text
stringlengths 2.5k
6.39M
| kind
stringclasses 3
values |
|---|---|
This notebook wants to make use of the evaluation techniques previously developed to select the best algorithms for this problem.
```
import pandas as pd
import numpy as np
import tubesml as tml
from sklearn.model_selection import KFold
from sklearn.pipeline import Pipeline
from sklearn.linear_model import Lasso, Ridge, SGDRegressor
from sklearn.ensemble import RandomForestRegressor, ExtraTreesRegressor
from sklearn.svm import SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
import xgboost as xgb
import lightgbm as lgb
from sklearn.metrics import mean_squared_error, mean_absolute_error
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
import sys
sys.path.append("..")
from source.clean import general_cleaner
from source.transf_category import recode_cat, make_ordinal
from source.transf_numeric import tr_numeric
import source.transf_univ as dfp
import source.utility as ut
import source.report as rp
import warnings
warnings.filterwarnings("ignore",
message="The dummies in this set do not match the ones in the train set, we corrected the issue.")
pd.set_option('max_columns', 500)
```
# Data preparation
Get the data ready to flow into the pipeline
```
df_train = pd.read_csv('../data/train.csv')
df_test = pd.read_csv('../data/test.csv')
df_train['Target'] = np.log1p(df_train.SalePrice)
df_train = df_train[df_train.GrLivArea < 4500].copy().reset_index()
del df_train['SalePrice']
train_set, test_set = ut.make_test(df_train,
test_size=0.2, random_state=654,
strat_feat='Neighborhood')
y = train_set['Target'].copy()
del train_set['Target']
y_test = test_set['Target']
del test_set['Target']
```
## Building the pipeline
This was introduced in another notebook and imported above
```
numeric_pipe = Pipeline([('fs', tml.DtypeSel(dtype='numeric')),
('imputer', tml.DfImputer(strategy='median')),
('transf', tr_numeric())])
cat_pipe = Pipeline([('fs', tml.DtypeSel(dtype='category')),
('imputer', tml.DfImputer(strategy='most_frequent')),
('ord', make_ordinal(['BsmtQual', 'KitchenQual',
'ExterQual', 'HeatingQC'])),
('recode', recode_cat()),
('dummies', tml.Dummify(drop_first=True))])
processing_pipe = tml.FeatureUnionDf(transformer_list=[('cat_pipe', cat_pipe),
('num_pipe', numeric_pipe)])
```
## Evaluation method
We have seen how it works in the previous notebook, we have thus imported the necessary functions above.
```
models = [('lasso', Lasso(alpha=0.01)), ('ridge', Ridge()), ('sgd', SGDRegressor()),
('forest', RandomForestRegressor(n_estimators=200)), ('xtree', ExtraTreesRegressor(n_estimators=200)),
('svr', SVR()),
('kneig', KNeighborsRegressor()),
('xgb', xgb.XGBRegressor(n_estimators=200, objective='reg:squarederror')),
('lgb', lgb.LGBMRegressor(n_estimators=200))]
mod_name = []
rmse_train = []
rmse_test = []
mae_train = []
mae_test = []
folds = KFold(5, shuffle=True, random_state=541)
for model in models:
train = train_set.copy()
test = test_set.copy()
print(model[0])
mod_name.append(model[0])
pipe = [('gen_cl', general_cleaner()),
('processing', processing_pipe),
('scl', dfp.df_scaler())] + [model]
model_pipe = Pipeline(pipe)
inf_preds = tml.cv_score(data=train, target=y, cv=folds, estimator=model_pipe)
model_pipe.fit(train, y)
preds = model_pipe.predict(test)
rp.plot_predictions(test, y_test, preds, savename=model[0]+'_preds.png')
rp.plot_predictions(train, y, inf_preds, savename=model[0]+'_inf_preds.png')
rmse_train.append(mean_squared_error(y, inf_preds))
rmse_test.append(mean_squared_error(y_test, preds))
mae_train.append(mean_absolute_error(np.expm1(y), np.expm1(inf_preds)))
mae_test.append(mean_absolute_error(np.expm1(y_test), np.expm1(preds)))
print(f'\tTrain set RMSE: {round(np.sqrt(mean_squared_error(y, inf_preds)), 4)}')
print(f'\tTrain set MAE: {round(mean_absolute_error(np.expm1(y), np.expm1(inf_preds)), 2)}')
print(f'\tTest set RMSE: {round(np.sqrt(mean_squared_error(y_test, preds)), 4)}')
print(f'\tTrain set MAE: {round(mean_absolute_error(np.expm1(y_test), np.expm1(preds)), 2)}')
print('_'*40)
print('\n')
results = pd.DataFrame({'model_name': mod_name,
'rmse_train': rmse_train, 'rmse_test': rmse_test,
'mae_train': mae_train, 'mae_test': mae_test})
results
results.sort_values(by='rmse_train').head(2)
results.sort_values(by='rmse_test').head(2)
results.sort_values(by='mae_train').head(2)
results.sort_values(by='mae_test').head(2)
```
|
github_jupyter
|
```
%matplotlib inline
```
분류기(Classifier) 학습하기
============================
지금까지 어떻게 신경망을 정의하고, 손실을 계산하며 또 가중치를 갱신하는지에
대해서 배웠습니다.
이제 아마도 이런 생각을 하고 계실텐데요,
데이터는 어떻게 하나요?
------------------------
일반적으로 이미지나 텍스트, 오디오나 비디오 데이터를 다룰 때는 표준 Python 패키지를
이용하여 NumPy 배열로 불러오면 됩니다. 그 후 그 배열을 ``torch.*Tensor`` 로 변환합니다.
- 이미지는 Pillow나 OpenCV 같은 패키지가 유용합니다.
- 오디오를 처리할 때는 SciPy와 LibROSA가 유용하고요.
- 텍스트의 경우에는 그냥 Python이나 Cython을 사용해도 되고, NLTK나 SpaCy도
유용합니다.
특별히 영상 분야를 위한 ``torchvision`` 이라는 패키지가 만들어져 있는데,
여기에는 Imagenet이나 CIFAR10, MNIST 등과 같이 일반적으로 사용하는 데이터셋을 위한
데이터 로더(data loader), 즉 ``torchvision.datasets`` 과 이미지용 데이터 변환기
(data transformer), 즉 ``torch.utils.data.DataLoader`` 가 포함되어 있습니다.
이러한 기능은 엄청나게 편리하며, 매번 유사한 코드(boilerplate code)를 반복해서
작성하는 것을 피할 수 있습니다.
이 튜토리얼에서는 CIFAR10 데이터셋을 사용합니다. 여기에는 다음과 같은 분류들이
있습니다: '비행기(airplane)', '자동차(automobile)', '새(bird)', '고양이(cat)',
'사슴(deer)', '개(dog)', '개구리(frog)', '말(horse)', '배(ship)', '트럭(truck)'.
그리고 CIFAR10에 포함된 이미지의 크기는 3x32x32로, 이는 32x32 픽셀 크기의 이미지가
3개 채널(channel)의 색상로 이뤄져 있다는 것을 뜻합니다.
.. figure:: /_static/img/cifar10.png
:alt: cifar10
cifar10
이미지 분류기 학습하기
----------------------------
다음과 같은 단계로 진행해보겠습니다:
1. ``torchvision`` 을 사용하여 CIFAR10의 학습용 / 시험용 데이터셋을
불러오고, 정규화(nomarlizing)합니다.
2. 합성곱 신경망(Convolution Neural Network)을 정의합니다.
3. 손실 함수를 정의합니다.
4. 학습용 데이터를 사용하여 신경망을 학습합니다.
5. 시험용 데이터를 사용하여 신경망을 검사합니다.
1. CIFAR10을 불러오고 정규화하기
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
``torchvision`` 을 사용하여 매우 쉽게 CIFAR10을 불러올 수 있습니다.
```
import torch
import torchvision
import torchvision.transforms as transforms
```
torchvision 데이터셋의 출력(output)은 [0, 1] 범위를 갖는 PILImage 이미지입니다.
이를 [-1, 1]의 범위로 정규화된 Tensor로 변환합니다.
<div class="alert alert-info"><h4>Note</h4><p>만약 Windows 환경에서 BrokenPipeError가 발생한다면,
torch.utils.data.DataLoader()의 num_worker를 0으로 설정해보세요.</p></div>
```
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
batch_size = 4
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=batch_size,
shuffle=True, num_workers=2)
testset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=batch_size,
shuffle=False, num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
```
재미삼아 학습용 이미지 몇 개를 보겠습니다.
```
import matplotlib.pyplot as plt
import numpy as np
# 이미지를 보여주기 위한 함수
def imshow(img):
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
# 학습용 이미지를 무작위로 가져오기
dataiter = iter(trainloader)
images, labels = dataiter.next()
# 이미지 보여주기
imshow(torchvision.utils.make_grid(images))
# 정답(label) 출력
print(' '.join('%5s' % classes[labels[j]] for j in range(batch_size)))
```
2. 합성곱 신경망(Convolution Neural Network) 정의하기
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
이전의 신경망 섹션에서 신경망을 복사한 후, (기존에 1채널 이미지만 처리하도록
정의된 것을) 3채널 이미지를 처리할 수 있도록 수정합니다.
```
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super().__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = torch.flatten(x, 1) # 배치를 제외한 모든 차원을 평탄화(flatten)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return x
net = Net()
```
3. 손실 함수와 Optimizer 정의하기
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
교차 엔트로피 손실(Cross-Entropy loss)과 모멘텀(momentum) 값을 갖는 SGD를 사용합니다.
```
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(net.parameters(), lr=0.001, momentum=0.9)
```
4. 신경망 학습하기
^^^^^^^^^^^^^^^^^^^^
이제 재미있는 부분이 시작됩니다.
단순히 데이터를 반복해서 신경망에 입력으로 제공하고, 최적화(Optimize)만 하면
됩니다.
```
for epoch in range(2): # 데이터셋을 수차례 반복합니다.
running_loss = 0.0
for i, data in enumerate(trainloader, 0):
# [inputs, labels]의 목록인 data로부터 입력을 받은 후;
inputs, labels = data
# 변화도(Gradient) 매개변수를 0으로 만들고
optimizer.zero_grad()
# 순전파 + 역전파 + 최적화를 한 후
outputs = net(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
# 통계를 출력합니다.
running_loss += loss.item()
if i % 2000 == 1999: # print every 2000 mini-batches
print('[%d, %5d] loss: %.3f' %
(epoch + 1, i + 1, running_loss / 2000))
running_loss = 0.0
print('Finished Training')
```
학습한 모델을 저장해보겠습니다:
```
PATH = './cifar_net.pth'
torch.save(net.state_dict(), PATH)
```
PyTorch 모델을 저장하는 자세한 방법은 `여기 <https://pytorch.org/docs/stable/notes/serialization.html>`_
를 참조해주세요.
5. 시험용 데이터로 신경망 검사하기
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
지금까지 학습용 데이터셋을 2회 반복하며 신경망을 학습시켰습니다.
신경망이 전혀 배운게 없을지도 모르니 확인해봅니다.
신경망이 예측한 출력과 진짜 정답(Ground-truth)을 비교하는 방식으로 확인합니다.
만약 예측이 맞다면 샘플을 '맞은 예측값(correct predictions)' 목록에 넣겠습니다.
첫번째로 시험용 데이터를 좀 보겠습니다.
```
dataiter = iter(testloader)
images, labels = dataiter.next()
# 이미지를 출력합니다.
imshow(torchvision.utils.make_grid(images))
print('GroundTruth: ', ' '.join('%5s' % classes[labels[j]] for j in range(4)))
```
이제, 저장했던 모델을 불러오도록 하겠습니다 (주: 모델을 저장하고 다시 불러오는
작업은 여기에서는 불필요하지만, 어떻게 하는지 설명을 위해 해보겠습니다):
```
net = Net()
net.load_state_dict(torch.load(PATH))
```
좋습니다, 이제 이 예제들을 신경망이 어떻게 예측했는지를 보겠습니다:
```
outputs = net(images)
```
출력은 10개 분류 각각에 대한 값으로 나타납니다. 어떤 분류에 대해서 더 높은 값이
나타난다는 것은, 신경망이 그 이미지가 해당 분류에 더 가깝다고 생각한다는 것입니다.
따라서, 가장 높은 값을 갖는 인덱스(index)를 뽑아보겠습니다:
```
_, predicted = torch.max(outputs, 1)
print('Predicted: ', ' '.join('%5s' % classes[predicted[j]]
for j in range(4)))
```
결과가 괜찮아보이네요.
그럼 전체 데이터셋에 대해서는 어떻게 동작하는지 보겠습니다.
```
correct = 0
total = 0
# 학습 중이 아니므로, 출력에 대한 변화도를 계산할 필요가 없습니다
with torch.no_grad():
for data in testloader:
images, labels = data
# 신경망에 이미지를 통과시켜 출력을 계산합니다
outputs = net(images)
# 가장 높은 값(energy)를 갖는 분류(class)를 정답으로 선택하겠습니다
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print('Accuracy of the network on the 10000 test images: %d %%' % (
100 * correct / total))
```
(10가지 분류 중에 하나를 무작위로) 찍었을 때의 정확도인 10% 보다는 나아보입니다.
신경망이 뭔가 배우긴 한 것 같네요.
그럼 어떤 것들을 더 잘 분류하고, 어떤 것들을 더 못했는지 알아보겠습니다:
```
# 각 분류(class)에 대한 예측값 계산을 위해 준비
correct_pred = {classname: 0 for classname in classes}
total_pred = {classname: 0 for classname in classes}
# 변화도는 여전히 필요하지 않습니다
with torch.no_grad():
for data in testloader:
images, labels = data
outputs = net(images)
_, predictions = torch.max(outputs, 1)
# 각 분류별로 올바른 예측 수를 모읍니다
for label, prediction in zip(labels, predictions):
if label == prediction:
correct_pred[classes[label]] += 1
total_pred[classes[label]] += 1
# 각 분류별 정확도(accuracy)를 출력합니다
for classname, correct_count in correct_pred.items():
accuracy = 100 * float(correct_count) / total_pred[classname]
print("Accuracy for class {:5s} is: {:.1f} %".format(classname,
accuracy))
```
자, 이제 다음으로 무엇을 해볼까요?
이러한 신경망들을 GPU에서 실행하려면 어떻게 해야 할까요?
GPU에서 학습하기
----------------
Tensor를 GPU로 이동했던 것처럼, 신경망 또한 GPU로 옮길 수 있습니다.
먼저 (CUDA를 사용할 수 있다면) 첫번째 CUDA 장치를 사용하도록 설정합니다:
```
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# CUDA 기기가 존재한다면, 아래 코드가 CUDA 장치를 출력합니다:
print(device)
```
이 섹션의 나머지 부분에서는 ``device`` 를 CUDA 장치라고 가정하겠습니다.
그리고 이 메소드(Method)들은 재귀적으로 모든 모듈의 매개변수와 버퍼를
CUDA tensor로 변경합니다:
.. code:: python
net.to(device)
또한, 각 단계에서 입력(input)과 정답(target)도 GPU로 보내야 한다는 것도 기억해야
합니다:
.. code:: python
inputs, labels = data[0].to(device), data[1].to(device)
CPU와 비교했을 때 어마어마한 속도 차이가 나지 않는 것은 왜 그럴까요?
그 이유는 바로 신경망이 너무 작기 때문입니다.
**연습:** 신경망의 크기를 키워보고, 얼마나 빨라지는지 확인해보세요.
(첫번째 ``nn.Conv2d`` 의 2번째 인자와 두번째 ``nn.Conv2d`` 의 1번째 인자는
같은 숫자여야 합니다.)
**다음 목표들을 달성했습니다**:
- 높은 수준에서 PyTorch의 Tensor library와 신경망을 이해합니다.
- 이미지를 분류하는 작은 신경망을 학습시킵니다.
여러개의 GPU에서 학습하기
-------------------------
모든 GPU를 활용해서 더욱 더 속도를 올리고 싶다면, :doc:`data_parallel_tutorial`
을 참고하세요.
이제 무엇을 해볼까요?
-----------------------
- :doc:`비디오 게임을 할 수 있는 신경망 학습시키기 </intermediate/reinforcement_q_learning>`
- `imagenet으로 최첨단(state-of-the-art) ResNet 신경망 학습시키기`_
- `적대적 생성 신경망으로 얼굴 생성기 학습시키기`_
- `순환 LSTM 네트워크를 사용해 단어 단위 언어 모델 학습시키기`_
- `다른 예제들 참고하기`_
- `더 많은 튜토리얼 보기`_
- `포럼에서 PyTorch에 대해 얘기하기`_
- `Slack에서 다른 사용자와 대화하기`_
```
# %%%%%%INVISIBLE_CODE_BLOCK%%%%%%
del dataiter
# %%%%%%INVISIBLE_CODE_BLOCK%%%%%%
```
|
github_jupyter
|
## TODO:
<ul>
<li>Usar o libreoffice e encontrar 2000 palavras erradas (80h)</li>
<li>Classificar as palavras por tipo (80h)</li>
</ul>
## <b>Italian Pipeline</b>
```
# load hunspell
import urllib
import json
import numpy as np
import pandas as pd
import itertools
from matplotlib import pyplot as plt
import re
suggestions = pd.DataFrame(data)
suggestions
suggestions.to_csv('suggestions.auto.csv')
import hunspell
it_spellchecker = hunspell.HunSpell('/home/rgomes/dictionaries/dictionaries/it/index.dic', '/home/rgomes/dictionaries/dictionaries/it/index.aff')
with open('../auto.spellchecker.results.filtered.json', encoding='utf-8') as data_file:
data = json.loads(data_file.read())
data = list(filter(lambda x: x,data))
a = map(lambda x: x['word'], data)
b = map(lambda x : (x,it_spellchecker.spell(x)), a)
asd = filter(lambda x: x[1] ,b)
errors_hunspell = list(filter(lambda x: x[1] == False , b))
ac_errors = filter(lambda x: re.search(r'[À-ž\'\`]', x[0]) ,errors_hunspell)
# for item in list(ac_errors):
#print(item[0] + '\n')
corrected_ac_errors = []
with open('../italian_accented_erros.txt', encoding='utf-8') as data_file2:
lines = data_file2.readlines()
corrected_ac_errors = list(filter(lambda y: y != '',map(lambda x: x.rstrip('\n'), lines)))
corrected_words = []
for index,x in enumerate(ac_errors):
if x[0] != corrected_ac_errors[index]:
corrected_words.append((x[0], corrected_ac_errors[index]))
all_words = []
with open('../italian_words_all.txt', encoding='utf-8') as data_file_all:
lines = data_file_all.readlines()
all_words = list(map(lambda x: x.rstrip('\n').lower(), lines))
all_words = list(map(lambda x: x.replace('!#$%&()*+,./:;<=>?@[\\]_{|}', ''), all_words))
def histogram(list):
d={}
for i in list:
if i in d:
d[i] += 1
else:
d[i] = 1
return d
def plotHistogram(data):
h = histogram(data)
h = sorted(h.items(), key=lambda x: x[1], reverse=True)
h = map(lambda x: x[1], h)
# remove the words that appears only once
h = filter(lambda x: x > 1, h)
plt.plot(list(h))
plt.show()
suggestions_csv = pd.read_csv('/home/rgomes/Downloads/suggestions filtered - suggestions.auto.csv')
suggestions_csv = suggestions_csv.replace(np.nan, '', regex=True)
suggestions_csv.drop(['is_italian_word', 'suggestions', 'HELPFUL LINK', 'Already removed words'], axis=1)
suggestions_corrected = []
for _, row in suggestions_csv.iterrows():
if row['spelling_correction']:
suggestions_corrected.append((row['word'], row['spelling_correction']))
suggestions_corrected
print(len(suggestions_corrected))
h = histogram(all_words)
h = sorted(h.items(), key=lambda x: x[1], reverse=True)
#######
# filtra apenas aquelas corrigidas com repeticao
combined_corrections_map = list(set(corrected_words + suggestions_corrected))
print('Total corrections {}'.format(len(combined_corrections_map)))
combined_words_list = list(map(lambda x : x[0].lower(), combined_corrections_map))
#print(combined_words_list)
mapped_combined_words = filter(lambda x : x[0].lower() in combined_words_list, h)
total_words = list(mapped_combined_words)
print(total_words[0])
count = 0
for w in total_words:
count = count + w[1]
print(count)
combined_corrections_map
print(len(corrected_words), len(suggestions_corrected))
a_ordered = filter(lambda x: re.search(r'[À-ž\'\`]', x[0]),h)
b_ordered = filter(lambda x: not it_spellchecker.spell(x[0]),a_ordered)
c_ordered = filter(lambda x: not(x[0] in combined_words_list),b_ordered)
d = list(c_ordered)
count2 = 0
for w in d:
count2 = count2 + w[1]
print(count2)
with open('../ordered_last_errors.txt', 'w') as ordered_last_errors:
for item in d:
ordered_last_errors.write(item[0] + '\n')
last_corrections = []
with open('../ordered_last_errors_corrected.txt') as ordered_last_corrections:
lines = list(map(lambda x: x.rstrip('\n').lower(), ordered_last_corrections))
for index, item in enumerate(d):
if item[0] != lines[index]:
last_corrections.append((item[0],lines[index]))
print(len(last_corrections))
h = histogram(all_words)
h = sorted(h.items(), key=lambda x: x[1], reverse=True)
# filtra apenas aquelas corrigidas com repeticao
combined_corrections_map = list(set(corrected_words + suggestions_corrected + last_corrections))
#combined_corrections_map = list(map(lambda x : (x[0].replace('!"#$%&\'()*+,-./:;<=>?@[\\]^_`{|}~', ''), combined_corrections_map)))
print('Total corrections {}'.format(len(combined_corrections_map)))
combined_words_list = list(map(lambda x : x[0].lower(), combined_corrections_map))
#print(combined_words_list)
mapped_combined_words = list(filter(lambda x : x[0].lower() in combined_words_list, h))
#remove rare cases and outliers
# todo: remove nonsense words verified by norton
total_words = list(filter(lambda x: x[1] > 1 and x[1] < 2200,mapped_combined_words))
print(total_words[0])
count = 0
for w in total_words:
count = count + w[1]
print(count)
all_count_dict = dict((a[0], a) for a in total_words)
all_corrections_dict = dict((a[0], a) for a in combined_corrections_map)
all_data = []
for item in all_count_dict:
if all_corrections_dict.get(item):
all_data.append((item, all_count_dict[item][1], all_corrections_dict[item][1]))
print(len(all_data))
df = pd.DataFrame(all_data)
df.to_csv('../final_corrections.csv')
```
|
github_jupyter
|
# Anna KaRNNa
In this notebook, I'll build a character-wise RNN trained on Anna Karenina, one of my all-time favorite books. It'll be able to generate new text based on the text from the book.
This network is based off of Andrej Karpathy's [post on RNNs](http://karpathy.github.io/2015/05/21/rnn-effectiveness/) and [implementation in Torch](https://github.com/karpathy/char-rnn). Also, some information [here at r2rt](http://r2rt.com/recurrent-neural-networks-in-tensorflow-ii.html) and from [Sherjil Ozair](https://github.com/sherjilozair/char-rnn-tensorflow) on GitHub. Below is the general architecture of the character-wise RNN.
<img src="assets/charseq.jpeg" width="500">
```
import time
from collections import namedtuple
import numpy as np
import tensorflow as tf
```
First we'll load the text file and convert it into integers for our network to use.
```
with open('anna.txt', 'r') as f:
text=f.read()
vocab = set(text)
vocab_to_int = {c: i for i, c in enumerate(vocab)}
int_to_vocab = dict(enumerate(vocab))
chars = np.array([vocab_to_int[c] for c in text], dtype=np.int32)
text[:100]
chars[:100]
```
Now I need to split up the data into batches, and into training and validation sets. I should be making a test set here, but I'm not going to worry about that. My test will be if the network can generate new text.
Here I'll make both input and target arrays. The targets are the same as the inputs, except shifted one character over. I'll also drop the last bit of data so that I'll only have completely full batches.
The idea here is to make a 2D matrix where the number of rows is equal to the number of batches. Each row will be one long concatenated string from the character data. We'll split this data into a training set and validation set using the `split_frac` keyword. This will keep 90% of the batches in the training set, the other 10% in the validation set.
```
def split_data(chars, batch_size, num_steps, split_frac=0.9):
"""
Split character data into training and validation sets, inputs and targets for each set.
Arguments
---------
chars: character array
batch_size: Size of examples in each of batch
num_steps: Number of sequence steps to keep in the input and pass to the network
split_frac: Fraction of batches to keep in the training set
Returns train_x, train_y, val_x, val_y
"""
slice_size = batch_size * num_steps
n_batches = int(len(chars) / slice_size)
# Drop the last few characters to make only full batches
x = chars[: n_batches*slice_size]
y = chars[1: n_batches*slice_size + 1]
# Split the data into batch_size slices, then stack them into a 2D matrix
x = np.stack(np.split(x, batch_size))
y = np.stack(np.split(y, batch_size))
# Now x and y are arrays with dimensions batch_size x n_batches*num_steps
# Split into training and validation sets, keep the virst split_frac batches for training
split_idx = int(n_batches*split_frac)
train_x, train_y= x[:, :split_idx*num_steps], y[:, :split_idx*num_steps]
val_x, val_y = x[:, split_idx*num_steps:], y[:, split_idx*num_steps:]
return train_x, train_y, val_x, val_y
train_x, train_y, val_x, val_y = split_data(chars, 10, 200)
train_x.shape
train_x[:,:10]
```
I'll write another function to grab batches out of the arrays made by split data. Here each batch will be a sliding window on these arrays with size `batch_size X num_steps`. For example, if we want our network to train on a sequence of 100 characters, `num_steps = 100`. For the next batch, we'll shift this window the next sequence of `num_steps` characters. In this way we can feed batches to the network and the cell states will continue through on each batch.
```
def get_batch(arrs, num_steps):
batch_size, slice_size = arrs[0].shape
n_batches = int(slice_size/num_steps)
for b in range(n_batches):
yield [x[:, b*num_steps: (b+1)*num_steps] for x in arrs]
def build_rnn(num_classes, batch_size=50, num_steps=50, lstm_size=128, num_layers=2,
learning_rate=0.001, grad_clip=5, sampling=False):
if sampling == True:
batch_size, num_steps = 1, 1
tf.reset_default_graph()
# Declare placeholders we'll feed into the graph
inputs = tf.placeholder(tf.int32, [batch_size, num_steps], name='inputs')
x_one_hot = tf.one_hot(inputs, num_classes, name='x_one_hot')
targets = tf.placeholder(tf.int32, [batch_size, num_steps], name='targets')
y_one_hot = tf.one_hot(targets, num_classes, name='y_one_hot')
y_reshaped = tf.reshape(y_one_hot, [-1, num_classes])
keep_prob = tf.placeholder(tf.float32, name='keep_prob')
# Build the RNN layers
lstm = tf.contrib.rnn.BasicLSTMCell(lstm_size)
drop = tf.contrib.rnn.DropoutWrapper(lstm, output_keep_prob=keep_prob)
cell = tf.contrib.rnn.MultiRNNCell([drop] * num_layers)
initial_state = cell.zero_state(batch_size, tf.float32)
# Run the data through the RNN layers
rnn_inputs = [tf.squeeze(i, squeeze_dims=[1]) for i in tf.split(x_one_hot, num_steps, 1)]
outputs, state = tf.contrib.rnn.static_rnn(cell, rnn_inputs, initial_state=initial_state)
final_state = tf.identity(state, name='final_state')
# Reshape output so it's a bunch of rows, one row for each cell output
seq_output = tf.concat(outputs, axis=1,name='seq_output')
output = tf.reshape(seq_output, [-1, lstm_size], name='graph_output')
# Now connect the RNN putputs to a softmax layer and calculate the cost
softmax_w = tf.Variable(tf.truncated_normal((lstm_size, num_classes), stddev=0.1),
name='softmax_w')
softmax_b = tf.Variable(tf.zeros(num_classes), name='softmax_b')
logits = tf.matmul(output, softmax_w) + softmax_b
preds = tf.nn.softmax(logits, name='predictions')
loss = tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=y_reshaped, name='loss')
cost = tf.reduce_mean(loss, name='cost')
# Optimizer for training, using gradient clipping to control exploding gradients
tvars = tf.trainable_variables()
grads, _ = tf.clip_by_global_norm(tf.gradients(cost, tvars), grad_clip)
train_op = tf.train.AdamOptimizer(learning_rate)
optimizer = train_op.apply_gradients(zip(grads, tvars))
# Export the nodes
export_nodes = ['inputs', 'targets', 'initial_state', 'final_state',
'keep_prob', 'cost', 'preds', 'optimizer']
Graph = namedtuple('Graph', export_nodes)
local_dict = locals()
graph = Graph(*[local_dict[each] for each in export_nodes])
return graph
```
## Hyperparameters
Here I'm defining the hyperparameters for the network. The two you probably haven't seen before are `lstm_size` and `num_layers`. These set the number of hidden units in the LSTM layers and the number of LSTM layers, respectively. Of course, making these bigger will improve the network's performance but you'll have to watch out for overfitting. If your validation loss is much larger than the training loss, you're probably overfitting. Decrease the size of the network or decrease the dropout keep probability.
```
batch_size = 100
num_steps = 100
lstm_size = 512
num_layers = 2
learning_rate = 0.001
```
## Write out the graph for TensorBoard
```
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
file_writer = tf.summary.FileWriter('./logs/1', sess.graph)
```
## Training
Time for training which is is pretty straightforward. Here I pass in some data, and get an LSTM state back. Then I pass that state back in to the network so the next batch can continue the state from the previous batch. And every so often (set by `save_every_n`) I calculate the validation loss and save a checkpoint.
```
!mkdir -p checkpoints/anna
epochs = 1
save_every_n = 200
train_x, train_y, val_x, val_y = split_data(chars, batch_size, num_steps)
model = build_rnn(len(vocab),
batch_size=batch_size,
num_steps=num_steps,
learning_rate=learning_rate,
lstm_size=lstm_size,
num_layers=num_layers)
saver = tf.train.Saver(max_to_keep=100)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
# Use the line below to load a checkpoint and resume training
#saver.restore(sess, 'checkpoints/anna20.ckpt')
n_batches = int(train_x.shape[1]/num_steps)
iterations = n_batches * epochs
for e in range(epochs):
# Train network
new_state = sess.run(model.initial_state)
loss = 0
for b, (x, y) in enumerate(get_batch([train_x, train_y], num_steps), 1):
iteration = e*n_batches + b
start = time.time()
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 0.5,
model.initial_state: new_state}
batch_loss, new_state, _ = sess.run([model.cost, model.final_state, model.optimizer],
feed_dict=feed)
loss += batch_loss
end = time.time()
print('Epoch {}/{} '.format(e+1, epochs),
'Iteration {}/{}'.format(iteration, iterations),
'Training loss: {:.4f}'.format(loss/b),
'{:.4f} sec/batch'.format((end-start)))
if (iteration%save_every_n == 0) or (iteration == iterations):
# Check performance, notice dropout has been set to 1
val_loss = []
new_state = sess.run(model.initial_state)
for x, y in get_batch([val_x, val_y], num_steps):
feed = {model.inputs: x,
model.targets: y,
model.keep_prob: 1.,
model.initial_state: new_state}
batch_loss, new_state = sess.run([model.cost, model.final_state], feed_dict=feed)
val_loss.append(batch_loss)
print('Validation loss:', np.mean(val_loss),
'Saving checkpoint!')
saver.save(sess, "checkpoints/anna/i{}_l{}_{:.3f}.ckpt".format(iteration, lstm_size, np.mean(val_loss)))
tf.train.get_checkpoint_state('checkpoints/anna')
```
## Sampling
Now that the network is trained, we'll can use it to generate new text. The idea is that we pass in a character, then the network will predict the next character. We can use the new one, to predict the next one. And we keep doing this to generate all new text. I also included some functionality to prime the network with some text by passing in a string and building up a state from that.
The network gives us predictions for each character. To reduce noise and make things a little less random, I'm going to only choose a new character from the top N most likely characters.
```
def pick_top_n(preds, vocab_size, top_n=5):
p = np.squeeze(preds)
p[np.argsort(p)[:-top_n]] = 0
p = p / np.sum(p)
c = np.random.choice(vocab_size, 1, p=p)[0]
return c
def sample(checkpoint, n_samples, lstm_size, vocab_size, prime="The "):
prime = "Far"
samples = [c for c in prime]
model = build_rnn(vocab_size, lstm_size=lstm_size, sampling=True)
saver = tf.train.Saver()
with tf.Session() as sess:
saver.restore(sess, checkpoint)
new_state = sess.run(model.initial_state)
for c in prime:
x = np.zeros((1, 1))
x[0,0] = vocab_to_int[c]
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
for i in range(n_samples):
x[0,0] = c
feed = {model.inputs: x,
model.keep_prob: 1.,
model.initial_state: new_state}
preds, new_state = sess.run([model.preds, model.final_state],
feed_dict=feed)
c = pick_top_n(preds, len(vocab))
samples.append(int_to_vocab[c])
return ''.join(samples)
checkpoint = "checkpoints/anna/i3560_l512_1.122.ckpt"
samp = sample(checkpoint, 2000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i200_l512_2.432.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i600_l512_1.750.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
checkpoint = "checkpoints/anna/i1000_l512_1.484.ckpt"
samp = sample(checkpoint, 1000, lstm_size, len(vocab), prime="Far")
print(samp)
```
|
github_jupyter
|
# Bayesian Estimation Supersedes the T-Test
```
%matplotlib inline
import numpy as np
import pymc3 as pm
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use('seaborn-darkgrid')
print('Running on PyMC3 v{}'.format(pm.__version__))
```
This model replicates the example used in:
Kruschke, John. (2012) **Bayesian estimation supersedes the t-test**. *Journal of Experimental Psychology*: General.
### The Problem
Several statistical inference procedures involve the comparison of two groups. We may be interested in whether one group is larger than another, or simply different from the other. We require a statistical model for this because true differences are usually accompanied by measurement or stochastic noise that prevent us from drawing conclusions simply from differences calculated from the observed data.
The *de facto* standard for statistically comparing two (or more) samples is to use a statistical test. This involves expressing a null hypothesis, which typically claims that there is no difference between the groups, and using a chosen test statistic to determine whether the distribution of the observed data is plausible under the hypothesis. This rejection occurs when the calculated test statistic is higher than some pre-specified threshold value.
Unfortunately, it is not easy to conduct hypothesis tests correctly, and their results are very easy to misinterpret. Setting up a statistical test involves several subjective choices (*e.g.* statistical test to use, null hypothesis to test, significance level) by the user that are rarely justified based on the problem or decision at hand, but rather, are usually based on traditional choices that are entirely arbitrary (Johnson 1999). The evidence that it provides to the user is indirect, incomplete, and typically overstates the evidence against the null hypothesis (Goodman 1999).
A more informative and effective approach for comparing groups is one based on **estimation** rather than **testing**, and is driven by Bayesian probability rather than frequentist. That is, rather than testing whether two groups are different, we instead pursue an estimate of how different they are, which is fundamentally more informative. Moreover, we include an estimate of uncertainty associated with that difference which includes uncertainty due to our lack of knowledge of the model parameters (epistemic uncertainty) and uncertainty due to the inherent stochasticity of the system (aleatory uncertainty).
## Example: Drug trial evaluation
To illustrate how this Bayesian estimation approach works in practice, we will use a fictitious example from Kruschke (2012) concerning the evaluation of a clinical trial for drug evaluation. The trial aims to evaluate the efficacy of a "smart drug" that is supposed to increase intelligence by comparing IQ scores of individuals in a treatment arm (those receiving the drug) to those in a control arm (those recieving a placebo). There are 47 individuals and 42 individuals in the treatment and control arms, respectively.
```
drug = (101,100,102,104,102,97,105,105,98,101,100,123,105,103,100,95,102,106,
109,102,82,102,100,102,102,101,102,102,103,103,97,97,103,101,97,104,
96,103,124,101,101,100,101,101,104,100,101)
placebo = (99,101,100,101,102,100,97,101,104,101,102,102,100,105,88,101,100,
104,100,100,100,101,102,103,97,101,101,100,101,99,101,100,100,
101,100,99,101,100,102,99,100,99)
y1 = np.array(drug)
y2 = np.array(placebo)
y = pd.DataFrame(dict(value=np.r_[y1, y2], group=np.r_[['drug']*len(drug), ['placebo']*len(placebo)]))
y.hist('value', by='group');
```
The first step in a Bayesian approach to inference is to specify the full probability model that corresponds to the problem. For this example, Kruschke chooses a Student-t distribution to describe the distributions of the scores in each group. This choice adds robustness to the analysis, as a T distribution is less sensitive to outlier observations, relative to a normal distribution. The three-parameter Student-t distribution allows for the specification of a mean $\mu$, a precision (inverse-variance) $\lambda$ and a degrees-of-freedom parameter $\nu$:
$$f(x|\mu,\lambda,\nu) = \frac{\Gamma(\frac{\nu + 1}{2})}{\Gamma(\frac{\nu}{2})} \left(\frac{\lambda}{\pi\nu}\right)^{\frac{1}{2}} \left[1+\frac{\lambda(x-\mu)^2}{\nu}\right]^{-\frac{\nu+1}{2}}$$
the degrees-of-freedom parameter essentially specifies the "normality" of the data, since larger values of $\nu$ make the distribution converge to a normal distribution, while small values (close to zero) result in heavier tails.
Thus, the likelihood functions of our model are specified as follows:
$$y^{(treat)}_i \sim T(\nu, \mu_1, \sigma_1)$$
$$y^{(placebo)}_i \sim T(\nu, \mu_2, \sigma_2)$$
As a simplifying assumption, we will assume that the degree of normality $\nu$ is the same for both groups. We will, of course, have separate parameters for the means $\mu_k, k=1,2$ and standard deviations $\sigma_k$.
Since the means are real-valued, we will apply normal priors on them, and arbitrarily set the hyperparameters to the pooled empirical mean of the data and twice the pooled empirical standard deviation, which applies very diffuse information to these quantities (and importantly, does not favor one or the other *a priori*).
$$\mu_k \sim N(\bar{x}, 2s)$$
```
μ_m = y.value.mean()
μ_s = y.value.std() * 2
with pm.Model() as model:
group1_mean = pm.Normal('group1_mean', μ_m, sigma=μ_s)
group2_mean = pm.Normal('group2_mean', μ_m, sigma=μ_s)
```
The group standard deviations will be given a uniform prior over a plausible range of values for the variability of the outcome variable, IQ.
In Kruschke's original model, he uses a very wide uniform prior for the group standard deviations, from the pooled empirical standard deviation divided by 1000 to the pooled standard deviation multiplied by 1000. This is a poor choice of prior, because very basic prior knowledge about measures of human coginition dictate that the variation cannot ever be as high as this upper bound. IQ is a standardized measure, and hence this constrains how variable a given population's IQ values can be. When you place such a wide uniform prior on these values, you are essentially giving a lot of prior weight on inadmissable values. In this example, there is little practical difference, but in general it is best to apply as much prior information that you have available to the parameterization of prior distributions.
We will instead set the group standard deviations to have a $\text{Uniform}(1,10)$ prior:
```
σ_low = 1
σ_high = 10
with model:
group1_std = pm.Uniform('group1_std', lower=σ_low, upper=σ_high)
group2_std = pm.Uniform('group2_std', lower=σ_low, upper=σ_high)
```
We follow Kruschke by making the prior for $\nu$ exponentially distributed with a mean of 30; this allocates high prior probability over the regions of the parameter that describe the range from normal to heavy-tailed data under the Student-T distribution.
```
with model:
ν = pm.Exponential('ν_minus_one', 1/29.) + 1
pm.kdeplot(np.random.exponential(30, size=10000), shade=0.5);
```
Since PyMC3 parameterizes the Student-T in terms of precision, rather than standard deviation, we must transform the standard deviations before specifying our likelihoods.
```
with model:
λ1 = group1_std**-2
λ2 = group2_std**-2
group1 = pm.StudentT('drug', nu=ν, mu=group1_mean, lam=λ1, observed=y1)
group2 = pm.StudentT('placebo', nu=ν, mu=group2_mean, lam=λ2, observed=y2)
```
Having fully specified our probabilistic model, we can turn our attention to calculating the comparisons of interest in order to evaluate the effect of the drug. To this end, we can specify deterministic nodes in our model for the difference between the group means and the difference between the group standard deviations. Wrapping them in named `Deterministic` objects signals to PyMC that we wish to record the sampled values as part of the output.
As a joint measure of the groups, we will also estimate the "effect size", which is the difference in means scaled by the pooled estimates of standard deviation. This quantity can be harder to interpret, since it is no longer in the same units as our data, but the quantity is a function of all four estimated parameters.
```
with model:
diff_of_means = pm.Deterministic('difference of means', group1_mean - group2_mean)
diff_of_stds = pm.Deterministic('difference of stds', group1_std - group2_std)
effect_size = pm.Deterministic('effect size',
diff_of_means / np.sqrt((group1_std**2 + group2_std**2) / 2))
```
Now, we can fit the model and evaluate its output.
```
with model:
trace = pm.sample(2000, cores=2)
```
We can plot the stochastic parameters of the model. PyMC's `plot_posterior` function replicates the informative histograms portrayed in Kruschke (2012). These summarize the posterior distributions of the parameters, and present a 95% credible interval and the posterior mean. The plots below are constructed with the final 1000 samples from each of the 2 chains, pooled together.
```
pm.plot_posterior(trace, varnames=['group1_mean','group2_mean', 'group1_std', 'group2_std', 'ν_minus_one'],
color='#87ceeb');
```
Looking at the group differences, we can conclude that there are meaningful differences between the two groups for all three measures. For these comparisons, it is useful to use zero as a reference value (`ref_val`); providing this reference value yields cumulative probabilities for the posterior distribution on either side of the value. Thus, for the difference in means, 99.4% of the posterior probability is greater than zero, which suggests the group means are credibly different. The effect size and differences in standard deviation are similarly positive.
These estimates suggest that the "smart drug" increased both the expected scores, but also the variability in scores across the sample. So, this does not rule out the possibility that some recipients may be adversely affected by the drug at the same time others benefit.
```
pm.plot_posterior(trace, varnames=['difference of means','difference of stds', 'effect size'],
ref_val=0,
color='#87ceeb');
```
When `forestplot` is called on a trace with more than one chain, it also plots the potential scale reduction parameter, which is used to reveal evidence for lack of convergence; values near one, as we have here, suggest that the model has converged.
```
pm.forestplot(trace, varnames=['group1_mean',
'group2_mean']);
pm.forestplot(trace, varnames=['group1_std',
'group2_std',
'ν_minus_one']);
pm.summary(trace,varnames=['difference of means', 'difference of stds', 'effect size'])
```
## References
1. Goodman SN. Toward evidence-based medical statistics. 1: The P value fallacy. Annals of Internal Medicine. 1999;130(12):995-1004. doi:10.7326/0003-4819-130-12-199906150-00008.
2. Johnson D. The insignificance of statistical significance testing. Journal of Wildlife Management. 1999;63(3):763-772.
3. Kruschke JK. Bayesian estimation supersedes the t test. J Exp Psychol Gen. 2013;142(2):573-603. doi:10.1037/a0029146.
The original pymc2 implementation was written by Andrew Straw and can be found here: https://github.com/strawlab/best
Ported to PyMC3 by [Thomas Wiecki](https://twitter.com/twiecki) (c) 2015, updated by Chris Fonnesbeck.
|
github_jupyter
|
# TensorFlow实战Titanic解析
## 一、数据读入及预处理
### 1. 使用pandas读入csv文件,读入为pands.DataFrame对象
```
import os
import numpy as np
import pandas as pd
import tensorflow as tf
# read data from file
data = pd.read_csv('data/train.csv')
print(data.info())
```
### 2. 预处理
1. 剔除空数据
2. 将'Sex'字段转换为int类型
3. 选取数值类型的字段,抛弃字符串类型字段
```
# fill nan values with 0
data = data.fillna(0)
# convert ['male', 'female'] values of Sex to [1, 0]
data['Sex'] = data['Sex'].apply(lambda s: 1 if s == 'male' else 0)
# 'Survived' is the label of one class,
# add 'Deceased' as the other class
data['Deceased'] = data['Survived'].apply(lambda s: 1 - s)
# select features and labels for training
dataset_X = data[['Sex', 'Age', 'Pclass', 'SibSp', 'Parch', 'Fare']]
dataset_Y = data[['Deceased', 'Survived']]
print(dataset_X)
print(dataset_Y)
```
### 3. 将训练数据切分为训练集(training set)和验证集(validation set)
```
from sklearn.model_selection import train_test_split
# split training data and validation set data
X_train, X_val, y_train, y_val = train_test_split(dataset_X.as_matrix(), dataset_Y.as_matrix(),
test_size=0.2,
random_state=42)
```
# 二、构建计算图
### 逻辑回归
逻辑回归是形式最简单,并且最容易理解的分类器之一。从数学上,逻辑回归的预测函数可以表示为如下公式:
*y = softmax(xW + b)*
其中,*x*为输入向量,是大小为*d×1*的列向量,*d*是特征数。*W*是大小为的*c×d*权重矩阵,*c*是分类类别数目。*b*是偏置向量,为*c×1*列向量。*softmax*在数学定义里,是指一种归一化指数函数。它将一个*k*维的向量*x*按照公式

的形式将向量中的元素转换为*(0, 1)*的区间。机器学习领域常使用这种方法将类似判别函数的置信度值转换为概率形式(如判别超平面的距离等)。*softmax*函数常用于输出层,用于指定唯一的分类输出。
### 1. 使用placeholder声明输入占位符
TensorFlow设计了数据Feed机制。也就是说计算程序并不会直接交互执行,而是在声明过程只做计算图的构建。所以,此时并不会触碰真实的数据,而只是通过placeholder算子声明一个输入数据的占位符,在后面真正运行计算时,才用数据替换占位符。
声明占位符placeholder需要给定三个参数,分别是输入数据的元素类型dtype、维度形状shape和占位符名称标识name。
```
# 声明输入数据占位符
# shape参数的第一个元素为None,表示可以同时放入任意条记录
X = tf.placeholder(tf.float32, shape=[None, 6], name='input')
y = tf.placeholder(tf.float32, shape=[None, 2], name='label')
```
### 2. 声明参数变量
变量的声明方式是直接定义tf.Variable()对象。
初始化变量对象有两种方式,一种是从protocol buffer结构VariableDef中反序列化,另一种是通过参数指定初始值。最简单的方式就是向下面程序这样,为变量传入初始值。初始值必须是一个tensor对象,或是可以通过convert_to_tensor()方法转换成tensor的Python对象。TensorFlow提供了多种构造随机tensor的方法,可以构造全零tensor、随机正态分布tensor等。定义变量会保留初始值的维度形状。
```
# 声明变量
weights = tf.Variable(tf.random_normal([6, 2]), name='weights')
bias = tf.Variable(tf.zeros([2]), name='bias')
```
### 3. 构造前向传播计算图
使用算子构建由输入计算出标签的计算过程。
在计算图的构建过程中,TensorFlow会自动推算每一个节点的输入输出形状。若无法运算,比如两个行列数不同的矩阵相加,则会直接报错。
```
y_pred = tf.nn.softmax(tf.matmul(X, weights) + bias)
```
### 4. 声明代价函数
使用交叉熵(cross entropy)作为代价函数。
```
# 使用交叉熵作为代价函数
cross_entropy = - tf.reduce_sum(y * tf.log(y_pred + 1e-10),
reduction_indices=1)
# 批量样本的代价值为所有样本交叉熵的平均值
cost = tf.reduce_mean(cross_entropy)
```
#### NOTE
在计算交叉熵的时候,对模型输出值 y_pred 加上了一个很小的误差值(在上面程序中是 1e-10),这是因为当 y_pred 十分接近真值 y_true 的时候,也就是 y_pred 的值非常接近 0 或 1 的取值时,计算会得到负无穷 -inf,从而导致输出非法,并进一步导致无法计算梯度,迭代陷入崩溃。要解决这个问题有三种办法:
1. 在计算时,直接加入一个极小的误差值,使计算合法。这样可以避免计算,但存在的问题是加入误差后相当于y_pred的值会突破1。在示例代码中使用了这种方案;
2. 使用 clip() 函数,当 y_pred 接近 0 时,将其赋值成为极小误差值。也就是将 y_pred 的取值范围限定在的范围内;
3. 当计算交叉熵的计算出现 nan 值时,显式地将cost设置为0。这种方式回避了 函数计算的问题,而是在最终的代价函数上进行容错处理。
### 5. 加入优化算法
TensorFlow内置了多种经典的优化算法,如随机梯度下降算法(SGD,Stochastic Gradient Descent)、动量算法(Momentum)、Adagrad算法、ADAM算法、RMSProp算法等。优化器内部会自动构建梯度计算和反向传播部分的计算图。
一般对于优化算法,最关键的参数是学习率(learning rate),对于学习率的设置是一门技术。同时,不同优化算法在不同问题上可能会有不同的收敛速度,在解决实际问题时可以做多种尝试。
```
# 使用随机梯度下降算法优化器来最小化代价,系统自动构建反向传播部分的计算图
train_op = tf.train.GradientDescentOptimizer(0.001).minimize(cost)
```
### 6. (optional) 计算准确率
```
# 计算准确率
correct_pred = tf.equal(tf.argmax(y, 1), tf.argmax(y_pred, 1))
acc_op = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
```
# 三、构建训练迭代 & 执行训练
### 启动Session,代入数据进行计算。训练结束后使用验证集评估训练效果
```
with tf.Session() as sess:
# variables have to be initialized at the first place
tf.global_variables_initializer().run()
# training loop
for epoch in range(10):
total_loss = 0.
for i in range(len(X_train)):
# prepare feed data and run
feed_dict = {X: [X_train[i]], y: [y_train[i]]}
_, loss = sess.run([train_op, cost], feed_dict=feed_dict)
total_loss += loss
# display loss per epoch
print('Epoch: %04d, total loss=%.9f' % (epoch + 1, total_loss))
print 'Training complete!'
# Accuracy calculated by TensorFlow
accuracy = sess.run(acc_op, feed_dict={X: X_val, y: y_val})
print("Accuracy on validation set: %.9f" % accuracy)
# Accuracy calculated by NumPy
pred = sess.run(y_pred, feed_dict={X: X_val})
correct = np.equal(np.argmax(pred, 1), np.argmax(y_val, 1))
numpy_accuracy = np.mean(correct.astype(np.float32))
print("Accuracy on validation set (numpy): %.9f" % numpy_accuracy)
```
# 四、存储和加载模型参数
变量的存储和读取是通过tf.train.Saver类来完成的。Saver对象在初始化时,为计算图加入了用于存储和加载变量的算子,并可以通过参数指定是要存储哪些变量。Saver对象的save()和restore()方法是触发图中算子的入口。
```
# 训练步数记录
global_step = tf.Variable(0, name='global_step', trainable=False)
# 存档入口
saver = tf.train.Saver()
# 在Saver声明之后定义的变量将不会被存储
# non_storable_variable = tf.Variable(777)
ckpt_dir = './ckpt_dir'
if not os.path.exists(ckpt_dir):
os.makedirs(ckpt_dir)
with tf.Session() as sess:
tf.global_variables_initializer().run()
# 加载模型存档
ckpt = tf.train.get_checkpoint_state(ckpt_dir)
if ckpt and ckpt.model_checkpoint_path:
print('Restoring from checkpoint: %s' % ckpt.model_checkpoint_path)
saver.restore(sess, ckpt.model_checkpoint_path)
start = global_step.eval()
for epoch in range(start, start + 10):
total_loss = 0.
for i in range(0, len(X_train)):
feed_dict = {
X: [X_train[i]],
y: [y_train[i]]
}
_, loss = sess.run([train_op, cost], feed_dict=feed_dict)
total_loss += loss
print('Epoch: %04d, loss=%.9f' % (epoch + 1, total_loss))
# 模型存档
global_step.assign(epoch).eval()
saver.save(sess, ckpt_dir + '/logistic.ckpt',
global_step=global_step)
print('Training complete!')
```
# TensorBoard
TensorBoard是TensorFlow配套的可视化工具,可以用来帮助理解复杂的模型和检查实现中的错误。
TensorBoard的工作方式是启动一个WEB服务,该服务进程从TensorFlow程序执行所得的事件日志文件(event files)中读取概要(summary)数据,然后将数据在网页中绘制成可视化的图表。概要数据主要包括以下几种类别:
1. 标量数据,如准确率、代价损失值,使用tf.summary.scalar加入记录算子;
2. 参数数据,如参数矩阵weights、偏置矩阵bias,一般使用tf.summary.histogram记录;
3. 图像数据,用tf.summary.image加入记录算子;
4. 音频数据,用tf.summary.audio加入记录算子;
5. 计算图结构,在定义tf.summary.FileWriter对象时自动记录。
可以通过TensorBoard展示的完整程序:
```
################################
# Constructing Dataflow Graph
################################
# arguments that can be set in command line
tf.app.flags.DEFINE_integer('epochs', 10, 'Training epochs')
tf.app.flags.DEFINE_integer('batch_size', 10, 'size of mini-batch')
FLAGS = tf.app.flags.FLAGS
with tf.name_scope('input'):
# create symbolic variables
X = tf.placeholder(tf.float32, shape=[None, 6])
y_true = tf.placeholder(tf.float32, shape=[None, 2])
with tf.name_scope('classifier'):
# weights and bias are the variables to be trained
weights = tf.Variable(tf.random_normal([6, 2]))
bias = tf.Variable(tf.zeros([2]))
y_pred = tf.nn.softmax(tf.matmul(X, weights) + bias)
# add histogram summaries for weights, view on tensorboard
tf.summary.histogram('weights', weights)
tf.summary.histogram('bias', bias)
# Minimise cost using cross entropy
# NOTE: add a epsilon(1e-10) when calculate log(y_pred),
# otherwise the result will be -inf
with tf.name_scope('cost'):
cross_entropy = - tf.reduce_sum(y_true * tf.log(y_pred + 1e-10),
reduction_indices=1)
cost = tf.reduce_mean(cross_entropy)
tf.summary.scalar('loss', cost)
# use gradient descent optimizer to minimize cost
train_op = tf.train.GradientDescentOptimizer(0.001).minimize(cost)
with tf.name_scope('accuracy'):
correct_pred = tf.equal(tf.argmax(y_true, 1), tf.argmax(y_pred, 1))
acc_op = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
# Add scalar summary for accuracy
tf.summary.scalar('accuracy', acc_op)
global_step = tf.Variable(0, name='global_step', trainable=False)
# use saver to save and restore model
saver = tf.train.Saver()
# this variable won't be stored, since it is declared after tf.train.Saver()
non_storable_variable = tf.Variable(777)
ckpt_dir = './ckpt_dir'
if not os.path.exists(ckpt_dir):
os.makedirs(ckpt_dir)
################################
# Training the model
################################
# use session to run the calculation
with tf.Session() as sess:
# create a log writer. run 'tensorboard --logdir=./logs'
writer = tf.summary.FileWriter('./logs', sess.graph)
merged = tf.summary.merge_all()
# variables have to be initialized at the first place
tf.global_variables_initializer().run()
# restore variables from checkpoint if exists
ckpt = tf.train.get_checkpoint_state(ckpt_dir)
if ckpt and ckpt.model_checkpoint_path:
print('Restoring from checkpoint: %s' % ckpt.model_checkpoint_path)
saver.restore(sess, ckpt.model_checkpoint_path)
start = global_step.eval()
# training loop
for epoch in range(start, start + FLAGS.epochs):
total_loss = 0.
for i in range(0, len(X_train), FLAGS.batch_size):
# train with mini-batch
feed_dict = {
X: X_train[i: i + FLAGS.batch_size],
y_true: y_train[i: i + FLAGS.batch_size]
}
_, loss = sess.run([train_op, cost], feed_dict=feed_dict)
total_loss += loss
# display loss per epoch
print('Epoch: %04d, loss=%.9f' % (epoch + 1, total_loss))
summary, accuracy = sess.run([merged, acc_op],
feed_dict={X: X_val, y_true: y_val})
writer.add_summary(summary, epoch) # Write summary
print('Accuracy on validation set: %.9f' % accuracy)
# set and update(eval) global_step with epoch
global_step.assign(epoch).eval()
saver.save(sess, ckpt_dir + '/logistic.ckpt',
global_step=global_step)
print('Training complete!')
```


|
github_jupyter
|
# The Stanford Sentiment Treebank
The Stanford Sentiment Treebank consists of sentences from movie reviews and human annotations of their sentiment. The task is to predict the sentiment of a given sentence. We use the two-way (positive/negative) class split, and use only sentence-level labels.
```
from IPython.display import display, Markdown
with open('../../doc/env_variables_setup.md', 'r') as fh:
content = fh.read()
display(Markdown(content))
```
## Import Packages
```
import tensorflow as tf
from transformers import (
BertConfig,
BertTokenizer,
XLMRobertaTokenizer,
TFBertModel,
TFXLMRobertaModel,
)
import os
from datetime import datetime
import tensorflow_datasets
from tensorboard import notebook
import math
#from google.cloud import storage
from googleapiclient import discovery
from googleapiclient import errors
import logging
import json
```
## Check configuration
```
print(tf.version.GIT_VERSION, tf.version.VERSION)
print(tf.keras.__version__)
gpus = tf.config.list_physical_devices('GPU')
if len(gpus)>0:
for gpu in gpus:
print('Name:', gpu.name, ' Type:', gpu.device_type)
else:
print('No GPU available !!!!')
```
## Define Paths
```
try:
data_dir=os.environ['PATH_DATASETS']
except KeyError:
print('missing PATH_DATASETS')
try:
tensorboard_dir=os.environ['PATH_TENSORBOARD']
except KeyError:
print('missing PATH_TENSORBOARD')
try:
savemodel_dir=os.environ['PATH_SAVE_MODEL']
except KeyError:
print('missing PATH_SAVE_MODEL')
```
# Import local packages
```
import utils.model_utils as mu
import importlib
importlib.reload(mu);
```
## Train the model on AI Platform Training (for production)
```
project_name = os.environ['PROJECT_ID']
project_id = 'projects/{}'.format(project_name)
ai_platform_training = discovery.build('ml', 'v1', cache_discovery=False)
# choose the model
model_name = 'tf_bert_classification'
#model_name = 'test_log_bert'
# variable used to build some variable's name
type_production = 'test' #'test', 'production'
hardware = 'cpu' #'cpu', 'gpu', 'tpu'
owner = os.environ['OWNER']
tier = 'basic' #'basic', 'custom'
python_version = '3.7'
runtime_version = '2.2'
hp_tuning= False
verbosity = 'INFO'
profiling = False
# use custom container
use_custom_container = False
tag='/test:v0.0.0'
# overwrite parameter for testing logging
test_logging = False
print(' modifying Tensorflow env variable')
# 0 = all messages are logged (default behavior)
# 1 = INFO messages are not printed
# 2 = INFO and WARNING messages are not printed
# 3 = INFO, WARNING, and ERROR messages are not printed
with open(os.environ['DIR_PROJ']+'/utils/env_variables.json', 'r') as outfile:
env_var = json.load(outfile)
if verbosity == 'DEBUG' or verbosity == 'VERBOSE' or verbosity == 'INFO':
env_var['TF_CPP_MIN_LOG_LEVEL'] = 0
env_var['TF_CPP_MIN_VLOG_LEVEL'] = 0
elif verbosity == 'WARNING':
env_var['TF_CPP_MIN_LOG_LEVEL'] = 1
env_var['TF_CPP_MIN_VLOG_LEVEL'] = 1
elif verbosity == 'ERROR':
env_var['TF_CPP_MIN_LOG_LEVEL'] = 2
env_var['TF_CPP_MIN_VLOG_LEVEL'] = 2
else:
env_var['TF_CPP_MIN_LOG_LEVEL'] = 3
env_var['TF_CPP_MIN_VLOG_LEVEL'] = 3
print("env_var['TF_CPP_MIN_LOG_LEVEL']=", env_var['TF_CPP_MIN_LOG_LEVEL'])
print("env_var['TF_CPP_MIN_VLOG_LEVEL']=", env_var['TF_CPP_MIN_VLOG_LEVEL'])
data={}
data['TF_CPP_MIN_LOG_LEVEL'] = env_var['TF_CPP_MIN_LOG_LEVEL']
data['TF_CPP_MIN_VLOG_LEVEL'] = env_var['TF_CPP_MIN_VLOG_LEVEL']
with open(os.environ['DIR_PROJ']+'/utils/env_variables.json', 'w') as outfile:
json.dump(data, outfile)
# define parameters for ai platform training
if not use_custom_container:
# delete old package version
for root, dirs, files in os.walk(os.environ['DIR_PROJ'] + '/dist/'):
for filename in files:
package_dist=os.environ['DIR_PROJ'] + '/dist/'+filename
if package_dist[-7:]=='.tar.gz':
print('removing package"', package_dist)
os.remove(package_dist)
package_gcs = mu.create_module_tar_archive(model_name)
else:
package_gcs = None
timestamp = datetime.now().strftime("%Y_%m_%d_%H%M%S")
if hp_tuning:
job_name = model_name+'_hp_tuning_'+hardware+'_'+timestamp
else:
job_name = model_name+'_'+hardware+'_'+timestamp
module_name = 'model.'+model_name+'.task'
if tier=='basic' and hardware=='cpu':
# CPU
region = 'europe-west1'
elif tier=='basic' and hardware=='gpu':
# GPU
region = 'europe-west1'
elif tier=='custom' and hardware=='gpu':
# Custom GPU
region = 'europe-west4'
elif tier=='basic' and hardware=='tpu':
# TPU
#region = 'us-central1'
region = 'europe-west4' # No zone in region europe-west4 has accelerators of all requested types
#region = 'europe-west6' # The request for 8 TPU_V2 accelerators exceeds the allowed maximum of 0 K80, 0 P100, 0 P4, 0 T4, 0 TPU_V2, 0 TPU_V2_POD, 0 TPU_V3, 0 TPU_V3_POD, 0 V100
#region = 'europe-west2' # No zone in region europe-west2 has accelerators of all requested types
elif tier=='custom' and hardware=='tpu':
# TPU
#region = 'us-central1'
region = 'europe-west4'
#region = 'europe-west6'
#region = 'europe-west2'
else:
# Default
region = 'europe-west1'
# define parameters for training of the model
if type_production=='production':
# reading metadata
_, info = tensorflow_datasets.load(name='glue/sst2',
data_dir=data_dir,
with_info=True)
# define parameters
epochs = 2
batch_size_train = 32
#batch_size_test = 32
batch_size_eval = 64
# Maxium length, becarefull BERT max length is 512!
max_length = 128
# extract parameters
size_train_dataset=info.splits['train'].num_examples
#size_test_dataset=info.splits['test'].num_examples
size_valid_dataset=info.splits['validation'].num_examples
# computer parameter
steps_per_epoch_train = math.ceil(size_train_dataset/batch_size_train)
#steps_per_epoch_test = math.ceil(size_test_dataset/batch_size_test)
steps_per_epoch_eval = math.ceil(size_valid_dataset/batch_size_eval)
#print('Dataset size: {:6}/{:6}/{:6}'.format(size_train_dataset, size_test_dataset, size_valid_dataset))
#print('Batch size: {:6}/{:6}/{:6}'.format(batch_size_train, batch_size_test, batch_size_eval))
#print('Step per epoch: {:6}/{:6}/{:6}'.format(steps_per_epoch_train, steps_per_epoch_test, steps_per_epoch_eval))
#print('Total number of batch: {:6}/{:6}/{:6}'.format(steps_per_epoch_train*(epochs+1), steps_per_epoch_test*(epochs+1), steps_per_epoch_eval*1))
print('Number of epoch: {:6}'.format(epochs))
print('Batch size: {:6}/{:6}'.format(batch_size_train, batch_size_eval))
print('Step per epoch: {:6}/{:6}'.format(steps_per_epoch_train, steps_per_epoch_eval))
else:
if hardware=='tpu':
epochs = 1
steps_per_epoch_train = 6 #5
batch_size_train = 32
steps_per_epoch_eval = 1
batch_size_eval = 64
else:
epochs = 1
steps_per_epoch_train = 6 #5
batch_size_train = 32
steps_per_epoch_eval = 1
batch_size_eval = 64
steps=epochs*steps_per_epoch_train
if steps<=5:
n_steps_history=4
elif steps>=5 and steps<1000:
n_steps_history=10
print('be carefull with profiling between step: 10-20')
else:
n_steps_history=int(steps/100)
print('be carefull with profiling between step: 10-20')
print('will compute accuracy on the test set every {} step so {} time'.format(n_steps_history, int(steps/n_steps_history)))
if profiling:
print(' profiling ...')
steps_per_epoch_train = 100
n_steps_history=25
input_eval_tfrecords = 'gs://'+os.environ['BUCKET_NAME']+'/tfrecord/sst2/bert-base-multilingual-uncased/valid' #'gs://public-test-data-gs/valid'
input_train_tfrecords = 'gs://'+os.environ['BUCKET_NAME']+'/tfrecord/sst2/bert-base-multilingual-uncased/train' #'gs://public-test-data-gs/train'
if hp_tuning:
output_dir = 'gs://'+os.environ['BUCKET_NAME']+'/training_model_gcp/'+model_name+'_hp_tuning_'+hardware+'_'+timestamp
else:
output_dir = 'gs://'+os.environ['BUCKET_NAME']+'/training_model_gcp/'+model_name+'_'+hardware+'_'+timestamp
pretrained_model_dir = 'gs://'+os.environ['BUCKET_NAME']+'/pretrained_model/bert-base-multilingual-uncased'
#epsilon = 1.7788921050163616e-06
#learning_rate= 0.0007763625134788308
epsilon = 1e-8
learning_rate= 5e-5
# bulding training_inputs
parameters = ['--epochs', str(epochs),
'--steps_per_epoch_train', str(steps_per_epoch_train),
'--batch_size_train', str(batch_size_train),
'--steps_per_epoch_eval', str(steps_per_epoch_eval),
'--n_steps_history', str(n_steps_history),
'--batch_size_eval', str(batch_size_eval),
'--input_eval_tfrecords', input_eval_tfrecords ,
'--input_train_tfrecords', input_train_tfrecords,
'--output_dir', output_dir,
'--pretrained_model_dir', pretrained_model_dir,
'--verbosity_level', verbosity,
'--epsilon', str(epsilon),
'--learning_rate', str(learning_rate)]
if hardware=='tpu':
parameters.append('--use_tpu')
parameters.append('True')
training_inputs = {
'args': parameters,
'region': region,
}
if not use_custom_container:
training_inputs['packageUris'] = [package_gcs]
training_inputs['pythonModule'] = module_name
training_inputs['runtimeVersion'] = runtime_version
training_inputs['pythonVersion'] = python_version
else:
accelerator_master = {'imageUri': image_uri}
training_inputs['masterConfig'] = accelerator_master
if tier=='basic' and hardware=='cpu':
# CPU
training_inputs['scaleTier'] = 'BASIC'
#training_inputs['scaleTier'] = 'STANDARD_1'
elif tier=='custom' and hardware=='cpu':
# CPU
training_inputs['scaleTier'] = 'CUSTOM'
training_inputs['masterType'] = 'n1-standard-16'
elif tier=='basic' and hardware=='gpu':
# GPU
training_inputs['scaleTier'] = 'BASIC_GPU'
elif tier=='custom' and hardware=='gpu':
# Custom GPU
training_inputs['scaleTier'] = 'CUSTOM'
training_inputs['masterType'] = 'n1-standard-8'
accelerator_master = {'acceleratorConfig': {
'count': '1',
'type': 'NVIDIA_TESLA_V100'}
}
training_inputs['masterConfig'] = accelerator_master
elif tier=='basic' and hardware=='tpu':
# TPU
training_inputs['scaleTier'] = 'BASIC_TPU'
elif tier=='custom' and hardware=='tpu':
# Custom TPU
training_inputs['scaleTier'] = 'CUSTOM'
training_inputs['masterType'] = 'n1-highcpu-16'
training_inputs['workerType'] = 'cloud_tpu'
training_inputs['workerCount'] = '1'
accelerator_master = {'acceleratorConfig': {
'count': '8',
'type': 'TPU_V3'}
}
training_inputs['workerConfig'] = accelerator_master
else:
# Default
training_inputs['scaleTier'] = 'BASIC'
print('======')
# add hyperparameter tuning to the job config.
if hp_tuning:
hyperparams = {
'algorithm': 'ALGORITHM_UNSPECIFIED',
'goal': 'MAXIMIZE',
'maxTrials': 3,
'maxParallelTrials': 2,
'maxFailedTrials': 1,
'enableTrialEarlyStopping': True,
'hyperparameterMetricTag': 'metric_accuracy_train_epoch',
'params': []}
hyperparams['params'].append({
'parameterName':'learning_rate',
'type':'DOUBLE',
'minValue': 1.0e-8,
'maxValue': 1.0,
'scaleType': 'UNIT_LOG_SCALE'})
hyperparams['params'].append({
'parameterName':'epsilon',
'type':'DOUBLE',
'minValue': 1.0e-9,
'maxValue': 1.0,
'scaleType': 'UNIT_LOG_SCALE'})
# Add hyperparameter specification to the training inputs dictionary.
training_inputs['hyperparameters'] = hyperparams
# building job_spec
labels = {'accelerator': hardware,
'prod_type': type_production,
'owner': owner}
if use_custom_container:
labels['type'] = 'custom_container'
else:
labels['type'] = 'gcp_runtime'
job_spec = {'jobId': job_name,
'labels': labels,
'trainingInput': training_inputs}
if test_logging:
# test
# variable used to build some variable's name
owner = os.environ['OWNER']
tier = 'basic'
verbosity = 'INFO'
# define parameters for ai platform training
if not use_custom_container:
package_gcs = package_gcs
else:
image_uri='gcr.io/'+os.environ['PROJECT_ID']+tag
job_name = 'debug_test_'+datetime.now().strftime("%Y_%m_%d_%H%M%S")
module_name = 'model.test-log.task'
#module_name = 'model.test.task'
region = 'europe-west1'
# bulding training_inputs
parameters = ['--verbosity_level', verbosity]
training_inputs = {
'args': parameters,
'region': region,
}
if not use_custom_container:
training_inputs['packageUris'] = [package_gcs]
training_inputs['pythonModule'] = module_name
training_inputs['runtimeVersion'] = runtime_version
training_inputs['pythonVersion'] = python_version
else:
accelerator_master = {'imageUri': image_uri}
#training_inputs['pythonModule'] = module_name # not working to overwrite the entrypoint
training_inputs['masterConfig'] = accelerator_master
training_inputs['scaleTier'] = 'BASIC'
# building job_spec
labels = {'accelerator': 'cpu',
'prod_type': 'debug',
'owner': owner}
if use_custom_container:
labels['type'] = 'custom_container'
else:
labels['type'] = 'gcp_runtime'
job_spec = {'jobId': job_name,
'labels': labels,
'trainingInput': training_inputs}
training_inputs, job_name
# submit the training job
request = ai_platform_training.projects().jobs().create(body=job_spec,
parent=project_id)
try:
response = request.execute()
print('Job status for {}:'.format(response['jobId']))
print(' state : {}'.format(response['state']))
print(' createTime: {}'.format(response['createTime']))
except errors.HttpError as err:
# For this example, just send some text to the logs.
# You need to import logging for this to work.
logging.error('There was an e0rror creating the training job.'
' Check the details:')
logging.error(err._get_reason())
# if you wnat to specify a specif job ID
#job_name = 'tf_bert_classification_2020_05_16_193551'
jobId = 'projects/{}/jobs/{}'.format(project_name, job_name)
request = ai_platform_training.projects().jobs().get(name=jobId)
response = None
try:
response = request.execute()
print('Job status for {}:'.format(response['jobId']))
print(' state : {}'.format(response['state']))
if 'trainingOutput' in response.keys():
if 'trials' in response['trainingOutput'].keys():
for sub_job in response['trainingOutput']['trials']:
print(' trials : {}'.format(sub_job))
if 'consumedMLUnits' in response.keys():
print(' consumedMLUnits : {}'.format(response['trainingOutput']['consumedMLUnits']))
if 'errorMessage' in response.keys():
print(' errorMessage : {}'.format(response['errorMessage']))
except errors.HttpError as err:
logging.error('There was an error getting the logs.'
