' :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test, baseline=baseline)
elif method_name == 'intgrad' :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test, baseline=baseline, steps=10)
else :
attributions = de.explain(method_name, target_tensor, input_tensor, x_test)
return attributions
#Specify deeplift attribution models
attribution_suffixes = [
'gradient',
'rescale',
'integrated_gradients',
#'eta_lrp'
]
attribution_method_names = [
'grad*input',
'deeplift',
'intgrad',
#'elrp'
]
#Gradient saliency/backprop visualization
import matplotlib.collections as collections
import operator
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import matplotlib.colors as colors
import matplotlib as mpl
from matplotlib.text import TextPath
from matplotlib.patches import PathPatch, Rectangle
from matplotlib.font_manager import FontProperties
from matplotlib import gridspec
from matplotlib.ticker import FormatStrFormatter
def plot_importance_scores(importance_scores, ref_seq, figsize=(12, 2), score_clip=None, sequence_template='', plot_start=0, plot_end=96) :
end_pos = ref_seq.find("#")
fig = plt.figure(figsize=figsize)
ax = plt.gca()
if score_clip is not None :
importance_scores = np.clip(np.copy(importance_scores), -score_clip, score_clip)
max_score = np.max(np.sum(importance_scores[:, :], axis=0)) + 0.01
for i in range(0, len(ref_seq)) :
mutability_score = np.sum(importance_scores[:, i])
dna_letter_at(ref_seq[i], i + 0.5, 0, mutability_score, ax)
plt.sca(ax)
plt.xlim((0, len(ref_seq)))
plt.ylim((0, max_score))
plt.axis('off')
plt.yticks([0.0, max_score], [0.0, max_score], fontsize=16)
plt.tight_layout()
plt.show()
#Run attribution methods
encoder = isol.OneHotEncoder(205)
score_clip = 1.5
for attr_suffix, attr_method_name in zip(attribution_suffixes, attribution_method_names) :
print("Attribution method = '" + attr_suffix + "'")
importance_scores_test = _deep_explain(attr_method_name)#[0]
importance_scores_test_signed = np.copy(importance_scores_test)
importance_scores_test = np.abs(importance_scores_test)
for plot_i in range(0, 5) :
print("Test sequence " + str(plot_i) + ":")
plot_dna_logo(x_test[plot_i, 0, :, :], sequence_template=sequence_template, figsize=(14, 0.65), plot_start=0, plot_end=205)
plot_importance_scores(importance_scores_test[plot_i, 0, :, :].T, encoder.decode(x_test[plot_i, 0, :, :]), figsize=(14, 0.65), score_clip=score_clip, sequence_template=sequence_template, plot_start=0, plot_end=205)
#Save predicted importance scores
model_name = "deepexplain_" + dataset_name + "_method_" + attr_suffix
np.save(model_name + "_importance_scores_test", importance_scores_test)
np.save(model_name + "_importance_scores_test_signed", importance_scores_test_signed)
| 0.813609 | 0.403009 |
```
import os
import google_auth_oauthlib.flow
import googleapiclient.discovery
import googleapiclient.errors
from oauth2client.tools import argparser
import pandas as pd
import datetime
from sqlalchemy import *
from sqlalchemy.ext.declarative import declarative_base
from sqlalchemy.orm import sessionmaker
### 1) API Key 인증
#get api key from local
f = open("data/apikey.txt", "r")
api_key = f.readline()
f.close()
scopes = ["https://www.googleapis.com/auth/youtube.readonly"]
DEVELOPER_KEY = api_key # Write down your api key here
YOUTUBE_API_SERVICE_NAME = "youtube"
YOUTUBE_API_VERSION = "v3"
youtube = googleapiclient.discovery.build(YOUTUBE_API_SERVICE_NAME, YOUTUBE_API_VERSION,
developerKey=DEVELOPER_KEY)
### 2) SQL에서 SELECT 채널ID 리스트
import login_mysql
mydb, cursor = login_mysql.login()
QUERY1 = """
SELECT distinct Playlist_Id
From Dimension_Playlist
"""
cursor.execute(QUERY1)
result1 = cursor.fetchall()
result1 = pd.DataFrame(result1)
result1["Playlist_Id"]
```
----
```
plistid = []
videoid = []
videotitle = []
channelid = []
published = []
for pid in result1["Playlist_Id"] :
DEVELOPER_KEY = "AIzaSyD3jF-8cTppgQQ2gCW7sOEqEgkmkolFA54"
YOUTUBE_API_SERVICE_NAME = "youtube"
YOUTUBE_API_VERSION = "v3"
youtube = googleapiclient.discovery.build(YOUTUBE_API_SERVICE_NAME, YOUTUBE_API_VERSION,
developerKey=DEVELOPER_KEY)
request = youtube.playlistItems().list(
part="contentDetails, id, snippet",
playlistId= pid,
maxResults=50
)
response = request.execute()
videoid.extend([each["snippet"]["resourceId"]["videoId"] for each in response["items"]])
videotitle.extend([each["snippet"]["title"] for each in response["items"]])
published.extend([each["snippet"]['publishedAt'] for each in response["items"]])
plistid.extend([each["snippet"]['playlistId'] for each in response["items"]])
channelid.extend([each["snippet"]['channelId'] for each in response["items"]])
while "nextPageToken" in response.keys():
npt = response["nextPageToken"]
request = youtube.playlistItems().list(
part="contentDetails, id, snippet",
playlistId= pid,
pageToken=npt,
maxResults=50
)
response = request.execute()
videoid.extend([each["snippet"]["resourceId"]["videoId"] for each in response["items"]])
videotitle.extend([each["snippet"]["title"] for each in response["items"]])
published.extend([each["snippet"]['publishedAt'] for each in response["items"]])
plistid.extend([each["snippet"]['playlistId'] for each in response["items"]])
channelid.extend([each["snippet"]['channelId'] for each in response["items"]])
if "nextPageToken" in response.keys():
npt = response["nextPageToken"]
else:
break
Dimension_Video = pd.DataFrame({"Published_date": published,
"Video_Id" : videoid, "Playlist_Id" : plistid, "Channel_Id" : channelid})
#Dimension_Video = pd.DataFrame({"Published_date": published, "Video_Title": videotitle ,
# "Video_Id" : videoid, "Playlist_Id" : plistid, "Channel_Id" : channelid})
```
-----
```
#3. 채널의 Fact_table 만들어서 DB에 insert하기
engine = create_engine("mysql://root:[email protected]/crwdb_yt?charset=utf8")
engine
Dimension_Video.reset_index(drop=True, inplace=True)
Dimension_Video.to_sql(name='Dimension_Video', if_exists = 'replace', con=engine, index=False)
```
|
github_jupyter
|
import os
import google_auth_oauthlib.flow
import googleapiclient.discovery
import googleapiclient.errors
from oauth2client.tools import argparser
import pandas as pd
import datetime
from sqlalchemy import *
from sqlalchemy.ext.declarative import declarative_base
from sqlalchemy.orm import sessionmaker
### 1) API Key 인증
#get api key from local
f = open("data/apikey.txt", "r")
api_key = f.readline()
f.close()
scopes = ["https://www.googleapis.com/auth/youtube.readonly"]
DEVELOPER_KEY = api_key # Write down your api key here
YOUTUBE_API_SERVICE_NAME = "youtube"
YOUTUBE_API_VERSION = "v3"
youtube = googleapiclient.discovery.build(YOUTUBE_API_SERVICE_NAME, YOUTUBE_API_VERSION,
developerKey=DEVELOPER_KEY)
### 2) SQL에서 SELECT 채널ID 리스트
import login_mysql
mydb, cursor = login_mysql.login()
QUERY1 = """
SELECT distinct Playlist_Id
From Dimension_Playlist
"""
cursor.execute(QUERY1)
result1 = cursor.fetchall()
result1 = pd.DataFrame(result1)
result1["Playlist_Id"]
plistid = []
videoid = []
videotitle = []
channelid = []
published = []
for pid in result1["Playlist_Id"] :
DEVELOPER_KEY = "AIzaSyD3jF-8cTppgQQ2gCW7sOEqEgkmkolFA54"
YOUTUBE_API_SERVICE_NAME = "youtube"
YOUTUBE_API_VERSION = "v3"
youtube = googleapiclient.discovery.build(YOUTUBE_API_SERVICE_NAME, YOUTUBE_API_VERSION,
developerKey=DEVELOPER_KEY)
request = youtube.playlistItems().list(
part="contentDetails, id, snippet",
playlistId= pid,
maxResults=50
)
response = request.execute()
videoid.extend([each["snippet"]["resourceId"]["videoId"] for each in response["items"]])
videotitle.extend([each["snippet"]["title"] for each in response["items"]])
published.extend([each["snippet"]['publishedAt'] for each in response["items"]])
plistid.extend([each["snippet"]['playlistId'] for each in response["items"]])
channelid.extend([each["snippet"]['channelId'] for each in response["items"]])
while "nextPageToken" in response.keys():
npt = response["nextPageToken"]
request = youtube.playlistItems().list(
part="contentDetails, id, snippet",
playlistId= pid,
pageToken=npt,
maxResults=50
)
response = request.execute()
videoid.extend([each["snippet"]["resourceId"]["videoId"] for each in response["items"]])
videotitle.extend([each["snippet"]["title"] for each in response["items"]])
published.extend([each["snippet"]['publishedAt'] for each in response["items"]])
plistid.extend([each["snippet"]['playlistId'] for each in response["items"]])
channelid.extend([each["snippet"]['channelId'] for each in response["items"]])
if "nextPageToken" in response.keys():
npt = response["nextPageToken"]
else:
break
Dimension_Video = pd.DataFrame({"Published_date": published,
"Video_Id" : videoid, "Playlist_Id" : plistid, "Channel_Id" : channelid})
#Dimension_Video = pd.DataFrame({"Published_date": published, "Video_Title": videotitle ,
# "Video_Id" : videoid, "Playlist_Id" : plistid, "Channel_Id" : channelid})
#3. 채널의 Fact_table 만들어서 DB에 insert하기
engine = create_engine("mysql://root:[email protected]/crwdb_yt?charset=utf8")
engine
Dimension_Video.reset_index(drop=True, inplace=True)
Dimension_Video.to_sql(name='Dimension_Video', if_exists = 'replace', con=engine, index=False)
| 0.187579 | 0.173533 |
<div class="alert alert-block alert-info" style="margin-top: 20px">
| Name | Description | Date
| :- |-------------: | :-:
|Reza Hashemi| Building Model Basics 2nd | On 23rd of August 2019 | width="750" align="center"></a></p>
</div>
# Building Blocks of Models
- ```nn.Linear```
- Nonlinear Activations
- Loss functions
- Optimizers
```
!pip3 install torch torchvision
import numpy as np
import pandas as pd
import torch, torchvision
torch.__version__
import torch.nn as nn
```
## 1. nn.Linear
```nn.Linear()``` is one of the basic building blocks of any neural network (NN) model
- Performs linear (or affine) transformation in the form of ```Wx (+ b)```. In NN terminology, generates a fully connected, or dense, layer.
- Two parameters, ```in_features``` and ```out_features``` should be specified
- Documentation: [linear_layers](https://pytorch.org/docs/stable/nn.html#linear-layers)
```python
torch.nn.Linear(in_features, # size of each input sample
out_features, # size of each output sample
bias = True) # whether bias (b) will be added or not
```
```
linear = nn.Linear(5, 1) # input dim = 5, output dim = 1
x = torch.FloatTensor([1, 2, 3, 4, 5]) # 1d tensor
print(linear(x))
y = torch.ones(3, 5) # 2d tensor
print(linear(y))
```
## 2. Nonlinear activations
PyTorch provides a number of nonlinear activation functions. Most commonly used ones are:
```python
torch.nn.ReLU() # relu
torch.nn.Sigmoid() # sigmoid
torch.nn.Tanh() # tangent hyperbolic
torch.nn.Softmax() # softmax
```
- Documentation: [nonlinear_activations](https://pytorch.org/docs/stable/nn.html#non-linear-activations-weighted-sum-nonlinearity)
```
relu = torch.nn.ReLU()
sigmoid = torch.nn.Sigmoid()
tanh = torch.nn.Tanh()
softmax = torch.nn.Softmax(dim = 0) # when using softmax, explicitly designate dimension
x = torch.randn(5) # five random numbers
print(x)
print(relu(x))
print(sigmoid(x))
print(tanh(x))
print(softmax(x))
```
## 3. Loss Functions
There are a number of loss functions that are already implemented in PyTorch. Common ones include:
- ```nn.MSELoss```: Mean squared error. Commonly used in regression tasks.
- ```nn.CrossEntropyLoss```: Cross entropy loss. Commonly used in classification tasks
```
a = torch.FloatTensor([2, 4, 5])
b = torch.FloatTensor([1, 3, 2])
mse = nn.MSELoss()
print(mse(a, b))
# note that when using CrossEntropyLoss, input has to have (N, C) shape, where
# N is the batch size
# C is the number of classes
a = torch.FloatTensor([[0.5, 0], [4.5, 0], [0, 0.4], [0, 0.1]]) # input
b = torch.LongTensor([1, 1, 1, 0]) # target
ce = nn.CrossEntropyLoss()
print(ce(a,b))
```
## 4. Optimizers
- ```torch.optim``` provides various optimization algorithms that are commonly used. Some of them are:
```python
optim.Adagrad
optim.Adam
optim.RMSprop
optim.SGD
```
- As arguments, (model) parameters and (optionally) learning rate are passed
- Model training process
- ```optimizer.zero_grad()```: sets all gradients to zero (for every training batches)
- ```loss_fn.backward()```: back propagate with respect to the loss function
- ```optimizer.step()```: update model parameters
```
## how pytorch models are trained with loss function and optimizers
# input and output data
x = torch.randn(5)
y = torch.ones(1)
model = nn.Linear(5, 1) # generate model
loss_fn = nn.MSELoss() # define loss function
optimizer = torch.optim.RMSprop(model.parameters(), lr = 0.01) # create optimizer
optimizer.zero_grad() # setting gradients to zero
loss_fn(model(x), y).backward() # back propagation
optimizer.step() # update parameters based on gradients computed
```
|
github_jupyter
|
- Nonlinear Activations
- Loss functions
- Optimizers
## 1. nn.Linear
torch.nn.Linear(in_features, # size of each input sample
out_features, # size of each output sample
bias = True) # whether bias (b) will be added or not
linear = nn.Linear(5, 1) # input dim = 5, output dim = 1
x = torch.FloatTensor([1, 2, 3, 4, 5]) # 1d tensor
print(linear(x))
y = torch.ones(3, 5) # 2d tensor
print(linear(y))
torch.nn.ReLU() # relu
torch.nn.Sigmoid() # sigmoid
torch.nn.Tanh() # tangent hyperbolic
torch.nn.Softmax() # softmax
relu = torch.nn.ReLU()
sigmoid = torch.nn.Sigmoid()
tanh = torch.nn.Tanh()
softmax = torch.nn.Softmax(dim = 0) # when using softmax, explicitly designate dimension
x = torch.randn(5) # five random numbers
print(x)
print(relu(x))
print(sigmoid(x))
print(tanh(x))
print(softmax(x))
a = torch.FloatTensor([2, 4, 5])
b = torch.FloatTensor([1, 3, 2])
mse = nn.MSELoss()
print(mse(a, b))
# note that when using CrossEntropyLoss, input has to have (N, C) shape, where
# N is the batch size
# C is the number of classes
a = torch.FloatTensor([[0.5, 0], [4.5, 0], [0, 0.4], [0, 0.1]]) # input
b = torch.LongTensor([1, 1, 1, 0]) # target
ce = nn.CrossEntropyLoss()
print(ce(a,b))
optim.Adagrad
optim.Adam
optim.RMSprop
optim.SGD
## how pytorch models are trained with loss function and optimizers
# input and output data
x = torch.randn(5)
y = torch.ones(1)
model = nn.Linear(5, 1) # generate model
loss_fn = nn.MSELoss() # define loss function
optimizer = torch.optim.RMSprop(model.parameters(), lr = 0.01) # create optimizer
optimizer.zero_grad() # setting gradients to zero
loss_fn(model(x), y).backward() # back propagation
optimizer.step() # update parameters based on gradients computed
| 0.768646 | 0.975946 |
```
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
```
# Least squares optimization
Many optimization problems involve minimization of a sum of squared residuals. We will take a look at finding the derivatives for least squares minimization.
In least squares problems, we usually have $m$ labeled observations $(x_i, y_i)$. We have a model that will predict $y_i$ given $x_i$ for some parameters $\beta$, $f(x) = X\beta$. We want to minimize the sum (or average) of squared residuals $r(x_i) = y_i - f(x_i)$. For example, the objective function is usually taken to be
$$
\frac{1}{2} \sum{r(x_i)^2}
$$
As a concrete example, suppose we want to fit a quadratic function to some observed data. We have
$$
f(x) = \beta_0 + \beta_1 x + \beta_2 x^2
$$
We want to minimize the objective function
$$
L = \frac{1}{2} \sum_{i=1}^m (y_i - f(x_i))^2
$$
Taking derivatives with respect to $\beta$, we get
$$
\frac{dL}{d\beta} =
\begin{bmatrix}
\sum_{i=1}^m f(x_i) - y_i \\
\sum_{i=1}^m x_i (f(x_i) - y_i) \\
\sum_{i=1}^m x_i^2 (f(x_i) - y_i)
\end{bmatrix}
$$
## Working with matrices
Writing the above system as a matrix, we have $f(x) = X\beta$, with
$$
X = \begin{bmatrix}
1 & x_1 & x_1^2 \\
1 & x_2 & x_2^2 \\
\vdots & \vdots & \vdots \\
1 & x_m & x_m^2
\end{bmatrix}
$$
and
$$
\beta = \begin{bmatrix}
\beta_0 \\
\beta_1 \\
\beta_2
\end{bmatrix}
$$
We want to find the derivative of $\Vert y - X\beta \Vert^2$, so
$$
\Vert y - X\beta \Vert^2 \\
= (y - X\beta)^T(y - X\beta) \\
= (y^T - \beta^TX^T)(y - X\beta) \\
= y^Ty - \beta^TX^Ty -y^TX\beta + \beta^TX^TX\beta
$$
Taking derivatives with respect to $\beta^T$ (we do this because the gradient is traditionally a row vector, and we want it as a column vector here), we get (after multiplying by $1/2$ for the residue function)
$$
\frac{dL}{d\beta^T} = X^TX\beta - X^Ty
$$
For example, if we are doing gradient descent, the update equation is
$$
\beta_{k+1} = \beta_k + \alpha (X^TX\beta - X^Ty)
$$
Note that if we set the derivative to zero and solve, we get
$$
X^TX\beta - X^Ty = 0
$$
and the normal equations
$$
\beta = (X^TX)^{-1}X^Ty
$$
For large $X$, solving the normal equations can be more expensive than simpler gradient descent. Note that the Levenberg-Marquadt algorithm is often used to optimize least squares problems.
## Example
You are given the following set of data to fit a quadratic polynomial to:
```python
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
```
Find the least squares solution using gradient descent.
```
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
def f(x, y, b):
return (b[0] + b[1]*x + b[2]*x**2 - y)
def res(x, y, b):
return sum(f(x,y, b)*f(x, y, b))
# Elementary form of gradient
def grad(x, y, b):
n = len(x)
return np.array([
sum(f(x, y, b)),
sum(x*f(x, y, b)),
sum(x**2*f(x, y, b))
])
# Matrix form of gradient
def grad_m(X, y, b):
return X.T@X@b- X.T@y
grad(x, y, np.zeros(3))
X = np.c_[np.ones(len(x)), x, x**2]
grad_m(X, y, np.zeros(3))
from scipy.linalg import solve
beta1 = solve(X.T@X, X.T@y)
beta1
max_iter = 10000
a = 0.0001 # learning rate
beta2 = np.zeros(3)
for i in range(max_iter):
beta2 -= a * grad(x, y, beta2)
beta2
a = 0.0001 # learning rate
beta3 = np.zeros(3)
for i in range(max_iter):
beta3 -= a * grad_m(X, y, beta3)
beta3
titles = ['svd', 'elementary', 'matrix']
plt.figure(figsize=(12,4))
for i, beta in enumerate([beta1, beta2, beta3], 1):
plt.subplot(1, 3, i)
plt.scatter(x, y, s=30)
plt.plot(x, beta[0] + beta[1]*x + beta[2]*x**2, color='red')
plt.title(titles[i-1])
```
### Curve fitting and least squares optimization
As shown above, least squares optimization is the technique most associated with curve fitting. For convenience, `scipy.optimize` provides a `curve_fit` function that uses Levenberg-Marquadt for minimization.
```
from scipy.optimize import curve_fit
def logistic4(x, a, b, c, d):
"""The four paramter logistic function is often used to fit dose-response relationships."""
return ((a-d)/(1.0+((x/c)**b))) + d
nobs = 24
xdata = np.linspace(0.5, 3.5, nobs)
ptrue = [10, 3, 1.5, 12]
ydata = logistic4(xdata, *ptrue) + 0.5*np.random.random(nobs)
popt, pcov = curve_fit(logistic4, xdata, ydata)
perr = yerr=np.sqrt(np.diag(pcov))
print('Param\tTrue\tEstim (+/- 1 SD)')
for p, pt, po, pe in zip('abcd', ptrue, popt, perr):
print('%s\t%5.2f\t%5.2f (+/-%5.2f)' % (p, pt, po, pe))
x = np.linspace(0, 4, 100)
y = logistic4(x, *popt)
plt.plot(xdata, ydata, 'o')
plt.plot(x, y)
pass
```
|
github_jupyter
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
x = np.arange(10)
y = np.array([ 1.58873597, 7.55101533, 10.71372171, 7.90123225,
-2.05877605, -12.40257359, -28.64568712, -46.39822281,
-68.15488905, -97.16032044])
def f(x, y, b):
return (b[0] + b[1]*x + b[2]*x**2 - y)
def res(x, y, b):
return sum(f(x,y, b)*f(x, y, b))
# Elementary form of gradient
def grad(x, y, b):
n = len(x)
return np.array([
sum(f(x, y, b)),
sum(x*f(x, y, b)),
sum(x**2*f(x, y, b))
])
# Matrix form of gradient
def grad_m(X, y, b):
return X.T@X@b- X.T@y
grad(x, y, np.zeros(3))
X = np.c_[np.ones(len(x)), x, x**2]
grad_m(X, y, np.zeros(3))
from scipy.linalg import solve
beta1 = solve(X.T@X, X.T@y)
beta1
max_iter = 10000
a = 0.0001 # learning rate
beta2 = np.zeros(3)
for i in range(max_iter):
beta2 -= a * grad(x, y, beta2)
beta2
a = 0.0001 # learning rate
beta3 = np.zeros(3)
for i in range(max_iter):
beta3 -= a * grad_m(X, y, beta3)
beta3
titles = ['svd', 'elementary', 'matrix']
plt.figure(figsize=(12,4))
for i, beta in enumerate([beta1, beta2, beta3], 1):
plt.subplot(1, 3, i)
plt.scatter(x, y, s=30)
plt.plot(x, beta[0] + beta[1]*x + beta[2]*x**2, color='red')
plt.title(titles[i-1])
from scipy.optimize import curve_fit
def logistic4(x, a, b, c, d):
"""The four paramter logistic function is often used to fit dose-response relationships."""
return ((a-d)/(1.0+((x/c)**b))) + d
nobs = 24
xdata = np.linspace(0.5, 3.5, nobs)
ptrue = [10, 3, 1.5, 12]
ydata = logistic4(xdata, *ptrue) + 0.5*np.random.random(nobs)
popt, pcov = curve_fit(logistic4, xdata, ydata)
perr = yerr=np.sqrt(np.diag(pcov))
print('Param\tTrue\tEstim (+/- 1 SD)')
for p, pt, po, pe in zip('abcd', ptrue, popt, perr):
print('%s\t%5.2f\t%5.2f (+/-%5.2f)' % (p, pt, po, pe))
x = np.linspace(0, 4, 100)
y = logistic4(x, *popt)
plt.plot(xdata, ydata, 'o')
plt.plot(x, y)
pass
| 0.580114 | 0.985663 |
```
%load_ext autoreload
%autoreload 2
```
# A simple sentiment prototype
```
import os # manipulate paths
import pandas as pd # SQL-like operations and convenience functions
import joblib # save and load models
```
Download the Sentiment140 data from [their website](http://help.sentiment140.com/for-students) or directly from [Standford site](http://cs.stanford.edu/people/alecmgo/trainingandtestdata.zip) and set `DATA_DIR` to the directory in which you have put the `CSV` files.
```
DATA_DIR = "./../data"
training_csv_file = os.path.join(DATA_DIR, 'training.1600000.processed.noemoticon.csv')
training_csv_file
```
## A peek at the data
```
names = ('polarity', 'id', 'date', 'query', 'author', 'text')
df = pd.read_csv(training_csv_file, encoding='latin1', names=names)
pd.options.display.max_colwidth = 140 # allow wide columns
df.head() # show first 5 rows
df.tail()
df['polarity'].replace({0: -1, 4: 1}, inplace=True)
text = df['text']
target = df['polarity'].values
print(len(target), len(text))
```
## Train the model
Set 20% of the data aside to test the trained model
```
from sklearn.cross_validation import train_test_split
text_train, text_validation, target_train, target_validation = (
train_test_split(text, target, test_size=0.2, random_state=42)
)
```
Build a pipeline
```
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_selection import SelectKBest, chi2
from sklearn.linear_model import LogisticRegressionCV
from sklearn.pipeline import Pipeline
vectorizer = CountVectorizer(ngram_range=(1, 2), max_features=100000)
feature_selector = SelectKBest(chi2, k=5000)
classifier = LogisticRegressionCV(n_jobs=4)
```
This next cell took ~3 minutes to run on my machine
```
if os.path.exists('model.pkl'):
sentiment_pipeline = joblib.load('model.pkl')
else:
sentiment_pipeline = Pipeline((
('v', vectorizer),
('f', feature_selector),
('c', classifier)
))
sentiment_pipeline.fit(text_train, target_train)
joblib.dump(sentiment_pipeline, 'model.pkl');
```
## Test the model
```
print(sentiment_pipeline.predict(['bad', 'good', "didnt like", "today was a good day", "i hate this product"]))
for text, target in zip(text_validation[:10], target_validation[:10]):
print(sentiment_pipeline.predict([text])[0], target, '\t', text)
sentiment_pipeline.score(text_validation, target_validation)
```
## What did the model learn?
```
feature_names = sentiment_pipeline.steps[0][1].get_feature_names()
feature_names = [feature_names[i] for i in
sentiment_pipeline.steps[1][1].get_support(indices=True)]
def show_most_informative_features(feature_names, clf, n=1000):
coefs_with_fns = sorted(zip(clf.coef_[0], feature_names))
top = zip(coefs_with_fns[:n], coefs_with_fns[:-(n + 1):-1])
for (coef_1, fn_1), (coef_2, fn_2) in top:
print("\t%.4f\t%-15s\t\t%.4f\t%-15s" % (coef_1, fn_1, coef_2, fn_2))
show_most_informative_features(feature_names, sentiment_pipeline.steps[2][1], n=500)
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
import os # manipulate paths
import pandas as pd # SQL-like operations and convenience functions
import joblib # save and load models
DATA_DIR = "./../data"
training_csv_file = os.path.join(DATA_DIR, 'training.1600000.processed.noemoticon.csv')
training_csv_file
names = ('polarity', 'id', 'date', 'query', 'author', 'text')
df = pd.read_csv(training_csv_file, encoding='latin1', names=names)
pd.options.display.max_colwidth = 140 # allow wide columns
df.head() # show first 5 rows
df.tail()
df['polarity'].replace({0: -1, 4: 1}, inplace=True)
text = df['text']
target = df['polarity'].values
print(len(target), len(text))
from sklearn.cross_validation import train_test_split
text_train, text_validation, target_train, target_validation = (
train_test_split(text, target, test_size=0.2, random_state=42)
)
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_selection import SelectKBest, chi2
from sklearn.linear_model import LogisticRegressionCV
from sklearn.pipeline import Pipeline
vectorizer = CountVectorizer(ngram_range=(1, 2), max_features=100000)
feature_selector = SelectKBest(chi2, k=5000)
classifier = LogisticRegressionCV(n_jobs=4)
if os.path.exists('model.pkl'):
sentiment_pipeline = joblib.load('model.pkl')
else:
sentiment_pipeline = Pipeline((
('v', vectorizer),
('f', feature_selector),
('c', classifier)
))
sentiment_pipeline.fit(text_train, target_train)
joblib.dump(sentiment_pipeline, 'model.pkl');
print(sentiment_pipeline.predict(['bad', 'good', "didnt like", "today was a good day", "i hate this product"]))
for text, target in zip(text_validation[:10], target_validation[:10]):
print(sentiment_pipeline.predict([text])[0], target, '\t', text)
sentiment_pipeline.score(text_validation, target_validation)
feature_names = sentiment_pipeline.steps[0][1].get_feature_names()
feature_names = [feature_names[i] for i in
sentiment_pipeline.steps[1][1].get_support(indices=True)]
def show_most_informative_features(feature_names, clf, n=1000):
coefs_with_fns = sorted(zip(clf.coef_[0], feature_names))
top = zip(coefs_with_fns[:n], coefs_with_fns[:-(n + 1):-1])
for (coef_1, fn_1), (coef_2, fn_2) in top:
print("\t%.4f\t%-15s\t\t%.4f\t%-15s" % (coef_1, fn_1, coef_2, fn_2))
show_most_informative_features(feature_names, sentiment_pipeline.steps[2][1], n=500)
| 0.359589 | 0.869493 |
## Is this just magic? What is Numba doing to make code run quickly?
Let's define a trivial example function.
```
from numba import jit
@jit
def add(a, b):
return a + b
add(1, 1)
```
Numba examines Python bytecode and then translates this into an 'intermediate representation'. To view this IR, run (compile) `add` and you can access the `inspect_types` method.
```
add.inspect_types()
```
Ok. Numba is has correctly inferred the type of the arguments, defining things as `int64` and running smoothly.
(What happens if you do `add(1., 1.)` and then `inspect_types`?)
```
add(1., 1.)
add.inspect_types()
```
### What about the actual LLVM code?
You can see the actual LLVM code generated by Numba using the `inspect_llvm()` method. Since it's a `dict`, doing the following will be slightly more visually friendly.
```
for k, v in add.inspect_llvm().items():
print(k, v)
```
## But there's a caveat
Now, watch what happens when the function we want to speed-up operates on object data types.
```
def add_object_n2_times(a, b, n):
cumsum = 0
for i in range(n):
for j in range(n):
cumsum += a.x + b.x
return cumsum
class MyInt(object):
def __init__(self, x):
self.x = x
a = MyInt(5)
b = MyInt(6)
%timeit add_object_n2_times(a, b, 500)
add_object_jit = jit()(add_object_n2_times)
%timeit add_object_jit(a, b, 500)
add_object_jit.inspect_types()
```
## What's all this pyobject business?
This means it has been compiled in `object` mode. This can be a faster than regular python if it can do loop lifting, but not that fast.
We want those `pyobjects` to be `int64` or another type that can be inferred by Numba. Your best bet is forcing `nopython` mode: this will throw an error if Numba finds itself in object mode, so that you _know_ that it can't give you speed.
For the full list of supported Python and NumPy features in `nopython` mode, see the Numba documentation here: http://numba.pydata.org/numba-doc/latest/reference/pysupported.html
## Figuring out what isn't working
```
%%file nopython_failure.py
from numba import jit
class MyInt(object):
def __init__(self, x):
self.x = x
@jit
def add_object(a, b):
for i in range(100):
c = i
f = i + 7
l = c + f
return a.x + b.x
a = MyInt(5)
b = MyInt(6)
add_object(a, b)
!numba --annotate-html fail.html nopython_failure.py
```
[fail.html](fail.html)
## Forcing `nopython` mode
```
add_object_jit = jit(nopython=True)(add_object_n2_times)
# This will fail
add_object_jit(a, b, 5)
from numba import njit
add_object_jit = njit(add_object_n2_times)
# This will also fail
add_object_jit(a, b, 5)
```
## Other compilation flags
There are two other main compilation flags for `@jit`
```python
cache=True
```
if you don't want to always want to get dinged by the compilation time for every run
```python
nogil=True
```
This releases the GIL. Note, however, that it doesn't do anything else, like make your program threadsafe. You have to manage all of those things on your own (use `concurrent.futures`).
|
github_jupyter
|
from numba import jit
@jit
def add(a, b):
return a + b
add(1, 1)
add.inspect_types()
add(1., 1.)
add.inspect_types()
for k, v in add.inspect_llvm().items():
print(k, v)
def add_object_n2_times(a, b, n):
cumsum = 0
for i in range(n):
for j in range(n):
cumsum += a.x + b.x
return cumsum
class MyInt(object):
def __init__(self, x):
self.x = x
a = MyInt(5)
b = MyInt(6)
%timeit add_object_n2_times(a, b, 500)
add_object_jit = jit()(add_object_n2_times)
%timeit add_object_jit(a, b, 500)
add_object_jit.inspect_types()
%%file nopython_failure.py
from numba import jit
class MyInt(object):
def __init__(self, x):
self.x = x
@jit
def add_object(a, b):
for i in range(100):
c = i
f = i + 7
l = c + f
return a.x + b.x
a = MyInt(5)
b = MyInt(6)
add_object(a, b)
!numba --annotate-html fail.html nopython_failure.py
add_object_jit = jit(nopython=True)(add_object_n2_times)
# This will fail
add_object_jit(a, b, 5)
from numba import njit
add_object_jit = njit(add_object_n2_times)
# This will also fail
add_object_jit(a, b, 5)
cache=True
nogil=True
| 0.399343 | 0.887205 |
# Parsing Inputs
In the chapter on [Grammars](Grammars.ipynb), we discussed how grammars can be
used to represent various languages. We also saw how grammars can be used to
generate strings of the corresponding language. Grammars can also perform the
reverse. That is, given a string, one can decompose the string into its
constituent parts that correspond to the parts of grammar used to generate it
– the _derivation tree_ of that string. These parts (and parts from other similar
strings) can later be recombined using the same grammar to produce new strings.
In this chapter, we use grammars to parse and decompose a given set of valid seed inputs into their corresponding derivation trees. This structural representation allows us to mutate, crossover, and recombine their parts in order to generate new valid, slightly changed inputs (i.e., fuzz)
**Prerequisites**
* You should have read the [chapter on grammars](Grammars.ipynb).
* An understanding of derivation trees from the [chapter on grammar fuzzer](GrammarFuzzer.ipynb)
is also required.
## Synopsis
<!-- Automatically generated. Do not edit. -->
To [use the code provided in this chapter](Importing.ipynb), write
```python
>>> from fuzzingbook.Parser import <identifier>
```
and then make use of the following features.
This chapter introduces `Parser` classes, parsing a string into a _derivation tree_ as introduced in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb). Two important parser classes are provided:
* [Parsing Expression Grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`), which are very efficient, but limited to specific grammar structure; and
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`), which accept any kind of context-free grammars.
Using any of these is fairly easy, though. First, instantiate them with a grammar:
```python
>>> from Grammars import US_PHONE_GRAMMAR
>>> us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
```
Then, use the `parse()` method to retrieve a list of possible derivation trees:
```python
>>> trees = us_phone_parser.parse("(555)987-6543")
>>> tree = list(trees)[0]
>>> display_tree(tree)
```

These derivation trees can then be used for test generation, notably for mutating and recombining existing inputs.
## Fuzzing a Simple Program
Here is a simple program that accepts a CSV file of vehicle details and processes this information.
```
def process_inventory(inventory):
res = []
for vehicle in inventory.split('\n'):
ret = process_vehicle(vehicle)
res.extend(ret)
return '\n'.join(res)
```
The CSV file contains details of one vehicle per line. Each row is processed in `process_vehicle()`.
```
def process_vehicle(vehicle):
year, kind, company, model, *_ = vehicle.split(',')
if kind == 'van':
return process_van(year, company, model)
elif kind == 'car':
return process_car(year, company, model)
else:
raise Exception('Invalid entry')
```
Depending on the kind of vehicle, the processing changes.
```
def process_van(year, company, model):
res = ["We have a %s %s van from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2010:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
def process_car(year, company, model):
res = ["We have a %s %s car from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2016:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
```
Here is a sample of inputs that the `process_inventory()` accepts.
```
mystring = """\
1997,van,Ford,E350
2000,car,Mercury,Cougar\
"""
print(process_inventory(mystring))
```
Let us try to fuzz this program. Given that the `process_inventory()` takes a CSV file, we can write a simple grammar for generating comma separated values, and generate the required CSV rows. For convenience, we fuzz `process_vehicle()` directly.
```
import string
CSV_GRAMMAR = {
'<start>': ['<csvline>'],
'<csvline>': ['<items>'],
'<items>': ['<item>,<items>', '<item>'],
'<item>': ['<letters>'],
'<letters>': ['<letter><letters>', '<letter>'],
'<letter>': list(string.ascii_letters + string.digits + string.punctuation + ' \t\n')
}
```
We need some infrastructure first for viewing the grammar.
```
import fuzzingbook_utils
from Grammars import EXPR_GRAMMAR, START_SYMBOL, RE_NONTERMINAL, is_valid_grammar, syntax_diagram
from Fuzzer import Fuzzer
from GrammarFuzzer import GrammarFuzzer, FasterGrammarFuzzer, display_tree, tree_to_string, dot_escape
from ExpectError import ExpectError
from Timer import Timer
syntax_diagram(CSV_GRAMMAR)
```
We generate `1000` values, and evaluate the `process_vehicle()` with each.
```
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 1000
valid = []
time = 0
for i in range(trials):
with Timer() as t:
vehicle_info = gf.fuzz()
try:
process_vehicle(vehicle_info)
valid.append(vehicle_info)
except:
pass
time += t.elapsed_time()
print("%d valid strings, that is GrammarFuzzer generated %f%% valid entries from %d inputs" %
(len(valid), len(valid) * 100.0 / trials, trials))
print("Total time of %f seconds" % time)
```
This is obviously not working. But why?
```
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 10
valid = []
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
```
None of the entries will get through unless the fuzzer can produce either `van` or `car`.
Indeed, the reason is that the grammar itself does not capture the complete information about the format. So here is another idea. We modify the `GrammarFuzzer` to know a bit about our format.
```
import copy
import random
class PooledGrammarFuzzer(GrammarFuzzer):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self._node_cache = {}
def update_cache(self, key, values):
self._node_cache[key] = values
def expand_node_randomly(self, node):
(symbol, children) = node
assert children is None
if symbol in self._node_cache:
if random.randint(0, 1) == 1:
return super().expand_node_randomly(node)
return copy.deepcopy(random.choice(self._node_cache[symbol]))
return super().expand_node_randomly(node)
```
Let us try again!
```
gf = PooledGrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
gf.update_cache('<item>', [
('<item>', [('car', [])]),
('<item>', [('van', [])]),
])
trials = 10
valid = []
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
```
At least we are getting somewhere! It would be really nice if _we could incorporate what we know about the sample data in our fuzzer._ In fact, it would be nice if we could _extract_ the template and valid values from samples, and use them in our fuzzing. How do we do that? The quick answer to this question is: Use a *parser*.
## Using a Parser
Generally speaking, a _parser_ is the part of a a program that processes (structured) input. The parsers we discuss in this chapter transform an input string into a _derivation tree_ (discussed in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb)). From a user's perspective, all it takes to parse an input is two steps:
1. Initialize the parser with a grammar, as in
```
parser = Parser(grammar)
```
2. Using the parser to retrieve a list of derivation trees:
```python
trees = parser.parse(input)
```
Once we have parsed a tree, we can use it just as the derivation trees produced from grammar fuzzing.
We discuss a number of such parsers, in particular
* [parsing expression grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`), which are very efficient, but limited to specific grammar structure; and
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`), which accept any kind of context-free grammars.
If you just want to _use_ parsers (say, because your main focus is testing), you can just stop here and move on [to the next chapter](LangFuzzer.ipynb), where we learn how to make use of parsed inputs to mutate and recombine them. If you want to _understand_ how parsers work, though, this chapter is right for you.
## An Ad Hoc Parser
As we saw in the previous section, programmers often have to extract parts of data that obey certain rules. For example, for *CSV* files, each element in a row is separated by *commas*, and multiple raws are used to store the data.
To extract the information, we write an ad hoc parser `parse_csv()`.
```
def parse_csv(mystring):
children = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in line.split(',')]))
return tree
```
We also change the default orientation of the graph to *left to right* rather than *top to bottom* for easier viewing using `lr_graph()`.
```
def lr_graph(dot):
dot.attr('node', shape='plain')
dot.graph_attr['rankdir'] = 'LR'
```
The `display_tree()` shows the structure of our CSV file after parsing.
```
tree = parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
```
This is of course simple. What if we encounter slightly more complexity? Again, another example from the Wikipedia.
```
mystring = '''\
1997,Ford,E350,"ac, abs, moon",3000.00\
'''
print(mystring)
```
We define a new annotation method `highlight_node()` to mark the nodes that are interesting.
```
def highlight_node(predicate):
def hl_node(dot, nid, symbol, ann):
if predicate(dot, nid, symbol, ann):
dot.node(repr(nid), dot_escape(symbol), fontcolor='red')
else:
dot.node(repr(nid), dot_escape(symbol))
return hl_node
```
Using `highlight_node()` we can highlight particular nodes that we were wrongly parsed.
```
tree = parse_csv(mystring)
bad_nodes = {5, 6, 7, 12, 13, 20, 22, 23, 24, 25}
def hl_predicate(_d, nid, _s, _a): return nid in bad_nodes
highlight_err_node = highlight_node(hl_predicate)
display_tree(tree, log=False, node_attr=highlight_err_node,
graph_attr=lr_graph)
```
The marked nodes indicate where our parsing went wrong. We can of course extend our parser to understand quotes. First we define some of the helper functions `parse_quote()`, `find_comma()` and `comma_split()`
```
def parse_quote(string, i):
v = string[i + 1:].find('"')
return v + i + 1 if v >= 0 else -1
def find_comma(string, i):
slen = len(string)
while i < slen:
if string[i] == '"':
i = parse_quote(string, i)
if i == -1:
return -1
if string[i] == ',':
return i
i += 1
return -1
def comma_split(string):
slen = len(string)
i = 0
while i < slen:
c = find_comma(string, i)
if c == -1:
yield string[i:]
return
else:
yield string[i:c]
i = c + 1
```
We can update our `parse_csv()` procedure to use our advanced quote parser.
```
def parse_csv(mystring):
children = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in comma_split(line)]))
return tree
```
Our new `parse_csv()` can now handle quotes correctly.
```
tree = parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
```
That of course does not survive long:
```
mystring = '''\
1999,Chevy,"Venture \\"Extended Edition, Very Large\\"",,5000.00\
'''
print(mystring)
```
A few embedded quotes are sufficient to confuse our parser again.
```
tree = parse_csv(mystring)
bad_nodes = {4, 5}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
```
Here is another record from that CSV file:
```
mystring = '''\
1996,Jeep,Grand Cherokee,"MUST SELL!
air, moon roof, loaded",4799.00
'''
print(mystring)
tree = parse_csv(mystring)
bad_nodes = {5, 6, 7, 8, 9, 10}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
```
Fixing this would require modifying both inner `parse_quote()` and the outer `parse_csv()` procedures. We note that each of these features actually documented in the CSV [RFC 4180](https://tools.ietf.org/html/rfc4180)
Indeed, each additional improvement falls apart even with a little extra complexity. The problem becomes severe when one encounters recursive expressions. For example, JSON is a common alternative to CSV files for saving data. Similarly, one may have to parse data from an HTML table instead of a CSV file if one is getting the data from the web.
One might be tempted to fix it with a little more ad hoc parsing, with a bit of *regular expressions* thrown in. However, that is the [path to insanity](https://stackoverflow.com/a/1732454).
It is here that _formal parsers_ shine. The main idea is that, any given set of strings belong to a language, and these languages can be specified by their grammars (as we saw in the [chapter on grammars](Grammars.ipynb)). The great thing about grammars is that they can be _composed_. That is, one can introduce finer and finer details into an internal structure without affecting the external structure, and similarly, one can change the external structure without much impact on the internal structure. We briefly describe grammars in the next section.
## Grammars
A grammar, as you have read from the [chapter on grammars](Grammars.ipynb) is a set of _rules_ that explain how the start symbol can be expanded. Each rule has a name, also called a _nonterminal_, and a set of _alternative choices_ in how the nonterminal can be expanded.
```
A1_GRAMMAR = {
"<start>": ["<expr>"],
"<expr>": ["<expr>+<expr>", "<expr>-<expr>", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A1_GRAMMAR)
```
In the above expression, the rule `<expr> : [<expr>+<expr>,<expr>-<expr>,<integer>]` corresponds to how the nonterminal `<expr>` might be expanded. The expression `<expr>+<expr>` corresponds to one of the alternative choices. We call this an _alternative_ expansion for the nonterminal `<expr>`. Finally, in an expression `<expr>+<expr>`, each of `<expr>`, `+`, and `<expr>` are _symbols_ in that expansion. A symbol could be either a nonterminal or a terminal symbol based on whether its expansion is available in the grammar.
Here is a string that represents an arithmetic expression that we would like to parse, which is specified by the grammar above:
```
mystring = '1+2'
```
The _derivation tree_ for our expression from this grammar is given by:
```
tree = ('<start>', [('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('2',
[])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
```
While a grammar can be used to specify a given language, there could be multiple
grammars that correspond to the same language. For example, here is another
grammar to describe the same addition expression.
```
A2_GRAMMAR = {
"<start>": ["<expr>"],
"<expr>": ["<integer><expr_>"],
"<expr_>": ["+<expr>", "-<expr>", ""],
"<integer>": ["<digit><integer_>"],
"<integer_>": ["<integer>", ""],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A2_GRAMMAR)
```
The corresponding derivation tree is given by:
```
tree = ('<start>', [('<expr>', [('<integer>', [('<digit>', [('1', [])]),
('<integer_>', [])]),
('<expr_>', [('+', []),
('<expr>',
[('<integer>',
[('<digit>', [('2', [])]),
('<integer_>', [])]),
('<expr_>', [])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
```
Indeed, there could be different classes of grammars that
describe the same language. For example, the first grammar `A1_GRAMMAR`
is a grammar that sports both _right_ and _left_ recursion, while the
second grammar `A2_GRAMMAR` does not have left recursion in the
nonterminals in any of its productions, but contains _epsilon_ productions.
(An epsilon production is a production that has empty string in its right
hand side.)
You would have noticed that we reuse the term `<expr>` in its own definition. Using the same nonterminal in its own definition is called *recursion*. There are two specific kinds of recursion one should be aware of in parsing, as we see in the next section.
#### Recursion
A grammar is _left recursive_ if any of its nonterminals are left recursive,
and a nonterminal is directly left-recursive if the left-most symbol of
any of its productions is itself.
```
LR_GRAMMAR = {
'<start>': ['<A>'],
'<A>': ['<A>a', ''],
}
syntax_diagram(LR_GRAMMAR)
mystring = 'aaaaaa'
display_tree(
('<start>', (('<A>', (('<A>', (('<A>', []), ('a', []))), ('a', []))), ('a', []))))
```
A grammar is indirectly left-recursive if any
of the left-most symbols can be expanded using their definitions to
produce the nonterminal as the left-most symbol of the expansion. The left
recursion is called a _hidden-left-recursion_ if during the series of
expansions of a nonterminal, one reaches a rule where the rule contains
the same nonterminal after a prefix of other symbols, and these symbols can
derive the empty string. For example, in `A1_GRAMMAR`, `<integer>` will be
considered hidden-left recursive if `<digit>` could derive an empty string.
Right recursive grammars are defined similarly.
Below is the derivation tree for the right recursive grammar that represents the same
language as that of `LR_GRAMMAR`.
```
RR_GRAMMAR = {
'<start>': ['<A>'],
'<A>': ['a<A>', ''],
}
syntax_diagram(RR_GRAMMAR)
display_tree(('<start>', ((
'<A>', (('a', []), ('<A>', (('a', []), ('<A>', (('a', []), ('<A>', []))))))),)))
```
#### Ambiguity
To complicate matters further, there could be
multiple derivation trees – also called _parses_ – corresponding to the
same string from the same grammar. For example, a string `1+2+3` can be parsed
in two ways as we see below using the `A1_GRAMMAR`
```
mystring = '1+2+3'
tree = ('<start>',
[('<expr>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>',
[('<digit>', [('2', [])])])])]), ('+', []),
('<expr>', [('<integer>', [('<digit>', [('3', [])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
tree = ('<start>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('2', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('3',
[])])])])])])])
assert tree_to_string(tree) == mystring
display_tree(tree)
```
There are many ways to resolve ambiguities. One approach taken by *Parsing Expression Grammars* explained in the next section is to specify a particular order of resolution, and choose the first one. Another approach is to simply return all possible derivation trees, which is the approach taken by *Earley parser* we develop later.
Next, we develop different parsers. To do that, we define a minimal interface for parsing that is obeyed by all parsers. There are two approaches to parsing a string using a grammar.
1. The traditional approach is to use a *lexer* (also called a *tokenizer* or a *scanner*) to first tokenize the incoming string, and feed the grammar one token at a time. The lexer is typically a smaller parser that accepts a *regular language*. The advantage of this approach is that the grammar used by the parser can eschew the details of tokenization. Further, one gets a shallow derivation tree at the end of the parsing which can be directly used for generating the *Abstract Syntax Tree*.
2. The second approach is to use a tree pruner after the complete parse. With this approach, one uses a grammar that incorporates complete details of the syntax. Next, the nodes corresponding to tokens are pruned and replaced with their corresponding strings as leaf nodes. The utility of this approach is that the parser is more powerful, and further there is no artificial distinction between *lexing* and *parsing*.
In this chapter, we use the second approach. This approach is implemented in the `prune_tree` method.
The *Parser* class we define below provides the minimal interface. The main methods that need to be implemented by the classes implementing this interface are `parse_prefix` and `parse`. The `parse_prefix` returns a tuple, which contains the index until which parsing was completed successfully, and the parse forest until that index. The method `parse` returns a list of derivation trees if the parse was successful.
```
class Parser(object):
def __init__(self, grammar, **kwargs):
self._grammar = grammar
self._start_symbol = kwargs.get('start_symbol', START_SYMBOL)
self.log = kwargs.get('log', False)
self.coalesce_tokens = kwargs.get('coalesce', True)
self.tokens = kwargs.get('tokens', set())
def grammar(self):
return self._grammar
def start_symbol(self):
return self._start_symbol
def parse_prefix(self, text):
"""Return pair (cursor, forest) for longest prefix of text"""
raise NotImplemented()
def parse(self, text):
cursor, forest = self.parse_prefix(text)
if cursor < len(text):
raise SyntaxError("at " + repr(text[cursor:]))
return [self.prune_tree(tree) for tree in forest]
def coalesce(self, children):
last = ''
new_lst = []
for cn, cc in children:
if cn not in self._grammar:
last += cn
else:
if last:
new_lst.append((last, []))
last = ''
new_lst.append((cn, cc))
if last:
new_lst.append((last, []))
return new_lst
def prune_tree(self, tree):
name, children = tree
if self.coalesce_tokens:
children = self.coalesce(children)
if name in self.tokens:
return (name, [(tree_to_string(tree), [])])
else:
return (name, [self.prune_tree(c) for c in children])
```
## Parsing Expression Grammars
A _[Parsing Expression Grammar](http://bford.info/pub/lang/peg)_ (*PEG*) \cite{Ford2004} is a type of _recognition based formal grammar_ that specifies the sequence of steps to take to parse a given string.
A _parsing expression grammar_ is very similar to a _context-free grammar_ (*CFG*) such as the ones we saw in the [chapter on grammars](Grammars.ipynb). As in a CFG, a parsing expression grammar is represented by a set of nonterminals and corresponding alternatives representing how to match each. For example, here is a PEG that matches `a` or `b`.
```
PEG1 = {
'<start>': ['a', 'b']
}
```
However, unlike the _CFG_, the alternatives represent *ordered choice*. That is, rather than choosing all rules that can potentially match, we stop at the first match that succeed. For example, the below _PEG_ can match `ab` but not `abc` unlike a _CFG_ which will match both. (We call the sequence of ordered choice expressions *choice expressions* rather than alternatives to make the distinction from _CFG_ clear.)
```
PEG2 = {
'<start>': ['ab', 'abc']
}
```
Each choice in a _choice expression_ represents a rule on how to satisfy that particular choice. The choice is a sequence of symbols (terminals and nonterminals) that are matched against a given text as in a _CFG_.
Beyond the syntax of grammar definitions we have seen so far, a _PEG_ can also contain a few additional elements. See the exercises at the end of the chapter for additional information.
The PEGs model the typical practice in handwritten recursive descent parsers, and hence it may be considered more intuitive to understand. We look at parsers for PEGs next.
### The Packrat Parser for Predicate Expression Grammars
Short of hand rolling a parser, _Packrat_ parsing is one of the simplest parsing techniques, and is one of the techniques for parsing PEGs.
The _Packrat_ parser is so named because it tries to cache all results from simpler problems in the hope that these solutions can be used to avoid re-computation later. We develop a minimal _Packrat_ parser next.
But before that, we need to implement a few supporting tools.
The `EXPR_GRAMMAR` we import from the [chapter on grammars](Grammars.ipynb) is oriented towards generation. In particular, the production rules are stored as strings. We need to massage this representation a little to conform to a canonical representation where each token in a rule is represented separately. The `canonical` format uses separate tokens to represent each symbol in an expansion.
```
import re
def canonical(grammar, letters=False):
def split(expansion):
if isinstance(expansion, tuple):
expansion = expansion[0]
return [token for token in re.split(
RE_NONTERMINAL, expansion) if token]
def tokenize(word):
return list(word) if letters else [word]
def canonical_expr(expression):
return [
token for word in split(expression)
for token in ([word] if word in grammar else tokenize(word))
]
return {
k: [canonical_expr(expression) for expression in alternatives]
for k, alternatives in grammar.items()
}
CE_GRAMMAR = canonical(EXPR_GRAMMAR); CE_GRAMMAR
```
We also provide a way to revert a canonical expression.
```
def non_canonical(grammar):
new_grammar = {}
for k in grammar:
rules = grammar[k]
new_rules = []
for rule in rules:
new_rules.append(''.join(rule))
new_grammar[k] = new_rules
return new_grammar
non_canonical(CE_GRAMMAR)
```
It is easier to work with the `canonical` representation during parsing. Hence, we update our parser class to store the `canonical` representation also.
```
class Parser(Parser):
def __init__(self, grammar, **kwargs):
self._grammar = grammar
self._start_symbol = kwargs.get('start_symbol', START_SYMBOL)
self.log = kwargs.get('log', False)
self.tokens = kwargs.get('tokens', set())
self.coalesce_tokens = kwargs.get('coalesce', True)
self.cgrammar = canonical(grammar)
```
### The Parser
We derive from the `Parser` base class first, and we accept the text to be parsed in the `parse()` method, which in turn calls `unify_key()` with the `start_symbol`.
__Note.__ While our PEG parser can produce only a single unambiguous parse tree, other parsers can produce multiple parses for ambiguous grammars. Hence, we return a list of trees (in this case with a single element).
```
class PEGParser(Parser):
def parse_prefix(self, text):
cursor, tree = self.unify_key(self.start_symbol(), text, 0)
return cursor, [tree]
```
#### Unify Key
The `unify_key()` algorithm is simple. If given a terminal symbol, it tries to match the symbol with the current position in the text. If the symbol and text match, it returns successfully with the new parse index `at`.
If on the other hand, it was given a nonterminal, it retrieves the choice expression corresponding to the key, and tries to match each choice *in order* using `unify_rule()`. If **any** of the rules succeed in being unified with the given text, the parse is considered a success, and we return with the new parse index returned by `unify_rule()`.
```
class PEGParser(PEGParser):
def unify_key(self, key, text, at=0):
if self.log:
print("unify_key: %s with %s" % (repr(key), repr(text[at:])))
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
mystring = "1"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_key('1', mystring)
mystring = "2"
peg.unify_key('1', mystring)
```
#### Unify Rule
The `unify_rule()` method is similar. It retrieves the tokens corresponding to the rule that it needs to unify with the text, and calls `unify_key()` on them in sequence. If **all** tokens are successfully unified with the text, the parse is a success.
```
class PEGParser(PEGParser):
def unify_rule(self, rule, text, at):
if self.log:
print('unify_rule: %s with %s' % (repr(rule), repr(text[at:])))
results = []
for token in rule:
at, res = self.unify_key(token, text, at)
if res is None:
return at, None
results.append(res)
return at, results
mystring = "0"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_rule(peg.cgrammar['<digit>'][0], mystring, 0)
mystring = "12"
peg.unify_rule(peg.cgrammar['<integer>'][0], mystring, 0)
mystring = "1 + 2"
peg = PEGParser(EXPR_GRAMMAR, log=False)
peg.parse(mystring)
```
The two methods are mutually recursive, and given that `unify_key()` tries each alternative until it succeeds, `unify_key` can be called multiple times with the same arguments. Hence, it is important to memoize the results of `unify_key`. Python provides a simple decorator `lru_cache` for memoizing any function call that has hashable arguments. We add that to our implementation so that repeated calls to `unify_key()` with the same argument get cached results.
This memoization gives the algorithm its name – _Packrat_.
```
from functools import lru_cache
class PEGParser(PEGParser):
@lru_cache(maxsize=None)
def unify_key(self, key, text, at=0):
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
```
We wrap initialization and calling of `PEGParser` in a method `parse()` already implemented in the `Parser` base class that accepts the text to be parsed along with the grammar.
Here are a few examples of our parser in action.
```
mystring = "1 + (2 * 3)"
peg = PEGParser(EXPR_GRAMMAR)
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = "1 * (2 + 3.35)"
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
```
One should be aware that while the grammar looks like a *CFG*, the language described by a *PEG* may be different. Indeed, only *LL(1)* grammars are guaranteed to represent the same language for both PEGs and other parsers. Behavior of PEGs for other classes of grammars could be surprising \cite{redziejowski2008}.
## Parsing Context-Free Grammars
### Problems with PEG
While _PEGs_ are simple at first sight, their behavior in some cases might be a bit unintuitive. For example, here is an example \cite{redziejowski2008}:
```
PEG_SURPRISE = {
"<A>": ["a<A>a", "aa"]
}
```
When interpreted as a *CFG* and used as a string generator, it will produce strings of the form `aa, aaaa, aaaaaa` that is, it produces strings where the number of `a` is $ 2*n $ where $ n > 0 $.
```
strings = []
for e in range(4):
f = GrammarFuzzer(PEG_SURPRISE, start_symbol='<A>')
tree = ('<A>', None)
for _ in range(e):
tree = f.expand_tree_once(tree)
tree = f.expand_tree_with_strategy(tree, f.expand_node_min_cost)
strings.append(tree_to_string(tree))
display_tree(tree)
strings
```
However, the _PEG_ parser can only recognize strings of the form $2^n$
```
peg = PEGParser(PEG_SURPRISE, start_symbol='<A>')
for s in strings:
with ExpectError():
for tree in peg.parse(s):
display_tree(tree)
print(s)
```
This is not the only problem with _Parsing Expression Grammars_. While *PEGs* are expressive and the *packrat* parser for parsing them is simple and intuitive, *PEGs* suffer from a major deficiency for our purposes. *PEGs* are oriented towards language recognition, and it is not clear how to translate an arbitrary *PEG* to a *CFG*. As we mentioned earlier, a naive re-interpretation of a *PEG* as a *CFG* does not work very well. Further, it is not clear what is the exact relation between the class of languages represented by *PEG* and the class of languages represented by *CFG*. Since our primary focus is *fuzzing* – that is _generation_ of strings – , we next look at _parsers that can accept context-free grammars_.
The general idea of *CFG* parser is the following: Peek at the input text for the allowed number of characters, and use these, and our parser state to determine which rules can be applied to complete parsing. We next look at a typical *CFG* parsing algorithm, the Earley Parser.
### The Earley Parser
The Earley parser is a general parser that is able to parse any arbitrary *CFG*. It was invented by Jay Earley \cite{Earley1970} for use in computational linguistics. While its computational complexity is $O(n^3)$ for parsing strings with arbitrary grammars, it can parse strings with unambiguous grammars in $O(n^2)$ time, and all *[LR(k)](https://en.wikipedia.org/wiki/LR_parser)* grammars in linear time ($O(n)$ \cite{Leo1991}). Further improvements – notably handling epsilon rules – were invented by Aycock et al. \cite{Aycock2002}.
Note that one restriction of our implementation is that the start symbol can have only one alternative in its alternative expressions. This is not a restriction in practice because any grammar with multiple alternatives for its start symbol can be extended with a new start symbol that has the original start symbol as its only choice. That is, given a grammar as below,
```
grammar = {
'<start>': ['<A>', '<B>'],
...
}
```
one may rewrite it as below to conform to the *single-alternative* rule.
```
grammar = {
'<start>': ['<start_>'],
'<start_>': ['<A>', '<B>'],
...
}
```
We first implement a simpler parser that is a parser for nearly all *CFGs*, but not quite. In particular, our parser does not understand _epsilon rules_ – rules that derive empty string. We show later how the parser can be extended to handle these.
We use the following grammar in our examples below.
```
SAMPLE_GRAMMAR = {
'<start>': ['<A><B>'],
'<A>': ['a<B>c', 'a<A>'],
'<B>': ['b<C>', '<D>'],
'<C>': ['c'],
'<D>': ['d']
}
C_SAMPLE_GRAMMAR = canonical(SAMPLE_GRAMMAR)
syntax_diagram(SAMPLE_GRAMMAR)
```
The basic idea of Earley parsing is the following:
* Start with the alternative expressions corresponding to the START_SYMBOL. These represent the possible ways to parse the string from a high level. Essentially each expression represents a parsing path. Queue each expression in our set of possible parses of the string. The parsed index of an expression is the part of expression that has already been recognized. In the beginning of parse, the parsed index of all expressions is at the beginning. Further, each letter gets a queue of expressions that recognizes that letter at that point in our parse.
* Examine our queue of possible parses and check if any of them start with a nonterminal. If it does, then that nonterminal needs to be recognized from the input before the given rule can be parsed. Hence, add the alternative expressions corresponding to the nonterminal to the queue. Do this recursively.
* At this point, we are ready to advance. Examine the current letter in the input, and select all expressions that have that particular letter at the parsed index. These expressions can now advance one step. Advance these selected expressions by incrementing their parsed index and add them to the queue of expressions in line for recognizing the next input letter.
* If while doing these things, we find that any of the expressions have finished parsing, we fetch its corresponding nonterminal, and advance all expressions that have that nonterminal at their parsed index.
* Continue this procedure recursively until all expressions that we have queued for the current letter have been processed. Then start processing the queue for the next letter.
We explain each step in detail with examples in the coming sections.
The parser uses dynamic programming to generate a table containing a _forest of possible parses_ at each letter index – the table contains as many columns as there are letters in the input, and each column contains different parsing rules at various stages of the parse.
For example, given an input `adcd`, the Column 0 would contain the following:
```
<start> : ● <A> <B>
```
which is the starting rule that indicates that we are currently parsing the rule `<start>`, and the parsing state is just before identifying the symbol `<A>`. It would also contain the following which are two alternative paths it could take to complete the parsing.
```
<A> : ● a <B> c
<A> : ● a <A>
```
Column 1 would contain the following, which represents the possible completion after reading `a`.
```
<A> : a ● <B> c
<A> : a ● <A>
<B> : ● b <C>
<B> : ● <D>
<A> : ● a <B> c
<A> : ● a <A>
<D> : ● d
```
Column 2 would contain the following after reading `d`
```
<D> : d ●
<B> : <D> ●
<A> : a <B> ● c
```
Similarly, Column 3 would contain the following after reading `c`
```
<A> : a <B> c ●
<start> : <A> ● <B>
<B> : ● b <C>
<B> : ● <D>
<D> : ● d
```
Finally, Column 4 would contain the following after reading `d`, with the `●` at the end of the `<start>` rule indicating that the parse was successful.
```
<D> : d ●
<B> : <D> ●
<start> : <A> <B> ●
```
As you can see from above, we are essentially filling a table (a table is also called a **chart**) of entries based on each letter we read, and the grammar rules that can be applied. This chart gives the parser its other name -- Chart parsing.
### Columns
We define the `Column` first. The `Column` is initialized by its own `index` in the input string, and the `letter` at that index. Internally, we also keep track of the states that are added to the column as the parsing progresses.
```
class Column(object):
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique = [], {}
def __str__(self):
return "%s chart[%d]\n%s" % (self.letter, self.index, "\n".join(
str(state) for state in self.states if state.finished()))
```
The `Column` only stores unique `states`. Hence, when a new `state` is `added` to our `Column`, we check whether it is already known.
```
class Column(Column):
def add(self, state):
if state in self._unique:
return self._unique[state]
self._unique[state] = state
self.states.append(state)
state.e_col = self
return self._unique[state]
```
### Items
An item represents a _parse in progress for a specific rule._ Hence the item contains the name of the nonterminal, and the corresponding alternative expression (`expr`) which together form the rule, and the current position of parsing in this expression -- `dot`.
**Note.** If you are familiar with [LR parsing](https://en.wikipedia.org/wiki/LR_parser), you will notice that an item is simply an `LR0` item.
```
class Item(object):
def __init__(self, name, expr, dot):
self.name, self.expr, self.dot = name, expr, dot
```
We also provide a few convenience methods. The method `finished()` checks if the `dot` has moved beyond the last element in `expr`. The method `advance()` produces a new `Item` with the `dot` advanced one token, and represents an advance of the parsing. The method `at_dot()` returns the current symbol being parsed.
```
class Item(Item):
def finished(self):
return self.dot >= len(self.expr)
def advance(self):
return Item(self.name, self.expr, self.dot + 1)
def at_dot(self):
return self.expr[self.dot] if self.dot < len(self.expr) else None
```
Here is how an item could be used. We first define our item
```
item_name = '<B>'
item_expr = C_SAMPLE_GRAMMAR[item_name][1]
an_item = Item(item_name, tuple(item_expr), 0)
```
To determine where the status of parsing, we use `at_dot()`
```
an_item.at_dot()
```
That is, the next symbol to be parsed is `<D>`
If we advance the item, we get another item that represents the finished parsing rule `<B>`.
```
another_item = an_item.advance()
another_item.finished()
```
### States
For `Earley` parsing, the state of the parsing is simply one `Item` along with some meta information such as the starting `s_col` and ending column `e_col` for each state. Hence we inherit from `Item` to create a `State`.
Since we are interested in comparing states, we define `hash()` and `eq()` with the corresponding methods.
```
class State(Item):
def __init__(self, name, expr, dot, s_col, e_col=None):
super().__init__(name, expr, dot)
self.s_col, self.e_col = s_col, e_col
def __str__(self):
def idx(var):
return var.index if var else -1
return self.name + ':= ' + ' '.join([
str(p)
for p in [*self.expr[:self.dot], '|', *self.expr[self.dot:]]
]) + "(%d,%d)" % (idx(self.s_col), idx(self.e_col))
def copy(self):
return State(self.name, self.expr, self.dot, self.s_col, self.e_col)
def _t(self):
return (self.name, self.expr, self.dot, self.s_col.index)
def __hash__(self):
return hash(self._t())
def __eq__(self, other):
return self._t() == other._t()
def advance(self):
return State(self.name, self.expr, self.dot + 1, self.s_col)
```
The usage of `State` is similar to that of `Item`. The only difference is that it is used along with the `Column` to track the parsing state. For example, we initialize the first column as follows:
```
col_0 = Column(0, None)
item_expr = tuple(*C_SAMPLE_GRAMMAR[START_SYMBOL])
start_state = State(START_SYMBOL, item_expr, 0, col_0)
col_0.add(start_state)
start_state.at_dot()
```
The first column is then updated by using `add()` method of `Column`
```
sym = start_state.at_dot()
for alt in C_SAMPLE_GRAMMAR[sym]:
col_0.add(State(sym, tuple(alt), 0, col_0))
for s in col_0.states:
print(s)
```
### The Parsing Algorithm
The _Earley_ algorithm starts by initializing the chart with columns (as many as there are letters in the input). We also seed the first column with a state representing the expression corresponding to the start symbol. In our case, the state corresponds to the start symbol with the `dot` at `0` is represented as below. The `●` symbol represents the parsing status. In this case, we have not parsed anything.
```
<start>: ● <A> <B>
```
We pass this partial chart to a method for filling the rest of the parse chart.
```
class EarleyParser(Parser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.cgrammar = canonical(grammar, letters=True)
```
Before starting to parse, we seed the chart with the state representing the ongoing parse of the start symbol.
```
class EarleyParser(EarleyParser):
def chart_parse(self, words, start):
alt = tuple(*self.cgrammar[start])
chart = [Column(i, tok) for i, tok in enumerate([None, *words])]
chart[0].add(State(start, alt, 0, chart[0]))
return self.fill_chart(chart)
```
The main parsing loop in `fill_chart()` has three fundamental operations. `predict()`, `scan()`, and `complete()`. We discuss `predict` next.
### Predicting States
We have already seeded `chart[0]` with a state `[<A>,<B>]` with `dot` at `0`. Next, given that `<A>` is a nonterminal, we `predict` the possible parse continuations of this state. That is, it could be either `a <B> c` or `A <A>`.
The general idea of `predict()` is as follows: Say you have a state with name `<A>` from the above grammar, and expression containing `[a,<B>,c]`. Imagine that you have seen `a` already, which means that the `dot` will be on `<B>`. Below, is a representation of our parse status. The left hand side of ● represents the portion already parsed (`a`), and the right hand side represents the portion yet to be parsed (`<B> c`).
```
<A>: a ● <B> c
```
To recognize `<B>`, we look at the definition of `<B>`, which has different alternative expressions. The `predict()` step adds each of these alternatives to the set of states, with `dot` at `0`.
```
<A>: a ● <B> c
<B>: ● b c
<B>: ● <D>
```
In essence, the `predict()` method, when called with the current nonterminal, fetches the alternative expressions corresponding to this nonterminal, and adds these as predicted _child_ states to the _current_ column.
```
class EarleyParser(EarleyParser):
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
```
To see how to use `predict`, we first construct the 0th column as before, and we assign the constructed column to an instance of the EarleyParser.
```
col_0 = Column(0, None)
col_0.add(start_state)
ep = EarleyParser(SAMPLE_GRAMMAR)
ep.chart = [col_0]
```
It should contain a single state -- `<start> at 0`
```
for s in ep.chart[0].states:
print(s)
```
We apply predict to fill out the 0th column, and the column should contain the possible parse paths.
```
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
```
### Scanning Tokens
What if rather than a nonterminal, the state contained a terminal symbol such as a letter? In that case, we are ready to make some progress. For example, consider the second state:
```
<B>: ● b c
```
We `scan` the next column's letter. Say the next token is `b`.
If the letter matches what we have, then create a new state by advancing the current state by one letter.
```
<B>: b ● c
```
This new state is added to the next column (i.e the column that has the matched letter).
```
class EarleyParser(EarleyParser):
def scan(self, col, state, letter):
if letter == col.letter:
col.add(state.advance())
```
As before, we construct the partial parse first, this time adding a new column so that we can observe the effects of `scan()`
```
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
ep.chart = [col_0, col_1]
new_state = ep.chart[0].states[1]
print(new_state)
ep.scan(col_1, new_state, 'a')
for s in ep.chart[1].states:
print(s)
```
### Completing Processing
When we advance, what if we actually `complete()` the processing of the current rule? If so, we want to update not just this state, but also all the _parent_ states from which this state was derived.
For example, say we have states as below.
```
<A>: a ● <B> c
<B>: b c ●
```
The state `<B>: b c ●` is now complete. So, we need to advance `<A>: a ● <B> c` one step forward.
How do we determine the parent states? Note from `predict` that we added the predicted child states to the _same_ column as that of the inspected state. Hence, we look at the starting column of the current state, with the same symbol `at_dot` as that of the name of the completed state.
For each such parent found, we advance that parent (because we have just finished parsing that non terminal for their `at_dot`) and add the new states to the current column.
```
class EarleyParser(EarleyParser):
def complete(self, col, state):
return self.earley_complete(col, state)
def earley_complete(self, col, state):
parent_states = [
st for st in state.s_col.states if st.at_dot() == state.name
]
for st in parent_states:
col.add(st.advance())
```
Here is an example of completed processing. First we complete the Column 0
```
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
col_2 = Column(2, 'd')
ep.chart = [col_0, col_1, col_2]
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
```
Then we use `scan()` to populate Column 1
```
for state in ep.chart[0].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_1, state, 'a')
for s in ep.chart[1].states:
print(s)
for state in ep.chart[1].states:
if state.at_dot() in SAMPLE_GRAMMAR:
ep.predict(col_1, state.at_dot(), state)
for s in ep.chart[1].states:
print(s)
```
Then we use `scan()` again to populate Column 2
```
for state in ep.chart[1].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_2, state, state.at_dot())
for s in ep.chart[2].states:
print(s)
```
Now, we can use `complete()`:
```
for state in ep.chart[2].states:
if state.finished():
ep.complete(col_2, state)
for s in ep.chart[2].states:
print(s)
```
### Filling the Chart
The main driving loop in `fill_chart()` essentially calls these operations in order. We loop over each column in order.
* For each column, fetch one state in the column at a time, and check if the state is `finished`.
* If it is, then we `complete()` all the parent states depending on this state.
* If the state was not finished, we check to see if the state's current symbol `at_dot` is a nonterminal.
* If it is a nonterminal, we `predict()` possible continuations, and update the current column with these states.
* If it was not, we `scan()` the next column and advance the current state if it matches the next letter.
```
class EarleyParser(EarleyParser):
def fill_chart(self, chart):
for i, col in enumerate(chart):
for state in col.states:
if state.finished():
self.complete(col, state)
else:
sym = state.at_dot()
if sym in self.cgrammar:
self.predict(col, sym, state)
else:
if i + 1 >= len(chart):
continue
self.scan(chart[i + 1], state, sym)
if self.log:
print(col, '\n')
return chart
```
We now can recognize a given string as belonging to a language represented by a grammar.
```
ep = EarleyParser(SAMPLE_GRAMMAR, log=True)
columns = ep.chart_parse('adcd', START_SYMBOL)
```
The chart we printed above only shows completed entries at each index. The parenthesized expression indicates the column just before the first character was recognized, and the ending column.
Notice how the `<start>` nonterminal shows fully parsed status.
```
last_col = columns[-1]
for s in last_col.states:
if s.name == '<start>':
print(s)
```
Since `chart_parse()` returns the completed table, we now need to extract the derivation trees.
### The Parse Method
For determining how far we have managed to parse, we simply look for the last index from `chart_parse()` where the `start_symbol` was found.
```
class EarleyParser(EarleyParser):
def parse_prefix(self, text):
self.table = self.chart_parse(text, self.start_symbol())
for col in reversed(self.table):
states = [
st for st in col.states if st.name == self.start_symbol()
]
if states:
return col.index, states
return -1, []
```
Here is the `parse_prefix()` in action.
```
ep = EarleyParser(SAMPLE_GRAMMAR)
cursor, last_states = ep.parse_prefix('adcd')
print(cursor, [str(s) for s in last_states])
```
The following is adapted from the excellent reference on Earley parsing by [Loup Vaillant](http://loup-vaillant.fr/tutorials/earley-parsing/).
Our `parse()` method is as follows. It depends on two methods `parse_forest()` and `extract_trees()` that will be defined next.
```
class EarleyParser(EarleyParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
forest = self.parse_forest(self.table, start)
for tree in self.extract_trees(forest):
yield self.prune_tree(tree)
```
### Parsing Paths
The `parse_paths()` method tries to unify the given expression in `named_expr` with the parsed string. For that, it extracts the last symbol in `named_expr` and checks if it is a terminal symbol. If it is, then it checks the chart at `til` to see if the letter corresponding to the position matches the terminal symbol. If it does, extend our start index by the length of the symbol.
If the symbol was a nonterminal symbol, then we retrieve the parsed states at the current end column index (`til`) that correspond to the nonterminal symbol, and collect the start index. These are the end column indexes for the remaining expression.
Given our list of start indexes, we obtain the parse paths from the remaining expression. If we can obtain any, then we return the parse paths. If not, we return an empty list.
```
class EarleyParser(EarleyParser):
def parse_paths(self, named_expr, chart, frm, til):
def paths(state, start, k, e):
if not e:
return [[(state, k)]] if start == frm else []
else:
return [[(state, k)] + r
for r in self.parse_paths(e, chart, frm, start)]
*expr, var = named_expr
starts = None
if var not in self.cgrammar:
starts = ([(var, til - len(var),
't')] if til > 0 and chart[til].letter == var else [])
else:
starts = [(s, s.s_col.index, 'n') for s in chart[til].states
if s.finished() and s.name == var]
return [p for s, start, k in starts for p in paths(s, start, k, expr)]
```
Here is the `parse_paths()` in action
```
print(SAMPLE_GRAMMAR['<start>'])
ep = EarleyParser(SAMPLE_GRAMMAR)
completed_start = last_states[0]
paths = ep.parse_paths(completed_start.expr, columns, 0, 4)
for path in paths:
print([list(str(s_) for s_ in s) for s in path])
```
That is, the parse path for `<start>` given the input `adcd` included recognizing the expression `<A><B>`. This was recognized by the two states: `<A>` from input(0) to input(2) which further involved recognizing the rule `a<B>c`, and the next state `<B>` from input(3) which involved recognizing the rule `<D>`.
### Parsing Forests
The `parse_forest()` method takes the state which represents the completed parse, and determines the possible ways that its expressions corresponded to the parsed expression. For example, say we are parsing `1+2+3`, and the state has `[<expr>,+,<expr>]` in `expr`. It could have been parsed as either `[{<expr>:1+2},+,{<expr>:3}]` or `[{<expr>:1},+,{<expr>:2+3}]`.
```
class EarleyParser(EarleyParser):
def forest(self, s, kind, chart):
return self.parse_forest(chart, s) if kind == 'n' else (s, [])
def parse_forest(self, chart, state):
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(v, k, chart) for v, k in reversed(pathexpr)]
for pathexpr in pathexprs]
ep = EarleyParser(SAMPLE_GRAMMAR)
result = ep.parse_forest(columns, last_states[0])
result
```
### Extracting Trees
What we have from `parse_forest()` is a forest of trees. We need to extract a single tree from that forest. That is accomplished as follows.
(For now, we return the first available derivation tree. To do that, we need to extract the parse forest from the state corresponding to `start`.)
```
class EarleyParser(EarleyParser):
def extract_a_tree(self, forest_node):
name, paths = forest_node
if not paths:
return (name, [])
return (name, [self.extract_a_tree(self.forest(*p)) for p in paths[0]])
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
```
We now verify that our parser can parse a given expression.
```
A3_GRAMMAR = {
"<start>": ["<bexpr>"],
"<bexpr>": [
"<aexpr><gt><aexpr>", "<aexpr><lt><aexpr>", "<aexpr>=<aexpr>",
"<bexpr>=<bexpr>", "<bexpr>&<bexpr>", "<bexpr>|<bexpr>", "(<bexrp>)"
],
"<aexpr>":
["<aexpr>+<aexpr>", "<aexpr>-<aexpr>", "(<aexpr>)", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
"<lt>": ['<'],
"<gt>": ['>']
}
syntax_diagram(A3_GRAMMAR)
mystring = '(1+24)=33'
parser = EarleyParser(A3_GRAMMAR)
for tree in parser.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
```
We now have a complete parser that can parse almost arbitrary *CFG*. There remains a small corner to fix -- the case of epsilon rules as we will see later.
### Ambiguous Parsing
Ambiguous grammars are grammars that can produce multiple derivation trees for some given string. For example, the `A3_GRAMMAR` can parse `1+2+3` in two different ways – `[1+2]+3` and `1+[2+3]`.
Extracting a single tree might be reasonable for unambiguous parses. However, what if the given grammar produces ambiguity when given a string? We need to extract all derivation trees in that case. We enhance our `extract_trees()` method to extract multiple derivation trees.
```
class EarleyParser(EarleyParser):
def extract_trees(self, forest_node):
name, paths = forest_node
if not paths:
yield (name, [])
results = []
for path in paths:
ptrees = [self.extract_trees(self.forest(*p)) for p in path]
for p in zip(*ptrees):
yield (name, p)
```
As before, we verify that everything works.
```
mystring = '1+2'
parser = EarleyParser(A1_GRAMMAR)
for tree in parser.parse(mystring):
assert mystring == tree_to_string(tree)
display_tree(tree)
```
One can also use a `GrammarFuzzer` to verify that everything works.
```
gf = GrammarFuzzer(A1_GRAMMAR)
for i in range(5):
s = gf.fuzz()
print(i, s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
```
### The Aycock Epsilon Fix
While parsing, one often requires to know whether a given nonterminal can derive an empty string. For example, in the following grammar A can derive an empty string, while B can't. The nonterminals that can derive an empty string are called _nullable_ nonterminals. For example, in the below grammar `E_GRAMMAR_1`, `<A>` is _nullable_, and since `<A>` is one of the alternatives of `<start>`, `<start>` is also _nullable_. But `<B>` is not _nullable_.
```
E_GRAMMAR_1 = {
'<start>': ['<A>', '<B>'],
'<A>': ['a', ''],
'<B>': ['b']
}
```
One of the problems with the original Earley implementation is that it does not handle rules that can derive empty strings very well. For example, the given grammar should match `a`
```
EPSILON = ''
E_GRAMMAR = {
'<start>': ['<S>'],
'<S>': ['<A><A><A><A>'],
'<A>': ['a', '<E>'],
'<E>': [EPSILON]
}
syntax_diagram(E_GRAMMAR)
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
with ExpectError():
trees = parser.parse(mystring)
```
Aycock et al.\cite{Aycock2002} suggests a simple fix. Their idea is to pre-compute the `nullable` set and use it to advance the `nullable` states. However, before we do that, we need to compute the `nullable` set. The `nullable` set consists of all nonterminals that can derive an empty string.
Computing the `nullable` set requires expanding each production rule in the grammar iteratively and inspecting whether a given rule can derive the empty string. Each iteration needs to take into account new terminals that have been found to be `nullable`. The procedure stops when we obtain a stable result. This procedure can be abstracted into a more general method `fixpoint`.
#### Fixpoint
A `fixpoint` of a function is an element in the function's domain such that it is mapped to itself. For example, 1 is a `fixpoint` of square root because `squareroot(1) == 1`.
(We use `str` rather than `hash` to check for equality in `fixpoint` because the data structure `set`, which we would like to use as an argument has a good string representation but is not hashable).
```
def fixpoint(f):
def helper(arg):
while True:
sarg = str(arg)
arg_ = f(arg)
if str(arg_) == sarg:
return arg
arg = arg_
return helper
```
Remember `my_sqrt()` from [the first chapter](Intro_Testing.ipynb)? We can define `my_sqrt()` using fixpoint.
```
def my_sqrt(x):
@fixpoint
def _my_sqrt(approx):
return (approx + x / approx) / 2
return _my_sqrt(1)
my_sqrt(2)
```
#### Nullable
Similarly, we can define `nullable` using `fixpoint`. We essentially provide the definition of a single intermediate step. That is, assuming that `nullables` contain the current `nullable` nonterminals, we iterate over the grammar looking for productions which are `nullable` -- that is, productions where the entire sequence can yield an empty string on some expansion.
We need to iterate over the different alternative expressions and their corresponding nonterminals. Hence we define a `rules()` method converts our dictionary representation to this pair format.
```
def rules(grammar):
return [(key, choice)
for key, choices in grammar.items()
for choice in choices]
```
The `terminals()` method extracts all terminal symbols from a `canonical` grammar representation.
```
def terminals(grammar):
return set(token
for key, choice in rules(grammar)
for token in choice if token not in grammar)
def nullable_expr(expr, nullables):
return all(token in nullables for token in expr)
def nullable(grammar):
productions = rules(grammar)
@fixpoint
def nullable_(nullables):
for A, expr in productions:
if nullable_expr(expr, nullables):
nullables |= {A}
return (nullables)
return nullable_({EPSILON})
for key, grammar in {
'E_GRAMMAR': E_GRAMMAR,
'E_GRAMMAR_1': E_GRAMMAR_1
}.items():
print(key, nullable(canonical(grammar)))
```
So, once we have the `nullable` set, all that we need to do is, after we have called `predict` on a state corresponding to a nonterminal, check if it is `nullable` and if it is, advance and add the state to the current column.
```
class EarleyParser(EarleyParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.cgrammar = canonical(grammar, letters=True)
self.epsilon = nullable(self.cgrammar)
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
if sym in self.epsilon:
col.add(state.advance())
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
for tree in parser.parse(mystring):
display_tree(tree)
```
To ensure that our parser does parse all kinds of grammars, let us try two more test cases.
```
DIRECTLY_SELF_REFERRING = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": [ "<expr>", "a"],
}
INDIRECTLY_SELF_REFERRING = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": [ "<aexpr>", "a"],
"<aexpr>": [ "<expr>"],
}
mystring = 'select a from a'
for grammar in [DIRECTLY_SELF_REFERRING, INDIRECTLY_SELF_REFERRING]:
trees = EarleyParser(grammar).parse(mystring)
for tree in trees:
assert mystring == tree_to_string(tree)
display_tree(tree)
```
### More Earley Parsing
A number of other optimizations exist for Earley parsers. A fast industrial strength Earley parser implementation is the [Marpa parser](https://jeffreykegler.github.io/Marpa-web-site/). Further, Earley parsing need not be restricted to character data. One may also parse streams (audio and video streams) \cite{qi2018generalized} using a generalized Earley parser.
## Testing the Parsers
While we have defined two parser variants, it would be nice to have some confirmation that our parses work well. While it is possible to formally prove that they work, it is much more satisfying to generate random grammars, their corresponding strings, and parse them using the same grammar.
```
def prod_line_grammar(nonterminals, terminals):
g = {
'<start>': ['<symbols>'],
'<symbols>': ['<symbol><symbols>', '<symbol>'],
'<symbol>': ['<nonterminals>', '<terminals>'],
'<nonterminals>': ['<lt><alpha><gt>'],
'<lt>': ['<'],
'<gt>': ['>'],
'<alpha>': nonterminals,
'<terminals>': terminals
}
if not nonterminals:
g['<nonterminals>'] = ['']
del g['<lt>']
del g['<alpha>']
del g['<gt>']
return g
syntax_diagram(prod_line_grammar(["A", "B", "C"], ["1", "2", "3"]))
def make_rule(nonterminals, terminals, num_alts):
prod_grammar = prod_line_grammar(nonterminals, terminals)
gf = GrammarFuzzer(prod_grammar, min_nonterminals=3, max_nonterminals=5)
name = "<%s>" % ''.join(random.choices(string.ascii_uppercase, k=3))
return (name, [gf.fuzz() for _ in range(num_alts)])
make_rule(["A", "B", "C"], ["1", "2", "3"], 3)
from Grammars import unreachable_nonterminals
def make_grammar(num_symbols=3, num_alts=3):
terminals = list(string.ascii_lowercase)
grammar = {}
name = None
for _ in range(num_symbols):
nonterminals = [k[1:-1] for k in grammar.keys()]
name, expansions = \
make_rule(nonterminals, terminals, num_alts)
grammar[name] = expansions
grammar[START_SYMBOL] = [name]
# Remove unused parts
for nonterminal in unreachable_nonterminals(grammar):
del grammar[nonterminal]
assert is_valid_grammar(grammar)
return grammar
make_grammar()
```
Now we verify if our arbitrary grammars can be used by the Earley parser.
```
for i in range(5):
my_grammar = make_grammar()
print(my_grammar)
parser = EarleyParser(my_grammar)
mygf = GrammarFuzzer(my_grammar)
s = mygf.fuzz()
print(s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
display_tree(tree)
```
With this, we have completed both implementation and testing of *arbitrary* CFG, which can now be used along with `LangFuzzer` to generate better fuzzing inputs.
## Background
Numerous parsing techniques exist that can parse a given string using a
given grammar, and produce corresponding derivation tree or trees. However,
some of these techniques work only on specific classes of grammars.
These classes of grammars are named after the specific kind of parser
that can accept grammars of that category. That is, the upper bound for
the capabilities of the parser defines the grammar class named after that
parser.
The *LL* and *LR* parsing are the main traditions in parsing. Here, *LL* means left-to-right, leftmost derivation, and it represents a top-down approach. On the other hand, and LR (left-to-right, rightmost derivation) represents a bottom-up approach. Another way to look at it is that LL parsers compute the derivation tree incrementally in *pre-order* while LR parsers compute the derivation tree in *post-order* \cite{pingali2015graphical}).
Different classes of grammars differ in the features that are available to
the user for writing a grammar of that class. That is, the corresponding
kind of parser will be unable to parse a grammar that makes use of more
features than allowed. For example, the `A2_GRAMMAR` is an *LL*
grammar because it lacks left recursion, while `A1_GRAMMAR` is not an
*LL* grammar. This is because an *LL* parser parses
its input from left to right, and constructs the leftmost derivation of its
input by expanding the nonterminals it encounters. If there is a left
recursion in one of these rules, an *LL* parser will enter an infinite loop.
Similarly, a grammar is LL(k) if it can be parsed by an LL parser with k lookahead token, and LR(k) grammar can only be parsed with LR parser with at least k lookahead tokens. These grammars are interesting because both LL(k) and LR(k) grammars have $O(n)$ parsers, and can be used with relatively restricted computational budget compared to other grammars.
The languages for which one can provide an *LL(k)* grammar is called *LL(k)* languages (where k is the minimum lookahead required). Similarly, *LR(k)* is defined as the set of languages that have an *LR(k)* grammar. In terms of languages, LL(k) $\subset$ LL(k+1) and LL(k) $\subset$ LR(k), and *LR(k)* $=$ *LR(1)*. All deterministic *CFLs* have an *LR(1)* grammar. However, there exist *CFLs* that are inherently ambiguous \cite{ogden1968helpful}, and for these, one can't provide an *LR(1)* grammar.
The other main parsing algorithms for *CFGs* are GLL \cite{scott2010gll}, GLR \cite{tomita1987efficient,tomita2012generalized}, and CYK \cite{grune2008parsing}.
The ALL(\*) (used by ANTLR) on the other hand is a grammar representation that uses *Regular Expression* like predicates (similar to advanced PEGs – see [Exercise](#Exercise-3:-PEG-Predicates)) rather than a fixed lookahead. Hence, ALL(\*) can accept a larger class of grammars than CFGs.
In terms of computational limits of parsing, the main CFG parsers have a complexity of $O(n^3)$ for arbitrary grammars. However, parsing with arbitrary *CFG* is reducible to boolean matrix multiplication \cite{Valiant1975} (and the reverse \cite{Lee2002}). This is at present bounded by $O(2^{23728639}$) \cite{LeGall2014}. Hence, worse case complexity for parsing arbitrary CFG is likely to remain close to cubic.
Regarding PEGs, the actual class of languages that is expressible in *PEG* is currently unknown. In particular, we know that *PEGs* can express certain languages such as $a^n b^n c^n$. However, we do not know if there exist *CFLs* that are not expressible with *PEGs*. In Section 2.3, we provided an instance of a counter-intuitive PEG grammar. While important for our purposes (we use grammars for generation of inputs) this is not a criticism of parsing with PEGs. PEG focuses on writing grammars for recognizing a given language, and not necessarily in interpreting what language an arbitrary PEG might yield. Given a Context-Free Language to parse, it is almost always possible to write a grammar for it in PEG, and given that 1) a PEG can parse any string in $O(n)$ time, and 2) at present we know of no CFL that can't be expressed as a PEG, and 3) compared with *LR* grammars, a PEG is often more intuitive because it allows top-down interpretation, when writing a parser for a language, PEGs should be under serious consideration.
## Synopsis
This chapter introduces `Parser` classes, parsing a string into a _derivation tree_ as introduced in the [chapter on efficient grammar fuzzing](GrammarFuzzer.ipynb). Two important parser classes are provided:
* [Parsing Expression Grammar parsers](#Parsing-Expression-Grammars) (`PEGParser`), which are very efficient, but limited to specific grammar structure; and
* [Earley parsers](#Parsing-Context-Free-Grammars) (`EarleyParser`), which accept any kind of context-free grammars.
Using any of these is fairly easy, though. First, instantiate them with a grammar:
```
from Grammars import US_PHONE_GRAMMAR
us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
```
Then, use the `parse()` method to retrieve a list of possible derivation trees:
```
trees = us_phone_parser.parse("(555)987-6543")
tree = list(trees)[0]
display_tree(tree)
```
These derivation trees can then be used for test generation, notably for mutating and recombining existing inputs.
## Lessons Learned
* Grammars can be used to generate derivation trees for a given string.
* Parsing Expression Grammars are intuitive, and easy to implement, but require care to write.
* Earley Parsers can parse arbitrary Context Free Grammars.
## Next Steps
* Use parsed inputs to [recombine existing inputs](LangFuzzer.ipynb)
## Exercises
### Exercise 1: An Alternative Packrat
In the _Packrat_ parser, we showed how one could implement a simple _PEG_ parser. That parser kept track of the current location in the text using an index. Can you modify the parser so that it simply uses the current substring rather than tracking the index? That is, it should no longer have the `at` parameter.
**Solution.** Here is a possible solution:
```
class PackratParser(Parser):
def parse_prefix(self, text):
txt, res = self.unify_key(self.start_symbol(), text)
return len(txt), [res]
def parse(self, text):
remain, res = self.parse_prefix(text)
if remain:
raise SyntaxError("at " + res)
return res
def unify_rule(self, rule, text):
results = []
for token in rule:
text, res = self.unify_key(token, text)
if res is None:
return text, None
results.append(res)
return text, results
def unify_key(self, key, text):
if key not in self.cgrammar:
if text.startswith(key):
return text[len(key):], (key, [])
else:
return text, None
for rule in self.cgrammar[key]:
text_, res = self.unify_rule(rule, text)
if res:
return (text_, (key, res))
return text, None
mystring = "1 + (2 * 3)"
for tree in PackratParser(EXPR_GRAMMAR).parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
```
### Exercise 2: More PEG Syntax
The _PEG_ syntax provides a few notational conveniences reminiscent of regular expressions. For example, it supports the following operators (letters `T` and `A` represents tokens that can be either terminal or nonterminal. `ε` is an empty string, and `/` is the ordered choice operator similar to the non-ordered choice operator `|`):
* `T?` represents an optional greedy match of T and `A := T?` is equivalent to `A := T/ε`.
* `T*` represents zero or more greedy matches of `T` and `A := T*` is equivalent to `A := T A/ε`.
* `T+` represents one or more greedy matches – equivalent to `TT*`
If you look at the three notations above, each can be represented in the grammar in terms of basic syntax.
Remember the exercise from [the chapter on grammars](Grammars.ipynb) that developed `define_ex_grammar()` that can represent grammars as Python code? extend `define_ex_grammar()` to `define_peg()` to support the above notational conveniences. The decorator should rewrite a given grammar that contains these notations to an equivalent grammar in basic syntax.
### Exercise 3: PEG Predicates
Beyond these notational conveniences, it also supports two predicates that can provide a powerful lookahead facility that does not consume any input.
* `T&A` represents an _And-predicate_ that matches `T` if `T` is matched, and it is immediately followed by `A`
* `T!A` represents a _Not-predicate_ that matches `T` if `T` is matched, and it is *not* immediately followed by `A`
Implement these predicates in our _PEG_ parser.
### Exercise 4: Earley Fill Chart
In the `Earley Parser`, `Column` class, we keep the states both as a `list` and also as a `dict` even though `dict` is ordered. Can you explain why?
**Hint**: see the `fill_chart` method.
**Solution.** Python allows us to append to a list in flight, while a dict, eventhough it is ordered does not allow that facility.
That is, the following will work
```python
values = [1]
for v in values:
values.append(v*2)
```
However, the following will result in an error
```python
values = {1:1}
for v in values:
values[v*2] = v*2
```
In the `fill_chart`, we make use of this facility to modify the set of states we are iterating on, on the fly.
### Exercise 5: Leo Parser
One of the problems with the original Earley parser is that while it can parse strings using arbitrary _Context Free Gramamrs_, its performance on right-recursive grammars is quadratic. That is, it takes $O(n^2)$ runtime and space for parsing with right-recursive grammars. For example, consider the parsing of the following string by two different grammars `LR_GRAMMAR` and `RR_GRAMMAR`.
```
mystring = 'aaaaaa'
```
To see the problem, we need to enable logging. Here is the logged version of parsing with the `LR_GRAMMAR`
```
result = EarleyParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass # consume the generator so that we can see the logs
```
Compare that to the parsing of `RR_GRAMMAR` as seen below:
```
result = EarleyParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
As can be seen from the parsing log for each letter, the number of states with representation `<A>: a <A> ● (i, j)` increases at each stage, and these are simply a left over from the previous letter. They do not contribute anything more to the parse other than to simply complete these entries. However, they take up space, and require resources for inspection, contributing a factor of `n` in analysis.
Joop Leo \cite{Leo1991} found that this inefficiency can be avoided by detecting right recursion. The idea is that before starting the `completion` step, check whether the current item has a _deterministic reduction path_. If such a path exists, add a copy of the topmost element of the _deteministic reduction path_ to the current column, and return. If not, perform the original `completion` step.
**Definition 2.1**: An item is said to be on the deterministic reduction path above $[A \rightarrow \gamma., i]$ if it is $[B \rightarrow \alpha A ., k]$ with $[B \rightarrow \alpha . A, k]$ being the only item in $ I_i $ with the dot in front of A, or if it is on the deterministic reduction path above $[B \rightarrow \alpha A ., k]$. An item on such a path is called *topmost* one if there is no item on the deterministic reduction path above it\cite{Leo1991}.
Finding a _deterministic reduction path_ is as follows:
Given a complete state, represented by `<A> : seq_1 ● (s, e)` where `s` is the starting column for this rule, and `e` the current column, there is a _deterministic reduction path_ **above** it if two constraints are satisfied.
1. There exist a *single* item in the form `<B> : seq_2 ● <A> (k, s)` in column `s`.
2. That should be the *single* item in s with dot in front of `<A>`
The resulting item is of the form `<B> : seq_2 <A> ● (k, e)`, which is simply item from (1) advanced, and is considered above `<A>:.. (s, e)` in the deterministic reduction path.
The `seq_1` and `seq_2` are arbitrary symbol sequences.
This forms the following chain of links, with `<A>:.. (s_1, e)` being the child of `<B>:.. (s_2, e)` etc.
Here is one way to visualize the chain:
```
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
```
Essentially, what we want to do is to identify potential deterministic right recursion candidates, perform completion on them, and *throw away the result*. We do this until we reach the top. See Grune et al.~\cite{grune2008parsing} for further information.
Note that the completions are in the same column (`e`), with each candidates with constraints satisfied
in further and further earlier columns (as shown below):
```
<C> : seq_3 ● <B> (s_3, s_2) --> <C> : seq_3 <B> ● (s_3, e)
|
<B> : seq_2 ● <A> (s_2, s_1) --> <B> : seq_2 <A> ● (s_2, e)
|
<A> : seq_1 ● (s_1, e)
```
Following this chain, the topmost item is the item `<C>:.. (s_3, e)` that does not have a parent. The topmost item needs to be saved is called a *transitive* item by Leo, and it is associated with the non-terminal symbol that started the lookup. The transitive item needs to be added to each column we inspect.
Here is the skeleton for the parser `LeoParser`.
```
class LeoParser(EarleyParser):
def complete(self, col, state):
return self.leo_complete(col, state)
def leo_complete(self, col, state):
detred = self.deterministic_reduction(state)
if detred:
col.add(detred.copy())
else:
self.earley_complete(col, state)
def deterministic_reduction(self, state):
raise NotImplemented()
```
Can you implement the `deterministic_reduction()` method to obtain the topmost element?
**Solution.** Here is a possible solution:
First, we update our `Column` class with the ability to add transitive items. Note that, while Leo asks the transitive to be added to the set $ I_k $ there is no actual requirement for the transitive states to be added to the `states` list. The transitive items are only intended for memoization and not for the `fill_chart()` method. Hence, we track them separately.
```
class Column(Column):
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique, self.transitives = [], {}, {}
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = state
return self.transitives[key]
```
Remember the picture we drew of the deterministic path?
```
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
```
We define a function `uniq_postdot()` that given the item `<A> := seq_1 ● (s_1, e)`, returns a `<B> : seq_2 ● <A> (s_2, s_1)` that satisfies the constraints mentioned in the above picture.
```
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
return matching_st_B[0] if matching_st_B else None
lp = LeoParser(RR_GRAMMAR)
[(str(s), str(lp.uniq_postdot(s))) for s in columns[-1].states]
```
We next define the function `get_top()` that is the core of deterministic reduction which gets the topmost state above the current state (`A`).
```
class LeoParser(LeoParser):
def get_top(self, state_A):
st_B_inc = self.uniq_postdot(state_A)
if not st_B_inc:
return None
t_name = st_B_inc.name
if t_name in st_B_inc.e_col.transitives:
return st_B_inc.e_col.transitives[t_name]
st_B = st_B_inc.advance()
top = self.get_top(st_B) or st_B
return st_B_inc.e_col.add_transitive(t_name, top)
```
Once we have the machinery in place, `deterministic_reduction()` itself is simply a wrapper to call `get_top()`
```
class LeoParser(LeoParser):
def deterministic_reduction(self, state):
return self.get_top(state)
lp = LeoParser(RR_GRAMMAR)
columns = lp.chart_parse(mystring, lp.start_symbol())
[(str(s), str(lp.get_top(s))) for s in columns[-1].states]
```
Now, both LR and RR grammars should work within $O(n)$ bounds.
```
result = LeoParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
We verify the Leo parser with a few more right recursive grammars.
```
RR_GRAMMAR2 = {
'<start>': ['<A>'],
'<A>': ['ab<A>', ''],
}
mystring2 = 'ababababab'
result = LeoParser(RR_GRAMMAR2, log=True).parse(mystring2)
for _ in result: pass
RR_GRAMMAR3 = {
'<start>': ['c<A>'],
'<A>': ['ab<A>', ''],
}
mystring3 = 'cababababab'
result = LeoParser(RR_GRAMMAR3, log=True).parse(mystring3)
for _ in result: pass
RR_GRAMMAR4 = {
'<start>': ['<A>c'],
'<A>': ['ab<A>', ''],
}
mystring4 = 'ababababc'
result = LeoParser(RR_GRAMMAR4, log=True).parse(mystring4)
for _ in result: pass
RR_GRAMMAR5 = {
'<start>': ['<A>'],
'<A>': ['ab<B>', ''],
'<B>': ['<A>'],
}
mystring5 = 'abababab'
result = LeoParser(RR_GRAMMAR5, log=True).parse(mystring5)
for _ in result: pass
RR_GRAMMAR6 = {
'<start>': ['<A>'],
'<A>': ['a<B>', ''],
'<B>': ['b<A>'],
}
mystring6 = 'abababab'
result = LeoParser(RR_GRAMMAR6, log=True).parse(mystring6)
for _ in result: pass
RR_GRAMMAR7 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a'],
}
mystring7 = 'aaaaaaaa'
result = LeoParser(RR_GRAMMAR7, log=True).parse(mystring7)
for _ in result: pass
```
We verify that our parser works correctly on `LR_GRAMMAR` too.
```
result = LeoParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
```
__Advanced:__ We have fixed the complexity bounds. However, because we are saving only the topmost item of a right recursion, we need to fix our parser to be aware of our fix while extracting parse trees. Can you fix it?
__Hint:__ Leo suggests simply transforming the Leo item sets to normal Earley sets, with the results from deterministic reduction expanded to their originals. For that, keep in mind the picture of constraint chain we drew earlier.
**Solution.** Here is a possible solution.
We first change the definition of `add_transitive()` so that results of deterministic reduction can be identified later.
```
class Column(Column):
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = TState(state.name, state.expr, state.dot,
state.s_col, state.e_col)
return self.transitives[key]
```
We also need a `back()` method to create the constraints.
```
class State(State):
def back(self):
return TState(self.name, self.expr, self.dot - 1, self.s_col, self.e_col)
```
We update `copy()` to make `TState` items instead.
```
class TState(State):
def copy(self):
return TState(self.name, self.expr, self.dot, self.s_col, self.e_col)
```
We now modify the `LeoParser` to keep track of the chain of constrains that we mentioned earlier.
```
class LeoParser(LeoParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self._postdots = {}
self.cgrammar = canonical(grammar, letters=True)
```
Next, we update the `uniq_postdot()` so that it tracks the chain of links.
```
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
if matching_st_B:
self._postdots[matching_st_B[0]._t()] = st_A
return matching_st_B[0]
return None
```
We next define a method `expand_tstate()` that, when given a `TState`, generates all the intermediate links that we threw away earlier for a given end column.
```
class LeoParser(LeoParser):
def expand_tstate(self, state, e):
if state._t() not in self._postdots:
return
c_C = self._postdots[state._t()]
e.add(c_C.advance())
self.expand_tstate(c_C.back(), e)
```
We define a `rearrange()` method to generate a reversed table where each column contains states that start at that column.
```
class LeoParser(LeoParser):
def rearrange(self, table):
f_table = [Column(c.index, c.letter) for c in table]
for col in table:
for s in col.states:
f_table[s.s_col.index].states.append(s)
return f_table
```
Here is the rearranged table. (Can you explain why the Column 0 has a large number of `<start>` items?)
```
ep = LeoParser(RR_GRAMMAR)
columns = ep.chart_parse(mystring, ep.start_symbol())
r_table = ep.rearrange(columns)
for col in r_table:
print(col, "\n")
```
We save the result of rearrange before going into `parse_forest()`.
```
class LeoParser(LeoParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
self.r_table = self.rearrange(self.table)
forest = self.extract_trees(self.parse_forest(self.table, start))
for tree in forest:
yield self.prune_tree(tree)
```
Finally, during `parse_forest()`, we first check to see if it is a transitive state, and if it is, expand it to the original sequence of states using `traverse_constraints()`.
```
class LeoParser(LeoParser):
def parse_forest(self, chart, state):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
return super().parse_forest(chart, state)
```
This completes our implementation of `LeoParser`.
We check whether the previously defined right recursive grammars parse and return the correct parse trees.
```
result = LeoParser(RR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR2).parse(mystring2)
for tree in result:
assert mystring2 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR3).parse(mystring3)
for tree in result:
assert mystring3 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR4).parse(mystring4)
for tree in result:
assert mystring4 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR5).parse(mystring5)
for tree in result:
assert mystring5 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR6).parse(mystring6)
for tree in result:
assert mystring6 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR7).parse(mystring7)
for tree in result:
assert mystring7 == tree_to_string(tree)
result = LeoParser(LR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
RR_GRAMMAR8 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a']
}
mystring8 = 'aa'
RR_GRAMMAR9 = {
'<start>': ['<A>'],
'<A>': ['<B><A>', '<B>'],
'<B>': ['b']
}
mystring9 = 'bbbbbbb'
result = LeoParser(RR_GRAMMAR8).parse(mystring8)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring8 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR9).parse(mystring9)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring9 == tree_to_string(tree)
```
### Exercise 6: Filtered Earley Parser
One of the problems with our Earley and Leo Parsers is that it can get stuck in infinite loops when parsing with grammars that contain token repetitions in alternatives. For example, consider the grammar below.
```
RECURSION_GRAMMAR = {
"<start>": ["<A>"],
"<A>": ["<A>", "<A>aa", "AA", "<B>"],
"<B>": ["<C>", "<C>cc" ,"CC"],
"<C>": ["<B>", "<B>bb", "BB"]
}
```
With this grammar, one can produce an infinite chain of derivations of `<A>`, (direct recursion) or an infinite chain of derivations of `<B> -> <C> -> <B> ...` (indirect recursion). The problem is that, our implementation can get stuck trying to derive one of these infinite chains.
```
from ExpectError import ExpectTimeout
with ExpectTimeout(1, print_traceback=False):
mystring = 'AA'
parser = LeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
```
Can you implement a solution such that any tree that contains such a chain is discarded?
**Solution.** Here is a possible solution.
```
class FilteredLeoParser(LeoParser):
def forest(self, s, kind, seen, chart):
return self.parse_forest(chart, s, seen) if kind == 'n' else (s, [])
def parse_forest(self, chart, state, seen=None):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
def was_seen(chain, s):
if isinstance(s, str):
return False
if len(s.expr) > 1:
return False
return s in chain
if len(state.expr) > 1: # things get reset if we have a non loop
seen = set()
elif seen is None: # initialization
seen = {state}
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(s, k, seen | {s}, chart)
for s, k in reversed(pathexpr)
if not was_seen(seen, s)] for pathexpr in pathexprs]
```
With the `FilteredLeoParser`, we should be able to recover minimal parse trees in reasonable time.
```
mystring = 'AA'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaaaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'CC'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBcc'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BB'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBccbb'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
```
As can be seen, we are able to recover minimal parse trees without hitting on infinite chains.
### Exercise 7: Iterative Earley Parser
Recursive algorithms are quite handy in some cases but sometimes we might want to have iteration instead of recursion due to memory or speed problems.
Can you implement an iterative version of the `EarleyParser`?
__Hint:__ In general, you can use a stack to replace a recursive algorithm with an iterative one. An easy way to do this is pushing the parameters onto a stack instead of passing them to the recursive function.
**Solution.** Here is a possible solution.
First, we define `parse_paths()` that extract paths from a parsed expression.
```
class IterativeEarleyParser(EarleyParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.shuffle = kwargs.get('shuffle_rules', True)
def parse_paths(self, named_expr, chart, frm, til):
if not named_expr:
return []
paths = []
# stack of (expr, index, path) tuples
path_build_stack = [(named_expr, til, [])]
def evaluate_path(path, index, expr):
if expr: # path is still being built
path_build_stack.append((expr, index, path))
elif index == frm: # path is complete
paths.append(path)
while path_build_stack:
expr, chart_index, path = path_build_stack.pop()
*expr, symbol = expr
if symbol in self.cgrammar:
for state in chart[chart_index].states:
if state.name == symbol and state.finished():
extended_path = path + [(state, 'n')]
evaluate_path(extended_path, state.s_col.index, expr)
elif chart_index > 0 and chart[chart_index].letter == symbol:
extended_path = path + [(symbol, 't')]
evaluate_path(extended_path, chart_index - len(symbol), expr)
return paths
```
Next we used these paths to recover the forest data structure using `parse_forest()`. Note from the previous exercise `FilteredEarleyParser` that grammars with token repetitions can cause the parser to get stuck in a loop. One alternative is to simply shuffle the order in which rules are expanded, which is governed by the `shuffle_rules` option.
```
class IterativeEarleyParser(EarleyParser):
def parse_forest(self, chart, state):
if not state.expr:
return (state.name, [])
outermost_forest = []
forest_build_stack = [(state, outermost_forest)]
while forest_build_stack:
st, forest = forest_build_stack.pop()
paths = self.parse_paths(st.expr, chart, st.s_col.index,
st.e_col.index)
if not paths:
continue
next_path = random.choice(paths) if self.shuffle else paths[0]
path_forest = []
for symbol_or_state, kind in reversed(next_path):
if kind == 'n':
new_forest = []
forest_build_stack.append((symbol_or_state, new_forest))
path_forest.append((symbol_or_state.name, new_forest))
else:
path_forest.append((symbol_or_state, []))
forest.append(path_forest)
return (state.name, outermost_forest)
```
Now we are ready to extract trees from the forest using `extract_a_tree()`
```
class IterativeEarleyParser(EarleyParser):
def extract_a_tree(self, forest_node):
outermost_tree = []
tree_build_stack = [(forest_node, outermost_tree)]
while tree_build_stack:
node, tree = tree_build_stack.pop()
name, node_paths = node
if node_paths:
for path in random.choice(node_paths):
new_tree = []
tree_build_stack.append((path, new_tree))
tree.append((path[0], new_tree))
else:
tree.append((name, []))
return (forest_node[0], outermost_tree)
```
For now, we simply extract the first tree found.
```
class IterativeEarleyParser(EarleyParser):
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
```
Let's see if it works with some of the grammars we have seen so far.
```
test_cases = [
(A1_GRAMMAR, '1-2-3+4-5'),
(A2_GRAMMAR, '1+2'),
(A3_GRAMMAR, '1+2+3-6=6-1-2-3'),
(LR_GRAMMAR, 'aaaaa'),
(RR_GRAMMAR, 'aa'),
(DIRECTLY_SELF_REFERRING, 'select a from a'),
(INDIRECTLY_SELF_REFERRING, 'select a from a'),
(RECURSION_GRAMMAR, 'AA'),
(RECURSION_GRAMMAR, 'AAaaaa'),
(RECURSION_GRAMMAR, 'BBccbb')
]
for i, (grammar, text) in enumerate(test_cases):
print(i, text)
tree, *_ = IterativeEarleyParser(grammar).parse(text)
assert text == tree_to_string(tree)
```
As can be seen, our `IterativeEarleyParser` is able to handle recursive grammars.
### Exercise 8: First Set of a Nonterminal
We previously gave a way to extract a the `nullable` (epsilon) set, which is often used for parsing.
Along with `nullable`, parsing algorithms often use two other sets [`first` and `follow`](https://en.wikipedia.org/wiki/Canonical_LR_parser#FIRST_and_FOLLOW_sets).
The first set of a terminal symbol is itself, and the first set of a nonterminal is composed of terminal symbols that can come at the beginning of any derivation
of that nonterminal. The first set of any nonterminal that can derive the empty string should contain `EPSILON`. For example, using our `A1_GRAMMAR`, the first set of both `<expr>` and `<start>` is `{0,1,2,3,4,5,6,7,8,9}`. The extraction first set for any self-recursive nonterminal is simple enough. One simply has to recursively compute the first set of the first element of its choice expressions. The computation of `first` set for a self-recursive nonterminal is tricky. One has to recursively compute the first set until one is sure that no more terminals can be added to the first set.
Can you implement the `first` set using our `fixpoint()` decorator?
**Solution.** The first set of all terminals is the set containing just themselves. So we initialize that first. Then we update the first set with rules that derive empty strings.
```
def firstset(grammar, nullable):
first = {i: {i} for i in terminals(grammar)}
for k in grammar:
first[k] = {EPSILON} if k in nullable else set()
return firstset_((rules(grammar), first, nullable))[1]
```
Finally, we rely on the `fixpoint` to update the first set with the contents of the current first set until the first set stops changing.
```
def first_expr(expr, first, nullable):
tokens = set()
for token in expr:
tokens |= first[token]
if token not in nullable:
break
return tokens
@fixpoint
def firstset_(arg):
(rules, first, epsilon) = arg
for A, expression in rules:
first[A] |= first_expr(expression, first, epsilon)
return (rules, first, epsilon)
firstset(canonical(A1_GRAMMAR), EPSILON)
```
### Exercise 9: Follow Set of a Nonterminal
The follow set definition is similar to the first set. The follow set of a nonterminal is the set of terminals that can occur just after that nonterminal is used in any derivation. The follow set of the start symbol is `EOF`, and the follow set of any nonterminal is the super set of first sets of all symbols that come after it in any choice expression.
For example, the follow set of `<expr>` in `A1_GRAMMAR` is the set `{EOF, +, -}`.
As in the previous exercise, implement the `followset()` using the `fixpoint()` decorator.
**Solution.** The implementation of `followset()` is similar to `firstset()`. We first initialize the follow set with `EOF`, get the epsilon and first sets, and use the `fixpoint()` decorator to iteratively compute the follow set until nothing changes.
```
EOF = '\0'
def followset(grammar, start):
follow = {i: set() for i in grammar}
follow[start] = {EOF}
epsilon = nullable(grammar)
first = firstset(grammar, epsilon)
return followset_((grammar, epsilon, first, follow))[-1]
```
Given the current follow set, one can update the follow set as follows:
```
@fixpoint
def followset_(arg):
grammar, epsilon, first, follow = arg
for A, expression in rules(grammar):
f_B = follow[A]
for t in reversed(expression):
if t in grammar:
follow[t] |= f_B
f_B = f_B | first[t] if t in epsilon else (first[t] - {EPSILON})
return (grammar, epsilon, first, follow)
followset(canonical(A1_GRAMMAR), START_SYMBOL)
```
### Exercise 10: A LL(1) Parser
As we mentioned previously, there exist other kinds of parsers that operate left-to-right with right most derivation (*LR(k)*) or left-to-right with left most derivation (*LL(k)*) with _k_ signifying the amount of lookahead the parser is permitted to use.
What should one do with the lookahead? That lookahead can be used to determine which rule to apply. In the case of an *LL(1)* parser, the rule to apply is determined by looking at the _first_ set of the different rules. We previously implemented `first_expr()` that takes a an expression, the set of `nullables`, and computes the first set of that rule.
If a rule can derive an empty set, then that rule may also be applicable if of sees the `follow()` set of the corresponding nonterminal.
#### Part 1: A LL(1) Parsing Table
The first part of this exercise is to implement the _parse table_ that describes what action to take for an *LL(1)* parser on seeing a terminal symbol on lookahead. The table should be in the form of a _dictionary_ such that the keys represent the nonterminal symbol, and the value should contain another dictionary with keys as terminal symbols and the particular rule to continue parsing as the value.
Let us illustrate this table with an example. The `parse_table()` method populates a `self.table` data structure that should conform to the following requirements:
```
class LL1Parser(Parser):
def parse_table(self):
self.my_rules = rules(self.cgrammar)
self.table = ... # fill in here to produce
def rules(self):
for i, rule in enumerate(self.my_rules):
print(i, rule)
def show_table(self):
ts = list(sorted(terminals(self.cgrammar)))
print('Rule Name\t| %s' % ' | '.join(t for t in ts))
for k in self.table:
pr = self.table[k]
actions = list(str(pr[t]) if t in pr else ' ' for t in ts)
print('%s \t| %s' % (k, ' | '.join(actions)))
```
On invocation of `LL1Parser(A2_GRAMMAR).show_table()`
It should result in the following table:
```
for i, r in enumerate(rules(canonical(A2_GRAMMAR))):
print("%d\t %s := %s" % (i, r[0], r[1]))
```
|Rule Name || + | - | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9|
|-----------||---|---|---|---|---|---|---|---|---|---|---|--|
|start || | | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0|
|expr || | | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1|
|expr_ || 2 | 3 | | | | | | | | | | |
|integer || | | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5|
|integer_ || 7 | 7 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6|
|digit || | | 8 | 9 |10 |11 |12 |13 |14 |15 |16 |17|
**Solution.** We define `predict()` as we explained before. Then we use the predicted rules to populate the parse table.
```
class LL1Parser(LL1Parser):
def predict(self, rulepair, first, follow, epsilon):
A, rule = rulepair
rf = first_expr(rule, first, epsilon)
if nullable_expr(rule, epsilon):
rf |= follow[A]
return rf
def parse_table(self):
self.my_rules = rules(self.cgrammar)
epsilon = nullable(self.cgrammar)
first = firstset(self.cgrammar, epsilon)
# inefficient, can combine the three.
follow = followset(self.cgrammar, self.start_symbol())
ptable = [(i, self.predict(rule, first, follow, epsilon))
for i, rule in enumerate(self.my_rules)]
parse_tbl = {k: {} for k in self.cgrammar}
for i, pvals in ptable:
(k, expr) = self.my_rules[i]
parse_tbl[k].update({v: i for v in pvals})
self.table = parse_tbl
ll1parser = LL1Parser(A2_GRAMMAR)
ll1parser.parse_table()
ll1parser.show_table()
```
#### Part 2: The Parser
Once we have the parse table, implementing the parser is as follows: Consider the first item from the sequence of tokens to parse, and seed the stack with the start symbol.
While the stack is not empty, extract the first symbol from the stack, and if the symbol is a terminal, verify that the symbol matches the item from the input stream. If the symbol is a nonterminal, use the symbol and input item to lookup the next rule from the parse table. Insert the rule thus found to the top of the stack. Keep track of the expressions being parsed to build up the parse table.
Use the parse table defined previously to implement the complete LL(1) parser.
**Solution.** Here is the complete parser:
```
class LL1Parser(LL1Parser):
def parse_helper(self, stack, inplst):
inp, *inplst = inplst
exprs = []
while stack:
val, *stack = stack
if isinstance(val, tuple):
exprs.append(val)
elif val not in self.cgrammar: # terminal
assert val == inp
exprs.append(val)
inp, *inplst = inplst or [None]
else:
if inp is not None:
i = self.table[val][inp]
_, rhs = self.my_rules[i]
stack = rhs + [(val, len(rhs))] + stack
return self.linear_to_tree(exprs)
def parse(self, inp):
self.parse_table()
k, _ = self.my_rules[0]
stack = [k]
return self.parse_helper(stack, inp)
def linear_to_tree(self, arr):
stack = []
while arr:
elt = arr.pop(0)
if not isinstance(elt, tuple):
stack.append((elt, []))
else:
# get the last n
sym, n = elt
elts = stack[-n:] if n > 0 else []
stack = stack[0:len(stack) - n]
stack.append((sym, elts))
assert len(stack) == 1
return stack[0]
ll1parser = LL1Parser(A2_GRAMMAR)
tree = ll1parser.parse('1+2')
display_tree(tree)
```
|
github_jupyter
|
>>> from fuzzingbook.Parser import <identifier>
>>> from Grammars import US_PHONE_GRAMMAR
>>> us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
>>> trees = us_phone_parser.parse("(555)987-6543")
>>> tree = list(trees)[0]
>>> display_tree(tree)
def process_inventory(inventory):
res = []
for vehicle in inventory.split('\n'):
ret = process_vehicle(vehicle)
res.extend(ret)
return '\n'.join(res)
def process_vehicle(vehicle):
year, kind, company, model, *_ = vehicle.split(',')
if kind == 'van':
return process_van(year, company, model)
elif kind == 'car':
return process_car(year, company, model)
else:
raise Exception('Invalid entry')
def process_van(year, company, model):
res = ["We have a %s %s van from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2010:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
def process_car(year, company, model):
res = ["We have a %s %s car from %s vintage." % (company, model, year)]
iyear = int(year)
if iyear > 2016:
res.append("It is a recent model!")
else:
res.append("It is an old but reliable model!")
return res
mystring = """\
1997,van,Ford,E350
2000,car,Mercury,Cougar\
"""
print(process_inventory(mystring))
import string
CSV_GRAMMAR = {
'<start>': ['<csvline>'],
'<csvline>': ['<items>'],
'<items>': ['<item>,<items>', '<item>'],
'<item>': ['<letters>'],
'<letters>': ['<letter><letters>', '<letter>'],
'<letter>': list(string.ascii_letters + string.digits + string.punctuation + ' \t\n')
}
import fuzzingbook_utils
from Grammars import EXPR_GRAMMAR, START_SYMBOL, RE_NONTERMINAL, is_valid_grammar, syntax_diagram
from Fuzzer import Fuzzer
from GrammarFuzzer import GrammarFuzzer, FasterGrammarFuzzer, display_tree, tree_to_string, dot_escape
from ExpectError import ExpectError
from Timer import Timer
syntax_diagram(CSV_GRAMMAR)
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 1000
valid = []
time = 0
for i in range(trials):
with Timer() as t:
vehicle_info = gf.fuzz()
try:
process_vehicle(vehicle_info)
valid.append(vehicle_info)
except:
pass
time += t.elapsed_time()
print("%d valid strings, that is GrammarFuzzer generated %f%% valid entries from %d inputs" %
(len(valid), len(valid) * 100.0 / trials, trials))
print("Total time of %f seconds" % time)
gf = GrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
trials = 10
valid = []
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
import copy
import random
class PooledGrammarFuzzer(GrammarFuzzer):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self._node_cache = {}
def update_cache(self, key, values):
self._node_cache[key] = values
def expand_node_randomly(self, node):
(symbol, children) = node
assert children is None
if symbol in self._node_cache:
if random.randint(0, 1) == 1:
return super().expand_node_randomly(node)
return copy.deepcopy(random.choice(self._node_cache[symbol]))
return super().expand_node_randomly(node)
gf = PooledGrammarFuzzer(CSV_GRAMMAR, min_nonterminals=4)
gf.update_cache('<item>', [
('<item>', [('car', [])]),
('<item>', [('van', [])]),
])
trials = 10
valid = []
time = 0
for i in range(trials):
vehicle_info = gf.fuzz()
try:
print(repr(vehicle_info), end="")
process_vehicle(vehicle_info)
except Exception as e:
print("\t", e)
else:
print()
parser = Parser(grammar)
trees = parser.parse(input)
def parse_csv(mystring):
children = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in line.split(',')]))
return tree
def lr_graph(dot):
dot.attr('node', shape='plain')
dot.graph_attr['rankdir'] = 'LR'
tree = parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
mystring = '''\
1997,Ford,E350,"ac, abs, moon",3000.00\
'''
print(mystring)
def highlight_node(predicate):
def hl_node(dot, nid, symbol, ann):
if predicate(dot, nid, symbol, ann):
dot.node(repr(nid), dot_escape(symbol), fontcolor='red')
else:
dot.node(repr(nid), dot_escape(symbol))
return hl_node
tree = parse_csv(mystring)
bad_nodes = {5, 6, 7, 12, 13, 20, 22, 23, 24, 25}
def hl_predicate(_d, nid, _s, _a): return nid in bad_nodes
highlight_err_node = highlight_node(hl_predicate)
display_tree(tree, log=False, node_attr=highlight_err_node,
graph_attr=lr_graph)
def parse_quote(string, i):
v = string[i + 1:].find('"')
return v + i + 1 if v >= 0 else -1
def find_comma(string, i):
slen = len(string)
while i < slen:
if string[i] == '"':
i = parse_quote(string, i)
if i == -1:
return -1
if string[i] == ',':
return i
i += 1
return -1
def comma_split(string):
slen = len(string)
i = 0
while i < slen:
c = find_comma(string, i)
if c == -1:
yield string[i:]
return
else:
yield string[i:c]
i = c + 1
def parse_csv(mystring):
children = []
tree = (START_SYMBOL, children)
for i, line in enumerate(mystring.split('\n')):
children.append(("record %d" % i, [(cell, [])
for cell in comma_split(line)]))
return tree
tree = parse_csv(mystring)
display_tree(tree, graph_attr=lr_graph)
mystring = '''\
1999,Chevy,"Venture \\"Extended Edition, Very Large\\"",,5000.00\
'''
print(mystring)
tree = parse_csv(mystring)
bad_nodes = {4, 5}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
mystring = '''\
1996,Jeep,Grand Cherokee,"MUST SELL!
air, moon roof, loaded",4799.00
'''
print(mystring)
tree = parse_csv(mystring)
bad_nodes = {5, 6, 7, 8, 9, 10}
display_tree(tree, node_attr=highlight_err_node, graph_attr=lr_graph)
A1_GRAMMAR = {
"<start>": ["<expr>"],
"<expr>": ["<expr>+<expr>", "<expr>-<expr>", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A1_GRAMMAR)
mystring = '1+2'
tree = ('<start>', [('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('2',
[])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
A2_GRAMMAR = {
"<start>": ["<expr>"],
"<expr>": ["<integer><expr_>"],
"<expr_>": ["+<expr>", "-<expr>", ""],
"<integer>": ["<digit><integer_>"],
"<integer_>": ["<integer>", ""],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"]
}
syntax_diagram(A2_GRAMMAR)
tree = ('<start>', [('<expr>', [('<integer>', [('<digit>', [('1', [])]),
('<integer_>', [])]),
('<expr_>', [('+', []),
('<expr>',
[('<integer>',
[('<digit>', [('2', [])]),
('<integer_>', [])]),
('<expr_>', [])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
LR_GRAMMAR = {
'<start>': ['<A>'],
'<A>': ['<A>a', ''],
}
syntax_diagram(LR_GRAMMAR)
mystring = 'aaaaaa'
display_tree(
('<start>', (('<A>', (('<A>', (('<A>', []), ('a', []))), ('a', []))), ('a', []))))
RR_GRAMMAR = {
'<start>': ['<A>'],
'<A>': ['a<A>', ''],
}
syntax_diagram(RR_GRAMMAR)
display_tree(('<start>', ((
'<A>', (('a', []), ('<A>', (('a', []), ('<A>', (('a', []), ('<A>', []))))))),)))
mystring = '1+2+3'
tree = ('<start>',
[('<expr>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>', [('<integer>',
[('<digit>', [('2', [])])])])]), ('+', []),
('<expr>', [('<integer>', [('<digit>', [('3', [])])])])])])
assert mystring == tree_to_string(tree)
display_tree(tree)
tree = ('<start>',
[('<expr>', [('<expr>', [('<integer>', [('<digit>', [('1', [])])])]),
('+', []),
('<expr>',
[('<expr>', [('<integer>', [('<digit>', [('2', [])])])]),
('+', []),
('<expr>', [('<integer>', [('<digit>', [('3',
[])])])])])])])
assert tree_to_string(tree) == mystring
display_tree(tree)
class Parser(object):
def __init__(self, grammar, **kwargs):
self._grammar = grammar
self._start_symbol = kwargs.get('start_symbol', START_SYMBOL)
self.log = kwargs.get('log', False)
self.coalesce_tokens = kwargs.get('coalesce', True)
self.tokens = kwargs.get('tokens', set())
def grammar(self):
return self._grammar
def start_symbol(self):
return self._start_symbol
def parse_prefix(self, text):
"""Return pair (cursor, forest) for longest prefix of text"""
raise NotImplemented()
def parse(self, text):
cursor, forest = self.parse_prefix(text)
if cursor < len(text):
raise SyntaxError("at " + repr(text[cursor:]))
return [self.prune_tree(tree) for tree in forest]
def coalesce(self, children):
last = ''
new_lst = []
for cn, cc in children:
if cn not in self._grammar:
last += cn
else:
if last:
new_lst.append((last, []))
last = ''
new_lst.append((cn, cc))
if last:
new_lst.append((last, []))
return new_lst
def prune_tree(self, tree):
name, children = tree
if self.coalesce_tokens:
children = self.coalesce(children)
if name in self.tokens:
return (name, [(tree_to_string(tree), [])])
else:
return (name, [self.prune_tree(c) for c in children])
PEG1 = {
'<start>': ['a', 'b']
}
PEG2 = {
'<start>': ['ab', 'abc']
}
import re
def canonical(grammar, letters=False):
def split(expansion):
if isinstance(expansion, tuple):
expansion = expansion[0]
return [token for token in re.split(
RE_NONTERMINAL, expansion) if token]
def tokenize(word):
return list(word) if letters else [word]
def canonical_expr(expression):
return [
token for word in split(expression)
for token in ([word] if word in grammar else tokenize(word))
]
return {
k: [canonical_expr(expression) for expression in alternatives]
for k, alternatives in grammar.items()
}
CE_GRAMMAR = canonical(EXPR_GRAMMAR); CE_GRAMMAR
def non_canonical(grammar):
new_grammar = {}
for k in grammar:
rules = grammar[k]
new_rules = []
for rule in rules:
new_rules.append(''.join(rule))
new_grammar[k] = new_rules
return new_grammar
non_canonical(CE_GRAMMAR)
class Parser(Parser):
def __init__(self, grammar, **kwargs):
self._grammar = grammar
self._start_symbol = kwargs.get('start_symbol', START_SYMBOL)
self.log = kwargs.get('log', False)
self.tokens = kwargs.get('tokens', set())
self.coalesce_tokens = kwargs.get('coalesce', True)
self.cgrammar = canonical(grammar)
class PEGParser(Parser):
def parse_prefix(self, text):
cursor, tree = self.unify_key(self.start_symbol(), text, 0)
return cursor, [tree]
class PEGParser(PEGParser):
def unify_key(self, key, text, at=0):
if self.log:
print("unify_key: %s with %s" % (repr(key), repr(text[at:])))
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
mystring = "1"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_key('1', mystring)
mystring = "2"
peg.unify_key('1', mystring)
class PEGParser(PEGParser):
def unify_rule(self, rule, text, at):
if self.log:
print('unify_rule: %s with %s' % (repr(rule), repr(text[at:])))
results = []
for token in rule:
at, res = self.unify_key(token, text, at)
if res is None:
return at, None
results.append(res)
return at, results
mystring = "0"
peg = PEGParser(EXPR_GRAMMAR, log=True)
peg.unify_rule(peg.cgrammar['<digit>'][0], mystring, 0)
mystring = "12"
peg.unify_rule(peg.cgrammar['<integer>'][0], mystring, 0)
mystring = "1 + 2"
peg = PEGParser(EXPR_GRAMMAR, log=False)
peg.parse(mystring)
from functools import lru_cache
class PEGParser(PEGParser):
@lru_cache(maxsize=None)
def unify_key(self, key, text, at=0):
if key not in self.cgrammar:
if text[at:].startswith(key):
return at + len(key), (key, [])
else:
return at, None
for rule in self.cgrammar[key]:
to, res = self.unify_rule(rule, text, at)
if res is not None:
return (to, (key, res))
return 0, None
mystring = "1 + (2 * 3)"
peg = PEGParser(EXPR_GRAMMAR)
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = "1 * (2 + 3.35)"
for tree in peg.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
PEG_SURPRISE = {
"<A>": ["a<A>a", "aa"]
}
strings = []
for e in range(4):
f = GrammarFuzzer(PEG_SURPRISE, start_symbol='<A>')
tree = ('<A>', None)
for _ in range(e):
tree = f.expand_tree_once(tree)
tree = f.expand_tree_with_strategy(tree, f.expand_node_min_cost)
strings.append(tree_to_string(tree))
display_tree(tree)
strings
peg = PEGParser(PEG_SURPRISE, start_symbol='<A>')
for s in strings:
with ExpectError():
for tree in peg.parse(s):
display_tree(tree)
print(s)
grammar = {
'<start>': ['<A>', '<B>'],
...
}
grammar = {
'<start>': ['<start_>'],
'<start_>': ['<A>', '<B>'],
...
}
SAMPLE_GRAMMAR = {
'<start>': ['<A><B>'],
'<A>': ['a<B>c', 'a<A>'],
'<B>': ['b<C>', '<D>'],
'<C>': ['c'],
'<D>': ['d']
}
C_SAMPLE_GRAMMAR = canonical(SAMPLE_GRAMMAR)
syntax_diagram(SAMPLE_GRAMMAR)
<start> : ● <A> <B>
<A> : ● a <B> c
<A> : ● a <A>
<A> : a ● <B> c
<A> : a ● <A>
<B> : ● b <C>
<B> : ● <D>
<A> : ● a <B> c
<A> : ● a <A>
<D> : ● d
<D> : d ●
<B> : <D> ●
<A> : a <B> ● c
<A> : a <B> c ●
<start> : <A> ● <B>
<B> : ● b <C>
<B> : ● <D>
<D> : ● d
<D> : d ●
<B> : <D> ●
<start> : <A> <B> ●
class Column(object):
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique = [], {}
def __str__(self):
return "%s chart[%d]\n%s" % (self.letter, self.index, "\n".join(
str(state) for state in self.states if state.finished()))
class Column(Column):
def add(self, state):
if state in self._unique:
return self._unique[state]
self._unique[state] = state
self.states.append(state)
state.e_col = self
return self._unique[state]
class Item(object):
def __init__(self, name, expr, dot):
self.name, self.expr, self.dot = name, expr, dot
class Item(Item):
def finished(self):
return self.dot >= len(self.expr)
def advance(self):
return Item(self.name, self.expr, self.dot + 1)
def at_dot(self):
return self.expr[self.dot] if self.dot < len(self.expr) else None
item_name = '<B>'
item_expr = C_SAMPLE_GRAMMAR[item_name][1]
an_item = Item(item_name, tuple(item_expr), 0)
an_item.at_dot()
another_item = an_item.advance()
another_item.finished()
class State(Item):
def __init__(self, name, expr, dot, s_col, e_col=None):
super().__init__(name, expr, dot)
self.s_col, self.e_col = s_col, e_col
def __str__(self):
def idx(var):
return var.index if var else -1
return self.name + ':= ' + ' '.join([
str(p)
for p in [*self.expr[:self.dot], '|', *self.expr[self.dot:]]
]) + "(%d,%d)" % (idx(self.s_col), idx(self.e_col))
def copy(self):
return State(self.name, self.expr, self.dot, self.s_col, self.e_col)
def _t(self):
return (self.name, self.expr, self.dot, self.s_col.index)
def __hash__(self):
return hash(self._t())
def __eq__(self, other):
return self._t() == other._t()
def advance(self):
return State(self.name, self.expr, self.dot + 1, self.s_col)
col_0 = Column(0, None)
item_expr = tuple(*C_SAMPLE_GRAMMAR[START_SYMBOL])
start_state = State(START_SYMBOL, item_expr, 0, col_0)
col_0.add(start_state)
start_state.at_dot()
sym = start_state.at_dot()
for alt in C_SAMPLE_GRAMMAR[sym]:
col_0.add(State(sym, tuple(alt), 0, col_0))
for s in col_0.states:
print(s)
<start>: ● <A> <B>
class EarleyParser(Parser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.cgrammar = canonical(grammar, letters=True)
class EarleyParser(EarleyParser):
def chart_parse(self, words, start):
alt = tuple(*self.cgrammar[start])
chart = [Column(i, tok) for i, tok in enumerate([None, *words])]
chart[0].add(State(start, alt, 0, chart[0]))
return self.fill_chart(chart)
<A>: a ● <B> c
<A>: a ● <B> c
<B>: ● b c
<B>: ● <D>
class EarleyParser(EarleyParser):
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
col_0 = Column(0, None)
col_0.add(start_state)
ep = EarleyParser(SAMPLE_GRAMMAR)
ep.chart = [col_0]
for s in ep.chart[0].states:
print(s)
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
<B>: ● b c
<B>: b ● c
class EarleyParser(EarleyParser):
def scan(self, col, state, letter):
if letter == col.letter:
col.add(state.advance())
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
ep.chart = [col_0, col_1]
new_state = ep.chart[0].states[1]
print(new_state)
ep.scan(col_1, new_state, 'a')
for s in ep.chart[1].states:
print(s)
<A>: a ● <B> c
<B>: b c ●
class EarleyParser(EarleyParser):
def complete(self, col, state):
return self.earley_complete(col, state)
def earley_complete(self, col, state):
parent_states = [
st for st in state.s_col.states if st.at_dot() == state.name
]
for st in parent_states:
col.add(st.advance())
ep = EarleyParser(SAMPLE_GRAMMAR)
col_1 = Column(1, 'a')
col_2 = Column(2, 'd')
ep.chart = [col_0, col_1, col_2]
ep.predict(col_0, '<A>', s)
for s in ep.chart[0].states:
print(s)
for state in ep.chart[0].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_1, state, 'a')
for s in ep.chart[1].states:
print(s)
for state in ep.chart[1].states:
if state.at_dot() in SAMPLE_GRAMMAR:
ep.predict(col_1, state.at_dot(), state)
for s in ep.chart[1].states:
print(s)
for state in ep.chart[1].states:
if state.at_dot() not in SAMPLE_GRAMMAR:
ep.scan(col_2, state, state.at_dot())
for s in ep.chart[2].states:
print(s)
for state in ep.chart[2].states:
if state.finished():
ep.complete(col_2, state)
for s in ep.chart[2].states:
print(s)
class EarleyParser(EarleyParser):
def fill_chart(self, chart):
for i, col in enumerate(chart):
for state in col.states:
if state.finished():
self.complete(col, state)
else:
sym = state.at_dot()
if sym in self.cgrammar:
self.predict(col, sym, state)
else:
if i + 1 >= len(chart):
continue
self.scan(chart[i + 1], state, sym)
if self.log:
print(col, '\n')
return chart
ep = EarleyParser(SAMPLE_GRAMMAR, log=True)
columns = ep.chart_parse('adcd', START_SYMBOL)
last_col = columns[-1]
for s in last_col.states:
if s.name == '<start>':
print(s)
class EarleyParser(EarleyParser):
def parse_prefix(self, text):
self.table = self.chart_parse(text, self.start_symbol())
for col in reversed(self.table):
states = [
st for st in col.states if st.name == self.start_symbol()
]
if states:
return col.index, states
return -1, []
ep = EarleyParser(SAMPLE_GRAMMAR)
cursor, last_states = ep.parse_prefix('adcd')
print(cursor, [str(s) for s in last_states])
class EarleyParser(EarleyParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
forest = self.parse_forest(self.table, start)
for tree in self.extract_trees(forest):
yield self.prune_tree(tree)
class EarleyParser(EarleyParser):
def parse_paths(self, named_expr, chart, frm, til):
def paths(state, start, k, e):
if not e:
return [[(state, k)]] if start == frm else []
else:
return [[(state, k)] + r
for r in self.parse_paths(e, chart, frm, start)]
*expr, var = named_expr
starts = None
if var not in self.cgrammar:
starts = ([(var, til - len(var),
't')] if til > 0 and chart[til].letter == var else [])
else:
starts = [(s, s.s_col.index, 'n') for s in chart[til].states
if s.finished() and s.name == var]
return [p for s, start, k in starts for p in paths(s, start, k, expr)]
print(SAMPLE_GRAMMAR['<start>'])
ep = EarleyParser(SAMPLE_GRAMMAR)
completed_start = last_states[0]
paths = ep.parse_paths(completed_start.expr, columns, 0, 4)
for path in paths:
print([list(str(s_) for s_ in s) for s in path])
class EarleyParser(EarleyParser):
def forest(self, s, kind, chart):
return self.parse_forest(chart, s) if kind == 'n' else (s, [])
def parse_forest(self, chart, state):
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(v, k, chart) for v, k in reversed(pathexpr)]
for pathexpr in pathexprs]
ep = EarleyParser(SAMPLE_GRAMMAR)
result = ep.parse_forest(columns, last_states[0])
result
class EarleyParser(EarleyParser):
def extract_a_tree(self, forest_node):
name, paths = forest_node
if not paths:
return (name, [])
return (name, [self.extract_a_tree(self.forest(*p)) for p in paths[0]])
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
A3_GRAMMAR = {
"<start>": ["<bexpr>"],
"<bexpr>": [
"<aexpr><gt><aexpr>", "<aexpr><lt><aexpr>", "<aexpr>=<aexpr>",
"<bexpr>=<bexpr>", "<bexpr>&<bexpr>", "<bexpr>|<bexpr>", "(<bexrp>)"
],
"<aexpr>":
["<aexpr>+<aexpr>", "<aexpr>-<aexpr>", "(<aexpr>)", "<integer>"],
"<integer>": ["<digit><integer>", "<digit>"],
"<digit>": ["0", "1", "2", "3", "4", "5", "6", "7", "8", "9"],
"<lt>": ['<'],
"<gt>": ['>']
}
syntax_diagram(A3_GRAMMAR)
mystring = '(1+24)=33'
parser = EarleyParser(A3_GRAMMAR)
for tree in parser.parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
class EarleyParser(EarleyParser):
def extract_trees(self, forest_node):
name, paths = forest_node
if not paths:
yield (name, [])
results = []
for path in paths:
ptrees = [self.extract_trees(self.forest(*p)) for p in path]
for p in zip(*ptrees):
yield (name, p)
mystring = '1+2'
parser = EarleyParser(A1_GRAMMAR)
for tree in parser.parse(mystring):
assert mystring == tree_to_string(tree)
display_tree(tree)
gf = GrammarFuzzer(A1_GRAMMAR)
for i in range(5):
s = gf.fuzz()
print(i, s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
E_GRAMMAR_1 = {
'<start>': ['<A>', '<B>'],
'<A>': ['a', ''],
'<B>': ['b']
}
EPSILON = ''
E_GRAMMAR = {
'<start>': ['<S>'],
'<S>': ['<A><A><A><A>'],
'<A>': ['a', '<E>'],
'<E>': [EPSILON]
}
syntax_diagram(E_GRAMMAR)
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
with ExpectError():
trees = parser.parse(mystring)
def fixpoint(f):
def helper(arg):
while True:
sarg = str(arg)
arg_ = f(arg)
if str(arg_) == sarg:
return arg
arg = arg_
return helper
def my_sqrt(x):
@fixpoint
def _my_sqrt(approx):
return (approx + x / approx) / 2
return _my_sqrt(1)
my_sqrt(2)
def rules(grammar):
return [(key, choice)
for key, choices in grammar.items()
for choice in choices]
def terminals(grammar):
return set(token
for key, choice in rules(grammar)
for token in choice if token not in grammar)
def nullable_expr(expr, nullables):
return all(token in nullables for token in expr)
def nullable(grammar):
productions = rules(grammar)
@fixpoint
def nullable_(nullables):
for A, expr in productions:
if nullable_expr(expr, nullables):
nullables |= {A}
return (nullables)
return nullable_({EPSILON})
for key, grammar in {
'E_GRAMMAR': E_GRAMMAR,
'E_GRAMMAR_1': E_GRAMMAR_1
}.items():
print(key, nullable(canonical(grammar)))
class EarleyParser(EarleyParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.cgrammar = canonical(grammar, letters=True)
self.epsilon = nullable(self.cgrammar)
def predict(self, col, sym, state):
for alt in self.cgrammar[sym]:
col.add(State(sym, tuple(alt), 0, col))
if sym in self.epsilon:
col.add(state.advance())
mystring = 'a'
parser = EarleyParser(E_GRAMMAR)
for tree in parser.parse(mystring):
display_tree(tree)
DIRECTLY_SELF_REFERRING = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": [ "<expr>", "a"],
}
INDIRECTLY_SELF_REFERRING = {
'<start>': ['<query>'],
'<query>': ['select <expr> from a'],
"<expr>": [ "<aexpr>", "a"],
"<aexpr>": [ "<expr>"],
}
mystring = 'select a from a'
for grammar in [DIRECTLY_SELF_REFERRING, INDIRECTLY_SELF_REFERRING]:
trees = EarleyParser(grammar).parse(mystring)
for tree in trees:
assert mystring == tree_to_string(tree)
display_tree(tree)
def prod_line_grammar(nonterminals, terminals):
g = {
'<start>': ['<symbols>'],
'<symbols>': ['<symbol><symbols>', '<symbol>'],
'<symbol>': ['<nonterminals>', '<terminals>'],
'<nonterminals>': ['<lt><alpha><gt>'],
'<lt>': ['<'],
'<gt>': ['>'],
'<alpha>': nonterminals,
'<terminals>': terminals
}
if not nonterminals:
g['<nonterminals>'] = ['']
del g['<lt>']
del g['<alpha>']
del g['<gt>']
return g
syntax_diagram(prod_line_grammar(["A", "B", "C"], ["1", "2", "3"]))
def make_rule(nonterminals, terminals, num_alts):
prod_grammar = prod_line_grammar(nonterminals, terminals)
gf = GrammarFuzzer(prod_grammar, min_nonterminals=3, max_nonterminals=5)
name = "<%s>" % ''.join(random.choices(string.ascii_uppercase, k=3))
return (name, [gf.fuzz() for _ in range(num_alts)])
make_rule(["A", "B", "C"], ["1", "2", "3"], 3)
from Grammars import unreachable_nonterminals
def make_grammar(num_symbols=3, num_alts=3):
terminals = list(string.ascii_lowercase)
grammar = {}
name = None
for _ in range(num_symbols):
nonterminals = [k[1:-1] for k in grammar.keys()]
name, expansions = \
make_rule(nonterminals, terminals, num_alts)
grammar[name] = expansions
grammar[START_SYMBOL] = [name]
# Remove unused parts
for nonterminal in unreachable_nonterminals(grammar):
del grammar[nonterminal]
assert is_valid_grammar(grammar)
return grammar
make_grammar()
for i in range(5):
my_grammar = make_grammar()
print(my_grammar)
parser = EarleyParser(my_grammar)
mygf = GrammarFuzzer(my_grammar)
s = mygf.fuzz()
print(s)
for tree in parser.parse(s):
assert tree_to_string(tree) == s
display_tree(tree)
from Grammars import US_PHONE_GRAMMAR
us_phone_parser = EarleyParser(US_PHONE_GRAMMAR)
trees = us_phone_parser.parse("(555)987-6543")
tree = list(trees)[0]
display_tree(tree)
class PackratParser(Parser):
def parse_prefix(self, text):
txt, res = self.unify_key(self.start_symbol(), text)
return len(txt), [res]
def parse(self, text):
remain, res = self.parse_prefix(text)
if remain:
raise SyntaxError("at " + res)
return res
def unify_rule(self, rule, text):
results = []
for token in rule:
text, res = self.unify_key(token, text)
if res is None:
return text, None
results.append(res)
return text, results
def unify_key(self, key, text):
if key not in self.cgrammar:
if text.startswith(key):
return text[len(key):], (key, [])
else:
return text, None
for rule in self.cgrammar[key]:
text_, res = self.unify_rule(rule, text)
if res:
return (text_, (key, res))
return text, None
mystring = "1 + (2 * 3)"
for tree in PackratParser(EXPR_GRAMMAR).parse(mystring):
assert tree_to_string(tree) == mystring
display_tree(tree)
values = [1]
for v in values:
values.append(v*2)
values = {1:1}
for v in values:
values[v*2] = v*2
mystring = 'aaaaaa'
result = EarleyParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass # consume the generator so that we can see the logs
result = EarleyParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
<C> : seq_3 ● <B> (s_3, s_2) --> <C> : seq_3 <B> ● (s_3, e)
|
<B> : seq_2 ● <A> (s_2, s_1) --> <B> : seq_2 <A> ● (s_2, e)
|
<A> : seq_1 ● (s_1, e)
class LeoParser(EarleyParser):
def complete(self, col, state):
return self.leo_complete(col, state)
def leo_complete(self, col, state):
detred = self.deterministic_reduction(state)
if detred:
col.add(detred.copy())
else:
self.earley_complete(col, state)
def deterministic_reduction(self, state):
raise NotImplemented()
class Column(Column):
def __init__(self, index, letter):
self.index, self.letter = index, letter
self.states, self._unique, self.transitives = [], {}, {}
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = state
return self.transitives[key]
<C> : seq_3 <B> ● (s_3, e)
| constraints satisfied by <C> : seq_3 ● <B> (s_3, s_2)
<B> : seq_2 <A> ● (s_2, e)
| constraints satisfied by <B> : seq_2 ● <A> (s_2, s_1)
<A> : seq_1 ● (s_1, e)
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
return matching_st_B[0] if matching_st_B else None
lp = LeoParser(RR_GRAMMAR)
[(str(s), str(lp.uniq_postdot(s))) for s in columns[-1].states]
class LeoParser(LeoParser):
def get_top(self, state_A):
st_B_inc = self.uniq_postdot(state_A)
if not st_B_inc:
return None
t_name = st_B_inc.name
if t_name in st_B_inc.e_col.transitives:
return st_B_inc.e_col.transitives[t_name]
st_B = st_B_inc.advance()
top = self.get_top(st_B) or st_B
return st_B_inc.e_col.add_transitive(t_name, top)
class LeoParser(LeoParser):
def deterministic_reduction(self, state):
return self.get_top(state)
lp = LeoParser(RR_GRAMMAR)
columns = lp.chart_parse(mystring, lp.start_symbol())
[(str(s), str(lp.get_top(s))) for s in columns[-1].states]
result = LeoParser(RR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
RR_GRAMMAR2 = {
'<start>': ['<A>'],
'<A>': ['ab<A>', ''],
}
mystring2 = 'ababababab'
result = LeoParser(RR_GRAMMAR2, log=True).parse(mystring2)
for _ in result: pass
RR_GRAMMAR3 = {
'<start>': ['c<A>'],
'<A>': ['ab<A>', ''],
}
mystring3 = 'cababababab'
result = LeoParser(RR_GRAMMAR3, log=True).parse(mystring3)
for _ in result: pass
RR_GRAMMAR4 = {
'<start>': ['<A>c'],
'<A>': ['ab<A>', ''],
}
mystring4 = 'ababababc'
result = LeoParser(RR_GRAMMAR4, log=True).parse(mystring4)
for _ in result: pass
RR_GRAMMAR5 = {
'<start>': ['<A>'],
'<A>': ['ab<B>', ''],
'<B>': ['<A>'],
}
mystring5 = 'abababab'
result = LeoParser(RR_GRAMMAR5, log=True).parse(mystring5)
for _ in result: pass
RR_GRAMMAR6 = {
'<start>': ['<A>'],
'<A>': ['a<B>', ''],
'<B>': ['b<A>'],
}
mystring6 = 'abababab'
result = LeoParser(RR_GRAMMAR6, log=True).parse(mystring6)
for _ in result: pass
RR_GRAMMAR7 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a'],
}
mystring7 = 'aaaaaaaa'
result = LeoParser(RR_GRAMMAR7, log=True).parse(mystring7)
for _ in result: pass
result = LeoParser(LR_GRAMMAR, log=True).parse(mystring)
for _ in result: pass
class Column(Column):
def add_transitive(self, key, state):
assert key not in self.transitives
self.transitives[key] = TState(state.name, state.expr, state.dot,
state.s_col, state.e_col)
return self.transitives[key]
class State(State):
def back(self):
return TState(self.name, self.expr, self.dot - 1, self.s_col, self.e_col)
class TState(State):
def copy(self):
return TState(self.name, self.expr, self.dot, self.s_col, self.e_col)
class LeoParser(LeoParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self._postdots = {}
self.cgrammar = canonical(grammar, letters=True)
class LeoParser(LeoParser):
def uniq_postdot(self, st_A):
col_s1 = st_A.s_col
parent_states = [
s for s in col_s1.states if s.expr and s.at_dot() == st_A.name
]
if len(parent_states) > 1:
return None
matching_st_B = [s for s in parent_states if s.dot == len(s.expr) - 1]
if matching_st_B:
self._postdots[matching_st_B[0]._t()] = st_A
return matching_st_B[0]
return None
class LeoParser(LeoParser):
def expand_tstate(self, state, e):
if state._t() not in self._postdots:
return
c_C = self._postdots[state._t()]
e.add(c_C.advance())
self.expand_tstate(c_C.back(), e)
class LeoParser(LeoParser):
def rearrange(self, table):
f_table = [Column(c.index, c.letter) for c in table]
for col in table:
for s in col.states:
f_table[s.s_col.index].states.append(s)
return f_table
ep = LeoParser(RR_GRAMMAR)
columns = ep.chart_parse(mystring, ep.start_symbol())
r_table = ep.rearrange(columns)
for col in r_table:
print(col, "\n")
class LeoParser(LeoParser):
def parse(self, text):
cursor, states = self.parse_prefix(text)
start = next((s for s in states if s.finished()), None)
if cursor < len(text) or not start:
raise SyntaxError("at " + repr(text[cursor:]))
self.r_table = self.rearrange(self.table)
forest = self.extract_trees(self.parse_forest(self.table, start))
for tree in forest:
yield self.prune_tree(tree)
class LeoParser(LeoParser):
def parse_forest(self, chart, state):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
return super().parse_forest(chart, state)
result = LeoParser(RR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR2).parse(mystring2)
for tree in result:
assert mystring2 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR3).parse(mystring3)
for tree in result:
assert mystring3 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR4).parse(mystring4)
for tree in result:
assert mystring4 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR5).parse(mystring5)
for tree in result:
assert mystring5 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR6).parse(mystring6)
for tree in result:
assert mystring6 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR7).parse(mystring7)
for tree in result:
assert mystring7 == tree_to_string(tree)
result = LeoParser(LR_GRAMMAR).parse(mystring)
for tree in result:
assert mystring == tree_to_string(tree)
RR_GRAMMAR8 = {
'<start>': ['<A>'],
'<A>': ['a<A>', 'a']
}
mystring8 = 'aa'
RR_GRAMMAR9 = {
'<start>': ['<A>'],
'<A>': ['<B><A>', '<B>'],
'<B>': ['b']
}
mystring9 = 'bbbbbbb'
result = LeoParser(RR_GRAMMAR8).parse(mystring8)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring8 == tree_to_string(tree)
result = LeoParser(RR_GRAMMAR9).parse(mystring9)
for tree in result:
print(repr(tree_to_string(tree)))
assert mystring9 == tree_to_string(tree)
RECURSION_GRAMMAR = {
"<start>": ["<A>"],
"<A>": ["<A>", "<A>aa", "AA", "<B>"],
"<B>": ["<C>", "<C>cc" ,"CC"],
"<C>": ["<B>", "<B>bb", "BB"]
}
from ExpectError import ExpectTimeout
with ExpectTimeout(1, print_traceback=False):
mystring = 'AA'
parser = LeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
class FilteredLeoParser(LeoParser):
def forest(self, s, kind, seen, chart):
return self.parse_forest(chart, s, seen) if kind == 'n' else (s, [])
def parse_forest(self, chart, state, seen=None):
if isinstance(state, TState):
self.expand_tstate(state.back(), state.e_col)
def was_seen(chain, s):
if isinstance(s, str):
return False
if len(s.expr) > 1:
return False
return s in chain
if len(state.expr) > 1: # things get reset if we have a non loop
seen = set()
elif seen is None: # initialization
seen = {state}
pathexprs = self.parse_paths(state.expr, chart, state.s_col.index,
state.e_col.index) if state.expr else []
return state.name, [[(s, k, seen | {s}, chart)
for s, k in reversed(pathexpr)
if not was_seen(seen, s)] for pathexpr in pathexprs]
mystring = 'AA'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'AAaaaa'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'CC'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBcc'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BB'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
mystring = 'BBccbb'
parser = FilteredLeoParser(RECURSION_GRAMMAR)
tree, *_ = parser.parse(mystring)
assert tree_to_string(tree) == mystring
display_tree(tree)
class IterativeEarleyParser(EarleyParser):
def __init__(self, grammar, **kwargs):
super().__init__(grammar, **kwargs)
self.shuffle = kwargs.get('shuffle_rules', True)
def parse_paths(self, named_expr, chart, frm, til):
if not named_expr:
return []
paths = []
# stack of (expr, index, path) tuples
path_build_stack = [(named_expr, til, [])]
def evaluate_path(path, index, expr):
if expr: # path is still being built
path_build_stack.append((expr, index, path))
elif index == frm: # path is complete
paths.append(path)
while path_build_stack:
expr, chart_index, path = path_build_stack.pop()
*expr, symbol = expr
if symbol in self.cgrammar:
for state in chart[chart_index].states:
if state.name == symbol and state.finished():
extended_path = path + [(state, 'n')]
evaluate_path(extended_path, state.s_col.index, expr)
elif chart_index > 0 and chart[chart_index].letter == symbol:
extended_path = path + [(symbol, 't')]
evaluate_path(extended_path, chart_index - len(symbol), expr)
return paths
class IterativeEarleyParser(EarleyParser):
def parse_forest(self, chart, state):
if not state.expr:
return (state.name, [])
outermost_forest = []
forest_build_stack = [(state, outermost_forest)]
while forest_build_stack:
st, forest = forest_build_stack.pop()
paths = self.parse_paths(st.expr, chart, st.s_col.index,
st.e_col.index)
if not paths:
continue
next_path = random.choice(paths) if self.shuffle else paths[0]
path_forest = []
for symbol_or_state, kind in reversed(next_path):
if kind == 'n':
new_forest = []
forest_build_stack.append((symbol_or_state, new_forest))
path_forest.append((symbol_or_state.name, new_forest))
else:
path_forest.append((symbol_or_state, []))
forest.append(path_forest)
return (state.name, outermost_forest)
class IterativeEarleyParser(EarleyParser):
def extract_a_tree(self, forest_node):
outermost_tree = []
tree_build_stack = [(forest_node, outermost_tree)]
while tree_build_stack:
node, tree = tree_build_stack.pop()
name, node_paths = node
if node_paths:
for path in random.choice(node_paths):
new_tree = []
tree_build_stack.append((path, new_tree))
tree.append((path[0], new_tree))
else:
tree.append((name, []))
return (forest_node[0], outermost_tree)
class IterativeEarleyParser(EarleyParser):
def extract_trees(self, forest):
yield self.extract_a_tree(forest)
test_cases = [
(A1_GRAMMAR, '1-2-3+4-5'),
(A2_GRAMMAR, '1+2'),
(A3_GRAMMAR, '1+2+3-6=6-1-2-3'),
(LR_GRAMMAR, 'aaaaa'),
(RR_GRAMMAR, 'aa'),
(DIRECTLY_SELF_REFERRING, 'select a from a'),
(INDIRECTLY_SELF_REFERRING, 'select a from a'),
(RECURSION_GRAMMAR, 'AA'),
(RECURSION_GRAMMAR, 'AAaaaa'),
(RECURSION_GRAMMAR, 'BBccbb')
]
for i, (grammar, text) in enumerate(test_cases):
print(i, text)
tree, *_ = IterativeEarleyParser(grammar).parse(text)
assert text == tree_to_string(tree)
def firstset(grammar, nullable):
first = {i: {i} for i in terminals(grammar)}
for k in grammar:
first[k] = {EPSILON} if k in nullable else set()
return firstset_((rules(grammar), first, nullable))[1]
def first_expr(expr, first, nullable):
tokens = set()
for token in expr:
tokens |= first[token]
if token not in nullable:
break
return tokens
@fixpoint
def firstset_(arg):
(rules, first, epsilon) = arg
for A, expression in rules:
first[A] |= first_expr(expression, first, epsilon)
return (rules, first, epsilon)
firstset(canonical(A1_GRAMMAR), EPSILON)
EOF = '\0'
def followset(grammar, start):
follow = {i: set() for i in grammar}
follow[start] = {EOF}
epsilon = nullable(grammar)
first = firstset(grammar, epsilon)
return followset_((grammar, epsilon, first, follow))[-1]
@fixpoint
def followset_(arg):
grammar, epsilon, first, follow = arg
for A, expression in rules(grammar):
f_B = follow[A]
for t in reversed(expression):
if t in grammar:
follow[t] |= f_B
f_B = f_B | first[t] if t in epsilon else (first[t] - {EPSILON})
return (grammar, epsilon, first, follow)
followset(canonical(A1_GRAMMAR), START_SYMBOL)
class LL1Parser(Parser):
def parse_table(self):
self.my_rules = rules(self.cgrammar)
self.table = ... # fill in here to produce
def rules(self):
for i, rule in enumerate(self.my_rules):
print(i, rule)
def show_table(self):
ts = list(sorted(terminals(self.cgrammar)))
print('Rule Name\t| %s' % ' | '.join(t for t in ts))
for k in self.table:
pr = self.table[k]
actions = list(str(pr[t]) if t in pr else ' ' for t in ts)
print('%s \t| %s' % (k, ' | '.join(actions)))
for i, r in enumerate(rules(canonical(A2_GRAMMAR))):
print("%d\t %s := %s" % (i, r[0], r[1]))
class LL1Parser(LL1Parser):
def predict(self, rulepair, first, follow, epsilon):
A, rule = rulepair
rf = first_expr(rule, first, epsilon)
if nullable_expr(rule, epsilon):
rf |= follow[A]
return rf
def parse_table(self):
self.my_rules = rules(self.cgrammar)
epsilon = nullable(self.cgrammar)
first = firstset(self.cgrammar, epsilon)
# inefficient, can combine the three.
follow = followset(self.cgrammar, self.start_symbol())
ptable = [(i, self.predict(rule, first, follow, epsilon))
for i, rule in enumerate(self.my_rules)]
parse_tbl = {k: {} for k in self.cgrammar}
for i, pvals in ptable:
(k, expr) = self.my_rules[i]
parse_tbl[k].update({v: i for v in pvals})
self.table = parse_tbl
ll1parser = LL1Parser(A2_GRAMMAR)
ll1parser.parse_table()
ll1parser.show_table()
class LL1Parser(LL1Parser):
def parse_helper(self, stack, inplst):
inp, *inplst = inplst
exprs = []
while stack:
val, *stack = stack
if isinstance(val, tuple):
exprs.append(val)
elif val not in self.cgrammar: # terminal
assert val == inp
exprs.append(val)
inp, *inplst = inplst or [None]
else:
if inp is not None:
i = self.table[val][inp]
_, rhs = self.my_rules[i]
stack = rhs + [(val, len(rhs))] + stack
return self.linear_to_tree(exprs)
def parse(self, inp):
self.parse_table()
k, _ = self.my_rules[0]
stack = [k]
return self.parse_helper(stack, inp)
def linear_to_tree(self, arr):
stack = []
while arr:
elt = arr.pop(0)
if not isinstance(elt, tuple):
stack.append((elt, []))
else:
# get the last n
sym, n = elt
elts = stack[-n:] if n > 0 else []
stack = stack[0:len(stack) - n]
stack.append((sym, elts))
assert len(stack) == 1
return stack[0]
ll1parser = LL1Parser(A2_GRAMMAR)
tree = ll1parser.parse('1+2')
display_tree(tree)
| 0.316053 | 0.980356 |
##### Import relevant packages
```
import pandas as pd
import matplotlib
```
##### Reference similarweb .csv files and read to DataFrames (need to be more programmatic)
```
feb_data_file = 'similarweb_feb.csv'
mar_data_file = 'similarweb_march.csv'
apr_data_file = 'similarweb_april.csv'
df_feb = pd.read_csv(feb_data_file)
df_mar = pd.read_csv(mar_data_file)
df_apr = pd.read_csv(apr_data_file, encoding='iso-8859-1')
```
##### Preview the top 5 rows of the February DataFrame
```
df_apr.head()
```
##### Reorder columns to most logical sequencing (not required)
```
columns_ordered = ['Account ID','Account Name', 'Name', 'Domain', 'Website Category','Category Rank',
'Global Rank', 'Top Traffic Country','2nd Traffic Country', 'Has Data','Total Monthly Visits',
'Monthly Unique Visitors','Bounce Rate', 'Pages Per Visit', 'Average Visit Duration',
'Total Visits MoM Growth','Desktop Visits Share', 'Mobile Web Visits Share',
'Direct Visits Share','Display Ads Visits Share','Mail Visits Share',
'Paid Search Visits Share', 'Social Visits Share','Snapshot Date']
df_feb = df_feb[columns_ordered]
df_mar = df_mar[columns_ordered]
df_apr = df_apr[columns_ordered]
```
##### Fill all #NAs with 0s for Monthly Visits and Paid Search/Display Ads columns
```
df_feb['Total Monthly Visits'].fillna(0, inplace=True)
df_feb['Paid Search Visits Share'].fillna(0, inplace=True)
df_feb['Display Ads Visits Share'].fillna(0, inplace=True)
df_mar['Total Monthly Visits'].fillna(0, inplace=True)
df_mar['Paid Search Visits Share'].fillna(0, inplace=True)
df_mar['Display Ads Visits Share'].fillna(0, inplace=True)
df_apr['Total Monthly Visits'].fillna(0, inplace=True)
df_apr['Paid Search Visits Share'].fillna(0, inplace=True)
df_apr['Display Ads Visits Share'].fillna(0, inplace=True)
```
##### Compute Total Ad Spend Visits Share (Paid Search + Display Ads) then calculate # of visits from Total Monthly Visits
```
df_feb['Total Ad Spend Visits Share'] = df_feb['Paid Search Visits Share'] + df_feb['Display Ads Visits Share']
df_feb['Total Ad Spend Visits'] = df_feb['Total Monthly Visits']*df_feb['Total Ad Spend Visits Share']/100
df_mar['Total Ad Spend Visits Share'] = df_mar['Paid Search Visits Share'] + df_mar['Display Ads Visits Share']
df_mar['Total Ad Spend Visits'] = df_mar['Total Monthly Visits']*df_mar['Total Ad Spend Visits Share']/100
df_apr['Total Ad Spend Visits Share'] = df_apr['Paid Search Visits Share'] + df_apr['Display Ads Visits Share']
df_apr['Total Ad Spend Visits'] = df_apr['Total Monthly Visits']*df_apr['Total Ad Spend Visits Share']/100
```
###### Slim down DataFrame for only relevant columns to be output to .csv
```
df_feb_slim = df_feb[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
df_mar_slim = df_mar[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
df_apr_slim = df_apr[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
```
###### Left join March and February, add suffixes to column names and drop redundant columns
```
merged_apr_mar = df_apr_slim.merge(df_mar_slim,on=['Account ID'],how='left',suffixes=('_apr', '_mar'))
df_feb_slim.columns = df_feb_slim.columns.map(lambda x: str(x) + '_feb' if x != 'Account ID' else x)
merged_apr_mar_feb = merged_apr_mar.merge(df_feb_slim,on=['Account ID'],how='left')
merged_apr_mar_feb.drop(columns=['Account Name_feb', 'Domain_feb', 'Website Category_feb','Account Name_mar', 'Domain_mar', 'Website Category_mar'], axis=1, inplace=True)
```
##### Convert Account ID (15-char) to 18-char ID
```
account_map_file = 'account_id_map.csv'
df_map = pd.read_csv(account_map_file, encoding='iso-8859-1')
df_map.drop(columns=['Parent Account ID'], axis=1, inplace=True)
merged_apr_mar_feb = merged_apr_mar_feb.merge(df_map, on=['Account ID'], how='left')
cols = list(merged_apr_mar_feb.columns)
cols.insert(0,cols.pop(cols.index('Account ID (18)')))
merged_apr_mar_feb = merged_apr_mar_feb.loc[:,cols]
```
##### Export merged DataFrame to .csv file on local repository
```
merged_apr_mar_feb.to_csv('feb_march_apr_similarweb.csv')
```
|
github_jupyter
|
import pandas as pd
import matplotlib
feb_data_file = 'similarweb_feb.csv'
mar_data_file = 'similarweb_march.csv'
apr_data_file = 'similarweb_april.csv'
df_feb = pd.read_csv(feb_data_file)
df_mar = pd.read_csv(mar_data_file)
df_apr = pd.read_csv(apr_data_file, encoding='iso-8859-1')
df_apr.head()
columns_ordered = ['Account ID','Account Name', 'Name', 'Domain', 'Website Category','Category Rank',
'Global Rank', 'Top Traffic Country','2nd Traffic Country', 'Has Data','Total Monthly Visits',
'Monthly Unique Visitors','Bounce Rate', 'Pages Per Visit', 'Average Visit Duration',
'Total Visits MoM Growth','Desktop Visits Share', 'Mobile Web Visits Share',
'Direct Visits Share','Display Ads Visits Share','Mail Visits Share',
'Paid Search Visits Share', 'Social Visits Share','Snapshot Date']
df_feb = df_feb[columns_ordered]
df_mar = df_mar[columns_ordered]
df_apr = df_apr[columns_ordered]
df_feb['Total Monthly Visits'].fillna(0, inplace=True)
df_feb['Paid Search Visits Share'].fillna(0, inplace=True)
df_feb['Display Ads Visits Share'].fillna(0, inplace=True)
df_mar['Total Monthly Visits'].fillna(0, inplace=True)
df_mar['Paid Search Visits Share'].fillna(0, inplace=True)
df_mar['Display Ads Visits Share'].fillna(0, inplace=True)
df_apr['Total Monthly Visits'].fillna(0, inplace=True)
df_apr['Paid Search Visits Share'].fillna(0, inplace=True)
df_apr['Display Ads Visits Share'].fillna(0, inplace=True)
df_feb['Total Ad Spend Visits Share'] = df_feb['Paid Search Visits Share'] + df_feb['Display Ads Visits Share']
df_feb['Total Ad Spend Visits'] = df_feb['Total Monthly Visits']*df_feb['Total Ad Spend Visits Share']/100
df_mar['Total Ad Spend Visits Share'] = df_mar['Paid Search Visits Share'] + df_mar['Display Ads Visits Share']
df_mar['Total Ad Spend Visits'] = df_mar['Total Monthly Visits']*df_mar['Total Ad Spend Visits Share']/100
df_apr['Total Ad Spend Visits Share'] = df_apr['Paid Search Visits Share'] + df_apr['Display Ads Visits Share']
df_apr['Total Ad Spend Visits'] = df_apr['Total Monthly Visits']*df_apr['Total Ad Spend Visits Share']/100
df_feb_slim = df_feb[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
df_mar_slim = df_mar[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
df_apr_slim = df_apr[['Account ID', 'Account Name', 'Domain', 'Website Category', 'Total Monthly Visits',
'Total Visits MoM Growth','Total Ad Spend Visits Share','Total Ad Spend Visits']]
merged_apr_mar = df_apr_slim.merge(df_mar_slim,on=['Account ID'],how='left',suffixes=('_apr', '_mar'))
df_feb_slim.columns = df_feb_slim.columns.map(lambda x: str(x) + '_feb' if x != 'Account ID' else x)
merged_apr_mar_feb = merged_apr_mar.merge(df_feb_slim,on=['Account ID'],how='left')
merged_apr_mar_feb.drop(columns=['Account Name_feb', 'Domain_feb', 'Website Category_feb','Account Name_mar', 'Domain_mar', 'Website Category_mar'], axis=1, inplace=True)
account_map_file = 'account_id_map.csv'
df_map = pd.read_csv(account_map_file, encoding='iso-8859-1')
df_map.drop(columns=['Parent Account ID'], axis=1, inplace=True)
merged_apr_mar_feb = merged_apr_mar_feb.merge(df_map, on=['Account ID'], how='left')
cols = list(merged_apr_mar_feb.columns)
cols.insert(0,cols.pop(cols.index('Account ID (18)')))
merged_apr_mar_feb = merged_apr_mar_feb.loc[:,cols]
merged_apr_mar_feb.to_csv('feb_march_apr_similarweb.csv')
| 0.283285 | 0.878262 |
# Histogram (automatic and manual methods)
How to plot a histogram in Python. This goes over automatic methods (which is what you'll use in practice) and also digs a bit deeper into how to manually calculate the bin edges and counts in a histogram, similar to what is described in Files → Labs → [Guidelines to plot a histogram.pdf](https://utah.instructure.com/courses/722066/files/folder/Labs?preview=119193793)
## Import Modules
```
%matplotlib inline
from matplotlib.pyplot import hist
import pandas as pd
import numpy as np
from math import ceil, sqrt
import os
```
## Read Data
```
df = pd.read_excel("test_data.xlsx")
```
## Plotting
### Using Defaults (10 bins)
```
df.plot.hist()
```
At this point, we could be done, as we've plotted a histogram. The instructions for the assignment talk about using the square root estimator, however.
### Using Square Root
```
hist(df, bins="sqrt")
```
At this point, we really are done, but let's take a look at other things we can do with the hist function and what's going on under the hood with determining the bin frequencies and bin edges.
### Using "Auto" Number of Bins
```
hist(df, bins="auto")
```
### Determine Bin Counts and Bin Edges using NumPy
```
counts, bins = np.histogram(df['column1'], bins="sqrt")
print(counts)
print(bins)
hist(df, bins=bins)
```
### Manually determine bin counts and edges
This mirrors the example on Canvas from Files → Labs → Lab1 → [Lab1-histogram.pdf](https://utah.instructure.com/courses/722066/files/folder/Labs/Lab1?preview=119193799)
#### Dataset min, max, and length
```
col1 = df['column1']
mn = min(col1)
mx = max(col1)
print(mn, mx)
n = len(col1)
print(n)
```
#### Number of bins (using square root), bin width, and bin edges
```
nbins = ceil(sqrt(n))
print(nbins)
binwidth = (mx-mn)/nbins
print(binwidth)
bins = np.linspace(mn, mx, num=nbins+1)
```
#### Histogram using manually determined bin edges
```
hist(df, bins=bins)
```
### Histogram without using hist()
Find how the data fits into the bins.
```
ids = np.searchsorted(bins, col1)
np.unique(ids)
ids[ids==0]=1
print(ids)
```
Count up the frequencies
```
freq = np.bincount(ids)[1:]
print(freq)
```
Change the bin edges into pairs.
```
ab = list(zip(bins[:-1], bins[1:]))
print(ab)
```
Find the center of each bin
```
bin_centers = [(a + b)/2 for a, b in ab]
```
Plot using the bin_centers, frequency values, and calculated bin width.
```
plt.bar(bin_centers, freq, width=binwidth)
```
Note that you should never really need to do this kind of "manual" plotting, but this illustrates how you would essentially calculate this "by hand", similar to the instructions shown on Canvas in the file linked above. In other words, plt.hist(df, bins="auto") is pretty much all you'd need.
## Print this notebook to PDF
```
os.system("jupyter nbconvert --to pdf histogram-auto-vs-manual.ipynb")
```
|
github_jupyter
|
%matplotlib inline
from matplotlib.pyplot import hist
import pandas as pd
import numpy as np
from math import ceil, sqrt
import os
df = pd.read_excel("test_data.xlsx")
df.plot.hist()
hist(df, bins="sqrt")
hist(df, bins="auto")
counts, bins = np.histogram(df['column1'], bins="sqrt")
print(counts)
print(bins)
hist(df, bins=bins)
col1 = df['column1']
mn = min(col1)
mx = max(col1)
print(mn, mx)
n = len(col1)
print(n)
nbins = ceil(sqrt(n))
print(nbins)
binwidth = (mx-mn)/nbins
print(binwidth)
bins = np.linspace(mn, mx, num=nbins+1)
hist(df, bins=bins)
ids = np.searchsorted(bins, col1)
np.unique(ids)
ids[ids==0]=1
print(ids)
freq = np.bincount(ids)[1:]
print(freq)
ab = list(zip(bins[:-1], bins[1:]))
print(ab)
bin_centers = [(a + b)/2 for a, b in ab]
plt.bar(bin_centers, freq, width=binwidth)
os.system("jupyter nbconvert --to pdf histogram-auto-vs-manual.ipynb")
| 0.26827 | 0.991058 |
<a href="https://colab.research.google.com/github/maegop/Exploratory-Data-Analysis-for-US-Accidents/blob/main/Pokemon_Stats.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
The dataset we'll be using is the compilation of stats and traits for the Pokémon video games. Pokémon is a popular game for generations of Nintendo handheld video game consoles where players collect and train animal-like creatures called Pokémon. We'll be creating a model to try to predict whether a Pokémon is a legendary Pokémon, a rare type of Pokémon who's the only one of its species.
There are a lot of existing compilations of Pokémon stats, but we'll be using a .CSV version found on Kaggle. There's a download button on the website, so save the file to your computer and we can begin.
Import data
```
!pip install opendatasets --upgrade --quiet
import opendatasets as od
download_url = 'https://www.kaggle.com/alopez247/pokemon'
od.download(download_url)
df = pd.read_csv('/content/pokemon/pokemon_alopez247.csv')
import tensorflow as tf
from tensorflow import keras
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
df.columns
```
We'll narrow our focus a little and only select categories we think will be relevant
```
df = df[['isLegendary','Generation', 'Type_1', 'Type_2', 'HP', 'Attack', 'Defense', 'Sp_Atk', 'Sp_Def', 'Speed','Color','Egg_Group_1','Height_m','Weight_kg','Body_Style']]
df
```
### Normalization
Format the data to be read by the model.
* Make sure all the data is numerical.
A few of the categories aren't numerical however. One example is the category that we'll be training our model to detect: the "isLegendary" column of data. These are the labels that we will eventually separate from the rest of the data and use as an answer key for the model's training. We'll convert this column from boolean "False" and "True" statements to the equivalent "0" and "1" integ
```
df['isLegendary'] = df['isLegendary'].astype(int)
df['isLegendary']
```
Converting data to numbers could be just assign a number to each category, such as: Water = 1, Fire = 2, Grass = 3 and so on. But this isn't a good idea because these numerical assignments aren't ordinal; they don't lie on a scale. By doing this, we would be implying that Water is closer to Fire than it is Grass, which doesn't really make sense.
The solution to this is to create dummy variables. By doing this we'll be creating a new column for each possible variable. There will be a column called "Water" that would be a 1 if it was a water Pokémon, and a 0 if it wasn't. Then there will be another column called "Fire" that would be a 1 if it was a fire Pokémon, and so forth for the rest of the types. This prevents us from implying any pattern or direction among the types.
```
def dummy_creation(df, dummy_categories):
for i in dummy_categories:
df_dummy = pd.get_dummies(df[i])
df = pd.concat([df,df_dummy],axis=1)
df = df.drop(i, axis=1)
return(df)
```
This function first uses pd.get_dummies to create a dummy DataFrame of that category. As it's a seperate DataFrame, we'll need to concatenate it to our original DataFrame. And since we now have the variables represented properly as separate columns, we drop the original column. Having this in a function is nice because we can quickly do this for many categories:
```
df = dummy_creation(df, ['Egg_Group_1', 'Body_Style', 'Color','Type_1', 'Type_2'])
df
```
The importance of creating dummy variables
* Some categories are not numerical.
* Converting multiple categories into numbers implies that they are on a scale.
* The categories for "Type_1" should all be boolean (0 or 1).
* All of the above are true.
```
df.info()
def train_test_splitter(DataFrame, column):
df_train = DataFrame.loc[df[column] != 1]
df_test = DataFrame.loc[df[column] == 1]
df_train = df_train.drop(column, axis=1)
df_test = df_test.drop(column, axis=1)
return(df_train, df_test)
df_train, df_test = train_test_splitter(df, 'Generation')
def label_delineator(df_train, df_test, label):
train_data = df_train.drop(label, axis=1).values
train_labels = df_train[label].values
test_data = df_test.drop(label,axis=1).values
test_labels = df_test[label].values
return(train_data, train_labels, test_data, test_labels)
train_data, train_labels, test_data, test_labels = label_delineator(df_train, df_test, 'isLegendary')
```
|
github_jupyter
|
!pip install opendatasets --upgrade --quiet
import opendatasets as od
download_url = 'https://www.kaggle.com/alopez247/pokemon'
od.download(download_url)
df = pd.read_csv('/content/pokemon/pokemon_alopez247.csv')
import tensorflow as tf
from tensorflow import keras
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import preprocessing
df.columns
df = df[['isLegendary','Generation', 'Type_1', 'Type_2', 'HP', 'Attack', 'Defense', 'Sp_Atk', 'Sp_Def', 'Speed','Color','Egg_Group_1','Height_m','Weight_kg','Body_Style']]
df
df['isLegendary'] = df['isLegendary'].astype(int)
df['isLegendary']
def dummy_creation(df, dummy_categories):
for i in dummy_categories:
df_dummy = pd.get_dummies(df[i])
df = pd.concat([df,df_dummy],axis=1)
df = df.drop(i, axis=1)
return(df)
df = dummy_creation(df, ['Egg_Group_1', 'Body_Style', 'Color','Type_1', 'Type_2'])
df
df.info()
def train_test_splitter(DataFrame, column):
df_train = DataFrame.loc[df[column] != 1]
df_test = DataFrame.loc[df[column] == 1]
df_train = df_train.drop(column, axis=1)
df_test = df_test.drop(column, axis=1)
return(df_train, df_test)
df_train, df_test = train_test_splitter(df, 'Generation')
def label_delineator(df_train, df_test, label):
train_data = df_train.drop(label, axis=1).values
train_labels = df_train[label].values
test_data = df_test.drop(label,axis=1).values
test_labels = df_test[label].values
return(train_data, train_labels, test_data, test_labels)
train_data, train_labels, test_data, test_labels = label_delineator(df_train, df_test, 'isLegendary')
| 0.420362 | 0.985356 |
```
from fastai.collab import *
import pandas as pd
import numpy as np
url_books = 'https://raw.githubusercontent.com/TannerGilbert/Tutorials/master/Recommendation%20System/books.csv'
url_ratings = 'https://raw.githubusercontent.com/TannerGilbert/Tutorials/master/Recommendation%20System/ratings.csv'
ratings = pd.read_csv(url_ratings, error_bad_lines=False, warn_bad_lines=False)
print(ratings.head(5))
print('---------------------')
print(ratings.shape)
print('---------------------')
print(ratings.isnull().sum())
data = CollabDataBunch.from_df(ratings, seed=42, valid_pct=0.1, user_name='user_id', item_name='book_id', rating_name='rating')
data.show_batch()
ratings.rating.min(), ratings.rating.max()
```
### EmbeddingDotBias Model
```
learn = collab_learner(data, n_factors=40, y_range=(1, 5), wd=1e-1, model_dir="/tmp/model/", path="/tmp/")
print(learn.summary())
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(5, 3e-4)
learn.save('goodbooks-dot-1')
```
### EmbeddingNN Model
```
learn = collab_learner(data, use_nn=True, emb_szs={'user_id': 40, 'book_id':40}, layers=[256, 128], y_range=(1, 5))
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(5, 1e-2)
learn.save('goodbooks-nn-1')
```
### Interpretation
```
learn = collab_learner(data, n_factors=40, y_range=(1, 5), wd=1e-1, model_dir="/tmp/model/", path="/tmp/")
learn.load('goodbooks-dot-1')
books = pd.read_csv(url_books, error_bad_lines=False, warn_bad_lines=False)
books.head()
g = ratings.groupby('book_id')['rating'].count()
top_books = g.sort_values(ascending=False).index.values[:1000]
top_books = top_books.astype(str)
top_books[:10]
top_books_with_name = []
for book in top_books:
top_books_with_name.append(books[(books['id']==int(book))]['title'].iloc[0])
top_books_with_name = np.array(top_books_with_name)
top_books_with_name
```
### Book Bias
```
learn.model
book_bias = learn.bias(top_books, is_item=True)
mean_ratings = ratings.groupby('book_id')['rating'].mean()
book_ratings = [(b, top_books_with_name[i], mean_ratings.loc[int(tb)]) for i, (tb, b) in enumerate(zip(top_books, book_bias))]
item0 = lambda o:o[0]
sorted(book_ratings, key=item0)[:15]
sorted(book_ratings, key=item0, reverse=True)[:15]
book_w = learn.weight(top_books, is_item=True)
book_w.shape
book_pca = book_w.pca(3)
book_pca.shape
fac0,fac1,fac2 = book_pca.t()
book_comp = [(f, i) for f,i in zip(fac0, top_books_with_name)]
sorted(book_comp, key=itemgetter(0), reverse=True)[:10]
sorted(book_comp, key=itemgetter(0))[:10]
book_comp = [(f, i) for f,i in zip(fac1, top_books_with_name)]
sorted(book_comp, key=itemgetter(0), reverse=True)[:10]
sorted(book_comp, key=itemgetter(0))[:10]
idxs = np.random.choice(len(top_books_with_name), 50, replace=False)
idxs = list(range(50))
X = fac0[idxs]
Y = fac2[idxs]
plt.figure(figsize=(15,15))
plt.scatter(X, Y)
for i, x, y in zip(top_books_with_name[idxs], X, Y):
plt.text(x,y,i, color=np.random.rand(3)*0.7, fontsize=11)
plt.show()
```
|
github_jupyter
|
from fastai.collab import *
import pandas as pd
import numpy as np
url_books = 'https://raw.githubusercontent.com/TannerGilbert/Tutorials/master/Recommendation%20System/books.csv'
url_ratings = 'https://raw.githubusercontent.com/TannerGilbert/Tutorials/master/Recommendation%20System/ratings.csv'
ratings = pd.read_csv(url_ratings, error_bad_lines=False, warn_bad_lines=False)
print(ratings.head(5))
print('---------------------')
print(ratings.shape)
print('---------------------')
print(ratings.isnull().sum())
data = CollabDataBunch.from_df(ratings, seed=42, valid_pct=0.1, user_name='user_id', item_name='book_id', rating_name='rating')
data.show_batch()
ratings.rating.min(), ratings.rating.max()
learn = collab_learner(data, n_factors=40, y_range=(1, 5), wd=1e-1, model_dir="/tmp/model/", path="/tmp/")
print(learn.summary())
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(5, 3e-4)
learn.save('goodbooks-dot-1')
learn = collab_learner(data, use_nn=True, emb_szs={'user_id': 40, 'book_id':40}, layers=[256, 128], y_range=(1, 5))
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(5, 1e-2)
learn.save('goodbooks-nn-1')
learn = collab_learner(data, n_factors=40, y_range=(1, 5), wd=1e-1, model_dir="/tmp/model/", path="/tmp/")
learn.load('goodbooks-dot-1')
books = pd.read_csv(url_books, error_bad_lines=False, warn_bad_lines=False)
books.head()
g = ratings.groupby('book_id')['rating'].count()
top_books = g.sort_values(ascending=False).index.values[:1000]
top_books = top_books.astype(str)
top_books[:10]
top_books_with_name = []
for book in top_books:
top_books_with_name.append(books[(books['id']==int(book))]['title'].iloc[0])
top_books_with_name = np.array(top_books_with_name)
top_books_with_name
learn.model
book_bias = learn.bias(top_books, is_item=True)
mean_ratings = ratings.groupby('book_id')['rating'].mean()
book_ratings = [(b, top_books_with_name[i], mean_ratings.loc[int(tb)]) for i, (tb, b) in enumerate(zip(top_books, book_bias))]
item0 = lambda o:o[0]
sorted(book_ratings, key=item0)[:15]
sorted(book_ratings, key=item0, reverse=True)[:15]
book_w = learn.weight(top_books, is_item=True)
book_w.shape
book_pca = book_w.pca(3)
book_pca.shape
fac0,fac1,fac2 = book_pca.t()
book_comp = [(f, i) for f,i in zip(fac0, top_books_with_name)]
sorted(book_comp, key=itemgetter(0), reverse=True)[:10]
sorted(book_comp, key=itemgetter(0))[:10]
book_comp = [(f, i) for f,i in zip(fac1, top_books_with_name)]
sorted(book_comp, key=itemgetter(0), reverse=True)[:10]
sorted(book_comp, key=itemgetter(0))[:10]
idxs = np.random.choice(len(top_books_with_name), 50, replace=False)
idxs = list(range(50))
X = fac0[idxs]
Y = fac2[idxs]
plt.figure(figsize=(15,15))
plt.scatter(X, Y)
for i, x, y in zip(top_books_with_name[idxs], X, Y):
plt.text(x,y,i, color=np.random.rand(3)*0.7, fontsize=11)
plt.show()
| 0.368747 | 0.745028 |
# Load previous results
```
import pickle
with open("results.pkl", "rb") as fh:
final_results = pickle.load(fh)
kw_threshholds = range(1, 21, 1)
with open("results.pkl", "wb") as fh:
pickle.dump(final_results, fh)
from collections import namedtuple
def get_data(algorithm, corpus):
Retrieval_scores = namedtuple("Retrieval_scores", "p r f a".split())
scores = final_results[corpus]
precision = [scores[i][algorithm]["precision"].mean() for i in kw_threshholds]
recall = [scores[i][algorithm]["recall"].mean() for i in kw_threshholds]
f1 = [scores[i][algorithm]["f1"].mean() for i in kw_threshholds]
return Retrieval_scores(precision, recall, f1, algorithm)
corpus = "semeval"
X = get_data("tfidf", corpus)
Y = get_data("tfidfed_textrank", corpus)
```
### Relative enhancement
```
def publication_name(n):
if n == "tfidfed_rake":
return "$Rake_s$"
elif n == "rake":
return "$Rake$"
elif n == "tfidfed_textrank":
return "$Textrank_s$"
elif n == "textrank":
return "$Textrank$"
elif n == "tfidf":
return "$tf-idf$"
elif n == "frankenrake":
return "$Ensemble$"
else:
raise Exception(f"No proper name substitution available for {n}")
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
plt.rcParams["font.family"] = 'serif'
colors = sns.color_palette("Set1", 6)
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(kw_threshholds, X.p, ':v', c=colors[0], label=f"$\\pi$ {publication_name(X.a)}")
ax.plot(kw_threshholds, X.r, ':D', c=colors[1], label=f"$\\rho$ {publication_name(X.a)}")
ax.plot(kw_threshholds, X.f, ':d', c=colors[2], label=f"F1 {publication_name(X.a)}")
ax.plot(kw_threshholds, Y.p, '-v', c=colors[0], alpha=.4, label=f"$\pi$ {publication_name(Y.a)}")
ax.plot(kw_threshholds, Y.r, '-D', c=colors[1], alpha=.4, label=f"$\\rho$ {publication_name(Y.a)}")
ax.plot(kw_threshholds, Y.f, '-d', c=colors[2], alpha=.4, label=f"F1 {publication_name(Y.a)}")
ax.set_ylim(0.0, .6)
ax.set_xlabel('Number of Keyphrases', fontsize=16)
ax.set_ylabel('Score', fontsize=16)
ax.legend(fontsize=14, frameon=False)
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=14)
ax.yaxis.set_major_formatter(FormatStrFormatter('%.1f'))
#ax.set_facecolor("white")
for spine in plt.gca().spines.values():
spine.set_visible(True)
#plt.title(f"{corpus_name} without Fuzzy Matching + KW Removal", fontsize=18)
plt.xticks(kw_threshholds)
fig.savefig(f"result_plots/{corpus}/{publication_name(X.a)}_vs_{publication_name(Y.a)}.pdf",
format="pdf", transparent=True, bbox_inches="tight")
plt.show()
```
# Plotting
```
def plot_ranking_stats(num_kwds, metric, corpus, algorithm_a, algorithm_b, algorithm_c, algorithm_d):
scores = final_results[corpus]
y_a = scores[num_kwds][algorithm_a].sort_values(by=metric)[::-1][metric]
mean_a = scores[num_kwds][algorithm_a][metric].mean()
y_b = scores[num_kwds][algorithm_b].sort_values(by=metric)[::-1][metric]
mean_b = scores[num_kwds][algorithm_b][metric].mean()
y_c = scores[num_kwds][algorithm_c].sort_values(by=metric)[::-1][metric]
mean_c = scores[num_kwds][algorithm_c][metric].mean()
y_d = scores[num_kwds][algorithm_d].sort_values(by=metric)[::-1][metric]
mean_d = scores[num_kwds][algorithm_d][metric].mean()
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(range(y_a.values.shape[0]), y_a.values, label=f"{publication_name(algorithm_a)}")
#ax.axhline(mean_a, color="red")
ax.plot(range(y_b.values.shape[0]), y_b.values, label=f"{publication_name(algorithm_b)}")
#ax.axhline(mean_b, color="red")
ax.plot(range(y_c.values.shape[0]), y_c.values, label=f"{publication_name(algorithm_c)}")
#ax.axhline(mean_b, color="red")
ax.plot(range(y_d.values.shape[0]), y_d.values, label=f"{publication_name(algorithm_d)}")
#ax.axhline(mean_b, color="red")
ax.set_xlabel("Rank", fontsize=16)
ax.set_ylabel("Score", fontsize=16)
ax.set_ylim(-0.02, 1)
ax.legend(fontsize=14, frameon=False)
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=14)
fig.savefig(f"result_plots/{corpus}/rank_plots_{metric}@{num_kwds}_Keywords.pdf",
format="pdf", transparent=True, bbox_inches="tight")
plt.show()
plot_ranking_stats(5, "f1", "semeval", "rake", "tfidfed_rake", "textrank", "tfidfed_textrank")
```
|
github_jupyter
|
import pickle
with open("results.pkl", "rb") as fh:
final_results = pickle.load(fh)
kw_threshholds = range(1, 21, 1)
with open("results.pkl", "wb") as fh:
pickle.dump(final_results, fh)
from collections import namedtuple
def get_data(algorithm, corpus):
Retrieval_scores = namedtuple("Retrieval_scores", "p r f a".split())
scores = final_results[corpus]
precision = [scores[i][algorithm]["precision"].mean() for i in kw_threshholds]
recall = [scores[i][algorithm]["recall"].mean() for i in kw_threshholds]
f1 = [scores[i][algorithm]["f1"].mean() for i in kw_threshholds]
return Retrieval_scores(precision, recall, f1, algorithm)
corpus = "semeval"
X = get_data("tfidf", corpus)
Y = get_data("tfidfed_textrank", corpus)
def publication_name(n):
if n == "tfidfed_rake":
return "$Rake_s$"
elif n == "rake":
return "$Rake$"
elif n == "tfidfed_textrank":
return "$Textrank_s$"
elif n == "textrank":
return "$Textrank$"
elif n == "tfidf":
return "$tf-idf$"
elif n == "frankenrake":
return "$Ensemble$"
else:
raise Exception(f"No proper name substitution available for {n}")
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import FormatStrFormatter
plt.rcParams["font.family"] = 'serif'
colors = sns.color_palette("Set1", 6)
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(kw_threshholds, X.p, ':v', c=colors[0], label=f"$\\pi$ {publication_name(X.a)}")
ax.plot(kw_threshholds, X.r, ':D', c=colors[1], label=f"$\\rho$ {publication_name(X.a)}")
ax.plot(kw_threshholds, X.f, ':d', c=colors[2], label=f"F1 {publication_name(X.a)}")
ax.plot(kw_threshholds, Y.p, '-v', c=colors[0], alpha=.4, label=f"$\pi$ {publication_name(Y.a)}")
ax.plot(kw_threshholds, Y.r, '-D', c=colors[1], alpha=.4, label=f"$\\rho$ {publication_name(Y.a)}")
ax.plot(kw_threshholds, Y.f, '-d', c=colors[2], alpha=.4, label=f"F1 {publication_name(Y.a)}")
ax.set_ylim(0.0, .6)
ax.set_xlabel('Number of Keyphrases', fontsize=16)
ax.set_ylabel('Score', fontsize=16)
ax.legend(fontsize=14, frameon=False)
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=14)
ax.yaxis.set_major_formatter(FormatStrFormatter('%.1f'))
#ax.set_facecolor("white")
for spine in plt.gca().spines.values():
spine.set_visible(True)
#plt.title(f"{corpus_name} without Fuzzy Matching + KW Removal", fontsize=18)
plt.xticks(kw_threshholds)
fig.savefig(f"result_plots/{corpus}/{publication_name(X.a)}_vs_{publication_name(Y.a)}.pdf",
format="pdf", transparent=True, bbox_inches="tight")
plt.show()
def plot_ranking_stats(num_kwds, metric, corpus, algorithm_a, algorithm_b, algorithm_c, algorithm_d):
scores = final_results[corpus]
y_a = scores[num_kwds][algorithm_a].sort_values(by=metric)[::-1][metric]
mean_a = scores[num_kwds][algorithm_a][metric].mean()
y_b = scores[num_kwds][algorithm_b].sort_values(by=metric)[::-1][metric]
mean_b = scores[num_kwds][algorithm_b][metric].mean()
y_c = scores[num_kwds][algorithm_c].sort_values(by=metric)[::-1][metric]
mean_c = scores[num_kwds][algorithm_c][metric].mean()
y_d = scores[num_kwds][algorithm_d].sort_values(by=metric)[::-1][metric]
mean_d = scores[num_kwds][algorithm_d][metric].mean()
fig, ax = plt.subplots(figsize=(8, 6))
ax.plot(range(y_a.values.shape[0]), y_a.values, label=f"{publication_name(algorithm_a)}")
#ax.axhline(mean_a, color="red")
ax.plot(range(y_b.values.shape[0]), y_b.values, label=f"{publication_name(algorithm_b)}")
#ax.axhline(mean_b, color="red")
ax.plot(range(y_c.values.shape[0]), y_c.values, label=f"{publication_name(algorithm_c)}")
#ax.axhline(mean_b, color="red")
ax.plot(range(y_d.values.shape[0]), y_d.values, label=f"{publication_name(algorithm_d)}")
#ax.axhline(mean_b, color="red")
ax.set_xlabel("Rank", fontsize=16)
ax.set_ylabel("Score", fontsize=16)
ax.set_ylim(-0.02, 1)
ax.legend(fontsize=14, frameon=False)
ax.tick_params(axis='x', labelsize=14)
ax.tick_params(axis='y', labelsize=14)
fig.savefig(f"result_plots/{corpus}/rank_plots_{metric}@{num_kwds}_Keywords.pdf",
format="pdf", transparent=True, bbox_inches="tight")
plt.show()
plot_ranking_stats(5, "f1", "semeval", "rake", "tfidfed_rake", "textrank", "tfidfed_textrank")
| 0.656438 | 0.638807 |
# Clustering Algorithms
1. K Means Clustering
2. Hierarchical Clustering
# Loading packages
```
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.cluster import KMeans
dataset = pd.read_csv('Mall_Customers.csv')
dataset.head(1)
dataset["Genre"].value_counts()
dataset["Age"].hist(bins = 100)
dataset["Annual Income (k$)"].value_counts()
dataset["Annual Income (k$)"].hist(bins = 75)
dataset["Spending Score (1-100)"].value_counts()
dataset["Spending Score (1-100)"].hist(bins = 100)
```
# K Means Clustering
```
# Importing the dataset
X = dataset.iloc[:, [3, 4]].values
# Using the elbow method to find the optimal number of clusters
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++', random_state = 42)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1, 11), wcss)
plt.title('The Elbow Method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS')
plt.show()
# Training the K-Means model on the dataset
kmeans = KMeans(n_clusters = 5, init = 'k-means++', random_state = 42)
y_kmeans = kmeans.fit_predict(X)
# Visualising the clusters
plt.scatter(X[y_kmeans == 0, 0], X[y_kmeans == 0, 1], s = 100, c = 'red', label = 'Cluster 1')
plt.scatter(X[y_kmeans == 1, 0], X[y_kmeans == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')
plt.scatter(X[y_kmeans == 2, 0], X[y_kmeans == 2, 1], s = 100, c = 'green', label = 'Cluster 3')
plt.scatter(X[y_kmeans == 3, 0], X[y_kmeans == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')
plt.scatter(X[y_kmeans == 4, 0], X[y_kmeans == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], s = 300, c = 'yellow', label = 'Centroids')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
```
# Hierarchical Clustering
```
# Using the dendrogram to find the optimal number of clusters
import scipy.cluster.hierarchy as sch
dendrogram = sch.dendrogram(sch.linkage(X, method = 'ward'))
plt.title('Dendrogram')
plt.xlabel('Customers')
plt.ylabel('Euclidean distances')
plt.show()
# Training the Hierarchical Clustering model on the dataset
from sklearn.cluster import AgglomerativeClustering
hc = AgglomerativeClustering(n_clusters = 5, affinity = 'euclidean', linkage = 'ward')
y_hc = hc.fit_predict(X)
# Visualising the clusters
plt.scatter(X[y_hc == 0, 0], X[y_hc == 0, 1], s = 100, c = 'red', label = 'Cluster 1')
plt.scatter(X[y_hc == 1, 0], X[y_hc == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')
plt.scatter(X[y_hc == 2, 0], X[y_hc == 2, 1], s = 100, c = 'green', label = 'Cluster 3')
plt.scatter(X[y_hc == 3, 0], X[y_hc == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')
plt.scatter(X[y_hc == 4, 0], X[y_hc == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from sklearn.cluster import KMeans
dataset = pd.read_csv('Mall_Customers.csv')
dataset.head(1)
dataset["Genre"].value_counts()
dataset["Age"].hist(bins = 100)
dataset["Annual Income (k$)"].value_counts()
dataset["Annual Income (k$)"].hist(bins = 75)
dataset["Spending Score (1-100)"].value_counts()
dataset["Spending Score (1-100)"].hist(bins = 100)
# Importing the dataset
X = dataset.iloc[:, [3, 4]].values
# Using the elbow method to find the optimal number of clusters
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters = i, init = 'k-means++', random_state = 42)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1, 11), wcss)
plt.title('The Elbow Method')
plt.xlabel('Number of clusters')
plt.ylabel('WCSS')
plt.show()
# Training the K-Means model on the dataset
kmeans = KMeans(n_clusters = 5, init = 'k-means++', random_state = 42)
y_kmeans = kmeans.fit_predict(X)
# Visualising the clusters
plt.scatter(X[y_kmeans == 0, 0], X[y_kmeans == 0, 1], s = 100, c = 'red', label = 'Cluster 1')
plt.scatter(X[y_kmeans == 1, 0], X[y_kmeans == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')
plt.scatter(X[y_kmeans == 2, 0], X[y_kmeans == 2, 1], s = 100, c = 'green', label = 'Cluster 3')
plt.scatter(X[y_kmeans == 3, 0], X[y_kmeans == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')
plt.scatter(X[y_kmeans == 4, 0], X[y_kmeans == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], s = 300, c = 'yellow', label = 'Centroids')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
# Using the dendrogram to find the optimal number of clusters
import scipy.cluster.hierarchy as sch
dendrogram = sch.dendrogram(sch.linkage(X, method = 'ward'))
plt.title('Dendrogram')
plt.xlabel('Customers')
plt.ylabel('Euclidean distances')
plt.show()
# Training the Hierarchical Clustering model on the dataset
from sklearn.cluster import AgglomerativeClustering
hc = AgglomerativeClustering(n_clusters = 5, affinity = 'euclidean', linkage = 'ward')
y_hc = hc.fit_predict(X)
# Visualising the clusters
plt.scatter(X[y_hc == 0, 0], X[y_hc == 0, 1], s = 100, c = 'red', label = 'Cluster 1')
plt.scatter(X[y_hc == 1, 0], X[y_hc == 1, 1], s = 100, c = 'blue', label = 'Cluster 2')
plt.scatter(X[y_hc == 2, 0], X[y_hc == 2, 1], s = 100, c = 'green', label = 'Cluster 3')
plt.scatter(X[y_hc == 3, 0], X[y_hc == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4')
plt.scatter(X[y_hc == 4, 0], X[y_hc == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
| 0.809427 | 0.950503 |
# **Amazon Lookout for Equipment** - Getting started
*Part 5 - Scheduling regular inference calls*
## Initialization
---
This repository is structured as follow:
```sh
. lookout-equipment-demo
|
├── data/
| ├── interim # Temporary intermediate data are stored here
| ├── processed # Finalized datasets are usually stored here
| | # before they are sent to S3 to allow the
| | # service to reach them
| └── raw # Immutable original data are stored here
|
├── getting_started/
| ├── 1_data_preparation.ipynb
| ├── 2_dataset_creation.ipynb
| ├── 3_model_training.ipynb
| ├── 4_model_evaluation.ipynb
| ├── 5_inference_scheduling.ipynb <<< THIS NOTEBOOK <<<
| └── 6_cleanup.ipynb
|
└── utils/
└── lookout_equipment_utils.py
```
### Notebook configuration update
```
!pip install --quiet --upgrade sagemaker lookoutequipment
```
### Imports
```
import boto3
import config
import datetime
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
import numpy as np
import os
import pandas as pd
import pytz
import sagemaker
import sys
import time
from matplotlib.gridspec import GridSpec
# SDK / toolbox for managing Lookout for Equipment API calls:
import lookoutequipment as lookout
```
### AWS Look & Feel definition for Matplotlib
```
from matplotlib import font_manager
# Load style sheet:
plt.style.use('../utils/aws_sagemaker_light.py')
# Get colors from custom AWS palette:
prop_cycle = plt.rcParams['axes.prop_cycle']
colors = prop_cycle.by_key()['color']
```
### Parameters
```
TMP_DATA = os.path.join('..', 'data', 'interim', 'getting-started')
PROCESSED_DATA = os.path.join('..', 'data', 'processed', 'getting-started')
INFERENCE_DATA = os.path.join(PROCESSED_DATA, 'inference-data')
TRAIN_DATA = os.path.join(PROCESSED_DATA, 'training-data', 'centrifugal-pump')
os.makedirs(INFERENCE_DATA, exist_ok=True)
os.makedirs(os.path.join(INFERENCE_DATA, 'input'), exist_ok=True)
os.makedirs(os.path.join(INFERENCE_DATA, 'output'), exist_ok=True)
ROLE_ARN = sagemaker.get_execution_role()
REGION_NAME = boto3.session.Session().region_name
BUCKET = config.BUCKET
PREFIX = config.PREFIX_INFERENCE
INFERENCE_SCHEDULER_NAME = config.INFERENCE_SCHEDULER_NAME
MODEL_NAME = config.MODEL_NAME
%matplotlib inline
```
## Create an inference scheduler
---
While navigating to the model details part of the console, you will see that you have no inference scheduled yet:

### Scheduler configuration
Let's create a new inference schedule: some parameters are mandatory, while others offer some added flexibility.
#### Mandatory Parameters
* Set `upload_frequency` at which the data will be uploaded for inference. Allowed values are `PT5M`, `PT10M`, `PT15M`, `PT30M` and `PT1H`.
* This is both the frequency of the inference scheduler and how often data are uploaded to the source bucket.
* **Note**: ***the upload frequency must be compatible with the sampling rate selected at training time.*** *For example, if a model was trained with a 30 minutes resampling, asking for 5 minutes won't work and you need to select either PT30M and PT1H for this parameter at inference time.*
* Set `input_bucket` to the S3 bucket of your inference data
* Set `input_prefix` to the S3 prefix of your inference data
* Set `output_bucket` to the S3 bucket where you want inference results
* Set `output_prefix` to the S3 prefix where you want inference results
* Set `role_arn` to the role to be used to **read** data to infer on and **write** inference output
#### Time zone parameter (optional)
You can set `INPUT_TIMEZONE_OFFSET` to the following allowed values: `+00:00`, `+00:30`, `+01:00`, ... `+11:30`, `+12:00`, `-00:00`, `-00:30`, `-01:00`, ... `-11:30`, `-12:00`.
This is the timezone the scheduler will use to find the input files to run inference for. A timezone's offset refers to how many hours the timezone is from Coordinated Universal Time (UTC).
Let's take an example:
* The current date April 5th, 2021 and time is 1pm UTC
* You're in India, which is 5 hour 30 ahead of UTC and you set the `INPUT_TIMEZONE_OFFSET` to `+05:30`
* If the scheduler wakes up at 1pm UTC, A filename called 20210405**1830**00 will be found (1pm + 5H30 = 6.30pm)
Use the following cell to convert time zone identifier (`Europe/Paris`, `US/Central`...) to a time zone offset. You can build a timezone object by leveraging the World Timezone Definition **[available here](https://gist.github.com/heyalexej/8bf688fd67d7199be4a1682b3eec7568)** or by listing the available ones using this code snippet:
```python
import pytz
for tz in pytz.all_timezones:
print tz
```
If you want to use universal time, replace the timezone string below (`Asia/Calcutta`) by `UTC`:
```
utc_timezone = pytz.timezone("UTC")
current_timezone = pytz.timezone("Asia/Calcutta")
tz_offset = datetime.datetime.now(current_timezone).strftime('%z')
tz_offset = tz_offset[:3] + ':' + tz_offset[3:]
tz_offset
```
#### Other optional parameters
* Set `delay_offset` to the number of minutes you expect the data to be delayed to upload. It's a time buffer to upload data.
* Set `timestamp_format`. The allowed values `EPOCH`, `yyyy-MM-dd-HH-mm-ss` or `yyyyMMddHHmmss`. This is the format of timestamp which is the suffix of the input data file name. This is used by Lookout Equipment to understand which files to run inference on (so that you don't need to remove previous files to let the scheduler finds which one to run on).
* Set `component_delimiter`. The allowed values `-`, `_` or ` `. This is the delimiter character that is used to separate the component from the timestamp in the input filename.
### Create the inference scheduler
The CreateInferenceScheduler API creates a scheduler. The following code prepares the configuration but does not create the scheduler just yet:
```
scheduler = lookout.LookoutEquipmentScheduler(
scheduler_name=INFERENCE_SCHEDULER_NAME,
model_name=MODEL_NAME
)
scheduler_params = {
'input_bucket': BUCKET,
'input_prefix': f'{PREFIX}/input/',
'output_bucket': BUCKET,
'output_prefix': f'{PREFIX}/output/',
'role_arn': ROLE_ARN,
'upload_frequency': 'PT5M',
'delay_offset': None,
'timezone_offset': tz_offset,
'component_delimiter': '_',
'timestamp_format': 'yyyyMMddHHmmss'
}
scheduler.set_parameters(**scheduler_params)
```
## Prepare the inference data
---
Let's prepare and send some data in the S3 input location our scheduler will monitor: we are going to extract 10 sequences of 5 minutes each (5 minutes being the minimum scheduler frequency). We assume that data are sampled at a rate of one data point per minute meaning that each sequence will be a CSV with 5 rows (to match the scheduler frequency). We have set aside a file we can use for inference. We need to update the timestamps to match the current time and date and then split the file in individual datasets of 5 rows each.
```
# Load the original inference data:
inference_fname = os.path.join(TMP_DATA, 'inference-data', 'inference.csv')
inference_df = pd.read_csv(inference_fname)
inference_df['Timestamp'] = pd.to_datetime(inference_df['Timestamp'])
inference_df = inference_df.set_index('Timestamp')
# How many sequences do we want to extract:
num_sequences = 12
# The scheduling frequency in minutes: this **MUST** match the
# resampling rate used to train the model:
frequency = 5
start = inference_df.index.min()
for i in range(num_sequences):
end = start + datetime.timedelta(minutes=+frequency - 1)
inference_input = inference_df.loc[start:end, :]
start = start + datetime.timedelta(minutes=+frequency)
# Rounding time to the previous X minutes
# where X is the selected frequency:
filename_tm = datetime.datetime.now(current_timezone)
filename_tm = filename_tm - datetime.timedelta(
minutes=filename_tm.minute % frequency,
seconds=filename_tm.second,
microseconds=filename_tm.microsecond
)
filename_tm = filename_tm + datetime.timedelta(minutes=+frequency * (i))
current_timestamp = (filename_tm).strftime(format='%Y%m%d%H%M%S')
# The timestamp inside the file are in UTC and are not linked to the current timezone:
timestamp_tm = datetime.datetime.now(utc_timezone)
timestamp_tm = timestamp_tm - datetime.timedelta(
minutes=timestamp_tm.minute % frequency,
seconds=timestamp_tm.second,
microseconds=timestamp_tm.microsecond
)
timestamp_tm = timestamp_tm + datetime.timedelta(minutes=+frequency * (i))
# We need to reset the index to match the time
# at which the scheduler will run inference:
new_index = pd.date_range(
start=timestamp_tm,
periods=inference_input.shape[0],
freq='1min'
)
inference_input.index = new_index
inference_input.index.name = 'Timestamp'
inference_input = inference_input.reset_index()
inference_input['Timestamp'] = inference_input['Timestamp'].dt.strftime('%Y-%m-%dT%H:%M:%S.%f')
# Export this file in CSV format:
scheduled_fname = os.path.join(INFERENCE_DATA, 'input', f'centrifugal-pump_{current_timestamp}.csv')
inference_input.to_csv(scheduled_fname, index=None)
# Upload the whole folder to S3, in the input location:
!aws s3 cp --recursive --quiet $INFERENCE_DATA/input s3://$BUCKET/$PREFIX/input
```
Our S3 bucket is now in the following state: this emulates what you could expect if your industrial information system was sending a new sample of data every five minutes.
Note how:
* Every files are located in the same folder
* Each file has the recorded timestamp in its name
* The timestamps are rounding to the closest 5 minutes (as our scheduler is configured to wake up every 5 minutes)

Now that we've prepared the data, we can create the scheduler by running:
```python
create_scheduler_response = lookout_client.create_inference_scheduler({
'ClientToken': uuid.uuid4().hex
})
```
The following method encapsulates the call to the [**CreateInferenceScheduler**](https://docs.aws.amazon.com/lookout-for-equipment/latest/ug/API_CreateInferenceScheduler.html) API:
```
create_scheduler_response = scheduler.create()
```
Our scheduler is now running and its inference history is currently empty:

## Get inference results
---
### List inference executions
**Let's now wait for 5-15 minutes to give some time to the scheduler to run its first inferences.** Once the wait is over, we can use the ListInferenceExecution API for our current inference scheduler. The only mandatory parameter is the scheduler name.
You can also choose a time period for which you want to query inference executions for. If you don't specify it, then all executions for an inference scheduler will be listed. If you want to specify the time range, you can do this:
```python
START_TIME_FOR_INFERENCE_EXECUTIONS = datetime.datetime(2010,1,3,0,0,0)
END_TIME_FOR_INFERENCE_EXECUTIONS = datetime.datetime(2010,1,5,0,0,0)
```
Which means the executions after `2010-01-03 00:00:00` and before `2010-01-05 00:00:00` will be listed.
You can also choose to query for executions in particular status, the allowed status are `IN_PROGRESS`, `SUCCESS` and `FAILED`.
The following cell use `scheduler.list_inference_executions()` as a wrapper around the [**ListInferenceExecutions**](https://docs.aws.amazon.com/lookout-for-equipment/latest/ug/API_ListInferenceExecutions.html) API:
```python
list_executions_response = lookout_client.list_inference_executions({
"MaxResults": 50,
"InferenceSchedulerName": INFERENCE_SCHEDULER_NAME,
"Status": EXECUTION_STATUS,
"DataStartTimeAfter": START_TIME_FOR_INFERENCE_EXECUTIONS,
"DataEndTimeBefore": END_TIME_FOR_INFERENCE_EXECUTIONS
})
```
```
START_TIME_FOR_INFERENCE_EXECUTIONS = None
END_TIME_FOR_INFERENCE_EXECUTIONS = None
EXECUTION_STATUS = None
execution_summaries = []
while len(execution_summaries) == 0:
execution_summaries = scheduler.list_inference_executions(
start_time=START_TIME_FOR_INFERENCE_EXECUTIONS,
end_time=END_TIME_FOR_INFERENCE_EXECUTIONS,
execution_status=EXECUTION_STATUS
)
if len(execution_summaries) == 0:
print('WAITING FOR THE FIRST INFERENCE EXECUTION')
time.sleep(60)
else:
print('FIRST INFERENCE EXECUTED\n')
break
print(len(execution_summaries), 'inference execution(s) found.')
print('Displaying the first three ones:')
execution_summaries[:3]
```
We have configured this scheduler to run every five minutes. After at least 5 minutes we can also see the history in the console populated with its first few executions: after an hour or so, we will see that the last ones failed as we only generated 12 files above:

When the scheduler starts (for example at `datetime.datetime(2021, 1, 27, 9, 15)`, it looks for **a single** CSV file located in the input location with a filename that contains a timestamp set to the previous step. For example, a file named:
* centrifugal-pump_2021012709**10**00.csv will be found and ingested
* centrifugal-pump_2021012708**15**00.csv will **not be** ingested (it will be ingested at the next inference execution however)
In addition, when opening the file `centrifugal-pump_20210127091000.csv`, it will also open one file before and after this execution time: it will then look for any row with a date that is between the `DataStartTime` and the `DataEndTime` of the inference execution. If it doesn't find such a row in any of these three files, an exception will be thrown: the status of the execution will be marked `Failed`.
### Download inference results
Let's have a look at the content now available in the scheduler output location: each inference execution creates a subfolder in the output directory. The subfolder name is the timestamp (GMT) at which the inference was executed and it contains a single [JSON lines](https://jsonlines.org/) file named `results.jsonl`:

Each execution summary is a JSON document that has the following format:
```
execution_summaries[0]
```
When the `Status` key from the previous JSON result is set to `SUCCESS`, you can collect the results location in the `CustomerResultObject` field. We are now going to loop through each execution result and download each JSON lines files generated by the scheduler. Then we will insert their results into an overall dataframe for further analysis:
```
results_df = scheduler.get_predictions()
results_df.head()
```
The content of each JSON lines file follows this format:
```json
[
{
'timestamp': '2022-03-30T10:46:00.000000',
'prediction': 1,
'prediction_reason': 'ANOMALY_DETECTED',
'anomaly_score': 0.91945,
'diagnostics': [
{'name': 'centrifugal-pump\\Sensor0', 'value': 0.12},
{'name': 'centrifugal-pump\\Sensor1', 'value': 0.0},
{'name': 'centrifugal-pump\\Sensor2', 'value': 0.0},
.
.
.
{'name': 'centrifugal-pump\\Sensor27', 'value': 0.08},
{'name': 'centrifugal-pump\\Sensor28', 'value': 0.02},
{'name': 'centrifugal-pump\\Sensor29', 'value': 0.02}
]
}
...
]
```
Each timestamp found in the file is associated to a prediction: 1 when an anomaly is detected an 0 otherwise. When the `prediction` field is 1 (an anomaly is detected), the `diagnostics` field contains each sensor (with the format `component`\\`tag`) and an associated percentage. This percentage corresponds to the magnitude of impact of a given sensor to the detected anomaly. For instance, in the example above, the tag `Sensor0` located on the `centrifugal-pump` component has an estimated 12% magnitude of impact to the anomaly detected at 8pm on April 7th 2021. This dataset has 23 sensors: if each sensor contributed the same way to this event, the impact of each of them would be `100 / 23 = 4.35%`, so 12% is indeed statistically significant.
### Visualizing the inference results
#### Single inference analysis
Each detected event have some detailed diagnostics. Let's unpack the details for the first event and plot a similar bar chart than what the console provides when it evaluates a trained model:
```
event_details = pd.DataFrame(results_df.iloc[0, 2:]).reset_index()
fig, ax = lookout.plot.plot_event_barh(event_details)
```
As we did in the previous notebook, the above bar chart is already of great help to pinpoint what might be going wrong with your asset. Let's load the initial tags description file we prepared in the first notebook and match the sensors with our initial components to group sensors by component:
```
# Agregate event diagnostics at the component level:
tags_description_fname = os.path.join(TMP_DATA, 'tags_description.csv')
tags_description_df = pd.read_csv(tags_description_fname)
event_details[['asset', 'sensor']] = event_details['name'].str.split('\\', expand=True)
component_diagnostics = pd.merge(event_details, tags_description_df, how='inner', left_on='sensor', right_on='Tag')[['name', 'value', 'Component']]
component_diagnostics = component_diagnostics.groupby(by='Component').sum().sort_values(by='value')
# Prepare Y position and values for bar chart:
y_pos = np.arange(component_diagnostics.shape[0])
values = list(component_diagnostics['value'])
# Plot the bar chart:
fig = plt.figure(figsize=(12,5))
ax = plt.subplot(1,1,1)
ax.barh(y_pos, component_diagnostics['value'], align='center')
ax.set_yticks(y_pos)
ax.set_yticklabels(list(component_diagnostics.index))
ax.xaxis.set_major_formatter(mtick.PercentFormatter(1.0))
# Add the values in each bar:
for i, v in enumerate(values):
ax.text(0.005, i, f'{v*100:.2f}%', color='#FFFFFF', fontweight='bold', verticalalignment='center')
# Show the final plot:
plt.show()
```
#### Multiple inferences analysis
We can also plot the contribution evolution for the top contributing signals over a period of time: the following graph gives an example of what a real time dashboard could look like to expose the results of an Amazon Lookout for Equipment scheduler.
**Note:** The plot stops after a while as there are no more anomaly detected.
```
fig = plt.figure(figsize=(24,9))
gs = GridSpec(nrows=3, ncols=1, height_ratios=[1.0, 0.2, 1.0], hspace=0.35)
df = results_df.loc[:, :].copy()
plot_start = np.min(df.index)
plot_end = np.max(df.index)
df = df.loc[plot_start:plot_end]
ax1 = fig.add_subplot(gs[0])
anomaly_plot = ax1.plot(results_df['anomaly_score'], marker='o', markersize=10)
anomaly_plot[0].set_markerfacecolor(colors[5] + '80')
ax1.set_xlim((plot_start, plot_end))
ax1.set_title(f'Centrifugal pump - Live anomaly detection')
ax1.set_xlabel('Raw anomaly score', fontsize=12)
ax3 = fig.add_subplot(gs[1])
lookout.plot.plot_range(results_df, 'Detected events', colors[5], ax3, column_name='prediction')
ax3.set_xlim((plot_start, plot_end))
bar_width = 0.0005
ax4 = fig.add_subplot(gs[2])
bottom_values = np.zeros((len(df.index),))
current_tags_list = list(df.iloc[:, 2:].sum().sort_values(ascending=False).head(8).index)
for tag in current_tags_list:
plt.bar(x=df.index, height=df[tag], bottom=bottom_values, alpha=0.8, width=bar_width, label=tag.split('\\')[1])
bottom_values += df[tag].values
all_other_tags = [t for t in df.columns if t not in current_tags_list][2:]
all_other_tags_contrib = df[all_other_tags].sum(axis='columns')
plt.bar(x=df.index, height=all_other_tags_contrib, bottom=bottom_values, alpha=0.8, width=bar_width, label='All the others', color='#CCCCCC')
ax4.legend(loc='lower center', ncol=5, bbox_to_anchor=(0.5, -0.45))
ax4.set_xlabel('Signal importance evolution', fontsize=12)
ax4.set_xlim((plot_start, plot_end))
plt.show()
```
## Inference scheduler operations
---
### Stop inference scheduler
**Be frugal**, running the scheduler is the main cost driver of Amazon Lookout for Equipment. Use the [**StopInferenceScheduler**](https://docs.aws.amazon.com/lookout-for-equipment/latest/ug/API_StopInferenceScheduler.html) API to stop an already running inference scheduler:
```python
stop_scheduler_response = lookout_client.stop_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
```
The following method is a wrapper around this API call and will stop the periodic inference executions:
```
scheduler.stop()
```
### Start an inference scheduler
You can restart any `STOPPED` inference scheduler using the [**StartInferenceScheduler**](https://docs.aws.amazon.com/lookout-for-equipment/latest/ug/API_StartInferenceScheduler.html) API:
```python
start_scheduler_response = lookout_client.start_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
```
The following method is a wrapper around this API call and will start the periodic inference executions:
```
scheduler.start()
```
### Delete an inference scheduler
You can delete a **stopped** scheduler you have no more use of: note that you can only have one scheduler per model.
```python
delete_scheduler_response = lookout_client.delete_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
```
The `scheduler.delete()` method is a wrapper around the [**DeleteInferenceScheduler**](https://docs.aws.amazon.com/lookout-for-equipment/latest/ug/API_DeleteInferenceScheduler.html) API and will delete a stopped scheduler:
```
scheduler.stop()
scheduler.delete()
```
## Conclusion
---
In this notebook, we used the model created in part 3 of this notebook, configured a scheduler and extracted the predictions obtained after it executed a few inferences.
We also showed how we could post-process the inference results to deliver better insights into the detected events.
|
github_jupyter
|
. lookout-equipment-demo
|
├── data/
| ├── interim # Temporary intermediate data are stored here
| ├── processed # Finalized datasets are usually stored here
| | # before they are sent to S3 to allow the
| | # service to reach them
| └── raw # Immutable original data are stored here
|
├── getting_started/
| ├── 1_data_preparation.ipynb
| ├── 2_dataset_creation.ipynb
| ├── 3_model_training.ipynb
| ├── 4_model_evaluation.ipynb
| ├── 5_inference_scheduling.ipynb <<< THIS NOTEBOOK <<<
| └── 6_cleanup.ipynb
|
└── utils/
└── lookout_equipment_utils.py
!pip install --quiet --upgrade sagemaker lookoutequipment
import boto3
import config
import datetime
import matplotlib.pyplot as plt
import matplotlib.ticker as mtick
import numpy as np
import os
import pandas as pd
import pytz
import sagemaker
import sys
import time
from matplotlib.gridspec import GridSpec
# SDK / toolbox for managing Lookout for Equipment API calls:
import lookoutequipment as lookout
from matplotlib import font_manager
# Load style sheet:
plt.style.use('../utils/aws_sagemaker_light.py')
# Get colors from custom AWS palette:
prop_cycle = plt.rcParams['axes.prop_cycle']
colors = prop_cycle.by_key()['color']
TMP_DATA = os.path.join('..', 'data', 'interim', 'getting-started')
PROCESSED_DATA = os.path.join('..', 'data', 'processed', 'getting-started')
INFERENCE_DATA = os.path.join(PROCESSED_DATA, 'inference-data')
TRAIN_DATA = os.path.join(PROCESSED_DATA, 'training-data', 'centrifugal-pump')
os.makedirs(INFERENCE_DATA, exist_ok=True)
os.makedirs(os.path.join(INFERENCE_DATA, 'input'), exist_ok=True)
os.makedirs(os.path.join(INFERENCE_DATA, 'output'), exist_ok=True)
ROLE_ARN = sagemaker.get_execution_role()
REGION_NAME = boto3.session.Session().region_name
BUCKET = config.BUCKET
PREFIX = config.PREFIX_INFERENCE
INFERENCE_SCHEDULER_NAME = config.INFERENCE_SCHEDULER_NAME
MODEL_NAME = config.MODEL_NAME
%matplotlib inline
import pytz
for tz in pytz.all_timezones:
print tz
utc_timezone = pytz.timezone("UTC")
current_timezone = pytz.timezone("Asia/Calcutta")
tz_offset = datetime.datetime.now(current_timezone).strftime('%z')
tz_offset = tz_offset[:3] + ':' + tz_offset[3:]
tz_offset
scheduler = lookout.LookoutEquipmentScheduler(
scheduler_name=INFERENCE_SCHEDULER_NAME,
model_name=MODEL_NAME
)
scheduler_params = {
'input_bucket': BUCKET,
'input_prefix': f'{PREFIX}/input/',
'output_bucket': BUCKET,
'output_prefix': f'{PREFIX}/output/',
'role_arn': ROLE_ARN,
'upload_frequency': 'PT5M',
'delay_offset': None,
'timezone_offset': tz_offset,
'component_delimiter': '_',
'timestamp_format': 'yyyyMMddHHmmss'
}
scheduler.set_parameters(**scheduler_params)
# Load the original inference data:
inference_fname = os.path.join(TMP_DATA, 'inference-data', 'inference.csv')
inference_df = pd.read_csv(inference_fname)
inference_df['Timestamp'] = pd.to_datetime(inference_df['Timestamp'])
inference_df = inference_df.set_index('Timestamp')
# How many sequences do we want to extract:
num_sequences = 12
# The scheduling frequency in minutes: this **MUST** match the
# resampling rate used to train the model:
frequency = 5
start = inference_df.index.min()
for i in range(num_sequences):
end = start + datetime.timedelta(minutes=+frequency - 1)
inference_input = inference_df.loc[start:end, :]
start = start + datetime.timedelta(minutes=+frequency)
# Rounding time to the previous X minutes
# where X is the selected frequency:
filename_tm = datetime.datetime.now(current_timezone)
filename_tm = filename_tm - datetime.timedelta(
minutes=filename_tm.minute % frequency,
seconds=filename_tm.second,
microseconds=filename_tm.microsecond
)
filename_tm = filename_tm + datetime.timedelta(minutes=+frequency * (i))
current_timestamp = (filename_tm).strftime(format='%Y%m%d%H%M%S')
# The timestamp inside the file are in UTC and are not linked to the current timezone:
timestamp_tm = datetime.datetime.now(utc_timezone)
timestamp_tm = timestamp_tm - datetime.timedelta(
minutes=timestamp_tm.minute % frequency,
seconds=timestamp_tm.second,
microseconds=timestamp_tm.microsecond
)
timestamp_tm = timestamp_tm + datetime.timedelta(minutes=+frequency * (i))
# We need to reset the index to match the time
# at which the scheduler will run inference:
new_index = pd.date_range(
start=timestamp_tm,
periods=inference_input.shape[0],
freq='1min'
)
inference_input.index = new_index
inference_input.index.name = 'Timestamp'
inference_input = inference_input.reset_index()
inference_input['Timestamp'] = inference_input['Timestamp'].dt.strftime('%Y-%m-%dT%H:%M:%S.%f')
# Export this file in CSV format:
scheduled_fname = os.path.join(INFERENCE_DATA, 'input', f'centrifugal-pump_{current_timestamp}.csv')
inference_input.to_csv(scheduled_fname, index=None)
# Upload the whole folder to S3, in the input location:
!aws s3 cp --recursive --quiet $INFERENCE_DATA/input s3://$BUCKET/$PREFIX/input
create_scheduler_response = lookout_client.create_inference_scheduler({
'ClientToken': uuid.uuid4().hex
})
create_scheduler_response = scheduler.create()
START_TIME_FOR_INFERENCE_EXECUTIONS = datetime.datetime(2010,1,3,0,0,0)
END_TIME_FOR_INFERENCE_EXECUTIONS = datetime.datetime(2010,1,5,0,0,0)
list_executions_response = lookout_client.list_inference_executions({
"MaxResults": 50,
"InferenceSchedulerName": INFERENCE_SCHEDULER_NAME,
"Status": EXECUTION_STATUS,
"DataStartTimeAfter": START_TIME_FOR_INFERENCE_EXECUTIONS,
"DataEndTimeBefore": END_TIME_FOR_INFERENCE_EXECUTIONS
})
START_TIME_FOR_INFERENCE_EXECUTIONS = None
END_TIME_FOR_INFERENCE_EXECUTIONS = None
EXECUTION_STATUS = None
execution_summaries = []
while len(execution_summaries) == 0:
execution_summaries = scheduler.list_inference_executions(
start_time=START_TIME_FOR_INFERENCE_EXECUTIONS,
end_time=END_TIME_FOR_INFERENCE_EXECUTIONS,
execution_status=EXECUTION_STATUS
)
if len(execution_summaries) == 0:
print('WAITING FOR THE FIRST INFERENCE EXECUTION')
time.sleep(60)
else:
print('FIRST INFERENCE EXECUTED\n')
break
print(len(execution_summaries), 'inference execution(s) found.')
print('Displaying the first three ones:')
execution_summaries[:3]
execution_summaries[0]
results_df = scheduler.get_predictions()
results_df.head()
[
{
'timestamp': '2022-03-30T10:46:00.000000',
'prediction': 1,
'prediction_reason': 'ANOMALY_DETECTED',
'anomaly_score': 0.91945,
'diagnostics': [
{'name': 'centrifugal-pump\\Sensor0', 'value': 0.12},
{'name': 'centrifugal-pump\\Sensor1', 'value': 0.0},
{'name': 'centrifugal-pump\\Sensor2', 'value': 0.0},
.
.
.
{'name': 'centrifugal-pump\\Sensor27', 'value': 0.08},
{'name': 'centrifugal-pump\\Sensor28', 'value': 0.02},
{'name': 'centrifugal-pump\\Sensor29', 'value': 0.02}
]
}
...
]
event_details = pd.DataFrame(results_df.iloc[0, 2:]).reset_index()
fig, ax = lookout.plot.plot_event_barh(event_details)
# Agregate event diagnostics at the component level:
tags_description_fname = os.path.join(TMP_DATA, 'tags_description.csv')
tags_description_df = pd.read_csv(tags_description_fname)
event_details[['asset', 'sensor']] = event_details['name'].str.split('\\', expand=True)
component_diagnostics = pd.merge(event_details, tags_description_df, how='inner', left_on='sensor', right_on='Tag')[['name', 'value', 'Component']]
component_diagnostics = component_diagnostics.groupby(by='Component').sum().sort_values(by='value')
# Prepare Y position and values for bar chart:
y_pos = np.arange(component_diagnostics.shape[0])
values = list(component_diagnostics['value'])
# Plot the bar chart:
fig = plt.figure(figsize=(12,5))
ax = plt.subplot(1,1,1)
ax.barh(y_pos, component_diagnostics['value'], align='center')
ax.set_yticks(y_pos)
ax.set_yticklabels(list(component_diagnostics.index))
ax.xaxis.set_major_formatter(mtick.PercentFormatter(1.0))
# Add the values in each bar:
for i, v in enumerate(values):
ax.text(0.005, i, f'{v*100:.2f}%', color='#FFFFFF', fontweight='bold', verticalalignment='center')
# Show the final plot:
plt.show()
fig = plt.figure(figsize=(24,9))
gs = GridSpec(nrows=3, ncols=1, height_ratios=[1.0, 0.2, 1.0], hspace=0.35)
df = results_df.loc[:, :].copy()
plot_start = np.min(df.index)
plot_end = np.max(df.index)
df = df.loc[plot_start:plot_end]
ax1 = fig.add_subplot(gs[0])
anomaly_plot = ax1.plot(results_df['anomaly_score'], marker='o', markersize=10)
anomaly_plot[0].set_markerfacecolor(colors[5] + '80')
ax1.set_xlim((plot_start, plot_end))
ax1.set_title(f'Centrifugal pump - Live anomaly detection')
ax1.set_xlabel('Raw anomaly score', fontsize=12)
ax3 = fig.add_subplot(gs[1])
lookout.plot.plot_range(results_df, 'Detected events', colors[5], ax3, column_name='prediction')
ax3.set_xlim((plot_start, plot_end))
bar_width = 0.0005
ax4 = fig.add_subplot(gs[2])
bottom_values = np.zeros((len(df.index),))
current_tags_list = list(df.iloc[:, 2:].sum().sort_values(ascending=False).head(8).index)
for tag in current_tags_list:
plt.bar(x=df.index, height=df[tag], bottom=bottom_values, alpha=0.8, width=bar_width, label=tag.split('\\')[1])
bottom_values += df[tag].values
all_other_tags = [t for t in df.columns if t not in current_tags_list][2:]
all_other_tags_contrib = df[all_other_tags].sum(axis='columns')
plt.bar(x=df.index, height=all_other_tags_contrib, bottom=bottom_values, alpha=0.8, width=bar_width, label='All the others', color='#CCCCCC')
ax4.legend(loc='lower center', ncol=5, bbox_to_anchor=(0.5, -0.45))
ax4.set_xlabel('Signal importance evolution', fontsize=12)
ax4.set_xlim((plot_start, plot_end))
plt.show()
stop_scheduler_response = lookout_client.stop_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
scheduler.stop()
start_scheduler_response = lookout_client.start_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
scheduler.start()
delete_scheduler_response = lookout_client.delete_inference_scheduler(
InferenceSchedulerName=INFERENCE_SCHEDULER_NAME
)
scheduler.stop()
scheduler.delete()
| 0.545528 | 0.884439 |
### TextCNN文本分类器
本程序使用pytorch构建并训练了可用于文本分类的TextCNN模型,请确认dataset目录中已存在kfold程序生成的csv文件,且已下载预训练词向量
附录中展示了测试过程中曾使用过的LSTM模型与较为朴素的TextCNN模型的代码
导入用到的库
```
import pandas as pd #用于数据处理
import numpy as np #用于矩阵计算
import torch #用于搭建及训练模型
import time #用于训练计时
import random #用于生成随机数
import os #用于文件操作
from torchtext import data #用于生成数据集
from torchtext.vocab import Vectors #用于载入预训练词向量
from tqdm import tqdm #用于绘制进度条
from torchtext.data import Iterator, BucketIterator #用于生成训练和测试所用的迭代器
import torch.nn as nn #用于搭建模型
import torch.optim as optim #用于生成优化函数
from matplotlib import pyplot as plt #用于绘制误差函数
torch.manual_seed(19260817) #设定随机数种子
torch.backends.cudnn.deterministic = True #保证可复现性
def tokenize(x): return x.split() #分词函数,后续操作中会用到
```
设定所用的训练&测试数据
定义MyDataset类并生成训练和测试所用数据集
```
TEXT = data.Field(sequential=True, tokenize=tokenize, fix_length=50) #设定句长为50
LABEL = data.Field(sequential=False, use_vocab=False)
# 定义Dataset类
class Dataset(data.Dataset):
name = 'Dataset'
def __init__(self, path, text_field, label_field):
fields = [("text", text_field), ("category", label_field)]
examples = []
csv_data = pd.read_csv(path) #从csv文件中读取数据
print('read data from {}'.format(path))
for text, label in tqdm(zip(csv_data['text'], csv_data['category'])):
examples.append(data.Example.fromlist([str(text), label], fields))
super(Dataset, self).__init__(examples, fields) #生成标准dataset
```
载入训练和测试所用的数据集
```
dataset_id = 2 #选择所使用的dataset组合
train_path = 'dataset/Train'+str(dataset_id)+'UTF8.csv' #训练数据文件路径
test_path = 'dataset/Test'+str(dataset_id)+'UTF8.csv' #测试数据文件路径
train = Dataset(train_path, text_field=TEXT, label_field=LABEL) #生成训练集
test = Dataset(test_path, text_field=TEXT, label_field=LABEL) #生成测试集
```
使用预训练词向量构建映射关系及权重矩阵
```
if not os.path.exists('.vector_cache'): #建立缓存文件夹以存储缓存文件
os.mkdir('.vector_cache')
vectors = Vectors(name='weibo') #使用微博数据集所训练好的词向量
TEXT.build_vocab(train, vectors=vectors, unk_init = torch.Tensor.normal_, min_freq=5) #构建映射,设定最低词频为5
weight_matrix = TEXT.vocab.vectors #构建权重矩阵
```
定义搭建模型使用的TextCNN类
```
class TextCNN(nn.Module):
def __init__(self, window_sizes, vocab_size = len(TEXT.vocab), pad_idx = TEXT.vocab.stoi[TEXT.pad_token], embedding_dim=300, text_len=50, output_dim=9, feature_size=100):
super().__init__() #调用nn.Module的构造函数进行初始化
self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=pad_idx) #使用embedding table构建语句到向量的映射
self.embedding.weight.data.copy_(weight_matrix) #载入由预训练词向量生成的权重矩阵
self.convs = nn.ModuleList([ #定义所使用的卷积操作
nn.Sequential(nn.Conv1d(in_channels=embedding_dim, out_channels=feature_size, kernel_size=h,), #1维卷积
nn.BatchNorm1d(num_features=feature_size), #正则化
nn.ReLU(), #ReLU
nn.MaxPool1d(kernel_size=text_len-h+1)) #Max Pooling
for h in window_sizes])
self.fc = nn.Linear(in_features=feature_size*len(window_sizes),out_features=9) #全连接层
self.dropout = nn.Dropout(0.4) #dropout
def forward(self, text): #前向传播
embedded = self.embedding(text)
embedded = embedded.permute(1, 2, 0) #[]
out = [conv(embedded) for conv in self.convs]
out = torch.cat(out, dim=1) #纵向拼接卷积操作输出的矩阵
out = out.view(-1, out.size(1)) #将矩阵拉直为向量
#out = self.dropout(out)
y = self.fc(out) #通过全连接层处理获得预测类别
return y #返回预测值
```
设定batch size并生成训练与测试所用的迭代器
```
batch_size = 256
train_iter = BucketIterator(dataset=train, batch_size=batch_size, shuffle=True)
test_iter = Iterator(dataset=test, batch_size=batch_size, shuffle=True)
```
模型训练函数
传入参数: 训练轮数
```
def fit(num_epoch=60):
torch.cuda.empty_cache() #清空gpu缓存
start = time.time() #记录训练开始时间
losses = [] #用于记录训练过程中loss
acc = [] #用于记录训练过程中准确率
for epoch in tqdm(range(1, num_epoch+1)):
for batch in train_iter: #轮流读取batch中元素进行训练
model.train() #将模型设为训练模式
model.zero_grad() #将上次计算得到的梯度值清零
optimizer.zero_grad()
predicted = model(batch.text.cuda()) #进行预测
loss = loss_function(predicted, batch.category.cuda()-1) #计算loss
loss.backward() #反向传播
optimizer.step() #修正模型
'''
if epoch%1==0: #每过一段时间进行一次预测
print('Epoch %d, Loss: %f' % (epoch, loss.item()))
model.eval() #将模型设为评估模式
with torch.no_grad():
acc.append(validation()) #对测试集进行预测评估模型效果
'''
losses.append(loss.item())
plt.plot(losses) #绘制训练过程中loss与训练次数的图像'''
end = time.time() #记录训练结束时间
print('Time used: %ds' %(end-start)) #打印训练所用时间
```
验证函数
通过对测试集中样本进行预测获得模型分类的准确率
```
def validation():
ans = 0 #用于记录训练集总样本数
correct = 0 #用于记录预测正确的样本数目
for batch in test_iter: #获得测试集准确率
predicted = model(batch.text.cuda()) #对batch内元素进行预测
label = batch.category.cuda()-1 #-1以使标签中的1-9变为0-8和预测类别相符
max_preds = predicted.argmax(dim = 1, keepdim = True) #取最大值下标获得所预测类别
correct += int(max_preds.squeeze(1).eq(label).sum()) #计算预测类别与正确类别相等的数目
ans += label.shape[0]
accuracy = correct/ans #计算准确率
print('Accuracy on testset: %.3f' %(accuracy))
torch.cuda.empty_cache() #清空gpu缓存
return accuracy #返回准确率
```
定义训练所需的模型,损失函数与优化器
```
model = TextCNN(window_sizes=[3,4,5,6]) #定义TextCNN模型
loss_function = nn.functional.cross_entropy #使用交叉熵损失函数
optimizer = optim.Adam(filter(lambda p: p.requires_grad, model.parameters()), lr=0.001) #使用Adam作为优化器
model.cuda() #将模型移至gpu
```
进行训练
```
torch.cuda.empty_cache() #清除gpu缓存
fit(15)
```
在测试集上进行预测
```
validation()
```
保存模型
```
torch.save(model.state_dict(), 'models/model.pth')
```
附录:
此处展示了模型测试过程中使用过的双向LSTM模型与朴素的TextCNN模型
朴素的TextCNN模型
预测准确率 84.6%
```
class OldTextCNN(nn.Module):
def __init__(self, vocab_size = len(TEXT.vocab), pad_idx = TEXT.vocab.stoi[TEXT.pad_token], embedding_dim=300, hidden_dim=50, output_dim=9):
super().__init__() #调用nn.Module的构造函数进行初始化
self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=pad_idx) #使用embedding table构建语句到向量的映射
self.embedding.weight.data.copy_(weight_matrix) #载入由预训练词向量生成的权重矩阵
self.fc1 = nn.Linear(embedding_dim, hidden_dim) #全连接层
self.relu = nn.ReLU() #ReLU
self.dropout = nn.Dropout(0.2) #dropout
self.fc2 = nn.Linear(hidden_dim, output_dim) #全连接层
def forward(self, text): #前向传播
embedded = self.embedding(text)
embedded = embedded.permute(1, 0, 2) #将batch_size移至第一维
pooled = nn.functional.avg_pool2d(embedded, (embedded.shape[1], 1)).squeeze(1) #对移至第二维的textlen进行average pooling获得二维张量(batch_size*embedding_dim)
droped = self.dropout(pooled)
fced = self.fc1(droped)
relued = self.relu(fced)
droped = self.dropout(relued)
y = self.fc2(droped) #通过全连接层处理获得预测类别
return y
```
双向LSTM
预测准确率: 83.2%
```
class LSTM(nn.Module):
def __init__(self, dropout1=0.2, dropout2=0.2, bidirectional=True, hidden_size=200, num_layers=2):
super(LSTM, self).__init__()
self.word_embeddings = nn.Embedding(len(TEXT.vocab), 300)
self.word_embeddings.weight.data.copy_(weight_matrix)
self.lstm = nn.LSTM(input_size=300, hidden_size=hidden_size, num_layers=num_layers, dropout=dropout1, bidirectional=bidirectional)
self.dropout = nn.Dropout(dropout2)
self.fc1 = nn.Linear(2*hidden_size, 50)
self.relu = nn.ReLU()
self.fc2 = nn.Linear(50,9)
self.sigmoid = nn.Sigmoid()
def forward(self, sentence):
embeds = self.word_embeddings(sentence)
_, (lstm_out, __) = self.lstm(embeds)
last = torch.cat((lstm_out[-2,:,:], lstm_out[-1,:,:]), dim=1)
fc1_in = self.dropout(last)
fc1_out = self.fc1(fc1_in)
fc2_in = self.relu(fc1_out)
y = self.fc2(fc2_in)
return y
```
|
github_jupyter
|
import pandas as pd #用于数据处理
import numpy as np #用于矩阵计算
import torch #用于搭建及训练模型
import time #用于训练计时
import random #用于生成随机数
import os #用于文件操作
from torchtext import data #用于生成数据集
from torchtext.vocab import Vectors #用于载入预训练词向量
from tqdm import tqdm #用于绘制进度条
from torchtext.data import Iterator, BucketIterator #用于生成训练和测试所用的迭代器
import torch.nn as nn #用于搭建模型
import torch.optim as optim #用于生成优化函数
from matplotlib import pyplot as plt #用于绘制误差函数
torch.manual_seed(19260817) #设定随机数种子
torch.backends.cudnn.deterministic = True #保证可复现性
def tokenize(x): return x.split() #分词函数,后续操作中会用到
TEXT = data.Field(sequential=True, tokenize=tokenize, fix_length=50) #设定句长为50
LABEL = data.Field(sequential=False, use_vocab=False)
# 定义Dataset类
class Dataset(data.Dataset):
name = 'Dataset'
def __init__(self, path, text_field, label_field):
fields = [("text", text_field), ("category", label_field)]
examples = []
csv_data = pd.read_csv(path) #从csv文件中读取数据
print('read data from {}'.format(path))
for text, label in tqdm(zip(csv_data['text'], csv_data['category'])):
examples.append(data.Example.fromlist([str(text), label], fields))
super(Dataset, self).__init__(examples, fields) #生成标准dataset
dataset_id = 2 #选择所使用的dataset组合
train_path = 'dataset/Train'+str(dataset_id)+'UTF8.csv' #训练数据文件路径
test_path = 'dataset/Test'+str(dataset_id)+'UTF8.csv' #测试数据文件路径
train = Dataset(train_path, text_field=TEXT, label_field=LABEL) #生成训练集
test = Dataset(test_path, text_field=TEXT, label_field=LABEL) #生成测试集
if not os.path.exists('.vector_cache'): #建立缓存文件夹以存储缓存文件
os.mkdir('.vector_cache')
vectors = Vectors(name='weibo') #使用微博数据集所训练好的词向量
TEXT.build_vocab(train, vectors=vectors, unk_init = torch.Tensor.normal_, min_freq=5) #构建映射,设定最低词频为5
weight_matrix = TEXT.vocab.vectors #构建权重矩阵
class TextCNN(nn.Module):
def __init__(self, window_sizes, vocab_size = len(TEXT.vocab), pad_idx = TEXT.vocab.stoi[TEXT.pad_token], embedding_dim=300, text_len=50, output_dim=9, feature_size=100):
super().__init__() #调用nn.Module的构造函数进行初始化
self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=pad_idx) #使用embedding table构建语句到向量的映射
self.embedding.weight.data.copy_(weight_matrix) #载入由预训练词向量生成的权重矩阵
self.convs = nn.ModuleList([ #定义所使用的卷积操作
nn.Sequential(nn.Conv1d(in_channels=embedding_dim, out_channels=feature_size, kernel_size=h,), #1维卷积
nn.BatchNorm1d(num_features=feature_size), #正则化
nn.ReLU(), #ReLU
nn.MaxPool1d(kernel_size=text_len-h+1)) #Max Pooling
for h in window_sizes])
self.fc = nn.Linear(in_features=feature_size*len(window_sizes),out_features=9) #全连接层
self.dropout = nn.Dropout(0.4) #dropout
def forward(self, text): #前向传播
embedded = self.embedding(text)
embedded = embedded.permute(1, 2, 0) #[]
out = [conv(embedded) for conv in self.convs]
out = torch.cat(out, dim=1) #纵向拼接卷积操作输出的矩阵
out = out.view(-1, out.size(1)) #将矩阵拉直为向量
#out = self.dropout(out)
y = self.fc(out) #通过全连接层处理获得预测类别
return y #返回预测值
batch_size = 256
train_iter = BucketIterator(dataset=train, batch_size=batch_size, shuffle=True)
test_iter = Iterator(dataset=test, batch_size=batch_size, shuffle=True)
def fit(num_epoch=60):
torch.cuda.empty_cache() #清空gpu缓存
start = time.time() #记录训练开始时间
losses = [] #用于记录训练过程中loss
acc = [] #用于记录训练过程中准确率
for epoch in tqdm(range(1, num_epoch+1)):
for batch in train_iter: #轮流读取batch中元素进行训练
model.train() #将模型设为训练模式
model.zero_grad() #将上次计算得到的梯度值清零
optimizer.zero_grad()
predicted = model(batch.text.cuda()) #进行预测
loss = loss_function(predicted, batch.category.cuda()-1) #计算loss
loss.backward() #反向传播
optimizer.step() #修正模型
'''
if epoch%1==0: #每过一段时间进行一次预测
print('Epoch %d, Loss: %f' % (epoch, loss.item()))
model.eval() #将模型设为评估模式
with torch.no_grad():
acc.append(validation()) #对测试集进行预测评估模型效果
'''
losses.append(loss.item())
plt.plot(losses) #绘制训练过程中loss与训练次数的图像'''
end = time.time() #记录训练结束时间
print('Time used: %ds' %(end-start)) #打印训练所用时间
def validation():
ans = 0 #用于记录训练集总样本数
correct = 0 #用于记录预测正确的样本数目
for batch in test_iter: #获得测试集准确率
predicted = model(batch.text.cuda()) #对batch内元素进行预测
label = batch.category.cuda()-1 #-1以使标签中的1-9变为0-8和预测类别相符
max_preds = predicted.argmax(dim = 1, keepdim = True) #取最大值下标获得所预测类别
correct += int(max_preds.squeeze(1).eq(label).sum()) #计算预测类别与正确类别相等的数目
ans += label.shape[0]
accuracy = correct/ans #计算准确率
print('Accuracy on testset: %.3f' %(accuracy))
torch.cuda.empty_cache() #清空gpu缓存
return accuracy #返回准确率
model = TextCNN(window_sizes=[3,4,5,6]) #定义TextCNN模型
loss_function = nn.functional.cross_entropy #使用交叉熵损失函数
optimizer = optim.Adam(filter(lambda p: p.requires_grad, model.parameters()), lr=0.001) #使用Adam作为优化器
model.cuda() #将模型移至gpu
torch.cuda.empty_cache() #清除gpu缓存
fit(15)
validation()
torch.save(model.state_dict(), 'models/model.pth')
class OldTextCNN(nn.Module):
def __init__(self, vocab_size = len(TEXT.vocab), pad_idx = TEXT.vocab.stoi[TEXT.pad_token], embedding_dim=300, hidden_dim=50, output_dim=9):
super().__init__() #调用nn.Module的构造函数进行初始化
self.embedding = nn.Embedding(vocab_size, embedding_dim, padding_idx=pad_idx) #使用embedding table构建语句到向量的映射
self.embedding.weight.data.copy_(weight_matrix) #载入由预训练词向量生成的权重矩阵
self.fc1 = nn.Linear(embedding_dim, hidden_dim) #全连接层
self.relu = nn.ReLU() #ReLU
self.dropout = nn.Dropout(0.2) #dropout
self.fc2 = nn.Linear(hidden_dim, output_dim) #全连接层
def forward(self, text): #前向传播
embedded = self.embedding(text)
embedded = embedded.permute(1, 0, 2) #将batch_size移至第一维
pooled = nn.functional.avg_pool2d(embedded, (embedded.shape[1], 1)).squeeze(1) #对移至第二维的textlen进行average pooling获得二维张量(batch_size*embedding_dim)
droped = self.dropout(pooled)
fced = self.fc1(droped)
relued = self.relu(fced)
droped = self.dropout(relued)
y = self.fc2(droped) #通过全连接层处理获得预测类别
return y
class LSTM(nn.Module):
def __init__(self, dropout1=0.2, dropout2=0.2, bidirectional=True, hidden_size=200, num_layers=2):
super(LSTM, self).__init__()
self.word_embeddings = nn.Embedding(len(TEXT.vocab), 300)
self.word_embeddings.weight.data.copy_(weight_matrix)
self.lstm = nn.LSTM(input_size=300, hidden_size=hidden_size, num_layers=num_layers, dropout=dropout1, bidirectional=bidirectional)
self.dropout = nn.Dropout(dropout2)
self.fc1 = nn.Linear(2*hidden_size, 50)
self.relu = nn.ReLU()
self.fc2 = nn.Linear(50,9)
self.sigmoid = nn.Sigmoid()
def forward(self, sentence):
embeds = self.word_embeddings(sentence)
_, (lstm_out, __) = self.lstm(embeds)
last = torch.cat((lstm_out[-2,:,:], lstm_out[-1,:,:]), dim=1)
fc1_in = self.dropout(last)
fc1_out = self.fc1(fc1_in)
fc2_in = self.relu(fc1_out)
y = self.fc2(fc2_in)
return y
| 0.577257 | 0.903379 |
<a href="https://colab.research.google.com/github/diem-ai/natural-language-processing/blob/master/spaCy_chapter2.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
Chapter 2: Large-scale data analysis with spaCy
In this chapter, you'll use your new skills to extract specific information from large volumes of text. You''ll learn how to make the most of spaCy's data structures, and how to effectively combine statistical and rule-based approaches for text analysis.
```
import spacy
# Import the Doc class
from spacy.tokens import Doc, Span
from spacy.lang.en import English
from spacy.matcher import Matcher, PhraseMatcher
nlp = spacy.load("en_core_web_sm")
```
**String to hashesh**
- Look up the string “cat” in nlp.vocab.strings to get the hash.
- Look up the hash to get back the string.
```
doc = nlp("I have a cat")
# Look up the hash for the word "cat"
cat_hash = nlp.vocab.strings["cat"]
print(cat_hash)
# Look up the cat_hash to get the string
cat_string = nlp.vocab.strings[cat_hash]
print(cat_string)
```
- Look up the string label “PERSON” in nlp.vocab.strings to get the hash.
- Look up the hash to get back the string.
```
doc = nlp("David Bowie is a PERSON")
# Look up the hash for the string label "PERSON"
person_hash = nlp.vocab.strings["PERSON"]
print(person_hash)
# Look up the person_hash to get the string
person_string = nlp.vocab.strings[person_hash]
print(person_string)
```
**Creating a Doc from scratch**
```
# Desired text: "spaCy is cool!"
words = ["spaCy", "is", "cool", "!"]
spaces = [True, True, False, False]
# Create a Doc from the words and spaces
doc = Doc(nlp.vocab, words=words, spaces=spaces)
print(doc.text)
```
Create a sentence: "Go, get started!"
```
# Desired text: "Go, get started!"
words = ["Go", ",", "get", "started", "!"]
spaces = [False, True, True, False, False]
doc = Doc(nlp.vocab, words=words, spaces=spaces)
print(doc.text)
```
Create a sentence: "Oh, really?!"
```
# Desired text: "Oh, really?!"
words = ["Oh", ",", "really", "?", "!"]
spaces = [False, True, False, False, False]
# Create a Doc from the words and spaces
doc = Doc(nlp.vocab, words = words, spaces = spaces)
print(doc.text)
```
**Doc, Span, Entities from Scratch**
- In this exercise, you’ll create the Doc and Span objects manually, and update the named entities – just like spaCy does behind the scenes. A shared nlp object has already been created.
```
nlp_en = English()
# Import the Doc and Span classes
from spacy.tokens import Doc, Span
words = ["I", "like", "David", "Bowie"]
spaces = [True, True, True, False]
# Create a doc from the words and spaces
doc = Doc(nlp_en.vocab, words=words, spaces=spaces)
print(doc.text)
# Create a span for "David Bowie" from the doc and assign it the label "PERSON"
span = Span(doc, 2, 4, label="PERSON")
print(span.text, span.label_)
# Add the span to the doc's entities
doc.ents = [span]
# Print entities' text and labels
print([(ent.text, ent.label_) for ent in doc.ents])
```
**Data Structure Best Practices**
```
nlp = spacy.load("en_core_web_sm")
doc = nlp("Berlin is a nice capital. Munich is historical city. We should stay there for a while. ")
# Get all tokens and part-of-speech tags
token_texts = [token.text for token in doc]
pos_tags = [token.pos_ for token in doc]
for index, pos in enumerate(pos_tags):
# Check if the current token is a proper noun
if pos == "PROPN":
# Check if the next token is a verb
if pos_tags[index + 1] == "VERB":
result = token_texts[index]
print("Found proper noun before a verb:", result)
print(pos_tags)
nlp = spacy.load("en_core_web_sm")
doc = nlp("Berlin is a nice capital. Munich is historical city. We should stay there for a while. ")
for token in doc:
if(token.pos_ == "PROPN"):
if( (token.i + 1) < len(doc) and (doc[token.i + 1].pos_ == "VERB") ):
print("Found proper noun before a verb:", token.text)
# print(token.tag_, token.pos_, token.ent_type_)
```
**Inspect Word Vector**
```
# Process a text
doc = nlp("Two bananas in pyjamas")
# Get the vector for the token "bananas"
bananas_vector = doc[1].vector
print(bananas_vector)
print(len(bananas_vector))
```
**Compare Similarity**
Use the doc.similarity method to compare doc1 to doc2 and print the result.
```
!python -m spacy download en_core_web_md
nlp_md = spacy.load("en_core_web_md")
doc1 = nlp_md("It's a warm summer day")
doc2 = nlp_md("It's sunny outside")
# Get the similarity of doc1 and doc2
similarity = doc1.similarity(doc2)
print(similarity)
```
Use the token.similarity method to compare token1 to token2 and print the result.
```
doc = nlp_md("TV and books")
token1, token2 = doc[0], doc[2]
# Get the similarity of the tokens "TV" and "books"
similarity = token1.similarity(token2)
print(similarity)
```
```
doc = nlp_md("This was a great restaurant. Afterwards, we went to a really nice bar.")
# Create spans for "great restaurant" and "really nice bar"
span1 = Span(doc, 3, 5)
span2 = Span(doc, 12, 15)
print(span1)
print(span2)
# Get the similarity of the spans
similarity = span1.similarity(span2)
print(similarity)
for token in doc:
print(token.text, token.i)
doc = nlp_md("This was a great restaurant. Afterwards, we went to a really nice bar.")
pattern1 = [{"TEXT": "great"}, {"TEXT": "restaurant"}]
matcher = Matcher(nlp_md.vocab)
matcher.add("PATTERN1", None, pattern1)
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Print pattern string name and text of matched span
print(doc.vocab.strings[match_id], doc[start:end].text)
```
**Combining models and rules**
- Both patterns in this exercise contain mistakes and won’t match as expected. Can you fix them? If you get stuck, try printing the tokens in the doc to see how the text will be split and adjust the pattern so that each dictionary represents one token.
- Edit pattern1 so that it correctly matches all case-insensitive mentions of "Amazon" plus a title-cased proper noun.
- Edit pattern2 so that it correctly matches all case-insensitive mentions of "ad-free", plus the following noun.
```
doc = nlp(
"Twitch Prime, the perks program for Amazon Prime members offering free "
"loot, games and other benefits, is ditching one of its best features: "
"ad-free viewing. According to an email sent out to Amazon Prime members "
"today, ad-free viewing will no longer be included as a part of Twitch "
"Prime for new members, beginning on September 14. However, members with "
"existing annual subscriptions will be able to continue to enjoy ad-free "
"viewing until their subscription comes up for renewal. Those with "
"monthly subscriptions will have access to ad-free viewing until October 15."
)
# Create the match patterns
pattern1 = [{"LOWER": "amazon"}, {"POS": "PROPN"}]
#pattern1 = [{"LOWER": "amazon"}, {"IS_TITLE": True, "POS": "PROPN"}]
pattern2 = [{"LOWER": "ad"}, {"POS": "PUNCT"}, {"LOWER": "free"}, {"POS": "NOUN"}]
# Initialize the Matcher and add the patterns
matcher = Matcher(nlp.vocab)
matcher.add("PATTERN1", None, pattern1)
matcher.add("PATTERN2", None, pattern2)
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Print pattern string name and text of matched span
print(doc.vocab.strings[match_id], start, end, doc[start:end].text)
```
**Efficient PharseMatcher**
- Sometimes it’s more efficient to match exact strings instead of writing patterns describing the individual tokens. This is especially true for finite categories of things – like all countries of the world. We already have a list of countries, so let’s use this as the basis of our information extraction script. A list of string names is available as the variable COUNTRIES.
- Import the PhraseMatcher and initialize it with the shared vocab as the variable matcher.
- Add the phrase patterns and call the matcher on the doc.
```
COUNTRIES = ['Afghanistan', 'Åland Islands', 'Albania', 'Algeria', 'American Samoa', 'Andorra', 'Angola', 'Anguilla', 'Antarctica', 'Antigua and Barbuda', 'Argentina', 'Armenia', 'Aruba', 'Australia', 'Austria', 'Azerbaijan', 'Bahamas', 'Bahrain', 'Bangladesh', 'Barbados', 'Belarus', 'Belgium', 'Belize', 'Benin', 'Bermuda', 'Bhutan', 'Bolivia (Plurinational State of)', 'Bonaire, Sint Eustatius and Saba', 'Bosnia and Herzegovina', 'Botswana', 'Bouvet Island', 'Brazil', 'British Indian Ocean Territory', 'United States Minor Outlying Islands', 'Virgin Islands (British)', 'Virgin Islands (U.S.)', 'Brunei Darussalam', 'Bulgaria', 'Burkina Faso', 'Burundi', 'Cambodia', 'Cameroon', 'Canada', 'Cabo Verde', 'Cayman Islands', 'Central African Republic', 'Chad', 'Chile', 'China', 'Christmas Island', 'Cocos (Keeling) Islands', 'Colombia', 'Comoros', 'Congo', 'Congo (Democratic Republic of the)', 'Cook Islands', 'Costa Rica', 'Croatia', 'Cuba', 'Curaçao', 'Cyprus', 'Czech Republic', 'Denmark', 'Djibouti', 'Dominica', 'Dominican Republic', 'Ecuador', 'Egypt', 'El Salvador', 'Equatorial Guinea', 'Eritrea', 'Estonia', 'Ethiopia', 'Falkland Islands (Malvinas)', 'Faroe Islands', 'Fiji', 'Finland', 'France', 'French Guiana', 'French Polynesia', 'French Southern Territories', 'Gabon', 'Gambia', 'Georgia', 'Germany', 'Ghana', 'Gibraltar', 'Greece', 'Greenland', 'Grenada', 'Guadeloupe', 'Guam', 'Guatemala', 'Guernsey', 'Guinea', 'Guinea-Bissau', 'Guyana', 'Haiti', 'Heard Island and McDonald Islands', 'Holy See', 'Honduras', 'Hong Kong', 'Hungary', 'Iceland', 'India', 'Indonesia', "Côte d'Ivoire", 'Iran (Islamic Republic of)', 'Iraq', 'Ireland', 'Isle of Man', 'Israel', 'Italy', 'Jamaica', 'Japan', 'Jersey', 'Jordan', 'Kazakhstan', 'Kenya', 'Kiribati', 'Kuwait', 'Kyrgyzstan', "Lao People's Democratic Republic", 'Latvia', 'Lebanon', 'Lesotho', 'Liberia', 'Libya', 'Liechtenstein', 'Lithuania', 'Luxembourg', 'Macao', 'Macedonia (the former Yugoslav Republic of)', 'Madagascar', 'Malawi', 'Malaysia', 'Maldives', 'Mali', 'Malta', 'Marshall Islands', 'Martinique', 'Mauritania', 'Mauritius', 'Mayotte', 'Mexico', 'Micronesia (Federated States of)', 'Moldova (Republic of)', 'Monaco', 'Mongolia', 'Montenegro', 'Montserrat', 'Morocco', 'Mozambique', 'Myanmar', 'Namibia', 'Nauru', 'Nepal', 'Netherlands', 'New Caledonia', 'New Zealand', 'Nicaragua', 'Niger', 'Nigeria', 'Niue', 'Norfolk Island', "Korea (Democratic People's Republic of)", 'Northern Mariana Islands', 'Norway', 'Oman', 'Pakistan', 'Palau', 'Palestine, State of', 'Panama', 'Papua New Guinea', 'Paraguay', 'Peru', 'Philippines', 'Pitcairn', 'Poland', 'Portugal', 'Puerto Rico', 'Qatar', 'Republic of Kosovo', 'Réunion', 'Romania', 'Russian Federation', 'Rwanda', 'Saint Barthélemy', 'Saint Helena, Ascension and Tristan da Cunha', 'Saint Kitts and Nevis', 'Saint Lucia', 'Saint Martin (French part)', 'Saint Pierre and Miquelon', 'Saint Vincent and the Grenadines', 'Samoa', 'San Marino', 'Sao Tome and Principe', 'Saudi Arabia', 'Senegal', 'Serbia', 'Seychelles', 'Sierra Leone', 'Singapore', 'Sint Maarten (Dutch part)', 'Slovakia', 'Slovenia', 'Solomon Islands', 'Somalia', 'South Africa', 'South Georgia and the South Sandwich Islands', 'Korea (Republic of)', 'South Sudan', 'Spain', 'Sri Lanka', 'Sudan', 'Suriname', 'Svalbard and Jan Mayen', 'Swaziland', 'Sweden', 'Switzerland', 'Syrian Arab Republic', 'Taiwan', 'Tajikistan', 'Tanzania, United Republic of', 'Thailand', 'Timor-Leste', 'Togo', 'Tokelau', 'Tonga', 'Trinidad and Tobago', 'Tunisia', 'Turkey', 'Turkmenistan', 'Turks and Caicos Islands', 'Tuvalu', 'Uganda', 'Ukraine', 'United Arab Emirates', 'United Kingdom of Great Britain and Northern Ireland', 'United States of America', 'Uruguay', 'Uzbekistan', 'Vanuatu', 'Venezuela (Bolivarian Republic of)', 'Viet Nam', 'Wallis and Futuna', 'Western Sahara', 'Yemen', 'Zambia', 'Zimbabwe']
doc = nlp_en("Czech Republic may help Slovakia protect its airspace. France is an ally of United States of America")
matcher = PhraseMatcher(nlp_en.vocab)
# Create pattern Doc objects and add them to the matcher
# This is the faster version of: [nlp(country) for country in COUNTRIES]
patterns = list(nlp_en.pipe(COUNTRIES))
matcher.add("COUNTRY", None, *patterns)
# Call the matcher on the test document and print the result
matches = matcher(doc)
print(matches)
print([doc[start:end] for match_id, start, end in matches])
```
**Extract Countries and Relationship**
- In the previous exercise, you wrote a script using spaCy’s PhraseMatcher to find country names in text. Let’s use that country matcher on a longer text, analyze the syntax and update the document’s entities with the matched countries.
- Iterate over the matches and create a Span with the label "GPE" (geopolitical entity).
- Overwrite the entities in doc.ents and add the matched span.
- Get the matched span’s root head token.
- Print the text of the head token and the span.
```
matcher = PhraseMatcher(nlp_en.vocab)
patterns = list(nlp_en.pipe(COUNTRIES))
matcher.add("COUNTRY", None, *patterns)
# Create a doc and find matches in it
doc = nlp_en("Czech Republic may help Slovakia protect its airspace. France is an ally of United States of America")
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Create a Span with the label for "GPE"
span = Span(doc, start, end, label="GPE")
#print(doc.ents)
# Overwrite the doc.ents and add the span
doc.ents = list(doc.ents) + [span]
# Get the span's root head token
span_root_head = span.root.head
# Print the text of the span root's head token and the span text
print(span_root_head.text, "-->", span.text)
# Print the entities in the document
print([(ent.text, ent.label_) for ent in doc.ents if ent.label_ == "GPE"])
```
|
github_jupyter
|
import spacy
# Import the Doc class
from spacy.tokens import Doc, Span
from spacy.lang.en import English
from spacy.matcher import Matcher, PhraseMatcher
nlp = spacy.load("en_core_web_sm")
doc = nlp("I have a cat")
# Look up the hash for the word "cat"
cat_hash = nlp.vocab.strings["cat"]
print(cat_hash)
# Look up the cat_hash to get the string
cat_string = nlp.vocab.strings[cat_hash]
print(cat_string)
doc = nlp("David Bowie is a PERSON")
# Look up the hash for the string label "PERSON"
person_hash = nlp.vocab.strings["PERSON"]
print(person_hash)
# Look up the person_hash to get the string
person_string = nlp.vocab.strings[person_hash]
print(person_string)
# Desired text: "spaCy is cool!"
words = ["spaCy", "is", "cool", "!"]
spaces = [True, True, False, False]
# Create a Doc from the words and spaces
doc = Doc(nlp.vocab, words=words, spaces=spaces)
print(doc.text)
# Desired text: "Go, get started!"
words = ["Go", ",", "get", "started", "!"]
spaces = [False, True, True, False, False]
doc = Doc(nlp.vocab, words=words, spaces=spaces)
print(doc.text)
# Desired text: "Oh, really?!"
words = ["Oh", ",", "really", "?", "!"]
spaces = [False, True, False, False, False]
# Create a Doc from the words and spaces
doc = Doc(nlp.vocab, words = words, spaces = spaces)
print(doc.text)
nlp_en = English()
# Import the Doc and Span classes
from spacy.tokens import Doc, Span
words = ["I", "like", "David", "Bowie"]
spaces = [True, True, True, False]
# Create a doc from the words and spaces
doc = Doc(nlp_en.vocab, words=words, spaces=spaces)
print(doc.text)
# Create a span for "David Bowie" from the doc and assign it the label "PERSON"
span = Span(doc, 2, 4, label="PERSON")
print(span.text, span.label_)
# Add the span to the doc's entities
doc.ents = [span]
# Print entities' text and labels
print([(ent.text, ent.label_) for ent in doc.ents])
nlp = spacy.load("en_core_web_sm")
doc = nlp("Berlin is a nice capital. Munich is historical city. We should stay there for a while. ")
# Get all tokens and part-of-speech tags
token_texts = [token.text for token in doc]
pos_tags = [token.pos_ for token in doc]
for index, pos in enumerate(pos_tags):
# Check if the current token is a proper noun
if pos == "PROPN":
# Check if the next token is a verb
if pos_tags[index + 1] == "VERB":
result = token_texts[index]
print("Found proper noun before a verb:", result)
print(pos_tags)
nlp = spacy.load("en_core_web_sm")
doc = nlp("Berlin is a nice capital. Munich is historical city. We should stay there for a while. ")
for token in doc:
if(token.pos_ == "PROPN"):
if( (token.i + 1) < len(doc) and (doc[token.i + 1].pos_ == "VERB") ):
print("Found proper noun before a verb:", token.text)
# print(token.tag_, token.pos_, token.ent_type_)
# Process a text
doc = nlp("Two bananas in pyjamas")
# Get the vector for the token "bananas"
bananas_vector = doc[1].vector
print(bananas_vector)
print(len(bananas_vector))
!python -m spacy download en_core_web_md
nlp_md = spacy.load("en_core_web_md")
doc1 = nlp_md("It's a warm summer day")
doc2 = nlp_md("It's sunny outside")
# Get the similarity of doc1 and doc2
similarity = doc1.similarity(doc2)
print(similarity)
doc = nlp_md("TV and books")
token1, token2 = doc[0], doc[2]
# Get the similarity of the tokens "TV" and "books"
similarity = token1.similarity(token2)
print(similarity)
doc = nlp_md("This was a great restaurant. Afterwards, we went to a really nice bar.")
# Create spans for "great restaurant" and "really nice bar"
span1 = Span(doc, 3, 5)
span2 = Span(doc, 12, 15)
print(span1)
print(span2)
# Get the similarity of the spans
similarity = span1.similarity(span2)
print(similarity)
for token in doc:
print(token.text, token.i)
doc = nlp_md("This was a great restaurant. Afterwards, we went to a really nice bar.")
pattern1 = [{"TEXT": "great"}, {"TEXT": "restaurant"}]
matcher = Matcher(nlp_md.vocab)
matcher.add("PATTERN1", None, pattern1)
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Print pattern string name and text of matched span
print(doc.vocab.strings[match_id], doc[start:end].text)
doc = nlp(
"Twitch Prime, the perks program for Amazon Prime members offering free "
"loot, games and other benefits, is ditching one of its best features: "
"ad-free viewing. According to an email sent out to Amazon Prime members "
"today, ad-free viewing will no longer be included as a part of Twitch "
"Prime for new members, beginning on September 14. However, members with "
"existing annual subscriptions will be able to continue to enjoy ad-free "
"viewing until their subscription comes up for renewal. Those with "
"monthly subscriptions will have access to ad-free viewing until October 15."
)
# Create the match patterns
pattern1 = [{"LOWER": "amazon"}, {"POS": "PROPN"}]
#pattern1 = [{"LOWER": "amazon"}, {"IS_TITLE": True, "POS": "PROPN"}]
pattern2 = [{"LOWER": "ad"}, {"POS": "PUNCT"}, {"LOWER": "free"}, {"POS": "NOUN"}]
# Initialize the Matcher and add the patterns
matcher = Matcher(nlp.vocab)
matcher.add("PATTERN1", None, pattern1)
matcher.add("PATTERN2", None, pattern2)
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Print pattern string name and text of matched span
print(doc.vocab.strings[match_id], start, end, doc[start:end].text)
COUNTRIES = ['Afghanistan', 'Åland Islands', 'Albania', 'Algeria', 'American Samoa', 'Andorra', 'Angola', 'Anguilla', 'Antarctica', 'Antigua and Barbuda', 'Argentina', 'Armenia', 'Aruba', 'Australia', 'Austria', 'Azerbaijan', 'Bahamas', 'Bahrain', 'Bangladesh', 'Barbados', 'Belarus', 'Belgium', 'Belize', 'Benin', 'Bermuda', 'Bhutan', 'Bolivia (Plurinational State of)', 'Bonaire, Sint Eustatius and Saba', 'Bosnia and Herzegovina', 'Botswana', 'Bouvet Island', 'Brazil', 'British Indian Ocean Territory', 'United States Minor Outlying Islands', 'Virgin Islands (British)', 'Virgin Islands (U.S.)', 'Brunei Darussalam', 'Bulgaria', 'Burkina Faso', 'Burundi', 'Cambodia', 'Cameroon', 'Canada', 'Cabo Verde', 'Cayman Islands', 'Central African Republic', 'Chad', 'Chile', 'China', 'Christmas Island', 'Cocos (Keeling) Islands', 'Colombia', 'Comoros', 'Congo', 'Congo (Democratic Republic of the)', 'Cook Islands', 'Costa Rica', 'Croatia', 'Cuba', 'Curaçao', 'Cyprus', 'Czech Republic', 'Denmark', 'Djibouti', 'Dominica', 'Dominican Republic', 'Ecuador', 'Egypt', 'El Salvador', 'Equatorial Guinea', 'Eritrea', 'Estonia', 'Ethiopia', 'Falkland Islands (Malvinas)', 'Faroe Islands', 'Fiji', 'Finland', 'France', 'French Guiana', 'French Polynesia', 'French Southern Territories', 'Gabon', 'Gambia', 'Georgia', 'Germany', 'Ghana', 'Gibraltar', 'Greece', 'Greenland', 'Grenada', 'Guadeloupe', 'Guam', 'Guatemala', 'Guernsey', 'Guinea', 'Guinea-Bissau', 'Guyana', 'Haiti', 'Heard Island and McDonald Islands', 'Holy See', 'Honduras', 'Hong Kong', 'Hungary', 'Iceland', 'India', 'Indonesia', "Côte d'Ivoire", 'Iran (Islamic Republic of)', 'Iraq', 'Ireland', 'Isle of Man', 'Israel', 'Italy', 'Jamaica', 'Japan', 'Jersey', 'Jordan', 'Kazakhstan', 'Kenya', 'Kiribati', 'Kuwait', 'Kyrgyzstan', "Lao People's Democratic Republic", 'Latvia', 'Lebanon', 'Lesotho', 'Liberia', 'Libya', 'Liechtenstein', 'Lithuania', 'Luxembourg', 'Macao', 'Macedonia (the former Yugoslav Republic of)', 'Madagascar', 'Malawi', 'Malaysia', 'Maldives', 'Mali', 'Malta', 'Marshall Islands', 'Martinique', 'Mauritania', 'Mauritius', 'Mayotte', 'Mexico', 'Micronesia (Federated States of)', 'Moldova (Republic of)', 'Monaco', 'Mongolia', 'Montenegro', 'Montserrat', 'Morocco', 'Mozambique', 'Myanmar', 'Namibia', 'Nauru', 'Nepal', 'Netherlands', 'New Caledonia', 'New Zealand', 'Nicaragua', 'Niger', 'Nigeria', 'Niue', 'Norfolk Island', "Korea (Democratic People's Republic of)", 'Northern Mariana Islands', 'Norway', 'Oman', 'Pakistan', 'Palau', 'Palestine, State of', 'Panama', 'Papua New Guinea', 'Paraguay', 'Peru', 'Philippines', 'Pitcairn', 'Poland', 'Portugal', 'Puerto Rico', 'Qatar', 'Republic of Kosovo', 'Réunion', 'Romania', 'Russian Federation', 'Rwanda', 'Saint Barthélemy', 'Saint Helena, Ascension and Tristan da Cunha', 'Saint Kitts and Nevis', 'Saint Lucia', 'Saint Martin (French part)', 'Saint Pierre and Miquelon', 'Saint Vincent and the Grenadines', 'Samoa', 'San Marino', 'Sao Tome and Principe', 'Saudi Arabia', 'Senegal', 'Serbia', 'Seychelles', 'Sierra Leone', 'Singapore', 'Sint Maarten (Dutch part)', 'Slovakia', 'Slovenia', 'Solomon Islands', 'Somalia', 'South Africa', 'South Georgia and the South Sandwich Islands', 'Korea (Republic of)', 'South Sudan', 'Spain', 'Sri Lanka', 'Sudan', 'Suriname', 'Svalbard and Jan Mayen', 'Swaziland', 'Sweden', 'Switzerland', 'Syrian Arab Republic', 'Taiwan', 'Tajikistan', 'Tanzania, United Republic of', 'Thailand', 'Timor-Leste', 'Togo', 'Tokelau', 'Tonga', 'Trinidad and Tobago', 'Tunisia', 'Turkey', 'Turkmenistan', 'Turks and Caicos Islands', 'Tuvalu', 'Uganda', 'Ukraine', 'United Arab Emirates', 'United Kingdom of Great Britain and Northern Ireland', 'United States of America', 'Uruguay', 'Uzbekistan', 'Vanuatu', 'Venezuela (Bolivarian Republic of)', 'Viet Nam', 'Wallis and Futuna', 'Western Sahara', 'Yemen', 'Zambia', 'Zimbabwe']
doc = nlp_en("Czech Republic may help Slovakia protect its airspace. France is an ally of United States of America")
matcher = PhraseMatcher(nlp_en.vocab)
# Create pattern Doc objects and add them to the matcher
# This is the faster version of: [nlp(country) for country in COUNTRIES]
patterns = list(nlp_en.pipe(COUNTRIES))
matcher.add("COUNTRY", None, *patterns)
# Call the matcher on the test document and print the result
matches = matcher(doc)
print(matches)
print([doc[start:end] for match_id, start, end in matches])
matcher = PhraseMatcher(nlp_en.vocab)
patterns = list(nlp_en.pipe(COUNTRIES))
matcher.add("COUNTRY", None, *patterns)
# Create a doc and find matches in it
doc = nlp_en("Czech Republic may help Slovakia protect its airspace. France is an ally of United States of America")
# Iterate over the matches
for match_id, start, end in matcher(doc):
# Create a Span with the label for "GPE"
span = Span(doc, start, end, label="GPE")
#print(doc.ents)
# Overwrite the doc.ents and add the span
doc.ents = list(doc.ents) + [span]
# Get the span's root head token
span_root_head = span.root.head
# Print the text of the span root's head token and the span text
print(span_root_head.text, "-->", span.text)
# Print the entities in the document
print([(ent.text, ent.label_) for ent in doc.ents if ent.label_ == "GPE"])
| 0.359701 | 0.968201 |
```
# default_exp models.deepim
```
# DeepIM
> Implementation of DeepIM model in PyTorch.
```
#hide
from nbdev.showdoc import *
from fastcore.nb_imports import *
from fastcore.test import *
#export
import torch
from torch import nn
from recohut.models.layers.embedding import EmbeddingLayer
from recohut.models.layers.common import MLP_Layer
from recohut.models.layers.interaction import InteractionMachine
from recohut.models.bases.ctr import CTRModel
#export
class DeepIM(CTRModel):
def __init__(self,
feature_map,
model_id="DeepIM",
task="binary_classification",
learning_rate=1e-3,
embedding_initializer="torch.nn.init.normal_(std=1e-4)",
embedding_dim=10,
im_order=2,
im_batch_norm=False,
hidden_units=[64, 64, 64],
hidden_activations="ReLU",
net_dropout=0,
batch_norm=False,
**kwargs):
super(DeepIM, self).__init__(feature_map,
model_id=model_id,
**kwargs)
self.embedding_layer = EmbeddingLayer(feature_map, embedding_dim)
self.im_layer = InteractionMachine(embedding_dim, im_order, im_batch_norm)
self.dnn = MLP_Layer(input_dim=embedding_dim * feature_map.num_fields,
output_dim=1,
hidden_units=hidden_units,
hidden_activations=hidden_activations,
output_activation=None,
dropout_rates=net_dropout,
batch_norm=batch_norm) \
if hidden_units is not None else None
self.output_activation = self.get_final_activation(task)
self.init_weights(embedding_initializer=embedding_initializer)
def forward(self, inputs):
feature_emb = self.embedding_layer(inputs)
y_pred = self.im_layer(feature_emb)
if self.dnn is not None:
y_pred += self.dnn(feature_emb.flatten(start_dim=1))
if self.output_activation is not None:
y_pred = self.output_activation(y_pred)
return y_pred
```
Example
```
params = {'model_id': 'DeepIM',
'data_dir': '/content/data',
'model_root': './checkpoints/',
'learning_rate': 1e-3,
'optimizer': 'adamw',
'task': 'binary_classification',
'loss': 'binary_crossentropy',
'metrics': ['logloss', 'AUC'],
'embedding_dim': 10,
'im_order': 2,
'im_batch_norm': False,
'hidden_units': [300, 300, 300],
'hidden_activations': 'relu',
'net_regularizer': 0,
'embedding_regularizer': 0,
'net_batch_norm': False,
'net_dropout': 0,
'batch_size': 64,
'epochs': 3,
'shuffle': True,
'seed': 2019,
'use_hdf5': True,
'workers': 1,
'verbose': 0}
model = DeepIM(ds.dataset.feature_map, **params)
pl_trainer(model, ds, max_epochs=5)
```
|
github_jupyter
|
# default_exp models.deepim
#hide
from nbdev.showdoc import *
from fastcore.nb_imports import *
from fastcore.test import *
#export
import torch
from torch import nn
from recohut.models.layers.embedding import EmbeddingLayer
from recohut.models.layers.common import MLP_Layer
from recohut.models.layers.interaction import InteractionMachine
from recohut.models.bases.ctr import CTRModel
#export
class DeepIM(CTRModel):
def __init__(self,
feature_map,
model_id="DeepIM",
task="binary_classification",
learning_rate=1e-3,
embedding_initializer="torch.nn.init.normal_(std=1e-4)",
embedding_dim=10,
im_order=2,
im_batch_norm=False,
hidden_units=[64, 64, 64],
hidden_activations="ReLU",
net_dropout=0,
batch_norm=False,
**kwargs):
super(DeepIM, self).__init__(feature_map,
model_id=model_id,
**kwargs)
self.embedding_layer = EmbeddingLayer(feature_map, embedding_dim)
self.im_layer = InteractionMachine(embedding_dim, im_order, im_batch_norm)
self.dnn = MLP_Layer(input_dim=embedding_dim * feature_map.num_fields,
output_dim=1,
hidden_units=hidden_units,
hidden_activations=hidden_activations,
output_activation=None,
dropout_rates=net_dropout,
batch_norm=batch_norm) \
if hidden_units is not None else None
self.output_activation = self.get_final_activation(task)
self.init_weights(embedding_initializer=embedding_initializer)
def forward(self, inputs):
feature_emb = self.embedding_layer(inputs)
y_pred = self.im_layer(feature_emb)
if self.dnn is not None:
y_pred += self.dnn(feature_emb.flatten(start_dim=1))
if self.output_activation is not None:
y_pred = self.output_activation(y_pred)
return y_pred
params = {'model_id': 'DeepIM',
'data_dir': '/content/data',
'model_root': './checkpoints/',
'learning_rate': 1e-3,
'optimizer': 'adamw',
'task': 'binary_classification',
'loss': 'binary_crossentropy',
'metrics': ['logloss', 'AUC'],
'embedding_dim': 10,
'im_order': 2,
'im_batch_norm': False,
'hidden_units': [300, 300, 300],
'hidden_activations': 'relu',
'net_regularizer': 0,
'embedding_regularizer': 0,
'net_batch_norm': False,
'net_dropout': 0,
'batch_size': 64,
'epochs': 3,
'shuffle': True,
'seed': 2019,
'use_hdf5': True,
'workers': 1,
'verbose': 0}
model = DeepIM(ds.dataset.feature_map, **params)
pl_trainer(model, ds, max_epochs=5)
| 0.85449 | 0.649912 |
```
import tensorflow as tf
print(tf.__version__)
# By the way, what is a server / service / API?
# Best way to learn is by example
# Here is a service that simply returns your IP address in a JSON
import requests
r = requests.get('https://api.ipify.org?format=json')
j = r.json()
print(j)
# Our Tensorflow model server is the same, except what it does is much more
# complex - it returns the predictions from a ML model!
# More imports
import numpy as np
import matplotlib.pyplot as plt
import os
import subprocess
from tensorflow.keras.layers import Input, Conv2D, Dense, Flatten, Dropout
from tensorflow.keras.models import Model
# Load in the data
fashion_mnist = tf.keras.datasets.fashion_mnist
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
print("x_train.shape:", x_train.shape)
print("x_test.shape:", x_test.shape)
# the data is only 2D!
# convolution expects height x width x color
x_train = np.expand_dims(x_train, -1)
x_test = np.expand_dims(x_test, -1)
print(x_train.shape)
# number of classes
K = len(set(y_train))
print("number of classes:", K)
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
x = Conv2D(64, (3, 3), strides=2, activation='relu')(x)
x = Conv2D(128, (3, 3), strides=2, activation='relu')(x)
x = Flatten()(x)
x = Dropout(0.2)(x)
x = Dense(512, activation='relu')(x)
x = Dropout(0.2)(x)
x = Dense(K, activation='softmax')(x)
model = Model(i, x)
model.summary()
# Compile and fit
# Note: make sure you are using the GPU for this!
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
r = model.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=15)
# Save the model to a temporary directory
import tempfile
MODEL_DIR = tempfile.gettempdir()
version = 1
export_path = os.path.join(MODEL_DIR, str(version))
print('export_path = {}\n'.format(export_path))
if os.path.isdir(export_path):
print('\nAlready saved a model, cleaning up\n')
!rm -r {export_path}
tf.saved_model.save(model, export_path)
print('\nSaved model:')
!ls -l {export_path}
!saved_model_cli show --dir {export_path} --all
# This is the same as you would do from your command line, but without the [arch=amd64], and no sudo
# You would instead do:
# echo "deb [arch=amd64] http://storage.googleapis.com/tensorflow-serving-apt stable tensorflow-model-server tensorflow-model-server-universal" | sudo tee /etc/apt/sources.list.d/tensorflow-serving.list && \
# curl https://storage.googleapis.com/tensorflow-serving-apt/tensorflow-serving.release.pub.gpg | sudo apt-key add -
!echo "deb http://storage.googleapis.com/tensorflow-serving-apt stable tensorflow-model-server tensorflow-model-server-universal" | tee /etc/apt/sources.list.d/tensorflow-serving.list && \
curl https://storage.googleapis.com/tensorflow-serving-apt/tensorflow-serving.release.pub.gpg | apt-key add -
!apt update
!apt-get install tensorflow-model-server
os.environ["MODEL_DIR"] = MODEL_DIR
%%bash --bg
nohup tensorflow_model_server \
--rest_api_port=8501 \
--model_name=fashion_model \
--model_base_path="${MODEL_DIR}" >server.log 2>&1
!tail server.log
# Label mapping
labels = '''T-shirt/top
Trouser
Pullover
Dress
Coat
Sandal
Shirt
Sneaker
Bag
Ankle boot'''.split("\n")
def show(idx, title):
plt.figure()
plt.imshow(x_test[idx].reshape(28,28), cmap='gray')
plt.axis('off')
plt.title('\n\n{}'.format(title), fontdict={'size': 16})
i = np.random.randint(0, len(x_test))
show(i, labels[y_test[i]])
# Format some data to pass to the server
# {
# "signature_name": "serving_default",
# "instances": [ an N x H x W x C list ],
# }
import json
data = json.dumps({"signature_name": "serving_default", "instances": x_test[0:3].tolist()})
print(data)
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model:predict', data=data, headers=headers)
j = r.json()
print(j.keys())
print(j)
# It looks like a 2-D array, let's check its shape
pred = np.array(j['predictions'])
print(pred.shape)
# This is the N x K output array from the model
# pred[n,k] is the probability that we believe the nth sample belongs to the kth class
# Get the predicted classes
pred = pred.argmax(axis=1)
# Map them back to strings
pred = [labels[i] for i in pred]
print(pred)
# Get the true labels
actual = [labels[i] for i in y_test[:3]]
print(actual)
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# Allows you to select a model by version
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/1:predict', data=data, headers=headers)
j = r.json()
pred = np.array(j['predictions'])
pred = pred.argmax(axis=1)
pred = [labels[i] for i in pred]
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# Let's make a new model version
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
x = Flatten()(x)
x = Dense(K, activation='softmax')(x)
model2 = Model(i, x)
model2.summary()
# Compile and fit
# Note: make sure you are using the GPU for this!
model2.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
r = model2.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=15)
# Save version 2 of the model
version = 2
export_path = os.path.join(MODEL_DIR, str(version))
print('export_path = {}\n'.format(export_path))
if os.path.isdir(export_path):
print('\nAlready saved a model, cleaning up\n')
!rm -r {export_path}
tf.saved_model.save(model2, export_path)
print('\nSaved model:')
!ls -l {export_path}
# Will Tensorflow serving know about the new model without restarting?
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/2:predict', data=data, headers=headers)
j = r.json()
pred = np.array(j['predictions'])
pred = pred.argmax(axis=1)
pred = [labels[i] for i in pred]
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# What if we use a version number that does not exist?
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/3:predict', data=data, headers=headers)
j = r.json()
print(j)
```
|
github_jupyter
|
import tensorflow as tf
print(tf.__version__)
# By the way, what is a server / service / API?
# Best way to learn is by example
# Here is a service that simply returns your IP address in a JSON
import requests
r = requests.get('https://api.ipify.org?format=json')
j = r.json()
print(j)
# Our Tensorflow model server is the same, except what it does is much more
# complex - it returns the predictions from a ML model!
# More imports
import numpy as np
import matplotlib.pyplot as plt
import os
import subprocess
from tensorflow.keras.layers import Input, Conv2D, Dense, Flatten, Dropout
from tensorflow.keras.models import Model
# Load in the data
fashion_mnist = tf.keras.datasets.fashion_mnist
(x_train, y_train), (x_test, y_test) = fashion_mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
print("x_train.shape:", x_train.shape)
print("x_test.shape:", x_test.shape)
# the data is only 2D!
# convolution expects height x width x color
x_train = np.expand_dims(x_train, -1)
x_test = np.expand_dims(x_test, -1)
print(x_train.shape)
# number of classes
K = len(set(y_train))
print("number of classes:", K)
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
x = Conv2D(64, (3, 3), strides=2, activation='relu')(x)
x = Conv2D(128, (3, 3), strides=2, activation='relu')(x)
x = Flatten()(x)
x = Dropout(0.2)(x)
x = Dense(512, activation='relu')(x)
x = Dropout(0.2)(x)
x = Dense(K, activation='softmax')(x)
model = Model(i, x)
model.summary()
# Compile and fit
# Note: make sure you are using the GPU for this!
model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
r = model.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=15)
# Save the model to a temporary directory
import tempfile
MODEL_DIR = tempfile.gettempdir()
version = 1
export_path = os.path.join(MODEL_DIR, str(version))
print('export_path = {}\n'.format(export_path))
if os.path.isdir(export_path):
print('\nAlready saved a model, cleaning up\n')
!rm -r {export_path}
tf.saved_model.save(model, export_path)
print('\nSaved model:')
!ls -l {export_path}
!saved_model_cli show --dir {export_path} --all
# This is the same as you would do from your command line, but without the [arch=amd64], and no sudo
# You would instead do:
# echo "deb [arch=amd64] http://storage.googleapis.com/tensorflow-serving-apt stable tensorflow-model-server tensorflow-model-server-universal" | sudo tee /etc/apt/sources.list.d/tensorflow-serving.list && \
# curl https://storage.googleapis.com/tensorflow-serving-apt/tensorflow-serving.release.pub.gpg | sudo apt-key add -
!echo "deb http://storage.googleapis.com/tensorflow-serving-apt stable tensorflow-model-server tensorflow-model-server-universal" | tee /etc/apt/sources.list.d/tensorflow-serving.list && \
curl https://storage.googleapis.com/tensorflow-serving-apt/tensorflow-serving.release.pub.gpg | apt-key add -
!apt update
!apt-get install tensorflow-model-server
os.environ["MODEL_DIR"] = MODEL_DIR
%%bash --bg
nohup tensorflow_model_server \
--rest_api_port=8501 \
--model_name=fashion_model \
--model_base_path="${MODEL_DIR}" >server.log 2>&1
!tail server.log
# Label mapping
labels = '''T-shirt/top
Trouser
Pullover
Dress
Coat
Sandal
Shirt
Sneaker
Bag
Ankle boot'''.split("\n")
def show(idx, title):
plt.figure()
plt.imshow(x_test[idx].reshape(28,28), cmap='gray')
plt.axis('off')
plt.title('\n\n{}'.format(title), fontdict={'size': 16})
i = np.random.randint(0, len(x_test))
show(i, labels[y_test[i]])
# Format some data to pass to the server
# {
# "signature_name": "serving_default",
# "instances": [ an N x H x W x C list ],
# }
import json
data = json.dumps({"signature_name": "serving_default", "instances": x_test[0:3].tolist()})
print(data)
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model:predict', data=data, headers=headers)
j = r.json()
print(j.keys())
print(j)
# It looks like a 2-D array, let's check its shape
pred = np.array(j['predictions'])
print(pred.shape)
# This is the N x K output array from the model
# pred[n,k] is the probability that we believe the nth sample belongs to the kth class
# Get the predicted classes
pred = pred.argmax(axis=1)
# Map them back to strings
pred = [labels[i] for i in pred]
print(pred)
# Get the true labels
actual = [labels[i] for i in y_test[:3]]
print(actual)
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# Allows you to select a model by version
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/1:predict', data=data, headers=headers)
j = r.json()
pred = np.array(j['predictions'])
pred = pred.argmax(axis=1)
pred = [labels[i] for i in pred]
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# Let's make a new model version
# Build the model using the functional API
i = Input(shape=x_train[0].shape)
x = Conv2D(32, (3, 3), strides=2, activation='relu')(i)
x = Flatten()(x)
x = Dense(K, activation='softmax')(x)
model2 = Model(i, x)
model2.summary()
# Compile and fit
# Note: make sure you are using the GPU for this!
model2.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy'])
r = model2.fit(x_train, y_train, validation_data=(x_test, y_test), epochs=15)
# Save version 2 of the model
version = 2
export_path = os.path.join(MODEL_DIR, str(version))
print('export_path = {}\n'.format(export_path))
if os.path.isdir(export_path):
print('\nAlready saved a model, cleaning up\n')
!rm -r {export_path}
tf.saved_model.save(model2, export_path)
print('\nSaved model:')
!ls -l {export_path}
# Will Tensorflow serving know about the new model without restarting?
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/2:predict', data=data, headers=headers)
j = r.json()
pred = np.array(j['predictions'])
pred = pred.argmax(axis=1)
pred = [labels[i] for i in pred]
for i in range(0,3):
show(i, f"True: {actual[i]}, Predicted: {pred[i]}")
# What if we use a version number that does not exist?
headers = {"content-type": "application/json"}
r = requests.post('http://localhost:8501/v1/models/fashion_model/versions/3:predict', data=data, headers=headers)
j = r.json()
print(j)
| 0.870446 | 0.513912 |
# Unconstrained convex optimization
*Selected Topics in Mathematical Optimization: 2017-2018*
**Michiel Stock** ([email]([email protected]))

```
import matplotlib.pyplot as plt
import numpy as np
from teachingtools import blue, green, yellow, orange, red, black
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
```
## Motivation
In this chapter we will study unconstrained convex problems, i.e. problems of the form
$$
\min_\mathbf{x}\, f(\mathbf{x})\,,
$$
in which $f$ is *convex*. Convex optimization problems are well understood. Their most attractive property is that when a minimizer exists, the minimizer is the unique global minimizer.
Most convex optimization problems do not have a closed-form solution, with the quadratic problems of the previous chapters as a notable exception. We will hence again have to resort to descent methods to find an (arbitrary accurate) approximate solution.
## Convex sets and functions
### Convex set
> **In words**: a set $\mathcal{C}$ is called *convex* if the line segment between any two points in $\mathcal{C}$ also lies in $\mathcal{C}$.
> **In symbols**: a set $\mathcal{C}$ is called *convex* if, for any $\mathbf{x}, \mathbf{x}' \in \mathcal{C}$ and any $\theta \in [0, 1]$, it holds that $\theta \mathbf{x} + (1 - \theta) \mathbf{x}' \in \mathcal{C}$.