' Check the details:')
logging.error(err._get_reason())
# how to stream logs
# --stream-logs
```
# TensorBoard for job running on GCP
```
# View open TensorBoard instance
#notebook.list()
# View pid
#!ps -ef|grep tensorboard
# Killed Tensorboard process by using pid
#!kill -9 pid
%load_ext tensorboard
#%reload_ext tensorboard
%tensorboard --logdir {output_dir+'/tensorboard'} \
#--host 0.0.0.0 \
#--port 6006 \
#--debugger_port 6006
%load_ext tensorboard
#%reload_ext tensorboard
%tensorboard --logdir {output_dir+'/hparams_tuning'} \
#--host 0.0.0.0 \
#--port 6006 \
#--debugger_port 6006
!tensorboard dev upload --logdir \
'gs://multilingual_text_classification/training_model_gcp/tf_bert_classification_cpu_2020_08_20_093837/tensorboard' --one_shot --yes
```
|
github_jupyter
|
# データサイエンス100本ノック(構造化データ加工編) - Python
## はじめに
- 初めに以下のセルを実行してください
- 必要なライブラリのインポートとデータベース(PostgreSQL)からのデータ読み込みを行います
- pandas等、利用が想定されるライブラリは以下セルでインポートしています
- その他利用したいライブラリがあれば適宜インストールしてください("!pip install ライブラリ名"でインストールも可能)
- 処理は複数回に分けても構いません
- 名前、住所等はダミーデータであり、実在するものではありません
```
import os
import pandas as pd
import numpy as np
from datetime import datetime, date
from dateutil.relativedelta import relativedelta
import math
import psycopg2
from sqlalchemy import create_engine
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from imblearn.under_sampling import RandomUnderSampler
pgconfig = {
'host': 'db',
'port': os.environ['PG_PORT'],
'database': os.environ['PG_DATABASE'],
'user': os.environ['PG_USER'],
'password': os.environ['PG_PASSWORD'],
}
# pd.read_sql用のコネクタ
conn = psycopg2.connect(**pgconfig)
df_customer = pd.read_sql(sql='select * from customer', con=conn)
df_category = pd.read_sql(sql='select * from category', con=conn)
df_product = pd.read_sql(sql='select * from product', con=conn)
df_receipt = pd.read_sql(sql='select * from receipt', con=conn)
df_store = pd.read_sql(sql='select * from store', con=conn)
df_geocode = pd.read_sql(sql='select * from geocode', con=conn)
```
# 演習問題
---
> P-001: レシート明細のデータフレーム(df_receipt)から全項目の先頭10件を表示し、どのようなデータを保有しているか目視で確認せよ。
```
df_receipt.head(10)
```
---
> P-002: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、10件表示させよ。
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']].head(10)
```
---
> P-003: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、10件表示させよ。ただし、sales_ymdはsales_dateに項目名を変更しながら抽出すること。
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']]. \
rename(columns={'sales_ymd': 'sales_date'}).head(10)
```
---
> P-004: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、以下の条件を満たすデータを抽出せよ。
> - 顧客ID(customer_id)が"CS018205000001"
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']]. \
query('customer_id == "CS018205000001"')
```
---
> P-005: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、以下の条件を満たすデータを抽出せよ。
> - 顧客ID(customer_id)が"CS018205000001"
> - 売上金額(amount)が1,000以上
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']] \
.query('customer_id == "CS018205000001" & amount >= 1000')
```
---
> P-006: レシート明細データフレーム「df_receipt」から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上数量(quantity)、売上金額(amount)の順に列を指定し、以下の条件を満たすデータを抽出せよ。
> - 顧客ID(customer_id)が"CS018205000001"
> - 売上金額(amount)が1,000以上または売上数量(quantity)が5以上
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'quantity', 'amount']].\
query('customer_id == "CS018205000001" & (amount >= 1000 | quantity >=5)')
```
---
> P-007: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、以下の条件を満たすデータを抽出せよ。
> - 顧客ID(customer_id)が"CS018205000001"
> - 売上金額(amount)が1,000以上2,000以下
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']] \
.query('customer_id == "CS018205000001" & 1000 <= amount <= 2000')
```
---
> P-008: レシート明細のデータフレーム(df_receipt)から売上日(sales_ymd)、顧客ID(customer_id)、商品コード(product_cd)、売上金額(amount)の順に列を指定し、以下の条件を満たすデータを抽出せよ。
> - 顧客ID(customer_id)が"CS018205000001"
> - 商品コード(product_cd)が"P071401019"以外
```
df_receipt[['sales_ymd', 'customer_id', 'product_cd', 'amount']] \
.query('customer_id == "CS018205000001" & product_cd != "P071401019"')
```
---
> P-009: 以下の処理において、出力結果を変えずにORをANDに書き換えよ。
`df_store.query('not(prefecture_cd == "13" | floor_area > 900)')`
```
df_store.query('prefecture_cd != "13" & floor_area <= 900')
```
---
> P-010: 店舗データフレーム(df_store)から、店舗コード(store_cd)が"S14"で始まるものだけ全項目抽出し、10件だけ表示せよ。
```
df_store.query("store_cd.str.startswith('S14')", engine='python').head(10)
```
---
> P-011: 顧客データフレーム(df_customer)から顧客ID(customer_id)の末尾が1のものだけ全項目抽出し、10件だけ表示せよ。
```
df_customer.query("customer_id.str.endswith('1')", engine='python').head(10)
```
---
> P-012: 店舗データフレーム(df_store)から横浜市の店舗だけ全項目表示せよ。
```
df_store.query("address.str.contains('横浜市')", engine='python')
```
---
> P-013: 顧客データフレーム(df_customer)から、ステータスコード(status_cd)の先頭がアルファベットのA〜Fで始まるデータを全項目抽出し、10件だけ表示せよ。
```
df_customer.query("status_cd.str.contains('^[A-F]', regex=True)",
engine='python').head(10)
```
---
> P-014: 顧客データフレーム(df_customer)から、ステータスコード(status_cd)の末尾が数字の1〜9で終わるデータを全項目抽出し、10件だけ表示せよ。
```
df_customer.query("status_cd.str.contains('[1-9]$', regex=True)", engine='python').head(10)
```
---
> P-015: 顧客データフレーム(df_customer)から、ステータスコード(status_cd)の先頭がアルファベットのA〜Fで始まり、末尾が数字の1〜9で終わるデータを全項目抽出し、10件だけ表示せよ。
```
df_customer.query("status_cd.str.contains('^[A-F].*[1-9]$', regex=True)",
engine='python').head(10)
```
---
> P-016: 店舗データフレーム(df_store)から、電話番号(tel_no)が3桁-3桁-4桁のデータを全項目表示せよ。
```
df_store.query("tel_no.str.contains('^[0-9]{3}-[0-9]{3}-[0-9]{4}$',regex=True)",
engine='python')
```
---
> P-17: 顧客データフレーム(df_customer)を生年月日(birth_day)で高齢順にソートし、先頭10件を全項目表示せよ。
```
df_customer.sort_values('birth_day', ascending=True).head(10)
```
---
> P-18: 顧客データフレーム(df_customer)を生年月日(birth_day)で若い順にソートし、先頭10件を全項目表示せよ。
```
df_customer.sort_values('birth_day', ascending=False).head(10)
```
---
> P-19: レシート明細データフレーム(df_receipt)に対し、1件あたりの売上金額(amount)が高い順にランクを付与し、先頭10件を抽出せよ。項目は顧客ID(customer_id)、売上金額(amount)、付与したランクを表示させること。なお、売上金額(amount)が等しい場合は同一順位を付与するものとする。
```
df_tmp = pd.concat([df_receipt[['customer_id', 'amount']]
,df_receipt['amount'].rank(method='min',
ascending=False)], axis=1)
df_tmp.columns = ['customer_id', 'amount', 'ranking']
df_tmp.sort_values('ranking', ascending=True).head(10)
```
---
> P-020: レシート明細データフレーム(df_receipt)に対し、1件あたりの売上金額(amount)が高い順にランクを付与し、先頭10件を抽出せよ。項目は顧客ID(customer_id)、売上金額(amount)、付与したランクを表示させること。なお、売上金額(amount)が等しい場合でも別順位を付与すること。
```
df_tmp = pd.concat([df_receipt[['customer_id', 'amount']]
,df_receipt['amount'].rank(method='first',
ascending=False)], axis=1)
df_tmp.columns = ['customer_id', 'amount', 'ranking']
df_tmp.sort_values('ranking', ascending=True).head(10)
```
---
> P-021: レシート明細データフレーム(df_receipt)に対し、件数をカウントせよ。
```
len(df_receipt)
```
---
> P-022: レシート明細データフレーム(df_receipt)の顧客ID(customer_id)に対し、ユニーク件数をカウントせよ。
```
len(df_receipt['customer_id'].unique())
```
---
> P-023: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)と売上数量(quantity)を合計せよ。
```
df_receipt.groupby('store_cd').agg({'amount':'sum',
'quantity':'sum'}).reset_index()
```
---
> P-024: レシート明細データフレーム(df_receipt)に対し、顧客ID(customer_id)ごとに最も新しい売上日(sales_ymd)を求め、10件表示せよ。
```
df_receipt.groupby('customer_id').sales_ymd.max().reset_index().head(10)
```
---
> P-025: レシート明細データフレーム(df_receipt)に対し、顧客ID(customer_id)ごとに最も古い売上日(sales_ymd)を求め、10件表示せよ。
```
df_receipt.groupby('customer_id').agg({'sales_ymd':'min'}).head(10)
```
---
> P-026: レシート明細データフレーム(df_receipt)に対し、顧客ID(customer_id)ごとに最も新しい売上日(sales_ymd)と古い売上日を求め、両者が異なるデータを10件表示せよ。
```
df_tmp = df_receipt.groupby('customer_id'). \
agg({'sales_ymd':['max','min']}).reset_index()
df_tmp.columns = ["_".join(pair) for pair in df_tmp.columns]
df_tmp.query('sales_ymd_max != sales_ymd_min').head(10)
```
---
> P-027: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)の平均を計算し、降順でTOP5を表示せよ。
```
df_receipt.groupby('store_cd').agg({'amount':'mean'}).reset_index(). \
sort_values('amount', ascending=False).head(5)
```
---
> P-028: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)の中央値を計算し、降順でTOP5を表示せよ。
```
df_receipt.groupby('store_cd').agg({'amount':'median'}).reset_index(). \
sort_values('amount', ascending=False).head(5)
```
---
> P-029: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに商品コード(product_cd)の最頻値を求めよ。
```
df_receipt.groupby('store_cd').product_cd. \
apply(lambda x: x.mode()).reset_index()
```
---
> P-030: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)の標本分散を計算し、降順でTOP5を表示せよ。
```
df_receipt.groupby('store_cd').amount.var(ddof=0).reset_index(). \
sort_values('amount', ascending=False).head(5)
```
---
> P-031: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)の標本標準偏差を計算し、降順でTOP5を表示せよ。
TIPS:
PandasとNumpyでddofのデフォルト値が異なることに注意しましょう
```
Pandas:
DataFrame.std(self, axis=None, skipna=None, level=None, ddof=1, numeric_only=None, **kwargs)
Numpy:
numpy.std(a, axis=None, dtype=None, out=None, ddof=0, keepdims=)
```
```
df_receipt.groupby('store_cd').amount.std(ddof=0).reset_index(). \
sort_values('amount', ascending=False).head(5)
```
---
> P-032: レシート明細データフレーム(df_receipt)の売上金額(amount)について、25%刻みでパーセンタイル値を求めよ。
```
# コード例1
np.percentile(df_receipt['amount'], q=[25, 50, 75,100])
# コード例2
df_receipt.amount.quantile(q=np.arange(5)/4)
```
---
> P-033: レシート明細データフレーム(df_receipt)に対し、店舗コード(store_cd)ごとに売上金額(amount)の平均を計算し、330以上のものを抽出せよ。
```
df_receipt.groupby('store_cd').amount.mean(). \
reset_index().query('amount >= 330')
```
---
> P-034: レシート明細データフレーム(df_receipt)に対し、顧客ID(customer_id)ごとに売上金額(amount)を合計して全顧客の平均を求めよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。
```
# queryを使わない書き方
df_receipt[~df_receipt['customer_id'].str.startswith("Z")]. \
groupby('customer_id').amount.sum().mean()
# queryを使う書き方
df_receipt.query('not customer_id.str.startswith("Z")',
engine='python').groupby('customer_id').amount.sum().mean()
```
---
> P-035: レシート明細データフレーム(df_receipt)に対し、顧客ID(customer_id)ごとに売上金額(amount)を合計して全顧客の平均を求め、平均以上に買い物をしている顧客を抽出せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。なお、データは10件だけ表示させれば良い。
```
df_receipt_tmp = df_receipt[~df_receipt['customer_id'].str.startswith("Z")]
amount_mean = df_receipt_tmp.groupby('customer_id').amount.sum().mean()
df_amount_sum = df_receipt_tmp.groupby('customer_id').amount.sum().reset_index()
df_amount_sum[df_amount_sum['amount'] >= amount_mean].head(10)
```
---
> P-036: レシート明細データフレーム(df_receipt)と店舗データフレーム(df_store)を内部結合し、レシート明細データフレームの全項目と店舗データフレームの店舗名(store_name)を10件表示させよ。
```
pd.merge(df_receipt, df_store[['store_cd','store_name']],
how='inner', on='store_cd').head(10)
```
---
> P-037: 商品データフレーム(df_product)とカテゴリデータフレーム(df_category)を内部結合し、商品データフレームの全項目とカテゴリデータフレームの小区分名(category_small_name)を10件表示させよ。
```
pd.merge(df_product
, df_category[['category_small_cd','category_small_name']]
, how='inner', on='category_small_cd').head(10)
```
---
> P-038: 顧客データフレーム(df_customer)とレシート明細データフレーム(df_receipt)から、各顧客ごとの売上金額合計を求めよ。ただし、売上実績がない顧客については売上金額を0として表示させること。また、顧客は性別コード(gender_cd)が女性(1)であるものを対象とし、非会員(顧客IDが"Z"から始まるもの)は除外すること。なお、結果は10件だけ表示させれば良い。
```
df_amount_sum = df_receipt.groupby('customer_id').amount.sum().reset_index()
df_tmp = df_customer. \
query('gender_cd == "1" and not customer_id.str.startswith("Z")',
engine='python')
pd.merge(df_tmp['customer_id'], df_amount_sum,
how='left', on='customer_id').fillna(0).head(10)
```
---
> P-039: レシート明細データフレーム(df_receipt)から売上日数の多い顧客の上位20件と、売上金額合計の多い顧客の上位20件を抽出し、完全外部結合せよ。ただし、非会員(顧客IDが"Z"から始まるもの)は除外すること。
```
df_sum = df_receipt.groupby('customer_id').amount.sum().reset_index()
df_sum = df_sum.query('not customer_id.str.startswith("Z")', engine='python')
df_sum = df_sum.sort_values('amount', ascending=False).head(20)
df_cnt = df_receipt[~df_receipt.duplicated(subset=['customer_id', 'sales_ymd'])]
df_cnt = df_cnt.query('not customer_id.str.startswith("Z")', engine='python')
df_cnt = df_cnt.groupby('customer_id').sales_ymd.count().reset_index()
df_cnt = df_cnt.sort_values('sales_ymd', ascending=False).head(20)
pd.merge(df_sum, df_cnt, how='outer', on='customer_id')
```
---
> P-040: 全ての店舗と全ての商品を組み合わせると何件のデータとなるか調査したい。店舗(df_store)と商品(df_product)を直積した件数を計算せよ。
```
df_store_tmp = df_store.copy()
df_product_tmp = df_product.copy()
df_store_tmp['key'] = 0
df_product_tmp['key'] = 0
len(pd.merge(df_store_tmp, df_product_tmp, how='outer', on='key'))
```
---
> P-041: レシート明細データフレーム(df_receipt)の売上金額(amount)を日付(sales_ymd)ごとに集計し、前日からの売上金額増減を計算せよ。なお、計算結果は10件表示すればよい。
```
df_sales_amount_by_date = df_receipt[['sales_ymd', 'amount']].\
groupby('sales_ymd').sum().reset_index()
df_sales_amount_by_date = pd.concat([df_sales_amount_by_date,
df_sales_amount_by_date.shift()], axis=1)
df_sales_amount_by_date.columns = ['sales_ymd','amount','lag_ymd','lag_amount']
df_sales_amount_by_date['diff_amount'] = \
df_sales_amount_by_date['amount'] - df_sales_amount_by_date['lag_amount']
df_sales_amount_by_date.head(10)
```
---
> P-042: レシート明細データフレーム(df_receipt)の売上金額(amount)を日付(sales_ymd)ごとに集計し、各日付のデータに対し、1日前、2日前、3日前のデータを結合せよ。結果は10件表示すればよい。
```
# コード例1:縦持ちケース
df_sales_amount_by_date = df_receipt[['sales_ymd', 'amount']]. \
groupby('sales_ymd').sum().reset_index()
for i in range(1, 4):
if i == 1:
df_lag = pd.concat([df_sales_amount_by_date,
df_sales_amount_by_date.shift(i)],axis=1)
else:
df_lag = df_lag.append(pd.concat([df_sales_amount_by_date,
df_sales_amount_by_date.shift(i)],
axis=1))
df_lag.columns = ['sales_ymd', 'amount', 'lag_ymd', 'lag_amount']
df_lag.dropna().sort_values(['sales_ymd','lag_ymd']).head(10)
# コード例2:横持ちケース
df_sales_amount_by_date = df_receipt[['sales_ymd', 'amount']].\
groupby('sales_ymd').sum().reset_index()
for i in range(1, 4):
if i == 1:
df_lag = pd.concat([df_sales_amount_by_date,
df_sales_amount_by_date.shift(i)],axis=1)
else:
df_lag = pd.concat([df_lag, df_sales_amount_by_date.shift(i)],axis=1)
df_lag.columns = ['sales_ymd', 'amount', 'lag_ymd_1', 'lag_amount_1',
'lag_ymd_2', 'lag_amount_2', 'lag_ymd_3', 'lag_amount_3']
df_lag.dropna().sort_values(['sales_ymd']).head(10)
```
---
> P-043: レシート明細データフレーム(df_receipt)と顧客データフレーム(df_customer)を結合し、性別(gender)と年代(ageから計算)ごとに売上金額(amount)を合計した売上サマリデータフレーム(df_sales_summary)を作成せよ。性別は0が男性、1が女性、9が不明を表すものとする。
>
> ただし、項目構成は年代、女性の売上金額、男性の売上金額、性別不明の売上金額の4項目とすること(縦に年代、横に性別のクロス集計)。また、年代は10歳ごとの階級とすること。
```
# コード例1
df_tmp = pd.merge(df_receipt, df_customer, how ='inner', on="customer_id")
df_tmp['era'] = df_tmp['age'].apply(lambda x: math.floor(x / 10) * 10)
df_sales_summary = pd.pivot_table(df_tmp, index='era', columns='gender_cd',
values='amount', aggfunc='sum').reset_index()
df_sales_summary.columns = ['era', 'male', 'female', 'unknown']
df_sales_summary
# コード例2
df_tmp = pd.merge(df_receipt, df_customer, how ='inner', on="customer_id")
df_tmp['era'] = np.floor(df_tmp['age'] / 10).astype(int) * 10
df_sales_summary = pd.pivot_table(df_tmp, index='era', columns='gender_cd',
values='amount', aggfunc='sum').reset_index()
df_sales_summary.columns = ['era', 'male', 'female', 'unknown']
df_sales_summary
```
---
> P-044: 前設問で作成した売上サマリデータフレーム(df_sales_summary)は性別の売上を横持ちさせたものであった。このデータフレームから性別を縦持ちさせ、年代、性別コード、売上金額の3項目に変換せよ。ただし、性別コードは男性を"00"、女性を"01"、不明を"99"とする。
```
df_sales_summary = df_sales_summary.set_index('era'). \
stack().reset_index().replace({'female':'01','male':'00','unknown':'99'}). \
rename(columns={'level_1':'gender_cd', 0: 'amount'})
df_sales_summary
```
---
> P-045: 顧客データフレーム(df_customer)の生年月日(birth_day)は日付型でデータを保有している。これをYYYYMMDD形式の文字列に変換し、顧客ID(customer_id)とともに抽出せよ。データは10件を抽出すれば良い。
```
pd.concat([df_customer['customer_id'],
pd.to_datetime(df_customer['birth_day']).dt.strftime('%Y%m%d')],
axis = 1).head(10)
```
---
> P-046: 顧客データフレーム(df_customer)の申し込み日(application_date)はYYYYMMDD形式の文字列型でデータを保有している。これを日付型に変換し、顧客ID(customer_id)とともに抽出せよ。データは10件を抽出すれば良い。
```
pd.concat([df_customer['customer_id'],
pd.to_datetime(df_customer['application_date'])], axis=1).head(10)
```
---
> P-047: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)はYYYYMMDD形式の数値型でデータを保有している。これを日付型に変換し、レシート番号(receipt_no)、レシートサブ番号(receipt_sub_no)とともに抽出せよ。データは10件を抽出すれば良い。
```
pd.concat([df_receipt[['receipt_no', 'receipt_sub_no']],
pd.to_datetime(df_receipt['sales_ymd'].astype('str'))],
axis=1).head(10)
```
---
> P-048: レシート明細データフレーム(df_receipt)の売上エポック秒(sales_epoch)は数値型のUNIX秒でデータを保有している。これを日付型に変換し、レシート番号(receipt_no)、レシートサブ番号(receipt_sub_no)とともに抽出せよ。データは10件を抽出すれば良い。
```
pd.concat([df_receipt[['receipt_no', 'receipt_sub_no']],
pd.to_datetime(df_receipt['sales_epoch'], unit='s')],
axis=1).head(10)
```
---
> P-049: レシート明細データフレーム(df_receipt)の売上エポック秒(sales_epoch)を日付型に変換し、「年」だけ取り出してレシート番号(receipt_no)、レシートサブ番号(receipt_sub_no)とともに抽出せよ。データは10件を抽出すれば良い。
```
pd.concat([df_receipt[['receipt_no', 'receipt_sub_no']],
pd.to_datetime(df_receipt['sales_epoch'], unit='s').dt.year],
axis=1).head(10)
```
---
> P-050: レシート明細データフレーム(df_receipt)の売上エポック秒(sales_epoch)を日付型に変換し、「月」だけ取り出してレシート番号(receipt_no)、レシートサブ番号(receipt_sub_no)とともに抽出せよ。なお、「月」は0埋め2桁で取り出すこと。データは10件を抽出すれば良い。
```
# dt.monthでも月を取得できるが、ここでは0埋め2桁で取り出すためstrftimeを利用している
pd.concat([df_receipt[['receipt_no', 'receipt_sub_no']],
pd.to_datetime(df_receipt['sales_epoch'], unit='s'). \
dt.strftime('%m')],axis=1).head(10)
```
---
> P-051: レシート明細データフレーム(df_receipt)の売上エポック秒を日付型に変換し、「日」だけ取り出してレシート番号(receipt_no)、レシートサブ番号(receipt_sub_no)とともに抽出せよ。なお、「日」は0埋め2桁で取り出すこと。データは10件を抽出すれば良い。
```
# dt.dayでも日を取得できるが、ここでは0埋め2桁で取り出すためstrftimeを利用している
pd.concat([df_receipt[['receipt_no', 'receipt_sub_no']],
pd.to_datetime(df_receipt['sales_epoch'], unit='s'). \
dt.strftime('%d')],axis=1).head(10)
```
---
> P-052: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計の上、売上金額合計に対して2,000円以下を0、2,000円より大きい金額を1に2値化し、顧客ID、売上金額合計とともに10件表示せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。
```
# コード例1
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python')
df_sales_amount = df_sales_amount[['customer_id', 'amount']]. \
groupby('customer_id').sum().reset_index()
df_sales_amount['sales_flg'] = df_sales_amount['amount']. \
apply(lambda x: 1 if x > 2000 else 0)
df_sales_amount.head(10)
# コード例2(np.whereの活用)
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python')
df_sales_amount = df_sales_amount[['customer_id', 'amount']]. \
groupby('customer_id').sum().reset_index()
df_sales_amount['sales_flg'] = np.where(df_sales_amount['amount'] > 2000, 1, 0)
df_sales_amount.head(10)
```
---
> P-053: 顧客データフレーム(df_customer)の郵便番号(postal_cd)に対し、東京(先頭3桁が100〜209のもの)を1、それ以外のものを0に2値化せよ。さらにレシート明細データフレーム(df_receipt)と結合し、全期間において売上実績がある顧客数を、作成した2値ごとにカウントせよ。
```
# コード例1
df_tmp = df_customer[['customer_id', 'postal_cd']].copy()
df_tmp['postal_flg'] = df_tmp['postal_cd']. \
apply(lambda x: 1 if 100 <= int(x[0:3]) <= 209 else 0)
pd.merge(df_tmp, df_receipt, how='inner', on='customer_id'). \
groupby('postal_flg').agg({'customer_id':'nunique'})
# コード例2(np.where、betweenの活用)
df_tmp = df_customer[['customer_id', 'postal_cd']].copy()
df_tmp['postal_flg'] = np.where(df_tmp['postal_cd'].str[0:3].astype(int)
.between(100, 209), 1, 0)
pd.merge(df_tmp, df_receipt, how='inner', on='customer_id'). \
groupby('postal_flg').agg({'customer_id':'nunique'})
```
---
> P-054: 顧客データフレーム(df_customer)の住所(address)は、埼玉県、千葉県、東京都、神奈川県のいずれかとなっている。都道府県毎にコード値を作成し、顧客ID、住所とともに抽出せよ。値は埼玉県を11、千葉県を12、東京都を13、神奈川県を14とすること。結果は10件表示させれば良い。
```
pd.concat([df_customer[['customer_id', 'address']],
df_customer['address'].str[0:3].map({'埼玉県': '11',
'千葉県':'12',
'東京都':'13',
'神奈川':'14'})],axis=1).head(10)
```
---
> P-055: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計し、その合計金額の四分位点を求めよ。その上で、顧客ごとの売上金額合計に対して以下の基準でカテゴリ値を作成し、顧客ID、売上金額合計とともに表示せよ。カテゴリ値は上から順に1〜4とする。結果は10件表示させれば良い。
>
> - 最小値以上第一四分位未満
> - 第一四分位以上第二四分位未満
> - 第二四分位以上第三四分位未満
> - 第三四分位以上
```
# コード例1
df_sales_amount = df_receipt[['customer_id', 'amount']]. \
groupby('customer_id').sum().reset_index()
pct25 = np.quantile(df_sales_amount['amount'], 0.25)
pct50 = np.quantile(df_sales_amount['amount'], 0.5)
pct75 = np.quantile(df_sales_amount['amount'], 0.75)
def pct_group(x):
if x < pct25:
return 1
elif pct25 <= x < pct50:
return 2
elif pct50 <= x < pct75:
return 3
elif pct75 <= x:
return 4
df_sales_amount['pct_group'] = df_sales_amount['amount'].apply(lambda x: pct_group(x))
df_sales_amount.head(10)
# 確認用
print('pct25:', pct25)
print('pct50:', pct50)
print('pct75:', pct75)
# コード例2
df_temp = df_receipt.groupby('customer_id')[['amount']].sum()
df_temp['quantile'], bins = \
pd.qcut(df_receipt.groupby('customer_id')['amount'].sum(), 4, retbins=True)
display(df_temp.head())
print('quantiles:', bins)
```
---
> P-056: 顧客データフレーム(df_customer)の年齢(age)をもとに10歳刻みで年代を算出し、顧客ID(customer_id)、生年月日(birth_day)とともに抽出せよ。ただし、60歳以上は全て60歳代とすること。年代を表すカテゴリ名は任意とする。先頭10件を表示させればよい。
```
# コード例1
df_customer_era = pd.concat([df_customer[['customer_id', 'birth_day']],
df_customer['age']. \
apply(lambda x: min(math.floor(x / 10) * 10, 60))],
axis=1)
df_customer_era.head(10)
# コード例2
df_customer['age_group'] = pd.cut(df_customer['age'],
bins=[0, 10, 20, 30, 40, 50, 60, np.inf],
right=False)
df_customer[['customer_id', 'birth_day', 'age_group']].head(10)
```
---
> P-057: 前問題の抽出結果と性別(gender)を組み合わせ、新たに性別×年代の組み合わせを表すカテゴリデータを作成せよ。組み合わせを表すカテゴリの値は任意とする。先頭10件を表示させればよい。
```
df_customer_era['era_gender'] = \
df_customer['gender_cd'] + df_customer_era['age'].astype('str')
df_customer_era.head(10)
```
---
> P-058: 顧客データフレーム(df_customer)の性別コード(gender_cd)をダミー変数化し、顧客ID(customer_id)とともに抽出せよ。結果は10件表示させれば良い。
```
pd.get_dummies(df_customer[['customer_id', 'gender_cd']],
columns=['gender_cd']).head(10)
```
---
> P-059: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計し、売上金額合計を平均0、標準偏差1に標準化して顧客ID、売上金額合計とともに表示せよ。標準化に使用する標準偏差は、不偏標準偏差と標本標準偏差のどちらでも良いものとする。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。結果は10件表示させれば良い。
TIPS:
- query()の引数engineで'python'か'numexpr'かを選択でき、デフォルトはインストールされていればnumexprが、無ければpythonが使われます。さらに、文字列メソッドはengine='python'でないとquery()メソッドで使えません。
```
# skleanのpreprocessing.scaleを利用するため、標本標準偏差で計算されている
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_sales_amount['amount_ss'] = preprocessing.scale(df_sales_amount['amount'])
df_sales_amount.head(10)
# コード例2(fitを行うことで、別のデータでも同じの平均・標準偏差で標準化を行える)
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
scaler = preprocessing.StandardScaler()
scaler.fit(df_sales_amount[['amount']])
df_sales_amount['amount_ss'] = scaler.transform(df_sales_amount[['amount']])
df_sales_amount.head(10)
```
---
> P-060: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計し、売上金額合計を最小値0、最大値1に正規化して顧客ID、売上金額合計とともに表示せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。結果は10件表示させれば良い。
```
# コード例1
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_sales_amount['amount_mm'] = \
preprocessing.minmax_scale(df_sales_amount['amount'])
df_sales_amount.head(10)
# コード例2(fitを行うことで、別のデータでも同じの平均・標準偏差で標準化を行える)
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
scaler = preprocessing.MinMaxScaler()
scaler.fit(df_sales_amount[['amount']])
df_sales_amount['amount_mm'] = scaler.transform(df_sales_amount[['amount']])
df_sales_amount.head(10)
```
---
> P-061: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計し、売上金額合計を常用対数化(底=10)して顧客ID、売上金額合計とともに表示せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。結果は10件表示させれば良い。
```
# skleanのpreprocessing.scaleを利用するため、標本標準偏差で計算されている
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_sales_amount['amount_log10'] = np.log10(df_sales_amount['amount'] + 0.5)
df_sales_amount.head(10)
```
---
> P-062: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客ID(customer_id)ごとに合計し、売上金額合計を自然対数化(底=e)して顧客ID、売上金額合計とともに表示せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。結果は10件表示させれば良い。
```
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_sales_amount['amount_loge'] = np.log(df_sales_amount['amount'] + 0.5)
df_sales_amount.head(10)
```
---
> P-063: 商品データフレーム(df_product)の単価(unit_price)と原価(unit_cost)から、各商品の利益額を算出せよ。結果は10件表示させれば良い。
```
df_tmp = df_product.copy()
df_tmp['unit_profit'] = df_tmp['unit_price'] - df_tmp['unit_cost']
df_tmp.head(10)
```
---
> P-064: 商品データフレーム(df_product)の単価(unit_price)と原価(unit_cost)から、各商品の利益率の全体平均を算出せよ。
ただし、単価と原価にはNULLが存在することに注意せよ。
```
df_tmp = df_product.copy()
df_tmp['unit_profit_rate'] = \
(df_tmp['unit_price'] - df_tmp['unit_cost']) / df_tmp['unit_price']
df_tmp['unit_profit_rate'].mean(skipna=True)
```
---
> P-065: 商品データフレーム(df_product)の各商品について、利益率が30%となる新たな単価を求めよ。ただし、1円未満は切り捨てること。そして結果を10件表示させ、利益率がおよそ30%付近であることを確認せよ。ただし、単価(unit_price)と原価(unit_cost)にはNULLが存在することに注意せよ。
```
df_tmp = df_product.copy()
df_tmp['new_price'] = np.floor(df_tmp['unit_cost'] / 0.7)
df_tmp['new_profit_rate'] = \
(df_tmp['new_price'] - df_tmp['unit_cost']) / df_tmp['new_price']
df_tmp.head(10)
```
---
> P-066: 商品データフレーム(df_product)の各商品について、利益率が30%となる新たな単価を求めよ。今回は、1円未満を丸めること(四捨五入または偶数への丸めで良い)。そして結果を10件表示させ、利益率がおよそ30%付近であることを確認せよ。ただし、単価(unit_price)と原価(unit_cost)にはNULLが存在することに注意せよ。
```
df_tmp = df_product.copy()
df_tmp['new_price'] = np.round(df_tmp['unit_cost'] / 0.7)
df_tmp['new_profit_rate'] = \
(df_tmp['new_price'] - df_tmp['unit_cost']) / df_tmp['new_price']
df_tmp.head(10)
```
---
> P-067: 商品データフレーム(df_product)の各商品について、利益率が30%となる新たな単価を求めよ。今回は、1円未満を切り上げること。そして結果を10件表示させ、利益率がおよそ30%付近であることを確認せよ。ただし、単価(unit_price)と原価(unit_cost)にはNULLが存在することに注意せよ。
```
df_tmp = df_product.copy()
df_tmp['new_price'] = np.ceil(df_tmp['unit_cost'] / 0.7)
df_tmp['new_profit_rate'] = \
(df_tmp['new_price'] - df_tmp['unit_cost']) / df_tmp['new_price']
df_tmp.head(10)
```
---
> P-068: 商品データフレーム(df_product)の各商品について、消費税率10%の税込み金額を求めよ。 1円未満の端数は切り捨てとし、結果は10件表示すれば良い。ただし、単価(unit_price)にはNULLが存在することに注意せよ。
```
df_tmp = df_product.copy()
df_tmp['price_tax'] = np.floor(df_tmp['unit_price'] * 1.1)
df_tmp.head(10)
```
---
> P-069: レシート明細データフレーム(df_receipt)と商品データフレーム(df_product)を結合し、顧客毎に全商品の売上金額合計と、カテゴリ大区分(category_major_cd)が"07"(瓶詰缶詰)の売上金額合計を計算の上、両者の比率を求めよ。抽出対象はカテゴリ大区分"07"(瓶詰缶詰)の売上実績がある顧客のみとし、結果は10件表示させればよい。
```
# コード例1
df_tmp_1 = pd.merge(df_receipt, df_product,
how='inner', on='product_cd').groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_tmp_2 = pd.merge(df_receipt, df_product.query('category_major_cd == "07"'),
how='inner', on='product_cd').groupby('customer_id').\
agg({'amount':'sum'}).reset_index()
df_tmp_3 = pd.merge(df_tmp_1, df_tmp_2, how='inner', on='customer_id')
df_tmp_3['rate_07'] = df_tmp_3['amount_y'] / df_tmp_3['amount_x']
df_tmp_3.head(10)
# コード例2
df_temp = df_receipt.merge(df_product, how='left', on='product_cd'). \
groupby(['customer_id', 'category_major_cd'])['amount'].sum().unstack()
df_temp = df_temp[df_temp['07'] > 0]
df_temp['sum'] = df_temp.sum(axis=1)
df_temp['07_rate'] = df_temp['07'] / df_temp['sum']
df_temp.head(10)
```
---
> P-070: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)に対し、顧客データフレーム(df_customer)の会員申込日(application_date)からの経過日数を計算し、顧客ID(customer_id)、売上日、会員申込日とともに表示せよ。結果は10件表示させれば良い(なお、sales_ymdは数値、application_dateは文字列でデータを保持している点に注意)。
```
df_tmp = pd.merge(df_receipt[['customer_id', 'sales_ymd']],
df_customer[['customer_id', 'application_date']],
how='inner', on='customer_id')
df_tmp = df_tmp.drop_duplicates()
df_tmp['sales_ymd'] = pd.to_datetime(df_tmp['sales_ymd'].astype('str'))
df_tmp['application_date'] = pd.to_datetime(df_tmp['application_date'])
df_tmp['elapsed_date'] = df_tmp['sales_ymd'] - df_tmp['application_date']
df_tmp.head(10)
```
---
> P-071: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)に対し、顧客データフレーム(df_customer)の会員申込日(application_date)からの経過月数を計算し、顧客ID(customer_id)、売上日、会員申込日とともに表示せよ。結果は10件表示させれば良い(なお、sales_ymdは数値、application_dateは文字列でデータを保持している点に注意)。1ヶ月未満は切り捨てること。
```
df_tmp = pd.merge(df_receipt[['customer_id', 'sales_ymd']],
df_customer[['customer_id', 'application_date']],
how='inner', on='customer_id')
df_tmp = df_tmp.drop_duplicates()
df_tmp['sales_ymd'] = pd.to_datetime(df_tmp['sales_ymd'].astype('str'))
df_tmp['application_date'] = pd.to_datetime(df_tmp['application_date'])
df_tmp['elapsed_date'] = df_tmp[['sales_ymd', 'application_date']]. \
apply(lambda x: relativedelta(x[0], x[1]).years * 12 + \
relativedelta(x[0], x[1]).months, axis=1)
df_tmp.sort_values('customer_id').head(10)
```
---
> P-072: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)に対し、顧客データフレーム(df_customer)の会員申込日(application_date)からの経過年数を計算し、顧客ID(customer_id)、売上日、会員申込日とともに表示せよ。結果は10件表示させれば良い(なお、sales_ymdは数値、application_dateは文字列でデータを保持している点に注意)。1年未満は切り捨てること。
```
df_tmp = pd.merge(df_receipt[['customer_id', 'sales_ymd']],
df_customer[['customer_id', 'application_date']],
how='inner', on='customer_id')
df_tmp = df_tmp.drop_duplicates()
df_tmp['sales_ymd'] = pd.to_datetime(df_tmp['sales_ymd'].astype('str'))
df_tmp['application_date'] = pd.to_datetime(df_tmp['application_date'])
df_tmp['elapsed_date'] = df_tmp[['sales_ymd', 'application_date']]. \
apply(lambda x: relativedelta(x[0], x[1]).years, axis=1)
df_tmp.head(10)
```
---
> P-073: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)に対し、顧客データフレーム(df_customer)の会員申込日(application_date)からのエポック秒による経過時間を計算し、顧客ID(customer_id)、売上日、会員申込日とともに表示せよ。結果は10件表示させれば良い(なお、sales_ymdは数値、application_dateは文字列でデータを保持している点に注意)。なお、時間情報は保有していないため各日付は0時0分0秒を表すものとする。
```
df_tmp = pd.merge(df_receipt[['customer_id', 'sales_ymd']],
df_customer[['customer_id', 'application_date']],
how='inner', on='customer_id')
df_tmp = df_tmp.drop_duplicates()
df_tmp['sales_ymd'] = pd.to_datetime(df_tmp['sales_ymd'].astype('str'))
df_tmp['application_date'] = pd.to_datetime(df_tmp['application_date'])
df_tmp['elapsed_date'] = \
(df_tmp['sales_ymd'].view(np.int64) / 10**9) - (df_tmp['application_date'].\
view(np.int64) / 10**9)
df_tmp.head(10)
```
---
> P-074: レシート明細データフレーム(df_receipt)の売上日(sales_ymd)に対し、当該週の月曜日からの経過日数を計算し、売上日、当該週の月曜日付とともに表示せよ。結果は10件表示させれば良い(なお、sales_ymdは数値でデータを保持している点に注意)。
```
df_tmp = df_receipt[['customer_id', 'sales_ymd']]
df_tmp = df_tmp.drop_duplicates()
df_tmp['sales_ymd'] = pd.to_datetime(df_tmp['sales_ymd'].astype('str'))
df_tmp['monday'] = df_tmp['sales_ymd']. \
apply(lambda x: x - relativedelta(days=x.weekday()))
df_tmp['elapsed_weekday'] = df_tmp['sales_ymd'] - df_tmp['monday']
df_tmp.head(10)
```
---
> P-075: 顧客データフレーム(df_customer)からランダムに1%のデータを抽出し、先頭から10件データを抽出せよ。
```
df_customer.sample(frac=0.01).head(10)
```
---
> P-076: 顧客データフレーム(df_customer)から性別(gender_cd)の割合に基づきランダムに10%のデータを層化抽出し、性別ごとに件数を集計せよ。
```
# sklearn.model_selection.train_test_splitを使用した例
_, df_tmp = train_test_split(df_customer, test_size=0.1,
stratify=df_customer['gender_cd'])
df_tmp.groupby('gender_cd').agg({'customer_id' : 'count'})
df_tmp.head(10)
```
---
> P-077: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客単位に合計し、合計した売上金額の外れ値を抽出せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。なお、ここでは外れ値を平均から3σ以上離れたものとする。結果は10件表示させれば良い。
```
# skleanのpreprocessing.scaleを利用するため、標本標準偏差で計算されている
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
df_sales_amount['amount_ss'] = preprocessing.scale(df_sales_amount['amount'])
df_sales_amount.query('abs(amount_ss) >= 3').head(10)
```
---
> P-078: レシート明細データフレーム(df_receipt)の売上金額(amount)を顧客単位に合計し、合計した売上金額の外れ値を抽出せよ。ただし、顧客IDが"Z"から始まるのものは非会員を表すため、除外して計算すること。なお、ここでは外れ値を第一四分位と第三四分位の差であるIQRを用いて、「第一四分位数-1.5×IQR」よりも下回るもの、または「第三四分位数+1.5×IQR」を超えるものとする。結果は10件表示させれば良い。
```
df_sales_amount = df_receipt.query('not customer_id.str.startswith("Z")',
engine='python'). \
groupby('customer_id'). \
agg({'amount':'sum'}).reset_index()
pct75 = np.percentile(df_sales_amount['amount'], q=75)
pct25 = np.percentile(df_sales_amount['amount'], q=25)
iqr = pct75 - pct25
amount_low = pct25 - (iqr * 1.5)
amount_hight = pct75 + (iqr * 1.5)
df_sales_amount.query('amount < @amount_low or @amount_hight < amount').head(10)
```
---
> P-079: 商品データフレーム(df_product)の各項目に対し、欠損数を確認せよ。
```
df_product.isnull().sum()
```
---
> P-080: 商品データフレーム(df_product)のいずれかの項目に欠損が発生しているレコードを全て削除した新たなdf_product_1を作成せよ。なお、削除前後の件数を表示させ、前設問で確認した件数だけ減少していることも確認すること。
```
df_product_1 = df_product.copy()
print('削除前:', len(df_product_1))
df_product_1.dropna(inplace=True)
print('削除後:', len(df_product_1))
```
---
> P-081: 単価(unit_price)と原価(unit_cost)の欠損値について、それぞれの平均値で補完した新たなdf_product_2を作成せよ。なお、平均値については1円未満を丸めること(四捨五入または偶数への丸めで良い)。補完実施後、各項目について欠損が生じていないことも確認すること。
```
df_product_2 = df_product.fillna({
'unit_price':np.round(np.nanmean(df_product['unit_price'])),
'unit_cost':np.round(np.nanmean(df_product['unit_cost']))})
df_product_2.isnull().sum()
```
---
> P-082: 単価(unit_price)と原価(unit_cost)の欠損値について、それぞれの中央値で補完した新たなdf_product_3を作成せよ。なお、中央値については1円未満を丸めること(四捨五入または偶数への丸めで良い)。補完実施後、各項目について欠損が生じていないことも確認すること。
```
df_product_3 = df_product.fillna({
'unit_price':np.round(np.nanmedian(df_product['unit_price'])),
'unit_cost':np.round(np.nanmedian(df_product['unit_cost']))})
df_product_3.isnull().sum()
```
---
> P-083: 単価(unit_price)と原価(unit_cost)の欠損値について、各商品の小区分(category_small_cd)ごとに算出した中央値で補完した新たなdf_product_4を作成せよ。なお、中央値については1円未満を丸めること(四捨五入または偶数への丸めで良い)。補完実施後、各項目について欠損が生じていないことも確認すること。
```
# コード例1
df_tmp = df_product.groupby('category_small_cd'). \
agg({'unit_price':'median', 'unit_cost':'median'}).reset_index()
df_tmp.columns = ['category_small_cd', 'median_price', 'median_cost']
df_product_4 = pd.merge(df_product, df_tmp, how='inner', on='category_small_cd')
df_product_4['unit_price'] = df_product_4[['unit_price', 'median_price']]. \
apply(lambda x: np.round(x[1]) if np.isnan(x[0]) else x[0], axis=1)
df_product_4['unit_cost'] = df_product_4[['unit_cost', 'median_cost']]. \
apply(lambda x: np.round(x[1]) if np.isnan(x[0]) else x[0], axis=1)
df_product_4.isnull().sum()
# コード例2(maskの活用)
df_tmp = (df_product
.groupby('category_small_cd')
.agg(median_price=('unit_price', 'median'),
median_cost=('unit_cost', 'median'))
.reset_index())
df_product_4 = df_product.merge(df_tmp,
how='inner',
on='category_small_cd')
df_product_4['unit_price'] = (df_product_4['unit_price']
.mask(df_product_4['unit_price'].isnull(),
df_product_4['median_price'].round()))
df_product_4['unit_cost'] = (df_product_4['unit_cost']
.mask(df_product_4['unit_cost'].isnull(),
df_product_4['median_cost'].round()))
df_product_4.isnull().sum()
# コード例3(fillna、transformの活用)
df_product_4 = df_product.copy()
for x in ['unit_price', 'unit_cost']:
df_product_4[x] = (df_product_4[x]
.fillna(df_product_4.groupby('category_small_cd')[x]
.transform('median')
.round()))
df_product_4.isnull().sum()
```
---
> P-084: 顧客データフレーム(df_customer)の全顧客に対し、全期間の売上金額に占める2019年売上金額の割合を計算せよ。ただし、売上実績がない場合は0として扱うこと。そして計算した割合が0超のものを抽出せよ。 結果は10件表示させれば良い。また、作成したデータにNAやNANが存在しないことを確認せよ。
```
df_tmp_1 = df_receipt.query('20190101 <= sales_ymd <= 20191231')
df_tmp_1 = pd.merge(df_customer['customer_id'],
df_tmp_1[['customer_id', 'amount']],
how='left', on='customer_id'). \
groupby('customer_id').sum().reset_index(). \
rename(columns={'amount':'amount_2019'})
df_tmp_2 = pd.merge(df_customer['customer_id'],
df_receipt[['customer_id', 'amount']],
how='left', on='customer_id'). \
groupby('customer_id').sum().reset_index()
df_tmp = pd.merge(df_tmp_1, df_tmp_2, how='inner', on='customer_id')
df_tmp['amount_2019'] = df_tmp['amount_2019'].fillna(0)
df_tmp['amount'] = df_tmp['amount'].fillna(0)
df_tmp['amount_rate'] = df_tmp['amount_2019'] / df_tmp['amount']
df_tmp['amount_rate'] = df_tmp['amount_rate'].fillna(0)
df_tmp.query('amount_rate > 0').head(10)
df_tmp.isnull().sum()
```
---
> P-085: 顧客データフレーム(df_customer)の全顧客に対し、郵便番号(postal_cd)を用いて経度緯度変換用データフレーム(df_geocode)を紐付け、新たなdf_customer_1を作成せよ。ただし、複数紐づく場合は経度(longitude)、緯度(latitude)それぞれ平均を算出すること。
```
df_customer_1 = pd.merge(df_customer[['customer_id', 'postal_cd']],
df_geocode[['postal_cd', 'longitude' ,'latitude']],
how='inner', on='postal_cd')
df_customer_1 = df_customer_1.groupby('customer_id'). \
agg({'longitude':'mean', 'latitude':'mean'}).reset_index(). \
rename(columns={'longitude':'m_longitude', 'latitude':'m_latitude'})
df_customer_1 = pd.merge(df_customer, df_customer_1,
how='inner', on='customer_id')
df_customer_1.head(3)
```
---
> P-086: 前設問で作成した緯度経度つき顧客データフレーム(df_customer_1)に対し、申込み店舗コード(application_store_cd)をキーに店舗データフレーム(df_store)と結合せよ。そして申込み店舗の緯度(latitude)・経度情報(longitude)と顧客の緯度・経度を用いて距離(km)を求め、顧客ID(customer_id)、顧客住所(address)、店舗住所(address)とともに表示せよ。計算式は簡易式で良いものとするが、その他精度の高い方式を利用したライブラリを利用してもかまわない。結果は10件表示すれば良い。
$$
緯度(ラジアン):\phi \\
経度(ラジアン):\lambda \\
距離L = 6371 * arccos(sin \phi_1 * sin \phi_2
+ cos \phi_1 * cos \phi_2 * cos(\lambda_1 − \lambda_2))
$$
```
# コード例1
def calc_distance(x1, y1, x2, y2):
distance = 6371 * math.acos(math.sin(math.radians(y1))
* math.sin(math.radians(y2))
+ math.cos(math.radians(y1))
* math.cos(math.radians(y2))
* math.cos(math.radians(x1) - math.radians(x2)))
return distance
df_tmp = pd.merge(df_customer_1, df_store, how='inner', left_on='application_store_cd', right_on='store_cd')
df_tmp['distance'] = df_tmp[['m_longitude', 'm_latitude','longitude', 'latitude']]. \
apply(lambda x: calc_distance(x[0], x[1], x[2], x[3]), axis=1)
df_tmp[['customer_id', 'address_x', 'address_y', 'distance']].head(10)
# コード例2
def calc_distance_numpy(x1, y1, x2, y2):
x1_r = np.radians(x1)
x2_r = np.radians(x2)
y1_r = np.radians(y1)
y2_r = np.radians(y2)
return 6371 * np.arccos(np.sin(y1_r) * np.sin(y2_r)
+ np.cos(y1_r) * np.cos(y2_r)
* np.cos(x1_r - x2_r))
df_tmp = df_customer_1.merge(df_store,
how='inner',
left_on='application_store_cd',
right_on='store_cd')
df_tmp['distance'] = calc_distance_numpy(df_tmp['m_longitude'],
df_tmp['m_latitude'],
df_tmp['longitude'],
df_tmp['latitude'])
df_tmp[['customer_id', 'address_x', 'address_y', 'distance']].head(10)
```
---
> P-087: 顧客データフレーム(df_customer)では、異なる店舗での申込みなどにより同一顧客が複数登録されている。名前(customer_name)と郵便番号(postal_cd)が同じ顧客は同一顧客とみなし、1顧客1レコードとなるように名寄せした名寄顧客データフレーム(df_customer_u)を作成せよ。ただし、同一顧客に対しては売上金額合計が最も高いものを残すものとし、売上金額合計が同一もしくは売上実績がない顧客については顧客ID(customer_id)の番号が小さいものを残すこととする。
```
df_receipt_tmp = df_receipt.groupby('customer_id') \
.agg(sum_amount=('amount','sum')).reset_index()
df_customer_u = pd.merge(df_customer, df_receipt_tmp,
how='left',
on='customer_id')
df_customer_u['sum_amount'] = df_customer_u['sum_amount'].fillna(0)
df_customer_u = df_customer_u.sort_values(['sum_amount', 'customer_id'],
ascending=[False, True])
df_customer_u.drop_duplicates(subset=['customer_name', 'postal_cd'],
keep='first', inplace=True)
print('df_customer:', len(df_customer),
'df_customer_u:', len(df_customer_u),
'diff:', len(df_customer) - len(df_customer_u))
```
---
> P-088: 前設問で作成したデータを元に、顧客データフレームに統合名寄IDを付与したデータフレーム(df_customer_n)を作成せよ。ただし、統合名寄IDは以下の仕様で付与するものとする。
>
> - 重複していない顧客:顧客ID(customer_id)を設定
> - 重複している顧客:前設問で抽出したレコードの顧客IDを設定
```
df_customer_n = pd.merge(df_customer,
df_customer_u[['customer_name',
'postal_cd', 'customer_id']],
how='inner', on =['customer_name', 'postal_cd'])
df_customer_n.rename(columns={'customer_id_x':'customer_id',
'customer_id_y':'integration_id'}, inplace=True)
print('ID数の差', len(df_customer_n['customer_id'].unique())
- len(df_customer_n['integration_id'].unique()))
```
---
> P-閑話: df_customer_1, df_customer_nは使わないので削除する。
```
del df_customer_1
del df_customer_n
```
---
> P-089: 売上実績がある顧客に対し、予測モデル構築のため学習用データとテスト用データに分割したい。それぞれ8:2の割合でランダムにデータを分割せよ。
```
df_sales= df_receipt.groupby('customer_id').agg({'amount':sum}).reset_index()
df_tmp = pd.merge(df_customer, df_sales['customer_id'],
how='inner', on='customer_id')
df_train, df_test = train_test_split(df_tmp, test_size=0.2, random_state=71)
print('学習データ割合: ', len(df_train) / len(df_tmp))
print('テストデータ割合: ', len(df_test) / len(df_tmp))
```
---
> P-090: レシート明細データフレーム(df_receipt)は2017年1月1日〜2019年10月31日までのデータを有している。売上金額(amount)を月次で集計し、学習用に12ヶ月、テスト用に6ヶ月のモデル構築用データを3セット作成せよ。
```
df_tmp = df_receipt[['sales_ymd', 'amount']].copy()
df_tmp['sales_ym'] = df_tmp['sales_ymd'].astype('str').str[0:6]
df_tmp = df_tmp.groupby('sales_ym').agg({'amount':'sum'}).reset_index()
# 関数化することで長期間データに対する多数のデータセットもループなどで処理できるようにする
def split_data(df, train_size, test_size, slide_window, start_point):
train_start = start_point * slide_window
test_start = train_start + train_size
return df[train_start : test_start], df[test_start : test_start + test_size]
df_train_1, df_test_1 = split_data(df_tmp, train_size=12,
test_size=6, slide_window=6, start_point=0)
df_train_2, df_test_2 = split_data(df_tmp, train_size=12,
test_size=6, slide_window=6, start_point=1)
df_train_3, df_test_3 = split_data(df_tmp, train_size=12,
test_size=6, slide_window=6, start_point=2)
df_train_1
df_test_1
```
---
> P-091: 顧客データフレーム(df_customer)の各顧客に対し、売上実績がある顧客数と売上実績がない顧客数が1:1となるようにアンダーサンプリングで抽出せよ。
```
# コード例1
#unbalancedのubUnderを使った例
df_tmp = df_receipt.groupby('customer_id').agg({'amount':'sum'}).reset_index()
df_tmp = pd.merge(df_customer, df_tmp, how='left', on='customer_id')
df_tmp['buy_flg'] = df_tmp['amount'].apply(lambda x: 0 if np.isnan(x) else 1)
print('0の件数', len(df_tmp.query('buy_flg == 0')))
print('1の件数', len(df_tmp.query('buy_flg == 1')))
positive_count = len(df_tmp.query('buy_flg == 1'))
rs = RandomUnderSampler(random_state=71)
df_sample, _ = rs.fit_resample(df_tmp, df_tmp.buy_flg)
print('0の件数', len(df_sample.query('buy_flg == 0')))
print('1の件数', len(df_sample.query('buy_flg == 1')))
# コード例2
#unbalancedのubUnderを使った例
df_tmp = df_customer.merge(df_receipt
.groupby('customer_id')['amount'].sum()
.reset_index(),
how='left',
on='customer_id')
df_tmp['buy_flg'] = np.where(df_tmp['amount'].isnull(), 0, 1)
print("サンプリング前のbuy_flgの件数")
print(df_tmp['buy_flg'].value_counts(), "\n")
positive_count = (df_tmp['buy_flg'] == 1).sum()
rs = RandomUnderSampler(random_state=71)
df_sample, _ = rs.fit_resample(df_tmp, df_tmp.buy_flg)
print("サンプリング後のbuy_flgの件数")
print(df_sample['buy_flg'].value_counts())
```
---
> P-092: 顧客データフレーム(df_customer)では、性別に関する情報が非正規化の状態で保持されている。これを第三正規化せよ。
```
df_gender = df_customer[['gender_cd', 'gender']].drop_duplicates()
df_customer_s = df_customer.drop(columns='gender')
```
---
> P-093: 商品データフレーム(df_product)では各カテゴリのコード値だけを保有し、カテゴリ名は保有していない。カテゴリデータフレーム(df_category)と組み合わせて非正規化し、カテゴリ名を保有した新たな商品データフレームを作成せよ。
```
df_product_full = pd.merge(df_product, df_category[['category_small_cd',
'category_major_name',
'category_medium_name',
'category_small_name']],
how = 'inner', on = 'category_small_cd')
```
---
> P-094: 先に作成したカテゴリ名付き商品データを以下の仕様でファイル出力せよ。なお、出力先のパスはdata配下とする。
>
> - ファイル形式はCSV(カンマ区切り)
> - ヘッダ有り
> - 文字コードはUTF-8
```
# コード例1
df_product_full.to_csv('../data/P_df_product_full_UTF-8_header.csv',
encoding='UTF-8', index=False)
# コード例2(BOM付きでExcelの文字化けを防ぐ)
df_product_full.to_csv('../data/P_df_product_full_UTF-8_header.csv',
encoding='utf_8_sig', index=False)
```
---
> P-095: 先に作成したカテゴリ名付き商品データを以下の仕様でファイル出力せよ。なお、出力先のパスはdata配下とする。
>
> - ファイル形式はCSV(カンマ区切り)
> - ヘッダ有り
> - 文字コードはCP932
```
df_product_full.to_csv('../data/P_df_product_full_CP932_header.csv',
encoding='CP932', index=False)
```
---
> P-096: 先に作成したカテゴリ名付き商品データを以下の仕様でファイル出力せよ。なお、出力先のパスはdata配下とする。
>
> - ファイル形式はCSV(カンマ区切り)
> - ヘッダ無し
> - 文字コードはUTF-8
```
df_product_full.to_csv('../data/P_df_product_full_UTF-8_noh.csv',
header=False ,encoding='UTF-8', index=False)
```
---
> P-097: 先に作成した以下形式のファイルを読み込み、データフレームを作成せよ。また、先頭3件を表示させ、正しくとりまれていることを確認せよ。
>
> - ファイル形式はCSV(カンマ区切り)
> - ヘッダ有り
> - 文字コードはUTF-8
```
df_tmp = pd.read_csv('../data/P_df_product_full_UTF-8_header.csv',
dtype={'category_major_cd':str,
'category_medium_cd':str,
'category_small_cd':str},
encoding='UTF-8')
df_tmp.head(3)
```
---
> P-098: 先に作成した以下形式のファイルを読み込み、データフレームを作成せよ。また、先頭3件を表示させ、正しくとりまれていることを確認せよ。
>
> - ファイル形式はCSV(カンマ区切り)
> - ヘッダ無し
> - 文字コードはUTF-8
```
df_tmp = pd.read_csv('../data/P_df_product_full_UTF-8_noh.csv',
dtype={1:str,
2:str,
3:str},
encoding='UTF-8', header=None)
df_tmp.head(3)
```
---
> P-099: 先に作成したカテゴリ名付き商品データを以下の仕様でファイル出力せよ。なお、出力先のパスはdata配下とする。
>
> - ファイル形式はTSV(タブ区切り)
> - ヘッダ有り
> - 文字コードはUTF-8
```
df_product_full.to_csv('../data/P_df_product_full_UTF-8_header.tsv',
sep='\t', encoding='UTF-8', index=False)
```
---
> P-100: 先に作成した以下形式のファイルを読み込み、データフレームを作成せよ。また、先頭3件を表示させ、正しくとりまれていることを確認せよ。
>
> - ファイル形式はTSV(タブ区切り)
> - ヘッダ有り
> - 文字コードはUTF-8
```
df_tmp = pd.read_table('../data/P_df_product_full_UTF-8_header.tsv',
dtype={'category_major_cd':str,
'category_medium_cd':str,
'category_small_cd':str},
encoding='UTF-8')
df_tmp.head(3)
```
# これで100本終わりです。おつかれさまでした!
|
github_jupyter
|
# Setup the ABSA Demo
### Step 1 - Install aditional pip packages on your Compute instance
```
!pip install git+https://github.com/hnky/nlp-architect.git@absa
!pip install spacy==2.1.8
```
### Step 2 - Download Notebooks, Training Data, Training / Inference scripts
```
import azureml
from azureml.core import Workspace, Datastore, Experiment, Environment, Model
import urllib.request
from pathlib import Path
# This will open an device login prompt. Login with your credentials that have access to the workspace.
# Connect to the workspace
ws = Workspace.from_config()
print("Using workspace:",ws.name,"in region", ws.location)
# Connect to the default datastore
ds = ws.get_default_datastore()
print("Datastore:",ds.name)
# Create directories
Path("dataset").mkdir(parents=True, exist_ok=True)
Path("notebooks").mkdir(parents=True, exist_ok=True)
Path("scripts").mkdir(parents=True, exist_ok=True)
Path("temp").mkdir(parents=True, exist_ok=True)
```
The cell below will take some time to run as it is downloading a large dataset plus code files. Please allow around 10-15 mins
```
# Download all files needed
base_link = "https://raw.githubusercontent.com/microsoft/ignite-learning-paths-training-aiml/main/aiml40/absa/"
# Download Data
if not Path("dataset/glove.840B.300d.zip").is_file():
urllib.request.urlretrieve('http://nlp.stanford.edu/data/glove.840B.300d.zip', 'dataset/glove.840B.300d.zip')
urllib.request.urlretrieve(base_link+'../dataset/clothing_absa_train.csv', 'dataset/clothing_absa_train.csv')
urllib.request.urlretrieve(base_link+'../dataset/clothing-absa-validation.json', 'dataset/clothing-absa-validation.json')
urllib.request.urlretrieve(base_link+'../dataset/clothing_absa_train_small.csv', 'dataset/clothing_absa_train_small.csv')
# Download Notebooks
urllib.request.urlretrieve(base_link+'notebooks/absa-hyperdrive.ipynb', 'notebooks/absa-hyperdrive.ipynb')
urllib.request.urlretrieve(base_link+'notebooks/absa.ipynb', 'notebooks/absa.ipynb')
# Download Scripts
urllib.request.urlretrieve(base_link+'scripts/score.py', 'scripts/score.py')
urllib.request.urlretrieve(base_link+'scripts/train.py', 'scripts/train.py')
# Upload data to the data store
ds.upload('dataset', target_path='clothing_data', overwrite=False, show_progress=True)
### Step 3 - Setup AMLS
from azureml.core.compute import ComputeTarget, AmlCompute
from azureml.core.compute_target import ComputeTargetException
cluster_name = "absa-cluster"
try:
cluster = ComputeTarget(workspace=ws, name=cluster_name)
print('Using compute cluster:', cluster_name)
except ComputeTargetException:
compute_config = AmlCompute.provisioning_configuration(vm_size='STANDARD_D3_V2',
vm_priority='lowpriority',
min_nodes=0,
max_nodes=8)
cluster = ComputeTarget.create(ws, cluster_name, compute_config)
cluster.wait_for_completion(show_output=True)
```
|
github_jupyter
|
```
# default_exp pds.utils
# default_cls_lvl 3
```
# PDS Utils
> Utilities used by the `pds` sub-package.
```
# hide
from nbverbose.showdoc import show_doc # noqa
# export
from typing import Union
from fastcore.utils import Path
import pandas as pd
import pvl
from planetarypy import utils
# export
class IndexLabel:
"Support working with label files of PDS Index tables."
def __init__(
self,
# Path to the labelfile for a PDS Indexfile.
# The actual table should reside in the same folder to be automatically parsed
# when calling the `read_index_data` method.
labelpath: Union[str, Path],
):
self.path = Path(labelpath)
"search for table name pointer and store key and fpath."
tuple = [i for i in self.pvl_lbl if i[0].startswith("^")][0]
self.tablename = tuple[0][1:]
self.index_name = tuple[1]
@property
def index_path(self):
p = self.path.parent / self.index_name
if not p.exists():
import warnings
warnings.warn(
"Fudging path name to lower case, opposing label value. (PDS data inconsistency)"
)
p = self.path.parent / self.index_name.lower()
if not p.exists():
warnings.warn("`index_path` still doesn't exist.")
return p
@property
def pvl_lbl(self):
return pvl.load(str(self.path))
@property
def table(self):
return self.pvl_lbl[self.tablename]
@property
def pvl_columns(self):
return self.table.getlist("COLUMN")
@property
def columns_dic(self):
return {col["NAME"]: col for col in self.pvl_columns}
@property
def colnames(self):
"""Read the columns in an PDS index label file.
The label file for the PDS indices describes the content
of the index files.
"""
colnames = []
for col in self.pvl_columns:
colnames.extend(PVLColumn(col).name_as_list)
return colnames
@property
def colspecs(self):
colspecs = []
columns = self.table.getlist("COLUMN")
for column in columns:
pvlcol = PVLColumn(column)
if pvlcol.items is None:
colspecs.append(pvlcol.colspecs)
else:
colspecs.extend(pvlcol.colspecs)
return colspecs
def read_index_data(self, convert_times=True):
return index_to_df(self.index_path, self, convert_times=convert_times)
# export
def index_to_df(
# Path to the index TAB file
indexpath: Union[str, Path],
# Label object that has both the column names and the columns widths as attributes
# 'colnames' and 'colspecs'
label: IndexLabel,
# Switch to control if to convert columns with "TIME" in name (unless COUNT is as well in name) to datetime
convert_times=True,
):
"""The main reader function for PDS Indexfiles.
In conjunction with an IndexLabel object that figures out the column widths,
this reader should work for all PDS TAB files.
"""
indexpath = Path(indexpath)
df = pd.read_fwf(
indexpath, header=None, names=label.colnames, colspecs=label.colspecs
)
if convert_times:
for column in [col for col in df.columns if "TIME" in col]:
if column in ["LOCAL_TIME", "DWELL_TIME"]:
continue
try:
df[column] = pd.to_datetime(df[column])
except ValueError:
df[column] = pd.to_datetime(
df[column], format=utils.nasa_dt_format_with_ms, errors="coerce"
)
except KeyError:
raise KeyError(f"{column} not in {df.columns}")
print("Done.")
return df
# export
class PVLColumn:
"Manages just one of the columns in a table that is described via PVL."
def __init__(self, pvlobj):
self.pvlobj = pvlobj
@property
def name(self):
return self.pvlobj["NAME"]
@property
def name_as_list(self):
"needs to return a list for consistency for cases when it's an array."
if self.items is None:
return [self.name]
else:
return [self.name + "_" + str(i + 1) for i in range(self.items)]
@property
def start(self):
"Decrease by one as Python is 0-indexed."
return self.pvlobj["START_BYTE"] - 1
@property
def stop(self):
return self.start + self.pvlobj["BYTES"]
@property
def items(self):
return self.pvlobj.get("ITEMS")
@property
def item_bytes(self):
return self.pvlobj.get("ITEM_BYTES")
@property
def item_offset(self):
return self.pvlobj.get("ITEM_OFFSET")
@property
def colspecs(self):
if self.items is None:
return (self.start, self.stop)
else:
i = 0
bucket = []
for _ in range(self.items):
off = self.start + self.item_offset * i
bucket.append((off, off + self.item_bytes))
i += 1
return bucket
def decode(self, linedata):
if self.items is None:
start, stop = self.colspecs
return linedata[start:stop]
else:
bucket = []
for (start, stop) in self.colspecs:
bucket.append(linedata[start:stop])
return bucket
def __repr__(self):
return self.pvlobj.__repr__()
# export
def decode_line(
linedata: str, # One line of a .tab data file
labelpath: Union[
str, Path
], # Path to the appropriate label that describes the data.
):
"Decode one line of tabbed data with the appropriate label file."
label = IndexLabel(labelpath)
for column in label.pvl_columns:
pvlcol = PVLColumn(column)
print(pvlcol.name, pvlcol.decode(linedata))
# export
def find_mixed_type_cols(
# Dataframe to be searched for mixed data-types
df: pd.DataFrame,
# Switch to control if NaN values in these problem columns should be replaced by the string 'UNKNOWN'
fix: bool = True,
) -> list: # List of column names that have data type changes within themselves.
"""For a given dataframe, find the columns that are of mixed type.
Tool to help with the performance warning when trying to save a pandas DataFrame as a HDF.
When a column changes datatype somewhere, pickling occurs, slowing down the reading process of the HDF file.
"""
result = []
for col in df.columns:
weird = (df[[col]].applymap(type) != df[[col]].iloc[0].apply(type)).any(axis=1)
if len(df[weird]) > 0:
print(col)
result.append(col)
if fix is True:
for col in result:
df[col].fillna("UNKNOWN", inplace=True)
return result
# export
def fix_hirise_edrcumindex(
infname: Union[str, Path], # Path to broken EDRCUMINDEX.TAB
outfname: Union[str, Path], # Path where to store the fixed TAB file
):
"""Fix HiRISE EDRCUMINDEX.
The HiRISE EDRCUMINDEX has some broken lines where the SCAN_EXPOSURE_DURATION is of format
F10.4 instead of the defined F9.4.
This function simply replaces those incidences with one less decimal fraction, so 20000.0000
becomes 20000.000.
"""
with open(str(infname)) as f:
with open(str(outfname, "w")) as newf:
for line in tqdm(f):
exp = line.split(",")[21]
if float(exp) > 9999.999:
# catching the return of write into dummy variable
_ = newf.write(line.replace(exp, exp[:9]))
else:
_ = newf.write(line)
```
|
github_jupyter
|
# **Built in Functions**
# **bool()**
Valores vazios ou zeros são considerado False, do contrário são considerados True (Truth Value Testing).
"Truth Value Testing". Isto é, decidir quando um valor é considerado True ou False
```
print(bool(0))
print(bool(""))
print(bool(None))
print(bool(1))
print(bool(-100))
print(bool(13.5))
print(bool("teste"))
print(bool(True))
```
# **f'' / .format()**
```
# antes da versão 3.6
a = ('Hello World!')
print('----> {} <----'.format(a))
# ou f''
print(f'----> {a} <----')
nome = 'José'
idade = 23
salario = 987.30
print(f'O {nome} tem {idade} anos e ganha R${salario:.2f}.') #Python 3.6+
print(f'O {nome:-^20} tem {idade} anos e ganha R${salario:.2f}.') #Python 3.6+
print(f'O {nome:-<20} tem {idade} anos e ganha R${salario:.2f}.') #Python 3.6+
print(f'O {nome:->20} tem {idade} anos e ganha R${salario:.2f}.') #Python 3.6+
print('O {} tem {} anos e ganha R${:.2f}.'.format(nome, idade, salario)) #Python 3
print('O %s tem %d anos.' % (nome, idade)) #Python 2
# formatação de traços + casas decimais
vlr = 120
vlr2 = 10.1
print(f'{vlr:->20.2f}')
print(f'{vlr2:-^20.2f}')
print(f'R${vlr:5.2f}')
print(f'R${vlr:6.2f}')
print(f'R${vlr:7.2f}')
print(f'R${vlr:8.2f}')
print(f'R${vlr:08.2f}')
print(f'R${vlr:010.4f}') # f para float
print(f'R${vlr:010d}')# d para inteiros
print(f'R${vlr:04d}')
print(f'R${vlr:4d}')
n1, n2, n3, n4 = 100, 1, 00.758, 15.77
print(f'n1 = {n1:6}\nn1 = {n1:06}') # total de casas = 6 com opção de colocar ou não zero
print(f'n2 = {n2:06}')
print(f'n2 = {n2: 6}')
print(f'n3 = {n3:06.3f}') # variavel + ':' + total de casas decimais + '.' + casas decimais a direita da ','
print(f'n4 = {n4:06.3f} ou {n4:.2f}')
# formatação com tab \t
for c in range(0,5):
print(f'O {c}º valor recebido é \t R$1000,00')
print('Agora sem o tab')
print(f'O {c}º valor recebido é R$1000,00')
print('-' * 35)
```
# **.find() .rfind**
```
frase = ' Curso em Vídeo Python '
print(frase.find('Curso'))
print('A letra "o" aparece a ultima vez na posição {}.'.format(frase.lower().rfind('o')+1))
```
# **print()**
value é o valor que queremos imprimir, as reticências indicam que a função pode receber mais de um valor, basta separá-los por vírgula.
sep é o separador entre os valores, por padrão o separador é um espaço em branco.
end é o que acontecerá ao final da função, por padrão há uma quebra de linha, uma nova linha (\n).
## fomatando o print
```
nome = 'Livio Alvarenga'
print(f'Prazer em te conhecer\n{nome}!') #\n executa um enter
print(f'Prazer em te conhecer {nome:20}!')
print(f'Prazer em te conhecer {nome:>20}!')
print(f'Prazer em te conhecer {nome:<20}!')
print(f'Prazer em te conhecer {nome:^20}!')
print(f'Prazer em te conhecer {nome:=^21}!')
print(f'{"FIM DO PROGRAMA":-^30}')
print(f'{"FIM DO PROGRAMA":^30}')
frase = ' Curso em Vídeo Python '
print(frase[3])
print(frase[:3])
print(frase[3:])
print(frase[0:10:2])
print("""imprimindo um texto longo!!! imprimindo um texto longo!!!
imprimindo um texto longo!!! imprimindo um texto longo!!!
imprimindo um texto longo!!! imprimindo um texto longo!!!""")
```
## print sep e end
```
# print com end
t1 = 't1'
t2 = 't2'
t3 = 't3'
print('{} --> {}'.format(t1, t2), end='')
print(f' --> {t3}', end='')
print(' --> FIM')
print("Brasil", "ganhou", 5, "titulos mundiais", sep="-")
```
## Imprimindo com pprint( )
```
from pprint import pprint
# ! Imprimindo com pprint + width
cliente = {'nome': 'Livio', 'Idade': 40, 'Cidade': 'Belo Horizonte'}
pprint(cliente, width=40)
```
# **round()**
```
# Retorna o valor com arredondamento
round(3.14151922,2)
```
# os.path.isdir
Este método vai nos retornar um booleano, True ou False, que vai dizer se o diretório existe ou não
```
from os.path import isdir
diretorio = "c:\\"
if isdir(diretorio):
print(f"O diretório {diretorio} existe!")
else:
print("O diretório não existe!")
diretorio = "xx:\\"
if isdir(diretorio):
print(f"O diretório {diretorio} existe!")
else:
print("O diretório não existe!")
```
|
github_jupyter
|
<h1>Table of Contents<span class="tocSkip"></span></h1>
<div class="toc" style="margin-top: 1em;"><ul class="toc-item"><li><ul class="toc-item"><li><span><a href="#(a)" data-toc-modified-id="(a)-0.1"><span class="toc-item-num">0.1 </span>(a)</a></span></li><li><span><a href="#(b)" data-toc-modified-id="(b)-0.2"><span class="toc-item-num">0.2 </span>(b)</a></span></li><li><span><a href="#(c)" data-toc-modified-id="(c)-0.3"><span class="toc-item-num">0.3 </span>(c)</a></span></li><li><span><a href="#(d)" data-toc-modified-id="(d)-0.4"><span class="toc-item-num">0.4 </span>(d)</a></span></li></ul></li></ul></div>
Use Newton’s method to find solutions accurate to within $10^{−4}$ for the following problems.
```
import numpy as np
from numpy import linalg
from abc import abstractmethod
import pandas as pd
import math
pd.options.display.float_format = '{:,.8f}'.format
np.set_printoptions(suppress=True, precision=8)
TOR = pow(10.0, -4)
MAX_ITR = 150
class NewtonMethod(object):
def __init__(self):
return
@abstractmethod
def f(self, x):
return NotImplementedError('Implement f()!')
@abstractmethod
def jacobian(self, x):
return NotImplementedError('Implement jacobian()!')
@abstractmethod
def run(self, x):
return NotImplementedError('Implement run()!')
```
## (a)
$$x^3 − 2x^2 − 5 = 0, [1, 4]$$
```
class Newton1D(NewtonMethod):
def __init__(self):
super(NewtonMethod, self).__init__()
def f(self, x):
return pow(x, 3) - 2 * pow(x, 2) - 5
def jacobian(self, x):
return 3 * pow(x, 2) - 4 * x
def run(self, x0):
df = pd.DataFrame(columns=['f(x)'])
row = len(df)
x = x0
df.loc[row] = [x]
for k in range(MAX_ITR):
try:
y = x - self.f(x) / self.jacobian(x)
except ValueError:
break
residual = math.fabs(x - y)
x = y
row = len(df)
df.loc[row] = [y]
if residual < TOR or x > 1e9:
break
return df
Newton1D().run(2.5).astype(np.float64)
```
## (b)
$$x^3 + 3x^2 − 1 = 0, [-3, -2]$$
```
class Newton1D(NewtonMethod):
def __init__(self):
super(NewtonMethod, self).__init__()
def f(self, x):
return pow(x, 3) + 3 * pow(x, 2) - 1
def jacobian(self, x):
return 3 * pow(x, 2) - 6 * x
def run(self, x0):
df = pd.DataFrame(columns=['f(x)'])
row = len(df)
x = x0
df.loc[row] = [x]
for k in range(MAX_ITR):
try:
y = x - self.f(x) / self.jacobian(x)
except ValueError:
break
residual = math.fabs(x - y)
x = y
row = len(df)
df.loc[row] = [y]
if residual < TOR or x > 1e9:
break
return df
Newton1D().run(-2.5).astype(np.float64)
```
## (c)
$$x−\cos x=0, [0, \frac{\pi}{2}]$$
```
class Newton1D(NewtonMethod):
def __init__(self):
super(NewtonMethod, self).__init__()
def f(self, x):
return x - math.cos(x)
def jacobian(self, x):
return 1 + math.sin(x)
def run(self, x0):
df = pd.DataFrame(columns=['f(x)'])
row = len(df)
x = x0
df.loc[row] = [x]
for k in range(MAX_ITR):
try:
y = x - self.f(x) / self.jacobian(x)
except ValueError:
break
residual = math.fabs(x - y)
x = y
row = len(df)
df.loc[row] = [y]
if residual < TOR or x > 1e9:
break
return df
Newton1D().run(math.pi / 4.0).astype(np.float64)
```
## (d)
$$x − 0.8 − 0.2 \sin x = 0, [0, \frac{\pi}{2}]$$
```
class Newton1D(NewtonMethod):
def __init__(self):
super(NewtonMethod, self).__init__()
def f(self, x):
return x - 0.8 - 0.2 * math.sin(x)
def jacobian(self, x):
return 1 - 0.2 * math.cos(x)
def run(self, x0):
df = pd.DataFrame(columns=['f(x)'])
row = len(df)
x = x0
df.loc[row] = [x]
for k in range(MAX_ITR):
try:
y = x - self.f(x) / self.jacobian(x)
except ValueError:
break
residual = math.fabs(x - y)
x = y
row = len(df)
df.loc[row] = [y]
if residual < TOR or x > 1e9:
break
return df
Newton1D().run(math.pi / 4.0).astype(np.float64)
```
|
github_jupyter
|
```
import torch
from torch.autograd import Variable
from torch import nn
import matplotlib.pyplot as plt
%matplotlib inline
torch.manual_seed(3)
```
# make data
```
x_train = torch.Tensor([[1],[2],[3]])
y_train = torch.Tensor([[1],[2],[3]])
x, y = Variable(x_train), Variable(y_train)
plt.scatter(x.data.numpy(), y.data.numpy())
plt.show()
```
# Naive Model
## Define Linear model
```
x, y
W = Variable(torch.rand(1,1))
W
x.mm(W)
```
## Define cost function
loss(x,y)=1/n∑|xi−yi|2loss(x,y)=1/n∑|xi−yi|2
```
cost_func = nn.MSELoss()
cost_func
```
## Training Linear Regression
```
plt.ion()
lr = 0.01
for step in range(300):
prediction = x.mm(W)
cost = cost_func(prediction, y)
gradient = (prediction-y).view(-1).dot(x.view(-1)) / len(x)
W -= lr * gradient
if step % 10 == 0:
plt.cla()
plt.scatter(x.data.numpy(), y.data.numpy())
plt.plot(x.data.numpy(), prediction.data.numpy(), 'r-')
plt.title('step %d, cost=%.4f, w=%.4f,grad=%.4f' % (step,cost.data, W.data[0], gradient.data))
plt.show()
# if step %10 == 0:
# print(step, "going cost")
# print(cost)
# print((prediction-y).view(-1))
# print((x.view(-1)))
# print(gradient)
# print(W)
plt.ioff()
x_test = Variable(torch.Tensor([[5]]))
y_test = x_test.mm(W)
y_test
```
# w/ nn Module
## Define Linear Model
```
model = nn.Linear(1, 1, bias=True)
print(model)
model.weight, model.bias
cost_func = nn.MSELoss()
for i in model.parameters():
print(i)
optimizer = torch.optim.SGD(model.parameters(), lr=0.01)
```
## Training w/ nn module
```
model(x)
plt.ion()
for step in range(300):
prediction = model(x)
cost = cost_func(prediction, y)
optimizer.zero_grad()
cost.backward()
optimizer.step()
if step % 10 == 0:
plt.cla()
plt.scatter(x.data.numpy(), y.data.numpy())
plt.plot(x.data.numpy(), prediction.data.numpy(), 'b--')
plt.title('cost=%.4f, w=%.4f, b=%.4f' % (cost.data,model.weight.data[0][0],model.bias.data))
plt.show()
plt.ioff()
x_test = Variable(torch.Tensor([[7]]))
y_test = model(x_test)
print('input : %.4f, output:%.4f' % (x_test.data[0][0], y_test.data[0][0]))
for step in range(300):
prediction = model(x)
cost = cost_func(prediction, y)
optimizer.zero_grad()
cost.backward()
optimizer.step()
x_test = Variable(torch.Tensor([[7]]))
y_test = model(x_test)
print('input : %.4f, output:%.4f' % (x_test.data[0][0], y_test.data[0][0]))
model.weight, model.bias
```
### Has "nn.MSELoss()" Convex Cost Space?
```
W_val, cost_val = [], []
for i in range(-30, 51):
W = i * 0.1
model.weight.data.fill_(W)
cost = cost_func(model(x),y)
W_val.append(W)
cost_val.append(cost.data)
plt.plot(W_val, cost_val, 'ro')
plt.show()
```
# Multivariate Linear model
```
import numpy as np
```
## make Data
```
xy = np.loadtxt('data-01-test-score.csv', delimiter=',', dtype=np.float32)
x_data = xy[:, 0:-1]
y_data = xy[:, [-1]]
print('shape: ', x_data.shape, '\nlength:', len(x_data), '\n', x_data )
print('shape: ', y_data.shape, '\nlength:', len(y_data), '\n', y_data )
x, y = Variable(torch.from_numpy(x_data)), Variable(torch.from_numpy(y_data))
x, y
```
## make Model
```
mv_model = nn.Linear(3, 1, bias=True)
print(mv_model)
print('weigh : ', mv_model.weight)
print('bias : ', mv_model.bias)
cost_func = nn.MSELoss()
optimizer = torch.optim.SGD(mv_model.parameters(), lr=1e-5)
```
## Training Model
```
for step in range(2000):
optimizer.zero_grad()
prediction = mv_model(x)
cost = cost_func(prediction, y)
cost.backward()
optimizer.step()
if step % 50 == 0:
print(step, "Cost: ", cost.data.numpy(), "\nPrediction:\n", prediction.data.t().numpy())
mv_model.state_dict()
```
## test
```
print("Model score : ",mv_model(Variable(torch.Tensor([[73,80,75]]))).data.numpy())
print("Real score : 73,80,75,152")
accuracy_list = []
for i,real_y in enumerate(y):
accuracy = (mv_model((x[i])).data.numpy() - real_y.data.numpy())
accuracy_list.append(np.absolute(accuracy))
for accuracy in accuracy_list:
print(accuracy)
print("sum accuracy : ",sum(accuracy_list))
print("avg accuracy : ",sum(accuracy_list)/len(y))
```
|
github_jupyter
|
# Import Dependencies
```
from config import api_key
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import requests
import datetime
import json
```
# Use API to get .json
```
endpoint = 'breweries'
page = 1
url = f"https://sandbox-api.brewerydb.com/v2/{endpoint}/?key={api_key}&p={page}&withLocations=Y&withSocialAccounts=Y"
brewery_data = requests.get(url).json()
#print(json.dumps(brewery_data, indent=4, sort_keys=True))
```
# Create DataFrame
- Initially, we pull just a few interesting columns for the dataframe, most importantly, the established dates and lat/lon coordinates for each brewery
- We will add distance columns later after doing some math
- Change the Established Date column to numeric in order to use in the scatter plot
```
brewery_dict = []
for result in range(0,19):
try:
brewery_info = {
'Brewery Name': brewery_data['data'][result]['name'],
'Brewery ID': brewery_data['data'][result]['id'],
'Established Date': brewery_data['data'][result]['established'],
'Is in business?': brewery_data['data'][result]['isInBusiness'],
'Website': brewery_data['data'][result]['website'],
'Country': brewery_data['data'][result]['locations'][0]['country']['isoCode'],
'City':brewery_data['data'][result]['locations'][0]['locality'],
'Latitude':brewery_data['data'][result]['locations'][0]['latitude'],
'Longitude':brewery_data['data'][result]['locations'][0]['longitude'],
'Primary Location':brewery_data['data'][result]['locations'][0]['isPrimary'],
'Distance from Chicago (km)':'',
'Distance from Pottsville (km)':''
}
except:
print('id not found')
brewery_dict.append(brewery_info)
brewery_df = pd.DataFrame(brewery_dict)
brewery_df['Established Date']=pd.to_numeric(brewery_df['Established Date'])
#brewery_df
```
# Determine Distances from Chicago
- use geopy to determine distances via lat/long data
- Chicago is one of the hot-spots for early American breweries, made possible by the German immigrant community
- Pottsville (Becky's hometown) is home to the oldest brewery in America - Yeungling!
- update the dataframe, clean it and export as a csv
```
#!pip install geopy
import geopy.distance
Chi_coords = (41.8781, -87.6298)
Pottsville_coords = (40.6856, -76.1955)
for x in range(0,19):
Brewery_coords = (brewery_df['Latitude'][x], brewery_df['Longitude'][x])
brewery_df['Distance from Chicago (km)'][x] = geopy.distance.distance(Chi_coords, Brewery_coords).km
brewery_df['Distance from Pottsville (km)'][x] = geopy.distance.distance(Pottsville_coords, Brewery_coords).km
brewery_df = brewery_df.drop_duplicates(subset=['Brewery ID'], keep='first')
brewery_df
brewery_df.to_csv("data/brewery_data.csv", encoding="utf-8", index=False)
```
# Figures
- I expect a greater number of older breweries closer to Chicago, given that some of the first instances of brewing in America occured here.
- With such few breweries available for free (boo sandbox), the scatter plot looks a little sparse. However, the general trend gives us preliminary data that shows that there may be a coorlation! If I wanted to do more with this, this would be good enough to convince me to splurge the $20 for full access
- plot for Pottsville is just for fun
```
#Chicago
plt.scatter(brewery_df['Distance from Chicago (km)'], brewery_df['Established Date'],
alpha=0.5, edgecolor ='black', color="blue",s=100)
#Chart elements
plt.title(f"Distance from Chicago vs. Established Year")
plt.xlabel('Distance from Chicago (km)')
plt.ylabel('Established Year')
plt.grid(True)
#Save and print
plt.savefig("images/Distance from Chicago vs. Established Year.png")
plt.show()
#Pottsville
plt.scatter(brewery_df['Distance from Pottsville (km)'], brewery_df['Established Date'], alpha=0.5, edgecolor ='black', color="red",s=100)
#Chart elements
plt.title(f"Distance from Pottsville vs. Established Year")
plt.xlabel('Distance from Pottsville (km)')
plt.ylabel('Established Year')
plt.grid(True)
#Save and print
#plt.savefig("images/Distance from Pottsville vs. Established Year.png")
plt.show()
#Empty Plot
plt.scatter(brewery_df['Distance from Chicago (km)'], brewery_df['Established Date'], alpha=0.5, edgecolor ='none', color="none",s=100)
#Chart elements
plt.title(f"Distance from Chicago vs. Established Year")
plt.xlabel('Distance from Chicago (km)')
plt.ylabel('Established Year')
plt.grid(True)
#Save and print
plt.savefig("images/Empty plot.png")
plt.show()
```
|
github_jupyter
|
# Image features exercise
*Complete and hand in this completed worksheet (including its outputs and any supporting code outside of the worksheet) with your assignment submission. For more details see the [assignments page](http://vision.stanford.edu/teaching/cs231n/assignments.html) on the course website.*
We have seen that we can achieve reasonable performance on an image classification task by training a linear classifier on the pixels of the input image. In this exercise we will show that we can improve our classification performance by training linear classifiers not on raw pixels but on features that are computed from the raw pixels.
All of your work for this exercise will be done in this notebook.
```
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
from __future__ import print_function
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# for auto-reloading extenrnal modules
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
```
## Load data
Similar to previous exercises, we will load CIFAR-10 data from disk.
```
from cs231n.features import color_histogram_hsv, hog_feature
def get_CIFAR10_data(num_training=49000, num_validation=1000, num_test=1000):
# Load the raw CIFAR-10 data
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# Subsample the data
mask = list(range(num_training, num_training + num_validation))
X_val = X_train[mask]
y_val = y_train[mask]
mask = list(range(num_training))
X_train = X_train[mask]
y_train = y_train[mask]
mask = list(range(num_test))
X_test = X_test[mask]
y_test = y_test[mask]
return X_train, y_train, X_val, y_val, X_test, y_test
X_train, y_train, X_val, y_val, X_test, y_test = get_CIFAR10_data()
```
## Extract Features
For each image we will compute a Histogram of Oriented
Gradients (HOG) as well as a color histogram using the hue channel in HSV
color space. We form our final feature vector for each image by concatenating
the HOG and color histogram feature vectors.
Roughly speaking, HOG should capture the texture of the image while ignoring
color information, and the color histogram represents the color of the input
image while ignoring texture. As a result, we expect that using both together
ought to work better than using either alone. Verifying this assumption would
be a good thing to try for the bonus section.
The `hog_feature` and `color_histogram_hsv` functions both operate on a single
image and return a feature vector for that image. The extract_features
function takes a set of images and a list of feature functions and evaluates
each feature function on each image, storing the results in a matrix where
each column is the concatenation of all feature vectors for a single image.
```
from cs231n.features import *
num_color_bins = 10 # Number of bins in the color histogram
feature_fns = [hog_feature, lambda img: color_histogram_hsv(img, nbin=num_color_bins)]
X_train_feats = extract_features(X_train, feature_fns, verbose=True)
X_val_feats = extract_features(X_val, feature_fns)
X_test_feats = extract_features(X_test, feature_fns)
# Preprocessing: Subtract the mean feature
mean_feat = np.mean(X_train_feats, axis=0, keepdims=True)
X_train_feats -= mean_feat
X_val_feats -= mean_feat
X_test_feats -= mean_feat
# Preprocessing: Divide by standard deviation. This ensures that each feature
# has roughly the same scale.
std_feat = np.std(X_train_feats, axis=0, keepdims=True)
X_train_feats /= std_feat
X_val_feats /= std_feat
X_test_feats /= std_feat
# Preprocessing: Add a bias dimension
X_train_feats = np.hstack([X_train_feats, np.ones((X_train_feats.shape[0], 1))])
X_val_feats = np.hstack([X_val_feats, np.ones((X_val_feats.shape[0], 1))])
X_test_feats = np.hstack([X_test_feats, np.ones((X_test_feats.shape[0], 1))])
```
## Train SVM on features
Using the multiclass SVM code developed earlier in the assignment, train SVMs on top of the features extracted above; this should achieve better results than training SVMs directly on top of raw pixels.
```
# Use the validation set to tune the learning rate and regularization strength
from cs231n.classifiers.linear_classifier import LinearSVM
learning_rates = [1e-9, 1e-8, 1e-7]
regularization_strengths = [5e4, 5e5, 5e6]
results = {}
best_val = -1
best_svm = None
pass
################################################################################
# TODO: #
# Use the validation set to set the learning rate and regularization strength. #
# This should be identical to the validation that you did for the SVM; save #
# the best trained classifer in best_svm. You might also want to play #
# with different numbers of bins in the color histogram. If you are careful #
# you should be able to get accuracy of near 0.44 on the validation set. #
################################################################################
for learning_rate in learning_rates:
for reg in regularization_strengths:
print('lr %e reg %e' % (learning_rate, reg,))
svm = LinearSVM()
loss_hist = svm.train(X_train_feats, y_train, learning_rate=learning_rate, reg=reg,
num_iters=1500, verbose=True)
y_train_pred = svm.predict(X_train_feats)
y_val_pred = svm.predict(X_val_feats)
accuracy_train = np.mean(y_train == y_train_pred)
accuracy_val = np.mean(y_val == y_val_pred)
results[(learning_rate, reg)] = (accuracy_train, accuracy_val)
if best_val < accuracy_val:
best_val = accuracy_val
best_svm = svm
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print('best validation accuracy achieved during cross-validation: %f' % best_val)
# Evaluate your trained SVM on the test set
y_test_pred = best_svm.predict(X_test_feats)
test_accuracy = np.mean(y_test == y_test_pred)
print(test_accuracy)
# An important way to gain intuition about how an algorithm works is to
# visualize the mistakes that it makes. In this visualization, we show examples
# of images that are misclassified by our current system. The first column
# shows images that our system labeled as "plane" but whose true label is
# something other than "plane".
examples_per_class = 8
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
for cls, cls_name in enumerate(classes):
idxs = np.where((y_test != cls) & (y_test_pred == cls))[0]
idxs = np.random.choice(idxs, examples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt.subplot(examples_per_class, len(classes), i * len(classes) + cls + 1)
plt.imshow(X_test[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls_name)
plt.show()
```
### Inline question 1:
Describe the misclassification results that you see. Do they make sense?
## Neural Network on image features
Earlier in this assigment we saw that training a two-layer neural network on raw pixels achieved better classification performance than linear classifiers on raw pixels. In this notebook we have seen that linear classifiers on image features outperform linear classifiers on raw pixels.
For completeness, we should also try training a neural network on image features. This approach should outperform all previous approaches: you should easily be able to achieve over 55% classification accuracy on the test set; our best model achieves about 60% classification accuracy.
```
print(X_train_feats.shape)
from cs231n.classifiers.neural_net import TwoLayerNet
input_dim = X_train_feats.shape[1]
hidden_dim = 1024
num_classes = 10
net = TwoLayerNet(input_dim, hidden_dim, num_classes)
best_net = None
################################################################################
# TODO: Train a two-layer neural network on image features. You may want to #
# cross-validate various parameters as in previous sections. Store your best #
# model in the best_net variable. #
################################################################################
# Train the network
_reg=0
_learning_rate=1e-4
_learning_rate_decay=0.95
_num_iters=1000
net = TwoLayerNet(input_dim, hidden_dim, num_classes)
# Train the network
stats = net.train(X_train_feats, y_train, X_val_feats, y_val,
num_iters=_num_iters, batch_size=200,
learning_rate=_learning_rate, learning_rate_decay=_learning_rate_decay,
reg=_reg, verbose=True)
# Predict on the validation set
val_acc = (net.predict(X_val_feats) == y_val).mean()
print('Validation accuracy: ', val_acc)
# Plot the loss function and train / validation accuracies
plt.subplot(2, 1, 1)
plt.plot(stats['loss_history'])
plt.title('Loss history')
plt.xlabel('Iteration')
plt.ylabel('Loss')
plt.subplot(2, 1, 2)
plt.plot(stats['train_acc_history'], label='train')
plt.plot(stats['val_acc_history'], label='val')
plt.title('Classification accuracy history')
plt.xlabel('Epoch')
plt.ylabel('Clasification accuracy')
plt.show()
################################################################################
# END OF YOUR CODE #
################################################################################
# Run your neural net classifier on the test set. You should be able to
# get more than 55% accuracy.
test_acc = (net.predict(X_test_feats) == y_test).mean()
print(test_acc)
```
# Bonus: Design your own features!
You have seen that simple image features can improve classification performance. So far we have tried HOG and color histograms, but other types of features may be able to achieve even better classification performance.
For bonus points, design and implement a new type of feature and use it for image classification on CIFAR-10. Explain how your feature works and why you expect it to be useful for image classification. Implement it in this notebook, cross-validate any hyperparameters, and compare its performance to the HOG + Color histogram baseline.
# Bonus: Do something extra!
Use the material and code we have presented in this assignment to do something interesting. Was there another question we should have asked? Did any cool ideas pop into your head as you were working on the assignment? This is your chance to show off!
|
github_jupyter
|
# Automate Retraining of Models using SageMaker Pipelines and Lambda
# Learning Objectives
1. Construct a [SageMaker Pipeline](https://aws.amazon.com/sagemaker/pipelines/) that consists of a data preprocessing step and a model training step.
2. Execute a SageMaker Pipeline manually
3. Build infrastructure, using [CloudFormation](https://aws.amazon.com/cloudformation/) and [AWS Lambda](https://aws.amazon.com/lambda/) to allow the Pipeline steps be executed in an event-driven manner when new data is dropped in S3.
## Introduction
This workshop shows how you can build and deploy SageMaker Pipelines for multistep processes. In this example, we will build a pipeline that:
1. Deduplicates the underlying data
2. Trains a built-in SageMaker algorithm (XGBoost)
A common workflow is that models need to be retrained when new data arrives. This notebook also shows how you can set up a Lambda function that will retrigger the retraining pipeline when new data comes in.
Please use the `Python 3 (Data Science)` kernel for this workshop.
```
import boto3
import json
import logging
import os
import pandas
import sagemaker
from sagemaker.workflow.parameters import ParameterString
from sagemaker.workflow.steps import ProcessingStep, TrainingStep
from sagemaker.sklearn.processing import SKLearnProcessor
from sagemaker.workflow.pipeline import Pipeline
from sagemaker.inputs import TrainingInput
from sagemaker.processing import ProcessingInput, ProcessingOutput
from sagemaker.estimator import Estimator
from time import gmtime, strftime
# set logs if not done already
logger = logging.getLogger("log")
if not logger.handlers:
logger.setLevel(logging.INFO)
logger.addHandler(logging.StreamHandler())
```
First, get permissions and other information. We will also create a pipeline name
```
session = sagemaker.Session()
default_bucket = session.default_bucket()
role = sagemaker.get_execution_role()
region = boto3.Session().region_name
s3_client = boto3.client("s3", region_name=region)
current_timestamp = strftime("%m-%d-%H-%M", gmtime())
pipeline_name = f"my-pipeline-{current_timestamp}"
prefix = f"pipeline-lab{current_timestamp}"
```
## Transfer Data into Your Account
```
copy_source = {
"Bucket": "aws-hcls-ml",
"Key": "workshop/immersion_day_workshop_data_DO_NOT_DELETE/data/ObesityDataSet_with_duplicates.csv",
}
s3_client.copy(
copy_source, default_bucket, f"{prefix}/ObesityDataSet_with_duplicates.csv"
)
copy_source = {
"Bucket": "aws-hcls-ml",
"Key": "workshop/immersion_day_workshop_data_DO_NOT_DELETE/kick_off_sagemaker_pipelines_lambda/other_material/lambda.zip",
}
s3_client.copy(copy_source, default_bucket, f"{prefix}/lambda.zip")
```
## Define the Pipeline
First we will create a preprocessing step. The preprocessing step simply removes duplicated rows from the dataset. The `preprocessing.py` script will be written locally, and then built as a SageMaker Pipelines step.
```
input_data = ParameterString(
name="InputData",
default_value=f"s3://{default_bucket}/{prefix}/ObesityDataSet_with_duplicates.csv",
)
%%writefile preprocessing.py
import pandas
import os
base_dir = "/opt/ml/processing/input"
the_files = os.listdir(base_dir)
the_file=[i for i in the_files if ".csv" in i][0] #get the first csv
print(the_file)
df_1=pandas.read_csv(f'{base_dir}/{the_file}',engine='python')
df_2=df_1.drop_duplicates()
df_2.to_csv(f'/opt/ml/processing/output/deduped_{the_file}.csv')
# Specify the container and framework options
sklearn_processor = SKLearnProcessor(
framework_version="0.23-1",
instance_type="ml.t3.medium",
instance_count=1,
base_job_name="sklearn-abalone-process",
role=role,
)
```
Now will will turn the preprocessing step as a SageMaker Processing Step with SageMaker Pipelines.
```
step_process = ProcessingStep(
name="deduplication-process",
processor=sklearn_processor,
inputs=[
ProcessingInput(source=input_data, destination="/opt/ml/processing/input"),
],
outputs=[
ProcessingOutput(output_name="deduplicated", source="/opt/ml/processing/output")
],
code="preprocessing.py",
)
```
## Define the Model
Now we will create a SageMaker model. We will use the SageMaker built-in XGBoost Algorithm.
```
# Define the model training parameters
model_path = f"s3://{default_bucket}/{prefix}/myPipelineTrain"
image_uri = sagemaker.image_uris.retrieve(
framework="xgboost",
region=region,
version="1.0-1",
py_version="py3",
instance_type="ml.m5.large",
)
xgb_train = Estimator(
image_uri=image_uri,
instance_type="ml.m5.large",
instance_count=1,
output_path=model_path,
role=role,
)
xgb_train.set_hyperparameters(
objective="reg:linear",
num_round=50,
max_depth=5,
eta=0.2,
gamma=4,
min_child_weight=6,
subsample=0.7,
silent=0,
)
```
Turn the model training into a SageMaker Pipeline Training Step.
```
# Define the training steps
step_train = TrainingStep(
name="model-training",
estimator=xgb_train,
inputs={
"train": TrainingInput(
s3_data=step_process.properties.ProcessingOutputConfig.Outputs[
"deduplicated"
].S3Output.S3Uri,
content_type="text/csv",
),
"validation": TrainingInput(
s3_data=step_process.properties.ProcessingOutputConfig.Outputs[
"deduplicated"
].S3Output.S3Uri,
content_type="text/csv",
),
},
)
```
## Create and Start the Pipeline
```
# Create a two-step data processing and model training pipeline
pipeline_name = "ObesityModelRetrainingPipeLine"
pipeline = Pipeline(
name=pipeline_name,
parameters=[
input_data,
],
steps=[step_process, step_train],
)
pipeline.upsert(role_arn=role)
pipeline_execution = pipeline.start()
# Wait 15 minutes for the pipeline to finish running. In the meantime, you can monitor its progress in SageMaker Studio
pipeline_execution.wait()
```
## Deploy a CloudFormation Template to retrain the Pipeline
Now we will deploy a cloudformation template that will allow for automated calling of the Pipeline when new files are dropped in an S3 bucket.
The architecture looks like this:

NOTE: In order to run the following steps you must first associate the following IAM policies to your SageMaker execution role:
- cloudformation:CreateStack
- cloudformation:DeleteStack
- cloudformation:DescribeStacks
- iam:CreateRole
- iam:DeleteRole
- iam:DeleteRolePolicy
- iam:GetRole
- iam:GetRolePolicy
- iam:PassRole
- iam:PutRolePolicy
- lambda:AddPermission
- lambda:CreateFunction
- lambda:GetFunction
- lambda:DeleteFuncton
```
# Create a new CloudFormation stack to trigger retraining with new data
stack_name = "sagemaker-automated-retraining"
with open("cfn_sagemaker_pipelines.yaml") as f:
template_str = f.read()
cfn = boto3.client("cloudformation")
cfn.create_stack(
StackName=stack_name,
TemplateBody=template_str,
Capabilities=["CAPABILITY_IAM"],
Parameters=[
{"ParameterKey": "StaticCodeBucket", "ParameterValue": default_bucket},
{"ParameterKey": "StaticCodeKey", "ParameterValue": f"{prefix}/lambda.zip"},
],
)
# Wait until stack creation is complete
waiter = cfn.get_waiter("stack_create_complete")
waiter.wait(StackName=stack_name)
# Identify the S3 bucket for triggering the training pipeline
input_bucket_name = cfn.describe_stacks(StackName=stack_name)["Stacks"][0]["Outputs"][0]["OutputValue"]
# Copy the training data to the input bucket to start a new pipeline execution
copy_source = {
"Bucket": default_bucket,
"Key": f"{prefix}/ObesityDataSet_with_duplicates.csv",
}
s3_client.copy(copy_source, input_bucket_name, "ObesityDataSet_with_duplicates.csv")
```
### (Optional)
1. Inspect that the `InputBucket` has new data
2. Examine the `SageMaker Pipelines` execution from the SageMaker Studio console
```
#!aws s3 rm --recursive s3://{input_bucket_name}
```
## Closing
In this notebook we demonstrated how to create a SageMaker pipeline for data processing and model training and triggered it using an S3 event.
|
github_jupyter
|
## Simple regression
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# Import relevant modules
import pymc
import numpy as np
def generateData(size, true_intercept, true_slope, order, noiseSigma):
x = np.linspace(0, 1, size)
# y = a + b*x
true_y = true_intercept + true_slope * (x ** order)
# add noise
y = true_y + np.random.normal(scale=noiseSigma, size=size)
return x, y, true_y
def plotData(x, y, true_y):
fig = plt.figure(figsize=(7, 7))
ax = fig.add_subplot(111, xlabel='x', ylabel='y', title='Generated data and underlying model')
ax.plot(x, y, 'x', label='sampled data')
ax.plot(x, true_y, label='true regression line', lw=2.)
plt.legend(loc=0);
```
### Fit linear model
```
(x, y, true_y) = generateData(size = 200, true_intercept = 1, true_slope = 20, order = 1, noiseSigma=1.0)
plotData(x, y, true_y)
#Fit linear model
sigma = pymc.HalfCauchy('sigma', 10, 1.)
intercept = pymc.Normal('Intercept', 0, 1)
x_coeff = pymc.Normal('x', 0, 1)
@pymc.deterministic
def m(intercept= intercept, x_coeff=x_coeff):
return intercept + (x ** 1) * x_coeff
likelihood = pymc.Normal(name='y', mu=m, tau=1.0/sigma, value=y, observed=True)
# Plot the model dependencies
import pymc.graph
from IPython.display import display_png
graph = pymc.graph.graph(S)
display_png(graph.create_png(), raw=True)
# Run inference
mcmc = pymc.MCMC([likelihood, sigma, intercept, x_coeff])
mcmc.sample(iter=10000, burn=500, thin=2)
pymc.Matplot.plot(mcmc)
```
### Exercise fit cubic model
```
# your code here
```
### Model selection
```
(x, y, true_y) = generateData(size = 200, true_intercept = 1, true_slope = 20, order = 3, noiseSigma=2.0)
plotData(x, y, true_y)
#Model selection
beta = pymc.Beta('beta', 1.0, 1.0)
ber = pymc.Bernoulli('ber', beta)
sigma = pymc.HalfCauchy('sigma', 10, 1.)
intercept = pymc.Normal('Intercept', 0, 1)
x_coeff = pymc.Normal('x', 0, 1)
@pymc.deterministic
def m(intercept= intercept, x_coeff=x_coeff, ber=ber):
if ber:
return intercept + (x ** 3) * x_coeff
else:
return intercept + (x ** 1) * x_coeff
likelihood = pymc.Normal(name='y', mu=m, tau=1.0/sigma, value=y, observed=True)
mcmc = pymc.MCMC([likelihood, sigma, intercept, x_coeff, beta, ber])
mcmc.sample(iter=10000, burn=500, thin=2)
pymc.Matplot.plot(mcmc)
plt.hist(np.array(mcmc.trace("ber")[:], dtype=np.int))
plt.xlim([0, 1.5])
```
### Exercise: find noise effect on the model linearity
```
# your code here
```
|
github_jupyter
|
# Module 5: Hierarchical Generators
This module covers writing layout/schematic generators that instantiate other generators. We will write a two-stage amplifier generator, which instatiates the common-source amplifier followed by the source-follower amplifier.
## AmpChain Layout Example
First, we will write a layout generator for the two-stage amplifier. The layout floorplan is drawn for you below:
<img src="bootcamp_pics/5_hierarchical_generator/hierachical_generator_1.PNG" alt="Drawing" style="width: 400px;"/>
This floorplan abuts the `AmpCS` instance next to `AmpSF` instance, the `VSS` ports are simply shorted together, and the top `VSS` port of `AmpSF` is ignored (they are connected together internally by dummy connections). The intermediate node of the two-stage amplifier is connected using a vertical routing track in the middle of the two amplifier blocks. `VDD` ports are connected to the top-most M6 horizontal track, and other ports are simply exported in-place.
The layout generator is reproduced below, with some parts missing (which you will fill out later). We will walk through the important sections of the code.
```python
class AmpChain(TemplateBase):
def __init__(self, temp_db, lib_name, params, used_names, **kwargs):
TemplateBase.__init__(self, temp_db, lib_name, params, used_names, **kwargs)
self._sch_params = None
@property
def sch_params(self):
return self._sch_params
@classmethod
def get_params_info(cls):
return dict(
cs_params='common source amplifier parameters.',
sf_params='source follower parameters.',
show_pins='True to draw pin geometries.',
)
def draw_layout(self):
"""Draw the layout of a transistor for characterization.
"""
# make copies of given dictionaries to avoid modifying external data.
cs_params = self.params['cs_params'].copy()
sf_params = self.params['sf_params'].copy()
show_pins = self.params['show_pins']
# disable pins in subcells
cs_params['show_pins'] = False
sf_params['show_pins'] = False
# create layout masters for subcells we will add later
cs_master = self.new_template(params=cs_params, temp_cls=AmpCS)
# TODO: create sf_master. Use AmpSFSoln class
sf_master = None
if sf_master is None:
return
# add subcell instances
cs_inst = self.add_instance(cs_master, 'XCS')
# add source follower to the right of common source
x0 = cs_inst.bound_box.right_unit
sf_inst = self.add_instance(sf_master, 'XSF', loc=(x0, 0), unit_mode=True)
# get VSS wires from AmpCS/AmpSF
cs_vss_warr = cs_inst.get_all_port_pins('VSS')[0]
sf_vss_warrs = sf_inst.get_all_port_pins('VSS')
# only connect bottom VSS wire of source follower
if sf_vss_warrs[0].track_id.base_index < sf_vss_warrs[1].track_id.base_index:
sf_vss_warr = sf_vss_warrs[0]
else:
sf_vss_warr = sf_vss_warrs[1]
# connect VSS of the two blocks together
vss = self.connect_wires([cs_vss_warr, sf_vss_warr])[0]
# get layer IDs from VSS wire
hm_layer = vss.layer_id
vm_layer = hm_layer + 1
top_layer = vm_layer + 1
# calculate template size
tot_box = cs_inst.bound_box.merge(sf_inst.bound_box)
self.set_size_from_bound_box(top_layer, tot_box, round_up=True)
# get subcell ports as WireArrays so we can connect them
vmid0 = cs_inst.get_all_port_pins('vout')[0]
vmid1 = sf_inst.get_all_port_pins('vin')[0]
vdd0 = cs_inst.get_all_port_pins('VDD')[0]
vdd1 = sf_inst.get_all_port_pins('VDD')[0]
# get vertical VDD TrackIDs
vdd0_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd0.middle))
vdd1_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd1.middle))
# connect VDD of each block to vertical M5
vdd0 = self.connect_to_tracks(vdd0, vdd0_tid)
vdd1 = self.connect_to_tracks(vdd1, vdd1_tid)
# connect M5 VDD to top M6 horizontal track
vdd_tidx = self.grid.get_num_tracks(self.size, top_layer) - 1
vdd_tid = TrackID(top_layer, vdd_tidx)
vdd = self.connect_to_tracks([vdd0, vdd1], vdd_tid)
# TODO: connect vmid0 and vmid1 to vertical track in the middle of two templates
# hint: use x0
vmid = None
if vmid is None:
return
# add pins on wires
self.add_pin('vmid', vmid, show=show_pins)
self.add_pin('VDD', vdd, show=show_pins)
self.add_pin('VSS', vss, show=show_pins)
# re-export pins on subcells.
self.reexport(cs_inst.get_port('vin'), show=show_pins)
self.reexport(cs_inst.get_port('vbias'), net_name='vb1', show=show_pins)
# TODO: reexport vout and vbias of source follower
# TODO: vbias should be renamed to vb2
# compute schematic parameters.
self._sch_params = dict(
cs_params=cs_master.sch_params,
sf_params=sf_master.sch_params,
)
```
## AmpChain Constructor
```python
class AmpChain(TemplateBase):
def __init__(self, temp_db, lib_name, params, used_names, **kwargs):
TemplateBase.__init__(self, temp_db, lib_name, params, used_names, **kwargs)
self._sch_params = None
@property
def sch_params(self):
return self._sch_params
@classmethod
def get_params_info(cls):
return dict(
cs_params='common source amplifier parameters.',
sf_params='source follower parameters.',
show_pins='True to draw pin geometries.',
)
```
First, notice that instead of subclassing `AnalogBase`, the `AmpChain` class subclasses `TemplateBase`. This is because we are not trying to draw transistor rows inside this layout generator; we just want to place and route multiple layout instances together. `TemplateBase` is the base class for all layout generators and it provides most placement and routing methods you need.
Next, notice that the parameters for `AmpChain` are simply parameter dictionaries for the two sub-generators. The ability to use complex data structures as generator parameters solves the parameter explosion problem when writing generators with many levels of hierarchy.
## Creating Layout Master
```python
# create layout masters for subcells we will add later
cs_master = self.new_template(params=cs_params, temp_cls=AmpCS)
# TODO: create sf_master. Use AmpSFSoln class
sf_master = None
```
Here, the `new_template()` function creates a new layout master, `cs_master`, which represents a generated layout cellview from the `AmpCS` layout generator. We can later instances of this master in the current layout, which are references to the generated `AmpCS` layout cellview, perhaps shifted and rotated. The main take away is that the `new_template()` function does not add any layout geometries to the current layout, but rather create a separate layout cellview which we may use later.
## Creating Layout Instance
```python
# add subcell instances
cs_inst = self.add_instance(cs_master, 'XCS')
# add source follower to the right of common source
x0 = cs_inst.bound_box.right_unit
sf_inst = self.add_instance(sf_master, 'XSF', loc=(x0, 0), unit_mode=True)
```
The `add_instance()` method adds an instance of the given layout master to the current cellview. By default, if no location or orientation is given, it puts the instance at the origin with no rotation. the `bound_box` attribute can then be used on the instance to get the bounding box of the instance. Here, the bounding box is used to determine the X coordinate of the source-follower.
## Get Instance Ports
```python
# get subcell ports as WireArrays so we can connect them
vmid0 = cs_inst.get_all_port_pins('vout')[0]
vmid1 = sf_inst.get_all_port_pins('vin')[0]
vdd0 = cs_inst.get_all_port_pins('VDD')[0]
vdd1 = sf_inst.get_all_port_pins('VDD')[0]
```
after adding an instance, the `get_all_port_pins()` function can be used to obtain a list of all pins as `WireArray` objects with the given name. In this case, we know that there's exactly one pin, so we use Python list indexing to obtain first element of the list.
## Routing Grid Object
```python
# get vertical VDD TrackIDs
vdd0_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd0.middle))
vdd1_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd1.middle))
```
the `self.grid` attribute of `TemplateBase` is a `RoutingGrid` objects, which provides many useful functions related to the routing grid. In this particular scenario, `coord_to_nearest_track()` is used to determine the vertical track index closest to the center of the `VDD` ports. These vertical tracks will be used later to connect the `VDD` ports together.
## Re-export Pins on Instances
```python
# re-export pins on subcells.
self.reexport(cs_inst.get_port('vin'), show=show_pins)
self.reexport(cs_inst.get_port('vbias'), net_name='vb1', show=show_pins)
# TODO: reexport vout and vbias of source follower
# TODO: vbias should be renamed to vb2
```
`TemplateBase` also provides a `reexport()` function, which is a convenience function to re-export an instance port in-place. The `net_name` optional parameter can be used to change the port name. In this example, the `vbias` port of common-source amplifier is renamed to `vb1`.
## Layout Exercises
Now you should know everything you need to finish the two-stage amplifier layout generator. Fill in the missing pieces to do the following:
1. Create layout master for `AmpSF` using the `AmpSFSoln` class.
2. Using `RoutingGrid`, determine the vertical track index in the middle of the two amplifier blocks, and connect `vmid` wires together using this track.
* Hint: variable `x0` is the X coordinate of the boundary between the two blocks.
3. Re-export `vout` and `vbias` of the source-follower. Rename `vbias` to `vb2`.
Once you're done, evaluate the cell below, which will generate the layout and run LVS. If everything is done correctly, a layout should be generated inthe `DEMO_AMP_CHAIN` library, and LVS should pass.
```
from bag.layout.routing import TrackID
from bag.layout.template import TemplateBase
from xbase_demo.demo_layout.core import AmpCS, AmpSFSoln
class AmpChain(TemplateBase):
def __init__(self, temp_db, lib_name, params, used_names, **kwargs):
TemplateBase.__init__(self, temp_db, lib_name, params, used_names, **kwargs)
self._sch_params = None
@property
def sch_params(self):
return self._sch_params
@classmethod
def get_params_info(cls):
return dict(
cs_params='common source amplifier parameters.',
sf_params='source follower parameters.',
show_pins='True to draw pin geometries.',
)
def draw_layout(self):
"""Draw the layout of a transistor for characterization.
"""
# make copies of given dictionaries to avoid modifying external data.
cs_params = self.params['cs_params'].copy()
sf_params = self.params['sf_params'].copy()
show_pins = self.params['show_pins']
# disable pins in subcells
cs_params['show_pins'] = False
sf_params['show_pins'] = False
# create layout masters for subcells we will add later
cs_master = self.new_template(params=cs_params, temp_cls=AmpCS)
# TODO: create sf_master. Use AmpSFSoln class
sf_master = None
if sf_master is None:
return
# add subcell instances
cs_inst = self.add_instance(cs_master, 'XCS')
# add source follower to the right of common source
x0 = cs_inst.bound_box.right_unit
sf_inst = self.add_instance(sf_master, 'XSF', loc=(x0, 0), unit_mode=True)
# get VSS wires from AmpCS/AmpSF
cs_vss_warr = cs_inst.get_all_port_pins('VSS')[0]
sf_vss_warrs = sf_inst.get_all_port_pins('VSS')
# only connect bottom VSS wire of source follower
if len(sf_vss_warrs) < 2 or sf_vss_warrs[0].track_id.base_index < sf_vss_warrs[1].track_id.base_index:
sf_vss_warr = sf_vss_warrs[0]
else:
sf_vss_warr = sf_vss_warrs[1]
# connect VSS of the two blocks together
vss = self.connect_wires([cs_vss_warr, sf_vss_warr])[0]
# get layer IDs from VSS wire
hm_layer = vss.layer_id
vm_layer = hm_layer + 1
top_layer = vm_layer + 1
# calculate template size
tot_box = cs_inst.bound_box.merge(sf_inst.bound_box)
self.set_size_from_bound_box(top_layer, tot_box, round_up=True)
# get subcell ports as WireArrays so we can connect them
vmid0 = cs_inst.get_all_port_pins('vout')[0]
vmid1 = sf_inst.get_all_port_pins('vin')[0]
vdd0 = cs_inst.get_all_port_pins('VDD')[0]
vdd1 = sf_inst.get_all_port_pins('VDD')[0]
# get vertical VDD TrackIDs
vdd0_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd0.middle))
vdd1_tid = TrackID(vm_layer, self.grid.coord_to_nearest_track(vm_layer, vdd1.middle))
# connect VDD of each block to vertical M5
vdd0 = self.connect_to_tracks(vdd0, vdd0_tid)
vdd1 = self.connect_to_tracks(vdd1, vdd1_tid)
# connect M5 VDD to top M6 horizontal track
vdd_tidx = self.grid.get_num_tracks(self.size, top_layer) - 1
vdd_tid = TrackID(top_layer, vdd_tidx)
vdd = self.connect_to_tracks([vdd0, vdd1], vdd_tid)
# TODO: connect vmid0 and vmid1 to vertical track in the middle of two templates
# hint: use x0
vmid = None
if vmid is None:
return
# add pins on wires
self.add_pin('vmid', vmid, show=show_pins)
self.add_pin('VDD', vdd, show=show_pins)
self.add_pin('VSS', vss, show=show_pins)
# re-export pins on subcells.
self.reexport(cs_inst.get_port('vin'), show=show_pins)
self.reexport(cs_inst.get_port('vbias'), net_name='vb1', show=show_pins)
# TODO: reexport vout and vbias of source follower
# TODO: vbias should be renamed to vb2
# compute schematic parameters.
self._sch_params = dict(
cs_params=cs_master.sch_params,
sf_params=sf_master.sch_params,
)
import os
# import bag package
import bag
from bag.io import read_yaml
# import BAG demo Python modules
import xbase_demo.core as demo_core
# load circuit specifications from file
spec_fname = os.path.join(os.environ['BAG_WORK_DIR'], 'specs_demo/demo.yaml')
top_specs = read_yaml(spec_fname)
# obtain BagProject instance
local_dict = locals()
if 'bprj' in local_dict:
print('using existing BagProject')
bprj = local_dict['bprj']
else:
print('creating BagProject')
bprj = bag.BagProject()
demo_core.run_flow(bprj, top_specs, 'amp_chain_soln', AmpChain, run_lvs=True, lvs_only=True)
```
## AmpChain Schematic Template
Now let's move on to schematic generator. As before, we need to create the schematic template first. A half-complete schematic template is provided for you in library `demo_templates`, cell `amp_chain`, shown below:
<img src="bootcamp_pics/5_hierarchical_generator/hierachical_generator_2.PNG" alt="Drawing" style="width: 400px;"/>
The schematic template for a hierarchical generator is very simple; you simply need to instantiate the schematic templates of the sub-blocks (***Not the generated schematic!***). For the exercise, instantiate the `amp_sf` schematic template from the `demo_templates` library, named it `XSF`, connect it, then evaluate the following cell to import the `amp_chain` netlist to Python.
```
import bag
# obtain BagProject instance
local_dict = locals()
if 'bprj' in local_dict:
print('using existing BagProject')
bprj = local_dict['bprj']
else:
print('creating BagProject')
bprj = bag.BagProject()
print('importing netlist from virtuoso')
bprj.import_design_library('demo_templates')
print('netlist import done')
```
## AmpChain Schematic Generator
With schematic template done, you are ready to write the schematic generator. It is also very simple, you just need to call the `design()` method, which you implemented previously, on each instance in the schematic. Complete the following schematic generator, then evaluate the cell to push it through the design flow.
```
%matplotlib inline
import os
from bag.design import Module
# noinspection PyPep8Naming
class demo_templates__amp_chain(Module):
"""Module for library demo_templates cell amp_chain.
Fill in high level description here.
"""
# hard coded netlist flie path to get jupyter notebook working.
yaml_file = os.path.join(os.environ['BAG_WORK_DIR'], 'BAG_XBase_demo',
'BagModules', 'demo_templates', 'netlist_info', 'amp_chain.yaml')
def __init__(self, bag_config, parent=None, prj=None, **kwargs):
Module.__init__(self, bag_config, self.yaml_file, parent=parent, prj=prj, **kwargs)
@classmethod
def get_params_info(cls):
# type: () -> Dict[str, str]
"""Returns a dictionary from parameter names to descriptions.
Returns
-------
param_info : Optional[Dict[str, str]]
dictionary from parameter names to descriptions.
"""
return dict(
cs_params='common-source amplifier parameters dictionary.',
sf_params='source-follwer amplifier parameters dictionary.',
)
def design(self, cs_params=None, sf_params=None):
self.instances['XCS'].design(**cs_params)
# TODO: design XSF
import os
# import bag package
import bag
from bag.io import read_yaml
# import BAG demo Python modules
import xbase_demo.core as demo_core
from xbase_demo.demo_layout.core import AmpChainSoln
# load circuit specifications from file
spec_fname = os.path.join(os.environ['BAG_WORK_DIR'], 'specs_demo/demo.yaml')
top_specs = read_yaml(spec_fname)
# obtain BagProject instance
local_dict = locals()
if 'bprj' in local_dict:
print('using existing BagProject')
bprj = local_dict['bprj']
else:
print('creating BagProject')
bprj = bag.BagProject()
demo_core.run_flow(bprj, top_specs, 'amp_chain', AmpChainSoln, sch_cls=demo_templates__amp_chain, run_lvs=True)
```
|
github_jupyter
|
# MNIST Convolutional Neural Network - Ensemble Learning
Gaetano Bonofiglio, Veronica Iovinella
In this notebook we will verify if our single-column architecture can get any advantage from using **ensemble learning**, so a multi-column architecture.
We will train multiple networks identical to the best one defined in notebook 03, feeding them with pre-processed images shuffled and distorted using a different pseudo-random seed. This should give us a good ensemble of networks that we can average for each classification.
A prediction doesn't take more time compared to a single-column, but training time scales by a factor of N, where N is the number of columns. Networks could be trained in parallel, but not on our current hardware that is saturated by the training of a single one.
## Imports
```
import os.path
from IPython.display import Image
from util import Util
u = Util()
import numpy as np
# Explicit random seed for reproducibility
np.random.seed(1337)
from keras.callbacks import ModelCheckpoint
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Convolution2D, MaxPooling2D
from keras.layers import Merge
from keras.utils import np_utils
from keras.preprocessing.image import ImageDataGenerator
from keras import backend as K
from keras.datasets import mnist
```
## Definitions
For this experiment we are using 5 networks, but usually a good number is in the range of 35 (but with more dataset alterations then we do).