### Convex functions
> **In words**: a function $f$ is *convex* if the line segment between $(\mathbf{x}, f(\mathbf{x}))$ and $(\mathbf{x}', f (\mathbf{x}'))$ lies above the graph of $f$.
> **In symbols**: a function $f : \mathbb{R}^n\rightarrow \mathbb{R}$ is *convex* if
> - dom($f$) is convex
> - for any $\mathbf{x}, \mathbf{x}' \in \text{dom}(f)$ and any $\theta \in [0, 1]$, it holds that $f(\theta \mathbf{x} + (1-\theta)\mathbf{x}') \leq\theta f(\mathbf{x}) +(1-\theta)f(\mathbf{x}')$.


From the definition, it follows that:
- If the function is differentiable, then $f(\mathbf{x})\geq f(\mathbf{x}')+\nabla f(\mathbf{x}')^\top(\mathbf{x}-\mathbf{x}')$ for all $\mathbf{x}$ and $\mathbf{x}' \in \text{dom}(f)$. **The first-order Taylor approximation is a global underestimator of $f$.**
- If the function is twice differentiable, then $\nabla^2 f(\mathbf{x})\succeq 0$ for any $\mathbf{x}\in\text{dom}(f)$.
Convex functions frequently arise:
- If $f$ and $g$ are both convex, then $m(x)=\max(f(x), g(x))$ and $h(x)=f(x)+g(x)$ are also convex.
- If $f$ and $g$ are convex functions and $g$ is non-decreasing over a univariate domain, then $h(x)=g(f(x))$ is convex. Example: $e^{f(x)}$ is convex if $f({x})$ is convex.
Note, the convexity of expected value in probability theory gives rise to *Jensen's inequality*. For any convex function $\varphi$, if holds that
$$
\varphi(\mathbb{E}[X]) \leq\mathbb{E}[\varphi(X)]\,.
$$
This implies for example that the square of an expected value of quantity is never greater than the expected square of that quantity.
### Strongly convex functions
> **In words**: a function $f$ is called *strongly convex* if it is at least as convex as a quadratic function.
> **In symbols**: a $f$ is called *strongly $m$-convex* (with $m>0$) if the function $f_m(\mathbf{x}) = f(\mathbf{x}) - \frac{m}{2}||\mathbf{x}||_2$ is convex.
If the first- and second order derivatives exists, a strongly $m$-convex satisfies:
- $f(\mathbf{x}') \geq f(\mathbf{x}) + \nabla f(\mathbf{x})^\top (\mathbf{x}'-\mathbf{x}) + \frac{m}{2}||\mathbf{x}'-\mathbf{x}||_2$
- $\nabla^2 f(\mathbf{x})-mI\succeq 0$ (all eigenvalues of the Hessian are greater than $m$)
If a function is $m$-strongly convex, this also implies that there exists an $M>m$ such that
$$
\nabla^2 f(\mathbf{x}) \preceq MI\,.
$$
Stated differently, for strongly convex functions the exist both a quadratic function with a smaller as well as a lower local curvature.

## Toy examples
To illustrate the algorithms, we introduce two toy functions to minimize:
- simple quadratic problem:
$$
f(x_1, x_2) = \frac{1}{2} (x_1^2 +\gamma x_2^2)\,,
$$
where $\gamma$ determines the condition number.
- a non-quadratic function:
$$
f(x_1, x_2) = \log(e^{x_1 +3x_2-0.1}+e^{x_1 -3x_2-0.1}+e^{-x_1 -0.1})\,.
$$
```
from teachingtools import plot_contour, add_path # functions for showing the toy examples
from teachingtools import quadratic, grad_quadratic, hessian_quadratic
from teachingtools import nonquadratic, grad_nonquadratic, hessian_nonquadratic
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
ax1.set_title('Quadratic')
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
ax2.set_title('Non-quadratic')
```
## General descent methods (recap)
Convex functions are usually minimized using descent methods. Again, line search is often used as a subroutine.
The outline of a general descent algorithm is given in the following pseudocode.
> **input** starting point $\mathbf{x}\in$ **dom** $f$.
>
> **repeat**
>
>> 1. Determine a descent direction $\Delta \mathbf{x}$.
>> 2. *Line seach*. Choose a step size $t>0$.
>> 3. *Update*. $\mathbf{x}:=\mathbf{x}+t\Delta \mathbf{x}$.
>
> **until** stopping criterion is satisfied.
>
> **output** $\mathbf{x}$
The specific optimization algorithms are hence determined by:
- method for determining the search direction $\Delta \mathbf{x}$, this is almost always based on the gradient of $f$
- method for choosing the step size $t$, may be fixed or adaptive
- the criterion used for terminating the descent, usually the algorithm stops when the improvement is smaller than a predefined value
## Backtracking line search
For quadratic optimization, as covered in Chapter 1, the optimal step size could be computed in closed form. In the general case, only an approximately optimal step size is used.
### Exact line search
As a subroutine of the general descent algorithm a line search has to be performed. A value for $t$ is chosen to minimize $f$ along the ray $\{\mathbf{x}+t\Delta \mathbf{x} \mid t\geq0\}$:
$$
t = \text{arg min}_{s\geq0}\ f(\mathbf{x}+s\Delta \mathbf{x})\,.
$$
Exact line search is used when the cost of solving the above minimization problem is small compared to the cost of calculating the search direction itself. This is sometimes the case when an analytical solution is available.
### Inexact line search
Often, the descent methods work well when the line search is done only approximately. This is because the computational resources are better spent to performing more *approximate* steps in the different directions because the direction of descent will change anyway.
Many methods exist for this, we will consider the *backtracking line search* (BTLS), described by the following pseudocode.
> **input** starting point $\mathbf{x}\in$ **dom** $f$, descent direction $\Delta \mathbf{x}$, gradient $\nabla f(\mathbf{x})$, $\alpha\in(0,0.5)$ and $\beta\in(0,1)$.
>
> $t:=1$
>
>**while** $f(\mathbf{x}+t\Delta \mathbf{x}) > f(x) +\alpha t \nabla f(\mathbf{x})^\top\Delta \mathbf{x}$
>
>> $t:=\beta t$
>
>
>**output** $t$
The backtracking line search has two parameters:
- $\alpha$: fraction of decrease in f predicted by linear interpolation we accept
- $\beta$: reduction of the step size in each iteration of the BLS
- typically, $0.01 \leq \alpha \leq 0.3$ and $0.1 \leq \beta < 1$

**Assignment 1**
1. Complete the code for the backtracking line search
2. Use this function find the step size $t$ to (approximately) minimize $f(x) = x^2 - 2x - 5$ starting from the point $0$. Choose a $\Delta x=10$.
```
def backtracking_line_search(f, x0, Dx, grad_f, alpha=0.1,
beta=0.7):
'''
Uses backtracking for finding the minimum over a line.
Inputs:
- f: function to be searched over a line
- x0: initial point
- Dx: direction to search
- grad_f: gradient of f
- alpha
- beta
Output:
- t: suggested stepsize
'''
# ...
while # ...
# ...
return t
```
```
function = lambda x : x**2 - 2*x - 5
gradient_function = lambda x : 2*x -2
backtracking_line_search(function, 0, 10, gradient_function)
```
**Question 1**
Describe the effect of $\alpha$, $\beta$ and $\Delta \mathbf{x}$. How can you perform a more precise search?
YOUR ANSWER HERE
## Gradient descent
A natural choice for the search direction is the negative gradient: $\Delta \mathbf{x} = -\nabla f(\mathbf{x})$. This algorithm is called the *gradient descent algorithm*.
### General gradient descent algorithm
>**input** starting point $\mathbf{x}\in$ **dom** $f$.
>
>**repeat**
>
>> 1. $\Delta \mathbf{x} := -\nabla f(\mathbf{x})$.
>> 2. *Line seach*. Choose a step size $t$ via exact or backtracking line search.
>> 3. *Update*. $\mathbf{x}:=\mathbf{x}+t\Delta \mathbf{x}$.
>
>**until** stopping criterion is satisfied.
>
>**output** $\mathbf{x}$
The stopping criterion is usually of the form $||\nabla f(\mathbf{x})||_2 \leq \nu$.
### Convergence analysis
The notion of strongly convexity allows us to bound the function $f$ by two quadratic functions. As such we can reuse the convergence analysis of the previous chapter.
If $f$ is strongly convex (constants $m$ and $M$ exist such that $mI\prec \nabla^2 f(\mathbf{x})\prec MI$), it holds that $f(\mathbf{x}^{(k)}) - p^*\leq \varepsilon$ after at most
$$
\frac{\log((f(\mathbf{x}^{(0)}) - p^*)/\varepsilon)}{\log(1/c)}
$$
iterations, where $c =1-\frac{m}{M}<1$.
We conclude:
- The number of steps needed for a given quality is proportional to the logarithm of the initial error.
- To increase the accuracy with an order of magnitude, only a few more steps are needed.
- Convergence is again determined by the *condition number* $M/m$. Note that for large condition numbers: $\log(1/c)=-\log(1-\frac{m}{M})\approx m/M$, so the number of required iterations increases linearly with increasing $M/m$.
**Assignment 2**
1. Complete the implementation of the gradient descent method.
2. Plot the paths for the two toy problems. Use $\mathbf{x}^{(0)}=[10,1]^\top$ for the quadratic function and $\mathbf{x}^{(0)}=[-0.5,0.9]^\top$ for the non-quadratic function as starting points.
3. Analyze the convergence.
```
def gradient_descent(f, x0, grad_f, alpha=0.2, beta=0.7,
nu=1e-3, trace=False):
'''
General gradient descent algorithm.
Inputs:
- f: function to be minimized
- x0: starting point
- grad_f: gradient of the function to be minimized
- alpha: parameter for btls
- beta: parameter for btls
- nu: parameter to determine if the algorithm is converged
- trace: (bool) store the path that is followed?
Outputs:
- xstar: the found minimum
- x_steps: path in the domain that is followed (if trace=True)
- f_steps: image of x_steps (if trace=True)
'''
x = x0 # initial value
if trace: x_steps = [x0.copy()]
if trace: f_steps = [f(x0)]
while True:
# ... # choose direction
if # ...
break # converged
# ...
if trace: x_steps.append(x.copy())
if trace: f_steps.append(f(x))
if trace: return x, x_steps, f_steps
else: return x
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = gradient_descent(quadratic, np.array([[10.0], [1.0]]),
grad_quadratic, nu=1e-5, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = gradient_descent(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, nu=1e-5, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(f_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(np.abs(f_steps_q))
ax1.semilogy()
ax2.plot(np.abs(f_steps_nq[:-1] - f_steps_nq[-1])) # error compared to last step
ax2.semilogy()
for ax in (ax1, ax2):
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
```
**Describe the convergence**
YOUR ANSWER HERE
Let us assess the effect of the hyperparameters of $\alpha$ and $\beta$ of the inexact line search.
```
alphas = np.linspace(0.01, 0.3, num=10) # alphas to explore
betas = np.linspace(0.01, 0.99, num=15) # betas to explore
steps_to_convergence = np.zeros((len(alphas), len(betas)))
for i, alpha in enumerate(alphas):
for j, beta in enumerate(betas):
xstar_q, x_steps_q, f_steps_q = gradient_descent(nonquadratic,
np.array([[10.0], [1.0]]),
grad_nonquadratic, nu=1e-5, trace=True,
alpha=alpha, beta=beta)
steps_to_convergence[i, j] = len(f_steps_q) - 1
A, B = np.meshgrid(alphas, betas)
fig, ax = plt.subplots()
cs = ax.contourf(A,B, np.log10(steps_to_convergence).T, cmap='hot')
ax.set_xlabel(r'$\alpha$')
ax.set_ylabel(r'$\beta$')
ax.set_title('$\log_{10}$ of number of steps until convergence\nnon-quadratic curve')
cbar = fig.colorbar(cs)
```
**Describe the effect of $\alpha$, $\beta$.**
YOUR ANSWER HERE
Furthermore let us see what the effect is of the condition number on the number of steps until convergence. We will explore this for the quadratic function.
```
condition_numbers = np.logspace(0, 3, num=25)
steps_to_convergence = np.zeros_like(condition_numbers)
for i, gamma in enumerate(condition_numbers):
xstar_q, x_steps_q, f_steps_q = gradient_descent(lambda x:quadratic(x, gamma=gamma),
np.array([[10.0], [1.0]]),
lambda x:grad_quadratic(x, gamma=gamma),
nu=1e-5, trace=True)
steps_to_convergence[i] = len(f_steps_q) - 1 # number of steps
fig, ax = plt.subplots()
ax.plot(condition_numbers, steps_to_convergence)
ax.loglog()
ax.set_ylabel('Number of steps\nuntil convergence')
ax.set_xlabel('Condition number')
```
**Describe the effect of the condition number.**
YOUR ANSWER HERE
## Steepest descent
Optimize the first-order Taylor approximation of a function:
$$
f(\mathbf{x}+\mathbf{v}) \approx \hat{f}(\mathbf{x}+\mathbf{v}) =f(\mathbf{x}) +\nabla f(\mathbf{x})^\top \mathbf{v}\,.
$$
The linear approximation $\hat{f}$ can be made arbitrary negative if we can freely choose $\mathbf{v}$! We have to constrain the *norm* of $\mathbf{v}$.
### Vector norms
A norm on $\mathbb{R}^n$ is a function $||\cdot||:\mathbb{R}^n\rightarrow \mathbb{R}$ with the following properties:
- $||\mathbf{x}||>0$, for any $\mathbf{x}\in\mathbb{R}^n$
- $||\mathbf{x}+\mathbf{y}|| \leq ||\mathbf{x}||+||\mathbf{y}||$, for any $\mathbf{x}, \mathbf{y}\in\mathbb{R}^n$
- $||\lambda \mathbf{x}|| = |\lambda|\, ||\mathbf{x}||$ for any $\lambda \in\mathbb{R}$ and any $\mathbf{x}\in\mathbb{R}^n$
- $||\mathbf{x}||=0$ if and only if $\mathbf{x}=0$
For example, for any $\mathbf{x}\in\mathbb{R}^n$ and $p\leq 1$:
$$
||\mathbf{x}||_p = \left(\sum_{i=1}^n |x_i|^p\right)^\frac{1}{p}\,.
$$
$||\cdot||_1$ is often called the $L_1$ norm and $||\cdot||_2$ the $L_2$ norm.
Consider $P\in \mathbb{R}^{n\times n}$ such that $P\succ 0$. The corresponding *quadratic norm*:
$$
||\mathbf{z}||_P = (\mathbf{z}^\top P\mathbf{z})^\frac{1}{2}=||P^\frac{1}{2}\mathbf{z}||_2\,.
$$
The matrix $P$ can be used to encode prior knowledge about the scales and dependencies in the space that we want to search.
### Dual norm
Let $|| \cdot ||$ be a norm on $\mathbb{R}^n$. The associated dual norm:
$$
||\mathbf{z}||_*=\sup_{\mathbf{x}} \{\mathbf{z}^\top\mathbf{x}\mid ||\mathbf{x}||\leq 1\}\,.
$$
Examples:
- The dual norm of $||\cdot||_1$ is $||\cdot||_\infty$
- The dual norm of $||\cdot||_2$ is $||\cdot||_2$
- The dual norm of $||\cdot||_P$ is defined by $||\mathbf{z}||_*=||P^{-\frac{1}{2}}\mathbf{z}||$
### Steepest descent directions
**Normalized steepest descent direction**:
$$
\Delta x_\text{nsd} = \text{arg min}_\mathbf{v}\, \{\nabla f(\mathbf{x})^T \mathbf{v} \mid ||\mathbf{v}||\leq 1 \}\,.
$$
**Unnormalized steepest descent direction**:
$$
\Delta x_\text{sd} = ||\nabla f(\mathbf{x})||_\star \Delta x_\text{nsd} \,.
$$
Note that we have
$$
\nabla f(\mathbf{x})^\top \Delta x_\text{sd} = ||\nabla f(\mathbf{x})||_\star \nabla f(\mathbf{x})^\top\Delta x_\text{nsd} = -||\nabla f(\mathbf{x})||^2_\star\,,
$$
so this is a valid descent method.

### Coordinate descent algorithm
Using the $L_1$ norm results in coordinate descent. For every iteration in this algorithm, we descent in the direction of the dimension where the absolute value of the gradient is largest.
>**input** starting point $\mathbf{x}\in$ **dom** $f$.
>
>**repeat**
>
>> 1. *Direction*. Choose $i$ such that $|\nabla f(\mathbf{x})_i|$ is maximal.
>> 2. *Choose direction*. $\Delta \mathbf{x} := -\nabla f(\mathbf{x})_i \mathbf{e}_i$
>> 3. *Line seach*. Choose a step size $t$ via exact or backtracking line search.
>> 4. *Update*. $\mathbf{x}:=\mathbf{x}+t\Delta \mathbf{x}$.
>
>**until** stopping criterion is satisfied.
>
>**output** $\mathbf{x}$
Here, $\mathbf{e}_i$ is the $i$-th basic vector.
The stopping criterion is usually of the form $||\nabla f(\mathbf{x})||_2 \leq \nu$.
Coordinate descent optimizes every dimension in turn, for this reason it is sometimes used in minimization problems which enforce sparseness (e.g. LASSO regression).
> *Optimizing one dimension at a time is usually a poor strategy. This is because different dimensions are often related.*
**Assignment 3**
1. Complete the implementation of the coordinate descent method.
2. Plot the paths for the two toy problems. Use the same stating points as before.
3. Analyze the convergence.
```
from unconstrained import coordinate_descent
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = coordinate_descent(quadratic, np.array([[10.1], [1.0]]),
grad_quadratic, nu=1e-5, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = coordinate_descent(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, nu=1e-5, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(f_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(np.abs(f_steps_q))
ax1.semilogy()
ax2.plot(np.abs(f_steps_nq[:-1] - f_steps_nq[-1])) # error compared to last step
ax2.semilogy()
for ax in (ax1, ax2):
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
```
**Describe the convergence of coordinate descent compared to gradient descent.**
YOUR ANSWER HERE
## Newton's method
### The Newton step
In Newton's method the descent direction is chosen as
$$
\Delta \mathbf{x}_\text{nt} = -(\nabla^2f(\mathbf{x}))^{-1} \nabla f(\mathbf{x})\,,
$$
which is called the *Newton step*.
If $f$ is convex, then $\nabla^2f(\mathbf{x})$ is positive definite and
$$
\nabla f(\mathbf{x})^\top \Delta \mathbf{\mathbf{x}}_\text{nt} \geq 0\,,
$$
hence the Newton step is a descent direction unless $\mathbf{x}$ is optimal.
This Newton step can be motivated in several ways.
**Minimizer of a second order approximation**
The second order Taylor approximation $\hat{f}$ of $f$ at $\mathbf{x}$ is
$$
f(\mathbf{x}+\mathbf{v})\approx\hat{f}(\mathbf{x}+\mathbf{v}) = f(\mathbf{x}) + \nabla f(\mathbf{x})^\top \mathbf{v} + \frac{1}{2} \mathbf{v}^\top \nabla^2 f(\mathbf{x}) \mathbf{v}\,
$$
which is a convex quadratic function of $\mathbf{v}$, and is minimized when $\mathbf{v}=\Delta \mathbf{x}_\text{nt}$.
This quadratic model will be particularly accurate when $\mathbf{x}$ is close to $\mathbf{x}^\star$.
**Steepest descent direction in Hessian norm**
The Newton step is the steepest descent step if a quadratic norm using the Hessian is used, i.e.
$$
||\mathbf{u}||_{\nabla^2f(\mathbf{x})}=(\mathbf{u}^\top\nabla^2f(\mathbf{x})\mathbf{u})^\frac{1}{2}\,.
$$
**Affine invariance of the Newton step**
> *A consistent algorithm should give the same results independent of the units in which quantities are measured.* ~ Donald Knuth
The Newton step is independent of linear or affine changes of coordinates. Consider a non-singular $n\times n$ transformation matrix $T$. If we apply a coordinate transformation $\mathbf{x}=T\mathbf{y}$ and define $\bar{f}(\mathbf{y}) = f(\mathbf{x})$, then
$$
\nabla \bar{f}(\mathbf{y}) = T^\top\nabla f(\mathbf{x})\,,\quad \nabla^2 \bar{f}(\mathbf{y}) = T^\top\nabla^2f(\mathbf{x})T\,.
$$
As such it follows that
$$
\mathbf{x} + \Delta \mathbf{x}_\text{nt} = T (\mathbf{y} + \Delta \mathbf{y}_\text{nt})\,.
$$
**Questions 2**
Does scaling and rotation affect the working of gradient descent and coordinate descent?
### Newton decrement
The Newton decrement is defined as
$$
\lambda(\mathbf{x}) = (\nabla f(\mathbf{x})^\top\nabla^2 f(x)^{-1}\nabla f(\mathbf{x}))^{1/2}\,.
$$
This can be related to the quantity $f(\mathbf{x})-\inf_\mathbf{y}\ \hat{f}(\mathbf{y})$:
$$
f(\mathbf{x})-\inf_\mathbf{y}\ \hat{f}(\mathbf{y}) = f(\mathbf{x}) - \hat{f}(\mathbf{x} +\Delta \mathbf{x}_\text{nt}) = \frac{1}{2} \lambda(\mathbf{x})^2\,.
$$
Thus $\frac{1}{2} \lambda(\mathbf{x})^2$ is an estimate of $f(\mathbf{x}) - p^*$, based on the quadratic approximation of $f$ at $\mathbf{x}$.
### Pseudocode of Newton's algortihm
>**input** starting point $\mathbf{x}\in$ **dom** $f$.
>
>**repeat**
>
>> 1. Compute the Newton step and decrement $\Delta \mathbf{x}_\text{nt} := -\nabla^2f(\mathbf{x})^{-1} \nabla f(\mathbf{x})$; $\lambda^2:=\nabla f(\mathbf{x})^\top\nabla^2 f(\mathbf{x})^{-1}\nabla f(\mathbf{x})$.
>> 2. *Stopping criterion* **break** if $\lambda^2/2 \leq \epsilon$.
>> 3. *Line seach*. Choose a step size $t$ via exact or backtracking line search.
>> 4. *Update*. $\mathbf{x}:=\mathbf{x}+t\Delta \mathbf{x}_\text{nt}$.
>
>**output** $\mathbf{x}$
The above algorithm is sometimes called the *damped* Newton method, as it uses a variable step size $t$. In practice, using a fixed step also works well. Here, one has to consider the computational cost of using BTLS versus performing a few extra Newton steps to attain the same accuracy.
### Convergence analysis
Iterations in Newton’s method fall into two stages:
- *damped Newton phase* $(t < 1)$ until $||\nabla f(\mathbf{x})||_2 \leq \eta$
- *pure Newton phase* $(t = 1)$: quadratic convergence
After a sufficiently large number of iterations, the number of correct digits doubles at each iteration.
**Assignment 4**
1. Complete the code for Newton's method.
2. Find the minima of the two toy problems. Use the same starting points as for gradient descent.
```
def newtons_method(f, x0, grad_f, hess_f, alpha=0.3,
beta=0.8, epsilon=1e-3, trace=False):
'''
Newton's method for minimizing functions.
Inputs:
- f: function to be minimized
- x0: starting point
- grad_f: gradient of the function to be minimized
- hess_f: hessian matrix of the function to be minimized
- alpha: parameter for btls
- beta: parameter for btls
- nu: parameter to determine if the algorithm is converged
- trace: (bool) store the path that is followed?
Outputs:
- xstar: the found minimum
- x_steps: path in the domain that is followed (if trace=True)
- f_steps: image of x_steps (if trace=True)
'''
x = x0 # initial value
if trace: x_steps = [x.copy()]
if trace: f_steps = [f(x0)]
while True:
# ...
if # ... # stopping criterion
break # converged
# ...
if trace: x_steps.append(x.copy())
if trace: f_steps.append(f(x))
if trace: return x, x_steps, f_steps
else: return x
```
```
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = newtons_method(quadratic, np.array([[10.0], [1.0]]),
grad_quadratic, hessian_quadratic, epsilon=1e-8, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = newtons_method(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, hessian_nonquadratic, epsilon=1e-8, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(x_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(f_steps_q)
ax1.semilogy()
ax2.plot(f_steps_nq)
for ax in (ax1, ax2):
ax.set_xlabel('iteration')
ax.set_ylabel('function value')
```
#### Effect of condtition number and rotations.
Below is an applet to demonstrate the effect of 1) changing the condition problem of the quadratic function and 2) rotating the function such that the axes of the ellipsis are no longer alligned with the coordinate axes.
```
def show_condition(gamma, theta):
quad_gamma = lambda x : quadratic(x, gamma, theta)
x0 = np.array([[10.], [1.]])
d_quad_gamma = lambda x : grad_quadratic(x, gamma, theta)
dd_quad_gamma = lambda x : hessian_quadratic(x, gamma, theta)
xstar_gd, x_steps_gd, f_steps_gd = gradient_descent(quad_gamma,
x0.copy(),
d_quad_gamma,
nu=1e-6, trace=True)
xstar_cd, x_steps_cd, f_steps_cd = coordinate_descent(quad_gamma,
x0.copy(),
d_quad_gamma,
nu=1e-6, trace=True)
xstar_nm, x_steps_nm, f_steps_nm = newtons_method(quad_gamma, x0.copy(),
d_quad_gamma, dd_quad_gamma, epsilon=1e-6, trace=True)
fig, ax1 = plt.subplots(ncols=1, figsize=(10, 10))
plot_contour(quad_gamma, [-10, 10], [-11, 11], ax1)
add_path(ax1, x_steps_gd, blue, label='GD')
add_path(ax1, x_steps_cd, red, label='CD')
add_path(ax1, x_steps_nm, green, label='NM')
ax1.legend(loc=3)
print('Gradient descent iterations: {}'.format(len(x_steps_gd) - 1 ))
print('Coordinate descent iterations: {}'.format(len(x_steps_cd) - 1 ))
print('Newton\'s iterations: {}'.format(len(x_steps_nm) - 1))
from ipywidgets import interact, FloatSlider
interact(show_condition, gamma=FloatSlider(min=0.1, max=20.0, step=0.1, value=1),
theta=FloatSlider(min=0, max=np.pi / 2, step=0.1, value=0))
```
**Describe the effect of the conditional number and rotations on the path of the different optimization algorithms.**
YOUR ANSWER HERE
### Scalability of the different algorithms
To study scaling towards higher dimensional problems, we minimize a function of the following form:
$$
f(\mathbf{x}) = \mathbf{x}^\top C \mathbf{x} - \sum_{i=1}^m \log(b_i - \mathbf{a}_i^\top\mathbf{x})
$$
with $C$ a positive-definite matrix and $b_i$ and $\mathbf{a}_i$ positive.
We will optimize such a function in 20, 100 and 250 dimensions.
```
from teachingtools import make_general_multidim_problem
n_sizes = [20, 100, 250]
m_sizes = [100, 300, 1000]
fig, axes = plt.subplots(ncols=3, figsize=(20, 7))
for (n, m, ax) in zip(n_sizes, m_sizes, axes):
f, g_f, h_f = make_general_multidim_problem(n, m)
xstar_gd, x_steps_gd, f_steps_gd = gradient_descent(f, np.zeros((n, 1)),
g_f, nu=1e-3, trace=True)
xstar_cd, x_steps_cd, f_steps_cd = coordinate_descent(f, np.zeros((n, 1)),
g_f, nu=1e-3, trace=True)
xstar_newton, x_steps_newton, f_steps_newton = newtons_method(f, np.zeros((n, 1)),
g_f, h_f, epsilon=1e-8, trace=True)
ax.plot(np.abs(f_steps_gd - f_steps_gd[-1]), color = blue, label='Gradient desc.')
ax.plot(np.abs(f_steps_cd - f_steps_cd[-1]), color = green, label='Coordinate desc.')
ax.plot(np.abs(f_steps_newton - f_steps_newton[-1]), color = red, label='Newton method')
ax.loglog()
ax.legend(loc=0)
ax.set_title('$n$={}, $m$={}'.format(n, m))
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
```
**Discuss the different algorithms with respect to their scalability.**
YOUR ANSWER HERE
### Summary Newton's method
- Convergence of Newton's algorithm is rapid and quadratic near $\mathbf{x}^\star$.
- Newton's algorithm is affine invariant, e.g. invariant to choice of coordinates or condition number.
- Newton's algorithm scales well with problem size. Computationally, computing and storing the Hessian might be prohibitive.
- The hyperparameters $\alpha$ and $\beta$ of BTLS do not influence the performance much.
## Quasi-Newton methods
Quasi-Newton methods try to emulate the success of the Newton method, but without the high computational burden of constructing the Hessian matrix every step. One of the most popular quasi-Newton algorithms is the *Broyden-Fletcher-Goldfarb-Shanno* (BFGS) algorithm. Here, the Hessian is approximated by a symmetric rank-one matrix.
## Numerical approximation of the gradient and Hessian
In many cases, there is no analytical expression for gradient and the Hessian. The finite difference method can motivate the following approximations for the gradient-vector product
$$
\nabla f(\mathbf{x})^\top\Delta\mathbf{x} \approx \frac{1}{2\epsilon} (f(\mathbf{x}+\epsilon\Delta\mathbf{x} ) - f(\mathbf{x}-\epsilon\Delta\mathbf{x} ))
$$
and the Hessian-vector product
$$
\nabla^2 f(\mathbf{x})^\top\Delta\mathbf{x} \approx \frac{1}{2\epsilon} (\nabla f(\mathbf{x}+\epsilon\Delta\mathbf{x} ) - \nabla f(\mathbf{x}-\epsilon\Delta\mathbf{x} ))\,
$$
with $\epsilon$ a small constant.
## Exercise: logistic regression

Consider the following problem: we have a dataset of $n$ instances: $T=\{(\mathbf{x}_i, y_i)\mid i=1\ldots n\}$. Here $\mathbf{x}_i\in \mathbb{R}^p$ is a $p$-dimensional feature vector and $y_i\in\{0,1\}$ is a binary label. This is a binary classification problem, we are interested in predicting the label of an instance based on its feature description. The goal of logistic regression is to find a function $f(\mathbf{x})$ that estimates the conditional probability of $Y$:
$$
\mathcal{P}(Y=1 \mid \mathbf{X} = \mathbf{x})\,.
$$
We will assume that this function $f(\mathbf{x})$ is of the form
$$
f(\mathbf{x}) = \sigma(\mathbf{w}^\top\mathbf{x})\,,
$$
with $\mathbf{w}$ a vector of parameters to be learned and $\sigma(.)$ the logistic map:
$$
\sigma(t) = \frac{e^{t}}{1+e^{t}}=\frac{1}{1+e^{-t}}\,.
$$
It is easy to see that the logistic mapping will ensure that $f(\mathbf{x})\in[0, 1]$, hence $f(\mathbf{x})$ can be interpreted as a probability.
Note that
$$
\frac{\text{d}\sigma(t)}{\text{d}t} = (1-\sigma(t))\sigma(t)\,.
$$
To find the best weights that separate the two classes, we can use the following structured loss function:
$$
\mathcal{L;\lambda}(\mathbf{w})=-\sum_{i=1}^n[y_i\log(\sigma(\mathbf{w}^\top\mathbf{x}_i))+(1-y_i)\log(1-\sigma(\mathbf{w}^\top\mathbf{x}_i))] +\lambda \mathbf{w}^\top\mathbf{w}\,.
$$
Here, the first part is the cross entropy, which penalizes disagreement between the prediction $f(\mathbf{x}_i)$ and the true label $y_i$, while the second term penalizes complex models in which $\mathbf{w}$ has a large norm. The trade-off between these two components is controlled by $\lambda$, a hyperparameter. In the course *Predictive modelling* of Willem Waegeman it is explained that by carefully tuning this parameter one can obtain an improved performance. **In this project we will study the influence $\lambda$ on the convergence of the optimization algorithms.**
> **Warning**: for this project there is a large risk of numerical problems when computing the loss function. This is because in the cross entropy $0\log(0)$ should by definition evaluate to its limit value of $0$. Numpy will evaluate this as `nan`. Use the provided function `cross_entropy` which safely computes $-\sum_{i=1}^n[y_i\log(\sigma_i)+(1-y_i)\log(1-\sigma_i)]$.

```
from teachingtools import logistic_toy
interact(logistic_toy, separation=FloatSlider(min=0, max=4, step=0.2, value=1),
log_lambda=FloatSlider(min=-5, max=5, step=1, value=1))
```
**Data overview**
Consider the data set in the file `BreastCancer.csv`. This dataset contains information about 569 female patients diagnosed with breast cancer. For each patient it was recorded wether the tumor was benign (B) or malignant (M), this is the response variable. Each tumor is described by 30 features, which encode some information about the tumor. We want to use logistic regression with regularization to predict wether a tumor is benign or malignant based on these features.
```
# pandas allows us to comfortably work with datasets in python
import pandas as pd
cancer_data = pd.read_csv('Data/BreastCancer.csv') # load data
cancer_data.head() # show first five rows
# extract response in binary encoding:
# 0 : B(enign)
# 1 : M(alignant)
binary_response = np.array(list(map(int, cancer_data.y == 'M')), dtype=float)
binary_response = binary_response.reshape((-1, 1)) # make column vector
# extract feature matrix X
features = cancer_data.select(lambda colname : colname[0] == 'x',
axis=1).values
# standarizing features
# this is needed for gradient descent to run faster
features -= features.mean(0)
features /= features.std(0)
```
**Assignments**
1. Derive and implement the loss function for logistic loss, the gradient and the Hessian of this loss function. These functions have as input the parameter vector $\mathbf{w}$, label vector $\mathbf{y}$, feature matrix $\mathbf{X}$ and $\lambda$. The logistic map and cross-entropy is already provided for you.
2. Consider $\lambda=0.1$, find the optimal parameter vector for this data using gradient descent, coordinate descent and Newton's method. Use standardized features. For each algorithm, give the number of steps the algorithm performed and the running time (use the [magic function](https://ipython.org/ipython-doc/3/interactive/magics.html) `%timeit`). Compare the loss for each of parameters obtained by the different algorithms.
3. How does regularization influence the optimization? Make a separate plot for gradient descent, coordinate descent and Newton's method with the the value of the loss as a function of the iteration of the given algorithm. Make separate the different methods and plot the convergence for $\lambda = [10^{-3}, 10^{-1}, 1, 10, 100]$. Does increased regularization make the optimization go faster or slower? Why does this make sense?
**Project assignment 1**
Complete the functions below.
```
# You can also use the implemented functions:
from logistic_regression import logistic_loss, grad_logistic_loss, hess_logistic_loss
# functions for first question
l_loss = lambda w : logistic_loss(w, binary_response, features, 0.1)
l_grad = lambda w : grad_logistic_loss(w, binary_response, features, 0.1)
l_hess = lambda w : hess_logistic_loss(w, binary_response, features, 0.1)
```
**Assignment 2**
Use gradient descent, coordinate descent and Newton's method to find the parameters of the logistic model ($\lambda=0.1$).
DISCUSS THE DIFFERENCE IN RUNNING TIME
**Project assignment 3**
Make a plot for each of the four optimization method in which you show the convergence for $\lambda = [10^{-3}, 10^{-1}, 1, 10, 100]$.
DISCUSS THE DIFFERENCE IN CONVERGENCE
## References
Boyd, S. and Vandenberghe, L. *'Convex Optimization'*. Cambridge University Press (2004) [link](https://stanford.edu/~boyd/cvxbook/)
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
from teachingtools import blue, green, yellow, orange, red, black
import warnings
warnings.filterwarnings("ignore")
%matplotlib inline
from teachingtools import plot_contour, add_path # functions for showing the toy examples
from teachingtools import quadratic, grad_quadratic, hessian_quadratic
from teachingtools import nonquadratic, grad_nonquadratic, hessian_nonquadratic
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
ax1.set_title('Quadratic')
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
ax2.set_title('Non-quadratic')
def backtracking_line_search(f, x0, Dx, grad_f, alpha=0.1,
beta=0.7):
'''
Uses backtracking for finding the minimum over a line.
Inputs:
- f: function to be searched over a line
- x0: initial point
- Dx: direction to search
- grad_f: gradient of f
- alpha
- beta
Output:
- t: suggested stepsize
'''
# ...
while # ...
# ...
return t
function = lambda x : x**2 - 2*x - 5
gradient_function = lambda x : 2*x -2
backtracking_line_search(function, 0, 10, gradient_function)
def gradient_descent(f, x0, grad_f, alpha=0.2, beta=0.7,
nu=1e-3, trace=False):
'''
General gradient descent algorithm.
Inputs:
- f: function to be minimized
- x0: starting point
- grad_f: gradient of the function to be minimized
- alpha: parameter for btls
- beta: parameter for btls
- nu: parameter to determine if the algorithm is converged
- trace: (bool) store the path that is followed?
Outputs:
- xstar: the found minimum
- x_steps: path in the domain that is followed (if trace=True)
- f_steps: image of x_steps (if trace=True)
'''
x = x0 # initial value
if trace: x_steps = [x0.copy()]
if trace: f_steps = [f(x0)]
while True:
# ... # choose direction
if # ...
break # converged
# ...
if trace: x_steps.append(x.copy())
if trace: f_steps.append(f(x))
if trace: return x, x_steps, f_steps
else: return x
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = gradient_descent(quadratic, np.array([[10.0], [1.0]]),
grad_quadratic, nu=1e-5, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = gradient_descent(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, nu=1e-5, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(f_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(np.abs(f_steps_q))
ax1.semilogy()
ax2.plot(np.abs(f_steps_nq[:-1] - f_steps_nq[-1])) # error compared to last step
ax2.semilogy()
for ax in (ax1, ax2):
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
alphas = np.linspace(0.01, 0.3, num=10) # alphas to explore
betas = np.linspace(0.01, 0.99, num=15) # betas to explore
steps_to_convergence = np.zeros((len(alphas), len(betas)))
for i, alpha in enumerate(alphas):
for j, beta in enumerate(betas):
xstar_q, x_steps_q, f_steps_q = gradient_descent(nonquadratic,
np.array([[10.0], [1.0]]),
grad_nonquadratic, nu=1e-5, trace=True,
alpha=alpha, beta=beta)
steps_to_convergence[i, j] = len(f_steps_q) - 1
A, B = np.meshgrid(alphas, betas)
fig, ax = plt.subplots()
cs = ax.contourf(A,B, np.log10(steps_to_convergence).T, cmap='hot')
ax.set_xlabel(r'$\alpha$')
ax.set_ylabel(r'$\beta$')
ax.set_title('$\log_{10}$ of number of steps until convergence\nnon-quadratic curve')
cbar = fig.colorbar(cs)
condition_numbers = np.logspace(0, 3, num=25)
steps_to_convergence = np.zeros_like(condition_numbers)
for i, gamma in enumerate(condition_numbers):
xstar_q, x_steps_q, f_steps_q = gradient_descent(lambda x:quadratic(x, gamma=gamma),
np.array([[10.0], [1.0]]),
lambda x:grad_quadratic(x, gamma=gamma),
nu=1e-5, trace=True)
steps_to_convergence[i] = len(f_steps_q) - 1 # number of steps
fig, ax = plt.subplots()
ax.plot(condition_numbers, steps_to_convergence)
ax.loglog()
ax.set_ylabel('Number of steps\nuntil convergence')
ax.set_xlabel('Condition number')
from unconstrained import coordinate_descent
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = coordinate_descent(quadratic, np.array([[10.1], [1.0]]),
grad_quadratic, nu=1e-5, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = coordinate_descent(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, nu=1e-5, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(f_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(np.abs(f_steps_q))
ax1.semilogy()
ax2.plot(np.abs(f_steps_nq[:-1] - f_steps_nq[-1])) # error compared to last step
ax2.semilogy()
for ax in (ax1, ax2):
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
def newtons_method(f, x0, grad_f, hess_f, alpha=0.3,
beta=0.8, epsilon=1e-3, trace=False):
'''
Newton's method for minimizing functions.
Inputs:
- f: function to be minimized
- x0: starting point
- grad_f: gradient of the function to be minimized
- hess_f: hessian matrix of the function to be minimized
- alpha: parameter for btls
- beta: parameter for btls
- nu: parameter to determine if the algorithm is converged
- trace: (bool) store the path that is followed?
Outputs:
- xstar: the found minimum
- x_steps: path in the domain that is followed (if trace=True)
- f_steps: image of x_steps (if trace=True)
'''
x = x0 # initial value
if trace: x_steps = [x.copy()]
if trace: f_steps = [f(x0)]
while True:
# ...
if # ... # stopping criterion
break # converged
# ...
if trace: x_steps.append(x.copy())
if trace: f_steps.append(f(x))
if trace: return x, x_steps, f_steps
else: return x
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
plot_contour(quadratic, (-11, 11), (-5, 5), ax1)
plot_contour(nonquadratic, (-2, 2), (-1, 1), ax2)
xstar_q, x_steps_q, f_steps_q = newtons_method(quadratic, np.array([[10.0], [1.0]]),
grad_quadratic, hessian_quadratic, epsilon=1e-8, trace=True)
add_path(ax1, x_steps_q, red)
print('Number of steps quadratic function: {}'.format(len(x_steps_q) - 1))
xstar_nq, x_steps_nq, f_steps_nq = newtons_method(nonquadratic, np.array([[-0.5], [0.9]]),
grad_nonquadratic, hessian_nonquadratic, epsilon=1e-8, trace=True)
add_path(ax2, x_steps_nq, red)
print('Number of steps non-quadratic function: {}'.format(len(x_steps_nq) - 1))
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(10, 5))
ax1.plot(f_steps_q)
ax1.semilogy()
ax2.plot(f_steps_nq)
for ax in (ax1, ax2):
ax.set_xlabel('iteration')
ax.set_ylabel('function value')
def show_condition(gamma, theta):
quad_gamma = lambda x : quadratic(x, gamma, theta)
x0 = np.array([[10.], [1.]])
d_quad_gamma = lambda x : grad_quadratic(x, gamma, theta)
dd_quad_gamma = lambda x : hessian_quadratic(x, gamma, theta)
xstar_gd, x_steps_gd, f_steps_gd = gradient_descent(quad_gamma,
x0.copy(),
d_quad_gamma,
nu=1e-6, trace=True)
xstar_cd, x_steps_cd, f_steps_cd = coordinate_descent(quad_gamma,
x0.copy(),
d_quad_gamma,
nu=1e-6, trace=True)
xstar_nm, x_steps_nm, f_steps_nm = newtons_method(quad_gamma, x0.copy(),
d_quad_gamma, dd_quad_gamma, epsilon=1e-6, trace=True)
fig, ax1 = plt.subplots(ncols=1, figsize=(10, 10))
plot_contour(quad_gamma, [-10, 10], [-11, 11], ax1)
add_path(ax1, x_steps_gd, blue, label='GD')
add_path(ax1, x_steps_cd, red, label='CD')
add_path(ax1, x_steps_nm, green, label='NM')
ax1.legend(loc=3)
print('Gradient descent iterations: {}'.format(len(x_steps_gd) - 1 ))
print('Coordinate descent iterations: {}'.format(len(x_steps_cd) - 1 ))
print('Newton\'s iterations: {}'.format(len(x_steps_nm) - 1))
from ipywidgets import interact, FloatSlider
interact(show_condition, gamma=FloatSlider(min=0.1, max=20.0, step=0.1, value=1),
theta=FloatSlider(min=0, max=np.pi / 2, step=0.1, value=0))
from teachingtools import make_general_multidim_problem
n_sizes = [20, 100, 250]
m_sizes = [100, 300, 1000]
fig, axes = plt.subplots(ncols=3, figsize=(20, 7))
for (n, m, ax) in zip(n_sizes, m_sizes, axes):
f, g_f, h_f = make_general_multidim_problem(n, m)
xstar_gd, x_steps_gd, f_steps_gd = gradient_descent(f, np.zeros((n, 1)),
g_f, nu=1e-3, trace=True)
xstar_cd, x_steps_cd, f_steps_cd = coordinate_descent(f, np.zeros((n, 1)),
g_f, nu=1e-3, trace=True)
xstar_newton, x_steps_newton, f_steps_newton = newtons_method(f, np.zeros((n, 1)),
g_f, h_f, epsilon=1e-8, trace=True)
ax.plot(np.abs(f_steps_gd - f_steps_gd[-1]), color = blue, label='Gradient desc.')
ax.plot(np.abs(f_steps_cd - f_steps_cd[-1]), color = green, label='Coordinate desc.')
ax.plot(np.abs(f_steps_newton - f_steps_newton[-1]), color = red, label='Newton method')
ax.loglog()
ax.legend(loc=0)
ax.set_title('$n$={}, $m$={}'.format(n, m))
ax.set_xlabel('Iteration')
ax.set_ylabel('Absolute error')
from teachingtools import logistic_toy
interact(logistic_toy, separation=FloatSlider(min=0, max=4, step=0.2, value=1),
log_lambda=FloatSlider(min=-5, max=5, step=1, value=1))
# pandas allows us to comfortably work with datasets in python
import pandas as pd
cancer_data = pd.read_csv('Data/BreastCancer.csv') # load data
cancer_data.head() # show first five rows
# extract response in binary encoding:
# 0 : B(enign)
# 1 : M(alignant)
binary_response = np.array(list(map(int, cancer_data.y == 'M')), dtype=float)
binary_response = binary_response.reshape((-1, 1)) # make column vector
# extract feature matrix X
features = cancer_data.select(lambda colname : colname[0] == 'x',
axis=1).values
# standarizing features
# this is needed for gradient descent to run faster
features -= features.mean(0)
features /= features.std(0)
# You can also use the implemented functions:
from logistic_regression import logistic_loss, grad_logistic_loss, hess_logistic_loss
# functions for first question
l_loss = lambda w : logistic_loss(w, binary_response, features, 0.1)
l_grad = lambda w : grad_logistic_loss(w, binary_response, features, 0.1)
l_hess = lambda w : hess_logistic_loss(w, binary_response, features, 0.1)
| 0.641535 | 0.990892 |
# Linear Algebra - Vectors
---
- Author: Diego Inácio
- GitHub: [github.com/diegoinacio](https://github.com/diegoinacio)
- Notebook: [linear-algebra_vectors.ipynb](https://github.com/diegoinacio/machine-learning-notebooks/blob/master/Mathematical-Foundations/linear-algebra_vectors.ipynb)
---
Linear Algebra topic about *Vectors*.
```
import numpy as np
```
## 1. What is a vector?
---
A vector $\large \vec{v}$ is a mathematical entity which has *magnitude* and *direction*.

## 2. Vector operations
---
All the following operations are applicable to any vector in $\large \mathbb{R}^n$.
### 2.1. Vector addition
---
The addition of two vectors $\large \vec{u}$ and $\large \vec{v}$ is done by the sum of their correspondent components, resulting in another vector.
$$ \large
\vec{u}+\vec{v} =
\begin{bmatrix}
u_1 \\ u_2 \\ \vdots \\ u_n
\end{bmatrix} +
\begin{bmatrix}
v_1 \\ v_2 \\ \vdots \\ v_n
\end{bmatrix} =
\begin{bmatrix}
v_1 + u_1 \\ v_2 + u_2 \\ \vdots \\ v_n + u_n
\end{bmatrix}
$$

For example:
$$ \large
\vec{u} =
\begin{bmatrix}
3 \\ 7
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
2 \\ 5
\end{bmatrix}
$$
$$ \large
\vec{u} + \vec{v} =
\begin{bmatrix}
3 + 2 \\ 7 + 5
\end{bmatrix} =
\begin{bmatrix}
5 \\ 12
\end{bmatrix}
$$
```
u = np.array([[3, 7]])
v = np.array([[2, 5]])
print(u.T, end=" = u\n\n")
print(v.T, end=" = v\n\n")
print((u + v).T, end=" = u + v")
```
### 2.2. Vector subtraction
---
Similarly to addition, the subtraction of two vectors $\large \vec{u}$ and $\large \vec{v}$ is done by the subtraction of their correspondent components, resulting in another vector.
$$ \large
\vec{u}-\vec{v} =
\begin{bmatrix}
u_1 \\ u_2 \\ \vdots \\ u_n
\end{bmatrix} -
\begin{bmatrix}
v_1 \\ v_2 \\ \vdots \\ v_n
\end{bmatrix} =
\begin{bmatrix}
v_1 - u_1 \\ v_2 - u_2 \\ \vdots \\ v_n - u_n
\end{bmatrix}
$$

For example:
$$ \large
\vec{u} =
\begin{bmatrix}
3 \\ 7
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
2 \\ 5
\end{bmatrix}
$$
$$ \large
\vec{u} - \vec{v} =
\begin{bmatrix}
3 - 2 \\ 7 - 5
\end{bmatrix} =
\begin{bmatrix}
1 \\ 2
\end{bmatrix}
$$
```
u = np.array([[3, 7]])
v = np.array([[2, 5]])
print(u.T, end=" = u\n\n")
print(v.T, end=" = v\n\n")
print((u - v).T, end=" = u - v")
```
### 2.3. Scalar multiplication
---
The scalar multiplication is the elementwise multiplication by a scalar number $\large \alpha$. The same rule can be applied to divisions.
$$ \large
\alpha\vec{u} =
\alpha \cdot
\begin{bmatrix}
u_1 \\ u_2 \\ \vdots \\ u_n
\end{bmatrix} =
\begin{bmatrix}
\alpha\cdot u_1 \\ \alpha\cdot u_2 \\ \vdots \\ \alpha\cdot u_n
\end{bmatrix}
$$

For example:
$$ \large
\alpha = 2
\quad , \quad
\vec{u} =
\begin{bmatrix}
3 \\ 7
\end{bmatrix}
$$
$$ \large
\alpha\cdot\vec{u} =
\begin{bmatrix}
2 \cdot 3 \\ 2 \cdot 7
\end{bmatrix} =
\begin{bmatrix}
6 \\ 14
\end{bmatrix}
$$
```
a = 2
u = np.array([[3, 7]])
print(a, end=" = a\n\n")
print(u.T, end=" = u\n\n")
print(a*u.T, end=" = au")
```
## 3. Dot product
---
Dot product is an algebraic operation, which has a huge number of applications. As a result, we have a scalar value.
$$ \large
\vec{u} \cdot \vec{v} =
\begin{bmatrix}
u_1 \\ u_2 \\ \vdots \\ u_n
\end{bmatrix} \cdot
\begin{bmatrix}
v_1 \\ v_2 \\ \vdots \\ v_n
\end{bmatrix} =
\sum_i^n u_i \cdot v_i
$$
$$ \large
\sum_i^n u_i \cdot v_i =
u_1 \cdot v_1 + u_2 \cdot v_2 + ... + u_n \cdot v_n
$$
For example:
$$ \large
\vec{u} =
\begin{bmatrix}
3 \\ 7
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
2 \\ 5
\end{bmatrix}
$$
$$ \large
\begin{aligned}
\vec{u} \cdot \vec{v} &= 3 \cdot 2 + 7 \cdot 5 \\
&= 6 + 35 \\
&= 41
\end{aligned}
$$
```
u = np.array([3, 7])
v = np.array([2, 5])
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(np.dot(u, v), end=" = u.v")
```
### 3.1 Unit vector
---
Unit vector (or versor) is a vector which has the magnitude equal to 1. The magnitude of a vector is based on the *euclidean norm* and can be found by:
$$ \large
\|\vec{u}\|_2 =
\left[ \sum_i^n u_i^2 \right]^{\frac{1}{2}}=\sqrt{u_1^2+u_2^2+...+u_n^2}
$$
Basically, a unit vector is a normalized vector, like:
$$ \large
\hat{u}=\frac{\vec{u}}{\|\vec{u}\|}
$$

For example:
$$ \large
\vec{u} = [3 \quad 4]
$$
$$ \large
\begin{aligned}
\|\vec{u}\| &= \sqrt{3^2+4^2} \\
&= \sqrt{9+16} \\
&= 5
\end{aligned}
$$
$$ \large
\frac{\vec{u}}{\|\vec{u}\|} =
\left[ \frac{3}{5} \quad \frac{4}{5} \right] =
[0.6 \quad 0.8]
$$
```
u = np.array([3, 4])
u_ = np.sum(u**2)**0.5
print(u, end=" = u\n\n")
print(u_, end=" = ||u||\n\n")
print(u/u_, end=" = û")
```
### 3.2. Angle between vectors
---
Given the geometric definition of *dot product*:
$$ \large
\vec{u} \cdot \vec{v} = \|\vec{u}\| \|\vec{v}\| \cos{\theta}
$$
so,
$$ \large
\theta=\cos^{-1} \frac{\vec{u} \cdot \vec{v}}{\|\vec{u}\| \|\vec{v}\|}
$$

For example:
$$ \large
\vec{u} =
\begin{bmatrix}
2 \\ 1
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
3 \\ 3
\end{bmatrix}
$$
$$ \large
\begin{aligned}
\angle (\vec{u}, \vec{v}) = \theta
&= \arccos \frac{2 \cdot 3 + 1 \cdot 3}{\sqrt{2^2+1^2} \sqrt{3^2+3^2}} \\
&= \arccos \frac{9}{\sqrt{5} \sqrt{18}} \\
&\approx 0.32 \text{rad} \quad \approx 18.43°
\end{aligned}
$$
```
u = np.array([2, 1])
v = np.array([3, 3])
uv = np.dot(u, v)
u_ = np.sum(u**2)**0.5
v_ = np.sum(v**2)**0.5
rad = np.arccos(uv/(u_*v_))
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(f'{np.rad2deg(rad):.02f}', end=" = θ")
```
### 3.3. Orthogonality and Parallelism
---
Two vectors are **parallel** or $\large \vec{u} // \vec{v}$ when there is a number $k$ which generalizes the relationship:
$$ \large
\vec{u} = k\vec{v}
$$
What it means that:
$$ \large
\frac{u_1}{v_1} = \frac{u_2}{v_2} = ... = \frac{u_n}{v_n} = k
$$
In other words, two vectors are parallel when their components are proportional.

For example:
$$ \large
\vec{u} =
\begin{bmatrix}
2 \\ 3
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
6 \\ 9
\end{bmatrix}
$$
$$ \large
\frac{2}{6} = \frac{3}{9} = \frac{1}{3}
$$
```
u = np.array([2, 3])
v = np.array([6, 9])
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(f'{u/v}', end=" = k")
```
Two vectors are **orthogonal** or $\large \vec{u} \bot \vec{v}$ when the angle $\theta$ between of them are 90° or their dot product is equal to 0.
$$ \large
\vec{u} \bot \vec{v} \Rightarrow \vec{u} \cdot \vec{v} = 0
$$

For example:
$$ \large
\vec{u} =
\begin{bmatrix}
2 \\ 1
\end{bmatrix}
\quad , \quad
\vec{v} =
\begin{bmatrix}
-2 \\ 4
\end{bmatrix}
$$
We can find it by calculating the dot product:
$$ \large
\vec{u} \cdot \vec{v} = 2 \cdot (-2) + 1 \cdot 4 = 0
$$
or by finding $\theta$:
$$ \large
\begin{aligned}
\angle (\vec{u}, \vec{v}) = \theta
&= \arccos \frac{2 \cdot (-2) + 1 \cdot 4}{\sqrt{2^2+1^2} \sqrt{(-2)^2+4^2}} \\
&= \arccos \frac{0}{\sqrt{5} \sqrt{20}} \\
&\approx 1.57 \text{rad} \quad = 90°
\end{aligned}
$$
```
u = np.array([2, 1])
v = np.array([-2, 4])
uv = np.dot(u, v)
u_ = np.sum(u**2)**0.5
v_ = np.sum(v**2)**0.5
rad = np.arccos(uv/(u_*v_))
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(uv, end=" = u.v\n\n")
print(f'{np.rad2deg(rad):.02f}', end=" = θ")
```
|
github_jupyter
|
import numpy as np
u = np.array([[3, 7]])
v = np.array([[2, 5]])
print(u.T, end=" = u\n\n")
print(v.T, end=" = v\n\n")
print((u + v).T, end=" = u + v")
u = np.array([[3, 7]])
v = np.array([[2, 5]])
print(u.T, end=" = u\n\n")
print(v.T, end=" = v\n\n")
print((u - v).T, end=" = u - v")
a = 2
u = np.array([[3, 7]])
print(a, end=" = a\n\n")
print(u.T, end=" = u\n\n")
print(a*u.T, end=" = au")
u = np.array([3, 7])
v = np.array([2, 5])
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(np.dot(u, v), end=" = u.v")
u = np.array([3, 4])
u_ = np.sum(u**2)**0.5
print(u, end=" = u\n\n")
print(u_, end=" = ||u||\n\n")
print(u/u_, end=" = û")
u = np.array([2, 1])
v = np.array([3, 3])
uv = np.dot(u, v)
u_ = np.sum(u**2)**0.5
v_ = np.sum(v**2)**0.5
rad = np.arccos(uv/(u_*v_))
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(f'{np.rad2deg(rad):.02f}', end=" = θ")
u = np.array([2, 3])
v = np.array([6, 9])
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(f'{u/v}', end=" = k")
u = np.array([2, 1])
v = np.array([-2, 4])
uv = np.dot(u, v)
u_ = np.sum(u**2)**0.5
v_ = np.sum(v**2)**0.5
rad = np.arccos(uv/(u_*v_))
print(u, end=" = u\n\n")
print(v, end=" = v\n\n")
print(uv, end=" = u.v\n\n")
print(f'{np.rad2deg(rad):.02f}', end=" = θ")
| 0.219421 | 0.987818 |
# Overview
This week's curriculum is a little bit of a mixed bag. We'll cover 2.33 topics that are not super-related, but both are _very useful_ (and there is some connection between them). The overview is
* Tricks for raw text (NLPP, Chapter 3) and finding the important words in a document (TF-IDF)
* Community Detection
In the first part, we will take a quick tour of NLPP1e's chapter 3, which is boring, but an amazing ressource that you'll keep returning to. Then we'll talk about how we can use simple statistics & machine learning to get text to show us what it's all about. We will even do a little visualization.
In the second part we will go back to network science, discussing community detection and trying it out on our very own dataset.
## The informal intro
You didn't think you'd be able to avoid hearing a little update from me, did you? I didn't think so :)
* Today, I'll go over the work we'll be focusing on today (spoiler, it's a brand new lecture which I'm very excited about), then
* And I'll talk about the stuff you'll be learning today is just now leading to break-throughs in our understanding of the brain!
* And finally, I'll talk a tiny amount about the next phase of the class (the project assignments).
(Next week, however, the informal intro will provide full details about the project phase of the class, so that's one not to miss.)
```
from IPython.display import YouTubeVideo, HTML, display
YouTubeVideo("XfDBYLA0q1I",width=800, height=450)
```
Video links
* Sune's community detection algorithm, for overlapping communities (that we're learning about today) is right now helping create break-throughs in brain science: https://news.iu.edu/stories/2020/10/iub/releases/19-neuroscientists-build-new-human-brain-network-model.html
**Questionnaire feedback**
Now for a second video, where I briefly address the second round of questionnaires. Thank you to those who filled one out! I also talk about the amazing TAs and give a great rant about active learning and why we keep doing it.
```
YouTubeVideo("T7qw4wbl_TQ",width=800, height=450)
```
Survey video links
* Active learning is better than class room learning https://www.pnas.org/content/111/23/8410
* Students think active learning is worse, but they actually learn more https://www.pnas.org/content/116/39/19251
* Sune's page on Employee of the month https://sunelehmann.com/employee-of-the-month/
# Processing real text (from out on the inter-webs)
Ok. So Chapter 3 in NLPP is all about working with text from the real world. Getting text from this internet, cleaning it, tokenizing, modifying (e.g. stemming, converting to lower case, etc) to get the text in shape to work with the NLTK tools you've already learned about - and many more.
In the process we'll learn more about regular expressions, as well as unicode; something we've already been struggling with a little bit will now be explained in more detail.
>
> **Video lecture**: Short overview of chapter 3 + a few words about kinds of language processing that we don't address in this class.
>
```
YouTubeVideo("Rwakh-HXPJk",width=800, height=450)
```
> *Reading*: NLPP Chapter 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.9, and 3.10\. It's not important that you go in depth with everything here - the key think is that you *know that Chapter 3 of this book exists*, and that it's a great place to return to if you're ever in need of an explanation of regular expressions, unicode, or other topics that you forget as soon as you stop using them (and don't worry, I forget about those things too).
>
# Words that characterize the heroes
In this section, we'll begin to play around with how far we can get with simple strategies for looking at text.
The video is basically just me talking about a fun paper, which shows you how little is needed in order to reveal something highly interesting about humans that produce text. But it's important. Don't miss this one!
> **Video lecture**: Simple methods reveal a lot. I talk a little bit about the paper: [Personality, Gender, and Age in the Language of Social Media: The Open-Vocabulary Approach](http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0073791).
```
YouTubeVideo("wkYvdfkVmlI",width=800, height=450)
```
Now, we'll use the wordcloud visualization techniques from the paper/video to learn about the wiki-pages we've downloaded.
In the exercises below, we'll use a technique from Sune's very own brain to do the Marvel vs. DC comparison (I call it "TF-TR"). Then proceed do some network analysis (!) to find network communities in one of the universes. Finally, jump back to NLP to used the communities we've just found to play with TF-IDF, a more standard technique.
We'll also do a bit of data cleaning along the way (using some of the stuff you've just read about in Chapter 3).
_Exercise_ 1: Comparing word-counts of Marvel and DC heroes. \[The longest exercise in the class 😱\]. It consists of four steps. And it's great.
**Step one: TF List**
The goal of this exercise is to create your own wordclouds, characterizing the two comic-book universes. Check out my awesome word-clouds below. I think it's pretty clear
Marvel | DC
:-------------------------:|:-------------------------:
 | 
*Setup*. All you need now is the wikipedia pages of the superheroes. If you didn't keep the wiki pages you previously downloaded, you can find them [here](https://github.com/SocialComplexityLab/socialgraphs2020/blob/master/files/character%20wiki%20pages.zip).
Once you have the texts down on your own computer, you will want to aggregate the text into two long lists. One based on all the text from the Marvel pages, and one based on all the text from the DC pages. In each list, you should keep all the words (or *tokens* to be more precise) that occur on the pages, and a count of how frequently each word occurs. For example, my Marvel list contains the entries:
```
...
wolverine 452
avenger 129
mutant 630
...
```
This list is called a ***Term Frequency*** (or TF) list. Let's build our own TF lists. Before you start counting, I am going to ask you do do a few things
> *Action items*
> * Tokenize the pages into individual strings
> * Remove all punctuation from your list of tokens
> * Set everything to lower case
> * (Optional) Lemmatize your words
If you are confused by the instructions for cleaning, go back and have a look Chapter 3 again.
> *Action item*:
> * Create your TF list for each universe.
**Step two: Word weights**
TF is not necessarily a good way of sorting a list, since many words are very common, so the most common words are not necessarily the most important ones. This is clear from the top counts in my marvel TF list
```
of 1623
comics 1577
marvel 1549
in 1528
and 1520
the 1518
to 1512
a 1511
is 1500
by 1498
was 1473
as 1466
ref 1457
with 1450
first 1447
```
You can fix some of this by removing stop-words (as is described in the book), but it's not enough. ***We want to pull out what's important.*** Thus, to create awesome and informative wordclouds like the ones I created above, we want to extract what's special about each of the two lists we're comparing.
The general way of doing that is using a strategy called TF-IDF. We will explore that strategy in execise 4 below. But out-of-the-box TF-IDF gets a bit weird when we only have two groups to compare. So we'll do something different (but related) here.
We want to use the information stored in TF, which is important since it says something about the most frequently occuring words. **But we want to weigh the TF information** using additional information about what is unique about each universe.
Specifically, we want to set things up such that - in the Marvel list, for example - words that are unique to Marvel get a high weight, while words that are unique to DC get a low weight, and words that occur at the same frequency in both lists are neutral. Vice versa for the DC list.
The way I chose to set up the weights here is simple. I use term ratios (TR), which I just made up, so don't look it up on the internet. What we care about is words that are used very differently in the two universes, so we just use their ratios.
Let me start with an example. The word `wolverine` occurs 452 in the marvel TF list and 9 times in the DC TF list, thus I set it's Marvel weight to
$$w_\textrm{wolverine}^{(m)} = \frac{452}{9 + c} = 13.29.$$
Similarly, its DC weight is
$$w_\textrm{wolverine}^{(d)} = \frac{9}{452 + c} = 0.02.$$
In both cases, I add the constant $c$ to the denominator in case a word occurs zero times. You can play around with the size of $c$ to understand the effect of chosing small/large values of it.
In general for some token $t$ with term frequency $\textrm{TF}^{(u)}_t$, in universe $u$ where $u \in \{m,d\}$, we define the weight as:
$$w_{t}^{(m)} = \frac{\textrm{TF}^{(m)}_t}{ \textrm{TF}^{(d)}_t + c}, \qquad \textrm{and} \qquad w_{t}^{(d)} = \frac{\textrm{TF}^{(d)}_t}{ \textrm{TF}^{(m)}_t + c}. $$
Thus, now we're ready to lists for each univers, where the ranking of token $t$ on list $u$ is given by $\textrm{TF}^{(u)}_t \times w_{t}^{(u)}$. I call this the *TF-TR* lists.
Note that the *TF-TR* lists have the properties that we requested above. The weight associated with a word is large when a word occurs much more frequently in the list we consider, compared to the other list. It's small when a word is rare in our list and frequent in the other. And it's approximately equal to one, when the word is equally frequent in both lists. (The downside is that it only works when you're finding important terms while two lists).
> *Action item*:
> * Create your TF-TR list for each universe.
> * Check out top 10 for each universe. Does it make sense?
The takehome here is that we get a good ranking by combining the term frequency with some weighting scheme. But, as we will see below, the TR weight that I created is not the only possible weight. There are many other options.
-----------
PS. Above in creating the ratios, I'm assuming that the universes have a comparable number of words in them. That's pretty much true. And while the approximation is OK for our purposes, it's not true in general. If you'd like an extra challenge, you can figure out how to account for differences in the size of each universe.
**Step three: Install the software**
First you must set up your system. The most difficult thing about creating the wordcloud is installing the `WordCloud` module. It's available on GitHub, check out the page [**here**](https://github.com/amueller/word_cloud).
If you're lucky, you can simply install using conda (and all dependencies, etc will be automatically fixed):
conda install -c conda-forge wordcloud
If you can't get that to work you can refer here https://anaconda.org/conda-forge/wordcloud.
Also, maybe the comments below are helpful:
* The module depends on the Python library PIL. Use `conda` to install that before you do anything else.
* On my system, the module needed the `gcc` compiler installed. If you're not already a programmer, you may have to install that. On Mac you get it by installing the [_command line tools_](http://osxdaily.com/2014/02/12/install-command-line-tools-mac-os-x/). On linux, it's probably already installed. And on Windows I'm not sure, but we'll figure it out during the exercises.
* Once that's all set up, you can use `pip` to install the `WordCloud` library, as [detailed on the GitHub page](https://github.com/amueller/word_cloud). But don't forget to use Anaconda's `pip` just as when you installed the communities library a few weeks ago.
* There are examples of how to use the module online, see [here](http://sebastianraschka.com/Articles/2014_twitter_wordcloud.html) and [here](https://bioinfoexpert.com/2015/05/26/generating-word-clouds-in-python/). If you're interested, you can read about how the package was put together [here](http://peekaboo-vision.blogspot.dk/2012/11/a-wordcloud-in-python.html).
**Step four: Draw the wordclouds**
> *Action items*
> * Get your lists ready for the word-cloud software
> - The package needs a single string to work on. The way that I converted my lists to a string was to simply combine all words together in one long string (separated by spaces), repeating each word according to its score (rounded up to the nearest integer value).
> - The `wordcloud` package looks for collocations in real texts, which is a problem when you make the list as above. The recommended fix is to simply set `collocations = False` as an option when you run the package.
> * Now, create a word-cloud for each universe. Feel free to make it as fancy or non-fancy as you like. Comment on the results. (If you'd like, you can remove stopwords/wiki-syntax - I did a bit of that for my own wordclouds.)
>
# Community detection
Now that we have worked hard on text analysis, it is time to go back to our network!
Before we begin, let's watch a great lecture to learn about communities. If you want all the details, I recommend you take a look at _Chapter 9_ in the _Network Science_ book ... but the video lecture below should be enough to get you started.
**Note**: For this and the next exercise, work on the _undirected_ version of the network.
> **_Video Lecture_**: Communities in networks.
You can watch the 2015 video [here](https://youtu.be/06GL_KGHdbE/).
```
YouTubeVideo("FSRoqXw28RI",width=800, height=450)
```
*Exercise 2*: Use the concept of modularity to explore how _community-like_ the universes are.
>
> * Explain the concept of modularity in your own words.
> * Consider the undirected version of the entire network, including both DC and Marvel.
> * Now create your own partition into communities, where all Marvel-characters are one community and all the DC-characters are another community. That's all you need, **now calculate the modularity of this partition**. Modularity is described in the _Network Science_ book, section 9.4.. Thus, use **equation 9.12** in the book to calculate the modularity _M_ of the partition described above. Are the universes good communities?
> * Would you expect this result in light of what we have found in the previous exercises?
*Exercise 3*: Community detection, considering each universe separately.
> * Consider the network of heroes for each universe separately.
> - **Note**. For this exercise, it's OK to just pick one of the universes.
> - If you want to work with the entire network. Then run community detection on each universe separately, then combine results to get a list containing all communities from both parts of the network.
> - (The reason for this is that the strong split between Marvel/DC universes can confuse the algorithm a bit).
> * Use [the Python Louvain-algorithm implementation](http://perso.crans.org/aynaud/communities/) to find communities each network. Report the value of modularity found by the algorithm. Is it higher or lower than what you found above for the universes as communities? What does this comparison reveal about them?
> * **Note**: This implementation is also available as Anaconda package. Install with `conda` as expained [here](https://anaconda.org/auto/python-louvain).
> * You can also try the *Infomap* algorithm instead if you're curious. Go to [this page]. (http://www.mapequation.org/code.html) and search for 'python'. It's harder to install, but a better community detection algorithm.
> Visualize the network, using the Force Atlas algorithm (see Lecture 5, exercise 2). This time assign each node a different color based on their _community_. Describe the structure you observe.
Now we jump back into the NLP work. **It's still OK to work with just one universe**. In this last exercise, we'll be creating word-clouds again. But this time, we'll be using the more standard method: TF-IDF.
*Exercise 4*: Wrap your brain around TF-IDF
First, let's learn about TF-IDF the way wikipedia explains it. Check out [the wikipedia page for TF-IDF](https://en.wikipedia.org/wiki/Tf%E2%80%93idf) and ***read the first part very carefully***. The idea is still to have a *term frequency* (TF) and a *weight* (IDF), but there are some additional things going on. For example, there are multiple definitions of TF. We just looked at the raw counts.
> *Action items*
> * Pick one of the alternative term frequency definitions. Explain why it might sometimes be prefereable to the raw count.
> * What does IDF stand for?
There are also multiple versions of IDF. Let's think about those for a moment.
> *Action items*
> * All of the IDF versions take the log of the calculated weight. Why do you think that is?
> * Explain why using IDF makes stopword removal less important.
> * In the TR weight that I defined in Exercise 1, we take into account how frequently each word appears inside each of the two documents. Is information of word counts inside each document used in the definition of IDF on Wikipedia?
I noted above that out-of-the box worked weirdly when you only have two documents. Let's see why that's the case. If we grab one of the simple definitions of IDF-weight from wikipedia
$$\log \left( \frac{N}{n_t+1} \right) + 1.$$
Where $N = 2$ is the number of documents and $n_t \in \{1,2\}$ is the number of documents containing the term $t$.
> *Action item*
> * What are the possible weights that a word can have?
> * Explain in your own words why TF-IDF might not result in ideal wordclods when you only have two documents.
*Pro-level consideration*: It is, of course, possible to define IDF weighting schemes that incorporate information of word-counts within each document, even if you have more than two documents. If you'd like to try to do that below, it's OK with me. If not, that's also fine.
**To be continued ...**. Next week, we'll look at sentiment and TFIDF for communities.
Thanks to TA Alexandra for helping create these exercises.
|
github_jupyter
|
from IPython.display import YouTubeVideo, HTML, display
YouTubeVideo("XfDBYLA0q1I",width=800, height=450)
YouTubeVideo("T7qw4wbl_TQ",width=800, height=450)
YouTubeVideo("Rwakh-HXPJk",width=800, height=450)
YouTubeVideo("wkYvdfkVmlI",width=800, height=450)
...
wolverine 452
avenger 129
mutant 630
...
of 1623
comics 1577
marvel 1549
in 1528
and 1520
the 1518
to 1512
a 1511
is 1500
by 1498
was 1473
as 1466
ref 1457
with 1450
first 1447
```
You can fix some of this by removing stop-words (as is described in the book), but it's not enough. ***We want to pull out what's important.*** Thus, to create awesome and informative wordclouds like the ones I created above, we want to extract what's special about each of the two lists we're comparing.
The general way of doing that is using a strategy called TF-IDF. We will explore that strategy in execise 4 below. But out-of-the-box TF-IDF gets a bit weird when we only have two groups to compare. So we'll do something different (but related) here.
We want to use the information stored in TF, which is important since it says something about the most frequently occuring words. **But we want to weigh the TF information** using additional information about what is unique about each universe.
Specifically, we want to set things up such that - in the Marvel list, for example - words that are unique to Marvel get a high weight, while words that are unique to DC get a low weight, and words that occur at the same frequency in both lists are neutral. Vice versa for the DC list.
The way I chose to set up the weights here is simple. I use term ratios (TR), which I just made up, so don't look it up on the internet. What we care about is words that are used very differently in the two universes, so we just use their ratios.
Let me start with an example. The word `wolverine` occurs 452 in the marvel TF list and 9 times in the DC TF list, thus I set it's Marvel weight to
$$w_\textrm{wolverine}^{(m)} = \frac{452}{9 + c} = 13.29.$$
Similarly, its DC weight is
$$w_\textrm{wolverine}^{(d)} = \frac{9}{452 + c} = 0.02.$$
In both cases, I add the constant $c$ to the denominator in case a word occurs zero times. You can play around with the size of $c$ to understand the effect of chosing small/large values of it.
In general for some token $t$ with term frequency $\textrm{TF}^{(u)}_t$, in universe $u$ where $u \in \{m,d\}$, we define the weight as:
$$w_{t}^{(m)} = \frac{\textrm{TF}^{(m)}_t}{ \textrm{TF}^{(d)}_t + c}, \qquad \textrm{and} \qquad w_{t}^{(d)} = \frac{\textrm{TF}^{(d)}_t}{ \textrm{TF}^{(m)}_t + c}. $$
Thus, now we're ready to lists for each univers, where the ranking of token $t$ on list $u$ is given by $\textrm{TF}^{(u)}_t \times w_{t}^{(u)}$. I call this the *TF-TR* lists.
Note that the *TF-TR* lists have the properties that we requested above. The weight associated with a word is large when a word occurs much more frequently in the list we consider, compared to the other list. It's small when a word is rare in our list and frequent in the other. And it's approximately equal to one, when the word is equally frequent in both lists. (The downside is that it only works when you're finding important terms while two lists).
> *Action item*:
> * Create your TF-TR list for each universe.
> * Check out top 10 for each universe. Does it make sense?
The takehome here is that we get a good ranking by combining the term frequency with some weighting scheme. But, as we will see below, the TR weight that I created is not the only possible weight. There are many other options.
-----------
PS. Above in creating the ratios, I'm assuming that the universes have a comparable number of words in them. That's pretty much true. And while the approximation is OK for our purposes, it's not true in general. If you'd like an extra challenge, you can figure out how to account for differences in the size of each universe.
**Step three: Install the software**
First you must set up your system. The most difficult thing about creating the wordcloud is installing the `WordCloud` module. It's available on GitHub, check out the page [**here**](https://github.com/amueller/word_cloud).
If you're lucky, you can simply install using conda (and all dependencies, etc will be automatically fixed):
conda install -c conda-forge wordcloud
If you can't get that to work you can refer here https://anaconda.org/conda-forge/wordcloud.
Also, maybe the comments below are helpful:
* The module depends on the Python library PIL. Use `conda` to install that before you do anything else.
* On my system, the module needed the `gcc` compiler installed. If you're not already a programmer, you may have to install that. On Mac you get it by installing the [_command line tools_](http://osxdaily.com/2014/02/12/install-command-line-tools-mac-os-x/). On linux, it's probably already installed. And on Windows I'm not sure, but we'll figure it out during the exercises.
* Once that's all set up, you can use `pip` to install the `WordCloud` library, as [detailed on the GitHub page](https://github.com/amueller/word_cloud). But don't forget to use Anaconda's `pip` just as when you installed the communities library a few weeks ago.
* There are examples of how to use the module online, see [here](http://sebastianraschka.com/Articles/2014_twitter_wordcloud.html) and [here](https://bioinfoexpert.com/2015/05/26/generating-word-clouds-in-python/). If you're interested, you can read about how the package was put together [here](http://peekaboo-vision.blogspot.dk/2012/11/a-wordcloud-in-python.html).
**Step four: Draw the wordclouds**
> *Action items*
> * Get your lists ready for the word-cloud software
> - The package needs a single string to work on. The way that I converted my lists to a string was to simply combine all words together in one long string (separated by spaces), repeating each word according to its score (rounded up to the nearest integer value).
> - The `wordcloud` package looks for collocations in real texts, which is a problem when you make the list as above. The recommended fix is to simply set `collocations = False` as an option when you run the package.
> * Now, create a word-cloud for each universe. Feel free to make it as fancy or non-fancy as you like. Comment on the results. (If you'd like, you can remove stopwords/wiki-syntax - I did a bit of that for my own wordclouds.)
>
# Community detection
Now that we have worked hard on text analysis, it is time to go back to our network!
Before we begin, let's watch a great lecture to learn about communities. If you want all the details, I recommend you take a look at _Chapter 9_ in the _Network Science_ book ... but the video lecture below should be enough to get you started.
**Note**: For this and the next exercise, work on the _undirected_ version of the network.
> **_Video Lecture_**: Communities in networks.
You can watch the 2015 video [here](https://youtu.be/06GL_KGHdbE/).
| 0.673621 | 0.957397 |
```
%load_ext autoreload
%autoreload 2
%matplotlib inline
from torch.utils.data import DataLoader
#export
from exp.nb_03 import *
x_train, y_train, x_valid, y_valid = get_data()
train_ds, valid_ds = Dataset(x_train, y_train), Dataset(x_valid, y_valid)
nh = 50
bs = 64
c = y_train.max().item() + 1
loss_func = F.cross_entropy
#export
class DataBunch():
def __init__(self, train_dl, valid_dl, c=None):
self.train_dl = train_dl
self.valid_dl = valid_dl
self.c = c
@property
def train_ds(self):
return self.train_dl.dataset
@property
def valid_ds(self):
return self.valid_dl.dataset
data = DataBunch(*get_dls(train_ds, valid_ds, bs), c)
#export
def get_model(data, lr=0.5, nh=50):
m = data.train_ds.x.shape[1]
model = nn.Sequential(nn.Linear(m, nh), nn.ReLU(), nn.Linear(nh, data.c))
return model, optim.SGD(model.parameters(), lr=lr)
class Learner():
def __init__(self, model, opt, loss_func, data):
self.model = model
self.opt = opt
self.loss_func = loss_func
self.data = data
learn = Learner(*get_model(data), loss_func, data)
def fit(epochs, learn):
for epoch in range(epochs):
learn.model.train()
for xb, yb in learn.data.train_dl:
loss = learn.loss_func(learn.model(xb), yb)
loss.backward()
learn.opt.step()
learn.opt.zero_grad()
learn.model.eval()
with torch.no_grad():
tot_loss, tot_acc = 0., 0.
for xb, yb in learn.data.valid_dl:
pred = learn.model(xb)
tot_loss += learn.loss_func(pred, yb)
tot_acc += accuracy(pred, yb)
nv = len(learn.data.valid_dl)
final_loss, final_acc = tot_loss / nv, tot_acc / nv
print(f'Epoch: {epoch} Loss: {final_loss} Accuracy: {final_acc}')
return final_loss, final_acc
loss,acc = fit(1, learn)
```
## CallbackHandler
This was our training loop (without validation) from the previous notebook, with the inner loop contents factored out:
```python
def one_batch(xb,yb):
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
def fit():
for epoch in range(epochs):
for b in train_dl: one_batch(*b)
```
Add callbacks so we can remove complexity from loop, and make it flexible:
```
def one_batch(xb, yb, cbh):
if not cbh.begin_batch(xb, yb):
return
loss = cbh.learn.loss_func(cbh.learn.model(xb), yb)
if not cbh.after_loss(loss):
return
loss.backward()
if cbh.after_backward():
cbh.learn.opt.step()
if cbh.after_step():
cbh.learn.opt.zero_grad()
def all_batches(dl, cbh):
for xb, yb in dl:
one_batch(xb, yb, cbh)
if cbh.do_stop():
return
def fit(epochs, learn, cbh):
if not cbh.begin_fit(learn):
return
for epoch in range(epochs):
if not cbh.begin_epoch(epoch):
continue
all_batches(learn.data.train_dl, cbh)
if cbh.begin_validate():
with torch.no_grad():
all_batches(learn.data.valid_dl, cbh)
if cbh.do_stop() or not cbh.after_epoch():
break
cbh.after_fit()
class Callback():
def begin_fit(self, learn):
self.learn = learn
return True
def after_fit(self):
return True
def begin_epoch(self, epoch):
self.epoch = epoch
return True
def begin_validate(self):
return True
def after_epoch(self):
return True
def begin_batch(self, xb, yb):
self.xb = xb
self.yb = yb
return True
def after_loss(self, loss):
self.loss = loss
return True
def after_backward(self):
return True
def after_step(self):
return True
class CallbackHandler():
def __init__(self, cbs=None):
self.cbs = cbs if cbs else []
def begin_fit(self, learn):
self.learn = learn
self.in_train = True
learn.stop = False
res = True
for cb in self.cbs:
res = res and cb.begin_fit(learn)
return res
def after_fit(self):
res = not self.in_train
for cb in self.cbs:
res = res and cb.after_fit()
return res
def begin_epoch(self, epoch):
self.learn.model.train()
self.in_train = True
res = True
for cb in self.cbs:
res = res and cb.begin_epoch(epoch)
return res
def begin_validate(self):
self.learn.model.eval()
self.in_train = False
res = True
for cb in self.cbs:
res = res and cb.begin_validate()
return res
def after_epoch(self):
res = True
for cb in self.cbs:
res = res and cb.after_epoch()
return res
def begin_batch(self, xb, yb):
res = True
for cb in self.cbs:
res = res and cb.begin_batch(xb, yb)
return res
def after_loss(self, loss):
res = self.in_train
for cb in self.cbs:
res = res and cb.after_loss(loss)
return res
def after_backward(self):
res = True
for cb in self.cbs:
res = res and cb.after_backward()
return res
def after_step(self):
res = True
for cb in self.cbs:
res = res and cb.after_step()
return res
def do_stop(self):
try:
return self.learn.stop
finally:
self.learn.stop = False
class TestCallback(Callback):
def __init__(self, limit):
super().__init__()
self.limit = limit
def begin_fit(self, learn):
super().begin_fit(learn)
self.n_iters = 0
return True
def after_step(self):
self.n_iters += 1
print(self.n_iters)
if (self.n_iters >= self.limit):
self.learn.stop = True
return True
fit(1, learn, cbh=CallbackHandler([TestCallback(10)]))
```
This is roughly how fastai does it now (except that the handler can also change and return `xb`, `yb`, and `loss`). But let's see if we can make things simpler and more flexible, so that a single class has access to everything and can change anything at any time. The fact that we're passing `cb` to so many functions is a strong hint they should all be in the same class!
## Runner
```
#export
import re
_camel_re1 = re.compile('(.)([A-Z][a-z]+)')
_camel_re2 = re.compile('([a-z0-9])([A-Z])')
def camel2snake(name):
s1 = re.sub(_camel_re1, r'\1_\2', name)
return re.sub(_camel_re2, r'\1_\2', s1).lower()
class Callback():
_order = 0
def set_runner(self, run):
self.run = run
def __getattr__(self, k):
return getattr(self.run, k)
@property
def name(self):
name = re.sub(r'Callback$', '', self.__class__.__name__)
return camel2snake(name or 'callback')
Callback().name
```
This first callback is reponsible to switch the model back and forth in training or validation mode, as well as maintaining a count of the iterations, or the percentage of iterations ellapsed in the epoch.
```
#export
class TrainEvalCallback(Callback):
def begin_fit(self):
self.run.n_epochs=0.
self.run.n_iter=0
def after_batch(self):
if not self.in_train: return
self.run.n_epochs += 1./self.iters
self.run.n_iter += 1
def begin_epoch(self):
self.run.n_epochs=self.epoch
self.model.train()
self.run.in_train=True
def begin_validate(self):
self.model.eval()
self.run.in_train=False
class TestCallback(Callback):
def after_step(self):
if self.train_eval.n_iters>=10: return True
cbname = 'TrainEvalCallback'
camel2snake(cbname)
TrainEvalCallback().name
#export
from typing import *
def listify(o):
if o is None:
return []
if isinstance(o, list):
return o
if isinstance(o, str):
return [o]
if isinstance(o, Iterable):
return list(o)
return [o]
#export
class Runner():
def __init__(self, cbs=None, cb_funcs=None):
cbs = listify(cbs)
for cbf in listify(cb_funcs):
cb = cbf()
setattr(self, cb.name, cb)
cbs.append(cb)
self.stop,self.cbs = False,[TrainEvalCallback()]+cbs
@property
def opt(self): return self.learn.opt
@property
def model(self): return self.learn.model
@property
def loss_func(self): return self.learn.loss_func
@property
def data(self): return self.learn.data
def one_batch(self, xb, yb):
self.xb,self.yb = xb,yb
if self('begin_batch'): return
self.pred = self.model(self.xb)
if self('after_pred'): return
self.loss = self.loss_func(self.pred, self.yb)
if self('after_loss') or not self.in_train: return
self.loss.backward()
if self('after_backward'): return
self.opt.step()
if self('after_step'): return
self.opt.zero_grad()
def all_batches(self, dl):
self.iters = len(dl)
for xb,yb in dl:
if self.stop: break
self.one_batch(xb, yb)
self('after_batch')
self.stop=False
def fit(self, epochs, learn):
self.epochs,self.learn = epochs,learn
try:
for cb in self.cbs: cb.set_runner(self)
if self('begin_fit'): return
for epoch in range(epochs):
self.epoch = epoch
if not self('begin_epoch'): self.all_batches(self.data.train_dl)
with torch.no_grad():
if not self('begin_validate'): self.all_batches(self.data.valid_dl)
if self('after_epoch'): break
finally:
self('after_fit')
self.learn = None
def __call__(self, cb_name):
for cb in sorted(self.cbs, key=lambda x: x._order):
f = getattr(cb, cb_name, None)
if f and f(): return True
return False
```
Third callback: how to compute metrics.
```
#export
class AvgStats():
def __init__(self, metrics, in_train): self.metrics,self.in_train = listify(metrics),in_train
def reset(self):
self.tot_loss,self.count = 0.,0
self.tot_mets = [0.] * len(self.metrics)
@property
def all_stats(self): return [self.tot_loss.item()] + self.tot_mets
@property
def avg_stats(self): return [o/self.count for o in self.all_stats]
def __repr__(self):
if not self.count: return ""
return f"{'train' if self.in_train else 'valid'}: {self.avg_stats}"
def accumulate(self, run):
bn = run.xb.shape[0]
self.tot_loss += run.loss * bn
self.count += bn
for i,m in enumerate(self.metrics):
self.tot_mets[i] += m(run.pred, run.yb) * bn
class AvgStatsCallback(Callback):
def __init__(self, metrics):
self.train_stats,self.valid_stats = AvgStats(metrics,True),AvgStats(metrics,False)
def begin_epoch(self):
self.train_stats.reset()
self.valid_stats.reset()
def after_loss(self):
stats = self.train_stats if self.in_train else self.valid_stats
with torch.no_grad(): stats.accumulate(self.run)
def after_epoch(self):
print(self.train_stats)
print(self.valid_stats)
learn = Learner(*get_model(data), loss_func, data)
stats = AvgStatsCallback([accuracy])
run = Runner(cbs = stats)
run.fit(2, learn)
loss, acc = stats.valid_stats.avg_stats
assert acc > 0.9
loss, acc
#export
from functools import partial
acc_cbf = partial(AvgStatsCallback, accuracy)
run = Runner(cb_funcs=acc_cbf)
run.fit(1, learn)
```
## Export
```
!python notebook2script.py 04_callbacks.ipynb
```
|
github_jupyter
|
%load_ext autoreload
%autoreload 2
%matplotlib inline
from torch.utils.data import DataLoader
#export
from exp.nb_03 import *
x_train, y_train, x_valid, y_valid = get_data()
train_ds, valid_ds = Dataset(x_train, y_train), Dataset(x_valid, y_valid)
nh = 50
bs = 64
c = y_train.max().item() + 1
loss_func = F.cross_entropy
#export
class DataBunch():
def __init__(self, train_dl, valid_dl, c=None):
self.train_dl = train_dl
self.valid_dl = valid_dl
self.c = c
@property
def train_ds(self):
return self.train_dl.dataset
@property
def valid_ds(self):
return self.valid_dl.dataset
data = DataBunch(*get_dls(train_ds, valid_ds, bs), c)
#export
def get_model(data, lr=0.5, nh=50):
m = data.train_ds.x.shape[1]
model = nn.Sequential(nn.Linear(m, nh), nn.ReLU(), nn.Linear(nh, data.c))
return model, optim.SGD(model.parameters(), lr=lr)
class Learner():
def __init__(self, model, opt, loss_func, data):
self.model = model
self.opt = opt
self.loss_func = loss_func
self.data = data
learn = Learner(*get_model(data), loss_func, data)
def fit(epochs, learn):
for epoch in range(epochs):
learn.model.train()
for xb, yb in learn.data.train_dl:
loss = learn.loss_func(learn.model(xb), yb)
loss.backward()
learn.opt.step()
learn.opt.zero_grad()
learn.model.eval()
with torch.no_grad():
tot_loss, tot_acc = 0., 0.
for xb, yb in learn.data.valid_dl:
pred = learn.model(xb)
tot_loss += learn.loss_func(pred, yb)
tot_acc += accuracy(pred, yb)
nv = len(learn.data.valid_dl)
final_loss, final_acc = tot_loss / nv, tot_acc / nv
print(f'Epoch: {epoch} Loss: {final_loss} Accuracy: {final_acc}')
return final_loss, final_acc
loss,acc = fit(1, learn)
def one_batch(xb,yb):
pred = model(xb)
loss = loss_func(pred, yb)
loss.backward()
opt.step()
opt.zero_grad()
def fit():
for epoch in range(epochs):
for b in train_dl: one_batch(*b)
def one_batch(xb, yb, cbh):
if not cbh.begin_batch(xb, yb):
return
loss = cbh.learn.loss_func(cbh.learn.model(xb), yb)
if not cbh.after_loss(loss):
return
loss.backward()
if cbh.after_backward():
cbh.learn.opt.step()
if cbh.after_step():
cbh.learn.opt.zero_grad()
def all_batches(dl, cbh):
for xb, yb in dl:
one_batch(xb, yb, cbh)
if cbh.do_stop():
return
def fit(epochs, learn, cbh):
if not cbh.begin_fit(learn):
return
for epoch in range(epochs):
if not cbh.begin_epoch(epoch):
continue
all_batches(learn.data.train_dl, cbh)
if cbh.begin_validate():
with torch.no_grad():
all_batches(learn.data.valid_dl, cbh)
if cbh.do_stop() or not cbh.after_epoch():
break
cbh.after_fit()
class Callback():
def begin_fit(self, learn):
self.learn = learn
return True
def after_fit(self):
return True
def begin_epoch(self, epoch):
self.epoch = epoch
return True
def begin_validate(self):
return True
def after_epoch(self):
return True
def begin_batch(self, xb, yb):
self.xb = xb
self.yb = yb
return True
def after_loss(self, loss):
self.loss = loss
return True
def after_backward(self):
return True
def after_step(self):
return True
class CallbackHandler():
def __init__(self, cbs=None):
self.cbs = cbs if cbs else []
def begin_fit(self, learn):
self.learn = learn
self.in_train = True
learn.stop = False
res = True
for cb in self.cbs:
res = res and cb.begin_fit(learn)
return res
def after_fit(self):
res = not self.in_train
for cb in self.cbs:
res = res and cb.after_fit()
return res
def begin_epoch(self, epoch):
self.learn.model.train()
self.in_train = True
res = True
for cb in self.cbs:
res = res and cb.begin_epoch(epoch)
return res
def begin_validate(self):
self.learn.model.eval()
self.in_train = False
res = True
for cb in self.cbs:
res = res and cb.begin_validate()
return res
def after_epoch(self):
res = True
for cb in self.cbs:
res = res and cb.after_epoch()
return res
def begin_batch(self, xb, yb):
res = True
for cb in self.cbs:
res = res and cb.begin_batch(xb, yb)
return res
def after_loss(self, loss):
res = self.in_train
for cb in self.cbs:
res = res and cb.after_loss(loss)
return res
def after_backward(self):
res = True
for cb in self.cbs:
res = res and cb.after_backward()
return res
def after_step(self):
res = True
for cb in self.cbs:
res = res and cb.after_step()
return res
def do_stop(self):
try:
return self.learn.stop
finally:
self.learn.stop = False
class TestCallback(Callback):
def __init__(self, limit):
super().__init__()
self.limit = limit
def begin_fit(self, learn):
super().begin_fit(learn)
self.n_iters = 0
return True
def after_step(self):
self.n_iters += 1
print(self.n_iters)
if (self.n_iters >= self.limit):
self.learn.stop = True
return True
fit(1, learn, cbh=CallbackHandler([TestCallback(10)]))
#export
import re
_camel_re1 = re.compile('(.)([A-Z][a-z]+)')
_camel_re2 = re.compile('([a-z0-9])([A-Z])')
def camel2snake(name):
s1 = re.sub(_camel_re1, r'\1_\2', name)
return re.sub(_camel_re2, r'\1_\2', s1).lower()
class Callback():
_order = 0
def set_runner(self, run):
self.run = run
def __getattr__(self, k):
return getattr(self.run, k)
@property
def name(self):
name = re.sub(r'Callback$', '', self.__class__.__name__)
return camel2snake(name or 'callback')
Callback().name
#export
class TrainEvalCallback(Callback):
def begin_fit(self):
self.run.n_epochs=0.
self.run.n_iter=0
def after_batch(self):
if not self.in_train: return
self.run.n_epochs += 1./self.iters
self.run.n_iter += 1
def begin_epoch(self):
self.run.n_epochs=self.epoch
self.model.train()
self.run.in_train=True
def begin_validate(self):
self.model.eval()
self.run.in_train=False
class TestCallback(Callback):
def after_step(self):
if self.train_eval.n_iters>=10: return True
cbname = 'TrainEvalCallback'
camel2snake(cbname)
TrainEvalCallback().name
#export
from typing import *
def listify(o):
if o is None:
return []
if isinstance(o, list):
return o
if isinstance(o, str):
return [o]
if isinstance(o, Iterable):
return list(o)
return [o]
#export
class Runner():
def __init__(self, cbs=None, cb_funcs=None):
cbs = listify(cbs)
for cbf in listify(cb_funcs):
cb = cbf()
setattr(self, cb.name, cb)
cbs.append(cb)
self.stop,self.cbs = False,[TrainEvalCallback()]+cbs
@property
def opt(self): return self.learn.opt
@property
def model(self): return self.learn.model
@property
def loss_func(self): return self.learn.loss_func
@property
def data(self): return self.learn.data
def one_batch(self, xb, yb):
self.xb,self.yb = xb,yb
if self('begin_batch'): return
self.pred = self.model(self.xb)
if self('after_pred'): return
self.loss = self.loss_func(self.pred, self.yb)
if self('after_loss') or not self.in_train: return
self.loss.backward()
if self('after_backward'): return
self.opt.step()
if self('after_step'): return
self.opt.zero_grad()
def all_batches(self, dl):
self.iters = len(dl)
for xb,yb in dl:
if self.stop: break
self.one_batch(xb, yb)
self('after_batch')
self.stop=False
def fit(self, epochs, learn):
self.epochs,self.learn = epochs,learn
try:
for cb in self.cbs: cb.set_runner(self)
if self('begin_fit'): return
for epoch in range(epochs):
self.epoch = epoch
if not self('begin_epoch'): self.all_batches(self.data.train_dl)
with torch.no_grad():
if not self('begin_validate'): self.all_batches(self.data.valid_dl)
if self('after_epoch'): break
finally:
self('after_fit')
self.learn = None
def __call__(self, cb_name):
for cb in sorted(self.cbs, key=lambda x: x._order):
f = getattr(cb, cb_name, None)
if f and f(): return True
return False
#export
class AvgStats():
def __init__(self, metrics, in_train): self.metrics,self.in_train = listify(metrics),in_train
def reset(self):
self.tot_loss,self.count = 0.,0
self.tot_mets = [0.] * len(self.metrics)
@property
def all_stats(self): return [self.tot_loss.item()] + self.tot_mets
@property
def avg_stats(self): return [o/self.count for o in self.all_stats]
def __repr__(self):
if not self.count: return ""
return f"{'train' if self.in_train else 'valid'}: {self.avg_stats}"
def accumulate(self, run):
bn = run.xb.shape[0]
self.tot_loss += run.loss * bn
self.count += bn
for i,m in enumerate(self.metrics):
self.tot_mets[i] += m(run.pred, run.yb) * bn
class AvgStatsCallback(Callback):
def __init__(self, metrics):
self.train_stats,self.valid_stats = AvgStats(metrics,True),AvgStats(metrics,False)
def begin_epoch(self):
self.train_stats.reset()
self.valid_stats.reset()
def after_loss(self):
stats = self.train_stats if self.in_train else self.valid_stats
with torch.no_grad(): stats.accumulate(self.run)
def after_epoch(self):
print(self.train_stats)
print(self.valid_stats)
learn = Learner(*get_model(data), loss_func, data)
stats = AvgStatsCallback([accuracy])
run = Runner(cbs = stats)
run.fit(2, learn)
loss, acc = stats.valid_stats.avg_stats
assert acc > 0.9
loss, acc
#export
from functools import partial
acc_cbf = partial(AvgStatsCallback, accuracy)
run = Runner(cb_funcs=acc_cbf)
run.fit(1, learn)
!python notebook2script.py 04_callbacks.ipynb
| 0.785925 | 0.793186 |
<img src="images/usm.jpg" width="480" height="240" align="left"/>
# MAT281 - Laboratorio N°01
## Objetivos de la clase
* Reforzar los conceptos básicos de python.
## Contenidos
* [Problema 01](#p1)
* [Problema 02](#p2)
* [Problema 03](#p3)
* [Problema 04](#p4)
<a id='p1'></a>
## Problema 01
### a) Calcular el número $\pi$
En los siglos XVII y XVIII, James Gregory y Gottfried Leibniz descubrieron una serie infinita que sirve para calcular $\pi$:
$$\displaystyle \pi = 4 \sum_{k=1}^{\infty}\dfrac{(-1)^{k+1}}{2k-1} = 4(1-\dfrac{1}{3}+\dfrac{1}{5}-\dfrac{1}{7} + ...) $$
Desarolle un programa para estimar el valor de $\pi$ ocupando el método de Leibniz, donde la entrada del programa debe ser un número entero $n$ que indique cuántos términos de la suma se utilizará.
* **Ejemplo**: *calcular_pi(3)* = 3.466666666666667, *calcular_pi(1000)* = 3.140592653839794
### Definir Función
```
def calcular_pi(n:int)->float:
"""
calcular_pi(n)
Aproximacion del valor de pi mediante el método de Leibniz
Parameters
----------
n : int
Numero de terminos.
Returns
-------
output : float
Valor aproximado de pi.
Examples
--------
>>> calcular_pi(3)
3.466666666666667
>>> calcular_pi(1000)
3.140592653839794
"""
pi = 0 # valor incial
for k in range(1,n+1):
numerador = (-1)**(k+1) # numerador de la iteracion i
denominador = 2*k-1 # denominador de la iteracion i
pi+=numerador/denominador # suma hasta el i-esimo termino
return 4*pi
# Acceso a la documentación
help(calcular_pi)
```
### Verificar ejemplos
```
# ejemplo 01
assert calcular_pi(3) == 3.466666666666667, "ejemplo 01 incorrecto"
# ejemplo 02
assert calcular_pi(1000) == 3.140592653839794, "ejemplo 02 incorrecto"
```
**Observación**:
* Note que si corre la línea de comando `calcular_pi(3.0)` le mandará un error ... ¿ por qué ?
* En los laboratorio, no se pide ser tan meticuloso con la documentacion.
* Lo primero es definir el código, correr los ejemplos y luego documentar correctamente.
### b) Calcular el número $e$
Euler realizó varios aportes en relación a $e$, pero no fue hasta 1748 cuando publicó su **Introductio in analysin infinitorum** que dio un tratamiento definitivo a las ideas sobre $e$. Allí mostró que:
En los siglos XVII y XVIII, James Gregory y Gottfried Leibniz descubrieron una serie infinita que sirve para calcular π:
$$\displaystyle e = \sum_{k=0}^{\infty}\dfrac{1}{k!} = 1+\dfrac{1}{2!}+\dfrac{1}{3!}+\dfrac{1}{4!} + ... $$
Desarolle un programa para estimar el valor de $e$ ocupando el método de Euler, donde la entrada del programa debe ser un número entero $n$ que indique cuántos términos de la suma se utilizará.
* **Ejemplo**: *calcular_e(3)* =2.5, *calcular_e(1000)* = 2.7182818284590455
### Definir función
### Verificar ejemplos
```
# ejemplo 01
assert calcular_e(3) == 2.5, "ejemplo 01 incorrecto"
# ejemplo 02
assert calcular_e(1000) == 2.7182818284590455, "ejemplo 02 incorrecto"
```
<a id='p2'></a>
## Problema 02
Sea $\sigma(n)$ definido como la suma de los divisores propios de $n$ (números menores que n que se dividen en $n$).
Los [números amigos](https://en.wikipedia.org/wiki/Amicable_numbers) son enteros positivos $n_1$ y $n_2$ tales que la suma de los divisores propios de uno es igual al otro número y viceversa, es decir, $\sigma(n_1)=\sigma(n_2)$ y $\sigma(n_2)=\sigma(n_1)$.
Por ejemplo, los números 220 y 284 son números amigos.
* los divisores propios de 220 son 1, 2, 4, 5, 10, 11, 20, 22, 44, 55 y 110; por lo tanto $\sigma(220) = 284$.
* los divisores propios de 284 son 1, 2, 4, 71 y 142; entonces $\sigma(284) = 220$.
Implemente una función llamada `amigos` cuyo input sean dos números naturales $n_1$ y $n_2$, cuyo output sea verifique si los números son amigos o no.
* **Ejemplo**: *amigos(220,284)* = True, *amigos(6,5)* = False
### Definir Función
### Verificar ejemplos
```
# ejemplo 01
assert amigos(220,284) == True, "ejemplo 01 incorrecto"
# ejemplo 02
assert amigos(6,5) == False, "ejemplo 02 incorrecto"
```
<a id='p3'></a>
## Problema 03
La [conjetura de Collatz](https://en.wikipedia.org/wiki/Collatz_conjecture), conocida también como conjetura $3n+1$ o conjetura de Ulam (entre otros nombres), fue enunciada por el matemático Lothar Collatz en 1937, y a la fecha no se ha resuelto.
Sea la siguiente operación, aplicable a cualquier número entero positivo:
* Si el número es par, se divide entre 2.
* Si el número es impar, se multiplica por 3 y se suma 1.
La conjetura dice que siempre alcanzaremos el 1 (y por tanto el ciclo 4, 2, 1) para cualquier número con el que comencemos.
Implemente una función llamada `collatz` cuyo input sea un número natural positivo $N$ y como output devulva la secuencia de números hasta llegar a 1.
* **Ejemplo**: *collatz(9)* = [9, 28, 14, 7, 22, 11, 34, 17, 52, 26, 13, 40, 20, 10, 5, 16, 8, 4, 2, 1]
### Definir Función
### Verificar ejemplos
```
# ejemplo 01
assert collatz(9) == [9, 28, 14, 7, 22, 11, 34, 17, 52, 26, 13, 40, 20, 10, 5, 16, 8, 4, 2, 1], "ejemplo 01 incorrecto"
```
<a id='p4'></a>
## Problema 04
La [conjetura de Goldbach](https://en.wikipedia.org/wiki/Goldbach%27s_conjecture) es uno de los problemas abiertos más antiguos en matemáticas. Concretamente, G.H. Hardy, en 1921, en su famoso discurso pronunciado en la Sociedad Matemática de Copenhague, comentó que probablemente la conjetura de Goldbach no es solo uno de los problemas no resueltos más difíciles de la teoría de números, sino de todas las matemáticas. Su enunciado es el siguiente:
$$\textrm{Todo número par mayor que 2 puede escribirse como suma de dos números primos - Christian Goldbach (1742)}$$
Implemente una función llamada `goldbach` cuyo input sea un número natural positivo $N$ y como output devuelva la suma de dos primos ($N1$ y $N2$) tal que: $N1+N2=N$.
* **Ejemplo**: goldbash(4) = (2,2), goldbash(6) = (3,3) , goldbash(8) = (3,5)
### Definir función
### Verificar ejemplos
```
# ejemplo 01
assert goldbash(4) == (2,2), "ejemplo 01 incorrecto"
# ejemplo 02
assert goldbash(6) == (3,3), "ejemplo 02 incorrecto"
# ejemplo 03
assert goldbash(8) == (3,5), "ejemplo 03 incorrecto"
```
|
github_jupyter
|
def calcular_pi(n:int)->float:
"""
calcular_pi(n)
Aproximacion del valor de pi mediante el método de Leibniz
Parameters
----------
n : int
Numero de terminos.
Returns
-------
output : float
Valor aproximado de pi.
Examples
--------
>>> calcular_pi(3)
3.466666666666667
>>> calcular_pi(1000)
3.140592653839794
"""
pi = 0 # valor incial
for k in range(1,n+1):
numerador = (-1)**(k+1) # numerador de la iteracion i
denominador = 2*k-1 # denominador de la iteracion i
pi+=numerador/denominador # suma hasta el i-esimo termino
return 4*pi
# Acceso a la documentación
help(calcular_pi)
# ejemplo 01
assert calcular_pi(3) == 3.466666666666667, "ejemplo 01 incorrecto"
# ejemplo 02
assert calcular_pi(1000) == 3.140592653839794, "ejemplo 02 incorrecto"
# ejemplo 01
assert calcular_e(3) == 2.5, "ejemplo 01 incorrecto"
# ejemplo 02
assert calcular_e(1000) == 2.7182818284590455, "ejemplo 02 incorrecto"
# ejemplo 01
assert amigos(220,284) == True, "ejemplo 01 incorrecto"
# ejemplo 02
assert amigos(6,5) == False, "ejemplo 02 incorrecto"
# ejemplo 01
assert collatz(9) == [9, 28, 14, 7, 22, 11, 34, 17, 52, 26, 13, 40, 20, 10, 5, 16, 8, 4, 2, 1], "ejemplo 01 incorrecto"
# ejemplo 01
assert goldbash(4) == (2,2), "ejemplo 01 incorrecto"
# ejemplo 02
assert goldbash(6) == (3,3), "ejemplo 02 incorrecto"
# ejemplo 03
assert goldbash(8) == (3,5), "ejemplo 03 incorrecto"
| 0.68342 | 0.980543 |
## Covert notebook to raw python
To run on ML-Engine, we strip all cells tagged "display" to remove the dependency on matplotlib.
The remote server/cluster does not have a display anyway, just logs.
(To view/edit tags on notebook cells: View>Cell Toolbar>Tags)
```
# Convert notebook to raw python format and remove all cells tagged "display"
NOTEBOOK='02_RNN_generator_temperatures_solution.ipynb'
jupyter nbconvert tutorial/${NOTEBOOK} \
--to python --TagRemovePreprocessor.remove_cell_tags={\"display\"} \
--output task.py
```
*** Windows users, please copy-paste this command into your command prompt: ***
```bash
jupyter nbconvert "tutorial/02_RNN_generator_temperatures_solution.ipynb" --to python --output task.py --TagRemovePreprocessor.remove_cell_tags={\"display\"}
```
## To run on ML-Engine
If you are using your own GCP account you have to first:
1. Create a Google Cloud Platform project
1. Enable billing
1. Create a Google Cloud Storage bucket (put in in region us-central1)
1. Enable the necessary APIs and request the necessary quotas
If you are using a lab account through Qwiklabs (Available soon):
1. Please [register your email address here](https://docs.google.com/forms/d/e/1FAIpQLScDruivAynhrL9XMyEozLZRRCuMLg-X0BFC3ct0VqHs_sW1cg/viewform?usp=sf_link) so that we can white-list you on Qwiklabs.
1. Go to Qwiklabs for the last part of the workshop. Qwiklabs will provision a free lab account on Google Cloud Platform with a GPU quota for you:
[https://events.qwiklabs.com/classrooms/<available soon>](https://events.qwiklabs.com/classrooms/XXXX)
1. Create a Google Cloud Storage bucket (put in in region us-central1)
And fill in the info in the variables below.
You can try running on a GPU by using --scale-tier=BASIC_GPU or a CPU using --scale-tier=BASIC
```
BUCKET='ml1-demo-martin'
PROJECT='cloudml-demo-martin'
REGION='us-central1'
JOBNAME=sines_$(date -u +%y%m%d_%H%M%S)
OUTDIR="gs://${BUCKET}/sinejobs/${JOBNAME}"
gcloud ml-engine jobs submit training $JOBNAME \
--region=$REGION \
--module-name=tutorial.task \
--package-path=tutorial \
--job-dir=$OUTDIR \
--scale-tier=BASIC_GPU \
--runtime-version=1.6 \
-- \
--data-dir="gs://good-temperatures-public"
```
*** Windows users, please copy-paste this command into your command prompt (replace <PROJECT> and <BUCKET> with your own values and adjust the job name job001 if needed): ***
```bash
gcloud ml-engine jobs submit training job001 --region="us-central1" --project=<PROJECT> --module-name="tutorial.task" --package-path="tutorial" --job-dir="gs://<BUCKET>/sinejobs/job001" --scale-tier=BASIC_GPU --runtime-version=1.6 -- --data-dir="gs://good-temperatures-public"
```
## To test-run locally as if the code was running on ML-Engine
Warning: this will use the "python" command to run (usually mapped to python 2 on mac, same as ML-Engine)
```
gcloud ml-engine local train \
--module-name=tutorial.task \
--package-path=tutorial \
--job-dir="checkpoints" \
-- \
--data-dir="gs://good-temperatures-public"
```
Copyright 2018 Google LLC
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
[http://www.apache.org/licenses/LICENSE-2.0](http://www.apache.org/licenses/LICENSE-2.0)
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|
github_jupyter
|
# Convert notebook to raw python format and remove all cells tagged "display"
NOTEBOOK='02_RNN_generator_temperatures_solution.ipynb'
jupyter nbconvert tutorial/${NOTEBOOK} \
--to python --TagRemovePreprocessor.remove_cell_tags={\"display\"} \
--output task.py
jupyter nbconvert "tutorial/02_RNN_generator_temperatures_solution.ipynb" --to python --output task.py --TagRemovePreprocessor.remove_cell_tags={\"display\"}
BUCKET='ml1-demo-martin'
PROJECT='cloudml-demo-martin'
REGION='us-central1'
JOBNAME=sines_$(date -u +%y%m%d_%H%M%S)
OUTDIR="gs://${BUCKET}/sinejobs/${JOBNAME}"
gcloud ml-engine jobs submit training $JOBNAME \
--region=$REGION \
--module-name=tutorial.task \
--package-path=tutorial \
--job-dir=$OUTDIR \
--scale-tier=BASIC_GPU \
--runtime-version=1.6 \
-- \
--data-dir="gs://good-temperatures-public"
gcloud ml-engine jobs submit training job001 --region="us-central1" --project=<PROJECT> --module-name="tutorial.task" --package-path="tutorial" --job-dir="gs://<BUCKET>/sinejobs/job001" --scale-tier=BASIC_GPU --runtime-version=1.6 -- --data-dir="gs://good-temperatures-public"
gcloud ml-engine local train \
--module-name=tutorial.task \
--package-path=tutorial \
--job-dir="checkpoints" \
-- \
--data-dir="gs://good-temperatures-public"
| 0.420481 | 0.894375 |
```
library(rjson)
library(data.table)
library(dplyr)
```
### Data repo: **STATSBOMB** data
```
repo <- '/Users/christian/Desktop/University/Birkbeck MSc Applied Statistics/Football/Data/Statsbomb/open-data-master/data'
```
### Loading in the **competitions** data
```
# reading in the data in JSON
competitions <- fromJSON(file = file.path(repo, 'competitions.json', fsep=.Platform$file.sep))
# loading data into data frame
competitions.df <- data.frame(do.call(rbind, competitions), stringsAsFactors = FALSE)
head(competitions.df)
```
### Loading in the **match** data
* `match.files`: First you need to span the tree to get all end files and the paths to those files
* Here the `recursive=TRUE` argument works wonders
```
match.files <- list.files(path= file.path(repo, 'matches', fsep='/'), recursive = TRUE, full.names = TRUE)
matches.list <- list()
# iterating through the files
for (i in 1:length(match.files)){
# creating a temporary variable that holds the raw JSON per file
# this is a list of dictionaries
match.temp <- fromJSON(file=match.files[i])
# using lapply (list apply) whereby for each json object, we unlist it (flattening it), and then transpose it
# we do that for all elements, and must use the function x as lapply expects a function
matches <- lapply(match.temp, function(x) data.frame(t(unlist(x)), stringsAsFactors = FALSE))
# we now stitch together all single row transposes into a dataframe of matches per JSON file
matches.df <- rbindlist(matches, fill=TRUE)
# and then we stick this in a list, so it'll be a list of data frames
matches.list[[i]] <- matches.df
}
# and now we stitch all of those data frames together in a master dataframe
all.matches.df <- data.frame(rbindlist(matches.list, fill=TRUE))
head(all.matches.df)
all.matches.df
```
### Cleaning up **matches**
> Want all columns that don't contain an N/A
> `which` provides index value of a list back when condition is true
> here we're counting cases **per row** where there's an N/A
> only interested in cases where columns have N/A zero times
```
columns.to.keep <- names(which(unlist(lapply(all.matches.df, function(x) length(which(is.na(x))))) == 0))
all.matches.clean <- all.matches.df[,columns.to.keep]
```
### And now transforming some columns from factors to numbers
> can call `str(all.matches.clean)` to learn the data type of the variables
```
str(all.matches.clean)
all.matches.clean$match_week <- as.numeric(all.matches.clean$match_week)
all.matches.clean$home_score <- as.numeric(all.matches.clean$home_score)
all.matches.clean$away_score <- as.numeric(all.matches.clean$away_score)
head(all.matches.clean)
```
### Loading in the **events** data
> There are 855 event files
```
event.files <- list.files(path= file.path(repo, 'events', fsep='/'), full.names = TRUE, recursive = TRUE)
event.list <- list()
l <- length(event.files)
# looping through all of the event files...
## takes a while
for (i in 1:l){
event.temp <- fromJSON(file = event.files[i])
team.id.clist <- c()
starting.x11.index <- which(unlist(lapply(event.temp, function(x) x$type$name)) == 'Starting XI')
starting.x11.list <- list()
# looping through the two indices for the two teams, populating the above list with two dataframes, one per team
for (s in starting.x11.index){
starting.x11.team1 <- data.frame(matrix(t(unlist(event.temp[[s]]$tactics$lineup)), ncol=5, byrow=TRUE), stringsAsFactors = FALSE)
# unlisting the event.temp element, getting the names of the tactics$lineup
colnames(starting.x11.team1) <- names(unlist(event.temp[[s]]$tactics$lineup))[1:5]
# adding three extra columns, one for formation, team_id, team_name
starting.x11.team1$formation <- event.temp[[s]]$tactics$formation
starting.x11.team1$team_id <- event.temp[[s]]$team$id
starting.x11.team1$team_name <- event.temp[[s]]$team$name
# update our clist of teamId's
team.id.clist <- c(team.id.clist, event.temp[[s]]$team$id)
# appending the starting.x11 data to the starting x11 list
## this will produce a list of two dataframes, one per team
starting.x11.list[[s]] <- starting.x11.team1
}
# now looking at passes
pass.index <- which(unlist(lapply(event.temp, function(x) x$type$name)) == 'Pass')
# and now filtering to get the pass indices for team 1, using the team 1 identifier from team.id.clist[1]
pass.team1 <- pass.index[which(unlist(lapply(pass.index, function(x) event.temp[[x]]$team$id)) == team.id.clist[1])]
pass.team1.df <- data.frame(matrix(NA, nrow=1, ncol=13))
colnames(pass.team1.df) <- c('Possession','Passer','X.Pass','Y.Pass','Pass.Type','Receiver','X.Receive','Y.Receive',
'Pass.Length','Pass.Angle','Body.Part','Pass.Pressure','Pass.Outcome')
for (p in 1:length(pass.team1)){
pass.temp <- event.temp[[pass.team1[p]]]
possession <- pass.temp$possession
passer <- pass.temp$player$id
pass.location <- pass.temp$location
pass.type <- pass.temp$pass$height$name
receiver <- pass.temp$pass$recipient$id
receive.location <- pass.temp$pass$end_location
pass.length <- pass.temp$pass$length
pass.angle <- pass.temp$pass$angle
body.part <- pass.temp$pass$body_part$name
pass.pressure <- pass.temp$under_pressure
pass.outcome <- pass.temp$pass$outcome$name
row.toadd <- c(possession, passer, pass.location, pass.type, receiver, receive.location
,pass.length, pass.angle, body.part, pass.pressure, pass.outcome)
pass.team1.df <- rbind(pass.team1.df, row.toadd)
}
# getting rid of the first empty row (this is minging)
pass.team1.df <- pass.team1.df[-1,]
pass.team1.df[,c(1:4,6:10)] <- lapply(pass.team1.df[,c(1:4,6:10)], as.numeric)
# this is basically a row number (partition by...)
# providing
pass.team1.df <- pass.team1.df %>% group_by(Possession) %>% mutate(seq = row_number())
pass.team1.df$team_id <- team.id.clist[1]
## AND NOW TO DO THE EXACT SAME THING FOR TEAM 2!
# and now filtering to get the pass indices for team 2, using the team 2 identifier from team.id.clist[2]
pass.team2 <- pass.index[which(unlist(lapply(pass.index, function(x) event.temp[[x]]$team$id)) == team.id.clist[2])]
pass.team2.df <- data.frame(matrix(NA, nrow=1, ncol=13))
colnames(pass.team2.df) <- c('Possession','Passer','X.Pass','Y.Pass','Pass.Type','Receiver','X.Receive','Y.Receive',
'Pass.Length','Pass.Angle','Body.Part','Pass.Pressure','Pass.Outcome')
for (p in 1:length(pass.team2)){
pass.temp <- event.temp[[pass.team2[p]]]
possession <- pass.temp$possession
passer <- pass.temp$player$id
pass.location <- pass.temp$location
pass.type <- pass.temp$pass$height$name
receiver <- pass.temp$pass$recipient$id
receive.location <- pass.temp$pass$end_location
pass.length <- pass.temp$pass$length
pass.angle <- pass.temp$pass$angle
body.part <- pass.temp$pass$body_part$name
pass.pressure <- pass.temp$under_pressure
pass.outcome <- pass.temp$pass$outcome$name
row.toadd <- c(possession, passer, pass.location, pass.type, receiver, receive.location
,pass.length, pass.angle, body.part, pass.pressure, pass.outcome)
pass.team2.df <- rbind(pass.team2.df, row.toadd)
}
# getting rid of the first empty row (this is minging)
pass.team2.df <- pass.team2.df[-1,]
pass.team2.df[,c(1:4,6:10)] <- lapply(pass.team2.df[,c(1:4,6:10)], as.numeric)
# this is basically a row number (partition by...)
# providing
pass.team2.df <- pass.team2.df %>% group_by(Possession) %>% mutate(seq = row_number())
pass.team2.df$team_id <- team.id.clist[2]
## AND NOW PUTTING IT ALL TOGETHER
pass.list <- list(pass.team1.df, pass.team2.df)
match.id <- strsplit(basename(event.files[i]), '[.]')[[1]][1]
event.list[[match.id]] <- list(starting.x11.list, pass.list)
}
event.list[[1]]
```
|
github_jupyter
|
library(rjson)
library(data.table)
library(dplyr)
repo <- '/Users/christian/Desktop/University/Birkbeck MSc Applied Statistics/Football/Data/Statsbomb/open-data-master/data'
# reading in the data in JSON
competitions <- fromJSON(file = file.path(repo, 'competitions.json', fsep=.Platform$file.sep))
# loading data into data frame
competitions.df <- data.frame(do.call(rbind, competitions), stringsAsFactors = FALSE)
head(competitions.df)
match.files <- list.files(path= file.path(repo, 'matches', fsep='/'), recursive = TRUE, full.names = TRUE)
matches.list <- list()
# iterating through the files
for (i in 1:length(match.files)){
# creating a temporary variable that holds the raw JSON per file
# this is a list of dictionaries
match.temp <- fromJSON(file=match.files[i])
# using lapply (list apply) whereby for each json object, we unlist it (flattening it), and then transpose it
# we do that for all elements, and must use the function x as lapply expects a function
matches <- lapply(match.temp, function(x) data.frame(t(unlist(x)), stringsAsFactors = FALSE))
# we now stitch together all single row transposes into a dataframe of matches per JSON file
matches.df <- rbindlist(matches, fill=TRUE)
# and then we stick this in a list, so it'll be a list of data frames
matches.list[[i]] <- matches.df
}
# and now we stitch all of those data frames together in a master dataframe
all.matches.df <- data.frame(rbindlist(matches.list, fill=TRUE))
head(all.matches.df)
all.matches.df
columns.to.keep <- names(which(unlist(lapply(all.matches.df, function(x) length(which(is.na(x))))) == 0))
all.matches.clean <- all.matches.df[,columns.to.keep]
str(all.matches.clean)
all.matches.clean$match_week <- as.numeric(all.matches.clean$match_week)
all.matches.clean$home_score <- as.numeric(all.matches.clean$home_score)
all.matches.clean$away_score <- as.numeric(all.matches.clean$away_score)
head(all.matches.clean)
event.files <- list.files(path= file.path(repo, 'events', fsep='/'), full.names = TRUE, recursive = TRUE)
event.list <- list()
l <- length(event.files)
# looping through all of the event files...
## takes a while
for (i in 1:l){
event.temp <- fromJSON(file = event.files[i])
team.id.clist <- c()
starting.x11.index <- which(unlist(lapply(event.temp, function(x) x$type$name)) == 'Starting XI')
starting.x11.list <- list()
# looping through the two indices for the two teams, populating the above list with two dataframes, one per team
for (s in starting.x11.index){
starting.x11.team1 <- data.frame(matrix(t(unlist(event.temp[[s]]$tactics$lineup)), ncol=5, byrow=TRUE), stringsAsFactors = FALSE)
# unlisting the event.temp element, getting the names of the tactics$lineup
colnames(starting.x11.team1) <- names(unlist(event.temp[[s]]$tactics$lineup))[1:5]
# adding three extra columns, one for formation, team_id, team_name
starting.x11.team1$formation <- event.temp[[s]]$tactics$formation
starting.x11.team1$team_id <- event.temp[[s]]$team$id
starting.x11.team1$team_name <- event.temp[[s]]$team$name
# update our clist of teamId's
team.id.clist <- c(team.id.clist, event.temp[[s]]$team$id)
# appending the starting.x11 data to the starting x11 list
## this will produce a list of two dataframes, one per team
starting.x11.list[[s]] <- starting.x11.team1
}
# now looking at passes
pass.index <- which(unlist(lapply(event.temp, function(x) x$type$name)) == 'Pass')
# and now filtering to get the pass indices for team 1, using the team 1 identifier from team.id.clist[1]
pass.team1 <- pass.index[which(unlist(lapply(pass.index, function(x) event.temp[[x]]$team$id)) == team.id.clist[1])]
pass.team1.df <- data.frame(matrix(NA, nrow=1, ncol=13))
colnames(pass.team1.df) <- c('Possession','Passer','X.Pass','Y.Pass','Pass.Type','Receiver','X.Receive','Y.Receive',
'Pass.Length','Pass.Angle','Body.Part','Pass.Pressure','Pass.Outcome')
for (p in 1:length(pass.team1)){
pass.temp <- event.temp[[pass.team1[p]]]
possession <- pass.temp$possession
passer <- pass.temp$player$id
pass.location <- pass.temp$location
pass.type <- pass.temp$pass$height$name
receiver <- pass.temp$pass$recipient$id
receive.location <- pass.temp$pass$end_location
pass.length <- pass.temp$pass$length
pass.angle <- pass.temp$pass$angle
body.part <- pass.temp$pass$body_part$name
pass.pressure <- pass.temp$under_pressure
pass.outcome <- pass.temp$pass$outcome$name
row.toadd <- c(possession, passer, pass.location, pass.type, receiver, receive.location
,pass.length, pass.angle, body.part, pass.pressure, pass.outcome)
pass.team1.df <- rbind(pass.team1.df, row.toadd)
}
# getting rid of the first empty row (this is minging)
pass.team1.df <- pass.team1.df[-1,]
pass.team1.df[,c(1:4,6:10)] <- lapply(pass.team1.df[,c(1:4,6:10)], as.numeric)
# this is basically a row number (partition by...)
# providing
pass.team1.df <- pass.team1.df %>% group_by(Possession) %>% mutate(seq = row_number())
pass.team1.df$team_id <- team.id.clist[1]
## AND NOW TO DO THE EXACT SAME THING FOR TEAM 2!
# and now filtering to get the pass indices for team 2, using the team 2 identifier from team.id.clist[2]
pass.team2 <- pass.index[which(unlist(lapply(pass.index, function(x) event.temp[[x]]$team$id)) == team.id.clist[2])]
pass.team2.df <- data.frame(matrix(NA, nrow=1, ncol=13))
colnames(pass.team2.df) <- c('Possession','Passer','X.Pass','Y.Pass','Pass.Type','Receiver','X.Receive','Y.Receive',
'Pass.Length','Pass.Angle','Body.Part','Pass.Pressure','Pass.Outcome')
for (p in 1:length(pass.team2)){
pass.temp <- event.temp[[pass.team2[p]]]
possession <- pass.temp$possession
passer <- pass.temp$player$id
pass.location <- pass.temp$location
pass.type <- pass.temp$pass$height$name
receiver <- pass.temp$pass$recipient$id
receive.location <- pass.temp$pass$end_location
pass.length <- pass.temp$pass$length
pass.angle <- pass.temp$pass$angle
body.part <- pass.temp$pass$body_part$name
pass.pressure <- pass.temp$under_pressure
pass.outcome <- pass.temp$pass$outcome$name
row.toadd <- c(possession, passer, pass.location, pass.type, receiver, receive.location
,pass.length, pass.angle, body.part, pass.pressure, pass.outcome)
pass.team2.df <- rbind(pass.team2.df, row.toadd)
}
# getting rid of the first empty row (this is minging)
pass.team2.df <- pass.team2.df[-1,]
pass.team2.df[,c(1:4,6:10)] <- lapply(pass.team2.df[,c(1:4,6:10)], as.numeric)
# this is basically a row number (partition by...)
# providing
pass.team2.df <- pass.team2.df %>% group_by(Possession) %>% mutate(seq = row_number())
pass.team2.df$team_id <- team.id.clist[2]
## AND NOW PUTTING IT ALL TOGETHER
pass.list <- list(pass.team1.df, pass.team2.df)
match.id <- strsplit(basename(event.files[i]), '[.]')[[1]][1]
event.list[[match.id]] <- list(starting.x11.list, pass.list)
}
event.list[[1]]
| 0.248079 | 0.846641 |
# Pandas support
This notebook provides a simple example of how to use Pint with Pandas. See the documentation for full details.
```
import pandas as pd
import pint
import numpy as np
from pint.pandas_interface import PintArray
ureg=pint.UnitRegistry()
Q_=ureg.Quantity
```
## Basic example
This example shows how the DataFrame works with Pint. However, it's not the most usual case so we also show how to read from a csv below.
```
df = pd.DataFrame({
"torque": PintArray(Q_([1, 2, 2, 3], "lbf ft")),
"angular_velocity": PintArray(Q_([1000, 2000, 2000, 3000], "rpm"))
})
df
df['power'] = df['torque'] * df['angular_velocity']
df
df.power
df.power.values
df.power.values.data
df.angular_velocity.values.data
df.power.pint.units
df.power.pint.to("kW").values
```
## Reading from csv
Reading from files is the far more standard way to use pandas. To facilitate this, DataFrame accessors are provided to make it easy to get to PintArrays.
## Setup
Here we create the DateFrame and save it to file, next we will show you how to load and read it.
We start with a DateFrame with column headers only.
```
df_init = pd.DataFrame({
"speed": [1000, 1100, 1200, 1200],
"mech power": [np.nan, np.nan, np.nan, np.nan],
"torque": [10, 10, 10, 10],
"rail pressure": [1000, 1000000000000, 1000, 1000],
"fuel flow rate": [10, 10, 10, 10],
"fluid power": [np.nan, np.nan, np.nan, np.nan],
})
df_init
```
Then we add a column header which contains units information
```
units = ["rpm", "kW", "N m", "bar", "l/min", "kW"]
df_to_save = df_init.copy()
df_to_save.columns = pd.MultiIndex.from_arrays([df_init.columns, units])
df_to_save
```
Now we save this to disk as a csv to give us our starting point.
```
test_csv_name = "pandas_test.csv"
df_to_save.to_csv(test_csv_name, index=False)
```
Now we are in a position to read the csv we just saved. Let's start by reading the file with units as a level in a multiindex column.
```
df = pd.read_csv(test_csv_name, header=[0,1])
df
```
Then use the DataFrame's pint accessor's quantify method to convert the columns from `np.ndarray`s to PintArrays, with units from the bottom column level.
```
df_ = df.pint.quantify(ureg, level=-1)
df_
```
As previously, operations between DataFrame columns are unit aware
```
df_['mech power'] = df_.speed*df_.torque
df_['fluid power'] = df_['fuel flow rate'] * df_['rail pressure']
df_
```
The DataFrame's `pint.dequantify` method then allows us to retrieve the units information as a header row once again.
```
df_.pint.dequantify()
```
This allows for some rather powerful abilities. For example, to change single column units
```
df_['fluid power'] = df_['fluid power'].pint.to("kW")
df_['mech power'] = df_['mech power'].pint.to("kW")
df_.pint.dequantify()
```
or the entire table's units
```
df_.pint.to_base_units().pint.dequantify()
```
|
github_jupyter
|
import pandas as pd
import pint
import numpy as np
from pint.pandas_interface import PintArray
ureg=pint.UnitRegistry()
Q_=ureg.Quantity
df = pd.DataFrame({
"torque": PintArray(Q_([1, 2, 2, 3], "lbf ft")),
"angular_velocity": PintArray(Q_([1000, 2000, 2000, 3000], "rpm"))
})
df
df['power'] = df['torque'] * df['angular_velocity']
df
df.power
df.power.values
df.power.values.data
df.angular_velocity.values.data
df.power.pint.units
df.power.pint.to("kW").values
df_init = pd.DataFrame({
"speed": [1000, 1100, 1200, 1200],
"mech power": [np.nan, np.nan, np.nan, np.nan],
"torque": [10, 10, 10, 10],
"rail pressure": [1000, 1000000000000, 1000, 1000],
"fuel flow rate": [10, 10, 10, 10],
"fluid power": [np.nan, np.nan, np.nan, np.nan],
})
df_init
units = ["rpm", "kW", "N m", "bar", "l/min", "kW"]
df_to_save = df_init.copy()
df_to_save.columns = pd.MultiIndex.from_arrays([df_init.columns, units])
df_to_save
test_csv_name = "pandas_test.csv"
df_to_save.to_csv(test_csv_name, index=False)
df = pd.read_csv(test_csv_name, header=[0,1])
df
df_ = df.pint.quantify(ureg, level=-1)
df_
df_['mech power'] = df_.speed*df_.torque
df_['fluid power'] = df_['fuel flow rate'] * df_['rail pressure']
df_
df_.pint.dequantify()
df_['fluid power'] = df_['fluid power'].pint.to("kW")
df_['mech power'] = df_['mech power'].pint.to("kW")
df_.pint.dequantify()
df_.pint.to_base_units().pint.dequantify()
| 0.286768 | 0.986546 |
# Exploring the Files and Analyzing Folder Structures
```
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:80% !important; }</style>"))
import ipywidgets as widgets
import ipython_blocking
from ipywidgets import interact
import sys
sys.path.append("../")
from jupyter_apis.Aux_fun import *
%%capture
!jupyter nbextension enable --py --sys-prefix qgrid
!jupyter nbextension enable --py --sys-prefix widgetsnbextension
%%capture
!conda install --yes --prefix {sys.prefix} python-graphviz
# had to install it from here, otherwise "dot" not found in path
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
```
**Important:** Set the path to the folder you are going to analyze
```
path_input = widgets.Text(
placeholder='/home/jovyan/test_data',
description='Folder Path',
disabled=False,
value='/home/jovyan/test_data',
)
button = widgets.Button(description='Run')
box = widgets.VBox(children=[path_input, button])
display(box)
%blockrun button
# get a Pandas dataframe with all files information needed for the visualizations
folder_path = path_input.value
df = get_df(folder_path)
```
## Treemap
The following treemap shows the size of each file extension.
This already gives some interesting insight into the way the files are distributed in the folder. Exploring your file system by size can be very helpful when looking for files or folders that you don’t need, but eat up a lot of memory.
```
Treemap(df)
```
According to Zipf's law (a discrete form of the continuous Pareto distribution), the second largest file type will be about half the size of the largest. The third largest, one third the size of the largest. The fourth, one fourth, and so on.
## Graphs
The folder structure can be represented with a graph using NetworkX package.
```
G = create_graph(df)
```
The total number of nodes is how many files and folders we have. The total number of edges is equal to the total number of nodes minus one, since every file and folder can be in only one folder and the first folder has no parent folder, this makes sense. The average degree is the average number of edges that a node can have.
Now we could apply all sorts of available [Algorithms](https://networkx.org/documentation/networkx-1.10/reference/algorithms.html) and [Functions](https://networkx.org/documentation/networkx-1.10/reference/functions.html) that can be applied to explore and analyze the folder structure.
```
show_graph(G)
# using a radial layout
show_graph(G, layout='radial')
```
Be aware though, that the time to compute these graph drawing can get quickly out of hand for large graphs (>10000)
## Table
It is very helpful to display the dataframe on a qgrid so we can filter it
```
qgrid_widget(df)
# If you filter the data, can get the new df with the following command
# qgrid_widget(df).get_changed_df()
```
### Some references
**size** describes the number of bytes by the file. (Note: Folders also have a size, although the size is excluding the files inside). The **folder** flag specifies whether this element is a folder, the **num_files** is the number of files within the folder, and the **depth** states how many layers of folders the file or folder is. Finally, **id** and **parent** are responsible to see the links between files and folders which can be used to create a graph.
##### Timestamps:
* **atime**: time of last access
* **mtime**: time of last modification
* **ctime**: time of last status (metadata) change like file permissions, file ownership, etc. (creation time in Windows)
Find more about what each field means on https://en.wikipedia.org/wiki/Stat_(system_call)
## Bar Charts
### Counting different types of file extensions
Let's explore the distribution of files by their file extensions.
In the first visualization, you will see the distribution sorted by their occurrence.
```
count_extentions_bar_chart(df)
```
### File extension by size
This showed us only the counts, but what about the sizes?
```
extetions_size_bar_chart(df)
```
### Folder sizes
Next, we can have a look now at the largest folders in the repository. The first thing to do, is to filter the data set to only have folders. The rest should be familiar to the previous visualizations
```
folder_sizes_bar_chart(df)
```
Reference: https://janakiev.com/blog/python-filesystem-analysis/
|
github_jupyter
|
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:80% !important; }</style>"))
import ipywidgets as widgets
import ipython_blocking
from ipywidgets import interact
import sys
sys.path.append("../")
from jupyter_apis.Aux_fun import *
%%capture
!jupyter nbextension enable --py --sys-prefix qgrid
!jupyter nbextension enable --py --sys-prefix widgetsnbextension
%%capture
!conda install --yes --prefix {sys.prefix} python-graphviz
# had to install it from here, otherwise "dot" not found in path
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
path_input = widgets.Text(
placeholder='/home/jovyan/test_data',
description='Folder Path',
disabled=False,
value='/home/jovyan/test_data',
)
button = widgets.Button(description='Run')
box = widgets.VBox(children=[path_input, button])
display(box)
%blockrun button
# get a Pandas dataframe with all files information needed for the visualizations
folder_path = path_input.value
df = get_df(folder_path)
Treemap(df)
G = create_graph(df)
show_graph(G)
# using a radial layout
show_graph(G, layout='radial')
qgrid_widget(df)
# If you filter the data, can get the new df with the following command
# qgrid_widget(df).get_changed_df()
count_extentions_bar_chart(df)
extetions_size_bar_chart(df)
folder_sizes_bar_chart(df)
| 0.181444 | 0.862815 |
```
import jax.numpy as jnp
poorHigh_w = jnp.array([ 5. , 25.489805 , 21.340055 , 18.866007 , 16.992601 ,
15.505475 , 14.555338 , 14.082295 , 13.727711 , 13.341486 ,
12.798906 , 12.521904 , 12.377323 , 12.404113 , 12.472523 ,
12.449293 , 12.425125 , 12.315584 , 12.28413 , 12.557176 ,
12.950956 , 13.28101 , 13.663953 , 14.085483 , 14.386494 ,
14.675861 , 15.288598 , 16.03489 , 16.875498 , 17.85997 ,
19.023191 , 20.684574 , 21.534988 , 21.85008 , 21.75778 ,
21.335236 , 20.909994 , 20.645237 , 20.527098 , 20.709085 ,
21.203526 , 22.163754 , 23.99394 , 25.21154 , 25.802986 ,
26.322763 , 19.94648 , 14.776118 , 10.781056 , 7.9825187,
6.2406187, 5.2877426, 5.018139 , 5.5149274, 6.5818796,
8.109399 , 9.689917 , 11.232063 , 11.937669 , 11.88838 ,
10.732118 ])
poorLow_w = jnp.array([ 5. , 25.489805, 23.686905, 23.583015, 25.342312,
28.00257 , 30.591097, 32.533695, 34.063004, 37.04034 ,
39.765858, 43.314583, 46.236256, 47.45725 , 52.702324,
54.02915 , 56.937912, 57.808475, 60.506454, 65.0779 ,
64.21815 , 69.055885, 71.45909 , 73.227325, 71.8414 ,
69.57499 , 70.19525 , 71.36607 , 70.757095, 71.88589 ,
77.26895 , 80.68532 , 83.56862 , 84.50874 , 85.76661 ,
84.835014, 84.37967 , 86.747314, 83.70736 , 82.37102 ,
80.50324 , 81.40658 , 79.05709 , 80.40889 , 75.73343 ,
71.24366 , 78.401115, 71.12398 , 69.43728 , 62.1735 ,
55.59825 , 49.64868 , 44.13118 , 39.437874, 35.924824,
33.57551 , 30.063377, 27.977575, 24.608301, 21.704208,
17.881044])
richHigh_w = jnp.array([ 5. , 44.169 , 31.724762 , 37.447975 ,
42.889927 , 48.366238 , 54.760906 , 62.743126 ,
71.45885 , 75.87016 , 81.989525 , 85.58819 ,
89.369 , 91.10194 , 92.69318 , 89.5441 ,
90.94471 , 93.78521 , 95.706245 , 97.30388 ,
98.64327 , 95.02311 , 96.78608 , 100.1138 ,
94.48128 , 98.134026 , 98.75603 , 101.81176 ,
104.17117 , 103.70764 , 102.2038 , 108.01187 ,
104.51686 , 105.77256 , 104.21921 , 98.54423 ,
94.70079 , 90.71106 , 84.16321 , 77.29525 ,
68.08416 , 62.34101 , 57.466663 , 51.467392 ,
44.139736 , 34.270702 , 23.744139 , 16.833946 ,
12.301145 , 9.27295 , 7.219111 , 5.966625 ,
5.492039 , 4.98194 , 4.834813 , 4.6924834,
4.852975 , 5.187632 , 5.0777164, 4.866398 ,
5.5222845])
richLow_w = jnp.array([ 5. , 44.169 , 47.22957 , 50.738888 ,
59.97472 , 69.64247 , 79.27276 , 89.62661 ,
90.6721 , 99.35745 , 102.28124 , 105.59956 ,
101.080925 , 105.09058 , 105.08204 , 105.55944 ,
104.844055 , 106.61257 , 108.10203 , 108.3012 ,
105.639084 , 108.951935 , 110.77622 , 112.92671 ,
113.83319 , 116.196465 , 117.862625 , 120.203285 ,
117.28213 , 114.9433 , 115.288536 , 118.71654 ,
117.59747 , 118.872314 , 112.656364 , 106.40605 ,
103.439674 , 97.335175 , 87.376045 , 81.003235 ,
71.80459 , 64.635086 , 58.837536 , 52.637325 ,
45.082092 , 35.56538 , 24.535505 , 17.934704 ,
12.570344 , 9.184089 , 6.821807 , 5.626688 ,
4.7433753, 4.529494 , 4.3361583, 4.325389 ,
4.406368 , 4.5582147, 4.5020976, 4.367129 ,
4.8314805])
%pylab inline
matplotlib.rcParams['figure.figsize'] = [16, 8]
plt.rcParams.update({'font.size': 15})
plt.title("wealth level at different age periods")
plt.plot(poorHigh_w, label = "poor high cost")
plt.plot(poorLow_w, label = "poor low cost")
plt.plot(richHigh_w, label = "rich high cost")
plt.plot(richLow_w, label = "rich low cost")
plt.legend()
```
|
github_jupyter
|
import jax.numpy as jnp
poorHigh_w = jnp.array([ 5. , 25.489805 , 21.340055 , 18.866007 , 16.992601 ,
15.505475 , 14.555338 , 14.082295 , 13.727711 , 13.341486 ,
12.798906 , 12.521904 , 12.377323 , 12.404113 , 12.472523 ,
12.449293 , 12.425125 , 12.315584 , 12.28413 , 12.557176 ,
12.950956 , 13.28101 , 13.663953 , 14.085483 , 14.386494 ,
14.675861 , 15.288598 , 16.03489 , 16.875498 , 17.85997 ,
19.023191 , 20.684574 , 21.534988 , 21.85008 , 21.75778 ,
21.335236 , 20.909994 , 20.645237 , 20.527098 , 20.709085 ,
21.203526 , 22.163754 , 23.99394 , 25.21154 , 25.802986 ,
26.322763 , 19.94648 , 14.776118 , 10.781056 , 7.9825187,
6.2406187, 5.2877426, 5.018139 , 5.5149274, 6.5818796,
8.109399 , 9.689917 , 11.232063 , 11.937669 , 11.88838 ,
10.732118 ])
poorLow_w = jnp.array([ 5. , 25.489805, 23.686905, 23.583015, 25.342312,
28.00257 , 30.591097, 32.533695, 34.063004, 37.04034 ,
39.765858, 43.314583, 46.236256, 47.45725 , 52.702324,
54.02915 , 56.937912, 57.808475, 60.506454, 65.0779 ,
64.21815 , 69.055885, 71.45909 , 73.227325, 71.8414 ,
69.57499 , 70.19525 , 71.36607 , 70.757095, 71.88589 ,
77.26895 , 80.68532 , 83.56862 , 84.50874 , 85.76661 ,
84.835014, 84.37967 , 86.747314, 83.70736 , 82.37102 ,
80.50324 , 81.40658 , 79.05709 , 80.40889 , 75.73343 ,
71.24366 , 78.401115, 71.12398 , 69.43728 , 62.1735 ,
55.59825 , 49.64868 , 44.13118 , 39.437874, 35.924824,
33.57551 , 30.063377, 27.977575, 24.608301, 21.704208,
17.881044])
richHigh_w = jnp.array([ 5. , 44.169 , 31.724762 , 37.447975 ,
42.889927 , 48.366238 , 54.760906 , 62.743126 ,
71.45885 , 75.87016 , 81.989525 , 85.58819 ,
89.369 , 91.10194 , 92.69318 , 89.5441 ,
90.94471 , 93.78521 , 95.706245 , 97.30388 ,
98.64327 , 95.02311 , 96.78608 , 100.1138 ,
94.48128 , 98.134026 , 98.75603 , 101.81176 ,
104.17117 , 103.70764 , 102.2038 , 108.01187 ,
104.51686 , 105.77256 , 104.21921 , 98.54423 ,
94.70079 , 90.71106 , 84.16321 , 77.29525 ,
68.08416 , 62.34101 , 57.466663 , 51.467392 ,
44.139736 , 34.270702 , 23.744139 , 16.833946 ,
12.301145 , 9.27295 , 7.219111 , 5.966625 ,
5.492039 , 4.98194 , 4.834813 , 4.6924834,
4.852975 , 5.187632 , 5.0777164, 4.866398 ,
5.5222845])
richLow_w = jnp.array([ 5. , 44.169 , 47.22957 , 50.738888 ,
59.97472 , 69.64247 , 79.27276 , 89.62661 ,
90.6721 , 99.35745 , 102.28124 , 105.59956 ,
101.080925 , 105.09058 , 105.08204 , 105.55944 ,
104.844055 , 106.61257 , 108.10203 , 108.3012 ,
105.639084 , 108.951935 , 110.77622 , 112.92671 ,
113.83319 , 116.196465 , 117.862625 , 120.203285 ,
117.28213 , 114.9433 , 115.288536 , 118.71654 ,
117.59747 , 118.872314 , 112.656364 , 106.40605 ,
103.439674 , 97.335175 , 87.376045 , 81.003235 ,
71.80459 , 64.635086 , 58.837536 , 52.637325 ,
45.082092 , 35.56538 , 24.535505 , 17.934704 ,
12.570344 , 9.184089 , 6.821807 , 5.626688 ,
4.7433753, 4.529494 , 4.3361583, 4.325389 ,
4.406368 , 4.5582147, 4.5020976, 4.367129 ,
4.8314805])
%pylab inline
matplotlib.rcParams['figure.figsize'] = [16, 8]
plt.rcParams.update({'font.size': 15})
plt.title("wealth level at different age periods")
plt.plot(poorHigh_w, label = "poor high cost")
plt.plot(poorLow_w, label = "poor low cost")
plt.plot(richHigh_w, label = "rich high cost")
plt.plot(richLow_w, label = "rich low cost")
plt.legend()
| 0.215846 | 0.123418 |
<div>
<a href="https://www.audiolabs-erlangen.de/fau/professor/mueller"><img src="data_layout/PCP_Teaser.png" width=100% style="float: right;" alt="PCP Teaser"></a>
</div>
# Unit 9: Discrete Fourier Transform (DFT)
<ul>
<li><a href='#learn'>Overview and Learning Objectives</a></li>
<li><a href='#inner'>Inner Product</a></li>
<li><a href='#dft'>Definition of DFT</a></li>
<li><a href='#phase'>DFT Phase</a></li>
<li><a href='#dftmatrix'>DFT Matrix</a></li>
<li><a href='#fft'>Fast Fourier Transform (FFT)</a></li>
<li><a href='#exercise_freq_index'>Exercise 1: Interpretation of Frequency Indices</a></li>
<li><a href='#exercise_missing_time'>Exercise 2: Missing Time Localization</a></li>
<li><a href='#exercise_chirp'>Exercise 3: Chirp Signal</a></li>
<li><a href='#exercise_inverse'>Exercise 4: Inverse DFT</a></li>
</ul>
<a id='learn'></a>
<div class="alert alert-block alert-warning">
<h2>Overview and Learning Objectives</h2>
The <strong>Fourier transform</strong> is one of the most important tools for a wide range of engineering and computer science applications. The general idea of <strong>Fourier analysis</strong> is to decompose a given signal into a weighted superposition of sinusoidal functions. Since these functions possess an explicit physical meaning regarding their frequencies, the decomposition is typically more accessible for subsequent processing steps than the original signal. Assuming that you are familiar with the Fourier transform and its applications in signal processing, we review in this unit the discrete variant of the Fourier transform known as <strong>Discrete Fourier Transform</strong> (DFT). We define the inner product that allows for comparing two vectors (e.g., discrete-time signals of finite length). The DFT can be thought of as comparing a given signal of finite length with a specific set of exponential signals (a complex variant of sinusoidal signals), each comparison yielding a complex-valued Fourier coefficient. Then, using suitable visualizations, we show how you can interpret the amplitudes and phases of these coefficients. Recall that one can express the DFT as a complex-valued square matrix. We show how separately plotting the real and imaginary parts leads to beautiful and insightful images. Applying a DFT boils down to computing a matrix–vector product, which we implement via the standard NumPy function <code>np.dot</code>. Since the number of operations for computing a DFT via a simple matrix–vector product is quadratic in the input length, the runtime of this approach becomes problematic with increasing length. This issue is exactly where the fast Fourier transform (FFT) comes into the game. We present this famous divide-and-conquer algorithm and provide a Python implementation. Furthermore, we compare the runtime behavior between the FFT implementation and the naive DFT implementation. We will further deepen your understanding of the Fourier transform by considering further examples and visualization in the exercises. In <a href='#exercise_freq_index'>Exercise 1</a>, you will learn how to interpret and plot frequency indices in a physically meaningful way. In <a href='#exercise_missing_time'>Exercise 2</a>, we discuss the issue of loosing time information when applying the Fourier transform, which is the main motivation for the <a href='https://www.audiolabs-erlangen.de/resources/MIR/FMP/C2/C2_STFT-Basic.html'>short-time Fourier transform</a>. In <a href='#exercise_chirp'>Exercise 3</a>, you will apply the DFT to a <strong>chirp signal</strong>, which yields another illustrative example of the DFT's properties. Finally, in <a href='#exercise_inverse'>Exercise 4</a>, we will invite you to explore the relationship between the DFT and its inverse. Again, an overarching goal of this unit is to apply and deepen your Python programming skills within the context of a central topic for signal processing.
</div>
<a id='inner'></a>
## Inner Product
In this notebook, we consider [discrete-time (DT) signals](PCP_08_signal.html) of finite length $N\in\mathbb{N}$, which we represent as vector
$$
x=(x(0),x(1),...,x(N-1))^\top\in\mathbb{R}^N
$$
with samples $x(n)\in\mathbb{R}^N$ for $n\in[0:N-1]$. Note that $\top$ indicates the transpose of a vector, thus converting a row vector into a column vector. Furthermore, note that we start indexing with the index $0$ (thus adapting our mathematical notation to Python conventions). A general concept for comparing two vectors (or signals) is the **inner product**. Given two vectors $x, y \in \mathbb{R}^N$, the inner product between $x$ and $y$ is defined as follows:
$$
\langle x | y \rangle := \sum_{n=0}^{N-1} x(n) y(n).
$$
The absolute value of the inner product may be interpreted as a measure of similarity between $x$ and $y$. If $x$ and $y$ are similar (i.e., if they point to more or less the same direction), the inner product $|\langle x | y \rangle|$ is large. If $x$ and $y$ are dissimilar (i.e., if $x$ and $y$ are more or less orthogonal to each other), the inner product $|\langle x | y \rangle|$ is close to zero.
One can extend this concept to **complex-valued** vectors $x,y\in\mathrm{C}^N$, where the inner product is defined as
$$
\langle x | y \rangle := \sum_{n=0}^{N-1} x(n) \overline{y(n)}.
$$
In the case of real-valued signals, the complex conjugate does not play any role and the definition of the complex-valued inner product reduces to the real-valued one. In the following code cell, we give some examples.
<div class="alert alert-block alert-warning">
<strong>Note:</strong>
One can use the NumPy function <code>np.vdot</code> to compute the inner product. However, opposed to the mathematical convention that conjugates the second argument, this function applies complex conjugation on the first argument. Therefore, for computing $\langle x | y \rangle$ as defined above, one has to call <code>np.vdot(y, x)</code>.
</div>
In the following, we generate and visualize three signals $x_1$, $x_2$, $x_3$. Then, we compute and discuss different inner products using the signals.
```
import numpy as np
from matplotlib import pyplot as plt
import libpcp.signal
%matplotlib inline
Fs = 64
dur = 1
x1, t = libpcp.signal.generate_example_signal(Fs=Fs, dur=dur)
x2, t = libpcp.signal.generate_sinusoid(dur=dur, Fs=Fs, amp=1, freq=2, phase=0.3)
x3, t = libpcp.signal.generate_sinusoid(dur=dur, Fs=Fs, amp=1, freq=6, phase=0.1)
def plot_inner_product(ax, t, x, y, color_x='k', color_y='r', label_x='x', label_y='y'):
"""Plot inner product
Notebook: PCP_09_dft.ipynb
Args:
ax: Axis handle
t: Time axis
x: Signal x
y: Signal y
color_x: Color of signal x (Default value = 'k')
color_y: Color of signal y (Default value = 'r')
label_x: Label of signal x (Default value = 'x')
label_y: Label of signal y (Default value = 'y')
"""
ax.plot(t, x, color=color_x, linewidth=1.0, linestyle='-', label=label_x)
ax.plot(t, y, color=color_y, linewidth=1.0, linestyle='-', label=label_y)
ax.set_xlim([0, t[-1]])
ax.set_ylim([-1.5, 1.5])
ax.set_xlabel('Time (seconds)')
ax.set_ylabel('Amplitude')
sim = np.vdot(y, x)
ax.set_title(r'$\langle$ %s $|$ %s $\rangle = %.1f$' % (label_x, label_y, sim))
ax.legend(loc='upper right')
plt.figure(figsize=(8, 5))
ax = plt.subplot(2, 2, 1)
plot_inner_product(ax, t, x1, x1, color_x='k', color_y='k', label_x='$x_1$', label_y='$x_1$')
ax = plt.subplot(2, 2, 2)
plot_inner_product(ax, t, x1, x2, color_x='k', color_y='r', label_x='$x_1$', label_y='$x_2$')
ax = plt.subplot(2, 2, 3)
plot_inner_product(ax, t, x1, x3, color_x='k', color_y='b', label_x='$x_1$', label_y='$x_3$')
ax = plt.subplot(2, 2, 4)
plot_inner_product(ax, t, x2, x3, color_x='r', color_y='b', label_x='$x_2$', label_y='$x_3$')
plt.tight_layout()
```
In the above example, one can make the following observations:
* The signal $x_1$ is similar to itself, leading to a large value of $\langle x_1 | x_1 \rangle=40.0$.
* The overall course of the signal $x_1$ strongly correlates with the sinusoid $x_2$, which is reflected by a relatively large value of $\langle x_1 | x_2 \rangle=29.9$.
* There are some finer oscillations of $x_1$ that are captured by $x_3$, leading to a still noticeable value of $\langle x_1 | x_3 \rangle=14.7$.
* The two sinusoids $x_2$ and $x_3$ are more or less uncorrelated, which is revealed by the value of $\langle x_2 | x_3 \rangle\approx 0$.
In other words, the above comparison reveals that the signal $x_1$ has a strong signal component of $2~\mathrm {Hz}$ (frequency of $x_2$) and $6~\mathrm {Hz}$ (frequency of $x_3$). Measuring correlations between an arbitrary signal and sinusoids of different frequencies is exactly the idea of performing a Fourier (or spectral) analysis.
<a id='dft'></a>
## Definition of DFT
Let $x\in \mathbb{C}^N$ be a vector of length $N\in\mathbb{N}$. The **discrete Fourier transform** (DFT) of $x$ is defined by:
$$ X(k) := \sum_{n=0}^{N-1} x(n) \exp(-2 \pi i k n / N) $$
for $k \in [0:N-1]$. The vector $X\in\mathbb{C}^N$ can be interpreted as a frequency representation of the time-domain signal $x$. To obtain a geometric interpretation of the DFT, we define the vector $\mathbf{e}_k \in\mathbb{C}^N$ with real part $\mathbf{c}_k=\mathrm{Re}(\mathbf{e}_k)$ and imaginary part $\mathbf{s}_k=\mathrm{Im}(\mathbf{e}_k)$ by
$$\mathbf{e}_k(n) := \exp(2 \pi i k n / N) = \cos(2 \pi i k n / N) + i \sin(2 \pi i k n / N)
= \mathbf{c}_k(n) + i \mathbf{s}_k(n)$$
for each $k \in [0:N-1]$.
This vector can be regarded as a [sampled version](PCP_08_signal.html) of the [exponential function](PCP_07_exp.html) of frequency $k/N$. Using inner products, the DFT can be expressed as
$$ X(k) = \sum_{n=0}^{N-1} x(n) \overline{\mathbf{e}_k}(n) = \langle x | \mathbf{e}_k \rangle,$$
thus measuring the similarity between the signal $x$ and the sampled exponential functions $\mathbf{e}_k$. The absolute value $|X(k)|$ indicates the degree of similarity between the signal $x$ and $\mathbf{e}_k$. In the case that $x\in \mathbb{R}^N$ is a real-valued vector (which is typically the case for audio signals), we obtain:
$$
X(k) = \langle x |\mathrm{Re}(\mathbf{e}_k) \rangle - i\langle x | \mathrm{Im}(\mathbf{e}_k) \rangle
= \langle x |\mathbf{c}_k \rangle - i\langle x | \mathbf{s}_k \rangle
$$
The following plot shows an example signal $x$ compared with functions $\overline{\mathbf{e}_k}$ for various frequency parameters $k$. The real and imaginary part of $\overline{\mathbf{e}_k}$ are shown in <font color='red'> red</font> and <font color='blue'> blue</font>, respectively.
```
def plot_signal_e_k(ax, x, k, show_e=True, show_opt=False):
"""Plot signal and k-th DFT sinusoid
Notebook: PCP_09_dft.ipynb
Args:
ax: Axis handle
x: Signal
k: Index of DFT
show_e: Shows cosine and sine (Default value = True)
show_opt: Shows cosine with optimal phase (Default value = False)
"""
N = len(x)
time_index = np.arange(N)
ax.plot(time_index, x, 'k', marker='.', markersize='10', linewidth=2.0, label='$x$')
plt.xlabel('Time (samples)')
e_k = np.exp(2 * np.pi * 1j * k * time_index / N)
c_k = np.real(e_k)
s_k = np.imag(e_k)
X_k = np.vdot(e_k, x)
plt.title(r'k = %d: Re($X(k)$) = %0.2f, Im($X(k)$) = %0.2f, $|X(k)|$=%0.2f' %
(k, X_k.real, X_k.imag, np.abs(X_k)))
if show_e is True:
ax.plot(time_index, c_k, 'r', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\mathrm{Re}(\overline{\mathbf{u}}_k)$')
ax.plot(time_index, s_k, 'b', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\mathrm{Im}(\overline{\mathbf{u}}_k)$')
if show_opt is True:
phase_k = - np.angle(X_k) / (2 * np.pi)
cos_k_opt = np.cos(2 * np.pi * (k * time_index / N - phase_k))
d_k = np.sum(x * cos_k_opt)
ax.plot(time_index, cos_k_opt, 'g', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\cos_{k, opt}$')
plt.grid()
plt.legend(loc='lower right')
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
plt.figure(figsize=(8, 15))
for k in range(1, 8):
ax = plt.subplot(7, 1, k)
plot_signal_e_k(ax, x, k=k)
plt.tight_layout()
```
<a id='phase'></a>
## DFT Phase
At first sight, the DFT may be a bit confusing: Why is a real-valued signal $x$ compared with a complex-valued sinusoid $\mathbf{e}_k$? What does the resulting complex-valued Fourier coefficient
$$
c_k:= X(k) := \langle x |\mathrm{Re}(\mathbf{e}_k) \rangle - i\langle x | \mathrm{Im}(\mathbf{e}_k) \rangle.
$$
encode? To understand this, we represent the complex number $c_k$ in form of its [polar representation](PCP_06_complex.html#polar)
$$
c_k = |c_k| \cdot \mathrm{exp}(i \gamma_k),
$$
where $\gamma_k$ is the [angle](PCP_06_complex.html) (given in radians). Furthermore, let $\mathbf{cos}_{k,\varphi}:[0:N-1]\to\mathbb{R}$ be a sampled sinusoid with frequency parameter $k$ and phase $\varphi\in[0,1)$, defined by
$$
\mathbf{cos}_{k,\varphi}(n) = \mathrm{cos}\big( 2\pi (kn/N - \varphi) \big)
$$
for $n\in[0,N-1]$. Defining $\varphi_k := - \frac{\gamma_k}{2 \pi}$, one obtains the following remarkable property of the Fourier coefficient $c_k$:
\begin{eqnarray}
|c_k| &=& \mathrm{max}_{\varphi\in[0,1)} \langle x | \mathbf{cos}_{k,\varphi} \rangle,\\
\varphi_k &=& \mathrm{argmax}_{\varphi\in[0,1)} \langle x | \mathbf{cos}_{k,\varphi} \rangle.
\end{eqnarray}
In other words, the phase $\varphi_k$ maximizes the correlation between $x$ and all possible sinusoids $\mathbf{cos}_{k,\varphi}$ with $\varphi\in[0,1)$. Furthermore, the magnitude $|c_k|$ yields this maximal value. Thus, computing a single correlation between $x$ and the complex-valued function $\mathbf{e}_k$ (which real part coincides with $\mathbf{cos}_{k,0}$, and its imaginary part with $\mathbf{cos}_{k,0.25}$) solves an optimization problem. In the following code cell, we demonstrate this optimality property, where the $\mathbf{cos}_{k,\varphi}$ with optimal phase $\varphi=\varphi_k$ is shown in <font color='green'>green</font>.
```
plt.figure(figsize=(8, 15))
for k in range(1, 8):
ax = plt.subplot(7, 1, k)
plot_signal_e_k(ax, x, k=k, show_e=False, show_opt=True)
plt.tight_layout()
```
<a id='dftmatrix'></a>
## DFT Matrix
Being a linear operator $\mathbb{C}^N \to \mathbb{C}^N$, the DFT can be expressed by some $N\times N$-matrix. This leads to the famous DFT matrix $\mathrm{DFT}_N \in \mathbb{C}^{N\times N}$ matrix, which is given by
$$\mathrm{DFT}_N(n, k) = \mathrm{exp}(-2 \pi i k n / N)$$
for $n\in[0:N-1]$ and $k\in[0:N-1]$. Let $\rho_N:=\exp(2 \pi i / N)$ be the primitive $N^\mathrm{th}$ [root of unity](PCP_07_exp.html#roots). Then
$$\sigma_N:= \overline{\rho_N} = \mathrm{exp}(-2 \pi i / N)$$
also defines a primitive $N^\mathrm{th}$ [root of unity](PCP_07_exp.html#roots). From the [properties of exponential functions](PCP_07_exp.html), one obtains that
$$ \sigma_N^{kn} = \mathrm{exp}(-2 \pi i / N)^{kn} = \mathrm{exp}(-2 \pi i k n / N)$$
From this, one obtains:
$$
\mathrm{DFT}_N =
\begin{pmatrix}
1 & 1 & 1 & \dots & 1 \\
1 & \sigma_N & \sigma_N^2 & \dots & \sigma_N^{N-1} \\
1 & \sigma_N^2 & \sigma_N^4 & \dots & \sigma_N^{2(N-1)} \\
\vdots & \vdots & \vdots & \ddots & \vdots \\
1 & \sigma_N^{N-1} & \sigma_N^{2(N-1)} & \dots & \sigma_N^{(N-1)(N-1)} \\
\end{pmatrix}
$$
In the following visualization, the real and imaginary part of $\mathrm{DFT}_N$ are shown, where the values are encoded by suitable colors. Note that the $k^\mathrm{th}$ row of $\mathrm{DFT}_N$ corresponds to the vector $\mathbf{e}_k$ as defined above.
```
def generate_matrix_dft(N, K):
"""Generate a DFT (discete Fourier transfrom) matrix
Notebook: PCP_09_dft.ipynb
Args:
N: Number of samples
K: Number of frequency bins
Returns:
dft: The DFT matrix
"""
dft = np.zeros((K, N), dtype=np.complex128)
time_index = np.arange(N)
for k in range(K):
dft[k, :] = np.exp(-2j * np.pi * k * time_index / N)
return dft
N = 32
dft_matrix = generate_matrix_dft(N, N)
plt.figure(figsize=(10, 4))
plt.subplot(1, 2, 1)
plt.title('$\mathrm{Re}(\mathrm{DFT}_N)$')
plt.imshow(np.real(dft_matrix), origin='lower', cmap='seismic', aspect='equal')
plt.xlabel('Time (sample, index $n$)')
plt.ylabel('Frequency (index $k$)')
plt.colorbar()
plt.subplot(1, 2, 2)
plt.title('$\mathrm{Im}(\mathrm{DFT}_N)$')
plt.imshow(np.imag(dft_matrix), origin='lower', cmap='seismic', aspect='equal')
plt.xlabel('Time (samples, index $n$)')
plt.ylabel('Frequency (index $k$)')
plt.colorbar()
plt.tight_layout()
```
We now write a function that computes the discrete Fourier transform $X = \mathrm{DFT}_N \cdot x$ of a signal $x\in\mathbb{C}^N$. We apply the function from above sampled at $N=64$ time points. The peaks of the magnitude Fourier transform $|X|$ correspond to the main frequency components the signal is composed of. Note that the magnitude Fourier transform is symmetrical around the center. Why? For the interpretation of the time and frequency axis, see also <a href='#exercise_freq_index'>Exercise 1: Interpretation of Frequency Indices</a></li>
```
def dft(x):
"""Compute the discete Fourier transfrom (DFT)
Notebook: PCP_09_dft.ipynb
Args:
x: Signal to be transformed
Returns:
X: Fourier transform of x
"""
x = x.astype(np.complex128)
N = len(x)
dft_mat = generate_matrix_dft(N, N)
return np.dot(dft_mat, x)
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
X = dft(x)
def plot_signal_dft(t, x, X, ax_sec=False, ax_Hz=False, freq_half=False, figsize=(10, 2)):
"""Plotting function for signals and its magnitude DFT
Notebook: PCP_09_dft.ipynb
Args:
t: Time axis (given in seconds)
x: Signal
X: DFT
ax_sec: Plots time axis in seconds (Default value = False)
ax_Hz: Plots frequency axis in Hertz (Default value = False)
freq_half: Plots only low half of frequency coefficients (Default value = False)
figsize: Size of figure (Default value = (10, 2))
"""
N = len(x)
if freq_half is True:
K = N // 2
X = X[:K]
else:
K = N
plt.figure(figsize=figsize)
ax = plt.subplot(1, 2, 1)
ax.set_title('$x$ with $N=%d$' % N)
if ax_sec is True:
ax.plot(t, x, 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Time (seconds)')
else:
ax.plot(x, 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Time (samples)')
ax.grid()
ax = plt.subplot(1, 2, 2)
ax.set_title('$|X|$')
if ax_Hz is True:
Fs = 1 / (t[1] - t[0])
ax_freq = Fs * np.arange(K) / N
ax.plot(ax_freq, np.abs(X), 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Frequency (Hz)')
else:
ax.plot(np.abs(X), 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Frequency (index)')
ax.grid()
plt.tight_layout()
plt.show()
plot_signal_dft(t, x, X)
plot_signal_dft(t, x, X, ax_sec=True, ax_Hz=True)
plot_signal_dft(t, x, X, ax_sec=True, ax_Hz=True, freq_half=True)
```
<a id='fft'></a>
## Fast Fourier Transform (FFT)
Next, we discuss the famous fast Fourier transform (FFT), which is a fast algorithm to compute the DFT. The FFT algorithm was originally found by Gauss in about 1805 and then rediscovered by Cooley and Tukey in 1965. The FFT algorithm is based on the observation that applying a DFT of even size $N=2M$ can be expressed in terms of applying two DFTs of half the size $M$. It exploits the fact that there are algebraic relations between the entries $\sigma_N^{kn} = \mathrm{exp}(-2 \pi i / N)^{kn}$ of DFT matrices. In particular, one has
$$\sigma_M = \sigma_N^2$$
In the FFT algorithm, one computes the DFT of the even-indexed and the uneven-indexed entries of $x$:
\begin{align}
(A(0), \dots, A(N/2-1)) &= \mathrm{DFT}_{N/2} \cdot (x(0), x(2), x(4), \dots, x(N-2))\\
(B(0), \dots, B(N/2-1)) &= \mathrm{DFT}_{N/2} \cdot (x(1), x(3), x(5), \dots, x(N-1))
\end{align}
With these two DFTs of size $N/2$, one can compute the full DFT of size $N$ via:
\begin{eqnarray}
C(k) &=& \sigma_N^k \cdot B(k)\\
X(k) &=& A(k) + C(k)\\
X(N/2 + k) &=& A(k) - C(k)\\
\end{eqnarray}
for $k \in [0: N/2 - 1]$. The numbers $\sigma_N^k$ are also called *twiddle factors*. If $N$ is a power of two, this idea can be applied recursively until one reaches the computation of $\mathrm{DFT}_{1}$ (the case $N=1$), which is simply multiplication by one (i.e. just returning the signal of length $N=1$). For further details, we refer to Section 2.4.3 of <a href="http://www.music-processing.de">[Müller, FMP, Springer 2015])</a> (see also Table 2.1).
In the following code, we provide a function `fft` that implements the FFT algorithm. We test the function `fft` by comparing its output with the one when applying the `dft` on a test signal `x`. For the comparison of result matrices, we use the NumPy functions [`np.array_equal`](https://numpy.org/doc/stable/reference/generated/numpy.array_equal.html) and [`np.allclose`](https://numpy.org/doc/stable/reference/generated/numpy.allclose.html#numpy.allclose).
```
def fft(x):
"""Compute the fast Fourier transform (FFT)
Notebook: PCP_09_dft.ipynb
Args:
x: Signal to be transformed
Returns:
X: Fourier transform of x
"""
x = x.astype(np.complex128)
N = len(x)
log2N = np.log2(N)
assert log2N == int(log2N), 'N must be a power of two!'
X = np.zeros(N, dtype=np.complex128)
if N == 1:
return x
else:
this_range = np.arange(N)
A = fft(x[this_range % 2 == 0])
B = fft(x[this_range % 2 == 1])
range_twiddle_k = np.arange(N // 2)
sigma = np.exp(-2j * np.pi * range_twiddle_k / N)
C = sigma * B
X[:N//2] = A + C
X[N//2:] = A - C
return X
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
X_via_dft = dft(x)
X_via_fft = fft(x)
X_via_fft_numpy = np.fft.fft(x)
is_equal = np.array_equal(X_via_dft, X_via_fft)
is_equal_tol = np.allclose(X_via_dft, X_via_fft)
is_equal_tol_np = np.allclose(X_via_dft, X_via_fft_numpy)
print('Equality test for dft(x) and fft(x) using np.array_equal: ', is_equal)
print('Equality test for dft(x) and fft(x) using np.allclose: ', is_equal_tol)
print('Equality test for dft(x) and np.fft.fft(x) using np.allclose:', is_equal_tol_np)
```
<div class="alert alert-block alert-warning">
<strong>Note:</strong> The test shows that our <code>dft</code> and <code>fft</code> implementations do not yield the same result (due to numerical issues). However, the results are numerically very close, which is verified by the test using <code>np.allclose</code>.
</div>
The FFT reduces the overall number of operations from the order of $N^2$ (needed when computing the usual matrix–vector product $\mathrm{DFT}_N \cdot x$) to the order of $N\log_2N$. The savings are enormous. For example, using $N=2^{10}=1024$, the FFT requires roughly $N\log_2N=10240$ instead of $N^2=1048576$ operations in the naive approach. Using the module `timeit`, which provides a simple way to time small bits of Python code, the following code compares the running time when using the naive approach and the FFT. Furthermore, we compare the running time with the highly optimized NumPy implementation <code>np.fft.fft</code>.
```
import timeit
rep = 3
for N in [256, 512, 1024, 2048, 4096]:
time_index = np.arange(N)
x = np.sin(2 * np.pi * time_index / N )
t_DFT = 1000 * timeit.timeit(lambda: dft(x), number=rep)/rep
t_FFT = timeit.timeit(lambda: fft(x), number=rep*5)/(rep*5)
t_FFT_np = timeit.timeit(lambda: np.fft.fft(x), number=rep*100)/(rep*100)
print(f'Runtime (ms) for N = {N:4d} : DFT {t_DFT:10.2f}, FFT {t_FFT:.5f}, FFT_np {t_FFT_np:.8f}')
```
## Exercises and Results
```
import libpcp.dft
show_result = True
```
<a id='exercise_freq_index'></a>
<div class="alert alert-block alert-info">
<strong>Exercise 1: Interpretation of Frequency Indices</strong><br>
Given a dimension $N\in\mathbb{N}$, the $\mathrm{DFT}_N$ transform a vector $x\in\mathbb{C}^N$ into another vector $X\in\mathbb{C}^N$. Assuming that $x$ represents a time-domain signal sampled with a sampling rate $F_\mathrm{s}$, one can associate the index $n\in[0:N-1]$ of the sample $x(n)$ with the physical time point $t = n/F_\mathrm{s}$ given in seconds. In case of the vector $X$, the index $k\in[0:N-1]$ of the coefficient $X(k)$ can be associated to a physical frequency value
$$
\omega=\frac{k \cdot F_\mathrm{s}}{N}.
$$
Furthermore, using a real-valued signal $x\in\mathbb{R}^N$, the upper part of $X\in\mathbb{C}^N$ becomes redundant, and it suffices to consider the first $K$ coefficients with $K=N/2$.
<ul>
<li>Find explanations why these properties apply.</li>
<li>Find out how the function <code>plot_signal_dft</code> uses these properties to convert and visualize the time and frequency axes.</li>
<li>Using the signal <code>x, t = libpcp.signal.generate_example_signal(Fs=64, dur=2)</code>, plot the signal and its magnitude Fourier transform once using axes given in indices and once using axes given in physical units (seconds, Hertz). Discuss the results.</li>
<li>Do the same for the signal <code>x, t = libpcp.signal.generate_example_signal(Fs=32, dur=2)</code>. What is going wrong and why?</li>
</ul>
</div>
```
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_freq_index(show_result=show_result)
```
<a id='exercise_missing_time'></a>
<div class="alert alert-block alert-info">
<strong>Exercise 2: Missing Time Localization</strong><br>
The Fourier transform yields frequency information that is averaged over the entire time axis. However, the information on when these frequencies occur is hidden in the transform. To demonstrate this phenomenon, construct the following two different signals defined on a common time axis $[0, T]$ with $T$ given in seconds (e.g., $T=6~\mathrm{sec}$).
<ul>
<li>A superposition of two sinusoids $f_1+f_2$ defined over the entire time interval $[0, T]$, where the first sinusoid $f_1$ has a frequency $\omega_1=1~\mathrm{Hz}$ and an amplitude of $1$, while the second sinusoid $f_2$ has a frequency $\omega_2=5~\mathrm{Hz}$ and an amplitude of $0.5$.</li>
<li>A concatenation of two sinusoids, where $f_1$ (specified as before) is now defined only on the subinterval $[0, T/2]$, and $f_2$ is defined on the subinterval $[T/2, T]$.
</ul>
Sample the interval $[0,T]$ to obtain $N$ samples (use <code>np.linspace</code>), with $N\in\mathbb{N}$ being power of two (e.g., $N=256$). Define DT-signals of the superposition and the concatenation and compute the DFT for each of the signals. Plot the signals as well as the resulting magnitude Fourier transforms and discuss the result.
</div>
```
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_missing_time(show_result=show_result)
```
<a id='exercise_chirp'></a>
<div class="alert alert-block alert-info">
<strong>Exercise 3: Chirp Signal</strong><br>
The function $f(t)=\sin\left(\pi t^2\right)$ defines a <strong>chirp signal</strong> (also called <strong>sweep signal</strong>), in which the frequency increases with time. The <strong>instantaneous frequency $\omega_t$</strong> of the chirp signal at time $t$ is the derivate of the sinusoid's argument divided by $2\pi$, thus $\omega_t = t$.
<ul>
<li>Let $[t_0,t_1]$ be a time interval (given in seconds) with $0\leq t_0<t_1$ and $N\in\mathbb{N}$ be power of two. Implement a function <code>generate_chirp</code> that outputs a sampled chirp signal <code>x</code> over the interval $[t_0,t_1]$ with $N$ samples (use <code>np.linspace</code>).</li>
<li>Compute the DFT of <code>x</code> for various input parameters $t_0$, $t_1$, and $N$. Plot the chirp signal as well as the resulting magnitude Fourier transform. Discuss the result.</li>
</ul>
</div>
```
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_chirp(show_result=show_result)
```
<a id='exercise_inverse'></a>
<div class="alert alert-block alert-info">
<strong>Exercise 4: Inverse DFT</strong><br>
The discrete Fourier transform given by the matrix $\mathrm{DFT}_N \in \mathbb{C}^{N\times N}$ is an invertible operation, given by the inverse DFT matrix $\mathrm{DFT}_N^{-1}$.
<ul>
<li>There is an explicit relation between $\mathrm{DFT}_N$ and its inverse $\mathrm{DFT}_N^{-1}$. Which one? </li>
<li>Write a function <code>generate_matrix_dft_inv</code> that explicitly generates $\mathrm{DFT}_N^{-1}$.
<li>Check your function by computing $\mathrm{DFT}_N \cdot \mathrm{DFT}_N^{-1}$ and $\mathrm{DFT}_N^{-1} \cdot \mathrm{DFT}_N$ (using <code>np.matmul</code>) and comparing these products with the identity matrix (using <code>np.eye</code> and <code>np.allclose</code>).</li>
<li>Furthermore, compute the inverse DFT by using <code>np.linalg.inv</code>. Compare the result with your function using <code>np.allclose</code>.
<li>Similar to <code>fft</code>, implement a fast inverse Fourier transform <code>fft_inv</code></li>
</ul>
</div>
```
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_inverse(show_result=show_result)
```
<div>
<a href="https://opensource.org/licenses/MIT"><img src="data_layout/PCP_License.png" width=100% style="float: right;" alt="PCP License"></a>
</div>
|
github_jupyter
|
import numpy as np
from matplotlib import pyplot as plt
import libpcp.signal
%matplotlib inline
Fs = 64
dur = 1
x1, t = libpcp.signal.generate_example_signal(Fs=Fs, dur=dur)
x2, t = libpcp.signal.generate_sinusoid(dur=dur, Fs=Fs, amp=1, freq=2, phase=0.3)
x3, t = libpcp.signal.generate_sinusoid(dur=dur, Fs=Fs, amp=1, freq=6, phase=0.1)
def plot_inner_product(ax, t, x, y, color_x='k', color_y='r', label_x='x', label_y='y'):
"""Plot inner product
Notebook: PCP_09_dft.ipynb
Args:
ax: Axis handle
t: Time axis
x: Signal x
y: Signal y
color_x: Color of signal x (Default value = 'k')
color_y: Color of signal y (Default value = 'r')
label_x: Label of signal x (Default value = 'x')
label_y: Label of signal y (Default value = 'y')
"""
ax.plot(t, x, color=color_x, linewidth=1.0, linestyle='-', label=label_x)
ax.plot(t, y, color=color_y, linewidth=1.0, linestyle='-', label=label_y)
ax.set_xlim([0, t[-1]])
ax.set_ylim([-1.5, 1.5])
ax.set_xlabel('Time (seconds)')
ax.set_ylabel('Amplitude')
sim = np.vdot(y, x)
ax.set_title(r'$\langle$ %s $|$ %s $\rangle = %.1f$' % (label_x, label_y, sim))
ax.legend(loc='upper right')
plt.figure(figsize=(8, 5))
ax = plt.subplot(2, 2, 1)
plot_inner_product(ax, t, x1, x1, color_x='k', color_y='k', label_x='$x_1$', label_y='$x_1$')
ax = plt.subplot(2, 2, 2)
plot_inner_product(ax, t, x1, x2, color_x='k', color_y='r', label_x='$x_1$', label_y='$x_2$')
ax = plt.subplot(2, 2, 3)
plot_inner_product(ax, t, x1, x3, color_x='k', color_y='b', label_x='$x_1$', label_y='$x_3$')
ax = plt.subplot(2, 2, 4)
plot_inner_product(ax, t, x2, x3, color_x='r', color_y='b', label_x='$x_2$', label_y='$x_3$')
plt.tight_layout()
def plot_signal_e_k(ax, x, k, show_e=True, show_opt=False):
"""Plot signal and k-th DFT sinusoid
Notebook: PCP_09_dft.ipynb
Args:
ax: Axis handle
x: Signal
k: Index of DFT
show_e: Shows cosine and sine (Default value = True)
show_opt: Shows cosine with optimal phase (Default value = False)
"""
N = len(x)
time_index = np.arange(N)
ax.plot(time_index, x, 'k', marker='.', markersize='10', linewidth=2.0, label='$x$')
plt.xlabel('Time (samples)')
e_k = np.exp(2 * np.pi * 1j * k * time_index / N)
c_k = np.real(e_k)
s_k = np.imag(e_k)
X_k = np.vdot(e_k, x)
plt.title(r'k = %d: Re($X(k)$) = %0.2f, Im($X(k)$) = %0.2f, $|X(k)|$=%0.2f' %
(k, X_k.real, X_k.imag, np.abs(X_k)))
if show_e is True:
ax.plot(time_index, c_k, 'r', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\mathrm{Re}(\overline{\mathbf{u}}_k)$')
ax.plot(time_index, s_k, 'b', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\mathrm{Im}(\overline{\mathbf{u}}_k)$')
if show_opt is True:
phase_k = - np.angle(X_k) / (2 * np.pi)
cos_k_opt = np.cos(2 * np.pi * (k * time_index / N - phase_k))
d_k = np.sum(x * cos_k_opt)
ax.plot(time_index, cos_k_opt, 'g', marker='.', markersize='5',
linewidth=1.0, linestyle=':', label='$\cos_{k, opt}$')
plt.grid()
plt.legend(loc='lower right')
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
plt.figure(figsize=(8, 15))
for k in range(1, 8):
ax = plt.subplot(7, 1, k)
plot_signal_e_k(ax, x, k=k)
plt.tight_layout()
plt.figure(figsize=(8, 15))
for k in range(1, 8):
ax = plt.subplot(7, 1, k)
plot_signal_e_k(ax, x, k=k, show_e=False, show_opt=True)
plt.tight_layout()
def generate_matrix_dft(N, K):
"""Generate a DFT (discete Fourier transfrom) matrix
Notebook: PCP_09_dft.ipynb
Args:
N: Number of samples
K: Number of frequency bins
Returns:
dft: The DFT matrix
"""
dft = np.zeros((K, N), dtype=np.complex128)
time_index = np.arange(N)
for k in range(K):
dft[k, :] = np.exp(-2j * np.pi * k * time_index / N)
return dft
N = 32
dft_matrix = generate_matrix_dft(N, N)
plt.figure(figsize=(10, 4))
plt.subplot(1, 2, 1)
plt.title('$\mathrm{Re}(\mathrm{DFT}_N)$')
plt.imshow(np.real(dft_matrix), origin='lower', cmap='seismic', aspect='equal')
plt.xlabel('Time (sample, index $n$)')
plt.ylabel('Frequency (index $k$)')
plt.colorbar()
plt.subplot(1, 2, 2)
plt.title('$\mathrm{Im}(\mathrm{DFT}_N)$')
plt.imshow(np.imag(dft_matrix), origin='lower', cmap='seismic', aspect='equal')
plt.xlabel('Time (samples, index $n$)')
plt.ylabel('Frequency (index $k$)')
plt.colorbar()
plt.tight_layout()
def dft(x):
"""Compute the discete Fourier transfrom (DFT)
Notebook: PCP_09_dft.ipynb
Args:
x: Signal to be transformed
Returns:
X: Fourier transform of x
"""
x = x.astype(np.complex128)
N = len(x)
dft_mat = generate_matrix_dft(N, N)
return np.dot(dft_mat, x)
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
X = dft(x)
def plot_signal_dft(t, x, X, ax_sec=False, ax_Hz=False, freq_half=False, figsize=(10, 2)):
"""Plotting function for signals and its magnitude DFT
Notebook: PCP_09_dft.ipynb
Args:
t: Time axis (given in seconds)
x: Signal
X: DFT
ax_sec: Plots time axis in seconds (Default value = False)
ax_Hz: Plots frequency axis in Hertz (Default value = False)
freq_half: Plots only low half of frequency coefficients (Default value = False)
figsize: Size of figure (Default value = (10, 2))
"""
N = len(x)
if freq_half is True:
K = N // 2
X = X[:K]
else:
K = N
plt.figure(figsize=figsize)
ax = plt.subplot(1, 2, 1)
ax.set_title('$x$ with $N=%d$' % N)
if ax_sec is True:
ax.plot(t, x, 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Time (seconds)')
else:
ax.plot(x, 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Time (samples)')
ax.grid()
ax = plt.subplot(1, 2, 2)
ax.set_title('$|X|$')
if ax_Hz is True:
Fs = 1 / (t[1] - t[0])
ax_freq = Fs * np.arange(K) / N
ax.plot(ax_freq, np.abs(X), 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Frequency (Hz)')
else:
ax.plot(np.abs(X), 'k', marker='.', markersize='3', linewidth=0.5)
ax.set_xlabel('Frequency (index)')
ax.grid()
plt.tight_layout()
plt.show()
plot_signal_dft(t, x, X)
plot_signal_dft(t, x, X, ax_sec=True, ax_Hz=True)
plot_signal_dft(t, x, X, ax_sec=True, ax_Hz=True, freq_half=True)
def fft(x):
"""Compute the fast Fourier transform (FFT)
Notebook: PCP_09_dft.ipynb
Args:
x: Signal to be transformed
Returns:
X: Fourier transform of x
"""
x = x.astype(np.complex128)
N = len(x)
log2N = np.log2(N)
assert log2N == int(log2N), 'N must be a power of two!'
X = np.zeros(N, dtype=np.complex128)
if N == 1:
return x
else:
this_range = np.arange(N)
A = fft(x[this_range % 2 == 0])
B = fft(x[this_range % 2 == 1])
range_twiddle_k = np.arange(N // 2)
sigma = np.exp(-2j * np.pi * range_twiddle_k / N)
C = sigma * B
X[:N//2] = A + C
X[N//2:] = A - C
return X
N = 64
x, t = libpcp.signal.generate_example_signal(Fs=N, dur=1)
X_via_dft = dft(x)
X_via_fft = fft(x)
X_via_fft_numpy = np.fft.fft(x)
is_equal = np.array_equal(X_via_dft, X_via_fft)
is_equal_tol = np.allclose(X_via_dft, X_via_fft)
is_equal_tol_np = np.allclose(X_via_dft, X_via_fft_numpy)
print('Equality test for dft(x) and fft(x) using np.array_equal: ', is_equal)
print('Equality test for dft(x) and fft(x) using np.allclose: ', is_equal_tol)
print('Equality test for dft(x) and np.fft.fft(x) using np.allclose:', is_equal_tol_np)
import timeit
rep = 3
for N in [256, 512, 1024, 2048, 4096]:
time_index = np.arange(N)
x = np.sin(2 * np.pi * time_index / N )
t_DFT = 1000 * timeit.timeit(lambda: dft(x), number=rep)/rep
t_FFT = timeit.timeit(lambda: fft(x), number=rep*5)/(rep*5)
t_FFT_np = timeit.timeit(lambda: np.fft.fft(x), number=rep*100)/(rep*100)
print(f'Runtime (ms) for N = {N:4d} : DFT {t_DFT:10.2f}, FFT {t_FFT:.5f}, FFT_np {t_FFT_np:.8f}')
import libpcp.dft
show_result = True
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_freq_index(show_result=show_result)
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_missing_time(show_result=show_result)
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_chirp(show_result=show_result)
#<solution>
# Your Solution
#</solution>
libpcp.dft.exercise_inverse(show_result=show_result)
| 0.831074 | 0.995558 |
# Visualization Notebook
In this notebook, the user can interact with trained models from the paper EpidemiOptim: A Toolbox for the Optimization of Control Policies in Epidemiological Models.
```
%matplotlib notebook
import sys
sys.path.append('../../')
from epidemioptim.analysis.notebook_utils import setup_visualization
from epidemioptim.utils import get_repo_path
```
Pick your parameters: seed, algorithm and epidemiological model determinism. Currently implemented:
### DQN
The user can control the mixing parameter $\beta$ that tunes the balance between economic $(C_e)$ and health $(C_h)$ costs: $C=(1-\beta)~C_h + \beta~C_e$. When it does so, the corresponding DQN policy is loaded and run in the model for one year. The user can then visualize the evolution of costs over a year. Lockdown enforcement are marked by red dots.
### GOAL_DQN
The user can control the mixing parameter $\beta$. There is only one policy. For each $\beta$, the optimal actions are selected according to the corresponding balance between Q-functions: $a^* = \text{argmax}_a (1-\beta)~Q_h + \beta~Q_e$. Each time $\beta$ is changed, the model is run over a year.
### GOAL_DQN_CONSTRAINTS
The user can control $\beta$ and the values of constraints on the maximum values of cumulative costs ($M_e$, $M_h$). Each time new parameters are selected, the resulting policy is run in the model over a year.
### NSGA
The user can observe the Pareto front produced by a run of the NSGA-II algorithm. When clicking on a solution from the Pareto front, the corresponding policy is loaded and run in the model over a year.
```
seed = None # None picks a random seed
algorithm = 'NSGA' # Pick from ['DQN', 'GOAL_DQN_CONSTRAINTS', 'GOAL_DQN', 'NSGA']
deterministic_model = False # whether the model is deterministic or not
```
The cell below launches the visualization.
```
valid_algorithms = ['DQN', 'GOAL_DQN_CONSTRAINTS', 'GOAL_DQN', 'NSGA']
assert algorithm in valid_algorithms, "Pick an algorithm from" + str(valid_algorithms)
if algorithm == 'DQN':
folder = get_repo_path() + "/data/data_for_visualization/DQN/"
elif algorithm == 'GOAL_DQN':
folder = get_repo_path() + "/data/data_for_visualization/GOAL_DQN/1/"
elif algorithm == 'GOAL_DQN_CONSTRAINTS':
folder = get_repo_path() + "/data/data_for_visualization/GOAL_DQN_CONST/1/"
elif algorithm == 'NSGA':
folder = get_repo_path() + "/data/data_for_visualization/NSGA/1/"
else:
raise NotImplementedError
setup_visualization(folder, algorithm, seed, deterministic_model)
```
|
github_jupyter
|
%matplotlib notebook
import sys
sys.path.append('../../')
from epidemioptim.analysis.notebook_utils import setup_visualization
from epidemioptim.utils import get_repo_path
seed = None # None picks a random seed
algorithm = 'NSGA' # Pick from ['DQN', 'GOAL_DQN_CONSTRAINTS', 'GOAL_DQN', 'NSGA']
deterministic_model = False # whether the model is deterministic or not
valid_algorithms = ['DQN', 'GOAL_DQN_CONSTRAINTS', 'GOAL_DQN', 'NSGA']
assert algorithm in valid_algorithms, "Pick an algorithm from" + str(valid_algorithms)
if algorithm == 'DQN':
folder = get_repo_path() + "/data/data_for_visualization/DQN/"
elif algorithm == 'GOAL_DQN':
folder = get_repo_path() + "/data/data_for_visualization/GOAL_DQN/1/"
elif algorithm == 'GOAL_DQN_CONSTRAINTS':
folder = get_repo_path() + "/data/data_for_visualization/GOAL_DQN_CONST/1/"
elif algorithm == 'NSGA':
folder = get_repo_path() + "/data/data_for_visualization/NSGA/1/"
else:
raise NotImplementedError
setup_visualization(folder, algorithm, seed, deterministic_model)
| 0.26218 | 0.959078 |
# Aerospace Design via Quasiconvex Optimization
Consider a triangle, or a wedge, located within a hypersonic flow. A standard aerospace design optimization problem is to design the wedge to maximize the lift-to-drag ratio (L/D) (or conversely minimize the D/L ratio), subject to certain geometric constraints. In this example, the wedge is known to have a constant hypotenuse, and our job is to choose its width and height.
The drag-to-lift ratio is given by
\begin{equation}
\frac{\mathrm{D}}{\mathrm{L}} = \frac{\mathrm{c_d}}{\mathrm{c_l}},
\end{equation}
where $\mathrm{c_d}$ and $\mathrm{c_l}$ are drag and lift coefficients, respectively, that are obtained by integrating the projection of the pressure coefficient in directions parallel to, and perpendicular to, the body.
It turns out that the the drag-to-lift ratio is a quasilinear function, as we'll now show. We will assume the pressure coefficient is given by the Newtonian sine-squared law for whetted areas of the body,
\begin{equation}
\mathrm{c_p} = 2(\hat{v}\cdot\hat{n})^2
\end{equation}
and elsewhere $\mathrm{c_p} = 0$. Here, $\hat{v}$ is the free stream direction, which for simplicity we will assume is parallel to the body so that, $\hat{v} = \langle 1, 0 \rangle$, and $\hat{n}$ is the local unit normal. For a wedge defined by width $\Delta x$, and height $\Delta y$,
\begin{equation}
\hat{n} = \langle -\Delta y/s,-\Delta x/s \rangle
\end{equation}
where $s$ is the hypotenuse length. Therefore,
\begin{equation}
\mathrm{c_p} = 2((1)(-\Delta y/s)+(0)(-\Delta x/s))^2 = \frac{2 \Delta y^2}{s^2}
\end{equation}
The lift and drag coefficients are given by
\begin{align*}
\mathrm{c_d} &= \frac{1}{c}\int_0^s -\mathrm{c_p}\hat{n}_x \mathrm{d}s \\
\mathrm{c_l} &= \frac{1}{c}\int_0^s -\mathrm{c_p}\hat{n}_y \mathrm{d}s
\end{align*}
Where $c$ is the reference chord length of the body. Given that $\hat{n}$, and therefore $\mathrm{c_p}$ are constant over the whetted surface of the body,
\begin{align*}
\mathrm{c_d} &= -\frac{s}{c}\mathrm{c_p}\hat{n}_x = \frac{s}{c}\frac{2 \Delta y^2}{s^2}\frac{\Delta y}{s} \\
\mathrm{c_l} &= -\frac{s}{c}\mathrm{c_p}\hat{n}_y = \frac{s}{c}\frac{2 \Delta y^2}{s^2}\frac{\Delta x}{s}
\end{align*}
Assuming $s=1$, so that $\Delta y = \sqrt{1-\Delta x^2}$, plugging in the above into the equation for $D/L$, we obtain
\begin{equation}
\frac{\mathrm{D}}{\mathrm{L}} = \frac{\Delta y}{\Delta x} = \frac{\sqrt{1-\Delta x^2}}{\Delta x} = \sqrt{\frac{1}{\Delta x^2}-1}.
\end{equation}
This function is representable as a DQCP, quasilinear function. We plot it below, and then we write it using DQCP.
```
%matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
import math
x = np.linspace(.25,1,num=201)
obj = []
for i in range(len(x)):
obj.append(math.sqrt(1/x[i]**2-1))
plt.plot(x,obj)
import cvxpy as cp
x = cp.Variable(pos=True)
obj = cp.sqrt(cp.inv_pos(cp.square(x))-1)
print("This objective function is", obj.curvature)
```
Minimizing this objective function subject to constraints representing payload requirements is a standard aerospace design problem. In this case we will consider the constraint that the wedge must be able to contain a rectangle of given length and width internally along its hypotenuse. This is representable as a convex constraint.
```
a = .05 # USER INPUT: height of rectangle, should be at most b
b = .65 # USER INPUT: width of rectangle
constraint = [a*cp.inv_pos(x)-(1-b)*cp.sqrt(1-cp.square(x))<=0]
print(constraint)
prob = cp.Problem(cp.Minimize(obj), constraint)
prob.solve(qcp=True, verbose=True)
print('Final L/D Ratio = ', 1/obj.value)
print('Final width of wedge = ', x.value)
print('Final height of wedge = ', math.sqrt(1-x.value**2))
```
Once the solution has been found, we can create a plot to verify that the rectangle is inscribed within the wedge.
```
y = math.sqrt(1-x.value**2)
lambda1 = a*x.value/y
lambda2 = a*x.value**2/y+a*y
lambda3 = a*x.value-y*(a*x.value/y-b)
plt.plot([0,x.value],[0,0],'b.-')
plt.plot([0,x.value],[0,-y],'b.-')
plt.plot([x.value,x.value],[0,-y],'b.-')
pt1 = [lambda1*x.value,-lambda1*y]
pt2 = [(lambda1+b)*x.value,-(lambda1+b)*y]
pt3 = [(lambda1+b)*x.value+a*y,-(lambda1+b)*y+a*x.value]
pt4 = [lambda1*x.value+a*y,-lambda1*y+a*x.value]
plt.plot([pt1[0],pt2[0]],[pt1[1],pt2[1]],'r.-')
plt.plot([pt2[0],pt3[0]],[pt2[1],pt3[1]],'r.-')
plt.plot([pt3[0],pt4[0]],[pt3[1],pt4[1]],'r.-')
plt.plot([pt4[0],pt1[0]],[pt4[1],pt1[1]],'r.-')
plt.axis('equal')
```
|
github_jupyter
|
%matplotlib inline
import matplotlib
import numpy as np
import matplotlib.pyplot as plt
import math
x = np.linspace(.25,1,num=201)
obj = []
for i in range(len(x)):
obj.append(math.sqrt(1/x[i]**2-1))
plt.plot(x,obj)
import cvxpy as cp
x = cp.Variable(pos=True)
obj = cp.sqrt(cp.inv_pos(cp.square(x))-1)
print("This objective function is", obj.curvature)
a = .05 # USER INPUT: height of rectangle, should be at most b
b = .65 # USER INPUT: width of rectangle
constraint = [a*cp.inv_pos(x)-(1-b)*cp.sqrt(1-cp.square(x))<=0]
print(constraint)
prob = cp.Problem(cp.Minimize(obj), constraint)
prob.solve(qcp=True, verbose=True)
print('Final L/D Ratio = ', 1/obj.value)
print('Final width of wedge = ', x.value)
print('Final height of wedge = ', math.sqrt(1-x.value**2))
y = math.sqrt(1-x.value**2)
lambda1 = a*x.value/y
lambda2 = a*x.value**2/y+a*y
lambda3 = a*x.value-y*(a*x.value/y-b)
plt.plot([0,x.value],[0,0],'b.-')
plt.plot([0,x.value],[0,-y],'b.-')
plt.plot([x.value,x.value],[0,-y],'b.-')
pt1 = [lambda1*x.value,-lambda1*y]
pt2 = [(lambda1+b)*x.value,-(lambda1+b)*y]
pt3 = [(lambda1+b)*x.value+a*y,-(lambda1+b)*y+a*x.value]
pt4 = [lambda1*x.value+a*y,-lambda1*y+a*x.value]
plt.plot([pt1[0],pt2[0]],[pt1[1],pt2[1]],'r.-')
plt.plot([pt2[0],pt3[0]],[pt2[1],pt3[1]],'r.-')
plt.plot([pt3[0],pt4[0]],[pt3[1],pt4[1]],'r.-')
plt.plot([pt4[0],pt1[0]],[pt4[1],pt1[1]],'r.-')
plt.axis('equal')
| 0.292191 | 0.992229 |
# Classificação de músicas do Spotify
# Decision tree
Árvores de decisão são modelos estatísticos que utilizam um treinamento supervisionado para a classificação e previsão de dados. Uma árvore de decisão é utilizada para representar visualmente e explicitamente as decisões e a tomada de decisão. Apesar de ser amplamente utilizada em mineração de dados com algumas variações pode e é amplamente utilizada em aprendizado de máquina.
### Representação de uma árvore de decisão:
Uma arvore de decisão como representada na figura abaixo é representada por nós e ramos, onde cada nó representa um atributo a ser testado e cada ramo descendente representa um possível valor para tal atributo.

+ Dataset: https://www.kaggle.com/geomack/spotifyclassification
+ Info sobre as colunas: https://developer.spotify.com/web-api/get-audio-features/
+ https://graphviz.gitlab.io/download/
+ conda install -c anaconda graphviz
+ conda install -c conda-forge pydotplus
```
import graphviz
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import pydotplus
import io
from scipy import misc
from sklearn import tree # pack age tree
from sklearn.metrics import accuracy_score # medir % acerto
from sklearn.model_selection import train_test_split # cortar dataset
from sklearn.tree import DecisionTreeClassifier, export_graphviz # arvore de decixao classificacao e graphviz para visualizar
```
*** Verificando os dados ***
```
df = pd.read_csv('data.csv')
df.head(3)
df.describe()
```
## Divisão treino e teste
### Divisão 70 - 30
```
train, test = train_test_split(df, test_size=round(len(df)*0.3))
print('Tamanho do set de treino: {},\nTamanho teste: {}'.format(len(train), len(test) ))
df.head()
```
O objetivo é dividir em grupos homogênioss com valor de 1 ou 0, dando uma serie de "caminhos" para determinar se o usuário gostou ou não da música.
```
# falar de overfitting comparando pagode funk e etc
# quanto maior o valor menor a arvore
tree = DecisionTreeClassifier(min_samples_split=100)
tree
features = ["danceability", "loudness", "valence", "energy", "instrumentalness", "acousticness", "key", "speechiness", "duration_ms"]
x_train = train[features]
y_train = train['target']
x_test = test[features]
y_test = test['target']
dct = tree.fit(x_train, y_train) # scikit fez uma decision tree
# visualizando
def showTree(tree, features, path):
file=io.StringIO()
export_graphviz(tree, out_file=file, feature_names=features)
pydotplus.graph_from_dot_data(file.getvalue()).write_png(path)
img = misc.imread(path)
plt.rcParams["figure.figsize"] = (20, 20)
plt.imshow(img)
%%time
showTree(dct, features, 'minhaprimeiradct.png')
y_pred = tree.predict(x_test)
y_pred
score = accuracy_score(y_test, y_pred)*100
print('Score = {}'.format(score))
```
# Overfitting e Underfitting
*** Underfitting:*** Quando você treina seu algoritmo e testa ele no próprio conjunto de treino e percebe que ele ainda tem uma tacha de erro considerável e então testa ele no conjunto de teste e percebe que a taxa de erro é semelhante mas ainda alta.
Isso quer dizer que estamos diante de um caso de Underfitting o algoritmo tem um alto Bias e ainda podemos melhorar sua classificação, para isso deveremos mexer em alguns parâmetros do algoritmo.
Claro que em nem todos os casos ira ocorrer dessa forma depende da natureza do algoritmo.
*** Overfitting: *** Agora você treinou seu algoritmo e depois disso resolveu aplicá-lo em seu conjunto de treino e fica feliz quando percebe que ele teve uma taxa de erro de 00,35% por exemplo. Mas quando aplica no conjunto de teste percebe que ele tem um desempenho horrível.
# Random Forest Classifier
Florestas aleatórias ou florestas de decisão aleatórias são um método de aprendizado conjunto para classificação, regressão e outras tarefas que operam construindo uma multiplicidade de árvores de decisão no momento do treinamento e gerando a classe que é o modo das classes (classificação) ou predição de média ) das árvores individuais. Florestas de decisão aleatórias corrigem o hábito das árvores de decisão de overfitting em seu conjunto de treinamento.
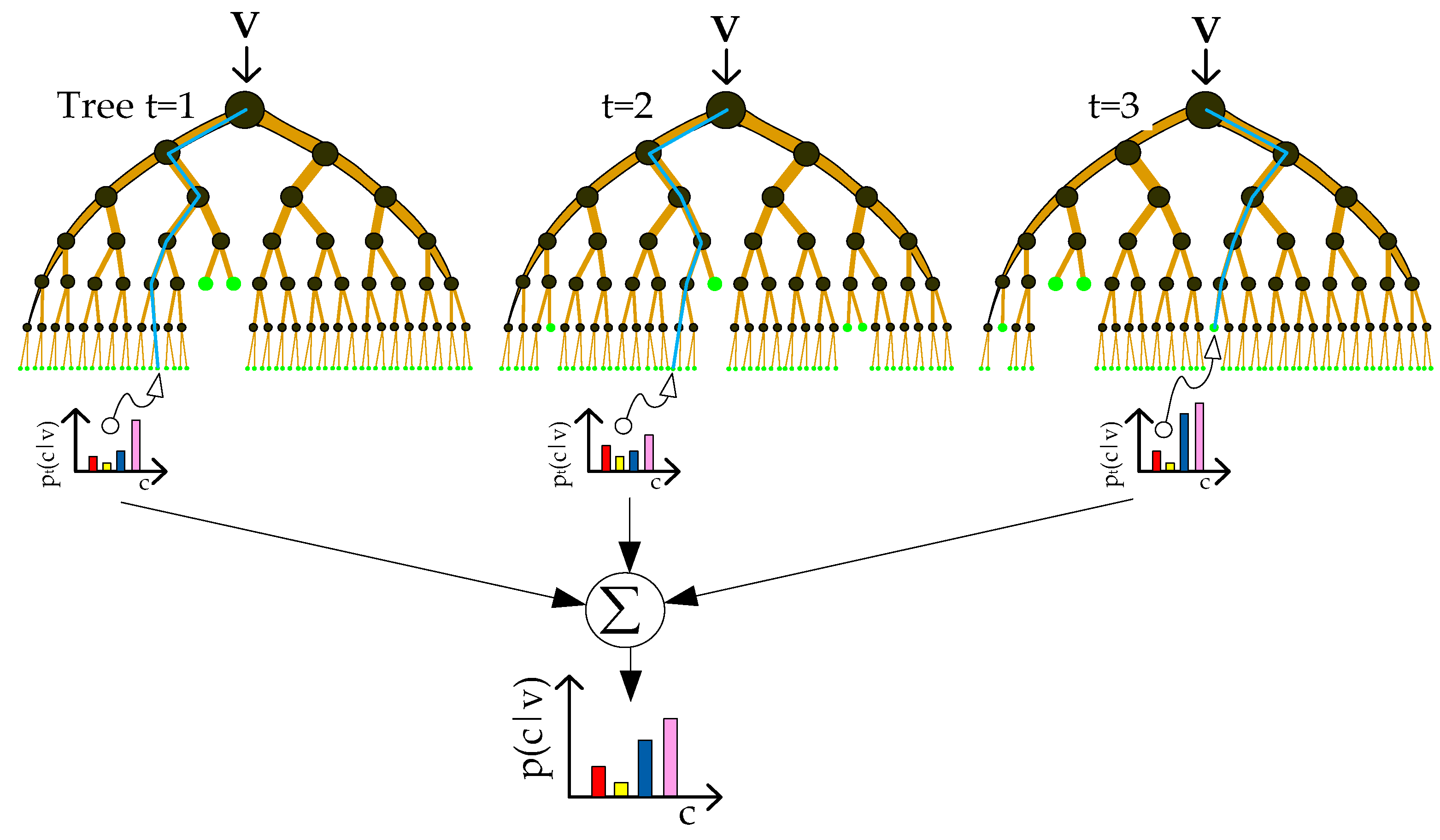
```
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators = 100)
clf.fit(x_train, y_train)
f_ypred = clf.predict(x_test)
score = accuracy_score(y_test, f_ypred) * 100
print('Score da decision tree: {}'.format(score))
```
# DESAFIO
https://www.kaggle.com/c/wine-quality-decision-tree
|
github_jupyter
|
import graphviz
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import pydotplus
import io
from scipy import misc
from sklearn import tree # pack age tree
from sklearn.metrics import accuracy_score # medir % acerto
from sklearn.model_selection import train_test_split # cortar dataset
from sklearn.tree import DecisionTreeClassifier, export_graphviz # arvore de decixao classificacao e graphviz para visualizar
df = pd.read_csv('data.csv')
df.head(3)
df.describe()
train, test = train_test_split(df, test_size=round(len(df)*0.3))
print('Tamanho do set de treino: {},\nTamanho teste: {}'.format(len(train), len(test) ))
df.head()
# falar de overfitting comparando pagode funk e etc
# quanto maior o valor menor a arvore
tree = DecisionTreeClassifier(min_samples_split=100)
tree
features = ["danceability", "loudness", "valence", "energy", "instrumentalness", "acousticness", "key", "speechiness", "duration_ms"]
x_train = train[features]
y_train = train['target']
x_test = test[features]
y_test = test['target']
dct = tree.fit(x_train, y_train) # scikit fez uma decision tree
# visualizando
def showTree(tree, features, path):
file=io.StringIO()
export_graphviz(tree, out_file=file, feature_names=features)
pydotplus.graph_from_dot_data(file.getvalue()).write_png(path)
img = misc.imread(path)
plt.rcParams["figure.figsize"] = (20, 20)
plt.imshow(img)
%%time
showTree(dct, features, 'minhaprimeiradct.png')
y_pred = tree.predict(x_test)
y_pred
score = accuracy_score(y_test, y_pred)*100
print('Score = {}'.format(score))
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators = 100)
clf.fit(x_train, y_train)
f_ypred = clf.predict(x_test)
score = accuracy_score(y_test, f_ypred) * 100
print('Score da decision tree: {}'.format(score))
| 0.481941 | 0.924279 |
```
# default_exp makers
```
# neos.makers
> Functions that define the workflow from parametric observable --> statistical model.
This module contains example workflows to go from the output of a neural network to a differentiable histogram, and to then use that as a basis for statistical modelling via the [HistFactory likelihood specification](https://scikit-hep.org/pyhf/intro.html#histfactory).
These functions are designed to be composed such that a final metric (e.g. expected p-value) is explicitly made a function of the parameters of the neural network. You can see this behaviour through the nested function design; one can specify all other hyperparameters ahead of time when initializing the functions, and the nn weights don't have to be specified until the inner function is called. Keep reading for examples!
## differentiable histograms from neural networks
```
#export
from functools import partial
import jax
import jax.numpy as jnp
import numpy as np
from relaxed import hist_kde as hist
#export
def hists_from_nn(
data_generator, predict, hpar_dict, method="softmax", LUMI=10, sig_scale=2, bkg_scale=10, reflect_infinities=False
):
"""Initialize a function `hist_maker` that returns a 'soft' histogram based
on a neural network with a softmax output. Choose which example problem to
try by setting the `example` argument.
Args:
data_generator: Callable that returns generated data (in jax array
format).
predict: Decision function for a parameterized observable, e.g. neural
network.
method: A string to specify the method to use for constructing soft
histograms. Either "softmax" or "kde".
LUMI: 'Luminosity' scaling factor for the yields.
sig_scale: Individual scaling factor for the signal yields.
bkg_scale: Individual scaling factor for the signal yields.
Returns:
hist_maker: A callable function that takes the parameters of the
observable (and optional hyperpars), then constructs signal,
background, and background uncertainty yields.
"""
data = data_generator()
if len(data) == 3:
if method == "softmax":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Two background
distributions are sampled from, which is meant to mimic the
situation in particle physics where one has a 'nominal'
prediction for a nuisance parameter (taken here as the mean of
two modes) and then alternate values (e.g. from varying up/down
by one standard deviation), which then modifies the background
pdf. Here, we take that effect to be a shift of the mean of the
distribution. The value for the background histogram is then
the mean of the resulting counts of the two modes, and the
uncertainty can be quantified through the count standard
deviation.
Arguments:
hm_params: a list containing:
nn: jax array of observable parameters.
"""
nn = hm_params
s, b_up, b_down = data
NMC = len(s)
s_hist = predict(nn, s).sum(axis=0) * sig_scale / NMC * LUMI
b_hists = [
predict(nn, b_up).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_down).sum(axis=0) * bkg_scale / NMC * LUMI,
]
b_mean = jnp.mean(jnp.asarray(b_hists), axis=0)
b_unc = jnp.std(jnp.asarray(b_hists), axis=0)
return s_hist, b_mean, b_unc
elif method == "kde":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data using a kde, all drawn from
multivariate normal distributions with different means. Two
background distributions are sampled from, which is meant to
mimic the situation in particle physics where one has a
'nominal' prediction for a nuisance parameter (taken here as
the mean of two modes) and then alternate values (e.g. from
varying up/down by one standard deviation), which then modifies
the background pdf. Here, we take that effect to be a shift of
the mean of the distribution. The value for the background
histogram is then the mean of the resulting counts of the two
modes, and the uncertainty can be quantified through the count
standard deviation.
Arguments:
hm_params: Array-like, consisting of:
nn: jax array of observable parameters.
bins: Array of bin edges, e.g. np.linspace(0,1,3)
defines a two-bin histogram with edges at 0, 0.5,
1.
bandwidth: Float that controls the 'smoothness' of the
kde. It's recommended to keep this fairly
similar to the bin width to avoid
oversmoothing the distribution. Going too low
will cause things to break, as the gradients
of the kde become unstable.
"""
nn = hm_params
bins, bandwidth = hpar_dict["bins"], hpar_dict["bandwidth"]
s, b_up, b_down = data
NMC = len(s)
nn_s, nn_b_up, nn_b_down = (
predict(nn, s).ravel(),
predict(nn, b_up).ravel(),
predict(nn, b_down).ravel(),
)
s_hist = hist(nn_s, bins, bandwidth, reflect_infinities=reflect_infinities) * sig_scale / NMC * LUMI
b_hists = jnp.asarray(
[
hist(nn_b_up, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_down, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
]
)
kde_counts = [
s_hist,
jnp.mean(b_hists, axis=0),
jnp.std(b_hists, axis=0),
]
return kde_counts
else:
assert False, (
f"Unsupported method: {method}"
" (only using kde or softmax for these examples)."
)
elif len(data) == 4:
if method == "softmax":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Three background
distributions are sampled from, which mimics the situation in
particle physics where one has a 'nominal' prediction for a
nuisance parameter (taken here as the mean of two modes) and
then alternate values (e.g. from varying up/down by one
standard deviation), which then modifies the background pdf.
Here, we take that effect to be a shift of the mean of the
distribution. The HistFactory 'histosys' nusiance parameter
will then be constructed from the yields downstream by
interpolating between them using pyhf.
Arguments:
hm_params: a list containing:
nn: jax array of observable parameters.
Returns:
Set of 4 counts for signal, background, and up/down modes.
"""
nn = hm_params
s, b_nom, b_up, b_down = data
NMC = len(s)
counts = [
predict(nn, s).sum(axis=0) * sig_scale / NMC * LUMI,
predict(nn, b_nom).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_up).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_down).sum(axis=0) * bkg_scale / NMC * LUMI,
]
return counts
elif method == "kde":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Three background
distributions are sampled from, which mimics the situation in
particle physics where one has a 'nominal' prediction for a
nuisance parameter (taken here as the mean of two modes) and
then alternate values (e.g. from varying up/down by one
standard deviation), which then modifies the background pdf.
Here, we take that effect to be a shift of the mean of the
distribution. The HistFactory 'histosys' nusiance parameter
will then be constructed from the yields downstream by
interpolating between them using pyhf.
Arguments:
hm_params: Array-like, consisting of:
nn: jax array of observable parameters.
bins: Array of bin edges, e.g. np.linspace(0,1,3)
defines a two-bin histogram with edges at 0, 0.5,
1.
bandwidth: Float that controls the 'smoothness' of the
kde. It's recommended to keep this fairly
similar to the bin width to avoid
oversmoothing the distribution. Going too low
will cause things to break, as the gradients
of the kde become unstable.
Returns:
Set of 4 counts for signal, background, and up/down modes.
"""
nn = hm_params
bins, bandwidth = hpar_dict["bins"], hpar_dict["bandwidth"]
s, b_nom, b_up, b_down = data
NMC = len(s)
nn_s, nn_b_nom, nn_b_up, nn_b_down = (
predict(nn, s).ravel(),
predict(nn, b_nom).ravel(),
predict(nn, b_up).ravel(),
predict(nn, b_down).ravel(),
)
kde_counts = [
hist(nn_s, bins, bandwidth, reflect_infinities=reflect_infinities) * sig_scale / NMC * LUMI,
hist(nn_b_nom, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_up, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_down, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