```
batch_size = 1024
nb_classes = 10
nb_epoch = 650
# checkpoint path
checkpoints_dir = "checkpoints"
# number of networks for ensamble learning
number_of_models = 5
# input image dimensions
img_rows, img_cols = 28, 28
# number of convolutional filters to use
nb_filters1 = 20
nb_filters2 = 40
# size of pooling area for max pooling
pool_size1 = (2, 2)
pool_size2 = (3, 3)
# convolution kernel size
kernel_size1 = (4, 4)
kernel_size2 = (5, 5)
# dense layer size
dense_layer_size1 = 200
# dropout rate
dropout = 0.15
# activation type
activation = 'relu'
```
## Data load
```
# the data, shuffled and split between train and test sets
(X_train, y_train), (X_test, y_test) = mnist.load_data()
u.plot_images(X_train[0:9], y_train[0:9])
if K.image_dim_ordering() == 'th':
X_train = X_train.reshape(X_train.shape[0], 1, img_rows, img_cols)
X_test = X_test.reshape(X_test.shape[0], 1, img_rows, img_cols)
input_shape = (1, img_rows, img_cols)
else:
X_train = X_train.reshape(X_train.shape[0], img_rows, img_cols, 1)
X_test = X_test.reshape(X_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
X_train = X_train.astype('float32')
X_test = X_test.astype('float32')
X_train /= 255
X_test /= 255
print('X_train shape:', X_train.shape)
print(X_train.shape[0], 'train samples')
print(X_test.shape[0], 'test samples')
# convert class vectors to binary class matrices
Y_train = np_utils.to_categorical(y_train, nb_classes)
Y_test = np_utils.to_categorical(y_test, nb_classes)
```
## Image preprocessing
```
datagen = ImageDataGenerator(
rotation_range=30,
width_shift_range=0.1,
height_shift_range=0.1,
zoom_range=0.1,
horizontal_flip=False)
# compute quantities required for featurewise normalization
# (std, mean, and principal components if ZCA whitening is applied)
datagen.fit(X_train)
```
## Model definition - Single column
This time we are going to define a helper functions to initialize the model, since we're going to use it on a list of models.
```
def initialize_network(model, dropout1=dropout, dropout2=dropout):
model.add(Convolution2D(nb_filters1, kernel_size1[0], kernel_size1[1],
border_mode='valid',
input_shape=input_shape, name='covolution_1_' + str(nb_filters1) + '_filters'))
model.add(Activation(activation, name='activation_1_' + activation))
model.add(MaxPooling2D(pool_size=pool_size1, name='max_pooling_1_' + str(pool_size1) + '_pool_size'))
model.add(Convolution2D(nb_filters2, kernel_size2[0], kernel_size2[1]))
model.add(Activation(activation, name='activation_2_' + activation))
model.add(MaxPooling2D(pool_size=pool_size2, name='max_pooling_1_' + str(pool_size2) + '_pool_size'))
model.add(Dropout(dropout))
model.add(Flatten())
model.add(Dense(dense_layer_size1, name='fully_connected_1_' + str(dense_layer_size1) + '_neurons'))
model.add(Activation(activation, name='activation_3_' + activation))
model.add(Dropout(dropout))
model.add(Dense(nb_classes, name='output_' + str(nb_classes) + '_neurons'))
model.add(Activation('softmax', name='softmax'))
model.compile(loss='categorical_crossentropy',
optimizer='adadelta',
metrics=['accuracy', 'precision', 'recall'])
# pseudo random generation of seeds
seeds = np.random.randint(10000, size=number_of_models)
# initializing all the models
models = [None] * number_of_models
for i in range(number_of_models):
models[i] = Sequential()
initialize_network(models[i])
```
## Training and evaluation - Single column
Again we are going to define a helper functions to train the model, since we're going to use them on a list.
```
def try_load_checkpoints(model, checkpoints_filepath, warn=False):
# loading weights from checkpoints
if os.path.exists(checkpoints_filepath):
model.load_weights(checkpoints_filepath)
elif warn:
print('Warning: ' + checkpoints_filepath + ' could not be loaded')
def fit(model, checkpoints_name='test', seed=1337, initial_epoch=0,
verbose=1, window_size=(-1), plot_history=False, evaluation=True):
if window_size == (-1):
window = 1 + np.random.randint(14)
else:
window = window_size
if window >= nb_epoch:
window = nb_epoch - 1
print("Not pre-processing " + str(window) + " epoch(s)")
checkpoints_filepath = os.path.join(checkpoints_dir, '04_MNIST_weights.best_' + checkpoints_name + '.hdf5')
try_load_checkpoints(model, checkpoints_filepath, True)
# checkpoint
checkpoint = ModelCheckpoint(checkpoints_filepath, monitor='val_precision', verbose=verbose, save_best_only=True, mode='max')
callbacks_list = [checkpoint]
# fits the model on batches with real-time data augmentation, for nb_epoch-100 epochs
history = model.fit_generator(datagen.flow(X_train, Y_train,
batch_size=batch_size,
# save_to_dir='distorted_data',
# save_format='png'
seed=1337),
samples_per_epoch=len(X_train), nb_epoch=(nb_epoch-window), verbose=0,
validation_data=(X_test, Y_test), callbacks=callbacks_list)
# ensuring best val_precision reached during training
try_load_checkpoints(model, checkpoints_filepath)
# fits the model on clear training set, for nb_epoch-700 epochs
history_cont = model.fit(X_train, Y_train, batch_size=batch_size, nb_epoch=window,
verbose=0, validation_data=(X_test, Y_test), callbacks=callbacks_list)
# ensuring best val_precision reached during training
try_load_checkpoints(model, checkpoints_filepath)
if plot_history:
print("History: ")
u.plot_history(history)
u.plot_history(history, 'precision')
print("Continuation of training with no pre-processing:")
u.plot_history(history_cont)
u.plot_history(history_cont, 'precision')
if evaluation:
print('Evaluating model ' + str(index))
score = model.evaluate(X_test, Y_test, verbose=0)
print('Test accuracy:', score[1]*100, '%')
print('Test error:', (1-score[2])*100, '%')
return history, history_cont
for index in range(number_of_models):
print("Training model " + str(index) + " ...")
if index == 0:
window_size = 20
plot_history = True
else:
window_size = (-1)
plot_history = False
history, history_cont = fit(models[index],
str(index),
seed=seeds[index],
initial_epoch=0,
verbose=0,
window_size=window_size,
plot_history=plot_history)
print("Done.\n\n")
```
Just by the different seeds, error changes **from 0.5% to 0.42%** (our best result so far with a single column). The training took 12 hours.
## Model definition - Multi column
The MCDNN is obtained by creating a new model that only has 1 layer, Merge, that does the average of the outputs of the models in the given list. No training is required since we're only doing the average.
```
merged_model = Sequential()
merged_model.add(Merge(models, mode='ave'))
merged_model.compile(loss='categorical_crossentropy',
optimizer='adadelta',
metrics=['accuracy', 'precision', 'recall'])
```
## Evaluation - Multi column
```
print('Evaluating ensemble')
score = merged_model.evaluate([np.asarray(X_test)] * number_of_models,
Y_test,
verbose=0)
print('Test accuracy:', score[1]*100, '%')
print('Test error:', (1-score[2])*100, '%')
```
The error improved from 0.42% with the best network of the ensemble, to 0.4%, that is out best result so far.
```
# The predict_classes function outputs the highest probability class
# according to the trained classifier for each input example.
predicted_classes = merged_model.predict_classes([np.asarray(X_test)] * number_of_models)
# Check which items we got right / wrong
correct_indices = np.nonzero(predicted_classes == y_test)[0]
incorrect_indices = np.nonzero(predicted_classes != y_test)[0]
u.plot_images(X_test[correct_indices[:9]], y_test[correct_indices[:9]],
predicted_classes[correct_indices[:9]])
u.plot_images(X_test[incorrect_indices[:9]], y_test[incorrect_indices[:9]],
predicted_classes[incorrect_indices[:9]])
u.plot_confusion_matrix(y_test, nb_classes, predicted_classes)
```
## Results
Training 5 networks took 12 hours, of course 5 times longer then a single one. The improvement was of 0.05% error, that is quite good considering this dataset (a human has 0.2% test error on MNIST).
To further increase the precision we would need over 30 columns trained on different widths.
|
github_jupyter
|
# PyCaret Fugue Integration
[Fugue](https://github.com/fugue-project/fugue) is a low-code unified interface for different computing frameworks such as Spark, Dask and Pandas. PyCaret is using Fugue to support distributed computing scenarios.
## Hello World
### Classification
Let's start with the most standard example, the code is exactly the same as the local version, there is no magic.
```
from pycaret.datasets import get_data
from pycaret.classification import *
setup(data=get_data("juice"), target = 'Purchase', n_jobs=1)
test_models = models().index.tolist()[:5]
```
`compare_model` is also exactly the same if you don't want to use a distributed system
```
compare_models(include=test_models, n_select=2)
```
Now let's make it distributed, as a toy case, on dask. The only thing changed is an additional parameter `parallel_backend`
```
from pycaret.parallel import FugueBackend
compare_models(include=test_models, n_select=2, parallel=FugueBackend("dask"))
```
In order to use Spark as the execution engine, you must have access to a Spark cluster, and you must have a `SparkSession`, let's initialize a local Spark session
```
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
```
Now just change `parallel_backend` to this session object, you make it run on Spark. You must understand this is a toy case. In the real situation, you need to have a SparkSession pointing to a real Spark cluster to enjoy the power of Spark
```
compare_models(include=test_models, n_select=2, parallel=FugueBackend(spark))
```
In the end, you can `pull` to get the metrics table
```
pull()
```
### Regression
It's follows the same pattern as classification.
```
from pycaret.datasets import get_data
from pycaret.regression import *
setup(data=get_data("insurance"), target = 'charges', n_jobs=1)
test_models = models().index.tolist()[:5]
```
`compare_model` is also exactly the same if you don't want to use a distributed system
```
compare_models(include=test_models, n_select=2)
```
Now let's make it distributed, as a toy case, on dask. The only thing changed is an additional parameter `parallel_backend`
```
from pycaret.parallel import FugueBackend
compare_models(include=test_models, n_select=2, parallel=FugueBackend("dask"))
```
In order to use Spark as the execution engine, you must have access to a Spark cluster, and you must have a `SparkSession`, let's initialize a local Spark session
```
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
```
Now just change `parallel_backend` to this session object, you make it run on Spark. You must understand this is a toy case. In the real situation, you need to have a SparkSession pointing to a real Spark cluster to enjoy the power of Spark
```
compare_models(include=test_models, n_select=2, parallel=FugueBackend(spark))
```
In the end, you can `pull` to get the metrics table
```
pull()
```
As you see, the results from the distributed versions can be different from your local versions. In the next section, we will show how to make them identical.
## A more practical case
The above examples are pure toys, to make things work perfectly in a distributed system you must be careful about a few things
### Use a lambda instead of a dataframe in setup
If you directly provide a dataframe in `setup`, this dataset will need to be sent to all worker nodes. If the dataframe is 1G, you have 100 workers, then it is possible your dirver machine will need to send out up to 100G data (depending on specific framework's implementation), then this data transfer becomes a bottleneck itself. Instead, if you provide a lambda function, it doesn't change the local compute scenario, but the driver will only send the function reference to workers, and each worker will be responsible to load the data by themselves, so there is no heavy traffic on the driver side.
### Be deterministic
You should always use `session_id` to make the distributed compute deterministic, otherwise, for the exactly same logic you could get drastically different selection for each run.
### Set n_jobs
It is important to be explicit on n_jobs when you want to run something distributedly, so it will not overuse the local/remote resources. This can also avoid resrouce contention, and make the compute faster.
```
from pycaret.classification import *
setup(data=lambda: get_data("juice", verbose=False, profile=False), target = 'Purchase', session_id=0, n_jobs=1);
```
### Set the appropriate batch_size
`batch_size` parameter helps adjust between load balence and overhead. For each batch, setup will be called only once. So
| Choice |Load Balance|Overhead|Best Scenario|
|---|---|---|---|
|Smaller batch size|Better|Worse|`training time >> data loading time` or `models ~= workers`|
|Larger batch size|Worse|Better|`training time << data loading time` or `models >> workers`|
The default value is set to `1`, meaning we want the best load balance.
### Display progress
In development, you can enable visual effect by `display_remote=True`, but meanwhile you must also enable [Fugue Callback](https://fugue-tutorials.readthedocs.io/tutorials/advanced/rpc.html) so that the driver can monitor worker progress. But it is recommended to turn off display in production.
```
fconf = {
"fugue.rpc.server": "fugue.rpc.flask.FlaskRPCServer", # keep this value
"fugue.rpc.flask_server.host": "0.0.0.0", # the driver ip address workers can access
"fugue.rpc.flask_server.port": "3333", # the open port on the dirver
"fugue.rpc.flask_server.timeout": "2 sec", # the timeout for worker to talk to driver
}
be = FugueBackend("dask", fconf, display_remote=True, batch_size=3, top_only=False)
compare_models(n_select=2, parallel=be)
```
## Notes
### Spark settings
It is highly recommended to have only 1 worker on each Spark executor, so the worker can fully utilize all cpus (set `spark.task.cpus`). Also when you do this you should explicitly set `n_jobs` in `setup` to the number of cpus of each executor.
```python
executor_cores = 4
spark = SparkSession.builder.config("spark.task.cpus", executor_cores).config("spark.executor.cores", executor_cores).getOrCreate()
setup(data=get_data("juice", verbose=False, profile=False), target = 'Purchase', session_id=0, n_jobs=executor_cores)
compare_models(n_select=2, parallel=FugueBackend(spark))
```
### Databricks
On Databricks, `spark` is the magic variable representing a SparkSession. But there is no difference to use. You do the exactly same thing as before:
```python
compare_models(parallel=FugueBackend(spark))
```
But Databricks, the visualization is difficult, so it may be a good idea to do two things:
* Set `verbose` to False in `setup`
* Set `display_remote` to False in `FugueBackend`
### Dask
Dask has fake distributed modes such as the default (multi-thread) and multi-process modes. The default mode will just work fine (but they are actually running sequentially), and multi-process doesn't work for PyCaret for now because it messes up with PyCaret's global variables. On the other hand, any Spark execution mode will just work fine.
### Local Parallelization
For practical use where you try non-trivial data and models, local parallelization (The eaiest way is to use local Dask as backend as shown above) normally doesn't have performance advantage. Because it's very easy to overload the CPUS on training, increasing the contention of resources. The value of local parallelization is to verify the code and give you confidence that the distributed environment will provide the expected result with much shorter time.
### How to develop
Distributed systems are powerful but you must follow some good practices to use them:
1. **From small to large:** initially, you must start with a small set of data, for example in `compare_model` limit the models you want to try to a small number of cheap models, and when you verify they work, you can change to a larger model collection.
2. **From local to distributed:** you should follow this sequence: verify small data locally then verify small data distributedly and then verify large data distributedly. The current design makes the transition seamless. You can do these sequentially: `parallel=None` -> `parallel=FugueBackend()` -> `parallel=FugueBackend(spark)`. In the second step, you can replace with a local SparkSession or local dask.
|
github_jupyter
|
We use Embeddings to represent text into a numerical form. Either into a one-hot encoding format called sparse vector or a fixed Dense representation called Dense Vector.
Every Word gets it meaning from the words it is surrounded by, So when we train our embeddings we want word with similar meaning or words used in similar context to be together.
For Example:-
1. Words like Aeroplane, chopper, Helicopter, Drone should be very close to each other because they share the same feature, they are flying object.
2. Words like Man and Women should be exact opposite to each other.
3. Sentences like "Coders are boring people." and "Programmers are boring." the word `coders` and `programmers` are used in similar context so they should be close to each other.
Word Embeddings are nothing but vectors in a vector space. And using some vector calculation we can easily find
1. Synonyms or similar words
2. Finding Analogies
3. Can be used as spell check (if trained on a large corpus)
4. Pretty Much Anything which you can do with vectors.
```
import torchtext
import numpy as np
import torch
glove = torchtext.vocab.GloVe(name = '6B', dim = 100)
print(f'There are {len(glove.itos)} words in the vocabulary')
glove.itos[:10]
glove.stoi["cat"]
def get_embedding(word):
return glove.vectors[glove.stoi[word]]
get_embedding("cat")
```
# Similar Context
To find words similar to input words. We have to first take the vector representation of all words and compute the eucledian distance of the input word with respect to all words and choose the n closest words by sorting the distance ascending order.
```
def get_closest_word(word,n=10):
input_vector = get_embedding(word).numpy() if isinstance(word,str) else word.numpy()
distance = np.linalg.norm(input_vector-glove.vectors.numpy(),axis=1)
sort_dis = np.argsort(distance)[:n]
return list(zip(np.array(glove.itos)[sort_dis] , distance[sort_dis]))
get_closest_word("sad",n=10)
def get_similarity_angle(word1,word2):
word1 = get_embedding(word1).view(1,-1)
word2 = get_embedding(word2).view(1,-1)
simi = torch.nn.CosineSimilarity(dim=1)(word1,word2).numpy()
return simi,np.rad2deg(np.arccos(simi))
get_similarity_angle("sad","awful")
```
# Analogies
```
def analogy( word1, word2, word3, n=5):
#get vectors for each word
word1_vector = get_embedding(word1)
word2_vector = get_embedding(word2)
word3_vector = get_embedding(word3)
#calculate analogy vector
analogy_vector = word2_vector - word1_vector + word3_vector
# #find closest words to analogy vector
candidate_words = get_closest_word( analogy_vector, n=n+3)
#filter out words already in analogy
candidate_words = [(word, dist) for (word, dist) in candidate_words
if word not in [word1, word2, word3]][:n]
print(f'{word1} is to {word2} as {word3} is to...')
return candidate_words
analogy('man', 'king', 'woman')
```
This is the canonical example which shows off this property of word embeddings. So why does it work? Why does the vector of 'woman' added to the vector of 'king' minus the vector of 'man' give us 'queen'?
If we think about it, the vector calculated from 'king' minus 'man' gives us a "royalty vector". This is the vector associated with traveling from a man to his royal counterpart, a king. If we add this "royality vector" to 'woman', this should travel to her royal equivalent, which is a queen!
```
analogy('india', 'delhi', 'australia')
get_closest_word("reliable")
```
# Case Studies
1. https://forums.fast.ai/t/nlp-any-libraries-dictionaries-out-there-for-fixing-common-spelling-errors/16411
2. Multilingual and Cross-lingual analysis: If you work on works in translation, or on the influence of writers who write in one language on those who write in another language, word vectors can valuable ways to study these kinds of cross-lingual relationships algorithmically.
[Case Study: Using word vectors to study endangered languages](https://raw.githubusercontent.com/YaleDHLab/lab-workshops/master/word-vectors/papers/coeckelbergs.pdf)
3. Studying Language Change over Time: If you want to study the way the meaning of a word has changed over time, word vectors provide an exceptional method for this kind of study.
[Case Study: Using word vectors to analyze the changing meaning of the word "gay" in the twentieth century.](https://nlp.stanford.edu/projects/histwords/)
4. Analyzing Historical Concept Formation: If you want to analyze the ways writers in a given historical period understood particular concepts like "honor" and "chivalry", then word vectors can provide excellent opportunities to uncover these hidden associations.
[Case Study: Using word vectors to study the ways eighteenth-century authors organized moral abstractions](https://raw.githubusercontent.com/YaleDHLab/lab-workshops/master/word-vectors/papers/heuser.pdf)
5. Uncovering Text Reuse: If you want to study text reuse or literary imitation (either within one language or across multiple languages), word vectors can provide excellent tools for identifying similar passages of text.
[Case Study: Using word vectors to uncover cross-lingual text reuse in eighteenth-century writing](https://douglasduhaime.com/posts/crosslingual-plagiarism-detection.html)
|
github_jupyter
|
# Document Embedding with Amazon SageMaker Object2Vec
1. [Introduction](#Introduction)
2. [Background](#Background)
1. [Embedding documents using Object2Vec](#Embedding-documents-using-Object2Vec)
3. [Download and preprocess Wikipedia data](#Download-and-preprocess-Wikipedia-data)
1. [Install and load dependencies](#Install-and-load-dependencies)
2. [Build vocabulary and tokenize datasets](#Build-vocabulary-and-tokenize-datasets)
3. [Upload preprocessed data to S3](#Upload-preprocessed-data-to-S3)
4. [Define SageMaker session, Object2Vec image, S3 input and output paths](#Define-SageMaker-session,-Object2Vec-image,-S3-input-and-output-paths)
5. [Train and deploy doc2vec](#Train-and-deploy-doc2vec)
1. [Learning performance boost with new features](#Learning-performance-boost-with-new-features)
2. [Training speedup with sparse gradient update](#Training-speedup-with-sparse-gradient-update)
6. [Apply learned embeddings to document retrieval task](#Apply-learned-embeddings-to-document-retrieval-task)
1. [Comparison with the StarSpace algorithm](#Comparison-with-the-StarSpace-algorithm)
## Introduction
In this notebook, we introduce four new features to Object2Vec, a general-purpose neural embedding algorithm: negative sampling, sparse gradient update, weight-sharing, and comparator operator customization. The new features together broaden the applicability of Object2Vec, improve its training speed and accuracy, and provide users with greater flexibility. See [Introduction to the Amazon SageMaker Object2Vec](https://aws.amazon.com/blogs/machine-learning/introduction-to-amazon-sagemaker-object2vec/) if you aren’t already familiar with Object2Vec.
We demonstrate how these new features extend the applicability of Object2Vec to a new Document Embedding use-case: A customer has a large collection of documents. Instead of storing these documents in its raw format or as sparse bag-of-words vectors, to achieve training efficiency in the various downstream tasks, she would like to instead embed all documents in a common low-dimensional space, so that the semantic distance between these documents are preserved.
## Background
Object2Vec is a highly customizable multi-purpose algorithm that can learn embeddings of pairs of objects. The embeddings are learned such that it preserves their pairwise similarities in the original space.
- Similarity is user-defined: users need to provide the algorithm with pairs of objects that they define as similar (1) or dissimilar (0); alternatively, the users can define similarity in a continuous sense (provide a real-valued similarity score).
- The learned embeddings can be used to efficiently compute nearest neighbors of objects, as well as to visualize natural clusters of related objects in the embedding space. In addition, the embeddings can also be used as features of the corresponding objects in downstream supervised tasks such as classification or regression.
### Embedding documents using Object2Vec
We demonstrate how, with the new features, Object2Vec can be used to embed a large collection of documents into vectors in the same latent space.
Similar to the widely used Word2Vec algorithm for word embedding, a natural approach to document embedding is to preprocess documents as (sentence, context) pairs, where the sentence and its matching context come from the same document. The matching context is the entire document with the given sentence removed. The idea is to embed both sentence and context into a low dimensional space such that their mutual similarity is maximized, since they belong to the same document and therefore should be semantically related. The learned encoder for the context can then be used to encode new documents into the same embedding space. In order to train the encoders for sentences and documents, we also need negative (sentence, context) pairs so that the model can learn to discriminate between semantically similar and dissimilar pairs. It is easy to generate such negatives by pairing sentences with documents that they do not belong to. Since there are many more negative pairs than positives in naturally occurring data, we typically resort to random sampling techniques to achieve a balance between positive and negative pairs in the training data. The figure below shows pictorially how the positive pairs and negative pairs are generated from unlabeled data for the purpose of learning embeddings for documents (and sentences).
We show how Object2Vec with the new *negative sampling feature* can be applied to the document embedding use-case. In addition, we show how the other new features, namely, *weight-sharing*, *customization of comparator operator*, and *sparse gradient update*, together enhance the algorithm's performance and user-experience in and beyond this use-case. Sections [Learning performance boost with new features](#Learning-performance-boost-with-new-features) and [Training speedup with sparse gradient update](#Training-speedup-with-sparse-gradient-update) in this notebook provide a detailed introduction to the new features.
## Download and preprocess Wikipedia data
Please be aware of the following requirements about the acknowledgment, copyright and availability, cited from the [data source description page](https://github.com/facebookresearch/StarSpace/blob/master/LICENSE.md).
> Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions: The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
```
%%bash
DATANAME="wikipedia"
DATADIR="/tmp/wiki"
mkdir -p "${DATADIR}"
if [ ! -f "${DATADIR}/${DATANAME}_train250k.txt" ]
then
echo "Downloading wikipedia data"
wget --quiet -c "https://s3-ap-northeast-1.amazonaws.com/dev.tech-sketch.jp/chakki/public/ja.wikipedia_250k.zip" -O "${DATADIR}/${DATANAME}_train.zip"
unzip "${DATADIR}/${DATANAME}_train.zip" -d "${DATADIR}"
fi
datadir = '/tmp/wiki'
!ls /tmp/wiki
```
### Install and load dependencies
```
!pip install keras tensorflow
import json
import os
import random
from itertools import chain
from keras.preprocessing.text import Tokenizer
from sklearn.preprocessing import normalize
## sagemaker api
import sagemaker, boto3
from sagemaker.session import s3_input
from sagemaker.predictor import json_serializer, json_deserializer
```
### Build vocabulary and tokenize datasets
```
def load_articles(filepath):
with open(filepath) as f:
for line in f:
yield map(str.split, line.strip().split('\t'))
def split_sents(article):
return [sent.split(' ') for sent in article.split('\t')]
def build_vocab(sents):
print('Build start...')
tok = Tokenizer(oov_token='<UNK>', filters='')
tok.fit_on_texts(sents)
print('Build end...')
return tok
def generate_positive_pairs_from_single_article(sents, tokenizer):
sents = list(sents)
idx = random.randrange(0, len(sents))
center = sents.pop(idx)
wrapper_tokens = tokenizer.texts_to_sequences(sents)
sent_tokens = tokenizer.texts_to_sequences([center])
wrapper_tokens = list(chain(*wrapper_tokens))
sent_tokens = list(chain(*sent_tokens))
yield {'in0': sent_tokens, 'in1': wrapper_tokens, 'label': 1}
def generate_positive_pairs_from_single_file(sents_per_article, tokenizer):
iter_list = [generate_positive_pairs_from_single_article(sents, tokenizer)
for sents in sents_per_article
]
return chain.from_iterable(iter_list)
filepath = os.path.join(datadir, 'ja.wikipedia_250k.txt')
sents_per_article = load_articles(filepath)
sents = chain(*sents_per_article)
tokenizer = build_vocab(sents)
# save
datadir = '.'
train_prefix = 'train250k'
fname = "wikipedia_{}.txt".format(train_prefix)
outfname = os.path.join(datadir, '{}_tokenized.jsonl'.format(train_prefix))
with open(outfname, 'w') as f:
sents_per_article = load_articles(filepath)
for sample in generate_positive_pairs_from_single_file(sents_per_article, tokenizer):
f.write('{}\n'.format(json.dumps(sample)))
# Shuffle training data
!shuf {outfname} > {train_prefix}_tokenized_shuf.jsonl
```
### Upload preprocessed data to S3
```
TRAIN_DATA="train250k_tokenized_shuf.jsonl"
# NOTE: define your s3 bucket and key here
S3_BUCKET = 'YOUR_BUCKET'
S3_KEY = 'object2vec-doc2vec'
%%bash -s "$TRAIN_DATA" "$S3_BUCKET" "$S3_KEY"
aws s3 cp "$1" s3://$2/$3/input/train/
```
## Define Sagemaker session, Object2Vec image, S3 input and output paths
```
from sagemaker import get_execution_role
from sagemaker.amazon.amazon_estimator import get_image_uri
region = boto3.Session().region_name
print("Your notebook is running on region '{}'".format(region))
sess = sagemaker.Session()
role = get_execution_role()
print("Your IAM role: '{}'".format(role))
container = get_image_uri(region, 'object2vec')
print("The image uri used is '{}'".format(container))
print("Using s3 buceket: {} and key prefix: {}".format(S3_BUCKET, S3_KEY))
## define input channels
s3_input_path = os.path.join('s3://', S3_BUCKET, S3_KEY, 'input')
s3_train = s3_input(os.path.join(s3_input_path, 'train', TRAIN_DATA),
distribution='ShardedByS3Key', content_type='application/jsonlines')
## define output path
output_path = os.path.join('s3://', S3_BUCKET, S3_KEY, 'models')
```
## Train and deploy doc2vec
We combine four new features into our training of Object2Vec:
- Negative sampling: With the new `negative_sampling_rate` hyperparameter, users of Object2Vec only need to provide positively labeled data pairs, and the algorithm automatically samples for negative data internally during training.
- Weight-sharing of embedding layer: The new `tied_token_embedding_weight` hyperparameter gives user the flexibility to share the embedding weights for both encoders, and it improves the performance of the algorithm in this use-case
- The new `comparator_list` hyperparameter gives users the flexibility to mix-and-match different operators so that they can tune the algorithm towards optimal performance for their applications.
```
# Define training hyperparameters
hyperparameters = {
"_kvstore": "device",
"_num_gpus": 'auto',
"_num_kv_servers": "auto",
"bucket_width": 0,
"dropout": 0.4,
"early_stopping_patience": 2,
"early_stopping_tolerance": 0.01,
"enc0_layers": "auto",
"enc0_max_seq_len": 50,
"enc0_network": "pooled_embedding",
"enc0_pretrained_embedding_file": "",
"enc0_token_embedding_dim": 300,
"enc0_vocab_size": len(tokenizer.word_index) + 1,
"enc1_network": "enc0",
"enc_dim": 300,
"epochs": 20,
"learning_rate": 0.01,
"mini_batch_size": 512,
"mlp_activation": "relu",
"mlp_dim": 512,
"mlp_layers": 2,
"num_classes": 2,
"optimizer": "adam",
"output_layer": "softmax",
"weight_decay": 0
}
hyperparameters['negative_sampling_rate'] = 3
hyperparameters['tied_token_embedding_weight'] = "true"
hyperparameters['comparator_list'] = "hadamard"
hyperparameters['token_embedding_storage_type'] = 'row_sparse'
# get estimator
doc2vec = sagemaker.estimator.Estimator(container,
role,
train_instance_count=1,
train_instance_type='ml.p2.xlarge',
output_path=output_path,
sagemaker_session=sess)
# set hyperparameters
doc2vec.set_hyperparameters(**hyperparameters)
# fit estimator with data
doc2vec.fit({'train': s3_train})
#doc2vec.fit({'train': s3_train, 'validation':s3_valid, 'test':s3_test})
# deploy model
doc2vec_model = doc2vec.create_model(
serializer=json_serializer,
deserializer=json_deserializer,
content_type='application/json')
predictor = doc2vec_model.deploy(initial_instance_count=1, instance_type='ml.m4.xlarge')
sent = '今日 の 昼食 は うどん だっ た'
sent_tokens = tokenizer.texts_to_sequences([sent])
payload = {'instances': [{'in0': sent_tokens[0]}]}
result = predictor.predict(payload)
print(result)
predictor.delete_endpoint()
```
|
github_jupyter
|
# Working with HEALPix data
[HEALPix](https://healpix.jpl.nasa.gov/) (Hierarchical Equal Area isoLatitude Pixelisation) is an algorithm that is often used to store data from all-sky surveys.
There are several tools in the Astropy ecosystem for working with HEALPix data, depending on what you need to do:
* The [astropy-healpix](https://astropy-healpix.readthedocs.io/en/latest/index.html) coordinated package is a BSD-licensed implementation of HEALPix which focuses on being able to convert celestial coordinates to HEALPix indices and vice-versa, as well as providing a few other low-level functions.
* The [reproject](https://reproject.readthedocs.io/en/stable/) coordinated package (which we've already looked at) includes functions for converting from/to HEALPix maps.
* The [HiPS](https://hips.readthedocs.io/en/latest/) affiliated package implements suport for the [HiPS](http://aladin.u-strasbg.fr/hips/) scheme for storing data that is based on HEALPix.
In this tutorial, we will take a look at the two first one of these, but we encourage you to learn more about HiPS too!
<section class="objectives panel panel-warning">
<div class="panel-heading">
<h2><span class="fa fa-certificate"></span> Objectives</h2>
</div>
<div class="panel-body">
<ul>
<li>Convert between celestial coordinates and HEALPix indices</li>
<li>Find the boundaries of HEALPix pixels</li>
<li>Find healpix pixels close to a position</li>
<li>Reproject a HEALPix map to a standard projection</li>
</ul>
</div>
</section>
## Documentation
This notebook only shows a subset of the functionality in astropy-healpix and reproject. For more information about the features presented below as well as other available features, you can read the
[astropy-healpix](https://astropy-healpix.readthedocs.io/en/latest/index.html) and the [reproject](https://reproject.readthedocs.io/en/stable/) documentation.
```
%matplotlib inline
import matplotlib.pyplot as plt
plt.rc('image', origin='lower')
plt.rc('figure', figsize=(10, 6))
```
## Data
For this tutorial, we will be using a downsampled version of the Planck HFI 857Ghz map which is stored as a HEALPix map ([data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits](data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits)).
## Using astropy-healpix
To start off, we can open the HEALPix file (which is a FITS file) with astropy.io.fits:
```
from astropy.io import fits
hdulist = fits.open('data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits')
hdulist.info()
```
The HEALPix map values are stored in HDU 1. This HDU also contains useful header information that helps us understand how to interpret the HEALPix values:
```
hdulist[1].header['NSIDE']
hdulist[1].header['ORDERING']
hdulist[1].header['COORDSYS']
```
With this information we can now construct a ``HEALPix`` object:
```
from astropy_healpix import HEALPix
from astropy.coordinates import Galactic
hp = HEALPix(nside=hdulist[1].header['NSIDE'],
order=hdulist[1].header['ORDERING'],
frame=Galactic())
```
We can then use this object to manipulate the HEALPix map. To start off, we can find out what the coordinates of specific pixels are:
```
hp.healpix_to_skycoord([13322, 2231, 66432])
```
and vice-versa:
```
from astropy.coordinates import SkyCoord
hp.skycoord_to_healpix(SkyCoord.from_name('M31'))
```
You can also find out what the boundaries of a pixel are:
```
edge = hp.boundaries_skycoord(649476, step=100)
edge
```
The ``step`` argument controls how many points to sample along the edge of the pixel. The result should be a polygon:
```
plt.plot(edge[0].l.deg, edge[0].b.deg)
```
You can find all HEALPix pixels within a certain radius of a known position:
```
from astropy import units as u
hp.cone_search_skycoord(SkyCoord.from_name('M31'), radius=1 * u.deg)
```
And finally you can interpolate the map at specific coordinates:
```
hp.interpolate_bilinear_skycoord(SkyCoord.from_name('M31'), hdulist[1].data['I_STOKES'])
```
<section class="challenge panel panel-success">
<div class="panel-heading">
<h2><span class="fa fa-pencil"></span> Challenge</h2>
</div>
<div class="panel-body">
<ol>
<li>Find the mean value of I_STOKES within 2 degrees of M42</li>
<li>Use astropy.coordinates to check that all the pixels returned by the cone search are indeed within 2 degrees of M42 (if not, why not? Hint: check the documentation of <a href="https://astropy-healpix.readthedocs.io/en/latest/api/astropy_healpix.HEALPix.html#astropy_healpix.HEALPix.cone_search_skycoord">cone_search_skycoord()</a>)</li>
</ol>
</div>
</section>
```
#1
import numpy as np
M42 = SkyCoord.from_name('M42')
m42_pixels = hp.cone_search_skycoord(M42, radius=2 * u.deg)
print(np.mean(hdulist[1].data['I_STOKES'][m42_pixels]))
#2
m42_cone_search_coords = hp.healpix_to_skycoord(m42_pixels)
separation = m42_cone_search_coords.separation(M42).degree
_ = plt.hist(separation, bins=50)
```
## Using reproject for HEALPix data
The reproject package is useful for HEALPix data to convert a HEALPix map to a regular projection, and vice-versa. For example, let's define a simple all-sky Plate-Caree WCS:
```
from astropy.wcs import WCS
wcs = WCS(naxis=2)
wcs.wcs.ctype = 'GLON-CAR', 'GLAT-CAR'
wcs.wcs.crval = 0, 0
wcs.wcs.crpix = 180.5, 90.5
wcs.wcs.cdelt = -1, 1
```
We can now use [reproject_from_healpix](https://reproject.readthedocs.io/en/stable/api/reproject.reproject_from_healpix.html#reproject.reproject_from_healpix) to convert the HEALPix map to this header:
```
from reproject import reproject_from_healpix
array, footprint = reproject_from_healpix('data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits',
wcs, shape_out=(180, 360))
plt.imshow(array, vmax=100)
```
You can also use [reproject_to_healpix](https://reproject.readthedocs.io/en/stable/api/reproject.reproject_to_healpix.html#reproject.reproject_to_healpix) to convert a regular map to a HEALPix array.
<section class="challenge panel panel-success">
<div class="panel-heading">
<h2><span class="fa fa-pencil"></span> Challenge</h2>
</div>
<div class="panel-body">
<ol>
<li>Reproject the HFI HEALPix map to the projection of the GAIA point source density map as well as the IRAS map that we used in previous tutorials.</li>
<li>Visualize the results using WCSAxes and optionally the image normalization options.</li>
</ol>
</div>
</section>
```
#1
header_gaia = fits.getheader('data/LMCDensFits1k.fits')
header_irsa = fits.getheader('data/ISSA_100_LMC.fits')
array_gaia, _ = reproject_from_healpix('data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits',
header_gaia)
array_irsa, _ = reproject_from_healpix('data/HFI_SkyMap_857_2048_R1.10_nominal_ZodiCorrected_lowres.fits',
header_irsa)
#2
from astropy.visualization import simple_norm
ax = plt.subplot(projection=WCS(header_gaia))
im =ax.imshow(array_gaia, cmap='plasma',
norm=simple_norm(array_gaia, stretch='sqrt', percent=99.5))
plt.colorbar(im)
ax.grid()
ax.set_xlabel('Galactic Longitude')
ax.set_ylabel('Galactic Latitude')
```
<center><i>This notebook was written by <a href="https://aperiosoftware.com/">Aperio Software Ltd.</a> © 2019, and is licensed under a <a href="https://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License (CC BY 4.0)</a></i></center>

|
github_jupyter
|
# Datasets for the book
Here we provide links to the datasets used in the book.
Important Notes:
1. Note that these datasets are provided on external servers by third parties
2. Due to security issues with github you will have to cut and paste FTP links (they are not provided as clickable URLs)
# Python and the Surrounding Software Ecology
### Interfacing with R via rpy2
* sequence.index
Please FTP from this URL(cut and paste)
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/historical_data/former_toplevel/sequence.index
# Next-generation Sequencing (NGS)
## Working with modern sequence formats
* SRR003265.filt.fastq.gz
Please FTP from this URL (cut and paste)
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA18489/sequence_read/SRR003265.filt.fastq.gz
## Working with BAM files
* NA18490_20_exome.bam
Please FTP from this URL (cut and paste)
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA18489/exome_alignment/NA18489.chrom20.ILLUMINA.bwa.YRI.exome.20121211.bam
* NA18490_20_exome.bam.bai
Please FTP from this URL (cut and paste)
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA18489/exome_alignment/NA18489.chrom20.ILLUMINA.bwa.YRI.exome.20121211.bam.bai
## Analyzing data in Variant Call Format (VCF)
* tabix link:
ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20130502/supporting/vcf_with_sample_level_annotation/ALL.chr22.phase3_shapeit2_mvncall_integrated_v5_extra_anno.20130502.genotypes.vcf.gz
# Genomics
### Working with high-quality reference genomes
* [falciparum.fasta](http://plasmodb.org/common/downloads/release-9.3/Pfalciparum3D7/fasta/data/PlasmoDB-9.3_Pfalciparum3D7_Genome.fasta)
### Dealing with low low-quality genome references
* gambiae.fa.gz
Please FTP from this URL (cut and paste)
ftp://ftp.vectorbase.org/public_data/organism_data/agambiae/Genome/agambiae.CHROMOSOMES-PEST.AgamP3.fa.gz
* [atroparvus.fa.gz](https://www.vectorbase.org/download/anopheles-atroparvus-ebroscaffoldsaatre1fagz)
### Traversing genome annotations
* [gambiae.gff3.gz](http://www.vectorbase.org/download/anopheles-gambiae-pestbasefeaturesagamp42gff3gz)
# PopGen
### Managing datasets with PLINK
* [hapmap.map.bz2](http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/hapmap3/plink_format/draft_2/hapmap3_r2_b36_fwd.consensus.qc.poly.map.bz2)
* [hapmap.ped.bz2](http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/hapmap3/plink_format/draft_2/hapmap3_r2_b36_fwd.consensus.qc.poly.ped.bz2)
* [relationships.txt](http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/hapmap3/plink_format/draft_2/relationships_w_pops_121708.txt)
# PDB
### Parsing mmCIF files with Biopython
* [1TUP.cif](http://www.rcsb.org/pdb/download/downloadFile.do?fileFormat=cif&compression=NO&structureId=1TUP)
# Python for Big genomics datasets
### Setting the stage for high-performance computing
These are the exact same files as _Managing datasets with PLINK_ above
### Programing with lazyness
* SRR003265_1.filt.fastq.gz Please ftp from this URL (cut and paste):
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA18489/sequence_read/SRR003265_1.filt.fastq.gz
* SRR003265_2.filt.fastq.gz Please ftp from this URL (cut and paste):
ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/data/NA18489/sequence_read/SRR003265_2.filt.fastq.gz
|
github_jupyter
|
# Table of Contents
<p><div class="lev1 toc-item"><a href="#Python-Basics-with-Numpy-(optional-assignment)" data-toc-modified-id="Python-Basics-with-Numpy-(optional-assignment)-1"><span class="toc-item-num">1 </span>Python Basics with Numpy (optional assignment)</a></div><div class="lev2 toc-item"><a href="#About-iPython-Notebooks" data-toc-modified-id="About-iPython-Notebooks-11"><span class="toc-item-num">1.1 </span>About iPython Notebooks</a></div><div class="lev2 toc-item"><a href="#1---Building-basic-functions-with-numpy" data-toc-modified-id="1---Building-basic-functions-with-numpy-12"><span class="toc-item-num">1.2 </span>1 - Building basic functions with numpy</a></div><div class="lev3 toc-item"><a href="#1.1---sigmoid-function,-np.exp()" data-toc-modified-id="1.1---sigmoid-function,-np.exp()-121"><span class="toc-item-num">1.2.1 </span>1.1 - sigmoid function, np.exp()</a></div><div class="lev3 toc-item"><a href="#1.2---Sigmoid-gradient" data-toc-modified-id="1.2---Sigmoid-gradient-122"><span class="toc-item-num">1.2.2 </span>1.2 - Sigmoid gradient</a></div><div class="lev3 toc-item"><a href="#1.3---Reshaping-arrays" data-toc-modified-id="1.3---Reshaping-arrays-123"><span class="toc-item-num">1.2.3 </span>1.3 - Reshaping arrays</a></div><div class="lev3 toc-item"><a href="#1.4---Normalizing-rows" data-toc-modified-id="1.4---Normalizing-rows-124"><span class="toc-item-num">1.2.4 </span>1.4 - Normalizing rows</a></div><div class="lev3 toc-item"><a href="#1.5---Broadcasting-and-the-softmax-function" data-toc-modified-id="1.5---Broadcasting-and-the-softmax-function-125"><span class="toc-item-num">1.2.5 </span>1.5 - Broadcasting and the softmax function</a></div><div class="lev2 toc-item"><a href="#2)-Vectorization" data-toc-modified-id="2)-Vectorization-13"><span class="toc-item-num">1.3 </span>2) Vectorization</a></div><div class="lev3 toc-item"><a href="#2.1-Implement-the-L1-and-L2-loss-functions" data-toc-modified-id="2.1-Implement-the-L1-and-L2-loss-functions-131"><span class="toc-item-num">1.3.1 </span>2.1 Implement the L1 and L2 loss functions</a></div>
# Python Basics with Numpy (optional assignment)
Welcome to your first assignment. This exercise gives you a brief introduction to Python. Even if you've used Python before, this will help familiarize you with functions we'll need.
**Instructions:**
- You will be using Python 3.
- Avoid using for-loops and while-loops, unless you are explicitly told to do so.
- Do not modify the (# GRADED FUNCTION [function name]) comment in some cells. Your work would not be graded if you change this. Each cell containing that comment should only contain one function.
- After coding your function, run the cell right below it to check if your result is correct.
**After this assignment you will:**
- Be able to use iPython Notebooks
- Be able to use numpy functions and numpy matrix/vector operations
- Understand the concept of "broadcasting"
- Be able to vectorize code
Let's get started!
## About iPython Notebooks ##
iPython Notebooks are interactive coding environments embedded in a webpage. You will be using iPython notebooks in this class. You only need to write code between the ### START CODE HERE ### and ### END CODE HERE ### comments. After writing your code, you can run the cell by either pressing "SHIFT"+"ENTER" or by clicking on "Run Cell" (denoted by a play symbol) in the upper bar of the notebook.
We will often specify "(≈ X lines of code)" in the comments to tell you about how much code you need to write. It is just a rough estimate, so don't feel bad if your code is longer or shorter.
**Exercise**: Set test to `"Hello World"` in the cell below to print "Hello World" and run the two cells below.
```
### START CODE HERE ### (≈ 1 line of code)
test = "Hello World"
### END CODE HERE ###
print ("test: " + test)
```
**Expected output**:
test: Hello World
<font color='blue'>
**What you need to remember**:
- Run your cells using SHIFT+ENTER (or "Run cell")
- Write code in the designated areas using Python 3 only
- Do not modify the code outside of the designated areas
## 1 - Building basic functions with numpy ##
Numpy is the main package for scientific computing in Python. It is maintained by a large community (www.numpy.org). In this exercise you will learn several key numpy functions such as np.exp, np.log, and np.reshape. You will need to know how to use these functions for future assignments.
### 1.1 - sigmoid function, np.exp() ###
Before using np.exp(), you will use math.exp() to implement the sigmoid function. You will then see why np.exp() is preferable to math.exp().
**Exercise**: Build a function that returns the sigmoid of a real number x. Use math.exp(x) for the exponential function.
**Reminder**:
$sigmoid(x) = \frac{1}{1+e^{-x}}$ is sometimes also known as the logistic function. It is a non-linear function used not only in Machine Learning (Logistic Regression), but also in Deep Learning.
<img src="images/Sigmoid.png" style="width:500px;height:228px;">
To refer to a function belonging to a specific package you could call it using package_name.function(). Run the code below to see an example with math.exp().
```
# GRADED FUNCTION: basic_sigmoid
import math
def basic_sigmoid(x):
"""
Compute sigmoid of x.
Arguments:
x -- A scalar
Return:
s -- sigmoid(x)
"""
### START CODE HERE ### (≈ 1 line of code)
s = 1 / (1 + math.exp(-x))
### END CODE HERE ###
return s
basic_sigmoid(3)
```
**Expected Output**:
<table style = "width:40%">
<tr>
<td>** basic_sigmoid(3) **</td>
<td>0.9525741268224334 </td>
</tr>
</table>
Actually, we rarely use the "math" library in deep learning because the inputs of the functions are real numbers. In deep learning we mostly use matrices and vectors. This is why numpy is more useful.
```
### One reason why we use "numpy" instead of "math" in Deep Learning ###
x = [1, 2, 3]
# basic_sigmoid(x) # you will see this give an error when you run it, because x is a vector.
```
In fact, if $ x = (x_1, x_2, ..., x_n)$ is a row vector then $np.exp(x)$ will apply the exponential function to every element of x. The output will thus be: $np.exp(x) = (e^{x_1}, e^{x_2}, ..., e^{x_n})$
```
import numpy as np
# example of np.exp
x = np.array([1, 2, 3])
print(np.exp(x)) # result is (exp(1), exp(2), exp(3))
```
Furthermore, if x is a vector, then a Python operation such as $s = x + 3$ or $s = \frac{1}{x}$ will output s as a vector of the same size as x.
```
# example of vector operation
x = np.array([1, 2, 3])
print (x + 3)
```
Any time you need more info on a numpy function, we encourage you to look at [the official documentation](https://docs.scipy.org/doc/numpy-1.10.1/reference/generated/numpy.exp.html).
You can also create a new cell in the notebook and write `np.exp?` (for example) to get quick access to the documentation.
**Exercise**: Implement the sigmoid function using numpy.
**Instructions**: x could now be either a real number, a vector, or a matrix. The data structures we use in numpy to represent these shapes (vectors, matrices...) are called numpy arrays. You don't need to know more for now.
$$ \text{For } x \in \mathbb{R}^n \text{, } sigmoid(x) = sigmoid\begin{pmatrix}
x_1 \\
x_2 \\
... \\
x_n \\
\end{pmatrix} = \begin{pmatrix}
\frac{1}{1+e^{-x_1}} \\
\frac{1}{1+e^{-x_2}} \\
... \\
\frac{1}{1+e^{-x_n}} \\
\end{pmatrix}\tag{1} $$
```
# GRADED FUNCTION: sigmoid
import numpy as np # this means you can access numpy functions by writing np.function() instead of numpy.function()
def sigmoid(x):
"""
Compute the sigmoid of x
Arguments:
x -- A scalar or numpy array of any size
Return:
s -- sigmoid(x)
"""
### START CODE HERE ### (≈ 1 line of code)
s = 1. / (1 + np.exp(-x))
### END CODE HERE ###
return s
x = np.array([1, 2, 3])
sigmoid(x)
```
**Expected Output**:
<table>
<tr>
<td> **sigmoid([1,2,3])**</td>
<td> array([ 0.73105858, 0.88079708, 0.95257413]) </td>
</tr>
</table>
### 1.2 - Sigmoid gradient
As you've seen in lecture, you will need to compute gradients to optimize loss functions using backpropagation. Let's code your first gradient function.
**Exercise**: Implement the function sigmoid_grad() to compute the gradient of the sigmoid function with respect to its input x. The formula is: $$sigmoid\_derivative(x) = \sigma'(x) = \sigma(x) (1 - \sigma(x))\tag{2}$$
You often code this function in two steps:
1. Set s to be the sigmoid of x. You might find your sigmoid(x) function useful.
2. Compute $\sigma'(x) = s(1-s)$
```
# GRADED FUNCTION: sigmoid_derivative
def sigmoid_derivative(x):
"""
Compute the gradient (also called the slope or derivative) of the sigmoid function with respect to its input x.
You can store the output of the sigmoid function into variables and then use it to calculate the gradient.
Arguments:
x -- A scalar or numpy array
Return:
ds -- Your computed gradient.
"""
### START CODE HERE ### (≈ 2 lines of code)
s = sigmoid(x)
ds = s * (1-s)
### END CODE HERE ###
return ds
x = np.array([1, 2, 3])
print ("sigmoid_derivative(x) = " + str(sigmoid_derivative(x)))
```
**Expected Output**:
<table>
<tr>
<td> **sigmoid_derivative([1,2,3])**</td>
<td> [ 0.19661193 0.10499359 0.04517666] </td>
</tr>
</table>
### 1.3 - Reshaping arrays ###
Two common numpy functions used in deep learning are [np.shape](https://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.shape.html) and [np.reshape()](https://docs.scipy.org/doc/numpy/reference/generated/numpy.reshape.html).
- X.shape is used to get the shape (dimension) of a matrix/vector X.
- X.reshape(...) is used to reshape X into some other dimension.
For example, in computer science, an image is represented by a 3D array of shape $(length, height, depth = 3)$. However, when you read an image as the input of an algorithm you convert it to a vector of shape $(length*height*3, 1)$. In other words, you "unroll", or reshape, the 3D array into a 1D vector.
<img src="images/image2vector_kiank.png" style="width:500px;height:300;">
**Exercise**: Implement `image2vector()` that takes an input of shape (length, height, 3) and returns a vector of shape (length\*height\*3, 1). For example, if you would like to reshape an array v of shape (a, b, c) into a vector of shape (a*b,c) you would do:
``` python
v = v.reshape((v.shape[0]*v.shape[1], v.shape[2])) # v.shape[0] = a ; v.shape[1] = b ; v.shape[2] = c
```
- Please don't hardcode the dimensions of image as a constant. Instead look up the quantities you need with `image.shape[0]`, etc.
```
# GRADED FUNCTION: image2vector
def image2vector(image):
"""
Argument:
image -- a numpy array of shape (length, height, depth)
Returns:
v -- a vector of shape (length*height*depth, 1)
"""
### START CODE HERE ### (≈ 1 line of code)
v = image.reshape(image.size, 1)
### END CODE HERE ###
return v
# This is a 3 by 3 by 2 array, typically images will be (num_px_x, num_px_y,3) where 3 represents the RGB values
image = np.array([[[ 0.67826139, 0.29380381],
[ 0.90714982, 0.52835647],
[ 0.4215251 , 0.45017551]],
[[ 0.92814219, 0.96677647],
[ 0.85304703, 0.52351845],
[ 0.19981397, 0.27417313]],
[[ 0.60659855, 0.00533165],
[ 0.10820313, 0.49978937],
[ 0.34144279, 0.94630077]]])
print ("image2vector(image) = " + str(image2vector(image)))
```
**Expected Output**:
<table style="width:100%">
<tr>
<td> **image2vector(image)** </td>
<td> [[ 0.67826139]
[ 0.29380381]
[ 0.90714982]
[ 0.52835647]
[ 0.4215251 ]
[ 0.45017551]
[ 0.92814219]
[ 0.96677647]
[ 0.85304703]
[ 0.52351845]
[ 0.19981397]
[ 0.27417313]
[ 0.60659855]
[ 0.00533165]
[ 0.10820313]
[ 0.49978937]
[ 0.34144279]
[ 0.94630077]]</td>
</tr>
</table>
### 1.4 - Normalizing rows
Another common technique we use in Machine Learning and Deep Learning is to normalize our data. It often leads to a better performance because gradient descent converges faster after normalization. Here, by normalization we mean changing x to $ \frac{x}{\| x\|} $ (dividing each row vector of x by its norm).
For example, if $$x =
\begin{bmatrix}
0 & 3 & 4 \\
2 & 6 & 4 \\
\end{bmatrix}\tag{3}$$ then $$\| x\| = np.linalg.norm(x, axis = 1, keepdims = True) = \begin{bmatrix}
5 \\
\sqrt{56} \\
\end{bmatrix}\tag{4} $$and $$ x\_normalized = \frac{x}{\| x\|} = \begin{bmatrix}
0 & \frac{3}{5} & \frac{4}{5} \\
\frac{2}{\sqrt{56}} & \frac{6}{\sqrt{56}} & \frac{4}{\sqrt{56}} \\
\end{bmatrix}\tag{5}$$ Note that you can divide matrices of different sizes and it works fine: this is called broadcasting and you're going to learn about it in part 5.
**Exercise**: Implement normalizeRows() to normalize the rows of a matrix. After applying this function to an input matrix x, each row of x should be a vector of unit length (meaning length 1).
```
# GRADED FUNCTION: normalizeRows
def normalizeRows(x):
"""
Implement a function that normalizes each row of the matrix x (to have unit length).
Argument:
x -- A numpy matrix of shape (n, m)
Returns:
x -- The normalized (by row) numpy matrix. You are allowed to modify x.
"""
### START CODE HERE ### (≈ 2 lines of code)
# Compute x_norm as the norm 2 of x. Use np.linalg.norm(..., ord = 2, axis = ..., keepdims = True)
x_norm = np.linalg.norm(x, ord=2, axis=1, keepdims=True)
# Divide x by its norm.
x = x / x_norm
### END CODE HERE ###
return x
x = np.array([
[0, 3, 4],
[2, 6, 4]])
print("normalizeRows(x) = " + str(normalizeRows(x)))
```
**Expected Output**:
<table style="width:60%">
<tr>
<td> **normalizeRows(x)** </td>
<td> [[ 0. 0.6 0.8 ]
[ 0.13736056 0.82416338 0.54944226]]</td>
</tr>
</table>
**Note**:
In normalizeRows(), you can try to print the shapes of x_norm and x, and then rerun the assessment. You'll find out that they have different shapes. This is normal given that x_norm takes the norm of each row of x. So x_norm has the same number of rows but only 1 column. So how did it work when you divided x by x_norm? This is called broadcasting and we'll talk about it now!
### 1.5 - Broadcasting and the softmax function ####
A very important concept to understand in numpy is "broadcasting". It is very useful for performing mathematical operations between arrays of different shapes. For the full details on broadcasting, you can read the official [broadcasting documentation](http://docs.scipy.org/doc/numpy/user/basics.broadcasting.html).
**Exercise**: Implement a softmax function using numpy. You can think of softmax as a normalizing function used when your algorithm needs to classify two or more classes. You will learn more about softmax in the second course of this specialization.
**Instructions**:
- $ \text{for } x \in \mathbb{R}^{1\times n} \text{, } softmax(x) = softmax(\begin{bmatrix}
x_1 &&
x_2 &&
... &&
x_n
\end{bmatrix}) = \begin{bmatrix}
\frac{e^{x_1}}{\sum_{j}e^{x_j}} &&
\frac{e^{x_2}}{\sum_{j}e^{x_j}} &&
... &&
\frac{e^{x_n}}{\sum_{j}e^{x_j}}
\end{bmatrix} $
- $\text{for a matrix } x \in \mathbb{R}^{m \times n} \text{, $x_{ij}$ maps to the element in the $i^{th}$ row and $j^{th}$ column of $x$, thus we have: }$ $$softmax(x) = softmax\begin{bmatrix}
x_{11} & x_{12} & x_{13} & \dots & x_{1n} \\
x_{21} & x_{22} & x_{23} & \dots & x_{2n} \\
\vdots & \vdots & \vdots & \ddots & \vdots \\
x_{m1} & x_{m2} & x_{m3} & \dots & x_{mn}
\end{bmatrix} = \begin{bmatrix}
\frac{e^{x_{11}}}{\sum_{j}e^{x_{1j}}} & \frac{e^{x_{12}}}{\sum_{j}e^{x_{1j}}} & \frac{e^{x_{13}}}{\sum_{j}e^{x_{1j}}} & \dots & \frac{e^{x_{1n}}}{\sum_{j}e^{x_{1j}}} \\
\frac{e^{x_{21}}}{\sum_{j}e^{x_{2j}}} & \frac{e^{x_{22}}}{\sum_{j}e^{x_{2j}}} & \frac{e^{x_{23}}}{\sum_{j}e^{x_{2j}}} & \dots & \frac{e^{x_{2n}}}{\sum_{j}e^{x_{2j}}} \\
\vdots & \vdots & \vdots & \ddots & \vdots \\
\frac{e^{x_{m1}}}{\sum_{j}e^{x_{mj}}} & \frac{e^{x_{m2}}}{\sum_{j}e^{x_{mj}}} & \frac{e^{x_{m3}}}{\sum_{j}e^{x_{mj}}} & \dots & \frac{e^{x_{mn}}}{\sum_{j}e^{x_{mj}}}
\end{bmatrix} = \begin{pmatrix}
softmax\text{(first row of x)} \\
softmax\text{(second row of x)} \\
... \\
softmax\text{(last row of x)} \\
\end{pmatrix} $$
```
# GRADED FUNCTION: softmax
def softmax(x):
"""Calculates the softmax for each row of the input x.
Your code should work for a row vector and also for matrices of shape (n, m).
Argument:
x -- A numpy matrix of shape (n,m)
Returns:
s -- A numpy matrix equal to the softmax of x, of shape (n,m)
"""
### START CODE HERE ### (≈ 3 lines of code)
# Apply exp() element-wise to x. Use np.exp(...).
x_exp = np.exp(x)
# Create a vector x_sum that sums each row of x_exp. Use np.sum(..., axis = 1, keepdims = True).
x_sum = np.sum(x_exp, axis=1, keepdims=True)
# Compute softmax(x) by dividing x_exp by x_sum. It should automatically use numpy broadcasting.
s = x_exp / x_sum
### END CODE HERE ###
return s
x = np.array([
[9, 2, 5, 0, 0],
[7, 5, 0, 0 ,0]])
print("softmax(x) = " + str(softmax(x)))
```
**Expected Output**:
<table style="width:60%">
<tr>
<td> **softmax(x)** </td>
<td> [[ 9.80897665e-01 8.94462891e-04 1.79657674e-02 1.21052389e-04
1.21052389e-04]
[ 8.78679856e-01 1.18916387e-01 8.01252314e-04 8.01252314e-04
8.01252314e-04]]</td>
</tr>
</table>
**Note**:
- If you print the shapes of x_exp, x_sum and s above and rerun the assessment cell, you will see that x_sum is of shape (2,1) while x_exp and s are of shape (2,5). **x_exp/x_sum** works due to python broadcasting.
Congratulations! You now have a pretty good understanding of python numpy and have implemented a few useful functions that you will be using in deep learning.
<font color='blue'>
**What you need to remember:**
- np.exp(x) works for any np.array x and applies the exponential function to every coordinate
- the sigmoid function and its gradient
- image2vector is commonly used in deep learning
- np.reshape is widely used. In the future, you'll see that keeping your matrix/vector dimensions straight will go toward eliminating a lot of bugs.
- numpy has efficient built-in functions
- broadcasting is extremely useful
## 2) Vectorization
In deep learning, you deal with very large datasets. Hence, a non-computationally-optimal function can become a huge bottleneck in your algorithm and can result in a model that takes ages to run. To make sure that your code is computationally efficient, you will use vectorization. For example, try to tell the difference between the following implementations of the dot/outer/elementwise product.
```
import time
x1 = [9, 2, 5, 0, 0, 7, 5, 0, 0, 0, 9, 2, 5, 0, 0]
x2 = [9, 2, 2, 9, 0, 9, 2, 5, 0, 0, 9, 2, 5, 0, 0]
### CLASSIC DOT PRODUCT OF VECTORS IMPLEMENTATION ###
tic = time.process_time()
dot = 0
for i in range(len(x1)):
dot+= x1[i]*x2[i]
toc = time.process_time()
print ("dot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC OUTER PRODUCT IMPLEMENTATION ###
tic = time.process_time()
outer = np.zeros((len(x1),len(x2))) # we create a len(x1)*len(x2) matrix with only zeros
for i in range(len(x1)):
for j in range(len(x2)):
outer[i,j] = x1[i]*x2[j]
toc = time.process_time()
print ("outer = " + str(outer) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC ELEMENTWISE IMPLEMENTATION ###
tic = time.process_time()
mul = np.zeros(len(x1))
for i in range(len(x1)):
mul[i] = x1[i]*x2[i]
toc = time.process_time()
print ("elementwise multiplication = " + str(mul) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### CLASSIC GENERAL DOT PRODUCT IMPLEMENTATION ###
W = np.random.rand(3,len(x1)) # Random 3*len(x1) numpy array
tic = time.process_time()
gdot = np.zeros(W.shape[0])
for i in range(W.shape[0]):
for j in range(len(x1)):
gdot[i] += W[i,j]*x1[j]
toc = time.process_time()
print ("gdot = " + str(gdot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
x1 = [9, 2, 5, 0, 0, 7, 5, 0, 0, 0, 9, 2, 5, 0, 0]
x2 = [9, 2, 2, 9, 0, 9, 2, 5, 0, 0, 9, 2, 5, 0, 0]
### VECTORIZED DOT PRODUCT OF VECTORS ###
tic = time.process_time()
dot = np.dot(x1,x2)
toc = time.process_time()
print ("dot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED OUTER PRODUCT ###
tic = time.process_time()
outer = np.outer(x1,x2)
toc = time.process_time()
print ("outer = " + str(outer) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED ELEMENTWISE MULTIPLICATION ###
tic = time.process_time()
mul = np.multiply(x1,x2)
toc = time.process_time()
print ("elementwise multiplication = " + str(mul) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
### VECTORIZED GENERAL DOT PRODUCT ###
tic = time.process_time()
dot = np.dot(W,x1)
toc = time.process_time()
print ("gdot = " + str(dot) + "\n ----- Computation time = " + str(1000*(toc - tic)) + "ms")
```
As you may have noticed, the vectorized implementation is much cleaner and more efficient. For bigger vectors/matrices, the differences in running time become even bigger.
**Note** that `np.dot()` performs a matrix-matrix or matrix-vector multiplication. This is different from `np.multiply()` and the `*` operator (which is equivalent to `.*` in Matlab/Octave), which performs an element-wise multiplication.
### 2.1 Implement the L1 and L2 loss functions
**Exercise**: Implement the numpy vectorized version of the L1 loss. You may find the function abs(x) (absolute value of x) useful.
**Reminder**:
- The loss is used to evaluate the performance of your model. The bigger your loss is, the more different your predictions ($ \hat{y} $) are from the true values ($y$). In deep learning, you use optimization algorithms like Gradient Descent to train your model and to minimize the cost.
- L1 loss is defined as:
$$\begin{align*} & L_1(\hat{y}, y) = \sum_{i=0}^m|y^{(i)} - \hat{y}^{(i)}| \end{align*}\tag{6}$$
```
# GRADED FUNCTION: L1
def L1(yhat, y):
"""
Arguments:
yhat -- vector of size m (predicted labels)
y -- vector of size m (true labels)
Returns:
loss -- the value of the L1 loss function defined above
"""
### START CODE HERE ### (≈ 1 line of code)
loss = np.sum(np.abs((y - yhat)))
### END CODE HERE ###
return loss
yhat = np.array([.9, 0.2, 0.1, .4, .9])
y = np.array([1, 0, 0, 1, 1])
print("L1 = " + str(L1(yhat,y)))
```
**Expected Output**:
<table style="width:20%">
<tr>
<td> **L1** </td>
<td> 1.1 </td>
</tr>
</table>
**Exercise**: Implement the numpy vectorized version of the L2 loss. There are several way of implementing the L2 loss but you may find the function np.dot() useful. As a reminder, if $x = [x_1, x_2, ..., x_n]$, then `np.dot(x,x)` = $\sum_{j=0}^n x_j^{2}$.
- L2 loss is defined as $$\begin{align*} & L_2(\hat{y},y) = \sum_{i=0}^m(y^{(i)} - \hat{y}^{(i)})^2 \end{align*}\tag{7}$$
```
# GRADED FUNCTION: L2
def L2(yhat, y):
"""
Arguments:
yhat -- vector of size m (predicted labels)
y -- vector of size m (true labels)
Returns:
loss -- the value of the L2 loss function defined above
"""
### START CODE HERE ### (≈ 1 line of code)
loss = np.sum(np.square(yhat - y))
### END CODE HERE ###
return loss
yhat = np.array([.9, 0.2, 0.1, .4, .9])
y = np.array([1, 0, 0, 1, 1])
print("L2 = " + str(L2(yhat,y)))
```
**Expected Output**:
<table style="width:20%">
<tr>
<td> **L2** </td>
<td> 0.43 </td>
</tr>
</table>
Congratulations on completing this assignment. We hope that this little warm-up exercise helps you in the future assignments, which will be more exciting and interesting!
<font color='blue'>
**What to remember:**
- Vectorization is very important in deep learning. It provides computational efficiency and clarity.
- You have reviewed the L1 and L2 loss.
- You are familiar with many numpy functions such as np.sum, np.dot, np.multiply, np.maximum, etc...
|
github_jupyter
|
```
import tensorflow as tf
from matplotlib import pylab
from tensorflow.examples.tutorials.mnist import input_data
import numpy as np
# Required for Data downaload and preparation
import struct
import gzip
import os
from six.moves.urllib.request import urlretrieve
```
## Defining Hyperparameters
Here we define the set of hyperparameters we're going to you in our example. These hyperparameters include `batch_size`, train dataset size (`n_train`), different layers in our CNN (`cnn_layer_ids`). You can find descriptions of each hyperparameter in comments.
```
batch_size = 100 # This is the typical batch size we've been using
image_size = 28 # This is the width/height of a single image
# Number of color channels in an image. These are black and white images
n_channels = 1
# Number of different digits we have images for (i.e. classes)
n_classes = 10
n_train = 55000 # Train dataset size
n_valid = 5000 # Validation dataset size
n_test = 10000 # Test dataset size
# Layers in the CNN in the order from input to output
cnn_layer_ids = ['conv1','pool1','conv2','pool2','fulcon1','softmax']
# Hyperparameters of each layer (e.g. filter size of each convolution layer)
layer_hyperparameters = {'conv1':{'weight_shape':[3,3,n_channels,16],'stride':[1,1,1,1],'padding':'SAME'},
'pool1':{'kernel_shape':[1,3,3,1],'stride':[1,2,2,1],'padding':'SAME'},
'conv2':{'weight_shape':[3,3,16,32],'stride':[1,1,1,1],'padding':'SAME'},
'pool2':{'kernel_shape':[1,3,3,1],'stride':[1,2,2,1],'padding':'SAME'},
'fulcon1':{'weight_shape':[7*7*32,128]},
'softmax':{'weight_shape':[128,n_classes]}
}
```
## Defining Inputs and Outputs
Here we define input and output placeholders required to process a batch of data. We will use the same placeholders for all training, validation and testing data as all of them are processed in same size batches.
```
# Inputs (Images) and Outputs (Labels) Placeholders
tf_inputs = tf.placeholder(shape=[batch_size, image_size, image_size, n_channels],dtype=tf.float32,name='tf_mnist_images')
tf_labels = tf.placeholder(shape=[batch_size, n_classes],dtype=tf.float32,name='tf_mnist_labels')
```
## Defining Model Parameters and Other Variables
Here we define various TensorFlow variables required for the following computations. These includes a global step variable (to decay learning rate) and weights and biases of each layer of the CNN.
```
# Global step for decaying the learning rate
global_step = tf.Variable(0,trainable=False)
# Initializing the variables
layer_weights = {}
layer_biases = {}
for layer_id in cnn_layer_ids:
if 'pool' not in layer_id:
layer_weights[layer_id] = tf.Variable(initial_value=tf.random_normal(shape=layer_hyperparameters[layer_id]['weight_shape'],
stddev=0.02,dtype=tf.float32),name=layer_id+'_weights')
layer_biases[layer_id] = tf.Variable(initial_value=tf.random_normal(shape=[layer_hyperparameters[layer_id]['weight_shape'][-1]],
stddev=0.01,dtype=tf.float32),name=layer_id+'_bias')
print('Variables initialized')
```
## Defining Inference of the CNN
Here we define the computations starting from input placeholder (`tf_inputs`) and then computing the hidden activations for each of the layers found in `cnn_layer_ids` (i.e. convolution/pooling and fulcon layers) and their respective parameters (`layer_hyperparamters`). At the final layer (`softmax`) we do not apply an activation function as for the rest of the layers, but obtain the unnormalized logit values without any activation function.
```
# Calculating Logits
h = tf_inputs
for layer_id in cnn_layer_ids:
if 'conv' in layer_id:
# For each convolution layer, compute the output by using conv2d function
# This operation results in a [batch_size, output_height, output_width, out_channels]
# sized 4 dimensional tensor
h = tf.nn.conv2d(h,layer_weights[layer_id],layer_hyperparameters[layer_id]['stride'],
layer_hyperparameters[layer_id]['padding']) + layer_biases[layer_id]
h = tf.nn.relu(h)
elif 'pool' in layer_id:
# For each pooling layer, compute the output by max pooling
# This operation results in a [batch_size, output_height, output_width, out_channels]
# sized 4 dimensional tensor
h = tf.nn.max_pool(h, layer_hyperparameters[layer_id]['kernel_shape'],layer_hyperparameters[layer_id]['stride'],
layer_hyperparameters[layer_id]['padding'])
elif layer_id == 'fulcon1':
# At the first fulcon layer we need to reshape the 4 dimensional output to a
# 2 dimensional output to be processed by fully connected layers
# Note this should only done once, before
# computing the output of the first fulcon layer
h = tf.reshape(h,[batch_size,-1])
h = tf.matmul(h,layer_weights[layer_id]) + layer_biases[layer_id]
h = tf.nn.relu(h)
elif layer_id == 'softmax':
# Note that here we do not perform the same reshaping we did for fulcon1
# We only perform the matrix multiplication on previous output
h = tf.matmul(h,layer_weights[layer_id]) + layer_biases[layer_id]
print('Calculated logits')
tf_logits = h
```
## Defining Loss
We use softmax cross entropy loss to optimize the parameters of the model.
```
# Calculating the softmax cross entropy loss with the computed logits and true labels (one hot encoded)
tf_loss = tf.nn.softmax_cross_entropy_with_logits_v2(logits=tf_logits,labels=tf_labels)
print('Loss defined')
```
## Model Parameter Optimizer
We define an exponentially decaying learning rate and an optimizer to optimize the parameters.
```
# Optimization
# Here we define the function to decay the learning rate exponentially.
# Everytime the global step increases the learning rate decreases
tf_learning_rate = tf.train.exponential_decay(learning_rate=0.001,global_step=global_step,decay_rate=0.5,decay_steps=1,staircase=True)
tf_loss_minimize = tf.train.RMSPropOptimizer(learning_rate=tf_learning_rate, momentum=0.9).minimize(tf_loss)
print('Loss minimization defined')
```
## Defining Predictions
We get the predictiosn out by applying a softmax activation to the logits. Additionally we define a global step increment function and will be increase every time the validation accuracy plateus.
```
tf_predictions = tf.nn.softmax(tf_logits)
print('Prediction defined')
tf_tic_toc = tf.assign(global_step, global_step + 1)
```
## Define Accuracy
A simple function to calculate accuracy for a given set of labels and predictions.
```
def accuracy(predictions,labels):
'''
Accuracy of a given set of predictions of size (N x n_classes) and
labels of size (N x n_classes)
'''
return np.sum(np.argmax(predictions,axis=1)==np.argmax(labels,axis=1))*100.0/labels.shape[0]
```
## Lolading Data
Here we download (if needed) the MNIST dataset and, perform reshaping and normalization. Also we conver the labels to one hot encoded vectors.
```
def maybe_download(url, filename, expected_bytes, force=False):
"""Download a file if not present, and make sure it's the right size."""
if force or not os.path.exists(filename):
print('Attempting to download:', filename)
filename, _ = urlretrieve(url + filename, filename)
print('\nDownload Complete!')
statinfo = os.stat(filename)
if statinfo.st_size == expected_bytes:
print('Found and verified', filename)
else:
raise Exception(
'Failed to verify ' + filename + '. Can you get to it with a browser?')
return filename
def read_mnist(fname_img, fname_lbl, one_hot=False):
print('\nReading files %s and %s'%(fname_img, fname_lbl))
# Processing images
with gzip.open(fname_img) as fimg:
magic, num, rows, cols = struct.unpack(">IIII", fimg.read(16))
print(num,rows,cols)
img = (np.frombuffer(fimg.read(num*rows*cols), dtype=np.uint8).reshape(num, rows, cols,1)).astype(np.float32)
print('(Images) Returned a tensor of shape ',img.shape)
#img = (img - np.mean(img)) /np.std(img)
img *= 1.0 / 255.0
# Processing labels
with gzip.open(fname_lbl) as flbl:
# flbl.read(8) reads upto 8 bytes
magic, num = struct.unpack(">II", flbl.read(8))
lbl = np.frombuffer(flbl.read(num), dtype=np.int8)
if one_hot:
one_hot_lbl = np.zeros(shape=(num,10),dtype=np.float32)
one_hot_lbl[np.arange(num),lbl] = 1.0
print('(Labels) Returned a tensor of shape: %s'%lbl.shape)
print('Sample labels: ',lbl[:10])
if not one_hot:
return img, lbl
else:
return img, one_hot_lbl
# Download data if needed
url = 'http://yann.lecun.com/exdb/mnist/'
# training data
maybe_download(url,'train-images-idx3-ubyte.gz',9912422)
maybe_download(url,'train-labels-idx1-ubyte.gz',28881)
# testing data
maybe_download(url,'t10k-images-idx3-ubyte.gz',1648877)
maybe_download(url,'t10k-labels-idx1-ubyte.gz',4542)
# Read the training and testing data
train_inputs, train_labels = read_mnist('train-images-idx3-ubyte.gz', 'train-labels-idx1-ubyte.gz',True)
test_inputs, test_labels = read_mnist('t10k-images-idx3-ubyte.gz', 't10k-labels-idx1-ubyte.gz',True)
valid_inputs, valid_labels = train_inputs[-n_valid:,:,:,:], train_labels[-n_valid:,:]
train_inputs, train_labels = train_inputs[:-n_valid,:,:,:], train_labels[:-n_valid,:]
print('\nTrain size: ', train_inputs.shape[0])
print('\nValid size: ', valid_inputs.shape[0])
print('\nTest size: ', test_inputs.shape[0])
```
## Data Generators for MNIST
Here we have the logic to iterate through each training, validation and testing datasets, in `batch_size` size strides.
```
train_index, valid_index, test_index = 0,0,0
def get_train_batch(images, labels, batch_size):
global train_index
batch = images[train_index:train_index+batch_size,:,:,:], labels[train_index:train_index+batch_size,:]
train_index = (train_index + batch_size)%(images.shape[0] - batch_size)
return batch
def get_valid_batch(images, labels, batch_size):
global valid_index
batch = images[valid_index:valid_index+batch_size,:,:,:], labels[valid_index:valid_index+batch_size,:]
valid_index = (valid_index + batch_size)%(images.shape[0] - batch_size)
return batch
def get_test_batch(images, labels, batch_size):
global test_index
batch = images[test_index:test_index+batch_size,:,:,:], labels[test_index:test_index+batch_size,:]
test_index = (test_index + batch_size)%(images.shape[0] - batch_size)
return batch
```
## Visualizing MNIST Results
Here we define a function to collect correctly and incorrectly classified samples to visualize later. Visualizing such samples will help us to understand why the CNN incorrectly classified certain samples.
```
# Makes sure we only collect 10 samples for each
correct_fill_index, incorrect_fill_index = 0,0
# Visualization purposes
correctly_predicted = np.empty(shape=(10,28,28,1),dtype=np.float32)
correct_predictions = np.empty(shape=(10,n_classes),dtype=np.float32)
incorrectly_predicted = np.empty(shape=(10,28,28,1),dtype=np.float32)
incorrect_predictions = np.empty(shape=(10,n_classes),dtype=np.float32)
def collect_samples(test_batch_predictions,test_images, test_labels):
global correctly_predicted, correct_predictions
global incorrectly_predicted, incorrect_predictions
global correct_fill_index, incorrect_fill_index
correct_indices = np.where(np.argmax(test_batch_predictions,axis=1)==np.argmax(test_labels,axis=1))[0]
incorrect_indices = np.where(np.argmax(test_batch_predictions,axis=1)!=np.argmax(test_labels,axis=1))[0]
if correct_indices.size>0 and correct_fill_index<10:
print('\nCollecting Correctly Predicted Samples')
chosen_index = np.random.choice(correct_indices)
correctly_predicted[correct_fill_index,:,:,:]=test_images[chosen_index,:].reshape(1,image_size,image_size,n_channels)
correct_predictions[correct_fill_index,:]=test_batch_predictions[chosen_index,:]
correct_fill_index += 1
if incorrect_indices.size>0 and incorrect_fill_index<10:
print('Collecting InCorrectly Predicted Samples')
chosen_index = np.random.choice(incorrect_indices)
incorrectly_predicted[incorrect_fill_index,:,:,:]=test_images[chosen_index,:].reshape(1,image_size,image_size,n_channels)
incorrect_predictions[incorrect_fill_index,:]=test_batch_predictions[chosen_index,:]
incorrect_fill_index += 1
```
## Running MNIST Classification
Here we train our CNN on MNIST data for `n_epochs` epochs. Each epoch we train the CNN with the full training dataset. Then we calculate the validation accuracy, according to which we decay the learning rate. Finally, each epoch we calculate the test accuracy which is computed using an independent test set. This code should run under 10 minutes if you run on a decent GPU and should reach to a test accuracy of about ~95%
```
# Parameters related to learning rate decay
# counts how many times the validation accuracy has not increased consecutively for
v_acc_not_increased_for = 0
# if the above count is above this value, decrease the learning rate
v_acc_threshold = 3
# currently recorded best validation accuracy
max_v_acc = 0.0
config = tf.ConfigProto(allow_soft_placement=True)
# Good practice to use this to avoid any surprising errors thrown by TensorFlow
config.gpu_options.allow_growth = True
config.gpu_options.per_process_gpu_memory_fraction = 0.9 # Making sure Tensorflow doesn't overflow the GPU
n_epochs = 25 # Number of epochs the training runs for
session = tf.InteractiveSession(config=config)
# Initialize all variables
tf.global_variables_initializer().run()
# Run training loop
for epoch in range(n_epochs):
loss_per_epoch = []
# Training phase. We train with all training data
# processing one batch at a time
for i in range(n_train//batch_size):
# Get the next batch of MNIST dataset
batch = get_train_batch(train_inputs, train_labels, batch_size)
# Run TensorFlow opeartions
l,_ = session.run([tf_loss,tf_loss_minimize],feed_dict={tf_inputs: batch[0].reshape(batch_size,image_size,image_size,n_channels),
tf_labels: batch[1]})
# Add the loss value to a list
loss_per_epoch.append(l)
print('Average loss in epoch %d: %.5f'%(epoch,np.mean(loss_per_epoch)))
# Validation phase. We compute validation accuracy
# processing one batch at a time
valid_accuracy_per_epoch = []
for i in range(n_valid//batch_size):
# Get the next validation data batch
vbatch_images,vbatch_labels = get_valid_batch(valid_inputs, valid_labels, batch_size)
# Compute validation predictions
valid_batch_predictions = session.run(
tf_predictions,feed_dict={tf_inputs: vbatch_images}
)
# Compute and add the validation accuracy to a python list
valid_accuracy_per_epoch.append(accuracy(valid_batch_predictions,vbatch_labels))
# Compute and print average validation accuracy
mean_v_acc = np.mean(valid_accuracy_per_epoch)
print('\tAverage Valid Accuracy in epoch %d: %.5f'%(epoch,np.mean(valid_accuracy_per_epoch)))
# Learning rate decay logic
if mean_v_acc > max_v_acc:
max_v_acc = mean_v_acc
else:
v_acc_not_increased_for += 1
# Time to decrease learning rate
if v_acc_not_increased_for >= v_acc_threshold:
print('\nDecreasing Learning rate\n')
session.run(tf_tic_toc) # Increase global_step
v_acc_not_increased_for = 0
# Testing phase. We compute test accuracy
# processing one batch at a time
accuracy_per_epoch = []
for i in range(n_test//batch_size):
btest_images, btest_labels = get_test_batch(test_inputs, test_labels, batch_size)
test_batch_predictions = session.run(tf_predictions,feed_dict={tf_inputs: btest_images})
accuracy_per_epoch.append(accuracy(test_batch_predictions,btest_labels))
# Collect samples for visualization only in the last epoch
if epoch==n_epochs-1:
collect_samples(test_batch_predictions, btest_images, btest_labels)
print('\tAverage Test Accuracy in epoch %d: %.5f\n'%(epoch,np.mean(accuracy_per_epoch)))
session.close()
```
## Visualizing Predictions
Let us see how when our CNN did when it comes to predictions
```
# Defining the plot related settings
pylab.figure(figsize=(25,20)) # in inches
width=0.5 # Width of a bar in the barchart
padding = 0.05 # Padding between two bars
labels = list(range(0,10)) # Class labels
# Defining X axis
x_axis = np.arange(0,10)
# We create 4 rows and 7 column set of subplots
# We choose these to put the titles in
# First row middle
pylab.subplot(4, 7, 4)
pylab.title('Correctly Classified Samples',fontsize=24)
# Second row middle
pylab.subplot(4, 7,11)
pylab.title('Softmax Predictions for Correctly Classified Samples',fontsize=24)
# For 7 steps
for sub_i in range(7):
# Draw the top row (digit images)
pylab.subplot(4, 7, sub_i + 1)
pylab.imshow(np.squeeze(correctly_predicted[sub_i]),cmap='gray')
pylab.axis('off')
# Draw the second row (prediction bar chart)
pylab.subplot(4, 7, 7 + sub_i + 1)
pylab.bar(x_axis + padding, correct_predictions[sub_i], width)
pylab.ylim([0.0,1.0])
pylab.xticks(x_axis, labels)
# Set titles for the third and fourth rows
pylab.subplot(4, 7, 18)
pylab.title('Incorrectly Classified Samples',fontsize=26)
pylab.subplot(4, 7,25)
pylab.title('Softmax Predictions for Incorrectly Classified Samples',fontsize=24)
# For 7 steps
for sub_i in range(7):
# Draw the third row (incorrectly classified digit images)
pylab.subplot(4, 7, 14 + sub_i + 1)
pylab.imshow(np.squeeze(incorrectly_predicted[sub_i]),cmap='gray')
pylab.axis('off')
# Draw the fourth row (incorrect predictions bar chart)
pylab.subplot(4, 7, 21 + sub_i + 1)
pylab.bar(x_axis + padding, incorrect_predictions[sub_i], width)
pylab.ylim([0.0,1.0])
pylab.xticks(x_axis, labels)
# Save the figure
pylab.savefig('mnist_results.png')
pylab.show()
```
|
github_jupyter
|
```
import numpy as np
import random
import sys
from scipy.special import expit as sigmoid
training_data_path = sys.argv[1]
testing_data_path = sys.argv[2]
output_path = sys.argv[3]
batch_size = int(sys.argv[4])
n0 = float(sys.argv[5])
activation = sys.argv[6]
hidden_layers_sizes = []
for i in range(7,len(sys.argv)):
hidden_layers_sizes.append(int(sys.argv[i]))
# training_data_path = "../data/devnagri_train.csv"
# testing_data_path = "../data/devnagri_test_public.csv"
# output_path = "../data/nn/a/cs1160328.txt"
# batch_size = 512
# n0 = 0.01
# activation = 'sigmoid'
# hidden_layers_sizes = [100]
def relu(x):
return (x>0) * x
def tanh(x):
return np.tanh(x)
def reluPrime(x):
return (x>0)+0
def tanhPrime(x):
return 1 - np.power(x,2)
def sigmoidPrime(x):
return x * (1 - x)
def exp_normalize(x):
b = np.amax(x,axis=1,keepdims = True)
y = np.exp(x - b)
return y / y.sum(axis=1,keepdims=True)
class NeuralNetwork:
def __init__(self,input_size,output_size,hidden_layers_sizes, activation):
self.weights = []
self.biases = []
if(activation == 'relu'):
self.activation = relu
self.activationPrime = reluPrime
elif(activation == 'tanh'):
self.activation = tanh
self.activationPrime = tanhPrime
else:
self.activation = sigmoid
self.activationPrime = sigmoidPrime
self.input_size = input_size
self.output_size = output_size
self.hiddent_layers_sizes = hidden_layers_sizes
prev_layer_count = input_size
for i in range(len(hidden_layers_sizes) + 1):
if i==len(hidden_layers_sizes):
self.weights.append(np.random.rand(prev_layer_count, output_size)/100)
self.biases.append(np.random.rand(1, output_size)/100)
else:
hidden_layer_count = hidden_layers_sizes[i]
self.weights.append(np.random.rand(prev_layer_count, hidden_layer_count)/100)
self.biases.append(np.random.rand(1, hidden_layer_count)/100)
prev_layer_count = hidden_layer_count
def train(self,inpX,inpY,batch_size,n0,max_iterations):
max_examples = inpX.shape[0]
max_possible_iterations = int(0.5 + max_examples / batch_size)
num_hidden_layers = len(self.weights) - 1
count = 0
lr = n0
totLoss = 0
prevAvgLoss = sys.float_info.max
epoch = 0
for n in range(max_iterations):
# Forming Mini Batches
i_eff = n%max_possible_iterations
# Updating Learning Rate
if (i_eff == 0 and n!=0):
avgLoss = totLoss/max_possible_iterations
if(np.absolute(avgLoss - prevAvgLoss) < 0.0001 * prevAvgLoss):
stopCount += 1
if stopCount > 1:
break
else:
stopCount = 0
if(avgLoss >= prevAvgLoss):
count += 1
lr = n0 / np.sqrt(count+1)
print("Epoch = ",epoch," Average Loss = ",avgLoss," New Learning Rate = ",lr)
epoch += 1
prevAvgLoss = avgLoss
totLoss = 0
outputs = []
if i_eff != max_possible_iterations - 1:
X = inpX[i_eff*batch_size: (i_eff+1)*batch_size]
Y = inpY[i_eff*batch_size: (i_eff+1)*batch_size]
else:
X = inpX[i_eff*batch_size:]
Y = inpY[i_eff*batch_size:]
# Neural Network Forward Propagation
outputs.append(X)
prev_layer_output = X
for i in range(num_hidden_layers + 1):
weight = self.weights[i]
bias = self.biases[i]
if i == num_hidden_layers:
prev_layer_output = sigmoid(prev_layer_output.dot(weight) + bias)
else:
prev_layer_output = self.activation(prev_layer_output.dot(weight) + bias)
outputs.append(prev_layer_output)
# Backpropagation
dWs = []
dbs = []
y_onehot = np.zeros((Y.shape[0],self.output_size))
y_onehot[range(Y.shape[0]),Y] = 1
for i in range(num_hidden_layers + 1,0,-1):
if i == num_hidden_layers + 1:
delta = (outputs[i] - y_onehot).dot(2/Y.shape[0]) * sigmoidPrime(outputs[i])
else:
delta = delta.dot(self.weights[i].T) * self.activationPrime(outputs[i])
dW = (outputs[i-1].T).dot(delta)
dWs.append(dW)
dbs.append(np.sum(delta,axis=0,keepdims=True))
if (n%100 == 0):
loss_ = np.sum(np.power(outputs[-1] - y_onehot,2) )/Y.shape[0]
labels_ = np.argmax(outputs[-1],axis = 1)
accuracy_ = 100 * np.sum(labels_ == Y)/Y.shape[0]
print("Iteration ",n,"\tLoss = ",loss_,"\tAccuracy = ",accuracy_,"%")
dWs.reverse()
dbs.reverse()
# Gradient Descent Parameter Update
for i in range(len(dWs)):
self.weights[i] += dWs[i].dot(-1 * lr)
self.biases[i] += dbs[i].dot(-1 * lr)
loss = np.sum(np.power(outputs[-1] - y_onehot,2) )/Y.shape[0]
totLoss += loss
def predict(self,X):
return self.forward_run(X)
def forward_run(self,X):
prev_layer_output = X
num_hidden_layers = len(self.weights) - 1
for i in range(num_hidden_layers + 1):
weight = self.weights[i]
bias = self.biases[i]
if i == num_hidden_layers:
probabilities = sigmoid(prev_layer_output.dot(weight) + bias)
labels = np.argmax(probabilities,axis = 1)
return labels
else:
prev_layer_output = self.activation(prev_layer_output.dot(weight) + bias)
def load_data(path,avg,std):
if avg is None:
input_data = np.loadtxt(open(path, "rb"), delimiter=",")
Y = input_data[:,0].copy()
X = input_data[:,1:].copy()
avg = np.average(X,axis=0)
X = X - avg
std = np.std(X,axis=0)
std[(std == 0)] = 1
X = X / std
return X,Y,avg,std
else:
input_data = np.loadtxt(open(path, "rb"), delimiter=",")
X = input_data[:,1:].copy()
X = (X - avg)/std
return X
inpX,Y,avg,std = load_data(training_data_path,None,None)
X = inpX.copy()
input_size = X.shape[1]
output_size = int(np.amax(Y))+1
num_examples = X.shape[0]
max_iterations = int(40*(num_examples/batch_size))
if(max_iterations < 25000):
max_iterations = 25000
network = NeuralNetwork(input_size,output_size,hidden_layers_sizes,activation)
network.train(X,Y.astype(int),batch_size,n0,max_iterations)
predictions = network.predict(X.copy())
print("Accuraccy on Training Data = ",100 * np.sum(predictions == Y)/Y.shape[0])
# print("Average of predictions on Training Data = ",np.average(predictions))
testX = load_data(testing_data_path,avg,std)
predictions = network.predict(testX)
np.savetxt(output_path,predictions,fmt="%i")
```
|
github_jupyter
|
```
from pyesasky import ESASkyWidget
from pyesasky import Catalogue
from pyesasky import CatalogueDescriptor
from pyesasky import MetadataDescriptor
from pyesasky import MetadataType
from pyesasky import CooFrame
# instantiating pyESASky instance
esasky = ESASkyWidget()
# loading pyESASky instance
esasky
# Go to the Cosmos field in ESASky (as resolved by SIMBAD):
esasky.goToTargetName('Cosmos Field')
#####################################################
# EX.1 creating a user defined catalogue on the fly #
#####################################################
catalogue = Catalogue('test catalogue name', CooFrame.FRAME_J2000, '#ee2345', 10)
# adding sources to the catalogue
catalogue.addSource('source name A', '150.44963', '2.24640', 1, [{"name":"Flux 1", "value":"10.5", "type":"STRING" },{"name":"Flux 2", "value":"1.7", "type":"STRING" }])
catalogue.addSource('source name B', '150.54963', '2.34640', 2, [{"name":"Flux 1", "value":"11.5", "type":"STRING" },{"name":"Flux 2", "value":"2.7", "type":"STRING" }])
catalogue.addSource('source name c', '150.34963', '2.44640', 3, [{"name":"Flux 1", "value":"12.5", "type":"STRING" },{"name":"Flux 2", "value":"0.7", "type":"STRING" }])
# overlay catalogue in pyESASky
esasky.overlayCatalogueWithDetails(catalogue)
############################################
# EX.2 importing a catalogue from CSV file #
############################################
# CatalogueDescriptor('<catName>', '<HTMLcolor>', <lineWidth>, '<idColumn>', '<nameColumn>', '<RAColumn>', '<DecColumn>', Metadata)
# where:
# - <catName> : name of the catalogue that will be used in pyESASky as label
# - <HTMLcolor> : HTML color. It could be a "Color name", "Hex color code" or "RGB color code"
# - <lineWidth> : width used to draw sources. From 1 to 10
# - <idColumn> : name of the column containing a unique identifier for sources if any. None if not applicable
# - <nameColumn> : name of the column with the name of the source
# - <RAColumn> : name of the RA column in degrees
# - <DecColumn> : name of the Dec column in degrees
# - Metadata : list of pyesasky.pyesasky.MetadataDescriptor in case it has been defined. [] otherwise.
catalogueDesc =CatalogueDescriptor('my test', 'yellow', 5, 'id', 'name', 'ra', 'dec', [])
# parse, import and overlay a catalogue from a CSV
esasky.overlayCatalogueFromCSV('./testcat', ',', catalogueDesc, 'J2000')
###################################################################
# EX.3 importing a catalogue from AstropyTable using Gaia archive #
###################################################################
from astroquery.gaia import Gaia
job = Gaia.launch_job("select top 10\
ra, dec, source_id, designation, ref_epoch,ra_dec_corr,astrometric_n_obs_al,matched_observations,duplicated_source,phot_variable_flag \
from gaiadr2.gaia_source order by source_id", verbose=True)
myGaiaData = job.get_results()
print(myGaiaData)
job.get_data()
# overlayCatalogueFromAstropyTable('<catName>', '<cooFrame>', <HTMLcolor>, '<(astropy.table)>', '<RAColumn>', '<DecColumn>', '<nameColumn>')
# where:
# - <catName> : name of the catalogue that will be used in pyESASky as label
# - <HTMLcolor> : HTML color. It could be a "Color name", "Hex color code" or "RGB color code"
# - <lineWidth> : width used to draw sources. From 1 to 10
# - <idColumn> : name of the column containing a unique identifier for sources if any. None if not applicable
# - <nameColumn> : name of the column with the name of the source
# - <RAColumn> : name of the RA column in degrees
# - <DecColumn> : name of the Dec column in degrees
esasky.overlayCatalogueFromAstropyTable('Gaia DR2', 'J2000', '#a343ff', 5, myGaiaData, '','','')
# Import the VizieR Astroquery module
from astroquery.vizier import Vizier
# Search for 'The XMM-Newton survey of the COSMOS field (Brusa+, 2010)':
catalog_list = Vizier.find_catalogs('Brusa+, 2010')
print({k:v.description for k,v in catalog_list.items()})
# Get the above list of catalogues:
Vizier.ROW_LIMIT = -1
catalogs = Vizier.get_catalogs(catalog_list.keys())
print(catalogs)
# Access one table:
Brusa = catalogs['J/ApJ/716/348/table2']
print(Brusa)
# Visualise the table in ESASky:
esasky.overlayCatalogueFromAstropyTable('Brusa', CooFrame.FRAME_J2000, '#00ff00', 5, Brusa, 'RAJ2000','DEJ2000','Name')
# Go to the LMC in ESASky (as resolved by SIMBAD):
esasky.goToTargetName('LMC')
# Search for 'The HIZOA-S survey':
catalog_list2 = Vizier.find_catalogs('HIZOA-S survey 2016') #HIZOA-S survey 2016
print({k:v.description for k,v in catalog_list2.items()})
# Get the above list of catalogues:
Vizier.ROW_LIMIT = -1
# Vizier.COLUMN_LIMIT = 20 Can't find the way to get all the columns rather than just the default columns. Going to try the TAP+ module
catalog = Vizier.get_catalogs(catalog_list2.keys())
print(catalog)
# Access the catalogue table:
HIZOA = catalog['J/AJ/151/52/table2'] #
print(HIZOA)
# Visualise the table in ESASky:
###### NOTE: NOT PLOTTING GALACTIC COORDS CORRECTLY
esasky.overlayCatalogueFromAstropyTable('HIZOA', CooFrame.FRAME_GALACTIC, '#0000ff', 7, HIZOA, 'GLON','GLAT','HIZOA')
# TRYING THE SAME BUT USING THE TAP/TAP+ ASTROQUERY MODULE:
# Import the TAP/TAP+ Astroquery module
from astroquery.utils.tap.core import TapPlus
vizier = TapPlus(url="http://tapvizier.u-strasbg.fr/TAPVizieR/tap")
tables = vizier.load_tables(only_names=True)
for table in (tables):
print(table.get_qualified_name())
#ONLY TAP+ compatible, so doesn't seem to work
table = vizier.load_table('viz7."J/AJ/128/16/table2"')
for column in (table.get_columns()):
print(column.get_name())
# This works in TOPCAT to download the whole table: SELECT * FROM "J/AJ/128/16/table2"
# This also works in TOPCAT : SELECT * FROM viz7."J/AJ/128/16/table2"
job = vizier.launch_job("SELECT * FROM "'viz7."J/AJ/128/16/table2"'"")
#This also works:
#job = vizier.launch_job("SELECT * FROM "+str('viz7."J/AJ/128/16/table2"')+"")
print(job)
Koribalski = job.get_results()
print(Koribalski['HIPASS', 'RAJ2000', 'DEJ2000'])
# Visualise the table in ESASky:
esasky.overlayCatalogueFromAstropyTable('Koribalski', CooFrame.FRAME_J2000, '#ff0000', 6, Koribalski, 'RAJ2000','DEJ2000','HIPASS')
```
|
github_jupyter
|
```
%pylab
%matplotlib inline
%run pdev notebook
```
# Radiosonde SONDE
```
ident = "SONDE"
plt.rcParams['figure.figsize'] = [12.0, 6.0]
plt.rcParams['lines.linewidth'] = 2
plt.rcParams['font.size'] = 15
yplevs = np.array([10,100,200,300,400,500,700,925])*100
save = True
!mkdir -p figures
rt.load_config()
rt.config
isonde = rt.cls.Radiosonde(ident)
#
# All the data available
#
isonde.list_store()
```
## Load Data
```
# close=False -> stay on disk,
# =True -> load to memory
close = False
```
### ERA5
```
if False:
isonde.add('ERA5', filename='ERA5_*.nc', cfunits=True, close=close, verbose=1)
if False:
isonde.add('ERA5_meta', filename='*_ERA5_station.nc', cfunits=True, close=close, verbose=1)
```
### ERA Interim
```
if False:
isonde.add('ERAI', filename='ERAI_*.nc', cfunits=True, close=close, verbose=1)
```
### IGRA v2
```
if False:
isonde.add('IGRAv2', cfunits=True, close=close, verbose=1)
```
### Upper Air Database (UADB)
```
if False:
isonde.add('UADB', cfunits=True, close=close, verbose=1)
```
### JRA-55
```
if False:
isonde.add('JRA55', close=close, verbose=1)
```
### CERA-20C
```
if False:
isonde.add('CERA20C', close=close, verbose=1)
```
### Standardized Combined Data
```
idata = None
#
# ERA5
#
if isonde.in_store('dataE5JC'):
isonde.add('dataE5JC', verbose=1)
idata = isonde.data.dataE5JC
#
# ERA Interim
#
if isonde.in_store('dataEIJC') and idata is None:
isonde.add('dataEIJC', verbose=1)
idata = isonde.data.dataEIJC
#
# IGRA
#
if isonde.in_store('dataIE5JC') and idata is None:
isonde.add('dataIE5JC', verbose=1)
idata = isonde.data.dataIE5JC
```
### Experiment Data
```
isonde.list_store(pattern='exp')
ivar = 'dpd'
version = 'v1'
isrc = 'mars5'
ires = 'era5'
expdata = None
#
# ERA5
#
if isonde.in_store('exp{}{}_{}_{}.nc'.format(ivar,version,isrc,ires)):
isonde.add('exp{}{}_{}_{}'.format(ivar,version,isrc,ires), verbose=1)
expdata = isonde.data['exp{}{}_{}_{}'.format(ivar,version,isrc,ires)]
#
# ERA Interim
#
if expdata is None:
isrc = 'marsi'
ires = 'erai'
if isonde.in_store('exp{}{}_{}_{}.nc'.format(ivar,version,isrc,ires)):
isonde.add('exp{}{}_{}_{}'.format(ivar,version,isrc,ires), verbose=1)
expdata = isonde.data['exp{}{}_{}_{}'.format(ivar,version,isrc,ires)]
#
# JRA55
#
if expdata is None:
isrc = 'mars5'
ires = 'jra55'
if isonde.in_store('exp{}{}_{}_{}.nc'.format(ivar,version,isrc,ires)):
isonde.add('exp{}{}_{}_{}'.format(ivar,version,isrc,ires), verbose=1)
expdata = isonde.data['exp{}{}_{}_{}'.format(ivar,version,isrc,ires)]
if idata is None:
print("No data ?")
exit()
#
# Some definitions
#
times = [0, 12]
start = '1979'
ende = '2019'
period = slice(start, ende)
period_str = "%s-%s" % (start, ende)
#
# Subset to only that period
#
idata = idata.sel(time=period, hour=times)
```
## Station Map
```
rt.plot.map.station_class(isonde, states=True, rivers=True, land=True, lakes=True)
if save:
savefig('figures/%s_station.png' % ident)
```
# Data Availability
```
dpdvars = []
tvars = []
for jvar in list(idata.data_vars):
if 'dpd_' in jvar:
if not any([i in jvar for i in ['err','_fg_','snht']]):
dpdvars.append(jvar)
if 't_' in jvar:
if not any([i in jvar for i in ['err','_fg_','snht']]):
tvars.append(jvar)
print(dpdvars)
print(tvars)
```
## Dewpoint depression
```
counts = idata.reset_coords()[dpdvars].count('time').sum('hour').to_dataframe()
counts.index /= 100.
counts.plot()
xticks(yplevs/100)
grid()
title("%s Counts %s" % (ident, period_str))
ylabel("Total counts [1]")
if save:
savefig('figures/%s_dpd_counts.png' % ident)
```
## Temperature
```
counts = idata.reset_coords()[tvars].count('time').sum('hour').to_dataframe()
counts.index /= 100.
counts.plot()
xticks(yplevs/100)
grid()
title("%s Counts %s" % (ident, period_str))
ylabel("Total counts [1]")
if save:
savefig('figures/%s_t_counts.png' % ident)
```
## Annual
```
counts = idata.reset_coords()[dpdvars].count('plev').resample(time='A').sum().to_dataframe()
n = len(idata.hour.values)
f, ax = subplots(n,1, sharex=True)
ax[0].set_title("%s Annual counts %s" % (ident, period_str))
for i,ihour in enumerate(idata.hour.values):
counts.xs(ihour, level=0).plot(grid=True, ax=ax[i], legend=True if i==0 else False)
ax[i].set_ylabel("%02d Z" % (ihour))
ax[i].set_xlabel('Years')
tight_layout()
if save:
savefig('figures/%s_dpd_ancounts.png' % (ident))
counts = idata.reset_coords()[tvars].count('plev').resample(time='A').sum().to_dataframe()
n = len(idata.hour.values)
f, ax = subplots(n,1, sharex=True)
ax[0].set_title("%s Annual counts %s" % (ident, period_str))
for i,ihour in enumerate(idata.hour.values):
counts.xs(ihour, level=0).plot(grid=True, ax=ax[i], legend=True if i==0 else False)
ax[i].set_ylabel("%02d Z" % (ihour))
ax[i].set_xlabel('Years')
tight_layout()
if save:
savefig('figures/%s_t_ancounts.png' % (ident))
```
# Dewpoint depression
```
obs = 'dpd_{}'.format(isrc)
hdim = 'hour'
for ihour in idata[hdim].values:
rt.plot.time.var(idata[obs].sel(**{hdim:ihour}), dim='time', lev='plev',
title='%s %s Radiosonde at %02d Z' % (ident, obs, ihour))
```
# Temperature
```
obs = 't_{}'.format(isrc)
hdim = 'hour'
for ihour in idata[hdim].values:
rt.plot.time.var(idata[obs].sel(**{hdim:ihour}), dim='time', lev='plev',
title='%s %s Radiosonde at %02d Z' % (ident, obs, ihour))
```
# Comparison with Reanalysis
```
dim = 'time'
hdim = 'hour'
lev = 'plev'
ivar = 'dpd'
obs = '{}_{}'.format(ivar, isrc)
plotvars = []
#
# Select Variables
#
for jvar in list(idata.data_vars):
if '_' in jvar:
iname = jvar.split('_')[1]
if jvar == "%s_%s" %(ivar, iname):
plotvars += [jvar]
print(plotvars)
#
# Select Level
#
ipres=10000
#
# Plot
#
ylims = (np.round(idata[obs].min()), np.round(idata[obs].max()))
for i,j in idata[plotvars].groupby(hdim):
m = j.sel(**{lev:ipres}).resample(**{dim:'M'}).mean(dim)
f, ax = plt.subplots(figsize=(16,4))
for jvar in plotvars:
rt.plot.time.var(m[jvar], ax=ax, dim=dim, label=jvar.replace(ivar+'_',''))
ax.set_ylabel("%s [%s]" % (ivar, idata[jvar].attrs['units']))
ax.set_xlabel('Time [M]')
ax.set_title('%s %s Comparison %s %02dZ at %d hPa' %(ident, ivar, period_str, i, ipres/100))
ax.legend(ncol=len(plotvars))
ax.set_ylim(ylims)
tight_layout()
if save:
savefig('figures/%s_%s_comparison_%04d_%02dZ.png' % (ident, ivar, ipres/100, i))
```
## Departures
```
dim = 'time'
hdim = 'hour'
lev = 'plev'
ivar = 'dpd'
obs = '{}_{}'.format(ivar, isrc)
plotvars = []
#
# Select Variables
#
for jvar in list(idata.data_vars):
if '_' in jvar:
iname = jvar.split('_')[1]
if jvar == "%s_%s" %(ivar, iname):
plotvars += [jvar]
print(plotvars)
#
# Select Level
#
ipres=30000
#
# Plot
#
ylims = (-10,10) # Manual
for i,j in idata[plotvars].groupby(hdim):
m = j.sel(**{lev:ipres}).resample(**{dim:'M'}).mean(dim)
f, ax = plt.subplots(figsize=(16,4))
for jvar in plotvars:
if jvar == obs:
continue
rt.plot.time.var(m[obs] - m[jvar], ax=ax, dim=dim, label=jvar.replace(ivar+'_',''))
ax.set_ylabel("%s [%s]" % (ivar, idata[jvar].attrs['units']))
ax.set_xlabel('Time [M]')
ax.set_title('%s Departures %s (OBS-BG) %s %02dZ at %d hPa' %(ident, ivar, period_str, i, ipres/100))
ax.legend(ncol=len(plotvars))
ax.set_ylim(ylims)
tight_layout()
if save:
savefig('figures/%s_%s_dep_%04d_%02dZ.png' % (ident, ivar, ipres/100, i))
```
# Adjustment Process
```
if expdata is None:
#
# Make Experiments
#
expdata = idata.copy()
else:
expdata = expdata.sel(**{dim: period})
```
## SNHT
```
dim = 'time'
hdim = 'hour'
ivar = 'dpd'
obs = '{}_{}'.format(ivar, isrc)
res = '{}_{}'.format(ivar, ires)
#
# Execute SNHT ?
#
if not '{}_snht'.format(obs) in expdata.data_vars:
#
# Calculate SNHT values with Parameters (window and missing)
#
expdata = rt.bp.snht(expdata, var=obs, dep=res, dim=dim,
window=1460,
missing=600,
verbose=1)
#
# Apply Threshold (threshold) and detect Peaks
# allowed distances between peaks (dist)
# minimum requires significant levels (min_levels)
#
expdata = expdata.groupby(hdim).apply(rt.bp.apply_threshold,
threshold=50,
dist=730,
min_levels=3,
var=obs + '_snht',
dim=dim)
#
# Plot SNHT
#
for i,j in expdata.groupby(hdim):
ax = rt.plot.time.threshold(j[obs + '_snht'], dim=dim, lev=lev, logy=False,
title=" %s SNHT %s at %02dZ" % (ident, period_str, i),
figsize=(12,4),
yticklabels=yplevs)
rt.plot.time.breakpoints(j[obs + '_snht_breaks'], ax=ax, startend=True)
tight_layout()
if save:
savefig('figures/%s_%s_snht_%s_%02dZ.png' % (ident, obs, ires, i))
```
## Breakpoints
```
#
# Give Breakpoint Information
#
for i,j in expdata.groupby(hdim):
_=rt.bp.get_breakpoints(j[obs + '_snht_breaks'], dim=dim, verbose=1)
```
## Adjustments
```
dim = 'time'
hdim = 'hour'
ivar = 'dpd'
obs = '{}_{}'.format(ivar, isrc)
res = '{}_{}'.format(ivar, ires)
# plotvars = [i for i in expdata.data_vars if '_dep' in i]
adjvars = "{obs},{obs}_m,{obs}_q,{obs}_qa".format(obs=obs)
adjvars = adjvars.split(',')
print(adjvars)
missing = False
for jvar in adjvars:
if jvar not in expdata.data_vars:
missing = True
```
### Run standard adjustment process
```
if missing:
from detect import run_standard
expdata = run_standard(idata, obs, res, meanadj=True, qadj=True, qqadj=True, verbose=1)
```
## Breakpoint Stats
```
ipres=85000
#
# MEAN ADJ
#
bins = np.round(np.nanpercentile(np.ravel(expdata[obs].sel(**{lev:ipres}).values), [1,99]))
bins = np.arange(bins[0]-2,bins[1]+2,1)
for i,j in expdata.groupby(hdim):
rt.plot.breakpoints_histograms(j.sel(**{lev:ipres}),
obs, '{}_m'.format(obs), '{}_snht_breaks'.format(obs),
figsize=(18,8),
other_var=res,
bins=bins);
if save:
savefig('figures/%s_bhist_m_%s_%02dZ_%04dhPa.png' % (ident, ivar, i, ipres/100))
ipres=85000
#
# QUANTIL ADJ
#
bins = np.round(np.nanpercentile(np.ravel(expdata[obs].sel(**{lev:ipres}).values), [1,99]))
bins = np.arange(bins[0]-2,bins[1]+2,1)
for i,j in expdata.groupby(hdim):
rt.plot.breakpoints_histograms(j.sel(**{lev:ipres}),
obs, '{}_q'.format(obs), '{}_snht_breaks'.format(obs),
figsize=(18,8),
other_var=res,
bins=bins);
if save:
savefig('figures/%s_bhist_q_%s_%02dZ_%04dhPa.png' % (ident, ivar, i, ipres/100))
ipres=85000
#
# QUANTIL ADJ
#
bins = np.round(np.nanpercentile(np.ravel(expdata[obs].sel(**{lev:ipres}).values), [1,99]))
bins = np.arange(bins[0]-2,bins[1]+2,1)
for i,j in expdata.groupby(hdim):
rt.plot.breakpoints_histograms(j.sel(**{lev:ipres}),
obs, '{}_qa'.format(obs), '{}_snht_breaks'.format(obs),
figsize=(18,8),
other_var=res,
bins=bins);
if save:
savefig('figures/%s_bhist_qa_%s_%02dZ_%04dhPa.png' % (ident, ivar, i, ipres/100))
```
## Adjustment methods
```
bvar = '{}_snht_breaks'.format(obs)
#
# Select Level
#
ipres=30000
#
# Plot
#
ylims = np.round(np.nanpercentile(np.ravel(expdata[obs].sel(**{lev:ipres}).rolling(**{dim:30, 'center':True, 'min_periods':10}).mean().values), [1,99]))
ylims += [-2,2]
for i,j in expdata[adjvars].groupby(hdim):
m = j.sel(**{lev:ipres}).rolling(**{dim:30, 'center':True, 'min_periods':10}).mean()
f, ax = plt.subplots(figsize=(16,4))
for jvar in adjvars:
rt.plot.time.var(m[jvar], ax=ax, dim=dim, label=jvar[-1:].upper() if jvar != obs else ivar, ls='-' if jvar == obs else '--')
if bvar in expdata.data_vars:
rt.plot.time.breakpoints(expdata[bvar].sel(**{hdim:i}), ax=ax, color='k', lw=2, ls='--')
ax.set_ylabel("%s [%s]" % (ivar, expdata[jvar].attrs['units']))
ax.set_xlabel('Time [M]')
ax.set_title('%s Adjustments %s %s %02dZ at %d hPa' %(ident, ivar, period_str, i, ipres/100))
ax.legend(ncol=len(plotvars))
ax.set_ylim(ylims)
tight_layout()
if save:
savefig('figures/%s_%s_adj_%04d_%02dZ.png' % (ident, ivar, ipres/100, i))
```
# Analysis
```
#
# Monthly Means
#
variables = list(unique(dpdvars + tvars + adjvars))
for jvar in variables[:]:
if jvar not in expdata.data_vars:
variables.remove(jvar)
print(variables)
mdata = expdata[variables].resample(**{dim:'M'}).mean(keep_attrs=True)
```
## Trends
```
trends = rt.met.time.trend(mdata, period=period, dim=dim, only_slopes=True)
with xr.set_options(keep_attrs=True):
trends = trends*3650. # Trends per Decade
for jvar in trends.data_vars:
trends[jvar].attrs['units'] = trends[jvar].attrs['units'].replace('day','decade')
xlims = (np.round(trends.min().to_array().min()), np.round(trends.max().to_array().max()))
n = mdata[hdim].size
f,ax = rt.plot.init_fig_horizontal(n=n, ratios=tuple([2]*n), sharey=True)
for i, ihour in enumerate(trends[hdim].values):
for jvar in variables:
rt.plot.profile.var(trends[jvar].sel(**{hdim:ihour}), ax=ax[i], label=jvar[-1:].upper() if jvar != obs else ivar)
ax[i].set_title('%02d' % ihour)
ax[i].set_xlim(xlims)
ax[i].set_xlabel("%s [%s]" % (mdata[obs].attrs['standard_name'], trends[jvar].attrs['units']))
f.suptitle('%s %s Trends %s' % (ident, ivar.upper(), period_str))
if save:
savefig('figures/%s_trends_%s.png' % (ident, ivar))
```
## Statistics
```
from detect import skills_table
for jvar in mdata.data_vars:
if jvar == obs or jvar == res:
continue
_ , ytable = skills_table(mdata[obs], mdata[res], mdata[jvar])
print("#"*50)
print(ident, obs, res, jvar)
print(ytable)
print("#"*50)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/mancunian1792/causal_scene_generation/blob/master/causal_model/game_characters/GameCharacter_ImageClassification.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from google.colab import drive
drive.mount("/content/gdrive", force_remount=True)
import keras
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from keras.utils import to_categorical
from keras.preprocessing import image
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from keras.utils import to_categorical
from tqdm import tqdm
from skimage.transform import rotate
from skimage.util import random_noise
from skimage.filters import gaussian
root_path = 'gdrive/My Drive/causal_scene_generation/game_characters/'
train_path = root_path + 'train/'
test_path = root_path + 'test/'
train_images = train_path + 'images/'
test_images = test_path + 'images/'
train_csv = train_path + 'train.csv'
test_csv = test_path + 'test.csv'
def preprocess(imgPath, filePath):
images = []
# Transform each image in the imgPath and add it to the input array
data = pd.read_csv(filePath)
for imgFile in tqdm(data["filename"]):
imgFullPath = imgPath + imgFile + ".png"
img = image.load_img(imgFullPath, target_size=(400,400,3), grayscale=False)
img = image.img_to_array(img)
img = img/255
images.append(img)
features = np.array(images)
# Get the labels for each
target = data.drop(["filename"], axis=1)
return features, target
def augmentData(features, target):
augmented_features = []
augmented_target = []
for idx in tqdm(range(features.shape[0])):
augmented_features.append(features[idx])
augmented_features.append(rotate(features[idx], angle=45, mode = 'wrap'))
augmented_features.append(np.fliplr(features[idx]))
augmented_features.append(np.flipud(features[idx]))
augmented_features.append(random_noise(features[idx],var=0.2**2))
for i in range(5):
augmented_target.append(target.iloc[idx, :])
return np.asarray(augmented_features), pd.DataFrame(augmented_target, columns= target.columns)
x_train, y_train = preprocess(train_images, train_csv)
x_train_augment, y_train_augment = augmentData(x_train, y_train)
del x_train, y_train
x_test, y_test = preprocess(test_images, test_csv)
x_test, x_validate, y_test, y_validate = train_test_split(x_test, y_test, random_state = 3000, test_size = 0.2)
plt.imshow(x_validate[2])
# Size of vector is 64 * 64 * 3 -> resize ((64 *64*3), 1)
# (/255 )
# Convert to grayscale.->
# The output shape
op_shape = y_train_augment.shape[1]
model = Sequential()
model.add(Conv2D(filters=16, kernel_size=(5, 5), activation="relu", input_shape=(400,400,3)))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=32, kernel_size=(10, 10), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(10, 10), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(5, 5), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(op_shape, activation='sigmoid'))
model.summary()
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
model.fit(x_train_augment, y_train_augment, epochs=10, validation_data=(x_test, y_test), batch_size=64)
model.save(root_path+"model-both-images.hdf5")
prediction = model.predict(x_validate)
prediction[0]
del x_train_augment, y_train_augment, x_test, y_test
```
### Attempt 2 - Image Classification
This time, i am splitting the images and modify the labels. The image classification will try to predict the entity (actor/reactor), character(satyr/golem), type(1/2/3) and entity_doing (action/reaction) and entity_doing_type(Idle/Attacking/Hurt/Die/Walking/Taunt)
```
# Modify the labels (Do - encoding)
splits_path = root_path + 'splits/'
splits_images = splits_path + 'images/'
splits_dataset = splits_path + 'split_dataset.csv'
df = pd.read_csv(splits_dataset)
df["type"] = df.type.str.extract('(\d+)')
images = df["img_name"]
target = df.drop(["img_name"], axis=1)
target = pd.get_dummies(target)
def processSplitImages(imgPath, filenames):
images_data = []
for img in tqdm(filenames):
imgFullPath = imgPath + img + ".png"
img = image.load_img(imgFullPath, target_size=(400,400,3), grayscale=False)
img = image.img_to_array(img)
img = img/255
images_data.append(img)
features = np.array(images_data)
return features
img_features = processSplitImages(splits_images, images)
# Split into train and test . And then augment the train data.
features_train, features_test, target_train, target_test = train_test_split(img_features, target, stratify=target, test_size=0.2)
del img_features, target
# Augmenting train data -> Not able to allocate enough RAM
#feature_train_augmented, target_augmented = augmentData(features_train, target_train)
op_shape = target_train.shape[1]
model = Sequential()
model.add(Conv2D(filters=16, kernel_size=(5, 5), activation="relu", input_shape=(400,400,3)))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=32, kernel_size=(10, 10), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(10, 10), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(5, 5), activation='relu'))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(64, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(op_shape, activation='sigmoid'))
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
from keras.callbacks import ModelCheckpoint
filepath=root_path + "weights-{epoch:02d}-{val_accuracy:.3f}.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='val_accuracy',
verbose=1, mode='max')
callbacks_list = [checkpoint]
model.fit(features_train, target_train, epochs=10, validation_data=(features_test, target_test), batch_size=64, callbacks=callbacks_list)
```
|
github_jupyter
|
```
%reload_ext autoreload
%autoreload 2
from fastai.tabular import *
```
# Rossmann
## Data preparation
To create the feature-engineered train_clean and test_clean from the Kaggle competition data, run `rossman_data_clean.ipynb`. One important step that deals with time series is this:
```python
add_datepart(train, "Date", drop=False)
add_datepart(test, "Date", drop=False)
```
```
path = Config().data_path()/'rossmann'
train_df = pd.read_pickle(path/'train_clean')
train_df.head().T
n = len(train_df); n
```
### Experimenting with a sample
```
idx = np.random.permutation(range(n))[:2000]
idx.sort()
small_train_df = train_df.iloc[idx[:1000]]
small_test_df = train_df.iloc[idx[1000:]]
small_cont_vars = ['CompetitionDistance', 'Mean_Humidity']
small_cat_vars = ['Store', 'DayOfWeek', 'PromoInterval']
small_train_df = small_train_df[small_cat_vars + small_cont_vars + ['Sales']]
small_test_df = small_test_df[small_cat_vars + small_cont_vars + ['Sales']]
small_train_df.head()
small_test_df.head()
categorify = Categorify(small_cat_vars, small_cont_vars)
categorify(small_train_df)
categorify(small_test_df, test=True)
small_test_df.head()
small_train_df.PromoInterval.cat.categories
small_train_df['PromoInterval'].cat.codes[:5]
fill_missing = FillMissing(small_cat_vars, small_cont_vars)
fill_missing(small_train_df)
fill_missing(small_test_df, test=True)
small_train_df[small_train_df['CompetitionDistance_na'] == True]
```
### Preparing full data set
```
train_df = pd.read_pickle(path/'train_clean')
test_df = pd.read_pickle(path/'test_clean')
len(train_df),len(test_df)
procs=[FillMissing, Categorify, Normalize]
cat_vars = ['Store', 'DayOfWeek', 'Year', 'Month', 'Day', 'StateHoliday', 'CompetitionMonthsOpen',
'Promo2Weeks', 'StoreType', 'Assortment', 'PromoInterval', 'CompetitionOpenSinceYear', 'Promo2SinceYear',
'State', 'Week', 'Events', 'Promo_fw', 'Promo_bw', 'StateHoliday_fw', 'StateHoliday_bw',
'SchoolHoliday_fw', 'SchoolHoliday_bw']
cont_vars = ['CompetitionDistance', 'Max_TemperatureC', 'Mean_TemperatureC', 'Min_TemperatureC',
'Max_Humidity', 'Mean_Humidity', 'Min_Humidity', 'Max_Wind_SpeedKm_h',
'Mean_Wind_SpeedKm_h', 'CloudCover', 'trend', 'trend_DE',
'AfterStateHoliday', 'BeforeStateHoliday', 'Promo', 'SchoolHoliday']
dep_var = 'Sales'
df = train_df[cat_vars + cont_vars + [dep_var,'Date']].copy()
test_df['Date'].min(), test_df['Date'].max()
cut = train_df['Date'][(train_df['Date'] == train_df['Date'][len(test_df)])].index.max()
cut
valid_idx = range(cut)
df[dep_var].head()
data = (TabularList.from_df(df, path=path, cat_names=cat_vars, cont_names=cont_vars, procs=procs,)
.split_by_idx(valid_idx)
.label_from_df(cols=dep_var, label_cls=FloatList, log=True)
.add_test(TabularList.from_df(test_df, path=path, cat_names=cat_vars, cont_names=cont_vars))
.databunch())
doc(FloatList)
```
## Model
```
max_log_y = np.log(np.max(train_df['Sales'])*1.2)
y_range = torch.tensor([0, max_log_y], device=defaults.device)
learn = tabular_learner(data, layers=[1000,500], ps=[0.001,0.01], emb_drop=0.04,
y_range=y_range, metrics=exp_rmspe)
learn.model
len(data.train_ds.cont_names)
learn.lr_find()
learn.recorder.plot()
learn.fit_one_cycle(5, 1e-3, wd=0.2)
learn.save('1')
learn.recorder.plot_losses(last=-1)
learn.load('1');
learn.fit_one_cycle(5, 3e-4)
learn.fit_one_cycle(5, 3e-4)
```
(10th place in the competition was 0.108)
```
test_preds=learn.get_preds(DatasetType.Test)
test_df["Sales"]=np.exp(test_preds[0].data).numpy().T[0]
test_df[["Id","Sales"]]=test_df[["Id","Sales"]].astype("int")
test_df[["Id","Sales"]].to_csv("rossmann_submission.csv",index=False)
```
|
github_jupyter
|
Code testing for https://github.com/pymc-devs/pymc3/pull/2986
```
import numpy as np
import pymc3 as pm
import pymc3.distributions.transforms as tr
import theano.tensor as tt
from theano.scan_module import until
import theano
import matplotlib.pylab as plt
import seaborn as sns
%matplotlib inline
```
# Polar transformation
```
# Polar to Cartesian
def backward(y):
# y = [r, theta]
x = tt.zeros(y.shape)
x = tt.inc_subtensor(x[0], y[0]*tt.cos(y[1]))
x = tt.inc_subtensor(x[1], y[0]*tt.sin(y[1]))
return x
def forward(x):
# y = [r, theta]
y = tt.zeros(x.shape)
y = tt.inc_subtensor(y[0], tt.sqrt(tt.square(x[0]) + tt.square(x[1])))
if y[0] != 0:
if x[1] < 0:
theta = -tt.arccos(x[0]/y[0])
else:
theta = tt.arccos(x[0]/y[0])
y = tt.inc_subtensor(y[1], theta)
return y
y = tt.vector('polar')
y.tag.test_value=np.asarray([1., np.pi/2])
f_inv = backward(y)
J, _ = theano.scan(lambda i, f, x: tt.grad(f[i], x),
sequences=tt.arange(f_inv.shape[0]),
non_sequences=[f_inv, y])
Jacob_f1 = theano.function([y], J)
Jacob_f1(np.asarray([1., np.pi/2]))
J2 = pm.theanof.jacobian(f_inv, [y])
Jacob_f2 = theano.function([y], J2)
Jacob_f2(np.asarray([1., np.pi/2]))
%timeit Jacob_f1(np.asarray([1., np.pi/2]))
%timeit Jacob_f2(np.asarray([1., np.pi/2]))
class VectorTransform(tr.Transform):
def jacobian_det(self, x):
f_inv = self.backward(x)
J, _ = theano.scan(lambda i, f, x: tt.grad(f[i], x),
sequences=tt.arange(f_inv.shape[0]),
non_sequences=[f_inv, x])
return tt.log(tt.abs_(tt.nlinalg.det(J)))
class Nealfun(VectorTransform):
name = "Neal_funnel"
def backward(self, y):
x = tt.zeros(y.shape)
x = tt.inc_subtensor(x[0], y[0] / 3.)
x = tt.inc_subtensor(x[1:], y[1:] / tt.exp(y[0] / 2))
return x
def forward(self, x):
y = tt.zeros(x.shape)
y = tt.inc_subtensor(y[0], x[0] * 3.)
y = tt.inc_subtensor(y[1:], tt.exp(x[0] * 3. / 2) * x[1:])
return y
y = tt.vector('y')
y.tag.test_value = np.zeros(101)
nealfun = Nealfun()
f_inv = nealfun.backward(y)
J1, _ = theano.scan(lambda i, f, x: tt.grad(f[i], x),
sequences=tt.arange(f_inv.shape[0]),
non_sequences=[f_inv, y])
Jacob_f1 = theano.function([y], J1)
J2 = pm.theanof.jacobian(f_inv, [y])
Jacob_f2 = theano.function([y], J2)
%timeit Jacob_f1(np.zeros(101))
%timeit Jacob_f2(np.zeros(101))
```
# Copulas
Background reading http://twiecki.github.io/blog/2018/05/03/copulas/
More information https://github.com/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/Gaussian_Copula.ipynb
```
import scipy.stats as st
norm = st.norm()
def norm_cdf(x):
return x_unif
def copulas_forward_func(nsample, cov, marg1_ppf, marg2_ppf):
mvnorm = st.multivariate_normal(mean=[0, 0], cov=cov)
# Generate random samples from multivariate normal with correlation .5
x = mvnorm.rvs(nsample)
x_unif = norm.cdf(x)
x_trans = np.vstack([marg1_ppf(x_unif[:, 0]), marg2_ppf(x_unif[:, 1])]).T
return x_trans, x_unif, x
cov = np.asarray([[1., 0.725], [0.725, 1.]])
marg1_ppf = st.gumbel_r().ppf
marg2_ppf = st.beta(a=10, b=2).ppf
x_trans, x_unif, x = copulas_forward_func(10000, cov, marg1_ppf, marg2_ppf)
sns.jointplot(x[:, 0], x[:, 1], kind='kde', stat_func=None)
sns.jointplot(x_unif[:, 0], x_unif[:, 1], kind='hex',
stat_func=None, joint_kws=dict(gridsize=50))
sns.jointplot(x_trans[:, 0], x_trans[:, 1], kind='kde',
stat_func=None, xlim=(-2, 6), ylim=(.6, 1.0),)
plt.tight_layout()
xrange = np.linspace(-2, 6, 200)
plt.hist(x_trans[:, 0], xrange, density='pdf')
plt.plot(xrange, st.gumbel_r.pdf(xrange));
def gumbel_cdf(value, mu, beta):
return tt.exp(-tt.exp(-(value-mu)/beta))
```
Beta CDF
```
from theano.scan_module import until
max_iter=200
value_, a, b = x_trans[:, 1], 10., 2.
value = theano.shared(np.reshape(value_, (1,len(value_))))
EPS = 3.0e-7
qab = a + b
qap = a + 1.0
qam = a - 1.0
def _step(i, az, bm, am, bz):
tem = i + i
d = i * (b - i) * value / ((qam + tem) * (a + tem))
d =- (a + i) * i * value / ((qap + tem) * (a + tem))
ap = az + d * am
bp = bz + d * bm
app = ap + d * az
bpp = bp + d * bz
aold = az
am = ap / bpp
bm = bp / bpp
az = app / bpp
bz = tt.ones_like(bz)
return (az, bm, am, bz), until(tt.sum(tt.lt(tt.abs_(az - aold), (EPS * tt.abs_(az)))))
(az, bm, am, bz), _ = theano.scan(_step,
sequences=[tt.arange(1, max_iter)],
outputs_info=[tt.ones_like(value),
tt.ones_like(value),
tt.ones_like(value),
1. - qab * value / qap])
def cont_fraction_beta(value_, a, b, max_iter=500):
'''Evaluates the continued fraction form of the incomplete Beta function.
Derived from implementation by Ali Shoaib (https://goo.gl/HxjIJx).
'''
EPS = 1.0e-20
qab = a + b
qap = a + 1.0
qam = a - 1.0
value = theano.shared(value_)
def _step(i, az, bm, am, bz):
tem = i + i
d = i * (b - i) * value / ((qam + tem) * (a + tem))
d = - (a + i) * i * value / ((qap + tem) * (a + tem))
ap = az + d * am
bp = bz + d * bm
app = ap + d * az
bpp = bp + d * bz
aold = az
am = ap / bpp
bm = bp / bpp
az = app / bpp
bz = tt.ones_like(bz)
return (az, bm, am, bz), until(tt.sum(tt.lt(tt.abs_(az - aold), (EPS * tt.abs_(az)))))
(az, bm, am, bz), _ = theano.scan(_step,
sequences=[tt.arange(1, max_iter)],
outputs_info=[tt.ones_like(value),
tt.ones_like(value),
tt.ones_like(value),
1. - qab * value / qap])
return az[-1]
def beta_cdf(value, a, b):
log_beta = tt.gammaln(a+b) - tt.gammaln(a) - tt.gammaln(b)
log_beta += a * tt.log(value) + b * tt.log(1 - value)
cdf = tt.switch(
tt.lt(value, (a + 1) / (a + b + 2)),
tt.exp(log_beta) * cont_fraction_beta(value, a, b) / a,
1. - tt.exp(log_beta) * cont_fraction_beta(1. - value, b, a) / b
)
return cdf
def normal_ppf(value):
return -np.sqrt(2.) * tt.erfcinv(2. * value)
functmp = theano.function([],
tt.stack([gumbel_cdf(x_trans[:, 0], 0., 1.),
beta_cdf(x_trans[:, 1], 10., 2.)]).T
)
x_ = functmp()
x_
x_unif
np.sum(~np.isfinite(x_))
with pm.Model() as model:
# r∼Uniform(−1,1)
r = pm.Uniform('r',lower=-1, upper=1)
cov = pm.Deterministic('cov',
tt.stacklists([[1., r],
[r, 1.]]))
a = pm.HalfNormal('alpha', 5., testval=10.)
b = pm.HalfNormal('beta', 2.5, testval=2.)
loc = pm.Normal('loc', 0., 5., testval=0.)
scale = pm.HalfNormal('scale', 2.5, testval=1.)
tr_func = normal_ppf(
tt.stack([gumbel_cdf(x_trans[:, 0], loc, scale),
beta_cdf(x_trans[:, 1], a, b)]).T
)
pm.MvNormal('obs', np.zeros(2), cov=cov, observed=tr_func)
pm.Gumbel('marg0', loc, scale, observed=x_trans[:, 0])
pm.Beta('marg1', a, b, observed=x_trans[:, 1])
```
The beta CDF does not quite work - use another distribution instead
```
from scipy.special import logit
xrange = np.linspace(0, 1, 200)
plt.hist(x_trans[:, 1], xrange, density='pdf')
logitnormpdf = st.norm.pdf(logit(xrange), loc=1.725, scale=.8) * 1/(xrange * (1-xrange))
plt.plot(xrange, logitnormpdf);
def logitnorm_cdf(value, mu, sd):
return .5 + .5*(tt.erf((pm.math.logit(value)-mu)/(np.sqrt(2)*sd)))
tr_func = normal_ppf(
tt.stack([gumbel_cdf(x_trans[:, 0], 0., 1.),
logitnorm_cdf(x_trans[:, 1], 1.725, .8)]).T
)
functmp = theano.function([], tr_func)
x_ = functmp()
sns.jointplot(x_[:, 0], x_[:, 1], kind='kde', stat_func=None);
np.sum(~np.isfinite(x_[:, 1]))
with pm.Model() as model:
# r∼Uniform(−1,1)
r = pm.Uniform('r',lower=-1, upper=1)
cov = pm.Deterministic('cov',
tt.stacklists([[1., r],
[r, 1.]]))
loc = pm.Normal('loc', 0., 5., testval=0.)
scale = pm.HalfNormal('scale', 2.5, testval=1.)
mu = pm.Normal('mu', 1., 1., testval=1.725)
sd = pm.HalfNormal('sd', .5, testval=.8)
tr_func = normal_ppf(
tt.stack([gumbel_cdf(x_trans[:, 0], loc, scale),
logitnorm_cdf(x_trans[:, 1], mu, sd)]).T
)
pm.MvNormal('obs', np.zeros(2), cov=cov, observed=tr_func)
pm.Gumbel('marg0', loc, scale, observed=x_trans[:, 0])
pm.LogitNormal('marg1', mu, sd, observed=x_trans[:, 1])
with model:
map1 = pm.find_MAP()
map1
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=map1['loc'], scale=map1['scale']))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
logitnormpdf = st.norm.pdf(logit(x1), loc=map1['mu'], scale=map1['sd']) * 1/(x1 * (1-x1))
ax[1].plot(x1, logitnormpdf);
with pm.Model() as model_marg:
loc = pm.Normal('loc', 0., 5., testval=0.)
scale = pm.HalfNormal('scale', 2.5, testval=1.)
mu = pm.Normal('mu', 1., 1., testval=1.725)
sd = pm.HalfNormal('sd', .5, testval=.8)
pm.Gumbel('marg0', loc, scale, observed=x_trans[:, 0])
pm.LogitNormal('marg1', mu, sd, observed=x_trans[:, 1])
map_ = pm.find_MAP()
map_
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=map_['loc'], scale=map_['scale']))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
logitnormpdf = st.norm.pdf(logit(x1), loc=map_['mu'], scale=map_['sd']) * 1/(x1 * (1-x1))
ax[1].plot(x1, logitnormpdf);
from pymc3.theanof import gradient
def jacobian_det(f_inv_x, x):
grad = tt.reshape(gradient(tt.sum(f_inv_x), [x]), x.shape)
return tt.log(tt.abs_(grad))
xt_0 = theano.shared(x_trans[:, 0])
xt_1 = theano.shared(x_trans[:, 1])
with pm.Model() as model2:
# r∼Uniform(−1,1)
r = pm.Uniform('r',lower=-1, upper=1)
cov = pm.Deterministic('cov',
tt.stacklists([[1., r],
[r, 1.]]))
loc = pm.Normal('loc', 0., 5., testval=0.)
scale = pm.HalfNormal('scale', 2.5, testval=1.)
mu = pm.Normal('mu', 1., .5, testval=1.725)
sd = pm.HalfNormal('sd', .5, testval=.8)
tr_func = normal_ppf(
tt.stack([gumbel_cdf(xt_0, loc, scale),
logitnorm_cdf(xt_1, mu, sd)]).T
)
pm.MvNormal('obs', np.zeros(2), cov=cov, observed=tr_func)
pm.Potential('jacob_det0', jacobian_det(normal_ppf(gumbel_cdf(xt_0, loc, scale)), xt_0))
pm.Potential('jacob_det1', jacobian_det(normal_ppf(logitnorm_cdf(xt_1, mu, sd)), xt_1))
map_ = pm.find_MAP()
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=map_['loc'], scale=map_['scale']))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
logitnormpdf = st.norm.pdf(logit(x1), loc=map_['mu'], scale=map_['sd']) * 1/(x1 * (1-x1))
ax[1].plot(x1, logitnormpdf);
```
Kumaraswamy distribution
```
from scipy.special import logit
xrange = np.linspace(0, 1, 200)
plt.hist(x_trans[:, 1], xrange, density='pdf')
Kumaraswamypdf = lambda x, a, b: a*b*np.power(x, a-1)*np.power(1-np.power(x, a), b-1)
plt.plot(xrange, Kumaraswamypdf(xrange, 8, 2));
def Kumaraswamy_cdf(value, a, b):
return 1 - tt.pow(1 - tt.pow(value, a), b)
tr_func = normal_ppf(
tt.stack([gumbel_cdf(x_trans[:, 0], 0., 1.),
Kumaraswamy_cdf(x_trans[:, 1], 8, 2)]).T
)
functmp = theano.function([], tr_func)
x_ = functmp()
sns.jointplot(x_[:, 0], x_[:, 1], kind='kde', stat_func=None);
np.sum(~np.isfinite(x_[:, 1]))
with pm.Model() as model_marg:
a = pm.HalfNormal('alpha', 5., testval=10.)
b = pm.HalfNormal('beta', 2.5, testval=2.)
loc = pm.Normal('loc', 0., 5., testval=0.)
scale = pm.HalfNormal('scale', 2.5, testval=1.)
pm.Gumbel('marg0', loc, scale, observed=x_trans[:, 0])
pm.Kumaraswamy('marg1', a, b, observed=x_trans[:, 1])
map_ = pm.find_MAP()
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=map_['loc'], scale=map_['scale']))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
ax[1].plot(x1, Kumaraswamypdf(x1, map_['alpha'], map_['beta']));
with pm.Model() as model2:
# r∼Uniform(−1,1)
r = pm.Uniform('r',lower=-1, upper=1)
cov = pm.Deterministic('cov',
tt.stacklists([[1., r],
[r, 1.]]))
a = pm.HalfNormal('alpha', 5.)
b = pm.HalfNormal('beta', 2.5)
loc = pm.Normal('loc', 0., 5.)
scale = pm.HalfNormal('scale', 2.5)
tr_func = normal_ppf(
tt.stack([gumbel_cdf(xt_0, loc, scale),
Kumaraswamy_cdf(xt_1, a, b)]).T
)
pm.MvNormal('obs', np.zeros(2), cov=cov, observed=tr_func)
pm.Potential('jacob_det0', jacobian_det(normal_ppf(gumbel_cdf(xt_0, loc, scale)), xt_0))
pm.Potential('jacob_det1', jacobian_det(normal_ppf(Kumaraswamy_cdf(xt_1, a, b)), xt_1))
map_ = pm.find_MAP()
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=map_['loc'], scale=map_['scale']))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
ax[1].plot(x1, Kumaraswamypdf(x1, map_['alpha'], map_['beta']));
map_
with model2:
trace = pm.sample()
_, ax = plt.subplots(1, 2, figsize=(10, 3))
x0 = np.linspace(-2, 6, 200)
ax[0].hist(x_trans[:, 0], x0, density='pdf')
ax[0].plot(x0, st.gumbel_r.pdf(x0, loc=trace['loc'].mean(), scale=trace['scale'].mean()))
x1 = np.linspace(0, 1, 200)
ax[1].hist(x_trans[:, 1], x1, density='pdf')
ax[1].plot(x1, Kumaraswamypdf(x1, trace['alpha'].mean(), trace['beta'].mean()));
```
|
github_jupyter
|
```
import pandas as pd
import os
import hashlib
import requests
from bs4 import BeautifulSoup
from bs4.element import Comment
import urllib.parse
from tqdm.notebook import tqdm
import random
from multiprocessing import Pool
import spacy
import numpy as np
industries = pd.read_csv("industry_categories.csv")
industries.head()
salary_industries = pd.read_csv("Salary-Industries.csv")
salary_industries.head()
GOOGLE_API_KEY = 'AIzaSyCqd-BAzUsp6a2ICBETWebYYwoA3d3EeWk'
class KeyValueCache:
def __init__(self, data_dir):
self.data_dir = data_dir
if not os.path.isdir(self.data_dir):
os.mkdir(self.data_dir)
def hash_of(self, key):
return hashlib.md5(key.encode('utf-8')).hexdigest()
def file_for(self, key):
return os.path.join(self.data_dir, self.hash_of(key) + '.html')
def contains(self, key):
"""Checks if there is content for the key"""
return os.path.isfile(self.file_for(key))
def get(self, key):
"""Returns the value of the key"""
with open(self.file_for(key)) as f:
return f.read()
def put(self, key, value):
"""Stores value at the key"""
with open(self.file_for(key), 'w') as f:
f.write(value)
return value
cache = KeyValueCache(os.path.join('.', '.cache'))
# print(cache.hash_of(b'abc'))
# print(cache.file_for(b'abc'))
# print(cache.contains(b'abc'))
# print(cache.put(b'abc', 'abc value'))
# print(cache.get(b'abc'))
# print(cache.contains(b'abc'))
```
requests quickstart: https://requests.kennethreitz.org/en/master/user/quickstart/
```
static_proxies = pd.read_csv("utils/trusted_proxies.csv")['proxy'].to_list()
def request_proxy(url):
proxies = static_proxies
return random.choice(proxies)
def request_user_agent(url):
agents = [
# 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/50.0.2661.102 Safari/537.36',
# 'Mozilla/5.0 (iPad; U; CPU OS 3_2_1 like Mac OS X; en-us) AppleWebKit/531.21.10 (KHTML, like Gecko) Mobile/7B405',
# 'Mozilla/5.0 (Linux; Android 8.0.0; SM-G960F Build/R16NW) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/62.0.3202.84 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 7.0; SM-G892A Build/NRD90M; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/60.0.3112.107 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 7.0; SM-G930VC Build/NRD90M; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/58.0.3029.83 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; SM-G935S Build/MMB29K; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/55.0.2883.91 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; SM-G920V Build/MMB29K) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/52.0.2743.98 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 5.1.1; SM-G928X Build/LMY47X) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/47.0.2526.83 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; Nexus 6P Build/MMB29P) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/47.0.2526.83 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 7.1.1; G8231 Build/41.2.A.0.219; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/59.0.3071.125 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; E6653 Build/32.2.A.0.253) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/52.0.2743.98 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0; HTC One X10 Build/MRA58K; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/61.0.3163.98 Mobile Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0; HTC One M9 Build/MRA58K) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/52.0.2743.98 Mobile Safari/537.3'
# 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/42.0.2311.135 Safari/537.36 Edge/12.246',
# 'Mozilla/5.0 (X11; CrOS x86_64 8172.45.0) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/51.0.2704.64 Safari/537.36',
# 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_11_2) AppleWebKit/601.3.9 (KHTML, like Gecko) Version/9.0.2 Safari/601.3.9',
# 'Mozilla/5.0 (Windows NT 6.1; WOW64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/47.0.2526.111 Safari/537.36',
# 'Mozilla/5.0 (X11; Ubuntu; Linux x86_64; rv:15.0) Gecko/20100101 Firefox/15.0.1',
# 'Mozilla/5.0 (Linux; Android 7.0; Pixel C Build/NRD90M; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/52.0.2743.98 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; SGP771 Build/32.2.A.0.253; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/52.0.2743.98 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 6.0.1; SHIELD Tablet K1 Build/MRA58K; wv) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/55.0.2883.91 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 7.0; SM-T827R4 Build/NRD90M) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/60.0.3112.116 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 5.0.2; SAMSUNG SM-T550 Build/LRX22G) AppleWebKit/537.36 (KHTML, like Gecko) SamsungBrowser/3.3 Chrome/38.0.2125.102 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 4.4.3; KFTHWI Build/KTU84M) AppleWebKit/537.36 (KHTML, like Gecko) Silk/47.1.79 like Chrome/47.0.2526.80 Safari/537.36',
# 'Mozilla/5.0 (Linux; Android 5.0.2; LG-V410/V41020c Build/LRX22G) AppleWebKit/537.36 (KHTML, like Gecko) Version/4.0 Chrome/34.0.1847.118 Safari/537.36',
# from Google desctop
# 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/61.0.3163.100 Safari/537.36',
# 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/60.0.3112.113 Safari/537.36'
# 'Mozilla/5.0 (Windows NT 6.1; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/61.0.3163.100 Safari/537.36',
# 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_12_6) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/61.0.3163.100 Safari/537.36',
# 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_12_6) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/60.0.3112.113 Safari/537.36',
# 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_12_6) AppleWebKit/604.1.38 (KHTML, like Gecko) Version/11.0 Safari/604.1.38',
# 'Mozilla/5.0 (Windows NT 6.1; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/60.0.3112.113 Safari/537.36',
# 'Mozilla/5.0 (Windows NT 10.0; WOW64; rv:55.0) Gecko/20100101 Firefox/55.0',
# 'Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/52.0.2743.116 Safari/537.36 Edge/15.15063'
# 'Mozilla/5.0 (Windows NT 6.1; WOW64; Trident/7.0; rv:11.0) like Gecko',
'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_13_6) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/77.0.3865.120 Safari/537.36'
]
return random.choice(agents)
def http_get(url):
headers = {
'User-Agent': request_user_agent(url),
'Accept': 'text/html,application/xhtml+xml,application/xml;q=1',
'Accept-Encoding': 'identity;q=1'
}
proxy = request_proxy(url)
proxies = {
"http": proxy,
"https": proxy
}
response = requests.get(url, headers=headers, proxies=proxies, timeout=10, allow_redirects=True)
# handle HTTPSConnectionPool, ProxyError to retry with a different proxy
return response
def cached_get(url, cache):
"""gets the content of a url either from cache or by making HTTP request"""
if cache.contains(url):
#print("Cached: {}".format(url))
return cache.get(url)
else:
#print("GET: {}".format(url))
try:
response = http_get(url)
if response: # check for 200
return cache.put(url, response.text)
else:
raise Exception(response.status_code)
except Exception as err:
print("ERROR: {}".format(err))
return None
serp = cached_get('https://www.google.com/search?q=Tech%20-%20IT%20department%20of%20national%20insurance%20company', cache)
def extract_links(serp):
def external_url_from_href(href):
if href.startswith('/url'):
return urllib.parse.parse_qs(urllib.parse.urlparse('https://www.google.com' + href).query).get('q')[0]
else:
return href
soup = BeautifulSoup(serp, 'html.parser')
hrefs = []
blocks_v1 = soup.select('#rso div.bkWMgd div.g:not(.g-blk):not(#ZpxfC):not(.gws-trips__outer-card):not(.knavi)')
#print("Elements found: {}".format(len(blocks_v1)))
for div in blocks_v1:
for a in div.select('a:not(.fl):not(.ab_button)'):
hrefs.append(external_url_from_href(a.get('href')))
blocks_v2 = soup.select('#main div.ZINbbc div.kCrYT > a')
#print("Elements found: {}".format(len(blocks_v2)))
for a in blocks_v2:
hrefs.append(external_url_from_href(a.get('href')))
return hrefs
#print(a)
#glinks = ['https://www.google.com' + l.get('href') for l in links]
#site_links = [urllib.parse.parse_qs(urllib.parse.urlparse(gl).query).get('q')[0] for gl in glinks]
#return site_links
serp = cached_get('https://www.google.com/search?q=Auto%20rental', cache)
#print(cache.file_for('https://www.google.com/search?q=Auto%20rental'))
# serp = cached_get('https://www.google.com/search?q=Accounting', cache)
extract_links(serp)
def splitup_to_queries(items, separator = None):
for i in items:
chunks = i.split(separator) if separator else [i]
for chunk in chunks:
yield (i, chunk.strip())
if len(chunks) > 1:
yield (i, " ".join(chunks))
def queries_to_links(items):
for industry, query in items:
search_url = "https://www.google.com/search?q={}".format(urllib.parse.quote(query))
serp = cached_get(search_url, cache)
if not serp:
continue
links = extract_links(serp)
yield (industry, search_url, links)
def download_links(items):
for industry, search_url, links in items:
for url in links[0:3]:
link_html = cached_get(url, cache)
if not link_html:
yield (industry, 'error', url)
else:
yield (industry, 'success', url)
def first_n_links(items):
for industry, search_url, links in items:
for url in links[0:3]:
yield url
def download_url(items):
c = cache
def download(u):
result = 'success' if cached_get(url, c) else 'error'
return (url, result)
with Pool(processes=10) as pool:
resutls = pool.imap(download, items, chunksize=8)
def download_data_for_list(list_to_download, separator=None):
it = download_links(queries_to_links(splitup_to_queries(list_to_download, separator=separator)))
successes = 0
errors = 0
progress = tqdm(it, desc='Downloading', miniters=1)
for industry, status, url in progress:
if status == 'error':
errors = errors + 1
print("ERROR: {}".format(url))
else:
successes = successes + 1
progress.set_postfix(insudtry = industry, successes = successes, errors = errors)
def download_serps_for_list(list_to_download, separator=None):
it = queries_to_links(splitup_to_queries(list_to_download, separator=separator))
for industry, search_url, urls in tqdm(it, desc='Downloading', miniters=1):
if len(urls) < 5:
print("{} {}".format(len(urls), search_url))
def download_pages_for_list_multiprocess(list_to_download, separator=None):
urls = first_n_links(queries_to_links(splitup_to_queries(list_to_download, separator=separator)))
def download(url):
print(url)
page = cached_get(url, cache)
result = 'success' if page else 'error'
return (url, result)
with Pool(processes=4) as pool:
resutls = pool.imap_unordered(download, list(urls), chunksize=4)
#download_data_for_list(industries['industry'].to_list(), separator='/')
download_serps_for_list(salary_industries['Industry Ref'].dropna().sort_values().to_list())
def industry_to_queries(industry, separator = None):
chunks = industry.split(separator) if separator else [industry]
result = [chunk.strip() for chunk in chunks]
if len(chunks) > 1:
result.append(" ".join(chunks))
return result
def queries_to_links(items):
for industry, query in items:
search_url = "https://www.google.com/search?q={}".format(urllib.parse.quote(query))
serp = cached_get(search_url, cache)
if not serp:
continue
links = extract_links(serp)
yield (industry, search_url, links)
def create_industry_term_url_map(items, separator=None):
records = []
for industry in items:
for search_term in industry_to_queries(industry, separator=separator):
search_url = "https://www.google.com/search?q={}".format(urllib.parse.quote(search_term))
serp = cached_get(search_url, cache)
if not serp:
continue
links = extract_links(serp)
for link in links:
records.append((industry, search_term, link))
return pd.DataFrame.from_records(records, columns=['industry', 'term', 'url'])
# industry_targets_urls = create_industry_term_url_map(industries['industry'].to_list(), separator='/')
# industry_targets_urls.to_csv("./utils/industry_targets_urls.csv", index=False)
#industry_inputs_urls = create_industry_term_url_map(salary_industries['Industry Ref'].dropna().sort_values().to_list())
#industry_inputs_urls.to_csv("./utils/industry_inputs_urls.csv", index=False)
industry_targets_urls = pd.read_csv("./utils/industry_targets_urls.csv")
industry_inputs_urls = pd.read_csv("./utils/industry_inputs_urls.csv")
industry_inputs_urls.head()
import re
def tag_visible(element):
if element.parent.name in ['style', 'script', 'head', 'title', 'meta', '[document]']:
return False
if isinstance(element, Comment):
return False
#if any(c.contains('hidden') for c in element.parent['class']):
# return False
return True
def text_from_html(body):
try:
soup = BeautifulSoup(body, 'html.parser')
texts = soup.find_all(text=True)
visible_texts = filter(tag_visible, texts)
visible_texts = map(lambda s: s.encode('utf-8', 'ignore').decode('utf-8'), visible_texts)
visible_texts = map(lambda s: re.sub(r'\s+', ' ', s).strip(), visible_texts)
visible_texts = filter(lambda s: len(s)>0, visible_texts)
visible_texts = filter(lambda s: len(s.split(' '))>5, visible_texts)
return ' '.join(visible_texts)
except:
return ''
print(cache.file_for('https://www.rightway.com/used-vehicles/'))
#page = cached_get('https://www.britannica.com/topic/finance', cache)
#text_from_html(page)
def extract_and_combine_text_from_urls(urls):
pages = [cache.get(url) for url in urls if cache.contains(url)]
texts = [text_from_html(page) for page in pages]
return " ".join(texts)
def is_downloaded(row):
return cache.contains(row['url'])
def file_for_url(row):
if cache.contains(row['url']):
return cache.file_for(row['url'])
else:
return None
def extract_text(row):
if cache.contains(row['url']):
return text_from_html(cache.get(row['url']))
else:
return None
def create_url_text_file(input_file, out_file):
df = pd.read_csv(input_file)
df['is_downloaded'] = df.apply(is_downloaded, axis=1)
df['file'] = df.apply(file_for_url, axis=1)
df['text'] = df.apply(extract_text, axis=1)
df.to_csv(out_file, index=False)
return df
industry_targets_url_text = create_url_text_file("./utils/industry_targets_urls.csv", "./utils/industry_targets_url_text.csv")
industry_targets_url_text.head()
industry_inputs_url_text = create_url_text_file("./utils/industry_inputs_urls.csv", "./utils/industry_inputs_url_text.csv")
industry_inputs_url_text.head()
industry_inputs_url_text['url'][industry_inputs_url_text['is_downloaded'] == False]
def combine_texts(series):
return " ".join([str(t) for t in series.values])
def create_text_file(input_file, out_file):
df = pd.read_csv(input_file)
df = df.groupby(['industry', 'term']).aggregate({'text': combine_texts}).reset_index()
df.to_csv(out_file, index=False)
return df
industry_targets_text = create_text_file("./utils/industry_targets_url_text.csv", "./utils/industry_targets_text.csv")
industry_targets_text.head()
industry_inputs_text = create_text_file("./utils/industry_inputs_url_text.csv", "./utils/industry_inputs_text.csv")
industry_inputs_text.head()
#industry_inputs_url_text['text'].apply(lambda r: len(r) if r else 0)
industry_inputs_url_text['file'].dropna()
industry_inputs_url_text['link_rank'] = (industry_inputs_url_text.groupby('term').cumcount()+1)
prioritized_urls = (
industry_inputs_url_text.loc[:,['term', 'url', 'link_rank', 'is_downloaded']]
.query('is_downloaded == False')
.sort_values(['link_rank', 'term'], ascending=[True, False])
#['url']
)
l = prioritized_urls.query('link_rank == 1')['term'].to_list()
prioritized_urls[prioritized_urls['term'].isin(l)]['url']
industry_inputs_url_text.groupby('term').aggregate({ 'is_downloaded': lambda g: any(g)}).query('is_downloaded == False')
def download(url):
page = cached_get(url, cache)
if page:
return ('success', url)
else:
return ('error', url)
def download_all(urls):
with Pool(10) as pool:
return pool.map(func, urls)
def first_few_url_for_each_term(term_url_df, n):
for g, rows in term_url_df.groupby('term'):
for url in rows['url'].to_list()[:n]:
yield url
l = [download(url) for url in tqdm(list(prioritized_urls[prioritized_urls['term'].isin(l)]['url']))]
s = 'asd\ud800sdf'.encode('utf-8', 'ignore').decode('utf-8')
print(s)
srs = pd.Series()
srs.loc[ 0 ] = s
srs.to_csv('testcase.csv')
```
|
github_jupyter
|
## Dependencies
```
import json, warnings, shutil, glob
from jigsaw_utility_scripts import *
from scripts_step_lr_schedulers import *
from transformers import TFXLMRobertaModel, XLMRobertaConfig
from tensorflow.keras.models import Model
from tensorflow.keras import optimizers, metrics, losses, layers
SEED = 0
seed_everything(SEED)
warnings.filterwarnings("ignore")
```
## TPU configuration
```
strategy, tpu = set_up_strategy()
print("REPLICAS: ", strategy.num_replicas_in_sync)
AUTO = tf.data.experimental.AUTOTUNE
```
# Load data
```
database_base_path = '/kaggle/input/jigsaw-data-split-roberta-192-ratio-2-upper/'
k_fold = pd.read_csv(database_base_path + '5-fold.csv')
valid_df = pd.read_csv("/kaggle/input/jigsaw-multilingual-toxic-comment-classification/validation.csv",
usecols=['comment_text', 'toxic', 'lang'])
print('Train samples: %d' % len(k_fold))
display(k_fold.head())
print('Validation samples: %d' % len(valid_df))
display(valid_df.head())
base_data_path = 'fold_3/'
fold_n = 3
# Unzip files
!tar -xvf /kaggle/input/jigsaw-data-split-roberta-192-ratio-2-upper/fold_3.tar.gz
```
# Model parameters
```
base_path = '/kaggle/input/jigsaw-transformers/XLM-RoBERTa/'
config = {
"MAX_LEN": 192,
"BATCH_SIZE": 128,
"EPOCHS": 4,
"LEARNING_RATE": 1e-5,
"ES_PATIENCE": None,
"base_model_path": base_path + 'tf-xlm-roberta-large-tf_model.h5',
"config_path": base_path + 'xlm-roberta-large-config.json'
}
with open('config.json', 'w') as json_file:
json.dump(json.loads(json.dumps(config)), json_file)
```
## Learning rate schedule
```
lr_min = 1e-7
lr_start = 1e-7
lr_max = config['LEARNING_RATE']
step_size = len(k_fold[k_fold[f'fold_{fold_n}'] == 'train']) // config['BATCH_SIZE']
total_steps = config['EPOCHS'] * step_size
hold_max_steps = 0
warmup_steps = step_size * 1
decay = .9997
rng = [i for i in range(0, total_steps, config['BATCH_SIZE'])]
y = [exponential_schedule_with_warmup(tf.cast(x, tf.float32), warmup_steps, hold_max_steps,
lr_start, lr_max, lr_min, decay) for x in rng]
sns.set(style="whitegrid")
fig, ax = plt.subplots(figsize=(20, 6))
plt.plot(rng, y)
print("Learning rate schedule: {:.3g} to {:.3g} to {:.3g}".format(y[0], max(y), y[-1]))
```
# Model
```
module_config = XLMRobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFXLMRobertaModel.from_pretrained(config['base_model_path'], config=module_config)
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
cls_token = last_hidden_state[:, 0, :]
output = layers.Dense(1, activation='sigmoid', name='output')(cls_token)
model = Model(inputs=[input_ids, attention_mask], outputs=output)
return model
```
# Train
```
# Load data
x_train = np.load(base_data_path + 'x_train.npy')
y_train = np.load(base_data_path + 'y_train_int.npy').reshape(x_train.shape[1], 1).astype(np.float32)
x_valid_ml = np.load(database_base_path + 'x_valid.npy')
y_valid_ml = np.load(database_base_path + 'y_valid.npy').reshape(x_valid_ml.shape[1], 1).astype(np.float32)
#################### ADD TAIL ####################
x_train = np.hstack([x_train, np.load(base_data_path + 'x_train_tail.npy')])
y_train = np.vstack([y_train, y_train])
step_size = x_train.shape[1] // config['BATCH_SIZE']
valid_step_size = x_valid_ml.shape[1] // config['BATCH_SIZE']
# Build TF datasets
train_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_train, y_train, config['BATCH_SIZE'], AUTO, seed=SEED))
valid_dist_ds = strategy.experimental_distribute_dataset(get_validation_dataset(x_valid_ml, y_valid_ml, config['BATCH_SIZE'], AUTO, repeated=True, seed=SEED))
train_data_iter = iter(train_dist_ds)
valid_data_iter = iter(valid_dist_ds)
# Step functions
@tf.function
def train_step(data_iter):
def train_step_fn(x, y):
with tf.GradientTape() as tape:
probabilities = model(x, training=True)
loss = loss_fn(y, probabilities)
grads = tape.gradient(loss, model.trainable_variables)
optimizer.apply_gradients(zip(grads, model.trainable_variables))
train_auc.update_state(y, probabilities)
train_loss.update_state(loss)
for _ in tf.range(step_size):
strategy.experimental_run_v2(train_step_fn, next(data_iter))
@tf.function
def valid_step(data_iter):
def valid_step_fn(x, y):
probabilities = model(x, training=False)
loss = loss_fn(y, probabilities)
valid_auc.update_state(y, probabilities)
valid_loss.update_state(loss)
for _ in tf.range(valid_step_size):
strategy.experimental_run_v2(valid_step_fn, next(data_iter))
# Train model
with strategy.scope():
model = model_fn(config['MAX_LEN'])
optimizer = optimizers.Adam(learning_rate=lambda:
exponential_schedule_with_warmup(tf.cast(optimizer.iterations, tf.float32),
warmup_steps, hold_max_steps, lr_start,
lr_max, lr_min, decay))
loss_fn = losses.binary_crossentropy
train_auc = metrics.AUC()
valid_auc = metrics.AUC()
train_loss = metrics.Sum()
valid_loss = metrics.Sum()
metrics_dict = {'loss': train_loss, 'auc': train_auc,
'val_loss': valid_loss, 'val_auc': valid_auc}
history = custom_fit(model, metrics_dict, train_step, valid_step, train_data_iter, valid_data_iter,
step_size, valid_step_size, config['BATCH_SIZE'], config['EPOCHS'],
config['ES_PATIENCE'], save_last=False)
# model.save_weights('model.h5')
# Make predictions
x_train = np.load(base_data_path + 'x_train.npy')
x_valid = np.load(base_data_path + 'x_valid.npy')
x_valid_ml_eval = np.load(database_base_path + 'x_valid.npy')
train_preds = model.predict(get_test_dataset(x_train, config['BATCH_SIZE'], AUTO))
valid_preds = model.predict(get_test_dataset(x_valid, config['BATCH_SIZE'], AUTO))
valid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))
k_fold.loc[k_fold[f'fold_{fold_n}'] == 'train', f'pred_{fold_n}'] = np.round(train_preds)
k_fold.loc[k_fold[f'fold_{fold_n}'] == 'validation', f'pred_{fold_n}'] = np.round(valid_preds)
valid_df[f'pred_{fold_n}'] = valid_ml_preds
# Fine-tune on validation set
#################### ADD TAIL ####################
x_valid_ml_tail = np.hstack([x_valid_ml, np.load(database_base_path + 'x_valid_tail.npy')])
y_valid_ml_tail = np.vstack([y_valid_ml, y_valid_ml])
valid_step_size_tail = x_valid_ml_tail.shape[1] // config['BATCH_SIZE']
# Build TF datasets
train_ml_dist_ds = strategy.experimental_distribute_dataset(get_training_dataset(x_valid_ml_tail, y_valid_ml_tail,
config['BATCH_SIZE'], AUTO, seed=SEED))
train_ml_data_iter = iter(train_ml_dist_ds)
history_ml = custom_fit(model, metrics_dict, train_step, valid_step, train_ml_data_iter, valid_data_iter,
valid_step_size_tail, valid_step_size, config['BATCH_SIZE'], 1,
config['ES_PATIENCE'], save_last=False)
# Join history
for key in history_ml.keys():
history[key] += history_ml[key]
model.save_weights('model_ml.h5')
# Make predictions
valid_ml_preds = model.predict(get_test_dataset(x_valid_ml_eval, config['BATCH_SIZE'], AUTO))
valid_df[f'pred_ml_{fold_n}'] = valid_ml_preds
### Delete data dir
shutil.rmtree(base_data_path)
```
## Model loss graph
```
plot_metrics(history)
```
# Model evaluation
```
display(evaluate_model_single_fold(k_fold, fold_n, label_col='toxic_int').style.applymap(color_map))
```
# Confusion matrix
```
train_set = k_fold[k_fold[f'fold_{fold_n}'] == 'train']
validation_set = k_fold[k_fold[f'fold_{fold_n}'] == 'validation']
plot_confusion_matrix(train_set['toxic_int'], train_set[f'pred_{fold_n}'],
validation_set['toxic_int'], validation_set[f'pred_{fold_n}'])
```
# Model evaluation by language
```
display(evaluate_model_single_fold_lang(valid_df, fold_n).style.applymap(color_map))
# ML fine-tunned preds
display(evaluate_model_single_fold_lang(valid_df, fold_n, pred_col='pred_ml').style.applymap(color_map))
```
# Visualize predictions
```
pd.set_option('max_colwidth', 120)
print('English validation set')
display(k_fold[['comment_text', 'toxic'] + [c for c in k_fold.columns if c.startswith('pred')]].head(10))
print('Multilingual validation set')
display(valid_df[['comment_text', 'toxic'] + [c for c in valid_df.columns if c.startswith('pred')]].head(10))
```
# Test set predictions
```
x_test = np.load(database_base_path + 'x_test.npy')
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE'], AUTO))
submission = pd.read_csv('/kaggle/input/jigsaw-multilingual-toxic-comment-classification/sample_submission.csv')
submission['toxic'] = test_preds
submission.to_csv('submission.csv', index=False)
display(submission.describe())
display(submission.head(10))
```
|
github_jupyter
|
# Plagiarism Detection Model
Now that you've created training and test data, you are ready to define and train a model. Your goal in this notebook, will be to train a binary classification model that learns to label an answer file as either plagiarized or not, based on the features you provide the model.
This task will be broken down into a few discrete steps:
* Upload your data to S3.
* Define a binary classification model and a training script.
* Train your model and deploy it.
* Evaluate your deployed classifier and answer some questions about your approach.
To complete this notebook, you'll have to complete all given exercises and answer all the questions in this notebook.
> All your tasks will be clearly labeled **EXERCISE** and questions as **QUESTION**.
It will be up to you to explore different classification models and decide on a model that gives you the best performance for this dataset.
---
## Load Data to S3
In the last notebook, you should have created two files: a `training.csv` and `test.csv` file with the features and class labels for the given corpus of plagiarized/non-plagiarized text data.
>The below cells load in some AWS SageMaker libraries and creates a default bucket. After creating this bucket, you can upload your locally stored data to S3.
Save your train and test `.csv` feature files, locally. To do this you can run the second notebook "2_Plagiarism_Feature_Engineering" in SageMaker or you can manually upload your files to this notebook using the upload icon in Jupyter Lab. Then you can upload local files to S3 by using `sagemaker_session.upload_data` and pointing directly to where the training data is saved.
```
import pandas as pd
import boto3
import sagemaker
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# session and role
sagemaker_session = sagemaker.Session()
role = sagemaker.get_execution_role()
# create an S3 bucket
bucket = sagemaker_session.default_bucket()
```
## EXERCISE: Upload your training data to S3
Specify the `data_dir` where you've saved your `train.csv` file. Decide on a descriptive `prefix` that defines where your data will be uploaded in the default S3 bucket. Finally, create a pointer to your training data by calling `sagemaker_session.upload_data` and passing in the required parameters. It may help to look at the [Session documentation](https://sagemaker.readthedocs.io/en/stable/session.html#sagemaker.session.Session.upload_data) or previous SageMaker code examples.
You are expected to upload your entire directory. Later, the training script will only access the `train.csv` file.
```
# should be the name of directory you created to save your features data
data_dir = 'plagiarism_data'
# set prefix, a descriptive name for a directory
prefix = 'sagemaker/plagiarism-data'
# upload all data to S3
input_data = sagemaker_session.upload_data(path=data_dir, bucket=bucket, key_prefix=prefix)
print(input_data)
```
### Test cell
Test that your data has been successfully uploaded. The below cell prints out the items in your S3 bucket and will throw an error if it is empty. You should see the contents of your `data_dir` and perhaps some checkpoints. If you see any other files listed, then you may have some old model files that you can delete via the S3 console (though, additional files shouldn't affect the performance of model developed in this notebook).
```
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# confirm that data is in S3 bucket
empty_check = []
for obj in boto3.resource('s3').Bucket(bucket).objects.all():
empty_check.append(obj.key)
print(obj.key)
assert len(empty_check) !=0, 'S3 bucket is empty.'
print('Test passed!')
```
---
# Modeling
Now that you've uploaded your training data, it's time to define and train a model!
The type of model you create is up to you. For a binary classification task, you can choose to go one of three routes:
* Use a built-in classification algorithm, like LinearLearner.
* Define a custom Scikit-learn classifier, a comparison of models can be found [here](https://scikit-learn.org/stable/auto_examples/classification/plot_classifier_comparison.html).
* Define a custom PyTorch neural network classifier.
It will be up to you to test out a variety of models and choose the best one. Your project will be graded on the accuracy of your final model.
---
## EXERCISE: Complete a training script
To implement a custom classifier, you'll need to complete a `train.py` script. You've been given the folders `source_sklearn` and `source_pytorch` which hold starting code for a custom Scikit-learn model and a PyTorch model, respectively. Each directory has a `train.py` training script. To complete this project **you only need to complete one of these scripts**; the script that is responsible for training your final model.
A typical training script:
* Loads training data from a specified directory
* Parses any training & model hyperparameters (ex. nodes in a neural network, training epochs, etc.)
* Instantiates a model of your design, with any specified hyperparams
* Trains that model
* Finally, saves the model so that it can be hosted/deployed, later
### Defining and training a model
Much of the training script code is provided for you. Almost all of your work will be done in the `if __name__ == '__main__':` section. To complete a `train.py` file, you will:
1. Import any extra libraries you need
2. Define any additional model training hyperparameters using `parser.add_argument`
2. Define a model in the `if __name__ == '__main__':` section
3. Train the model in that same section
Below, you can use `!pygmentize` to display an existing `train.py` file. Read through the code; all of your tasks are marked with `TODO` comments.
**Note: If you choose to create a custom PyTorch model, you will be responsible for defining the model in the `model.py` file,** and a `predict.py` file is provided. If you choose to use Scikit-learn, you only need a `train.py` file; you may import a classifier from the `sklearn` library.
```
# directory can be changed to: source_sklearn or source_pytorch
!pygmentize source_sklearn/train.py
```
### Provided code
If you read the code above, you can see that the starter code includes a few things:
* Model loading (`model_fn`) and saving code
* Getting SageMaker's default hyperparameters
* Loading the training data by name, `train.csv` and extracting the features and labels, `train_x`, and `train_y`
If you'd like to read more about model saving with [joblib for sklearn](https://scikit-learn.org/stable/modules/model_persistence.html) or with [torch.save](https://pytorch.org/tutorials/beginner/saving_loading_models.html), click on the provided links.
---
# Create an Estimator
When a custom model is constructed in SageMaker, an entry point must be specified. This is the Python file which will be executed when the model is trained; the `train.py` function you specified above. To run a custom training script in SageMaker, construct an estimator, and fill in the appropriate constructor arguments:
* **entry_point**: The path to the Python script SageMaker runs for training and prediction.
* **source_dir**: The path to the training script directory `source_sklearn` OR `source_pytorch`.
* **entry_point**: The path to the Python script SageMaker runs for training and prediction.
* **source_dir**: The path to the training script directory `train_sklearn` OR `train_pytorch`.
* **entry_point**: The path to the Python script SageMaker runs for training.
* **source_dir**: The path to the training script directory `train_sklearn` OR `train_pytorch`.
* **role**: Role ARN, which was specified, above.
* **train_instance_count**: The number of training instances (should be left at 1).
* **train_instance_type**: The type of SageMaker instance for training. Note: Because Scikit-learn does not natively support GPU training, Sagemaker Scikit-learn does not currently support training on GPU instance types.
* **sagemaker_session**: The session used to train on Sagemaker.
* **hyperparameters** (optional): A dictionary `{'name':value, ..}` passed to the train function as hyperparameters.
Note: For a PyTorch model, there is another optional argument **framework_version**, which you can set to the latest version of PyTorch, `1.0`.
## EXERCISE: Define a Scikit-learn or PyTorch estimator
To import your desired estimator, use one of the following lines:
```
from sagemaker.sklearn.estimator import SKLearn
```
```
from sagemaker.pytorch import PyTorch
```
```
# your import and estimator code, here
# import a PyTorch wrapper
from sagemaker.pytorch import PyTorch
# specify an output path
output_path = f"s3://{bucket}/{prefix}"
# instantiate a pytorch estimator
estimator = PyTorch(
entry_point="train.py",
source_dir="source_pytorch",
role=role,
framework_version="1.0",
train_instance_count=1,
train_instance_type="ml.c4.xlarge",
output_path=output_path,
sagemaker_session=sagemaker_session,
hyperparameters={
"input_features": 2,
"hidden_dim": 20,
"output_dim": 1,
"epochs": 160
})
```
## EXERCISE: Train the estimator
Train your estimator on the training data stored in S3. This should create a training job that you can monitor in your SageMaker console.
```
train_data_path = input_data + "/train.csv"
print(train_data_path)
%%time
# Train your estimator on S3 training data
estimator.fit({'train': train_data_path})
```
## EXERCISE: Deploy the trained model
After training, deploy your model to create a `predictor`. If you're using a PyTorch model, you'll need to create a trained `PyTorchModel` that accepts the trained `<model>.model_data` as an input parameter and points to the provided `source_pytorch/predict.py` file as an entry point.
To deploy a trained model, you'll use `<model>.deploy`, which takes in two arguments:
* **initial_instance_count**: The number of deployed instances (1).
* **instance_type**: The type of SageMaker instance for deployment.
Note: If you run into an instance error, it may be because you chose the wrong training or deployment instance_type. It may help to refer to your previous exercise code to see which types of instances we used.
```
%%time
# uncomment, if needed
from sagemaker.pytorch import PyTorchModel
model = PyTorchModel(
entry_point="predict.py",
role=role,
framework_version="1.0",
model_data=estimator.model_data,
source_dir="source_pytorch"
)
# deploy your model to create a predictor
predictor = model.deploy(initial_instance_count=1, instance_type="ml.t2.medium")
```
---
# Evaluating Your Model
Once your model is deployed, you can see how it performs when applied to our test data.
The provided cell below, reads in the test data, assuming it is stored locally in `data_dir` and named `test.csv`. The labels and features are extracted from the `.csv` file.
```
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
import os
# read in test data, assuming it is stored locally
test_data = pd.read_csv(os.path.join(data_dir, "test.csv"), header=None, names=None)
# labels are in the first column
test_y = test_data.iloc[:,0]
test_x = test_data.iloc[:,1:]
```
## EXERCISE: Determine the accuracy of your model
Use your deployed `predictor` to generate predicted, class labels for the test data. Compare those to the *true* labels, `test_y`, and calculate the accuracy as a value between 0 and 1.0 that indicates the fraction of test data that your model classified correctly. You may use [sklearn.metrics](https://scikit-learn.org/stable/modules/classes.html#module-sklearn.metrics) for this calculation.
**To pass this project, your model should get at least 90% test accuracy.**
```
# First: generate predicted, class labels
test_y_preds = predictor.predict(test_x)
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# test that your model generates the correct number of labels
assert len(test_y_preds)==len(test_y), 'Unexpected number of predictions.'
print('Test passed!')
# Second: calculate the test accuracy
from sklearn.metrics import accuracy_score
accuracy = accuracy_score(test_y, test_y_preds)
print(accuracy)
## print out the array of predicted and true labels, if you want
print('\nPredicted class labels: ')
print(test_y_preds)
print('\nTrue class labels: ')
print(test_y.values)
```
### Question 1: How many false positives and false negatives did your model produce, if any? And why do you think this is?
** Answer**:
```
# code to evaluate the endpoint on test data
# returns a variety of model metrics
def evaluate(test_preds, test_labels, verbose=True):
# rounding and squeezing array
test_preds = np.squeeze(np.round(test_preds))
# calculate true positives, false positives, true negatives, false negatives
tp = np.logical_and(test_labels, test_preds).sum()
fp = np.logical_and(1-test_labels, test_preds).sum()
tn = np.logical_and(1-test_labels, 1-test_preds).sum()
fn = np.logical_and(test_labels, 1-test_preds).sum()
# calculate binary classification metrics
recall = tp / (tp + fn)
precision = tp / (tp + fp)
accuracy = (tp + tn) / (tp + fp + tn + fn)
# print metrics
if verbose:
print(pd.crosstab(test_labels, test_preds, rownames=['actuals'], colnames=['predictions']))
print("\n{:<11} {:.3f}".format('Recall:', recall))
print("{:<11} {:.3f}".format('Precision:', precision))
print("{:<11} {:.3f}".format('Accuracy:', accuracy))
print()
return {'TP': tp, 'FP': fp, 'FN': fn, 'TN': tn,
'Precision': precision, 'Recall': recall, 'Accuracy': accuracy}
metrics = evaluate(test_y_preds, test_y.values, True)
```
false positives is 1 and false negatives is 0. The result is pretty good. The reason may be 1. sample is small, 2. features is not enough and didn't describe too much characters of the text.
### Question 2: How did you decide on the type of model to use?
** Answer**:
The basic model of sklearn and pytorch are all linear model. The problem is linear inseparable. Thus, deep learning is better for this problem because the pytorch model stacks two layer linear models.
----
## EXERCISE: Clean up Resources
After you're done evaluating your model, **delete your model endpoint**. You can do this with a call to `.delete_endpoint()`. You need to show, in this notebook, that the endpoint was deleted. Any other resources, you may delete from the AWS console, and you will find more instructions on cleaning up all your resources, below.
```
# uncomment and fill in the line below!
predictor.delete_endpoint()
```
### Deleting S3 bucket
When you are *completely* done with training and testing models, you can also delete your entire S3 bucket. If you do this before you are done training your model, you'll have to recreate your S3 bucket and upload your training data again.
```
# deleting bucket, uncomment lines below
# bucket_to_delete = boto3.resource('s3').Bucket(bucket)
# bucket_to_delete.objects.all().delete()
```
### Deleting all your models and instances
When you are _completely_ done with this project and do **not** ever want to revisit this notebook, you can choose to delete all of your SageMaker notebook instances and models by following [these instructions](https://docs.aws.amazon.com/sagemaker/latest/dg/ex1-cleanup.html). Before you delete this notebook instance, I recommend at least downloading a copy and saving it, locally.
---
## Further Directions
There are many ways to improve or add on to this project to expand your learning or make this more of a unique project for you. A few ideas are listed below:
* Train a classifier to predict the *category* (1-3) of plagiarism and not just plagiarized (1) or not (0).
* Utilize a different and larger dataset to see if this model can be extended to other types of plagiarism.
* Use language or character-level analysis to find different (and more) similarity features.
* Write a complete pipeline function that accepts a source text and submitted text file, and classifies the submitted text as plagiarized or not.
* Use API Gateway and a lambda function to deploy your model to a web application.
These are all just options for extending your work. If you've completed all the exercises in this notebook, you've completed a real-world application, and can proceed to submit your project. Great job!
|
github_jupyter
|
# Batch Processing!
#### A notebook to show some of the capilities available through the pCunch package
This is certainly not an exhaustive look at everything that the pCrunch module can do, but should hopefully provide some insight.
...or, maybe I'm just procrastinating doing more useful work.
```
# Python Modules and instantiation
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import time
import os
# %matplotlib widget
# ROSCO toolbox modules
from ROSCO_toolbox import utilities as rosco_utilities
# WISDEM modules
from wisdem.aeroelasticse.Util import FileTools
# Batch Analysis tools
from pCrunch import Processing, Analysis
from pCrunch import pdTools
# Instantiate fast_IO
fast_io = rosco_utilities.FAST_IO()
fast_pl = rosco_utilities.FAST_Plots()
import importlib
Processing = importlib.reload(Processing)
Analysis = importlib.reload(Analysis)
```
## Define file paths and filenames
I'm loading a case matrix that is output when using wisdem.aeroelasticse.CaseGen_General to run a series of batch runs to initialize the output files here.
Note that this isn't necessary, just my workflow in this notebook.
```
# point to some file paths
outfile_base = '/Users/nabbas/Documents/Projects/ROSCO_dev/DLC_Analysis/DLC_Outputs/5MW_Land_DLC11/'
fname_case_matrix = os.path.join(outfile_base,'case_matrix.yaml')
# Load case matrix into datafraome
case_matrix = FileTools.load_yaml(fname_case_matrix, package=1)
cm = pd.DataFrame(case_matrix)
# pull wind speed values from InflowWind filenames
windspeeds, seed, IECtype, cmw = Processing.get_windspeeds(cm, return_df=True)
cmw.head()
```
#### Comparison cases
I'm comparing two different controllers here, so I'm going to define two lists of output filenames, each corresponding to the output files from each controller
```
# Define controllers we care to separate things by
controllers = list(set(cmw[('ServoDyn', 'DLL_FileName')]))
controllers
# Parse find outfiles names
outfiles = []
for cont in controllers:
case_names = cmw[cmw[('ServoDyn','DLL_FileName')]==cont]['Case_Name']
outnames = list( outfile_base + case_names + '.outb' )
outfiles.append(outnames)
```
### outfiles
In the end, we just need a list of OpenFAST output files. Here, we have a structure that looks something like `[[], []]`. This could be extended any amount like `[[],[],...,[], []]`, or just be one list of strings `[]`.
## Now we can do some processing!
First, let's load the FAST_Processing class and initialize some parameters.
```
fp = Processing.FAST_Processing()
fp.OpenFAST_outfile_list = outfiles
fp.dataset_names = ['DLC1.1', 'DLC1.3']
fp.to = 30
fp.parallel_analysis = True
fp.save_LoadRanking = False
fp.save_SummaryStats = False
fp.verbose=True
# # Can defined specific variables for load ranking if desired
# fp.ranking_vars = [["RotSpeed"],
# ["OoPDefl1", "OoPDefl2", "OoPDefl3"],
# ['RootMxc1', 'RootMxc2', 'RootMxc3'],
# ['TwrBsFyt'],
# ]
```
#### The fast way to compare things.
We could now collect all of the summary stats and load rankings using:
```
stats,load_rankings = fp.batch_processing()
```
In `fp.batch_processing()` most of the analysis is done for any structure of data. I'm going to step through things a bit more piecewise in this notebook, however.
NOTE: The goal in `batch_processing` is to have a "do anything" script. It is a work in progress, but getting there...
```
# stats,load_rankings = fp.batch_processing()
```
## Design Comparisons
We can use fp.design_comparison to compare multiple sets of runs (like we are in this case...). This will generate summary stats and load rankings, running in parrallel when it can and is told to. `fp.batch_processing()` functionally does the same thing if we give it an outfile matrix with equal size lists. We'll show the design comparison here to show a break down
```
stats, load_ranking = fp.design_comparison(outfiles)
```
#### Breaking it down further...
`fp.batch_processing()` calls `Analysis.Loads_Analysls.full_loads_analysis()` to load openfast data, generate stats, and calculate load rankings. Because we defined `fp.parallel_analysis=True` this process was parallelized. This helps for speed and memory reasons, because now every openfast run is not saved. `fp.batch_processing()` then takes all of the output data and parses it back together.
Separately, we call call `Analysis.Loads_Analysls.full_loads_analysis()` with `return_FastData=True` and all of the fast data will be returned. Because we are comparing data though, we'll stick with the design comparison tools.
#### Loading data
We can also just load previously parsed data if we ran `FAST_Processing` with the `save_LoadRankings` and `save_SummaryStates` flags as True.
```
# Or load stats and load rankings
root = '/Users/nabbas/Documents/Projects/ROSCO_dev/DLC_Analysis/DLC_Outputs/5MW_Land_DLC11/stats/'
lrfile = [root+'dataset0_LoadRanking.yaml', root+'dataset1_LoadRanking.yaml']
sfile = [root+'dataset0_stats.yaml', root+'dataset1_stats.yaml']
fname_case_matrix = root+'../case_matrix.yaml'
stats = [FileTools.load_yaml(sf, package=1) for sf in sfile]
load_rankings = [FileTools.load_yaml(lf, package=1) for lf in lrfile]
case_matrix = FileTools.load_yaml(fname_case_matrix, package=1)
cm = pd.DataFrame(case_matrix)
```
### We can look at our data a bit further with pandas dataframes
The data here is just for a few runs for simplicity. Usually you'd do this for a LOT more cases...
```
stats_df = pdTools.dict2df(stats, names=['ROSCO', 'Legacy'])
stats_df.head()
```
### Load Ranking
Lets re-run the load ranking for the sake of example. We'll have to load the analysis tools, and then run the load ranking for the stats we just found
```
fa = Analysis.Loads_Analysis()
fa.t0 = 30
fa.verbose = False
```
Define the ranking variables and statiscits of interest. Note that `len(ranking_vars) == len(ranking_stats)`! We can pass this a list of stats (multiple runs), a dictionary with one run of stats, or a pandas dataframe with the requisite stats. If the inner list contains multiple OpenFAST channels, the load_rankings function will find the min/max/mean of the collection of the channels (e.g., max out of plane tip deflection of all three blades).
We'll also output a dictionary and a pandas DataFrame from `fa.load_ranking()`
```
fa.ranking_vars = [['TwrBsFxt'], ['OoPDefl1', 'OoPDefl2', 'OoPDefl3']]
fa.ranking_stats = ['max', 'min']
load_ranking, load_ranking_df = fa.load_ranking(stats_df, get_df=True)
load_ranking_df.head()
```
This is organized for each iteration of `[ranking_vars, ranking_stats]`. The stats are ordered accordingly, and `(stat)_case_idx` refers to the case name index of each load.
## Wind speed related analysis
We often want to make sense of some batch output data with data binned by windspeed. We can leverage the case-matrix from our output data to figure out the input wind speeds. Of course, `('InflowWind', 'Filename')` must exist in the case matrix. Lets load the wind speeds, save them, and append them to the case matrix as `('InflowWind', 'WindSpeed')`.
```
windspeed, seed, IECtype, cmw = Processing.get_windspeeds(cm, return_df=True)
cmw
```
### AEP
Now that we know the wind speeds that we were operating at, we can find the AEP. We define the turbine class here, and the cumulative distribution or probability density function
for the Weibull distribution per IEC 61400 is generated. We can then calculate the AEP.
If we first want to verify the PDF, we initialize the `power_production` function, define the turbine class, and can plot a PDF (or CDF) for a given range of wind speeds:
```
pp = Analysis.Power_Production()
pp.turbine_class = 2
Vrange = np.arange(2,26) # Range of wind speeds being considered
weib_prob = pp.prob_WindDist(Vrange,disttype='pdf')
plt.close('all')
plt.plot(Vrange, weib_prob)
plt.grid(True)
plt.xlabel("Wind Speed m/s")
plt.ylabel('Probability')
plt.title('Probability Density Function \n IEC Class 2 Wind Speeds ')
plt.show()
```
To get the AEP, we need to provide the wind speeds that the simulations were run for, and the corresponding average power results. Internally, in power_production.AEP, the mean power for a given average wind sped is multiplied times the wind speed's probability, then extrapolated to represent yearly production.
Note: this might throw a python warning due to some poor pandas indexing practices - to be cleaned up eventually!
To get the AEP for each, the process is simple:
```
AEP = pp.AEP(stats, windspeeds)
print('AEP = {}'.format(AEP))
```
##### About the wind speed warning:
Here, we get a warning about the input windspeed array. This is because we passed the complete array output from Processing.get_windspeeds to the AEP function. The input windspeeds to power_production.AEP must satisfy either of the following two conditions:
- each wind speed value corresponds to each each statistic value, so `len(windspeeds) = len(stats_df)`
- each wind speed value corresponds to each run in the case matrix, so `len(windspeeds) = len(cm)`
If the second of these conditions is satisfied, it is assumed that each dataset has the same wind speeds corresponding to each run. So, in this case, the wind speeds corresponding to DLC_1.1 and DLC_1.3 should be the same.
## Plotting
Finally, we can make some plots. There are a few tools we have at our disposal here. First, we can look at more plots that show our design performance as a function of wind speed. Notably, we can pass the stats dictionary or dataframe to these statistics-related scripts.
Currently, `an_plts.stat_curve()` can plot a "statistics curve" for of two types, a bar or a line graph.
A bar graph is useful to compare design cases easily:
```
plt.close()
an_plts = Analysis.wsPlotting()
an_plts.stat_curve(windspeed, stats, 'TwrBsFxt', 'bar', names=['ROSCO', 'Legacy'])
plt.show()
```
A line graph can be useful to show turbulent wind curves. Here we show the means with a first level of errorbars corresponding to standard deviations, and a second level showing minimums and maximums.
```
an_plts.stat_curve(windspeed, stats, 'GenPwr', 'line', stat_idx=0, names=['ROSCO'])
plt.show()
```
### Load Ranking (soon)
We can plot the load rankings...
... pulling this into `Analysis.py` is in progress.
First, we define how we will classify our comparisons. Most commonly this would be `('IEC','DLC')`, but I'm comparing controllers here. The `classifier_type` functionally refers to the channel of the case matrix to separate the data by, and the `classifier_names` are simply labels for the classifiers.
```
# Define a classification channel from the case-matrix
classifier_type = ('ServoDyn', 'DLL_FileName')
classifier_names = ['ROSCO', 'legacy']
# Plot load rankings
fig_list, ax_list = an_plts.plot_load_ranking(load_ranking, cm, classifier_type, classifier_names=classifier_names, n_rankings=10, caseidx_labels=True)
# modify axis labels
for ax in ax_list:
ax.set_xlabel('Controller [-]', fontsize=10, fontweight='bold')
plt.show()
```
### Time domain plotting
We can also look at our data from the time domain results.
We can compare any number of channels using the ROSCO toolbox plotting tools. First we'll load two cases to plot together, then plot the time histories.
```
# Load some time domain cases
filenames = [outfiles[0][70], outfiles[1][70]] # select the 70th run from each dataset
fast_data = fast_io.load_FAST_out(filenames, tmin=30)
# Change names so the legends make sense
fast_data[0]['meta']['name'] = 'ROSCO'
fast_data[1]['meta']['name'] = 'Legacy'
# Define the plots we want to make (can be as many or as few channels and plots as you would like...)
cases = {'Baseline': ['Wind1VelX', 'GenPwr', 'BldPitch1', 'GenTq', 'RotSpeed'],
'Blade' : ['OoPDefl1', 'RootMyb1']}
# plot
fast_pl.plot_fast_out(cases, fast_data)
plt.show()
```
### Spectral Analysis
We can additionally do some frequency domain analysis. Here, `spec_cases` is defined by `(channel, run)` where the run index corresponds to the desired plotting index in the loaded fast data.
```
spec_cases = [('RootMyb1', 0), ('TwrBsFxt', 1)]
twrfreq = .0716
twrfreq_label = ['Tower']
fig, ax = fast_pl.plot_spectral(fast_data, spec_cases,
show_RtSpeed=True, RtSpeed_idx=[0],
add_freqs=[twrfreq], add_freq_labels=twrfreq_label,
averaging='Welch')
ax.set_title('DLC_1.1')
plt.show()
```
### Other fun plots
Finally, we can plot the data distribution of any channels from our fast output data
```
channels = ['GenPwr']
caseid = [0,1]
an_plts.distribution(fast_data, channels, caseid, names=['ROSCO', 'Legacy'])
plt.show()
```
## In conclusion...
If you made it this far, thanks for reading...
There are a number of smaller subfunctionalities that are also available within these tools shows above. Perhaps most importantly, everything is fairly modularar - the hope being that these can provide some high-level tools that everyone can assimilate into their own workflows without too much disruption.
Please add, contribute, fix, etc... That would be great for everyone involved!
|
github_jupyter
|
# Deep learning for computer vision
This notebook will teach you to build and train convolutional networks for image recognition. Brace yourselves.
# CIFAR dataset
This week, we shall focus on the image recognition problem on cifar10 dataset
* 60k images of shape 3x32x32
* 10 different classes: planes, dogs, cats, trucks, etc.
<img src="cifar10.jpg" style="width:80%">
```
import numpy as np
from cifar import load_cifar10
X_train,y_train,X_val,y_val,X_test,y_test = load_cifar10("cifar_data")
class_names = np.array(['airplane','automobile ','bird ','cat ','deer ','dog ','frog ','horse ','ship ','truck'])
print (X_train.shape,y_train.shape)
import matplotlib.pyplot as plt
%matplotlib inline
plt.figure(figsize=[12,10])
for i in range(12):
plt.subplot(3,4,i+1)
plt.xlabel(class_names[y_train[i]])
plt.imshow(np.transpose(X_train[i],[1,2,0]))
```
# Building a network
Simple neural networks with layers applied on top of one another can be implemented as `torch.nn.Sequential` - just add a list of pre-built modules and let it train.
```
import torch, torch.nn as nn
import torch.nn.functional as F
from torch.autograd import Variable
# a special module that converts [batch, channel, w, h] to [batch, units]
class Flatten(nn.Module):
def forward(self, input):
return input.view(input.size(0), -1)
```
Let's start with a dense network for our baseline:
```
model = nn.Sequential()
# reshape from "images" to flat vectors
model.add_module('flatten', Flatten())
# dense "head"
model.add_module('dense1', nn.Linear(3 * 32 * 32, 64))
model.add_module('dense1_relu', nn.ReLU())
model.add_module('dense2_logits', nn.Linear(64, 10)) # logits for 10 classes
```
As in our basic tutorial, we train our model with negative log-likelihood aka crossentropy.
```
def compute_loss(X_batch, y_batch):
X_batch = Variable(torch.FloatTensor(X_batch))
y_batch = Variable(torch.LongTensor(y_batch))
logits = model(X_batch)
return F.cross_entropy(logits, y_batch).mean()
# example
compute_loss(X_train[:5], y_train[:5])
```
### Training on minibatches
* We got 40k images, that's way too many for a full-batch SGD. Let's train on minibatches instead
* Below is a function that splits the training sample into minibatches
```
# An auxilary function that returns mini-batches for neural network training
def iterate_minibatches(X, y, batchsize):
indices = np.random.permutation(np.arange(len(X)))
for start in range(0, len(indices), batchsize):
ix = indices[start: start + batchsize]
yield X[ix], y[ix]
opt = torch.optim.SGD(model.parameters(), lr=0.01)
train_loss = []
val_accuracy = []
import time
num_epochs = 100 # total amount of full passes over training data
batch_size = 50 # number of samples processed in one SGD iteration
for epoch in range(num_epochs):
# In each epoch, we do a full pass over the training data:
start_time = time.time()
model.train(True) # enable dropout / batch_norm training behavior
for X_batch, y_batch in iterate_minibatches(X_train, y_train, batch_size):
# train on batch
loss = compute_loss(X_batch, y_batch)
loss.backward()
opt.step()
opt.zero_grad()
train_loss.append(loss.data.numpy())
# And a full pass over the validation data:
model.train(False) # disable dropout / use averages for batch_norm
for X_batch, y_batch in iterate_minibatches(X_val, y_val, batch_size):
logits = model(Variable(torch.FloatTensor(X_batch)))
y_pred = logits.max(1)[1].data.numpy()
val_accuracy.append(np.mean(y_batch == y_pred))
# Then we print the results for this epoch:
print("Epoch {} of {} took {:.3f}s".format(
epoch + 1, num_epochs, time.time() - start_time))
print(" training loss (in-iteration): \t{:.6f}".format(
np.mean(train_loss[-len(X_train) // batch_size :])))
print(" validation accuracy: \t\t\t{:.2f} %".format(
np.mean(val_accuracy[-len(X_val) // batch_size :]) * 100))
```
Don't wait for full 100 epochs. You can interrupt training after 5-20 epochs once validation accuracy stops going up.
```
```
```
```
```
```
```
```
```
```
### Final test
```
model.train(False) # disable dropout / use averages for batch_norm
test_batch_acc = []
for X_batch, y_batch in iterate_minibatches(X_test, y_test, 500):
logits = model(Variable(torch.FloatTensor(X_batch)))
y_pred = logits.max(1)[1].data.numpy()
test_batch_acc.append(np.mean(y_batch == y_pred))
test_accuracy = np.mean(test_batch_acc)
print("Final results:")
print(" test accuracy:\t\t{:.2f} %".format(
test_accuracy * 100))
if test_accuracy * 100 > 95:
print("Double-check, than consider applying for NIPS'17. SRSly.")
elif test_accuracy * 100 > 90:
print("U'r freakin' amazin'!")
elif test_accuracy * 100 > 80:
print("Achievement unlocked: 110lvl Warlock!")
elif test_accuracy * 100 > 70:
print("Achievement unlocked: 80lvl Warlock!")
elif test_accuracy * 100 > 60:
print("Achievement unlocked: 70lvl Warlock!")
elif test_accuracy * 100 > 50:
print("Achievement unlocked: 60lvl Warlock!")
else:
print("We need more magic! Follow instructons below")
```
## Task I: small convolution net
### First step
Let's create a mini-convolutional network with roughly such architecture:
* Input layer
* 3x3 convolution with 10 filters and _ReLU_ activation
* 2x2 pooling (or set previous convolution stride to 3)
* Flatten
* Dense layer with 100 neurons and _ReLU_ activation
* 10% dropout
* Output dense layer.