]
return [k + 1e-8 for k in kde_counts]
else:
assert False, (
f"Unsupported method: {method}"
" (only using kde or softmax for these examples)."
)
else:
assert False, (
f"Unsupported number of blobs: {blobs}"
" (only using 3 or 4 blobs for these examples)."
)
return hist_maker
```
### Usage:
Begin by instantiating `hists_from_nn` with a function that generates a 3 or 4-tuple of data (we have `generate_blobs` for this!), and a neural network `predict` method (takes inputs & weights, returns output)
```
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn
from neos.data import generate_blobs
# data generator
gen_data = generate_blobs(rng=PRNGKey(1),blobs=4)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
)
```
Now, when we initialize our neural network's weights and pass them to `hist_maker` along with some hyperparameters for the histogram (binning, bandwidth), we should get back a set of event yields:
```
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
hist_maker(network)
```
## statistical models
```
#export
import pyhf
jax_backend = pyhf.tensor.jax_backend(precision="64b")
pyhf.set_backend(jax_backend)
from neos.models import hepdata_like
#export
def hepdata_like_from_hists(histogram_maker):
"""Returns a function that constructs a typical 'hepdata-like' statistical
model with signal, background, and background uncertainty yields when
evaluated at the parameters of the observable.
Args:
histogram_maker: A function that, when called, returns a secondary function
that takes the observable's parameters as argument, and returns yields.
Returns:
nn_model_maker: A function that returns a Model object (either from
`neos.models` or from `pyhf`) when evaluated at the observable's parameters,
along with the background-only parameters for use in downstream inference.
"""
def nn_model_maker(hm_params):
s, b, db = histogram_maker(hm_params)
m = hepdata_like(s, b, db) # neos 'pyhf' model
nompars = m.config.suggested_init()
bonlypars = jnp.asarray([x for x in nompars])
bonlypars = jax.ops.index_update(bonlypars, m.config.poi_index, 0.0)
return m, bonlypars
return nn_model_maker
```
### Usage:
```
# define a hist_maker as above
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn, hepdata_like_from_hists
from neos.data import generate_blobs
# data generator, three blobs only for this model
gen_data = generate_blobs(rng=PRNGKey(1),blobs=3)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
)
# then use this to define your model:
model = hepdata_like_from_hists(hist_maker)
```
Similar to above, we can get output at this stage by initializing the neural network. `hepdata_like_from_hists` will return a `Model` object with callable `logpdf` method, as well as the model parameters in the background-only scenario for convenience. See [this link](https://scikit-hep.org/pyhf/_generated/pyhf.simplemodels.hepdata_like.html) for more about the type of model being used here, as well as the rest of the `pyhf` docs for added physics context.
```
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
m, bkg_only_pars = model(network)
m.logpdf(bkg_only_pars,data=[1,1])
#export
import sys
from unittest.mock import patch
jax_backend = pyhf.tensor.jax_backend(precision='64b')
pyhf.set_backend(jax_backend)
def histosys_model_from_hists(histogram_maker):
"""Returns a function that constructs a HEP statistical model using a
'histosys' uncertainty for the background (nominal background, up and down
systematic variations) when evaluated at the parameters of the observable.
Args:
histogram_maker: A function that, when called, returns a secondary function
that takes the observable's parameters as argument, and returns yields.
Returns:
nn_model_maker: A function that returns a `pyhf.Model` object when
evaluated at the observable's parameters (nn weights), along with the
background-only parameters for use in downstream inference.
"""
@patch('pyhf.default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code0'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code1'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code2'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code4'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code4p'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.shapefactor'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.shapesys'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.staterror'], 'default_backend', new=jax_backend)
def from_spec(yields):
s, b, bup, bdown = yields
spec = {
"channels": [
{
"name": "nn",
"samples": [
{
"name": "signal",
"data": s,
"modifiers": [
{"name": "mu", "type": "normfactor", "data": None}
],
},
{
"name": "bkg",
"data": b,
"modifiers": [
{
"name": "nn_histosys",
"type": "histosys",
"data": {
"lo_data": bdown,
"hi_data": bup,
},
}
],
},
],
},
],
}
return pyhf.Model(spec)
def nn_model_maker(hm_params):
yields = histogram_maker(hm_params)
m = from_spec(yields)
nompars = m.config.suggested_init()
bonlypars = jnp.asarray([x for x in nompars])
bonlypars = jax.ops.index_update(bonlypars, m.config.poi_index, 0.0)
return m, bonlypars
return nn_model_maker
```
### Usage:
```
# define a hist_maker as above
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn, histosys_model_from_hists
from neos.data import generate_blobs
# data generator, four blobs only for this model
gen_data = generate_blobs(rng=PRNGKey(1),blobs=4)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hyperpars = dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=hyperpars)
# then use this to define your model:
model = histosys_model_from_hists(hist_maker)
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
# instantiate model and eval logpdf
m, bkg_only_pars = model(network)
m.logpdf(bkg_only_pars,data=[1,1,1])
```
|
github_jupyter
|
# default_exp makers
#export
from functools import partial
import jax
import jax.numpy as jnp
import numpy as np
from relaxed import hist_kde as hist
#export
def hists_from_nn(
data_generator, predict, hpar_dict, method="softmax", LUMI=10, sig_scale=2, bkg_scale=10, reflect_infinities=False
):
"""Initialize a function `hist_maker` that returns a 'soft' histogram based
on a neural network with a softmax output. Choose which example problem to
try by setting the `example` argument.
Args:
data_generator: Callable that returns generated data (in jax array
format).
predict: Decision function for a parameterized observable, e.g. neural
network.
method: A string to specify the method to use for constructing soft
histograms. Either "softmax" or "kde".
LUMI: 'Luminosity' scaling factor for the yields.
sig_scale: Individual scaling factor for the signal yields.
bkg_scale: Individual scaling factor for the signal yields.
Returns:
hist_maker: A callable function that takes the parameters of the
observable (and optional hyperpars), then constructs signal,
background, and background uncertainty yields.
"""
data = data_generator()
if len(data) == 3:
if method == "softmax":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Two background
distributions are sampled from, which is meant to mimic the
situation in particle physics where one has a 'nominal'
prediction for a nuisance parameter (taken here as the mean of
two modes) and then alternate values (e.g. from varying up/down
by one standard deviation), which then modifies the background
pdf. Here, we take that effect to be a shift of the mean of the
distribution. The value for the background histogram is then
the mean of the resulting counts of the two modes, and the
uncertainty can be quantified through the count standard
deviation.
Arguments:
hm_params: a list containing:
nn: jax array of observable parameters.
"""
nn = hm_params
s, b_up, b_down = data
NMC = len(s)
s_hist = predict(nn, s).sum(axis=0) * sig_scale / NMC * LUMI
b_hists = [
predict(nn, b_up).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_down).sum(axis=0) * bkg_scale / NMC * LUMI,
]
b_mean = jnp.mean(jnp.asarray(b_hists), axis=0)
b_unc = jnp.std(jnp.asarray(b_hists), axis=0)
return s_hist, b_mean, b_unc
elif method == "kde":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data using a kde, all drawn from
multivariate normal distributions with different means. Two
background distributions are sampled from, which is meant to
mimic the situation in particle physics where one has a
'nominal' prediction for a nuisance parameter (taken here as
the mean of two modes) and then alternate values (e.g. from
varying up/down by one standard deviation), which then modifies
the background pdf. Here, we take that effect to be a shift of
the mean of the distribution. The value for the background
histogram is then the mean of the resulting counts of the two
modes, and the uncertainty can be quantified through the count
standard deviation.
Arguments:
hm_params: Array-like, consisting of:
nn: jax array of observable parameters.
bins: Array of bin edges, e.g. np.linspace(0,1,3)
defines a two-bin histogram with edges at 0, 0.5,
1.
bandwidth: Float that controls the 'smoothness' of the
kde. It's recommended to keep this fairly
similar to the bin width to avoid
oversmoothing the distribution. Going too low
will cause things to break, as the gradients
of the kde become unstable.
"""
nn = hm_params
bins, bandwidth = hpar_dict["bins"], hpar_dict["bandwidth"]
s, b_up, b_down = data
NMC = len(s)
nn_s, nn_b_up, nn_b_down = (
predict(nn, s).ravel(),
predict(nn, b_up).ravel(),
predict(nn, b_down).ravel(),
)
s_hist = hist(nn_s, bins, bandwidth, reflect_infinities=reflect_infinities) * sig_scale / NMC * LUMI
b_hists = jnp.asarray(
[
hist(nn_b_up, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_down, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
]
)
kde_counts = [
s_hist,
jnp.mean(b_hists, axis=0),
jnp.std(b_hists, axis=0),
]
return kde_counts
else:
assert False, (
f"Unsupported method: {method}"
" (only using kde or softmax for these examples)."
)
elif len(data) == 4:
if method == "softmax":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Three background
distributions are sampled from, which mimics the situation in
particle physics where one has a 'nominal' prediction for a
nuisance parameter (taken here as the mean of two modes) and
then alternate values (e.g. from varying up/down by one
standard deviation), which then modifies the background pdf.
Here, we take that effect to be a shift of the mean of the
distribution. The HistFactory 'histosys' nusiance parameter
will then be constructed from the yields downstream by
interpolating between them using pyhf.
Arguments:
hm_params: a list containing:
nn: jax array of observable parameters.
Returns:
Set of 4 counts for signal, background, and up/down modes.
"""
nn = hm_params
s, b_nom, b_up, b_down = data
NMC = len(s)
counts = [
predict(nn, s).sum(axis=0) * sig_scale / NMC * LUMI,
predict(nn, b_nom).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_up).sum(axis=0) * bkg_scale / NMC * LUMI,
predict(nn, b_down).sum(axis=0) * bkg_scale / NMC * LUMI,
]
return counts
elif method == "kde":
def hist_maker(hm_params):
"""Uses the nn decision function `predict` to form histograms
from signal and background data, all drawn from multivariate
normal distributions with different means. Three background
distributions are sampled from, which mimics the situation in
particle physics where one has a 'nominal' prediction for a
nuisance parameter (taken here as the mean of two modes) and
then alternate values (e.g. from varying up/down by one
standard deviation), which then modifies the background pdf.
Here, we take that effect to be a shift of the mean of the
distribution. The HistFactory 'histosys' nusiance parameter
will then be constructed from the yields downstream by
interpolating between them using pyhf.
Arguments:
hm_params: Array-like, consisting of:
nn: jax array of observable parameters.
bins: Array of bin edges, e.g. np.linspace(0,1,3)
defines a two-bin histogram with edges at 0, 0.5,
1.
bandwidth: Float that controls the 'smoothness' of the
kde. It's recommended to keep this fairly
similar to the bin width to avoid
oversmoothing the distribution. Going too low
will cause things to break, as the gradients
of the kde become unstable.
Returns:
Set of 4 counts for signal, background, and up/down modes.
"""
nn = hm_params
bins, bandwidth = hpar_dict["bins"], hpar_dict["bandwidth"]
s, b_nom, b_up, b_down = data
NMC = len(s)
nn_s, nn_b_nom, nn_b_up, nn_b_down = (
predict(nn, s).ravel(),
predict(nn, b_nom).ravel(),
predict(nn, b_up).ravel(),
predict(nn, b_down).ravel(),
)
kde_counts = [
hist(nn_s, bins, bandwidth, reflect_infinities=reflect_infinities) * sig_scale / NMC * LUMI,
hist(nn_b_nom, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_up, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
hist(nn_b_down, bins, bandwidth, reflect_infinities=reflect_infinities) * bkg_scale / NMC * LUMI,
]
return [k + 1e-8 for k in kde_counts]
else:
assert False, (
f"Unsupported method: {method}"
" (only using kde or softmax for these examples)."
)
else:
assert False, (
f"Unsupported number of blobs: {blobs}"
" (only using 3 or 4 blobs for these examples)."
)
return hist_maker
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn
from neos.data import generate_blobs
# data generator
gen_data = generate_blobs(rng=PRNGKey(1),blobs=4)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
)
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
hist_maker(network)
#export
import pyhf
jax_backend = pyhf.tensor.jax_backend(precision="64b")
pyhf.set_backend(jax_backend)
from neos.models import hepdata_like
#export
def hepdata_like_from_hists(histogram_maker):
"""Returns a function that constructs a typical 'hepdata-like' statistical
model with signal, background, and background uncertainty yields when
evaluated at the parameters of the observable.
Args:
histogram_maker: A function that, when called, returns a secondary function
that takes the observable's parameters as argument, and returns yields.
Returns:
nn_model_maker: A function that returns a Model object (either from
`neos.models` or from `pyhf`) when evaluated at the observable's parameters,
along with the background-only parameters for use in downstream inference.
"""
def nn_model_maker(hm_params):
s, b, db = histogram_maker(hm_params)
m = hepdata_like(s, b, db) # neos 'pyhf' model
nompars = m.config.suggested_init()
bonlypars = jnp.asarray([x for x in nompars])
bonlypars = jax.ops.index_update(bonlypars, m.config.poi_index, 0.0)
return m, bonlypars
return nn_model_maker
# define a hist_maker as above
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn, hepdata_like_from_hists
from neos.data import generate_blobs
# data generator, three blobs only for this model
gen_data = generate_blobs(rng=PRNGKey(1),blobs=3)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
)
# then use this to define your model:
model = hepdata_like_from_hists(hist_maker)
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
m, bkg_only_pars = model(network)
m.logpdf(bkg_only_pars,data=[1,1])
#export
import sys
from unittest.mock import patch
jax_backend = pyhf.tensor.jax_backend(precision='64b')
pyhf.set_backend(jax_backend)
def histosys_model_from_hists(histogram_maker):
"""Returns a function that constructs a HEP statistical model using a
'histosys' uncertainty for the background (nominal background, up and down
systematic variations) when evaluated at the parameters of the observable.
Args:
histogram_maker: A function that, when called, returns a secondary function
that takes the observable's parameters as argument, and returns yields.
Returns:
nn_model_maker: A function that returns a `pyhf.Model` object when
evaluated at the observable's parameters (nn weights), along with the
background-only parameters for use in downstream inference.
"""
@patch('pyhf.default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code0'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code1'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code2'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code4'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.interpolators.code4p'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.shapefactor'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.shapesys'], 'default_backend', new=jax_backend)
@patch.object(sys.modules['pyhf.modifiers.staterror'], 'default_backend', new=jax_backend)
def from_spec(yields):
s, b, bup, bdown = yields
spec = {
"channels": [
{
"name": "nn",
"samples": [
{
"name": "signal",
"data": s,
"modifiers": [
{"name": "mu", "type": "normfactor", "data": None}
],
},
{
"name": "bkg",
"data": b,
"modifiers": [
{
"name": "nn_histosys",
"type": "histosys",
"data": {
"lo_data": bdown,
"hi_data": bup,
},
}
],
},
],
},
],
}
return pyhf.Model(spec)
def nn_model_maker(hm_params):
yields = histogram_maker(hm_params)
m = from_spec(yields)
nompars = m.config.suggested_init()
bonlypars = jnp.asarray([x for x in nompars])
bonlypars = jax.ops.index_update(bonlypars, m.config.poi_index, 0.0)
return m, bonlypars
return nn_model_maker
# define a hist_maker as above
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental import stax
import neos
from neos.makers import hists_from_nn, histosys_model_from_hists
from neos.data import generate_blobs
# data generator, four blobs only for this model
gen_data = generate_blobs(rng=PRNGKey(1),blobs=4)
# nn
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1),
stax.Sigmoid
)
hyperpars = dict(bandwidth=0.5, bins=jnp.linspace(0,1,3))
hist_maker = hists_from_nn(gen_data, predict, method='kde', hpar_dict=hyperpars)
# then use this to define your model:
model = histosys_model_from_hists(hist_maker)
_, network = init_random_params(jax.random.PRNGKey(13), (-1, 2))
# instantiate model and eval logpdf
m, bkg_only_pars = model(network)
m.logpdf(bkg_only_pars,data=[1,1,1])
| 0.880592 | 0.968381 |
```
import numpy as np
import torch
import matplotlib.pyplot as plt
import conv_autoenc
import torch.nn as nn
import h5py
import umap
import scipy.spatial as spat
dset_path="./data/data_temp/"
batch_size=20
n_z=20
n_channels=2
visual=True
h5=h5py.File(dset_path,"r+")
T,W,H=h5.attrs["T"],h5.attrs["W"],h5.attrs["H"]
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
def to_np(ten):
return ten.cpu().detach().numpy()
class DS(torch.utils.data.Dataset):
def __init__(self):
super(DS,self).__init__()
def __getitem__(self,i):
return torch.tensor(np.max(np.array(h5[str(i)+"/frame"]),axis=3)/255)
def __len__(self):
return T
ds=DS()
dl=torch.utils.data.DataLoader(ds,batch_size=batch_size,shuffle=True,pin_memory=True)
net=conv_autoenc.Net(n_channels=n_channels,n_z=n_z)
net.to(device=device)
None
%matplotlib notebook
if visual:
fig=plt.figure(figsize=(8,5))
ax1=fig.add_subplot(2,1,1)
lplot=ax1.plot([],[],label="Loss")[0]
ax1.legend()
ax1.set_yscale("log")
ax2=fig.add_subplot(2,2,3)
im=ax2.imshow(np.zeros((W,H)).T,vmin=0,vmax=0.8)
ax3=fig.add_subplot(2,2,4)
imans=ax3.imshow(np.zeros((W,H)).T,vmin=0,vmax=0.8)
def update():
if len(losses)<2:
return
ax1.set_ylim(np.min(losses),np.max(losses))
ax1.set_xlim(1,len(losses))
ts=np.arange(1,len(losses)+1)
lplot.set_data(np.stack([ts,np.array(losses)]))
im.set_array(to_np(res[0,0]).T)
imans.set_array(to_np(ims[0,0]).T)
fig.canvas.draw()
num_epochs=30
opt=torch.optim.Adam(net.parameters())
losses=[]
for epoch in range(num_epochs):
print("\r Epoch "+str(epoch+1)+"/"+str(num_epochs),end="")
for i,ims in enumerate(dl):
ims=ims.to(device=device,dtype=torch.float32)
res,latent=net(ims)
loss=nn.functional.mse_loss(res,ims)
opt.zero_grad()
loss.backward()
opt.step()
losses.append(loss.item())
if visual:
update()
net.eval()
vecs=[]
with torch.no_grad():
for i in range(T):
if (i+1)%100==0:
print("\r"+str(i)+"/"+str(T),end="")
_,latent=net(ds[i].unsqueeze(0).to(device=device,dtype=torch.float32))
vecs.append(to_np(latent[0]))
vecs=np.array(vecs)
key="vecs"
if key in h5.keys():
del h5[key]
ds=h5.create_dataset(key,shape=(T,n_z),dtype="f4")
ds[...]=vecs.astype(np.float32)
def standardize(vecs):
m=np.mean(vecs,axis=0)
s=np.std(vecs,axis=0)
return (vecs-m)/(s+1e-8)
vecs=standardize(vecs)
u_map=umap.UMAP(n_components=2)
res=u_map.fit_transform(vecs)
distmat=spat.distance_matrix(res,res)
plt.subplot(121)
plt.scatter(res[:,0],res[:,1],s=1)
plt.subplot(122)
plt.imshow(distmat)
key="distmat"
if key in h5.keys():
del h5[key]
ds=h5.create_dataset(key,shape=(T,T),dtype="f4")
ds[...]=distmat.astype(np.float32)
#check
plt.figure(figsize=(8,4))
exs=np.random.choice(T,4,replace=False)
for i,ex in enumerate(exs):
plt.subplot(4,5,i*5+1)
plt.imshow(np.max(np.array(h5[str(ex)+"/frame"][0]),axis=2).T)
close=np.argsort(distmat[ex])[1:]
for b in range(4):
plt.subplot(4,5,i*5+b+2)
plt.imshow(np.max(np.array(h5[str(close[b])+"/frame"][0]),axis=2).T)
h5.close()
```
|
github_jupyter
|
import numpy as np
import torch
import matplotlib.pyplot as plt
import conv_autoenc
import torch.nn as nn
import h5py
import umap
import scipy.spatial as spat
dset_path="./data/data_temp/"
batch_size=20
n_z=20
n_channels=2
visual=True
h5=h5py.File(dset_path,"r+")
T,W,H=h5.attrs["T"],h5.attrs["W"],h5.attrs["H"]
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
def to_np(ten):
return ten.cpu().detach().numpy()
class DS(torch.utils.data.Dataset):
def __init__(self):
super(DS,self).__init__()
def __getitem__(self,i):
return torch.tensor(np.max(np.array(h5[str(i)+"/frame"]),axis=3)/255)
def __len__(self):
return T
ds=DS()
dl=torch.utils.data.DataLoader(ds,batch_size=batch_size,shuffle=True,pin_memory=True)
net=conv_autoenc.Net(n_channels=n_channels,n_z=n_z)
net.to(device=device)
None
%matplotlib notebook
if visual:
fig=plt.figure(figsize=(8,5))
ax1=fig.add_subplot(2,1,1)
lplot=ax1.plot([],[],label="Loss")[0]
ax1.legend()
ax1.set_yscale("log")
ax2=fig.add_subplot(2,2,3)
im=ax2.imshow(np.zeros((W,H)).T,vmin=0,vmax=0.8)
ax3=fig.add_subplot(2,2,4)
imans=ax3.imshow(np.zeros((W,H)).T,vmin=0,vmax=0.8)
def update():
if len(losses)<2:
return
ax1.set_ylim(np.min(losses),np.max(losses))
ax1.set_xlim(1,len(losses))
ts=np.arange(1,len(losses)+1)
lplot.set_data(np.stack([ts,np.array(losses)]))
im.set_array(to_np(res[0,0]).T)
imans.set_array(to_np(ims[0,0]).T)
fig.canvas.draw()
num_epochs=30
opt=torch.optim.Adam(net.parameters())
losses=[]
for epoch in range(num_epochs):
print("\r Epoch "+str(epoch+1)+"/"+str(num_epochs),end="")
for i,ims in enumerate(dl):
ims=ims.to(device=device,dtype=torch.float32)
res,latent=net(ims)
loss=nn.functional.mse_loss(res,ims)
opt.zero_grad()
loss.backward()
opt.step()
losses.append(loss.item())
if visual:
update()
net.eval()
vecs=[]
with torch.no_grad():
for i in range(T):
if (i+1)%100==0:
print("\r"+str(i)+"/"+str(T),end="")
_,latent=net(ds[i].unsqueeze(0).to(device=device,dtype=torch.float32))
vecs.append(to_np(latent[0]))
vecs=np.array(vecs)
key="vecs"
if key in h5.keys():
del h5[key]
ds=h5.create_dataset(key,shape=(T,n_z),dtype="f4")
ds[...]=vecs.astype(np.float32)
def standardize(vecs):
m=np.mean(vecs,axis=0)
s=np.std(vecs,axis=0)
return (vecs-m)/(s+1e-8)
vecs=standardize(vecs)
u_map=umap.UMAP(n_components=2)
res=u_map.fit_transform(vecs)
distmat=spat.distance_matrix(res,res)
plt.subplot(121)
plt.scatter(res[:,0],res[:,1],s=1)
plt.subplot(122)
plt.imshow(distmat)
key="distmat"
if key in h5.keys():
del h5[key]
ds=h5.create_dataset(key,shape=(T,T),dtype="f4")
ds[...]=distmat.astype(np.float32)
#check
plt.figure(figsize=(8,4))
exs=np.random.choice(T,4,replace=False)
for i,ex in enumerate(exs):
plt.subplot(4,5,i*5+1)
plt.imshow(np.max(np.array(h5[str(ex)+"/frame"][0]),axis=2).T)
close=np.argsort(distmat[ex])[1:]
for b in range(4):
plt.subplot(4,5,i*5+b+2)
plt.imshow(np.max(np.array(h5[str(close[b])+"/frame"][0]),axis=2).T)
h5.close()
| 0.52756 | 0.411229 |
```
# Imports gerais
import pandas as pd
import numpy as np
import os
import pickle
# Imports de dataviz
import seaborn as sns
import matplotlib.pyplot as plt
# Imports do SKLEARN
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_validate
from sklearn.metrics import classification_report
from sklearn import preprocessing
from sklearn.metrics import confusion_matrix
```
# Extração dos dados
Nesta etapa foi realizado o carregamento dos dados e a filtragem dos dados relevantes, desta forma, somente as colunas que serão utilizadas (title e category) foram lidas e transformadas em Dataframe.
```
# Data extraction
dataset_path = os.getenv("DATASET_PATH")
col_list=['title', 'category']
df = pd.read_csv(dataset_path, usecols=col_list, encoding='utf-8')
print(df.info())
```
# Modelagem dos dados
A modelagem dos dados seguiu os seguintes passos:
1. Transformação das categorias (df['category']) em *labels*, utilizando a classe LabelEncoder do Sklearn.
2. Os dados foram divididos entre dados de treino e dados de teste, utilizando a função train_test_split do sklearn. A proporção de dados destinados para teste/treino foi de 0.3/0.7.
3. Um pipeline para o processamento dos dados foi criado, fazendo uso da função make_pipeline do sklearn. Neste processo, como modelagem de linguage, foi escolhida a abordagem Bag of Words, que, não obstante simples, obteve uma performance satisfatória. Ademais, foi escolhido o algoritmo *LogisticRegression*, pois ao compararmos com outros algoritmos (como tf-idf com NB que obteve 0.82 de acuracia geral), o seu desempenho foi melhor.
4. Foi realizado uma série de testes para verificar métricas relacionadas a performance (acurácia, precision, recall e f1-score) do modelo.
```
le = preprocessing.LabelEncoder()
df['category_encoded'] = le.fit_transform(df['category'])
X_train, X_test = train_test_split(df['title'], test_size=0.3, random_state=42)
y_train, y_test = train_test_split(df['category_encoded'], test_size=0.3, random_state=42)
# Pipeline
model = make_pipeline(CountVectorizer(), LogisticRegression(fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=1000,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False))
LogisticRegression = model.fit(X_train,y_train) # apply scaling on training data
print(f'train score:', LogisticRegression.score(X_train, y_train))
LogisticRegression.score(X_test, y_test) #0.8644 com tfidf
print(f'test score:',LogisticRegression.score(X_test, y_test))
predictions_LogisticRegression = LogisticRegression.predict(X_test)
metrics_results = classification_report(y_test, predictions_LogisticRegression)
print(metrics_results)
print('Nome das classes: ',{n:i for n, i in enumerate(le.classes_)})
cm = confusion_matrix(y_test,predictions_LogisticRegression)
df_cm = pd.DataFrame(cm)
plt.figure(figsize=(10,8))
ax = sns.heatmap(df_cm)
print('confusion matrix: \n',df_cm)
skf = StratifiedKFold(n_splits=9, random_state=42, shuffle=True)
print(skf)
X = df['title']
y = df['category_encoded']
for i, [train_index, test_index] in enumerate(skf.split(X, y)):
print('Validation set #%d' % (i+1), "TRAIN:", train_index, "TEST:", test_index)
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
LogisticRegression = model.fit(X_train,y_train) # apply scaling on training data
print(f'train score:', LogisticRegression.score(X_train, y_train))
LogisticRegression.score(X_test, y_test) #0.87035 com count e 0.8644 com tfidf
print(f'test score:',LogisticRegression.score(X_test, y_test))
```
# Export model and results
```
model_path = os.getenv("MODEL_PATH")
print(model_path)
metrics_path = os.getenv("METRICS_PATH")
sns.heatmap(df_cm).figure.savefig(os.path.join('/usr/src/data/', 'confusion_matrix_heatmap.png'))
pkl_filename = "model.pkl"
with open(model_path, 'wb') as file:
pickle.dump(model, file)
with open(metrics_path, 'w') as file:
file.write(metrics_results)
# Verificando se o arquivo pickle foi escrito corretamente
with open(pkl_filename, 'rb') as file:
pickle_model = pickle.load(file)
score = pickle_model.score(X_test, y_test)
print("Test score: {0:.2f} %".format(100 * score))
Ypredict = pickle_model.predict(X_test)
```
|
github_jupyter
|
# Imports gerais
import pandas as pd
import numpy as np
import os
import pickle
# Imports de dataviz
import seaborn as sns
import matplotlib.pyplot as plt
# Imports do SKLEARN
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_validate
from sklearn.metrics import classification_report
from sklearn import preprocessing
from sklearn.metrics import confusion_matrix
# Data extraction
dataset_path = os.getenv("DATASET_PATH")
col_list=['title', 'category']
df = pd.read_csv(dataset_path, usecols=col_list, encoding='utf-8')
print(df.info())
le = preprocessing.LabelEncoder()
df['category_encoded'] = le.fit_transform(df['category'])
X_train, X_test = train_test_split(df['title'], test_size=0.3, random_state=42)
y_train, y_test = train_test_split(df['category_encoded'], test_size=0.3, random_state=42)
# Pipeline
model = make_pipeline(CountVectorizer(), LogisticRegression(fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=1000,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=None, solver='lbfgs', tol=0.0001, verbose=0,
warm_start=False))
LogisticRegression = model.fit(X_train,y_train) # apply scaling on training data
print(f'train score:', LogisticRegression.score(X_train, y_train))
LogisticRegression.score(X_test, y_test) #0.8644 com tfidf
print(f'test score:',LogisticRegression.score(X_test, y_test))
predictions_LogisticRegression = LogisticRegression.predict(X_test)
metrics_results = classification_report(y_test, predictions_LogisticRegression)
print(metrics_results)
print('Nome das classes: ',{n:i for n, i in enumerate(le.classes_)})
cm = confusion_matrix(y_test,predictions_LogisticRegression)
df_cm = pd.DataFrame(cm)
plt.figure(figsize=(10,8))
ax = sns.heatmap(df_cm)
print('confusion matrix: \n',df_cm)
skf = StratifiedKFold(n_splits=9, random_state=42, shuffle=True)
print(skf)
X = df['title']
y = df['category_encoded']
for i, [train_index, test_index] in enumerate(skf.split(X, y)):
print('Validation set #%d' % (i+1), "TRAIN:", train_index, "TEST:", test_index)
X_train, X_test = X[train_index], X[test_index]
y_train, y_test = y[train_index], y[test_index]
LogisticRegression = model.fit(X_train,y_train) # apply scaling on training data
print(f'train score:', LogisticRegression.score(X_train, y_train))
LogisticRegression.score(X_test, y_test) #0.87035 com count e 0.8644 com tfidf
print(f'test score:',LogisticRegression.score(X_test, y_test))
model_path = os.getenv("MODEL_PATH")
print(model_path)
metrics_path = os.getenv("METRICS_PATH")
sns.heatmap(df_cm).figure.savefig(os.path.join('/usr/src/data/', 'confusion_matrix_heatmap.png'))
pkl_filename = "model.pkl"
with open(model_path, 'wb') as file:
pickle.dump(model, file)
with open(metrics_path, 'w') as file:
file.write(metrics_results)
# Verificando se o arquivo pickle foi escrito corretamente
with open(pkl_filename, 'rb') as file:
pickle_model = pickle.load(file)
score = pickle_model.score(X_test, y_test)
print("Test score: {0:.2f} %".format(100 * score))
Ypredict = pickle_model.predict(X_test)
| 0.399929 | 0.748053 |
# Time normalization of data
> Marcos Duarte
> Laboratory of Biomechanics and Motor Control ([http://demotu.org/](http://demotu.org/))
> Federal University of ABC, Brazil
Time normalization is usually employed for the temporal alignment of cyclic data obtained from different trials with different duration (number of points). The most simple and common procedure for time normalization used in Biomechanics and Motor Control is known as the normalization to percent cycle (although it might not be the most adequate procedure in certain cases ([Helwig et al., 2011](http://www.sciencedirect.com/science/article/pii/S0021929010005038)).
In the percent cycle, a fixed number (typically a temporal base from 0 to 100%) of new equally spaced data is created based on the old data with a mathematical procedure known as interpolation.
**Interpolation** is the estimation of new data points within the range of known data points. This is different from **extrapolation**, the estimation of data points outside the range of known data points.
Time normalization of data using interpolation is a simple procedure and it doesn't matter if the original data have more or less data points than desired.
The Python function `tnorm.py` (code at the end of this text) implements the normalization to percent cycle procedure for time normalization. The function signature is:
```python
yn, tn, indie = tnorm(y, axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
```
Let's see now how to perform interpolation and time normalization; first let's import the necessary Python libraries and configure the environment:
```
# Import the necessary libraries
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import sys
sys.path.insert(1, r'./../functions') # add to pythonpath
```
For instance, consider the data shown next. The time normalization of these data to represent a cycle from 0 to 100%, with a step of 1% (101 data points) is:
```
y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
print("y data:")
y
t = np.linspace(0, 100, len(y)) # time vector for the original data
tn = np.linspace(0, 100, 101) # new time vector for the new time-normalized data
yn = np.interp(tn, t, y) # new time-normalized data
print("y data interpolated to 101 points:")
yn
```
The key is the Numpy `interp` function, from its help:
>interp(x, xp, fp, left=None, right=None)
>One-dimensional linear interpolation.
>Returns the one-dimensional piecewise linear interpolant to a function with given values at discrete data-points.
A plot of the data will show what we have done:
```
plt.figure(figsize=(10,5))
plt.plot(t, y, 'bo-', lw=2, label='original data')
plt.plot(tn, yn, '.-', color=[1, 0, 0, .5], lw=2, label='time normalized')
plt.legend(loc='best', framealpha=.5)
plt.xlabel('Cycle [%]')
plt.show()
```
The function `tnorm.py` implements this kind of normalization with option for a different interpolation than the linear one used, deal with missing points in the data (if these missing points are not at the extremities of the data because the interpolation function can not extrapolate data), other things.
Let's see the `tnorm.py` examples:
```
from tnorm import tnorm
>>> # Default options: cubic spline interpolation passing through
>>> # each datum, 101 points, and no plot
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> tnorm(y)
>>> # Linear interpolation passing through each datum
>>> yn, tn, indie = tnorm(y, k=1, smooth=0, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing
>>> yn, tn, indie = tnorm(y, k=3, smooth=1, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing and 50 points
>>> x = np.linspace(-3, 3, 60)
>>> y = np.exp(-x**2) + np.random.randn(60)/10
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data (use NaN as mask)
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[:10] = np.NaN # first ten points are missing
>>> y[30: 41] = np.NaN # make other 10 missing points
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, nan_at_ext='replace', show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=1, smooth=0, nan_at_ext='replace', show=True)
>>> # Deal with 2-D array
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y = np.vstack((y-1, y[::-1])).T
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
```
## Function tnorm.py
```
# %load './../functions/tnorm.py'
"""Time normalization (from 0 to 100% with step interval)."""
import numpy as np
__author__ = 'Marcos Duarte, https://github.com/demotu/BMC'
__version__ = "1.0.6"
__license__ = "MIT"
def tnorm(y, axis=0, step=1, k=3, smooth=0, mask=None, nan_at_ext='delete',
show=False, ax=None):
"""Time normalization (from 0 to 100% with step interval).
Time normalization is usually employed for the temporal alignment of data
obtained from different trials with different duration (number of points).
This code implements a procedure knwown as the normalization to percent
cycle.
This code can perform simple linear interpolation passing through each
datum or spline interpolation (up to quintic splines) passing through each
datum (knots) or not (in case a smoothing parameter > 0 is inputted).
NaNs and any value inputted as a mask parameter and that appears at the
extremities might be removed or replaced by the first/last not-NaN value
before the interpolation because this code does not perform extrapolation.
For a 2D array, the entire row with NaN or a mask value at the extermity
might be removed because of alignment issues with the data from different
columns. As result, if there is a column of only NaNs in the data, the
time normalization can't be performed (an empty NaNs and any value
inputted as a mask parameter and that appears in the middle of the data
(which may represent missing data) are ignored and the interpolation is
performed through these points.
See this IPython notebook [2]_.
Parameters
----------
y : 1-D or 2-D array_like
Array of independent input data. Must be increasing.
If 2-D array, the data in each axis will be interpolated.
axis : int, 0 or 1, optional (default = 0)
Axis along which the interpolation is performed.
0: data in each column are interpolated; 1: for row interpolation
step : float or int, optional (default = 1)
Interval from 0 to 100% to resample y or the number of points y
should be interpolated. In the later case, the desired number of
points should be expressed with step as a negative integer.
For instance, step = 1 or step = -101 will result in the same
number of points at the interpolation (101 points).
If step == 0, the number of points will be the number of data in y.
k : int, optional (default = 3)
Degree of the smoothing spline. Must be 1 <= k <= 5.
If 3, a cubic spline is used.
The number of data points must be larger than k.
smooth : float or None, optional (default = 0)
Positive smoothing factor used to choose the number of knots.
If 0, spline will interpolate through all data points.
If None, smooth=len(y).
mask : None or float, optional (default = None)
Mask to identify missing values which will be ignored.
It can be a list of values.
NaN values will be ignored and don't need to be in the mask.
nan_at_ext : string, optional (default = 'delete')
Method to deal with NaNs at the extremities.
'delete' will delete any NaN at the extremities (the corresponding
entire row in `y` for a 2-D array).
'replace' will replace any NaN at the extremities by first/last
not-NaN value in `y`.
show : bool, optional (default = False)
True (1) plot data in a matplotlib figure.
False (0) to not plot.
ax : a matplotlib.axes.Axes instance, optional (default = None).
Returns
-------
yn : 1-D or 2-D array
Interpolated data (if axis == 0, column oriented for 2-D array).
tn : 1-D array
New x values (from 0 to 100) for the interpolated data.
inds : list
Indexes of first and last rows without NaNs at the extremities of `y`.
If there is no NaN in the data, this list is [0, y.shape[0]-1].
Notes
-----
This code performs interpolation to create data with the desired number of
points using a one-dimensional smoothing spline fit to a given set of data
points (scipy.interpolate.UnivariateSpline function).
References
----------
.. [1] http://www.sciencedirect.com/science/article/pii/S0021929010005038
.. [2] http://nbviewer.ipython.org/github/demotu/BMC/blob/master/notebooks/TimeNormalization.ipynb
See Also
--------
scipy.interpolate.UnivariateSpline:
One-dimensional smoothing spline fit to a given set of data points.
Examples
--------
>>> # Default options: cubic spline interpolation passing through
>>> # each datum, 101 points, and no plot
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> tnorm(y)
>>> # Linear interpolation passing through each datum
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> yn, tn, indie = tnorm(y, k=1, smooth=0, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> yn, tn, indie = tnorm(y, k=3, smooth=1, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing and 50 points
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data (use NaN as mask)
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[:10] = np.NaN # first ten points are missing
>>> y[30: 41] = np.NaN # make other 10 missing points
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, nan_at_ext='replace', show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=1, smooth=0, nan_at_ext='replace', show=True)
>>> # Deal with 2-D array
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y = np.vstack((y-1, y[::-1])).T
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
Version history
---------------
'1.0.6':
Deleted 'from __future__ import ...'
Added parameter `nan_at_ext`
Adjusted outputs to have always the same type
"""
from scipy.interpolate import UnivariateSpline
y = np.asarray(y)
if axis:
y = y.T
if y.ndim == 1:
y = np.reshape(y, (-1, 1))
# turn mask into NaN
if mask is not None:
y[y == mask] = np.NaN
iini = 0
iend = y.shape[0]-1
if nan_at_ext.lower() == 'delete':
# delete rows with missing values at the extremities
while y.size and np.isnan(np.sum(y[0])):
y = np.delete(y, 0, axis=0)
iini += 1
while y.size and np.isnan(np.sum(y[-1])):
y = np.delete(y, -1, axis=0)
iend -= 1
else:
# replace NaN at the extremities by first/last not-NaN
if np.any(np.isnan(y[0])):
for col in range(y.shape[1]):
ind_not_nan = np.nonzero(~np.isnan(y[:, col]))[0]
if ind_not_nan.size:
y[0, col] = y[ind_not_nan[0], col]
else:
y = np.empty((0, 0))
break
if np.any(np.isnan(y[-1])):
for col in range(y.shape[1]):
ind_not_nan = np.nonzero(~np.isnan(y[:, col]))[0]
if ind_not_nan.size:
y[-1, col] = y[ind_not_nan[-1], col]
else:
y = np.empty((0, 0))
break
# check if there are still data
if not y.size:
return np.empty((0, 0)), np.empty(0), []
if y.size == 1:
return y.flatten(), np.array(0), [0, 0]
indie = [iini, iend]
t = np.linspace(0, 100, y.shape[0])
if step == 0:
tn = t
elif step > 0:
tn = np.linspace(0, 100, np.round(100 / step + 1))
else:
tn = np.linspace(0, 100, -step)
yn = np.empty([tn.size, y.shape[1]]) * np.NaN
for col in np.arange(y.shape[1]):
# ignore NaNs inside data for the interpolation
ind = np.isfinite(y[:, col])
if np.sum(ind) > 1: # at least two points for the interpolation
spl = UnivariateSpline(t[ind], y[ind, col], k=k, s=smooth)
yn[:, col] = spl(tn)
if show:
_plot(t, y, ax, tn, yn)
if axis:
y = y.T
if yn.shape[1] == 1:
yn = yn.flatten()
return yn, tn, indie
def _plot(t, y, ax, tn, yn):
"""Plot results of the tnorm function, see its help."""
try:
import matplotlib.pyplot as plt
except ImportError:
print('matplotlib is not available.')
else:
if ax is None:
_, ax = plt.subplots(1, 1, figsize=(8, 5))
ax.set_prop_cycle('color', ['b', 'r', 'b', 'g', 'b', 'y', 'b', 'c', 'b', 'm'])
#ax.set_color_cycle(['b', 'r', 'b', 'g', 'b', 'y', 'b', 'c', 'b', 'm'])
for col in np.arange(y.shape[1]):
if y.shape[1] == 1:
ax.plot(t, y[:, col], 'o-', lw=1, label='Original data')
ax.plot(tn, yn[:, col], '.-', lw=2,
label='Interpolated')
else:
ax.plot(t, y[:, col], 'o-', lw=1)
ax.plot(tn, yn[:, col], '.-', lw=2, label='Col= %d' % col)
ax.locator_params(axis='y', nbins=7)
ax.legend(fontsize=12, loc='best', framealpha=.5, numpoints=1)
plt.xlabel('[%]')
plt.tight_layout()
plt.show()
```
|
github_jupyter
|
yn, tn, indie = tnorm(y, axis=0, step=1, k=3, smooth=0, mask=None,
nan_at_ext='delete', show=False, ax=None)
# Import the necessary libraries
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
import sys
sys.path.insert(1, r'./../functions') # add to pythonpath
y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
print("y data:")
y
t = np.linspace(0, 100, len(y)) # time vector for the original data
tn = np.linspace(0, 100, 101) # new time vector for the new time-normalized data
yn = np.interp(tn, t, y) # new time-normalized data
print("y data interpolated to 101 points:")
yn
plt.figure(figsize=(10,5))
plt.plot(t, y, 'bo-', lw=2, label='original data')
plt.plot(tn, yn, '.-', color=[1, 0, 0, .5], lw=2, label='time normalized')
plt.legend(loc='best', framealpha=.5)
plt.xlabel('Cycle [%]')
plt.show()
from tnorm import tnorm
>>> # Default options: cubic spline interpolation passing through
>>> # each datum, 101 points, and no plot
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> tnorm(y)
>>> # Linear interpolation passing through each datum
>>> yn, tn, indie = tnorm(y, k=1, smooth=0, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing
>>> yn, tn, indie = tnorm(y, k=3, smooth=1, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing and 50 points
>>> x = np.linspace(-3, 3, 60)
>>> y = np.exp(-x**2) + np.random.randn(60)/10
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data (use NaN as mask)
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[:10] = np.NaN # first ten points are missing
>>> y[30: 41] = np.NaN # make other 10 missing points
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, nan_at_ext='replace', show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=1, smooth=0, nan_at_ext='replace', show=True)
>>> # Deal with 2-D array
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y = np.vstack((y-1, y[::-1])).T
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
# %load './../functions/tnorm.py'
"""Time normalization (from 0 to 100% with step interval)."""
import numpy as np
__author__ = 'Marcos Duarte, https://github.com/demotu/BMC'
__version__ = "1.0.6"
__license__ = "MIT"
def tnorm(y, axis=0, step=1, k=3, smooth=0, mask=None, nan_at_ext='delete',
show=False, ax=None):
"""Time normalization (from 0 to 100% with step interval).
Time normalization is usually employed for the temporal alignment of data
obtained from different trials with different duration (number of points).
This code implements a procedure knwown as the normalization to percent
cycle.
This code can perform simple linear interpolation passing through each
datum or spline interpolation (up to quintic splines) passing through each
datum (knots) or not (in case a smoothing parameter > 0 is inputted).
NaNs and any value inputted as a mask parameter and that appears at the
extremities might be removed or replaced by the first/last not-NaN value
before the interpolation because this code does not perform extrapolation.
For a 2D array, the entire row with NaN or a mask value at the extermity
might be removed because of alignment issues with the data from different
columns. As result, if there is a column of only NaNs in the data, the
time normalization can't be performed (an empty NaNs and any value
inputted as a mask parameter and that appears in the middle of the data
(which may represent missing data) are ignored and the interpolation is
performed through these points.
See this IPython notebook [2]_.
Parameters
----------
y : 1-D or 2-D array_like
Array of independent input data. Must be increasing.
If 2-D array, the data in each axis will be interpolated.
axis : int, 0 or 1, optional (default = 0)
Axis along which the interpolation is performed.
0: data in each column are interpolated; 1: for row interpolation
step : float or int, optional (default = 1)
Interval from 0 to 100% to resample y or the number of points y
should be interpolated. In the later case, the desired number of
points should be expressed with step as a negative integer.
For instance, step = 1 or step = -101 will result in the same
number of points at the interpolation (101 points).
If step == 0, the number of points will be the number of data in y.
k : int, optional (default = 3)
Degree of the smoothing spline. Must be 1 <= k <= 5.
If 3, a cubic spline is used.
The number of data points must be larger than k.
smooth : float or None, optional (default = 0)
Positive smoothing factor used to choose the number of knots.
If 0, spline will interpolate through all data points.
If None, smooth=len(y).
mask : None or float, optional (default = None)
Mask to identify missing values which will be ignored.
It can be a list of values.
NaN values will be ignored and don't need to be in the mask.
nan_at_ext : string, optional (default = 'delete')
Method to deal with NaNs at the extremities.
'delete' will delete any NaN at the extremities (the corresponding
entire row in `y` for a 2-D array).
'replace' will replace any NaN at the extremities by first/last
not-NaN value in `y`.
show : bool, optional (default = False)
True (1) plot data in a matplotlib figure.
False (0) to not plot.
ax : a matplotlib.axes.Axes instance, optional (default = None).
Returns
-------
yn : 1-D or 2-D array
Interpolated data (if axis == 0, column oriented for 2-D array).
tn : 1-D array
New x values (from 0 to 100) for the interpolated data.
inds : list
Indexes of first and last rows without NaNs at the extremities of `y`.
If there is no NaN in the data, this list is [0, y.shape[0]-1].
Notes
-----
This code performs interpolation to create data with the desired number of
points using a one-dimensional smoothing spline fit to a given set of data
points (scipy.interpolate.UnivariateSpline function).
References
----------
.. [1] http://www.sciencedirect.com/science/article/pii/S0021929010005038
.. [2] http://nbviewer.ipython.org/github/demotu/BMC/blob/master/notebooks/TimeNormalization.ipynb
See Also
--------
scipy.interpolate.UnivariateSpline:
One-dimensional smoothing spline fit to a given set of data points.
Examples
--------
>>> # Default options: cubic spline interpolation passing through
>>> # each datum, 101 points, and no plot
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> tnorm(y)
>>> # Linear interpolation passing through each datum
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> yn, tn, indie = tnorm(y, k=1, smooth=0, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing
>>> y = [5, 4, 10, 8, 1, 10, 2, 7, 1, 3]
>>> yn, tn, indie = tnorm(y, k=3, smooth=1, mask=None, show=True)
>>> # Cubic spline interpolation with smoothing and 50 points
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data (use NaN as mask)
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[:10] = np.NaN # first ten points are missing
>>> y[30: 41] = np.NaN # make other 10 missing points
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, nan_at_ext='replace', show=True)
>>> # Deal with missing data at the extremities replacing by first/last not-NaN
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y[0:10] = np.NaN # first ten points are missing
>>> y[-10:] = np.NaN # last ten points are missing
>>> yn, tn, indie = tnorm(y, step=-50, k=1, smooth=0, nan_at_ext='replace', show=True)
>>> # Deal with 2-D array
>>> x = np.linspace(-3, 3, 100)
>>> y = np.exp(-x**2) + np.random.randn(100)/10
>>> y = np.vstack((y-1, y[::-1])).T
>>> yn, tn, indie = tnorm(y, step=-50, k=3, smooth=1, show=True)
Version history
---------------
'1.0.6':
Deleted 'from __future__ import ...'
Added parameter `nan_at_ext`
Adjusted outputs to have always the same type
"""
from scipy.interpolate import UnivariateSpline
y = np.asarray(y)
if axis:
y = y.T
if y.ndim == 1:
y = np.reshape(y, (-1, 1))
# turn mask into NaN
if mask is not None:
y[y == mask] = np.NaN
iini = 0
iend = y.shape[0]-1
if nan_at_ext.lower() == 'delete':
# delete rows with missing values at the extremities
while y.size and np.isnan(np.sum(y[0])):
y = np.delete(y, 0, axis=0)
iini += 1
while y.size and np.isnan(np.sum(y[-1])):
y = np.delete(y, -1, axis=0)
iend -= 1
else:
# replace NaN at the extremities by first/last not-NaN
if np.any(np.isnan(y[0])):
for col in range(y.shape[1]):
ind_not_nan = np.nonzero(~np.isnan(y[:, col]))[0]
if ind_not_nan.size:
y[0, col] = y[ind_not_nan[0], col]
else:
y = np.empty((0, 0))
break
if np.any(np.isnan(y[-1])):
for col in range(y.shape[1]):
ind_not_nan = np.nonzero(~np.isnan(y[:, col]))[0]
if ind_not_nan.size:
y[-1, col] = y[ind_not_nan[-1], col]
else:
y = np.empty((0, 0))
break
# check if there are still data
if not y.size:
return np.empty((0, 0)), np.empty(0), []
if y.size == 1:
return y.flatten(), np.array(0), [0, 0]
indie = [iini, iend]
t = np.linspace(0, 100, y.shape[0])
if step == 0:
tn = t
elif step > 0:
tn = np.linspace(0, 100, np.round(100 / step + 1))
else:
tn = np.linspace(0, 100, -step)
yn = np.empty([tn.size, y.shape[1]]) * np.NaN
for col in np.arange(y.shape[1]):
# ignore NaNs inside data for the interpolation
ind = np.isfinite(y[:, col])
if np.sum(ind) > 1: # at least two points for the interpolation
spl = UnivariateSpline(t[ind], y[ind, col], k=k, s=smooth)
yn[:, col] = spl(tn)
if show:
_plot(t, y, ax, tn, yn)
if axis:
y = y.T
if yn.shape[1] == 1:
yn = yn.flatten()
return yn, tn, indie
def _plot(t, y, ax, tn, yn):
"""Plot results of the tnorm function, see its help."""
try:
import matplotlib.pyplot as plt
except ImportError:
print('matplotlib is not available.')
else:
if ax is None:
_, ax = plt.subplots(1, 1, figsize=(8, 5))
ax.set_prop_cycle('color', ['b', 'r', 'b', 'g', 'b', 'y', 'b', 'c', 'b', 'm'])
#ax.set_color_cycle(['b', 'r', 'b', 'g', 'b', 'y', 'b', 'c', 'b', 'm'])
for col in np.arange(y.shape[1]):
if y.shape[1] == 1:
ax.plot(t, y[:, col], 'o-', lw=1, label='Original data')
ax.plot(tn, yn[:, col], '.-', lw=2,
label='Interpolated')
else:
ax.plot(t, y[:, col], 'o-', lw=1)
ax.plot(tn, yn[:, col], '.-', lw=2, label='Col= %d' % col)
ax.locator_params(axis='y', nbins=7)
ax.legend(fontsize=12, loc='best', framealpha=.5, numpoints=1)
plt.xlabel('[%]')
plt.tight_layout()
plt.show()
| 0.667148 | 0.985496 |
# Example - Reading COGs in Parallel
Cloud Optimized Geotiffs (COGs) can be internally chunked, which makes it possible to read them in parallel from multiple threads. However, the libraries `rioxarray` builds on, `rasterio` and `GDAL`, require some care to be used safely from multiple threads within a single process. By default, [rioxarray.open_rasterio](../rioxarray.rst#rioxarray-open-rasterio) will acquire a per-process lock when reading a chunk of a COG.
If you're using `rioxarray` with [Dask](http://docs.dask.org/) through the `chunks` keyword, you can also specify the `lock=False` keyword to ensure that reading *and* operating on your data happen in parallel.
Note: Also see [Reading and Writing with Dask](dask_read_write.ipynb)
## Scheduler Choice
Dask has [several schedulers](https://docs.dask.org/en/latest/scheduling.html) which run computations in parallel. Which scheduler is best depends on a variety of factors, including whether your computation holds Python's Global Interpreter Lock, whether how much data needs to be moved around, and whether you need more than one machine's computational power. This section about read-locks only applies if you have more than one thread in a process. This will happen with Dask's [local threaded scheduler](https://docs.dask.org/en/latest/scheduling.html#local-threads) and its [distributed scheduler](https://distributed.dask.org/en/latest/) when configured to use more than one thread per worker.
By default, `xarray` objects will use the local `threaded` scheduler.
## Reading without Locks
To read a COG without any locks, you'd specify `lock=False`. This tells `rioxarray` to open a new `rasterio.DatasetReader` in each thread, rather than trying to share one amongst multiple threads.
```
import rioxarray
url = (
"https://naipeuwest.blob.core.windows.net/naip/v002/md/2013/md_100cm_2013/"
"39076/m_3907617_ne_18_1_20130924.tif"
)
ds = rioxarray.open_rasterio(url, lock=False, chunks=(4, "auto", -1))
%time _ = ds.mean().compute()
```
Note: these timings are from a VM in the same Azure data center that's hosting the COG. Running this locally will give different times.
## Chunking
For maximum read performance, the chunking pattern you request should align with the internal chunking of the COG. Typically this means reading the data in a "row major" format: your chunks should be as wide as possible along the columns. We did that above with the chunks of `(4, "auto", -1)`. The `-1` says "include all the columns", and the `"auto"` will make the chunking along the rows as large as possible while staying in a reasonable limit (specified in `dask.config.get("array.chunk-size")`).
If we flipped that, and instead read as much of the rows as possible, we'll see slower performance.
```
ds = rioxarray.open_rasterio(url, lock=False, chunks=(1, -1, "auto"))
%time _ = ds.mean().compute()
```
That said, reading is typically just the first step in a larger computation. You'd want to consider what chunking is best for your whole computation. See https://docs.dask.org/en/latest/array-chunks.html for more on choosing chunks.
## Caching Considerations
Specifying `lock=False` will disable some internal caching done by xarray or rasterio. For example, the first and second reads here are roughly the same, since nothing is cached.
```
ds = rioxarray.open_rasterio(url, lock=False, chunks=(4, "auto", -1))
%time _ = ds.mean().compute()
%time _ = ds.mean().compute()
```
By default and when a lock is passed in, the initial read is slower (since some threads are waiting around for a lock).
```
ds = rioxarray.open_rasterio(url, chunks=(4, "auto", -1)) # use the default locking
%time _ = ds.mean().compute()
```
But thanks to caching, subsequent reads are much faster.
```
%time _ = ds.mean().compute()
```
If you're reapeatedly reading subsets of the data, using the default lock or `lock=some_lock_object` to benefit from the caching.
|
github_jupyter
|
import rioxarray
url = (
"https://naipeuwest.blob.core.windows.net/naip/v002/md/2013/md_100cm_2013/"
"39076/m_3907617_ne_18_1_20130924.tif"
)
ds = rioxarray.open_rasterio(url, lock=False, chunks=(4, "auto", -1))
%time _ = ds.mean().compute()
ds = rioxarray.open_rasterio(url, lock=False, chunks=(1, -1, "auto"))
%time _ = ds.mean().compute()
ds = rioxarray.open_rasterio(url, lock=False, chunks=(4, "auto", -1))
%time _ = ds.mean().compute()
%time _ = ds.mean().compute()
ds = rioxarray.open_rasterio(url, chunks=(4, "auto", -1)) # use the default locking
%time _ = ds.mean().compute()
%time _ = ds.mean().compute()
| 0.513668 | 0.96707 |
# Exercise 3 - Building Model
Write a class ``Building`` in a file ``Building.py`` in the same directory that contains the data folder and this exercise.

The class reads a building.xlsx, populates the relevant attributes from two sheets "params" and "thermal_hull", and calculates the LT! To do this, it will reuse the functions you wrote in Exercise 2 and use it as its class methods.
> More on Classes and Methods: https://towardsdatascience.com/get-started-with-object-oriented-programming-in-python-classes-and-instances-2c1849e0b411
The class' ``__init__`` method shoud take the following Inputs
* ``path`` the path of the excel
### Attributes
The class needs to have the following attributes that we can later use in the simulation model, so take care when naming:
* ``self.bgf`` Bruttogeschoßfläche aus dem Blatt params
* ``self.heat_capacity`` Spez. Wirksame Speicherkapazität des GEbäudes in Wh/m²K im Blatt params
* ``self.net_storey_height`` Netto Raumhöhe
* ``self.LT`` Leitwert
You can and should use additional variables and attributes to make the programming more understandable.
### Methods
Build upon exercise 2 and repurpose the functions you developed to read the excel, calculate the relevant additional parameters and call all methods in the right order in the ``__init__()`` function to build the Building model. As a suggestion, the building Class could have the following methods to calculate the Attributes:
* ``load_params(self, path): `` loads the sheet "params" of a excel at path and returns it as a dataframe
* ``load_hull(self, path):`` loads the sheet "thermal_ hull" of a excel at path and returns it as a dataframe
* ``insert_windows(self, hull_df, u_f, ff_anteil):`` takes a hull dataframe from load_hull() and replaces an opak wall with a wall and a window entry, taking the window share and u-value as inputs. **Add the u_f, and ff_anteil variables also to the ``__init__`` parameters and pass it there to the insert_windows() call.**
* ``L_T(self, hull_df):`` calculates the LT from a Hull Dataframe
## Test
Test the Building class by creating an instance of the class with all available building exels.
see that all required attribues work as expected and compare the LT of the buildings.
Try different "Fensterflächenanteile"
```
from Building import Building
test = Building()
print(test.bgf)
print(test.heat_capacity)
print(test.net_storey_height)
print(test.LT)
test.params_df
test.hull_df
#note that the original excel only had the
ph = Building("data/building_ph.xlsx")
ph.LT
ph.hull_df
oib = Building("data/building_oib_16linie.xlsx")
oib.LT
oib.hull_df
```
|
github_jupyter
|
from Building import Building
test = Building()
print(test.bgf)
print(test.heat_capacity)
print(test.net_storey_height)
print(test.LT)
test.params_df
test.hull_df
#note that the original excel only had the
ph = Building("data/building_ph.xlsx")
ph.LT
ph.hull_df
oib = Building("data/building_oib_16linie.xlsx")
oib.LT
oib.hull_df
| 0.282097 | 0.959039 |
```
import time
import matplotlib.pyplot as plt
import numpy as np
import torch
import syft as sy
from syft.frameworks.torch.federated import FederatedDataset, FederatedDataLoader, BaseDataset
from torchvision import transforms
from torchvision import datasets
transforms_image = transforms.Compose([transforms.Resize(32),
transforms.CenterCrop(32),
transforms.ToTensor()])
train_xray = torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/train',
transform=transforms_image),
batch_size=20, shuffle=True)
def imshow(img):
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# obtain one batch of training images
dataiter = iter(train_xray)
images, _ = dataiter.next() # _ for no labels
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(20, 4))
plot_size=20
for idx in np.arange(plot_size):
ax = fig.add_subplot(2, plot_size/2, idx+1, xticks=[], yticks=[])
imshow(images[idx])
hook = sy.TorchHook(torch)
hook = sy.TorchHook(torch)
bob = sy.VirtualWorker(hook, id="bob")
alice = sy.VirtualWorker(hook, id="alice")
image_transforms = {'train': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])]),
'valid': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])]),
'test': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])])
}
loaders = {'train': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/train',
transform=image_transforms['train']),
batch_size=128, shuffle=True),
'valid': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/val',
transform=image_transforms['valid']),
batch_size=128, shuffle=True),
'test': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/test',
transform=image_transforms['test']),
batch_size=128, shuffle=True)}
import math
import logging
import torch
from torch.utils.data import Dataset
logger = logging.getLogger(__name__)
def dataset_federate(dataset, workers):
"""
Add a method to easily transform a torch.Dataset or a sy.BaseDataset
into a sy.FederatedDataset. The dataset given is split in len(workers)
part and sent to each workers
"""
logger.info("Scanning and sending data to {}...".format(", ".join([w.id for w in workers])))
# take ceil to have exactly len(workers) sets after splitting
data_size = math.ceil(len(dataset) / len(workers))
# Fix for old versions of torchvision
# if not hasattr(dataset, "data"):
# if hasattr(dataset, "train_data"):
# dataset.data = dataset.train_data
# elif hasattr(dataset, "test_data"):
# dataset.data = dataset.test_data
# else:
# raise AttributeError("Could not find inputs in dataset")
# if not hasattr(dataset, "targets"):
# if hasattr(dataset, "train_labels"):
# dataset.targets = dataset.train_labels
# elif hasattr(dataset, "test_labels"):
# dataset.targets = dataset.test_labels
# else:
# raise AttributeError("Could not find targets in dataset")
datasets = []
data_loader = torch.utils.data.DataLoader(dataset, batch_size=data_size, drop_last=True)
for dataset_idx, (data, targets) in enumerate(data_loader):
worker = workers[dataset_idx % len(workers)]
logger.debug("Sending data to worker %s", worker.id)
data = data.send(worker)
targets = targets.send(worker)
datasets.append(BaseDataset(data, targets)) # .send(worker)
logger.debug("Done!")
return FederatedDataset(datasets)
datasets.ImageFolder.federate = dataset_federate
federated_train_loader = sy.FederatedDataLoader(datasets.ImageFolder('chest_xray/train',
transform=image_transforms['train']).federate((bob, alice)),
batch_size=128, shuffle=True)
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3, 20, 5, 1)
self.conv2 = nn.Conv2d(20, 50, 5, 1)
self.fc1 = nn.Linear(140450, 500)
self.fc2 = nn.Linear(500, 10)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(x, 2, 2)
x = F.relu(self.conv2(x))
x = F.max_pool2d(x, 2, 2)
x = x.view(x.shape[0], -1)
x = F.relu(self.fc1(x))
x = self.fc2(x)
return F.log_softmax(x, dim=1)
model = Net()
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr = 0.0005)
for epoch in range(10):
model.train()
for batch_idx, (data, target) in enumerate(federated_train_loader):
model.send(data.location)
output = model(data)
data, target = data.to('cpu'), target.to('cpu')
loss = criterion(output, target)
optimizer.zero_grad()
loss.backward()
optimizer.step()
model.get()
if batch_idx % 30 == 0:
loss = loss.get()
print('Train Epoch: {} Loss: {:.6f}'.format(
epoch, loss.item()))
model.eval()
print('Epoch: ', epoch)
total_correct = 0
total = 0
for batch_idx, (data, target) in enumerate(loaders['test']):
output = model(data)
loss = criterion(output, target)
print('Loss: ', loss.item())
max_arg_output = torch.argmax(output, dim=1)
total_correct += int(torch.sum(max_arg_output == target))
total += data.shape[0]
print('Testing data accuracy: {:.0%}'.format(total_correct/total))
if total_correct/total > 0.5:
torch.save(model.state_dict(), 'pt_federated/XRP_' + str(time.strftime("%Y%m%d_%H%M%S"))+'.pt')
```
|
github_jupyter
|
import time
import matplotlib.pyplot as plt
import numpy as np
import torch
import syft as sy
from syft.frameworks.torch.federated import FederatedDataset, FederatedDataLoader, BaseDataset
from torchvision import transforms
from torchvision import datasets
transforms_image = transforms.Compose([transforms.Resize(32),
transforms.CenterCrop(32),
transforms.ToTensor()])
train_xray = torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/train',
transform=transforms_image),
batch_size=20, shuffle=True)
def imshow(img):
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# obtain one batch of training images
dataiter = iter(train_xray)
images, _ = dataiter.next() # _ for no labels
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(20, 4))
plot_size=20
for idx in np.arange(plot_size):
ax = fig.add_subplot(2, plot_size/2, idx+1, xticks=[], yticks=[])
imshow(images[idx])
hook = sy.TorchHook(torch)
hook = sy.TorchHook(torch)
bob = sy.VirtualWorker(hook, id="bob")
alice = sy.VirtualWorker(hook, id="alice")
image_transforms = {'train': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])]),
'valid': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])]),
'test': transforms.Compose([transforms.Resize(224),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])])
}
loaders = {'train': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/train',
transform=image_transforms['train']),
batch_size=128, shuffle=True),
'valid': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/val',
transform=image_transforms['valid']),
batch_size=128, shuffle=True),
'test': torch.utils.data.DataLoader(datasets.ImageFolder('chest_xray/test',
transform=image_transforms['test']),
batch_size=128, shuffle=True)}
import math
import logging
import torch
from torch.utils.data import Dataset
logger = logging.getLogger(__name__)
def dataset_federate(dataset, workers):
"""
Add a method to easily transform a torch.Dataset or a sy.BaseDataset
into a sy.FederatedDataset. The dataset given is split in len(workers)
part and sent to each workers
"""
logger.info("Scanning and sending data to {}...".format(", ".join([w.id for w in workers])))
# take ceil to have exactly len(workers) sets after splitting
data_size = math.ceil(len(dataset) / len(workers))
# Fix for old versions of torchvision
# if not hasattr(dataset, "data"):
# if hasattr(dataset, "train_data"):
# dataset.data = dataset.train_data
# elif hasattr(dataset, "test_data"):
# dataset.data = dataset.test_data
# else:
# raise AttributeError("Could not find inputs in dataset")
# if not hasattr(dataset, "targets"):
# if hasattr(dataset, "train_labels"):
# dataset.targets = dataset.train_labels
# elif hasattr(dataset, "test_labels"):
# dataset.targets = dataset.test_labels
# else:
# raise AttributeError("Could not find targets in dataset")
datasets = []
data_loader = torch.utils.data.DataLoader(dataset, batch_size=data_size, drop_last=True)
for dataset_idx, (data, targets) in enumerate(data_loader):
worker = workers[dataset_idx % len(workers)]
logger.debug("Sending data to worker %s", worker.id)
data = data.send(worker)
targets = targets.send(worker)
datasets.append(BaseDataset(data, targets)) # .send(worker)
logger.debug("Done!")
return FederatedDataset(datasets)
datasets.ImageFolder.federate = dataset_federate
federated_train_loader = sy.FederatedDataLoader(datasets.ImageFolder('chest_xray/train',
transform=image_transforms['train']).federate((bob, alice)),
batch_size=128, shuffle=True)
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.conv1 = nn.Conv2d(3, 20, 5, 1)
self.conv2 = nn.Conv2d(20, 50, 5, 1)
self.fc1 = nn.Linear(140450, 500)
self.fc2 = nn.Linear(500, 10)
def forward(self, x):
x = F.relu(self.conv1(x))
x = F.max_pool2d(x, 2, 2)
x = F.relu(self.conv2(x))
x = F.max_pool2d(x, 2, 2)
x = x.view(x.shape[0], -1)
x = F.relu(self.fc1(x))
x = self.fc2(x)
return F.log_softmax(x, dim=1)
model = Net()
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr = 0.0005)
for epoch in range(10):
model.train()
for batch_idx, (data, target) in enumerate(federated_train_loader):
model.send(data.location)
output = model(data)
data, target = data.to('cpu'), target.to('cpu')
loss = criterion(output, target)
optimizer.zero_grad()
loss.backward()
optimizer.step()
model.get()
if batch_idx % 30 == 0:
loss = loss.get()
print('Train Epoch: {} Loss: {:.6f}'.format(
epoch, loss.item()))
model.eval()
print('Epoch: ', epoch)
total_correct = 0
total = 0
for batch_idx, (data, target) in enumerate(loaders['test']):
output = model(data)
loss = criterion(output, target)
print('Loss: ', loss.item())
max_arg_output = torch.argmax(output, dim=1)
total_correct += int(torch.sum(max_arg_output == target))
total += data.shape[0]
print('Testing data accuracy: {:.0%}'.format(total_correct/total))
if total_correct/total > 0.5:
torch.save(model.state_dict(), 'pt_federated/XRP_' + str(time.strftime("%Y%m%d_%H%M%S"))+'.pt')
| 0.810404 | 0.635618 |
# Basic Classification using the Keras Sequential API
## Learning Objectives
1. Build a model
2. Train this model on example data
3. Use the model to make predictions about unknown data
## Introduction
In this notebook, you use machine learning to *categorize* Iris flowers by species. It uses TensorFlow to:
* Use TensorFlow's default eager execution development environment
* Import data with the Datasets API
* Build models and layers with TensorFlow's Keras API
Here firstly we will Import and parse the dataset, then select the type of model. After that Train the model.
At last we will Evaluate the model's effectiveness and then use the trained model to make predictions.
Each learning objective will correspond to a __#TODO__ in the [student lab notebook](https://github.com/GoogleCloudPlatform/training-data-analyst/blob/master/courses/machine_learning/deepdive2/introduction_to_tensorflow/labs/basic_intro_logistic_regression.ipynb) -- try to complete that notebook first before reviewing this solution notebook.
```
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
# Ensure the right version of Tensorflow is installed.
!pip freeze | grep tensorflow==2.1 || pip install tensorflow==2.1
```
### Configure imports
Import TensorFlow and the other required Python modules. By default, TensorFlow uses eager execution to evaluate operations immediately, returning concrete values instead of creating a computational graph that is executed later. If you are used to a REPL or the `python` interactive console, this feels familiar.
```
import os
import matplotlib.pyplot as plt
import tensorflow as tf
print("TensorFlow version: {}".format(tf.__version__))
print("Eager execution: {}".format(tf.executing_eagerly()))
```
## The Iris classification problem
Imagine you are a botanist seeking an automated way to categorize each Iris flower you find. Machine learning provides many algorithms to classify flowers statistically. For instance, a sophisticated machine learning program could classify flowers based on photographs. Our ambitions are more modest—we're going to classify Iris flowers based on the length and width measurements of their [sepals](https://en.wikipedia.org/wiki/Sepal) and [petals](https://en.wikipedia.org/wiki/Petal).
The Iris genus entails about 300 species, but our program will only classify the following three:
* Iris setosa
* Iris virginica
* Iris versicolor
<table>
<tr><td>
<img src="https://www.tensorflow.org/images/iris_three_species.jpg"
alt="Petal geometry compared for three iris species: Iris setosa, Iris virginica, and Iris versicolor">
</td></tr>
<tr><td align="center">
<b>Figure 1.</b> <a href="https://commons.wikimedia.org/w/index.php?curid=170298">Iris setosa</a> (by <a href="https://commons.wikimedia.org/wiki/User:Radomil">Radomil</a>, CC BY-SA 3.0), <a href="https://commons.wikimedia.org/w/index.php?curid=248095">Iris versicolor</a>, (by <a href="https://commons.wikimedia.org/wiki/User:Dlanglois">Dlanglois</a>, CC BY-SA 3.0), and <a href="https://www.flickr.com/photos/33397993@N05/3352169862">Iris virginica</a> (by <a href="https://www.flickr.com/photos/33397993@N05">Frank Mayfield</a>, CC BY-SA 2.0).<br/>
</td></tr>
</table>
Fortunately, someone has already created a [dataset of 120 Iris flowers](https://en.wikipedia.org/wiki/Iris_flower_data_set) with the sepal and petal measurements. This is a classic dataset that is popular for beginner machine learning classification problems.
## Import and parse the training dataset
Download the dataset file and convert it into a structure that can be used by this Python program.
### Download the dataset
Download the training dataset file using the `tf.keras.utils.get_file` function. This returns the file path of the downloaded file:
```
train_dataset_url = "https://storage.googleapis.com/download.tensorflow.org/data/iris_training.csv"
train_dataset_fp = tf.keras.utils.get_file(fname=os.path.basename(train_dataset_url),
origin=train_dataset_url)
print("Local copy of the dataset file: {}".format(train_dataset_fp))
```
### Inspect the data
This dataset, `iris_training.csv`, is a plain text file that stores tabular data formatted as comma-separated values (CSV). Use the `head -n5` command to take a peek at the first five entries:
```
!head -n5 {train_dataset_fp}
```
From this view of the dataset, notice the following:
1. The first line is a header containing information about the dataset:
* There are 120 total examples. Each example has four features and one of three possible label names.
2. Subsequent rows are data records, one [example](https://developers.google.com/machine-learning/glossary/#example) per line, where:
* The first four fields are [features](https://developers.google.com/machine-learning/glossary/#feature): these are the characteristics of an example. Here, the fields hold float numbers representing flower measurements.
* The last column is the [label](https://developers.google.com/machine-learning/glossary/#label): this is the value we want to predict. For this dataset, it's an integer value of 0, 1, or 2 that corresponds to a flower name.
Let's write that out in code:
```
# column order in CSV file
column_names = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'species']
feature_names = column_names[:-1]
label_name = column_names[-1]
print("Features: {}".format(feature_names))
print("Label: {}".format(label_name))
```
Each label is associated with string name (for example, "setosa"), but machine learning typically relies on numeric values. The label numbers are mapped to a named representation, such as:
* `0`: Iris setosa
* `1`: Iris versicolor
* `2`: Iris virginica
For more information about features and labels, see the [ML Terminology section of the Machine Learning Crash Course](https://developers.google.com/machine-learning/crash-course/framing/ml-terminology).
```
class_names = ['Iris setosa', 'Iris versicolor', 'Iris virginica']
```
### Create a `tf.data.Dataset`
TensorFlow's Dataset API handles many common cases for loading data into a model. This is a high-level API for reading data and transforming it into a form used for training.
Since the dataset is a CSV-formatted text file, use the `tf.data.experimental.make_csv_dataset` function to parse the data into a suitable format. Since this function generates data for training models, the default behavior is to shuffle the data (`shuffle=True, shuffle_buffer_size=10000`), and repeat the dataset forever (`num_epochs=None`). We also set the [batch_size](https://developers.google.com/machine-learning/glossary/#batch_size) parameter:
```
batch_size = 32
train_dataset = tf.data.experimental.make_csv_dataset(
train_dataset_fp,
batch_size,
column_names=column_names,
label_name=label_name,
num_epochs=1)
```
The `make_csv_dataset` function returns a `tf.data.Dataset` of `(features, label)` pairs, where `features` is a dictionary: `{'feature_name': value}`
These `Dataset` objects are iterable. Let's look at a batch of features:
```
features, labels = next(iter(train_dataset))
print(features)
```
Notice that like-features are grouped together, or *batched*. Each example row's fields are appended to the corresponding feature array. Change the `batch_size` to set the number of examples stored in these feature arrays.
You can start to see some clusters by plotting a few features from the batch:
```
plt.scatter(features['petal_length'],
features['sepal_length'],
c=labels,
cmap='viridis')
plt.xlabel("Petal length")
plt.ylabel("Sepal length")
plt.show()
```
To simplify the model building step, create a function to repackage the features dictionary into a single array with shape: `(batch_size, num_features)`.
This function uses the `tf.stack` method which takes values from a list of tensors and creates a combined tensor at the specified dimension:
```
def pack_features_vector(features, labels):
"""Pack the features into a single array."""
features = tf.stack(list(features.values()), axis=1)
return features, labels
```
Then use the `tf.data.Dataset#map` method to pack the `features` of each `(features,label)` pair into the training dataset:
```
train_dataset = train_dataset.map(pack_features_vector)
```
The features element of the `Dataset` are now arrays with shape `(batch_size, num_features)`. Let's look at the first few examples:
```
features, labels = next(iter(train_dataset))
print(features[:5])
```
## Select the type of model
### Why model?
A [model](https://developers.google.com/machine-learning/crash-course/glossary#model) is a relationship between features and the label. For the Iris classification problem, the model defines the relationship between the sepal and petal measurements and the predicted Iris species. Some simple models can be described with a few lines of algebra, but complex machine learning models have a large number of parameters that are difficult to summarize.
Could you determine the relationship between the four features and the Iris species *without* using machine learning? That is, could you use traditional programming techniques (for example, a lot of conditional statements) to create a model? Perhaps—if you analyzed the dataset long enough to determine the relationships between petal and sepal measurements to a particular species. And this becomes difficult—maybe impossible—on more complicated datasets. A good machine learning approach *determines the model for you*. If you feed enough representative examples into the right machine learning model type, the program will figure out the relationships for you.
### Select the model
We need to select the kind of model to train. There are many types of models and picking a good one takes experience. This tutorial uses a neural network to solve the Iris classification problem. [Neural networks](https://developers.google.com/machine-learning/glossary/#neural_network) can find complex relationships between features and the label. It is a highly-structured graph, organized into one or more [hidden layers](https://developers.google.com/machine-learning/glossary/#hidden_layer). Each hidden layer consists of one or more [neurons](https://developers.google.com/machine-learning/glossary/#neuron). There are several categories of neural networks and this program uses a dense, or [fully-connected neural network](https://developers.google.com/machine-learning/glossary/#fully_connected_layer): the neurons in one layer receive input connections from *every* neuron in the previous layer. For example, Figure 2 illustrates a dense neural network consisting of an input layer, two hidden layers, and an output layer:
<table>
<tr><td>
<img src="https://www.tensorflow.org/images/custom_estimators/full_network.png"
alt="A diagram of the network architecture: Inputs, 2 hidden layers, and outputs">
</td></tr>
<tr><td align="center">
<b>Figure 2.</b> A neural network with features, hidden layers, and predictions.<br/>
</td></tr>
</table>
When the model from Figure 2 is trained and fed an unlabeled example, it yields three predictions: the likelihood that this flower is the given Iris species. This prediction is called [inference](https://developers.google.com/machine-learning/crash-course/glossary#inference). For this example, the sum of the output predictions is 1.0. In Figure 2, this prediction breaks down as: `0.02` for *Iris setosa*, `0.95` for *Iris versicolor*, and `0.03` for *Iris virginica*. This means that the model predicts—with 95% probability—that an unlabeled example flower is an *Iris versicolor*.
### Create a model using Keras
The TensorFlow `tf.keras` API is the preferred way to create models and layers. This makes it easy to build models and experiment while Keras handles the complexity of connecting everything together.
The `tf.keras.Sequential` model is a linear stack of layers. Its constructor takes a list of layer instances, in this case, two `tf.keras.layers.Dense` layers with 10 nodes each, and an output layer with 3 nodes representing our label predictions. The first layer's `input_shape` parameter corresponds to the number of features from the dataset, and is required:
```
# TODO 1
model = tf.keras.Sequential([
tf.keras.layers.Dense(10, activation=tf.nn.relu, input_shape=(4,)), # input shape required
tf.keras.layers.Dense(10, activation=tf.nn.relu),
tf.keras.layers.Dense(3)
])
```
The [activation function](https://developers.google.com/machine-learning/crash-course/glossary#activation_function) determines the output shape of each node in the layer. These non-linearities are important—without them the model would be equivalent to a single layer. There are many `tf.keras.activations`, but [ReLU](https://developers.google.com/machine-learning/crash-course/glossary#ReLU) is common for hidden layers.
The ideal number of hidden layers and neurons depends on the problem and the dataset. Like many aspects of machine learning, picking the best shape of the neural network requires a mixture of knowledge and experimentation. As a rule of thumb, increasing the number of hidden layers and neurons typically creates a more powerful model, which requires more data to train effectively.
### Using the model
Let's have a quick look at what this model does to a batch of features:
```
predictions = model(features)
predictions[:5]
```
Here, each example returns a [logit](https://developers.google.com/machine-learning/crash-course/glossary#logits) for each class.
To convert these logits to a probability for each class, use the [softmax](https://developers.google.com/machine-learning/crash-course/glossary#softmax) function:
```
tf.nn.softmax(predictions[:5])
```
Taking the `tf.argmax` across classes gives us the predicted class index. But, the model hasn't been trained yet, so these aren't good predictions:
```
print("Prediction: {}".format(tf.argmax(predictions, axis=1)))
print("Labels: {}".format(labels))
```
## Train the model
[Training](https://developers.google.com/machine-learning/crash-course/glossary#training) is the stage of machine learning when the model is gradually optimized, or the model *learns* the dataset. The goal is to learn enough about the structure of the training dataset to make predictions about unseen data. If you learn *too much* about the training dataset, then the predictions only work for the data it has seen and will not be generalizable. This problem is called [overfitting](https://developers.google.com/machine-learning/crash-course/glossary#overfitting)—it's like memorizing the answers instead of understanding how to solve a problem.
The Iris classification problem is an example of [supervised machine learning](https://developers.google.com/machine-learning/glossary/#supervised_machine_learning): the model is trained from examples that contain labels. In [unsupervised machine learning](https://developers.google.com/machine-learning/glossary/#unsupervised_machine_learning), the examples don't contain labels. Instead, the model typically finds patterns among the features.
### Define the loss and gradient function
Both training and evaluation stages need to calculate the model's [loss](https://developers.google.com/machine-learning/crash-course/glossary#loss). This measures how off a model's predictions are from the desired label, in other words, how bad the model is performing. We want to minimize, or optimize, this value.
Our model will calculate its loss using the `tf.keras.losses.SparseCategoricalCrossentropy` function which takes the model's class probability predictions and the desired label, and returns the average loss across the examples.
```
loss_object = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
def loss(model, x, y, training):
# TODO 2
# training=training is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
y_ = model(x, training=training)
return loss_object(y_true=y, y_pred=y_)
l = loss(model, features, labels, training=False)
print("Loss test: {}".format(l))
```
Use the `tf.GradientTape` context to calculate the [gradients](https://developers.google.com/machine-learning/crash-course/glossary#gradient) used to optimize your model:
```
def grad(model, inputs, targets):
with tf.GradientTape() as tape:
loss_value = loss(model, inputs, targets, training=True)
return loss_value, tape.gradient(loss_value, model.trainable_variables)
```
### Create an optimizer
An [optimizer](https://developers.google.com/machine-learning/crash-course/glossary#optimizer) applies the computed gradients to the model's variables to minimize the `loss` function. You can think of the loss function as a curved surface (see Figure 3) and we want to find its lowest point by walking around. The gradients point in the direction of steepest ascent—so we'll travel the opposite way and move down the hill. By iteratively calculating the loss and gradient for each batch, we'll adjust the model during training. Gradually, the model will find the best combination of weights and bias to minimize loss. And the lower the loss, the better the model's predictions.
<table>
<tr><td>
<img src="https://cs231n.github.io/assets/nn3/opt1.gif" width="70%"
alt="Optimization algorithms visualized over time in 3D space.">
</td></tr>
<tr><td align="center">
<b>Figure 3.</b> Optimization algorithms visualized over time in 3D space.<br/>(Source: <a href="http://cs231n.github.io/neural-networks-3/">Stanford class CS231n</a>, MIT License, Image credit: <a href="https://twitter.com/alecrad">Alec Radford</a>)
</td></tr>
</table>
TensorFlow has many optimization algorithms available for training. This model uses the `tf.keras.optimizers.SGD` that implements the [stochastic gradient descent](https://developers.google.com/machine-learning/crash-course/glossary#gradient_descent) (SGD) algorithm. The `learning_rate` sets the step size to take for each iteration down the hill. This is a *hyperparameter* that you'll commonly adjust to achieve better results.
Let's setup the optimizer:
```
optimizer = tf.keras.optimizers.SGD(learning_rate=0.01)
```
We'll use this to calculate a single optimization step:
```
loss_value, grads = grad(model, features, labels)
print("Step: {}, Initial Loss: {}".format(optimizer.iterations.numpy(),
loss_value.numpy()))
optimizer.apply_gradients(zip(grads, model.trainable_variables))
print("Step: {},Loss: {}".format(optimizer.iterations.numpy(),
loss(model, features, labels, training=True).numpy()))
```
### Training loop
With all the pieces in place, the model is ready for training! A training loop feeds the dataset examples into the model to help it make better predictions. The following code block sets up these training steps:
1. Iterate each *epoch*. An epoch is one pass through the dataset.
2. Within an epoch, iterate over each example in the training `Dataset` grabbing its *features* (`x`) and *label* (`y`).
3. Using the example's features, make a prediction and compare it with the label. Measure the inaccuracy of the prediction and use that to calculate the model's loss and gradients.
4. Use an `optimizer` to update the model's variables.
5. Keep track of some stats for visualization.
6. Repeat for each epoch.
The `num_epochs` variable is the number of times to loop over the dataset collection. Counter-intuitively, training a model longer does not guarantee a better model. `num_epochs` is a [hyperparameter](https://developers.google.com/machine-learning/glossary/#hyperparameter) that you can tune. Choosing the right number usually requires both experience and experimentation:
```
## Note: Rerunning this cell uses the same model variables
# Keep results for plotting
train_loss_results = []
train_accuracy_results = []
num_epochs = 201
for epoch in range(num_epochs):
epoch_loss_avg = tf.keras.metrics.Mean()
epoch_accuracy = tf.keras.metrics.SparseCategoricalAccuracy()
# Training loop - using batches of 32
for x, y in train_dataset:
# Optimize the model
loss_value, grads = grad(model, x, y)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
# Track progress
epoch_loss_avg.update_state(loss_value) # Add current batch loss
# Compare predicted label to actual label
# training=True is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
epoch_accuracy.update_state(y, model(x, training=True))
# End epoch
train_loss_results.append(epoch_loss_avg.result())
train_accuracy_results.append(epoch_accuracy.result())
if epoch % 50 == 0:
print("Epoch {:03d}: Loss: {:.3f}, Accuracy: {:.3%}".format(epoch,
epoch_loss_avg.result(),
epoch_accuracy.result()))
```
### Visualize the loss function over time
While it's helpful to print out the model's training progress, it's often *more* helpful to see this progress. [TensorBoard](https://www.tensorflow.org/tensorboard) is a nice visualization tool that is packaged with TensorFlow, but we can create basic charts using the `matplotlib` module.
Interpreting these charts takes some experience, but you really want to see the *loss* go down and the *accuracy* go up:
```
fig, axes = plt.subplots(2, sharex=True, figsize=(12, 8))
fig.suptitle('Training Metrics')
axes[0].set_ylabel("Loss", fontsize=14)
axes[0].plot(train_loss_results)
axes[1].set_ylabel("Accuracy", fontsize=14)
axes[1].set_xlabel("Epoch", fontsize=14)
axes[1].plot(train_accuracy_results)
plt.show()
```
## Evaluate the model's effectiveness
Now that the model is trained, we can get some statistics on its performance.
*Evaluating* means determining how effectively the model makes predictions. To determine the model's effectiveness at Iris classification, pass some sepal and petal measurements to the model and ask the model to predict what Iris species they represent. Then compare the model's predictions against the actual label. For example, a model that picked the correct species on half the input examples has an [accuracy](https://developers.google.com/machine-learning/glossary/#accuracy) of `0.5`. Figure 4 shows a slightly more effective model, getting 4 out of 5 predictions correct at 80% accuracy:
<table cellpadding="8" border="0">
<colgroup>
<col span="4" >
<col span="1" bgcolor="lightblue">
<col span="1" bgcolor="lightgreen">
</colgroup>
<tr bgcolor="lightgray">
<th colspan="4">Example features</th>
<th colspan="1">Label</th>
<th colspan="1" >Model prediction</th>
</tr>
<tr>
<td>5.9</td><td>3.0</td><td>4.3</td><td>1.5</td><td align="center">1</td><td align="center">1</td>
</tr>
<tr>
<td>6.9</td><td>3.1</td><td>5.4</td><td>2.1</td><td align="center">2</td><td align="center">2</td>
</tr>
<tr>
<td>5.1</td><td>3.3</td><td>1.7</td><td>0.5</td><td align="center">0</td><td align="center">0</td>
</tr>
<tr>
<td>6.0</td> <td>3.4</td> <td>4.5</td> <td>1.6</td> <td align="center">1</td><td align="center" bgcolor="red">2</td>
</tr>
<tr>
<td>5.5</td><td>2.5</td><td>4.0</td><td>1.3</td><td align="center">1</td><td align="center">1</td>
</tr>
<tr><td align="center" colspan="6">
<b>Figure 4.</b> An Iris classifier that is 80% accurate.<br/>
</td></tr>
</table>
### Setup the test dataset
Evaluating the model is similar to training the model. The biggest difference is the examples come from a separate [test set](https://developers.google.com/machine-learning/crash-course/glossary#test_set) rather than the training set. To fairly assess a model's effectiveness, the examples used to evaluate a model must be different from the examples used to train the model.
The setup for the test `Dataset` is similar to the setup for training `Dataset`. Download the CSV text file and parse that values, then give it a little shuffle:
```
test_url = "https://storage.googleapis.com/download.tensorflow.org/data/iris_test.csv"
test_fp = tf.keras.utils.get_file(fname=os.path.basename(test_url),
origin=test_url)
test_dataset = tf.data.experimental.make_csv_dataset(
test_fp,
batch_size,
column_names=column_names,
label_name='species',
num_epochs=1,
shuffle=False)
test_dataset = test_dataset.map(pack_features_vector)
```
### Evaluate the model on the test dataset
Unlike the training stage, the model only evaluates a single [epoch](https://developers.google.com/machine-learning/glossary/#epoch) of the test data. In the following code cell, we iterate over each example in the test set and compare the model's prediction against the actual label. This is used to measure the model's accuracy across the entire test set:
```
test_accuracy = tf.keras.metrics.Accuracy()
for (x, y) in test_dataset:
# training=False is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
logits = model(x, training=False)
prediction = tf.argmax(logits, axis=1, output_type=tf.int32)
test_accuracy(prediction, y)
print("Test set accuracy: {:.3%}".format(test_accuracy.result()))
```
We can see on the last batch, for example, the model is usually correct:
```
tf.stack([y,prediction],axis=1)
```
## Use the trained model to make predictions
We've trained a model and "proven" that it's good—but not perfect—at classifying Iris species. Now let's use the trained model to make some predictions on [unlabeled examples](https://developers.google.com/machine-learning/glossary/#unlabeled_example); that is, on examples that contain features but not a label.
In real-life, the unlabeled examples could come from lots of different sources including apps, CSV files, and data feeds. For now, we're going to manually provide three unlabeled examples to predict their labels. Recall, the label numbers are mapped to a named representation as:
* `0`: Iris setosa
* `1`: Iris versicolor
* `2`: Iris virginica
```
# TODO 3
predict_dataset = tf.convert_to_tensor([
[5.1, 3.3, 1.7, 0.5,],
[5.9, 3.0, 4.2, 1.5,],
[6.9, 3.1, 5.4, 2.1]
])
# training=False is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
predictions = model(predict_dataset, training=False)
for i, logits in enumerate(predictions):
class_idx = tf.argmax(logits).numpy()
p = tf.nn.softmax(logits)[class_idx]
name = class_names[class_idx]
print("Example {} prediction: {} ({:4.1f}%)".format(i, name, 100*p))
```
|
github_jupyter
|
!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst
# Ensure the right version of Tensorflow is installed.
!pip freeze | grep tensorflow==2.1 || pip install tensorflow==2.1
import os
import matplotlib.pyplot as plt
import tensorflow as tf
print("TensorFlow version: {}".format(tf.__version__))
print("Eager execution: {}".format(tf.executing_eagerly()))
train_dataset_url = "https://storage.googleapis.com/download.tensorflow.org/data/iris_training.csv"
train_dataset_fp = tf.keras.utils.get_file(fname=os.path.basename(train_dataset_url),
origin=train_dataset_url)
print("Local copy of the dataset file: {}".format(train_dataset_fp))
!head -n5 {train_dataset_fp}
# column order in CSV file
column_names = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width', 'species']
feature_names = column_names[:-1]
label_name = column_names[-1]
print("Features: {}".format(feature_names))
print("Label: {}".format(label_name))
class_names = ['Iris setosa', 'Iris versicolor', 'Iris virginica']
batch_size = 32
train_dataset = tf.data.experimental.make_csv_dataset(
train_dataset_fp,
batch_size,
column_names=column_names,
label_name=label_name,
num_epochs=1)
features, labels = next(iter(train_dataset))
print(features)
plt.scatter(features['petal_length'],
features['sepal_length'],
c=labels,
cmap='viridis')
plt.xlabel("Petal length")
plt.ylabel("Sepal length")
plt.show()
def pack_features_vector(features, labels):
"""Pack the features into a single array."""
features = tf.stack(list(features.values()), axis=1)
return features, labels
train_dataset = train_dataset.map(pack_features_vector)
features, labels = next(iter(train_dataset))
print(features[:5])
# TODO 1
model = tf.keras.Sequential([
tf.keras.layers.Dense(10, activation=tf.nn.relu, input_shape=(4,)), # input shape required
tf.keras.layers.Dense(10, activation=tf.nn.relu),
tf.keras.layers.Dense(3)
])
predictions = model(features)
predictions[:5]
tf.nn.softmax(predictions[:5])
print("Prediction: {}".format(tf.argmax(predictions, axis=1)))
print("Labels: {}".format(labels))
loss_object = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
def loss(model, x, y, training):
# TODO 2
# training=training is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
y_ = model(x, training=training)
return loss_object(y_true=y, y_pred=y_)
l = loss(model, features, labels, training=False)
print("Loss test: {}".format(l))
def grad(model, inputs, targets):
with tf.GradientTape() as tape:
loss_value = loss(model, inputs, targets, training=True)
return loss_value, tape.gradient(loss_value, model.trainable_variables)
optimizer = tf.keras.optimizers.SGD(learning_rate=0.01)
loss_value, grads = grad(model, features, labels)
print("Step: {}, Initial Loss: {}".format(optimizer.iterations.numpy(),
loss_value.numpy()))
optimizer.apply_gradients(zip(grads, model.trainable_variables))
print("Step: {},Loss: {}".format(optimizer.iterations.numpy(),
loss(model, features, labels, training=True).numpy()))
## Note: Rerunning this cell uses the same model variables
# Keep results for plotting
train_loss_results = []
train_accuracy_results = []
num_epochs = 201
for epoch in range(num_epochs):
epoch_loss_avg = tf.keras.metrics.Mean()
epoch_accuracy = tf.keras.metrics.SparseCategoricalAccuracy()
# Training loop - using batches of 32
for x, y in train_dataset:
# Optimize the model
loss_value, grads = grad(model, x, y)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
# Track progress
epoch_loss_avg.update_state(loss_value) # Add current batch loss
# Compare predicted label to actual label
# training=True is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
epoch_accuracy.update_state(y, model(x, training=True))
# End epoch
train_loss_results.append(epoch_loss_avg.result())
train_accuracy_results.append(epoch_accuracy.result())
if epoch % 50 == 0:
print("Epoch {:03d}: Loss: {:.3f}, Accuracy: {:.3%}".format(epoch,
epoch_loss_avg.result(),
epoch_accuracy.result()))
fig, axes = plt.subplots(2, sharex=True, figsize=(12, 8))
fig.suptitle('Training Metrics')
axes[0].set_ylabel("Loss", fontsize=14)
axes[0].plot(train_loss_results)
axes[1].set_ylabel("Accuracy", fontsize=14)
axes[1].set_xlabel("Epoch", fontsize=14)
axes[1].plot(train_accuracy_results)
plt.show()
test_url = "https://storage.googleapis.com/download.tensorflow.org/data/iris_test.csv"
test_fp = tf.keras.utils.get_file(fname=os.path.basename(test_url),
origin=test_url)
test_dataset = tf.data.experimental.make_csv_dataset(
test_fp,
batch_size,
column_names=column_names,
label_name='species',
num_epochs=1,
shuffle=False)
test_dataset = test_dataset.map(pack_features_vector)
test_accuracy = tf.keras.metrics.Accuracy()
for (x, y) in test_dataset:
# training=False is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
logits = model(x, training=False)
prediction = tf.argmax(logits, axis=1, output_type=tf.int32)
test_accuracy(prediction, y)
print("Test set accuracy: {:.3%}".format(test_accuracy.result()))
tf.stack([y,prediction],axis=1)
# TODO 3
predict_dataset = tf.convert_to_tensor([
[5.1, 3.3, 1.7, 0.5,],
[5.9, 3.0, 4.2, 1.5,],
[6.9, 3.1, 5.4, 2.1]
])
# training=False is needed only if there are layers with different
# behavior during training versus inference (e.g. Dropout).
predictions = model(predict_dataset, training=False)
for i, logits in enumerate(predictions):
class_idx = tf.argmax(logits).numpy()
p = tf.nn.softmax(logits)[class_idx]
name = class_names[class_idx]
print("Example {} prediction: {} ({:4.1f}%)".format(i, name, 100*p))
| 0.651798 | 0.989327 |
### Note
* Instructions have been included for each segment. You do not have to follow them exactly, but they are included to help you think through the steps.
```
# Dependencies and Setup
import pandas as pd
import numpy as np
import os
import locale
# Set number and currency formatting
locale.setlocale(locale.LC_ALL,'en_US')
# File to Load (Remember to Change These)
schools_data = os.path.join('Resources', 'schools_complete.csv')
student_data = os.path.join('Resources', 'students_complete.csv')
# Read School and Student Data File and store into Pandas DataFrames
schools_df = pd.read_csv(schools_data)
students_df = pd.read_csv(student_data)
# Combine the data into a single dataset.
schools_df.rename(columns = {'name':'school_name'}, inplace = True)
merged_df = students_df.merge(schools_df, how = 'left', on = 'school_name')
merged_df.head
```
## District Summary
* Calculate the total number of schools
* Calculate the total number of students
* Calculate the total budget
* Calculate the average math score
* Calculate the average reading score
* Calculate the percentage of students with a passing math score (70 or greater)
* Calculate the percentage of students with a passing reading score (70 or greater)
* Calculate the percentage of students who passed math **and** reading (% Overall Passing)
* Create a dataframe to hold the above results
* Optional: give the displayed data cleaner formatting
```
#create array of unique school names
unique_school_names = schools_df['school_name'].unique()
#gives the length of unique school names to give us how many schools
school_count = len(unique_school_names)
#district student count
dist_student_count = schools_df['size'].sum()
#student count from student file (to verify with district student count)
total_student_rec = students_df['student_name'].count()
#total budget
total_budget = schools_df['budget'].sum()
#calculations for number and % passing reading
num_passing_reading = students_df.loc[students_df['reading_score'] >= 70]['reading_score'].count()
perc_pass_reading = num_passing_reading/total_student_rec
#calculations for number and % passing math
num_passing_math = students_df.loc[students_df['math_score'] >= 70]['math_score'].count()
perc_pass_math = num_passing_math/total_student_rec
#average math score calculation
avg_math_score = students_df['math_score'].mean()
#average reading score calculation
avg_reading_score = students_df['reading_score'].mean()
#Overall Passing Rate Calculations
overall_pass = students_df[(students_df['math_score'] >= 70) & (students_df['reading_score'] >= 70)]['student_name'].count()/total_student_rec
# district dataframe from dictionary
district_summary = pd.DataFrame({
"Total Schools": [school_count],
"Total Students": [dist_student_count],
"Total Budget": [total_budget],
"Average Reading Score": [avg_reading_score],
"Average Math Score": [avg_math_score],
"% Passing Reading":[perc_pass_reading],
"% Passing Math": [perc_pass_math],
"Overall Passing Rate": [overall_pass]
})
#store as different df to change order
dist_sum = district_summary[["Total Schools",
"Total Students",
"Total Budget",
"Average Reading Score",
"Average Math Score",
'% Passing Reading',
'% Passing Math',
'Overall Passing Rate']]
# Create a dataframe to hold the above results
results = pd.DataFrame({"Total Schools":[school_count],"Total Students":[dist_student_count],"Total Budget":[total_budget],"Average Math Score":[avg_math_score],"Average Reading Score":[avg_reading_score],"% Passing Math":[perc_pass_math],"% Passing Reading":[perc_pass_reading], "% Overall Passing":[overall_pass]})
#format cells
results['Total Students'] = results['Total Students'].map("{:,}".format)
results['Total Budget'] = results['Total Budget'].map("${:,.2f}".format)
results['Average Math Score'] = results['Average Math Score'].map("{:.2f}".format)
results['Average Reading Score'] = results['Average Reading Score'].map("{:.2f}".format)
results['% Passing Math'] = results['% Passing Math'].map("{:.2f}".format)
results['% Passing Reading'] = results['% Passing Reading'].map("{:.2f}".format)
results['% Overall Passing'] = results['% Overall Passing'].map("{:.2f}".format)
dist_sum
```
## School Summary
* Create an overview table that summarizes key metrics about each school, including:
* School Name
* School Type
* Total Students
* Total School Budget
* Per Student Budget
* Average Math Score
* Average Reading Score
* % Passing Math
* % Passing Reading
* % Overall Passing (The percentage of students that passed math **and** reading.)
* Create a dataframe to hold the above results
```
#groups by school
by_school = merged_df.set_index('school_name').groupby(['school_name'])
#school types
sch_types = schools_df.set_index('school_name')['type']
# total students by school
stu_per_sch = by_school['Student ID'].count()
# school budget
sch_budget = schools_df.set_index('school_name')['budget']
#per student budget
stu_budget = schools_df.set_index('school_name')['budget']/schools_df.set_index('school_name')['size']
#avg scores by school
avg_math = by_school['math_score'].mean()
avg_read = by_school['reading_score'].mean()
# % passing scores
pass_math = merged_df[merged_df['math_score'] >= 70].groupby('school_name')['Student ID'].count()/stu_per_sch
pass_read = merged_df[merged_df['reading_score'] >= 70].groupby('school_name')['Student ID'].count()/stu_per_sch
overall = merged_df[(merged_df['reading_score'] >= 70) & (merged_df['math_score'] >= 70)].groupby('school_name')['Student ID'].count()/stu_per_sch
sch_summary = pd.DataFrame({
"School Type": sch_types,
"Total Students": stu_per_sch,
"Per Student Budget": stu_budget,
"Total School Budget": sch_budget,
"Average Math Score": avg_math,
"Average Reading Score": avg_read,
'% Passing Math': pass_math,
'% Passing Reading': pass_read,
"Overall Passing Rate": overall
})
#munging
sch_summary = sch_summary[['School Type',
'Total Students',
'Total School Budget',
'Per Student Budget',
'Average Math Score',
'Average Reading Score',
'% Passing Math',
'% Passing Reading',
'Overall Passing Rate']]
#formatting
sch_summary = sch_summary[["School Type", "Total Students", "Total School Budget", "Per Student Budget",
"Average Math Score", "Average Reading Score", "% Passing Math",
"% Passing Reading", "Overall Passing Rate"]]
sch_summary["Total School Budget"] = sch_summary["Total School Budget"].map("$(:,2f)".format)
sch_summary
```
## Top Performing Schools (By % Overall Passing)
* Sort and display the top five performing schools by % overall passing.
```
# Top Performing Schools (By % Overall Passing)
top_performing = sch_summary.sort_values(["Overall Passing Rate"], ascending = False)
top_performing.head()
```
## Bottom Performing Schools (By % Overall Passing)
* Sort and display the five worst-performing schools by % overall passing.
```
# Bottom Performing Schools (By % Overall Passing)
bottom_performing = sch_summary.sort_values(["Overall Passing Rate"], ascending = True)
bottom_performing.head()
```
## Math Scores by Grade
* Create a table that lists the average Reading Score for students of each grade level (9th, 10th, 11th, 12th) at each school.
* Create a pandas series for each grade. Hint: use a conditional statement.
* Group each series by school
* Combine the series into a dataframe
* Optional: give the displayed data cleaner formatting
```
#creates grade level average math scores for each school
ninth_math = students_df.loc[students_df['grade'] == '9th'].groupby('school_name')["math_score"].mean()
tenth_math = students_df.loc[students_df['grade'] == '10th'].groupby('school_name')["math_score"].mean()
eleventh_math = students_df.loc[students_df['grade'] == '11th'].groupby('school_name')["math_score"].mean()
twelfth_math = students_df.loc[students_df['grade'] == '12th'].groupby('school_name')["math_score"].mean()
math_scores = pd.DataFrame({
"9th": ninth_math,
"10th": tenth_math,
"11th": eleventh_math,
"12th": twelfth_math
})
math_scores = math_scores[['9th', '10th', '11th', '12th']]
math_scores.index.name = "School"
#show and format
math_scores
```
## Reading Score by Grade
* Perform the same operations as above for reading scores
```
#creates grade level average reading scores for each school
ninth_reading = students_df.loc[students_df['grade'] == '9th'].groupby('school_name')["reading_score"].mean()
tenth_reading = students_df.loc[students_df['grade'] == '10th'].groupby('school_name')["reading_score"].mean()
eleventh_reading = students_df.loc[students_df['grade'] == '11th'].groupby('school_name')["reading_score"].mean()
twelfth_reading = students_df.loc[students_df['grade'] == '12th'].groupby('school_name')["reading_score"].mean()
#merges the reading score averages by school and grade together
reading_scores = pd.DataFrame({
"9th": ninth_reading,
"10th": tenth_reading,
"11th": eleventh_reading,
"12th": twelfth_reading
})
reading_scores = reading_scores[['9th', '10th', '11th', '12th']]
reading_scores.index.name = "School"
reading_scores
```
## Scores by School Spending
* Create a table that breaks down school performances based on average Spending Ranges (Per Student). Use 4 reasonable bins to group school spending. Include in the table each of the following:
* Average Math Score
* Average Reading Score
* % Passing Math
* % Passing Reading
* Overall Passing Rate (Average of the above two)
## Scores by School Size
* Perform the same operations as above, based on school size.
## Scores by School Type
* Perform the same operations as above, based on school type
|
github_jupyter
|
# Dependencies and Setup
import pandas as pd
import numpy as np
import os
import locale
# Set number and currency formatting
locale.setlocale(locale.LC_ALL,'en_US')
# File to Load (Remember to Change These)
schools_data = os.path.join('Resources', 'schools_complete.csv')
student_data = os.path.join('Resources', 'students_complete.csv')
# Read School and Student Data File and store into Pandas DataFrames
schools_df = pd.read_csv(schools_data)
students_df = pd.read_csv(student_data)
# Combine the data into a single dataset.
schools_df.rename(columns = {'name':'school_name'}, inplace = True)
merged_df = students_df.merge(schools_df, how = 'left', on = 'school_name')
merged_df.head
#create array of unique school names
unique_school_names = schools_df['school_name'].unique()
#gives the length of unique school names to give us how many schools
school_count = len(unique_school_names)
#district student count
dist_student_count = schools_df['size'].sum()
#student count from student file (to verify with district student count)
total_student_rec = students_df['student_name'].count()
#total budget
total_budget = schools_df['budget'].sum()
#calculations for number and % passing reading
num_passing_reading = students_df.loc[students_df['reading_score'] >= 70]['reading_score'].count()
perc_pass_reading = num_passing_reading/total_student_rec
#calculations for number and % passing math
num_passing_math = students_df.loc[students_df['math_score'] >= 70]['math_score'].count()
perc_pass_math = num_passing_math/total_student_rec
#average math score calculation
avg_math_score = students_df['math_score'].mean()
#average reading score calculation
avg_reading_score = students_df['reading_score'].mean()
#Overall Passing Rate Calculations
overall_pass = students_df[(students_df['math_score'] >= 70) & (students_df['reading_score'] >= 70)]['student_name'].count()/total_student_rec
# district dataframe from dictionary
district_summary = pd.DataFrame({
"Total Schools": [school_count],
"Total Students": [dist_student_count],
"Total Budget": [total_budget],
"Average Reading Score": [avg_reading_score],
"Average Math Score": [avg_math_score],
"% Passing Reading":[perc_pass_reading],
"% Passing Math": [perc_pass_math],
"Overall Passing Rate": [overall_pass]
})
#store as different df to change order
dist_sum = district_summary[["Total Schools",
"Total Students",
"Total Budget",
"Average Reading Score",
"Average Math Score",
'% Passing Reading',
'% Passing Math',
'Overall Passing Rate']]
# Create a dataframe to hold the above results
results = pd.DataFrame({"Total Schools":[school_count],"Total Students":[dist_student_count],"Total Budget":[total_budget],"Average Math Score":[avg_math_score],"Average Reading Score":[avg_reading_score],"% Passing Math":[perc_pass_math],"% Passing Reading":[perc_pass_reading], "% Overall Passing":[overall_pass]})
#format cells
results['Total Students'] = results['Total Students'].map("{:,}".format)
results['Total Budget'] = results['Total Budget'].map("${:,.2f}".format)
results['Average Math Score'] = results['Average Math Score'].map("{:.2f}".format)
results['Average Reading Score'] = results['Average Reading Score'].map("{:.2f}".format)
results['% Passing Math'] = results['% Passing Math'].map("{:.2f}".format)
results['% Passing Reading'] = results['% Passing Reading'].map("{:.2f}".format)
results['% Overall Passing'] = results['% Overall Passing'].map("{:.2f}".format)
dist_sum
#groups by school
by_school = merged_df.set_index('school_name').groupby(['school_name'])
#school types
sch_types = schools_df.set_index('school_name')['type']
# total students by school
stu_per_sch = by_school['Student ID'].count()
# school budget
sch_budget = schools_df.set_index('school_name')['budget']
#per student budget
stu_budget = schools_df.set_index('school_name')['budget']/schools_df.set_index('school_name')['size']
#avg scores by school
avg_math = by_school['math_score'].mean()
avg_read = by_school['reading_score'].mean()
# % passing scores
pass_math = merged_df[merged_df['math_score'] >= 70].groupby('school_name')['Student ID'].count()/stu_per_sch
pass_read = merged_df[merged_df['reading_score'] >= 70].groupby('school_name')['Student ID'].count()/stu_per_sch
overall = merged_df[(merged_df['reading_score'] >= 70) & (merged_df['math_score'] >= 70)].groupby('school_name')['Student ID'].count()/stu_per_sch
sch_summary = pd.DataFrame({
"School Type": sch_types,
"Total Students": stu_per_sch,
"Per Student Budget": stu_budget,
"Total School Budget": sch_budget,
"Average Math Score": avg_math,
"Average Reading Score": avg_read,
'% Passing Math': pass_math,
'% Passing Reading': pass_read,
"Overall Passing Rate": overall
})
#munging
sch_summary = sch_summary[['School Type',
'Total Students',
'Total School Budget',
'Per Student Budget',
'Average Math Score',
'Average Reading Score',
'% Passing Math',
'% Passing Reading',
'Overall Passing Rate']]
#formatting
sch_summary = sch_summary[["School Type", "Total Students", "Total School Budget", "Per Student Budget",
"Average Math Score", "Average Reading Score", "% Passing Math",
"% Passing Reading", "Overall Passing Rate"]]
sch_summary["Total School Budget"] = sch_summary["Total School Budget"].map("$(:,2f)".format)
sch_summary
# Top Performing Schools (By % Overall Passing)
top_performing = sch_summary.sort_values(["Overall Passing Rate"], ascending = False)
top_performing.head()
# Bottom Performing Schools (By % Overall Passing)
bottom_performing = sch_summary.sort_values(["Overall Passing Rate"], ascending = True)
bottom_performing.head()
#creates grade level average math scores for each school
ninth_math = students_df.loc[students_df['grade'] == '9th'].groupby('school_name')["math_score"].mean()
tenth_math = students_df.loc[students_df['grade'] == '10th'].groupby('school_name')["math_score"].mean()
eleventh_math = students_df.loc[students_df['grade'] == '11th'].groupby('school_name')["math_score"].mean()
twelfth_math = students_df.loc[students_df['grade'] == '12th'].groupby('school_name')["math_score"].mean()
math_scores = pd.DataFrame({
"9th": ninth_math,
"10th": tenth_math,
"11th": eleventh_math,
"12th": twelfth_math
})
math_scores = math_scores[['9th', '10th', '11th', '12th']]
math_scores.index.name = "School"
#show and format
math_scores
#creates grade level average reading scores for each school
ninth_reading = students_df.loc[students_df['grade'] == '9th'].groupby('school_name')["reading_score"].mean()
tenth_reading = students_df.loc[students_df['grade'] == '10th'].groupby('school_name')["reading_score"].mean()
eleventh_reading = students_df.loc[students_df['grade'] == '11th'].groupby('school_name')["reading_score"].mean()
twelfth_reading = students_df.loc[students_df['grade'] == '12th'].groupby('school_name')["reading_score"].mean()
#merges the reading score averages by school and grade together
reading_scores = pd.DataFrame({
"9th": ninth_reading,
"10th": tenth_reading,
"11th": eleventh_reading,
"12th": twelfth_reading
})
reading_scores = reading_scores[['9th', '10th', '11th', '12th']]
reading_scores.index.name = "School"
reading_scores
| 0.461502 | 0.766512 |
```
import sympy
from sympy.parsing.sympy_parser import parse_expr
from ipywidgets import interact, FloatRangeSlider
import plotnine as p9
import pandas as pd
from math import pi
x = sympy.Symbol('x')
def factorial(n):
"""
Calculates factorial
"""
result = n
if n == 1 or n == 0:
return 1
while n > 1:
result *= (n-1)
n -= 1
return result
def n_derivative(expression, n: int):
"""
Calculates n'th derivative
"""
derivative = expression
while n > 0:
derivative = sympy.diff(derivative)
n -= 1
return derivative
def taylor_series(expression, n: int, around: float):
"""
Calculates Taylor series
n - order / number of terms
around - approximation around certain point; real or complex number
"""
derr_sum = 0
while n >= 0:
derr_sum += (
n_derivative(expression, n).evalf(subs={'x': around})
* ((x - around) ** n)
/ factorial(n)
)
n -= 1
return derr_sum
taylor_series(sympy.cos(x), 4, 0)
```
```
type(type(sympy.sin(x)))
def create_dataset(expression, n, around):
"""
"""
a = pd.Series(range(0, 1000+1, 1))/1000 * 4 * pi - 2 * pi
expr = parse_expr(expression)
def evaluate(x):
return float(expr.evalf(subs={'x': x}))
sin_data = pd.DataFrame(data={
# create sample data
'x': a,
'f(x)': a.apply(evaluate),
"f_approx": a.apply(
sympy.lambdify(
x,
taylor_series(
expression=expr,
n=n,
around=around
)
)
)
})
return sin_data
@interact(
expression='sin(x)',
n=(0, 10, 1),
around=(-4, 4, 0.1),
y_display_range=FloatRangeSlider(
value=[-1.5, 1.5],
min=-10.0,
max=10.0,
step=0.5,
description='Y axis range'
)
)
def create_plot(expression, n, around, y_display_range):
data = create_dataset(expression, n, around)
y_values = data[['f_approx', 'f(x)']]
data = data[
(y_values.min(axis=1) > y_display_range[0])
&
(y_values.max(axis=1) < y_display_range[1])
]
plot = (
p9.ggplot(data=data)
+ p9.aes(x="x")
+ p9.geom_point(color="red", mapping=p9.aes(y="f(x)"))
+ p9.geom_point(color="blue", alpha=0.15, mapping=p9.aes(y="f_approx"))
+ p9.ylim(y_display_range)
)
plot.draw()
```
|
github_jupyter
|
import sympy
from sympy.parsing.sympy_parser import parse_expr
from ipywidgets import interact, FloatRangeSlider
import plotnine as p9
import pandas as pd
from math import pi
x = sympy.Symbol('x')
def factorial(n):
"""
Calculates factorial
"""
result = n
if n == 1 or n == 0:
return 1
while n > 1:
result *= (n-1)
n -= 1
return result
def n_derivative(expression, n: int):
"""
Calculates n'th derivative
"""
derivative = expression
while n > 0:
derivative = sympy.diff(derivative)
n -= 1
return derivative
def taylor_series(expression, n: int, around: float):
"""
Calculates Taylor series
n - order / number of terms
around - approximation around certain point; real or complex number
"""
derr_sum = 0
while n >= 0:
derr_sum += (
n_derivative(expression, n).evalf(subs={'x': around})
* ((x - around) ** n)
/ factorial(n)
)
n -= 1
return derr_sum
taylor_series(sympy.cos(x), 4, 0)
type(type(sympy.sin(x)))
def create_dataset(expression, n, around):
"""
"""
a = pd.Series(range(0, 1000+1, 1))/1000 * 4 * pi - 2 * pi
expr = parse_expr(expression)
def evaluate(x):
return float(expr.evalf(subs={'x': x}))
sin_data = pd.DataFrame(data={
# create sample data
'x': a,
'f(x)': a.apply(evaluate),
"f_approx": a.apply(
sympy.lambdify(
x,
taylor_series(
expression=expr,
n=n,
around=around
)
)
)
})
return sin_data
@interact(
expression='sin(x)',
n=(0, 10, 1),
around=(-4, 4, 0.1),
y_display_range=FloatRangeSlider(
value=[-1.5, 1.5],
min=-10.0,
max=10.0,
step=0.5,
description='Y axis range'
)
)
def create_plot(expression, n, around, y_display_range):
data = create_dataset(expression, n, around)
y_values = data[['f_approx', 'f(x)']]
data = data[
(y_values.min(axis=1) > y_display_range[0])
&
(y_values.max(axis=1) < y_display_range[1])
]
plot = (
p9.ggplot(data=data)
+ p9.aes(x="x")
+ p9.geom_point(color="red", mapping=p9.aes(y="f(x)"))
+ p9.geom_point(color="blue", alpha=0.15, mapping=p9.aes(y="f_approx"))
+ p9.ylim(y_display_range)
)
plot.draw()
| 0.702938 | 0.822759 |
## Visualization of Airport Data on Map.
The idea and the airport's data for this demo was found in the "Towards Data Science" article [How to Visualize Data on top of a Map in Python using the Geoviews library](https://towardsdatascience.com/how-to-visualize-data-on-top-of-a-map-in-python-using-the-geoviews-library-c4f444ca2929) by [Christos Zeglis](https://medium.com/@christoszeglis).
The paper demonstrates how to make a plot to visualize the passengers volume for the busiest airports in Greece, and the neighbor country, Turkey, for comparison reasons.
The author uses Geoviews and Bokeh libraries to achieve such visualisation. We are going to use the [Lets-Plot](https://github.com/JetBrains/lets-plot/blob/master/README.md) library solely to do the same job and, on top of it, fill both countries boundaries with semi-transparent colors.
The tasks completed in this tutorial:
- Configuring map-tiles for the interactive base-map layer.
- Obtaining boundaries of Greece and Turkey using Lets-Plot geo-coding module.
- Creating a proportional symbols map by combining base-map, polygons and point layers.
- Castomizing the tooltip contents.
```
import numpy as np
import pandas as pd
from lets_plot import *
LetsPlot.setup_html()
```
### Configuring the basemap.
For the purposes of this tutorial, we are going to use "CityLights 2012" map-tiles [© NASA Global Imagery Browse Services (GIBS)](https://earthdata.nasa.gov/eosdis/science-system-description/eosdis-components/gibs).
```
LetsPlot.set(maptiles_zxy(
url='https://gibs.earthdata.nasa.gov/wmts/epsg3857/best/VIIRS_CityLights_2012/default//GoogleMapsCompatible_Level8/{z}/{y}/{x}.jpg',
attribution='<a href="https://earthdata.nasa.gov/eosdis/science-system-description/eosdis-components/gibs">© NASA Global Imagery Browse Services (GIBS)</a>',
max_zoom=8
))
```
### Loading the "airports" dataset.
The "airports" dataset is already cleaned and only contains data on Greece and Turkey airports.
```
airports = pd.read_csv("../data/airports.csv")
airports.head(3)
```
### Obtaining boundaries (or polygons) of Greece and Turkey.
On this step we are using built-in geo-coding capabilities of the Lets-Plot library.
```
from lets_plot.geo_data import *
countries_gcoder = geocode_countries(['GR', 'TR'])
countries_gcoder.get_geocodes()
```
### Showing the data on map.
- Add an interactive base-map layer with custom initial location and zoom level.
- Add polygons layer to fill the country boundaries with semi-transparent colors.
- Add points layer marking the airports location with the point size proportional to the airport's passengers volume.
- Customize tooltip on the points layer to show all the airport data.
- Use the 'scale_fill_manual()' function in order to fill polygons and points with blue and red colors.
- Setup the desired marker's size using the 'scale_size()' function.
```
(ggplot()
+ geom_livemap(location=[26.65, 38.61],
zoom=6)
+ geom_polygon(aes(fill='country'),
data=countries_gcoder.get_boundaries(),
alpha=.2)
+ geom_point(aes('longitude', 'latitude', fill='country', size='passengers'),
data=airports,
shape=21,
alpha=.7,
color='white',
tooltips=layer_tooltips()
.format('passengers', '{.1f} m' )
.format('^x', '.2f').format('^y', '.2f')
.line('@|@IATA')
.line('Passengers|@passengers')
.line('City|@city')
.line('Country|@country')
.line('Longitude|^x')
.line('Latitude|^y'))
+ scale_fill_manual(values=['#30a2da', '#fc4f30'])
+ scale_size(range=[10, 40], trans='sqrt')
+ theme(legend_position='none')
+ ggsize(900, 520)
)
```
|
github_jupyter
|
import numpy as np
import pandas as pd
from lets_plot import *
LetsPlot.setup_html()
LetsPlot.set(maptiles_zxy(
url='https://gibs.earthdata.nasa.gov/wmts/epsg3857/best/VIIRS_CityLights_2012/default//GoogleMapsCompatible_Level8/{z}/{y}/{x}.jpg',
attribution='<a href="https://earthdata.nasa.gov/eosdis/science-system-description/eosdis-components/gibs">© NASA Global Imagery Browse Services (GIBS)</a>',
max_zoom=8
))
airports = pd.read_csv("../data/airports.csv")
airports.head(3)
from lets_plot.geo_data import *
countries_gcoder = geocode_countries(['GR', 'TR'])
countries_gcoder.get_geocodes()
(ggplot()
+ geom_livemap(location=[26.65, 38.61],
zoom=6)
+ geom_polygon(aes(fill='country'),
data=countries_gcoder.get_boundaries(),
alpha=.2)
+ geom_point(aes('longitude', 'latitude', fill='country', size='passengers'),
data=airports,
shape=21,
alpha=.7,
color='white',
tooltips=layer_tooltips()
.format('passengers', '{.1f} m' )
.format('^x', '.2f').format('^y', '.2f')
.line('@|@IATA')
.line('Passengers|@passengers')
.line('City|@city')
.line('Country|@country')
.line('Longitude|^x')
.line('Latitude|^y'))
+ scale_fill_manual(values=['#30a2da', '#fc4f30'])
+ scale_size(range=[10, 40], trans='sqrt')
+ theme(legend_position='none')
+ ggsize(900, 520)
)
| 0.383295 | 0.9814 |
```
# default_exp optimizer
#export
from fastai2.torch_basics import *
from nbdev.showdoc import *
```
# Optimizer
> Define the general fastai optimizer and the variants
## `_BaseOptimizer` -
```
#export
class _BaseOptimizer():
"Common functionality between `Optimizer` and `OptimWrapper`"
def all_params(self, n=slice(None), with_grad=False):
res = L((p,pg,self.state[p],hyper) for pg,hyper in zip(self.param_lists[n],self.hypers[n]) for p in pg)
return L(o for o in res if o[0].grad is not None) if with_grad else res
def _set_require_grad(self, rg, p,pg,state,h): p.requires_grad_(rg or state.get('force_train', False))
def freeze_to(self, n):
self.frozen_idx = n if n >= 0 else len(self.param_lists) + n
if self.frozen_idx >= len(self.param_lists):
warn(f"Freezing {self.frozen_idx} groups; model has {len(self.param_lists)}; whole model is frozen.")
for o in self.all_params(slice(n, None)): self._set_require_grad(True, *o)
for o in self.all_params(slice(None, n)): self._set_require_grad(False, *o)
def freeze(self):
assert(len(self.param_lists)>1)
self.freeze_to(-1)
def set_freeze(self, n, rg, ignore_force_train=False):
for p in self.param_lists[n]: p.requires_grad_(rg or (state.get('force_train', False) and not ignore_force_train))
def unfreeze(self): self.freeze_to(0)
def set_hypers(self, **kwargs): L(kwargs.items()).starmap(self.set_hyper)
def _set_hyper(self, k, v):
for v_,h in zip(v, self.hypers): h[k] = v_
def set_hyper(self, k, v):
if isinstance(v, slice):
if v.start: v = even_mults(v.start, v.stop, len(self.param_lists))
else: v = [v.stop/10]*(len(self.param_lists)-1) + [v.stop]
v = L(v, use_list=None)
if len(v)==1: v = v*len(self.param_lists)
assert len(v) == len(self.hypers), f"Trying to set {len(v)} values for {k} but there are {len(self.param_lists)} parameter groups."
self._set_hyper(k, v)
@property
def param_groups(self): return [{**{'params': pg}, **hp} for pg,hp in zip(self.param_lists, self.hypers)]
@param_groups.setter
def param_groups(self, v):
for pg,v_ in zip(self.param_lists,v): pg = v_['params']
for hyper,v_ in zip(self.hypers,v):
for k,t in v_.items():
if k != 'params': hyper[k] = t
add_docs(_BaseOptimizer,
all_params="List of param_groups, parameters, and hypers",
freeze_to="Freeze parameter groups up to `n`",
freeze="Freeze up to last parameter group",
set_freeze="Set `rg` for parameter group `n` only",
unfreeze="Unfreeze the entire model",
set_hypers="`set_hyper` for all `kwargs`",
set_hyper="Set the value(s) in `v` for hyper-parameter `k`")
#export
def _update(state, new=None):
if new is None: return state
if isinstance(new, dict): state.update(new)
return state
```
## `Optimizer` -
```
# export
class Optimizer(_BaseOptimizer):
"Base optimizer class for the fastai library, updating `params` with `cbs`"
_keep_on_clear = ['force_train', 'do_wd']
def __init__(self, params, cbs, train_bn=True, **defaults):
params = L(params)
self.cbs,self.state,self.train_bn = L(cbs),defaultdict(dict),train_bn
defaults = merge(*self.cbs.attrgot('defaults'), defaults)
self.param_lists = L(L(p) for p in params) if isinstance(params[0], (L,list)) else L([params])
self.hypers = L({} for _ in range_of(self.param_lists))
self.set_hypers(**defaults)
self.frozen_idx = 0
def zero_grad(self):
for p,*_ in self.all_params(with_grad=True):
p.grad.detach_()
p.grad.zero_()
def step(self):
for p,pg,state,hyper in self.all_params(with_grad=True):
for cb in self.cbs: state = _update(state, cb(p, **{**state, **hyper}))
self.state[p] = state
def clear_state(self):
for p,pg,state,hyper in self.all_params():
self.state[p] = {k: state[k] for k in self._keep_on_clear if k in state}
def state_dict(self):
state = [self.state[p] for p,*_ in self.all_params()]
return {'state': state, 'hypers': self.hypers}
def load_state_dict(self, sd):
assert len(sd["hypers"]) == len(self.param_lists)
assert len(sd["state"]) == sum([len(pg) for pg in self.param_lists])
self.hypers = sd['hypers']
self.state = {p: s for p,s in zip(self.all_params().itemgot(0), sd['state'])}
add_docs(Optimizer,
zero_grad="Standard PyTorch API: Zero all the grad attributes of the parameters",
step="Standard PyTorch API: Update the stats and execute the steppers in on all parameters that have a grad",
state_dict="Return the state of the optimizer in a dictionary",
load_state_dict="Load the content of `sd`",
clear_state="Reset the state of the optimizer")
```
### Initializing an Optimizer
`params` will be used to create the `param_groups` of the optimizer. If it's a collection (or a generator) of parameters, it will be a `L` containing one `L` with all the parameters. To define multiple parameter groups `params` should be passed as a collection (or a generator) of `L`s.
> Note: In PyTorch, <code>model.parameters()</code> returns a generator with all the parameters, that you can directly pass to <code>Optimizer</code>.
```
opt = Optimizer([1,2,3], noop)
test_eq(opt.param_lists, [[1,2,3]])
opt = Optimizer(range(3), noop)
test_eq(opt.param_lists, [[0,1,2]])
opt = Optimizer([[1,2],[3]], noop)
test_eq(opt.param_lists, [[1,2],[3]])
opt = Optimizer(([o,o+1] for o in range(0,4,2)), noop)
test_eq(opt.param_lists, [[0,1],[2,3]])
```
`cbs` is a list of functions that will be composed when applying the step. For instance, you can compose a function making the SGD step, with another one applying weight decay. Additionally, each `cb` can have a `defaults` attribute that contains hyper-parameters and their default value. Those are all gathered at initialization, and new values can be passed to override those defaults with the `defaults` kwargs. The steppers will be called by `Optimizer.step` (which is the standard PyTorch name), and gradients can be cleared with `Optimizer.zero_grad` (also a standard PyTorch name).
Once the defaults have all been pulled off, they are copied as many times as there are `param_groups` and stored in `hypers`. To apply different hyper-parameters to different groups (differential learning rates, or no weight decay for certain layers for instance), you will need to adjsut those values after the init.
```
def tst_arg(p, lr=0, **kwargs): return p
tst_arg.defaults = dict(lr=1e-2)
def tst_arg2(p, lr2=0, **kwargs): return p
tst_arg2.defaults = dict(lr2=1e-3)
def tst_arg3(p, mom=0, **kwargs): return p
tst_arg3.defaults = dict(mom=0.9)
def tst_arg4(p, **kwargs): return p
opt = Optimizer([1,2,3], [tst_arg,tst_arg2, tst_arg3])
test_eq(opt.hypers, [{'lr2': 1e-3, 'mom': 0.9, 'lr': 1e-2}])
opt = Optimizer([1,2,3], tst_arg, lr=0.1)
test_eq(opt.hypers, [{'lr': 0.1}])
opt = Optimizer([[1,2],[3]], tst_arg)
test_eq(opt.hypers, [{'lr': 1e-2}, {'lr': 1e-2}])
opt = Optimizer([[1,2],[3]], tst_arg, lr=0.1)
test_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.1}])
```
For each hyper-parameter, you can pass a slice or a collection to set them, if there are multiple parameter groups. A slice will be converted to a log-uniform collection from its beginning to its end, or if it only has an end `e`, to a collection of as many values as there are parameter groups that are `...,e/10,e/10,e`.
Setting an yper-paramter with a collection that has a different number of elements than the optimizer has paramter groups will raise an error.
```
opt = Optimizer([[1,2],[3]], tst_arg, lr=[0.1,0.2])
test_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.2}])
opt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-2))
test_eq(opt.hypers, [{'lr': 1e-3}, {'lr': 1e-3}, {'lr': 1e-2}])
opt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-4,1e-2))
test_eq(opt.hypers, [{'lr': 1e-4}, {'lr': 1e-3}, {'lr': 1e-2}])
test_eq(opt.param_groups, [{'params': [1,2], 'lr': 1e-4}, {'params': [3], 'lr': 1e-3}, {'params': [4], 'lr': 1e-2}])
test_fail(lambda: Optimizer([[1,2],[3],[4]], tst_arg, lr=np.array([0.1,0.2])))
```
### Basic steppers
To be able to give examples of optimizer steps, we will need some steppers, like the following:
```
#export
def sgd_step(p, lr, **kwargs):
p.data.add_(p.grad.data, alpha=-lr)
def tst_param(val, grad=None):
"Create a tensor with `val` and a gradient of `grad` for testing"
res = tensor([val]).float()
res.grad = tensor([val/10 if grad is None else grad]).float()
return res
p = tst_param(1., 0.1)
sgd_step(p, 1.)
test_eq(p, tensor([0.9]))
test_eq(p.grad, tensor([0.1]))
#export
def weight_decay(p, lr, wd, do_wd=True, **kwargs):
"Weight decay as decaying `p` with `lr*wd`"
if do_wd and wd!=0: p.data.mul_(1 - lr*wd)
weight_decay.defaults = dict(wd=0.)
p = tst_param(1., 0.1)
weight_decay(p, 1., 0.1)
test_eq(p, tensor([0.9]))
test_eq(p.grad, tensor([0.1]))
#export
def l2_reg(p, lr, wd, do_wd=True, **kwargs):
"L2 regularization as adding `wd*p` to `p.grad`"
if do_wd and wd!=0: p.grad.data.add_(p.data, alpha=wd)
l2_reg.defaults = dict(wd=0.)
p = tst_param(1., 0.1)
l2_reg(p, 1., 0.1)
test_eq(p, tensor([1.]))
test_eq(p.grad, tensor([0.2]))
```
> Warning: Weight decay and L2 regularization is the same thing for basic SGD, but for more complex optimizers, they are very different.
### Making the step
```
show_doc(Optimizer.step)
```
This method will loop over all param groups, then all parameters for which `grad` is not None and call each function in `stepper`, passing it the parameter `p` with the hyper-parameters in the corresponding dict in `hypers`.
```
#test basic step
r = L.range(4)
def tst_params(): return r.map(tst_param)
params = tst_params()
opt = Optimizer(params, sgd_step, lr=0.1)
opt.step()
test_close([p.item() for p in params], r.map(mul(0.99)))
#test two steps
params = tst_params()
opt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)
opt.step()
test_close([p.item() for p in params], r.map(mul(0.98)))
#test None gradients are ignored
params = tst_params()
opt = Optimizer(params, sgd_step, lr=0.1)
params[-1].grad = None
opt.step()
test_close([p.item() for p in params], [0., 0.99, 1.98, 3.])
#test discriminative lrs
params = tst_params()
opt = Optimizer([params[:2], params[2:]], sgd_step, lr=0.1)
opt.hypers[0]['lr'] = 0.01
opt.step()
test_close([p.item() for p in params], [0., 0.999, 1.98, 2.97])
show_doc(Optimizer.zero_grad)
params = tst_params()
opt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)
opt.zero_grad()
[test_eq(p.grad, tensor([0.])) for p in params];
```
Some of the `Optimizer` `cbs` can be functions updating the state associated with a parameter. That state can then be used by any stepper. The best example is a momentum calculation.
```
def tst_stat(p, **kwargs):
s = kwargs.get('sum', torch.zeros_like(p)) + p.data
return {'sum': s}
tst_stat.defaults = {'mom': 0.9}
#Test Optimizer init
opt = Optimizer([1,2,3], tst_stat)
test_eq(opt.hypers, [{'mom': 0.9}])
opt = Optimizer([1,2,3], tst_stat, mom=0.99)
test_eq(opt.hypers, [{'mom': 0.99}])
#Test stat
x = torch.randn(4,5)
state = tst_stat(x)
assert 'sum' in state
test_eq(x, state['sum'])
state = tst_stat(x, **state)
test_eq(state['sum'], 2*x)
```
## Statistics
```
# export
def average_grad(p, mom, dampening=False, grad_avg=None, **kwargs):
"Keeps track of the avg grads of `p` in `state` with `mom`."
if grad_avg is None: grad_avg = torch.zeros_like(p.grad.data)
damp = 1-mom if dampening else 1.
grad_avg.mul_(mom).add_(p.grad.data, alpha=damp)
return {'grad_avg': grad_avg}
average_grad.defaults = dict(mom=0.9)
```
`dampening=False` gives the classical formula for momentum in SGD:
```
new_val = old_val * mom + grad
```
whereas `dampening=True` makes it an exponential moving average:
```
new_val = old_val * mom + grad * (1-mom)
```
```
p = tst_param([1,2,3], [4,5,6])
state = {}
state = average_grad(p, mom=0.9, **state)
test_eq(state['grad_avg'], p.grad)
state = average_grad(p, mom=0.9, **state)
test_eq(state['grad_avg'], p.grad * 1.9)
#Test dampening
state = {}
state = average_grad(p, mom=0.9, dampening=True, **state)
test_eq(state['grad_avg'], 0.1*p.grad)
state = average_grad(p, mom=0.9, dampening=True, **state)
test_close(state['grad_avg'], (0.1*0.9+0.1)*p.grad)
# export
def average_sqr_grad(p, sqr_mom, dampening=True, sqr_avg=None, **kwargs):
if sqr_avg is None: sqr_avg = torch.zeros_like(p.grad.data)
damp = 1-sqr_mom if dampening else 1.
sqr_avg.mul_(sqr_mom).addcmul_(p.grad.data, p.grad.data, value=damp)
return {'sqr_avg': sqr_avg}
average_sqr_grad.defaults = dict(sqr_mom=0.99)
```
`dampening=False` gives the classical formula for momentum in SGD:
```
new_val = old_val * mom + grad**2
```
whereas `dampening=True` makes it an exponential moving average:
```
new_val = old_val * mom + (grad**2) * (1-mom)
```
```
p = tst_param([1,2,3], [4,5,6])
state = {}
state = average_sqr_grad(p, sqr_mom=0.99, dampening=False, **state)
test_eq(state['sqr_avg'], p.grad.pow(2))
state = average_sqr_grad(p, sqr_mom=0.99, dampening=False, **state)
test_eq(state['sqr_avg'], p.grad.pow(2) * 1.99)
#Test dampening
state = {}
state = average_sqr_grad(p, sqr_mom=0.99, **state)
test_close(state['sqr_avg'], 0.01*p.grad.pow(2))
state = average_sqr_grad(p, sqr_mom=0.99, **state)
test_close(state['sqr_avg'], (0.01*0.99+0.01)*p.grad.pow(2))
```
### Freezing part of the model
```
show_doc(Optimizer.freeze, name="Optimizer.freeze")
show_doc(Optimizer.freeze_to, name="Optimizer.freeze_to")
show_doc(Optimizer.unfreeze, name="Optimizer.unfreeze")
#Freezing the first layer
params = [tst_params(), tst_params(), tst_params()]
opt = Optimizer(params, sgd_step, lr=0.1)
opt.freeze_to(1)
req_grad = Self.requires_grad()
test_eq(L(params[0]).map(req_grad), [False]*4)
for i in {1,2}: test_eq(L(params[i]).map(req_grad), [True]*4)
#Unfreezing
opt.unfreeze()
for i in range(2): test_eq(L(params[i]).map(req_grad), [True]*4)
#TODO: test warning
# opt.freeze_to(3)
```
Parameters such as batchnorm weights/bias can be marked to always be in training mode, just put `force_train=true` in their state.
```
params = [tst_params(), tst_params(), tst_params()]
opt = Optimizer(params, sgd_step, lr=0.1)
for p in L(params[1])[[1,3]]: opt.state[p] = {'force_train': True}
opt.freeze()
test_eq(L(params[0]).map(req_grad), [False]*4)
test_eq(L(params[1]).map(req_grad), [False, True, False, True])
test_eq(L(params[2]).map(req_grad), [True]*4)
```
### Serializing
```
show_doc(Optimizer.state_dict)
show_doc(Optimizer.load_state_dict)
p = tst_param([1,2,3], [4,5,6])
opt = Optimizer(p, average_grad)
opt.step()
test_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))
sd = opt.state_dict()
p1 = tst_param([10,20,30], [40,50,60])
opt = Optimizer(p1, average_grad, mom=0.99)
test_eq(opt.hypers[0]['mom'], 0.99)
test_eq(opt.state, {})
opt.load_state_dict(sd)
test_eq(opt.hypers[0]['mom'], 0.9)
test_eq(opt.state[p1]['grad_avg'], tensor([[4., 5., 6.]]))
show_doc(Optimizer.clear_state)
p = tst_param([1,2,3], [4,5,6])
opt = Optimizer(p, average_grad)
opt.state[p] = {'force_train': True}
opt.step()
test_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))
opt.clear_state()
test_eq(opt.state[p], {'force_train': True})
```
## Optimizers
### SGD with momentum
```
#export
def momentum_step(p, lr, grad_avg, **kwargs):
"Step for SGD with momentum with `lr`"
p.data.add_(grad_avg, alpha=-lr)
#export
@log_args(to_return=True, but='params')
def SGD(params, lr, mom=0., wd=0., decouple_wd=True):
"A `Optimizer` for SGD with `lr` and `mom` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
if mom != 0: cbs.append(average_grad)
cbs.append(sgd_step if mom==0 else momentum_step)
return Optimizer(params, cbs, lr=lr, mom=mom, wd=wd)
```
Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
```
#Vanilla SGD
params = tst_params()
opt = SGD(params, lr=0.1)
opt.step()
test_close([p.item() for p in params], [i*0.99 for i in range(4)])
opt.step()
[p.item() for p in params]
test_close([p.item() for p in params], [i*0.98 for i in range(4)])
#SGD with momentum
params = tst_params()
opt = SGD(params, lr=0.1, mom=0.9)
assert isinstance(opt, Optimizer)
opt.step()
test_close([p.item() for p in params], [i*0.99 for i in range(4)])
opt.step()
[p.item() for p in params]
test_close([p.item() for p in params], [i*(1 - 0.1 * (0.1 + 0.1*1.9)) for i in range(4)])
for i,p in enumerate(params): test_close(opt.state[p]['grad_avg'].item(), i*0.19)
```
Test weight decay, notice how we can see that L2 regularization is different from weight decay even for simple SGD with momentum.
```
params = tst_params()
#Weight decay
opt = SGD(params, lr=0.1, mom=0.9, wd=0.1)
opt.step()
test_close([p.item() for p in params], [i*0.98 for i in range(4)])
#L2 reg
opt = SGD(params, lr=0.1, mom=0.9, wd=0.1, decouple_wd=False)
opt.step()
#TODO: fix cause this formula was wrong
#test_close([p.item() for p in params], [i*0.97 for i in range(4)])
```
### RMSProp
```
#export
def rms_prop_step(p, lr, sqr_avg, eps, grad_avg=None, **kwargs):
"Step for SGD with momentum with `lr`"
denom = sqr_avg.sqrt().add_(eps)
p.data.addcdiv_((grad_avg if grad_avg is not None else p.grad), denom, value=-lr)
rms_prop_step.defaults = dict(eps=1e-8)
#export
@log_args(to_return=True, but='params')
def RMSProp(params, lr, sqr_mom=0.99, mom=0., wd=0., decouple_wd=True):
"A `Optimizer` for RMSProp with `lr`, `sqr_mom`, `mom` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += ([average_sqr_grad] if mom==0. else [average_grad, average_sqr_grad])
cbs.append(rms_prop_step)
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, wd=wd)
```
RMSProp was introduced by Geoffrey Hinton in his [course](http://www.cs.toronto.edu/~tijmen/csc321/slides/lecture_slides_lec6.pdf). What is named `sqr_mom` here is the `alpha` in the course. Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
```
#Without momentum
import math
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RMSProp(params, lr=0.1)
opt.step()
test_close(params[0], tensor([0.,1.,2.]))
opt.step()
step = - 0.1 * 0.1 / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)
test_close(params[0], tensor([step, 1+step, 2+step]))
#With momentum
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RMSProp(params, lr=0.1, mom=0.9)
opt.step()
test_close(params[0], tensor([0.,1.,2.]))
opt.step()
step = - 0.1 * (0.1 + 0.9*0.1) / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)
test_close(params[0], tensor([step, 1+step, 2+step]))
```
### Adam
```
#export
def step_stat(p, step=0, **kwargs):
"Register the number of steps done in `state` for `p`"
step += 1
return {'step' : step}
p = tst_param(1,0.1)
state = {}
state = step_stat(p, **state)
test_eq(state['step'], 1)
for _ in range(5): state = step_stat(p, **state)
test_eq(state['step'], 6)
#export
def debias(mom, damp, step): return damp * (1 - mom**step) / (1-mom)
#export
def adam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):
"Step for Adam with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
p.data.addcdiv_(grad_avg, (sqr_avg/debias2).sqrt() + eps, value = -lr / debias1)
return p
adam_step._defaults = dict(eps=1e-5)
#export
@log_args(to_return=True, but='params')
def Adam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0.01, decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), average_sqr_grad, step_stat, adam_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
```
Adam was introduced by Diederik P. Kingma and Jimmy Ba in [Adam: A Method for Stochastic Optimization](https://arxiv.org/abs/1412.6980). For consistency accross optimizers, we renamed `beta1` and `beta2` in the paper to `mom` and `sqr_mom`. Note that our defaults also differ from the paper (0.99 for `sqr_mom` or `beta2`, 1e-5 for `eps`). Those values seem to be better from our experiments in a wide range of situations.
Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
> Note: Don't forget that `eps` is an hyper-parameter you can change. Some models won't train without a very high `eps` like 0.1 (intuitively, the higher `eps` is, the closer we are to normal SGD). The usual default of 1e-8 is often too extreme in the sense we don't manage to get as good results as with SGD.
```
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = Adam(params, lr=0.1, wd=0)
opt.step()
step = -0.1 * 0.1 / (math.sqrt(0.1**2) + 1e-8)
test_close(params[0], tensor([1+step, 2+step, 3+step]))
opt.step()
test_close(params[0], tensor([1+2*step, 2+2*step, 3+2*step]), eps=1e-3)