__Convolutional layers__ in torch are just like all other layers, but with a specific set of parameters:
__`...`__
__`model.add_module('conv1', nn.Conv2d(in_channels=3, out_channels=10, kernel_size=3)) # convolution`__
__`model.add_module('pool1', nn.MaxPool2d(2)) # max pooling 2x2`__
__`...`__
Once you're done (and compute_loss no longer raises errors), train it with __Adam__ optimizer with default params (feel free to modify the code above).
If everything is right, you should get at least __50%__ validation accuracy.
```
```
```
```
```
```
```
```
```
```
__Hint:__ If you don't want to compute shapes by hand, just plug in any shape (e.g. 1 unit) and run compute_loss. You will see something like this:
__`RuntimeError: size mismatch, m1: [5 x 1960], m2: [1 x 64] at /some/long/path/to/torch/operation`__
See the __1960__ there? That's your actual input shape.
## Task 2: adding normalization
* Add batch norm (with default params) between convolution and ReLU
* nn.BatchNorm*d (1d for dense, 2d for conv)
* usually better to put them after linear/conv but before nonlinearity
* Re-train the network with the same optimizer, it should get at least 60% validation accuracy at peak.
```
```
```
```
```
```
```
```
```
```
```
```
```
```
## Task 3: Data Augmentation
There's a powerful torch tool for image preprocessing useful to do data preprocessing and augmentation.
Here's how it works: we define a pipeline that
* makes random crops of data (augmentation)
* randomly flips image horizontally (augmentation)
* then normalizes it (preprocessing)
```
from torchvision import transforms
means = np.array((0.4914, 0.4822, 0.4465))
stds = np.array((0.2023, 0.1994, 0.2010))
transform_augment = transforms.Compose([
transforms.RandomCrop(32, padding=4),
transforms.RandomRotation([-30, 30]),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(means, stds),
])
from torchvision.datasets import CIFAR10
train_loader = CIFAR10("./cifar_data/", train=True, transform=transform_augment)
train_batch_gen = torch.utils.data.DataLoader(train_loader,
batch_size=32,
shuffle=True,
num_workers=1)
for (x_batch, y_batch) in train_batch_gen:
print('X:', type(x_batch), x_batch.shape)
print('y:', type(y_batch), y_batch.shape)
for i, img in enumerate(x_batch.numpy()[:8]):
plt.subplot(2, 4, i+1)
plt.imshow(img.transpose([1,2,0]) * stds + means )
raise NotImplementedError("Plese use this code in your training loop")
# TODO use this in your training loop
```
When testing, we don't need random crops, just normalize with same statistics.
```
transform_test = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(means, stds),
])
test_loader = <YOUR CODE>
```
# Homework 2.2: The Quest For A Better Network
In this assignment you will build a monster network to solve CIFAR10 image classification.
This notebook is intended as a sequel to seminar 3, please give it a try if you haven't done so yet.
(please read it at least diagonally)
* The ultimate quest is to create a network that has as high __accuracy__ as you can push it.
* There is a __mini-report__ at the end that you will have to fill in. We recommend reading it first and filling it while you iterate.
## Grading
* starting at zero points
* +20% for describing your iteration path in a report below.
* +20% for building a network that gets above 20% accuracy
* +10% for beating each of these milestones on __TEST__ dataset:
* 50% (50% points)
* 60% (60% points)
* 65% (70% points)
* 70% (80% points)
* 75% (90% points)
* 80% (full points)
## Restrictions
* Please do NOT use pre-trained networks for this assignment until you reach 80%.
* In other words, base milestones must be beaten without pre-trained nets (and such net must be present in the e-mail). After that, you can use whatever you want.
* you __can__ use validation data for training, but you __can't'__ do anything with test data apart from running the evaluation procedure.
## Tips on what can be done:
* __Network size__
* MOAR neurons,
* MOAR layers, ([torch.nn docs](http://pytorch.org/docs/master/nn.html))
* Nonlinearities in the hidden layers
* tanh, relu, leaky relu, etc
* Larger networks may take more epochs to train, so don't discard your net just because it could didn't beat the baseline in 5 epochs.
* Ph'nglui mglw'nafh Cthulhu R'lyeh wgah'nagl fhtagn!
### The main rule of prototyping: one change at a time
* By now you probably have several ideas on what to change. By all means, try them out! But there's a catch: __never test several new things at once__.
### Optimization
* Training for 100 epochs regardless of anything is probably a bad idea.
* Some networks converge over 5 epochs, others - over 500.
* Way to go: stop when validation score is 10 iterations past maximum
* You should certainly use adaptive optimizers
* rmsprop, nesterov_momentum, adam, adagrad and so on.
* Converge faster and sometimes reach better optima
* It might make sense to tweak learning rate/momentum, other learning parameters, batch size and number of epochs
* __BatchNormalization__ (nn.BatchNorm2d) for the win!
* Sometimes more batch normalization is better.
* __Regularize__ to prevent overfitting
* Add some L2 weight norm to the loss function, PyTorch will do the rest
* Can be done manually or like [this](https://discuss.pytorch.org/t/simple-l2-regularization/139/2).
* Dropout (`nn.Dropout`) - to prevent overfitting
* Don't overdo it. Check if it actually makes your network better
### Convolution architectures
* This task __can__ be solved by a sequence of convolutions and poolings with batch_norm and ReLU seasoning, but you shouldn't necessarily stop there.
* [Inception family](https://hacktilldawn.com/2016/09/25/inception-modules-explained-and-implemented/), [ResNet family](https://towardsdatascience.com/an-overview-of-resnet-and-its-variants-5281e2f56035?gi=9018057983ca), [Densely-connected convolutions (exotic)](https://arxiv.org/abs/1608.06993), [Capsule networks (exotic)](https://arxiv.org/abs/1710.09829)
* Please do try a few simple architectures before you go for resnet-152.
* Warning! Training convolutional networks can take long without GPU. That's okay.
* If you are CPU-only, we still recomment that you try a simple convolutional architecture
* a perfect option is if you can set it up to run at nighttime and check it up at the morning.
* Make reasonable layer size estimates. A 128-neuron first convolution is likely an overkill.
* __To reduce computation__ time by a factor in exchange for some accuracy drop, try using __stride__ parameter. A stride=2 convolution should take roughly 1/4 of the default (stride=1) one.
### Data augmemntation
* getting 5x as large dataset for free is a great
* Zoom-in+slice = move
* Rotate+zoom(to remove black stripes)
* Add Noize (gaussian or bernoulli)
* Simple way to do that (if you have PIL/Image):
* ```from scipy.misc import imrotate,imresize```
* and a few slicing
* Other cool libraries: cv2, skimake, PIL/Pillow
* A more advanced way is to use torchvision transforms:
```
transform_train = transforms.Compose([
transforms.RandomCrop(32, padding=4),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010)),
])
trainset = torchvision.datasets.CIFAR10(root=path_to_cifar_like_in_seminar, train=True, download=True, transform=transform_train)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=128, shuffle=True, num_workers=2)
```
* Or use this tool from Keras (requires theano/tensorflow): [tutorial](https://blog.keras.io/building-powerful-image-classification-models-using-very-little-data.html), [docs](https://keras.io/preprocessing/image/)
* Stay realistic. There's usually no point in flipping dogs upside down as that is not the way you usually see them.
```
```
```
```
```
```
```
```
There is a template for your solution below that you can opt to use or throw away and write it your way.
|
github_jupyter
|
# Load MXNet model
In this tutorial, you learn how to load an existing MXNet model and use it to run a prediction task.
## Preparation
This tutorial requires the installation of Java Kernel. For more information on installing the Java Kernel, see the [README](https://github.com/awslabs/djl/blob/master/jupyter/README.md) to install Java Kernel.
```
%mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/
%maven ai.djl:api:0.3.0-SNAPSHOT
%maven ai.djl:repository:0.3.0-SNAPSHOT
%maven ai.djl:model-zoo:0.3.0-SNAPSHOT
%maven ai.djl.mxnet:mxnet-engine:0.3.0-SNAPSHOT
%maven ai.djl.mxnet:mxnet-model-zoo:0.3.0-SNAPSHOT
%maven org.slf4j:slf4j-api:1.7.26
%maven org.slf4j:slf4j-simple:1.7.26
%maven net.java.dev.jna:jna:5.3.0
// See https://github.com/awslabs/djl/blob/master/mxnet/mxnet-engine/README.md
// for more MXNet library selection options
%maven ai.djl.mxnet:mxnet-native-auto:1.6.0-SNAPSHOT
import java.awt.image.*;
import java.nio.file.*;
import java.util.*;
import java.util.stream.*;
import ai.djl.*;
import ai.djl.inference.*;
import ai.djl.ndarray.*;
import ai.djl.ndarray.index.*;
import ai.djl.modality.*;
import ai.djl.modality.cv.*;
import ai.djl.modality.cv.util.*;
import ai.djl.modality.cv.transform.*;
import ai.djl.mxnet.zoo.*;
import ai.djl.translate.*;
import ai.djl.training.util.*;
import ai.djl.util.*;
import ai.djl.basicmodelzoo.cv.classification.*;
```
## Step 1: Prepare your MXNet model
This tutorial assumes that you have a MXNet model trained using Python. A MXNet symbolic model usually contains the following files:
* Symbol file: {MODEL_NAME}-symbol.json - a json file that contains network information about the model
* Parameters file: {MODEL_NAME}-{EPOCH}.params - a binary file that stores the parameter weight and bias
* Synset file: synset.txt - an optional text file that stores classification classes labels
This tutorial uses a pre-trained MXNet `resnet18_v1` model.
We use [DownloadUtils.java] for downloading files from internet.
```
%load DownloadUtils.java
DownloadUtils.download("https://mlrepo.djl.ai/model/cv/image_classification/ai/djl/mxnet/resnet/0.0.1/resnet18_v1-symbol.json", "build/resnet/resnet18_v1-symbol.json", new ProgressBar());
DownloadUtils.download("https://mlrepo.djl.ai/model/cv/image_classification/ai/djl/mxnet/resnet/0.0.1/resnet18_v1-0000.params.gz", "build/resnet/resnet18_v1-0000.params", new ProgressBar());
DownloadUtils.download("https://mlrepo.djl.ai/model/cv/image_classification/ai/djl/mxnet/synset.txt", "build/resnet/synset.txt", new ProgressBar());
```
## Step 2: Load your model
```
Path modelDir = Paths.get("build/resnet");
Model model = Model.newInstance();
model.load(modelDir, "resnet18_v1");
```
## Step 3: Create a `Translator`
```
Pipeline pipeline = new Pipeline();
pipeline.add(new CenterCrop()).add(new Resize(224, 224)).add(new ToTensor());
Translator<BufferedImage, Classifications> translator = ImageClassificationTranslator.builder()
.setPipeline(pipeline)
.setSynsetArtifactName("synset.txt")
.build();
```
## Step 4: Load image for classification
```
var img = BufferedImageUtils.fromUrl("https://djl-ai.s3.amazonaws.com/resources/images/kitten.jpg");
img
```
## Step 5: Run inference
```
Predictor<BufferedImage, Classifications> predictor = model.newPredictor(translator);
Classifications classifications = predictor.predict(img);
classifications
```
## Summary
Now, you can load any MXNet symbolic model and run inference.
|
github_jupyter
|
# Modeling and Simulation in Python
Chapter 18
Copyright 2017 Allen Downey
License: [Creative Commons Attribution 4.0 International](https://creativecommons.org/licenses/by/4.0)
```
# Configure Jupyter so figures appear in the notebook
%matplotlib inline
# Configure Jupyter to display the assigned value after an assignment
%config InteractiveShell.ast_node_interactivity='last_expr_or_assign'
# import functions from the modsim.py module
from modsim import *
```
### Code from the previous chapter
Read the data.
```
data = pd.read_csv('data/glucose_insulin.csv', index_col='time');
```
Interpolate the insulin data.
```
I = interpolate(data.insulin)
```
Initialize the parameters
```
G0 = 290
k1 = 0.03
k2 = 0.02
k3 = 1e-05
```
To estimate basal levels, we'll use the concentrations at `t=0`.
```
Gb = data.glucose[0]
Ib = data.insulin[0]
```
Create the initial condtions.
```
init = State(G=G0, X=0)
```
Make the `System` object.
```
t_0 = get_first_label(data)
t_end = get_last_label(data)
system = System(init=init,
k1=k1, k2=k2, k3=k3,
I=I, Gb=Gb, Ib=Ib,
t_0=t_0, t_end=t_end, dt=2)
def update_func(state, t, system):
"""Updates the glucose minimal model.
state: State object
t: time in min
system: System object
returns: State object
"""
G, X = state
unpack(system)
dGdt = -k1 * (G - Gb) - X*G
dXdt = k3 * (I(t) - Ib) - k2 * X
G += dGdt * dt
X += dXdt * dt
return State(G=G, X=X)
def run_simulation(system, update_func):
"""Runs a simulation of the system.
system: System object
update_func: function that updates state
returns: TimeFrame
"""
unpack(system)
frame = TimeFrame(columns=init.index)
frame.row[t_0] = init
ts = linrange(t_0, t_end, dt)
for t in ts:
frame.row[t+dt] = update_func(frame.row[t], t, system)
return frame
%time results = run_simulation(system, update_func);
```
### Numerical solution
In the previous chapter, we approximated the differential equations with difference equations, and solved them using `run_simulation`.
In this chapter, we solve the differential equation numerically using `run_ode_solver`, which is a wrapper for the SciPy ODE solver.
Instead of an update function, we provide a slope function that evaluates the right-hand side of the differential equations. We don't have to do the update part; the solver does it for us.
```
def slope_func(state, t, system):
"""Computes derivatives of the glucose minimal model.
state: State object
t: time in min
system: System object
returns: derivatives of G and X
"""
G, X = state
unpack(system)
dGdt = -k1 * (G - Gb) - X*G
dXdt = k3 * (I(t) - Ib) - k2 * X
return dGdt, dXdt
```
We can test the slope function with the initial conditions.
```
slope_func(init, 0, system)
```
Here's how we run the ODE solver.
```
%time results2, details = run_ode_solver(system, slope_func, t_eval=data.index);
```
`details` is a `ModSimSeries` object with information about how the solver worked.
```
details
```
`results` is a `TimeFrame` with one row for each time step and one column for each state variable:
```
results2
```
Plotting the results from `run_simulation` and `run_ode_solver`, we can see that they are not very different.
```
plot(results.G, 'g-')
plot(results2.G, 'b-')
plot(data.glucose, 'bo')
```
The differences in `G` are less than 1%.
```
diff = results.G - results2.G
percent_diff = diff / results2.G * 100
percent_diff.dropna()
```
### Optimization
Now let's find the parameters that yield the best fit for the data.
We'll use these values as an initial estimate and iteratively improve them.
```
params = Params(G0 = 290,
k1 = 0.03,
k2 = 0.02,
k3 = 1e-05)
```
`make_system` takes the parameters and actual data and returns a `System` object.
```
def make_system(params, data):
"""Makes a System object with the given parameters.
params: sequence of G0, k1, k2, k3
data: DataFrame with `glucose` and `insulin`
returns: System object
"""
G0, k1, k2, k3 = params
Gb = data.glucose[0]
Ib = data.insulin[0]
t_0 = get_first_label(data)
t_end = get_last_label(data)
init = State(G=G0, X=0)
return System(G0=G0, k1=k1, k2=k2, k3=k3,
init=init, Gb=Gb, Ib=Ib,
t_0=t_0, t_end=t_end)
system = make_system(params, data)
```
`error_func` takes the parameters and actual data, makes a `System` object, and runs `odeint`, then compares the results to the data. It returns an array of errors.
```
def error_func(params, data):
"""Computes an array of errors to be minimized.
params: sequence of parameters
data: DataFrame of values to be matched
returns: array of errors
"""
print(params)
# make a System with the given parameters
system = make_system(params, data)
# solve the ODE
results, details = run_ode_solver(system, slope_func, t_eval=data.index)
# compute the difference between the model
# results and actual data
errors = results.G - data.glucose
return errors
```
When we call `error_func`, we provide a sequence of parameters as a single object.
Here's how that works:
```
error_func(params, data)
```
`fit_leastsq` is a wrapper for `scipy.optimize.leastsq`
Here's how we call it.
```
best_params, fit_details = fit_leastsq(error_func, params, data)
```
The first return value is a `Params` object with the best parameters:
```
best_params
```
The second return value is a `ModSimSeries` object with information about the results.
```
fit_details
fit_details
```
Now that we have `best_params`, we can use it to make a `System` object and run it.
```
system = make_system(best_params, data)
results, details = run_ode_solver(system, slope_func, t_eval=data.index)
details.message
```
Here are the results, along with the data. The first few points of the model don't fit the data, but we don't expect them to.
```
plot(results.G, label='simulation')
plot(data.glucose, 'bo', label='glucose data')
decorate(xlabel='Time (min)',
ylabel='Concentration (mg/dL)')
savefig('figs/chap08-fig04.pdf')
```
### Interpreting parameters
Based on the parameters of the model, we can estimate glucose effectiveness and insulin sensitivity.
```
def indices(params):
"""Compute glucose effectiveness and insulin sensitivity.
params: sequence of G0, k1, k2, k3
data: DataFrame with `glucose` and `insulin`
returns: State object containing S_G and S_I
"""
G0, k1, k2, k3 = params
return State(S_G=k1, S_I=k3/k2)
```
Here are the results.
```
indices(best_params)
```
### Under the hood
Here's the source code for `run_ode_solver` and `fit_leastsq`, if you'd like to know how they work.
```
%psource run_ode_solver
%psource fit_leastsq
```
## Exercises
**Exercise:** Since we don't expect the first few points to agree, it's probably better not to make them part of the optimization process. We can ignore them by leaving them out of the `Series` returned by `error_func`. Modify the last line of `error_func` to return `errors.loc[8:]`, which includes only the elements of the `Series` from `t=8` and up.
Does that improve the quality of the fit? Does it change the best parameters by much?
Note: You can read more about this use of `loc` [in the Pandas documentation](https://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-integer).
**Exercise:** How sensitive are the results to the starting guess for the parameters. If you try different values for the starting guess, do we get the same values for the best parameters?
**Related reading:** You might be interested in this article about [people making a DIY artificial pancreas](https://www.bloomberg.com/news/features/2018-08-08/the-250-biohack-that-s-revolutionizing-life-with-diabetes).
|
github_jupyter
|
```
%load_ext autoreload
%autoreload 2
import ambry
l = ambry.get_library()
b = l.bundle('d04w001') # Geoschemas
sumlevels_p = l.partition('census.gov-acs_geofile-schemas-2009e-sumlevels')
sumlevels = {}
for row in sumlevels_p.stream(as_dict=True):
sumlevels[row['sumlevel']] = row['description']
from collections import defaultdict, Counter
from geoid import base62_encode
collector = {}
geoids = {}
descriptions = {}
for p in b.partitions:
#print "=====", p.identity.name
l = {}
for i, c in enumerate(p.table.columns):
if i > 5 and c.name not in ('name','geoid', 'memi'):
l[c.name] = [Counter(), 0]
descriptions[c.name] = c.description
for i, row in enumerate(p.stream(as_dict=True)):
if i >= 500:
break
geoid = row['geoid']
for k in l:
v = row[k]
if not str(v).strip():
continue
try:
# The index is not guarantted to be found in the right position; it could be at the start of the
# geoid, so we keep track of the most common place it is found
idx = geoid.index(str(v))
size = len(str(v))
# Kepp tract of the right end position, not the start, since the end pos is independent of the length
l[k][0][idx+size] += 1
l[k][1] = max(l[k][1], size)
except ValueError:
pass
ordered = []
for k, v in l.items():
most = v[0].most_common(1)
if most:
size = v[1]
start = most[0][0] - size
ordered.append((k, start, size))
ordered = sorted(ordered, key = lambda r: r[1])
#for e in ordered:
# print " ", e, len(base62_encode(10**e[2]))
geoids[int(p.grain)] = ordered
for e in ordered:
collector[e[0]]=(e[2],len(base62_encode(10**e[2])) )
# Print out the lengths array
out = []
for k, v in collector.items():
out.append('\'{}\': {}, # {}'.format(k, v[0], descriptions[k]))
print '\n'.join(sorted(out))
for sl in sorted(geoids):
ordered = geoids[sl]
print str(sl)+':', str([ str(e[0]) for e in ordered ])+',', "#", sumlevels[sl]
from geoid import names, segments
names_map = {v:k for k, v in names.items()}
seen = set()
for k, v in segments.items():
if k in names_map:
pass
else:
name = '_'.join( e for e in v)
name = name[0].lower() + name[1:]
if name in seen:
name += str(k)
seen.add(name)
print "'{}': {},".format(name, k)
%load_ext autoreload
%autoreload 2
from geoid.acs import AcsGeoid
for p in b.partitions:
for i, row in enumerate(p.stream(as_dict=True)):
if i >= 500:
break
geoid = row['geoid']
try:
AcsGeoid.parse(geoid)
except Exception as e:
print geoid, e
raise
```
|
github_jupyter
|
```
import sys
from pathlib import Path
sys.path.append(str(Path.cwd().parent.parent))
import numpy as np
from kymatio.scattering2d.core.scattering2d import scattering2d
import matplotlib.pyplot as plt
import torch
import torchvision
from kymatio import Scattering2D
from PIL import Image
from IPython.display import display
from torchvision.transforms import *
#img = Image.open('/NOBACKUP/gauthiers/KTH/sample_a/wood/54a-scale_10_im_10_col.png')
img = Image.open('/NOBACKUP/gauthiers/chest_xrays_preprocess/train/positive/MIDRC-RICORD-1C-SITE2-000216-21074-0.png')
rsz_transf = torchvision.transforms.Resize((128,128))
img = rsz_transf(img)
display(img)
```
Rotation
```
transformation = torchvision.transforms.RandomRotation(degrees = 45)
transformation.degrees = [45,45]
img_rot2 = transformation(img)
display(img_rot2)
```
Blur
```
transformation = torchvision.transforms.GaussianBlur(3)
img_blur = transformation(img)
display(img_blur)
```
Perspective
```
transformation = torchvision.transforms.RandomPerspective()
img_rdmPersp = transformation(img)
display(img_rdmPersp)
transforms = torchvision.transforms.RandomPerspective(distortion_scale=0.5,p=1)
transforms.distortion_scale = 0.9
img_1 = transforms(img)
display(img_1)
transforms = torchvision.transforms.RandomAffine(degrees = 0, shear=90)
img_2 = transforms(img)
display(img_2)
```
À la Mallat
```
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
device = torch.device('cpu')
import time
t0 = time.time()
# Function \tau in Mallat's. Deform the index u. The function is chosen arbitrary as an example.
tau = lambda u : (0.5*u[0]+0.3*u[1]**2,0.3*u[1])
# Deform the index u for all u of the image.
tau_mat = lambda grid : torch.tensor([[tau(grid[i,j,:]) for j in range(len(grid))] for i in range(len(grid))],device = device)
tauV = lambda u : torch.stack([0.5*u[:,0]+0.3*u[:,1]**2,0.3*u[:,1]]).T
# Deforms the image given a function \tau.
def diffeo(img,tau):
# Image to tensor
transf = torchvision.transforms.ToTensor()
img = transf(img).unsqueeze(0).to(device)
# Number of pixels. Suppose square image.
dim = img.shape[-1]
# Create a (dim x dim) matrix of 2d vectors. Each vector represents the normalized position in the grid.
# Normalized means (-1,-1) is top left and (1,1) is bottom right.
grid = torch.tensor([[[x,y] for x in torch.linspace(-1,1,dim,device = device)] for y in torch.linspace(-1,1,dim,device = device)],device = device)
# Apply u-tau(u) in Mallat's.
grid_transf = (grid - tau_mat(grid)).unsqueeze(0)
# Apply x(u-tau(u)) by interpolating the image at the index points given by grid_transf.
img_transf = torch.nn.functional.grid_sample(img,grid_transf).squeeze(0)
# Tensor to image
transf = torchvision.transforms.ToPILImage()
return transf(img_transf)
# Calculate the deformation size : sup |J_{tau}(u)| over u.
def deformation_size(tau):
# Set a precision. This is arbitrary.
precision = 128
# Create a (flatten) grid of points between (-1,-1) and (1,1). This is the same grid as in the previous
# function (but flatten), but it feels arbitrary also.
points = [torch.tensor([x,y],device = device) for x in torch.linspace(-1,1,precision,device = device) for y in torch.linspace(-1,1,precision,device = device)]
# Evaluate the Jacobian of tau in each of those points. Returns a tensor of precision^2 x 2 x 2, i.e.
# for each point in points the 2 x 2 jacobian. Is it necessary to compute on all points, or only on the
# boundary would be sufficient?
t1 = time.time()
jac = torch.stack(list(map(lambda point : torch.stack(torch.autograd.functional.jacobian(tau,point)), points)))
print("grad calc +", (time.time()-t1))
# Find the norm of those jacobians.
norm_jac = torch.linalg.matrix_norm(jac,ord=2,dim=(1, 2))
# Return the Jacobian with the biggest norm.
return torch.max(norm_jac)
img_diffeo = diffeo(img,tau)
display(img_diffeo)
deformation_size(tau)
print("full notebook +", (time.time()-t0))
tau(torch.randn((64,2)))
points = [torch.tensor([0.,0.]),torch.tensor([1.,2.])]
jac = torch.autograd.functional.jacobian(tau,points[0])
jac2 = torch.stack(jac)
jac = torch.autograd.functional.jacobian(tau,points[1])
jac3 = torch.stack(jac)
n = 0
jac4 = torch.cat([jac2.unsqueeze(n),jac3.unsqueeze(n)],dim = n)
print(jac2)
print(jac3)
print(jac4)
print(jac4.shape)
jac5 = torch.cat([torch.stack(torch.autograd.functional.jacobian(tau,point)).unsqueeze(0) for point in points], dim = 0)
print(jac5)
points = [torch.tensor([0.,0.]),torch.tensor([1.,2.])]
jac = torch.stack(list(map(lambda point : torch.stack(torch.autograd.functional.jacobian(tau,point)), points)))
print(jac)
print(jac.shape)
points = [torch.tensor([0.,0.]),torch.tensor([1.,2.])]
jac = torch.cat([torch.cat([x.unsqueeze(1) for x in torch.autograd.functional.jacobian(tau,point)],dim =1).unsqueeze(2) for point in points],dim = 2)
print(jac)
print(jac.shape)
eps = 0.3
tau = lambda u : (eps*u[0],eps*u[1])
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (eps*u[1],eps*u[0])
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (eps*(u[0]+u[1]),eps*(u[0]+u[1]))
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (eps*(u[0]+u[1]),eps*(u[0]-u[1]))
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (eps*(u[0]**2+u[1]**2),eps*(2*u[0]*u[1]))
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (eps*(u[0]**2+u[1]**2),-eps*(2*u[0]*u[1]))
display(diffeo(img,tau))
eps = 0.3
tau = lambda u : (torch.exp(eps*u[0])-1,torch.exp(eps*u[1])-1)
display(diffeo(img,tau))
```
|
github_jupyter
|
# Todoist Data Analysis
This notebook processed the downloaded history of your todoist tasks. See [todoist_downloader.ipynb](https://github.com/markwk/qs_ledger/blob/master/todoist/todoist_downloader.ipynb) to export and download your task history from Todoist.
---
```
from datetime import date, datetime as dt, timedelta as td
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
%matplotlib inline
# supress warnings
import warnings
warnings.filterwarnings('ignore')
```
---
# General Data Analysis of Todoist Tasks
```
# import raw data
raw_tasks = pd.read_csv("data/todost-raw-tasks-completed.csv")
len(raw_tasks)
# import processed data
tasks = pd.read_csv("data/todost-tasks-completed.csv")
len(tasks)
```
----
### Simple Data Analysis: Completed Tasks Per Year
```
year_data = tasks['year'].value_counts().sort_index()
# Chart Monthly Tasks Count
dataset = year_data
chart_title = 'Number of Tasks Completed Per Year'
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.bar(figsize=(14, 5), rot=0, legend=False)
ax.set_ylabel('Tasks Completed')
ax.set_xlabel('')
ax.set_title(chart_title)
plt.show()
```
### Simple Data Analysis: Completed Tasks Per Month
```
# simple breakdown by month
totals_by_month = tasks['month'].value_counts().sort_index()
# Chart Monthly Tasks Count
dataset = totals_by_month.tail(24)
chart_title = 'Monthly Number of Tasks Completed (Last 24 Months)'
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.bar(figsize=(14, 5), rot=90, colormap='spring', stacked=True, legend=False)
ax.set_ylabel('Tasks Completed')
ax.set_xlabel('')
ax.set_title(chart_title)
plt.show()
```
------
### Simple Data Analysis: Completed Tasks by Day of Week
```
totals_dow = tasks['dow'].value_counts().sort_index()
dataset = totals_dow
chart_title = 'Completed Tasks by Day of Week'
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.bar(figsize=(14, 5), rot=0, colormap='autumn', stacked=True, legend=False)
ax.set_ylabel('# Completed')
ax.set_xlabel('')
ax.set_title(chart_title)
plt.show()
```
-----
### Simple Data Analysis: Completed Tasks by Hour of the Day
```
hour_counts = tasks['hour'].value_counts().sort_index()
ax = hour_counts.plot(kind='line', figsize=[10, 4], linewidth=4, alpha=1, marker='o', color='#6684c1',
markeredgecolor='#6684c1', markerfacecolor='w', markersize=8, markeredgewidth=2)
xlabels = hour_counts.index.map(lambda x: '{:02}:00'.format(x))
ax.set_xticks(range(len(xlabels)))
ax.set_xticklabels(xlabels, rotation=45, rotation_mode='anchor', ha='right')
ax.set_xlim((hour_counts.index[0], hour_counts.index[-1]))
ax.yaxis.grid(True)
hour_max = hour_counts.max()
ax.set_ylim((0, hour_max+20))
ax.set_ylabel('Number of Tasks')
ax.set_xlabel('', )
ax.set_title('Number of Tasks Completed per hour of the day', )
plt.show()
```
----
## Daily Count of Tasks Completed
```
daily_counts = tasks['date'].value_counts().sort_index()
dataset = daily_counts.tail(30)
chart_title = 'Number of Tasks Completed per Day'
n_groups = len(dataset)
index = np.arange(n_groups)
ax = dataset.plot(kind='line', figsize=[12, 5], linewidth=4, alpha=1, marker='o', color='#6684c1',
markeredgecolor='#6684c1', markerfacecolor='w', markersize=8, markeredgewidth=2)
ax.yaxis.grid(True)
ax.xaxis.grid(True)
ax.set_xticks(index)
ax.set_ylabel('Tasks Completed Count')
# ax.set_xlabel('')
plt.xticks(index, dataset.index, rotation=90)
ax.set_title(chart_title)
plt.show()
# Export
daily_counts.to_csv("data/todoist-daily-completed.csv", index=True)
```
-----
### Projects Breakdown
```
# Optionally pass a list of projects to exclude
exclude_proj = ['Project1', 'Project2']
tasks_data = tasks[~tasks.project_name.isin(exclude_proj)]
project_counts = tasks_data['project_name'].value_counts().sort_values(ascending=False)
# Chart Project Tasks
dataset = project_counts.sort_values(ascending=True).tail(15)
chart_title = 'Project Tasks Breakdown'
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.barh(y='Hours', figsize=(8, 8), colormap='plasma', legend=False)
ax.set_ylabel('')
ax.set_xlabel('Task #')
ax.set_title(chart_title)
plt.show()
```
-----
## General Summary of Todoist Tasks
```
# Life-time Project Time Summary
print('====== Todoist Lifetime Summary ====== ')
print('Total Tasks Completed: {:,}'.format(len(tasks)))
daily_average = round(daily_counts.mean(),1)
print('Daily Task Average: {:,}'.format(daily_average))
print(' ')
print('Top 5 Days with Most Tasks Completed:')
for i, v in daily_counts.sort_values(ascending=False).head(5).items():
print(v, 'tasks on ', i)
```
------
# Year in Review
```
# Set Year
target_year = 2018
```
### Year: Top Projects
```
def yearly_top_projects_chart(year, exclude_projects=[]):
year_data = tasks[tasks['year'] == year]
# Optionally pass a list of projects to exclude
if exclude_projects:
exclude_proj = exclude_projects
year_data = year_data[~tasks.project_name.isin(exclude_proj)]
project_counts = year_data['project_name'].value_counts().sort_values(ascending=False)
project_counts = year_data['project_name'].value_counts().sort_values(ascending=False)
# Chart Project Tasks
dataset = project_counts.sort_values(ascending=True).tail(10)
chart_title = '{} Project Tasks Breakdown'.format(year)
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.barh(y='Hours', figsize=(8, 8), colormap='plasma', legend=False)
ax.set_ylabel('')
ax.set_xlabel('Task #')
ax.set_title(chart_title)
plt.show()
# yearly_top_projects_chart(year=target_year, exclude_projects=['ProjectName', 'ProjectName2''])
yearly_top_projects_chart(year=target_year)
```
### Year: Day of Week Comparison
```
def yearly_dow_chart(year):
year_data = tasks[tasks['year'] == year]
yearly_dow = year_data['dow'].value_counts().sort_index()
days_of_week_list = ['Mon', 'Tues', 'Wed', 'Thurs', 'Friday', 'Sat', 'Sun']
yearly_dow.index = days_of_week_list
chart_title = '{} Tasks Completed by Day of Week | Yearly Total: {:,}'.format(year, yearly_dow.sum())
plt.style.use('seaborn-darkgrid')
ax = yearly_dow.plot.bar(stacked=True, rot=0, figsize=(12,4))
ax.set_xlabel('')
ax.set_ylabel('Hours')
ax.set_title(chart_title)
plt.show()
yearly_dow_chart(year=target_year)
```
### Year: Monthly Tasks Completed Chart
```
def yearly_months_chart(year):
year_data = tasks[tasks['year'] == year]
yearly_months = year_data['month'].value_counts().sort_index()
months_of_year = ['Jan', 'Feb', 'March', 'April', 'May', 'June', 'July',
'Aug', 'Sept', 'Oct', 'Nov', 'Dec']
yearly_months.index = months_of_year
# Chart Monthly Tasks Count
dataset = yearly_months
chart_title = 'Monthly Number of Tasks Completed'
plt.style.use('seaborn-darkgrid')
ax = dataset.plot.bar(figsize=(14, 5), rot=0, colormap='spring', stacked=True, legend=False)
ax.set_ylabel('Tasks Completed')
ax.set_xlabel('')
ax.set_title(chart_title)
plt.show()
yearly_months_chart(year=target_year)
```
#### Year: Tasks Heat Map
```
# Helper Function to Create Heat Map from Data
# Adapted from https://stackoverflow.com/questions/32485907/matplotlib-and-numpy-create-a-calendar-heatmap
DAYS = ['Sun.', 'Mon.', 'Tues.', 'Wed.', 'Thurs.', 'Fri.', 'Sat.']
MONTHS = ['Jan.', 'Feb.', 'Mar.', 'Apr.', 'May', 'June', 'July', 'Aug.', 'Sept.', 'Oct.', 'Nov.', 'Dec.']
def date_heatmap(series, start=None, end=None, mean=False, ax=None, **kwargs):
'''Plot a calendar heatmap given a datetime series.
Arguments:
series (pd.Series):
A series of numeric values with a datetime index. Values occurring
on the same day are combined by sum.
start (Any):
The first day to be considered in the plot. The value can be
anything accepted by :func:`pandas.to_datetime`. The default is the
earliest date in the data.
end (Any):
The last day to be considered in the plot. The value can be
anything accepted by :func:`pandas.to_datetime`. The default is the
latest date in the data.
mean (bool):
Combine values occurring on the same day by mean instead of sum.
ax (matplotlib.Axes or None):
The axes on which to draw the heatmap. The default is the current
axes in the :module:`~matplotlib.pyplot` API.
**kwargs:
Forwarded to :meth:`~matplotlib.Axes.pcolormesh` for drawing the
heatmap.
Returns:
matplotlib.collections.Axes:
The axes on which the heatmap was drawn. This is set as the current
axes in the `~matplotlib.pyplot` API.
'''
# Combine values occurring on the same day.
dates = series.index.floor('D')
group = series.groupby(dates)
series = group.mean() if mean else group.sum()
# Parse start/end, defaulting to the min/max of the index.
start = pd.to_datetime(start or series.index.min())
end = pd.to_datetime(end or series.index.max())
# We use [start, end) as a half-open interval below.
end += np.timedelta64(1, 'D')
# Get the previous/following Sunday to start/end.
# Pandas and numpy day-of-week conventions are Monday=0 and Sunday=6.
start_sun = start - np.timedelta64((start.dayofweek + 1) % 7, 'D')
end_sun = end + np.timedelta64(7 - end.dayofweek - 1, 'D')
# Create the heatmap and track ticks.
num_weeks = (end_sun - start_sun).days // 7
heatmap = np.zeros((7, num_weeks))
ticks = {} # week number -> month name
for week in range(num_weeks):
for day in range(7):
date = start_sun + np.timedelta64(7 * week + day, 'D')
if date.day == 1:
ticks[week] = MONTHS[date.month - 1]
if date.dayofyear == 1:
ticks[week] += f'\n{date.year}'
if start <= date < end:
heatmap[day, week] = series.get(date, 0)
# Get the coordinates, offset by 0.5 to align the ticks.
y = np.arange(8) - 0.5
x = np.arange(num_weeks + 1) - 0.5
# Plot the heatmap. Prefer pcolormesh over imshow so that the figure can be
# vectorized when saved to a compatible format. We must invert the axis for
# pcolormesh, but not for imshow, so that it reads top-bottom, left-right.
ax = ax or plt.gca()
mesh = ax.pcolormesh(x, y, heatmap, **kwargs)
ax.invert_yaxis()
# Set the ticks.
ax.set_xticks(list(ticks.keys()))
ax.set_xticklabels(list(ticks.values()))
ax.set_yticks(np.arange(7))
ax.set_yticklabels(DAYS)
# Set the current image and axes in the pyplot API.
plt.sca(ax)
plt.sci(mesh)
return ax
def year_heat_chart(year):
# Filter by Year
year_data = tasks[(tasks['year'] == year)]
# daily count
year_dates_data = year_data['date'].value_counts().reset_index()
year_dates_data.columns = ['date', 'count']
year_dates_data['date'] = pd.to_datetime(year_dates_data['date'])
# Generate all dates in that year
first_date = str(year)+'-01-01'
last_date = str(year)+'-12-31'
all_dates = pd.date_range(start=first_date, end=last_date)
all_dates = pd.DataFrame(all_dates, columns=['date'])
# combine actual runs by date with total dates possible
year_data = pd.merge(left=all_dates, right=year_dates_data,
left_on="date", right_on="date", how="outer")
year_data['count'].fillna(0, inplace=True)
year_data = year_data.set_index(pd.DatetimeIndex(year_data['date']))
max_daily_count = round(year_data['count'].max(),2)
# key stat and title
total_tasks = round(year_data['count'].sum())
chart_title = '{} Todoist Tasks Heatmap | Total Tasks: {:,}'.format(year, total_tasks)
# set chart data
data = year_data['count']
data.index = year_data.index
# plot data
figsize = plt.figaspect(7 / 56)
fig = plt.figure(figsize=figsize)
ax = date_heatmap(data, edgecolor='black')
max_count = int(round(data.max(),0))
steps = int(round(max_count / 6, 0))
plt.colorbar(ticks=range(0, max_count, steps), pad=0.02)
cmap = mpl.cm.get_cmap('Purples', max_daily_count)
plt.set_cmap(cmap)
plt.clim(0, max_daily_count)
ax.set_aspect('equal')
ax.set_title(chart_title)
plt.show()
year_heat_chart(year=target_year)
# compare previous year:
year_heat_chart(year=2017)
```
### Yearly Summary
```
def yearly_summary(year):
print('====== {} Todoist Summary ======'.format(year))
# Data Setup
year_data = tasks[(tasks['year'] == year)]
print('Total Tasks Completed: {:,}'.format(len(year_data)))
daily_counts = year_data['date'].value_counts().sort_index()
daily_average = round(daily_counts.mean(),1)
print('Daily Task Average: {:,}'.format(daily_average))
print(' ')
project_counts = year_data['project_name'].value_counts()
print('=== Top Projects ===')
for i, v in project_counts.sort_values(ascending=False).head(7).items():
print("* ", v, 'tasks on ', i)
print(' ')
print('=== Monthly Breakdown ===')
monthly_counts = year_data['month'].value_counts().sort_index()
print('Monthly Task Average: {:,}'.format(round(monthly_counts.mean(),1)))
print('> Top 3 Months:')
for i, v in monthly_counts.sort_values(ascending=False).head(3).items():
print("* ", v, 'tasks on ', i)
print('> Bottom 3 Months:')
for i, v in monthly_counts.sort_values(ascending=True).head(3).items():
print("* ", v, 'tasks on ', i)
print(' ')
print('Top 5 Days with Most Tasks Completed:')
for i, v in daily_counts.sort_values(ascending=False).head(5).items():
print("* ", v, 'tasks on ', i)
yearly_summary(year=target_year)
```
|
github_jupyter
|
```
import scanpy as sc
import pandas as pd
import numpy as np
import scipy as sp
from statsmodels.stats.multitest import multipletests
import matplotlib.pyplot as plt
import seaborn as sns
import os
from os.path import join
import time
plt.rcParams['pdf.fonttype'] = 42
plt.rcParams['ps.fonttype'] = 42
# scTRS tools
import scdrs.util as util
import scdrs.data_loader as dl
import scdrs.method as md
# autoreload
%load_ext autoreload
%autoreload 2
# Constants
DATA_PATH='/n/holystore01/LABS/price_lab/Users/mjzhang/scDRS_data'
OUT_PATH=DATA_PATH+'/results/fig_simu'
# GS
GS_LIST = ['%s_ngene%d'%(prefix, size) for prefix in ['all', 'highmean', 'highvar', 'highbvar']
for size in [100, 500, 1000]]
temp_dic = {'all': 'random genes', 'highmean': 'random high mean-expr genes',
'highvar': 'random high variance genes', 'highbvar': 'random overdispersed genes'}
DIC_GS_NAME = {x:x.split('_')[1].replace('ngene','')+' '+temp_dic[x.split('_')[0]]
for x in GS_LIST}
# DATA_LIST
DATA_LIST = ['tms_facs.ncell_10k']
# Results
DIC_RES_PATH = {'sctrs': DATA_PATH+'/simulation_data/score_file/@d.@g',
'seurat': DATA_PATH+'/simulation_data/score_file/result_scanpy/@d.@g',
'vision': DATA_PATH+'/simulation_data/score_file/result_vision/@d.@g',
'vam': DATA_PATH+'/simulation_data/score_file/result_vam/@[email protected]'}
METHOD_LIST = list(DIC_RES_PATH.keys())
DIC_METHOD_NAME = {'sctrs':'scDRS', 'seurat': 'Seurat', 'vision':'Vision', 'vam':'VAM'}
DIC_METHOD_COLOR = {'sctrs':'C0', 'seurat': 'C1', 'vision':'C2', 'vam':'C3'}
for method in METHOD_LIST:
if method not in DIC_METHOD_NAME.keys():
DIC_METHOD_NAME[method] = method
if method not in DIC_METHOD_COLOR.keys():
DIC_METHOD_COLOR[method] = 'C%d'%len(DIC_METHOD_COLOR)
# Read results
import itertools
q_list = 10**np.linspace(-3,0,30)
dic_res = {}
for gs,dname,method in itertools.product(GS_LIST, DATA_LIST, METHOD_LIST):
print(gs,dname,method)
df_gs = pd.read_csv(DATA_PATH+'/simulation_data/gs_file/%s.gs'%gs, sep='\t', index_col=0)
df_res = pd.DataFrame(index=df_gs.index, columns=q_list, data=-1)
# load scTRS results
if method=='sctrs':
for trait in df_gs.index:
score_file = DIC_RES_PATH[method].replace('@d',dname).replace('@g',gs) + '/%s.score.gz'%trait
if os.path.exists(score_file):
temp_df = pd.read_csv(score_file, sep='\t')
df_res.loc[trait, q_list] = np.quantile(temp_df['pval'], q_list)
else:
print('# file missing: ', score_file)
dic_res['%s:%s:%s'%(dname,gs,method)] = df_res.copy()
# load vam results
if method=='vam':
score_file = DIC_RES_PATH[method].replace('@d',dname).replace('@g',gs)
if os.path.exists(score_file):
temp_df = pd.read_csv(score_file, sep='\t')
temp_df.columns = [x.replace('.','_') for x in temp_df.columns]
drop_list = temp_df.columns[temp_df.mean(axis=0)>0.99]
for trait in df_gs.index:
if trait in drop_list:
print('# %s dropped'%trait)
continue
df_res.loc[trait, q_list] = np.quantile(temp_df[trait], q_list)
df_res = df_res.loc[(df_res==-1).sum(axis=1)==0]
dic_res['%s:%s:%s'%(dname,gs,method)] = df_res.copy()
else:
print('# file missing: ', score_file)
# load vision result
if method=='vision':
for trait in df_gs.index:
score_file = DIC_RES_PATH[method].replace('@d',dname).replace('@g',gs) + '/%s.score.gz'%trait
if os.path.exists(score_file):
temp_df = pd.read_csv(score_file, sep='\t')
df_res.loc[trait, q_list] = np.quantile(temp_df['norm_pval'], q_list)
else:
print('# file missing: ', score_file)
dic_res['%s:%s:%s'%(dname,gs,method)] = df_res.copy()
# load seurat results
if method=='seurat':
for trait in df_gs.index:
score_file = DIC_RES_PATH[method].replace('@d',dname).replace('@g',gs) + '/%s.score.gz'%trait
if os.path.exists(score_file):
temp_df = pd.read_csv(score_file, sep='\t')
df_res.loc[trait, q_list] = np.quantile(temp_df['pval'], q_list)
else:
print('# file missing: ', score_file)
dic_res['%s:%s:%s'%(dname,gs,method)] = df_res.copy()
# Q-Q plot
dname = 'tms_facs.ncell_10k'
plot_list = ['%s:%s'%(dname, x) for x in GS_LIST]
plot_method_list = ['sctrs', 'vision', 'seurat', 'vam']
for plot_name in plot_list:
dname,gs=plot_name.split(':')
df_plot_mean = pd.DataFrame(index=q_list, columns=plot_method_list, data=-1)
df_plot_se = pd.DataFrame(index=q_list, columns=plot_method_list, data=-1)
for method in plot_method_list:
res = '%s:%s'%(plot_name,method)
temp_df = dic_res[res][q_list].loc[(dic_res[res][q_list]==-1).sum(axis=1)==0]
df_plot_mean.loc[q_list, method] = temp_df.mean(axis=0)
df_plot_se.loc[q_list, method] = temp_df.std(axis=0)/np.sqrt(temp_df.shape[0])
df_plot_mean = df_plot_mean.clip(lower=1e-4)
df_plot_se = df_plot_se.clip(lower=1e-10)
# Compute distance and p-value
df_plot_dist = np.absolute(np.log10(df_plot_mean.T)-np.log10(df_plot_mean.index)).T
df_plot_dist.drop(1, axis=0, inplace=True)
df_plot_dist = df_plot_dist.max(axis=0)
temp_df = np.absolute(df_plot_mean.T-df_plot_mean.index).T / df_plot_se
df_plot_p = pd.DataFrame(index=df_plot_mean.index, columns=df_plot_mean.columns,
data=(1-sp.stats.norm.cdf(temp_df))*2)
df_plot_p.drop(1, axis=0, inplace=True)
df_plot_p = df_plot_p.median(axis=0)
# Plot
plt.figure(figsize=[4.2,4])
df_plot_logerr = np.log10(df_plot_mean+1.96*df_plot_se) - np.log10(df_plot_mean)
for i_method,method in enumerate(plot_method_list):
plt.errorbar(-np.log10(df_plot_mean.index), -np.log10(df_plot_mean[method]),
yerr = df_plot_logerr[method], label=DIC_METHOD_NAME[method],
fmt='.', markersize=4, elinewidth=1, color=DIC_METHOD_COLOR[method], zorder=8-i_method)
plt.plot([0, 3], [0, 3], linestyle='--', linewidth=1, color='k', zorder=0)
plt.xlabel('Theoretical -log10(p) quantiles')
plt.ylabel('Actual -log10(p) quantiles')
plt.yticks([0,0.5,1,1.5,2,2.5,3,3.5,4],[0,0.5,1,1.5,2,2.5,3,3.5,'>4'])
plt.grid(linestyle='--', linewidth=0.5)
if 'all' in gs:
plt.title('Null simulations (%s)'%DIC_GS_NAME[gs])
else:
plt.title('Null simulations\n(%s)'%DIC_GS_NAME[gs])
plt.legend()
plt.tight_layout()
plt.savefig(OUT_PATH+'/%s.%s.svg'%(dname,gs))
plt.show()
# Store data for the main figure 'tms_facs.ncell_10k:all_ngene1000'
if plot_name=='tms_facs.ncell_10k:all_ngene1000':
SUPP_TAB_PATH='/n/holystore01/LABS/price_lab/Users/mjzhang/scDRS_data/supp_table'
df_plot_mean.columns = ['%s.mean'%x for x in df_plot_mean]
df_plot_se.columns = ['%s.se'%x for x in df_plot_se]
df_out = df_plot_mean.join(df_plot_se)
df_out.index.name='quantile'
df_out = df_out[['%s.%s'%(x,y) for x in plot_method_list for y in ['mean', 'se']]]
df_out.to_csv(SUPP_TAB_PATH+'/supp_tab_fig2a.tsv', sep='\t')
for method in plot_method_list:
print(method, (np.log10(df_out['%s.mean'%method]+1.96*df_out['%s.se'%method])
- np.log10(df_out['%s.mean'%method])).max())
```
### Cell type-disease association
```
# Load single-cell data
adata = sc.read_h5ad(DATA_PATH+'/simulation_data/single_cell_data/tms_facs.ncell_10k.h5ad')
# Read full score
dic_res_full = {}
score_file=DATA_PATH+'/simulation_data/score_file/tms_facs.ncell_10k.all_ngene1000'
df_gs = pd.read_csv(DATA_PATH+'/simulation_data/gs_file/all_ngene1000.gs', sep='\t', index_col=0)
for trait in df_gs.index:
if os.path.exists(score_file+'/%s.full_score.gz'%trait):
dic_res_full[trait] = pd.read_csv(score_file+'/%s.full_score.gz'%trait, sep='\t', index_col=0)
else:
print('# file missing: ', score_file)
# Cell type-disease association
celltype_list = sorted(set(adata.obs['cell_ontology_class']))
trait_list = list(df_gs.index)
df_stats = pd.DataFrame(index=celltype_list, columns=trait_list, dtype=float)
for trait in trait_list:
for ct in celltype_list:
cell_list = adata.obs_names[adata.obs['cell_ontology_class']==ct]
temp_df = dic_res_full[trait].loc[cell_list].copy()
score_q95 = np.quantile(temp_df['norm_score'], 0.95)
temp_df = temp_df[[x for x in temp_df.columns if x.startswith('ctrl_norm_score')]]
v_ctrl_score_q95 = np.quantile(temp_df, 0.95, axis=0)
df_stats.loc[ct,trait] = ((v_ctrl_score_q95>=score_q95).sum()+1) / (v_ctrl_score_q95.shape[0]+1)
df_stats_fdr = df_stats.copy()
print('# n_celltype=%d, n_rep=%d'%df_stats_fdr.shape)
for col in df_stats_fdr:
df_stats_fdr[col] = multipletests(df_stats[col], method='fdr_bh')[1]
for alpha in [0.05, 0.1, 0.2]:
v_fd = (df_stats_fdr<alpha).sum(axis=0)
v_d = v_fd.clip(lower=1)
v_fdp = v_fd / v_d
print('# alpha=%0.2f, FDP=%0.3f (SE=%0.3f)'
%(alpha, v_fdp.mean(), 1.96*v_fdp.std()/np.sqrt(df_stats_fdr.shape[1])))
```
|
github_jupyter
|
# Properties of drugs
Find various properties of the individual drugs
1.) ATC
2.) GO Annotations
3.) Disease
4.) KeGG Pathways
5.) SIDER (known effects)
6.) Offside (known off sides)
7.) TwoSides
8.) Drug Properties (physico-chemical properties)
9.) Enzymes, Transporters and Carriers
10.) Chemical_Gentic Perturbations (MsigDB)
## 1. ATC
Extract information about the anatomical as well as therapeutic group a drug is associated to using DrugBank as main source
```
import networkx as nx
#The the ATC classification from drugbank (see python file: 2a_Create_DrugBank_Network.ipynb)
DrugBankInfo = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
print 'DrugBank Network loaded'
#Create output file
fp_out = open('../results/Drug_Properties/CLOUD_to_ATC.csv','w')
fp_out.write('CLOUD,DrugBankID,First_Level_ATCs,Second_Level_ATCs\n')
#Dictionary containing DrugBank to CLOUD identifier
DrugBank_to_CLOUD = {}
#parse through all CLOUD drugs and check for ATC code annotation in drugbank (Use first and second level; third level and below too specific)
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv','r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
DrugBank_to_CLOUD[tmp[1]] = tmp[0]
first_level = set()
fist_second_level = set()
if DrugBankInfo.has_node(tmp[1]):
if DrugBankInfo.node[tmp[1]].has_key('ATCcode'):
atc_codes = DrugBankInfo.node[tmp[1]]['ATCcode'].split(',')
if '' in atc_codes:
atc_codes.remove('')
for atc in atc_codes:
atc = atc.strip()
first_level.add(atc[0])
fist_second_level.add(atc[0:3])
fp_out.write(tmp[0]+','+tmp[1]+','+';'.join(first_level)+','+';'.join(fist_second_level)+'\n')
fp.close()
fp_out.close()
print 'Finished ATC annotations'
```
## 2. GO Annotations
Extract GO annotations from GeneOntology for the targets of the individual drugs. Not only leaf but also upstream term information is collected for the three branches (i) Function, (ii) Component, (iii) Process
```
#use our inhouse database and the corresponding python file to create the upward ontology for every leaf GO term (all get included)
#Download (http://www.geneontology.org/page/downloads)
import gene2terms_addupstream as GO
#Include all threee GO branches
go_branches = ['Function','Process','Component']
#Find all the targets for the individual cloud drugs
cloud_targets = {}
fp = open('../data/Drug_Properties/CLOUD_All_Targets.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_targets[tmp[0]] = tmp[2].split(';')
fp.close()
#contain all CLOUD identifier
all_clouds = cloud_targets.keys()
all_clouds.sort()
#Go throug the GO branches and find GO terms for a specific drug via: Drug --> Targets --> Associated GO-Terms
drug_to_GO = {}
for go_branch in go_branches:
print go_branch
drug_to_GO[go_branch] = {}
GO_Association_UP, GO_genes_annotation = GO.getAllGene_Annotation(go_branch)
for drug in all_clouds:
drug_to_GO[go_branch][drug] = []
for target in cloud_targets[drug]:
drug_to_GO[go_branch][drug].extend(GO_Association_UP[target])
drug_to_GO[go_branch][drug] = list(set(drug_to_GO[go_branch][drug]))
#Save CLOUD drug to GO term annotations
fp_out = open('../results/Drug_Properties/CLOUD_to_GOterms.csv','w')
fp_out.write('CLOUD,GO_Function,GO_Process,GO_Component\n')
for cloud in all_clouds:
fp_out.write(cloud+','+';'.join(drug_to_GO['Function'][cloud])+','+';'.join(drug_to_GO['Process'][cloud])+','+';'.join(drug_to_GO['Component'][cloud])+'\n')
fp_out.close()
print 'Finished GO'
```
## 3. Diseases
Extract Disesase annotations from DiseaseOntology for the targets of the individual drugs. Not only leaf but also upstream term information is collected.
```
# Download from http://www.disgenet.org/web/DisGeNET/menu/downloads and http://disease-ontology.org/downloads/
# Again use inhouse database (manually curated), and corresponding scripts
# Get all cloud drug targets
fp = open('../data/Drug_Properties/CLOUD_All_Targets.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_targets[tmp[0]] = tmp[2].split(';')
fp.close()
all_clouds = cloud_targets.keys()
all_clouds.sort()
#Extrate the upward disease ontology (find all disease associated leaf plus upwards ontology terms for a specific gene)
Disease_Association_UP,d_diseases_annotation = GO.getAllGene_Disease_Annotation()
all_proteins = Disease_Association_UP.keys()
all_proteins = [int(x) for x in all_proteins]
all_proteins.sort()
fp_out = open('../results/Drug_Properties/Gene_to_Disease.csv','w')
fp_out.write('Gene,Disease_ID\n')
for protein in all_proteins:
fp_out.write(str(protein)+','+';'.join(Disease_Association_UP[str(protein)])+'\n')
fp_out.close()
break
#associated drug with diseaes
drug_to_Diseases = {}
for drug in all_clouds:
drug_to_Diseases[drug] = []
for target in cloud_targets[drug]:
drug_to_Diseases[drug].extend(Disease_Association_UP[target])
drug_to_Diseases[drug] = list(set(drug_to_Diseases[drug]))
fp_out = open('../results/Drug_Properties/CLOUD_to_Disease.csv','w')
fp_out.write('CLOUD,Disease_ID\n')
for cloud in all_clouds:
fp_out.write(cloud+','+';'.join(drug_to_Diseases[cloud])+'\n')
fp_out.close()
print 'Finished Diseases'
```
## 4. KeGG Pathways
Extract information about pathways being annotated to (i) the drug itself, as well as (ii) pathways associated to the target of drugs
```
'''
Extract direct drug <--> pathway annotations
'''
#Get KeGG pathways via the biopython.KEGG REST
from Bio.KEGG import REST
#Find the KeGG identifiers via the drugbank annotations
DrugBankInfo = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
print 'DrugBank Network loaded'
#parse through all CLOUD targets
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv','r')
fp.next()
drug_to_pathways = {}
all_targeted_Pathways = set()
all_clouds = []
kegg_IDs = {}
#find the KeGG Drug page and find PATHWAY informations (direct drug to pathway)
for line in fp:
tmp = line.strip().split(',')
drug_to_pathways[tmp[0]] = []
all_clouds.append(tmp[0])
if DrugBankInfo.has_node(tmp[1]):
if DrugBankInfo.node[tmp[1]].has_key('KEGGDrug'):
kegg_ID = DrugBankInfo.node[tmp[1]]['KEGGDrug']
kegg_IDs[tmp[0]] = kegg_ID
drug_file = REST.kegg_get(kegg_ID).read()
for line in drug_file.rstrip().split("\n"):
section = line[:12].strip() # section names are within 12 columns
if not section == "":
current_section = section
if current_section == "PATHWAY":
tmp2 = line[12:].split(' ')
pathwayID = tmp2[0].split('(')[0]
drug_to_pathways[tmp[0]].append(pathwayID)
all_targeted_Pathways.add(pathwayID)
print 'Number of pathways directed targeted: %d' %len(all_targeted_Pathways)
all_clouds.sort()
'''
Additonally to finding the direct annotations, also find drug <--> targets <--> pathways associated to those target annotations
'''
#Get all targets
cloud_targets = {}
fp = open('../data/Drug_Properties/CLOUD_All_Targets.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_targets[tmp[0]] = tmp[2].split(';')
fp.close()
# find human pahtways
human_pathways = REST.kegg_list("pathway", "hsa").read()
# get all human pathways, and add the dictionary
pathways = {}
for line in human_pathways.rstrip().split("\n"):
entry, description = line.split("\t")
pathways[entry] = {'Description' :description, 'IDs':None,'Symbols':None}
print len(pathways)
# Get the genes for pathways and add them to a list
for pathway in pathways.keys():
pathway_file = REST.kegg_get(pathway).read() # query and read each pathway
# iterate through each KEGG pathway file, keeping track of which section
# of the file we're in, only read the gene in each pathway
current_section = None
genesSymbols = []
genesIDs = []
for line in pathway_file.rstrip().split("\n"):
section = line[:12].strip() # section names are within 12 columns
if not section == "":
current_section = section
if current_section == "GENE":
if ';' in line:
gene_identifiers, gene_description = line[12:].split("; ")
gene_id, gene_symbol = gene_identifiers.split()
if not gene_id in genesIDs:
genesIDs.append(gene_id)
genesSymbols.append(gene_symbol)
pathways[pathway] = genesIDs
via_target_assigned_Pathways = {}
second_assigned_pathways = set()
for cloud in all_clouds:
via_target_assigned_Pathways[cloud] = []
targets = cloud_targets[cloud]
for p in pathways:
if len(set(targets).intersection(set(pathways[p]))) > 0:
via_target_assigned_Pathways[cloud].append(p)
second_assigned_pathways.add(p)
print 'Number of pathways indirected targeted: %d' %len(second_assigned_pathways)
fp_out = open('../results/Drug_Properties/CLOUD_to_KeGG_Pathways.csv','w')
fp_out.write('CLOUD,KeGG_DrugID,KeGG_Assigned_Pathways,Via_Target_Assigned\n')
for cloud in all_clouds:
if kegg_IDs.has_key(cloud):
fp_out.write(cloud+','+kegg_IDs[cloud]+','+';'.join(drug_to_pathways[cloud])+','+';'.join(via_target_assigned_Pathways[cloud])+'\n')
else:
fp_out.write(cloud+',,'+';'.join(drug_to_pathways[cloud])+','+';'.join(via_target_assigned_Pathways[cloud])+'\n')
fp_out.close()
print 'Finished Pathways'
```
## 5. SIDER
Extract information about known adverse reaction of drugs using the Sider database
```
def ATC_To_PubChem(isOffsides = 'None'):
'''
Sider offerst a direct conversion from ATC code to the internally used PubChem ID.
Offers a better coverage.
Download: http://sideeffects.embl.de/download/ [Nov. 2018] drug_atc.tsv file
(here named: Pubchem_To_ATC)
'''
dic_ATc_To_Pubchem = {}
fp = open('../data/Drug_Properties/Pubchem_To_ATC.tsv')
for line in fp:
tmp = line.strip().split('\t')
dic_ATc_To_Pubchem[tmp[1]] = tmp[0]
cloud_drugs = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
#find pubchem identifiers via ATC identifiers (as pubchem identifiers sometimes not unique neithers SID nor CID)
cloud_to_Pubchem = {}
PubChem_to_cloud = {}
found_PubChems = []
for drugBankID in cloud_drugs.nodes():
if cloud_drugs.node[drugBankID].has_key('ATCcode'):
all_codes = [x.strip() for x in cloud_drugs.node[drugBankID]['ATCcode'].split(',') if x != '']
for code in all_codes:
if dic_ATc_To_Pubchem.has_key(code):
pubChemID = dic_ATc_To_Pubchem[code][3:]
if isOffsides == 'offsides':
tmp = list(pubChemID)
tmp[0] = '0'
pubChemID = ''.join(tmp)
cloud_to_Pubchem[drugBankID] = pubChemID
PubChem_to_cloud[pubChemID] = drugBankID
found_PubChems.append(pubChemID)
return cloud_to_Pubchem, PubChem_to_cloud,found_PubChems
'''
Download SIDER.tsv from http://sideeffects.embl.de/download/ [Nov. 2018]
'''
#get the different identifiers of a drug
DrugBank_To_CLOUD = {}
CLOUD_To_DrugBank = {}
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv')
fp.next()
all_clouds = []
for line in fp:
tmp = line.strip().split(',')
all_clouds.append(tmp[0])
DrugBank_To_CLOUD[tmp[1]] = tmp[0]
CLOUD_To_DrugBank[tmp[0]] = tmp[1]
fp.close()
all_clouds.sort()
#extract pubchem identifier via ATC codes
DrugBank_to_Pubchem_viaATC, PubChem_to_cloud_viaATC,found_PubChems_viaATC = ATC_To_PubChem()
#further use drugbank to find additional pubchem identifiers for the cloud drugs
cloud_drugs = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
#associate cloud with the different pubchem identifiers
pubchemCompound_To_DrugBank = {}
DrugBank_to_PubChem = {}
pubchemCompound = []
pubchemSubstance = []
for node in cloud_drugs.nodes():
if cloud_drugs.node[node].has_key('PubChemCompound'):
pubchemCompound.append(cloud_drugs.node[node]['PubChemCompound'])
pubchemCompound_To_DrugBank[cloud_drugs.node[node]['PubChemCompound']] = node
DrugBank_to_PubChem[node] = cloud_drugs.node[node]['PubChemCompound']
#Combine both dictionaries together
for key in DrugBank_to_Pubchem_viaATC:
DrugBank_to_PubChem[key] = DrugBank_to_Pubchem_viaATC[key]
#check the SIDER database for given sideeffect of a given drug (once via the ATC to pubchem identfiers; once via drugbank to pubchem)
compund_sideEffect = {}
fp = open('../data/Drug_Properties/SIDER.tsv','r')
for line in fp:
tmp = line.strip().split('\t')
id1 = tmp[1][3:]
id2 = tmp[2][3:]
if id1 in found_PubChems_viaATC:
if compund_sideEffect.has_key(PubChem_to_cloud_viaATC[id1]):
compund_sideEffect[PubChem_to_cloud_viaATC[id1]].append(tmp[3])
else:
compund_sideEffect[PubChem_to_cloud_viaATC[id1]] = [tmp[3]]
if id1 in pubchemCompound:
if compund_sideEffect.has_key(pubchemCompound_To_DrugBank[id1]):
compund_sideEffect[pubchemCompound_To_DrugBank[id1]].append(tmp[3])
else:
compund_sideEffect[pubchemCompound_To_DrugBank[id1]] = [tmp[3]]
if id2 in found_PubChems_viaATC:
if compund_sideEffect.has_key(PubChem_to_cloud_viaATC[id2]):
compund_sideEffect[PubChem_to_cloud_viaATC[id2]].append(tmp[3])
else:
compund_sideEffect[PubChem_to_cloud_viaATC[id2]] = [tmp[3]]
if id2 in pubchemCompound:
if compund_sideEffect.has_key(pubchemCompound_To_DrugBank[id2]):
compund_sideEffect[pubchemCompound_To_DrugBank[id2]].append(tmp[3])
else:
compund_sideEffect[pubchemCompound_To_DrugBank[id2]] = [tmp[3]]
##
# Save results
##
fp = open('../results/Drug_Properties/CLOUD_to_SIDER.csv','w')
fp.write('CLOUD,PubChem,SIDER_Ids\n')
for key in all_clouds:
if compund_sideEffect.has_key(CLOUD_To_DrugBank[key]):
fp.write(key +','+DrugBank_to_PubChem[CLOUD_To_DrugBank[key]]+','+';'.join(list(set(compund_sideEffect[CLOUD_To_DrugBank[key]])))+'\n')
elif DrugBank_to_PubChem.has_key(CLOUD_To_DrugBank[key]):
fp.write(key +','+DrugBank_to_PubChem[CLOUD_To_DrugBank[key]]+',' + '\n')
else:
fp.write(key + ',,\n')
fp.close()
print 'Finish with SIDER'
```
## 6. Offsides
Extract information about known adverse reaction of drugs using the Offside database (Tantonetti)
```
'''
Download Offsides.tsv from http://tatonettilab.org/resources/tatonetti-stm.html [Nov. 2018]
'''
#get the different identifiers of a drug
DrugBank_To_CLOUD = {}
CLOUD_To_DrugBank = {}
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv')
fp.next()
for line in fp:
tmp = line.strip().split(',')
DrugBank_To_CLOUD[tmp[1]] = tmp[0]
CLOUD_To_DrugBank[tmp[0]] = tmp[1]
fp.close()
#extract pubchem identifier via ATC codes
DrugBank_to_Pubchem_viaATC, PubChem_to_cloud_viaATC, found_PubChems_viaATC = ATC_To_PubChem('offsides')
#further use drugbank to find additional pubchem identifiers for the cloud drugs
cloud_drugs = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
#associate cloud with the different pubchem identifiers
pubchemCompound_To_DrugBank = {}
DrugBank_to_PubChem = {}
pubchemCompound = []
pubchemSubstance = []
for node in cloud_drugs.nodes():
if cloud_drugs.node[node].has_key('PubChemCompound'):
pubchemCompound.append(cloud_drugs.node[node]['PubChemCompound'].zfill(9))
pubchemCompound_To_DrugBank[cloud_drugs.node[node]['PubChemCompound'].zfill(9)] = node
DrugBank_to_PubChem[node] = cloud_drugs.node[node]['PubChemCompound'].zfill(9)
# Combine both dictionaries together
for key in DrugBank_to_Pubchem_viaATC:
DrugBank_to_PubChem[key] = DrugBank_to_Pubchem_viaATC[key]
#check the OFFSIDES database for given sideeffect of a given drug (once via the ATC to pubchem identfiers; once via drugbank to pubchem)
compund_sideEffect = {}
fp = open('../data/Drug_Properties/Offsides.tsv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split('\t')
id1 = tmp[0].replace('"','')[3:]
sideEffect = tmp[2].replace('"','')
#print id1
if id1 in found_PubChems_viaATC:
if compund_sideEffect.has_key(PubChem_to_cloud_viaATC[id1]):
compund_sideEffect[PubChem_to_cloud_viaATC[id1]].append(sideEffect)
else:
compund_sideEffect[PubChem_to_cloud_viaATC[id1]] = [sideEffect]
print len(compund_sideEffect.keys())
# print compund_sideEffect.keys()
if id1 in pubchemCompound:
if compund_sideEffect.has_key(pubchemCompound_To_DrugBank[id1]):
compund_sideEffect[pubchemCompound_To_DrugBank[id1]].append(sideEffect)
else:
compund_sideEffect[pubchemCompound_To_DrugBank[id1]] = [sideEffect]
print len(compund_sideEffect.keys())
# print compund_sideEffect.keys()
fp = open('../results/Drug_Properties/CLOUD_to_Offsides.csv', 'w')
fp.write('CLOUD,PubChem,OFFSIDE_Ids\n')
for key in all_clouds:
if compund_sideEffect.has_key(CLOUD_To_DrugBank[key]):
fp.write(key +','+DrugBank_to_PubChem[CLOUD_To_DrugBank[key]]+','+';'.join(list(set(compund_sideEffect[CLOUD_To_DrugBank[key]])))+'\n')
elif DrugBank_to_PubChem.has_key(CLOUD_To_DrugBank[key]):
fp.write(key + ',' +DrugBank_to_PubChem[CLOUD_To_DrugBank[key]]+',' + '\n')
else:
fp.write(key + ',,\n')
fp.close()
print 'Finish with OFFSIDES'
```
## 7. TwoSides
Extract information about side effects for drug combinations using TwoSide (Tantonetti))
```
'''
Download Offsides.tsv from http://tatonettilab.org/resources/tatonetti-stm.html [Nov. 2018]
'''
#get the different identifiers of a drug
DrugBank_To_CLOUD = {}
CLOUD_To_DrugBank = {}
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv')
fp.next()
for line in fp:
tmp = line.strip().split(',')
DrugBank_To_CLOUD[tmp[1]] = tmp[0]
CLOUD_To_DrugBank[tmp[0]] = tmp[1]
fp.close()
#extract pubchem identifier via ATC codes
DrugBank_to_Pubchem_viaATC, PubChem_to_cloud_viaATC, found_PubChems_viaATC = ATC_To_PubChem('offsides')
#further use drugbank to find additional pubchem identifiers for the cloud drugs
cloud_drugs = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
pubchemCompound_To_DrugBank = {}
DrugBank_to_PubChem = {}
pubchemCompound = []
pubchemSubstance = []
for node in cloud_drugs.nodes():
if cloud_drugs.node[node].has_key('PubChemCompound'):
pubchemCompound.append(cloud_drugs.node[node]['PubChemCompound'].zfill(9))
pubchemCompound_To_DrugBank[cloud_drugs.node[node]['PubChemCompound'].zfill(9)] = node
DrugBank_to_PubChem[node] = cloud_drugs.node[node]['PubChemCompound'].zfill(9)
# Combine both dictionaries together
for key in DrugBank_to_Pubchem_viaATC:
DrugBank_to_PubChem[key] = DrugBank_to_Pubchem_viaATC[key]
#check the SIDER database for given sideeffect of a given drug (once via the ATC to pubchem identfiers; once via drugbank to pubchem)
TwoSide_Network = nx.Graph()
fp = open('../data/Drug_Properties/TwoSides.tsv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split('\t')
id1 = tmp[0][3:]
id2 = tmp[1][3:]
sideEffect = tmp[4]
#print id1
found_id1 = None
found_id2 = None
if id1 in found_PubChems_viaATC:
found_id1 = PubChem_to_cloud_viaATC[id1]
elif id1 in pubchemCompound:
found_id1 = pubchemCompound_To_DrugBank[id1]
if found_id1 != None:
if id2 in found_PubChems_viaATC:
found_id2 = PubChem_to_cloud_viaATC[id2]
elif id2 in pubchemCompound:
found_id2 = pubchemCompound_To_DrugBank[id2]
if found_id2 != None:
if TwoSide_Network.has_edge(found_id1,found_id2) == False:
TwoSide_Network.add_edge(found_id1,found_id2)
TwoSide_Network[found_id1][found_id2]['SideEffect'] = sideEffect
else:
TwoSide_Network[found_id1][found_id2]['SideEffect'] = TwoSide_Network[found_id1][found_id2]['SideEffect'] +',' + sideEffect
nx.write_gml(TwoSide_Network,'../results/Drug_Properties/TwoSide_CLOUDs.gml')
print 'Finish with TwoSides'
```
## 8. Drug Properties
Extract Physicochemical properties of the drugs e.g. Lipinski Rule of 5, LogS, LogP etc. Use DrugBank as main source of information
```
'''
Physicochemical properties (calculated) offered by DrugBank
'''
#List of interesting physicochemical properties (continues)
Continuesfeatures = ['Polarizability','logS','logP','NumberofRings','PhysiologicalCharge',
'PolarSurfaceAreaPSA','pKastrongestbasic','pKastrongestacidic',
'Refractivity','MonoisotopicWeight','HBondDonorCount',
'RotatableBondCount','WaterSolubility']
##List of interesting physicochemical properties (discrete)
discreteFeatures = ['DrugSubClass','DrugClass','Family']
#Drugbank file
DrugBankInfo = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03_CLOUD_Only.gml')
print 'DrugBank Network loaded'
#output file
fp = open('../data/Drug_Properties/CLOUD_DrugBank_PubChem_Chembl.csv','r')
fp.next()
#parse through all cloud drugs and find physicochemical propterties
CLOUD_Chemical_properties = {}
all_clouds = []
kegg_IDs = {}
for line in fp:
tmp = line.strip().split(',')
all_clouds.append(tmp[0])
CLOUD_Chemical_properties[tmp[0]] = {}
if DrugBankInfo.has_node(tmp[1]):
CLOUD_Chemical_properties[tmp[0]]['DrugBankID'] = tmp[1]
for c in Continuesfeatures:
if DrugBankInfo.node[tmp[1]].has_key(c):
CLOUD_Chemical_properties[tmp[0]][c] = str(DrugBankInfo.node[tmp[1]][c])
else:
CLOUD_Chemical_properties[tmp[0]][c] = 'None'
for d in discreteFeatures:
if DrugBankInfo.node[tmp[1]].has_key(d):
CLOUD_Chemical_properties[tmp[0]][d] = str(DrugBankInfo.node[tmp[1]][d])
else:
CLOUD_Chemical_properties[tmp[0]][d] = 'None'
else:
CLOUD_Chemical_properties[tmp[0]]['DrugBankID'] = 'None'
for c in Continuesfeatures:
CLOUD_Chemical_properties[tmp[0]][c] = 'None'
for d in discreteFeatures:
CLOUD_Chemical_properties[tmp[0]][d] = 'None'
##
# Save results
##
fp = open('../results/Drug_Properties/CLOUD_to_ChemicalProperties.tsv', 'w')
fp.write('CLOUD\tDrugBankID\t')
fp.write('\t'.join(Continuesfeatures)+'\t'+'\t'.join(discreteFeatures)+'\n')
for cloud in all_clouds:
fp.write(cloud+'\t'+CLOUD_Chemical_properties[cloud]['DrugBankID'])
for c in Continuesfeatures:
fp.write('\t'+CLOUD_Chemical_properties[cloud][c])
for d in discreteFeatures:
fp.write('\t'+CLOUD_Chemical_properties[cloud][d])
fp.write('\n')
fp.close()
print 'Finish with Chemical Properties'
```
## 9. Targets, Enzymes, Transporters and Carriers
Split the full lust of targets into targets, enzymes, transporters and carriers
Therefore use the DrugBank annotations of what a target, transporter, carrier and enzyme is. Go trough all drugbank targets and take the corresponding annotations.
Then go trough the CLOUD targets and assign the targets accordingly. If drugbank does not show any annotation the gene is assumed to be a target.
Enzymes: e.g. CYP3A1
Transporter: e.g. MDR5
Carriers: e.g. ALB
```
DrugBankInfo = nx.read_gml('../data/Drug_Properties/Drugbank_2018-07-03.gml')
print 'Full DrugBank Network loaded'
annotated_enzyme_symbols = set()
annotated_transporters_symbols = set()
annotated_carriers_symbols = set()
#Go through all drugs in drugbank and extract target information; bin it correctly into one of the three classes
for drug in list(DrugBankInfo.nodes()):
if DrugBankInfo.node[drug].has_key('Enzymes'):
enzymes = [x for x in DrugBankInfo.node[drug]['Enzymes'].strip().split(',') if x != '']
for e in enzymes:
annotated_enzyme_symbols.add(e.split('_')[0])
if DrugBankInfo.node[drug].has_key('Transporters'):
transporters = [x for x in DrugBankInfo.node[drug]['Transporters'].strip().split(',') if x != '']
for t in transporters:
annotated_transporters_symbols.add(t.split('_')[0])
if DrugBankInfo.node[drug].has_key('Carriers'):
carriers = [x for x in DrugBankInfo.node[drug]['Carriers'].strip().split(',') if x != '']
for c in carriers:
annotated_carriers_symbols.add(c.split('_')[0])
#Plot the number of found Enzymes, Transporters, Carriers
print len(annotated_enzyme_symbols)
print len(annotated_transporters_symbols)
print len(annotated_carriers_symbols)
'''
Parse the enzyme, carriers and transporter SYMBOLS to EntrezIDs using mygeneinfo
'''
import mygene
mg = mygene.MyGeneInfo()
#Enzymes
query = mg.querymany(annotated_enzyme_symbols, scope='symbol', species='human',verbose=False)
final_annotated_enzyme_symbols = []
final_annotated_enzyme_IDs = []
for result in query:
if result.has_key('entrezgene'):
final_annotated_enzyme_symbols.append(result['symbol'])
final_annotated_enzyme_IDs.append(str(result['_id']))
#Transporters
query = mg.querymany(annotated_transporters_symbols, scope='symbol', species='human',verbose=False)
final_annotated_transporters_symbols = []
final_annotated_transporters_IDs = []
for result in query:
if result.has_key('entrezgene'):
final_annotated_transporters_symbols.append(result['symbol'])
final_annotated_transporters_IDs.append(str(result['_id']))
#Carriers
query = mg.querymany(annotated_carriers_symbols, scope='symbol', species='human',verbose=False)
final_annotated_carriers_symbols = []
final_annotated_carriers_IDs = []
for result in query:
if result.has_key('entrezgene'):
final_annotated_carriers_symbols.append(result['symbol'])
final_annotated_carriers_IDs.append(str(result['_id']))
print len(final_annotated_enzyme_IDs)
print len(final_annotated_transporters_IDs)
print len(final_annotated_carriers_IDs)
'''
Create an output file with the various transporters/enzymes/targets etc. being split.
'''
#Get the DrugBank targets
cloud_DrugBanktargets = {}
fp = open('../data/Drug_Properties/CLOUD_DrugBank_Targets_ONLY.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_DrugBanktargets[tmp[0]] = tmp[2].split(';')
fp.close()
#Get all targets accociated to the individual CLOUDS (including CYP etc.)
cloud_targets = {}
fp = open('../data/Drug_Properties/CLOUD_All_Targets.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_targets[tmp[0]] = tmp[2].split(';')
fp.close()
#List containing all CLOUD identifiers
all_clouds = cloud_targets.keys()
all_clouds.sort()
#Create output file
fp_out = open('../results/Drug_Properties/CLOUD_to_TargetsSplit.csv', 'w')
fp_out.write('CLOUD,Targets,Transporters,Enzymes,Carriers\n')
#save the per drug annotations of CLOUD drugs
targets_number = []
enzymes_number = []
transporters_number = []
carriers_number = []
#save total amount of distinct targets, enzymes etc. targeted by CLOUD
different_targets = set()
different_enzymes = set()
different_transporters = set()
different_carriers = set()
#save total amount of targets found
all_targets = 0
#Go through all CLOUDS
for cloud in all_clouds:
targets = []
enzymes = []
carriers = []
transporters = []
for target in cloud_targets[cloud]:
#First check if the target is annoated in DrugBank to be a target of this drug! (sometimes CYP or other can be main targets)
if target in cloud_DrugBanktargets[cloud]:
targets.append(target)
else:
#If it is not the main target of this drug bin it correctly according to drugbank standards
not_associated = False
if target in final_annotated_enzyme_IDs:
enzymes.append(target)
not_associated = True
if target in final_annotated_transporters_IDs:
transporters.append(target)
not_associated = True
if target in final_annotated_carriers_IDs:
carriers.append(target)
not_associated = True
if not_associated == False:
targets.append(target)
fp_out.write(cloud+','+';'.join(targets)+','+';'.join(transporters)+','+';'.join(enzymes)+','+';'.join(carriers)+'\n')
#Save the results
all_targets += len(targets)
targets_number.append(len(targets))
enzymes_number.append(len(enzymes))
transporters_number.append(len(transporters))
carriers_number.append(len(carriers))
different_targets = different_targets.union(set(targets))
different_enzymes = different_enzymes.union(set(enzymes))
different_transporters = different_transporters.union(set(transporters))
different_carriers = different_carriers.union(set(carriers))
fp_out.close()
'''
CREATE OUTPUT OVERVIEW OVER DRUG TARGETS/ANNOTATIONS
'''
import numpy as np
from matplotlib import pylab as plt
print'Mean number of targets: %.2f' %np.mean(targets_number)
print'Median number of targets: %.2f' %np.median(targets_number)
print'Mean number of enzymes: %.2f' %np.mean(enzymes_number)
print'Mean number of carriers: %.2f' %np.mean(carriers_number)
print'Mean number of transporters: %.2f' %np.mean(transporters_number)
print 'Total number of targets: %d' %all_targets
print 'Number of distinct targets: %d' %len(different_targets)
print'Number of distinct enzymes: %d' %len(different_enzymes)
print'Number of distinct carriers: %d' %len(different_carriers)
print'Number of distinct transporters: %d' %len(different_transporters)
plt.hist(targets_number,bins=22, color='#40B9D4')
plt.axvline(np.mean(targets_number),ls='--', color='grey')
plt.savefig('../results/Drug_Properties/CLOUD_TargetsFiltered.pdf')
plt.close()
```
## 10. Chemical Genetic perturbations
Use the msigDB Chemical_Genetic_Perturbations set to annotate the CLOUD target respetively
```
'''
Download from http://software.broadinstitute.org/gsea/msigdb/collections.jsp#C5 [December 17. 2018]
'''
#Get all CLOUD targets
cloud_targets = {}
fp = open('../data/Drug_Properties/CLOUD_All_Targets.csv', 'r')
fp.next()
for line in fp:
tmp = line.strip().split(',')
cloud_targets[tmp[0]] = tmp[2].split(';')
fp.close()
#Find the gene to perturbation associated (one gene can have various associated perturbations)
fp = open('../data/Drug_Properties/Msig_ChemGen_Perturbation.gmt','r')
gene_to_perturbation = {}
for line in fp:
tmp = line.strip().split('\t')
for gene in tmp[2:]:
if gene_to_perturbation.has_key(gene):
gene_to_perturbation[gene].append(tmp[0])
else:
gene_to_perturbation[gene] = [tmp[0]]
fp.close()
#find cloud associations via CLOUD --> Targets ===> Perturbations associated with certain targets
fp_out = open('../results/Drug_Properties/CLOUD_to_Perturbations.csv', 'w')
fp_out.write('CLOUD,Perturbations\n')
for cloud in all_clouds:
perturbations = []
for gene in cloud_targets[cloud]:
if gene_to_perturbation.has_key(gene):
perturbations.extend(gene_to_perturbation[gene])
fp_out.write(cloud+','+';'.join(perturbations)+'\n')
fp_out.close()
```
|
github_jupyter
|
# Graded Programming Assignment
In this assignment, you will implement re-use the unsupervised anomaly detection algorithm but turn it into a simpler feed forward neural network for supervised classification.
You are training the neural network from healthy and broken samples and at later stage hook it up to a message queue for real-time anomaly detection.
We've provided a skeleton for you containing all the necessary code but left out some important parts indicated with ### your code here ###
After you’ve completed the implementation please submit it to the autograder
```
!pip install tensorflow==2.2.0rc0
import tensorflow as tf
if not tf.__version__ == '2.2.0-rc0':
print(tf.__version__)
raise ValueError('please upgrade to TensorFlow 2.2.0-rc0, or restart your Kernel (Kernel->Restart & Clear Output)')
```
Now we import all the dependencies
```
import numpy as np
from numpy import concatenate
from matplotlib import pyplot
from pandas import read_csv
from pandas import DataFrame
from pandas import concat
import sklearn
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_squared_error
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Dropout
from tensorflow.keras.layers import LSTM
from tensorflow.keras.callbacks import Callback
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense, Activation
import pickle
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
import sys
from queue import Queue
import pandas as pd
import json
%matplotlib inline
```
We grab the files necessary for taining. Those are sampled from the lorenz attractor model implemented in NodeRED. Those are two serialized pickle numpy arrays. In case you are interested in how these data has been generated please have a look at the following tutorial. https://developer.ibm.com/tutorials/iot-deep-learning-anomaly-detection-2/
```
!rm watsoniotp.*
!wget https://raw.githubusercontent.com/romeokienzler/developerWorks/master/lorenzattractor/watsoniotp.healthy.phase_aligned.pickle
!wget https://raw.githubusercontent.com/romeokienzler/developerWorks/master/lorenzattractor/watsoniotp.broken.phase_aligned.pickle
!mv watsoniotp.healthy.phase_aligned.pickle watsoniotp.healthy.pickle
!mv watsoniotp.broken.phase_aligned.pickle watsoniotp.broken.pickle
```
De-serialize the numpy array containing the training data
```
data_healthy = pickle.load(open('watsoniotp.healthy.pickle', 'rb'), encoding='latin1')
data_broken = pickle.load(open('watsoniotp.broken.pickle', 'rb'), encoding='latin1')
```
Reshape to three columns and 3000 rows. In other words three vibration sensor axes and 3000 samples
Since this data is sampled from the Lorenz Attractor Model, let's plot it with a phase lot to get the typical 2-eyed plot. First for the healthy data
```
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot(data_healthy[:,0], data_healthy[:,1], data_healthy[:,2],lw=0.5)
ax.set_xlabel("X Axis")
ax.set_ylabel("Y Axis")
ax.set_zlabel("Z Axis")
ax.set_title("Lorenz Attractor")
```
Then for the broken one
```
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot(data_broken[:,0], data_broken[:,1], data_broken[:,2],lw=0.5)
ax.set_xlabel("X Axis")
ax.set_ylabel("Y Axis")
ax.set_zlabel("Z Axis")
ax.set_title("Lorenz Attractor")
```
In the previous examples, we fed the raw data into an LSTM. Now we want to use an ordinary feed-forward network. So we need to do some pre-processing of this time series data
A widely-used method in traditional data science and signal processing is called Discrete Fourier Transformation. This algorithm transforms from the time to the frequency domain, or in other words, it returns the frequency spectrum of the signals.
The most widely used implementation of the transformation is called FFT, which stands for Fast Fourier Transformation, let’s run it and see what it returns
```
data_healthy_fft = np.fft.fft(data_healthy).real
data_broken_fft = np.fft.fft(data_broken).real
```
Let’s first have a look at the shape and contents of the arrays.
```
print (data_healthy_fft.shape)
print (data_healthy_fft)
```
First, we notice that the shape is the same as the input data. So if we have 3000 samples, we get back 3000 spectrum values, or in other words 3000 frequency bands with the intensities.
The second thing we notice is that the data type of the array entries is not float anymore, it is complex. So those are not complex numbers, it is just a means for the algorithm the return two different frequency compositions in one go. The real part returns a sine decomposition and the imaginary part a cosine. We will ignore the cosine part in this example since it turns out that the sine part already gives us enough information to implement a good classifier.
But first let’s plot the two arrays to get an idea how a healthy and broken frequency spectrum differ
```
fig, ax = plt.subplots(num=None, figsize=(14, 6), dpi=80, facecolor='w', edgecolor='k')
size = len(data_healthy_fft)
ax.plot(range(0,size), data_healthy_fft[:,0].real, '-', color='blue', animated = True, linewidth=1)
ax.plot(range(0,size), data_healthy_fft[:,1].real, '-', color='red', animated = True, linewidth=1)
ax.plot(range(0,size), data_healthy_fft[:,2].real, '-', color='green', animated = True, linewidth=1)
fig, ax = plt.subplots(num=None, figsize=(14, 6), dpi=80, facecolor='w', edgecolor='k')
size = len(data_healthy_fft)
ax.plot(range(0,size), data_broken_fft[:,0].real, '-', color='blue', animated = True, linewidth=1)
ax.plot(range(0,size), data_broken_fft[:,1].real, '-', color='red', animated = True, linewidth=1)
ax.plot(range(0,size), data_broken_fft[:,2].real, '-', color='green', animated = True, linewidth=1)
```
So, what we've been doing is so called feature transformation step. We’ve transformed the data set in a way that our machine learning algorithm – a deep feed forward neural network implemented as binary classifier – works better. So now let's scale the data to a 0..1
```
def scaleData(data):
# normalize features
scaler = MinMaxScaler(feature_range=(0, 1))
return scaler.fit_transform(data)
```
And please don’t worry about the warnings. As explained before we don’t need the imaginary part of the FFT
```
data_healthy_scaled = scaleData(data_healthy_fft)
data_broken_scaled = scaleData(data_broken_fft)
data_healthy_scaled = data_healthy_scaled.T
data_broken_scaled = data_broken_scaled.T
```
Now we reshape again to have three examples (rows) and 3000 features (columns). It's important that you understand this. We have turned our initial data set which containd 3 columns (dimensions) of 3000 samples. Since we applied FFT on each column we've obtained 3000 spectrum values for each of the 3 three columns. We are now using each column with the 3000 spectrum values as one row (training example) and each of the 3000 spectrum values becomes a column (or feature) in the training data set
```
data_healthy_scaled.reshape(3, 3000)
data_broken_scaled.reshape(3, 3000)
```
# Start of Assignment
The first thing we need to do is to install a little helper library for submitting the solutions to the coursera grader:
```
!rm -f rklib.py
!wget https://raw.githubusercontent.com/IBM/coursera/master/rklib.py
```
Please specify you email address you are using with cousera here:
```
from rklib import submit, submitAll
key = "4vkB9vnrEee8zg4u9l99rA"
all_parts = ["O5cR9","0dXlH","ZzEP8"]
email = #### your code here ###
```
## Task
Given, the explanation above, please fill in the following two constants in order to make the neural network work properly
```
#### your code here ###
dim = #### your code here ###
samples = #### your code here ###
```
### Submission
Now it’s time to submit your first solution. Please make sure that the secret variable contains a valid submission token. You can obtain it from the courser web page of the course using the grader section of this assignment.
```
part = "O5cR9"
token = #### your code here ### (have a look here if you need more information on how to obtain the token https://youtu.be/GcDo0Rwe06U?t=276)
parts_data = {}
parts_data["0dXlH"] = json.dumps({"number_of_neurons_layer1": 0, "number_of_neurons_layer2": 0, "number_of_neurons_layer3": 0, "number_of_epochs": 0})
parts_data["O5cR9"] = json.dumps({"dim": dim, "samples": samples})
parts_data["ZzEP8"] = None
submitAll(email, token, key, parts_data)
```
To observe how training works we just print the loss during training
```
class LossHistory(Callback):
def on_train_begin(self, logs={}):
self.losses = []
def on_batch_end(self, batch, logs={}):
sys.stdout.write(str(logs.get('loss'))+str(', '))
sys.stdout.flush()
self.losses.append(logs.get('loss'))
lr = LossHistory()
```
## Task
Please fill in the following constants to properly configure the neural network. For some of them you have to find out the precise value, for others you can try and see how the neural network is performing at a later stage. The grader only looks at the values which need to be precise
```
number_of_neurons_layer1 = #### your code here ###
number_of_neurons_layer2 = #### your code here ###
number_of_neurons_layer3 = #### your code here ###
number_of_epochs = #### your code here ###
```
### Submission
Please submit your constants to the grader
```
parts_data = {}
parts_data["0dXlH"] = json.dumps({"number_of_neurons_layer1": number_of_neurons_layer1, "number_of_neurons_layer2": number_of_neurons_layer2, "number_of_neurons_layer3": number_of_neurons_layer3, "number_of_epochs": number_of_epochs})
parts_data["O5cR9"] = json.dumps({"dim": dim, "samples": samples})
parts_data["ZzEP8"] = None
token = #### your code here ###
submitAll(email, token, key, parts_data)
```
## Task
Now it’s time to create the model. Please fill in the placeholders. Please note since this is only a toy example, we don't use a separate corpus for training and testing. Just use the same data for fitting and scoring
```
# design network
from tensorflow.keras import optimizers
sgd = optimizers.SGD(lr=0.01, clipnorm=1.)
model = Sequential()
model.add(Dense(number_of_neurons_layer1,input_shape=(dim, ), activation='relu'))
model.add(Dense(number_of_neurons_layer2, activation='relu'))
model.add(Dense(number_of_neurons_layer3, activation='sigmoid'))
model.compile(loss='binary_crossentropy', optimizer=sgd)
def train(data,label):
model.fit(#### your code here ###, #### your code here ###, epochs=number_of_epochs, batch_size=72, validation_data=(data, label), verbose=0, shuffle=True,callbacks=[lr])
def score(data):
return model.predict(data)
```
We prepare the training data by concatenating a label “0” for the broken and a label “1” for the healthy data. Finally we union the two data sets together
```
label_healthy = np.repeat(1,3)
label_healthy.shape = (3,1)
label_broken = np.repeat(0,3)
label_broken.shape = (3,1)
train_healthy = np.hstack((data_healthy_scaled,label_healthy))
train_broken = np.hstack((data_broken_scaled,label_broken))
train_both = np.vstack((train_healthy,train_broken))
```
Let’s have a look at the two training sets for broken and healthy and at the union of them. Note that the last column is the label
```
pd.DataFrame(train_healthy)
pd.DataFrame(train_broken)
pd.DataFrame(train_both)
```
So those are frequency bands. Notice that although many frequency bands are having nearly the same energy, the neural network algorithm still can work those out which are significantly different.
## Task
Now it’s time to do the training. Please provide the first 3000 columns of the array as the 1st parameter and column number 3000 containing the label as 2nd parameter. Please use the python array slicing syntax to obtain those.
The following link tells you more about the numpy array slicing syntax
https://docs.scipy.org/doc/numpy-1.13.0/reference/arrays.indexing.html
```
features = train_both[:,#### your code here ###]
labels = train_both[:,#### your code here ###]
```
Now it’s time to do the training. You should see the loss trajectory go down, we will also plot it later. Note: We also could use TensorBoard for this but for this simple scenario we skip it. In some rare cases training doesn’t converge simply because random initialization of the weights caused gradient descent to start at a sub-optimal spot on the cost hyperplane. Just recreate the model (the cell which contains *model = Sequential()*) and re-run all subsequent steps and train again
```
train(features,labels)
```
Let's plot the losses
```
fig, ax = plt.subplots(num=None, figsize=(14, 6), dpi=80, facecolor='w', edgecolor='k')
size = len(lr.losses)
ax.plot(range(0,size), lr.losses, '-', color='blue', animated = True, linewidth=1)
```
Now let’s examine whether we are getting good results. Note: best practice is to use a training and a test data set for this which we’ve omitted here for simplicity
```
score(data_healthy_scaled)
score(data_broken_scaled)
```
### Submission
In case you feel confident that everything works as it should (getting values close to one for the healthy and close to zero for the broken case) you can make sure that the secret variable contains a valid submission token and submit your work to the grader
```
parts_data = {}
parts_data["0dXlH"] = json.dumps({"number_of_neurons_layer1": number_of_neurons_layer1, "number_of_neurons_layer2": number_of_neurons_layer2, "number_of_neurons_layer3": number_of_neurons_layer3, "number_of_epochs": number_of_epochs})
parts_data["O5cR9"] = json.dumps({"dim": dim, "samples": samples})
token = #### your code here ###
prediction = str(np.sum(score(data_healthy_scaled))/3)
myData={'healthy' : prediction}
myData
parts_data["ZzEP8"] = json.dumps(myData)
submitAll(email, token, key, parts_data)
```
|
github_jupyter
|
# Reproduce Allen smFISH results with Starfish
This notebook walks through a work flow that reproduces the smFISH result for one field of view using the starfish package.
```
from copy import deepcopy
from glob import glob
import json
import os
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import ndimage as ndi
from scipy import stats
from skimage import (exposure, feature, filters, io, measure,
morphology, restoration, segmentation, transform,
util, img_as_float)
from starfish.io import Stack
from starfish.constants import Indices
# # developer note: for rapid iteration, it may be better to run this cell, download the data once, and load
# # the data from the local disk. If so, uncomment this cell and run this instead of the above.
# !aws s3 sync s3://czi.starfish.data.public/20180606/allen_smFISH ./allen_smFISH
# experiment_json = os.path.abspath("./allen_smFISH/fov_001/experiment.json")
# this is a large (1.1GB) FOV, so the download may take some time
experiment_json = 'https://dmf0bdeheu4zf.cloudfront.net/20180606/allen_smFISH/fov_001/experiment.json'
```
Load the Stack object, which while not well-named right now, should be thought of as an access point to an "ImageDataSet". In practice, we expect the Stack object or something similar to it to be an access point for _multiple_ fields of view. In practice, the thing we talk about as a "TileSet" is the `Stack.image` object. The data are currently stored in-memory in a `numpy.ndarray`, and that is where most of our operations are done.