```
### RAdam
RAdam (for rectified Adam) was introduced by Zhang et al. in [On the Variance of the Adaptive Learning Rate and Beyond](https://arxiv.org/abs/1907.08610) to slightly modify the Adam optimizer to be more stable at the beginning of training (and thus not require a long warmup). They use an estimate of the variance of the moving average of the squared gradients (the term in the denominator of traditional Adam) and rescale this moving average by this term before performing the update.
This version also incorporates [SAdam](https://arxiv.org/abs/1908.00700); set `beta` to enable this (definition same as in the paper).
```
#export
def radam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, beta, **kwargs):
"Step for RAdam with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
r_inf = 2/(1-sqr_mom) - 1
r = r_inf - 2*step*sqr_mom**step/(1-sqr_mom**step)
if r > 5:
v = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))
denom = (sqr_avg/debias2).sqrt()
if eps: denom += eps
if beta: denom = F.softplus(denom, beta)
p.data.addcdiv_(grad_avg, denom, value = -lr*v / debias1)
else: p.data.add_(grad_avg, alpha=-lr / debias1)
return p
radam_step._defaults = dict(eps=1e-5)
#export
@log_args(to_return=True, but='params')
def RAdam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., beta=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs = [partial(average_grad, dampening=True), average_sqr_grad, step_stat, radam_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd, beta=beta)
```
This is the effective correction apported to the adam step for 500 iterations in RAdam. We can see how it goes from 0 to 1, mimicking the effect of a warm-up.
```
beta = 0.99
r_inf = 2/(1-beta) - 1
rs = np.array([r_inf - 2*s*beta**s/(1-beta**s) for s in range(5,500)])
v = np.sqrt(((rs-4) * (rs-2) * r_inf)/((r_inf-4)*(r_inf-2)*rs))
plt.plot(v);
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RAdam(params, lr=0.1)
#The r factor is lower than 5 during the first 5 steps so updates use the aveage of gradients (all the same)
r_inf = 2/(1-0.99) - 1
for i in range(5):
r = r_inf - 2*(i+1)*0.99**(i+1)/(1-0.99**(i+1))
assert r <= 5
opt.step()
p = tensor([0.95, 1.9, 2.85])
test_close(params[0], p)
#The r factor is greater than 5 for the sixth step so we update with RAdam
r = r_inf - 2*6*0.99**6/(1-0.99**6)
assert r > 5
opt.step()
v = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))
step = -0.1*0.1*v/(math.sqrt(0.1**2) + 1e-8)
test_close(params[0], p+step)
```
### QHAdam
QHAdam (for Quasi-Hyperbolic Adam) was introduced by Ma & Yarats in [Quasi-Hyperbolic Momentum and Adam for Deep Learning](https://arxiv.org/pdf/1810.06801.pdf) as a *"computationally cheap, intuitive to interpret, and simple to implement"* optimizer. Additional code can be found in their [qhoptim repo](https://github.com/facebookresearch/qhoptim). QHAdam is based on QH-Momentum, which introduces the immediate discount factor `nu`, encapsulating plain SGD (`nu = 0`) and momentum (`nu = 1`). QH-Momentum is defined below, where g_t+1 is the update of the moment. An interpretation of QHM is as a nu-weighted average of the momentum update step and the plain SGD update step.
> θ_t+1 ← θ_t − lr * [(1 − nu) · ∇L_t(θ_t) + nu · g_t+1]
QHAdam takes the concept behind QHM above and applies it to Adam, replacing both of Adam’s moment estimators with quasi-hyperbolic terms.
The paper's suggested default parameters are `mom = 0.999`, `sqr_mom = 0.999`, `nu_1 = 0.7` and `and nu_2 = 1.0`. When training is not stable, it is possible that setting `nu_2 < 1` can improve stability by imposing a tighter step size bound. Note that QHAdam recovers Adam when `nu_1 = nu_2 = 1.0`. QHAdam recovers RMSProp (Hinton et al., 2012) when `nu_1 = 0` and `nu_2 = 1`, and NAdam (Dozat, 2016) when `nu_1 = mom` and `nu_2 = 1`.
Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
```
#export
def qhadam_step(p, lr, mom, sqr_mom, sqr_avg, nu_1, nu_2, step, grad_avg, eps, **kwargs):
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
p.data.addcdiv_(((1-nu_1) * p.grad.data) + (nu_1 * (grad_avg / debias1)),
(((1 - nu_2) * (p.grad.data)**2) + (nu_2 * (sqr_avg / debias2))).sqrt() + eps,
value = -lr)
return p
qhadam_step._defaults = dict(eps=1e-8)
#export
@log_args(to_return=True, but='params')
def QHAdam(params, lr, mom=0.999, sqr_mom=0.999, nu_1=0.7, nu_2 = 1.0, eps=1e-8, wd=0., decouple_wd=True):
"An `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `nus`, eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), partial(average_sqr_grad, dampening=True), step_stat, qhadam_step]
return Optimizer(params, cbs, lr=lr, nu_1=nu_1, nu_2=nu_2 ,
mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = QHAdam(params, lr=0.1)
opt.step()
step = -0.1 * (((1-0.7) * 0.1) + (0.7 * 0.1)) / (
math.sqrt(((1-1.0) * 0.1**2) + (1.0 * 0.1**2)) + 1e-8)
test_close(params[0], tensor([1+step, 2+step, 3+step]))
opt.step()
test_close(params[0], tensor([1+2*step, 2+2*step, 3+2*step]), eps=1e-3)
```
### LARS/LARC
```
#export
def larc_layer_lr(p, lr, trust_coeff, wd, eps, clip=True, **kwargs):
"Computes the local lr before weight decay is applied"
p_norm,g_norm = torch.norm(p.data),torch.norm(p.grad.data)
local_lr = lr*trust_coeff * (p_norm) / (g_norm + p_norm * wd + eps)
return {'local_lr': min(lr, local_lr) if clip else local_lr}
larc_layer_lr.defaults = dict(trust_coeff=0.02, wd=0., eps=1e-8)
#export
def larc_step(p, local_lr, grad_avg=None, **kwargs):
"Step for LARC `local_lr` on `p`"
p.data.add_(p.grad.data if grad_avg is None else grad_avg, alpha = -local_lr)
#export
@log_args(to_return=True, but='params')
def Larc(params, lr, mom=0.9, clip=True, trust_coeff=0.02, eps=1e-8, wd=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
if mom!=0.: cbs.append(average_grad)
cbs += [partial(larc_layer_lr, clip=clip), larc_step]
return Optimizer(params, cbs, lr=lr, mom=mom, trust_coeff=trust_coeff, eps=eps, wd=wd)
```
The LARS optimizer was first introduced in [Large Batch Training of Convolutional Networks](https://arxiv.org/abs/1708.03888) then refined in its LARC variant (original LARS is with `clip=False`). A learning rate is computed for each individual layer with a certain `trust_coefficient`, then clipped to be always less than `lr`.
Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
```
params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]
opt = Larc(params, lr=0.1)
opt.step()
#First param local lr is 0.02 < lr so it's not clipped
test_close(opt.state[params[0]]['local_lr'], 0.02)
#Second param local lr is 0.2 > lr so it's clipped
test_eq(opt.state[params[1]]['local_lr'], 0.1)
test_close(params[0], tensor([0.998,1.996,2.994]))
test_close(params[1], tensor([0.999,1.998,2.997]))
params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]
opt = Larc(params, lr=0.1, clip=False)
opt.step()
#No clipping
test_close(opt.state[params[0]]['local_lr'], 0.02)
test_close(opt.state[params[1]]['local_lr'], 0.2)
test_close(params[0], tensor([0.998,1.996,2.994]))
test_close(params[1], tensor([0.998,1.996,2.994]))
```
### LAMB
```
#export
def lamb_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):
"Step for LAMB with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
r1 = p.data.pow(2).mean().sqrt()
step = (grad_avg/debias1) / ((sqr_avg/debias2).sqrt()+eps)
r2 = step.pow(2).mean().sqrt()
q = 1 if r1 == 0 or r2 == 0 else min(r1/r2,10)
p.data.add_(step, alpha = -lr * q)
lamb_step._defaults = dict(eps=1e-6, wd=0.)
#export
@log_args(to_return=True, but='params')
def Lamb(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), average_sqr_grad, step_stat, lamb_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
```
LAMB was introduced in [Large Batch Optimization for Deep Learning: Training BERT in 76 minutes](https://arxiv.org/abs/1904.00962). Intuitively, it's LARC applied to Adam. As in `Adam`, we renamed `beta1` and `beta2` in the paper to `mom` and `sqr_mom`. Note that our defaults also differ from the paper (0.99 for `sqr_mom` or `beta2`, 1e-5 for `eps`). Those values seem to be better from our experiments in a wide range of situations.
Optional weight decay of `wd` is applied, as true weight decay (decay the weights directly) if `decouple_wd=True` else as L2 regularization (add the decay to the gradients).
```
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = Lamb(params, lr=0.1)
opt.step()
test_close(params[0], tensor([0.7840,1.7840,2.7840]), eps=1e-3)
```
## Lookahead -
Lookahead was introduced by Zhang et al. in [Lookahead Optimizer: k steps forward, 1 step back](https://arxiv.org/abs/1907.08610). It can be run on top of any optimizer and consists in having the final weights of the model be a moving average. In practice, we update our model using the internal optimizer but keep a copy of old weights that and every `k` steps, we change the weghts by a moving average of the *fast weights* (the ones updated by the inner optimizer) with the *slow weights* (the copy of old weights). Those *slow weights* act like a stability mechanism.
```
#export
@log_args(but='opt')
class Lookahead(Optimizer, GetAttr):
"Wrap `opt` in a lookahead optimizer"
_default='opt'
def __init__(self, opt, k=6, alpha=0.5):
store_attr(self, 'opt,k,alpha')
self._init_state()
def step(self):
if self.slow_weights is None: self._copy_weights()
self.opt.step()
self.count += 1
if self.count%self.k != 0: return
for slow_pg,fast_pg in zip(self.slow_weights,self.param_lists):
for slow_p,fast_p in zip(slow_pg,fast_pg):
slow_p.data.add_(fast_p.data-slow_p.data, alpha=self.alpha)
fast_p.data.copy_(slow_p.data)
def clear_state(self):
self.opt.clear_state()
self._init_state()
def state_dict(self):
state = self.opt.state_dict()
state.update({'count': self.count, 'slow_weights': self.slow_weights})
return state
def load_state_dict(self, sd):
self.count = sd.pop('count')
self.slow_weights = sd.pop('slow_weights')
self.opt.load_state_dict(sd)
def _init_state(self): self.count,self.slow_weights = 0,None
def _copy_weights(self): self.slow_weights = L(L(p.clone().detach() for p in pg) for pg in self.param_lists)
@property
def param_lists(self): return self.opt.param_lists
@param_lists.setter
def param_lists(self, v): self.opt.param_lists = v
params = tst_param([1,2,3], [0.1,0.2,0.3])
p,g = params[0].data.clone(),tensor([0.1,0.2,0.3])
opt = Lookahead(SGD(params, lr=0.1))
for k in range(5): opt.step()
#first 5 steps are normal SGD steps
test_close(params[0], p - 0.5*g)
#Since k=6, sixth step is a moving average of the 6 SGD steps with the intial weight
opt.step()
test_close(params[0], p * 0.5 + (p-0.6*g) * 0.5)
#export
@delegates(RAdam)
def ranger(p, lr, mom=0.95, wd=0.01, eps=1e-6, **kwargs):
"Convenience method for `Lookahead` with `RAdam`"
return Lookahead(RAdam(p, lr=lr, mom=mom, wd=wd, eps=eps, **kwargs))
```
## OptimWrapper -
```
#export
def detuplify_pg(d):
res = {}
for k,v in d.items():
if k == 'params': continue
if is_listy(v): res.update(**{f'{k}__{i}': v_ for i,v_ in enumerate(v)})
else: res[k] = v
return res
tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}
test_eq(detuplify_pg(tst), {'lr': 1e-2, 'mom': 0.9})
tst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}
test_eq(detuplify_pg(tst), {'lr': 1e-2, 'betas__0': 0.9, 'betas__1': 0.999})
#export
def set_item_pg(pg, k, v):
if '__' not in k: pg[k] = v
else:
name,idx = k.split('__')
pg[name] = tuple(v if i==int(idx) else pg[name][i] for i in range_of(pg[name]))
return pg
tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}
test_eq(set_item_pg(tst, 'lr', 1e-3), {'lr': 1e-3, 'mom': 0.9, 'params':[0,1,2]})
tst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}
test_eq(set_item_pg(tst, 'betas__0', 0.95), {'lr': 1e-2, 'betas': (0.95,0.999), 'params':[0,1,2]})
#export
pytorch_hp_map = {'momentum': 'mom', 'weight_decay': 'wd', 'alpha': 'sqr_mom', 'betas__0': 'mom', 'betas__1': 'sqr_mom'}
#export
class OptimWrapper(_BaseOptimizer, GetAttr):
_xtra=['zero_grad', 'step', 'state_dict', 'load_state_dict']
_default='opt'
def __init__(self, opt, hp_map=None):
self.opt = opt
if hp_map is None: hp_map = pytorch_hp_map
self.fwd_map = {k: hp_map[k] if k in hp_map else k for k in detuplify_pg(opt.param_groups[0]).keys()}
self.bwd_map = {v:k for k,v in self.fwd_map.items()}
self.state = defaultdict(dict, {})
self.frozen_idx = 0
@property
def hypers(self):
return [{self.fwd_map[k]:v for k,v in detuplify_pg(pg).items() if k != 'params'} for pg in self.opt.param_groups]
def _set_hyper(self, k, v):
for pg,v_ in zip(self.opt.param_groups,v): pg = set_item_pg(pg, self.bwd_map[k], v_)
def clear_state(self): self.opt.state = defaultdict(dict, {})
@property
def param_lists(self): return [pg['params'] for pg in self.opt.param_groups]
@param_lists.setter
def param_lists(self, v):
for pg,v_ in zip(self.opt.param_groups,v): pg['params'] = v_
sgd = SGD([tensor([1,2,3])], lr=1e-3, mom=0.9, wd=1e-2)
tst_sgd = OptimWrapper(torch.optim.SGD([tensor([1,2,3])], lr=1e-3, momentum=0.9, weight_decay=1e-2))
#Access to param_groups
test_eq(tst_sgd.param_lists, sgd.param_lists)
#Set param_groups
tst_sgd.param_lists = [[tensor([4,5,6])]]
test_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])
#Access to hypers
test_eq(tst_sgd.hypers, [{**sgd.hypers[0], 'dampening': 0., 'nesterov': False}])
#Set hypers
tst_sgd.set_hyper('mom', 0.95)
test_eq(tst_sgd.opt.param_groups[0]['momentum'], 0.95)
tst_sgd = OptimWrapper(torch.optim.SGD([{'params': [tensor([1,2,3])], 'lr': 1e-3},
{'params': [tensor([4,5,6])], 'lr': 1e-2}], momentum=0.9, weight_decay=1e-2))
sgd = SGD([[tensor([1,2,3])], [tensor([4,5,6])]], lr=[1e-3, 1e-2], mom=0.9, wd=1e-2)
#Access to param_groups
test_eq(tst_sgd.param_lists, sgd.param_lists)
#Set param_groups
tst_sgd.param_lists = [[tensor([4,5,6])], [tensor([1,2,3])]]
test_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])
test_eq(tst_sgd.opt.param_groups[1]['params'], [tensor(1,2,3)])
#Access to hypers
test_eq(tst_sgd.hypers, [{**sgd.hypers[i], 'dampening': 0., 'nesterov': False} for i in range(2)])
#Set hypers
tst_sgd.set_hyper('mom', 0.95)
test_eq([pg['momentum'] for pg in tst_sgd.opt.param_groups], [0.95,0.95])
tst_sgd.set_hyper('lr', [1e-4,1e-3])
test_eq([pg['lr'] for pg in tst_sgd.opt.param_groups], [1e-4,1e-3])
#hide
#check it works with tuply hp names like in Adam
tst_adam = OptimWrapper(torch.optim.Adam([tensor([1,2,3])], lr=1e-2, betas=(0.9, 0.99)))
test_eq(tst_adam.hypers, [{'lr': 0.01, 'mom': 0.9, 'sqr_mom': 0.99, 'eps': 1e-08, 'wd': 0, 'amsgrad': False}])
tst_adam.set_hyper('mom', 0.95)
test_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.99))
tst_adam.set_hyper('sqr_mom', 0.9)
test_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.9))
def _mock_train(m, x, y, opt):
m.train()
for i in range(0, 100, 25):
z = m(x[i:i+25])
loss = F.mse_loss(z, y[i:i+25])
loss.backward()
opt.step()
opt.zero_grad()
m = nn.Linear(4,5)
x = torch.randn(100, 3, 4)
y = torch.randn(100, 3, 5)
try:
torch.save(m.state_dict(), 'tmp.pth')
wgt,bias = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt1 = OptimWrapper(torch.optim.AdamW(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))
_mock_train(m, x.clone(), y.clone(), opt1)
wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt2 = Adam(m.parameters(), 1e-3, wd=1e-2)
_mock_train(m, x.clone(), y.clone(), opt2)
wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()
test_close(wgt1,wgt2,eps=1e-3)
test_close(bias1,bias2,eps=1e-3)
finally: os.remove('tmp.pth')
m = nn.Linear(4,5)
x = torch.randn(100, 3, 4)
y = torch.randn(100, 3, 5)
try:
torch.save(m.state_dict(), 'tmp.pth')
wgt,bias = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt1 = OptimWrapper(torch.optim.Adam(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))
_mock_train(m, x.clone(), y.clone(), opt1)
wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt2 = Adam(m.parameters(), 1e-3, wd=1e-2, decouple_wd=False)
_mock_train(m, x.clone(), y.clone(), opt2)
wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()
test_close(wgt1,wgt2,eps=1e-3)
test_close(bias1,bias2,eps=1e-3)
finally: os.remove('tmp.pth')
```
## Export -
```
#hide
from nbdev.export import *
notebook2script()
```
|
github_jupyter
|
# default_exp optimizer
#export
from fastai2.torch_basics import *
from nbdev.showdoc import *
#export
class _BaseOptimizer():
"Common functionality between `Optimizer` and `OptimWrapper`"
def all_params(self, n=slice(None), with_grad=False):
res = L((p,pg,self.state[p],hyper) for pg,hyper in zip(self.param_lists[n],self.hypers[n]) for p in pg)
return L(o for o in res if o[0].grad is not None) if with_grad else res
def _set_require_grad(self, rg, p,pg,state,h): p.requires_grad_(rg or state.get('force_train', False))
def freeze_to(self, n):
self.frozen_idx = n if n >= 0 else len(self.param_lists) + n
if self.frozen_idx >= len(self.param_lists):
warn(f"Freezing {self.frozen_idx} groups; model has {len(self.param_lists)}; whole model is frozen.")
for o in self.all_params(slice(n, None)): self._set_require_grad(True, *o)
for o in self.all_params(slice(None, n)): self._set_require_grad(False, *o)
def freeze(self):
assert(len(self.param_lists)>1)
self.freeze_to(-1)
def set_freeze(self, n, rg, ignore_force_train=False):
for p in self.param_lists[n]: p.requires_grad_(rg or (state.get('force_train', False) and not ignore_force_train))
def unfreeze(self): self.freeze_to(0)
def set_hypers(self, **kwargs): L(kwargs.items()).starmap(self.set_hyper)
def _set_hyper(self, k, v):
for v_,h in zip(v, self.hypers): h[k] = v_
def set_hyper(self, k, v):
if isinstance(v, slice):
if v.start: v = even_mults(v.start, v.stop, len(self.param_lists))
else: v = [v.stop/10]*(len(self.param_lists)-1) + [v.stop]
v = L(v, use_list=None)
if len(v)==1: v = v*len(self.param_lists)
assert len(v) == len(self.hypers), f"Trying to set {len(v)} values for {k} but there are {len(self.param_lists)} parameter groups."
self._set_hyper(k, v)
@property
def param_groups(self): return [{**{'params': pg}, **hp} for pg,hp in zip(self.param_lists, self.hypers)]
@param_groups.setter
def param_groups(self, v):
for pg,v_ in zip(self.param_lists,v): pg = v_['params']
for hyper,v_ in zip(self.hypers,v):
for k,t in v_.items():
if k != 'params': hyper[k] = t
add_docs(_BaseOptimizer,
all_params="List of param_groups, parameters, and hypers",
freeze_to="Freeze parameter groups up to `n`",
freeze="Freeze up to last parameter group",
set_freeze="Set `rg` for parameter group `n` only",
unfreeze="Unfreeze the entire model",
set_hypers="`set_hyper` for all `kwargs`",
set_hyper="Set the value(s) in `v` for hyper-parameter `k`")
#export
def _update(state, new=None):
if new is None: return state
if isinstance(new, dict): state.update(new)
return state
# export
class Optimizer(_BaseOptimizer):
"Base optimizer class for the fastai library, updating `params` with `cbs`"
_keep_on_clear = ['force_train', 'do_wd']
def __init__(self, params, cbs, train_bn=True, **defaults):
params = L(params)
self.cbs,self.state,self.train_bn = L(cbs),defaultdict(dict),train_bn
defaults = merge(*self.cbs.attrgot('defaults'), defaults)
self.param_lists = L(L(p) for p in params) if isinstance(params[0], (L,list)) else L([params])
self.hypers = L({} for _ in range_of(self.param_lists))
self.set_hypers(**defaults)
self.frozen_idx = 0
def zero_grad(self):
for p,*_ in self.all_params(with_grad=True):
p.grad.detach_()
p.grad.zero_()
def step(self):
for p,pg,state,hyper in self.all_params(with_grad=True):
for cb in self.cbs: state = _update(state, cb(p, **{**state, **hyper}))
self.state[p] = state
def clear_state(self):
for p,pg,state,hyper in self.all_params():
self.state[p] = {k: state[k] for k in self._keep_on_clear if k in state}
def state_dict(self):
state = [self.state[p] for p,*_ in self.all_params()]
return {'state': state, 'hypers': self.hypers}
def load_state_dict(self, sd):
assert len(sd["hypers"]) == len(self.param_lists)
assert len(sd["state"]) == sum([len(pg) for pg in self.param_lists])
self.hypers = sd['hypers']
self.state = {p: s for p,s in zip(self.all_params().itemgot(0), sd['state'])}
add_docs(Optimizer,
zero_grad="Standard PyTorch API: Zero all the grad attributes of the parameters",
step="Standard PyTorch API: Update the stats and execute the steppers in on all parameters that have a grad",
state_dict="Return the state of the optimizer in a dictionary",
load_state_dict="Load the content of `sd`",
clear_state="Reset the state of the optimizer")
opt = Optimizer([1,2,3], noop)
test_eq(opt.param_lists, [[1,2,3]])
opt = Optimizer(range(3), noop)
test_eq(opt.param_lists, [[0,1,2]])
opt = Optimizer([[1,2],[3]], noop)
test_eq(opt.param_lists, [[1,2],[3]])
opt = Optimizer(([o,o+1] for o in range(0,4,2)), noop)
test_eq(opt.param_lists, [[0,1],[2,3]])
def tst_arg(p, lr=0, **kwargs): return p
tst_arg.defaults = dict(lr=1e-2)
def tst_arg2(p, lr2=0, **kwargs): return p
tst_arg2.defaults = dict(lr2=1e-3)
def tst_arg3(p, mom=0, **kwargs): return p
tst_arg3.defaults = dict(mom=0.9)
def tst_arg4(p, **kwargs): return p
opt = Optimizer([1,2,3], [tst_arg,tst_arg2, tst_arg3])
test_eq(opt.hypers, [{'lr2': 1e-3, 'mom': 0.9, 'lr': 1e-2}])
opt = Optimizer([1,2,3], tst_arg, lr=0.1)
test_eq(opt.hypers, [{'lr': 0.1}])
opt = Optimizer([[1,2],[3]], tst_arg)
test_eq(opt.hypers, [{'lr': 1e-2}, {'lr': 1e-2}])
opt = Optimizer([[1,2],[3]], tst_arg, lr=0.1)
test_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.1}])
opt = Optimizer([[1,2],[3]], tst_arg, lr=[0.1,0.2])
test_eq(opt.hypers, [{'lr': 0.1}, {'lr': 0.2}])
opt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-2))
test_eq(opt.hypers, [{'lr': 1e-3}, {'lr': 1e-3}, {'lr': 1e-2}])
opt = Optimizer([[1,2],[3],[4]], tst_arg, lr=slice(1e-4,1e-2))
test_eq(opt.hypers, [{'lr': 1e-4}, {'lr': 1e-3}, {'lr': 1e-2}])
test_eq(opt.param_groups, [{'params': [1,2], 'lr': 1e-4}, {'params': [3], 'lr': 1e-3}, {'params': [4], 'lr': 1e-2}])
test_fail(lambda: Optimizer([[1,2],[3],[4]], tst_arg, lr=np.array([0.1,0.2])))
#export
def sgd_step(p, lr, **kwargs):
p.data.add_(p.grad.data, alpha=-lr)
def tst_param(val, grad=None):
"Create a tensor with `val` and a gradient of `grad` for testing"
res = tensor([val]).float()
res.grad = tensor([val/10 if grad is None else grad]).float()
return res
p = tst_param(1., 0.1)
sgd_step(p, 1.)
test_eq(p, tensor([0.9]))
test_eq(p.grad, tensor([0.1]))
#export
def weight_decay(p, lr, wd, do_wd=True, **kwargs):
"Weight decay as decaying `p` with `lr*wd`"
if do_wd and wd!=0: p.data.mul_(1 - lr*wd)
weight_decay.defaults = dict(wd=0.)
p = tst_param(1., 0.1)
weight_decay(p, 1., 0.1)
test_eq(p, tensor([0.9]))
test_eq(p.grad, tensor([0.1]))
#export
def l2_reg(p, lr, wd, do_wd=True, **kwargs):
"L2 regularization as adding `wd*p` to `p.grad`"
if do_wd and wd!=0: p.grad.data.add_(p.data, alpha=wd)
l2_reg.defaults = dict(wd=0.)
p = tst_param(1., 0.1)
l2_reg(p, 1., 0.1)
test_eq(p, tensor([1.]))
test_eq(p.grad, tensor([0.2]))
show_doc(Optimizer.step)
#test basic step
r = L.range(4)
def tst_params(): return r.map(tst_param)
params = tst_params()
opt = Optimizer(params, sgd_step, lr=0.1)
opt.step()
test_close([p.item() for p in params], r.map(mul(0.99)))
#test two steps
params = tst_params()
opt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)
opt.step()
test_close([p.item() for p in params], r.map(mul(0.98)))
#test None gradients are ignored
params = tst_params()
opt = Optimizer(params, sgd_step, lr=0.1)
params[-1].grad = None
opt.step()
test_close([p.item() for p in params], [0., 0.99, 1.98, 3.])
#test discriminative lrs
params = tst_params()
opt = Optimizer([params[:2], params[2:]], sgd_step, lr=0.1)
opt.hypers[0]['lr'] = 0.01
opt.step()
test_close([p.item() for p in params], [0., 0.999, 1.98, 2.97])
show_doc(Optimizer.zero_grad)
params = tst_params()
opt = Optimizer(params, [weight_decay, sgd_step], lr=0.1, wd=0.1)
opt.zero_grad()
[test_eq(p.grad, tensor([0.])) for p in params];
def tst_stat(p, **kwargs):
s = kwargs.get('sum', torch.zeros_like(p)) + p.data
return {'sum': s}
tst_stat.defaults = {'mom': 0.9}
#Test Optimizer init
opt = Optimizer([1,2,3], tst_stat)
test_eq(opt.hypers, [{'mom': 0.9}])
opt = Optimizer([1,2,3], tst_stat, mom=0.99)
test_eq(opt.hypers, [{'mom': 0.99}])
#Test stat
x = torch.randn(4,5)
state = tst_stat(x)
assert 'sum' in state
test_eq(x, state['sum'])
state = tst_stat(x, **state)
test_eq(state['sum'], 2*x)
# export
def average_grad(p, mom, dampening=False, grad_avg=None, **kwargs):
"Keeps track of the avg grads of `p` in `state` with `mom`."
if grad_avg is None: grad_avg = torch.zeros_like(p.grad.data)
damp = 1-mom if dampening else 1.
grad_avg.mul_(mom).add_(p.grad.data, alpha=damp)
return {'grad_avg': grad_avg}
average_grad.defaults = dict(mom=0.9)
new_val = old_val * mom + grad
new_val = old_val * mom + grad * (1-mom)
p = tst_param([1,2,3], [4,5,6])
state = {}
state = average_grad(p, mom=0.9, **state)
test_eq(state['grad_avg'], p.grad)
state = average_grad(p, mom=0.9, **state)
test_eq(state['grad_avg'], p.grad * 1.9)
#Test dampening
state = {}
state = average_grad(p, mom=0.9, dampening=True, **state)
test_eq(state['grad_avg'], 0.1*p.grad)
state = average_grad(p, mom=0.9, dampening=True, **state)
test_close(state['grad_avg'], (0.1*0.9+0.1)*p.grad)
# export
def average_sqr_grad(p, sqr_mom, dampening=True, sqr_avg=None, **kwargs):
if sqr_avg is None: sqr_avg = torch.zeros_like(p.grad.data)
damp = 1-sqr_mom if dampening else 1.
sqr_avg.mul_(sqr_mom).addcmul_(p.grad.data, p.grad.data, value=damp)
return {'sqr_avg': sqr_avg}
average_sqr_grad.defaults = dict(sqr_mom=0.99)
new_val = old_val * mom + grad**2
new_val = old_val * mom + (grad**2) * (1-mom)
p = tst_param([1,2,3], [4,5,6])
state = {}
state = average_sqr_grad(p, sqr_mom=0.99, dampening=False, **state)
test_eq(state['sqr_avg'], p.grad.pow(2))
state = average_sqr_grad(p, sqr_mom=0.99, dampening=False, **state)
test_eq(state['sqr_avg'], p.grad.pow(2) * 1.99)
#Test dampening
state = {}
state = average_sqr_grad(p, sqr_mom=0.99, **state)
test_close(state['sqr_avg'], 0.01*p.grad.pow(2))
state = average_sqr_grad(p, sqr_mom=0.99, **state)
test_close(state['sqr_avg'], (0.01*0.99+0.01)*p.grad.pow(2))
show_doc(Optimizer.freeze, name="Optimizer.freeze")
show_doc(Optimizer.freeze_to, name="Optimizer.freeze_to")
show_doc(Optimizer.unfreeze, name="Optimizer.unfreeze")
#Freezing the first layer
params = [tst_params(), tst_params(), tst_params()]
opt = Optimizer(params, sgd_step, lr=0.1)
opt.freeze_to(1)
req_grad = Self.requires_grad()
test_eq(L(params[0]).map(req_grad), [False]*4)
for i in {1,2}: test_eq(L(params[i]).map(req_grad), [True]*4)
#Unfreezing
opt.unfreeze()
for i in range(2): test_eq(L(params[i]).map(req_grad), [True]*4)
#TODO: test warning
# opt.freeze_to(3)
params = [tst_params(), tst_params(), tst_params()]
opt = Optimizer(params, sgd_step, lr=0.1)
for p in L(params[1])[[1,3]]: opt.state[p] = {'force_train': True}
opt.freeze()
test_eq(L(params[0]).map(req_grad), [False]*4)
test_eq(L(params[1]).map(req_grad), [False, True, False, True])
test_eq(L(params[2]).map(req_grad), [True]*4)
show_doc(Optimizer.state_dict)
show_doc(Optimizer.load_state_dict)
p = tst_param([1,2,3], [4,5,6])
opt = Optimizer(p, average_grad)
opt.step()
test_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))
sd = opt.state_dict()
p1 = tst_param([10,20,30], [40,50,60])
opt = Optimizer(p1, average_grad, mom=0.99)
test_eq(opt.hypers[0]['mom'], 0.99)
test_eq(opt.state, {})
opt.load_state_dict(sd)
test_eq(opt.hypers[0]['mom'], 0.9)
test_eq(opt.state[p1]['grad_avg'], tensor([[4., 5., 6.]]))
show_doc(Optimizer.clear_state)
p = tst_param([1,2,3], [4,5,6])
opt = Optimizer(p, average_grad)
opt.state[p] = {'force_train': True}
opt.step()
test_eq(opt.state[p]['grad_avg'], tensor([[4., 5., 6.]]))
opt.clear_state()
test_eq(opt.state[p], {'force_train': True})
#export
def momentum_step(p, lr, grad_avg, **kwargs):
"Step for SGD with momentum with `lr`"
p.data.add_(grad_avg, alpha=-lr)
#export
@log_args(to_return=True, but='params')
def SGD(params, lr, mom=0., wd=0., decouple_wd=True):
"A `Optimizer` for SGD with `lr` and `mom` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
if mom != 0: cbs.append(average_grad)
cbs.append(sgd_step if mom==0 else momentum_step)
return Optimizer(params, cbs, lr=lr, mom=mom, wd=wd)
#Vanilla SGD
params = tst_params()
opt = SGD(params, lr=0.1)
opt.step()
test_close([p.item() for p in params], [i*0.99 for i in range(4)])
opt.step()
[p.item() for p in params]
test_close([p.item() for p in params], [i*0.98 for i in range(4)])
#SGD with momentum
params = tst_params()
opt = SGD(params, lr=0.1, mom=0.9)
assert isinstance(opt, Optimizer)
opt.step()
test_close([p.item() for p in params], [i*0.99 for i in range(4)])
opt.step()
[p.item() for p in params]
test_close([p.item() for p in params], [i*(1 - 0.1 * (0.1 + 0.1*1.9)) for i in range(4)])
for i,p in enumerate(params): test_close(opt.state[p]['grad_avg'].item(), i*0.19)
params = tst_params()
#Weight decay
opt = SGD(params, lr=0.1, mom=0.9, wd=0.1)
opt.step()
test_close([p.item() for p in params], [i*0.98 for i in range(4)])
#L2 reg
opt = SGD(params, lr=0.1, mom=0.9, wd=0.1, decouple_wd=False)
opt.step()
#TODO: fix cause this formula was wrong
#test_close([p.item() for p in params], [i*0.97 for i in range(4)])
#export
def rms_prop_step(p, lr, sqr_avg, eps, grad_avg=None, **kwargs):
"Step for SGD with momentum with `lr`"
denom = sqr_avg.sqrt().add_(eps)
p.data.addcdiv_((grad_avg if grad_avg is not None else p.grad), denom, value=-lr)
rms_prop_step.defaults = dict(eps=1e-8)
#export
@log_args(to_return=True, but='params')
def RMSProp(params, lr, sqr_mom=0.99, mom=0., wd=0., decouple_wd=True):
"A `Optimizer` for RMSProp with `lr`, `sqr_mom`, `mom` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += ([average_sqr_grad] if mom==0. else [average_grad, average_sqr_grad])
cbs.append(rms_prop_step)
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, wd=wd)
#Without momentum
import math
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RMSProp(params, lr=0.1)
opt.step()
test_close(params[0], tensor([0.,1.,2.]))
opt.step()
step = - 0.1 * 0.1 / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)
test_close(params[0], tensor([step, 1+step, 2+step]))
#With momentum
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RMSProp(params, lr=0.1, mom=0.9)
opt.step()
test_close(params[0], tensor([0.,1.,2.]))
opt.step()
step = - 0.1 * (0.1 + 0.9*0.1) / (math.sqrt((0.01*0.99+0.01) * 0.1**2) + 1e-8)
test_close(params[0], tensor([step, 1+step, 2+step]))
#export
def step_stat(p, step=0, **kwargs):
"Register the number of steps done in `state` for `p`"
step += 1
return {'step' : step}
p = tst_param(1,0.1)
state = {}
state = step_stat(p, **state)
test_eq(state['step'], 1)
for _ in range(5): state = step_stat(p, **state)
test_eq(state['step'], 6)
#export
def debias(mom, damp, step): return damp * (1 - mom**step) / (1-mom)
#export
def adam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):
"Step for Adam with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
p.data.addcdiv_(grad_avg, (sqr_avg/debias2).sqrt() + eps, value = -lr / debias1)
return p
adam_step._defaults = dict(eps=1e-5)
#export
@log_args(to_return=True, but='params')
def Adam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0.01, decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), average_sqr_grad, step_stat, adam_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = Adam(params, lr=0.1, wd=0)
opt.step()
step = -0.1 * 0.1 / (math.sqrt(0.1**2) + 1e-8)
test_close(params[0], tensor([1+step, 2+step, 3+step]))
opt.step()
test_close(params[0], tensor([1+2*step, 2+2*step, 3+2*step]), eps=1e-3)
#export
def radam_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, beta, **kwargs):
"Step for RAdam with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
r_inf = 2/(1-sqr_mom) - 1
r = r_inf - 2*step*sqr_mom**step/(1-sqr_mom**step)
if r > 5:
v = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))
denom = (sqr_avg/debias2).sqrt()
if eps: denom += eps
if beta: denom = F.softplus(denom, beta)
p.data.addcdiv_(grad_avg, denom, value = -lr*v / debias1)
else: p.data.add_(grad_avg, alpha=-lr / debias1)
return p
radam_step._defaults = dict(eps=1e-5)
#export
@log_args(to_return=True, but='params')
def RAdam(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., beta=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs = [partial(average_grad, dampening=True), average_sqr_grad, step_stat, radam_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd, beta=beta)
beta = 0.99
r_inf = 2/(1-beta) - 1
rs = np.array([r_inf - 2*s*beta**s/(1-beta**s) for s in range(5,500)])
v = np.sqrt(((rs-4) * (rs-2) * r_inf)/((r_inf-4)*(r_inf-2)*rs))
plt.plot(v);
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = RAdam(params, lr=0.1)
#The r factor is lower than 5 during the first 5 steps so updates use the aveage of gradients (all the same)
r_inf = 2/(1-0.99) - 1
for i in range(5):
r = r_inf - 2*(i+1)*0.99**(i+1)/(1-0.99**(i+1))
assert r <= 5
opt.step()
p = tensor([0.95, 1.9, 2.85])
test_close(params[0], p)
#The r factor is greater than 5 for the sixth step so we update with RAdam
r = r_inf - 2*6*0.99**6/(1-0.99**6)
assert r > 5
opt.step()
v = math.sqrt(((r-4) * (r-2) * r_inf)/((r_inf-4)*(r_inf-2)*r))
step = -0.1*0.1*v/(math.sqrt(0.1**2) + 1e-8)
test_close(params[0], p+step)
#export
def qhadam_step(p, lr, mom, sqr_mom, sqr_avg, nu_1, nu_2, step, grad_avg, eps, **kwargs):
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
p.data.addcdiv_(((1-nu_1) * p.grad.data) + (nu_1 * (grad_avg / debias1)),
(((1 - nu_2) * (p.grad.data)**2) + (nu_2 * (sqr_avg / debias2))).sqrt() + eps,
value = -lr)
return p
qhadam_step._defaults = dict(eps=1e-8)
#export
@log_args(to_return=True, but='params')
def QHAdam(params, lr, mom=0.999, sqr_mom=0.999, nu_1=0.7, nu_2 = 1.0, eps=1e-8, wd=0., decouple_wd=True):
"An `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `nus`, eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), partial(average_sqr_grad, dampening=True), step_stat, qhadam_step]
return Optimizer(params, cbs, lr=lr, nu_1=nu_1, nu_2=nu_2 ,
mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = QHAdam(params, lr=0.1)
opt.step()
step = -0.1 * (((1-0.7) * 0.1) + (0.7 * 0.1)) / (
math.sqrt(((1-1.0) * 0.1**2) + (1.0 * 0.1**2)) + 1e-8)
test_close(params[0], tensor([1+step, 2+step, 3+step]))
opt.step()
test_close(params[0], tensor([1+2*step, 2+2*step, 3+2*step]), eps=1e-3)
#export
def larc_layer_lr(p, lr, trust_coeff, wd, eps, clip=True, **kwargs):
"Computes the local lr before weight decay is applied"
p_norm,g_norm = torch.norm(p.data),torch.norm(p.grad.data)
local_lr = lr*trust_coeff * (p_norm) / (g_norm + p_norm * wd + eps)
return {'local_lr': min(lr, local_lr) if clip else local_lr}
larc_layer_lr.defaults = dict(trust_coeff=0.02, wd=0., eps=1e-8)
#export
def larc_step(p, local_lr, grad_avg=None, **kwargs):
"Step for LARC `local_lr` on `p`"
p.data.add_(p.grad.data if grad_avg is None else grad_avg, alpha = -local_lr)
#export
@log_args(to_return=True, but='params')
def Larc(params, lr, mom=0.9, clip=True, trust_coeff=0.02, eps=1e-8, wd=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
if mom!=0.: cbs.append(average_grad)
cbs += [partial(larc_layer_lr, clip=clip), larc_step]
return Optimizer(params, cbs, lr=lr, mom=mom, trust_coeff=trust_coeff, eps=eps, wd=wd)
params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]
opt = Larc(params, lr=0.1)
opt.step()
#First param local lr is 0.02 < lr so it's not clipped
test_close(opt.state[params[0]]['local_lr'], 0.02)
#Second param local lr is 0.2 > lr so it's clipped
test_eq(opt.state[params[1]]['local_lr'], 0.1)
test_close(params[0], tensor([0.998,1.996,2.994]))
test_close(params[1], tensor([0.999,1.998,2.997]))
params = [tst_param([1,2,3], [0.1,0.2,0.3]), tst_param([1,2,3], [0.01,0.02,0.03])]
opt = Larc(params, lr=0.1, clip=False)
opt.step()
#No clipping
test_close(opt.state[params[0]]['local_lr'], 0.02)
test_close(opt.state[params[1]]['local_lr'], 0.2)
test_close(params[0], tensor([0.998,1.996,2.994]))
test_close(params[1], tensor([0.998,1.996,2.994]))
#export
def lamb_step(p, lr, mom, step, sqr_mom, grad_avg, sqr_avg, eps, **kwargs):
"Step for LAMB with `lr` on `p`"
debias1 = debias(mom, 1-mom, step)
debias2 = debias(sqr_mom, 1-sqr_mom, step)
r1 = p.data.pow(2).mean().sqrt()
step = (grad_avg/debias1) / ((sqr_avg/debias2).sqrt()+eps)
r2 = step.pow(2).mean().sqrt()
q = 1 if r1 == 0 or r2 == 0 else min(r1/r2,10)
p.data.add_(step, alpha = -lr * q)
lamb_step._defaults = dict(eps=1e-6, wd=0.)
#export
@log_args(to_return=True, but='params')
def Lamb(params, lr, mom=0.9, sqr_mom=0.99, eps=1e-5, wd=0., decouple_wd=True):
"A `Optimizer` for Adam with `lr`, `mom`, `sqr_mom`, `eps` and `params`"
cbs = [weight_decay] if decouple_wd else [l2_reg]
cbs += [partial(average_grad, dampening=True), average_sqr_grad, step_stat, lamb_step]
return Optimizer(params, cbs, lr=lr, mom=mom, sqr_mom=sqr_mom, eps=eps, wd=wd)
params = tst_param([1,2,3], [0.1,0.2,0.3])
opt = Lamb(params, lr=0.1)
opt.step()
test_close(params[0], tensor([0.7840,1.7840,2.7840]), eps=1e-3)
#export
@log_args(but='opt')
class Lookahead(Optimizer, GetAttr):
"Wrap `opt` in a lookahead optimizer"
_default='opt'
def __init__(self, opt, k=6, alpha=0.5):
store_attr(self, 'opt,k,alpha')
self._init_state()
def step(self):
if self.slow_weights is None: self._copy_weights()
self.opt.step()
self.count += 1
if self.count%self.k != 0: return
for slow_pg,fast_pg in zip(self.slow_weights,self.param_lists):
for slow_p,fast_p in zip(slow_pg,fast_pg):
slow_p.data.add_(fast_p.data-slow_p.data, alpha=self.alpha)
fast_p.data.copy_(slow_p.data)
def clear_state(self):
self.opt.clear_state()
self._init_state()
def state_dict(self):
state = self.opt.state_dict()
state.update({'count': self.count, 'slow_weights': self.slow_weights})
return state
def load_state_dict(self, sd):
self.count = sd.pop('count')
self.slow_weights = sd.pop('slow_weights')
self.opt.load_state_dict(sd)
def _init_state(self): self.count,self.slow_weights = 0,None
def _copy_weights(self): self.slow_weights = L(L(p.clone().detach() for p in pg) for pg in self.param_lists)
@property
def param_lists(self): return self.opt.param_lists
@param_lists.setter
def param_lists(self, v): self.opt.param_lists = v
params = tst_param([1,2,3], [0.1,0.2,0.3])
p,g = params[0].data.clone(),tensor([0.1,0.2,0.3])
opt = Lookahead(SGD(params, lr=0.1))
for k in range(5): opt.step()
#first 5 steps are normal SGD steps
test_close(params[0], p - 0.5*g)
#Since k=6, sixth step is a moving average of the 6 SGD steps with the intial weight
opt.step()
test_close(params[0], p * 0.5 + (p-0.6*g) * 0.5)
#export
@delegates(RAdam)
def ranger(p, lr, mom=0.95, wd=0.01, eps=1e-6, **kwargs):
"Convenience method for `Lookahead` with `RAdam`"
return Lookahead(RAdam(p, lr=lr, mom=mom, wd=wd, eps=eps, **kwargs))
#export
def detuplify_pg(d):
res = {}
for k,v in d.items():
if k == 'params': continue
if is_listy(v): res.update(**{f'{k}__{i}': v_ for i,v_ in enumerate(v)})
else: res[k] = v
return res
tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}
test_eq(detuplify_pg(tst), {'lr': 1e-2, 'mom': 0.9})
tst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}
test_eq(detuplify_pg(tst), {'lr': 1e-2, 'betas__0': 0.9, 'betas__1': 0.999})
#export
def set_item_pg(pg, k, v):
if '__' not in k: pg[k] = v
else:
name,idx = k.split('__')
pg[name] = tuple(v if i==int(idx) else pg[name][i] for i in range_of(pg[name]))
return pg
tst = {'lr': 1e-2, 'mom': 0.9, 'params':[0,1,2]}
test_eq(set_item_pg(tst, 'lr', 1e-3), {'lr': 1e-3, 'mom': 0.9, 'params':[0,1,2]})
tst = {'lr': 1e-2, 'betas': (0.9,0.999), 'params':[0,1,2]}
test_eq(set_item_pg(tst, 'betas__0', 0.95), {'lr': 1e-2, 'betas': (0.95,0.999), 'params':[0,1,2]})
#export
pytorch_hp_map = {'momentum': 'mom', 'weight_decay': 'wd', 'alpha': 'sqr_mom', 'betas__0': 'mom', 'betas__1': 'sqr_mom'}
#export
class OptimWrapper(_BaseOptimizer, GetAttr):
_xtra=['zero_grad', 'step', 'state_dict', 'load_state_dict']
_default='opt'
def __init__(self, opt, hp_map=None):
self.opt = opt
if hp_map is None: hp_map = pytorch_hp_map
self.fwd_map = {k: hp_map[k] if k in hp_map else k for k in detuplify_pg(opt.param_groups[0]).keys()}
self.bwd_map = {v:k for k,v in self.fwd_map.items()}
self.state = defaultdict(dict, {})
self.frozen_idx = 0
@property
def hypers(self):
return [{self.fwd_map[k]:v for k,v in detuplify_pg(pg).items() if k != 'params'} for pg in self.opt.param_groups]
def _set_hyper(self, k, v):
for pg,v_ in zip(self.opt.param_groups,v): pg = set_item_pg(pg, self.bwd_map[k], v_)
def clear_state(self): self.opt.state = defaultdict(dict, {})
@property
def param_lists(self): return [pg['params'] for pg in self.opt.param_groups]
@param_lists.setter
def param_lists(self, v):
for pg,v_ in zip(self.opt.param_groups,v): pg['params'] = v_
sgd = SGD([tensor([1,2,3])], lr=1e-3, mom=0.9, wd=1e-2)
tst_sgd = OptimWrapper(torch.optim.SGD([tensor([1,2,3])], lr=1e-3, momentum=0.9, weight_decay=1e-2))
#Access to param_groups
test_eq(tst_sgd.param_lists, sgd.param_lists)
#Set param_groups
tst_sgd.param_lists = [[tensor([4,5,6])]]
test_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])
#Access to hypers
test_eq(tst_sgd.hypers, [{**sgd.hypers[0], 'dampening': 0., 'nesterov': False}])
#Set hypers
tst_sgd.set_hyper('mom', 0.95)
test_eq(tst_sgd.opt.param_groups[0]['momentum'], 0.95)
tst_sgd = OptimWrapper(torch.optim.SGD([{'params': [tensor([1,2,3])], 'lr': 1e-3},
{'params': [tensor([4,5,6])], 'lr': 1e-2}], momentum=0.9, weight_decay=1e-2))
sgd = SGD([[tensor([1,2,3])], [tensor([4,5,6])]], lr=[1e-3, 1e-2], mom=0.9, wd=1e-2)
#Access to param_groups
test_eq(tst_sgd.param_lists, sgd.param_lists)
#Set param_groups
tst_sgd.param_lists = [[tensor([4,5,6])], [tensor([1,2,3])]]
test_eq(tst_sgd.opt.param_groups[0]['params'], [tensor(4,5,6)])
test_eq(tst_sgd.opt.param_groups[1]['params'], [tensor(1,2,3)])
#Access to hypers
test_eq(tst_sgd.hypers, [{**sgd.hypers[i], 'dampening': 0., 'nesterov': False} for i in range(2)])
#Set hypers
tst_sgd.set_hyper('mom', 0.95)
test_eq([pg['momentum'] for pg in tst_sgd.opt.param_groups], [0.95,0.95])
tst_sgd.set_hyper('lr', [1e-4,1e-3])
test_eq([pg['lr'] for pg in tst_sgd.opt.param_groups], [1e-4,1e-3])
#hide
#check it works with tuply hp names like in Adam
tst_adam = OptimWrapper(torch.optim.Adam([tensor([1,2,3])], lr=1e-2, betas=(0.9, 0.99)))
test_eq(tst_adam.hypers, [{'lr': 0.01, 'mom': 0.9, 'sqr_mom': 0.99, 'eps': 1e-08, 'wd': 0, 'amsgrad': False}])
tst_adam.set_hyper('mom', 0.95)
test_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.99))
tst_adam.set_hyper('sqr_mom', 0.9)
test_eq(tst_adam.opt.param_groups[0]['betas'], (0.95, 0.9))
def _mock_train(m, x, y, opt):
m.train()
for i in range(0, 100, 25):
z = m(x[i:i+25])
loss = F.mse_loss(z, y[i:i+25])
loss.backward()
opt.step()
opt.zero_grad()
m = nn.Linear(4,5)
x = torch.randn(100, 3, 4)
y = torch.randn(100, 3, 5)
try:
torch.save(m.state_dict(), 'tmp.pth')
wgt,bias = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt1 = OptimWrapper(torch.optim.AdamW(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))
_mock_train(m, x.clone(), y.clone(), opt1)
wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt2 = Adam(m.parameters(), 1e-3, wd=1e-2)
_mock_train(m, x.clone(), y.clone(), opt2)
wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()
test_close(wgt1,wgt2,eps=1e-3)
test_close(bias1,bias2,eps=1e-3)
finally: os.remove('tmp.pth')
m = nn.Linear(4,5)
x = torch.randn(100, 3, 4)
y = torch.randn(100, 3, 5)
try:
torch.save(m.state_dict(), 'tmp.pth')
wgt,bias = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt1 = OptimWrapper(torch.optim.Adam(m.parameters(), betas=(0.9, 0.99), eps=1e-5, weight_decay=1e-2))
_mock_train(m, x.clone(), y.clone(), opt1)
wgt1,bias1 = m.weight.data.clone(),m.bias.data.clone()
m.load_state_dict(torch.load('tmp.pth'))
opt2 = Adam(m.parameters(), 1e-3, wd=1e-2, decouple_wd=False)
_mock_train(m, x.clone(), y.clone(), opt2)
wgt2,bias2 = m.weight.data.clone(),m.bias.data.clone()
test_close(wgt1,wgt2,eps=1e-3)
test_close(bias1,bias2,eps=1e-3)
finally: os.remove('tmp.pth')
#hide
from nbdev.export import *
notebook2script()
| 0.606964 | 0.833223 |
Taken from [second part of Speech processing series](https://towardsdatascience.com/audio-deep-learning-made-simple-part-2-why-mel-spectrograms-perform-better-aad889a93505)
- We already know how sound is represented digitally, and that we need to convert it into a spectrogram for use in deep learning architectures. This part dives into detail how that is done and how we can tune that conversion to get better performance.
#### Audio file formats and Python libraries
- Audio data for deep learning models will usually start out as digital audio files
- Different types of audio formats: .wav, .mp3, .wma, .aac, .flac
- Commonly used libraries: librosa, scipy, torchaudio (pytorch), ...
### Audio signal data
- Audio data is obtained by sampling the sound wave at regular time intervals and measuring the intensity or amplitude of the wave at each sample. The metadata for that audio tells us the sampling rate which is the number of samples per second.
- In memory, audio is represented as a time series of numbers, representing the amplitude at each timestep. For instance, if the sample rate was 16800, a one-second clip of audio would have 16800 numbers. Since the measurements are taken at fixed intervals of time, the data contains only the amplitude numbers and not the time values. Given the sample rate, we can figure out at what time instant each amplitude number measurement was taken.
```
import librosa
import librosa.display
import numpy as np
import matplotlib.pyplot as plt
sr = 16800
x, _ = librosa.load('./audio/SA1.WAV', sr = sr)
x.shape
x
max(x), min(x), np.mean(x)
```
### Spectrograms
- Deep learning models rarely take this raw audio directly as input. The common practice is to convert the audio into a spectrogram. The spectrogram is a concise 'snapshot' of an audio wave in "image" format.
- Spectrograms are generated from sound signals using Fourier Transforms. A Fourier Transform decomposes the signal into its constituent frequencies and displays the amplitude of each frequency present in the signal.
- A Spectrogram chops up the duration of the sound signal into smaller time segments and then applies the Fourier Transform to each segment, to determine the frequencies contained in that segment. It then combines the Fourier Transforms for all those segments into a single plot.
- It plots Frequency (y-axis) vs Time (x-axis) and uses different colors to indicate the Amplitude of each frequency. The brighter the color, the higher the energy of the signal.
### How do humans hear frequencies?
- The way we hear frequencies in sound is known as 'pitch'. It is a subjective impression of the frequency. So a high-pitched sound has a higher frequency than a low-pitched sound. Humans do not percieve frequencies linearly.
- 200 Hz vs 100 Hz is percieved differently from 1000 Hz vs 1100 Hz.
- ``We hear sound in a logarithmic scale rather than a linear scale.``
### Mel scale
- Mel scale is a scale of pitches, such that each unit is judged by listeners to be equal in pitch distance from the next.
``Amplitude of a sound is rather logarithmical than linear``
### Mel Spectrograms
- A Mel Spectrogram makes two important changes relative to a regular Spectrogram that plots Frequency vs Time.
* It uses the Mel Scale instead of Frequency on the y-axis
* It uses the Decibel Scale instead of Amplitude to indicate colors
- For deep learning models, we usually use this rather than a simple Spectrogram.
```
sgram = librosa.stft(x)
librosa.display.specshow(sgram)
sgram_mag, _ = librosa.magphase(sgram)
mel_scale_sgram = librosa.feature.melspectrogram(S = sgram_mag, sr = sr)
librosa.display.specshow(mel_scale_sgram)
mel_sgram = librosa.amplitude_to_db(mel_scale_sgram, ref = np.min)
librosa.display.specshow(mel_sgram, sr = sr, x_axis ='time', y_axis ='mel')
plt.colorbar(format = '%+2.0f dB')
```
|
github_jupyter
|
import librosa
import librosa.display
import numpy as np
import matplotlib.pyplot as plt
sr = 16800
x, _ = librosa.load('./audio/SA1.WAV', sr = sr)
x.shape
x
max(x), min(x), np.mean(x)
sgram = librosa.stft(x)
librosa.display.specshow(sgram)
sgram_mag, _ = librosa.magphase(sgram)
mel_scale_sgram = librosa.feature.melspectrogram(S = sgram_mag, sr = sr)
librosa.display.specshow(mel_scale_sgram)
mel_sgram = librosa.amplitude_to_db(mel_scale_sgram, ref = np.min)
librosa.display.specshow(mel_sgram, sr = sr, x_axis ='time', y_axis ='mel')
plt.colorbar(format = '%+2.0f dB')
| 0.214362 | 0.987326 |
#### Bibliotecas
```
# Manipulação de dados
import pandas as pd
# Visualização de dados
%matplotlib inline
import matplotlib.pyplot as plt
```
#### Importando a base de dados
```
dados = pd.read_csv('aluguel.csv', sep=';')
# Mostrando a parte inicial dos dados
dados.head()
```
#### Analisando informações gerais sobre a base de dados
```
# Quantidades de dados não-nulos e tipos de variáveis em cada coluna
dados.info()
# Tamanho da base de dados
dados.shape
print('A base de dados apresenta {} imóveis registrados e {} variáveis'.format(dados.shape[0], dados.shape[1]))
```
#### Analisando os tipos de imóveis presentes na base de dados
```
# Visualizandoo os tipos de imóveis e a quantidade de cada um deles na base de dados
dados['Tipo'].value_counts()
# Organizando a visualização dos tipos de imóveis
tipo_de_imovel = pd.DataFrame(dados['Tipo'].value_counts())
# Ajustando o index
tipo_de_imovel.reset_index(inplace=True)
tipo_de_imovel.head()
# Renomeando as colunas
tipo_de_imovel.rename(columns={'index': 'Tipo', 'Tipo': 'Quantidade'}, inplace=True)
# Visualização do DataFrame ajustado
tipo_de_imovel
```
#### Analisando somente os imóveis residenciais
```
# Listando todos os tipos de imóveis da base de dados
tipo_de_imovel['Tipo'].values
```
Os imóveis considerados residenciais dentre os presentes na base de dados são: Apartamento, Casa de Condomínio, Casa, Quitinete e Casa de Vila.
```
# Salvando os imóveis residenciais em uma variavel
imoveis_residenciais = ['Apartamento', 'Casa de Condomínio', 'Casa', 'Quitinete', 'Casa de Vila']
# Salvando em uma variável quais dados da base são referentes aos imóveis residenciais selecionados
selecao = dados['Tipo'].isin(imoveis_residenciais)
# Filtrando os dados da base de acordo com a seleção feita
dados_residencial = dados[selecao]
# Visualizando os 10 primeiros valores do novo DataFrame
dados_residencial.head(10)
# Verificando se a seleção feita está correta
list(dados_residencial['Tipo'].drop_duplicates())
# Quantidade de imóveis residenciais da base
dados_residencial.shape[0]
# Ajustando o index do DataFrame que contém somente os imóveis residencias
dados_residencial.reset_index(inplace=True)
dados_residencial.head()
# Excluindo a coluna 'index'
dados_residencial = dados_residencial.drop(columns=['index'])
```
#### Exportando o novo DataFrame com os dados dos imóveis residenciais
```
dados_residencial.to_csv('aluguel_residencial.csv', sep=';')
```
#### Explorando os dados dos imóveis residenciais
```
# Visualizando os dados
dados_residencial.head(10)
```
Quantidade de imóveis classificados com tipo 'Apartamento'
```
# Criando uma seleção
selecao = dados_residencial['Tipo'] == 'Apartamento'
# Filtrando os dados de acordo com a seleção
quantidade_apartamentos = dados_residencial[selecao].shape[0]
# Printando a quantidade de apartamentos
print('A base de dados contém {} apartamentos'.format(quantidade_apartamentos))
```
Quantidade de imóveis classificados como 'Casa', 'Casa de Condomínio' e 'Casa de Vila'
```
# Criando uma seleção
selecao = ((dados_residencial['Tipo'] == 'Casa') | (dados_residencial['Tipo'] == 'Casa de Condomínio') | (dados_residencial['Tipo'] == 'Casa de Vila'))
# Filtrando os dados de acordo com a seleção
quantidade_casas = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} casas'.format(quantidade_casas))
```
Quantidade de imóveis com área entre 60 e 100 m²
```
# Criando uma seleção
selecao = (dados_residencial['Area'] >= 60) & (dados_residencial['Area'] <= 100)
# Filtrando os dados de acordo com a seleção
quantidade_area_60_a_100 = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} imóveis com área entre 60 e 100 m²'.format(quantidade_area_60_a_100))
```
Quantidade de imóveis com pelo menos 4 quartos e aluguel menor que R$ 2.000,00
```
# Criando uma seleção
selecao = (dados_residencial['Quartos'] >= 4) & (dados_residencial['Valor'] < 2000.0)
# Filtrando os dados de acordo com a seleção
quantidade_4_quartos_menor_2000 = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} imóveis com pelo menos 4 quartos e aluguel menor R$ 2.000,00'.format(quantidade_4_quartos_menor_2000))
```
#### Tratamento de dados faltantes
```
# DataFrame mostrando onde os dados são nulos e onde não são
dados_residencial.isnull()
# Visualizando um resumo das informações do DataFrame
dados_residencial.info()
```
Observa-se que o DataFrame tem um total de 22580 entradas e que, portanto, as colunas Valor, Condomínio e IPTU apresentam dados nulos. Então, é necessário investigar mais a fundo esses dados faltantes para aplicar o tratamento mais adequado a eles.
O valor do aluguel é uma informação muito relevante, por isso, vou começar investigando ela.
```
# Visualizando todas as entradas com o Valor nulo
dados_residencial[dados_residencial['Valor'].isnull()]
```
Como são poucas as entradas que faltam os dados do valor do aluguel, isso não causará impactos significativos nas análises, por isso, esses imóveis serão removidos.
```
# Removendo as linhas que apresentam dados nulos na coluna Valor
dados_residencial.dropna(subset=['Valor'], inplace=True)
```
Quanto ao condomínio, se o imóvel não é um apartamento ou uma casa de condomínio, o preço do condomínio é igual a zero. Portanto, vou investigar se esses dados nulos são apenas referentes a esses tipos de imóveis.
```
# Selecionando apartamento e casa de condomínio com condomínio nulo
selecao = ((dados_residencial['Tipo'] == 'Apartamento') | (dados_residencial['Tipo'] == 'Casa de Condomínio')) & (dados_residencial['Condominio'].isnull())
dados_residencial[selecao]
```
Há 840 imóveis do tipo apartamento e casa de condomínio que não possuem informação quanto ao preço do condomínio. Como essa informação é relevante para esse tipo de imóvel e ela está faltando, esses imóveis serão removidos da base.
```
# Invertendo a seleção para pegar todos os dados desejados
dados_residencial = dados_residencial[~selecao]
# Visualizando os dados
dados_residencial.head(10)
```
Verificando quantos dados faltantes restaram na coluna condomínio
```
dados_residencial[dados_residencial['Condominio'].isnull()].shape[0]
```
Verificando quantos dados nulos temos no IPTU
```
dados_residencial[dados_residencial['IPTU'].isnull()].shape[0]
```
O tratamento feito para esses dados faltantes agora será de substituir os seus valores nulos por zero. No caso do condomínio, porque os imóveis que permanecem com o valor nulo não tem condomínio a ser pago e, para o IPTU, há uma grande quantidade de dados faltantes, o que indica que esse valor não costuma ser informado e não é interessante perder todos esses dados, por isso, será aplicado esse tratamento.
```
# Substituindo os valores nulos por zero
dados_residencial = dados_residencial.fillna({'Condominio': 0, 'IPTU': 0})
# Visualizando os dados
dados_residencial.head(10)
# Verificando agora o resumo das informações do DataFrame novamente
dados_residencial.info()
```
Agora tem-se a mesma quantidade de entradas e de dados não nulos em todas as colunas
```
# Ajustando o index do DataFrame após o tratamento dos dados faltantes
dados_residencial.reset_index(inplace=True)
dados_residencial = dados_residencial.drop(columns=['index'])
dados_residencial.head()
```
#### Criando novas variáveis
Vamos criar uma variável para apresenta o valor bruto, que é o total a ser pago por mês, somando o valor do aluguel, condomínio e IPTU
```
dados_residencial['Valor Bruto'] = dados_residencial['Valor'] + dados_residencial['Condominio'] + dados_residencial['IPTU']
dados_residencial.head(10)
```
Valor do m²
```
dados_residencial['Valor/m²'] = round((dados_residencial['Valor'] / dados_residencial['Area']), 2)
dados_residencial.head(10)
```
Valor bruto do m²
```
dados_residencial['Valor Bruto/m²'] = round((dados_residencial['Valor Bruto'] / dados_residencial['Area']), 2)
dados_residencial.head(10)
```
Separando os tipos de imóveis em dois grandes grupos Apartamento e Casa e salvando em uma nova variável chamada Tipo Agregado
```
# Definindo quais tipos de imóveis farão parte do grande grupo Casa
casa = ['Casa', 'Casa de Condomínio', 'Casa de Vila']
# Fazendo uma iteração para verificar quais imóveis são casa e quais são apartamento e salvar em Tipo Agregado
dados_residencial['Tipo Agregado'] = dados_residencial['Tipo'].apply(lambda x: 'Casa' if x in casa else 'Apartamento')
dados_residencial.head(10)
```
#### Excluindo variáveis
```
# Salvando as variáveis novas em um novo DataFrame
dados_aux = pd.DataFrame(dados_residencial[['Tipo Agregado', 'Valor/m²', 'Valor Bruto', 'Valor Bruto/m²']])
# Visualizando
dados_aux.head(10)
# Deletando a variável Valor Bruto
del dados_aux['Valor Bruto']
dados_aux.head(10)
# Deletando a variável Valor Bruto/m²
dados_aux.pop('Valor Bruto/m²')
dados_aux.head(10)
# Removendo esses dados do DataFrame original
dados_residencial.drop(['Valor Bruto', 'Valor Bruto/m²'], axis=1, inplace=True)
dados_residencial
```
#### Criando agrupamentos
Cálculo da média dos valores de aluguel
```
dados['Valor'].mean()
```
Agrupando os dados por bairro
```
# Método groupby
grupo_bairros = dados_residencial.groupby('Bairro')
# Tipo de dados
type(grupo_bairros)
# Visualizando os grupos criados
grupo_bairros.groups
# Melhorando essa visualização dos grupos
for bairro, data in grupo_bairros:
print('Bairro: {} - Valor médio de aluguel: R$ {}'.format(bairro, round(data['Valor'].mean(), 2)))
# Visualizando de uma forma melhor
grupo_bairros['Valor'].mean().round(2)
# Visualizando valor e condomínio médio
grupo_bairros[['Valor', 'Condominio']].mean().round(2)
```
#### Fazendo análises de estatística descritiva
```
grupo_bairros['Valor'].describe().round(2)
# Selecionando as informações desejadas
grupo_bairros['Valor'].aggregate(['min', 'max', 'sum'])
# Renomeando as colunas
grupo_bairros['Valor'].aggregate(['min', 'max', 'sum']).rename(columns={'min': 'Mínimo', 'max': 'Máximo', 'sum': 'Soma'})
# Visualizando os dados em um gráfico
ax = grupo_bairros['Valor'].mean().plot.bar(color='blue')
ax.figure.set_size_inches(30, 10)
ax.set_title('Valor Médio do Aluguel por Bairro', fontsize=22)
ax.set_xlabel('')
ax.set_ylabel('Valor do Aluguel', fontsize=14)
```
A visualização do gráfico não está muito boa porque tem muitas informações no eixo x (grande quantidade de bairros)
```
# Visualizando os dados em um gráfico
ax = grupo_bairros['Valor'].max().plot.bar(color='blue')
ax.figure.set_size_inches(30, 10)
ax.set_title('Valor Máximo do Aluguel por Bairro', fontsize=22)
ax.set_xlabel('')
ax.set_ylabel('Valor do Aluguel', fontsize=14)
```
A visualização do gráfico está ruim também porque tem muitos dados, mas também porque tem quatro bairros com valores máximos muito maiores que os demais, o que deixa as barras dos outros bairros muito pequenas e difícil de visualizar.
#### Identificando e removendo Outliers
```
dados_residencial.boxplot(['Valor'])
```
Não é possível analisar a distribuição dos valores de aluguel por conta dos valores discrepantes, muito acima dos demais valores. Pode-se observar que os dados indicam que há imóveis com valores de aluguel acima de 500000, o que é absurdo. Esses valores possivelmente devem ser referentes ao valor de compra dos imóveis. Mas vamos analisar essas informações.
```
dados_residencial[dados_residencial['Valor'] >= 500000]
```
Montando um novo boxplot com os valores removendo os outliers
```
# Salvando os valores em uma variável
valor = dados['Valor']
# Estabelecendo os intervalos de valores do boxplot
Q1 = valor.quantile(.25)
Q3 = valor.quantile(.75)
IIQ = Q3 - Q1
# Estabelecendo os valores limites do boxplot
limite_inferior = Q1 - 1.5 * IIQ
limite_superior = Q3 + 1.5 * IIQ
# Selecionando os valores de acordo com os limites
selecao = (valor >= limite_inferior) & (valor <= limite_superior)
novos_dados = dados[selecao]
```
Fazendo um novo boxplot com os dados selecionados
```
ax = novos_dados.boxplot(['Valor'])
ax.figure.set_size_inches(16, 8)
```
Construindo um histograma com os valores antes e depois da seleção
```
# Histograma antes da seleção
ax = dados_residencial.hist(['Valor'], figsize=(12,6))
# Histograma depois da seleção
ax = novos_dados.hist(['Valor'], figsize=(12,6))
```
Fazendo o mesmo procedimento de remoção de outliers para cada tipo de imóvel
```
ax = dados_residencial.boxplot(['Valor'], by=['Tipo'])
ax.figure.set_size_inches(12, 6)
# Agrupando por tipo de imóvel
grupo_tipo = dados_residencial.groupby('Tipo')['Valor']
# Estabelecendo os intervalos de valores do boxplot
Q1 = grupo_tipo.quantile(.25)
Q3 = grupo_tipo.quantile(.75)
IIQ = Q3 - Q1
# Estabelecendo os valores limites do boxplot
limite_inferior = Q1 - 1.5 * IIQ
limite_superior = Q3 + 1.5 * IIQ
# Criando um novo DataFrame com os dados selecionados
# DataFrame vazio
novos_dados = pd.DataFrame()
# Iterando por cada tipo de imóvel
for tipo in grupo_tipo.groups.keys():
# Cria uma Series marcando True se for o tipo atual da iteração e False se não for
eh_tipo = dados_residencial['Tipo'] == tipo
# Cria uma Series marcando True se estiver dentro do limite e False se estiver fora
eh_dentro_limite = (dados_residencial['Valor'] >= limite_inferior[tipo]) & (dados_residencial['Valor'] <= limite_superior[tipo])
# Cria uma Series marcando True para quando as duas variáveis forem True e False caso contrário
selecao = eh_tipo & eh_dentro_limite
# Faz a seleção dos dados marcando True e salva em uma outra variável
dados_selecao = dados_residencial[selecao]
# Adiciona os dados selecionado no DataFrame criado
novos_dados = pd.concat([novos_dados, dados_selecao])
# Visualizando o DataFrame criado
novos_dados.head()
# Fazendo o boxplot com os dados tratados
ax = novos_dados.boxplot(['Valor'], by=['Tipo'])
ax.figure.set_size_inches(16, 8)
```
|
github_jupyter
|
# Manipulação de dados
import pandas as pd
# Visualização de dados
%matplotlib inline
import matplotlib.pyplot as plt
dados = pd.read_csv('aluguel.csv', sep=';')
# Mostrando a parte inicial dos dados
dados.head()
# Quantidades de dados não-nulos e tipos de variáveis em cada coluna
dados.info()
# Tamanho da base de dados
dados.shape
print('A base de dados apresenta {} imóveis registrados e {} variáveis'.format(dados.shape[0], dados.shape[1]))
# Visualizandoo os tipos de imóveis e a quantidade de cada um deles na base de dados
dados['Tipo'].value_counts()
# Organizando a visualização dos tipos de imóveis
tipo_de_imovel = pd.DataFrame(dados['Tipo'].value_counts())
# Ajustando o index
tipo_de_imovel.reset_index(inplace=True)
tipo_de_imovel.head()
# Renomeando as colunas
tipo_de_imovel.rename(columns={'index': 'Tipo', 'Tipo': 'Quantidade'}, inplace=True)
# Visualização do DataFrame ajustado
tipo_de_imovel
# Listando todos os tipos de imóveis da base de dados
tipo_de_imovel['Tipo'].values
# Salvando os imóveis residenciais em uma variavel
imoveis_residenciais = ['Apartamento', 'Casa de Condomínio', 'Casa', 'Quitinete', 'Casa de Vila']
# Salvando em uma variável quais dados da base são referentes aos imóveis residenciais selecionados
selecao = dados['Tipo'].isin(imoveis_residenciais)
# Filtrando os dados da base de acordo com a seleção feita
dados_residencial = dados[selecao]
# Visualizando os 10 primeiros valores do novo DataFrame
dados_residencial.head(10)
# Verificando se a seleção feita está correta
list(dados_residencial['Tipo'].drop_duplicates())
# Quantidade de imóveis residenciais da base
dados_residencial.shape[0]
# Ajustando o index do DataFrame que contém somente os imóveis residencias
dados_residencial.reset_index(inplace=True)
dados_residencial.head()
# Excluindo a coluna 'index'
dados_residencial = dados_residencial.drop(columns=['index'])
dados_residencial.to_csv('aluguel_residencial.csv', sep=';')
# Visualizando os dados
dados_residencial.head(10)
# Criando uma seleção
selecao = dados_residencial['Tipo'] == 'Apartamento'
# Filtrando os dados de acordo com a seleção
quantidade_apartamentos = dados_residencial[selecao].shape[0]
# Printando a quantidade de apartamentos
print('A base de dados contém {} apartamentos'.format(quantidade_apartamentos))
# Criando uma seleção
selecao = ((dados_residencial['Tipo'] == 'Casa') | (dados_residencial['Tipo'] == 'Casa de Condomínio') | (dados_residencial['Tipo'] == 'Casa de Vila'))
# Filtrando os dados de acordo com a seleção
quantidade_casas = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} casas'.format(quantidade_casas))
# Criando uma seleção
selecao = (dados_residencial['Area'] >= 60) & (dados_residencial['Area'] <= 100)
# Filtrando os dados de acordo com a seleção
quantidade_area_60_a_100 = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} imóveis com área entre 60 e 100 m²'.format(quantidade_area_60_a_100))
# Criando uma seleção
selecao = (dados_residencial['Quartos'] >= 4) & (dados_residencial['Valor'] < 2000.0)
# Filtrando os dados de acordo com a seleção
quantidade_4_quartos_menor_2000 = dados_residencial[selecao].shape[0]
# Printando a quantidade de casas
print('A base de dados contém {} imóveis com pelo menos 4 quartos e aluguel menor R$ 2.000,00'.format(quantidade_4_quartos_menor_2000))
# DataFrame mostrando onde os dados são nulos e onde não são
dados_residencial.isnull()
# Visualizando um resumo das informações do DataFrame
dados_residencial.info()
# Visualizando todas as entradas com o Valor nulo
dados_residencial[dados_residencial['Valor'].isnull()]
# Removendo as linhas que apresentam dados nulos na coluna Valor
dados_residencial.dropna(subset=['Valor'], inplace=True)
# Selecionando apartamento e casa de condomínio com condomínio nulo
selecao = ((dados_residencial['Tipo'] == 'Apartamento') | (dados_residencial['Tipo'] == 'Casa de Condomínio')) & (dados_residencial['Condominio'].isnull())
dados_residencial[selecao]
# Invertendo a seleção para pegar todos os dados desejados
dados_residencial = dados_residencial[~selecao]
# Visualizando os dados
dados_residencial.head(10)
dados_residencial[dados_residencial['Condominio'].isnull()].shape[0]
dados_residencial[dados_residencial['IPTU'].isnull()].shape[0]
# Substituindo os valores nulos por zero
dados_residencial = dados_residencial.fillna({'Condominio': 0, 'IPTU': 0})
# Visualizando os dados
dados_residencial.head(10)
# Verificando agora o resumo das informações do DataFrame novamente
dados_residencial.info()
# Ajustando o index do DataFrame após o tratamento dos dados faltantes
dados_residencial.reset_index(inplace=True)
dados_residencial = dados_residencial.drop(columns=['index'])
dados_residencial.head()
dados_residencial['Valor Bruto'] = dados_residencial['Valor'] + dados_residencial['Condominio'] + dados_residencial['IPTU']
dados_residencial.head(10)
dados_residencial['Valor/m²'] = round((dados_residencial['Valor'] / dados_residencial['Area']), 2)
dados_residencial.head(10)
dados_residencial['Valor Bruto/m²'] = round((dados_residencial['Valor Bruto'] / dados_residencial['Area']), 2)
dados_residencial.head(10)
# Definindo quais tipos de imóveis farão parte do grande grupo Casa
casa = ['Casa', 'Casa de Condomínio', 'Casa de Vila']
# Fazendo uma iteração para verificar quais imóveis são casa e quais são apartamento e salvar em Tipo Agregado
dados_residencial['Tipo Agregado'] = dados_residencial['Tipo'].apply(lambda x: 'Casa' if x in casa else 'Apartamento')
dados_residencial.head(10)
# Salvando as variáveis novas em um novo DataFrame
dados_aux = pd.DataFrame(dados_residencial[['Tipo Agregado', 'Valor/m²', 'Valor Bruto', 'Valor Bruto/m²']])
# Visualizando
dados_aux.head(10)
# Deletando a variável Valor Bruto
del dados_aux['Valor Bruto']
dados_aux.head(10)
# Deletando a variável Valor Bruto/m²
dados_aux.pop('Valor Bruto/m²')
dados_aux.head(10)
# Removendo esses dados do DataFrame original
dados_residencial.drop(['Valor Bruto', 'Valor Bruto/m²'], axis=1, inplace=True)
dados_residencial
dados['Valor'].mean()
# Método groupby
grupo_bairros = dados_residencial.groupby('Bairro')
# Tipo de dados
type(grupo_bairros)
# Visualizando os grupos criados
grupo_bairros.groups
# Melhorando essa visualização dos grupos
for bairro, data in grupo_bairros:
print('Bairro: {} - Valor médio de aluguel: R$ {}'.format(bairro, round(data['Valor'].mean(), 2)))
# Visualizando de uma forma melhor
grupo_bairros['Valor'].mean().round(2)
# Visualizando valor e condomínio médio
grupo_bairros[['Valor', 'Condominio']].mean().round(2)
grupo_bairros['Valor'].describe().round(2)
# Selecionando as informações desejadas
grupo_bairros['Valor'].aggregate(['min', 'max', 'sum'])
# Renomeando as colunas
grupo_bairros['Valor'].aggregate(['min', 'max', 'sum']).rename(columns={'min': 'Mínimo', 'max': 'Máximo', 'sum': 'Soma'})
# Visualizando os dados em um gráfico
ax = grupo_bairros['Valor'].mean().plot.bar(color='blue')
ax.figure.set_size_inches(30, 10)
ax.set_title('Valor Médio do Aluguel por Bairro', fontsize=22)
ax.set_xlabel('')
ax.set_ylabel('Valor do Aluguel', fontsize=14)
# Visualizando os dados em um gráfico
ax = grupo_bairros['Valor'].max().plot.bar(color='blue')
ax.figure.set_size_inches(30, 10)
ax.set_title('Valor Máximo do Aluguel por Bairro', fontsize=22)
ax.set_xlabel('')
ax.set_ylabel('Valor do Aluguel', fontsize=14)
dados_residencial.boxplot(['Valor'])
dados_residencial[dados_residencial['Valor'] >= 500000]
# Salvando os valores em uma variável
valor = dados['Valor']
# Estabelecendo os intervalos de valores do boxplot
Q1 = valor.quantile(.25)
Q3 = valor.quantile(.75)
IIQ = Q3 - Q1
# Estabelecendo os valores limites do boxplot
limite_inferior = Q1 - 1.5 * IIQ
limite_superior = Q3 + 1.5 * IIQ
# Selecionando os valores de acordo com os limites
selecao = (valor >= limite_inferior) & (valor <= limite_superior)
novos_dados = dados[selecao]
ax = novos_dados.boxplot(['Valor'])
ax.figure.set_size_inches(16, 8)
# Histograma antes da seleção
ax = dados_residencial.hist(['Valor'], figsize=(12,6))
# Histograma depois da seleção
ax = novos_dados.hist(['Valor'], figsize=(12,6))
ax = dados_residencial.boxplot(['Valor'], by=['Tipo'])
ax.figure.set_size_inches(12, 6)
# Agrupando por tipo de imóvel
grupo_tipo = dados_residencial.groupby('Tipo')['Valor']
# Estabelecendo os intervalos de valores do boxplot
Q1 = grupo_tipo.quantile(.25)
Q3 = grupo_tipo.quantile(.75)
IIQ = Q3 - Q1
# Estabelecendo os valores limites do boxplot
limite_inferior = Q1 - 1.5 * IIQ
limite_superior = Q3 + 1.5 * IIQ
# Criando um novo DataFrame com os dados selecionados
# DataFrame vazio
novos_dados = pd.DataFrame()
# Iterando por cada tipo de imóvel
for tipo in grupo_tipo.groups.keys():
# Cria uma Series marcando True se for o tipo atual da iteração e False se não for
eh_tipo = dados_residencial['Tipo'] == tipo
# Cria uma Series marcando True se estiver dentro do limite e False se estiver fora
eh_dentro_limite = (dados_residencial['Valor'] >= limite_inferior[tipo]) & (dados_residencial['Valor'] <= limite_superior[tipo])
# Cria uma Series marcando True para quando as duas variáveis forem True e False caso contrário
selecao = eh_tipo & eh_dentro_limite
# Faz a seleção dos dados marcando True e salva em uma outra variável
dados_selecao = dados_residencial[selecao]
# Adiciona os dados selecionado no DataFrame criado
novos_dados = pd.concat([novos_dados, dados_selecao])
# Visualizando o DataFrame criado
novos_dados.head()
# Fazendo o boxplot com os dados tratados
ax = novos_dados.boxplot(['Valor'], by=['Tipo'])
ax.figure.set_size_inches(16, 8)
| 0.359252 | 0.936518 |
<a href="https://colab.research.google.com/github/waltz2u/HuskyCoin/blob/master/HuskyCoin.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
import time
import hashlib
import json
import requests
import base64
from flask import Flask, request
from multiprocessing import Process, Pipe
import ecdsa
from miner_config import MINER_ADDRESS, MINER_NODE_URL, PEER_NODES
node = Flask(__name__)
class Block:
def __init__(self, index, timestamp, data, previous_hash):
"""Returns a new Block object. Each block is "chained" to its previous
by calling its unique hash.
Args:
index (int): Block number.
timestamp (int): Block creation timestamp.
data (str): Data to be sent.
previous_hash(str): String representing previous block unique hash.
Attrib:
index (int): Block number.
timestamp (int): Block creation timestamp.
data (str): Data to be sent.
previous_hash(str): String representing previous block unique hash.
hash(str): Current block unique hash.
"""
self.index = index
self.timestamp = timestamp
self.data = data
self.previous_hash = previous_hash
self.hash = self.hash_block()
def hash_block(self):
"""Creates the unique hash for the block. It uses sha256."""
sha = hashlib.sha256()
sha.update((str(self.index) + str(self.timestamp) + str(self.data) + str(self.previous_hash)).encode('utf-8'))
return sha.hexdigest()
def create_genesis_block():
"""To create each block, it needs the hash of the previous one. First
block has no previous, so it must be created manually (with index zero
and arbitrary previous hash)"""
return Block(0, time.time(), {
"proof-of-work": 9,
"transactions": None},
"0")
# Node's blockchain copy
BLOCKCHAIN = [create_genesis_block()]
""" Stores the transactions that this node has in a list.
If the node you sent the transaction adds a block
it will get accepted, but there is a chance it gets
discarded and your transaction goes back as if it was never
processed"""
NODE_PENDING_TRANSACTIONS = []
def proof_of_work(last_proof, blockchain):
# Creates a variable that we will use to find our next proof of work
incrementer = last_proof + 1
# Keep incrementing the incrementer until it's equal to a number divisible by 9
# and the proof of work of the previous block in the chain
start_time = time.time()
while not (incrementer % 7919 == 0 and incrementer % last_proof == 0):
incrementer += 1
# Check if any node found the solution every 60 seconds
if int((time.time()-start_time) % 60) == 0:
# If any other node got the proof, stop searching
new_blockchain = consensus(blockchain)
if new_blockchain:
# (False: another node got proof first, new blockchain)
return False, new_blockchain
# Once that number is found, we can return it as a proof of our work
return incrementer, blockchain
def mine(a, blockchain, node_pending_transactions):
BLOCKCHAIN = blockchain
NODE_PENDING_TRANSACTIONS = node_pending_transactions
while True:
"""Mining is the only way that new coins can be created.
In order to prevent too many coins to be created, the process
is slowed down by a proof of work algorithm.
"""
# Get the last proof of work
last_block = BLOCKCHAIN[-1]
last_proof = last_block.data['proof-of-work']
# Find the proof of work for the current block being mined
# Note: The program will hang here until a new proof of work is found
proof = proof_of_work(last_proof, BLOCKCHAIN)
# If we didn't guess the proof, start mining again
if not proof[0]:
# Update blockchain and save it to file
BLOCKCHAIN = proof[1]
a.send(BLOCKCHAIN)
continue
else:
# Once we find a valid proof of work, we know we can mine a block so
# ...we reward the miner by adding a transaction
# First we load all pending transactions sent to the node server
NODE_PENDING_TRANSACTIONS = requests.get(url = MINER_NODE_URL + '/txion', params = {'update':MINER_ADDRESS}).content
NODE_PENDING_TRANSACTIONS = json.loads(NODE_PENDING_TRANSACTIONS)
# Then we add the mining reward
NODE_PENDING_TRANSACTIONS.append({
"from": "network",
"to": MINER_ADDRESS,
"amount": 1})
# Now we can gather the data needed to create the new block
new_block_data = {
"proof-of-work": proof[0],
"transactions": list(NODE_PENDING_TRANSACTIONS)
}
new_block_index = last_block.index + 1
new_block_timestamp = time.time()
last_block_hash = last_block.hash
# Empty transaction list
NODE_PENDING_TRANSACTIONS = []
# Now create the new block
mined_block = Block(new_block_index, new_block_timestamp, new_block_data, last_block_hash)
BLOCKCHAIN.append(mined_block)
# Let the client know this node mined a block
print(json.dumps({
"index": new_block_index,
"timestamp": str(new_block_timestamp),
"data": new_block_data,
"hash": last_block_hash
}) + "\n")
a.send(BLOCKCHAIN)
requests.get(url = MINER_NODE_URL + '/blocks', params = {'update':MINER_ADDRESS})
def find_new_chains():
# Get the blockchains of every other node
other_chains = []
for node_url in PEER_NODES:
# Get their chains using a GET request
block = requests.get(url = node_url + "/blocks").content
# Convert the JSON object to a Python dictionary
block = json.loads(block)
# Verify other node block is correct
validated = validate_blockchain(block)
if validated:
# Add it to our list
other_chains.append(block)
return other_chains
def consensus(blockchain):
# Get the blocks from other nodes
other_chains = find_new_chains()
# If our chain isn't longest, then we store the longest chain
BLOCKCHAIN = blockchain
longest_chain = BLOCKCHAIN
for chain in other_chains:
if len(longest_chain) < len(chain):
longest_chain = chain
# If the longest chain wasn't ours, then we set our chain to the longest
if longest_chain == BLOCKCHAIN:
# Keep searching for proof
return False
else:
# Give up searching proof, update chain and start over again
BLOCKCHAIN = longest_chain
return BLOCKCHAIN
def validate_blockchain(block):
"""Validate the submitted chain. If hashes are not correct, return false
block(str): json
"""
return True
@node.route('/blocks', methods=['GET'])
def get_blocks():
# Load current blockchain. Only you should update your blockchain
if request.args.get("update") == MINER_ADDRESS:
global BLOCKCHAIN
BLOCKCHAIN = b.recv()
chain_to_send = BLOCKCHAIN
# Converts our blocks into dictionaries so we can send them as json objects later
chain_to_send_json = []
for block in chain_to_send:
block = {
"index": str(block.index),
"timestamp": str(block.timestamp),
"data": str(block.data),
"hash": block.hash
}
chain_to_send_json.append(block)
# Send our chain to whomever requested it
chain_to_send = json.dumps(chain_to_send_json)
return chain_to_send
@node.route('/txion', methods=['GET', 'POST'])
def transaction():
"""Each transaction sent to this node gets validated and submitted.
Then it waits to be added to the blockchain. Transactions only move
coins, they don't create it.
"""
if request.method == 'POST':
# On each new POST request, we extract the transaction data
new_txion = request.get_json()
# Then we add the transaction to our list
if validate_signature(new_txion['from'], new_txion['signature'], new_txion['message']):
NODE_PENDING_TRANSACTIONS.append(new_txion)
# Because the transaction was successfully
# submitted, we log it to our console
print("New transaction")
print("FROM: {0}".format(new_txion['from']))
print("TO: {0}".format(new_txion['to']))
print("AMOUNT: {0}\n".format(new_txion['amount']))
# Then we let the client know it worked out
return "Transaction submission successful\n"
else:
return "Transaction submission failed. Wrong signature\n"
# Send pending transactions to the mining process
elif request.method == 'GET' and request.args.get("update") == MINER_ADDRESS:
pending = json.dumps(NODE_PENDING_TRANSACTIONS)
# Empty transaction list
NODE_PENDING_TRANSACTIONS[:] = []
return pending
def validate_signature(public_key, signature, message):
"""Verifies if the signature is correct. This is used to prove
it's you (and not someone else) trying to do a transaction with your
address. Called when a user tries to submit a new transaction.
"""
public_key = (base64.b64decode(public_key)).hex()
signature = base64.b64decode(signature)
vk = ecdsa.VerifyingKey.from_string(bytes.fromhex(public_key), curve=ecdsa.SECP256k1)
# Try changing into an if/else statement as except is too broad.
try:
return vk.verify(signature, message.encode())
except:
return False
def welcome_msg():
print(""" =========================================\n
SIMPLE COIN v1.0.0 - BLOCKCHAIN SYSTEM\n
=========================================\n\n
You can find more help at: https://github.com/cosme12/SimpleCoin\n
Make sure you are using the latest version or you may end in
a parallel chain.\n\n\n""")
if __name__ == '__main__':
welcome_msg()
# Start mining
a, b = Pipe()
p1 = Process(target=mine, args=(a, BLOCKCHAIN, NODE_PENDING_TRANSACTIONS))
p1.start()
# Start server to receive transactions
p2 = Process(target=node.run(), args=b)
p2.start()
"""This is going to be your wallet. Here you can do several things:
- Generate a new address (public and private key). You are going
to use this address (public key) to send or receive any transactions. You can
have as many addresses as you wish, but keep in mind that if you
lose its credential data, you will not be able to retrieve it.
- Send coins to another address
- Retrieve the entire blockchain and check your balance
If this is your first time using this script don't forget to generate
a new address and edit miner config file with it (only if you are
going to mine).
Timestamp in hashed message. When you send your transaction it will be received
by several nodes. If any node mine a block, your transaction will get added to the
blockchain but other nodes still will have it pending. If any node see that your
transaction with same timestamp was added, they should remove it from the
node_pending_transactions list to avoid it get processed more than 1 time.
"""
import requests
import time
import base64
import ecdsa
def wallet():
response = None
while response not in ["1", "2", "3"]:
response = input("""What do you want to do?
1. Generate new wallet
2. Send coins to another wallet
3. Check transactions\n""")
if response == "1":
# Generate new wallet
print("""=========================================\n
IMPORTANT: save this credentials or you won't be able to recover your wallet\n
=========================================\n""")
generate_ECDSA_keys()
elif response == "2":
addr_from = input("From: introduce your wallet address (public key)\n")
private_key = input("Introduce your private key\n")
addr_to = input("To: introduce destination wallet address\n")
amount = input("Amount: number stating how much do you want to send\n")
print("=========================================\n\n")
print("Is everything correct?\n")
print("From: {0}\nPrivate Key: {1}\nTo: {2}\nAmount: {3}\n".format(addr_from, private_key, addr_to, amount))
response = input("y/n\n")
if response.lower() == "y":
send_transaction(addr_from, private_key, addr_to, amount)
else: # Will always occur when response == 3.
check_transactions()
def send_transaction(addr_from, private_key, addr_to, amount):
"""Sends your transaction to different nodes. Once any of the nodes manage
to mine a block, your transaction will be added to the blockchain. Despite
that, there is a low chance your transaction gets canceled due to other nodes
having a longer chain. So make sure your transaction is deep into the chain
before claiming it as approved!
"""
# For fast debugging REMOVE LATER
# private_key="181f2448fa4636315032e15bb9cbc3053e10ed062ab0b2680a37cd8cb51f53f2"
# amount="3000"
# addr_from="SD5IZAuFixM3PTmkm5ShvLm1tbDNOmVlG7tg6F5r7VHxPNWkNKbzZfa+JdKmfBAIhWs9UKnQLOOL1U+R3WxcsQ=="
# addr_to="SD5IZAuFixM3PTmkm5ShvLm1tbDNOmVlG7tg6F5r7VHxPNWkNKbzZfa+JdKmfBAIhWs9UKnQLOOL1U+R3WxcsQ=="
if len(private_key) == 64:
signature, message = sign_ECDSA_msg(private_key)
url = 'http://localhost:5000/txion'
payload = {"from": addr_from,
"to": addr_to,
"amount": amount,
"signature": signature.decode(),
"message": message}
headers = {"Content-Type": "application/json"}
res = requests.post(url, json=payload, headers=headers)
print(res.text)
else:
print("Wrong address or key length! Verify and try again.")
def check_transactions():
"""Retrieve the entire blockchain. With this you can check your
wallets balance. If the blockchain is to long, it may take some time to load.
"""
res = requests.get('http://localhost:5000/blocks')
print(res.text)
def generate_ECDSA_keys():
"""This function takes care of creating your private and public (your address) keys.
It's very important you don't lose any of them or those wallets will be lost
forever. If someone else get access to your private key, you risk losing your coins.
private_key: str
public_ley: base64 (to make it shorter)
"""
sk = ecdsa.SigningKey.generate(curve=ecdsa.SECP256k1) #this is your sign (private key)
private_key = sk.to_string().hex() #convert your private key to hex
vk = sk.get_verifying_key() #this is your verification key (public key)
public_key = vk.to_string().hex()
#we are going to encode the public key to make it shorter
public_key = base64.b64encode(bytes.fromhex(public_key))
filename = input("Write the name of your new address: ") + ".txt"
with open(filename, "w") as f:
f.write("Private key: {0}\nWallet address / Public key: {1}".format(private_key, public_key.decode()))
print("Your new address and private key are now in the file {0}".format(filename))
def sign_ECDSA_msg(private_key):
"""Sign the message to be sent
private_key: must be hex
return
signature: base64 (to make it shorter)
message: str
"""
# Get timestamp, round it, make it into a string and encode it to bytes
message = str(round(time.time()))
bmessage = message.encode()
sk = ecdsa.SigningKey.from_string(bytes.fromhex(private_key), curve=ecdsa.SECP256k1)
signature = base64.b64encode(sk.sign(bmessage))
return signature, message
if __name__ == '__main__':
print(""" =========================================\n
SIMPLE COIN v1.0.0 - BLOCKCHAIN SYSTEM\n
=========================================\n\n
You can find more help at: https://github.com/cosme12/SimpleCoin\n
Make sure you are using the latest version or you may end in
a parallel chain.\n\n\n""")
wallet()
input("Press ENTER to exit...")
```
|
github_jupyter
|
import time
import hashlib
import json
import requests
import base64
from flask import Flask, request
from multiprocessing import Process, Pipe
import ecdsa
from miner_config import MINER_ADDRESS, MINER_NODE_URL, PEER_NODES
node = Flask(__name__)
class Block:
def __init__(self, index, timestamp, data, previous_hash):
"""Returns a new Block object. Each block is "chained" to its previous
by calling its unique hash.
Args:
index (int): Block number.
timestamp (int): Block creation timestamp.
data (str): Data to be sent.
previous_hash(str): String representing previous block unique hash.
Attrib:
index (int): Block number.
timestamp (int): Block creation timestamp.
data (str): Data to be sent.
previous_hash(str): String representing previous block unique hash.
hash(str): Current block unique hash.
"""
self.index = index
self.timestamp = timestamp
self.data = data
self.previous_hash = previous_hash
self.hash = self.hash_block()
def hash_block(self):
"""Creates the unique hash for the block. It uses sha256."""
sha = hashlib.sha256()
sha.update((str(self.index) + str(self.timestamp) + str(self.data) + str(self.previous_hash)).encode('utf-8'))
return sha.hexdigest()
def create_genesis_block():
"""To create each block, it needs the hash of the previous one. First
block has no previous, so it must be created manually (with index zero
and arbitrary previous hash)"""
return Block(0, time.time(), {
"proof-of-work": 9,
"transactions": None},
"0")
# Node's blockchain copy
BLOCKCHAIN = [create_genesis_block()]
""" Stores the transactions that this node has in a list.
If the node you sent the transaction adds a block
it will get accepted, but there is a chance it gets
discarded and your transaction goes back as if it was never
processed"""
NODE_PENDING_TRANSACTIONS = []
def proof_of_work(last_proof, blockchain):
# Creates a variable that we will use to find our next proof of work
incrementer = last_proof + 1
# Keep incrementing the incrementer until it's equal to a number divisible by 9
# and the proof of work of the previous block in the chain
start_time = time.time()
while not (incrementer % 7919 == 0 and incrementer % last_proof == 0):
incrementer += 1
# Check if any node found the solution every 60 seconds
if int((time.time()-start_time) % 60) == 0:
# If any other node got the proof, stop searching
new_blockchain = consensus(blockchain)
if new_blockchain:
# (False: another node got proof first, new blockchain)
return False, new_blockchain
# Once that number is found, we can return it as a proof of our work
return incrementer, blockchain
def mine(a, blockchain, node_pending_transactions):
BLOCKCHAIN = blockchain
NODE_PENDING_TRANSACTIONS = node_pending_transactions
while True:
"""Mining is the only way that new coins can be created.
In order to prevent too many coins to be created, the process
is slowed down by a proof of work algorithm.
"""
# Get the last proof of work
last_block = BLOCKCHAIN[-1]
last_proof = last_block.data['proof-of-work']
# Find the proof of work for the current block being mined
# Note: The program will hang here until a new proof of work is found
proof = proof_of_work(last_proof, BLOCKCHAIN)
# If we didn't guess the proof, start mining again
if not proof[0]:
# Update blockchain and save it to file
BLOCKCHAIN = proof[1]
a.send(BLOCKCHAIN)
continue
else:
# Once we find a valid proof of work, we know we can mine a block so
# ...we reward the miner by adding a transaction
# First we load all pending transactions sent to the node server
NODE_PENDING_TRANSACTIONS = requests.get(url = MINER_NODE_URL + '/txion', params = {'update':MINER_ADDRESS}).content
NODE_PENDING_TRANSACTIONS = json.loads(NODE_PENDING_TRANSACTIONS)
# Then we add the mining reward
NODE_PENDING_TRANSACTIONS.append({
"from": "network",
"to": MINER_ADDRESS,
"amount": 1})
# Now we can gather the data needed to create the new block
new_block_data = {
"proof-of-work": proof[0],
"transactions": list(NODE_PENDING_TRANSACTIONS)
}
new_block_index = last_block.index + 1
new_block_timestamp = time.time()
last_block_hash = last_block.hash
# Empty transaction list
NODE_PENDING_TRANSACTIONS = []
# Now create the new block
mined_block = Block(new_block_index, new_block_timestamp, new_block_data, last_block_hash)
BLOCKCHAIN.append(mined_block)
# Let the client know this node mined a block
print(json.dumps({
"index": new_block_index,
"timestamp": str(new_block_timestamp),
"data": new_block_data,
"hash": last_block_hash
}) + "\n")
a.send(BLOCKCHAIN)
requests.get(url = MINER_NODE_URL + '/blocks', params = {'update':MINER_ADDRESS})
def find_new_chains():
# Get the blockchains of every other node
other_chains = []
for node_url in PEER_NODES:
# Get their chains using a GET request
block = requests.get(url = node_url + "/blocks").content
# Convert the JSON object to a Python dictionary
block = json.loads(block)
# Verify other node block is correct
validated = validate_blockchain(block)
if validated:
# Add it to our list
other_chains.append(block)
return other_chains
def consensus(blockchain):
# Get the blocks from other nodes
other_chains = find_new_chains()
# If our chain isn't longest, then we store the longest chain
BLOCKCHAIN = blockchain
longest_chain = BLOCKCHAIN
for chain in other_chains:
if len(longest_chain) < len(chain):
longest_chain = chain
# If the longest chain wasn't ours, then we set our chain to the longest
if longest_chain == BLOCKCHAIN:
# Keep searching for proof
return False
else:
# Give up searching proof, update chain and start over again
BLOCKCHAIN = longest_chain
return BLOCKCHAIN
def validate_blockchain(block):
"""Validate the submitted chain. If hashes are not correct, return false
block(str): json
"""
return True
@node.route('/blocks', methods=['GET'])
def get_blocks():
# Load current blockchain. Only you should update your blockchain
if request.args.get("update") == MINER_ADDRESS:
global BLOCKCHAIN
BLOCKCHAIN = b.recv()
chain_to_send = BLOCKCHAIN
# Converts our blocks into dictionaries so we can send them as json objects later
chain_to_send_json = []
for block in chain_to_send:
block = {
"index": str(block.index),
"timestamp": str(block.timestamp),
"data": str(block.data),
"hash": block.hash
}
chain_to_send_json.append(block)
# Send our chain to whomever requested it
chain_to_send = json.dumps(chain_to_send_json)
return chain_to_send
@node.route('/txion', methods=['GET', 'POST'])
def transaction():
"""Each transaction sent to this node gets validated and submitted.
Then it waits to be added to the blockchain. Transactions only move
coins, they don't create it.
"""
if request.method == 'POST':
# On each new POST request, we extract the transaction data
new_txion = request.get_json()
# Then we add the transaction to our list
if validate_signature(new_txion['from'], new_txion['signature'], new_txion['message']):
NODE_PENDING_TRANSACTIONS.append(new_txion)
# Because the transaction was successfully
# submitted, we log it to our console
print("New transaction")
print("FROM: {0}".format(new_txion['from']))
print("TO: {0}".format(new_txion['to']))
print("AMOUNT: {0}\n".format(new_txion['amount']))
# Then we let the client know it worked out
return "Transaction submission successful\n"
else:
return "Transaction submission failed. Wrong signature\n"
# Send pending transactions to the mining process
elif request.method == 'GET' and request.args.get("update") == MINER_ADDRESS:
pending = json.dumps(NODE_PENDING_TRANSACTIONS)
# Empty transaction list
NODE_PENDING_TRANSACTIONS[:] = []
return pending
def validate_signature(public_key, signature, message):
"""Verifies if the signature is correct. This is used to prove
it's you (and not someone else) trying to do a transaction with your
address. Called when a user tries to submit a new transaction.
"""
public_key = (base64.b64decode(public_key)).hex()
signature = base64.b64decode(signature)
vk = ecdsa.VerifyingKey.from_string(bytes.fromhex(public_key), curve=ecdsa.SECP256k1)
# Try changing into an if/else statement as except is too broad.
try:
return vk.verify(signature, message.encode())
except:
return False
def welcome_msg():
print(""" =========================================\n
SIMPLE COIN v1.0.0 - BLOCKCHAIN SYSTEM\n
=========================================\n\n
You can find more help at: https://github.com/cosme12/SimpleCoin\n
Make sure you are using the latest version or you may end in
a parallel chain.\n\n\n""")
if __name__ == '__main__':
welcome_msg()
# Start mining
a, b = Pipe()
p1 = Process(target=mine, args=(a, BLOCKCHAIN, NODE_PENDING_TRANSACTIONS))
p1.start()
# Start server to receive transactions
p2 = Process(target=node.run(), args=b)
p2.start()
"""This is going to be your wallet. Here you can do several things:
- Generate a new address (public and private key). You are going
to use this address (public key) to send or receive any transactions. You can
have as many addresses as you wish, but keep in mind that if you
lose its credential data, you will not be able to retrieve it.
- Send coins to another address
- Retrieve the entire blockchain and check your balance
If this is your first time using this script don't forget to generate
a new address and edit miner config file with it (only if you are
going to mine).
Timestamp in hashed message. When you send your transaction it will be received
by several nodes. If any node mine a block, your transaction will get added to the
blockchain but other nodes still will have it pending. If any node see that your
transaction with same timestamp was added, they should remove it from the
node_pending_transactions list to avoid it get processed more than 1 time.
"""
import requests
import time
import base64
import ecdsa
def wallet():
response = None
while response not in ["1", "2", "3"]:
response = input("""What do you want to do?
1. Generate new wallet
2. Send coins to another wallet
3. Check transactions\n""")
if response == "1":
# Generate new wallet
print("""=========================================\n
IMPORTANT: save this credentials or you won't be able to recover your wallet\n
=========================================\n""")
generate_ECDSA_keys()
elif response == "2":
addr_from = input("From: introduce your wallet address (public key)\n")
private_key = input("Introduce your private key\n")
addr_to = input("To: introduce destination wallet address\n")
amount = input("Amount: number stating how much do you want to send\n")
print("=========================================\n\n")
print("Is everything correct?\n")
print("From: {0}\nPrivate Key: {1}\nTo: {2}\nAmount: {3}\n".format(addr_from, private_key, addr_to, amount))
response = input("y/n\n")
if response.lower() == "y":
send_transaction(addr_from, private_key, addr_to, amount)
else: # Will always occur when response == 3.
check_transactions()
def send_transaction(addr_from, private_key, addr_to, amount):
"""Sends your transaction to different nodes. Once any of the nodes manage
to mine a block, your transaction will be added to the blockchain. Despite
that, there is a low chance your transaction gets canceled due to other nodes
having a longer chain. So make sure your transaction is deep into the chain
before claiming it as approved!
"""
# For fast debugging REMOVE LATER
# private_key="181f2448fa4636315032e15bb9cbc3053e10ed062ab0b2680a37cd8cb51f53f2"
# amount="3000"
# addr_from="SD5IZAuFixM3PTmkm5ShvLm1tbDNOmVlG7tg6F5r7VHxPNWkNKbzZfa+JdKmfBAIhWs9UKnQLOOL1U+R3WxcsQ=="
# addr_to="SD5IZAuFixM3PTmkm5ShvLm1tbDNOmVlG7tg6F5r7VHxPNWkNKbzZfa+JdKmfBAIhWs9UKnQLOOL1U+R3WxcsQ=="
if len(private_key) == 64:
signature, message = sign_ECDSA_msg(private_key)
url = 'http://localhost:5000/txion'
payload = {"from": addr_from,
"to": addr_to,
"amount": amount,
"signature": signature.decode(),
"message": message}
headers = {"Content-Type": "application/json"}
res = requests.post(url, json=payload, headers=headers)
print(res.text)
else:
print("Wrong address or key length! Verify and try again.")
def check_transactions():
"""Retrieve the entire blockchain. With this you can check your
wallets balance. If the blockchain is to long, it may take some time to load.
"""
res = requests.get('http://localhost:5000/blocks')
print(res.text)
def generate_ECDSA_keys():
"""This function takes care of creating your private and public (your address) keys.
It's very important you don't lose any of them or those wallets will be lost
forever. If someone else get access to your private key, you risk losing your coins.
private_key: str
public_ley: base64 (to make it shorter)
"""
sk = ecdsa.SigningKey.generate(curve=ecdsa.SECP256k1) #this is your sign (private key)
private_key = sk.to_string().hex() #convert your private key to hex
vk = sk.get_verifying_key() #this is your verification key (public key)
public_key = vk.to_string().hex()
#we are going to encode the public key to make it shorter
public_key = base64.b64encode(bytes.fromhex(public_key))
filename = input("Write the name of your new address: ") + ".txt"
with open(filename, "w") as f:
f.write("Private key: {0}\nWallet address / Public key: {1}".format(private_key, public_key.decode()))
print("Your new address and private key are now in the file {0}".format(filename))
def sign_ECDSA_msg(private_key):
"""Sign the message to be sent
private_key: must be hex
return
signature: base64 (to make it shorter)
message: str
"""
# Get timestamp, round it, make it into a string and encode it to bytes
message = str(round(time.time()))
bmessage = message.encode()
sk = ecdsa.SigningKey.from_string(bytes.fromhex(private_key), curve=ecdsa.SECP256k1)
signature = base64.b64encode(sk.sign(bmessage))
return signature, message
if __name__ == '__main__':
print(""" =========================================\n
SIMPLE COIN v1.0.0 - BLOCKCHAIN SYSTEM\n
=========================================\n\n
You can find more help at: https://github.com/cosme12/SimpleCoin\n
Make sure you are using the latest version or you may end in
a parallel chain.\n\n\n""")
wallet()
input("Press ENTER to exit...")
| 0.684686 | 0.731574 |
```
from google.colab import drive
drive.mount("/content/gdrive")
import numpy as np
import pandas as pd
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
import re
import torch
df = pd.read_csv("/content/gdrive/MyDrive/tidydata.csv")
df['label'] = df['recommended'].apply(lambda x: 0 if x== True else 1)
X = df[['review']]
y = df['label']
X_train, X_test, y_train, y_test = train_test_split(df.index.values, df.label.values, test_size=0.2, random_state=40, stratify=df.label.values)
df['type'] = ['tmp']*df.shape[0]
df.loc[X_train, 'type'] = 'train'
df.loc[X_test, 'type'] = 'test'
X_train_list = list(df[df.type=='train'].review.values)
Y_train_list = list(df[df.type=='train'].label.values)
tmp1 = []
tmp2 = []
for i in range(len(X_train_list)):
if X_train_list[i]==X_train_list[i]:
tmp1.append(X_train_list[i])
tmp2.append(Y_train_list[i])
X_train_list = tmp1
Y_train_list = tmp2
X_test_list = list(df[df.type=='test'].review.values)
Y_test_list = list(df[df.type=='test'].label.values)
tmp1 = []
tmp2 = []
for i in range(len(X_test_list)):
if X_test_list[i]==X_test_list[i]:
tmp1.append(X_test_list[i])
tmp2.append(Y_test_list[i])
X_test_list = tmp1
Y_test_list = tmp2
from nltk.corpus import stopwords
import nltk
import re
nltk.download("stopwords")
stopwords_set = set(stopwords.words('english'))
sentences = []
for single_des in df[df.type=='train'].review.values:
for s in re.split("\.", str(single_des)):
if len(s) > 2:
sentence = []
for word in s.split(" "):
if len(word)>1 and word not in stopwords_set:
sentence.append(word.strip().lower())
sentences.append(sentence)
pip install gensim --upgrade
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler,OrdinalEncoder,OneHotEncoder
from sklearn.pipeline import make_pipeline
from sklearn.compose import make_column_transformer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
import gensim
from gensim.models import Word2Vec
import re
VECTOR_SIZE = 300
model = Word2Vec(sentences, vector_size=VECTOR_SIZE, window=15, min_count=5, workers=4)
words_vob = list(model.wv.index_to_key)
w2v_vector_train = np.zeros((len(X_train_list), VECTOR_SIZE))
for i in range(len(X_train_list)):
word_list = X_train_list[i].split(" ")
single_vector = np.zeros(VECTOR_SIZE)
cnt = 0
for word in word_list:
word = word.strip().lower()
if word in words_vob:
single_vector += model.wv[word]
cnt += 1
if cnt > 0:
w2v_vector_train[i] = single_vector / cnt
words_vob = list(model.wv.index_to_key)
w2v_vector_test = np.zeros((len(X_test_list), VECTOR_SIZE))
for i in range(len(X_test_list)):
word_list = X_test_list[i].split(" ")
single_vector = np.zeros(VECTOR_SIZE)
cnt = 0
for word in word_list:
word = word.strip().lower()
if word in words_vob:
single_vector += model.wv[word]
cnt += 1
if cnt > 0:
w2v_vector_test[i] = single_vector / cnt
train_emb = []
test_emb = []
for c in w2v_vector_train:
train_emb.append(list(c))
for c in w2v_vector_test:
test_emb.append(list(c))
import json
with open("/content/gdrive/MyDrive/df_train_w2v_win15.json", "w") as f:
json.dump(train_emb, f)
with open("/content/gdrive/MyDrive/df_test_w2v_win15.json", "w") as f:
json.dump(test_emb, f)
```
|
github_jupyter
|
from google.colab import drive
drive.mount("/content/gdrive")
import numpy as np
import pandas as pd
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
import re
import torch
df = pd.read_csv("/content/gdrive/MyDrive/tidydata.csv")
df['label'] = df['recommended'].apply(lambda x: 0 if x== True else 1)
X = df[['review']]
y = df['label']
X_train, X_test, y_train, y_test = train_test_split(df.index.values, df.label.values, test_size=0.2, random_state=40, stratify=df.label.values)
df['type'] = ['tmp']*df.shape[0]
df.loc[X_train, 'type'] = 'train'
df.loc[X_test, 'type'] = 'test'
X_train_list = list(df[df.type=='train'].review.values)
Y_train_list = list(df[df.type=='train'].label.values)
tmp1 = []
tmp2 = []
for i in range(len(X_train_list)):
if X_train_list[i]==X_train_list[i]:
tmp1.append(X_train_list[i])
tmp2.append(Y_train_list[i])
X_train_list = tmp1
Y_train_list = tmp2
X_test_list = list(df[df.type=='test'].review.values)
Y_test_list = list(df[df.type=='test'].label.values)
tmp1 = []
tmp2 = []
for i in range(len(X_test_list)):
if X_test_list[i]==X_test_list[i]:
tmp1.append(X_test_list[i])
tmp2.append(Y_test_list[i])
X_test_list = tmp1
Y_test_list = tmp2
from nltk.corpus import stopwords
import nltk
import re
nltk.download("stopwords")
stopwords_set = set(stopwords.words('english'))
sentences = []
for single_des in df[df.type=='train'].review.values:
for s in re.split("\.", str(single_des)):
if len(s) > 2:
sentence = []
for word in s.split(" "):
if len(word)>1 and word not in stopwords_set:
sentence.append(word.strip().lower())
sentences.append(sentence)
pip install gensim --upgrade
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler,OrdinalEncoder,OneHotEncoder
from sklearn.pipeline import make_pipeline
from sklearn.compose import make_column_transformer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
import gensim
from gensim.models import Word2Vec
import re
VECTOR_SIZE = 300
model = Word2Vec(sentences, vector_size=VECTOR_SIZE, window=15, min_count=5, workers=4)
words_vob = list(model.wv.index_to_key)
w2v_vector_train = np.zeros((len(X_train_list), VECTOR_SIZE))
for i in range(len(X_train_list)):
word_list = X_train_list[i].split(" ")
single_vector = np.zeros(VECTOR_SIZE)
cnt = 0
for word in word_list:
word = word.strip().lower()
if word in words_vob:
single_vector += model.wv[word]
cnt += 1
if cnt > 0:
w2v_vector_train[i] = single_vector / cnt
words_vob = list(model.wv.index_to_key)
w2v_vector_test = np.zeros((len(X_test_list), VECTOR_SIZE))
for i in range(len(X_test_list)):
word_list = X_test_list[i].split(" ")
single_vector = np.zeros(VECTOR_SIZE)
cnt = 0
for word in word_list:
word = word.strip().lower()
if word in words_vob:
single_vector += model.wv[word]
cnt += 1
if cnt > 0:
w2v_vector_test[i] = single_vector / cnt
train_emb = []
test_emb = []
for c in w2v_vector_train:
train_emb.append(list(c))
for c in w2v_vector_test:
test_emb.append(list(c))
import json
with open("/content/gdrive/MyDrive/df_train_w2v_win15.json", "w") as f:
json.dump(train_emb, f)
with open("/content/gdrive/MyDrive/df_test_w2v_win15.json", "w") as f:
json.dump(test_emb, f)
| 0.101947 | 0.220783 |
```
from os.path import join
import json
import csv
import glob
import os
from pandas import read_csv, DataFrame, Series
from qiime2 import Artifact
from statsmodels.stats.weightstats import DescrStatsW
from scipy.stats import t, sem, ttest_rel
from IPython.display import Image
empo3_dir = '/Users/benkaehler/Data/empo_3/'
columns = ['sample type', 'sample', 'fold']
data = {c:[] for c in columns}
for type_dir in glob.glob(join(empo3_dir, '*')):
if not os.path.exists(join(type_dir, 'results', 'weights.qza')):
continue
type_ = os.path.basename(type_dir)
if type_ in ('sterile-water-blank', 'single-strain', 'mock-community', 'nick', 'plant-surface'):
continue
for fold_dir in glob.glob(join(type_dir, 'tmp', 'fold-*')):
_, fold = fold_dir.rsplit('-', 1)
with open(join(fold_dir, 'sample_test.json')) as fh:
try:
samples = json.load(fh)
except UnicodeDecodeError:
print(join(fold_dir, 'sample_test.json'), 'is corrupted')
data['sample'].extend(samples)
data['sample type'].extend([type_]*len(samples))
data['fold'].extend([fold]*len(samples))
folds = DataFrame(data)
er = read_csv(join(empo3_dir, 'eval_taxa_er.tsv'), sep='\t')
folds = folds.set_index(['sample type', 'sample'])
er = er.join(folds, ['sample type', 'sample'])
er = er[(er['class weights'] == 'bespoke70') |
(er['class weights'] == 'uniform70') |
(er['class weights'] == 'average')]
level7 = er[(er['level'] == 7) |
(er['level'] == 6)]
grouped = level7.groupby(['sample type', 'class weights', 'level', 'fold'])
def weighted_stats(x):
errors = x['errors'].sum()
abundance = x['reads'].sum()
return Series([errors/abundance*100], index=['mean'])
table2 = grouped.apply(weighted_stats)
table2.reset_index(inplace=True)
grouped = table2.groupby(['sample type', 'class weights', 'level'])
def weighted_stats(x):
mu = x['mean'].mean()
se = sem(x['mean'])
lower, upper = t.interval(0.95, 5, mu, se)
return Series([mu, lower, upper], index=['mean', 'lower', 'upper'])
table2 = grouped.apply(weighted_stats)
table2.reset_index(inplace=True)
old_labels = 'average', 'uniform70', 'bespoke70'
new_labels = 'Average', 'Uniform', 'Bespoke'
for old, new in zip(old_labels, new_labels):
table2.loc[table2['class weights'] == old, 'class weights'] = new
old_labels = [
'animal-corpus',
'animal-distal-gut',
'animal-proximal-gut',
'animal-secretion',
'animal-surface',
'plant-corpus',
'plant-rhizosphere',
'plant-surface',
'sediment-non-saline',
'soil-non-saline',
'surface-non-saline',
'water-non-saline',
'sediment-saline',
'surface-saline',
'water-saline'
]
new_labels = [
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
]
for old, new in zip(old_labels, new_labels):
table2.loc[table2['sample type'] == old, 'sample type'] = new
old_labels = 6, 7
new_labels = 'Genus', 'Species'
for old, new in zip(old_labels, new_labels):
table2.loc[table2['level'] == old, 'level'] = new
table2 = table2[['sample type', 'level', 'class weights', 'mean']]
table2 = table2.pivot_table(values='mean', index=['class weights', 'level'], columns=['sample type']).T
table2 = table2.reindex([
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
])
table2.round(1)
species_less_genus = table2['Bespoke']['Species'] - table2['Uniform']['Genus']
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Uniform']['Genus'].mean(), sem(table2['Uniform']['Genus']))
mu = species_less_genus.mean()
se = sem(species_less_genus)
print(mu, se)
ttest_rel(table2['Bespoke']['Species'], table2['Uniform']['Genus'])
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Average']['Species'].mean(), sem(table2['Average']['Species']))
print(table2['Uniform']['Species'].mean(), sem(table2['Uniform']['Species']))
ttest_rel(table2['Bespoke']['Species'], table2['Average']['Species'])
ttest_rel(table2['Average']['Species'], table2['Uniform']['Species'])
species_less_species = table2['Bespoke']['Species'] - table2['Uniform']['Species']
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Uniform']['Species'].mean(), sem(table2['Uniform']['Species']))
mu = species_less_genus.mean()
se = sem(species_less_genus)
print(mu, se)
table2
table2.index
bespoke = level7[level7['class weights'] == 'bespoke70']
bespoke.rename(columns={'F-measure':'bespoke'}, inplace=True)
uniform = level7[level7['class weights'] == 'uniform70']
uniform.rename(columns={'F-measure':'uniform'}, inplace=True)
uniform = uniform.set_index(['sample type', 'sample', 'fold'])
for_diff = bespoke.join(uniform, ['sample type', 'sample', 'fold'], rsuffix='_uniform')
grouped = for_diff.groupby(['sample type', 'class weights', 'fold'])
def weighted_stats(x):
d1 = DescrStatsW(x['bespoke'] - x['uniform'], weights=x['abundance'])
return Series([d1.mean], index=['mean'])
figure1 = grouped.apply(weighted_stats)
figure1.reset_index(inplace=True)
grouped = figure1.groupby(['sample type', 'class weights'])
def weighted_stats(x):
mu = x['mean'].mean()
se = sem(x['mean'])
lower, upper = t.interval(0.95, 5, mu, se)
return Series([mu, lower, upper], index=['mean', 'lower', 'upper'])
figure1 = grouped.apply(weighted_stats)
figure1.reset_index(inplace=True)
old_labels = 'uniform70', 'bespoke70'
new_labels = 'Uniform Genus Level', 'Bespoke Species Level'
for old, new in zip(old_labels, new_labels):
figure1.loc[figure1['class weights'] == old, 'class weights'] = new
old_labels = [
'animal-corpus',
'animal-distal-gut',
'animal-proximal-gut',
'animal-secretion',
'animal-surface',
'plant-corpus',
'plant-rhizosphere',
'plant-surface',
'sediment-non-saline',
'soil-non-saline',
'surface-non-saline',
'water-non-saline',
'sediment-saline',
'surface-saline',
'water-saline'
]
new_labels = [
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
]
for old, new in zip(old_labels, new_labels):
figure1.loc[figure1['sample type'] == old, 'sample type'] = new
figure1['direction'] = ['none']*len(figure1)
figure1.loc[figure1['mean'] >= 0., 'direction'] = 'Bespoke Species Level'
figure1.loc[figure1['mean'] < 0., 'direction'] = 'Uniform Genus Level'
%%R -i figure1
figure1$sample.type = factor(figure1$sample.type, levels=c(
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'))
figure1$class.weights = factor(figure1$class.weights, levels=c(
'Uniform Genus Level', 'Bespoke Species Level'
))
cbPalette <- c("#009E73", "#E69F00")
ggplot(data=figure1, aes(x=sample.type, y=mean, fill=direction)) +
geom_bar(stat="identity", position="identity") +
coord_flip() +
theme_bw() +
theme(legend.position="bottom",
legend.direction="horizontal") +
labs(x='EMPO3 Habitat', y='Unfair F-measure Difference') +
geom_errorbar(aes(ymin=lower, ymax=upper), position=position_dodge()) +
scale_fill_manual(values=cbPalette) +
guides(fill=guide_legend(title="More Accurate", nrow=2, byrow=TRUE))
ggsave(file="figure3.png", width=5, height=5, dpi=300)
Image("figure3.png")
max(figure1['upper'] - figure1['lower'])
```
|
github_jupyter
|
from os.path import join
import json
import csv
import glob
import os
from pandas import read_csv, DataFrame, Series
from qiime2 import Artifact
from statsmodels.stats.weightstats import DescrStatsW
from scipy.stats import t, sem, ttest_rel
from IPython.display import Image
empo3_dir = '/Users/benkaehler/Data/empo_3/'
columns = ['sample type', 'sample', 'fold']
data = {c:[] for c in columns}
for type_dir in glob.glob(join(empo3_dir, '*')):
if not os.path.exists(join(type_dir, 'results', 'weights.qza')):
continue
type_ = os.path.basename(type_dir)
if type_ in ('sterile-water-blank', 'single-strain', 'mock-community', 'nick', 'plant-surface'):
continue
for fold_dir in glob.glob(join(type_dir, 'tmp', 'fold-*')):
_, fold = fold_dir.rsplit('-', 1)
with open(join(fold_dir, 'sample_test.json')) as fh:
try:
samples = json.load(fh)
except UnicodeDecodeError:
print(join(fold_dir, 'sample_test.json'), 'is corrupted')
data['sample'].extend(samples)
data['sample type'].extend([type_]*len(samples))
data['fold'].extend([fold]*len(samples))
folds = DataFrame(data)
er = read_csv(join(empo3_dir, 'eval_taxa_er.tsv'), sep='\t')
folds = folds.set_index(['sample type', 'sample'])
er = er.join(folds, ['sample type', 'sample'])
er = er[(er['class weights'] == 'bespoke70') |
(er['class weights'] == 'uniform70') |
(er['class weights'] == 'average')]
level7 = er[(er['level'] == 7) |
(er['level'] == 6)]
grouped = level7.groupby(['sample type', 'class weights', 'level', 'fold'])
def weighted_stats(x):
errors = x['errors'].sum()
abundance = x['reads'].sum()
return Series([errors/abundance*100], index=['mean'])
table2 = grouped.apply(weighted_stats)
table2.reset_index(inplace=True)
grouped = table2.groupby(['sample type', 'class weights', 'level'])
def weighted_stats(x):
mu = x['mean'].mean()
se = sem(x['mean'])
lower, upper = t.interval(0.95, 5, mu, se)
return Series([mu, lower, upper], index=['mean', 'lower', 'upper'])
table2 = grouped.apply(weighted_stats)
table2.reset_index(inplace=True)
old_labels = 'average', 'uniform70', 'bespoke70'
new_labels = 'Average', 'Uniform', 'Bespoke'
for old, new in zip(old_labels, new_labels):
table2.loc[table2['class weights'] == old, 'class weights'] = new
old_labels = [
'animal-corpus',
'animal-distal-gut',
'animal-proximal-gut',
'animal-secretion',
'animal-surface',
'plant-corpus',
'plant-rhizosphere',
'plant-surface',
'sediment-non-saline',
'soil-non-saline',
'surface-non-saline',
'water-non-saline',
'sediment-saline',
'surface-saline',
'water-saline'
]
new_labels = [
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
]
for old, new in zip(old_labels, new_labels):
table2.loc[table2['sample type'] == old, 'sample type'] = new
old_labels = 6, 7
new_labels = 'Genus', 'Species'
for old, new in zip(old_labels, new_labels):
table2.loc[table2['level'] == old, 'level'] = new
table2 = table2[['sample type', 'level', 'class weights', 'mean']]
table2 = table2.pivot_table(values='mean', index=['class weights', 'level'], columns=['sample type']).T
table2 = table2.reindex([
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
])
table2.round(1)
species_less_genus = table2['Bespoke']['Species'] - table2['Uniform']['Genus']
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Uniform']['Genus'].mean(), sem(table2['Uniform']['Genus']))
mu = species_less_genus.mean()
se = sem(species_less_genus)
print(mu, se)
ttest_rel(table2['Bespoke']['Species'], table2['Uniform']['Genus'])
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Average']['Species'].mean(), sem(table2['Average']['Species']))
print(table2['Uniform']['Species'].mean(), sem(table2['Uniform']['Species']))
ttest_rel(table2['Bespoke']['Species'], table2['Average']['Species'])
ttest_rel(table2['Average']['Species'], table2['Uniform']['Species'])
species_less_species = table2['Bespoke']['Species'] - table2['Uniform']['Species']
print(table2['Bespoke']['Species'].mean(), sem(table2['Bespoke']['Species']))
print(table2['Uniform']['Species'].mean(), sem(table2['Uniform']['Species']))
mu = species_less_genus.mean()
se = sem(species_less_genus)
print(mu, se)
table2
table2.index
bespoke = level7[level7['class weights'] == 'bespoke70']
bespoke.rename(columns={'F-measure':'bespoke'}, inplace=True)
uniform = level7[level7['class weights'] == 'uniform70']
uniform.rename(columns={'F-measure':'uniform'}, inplace=True)
uniform = uniform.set_index(['sample type', 'sample', 'fold'])
for_diff = bespoke.join(uniform, ['sample type', 'sample', 'fold'], rsuffix='_uniform')
grouped = for_diff.groupby(['sample type', 'class weights', 'fold'])
def weighted_stats(x):
d1 = DescrStatsW(x['bespoke'] - x['uniform'], weights=x['abundance'])
return Series([d1.mean], index=['mean'])
figure1 = grouped.apply(weighted_stats)
figure1.reset_index(inplace=True)
grouped = figure1.groupby(['sample type', 'class weights'])
def weighted_stats(x):
mu = x['mean'].mean()
se = sem(x['mean'])
lower, upper = t.interval(0.95, 5, mu, se)
return Series([mu, lower, upper], index=['mean', 'lower', 'upper'])
figure1 = grouped.apply(weighted_stats)
figure1.reset_index(inplace=True)
old_labels = 'uniform70', 'bespoke70'
new_labels = 'Uniform Genus Level', 'Bespoke Species Level'
for old, new in zip(old_labels, new_labels):
figure1.loc[figure1['class weights'] == old, 'class weights'] = new
old_labels = [
'animal-corpus',
'animal-distal-gut',
'animal-proximal-gut',
'animal-secretion',
'animal-surface',
'plant-corpus',
'plant-rhizosphere',
'plant-surface',
'sediment-non-saline',
'soil-non-saline',
'surface-non-saline',
'water-non-saline',
'sediment-saline',
'surface-saline',
'water-saline'
]
new_labels = [
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'
]
for old, new in zip(old_labels, new_labels):
figure1.loc[figure1['sample type'] == old, 'sample type'] = new
figure1['direction'] = ['none']*len(figure1)
figure1.loc[figure1['mean'] >= 0., 'direction'] = 'Bespoke Species Level'
figure1.loc[figure1['mean'] < 0., 'direction'] = 'Uniform Genus Level'
%%R -i figure1
figure1$sample.type = factor(figure1$sample.type, levels=c(
'Animal corpus',
'Animal distal gut',
'Animal proximal gut',
'Animal secretion',
'Animal surface',
'Plant corpus',
'Plant rhizosphere',
'Plant surface',
'Sediment (non-saline)',
'Soil (non-saline)',
'Surface (non-saline)',
'Water (non-saline)',
'Sediment (saline)',
'Surface (saline)',
'Water (saline)'))
figure1$class.weights = factor(figure1$class.weights, levels=c(
'Uniform Genus Level', 'Bespoke Species Level'
))
cbPalette <- c("#009E73", "#E69F00")
ggplot(data=figure1, aes(x=sample.type, y=mean, fill=direction)) +
geom_bar(stat="identity", position="identity") +
coord_flip() +
theme_bw() +
theme(legend.position="bottom",
legend.direction="horizontal") +
labs(x='EMPO3 Habitat', y='Unfair F-measure Difference') +
geom_errorbar(aes(ymin=lower, ymax=upper), position=position_dodge()) +
scale_fill_manual(values=cbPalette) +
guides(fill=guide_legend(title="More Accurate", nrow=2, byrow=TRUE))
ggsave(file="figure3.png", width=5, height=5, dpi=300)
Image("figure3.png")
max(figure1['upper'] - figure1['lower'])
| 0.33231 | 0.238672 |
```
# Create code to answer each of the following questions.
# Hint: You will need multiple target URLs and multiple API requests.
# Dependencies
import requests
import json
# Google API Key
from config import gkey
# 1. What are the geocoordinates (latitude and longitude) of Seattle,
# Washington?
target_city = "Seattle, Washington"
params = {"address": target_city, "key": gkey}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/geocode/json"
print("Drill #1: The Geocoordinates of Seattle, WA")
# Run request
response = requests.get(base_url, params=params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
# Convert to JSON
seattle_geo = response.json()
# Extract lat/lng
lat = seattle_geo["results"][0]["geometry"]["location"]["lat"]
lng = seattle_geo["results"][0]["geometry"]["location"]["lng"]
# Print results
print(f"{target_city}: {lat}, {lng}")
# 2. What are the geocoordinates (latitude and longitude) of The White House?
# update params dict
target_city = "The White House"
params["address"] = target_city
print("Drill #2: The Geocoordinates of the White House")
# Run request
response = requests.get(base_url, params=params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
dc_geo = response.json()
# Extract lat/lng
lat = dc_geo["results"][0]["geometry"]["location"]["lat"]
lng = dc_geo["results"][0]["geometry"]["location"]["lng"]
# Print results
print(f"{target_city}: {lat}, {lng}")
# 3. Find the name and address of a bike store in Seattle, Washington.
# Hint: See https://developers.google.com/places/web-service/supported_types
target_type = "bicycle_store"
seattle_coords = "47.6062095,-122.3320708"
radius = 8000
# rewrite params dict
params = {
"location": seattle_coords,
"types": target_type,
"radius": radius,
"key": gkey
}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
print("Drill #3: A Bike Store in Seattle, WA")
# Run request
response = requests.get(base_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
seattle_bikes = response.json()
# Print the JSON (pretty printed)
# print(json.dumps(seattle_bikes, indent=4, sort_keys=True))
# Print the name and address of the first bike shop to appear
print(seattle_bikes["results"][0]["name"])
print(seattle_bikes["results"][0]["vicinity"])
# 4. Find a balloon store near the White House.
target_search = "Balloon Store"
dc_coords = "38.8976763,-77.0365298"
# redefine params
params = {
"location": dc_coords,
"keyword": target_search,
"radius": radius,
"key": gkey
}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
print("Drill #4: A Balloon Store Near the White House")
# Run request
dc_balloons = requests.get(base_url, params).json()
# Print the JSON (pretty printed)
# print(json.dumps(dc_balloons, indent=4, sort_keys=True))
# Print the name and address of the first balloon shop that appears
print(dc_balloons["results"][0]["name"])
print(dc_balloons["results"][0]["vicinity"])
# 5. Find the nearest dentist to your house.
# Hint: Use Google Maps to find your latitude and Google Places to find
# the dentist. You may also need the rankby property.
# Google geocode to find lat, lng
my_address = "151 Sip Ave"
params = {
"address": my_address,
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/geocode/json"
print("Drill #5: A Dentist Near My House")
my_geo = requests.get(base_url, params).json()
lat = my_geo["results"][0]["geometry"]["location"]["lat"]
lng = my_geo["results"][0]["geometry"]["location"]["lng"]
# Use lat, lng to use places API to find nearest dentist
target_search = "dentist"
params = {
"location": f"{lat},{lng}",
"types": target_search,
"rankby": "distance",
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
# Run request
response = requests.get(base_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
my_dentist = response.json()
# Print the JSON (pretty printed)
# print(json.dumps(my_dentist, indent=4, sort_keys=True))
# Print the name and address of the first dentist that appears
print(my_dentist["results"][0]["name"])
print(my_dentist["results"][0]["vicinity"])
# 6. Bonus: Find the names and addresses of the top five restaurants in your home city.
# Hint: Read about "Text Search Results"
# (https://developers.google.com/places/web-service/search#TextSearchRequests)
my_phrase = "best restaurant in Washington, DC"
target_url = "https://maps.googleapis.com/maps/api/place/textsearch/json"
params = {
"query": my_phrase,
"key": gkey
}
print("Drill #6: Bonus")
response = requests.get(target_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
happy_places = response.json()
# print(json.dumps(happy_places, indent=4, sort_keys=True))
counter = 0
for place in happy_places["results"]:
print(place["name"])
print(place["formatted_address"])
counter += 1
if counter == 5:
break
```
|
github_jupyter
|
# Create code to answer each of the following questions.
# Hint: You will need multiple target URLs and multiple API requests.
# Dependencies
import requests
import json
# Google API Key
from config import gkey
# 1. What are the geocoordinates (latitude and longitude) of Seattle,
# Washington?
target_city = "Seattle, Washington"
params = {"address": target_city, "key": gkey}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/geocode/json"
print("Drill #1: The Geocoordinates of Seattle, WA")
# Run request
response = requests.get(base_url, params=params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
# Convert to JSON
seattle_geo = response.json()
# Extract lat/lng
lat = seattle_geo["results"][0]["geometry"]["location"]["lat"]
lng = seattle_geo["results"][0]["geometry"]["location"]["lng"]
# Print results
print(f"{target_city}: {lat}, {lng}")
# 2. What are the geocoordinates (latitude and longitude) of The White House?
# update params dict
target_city = "The White House"
params["address"] = target_city
print("Drill #2: The Geocoordinates of the White House")
# Run request
response = requests.get(base_url, params=params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
dc_geo = response.json()
# Extract lat/lng
lat = dc_geo["results"][0]["geometry"]["location"]["lat"]
lng = dc_geo["results"][0]["geometry"]["location"]["lng"]
# Print results
print(f"{target_city}: {lat}, {lng}")
# 3. Find the name and address of a bike store in Seattle, Washington.
# Hint: See https://developers.google.com/places/web-service/supported_types
target_type = "bicycle_store"
seattle_coords = "47.6062095,-122.3320708"
radius = 8000
# rewrite params dict
params = {
"location": seattle_coords,
"types": target_type,
"radius": radius,
"key": gkey
}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
print("Drill #3: A Bike Store in Seattle, WA")
# Run request
response = requests.get(base_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
seattle_bikes = response.json()
# Print the JSON (pretty printed)
# print(json.dumps(seattle_bikes, indent=4, sort_keys=True))
# Print the name and address of the first bike shop to appear
print(seattle_bikes["results"][0]["name"])
print(seattle_bikes["results"][0]["vicinity"])
# 4. Find a balloon store near the White House.
target_search = "Balloon Store"
dc_coords = "38.8976763,-77.0365298"
# redefine params
params = {
"location": dc_coords,
"keyword": target_search,
"radius": radius,
"key": gkey
}
# Build URL using the Google Maps API
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
print("Drill #4: A Balloon Store Near the White House")
# Run request
dc_balloons = requests.get(base_url, params).json()
# Print the JSON (pretty printed)
# print(json.dumps(dc_balloons, indent=4, sort_keys=True))
# Print the name and address of the first balloon shop that appears
print(dc_balloons["results"][0]["name"])
print(dc_balloons["results"][0]["vicinity"])
# 5. Find the nearest dentist to your house.
# Hint: Use Google Maps to find your latitude and Google Places to find
# the dentist. You may also need the rankby property.
# Google geocode to find lat, lng
my_address = "151 Sip Ave"
params = {
"address": my_address,
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/geocode/json"
print("Drill #5: A Dentist Near My House")
my_geo = requests.get(base_url, params).json()
lat = my_geo["results"][0]["geometry"]["location"]["lat"]
lng = my_geo["results"][0]["geometry"]["location"]["lng"]
# Use lat, lng to use places API to find nearest dentist
target_search = "dentist"
params = {
"location": f"{lat},{lng}",
"types": target_search,
"rankby": "distance",
"key": gkey
}
base_url = "https://maps.googleapis.com/maps/api/place/nearbysearch/json"
# Run request
response = requests.get(base_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
my_dentist = response.json()
# Print the JSON (pretty printed)
# print(json.dumps(my_dentist, indent=4, sort_keys=True))
# Print the name and address of the first dentist that appears
print(my_dentist["results"][0]["name"])
print(my_dentist["results"][0]["vicinity"])
# 6. Bonus: Find the names and addresses of the top five restaurants in your home city.
# Hint: Read about "Text Search Results"
# (https://developers.google.com/places/web-service/search#TextSearchRequests)
my_phrase = "best restaurant in Washington, DC"
target_url = "https://maps.googleapis.com/maps/api/place/textsearch/json"
params = {
"query": my_phrase,
"key": gkey
}
print("Drill #6: Bonus")
response = requests.get(target_url, params)
# print the response URL, avoid doing for public GitHub repos in order to avoid exposing key
# print(response.url)
happy_places = response.json()
# print(json.dumps(happy_places, indent=4, sort_keys=True))
counter = 0
for place in happy_places["results"]:
print(place["name"])
print(place["formatted_address"])
counter += 1
if counter == 5:
break
| 0.390127 | 0.372248 |
Deep Learning Models -- A collection of various deep learning architectures, models, and tips for TensorFlow and PyTorch in Jupyter Notebooks.
- Author: Sebastian Raschka
- GitHub Repository: https://github.com/rasbt/deeplearning-models
```
%load_ext watermark
%watermark -a 'Sebastian Raschka' -v -p torch
```
# Model Zoo -- Using PyTorch Dataset Loading Utilities for Custom Datasets (Images from Quickdraw)
This notebook provides an example for how to load an image dataset, stored as individual PNG files, using PyTorch's data loading utilities. For a more in-depth discussion, please see the official
- [Data Loading and Processing Tutorial](http://pytorch.org/tutorials/beginner/data_loading_tutorial.html)
- [torch.utils.data](http://pytorch.org/docs/master/data.html) API documentation
In this example, we are using the Quickdraw dataset consisting of handdrawn objects, which is available at https://quickdraw.withgoogle.com.
To execute the following examples, you need to download the ".npy" (bitmap files in NumPy). You don't need to download all of the 345 categories but only a subset you are interested in. The groups/subsets can be individually downloaded from https://console.cloud.google.com/storage/browser/quickdraw_dataset/full/numpy_bitmap
Unfortunately, the Google cloud storage currently does not support selecting and downloading multiple groups at once. Thus, in order to download all groups most coneniently, we need to use their `gsutil` (https://cloud.google.com/storage/docs/gsutil_install) tool. If you want to install that, you can then use
mkdir quickdraw-npy
gsutil -m cp gs://quickdraw_dataset/full/numpy_bitmap/*.npy quickdraw-npy
Note that if you download the whole dataset, this will take up 37 Gb of storage space.
## Imports
```
import pandas as pd
import numpy as np
import os
import torch
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from torchvision import transforms
%matplotlib inline
import matplotlib.pyplot as plt
from PIL import Image
```
## Dataset
After downloading the dataset to a local directory, `quickdraw-npy`, the next step is to select certain groups we are interested in analyzing. Let's say we are interested in the following groups defined in the `label_dict` in the next code cell:
```
label_dict = {
"lollipop": 0,
"binoculars": 1,
"mouse": 2,
"basket": 3,
"penguin": 4,
"washing machine": 5,
"canoe": 6,
"eyeglasses": 7,
"beach": 8,
"screwdriver": 9,
}
```
The dictionary values shall represent class labels that we could use for a classification task, for example.
### Conversion to PNG files
Next we are going to convert the groups we are interested in (specified in the dictionary above) to individual PNG files using a helper function (note that this might take a while):
```
# load utilities from ../helper.py
import sys
sys.path.insert(0, '..')
from helper import quickdraw_npy_to_imagefile
quickdraw_npy_to_imagefile(inpath='quickdraw-npy',
outpath='quickdraw-png_set1',
subset=label_dict.keys())
```
### Preprocessing into train/valid/test subsets and creating a label files
For convenience, let's create a CSV file mapping file names to class labels. First, let's collect the files and labels.
```
paths, labels = [], []
main_dir = 'quickdraw-png_set1/'
for d in os.listdir(main_dir):
subdir = os.path.join(main_dir, d)
if not os.path.isdir(subdir):
continue
for f in os.listdir(subdir):
path = os.path.join(d, f)
paths.append(path)
labels.append(label_dict[d])
print('Num paths:', len(paths))
print('Num labels:', len(labels))
```
Next, we shuffle the dataset and assign 70% of the dataset for training, 10% for validation, and 20% for testing.
```
from mlxtend.preprocessing import shuffle_arrays_unison
paths2, labels2 = shuffle_arrays_unison(arrays=[np.array(paths), np.array(labels)], random_seed=3)
cut1 = int(len(paths)*0.7)
cut2 = int(len(paths)*0.8)
paths_train, labels_train = paths2[:cut1], labels2[:cut1]
paths_valid, labels_valid = paths2[cut1:cut2], labels2[cut1:cut2]
paths_test, labels_test = paths2[cut2:], labels2[cut2:]
```
Finally, let us create a CSV file that maps the file paths to the class labels (here only shown for the training set for simplicity):
```
df = pd.DataFrame(
{'Path': paths_train,
'Label': labels_train,
})
df = df.set_index('Path')
df.to_csv('quickdraw_png_set1_train.csv')
df.head()
```
Finally, let's open one of the images to make sure they look ok:
```
main_dir = 'quickdraw-png_set1/'
img = Image.open(os.path.join(main_dir, df.index[99]))
img = np.asarray(img, dtype=np.uint8)
print(img.shape)
plt.imshow(np.array(img), cmap='binary');
```
## Implementing a Custom Dataset Class
Now, we implement a custom `Dataset` for reading the images. The `__getitem__` method will
1. read a single image from disk based on an `index` (more on batching later)
2. perform a custom image transformation (if a `transform` argument is provided in the `__init__` construtor)
3. return a single image and it's corresponding label
```
class QuickdrawDataset(Dataset):
"""Custom Dataset for loading Quickdraw images"""
def __init__(self, txt_path, img_dir, transform=None):
df = pd.read_csv(txt_path, sep=",", index_col=0)
self.img_dir = img_dir
self.txt_path = txt_path
self.img_names = df.index.values
self.y = df['Label'].values
self.transform = transform
def __getitem__(self, index):
img = Image.open(os.path.join(self.img_dir,
self.img_names[index]))
if self.transform is not None:
img = self.transform(img)
label = self.y[index]
return img, label
def __len__(self):
return self.y.shape[0]
```
Now that we have created our custom Dataset class, let us add some custom transformations via the `transforms` utilities from `torchvision`, we
1. normalize the images (here: dividing by 255)
2. converting the image arrays into PyTorch tensors
Then, we initialize a Dataset instance for the training images using the 'quickdraw_png_set1_train.csv' label file (we omit the test set, but the same concepts apply).
Finally, we initialize a `DataLoader` that allows us to read from the dataset.
```
# Note that transforms.ToTensor()
# already divides pixels by 255. internally
custom_transform = transforms.Compose([#transforms.Lambda(lambda x: x/255.),
transforms.ToTensor()])
train_dataset = QuickdrawDataset(txt_path='quickdraw_png_set1_train.csv',
img_dir='quickdraw-png_set1/',
transform=custom_transform)
train_loader = DataLoader(dataset=train_dataset,
batch_size=128,
shuffle=True,
num_workers=4)
```
That's it, now we can iterate over an epoch using the train_loader as an iterator and use the features and labels from the training dataset for model training:
## Iterating Through the Custom Dataset
```
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
torch.manual_seed(0)
num_epochs = 2
for epoch in range(num_epochs):
for batch_idx, (x, y) in enumerate(train_loader):
print('Epoch:', epoch+1, end='')
print(' | Batch index:', batch_idx, end='')
print(' | Batch size:', y.size()[0])
x = x.to(device)
y = y.to(device)
break
```
Just to make sure that the batches are being loaded correctly, let's print out the dimensions of the last batch:
```
x.shape
```
As we can see, each batch consists of 128 images, just as specified. However, one thing to keep in mind though is that
PyTorch uses a different image layout (which is more efficient when working with CUDA); here, the image axes are "num_images x channels x height x width" (NCHW) instead of "num_images height x width x channels" (NHWC):
To visually check that the images that coming of the data loader are intact, let's swap the axes to NHWC and convert an image from a Torch Tensor to a NumPy array so that we can visualize the image via `imshow`:
```
one_image = x[0].permute(1, 2, 0)
one_image.shape
# note that imshow also works fine with scaled
# images in [0, 1] range.
plt.imshow(one_image.to(torch.device('cpu')).squeeze(), cmap='binary');
%watermark -iv
```
|
github_jupyter
|
%load_ext watermark
%watermark -a 'Sebastian Raschka' -v -p torch
import pandas as pd
import numpy as np
import os
import torch
from torch.utils.data import Dataset
from torch.utils.data import DataLoader
from torchvision import transforms
%matplotlib inline
import matplotlib.pyplot as plt
from PIL import Image
label_dict = {
"lollipop": 0,
"binoculars": 1,
"mouse": 2,
"basket": 3,
"penguin": 4,
"washing machine": 5,
"canoe": 6,
"eyeglasses": 7,
"beach": 8,
"screwdriver": 9,
}
# load utilities from ../helper.py
import sys
sys.path.insert(0, '..')
from helper import quickdraw_npy_to_imagefile
quickdraw_npy_to_imagefile(inpath='quickdraw-npy',
outpath='quickdraw-png_set1',
subset=label_dict.keys())
paths, labels = [], []
main_dir = 'quickdraw-png_set1/'
for d in os.listdir(main_dir):
subdir = os.path.join(main_dir, d)
if not os.path.isdir(subdir):
continue
for f in os.listdir(subdir):
path = os.path.join(d, f)
paths.append(path)
labels.append(label_dict[d])
print('Num paths:', len(paths))
print('Num labels:', len(labels))
from mlxtend.preprocessing import shuffle_arrays_unison
paths2, labels2 = shuffle_arrays_unison(arrays=[np.array(paths), np.array(labels)], random_seed=3)
cut1 = int(len(paths)*0.7)
cut2 = int(len(paths)*0.8)
paths_train, labels_train = paths2[:cut1], labels2[:cut1]
paths_valid, labels_valid = paths2[cut1:cut2], labels2[cut1:cut2]
paths_test, labels_test = paths2[cut2:], labels2[cut2:]
df = pd.DataFrame(
{'Path': paths_train,
'Label': labels_train,
})
df = df.set_index('Path')
df.to_csv('quickdraw_png_set1_train.csv')
df.head()
main_dir = 'quickdraw-png_set1/'
img = Image.open(os.path.join(main_dir, df.index[99]))
img = np.asarray(img, dtype=np.uint8)
print(img.shape)
plt.imshow(np.array(img), cmap='binary');
class QuickdrawDataset(Dataset):
"""Custom Dataset for loading Quickdraw images"""
def __init__(self, txt_path, img_dir, transform=None):
df = pd.read_csv(txt_path, sep=",", index_col=0)
self.img_dir = img_dir
self.txt_path = txt_path
self.img_names = df.index.values
self.y = df['Label'].values
self.transform = transform
def __getitem__(self, index):
img = Image.open(os.path.join(self.img_dir,
self.img_names[index]))
if self.transform is not None:
img = self.transform(img)
label = self.y[index]
return img, label
def __len__(self):
return self.y.shape[0]
# Note that transforms.ToTensor()
# already divides pixels by 255. internally
custom_transform = transforms.Compose([#transforms.Lambda(lambda x: x/255.),
transforms.ToTensor()])
train_dataset = QuickdrawDataset(txt_path='quickdraw_png_set1_train.csv',
img_dir='quickdraw-png_set1/',
transform=custom_transform)
train_loader = DataLoader(dataset=train_dataset,
batch_size=128,
shuffle=True,
num_workers=4)
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
torch.manual_seed(0)
num_epochs = 2
for epoch in range(num_epochs):
for batch_idx, (x, y) in enumerate(train_loader):
print('Epoch:', epoch+1, end='')
print(' | Batch index:', batch_idx, end='')
print(' | Batch size:', y.size()[0])
x = x.to(device)
y = y.to(device)
break
x.shape
one_image = x[0].permute(1, 2, 0)
one_image.shape
# note that imshow also works fine with scaled
# images in [0, 1] range.
plt.imshow(one_image.to(torch.device('cpu')).squeeze(), cmap='binary');
%watermark -iv
| 0.547706 | 0.967778 |
### Get the Personalize boto3 Client
All of the code that we are using in this lab is Python, but any langugage supported by SageMaker could be used. In this initial piece of code we are loading in the library dependencies that we need for the rest of the lab:
- **boto3** - standard Python SDK that wraps the AWS CLI
- **json** - used to manipulate JSON structures used by our API calls
- **numpy** and **pandas** - standard libraries used by Data Scientists everywhere
- **time** - used for some time manipulation calls
```
import boto3
import json
import numpy as np
import pandas as pd
import time
personalize = boto3.client(service_name='personalize')
personalize_runtime = boto3.client(service_name='personalize-runtime')
```
### Specify a Bucket and Data Output Location
For this demo, we'll choose to upload data to Amazon Personalize directly from an S3 bucket. Hence, you need to create a new bucket - please name your bucket before running this Code cell, overriding what is shown in the code cell, and you need to ensure that the bucket name is globally unique; for this lab we recommend using your name or initials, followed by *-jnb-personalize-lab*, as that is likely to be unique.
If the bucket already exists - such as if you execute this code cell a second time - then it will not create a new bucket, and will not make any changes to the exsting bucket. Whilst we're here, we also define the name of the file in S3 that will eventually hold our model training data.
```
bucket = "{your-prefix}-summit-personalize-lab" # replace with the name of your S3 bucket
filename = "DEMO-movie-lens-100k.csv"
s3 = boto3.client('s3')
if boto3.resource('s3').Bucket(bucket).creation_date is None:
s3.create_bucket(ACL = "private", Bucket = bucket)
print("Creating bucket: {}".format(bucket))
```
### Download, Prepare, and Upload Training Data
#### Download and Explore the Dataset
In this step we download the entire Movie Lens data set zip-file and unzip it - it will go in the same location as the physical notebook *.ipynb* file and the *u.item* file that you downloaded earlier. We use the **pandas** library to read in the *u.data* file, which contains all of the movie reviews; this file consists of a movie ID, a user ID, a timestamp and a rating of between 1 and 5, and there are 100,000 unique reviews.
```
!wget -N http://files.grouplens.org/datasets/movielens/ml-100k.zip
!unzip -o ml-100k.zip
data = pd.read_csv('./ml-100k/u.data', sep='\t', names=['USER_ID', 'ITEM_ID', 'RATING', 'TIMESTAMP'])
pd.set_option('display.max_rows', 5)
data
```
#### Prepare and Upload Data
We don't actually need all of the review data. We would like to recommend movies that a user would actually watch based upon what they and others have liked - we don't want to provide a rating between 1 and 5 for every movie in the data set! Hence, we're going to use **pandas** to drop any reviews that are not > 3.6, and once we've done that also drop the review column - we're left with a subset of the original that contain 4- and 5-star ratings, which is basically indicating movies that the user really liked. Of course, we could use all of the data, but this would lead to far greater data import and model training time.
Additionally, the reviews are quite old - they are from August 1997 to April 1998. Some of the Amazon Personalize recipies react differently depending upon the age of the interactions - for instance, the _Similar Items_ recipe has several hyperparameters around how to handle 'decaying' interactions. In order to make this lab easier, and not have to worry about these hyperparameters, we are shifting all of the review timestamps to be from August 2018 up until April 2019.
We then write that out to a file named as per that defined two steps previously, and upload it into our S3 bucket.
This is the minimum amount of data that Amazon Personalize needs to train a model on - you need just 1000 rows of user/item/timestamp interactions, but we still have many 10s of thousands of entries left from our original 100,000 review dataset. This file is known in Amazon Personalize as an **Interactions** data file. Other data files are usable, such as ones that define additional metadata about the movies (such as year and genre) and another that defines demographic data about the user (such as age, gender and location). In this lab we do not need them, but this information is available in the Movie Lens data set files that you have downloaded - you can create your own models based upon those at a later date.
```
data = data[data['RATING'] > 3.6] # keep only movies rated 3.6 and above
data = data[['USER_ID', 'ITEM_ID', 'TIMESTAMP']] # select columns that match the columns in the schema below
data['TIMESTAMP'] = data['TIMESTAMP'] + 660833618 # make reviews end 1st April 2019 rather than 23rd April 1998
data.to_csv(filename, index=False)
boto3.Session().resource('s3').Bucket(bucket).Object(filename).upload_file(filename)
```
### Create Schema
Amazon Personalize uses *Schemas* to tell it how to interpret your data files. This step defines the schema for our Interations file, which consists solely of a `USER_ID`, `ITEM_ID` and `TIMESTAMP`. Once defined we pass it into Personalize for use.
```
schema = {
"type": "record",
"name": "Interactions",
"namespace": "com.amazonaws.personalize.schema",
"fields": [
{
"name": "USER_ID",
"type": "string"
},
{
"name": "ITEM_ID",
"type": "string"
},
{
"name": "TIMESTAMP",
"type": "long"
}
],
"version": "1.0"
}
create_schema_response = personalize.create_schema(
name = "summit-lab-recs-schema",
schema = json.dumps(schema)
)
schema_arn = create_schema_response['schemaArn']
print(json.dumps(create_schema_response, indent=2))
```
### Create and Wait for Dataset Group
Now that we have defined a schema, and we have our Interactions data file, we can import the data into Personalize. But first we have to define a *Dataset Group*, which is essentially a collection of imported data files, trained models and campaigns - each Dataset Group can contain one, and only one, Interaction, Item Metadata and User Demographic file. When you train a model Personalize will use **all** data files present within its Dataset Group.
#### Create Dataset Group
```
create_dataset_group_response = personalize.create_dataset_group(
name = "summit-recs-dataset-group"
)
dataset_group_arn = create_dataset_group_response['datasetGroupArn']
print(json.dumps(create_dataset_group_response, indent=2))
```
#### Wait for Dataset Group to Have ACTIVE Status
A number of Personalize API calls do take time, hence the calls are asynchronous. Before we can continue with the next stage we need to poll the status of the `create_dataset_group()` call from the previous code cell - once the Dataset Group is active then we can continue. **NOTE: this step should not take more than 1-2 minutes to complete**
```
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_dataset_group_response = personalize.describe_dataset_group(
datasetGroupArn = dataset_group_arn
)
status = describe_dataset_group_response["datasetGroup"]["status"]
print("DatasetGroup: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(15)
```
### Create Dataset
We now have to create our dataset for the Interactions file. This step does not actually import any data, rather it creates an internal structure for the data to be imported into.
```
dataset_type = "INTERACTIONS"
create_dataset_response = personalize.create_dataset(
datasetType = dataset_type,
datasetGroupArn = dataset_group_arn,
schemaArn = schema_arn,
name="summit-recs-dataset"
)
dataset_arn = create_dataset_response['datasetArn']
print(json.dumps(create_dataset_response, indent=2))
```
### Prepare, Create, and Wait for Dataset Import Job
#### Attach policy to S3 bucket
Whilst we have created an S3 bucket, and our Interactions data file is sat there waiting to be imported, we have a problem - you may have full access to the bucket via the AWS console or APIs, but the Amazon Personalize service does not. Hence, you have to create an S3 bucket policy that explicitly grants the service access to the `GetObject` and `ListBucket` commands in S3. This code step creates such a policy and attaches it to your S3 bucket.
Note, any Personalize API calls that need to access you S3 bucket need to be done using an IAM role that gives it permission - this step simply allows the service to access the bucket if, and only if, roles with appropriate permissions are used.
```
s3 = boto3.client("s3")
policy = {
"Version": "2012-10-17",
"Id": "PersonalizeS3BucketAccessPolicy",
"Statement": [
{
"Sid": "PersonalizeS3BucketAccessPolicy",
"Effect": "Allow",
"Principal": {
"Service": "personalize.amazonaws.com"
},
"Action": [
"s3:GetObject",
"s3:ListBucket"
],
"Resource": [
"arn:aws:s3:::{}".format(bucket),
"arn:aws:s3:::{}/*".format(bucket)
]
}
]
}
s3.put_bucket_policy(Bucket=bucket, Policy=json.dumps(policy));
```
#### Find Personalize S3 Role ARN
As part of the AWS Event Engine process we have defined an IAM role that gives Personalize the ability to access S3 buckets - as mentioned this is needed as well as the S3 bucket policy. As the Event Engine creates the IAM role via CloudFormation it will always have an essentially random numeric suffix, so we cannot hard-code it into the lab. This code cell looks for any any service role that has the name _PersonalizeS3RoleForLab_ in it and selects it as the ARN that we need.
```
iam = boto3.client("iam")
role_name = "PersonalizeS3RoleForLab"
prefix_name = "/service-role"
role_list = iam.list_roles(PathPrefix=prefix_name)
for role in role_list['Roles']:
if role_name in (role['Arn']):
role_arn = (role['Arn'])
role_arn
```
#### Create Dataset Import Job
This pulls together the information that we have on our Dataset, on our S3 bucket, on our Interactions file and a suitable role for Personalize, and then triggers the actual data import process.
```
create_dataset_import_job_response = personalize.create_dataset_import_job(
jobName = "summit-recs-dataset-import-job",
datasetArn = dataset_arn,
dataSource = {
"dataLocation": "s3://{}/{}".format(bucket, filename)
},
roleArn = role_arn
)
dataset_import_job_arn = create_dataset_import_job_response['datasetImportJobArn']
print(json.dumps(create_dataset_import_job_response, indent=2))
```
#### Wait for Dataset Import Job and Dataset Import Job Run to Have ACTIVE Status
We now poll the status of Interactions file import job, as until it is complete we cannot continue. **Note: this can take anything between 12-25 minutes to complete**
```
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_dataset_import_job_response = personalize.describe_dataset_import_job(
datasetImportJobArn = dataset_import_job_arn
)
dataset_import_job = describe_dataset_import_job_response["datasetImportJob"]
if "latestDatasetImportJobRun" not in dataset_import_job:
status = dataset_import_job["status"]
print("DatasetImportJob: {}".format(status))
else:
status = dataset_import_job["latestDatasetImportJobRun"]["status"]
print("LatestDatasetImportJobRun: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
```
### Select Recipe
There are many different algorithm recipes available within Personalize, this is a list of all supported algorithms at the time of the workshop. We are going to select the standard HRNN recipe, which only needs the Interactions file and not the Item metadata or User demographic files.
```
recipe_list = [
"arn:aws:personalize:::recipe/aws-hrnn",
"arn:aws:personalize:::recipe/aws-hrnn-coldstart",
"arn:aws:personalize:::recipe/aws-hrnn-metadata",
"arn:aws:personalize:::recipe/aws-personalized-ranking",
"arn:aws:personalize:::recipe/aws-popularity-count",
"arn:aws:personalize:::recipe/aws-sims"
]
recipe_arn = recipe_list[0]
print(recipe_arn)
```
### Create and Wait for Solution
With our data imported we can now train our ML solution. This consists of just a single API to Personalize, where we specify the Dataset to use.
#### Create Solution
```
create_solution_response = personalize.create_solution(
name = "summit-lab-recs-solution",
datasetGroupArn = dataset_group_arn,
recipeArn = recipe_arn
)
solution_arn = create_solution_response['solutionArn']
print(json.dumps(create_solution_response, indent=2))
```
#### Create Solution Version
```
create_solution_version_response = personalize.create_solution_version(
solutionArn = solution_arn
)
solution_version_arn = create_solution_version_response['solutionVersionArn']
print(json.dumps(create_solution_version_response, indent=2))
```
#### Wait for Solution to Have ACTIVE Status
We now poll the status of solution creation job, as until it is complete we cannot continue. **Note: this can take anything between 25-50 minutes to complete**
```
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_solution_version_response = personalize.describe_solution_version(
solutionVersionArn = solution_version_arn
)
status = describe_solution_version_response["solutionVersion"]["status"]
print("SolutionVersion: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
```
#### Create Additional Solutions in the Console
Whilst you're waiting for this to complete, jump back into the original Lab Guidebook - there we will walk you through creating two additional solutions in parallel using the same dataset; one for Personalized Rankings and one for Item-to-Item Similarities (or SIMS), both of which can be used in the final application. Once you've begun to create both additional solutions you can come back here and continue.
#### Get Metrics of Solution
Once the soluition is built you can look up the various metrics that Personalize provides - this allows you to see how well a model has been trained. If you are re-training models after the acquisition of new data then these metrics can tell you if the models are training equally as well as before, better than before or worse than before, giving you the information that you need in order to decide whether or not to push a new model into Production. You can also compare results across multiple different algorithm recipes, helping you choose the best performing one for you particular dataset.
You can find details on each of the metrics in our [documentation](https://docs.aws.amazon.com/personalize/latest/dg/working-with-training-metrics.html).
```
get_solution_metrics_response = personalize.get_solution_metrics(
solutionVersionArn = solution_version_arn
)
print(json.dumps(get_solution_metrics_response, indent=2))
```
### Create and Wait for Campaign
A trained model is exactly that - just a model. In order to use it you need to create an API endpoint, and you do this by creating a *Campaign*. A Campaign simply provides the endpoint for a specific version of your model, and as such you are able to host endpoint for multiple versions of your models simultaneously, allowing you to do things like A/B testing of new models.
At the campaign level we specify the the minimum deployed size of the inference engine in terms of transactions per second - whilst this engine can scale up and down dynamically it will never scale below this level, but please note that pricing for Personalize is heavily based around the number of TPS currently deployed.
#### Create campaign
```
create_campaign_response = personalize.create_campaign(
name = "summit-lab-recs-campaign",
solutionVersionArn = solution_version_arn,
minProvisionedTPS = 1
)
campaign_arn = create_campaign_response['campaignArn']
print(json.dumps(create_campaign_response, indent=2))
```
#### Wait for Campaign to Have ACTIVE Status
We now poll the status of Campaign creation job, as until it is complete we cannot continue. **Note: this can take anything between 3-15 minutes to complete**
```
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_campaign_response = personalize.describe_campaign(
campaignArn = campaign_arn
)
status = describe_campaign_response["campaign"]["status"]
print("Campaign: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
```
### Get Recommendations
Finally, we have a deployed Campaign end-point, which hosts a specific version of our trained model - we are now able to make recommendation inference requests against it. However, the Personalize recommendation calls just return itemID values - it returns no context around the title of the movie, Hence, we use our pre-loaded version of the *u.item* file that contains the movie titles - we load in the file via **pandas** library calls and pick out a random userID and movieID from our training date. This info is displayed, along with the name of the movie.
#### Select a User and an Item
```
items = pd.read_csv('./u.item', sep='\t', usecols=[0,1], header=None)
items.columns = ['ITEM_ID', 'TITLE']
user_id, item_id, _ = data.sample().values[0]
item_title = items.loc[items['ITEM_ID'] == item_id].values[0][-1]
print("USER: {}".format(user_id))
print("ITEM: {}".format(item_title))
items
```
#### Call GetRecommendations
The last thing we do is actually make a Personalize recommendations inference call - as you can see from the Code cell this is literally a single line of code with a userID and itemID as input variables (although, strictly speaking, you only need the userID for the datasets that we have).
The inference call returns a list of up to 25 itemIDs from the training set - we take that and look up the corresponding movie titles from the *u.item* file and display them; this is far more useful than just a list of ID values.
```
get_recommendations_response = personalize_runtime.get_recommendations(
campaignArn = campaign_arn,
userId = str(user_id)
)
item_list = get_recommendations_response['itemList']
title_list = [items.loc[items['ITEM_ID'] == np.int(item['itemId'])].values[0][-1] for item in item_list]
print("Recommendations: {}".format(json.dumps(title_list, indent=2)))
```
|
github_jupyter
|
import boto3
import json
import numpy as np
import pandas as pd
import time
personalize = boto3.client(service_name='personalize')
personalize_runtime = boto3.client(service_name='personalize-runtime')
bucket = "{your-prefix}-summit-personalize-lab" # replace with the name of your S3 bucket
filename = "DEMO-movie-lens-100k.csv"
s3 = boto3.client('s3')
if boto3.resource('s3').Bucket(bucket).creation_date is None:
s3.create_bucket(ACL = "private", Bucket = bucket)
print("Creating bucket: {}".format(bucket))
!wget -N http://files.grouplens.org/datasets/movielens/ml-100k.zip
!unzip -o ml-100k.zip
data = pd.read_csv('./ml-100k/u.data', sep='\t', names=['USER_ID', 'ITEM_ID', 'RATING', 'TIMESTAMP'])
pd.set_option('display.max_rows', 5)
data
data = data[data['RATING'] > 3.6] # keep only movies rated 3.6 and above
data = data[['USER_ID', 'ITEM_ID', 'TIMESTAMP']] # select columns that match the columns in the schema below
data['TIMESTAMP'] = data['TIMESTAMP'] + 660833618 # make reviews end 1st April 2019 rather than 23rd April 1998
data.to_csv(filename, index=False)
boto3.Session().resource('s3').Bucket(bucket).Object(filename).upload_file(filename)
schema = {
"type": "record",
"name": "Interactions",
"namespace": "com.amazonaws.personalize.schema",
"fields": [
{
"name": "USER_ID",
"type": "string"
},
{
"name": "ITEM_ID",
"type": "string"
},
{
"name": "TIMESTAMP",
"type": "long"
}
],
"version": "1.0"
}
create_schema_response = personalize.create_schema(
name = "summit-lab-recs-schema",
schema = json.dumps(schema)
)
schema_arn = create_schema_response['schemaArn']
print(json.dumps(create_schema_response, indent=2))
create_dataset_group_response = personalize.create_dataset_group(
name = "summit-recs-dataset-group"
)
dataset_group_arn = create_dataset_group_response['datasetGroupArn']
print(json.dumps(create_dataset_group_response, indent=2))
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_dataset_group_response = personalize.describe_dataset_group(
datasetGroupArn = dataset_group_arn
)
status = describe_dataset_group_response["datasetGroup"]["status"]
print("DatasetGroup: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(15)
dataset_type = "INTERACTIONS"
create_dataset_response = personalize.create_dataset(
datasetType = dataset_type,
datasetGroupArn = dataset_group_arn,
schemaArn = schema_arn,
name="summit-recs-dataset"
)
dataset_arn = create_dataset_response['datasetArn']
print(json.dumps(create_dataset_response, indent=2))
s3 = boto3.client("s3")
policy = {
"Version": "2012-10-17",
"Id": "PersonalizeS3BucketAccessPolicy",
"Statement": [
{
"Sid": "PersonalizeS3BucketAccessPolicy",
"Effect": "Allow",
"Principal": {
"Service": "personalize.amazonaws.com"
},
"Action": [
"s3:GetObject",
"s3:ListBucket"
],
"Resource": [
"arn:aws:s3:::{}".format(bucket),
"arn:aws:s3:::{}/*".format(bucket)
]
}
]
}
s3.put_bucket_policy(Bucket=bucket, Policy=json.dumps(policy));
iam = boto3.client("iam")
role_name = "PersonalizeS3RoleForLab"
prefix_name = "/service-role"
role_list = iam.list_roles(PathPrefix=prefix_name)
for role in role_list['Roles']:
if role_name in (role['Arn']):
role_arn = (role['Arn'])
role_arn
create_dataset_import_job_response = personalize.create_dataset_import_job(
jobName = "summit-recs-dataset-import-job",
datasetArn = dataset_arn,
dataSource = {
"dataLocation": "s3://{}/{}".format(bucket, filename)
},
roleArn = role_arn
)
dataset_import_job_arn = create_dataset_import_job_response['datasetImportJobArn']
print(json.dumps(create_dataset_import_job_response, indent=2))
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_dataset_import_job_response = personalize.describe_dataset_import_job(
datasetImportJobArn = dataset_import_job_arn
)
dataset_import_job = describe_dataset_import_job_response["datasetImportJob"]
if "latestDatasetImportJobRun" not in dataset_import_job:
status = dataset_import_job["status"]
print("DatasetImportJob: {}".format(status))
else:
status = dataset_import_job["latestDatasetImportJobRun"]["status"]
print("LatestDatasetImportJobRun: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
recipe_list = [
"arn:aws:personalize:::recipe/aws-hrnn",
"arn:aws:personalize:::recipe/aws-hrnn-coldstart",
"arn:aws:personalize:::recipe/aws-hrnn-metadata",
"arn:aws:personalize:::recipe/aws-personalized-ranking",
"arn:aws:personalize:::recipe/aws-popularity-count",
"arn:aws:personalize:::recipe/aws-sims"
]
recipe_arn = recipe_list[0]
print(recipe_arn)
create_solution_response = personalize.create_solution(
name = "summit-lab-recs-solution",
datasetGroupArn = dataset_group_arn,
recipeArn = recipe_arn
)
solution_arn = create_solution_response['solutionArn']
print(json.dumps(create_solution_response, indent=2))
create_solution_version_response = personalize.create_solution_version(
solutionArn = solution_arn
)
solution_version_arn = create_solution_version_response['solutionVersionArn']
print(json.dumps(create_solution_version_response, indent=2))
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_solution_version_response = personalize.describe_solution_version(
solutionVersionArn = solution_version_arn
)
status = describe_solution_version_response["solutionVersion"]["status"]
print("SolutionVersion: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
get_solution_metrics_response = personalize.get_solution_metrics(
solutionVersionArn = solution_version_arn
)
print(json.dumps(get_solution_metrics_response, indent=2))
create_campaign_response = personalize.create_campaign(
name = "summit-lab-recs-campaign",
solutionVersionArn = solution_version_arn,
minProvisionedTPS = 1
)
campaign_arn = create_campaign_response['campaignArn']
print(json.dumps(create_campaign_response, indent=2))
status = None
max_time = time.time() + 3*60*60 # 3 hours
while time.time() < max_time:
describe_campaign_response = personalize.describe_campaign(
campaignArn = campaign_arn
)
status = describe_campaign_response["campaign"]["status"]
print("Campaign: {}".format(status))
if status == "ACTIVE" or status == "CREATE FAILED":
break
time.sleep(60)
items = pd.read_csv('./u.item', sep='\t', usecols=[0,1], header=None)
items.columns = ['ITEM_ID', 'TITLE']
user_id, item_id, _ = data.sample().values[0]
item_title = items.loc[items['ITEM_ID'] == item_id].values[0][-1]
print("USER: {}".format(user_id))
print("ITEM: {}".format(item_title))
items
get_recommendations_response = personalize_runtime.get_recommendations(
campaignArn = campaign_arn,
userId = str(user_id)
)
item_list = get_recommendations_response['itemList']
title_list = [items.loc[items['ITEM_ID'] == np.int(item['itemId'])].values[0][-1] for item in item_list]
print("Recommendations: {}".format(json.dumps(title_list, indent=2)))
| 0.182608 | 0.952882 |
# Lab: Titanic Survival Exploration with Decision Trees
## Getting Started
In this lab, you will see how decision trees work by implementing a decision tree in sklearn.
We'll start by loading the dataset and displaying some of its rows.
```
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from IPython.display import display # Allows the use of display() for DataFrames
# Pretty display for notebooks
%matplotlib inline
# Set a random seed
import random
random.seed(42)
# Load the dataset
in_file = 'titanic_data.csv'
full_data = pd.read_csv(in_file)
# Print the first few entries of the RMS Titanic data
display(full_data.head())
```
Recall that these are the various features present for each passenger on the ship:
- **Survived**: Outcome of survival (0 = No; 1 = Yes)
- **Pclass**: Socio-economic class (1 = Upper class; 2 = Middle class; 3 = Lower class)
- **Name**: Name of passenger
- **Sex**: Sex of the passenger
- **Age**: Age of the passenger (Some entries contain `NaN`)
- **SibSp**: Number of siblings and spouses of the passenger aboard
- **Parch**: Number of parents and children of the passenger aboard
- **Ticket**: Ticket number of the passenger
- **Fare**: Fare paid by the passenger
- **Cabin** Cabin number of the passenger (Some entries contain `NaN`)
- **Embarked**: Port of embarkation of the passenger (C = Cherbourg; Q = Queenstown; S = Southampton)
Since we're interested in the outcome of survival for each passenger or crew member, we can remove the **Survived** feature from this dataset and store it as its own separate variable `outcomes`. We will use these outcomes as our prediction targets.
Run the code cell below to remove **Survived** as a feature of the dataset and store it in `outcomes`.
```
# Store the 'Survived' feature in a new variable and remove it from the dataset
outcomes = full_data['Survived']
features_raw = full_data.drop('Survived', axis = 1)
# Show the new dataset with 'Survived' removed
display(features_raw.head())
```
The very same sample of the RMS Titanic data now shows the **Survived** feature removed from the DataFrame. Note that `data` (the passenger data) and `outcomes` (the outcomes of survival) are now *paired*. That means for any passenger `data.loc[i]`, they have the survival outcome `outcomes[i]`.
## Preprocessing the data
Now, let's do some data preprocessing. First, we'll remove the names of the passengers, and then one-hot encode the features.
One-Hot encoding is useful for changing over categorical data into numerical data, with each different option within a category changed into either a 0 or 1 in a separate *new* category as to whether it is that option or not (e.g. Queenstown port or not Queenstown port). Check out [this article](https://hackernoon.com/what-is-one-hot-encoding-why-and-when-do-you-have-to-use-it-e3c6186d008f) before continuing.
**Question:** Why would it be a terrible idea to one-hot encode the data without removing the names?
```
# Removing the names
features_no_names = features_raw.drop(['Name'], axis=1)
# One-hot encoding
features = pd.get_dummies(features_no_names)
```
And now we'll fill in any blanks with zeroes.
```
features = features.fillna(0.0)
display(features.head())
```
## (TODO) Training the model
Now we're ready to train a model in sklearn. First, let's split the data into training and testing sets. Then we'll train the model on the training set.
```
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(features, outcomes, test_size=0.2, random_state=42)
# Import the classifier from sklearn
from sklearn.tree import DecisionTreeClassifier
# TODO: Define the classifier, and fit it to the data
model = DecisionTreeClassifier()
model.fit(X_train,y_train)
```
## Testing the model
Now, let's see how our model does, let's calculate the accuracy over both the training and the testing set.
```
# Making predictions
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
# Calculate the accuracy
from sklearn.metrics import accuracy_score
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred)
print('The training accuracy is', train_accuracy)
print('The test accuracy is', test_accuracy)
```
# Exercise: Improving the model
Ok, high training accuracy and a lower testing accuracy. We may be overfitting a bit.
So now it's your turn to shine! Train a new model, and try to specify some parameters in order to improve the testing accuracy, such as:
- `max_depth`
- `min_samples_leaf`
- `min_samples_split`
You can use your intuition, trial and error, or even better, feel free to use Grid Search!
**Challenge:** Try to get to 85% accuracy on the testing set. If you'd like a hint, take a look at the solutions notebook next.
```
# TODO: Train the model
model2= DecisionTreeClassifier(max_depth=6,min_samples_leaf=5,min_samples_split=11)
model2.fit(X_train,y_train)
# TODO: Make predictions
y_train_pred2= model2.predict(X_train)
y_test_pred2 = model2.predict(X_test)
# TODO: Calculate the accuracy
from sklearn.metrics import accuracy_score
train_accuracy2 = accuracy_score(y_train, y_train_pred2)
test_accuracy2 = accuracy_score(y_test, y_test_pred2)
print('The training accuracy is', train_accuracy2)
print('The test accuracy is', test_accuracy2)
```
|
github_jupyter
|
# Import libraries necessary for this project
import numpy as np
import pandas as pd
from IPython.display import display # Allows the use of display() for DataFrames
# Pretty display for notebooks
%matplotlib inline
# Set a random seed
import random
random.seed(42)
# Load the dataset
in_file = 'titanic_data.csv'
full_data = pd.read_csv(in_file)
# Print the first few entries of the RMS Titanic data
display(full_data.head())
# Store the 'Survived' feature in a new variable and remove it from the dataset
outcomes = full_data['Survived']
features_raw = full_data.drop('Survived', axis = 1)
# Show the new dataset with 'Survived' removed
display(features_raw.head())
# Removing the names
features_no_names = features_raw.drop(['Name'], axis=1)
# One-hot encoding
features = pd.get_dummies(features_no_names)
features = features.fillna(0.0)
display(features.head())
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(features, outcomes, test_size=0.2, random_state=42)
# Import the classifier from sklearn
from sklearn.tree import DecisionTreeClassifier
# TODO: Define the classifier, and fit it to the data
model = DecisionTreeClassifier()
model.fit(X_train,y_train)
# Making predictions
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
# Calculate the accuracy
from sklearn.metrics import accuracy_score
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred)
print('The training accuracy is', train_accuracy)
print('The test accuracy is', test_accuracy)
# TODO: Train the model
model2= DecisionTreeClassifier(max_depth=6,min_samples_leaf=5,min_samples_split=11)
model2.fit(X_train,y_train)
# TODO: Make predictions
y_train_pred2= model2.predict(X_train)
y_test_pred2 = model2.predict(X_test)
# TODO: Calculate the accuracy
from sklearn.metrics import accuracy_score
train_accuracy2 = accuracy_score(y_train, y_train_pred2)
test_accuracy2 = accuracy_score(y_test, y_test_pred2)
print('The training accuracy is', train_accuracy2)
print('The test accuracy is', test_accuracy2)
| 0.583559 | 0.988245 |
# Time-series outlier detection using Prophet on weather data
## Method
The Prophet outlier detector uses the [Prophet](https://facebook.github.io/prophet/) time series forecasting package explained in [this excellent paper](https://peerj.com/preprints/3190/). The underlying Prophet model is a decomposable univariate time series model combining trend, seasonality and holiday effects. The model forecast also includes an uncertainty interval around the estimated trend component using the [MAP estimate](https://en.wikipedia.org/wiki/Maximum_a_posteriori_estimation) of the extrapolated model. Alternatively, full Bayesian inference can be done at the expense of increased compute. The upper and lower values of the uncertainty interval can then be used as outlier thresholds for each point in time. First, the distance from the observed value to the nearest uncertainty boundary (upper or lower) is computed. If the observation is within the boundaries, the outlier score equals the negative distance. As a result, the outlier score is the lowest when the observation equals the model prediction. If the observation is outside of the boundaries, the score equals the distance measure and the observation is flagged as an outlier. One of the main drawbacks of the method however is that you need to refit the model as new data comes in. This is undesirable for applications with high throughput and real-time detection.
To use this detector, first install Prophet by running `pip install alibi-detect[prophet]`.
## Dataset
The example uses a weather time series dataset recorded by the [Max-Planck-Institute for Biogeochemistry](https://www.bgc-jena.mpg.de/wetter/). The dataset contains 14 different features such as air temperature, atmospheric pressure, and humidity. These were collected every 10 minutes, beginning in 2003. Like the [TensorFlow time-series tutorial](https://www.tensorflow.org/tutorials/structured_data/time_series), we only use data collected between 2009 and 2016.
```
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import tensorflow as tf
from alibi_detect.od import OutlierProphet
from alibi_detect.utils.fetching import fetch_detector
from alibi_detect.utils.saving import save_detector, load_detector
```
## Load dataset
```
zip_path = tf.keras.utils.get_file(
origin='https://storage.googleapis.com/tensorflow/tf-keras-datasets/jena_climate_2009_2016.csv.zip',
fname='jena_climate_2009_2016.csv.zip',
extract=True
)
csv_path, _ = os.path.splitext(zip_path)
df = pd.read_csv(csv_path)
df['Date Time'] = pd.to_datetime(df['Date Time'], format='%d.%m.%Y %H:%M:%S')
print(df.shape)
df.head()
```
Select subset to test Prophet model on:
```
n_prophet = 10000
```
Prophet model expects a DataFrame with 2 columns: one named ```ds``` with the timestamps and one named ```y``` with the time series to be evaluated. We will just look at the temperature data:
```
d = {'ds': df['Date Time'][:n_prophet], 'y': df['T (degC)'][:n_prophet]}
df_T = pd.DataFrame(data=d)
print(df_T.shape)
df_T.head()
plt.plot(df_T['ds'], df_T['y'])
plt.title('T (in °C) over time')
plt.xlabel('Time')
plt.ylabel('T (in °C)')
plt.show()
```
## Load or define outlier detector
The pretrained outlier and adversarial detectors used in the example notebooks can be found [here](https://console.cloud.google.com/storage/browser/seldon-models/alibi-detect). You can use the built-in ```fetch_detector``` function which saves the pre-trained models in a local directory ```filepath``` and loads the detector. Alternatively, you can train a detector from scratch:
```
load_outlier_detector = False
filepath = 'my_path' # change to directory where model is downloaded
if load_outlier_detector: # load pretrained outlier detector
detector_type = 'outlier'
dataset = 'weather'
detector_name = 'OutlierProphet'
od = fetch_detector(filepath, detector_type, dataset, detector_name)
filepath = os.path.join(filepath, detector_name)
else: # initialize, fit and save outlier detector
od = OutlierProphet(threshold=.9)
od.fit(df_T)
save_detector(od, filepath)
```
Please check out the [documentation](https://docs.seldon.io/projects/alibi-detect/en/latest/methods/prophet.html) as well as the original [Prophet documentation](https://facebook.github.io/prophet/) on how to customize the Prophet-based outlier detector and add seasonalities, holidays, opt for a saturating logistic growth model or apply parameter regularization.
## Predict outliers on test data
Define the test data. It is important that the timestamps of the test data follow the training data. We check this below by comparing the first few rows of the test DataFrame with the last few of the training DataFrame:
```
n_periods = 1000
d = {'ds': df['Date Time'][n_prophet:n_prophet+n_periods],
'y': df['T (degC)'][n_prophet:n_prophet+n_periods]}
df_T_test = pd.DataFrame(data=d)
df_T_test.head()
df_T.tail()
```
Predict outliers on test data:
```
od_preds = od.predict(
df_T_test,
return_instance_score=True,
return_forecast=True
)
```
## Visualize results
We can first visualize our predictions with Prophet's built in plotting functionality. This also allows us to include historical predictions:
```
future = od.model.make_future_dataframe(periods=n_periods, freq='10T', include_history=True)
forecast = od.model.predict(future)
fig = od.model.plot(forecast)
```
We can also plot the breakdown of the different components in the forecast. Since we did not do full Bayesian inference with `mcmc_samples`, the uncertaintly intervals of the forecast are determined by the [MAP estimate](https://en.wikipedia.org/wiki/Maximum_a_posteriori_estimation) of the extrapolated trend.
```
fig = od.model.plot_components(forecast)
```
It is clear that the further we predict in the future, the wider the uncertainty intervals which determine the outlier threshold.
Let's overlay the actual data with the upper and lower outlier thresholds predictions and check where we predicted outliers:
```
forecast['y'] = df['T (degC)'][:n_prophet+n_periods]
pd.plotting.register_matplotlib_converters() # needed to plot timestamps
forecast[-n_periods:].plot(x='ds', y=['y', 'yhat', 'yhat_upper', 'yhat_lower'])
plt.title('Predicted T (in °C) over time')
plt.xlabel('Time')
plt.ylabel('T (in °C)')
plt.show()
```
Outlier scores and predictions:
```
od_preds['data']['forecast']['threshold'] = np.zeros(n_periods)
od_preds['data']['forecast'][-n_periods:].plot(x='ds', y=['score', 'threshold'])
plt.title('Outlier score over time')
plt.xlabel('Time')
plt.ylabel('Outlier score')
plt.show()
```
The outlier scores naturally trend down as uncertainty increases when we predict further in the future.
Let's look at some individual outliers:
```
df_fcst = od_preds['data']['forecast']
df_outlier = df_fcst.loc[df_fcst['score'] > 0]
print('Number of outliers: {}'.format(df_outlier.shape[0]))
df_outlier[['ds', 'yhat', 'yhat_lower', 'yhat_upper', 'y']]
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import tensorflow as tf
from alibi_detect.od import OutlierProphet
from alibi_detect.utils.fetching import fetch_detector
from alibi_detect.utils.saving import save_detector, load_detector
zip_path = tf.keras.utils.get_file(
origin='https://storage.googleapis.com/tensorflow/tf-keras-datasets/jena_climate_2009_2016.csv.zip',
fname='jena_climate_2009_2016.csv.zip',
extract=True
)
csv_path, _ = os.path.splitext(zip_path)
df = pd.read_csv(csv_path)
df['Date Time'] = pd.to_datetime(df['Date Time'], format='%d.%m.%Y %H:%M:%S')
print(df.shape)
df.head()
n_prophet = 10000
d = {'ds': df['Date Time'][:n_prophet], 'y': df['T (degC)'][:n_prophet]}
df_T = pd.DataFrame(data=d)
print(df_T.shape)
df_T.head()
plt.plot(df_T['ds'], df_T['y'])
plt.title('T (in °C) over time')
plt.xlabel('Time')
plt.ylabel('T (in °C)')
plt.show()
load_outlier_detector = False
filepath = 'my_path' # change to directory where model is downloaded
if load_outlier_detector: # load pretrained outlier detector
detector_type = 'outlier'
dataset = 'weather'
detector_name = 'OutlierProphet'
od = fetch_detector(filepath, detector_type, dataset, detector_name)
filepath = os.path.join(filepath, detector_name)
else: # initialize, fit and save outlier detector
od = OutlierProphet(threshold=.9)
od.fit(df_T)
save_detector(od, filepath)
n_periods = 1000
d = {'ds': df['Date Time'][n_prophet:n_prophet+n_periods],
'y': df['T (degC)'][n_prophet:n_prophet+n_periods]}
df_T_test = pd.DataFrame(data=d)
df_T_test.head()
df_T.tail()
od_preds = od.predict(
df_T_test,
return_instance_score=True,
return_forecast=True
)
future = od.model.make_future_dataframe(periods=n_periods, freq='10T', include_history=True)
forecast = od.model.predict(future)
fig = od.model.plot(forecast)
fig = od.model.plot_components(forecast)
forecast['y'] = df['T (degC)'][:n_prophet+n_periods]
pd.plotting.register_matplotlib_converters() # needed to plot timestamps
forecast[-n_periods:].plot(x='ds', y=['y', 'yhat', 'yhat_upper', 'yhat_lower'])
plt.title('Predicted T (in °C) over time')
plt.xlabel('Time')
plt.ylabel('T (in °C)')
plt.show()
od_preds['data']['forecast']['threshold'] = np.zeros(n_periods)
od_preds['data']['forecast'][-n_periods:].plot(x='ds', y=['score', 'threshold'])
plt.title('Outlier score over time')
plt.xlabel('Time')
plt.ylabel('Outlier score')
plt.show()
df_fcst = od_preds['data']['forecast']
df_outlier = df_fcst.loc[df_fcst['score'] > 0]
print('Number of outliers: {}'.format(df_outlier.shape[0]))
df_outlier[['ds', 'yhat', 'yhat_lower', 'yhat_upper', 'y']]
| 0.634996 | 0.992123 |
# Test cases requiring or benefiting from the context of a notebook
If the notebook runs successfully from start to finish, the test is successful!
TODO(all): Add additional tests and/or tests with particular assertions, as we encounter Python package version incompatibilities not currently detected by these tests.
In general, only add test cases here that require the context of a notebook. This is because this notebook, as currently written, will abort at the **first** failure. Compare this to a proper test suite where all cases are run, giving much more information about the full extent of any problems encountered.
# Package versions
```
!pip3 freeze
```
# Test cases requiring the context of a notebook
## Test package installations
NOTE: installing packages via `%pip` installs them into the running kernel - no kernel restart needed.
```
import sys
sys.path
!env | grep PIP
```
### Install a package we do not anticipate already being installed on the base image
```
output = !pip3 show rich
print(output) # Should show not yet installed.
assert(0 == output.count('Name: rich'))
%pip install rich==10.16.1
output = !pip3 show rich
print(output) # Should show that it is now installed!
assert(1 == output.count('Name: rich'))
```
### Install a package **from source** that we do not anticipate already being installed on the base image
```
output = !pip3 show docstring-parser
print(output) # Should show not yet installed.
assert(0 == output.count('Name: docstring-parser'))
%pip install docstring-parser==0.13
output = !pip3 show docstring-parser
print(output) # Should show that it is now installed!
assert(1 == output.count('Name: docstring-parser'))
```
## Test ipython widgets
```
import ipywidgets as widgets
widgets.IntSlider()
## Test python images come with base google image
from markdown import *
markdown
import readline
readline.parse_and_bind('tab: complete')
# Teste scipy
from scipy import misc
import matplotlib.pyplot as plt
face = misc.face()
plt.imshow(face)
plt.show()
```
## Test BigQuery magic
* As of release [google-cloud-bigquery 1.26.0 (2020-07-20)](https://github.com/googleapis/python-bigquery/blob/master/CHANGELOG.md#1260-2020-07-20) the BigQuery Python client uses the BigQuery Storage client by default.
* This currently causes an error on Terra Cloud Runtimes `the user does not have 'bigquery.readsessions.create' permission for '<Terra billing project id>'`.
* To work around this, we do two things:
1. remove the dependency `google-cloud-bigquery-storage` from the `terra-jupyter-python` image
1. use flag `--use_rest_api` with `%%bigquery`
```
%load_ext google.cloud.bigquery
%%bigquery --use_rest_api
SELECT country_name, alpha_2_code
FROM `bigquery-public-data.utility_us.country_code_iso`
WHERE alpha_2_code LIKE 'A%'
LIMIT 5
```
## Test pandas profiling
```
import numpy as np
import pandas as pd
from pandas_profiling import ProfileReport
df = pd.DataFrame(
np.random.rand(100, 5),
columns=['a', 'b', 'c', 'd', 'e']
)
profile = ProfileReport(df, title='Pandas Profiling Report')
profile
```
# Test cases benefiting from the context of a notebook
Strictly speaking, these could be moved into the Python test cases, if desired.
## Test matplotlib
```
from __future__ import print_function, division
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
x = np.random.randn(10000) # example data, random normal distribution
num_bins = 50
n, bins, patches = plt.hist(x, num_bins, facecolor="green", alpha=0.5)
plt.xlabel(r"Description of $x$ coordinate (units)")
plt.ylabel(r"Description of $y$ coordinate (units)")
plt.title(r"Histogram title here (remove for papers)")
plt.show();
```
## Test plotnine
```
from plotnine import ggplot, geom_point, aes, stat_smooth, facet_wrap
from plotnine.data import mtcars
(ggplot(mtcars, aes('wt', 'mpg', color='factor(gear)'))
+ geom_point()
+ stat_smooth(method='lm')
+ facet_wrap('~gear'))
```
## Test ggplot
```
from ggplot import *
ggplot
```
## Test source control tool availability
```
%%bash
which git
which ssh-agent
which ssh-add
```
## Test gcloud tools
```
%%bash
gcloud version
%%bash
gcloud auth activate-service-account --key-file $GOOGLE_APPLICATION_CREDENTIALS
%%bash
gsutil ls gs://gcp-public-data--gnomad
%%bash
bq --project_id bigquery-public-data ls gnomAD
```
## Test Google Libraries
```
from google.cloud import datastore
datastore_client = datastore.Client()
from google.api_core import operations_v1
from google.cloud import storage
%%bash
# test composite object, requires python crcmod to be installed
gsutil cp gs://terra-docker-image-documentation/test-composite.cram .
from google.cloud import bigquery
```
## Test TensorFlow
### See https://www.tensorflow.org/tutorials/quickstart/beginner
The oneAPI Deep Neural Network Library (oneDNN) optimizations are also now available in the official x86-64 TensorFlow after v2.5. Users can enable those CPU optimizations by setting the the environment variable TF_ENABLE_ONEDNN_OPTS=1 for the official x86-64 TensorFlow after v2.5.
We enable oneDNN Verbose log to validate the existenance of oneDNN optimization via DNNL_VERBSOE environemnt variable, and also set CUDA_VISIBLE_DEVCIES to -1 to run the workload on CPU.
```
import os
os.environ['TF_ENABLE_ONEDNN_OPTS'] = '1'
os.environ['DNNL_VERBOSE'] = '1'
os.environ['CUDA_VISIBLE_DEVICES']="-1"
```
Set up TensorFlow
```
import tensorflow as tf
import keras
print("TensorFlow version:", tf.__version__)
print("Keras version:", keras.__version__)
print("TensorFlow executing_eagerly:", tf.executing_eagerly())
```
Load a dataset
```
# Load a dataset
mnist = tf.keras.datasets.mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
```
Build a machine learning model
```
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(input_shape=(28, 28)),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10)
])
predictions = model(x_train[:1]).numpy()
predictions
```
Define a loss function for training
```
tf.nn.softmax(predictions).numpy()
loss_fn = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
loss_fn(y_train[:1], predictions).numpy()
model.compile(optimizer='adam',
loss=loss_fn,
metrics=['accuracy'])
```
Train and evaluate your model
```
model.fit(x_train, y_train, epochs=5)
model.evaluate(x_test, y_test, verbose=2)
probability_model = tf.keras.Sequential([
model,
tf.keras.layers.Softmax()
])
probability_model(x_test[:5])
```
### Validate usage of oneDNN optimization
>Please redirect standard outputs and errors to stdout.txt and stderr.txt files by starting jupyter notebook with below command.
```
jupyter notebook --ip=0.0.0.0 > stdout.txt 2>stderr.txt
```
First, we could check whether we have dnnl verose log or not while we test TensorFlow in the previous section.
```
!cat /tmp/stdout.txt | grep dnnl
```
Second, we could further analyze what oneDNN primitives are used while we run the workload by using a profile_utils.py script.
```
!wget https://raw.githubusercontent.com/oneapi-src/oneAPI-samples/master/Libraries/oneDNN/tutorials/profiling/profile_utils.py
```
```
import warnings
warnings.filterwarnings('ignore')
```
Finally, users should be able to see that inner_product oneDNN primitive is used for the workload.
```
run profile_utils.py /tmp/stdout.txt
```
### Validate Intel® Extension for Scikit-Learn Optimization
Let's test that [Intel® Extension for Scikit-Learn](https://www.intel.com/content/www/us/en/developer/articles/guide/intel-extension-for-scikit-learn-getting-started.html) is installed properly by successfully running the following cell. If it is on, a warning should print saying that Intel® Extension for Scikit-Learn has been enabled.
```
from sklearnex import patch_sklearn, unpatch_sklearn
patch_sklearn()
from sklearn import datasets, svm, metrics, preprocessing
from sklearn.model_selection import train_test_split
#should print warning
```
Now let's just run some regular scikit-learn code with the optimization enabled to ensure everything is working properly with Intel® Extension for Scikit-Learn enabled.
```
digits = datasets.load_digits()
X,Y = digits.data, digits.target
# Split dataset into 80% train images and 20% test images
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, shuffle=True)
# normalize the input values by scaling each feature by its maximum absolute value
X_train = preprocessing.maxabs_scale(X_train)
X_test = preprocessing.maxabs_scale(X_test)
# Create a classifier: a support vector classifier
model = svm.SVC(gamma=0.001, C=100)
# Learn the digits on the train subset
model.fit(X_train, Y_train)
# Now predicting the digit for test images using the trained model
Y_pred = model.predict(X_test)
result = model.score(X_test, Y_test)
print(f"Model accuracy on test data: {result}")
```
Then turn off the Intel Extension for Scikit-Learn optimizations through the unpatch method.
```
unpatch_sklearn() # then unpatch optimizations
```
### Validate Intel XGBoost Optimizations
Starting with [XGBoost](https://xgboost.readthedocs.io/en/latest/index.html) 0.81 version onward, Intel has been directly upstreaming many training optimizations to provide superior performance on Intel® CPUs.
Starting with XGBoost 1.3 version onward, Intel has been upstreaming inference optimizations to provide even more performance on Intel® CPUs.
This well-known, machine-learning package for gradient-boosted decision trees now includes seamless, drop-in acceleration for Intel® architectures to significantly speed up model training and improve accuracy for better predictions.
We will use the following cell to validate the XGBoost version to determine if these optimizations are enabled.
```
import xgboost as xgb
import os
major_version = int(xgb.__version__.split(".")[0])
minor_version = int(xgb.__version__.split(".")[1])
print("XGBoost version installed: ", xgb.__version__)
if major_version >= 0:
if major_version == 0:
if minor_version >= 81:
print("Intel optimizations for XGBoost training enabled in hist method!")
if major_version >= 1:
print("Intel optimizations for XGBoost training enabled in hist method!")
if minor_version >= 3:
print("Intel optimizations for XGBoost inference enabled!")
else:
print("Intel XGBoost optimizations are disabled! Please install or update XGBoost version to 0.81+ to enable Intel's optimizations that have been upstreamed to the main XGBoost project.")
```
|
github_jupyter
|
!pip3 freeze
import sys
sys.path
!env | grep PIP
output = !pip3 show rich
print(output) # Should show not yet installed.
assert(0 == output.count('Name: rich'))
%pip install rich==10.16.1
output = !pip3 show rich
print(output) # Should show that it is now installed!
assert(1 == output.count('Name: rich'))
output = !pip3 show docstring-parser
print(output) # Should show not yet installed.
assert(0 == output.count('Name: docstring-parser'))
%pip install docstring-parser==0.13
output = !pip3 show docstring-parser
print(output) # Should show that it is now installed!
assert(1 == output.count('Name: docstring-parser'))
import ipywidgets as widgets
widgets.IntSlider()
## Test python images come with base google image
from markdown import *
markdown
import readline
readline.parse_and_bind('tab: complete')
# Teste scipy
from scipy import misc
import matplotlib.pyplot as plt
face = misc.face()
plt.imshow(face)
plt.show()
%load_ext google.cloud.bigquery
%%bigquery --use_rest_api
SELECT country_name, alpha_2_code
FROM `bigquery-public-data.utility_us.country_code_iso`
WHERE alpha_2_code LIKE 'A%'
LIMIT 5
import numpy as np
import pandas as pd
from pandas_profiling import ProfileReport
df = pd.DataFrame(
np.random.rand(100, 5),
columns=['a', 'b', 'c', 'd', 'e']
)
profile = ProfileReport(df, title='Pandas Profiling Report')
profile
from __future__ import print_function, division
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
%matplotlib inline
x = np.random.randn(10000) # example data, random normal distribution
num_bins = 50
n, bins, patches = plt.hist(x, num_bins, facecolor="green", alpha=0.5)
plt.xlabel(r"Description of $x$ coordinate (units)")
plt.ylabel(r"Description of $y$ coordinate (units)")
plt.title(r"Histogram title here (remove for papers)")
plt.show();
from plotnine import ggplot, geom_point, aes, stat_smooth, facet_wrap
from plotnine.data import mtcars
(ggplot(mtcars, aes('wt', 'mpg', color='factor(gear)'))
+ geom_point()
+ stat_smooth(method='lm')
+ facet_wrap('~gear'))
from ggplot import *
ggplot
%%bash
which git
which ssh-agent
which ssh-add
%%bash
gcloud version
%%bash
gcloud auth activate-service-account --key-file $GOOGLE_APPLICATION_CREDENTIALS
%%bash
gsutil ls gs://gcp-public-data--gnomad
%%bash
bq --project_id bigquery-public-data ls gnomAD
from google.cloud import datastore
datastore_client = datastore.Client()
from google.api_core import operations_v1
from google.cloud import storage
%%bash
# test composite object, requires python crcmod to be installed
gsutil cp gs://terra-docker-image-documentation/test-composite.cram .
from google.cloud import bigquery
import os
os.environ['TF_ENABLE_ONEDNN_OPTS'] = '1'
os.environ['DNNL_VERBOSE'] = '1'
os.environ['CUDA_VISIBLE_DEVICES']="-1"
import tensorflow as tf
import keras
print("TensorFlow version:", tf.__version__)
print("Keras version:", keras.__version__)
print("TensorFlow executing_eagerly:", tf.executing_eagerly())
# Load a dataset
mnist = tf.keras.datasets.mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x_train, x_test = x_train / 255.0, x_test / 255.0
model = tf.keras.models.Sequential([
tf.keras.layers.Flatten(input_shape=(28, 28)),
tf.keras.layers.Dense(128, activation='relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(10)
])
predictions = model(x_train[:1]).numpy()
predictions
tf.nn.softmax(predictions).numpy()
loss_fn = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
loss_fn(y_train[:1], predictions).numpy()
model.compile(optimizer='adam',
loss=loss_fn,
metrics=['accuracy'])
model.fit(x_train, y_train, epochs=5)
model.evaluate(x_test, y_test, verbose=2)
probability_model = tf.keras.Sequential([
model,
tf.keras.layers.Softmax()
])
probability_model(x_test[:5])
jupyter notebook --ip=0.0.0.0 > stdout.txt 2>stderr.txt
!cat /tmp/stdout.txt | grep dnnl
!wget https://raw.githubusercontent.com/oneapi-src/oneAPI-samples/master/Libraries/oneDNN/tutorials/profiling/profile_utils.py
import warnings
warnings.filterwarnings('ignore')
run profile_utils.py /tmp/stdout.txt
from sklearnex import patch_sklearn, unpatch_sklearn
patch_sklearn()
from sklearn import datasets, svm, metrics, preprocessing
from sklearn.model_selection import train_test_split
#should print warning
digits = datasets.load_digits()
X,Y = digits.data, digits.target
# Split dataset into 80% train images and 20% test images
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, shuffle=True)
# normalize the input values by scaling each feature by its maximum absolute value
X_train = preprocessing.maxabs_scale(X_train)
X_test = preprocessing.maxabs_scale(X_test)
# Create a classifier: a support vector classifier
model = svm.SVC(gamma=0.001, C=100)
# Learn the digits on the train subset
model.fit(X_train, Y_train)
# Now predicting the digit for test images using the trained model
Y_pred = model.predict(X_test)
result = model.score(X_test, Y_test)
print(f"Model accuracy on test data: {result}")
unpatch_sklearn() # then unpatch optimizations
import xgboost as xgb
import os
major_version = int(xgb.__version__.split(".")[0])
minor_version = int(xgb.__version__.split(".")[1])
print("XGBoost version installed: ", xgb.__version__)
if major_version >= 0:
if major_version == 0:
if minor_version >= 81:
print("Intel optimizations for XGBoost training enabled in hist method!")
if major_version >= 1:
print("Intel optimizations for XGBoost training enabled in hist method!")
if minor_version >= 3:
print("Intel optimizations for XGBoost inference enabled!")
else:
print("Intel XGBoost optimizations are disabled! Please install or update XGBoost version to 0.81+ to enable Intel's optimizations that have been upstreamed to the main XGBoost project.")
| 0.569853 | 0.954478 |
```
import warnings
warnings.filterwarnings("ignore")
from IPython.core.display import display, HTML
display(HTML("<style>.container {width: 80% !important; }</style>"))
import sys
import time
import scanpy as sc
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
from matplotlib import colors
myColors = ['#e6194b', '#3cb44b', '#ffe119', '#4363d8', '#f58231',
'#911eb4', '#46f0f0', '#f032e6', '#bcf60c', '#fabebe',
'#008080', '#e6beff', '#9a6324', '#fffac8', '#800000',
'#aaffc3', '#808000', '#ffd8b1', '#000075', '#808080',
'#307D7E', '#000000', "#DDEFFF", "#000035", "#7B4F4B",
"#A1C299", "#300018", "#C2FF99", "#0AA6D8", "#013349",
"#00846F", "#8CD0FF", "#3B9700", "#04F757", "#C8A1A1",
"#1E6E00", "#DFFB71", "#868E7E", "#513A01", "#CCAA35"]
colors2 = plt.cm.Reds(np.linspace(0, 1, 128))
colors3 = plt.cm.Greys_r(np.linspace(0.7,0.8,20))
colorsComb = np.vstack([colors3, colors2])
mymap = colors.LinearSegmentedColormap.from_list('my_colormap', colorsComb)
sys.path.append("../../../functions")
from SMaSH_functions import SMaSH_functions
sf = SMaSH_functions()
sys.path.append("/home/ubuntu/Taneda/Functions")
from scRNA_functions import scRNA_functions
fc = scRNA_functions()
```
# Loading annData object
```
obj = sc.read_h5ad('../../../../../External_datasets/mouse_brain_all_cells_20200625_with_annotations.h5ad')
obj.X = obj.X.toarray()
obj = obj[obj.obs["Cell broad annotation"]=="Inh"]
print("%d genes across %s cells"%(obj.n_vars, obj.n_obs))
new_sub_annotation = []
for c in obj.obs["Cell sub annotation"].tolist():
if c in ['Inh_1', 'Inh_2', 'Inh_3', 'Inh_4', 'Inh_5', 'Inh_6']:
new_sub_annotation.append('Inh')
elif c in ['Inh_Meis2_1', 'Inh_Meis2_2', 'Inh_Meis2_3', 'Inh_Meis2_4']:
new_sub_annotation.append('Inh_Meis2')
else:
new_sub_annotation.append(c)
obj.obs["Cell sub annotation"] = new_sub_annotation
obj.obs["Cell sub annotation"] = obj.obs["Cell sub annotation"].astype("category")
```
#### Data preparation
```
sf.data_preparation(obj)
```
#### Removing general genes
```
s1 = time.time()
obj = sf.remove_general_genes(obj, species='mouse')
```
#### Removing house-keeping genes
http://www.housekeeping.unicamp.br/?homePageGlobal
```
obj = sf.remove_housekeepingenes(obj, path="../../../data/house_keeping_genes_Mouse_cortex.txt")
```
#### Removing genes expressed in less than 30% within groups
```
obj = sf.remove_features_pct(obj, group_by="Cell sub annotation", pct=0.3)
```
#### Removing genes expressed in more than 50% in a given group where genes are expressed for more 75% within a given group
```
obj = sf.remove_features_pct_2groups(obj, group_by="Cell sub annotation", pct1=0.75, pct2=0.5)
```
#### Revert PCA
```
obj = sf.scale_filter_features(obj, n_components=None, filter_expression=True)
obj.var.set_index(obj.var["SYMBOL"], inplace=True, drop=False)
obj.var.index.name = None
```
#### ensemble_learning
```
s2 = time.time()
clf = sf.ensemble_learning(obj, group_by="Cell sub annotation", classifier="BalancedRandomForest", balance=True, verbose=True)
```
#### gini_importance
```
selectedGenes, selectedGenes_dict = sf.gini_importance(obj, clf, group_by="Cell sub annotation", verbose=True, restrict_top=("local", 20))
e2 = time.time()
```
#### Classifiers
```
sf.run_classifiers(obj, group_by="Cell sub annotation", genes=selectedGenes, classifier="KNN", balance=True, title="BRF-KNN")
```
#### Sorting genes per cluster
```
axs, selectedGenes_top_dict = sf.sort_and_plot(obj, selectedGenes, group_by="Cell sub annotation", group_by2=None, top=3, figsize=(8,14))
e1 = time.time()
axs.savefig("Figures/BRF_top5_perGroup.pdf")
```
# Elapsed time
```
print("%d genes across %s cells"%(obj.n_vars, obj.n_obs))
print('Elapsed time (s): ', e1-s1)
print('Elapsed time (s): ', e2-s2)
```
|
github_jupyter
|
import warnings
warnings.filterwarnings("ignore")
from IPython.core.display import display, HTML
display(HTML("<style>.container {width: 80% !important; }</style>"))
import sys
import time
import scanpy as sc
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
from matplotlib import colors
myColors = ['#e6194b', '#3cb44b', '#ffe119', '#4363d8', '#f58231',
'#911eb4', '#46f0f0', '#f032e6', '#bcf60c', '#fabebe',
'#008080', '#e6beff', '#9a6324', '#fffac8', '#800000',
'#aaffc3', '#808000', '#ffd8b1', '#000075', '#808080',
'#307D7E', '#000000', "#DDEFFF", "#000035", "#7B4F4B",
"#A1C299", "#300018", "#C2FF99", "#0AA6D8", "#013349",
"#00846F", "#8CD0FF", "#3B9700", "#04F757", "#C8A1A1",
"#1E6E00", "#DFFB71", "#868E7E", "#513A01", "#CCAA35"]
colors2 = plt.cm.Reds(np.linspace(0, 1, 128))
colors3 = plt.cm.Greys_r(np.linspace(0.7,0.8,20))
colorsComb = np.vstack([colors3, colors2])
mymap = colors.LinearSegmentedColormap.from_list('my_colormap', colorsComb)
sys.path.append("../../../functions")
from SMaSH_functions import SMaSH_functions
sf = SMaSH_functions()
sys.path.append("/home/ubuntu/Taneda/Functions")
from scRNA_functions import scRNA_functions
fc = scRNA_functions()
obj = sc.read_h5ad('../../../../../External_datasets/mouse_brain_all_cells_20200625_with_annotations.h5ad')
obj.X = obj.X.toarray()
obj = obj[obj.obs["Cell broad annotation"]=="Inh"]
print("%d genes across %s cells"%(obj.n_vars, obj.n_obs))
new_sub_annotation = []
for c in obj.obs["Cell sub annotation"].tolist():
if c in ['Inh_1', 'Inh_2', 'Inh_3', 'Inh_4', 'Inh_5', 'Inh_6']:
new_sub_annotation.append('Inh')
elif c in ['Inh_Meis2_1', 'Inh_Meis2_2', 'Inh_Meis2_3', 'Inh_Meis2_4']:
new_sub_annotation.append('Inh_Meis2')
else:
new_sub_annotation.append(c)
obj.obs["Cell sub annotation"] = new_sub_annotation
obj.obs["Cell sub annotation"] = obj.obs["Cell sub annotation"].astype("category")
sf.data_preparation(obj)
s1 = time.time()
obj = sf.remove_general_genes(obj, species='mouse')
obj = sf.remove_housekeepingenes(obj, path="../../../data/house_keeping_genes_Mouse_cortex.txt")
obj = sf.remove_features_pct(obj, group_by="Cell sub annotation", pct=0.3)
obj = sf.remove_features_pct_2groups(obj, group_by="Cell sub annotation", pct1=0.75, pct2=0.5)
obj = sf.scale_filter_features(obj, n_components=None, filter_expression=True)
obj.var.set_index(obj.var["SYMBOL"], inplace=True, drop=False)
obj.var.index.name = None
s2 = time.time()
clf = sf.ensemble_learning(obj, group_by="Cell sub annotation", classifier="BalancedRandomForest", balance=True, verbose=True)
selectedGenes, selectedGenes_dict = sf.gini_importance(obj, clf, group_by="Cell sub annotation", verbose=True, restrict_top=("local", 20))
e2 = time.time()
sf.run_classifiers(obj, group_by="Cell sub annotation", genes=selectedGenes, classifier="KNN", balance=True, title="BRF-KNN")
axs, selectedGenes_top_dict = sf.sort_and_plot(obj, selectedGenes, group_by="Cell sub annotation", group_by2=None, top=3, figsize=(8,14))
e1 = time.time()
axs.savefig("Figures/BRF_top5_perGroup.pdf")
print("%d genes across %s cells"%(obj.n_vars, obj.n_obs))
print('Elapsed time (s): ', e1-s1)
print('Elapsed time (s): ', e2-s2)
| 0.248261 | 0.63672 |
<a href="https://colab.research.google.com/github/wooohoooo/Thompson-Sampling-Examples/blob/master/Handmade_Bandit_solvers_Non_Linear_Context_Normal_Distribution.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# Intro
This notebook continues the series about multi-armed bandit (MAB) solutions.
This time we will look at MAB with nonlinear contexts. To understand this, you should at least have an understanding of non-contextual Thompson Sampling, as presented e.g. here [link other notebook], even better if you already looked at the linear counterpart of this notebook: [link to other other notebook]
Like in the previous posts, we will first look at the problem at hand and how we can emulate a an environment that can benefit from a nonlinear contextual MAB solver.
In the previous entries, [we looked into the problem of how to find out which arm is better, such as whether a red button or a blue button is likely to generate more traffic to our website, or whether Neo is more likely to choose the red or the blue pill so we can hide the tracer inside (I rewatched the move in the mean time and am now more up to date)] as well as the same question but this time we know the users/Neo's age.
# MOVE TO LINEAR CASE!
This time, we will add context to the decision, this could be the users age, the users gender, the users education, any other information about the user or any combination thereof. In general, we want to use all the information available to us to make sure our decision is well informed; but in this case we will go for a context of two variables to make sure we can visualize it nicely.
# END
We're now going to look at nonlinear interaction between features.
In our case, the context we have is the users age, the decision we have to make is whether to show them a dog or not. Other than last time, our reward isn't a click tho; instead it's the amount a user will give when asked for a donation to the corona relief fund.

(I couldn't find an image of a dog as bandit, so this pawrate will have to be enought)
As we learned before, in order to use Thompson Sampling, we need distributions over the expected outcome of each arm; while we were using simple beta binomials for the non-contextual case, and a sigmoid distributin in the linear contextual case. Now we will be using Normal Distributions.
We will be doing something very similar to last time, where we used a number (the age) to predict the likelihood or a user clicking a red vs a green button; but this time, we will do it 'the other way': Where last time we converted a line into a click (so either 0 or 1), we will this time convert a line into two lines (age mapped to two responses), thus creating a non-linear reward function.
For this simple notebook, we will walk through a contextual example using a normal distribution derived via Bayesian Linear Regression. More complex solutions exist, notably one introduced in this paper: .
Again, this is aimed at application designers rather than students of th mathematicl descriptions behind Thompson Sampling. A primer and possibly a deeper explanation on these topics migh follow in the future.
But let's take a step back and look at the problem we're trying to solve.
# getting ready
```
#maths
import numpy as np
import scipy.stats as stats
from sklearn.utils import shuffle
import random
from fastprogress.fastprogress import master_bar, progress_bar
random.seed(4122)
np.random.seed(4122)
#pretty things
matplotlib_style = 'seaborn-notebook' #@param ['fivethirtyeight', 'bmh', 'ggplot', 'seaborn', 'default', 'Solarize_Light2', 'classic', 'dark_background', 'seaborn-colorblind', 'seaborn-notebook']
import matplotlib.pyplot as plt; plt.style.use(matplotlib_style)
import matplotlib.axes as axes;
from matplotlib.patches import Ellipse
import matplotlib.cm as cm
%matplotlib inline
import seaborn as sns; sns.set_context('notebook')
from IPython.core.pylabtools import figsize
#@markdown This sets the resolution of the plot outputs (`retina` is the highest resolution)
notebook_screen_res = 'retina' #@param ['retina', 'png', 'jpeg', 'svg', 'pdf']
%config InlineBackend.figure_format = notebook_screen_res
width = "16" #@param [16,8,25]
height = "4.5" #@param [4.5, 9,16,8,25]
width = int(width)
height = float(height)
plt.rcParams['figure.figsize'] = width,height
#set some parameters
# number of features
num_features = "1" #@param [1, 2,3,50]
num_features = int(num_features)
# data size
num_data = "1000" #@param[25,100, 1000,10000]
num_data = int(num_data)
# slopes of reward depending for arm 1 and arm 2
theta1 = 0
theta2 = 10
theta = [int(theta1), int(theta2)]
# noise in the data
noise = "1" #@param [0.0, 0.1, 0.5, 1,5,10,100]
noise = float(noise)
# scale of uncertainty
v = "50"#@param [1,10, 50, 100, 1000]
v = int(v)
# number of hidden units to use
num_hidden_units = "2" #@param[1,2,10,100]
num_hidden_units = int(num_hidden_units)
```
# What is a contextual multi armed bandit situation?
The contextual multi armed bandit situation is similar to the one we encountered previously. We have a deicsion to make, and depending on that decision, we will get a reward. Not much new, so far. We want to know whether a dog on our website makes the user pay more for our product, think Humble Bundle.
As we did last time, we have to define a 'user' that will react to the arms we play. And as last time, we do this via a function called 'customer_reaction'.
However, this time we have have some information about the user, formally known as context. In this case, it is the users age (1). We think that depending on how old the users are, they will react DIFFERENTLY to whether a dog is present or not. Here it is 'the older you are, the more you spending increases when you see a dog', meaning showing people a dog is the best decision above a certain age; below that age we're more likely to receive more money if we DO NOT SHOW A DOG.
This we'll model as a linear slope with
$y = x \cdot m + t + \epsilon $
where
$y$ is the target, i.e. how much the user spends
$x$ is the context, i.e. the users age
$m$ is a linear factor decided by whether a dog is present or not
$t$ is the onset; in our case that is 0
$\epsilon$ is gaussian noise, since we're still in a probabilistic setting
(1) the users age is between -10 and 10 in this case. If you have a problem with this... redo this notebook with more sensible numbers. Have fun!
```
X_ = np.array([1,2,3])
arm = np.array([1,2])
num_arms = 2
num_contexts = 1
#num_data = 1000
#create X
X = np.zeros((num_data,num_contexts + num_arms))
for i in range(num_contexts):
X[:,i] = np.linspace(-1,1,num_data).T
for i in range(num_arms):
X[:,i+num_contexts] = np.random.binomial(1,.6,num_data)
X[:5,:]
X = shuffle(X)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
#X = scaler.fit_transform(X)
X[:,2] = abs(1-X[:,1])
X
def customer_reaction(context, choice, theta = theta, noise=noise):
features = np.append(context,choice)
# decide on which slope to use depending on whether a dog is present or not
m = theta[0] if features[1] else theta[1]
intercept = 0 if features[1] else 0
# $y = x \cdot m + t + \epsilon $
y = features[0] * m + intercept + np.random.normal(0,noise)
return y
customer_reaction([9.0],[1,0])
customer_reaction([9.0],[0,1])
y = []
for i in range(num_data):
context = X[i][0]
choice = X[i][1]
outcome = customer_reaction(context,choice)
y.append(outcome)
y = np.array(y).flatten()
index = X[:,1] == True
plt.scatter(X[:,0][index],y[index],label='with dog',c='red')
plt.scatter(X[:,0][~index],y[~index],label='without dog',c='black')
plt.legend()
```
# Now that we have defined the Problem, let's solve it!
Remember the simple API we defined last time. We will reuse it.
We will implement a way of choosing an arm to play according to some policy, and we will implement a way of updating the policy with the observations. Note that this time, we add context to both methods, and we explicity pass a model that will help us solve the problem.
```
class AbstractContextualSolver(object):
def __init__(self, model, num_arms):
self.model = model
self.num_arms = num_arms
def choose_arm(self,context):
"""choose an arm to play according to internal policy"""
raise NotImplementedError
def update(self, arm, context, reward):
""" update internal policy to reflect changed knowledge"""
raise NotImplementedError
```
But wait!
We learned last time that in order to do thompson sampling, we need a distribution to draw from per arm we want to play; this distribution reflects the likelihood for the reward we're likely to receive.
We also know that distributions usually don't use context; $ \mathcal{N}(\mu,\sigma)$ relies only on the mean and the standard deviation of the data we have observed - but in our case, $\mu$ changes rapidly with depending on whether a dog is present or not.
What do we do?
We become Bayesian, that's what we do. Honestly, that should always be the first thing you do when facing a problem.

More specifically, we will use a technique known as bayesian linear regression, which you can read about more about [here](https://en.wikipedia.org/wiki/Bayesian_linear_regression) and [here](https://towardsdatascience.com/introduction-to-bayesian-linear-regression-e66e60791ea7). Let's meet again after you checked those out.
Hi! So now that you read that, you know how Bayesian Linear Regression can be used to derive a normal distribution depending on a linear model that we can train to give us everything we need.
Let's do that.
```
import torch
from torch.autograd import Variable
class SimpleModel(torch.nn.Module):
def __init__(self,num_input, p=0.05, decay=0.001, non_linearity=torch.nn.LeakyReLU):
super(SimpleModel, self).__init__()
self.dropout_p = p
self.decay = decay
self.f = torch.nn.Sequential(
torch.nn.Linear(num_input,num_hidden_units),
torch.nn.ReLU(),
# torch.nn.Dropout(p=self.dropout_p),
# torch.nn.Linear(20,20),
# torch.nn.ReLU(),
# torch.nn.Dropout(p=self.dropout_p),
# torch.nn.Linear(20, 10),
# non_linearity(),
# torch.nn.Dropout(p=self.dropout_p),
torch.nn.Linear(num_hidden_units,1)
)
def forward(self, X):
X = Variable(torch.Tensor(X), requires_grad=False)
return self.f(X)
class VanillaEnsemble(object):
def __init__(self, p=0.00, decay=0.001, non_linearity=torch.nn.LeakyReLU, n_models=10, model_list=None):
self.models = [SimpleModel(p, decay, non_linearity) for model in range(n_models)]
self.optimizers = [torch.optim.SGD(
model.parameters(),
weight_decay=decay,
lr=0.01) for model in self.models]
self.criterion = torch.nn.MSELoss()
self.dropout_p = p
self.decay = decay
def fit_model(self, model, optimizer, X_obs,y_obs):
y = Variable(torch.Tensor(y_obs[:, np.newaxis]), requires_grad=False)
y_pred = model(X_obs[:, np.newaxis])
optimizer.zero_grad()
loss = self.criterion(y_pred, y)
loss.backward()
optimizer.step()
return loss
def observe(self,X,y):
return self.fit_ensemble(X,y)
def fit_ensemble(self, X_obs, y_obs):
losslist = []
for model, optimizer in zip(self.models, self.optimizers):
losslist.append(self.fit_model(model, optimizer, X_obs,y_obs))
return losslist
def predict_distribution(self, X, ):
outputs = np.hstack([model(X[:, np.newaxis]).data.numpy() for model in self.models])
y_mean = outputs.mean(axis=1)
y_std = outputs.std(axis=1)
return {'means':y_mean,
'stds':y_std}
class OnlineBootstrapEnsemble(object):
def __init__(self, p=0.00, decay=0.001, non_linearity=torch.nn.LeakyReLU, n_models=10, model_list=None):
self.models = [SimpleModel(p, decay, non_linearity) for model in range(n_models)]
self.optimizers = [torch.optim.SGD(
model.parameters(),
weight_decay=decay,
lr=0.01) for model in self.models]
self.criterion = torch.nn.MSELoss()
self.dropout_p = p
self.decay = decay
def fit_model(self, model, optimizer, X_obs,y_obs):
y = Variable(torch.Tensor(y_obs[:, np.newaxis]), requires_grad=False)
y_pred = model(X_obs[:, np.newaxis])
optimizer.zero_grad()
loss = self.criterion(y_pred, y)
loss.backward()
optimizer.step()
return loss
def observe(self,X,y):
return self.fit_ensemble(X,y)
def fit_ensemble(self, X_obs, y_obs):
losslist = []
for model, optimizer in zip(self.models, self.optimizers):
if random.random() > 0.5:
losslist.append(self.fit_model(model, optimizer, X_obs,y_obs))
self.fit_model(model, optimizer, X_obs,y_obs)
return losslist
def predict_distribution(self, X, ):
outputs = np.hstack([model(X[:, np.newaxis]).data.numpy() for model in self.models])
y_mean = outputs.mean(axis=1)
y_std = outputs.std(axis=1)
return {'means':y_mean,
'stds':y_std}
ensemble = VanillaEnsemble(3)#OnlineBootstrapEnsemble(3)
means = ensemble.predict_distribution(X[~index])['means']
stds = ensemble.predict_distribution(X[~index])['stds']
plt.scatter(X[:,0][~index],means)
X_train = X[~index]
y_train = np.atleast_2d(y[~index])
np.atleast_2d(y_train[:,:5])
ensemble.fit_ensemble(X_train,y_train.T)
means = ensemble.predict_distribution(X[~index])['means']
stds = ensemble.predict_distribution(X[~index])['stds']
plt.scatter(X[:,0][~index],means)
plt.figure()
plt.scatter(X[:,0][~index],stds)
class GaussianThompsonSampler(AbstractContextualSolver):
"""solves the contextual MAB problem with normal reward distribution
by utilising a model to predict the distribution
for each context and each arm individually"""
def choose_arm(self,context):
reward_list = []
dist = stats.norm
for i in range(self.num_arms):
arm = np.array([0,0])
arm[i] = 1
X = np.atleast_2d(np.append(arm, context))
model_distribution = self.model.predict_distribution(X)
reward_sample = dist.rvs(model_distribution['means'],model_distribution['stds'])
reward_list += [reward_sample]
arms = np.array([0,0])
arms[np.argmax(reward_list)] = 1
return arms
def update(self,arm,context,reward):
X = np.atleast_2d(np.append(arm, context))
reward = np.atleast_2d(reward).T
self.model.observe(X, reward)
for num_hidden_units in [1,2,5,10,100]:
breg = VanillaEnsemble(3)
gts = GaussianThompsonSampler(breg,num_arms = 2)
context = X[:,0]
y_hat_list = []
best_arms = []
for i in progress_bar(range(num_data)):
arm = gts.choose_arm(context[i])
y_hat = customer_reaction(context[i],arm)
y_hat_list += [y_hat]
gts.update(arm, context[i], y_hat)
#best_arms.append(best_arm_chosen(context[i], y_hat))
plt.figure()
plt.scatter(context,np.array(y_hat_list))
best_arms[:5], best_arms[-5:]
```
|
github_jupyter
|
#maths
import numpy as np
import scipy.stats as stats
from sklearn.utils import shuffle
import random
from fastprogress.fastprogress import master_bar, progress_bar
random.seed(4122)
np.random.seed(4122)
#pretty things
matplotlib_style = 'seaborn-notebook' #@param ['fivethirtyeight', 'bmh', 'ggplot', 'seaborn', 'default', 'Solarize_Light2', 'classic', 'dark_background', 'seaborn-colorblind', 'seaborn-notebook']
import matplotlib.pyplot as plt; plt.style.use(matplotlib_style)
import matplotlib.axes as axes;
from matplotlib.patches import Ellipse
import matplotlib.cm as cm
%matplotlib inline
import seaborn as sns; sns.set_context('notebook')
from IPython.core.pylabtools import figsize
#@markdown This sets the resolution of the plot outputs (`retina` is the highest resolution)
notebook_screen_res = 'retina' #@param ['retina', 'png', 'jpeg', 'svg', 'pdf']
%config InlineBackend.figure_format = notebook_screen_res
width = "16" #@param [16,8,25]
height = "4.5" #@param [4.5, 9,16,8,25]
width = int(width)
height = float(height)
plt.rcParams['figure.figsize'] = width,height
#set some parameters
# number of features
num_features = "1" #@param [1, 2,3,50]
num_features = int(num_features)
# data size
num_data = "1000" #@param[25,100, 1000,10000]
num_data = int(num_data)
# slopes of reward depending for arm 1 and arm 2
theta1 = 0
theta2 = 10
theta = [int(theta1), int(theta2)]
# noise in the data
noise = "1" #@param [0.0, 0.1, 0.5, 1,5,10,100]
noise = float(noise)
# scale of uncertainty
v = "50"#@param [1,10, 50, 100, 1000]
v = int(v)
# number of hidden units to use
num_hidden_units = "2" #@param[1,2,10,100]
num_hidden_units = int(num_hidden_units)
X_ = np.array([1,2,3])
arm = np.array([1,2])
num_arms = 2
num_contexts = 1
#num_data = 1000
#create X
X = np.zeros((num_data,num_contexts + num_arms))
for i in range(num_contexts):
X[:,i] = np.linspace(-1,1,num_data).T
for i in range(num_arms):
X[:,i+num_contexts] = np.random.binomial(1,.6,num_data)
X[:5,:]
X = shuffle(X)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
#X = scaler.fit_transform(X)
X[:,2] = abs(1-X[:,1])
X
def customer_reaction(context, choice, theta = theta, noise=noise):
features = np.append(context,choice)
# decide on which slope to use depending on whether a dog is present or not
m = theta[0] if features[1] else theta[1]
intercept = 0 if features[1] else 0
# $y = x \cdot m + t + \epsilon $
y = features[0] * m + intercept + np.random.normal(0,noise)
return y
customer_reaction([9.0],[1,0])
customer_reaction([9.0],[0,1])
y = []
for i in range(num_data):
context = X[i][0]
choice = X[i][1]
outcome = customer_reaction(context,choice)
y.append(outcome)
y = np.array(y).flatten()
index = X[:,1] == True
plt.scatter(X[:,0][index],y[index],label='with dog',c='red')
plt.scatter(X[:,0][~index],y[~index],label='without dog',c='black')
plt.legend()
class AbstractContextualSolver(object):
def __init__(self, model, num_arms):
self.model = model
self.num_arms = num_arms
def choose_arm(self,context):
"""choose an arm to play according to internal policy"""
raise NotImplementedError
def update(self, arm, context, reward):
""" update internal policy to reflect changed knowledge"""
raise NotImplementedError
import torch
from torch.autograd import Variable
class SimpleModel(torch.nn.Module):
def __init__(self,num_input, p=0.05, decay=0.001, non_linearity=torch.nn.LeakyReLU):
super(SimpleModel, self).__init__()
self.dropout_p = p
self.decay = decay
self.f = torch.nn.Sequential(
torch.nn.Linear(num_input,num_hidden_units),
torch.nn.ReLU(),
# torch.nn.Dropout(p=self.dropout_p),
# torch.nn.Linear(20,20),
# torch.nn.ReLU(),
# torch.nn.Dropout(p=self.dropout_p),
# torch.nn.Linear(20, 10),
# non_linearity(),
# torch.nn.Dropout(p=self.dropout_p),
torch.nn.Linear(num_hidden_units,1)
)
def forward(self, X):
X = Variable(torch.Tensor(X), requires_grad=False)
return self.f(X)
class VanillaEnsemble(object):
def __init__(self, p=0.00, decay=0.001, non_linearity=torch.nn.LeakyReLU, n_models=10, model_list=None):
self.models = [SimpleModel(p, decay, non_linearity) for model in range(n_models)]
self.optimizers = [torch.optim.SGD(
model.parameters(),
weight_decay=decay,
lr=0.01) for model in self.models]
self.criterion = torch.nn.MSELoss()
self.dropout_p = p
self.decay = decay
def fit_model(self, model, optimizer, X_obs,y_obs):
y = Variable(torch.Tensor(y_obs[:, np.newaxis]), requires_grad=False)
y_pred = model(X_obs[:, np.newaxis])
optimizer.zero_grad()
loss = self.criterion(y_pred, y)
loss.backward()
optimizer.step()
return loss
def observe(self,X,y):
return self.fit_ensemble(X,y)
def fit_ensemble(self, X_obs, y_obs):
losslist = []
for model, optimizer in zip(self.models, self.optimizers):
losslist.append(self.fit_model(model, optimizer, X_obs,y_obs))
return losslist
def predict_distribution(self, X, ):
outputs = np.hstack([model(X[:, np.newaxis]).data.numpy() for model in self.models])
y_mean = outputs.mean(axis=1)
y_std = outputs.std(axis=1)
return {'means':y_mean,
'stds':y_std}
class OnlineBootstrapEnsemble(object):
def __init__(self, p=0.00, decay=0.001, non_linearity=torch.nn.LeakyReLU, n_models=10, model_list=None):
self.models = [SimpleModel(p, decay, non_linearity) for model in range(n_models)]
self.optimizers = [torch.optim.SGD(
model.parameters(),
weight_decay=decay,
lr=0.01) for model in self.models]
self.criterion = torch.nn.MSELoss()
self.dropout_p = p
self.decay = decay
def fit_model(self, model, optimizer, X_obs,y_obs):
y = Variable(torch.Tensor(y_obs[:, np.newaxis]), requires_grad=False)
y_pred = model(X_obs[:, np.newaxis])
optimizer.zero_grad()
loss = self.criterion(y_pred, y)
loss.backward()
optimizer.step()
return loss
def observe(self,X,y):
return self.fit_ensemble(X,y)
def fit_ensemble(self, X_obs, y_obs):
losslist = []
for model, optimizer in zip(self.models, self.optimizers):
if random.random() > 0.5:
losslist.append(self.fit_model(model, optimizer, X_obs,y_obs))
self.fit_model(model, optimizer, X_obs,y_obs)
return losslist
def predict_distribution(self, X, ):
outputs = np.hstack([model(X[:, np.newaxis]).data.numpy() for model in self.models])
y_mean = outputs.mean(axis=1)
y_std = outputs.std(axis=1)
return {'means':y_mean,
'stds':y_std}
ensemble = VanillaEnsemble(3)#OnlineBootstrapEnsemble(3)
means = ensemble.predict_distribution(X[~index])['means']
stds = ensemble.predict_distribution(X[~index])['stds']
plt.scatter(X[:,0][~index],means)
X_train = X[~index]
y_train = np.atleast_2d(y[~index])
np.atleast_2d(y_train[:,:5])
ensemble.fit_ensemble(X_train,y_train.T)
means = ensemble.predict_distribution(X[~index])['means']
stds = ensemble.predict_distribution(X[~index])['stds']
plt.scatter(X[:,0][~index],means)
plt.figure()
plt.scatter(X[:,0][~index],stds)
class GaussianThompsonSampler(AbstractContextualSolver):
"""solves the contextual MAB problem with normal reward distribution
by utilising a model to predict the distribution
for each context and each arm individually"""
def choose_arm(self,context):
reward_list = []
dist = stats.norm
for i in range(self.num_arms):
arm = np.array([0,0])
arm[i] = 1
X = np.atleast_2d(np.append(arm, context))
model_distribution = self.model.predict_distribution(X)
reward_sample = dist.rvs(model_distribution['means'],model_distribution['stds'])
reward_list += [reward_sample]
arms = np.array([0,0])
arms[np.argmax(reward_list)] = 1
return arms
def update(self,arm,context,reward):
X = np.atleast_2d(np.append(arm, context))
reward = np.atleast_2d(reward).T
self.model.observe(X, reward)
for num_hidden_units in [1,2,5,10,100]:
breg = VanillaEnsemble(3)
gts = GaussianThompsonSampler(breg,num_arms = 2)
context = X[:,0]
y_hat_list = []
best_arms = []
for i in progress_bar(range(num_data)):
arm = gts.choose_arm(context[i])
y_hat = customer_reaction(context[i],arm)
y_hat_list += [y_hat]
gts.update(arm, context[i], y_hat)
#best_arms.append(best_arm_chosen(context[i], y_hat))
plt.figure()
plt.scatter(context,np.array(y_hat_list))
best_arms[:5], best_arms[-5:]
| 0.752377 | 0.98302 |
```
import os
from IPython.display import display,Audio,HTML
import scipy.io.wavfile as wav
import numpy as np
import speechpy
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
from sklearn.cross_validation import train_test_split
from sklearn.preprocessing import LabelEncoder
import time
import xgboost as xgb
from sklearn import metrics
sns.set()
def extract_features(signal, fs):
frames = speechpy.processing.stack_frames(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01, filter=lambda x: np.ones((x,)),zero_padding=True)
power_spectrum = speechpy.processing.power_spectrum(frames, fft_points=1)
logenergy = speechpy.feature.lmfe(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01,num_filters=1, fft_length=512, low_frequency=0, high_frequency=None)
mfcc = speechpy.feature.mfcc(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01,num_filters=1, fft_length=512, low_frequency=0, high_frequency=None)
mfcc_cmvn = speechpy.processing.cmvnw(mfcc,win_size=301,variance_normalization=True)
mfcc_feature_cube = speechpy.feature.extract_derivative_feature(mfcc)
return np.hstack([power_spectrum[:,0],logenergy[:,0],mfcc_cmvn[:,0],mfcc_feature_cube[:,0,1]])
def extract_files(folder):
location = folder + '/'
elements = os.listdir(location)
results = []
for i in elements:
try:
fs, signal = wav.read(location+i)
results.append([folder]+extract_features(signal, fs).tolist())
except:
continue
return results
folders = [i for i in os.listdir(os.getcwd())if i.find('.md') < 0 and i.find('.txt') < 0 and i.find('ipynb') < 0 and i.find('LICENSE') < 0]
output = []
for i in folders:
print(i)
output += extract_files(i)
output = [i for i in output if len(i) == 397]
dataset=np.array(output)
np.random.shuffle(dataset)
print(np.unique(dataset[:,0]).tolist())
target = LabelEncoder().fit_transform(dataset[:,0])
train_X, test_X, train_Y, test_Y = train_test_split(dataset[:, 1:], target, test_size = 0.2)
params_lgd = {
'boosting_type': 'dart',
'objective': 'multiclass',
'colsample_bytree': 0.4,
'subsample': 0.8,
'learning_rate': 0.1,
'silent': False,
'n_estimators': 10000,
'reg_lambda': 0.0005,
'device':'gpu'
}
clf = lgb.LGBMClassifier(**params_lgd)
lasttime = time.time()
clf.fit(train_X,train_Y, eval_set=[(train_X,train_Y), (test_X,test_Y)],
eval_metric='logloss', early_stopping_rounds=20, verbose=False)
print('time taken to fit lgb:', time.time()-lasttime, 'seconds ')
fig, ax = plt.subplots(figsize=(12,70))
lgb.plot_importance(clf, ax=ax)
plt.show()
predicted = clf.predict(test_X)
print('accuracy validation set: ', np.mean(predicted == test_Y))
# print scores
print(metrics.classification_report(test_Y, predicted, target_names = np.unique(dataset[:,0]).tolist()))
params_xgd = {
'min_child_weight': 10.0,
'max_depth': 7,
'objective': 'multi:softprob',
'max_delta_step': 1.8,
'colsample_bytree': 0.4,
'subsample': 0.8,
'learning_rate': 0.1,
'gamma': 0.65,
'nthread': -1,
'silent': False,
'n_estimators': 10000
}
clf = xgb.XGBClassifier(**params_xgd)
lasttime = time.time()
clf.fit(train_X,train_Y, eval_set=[(train_X,train_Y), (test_X,test_Y)],
eval_metric='mlogloss', early_stopping_rounds=20, verbose=False)
print('time taken to fit xgb:', time.time()-lasttime, 'seconds ')
predicted = clf.predict(test_X)
print('accuracy validation set: ', np.mean(predicted == test_Y))
# print scores
print(metrics.classification_report(test_Y, predicted, target_names = np.unique(dataset[:,0]).tolist()))
fig, ax = plt.subplots(figsize=(12,70))
xgb.plot_importance(clf, ax=ax)
plt.show()
```
|
github_jupyter
|
import os
from IPython.display import display,Audio,HTML
import scipy.io.wavfile as wav
import numpy as np
import speechpy
import matplotlib.pyplot as plt
import seaborn as sns
import lightgbm as lgb
from sklearn.cross_validation import train_test_split
from sklearn.preprocessing import LabelEncoder
import time
import xgboost as xgb
from sklearn import metrics
sns.set()
def extract_features(signal, fs):
frames = speechpy.processing.stack_frames(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01, filter=lambda x: np.ones((x,)),zero_padding=True)
power_spectrum = speechpy.processing.power_spectrum(frames, fft_points=1)
logenergy = speechpy.feature.lmfe(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01,num_filters=1, fft_length=512, low_frequency=0, high_frequency=None)
mfcc = speechpy.feature.mfcc(signal, sampling_frequency=fs, frame_length=0.020, frame_stride=0.01,num_filters=1, fft_length=512, low_frequency=0, high_frequency=None)
mfcc_cmvn = speechpy.processing.cmvnw(mfcc,win_size=301,variance_normalization=True)
mfcc_feature_cube = speechpy.feature.extract_derivative_feature(mfcc)
return np.hstack([power_spectrum[:,0],logenergy[:,0],mfcc_cmvn[:,0],mfcc_feature_cube[:,0,1]])
def extract_files(folder):
location = folder + '/'
elements = os.listdir(location)
results = []
for i in elements:
try:
fs, signal = wav.read(location+i)
results.append([folder]+extract_features(signal, fs).tolist())
except:
continue
return results
folders = [i for i in os.listdir(os.getcwd())if i.find('.md') < 0 and i.find('.txt') < 0 and i.find('ipynb') < 0 and i.find('LICENSE') < 0]
output = []
for i in folders:
print(i)
output += extract_files(i)
output = [i for i in output if len(i) == 397]
dataset=np.array(output)
np.random.shuffle(dataset)
print(np.unique(dataset[:,0]).tolist())
target = LabelEncoder().fit_transform(dataset[:,0])
train_X, test_X, train_Y, test_Y = train_test_split(dataset[:, 1:], target, test_size = 0.2)
params_lgd = {
'boosting_type': 'dart',
'objective': 'multiclass',
'colsample_bytree': 0.4,
'subsample': 0.8,
'learning_rate': 0.1,
'silent': False,
'n_estimators': 10000,
'reg_lambda': 0.0005,
'device':'gpu'
}
clf = lgb.LGBMClassifier(**params_lgd)
lasttime = time.time()
clf.fit(train_X,train_Y, eval_set=[(train_X,train_Y), (test_X,test_Y)],
eval_metric='logloss', early_stopping_rounds=20, verbose=False)
print('time taken to fit lgb:', time.time()-lasttime, 'seconds ')
fig, ax = plt.subplots(figsize=(12,70))
lgb.plot_importance(clf, ax=ax)
plt.show()
predicted = clf.predict(test_X)
print('accuracy validation set: ', np.mean(predicted == test_Y))
# print scores
print(metrics.classification_report(test_Y, predicted, target_names = np.unique(dataset[:,0]).tolist()))
params_xgd = {
'min_child_weight': 10.0,
'max_depth': 7,
'objective': 'multi:softprob',
'max_delta_step': 1.8,
'colsample_bytree': 0.4,
'subsample': 0.8,
'learning_rate': 0.1,
'gamma': 0.65,
'nthread': -1,
'silent': False,
'n_estimators': 10000
}
clf = xgb.XGBClassifier(**params_xgd)
lasttime = time.time()
clf.fit(train_X,train_Y, eval_set=[(train_X,train_Y), (test_X,test_Y)],
eval_metric='mlogloss', early_stopping_rounds=20, verbose=False)
print('time taken to fit xgb:', time.time()-lasttime, 'seconds ')
predicted = clf.predict(test_X)
print('accuracy validation set: ', np.mean(predicted == test_Y))
# print scores
print(metrics.classification_report(test_Y, predicted, target_names = np.unique(dataset[:,0]).tolist()))
fig, ax = plt.subplots(figsize=(12,70))
xgb.plot_importance(clf, ax=ax)
plt.show()
| 0.344554 | 0.34275 |
- https://www.kaggle.com/tanlikesmath/intro-aptos-diabetic-retinopathy-eda-starter
- https://medium.com/@btahir/a-quick-guide-to-using-regression-with-image-data-in-fastai-117304c0af90
# params
```
PRFX = 'CvCrop070314'
p_prp = '../output/Prep0703'
p_o = f'../output/{PRFX}'
SEED = 111
dbg = False
if dbg:
dbgsz = 500
BS = 256
SZ = 224
FP16 = True
import multiprocessing
multiprocessing.cpu_count() # 2
from fastai.vision import *
xtra_tfms = []
# xtra_tfms += [rgb_randomize(channel=i, thresh=1e-4) for i in range(3)]
params_tfms = dict(
do_flip=True,
flip_vert=False,
max_rotate=10,
max_warp=0,
max_zoom=1.1,
p_affine=0.5,
max_lighting=0.2,
p_lighting=0.5,
xtra_tfms=xtra_tfms)
resize_method = ResizeMethod.CROP
padding_mode = 'zeros'
USE_TTA = True
```
# setup
```
import fastai
print('fastai.__version__: ', fastai.__version__)
import random
import numpy as np
import torch
import os
def set_torch_seed(seed=SEED):
os.environ['PYTHONHASHSEED'] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
set_torch_seed()
from fastai import *
from fastai.vision import *
import pandas as pd
import scipy as sp
from sklearn.metrics import cohen_kappa_score
def quadratic_weighted_kappa(y1, y2):
return cohen_kappa_score(y1, y2, weights='quadratic')
```
# preprocess
```
img2grd = []
p = '../input/aptos2019-blindness-detection'
pp = Path(p)
train = pd.read_csv(pp/'train.csv')
test = pd.read_csv(pp/'test.csv')
len_blnd = len(train)
len_blnd_test = len(test)
img2grd_blnd = [(f'{p_prp}/aptos2019-blindness-detection/train_images/{o[0]}.png',o[1]) for o in train.values]
len_blnd, len_blnd_test
img2grd += img2grd_blnd
display(len(img2grd))
display(Counter(o[1] for o in img2grd).most_common())
if np.all([Path(o[0]).exists() for o in img2grd]): print('All files are here!')
df = pd.DataFrame(img2grd)
df.columns = ['fnm', 'target']
df.shape
set_torch_seed()
idx_blnd_train = np.where(df.fnm.str.contains('aptos2019-blindness-detection/train_images'))[0]
idx_val = np.random.choice(idx_blnd_train, len_blnd_test, replace=False)
df['is_val']=False
df.loc[idx_val, 'is_val']=True
if dbg:
df=df.head(dbgsz)
```
# dataset
```
tfms = get_transforms(**params_tfms)
def get_data(sz, bs):
src = (ImageList.from_df(df=df,path='./',cols='fnm')
.split_from_df(col='is_val')
.label_from_df(cols='target',
label_cls=FloatList)
)
data= (src.transform(tfms,
size=sz,
resize_method=resize_method,
padding_mode=padding_mode) #Data augmentation
.databunch(bs=bs) #DataBunch
.normalize(imagenet_stats) #Normalize
)
return data
bs = BS
sz = SZ
set_torch_seed()
data = get_data(sz, bs)
data.show_batch(rows=3, figsize=(7,6))
```
# model
```
%%time
# Downloading: "https://download.pytorch.org/models/resnet50-19c8e357.pth" to /tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth
# Making pretrained weights work without needing to find the default filename
if not os.path.exists('/tmp/.cache/torch/checkpoints/'):
os.makedirs('/tmp/.cache/torch/checkpoints/')
!cp '../input/pytorch-vision-pretrained-models/resnet50-19c8e357.pth' '/tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth'
learn = cnn_learner(data,
base_arch = models.resnet50,
path=p_o)
learn.loss = MSELossFlat
if FP16: learn = learn.to_fp16()
%%time
learn.freeze()
learn.lr_find()
```
```
learn.recorder.plot()
set_torch_seed()
learn.fit_one_cycle(4, max_lr = 1e-2)
learn.recorder.plot_losses()
# learn.recorder.plot_metrics()
learn.save('mdl-frozen')
learn.unfreeze()
%%time
learn.lr_find()
learn.recorder.plot(suggestion=True)
set_torch_seed()
learn.fit_one_cycle(6, max_lr=slice(1e-6,1e-3))
!nvidia-smi
learn.recorder.plot_losses()
# learn.recorder.plot_metrics()
learn.save('mdl')
```
# validate and thresholding
```
learn = learn.to_fp32()
learn = learn.load('mdl')
%%time
set_torch_seed()
preds_val_tta, y_val = learn.TTA(ds_type=DatasetType.Valid)
%%time
set_torch_seed()
preds_val, y_val = learn.get_preds(ds_type=DatasetType.Valid)
preds_val = preds_val.numpy().squeeze()
preds_val_tta = preds_val_tta.numpy().squeeze()
y_val= y_val.numpy()
np.save(f'{p_o}/preds_val.npy', preds_val)
np.save(f'{p_o}/preds_val_tta.npy', preds_val_tta)
np.save(f'{p_o}/y_val.npy', y_val)
# https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/88773#latest-515044
# We used OptimizedRounder given by hocop1. https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/76107#480970
# put numerical value to one of bins
def to_bins(x, borders):
for i in range(len(borders)):
if x <= borders[i]:
return i
return len(borders)
class Hocop1OptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _loss(self, coef, X, y, idx):
X_p = np.array([to_bins(pred, coef) for pred in X])
ll = -quadratic_weighted_kappa(y, X_p)
return ll
def fit(self, X, y):
coef = [1.5, 2.0, 2.5, 3.0]
golden1 = 0.618
golden2 = 1 - golden1
ab_start = [(1, 2), (1.5, 2.5), (2, 3), (2.5, 3.5)]
for it1 in range(10):
for idx in range(4):
# golden section search
a, b = ab_start[idx]
# calc losses
coef[idx] = a
la = self._loss(coef, X, y, idx)
coef[idx] = b
lb = self._loss(coef, X, y, idx)
for it in range(20):
# choose value
if la > lb:
a = b - (b - a) * golden1
coef[idx] = a
la = self._loss(coef, X, y, idx)
else:
b = b - (b - a) * golden2
coef[idx] = b
lb = self._loss(coef, X, y, idx)
self.coef_ = {'x': coef}
def predict(self, X, coef):
X_p = np.array([to_bins(pred, coef) for pred in X])
return X_p
def coefficients(self):
return self.coef_['x']
# https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/76107#480970
class AbhishekOptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _kappa_loss(self, coef, X, y):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
ll = quadratic_weighted_kappa(y, X_p)
return -ll
def fit(self, X, y):
loss_partial = partial(self._kappa_loss, X=X, y=y)
initial_coef = [0.5, 1.5, 2.5, 3.5]
self.coef_ = sp.optimize.minimize(loss_partial, initial_coef, method='nelder-mead')
def predict(self, X, coef):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
return X_p
def coefficients(self):
return self.coef_['x']
def bucket(preds_raw, coef = [0.5, 1.5, 2.5, 3.5]):
preds = np.zeros(preds_raw.shape)
for i, pred in enumerate(preds_raw):
if pred < coef[0]:
preds[i] = 0
elif pred >= coef[0] and pred < coef[1]:
preds[i] = 1
elif pred >= coef[1] and pred < coef[2]:
preds[i] = 2
elif pred >= coef[2] and pred < coef[3]:
preds[i] = 3
else:
preds[i] = 4
return preds
optnm2coefs = {'simple': [0.5, 1.5, 2.5, 3.5]}
%%time
set_torch_seed()
optR = Hocop1OptimizedRounder()
optR.fit(preds_val_tta, y_val)
optnm2coefs['hocop1_tta'] = optR.coefficients()
%%time
set_torch_seed()
optR = Hocop1OptimizedRounder()
optR.fit(preds_val, y_val)
optnm2coefs['hocop1'] = optR.coefficients()
%%time
set_torch_seed()
optR = AbhishekOptimizedRounder()
optR.fit(preds_val_tta, y_val)
optnm2coefs['abhishek_tta'] = optR.coefficients()
%%time
set_torch_seed()
optR = AbhishekOptimizedRounder()
optR.fit(preds_val, y_val)
optnm2coefs['abhishek'] = optR.coefficients()
optnm2coefs
optnm2preds_val_grd = {k: bucket(preds_val, coef) for k,coef in optnm2coefs.items()}
optnm2qwk = {k: quadratic_weighted_kappa(y_val, preds) for k,preds in optnm2preds_val_grd.items()}
optnm2qwk
Counter(y_val).most_common()
preds_val_grd = optnm2preds_val_grd['abhishek'].squeeze()
preds_val_grd.mean()
Counter(preds_val_grd).most_common()
list(zip(preds_val_grd, y_val))[:10]
(preds_val_grd== y_val.squeeze()).mean()
pickle.dump(optnm2qwk, open(f'{p_o}/optnm2qwk.p', 'wb'))
pickle.dump(optnm2preds_val_grd, open(f'{p_o}/optnm2preds_val_grd.p', 'wb'))
pickle.dump(optnm2coefs, open(f'{p_o}/optnm2coefs.p', 'wb'))
```
# testing
This goes to Kernel!!
## params
```
PRFX = 'CvCrop070314'
p_o = f'../output/{PRFX}'
SEED = 111
dbg = False
if dbg:
dbgsz = 500
BS = 128
SZ = 224
from fastai.vision import *
xtra_tfms = []
# xtra_tfms += [rgb_randomize(channel=i, thresh=1e-4) for i in range(3)]
params_tfms = dict(
do_flip=True,
flip_vert=False,
max_rotate=10,
max_warp=0,
max_zoom=1.1,
p_affine=0.5,
max_lighting=0.2,
p_lighting=0.5,
xtra_tfms=xtra_tfms)
resize_method = ResizeMethod.CROP
padding_mode = 'zeros'
USE_TTA = True
import fastai
print(fastai.__version__)
```
## setup
```
import fastai
print('fastai.__version__: ', fastai.__version__)
import random
import numpy as np
import torch
import os
def set_torch_seed(seed=SEED):
os.environ['PYTHONHASHSEED'] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
set_torch_seed()
from fastai import *
from fastai.vision import *
import pandas as pd
```
## preprocess
```
img2grd = []
p = '../input/aptos2019-blindness-detection'
pp = Path(p)
train = pd.read_csv(pp/'train.csv')
test = pd.read_csv(pp/'test.csv')
len_blnd = len(train)
len_blnd_test = len(test)
img2grd_blnd = [(f'{p_prp}/aptos2019-blindness-detection/train_images/{o[0]}.png',o[1]) for o in train.values]
len_blnd, len_blnd_test
img2grd += img2grd_blnd
display(len(img2grd))
display(Counter(o[1] for o in img2grd).most_common())
if np.all([Path(o[0]).exists() for o in img2grd]): print('All files are here!')
df = pd.DataFrame(img2grd)
df.columns = ['fnm', 'target']
df.shape
df.head()
set_torch_seed()
idx_blnd_train = np.where(df.fnm.str.contains('aptos2019-blindness-detection/train_images'))[0]
idx_val = np.random.choice(idx_blnd_train, len_blnd_test, replace=False)
df['is_val']=False
df.loc[idx_val, 'is_val']=True
if dbg:
df=df.head(dbgsz)
```
## dataset
```
tfms = get_transforms(**params_tfms)
def get_data(sz, bs):
src = (ImageList.from_df(df=df,path='./',cols='fnm')
.split_from_df(col='is_val')
.label_from_df(cols='target',
label_cls=FloatList)
)
data= (src.transform(tfms,
size=sz,
resize_method=resize_method,
padding_mode=padding_mode) #Data augmentation
.databunch(bs=bs,num_workers=2) #DataBunch
.normalize(imagenet_stats) #Normalize
)
return data
bs = BS
sz = SZ
set_torch_seed()
data = get_data(sz, bs)
```
## model
```
%%time
# Downloading: "https://download.pytorch.org/models/resnet50-19c8e357.pth" to /tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth
# Making pretrained weights work without needing to find the default filename
if not os.path.exists('/tmp/.cache/torch/checkpoints/'):
os.makedirs('/tmp/.cache/torch/checkpoints/')
!cp '../input/pytorch-vision-pretrained-models/resnet50-19c8e357.pth' '/tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth'
set_torch_seed()
learn = cnn_learner(data,
base_arch = models.resnet50,
path=p_o)
learn.loss = MSELossFlat
learn = learn.load('mdl')
df_test = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')
df_test.head()
learn.data.add_test(
ImageList.from_df(df_test,
f'{p_prp}/aptos2019-blindness-detection/',
folder='test_images',
suffix='.png'))
%%time
# Predictions for test set
set_torch_seed()
preds_tst_tta, _ = learn.TTA(ds_type=DatasetType.Test)
%%time
# Predictions for test set
set_torch_seed()
preds_tst, _ = learn.get_preds(ds_type=DatasetType.Test)
preds_tst = preds_tst.numpy().squeeze()
preds_tst_tta = preds_tst_tta.numpy().squeeze()
np.save(f'{p_o}/preds_tst.npy', preds_tst)
np.save(f'{p_o}/preds_tst_tta.npy', preds_tst_tta)
preds_tst2use = preds_tst
def bucket(preds_raw, coef = [0.5, 1.5, 2.5, 3.5]):
preds = np.zeros(preds_raw.shape)
for i, pred in enumerate(preds_raw):
if pred < coef[0]:
preds[i] = 0
elif pred >= coef[0] and pred < coef[1]:
preds[i] = 1
elif pred >= coef[1] and pred < coef[2]:
preds[i] = 2
elif pred >= coef[2] and pred < coef[3]:
preds[i] = 3
else:
preds[i] = 4
return preds
optnm2qwk = pickle.load(open(f'{p_o}/optnm2qwk.p','rb'))
optnm2coefs = pickle.load(open(f'{p_o}/optnm2coefs.p','rb'))
optnm2qwk
coef = optnm2coefs['abhishek']
preds_tst_grd = bucket(preds_tst2use, coef)
Counter(preds_tst_grd.squeeze()).most_common()
```
## submit
```
subm = pd.read_csv("../input/aptos2019-blindness-detection/test.csv")
subm['diagnosis'] = preds_tst_grd.squeeze().astype(int)
subm.head()
subm.diagnosis.value_counts()
subm.to_csv(f"{p_o}/submission.csv", index=False)
```
|
github_jupyter
|
PRFX = 'CvCrop070314'
p_prp = '../output/Prep0703'
p_o = f'../output/{PRFX}'
SEED = 111
dbg = False
if dbg:
dbgsz = 500
BS = 256
SZ = 224
FP16 = True
import multiprocessing
multiprocessing.cpu_count() # 2
from fastai.vision import *
xtra_tfms = []
# xtra_tfms += [rgb_randomize(channel=i, thresh=1e-4) for i in range(3)]
params_tfms = dict(
do_flip=True,
flip_vert=False,
max_rotate=10,
max_warp=0,
max_zoom=1.1,
p_affine=0.5,
max_lighting=0.2,
p_lighting=0.5,
xtra_tfms=xtra_tfms)
resize_method = ResizeMethod.CROP
padding_mode = 'zeros'
USE_TTA = True
import fastai
print('fastai.__version__: ', fastai.__version__)
import random
import numpy as np
import torch
import os
def set_torch_seed(seed=SEED):
os.environ['PYTHONHASHSEED'] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
set_torch_seed()
from fastai import *
from fastai.vision import *
import pandas as pd
import scipy as sp
from sklearn.metrics import cohen_kappa_score
def quadratic_weighted_kappa(y1, y2):
return cohen_kappa_score(y1, y2, weights='quadratic')
img2grd = []
p = '../input/aptos2019-blindness-detection'
pp = Path(p)
train = pd.read_csv(pp/'train.csv')
test = pd.read_csv(pp/'test.csv')
len_blnd = len(train)
len_blnd_test = len(test)
img2grd_blnd = [(f'{p_prp}/aptos2019-blindness-detection/train_images/{o[0]}.png',o[1]) for o in train.values]
len_blnd, len_blnd_test
img2grd += img2grd_blnd
display(len(img2grd))
display(Counter(o[1] for o in img2grd).most_common())
if np.all([Path(o[0]).exists() for o in img2grd]): print('All files are here!')
df = pd.DataFrame(img2grd)
df.columns = ['fnm', 'target']
df.shape
set_torch_seed()
idx_blnd_train = np.where(df.fnm.str.contains('aptos2019-blindness-detection/train_images'))[0]
idx_val = np.random.choice(idx_blnd_train, len_blnd_test, replace=False)
df['is_val']=False
df.loc[idx_val, 'is_val']=True
if dbg:
df=df.head(dbgsz)
tfms = get_transforms(**params_tfms)
def get_data(sz, bs):
src = (ImageList.from_df(df=df,path='./',cols='fnm')
.split_from_df(col='is_val')
.label_from_df(cols='target',
label_cls=FloatList)
)
data= (src.transform(tfms,
size=sz,
resize_method=resize_method,
padding_mode=padding_mode) #Data augmentation
.databunch(bs=bs) #DataBunch
.normalize(imagenet_stats) #Normalize
)
return data
bs = BS
sz = SZ
set_torch_seed()
data = get_data(sz, bs)
data.show_batch(rows=3, figsize=(7,6))
%%time
# Downloading: "https://download.pytorch.org/models/resnet50-19c8e357.pth" to /tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth
# Making pretrained weights work without needing to find the default filename
if not os.path.exists('/tmp/.cache/torch/checkpoints/'):
os.makedirs('/tmp/.cache/torch/checkpoints/')
!cp '../input/pytorch-vision-pretrained-models/resnet50-19c8e357.pth' '/tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth'
learn = cnn_learner(data,
base_arch = models.resnet50,
path=p_o)
learn.loss = MSELossFlat
if FP16: learn = learn.to_fp16()
%%time
learn.freeze()
learn.lr_find()
learn.recorder.plot()
set_torch_seed()
learn.fit_one_cycle(4, max_lr = 1e-2)
learn.recorder.plot_losses()
# learn.recorder.plot_metrics()
learn.save('mdl-frozen')
learn.unfreeze()
%%time
learn.lr_find()
learn.recorder.plot(suggestion=True)
set_torch_seed()
learn.fit_one_cycle(6, max_lr=slice(1e-6,1e-3))
!nvidia-smi
learn.recorder.plot_losses()
# learn.recorder.plot_metrics()
learn.save('mdl')
learn = learn.to_fp32()
learn = learn.load('mdl')
%%time
set_torch_seed()
preds_val_tta, y_val = learn.TTA(ds_type=DatasetType.Valid)
%%time
set_torch_seed()
preds_val, y_val = learn.get_preds(ds_type=DatasetType.Valid)
preds_val = preds_val.numpy().squeeze()
preds_val_tta = preds_val_tta.numpy().squeeze()
y_val= y_val.numpy()
np.save(f'{p_o}/preds_val.npy', preds_val)
np.save(f'{p_o}/preds_val_tta.npy', preds_val_tta)
np.save(f'{p_o}/y_val.npy', y_val)
# https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/88773#latest-515044
# We used OptimizedRounder given by hocop1. https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/76107#480970
# put numerical value to one of bins
def to_bins(x, borders):
for i in range(len(borders)):
if x <= borders[i]:
return i
return len(borders)
class Hocop1OptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _loss(self, coef, X, y, idx):
X_p = np.array([to_bins(pred, coef) for pred in X])
ll = -quadratic_weighted_kappa(y, X_p)
return ll
def fit(self, X, y):
coef = [1.5, 2.0, 2.5, 3.0]
golden1 = 0.618
golden2 = 1 - golden1
ab_start = [(1, 2), (1.5, 2.5), (2, 3), (2.5, 3.5)]
for it1 in range(10):
for idx in range(4):
# golden section search
a, b = ab_start[idx]
# calc losses
coef[idx] = a
la = self._loss(coef, X, y, idx)
coef[idx] = b
lb = self._loss(coef, X, y, idx)
for it in range(20):
# choose value
if la > lb:
a = b - (b - a) * golden1
coef[idx] = a
la = self._loss(coef, X, y, idx)
else:
b = b - (b - a) * golden2
coef[idx] = b
lb = self._loss(coef, X, y, idx)
self.coef_ = {'x': coef}
def predict(self, X, coef):
X_p = np.array([to_bins(pred, coef) for pred in X])
return X_p
def coefficients(self):
return self.coef_['x']
# https://www.kaggle.com/c/petfinder-adoption-prediction/discussion/76107#480970
class AbhishekOptimizedRounder(object):
def __init__(self):
self.coef_ = 0
def _kappa_loss(self, coef, X, y):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
ll = quadratic_weighted_kappa(y, X_p)
return -ll
def fit(self, X, y):
loss_partial = partial(self._kappa_loss, X=X, y=y)
initial_coef = [0.5, 1.5, 2.5, 3.5]
self.coef_ = sp.optimize.minimize(loss_partial, initial_coef, method='nelder-mead')
def predict(self, X, coef):
X_p = np.copy(X)
for i, pred in enumerate(X_p):
if pred < coef[0]:
X_p[i] = 0
elif pred >= coef[0] and pred < coef[1]:
X_p[i] = 1
elif pred >= coef[1] and pred < coef[2]:
X_p[i] = 2
elif pred >= coef[2] and pred < coef[3]:
X_p[i] = 3
else:
X_p[i] = 4
return X_p
def coefficients(self):
return self.coef_['x']
def bucket(preds_raw, coef = [0.5, 1.5, 2.5, 3.5]):
preds = np.zeros(preds_raw.shape)
for i, pred in enumerate(preds_raw):
if pred < coef[0]:
preds[i] = 0
elif pred >= coef[0] and pred < coef[1]:
preds[i] = 1
elif pred >= coef[1] and pred < coef[2]:
preds[i] = 2
elif pred >= coef[2] and pred < coef[3]:
preds[i] = 3
else:
preds[i] = 4
return preds
optnm2coefs = {'simple': [0.5, 1.5, 2.5, 3.5]}
%%time
set_torch_seed()
optR = Hocop1OptimizedRounder()
optR.fit(preds_val_tta, y_val)
optnm2coefs['hocop1_tta'] = optR.coefficients()
%%time
set_torch_seed()
optR = Hocop1OptimizedRounder()
optR.fit(preds_val, y_val)
optnm2coefs['hocop1'] = optR.coefficients()
%%time
set_torch_seed()
optR = AbhishekOptimizedRounder()
optR.fit(preds_val_tta, y_val)
optnm2coefs['abhishek_tta'] = optR.coefficients()
%%time
set_torch_seed()
optR = AbhishekOptimizedRounder()
optR.fit(preds_val, y_val)
optnm2coefs['abhishek'] = optR.coefficients()
optnm2coefs
optnm2preds_val_grd = {k: bucket(preds_val, coef) for k,coef in optnm2coefs.items()}
optnm2qwk = {k: quadratic_weighted_kappa(y_val, preds) for k,preds in optnm2preds_val_grd.items()}
optnm2qwk
Counter(y_val).most_common()
preds_val_grd = optnm2preds_val_grd['abhishek'].squeeze()
preds_val_grd.mean()
Counter(preds_val_grd).most_common()
list(zip(preds_val_grd, y_val))[:10]
(preds_val_grd== y_val.squeeze()).mean()
pickle.dump(optnm2qwk, open(f'{p_o}/optnm2qwk.p', 'wb'))
pickle.dump(optnm2preds_val_grd, open(f'{p_o}/optnm2preds_val_grd.p', 'wb'))
pickle.dump(optnm2coefs, open(f'{p_o}/optnm2coefs.p', 'wb'))
PRFX = 'CvCrop070314'
p_o = f'../output/{PRFX}'
SEED = 111
dbg = False
if dbg:
dbgsz = 500
BS = 128
SZ = 224
from fastai.vision import *
xtra_tfms = []
# xtra_tfms += [rgb_randomize(channel=i, thresh=1e-4) for i in range(3)]
params_tfms = dict(
do_flip=True,
flip_vert=False,
max_rotate=10,
max_warp=0,
max_zoom=1.1,
p_affine=0.5,
max_lighting=0.2,
p_lighting=0.5,
xtra_tfms=xtra_tfms)
resize_method = ResizeMethod.CROP
padding_mode = 'zeros'
USE_TTA = True
import fastai
print(fastai.__version__)
import fastai
print('fastai.__version__: ', fastai.__version__)
import random
import numpy as np
import torch
import os
def set_torch_seed(seed=SEED):
os.environ['PYTHONHASHSEED'] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
set_torch_seed()
from fastai import *
from fastai.vision import *
import pandas as pd
img2grd = []
p = '../input/aptos2019-blindness-detection'
pp = Path(p)
train = pd.read_csv(pp/'train.csv')
test = pd.read_csv(pp/'test.csv')
len_blnd = len(train)
len_blnd_test = len(test)
img2grd_blnd = [(f'{p_prp}/aptos2019-blindness-detection/train_images/{o[0]}.png',o[1]) for o in train.values]
len_blnd, len_blnd_test
img2grd += img2grd_blnd
display(len(img2grd))
display(Counter(o[1] for o in img2grd).most_common())
if np.all([Path(o[0]).exists() for o in img2grd]): print('All files are here!')
df = pd.DataFrame(img2grd)
df.columns = ['fnm', 'target']
df.shape
df.head()
set_torch_seed()
idx_blnd_train = np.where(df.fnm.str.contains('aptos2019-blindness-detection/train_images'))[0]
idx_val = np.random.choice(idx_blnd_train, len_blnd_test, replace=False)
df['is_val']=False
df.loc[idx_val, 'is_val']=True
if dbg:
df=df.head(dbgsz)
tfms = get_transforms(**params_tfms)
def get_data(sz, bs):
src = (ImageList.from_df(df=df,path='./',cols='fnm')
.split_from_df(col='is_val')
.label_from_df(cols='target',
label_cls=FloatList)
)
data= (src.transform(tfms,
size=sz,
resize_method=resize_method,
padding_mode=padding_mode) #Data augmentation
.databunch(bs=bs,num_workers=2) #DataBunch
.normalize(imagenet_stats) #Normalize
)
return data
bs = BS
sz = SZ
set_torch_seed()
data = get_data(sz, bs)
%%time
# Downloading: "https://download.pytorch.org/models/resnet50-19c8e357.pth" to /tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth
# Making pretrained weights work without needing to find the default filename
if not os.path.exists('/tmp/.cache/torch/checkpoints/'):
os.makedirs('/tmp/.cache/torch/checkpoints/')
!cp '../input/pytorch-vision-pretrained-models/resnet50-19c8e357.pth' '/tmp/.cache/torch/checkpoints/resnet50-19c8e357.pth'
set_torch_seed()
learn = cnn_learner(data,
base_arch = models.resnet50,
path=p_o)
learn.loss = MSELossFlat
learn = learn.load('mdl')
df_test = pd.read_csv('../input/aptos2019-blindness-detection/test.csv')
df_test.head()
learn.data.add_test(
ImageList.from_df(df_test,
f'{p_prp}/aptos2019-blindness-detection/',
folder='test_images',
suffix='.png'))
%%time
# Predictions for test set
set_torch_seed()
preds_tst_tta, _ = learn.TTA(ds_type=DatasetType.Test)
%%time
# Predictions for test set
set_torch_seed()
preds_tst, _ = learn.get_preds(ds_type=DatasetType.Test)
preds_tst = preds_tst.numpy().squeeze()
preds_tst_tta = preds_tst_tta.numpy().squeeze()
np.save(f'{p_o}/preds_tst.npy', preds_tst)
np.save(f'{p_o}/preds_tst_tta.npy', preds_tst_tta)
preds_tst2use = preds_tst
def bucket(preds_raw, coef = [0.5, 1.5, 2.5, 3.5]):
preds = np.zeros(preds_raw.shape)
for i, pred in enumerate(preds_raw):
if pred < coef[0]:
preds[i] = 0
elif pred >= coef[0] and pred < coef[1]:
preds[i] = 1
elif pred >= coef[1] and pred < coef[2]:
preds[i] = 2
elif pred >= coef[2] and pred < coef[3]:
preds[i] = 3
else:
preds[i] = 4
return preds
optnm2qwk = pickle.load(open(f'{p_o}/optnm2qwk.p','rb'))
optnm2coefs = pickle.load(open(f'{p_o}/optnm2coefs.p','rb'))
optnm2qwk
coef = optnm2coefs['abhishek']
preds_tst_grd = bucket(preds_tst2use, coef)
Counter(preds_tst_grd.squeeze()).most_common()
subm = pd.read_csv("../input/aptos2019-blindness-detection/test.csv")
subm['diagnosis'] = preds_tst_grd.squeeze().astype(int)
subm.head()
subm.diagnosis.value_counts()
subm.to_csv(f"{p_o}/submission.csv", index=False)
| 0.449151 | 0.466056 |
# PyTorch quickstart
## Welcome to PrimeHub!
In this quickstart, we will perfome following actions to train and deploy the model:
1. Train a neural network that classifies images.
1. Move trained model to <a target="_blank" href="https://docs.primehub.io/docs/quickstart/nb-data-store#phfs-storage">PHFS Storage</a>.
1. Deploy the trained model on PrimeHub <a target="_blank" href="https://docs.primehub.io/docs/model-deployment-feature">Model Deployments</a>.
1. Test deployed model.
1. (Advanced) Use PrimeHub <a target="_blank" href="https://github.com/InfuseAI/primehub-python-sdk">Python SDK</a> to deploy the trained model.
### Prerequisites
1. Enable <a target="_blank" href="https://docs.primehub.io/docs/model-deployment-feature">Model Deployments</a>.
1. Enable <a target="_blank" href="https://docs.primehub.io/docs/quickstart/nb-data-store#phfs-storage">PHFS Storage</a>.
**Contact your admin if any prerequisite is not enabled yet.**
## 1. Train a neural network that classifies images
Firstly, let's import libraries.
```
import os
import json
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch.nn import functional as F
from torch import nn
from torch.utils.data import DataLoader
from torchvision.datasets import MNIST
from torchvision import transforms
```
Load and prepare the MNIST dataset. Convert the samples to tensor and normalize them.
```
!wget www.di.ens.fr/~lelarge/MNIST.tar.gz
!tar -zxvf MNIST.tar.gz
!rm MNIST.tar.gz
transform=transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
mnist_train = MNIST(os.getcwd(), train=True, download=True, transform=transform)
mnist_train = DataLoader(mnist_train, batch_size=64, shuffle=True)
mnist_test = MNIST(os.getcwd(), train=False, download=True, transform=transform)
mnist_test = DataLoader(mnist_test, batch_size=64)
```
Set device on GPU if available, else CPU.
```
if torch.cuda.is_available():
device = torch.cuda.current_device()
print(torch.cuda.device(device))
print('Device Count:', torch.cuda.device_count())
print('Device Name: {}'.format(torch.cuda.get_device_name(device)))
else:
device = 'cpu'
```
Build the model class.
```
class PyTorchModel(nn.Module):
def __init__(self):
super().__init__()
# mnist images are (1, 28, 28) (channels, width, height)
self.layer_1 = nn.Linear(28 * 28, 128)
self.layer_2 = nn.Linear(128, 256)
self.layer_3 = nn.Linear(256, 10)
def forward(self, x):
batch_size, channels, width, height = x.size()
# (b, 1, 28, 28) -> (b, 1*28*28)
x = x.view(batch_size, -1)
x = self.layer_1(x)
x = F.relu(x)
x = self.layer_2(x)
x = F.relu(x)
x = self.layer_3(x)
x = F.softmax(x, dim=1)
return x
```
Create the model instance.
```
net = PyTorchModel().to(device)
```
Choose an optimizer, loss function.
```
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-3)
```
Train the model to minimize the loss. It prints the loss every 200 mini-batches.
```
for epoch in range(2):
running_loss = 0.0
for i, data in enumerate(mnist_train, 0):
inputs, labels = data
optimizer.zero_grad()
outputs = net(inputs.to(device))
loss = criterion(outputs, labels.to(device))
loss.backward()
optimizer.step()
running_loss += loss.item()
if i % 200 == 199:
print("[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 199))
running_loss = 0.0
```
Use the test data to check the model performance.
```
correct = 0
total = 0
with torch.no_grad():
for data in mnist_test:
images, labels = data
outputs = net(images.to(device))
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels.to(device)).sum().item()
print("Accuracy of the network on the %d test images: %d %%" % (total, 100 * correct / total))
```
Save the trained model.
```
now = datetime.now()
date_time = now.strftime("%Y%m%d-%H%M%S")
SAVED_DIR = f"pytorch-model-{date_time}"
os.makedirs(SAVED_DIR, exist_ok=True)
torch.save(net.state_dict(), os.path.join(SAVED_DIR, "model.pt"))
print(f"We successfully saved the model in {SAVED_DIR}.")
```
Save the model class file. The class name must be `PyTorchModel`. The content is the model class content and the imports used in the class.
```
model_class_file_content = """
import torch
from torch.nn import functional as F
from torch import nn
class PyTorchModel(nn.Module):
def __init__(self):
super().__init__()
# mnist images are (1, 28, 28) (channels, width, height)
self.layer_1 = nn.Linear(28 * 28, 128)
self.layer_2 = nn.Linear(128, 256)
self.layer_3 = nn.Linear(256, 10)
def forward(self, x):
batch_size, channels, width, height = x.size()
# (b, 1, 28, 28) -> (b, 1*28*28)
x = x.view(batch_size, -1)
x = self.layer_1(x)
x = F.relu(x)
x = self.layer_2(x)
x = F.relu(x)
x = self.layer_3(x)
x = F.softmax(x, dim=1)
return x
"""
model_class_file = open(os.path.join(SAVED_DIR, "ModelClass.py"), "w")
model_class_file.write(model_class_file_content)
model_class_file.close()
print(f"We successfully saved the model class file in {SAVED_DIR}.")
```
## 2. Move trained model to <a target="_blank" href="https://docs.primehub.io/docs/quickstart/nb-data-store#phfs-storage">PHFS Storage</a>
To deploy our model, we need to move model to PHFS storage.
```
!mv $SAVED_DIR ~/phfs
```
Check the model under PHFS storage.
```
!ls -l ~/phfs/$SAVED_DIR
```
## 3. Deploy the trained model on PrimeHub <a target="_blank" href="https://docs.primehub.io/docs/model-deployment-feature">Model Deployments</a>
```
print(f"Now, we've already prepared the ready-to-deploy model in ~/phfs/{SAVED_DIR}.")
```
Next, back to PrimeHub and select `Deployments`.
<img src="img/pytorch/3-1-menu.png"/>
In the `Deployments` page, click on `Create Deployment` button.
<img src="img/pytorch/3-2-create-deployment.png"/>
1. Fill in the [Deployment Name] field with [pytorch-quickstart].
1. Select the [Model Image] field with [PyTorch server]; this is a pre-packaged model server image that can serve PyTorch model.
<img src="img/pytorch/3-3-name-model-image.png"/>
```
print(f"Fill in the [Model URI] field with [phfs:///{SAVED_DIR}].")
```
<img src="img/pytorch/3-4-model-uri.png"/>
Choose the Instance Type, the minimal requirements in this quickstart is `CPU: 0.5 / Memory: 1 G / GPU: 0`.
<img src="img/pytorch/3-5-resource.png"/>
Then, click on `Deploy` button.
Our model is deploying, let's click on the `Manage` button of `pytorch-quickstart` deployment cell.
<img src="img/pytorch/3-6-deployment-list.png"/>
In the deployment detail page, we can see the `Status` is `Deploying`.
<img src="img/pytorch/3-7-deploying.png"/>
Wait for a while and our model is `Deployed` now!
To test our deployment, let's copy the value of `Endpoint` (`https://.../predictions`).
<img src="img/pytorch/3-8-deployed.png"/>
## 4. Test deployed model
Before testing, let's display the test image. It is a 28x28 grayscale image represented as numpy.ndarray.
```
array = np.array([[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]])
plt.imshow(array, cmap='gray')
plt.show()
```
Then, replace the `<MODEL_DEPLOYMENT_ENDPOINT>` in the below cell with `https://.../predictions` (Endpoint value copied from deployment detail page).
```
%env ENDPOINT=<MODEL_DEPLOYMENT_ENDPOINT>
```
Run below cell to send request to deployed model endpoint.
```
response=!curl -X POST $ENDPOINT \
-H 'Content-Type: application/json' \
-d '{ "data": {"ndarray": [[[[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]]] } }'
```
In the following cells, we will parse the model response to find the highest prediction probability.
```
result = json.loads(response[-1])
```
Print out response data.
```
result
print(f"The model predicts the number is {np.argmax(result['data']['ndarray'][0])}")
```
We have trained a model in Notebook and deployed it as an endpoint service that can respond to requests anytime from everywhere!
## 5. (Advanced) Use PrimeHub <a target="_blank" href="https://github.com/InfuseAI/primehub-python-sdk">Python SDK</a> to deploy the trained model
In addition to deploy the trained model via PrimeHub UI, we can simply use PrimeHub `Python SDK` to perform the same operations.
Let's install PrimeHub Python SDK with pip.
```
!pip install primehub-python-sdk
```
After installed, the SDK needs a configuration file stored in the `~/.primehub/config.json`.
Hence, back to PrimeHub UI and select `API Token` from the upper-right menu.
<img src="../tensorflow2/img/tensorflow2/5-1-menu.png"/>
In the `API Token` page, click on the `Request API Token` button.
<img src="../tensorflow2/img/tensorflow2/5-2-request-api-token.png"/>
After the token is granted, we can click on the `Download Config` button to save the `config.json` file locally.
<img src="../tensorflow2/img/tensorflow2/5-3-download-config.png"/>
Next, uploading the downloaded `config.json` file and moving it to the `~/.primehub/config.json`.
Verify configration with PrimeHub Python SDK.
```
from primehub import PrimeHub, PrimeHubConfig
ph = PrimeHub(PrimeHubConfig())
if ph.is_ready():
print("PrimeHub Python SDK setup successfully")
else:
print("PrimeHub Python SDK couldn't get the information, please ensure your configuration is stored in the ~/.primehub/config.json")
```
We can refer to <a target="_blank" href="https://github.com/InfuseAI/primehub-python-sdk/blob/main/docs/notebook/deployments.ipynb">Deployments command</a> to find out more useful manipulations of PrimeHub Python SDK.
List all deployments.
```
ph.deployments.list()
```
Delete the `pytorch-quickstart` deployment that we created early.
```
ph.deployments.delete(ph.deployments.list()[0]["id"])
```
Create a new deployment by specifing its config information:
- `id`: the folder name we stored in PHFS
- `name`: pytorch-sdk-quickstart
- `modelImage`: a pre-packaged model server image that can serve PyTorch model
- `modelURI`: the trained model stored in PHFS
- `instanceType`: cpu-1 (CPU: 1 / Memory: 2G / GPU: 0)
- `replicas`: 1
```
config = {
"id": f"{SAVED_DIR}",
"name": "pytorch-sdk-quickstart",
"modelImage": "infuseai/pytorch-prepackaged:v0.2.0",
"modelURI": f"phfs:///{SAVED_DIR}",
"instanceType": "cpu-1",
"replicas": 1
}
deployment = ph.deployments.create(config)
print(deployment)
```
Keep waiting until deployed.
```
ph.deployments.wait(deployment["id"])
```
After deployed, parsing the deployment endpoint.
```
endpoint = ph.deployments.get(deployment["id"])["endpoint"]
%env ENDPOINT=$endpoint
```
Run below cell to send request to deployed model endpoint.
```
response=!curl -X POST $ENDPOINT \
-H 'Content-Type: application/json' \
-d '{ "data": {"ndarray": [[[[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]]] } }'
```
In the following cells, we will parse the model response to find the highest prediction probability.
```
result = json.loads(response[-1])
```
Print out response data.
```
result
print(f"The model predicts the number is {np.argmax(result['data']['ndarray'][0])}")
```
Congratulations! We have efficiently deployed our model via PrimeHub Python SDK!
In the last, we can delete our deployment to ensure no resources will be wasted after this quickstart tutorial!
```
ph.deployments.delete(deployment["id"])
```
|
github_jupyter
|
import os
import json
from datetime import datetime
import numpy as np
import matplotlib.pyplot as plt
import torch
from torch.nn import functional as F
from torch import nn
from torch.utils.data import DataLoader
from torchvision.datasets import MNIST
from torchvision import transforms
!wget www.di.ens.fr/~lelarge/MNIST.tar.gz
!tar -zxvf MNIST.tar.gz
!rm MNIST.tar.gz
transform=transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))])
mnist_train = MNIST(os.getcwd(), train=True, download=True, transform=transform)
mnist_train = DataLoader(mnist_train, batch_size=64, shuffle=True)
mnist_test = MNIST(os.getcwd(), train=False, download=True, transform=transform)
mnist_test = DataLoader(mnist_test, batch_size=64)
if torch.cuda.is_available():
device = torch.cuda.current_device()
print(torch.cuda.device(device))
print('Device Count:', torch.cuda.device_count())
print('Device Name: {}'.format(torch.cuda.get_device_name(device)))
else:
device = 'cpu'
class PyTorchModel(nn.Module):
def __init__(self):
super().__init__()
# mnist images are (1, 28, 28) (channels, width, height)
self.layer_1 = nn.Linear(28 * 28, 128)
self.layer_2 = nn.Linear(128, 256)
self.layer_3 = nn.Linear(256, 10)
def forward(self, x):
batch_size, channels, width, height = x.size()
# (b, 1, 28, 28) -> (b, 1*28*28)
x = x.view(batch_size, -1)
x = self.layer_1(x)
x = F.relu(x)
x = self.layer_2(x)
x = F.relu(x)
x = self.layer_3(x)
x = F.softmax(x, dim=1)
return x
net = PyTorchModel().to(device)
criterion = nn.NLLLoss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-3)
for epoch in range(2):
running_loss = 0.0
for i, data in enumerate(mnist_train, 0):
inputs, labels = data
optimizer.zero_grad()
outputs = net(inputs.to(device))
loss = criterion(outputs, labels.to(device))
loss.backward()
optimizer.step()
running_loss += loss.item()
if i % 200 == 199:
print("[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 199))
running_loss = 0.0
correct = 0
total = 0
with torch.no_grad():
for data in mnist_test:
images, labels = data
outputs = net(images.to(device))
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels.to(device)).sum().item()
print("Accuracy of the network on the %d test images: %d %%" % (total, 100 * correct / total))
now = datetime.now()
date_time = now.strftime("%Y%m%d-%H%M%S")
SAVED_DIR = f"pytorch-model-{date_time}"
os.makedirs(SAVED_DIR, exist_ok=True)
torch.save(net.state_dict(), os.path.join(SAVED_DIR, "model.pt"))
print(f"We successfully saved the model in {SAVED_DIR}.")
model_class_file_content = """
import torch
from torch.nn import functional as F
from torch import nn
class PyTorchModel(nn.Module):
def __init__(self):
super().__init__()
# mnist images are (1, 28, 28) (channels, width, height)
self.layer_1 = nn.Linear(28 * 28, 128)
self.layer_2 = nn.Linear(128, 256)
self.layer_3 = nn.Linear(256, 10)
def forward(self, x):
batch_size, channels, width, height = x.size()
# (b, 1, 28, 28) -> (b, 1*28*28)
x = x.view(batch_size, -1)
x = self.layer_1(x)
x = F.relu(x)
x = self.layer_2(x)
x = F.relu(x)
x = self.layer_3(x)
x = F.softmax(x, dim=1)
return x
"""
model_class_file = open(os.path.join(SAVED_DIR, "ModelClass.py"), "w")
model_class_file.write(model_class_file_content)
model_class_file.close()
print(f"We successfully saved the model class file in {SAVED_DIR}.")
!mv $SAVED_DIR ~/phfs
!ls -l ~/phfs/$SAVED_DIR
print(f"Now, we've already prepared the ready-to-deploy model in ~/phfs/{SAVED_DIR}.")
print(f"Fill in the [Model URI] field with [phfs:///{SAVED_DIR}].")
array = np.array([[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]])
plt.imshow(array, cmap='gray')
plt.show()
%env ENDPOINT=<MODEL_DEPLOYMENT_ENDPOINT>
response=!curl -X POST $ENDPOINT \
-H 'Content-Type: application/json' \
-d '{ "data": {"ndarray": [[[[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]]] } }'
result = json.loads(response[-1])
result
print(f"The model predicts the number is {np.argmax(result['data']['ndarray'][0])}")
!pip install primehub-python-sdk
from primehub import PrimeHub, PrimeHubConfig
ph = PrimeHub(PrimeHubConfig())
if ph.is_ready():
print("PrimeHub Python SDK setup successfully")
else:
print("PrimeHub Python SDK couldn't get the information, please ensure your configuration is stored in the ~/.primehub/config.json")
ph.deployments.list()
ph.deployments.delete(ph.deployments.list()[0]["id"])
config = {
"id": f"{SAVED_DIR}",
"name": "pytorch-sdk-quickstart",
"modelImage": "infuseai/pytorch-prepackaged:v0.2.0",
"modelURI": f"phfs:///{SAVED_DIR}",
"instanceType": "cpu-1",
"replicas": 1
}
deployment = ph.deployments.create(config)
print(deployment)
ph.deployments.wait(deployment["id"])
endpoint = ph.deployments.get(deployment["id"])["endpoint"]
%env ENDPOINT=$endpoint
response=!curl -X POST $ENDPOINT \
-H 'Content-Type: application/json' \
-d '{ "data": {"ndarray": [[[[0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.32941176470588235, 0.7254901960784313, 0.6235294117647059, 0.592156862745098, 0.23529411764705882, 0.1411764705882353, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.8705882352941177, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9450980392156862, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.7764705882352941, 0.6666666666666666, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.2627450980392157, 0.4470588235294118, 0.2823529411764706, 0.4470588235294118, 0.6392156862745098, 0.8901960784313725, 0.996078431372549, 0.8823529411764706, 0.996078431372549, 0.996078431372549, 0.996078431372549, 0.9803921568627451, 0.8980392156862745, 0.996078431372549, 0.996078431372549, 0.5490196078431373, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.06666666666666667, 0.25882352941176473, 0.054901960784313725, 0.2627450980392157, 0.2627450980392157, 0.2627450980392157, 0.23137254901960785, 0.08235294117647059, 0.9254901960784314, 0.996078431372549, 0.41568627450980394, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.3254901960784314, 0.9921568627450981, 0.8196078431372549, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.08627450980392157, 0.9137254901960784, 1.0, 0.3254901960784314, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5058823529411764, 0.996078431372549, 0.9333333333333333, 0.17254901960784313, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23137254901960785, 0.9764705882352941, 0.996078431372549, 0.24313725490196078, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.7333333333333333, 0.0196078431372549, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.03529411764705882, 0.803921568627451, 0.9725490196078431, 0.22745098039215686, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.49411764705882355, 0.996078431372549, 0.7137254901960784, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.29411764705882354, 0.984313725490196, 0.9411764705882353, 0.2235294117647059, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.07450980392156863, 0.8666666666666667, 0.996078431372549, 0.6509803921568628, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.011764705882352941, 0.796078431372549, 0.996078431372549, 0.8588235294117647, 0.13725490196078433, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.14901960784313725, 0.996078431372549, 0.996078431372549, 0.30196078431372547, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.12156862745098039, 0.8784313725490196, 0.996078431372549, 0.45098039215686275, 0.00392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.5215686274509804, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.23921568627450981, 0.9490196078431372, 0.996078431372549, 0.996078431372549, 0.20392156862745098, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.996078431372549, 0.8588235294117647, 0.1568627450980392, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.4745098039215686, 0.996078431372549, 0.8117647058823529, 0.07058823529411765, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]]] } }'
result = json.loads(response[-1])
result
print(f"The model predicts the number is {np.argmax(result['data']['ndarray'][0])}")
ph.deployments.delete(deployment["id"])
| 0.859958 | 0.984291 |
```
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
data = pd.read_csv('https://raw.githubusercontent.com/PacktWorkshops/The-Data-Analysis-Workshop/master/Chapter09/Datasets/energydata_complete.csv')
data.head()
data.isnull().sum()
df1 = data.rename(columns = {
'date' : 'date_time',
'Appliances' : 'a_energy',
'lights' : 'l_energy',
'T1' : 'kitchen_temp',
'RH_1' : 'kitchen_hum',
'T2' : 'liv_temp',
'RH_2' : 'liv_hum',
'T3' : 'laun_temp',
'RH_3' : 'laun_hum',
'T4' : 'off_temp',
'RH_4' : 'off_hum',
'T5' : 'bath_temp',
'RH_5' : 'bath_hum',
'T6' : 'out_b_temp',
'RH_6' : 'out_b_hum',
'T7' : 'iron_temp',
'RH_7' : 'iron_hum',
'T8' : 'teen_temp',
'RH_8' : 'teen_hum',
'T9' : 'par_temp',
'RH_9' : 'par_hum',
'T_out' : 'out_temp',
'Press_mm_hg' : 'out_press',
'RH_out' : 'out_hum',
'Windspeed' : 'wind',
'Visibility' : 'visibility',
'Tdewpoint' : 'dew_point',
'rv1' : 'rv1',
'rv2' : 'rv2'
})
df1.head()
df1.tail()
df1.describe()
lights_box = sns.boxplot(df1.l_energy)
l = [0, 10, 20, 30, 40, 50, 60, 70]
counts = []
for i in l:
a = (df1.l_energy == i).sum()
counts.append(a)
counts
lights = sns.barplot(x = l, y = counts)
lights.set_xlabel('Energy Consumed by Lights')
lights.set_ylabel('Number of Lights')
lights.set_title('Distribution of Energy Consumed by Lights')
((df1.l_energy == 0).sum() / (df1.shape[0])) * 100
new_data = df1
new_data.drop(['l_energy'], axis = 1, inplace = True)
new_data.head()
app_box = sns.boxplot(new_data.a_energy)
out = (new_data['a_energy'] > 200).sum()
out
(out/19735) * 100
out_e = (new_data['a_energy'] > 950).sum()
out_e
(out_e/19735) * 100
energy = new_data[(new_data['a_energy'] <= 200)]
energy.describe()
new_en = energy
new_en['date_time'] = pd.to_datetime(new_en.date_time, format = '%Y-%m-%d %H:%M:%S')
new_en.head()
new_en.insert(loc = 1, column = 'month', value = new_en.date_time.dt.month)
new_en.insert(loc = 2, column = 'day', value = (new_en.date_time.dt.dayofweek)+1)
new_en.head()
import plotly.graph_objs as go
app_date = go.Scatter(x = new_en.date_time, mode = "lines", y = new_en.a_energy)
layout = go.Layout(title = 'Appliance Energy Consumed by Date', xaxis = dict(title='Date'), yaxis = dict(title='Wh'))
fig = go.Figure(data = [app_date], layout = layout)
fig.show()
app_mon = new_en.groupby(by = ['month'], as_index = False)['a_energy'].sum()
app_mon
app_mon.sort_values(by = 'a_energy', ascending = False).head()
plt.subplots(figsize = (15, 6))
am = sns.barplot(app_mon.month, app_mon.a_energy)
plt.xlabel('Month')
plt.ylabel('Energy Consumed by Appliances')
plt.title('Total Energy Consumed by Appliances per Month')
plt.show()
```
**Activity 9.02**
```
app_day = new_en.groupby(by = ['day'], as_index = False)['a_energy'].sum()
app_day
app_day.sort_values(by = 'a_energy', ascending = False)
plt.subplots(figsize = (15, 6))
ad = sns.barplot(app_day.day, app_day.a_energy)
plt.xlabel('Day of the Week')
plt.ylabel('Energy Consumed by Appliances')
plt.title('Total Energy Consumed by Appliances')
plt.show()
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
data = pd.read_csv('https://raw.githubusercontent.com/PacktWorkshops/The-Data-Analysis-Workshop/master/Chapter09/Datasets/energydata_complete.csv')
data.head()
data.isnull().sum()
df1 = data.rename(columns = {
'date' : 'date_time',
'Appliances' : 'a_energy',
'lights' : 'l_energy',
'T1' : 'kitchen_temp',
'RH_1' : 'kitchen_hum',
'T2' : 'liv_temp',
'RH_2' : 'liv_hum',
'T3' : 'laun_temp',
'RH_3' : 'laun_hum',
'T4' : 'off_temp',
'RH_4' : 'off_hum',
'T5' : 'bath_temp',
'RH_5' : 'bath_hum',
'T6' : 'out_b_temp',
'RH_6' : 'out_b_hum',
'T7' : 'iron_temp',
'RH_7' : 'iron_hum',
'T8' : 'teen_temp',
'RH_8' : 'teen_hum',
'T9' : 'par_temp',
'RH_9' : 'par_hum',
'T_out' : 'out_temp',
'Press_mm_hg' : 'out_press',
'RH_out' : 'out_hum',
'Windspeed' : 'wind',
'Visibility' : 'visibility',
'Tdewpoint' : 'dew_point',
'rv1' : 'rv1',
'rv2' : 'rv2'
})
df1.head()
df1.tail()
df1.describe()
lights_box = sns.boxplot(df1.l_energy)
l = [0, 10, 20, 30, 40, 50, 60, 70]
counts = []
for i in l:
a = (df1.l_energy == i).sum()
counts.append(a)
counts
lights = sns.barplot(x = l, y = counts)
lights.set_xlabel('Energy Consumed by Lights')
lights.set_ylabel('Number of Lights')
lights.set_title('Distribution of Energy Consumed by Lights')
((df1.l_energy == 0).sum() / (df1.shape[0])) * 100
new_data = df1
new_data.drop(['l_energy'], axis = 1, inplace = True)
new_data.head()
app_box = sns.boxplot(new_data.a_energy)
out = (new_data['a_energy'] > 200).sum()
out
(out/19735) * 100
out_e = (new_data['a_energy'] > 950).sum()
out_e
(out_e/19735) * 100
energy = new_data[(new_data['a_energy'] <= 200)]
energy.describe()
new_en = energy
new_en['date_time'] = pd.to_datetime(new_en.date_time, format = '%Y-%m-%d %H:%M:%S')
new_en.head()
new_en.insert(loc = 1, column = 'month', value = new_en.date_time.dt.month)
new_en.insert(loc = 2, column = 'day', value = (new_en.date_time.dt.dayofweek)+1)
new_en.head()
import plotly.graph_objs as go
app_date = go.Scatter(x = new_en.date_time, mode = "lines", y = new_en.a_energy)
layout = go.Layout(title = 'Appliance Energy Consumed by Date', xaxis = dict(title='Date'), yaxis = dict(title='Wh'))
fig = go.Figure(data = [app_date], layout = layout)
fig.show()
app_mon = new_en.groupby(by = ['month'], as_index = False)['a_energy'].sum()
app_mon
app_mon.sort_values(by = 'a_energy', ascending = False).head()
plt.subplots(figsize = (15, 6))
am = sns.barplot(app_mon.month, app_mon.a_energy)
plt.xlabel('Month')
plt.ylabel('Energy Consumed by Appliances')
plt.title('Total Energy Consumed by Appliances per Month')
plt.show()
app_day = new_en.groupby(by = ['day'], as_index = False)['a_energy'].sum()
app_day
app_day.sort_values(by = 'a_energy', ascending = False)
plt.subplots(figsize = (15, 6))
ad = sns.barplot(app_day.day, app_day.a_energy)
plt.xlabel('Day of the Week')
plt.ylabel('Energy Consumed by Appliances')
plt.title('Total Energy Consumed by Appliances')
plt.show()
| 0.452536 | 0.579311 |
<a href="https://colab.research.google.com/github/anadiedrichs/time-series-analysis/blob/master/parcial_2_2018_consigna.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
## Parcial Teleinformática: consignas apartado análisis de datos
**Legajo**: COMPLETE
**Nombre**: COMPLETE
### Indicaciones
* Copie este notebook en su propio google drive
* Trabaje sobre el mismo completando respuestas con bloques de texto y código dónde sea necesario
* Puede agregar todos los bloques de código que crea necesario
* Puede acceder y utilizar código brindado por la cátedra
* Puede utilizar otras librerías o formatos no dados en la cátedra (aunque no es necesario para resolver las consignas ni da puntos extras) o realizar consultas en internet.
* Las conclusiones o respuestas son suyas, no copie ni debe copiar a sus compañeros ni copie o pegue conclusiones o texto de Internet.
* Lea atentamente todas las consignas
* Complete las respuestas a las preguntas o interpretaciones de la forma más completa posible
### ENTREGA
Para confirmar la entrega realice **todos** estos pasos:
1. Compartir el notebook al mail [email protected]
2. Copiar el link de acceso al notebook en la tarea de entrega, creada en el campus virtual unidad nro 8. Para ello diríjase a Share --> get shareable link o Compartir --> obtener enlace para compartir.
3. Subir el archivo del notebook en la tarea en la tarea de entrega, creada en el campus virtual unidad nro 8.
¿Cómo descargar el notebook? Ir a File --> Download .ipynb para descargar el notebook. Comprima en formato .zip o .tar y luego suba a la plataforma campus virtual en la tarea asignada al parcial.
*Fecha y hora límite para realizar entrega: jueves 14/11/2018 hasta las 23:30 hs en el salón de clases*
### Importar dataset
#### Descarga del dataset
La fuente u origen del dataset es este enlace
https://archive.ics.uci.edu/ml/datasets/individual+household+electric+power+consumption
Ejecute los siguientes bloques de código para importar el dataset
```
from io import StringIO
from zipfile import ZipFile
from urllib.request import urlopen
URL = "https://archive.ics.uci.edu/ml/machine-learning-databases/00235/household_power_consumption.zip"
import requests, zipfile, io
r = requests.get(URL)
z = zipfile.ZipFile(io.BytesIO(r.content))
z.extractall("/content")
z.close()
!ls
# visualizar algunas líneas de un archivo
import os
f = open("/content/household_power_consumption.txt")
data = f.read()
f.close()
lines = data.split('\n')
header = lines[0].split(',')
lines = lines[1:]
print(header)
print(len(lines))
from numpy import nan
from numpy import isnan
from pandas import read_csv
from pandas import to_numeric
dataset = read_csv('household_power_consumption.txt', sep=';', header=0, low_memory=False, infer_datetime_format=True, parse_dates={'datetime':[0,1]}, index_col=['datetime'])
```
### Describir dataset
Evaluamos el tamaño del dataset: ¿Cuántas variables y cuántas mediciones tiene el dataset? Ejecute en el bloque de código las sentencias para averiguarlo
**Respuesta:**
```
```
¿Cuáles son las variables (nombres y de una breve descripción?)
Escriba sus nombres y de una **completa** descripción de cada una de las variables (campos o columnas).
**Respuesta**
### Valores perdidos
¿Cómo están etiquetados los valores perdidos en el dataset? Ayuda visualizar el dataset con el siguiente bloque de código. Indique con qué caracter se marcan los valores perdidos.
**Respuesta:**
```
dataset.iloc[6832:6900,]
```
¿Cuántas filas del dataset (mediciones) tienen valores perdidos? Modifique la siguiente sentencia de código que recupera todas las filas con el valor perdido para dar la respuesta.
**Respuesta**
```
dataset.loc[dataset["Sub_metering_1"] == "CARACTER"].shape
```
**Reemplace el texto CARACTER por el caracter correcto que indica valor perdido y ejecute.**
```
# INDICAmos cuales son valores perdidos
dataset.replace('CARACTER', nan, inplace=True)
```
La función fill_missing reemplaza los valores perdidos. Ejecute el siguiente bloque de código.
```
# funcion para reemplazar valores perdidos
def fill_missing(values):
one_day = 60 * 24
for row in range(values.shape[0]):
for col in range(values.shape[1]):
if isnan(values[row, col]):
values[row, col] = values[row - one_day, col]
dataset = dataset.astype('float32')
fill_missing(dataset.values)
```
Explique que es lo que realiza o qué criterio toma para reemplazar los valores perdidos la función *fill_missing*.
**Respuesta**
### Interpretaciones
El siguiente código agrega una columna nueva llamada submetering_4. ¿Qué es lo que representa?
sub_metering_4 = (global_active_power * 1000 / 60) - (sub_metering_1 + sub_metering_2 + sub_metering_3)
**Respuesta**
```
values = dataset.values
dataset['sub_metering_4'] = (values[:,0] * 1000 / 60) - (values[:,4] + values[:,5] + values[:,6])
```
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
```
# line plots
from pandas import read_csv
from matplotlib import pyplot
# line plot for each variable
pyplot.figure()
for i in range(len(dataset.columns)):
pyplot.subplot(len(dataset.columns), 1, i+1)
name = dataset.columns[i]
pyplot.plot(dataset[name])
pyplot.title(name, y=0)
pyplot.show()
```
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
```
# daily line plots
from pandas import read_csv
from matplotlib import pyplot
# plot active power for each year
days = [x for x in range(1, 10)]
pyplot.figure()
for i in range(len(days)):
# prepare subplot
ax = pyplot.subplot(len(days), 1, i+1)
# determine the day to plot
day = '2007-01-' + str(days[i])
# get all observations for the day
result = dataset[day]
# plot the active power for the day
pyplot.plot(result['Global_active_power'])
# add a title to the subplot
pyplot.title(day, y=0, loc='left')
pyplot.show()
```
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
```
# monthly histogram plots
from pandas import read_csv
from matplotlib import pyplot
# plot active power for each year
months = [x for x in range(1, 13)]
pyplot.figure()
for i in range(len(months)):
# prepare subplot
ax = pyplot.subplot(len(months), 1, i+1)
# determine the month to plot
month = '2007-' + str(months[i])
# get all observations for the month
result = dataset[month]
# plot the active power for the month
result['Global_active_power'].hist(bins=100)
# zoom in on the distribution
ax.set_xlim(0, 5)
# add a title to the subplot
pyplot.title(month, y=0, loc='right')
pyplot.show()
```
¿Qué nos muestran los siguientes gráficos? Ejecute los bloques de código. Explique e interprete.
**Respuesta**
```
dataset.iloc[0:50000,4:7].plot()
```
### PARA GLOBAL Procesamiento de la señal
Evalue un par de señales con aplicando métodos matemáticos vistos (FFT y otros) para realizar un análisis preciso. Cree los bloques de código necesarios y muestre el gráfico con el resultado. Escriba conclusiones e interpretación de lo elaborado.
```
```
|
github_jupyter
|
from io import StringIO
from zipfile import ZipFile
from urllib.request import urlopen
URL = "https://archive.ics.uci.edu/ml/machine-learning-databases/00235/household_power_consumption.zip"
import requests, zipfile, io
r = requests.get(URL)
z = zipfile.ZipFile(io.BytesIO(r.content))
z.extractall("/content")
z.close()
!ls
# visualizar algunas líneas de un archivo
import os
f = open("/content/household_power_consumption.txt")
data = f.read()
f.close()
lines = data.split('\n')
header = lines[0].split(',')
lines = lines[1:]
print(header)
print(len(lines))
from numpy import nan
from numpy import isnan
from pandas import read_csv
from pandas import to_numeric
dataset = read_csv('household_power_consumption.txt', sep=';', header=0, low_memory=False, infer_datetime_format=True, parse_dates={'datetime':[0,1]}, index_col=['datetime'])
```
¿Cuáles son las variables (nombres y de una breve descripción?)
Escriba sus nombres y de una **completa** descripción de cada una de las variables (campos o columnas).
**Respuesta**
### Valores perdidos
¿Cómo están etiquetados los valores perdidos en el dataset? Ayuda visualizar el dataset con el siguiente bloque de código. Indique con qué caracter se marcan los valores perdidos.
**Respuesta:**
¿Cuántas filas del dataset (mediciones) tienen valores perdidos? Modifique la siguiente sentencia de código que recupera todas las filas con el valor perdido para dar la respuesta.
**Respuesta**
**Reemplace el texto CARACTER por el caracter correcto que indica valor perdido y ejecute.**
La función fill_missing reemplaza los valores perdidos. Ejecute el siguiente bloque de código.
Explique que es lo que realiza o qué criterio toma para reemplazar los valores perdidos la función *fill_missing*.
**Respuesta**
### Interpretaciones
El siguiente código agrega una columna nueva llamada submetering_4. ¿Qué es lo que representa?
sub_metering_4 = (global_active_power * 1000 / 60) - (sub_metering_1 + sub_metering_2 + sub_metering_3)
**Respuesta**
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
¿Qué nos muestra el siguiente gráfico? Ejecute el siguiente bloque de código. Explique e interprete.
**Respuesta**
¿Qué nos muestran los siguientes gráficos? Ejecute los bloques de código. Explique e interprete.
**Respuesta**
### PARA GLOBAL Procesamiento de la señal
Evalue un par de señales con aplicando métodos matemáticos vistos (FFT y otros) para realizar un análisis preciso. Cree los bloques de código necesarios y muestre el gráfico con el resultado. Escriba conclusiones e interpretación de lo elaborado.
| 0.348867 | 0.925365 |
```
import numpy as np
import matplotlib.pyplot as plt
import keras
import tensorflow as tf
from imageai.Detection import ObjectDetection
import os
```
### object detection source:
https://github.com/OlafenwaMoses/ImageAI/blob/master/imageai/Detection/README.md#objectextraction
```
img_b = 'h_40_v_-127.jpg'
img_a = 'detected_' + img_b
```
### load a pre-trained model and excuting obj detection
```
execution_path = os.getcwd()
detector = ObjectDetection()
detector.setModelTypeAsRetinaNet()
detector.setModelPath(os.path.join(execution_path , "resnet50_v2.1.0.h5"))
```
### load model, normal mode
```
detector.loadModel()
```
### mode: "normal"(default), "fast", "faster" , "fastest" and "flash".
```
detector.loadModel(detection_speed="fast")
```
### run detection, full detection
```
detections = detector.detectObjectsFromImage(input_image=os.path.join(execution_path ,img_b),
output_image_path=os.path.join(execution_path , img_a))
```
### Specify which object type need to be detect, optional
#### customized detection
#### hide name on output image
#### hide probability
#### availiable object type:
person, bicycle, car, motorcycle, airplane, bus, train, truck, boat, traffic light, fire hydrant, stop_sign,
parking meter, bench, bird, cat, dog, horse, sheep, cow, elephant, bear, zebra,
giraffe, backpack, umbrella, handbag, tie, suitcase, frisbee, skis, snowboard,
sports ball, kite, baseball bat, baseball glove, skateboard, surfboard, tennis racket,
bottle, wine glass, cup, fork, knife, spoon, bowl, banana, apple, sandwich, orange,
broccoli, carrot, hot dog, pizza, donot, cake, chair, couch, potted plant, bed,
dining table, toilet, tv, laptop, mouse, remote, keyboard, cell phone, microwave, oven,
toaster, sink, refrigerator, book, clock, vase, scissors, teddy bear, hair dryer, toothbrush.
```
custom_objects = detector.CustomObjects(person=True)
detections = detector.detectObjectsFromImage(custom_objects = custom_objects,
input_image = os.path.join(execution_path ,img_b),
output_image_path = os.path.join(execution_path , img_a),
minimum_percentage_probability = 30, display_object_name=False,
display_percentage_probability=False)