The numpy array can be accessed through Stack.image.numpy\_array (public method, read only) or Stack.image.\_data (read and write)
```
codebook = pd.read_json('https://dmf0bdeheu4zf.cloudfront.net/20180606/allen_smFISH/fov_001/codebook.json')
codebook
```
We're ready now to load the experiment into starfish (This experiment is big, it takes a few minutes):
```
s = Stack()
s.read(experiment_json)
```
All of our implemented operations leverage the `Stack.image.apply` method to apply a single function over each of the tiles or volumes in the FOV, depending on whether the method accepts a 2d or 3d array. Below, we're clipping each image independently at the 10th percentile. I've placed the imports next to the methods so that you can easily locate the code, should you want to look under the hood and understand what parameters have been chosen.
The verbose flag for our apply loops could use a bit more refinement. We should be able to tell it how many images it needs to process from looking at the image stack, but for now it's dumb so just reports the number of tiles or volumes it's processed. This FOV has 102 images over 3 volumes.
```
from starfish.pipeline.filter import Filter
s_clip = Filter.Clip(p_min=10, p_max=100, verbose=True)
s_clip.filter(s.image)
```
We're still working through the backing of the Stack.image object with the on-disk or on-cloud Tile spec. As a result, most of our methods work in-place. For now, we can hack around this by deepcopying the data before administering the operation. This notebook was developed on a 64gb workstation, so be aware of the memory usage when copying!
```
# filtered_backup = deepcopy(s)
```
If you ever want to visualize the image in the notebook, we've added a widget to do that. The first parameter is an indices dict that specifies which hybridization round, channel, z-slice you want to view. The result is a pageable visualization across that arbitrary set of slices. Below I'm visualizing the first channel, which your codebook tells me is Nmnt.
[N.B. once you click on the slider, you can page with the arrow keys on the keyboard.]
```
s.image.show_stack({Indices.CH: 0});
s_bandpass = Filter.Bandpass(lshort=0.5, llong=7, threshold=None, truncate=4, verbose=True)
s_bandpass.filter(s.image)
```
For bandpass, there's a point where things get weird, at `c == 0; z <= 14`. In that range the images look mostly like noise. However, _above_ that, they look great + background subtracted! The later stages of the pipeline appear robust to this, though, as no spots are called for the noisy sections.
```
# I wasn't sure if this clipping was supposed to be by volume or tile. I've done tile here, but it can be easily
# switched to volume.
s_clip = Filter.Clip(p_min=10, p_max=100, is_volume=False, verbose=True)
s_clip.filter(s.image)
sigma=(1, 0, 0) # filter only in z, do nothing in x, y
glp = Filter.GaussianLowPass(sigma=sigma, is_volume=True, verbose=True)
glp.filter(s.image)
```
Below, because spot finding is so slow when single-plex, we'll pilot this on a max projection to show that the parameters work. Here's what trackpy.locate, which we wrap, produces for a z-projection of channel 1. To do use our plotting methods on z-projections we have to expose some of the starfish internals, which will be improved upon.
```
from showit import image
from trackpy import locate
# grab a section from the tensor.
ch1 = s.image.max_proj(Indices.Z)[0, 1]
results = locate(ch1, diameter=3, minmass=250, maxsize=3, separation=5, preprocess=False, percentile=10)
results.columns = ['x', 'y', 'intensity', 'r', 'eccentricity', 'signal', 'raw_mass', 'ep']
# plot the z-projection
f, ax = plt.subplots(figsize=(20, 20))
ax.imshow(ch1, vmin=15, vmax=52, cmap=plt.cm.gray)
# draw called spots on top as red circles
# scale radius plots the red circle at scale_radius * spot radius
s.image._show_spots(results, ax=plt.gca(), scale_radius=7)
```
Below spot finding is on the _volumes_ for each channel. This will take about `11m30s`
```
from starfish.pipeline.features.spots.detector import SpotFinder
# I've guessed at these parameters from the allen_smFISH code, but you might want to tweak these a bit.
# as you can see, this function takes a while. It will be great to parallelize this. That's also coming,
# although we haven't figured out where it fits in the priority list.
kwargs = dict(
spot_diameter=3, # must be odd integer
min_mass=300,
max_size=3, # this is max _radius_
separation=5,
noise_size=0.65, # this is not used because preprocess is False
preprocess=False,
percentile=10, # this is irrelevant when min_mass, spot_diameter, and max_size are set properly
verbose=True,
is_volume=True,
)
lmpf = SpotFinder.LocalMaxPeakFinder(**kwargs)
spot_attributes = lmpf.find(s.image)
# save the results to disk as json
for attrs, (hyb, ch) in spot_attributes:
attrs.save(f'spot_attributes_c{ch.value}.json')
# # if you want to load them back in the same shape, here's how:
# from starfish.pipeline.features.spot_attributes import SpotAttributes
# spot_attributes = [SpotAttributes.load(attrs) for attrs in glob('spot_attributes_c*.json')]
# this is not a very performant function because of how matplotlib renders circles as individual artists,
# but I think it's useful for debugging the spot detection.
# Note that in places where spots are "missed" it is often because they've been localized to individual
# nearby z-planes, whereas most spots exist across several layers of z.
s.image.show_stack({Indices.CH: 1, Indices.HYB: 0}, show_spots=spot_attributes[1][0], figure_size=(20, 20), p_min=60, p_max=99.9);
```
|
github_jupyter
|
```
# default_exp helpers
```
# helpers
> this didn't fit anywhere else
```
#export
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
import matplotlib.pyplot as plt
#ToDo: Propagate them through the methods
iters = 10
l2 = 1
n_std = 4
from pygments import highlight
from pygments.lexers import PythonLexer
from pygments.formatters import HtmlFormatter
import IPython
def plot_regret(y_optimal_list,y_hat_list):
y_optimal_array = np.array(y_optimal_list)
y_hat_array = np.array(y_hat_list)
regret_list = []
regret = np.cumsum(y_optimal_array - y_hat_array)
plt.plot(regret)
def showcase_code(pyfile,class_name = False, method_name = False, end_string = False):
"""shows content of py file"""
with open(pyfile) as f:
code = f.read()
if class_name:
#1. find beginning (class + <name>)
index = code.find(f'class {class_name}')
code = code[index:]
#2. find end (class (new class!) or end of script)
end_index = code[7:].find('class')
if method_name:
#1. find beginning (class + <name>)
index = code.find(f'def {method_name}')
code = code[index:]
#2. find end (class (new class!) or end of script)
end_index = code[7:].find('def')
if end_string:
end_index = code[7:].find('# helpers')
code = code[:end_index]
formatter = HtmlFormatter()
return IPython.display.HTML('<style type="text/css">{}</style>{}'.format(
formatter.get_style_defs('.highlight'),
highlight(code, PythonLexer(), formatter)))
showcase_code('thompson_sampling/helpers.py',method_name='showcase_code')
showcase_code('thompson_sampling/solvers.py',class_name='BetaBandit', end_string = True)
#export
import scipy.stats as stats
def plot_online_logreg(online_lr, wee_x, wee_y):
# closing other figures
plt.close('all')
plt.figure(figsize=[9,3.5], dpi=150)
# let us check the distribution of weights and uncertainty bounds
plt.figure(figsize=[9,3.5], dpi=150)
# plotting the pdf of the weight distribution
X_pdf = np.linspace(-4, 4, 1000)
pdf = stats.norm(loc=online_lr.m, scale=online_lr.q**(-1.0)).pdf(X_pdf)
# range and resolution of probability plot
X_prob = np.linspace(-6, 6, 1000)
p_dist = 1/(1 + np.exp(-X_prob * online_lr.m))
p_dist_plus = 1/(1 + np.exp(-X_prob * (online_lr.m + 2*online_lr.q**(-1.0))))
p_dist_minus = 1/(1 + np.exp(-X_prob * (online_lr.m - 2*online_lr.q**(-1.0))))
# opening subplots
ax1 = plt.subplot2grid((1, 5), (0, 0), colspan=2, rowspan=1)
ax2 = plt.subplot2grid((1, 5), (0, 2), colspan=3, rowspan=1)
# plotting distriution of weights
ax1.plot(X_pdf, pdf, color='b', linewidth=2, alpha=0.5)
#ax1.plot([cmab.weights[0][1], cmab.weights[0][1]], [0, max(pdf)], 'k--', label='True $\\beta$', linewidth=1)
ax1.fill_between(X_pdf, pdf, 0, color='b', alpha=0.2)
# plotting probabilities
ax2.plot(X_prob, p_dist, color='b', linewidth=2, alpha=0.5)
ax2.fill_between(X_prob, p_dist_plus, p_dist_minus, color='b', alpha=0.2)
ax2.scatter(wee_x, wee_y, c='k')
# title and comments
ax1.set_title('OLR estimate for $\\beta$', fontsize=10)
ax1.set_xlabel('$\\beta$', fontsize=10); ax1.set_ylabel('$density$', fontsize=10)
ax2.set_title('OLR estimate for $\\theta(x)$', fontsize=10)
ax2.set_xlabel('$x$', fontsize=10); ax2.set_ylabel('$\\theta(x)$', fontsize=10)
ax1.legend(fontsize=10)
plt.tight_layout()
plt.show()
import numpy as np
from thompson_sampling.models import OnlineLogisticRegression, BatchBayesLinReg
from thompson_sampling.multi_armed_bandits import contextual_categorical_bandit
theta = [0.6,1.0]
noise = 0.1
wee_x = np.random.uniform(-6,6,10)
wee_y = np.array([contextual_categorical_bandit(x,0, theta, noise)[0] for x in wee_x])
# OLR object
online_lr = OnlineLogisticRegression(1, .5, 1)
for i in range(len(wee_y)):
online_lr.fit(wee_x[i].reshape(-1,1), wee_y[i].reshape(-1,1))
plot_online_logreg(online_lr, wee_x, wee_y)
#export
from mpl_toolkits.axes_grid1 import ImageGrid
def prettify_ax(ax):
ax.set_xlim(-1, 1)
ax.set_ylim(-1, 1)
ax.set_xlabel('$w_1$')
ax.set_ylabel('$w_2$')
return ax
def sample(n, weights):
for _ in range(n):
x = np.array([1, np.random.uniform(-1, 1)])
y = np.dot(weights, x) + np.random.normal(0, .2)
yield x, y
def sample(n, weights):
X = np.array([[1, np.random.uniform(-1, 1)] for i in range(n)])
y = [np.dot(weights, x) + np.random.normal(0, .2) for x in X]
return X, y
def plot_param_through_time(model,N,n_samples,X,y,):
w = np.linspace(-1, 1, 100)
W = np.dstack(np.meshgrid(w, w))
n_samples = 5
fig = plt.figure(figsize=(7 * n_samples, 21))
grid = ImageGrid(
fig, 111, # similar to subplot(111)
nrows_ncols=(n_samples, 3), # creates a n_samplesx3 grid of axes
axes_pad=.5 # pad between axes in inch.
)
# We'll store the features and targets for plotting purposes
xs = []
ys = []
for i, (xi, yi) in enumerate(zip(X,y)):
pred_dist = model.predict(xi)
# Prior weight distribution
ax = prettify_ax(grid[3 * i])
ax.set_title(f'Prior weight distribution #{i + 1}')
ax.contourf(w, w, model.weights_dist.pdf(W), N, cmap='viridis')
ax.scatter(*weights, color='red') # true weights the model has to find
# Update model
model.learn(xi, yi)
# Prior weight distribution
ax = prettify_ax(grid[3 * i + 1])
ax.set_title(f'Posterior weight distribution #{i + 1}')
ax.contourf(w, w, model.weights_dist.pdf(W), N, cmap='viridis')
ax.scatter(*weights, color='red') # true weights the model has to find
# Posterior target distribution
xs.append(xi)
ys.append(yi)
posteriors = [model.predict(np.array([1, wi])) for wi in w]
ax = prettify_ax(grid[3 * i + 2])
ax.set_title(f'Posterior target distribution #{i + 1}')
# Plot the old points and the new points
ax.scatter([xi[1] for xi in xs[:-1]], ys[:-1])
ax.scatter(xs[-1][1], ys[-1], marker='*')
# Plot the predictive mean along with the predictive interval
ax.plot(w, [p.mean() for p in posteriors], linestyle='--')
cis = [p.interval(.95) for p in posteriors]
ax.fill_between(
x=w,
y1=[ci[0] for ci in cis],
y2=[ci[1] for ci in cis],
alpha=.1
)
# Plot the true target distribution
ax.plot(w, [np.dot(weights, [1, xi]) for xi in w], color='red')
def sample(n, weights):
for _ in range(n):
x = np.array([1, np.random.uniform(-1, 1)])
y = np.dot(weights, x) + np.random.normal(0, .2)
yield x, y
def sample(n, weights):
X = np.array([[1, np.random.uniform(-1, 1)] for i in range(n)])
y = [np.dot(weights, x) + np.random.normal(0, .2) for x in X]
return X, y
model = BatchBayesLinReg(n_features=2, alpha=2, beta=25)
np.random.seed(42)
# Pick some true parameters that the model has to find
weights = np.array([-.3, .5])
n_samples = 5
N = 100
X,y = sample(n_samples, weights)
plot_param_through_time(model,N,n_samples,X,y)
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/findingfoot/ML_practice-codes/blob/master/principal_component_analysis_.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
```
from sklearn import datasets
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
cancer = datasets.load_breast_cancer()
print(cancer.DESCR)
#checking if 0 represents malignant or benign
# we already know that there are 357 benign values. we count the count of data points that are classified as 1 and cross check with the information we already have
len(cancer.data[cancer.target == 1])
# How features affect the target
fig, axes = plt.subplots(10,3, figsize = (12,9))
malignant = cancer.data[cancer.target == 0]
benign = cancer.data[cancer.target ==1]
ax = axes.ravel()
for i in range(30):
_, bins = np.histogram(cancer.data[:,i], bins = 40)
ax[i].hist(malignant[:,i], bins = bins, color = 'r', alpha = 0.5)
ax[i].hist(benign[:,i], bins = bins, color = 'y', alpha = 0.8)
ax[i].set_title(cancer.feature_names[i], fontsize = 8 )
ax[i].axes.get_xaxis().set_visible(False)
ax[i].set_yticks(())
ax[0].legend(['Malignant', 'Benign'], loc = "best")
plt.tight_layout()
plt.show()
cancer_df = pd.DataFrame(cancer.data, columns = cancer.feature_names)
cancer_df.head()
plt.subplot(1,2,1)
plt.scatter(cancer_df['worst symmetry'], cancer_df['worst texture'], s = cancer_df['worst area']*0.05,color = 'teal', label = 'check', alpha = 0.3)
plt.xlabel('Worst Symmetry', fontsize = 12)
plt.ylabel('Worst Texture', fontsize = 12)
plt.subplot(1,2,2)
plt.scatter(cancer_df['mean radius'], cancer_df['mean concave points'], s = cancer_df['mean area']*0.05,color = 'teal', label = 'check', alpha = 0.3)
plt.xlabel('Mean Radius', fontsize = 12)
plt.ylabel('Mean Concave', fontsize = 12)
plt.subplot(1,2,2)
# we need to scale the data before the fitting algorithm is implemented.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(cancer.data)
scaled_x = scaler.transform(cancer.data)
scaled_x.max(axis=0)
from sklearn.decomposition import PCA
pca = PCA(n_components = 3)
pca.fit(scaled_x)
x_pca = pca.transform(scaled_x)
x_pca.shape
variance_test = np.var(x_pca, axis =0)
variance_ratio = variance_test/np.sum(variance_test)
print(variance_ratio)
Xax=x_pca[:,0]
Yax=x_pca[:,1]
labels=cancer.target
cdict={0:'red',1:'green'}
labl={0:'Malignant',1:'Benign'}
marker={0:'*',1:'o'}
alpha={0:.3, 1:.5}
fig,ax=plt.subplots(figsize=(7,5))
fig.patch.set_facecolor('white')
for l in np.unique(labels):
ix=np.where(labels==l)
ax.scatter(Xax[ix],Yax[ix],c=cdict[l],s=40,
label=labl[l],marker=marker[l],alpha=alpha[l])
# for loop ends
plt.xlabel("First Principal Component",fontsize=14)
plt.ylabel("Second Principal Component",fontsize=14)
plt.legend()
plt.show()
plt.matshow(pca.components_,cmap='viridis')
plt.yticks([0,1,2],['1st Comp','2nd Comp','3rd Comp'],fontsize=10)
plt.colorbar()
plt.xticks(range(len(cancer.feature_names)),cancer.feature_names,rotation=65,ha='left')
plt.tight_layout()
plt.show()#
feature_worst=list(cancer_df.columns[20:31]) # select the 'worst' features
import seaborn as sns
s=sns.heatmap(cancer_df[feature_worst].corr(),cmap='coolwarm')
s.set_yticklabels(s.get_yticklabels(),rotation=30,fontsize=7)
s.set_xticklabels(s.get_xticklabels(),rotation=30,fontsize=7)
plt.show()
```
|
github_jupyter
|
What you should know about C
----
- Write, compile and run a simple program in C
- Static types
- Control flow especially `for` loop
- Using functions
- Using structs
- Pointers and arrays
- Function pointers
- Dynamic memory allocation
- Separate compilation and `make`
### Structs
**Exercise 1**
Write and use a `struct` to represent dates.
```
```
**Solution**
```
%%file ex1.c
#include <stdio.h>
typedef struct {
int day;
int month;
int year;
} date;
int main(int argc, char* argv[])
{
date d1;
d1.day = 29;
d1.month = 3;
d1.year = 2016;
date d2 = {30, 3, 2016};
date d3 = {.year = 2016, .month = 3, .day = 31};
printf("%d-%d-%d\n", d1.month, d1.day, d1.year);
printf("%d-%d-%d\n", d2.month, d2.day, d2.year);
printf("%d-%d-%d\n", d3.month, d3.day, d3.year);
}
%%bash
gcc -std=c99 -o ex1 ex1.c
%%bash
./ex1
```
### Pointers
**Exercise 2**
Write and use pointers for working with
- (a) doubles
- (b) the date struct
- (c) vector of doubles
- (d) 2D array of doubles
```
```
**Solution**
```
%%file ex2a.c
#include <stdio.h>
#include <stdlib.h>
int main(int argc, char* argv[])
{
double x1 = 2.78;
double x2 = 3.14;
double *p1 = malloc(sizeof(double));
if (p1 == NULL) return -1;
double *p2 = calloc(sizeof(double), 1);
if (p2 == NULL) return -1;
printf("%p: %.2f\n", p1, *p1);
printf("%p: %.2f\n\n", p2, *p2);
p1 = &x1;
*p2 = x2;
printf("%p: %.2f\n", p1, *p1);
printf("%p: %.2f\n", p2, *p2);
// free(p1);
// free(p2);
}
%%bash
gcc -std=c99 -o ex2a ex2a.c
%%bash
./ex2a
```
**Solution**
```
%%file ex2b.c
#include <stdio.h>
#include <stdlib.h>
typedef struct {
int day;
int month;
int year;
} date;
int main(int argc, char* argv[])
{
date *d1 = malloc(sizeof(date));
if (d1 == NULL) return -1;
d1->day = 29;
d1->month = 3;
d1->year = 2016;
printf("%d-%d-%d\n", d1->month, d1->day, d1->year);
printf("%d-%d-%d\n", (*d1).month, (*d1).day, (*d1).year);
free(d1);
}
%%bash
gcc -std=c99 -o ex2b ex2b.c
%%bash
./ex2b
```
**Solution**
```
%%file ex2c.c
#include <stdio.h>
#include <stdlib.h>
int main(int argc, char* argv[])
{
int n = atoi(argv[1]);
double *xs = calloc(sizeof(double), n);
if (xs == NULL) return -1;
for (int i=0; i<n; i++) {
xs[i] = i*i;
}
printf("%.2f\n", *(xs));
printf("%.2f\n", *(xs + 2));
printf("%.2f\n", xs[0]);
printf("%.2f\n", xs[2]);
free(xs);
}
%%bash
gcc -std=c99 -o ex2c ex2c.c
%%bash
./ex2c 10
```
**Solution**
```
%%file ex2d.c
#include <stdio.h>
#include <stdlib.h>
int main(int argc, char* argv[])
{
int rows = 2;;
int cols = 3;
double **xs = malloc(sizeof(double) * rows);
for (int i=0; i < rows; i++) {
xs[i] = calloc(sizeof(double), cols);
}
for (int i=0; i<rows; i++) {
for (int j=0; j<cols; j++) {
xs[i][j] = i+j;
}
}
printf("%.2f\n", xs[0][0]);
printf("%.2f\n", xs[1][2]);
for (int i=0; i<rows; i++) {
free(xs[i]);
}
free(xs);
}
%%bash
gcc -std=c99 -o ex2d ex2d.c
%%bash
./ex2d
```
### Function pointers
**Exercise 3**
Write and use a function pointer.
**Solution**
```
%%file ex3.c
#include <stdio.h>
#include <stdlib.h>
double add(double x, double y) {
return x + y;
}
double mult(double x, double y) {
return x * y;
}
int main(int argc, char* argv[])
{
double a = 3.0;
double b = 4.0;
double (*f)(double, double) = add;
typedef double (*fp)(double, double);
fp g = mult;
printf("%.2f\n", add(a, b));
printf("%.2f\n", f(a, b));
printf("%.2f\n", g(a, b));
}
%%bash
gcc -std=c99 -o ex3 ex3.c
%%bash
./ex3
```
### Separate compilation
**Exercise 4**
Write header and implementation files for the add function, and use the function in a separate driver file. Use a makefile to compile the executable.
```
```
**Solution**
```
%%file ex4.h
#pragma once
double add(double x, double y);
%%file ex4.c
#include "ex4.h"
double add(double x, double y) {
return x + y;
}
%%file ex4_main.c
#include <stdio.h>
#include "ex4.h"
int main() {
double a = 3.0;
double b = 4.0;
printf("%.2f\n", add(a, b));
}
%%file makefile
ex4_main: ex4_main.c ex4.o
gcc -std=c99 -o ex4_main ex4_main.c ex4.o
ex4.o: ex4.c
gcc -std=c99 -c ex4.c
%%bash
make
%%bash
./ex4_main
%%file makefile
TARGET = ex4_main
OBJECTS = ex4.o
CFLAGS = -O3 -std=c99
LDLIBS = -lm
CC = gcc
all: $(TARGET)
clean:
rm $(TARGET) $(OBJECTS)
$(TARGET): $(OBJECTS)
%%bash
make clean
make
%%bash
./ex4_main
```
What you should know about C++
----
- Anonymous functions
- Generalized function pointers
- Ranged for
- Using the standard template library
- Iterators
- Containers
- Algorithms
- The `random` library
- Using `amradillo`
**Exercise 5**
Implement Newton's method in 1D for root finding. Pass in the function and gradient as generalized function pointers. Use the method to find all roots of the polynomial equation $f(x) = x^3 - 7x - 6$
```
```
**Solution**
```
%%file ex5.cpp
#include <iostream>
#include <vector>
#include <iomanip>
#include <cmath>
#include <functional>
using std::vector;
using std::cout;
using std::function;
using func = function<double(double)>;
double newton(double x, func f, func fprime, int max_iter=10) {
for (int i=0; i<max_iter; i++) {
x -= f(x)/fprime(x);
}
return x;
};
int main()
{
auto f = [](double x) { return pow(x, 3) - 7*x - 6; };
auto fprime = [](double x) { return 3.0*pow(x, 2) - 7; };
vector<double> x = {-5, 0, 5};
for (auto x_: x) {
cout << std::setw(2) << x_ << ": "
<< std::setw(3) << newton(x_, f, fprime) << "\n";
}
}
%%bash
g++ -std=c++11 ex5.cpp -o ex5
%%bash
./ex5
```
**Exercise 6**
Use the armadillo library to
- Generate 10 x-coordinates linearly spaced between 10 and 15
- Generate 10 random y-values as $y = 3x^2 - 7x + 2 + \epsilon$ where $\epsilon \sim 10 N(0,1)$
- Find the length of $x$ and $y$ and the Euclidean distance between $x$ and $y$
- Find the correlation between $x$ and $y$
- Solve the linear system to find a quadratic fit for this data
```
```
**Solution**
```
%%file ex6.cpp
#include <iostream>
#include <fstream>
#include <armadillo>
using std::cout;
using std::ofstream;
using namespace arma;
int main()
{
vec x = linspace<vec>(10.0,15.0,10);
vec eps = 10*randn<vec>(10);
vec y = 3*x%x - 7*x + 2 + eps;
cout << "x:\n" << x << "\n";
cout << "y:\n" << y << "\n";
cout << "Lenght of x is: " << norm(x) << "\n";
cout << "Lenght of y is: " << norm(y) << "\n";
cout << "Distance(x, y) is: " << norm(x-y) << "\n";
cout << "Correlation(x, y) is: " << cor(x, y) << "\n";
mat A = join_rows(ones<vec>(10), x);
A = join_rows(A, x%x);
cout << "A:\n" << A << "\n";
vec b = solve(A, y);
cout << "b:\n" << b << "\n";
ofstream fout1("x.txt");
x.print(fout1);
ofstream fout2("y.txt");
y.print(fout2);
ofstream fout3("b.txt");
b.print(fout3);
}
%%bash
g++ -std=c++11 ex6.cpp -o ex6 -larmadillo
%%bash
./ex6
x = np.loadtxt('x.txt')
y = np.loadtxt('y.txt')
b = np.loadtxt('b.txt')
plt.scatter(x, y, s=40)
plt.plot(x, b[0] + b[1]*x + b[2]*x**2, c='red')
pass
```
|
github_jupyter
|
## Training Network
In supervised training, the network processes inputs and compares its resulting outputs against the desired outputs.
Errors are propagated back through the system, causing the system to adjust the weights which control the network. This is done using the Backpropagation algorithm, also called backprop. This process occurs over and over as the weights are continually tweaked.
The set of data which enables the training is called the "training set."
During the training of a network the same set of data is processed many times as the connection weights are ever refined. Iteratively passing batches of data through the network and updating the weights, so that the error is decreased, is known as Stochastic Gradient Descent (SGD).
Training refers to determining the best set of weights for maximizing a neural network’s accuracy.
The amount by which the weights are changed is determined by a parameter called Learning rate.
Neural networks can be used without knowing precisely how training works. Most modern machine learning libraries have greatly automated the training process.
### NOTE:
Basicaly this notebook prepared to use within **Google Colab**: https://colab.research.google.com/.
The Google Colabatory has **free Tesla K80 GPU** and already prepared to develop deep learning applications.
First time opens this notebook, do not forget to enable **Python 3** runtime and **GPU** accelerator in Google Colab **Notebook Settings**.
### Setup Project
Create workspace and change directory.
```
PROJECT_HOME = '/content/keras-movie-reviews-classification'
import os.path
if not os.path.exists(PROJECT_HOME):
os.makedirs(PROJECT_HOME)
os.chdir(PROJECT_HOME)
!pwd
```
### Import Project
Import GitHub project to workspace.
```
# Import project and override existing data.
!git init .
!git remote add -t \* -f origin https://github.com/alex-agency/keras-movie-reviews-classification.git
!git reset --hard origin/master
!git checkout
!ls -la input
```
### Keras
Keras is a high-level API, written in Python and capable of running on top of TensorFlow, Theano, or CNTK deep learning frameworks.
Keras provides a simple and modular API to create and train Neural Networks, hiding most of the complicated details under the hood.
By default, Keras is configured to use Tensorflow as the backend since it is the most popular choice.
Keras is becoming super popular recently because of its simplicity.
### Keras workflow
<img src="https://www.learnopencv.com/wp-content/uploads/2017/09/keras-workflow.jpg" width="700px">
```
# Load Keras libraries
from keras.models import load_model
from keras import callbacks
```
### Load model and dataset
Loading model definition from HDF5 file.
```
import numpy as np
# Load data from numpy array
loaded = np.load('input/dataset.npz')
(X_train, Y_train), (X_test, Y_test) = loaded['dataset']
# Load model from HDF5 file.
model = load_model('input/mlps-model-definition.h5') # model with MLP network
print("Model Summary")
print(model.summary())
```
### Configuring the training process
Once the model is ready, we need to configure the learning process.
Compile the model means that Keras will generate a computation graph in TensorFlow.
### Loss functions
In a supervised learning problem, we have to find the error between the actual values and the predicted value. There can be different metrics which can be used to evaluate this error. This metric is often called loss function or cost function or objective function. There can be more than one loss function depending on what you are doing with the error. In general, we use:
* binary-cross-entropy for a binary classification problem
* categorical-cross-entropy for a multi-class classification problem
* mean-squared-error for a regression problem and so on
### Optimizers
An Optimizer determines how the network weights are updated.
Keras provides a lot of optimizers to choose from.
RMSprop and Adam is a good choice of optimizer for most problems.
### Overfitting
Overfitting describes the situation in which your model is over-optimized to accurately predict the training set, at the expense of generalizing to unknown data (which is the objective of learning in the first place). This can happen because the model greatly twists itself to perfectly conform to the training set, even capturing its underlying noise.
How can we avoid overfitting? The simplest solution is to split our dataset into a training set and a test set. The training set is used for the optimization procedure, but we evaluate the accuracy of our model by forwarding the test set to the trained model and measuring its accuracy.
During training, we can monitor the accuracy of the model on the training set and test set. The longer we train, the more likely our training accuracy is to go higher and higher, but at some point, it is likely the test set will stop improving. This is a cue to stop training at that point. We should generally expect that training accuracy is higher than test accuracy, but if it is much higher, that is a clue that we have overfit.
```
# Compile model
model.compile(loss='binary_crossentropy', # cross-entropy loss function for binary classification
optimizer='adam', # Adam optimiser one of the most popular optimization method
metrics=['accuracy']) # print the accuracy during training
# Early stopping callback
# Stop training when a monitored quantity has stopped improving.
# Using held-out validation set, to determine when to terminate the training process to avoid overfitting.
early_stopping = callbacks.EarlyStopping(monitor='val_loss', # quantity to be monitored
min_delta=0, # minimum change in the monitored quantity to qualify as an improvement
patience=2, # number of epochs with no improvement after which training will be stopped
verbose=1, mode='auto')
# Train model
history = model.fit(X_train, Y_train, # train the model using the training set
batch_size=8, # in each iteration, use size of training examples at once
epochs=20, # iterate amount of times over the entire training set
callbacks=[early_stopping], # called after each epoch
validation_split=0.2, # use 20% of the data for validation
verbose=2) # enables detailed logs, where 2 is print some information after each epoch
# Evaluate model
score = model.evaluate(X_test, Y_test, verbose=0) # evaluate the trained model on the test set
print('Test loss:', score[0])
print('Test accuracy:', score[1])
import matplotlib.pyplot as plt
# Plot the loss over each epochs.
plt.plot(history.history['loss'], label='training')
plt.plot(history.history['val_loss'], label='validation')
plt.legend()
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.show()
# Plot the accuracy evaluated on the training set.
plt.plot(history.history['acc'], label='training');
plt.plot(history.history['val_acc'], label='validation');
plt.legend()
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.show()
```
### Export trained model to file
Saving whole Keras model into a single HDF5 file which will contain:
* the architecture of the model, allowing to re-create the model
* the weights of the model
* the training configuration (loss, optimizer)
* the state of the optimizer, allowing to resume training exactly where you left off.
```
# Model filename
model_filename = 'mlps-model.h5'
# Create output directory
output_dir = 'output'
if not os.path.exists(output_dir):
os.makedirs(output_dir)
model_file = os.path.join(output_dir, model_filename)
# Export model into HDF5 file.
model.save(model_file)
!ls -la output
```
### Downloading file to your local file system
It will invoke a browser download of the file to your local computer.
```
from google.colab import files
# Download file
files.download(model_file)
```
|
github_jupyter
|
# Basic Motion
Welcome to JetBot's browser based programming interface! This document is
called a *Jupyter Notebook*, which combines text, code, and graphic
display all in one! Prett neat, huh? If you're unfamiliar with *Jupyter* we suggest clicking the
``Help`` drop down menu in the top toolbar. This has useful references for
programming with *Jupyter*.
In this notebook, we'll cover the basics of controlling JetBot.
### Importing the Robot class
To get started programming JetBot, we'll need to import the ``Robot`` class. This class
allows us to easily control the robot's motors! This is contained in the ``jetbot`` package.
> If you're new to Python, a *package* is essentially a folder containing
> code files. These code files are called *modules*.
To import the ``Robot`` class, highlight the cell below and press ``ctrl + enter`` or the ``play`` icon above.
This will execute the code contained in the cell
```
from jetbot import Robot
```
Now that we've imported the ``Robot`` class we can initialize the class *instance* as follows.
```
robot = Robot()
```
### Commanding the robot
Now that we've created our ``Robot`` instance we named "robot", we can use this instance
to control the robot. To make the robot spin counterclockwise at 30% of it's max speed
we can call the following
> WARNING: This next command will make the robot move! Please make sure the robot has clearance.
```
robot.left(speed=0.3)
```
Cool, you should see the robot spin counterclockwise!
> If your robot didn't turn left, that means one of the motors is wired backwards! Try powering down your
> robot and swapping the terminals that the ``red`` and ``black`` cables of the incorrect motor.
>
> REMINDER: Always be careful to check your wiring, and don't change the wiring on a running system!
Now, to stop the robot you can call the ``stop`` method.
```
robot.stop()
```
Maybe we only want to run the robot for a set period of time. For that, we can use the Python ``time`` package.
```
import time
```
This package defines the ``sleep`` function, which causes the code execution to block for the specified number of seconds
before running the next command. Try the following to make the robot turn left only for half a second.
```
robot.left(0.3)
time.sleep(0.5)
robot.stop()
```
Great. You should see the robot turn left for a bit and then stop.
> Wondering what happened to the ``speed=`` inside the ``left`` method? Python allows
> us to set function parameters by either their name, or the order that they are defined
> (without specifying the name).
The ``BasicJetbot`` class also has the methods ``right``, ``forward``, and ``backwards``. Try creating your own cell to make
the robot move forward at 50% speed for one second.
Create a new cell by highlighting an existing cell and pressing ``b`` or the ``+`` icon above. Once you've done that, type in the code that you think will make the robot move forward at 50% speed for one second.
### Controlling motors individually
Above we saw how we can control the robot using commands like ``left``, ``right``, etc. But what if we want to set each motor speed
individually? Well, there are two ways you can do this
The first way is to call the ``set_motors`` method. For example, to turn along a left arch for a second we could set the left motor to 30% and the right motor to 60% like follows.
```
robot.set_motors(0.3, 0.6)
time.sleep(1.0)
robot.stop()
```
Great! You should see the robot move along a left arch. But actually, there's another way that we could accomplish the same thing.
The ``Robot`` class has two attributes named ``left_motor`` and ``right_motor`` that represent each motor individually.
These attributes are ``Motor`` class instances, each which contains a ``value`` attribute. This ``value`` attribute
is a [traitlet](https://github.com/ipython/traitlets) which generates ``events`` when assigned a new value. In the motor
class, we attach a function that updates the motor commands whenever the value changes.
So, to accomplish the exact same thing we did above, we could execute the following.
```
robot.left_motor.value = 0.34
robot.left_motor.alpha = 0.9
robot.right_motor.value = 0.34
robot.right_motor.alpha = 0.81
time.sleep(3)
robot.left_motor.value = 0.0
robot.right_motor.value = 0.0
```
You should see the robot move in the same exact way!
### Link motors to traitlets
A really cool feature about these [traitlets](https://github.com/ipython/traitlets) is that we can
also link them to other traitlets! This is super handy because Jupyter Notebooks allow us
to make graphical ``widgets`` that use traitlets under the hood. This means we can attach
our motors to ``widgets`` to control them from the browser, or just visualize the value.
To show how to do this, let's create and display two sliders that we'll use to control our motors.
```
import ipywidgets.widgets as widgets
from IPython.display import display
# create two sliders with range [-1.0, 1.0]
left_slider = widgets.FloatSlider(description='left', min=-1.0, max=1.0, step=0.01, orientation='vertical')
right_slider = widgets.FloatSlider(description='right', min=-1.0, max=1.0, step=0.01, orientation='vertical')
# create a horizontal box container to place the sliders next to eachother
slider_container = widgets.HBox([left_slider, right_slider])
# display the container in this cell's output
display(slider_container)
```
You should see two ``vertical`` sliders displayed above.
> HELPFUL TIP: In Jupyter Lab, you can actually "pop" the output of cells into entirely separate window! It will still be
> connected to the notebook, but displayed separately. This is helpful if we want to pin the output of code we executed elsewhere.
> To do this, right click the output of the cell and select ``Create New View for Output``. You can then drag the new window
> to a location you find pleasing.
Try clicking and dragging the sliders up and down. Notice nothing happens when we move the sliders currently. That's because we haven't connected them to motors yet! We'll do that by using the ``link`` function from the traitlets package.
```
import traitlets
left_link = traitlets.link((left_slider, 'value'), (robot.left_motor, 'value'))
right_link = traitlets.link((right_slider, 'value'), (robot.right_motor, 'value'))
```
Now try dragging the sliders (slowly at first). You should see the respective motor turn!
The ``link`` function that we created above actually creates a bi-directional link! That means,
if we set the motor values elsewhere, the sliders will update! Try executing the code block below
```
robot.forward(0.3)
time.sleep(0.5)
robot.stop()
```
You should see the sliders respond to the motor commands! If we want to remove this connection we can call the
``unlink`` method of each link.
```
left_link.unlink()
right_link.unlink()
```
But what if we don't want a *bi-directional* link, let's say we only want to use the sliders to display the motor values,
but not control them. For that we can use the ``dlink`` function. The left input is the ``source`` and the right input is the ``target``
```
left_link = traitlets.dlink((robot.left_motor, 'value'), (left_slider, 'value'))
right_link = traitlets.dlink((robot.right_motor, 'value'), (right_slider, 'value'))
```
Now try moving the sliders. You should see that the robot doesn't respond. But when set the motors using a different method,
the sliders will update and display the value!
### Attach functions to events
Another way to use traitlets, is by attaching functions (like ``forward``) to events. These
functions will get called whenever a change to the object occurs, and will be passed some information about that change
like the ``old`` value and the ``new`` value.
Let's create and display some buttons that we'll use to control the robot.
```
# create buttons
button_layout = widgets.Layout(width='100px', height='80px', align_self='center')
stop_button = widgets.Button(description='stop', button_style='danger', layout=button_layout)
forward_button = widgets.Button(description='forward', layout=button_layout)
backward_button = widgets.Button(description='backward', layout=button_layout)
left_button = widgets.Button(description='left', layout=button_layout)
right_button = widgets.Button(description='right', layout=button_layout)
# display buttons
middle_box = widgets.HBox([left_button, stop_button, right_button], layout=widgets.Layout(align_self='center'))
controls_box = widgets.VBox([forward_button, middle_box, backward_button])
display(controls_box)
```
You should see a set of robot controls displayed above! But right now they wont do anything. To do that
we'll need to create some functions that we'll attach to the button's ``on_click`` event.
```
def stop(change):
robot.stop()
def step_forward(change):
robot.forward(0.3)
time.sleep(0.5)
robot.stop()
def step_backward(change):
robot.backward(0.3)
time.sleep(0.5)
robot.stop()
def step_left(change):
robot.left(0.3)
time.sleep(0.5)
robot.stop()
def step_right(change):
robot.right(0.3)
time.sleep(0.5)
robot.stop()
```
Now that we've defined the functions, let's attach them to the on-click events of each button
```
# link buttons to actions
stop_button.on_click(stop)
forward_button.on_click(step_forward)
backward_button.on_click(step_backward)
left_button.on_click(step_left)
right_button.on_click(step_right)
```
Now when you click each button, you should see the robot move!
### Heartbeat Killswitch
Here we show how to connect a 'heartbeat' to stop the robot from moving. This is a simple way to detect if the robot connection is alive. You can lower the slider below to reduce the period (in seconds) of the heartbeat. If a round-trip communication between broswer cannot be made within two heartbeats, the '`status`' attribute of the heartbeat will be set ``dead``. As soon as the connection is restored, the ``status`` attribute will return to ``alive``.
```
from jetbot import Heartbeat
heartbeat = Heartbeat()
# this function will be called when heartbeat 'alive' status changes
def handle_heartbeat_status(change):
if change['new'] == Heartbeat.Status.dead:
robot.stop()
heartbeat.observe(handle_heartbeat_status, names='status')
period_slider = widgets.FloatSlider(description='period', min=0.001, max=0.5, step=0.01, value=0.5)
traitlets.dlink((period_slider, 'value'), (heartbeat, 'period'))
display(period_slider, heartbeat.pulseout)
```
Try executing the code below to start the motors, and then lower the slider to see what happens. You can also try disconnecting your robot or PC.
```
robot.left(0.2)
# now lower the `period` slider above until the network heartbeat can't be satisfied
```
### Conclusion
That's it for this example notebook! Hopefully you feel confident that you can program your robot to move around now :)
|
github_jupyter
|
# 머신 러닝 교과서 3판
# 14장 - 텐서플로의 구조 자세히 알아보기 (2/3)
**아래 링크를 통해 이 노트북을 주피터 노트북 뷰어(nbviewer.jupyter.org)로 보거나 구글 코랩(colab.research.google.com)에서 실행할 수 있습니다.**
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://nbviewer.jupyter.org/github/rickiepark/python-machine-learning-book-3rd-edition/blob/master/ch14/ch14_part2.ipynb"><img src="https://jupyter.org/assets/main-logo.svg" width="28" />주피터 노트북 뷰어로 보기</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/rickiepark/python-machine-learning-book-3rd-edition/blob/master/ch14/ch14_part2.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />구글 코랩(Colab)에서 실행하기</a>
</td>
</table>
### 목차
- 텐서플로 추정기
- 특성 열 사용하기
- 사전에 준비된 추정기로 머신 러닝 수행하기
```
import numpy as np
import tensorflow as tf
import pandas as pd
from IPython.display import Image
tf.__version__
```
## 텐서플로 추정기
##### 사전에 준비된 추정기 사용하는 단계
* **단계 1:** 데이터 로딩을 위해 입력 함수 정의하기
* **단계 2:** 추정기와 데이터 사이를 연결하기 위해 특성 열 정의하기
* **단계 3:** 추정기 객체를 만들거나 케라스 모델을 추정기로 바꾸기
* **단계 4:** 추정기 사용하기: train() evaluate() predict()
```
tf.random.set_seed(1)
np.random.seed(1)
```
### 특성 열 사용하기
* 정의: https://developers.google.com/machine-learning/glossary/#feature_columns
* 문서: https://www.tensorflow.org/api_docs/python/tf/feature_column
```
Image(url='https://git.io/JL56E', width=700)
dataset_path = tf.keras.utils.get_file("auto-mpg.data",
("http://archive.ics.uci.edu/ml/machine-learning-databases"
"/auto-mpg/auto-mpg.data"))
column_names = ['MPG', 'Cylinders', 'Displacement', 'Horsepower',
'Weight', 'Acceleration', 'ModelYear', 'Origin']
df = pd.read_csv(dataset_path, names=column_names,
na_values = "?", comment='\t',
sep=" ", skipinitialspace=True)
df.tail()
print(df.isna().sum())
df = df.dropna()
df = df.reset_index(drop=True)
df.tail()
import sklearn
import sklearn.model_selection
df_train, df_test = sklearn.model_selection.train_test_split(df, train_size=0.8)
train_stats = df_train.describe().transpose()
train_stats
numeric_column_names = ['Cylinders', 'Displacement', 'Horsepower', 'Weight', 'Acceleration']
df_train_norm, df_test_norm = df_train.copy(), df_test.copy()
for col_name in numeric_column_names:
mean = train_stats.loc[col_name, 'mean']
std = train_stats.loc[col_name, 'std']
df_train_norm.loc[:, col_name] = (df_train_norm.loc[:, col_name] - mean)/std
df_test_norm.loc[:, col_name] = (df_test_norm.loc[:, col_name] - mean)/std
df_train_norm.tail()
```
#### 수치형 열
```
numeric_features = []
for col_name in numeric_column_names:
numeric_features.append(tf.feature_column.numeric_column(key=col_name))
numeric_features
feature_year = tf.feature_column.numeric_column(key="ModelYear")
bucketized_features = []
bucketized_features.append(tf.feature_column.bucketized_column(
source_column=feature_year,
boundaries=[73, 76, 79]))
print(bucketized_features)
feature_origin = tf.feature_column.categorical_column_with_vocabulary_list(
key='Origin',
vocabulary_list=[1, 2, 3])
categorical_indicator_features = []
categorical_indicator_features.append(tf.feature_column.indicator_column(feature_origin))
print(categorical_indicator_features)
```
### 사전에 준비된 추정기로 머신러닝 수행하기
```
def train_input_fn(df_train, batch_size=8):
df = df_train.copy()
train_x, train_y = df, df.pop('MPG')
dataset = tf.data.Dataset.from_tensor_slices((dict(train_x), train_y))
# 셔플, 반복, 배치
return dataset.shuffle(1000).repeat().batch(batch_size)
## 조사
ds = train_input_fn(df_train_norm)
batch = next(iter(ds))
print('키:', batch[0].keys())
print('ModelYear:', batch[0]['ModelYear'])
all_feature_columns = (numeric_features +
bucketized_features +
categorical_indicator_features)
print(all_feature_columns)
regressor = tf.estimator.DNNRegressor(
feature_columns=all_feature_columns,
hidden_units=[32, 10],
model_dir='models/autompg-dnnregressor/')
EPOCHS = 1000
BATCH_SIZE = 8
total_steps = EPOCHS * int(np.ceil(len(df_train) / BATCH_SIZE))
print('훈련 스텝:', total_steps)
regressor.train(
input_fn=lambda:train_input_fn(df_train_norm, batch_size=BATCH_SIZE),
steps=total_steps)
reloaded_regressor = tf.estimator.DNNRegressor(
feature_columns=all_feature_columns,
hidden_units=[32, 10],
warm_start_from='models/autompg-dnnregressor/',
model_dir='models/autompg-dnnregressor/')
def eval_input_fn(df_test, batch_size=8):
df = df_test.copy()
test_x, test_y = df, df.pop('MPG')
dataset = tf.data.Dataset.from_tensor_slices((dict(test_x), test_y))
return dataset.batch(batch_size)
eval_results = reloaded_regressor.evaluate(
input_fn=lambda:eval_input_fn(df_test_norm, batch_size=8))
for key in eval_results:
print('{:15s} {}'.format(key, eval_results[key]))
print('평균 손실 {:.4f}'.format(eval_results['average_loss']))
pred_res = regressor.predict(input_fn=lambda: eval_input_fn(df_test_norm, batch_size=8))
print(next(iter(pred_res)))
```
#### Boosted Tree Regressor
```
boosted_tree = tf.estimator.BoostedTreesRegressor(
feature_columns=all_feature_columns,
n_batches_per_layer=20,
n_trees=200)
boosted_tree.train(
input_fn=lambda:train_input_fn(df_train_norm, batch_size=BATCH_SIZE))
eval_results = boosted_tree.evaluate(
input_fn=lambda:eval_input_fn(df_test_norm, batch_size=8))
print(eval_results)
print('평균 손실 {:.4f}'.format(eval_results['average_loss']))
```
|
github_jupyter
|
<h1 align="center">Welcome to SimpleITK Jupyter Notebooks</h1>
## Newcomers to Jupyter Notebooks:
1. We use two types of cells, code and markdown.
2. To run a code cell, select it (mouse or arrow key so that it is highlighted) and then press shift+enter which also moves focus to the next cell or ctrl+enter which doesn't.
3. Closing the browser window does not close the Jupyter server. To close the server, go to the terminal where you ran it and press ctrl+c twice.
For additional details see the [Jupyter Notebook Quick Start Guide](https://jupyter-notebook-beginner-guide.readthedocs.io/en/latest/index.html).
## SimpleITK Environment Setup
Check that SimpleITK and auxiliary program(s) are correctly installed in your environment, and that you have the SimpleITK version which you expect (<b>requires network connectivity</b>).
You can optionally download all of the data used in the notebooks in advance. This step is only necessary if you expect to run the notebooks without network connectivity.
The following cell checks that all expected packages are installed.
```
from __future__ import print_function
import importlib
from distutils.version import LooseVersion
# check that all packages are installed (see requirements.txt file)
required_packages = {'jupyter',
'numpy',
'matplotlib',
'ipywidgets',
'scipy',
'pandas',
'SimpleITK'
}
problem_packages = list()
# Iterate over the required packages: If the package is not installed
# ignore the exception.
for package in required_packages:
try:
p = importlib.import_module(package)
except ImportError:
problem_packages.append(package)
if len(problem_packages) is 0:
print('All is well.')
else:
print('The following packages are required but not installed: ' \
+ ', '.join(problem_packages))
import SimpleITK as sitk
%run update_path_to_download_script
from downloaddata import fetch_data, fetch_data_all
from ipywidgets import interact
print(sitk.Version())
```
We expect that you have an external image viewer installed. The default viewer is <a href="https://fiji.sc/#download">Fiji</a>. If you have another viewer (i.e. ITK-SNAP or 3D Slicer) you will need to set an environment variable to point to it. This can be done from within a notebook as shown below.
```
# Uncomment the line below to change the default external viewer to your viewer of choice and test that it works.
#%env SITK_SHOW_COMMAND /Applications/ITK-SNAP.app/Contents/MacOS/ITK-SNAP
# Retrieve an image from the network, read it and display using the external viewer.
# The show method will also set the display window's title and by setting debugOn to True,
# will also print information with respect to the command it is attempting to invoke.
# NOTE: The debug information is printed to the terminal from which you launched the notebook
# server.
sitk.Show(sitk.ReadImage(fetch_data("SimpleITK.jpg")), "SimpleITK Logo", debugOn=True)
```
Now we check that the ipywidgets will display correctly. When you run the following cell you should see a slider.
If you don't see a slider please shutdown the Jupyter server, at the command line prompt press Control-c twice, and then run the following command:
```jupyter nbextension enable --py --sys-prefix widgetsnbextension```
```
interact(lambda x: x, x=(0,10));
```
Download all of the data in advance if you expect to be working offline (may take a couple of minutes).
```
fetch_data_all(os.path.join('..','Data'), os.path.join('..','Data','manifest.json'))
```
|
github_jupyter
|
[View in Colaboratory](https://colab.research.google.com/github/PranY/FastAI_projects/blob/master/TSG.ipynb)
```
!pip install fastai
!pip install torch_nightly -f https://download.pytorch.org/whl/nightly/cu92/torch_nightly.html
! pip install kaggle
! pip install tqdm
from google.colab import drive
drive.mount('/content/drive')
! ls "drive/My Drive"
! cp drive/My\ Drive/kaggle.json ~/.kaggle/
! kaggle competitions download -c tgs-salt-identification-challenge
! python -c 'import fastai; print(fastai.__version__)'
! python -c 'import fastai; fastai.show_install(0)'
!ls
# ! rm -r train/
# !rm -r test/
! mkdir train
! mkdir test
! unzip train.zip -d train
! unzip test.zip -d test
! ls train/images | wc -l
! ls train/masks | wc -l
! ls test/images | wc -l
%matplotlib inline
%reload_ext autoreload
%autoreload 2
from tqdm import tqdm_notebook
from fastai import *
from fastai.vision import *
#from fastai.docs import *
import PIL
# Loading of training/testing ids and depths
train_df = pd.read_csv("train.csv", index_col="id", usecols=[0])
depths_df = pd.read_csv("depths.csv", index_col="id")
train_df = train_df.join(depths_df)
test_df = depths_df[~depths_df.index.isin(train_df.index)]
num_workers=0
len(train_df)
PATH_X = Path('train/images')
PATH_Y = Path('train/masks')
# def resize2d(fn:PathOrStr, sz) -> Image:
# img = PIL.Image.open(fn)
# img = img.resize((sz,sz), PIL.Image.BILINEAR)
# img.save(fn)
# for l in list(PATH_X.iterdir()):
# resize2d(l,128)
# for l in list(PATH_Y.iterdir()):
# resize2d(l,128)
# Reducing mask images to {0,1}
def FormatMask(fn:PathOrStr) -> Image:
img = PIL.Image.open(fn).convert('L')
# Let numpy do the heavy lifting for converting pixels to pure black or white
bw = np.asarray(img).copy()
# Pixel range is 0...255, 256/2 = 128
bw[bw < 128] = 0 # Black
bw[bw >= 128] = 1 # White
# Now we put it back in Pillow/PIL land
imfile = PIL.Image.fromarray(bw)
imfile.save(fn)
for l in list(PATH_Y.iterdir()):
FormatMask(l)
class ImageMask(Image):
"Class for image segmentation target."
def lighting(self, func:LightingFunc, *args:Any, **kwargs:Any)->'Image': return self
def refresh(self):
self.sample_kwargs['mode'] = 'bilinear'
return super().refresh()
@property
def data(self)->TensorImage:
"Return this image pixels as a `LongTensor`."
return self.px.long()
def show(self, ax:plt.Axes=None, figsize:tuple=(3,3), title:Optional[str]=None, hide_axis:bool=True,
cmap:str='viridis', alpha:float=0.5):
ax = _show_image(self, ax=ax, hide_axis=hide_axis, cmap=cmap, figsize=figsize, alpha=alpha)
if title: ax.set_title(title)
def open_mask(fn:PathOrStr)->ImageMask:
"Return `ImageMask` object create from mask in file `fn`."
x = PIL.Image.open(fn).convert('L')
return ImageMask(pil2tensor(x).float().div_(255))
def _show_image(img:Image, ax:plt.Axes=None, figsize:tuple=(3,3), hide_axis:bool=True, cmap:str='binary',
alpha:float=None)->plt.Axes:
if ax is None: fig,ax = plt.subplots(figsize=figsize)
ax.imshow(image2np(img.data), cmap=cmap, alpha=alpha)
if hide_axis: ax.axis('off')
return ax
img = next(PATH_X.iterdir())
open_image(img).show()
open_image(img).size
def get_y_fn(x_fn): return PATH_Y/f'{x_fn.name[:-4]}.png'
img_y_f = get_y_fn(img)
open_mask(img_y_f).show()
open_mask(img_y_f).size
x = open_image(img)
x.show(y=open_mask(img_y_f))
x.shape
open_image(img).shape, open_mask(img_y_f).shape
def get_datasets(path):
x_fns = [o for o in path.iterdir() if o.is_file()]
y_fns = [get_y_fn(o) for o in x_fns]
mask = [o>=1000 for o in range(len(x_fns))]
arrs = arrays_split(mask, x_fns, y_fns)
return [SegmentationDataset(*o) for o in arrs]
train_ds,valid_ds = get_datasets(PATH_X)
train_ds,valid_ds
x,y = next(iter(train_ds))
x.shape, y.shape, type(x), type(y)
size = 128
def get_tfm_datasets(size):
datasets = get_datasets(PATH_X)
tfms = get_transforms(do_flip=True, max_rotate=4, max_lighting=0.2)
return transform_datasets(train_ds, valid_ds, tfms=tfms, tfm_y=True, size=size, padding_mode='border')
train_tds, *_ = get_tfm_datasets(size)
for i in range(0,3):
train_tds[i][0].show()
for i in range(0,3):
train_tds[i][1].show()
_,axes = plt.subplots(1,4, figsize=(12,6))
for i, ax in enumerate(axes.flat):
imgx,imgy = train_tds[i]
imgx.show(ax, y=imgy)
default_norm,default_denorm = normalize_funcs( mean=tensor([0.4850, 0.4560, 0.4060]), std=tensor([0.2290, 0.2240, 0.2250]))
bs = 32
def get_data(size, bs):
return DataBunch.create(*get_tfm_datasets(size), bs=bs, tfms=default_norm)
data = get_data(size, bs)
#export
def show_xy_images(x:Tensor,y:Tensor,rows:int,figsize:tuple=(9,9)):
"Shows a selection of images and targets from a given batch."
fig, axs = plt.subplots(rows,rows,figsize=figsize)
for i, ax in enumerate(axs.flatten()): show_image(x[i], y=y[i], ax=ax)
plt.tight_layout()
x,y = next(iter(data.train_dl))
x,y = x.cpu(),y.cpu()
x = default_denorm(x)
show_xy_images(x,y,4, figsize=(9,9))
x.shape, y.shape
head = std_upsample_head(2, 512,256,256,256,256)
head
def dice(input:Tensor, targs:Tensor) -> Rank0Tensor:
"Dice coefficient metric for binary target"
n = targs.shape[0]
input = input.argmax(dim=1).view(n,-1)
targs = targs.view(n,-1)
intersect = (input*targs).sum().float()
union = (input+targs).sum().float()
return 2. * intersect / union
def accuracy(input:Tensor, targs:Tensor) -> Rank0Tensor:
"Accuracy"
n = targs.shape[0]
input = input.argmax(dim=1).view(n,-1)
targs = targs.view(n,-1)
return (input==targs).float().mean()
metrics=[accuracy, dice]
learn = ConvLearner(data, models.resnet34, custom_head=head,
metrics=metrics)
lr_find(learn)
learn.recorder.plot()
learn.loss_func
lr = 1e-1
learn.fit_one_cycle(10, slice(lr))
# memory footprint support libraries/code
!ln -sf /opt/bin/nvidia-smi /usr/bin/nvidia-smi
!pip install gputil
!pip install psutil
!pip install humanize
import psutil
import humanize
import os
import GPUtil as GPU
GPUs = GPU.getGPUs()
# XXX: only one GPU on Colab and isn’t guaranteed
gpu = GPUs[0]
def printm():
process = psutil.Process(os.getpid())
print("Gen RAM Free: " + humanize.naturalsize( psutil.virtual_memory().available ), " | Proc size: " + humanize.naturalsize( process.memory_info().rss))
print("GPU RAM Free: {0:.0f}MB | Used: {1:.0f}MB | Util {2:3.0f}% | Total {3:.0f}MB".format(gpu.memoryFree, gpu.memoryUsed, gpu.memoryUtil*100, gpu.memoryTotal))
printm()
```
|
github_jupyter
|
# Measuring Monotonic Relationships
By Evgenia "Jenny" Nitishinskaya and Delaney Granizo-Mackenzie with example algorithms by David Edwards
Reference: DeFusco, Richard A. "Tests Concerning Correlation: The Spearman Rank Correlation Coefficient." Quantitative Investment Analysis. Hoboken, NJ: Wiley, 2007
Part of the Quantopian Lecture Series:
* [www.quantopian.com/lectures](https://www.quantopian.com/lectures)
* [github.com/quantopian/research_public](https://github.com/quantopian/research_public)
---
The Spearman Rank Correlation Coefficient allows us to determine whether or not two data series move together; that is, when one increases (decreases) the other also increases (decreases). This is more general than a linear relationship; for instance, $y = e^x$ is a monotonic function, but not a linear one. Therefore, in computing it we compare not the raw data but the ranks of the data.
This is useful when your data sets may be in different units, and therefore not linearly related (for example, the price of a square plot of land and its side length, since the price is more likely to be linear in the area). It's also suitable for data sets which not satisfy the assumptions that other tests require, such as the observations being normally distributed as would be necessary for a t-test.
```
import numpy as np
import scipy.stats as stats
import matplotlib.pyplot as plt
import math
# Example of ranking data
l = [10, 9, 5, 7, 5]
print 'Raw data: ', l
print 'Ranking: ', list(stats.rankdata(l, method='average'))
```
## Spearman Rank Correlation
### Intuition
The intution is now that instead of looking at the relationship between the two variables, we look at the relationship between the ranks. This is robust to outliers and the scale of the data.
### Definition
The argument `method='average'` indicates that when we have a tie, we average the ranks that the numbers would occupy. For example, the two 5's above, which would take up ranks 1 and 2, each get assigned a rank of $1.5$.
To compute the Spearman rank correlation for two data sets $X$ and $Y$, each of size $n$, we use the formula
$$r_S = 1 - \frac{6 \sum_{i=1}^n d_i^2}{n(n^2 - 1)}$$
where $d_i$ is the difference between the ranks of the $i$th pair of observations, $X_i - Y_i$.
The result will always be between $-1$ and $1$. A positive value indicates a positive relationship between the variables, while a negative value indicates an inverse relationship. A value of 0 implies the absense of any monotonic relationship. This does not mean that there is no relationship; for instance, if $Y$ is equal to $X$ with a delay of 2, they are related simply and precisely, but their $r_S$ can be close to zero:
##Experiment
Let's see what happens if we draw $X$ from a poisson distribution (non-normal), and then set $Y = e^X + \epsilon$ where $\epsilon$ is drawn from another poisson distribution. We'll take the spearman rank and the correlation coefficient on this data and then run the entire experiment many times. Because $e^X$ produces many values that are far away from the rest, we can this of this as modeling 'outliers' in our data. Spearman rank compresses the outliers and does better at measuring correlation. Normal correlation is confused by the outliers and on average will measure less of a relationship than is actually there.
```
## Let's see an example of this
n = 100
def compare_correlation_and_spearman_rank(n, noise):
X = np.random.poisson(size=n)
Y = np.exp(X) + noise * np.random.normal(size=n)
Xrank = stats.rankdata(X, method='average')
# n-2 is the second to last element
Yrank = stats.rankdata(Y, method='average')
diffs = Xrank - Yrank # order doesn't matter since we'll be squaring these values
r_s = 1 - 6*sum(diffs*diffs)/(n*(n**2 - 1))
c_c = np.corrcoef(X, Y)[0,1]
return r_s, c_c
experiments = 1000
spearman_dist = np.ndarray(experiments)
correlation_dist = np.ndarray(experiments)
for i in range(experiments):
r_s, c_c = compare_correlation_and_spearman_rank(n, 1.0)
spearman_dist[i] = r_s
correlation_dist[i] = c_c
print 'Spearman Rank Coefficient: ' + str(np.mean(spearman_dist))
# Compare to the regular correlation coefficient
print 'Correlation coefficient: ' + str(np.mean(correlation_dist))
```
Let's take a look at the distribution of measured correlation coefficients and compare the spearman with the regular metric.
```
plt.hist(spearman_dist, bins=50, alpha=0.5)
plt.hist(correlation_dist, bins=50, alpha=0.5)
plt.legend(['Spearman Rank', 'Regular Correlation'])
plt.xlabel('Correlation Coefficient')
plt.ylabel('Frequency');
```
Now let's see how the Spearman rank and Regular coefficients cope when we add more noise to the situation.
```
n = 100
noises = np.linspace(0, 3, 30)
experiments = 100
spearman = np.ndarray(len(noises))
correlation = np.ndarray(len(noises))
for i in range(len(noises)):
# Run many experiments for each noise setting
rank_coef = 0.0
corr_coef = 0.0
noise = noises[i]
for j in range(experiments):
r_s, c_c = compare_correlation_and_spearman_rank(n, noise)
rank_coef += r_s
corr_coef += c_c
spearman[i] = rank_coef/experiments
correlation[i] = corr_coef/experiments
plt.scatter(noises, spearman, color='r')
plt.scatter(noises, correlation)
plt.legend(['Spearman Rank', 'Regular Correlation'])
plt.xlabel('Amount of Noise')
plt.ylabel('Average Correlation Coefficient')
```
We can see that the Spearman rank correlation copes with the non-linear relationship much better at most levels of noise. Interestingly, at very high levels, it seems to do worse than regular correlation.
##Delay in correlation
Of you might have the case that one process affects another, but after a time lag. Now let's see what happens if we add the delay.
```
n = 100
X = np.random.rand(n)
Xrank = stats.rankdata(X, method='average')
# n-2 is the second to last element
Yrank = stats.rankdata([1,1] + list(X[:(n-2)]), method='average')
diffs = Xrank - Yrank # order doesn't matter since we'll be squaring these values
r_s = 1 - 6*sum(diffs*diffs)/(n*(n**2 - 1))
print r_s
```
Sure enough, the relationship is not detected. It is important when using both regular and spearman correlation to check for lagged relationships by offsetting your data and testing for different offset values.
##Built-In Function
We can also use the `spearmanr` function in the `scipy.stats` library:
```
# Generate two random data sets
np.random.seed(161)
X = np.random.rand(10)
Y = np.random.rand(10)
r_s = stats.spearmanr(X, Y)
print 'Spearman Rank Coefficient: ', r_s[0]
print 'p-value: ', r_s[1]
```
We now have ourselves an $r_S$, but how do we interpret it? It's positive, so we know that the variables are not anticorrelated. It's not very large, so we know they aren't perfectly positively correlated, but it's hard to say from a glance just how significant the correlation is. Luckily, `spearmanr` also computes the p-value for this coefficient and sample size for us. We can see that the p-value here is above 0.05; therefore, we cannot claim that $X$ and $Y$ are correlated.
##Real World Example: Mutual Fund Expense Ratio
Now that we've seen how Spearman rank correlation works, we'll quickly go through the process again with some real data. For instance, we may wonder whether the expense ratio of a mutual fund is indicative of its three-year Sharpe ratio. That is, does spending more money on administration, management, etc. lower the risk or increase the returns? Quantopian does not currently support mutual funds, so we will pull the data from Yahoo Finance. Our p-value cutoff will be the usual default of 0.05.
### Data Source
Thanks to [Matthew Madurski](https://github.com/dursk) for the data. To obtain the same data:
1. Download the csv from this link. https://gist.github.com/dursk/82eee65b7d1056b469ab
2. Upload it to the 'data' folder in your research account.
```
mutual_fund_data = local_csv('mutual_fund_data.csv')
expense = mutual_fund_data['Annual Expense Ratio'].values
sharpe = mutual_fund_data['Three Year Sharpe Ratio'].values
plt.scatter(expense, sharpe)
plt.xlabel('Expense Ratio')
plt.ylabel('Sharpe Ratio')
r_S = stats.spearmanr(expense, sharpe)
print 'Spearman Rank Coefficient: ', r_S[0]
print 'p-value: ', r_S[1]
```
Our p-value is below the cutoff, which means we accept the hypothesis that the two are correlated. The negative coefficient indicates that there is a negative correlation, and that more expensive mutual funds have worse sharpe ratios. However, there is some weird clustering in the data, it seems there are expensive groups with low sharpe ratios, and a main group whose sharpe ratio is unrelated to the expense. Further analysis would be required to understand what's going on here.
## Real World Use Case: Evaluating a Ranking Model
### NOTE: [Factor Analysis](https://www.quantopian.com/lectures/factor-analysis) now covers this topic in much greater detail
Let's say that we have some way of ranking securities and that we'd like to test how well our ranking performs in practice. In this case our model just takes the mean daily return for the last month and ranks the stocks by that metric.
We hypothesize that this will be predictive of the mean returns over the next month. To test this we score the stocks based on a lookback window, then take the spearman rank correlation of the score and the mean returns over the walk forward month.
```
symbol_list = ['A', 'AA', 'AAC', 'AAL', 'AAMC', 'AAME', 'AAN', 'AAOI', 'AAON', 'AAP', 'AAPL', 'AAT', 'AAU', 'AAV', 'AAVL', 'AAWW', 'AB', 'ABAC', 'ABAX', 'ABB', 'ABBV', 'ABC', 'ABCB', 'ABCD', 'ABCO', 'ABCW', 'ABDC', 'ABEV', 'ABG', 'ABGB']
# Get the returns over the lookback window
start = '2014-12-01'
end = '2015-01-01'
historical_returns = get_pricing(symbol_list, fields='price', start_date=start, end_date=end).pct_change()[1:]
# Compute our stock score
scores = np.mean(historical_returns)
print 'Our Scores\n'
print scores
print '\n'
start = '2015-01-01'
end = '2015-02-01'
walk_forward_returns = get_pricing(symbol_list, fields='price', start_date=start, end_date=end).pct_change()[1:]
walk_forward_returns = np.mean(walk_forward_returns)
print 'The Walk Forward Returns\n'
print walk_forward_returns
print '\n'
plt.scatter(scores, walk_forward_returns)
plt.xlabel('Scores')
plt.ylabel('Walk Forward Returns')
r_s = stats.spearmanr(scores, walk_forward_returns)
print 'Correlation Coefficient: ' + str(r_s[0])
print 'p-value: ' + str(r_s[1])
```
The p-value indicates that our hypothesis is false and we accept the null hypothesis that our ranking was no better than random. This is a really good check of any ranking system one devises for constructing a long-short equity portfolio.
*This presentation is for informational purposes only and does not constitute an offer to sell, a solicitation to buy, or a recommendation for any security; nor does it constitute an offer to provide investment advisory or other services by Quantopian, Inc. ("Quantopian"). Nothing contained herein constitutes investment advice or offers any opinion with respect to the suitability of any security, and any views expressed herein should not be taken as advice to buy, sell, or hold any security or as an endorsement of any security or company. In preparing the information contained herein, Quantopian, Inc. has not taken into account the investment needs, objectives, and financial circumstances of any particular investor. Any views expressed and data illustrated herein were prepared based upon information, believed to be reliable, available to Quantopian, Inc. at the time of publication. Quantopian makes no guarantees as to their accuracy or completeness. All information is subject to change and may quickly become unreliable for various reasons, including changes in market conditions or economic circumstances.*
|
github_jupyter
|
# In this notebook an estimator for the Volume will be trained. No hyperparameters will be searched for, and the ones from the 'Close' values estimator will be used instead.
```
# Basic imports
import os
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import datetime as dt
import scipy.optimize as spo
import sys
from time import time
from sklearn.metrics import r2_score, median_absolute_error
%matplotlib inline
%pylab inline
pylab.rcParams['figure.figsize'] = (20.0, 10.0)
%load_ext autoreload
%autoreload 2
sys.path.append('../../')
from sklearn.externals import joblib
import utils.preprocessing as pp
import predictor.feature_extraction as fe
```
## Let's generate the datasets
```
def generate_one_set(params):
# print(('-'*70 + '\n {}, {} \n' + '-'*70).format(params['base_days'].values, params['ahead_days'].values))
tic = time()
train_val_time = int(params['train_val_time'])
base_days = int(params['base_days'])
step_days = int(params['step_days'])
ahead_days = int(params['ahead_days'])
print('Generating: base{}_ahead{}'.format(base_days, ahead_days))
pid = 'base{}_ahead{}'.format(base_days, ahead_days)
# Getting the data
data_df = pd.read_pickle('../../data/data_train_val_df.pkl')
today = data_df.index[-1] # Real date
print(pid + ') data_df loaded')
# Drop symbols with many missing points
data_df = pp.drop_irrelevant_symbols(data_df, params['GOOD_DATA_RATIO'])
print(pid + ') Irrelevant symbols dropped.')
# Generate the intervals for the predictor
x, y = fe.generate_train_intervals(data_df,
train_val_time,
base_days,
step_days,
ahead_days,
today,
fe.feature_volume_one_to_one,
target_feature=fe.VOLUME_FEATURE)
print(pid + ') Intervals generated')
# Drop "bad" samples and fill missing data
x_y_df = pd.concat([x, y], axis=1)
x_y_df = pp.drop_irrelevant_samples(x_y_df, params['SAMPLES_GOOD_DATA_RATIO'])
x = x_y_df.iloc[:, :-1]
y = x_y_df.iloc[:, -1]
x = pp.fill_missing(x)
print(pid + ') Irrelevant samples dropped and missing data filled.')
# Pickle that
x.to_pickle('../../data/x_volume_{}.pkl'.format(pid))
y.to_pickle('../../data/y_volume_{}.pkl'.format(pid))
toc = time()
print('%s) %i intervals generated in: %i seconds.' % (pid, x.shape[0], (toc-tic)))
return pid, x, y
best_params_df = pd.read_pickle('../../data/best_params_final_df.pkl').loc[1,:]
to_drop = [
'model',
'mre',
'r2',
'x_filename',
'y_filename',
'train_days'
]
best_params_df.drop(to_drop, inplace=True)
best_params_df
generate_one_set(best_params_df)
x_volume = pd.read_pickle('../../data/x_volume_base112_ahead1.pkl')
print(x_volume.shape)
x_volume.head()
y_volume = pd.read_pickle('../../data/y_volume_base112_ahead1.pkl')
print(y_volume.shape)
y_volume.head()
```
## Let's generate the test dataset, also
```
def generate_one_test_set(params, data_df):
# print(('-'*70 + '\n {}, {} \n' + '-'*70).format(params['base_days'].values, params['ahead_days'].values))
tic = time()
train_val_time = int(params['train_val_time'])
base_days = int(params['base_days'])
step_days = int(params['step_days'])
ahead_days = int(params['ahead_days'])
print('Generating: base{}_ahead{}'.format(base_days, ahead_days))
pid = 'base{}_ahead{}'.format(base_days, ahead_days)
# Getting the data
today = data_df.index[-1] # Real date
print(pid + ') data_df loaded')
# Drop symbols with many missing points
y_train_df = pd.read_pickle('../../data/y_volume_{}.pkl'.format(pid))
kept_symbols = y_train_df.index.get_level_values(1).unique().tolist()
data_df = data_df.loc[:, (slice(None), kept_symbols)]
print(pid + ') Irrelevant symbols dropped.')
# Generate the intervals for the predictor
x, y = fe.generate_train_intervals(data_df,
train_val_time,
base_days,
step_days,
ahead_days,
today,
fe.feature_volume_one_to_one,
target_feature=fe.VOLUME_FEATURE)
print(pid + ') Intervals generated')
# Drop "bad" samples and fill missing data
x_y_df = pd.concat([x, y], axis=1)
x_y_df = pp.drop_irrelevant_samples(x_y_df, params['SAMPLES_GOOD_DATA_RATIO'])
x = x_y_df.iloc[:, :-1]
y = x_y_df.iloc[:, -1]
x = pp.fill_missing(x)
print(pid + ') Irrelevant samples dropped and missing data filled.')
# Pickle that
x.to_pickle('../../data/x_volume_{}_test.pkl'.format(pid))
y.to_pickle('../../data/y_volume_{}_test.pkl'.format(pid))
toc = time()
print('%s) %i intervals generated in: %i seconds.' % (pid, x.shape[0], (toc-tic)))
return pid, x,
data_test_df = pd.read_pickle('../../data/data_test_df.pkl')
generate_one_test_set(best_params_df, data_test_df)
x_volume_test = pd.read_pickle('../../data/x_volume_base112_ahead1_test.pkl')
print(x_volume_test.shape)
x_volume_test.head()
y_volume_test = pd.read_pickle('../../data/y_volume_base112_ahead1_test.pkl')
print(y_volume_test.shape)
y_volume_test.head()
```
## Let's train a predictor for the 'Volume' with the same hyperparameters as for the 'Close' one.
```
best_params_df = pd.read_pickle('../../data/best_params_final_df.pkl')
import predictor.feature_extraction as fe
from predictor.linear_predictor import LinearPredictor
import utils.misc as misc
import predictor.evaluation as ev
ahead_days = 1
# Get some parameters
train_days = int(best_params_df.loc[ahead_days, 'train_days'])
GOOD_DATA_RATIO, \
train_val_time, \
base_days, \
step_days, \
ahead_days, \
SAMPLES_GOOD_DATA_RATIO, \
x_filename, \
y_filename = misc.unpack_params(best_params_df.loc[ahead_days,:])
pid = 'base{}_ahead{}'.format(base_days, ahead_days)
# Get the datasets
x_train = pd.read_pickle('../../data/x_volume_{}.pkl'.format(pid))
y_train = pd.read_pickle('../../data/y_volume_{}.pkl'.format(pid))
x_test = pd.read_pickle('../../data/x_volume_{}_test.pkl'.format(pid)).sort_index()
y_test = pd.DataFrame(pd.read_pickle('../../data/y_volume_{}_test.pkl'.format(pid))).sort_index()
# Let's cut the training set to use only the required number of samples
end_date = x_train.index.levels[0][-1]
start_date = fe.add_market_days(end_date, -train_days)
x_sub_df = x_train.loc[(slice(start_date,None),slice(None)),:]
y_sub_df = pd.DataFrame(y_train.loc[(slice(start_date,None),slice(None))])
# Create the estimator and train
estimator = LinearPredictor()
estimator.fit(x_sub_df, y_sub_df)
# Get the training and test predictions
y_train_pred = estimator.predict(x_sub_df)
y_test_pred = estimator.predict(x_test)
# Get the training and test metrics for each symbol
metrics_train = ev.get_metrics_df(y_sub_df, y_train_pred)
metrics_test = ev.get_metrics_df(y_test, y_test_pred)
# Show the mean metrics
metrics_df = pd.DataFrame(columns=['train', 'test'])
metrics_df['train'] = metrics_train.mean()
metrics_df['test'] = metrics_test.mean()
print('Mean metrics: \n{}\n{}'.format(metrics_df,'-'*70))
# Plot the metrics in time
metrics_train_time = ev.get_metrics_in_time(y_sub_df, y_train_pred, base_days + ahead_days)
metrics_test_time = ev.get_metrics_in_time(y_test, y_test_pred, base_days + ahead_days)
plt.plot(metrics_train_time[2], metrics_train_time[0], label='train', marker='.')
plt.plot(metrics_test_time[2], metrics_test_time[0], label='test', marker='.')
plt.title('$r^2$ metrics')
plt.legend()
plt.figure()
plt.plot(metrics_train_time[2], metrics_train_time[1], label='train', marker='.')
plt.plot(metrics_test_time[2], metrics_test_time[1], label='test', marker='.')
plt.title('MRE metrics')
plt.legend()
joblib.dump(estimator, '../../data/best_volume_predictor.pkl')
```
|
github_jupyter
|
## Dependencies
```
import json, glob
from tweet_utility_scripts import *
from tweet_utility_preprocess_roberta_scripts_aux import *
from transformers import TFRobertaModel, RobertaConfig
from tokenizers import ByteLevelBPETokenizer
from tensorflow.keras import layers
from tensorflow.keras.models import Model
```
# Load data
```
test = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/test.csv')
print('Test samples: %s' % len(test))
display(test.head())
```
# Model parameters
```
input_base_path = '/kaggle/input/166-robertabase-last/'
with open(input_base_path + 'config.json') as json_file:
config = json.load(json_file)
config
base_path = '/kaggle/input/qa-transformers/roberta/'
vocab_path = base_path + 'roberta-base-vocab.json'
merges_path = base_path + 'roberta-base-merges.txt'
config['base_model_path'] = base_path + 'roberta-base-tf_model.h5'
config['config_path'] = base_path + 'roberta-base-config.json'
model_path_list = glob.glob(input_base_path + '*.h5')
model_path_list.sort()
print('Models to predict:')
print(*model_path_list, sep = "\n")
```
# Tokenizer
```
tokenizer = ByteLevelBPETokenizer(vocab_file=vocab_path, merges_file=merges_path,
lowercase=True, add_prefix_space=True)
```
# Pre process
```
test['text'].fillna('', inplace=True)
test["text"] = test["text"].apply(lambda x: x.lower())
test["text"] = test["text"].apply(lambda x: x.strip())
x_test, x_test_aux, x_test_aux_2 = get_data_test(test, tokenizer, config['MAX_LEN'], preprocess_fn=preprocess_roberta_test)
```
# Model
```
module_config = RobertaConfig.from_pretrained(config['config_path'], output_hidden_states=False)
def model_fn(MAX_LEN):
input_ids = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='input_ids')
attention_mask = layers.Input(shape=(MAX_LEN,), dtype=tf.int32, name='attention_mask')
base_model = TFRobertaModel.from_pretrained(config['base_model_path'], config=module_config, name="base_model")
last_hidden_state, _ = base_model({'input_ids': input_ids, 'attention_mask': attention_mask})
x_start = layers.Dropout(.1)(last_hidden_state)
x_start = layers.Dense(1)(x_start)
x_start = layers.Flatten()(x_start)
y_start = layers.Activation('softmax', name='y_start')(x_start)
x_end = layers.Dropout(.1)(last_hidden_state)
x_end = layers.Dense(1)(x_end)
x_end = layers.Flatten()(x_end)
y_end = layers.Activation('softmax', name='y_end')(x_end)
model = Model(inputs=[input_ids, attention_mask], outputs=[y_start, y_end])
return model
```
# Make predictions
```
NUM_TEST_IMAGES = len(test)
test_start_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
test_end_preds = np.zeros((NUM_TEST_IMAGES, config['MAX_LEN']))
for model_path in model_path_list:
print(model_path)
model = model_fn(config['MAX_LEN'])
model.load_weights(model_path)
test_preds = model.predict(get_test_dataset(x_test, config['BATCH_SIZE']))
test_start_preds += test_preds[0]
test_end_preds += test_preds[1]
```
# Post process
```
test['start'] = test_start_preds.argmax(axis=-1)
test['end'] = test_end_preds.argmax(axis=-1)
test['text_len'] = test['text'].apply(lambda x : len(x))
test['text_wordCnt'] = test['text'].apply(lambda x : len(x.split(' ')))
test["end"].clip(0, test["text_len"], inplace=True)
test["start"].clip(0, test["end"], inplace=True)
test['selected_text'] = test.apply(lambda x: decode(x['start'], x['end'], x['text'], config['question_size'], tokenizer), axis=1)
test["selected_text"].fillna(test["text"], inplace=True)
```
# Visualize predictions
```
display(test.head(10))
```
# Test set predictions
```
submission = pd.read_csv('/kaggle/input/tweet-sentiment-extraction/sample_submission.csv')
submission['selected_text'] = test["selected_text"]
submission.to_csv('submission.csv', index=False)
submission.head(10)
```
|
github_jupyter
|
[View in Colaboratory](https://colab.research.google.com/github/thonic92/chal_TM/blob/master/model_tweets.ipynb)
```
import json
import numpy as np
import pandas as pd
import tensorflow as tf
from keras.models import Sequential
from keras.layers import Dense
from keras.layers import Dropout
from keras.layers import LSTM
from keras.callbacks import ModelCheckpoint
from keras.utils import np_utils
import sys
import re
import unicodedata
from collections import Counter
import nltk
with open("/content/gdrive/My Drive/json_datas_full.json", "r", encoding="latin1",errors='ignore' ) as read_file:
data = json.load(read_file)
tweets = []
for i in range(len(data)):
tweets.append(data[i]['text'].lower())
print(tweets[0:2])
tweets_str = ' '.join(tweets)
tweets_str=unicodedata.normalize('NFD',tweets_str).encode('ascii', 'ignore').decode("utf-8")
print(tweets_str[0:1000])
tweets_words = tweets_str.split(' ')
#print(tweets_words[1:100])
type(tweets_words)
#print(sorted(set(tweets_words)))
print(Counter(tweets_words).most_common()[0:100])
print(Counter(list(nltk.bigrams(tweets_words))).most_common()[0:100])
# on retire les urls
tweets_str = re.sub("(@[A-Za-z0-9]+)|([^0-9A-Za-z \t])|(\w+:\/\/\S+)"," ", tweets_str)
tweets_str = re.sub("paris2024"," ", tweets_str)
tweets_str = re.sub("jo2024"," ", tweets_str)
tweets_str = re.sub("jo 2024"," ", tweets_str)
tweets_str = re.sub("jo de 2024"," ", tweets_str)
tweets_str = re.sub("paris"," ", tweets_str)
tweets_str = re.sub("sport"," ", tweets_str)
tweets_str = re.sub("olympicday"," ", tweets_str)
tweets_str = re.sub("enmodejo"," ", tweets_str)
tweets_str = re.sub("grandparis"," ", tweets_str)
tweets_str = re.sub("cio"," ", tweets_str)
tweets_str = re.sub("jouerlejeu"," ", tweets_str)
tweets_str = re.sub("jeuxolympiques"," ", tweets_str)
tweets_str = re.sub("venezpartager"," ", tweets_str)
tweets_str = re.sub("jo2024paris"," ", tweets_str)
tweets_str = re.sub("jerevedesjeux"," ", tweets_str)
tweets_str = re.sub("france"," ", tweets_str)
tweets_str = re.sub("madeforsharing"," ", tweets_str)
tweets_str = re.sub("rio2016"," ", tweets_str)
tweets_str = re.sub("generation2024"," ", tweets_str)
tweets_str = re.sub("gagnonsensemble"," ", tweets_str)
tweets_str = re.sub("engager"," ", tweets_str)
tweets_str = re.sub("pleinement"," ", tweets_str)
tweets_str = re.sub("candidature"," ", tweets_str)
tweets_str = re.sub("nouvelle etape"," ", tweets_str)
tweets_str = re.sub("hidalgo veut"," ", tweets_str)
tweets_str = re.sub("favorable"," ", tweets_str)
tweets_str = re.sub("s engage"," ", tweets_str)
tweets_str = ' '.join(tweets_str.split())
tweets_str = tweets_str[0:300000]
print(tweets_str[0:1000])
len(tweets_str)
chars = sorted(list(set(tweets_str)))
char_to_int = dict((c, i) for i, c in enumerate(chars))
print(chars)
len(char_to_int)
n_chars = len(tweets_str.split(sep=' '))
n_vocab = len(chars)
print(n_chars)
print(n_vocab)
seq_length = 100
dataX = []
dataY = []
for i in range(0, n_chars - seq_length, 1):
seq_in = tweets_str[i:i + seq_length]
seq_out = tweets_str[i + seq_length]
dataX.append([char_to_int[char] for char in seq_in])
dataY.append(char_to_int[seq_out])
n_patterns = len(dataX)
print(n_patterns)
# reshape X to be [samples, time steps, features]
X = np.reshape(dataX, (n_patterns, seq_length, 1))
# normalize
X = X / float(n_vocab)
# one hot encode the output variable
y = np_utils.to_categorical(dataY)
model = Sequential()
model.add(LSTM(256, input_shape=(X.shape[1], X.shape[2]), return_sequences=True))
model.add(Dropout(0.2))
model.add(LSTM(256))
model.add(Dropout(0.2))
model.add(Dense(y.shape[1], activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam')
filepath="/content/gdrive/My Drive/weights-improvement-{epoch:02d}-{loss:.4f}-bigger3.hdf5"
checkpoint = ModelCheckpoint(filepath, monitor='loss', verbose=1, save_best_only=True, mode='min')
callbacks_list = [checkpoint]
model.fit(X, y, epochs=30, batch_size=128, callbacks=callbacks_list)
```
|
github_jupyter
|
<div align="center">
<h1>Homework 7</h1>
<p>
<div align="center">
<h2>Yutong Dai [email protected]</h2>
</div>
</p>
</div>
## 6.33
The dual problem is
$$
\begin{align}
& \min \quad 3 w_1 + 6 w_2\\
& s.t \quad w_1 + 2w_2 \geq 2\\
& \qquad w_1 + 3w_2 \geq -3\\
& \qquad w_1\leq 0,w_2\geq 0
\end{align}
$$
It's easy to verify $(w_1^*,w_2^*)=(\frac{11}{-2}, \frac{5}{2})$ is a feasible solution to the dual and satisfy the KKT condition. Therefore, $(x_1^*,x_2^*)=(3/2, 3/2)$ is the optimal solution to the dual.
---
* The first method is "Big-M" method. First convert the problem to the standard form and adding the artificial variables, where they serve as the initial basis.
* The second method is the artificial constraints technique, where we adding a upper bound on the summation of all non-basic variables.
I will use the second method.
The tableau for the primal is as follow, where the dual is not feasible.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | RHS |
| --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 2 | -3 | 0 | 0 | 0 |
| $x_3$ | 0 | -1 | -1 | 1 | 0 | -3 |
| $x_4$ | 0 | 3 | 1 | 0 | 1 | 6 |
Adding constrain $x_1 + x_2 \leq M$, we have the following tableau.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 2 | -3 | 0 | 0 | 0 | 0 |
| $x_5$ | 0 | 1 | 1 | 0 | 0 | 1 | M |
| $x_3$ | 0 | -1 | -1 | 1 | 0 | 0 | -3 |
| $x_4$ | 0 | 3 | 1 | 0 | 1 | 0 | 6 |
* The first iteration:
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -5 | 0 | 0 | -2 | -2M |
| $x_1$ | 0 | 1 | 1 | 0 | 0 | 1 | M |
| $x_3$ | 0 | 0 | 0 | 1 | 0 | 1 | -3 + M |
| $x_4$ | 0 | 0 | -2 | 0 | 1 | -3 | 6 -3M |
* The second iteration:
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -11/3 | 0 | -2/3 | 0 | -4 |
| $x_1$ | 0 | 1 | 1/3 | 0 | 1/3 | 0 | 2 |
| $x_3$ | 0 | 0 | -2/3 | 1 | 1/3 | 0 | -1 |
| $x_5$ | 0 | 0 | 2/3 | 0 | -1/3 | 1 | M-2 |
* The third iteration:
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | -11/2 | -5/2 | 0 | 3/2 |
| $x_1$ | 0 | 1 | 0 | 1/2 | 1/2 | 0 | 3/2 |
| $x_2$ | 0 | 0 | 1 | -3/2 | -1/2 | 0 | 3/2 |
| $x_4$ | 0 | 0 | 0 | 1 | 0 | 1 | M-2 |
So the optimal solution for the primal is $(3/2, 3/2)$.
## 6.54
**a)**
The dual problem is
$$
\begin{align}
& \min \quad 8w_1 + 4w_2\\
& s.t \quad w_1 - w_2 \geq 2\\
& \qquad 2w_1 - w_2 \geq 1\\
& \qquad 3w_1 - 2w_2 \geq -1\\
& \qquad w_1\leq 0,w_2\geq 0
\end{align}
$$
Since the constraints in the primal are of $\leq$ type, we know that the optimal solution for the dual is $(2,0)$ .
**b)**
Note $x_2$ is a non-basic feasible solution and $c_2' - z_2=1>0$, therefore $x_2$ will enter the basis and change the optimal solution.
The tableau becomes
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 1 | -3 | -2 | 0 | -16 |
| $x_1$ | 0 | 1 | 2 | 1 | 1 | 0 | 8 |
| $x_5$ | 0 | 0 | 3 | -1 | 1 | 1 | 12 |
After one iteration, we reach the optimal tableau.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | -3/2 | -3/2 | 0 | -20 |
| $x_2$ | 0 | 1/2 | 0 | 1/2 | 1/2 | 0 | 4 |
| $x_5$ | 0 | -1/2 | 0 | -3/2 | 1/2 | 1 | 0 |
The new optimal solution becomes $(x_1, x_2, x_3)=(0,4,0)$
**c)**
Note $x_2$ is a non-basic feasible solution and $c_2 - c_B^TB^{-1}A_j'=1-1/3=2/3>0$, therefore $x_2$ will enter the basis and change the optimal solution.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 2/3 | -3 | -2 | 0 | -16 |
| $x_1$ | 0 | 1 | 1/6 | 1 | 1 | 0 | 8 |
| $x_5$ | 0 | 0 | 7/6 | -1 | 1 | 1 | 12 |
After one iteration, we reach the optimal tableau.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | -17/7 | -18/7 | -4/7 | -28 |
| $x_1$ | 0 | 1 | 0 | 8/7 | 6/7 | -1/7 | 44/7 |
| $x_2$ | 0 | 0 | 1 | -6/7 | 1/7 | 6/7 | 72/7 |
The new optimal solution becomes $(x_1, x_2, x_3)=(44/7,72/7,0)$
**d)**
Set up the tableau as
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -3 | -3 | -2 | 0 | 0 | -16 |
| $M$ | -1 | 0 | 0 | 0 | 0 | 0 | -1 | 0 |
| $x_1$ | 0 | 1 | 2 | 1 | 1 | 0 | 0 | 8 |
| $x_5$ | 0 | 0 | 3 | -1 | 1 | 1 | 0 | 12 |
| $x_6$ | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 3 |
and make $x_6$ as true basic variable by adding the last row to the zero row. We obtain
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -3 | -3 | -2 | 0 | 0 | -16 |
| $M$ | -1 | 1 | 2 | 0 | 0 | 0 | 0 | 3 |
| $x_1$ | 0 | 1 | 2 | 1 | 1 | 0 | 0 | 8 |
| $x_5$ | 0 | 0 | 3 | -1 | 1 | 1 | 0 | 12 |
| $x_6$ | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 3 |
After one iteration,
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -3/2 | 0 | -2 | 0 | 3/2 | -23/2 |
| $M$ | -1 | 0 | 0 | 0 | 0 | 0 | -1 | 0 |
| $x_1$ | 0 | 1 | 3/2 | 0 | 1 | 0 | -1/2 | 13/2 |
| $x_5$ | 0 | 0 | 7/2 | 0 | 1 | 1 | 1/2 | 27/2 |
| $x_6$ | 0 | 0 | 1/2 | 1 | 0 | 0 | 1/2 | 3/2 |
we reach the optimal. The new optimal solution becomes $(x_1, x_2, x_3)=(13/2, 0, 3/2)$
**e)**
Suppose the new right-hand-side is $b'$. Then $B^{-1}b'=(b_1' , b_1'+ b_2')^T$. As we will increase 8 or 4 to $b_1'$ or $b_2'$. Either way will ensure $B^{-1}b'\geq 0$, therefore, the same basis is still optimal.
- If we change $b_1$ then, we will change the optimal solution from $(b_1,0,0)$ to $(b_1',0,0)$. It will increase the objective value by $2(b_1' -b_1)$
- If we change $b_2$ then, we won't change the optimal solution $(b_1,0,0)$, hence the objective value.
**f)**
As $c_6 -x_6=6-wA_6=2>0$, $x_6$ will enter the basis.
The initial tableau is
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -3 | -3 | -2 | 0 | 2 | -16 |
| $x_1$ | 0 | 1 | 2 | 1 | 1 | 0 | 2 | 8 |
| $x_5$ | 0 | 0 | 3 | -1 | 1 | 1 | 3 | 12 |
After one iteration, the tableau becomes
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | -1 | -5 | -4 | -3 | 0 | 0 | -24 |
| $x_6$ | 0 | 1/2 | 1 | 1/2 | 1/2 | 0 | 1 | 4 |
| $x_5$ | 0 | -3/2 | 0 | -5/2 | -1/2 | 1 | 0 | 0 |
The optimal solution is $(x_1, x_2, x_3,x_6)=(0,0,0,4)$
## 6.68
Before we proceed, we need to calculate a few quantity:
* $(c_6,c_7,c_8)-(c_1,c_2,c_3)B^{-1}A_{[:,(6,7,8)]} = (\bar c_6,\bar c_7, \bar c_8)=(-2,-1/10,-2) \Rightarrow (c_1,c_2,c_3)=(2,4,1)$, where $A_{[:,(6,7,8)]}$ is $I_3$.
* $(c_4,c_5) - (c_1,c_2,c_3)B^{-1}A_{[:,(3,4)]}=(\bar c_4,\bar c_5)=(-2,0)\Rightarrow (c_4,c_5)=(3,2)$
* $b=B\bar b=(14/9, 110/3, 46/9)^T$
We perturbe the $b$ along the direction $d=(-1,0,0)^T$.
**Iteration 1:**
* Calculate $B^{-1}d = (-0.5, 1 , -5)^T$, So $S=\{1,3\}$.
* Calculate the minimal ration $\theta=7/5$.
* If $\theta\in [0,7/5]$, the current basis $(A_1,A_2,A_3)$ is always optimal. Further, the objective value and right hand side will be
$$
z(\theta) = 17 - 2\theta \qquad \bar b = (3-\frac{1}{2}\theta, 1 + \theta, 7-5\theta)^T.
$$
* When $\theta =7/5$, then $x_3=0$, therefore we perform dual simplex method on the tableau below.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | $x_7$ | $x_8$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | 0 | -2 | 0 | -2 | -1/10 | -2 | -71/5 |
| $x_1$ | 0 | 1 | 0 | 0 | -1 | 0 | 1/2 | 1/5 | -1 | 23/10 |
| $x_2$ | 0 | 0 | 1 | 0 | 2 | 1 | -1 | 0 | 1/2 | 12/5 |
| $x_3$ | 0 | 0 | 0 | 1 | -1 | -2 | 5 | -3/10 | 2 | 0 |
So $x_3$ will leave and $x_5$ will enter.
The tableau becomes
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | $x_7$ | $x_8$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | 0 | -2 | 0 | -2 | -1/10 | -2 | -71/5 |
| $x_1$ | 0 | 1 | 0 | 0 | -1 | 0 | 1/2 | 1/5 | -1 | 23/10 |
| $x_2$ | 0 | 0 | 1 | 1/2 | 3/2 | 0 | 3/2 | -3/20 | 3/2 | 12/5 |
| $x_5$ | 0 | 0 | 0 | -1/2 | 1/2 | 1 | -5/2 | 3/20 | -1 | 0 |
**Iteration 2:**
* Calculate $B^{-1}d = (-0.5, -1.5 , 2.5)^T$, $B^{-1}b=(3,4.5, -3.5)$So $S=\{1,2\}$.
* Calculate the minimal ration $\theta=3$.
* If $\theta\in [7/5, 3]$, the current basis $(A_1,A_2,A_5)$ is always optimal. Further, the objective value and right hand side will be
$$
z(\theta) = 17 - 2\theta \qquad \bar b = (3-\frac{1}{2}\theta, \frac{9}{2} - \frac{3}{2} \theta, \frac{-7}{2}+\frac{5}{2}\theta)^T.
$$
* When $\theta =3$, then $x_2=0$, therefore we perform dual simplex method on the tableau below.
The tableau becomes
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | $x_7$ | $x_8$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | 0 | 0 | -2 | 0 | -2 | -1/10 | -2 | -11 |
| $x_1$ | 0 | 1 | 0 | 0 | -1 | 0 | 1/2 | 1/5 | -1 | 3/2 |
| $x_2$ | 0 | 0 | 1 | 1/2 | 3/2 | 0 | 3/2 | -3/20 | 3/2 | 0 |
| $x_5$ | 0 | 0 | 0 | -1/2 | 1/2 | 1 | -5/2 | 3/20 | -1 | 4 |
So $x_2$ will leave and $x_7$ will enter.
The tableau becomes
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | $x_7$ | $x_8$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -2/3 | -1/3 | -3 | 0 | -3 | 0 | -3 | -11 |
| $x_1$ | 0 | 0 | 1 | 4/3 | 2/3 | 0 | 5/2 | 0 | 1 | 3/2 |
| $x_7$ | 0 | 0 | -20/3 | -10/3 | -10 | 0 | -10 | 1 | -10 | 0 |
| $x_5$ | 0 | 0 | 1 | 0 | 2 | 1 | -1 | 0 | 1/2 | 4 |
**Iteration 3:**
* Calculate $B^{-1}d = (-2.5, 10 , 1)^T$, $B^{-1}b=(9,-30,1)$So $S=\{1\}$.
* Calculate the minimal ration $\theta=18/5$.
* If $\theta\in [3,18/5]$, the current basis $(A_1,A_7,A_5)$ is always optimal. Further, the objective value and right hand side will be
$$
z(\theta) = 20 - 3\theta \qquad \bar b = (9-\frac{5}{2}\theta, -30 + 10 \theta, 1+\theta)^T.
$$
* When $\theta =18/5$, then $x_1=0$, therefore we perform dual simplex method on the tableau below.
| | $z$ | $x_1$ | $x_2$ | $x_3$ | $x_4$ | $x_5$ | $x_6$ | $x_7$ | $x_8$ | RHS |
| --- | --- | --- | --- | --- | --- | --- | --- | --- | --- | --- |
| $z$ | -1 | 0 | -2/3 | -1/3 | -3 | 0 | -3 | 0 | -3 | -46/5 |
| $x_1$ | 0 | 0 | 1 | 4/3 | 2/3 | 0 | 5/2 | 0 | 1 | 0 |
| $x_7$ | 0 | 0 | -20/3 | -10/3 | -10 | 0 | -10 | 1 | -10 | 6 |
| $x_5$ | 0 | 0 | 1 | 0 | 2 | 1 | -1 | 0 | 1/2 | 23/5 |
We can not pivot anymore. Hence the algorithm terminates, which means the problem is infeasible for $\theta > 18/5$.