```
### Print detected items and probability
```
for eachObject in detections:
print(eachObject["name"] , " : " , eachObject["percentage_probability"] )
```
|
github_jupyter
|
import numpy as np
import matplotlib.pyplot as plt
import keras
import tensorflow as tf
from imageai.Detection import ObjectDetection
import os
img_b = 'h_40_v_-127.jpg'
img_a = 'detected_' + img_b
execution_path = os.getcwd()
detector = ObjectDetection()
detector.setModelTypeAsRetinaNet()
detector.setModelPath(os.path.join(execution_path , "resnet50_v2.1.0.h5"))
detector.loadModel()
detector.loadModel(detection_speed="fast")
detections = detector.detectObjectsFromImage(input_image=os.path.join(execution_path ,img_b),
output_image_path=os.path.join(execution_path , img_a))
custom_objects = detector.CustomObjects(person=True)
detections = detector.detectObjectsFromImage(custom_objects = custom_objects,
input_image = os.path.join(execution_path ,img_b),
output_image_path = os.path.join(execution_path , img_a),
minimum_percentage_probability = 30, display_object_name=False,
display_percentage_probability=False)
for eachObject in detections:
print(eachObject["name"] , " : " , eachObject["percentage_probability"] )
| 0.432063 | 0.698284 |
## Overview
This is a technique outlined in [Leon A. Gatys' paper, A Neural Algorithm of Artistic Style](https://arxiv.org/abs/1508.06576), which is a great read, and you should definitely check it out.
Neural style transfer is an optimization technique used to take three images, a **content** image, a **style reference** image (such as an artwork by a famous painter), and the **input** image you want to style -- and blend them together such that the input image is transformed to look like the content image, but “painted” in the style of the style image.
```
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['figure.figsize'] = (10,10)
mpl.rcParams['axes.grid'] = False
import numpy as np
from PIL import Image
import time
import functools
import tensorflow as tf
from tensorflow.python.keras.preprocessing import image as kp_image
from tensorflow.python.keras import models
from tensorflow.python.keras import losses
from tensorflow.python.keras import layers
from tensorflow.python.keras import backend as K
## Enable eager execution
tf.enable_eager_execution()
tf.device('/gpu:0')
print(f'Eager execution: {tf.executing_eagerly()}')
#assign images path
content_path= 'panda.jpg'
style_path = 'picasso_animal.jpg'
def load_img(path):
max_dim = 512
img = Image.open(path)
long = max(img.size)
scale = max_dim/long
img = img.resize((round(img.size[0]*scale), round(img.size[1]*scale)), Image.ANTIALIAS)
img = kp_image.img_to_array(img)
img = np.expand_dims(img, axis=0)
return img
def imshow(img,title=''):
if len(img.shape)>3:
out = np.squeeze(img, axis=0)
out = out.astype('uint8')
plt.imshow(out)
plt.title = title
content_image = load_img(content_path)
style_image = load_img(style_path)
plt.subplot(1,2,1)
imshow(content_image, 'Content Image')
plt.subplot(1,2,2)
imshow(style_image, 'Style Image')
def load_process_img(path):
img = load_img(path)
img = tf.keras.applications.vgg19.preprocess_input(img)
return img
def deprocess_img(processed_img):
x = processed_img.copy()
if len(x.shape) == 4:
x = np.squeeze(x, 0)
assert len(x.shape) == 3, ("Input to deprocess image must be an image of "
"dimension [1, height, width, channel] or [height, width, channel]")
if len(x.shape) != 3:
raise ValueError("Invalid input to deprocessing image")
# perform the inverse of the preprocessiing step
x[:, :, 0] += 103.939
x[:, :, 1] += 116.779
x[:, :, 2] += 123.68
x = x[:, :, ::-1]
x = np.clip(x, 0, 255).astype('uint8')
return x
vgg_model_test = tf.keras.applications.VGG19()
vgg_model_test.summary()
# we will select intermediate layer to perform loss function
content_layers = ['block5_conv2']
style_layers = ['block1_conv1',
'block2_conv1',
'block3_conv1',
'block4_conv1',
'block5_conv1'
]
def get_model_with_selected_layers():
vgg_model = tf.keras.applications.VGG19(include_top=False, weights='imagenet')
style_outputs = [ vgg_model.get_layer(name).output for name in style_layers]
content_outputs = [vgg_model.get_layer(name).output for name in content_layers]
model_outputs = style_outputs + content_outputs
return models.Model(vgg_model.input, model_outputs )
def get_content_loss(gen_content, target):
return tf.reduce_sum(tf.square(gen_content - target))
def gram_matrix(input_tensor):
channel = input_tensor.shape[-1]
a = tf.reshape(input_tensor, [-1,channel])
n = tf.shape(a)[0]
gram = tf.matmul(a,a, transpose_b = True)
return gram/tf.cast(n, float32)
def get_style_cost(gen_style, gram_target):
return tf.reduce_sum(tf.square(gram_matrix(gen_style) - gram_target))
def feature_extractor(model, content_path, style_path):
content_image = load_process_img(content_path)
style_image = load_process_img(style_path)
style_outputs = model(style_image)
content_outputs = model(content_image)
style_features = [layer[0] for layer in style_outputs[:len(style_layers)]]
content_features = [layer[0] for layer in content_outputs[len(style_layers):]]
return style_features, content_features
def compute_loss(model, loss_weights, gen_image, gram_style_features, content_features):
style_weight, content_weight = loss_weights
outputs = model(gen_image)
style_output_features = outputs[:len(style_layers)]
content_output_features = outputs[len(style_layers)]
style_loss = 0.
content_loss = 0.
style_layers_weight = 1.0/float(len(style_layers))
for gen_style, gram_target in zip(style_output_features,gram_style_features):
style_loss += style_layers_weight * get_style_cost(gen_style[0], gram_target)
content_layers_weights = 1.0/float(len(content_layers))
for gen_content, target in zip(content_output_features,content_features):
content_loss += content_layers_weights * get_content_loss(gen_content[0], target)
loss = style_score + content_score
return loss, style_loss, content_loss
def compute_grads(cfg):
with tf.GradientTape() as tape:
all_loss = compute_loss(**cfg)
# Compute gradients wrt input image
total_loss = all_loss[0]
return tape.gradient(total_loss, cfg['init_image']), all_loss
import IPython.display
def run_style_transfer(content_path,
style_path,
num_iterations=1000,
content_weight=1e3,
style_weight=1e-2):
# We don't need to (or want to) train any layers of our model, so we set their
# trainable to false.
model = get_model_with_selected_layers()
for layer in model.layers:
layer.trainable = False
# Get the style and content feature representations (from our specified intermediate layers)
style_features, content_features = feature_extractor(model, content_path, style_path)
gram_style_features = [gram_matrix(style_feature) for style_feature in style_features]
# Set initial image
init_image = load_and_process_img(content_path)
init_image = tf.Variable(init_image, dtype=tf.float32)
# Create our optimizer
opt = tf.train.AdamOptimizer(learning_rate=5, beta1=0.99, epsilon=1e-1)
# For displaying intermediate images
iter_count = 1
# Store our best result
best_loss, best_img = float('inf'), None
# Create a nice config
loss_weights = (style_weight, content_weight)
cfg = {
'model': model,
'loss_weights': loss_weights,
'init_image': init_image,
'gram_style_features': gram_style_features,
'content_features': content_features
}
# For displaying
num_rows = 2
num_cols = 5
display_interval = num_iterations/(num_rows*num_cols)
start_time = time.time()
global_start = time.time()
norm_means = np.array([103.939, 116.779, 123.68])
min_vals = -norm_means
max_vals = 255 - norm_means
imgs = []
for i in range(num_iterations):
grads, all_loss = compute_grads(cfg)
loss, style_score, content_score = all_loss
opt.apply_gradients([(grads, init_image)])
clipped = tf.clip_by_value(init_image, min_vals, max_vals)
init_image.assign(clipped)
end_time = time.time()
if loss < best_loss:
# Update best loss and best image from total loss.
best_loss = loss
best_img = deprocess_img(init_image.numpy())
if i % display_interval== 0:
start_time = time.time()
# Use the .numpy() method to get the concrete numpy array
plot_img = init_image.numpy()
plot_img = deprocess_img(plot_img)
imgs.append(plot_img)
IPython.display.clear_output(wait=True)
IPython.display.display_png(Image.fromarray(plot_img))
print('Iteration: {}'.format(i))
print('Total loss: {:.4e}, '
'style loss: {:.4e}, '
'content loss: {:.4e}, '
'time: {:.4f}s'.format(loss, style_score, content_score, time.time() - start_time))
print('Total time: {:.4f}s'.format(time.time() - global_start))
IPython.display.clear_output(wait=True)
plt.figure(figsize=(14,4))
for i,img in enumerate(imgs):
plt.subplot(num_rows,num_cols,i+1)
plt.imshow(img)
plt.xticks([])
plt.yticks([])
return best_img, best_loss
best, best_loss = run_style_transfer(content_path,
style_path, num_iterations=100)
def show_results(best_img, content_path, style_path, show_large_final=True):
plt.figure(figsize=(10, 5))
content = load_img(content_path)
style = load_img(style_path)
plt.subplot(1, 2, 1)
imshow(content, 'Content Image')
plt.subplot(1, 2, 2)
imshow(style, 'Style Image')
if show_large_final:
plt.figure(figsize=(10, 10))
plt.imshow(best_img)
plt.title('Output Image')
plt.show()
show_results(best, content_path, style_path)
```
|
github_jupyter
|
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['figure.figsize'] = (10,10)
mpl.rcParams['axes.grid'] = False
import numpy as np
from PIL import Image
import time
import functools
import tensorflow as tf
from tensorflow.python.keras.preprocessing import image as kp_image
from tensorflow.python.keras import models
from tensorflow.python.keras import losses
from tensorflow.python.keras import layers
from tensorflow.python.keras import backend as K
## Enable eager execution
tf.enable_eager_execution()
tf.device('/gpu:0')
print(f'Eager execution: {tf.executing_eagerly()}')
#assign images path
content_path= 'panda.jpg'
style_path = 'picasso_animal.jpg'
def load_img(path):
max_dim = 512
img = Image.open(path)
long = max(img.size)
scale = max_dim/long
img = img.resize((round(img.size[0]*scale), round(img.size[1]*scale)), Image.ANTIALIAS)
img = kp_image.img_to_array(img)
img = np.expand_dims(img, axis=0)
return img
def imshow(img,title=''):
if len(img.shape)>3:
out = np.squeeze(img, axis=0)
out = out.astype('uint8')
plt.imshow(out)
plt.title = title
content_image = load_img(content_path)
style_image = load_img(style_path)
plt.subplot(1,2,1)
imshow(content_image, 'Content Image')
plt.subplot(1,2,2)
imshow(style_image, 'Style Image')
def load_process_img(path):
img = load_img(path)
img = tf.keras.applications.vgg19.preprocess_input(img)
return img
def deprocess_img(processed_img):
x = processed_img.copy()
if len(x.shape) == 4:
x = np.squeeze(x, 0)
assert len(x.shape) == 3, ("Input to deprocess image must be an image of "
"dimension [1, height, width, channel] or [height, width, channel]")
if len(x.shape) != 3:
raise ValueError("Invalid input to deprocessing image")
# perform the inverse of the preprocessiing step
x[:, :, 0] += 103.939
x[:, :, 1] += 116.779
x[:, :, 2] += 123.68
x = x[:, :, ::-1]
x = np.clip(x, 0, 255).astype('uint8')
return x
vgg_model_test = tf.keras.applications.VGG19()
vgg_model_test.summary()
# we will select intermediate layer to perform loss function
content_layers = ['block5_conv2']
style_layers = ['block1_conv1',
'block2_conv1',
'block3_conv1',
'block4_conv1',
'block5_conv1'
]
def get_model_with_selected_layers():
vgg_model = tf.keras.applications.VGG19(include_top=False, weights='imagenet')
style_outputs = [ vgg_model.get_layer(name).output for name in style_layers]
content_outputs = [vgg_model.get_layer(name).output for name in content_layers]
model_outputs = style_outputs + content_outputs
return models.Model(vgg_model.input, model_outputs )
def get_content_loss(gen_content, target):
return tf.reduce_sum(tf.square(gen_content - target))
def gram_matrix(input_tensor):
channel = input_tensor.shape[-1]
a = tf.reshape(input_tensor, [-1,channel])
n = tf.shape(a)[0]
gram = tf.matmul(a,a, transpose_b = True)
return gram/tf.cast(n, float32)
def get_style_cost(gen_style, gram_target):
return tf.reduce_sum(tf.square(gram_matrix(gen_style) - gram_target))
def feature_extractor(model, content_path, style_path):
content_image = load_process_img(content_path)
style_image = load_process_img(style_path)
style_outputs = model(style_image)
content_outputs = model(content_image)
style_features = [layer[0] for layer in style_outputs[:len(style_layers)]]
content_features = [layer[0] for layer in content_outputs[len(style_layers):]]
return style_features, content_features
def compute_loss(model, loss_weights, gen_image, gram_style_features, content_features):
style_weight, content_weight = loss_weights
outputs = model(gen_image)
style_output_features = outputs[:len(style_layers)]
content_output_features = outputs[len(style_layers)]
style_loss = 0.
content_loss = 0.
style_layers_weight = 1.0/float(len(style_layers))
for gen_style, gram_target in zip(style_output_features,gram_style_features):
style_loss += style_layers_weight * get_style_cost(gen_style[0], gram_target)
content_layers_weights = 1.0/float(len(content_layers))
for gen_content, target in zip(content_output_features,content_features):
content_loss += content_layers_weights * get_content_loss(gen_content[0], target)
loss = style_score + content_score
return loss, style_loss, content_loss
def compute_grads(cfg):
with tf.GradientTape() as tape:
all_loss = compute_loss(**cfg)
# Compute gradients wrt input image
total_loss = all_loss[0]
return tape.gradient(total_loss, cfg['init_image']), all_loss
import IPython.display
def run_style_transfer(content_path,
style_path,
num_iterations=1000,
content_weight=1e3,
style_weight=1e-2):
# We don't need to (or want to) train any layers of our model, so we set their
# trainable to false.
model = get_model_with_selected_layers()
for layer in model.layers:
layer.trainable = False
# Get the style and content feature representations (from our specified intermediate layers)
style_features, content_features = feature_extractor(model, content_path, style_path)
gram_style_features = [gram_matrix(style_feature) for style_feature in style_features]
# Set initial image
init_image = load_and_process_img(content_path)
init_image = tf.Variable(init_image, dtype=tf.float32)
# Create our optimizer
opt = tf.train.AdamOptimizer(learning_rate=5, beta1=0.99, epsilon=1e-1)
# For displaying intermediate images
iter_count = 1
# Store our best result
best_loss, best_img = float('inf'), None
# Create a nice config
loss_weights = (style_weight, content_weight)
cfg = {
'model': model,
'loss_weights': loss_weights,
'init_image': init_image,
'gram_style_features': gram_style_features,
'content_features': content_features
}
# For displaying
num_rows = 2
num_cols = 5
display_interval = num_iterations/(num_rows*num_cols)
start_time = time.time()
global_start = time.time()
norm_means = np.array([103.939, 116.779, 123.68])
min_vals = -norm_means
max_vals = 255 - norm_means
imgs = []
for i in range(num_iterations):
grads, all_loss = compute_grads(cfg)
loss, style_score, content_score = all_loss
opt.apply_gradients([(grads, init_image)])
clipped = tf.clip_by_value(init_image, min_vals, max_vals)
init_image.assign(clipped)
end_time = time.time()
if loss < best_loss:
# Update best loss and best image from total loss.
best_loss = loss
best_img = deprocess_img(init_image.numpy())
if i % display_interval== 0:
start_time = time.time()
# Use the .numpy() method to get the concrete numpy array
plot_img = init_image.numpy()
plot_img = deprocess_img(plot_img)
imgs.append(plot_img)
IPython.display.clear_output(wait=True)
IPython.display.display_png(Image.fromarray(plot_img))
print('Iteration: {}'.format(i))
print('Total loss: {:.4e}, '
'style loss: {:.4e}, '
'content loss: {:.4e}, '
'time: {:.4f}s'.format(loss, style_score, content_score, time.time() - start_time))
print('Total time: {:.4f}s'.format(time.time() - global_start))
IPython.display.clear_output(wait=True)
plt.figure(figsize=(14,4))
for i,img in enumerate(imgs):
plt.subplot(num_rows,num_cols,i+1)
plt.imshow(img)
plt.xticks([])
plt.yticks([])
return best_img, best_loss
best, best_loss = run_style_transfer(content_path,
style_path, num_iterations=100)
def show_results(best_img, content_path, style_path, show_large_final=True):
plt.figure(figsize=(10, 5))
content = load_img(content_path)
style = load_img(style_path)
plt.subplot(1, 2, 1)
imshow(content, 'Content Image')
plt.subplot(1, 2, 2)
imshow(style, 'Style Image')
if show_large_final:
plt.figure(figsize=(10, 10))
plt.imshow(best_img)
plt.title('Output Image')
plt.show()
show_results(best, content_path, style_path)
| 0.631594 | 0.850096 |

# Principal Component Analysis
This Jupyter notebook has been written to partner with the [Principal components blog post](https://tinhatben.com/2016/04/13/principal-component-analysis/)
Copyright (c) 2016, tinhatben
All rights reserved.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions are met:
* Redistributions of source code must retain the above copyright notice, this
list of conditions and the following disclaimer.
* Redistributions in binary form must reproduce the above copyright notice,
this list of conditions and the following disclaimer in the documentation
and/or other materials provided with the distribution.
* Neither the name of tinhatben nor the names of its
contributors may be used to endorse or promote products derived from
this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE
DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE
FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL
DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR
SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER
CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE
OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
```
# imports
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
```
-----------------------------------------------------------------------------------------------------------------------
-----------------------------------------------------------------------------------------------------------------------
# Components of PCA
## Mean
In this example we will show an implementation of an equation to calculate the mean, as well as using Python and numpy's interal functions. We recall from the blog post:
$$ \large{\mu = \frac{1}{n}\sum_{k=1}^n x_k }$$
The numpy documentation for numpy.mean can be found [here](https://docs.scipy.org/doc/numpy/reference/generated/numpy.mean.html)
```
# Calculating the mean by hand on some very simple data
very_basic_data = np.array([1,2,3,4,5])
very_basic_data_2d = np.array([[1,2,3,4,5],[6,7,8,9,10]])
print("Very basic data: %s" % very_basic_data.__str__())
print("Very basic data 2D:\n %s\n" % very_basic_data_2d.__str__())
n = very_basic_data.shape[0]
n_2d = very_basic_data_2d.shape[1]
mu = (np.sum(very_basic_data)) / n
mu_2d = (np.sum(very_basic_data_2d, axis=1)) / n_2d
print("----Calculating By Hand----")
print("\nThe mean of very_basic data is : %0.2f" % mu )
print("The mean of very_basic data - 2D is : %0.2f and %0.2f\n" % (mu_2d[0], mu_2d[1]) )
print("----Calculating Using NumPy----")
# Calculating using numpy
mu = np.mean(very_basic_data)
mu_2d = np.mean(very_basic_data_2d, axis=1)
print("\nThe mean of very_basic data is (calculated using NumPy): %0.2f" % mu )
```
# Standard Deviation
Recall that he standard deviation is:
$$ \large{\sigma = \sqrt{\frac{\sum_{k=1}^n (x_k - \mu)^2}{n - 1}}}$$
The numpy documentation to the standard deviation (numpy.std) can be found [here](https://docs.scipy.org/doc/numpy/reference/generated/numpy.std.html)
```
# Add some noise to very_basic_data to ensure
from random import randint
very_basic_data *= randint(1,5)
very_basic_data_2d[0,:] *= randint(1,5)
very_basic_data_2d[1,:] *= randint(1,5)
mu = np.mean(very_basic_data)
mu_2d = np.mean(very_basic_data_2d, axis=1)
print("\n" + 10 *"----" + "1D Data" + 10*"----")
print("The very basic data set with some random noise: %s" % str(very_basic_data))
print("The new mean (mu) = %0.2f" % mu)
# Using the very basic data
sigma_squared = np.sum((very_basic_data - mu) ** 2) / (n - 1)
print("Variance (sigma^2) = %0.2f" % sigma_squared)
# Standard deviation
sigma = np.sqrt(sigma_squared)
print("Standard deviation (sigma) = %0.2f" % sigma)
# You can also calculate the standard deviation using numpy
sigma = np.std(very_basic_data, ddof = 1) # ddof = 1, is used to ensure the denominator used is n - 1, not n
print("Standard deviation using NumPy (sigma) = %0.2f" % sigma)
print("\n" + 10 *"----" + "2D Data" + 10*"----")
print("The very basic data set (2D) with some random noise: %s" % str(very_basic_data_2d))
print("The new 2D mean (mu) = %0.2f and %0.2f" % (mu_2d[0], mu_2d[1]))
# Using the very basic data
sigma_squared_2d = np.zeros(2)
sigma_squared_2d[0] = np.sum((very_basic_data_2d[0,:] - mu_2d[0]) ** 2) / (n_2d - 1)
sigma_squared_2d[1] = np.sum((very_basic_data_2d[1,:] - mu_2d[1]) ** 2) / (n_2d - 1)
print("Variance (sigma^2) = %0.2f and %0.2f" % (sigma_squared_2d[0], sigma_squared_2d[1]))
# Standard deviation
sigma_2d = np.sqrt(sigma_squared_2d)
print("Standard deviation (sigma) = %0.2f and %0.2f" % (sigma_2d[0], sigma_2d[1]))
# You can also calculate the standard deviation using numpy
sigma_2d = np.std(very_basic_data_2d, ddof = 1, axis=1)
print("Standard deviation using NumPy (sigma) = %0.2f and %0.2f" % (sigma_2d[0], sigma_2d[1]))
print("\n\n**Note: The variance calculated for the 2D data sets, considers each row or array as an independant data set")
```
# Covariance
The covariance of X and Y is defined as:
$$\large{cov(X,Y) = \frac{\sum_{k=1}^n(X_k - \mu_X)(Y_k - \mu_Y)}{n-1}}$$
At this point we will introduce a new fake data set. This is a set of sample data I <i>made up</i>, the data consists of two columns and 10 rows. The first column represents the number of coffees consumed and the second column represents pages of a PhD thesis written.
```
# Defining the data
coffee_v_thesis = np.array([
[0, 2.94],
[2.3, 5.45],
[1.2, 5.41],
[3.5, 12.10],
[5.1, 20.75],
[5.2, 21.48],
[10, 31.60],
[10.2, 40.84],
[6.2, 15.12],
])
# Visualising the data
plt.figure(figsize=(8,8))
plt.scatter(x=coffee_v_thesis[:,0], y=coffee_v_thesis[:,1], color="#7a7a7a", s=30)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis")
```
Where P is the number of pages written and C is the number of coffees consumed: computing
$$\large{cov(P, C)}$$
The documentation for `numpy.cov` can be found [here](https://docs.scipy.org/doc/numpy/reference/generated/numpy.cov.html)
```
C = coffee_v_thesis[:,0] # Select only the first column for coffee
P = coffee_v_thesis[:,1] # Second column for pages
n = coffee_v_thesis[:,0].shape[0] # Number of elements in the samples
mean_C = np.mean(C)
mean_P = np.mean(P)
cov_P_C = np.sum((C - mean_C) * (P - mean_P)) / (n - 1)
print ("cov(P,C) = %0.2f" % cov_P_C)
# Computing the covariance using numpy
# The first row, second element of the matrix corresponds to cov(P, C)
cov_P_C = np.cov(coffee_v_thesis[:,0], coffee_v_thesis[:,1])[0][1]
print ("cov(P,C) (calculated using Numpy = %0.2f" % cov_P_C)
print("**Notice the positive covariance, more coffee = more thesis pages!")
```
## Covariance matrix
$$\large{\overline{cov} = \begin{bmatrix}
cov(X, X) & cov(X,Y) & cov(X,Z)\\
cov(Y, X) & cov(Y, Y) & cov(Y, Z)\\
cov(Z, X) & cov(Z, Y) & cov(Z, Z)
\end{bmatrix}}$$
Computing the covariance matrix of `coffee_v_thesis`. Notice the symmetry of the matrix
```
cov_P_C = np.cov(coffee_v_thesis[:,0], coffee_v_thesis[:,1])
print("The covariance matrix of coffee_v_thesis:\n%s" % cov_P_C.__str__())
```
## Eigenvalues and Eigenvectors
Calculate the eigenvalues and eigevectors of the covariance matrix `cov_P_C`
There are a couple of different ways of calculating the eigenvalues and eigenvectors. The most straight-forward is to use `numpy.linalg.eig`. The documentation for numpy.linalg.eig can be found [here](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.eig.html#numpy.linalg.eig).
```
eigenvalues, eigenvectors = np.linalg.eig(cov_P_C)
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
```
Another way of calculating the eigenvalues and eigenvectors is to use the singular value decomposition `numpy.linalg.svd` function within numpy (click [here](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.svd.html) for the documentation). One convenient difference compared to `numpy.linalg.eig` is that singular value decomposition returnes the eigenvalues in ascending order which is useful to PCA
The function `numpy.linalg.svd` takes a matrix `a` and returns two matrices `u` and `v` and an array of scalars `s` such that:
`u * np.diag(s) * v = a`
$$ a = U S V^T $$
Where `np.diag` computes the diagonal matrix of `s`.
The eigenvalues of the matrix `a` can be calculated as a multiple of `S`.
The eigenvectors of the matrix `a` for the eigenvalues, correspond to the rows of `V` of the columns of `U`
```
U, S, V = np.linalg.svd(cov_P_C, full_matrices=False)
eigenvalues = S
eigevectors = U
# You can check the decomposition occurred correctly if the following statement returns True
print("Correct transform: ", np.allclose(cov_P_C, np.dot(U, np.dot(np.diag(S), V))))
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
```
-----------------------------------------------------------------------------------------------------------------------
-----------------------------------------------------------------------------------------------------------------------
# Executing PCA
## 1. Subtract the means
This ensures that the data is centered around the origin, looking at the graph below you can see the centre at (0,0)
```
# Make a copy of the data
X = np.copy(coffee_v_thesis)
x_mean, y_mean = np.mean(X, axis=0)
X[:,0] -= x_mean
X[:,1] -= y_mean
x_mean, y_mean = np.mean(X, axis=0)
# Confirm the data is now centered about the mean i.e. the mean is close to 0
print("Confirm the data is now centered about the mean: mu_x = %0.2f, mu_y = %0.2f" % (x_mean, y_mean))
# Visualising the data
plt.figure(figsize=(8,8))
plt.scatter(x=X[:,0], y=X[:,1], color="#7a7a7a", s=30)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis (mean adjusted)")
```
## 2. Calculate the Covariance Matrix
Calculate the covariance using the mean adjusted data
```
cov_P_C = np.cov(X[:,0], X[:,1])
print("Covariance of mean adjusted data\n%s" % cov_P_C.__str__())
```
## 3. Calculate the eigenvalues and eigenvectors
Calculate the eigenvalues and eigenvectors of the covariance matrix. We will use the singular value decomposition `np.linalg.svd` function to ensure the eigenvalue are ordered from largest to smallest
```
U, S, V = np.linalg.svd(cov_P_C, full_matrices=False)
eigenvalues = S
eigevectors = U
# You can check the decomposition occurred correctly if the following statement returns True
print("Correct transform: %s" % str(np.allclose(cov_P_C, np.dot(U, np.dot(np.diag(S), V)))))
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
```
Determine the percentage of the variance represented by each eigenvalue
```
# Calculate the total variance
V_t = np.sum(eigenvalues)
print("Total variance V_t = %0.4f" % V_t)
# Determine the proportion
V_prop = eigenvalues / V_t
print("Proportion of variance V_prop = %s" % V_prop.__str__())
# Determine the proportion represented by the principal component
print("Proportion of variance represented by principal component %0.2f %%" % (V_prop[0] * 100))
```
We will choose to use eigenvalues an eigenvectors that represent 98% of variance.
$$\large{P \subset E \ni
P = E[..t]}$$
In this example it is only the principal component that provides sufficient variance. One option for PCA is to select only the principal component as it represents the most variance for any single eigenvalue.
```
f_v = 0.99 # Proportion of total variation 98%
#Select only the components that account for 98% of the variance
cum_sum = np.cumsum(eigenvalues)
selected_eigenvalues = np.nonzero(cum_sum > (f_v * V_t))
print("Eigenvalue cumulative sum: %s" % cum_sum.__str__())
print("Cumulative sum > f_v (%0.2f) = %s" % (f_v, selected_eigenvalues))
# Filter out the negligable components
t = filter(lambda x: x!=0, selected_eigenvalues[0])[0]
print("Number of selected eigenvalues = %d" % t)
print("Selected eignvalues = %s" % str(eigenvalues[0:t]))
# Assign the selected eigenvectors to P
P = eigenvectors[0:t]
print("P = %s" % P.__str__())
```
## 4. Transform the data
Now that we have the selected eigenvectors we can transform the mean adjusted data.
$$\large{x_{transformed} = Px^T}$$
The transformed data is no longer in terms of number of coffees consumed and pages written, but rather in terms of the selected vector. The data lies in this new space and cannot be thought of directly in terms of the original parameters.
```
data_transformed = P.dot(X.T).transpose()
# Return the transformed data
print("Transformed data: \n%s" % data_transformed.__str__())
```
Notice now the data set is one dimensional. This is because we only selected one of the eigenvectors and the other axes has been "thrown out". In this example there are only two eigenvectors, so by selecting both we would have included all the variance of the data set and not have gained much benefit from completing PCA. We now have a representation of the original data set that is in terms of the *P*, the mean and is now smaller in size than the original.
## 5. Recovering the Data
Now say we have done some processing on the data and we wish to return it to the original *coffees consumed* vs *pages written space*. To recover the data we use:
```
data_recovered = np.linalg.pinv(P).dot(data_transformed.T).T
x_mean, y_mean = np.mean(coffee_v_thesis, axis=0)
# Add the means
data_recovered[:,0] += x_mean
data_recovered[:,1] += y_mean
# Visualising the data
plt.figure(figsize=(10,10))
original_data_fig = plt.scatter(x=coffee_v_thesis[:,0], y=coffee_v_thesis[:,1], color="#7a7a7a", s=40)
recovered_data_fig = plt.scatter(x=data_recovered[:,0], y=data_recovered[:,1], color="#ffaf1f", s=40)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis (mean adjusted)")
plt.legend([original_data_fig, recovered_data_fig], ["Original Data Set", "Recovered Data Set"])
plt.savefig("recovered_data.jpg", dpi=500)
```
Note that we have not completely recreated the data set, as we only selected one of the eigenvalues we have lost some of the information describing the data. You can see that most of the data lines along a diagonal line, it is this line that is represented by the principal component.
```
#End of notebook
```
|
github_jupyter
|
# imports
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Calculating the mean by hand on some very simple data
very_basic_data = np.array([1,2,3,4,5])
very_basic_data_2d = np.array([[1,2,3,4,5],[6,7,8,9,10]])
print("Very basic data: %s" % very_basic_data.__str__())
print("Very basic data 2D:\n %s\n" % very_basic_data_2d.__str__())
n = very_basic_data.shape[0]
n_2d = very_basic_data_2d.shape[1]
mu = (np.sum(very_basic_data)) / n
mu_2d = (np.sum(very_basic_data_2d, axis=1)) / n_2d
print("----Calculating By Hand----")
print("\nThe mean of very_basic data is : %0.2f" % mu )
print("The mean of very_basic data - 2D is : %0.2f and %0.2f\n" % (mu_2d[0], mu_2d[1]) )
print("----Calculating Using NumPy----")
# Calculating using numpy
mu = np.mean(very_basic_data)
mu_2d = np.mean(very_basic_data_2d, axis=1)
print("\nThe mean of very_basic data is (calculated using NumPy): %0.2f" % mu )
# Add some noise to very_basic_data to ensure
from random import randint
very_basic_data *= randint(1,5)
very_basic_data_2d[0,:] *= randint(1,5)
very_basic_data_2d[1,:] *= randint(1,5)
mu = np.mean(very_basic_data)
mu_2d = np.mean(very_basic_data_2d, axis=1)
print("\n" + 10 *"----" + "1D Data" + 10*"----")
print("The very basic data set with some random noise: %s" % str(very_basic_data))
print("The new mean (mu) = %0.2f" % mu)
# Using the very basic data
sigma_squared = np.sum((very_basic_data - mu) ** 2) / (n - 1)
print("Variance (sigma^2) = %0.2f" % sigma_squared)
# Standard deviation
sigma = np.sqrt(sigma_squared)
print("Standard deviation (sigma) = %0.2f" % sigma)
# You can also calculate the standard deviation using numpy
sigma = np.std(very_basic_data, ddof = 1) # ddof = 1, is used to ensure the denominator used is n - 1, not n
print("Standard deviation using NumPy (sigma) = %0.2f" % sigma)
print("\n" + 10 *"----" + "2D Data" + 10*"----")
print("The very basic data set (2D) with some random noise: %s" % str(very_basic_data_2d))
print("The new 2D mean (mu) = %0.2f and %0.2f" % (mu_2d[0], mu_2d[1]))
# Using the very basic data
sigma_squared_2d = np.zeros(2)
sigma_squared_2d[0] = np.sum((very_basic_data_2d[0,:] - mu_2d[0]) ** 2) / (n_2d - 1)
sigma_squared_2d[1] = np.sum((very_basic_data_2d[1,:] - mu_2d[1]) ** 2) / (n_2d - 1)
print("Variance (sigma^2) = %0.2f and %0.2f" % (sigma_squared_2d[0], sigma_squared_2d[1]))
# Standard deviation
sigma_2d = np.sqrt(sigma_squared_2d)
print("Standard deviation (sigma) = %0.2f and %0.2f" % (sigma_2d[0], sigma_2d[1]))
# You can also calculate the standard deviation using numpy
sigma_2d = np.std(very_basic_data_2d, ddof = 1, axis=1)
print("Standard deviation using NumPy (sigma) = %0.2f and %0.2f" % (sigma_2d[0], sigma_2d[1]))
print("\n\n**Note: The variance calculated for the 2D data sets, considers each row or array as an independant data set")
# Defining the data
coffee_v_thesis = np.array([
[0, 2.94],
[2.3, 5.45],
[1.2, 5.41],
[3.5, 12.10],
[5.1, 20.75],
[5.2, 21.48],
[10, 31.60],
[10.2, 40.84],
[6.2, 15.12],
])
# Visualising the data
plt.figure(figsize=(8,8))
plt.scatter(x=coffee_v_thesis[:,0], y=coffee_v_thesis[:,1], color="#7a7a7a", s=30)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis")
C = coffee_v_thesis[:,0] # Select only the first column for coffee
P = coffee_v_thesis[:,1] # Second column for pages
n = coffee_v_thesis[:,0].shape[0] # Number of elements in the samples
mean_C = np.mean(C)
mean_P = np.mean(P)
cov_P_C = np.sum((C - mean_C) * (P - mean_P)) / (n - 1)
print ("cov(P,C) = %0.2f" % cov_P_C)
# Computing the covariance using numpy
# The first row, second element of the matrix corresponds to cov(P, C)
cov_P_C = np.cov(coffee_v_thesis[:,0], coffee_v_thesis[:,1])[0][1]
print ("cov(P,C) (calculated using Numpy = %0.2f" % cov_P_C)
print("**Notice the positive covariance, more coffee = more thesis pages!")
cov_P_C = np.cov(coffee_v_thesis[:,0], coffee_v_thesis[:,1])
print("The covariance matrix of coffee_v_thesis:\n%s" % cov_P_C.__str__())
eigenvalues, eigenvectors = np.linalg.eig(cov_P_C)
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
U, S, V = np.linalg.svd(cov_P_C, full_matrices=False)
eigenvalues = S
eigevectors = U
# You can check the decomposition occurred correctly if the following statement returns True
print("Correct transform: ", np.allclose(cov_P_C, np.dot(U, np.dot(np.diag(S), V))))
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
# Make a copy of the data
X = np.copy(coffee_v_thesis)
x_mean, y_mean = np.mean(X, axis=0)
X[:,0] -= x_mean
X[:,1] -= y_mean
x_mean, y_mean = np.mean(X, axis=0)
# Confirm the data is now centered about the mean i.e. the mean is close to 0
print("Confirm the data is now centered about the mean: mu_x = %0.2f, mu_y = %0.2f" % (x_mean, y_mean))
# Visualising the data
plt.figure(figsize=(8,8))
plt.scatter(x=X[:,0], y=X[:,1], color="#7a7a7a", s=30)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis (mean adjusted)")
cov_P_C = np.cov(X[:,0], X[:,1])
print("Covariance of mean adjusted data\n%s" % cov_P_C.__str__())
U, S, V = np.linalg.svd(cov_P_C, full_matrices=False)
eigenvalues = S
eigevectors = U
# You can check the decomposition occurred correctly if the following statement returns True
print("Correct transform: %s" % str(np.allclose(cov_P_C, np.dot(U, np.dot(np.diag(S), V)))))
print("eigenvalues: %s" % eigenvalues.__str__())
print("eigenvectors: \n%s" % eigenvectors.__str__())
# Calculate the total variance
V_t = np.sum(eigenvalues)
print("Total variance V_t = %0.4f" % V_t)
# Determine the proportion
V_prop = eigenvalues / V_t
print("Proportion of variance V_prop = %s" % V_prop.__str__())
# Determine the proportion represented by the principal component
print("Proportion of variance represented by principal component %0.2f %%" % (V_prop[0] * 100))
f_v = 0.99 # Proportion of total variation 98%
#Select only the components that account for 98% of the variance
cum_sum = np.cumsum(eigenvalues)
selected_eigenvalues = np.nonzero(cum_sum > (f_v * V_t))
print("Eigenvalue cumulative sum: %s" % cum_sum.__str__())
print("Cumulative sum > f_v (%0.2f) = %s" % (f_v, selected_eigenvalues))
# Filter out the negligable components
t = filter(lambda x: x!=0, selected_eigenvalues[0])[0]
print("Number of selected eigenvalues = %d" % t)
print("Selected eignvalues = %s" % str(eigenvalues[0:t]))
# Assign the selected eigenvectors to P
P = eigenvectors[0:t]
print("P = %s" % P.__str__())
data_transformed = P.dot(X.T).transpose()
# Return the transformed data
print("Transformed data: \n%s" % data_transformed.__str__())
data_recovered = np.linalg.pinv(P).dot(data_transformed.T).T
x_mean, y_mean = np.mean(coffee_v_thesis, axis=0)
# Add the means
data_recovered[:,0] += x_mean
data_recovered[:,1] += y_mean
# Visualising the data
plt.figure(figsize=(10,10))
original_data_fig = plt.scatter(x=coffee_v_thesis[:,0], y=coffee_v_thesis[:,1], color="#7a7a7a", s=40)
recovered_data_fig = plt.scatter(x=data_recovered[:,0], y=data_recovered[:,1], color="#ffaf1f", s=40)
plt.ylabel("Pages of thesis written")
plt.xlabel("Coffees consumed")
plt.title("Coffee vs Thesis (mean adjusted)")
plt.legend([original_data_fig, recovered_data_fig], ["Original Data Set", "Recovered Data Set"])
plt.savefig("recovered_data.jpg", dpi=500)
#End of notebook
| 0.612541 | 0.801975 |
```
# look at tools/set_up_magics.ipynb
yandex_metrica_allowed = True ; get_ipython().run_cell('# one_liner_str\n\nget_ipython().run_cell_magic(\'javascript\', \'\', \'// setup cpp code highlighting\\nIPython.CodeCell.options_default.highlight_modes["text/x-c++src"] = {\\\'reg\\\':[/^%%cpp/]} ;\')\n\n# creating magics\nfrom IPython.core.magic import register_cell_magic, register_line_magic\nfrom IPython.display import display, Markdown, HTML\nimport argparse\nfrom subprocess import Popen, PIPE\nimport random\nimport sys\nimport os\nimport re\nimport signal\nimport shutil\nimport shlex\nimport glob\n\n@register_cell_magic\ndef save_file(args_str, cell, line_comment_start="#"):\n parser = argparse.ArgumentParser()\n parser.add_argument("fname")\n parser.add_argument("--ejudge-style", action="store_true")\n args = parser.parse_args(args_str.split())\n \n cell = cell if cell[-1] == \'\\n\' or args.no_eof_newline else cell + "\\n"\n cmds = []\n with open(args.fname, "w") as f:\n f.write(line_comment_start + " %%cpp " + args_str + "\\n")\n for line in cell.split("\\n"):\n line_to_write = (line if not args.ejudge_style else line.rstrip()) + "\\n"\n if line.startswith("%"):\n run_prefix = "%run "\n if line.startswith(run_prefix):\n cmds.append(line[len(run_prefix):].strip())\n f.write(line_comment_start + " " + line_to_write)\n continue\n run_prefix = "%# "\n if line.startswith(run_prefix):\n f.write(line_comment_start + " " + line_to_write)\n continue\n raise Exception("Unknown %%save_file subcommand: \'%s\'" % line)\n else:\n f.write(line_to_write)\n f.write("" if not args.ejudge_style else line_comment_start + r" line without \\n")\n for cmd in cmds:\n display(Markdown("Run: `%s`" % cmd))\n get_ipython().system(cmd)\n\n@register_cell_magic\ndef cpp(fname, cell):\n save_file(fname, cell, "//")\n\n@register_cell_magic\ndef asm(fname, cell):\n save_file(fname, cell, "//")\n \n@register_cell_magic\ndef makefile(fname, cell):\n assert not fname\n save_file("makefile", cell.replace(" " * 4, "\\t"))\n \n@register_line_magic\ndef p(line):\n try:\n expr, comment = line.split(" #")\n display(Markdown("`{} = {}` # {}".format(expr.strip(), eval(expr), comment.strip())))\n except:\n display(Markdown("{} = {}".format(line, eval(line))))\n \ndef show_file(file, clear_at_begin=True, return_html_string=False):\n if clear_at_begin:\n get_ipython().system("truncate --size 0 " + file)\n obj = file.replace(\'.\', \'_\').replace(\'/\', \'_\') + "_obj"\n html_string = \'\'\'\n <!--MD_BEGIN_FILTER-->\n <script type=text/javascript>\n var entrance___OBJ__ = 0;\n var errors___OBJ__ = 0;\n function refresh__OBJ__()\n {\n entrance___OBJ__ -= 1;\n var elem = document.getElementById("__OBJ__");\n if (elem) {\n var xmlhttp=new XMLHttpRequest();\n xmlhttp.onreadystatechange=function()\n {\n var elem = document.getElementById("__OBJ__");\n console.log(!!elem, xmlhttp.readyState, xmlhttp.status, entrance___OBJ__);\n if (elem && xmlhttp.readyState==4) {\n if (xmlhttp.status==200)\n {\n errors___OBJ__ = 0;\n if (!entrance___OBJ__) {\n elem.innerText = xmlhttp.responseText;\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n return xmlhttp.responseText;\n } else {\n errors___OBJ__ += 1;\n if (errors___OBJ__ < 10 && !entrance___OBJ__) {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n }\n }\n }\n xmlhttp.open("GET", "__FILE__", true);\n xmlhttp.setRequestHeader("Cache-Control", "no-cache");\n xmlhttp.send(); \n }\n }\n \n if (!entrance___OBJ__) {\n entrance___OBJ__ += 1;\n refresh__OBJ__(); \n }\n </script>\n \n <font color="white"> <tt>\n <p id="__OBJ__" style="font-size: 16px; border:3px #333333 solid; background: #333333; border-radius: 10px; padding: 10px; "></p>\n </tt> </font>\n <!--MD_END_FILTER-->\n <!--MD_FROM_FILE __FILE__ -->\n \'\'\'.replace("__OBJ__", obj).replace("__FILE__", file)\n if return_html_string:\n return html_string\n display(HTML(html_string))\n \nBASH_POPEN_TMP_DIR = "./bash_popen_tmp"\n \ndef bash_popen_terminate_all():\n for p in globals().get("bash_popen_list", []):\n print("Terminate pid=" + str(p.pid), file=sys.stderr)\n p.terminate()\n globals()["bash_popen_list"] = []\n if os.path.exists(BASH_POPEN_TMP_DIR):\n shutil.rmtree(BASH_POPEN_TMP_DIR)\n\nbash_popen_terminate_all() \n\ndef bash_popen(cmd):\n if not os.path.exists(BASH_POPEN_TMP_DIR):\n os.mkdir(BASH_POPEN_TMP_DIR)\n h = os.path.join(BASH_POPEN_TMP_DIR, str(random.randint(0, 1e18)))\n stdout_file = h + ".out.html"\n stderr_file = h + ".err.html"\n run_log_file = h + ".fin.html"\n \n stdout = open(stdout_file, "wb")\n stdout = open(stderr_file, "wb")\n \n html = """\n <table width="100%">\n <colgroup>\n <col span="1" style="width: 70px;">\n <col span="1">\n </colgroup> \n <tbody>\n <tr> <td><b>STDOUT</b></td> <td> {stdout} </td> </tr>\n <tr> <td><b>STDERR</b></td> <td> {stderr} </td> </tr>\n <tr> <td><b>RUN LOG</b></td> <td> {run_log} </td> </tr>\n </tbody>\n </table>\n """.format(\n stdout=show_file(stdout_file, return_html_string=True),\n stderr=show_file(stderr_file, return_html_string=True),\n run_log=show_file(run_log_file, return_html_string=True),\n )\n \n cmd = """\n bash -c {cmd} &\n pid=$!\n echo "Process started! pid=${{pid}}" > {run_log_file}\n wait ${{pid}}\n echo "Process finished! exit_code=$?" >> {run_log_file}\n """.format(cmd=shlex.quote(cmd), run_log_file=run_log_file)\n # print(cmd)\n display(HTML(html))\n \n p = Popen(["bash", "-c", cmd], stdin=PIPE, stdout=stdout, stderr=stdout)\n \n bash_popen_list.append(p)\n return p\n\n\n@register_line_magic\ndef bash_async(line):\n bash_popen(line)\n \n \ndef show_log_file(file, return_html_string=False):\n obj = file.replace(\'.\', \'_\').replace(\'/\', \'_\') + "_obj"\n html_string = \'\'\'\n <!--MD_BEGIN_FILTER-->\n <script type=text/javascript>\n var entrance___OBJ__ = 0;\n var errors___OBJ__ = 0;\n function halt__OBJ__(elem, color)\n {\n elem.setAttribute("style", "font-size: 14px; background: " + color + "; padding: 10px; border: 3px; border-radius: 5px; color: white; "); \n }\n function refresh__OBJ__()\n {\n entrance___OBJ__ -= 1;\n if (entrance___OBJ__ < 0) {\n entrance___OBJ__ = 0;\n }\n var elem = document.getElementById("__OBJ__");\n if (elem) {\n var xmlhttp=new XMLHttpRequest();\n xmlhttp.onreadystatechange=function()\n {\n var elem = document.getElementById("__OBJ__");\n console.log(!!elem, xmlhttp.readyState, xmlhttp.status, entrance___OBJ__);\n if (elem && xmlhttp.readyState==4) {\n if (xmlhttp.status==200)\n {\n errors___OBJ__ = 0;\n if (!entrance___OBJ__) {\n if (elem.innerHTML != xmlhttp.responseText) {\n elem.innerHTML = xmlhttp.responseText;\n }\n if (elem.innerHTML.includes("Process finished.")) {\n halt__OBJ__(elem, "#333333");\n } else {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n }\n return xmlhttp.responseText;\n } else {\n errors___OBJ__ += 1;\n if (!entrance___OBJ__) {\n if (errors___OBJ__ < 6) {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n } else {\n halt__OBJ__(elem, "#994444");\n }\n }\n }\n }\n }\n xmlhttp.open("GET", "__FILE__", true);\n xmlhttp.setRequestHeader("Cache-Control", "no-cache");\n xmlhttp.send(); \n }\n }\n \n if (!entrance___OBJ__) {\n entrance___OBJ__ += 1;\n refresh__OBJ__(); \n }\n </script>\n\n <p id="__OBJ__" style="font-size: 14px; background: #000000; padding: 10px; border: 3px; border-radius: 5px; color: white; ">\n </p>\n \n </font>\n <!--MD_END_FILTER-->\n <!--MD_FROM_FILE __FILE__.md -->\n \'\'\'.replace("__OBJ__", obj).replace("__FILE__", file)\n if return_html_string:\n return html_string\n display(HTML(html_string))\n\n \nclass TInteractiveLauncher:\n tmp_path = "./interactive_launcher_tmp"\n def __init__(self, cmd):\n try:\n os.mkdir(TInteractiveLauncher.tmp_path)\n except:\n pass\n name = str(random.randint(0, 1e18))\n self.inq_path = os.path.join(TInteractiveLauncher.tmp_path, name + ".inq")\n self.log_path = os.path.join(TInteractiveLauncher.tmp_path, name + ".log")\n \n os.mkfifo(self.inq_path)\n open(self.log_path, \'w\').close()\n open(self.log_path + ".md", \'w\').close()\n\n self.pid = os.fork()\n if self.pid == -1:\n print("Error")\n if self.pid == 0:\n exe_cands = glob.glob("../tools/launcher.py") + glob.glob("../../tools/launcher.py")\n assert(len(exe_cands) == 1)\n assert(os.execvp("python3", ["python3", exe_cands[0], "-l", self.log_path, "-i", self.inq_path, "-c", cmd]) == 0)\n self.inq_f = open(self.inq_path, "w")\n interactive_launcher_opened_set.add(self.pid)\n show_log_file(self.log_path)\n\n def write(self, s):\n s = s.encode()\n assert len(s) == os.write(self.inq_f.fileno(), s)\n \n def get_pid(self):\n n = 100\n for i in range(n):\n try:\n return int(re.findall(r"PID = (\\d+)", open(self.log_path).readline())[0])\n except:\n if i + 1 == n:\n raise\n time.sleep(0.1)\n \n def input_queue_path(self):\n return self.inq_path\n \n def close(self):\n self.inq_f.close()\n os.waitpid(self.pid, 0)\n os.remove(self.inq_path)\n # os.remove(self.log_path)\n self.inq_path = None\n self.log_path = None \n interactive_launcher_opened_set.remove(self.pid)\n self.pid = None\n \n @staticmethod\n def terminate_all():\n if "interactive_launcher_opened_set" not in globals():\n globals()["interactive_launcher_opened_set"] = set()\n global interactive_launcher_opened_set\n for pid in interactive_launcher_opened_set:\n print("Terminate pid=" + str(pid), file=sys.stderr)\n os.kill(pid, signal.SIGKILL)\n os.waitpid(pid, 0)\n interactive_launcher_opened_set = set()\n if os.path.exists(TInteractiveLauncher.tmp_path):\n shutil.rmtree(TInteractiveLauncher.tmp_path)\n \nTInteractiveLauncher.terminate_all()\n \nyandex_metrica_allowed = bool(globals().get("yandex_metrica_allowed", False))\nif yandex_metrica_allowed:\n display(HTML(\'\'\'<!-- YANDEX_METRICA_BEGIN -->\n <script type="text/javascript" >\n (function(m,e,t,r,i,k,a){m[i]=m[i]||function(){(m[i].a=m[i].a||[]).push(arguments)};\n m[i].l=1*new Date();k=e.createElement(t),a=e.getElementsByTagName(t)[0],k.async=1,k.src=r,a.parentNode.insertBefore(k,a)})\n (window, document, "script", "https://mc.yandex.ru/metrika/tag.js", "ym");\n\n ym(59260609, "init", {\n clickmap:true,\n trackLinks:true,\n accurateTrackBounce:true\n });\n </script>\n <noscript><div><img src="https://mc.yandex.ru/watch/59260609" style="position:absolute; left:-9999px;" alt="" /></div></noscript>\n <!-- YANDEX_METRICA_END -->\'\'\'))\n\ndef make_oneliner():\n html_text = \'("В этот ноутбук встроен код Яндекс Метрики для сбора статистики использований. Если вы не хотите, чтобы по вам собиралась статистика, исправьте: yandex_metrica_allowed = False" if yandex_metrica_allowed else "")\'\n html_text += \' + "<""!-- MAGICS_SETUP_PRINTING_END -->"\'\n return \'\'.join([\n \'# look at tools/set_up_magics.ipynb\\n\',\n \'yandex_metrica_allowed = True ; get_ipython().run_cell(%s);\' % repr(one_liner_str),\n \'display(HTML(%s))\' % html_text,\n \' #\'\'MAGICS_SETUP_END\'\n ])\n \n\n');display(HTML(("В этот ноутбук встроен код Яндекс Метрики для сбора статистики использований. Если вы не хотите, чтобы по вам собиралась статистика, исправьте: yandex_metrica_allowed = False" if yandex_metrica_allowed else "") + "<""!-- MAGICS_SETUP_PRINTING_END -->")) #MAGICS_SETUP_END
```
# Жизнь без стандартной библиотеки
Что это значит? Значит, что функции взаимодействия с внещним миром (чтение, запись файлов и т. д.) будут реализованы в самом бинаре программы. Возможно вы даже лично напишите их код.
## Компилим как обычно
```
%%cpp main.c
%run gcc -m32 -masm=intel -fno-asynchronous-unwind-tables -O3 main.c -S -o main.S
%run gcc -m32 -masm=intel -O3 main.c -o main.exe
%run ls -la main.exe
%run ldd main.exe # Выводим зависимости по динамическим библиотекам
%run cat main.S
%run objdump -M intel -d main.exe
int main() {
return 0;
}
```
## Компилим, статически линкуя libc
```
%%cpp main2.c
%run gcc -m32 -masm=intel -fno-asynchronous-unwind-tables -static -flto -O3 main2.c -S -o main2.S
%run gcc -m32 -masm=intel -static -flto -O3 main2.c -o main2.exe
%run ls -la main2.exe # Заметьте, что размер стал сильно больше
%run ldd main2.exe
//%run objdump -M intel -d main2.exe
%run ./main2.exe
int main() {
return 0;
}
!objdump -M intel -d main2.exe | grep -A 30 "<main>:"
#!objdump -M intel -d main2.exe | grep -A 30 "s80ea9f0"
```
# Пишем сами без libc
```
%%cpp minimal.c
%run gcc -m32 -masm=intel -nostdlib -O3 minimal.c -o minimal.exe
%run gcc -m32 -masm=intel -nostdlib -fno-asynchronous-unwind-tables -O3 minimal.c -S -o minimal.S
%run ls -la minimal.exe # Заметьте, что размер стал очень маленьким :)
//%run ldd minimal.exe
//%run cat minimal.S
//%run objdump -d minimal.exe
%run ./minimal.exe ; echo $?
#include <sys/syscall.h>
// Универсальная функция для совершения системных вызовов
int syscall(int code, ...);
__asm__(R"(
syscall:
push ebx
push ebp
push esi
push edi
mov eax, DWORD PTR [esp + 20]
mov ebx, DWORD PTR [esp + 24]
mov ecx, DWORD PTR [esp + 28]
mov edx, DWORD PTR [esp + 32]
mov esi, DWORD PTR [esp + 36]
mov edi, DWORD PTR [esp + 40]
int 0x80
pop edi
pop esi
pop ebp
pop ebx
ret
)");
void int_to_s(unsigned int i, char* s, int* len) {
int clen = 0;
for (int ic = i; ic; ic /= 10, ++clen);
clen = clen ?: 1;
s[clen] = '\0';
for (int j = 0; j < clen; ++j, i /= 10) {
s[clen - j - 1] = '0' + i % 10;
}
*len = clen;
}
unsigned int s_to_int(char* s) {
unsigned int res = 0;
while ('0' <= *s && *s <= '9') {
res *= 10;
res += *s - '0';
++s;
}
return res;
}
int print_int(int fd, unsigned int i) {
char s[20];
int len;
int_to_s(i, s, &len);
return syscall(SYS_write, fd, s, len);
}
int print_s(int fd, const char* s) {
int len = 0;
while (s[len]) ++len;
return syscall(SYS_write, fd, s, len);
}
// Пример использования системного вызова для завершения работы программы
void _exit(int code);
__asm__(R"(
_exit:
mov eax, 1
mov ebx, [esp + 4]
int 0x80
)");
const char hello_s[] = "Hello world from function 'write'!\n";
const int hello_s_size = sizeof(hello_s);
// Пример использования системного вызова для вывода в stdout
int write();
__asm__(R"(
write:
push ebx
mov eax, 4
mov ebx, 1
lea ecx, [hello_s]
mov edx, hello_s_size
int 0x80
pop ebx
ret
)");
// Именно с этой функции всегда начинается выполнение программы
void _start() {
const char hello_s_2[] = "Hello world from 'syscall'!\n";
write();
syscall(SYS_write, 1, hello_s_2, sizeof(hello_s_2));
print_s(1, "Look at this value: "); print_int(1, 10050042); print_s(1, "\n");
print_s(1, "Look at this value: "); print_int(1, s_to_int("123456")); print_s(1, "\n");
syscall(SYS_exit, 0);
_exit(-1);
}
```
# Смотрим на адреса различных переменных. Проверяем, что секции памяти расположены так, как мы ожидаем
```
%%cpp look_at_addresses.c
%run gcc -m32 -masm=intel -O0 look_at_addresses.c -o look_at_addresses.exe
%run ./look_at_addresses.exe
#include <stdio.h>
#include <stdlib.h>
int func(int a) {
return a;
}
int* func_s() {
static int a;
return &a;
}
int data[123] = {1, 2, 3};
int main2() {
int local2 = 5;
printf("Local 'local2' addr = %p\n", &local2);
}
int main() {
int local = 1;
static int st = 2;
int* all = malloc(12);
printf("Func func addr = %p\n", (void*)func);
printf("Func func_s addr = %p\n", (void*)func_s);
printf("Global var addr = %p\n", data);
printf("Static 'st' addr = %p\n", &st);
printf("Static 'func_s.a' addr = %p\n", func_s());
printf("Local 'local' addr = %p\n", &local);
main2();
printf("Heap 'all' addr = %p\n", all);
return 0;
}
```
# Разбираемся в системным вызовом brk
`void *sbrk(intptr_t increment);`
```
%%cpp minimal.c
%run gcc -m32 -masm=intel -nostdlib -O3 minimal.c -o minimal.exe
%run gcc -m32 -masm=intel -nostdlib -fno-asynchronous-unwind-tables -O3 minimal.c -S -o minimal.S
//%run cat minimal.S
//%run objdump -d minimal.exe
%run ./minimal.exe ; echo $?
#include <sys/syscall.h>
// Универсальная функция для совершения системных вызовов
int syscall(int code, ...);
__asm__(R"(
syscall:
push ebx
push ebp
push esi
push edi
mov eax, DWORD PTR [esp + 20]
mov ebx, DWORD PTR [esp + 24]
mov ecx, DWORD PTR [esp + 28]
mov edx, DWORD PTR [esp + 32]
mov esi, DWORD PTR [esp + 36]
mov edi, DWORD PTR [esp + 40]
int 0x80
pop edi
pop esi
pop ebp
pop ebx
ret
)");
void int_to_s(unsigned int i, char* s, int* len) {
int clen = 0;
for (int ic = i; ic; ic /= 10, ++clen);
clen = clen ?: 1;
s[clen] = '\0';
for (int j = 0; j < clen; ++j, i /= 10) {
s[clen - j - 1] = '0' + i % 10;
}
*len = clen;
}
unsigned int s_to_int(char* s) {
unsigned int res = 0;
while ('0' <= *s && *s <= '9') {
res *= 10;
res += *s - '0';
++s;
}
return res;
}
int print_int(int fd, unsigned int i) {
char s[20];
int len;
int_to_s(i, s, &len);
return syscall(SYS_write, fd, s, len);
}
int print_s(int fd, const char* s) {
int len = 0;
while (s[len]) ++len;
return syscall(SYS_write, fd, s, len);
}
// Пример использования системного вызова для завершения работы программы
void _exit(int code);
__asm__(R"(
_exit:
mov eax, 1
mov ebx, [esp + 4]
int 0x80
)");
const char hello_s[] = "Hello world from function 'write'!\n";
const int hello_s_size = sizeof(hello_s);
// Пример использования системного вызова для вывода в stdout
int write();
__asm__(R"(
write:
push ebx
mov eax, 4
mov ebx, 1
lea ecx, [hello_s]
mov edx, hello_s_size
int 0x80
pop ebx
ret
)");
// Именно с этой функции всегда начинается выполнение программы
void _start() {
const int size = 100 * 1000 * 1000;
int* data_start = (void*)syscall(SYS_brk, 0);
int* data_end = (void*)syscall(SYS_brk, (int)data_start + size);
print_s(1, "Data begin: "); print_int(1, (int)(void*)data_start); print_s(1, "\n");
print_s(1, "Data end: "); print_int(1, (int)(void*)data_end); print_s(1, "\n");
data_start[0] = 1;
for (int i = 1; i < (data_end - data_start); ++i) {
data_start[i] = data_start[i - 1] + 1;
}
print_int(1, data_end[-1]); print_s(1, "\n");
_exit(0);
}
hex(146067456)
hex(100500000)
%%asm asm.S
%run gcc -m32 -nostdlib asm.S -o asm.exe
%run ./asm.exe
.intel_syntax noprefix
.text
.global _start
_start:
mov eax, 4
mov ebx, 1
mov ecx, hello_world_ptr
mov edx, 14
int 0x80
mov eax, 1
mov ebx, 1
int 0x80
.data
hello_world:
.string "Hello, World!\n"
hello_world_ptr:
.long hello_world
```
|
github_jupyter
|
# look at tools/set_up_magics.ipynb
yandex_metrica_allowed = True ; get_ipython().run_cell('# one_liner_str\n\nget_ipython().run_cell_magic(\'javascript\', \'\', \'// setup cpp code highlighting\\nIPython.CodeCell.options_default.highlight_modes["text/x-c++src"] = {\\\'reg\\\':[/^%%cpp/]} ;\')\n\n# creating magics\nfrom IPython.core.magic import register_cell_magic, register_line_magic\nfrom IPython.display import display, Markdown, HTML\nimport argparse\nfrom subprocess import Popen, PIPE\nimport random\nimport sys\nimport os\nimport re\nimport signal\nimport shutil\nimport shlex\nimport glob\n\n@register_cell_magic\ndef save_file(args_str, cell, line_comment_start="#"):\n parser = argparse.ArgumentParser()\n parser.add_argument("fname")\n parser.add_argument("--ejudge-style", action="store_true")\n args = parser.parse_args(args_str.split())\n \n cell = cell if cell[-1] == \'\\n\' or args.no_eof_newline else cell + "\\n"\n cmds = []\n with open(args.fname, "w") as f:\n f.write(line_comment_start + " %%cpp " + args_str + "\\n")\n for line in cell.split("\\n"):\n line_to_write = (line if not args.ejudge_style else line.rstrip()) + "\\n"\n if line.startswith("%"):\n run_prefix = "%run "\n if line.startswith(run_prefix):\n cmds.append(line[len(run_prefix):].strip())\n f.write(line_comment_start + " " + line_to_write)\n continue\n run_prefix = "%# "\n if line.startswith(run_prefix):\n f.write(line_comment_start + " " + line_to_write)\n continue\n raise Exception("Unknown %%save_file subcommand: \'%s\'" % line)\n else:\n f.write(line_to_write)\n f.write("" if not args.ejudge_style else line_comment_start + r" line without \\n")\n for cmd in cmds:\n display(Markdown("Run: `%s`" % cmd))\n get_ipython().system(cmd)\n\n@register_cell_magic\ndef cpp(fname, cell):\n save_file(fname, cell, "//")\n\n@register_cell_magic\ndef asm(fname, cell):\n save_file(fname, cell, "//")\n \n@register_cell_magic\ndef makefile(fname, cell):\n assert not fname\n save_file("makefile", cell.replace(" " * 4, "\\t"))\n \n@register_line_magic\ndef p(line):\n try:\n expr, comment = line.split(" #")\n display(Markdown("`{} = {}` # {}".format(expr.strip(), eval(expr), comment.strip())))\n except:\n display(Markdown("{} = {}".format(line, eval(line))))\n \ndef show_file(file, clear_at_begin=True, return_html_string=False):\n if clear_at_begin:\n get_ipython().system("truncate --size 0 " + file)\n obj = file.replace(\'.\', \'_\').replace(\'/\', \'_\') + "_obj"\n html_string = \'\'\'\n <!--MD_BEGIN_FILTER-->\n <script type=text/javascript>\n var entrance___OBJ__ = 0;\n var errors___OBJ__ = 0;\n function refresh__OBJ__()\n {\n entrance___OBJ__ -= 1;\n var elem = document.getElementById("__OBJ__");\n if (elem) {\n var xmlhttp=new XMLHttpRequest();\n xmlhttp.onreadystatechange=function()\n {\n var elem = document.getElementById("__OBJ__");\n console.log(!!elem, xmlhttp.readyState, xmlhttp.status, entrance___OBJ__);\n if (elem && xmlhttp.readyState==4) {\n if (xmlhttp.status==200)\n {\n errors___OBJ__ = 0;\n if (!entrance___OBJ__) {\n elem.innerText = xmlhttp.responseText;\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n return xmlhttp.responseText;\n } else {\n errors___OBJ__ += 1;\n if (errors___OBJ__ < 10 && !entrance___OBJ__) {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n }\n }\n }\n xmlhttp.open("GET", "__FILE__", true);\n xmlhttp.setRequestHeader("Cache-Control", "no-cache");\n xmlhttp.send(); \n }\n }\n \n if (!entrance___OBJ__) {\n entrance___OBJ__ += 1;\n refresh__OBJ__(); \n }\n </script>\n \n <font color="white"> <tt>\n <p id="__OBJ__" style="font-size: 16px; border:3px #333333 solid; background: #333333; border-radius: 10px; padding: 10px; "></p>\n </tt> </font>\n <!--MD_END_FILTER-->\n <!--MD_FROM_FILE __FILE__ -->\n \'\'\'.replace("__OBJ__", obj).replace("__FILE__", file)\n if return_html_string:\n return html_string\n display(HTML(html_string))\n \nBASH_POPEN_TMP_DIR = "./bash_popen_tmp"\n \ndef bash_popen_terminate_all():\n for p in globals().get("bash_popen_list", []):\n print("Terminate pid=" + str(p.pid), file=sys.stderr)\n p.terminate()\n globals()["bash_popen_list"] = []\n if os.path.exists(BASH_POPEN_TMP_DIR):\n shutil.rmtree(BASH_POPEN_TMP_DIR)\n\nbash_popen_terminate_all() \n\ndef bash_popen(cmd):\n if not os.path.exists(BASH_POPEN_TMP_DIR):\n os.mkdir(BASH_POPEN_TMP_DIR)\n h = os.path.join(BASH_POPEN_TMP_DIR, str(random.randint(0, 1e18)))\n stdout_file = h + ".out.html"\n stderr_file = h + ".err.html"\n run_log_file = h + ".fin.html"\n \n stdout = open(stdout_file, "wb")\n stdout = open(stderr_file, "wb")\n \n html = """\n <table width="100%">\n <colgroup>\n <col span="1" style="width: 70px;">\n <col span="1">\n </colgroup> \n <tbody>\n <tr> <td><b>STDOUT</b></td> <td> {stdout} </td> </tr>\n <tr> <td><b>STDERR</b></td> <td> {stderr} </td> </tr>\n <tr> <td><b>RUN LOG</b></td> <td> {run_log} </td> </tr>\n </tbody>\n </table>\n """.format(\n stdout=show_file(stdout_file, return_html_string=True),\n stderr=show_file(stderr_file, return_html_string=True),\n run_log=show_file(run_log_file, return_html_string=True),\n )\n \n cmd = """\n bash -c {cmd} &\n pid=$!\n echo "Process started! pid=${{pid}}" > {run_log_file}\n wait ${{pid}}\n echo "Process finished! exit_code=$?" >> {run_log_file}\n """.format(cmd=shlex.quote(cmd), run_log_file=run_log_file)\n # print(cmd)\n display(HTML(html))\n \n p = Popen(["bash", "-c", cmd], stdin=PIPE, stdout=stdout, stderr=stdout)\n \n bash_popen_list.append(p)\n return p\n\n\n@register_line_magic\ndef bash_async(line):\n bash_popen(line)\n \n \ndef show_log_file(file, return_html_string=False):\n obj = file.replace(\'.\', \'_\').replace(\'/\', \'_\') + "_obj"\n html_string = \'\'\'\n <!--MD_BEGIN_FILTER-->\n <script type=text/javascript>\n var entrance___OBJ__ = 0;\n var errors___OBJ__ = 0;\n function halt__OBJ__(elem, color)\n {\n elem.setAttribute("style", "font-size: 14px; background: " + color + "; padding: 10px; border: 3px; border-radius: 5px; color: white; "); \n }\n function refresh__OBJ__()\n {\n entrance___OBJ__ -= 1;\n if (entrance___OBJ__ < 0) {\n entrance___OBJ__ = 0;\n }\n var elem = document.getElementById("__OBJ__");\n if (elem) {\n var xmlhttp=new XMLHttpRequest();\n xmlhttp.onreadystatechange=function()\n {\n var elem = document.getElementById("__OBJ__");\n console.log(!!elem, xmlhttp.readyState, xmlhttp.status, entrance___OBJ__);\n if (elem && xmlhttp.readyState==4) {\n if (xmlhttp.status==200)\n {\n errors___OBJ__ = 0;\n if (!entrance___OBJ__) {\n if (elem.innerHTML != xmlhttp.responseText) {\n elem.innerHTML = xmlhttp.responseText;\n }\n if (elem.innerHTML.includes("Process finished.")) {\n halt__OBJ__(elem, "#333333");\n } else {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n }\n }\n return xmlhttp.responseText;\n } else {\n errors___OBJ__ += 1;\n if (!entrance___OBJ__) {\n if (errors___OBJ__ < 6) {\n entrance___OBJ__ += 1;\n console.log("req");\n window.setTimeout("refresh__OBJ__()", 300); \n } else {\n halt__OBJ__(elem, "#994444");\n }\n }\n }\n }\n }\n xmlhttp.open("GET", "__FILE__", true);\n xmlhttp.setRequestHeader("Cache-Control", "no-cache");\n xmlhttp.send(); \n }\n }\n \n if (!entrance___OBJ__) {\n entrance___OBJ__ += 1;\n refresh__OBJ__(); \n }\n </script>\n\n <p id="__OBJ__" style="font-size: 14px; background: #000000; padding: 10px; border: 3px; border-radius: 5px; color: white; ">\n </p>\n \n </font>\n <!--MD_END_FILTER-->\n <!--MD_FROM_FILE __FILE__.md -->\n \'\'\'.replace("__OBJ__", obj).replace("__FILE__", file)\n if return_html_string:\n return html_string\n display(HTML(html_string))\n\n \nclass TInteractiveLauncher:\n tmp_path = "./interactive_launcher_tmp"\n def __init__(self, cmd):\n try:\n os.mkdir(TInteractiveLauncher.tmp_path)\n except:\n pass\n name = str(random.randint(0, 1e18))\n self.inq_path = os.path.join(TInteractiveLauncher.tmp_path, name + ".inq")\n self.log_path = os.path.join(TInteractiveLauncher.tmp_path, name + ".log")\n \n os.mkfifo(self.inq_path)\n open(self.log_path, \'w\').close()\n open(self.log_path + ".md", \'w\').close()\n\n self.pid = os.fork()\n if self.pid == -1:\n print("Error")\n if self.pid == 0:\n exe_cands = glob.glob("../tools/launcher.py") + glob.glob("../../tools/launcher.py")\n assert(len(exe_cands) == 1)\n assert(os.execvp("python3", ["python3", exe_cands[0], "-l", self.log_path, "-i", self.inq_path, "-c", cmd]) == 0)\n self.inq_f = open(self.inq_path, "w")\n interactive_launcher_opened_set.add(self.pid)\n show_log_file(self.log_path)\n\n def write(self, s):\n s = s.encode()\n assert len(s) == os.write(self.inq_f.fileno(), s)\n \n def get_pid(self):\n n = 100\n for i in range(n):\n try:\n return int(re.findall(r"PID = (\\d+)", open(self.log_path).readline())[0])\n except:\n if i + 1 == n:\n raise\n time.sleep(0.1)\n \n def input_queue_path(self):\n return self.inq_path\n \n def close(self):\n self.inq_f.close()\n os.waitpid(self.pid, 0)\n os.remove(self.inq_path)\n # os.remove(self.log_path)\n self.inq_path = None\n self.log_path = None \n interactive_launcher_opened_set.remove(self.pid)\n self.pid = None\n \n @staticmethod\n def terminate_all():\n if "interactive_launcher_opened_set" not in globals():\n globals()["interactive_launcher_opened_set"] = set()\n global interactive_launcher_opened_set\n for pid in interactive_launcher_opened_set:\n print("Terminate pid=" + str(pid), file=sys.stderr)\n os.kill(pid, signal.SIGKILL)\n os.waitpid(pid, 0)\n interactive_launcher_opened_set = set()\n if os.path.exists(TInteractiveLauncher.tmp_path):\n shutil.rmtree(TInteractiveLauncher.tmp_path)\n \nTInteractiveLauncher.terminate_all()\n \nyandex_metrica_allowed = bool(globals().get("yandex_metrica_allowed", False))\nif yandex_metrica_allowed:\n display(HTML(\'\'\'<!-- YANDEX_METRICA_BEGIN -->\n <script type="text/javascript" >\n (function(m,e,t,r,i,k,a){m[i]=m[i]||function(){(m[i].a=m[i].a||[]).push(arguments)};\n m[i].l=1*new Date();k=e.createElement(t),a=e.getElementsByTagName(t)[0],k.async=1,k.src=r,a.parentNode.insertBefore(k,a)})\n (window, document, "script", "https://mc.yandex.ru/metrika/tag.js", "ym");\n\n ym(59260609, "init", {\n clickmap:true,\n trackLinks:true,\n accurateTrackBounce:true\n });\n </script>\n <noscript><div><img src="https://mc.yandex.ru/watch/59260609" style="position:absolute; left:-9999px;" alt="" /></div></noscript>\n <!-- YANDEX_METRICA_END -->\'\'\'))\n\ndef make_oneliner():\n html_text = \'("В этот ноутбук встроен код Яндекс Метрики для сбора статистики использований. Если вы не хотите, чтобы по вам собиралась статистика, исправьте: yandex_metrica_allowed = False" if yandex_metrica_allowed else "")\'\n html_text += \' + "<""!-- MAGICS_SETUP_PRINTING_END -->"\'\n return \'\'.join([\n \'# look at tools/set_up_magics.ipynb\\n\',\n \'yandex_metrica_allowed = True ; get_ipython().run_cell(%s);\' % repr(one_liner_str),\n \'display(HTML(%s))\' % html_text,\n \' #\'\'MAGICS_SETUP_END\'\n ])\n \n\n');display(HTML(("В этот ноутбук встроен код Яндекс Метрики для сбора статистики использований. Если вы не хотите, чтобы по вам собиралась статистика, исправьте: yandex_metrica_allowed = False" if yandex_metrica_allowed else "") + "<""!-- MAGICS_SETUP_PRINTING_END -->")) #MAGICS_SETUP_END
%%cpp main.c
%run gcc -m32 -masm=intel -fno-asynchronous-unwind-tables -O3 main.c -S -o main.S
%run gcc -m32 -masm=intel -O3 main.c -o main.exe
%run ls -la main.exe
%run ldd main.exe # Выводим зависимости по динамическим библиотекам
%run cat main.S
%run objdump -M intel -d main.exe
int main() {
return 0;
}
%%cpp main2.c
%run gcc -m32 -masm=intel -fno-asynchronous-unwind-tables -static -flto -O3 main2.c -S -o main2.S
%run gcc -m32 -masm=intel -static -flto -O3 main2.c -o main2.exe
%run ls -la main2.exe # Заметьте, что размер стал сильно больше
%run ldd main2.exe
//%run objdump -M intel -d main2.exe
%run ./main2.exe
int main() {
return 0;
}
!objdump -M intel -d main2.exe | grep -A 30 "<main>:"
#!objdump -M intel -d main2.exe | grep -A 30 "s80ea9f0"
%%cpp minimal.c
%run gcc -m32 -masm=intel -nostdlib -O3 minimal.c -o minimal.exe
%run gcc -m32 -masm=intel -nostdlib -fno-asynchronous-unwind-tables -O3 minimal.c -S -o minimal.S
%run ls -la minimal.exe # Заметьте, что размер стал очень маленьким :)
//%run ldd minimal.exe
//%run cat minimal.S
//%run objdump -d minimal.exe
%run ./minimal.exe ; echo $?
#include <sys/syscall.h>
// Универсальная функция для совершения системных вызовов
int syscall(int code, ...);
__asm__(R"(
syscall:
push ebx
push ebp
push esi
push edi
mov eax, DWORD PTR [esp + 20]
mov ebx, DWORD PTR [esp + 24]
mov ecx, DWORD PTR [esp + 28]
mov edx, DWORD PTR [esp + 32]
mov esi, DWORD PTR [esp + 36]
mov edi, DWORD PTR [esp + 40]
int 0x80
pop edi
pop esi
pop ebp
pop ebx
ret
)");
void int_to_s(unsigned int i, char* s, int* len) {
int clen = 0;
for (int ic = i; ic; ic /= 10, ++clen);
clen = clen ?: 1;
s[clen] = '\0';
for (int j = 0; j < clen; ++j, i /= 10) {
s[clen - j - 1] = '0' + i % 10;
}
*len = clen;
}
unsigned int s_to_int(char* s) {
unsigned int res = 0;
while ('0' <= *s && *s <= '9') {
res *= 10;
res += *s - '0';
++s;
}
return res;
}
int print_int(int fd, unsigned int i) {
char s[20];
int len;
int_to_s(i, s, &len);
return syscall(SYS_write, fd, s, len);
}
int print_s(int fd, const char* s) {
int len = 0;
while (s[len]) ++len;
return syscall(SYS_write, fd, s, len);
}
// Пример использования системного вызова для завершения работы программы
void _exit(int code);
__asm__(R"(
_exit:
mov eax, 1
mov ebx, [esp + 4]
int 0x80
)");
const char hello_s[] = "Hello world from function 'write'!\n";
const int hello_s_size = sizeof(hello_s);
// Пример использования системного вызова для вывода в stdout
int write();
__asm__(R"(
write:
push ebx
mov eax, 4
mov ebx, 1
lea ecx, [hello_s]
mov edx, hello_s_size
int 0x80
pop ebx
ret
)");
// Именно с этой функции всегда начинается выполнение программы
void _start() {
const char hello_s_2[] = "Hello world from 'syscall'!\n";
write();
syscall(SYS_write, 1, hello_s_2, sizeof(hello_s_2));
print_s(1, "Look at this value: "); print_int(1, 10050042); print_s(1, "\n");
print_s(1, "Look at this value: "); print_int(1, s_to_int("123456")); print_s(1, "\n");
syscall(SYS_exit, 0);
_exit(-1);
}
%%cpp look_at_addresses.c
%run gcc -m32 -masm=intel -O0 look_at_addresses.c -o look_at_addresses.exe
%run ./look_at_addresses.exe
#include <stdio.h>
#include <stdlib.h>
int func(int a) {
return a;
}
int* func_s() {
static int a;
return &a;
}
int data[123] = {1, 2, 3};
int main2() {
int local2 = 5;
printf("Local 'local2' addr = %p\n", &local2);
}
int main() {
int local = 1;
static int st = 2;
int* all = malloc(12);
printf("Func func addr = %p\n", (void*)func);
printf("Func func_s addr = %p\n", (void*)func_s);
printf("Global var addr = %p\n", data);
printf("Static 'st' addr = %p\n", &st);
printf("Static 'func_s.a' addr = %p\n", func_s());
printf("Local 'local' addr = %p\n", &local);
main2();
printf("Heap 'all' addr = %p\n", all);
return 0;
}
%%cpp minimal.c
%run gcc -m32 -masm=intel -nostdlib -O3 minimal.c -o minimal.exe
%run gcc -m32 -masm=intel -nostdlib -fno-asynchronous-unwind-tables -O3 minimal.c -S -o minimal.S
//%run cat minimal.S
//%run objdump -d minimal.exe
%run ./minimal.exe ; echo $?
#include <sys/syscall.h>
// Универсальная функция для совершения системных вызовов
int syscall(int code, ...);
__asm__(R"(
syscall:
push ebx
push ebp
push esi
push edi
mov eax, DWORD PTR [esp + 20]
mov ebx, DWORD PTR [esp + 24]
mov ecx, DWORD PTR [esp + 28]
mov edx, DWORD PTR [esp + 32]
mov esi, DWORD PTR [esp + 36]
mov edi, DWORD PTR [esp + 40]
int 0x80
pop edi
pop esi
pop ebp
pop ebx
ret
)");
void int_to_s(unsigned int i, char* s, int* len) {
int clen = 0;
for (int ic = i; ic; ic /= 10, ++clen);
clen = clen ?: 1;
s[clen] = '\0';
for (int j = 0; j < clen; ++j, i /= 10) {
s[clen - j - 1] = '0' + i % 10;
}
*len = clen;
}
unsigned int s_to_int(char* s) {
unsigned int res = 0;
while ('0' <= *s && *s <= '9') {
res *= 10;
res += *s - '0';
++s;
}
return res;
}
int print_int(int fd, unsigned int i) {
char s[20];
int len;
int_to_s(i, s, &len);
return syscall(SYS_write, fd, s, len);
}
int print_s(int fd, const char* s) {
int len = 0;
while (s[len]) ++len;
return syscall(SYS_write, fd, s, len);
}
// Пример использования системного вызова для завершения работы программы
void _exit(int code);
__asm__(R"(
_exit:
mov eax, 1
mov ebx, [esp + 4]
int 0x80
)");
const char hello_s[] = "Hello world from function 'write'!\n";
const int hello_s_size = sizeof(hello_s);
// Пример использования системного вызова для вывода в stdout
int write();
__asm__(R"(
write:
push ebx
mov eax, 4
mov ebx, 1
lea ecx, [hello_s]
mov edx, hello_s_size
int 0x80
pop ebx
ret
)");
// Именно с этой функции всегда начинается выполнение программы
void _start() {
const int size = 100 * 1000 * 1000;
int* data_start = (void*)syscall(SYS_brk, 0);
int* data_end = (void*)syscall(SYS_brk, (int)data_start + size);
print_s(1, "Data begin: "); print_int(1, (int)(void*)data_start); print_s(1, "\n");
print_s(1, "Data end: "); print_int(1, (int)(void*)data_end); print_s(1, "\n");
data_start[0] = 1;
for (int i = 1; i < (data_end - data_start); ++i) {
data_start[i] = data_start[i - 1] + 1;
}
print_int(1, data_end[-1]); print_s(1, "\n");
_exit(0);
}
hex(146067456)
hex(100500000)
%%asm asm.S
%run gcc -m32 -nostdlib asm.S -o asm.exe
%run ./asm.exe
.intel_syntax noprefix
.text
.global _start
_start:
mov eax, 4
mov ebx, 1
mov ecx, hello_world_ptr
mov edx, 14
int 0x80
mov eax, 1
mov ebx, 1
int 0x80
.data
hello_world:
.string "Hello, World!\n"
hello_world_ptr:
.long hello_world
| 0.133331 | 0.227491 |
<h2 align=center>Data Visualization and Analysis of Worldwide Box Office Revenue (Part 2)</h2>
<img src="revenue.png">
### (Part 1) Libraries
```
import numpy as np
import pandas as pd
pd.set_option('max_columns', None)
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
plt.style.use('ggplot')
import datetime
import lightgbm as lgb
from scipy import stats
from scipy.sparse import hstack, csr_matrix
from sklearn.model_selection import train_test_split, KFold
from wordcloud import WordCloud
from collections import Counter
from nltk.corpus import stopwords
from nltk.util import ngrams
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.preprocessing import StandardScaler
import nltk
nltk.download('stopwords')
stop = set(stopwords.words('english'))
import os
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.tools as tls
import xgboost as xgb
import lightgbm as lgb
from sklearn import model_selection
from sklearn.metrics import accuracy_score
import json
import ast
from urllib.request import urlopen
from PIL import Image
from sklearn.preprocessing import LabelEncoder
import time
from sklearn.metrics import mean_squared_error
from sklearn.linear_model import LinearRegression
from sklearn import linear_model
```
### (Part 1) Data Loading and Exploration
```
train = pd.read_csv('data/train.csv')
test = pd.read_csv('data/test.csv')
train.head()
```
### (Part 1) Visualizing the Target Distribution
```
fig, ax = plt.subplots(figsize = (16, 6))
plt.subplot(1, 2, 1)
plt.hist(train['revenue']);
plt.title('Distribution of revenue');
plt.subplot(1, 2, 2)
plt.hist(np.log1p(train['revenue']));
plt.title('Distribution of log of revenue');
train['log_revenue'] = np.log1p(train['revenue'])
```
### (Part 1) Relationship between Film Revenue and Budget
```
fig, ax = plt.subplots(figsize = (16, 6))
plt.subplot(1, 2, 1)
plt.hist(train['budget']);
plt.title('Distribution of budget');
plt.subplot(1, 2, 2)
plt.hist(np.log1p(train['budget']));
plt.title('Distribution of log of budget');
plt.figure(figsize=(16, 8))
plt.subplot(1, 2, 1)
plt.scatter(train['budget'], train['revenue'])
plt.title('Revenue vs budget');
plt.subplot(1, 2, 2)
plt.scatter(np.log1p(train['budget']), train['log_revenue'])
plt.title('Log Revenue vs log budget');
train['log_budget'] = np.log1p(train['budget'])
test['log_budget'] = np.log1p(test['budget'])
```
### (Part 1) Does having an Official Homepage Affect Revenue?
```
train['homepage'].value_counts().head(10)
train['has_homepage'] = 0
train.loc[train['homepage'].isnull() == False, 'has_homepage'] = 1
test['has_homepage'] = 0
test.loc[test['homepage'].isnull() == False, 'has_homepage'] = 1
sns.catplot(x='has_homepage', y='revenue', data=train);
plt.title('Revenue for film with and without homepage');
```
### (Part 1) Distribution of Languages in Film
```
plt.figure(figsize=(16, 8))
plt.subplot(1, 2, 1)
sns.boxplot(x='original_language', y='revenue', data=train.loc[train['original_language'].isin(train['original_language'].value_counts().head(10).index)]);
plt.title('Mean revenue per language');
plt.subplot(1, 2, 2)
sns.boxplot(x='original_language', y='log_revenue', data=train.loc[train['original_language'].isin(train['original_language'].value_counts().head(10).index)]);
plt.title('Mean log revenue per language');
```
### (Part 1) Frequent Words in Film Titles and Discriptions
```
plt.figure(figsize = (12, 12))
text = ' '.join(train['original_title'].values)
wordcloud = WordCloud(max_font_size=None, background_color='white', width=1200, height=1000).generate(text)
plt.imshow(wordcloud)
plt.title('Top words in titles')
plt.axis("off")
plt.show()
plt.figure(figsize = (12, 12))
text = ' '.join(train['overview'].fillna('').values)
wordcloud = WordCloud(max_font_size=None, background_color='white', width=1200, height=1000).generate(text)
plt.imshow(wordcloud)
plt.title('Top words in overview')
plt.axis("off")
plt.show()
```
### (Part 1) Do Film Descriptions Impact Revenue?
```
import eli5
vectorizer = TfidfVectorizer(
sublinear_tf=True,
analyzer='word',
token_pattern=r'\w{1,}',
ngram_range=(1, 2),
min_df=5)
overview_text = vectorizer.fit_transform(train['overview'].fillna(''))
linreg = LinearRegression()
linreg.fit(overview_text, train['log_revenue'])
eli5.show_weights(linreg, vec=vectorizer, top=20, feature_filter=lambda x: x != '<BIAS>')
print('Target value:', train['log_revenue'][1000])
eli5.show_prediction(linreg, doc=train['overview'].values[1000], vec=vectorizer)
```
### Task 1: Analyzing Movie Release Dates
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
test.loc[test['release_date'].isnull() == False, 'release_date'].head()
```
### Task 2: Preprocessing Features
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
def fix_date(x):
year = x.split('/')[2]
if int(year) <= 19:
return x[:-2] + '20' + year
else:
return x[:-2] + '19' + year
test.loc[test['release_date'].isnull() == True].head()
test.loc[test['release_date'].isnull() == True, 'release_date'] = '05/01/00'
train['release_date'] = train['release_date'].apply(lambda x: fix_date(x))
test['release_date'] = test['release_date'].apply(lambda x: fix_date(x))
```
### Task 3: Creating Features Based on Release Date
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
train['release_date'] = pd.to_datetime(train['release_date'])
test['release_date'] = pd.to_datetime(test['release_date'])
def process_date(df):
date_parts = ["year", "weekday", "month", 'weekofyear', 'day', 'quarter']
for part in date_parts:
part_col = 'release_date' + "_" + part
df[part_col] = getattr(df['release_date'].dt, part).astype(int)
return df
train = process_date(train)
test = process_date(test)
```
### Task 4: Using Plotly to Visualize the Number of Films Per Year
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
# Count no.of films released per year and sort the years in ascending order
# Do this for both Train and Test Sets
d1 = train['release_date_year'].value_counts().sort_index()
d2 = test['release_date_year'].value_counts().sort_index()
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
# x values are years, and y values are movie counts, name=legend
data = [go.Scatter(x=d1.index, y=d1.values, name='train'),
go.Scatter(x=d2.index, y=d2.values, name='test')]
layout = go.Layout(dict(title = "Number of films per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
),legend=dict(
orientation="v"))
py.iplot(dict(data=data, layout=layout))
```
### Task 5: Number of Films and Revenue Per Year
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
d1 = train['release_date_year'].value_counts().sort_index()
d2 = train.groupby(['release_date_year'])['revenue'].sum()
data = [go.Scatter(x=d1.index, y=d1.values, name='film count'),
go.Scatter(x=d2.index, y=d2.values, name='total revenue', yaxis='y2')]
layout = go.Layout(dict(title = "Number of films and total revenue per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
yaxis2=dict(title='Total revenue', overlaying='y', side='right')),
legend=dict(orientation="v"))
py.iplot(dict(data=data, layout=layout))
d1 = train['release_date_year'].value_counts().sort_index()
d2 = train.groupby(['release_date_year'])['revenue'].mean()
data = [go.Scatter(x=d1.index, y=d1.values, name='film count'),
go.Scatter(x=d2.index, y=d2.values, name='mean revenue', yaxis='y2')]
layout = go.Layout(dict(title = "Number of films and average revenue per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
yaxis2=dict(title='Average revenue', overlaying='y', side='right')
),legend=dict(
orientation="v"))
py.iplot(dict(data=data, layout=layout))
```
### Task 6: Do Release Days Impact Revenue?
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
sns.catplot(x='release_date_weekday', y='revenue', data=train);
plt.title('Revenue on different days of week of release');
```
### Task 7: Relationship between Runtime and Revenue
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
```
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
sns.distplot(train['runtime'].fillna(0) / 60, bins=40, kde=False);
plt.title('Distribution of length of film in hours');
plt.subplot(1, 2, 2)
sns.scatterplot(train['runtime'].fillna(0)/60, train['revenue'])
plt.title('runtime vs revenue');
```
### Task 8: Highest Grossing Genres
***
Note: If you are starting the notebook from this task, you can run cells from all the previous tasks in the kernel by going to the top menu and Kernel > Restart and Run All
***
|
github_jupyter
|
import numpy as np
import pandas as pd
pd.set_option('max_columns', None)
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
plt.style.use('ggplot')
import datetime
import lightgbm as lgb
from scipy import stats
from scipy.sparse import hstack, csr_matrix
from sklearn.model_selection import train_test_split, KFold
from wordcloud import WordCloud
from collections import Counter
from nltk.corpus import stopwords
from nltk.util import ngrams
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.preprocessing import StandardScaler
import nltk
nltk.download('stopwords')
stop = set(stopwords.words('english'))
import os
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.tools as tls
import xgboost as xgb
import lightgbm as lgb
from sklearn import model_selection
from sklearn.metrics import accuracy_score
import json
import ast
from urllib.request import urlopen
from PIL import Image
from sklearn.preprocessing import LabelEncoder
import time
from sklearn.metrics import mean_squared_error
from sklearn.linear_model import LinearRegression
from sklearn import linear_model
train = pd.read_csv('data/train.csv')
test = pd.read_csv('data/test.csv')
train.head()
fig, ax = plt.subplots(figsize = (16, 6))
plt.subplot(1, 2, 1)
plt.hist(train['revenue']);
plt.title('Distribution of revenue');
plt.subplot(1, 2, 2)
plt.hist(np.log1p(train['revenue']));
plt.title('Distribution of log of revenue');
train['log_revenue'] = np.log1p(train['revenue'])
fig, ax = plt.subplots(figsize = (16, 6))
plt.subplot(1, 2, 1)
plt.hist(train['budget']);
plt.title('Distribution of budget');
plt.subplot(1, 2, 2)
plt.hist(np.log1p(train['budget']));
plt.title('Distribution of log of budget');
plt.figure(figsize=(16, 8))
plt.subplot(1, 2, 1)
plt.scatter(train['budget'], train['revenue'])
plt.title('Revenue vs budget');
plt.subplot(1, 2, 2)
plt.scatter(np.log1p(train['budget']), train['log_revenue'])
plt.title('Log Revenue vs log budget');
train['log_budget'] = np.log1p(train['budget'])
test['log_budget'] = np.log1p(test['budget'])
train['homepage'].value_counts().head(10)
train['has_homepage'] = 0
train.loc[train['homepage'].isnull() == False, 'has_homepage'] = 1
test['has_homepage'] = 0
test.loc[test['homepage'].isnull() == False, 'has_homepage'] = 1
sns.catplot(x='has_homepage', y='revenue', data=train);
plt.title('Revenue for film with and without homepage');
plt.figure(figsize=(16, 8))
plt.subplot(1, 2, 1)
sns.boxplot(x='original_language', y='revenue', data=train.loc[train['original_language'].isin(train['original_language'].value_counts().head(10).index)]);
plt.title('Mean revenue per language');
plt.subplot(1, 2, 2)
sns.boxplot(x='original_language', y='log_revenue', data=train.loc[train['original_language'].isin(train['original_language'].value_counts().head(10).index)]);
plt.title('Mean log revenue per language');
plt.figure(figsize = (12, 12))
text = ' '.join(train['original_title'].values)
wordcloud = WordCloud(max_font_size=None, background_color='white', width=1200, height=1000).generate(text)
plt.imshow(wordcloud)
plt.title('Top words in titles')
plt.axis("off")
plt.show()
plt.figure(figsize = (12, 12))
text = ' '.join(train['overview'].fillna('').values)
wordcloud = WordCloud(max_font_size=None, background_color='white', width=1200, height=1000).generate(text)
plt.imshow(wordcloud)
plt.title('Top words in overview')
plt.axis("off")
plt.show()
import eli5
vectorizer = TfidfVectorizer(
sublinear_tf=True,
analyzer='word',
token_pattern=r'\w{1,}',
ngram_range=(1, 2),
min_df=5)
overview_text = vectorizer.fit_transform(train['overview'].fillna(''))
linreg = LinearRegression()
linreg.fit(overview_text, train['log_revenue'])
eli5.show_weights(linreg, vec=vectorizer, top=20, feature_filter=lambda x: x != '<BIAS>')
print('Target value:', train['log_revenue'][1000])
eli5.show_prediction(linreg, doc=train['overview'].values[1000], vec=vectorizer)
test.loc[test['release_date'].isnull() == False, 'release_date'].head()
def fix_date(x):
year = x.split('/')[2]
if int(year) <= 19:
return x[:-2] + '20' + year
else:
return x[:-2] + '19' + year
test.loc[test['release_date'].isnull() == True].head()
test.loc[test['release_date'].isnull() == True, 'release_date'] = '05/01/00'
train['release_date'] = train['release_date'].apply(lambda x: fix_date(x))
test['release_date'] = test['release_date'].apply(lambda x: fix_date(x))
train['release_date'] = pd.to_datetime(train['release_date'])
test['release_date'] = pd.to_datetime(test['release_date'])
def process_date(df):
date_parts = ["year", "weekday", "month", 'weekofyear', 'day', 'quarter']
for part in date_parts:
part_col = 'release_date' + "_" + part
df[part_col] = getattr(df['release_date'].dt, part).astype(int)
return df
train = process_date(train)
test = process_date(test)
# Count no.of films released per year and sort the years in ascending order
# Do this for both Train and Test Sets
d1 = train['release_date_year'].value_counts().sort_index()
d2 = test['release_date_year'].value_counts().sort_index()
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
# x values are years, and y values are movie counts, name=legend
data = [go.Scatter(x=d1.index, y=d1.values, name='train'),
go.Scatter(x=d2.index, y=d2.values, name='test')]
layout = go.Layout(dict(title = "Number of films per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
),legend=dict(
orientation="v"))
py.iplot(dict(data=data, layout=layout))
d1 = train['release_date_year'].value_counts().sort_index()
d2 = train.groupby(['release_date_year'])['revenue'].sum()
data = [go.Scatter(x=d1.index, y=d1.values, name='film count'),
go.Scatter(x=d2.index, y=d2.values, name='total revenue', yaxis='y2')]
layout = go.Layout(dict(title = "Number of films and total revenue per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
yaxis2=dict(title='Total revenue', overlaying='y', side='right')),
legend=dict(orientation="v"))
py.iplot(dict(data=data, layout=layout))
d1 = train['release_date_year'].value_counts().sort_index()
d2 = train.groupby(['release_date_year'])['revenue'].mean()
data = [go.Scatter(x=d1.index, y=d1.values, name='film count'),
go.Scatter(x=d2.index, y=d2.values, name='mean revenue', yaxis='y2')]
layout = go.Layout(dict(title = "Number of films and average revenue per year",
xaxis = dict(title = 'Year'),
yaxis = dict(title = 'Count'),
yaxis2=dict(title='Average revenue', overlaying='y', side='right')
),legend=dict(
orientation="v"))
py.iplot(dict(data=data, layout=layout))
sns.catplot(x='release_date_weekday', y='revenue', data=train);
plt.title('Revenue on different days of week of release');
plt.figure(figsize=(15, 6))
plt.subplot(1, 2, 1)
sns.distplot(train['runtime'].fillna(0) / 60, bins=40, kde=False);
plt.title('Distribution of length of film in hours');
plt.subplot(1, 2, 2)
sns.scatterplot(train['runtime'].fillna(0)/60, train['revenue'])
plt.title('runtime vs revenue');
| 0.588534 | 0.811713 |
```
import dash_mdc_neptune as mdc
from jupyter_dash import JupyterDash
from dash.dependencies import Input, Output
# Test toggle
toggle = mdc.Toggle(
id="toggle",
orientation="vertical",
options=["test a", "test b"],
selected="test b",
)
text = mdc.Typography(
id="text",
text="Content...",
variant="body2",
)
box = mdc.Box(children=[toggle, text])
# Test dropdown
dropdown = mdc.Dropdown(
id="dropdown",
width=150,
labelText="test",
helperText="test",
options=["test 1", "test 2"],
multiple=True,
selected=["test 2"],
)
text_2 = mdc.Typography(
id="text-2",
text="Content...",
variant="body2",
)
box_2 = mdc.Box(children=[dropdown, text_2])
# Test calendar
calendar = mdc.Calendar(
id="calendar",
width=150,
labelText="test",
helperText="test",
minDate="2022-01-01",
disableFuture=True,
selected="2022-05-01",
)
text_3 = mdc.Typography(
id="text-3",
text="Content...",
variant="body2",
)
box_3 = mdc.Box(children=[calendar, text_3])
# Test slider
slider = mdc.Slider(
id="slider",
width=200,
labelText="test",
minValue=-100,
maxValue=100,
stepValue=10,
selected=50,
marks=[
{"label": "-100", "value": -100},
{"label": "-50", "value": -50},
{"label": "0", "value": 0},
{"label": "50", "value": 50},
{"label": "100", "value": 100},
],
)
text_4 = mdc.Typography(
id="text-4",
text="Content...",
variant="body2",
)
box_4 = mdc.Box(children=[slider, text_4])
# Dashboard layout
tab = mdc.Tab(
children=[box, box_2, box_3, box_4],
tabs=[
{"label": "Toggle"},
{"label": "Dropdown"},
{"label": "Calendar"},
{"label": "Slider"},
]
)
section = mdc.Section(
orientation="columns",
children=tab,
cards=[{"title": "Card 1"}]
)
page = mdc.Page(orientation="columns", children=section)
navbar = mdc.NavBar(title="Custom dash")
layout = mdc.Dashboard(children=[navbar, page])
# Dash app
app = JupyterDash(__name__)
app.layout = layout
@app.callback(
Output(component_id='text', component_property='text'),
Input(component_id='toggle', component_property='selected'),
)
def update_toggle(value):
return value
@app.callback(
Output(component_id='text-2', component_property='text'),
Input(component_id='dropdown', component_property='selected'),
)
def update_dropdown(value):
if not value:
return "No content..."
return ", ".join(value)
@app.callback(
Output(component_id='text-3', component_property='text'),
Input(component_id='calendar', component_property='selected'),
)
def update_calendar(value):
if not value:
return "No content..."
return value
@app.callback(
Output(component_id='text-4', component_property='text'),
Input(component_id='slider', component_property='selected'),
)
def update_slider(value):
return value
app.run_server(mode='jupyterlab', host="0.0.0.0", port=8001, debug=False)
```
|
github_jupyter
|
import dash_mdc_neptune as mdc
from jupyter_dash import JupyterDash
from dash.dependencies import Input, Output
# Test toggle
toggle = mdc.Toggle(
id="toggle",
orientation="vertical",
options=["test a", "test b"],
selected="test b",
)
text = mdc.Typography(
id="text",
text="Content...",
variant="body2",
)
box = mdc.Box(children=[toggle, text])
# Test dropdown
dropdown = mdc.Dropdown(
id="dropdown",
width=150,
labelText="test",
helperText="test",
options=["test 1", "test 2"],
multiple=True,
selected=["test 2"],
)
text_2 = mdc.Typography(
id="text-2",
text="Content...",
variant="body2",
)
box_2 = mdc.Box(children=[dropdown, text_2])
# Test calendar
calendar = mdc.Calendar(
id="calendar",
width=150,
labelText="test",
helperText="test",
minDate="2022-01-01",
disableFuture=True,
selected="2022-05-01",
)
text_3 = mdc.Typography(
id="text-3",
text="Content...",
variant="body2",
)
box_3 = mdc.Box(children=[calendar, text_3])
# Test slider
slider = mdc.Slider(
id="slider",
width=200,
labelText="test",
minValue=-100,
maxValue=100,
stepValue=10,
selected=50,
marks=[
{"label": "-100", "value": -100},
{"label": "-50", "value": -50},
{"label": "0", "value": 0},
{"label": "50", "value": 50},
{"label": "100", "value": 100},
],
)
text_4 = mdc.Typography(
id="text-4",
text="Content...",
variant="body2",
)
box_4 = mdc.Box(children=[slider, text_4])
# Dashboard layout
tab = mdc.Tab(
children=[box, box_2, box_3, box_4],
tabs=[
{"label": "Toggle"},
{"label": "Dropdown"},
{"label": "Calendar"},
{"label": "Slider"},
]
)
section = mdc.Section(
orientation="columns",
children=tab,
cards=[{"title": "Card 1"}]
)
page = mdc.Page(orientation="columns", children=section)
navbar = mdc.NavBar(title="Custom dash")
layout = mdc.Dashboard(children=[navbar, page])
# Dash app
app = JupyterDash(__name__)
app.layout = layout
@app.callback(
Output(component_id='text', component_property='text'),
Input(component_id='toggle', component_property='selected'),
)
def update_toggle(value):
return value
@app.callback(
Output(component_id='text-2', component_property='text'),
Input(component_id='dropdown', component_property='selected'),
)
def update_dropdown(value):
if not value:
return "No content..."
return ", ".join(value)
@app.callback(
Output(component_id='text-3', component_property='text'),
Input(component_id='calendar', component_property='selected'),
)
def update_calendar(value):
if not value:
return "No content..."
return value
@app.callback(
Output(component_id='text-4', component_property='text'),
Input(component_id='slider', component_property='selected'),
)
def update_slider(value):
return value
app.run_server(mode='jupyterlab', host="0.0.0.0", port=8001, debug=False)
| 0.534127 | 0.398114 |
---
_You are currently looking at **version 1.0** of this notebook. To download notebooks and datafiles, as well as get help on Jupyter notebooks in the Coursera platform, visit the [Jupyter Notebook FAQ](https://www.coursera.org/learn/python-data-analysis/resources/0dhYG) course resource._
---
# The Series Data Structure
```
import pandas as pd
pd.Series?
animals = ['Tiger', 'Bear', 'Moose']
pd.Series(animals)
numbers = [1, 2, 3]
pd.Series(numbers)
animals = ['Tiger', 'Bear', None]
pd.Series(animals)
numbers = [1, 2, None]
pd.Series(numbers)
import numpy as np
np.nan == None
np.nan == np.nan
np.isnan(np.nan)
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports)
s
s.index
s = pd.Series(['Tiger', 'Bear', 'Moose'], index=['India', 'America', 'Canada'])
s
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports, index=['Golf', 'Sumo', 'Hockey'])
s
```
# Querying a Series
```
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports)
s
s.iloc[3]
s.loc['Golf']
s[3]
s['Golf']
sports = {99: 'Bhutan',
100: 'Scotland',
101: 'Japan',
102: 'South Korea'}
s = pd.Series(sports)
s[0] #This won't call s.iloc[0] as one might expect, it generates an error instead
s = pd.Series([100.00, 120.00, 101.00, 3.00])
s
total = 0
for item in s:
total+=item
print(total)
import numpy as np
total = np.sum(s)
print(total)
#this creates a big series of random numbers
s = pd.Series(np.random.randint(0,1000,10000))
s.head()
len(s)
%%timeit -n 100
summary = 0
for item in s:
summary+=item
%%timeit -n 100
summary = np.sum(s)
s+=2 #adds two to each item in s using broadcasting
s.head()
for label, value in s.iteritems():
s.set_value(label, value+2)
s.head()
%%timeit -n 10
s = pd.Series(np.random.randint(0,1000,10000))
for label, value in s.iteritems():
s.loc[label]= value+2
%%timeit -n 10
s = pd.Series(np.random.randint(0,1000,10000))
s+=2
s = pd.Series([1, 2, 3])
s.loc['Animal'] = 'Bears'
s
original_sports = pd.Series({'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'})
cricket_loving_countries = pd.Series(['Australia',
'Barbados',
'Pakistan',
'England'],
index=['Cricket',
'Cricket',
'Cricket',
'Cricket'])
all_countries = original_sports.append(cricket_loving_countries)
original_sports
cricket_loving_countries
all_countries
all_countries.loc['Cricket']
```
# The DataFrame Data Structure
```
import pandas as pd
purchase_1 = pd.Series({'Name': 'Chris',
'Item Purchased': 'Dog Food',
'Cost': 22.50})
purchase_2 = pd.Series({'Name': 'Kevyn',
'Item Purchased': 'Kitty Litter',
'Cost': 2.50})
purchase_3 = pd.Series({'Name': 'Vinod',
'Item Purchased': 'Bird Seed',
'Cost': 5.00})
df = pd.DataFrame([purchase_1, purchase_2, purchase_3], index=['Store 1', 'Store 1', 'Store 2'])
df.head()
df.loc['Store 2']
type(df.loc['Store 2'])
df.loc['Store 1']
df.loc['Store 1', 'Cost']
df.T
df.T.loc['Cost']
df['Cost']
df.loc['Store 1']['Cost']
df.loc[:,['Name', 'Cost']]
df.drop('Store 1')
df
copy_df = df.copy()
copy_df = copy_df.drop('Store 1')
copy_df
copy_df.drop?
del copy_df['Name']
copy_df
df['Location'] = None
df
```
# Dataframe Indexing and Loading
```
costs = df['Cost']
costs
costs+=2
costs
df
!cat olympics.csv
df = pd.read_csv('olympics.csv')
df.head()
df = pd.read_csv('olympics.csv', index_col = 0, skiprows=1)
df.head()
df.columns
for col in df.columns:
if col[:2]=='01':
df.rename(columns={col:'Gold' + col[4:]}, inplace=True)
if col[:2]=='02':
df.rename(columns={col:'Silver' + col[4:]}, inplace=True)
if col[:2]=='03':
df.rename(columns={col:'Bronze' + col[4:]}, inplace=True)
if col[:1]=='№':
df.rename(columns={col:'#' + col[1:]}, inplace=True)
df.head()
```
# Querying a DataFrame
```
df['Gold'] > 0
only_gold = df.where(df['Gold'] > 0)
only_gold.head()
only_gold['Gold'].count()
df['Gold'].count()
only_gold = only_gold.dropna()
only_gold.head()
only_gold = df[df['Gold'] > 0]
only_gold.head()
len(df[(df['Gold'] > 0) | (df['Gold.1'] > 0)])
df[(df['Gold.1'] > 0) & (df['Gold'] == 0)]
```
# Indexing Dataframes
```
df.head()
df['country'] = df.index
df = df.set_index('Gold')
df.head()
df = df.reset_index()
df.head()
df = pd.read_csv('census.csv')
df.head()
df['SUMLEV'].unique()
df=df[df['SUMLEV'] == 50]
df.head()
columns_to_keep = ['STNAME',
'CTYNAME',
'BIRTHS2010',
'BIRTHS2011',
'BIRTHS2012',
'BIRTHS2013',
'BIRTHS2014',
'BIRTHS2015',
'POPESTIMATE2010',
'POPESTIMATE2011',
'POPESTIMATE2012',
'POPESTIMATE2013',
'POPESTIMATE2014',
'POPESTIMATE2015']
df = df[columns_to_keep]
df.head()
df = df.set_index(['STNAME', 'CTYNAME'])
df.head()
df.loc['Michigan', 'Washtenaw County']
df.loc[ [('Michigan', 'Washtenaw County'),
('Michigan', 'Wayne County')] ]
```
# Missing values
```
df = pd.read_csv('log.csv')
df
df.fillna?
df = df.set_index('time')
df = df.sort_index()
df
df = df.reset_index()
df = df.set_index(['time', 'user'])
df
df = df.fillna(method='ffill')
df.head()
```
|
github_jupyter
|
import pandas as pd
pd.Series?
animals = ['Tiger', 'Bear', 'Moose']
pd.Series(animals)
numbers = [1, 2, 3]
pd.Series(numbers)
animals = ['Tiger', 'Bear', None]
pd.Series(animals)
numbers = [1, 2, None]
pd.Series(numbers)
import numpy as np
np.nan == None
np.nan == np.nan
np.isnan(np.nan)
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports)
s
s.index
s = pd.Series(['Tiger', 'Bear', 'Moose'], index=['India', 'America', 'Canada'])
s
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports, index=['Golf', 'Sumo', 'Hockey'])
s
sports = {'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'}
s = pd.Series(sports)
s
s.iloc[3]
s.loc['Golf']
s[3]
s['Golf']
sports = {99: 'Bhutan',
100: 'Scotland',
101: 'Japan',
102: 'South Korea'}
s = pd.Series(sports)
s[0] #This won't call s.iloc[0] as one might expect, it generates an error instead
s = pd.Series([100.00, 120.00, 101.00, 3.00])
s
total = 0
for item in s:
total+=item
print(total)
import numpy as np
total = np.sum(s)
print(total)
#this creates a big series of random numbers
s = pd.Series(np.random.randint(0,1000,10000))
s.head()
len(s)
%%timeit -n 100
summary = 0
for item in s:
summary+=item
%%timeit -n 100
summary = np.sum(s)
s+=2 #adds two to each item in s using broadcasting
s.head()
for label, value in s.iteritems():
s.set_value(label, value+2)
s.head()
%%timeit -n 10
s = pd.Series(np.random.randint(0,1000,10000))
for label, value in s.iteritems():
s.loc[label]= value+2
%%timeit -n 10
s = pd.Series(np.random.randint(0,1000,10000))
s+=2
s = pd.Series([1, 2, 3])
s.loc['Animal'] = 'Bears'
s
original_sports = pd.Series({'Archery': 'Bhutan',
'Golf': 'Scotland',
'Sumo': 'Japan',
'Taekwondo': 'South Korea'})
cricket_loving_countries = pd.Series(['Australia',
'Barbados',
'Pakistan',
'England'],
index=['Cricket',
'Cricket',
'Cricket',
'Cricket'])
all_countries = original_sports.append(cricket_loving_countries)
original_sports
cricket_loving_countries
all_countries
all_countries.loc['Cricket']
import pandas as pd
purchase_1 = pd.Series({'Name': 'Chris',
'Item Purchased': 'Dog Food',
'Cost': 22.50})
purchase_2 = pd.Series({'Name': 'Kevyn',
'Item Purchased': 'Kitty Litter',
'Cost': 2.50})
purchase_3 = pd.Series({'Name': 'Vinod',
'Item Purchased': 'Bird Seed',
'Cost': 5.00})
df = pd.DataFrame([purchase_1, purchase_2, purchase_3], index=['Store 1', 'Store 1', 'Store 2'])
df.head()
df.loc['Store 2']
type(df.loc['Store 2'])
df.loc['Store 1']
df.loc['Store 1', 'Cost']
df.T
df.T.loc['Cost']
df['Cost']
df.loc['Store 1']['Cost']
df.loc[:,['Name', 'Cost']]
df.drop('Store 1')
df
copy_df = df.copy()
copy_df = copy_df.drop('Store 1')
copy_df
copy_df.drop?
del copy_df['Name']
copy_df
df['Location'] = None
df
costs = df['Cost']
costs
costs+=2
costs
df
!cat olympics.csv
df = pd.read_csv('olympics.csv')
df.head()
df = pd.read_csv('olympics.csv', index_col = 0, skiprows=1)
df.head()
df.columns
for col in df.columns:
if col[:2]=='01':
df.rename(columns={col:'Gold' + col[4:]}, inplace=True)
if col[:2]=='02':
df.rename(columns={col:'Silver' + col[4:]}, inplace=True)
if col[:2]=='03':
df.rename(columns={col:'Bronze' + col[4:]}, inplace=True)
if col[:1]=='№':
df.rename(columns={col:'#' + col[1:]}, inplace=True)
df.head()
df['Gold'] > 0
only_gold = df.where(df['Gold'] > 0)
only_gold.head()
only_gold['Gold'].count()
df['Gold'].count()
only_gold = only_gold.dropna()
only_gold.head()
only_gold = df[df['Gold'] > 0]
only_gold.head()
len(df[(df['Gold'] > 0) | (df['Gold.1'] > 0)])
df[(df['Gold.1'] > 0) & (df['Gold'] == 0)]
df.head()
df['country'] = df.index
df = df.set_index('Gold')
df.head()
df = df.reset_index()
df.head()
df = pd.read_csv('census.csv')
df.head()
df['SUMLEV'].unique()
df=df[df['SUMLEV'] == 50]
df.head()
columns_to_keep = ['STNAME',
'CTYNAME',
'BIRTHS2010',
'BIRTHS2011',
'BIRTHS2012',
'BIRTHS2013',
'BIRTHS2014',
'BIRTHS2015',
'POPESTIMATE2010',
'POPESTIMATE2011',
'POPESTIMATE2012',
'POPESTIMATE2013',
'POPESTIMATE2014',
'POPESTIMATE2015']
df = df[columns_to_keep]
df.head()
df = df.set_index(['STNAME', 'CTYNAME'])
df.head()
df.loc['Michigan', 'Washtenaw County']
df.loc[ [('Michigan', 'Washtenaw County'),
('Michigan', 'Wayne County')] ]
df = pd.read_csv('log.csv')
df
df.fillna?
df = df.set_index('time')
df = df.sort_index()
df
df = df.reset_index()
df = df.set_index(['time', 'user'])
df
df = df.fillna(method='ffill')
df.head()
| 0.129788 | 0.820721 |
```
from lxml import etree
from typing import List, Tuple
def load_sentirueval_2016(file_name: str) -> Tuple[List[str], List[str]]:
texts = []
labels = []
with open(file_name, mode='rb') as fp:
xml_data = fp.read()
root = etree.fromstring(xml_data)
for database in root.getchildren():
if database.tag == 'database':
for table in database.getchildren():
if table.tag != 'table':
continue
new_text = None
new_label = None
for column in table.getchildren():
if column.get('name') == 'text':
new_text = str(column.text).strip()
if new_label is not None:
break
elif column.get('name') not in {'id', 'twitid', 'date'}:
if new_label is None:
label_candidate = str(column.text).strip()
if label_candidate in {'0', '1', '-1'}:
new_label = 'negative' if label_candidate == '-1' else \
('positive' if label_candidate == '1' else 'neutral')
if new_text is not None:
break
if (new_text is None) or (new_label is None):
raise ValueError('File `{0}` contains some error!'.format(file_name))
texts.append(new_text)
labels.append(new_label)
break
return texts, labels
texts, labels = load_sentirueval_2016('bank_train_2016.xml')
print('Number of texts is {0}, number of labels is {1}.'.format(len(texts), len(labels)))
import random
for idx in random.choices(list(range(len(texts))), k=20):
print('{0} => {1}'.format(labels[idx], texts[idx]))
positive_tweets = [texts[idx] for idx in range(len(texts)) if labels[idx] == 'positive']
negative_tweets = [texts[idx] for idx in range(len(texts)) if labels[idx] == 'negative']
for cur in positive_tweets[:5]: print(cur)
for cur in negative_tweets[:5]: print(cur)
from nltk import word_tokenize
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer(lowercase=True, tokenizer=word_tokenize)
vectorizer.fit(texts)
print(vectorizer.get_feature_names()[0:20])
print(len(vectorizer.get_feature_names()))
X = vectorizer.transform(texts)
print(type(X))
print(texts[0])
print(X[0])
print(vectorizer.get_feature_names()[6321])
print(vectorizer.get_feature_names()[9866])
print(vectorizer.get_feature_names()[19056])
from sklearn.feature_extraction.text import TfidfTransformer
transformer = TfidfTransformer().fit(X)
X_transformed = transformer.transform(X)
print(X_transformed[0])
print(vectorizer.get_feature_names()[19056])
print(vectorizer.get_feature_names()[7199])
tokens_with_IDF = list(zip(vectorizer.get_feature_names(), transformer.idf_))
for feature, idf in tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
sorted_tokens_with_IDF = sorted(tokens_with_IDF, key=lambda it: (-it[1], it[0]))
for feature, idf in sorted_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
from sklearn.feature_selection import SelectPercentile, chi2
selector = SelectPercentile(chi2, percentile=20)
selector.fit(X_transformed, labels)
selected_tokens_with_IDF = [tokens_with_IDF[idx] for idx in selector.get_support(indices=True)]
print(len(selected_tokens_with_IDF))
for feature, idf in selected_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
selected_and_sorted_tokens_with_IDF = sorted(selected_tokens_with_IDF, key=lambda it: (-it[1], it[0]))
for feature, idf in selected_and_sorted_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
from sklearn.linear_model import LogisticRegression
cls = LogisticRegression(solver='liblinear', verbose=True)
X_transformed_and_selected = selector.transform(X_transformed)
cls.fit(X_transformed_and_selected, labels)
classes_list = list(cls.classes_)
print(classes_list)
texts_for_testing, labels_for_testing = load_sentirueval_2016('banks_test_etalon.xml')
from sklearn.pipeline import Pipeline
pipeline_for_se = Pipeline(
[
('vectorizer', vectorizer),
('tfidf', transformer),
('selector', selector),
('final_classifier', cls)
]
)
from sklearn.metrics import roc_curve
import numpy as np
probabilities = pipeline_for_se.predict_proba(texts_for_testing)
y_true_positives = list(map(lambda it: 1 if it == 'positive' else 0, labels_for_testing))
y_score_positives = probabilities[:, classes_list.index('positive')]
fpr_positives, tpr_positives, _, = roc_curve(y_true_positives, y_score_positives)
y_true_negatives = list(map(lambda it: 1 if it == 'negative' else 0, labels_for_testing))
y_score_negatives = probabilities[:, classes_list.index('negative')]
fpr_negatives, tpr_negatives, _, = roc_curve(y_true_negatives, y_score_negatives)
y_true_neutrals = list(map(lambda it: 1 if it == 'neutral' else 0, labels_for_testing))
y_score_neutrals = probabilities[:, classes_list.index('neutral')]
fpr_neutrals, tpr_neutrals, _, = roc_curve(y_true_neutrals, y_score_neutrals)
import matplotlib.pyplot as plt
plt.figure(1)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr_positives, tpr_positives, label='Positive Sentiment')
plt.plot(fpr_negatives, tpr_negatives, label='Negative Sentiment')
plt.plot(fpr_neutrals, tpr_neutrals, label='Neutral Sentiment')
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
from sklearn.metrics import f1_score
predicted_labels = pipeline_for_se.predict(texts_for_testing)
print('F1-micro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='micro')))
print('F1-macro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='macro')))
from sklearn.metrics import classification_report
print(classification_report(labels_for_testing, predicted_labels, digits=6))
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
f1_macro_scorer = make_scorer(f1_score, average='macro')
cv = GridSearchCV(
estimator=LogisticRegression(solver='liblinear'),
param_grid={'C': [1e-2, 1e-1, 1, 1e+1, 1e+2, 1e+3], 'penalty': ['l1', 'l2']},
scoring=f1_macro_scorer,
n_jobs=-1
)
cv.fit(X_transformed_and_selected, labels)
print(cv.best_params_)
best_cls = LogisticRegression(solver='liblinear', verbose=True, C=10.0, penalty='l1')
best_cls.fit(X_transformed_and_selected, labels)
best_pipeline_for_se = Pipeline(
[
('vectorizer', vectorizer),
('tfidf', transformer),
('selector', selector),
('final_classifier', best_cls)
]
)
probabilities = best_pipeline_for_se.predict_proba(texts_for_testing)
y_true_positives = list(map(lambda it: 1 if it == 'positive' else 0, labels_for_testing))
y_score_positives = probabilities[:, classes_list.index('positive')]
fpr_positives, tpr_positives, _, = roc_curve(y_true_positives, y_score_positives)
y_true_negatives = list(map(lambda it: 1 if it == 'negative' else 0, labels_for_testing))
y_score_negatives = probabilities[:, classes_list.index('negative')]
fpr_negatives, tpr_negatives, _, = roc_curve(y_true_negatives, y_score_negatives)
y_true_neutrals = list(map(lambda it: 1 if it == 'neutral' else 0, labels_for_testing))
y_score_neutrals = probabilities[:, classes_list.index('neutral')]
fpr_neutrals, tpr_neutrals, _, = roc_curve(y_true_neutrals, y_score_neutrals)
plt.figure(2)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr_positives, tpr_positives, label='Positive Sentiment')
plt.plot(fpr_negatives, tpr_negatives, label='Negative Sentiment')
plt.plot(fpr_neutrals, tpr_neutrals, label='Neutral Sentiment')
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
predicted_labels = best_pipeline_for_se.predict(texts_for_testing)
print('F1-micro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='micro')))
print('F1-macro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='macro')))
print(classification_report(labels_for_testing, predicted_labels, digits=6))
```
|
github_jupyter
|
from lxml import etree
from typing import List, Tuple
def load_sentirueval_2016(file_name: str) -> Tuple[List[str], List[str]]:
texts = []
labels = []
with open(file_name, mode='rb') as fp:
xml_data = fp.read()
root = etree.fromstring(xml_data)
for database in root.getchildren():
if database.tag == 'database':
for table in database.getchildren():
if table.tag != 'table':
continue
new_text = None
new_label = None
for column in table.getchildren():
if column.get('name') == 'text':
new_text = str(column.text).strip()
if new_label is not None:
break
elif column.get('name') not in {'id', 'twitid', 'date'}:
if new_label is None:
label_candidate = str(column.text).strip()
if label_candidate in {'0', '1', '-1'}:
new_label = 'negative' if label_candidate == '-1' else \
('positive' if label_candidate == '1' else 'neutral')
if new_text is not None:
break
if (new_text is None) or (new_label is None):
raise ValueError('File `{0}` contains some error!'.format(file_name))
texts.append(new_text)
labels.append(new_label)
break
return texts, labels
texts, labels = load_sentirueval_2016('bank_train_2016.xml')
print('Number of texts is {0}, number of labels is {1}.'.format(len(texts), len(labels)))
import random
for idx in random.choices(list(range(len(texts))), k=20):
print('{0} => {1}'.format(labels[idx], texts[idx]))
positive_tweets = [texts[idx] for idx in range(len(texts)) if labels[idx] == 'positive']
negative_tweets = [texts[idx] for idx in range(len(texts)) if labels[idx] == 'negative']
for cur in positive_tweets[:5]: print(cur)
for cur in negative_tweets[:5]: print(cur)
from nltk import word_tokenize
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer(lowercase=True, tokenizer=word_tokenize)
vectorizer.fit(texts)
print(vectorizer.get_feature_names()[0:20])
print(len(vectorizer.get_feature_names()))
X = vectorizer.transform(texts)
print(type(X))
print(texts[0])
print(X[0])
print(vectorizer.get_feature_names()[6321])
print(vectorizer.get_feature_names()[9866])
print(vectorizer.get_feature_names()[19056])
from sklearn.feature_extraction.text import TfidfTransformer
transformer = TfidfTransformer().fit(X)
X_transformed = transformer.transform(X)
print(X_transformed[0])
print(vectorizer.get_feature_names()[19056])
print(vectorizer.get_feature_names()[7199])
tokens_with_IDF = list(zip(vectorizer.get_feature_names(), transformer.idf_))
for feature, idf in tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
sorted_tokens_with_IDF = sorted(tokens_with_IDF, key=lambda it: (-it[1], it[0]))
for feature, idf in sorted_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
from sklearn.feature_selection import SelectPercentile, chi2
selector = SelectPercentile(chi2, percentile=20)
selector.fit(X_transformed, labels)
selected_tokens_with_IDF = [tokens_with_IDF[idx] for idx in selector.get_support(indices=True)]
print(len(selected_tokens_with_IDF))
for feature, idf in selected_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
selected_and_sorted_tokens_with_IDF = sorted(selected_tokens_with_IDF, key=lambda it: (-it[1], it[0]))
for feature, idf in selected_and_sorted_tokens_with_IDF[0:20]: print('{0:.6f} => {1}'.format(idf, feature))
from sklearn.linear_model import LogisticRegression
cls = LogisticRegression(solver='liblinear', verbose=True)
X_transformed_and_selected = selector.transform(X_transformed)
cls.fit(X_transformed_and_selected, labels)
classes_list = list(cls.classes_)
print(classes_list)
texts_for_testing, labels_for_testing = load_sentirueval_2016('banks_test_etalon.xml')
from sklearn.pipeline import Pipeline
pipeline_for_se = Pipeline(
[
('vectorizer', vectorizer),
('tfidf', transformer),
('selector', selector),
('final_classifier', cls)
]
)
from sklearn.metrics import roc_curve
import numpy as np
probabilities = pipeline_for_se.predict_proba(texts_for_testing)
y_true_positives = list(map(lambda it: 1 if it == 'positive' else 0, labels_for_testing))
y_score_positives = probabilities[:, classes_list.index('positive')]
fpr_positives, tpr_positives, _, = roc_curve(y_true_positives, y_score_positives)
y_true_negatives = list(map(lambda it: 1 if it == 'negative' else 0, labels_for_testing))
y_score_negatives = probabilities[:, classes_list.index('negative')]
fpr_negatives, tpr_negatives, _, = roc_curve(y_true_negatives, y_score_negatives)
y_true_neutrals = list(map(lambda it: 1 if it == 'neutral' else 0, labels_for_testing))
y_score_neutrals = probabilities[:, classes_list.index('neutral')]
fpr_neutrals, tpr_neutrals, _, = roc_curve(y_true_neutrals, y_score_neutrals)
import matplotlib.pyplot as plt
plt.figure(1)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr_positives, tpr_positives, label='Positive Sentiment')
plt.plot(fpr_negatives, tpr_negatives, label='Negative Sentiment')
plt.plot(fpr_neutrals, tpr_neutrals, label='Neutral Sentiment')
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
from sklearn.metrics import f1_score
predicted_labels = pipeline_for_se.predict(texts_for_testing)
print('F1-micro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='micro')))
print('F1-macro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='macro')))
from sklearn.metrics import classification_report
print(classification_report(labels_for_testing, predicted_labels, digits=6))
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer
f1_macro_scorer = make_scorer(f1_score, average='macro')
cv = GridSearchCV(
estimator=LogisticRegression(solver='liblinear'),
param_grid={'C': [1e-2, 1e-1, 1, 1e+1, 1e+2, 1e+3], 'penalty': ['l1', 'l2']},
scoring=f1_macro_scorer,
n_jobs=-1
)
cv.fit(X_transformed_and_selected, labels)
print(cv.best_params_)
best_cls = LogisticRegression(solver='liblinear', verbose=True, C=10.0, penalty='l1')
best_cls.fit(X_transformed_and_selected, labels)
best_pipeline_for_se = Pipeline(
[
('vectorizer', vectorizer),
('tfidf', transformer),
('selector', selector),
('final_classifier', best_cls)
]
)
probabilities = best_pipeline_for_se.predict_proba(texts_for_testing)
y_true_positives = list(map(lambda it: 1 if it == 'positive' else 0, labels_for_testing))
y_score_positives = probabilities[:, classes_list.index('positive')]
fpr_positives, tpr_positives, _, = roc_curve(y_true_positives, y_score_positives)
y_true_negatives = list(map(lambda it: 1 if it == 'negative' else 0, labels_for_testing))
y_score_negatives = probabilities[:, classes_list.index('negative')]
fpr_negatives, tpr_negatives, _, = roc_curve(y_true_negatives, y_score_negatives)
y_true_neutrals = list(map(lambda it: 1 if it == 'neutral' else 0, labels_for_testing))
y_score_neutrals = probabilities[:, classes_list.index('neutral')]
fpr_neutrals, tpr_neutrals, _, = roc_curve(y_true_neutrals, y_score_neutrals)
plt.figure(2)
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr_positives, tpr_positives, label='Positive Sentiment')
plt.plot(fpr_negatives, tpr_negatives, label='Negative Sentiment')
plt.plot(fpr_neutrals, tpr_neutrals, label='Neutral Sentiment')
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
predicted_labels = best_pipeline_for_se.predict(texts_for_testing)
print('F1-micro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='micro')))
print('F1-macro = {0:.6f}'.format(f1_score(labels_for_testing, predicted_labels, average='macro')))
print(classification_report(labels_for_testing, predicted_labels, digits=6))
| 0.640973 | 0.286937 |
```
import cartopy.crs
import os
from glob import glob
from cartopy import feature
import matplotlib.pyplot as plt
import matplotlib as mpl
from matplotlib.ticker import FormatStrFormatter
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
import numpy
import xarray
import pandas
import pathlib
import yaml
import math
from haversine import haversine
import matplotlib.gridspec as gridspec
```
## Spill locations
```
lat = {}
lon = {}
sample_radius = 20 #km
# BP
lat['BP'] = 48.86111
lon['BP'] = -122.758
# Haro Strait, between Stuart Is. and Moresby Is.
# US/CAD border at Turn Point
lat['StuartMoresby'] = 48 + 39/60 + 58/3600
lon['StuartMoresby'] = -123 -12/60 - 26/3600
# EJDF (this ended up being too NW to capture the central voluntary traffic route region)
# lat['EJDF'] = 48 + 26/60 + 7/3600
# lon['EJDF'] = -122 - 54/60 - 35/3600
# EJDF on CAD/US border
lat['EJDF'] = 48 + 16/60 + 39/3600
lon['EJDF'] = -123 - 6/60 - 30/3600
# EJDF on CAD/US border
lat['Salmon Bank'] = 48 + 16/60 + 39/3600
lon['Salmon Bank'] = -123
SM = (lat['StuartMoresby'],lon['StuartMoresby'])
[*lat]
```
## Graphic specifications
```
mpl.rc('font', size=11)
# some of the following may be repetetive but can also be set relative to the font value above
# (eg "xx-small, x-small,small, medium, large, x-large, xx-large, larger, or smaller"; see link above for details)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
mpl.rc('legend', fontsize=12)
mpl.rc('axes', titlesize=16)
mpl.rc('axes', labelsize=12)
mpl.rc('figure', titlesize=16)
mpl.rc('text', usetex=False)
mpl.rc('font', family='sans-serif', weight='normal', style='normal')
def concatenate_spillcsv_files():
"""
Concatenates all .csv spill files in directory
and changes the oil type names to those used in our study
"""
df={}
data_directory = '/ocean/rmueller/MIDOSS/spill_files_mctest/'
filenames = sorted(glob(os.path.join(data_directory,"*.csv")))
for index,fn in enumerate(filenames):
print(index,fn)
df[index] = pandas.read_csv(fn)
# rename lagrangian files as oil types (for plotting)
df[index]['Lagrangian_template'] = df[index]['Lagrangian_template'].replace(
['Lagrangian_akns.dat','Lagrangian_bunker.dat',
'Lagrangian_diesel.dat','Lagrangian_gas.dat',
'Lagrangian_jet.dat','Lagrangian_dilbit.dat',
'Lagrangian_other.dat'],
['ANS','Bunker-C','Diesel','Diesel',
'Diesel', 'Dilbit', 'Bunker-C']
)
if index == 0:
df_combined = df[index].copy()
else:
df_combined = pandas.concat([df_combined, df[index]])
return df, df_combined
def bins_labels(bins, **kwargs):
"""
Labels x-axis of histogram with labels in center of bar
Inputs:
- bins [vector]: e.g. bins = numpy.arange(0,1.7e8,1e7)
"""
bin_w = (max(bins) - min(bins)) / (len(bins) - 1)
plt.xticks(numpy.arange(min(bins)+bin_w/2, max(bins)+2*bin_w/2, bin_w), bins, **kwargs, rotation=90)
plt.xlim(bins[0], bins[-1])
```
### Load 100,000 spills files
```
df, df_combined = concatenate_spillcsv_files()
print(f'{max(df_combined.spill_volume):.2e}')
```
#### Create histogram of spill volumes for all 70,000 iterations
```
fig, ax = plt.subplots(figsize=(10, 5))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(df_combined.spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of spill volume (liters)\n Bin width = {bin_width:1.1e}")
plt.show()
```
### Retrieve values within 20 km of TP and EJDF
```
for region in [*lat]:
distance = []
# loop through spill location and calculate distance from region epicenter
for idx,spill_lats in enumerate(df_combined['spill_lat']):
spill_site = (df_combined.spill_lat.iloc[idx], df_combined.spill_lon.iloc[idx])
distance.append(haversine(
(lat[region],lon[region]),
spill_site
))
# I tried doing a directly assign but couldn't get it to work for some reason
df_combined[f'distance_{region}'] = distance
subsampled_spills = {}
max_spill_volume = 0
for region in [*lat]:
subsampled_spills[region] = df_combined.loc[
(df_combined[f'distance_{region}'] < sample_radius)
]
fig, ax = plt.subplots(figsize=(7,4))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(subsampled_spills[region].spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of {subsampled_spills[region].shape[0]} spill volumes (liters) in {region} \n Bin width = {bin_width:1.1e}")
plt.show()
# Determing max spill volume across all regions
max_spill_volume = max(max_spill_volume,max(subsampled_spills[region].spill_volume))
subsampled_spills[region].head(1)
oil_types = subsampled_spills[region].groupby('Lagrangian_template').count()
oil_types.head(1)
```
### plot oil type count by spill region
```
oil_types = {}
for region in [*lat]:
oil_types[region]=subsampled_spills[region]
fig, ax = plt.subplots(figsize=(7,4))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(subsampled_spills[region].spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of {subsampled_spills[region].shape[0]} spill volumes (liters) in {region} \n Bin width = {bin_width:1.1e}")
plt.show()
# Determing max spill volume across all regions
max_spill_volume = max(max_spill_volume,max(subsampled_spills[region].spill_volume))
# define axis limits
axlim = numpy.zeros(4)
axlim[0] = -122-17/60-29/3600
axlim[1] = -124-44/60-40/3600
axlim[2] = 47+47/60+36/3600
axlim[3] =49+34/60+54/3600
# axlim[0] = df_combined.spill_lon.min()
# axlim[1] = df_combined.spill_lon.max()
# axlim[2] = df_combined.spill_lat.min()
# axlim[3] = df_combined.spill_lat.max()
spill_volume_fraction = {}
region_colors = {
'StuartMoresby':'darkslateblue',
'EJDF':'darkslategrey',
'Salmon Bank':'salmon',
'BP':'limegreen'
}
ms = 300
rotated_crs = cartopy.crs.RotatedPole(
pole_longitude=120.0,
pole_latitude=63.75
)
plain_crs = cartopy.crs.PlateCarree()
%matplotlib inline
fig,axs = plt.subplots(figsize=(30, 15),
subplot_kw={"projection": rotated_crs, "facecolor": "white"}
)
terminal_loc = 0
axs.add_feature(feature.GSHHSFeature(
'auto',
edgecolor='darkgrey',
facecolor='none'
), zorder=1)
for region in [*lat]:
spill_volume_fraction[region] = (
subsampled_spills[region].spill_volume/max_spill_volume
)
scatter_bunker = axs.scatter(
subsampled_spills[region].spill_lon,
subsampled_spills[region].spill_lat,
s = ms * spill_volume_fraction[region],
color=region_colors[region],
edgecolors='grey',
linewidth=0.7,
transform=plain_crs,
zorder=2)
axs.scatter(
lon.values(),
lat.values(),
color='yellow',
marker=(5, 1),
s = 150,
alpha=0.5,
edgecolors=region_colors[region],
linewidth=0.7,
transform=plain_crs,
zorder=2)
axs.set_extent(axlim, crs=plain_crs)
axs.set_title('20 km spill radius examples')
plt.savefig('20km_examples.jpeg')
plt.show()
lon.values()
```
|
github_jupyter
|
import cartopy.crs
import os
from glob import glob
from cartopy import feature
import matplotlib.pyplot as plt
import matplotlib as mpl
from matplotlib.ticker import FormatStrFormatter
from mpl_toolkits.axes_grid1.inset_locator import inset_axes
import numpy
import xarray
import pandas
import pathlib
import yaml
import math
from haversine import haversine
import matplotlib.gridspec as gridspec
lat = {}
lon = {}
sample_radius = 20 #km
# BP
lat['BP'] = 48.86111
lon['BP'] = -122.758
# Haro Strait, between Stuart Is. and Moresby Is.
# US/CAD border at Turn Point
lat['StuartMoresby'] = 48 + 39/60 + 58/3600
lon['StuartMoresby'] = -123 -12/60 - 26/3600
# EJDF (this ended up being too NW to capture the central voluntary traffic route region)
# lat['EJDF'] = 48 + 26/60 + 7/3600
# lon['EJDF'] = -122 - 54/60 - 35/3600
# EJDF on CAD/US border
lat['EJDF'] = 48 + 16/60 + 39/3600
lon['EJDF'] = -123 - 6/60 - 30/3600
# EJDF on CAD/US border
lat['Salmon Bank'] = 48 + 16/60 + 39/3600
lon['Salmon Bank'] = -123
SM = (lat['StuartMoresby'],lon['StuartMoresby'])
[*lat]
mpl.rc('font', size=11)
# some of the following may be repetetive but can also be set relative to the font value above
# (eg "xx-small, x-small,small, medium, large, x-large, xx-large, larger, or smaller"; see link above for details)
mpl.rc('xtick', labelsize=12)
mpl.rc('ytick', labelsize=12)
mpl.rc('legend', fontsize=12)
mpl.rc('axes', titlesize=16)
mpl.rc('axes', labelsize=12)
mpl.rc('figure', titlesize=16)
mpl.rc('text', usetex=False)
mpl.rc('font', family='sans-serif', weight='normal', style='normal')
def concatenate_spillcsv_files():
"""
Concatenates all .csv spill files in directory
and changes the oil type names to those used in our study
"""
df={}
data_directory = '/ocean/rmueller/MIDOSS/spill_files_mctest/'
filenames = sorted(glob(os.path.join(data_directory,"*.csv")))
for index,fn in enumerate(filenames):
print(index,fn)
df[index] = pandas.read_csv(fn)
# rename lagrangian files as oil types (for plotting)
df[index]['Lagrangian_template'] = df[index]['Lagrangian_template'].replace(
['Lagrangian_akns.dat','Lagrangian_bunker.dat',
'Lagrangian_diesel.dat','Lagrangian_gas.dat',
'Lagrangian_jet.dat','Lagrangian_dilbit.dat',
'Lagrangian_other.dat'],
['ANS','Bunker-C','Diesel','Diesel',
'Diesel', 'Dilbit', 'Bunker-C']
)
if index == 0:
df_combined = df[index].copy()
else:
df_combined = pandas.concat([df_combined, df[index]])
return df, df_combined
def bins_labels(bins, **kwargs):
"""
Labels x-axis of histogram with labels in center of bar
Inputs:
- bins [vector]: e.g. bins = numpy.arange(0,1.7e8,1e7)
"""
bin_w = (max(bins) - min(bins)) / (len(bins) - 1)
plt.xticks(numpy.arange(min(bins)+bin_w/2, max(bins)+2*bin_w/2, bin_w), bins, **kwargs, rotation=90)
plt.xlim(bins[0], bins[-1])
df, df_combined = concatenate_spillcsv_files()
print(f'{max(df_combined.spill_volume):.2e}')
fig, ax = plt.subplots(figsize=(10, 5))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(df_combined.spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of spill volume (liters)\n Bin width = {bin_width:1.1e}")
plt.show()
for region in [*lat]:
distance = []
# loop through spill location and calculate distance from region epicenter
for idx,spill_lats in enumerate(df_combined['spill_lat']):
spill_site = (df_combined.spill_lat.iloc[idx], df_combined.spill_lon.iloc[idx])
distance.append(haversine(
(lat[region],lon[region]),
spill_site
))
# I tried doing a directly assign but couldn't get it to work for some reason
df_combined[f'distance_{region}'] = distance
subsampled_spills = {}
max_spill_volume = 0
for region in [*lat]:
subsampled_spills[region] = df_combined.loc[
(df_combined[f'distance_{region}'] < sample_radius)
]
fig, ax = plt.subplots(figsize=(7,4))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(subsampled_spills[region].spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of {subsampled_spills[region].shape[0]} spill volumes (liters) in {region} \n Bin width = {bin_width:1.1e}")
plt.show()
# Determing max spill volume across all regions
max_spill_volume = max(max_spill_volume,max(subsampled_spills[region].spill_volume))
subsampled_spills[region].head(1)
oil_types = subsampled_spills[region].groupby('Lagrangian_template').count()
oil_types.head(1)
oil_types = {}
for region in [*lat]:
oil_types[region]=subsampled_spills[region]
fig, ax = plt.subplots(figsize=(7,4))
bin_width = 1e7
bins = numpy.arange(0, 1.8e8,bin_width)
binned_data = plt.hist(subsampled_spills[region].spill_volume, bins=bins, histtype = 'bar', facecolor = 'blue')
bins_labels(bins, fontsize=12)
ax.xaxis.set_major_formatter(FormatStrFormatter('%.2e'))
plt.yscale("log")
plt.ylabel("spill counts")
plt.title(f"Histogram of {subsampled_spills[region].shape[0]} spill volumes (liters) in {region} \n Bin width = {bin_width:1.1e}")
plt.show()
# Determing max spill volume across all regions
max_spill_volume = max(max_spill_volume,max(subsampled_spills[region].spill_volume))
# define axis limits
axlim = numpy.zeros(4)
axlim[0] = -122-17/60-29/3600
axlim[1] = -124-44/60-40/3600
axlim[2] = 47+47/60+36/3600
axlim[3] =49+34/60+54/3600
# axlim[0] = df_combined.spill_lon.min()
# axlim[1] = df_combined.spill_lon.max()
# axlim[2] = df_combined.spill_lat.min()
# axlim[3] = df_combined.spill_lat.max()
spill_volume_fraction = {}
region_colors = {
'StuartMoresby':'darkslateblue',
'EJDF':'darkslategrey',
'Salmon Bank':'salmon',
'BP':'limegreen'
}
ms = 300
rotated_crs = cartopy.crs.RotatedPole(
pole_longitude=120.0,
pole_latitude=63.75
)
plain_crs = cartopy.crs.PlateCarree()
%matplotlib inline
fig,axs = plt.subplots(figsize=(30, 15),
subplot_kw={"projection": rotated_crs, "facecolor": "white"}
)
terminal_loc = 0
axs.add_feature(feature.GSHHSFeature(
'auto',
edgecolor='darkgrey',
facecolor='none'
), zorder=1)
for region in [*lat]:
spill_volume_fraction[region] = (
subsampled_spills[region].spill_volume/max_spill_volume
)
scatter_bunker = axs.scatter(
subsampled_spills[region].spill_lon,
subsampled_spills[region].spill_lat,
s = ms * spill_volume_fraction[region],
color=region_colors[region],
edgecolors='grey',
linewidth=0.7,
transform=plain_crs,
zorder=2)
axs.scatter(
lon.values(),
lat.values(),
color='yellow',
marker=(5, 1),
s = 150,
alpha=0.5,
edgecolors=region_colors[region],
linewidth=0.7,
transform=plain_crs,
zorder=2)
axs.set_extent(axlim, crs=plain_crs)
axs.set_title('20 km spill radius examples')
plt.savefig('20km_examples.jpeg')
plt.show()
lon.values()
| 0.393735 | 0.698728 |
```
import discretisedfield as df
import xarray as xr
import numpy as np
mesh = df.Mesh(p1=(0, 0, 0), p2=(20, 10, 5), cell=(1, 1, 1))
field0 = df.Field(mesh, dim=3, value=(0, 0, 1), norm=1)
```
Four additional fields with changed value, to resemble a time series.
```
field1 = df.Field(mesh, dim=3, value=(1, 0, 2), norm=1)
field2 = df.Field(mesh, dim=3, value=(1, 0, 1), norm=1)
field3 = df.Field(mesh, dim=3, value=(2, 0, 1), norm=1)
field4 = df.Field(mesh, dim=3, value=(1, 0, 0), norm=1)
field0.plane('y').mpl()
field1.plane('y').mpl()
```
### Coordinates of the mesh/in time
Mesh coordinates shoud at some point be available from `df.Mesh`.
```
x = np.linspace(0.5, 19.5, 20)
y = np.linspace(0.5, 9.5, 10)
z = np.linspace(0.5, 4.5, 5)
t = np.linspace(0, 4, 5)
```
---
# Single field
### DataArray
```
data = xr.DataArray(field0.array,
dims=('x', 'y', 'z', 'vector'),
coords={'x': x, 'y': y, 'z': z, 'vector': ['mx', 'my', 'mz']})
data
```
4 different access methods (for the same point):
```
data[0, 0, 0]
data.loc[.5, .5, .5]
data.isel(x=0, y=0, z=0)
data.sel(x=.5, y=.5, z=.5, vector='mx')#.sel(vector='mx')
```
Getting one vector component
```
data.sel(vector='mx')
data['vector']
data.sel(z=.5, vector='mz').plot()
```
### Dataset
```
dataset = xr.Dataset(
{
'mx': (['x', 'y', 'z'], field0.array[..., 0]), # field0.x
'my': (['x', 'y', 'z'], field0.array[..., 1]),
'mz': (['x', 'y', 'z'], field0.array[..., 2]),
},
coords={
'x': x,
'y': y,
'z': z,
},
)
dataset
dataset.loc[dict(x=.5, y=.5, z=.5)]
dataset[{'x': 0, 'y': 0, 'z': 0}]
dataset.mx
```
Access to the underlying numpy array
```
type(dataset.to_array().data)
dataset.sel(x=.5, y=.5, z=.5).to_array().data
dataset.mx.sel(z=.5).plot()
```
---
# Time series
```
data = xr.DataArray(np.stack([field0.array,
field1.array,
field2.array,
field3.array,
field4.array]),
dims=('t', 'x', 'y', 'z', 'vector'),
coords={'t': t, 'x': x, 'y': y, 'z': z, 'vector': ['mx', 'my', 'mz']})
data
dataset = xr.Dataset(
{
'mx': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 0],
field1.array[..., 0],
field2.array[..., 0],
field3.array[..., 0],
field4.array[..., 0]])),
'my': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 1],
field1.array[..., 1],
field2.array[..., 1],
field3.array[..., 1],
field4.array[..., 1]])),
'mz': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 2],
field1.array[..., 2],
field2.array[..., 2],
field3.array[..., 2],
field4.array[..., 2]])),
},
coords={
't': t,
'x': x,
'y': y,
'z': z,
},
)
dataset
```
|
github_jupyter
|
import discretisedfield as df
import xarray as xr
import numpy as np
mesh = df.Mesh(p1=(0, 0, 0), p2=(20, 10, 5), cell=(1, 1, 1))
field0 = df.Field(mesh, dim=3, value=(0, 0, 1), norm=1)
field1 = df.Field(mesh, dim=3, value=(1, 0, 2), norm=1)
field2 = df.Field(mesh, dim=3, value=(1, 0, 1), norm=1)
field3 = df.Field(mesh, dim=3, value=(2, 0, 1), norm=1)
field4 = df.Field(mesh, dim=3, value=(1, 0, 0), norm=1)
field0.plane('y').mpl()
field1.plane('y').mpl()
x = np.linspace(0.5, 19.5, 20)
y = np.linspace(0.5, 9.5, 10)
z = np.linspace(0.5, 4.5, 5)
t = np.linspace(0, 4, 5)
data = xr.DataArray(field0.array,
dims=('x', 'y', 'z', 'vector'),
coords={'x': x, 'y': y, 'z': z, 'vector': ['mx', 'my', 'mz']})
data
data[0, 0, 0]
data.loc[.5, .5, .5]
data.isel(x=0, y=0, z=0)
data.sel(x=.5, y=.5, z=.5, vector='mx')#.sel(vector='mx')
data.sel(vector='mx')
data['vector']
data.sel(z=.5, vector='mz').plot()
dataset = xr.Dataset(
{
'mx': (['x', 'y', 'z'], field0.array[..., 0]), # field0.x
'my': (['x', 'y', 'z'], field0.array[..., 1]),
'mz': (['x', 'y', 'z'], field0.array[..., 2]),
},
coords={
'x': x,
'y': y,
'z': z,
},
)
dataset
dataset.loc[dict(x=.5, y=.5, z=.5)]
dataset[{'x': 0, 'y': 0, 'z': 0}]
dataset.mx
type(dataset.to_array().data)
dataset.sel(x=.5, y=.5, z=.5).to_array().data
dataset.mx.sel(z=.5).plot()
data = xr.DataArray(np.stack([field0.array,
field1.array,
field2.array,
field3.array,
field4.array]),
dims=('t', 'x', 'y', 'z', 'vector'),
coords={'t': t, 'x': x, 'y': y, 'z': z, 'vector': ['mx', 'my', 'mz']})
data
dataset = xr.Dataset(
{
'mx': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 0],
field1.array[..., 0],
field2.array[..., 0],
field3.array[..., 0],
field4.array[..., 0]])),
'my': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 1],
field1.array[..., 1],
field2.array[..., 1],
field3.array[..., 1],
field4.array[..., 1]])),
'mz': (['t', 'x', 'y', 'z'], np.stack([field0.array[..., 2],
field1.array[..., 2],
field2.array[..., 2],
field3.array[..., 2],
field4.array[..., 2]])),
},
coords={
't': t,
'x': x,
'y': y,
'z': z,
},
)
dataset
| 0.408041 | 0.943138 |
### Classify Ship images using CNN
```
import pandas as pd
import numpy as np
import keras
from keras.models import Sequential
from keras.layers import Dense, Flatten, Dropout
from keras.layers import Conv2D,MaxPooling2D
from keras.callbacks import ModelCheckpoint
from keras import regularizers
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras.preprocessing import image
from sklearn.model_selection import train_test_split
from tqdm import tqdm
import os
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning v1\\train")
train = pd.read_csv('train.csv')
train_image = []
for i in tqdm(range(train.shape[0])):
img = image.load_img('images/'+train['image'][i], target_size=(28,28,3), grayscale=False)
img = image.img_to_array(img)
#img = img/255
train_image.append(img)
X = np.array(train_image)
from keras.applications.resnet50 import ResNet50, preprocess_input
HEIGHT = 197
WIDTH = 197
base_model = ResNet50(weights='imagenet',
include_top=False,
input_shape=(HEIGHT, WIDTH, 3))
from keras.preprocessing.image import ImageDataGenerator
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning v1")
TRAIN_DIR = "ImgDir"
HEIGHT = 197
WIDTH = 197
BATCH_SIZE = 8
train_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input,
rotation_range=90,
horizontal_flip=True,
vertical_flip=True
)
train_generator = train_datagen.flow_from_directory(TRAIN_DIR,
target_size=(HEIGHT, WIDTH),
batch_size=BATCH_SIZE)
from keras.layers import Dense, Activation, Flatten, Dropout
from keras.models import Sequential, Model
def build_finetune_model(base_model, dropout, fc_layers, num_classes):
for layer in base_model.layers:
layer.trainable = False
x = base_model.output
x = Flatten()(x)
for fc in fc_layers:
# New FC layer, random init
x = Dense(fc, activation='relu')(x)
x = Dropout(dropout)(x)
# New softmax layer
predictions = Dense(num_classes, activation='softmax')(x)
finetune_model = Model(inputs=base_model.input, outputs=predictions)
return finetune_model
class_list = ["Cargo", "Military", "Cruise"]
FC_LAYERS = [1024, 1024]
dropout = 0.5
finetune_model = build_finetune_model(base_model,
dropout=dropout,
fc_layers=FC_LAYERS,
num_classes=len(class_list))
from keras.optimizers import SGD, Adam
NUM_EPOCHS = 1
BATCH_SIZE = 8
num_train_images = 100
adam = Adam(lr=0.00001)
finetune_model.compile(adam, loss='categorical_crossentropy', metrics=['accuracy'])
filepath="checkpoints"
checkpoint = ModelCheckpoint(filepath, monitor=["acc"], verbose=1, mode='max')
callbacks_list = [checkpoint]
history = finetune_model.fit_generator(train_generator, epochs=NUM_EPOCHS, workers=8,
steps_per_epoch=num_train_images // BATCH_SIZE,
shuffle=True, callbacks=callbacks_list)
plot_training(history)
# Plot the training and validation loss + accuracy
def plot_training(history):
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r.')
plt.plot(epochs, val_acc, 'r')
plt.title('Training and validation accuracy')
# plt.figure()
# plt.plot(epochs, loss, 'r.')
# plt.plot(epochs, val_loss, 'r-')
# plt.title('Training and validation loss')
plt.show()
finetune_model.predict()
y=train['category'].values
#y = to_categorical(y)
x_train, x_test, y_train, y_test = train_test_split(X, y, random_state=42, test_size=0.2)
print(x_train.shape)
print(y_train.shape)
print(x_test.shape)
print(y_test.shape)
from matplotlib import pyplot
from scipy.misc import toimage
def show_imgs(X):
pyplot.figure(1)
k = 0
for i in range(0,4):
for j in range(0,4):
pyplot.subplot2grid((4,4),(i,j))
pyplot.imshow(toimage(X[k]))
k = k+1
# show the plot
pyplot.show()
show_imgs(x_test[:16])
x_train=x_train/255
x_test = x_test/255
n_class=6
y_train = keras.utils.to_categorical(y_train,n_class)
y_test = keras.utils.to_categorical(y_test,n_class)
n_conv = 64
k_conv = (3,3)
y_train.shape
weight_decay = 1e-4
model = Sequential()
model.add(Conv2D(32,(3,3),padding='same', activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(Dropout(0.2))
model.add(Conv2D(64,(3,3),padding='same', activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Conv2D(128,(3,3), padding='same',activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(128,activation='relu'))
model.add(Dense(6,activation='softmax'))
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
model.fit(x_train,y_train,epochs=1,verbose=1, validation_data=(x_test,y_test))
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning\\train")
test = pd.read_csv('test_ApKoW4T.csv')
test_image = []
for i in tqdm(range(test.shape[0])):
img = image.load_img('images/'+test['image'][i], target_size=(197,197,3), grayscale=False)
img = image.img_to_array(img)
img = img/255
test_image.append(img)
test = np.array(test_image)
prediction = finetune_model.predict(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
model.save('trained_epoch_5.h5')
filepath='trained_epoch_5.h5'
from keras.callbacks import ModelCheckpoint
from keras.models import load_model
new_model = load_model('trained_epoch_5.h5')
checkpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')
callback_list = [checkpoint]
new_model.fit(x_train,y_train, epochs=20, validation_data=(x_test,y_test),callbacks = callback_list)
new_model.save('trained_epoch_25.h5')
prediction = new_model.predict_classes(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
filepath='trained_epoch_25.h5'
new_model1 = load_model('trained_epoch_25.h5')
checkpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')
callback_list = [checkpoint]
new_model1.fit(x_train,y_train, epochs=20, validation_data=(x_test,y_test),callbacks = callback_list)
prediction=new_model1.predict_classes(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
```
|
github_jupyter
|
import pandas as pd
import numpy as np
import keras
from keras.models import Sequential
from keras.layers import Dense, Flatten, Dropout
from keras.layers import Conv2D,MaxPooling2D
from keras.callbacks import ModelCheckpoint
from keras import regularizers
from keras.layers.normalization import BatchNormalization
from keras.utils import to_categorical
from keras.preprocessing import image
from sklearn.model_selection import train_test_split
from tqdm import tqdm
import os
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning v1\\train")
train = pd.read_csv('train.csv')
train_image = []
for i in tqdm(range(train.shape[0])):
img = image.load_img('images/'+train['image'][i], target_size=(28,28,3), grayscale=False)
img = image.img_to_array(img)
#img = img/255
train_image.append(img)
X = np.array(train_image)
from keras.applications.resnet50 import ResNet50, preprocess_input
HEIGHT = 197
WIDTH = 197
base_model = ResNet50(weights='imagenet',
include_top=False,
input_shape=(HEIGHT, WIDTH, 3))
from keras.preprocessing.image import ImageDataGenerator
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning v1")
TRAIN_DIR = "ImgDir"
HEIGHT = 197
WIDTH = 197
BATCH_SIZE = 8
train_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input,
rotation_range=90,
horizontal_flip=True,
vertical_flip=True
)
train_generator = train_datagen.flow_from_directory(TRAIN_DIR,
target_size=(HEIGHT, WIDTH),
batch_size=BATCH_SIZE)
from keras.layers import Dense, Activation, Flatten, Dropout
from keras.models import Sequential, Model
def build_finetune_model(base_model, dropout, fc_layers, num_classes):
for layer in base_model.layers:
layer.trainable = False
x = base_model.output
x = Flatten()(x)
for fc in fc_layers:
# New FC layer, random init
x = Dense(fc, activation='relu')(x)
x = Dropout(dropout)(x)
# New softmax layer
predictions = Dense(num_classes, activation='softmax')(x)
finetune_model = Model(inputs=base_model.input, outputs=predictions)
return finetune_model
class_list = ["Cargo", "Military", "Cruise"]
FC_LAYERS = [1024, 1024]
dropout = 0.5
finetune_model = build_finetune_model(base_model,
dropout=dropout,
fc_layers=FC_LAYERS,
num_classes=len(class_list))
from keras.optimizers import SGD, Adam
NUM_EPOCHS = 1
BATCH_SIZE = 8
num_train_images = 100
adam = Adam(lr=0.00001)
finetune_model.compile(adam, loss='categorical_crossentropy', metrics=['accuracy'])
filepath="checkpoints"
checkpoint = ModelCheckpoint(filepath, monitor=["acc"], verbose=1, mode='max')
callbacks_list = [checkpoint]
history = finetune_model.fit_generator(train_generator, epochs=NUM_EPOCHS, workers=8,
steps_per_epoch=num_train_images // BATCH_SIZE,
shuffle=True, callbacks=callbacks_list)
plot_training(history)
# Plot the training and validation loss + accuracy
def plot_training(history):
acc = history.history['acc']
val_acc = history.history['val_acc']
loss = history.history['loss']
val_loss = history.history['val_loss']
epochs = range(len(acc))
plt.plot(epochs, acc, 'r.')
plt.plot(epochs, val_acc, 'r')
plt.title('Training and validation accuracy')
# plt.figure()
# plt.plot(epochs, loss, 'r.')
# plt.plot(epochs, val_loss, 'r-')
# plt.title('Training and validation loss')
plt.show()
finetune_model.predict()
y=train['category'].values
#y = to_categorical(y)
x_train, x_test, y_train, y_test = train_test_split(X, y, random_state=42, test_size=0.2)
print(x_train.shape)
print(y_train.shape)
print(x_test.shape)
print(y_test.shape)
from matplotlib import pyplot
from scipy.misc import toimage
def show_imgs(X):
pyplot.figure(1)
k = 0
for i in range(0,4):
for j in range(0,4):
pyplot.subplot2grid((4,4),(i,j))
pyplot.imshow(toimage(X[k]))
k = k+1
# show the plot
pyplot.show()
show_imgs(x_test[:16])
x_train=x_train/255
x_test = x_test/255
n_class=6
y_train = keras.utils.to_categorical(y_train,n_class)
y_test = keras.utils.to_categorical(y_test,n_class)
n_conv = 64
k_conv = (3,3)
y_train.shape
weight_decay = 1e-4
model = Sequential()
model.add(Conv2D(32,(3,3),padding='same', activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(Dropout(0.2))
model.add(Conv2D(64,(3,3),padding='same', activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Conv2D(128,(3,3), padding='same',activation='relu',kernel_regularizer=regularizers.l2(weight_decay)))
model.add(MaxPooling2D())
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(128,activation='relu'))
model.add(Dense(6,activation='softmax'))
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])
model.fit(x_train,y_train,epochs=1,verbose=1, validation_data=(x_test,y_test))
os.chdir("D:\My Personal Documents\Learnings\Data Science\Hackathan - Game of Deep Learning\\train")
test = pd.read_csv('test_ApKoW4T.csv')
test_image = []
for i in tqdm(range(test.shape[0])):
img = image.load_img('images/'+test['image'][i], target_size=(197,197,3), grayscale=False)
img = image.img_to_array(img)
img = img/255
test_image.append(img)
test = np.array(test_image)
prediction = finetune_model.predict(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
model.save('trained_epoch_5.h5')
filepath='trained_epoch_5.h5'
from keras.callbacks import ModelCheckpoint
from keras.models import load_model
new_model = load_model('trained_epoch_5.h5')
checkpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')
callback_list = [checkpoint]
new_model.fit(x_train,y_train, epochs=20, validation_data=(x_test,y_test),callbacks = callback_list)
new_model.save('trained_epoch_25.h5')
prediction = new_model.predict_classes(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
filepath='trained_epoch_25.h5'
new_model1 = load_model('trained_epoch_25.h5')
checkpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')
callback_list = [checkpoint]
new_model1.fit(x_train,y_train, epochs=20, validation_data=(x_test,y_test),callbacks = callback_list)
prediction=new_model1.predict_classes(test)
pd.DataFrame(prediction).to_csv('test_pred.csv')
| 0.748536 | 0.655102 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.