## 6.72
**a)**
$$
\begin{align}
& \max \quad 6w + \min_{(x_1,x_2) \in X} \{(1-3w)x_1 + (2-w)x_2\}\\
& s.t \quad w\geq 0
\end{align}
$$
**b)**
The minimal of $\min_{(x_1,x_2) \in X} \{(1-3w)x_1 + (2-w)x_2\}$ is obtained on one of the following extreme points
$$(0,0), (8,0), (3,5), (0,2).$$
Plug these four points into $f(w)$, we end up with
$$f(w)=6w + \min\{0, 4-2w, 13-14w, 8-24w\}.$$
**c)**
$$
f(w)=
\begin{cases}
6w, & 0 \leq w \leq 1/3 \\
8-18w, & w \geq 1/3
\end{cases}
$$
```
import numpy as np
import matplotlib.pyplot as plt
plt.plot(1/3,2,'ro',markersize=10)
x1 = np.linspace(0,1/3,10)
x2 = np.linspace(1/3,1,10)
plt.plot(x1,6*x1,'k-',label=r"$z=6w$")
plt.plot(x2,8 - 18*x2,'k-', label=r"$z=8-18w$")
plt.legend()
plt.show()
```
**d)**
The optimal solution for the Lagrangian dual problem is $w=1/3$.
**e)**
Since $w=1/3$, $f(w)=2 + \min_{(x_1,x_2) \in X}5/3 x_2=2$, we know $x_2=0$ and therefore $x_1=2$.
So the optimal solution for the primal $(x_1, x_2)=(2,0)$.
## Exercise 5.14
**a)**
$$(c - 10d)' x = (c + 10d)' x \Rightarrow d'x = 0.$$
The same holds for $Ax = b + \theta f$. Therefore, $5 d'x = 0$. As the optimality and feasibility conditions hold, the same basis remains optimal.
**b)**
For fixed $\theta$, let $B$ be an arbitrary basis. Then we have $x=(X_B,X_N)=(B^{-1}(b+\theta f),0)$. Suppose $\{B^j\}$ are all possible basis derived from A. Then our problem becomes
$$f(\theta) = \underset{j}{\text{min}} \{(c+ \theta d)' {B^j}^{-1}(b + \theta f)\}, $$
where ${B^j}^{-1}(b + \theta f) \geq 0$.
Clearly, $f(\theta)$ is a piecewise quadratic function of $\theta$ if $f\neq 0$
Let $K$ be the number of possible bases, then the upper bound on the number of pieces is $2K$.
**c)**
\begin{aligned}
& \text{minimize} && \theta d'x \\
& \text{subject to} && Ax = \theta f \\
& && x \geq 0
\end{aligned}
Let $B$ be an optimal basis for $\theta = 1$ and assume that $\theta > 0$. $d' - d'_B B^{-1} A \geq 0 \text{ and } B^{-1}f \geq 0$. Hence for nonnegative $\theta$ satisfying $\theta d' - d'_B B^{-1} A \geq 0 \text{ and } \theta B^{-1}f \geq 0$ keeps this same basis optimal.
**d)**
Consider $b, f = 0$, $f(\theta)$ is constant in $\theta$, hence both convex and concave.
|
github_jupyter
|
# Assignment: Global average budgets in the CESM pre-industrial control simulation
## Learning goals
Students completing this assignment will gain the following skills and concepts:
- Continued practice working with the Jupyter notebook
- Familiarity with atmospheric output from the CESM simulation
- More complete comparison of the global energy budget in the CESM control simulation to the observations
- Validation of the annual cycle of surface temperature against observations
- Opportunity to formulate a hypothesis about these global temperature variations
- Python programming skills: basic xarray usage: opening gridded dataset and taking averages
## Instructions
- In a local copy of this notebook (on the JupyterHub or your own device) **add your answers in additional cells**.
- **Complete the required problems** below.
- Remember to set your cell types to `Markdown` for text, and `Code` for Python code!
- **Include comments** in your code to explain your method as necessary.
- Remember to actually answer the questions. **Written answers are required** (not just code and figures!)
- Submit your solutions in **a single Jupyter notebook** that contains your text, your code, and your figures.
- *Make sure that your notebook* ***runs cleanly without errors:***
- Save your notebook
- From the `Kernel` menu, select `Restart & Run All`
- Did the notebook run from start to finish without error and produce the expected output?
- If yes, save again and submit your notebook file
- If no, fix the errors and try again.
## Problem 1: The global energy budget in the CESM control simulation
Compute the **global, time average** of each of the following quantities, and **compare them to the observed values** from the Trenberth and Fasullo (2012) figure in the course notes. Recall that when you want to repeat an operation, you should write a function for it!
- Solar Radiation budget:
- Incoming Solar Radiation, or Insolation
- Reflected Solar Radiation at the top of atmosphere
- Solar Radiation Reflected by Surface
- Solar Radiation Absorbed by Surface
- Solar Radiation Refelected by Clouds and Atmosphere *(you can calculate this as the difference between the reflected radiation at the top of atmosphere and reflected radiation at the surface)*
- Total Absorbed Solar Radiation (ASR) at the top of atmosphere
- Solar Radiation Absorbed by Atmosphere *(you can calculate this as the residual of your budget, i.e. what's left over after accounting for all other absorption and reflection)*
- Longwave Radiation budget:
- Outgoing Longwave Radiation
- Upward emission from the surface
- Downwelling radiation at the surface
- Other surface fluxes:
- "Thermals", or *sensible heat flux*. *You will find this in the field called `SHFLX` in your dataset.*
- "Evapotranspiration", or *latent heat flux*. *You will find this in the field called `LHFLX` in your dataset.*
*Note we will look more carefully at atmospheric absorption and emission processes later. You do not need to try to calculate terms such as "Emitted by Atmosphere" or "Atmospheric Window"*
**Based on your results above, answer the following questions:**
- Is the CESM control simulation at (or near) **energy balance**?
- Do you think this simulation is near equilibrium?
- Summarize in your own words what you think are the most important similarities and differences of the global energy budgets in the CESM simulation and the observations.
## Problem 2: Verifying the annual cycle in global mean surface temperature against observations
In the class notes we plotted the **timeseries of global mean surface temperature** in the CESM control simulation, and found an **annual cycle**. The purpose of this exercise is to verify that this phenomenon is also found in the observed temperature record. If so, then we can conclude that it is a real feature of Earth's climate and not an artifact of the numerical model.
For observations, we will use the **NCEP Reanalysis data**.
*Reanalysis data is really a blend of observations and output from numerical weather prediction models. It represents our “best guess” at conditions over the whole globe, including regions where observations are very sparse.*
The necessary data are all served up over the internet. We will look at monthly climatologies averaged over the 30 year period 1981 - 2010.
You can browse the available data here:
https://psl.noaa.gov/thredds/catalog/Datasets/ncep.reanalysis.derived/catalog.html
**Surface air temperature** is contained in a file called `air.2m.mon.ltm.nc`, which is found in the collection called `surface_gauss`.
Here's a link directly to the catalog page for this data file:
https://psl.noaa.gov/thredds/catalog/Datasets/ncep.reanalysis.derived/surface_gauss/catalog.html?dataset=Datasets/ncep.reanalysis.derived/surface_gauss/air.2m.mon.ltm.nc
Now click on the `OPeNDAP` link. A page opens up with lots of information about the contents of the file. The `Data URL` is what we need to read the data into our Python session. For example, this code opens the file and displays a list of the variables it contains:
```
import xarray as xr
url = 'https://psl.noaa.gov/thredds/dodsC/Datasets/ncep.reanalysis.derived/surface_gauss/air.2m.mon.ltm.nc'
ncep_air2m = xr.open_dataset(url, decode_times=False)
print(ncep_air2m)
```
The temperature data is called `air`. Take a look at the details:
```
print(ncep_air2m.air)
```
Notice that the dimensions are `(time: 12, lat: 94, lon: 192)`. The time dimension is calendar months. But note that the lat/lon grid is not the same as our model output!
*Think about how you will handle calculating the global average of these data.*
### Your task:
- Make a well-labeled timeseries graph of the global-averaged observed average surface air temperature climatology.
- Verify that the annual cycle we found in the CESM simulation also exists in the observations.
- In your own words, suggest a plausible physical explanation for why this annual cycle exists.
|
github_jupyter
|
# SentencePiece and BPE
## Introduction to Tokenization
In order to process text in neural network models it is first required to **encode** text as numbers with ids, since the tensor operations act on numbers. Finally, if the output of the network is to be words, it is required to **decode** the predicted tokens ids back to text.
To encode text, the first decision that has to be made is to what level of graularity are we going to consider the text? Because ultimately, from these **tokens**, features are going to be created about them. Many different experiments have been carried out using *words*, *morphological units*, *phonemic units*, *characters*. For example,
- Tokens are tricky. (raw text)
- Tokens are tricky . ([words](https://arxiv.org/pdf/1301.3781))
- Token s _ are _ trick _ y . ([morphemes](https://arxiv.org/pdf/1907.02423.pdf))
- t oʊ k ə n z _ ɑː _ ˈt r ɪ k i. ([phonemes](https://www.aclweb.org/anthology/W18-5812.pdf), for STT)
- T o k e n s _ a r e _ t r i c k y . ([character](https://www.aclweb.org/anthology/C18-1139/))
But how to identify these units, such as words, is largely determined by the language they come from. For example, in many European languages a space is used to separate words, while in some Asian languages there are no spaces between words. Compare English and Mandarin.
- Tokens are tricky. (original sentence)
- 标记很棘手 (Mandarin)
- Biāojì hěn jíshǒu (pinyin)
- 标记 很 棘手 (Mandarin with spaces)
So, the ability to **tokenize**, i.e. split text into meaningful fundamental units is not always straight-forward.
Also, there are practical issues of how large our *vocabulary* of words, `vocab_size`, should be, considering memory limitations vs. coverage. A compromise may be need to be made between:
* the finest-grained models employing characters which can be memory intensive and
* more computationally efficient *subword* units such as [n-grams](https://arxiv.org/pdf/1712.09405) or larger units.
In [SentencePiece](https://www.aclweb.org/anthology/D18-2012.pdf) unicode characters are grouped together using either a [unigram language model](https://www.aclweb.org/anthology/P18-1007.pdf) (used in this week's assignment) or [BPE](https://arxiv.org/pdf/1508.07909.pdf), **byte-pair encoding**. We will discuss BPE, since BERT and many of its variants use a modified version of BPE and its pseudocode is easy to implement and understand... hopefully!
## SentencePiece Preprocessing
### NFKC Normalization
Unsurprisingly, even using unicode to initially tokenize text can be ambiguous, e.g.,
```
eaccent = '\u00E9'
e_accent = '\u0065\u0301'
print(f'{eaccent} = {e_accent} : {eaccent == e_accent}')
```
SentencePiece uses the Unicode standard normalization form, [NFKC](https://en.wikipedia.org/wiki/Unicode_equivalence), so this isn't an issue. Looking at our example from above but with normalization:
```
from unicodedata import normalize
norm_eaccent = normalize('NFKC', '\u00E9')
norm_e_accent = normalize('NFKC', '\u0065\u0301')
print(f'{norm_eaccent} = {norm_e_accent} : {norm_eaccent == norm_e_accent}')
```
Normalization has actually changed the unicode code point (unicode unique id) for one of these two characters.
```
def get_hex_encoding(s):
return ' '.join(hex(ord(c)) for c in s)
def print_string_and_encoding(s):
print(f'{s} : {get_hex_encoding(s)}')
for s in [eaccent, e_accent, norm_eaccent, norm_e_accent]:
print_string_and_encoding(s)
```
This normalization has other side effects which may be considered useful such as converting curly quotes “ to " their ASCII equivalent. (<sup>*</sup>Although we *now* lose directionality of the quote...)
### Lossless Tokenization<sup>*</sup>
SentencePiece also ensures that when you tokenize your data and detokenize your data the original position of white space is preserved. <sup>*</sup>However, tabs and newlines are converted to spaces, please try this experiment yourself later below.
To ensure this **lossless tokenization**, SentencePiece replaces white space with _ (U+2581). So that a simple join of the tokens by replace underscores with spaces can restore the white space, even if there are consecutive symbols. But remember first to normalize and then replace spaces with _ (U+2581). As the following example shows.
```
s = 'Tokenization is hard.'
s_ = s.replace(' ', '\u2581')
s_n = normalize('NFKC', 'Tokenization is hard.')
print(get_hex_encoding(s))
print(get_hex_encoding(s_))
print(get_hex_encoding(s_n))
```
So the special unicode underscore was replaced by the ASCII unicode. Reversing the order of the second and third operations, we that the special unicode underscore was retained.
```
s = 'Tokenization is hard.'
sn = normalize('NFKC', 'Tokenization is hard.')
sn_ = s.replace(' ', '\u2581')
print(get_hex_encoding(s))
print(get_hex_encoding(sn))
print(get_hex_encoding(sn_))
```
## BPE Algorithm
Now that we have discussed the preprocessing that SentencePiece performs, we will go get our data, preprocess, and apply the BPE algorithm. We will show how this reproduces the tokenization produced by training SentencePiece on our example dataset (from this week's assignment).
### Preparing our Data
First, we get our Squad data and process as above.
```
import ast
def convert_json_examples_to_text(filepath):
example_jsons = list(map(ast.literal_eval, open(filepath))) # Read in the json from the example file
texts = [example_json['text'].decode('utf-8') for example_json in example_jsons] # Decode the byte sequences
text = '\n\n'.join(texts) # Separate different articles by two newlines
text = normalize('NFKC', text) # Normalize the text
with open('example.txt', 'w') as fw:
fw.write(text)
return text
text = convert_json_examples_to_text('./data/data.txt')
print(text[:900])
```
In the algorithm the `vocab` variable is actually a frequency dictionary of the words. Further, those words have been prepended with an *underscore* to indicate that they are the beginning of a word. Finally, the characters have been delimited by spaces so that the BPE algorithm can group the most common characters together in the dictionary in a greedy fashion. We will see how that is done shortly.
```
from collections import Counter
vocab = Counter(['\u2581' + word for word in text.split()])
vocab = {' '.join([l for l in word]): freq for word, freq in vocab.items()}
def show_vocab(vocab, end='\n', limit=20):
"""Show word frequencys in vocab up to the limit number of words"""
shown = 0
for word, freq in vocab.items():
print(f'{word}: {freq}', end=end)
shown +=1
if shown > limit:
break
show_vocab(vocab)
```
We check the size of the vocabulary (frequency dictionary) because this is the one hyperparameter that BPE depends on crucially on how far it breaks up a word into SentencePieces. It turns out that for our trained model on our small dataset that 60% of 455 merges of the most frequent characters need to be done to reproduce the upperlimit of a 32K `vocab_size` over the entire corpus of examples.
```
print(f'Total number of unique words: {len(vocab)}')
print(f'Number of merges required to reproduce SentencePiece training on the whole corpus: {int(0.60*len(vocab))}')
```
### BPE Algorithm
Directly from the BPE paper we have the following algorithm.
```
import re, collections
def get_stats(vocab):
pairs = collections.defaultdict(int)
for word, freq in vocab.items():
symbols = word.split()
for i in range(len(symbols) - 1):
pairs[symbols[i], symbols[i+1]] += freq
return pairs
def merge_vocab(pair, v_in):
v_out = {}
bigram = re.escape(' '.join(pair))
p = re.compile(r'(?<!\S)' + bigram + r'(?!\S)')
for word in v_in:
w_out = p.sub(''.join(pair), word)
v_out[w_out] = v_in[word]
return v_out
def get_sentence_piece_vocab(vocab, frac_merges=0.60):
sp_vocab = vocab.copy()
num_merges = int(len(sp_vocab)*frac_merges)
for i in range(num_merges):
pairs = get_stats(sp_vocab)
best = max(pairs, key=pairs.get)
sp_vocab = merge_vocab(best, sp_vocab)
return sp_vocab
```
To understand what's going on first take a look at the third function `get_sentence_piece_vocab`. It takes in the current `vocab` word-frequency dictionary and the fraction, `frac_merges`, of the total `vocab_size` to merge characters in the words of the dictionary, `num_merges` times. Then for each *merge* operation it `get_stats` on how many of each pair of character sequences there are. It gets the most frequent *pair* of symbols as the `best` pair. Then it merges that pair of symbols (removes the space between them) in each word in the `vocab` that contains this `best` (= `pair`). Consequently, `merge_vocab` creates a new `vocab`, `v_out`. This process is repeated `num_merges` times and the result is the set of SentencePieces (keys of the final `sp_vocab`).
### Additional Discussion of BPE Algorithm
Please feel free to skip the below if the above description was enough.
In a little more detail then, we can see in `get_stats` we initially create a list of bigram (two character sequence) frequencies from our vocabulary. Later, this may include trigrams, quadgrams, etc. Note that the key of the `pairs` frequency dictionary is actually a 2-tuple, which is just shorthand notation for a pair.
In `merge_vocab` we take in an individual `pair` (of character sequences, note this is the most frequency `best` pair) and the current `vocab` as `v_in`. We create a new `vocab`, `v_out`, from the old by joining together the characters in the pair (removing the space), if they are present in a word of the dictionary.
[Warning](https://regex101.com/): the expression `(?<!\S)` means that either a whitespace character follows before the `bigram` or there is nothing before the bigram (it is the beginning of the word), similarly for `(?!\S)` for preceding whitespace or the end of the word.
```
sp_vocab = get_sentence_piece_vocab(vocab)
show_vocab(sp_vocab)
```
## Train SentencePiece BPE Tokenizer on Example Data
### Explore SentencePiece Model
First let us explore the SentencePiece model provided with this week's assignment. Remember you can always use Python's built in `help` command to see the documentation for any object or method.
```
import sentencepiece as spm
sp = spm.SentencePieceProcessor(model_file='./data/sentencepiece.model')
# help(sp)
```
Let's work with the first sentence of our example text.
```
s0 = 'Beginners BBQ Class Taking Place in Missoula!'
# encode: text => id
print(sp.encode_as_pieces(s0))
print(sp.encode_as_ids(s0))
# decode: id => text
print(sp.decode_pieces(sp.encode_as_pieces(s0)))
print(sp.decode_ids([12847, 277]))
```
Notice how SentencePiece breaks the words into seemingly odd parts, but we've seen something similar from our work with BPE. But how close were we to this model trained on the whole corpus of examples with a `vocab_size` of 32,000 instead of 455? Here you can also test what happens to white space, like '\n'.
But first let us note that SentencePiece encodes the SentencePieces, the tokens, and has reserved some of the ids as can be seen in this week's assignment.
```
uid = 15068
spiece = "\u2581BBQ"
unknown = "__MUST_BE_UNKNOWN__"
# id <=> piece conversion
print(f'SentencePiece for ID {uid}: {sp.id_to_piece(uid)}')
print(f'ID for Sentence Piece {spiece}: {sp.piece_to_id(spiece)}')
# returns 0 for unknown tokens (we can change the id for UNK)
print(f'ID for unknown text {unknown}: {sp.piece_to_id(unknown)}')
print(f'Beginning of sentence id: {sp.bos_id()}')
print(f'Pad id: {sp.pad_id()}')
print(f'End of sentence id: {sp.eos_id()}')
print(f'Unknown id: {sp.unk_id()}')
print(f'Vocab size: {sp.vocab_size()}')
```
We can also check what are the ids for the first part and last part of the vocabulary.
```
print('\nId\tSentP\tControl?')
print('------------------------')
# <unk>, <s>, </s> are defined by default. Their ids are (0, 1, 2)
# <s> and </s> are defined as 'control' symbol.
for uid in range(10):
print(uid, sp.id_to_piece(uid), sp.is_control(uid), sep='\t')
# for uid in range(sp.vocab_size()-10,sp.vocab_size()):
# print(uid, sp.id_to_piece(uid), sp.is_control(uid), sep='\t')
```
### Train SentencePiece BPE model with our example.txt
Finally, let's train our own BPE model directly from the SentencePiece library and compare it to the results of our implemention of the algorithm from the BPE paper itself.
```
spm.SentencePieceTrainer.train('--input=example.txt --model_prefix=example_bpe --vocab_size=450 --model_type=bpe')
sp_bpe = spm.SentencePieceProcessor()
sp_bpe.load('example_bpe.model')
print('*** BPE ***')
print(sp_bpe.encode_as_pieces(s0))
show_vocab(sp_vocab, end = ', ')
```
Our implementation of BPE's code from the paper matches up pretty well with the library itself! The differences are probably accounted for by the `vocab_size`. There is also another technical difference in that in the SentencePiece implementation of BPE a priority queue is used to more efficiently keep track of the *best pairs*. Actually, there is a priority queue in the Python standard library called `heapq` if you would like to give that a try below!
## Optionally try to implement BPE using a priority queue below
```
from heapq import heappush, heappop
def heapsort(iterable):
h = []
for value in iterable:
heappush(h, value)
return [heappop(h) for i in range(len(h))]
a = [1,4,3,1,3,2,1,4,2]
heapsort(a)
```
For a more extensive example consider looking at the [SentencePiece repo](https://github.com/google/sentencepiece/blob/master/python/sentencepiece_python_module_example.ipynb). The last few sections of this code was repurposed from that tutorial. Thanks for your participation! Next stop BERT and T5!
|
github_jupyter
|
### *IPCC SR15 scenario assessment*
<img style="float: right; height: 80px; padding-left: 20px;" src="../_static/IIASA_logo.png">
<img style="float: right; height: 80px;" src="../_static/IAMC_logo.jpg">
# Characteristics of four illustrative model pathways
## Figure 3b of the *Summary for Policymakers*
This notebook derives the figure panels and indicators for the table in Figure 3b in the Summary for Policymakers
of the IPCC's _"Special Report on Global Warming of 1.5°C"_.
The scenario data used in this analysis can be accessed and downloaded at [https://data.ene.iiasa.ac.at/iamc-1.5c-explorer](https://data.ene.iiasa.ac.at/iamc-1.5c-explorer).
## Load `pyam` package and other dependencies
```
import pandas as pd
import numpy as np
import io
import itertools
import yaml
import math
import matplotlib.pyplot as plt
plt.style.use('style_sr15.mplstyle')
%matplotlib inline
import pyam
```
## Import scenario data, categorization and specifications files
The metadata file with scenario categorisation and quantitative indicators can be downloaded at [https://data.ene.iiasa.ac.at/iamc-1.5c-explorer](https://data.ene.iiasa.ac.at/iamc-1.5c-explorer).
Alternatively, it can be re-created using the notebook `sr15_2.0_categories_indicators`.
The last cell of this section loads and assigns a number of auxiliary lists as defined in the categorization notebook.
```
sr1p5 = pyam.IamDataFrame(data='../data/iamc15_scenario_data_world_r2.0.xlsx')
sr1p5.load_meta('sr15_metadata_indicators.xlsx')
with open("sr15_specs.yaml", 'r') as stream:
specs = yaml.load(stream, Loader=yaml.FullLoader)
rc = pyam.run_control()
for item in specs.pop('run_control').items():
rc.update({item[0]: item[1]})
cats_15 = specs.pop('cats_15')
cats_15_no_lo = specs.pop('cats_15_no_lo')
marker = specs.pop('marker')
```
## Downselect scenario ensemble to categories of interest for this assessment
```
sr1p5.meta.rename(columns={'Kyoto-GHG|2010 (SAR)': 'kyoto_ghg_2010'}, inplace=True)
df = sr1p5.filter(category=cats_15)
```
## Global carbon dioxide emissions in four illustrative pathways
Figure SPM3b shows the contribution to CO2 emissions and removal by three categories in the four illustrative pathways.
This illustration does not use the emissions timeseries as reported by the models. This is because the variable `Emissions|CO2|Energy and Industrial Processes` represents net emissions, incorporating carbon dioxide removal in this sector.
The steps below compute the gross emissions. The long variable names are mapped to short variables for easier readibility.
```
afolu_var = 'Emissions|CO2|AFOLU'
ene_ind_var = 'Emissions|CO2|Energy and Industrial Processes'
beccs_var ='Carbon Sequestration|CCS|Biomass'
```
We downselect the entire data to the four illustrative pathways (`marker` scenarios) and the three variables of interest. For consistency with the figure in the SPM, the units are converted to Gt CO2.
```
pw = df.filter(marker=marker, variable=[afolu_var, ene_ind_var, beccs_var],
year=range(2010, 2101, 10))
pw.convert_unit('Mt CO2/yr', 'Gt CO2/yr', inplace=True)
```
As a first step, we extract the timeseries for the AFOLU emissions and rename the variable for brevity. This data will be used as is in this figure.
```
afolu = (
pw.filter(variable=afolu_var)
.rename(variable={afolu_var: 'AFOLU'})
)
```
The energy-and-industry and BECCS timeseries data needs some processing. It is first separated into two distinct dataframes, and the BECCS variable is renamed for brevity.
```
ene_ind = pw.filter(variable=ene_ind_var)
beccs = (
pw.filter(variable=beccs_var)
.rename(variable={beccs_var: 'BECCS'})
)
```
The variable `Carbon Sequestration|CCS|Biomass` reports removed carbon dioxide as positive values. For use in this figure, the sign needs to be reversed.
```
beccs.data.value = - beccs.data.value
```
The `LED` marker scenario does not use any BECCS by assumption of the scenario design. For this reason, the variable `Carbon Sequestration|CCS|Biomass` was not defined when the MESSAGE team submitted the scenario results to the IAMC 1.5°C Scenario Data ensemble.
For easier computation, we add this data series manually here.
```
years = beccs.timeseries().columns
beccs.append(
pyam.IamDataFrame(
pd.DataFrame([0] * len(years), index=years).T,
model='MESSAGEix-GLOBIOM 1.0', scenario='LowEnergyDemand',
region='World', variable='BECCS', unit='Gt CO2/yr'),
inplace=True
)
```
As a third step, we compute the difference between net CO2 emissions from the energy sector & industry and BECCS to obtain gross CO2 emissions in that sector.
```
def get_value(df):
cols = ['model', 'scenario', 'region', 'year', 'unit']
return df.data.set_index(cols)['value']
diff = get_value(ene_ind) - get_value(beccs)
ene_ind_gross = pyam.IamDataFrame(diff, variable='Fossil fuel and industry')
```
We now combine the three contribution dataframes into one joint dataframe for plotting. Because the `beccs` IamDataFrame was partially altered, concatenating directly causes an issue, so we remove all `meta` columns from that dataframe beforehand.
```
beccs.meta = beccs.meta.drop(columns=beccs.meta.columns)
co2 = pyam.concat([ene_ind_gross, afolu, beccs])
```
We now proceed to plot the four illustrative pathways.
```
fig, ax = plt.subplots(1, 4, figsize=(14, 4), sharey=True)
for i, m in enumerate(['LED', 'S1', 'S2', 'S5']):
co2.filter(marker=m).stack_plot(ax=ax[i], total=True, legend=False)
ax[i].title.set_text(m)
ax[3].legend(loc=1)
```
## Collecting indicators across illustrative pathways
### Initialize a `pyam.Statistics` instance
```
base_year = 2010
compare_years = [2030, 2050]
years = [base_year] + compare_years
stats = pyam.Statistics(df=df, groupby={'marker': ['LED', 'S1', 'S2', 'S5']},
filters=[(('pathways', 'no & lo os 1.5'), {'category': cats_15_no_lo})])
```
### CO2 and Kyoto GHG emissions reductions
```
co2 = (
df.filter(kyoto_ghg_2010='in range', variable='Emissions|CO2', year=years)
.convert_unit('Mt CO2/yr', 'Gt CO2/yr')
.timeseries()
)
for y in compare_years:
stats.add((co2[y] / co2[2010] - 1) * 100,
'CO2 emission reduction (% relative to 2010)',
subheader=y)
kyoto_ghg = (
df.filter(kyoto_ghg_2010='in range', variable='Emissions|Kyoto Gases (SAR-GWP100)', year=years)
.rename(unit={'Mt CO2-equiv/yr': 'Mt CO2e/yr'})
.convert_unit('Mt CO2e/yr','Gt CO2e/yr')
.timeseries()
)
for y in compare_years:
stats.add((kyoto_ghg[y] / kyoto_ghg[base_year] - 1) * 100,
'Kyoto-GHG emission reduction (SAR-GWP100), % relative to {})'.format(base_year),
subheader=y)
```
### Final energy demand reduction relative to 2010
```
fe = df.filter(variable='Final Energy', year=years).timeseries()
for y in compare_years:
stats.add((fe[y] / fe[base_year] - 1) * 100,
'Final energy demand reduction relative to {} (%)'.format(base_year),
subheader=y)
```
### Share of renewables in electricity generation
```
def add_stats_share(stats, var_list, name, total, total_name, years, df=df):
_df = df.filter(variable=var_list)
for v in var_list:
_df.require_variable(v, exclude_on_fail=True)
_df.filter(exclude=False, inplace=True)
component = (
_df.timeseries()
.groupby(['model', 'scenario']).sum()
)
share = component / total * 100
for y in years:
stats.add(share[y], header='Share of {} in {} (%)'.format(name, total_name),
subheader=y)
ele = df.filter(variable='Secondary Energy|Electricity', year=compare_years).timeseries()
ele.index = ele.index.droplevel([2, 3, 4])
ele_re_vars = [
'Secondary Energy|Electricity|Biomass',
'Secondary Energy|Electricity|Non-Biomass Renewables'
]
add_stats_share(stats, ele_re_vars, 'renewables', ele, 'electricity', compare_years)
```
### Changes in primary energy mix
```
mapping = [
('coal', 'Coal'),
('oil', 'Oil'),
('gas', 'Gas'),
('nuclear', 'Nuclear'),
('bioenergy', 'Biomass'),
('non-biomass renewables', 'Non-Biomass Renewables')
]
for (n, v) in mapping:
data = df.filter(variable='Primary Energy|{}'.format(v), year=years).timeseries()
for y in compare_years:
stats.add((data[y] / data[base_year] - 1) * 100,
header='Primary energy from {} (% rel to {})'.format(n, base_year),
subheader=y)
```
### Cumulative carbon capture and sequestration until the end of the century
```
def cumulative_ccs(variable, name, first_year=2016, last_year=2100):
data = (
df.filter(variable=variable)
.convert_unit('Mt CO2/yr', 'Gt CO2/yr')
.timeseries()
)
stats.add(
data.apply(pyam.cumulative, raw=False, axis=1,
first_year=first_year, last_year=last_year),
header='Cumulative {} until {} (GtCO2)'.format(name, last_year), subheader='')
cumulative_ccs('Carbon Sequestration|CCS', 'CCS')
cumulative_ccs('Carbon Sequestration|CCS|Biomass', 'BECCS')
```
### Land cover for energy crops
Convert unit to SI unit (million square kilometers).
```
energy_crops = (
df.filter(variable='Land Cover|Cropland|Energy Crops', year=2050)
.convert_unit('million ha', 'million km2', factor=0.01)
.timeseries()
)
stats.add(energy_crops[2050], header='Land are for energy crops (million km2)')
```
### Emissions from land use
```
species = ['CH4', 'N2O']
for n in species:
data = df.filter(kyoto_ghg_2010='in range', variable='Emissions|{}|AFOLU'.format(n), year=years).timeseries()
for y in compare_years:
stats.add((data[y] / data[base_year] - 1) * 100,
header='Agricultural {} emissions (% rel to {})'.format(n, base_year),
subheader=y)
```
## Display summary statistics and export to `xlsx`
```
summary = stats.summarize(interquartile=True, custom_format='{:.0f}').T
summary
summary.to_excel('output/spm_sr15_figure3b_indicators_table.xlsx')
```
|
github_jupyter
|
# MaterialsCoord benchmarking – sensitivity to perturbation analysis
This notebook demonstrates how to use MaterialsCoord to benchmark the sensitivity of bonding algorithms to structural perturbations. Perturbations are introduced according the Einstein crystal test rig, in which site is perturbed so that the distribution around the equilibrium position yields a normal distribution for each Cartesian component.
The perturbation complies thus with the expectation for an Einstein crystal,
in which the potential is given by $V(\delta r) = 0.5 k_\mathrm{spring} \delta r^2$, where
$k_\mathrm{spring}$ denotes the spring constant with which the sites are tethered to
their equilibrium position, and $\delta r$ is the distance of the site under
consideration from its equilibrium position.
The MaterialsCoord `Benchmark` class accepts a `perturb_sigma` option, which is equal to $(k_\mathrm{B}T/k_\mathrm{spring})^{0.5}$.
*Written using:*
- MaterialsCoord==0.1.0
*Authors: Hillary Pan, Alex Ganose (10/12/19)*
---
First, lets initialize the near neighbor methods we are interested in.
```
from pymatgen.analysis.local_env import BrunnerNN_reciprocal, EconNN, JmolNN, \
MinimumDistanceNN, MinimumOKeeffeNN, MinimumVIRENN, \
VoronoiNN, CrystalNN
nn_methods = [
BrunnerNN_reciprocal(), EconNN(tol=0.5), JmolNN(), CrystalNN(), VoronoiNN(tol=0.5),
MinimumDistanceNN(), MinimumOKeeffeNN(), MinimumVIRENN()
]
```
Next, import the benchmark and choose which structures we are interested in.
```
from materialscoord.core import Benchmark
structure_groups = ["common_binaries", "elemental", "A2BX4", "ABX3", "ABX4"]
```
Choose the initial and final perturbation sigma values to include, as well as the number of steps inbetween.
```
import numpy as np
initial_sigma = 0
final_sigma = 0.2
nsteps = 51
sigmas = np.linspace(initial_sigma, final_sigma, nsteps)
```
Run the benchmark with the perturbation turned on. Note we have disabled symmetry so that each perturbed site is treated separately. Due to the absence of symmetry and the slow speed of `MinimumVIRENN`, this can take a long time (14 hours on a 2017 MacBook Pro).
```
from tqdm import tqdm_notebook
results = []
for sigma in tqdm_notebook(sigmas):
bm = Benchmark.from_structure_group(structure_groups, perturb_sigma=sigma, symprec=None)
sigma_scores = bm.score(nn_methods)
results.append(sigma_scores.iloc[-1].values)
```
Finally, plot the results.
```
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib import ticker
import os
from scipy.signal import savgol_filter
import seaborn as sns
plt_results = np.array(results).T
# define matplotlib style settings
style = {
"font.sans-serif": ["Helvetica", "Arial"], "axes.labelsize": 16,
"xtick.labelsize": 16, "ytick.labelsize": 16, "xtick.direction": "in",
"ytick.direction": "in", "xtick.major.size": 8, "xtick.minor.size": 4,
"ytick.major.size": 8, "ytick.minor.size": 4, "lines.linewidth": 2.5,
"lines.markersize": 10, "axes.linewidth": 1.2, "xtick.major.width": 1.2,
"xtick.minor.width": 1.2, "ytick.major.width": 1.2, "ytick.minor.width": 1.2,
"pdf.fonttype":42
}
nn_method_mapping = {"BrunnerNN_reciprocal": "BrunnerNN"}
colors = sns.color_palette("deep")
order = [5, 6, 7, 2, 1, 0, 4, 3]
plt.style.use(style)
fig = plt.figure(figsize=(6, 6))
ax = plt.gca()
for i, x in enumerate(order):
method = nn_methods[x]
y_vals = plt_results[x]
name = method.__class__.__name__
c = colors[i]
name = nn_method_mapping.get(name, name)
# smooth the lines with a double pass through a savgol filter
# more ideal would be to take averages accross multiple runs
# but due to the time taken to generate the data this is impractical
y_vals = savgol_filter(y_vals, 27, 2)
y_vals = savgol_filter(y_vals, 27, 2)
ax.plot(sigmas, y_vals, label=name, c=c)
ax.set(ylabel="Benchmark score", xlabel="Sigma (Å)")
ax.set_xlim((0, 0.2))
ax.yaxis.set_major_locator(ticker.MaxNLocator(5))
plt.legend(loc='upper left', bbox_to_anchor=(1, 1), frameon=False, fontsize=15)
plt.savefig(os.path.join("plots", "perturbation-tolerance.pdf"), bbox_inches="tight")
plt.show()
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/jeffheaton/t81_558_deep_learning/blob/master/t81_558_class_12_02_qlearningreinforcement.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# T81-558: Applications of Deep Neural Networks
**Module 12: Reinforcement Learning**
* Instructor: [Jeff Heaton](https://sites.wustl.edu/jeffheaton/), McKelvey School of Engineering, [Washington University in St. Louis](https://engineering.wustl.edu/Programs/Pages/default.aspx)
* For more information visit the [class website](https://sites.wustl.edu/jeffheaton/t81-558/).
# Module 12 Video Material
* Part 12.1: Introduction to the OpenAI Gym [[Video]](https://www.youtube.com/watch?v=_KbUxgyisjM&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_12_01_ai_gym.ipynb)
* **Part 12.2: Introduction to Q-Learning** [[Video]](https://www.youtube.com/watch?v=A3sYFcJY3lA&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_12_02_qlearningreinforcement.ipynb)
* Part 12.3: Keras Q-Learning in the OpenAI Gym [[Video]](https://www.youtube.com/watch?v=qy1SJmsRhvM&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_12_03_keras_reinforce.ipynb)
* Part 12.4: Atari Games with Keras Neural Networks [[Video]](https://www.youtube.com/watch?v=co0SwPWoZh0&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_12_04_atari.ipynb)
* Part 12.5: Application of Reinforcement Learning [[Video]](https://www.youtube.com/watch?v=1jQPP3RfwMI&list=PLjy4p-07OYzulelvJ5KVaT2pDlxivl_BN) [[Notebook]](t81_558_class_12_05_apply_rl.ipynb)
# Google CoLab Instructions
The following code ensures that Google CoLab is running the correct version of TensorFlow.
```
try:
from google.colab import drive
%tensorflow_version 2.x
COLAB = True
print("Note: using Google CoLab")
except:
print("Note: not using Google CoLab")
COLAB = False
if COLAB:
!sudo apt-get install -y xvfb ffmpeg x11-utils
!pip install -q 'gym==0.10.11'
!pip install -q 'imageio==2.4.0'
!pip install -q PILLOW
!pip install -q 'pyglet==1.3.2'
!pip install -q pyvirtualdisplay
!pip install -q tf-agents
```
# Part 12.2: Introduction to Q-Learning
Q-Learning is a foundational technique upon which deep reinforcement learning is based. Before we explore deep reinforcement learning, it is essential to understand Q-Learning. Several components make up any Q-Learning system.
* **Agent** - The agent is an entity that exists in an environment that takes actions to affect the state of the environment, to receive rewards.
* **Environment** - The environment is the universe that the agent exists in. The environment is always in a specific state that is changed by the actions of the agent.
* **Actions** - Steps that can be performed by the agent to alter the environment
* **Step** - A step occurs each time that the agent performs an action and potentially changes the environment state.
* **Episode** - A chain of steps that ultimately culminates in the environment entering a terminal state.
* **Epoch** - A training iteration of the agent that contains some number of episodes.
* **Terminal State** - A state in which further actions do not make sense. In many environments, a terminal state occurs when the agent has one, lost, or the environment exceeding the maximum number of steps.
Q-Learning works by building a table that suggests an action for every possible state. This approach runs into several problems. First, the environment is usually composed of several continuous numbers, resulting in an infinite number of states. Q-Learning handles continuous states by binning these numeric values into ranges.
Additionally, Q-Learning primarily deals with discrete actions, such as pressing a joystick up or down. Out of the box, Q-Learning does not deal with continuous inputs, such as a car's accelerator that can be in a range of positions from released to fully engaged. Researchers have come up with clever tricks to allow Q-Learning to accommodate continuous actions.
In the next chapter, we will learn more about deep reinforcement learning. Deep neural networks can help to solve the problems of continuous environments and action spaces. For now, we will apply regular Q-Learning to the Mountain Car problem from OpenAI Gym.
### Introducing the Mountain Car
This section will demonstrate how Q-Learning can create a solution to the mountain car gym environment. The Mountain car is an environment where a car must climb a mountain. Because gravity is stronger than the car's engine, even with full throttle, it cannot merely accelerate up the steep slope. The vehicle is situated in a valley and must learn to utilize potential energy by driving up the opposite hill before the car can make it to the goal at the top of the rightmost hill.
First, it might be helpful to visualize the mountain car environment. The following code shows this environment. This code makes use of TF-Agents to perform this render. Usually, we use TF-Agents for the type of deep reinforcement learning that we will see in the next module. However, for now, TF-Agents is just used to render the mountain care environment.
```
import tf_agents
from tf_agents.environments import suite_gym
import PIL.Image
import pyvirtualdisplay
display = pyvirtualdisplay.Display(visible=0, size=(1400, 900)).start()
env_name = 'MountainCar-v0'
env = suite_gym.load(env_name)
env.reset()
PIL.Image.fromarray(env.render())
```
The mountain car environment provides the following discrete actions:
* 0 - Apply left force
* 1 - Apply no force
* 2 - Apply right force
The mountain car environment is made up of the following continuous values:
* state[0] - Position
* state[1] - Velocity
The following code shows an agent that applies full throttle to climb the hill. The cart is not strong enough. It will need to use potential energy from the mountain behind it.
```
import gym
from gym.wrappers import Monitor
import glob
import io
import base64
from IPython.display import HTML
from pyvirtualdisplay import Display
from IPython import display as ipythondisplay
display = Display(visible=0, size=(1400, 900))
display.start()
"""
Utility functions to enable video recording of gym environment
and displaying it.
To enable video, just do "env = wrap_env(env)""
"""
def show_video():
mp4list = glob.glob('video/*.mp4')
if len(mp4list) > 0:
mp4 = mp4list[0]
video = io.open(mp4, 'r+b').read()
encoded = base64.b64encode(video)
ipythondisplay.display(HTML(data='''<video alt="test" autoplay
loop controls style="height: 400px;">
<source src="data:video/mp4;base64,{0}" type="video/mp4" />
</video>'''.format(encoded.decode('ascii'))))
else:
print("Could not find video")
def wrap_env(env):
env = Monitor(env, './video', force=True)
return env
import gym
if COLAB:
env = wrap_env(gym.make("MountainCar-v0"))
else:
env = gym.make("MountainCar-v0")
env.reset()
done = False
i = 0
while not done:
i += 1
state, reward, done, _ = env.step(2)
env.render()
print(f"Step {i}: State={state}, Reward={reward}")
env.close()
show_video()
```
### Programmed Car
Now we will look at a car that I hand-programmed. This car is straightforward; however, it solves the problem. The programmed car always applies force to one direction or another. It does not break. Whatever direction the vehicle is currently rolling, the agent uses power in that direction. Therefore, the car begins to climb a hill, is overpowered, and turns backward. However, once it starts to roll backward force is immediately applied in this new direction.
The following code implements this preprogrammed car.
```
import gym
if COLAB:
env = wrap_env(gym.make("MountainCar-v0"))
else:
env = gym.make("MountainCar-v0")
state = env.reset()
done = False
i = 0
while not done:
i += 1
if state[1]>0:
action = 2
else:
action = 0
state, reward, done, _ = env.step(action)
env.render()
print(f"Step {i}: State={state}, Reward={reward}")
env.close()
```
We now visualize the preprogrammed car solving the problem.
```
show_video()
```
### Reinforcement Learning
Q-Learning is a system of rewards that the algorithm gives an agent for successfully moving the environment into a state considered successful. These rewards are the Q-values from which this algorithm takes its name. The final output from the Q-Learning algorithm is a table of Q-values that indicate the reward value of every action that the agent can take, given every possible environment state. The agent must bin continuous state values into a fixed finite number of columns.
Learning occurs when the algorithm runs the agent and environment through a series of episodes and updates the Q-values based on the rewards received from actions taken; Figure 12.REINF provides a high-level overview of this reinforcement or Q-Learning loop.
**Figure 12.REINF:Reinforcement/Q Learning**

The Q-values can dictate action by selecting the action column with the highest Q-value for the current environment state. The choice between choosing a random action and a Q-value driven action is governed by the epsilon ($\epsilon$) parameter, which is the probability of random action.
Each time through the training loop, the training algorithm updates the Q-values according to the following equation.
$Q^{new}(s_{t},a_{t}) \leftarrow \underbrace{Q(s_{t},a_{t})}_{\text{old value}} + \underbrace{\alpha}_{\text{learning rate}} \cdot \overbrace{\bigg( \underbrace{\underbrace{r_{t}}_{\text{reward}} + \underbrace{\gamma}_{\text{discount factor}} \cdot \underbrace{\max_{a}Q(s_{t+1}, a)}_{\text{estimate of optimal future value}}}_{\text{new value (temporal difference target)}} - \underbrace{Q(s_{t},a_{t})}_{\text{old value}} \bigg) }^{\text{temporal difference}}$
There are several parameters in this equation:
* alpha ($\alpha$) - The learning rate, how much should the current step cause the Q-values to be updated.
* lambda ($\lambda$) - The discount factor is the percentage of future reward that the algorithm should consider in this update.
This equation modifies several values:
* $Q(s_t,a_t)$ - The Q-table. For each combination of states, what reward would the agent likely receive for performing each action?
* $s_t$ - The current state.
* $r_t$ - The last reward received.
* $a_t$ - The action that the agent will perform.
The equation works by calculating a delta (temporal difference) that the equation should apply to the old state. This learning rate ($\alpha$) scales this delta. A learning rate of 1.0 would fully implement the temporal difference to the Q-values each iteration and would likely be very chaotic.
There are two parts to the temporal difference: the new and old values. The new value is subtracted from the old value to provide a delta; the full amount that we would change the Q-value by if the learning rate did not scale this value. The new value is a summation of the reward received from the last action and the maximum of the Q-values from the resulting state when the client takes this action. It is essential to add the maximum of action Q-values for the new state because it estimates the optimal future values from proceeding with this action.
### Q-Learning Car
We will now use Q-Learning to produce a car that learns to drive itself. Look out, Tesla! We begin by defining two essential functions.
```
import gym
import numpy as np
# This function converts the floating point state values into
# discrete values. This is often called binning. We divide
# the range that the state values might occupy and assign
# each region to a bucket.
def calc_discrete_state(state):
discrete_state = (state - env.observation_space.low)/buckets
return tuple(discrete_state.astype(np.int))
# Run one game. The q_table to use is provided. We also
# provide a flag to indicate if the game should be
# rendered/animated. Finally, we also provide
# a flag to indicate if the q_table should be updated.
def run_game(q_table, render, should_update):
done = False
discrete_state = calc_discrete_state(env.reset())
success = False
while not done:
# Exploit or explore
if np.random.random() > epsilon:
# Exploit - use q-table to take current best action
# (and probably refine)
action = np.argmax(q_table[discrete_state])
else:
# Explore - t
action = np.random.randint(0, env.action_space.n)
# Run simulation step
new_state, reward, done, _ = env.step(action)
# Convert continuous state to discrete
new_state_disc = calc_discrete_state(new_state)
# Have we reached the goal position (have we won?)?
if new_state[0] >= env.unwrapped.goal_position:
success = True
# Update q-table
if should_update:
max_future_q = np.max(q_table[new_state_disc])
current_q = q_table[discrete_state + (action,)]
new_q = (1 - LEARNING_RATE) * current_q + LEARNING_RATE * \
(reward + DISCOUNT * max_future_q)
q_table[discrete_state + (action,)] = new_q
discrete_state = new_state_disc
if render:
env.render()
return success
```
Several hyperparameters are very important for Q-Learning. These parameters will likely need adjustment as you apply Q-Learning to other problems. Because of this, it is crucial to understand the role of each parameter.
* **LEARNING_RATE** The rate at which previous Q-values are updated based on new episodes run during training.
* **DISCOUNT** The amount of significance to give estimates of future rewards when added to the reward for the current action taken. A value of 0.95 would indicate a discount of 5% to the future reward estimates.
* **EPISODES** The number of episodes to train over. Increase this for more complex problems; however, training time also increases.
* **SHOW_EVERY** How many episodes to allow to elapse before showing an update.
* **DISCRETE_GRID_SIZE** How many buckets to use when converting each of the continuous state variables. For example, [10, 10] indicates that the algorithm should use ten buckets for the first and second state variables.
* **START_EPSILON_DECAYING** Epsilon is the probability that the agent will select a random action over what the Q-Table suggests. This value determines the starting probability of randomness.
* **END_EPSILON_DECAYING** How many episodes should elapse before epsilon goes to zero and no random actions are permitted. For example, EPISODES//10 means only the first 1/10th of the episodes might have random actions.
```
LEARNING_RATE = 0.1
DISCOUNT = 0.95
EPISODES = 50000
SHOW_EVERY = 1000
DISCRETE_GRID_SIZE = [10, 10]
START_EPSILON_DECAYING = 0.5
END_EPSILON_DECAYING = EPISODES//10
```
We can now make the environment. If we are running in Google COLAB then we wrap the environment to be displayed inside the web browser. Next create the discrete buckets for state and build Q-table.
```
if COLAB:
env = wrap_env(gym.make("MountainCar-v0"))
else:
env = gym.make("MountainCar-v0")
epsilon = 1
epsilon_change = epsilon/(END_EPSILON_DECAYING - START_EPSILON_DECAYING)
buckets = (env.observation_space.high - env.observation_space.low) \
/DISCRETE_GRID_SIZE
q_table = np.random.uniform(low=-3, high=0, size=(DISCRETE_GRID_SIZE \
+ [env.action_space.n]))
success = False
```
We can now make the environment. If we are running in Google COLAB then we wrap the environment to be displayed inside the web browser. Next, create the discrete buckets for state and build Q-table.
```
episode = 0
success_count = 0
# Loop through the required number of episodes
while episode<EPISODES:
episode+=1
done = False
# Run the game. If we are local, display render animation at SHOW_EVERY
# intervals.
if episode % SHOW_EVERY == 0:
print(f"Current episode: {episode}, success: {success_count}" +\
" ({float(success_count)/SHOW_EVERY})")
success = run_game(q_table, True, False)
success_count = 0
else:
success = run_game(q_table, False, True)
# Count successes
if success:
success_count += 1
# Move epsilon towards its ending value, if it still needs to move
if END_EPSILON_DECAYING >= episode >= START_EPSILON_DECAYING:
epsilon = max(0, epsilon - epsilon_change)
print(success)
```
As you can see, the number of successful episodes generally increases as training progresses. It is not advisable to stop the first time that we observe 100% success over 1,000 episodes. There is a randomness to most games, so it is not likely that an agent would retain its 100% success rate with a new run. Once you observe that the agent has gotten 100% for several update intervals, it might be safe to stop training.
# Running and Observing the Agent
Now that the algorithm has trained the agent, we can observe the agent in action. You can use the following code to see the agent in action.
```
run_game(q_table, True, False)
show_video()
```
# Inspecting the Q-Table
We can also display the Q-table. The following code shows the action that the agent would perform for each environment state. As the weights of a neural network, this table is not straightforward to interpret. Some patterns do emerge in that directions do arise, as seen by calculating the means of rows and columns. The actions seem consistent at upper and lower halves of both velocity and position.
```
import pandas as pd
df = pd.DataFrame(q_table.argmax(axis=2))
df.columns = [f'v-{x}' for x in range(DISCRETE_GRID_SIZE[0])]
df.index = [f'p-{x}' for x in range(DISCRETE_GRID_SIZE[1])]
df
df.mean(axis=0)
df.mean(axis=1)
```
|
github_jupyter
|
```
"""
Update Parameters Here
"""
COLLECTION_NAME = "Quaks"
CONTRACT = "0x07bbdaf30e89ea3ecf6cadc80d6e7c4b0843c729"
BEFORE_TIME = "2021-09-02T00:00:00" # One day after the last mint (e.g. https://etherscan.io/tx/0x206c846d0d1739faa9835e16ff419d15708a558357a9413619e65dacf095ac7a)
# these should usually stay the same
METHOD = "raritytools"
"""
Created on Tue Sep 14 20:17:07 2021
mint data. Doesn't work when Opensea's API is being shitty
@author: nbax1, slight modifications by mdigi14
"""
import pandas as pd
from utils import config
from utils import constants
from utils import opensea
"""
Helper Functions
"""
def get_mint_events(contract, before_time, rarity_db):
data = opensea.get_opensea_events(
contract_address=contract,
account_address=constants.MINT_ADDRESS,
event_type="transfer",
occurred_before=before_time,
)
df = pd.json_normalize(data)
df = df.loc[df["from_account.address"] == constants.MINT_ADDRESS]
df_rar = pd.DataFrame(rarity_db)
os_tokens = df["asset.token_id"].astype(int).tolist()
rar_tokens = df_rar["TOKEN_ID"].astype(int).tolist()
set1 = set(rar_tokens)
set2 = set(os_tokens)
missing_tokens = list(sorted(set1 - set2))
if missing_tokens:
print(
f"Missing tokens: {missing_tokens}\nTrying to fetch event for missing tokens..."
)
missing_data = []
for token in missing_tokens:
missing_data.extend(
opensea.get_opensea_events(
contract_address=contract,
account_address=constants.MINT_ADDRESS,
event_type="transfer",
occurred_before=before_time,
token_id=token,
)
)
df_missing_data = pd.json_normalize(missing_data)
# Merge missing data with rest of data
df_all = pd.concat([df, df_missing_data])
# make sure token_id is an integer
df_all["asset.token_id"] = df_all["asset.token_id"].astype(int)
RARITY_DB["TOKEN_ID"] = RARITY_DB["TOKEN_ID"].astype(int)
# add rarity rank to minting data
df_all = df_all.merge(RARITY_DB, left_on="asset.token_id", right_on="TOKEN_ID")
# Keep only the columns we want
df_all = df_all[
[
"transaction.transaction_hash",
"to_account.address",
"asset.token_id",
"asset.owner.address",
"Rank",
"transaction.timestamp",
]
]
# Rename columns
df_all.columns = [
"txid",
"to_account",
"TOKEN_ID",
"current_owner",
"rank",
"time",
]
print(f"Downloaded {df_all.shape[0]} events")
return df_all
"""
Gerenerate Dataset
"""
RARITY_CSV = f"{config.RARITY_FOLDER}/{COLLECTION_NAME}_{METHOD}.csv"
RARITY_DB = pd.read_csv(RARITY_CSV)
mint_db = get_mint_events(CONTRACT, BEFORE_TIME, RARITY_DB)
mint_db = mint_db.sort_values(by=["TOKEN_ID"])
mint_db.to_csv(f"{config.MINTING_FOLDER}/{COLLECTION_NAME}_minting.csv", index=False)
```
|
github_jupyter
|
## INTRODUCTION
- It’s a Python based scientific computing package targeted at two sets of audiences:
- A replacement for NumPy to use the power of GPUs
- Deep learning research platform that provides maximum flexibility and speed
- pros:
- Iinteractively debugging PyTorch. Many users who have used both frameworks would argue that makes pytorch significantly easier to debug and visualize.
- Clean support for dynamic graphs
- Organizational backing from Facebook
- Blend of high level and low level APIs
- cons:
- Much less mature than alternatives
- Limited references / resources outside of the official documentation
- I accept you know neural network basics. If you do not know check my tutorial. Because I will not explain neural network concepts detailed, I only explain how to use pytorch for neural network
- Neural Network tutorial: https://www.kaggle.com/kanncaa1/deep-learning-tutorial-for-beginners
- The most important parts of this tutorial from matrices to ANN. If you learn these parts very well, implementing remaining parts like CNN or RNN will be very easy.
<br>
<br>**Content:**
1. Basics of Pytorch, Linear Regression, Logistic Regression, Artificial Neural Network (ANN), Concolutional Neural Network (CNN)
- https://www.kaggle.com/kanncaa1/pytorch-tutorial-for-deep-learning-lovers/code
1. [Recurrent Neural Network (RNN)](#1)
```
# This Python 3 environment comes with many helpful analytics libraries installed
# It is defined by the kaggle/python docker image: https://github.com/kaggle/docker-python
# For example, here's several helpful packages to load in
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
```
<a id="1"></a> <br>
### Recurrent Neural Network (RNN)
- RNN is essentially repeating ANN but information get pass through from previous non-linear activation function output.
- **Steps of RNN:**
1. Import Libraries
1. Prepare Dataset
1. Create RNN Model
- hidden layer dimension is 100
- number of hidden layer is 1
1. Instantiate Model Class
1. Instantiate Loss Class
- Cross entropy loss
- It also has softmax(logistic function) in it.
1. Instantiate Optimizer Class
- SGD Optimizer
1. Traning the Model
1. Prediction
```
# Import Libraries
import torch
import torch.nn as nn
import torchvision.transforms as transforms
from torch.autograd import Variable
from sklearn.model_selection import train_test_split
# Prepare Dataset
# load data
train = pd.read_csv(r"../input/train.csv",dtype = np.float32)
# split data into features(pixels) and labels(numbers from 0 to 9)
targets_numpy = train.label.values
features_numpy = train.loc[:,train.columns != "label"].values/255 # normalization
# train test split. Size of train data is 80% and size of test data is 20%.
features_train, features_test, targets_train, targets_test = train_test_split(features_numpy,
targets_numpy,
test_size = 0.2,
random_state = 42)
# create feature and targets tensor for train set. As you remember we need variable to accumulate gradients. Therefore first we create tensor, then we will create variable
featuresTrain = torch.from_numpy(features_train)
targetsTrain = torch.from_numpy(targets_train).type(torch.LongTensor) # data type is long
# create feature and targets tensor for test set.
featuresTest = torch.from_numpy(features_test)
targetsTest = torch.from_numpy(targets_test).type(torch.LongTensor) # data type is long
# batch_size, epoch and iteration
batch_size = 100
n_iters = 10000
num_epochs = n_iters / (len(features_train) / batch_size)
num_epochs = int(num_epochs)
# Pytorch train and test sets
train = torch.utils.data.TensorDataset(featuresTrain,targetsTrain)
test = torch.utils.data.TensorDataset(featuresTest,targetsTest)
# data loader
train_loader = torch.utils.data.DataLoader(train, batch_size = batch_size, shuffle = False)
test_loader = torch.utils.data.DataLoader(test, batch_size = batch_size, shuffle = False)
# visualize one of the images in data set
plt.imshow(features_numpy[10].reshape(28,28))
plt.axis("off")
plt.title(str(targets_numpy[10]))
plt.savefig('graph.png')
plt.show()
# Create RNN Model
class RNNModel(nn.Module):
def __init__(self, input_dim, hidden_dim, layer_dim, output_dim):
super(RNNModel, self).__init__()
# Number of hidden dimensions
self.hidden_dim = hidden_dim
# Number of hidden layers
self.layer_dim = layer_dim
# RNN
self.rnn = nn.RNN(input_dim, hidden_dim, layer_dim, batch_first=True,
nonlinearity='relu')
# Readout layer
self.fc = nn.Linear(hidden_dim, output_dim)
def forward(self, x):
# Initialize hidden state with zeros
h0 = Variable(torch.zeros(self.layer_dim, x.size(0), self.hidden_dim))
# One time step
out, hn = self.rnn(x, h0)
out = self.fc(out[:, -1, :])
return out
# batch_size, epoch and iteration
batch_size = 100
n_iters = 2500
num_epochs = n_iters / (len(features_train) / batch_size)
num_epochs = int(num_epochs)
# Pytorch train and test sets
train = torch.utils.data.TensorDataset(featuresTrain,targetsTrain)
test = torch.utils.data.TensorDataset(featuresTest,targetsTest)
# data loader
train_loader = torch.utils.data.DataLoader(train, batch_size = batch_size, shuffle = False)
test_loader = torch.utils.data.DataLoader(test, batch_size = batch_size, shuffle = False)
# Create RNN
input_dim = 28 # input dimension
hidden_dim = 100 # hidden layer dimension
layer_dim = 2 # number of hidden layers
output_dim = 10 # output dimension
model = RNNModel(input_dim, hidden_dim, layer_dim, output_dim)
# Cross Entropy Loss
error = nn.CrossEntropyLoss()
# SGD Optimizer
learning_rate = 0.05
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
seq_dim = 28
loss_list = []
iteration_list = []
accuracy_list = []
count = 0
for epoch in range(num_epochs):
for i, (images, labels) in enumerate(train_loader):
train = Variable(images.view(-1, seq_dim, input_dim))
labels = Variable(labels )
# Clear gradients
optimizer.zero_grad()
# Forward propagation
outputs = model(train)
# Calculate softmax and ross entropy loss
loss = error(outputs, labels)
# Calculating gradients
loss.backward()
# Update parameters
optimizer.step()
count += 1
if count % 250 == 0:
# Calculate Accuracy
correct = 0
total = 0
# Iterate through test dataset
for images, labels in test_loader:
images = Variable(images.view(-1, seq_dim, input_dim))
# Forward propagation
outputs = model(images)
# Get predictions from the maximum value
predicted = torch.max(outputs.data, 1)[1]
# Total number of labels
total += labels.size(0)
correct += (predicted == labels).sum()
accuracy = 100 * correct / float(total)
# store loss and iteration
loss_list.append(loss.data)
iteration_list.append(count)
accuracy_list.append(accuracy)
if count % 500 == 0:
# Print Loss
print('Iteration: {} Loss: {} Accuracy: {} %'.format(count, loss.data[0], accuracy))
# visualization loss
plt.plot(iteration_list,loss_list)
plt.xlabel("Number of iteration")
plt.ylabel("Loss")
plt.title("RNN: Loss vs Number of iteration")
plt.show()
# visualization accuracy
plt.plot(iteration_list,accuracy_list,color = "red")
plt.xlabel("Number of iteration")
plt.ylabel("Accuracy")
plt.title("RNN: Accuracy vs Number of iteration")
plt.savefig('graph.png')
plt.show()
```
### Conclusion
In this tutorial, we learn:
1. Basics of pytorch
1. Linear regression with pytorch
1. Logistic regression with pytorch
1. Artificial neural network with with pytorch
1. Convolutional neural network with pytorch
- https://www.kaggle.com/kanncaa1/pytorch-tutorial-for-deep-learning-lovers/code
1. Recurrent neural network with pytorch
<br> If you have any question or suggest, I will be happy to hear it
|
github_jupyter
|
# TTV Retrieval for Kepler-36 (a well-studied, dynamically-interacting system)
In this notebook, we will perform a dynamical retrieval for Kepler-36 = KOI-277. With two neighboring planets of drastically different densities (the inner planet is rocky and the outer planet is gaseous; see [Carter et al. 2012](https://ui.adsabs.harvard.edu/abs/2012Sci...337..556C/abstract)), this is one of the more well-studied TTV systems in existence. First, let's import packages and download data from the Rowe et al. (2015) TTV catalog:
```
%matplotlib inline
import ttvnest
import numpy as np
koi = 277
nplanets = 2
data, errs, epochs = ttvnest.load_data.get_data(koi, nplanets)
```
Now, let's set up the ttvnest system:
```
kepler36_b = ttvnest.TTVPlanet(data[1], errs[1], epochs[1], mass_prior = ('Uniform', 0, 100.),
period_prior = ('Normal', 13.84, 0.01)
)
kepler36_c = ttvnest.TTVPlanet(data[0], errs[0], epochs[0], mass_prior = ('Uniform', 0, 100.),
period_prior = ('Normal', 16.23, 0.01)
)
kepler36 = ttvnest.TTVSystem(kepler36_b, kepler36_c)
```
Before retrieval, let's plot the data alone to see what they look like:
```
ttvnest.plot_utils.plot_ttv_data(kepler36)
```
Clear, anticorrelated signals! Let's retrieve:
```
results = kepler36.retrieve()
```
Let's check out our results. I'm not going to work out the Carter et al. (2012) posterior distribution on the eccentricity vectors since they use a different basis than I choose here. But it's probably worth converting their mass ratio constraints to what we should expect here. They get a mass ratio sum $q_+ = (M_1 + M_2)/M_\star= 3.51\times10^{-5}$. In ttvnest dynamical masses are normalized by $3\times10^{-6} = M_\mathrm{Earth}/M_\mathrm{Sun}$, so this gives $q_+ = 11.7$ in our units. Their planetary mass ratio is $q_p = M_1/M_2 = 0.55$. Taken together, this gives dynamical masses of $M_1/M_\star = 4.15$ and $M_2/M_\star = 7.55$.
Let's see if we get there...
```
kepler36.posterior_summary()
ttvnest.plot_utils.plot_results(kepler36, uncertainty_curves = 100,
sim_length = 365.25*10, outname = 'kepler36')
```
We are a little on the low side, but that's apparently to be expected from other works like Hadden & Lithwick (2017). Let's make the dynesty plots for good measure:
```
ttvnest.plot_utils.dynesty_plots(kepler36, outname = 'kepler36')
```
Wow, what a nice system. Let's save our results for later:
```
ttvnest.io_utils.save_results(kepler36, 'kepler36.p')
```
|
github_jupyter
|
<a href="https://colab.research.google.com/github/AmberLJC/FedScale/blob/master/dataset/Femnist_stats.ipynb" target="_parent"><img src="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"/></a>
# **[Jupyter notebook] Understand the heterogeneous FL data.**
# Download the Femnist dataset and FedScale
Follow the sownload instruction in /content/FedScale/dataset/download.sh
```
# Download Fedscale and femnist dataset
!pwd
!wget -O /content/femnist.tar.gz https://fedscale.eecs.umich.edu/dataset/femnist.tar.gz
!tar -xf /content/femnist.tar.gz -C /content/
!rm -f /content/femnist.tar.gz
!echo -e "${GREEN}FEMNIST dataset downloaded!${NC}"
!git clone https://github.com/AmberLJC/FedScale.git
from torch.utils.data import DataLoader
import matplotlib.pyplot as plt
import numpy as np
from FedScale.core.utils.femnist import FEMNIST
from FedScale.core.utils.utils_data import get_data_transform
from FedScale.core.utils.divide_data import DataPartitioner
from FedScale.core.argParser import args
```
# Data Loader
```
train_transform, test_transform = get_data_transform('mnist')
train_dataset = FEMNIST('/content/femnist', dataset='train', transform=train_transform)
test_dataset = FEMNIST('/content/femnist', dataset='test', transform=test_transform)
```
Partition the dataset by the `clientclient_data_mapping` file, which gives the real-world client-level heterogeneoity.
```
args.task = 'cv'
training_sets = DataPartitioner(data=train_dataset, args=args, numOfClass=62)
training_sets.partition_data_helper(num_clients=None, data_map_file='/content/femnist/client_data_mapping/train.csv')
#testing_sets = DataPartitioner(data=test_dataset, args=args, numOfClass=62, isTest=True)
#testing_sets.partition_data_helper(num_clients=None, data_map_file='/content/femnist/client_data_mapping/train.csv')
```
# Print and plot statistics of the dataset.
```
print(f'Total number of data smaples: {training_sets.getDataLen()}')
print(f'Total number of clients: {training_sets.getClientLen()}')
print(f'The number of data smaples of each clients: {training_sets.getSize()}')
print(f'The number of unique labels of each clients: {training_sets.getClientLabel()}')
fig, axs = plt.subplots(1, 2, sharey=True, tight_layout=True)
size_dist = training_sets.getSize()['size']
n_bins = 20
axs[0].hist(size_dist, bins=n_bins)
axs[0].set_title('Client data size distribution')
label_dist = training_sets.getClientLabel()
axs[1].hist(label_dist, bins=n_bins)
axs[1].set_title('Client label distribution')
```
# Visiualize the clients' data.
```
rank=1
isTest = False
dropLast = True
partition = training_sets.use(rank - 1, isTest)
num_loaders = min(int(len(partition)/ args.batch_size/2), args.num_loaders)
dataloader = DataLoader(partition, batch_size=16, shuffle=True, pin_memory=True, timeout=60, num_workers=num_loaders, drop_last=dropLast)
for data in iter(dataloader):
plt.imshow(np.transpose(data[0][0].numpy(), (1, 2, 0)))
break
```
|
github_jupyter
|
## Code for policy section
```
# Load libraries
import numpy as np
import matplotlib.pyplot as plt
import matplotlib as mlp
# Ensure type 1 fonts are used
mlp.rcParams['ps.useafm'] = True
mlp.rcParams['pdf.use14corefonts'] = True
mlp.rcParams['text.usetex'] = True
import seaborn as sns
import pandas as pd
import pickle
import itertools as it
```
## Solve for the final size of the outbreak in Lombardy, Italy
```
# Estimate based on the value of the basic reproduction number as provided by best fit
# For formula, see here: https://web.stanford.edu/~jhj1/teachingdocs/Jones-on-R0.pdf
from sympy import Symbol, solve, log
x = Symbol('x')
r0 = 3.16
s_inf = solve(log(x)-r0_max*(x-1),x)[0]
print("% of the population that is still susceptible by the end of the outbreak in Lombardy, Italy: {0:10.4f}".format(s_inf*100))
print("% of the population that has ever been infected by the end of the outbreak in Lombardy, Italy: {0:10.4f}".format(100-s_inf*100))
# Set of colors
# For age group policies
color_list_shahin = ['orange','green','blue','purple','black']
# For additional baseline policies (50% or 100% of the population being asked to shelter-in-place)
color_list_add = ['dodgerblue','hotpink']
# Number of distinct ages in the UN age distribution
# Currently ages 0-100, with each age counted separately
n_ages = 101
# Shelter-in-place probabilities per age group, equivalent to 1 million of the considered generation in each case
age_ranges = [(0,14), (15,29), (30,49), (50,69), (70,100)]
isolation_rates_by_age = [0.803689, 0.713332, 0.380842, 0.358301, 0.516221]
# Learn about the structure of the folder containing the simulation results
all_possible_combos = []
for a, iso_rate in zip(age_ranges, isolation_rates_by_age):
combo = np.zeros(n_ages)
combo[a[0]:a[1]+1] = iso_rate
all_possible_combos.append(combo)
# Two possibilities for mean time to isolation: either 4.6 days (default value) or a large number to mimic no isolation in place
mean_time_to_isolations = [4.6, 10000]
all_possible_combos = list(it.product(mean_time_to_isolations, all_possible_combos))
NUM_COMBOS = len(all_possible_combos)
print("NUM COMBOS:",NUM_COMBOS)
mtti_val_even = all_possible_combos[0][0]
combo_frac_stay_home_even = all_possible_combos[0][1]
mtti_val_odd = all_possible_combos[1][0]
combo_frac_stay_home_odd = all_possible_combos[1][1]
print("Value of mean time to isolation - even index: ", mtti_val_even)
print("Combo fraction stay home - even index", combo_frac_stay_home_even)
print("Value of mean time to isolation - odd index: ", mtti_val_odd)
print("Combo fraction stay home - odd index: ", combo_frac_stay_home_odd)
# Learn about the structure of the folder containing the simulation results
all_possible_combos = []
for a in age_ranges:
# Either 50% or 100% of the population in each age group is asked to shelter-in-place
for val in [0.5, 1.0]:
combo = np.zeros(n_ages)
combo[a[0]:a[1]+1]=val
all_possible_combos.append(combo)
# Two possibilities for mean time to isolation: either 4.6 days (default value) or a large number to mimic no isolation in place
mean_time_to_isolations = [4.6, 10000]
all_possible_combos = list(it.product(mean_time_to_isolations, all_possible_combos))
NUM_COMBOS = len(all_possible_combos)
print("NUM COMBOS:",NUM_COMBOS)
mtti_val_even = all_possible_combos[0][0]
combo_frac_stay_home_even = all_possible_combos[0][1]
mtti_val_odd = all_possible_combos[1][0]
combo_frac_stay_home_odd = all_possible_combos[1][1]
print("Value of mean time to isolation - even index: ", mtti_val_even)
print("Combo fraction stay home - even index: ", combo_frac_stay_home_even)
print("Value of mean time to isolation - odd index: ", mtti_val_even)
print("Combo fraction stay home - odd index: ", combo_frac_stay_home_even)
# Set font sizes for plots
legend_fontsize = 13
title_fontsize = 15
xlab_fontsize = 23
ylab_fontsize = 23
xtick_fontsize = 17
ytick_fontsize = 17
```
## Functions to be used to plot four subgraphs in Figure 8
### Function to be used to plot the projected percentage of infected people in the population over time, in the absence of physical distancing
### Figures 8(a) and 8(b)
```
def perc_infected_age_group_node_removal(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder1, folder2, filename1, filename2, option, specific_title):
if option == 2:
nb = 0
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start,combo_end,2):
nb +=1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Mild = pd.read_csv( folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Age group: ", group_vec_age[nb-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline: ", j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,color='red',linestyle='--')
plt.legend(['Absence of\n intervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize = 13)
plt.ylim(0,100)
plt.title(specific_title,fontsize=15)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.ylabel('Percentage of infected', fontsize=23)
plt.xlabel('Days since patient zero', fontsize=23)
return(plt)
elif option == 1:
nb = 0
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
Infected_Trials=np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start+1,combo_end,2):
nb +=1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Mild = pd.read_csv( folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Age group: ", group_vec_age[nb-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline: ", j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,color='red',linestyle='--')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize = 13)
plt.ylim(0,100)
plt.title(specific_title,fontsize=15)
plt.ylabel('Percentage of infected', fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero', fontsize=23)
return(plt)
else:
nb = 0
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
Infected_Trials=np.zeros((100,sim_end+1))
for i in range(100):
Mild = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Documented = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_documented.csv',delimiter=' ',header=None)
Severe = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start,combo_end):
nb +=1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Mild = pd.read_csv( folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size
print("Age group: ", group_vec_age[nb-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline: ", j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,color='red',linestyle='--')
plt.ylim(0,100)
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize = 13)
plt.title(specific_title, fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.ylabel('Percentage of infected', fontsize=23)
plt.xlabel('Days since patient zero', fontsize=23)
return(plt)
```
### Function to be used to plot the projected number of deaths over time, in the absence of physical distancing
### Figures 8(c) and 8(d)
```
def death_age_group_node_removal(group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end,
folder1, folder2, filename1, filename2, option, specific_title):
if option == 2:
nb = 0
# Baseline - No intervention
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
D=np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv', delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline 0: No intervention")
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
for j in range(combo_start,combo_end,2):
nb +=1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv', delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today : ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# Additional baselines - 50% and 100% of population stays home
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv', delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline: ", j-1)
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_add[j-2], linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0, color='red', linestyle='--')
plt.legend(['Absence of\n intervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize=legend_fontsize)
plt.ylim(0,400)
plt.title(specific_title, fontsize=title_fontsize)
plt.xlabel('Days since patient zero', fontsize=xlab_fontsize)
plt.ylabel('Total deaths (thousands)', fontsize=ylab_fontsize)
plt.xticks(fontsize=xtick_fontsize)
plt.yticks(fontsize=ytick_fontsize)
return(plt)
elif option == 1:
nb = 0
# Baseline - No intervention
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv', delimiter=' ', header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline 0: No intervention")
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
# Average simulations per age group over n_sims random seeds
for j in range(combo_start+1,combo_end,2):
nb +=1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv', delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulatuon: ", D[today])
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# Additional baselines - 50% and 100% of population stays home
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline: ", j-1)
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,color='red',linestyle='--')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize = 13)
plt.ylim(0,400)
plt.title(specific_title,fontsize=15)
plt.ylabel('Total deaths (thousands)', fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero', fontsize=23)
return(plt)
else:
nb = 0
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
D=np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline 0: No intervention")
print("# of deaths on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", Infected_Trials[sim_end])
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
for j in range(combo_start,combo_end):
nb = nb+1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
if i < 50:
folder = folder1
filename = filename1
else:
folder = folder2
filename = filename2
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i%50) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline: ", j-1)
print("% infected on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,color='red',linestyle='--')
plt.ylim(0,400)
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'], fontsize = 13)
plt.title(specific_title, fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.ylabel('Total deaths (thousands)', fontsize=23)
plt.xlabel('Days since patient zero', fontsize=23)
return(plt)
```
## Functions to be used to plot four subgraphs in Figure 9
### Function to be used to plot the projected percentage of infected people in the population over time, when physical distancing is in place
### Figures 9(a) and 9(b)
```
def perc_infected_age_group_node_removal_lockdown(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title):
if option == 2:
nb = 0
Infected_Trials = np.zeros((n_sims,sim_end+1))
# Baseline - "No intervention" scenario
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Mild = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv', delimiter=' ', header=None)
Severe = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv', delimiter=' ', header=None)
Critical = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv', delimiter=' ',header=None)
R = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv', delimiter=' ', header=None)
D = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv', delimiter=' ', header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start,combo_end,2):
nb +=1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv', delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv', delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv', delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv', delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv', delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Age group: ", group_vec_age[nb-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_shahin[nb-1])
# new baseline - 50% or 100% of the population of an age group is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials/pop_size*100.
Infected_Trials = Infected_Trials.mean(axis=0)
print("Baseline: ", j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,100)
plt.title(specific_title)
plt.ylabel('Percentage of infected',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
elif option == 1:
nb = 0
Infected_Trials = np.zeros((n_sims,sim_end+1))
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Mild = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start+1,combo_end,2):
nb = nb+1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Age group: ", group_vec_age[nb-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv( base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline: ", j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,100)
plt.title(specific_title)
plt.ylabel('Percentage of infected',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
else:
nb = 0
Infected_Trials = np.zeros((n_sims,sim+end+1))
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Mild = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Documented = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_documented.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline 0: No intervention")
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color='gray',linestyle='-.')
for j in range(combo_start,combo_end):
nb +=1
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Age group: ", group_vec_age[j-1])
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials, color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
Infected_Trials = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Mild = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_mild.csv',delimiter=' ',header=None)
Severe = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_severe.csv',delimiter=' ',header=None)
Critical = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_critical.csv',delimiter=' ',header=None)
R = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_recovered.csv',delimiter=' ',header=None)
D = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
Infected_Trials[i,:] = Mild+Severe+Critical+R+D
Infected_Trials = Infected_Trials.mean(axis=0)
Infected_Trials = Infected_Trials/pop_size*100.
print("Baseline ",j-1)
print("% infected on lockdown day: ", Infected_Trials[t_lockdown_vec[0]])
print("% infected today: ", Infected_Trials[today])
print("% infected at the end of the simulation: ", Infected_Trials[sim_end])
plt.plot(Infected_Trials,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,100)
plt.title(specific_title)
plt.ylabel('Percentage of infected',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
```
### Function to be used to plot the projected number of deaths over time, when physical distancing is in place
### Figures 9(c) and 9(d)
```
def death_age_group_node_removal_lockdown(group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title):
if option == 2:
nb = 0
D=np.zeros((n_sims,sim_end+1))
# baseline
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Deaths = pd.read_csv(base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Baseline 0: No intervention")
print("# of deaths on lockdown day", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
# not baseline
for j in range(combo_start,combo_end,2):
nb +=1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# new baseline - 50% or 100% of the population of an age group is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Baseline: ",j-1)
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0, linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,400)
plt.title(specific_title)
plt.ylabel('Total deaths (thousands)',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
elif option == 1:
nb = 0
# Baseline
D=np.zeros((n_sims,sim_end+1))
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Deaths = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Baseline: No intervention")
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
# Per age group
for j in range(combo_start+1,combo_end,2):
nb = nb +1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: ", D[t_lockdown_vec[0]])
print("# of deaths today: ", D[today])
print("# of deaths at the end of the simulation: ", D[sim_end])
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Baseline: ", j-1)
print("# of deaths on lockdown day: " + str(D[t_lockdown_vec[0]]))
print("# of deaths today: " + str(D[today]))
print("# of deaths at the end of the simulation: " + str(D[sim_end]))
D = D/1000.
plt.plot(D,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(5)]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,400)
plt.title(specific_title)
plt.ylabel('Total deaths (thousands)',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
else:
nb = 0
# baseline
D = np.zeros((n_sims,sim_end+1))
base_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline_0_paramsweep_n10000000.0_i0_N'
base_folder = 'nolockdown_noage/'
for i in range(n_sims):
Deaths = pd.read_csv( base_folder + base_filename + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Baseline: No intervention")
print("# of deaths on lockdown day: " + str(D[t_lockdown_vec[0]]))
print("# of deaths today: " + str(D[today]))
print("# of deaths at the end of the simulation: " + str(D[sim_end]))
D = D/1000.
plt.plot(D,color='gray',linestyle='-.')
# Per age group
for j in range(combo_start,combo_end):
nb +=1
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(folder + filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:]=Deaths
D = D.mean(axis=0)
print("Age group: ", group_vec_age[nb-1])
print("# of deaths on lockdown day: " + str(D[t_lockdown_vec[0]]))
print("# of deaths today: " + str(D[today]))
print("# of deaths at the end of the simulation: " + str(D[sim_end]))
D = D/1000.
plt.plot(D,color=color_list_shahin[nb-1])
# new baseline - 50% population is isolated
base2_filename = 'lombardy_distributed_agepolicy_nolockdown_baseline2_0_paramsweep_n10000000.0_i'
base2_folder = 'nolockdown_fullisolation/'
for j in range(2,4):
D = np.zeros((n_sims,sim_end+1))
for i in range(n_sims):
Deaths = pd.read_csv(base2_folder + base2_filename + str(j) + '_N' + str(i) + '_p0.029_m4_s22_deaths.csv',delimiter=' ',header=None)
D[i,:] = Deaths
D = D.mean(axis=0)
print("Baseline: ",j-1)
print("# of deaths on lockdown day:" + str(t_lockdown_vec[0]))
print("# of deaths today: " + str(D[today]))
print("# of deaths at the end of the simulation: "+ str(D[sim_end]))
D = D/1000.
plt.plot(D,color=color_list_add[j-2],linestyle='-.')
plt.axvline(t_lockdown_vec[0], 0,linestyle='--',color='red')
plt.legend(['Absence of\nintervention']+['Ages ' + str(group_vec_age[i]) for i in range(len(group_vec_age))]+['All ages\n50\% confined','All ages\n100\% confined'],fontsize=13)
plt.ylim(0,400)
plt.title(specific_title)
plt.ylabel('Total deaths (thousands)',fontsize=23)
plt.xticks(fontsize=17)
plt.yticks(fontsize=17)
plt.xlabel('Days since patient zero',fontsize=23)
return(plt)
```
## Figure 8(a)
```
# Mean time to isolation 4.6 and 50% of age category removed
t_lockdown_vec = [46]
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename1 = 'lombardy_distributed_agepolicy_0_paramsweep_n10000000.0_i'
filename2 = 'lombardy_distributed_agepolicy_1_paramsweep_n10000000.0_i'
folder1 = 'perc_policy_results/run1/'
folder2 = 'perc_policy_results/run2/'
option = 2
specific_title = ''
perc_infected_age_group_node_removal(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder1, folder2, filename1, filename2, option, specific_title)
```
## Figure 8(c)
```
# Mean time to isolation 4.6 and 50% of age category removed
t_lockdown_vec = [46]
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename1 = 'lombardy_distributed_agepolicy_0_paramsweep_n10000000.0_i'
filename2 = 'lombardy_distributed_agepolicy_1_paramsweep_n10000000.0_i'
folder1 = 'perc_policy_results/run1/'
folder2 = 'perc_policy_results/run2/'
option = 2
#specific_title = 'Mean Time to Isolation = 4.6 days for all' + '\n50% stay home, per age group'
specific_title = ''
death_age_group_node_removal( group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder1, folder2, filename1, filename2, option, specific_title)
```
## Figure 8(b)
```
# Mean time to isolation 4.6 and 100% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename1 = 'lombardy_distributed_agepolicy_0_paramsweep_n10000000.0_i'
filename2 = 'lombardy_distributed_agepolicy_1_paramsweep_n10000000.0_i'
folder1 = 'perc_policy_results/run1/'
folder2 = 'perc_policy_results/run2/'
option = 1
specific_title = ''
perc_infected_age_group_node_removal(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder1, folder2, filename1, filename2, option, specific_title)
```
## Figure 8(d)
```
# Mean time to isolation 4.6 and 100% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename1 = 'lombardy_distributed_agepolicy_0_paramsweep_n10000000.0_i'
filename2 = 'lombardy_distributed_agepolicy_1_paramsweep_n10000000.0_i'
folder1 = 'perc_policy_results/run1/'
folder2 = 'perc_policy_results/run2/'
option = 1
specific_title = ''
death_age_group_node_removal(group_vec_age,t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder1, folder2, filename1, filename2, option, specific_title)
```
## Figure 9(a)
```
# Mean time to isolation 4.6 and 50% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
# As of March 29 of 2020
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename = 'lombardy_distributed_agepolicy_yeslockdown_0_paramsweep_n10000000.0_i'
folder = 'lockdown_perc_policy_results/'
option = 2
specific_title = ''
perc_infected_age_group_node_removal_lockdown(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title)
```
## Figure 9(c)
```
# Mean time to isolation 4.6 and 50% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
# As of March 29 of 2020
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename = 'lombardy_distributed_agepolicy_yeslockdown_0_paramsweep_n10000000.0_i'
folder = 'lockdown_perc_policy_results/'
option = 2
specific_title = ''
death_age_group_node_removal_lockdown(group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title)
```
## Figure 9(b)
```
# Mean time to isolation 4.6 and 100% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename = 'lombardy_distributed_agepolicy_yeslockdown_0_paramsweep_n10000000.0_i'
folder = 'lockdown_perc_policy_results/'
option = 1
# Lombardy - Time of Lockdown = 46 days\n, \nInfected = Mild+Severe+Critical+R+D
#specific_title = 'Mean Time to Isolation = 4.6 days for all' + '\n100% stay home, per age group' + '\n+ Social distance increased by a factor of 2'
specific_title = ''
perc_infected_age_group_node_removal_lockdown(pop_size, group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title)
```
## Figure 9(d)
```
# Mean time to isolation 4.6 and 100% of age category removed
t_lockdown_vec = [46]
n_sims = 100
sim_end = 119
today = 67
group_vec_age = ['0-14','15-29','30-49','50-69','70+']
combo_start = 0
combo_end = 10
pop_size = 10000000
filename = 'lombardy_distributed_agepolicy_yeslockdown_0_paramsweep_n10000000.0_i'
folder = 'lockdown_perc_policy_results/'
option = 1
specific_title = ''
death_age_group_node_removal_lockdown(group_vec_age, t_lockdown_vec, n_sims, sim_end, today, combo_start, combo_end, folder, filename, option, specific_title)
```
|
github_jupyter
|
```
%load_ext autoreload
%autoreload 2
```
# Sampling from a Bayesian network: an open problem
A Bayesian network encodes a probability distribution. It is often desirable to be able to sample from a Bayesian network. The most common way to do this is via forward sampling (also called prior sampling). It's a really dumb algorithm that is trivial to implement. You just loop over the nodes in breadth-first order and sample a value each node, conditioning on the parents (which have already been sampled).
The problem with forward sampling is that impossible situations can arise for some networks. Basically, forward sampling doesn't ensure that the produced samples are *valid*. The easiest way to grok this is via some examples.
## Example 1
```
import hedgehog as hh
import pandas as pd
def example_1():
X = pd.DataFrame(
[
[True, True, True],
[False, False, False]
],
columns=['A', 'B', 'C']
)
bn = hh.BayesNet(
(['A', 'B'], 'C')
)
bn.fit(X)
return bn
bn = example_1()
bn
bn.full_joint_dist()
```
The problem with forward sampling is this case is that if we sample from A and then B independently, then we can end up by sampling pairs (A, B) that don't exist. This will raise an error when we condition P(C) on its parents.
In `hedhehog`, this will raise a `KeyError` when `sample` is called because the distribution that corresponds to `(A=False, B=True)` doesn't exist.
```
while True:
try:
bn.sample()
except KeyError:
print('Yep, told you.')
break
```
## Example 2
```
import hedgehog as hh
import pandas as pd
def example_2():
X = pd.DataFrame(
[
[1, 1, 1, 1],
[2, 1, 2, 1]
],
columns=['A', 'B', 'C', 'D']
)
bn = hh.BayesNet(
('A', 'B'),
('B', 'C'),
(['A', 'C'], 'D')
)
bn.fit(X)
return bn
bn = example_2()
bn
```
In this case, a problem will occur if we sample `(A, 1)`, then `(B, 1)`, then `(C, 2)`. Indeed, `(A, 1)` and `(C, 1)` have never been seen so there's now way of sampling `D`.
```
while True:
try:
bn.sample()
except KeyError:
print('Yep, told you.')
break
```
One way to circumvent these issues would be to sample from the full joint distribution. But this is too costly. Another way is to add a prior distribution by supposing that every combination occurred once, but that's not elegant.
Ideally we would like to have some way of doing forward sampling that only produces valid data. This is still an open question for me.
|
github_jupyter
|
```
import time
start = time.perf_counter()
import tensorflow as tf
import pickle
import import_ipynb
import os
from model import Model
from utils import build_dict, build_dataset, batch_iter
embedding_size=300
num_hidden = 300
num_layers = 3
learning_rate = 0.001
beam_width = 10
keep_prob = 0.8
glove = True
batch_size=256
num_epochs=10
if not os.path.exists("saved_model"):
os.mkdir("saved_model")
else:
old_model_checkpoint_path = open('saved_model/checkpoint', 'r')
old_model_checkpoint_path = "".join(["saved_model/",old_model_checkpoint_path.read().splitlines()[0].split('"')[1]])
print("Building dictionary...")
word_dict, reversed_dict, article_max_len, summary_max_len = build_dict("train", toy=True)
print("Loading training dataset...")
train_x, train_y = build_dataset("train", word_dict, article_max_len, summary_max_len, toy=True)
with tf.Session() as sess:
model = Model(reversed_dict, article_max_len, summary_max_len, embedding_size, num_hidden, num_layers, learning_rate, beam_width, keep_prob, glove)
sess.run(tf.global_variables_initializer())
saver = tf.train.Saver(tf.global_variables())
if 'old_model_checkpoint_path' in globals():
print("Continuing from previous trained model:" , old_model_checkpoint_path , "...")
saver.restore(sess, old_model_checkpoint_path )
batches = batch_iter(train_x, train_y, batch_size, num_epochs)
num_batches_per_epoch = (len(train_x) - 1) // batch_size + 1
print("\nIteration starts.")
print("Number of batches per epoch :", num_batches_per_epoch)
for batch_x, batch_y in batches:
batch_x_len = list(map(lambda x: len([y for y in x if y != 0]), batch_x))
batch_decoder_input = list(map(lambda x: [word_dict["<s>"]] + list(x), batch_y))
batch_decoder_len = list(map(lambda x: len([y for y in x if y != 0]), batch_decoder_input))
batch_decoder_output = list(map(lambda x: list(x) + [word_dict["</s>"]], batch_y))
batch_decoder_input = list(
map(lambda d: d + (summary_max_len - len(d)) * [word_dict["<padding>"]], batch_decoder_input))
batch_decoder_output = list(
map(lambda d: d + (summary_max_len - len(d)) * [word_dict["<padding>"]], batch_decoder_output))
train_feed_dict = {
model.batch_size: len(batch_x),
model.X: batch_x,
model.X_len: batch_x_len,
model.decoder_input: batch_decoder_input,
model.decoder_len: batch_decoder_len,
model.decoder_target: batch_decoder_output
}
_, step, loss = sess.run([model.update, model.global_step, model.loss], feed_dict=train_feed_dict)
if step % 1000 == 0:
print("step {0}: loss = {1}".format(step, loss))
if step % num_batches_per_epoch == 0:
hours, rem = divmod(time.perf_counter() - start, 3600)
minutes, seconds = divmod(rem, 60)
saver.save(sess, "./saved_model/model.ckpt", global_step=step)
print(" Epoch {0}: Model is saved.".format(step // num_batches_per_epoch),
"Elapsed: {:0>2}:{:0>2}:{:05.2f}".format(int(hours),int(minutes),seconds) , "\n")
```
|
github_jupyter
|
# Python Bindings Demo
This is a very simple demo / playground / testing site for the Python Bindings for BART.
This is mainly used to show off Numpy interoperability and give a basic sense for how more complex tools will look in Python.
## Overview
Currently, Python users can interact with BART via a command-line wrapper. For example, the following line of Python code generates a simple Shepp-Logan phantom in K-Space and reconstructs the original image via inverse FFT.
```
shepp_ksp = bart(1, 'phantom -k -x 128')
shepp_recon = bart(1, 'fft -i 3' shepp_recon)
```
#### The Python bindings, `bartpy`, build on this wrapper in the following ways:
- 'Pythonic' interface with explicit functions and objects
- (Mostly) automated generation to minimize the maintenance burden
- Access to lower-level operators (e.g., `linops` submodule) to allow users to use BART functions seamlessly alongside Python libraries like Numpy, Sigpy, SciPy, Tensorflow or pyTorch
- RAM-based memory management
- Current wrapper writes data to disk, invokes the BART tools from the command line, and then reads data from disk
- Memory-based approach is ostensibly faster
## Getting Started
To begin, we import `numpy` and `matplotlib` for array manipulation and data visualization. We will then import the Python bindings
```
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits import mplot3d
%matplotlib inline
```
### BART-related Imports
We will now import `bartpy` tools for generating phantoms and performing the Fast Fourier Transform (FFT), as well as utilities for interacting with `.cfl` files.
```
from bartpy.simu.phantom import phantom
from bartpy.num.fft import fft, ifft
from bartpy.utils.cfl import readcfl, writecfl
```
## A closer look
<span style="font-size: 1.3em;">`phantom(dims, ksp, d3, ptype)`</span>
- `dims`: iterable specifying dimensions of the phantom. Cannot exceed 16, and follows BART dimension conventions
- `ksp`: boolean value indicating whether or not to generate the phantom in k-space
- `d3`: boolean value indicating whether or not to generate a 3D phantom
- `ptype`: Specifies type of phantom.
```
phantom
shepp = phantom([128, 128], ksp=False, d3=False)
plt.imshow(shepp))
```
## Reconstruction via FFT
### Command Line
Here is a simple recon task completed with BART on the command line.
```
!bart phantom -x 128 -k -B logo
!bart fft -i 3 logo logo_recon
gnd = readcfl('logo_recon')
plt.imshow(abs(gnd.T))
```
### Pure Python
Now here is our task completed entirely in Python, using `bartpy`
<span style="color: red">FIXME: The order of dimensions
is wrong</span>
```
logo_ksp = phantom([128, 128], ksp=True, ptype='bart')
plt.imshow(np.log(abs(logo_ksp)))
logo_recon = ifft(logo_ksp, flags=3)
plt.imshow(abs(logo_recon))
```
This is a brief example of the more 'Pythonic' approach offered by the Python bindings.
|
github_jupyter
|
## Scrape Archived Mini Normals from Mafiascum.net
#### Scrapy Structure/Lingo:
**Spiders** extract data **items**, which Scrapy send one by one to a configured **item pipeline** (if there is possible) to do post-processing on the items.)
## Import relevant packages...
```
import scrapy
import math
import logging
import json
from scrapy.crawler import CrawlerProcess
from scrapy.spiders import CrawlSpider, Rule
from scrapy.item import Item, Field
from scrapy.selector import Selector
```
## Initial variables...
```
perpage = 25
class PostItem(scrapy.Item):
pagelink = scrapy.Field()
forum = scrapy.Field()
thread = scrapy.Field()
number = scrapy.Field()
timestamp = scrapy.Field()
user = scrapy.Field()
content = scrapy.Field()
```
## Define what happens to scrape output...
```
# The following pipeline stores all scraped items (from all spiders)
# into a single items.jl file, containing one item per line serialized
# in JSON format:
class JsonWriterPipeline(object):
# operations performed when spider starts
def open_spider(self, spider):
self.file = open('posts.jl', 'w')
# when the spider finishes
def close_spider(self, spider):
self.file.close()
# when the spider yields an item
def process_item(self, item, spider):
line = json.dumps(dict(item)) + "\n"
self.file.write(line)
return item
```
## Define spider...
```
class MafiaScumSpider(scrapy.Spider):
name = 'mafiascum'
# define set of threads we're going to scrape from (ie all of them)
start_urls = [each[:each.find('\n')] for each in open('archive.txt').read().split('\n\n\n')]
# settings
custom_settings = {'LOG_LEVEL': logging.WARNING,
'ITEM_PIPELINES': {'__main__.JsonWriterPipeline': 1}}
# get page counts and then do the REAL parse on every single page
def parse(self, response):
# find page count
try:
postcount = Selector(response).xpath(
'//div[@class="pagination"]/text()').extract()
postcount = int(postcount[0][4:postcount[0].find(' ')])
# yield parse for every page of thread
for i in range(math.ceil(postcount/perpage)):
yield scrapy.Request(response.url+'&start='+str(i*perpage),
callback=self.parse_page)
except IndexError: # if can't, the thread probably doesn't exist
return
def parse_page(self, response):
# scan through posts on page and yield Post items for each
sel = Selector(response)
location = sel.xpath('//div[@id="page-body"]/h2/a/@href').extract()[0]
forum = location[location.find('f=')+2:location.find('&t=')]
if location.count('&') == 1:
thread = location[location.find('&t=')+3:]
elif location.count('&') == 2:
thread = location[
location.find('&t=')+3:location.rfind('&')]
posts = (sel.xpath('//div[@class="post bg1"]') +
sel.xpath('//div[@class="post bg2"]'))
for p in posts:
post = PostItem()
post['forum'] = forum
post['thread'] = thread
post['pagelink'] = response.url
try:
post['number'] = p.xpath(
'div/div[@class="postbody"]/p/a[2]/strong/text()').extract()[0][1:]
except IndexError:
post['number'] = p.xpath(
'div[@class="postbody"]/p/a[2]/strong/text()').extract()[0][1:]
try:
post['timestamp'] = p.xpath(
'div/div/p/text()[4]').extract()[0][23:-4]
except IndexError:
post['timestamp'] = p.xpath(
'div[@class="postbody"]/p/text()[4]').extract()[0][23:-4]
try:
post['user'] = p.xpath('div/div/dl/dt/a/text()').extract()[0]
except IndexError:
post['user'] = '<<DELETED_USER>>'
try:
post['content'] = p.xpath(
'div/div/div[@class="content"]').extract()[0][21:-6]
except IndexError:
post['content'] = p.xpath(
'div[@class="postbody"]/div[@class="content"]').extract()[0][21:-6]
yield post
```
## Start scraping...
```
process = CrawlerProcess({
'USER_AGENT': 'Mozilla/4.0 (compatible; MSIE 7.0; Windows NT 5.1)'
})
process.crawl(MafiaScumSpider)
process.start()
```
...and output should be a json file in same directory as this notebook!
## Leftover Code...
```
# open mini normal archive
# ??? i don't remember what this does; probably helped me collect archive links some time ago
runthis = False
if runthis:
# relevant packages
from selenium import webdriver
from scrapy.selector import Selector
import re
# configure browser
options = webdriver.ChromeOptions()
options.binary_location = '/Applications/Google Chrome Canary.app/Contents/MacOS/Google Chrome Canary'
options.add_argument('window-size=800x841')
driver = webdriver.Chrome(chrome_options=options)
# get the thread titles and links
links = []
titles = []
for i in range(0, 400, 100):
driver.get('https://forum.mafiascum.net/viewforum.php?f=53&start=' + str(i))
sel = Selector(text=driver.page_source)
links += sel.xpath('//div[@class="forumbg"]/div/ul[@class="topiclist topics"]/li/dl/dt/a[1]/@href').extract()
titles += sel.xpath('//div[@class="forumbg"]/div/ul[@class="topiclist topics"]/li/dl/dt/a[1]/text()').extract()
# formatting, excluding needless threads...
titles = titles[1:]
links = links[1:]
del links[titles.index('Mini Normal Archives')]
del titles[titles.index('Mini Normal Archives')]
titles = [re.search(r'\d+', each).group(0) for each in titles]
# match txt archive game numbers with forum archive game numbers to find links
f = open('archive.txt', 'r')
txtarchives = f.read().split('\n\n\n')
numbers = [re.search(r'\d+', each[:each.find('\n')]).group(0) for each in txtarchives]
f.close()
# store the result...
for i, n in enumerate(numbers):
txtarchives[i] = 'http://forum.mafiascum.net' + links[titles.index(n)][1:] + '\n' + txtarchives[i]
f = open('archive2.txt', 'w')
f.write('\n\n\n'.join(txtarchives))
f.close()
```
|
github_jupyter
|
While going through our script we will gradually understand the use of this packages
```
import tensorflow as tf #no need to describe ;)
import numpy as np #allows array operation
import pandas as pd #we will use it to read and manipulate files and columns content
from nltk.corpus import stopwords #provides list of english stopwords
stop = stopwords.words('english')
#PRINT VERSION!!
tf.__version__
```
To do this notebook we will use New York Times user comments (from Kaggle Datasets).
When we will create the language classifier we will use other data but now let's rely on an english natural language source, so now we read the data.
```
#PLEASE DOWNLOAD THE FILE HERE: https://www.kaggle.com/aashita/nyt-comments
train = pd.read_csv('CommentsApril2017.csv')
```
Let's have a quick look at the data trying to find what is the column that we need.
Looks like commentBody is the right candidate.
```
train.head()
```
now we first put everything to lowercase and then replace undesired characters
```
train['commentBody_lower'] = train["commentBody"].str.lower()
train['commentBody_no_punctiation'] = train['commentBody_lower'].str.replace('[^\w\s]','')
```
let's check how the text looks like now!
Well everything is lowercase and no "ugly characters"
```
train['commentBody_no_punctiation'].head()
```
Now we remove stopwords and then fill empy cells with "fillna" word.
```
train['commentBody_no_stopwords'] = train['commentBody_no_punctiation'].apply(lambda x: ' '.join([word for word in x.split() if word not in (stop)]))
train["commentBody_no_stopwords"] = train["commentBody_no_stopwords"].fillna("fillna")
```
This is how our cleaned text looks like we can see that everything is lowercase and the stopwords are missing, for example "this". Now let's go back to slides.
```
train['commentBody_no_stopwords'].head()
tf_train = train
```
We first assign our current data frame to another to keep track of our work then we read the first sentence and count words that result to be 21
```
tf_train['commentBody_no_stopwords'][1]
tf_train['commentBody_no_stopwords'][1].count(' ')
max_features=5000 #we set maximum number of words to 5000
maxlen=100 #and maximum sequence length to 100
tok = tf.keras.preprocessing.text.Tokenizer(num_words=max_features) #tokenizer step
tok.fit_on_texts(list(tf_train['commentBody_no_stopwords'])) #fit to cleaned text
tf_train=tok.texts_to_sequences(list(tf_train['commentBody_no_stopwords'])) #this is how we create sequences
print(type(tf_train)) #we see that the type is now list
print(len(tf_train[1])) #we see that the number of words of the sentence is decreased to 16
tf_train[1] #and this is how our sentece looks like now, exactly a sequence of integers
tf_train=tf.keras.preprocessing.sequence.pad_sequences(tf_train, maxlen=maxlen) #let's execute pad step
print(len(tf_train[1]))
tf_train[1] #this is how our sentece looks like after the pad step we don't have anymore 16 words but 100 (equivalent to maxlen)
train['commentBody_no_stopwords'][1] #let's look at the input text
tf_train = pd.DataFrame(tf_train)
tf_train.head() #let's look at the final matrix that will use as an input for our deep learning algorithms, do you remember
#how original text looked like?
```
|
github_jupyter
|
# Autonomous driving - Car detection
Welcome to your week 3 programming assignment. You will learn about object detection using the very powerful YOLO model. Many of the ideas in this notebook are described in the two YOLO papers: Redmon et al., 2016 (https://arxiv.org/abs/1506.02640) and Redmon and Farhadi, 2016 (https://arxiv.org/abs/1612.08242).
**You will learn to**:
- Use object detection on a car detection dataset
- Deal with bounding boxes
Run the following cell to load the packages and dependencies that are going to be useful for your journey!
```
import argparse
import os
import matplotlib.pyplot as plt
from matplotlib.pyplot import imshow
import scipy.io
import scipy.misc
import numpy as np
import pandas as pd
import PIL
import tensorflow as tf
from keras import backend as K
from keras.layers import Input, Lambda, Conv2D
from keras.models import load_model, Model
from yolo_utils import read_classes, read_anchors, generate_colors, preprocess_image, draw_boxes, scale_boxes
from yad2k.models.keras_yolo import yolo_head, yolo_boxes_to_corners, preprocess_true_boxes, yolo_loss, yolo_body
%matplotlib inline
```
**Important Note**: As you can see, we import Keras's backend as K. This means that to use a Keras function in this notebook, you will need to write: `K.function(...)`.
## 1 - Problem Statement
You are working on a self-driving car. As a critical component of this project, you'd like to first build a car detection system. To collect data, you've mounted a camera to the hood (meaning the front) of the car, which takes pictures of the road ahead every few seconds while you drive around.
<center>
<video width="400" height="200" src="nb_images/road_video_compressed2.mp4" type="video/mp4" controls>
</video>
</center>
<caption><center> Pictures taken from a car-mounted camera while driving around Silicon Valley. <br> We would like to especially thank [drive.ai](https://www.drive.ai/) for providing this dataset! Drive.ai is a company building the brains of self-driving vehicles.
</center></caption>
<img src="nb_images/driveai.png" style="width:100px;height:100;">
You've gathered all these images into a folder and have labelled them by drawing bounding boxes around every car you found. Here's an example of what your bounding boxes look like.
<img src="nb_images/box_label.png" style="width:500px;height:250;">
<caption><center> <u> **Figure 1** </u>: **Definition of a box**<br> </center></caption>
If you have 80 classes that you want YOLO to recognize, you can represent the class label $c$ either as an integer from 1 to 80, or as an 80-dimensional vector (with 80 numbers) one component of which is 1 and the rest of which are 0. The video lectures had used the latter representation; in this notebook, we will use both representations, depending on which is more convenient for a particular step.
In this exercise, you will learn how YOLO works, then apply it to car detection. Because the YOLO model is very computationally expensive to train, we will load pre-trained weights for you to use.
## 2 - YOLO
YOLO ("you only look once") is a popular algoritm because it achieves high accuracy while also being able to run in real-time. This algorithm "only looks once" at the image in the sense that it requires only one forward propagation pass through the network to make predictions. After non-max suppression, it then outputs recognized objects together with the bounding boxes.
### 2.1 - Model details
First things to know:
- The **input** is a batch of images of shape (m, 608, 608, 3)
- The **output** is a list of bounding boxes along with the recognized classes. Each bounding box is represented by 6 numbers $(p_c, b_x, b_y, b_h, b_w, c)$ as explained above. If you expand $c$ into an 80-dimensional vector, each bounding box is then represented by 85 numbers.
We will use 5 anchor boxes. So you can think of the YOLO architecture as the following: IMAGE (m, 608, 608, 3) -> DEEP CNN -> ENCODING (m, 19, 19, 5, 85).
Lets look in greater detail at what this encoding represents.
<img src="nb_images/architecture.png" style="width:700px;height:400;">
<caption><center> <u> **Figure 2** </u>: **Encoding architecture for YOLO**<br> </center></caption>
If the center/midpoint of an object falls into a grid cell, that grid cell is responsible for detecting that object.
Since we are using 5 anchor boxes, each of the 19 x19 cells thus encodes information about 5 boxes. Anchor boxes are defined only by their width and height.
For simplicity, we will flatten the last two last dimensions of the shape (19, 19, 5, 85) encoding. So the output of the Deep CNN is (19, 19, 425).
<img src="nb_images/flatten.png" style="width:700px;height:400;">
<caption><center> <u> **Figure 3** </u>: **Flattening the last two last dimensions**<br> </center></caption>
Now, for each box (of each cell) we will compute the following elementwise product and extract a probability that the box contains a certain class.
<img src="nb_images/probability_extraction.png" style="width:700px;height:400;">
<caption><center> <u> **Figure 4** </u>: **Find the class detected by each box**<br> </center></caption>
Here's one way to visualize what YOLO is predicting on an image:
- For each of the 19x19 grid cells, find the maximum of the probability scores (taking a max across both the 5 anchor boxes and across different classes).
- Color that grid cell according to what object that grid cell considers the most likely.
Doing this results in this picture:
<img src="nb_images/proba_map.png" style="width:300px;height:300;">
<caption><center> <u> **Figure 5** </u>: Each of the 19x19 grid cells colored according to which class has the largest predicted probability in that cell.<br> </center></caption>
Note that this visualization isn't a core part of the YOLO algorithm itself for making predictions; it's just a nice way of visualizing an intermediate result of the algorithm.
Another way to visualize YOLO's output is to plot the bounding boxes that it outputs. Doing that results in a visualization like this:
<img src="nb_images/anchor_map.png" style="width:200px;height:200;">
<caption><center> <u> **Figure 6** </u>: Each cell gives you 5 boxes. In total, the model predicts: 19x19x5 = 1805 boxes just by looking once at the image (one forward pass through the network)! Different colors denote different classes. <br> </center></caption>
In the figure above, we plotted only boxes that the model had assigned a high probability to, but this is still too many boxes. You'd like to filter the algorithm's output down to a much smaller number of detected objects. To do so, you'll use non-max suppression. Specifically, you'll carry out these steps:
- Get rid of boxes with a low score (meaning, the box is not very confident about detecting a class)
- Select only one box when several boxes overlap with each other and detect the same object.
### 2.2 - Filtering with a threshold on class scores
You are going to apply a first filter by thresholding. You would like to get rid of any box for which the class "score" is less than a chosen threshold.
The model gives you a total of 19x19x5x85 numbers, with each box described by 85 numbers. It'll be convenient to rearrange the (19,19,5,85) (or (19,19,425)) dimensional tensor into the following variables:
- `box_confidence`: tensor of shape $(19 \times 19, 5, 1)$ containing $p_c$ (confidence probability that there's some object) for each of the 5 boxes predicted in each of the 19x19 cells.
- `boxes`: tensor of shape $(19 \times 19, 5, 4)$ containing $(b_x, b_y, b_h, b_w)$ for each of the 5 boxes per cell.
- `box_class_probs`: tensor of shape $(19 \times 19, 5, 80)$ containing the detection probabilities $(c_1, c_2, ... c_{80})$ for each of the 80 classes for each of the 5 boxes per cell.
**Exercise**: Implement `yolo_filter_boxes()`.
1. Compute box scores by doing the elementwise product as described in Figure 4. The following code may help you choose the right operator:
```python
a = np.random.randn(19*19, 5, 1)
b = np.random.randn(19*19, 5, 80)
c = a * b # shape of c will be (19*19, 5, 80)
```
2. For each box, find:
- the index of the class with the maximum box score ([Hint](https://keras.io/backend/#argmax)) (Be careful with what axis you choose; consider using axis=-1)
- the corresponding box score ([Hint](https://keras.io/backend/#max)) (Be careful with what axis you choose; consider using axis=-1)
3. Create a mask by using a threshold. As a reminder: `([0.9, 0.3, 0.4, 0.5, 0.1] < 0.4)` returns: `[False, True, False, False, True]`. The mask should be True for the boxes you want to keep.
4. Use TensorFlow to apply the mask to box_class_scores, boxes and box_classes to filter out the boxes we don't want. You should be left with just the subset of boxes you want to keep. ([Hint](https://www.tensorflow.org/api_docs/python/tf/boolean_mask))
Reminder: to call a Keras function, you should use `K.function(...)`.
```
# GRADED FUNCTION: yolo_filter_boxes
def yolo_filter_boxes(box_confidence, boxes, box_class_probs, threshold = .6):
"""Filters YOLO boxes by thresholding on object and class confidence.
Arguments:
box_confidence -- tensor of shape (19, 19, 5, 1)
boxes -- tensor of shape (19, 19, 5, 4)
box_class_probs -- tensor of shape (19, 19, 5, 80)
threshold -- real value, if [ highest class probability score < threshold], then get rid of the corresponding box
Returns:
scores -- tensor of shape (None,), containing the class probability score for selected boxes
boxes -- tensor of shape (None, 4), containing (b_x, b_y, b_h, b_w) coordinates of selected boxes
classes -- tensor of shape (None,), containing the index of the class detected by the selected boxes
Note: "None" is here because you don't know the exact number of selected boxes, as it depends on the threshold.
For example, the actual output size of scores would be (10,) if there are 10 boxes.
"""
# Step 1: Compute box scores
### START CODE HERE ### (≈ 1 line)
box_scores = box_confidence * box_class_probs
### END CODE HERE ###
# Step 2: Find the box_classes thanks to the max box_scores, keep track of the corresponding score
### START CODE HERE ### (≈ 2 lines)
box_classes = K.argmax(box_scores, axis=-1)
box_class_scores = K.max(box_scores,axis=-1)
### END CODE HERE ###
# Step 3: Create a filtering mask based on "box_class_scores" by using "threshold". The mask should have the
# same dimension as box_class_scores, and be True for the boxes you want to keep (with probability >= threshold)
### START CODE HERE ### (≈ 1 line)
filtering_mask = box_class_scores >= threshold
### END CODE HERE ###
# Step 4: Apply the mask to scores, boxes and classes
### START CODE HERE ### (≈ 3 lines)
scores = tf.boolean_mask(box_class_scores, filtering_mask)
boxes = tf.boolean_mask(boxes, filtering_mask)
classes = tf.boolean_mask(box_classes, filtering_mask)
### END CODE HERE ###
return scores, boxes, classes
with tf.Session() as test_a:
box_confidence = tf.random_normal([19, 19, 5, 1], mean=1, stddev=4, seed = 1)
boxes = tf.random_normal([19, 19, 5, 4], mean=1, stddev=4, seed = 1)
box_class_probs = tf.random_normal([19, 19, 5, 80], mean=1, stddev=4, seed = 1)
scores, boxes, classes = yolo_filter_boxes(box_confidence, boxes, box_class_probs, threshold = 0.5)
print("scores[2] = " + str(scores[2].eval()))
print("boxes[2] = " + str(boxes[2].eval()))
print("classes[2] = " + str(classes[2].eval()))
print("scores.shape = " + str(scores.shape))
print("boxes.shape = " + str(boxes.shape))
print("classes.shape = " + str(classes.shape))
```
**Expected Output**:
<table>
<tr>
<td>
**scores[2]**
</td>
<td>
10.7506
</td>
</tr>
<tr>
<td>
**boxes[2]**
</td>
<td>
[ 8.42653275 3.27136683 -0.5313437 -4.94137383]
</td>
</tr>
<tr>
<td>
**classes[2]**
</td>
<td>
7
</td>
</tr>
<tr>
<td>
**scores.shape**
</td>
<td>
(?,)
</td>
</tr>
<tr>
<td>
**boxes.shape**
</td>
<td>
(?, 4)
</td>
</tr>
<tr>
<td>
**classes.shape**
</td>
<td>
(?,)
</td>
</tr>
</table>
### 2.3 - Non-max suppression ###
Even after filtering by thresholding over the classes scores, you still end up a lot of overlapping boxes. A second filter for selecting the right boxes is called non-maximum suppression (NMS).
<img src="nb_images/non-max-suppression.png" style="width:500px;height:400;">
<caption><center> <u> **Figure 7** </u>: In this example, the model has predicted 3 cars, but it's actually 3 predictions of the same car. Running non-max suppression (NMS) will select only the most accurate (highest probabiliy) one of the 3 boxes. <br> </center></caption>
Non-max suppression uses the very important function called **"Intersection over Union"**, or IoU.
<img src="nb_images/iou.png" style="width:500px;height:400;">
<caption><center> <u> **Figure 8** </u>: Definition of "Intersection over Union". <br> </center></caption>
**Exercise**: Implement iou(). Some hints:
- In this exercise only, we define a box using its two corners (upper left and lower right): (x1, y1, x2, y2) rather than the midpoint and height/width.
- To calculate the area of a rectangle you need to multiply its height (y2 - y1) by its width (x2 - x1)
- You'll also need to find the coordinates (xi1, yi1, xi2, yi2) of the intersection of two boxes. Remember that:
- xi1 = maximum of the x1 coordinates of the two boxes
- yi1 = maximum of the y1 coordinates of the two boxes
- xi2 = minimum of the x2 coordinates of the two boxes
- yi2 = minimum of the y2 coordinates of the two boxes
In this code, we use the convention that (0,0) is the top-left corner of an image, (1,0) is the upper-right corner, and (1,1) the lower-right corner.
```
# GRADED FUNCTION: iou
def iou(box1, box2):
"""Implement the intersection over union (IoU) between box1 and box2
Arguments:
box1 -- first box, list object with coordinates (x1, y1, x2, y2)
box2 -- second box, list object with coordinates (x1, y1, x2, y2)
"""
# Calculate the (y1, x1, y2, x2) coordinates of the intersection of box1 and box2. Calculate its Area.
### START CODE HERE ### (≈ 5 lines)
xi1 = np.maximum(box1[0],box2[0])
yi1 = np.maximum(box1[1],box2[1])
xi2 = np.minimum(box1[2],box2[2])
yi2 = np.minimum(box1[3],box2[3])
inter_area = (yi2-yi1) * (xi2-xi1)
### END CODE HERE ###
# Calculate the Union area by using Formula: Union(A,B) = A + B - Inter(A,B)
### START CODE HERE ### (≈ 3 lines)
box1_area = (box1[3]-box1[1]) * (box1[2]-box1[0])
box2_area = (box2[3]-box2[1]) * (box2[2]-box2[0])
union_area = box1_area + box2_area - inter_area
### END CODE HERE ###
# compute the IoU
### START CODE HERE ### (≈ 1 line)
iou = inter_area / union_area
### END CODE HERE ###
return iou
box1 = (2, 1, 4, 3)
box2 = (1, 2, 3, 4)
print("iou = " + str(iou(box1, box2)))
```
**Expected Output**:
<table>
<tr>
<td>
**iou = **
</td>
<td>
0.14285714285714285
</td>
</tr>
</table>
You are now ready to implement non-max suppression. The key steps are:
1. Select the box that has the highest score.
2. Compute its overlap with all other boxes, and remove boxes that overlap it more than `iou_threshold`.
3. Go back to step 1 and iterate until there's no more boxes with a lower score than the current selected box.
This will remove all boxes that have a large overlap with the selected boxes. Only the "best" boxes remain.
**Exercise**: Implement yolo_non_max_suppression() using TensorFlow. TensorFlow has two built-in functions that are used to implement non-max suppression (so you don't actually need to use your `iou()` implementation):
- [tf.image.non_max_suppression()](https://www.tensorflow.org/api_docs/python/tf/image/non_max_suppression)
- [K.gather()](https://www.tensorflow.org/api_docs/python/tf/gather)
```
# GRADED FUNCTION: yolo_non_max_suppression
def yolo_non_max_suppression(scores, boxes, classes, max_boxes = 10, iou_threshold = 0.5):
"""
Applies Non-max suppression (NMS) to set of boxes
Arguments:
scores -- tensor of shape (None,), output of yolo_filter_boxes()
boxes -- tensor of shape (None, 4), output of yolo_filter_boxes() that have been scaled to the image size (see later)
classes -- tensor of shape (None,), output of yolo_filter_boxes()
max_boxes -- integer, maximum number of predicted boxes you'd like
iou_threshold -- real value, "intersection over union" threshold used for NMS filtering
Returns:
scores -- tensor of shape (, None), predicted score for each box
boxes -- tensor of shape (4, None), predicted box coordinates
classes -- tensor of shape (, None), predicted class for each box
Note: The "None" dimension of the output tensors has obviously to be less than max_boxes. Note also that this
function will transpose the shapes of scores, boxes, classes. This is made for convenience.
"""
max_boxes_tensor = K.variable(max_boxes, dtype='int32') # tensor to be used in tf.image.non_max_suppression()
K.get_session().run(tf.variables_initializer([max_boxes_tensor])) # initialize variable max_boxes_tensor
# Use tf.image.non_max_suppression() to get the list of indices corresponding to boxes you keep
### START CODE HERE ### (≈ 1 line)
nms_indices = tf.image.non_max_suppression(boxes,scores,max_boxes,iou_threshold)
### END CODE HERE ###
# Use K.gather() to select only nms_indices from scores, boxes and classes
### START CODE HERE ### (≈ 3 lines)
scores = K.gather(scores,nms_indices)
boxes = K.gather(boxes,nms_indices)
classes = K.gather(classes,nms_indices)
### END CODE HERE ###
return scores, boxes, classes
with tf.Session() as test_b:
scores = tf.random_normal([54,], mean=1, stddev=4, seed = 1)
boxes = tf.random_normal([54, 4], mean=1, stddev=4, seed = 1)
classes = tf.random_normal([54,], mean=1, stddev=4, seed = 1)
scores, boxes, classes = yolo_non_max_suppression(scores, boxes, classes)
print("scores[2] = " + str(scores[2].eval()))
print("boxes[2] = " + str(boxes[2].eval()))
print("classes[2] = " + str(classes[2].eval()))
print("scores.shape = " + str(scores.eval().shape))
print("boxes.shape = " + str(boxes.eval().shape))
print("classes.shape = " + str(classes.eval().shape))
```
**Expected Output**:
<table>
<tr>
<td>
**scores[2]**
</td>
<td>
6.9384
</td>
</tr>
<tr>
<td>
**boxes[2]**
</td>
<td>
[-5.299932 3.13798141 4.45036697 0.95942086]
</td>
</tr>
<tr>
<td>
**classes[2]**
</td>
<td>
-2.24527
</td>
</tr>
<tr>
<td>
**scores.shape**
</td>
<td>
(10,)
</td>
</tr>
<tr>
<td>
**boxes.shape**
</td>
<td>
(10, 4)
</td>
</tr>
<tr>
<td>
**classes.shape**
</td>
<td>
(10,)
</td>
</tr>
</table>
### 2.4 Wrapping up the filtering
It's time to implement a function taking the output of the deep CNN (the 19x19x5x85 dimensional encoding) and filtering through all the boxes using the functions you've just implemented.
**Exercise**: Implement `yolo_eval()` which takes the output of the YOLO encoding and filters the boxes using score threshold and NMS. There's just one last implementational detail you have to know. There're a few ways of representing boxes, such as via their corners or via their midpoint and height/width. YOLO converts between a few such formats at different times, using the following functions (which we have provided):
```python
boxes = yolo_boxes_to_corners(box_xy, box_wh)
```
which converts the yolo box coordinates (x,y,w,h) to box corners' coordinates (x1, y1, x2, y2) to fit the input of `yolo_filter_boxes`
```python
boxes = scale_boxes(boxes, image_shape)
```
YOLO's network was trained to run on 608x608 images. If you are testing this data on a different size image--for example, the car detection dataset had 720x1280 images--this step rescales the boxes so that they can be plotted on top of the original 720x1280 image.
Don't worry about these two functions; we'll show you where they need to be called.
```
# GRADED FUNCTION: yolo_eval
def yolo_eval(yolo_outputs, image_shape = (720., 1280.), max_boxes=10, score_threshold=.6, iou_threshold=.5):
"""
Converts the output of YOLO encoding (a lot of boxes) to your predicted boxes along with their scores, box coordinates and classes.
Arguments:
yolo_outputs -- output of the encoding model (for image_shape of (608, 608, 3)), contains 4 tensors:
box_confidence: tensor of shape (None, 19, 19, 5, 1)
box_xy: tensor of shape (None, 19, 19, 5, 2)
box_wh: tensor of shape (None, 19, 19, 5, 2)
box_class_probs: tensor of shape (None, 19, 19, 5, 80)
image_shape -- tensor of shape (2,) containing the input shape, in this notebook we use (608., 608.) (has to be float32 dtype)
max_boxes -- integer, maximum number of predicted boxes you'd like
score_threshold -- real value, if [ highest class probability score < threshold], then get rid of the corresponding box
iou_threshold -- real value, "intersection over union" threshold used for NMS filtering
Returns:
scores -- tensor of shape (None, ), predicted score for each box
boxes -- tensor of shape (None, 4), predicted box coordinates
classes -- tensor of shape (None,), predicted class for each box
"""
### START CODE HERE ###
# Retrieve outputs of the YOLO model (≈1 line)
box_confidence, box_xy, box_wh, box_class_probs = yolo_outputs
# Convert boxes to be ready for filtering functions
boxes = yolo_boxes_to_corners(box_xy, box_wh)
# Use one of the functions you've implemented to perform Score-filtering with a threshold of score_threshold (≈1 line)
scores, boxes, classes = yolo_filter_boxes(box_confidence, boxes, box_class_probs, score_threshold)
# Scale boxes back to original image shape.
boxes = scale_boxes(boxes, image_shape)
# Use one of the functions you've implemented to perform Non-max suppression with a threshold of iou_threshold (≈1 line)
scores, boxes, classes = yolo_non_max_suppression(scores, boxes, classes, max_boxes, iou_threshold)
### END CODE HERE ###
return scores, boxes, classes
with tf.Session() as test_b:
yolo_outputs = (tf.random_normal([19, 19, 5, 1], mean=1, stddev=4, seed = 1),
tf.random_normal([19, 19, 5, 2], mean=1, stddev=4, seed = 1),
tf.random_normal([19, 19, 5, 2], mean=1, stddev=4, seed = 1),
tf.random_normal([19, 19, 5, 80], mean=1, stddev=4, seed = 1))
scores, boxes, classes = yolo_eval(yolo_outputs)
print("scores[2] = " + str(scores[2].eval()))
print("boxes[2] = " + str(boxes[2].eval()))
print("classes[2] = " + str(classes[2].eval()))
print("scores.shape = " + str(scores.eval().shape))
print("boxes.shape = " + str(boxes.eval().shape))
print("classes.shape = " + str(classes.eval().shape))
```
**Expected Output**:
<table>
<tr>
<td>
**scores[2]**
</td>
<td>
138.791
</td>
</tr>
<tr>
<td>
**boxes[2]**
</td>
<td>
[ 1292.32971191 -278.52166748 3876.98925781 -835.56494141]
</td>
</tr>
<tr>
<td>
**classes[2]**
</td>
<td>
54
</td>
</tr>
<tr>
<td>
**scores.shape**
</td>
<td>
(10,)
</td>
</tr>
<tr>
<td>
**boxes.shape**
</td>
<td>
(10, 4)
</td>
</tr>
<tr>
<td>
**classes.shape**
</td>
<td>
(10,)
</td>
</tr>
</table>
<font color='blue'>
**Summary for YOLO**:
- Input image (608, 608, 3)
- The input image goes through a CNN, resulting in a (19,19,5,85) dimensional output.
- After flattening the last two dimensions, the output is a volume of shape (19, 19, 425):
- Each cell in a 19x19 grid over the input image gives 425 numbers.
- 425 = 5 x 85 because each cell contains predictions for 5 boxes, corresponding to 5 anchor boxes, as seen in lecture.
- 85 = 5 + 80 where 5 is because $(p_c, b_x, b_y, b_h, b_w)$ has 5 numbers, and and 80 is the number of classes we'd like to detect
- You then select only few boxes based on:
- Score-thresholding: throw away boxes that have detected a class with a score less than the threshold
- Non-max suppression: Compute the Intersection over Union and avoid selecting overlapping boxes
- This gives you YOLO's final output.
## 3 - Test YOLO pretrained model on images
In this part, you are going to use a pretrained model and test it on the car detection dataset. As usual, you start by **creating a session to start your graph**. Run the following cell.
```
sess = K.get_session()
```
### 3.1 - Defining classes, anchors and image shape.
Recall that we are trying to detect 80 classes, and are using 5 anchor boxes. We have gathered the information about the 80 classes and 5 boxes in two files "coco_classes.txt" and "yolo_anchors.txt". Let's load these quantities into the model by running the next cell.
The car detection dataset has 720x1280 images, which we've pre-processed into 608x608 images.
```
class_names = read_classes("model_data/coco_classes.txt")
anchors = read_anchors("model_data/yolo_anchors.txt")
image_shape = (720., 1280.)
```
### 3.2 - Loading a pretrained model
Training a YOLO model takes a very long time and requires a fairly large dataset of labelled bounding boxes for a large range of target classes. You are going to load an existing pretrained Keras YOLO model stored in "yolo.h5". (These weights come from the official YOLO website, and were converted using a function written by Allan Zelener. References are at the end of this notebook. Technically, these are the parameters from the "YOLOv2" model, but we will more simply refer to it as "YOLO" in this notebook.) Run the cell below to load the model from this file.
```
yolo_model = load_model("model_data/yolo.h5")
```
This loads the weights of a trained YOLO model. Here's a summary of the layers your model contains.
```
yolo_model.summary()
```
**Note**: On some computers, you may see a warning message from Keras. Don't worry about it if you do--it is fine.
**Reminder**: this model converts a preprocessed batch of input images (shape: (m, 608, 608, 3)) into a tensor of shape (m, 19, 19, 5, 85) as explained in Figure (2).
### 3.3 - Convert output of the model to usable bounding box tensors
The output of `yolo_model` is a (m, 19, 19, 5, 85) tensor that needs to pass through non-trivial processing and conversion. The following cell does that for you.
```
yolo_outputs = yolo_head(yolo_model.output, anchors, len(class_names))
```
You added `yolo_outputs` to your graph. This set of 4 tensors is ready to be used as input by your `yolo_eval` function.
### 3.4 - Filtering boxes
`yolo_outputs` gave you all the predicted boxes of `yolo_model` in the correct format. You're now ready to perform filtering and select only the best boxes. Lets now call `yolo_eval`, which you had previously implemented, to do this.
```
scores, boxes, classes = yolo_eval(yolo_outputs, image_shape)
```
### 3.5 - Run the graph on an image
Let the fun begin. You have created a (`sess`) graph that can be summarized as follows:
1. <font color='purple'> yolo_model.input </font> is given to `yolo_model`. The model is used to compute the output <font color='purple'> yolo_model.output </font>
2. <font color='purple'> yolo_model.output </font> is processed by `yolo_head`. It gives you <font color='purple'> yolo_outputs </font>
3. <font color='purple'> yolo_outputs </font> goes through a filtering function, `yolo_eval`. It outputs your predictions: <font color='purple'> scores, boxes, classes </font>
**Exercise**: Implement predict() which runs the graph to test YOLO on an image.
You will need to run a TensorFlow session, to have it compute `scores, boxes, classes`.
The code below also uses the following function:
```python
image, image_data = preprocess_image("images/" + image_file, model_image_size = (608, 608))
```
which outputs:
- image: a python (PIL) representation of your image used for drawing boxes. You won't need to use it.
- image_data: a numpy-array representing the image. This will be the input to the CNN.
**Important note**: when a model uses BatchNorm (as is the case in YOLO), you will need to pass an additional placeholder in the feed_dict {K.learning_phase(): 0}.
```
def predict(sess, image_file):
"""
Runs the graph stored in "sess" to predict boxes for "image_file". Prints and plots the preditions.
Arguments:
sess -- your tensorflow/Keras session containing the YOLO graph
image_file -- name of an image stored in the "images" folder.
Returns:
out_scores -- tensor of shape (None, ), scores of the predicted boxes
out_boxes -- tensor of shape (None, 4), coordinates of the predicted boxes
out_classes -- tensor of shape (None, ), class index of the predicted boxes
Note: "None" actually represents the number of predicted boxes, it varies between 0 and max_boxes.
"""
# Preprocess your image
image, image_data = preprocess_image("images/" + image_file, model_image_size = (608, 608))
# Run the session with the correct tensors and choose the correct placeholders in the feed_dict.
# You'll need to use feed_dict={yolo_model.input: ... , K.learning_phase(): 0})
### START CODE HERE ### (≈ 1 line)
out_scores, out_boxes, out_classes = sess.run(
[scores, boxes, classes],
feed_dict={
yolo_model.input: image_data,
K.learning_phase(): 0
})
### END CODE HERE ###
# Print predictions info
print('Found {} boxes for {}'.format(len(out_boxes), image_file))
# Generate colors for drawing bounding boxes.
colors = generate_colors(class_names)
# Draw bounding boxes on the image file
draw_boxes(image, out_scores, out_boxes, out_classes, class_names, colors)
# Save the predicted bounding box on the image
image.save(os.path.join("out", image_file), quality=90)
# Display the results in the notebook
output_image = scipy.misc.imread(os.path.join("out", image_file))
imshow(output_image)
return out_scores, out_boxes, out_classes
```
Run the following cell on the "test.jpg" image to verify that your function is correct.
```
out_scores, out_boxes, out_classes = predict(sess, "test.jpg")
```
**Expected Output**:
<table>
<tr>
<td>
**Found 7 boxes for test.jpg**
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.60 (925, 285) (1045, 374)
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.66 (706, 279) (786, 350)
</td>
</tr>
<tr>
<td>
**bus**
</td>
<td>
0.67 (5, 266) (220, 407)
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.70 (947, 324) (1280, 705)
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.74 (159, 303) (346, 440)
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.80 (761, 282) (942, 412)
</td>
</tr>
<tr>
<td>
**car**
</td>
<td>
0.89 (367, 300) (745, 648)
</td>
</tr>
</table>
The model you've just run is actually able to detect 80 different classes listed in "coco_classes.txt". To test the model on your own images:
1. Click on "File" in the upper bar of this notebook, then click "Open" to go on your Coursera Hub.
2. Add your image to this Jupyter Notebook's directory, in the "images" folder
3. Write your image's name in the cell above code
4. Run the code and see the output of the algorithm!
If you were to run your session in a for loop over all your images. Here's what you would get:
<center>
<video width="400" height="200" src="nb_images/pred_video_compressed2.mp4" type="video/mp4" controls>
</video>
</center>
<caption><center> Predictions of the YOLO model on pictures taken from a camera while driving around the Silicon Valley <br> Thanks [drive.ai](https://www.drive.ai/) for providing this dataset! </center></caption>
<font color='blue'>
**What you should remember**:
- YOLO is a state-of-the-art object detection model that is fast and accurate
- It runs an input image through a CNN which outputs a 19x19x5x85 dimensional volume.
- The encoding can be seen as a grid where each of the 19x19 cells contains information about 5 boxes.
- You filter through all the boxes using non-max suppression. Specifically:
- Score thresholding on the probability of detecting a class to keep only accurate (high probability) boxes
- Intersection over Union (IoU) thresholding to eliminate overlapping boxes
- Because training a YOLO model from randomly initialized weights is non-trivial and requires a large dataset as well as lot of computation, we used previously trained model parameters in this exercise. If you wish, you can also try fine-tuning the YOLO model with your own dataset, though this would be a fairly non-trivial exercise.
**References**: The ideas presented in this notebook came primarily from the two YOLO papers. The implementation here also took significant inspiration and used many components from Allan Zelener's github repository. The pretrained weights used in this exercise came from the official YOLO website.
- Joseph Redmon, Santosh Divvala, Ross Girshick, Ali Farhadi - [You Only Look Once: Unified, Real-Time Object Detection](https://arxiv.org/abs/1506.02640) (2015)
- Joseph Redmon, Ali Farhadi - [YOLO9000: Better, Faster, Stronger](https://arxiv.org/abs/1612.08242) (2016)
- Allan Zelener - [YAD2K: Yet Another Darknet 2 Keras](https://github.com/allanzelener/YAD2K)
- The official YOLO website (https://pjreddie.com/darknet/yolo/)
**Car detection dataset**:
<a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Creative Commons License" style="border-width:0" src="https://i.creativecommons.org/l/by/4.0/88x31.png" /></a><br /><span xmlns:dct="http://purl.org/dc/terms/" property="dct:title">The Drive.ai Sample Dataset</span> (provided by drive.ai) is licensed under a <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>. We are especially grateful to Brody Huval, Chih Hu and Rahul Patel for collecting and providing this dataset.
|
github_jupyter
|
```
import os
import csv
import platform
import pandas as pd
import networkx as nx
from graph_partitioning import GraphPartitioning, utils
run_metrics = True
cols = ["WASTE", "CUT RATIO", "EDGES CUT", "TOTAL COMM VOLUME", "Qds", "CONDUCTANCE", "MAXPERM", "NMI", "FSCORE", "FSCORE RELABEL IMPROVEMENT", "LONELINESS"]
#cols = ["WASTE", "CUT RATIO", "EDGES CUT", "TOTAL COMM VOLUME", "Q", "Qds", "CONDUCTANCE", "LONELINESS", "NETWORK PERMANENCE", "NORM. MUTUAL INFO", "EDGE CUT WEIGHT", "FSCORE", "FSCORE RELABEL IMPROVEMENT"]
#cols = ["WASTE", "CUT RATIO", "EDGES CUT", "TOTAL COMM VOLUME", "MODULARITY", "LONELINESS", "NETWORK PERMANENCE", "NORM. MUTUAL INFO", "EDGE CUT WEIGHT", "FSCORE", "FSCORE RELABEL IMPROVEMENT"]
pwd = %pwd
config = {
"DATA_FILENAME": os.path.join(pwd, "data", "predition_model_tests", "network", "network_$$.txt"),
"OUTPUT_DIRECTORY": os.path.join(pwd, "output"),
# Set which algorithm is run for the PREDICTION MODEL.
# Either: 'FENNEL' or 'SCOTCH'
"PREDICTION_MODEL_ALGORITHM": "FENNEL",
# Alternativly, read input file for prediction model.
# Set to empty to generate prediction model using algorithm value above.
"PREDICTION_MODEL": "",
"PARTITIONER_ALGORITHM": "FENNEL",
# File containing simulated arrivals. This is used in simulating nodes
# arriving at the shelter. Nodes represented by line number; value of
# 1 represents a node as arrived; value of 0 represents the node as not
# arrived or needing a shelter.
"SIMULATED_ARRIVAL_FILE": os.path.join(pwd,
"data",
"predition_model_tests",
"dataset_1_shift_rotate",
"simulated_arrival_list",
"percentage_of_prediction_correct_100",
"arrival_100_$$.txt"
),
# File containing the prediction of a node arriving. This is different to the
# simulated arrivals, the values in this file are known before the disaster.
"PREDICTION_LIST_FILE": os.path.join(pwd,
"data",
"predition_model_tests",
"dataset_1_shift_rotate",
"prediction_list",
"prediction_$$.txt"
),
# File containing the geographic location of each node, in "x,y" format.
"POPULATION_LOCATION_FILE": os.path.join(pwd,
"data",
"predition_model_tests",
"coordinates",
"coordinates_$$.txt"
),
# Number of shelters
"num_partitions": 4,
# The number of iterations when making prediction model
"num_iterations": 1,
# Percentage of prediction model to use before discarding
# When set to 0, prediction model is discarded, useful for one-shot
"prediction_model_cut_off": .0,
# Alpha value used in one-shot (when restream_batches set to 1)
"one_shot_alpha": 0.5,
"use_one_shot_alpha": False,
# Number of arrivals to batch before recalculating alpha and restreaming.
"restream_batches": 50,
# When the batch size is reached: if set to True, each node is assigned
# individually as first in first out. If set to False, the entire batch
# is processed and empty before working on the next batch.
"sliding_window": False,
# Create virtual nodes based on prediction model
"use_virtual_nodes": False,
# Virtual nodes: edge weight
"virtual_edge_weight": 1.0,
# Loneliness score parameter. Used when scoring a partition by how many
# lonely nodes exist.
"loneliness_score_param": 1.2,
####
# GRAPH MODIFICATION FUNCTIONS
# Also enables the edge calculation function.
"graph_modification_functions": True,
# If set, the node weight is set to 100 if the node arrives at the shelter,
# otherwise the node is removed from the graph.
"alter_arrived_node_weight_to_100": False,
# Uses generalized additive models from R to generate prediction of nodes not
# arrived. This sets the node weight on unarrived nodes the the prediction
# given by a GAM.
# Needs POPULATION_LOCATION_FILE to be set.
"alter_node_weight_to_gam_prediction": False,
# Enables edge expansion when graph_modification_functions is set to true
"edge_expansion_enabled": True,
# The value of 'k' used in the GAM will be the number of nodes arrived until
# it reaches this max value.
"gam_k_value": 100,
# Alter the edge weight for nodes that haven't arrived. This is a way to
# de-emphasise the prediction model for the unknown nodes.
"prediction_model_emphasis": 1.0,
# This applies the prediction_list_file node weights onto the nodes in the graph
# when the prediction model is being computed and then removes the weights
# for the cutoff and batch arrival modes
"apply_prediction_model_weights": True,
"compute_metrics_enabled": True,
"SCOTCH_LIB_PATH": os.path.join(pwd, "libs/scotch/macOS/libscotch.dylib")
if 'Darwin' in platform.system()
else "/usr/local/lib/libscotch.so",
# Path to the PaToH shared library
"PATOH_LIB_PATH": os.path.join(pwd, "libs/patoh/lib/macOS/libpatoh.dylib")
if 'Darwin' in platform.system()
else os.path.join(pwd, "libs/patoh/lib/linux/libpatoh.so"),
"PATOH_ITERATIONS": 5,
# Expansion modes: 'avg_node_weight', 'total_node_weight', 'smallest_node_weight'
# 'largest_node_weight'
# add '_squared' or '_sqrt' at the end of any of the above for ^2 or sqrt(weight)
# i.e. 'avg_node_weight_squared
"PATOH_HYPEREDGE_EXPANSION_MODE": 'no_expansion',
# Edge Expansion: average, total, minimum, maximum, product, product_squared, sqrt_product
"EDGE_EXPANSION_MODE" : 'total',
# Whether nodes should be reordered using a centrality metric for optimal node assignments in batch mode
# This is specific to FENNEL and at the moment Leverage Centrality is used to compute new noder orders
"FENNEL_NODE_REORDERING_ENABLED": False,
# The node ordering scheme: PII_LH (political index), LEVERAGE_HL, DEGREE_HL, BOTTLENECK_HL
"FENNEL_NODE_REODERING_SCHEME": 'BOTTLENECK_HL',
# Whether the Friend of a Friend scoring system is active during FENNEL partitioning.
# FOAF employs information about a node's friends to determine the best partition when
# this node arrives at a shelter and no shelter has friends already arrived
"FENNEL_FRIEND_OF_A_FRIEND_ENABLED": False,
# Alters how much information to print. Keep it at 1 for this notebook.
# 0 - will print nothing, useful for batch operations.
# 1 - prints basic information on assignments and operations.
# 2 - prints more information as it batches arrivals.
"verbose": 1
}
#gp = GraphPartitioning(config)
# Optional: shuffle the order of nodes arriving
# Arrival order should not be shuffled if using GAM to alter node weights
#random.shuffle(gp.arrival_order)
%pylab inline
import scipy
from copy import deepcopy
iterations = 1000 # the number of individual networks to run
# BOTTLENECK 1 Restream, no FOAF, Lonely after
# change these variables:
ordering_enabled_mode = [True]#[False, True]
for mode in ordering_enabled_mode:
#for mode in range(1, 51):
metricsDataPrediction = []
metricsDataAssign = []
config['FENNEL_NODE_REORDERING_ENABLED'] = mode
config['FENNEL_NODE_REORDERING_SCHEME'] = 'BOTTLENECK_HL'
config['FENNEL_FRIEND_OF_A_FRIEND_ENABLED'] = False
print('Mode', mode)
for i in range(0, iterations):
if (i % 50) == 0:
print('Mode', mode, 'Iteration', str(i))
conf = deepcopy(config)
#if mode == 'no_expansion':
# config['edge_expansion_enabled'] = False
#conf["DATA_FILENAME"] = os.path.join(pwd, "data", "predition_model_tests", "network", "network_" + str(i + 1) + ".txt")
conf["DATA_FILENAME"] = conf["DATA_FILENAME"].replace('$$', str(i + 1))
conf["SIMULATED_ARRIVAL_FILE"] = conf["SIMULATED_ARRIVAL_FILE"].replace('$$', str(i + 1))
conf["PREDICTION_LIST_FILE"] = conf["PREDICTION_LIST_FILE"].replace('$$', str(i + 1))
conf["POPULATION_LOCATION_FILE"] = conf["POPULATION_LOCATION_FILE"].replace('$$', str(i + 1))
conf["compute_metrics_enabled"] = False
conf['PREDICTION_MODEL'] = conf['PREDICTION_MODEL'].replace('$$', str(i + 1))
#print(i, conf)
#print('config', config)
with GraphPartitioning(conf) as gp:
#gp = GraphPartitioning(config)
gp.verbose = 0
gp.load_network()
gp.init_partitioner()
m = gp.prediction_model()
m = gp.assign_cut_off()
m = gp.batch_arrival()
Gsub = gp.G.subgraph(gp.nodes_arrived)
gp.compute_metrics_enabled = True
m = [gp._print_score(Gsub)]
gp.compute_metrics_enabled = False
totalM = len(m)
metricsDataPrediction.append(m[totalM - 1])
waste = ''
cutratio = ''
ec = ''
tcv = ''
qds = ''
conductance = ''
maxperm = ''
nmi = ''
lonliness = ''
fscore = ''
fscoreimprove = ''
qdsOv = ''
condOv = ''
dataWaste = []
dataCutRatio = []
dataEC = []
dataTCV = []
dataQDS = []
dataCOND = []
dataMAXPERM = []
dataNMI = []
dataLonliness = []
dataFscore = []
dataFscoreImprove = []
for i in range(0, iterations):
dataWaste.append(metricsDataPrediction[i][0])
dataCutRatio.append(metricsDataPrediction[i][1])
dataEC.append(metricsDataPrediction[i][2])
dataTCV.append(metricsDataPrediction[i][3])
dataQDS.append(metricsDataPrediction[i][4])
dataCOND.append(metricsDataPrediction[i][5])
dataMAXPERM.append(metricsDataPrediction[i][6])
dataNMI.append(metricsDataPrediction[i][7])
dataFscore.append(metricsDataPrediction[i][8])
dataFscoreImprove.append(metricsDataPrediction[i][9])
dataLonliness.append(metricsDataPrediction[i][10])
if(len(waste)):
waste = waste + ','
waste = waste + str(metricsDataPrediction[i][0])
if(len(cutratio)):
cutratio = cutratio + ','
cutratio = cutratio + str(metricsDataPrediction[i][1])
if(len(ec)):
ec = ec + ','
ec = ec + str(metricsDataPrediction[i][2])
if(len(tcv)):
tcv = tcv + ','
tcv = tcv + str(metricsDataPrediction[i][3])
if(len(qds)):
qds = qds + ','
qds = qds + str(metricsDataPrediction[i][4])
if(len(conductance)):
conductance = conductance + ','
conductance = conductance + str(metricsDataPrediction[i][5])
if(len(maxperm)):
maxperm = maxperm + ','
maxperm = maxperm + str(metricsDataPrediction[i][6])
if(len(nmi)):
nmi = nmi + ','
nmi = nmi + str(metricsDataPrediction[i][7])
if(len(fscore)):
fscore = fscore + ','
fscore = fscore + str(metricsDataPrediction[i][8])
if(len(fscoreimprove)):
fscoreimprove = fscoreimprove + ','
fscoreimprove = fscoreimprove + str(metricsDataPrediction[i][8])
if(len(lonliness)):
lonliness = lonliness + ','
lonliness = lonliness + str(dataLonliness[i])
waste = 'WASTE,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataWaste)) + ',' + str(scipy.std(dataWaste)) + ',' + waste
cutratio = 'CUT_RATIO,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataCutRatio)) + ',' + str(scipy.std(dataCutRatio)) + ',' + cutratio
ec = 'EC,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataEC)) + ',' + str(scipy.std(dataEC)) + ',' + ec
tcv = 'TCV,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataTCV)) + ',' + str(scipy.std(dataTCV)) + ',' + tcv
lonliness = "LONELINESS," + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataLonliness)) + ',' + str(scipy.std(dataLonliness)) + ',' + lonliness
qds = 'QDS,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataQDS)) + ',' + str(scipy.std(dataQDS)) + ',' + qds
conductance = 'CONDUCTANCE,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataCOND)) + ',' + str(scipy.std(dataCOND)) + ',' + conductance
maxperm = 'MAXPERM,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataMAXPERM)) + ',' + str(scipy.std(dataMAXPERM)) + ',' + maxperm
nmi = 'NMI,' + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataNMI)) + ',' + str(scipy.std(dataNMI)) + ',' + nmi
fscore = "FSCORE," + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataFscore)) + ',' + str(scipy.std(dataFscore)) + ',' + fscore
fscoreimprove = "FSCORE_IMPROVE," + 'centrality_enabled_mode_' + str(config['FENNEL_NODE_REORDERING_ENABLED']) + ',' + str(scipy.mean(dataFscoreImprove)) + ',' + str(scipy.std(dataFscoreImprove)) + ',' + fscoreimprove
print(waste)
print(cutratio)
print(ec)
print(tcv)
print(lonliness)
print(qds)
print(conductance)
print(maxperm)
print(fscore)
print(fscoreimprove)
```
|
github_jupyter
|
# Class activation map evaluation
```
import cv2
import numpy as np
import matplotlib.pyplot as plt
import json
import os
import pandas as pd
from pocovidnet.evaluate_covid19 import Evaluator
from pocovidnet.grad_cam import GradCAM
from pocovidnet.cam import get_class_activation_map
from pocovidnet.model import get_model
```
## Code to crop ICLUS videos automatically
```
with open(os.path.join("../../../../data_pocovid/results_oct_wrong_crossval/iclus/", 'ICLUS_cropping.json'), "r") as infile:
frame_cut = json.load(infile)
bottom = 70 # 90
top = 570 # 542
left = 470 # 480
right = 970 # 932
# [70:570, 470:970]
crop = [bottom, top, left, right]
data_dir = "../../../data/ICLUS"
for subfolder in os.listdir(data_dir):
if "linear" in subfolder.lower() or subfolder.startswith(".") or not os.path.isdir(os.path.join(data_dir,subfolder)):
continue
for vid in os.listdir(os.path.join(data_dir, subfolder)):
vid_id = vid.split(".")[0]
if vid.startswith("."):
continue
print("process next file ", vid)
if vid_id not in ["40", "42"]: # frame_cut.keys():
continue
video_path = os.path.join(data_dir, subfolder, vid)
crop = frame_cut[vid_id]
while True:
bottom, top, left, right = crop
cap = cv2.VideoCapture(video_path)
# count = 0
# while cap.isOpened() and count< 1:
for _ in range(3):
ret, frame = cap.read()
plt.imshow(frame[bottom:top, left:right])
plt.show()
crop_in = input("okay?")
if crop_in == 1 or crop_in =="1":
frame_cut[vid_id] = crop
break
crop_in = input("input list " + str(crop))
crop = eval(crop_in)
print(crop)
# out_iclus_data = "../results_oct/iclus"
with open(os.path.join(data_dir, 'ICLUS_cropping.json'), "w") as outfile:
json.dump(frame_cut, outfile)
```
### ICLUS Auswertung:
```
severity = pd.read_csv("../../../data/iclus_severity.csv", delimiter=";")
convex_table = severity[severity["filename"].str.contains("convex")]
convex_vids = convex_table["Video"]
# Make list of IDs that we analyze
data_dir = "../../../data/ICLUS"
process_vid_numbers = []
for subfolder in os.listdir(data_dir):
if "linear" in subfolder.lower() or subfolder.startswith(".") or os.path.isfile(os.path.join(data_dir,subfolder)):
continue
for vid in os.listdir(os.path.join(data_dir, subfolder)):
vid_id = vid.split(".")[0]
if vid.startswith("."):
continue
video_path = os.path.join(data_dir, subfolder, vid)
# print(int(vid.split(".")[0]) in convex_vids)
process_vid_numbers.append(int(vid.split(".")[0]) )
# Check whether we cover all videos
for vid in convex_vids.values:
if vid not in process_vid_numbers:
print("In ICLUS tabelle but not in our folder:", vid)
if str(vid) not in frame_cut.keys():
print("not in crop dict:", vid)
for vid in process_vid_numbers:
if vid not in convex_vids.values:
print("In our folder but not in ICLUS:", vid)
# Make label dict:
iclus_labels = dict(zip(convex_table["Video"], convex_table["Score"]))
in_path = os.path.join(res_dir, f"cam_{vid_id}.npy")
os.path.exists(in_path)
```
### Analyze results
```
# 6 normal (Gabriel, but here 1), 25 normal (Gabriel), but here 3
iclus_labels
# directory with numpy files
len(iclus_labels.keys())
res_dir = "../../../../data_pocovid/results_oct_wrong_crossval/iclus/base"
gt, preds, pred_probs = list(), list(), list()
print("gt pred")
for vid_id in iclus_labels.keys():
in_path = os.path.join(res_dir, f"cam_{vid_id}.npy")
if not os.path.exists(in_path):
print("Warning: logits do not exist", in_path)
continue
logits = np.load(in_path)
prob = np.mean(logits[:, 0])
avg_covid_prob = np.argmax(np.mean(logits, axis=0)) #
# print(avg_covid_prob)
gt.append(iclus_labels[vid_id])
pred_probs.append(prob)
preds.append(avg_covid_prob)
if iclus_labels[vid_id]>2 and avg_covid_prob==2 or iclus_labels[vid_id]==0 and avg_covid_prob==0:
print("wrong, severity is ", iclus_labels[vid_id], "pred is", avg_covid_prob,"video:", vid_id)
# print(gt[-1], preds[-1])
plt.scatter(gt, pred_probs)
plt.plot([0,3], [0,1])
plt.show()
check = "../../models/cross_validation_neurips/"
file_list = list()
for folder in os.listdir(check):
if folder[0] == ".":
continue
for classe in os.listdir(os.path.join(check, folder)):
if classe[0] == "." or classe[0] == "u":
continue
uni = []
is_image = 0
for file in os.listdir(os.path.join(check, folder, classe)):
if file[0] == ".":
continue
if len(file.split(".")) == 2:
is_image += 1
uni.append(file.split(".")[0])
file_list.extend(np.unique(uni).tolist())
with open("../../models/in_neurips.json", "w") as outfile:
json.dump(file_list, outfile)
```
### Old video evaluator
```
from skvideo import io
class VideoEvaluator(Evaluator):
def __init__(self, weights_dir="../trained_models_cam", ensemble=True, split=None, model_id=None, num_classes=3):
Evaluator.__init__(
self, ensemble=ensemble, split=split, model_id=model_id, num_classes=num_classes
)
def __call__(self, video_path):
"""Performs a forward pass through the restored model
Arguments:
video_path: str -- file path to a video to process. Possibly types are mp4, gif, mpeg
return_cams: int -- number of frames to return with activation maps overlayed. If zero,
only the predictions will be returned. Always selects the frames with
highest probability for the predicted class
Returns:
cams: if return_cams>0, images with overlay are returned as a np.array of shape
{number models} x {return_cams} x 224 x 224 x 3
mean_preds: np array of shape {video length} x {number classes}. Contains
class probabilities per frame
"""
self.image_arr = self.read_video(video_path)
self.predictions = np.stack([model.predict(self.image_arr) for model in self.models])
mean_preds = np.mean(self.predictions, axis=0, keepdims=False)
class_idx = np.argmax(np.mean(np.array(mean_preds), axis=0))
return mean_preds
def cam_important_frames(self, class_idx, threshold=0.5, nr_cams=None, zeroing=0.65, save_video_path=None): # "out_video.mp4"):
mean_preds = np.mean(self.predictions, axis=0, keepdims=False)
# compute general video class
# class_idx = np.argmax(np.mean(np.array(mean_preds), axis=0))
prediction = np.argmax(np.mean(np.array(mean_preds), axis=0))
print("predicted", prediction, "gt", class_idx)
print("pred probs covid", [round(m, 2) for m in mean_preds[:,0]])
# get most important frames (the ones above threshold)
if nr_cams is not None:
best_frames = np.argsort(mean_preds[:, class_idx])[-nr_cams:]
else:
best_frames = np.where(mean_preds[:, class_idx]>threshold)[0]
# best_frames = np.arange(len(mean_preds))
print("frames above threshold", best_frames)
return_cams = len(best_frames)
if len(best_frames)==0:
print("no frame above threshold")
return 0
# copy image arr - need values between 0 and 255
copied_arr = (self.image_arr.copy() * 255).astype(int)
cams = np.zeros((return_cams, 224, 224, 3))
for j, b_frame in enumerate(best_frames):
# get highest prob model for these frames
model_idx = np.argmax(self.predictions[:, b_frame, class_idx], axis=0)
take_model = self.models[model_idx]
if "cam" in self.model_id:
in_img = np.expand_dims(self.image_arr[b_frame], 0)
# print(in_img.shape)
cams[j] = get_class_activation_map(take_model, in_img, class_idx, image_weight=1, zeroing=zeroing).astype(int)
else:
# run grad cam for other models
gradcam = GradCAM()
cams[j] = gradcam.explain(self.image_arr[b_frame], take_model, class_idx, return_map=False,image_weight=1, layer_name="block5_conv3", zeroing=zeroing, heatmap_weight=0.25)
if save_video_path is None:
return cams
else:
for j in range(return_cams):
copied_arr[best_frames[j]] = cams[j]
copied_arr = np.repeat(copied_arr, 3, axis=0)
io.vwrite(save_video_path+".mpeg", copied_arr, outputdict={"-vcodec":"mpeg2video"})
def read_video(self, video_path):
assert os.path.exists(video_path), "video file not found"
cap = cv2.VideoCapture(video_path)
images = []
counter = 0
while cap.isOpened():
ret, frame = cap.read()
if (ret != True):
break
if counter<1:
plt.imshow(frame[30:360, 100:430]) # ICLUS: [70:570, 470:970]) # [25:350, 100:425]) # LOTTE:[30:400, 80:450]
plt.show()
counter += 1
continue
counter += 1
img_processed = self.preprocess(frame)[0]
images.append(img_processed)
cap.release()
return np.array(images)
def preprocess(self, image, cut=True):
"""Apply image preprocessing pipeline
Arguments:
image {np.array} -- Arbitrary shape, quadratic preferred
Returns:
np.array -- Shape 224,224. Normalized to [0, 1].
"""
if cut:
image = image[30:360, 100:430]
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = cv2.resize(image, (224, 224))
image = np.expand_dims(np.array(image), 0) / 255.0
return image
def important_frames(self, preds, predicted_class, n_return=5):
preds_arr = np.array(preds)
frame_scores = preds_arr[:, predicted_class]
best_frames = np.argsort(frame_scores)[-n_return:]
return best_frames
evaluator = VideoEvaluator(ensemble=True, model_id="vgg_cam", num_classes=4)
```
### Run ICLUS data
```
def pred_plot(preds, save_path):
plt.figure(figsize=(15,8))
plt.plot(preds[:,0], label="covid")
plt.plot(preds[:,1], label="pneu")
plt.plot(preds[:,2], label="healthy")
plt.legend()
plt.savefig(save_path+".png")
plt.show()
# plt.plot(preds[:,1], label="pneu")
# iclus_dir = "Videos_31_to_40"
iclus_dir = "test_data_regular/pat2"
# iclus_dir = "data/pocus_videos/convex/"
# out_iclus_data = "vids_preds_regular_test"
# out_iclus_data = "vids_preds_iclus"
out_iclus_data = "reg_test/pat2"
GT_CLASS = 2
for vid in os.listdir(iclus_dir):
vid_id = vid.split(".")[0]
if vid.startswith(".") or os.path.exists(os.path.join(out_iclus_data,"cam_"+vid_id+".npy")):
print("already done", vid)
continue
print("process next file ", vid)
preds = evaluator(os.path.join(iclus_dir, vid))
np.save(os.path.join(out_iclus_data,"cam_"+vid_id+".npy"), preds)
plt.imshow(evaluator.image_arr[0])
plt.savefig(os.path.join(out_iclus_data,"cam_"+vid_id+"expl_img.png"))
print("saved predictions")
pred_plot(preds, os.path.join(out_iclus_data,"cam_"+vid_id))
print("saved plot")
evaluator.cam_important_frames(GT_CLASS, save_video_path=os.path.join(out_iclus_data, "cam_"+vid_id))
```
#### ICLUS notes:
47 falsch predicted aber passt
schaut weird aus: 48, 49, 50 (linear or what is this? alle als healthy predicted)
Must do again: 36
13, 11, 31, 32: linear probes that are deleted, 22, 24, 26 (they are all kept), 28
12, 15, 16, 17, 18, 19, 20 were fine already with bad cropping
1, 3, 9, 10 is fine already
NEW PROCESSED: 14, 8, 7, 6, 4, 5, 2
CODE TO PROCESS SOME AGAIN:
if os.path.exists("vids_preds_iclus/cam_vid"+vid_id+".npy"):
preds_prev = np.load("vids_preds_iclus/cam_vid"+vid_id+".npy")
predicted_class = np.argmax(np.mean(np.array(preds_prev), axis=0))
print(predicted_class, np.mean(np.array(preds_prev), axis=0))
if predicted_class==0:
print("file is already predicted covid", vid)
continue
### Evaluate on train data
```
vid_in_path = "../../data/pocus_videos/Convex/"
gt_dict = {"Cov":0, "Reg":2, "Pne":1, "pne":1}
out_path="vid_outputs_cam"
for vid in os.listdir(vid_in_path):
if vid[:3] not in ["Pne", "pne", "Cov", "Reg"]:
print(vid)
continue
if os.path.exists(os.path.join(out_path, vid.split(".")[0]+".mpeg")):
print("already done", vid)
continue
vid_in = os.path.join(vid_in_path, vid)
print(vid_in)
preds = evaluator(vid_in)
gt = gt_dict[vid[:3]]
evaluator.cam_important_frames(gt, save_video_path=os.path.join(out_path, vid.split(".")[0]))
```
### Evaluate on test data
```
out_path_overall="vid_outputs_cam_test/"
path_crossval = "../../data/cross_validation"
per_split = [[] for _ in range(5)]
for fold in range(5):
out_path = os.path.join(out_path_overall, "fold"+str(fold))
# load weights of the respective fold model
print("NEW FOLD", fold)
# make sure the variable is cleared
evaluator = None
# load weights
evaluator = VideoEvaluator(ensemble=False, split=fold, model_id="vgg_cam", num_classes=4)
# get all names belonging to this fold
vidnames = []
for mod in ["covid", "pneumonia", "regular"]:
for f in os.listdir(os.path.join(path_crossval, "split"+str(fold), mod)):
if f[0]!=".":
fparts = f.split(".")
vidnames.append(fparts[0]+"."+fparts[1][:3])
# iterate over the relevant files
names = np.unique(vidnames)
for name in names:
if name[-3:] in ["mp4", "mov", "gif"]:
print(name)
vid_in = os.path.join(vid_in_path, name)
if not os.path.exists(vid_in):
print("does not exist! - butterfly?", vid_in)
continue
if os.path.exists(os.path.join(out_path, name.split(".")[0]+".mpeg")):
print("already done", name)
continue
print(vid_in)
preds = evaluator(vid_in)
gt = gt_dict[name[:3]]
evaluator.cam_important_frames(gt, save_video_path=os.path.join(out_path, name.split(".")[0]))
```
## Make point plot for CAMs
```
def max_kernel(heatmap, kernel_size=9):
k2 = kernel_size//2
# pad array
arr = np.pad(heatmap, ((k2,k2),(k2,k2)), 'constant', constant_values=0)
# get coordinates of maximum
x_coords, y_coords = divmod(np.argmax(arr.flatten()), len(arr[0]))
patch = arr[x_coords-k2:x_coords+k2+1, y_coords-k2:y_coords+k2+1]
# print(x_coords, y_coords)
# plt.imshow(arr)
# plt.show()
res_out = np.zeros((kernel_size-2,kernel_size-2))
for i in range(kernel_size-2):
for j in range(kernel_size-2):
res_out[i,j] = np.mean(patch[i:i+3, j:j+3])
max_x, max_y = divmod(np.argmax(res_out.flatten()), kernel_size-2)
# print(max_x, max_y)
# print(x_coords+max_x-k2+1, y_coords+max_y-k2+1)
# plt.imshow(res_out)
# plt.show()
return x_coords+max_x-2*k2+1, y_coords+max_y-2*k2+1
# max_kernel((np.random.rand(10,10)*20).astype(int))
def convolve_faster(img, kernel):
"""
Convolve a 2d img with a kernel, storing the output in the cell
corresponding the the left or right upper corner
:param img: 2d numpy array
:param kernel: kernel (must have equal size and width)
:param neg: if neg=0, store in upper left corner, if neg=1,
store in upper right corner
:return convolved image of same size
"""
k_size = len(kernel)
# a = np.pad(img, ((0, k_size-1), (0, k_size-1)))
padded = np.pad(img, ((k_size//2, k_size//2), (k_size//2, k_size//2)))
s = kernel.shape + tuple(np.subtract(padded.shape, kernel.shape) + 1)
strd = np.lib.stride_tricks.as_strided
subM = strd(padded, shape=s, strides=padded.strides * 2)
return np.einsum('ij,ijkl->kl', kernel, subM)
# in_img = np.random.rand(20,20)
# plt.imshow(in_img)
# plt.show()
# out = convolve_faster(in_img, np.ones((7,7)))
# plt.imshow(out)
# plt.show()
# print(in_img.shape, out.shape)
```
### Process all test data
```
path_crossval = "../../data/cross_validation"
gt_dict = {"Reg":2, "Pne":1, "pne":1, "Cov":0}
gradcam = GradCAM()
all_predictions = []
heatmap_points, predicted, gt_class, overlays, fnames = [], [], [], [], []
for fold in range(5):
# load weights of the respective fold model
print("NEW FOLD", fold)
# make sure the variable is cleared
evaluator = None
# load weights
evaluator = Evaluator(ensemble=False, split=fold, model_id="vgg_base", num_classes=4)
# get all names belonging to this fold
all_images_arr = []
gt, name = [], []
for mod in ["covid", "pneumonia", "regular"]:
for f in os.listdir(os.path.join(path_crossval, "split"+str(fold), mod)):
if f[0]!=".":
# fparts = f.split(".")
# vidnames.append(fparts[0]+"."+fparts[1][:3])
img_loaded = cv2.imread(os.path.join(path_crossval, "split"+str(fold), mod, f))
img_preprocc = evaluator.preprocess(img_loaded)[0]
gt.append(gt_dict[f[:3]])
all_images_arr.append(img_preprocc)
name.append(f)
all_images_arr = np.array(all_images_arr)
# get predictions
print("process all images in fold", fold, "with shape", all_images_arr.shape)
fold_preds = evaluator.models[0].predict(all_images_arr)
class_idx_per_img = np.argmax(fold_preds, axis=1)
all_predictions.append(fold_preds)
# get heatmap
for i, img in enumerate(all_images_arr):
# plt.imshow(img)
# plt.show()
# overlay, heatmap = get_class_activation_map(evaluator.models[0], img, gt[i], image_weight=1, return_map=True, zeroing=0.65)
overlay, heatmap = gradcam.explain(img, evaluator.models[0], gt[i], return_map=True, image_weight=1, layer_name="block5_conv3", zeroing=0.65, heatmap_weight=0.25)
# plt.figure(figsize=(10,10))
# plt.imshow(overlay.astype(int))
# plt.show()
overlays.append(overlay.astype(int))
# convolve with big kernel
convolved_overlay = convolve_faster(heatmap, np.ones((19,19)))
# print("previously:", divmod(np.argmax(heatmap.flatten()), len(heatmap[0])))
x_coord, y_coord = divmod(np.argmax(convolved_overlay.flatten()), len(convolved_overlay[0]))
## previous version: 9x9 umkreis and 3x3 kernel
# x_coord, y_coord = max_kernel(heatmap) # np.where(heatmap==np.max(heatmap))
# print(x_coord, y_coord)
heatmap_points.append([x_coord, y_coord])
predicted.append(class_idx_per_img[i])
gt_class.append(gt[i])
fnames.append(name[i])
# print([x_coord, y_coord], class_idx_per_img[i], gt[i])
len(predicted), len(gt_class), len(heatmap_points), np.asarray(overlays).shape
np.where(np.asarray(predicted)==3)
hm_p = np.array(heatmap_points)
print(hm_p.shape)
# plt.figure(figsize=(20,20))
plt.imshow(overlays[1])
plt.scatter(hm_p[:,1], hm_p[:,0], c=predicted)
plt.show()
hm_p = np.array(heatmap_points)
print(hm_p.shape)
# plt.figure(figsize=(20,20))
plt.imshow(overlays[1])
plt.scatter(hm_p[:,1], hm_p[:,0], c=predicted)
plt.show()
df = pd.DataFrame()
df["file"] = fnames
df["predicted"] = predicted
df["gt"] = gt_class
df["max_x"] = np.asarray(heatmap_points)[:,0].tolist()
df["max_y"] = np.asarray(heatmap_points)[:,1].tolist()
df.to_csv("heatmap_points_grad.csv")
np.save("overlayed_hm.npy", overlays)
df
```
## ICLUS evaluation
```
# out_iclus_data = "vids_preds_regular_test"
out_iclus_data = "vids_preds_iclus"
all_class_preds = []
correct_frames = 0
wrong_frames = 0
avg_corr_frames = []
all_frames = 0
# plt.figure(figsize=(20,10))
for f in os.listdir(out_iclus_data):
if f[-3:]=="npy":
preds = np.load(os.path.join(out_iclus_data, f))
# plt.plot(preds[:,0])
# print(preds.shape)
# frame based
frame_pred = np.argmax(preds, axis=1)
all_frames += len(frame_pred)
correct_frames += np.sum(frame_pred==0)
wrong_frames += np.sum(frame_pred!=0)
avg_corr_frames.append(np.sum(frame_pred==0)/len(frame_pred))
# video classification - majority vote
uni, counts = np.unique(frame_pred, return_counts=True)
# all_class_preds.append(uni[np.argmax(counts)])
# version with probabilities and not majority vote:
vid_class_pred = np.argmax(np.mean(preds, axis=0))
all_class_preds.append(vid_class_pred)
if all_class_preds[-1]!=0:
print("wrongly classified", f)
# print(wrong_frames+ correct_frames, all_frames)
print("Included in total ICLUS videos (without linear probes):", len(all_class_preds))
assert all_frames==wrong_frames+correct_frames
print("Frame accuracy:", correct_frames/float(all_frames))
print("video class accuracy (max avg probability): ", np.sum(np.array(all_class_preds)==0)/len(all_class_preds))
print("Mean and std of ratio of correctly classified frames per video:", np.mean(avg_corr_frames), np.std(avg_corr_frames))
# plt.show()
iclus_preds = all_class_preds
```
## Evaluation Lotte's test data
```
reg_test_data = "vid_outputs_REGULAR"
all_class_preds = []
correct_frames = 0
wrong_frames = 0
avg_corr_frames = []
all_frames = 0
# plt.figure(figsize=(20,10))
for subdir in os.listdir(reg_test_data):
if subdir[0]==".":
continue
print(subdir)
for f in os.listdir(os.path.join(reg_test_data, subdir)):
if f[-3:]=="npy":
preds = np.load(os.path.join(reg_test_data, subdir, f))
print(os.path.join(reg_test_data, subdir, f))
# print(preds.shape)
# frame based
frame_pred = np.argmax(preds, axis=1)
all_frames += len(frame_pred)
correct_frames += np.sum(frame_pred==2)
wrong_frames += np.sum(frame_pred!=2)
avg_corr_frames.append(np.sum(frame_pred==2)/len(frame_pred))
# video classification - majority vote
vid_class_pred = np.argmax(np.mean(preds, axis=0))
all_class_preds.append(vid_class_pred)
# print(frame_pred)
if all_class_preds[-1]!=2:
print("wrongly classified", f)
# version with probabilities and not majority vote:
# vid_class_pred = np.argmax(np.mean(preds, axis=0))
# all_class_preds.append(vid_class_pred)
# print(wrong_frames+ correct_frames, all_frames)
print("Included in total ICLUS videos (without linear probes):", len(all_class_preds))
assert all_frames==wrong_frames+correct_frames
print("Frame accuracy:", correct_frames/float(all_frames))
print("video class accuracy (max avg probability): ", np.sum(np.array(all_class_preds)==2)/len(all_class_preds))
print("Mean and std of ratio of correctly classified frames per video:", np.mean(avg_corr_frames), np.std(avg_corr_frames))
# plt.show()
reg_preds = all_class_preds
# sensitivity of both together
all_gt = np.asarray([1 for _ in range(len(iclus_preds))] + [0 for _ in range(len(reg_preds))])
all_preds = np.asarray(iclus_preds + reg_preds)
all_preds = np.absolute(all_preds/2 - 1).astype(int)
print(all_preds)
print(len(all_preds), len(all_gt))
print(recall_score(all_gt, all_preds))
print(precision_score(all_gt, all_preds))
from sklearn.metrics import recall_score, precision_score, accuracy_score
accuracy_score(all_gt, all_preds)
```
## MD comments evaluation
### Read in and merge
```
mapping = pd.read_csv("mapping.csv").drop(columns=["Unnamed: 0"])
gb_comments = pd.read_csv("CAM_scores_GB.csv")
gb_comments = gb_comments.drop([0,1])
lotte_comments = pd.read_csv("CAM_scores_lotte.csv")
lotte_comments = lotte_comments.rename(columns={'Score - how helpful is the heatmap (0=only distracting, 5=very helpful)': 'lotte_score',
'Better one (put 1 if this one is the better one)': "lotte_better",
'Class (Your guess)': 'lotte_class',
'Patterns that can be seen':'lotte_patterns',
'Patterns the heatmap highlights':'lotte_heatmap_patterns'}).drop(columns=["Unnamed: 6"])
gb_comments = gb_comments.rename(columns={'Score - how helpful is the heatmap (0=only distracting, 5=very helpful)': 'gb_score',
'Better one (put 1 if this one is the better one)': "gb_better",
'Class (Your guess)': 'gb_class',
'Patterns that can be seen':'gb_patterns',
'Patterns the heatmap highlights':'gb_heatmap_patterns'})
lotte_comments['lotte_score'] = lotte_comments['lotte_score'].apply(lambda x: x-3 + int(x>=3))
merge_map_gb = pd.merge(mapping, gb_comments, how="inner", left_on="new_filename", right_on="Filename")
merge_map_lotte = pd.merge(merge_map_gb, lotte_comments, how="inner", left_on="new_filename", right_on="Filename")
merge_map_lotte.to_csv("CAM_scores_MDs.csv")
```
### Clean
```
final_table.columns
# after manual cleaning:
final_table = pd.read_csv("CAM_scores_MDs.csv")
train_score_gb = 0
test_score_gb = 0
train_score_lo = 0
test_score_lo = 0
train_better_gb = []
train_better_lo = []
for group_name, group_df in final_table.groupby("previous_filename"):
print("--------")
print(group_df[["gb_better", "lotte_better", "is_train"]])
if np.all(pd.isnull(group_df["gb_better"])) or len(np.where(group_df["gb_better"].values=="1")[0])==0:
train_score_gb += 0.5
test_score_gb += 0.5
print("gb: equally good")
train_better_gb.append(0.5)
else:
# if len(np.where(group_df["gb_better"].values=="1")[0])==0:
# raise RuntimeError("no valid value found")
if np.where(group_df["gb_better"].values=="1")==np.where(group_df["is_train"].values==1):
print("gb: train better")
train_score_gb += 1
train_better_gb.append(1)
else:
test_score_gb += 1
train_better_gb.append(0)
print("gb: test better")
# get lotte score
if np.all(pd.isnull(group_df["lotte_better"])):
train_score_gb += 0.5
test_score_gb += 0.5
train_better_lo.append(0.5)
print("lotte: equally good")
else:
if len(np.where(group_df["lotte_better"].values==1)[0])==0:
raise RuntimeError("no valid value found")
if np.where(group_df["lotte_better"].values==1)==np.where(group_df["is_train"].values==1):
print("lotte: train better")
train_score_lo += 1
train_better_lo.append(1)
else:
test_score_lo += 1
train_better_lo.append(0)
print("lotte: test better")
for i, row in group_df.iterrows():
if int(row["is_train"])==1:
print(row["gb_better"], row["lotte_better"], row["is_train"])
# gb_scores = group_df["gb_better"]
# lotte_scores = group_df["lotte_better"]
# train_test = group_df["is_train"]
len(train_better_lo), len(train_better_gb)
better_arr = np.swapaxes(np.stack([train_better_lo, train_better_gb]), 1, 0)
agree = np.sum(better_arr[:,0]==better_arr[:,1])
print("agreement (both exactly same)", agree/len(better_arr))
print("disagreement (one 1 one 0)", len(np.where(np.absolute(better_arr[:,0]-better_arr[:,1])==1)[0])/len(better_arr))
print("average score for train better:", np.mean(train_better_lo), np.mean(train_better_gb))
print("numbers unique",np.unique(train_better_lo, return_counts=True), np.unique(train_better_gb, return_counts=True))
```
#### Evaluate scores - Add label
```
label = [val[:3].lower() for val in final_table["previous_filename"].values]
np.unique(label, return_counts=True)
np.mean(final_table[final_table["is_train"]==0]["gb_score"])
final_table["label"] = label
```
#### Get average score of Lotte and Gabriel together
```
only_test = final_table[final_table["is_train"]==0]
all_scores = only_test["gb_score"].values.tolist() + only_test["lotte_score"].values.tolist()
print("Mean score lotte and gabriel together (test):", np.mean(all_scores))
# other method: average per video scores first:
mean_scores = 0.5* (only_test["gb_score"].values + only_test["lotte_score"].values)
print("Mean score lotte and gabriel together (test) - other method:", np.mean(mean_scores))
print(np.vstack([only_test["gb_score"].values, only_test["lotte_score"].values]))
only_test["mean_scores"] = mean_scores.tolist()
only_test.groupby("label").agg({"mean_scores":"mean"})
```
#### Test whether test better train significant
```
from scipy.stats import ttest_ind, ttest_rel, wilcoxon, mannwhitneyu
only_train = final_table[final_table["is_train"]==1]
all_train_scores = only_train["gb_score"].values.tolist() + only_train["lotte_score"].values.tolist()
only_test = final_table[final_table["is_train"]==0]
all_test_scores = only_test["gb_score"].values.tolist() + only_test["lotte_score"].values.tolist()
print("means", np.mean(all_train_scores), np.mean(all_test_scores))
print("Ttest ind:", ttest_ind(all_train_scores,all_test_scores, equal_var=False))
print("ttest related:", ttest_rel(all_train_scores,all_test_scores))
print("Wilcoxon:", wilcoxon(all_train_scores,all_test_scores))
print("mannwhitneyu", mannwhitneyu(all_train_scores,all_test_scores))
# Ttest related
# Examples for use are scores of the same set of student in different exams,
# or repeated sampling from the same units. The test measures whether the average score
# differs significantly across samples (e.g. exams). If we observe a large p-value, for
# example greater than 0.05 or 0.1 then we cannot reject the null hypothesis of identical average scores
print(len(all_train_scores), len(all_test_scores))
plt.scatter(range(len(all_test_scores)), all_test_scores)
plt.scatter(range(len(all_train_scores)), all_train_scores)
```
#### Grouped for separate scores
```
only_test = final_table[final_table["is_train"]==0]
grouped = only_test.groupby("label").agg({"lotte_score":"mean", "gb_score":"mean"})
grouped
only_test = only_test.fillna("none")
gb_all_with_consolidations = only_test[only_test["gb_patterns"].str.contains("onsolida")]
print("number of videos with consolidations", len(gb_all_with_consolidations))
print("GB heatmap highlights consolidation", len(gb_all_with_consolidations[gb_all_with_consolidations["gb_heatmap_patterns"].str.contains("onsolida")]))
print("Lotte heatmap highlights consolidation", len(gb_all_with_consolidations[gb_all_with_consolidations["lotte_heatmap_patterns"].str.contains("onsolida")]))
gb_all_with_alines = only_test[only_test["gb_patterns"].str.contains("A")]
print("number of videos with A lines", len(gb_all_with_alines))
print("GB heatmap highlights A lines", len(gb_all_with_alines[gb_all_with_alines["gb_heatmap_patterns"].str.contains("A")]))
print("Lotte heatmap highlights A lines", len(gb_all_with_alines[gb_all_with_alines["lotte_heatmap_patterns"].str.contains("A")]))
gb_all_with_blines = only_test[only_test["gb_patterns"].str.contains("B")]
print("number of videos with B lines", len(gb_all_with_blines))
print("GB heatmap highlights B lines", len(gb_all_with_blines[gb_all_with_blines["gb_heatmap_patterns"].str.contains("B")]))
print("Lotte heatmap highlights B lines", len(gb_all_with_blines[gb_all_with_blines["lotte_heatmap_patterns"].str.contains("B")]))
print("Note: Lotte usually writes that it catches ONE bline in the video, or beginning of bline")
class_wise = []
for pattern in ["onsol", "B", "A"]:
print("--------", pattern)
gb_all_with_pattern = only_test[only_test["gb_patterns"].str.contains(pattern)]
for classe in ["cov", "pne", "reg"]:
class_filtered = gb_all_with_pattern[gb_all_with_pattern["label"]==classe]
print(classe, pattern, len(class_filtered))
# gb_all_with_pattern = class_filtered[class_filtered["gb_patterns"].str.contains(pattern)]
number_found = 0.5*(len(class_filtered[class_filtered["gb_heatmap_patterns"].str.contains(pattern)])
+ len(class_filtered[class_filtered["lotte_heatmap_patterns"].str.contains(pattern)]))
if len(class_filtered)>0:
print(classe, number_found/len(class_filtered))
# print(gb_all_with_pattern["label"])
from matplotlib import rc
rc('text', usetex=False)
fig, ax = plt.subplots()
rects = ax.barh(["Consolidations \n (pneumonia)", "A-lines \n (healthy)", "Pleural line \n (healthy if regular)", "B-lines \n (COVID-19)"], [17/18, 8/13, 9/20, 3/12], width
, color = ["palegreen","greenyellow","sandybrown", "indianred"])
ax.set_xlim(0,1)
# Add some text for labels, title and custom x-axis tick labels, etc.
# ax.set_ylabel('Scores')
# ax.set_title('Scores by group and gender')
# ax.set_yticks(["Consolidations \n (pneumonia)", "A-lines \n (healthy)", "Pleural line \n (healthy if regular)", "B-lines \n (COVID-19)"], fontsize=13)
ax.set_xlabel("Ratio of samples highlighted by CAM", fontsize=13)
ax.legend()
def autolabel(rects):
"""Attach a text label above each bar in *rects*, displaying its height."""
for rect in rects:
height = rect.get_height()
ax.annotate('{}'.format(height),
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3), # 3 points vertical offset
textcoords="offset points",
ha='center', va='bottom')
autolabel(rects)
fig.tight_layout()
plt.figure(figsize=(6,3))
width=0.5
plt.barh(["Consolidations \n (pneumonia)", "A-lines \n (healthy)", "Pleural line", "B-lines \n (COVID-19)"], [17/18, 8/13, 9/20, 3/12], width
, color = ["palegreen","greenyellow","sandybrown", "indianred"])
plt.xlim(0,1)
plt.yticks(fontsize=13)
plt.xlabel("Ratio of samples highlighted by CAM", fontsize=13)
plt.tight_layout()
plt.savefig("barplot_cam.pdf")
print("FROM GABRIELS PATTERNS:")
for pattern in ["onsolida", "A", "B", "ronchogram", "ffusion"]:
print("-------------------")
gb_all_with_pattern = only_test[only_test["gb_patterns"].str.contains(pattern)]
print("number of videos with ", pattern, len(gb_all_with_pattern))
print("GB heatmap highlights ", pattern, len(gb_all_with_pattern[gb_all_with_pattern["gb_heatmap_patterns"].str.contains(pattern)]))
print("Lotte heatmap highlights ", pattern, len(gb_all_with_pattern[gb_all_with_pattern["lotte_heatmap_patterns"].str.contains(pattern)]))
print("---------------")
print("Note: observed that both MDs agreed where consolidations are found")
print("Note: Lotte usually writes that it catches ONE bline in the video, or beginning of bline")
print("FROM LOTTES PATTERNS:")
for pattern in ["onsolida", "A", "B", "ffusion", "leura"]:
print("-------------------")
gb_all_with_pattern = only_test[only_test["lotte_patterns"].str.contains(pattern)]
print("number of videos with ", pattern, len(gb_all_with_pattern))
print("GB heatmap highlights ", pattern, len(gb_all_with_pattern[gb_all_with_pattern["gb_heatmap_patterns"].str.contains(pattern)]))
print("Lotte heatmap highlights ", pattern, len(gb_all_with_pattern[gb_all_with_pattern["lotte_heatmap_patterns"].str.contains(pattern)]))
print("---------------")
print("Note: observed that both MDs agreed where consolidations are found")
print("Note: Lotte usually writes that it catches ONE bline in the video, or beginning of bline")
print("overall number of videos", len(only_test))
for name in ["gb", "lotte"]:
print("---------- "+name+" --------------")
for pattern in ["uscle", "fat", "skin"]:
print(pattern, np.sum(only_test[name+"_heatmap_patterns"].str.contains(pattern)))
```
#### Notes:
GB: 1 time "Avoids the liver, I'm impressed, but several times "tricked by the liver"
## Backups
## Test gradcam
```
normal_eval = Evaluator(ensemble=False, split=0) # , model_id="")
vid_in = vid_in_path + "Pneu_liftl_pneu_case3_clip5.mp4"
img = cv2.imread("../../data/my_found_data/Cov_efsumb1_2.png")
img = cv2.imread("../../data/pocus_images/convex/Cov_blines_covidmanifestation_paper2.png")
img = evaluator.preprocess(img)
grad = GradCAM()
out_map = grad.explain(img[0], evaluator.models[0], 0, return_map=False, layer_name="block5_conv3", zeroing=0.6)
plt.imshow(out_map.astype(int))
out_cam = get_class_activation_map(evaluator.models[0], img, 1, heatmap_weight=0.1, zeroing=0.8)
```
### Check cross val
```
check = "../../data/cross_validation"
file_list = []
for folder in os.listdir(check):
if folder[0]==".":
continue
for classe in os.listdir(os.path.join(check, folder)):
if classe[0]==".": # or classe[0]=="u":
continue
uni = []
is_image = 0
for file in os.listdir(os.path.join(check, folder, classe)):
if file[0]==".":
continue
if len(file.split("."))==2:
is_image+=1
file_list.append(file)
uni.append(file.split(".")[0])
# assert file[:3].lower()==classe[:3], "wrong label"+file[:3]+classe[:3]
print(folder, classe, len(np.unique(uni)), len(uni), is_image)
assert len(file_list)==len(np.unique(file_list))
print(len(file_list))
```
## Copy from train and test folders, give new ideas, and construct mapping
```
testcam = "vid_outputs_cam_test"
files_to_process = []
for subdir in os.listdir(testcam):
if subdir[0]=="." or subdir=="not taken" :
continue
for f in os.listdir(os.path.join(testcam, subdir)):
if f[0]==".":
continue
if not os.path.exists(os.path.join("vid_outputs_cam", f)):
print("does not exist in train", subdir, f)
# if not "RUQ" in f:
# todo.append(f.split(".")[0])
else:
files_to_process.append(os.path.join(subdir, f))
# print(todo)
# code to copy files to randomized thing
import shutil
drop_cams_dir = "vids_to_check"
test_cam_dir = "vid_outputs_cam_test"
train_cam_dir = "vid_outputs_cam"
# create directory
if not os.path.exists(drop_cams_dir):
os.makedirs(drop_cams_dir)
# give random ids
ids = np.random.permutation(len(files_to_process))
# define dataframe columns
new_fname = []
old_fname = []
is_train = []
fold = []
for i, f_name_path in enumerate(files_to_process):
split_name, f_name = tuple(f_name_path.split(os.sep))
split = int(split_name[-1])
# randomly add to model2
out_f_name = "video_"+str(ids[i])+"_model_"
old_fname.append(f_name)
old_fname.append(f_name)
rand_folder_train = np.random.rand()<0.5
print("train gets 1?", rand_folder_train)
# copy train data
train_outfname = out_f_name + str(int(rand_folder_train)) + ".mpeg"
train_to_path = os.path.join(drop_cams_dir, train_outfname)
cp_from_path = os.path.join(train_cam_dir, f_name)
# append for df
is_train.append(1)
fold.append(split)
new_fname.append(train_outfname)
print("TRAIN:", cp_from_path, train_to_path)
shutil.copy(cp_from_path, train_to_path)
# copy test
test_outfname = out_f_name + str(int(not rand_folder_train)) + ".mpeg"
test_to_path = os.path.join(drop_cams_dir, test_outfname)
cp_from_path = os.path.join(test_cam_dir, split_name, f_name)
# append for df
fold.append(split)
is_train.append(0)
new_fname.append(test_outfname)
print("TEST:", cp_from_path, test_to_path)
shutil.copy(cp_from_path, test_to_path)
df = pd.DataFrame()
df["previous_filename"] = old_fname
df["new_filename"] = new_fname
df["is_train"] = is_train
df["fold"] = fold
df.head(30)
df.to_csv(drop_cams_dir+"/mapping.csv")
iclus_dir = "test_data_regular/pat1"
# out_path = "iclus_videos_processed"
FRAMERATE = 3
MAX_FRAMES = 30
for fn in os.listdir(iclus_dir):
if fn[0]==".":
continue
cap = cv2.VideoCapture(os.path.join(iclus_dir, fn))
n_frames = cap.get(7)
frameRate = cap.get(5)
nr_selected = 0
every_x_image = int(frameRate / FRAMERATE)
while cap.isOpened() and nr_selected < MAX_FRAMES:
ret, frame = cap.read()
if (ret != True):
break
print(cap.get(1), cap.get(2), cap.get(3), cap.get(4), cap.get(5), cap.get(6), cap.get(7))
h, w, _ = frame.shape
# print(h,w)
plt.imshow(frame[30:400, 80:450])
plt.show()
# SAVE
# if ((frameId+1) % every_x_image == 0):
# # storing the frames in a new folder named test_1
# filename = out_path + fn + "_frame%d.jpg" % frameId
# cv2.imwrite(filename, frame)
# nr_selected += 1
# print(frameId, nr_selected)
cap.release()
import shutil
check = "../../data/cross_validation_segmented"
out = "../../data/cross_validation_segmented_new"
for folder in os.listdir(check):
if folder[0]==".":
continue
os.makedirs(os.path.join(out, folder))
for classe in os.listdir(os.path.join(check, folder)):
os.makedirs(os.path.join(out, folder, classe))
if classe[0]==".": # or classe[0]=="u":
continue
for f in os.listdir(os.path.join(check, folder, classe)):
if f[-3:]=="gif":
shutil.copy(os.path.join(check, folder, classe, f), os.path.join(out, folder, classe, f[:-4]))
elif f[-3:] =="npz":
shutil.copy(os.path.join(check, folder, classe, f), os.path.join(out, folder, classe, f))
```
### Cut Lotte's videos
```
file_list = ["pat1Image_132943.mpeg",
"pat1Image_133043.mpeg",
"pat1Image_133138.mpeg",
"pat1Image_133232.mpeg",
"pat1Image_133327.mpeg",
"pat1Image_133410.mpeg",
"pat2Image_133824.mpeg",
"pat2Image_133952.mpeg",
"pat2Image_134138.mpeg",
"pat2Image_134240.mpeg",
"pat2Image_134348.mpeg",
"pat2Image_134441.mpeg",
"pat3Image_134711.mpeg",
"pat3Image_134811.mpeg",
"pat3Image_134904.mpeg",
"pat3Image_135026.mpeg",
"pat3Image_135128.mpeg",
"pat3Image_135215.mpeg",
"pat4Image_135904.mpeg",
"pat4Image_140024.mpeg",
"pat4Image_140238.mpeg",
"pat4Image_140434.mpeg",
"pat4Image_140606.mpeg",
"pat4Image_140705.mpeg"]
copy_path = "../../data/pocus_videos/convex/"
for f in file_list:
video_path = "reg_propro/"+f
# cap = cv2.VideoCapture(video_path)
# print(cap.get(7))
# cap.release()
print("Reg_"+f)
shutil.copy(video_path, copy_path+"Reg_"+f)
out_dir = "reg_propro/pat4"
in_dir = "test_data_regular/pat4"
for vid in os.listdir(in_dir):
if vid[0]==".":
continue
video_path = os.path.join(in_dir, vid)
cap = cv2.VideoCapture(video_path)
images = []
counter = 0
while cap.isOpened():
ret, frame = cap.read()
if (ret != True):
break
if counter<1:
plt.imshow(frame[30:400, 80:450]) # ICLUS: [70:570, 470:970]) # [25:350, 100:425]) # LOTTE:[30:400, 80:450]
plt.show()
counter += 1
continue
counter += 1
img_processed = frame[30:400, 80:450]
images.append(img_processed)
cap.release()
images = np.asarray(images)
print(images.shape)
io.vwrite(out_dir+ vid.split(".")[0]+".mpeg", images, outputdict={"-vcodec":"mpeg2video"})
```
### Display logo on frames
```
test_vid = "../../data/pocus_videos/convex/Pneu-Atlas-pneumonia2.gif"
cap = cv2.VideoCapture(test_vid)
ret, frame = cap.read()
cap.release()
plt.imshow(frame)
plt.show()
logo = plt.imread("Logo.png")
logo = cv2.resize(logo, (50,50), )
plt.imshow(logo)
```
|
github_jupyter
|
# Collect Physicists Raw Data
The goal of this notebook is to collect demographic data on the list of [physicists notable for their achievements](../data/raw/physicists.txt). Wikipedia contains this semi-structured data in an *Infobox* on the top right side of the article for each physicist. However, similar data is available in a more machine readable, [JSON](https://www.json.org/) format from [DBpedia](https://wiki.dbpedia.org/about). We will need to send HTTP requests to DBpedia to get the JSON data. For an example, compare *Albert Einstein's* [Wikipedia infobox](https://en.wikipedia.org/wiki/Albert_Einstein) to his [DBPedia JSON](http://dbpedia.org/data/Albert_Einstein.json). It is important to realize, that although the data is similar, it is not identical.
The shortcomings of Wikipedia infoboxes and the advantages of DBpedia datasets are explained in section 4.3 of [DBpedia datasets](https://wiki.dbpedia.org/services-resources/datasets/dbpedia-datasets#h434-10). But basically the summary is that DBpedia data is much cleaner and better structured than Wikipedia Infoboxes as it is based on hand-generated mappings of Wikipedia infoboxes / templates to a [DBpedia ontology](https://wiki.dbpedia.org/services-resources/ontology). Consequently, we will be using DBpedia as the data source for this project.
However, DBpedia does have the disadvantage that its content is roughly 6-18 months behind updates applied to Wikipedia content. This is due to its data being generated from a [static dump of Wikipedia content](https://wiki.dbpedia.org/online-access/DBpediaLive) in a process that takes approximately 6 months. The fact that the data is not in sync with the latest Wikipedia content is not of great significance for this project as the data is edited infrequently. Also when edits are made, they tend to be only minor.
## Setting the Environment
A few initialization steps are needed to setup the environment:
- The locale needs to be set for all categories to the user’s default setting (typically specified in the LANG environment variable) to enable correct sorting of physicists names with accents.
- A bool constant `FETCH_JSON_DATA` needs to be set to decide whether to fetch the json data. Set to False so that the previously fetched data is used. In this case the results of the study are guaranteed be reproducible. Set to True so that the latest data is fetched. In this case it is possible that the results of the study will change.
```
import locale
locale.setlocale(locale.LC_ALL, '')
FETCH_JSON_DATA = False
```
## Constructing the URLs
To make the HTTP requests, we will need a list of URLs representing the resources (i.e the physicists). It's fairly easy to construct these URLs from the list of notable physicists. However, it's important to "quote" any physicist name in unicode since unicode characters are not allowed in URLs. OK let's create the list now.
```
import gzip
import os
import shutil
from collections import OrderedDict
import jsonlines
import pandas as pd
from src.data.jsonl_utils import read_jsonl
from src.data.url_utils import DBPEDIA_DATA_URL
from src.data.url_utils import fetch_json_data
from src.data.url_utils import urls_progress_bar
def construct_urls(file='../data/raw/physicists.txt'):
"""Construct DBpedia data URLs from list in file.
Args:
file (str): File containing a list of url filepaths
with spaces replacing underscores.
Returns:
list(str): List of URLs.
"""
with open(file, encoding='utf-8') as file:
names = [line.rstrip('\n') for line in file]
urls = [DBPEDIA_DATA_URL + name.replace(' ', '_') + '.json'
for name in names]
return urls
urls_to_fetch = construct_urls()
assert(len(urls_to_fetch) == 1069)
```
## Fetching the Data
Now we have the list of URLs, it's time to make the HTTP requests to acquire the data. The code is asynchronous, which dramatically helps with performance. It is important to set the `max_workers` parameter sensibly in order to crawl responsibly and not hammer the site's server. Although the site seems to be rate limited, it's still good etiquette.
```
jsonl_file = '../data/raw/physicists.jsonl'
if FETCH_JSON_DATA:
json_data = fetch_json_data(urls_to_fetch, max_workers=20, timeout=30,
progress_bar=urls_progress_bar(len(urls_to_fetch)))
else:
json_data = read_jsonl('../data/raw/physicists.jsonl' + '.gz')
```
Let's sort the data alphabetically by URL, confirm that all the data was fetched and take a look at the first JSON response.
```
if FETCH_JSON_DATA:
json_data = OrderedDict(sorted(json_data.items(), key=lambda x: locale.strxfrm(x[0])))
assert(len(json_data) == 1069)
print(list(json_data.keys())[0])
print(list(json_data.values())[0])
else:
assert(len(json_data) == 1058)
print(json_data[0])
```
It is clear that every request successfully received a response. However, we see that some responses came back empty from the server. Basically, although there are Wikipedia pages for these physicists, they do not have a corresponding page in DBpedia (or the page in DBpedia has a different name). Not to worry, there are only 11 and they are not so famous, so we will just exclude these "Z-listers" from the analysis.
```
if FETCH_JSON_DATA:
urls_to_drop = [url for (url, data) in json_data.items() if not data]
assert(len(urls_to_drop) == 11)
display(urls_to_drop)
if FETCH_JSON_DATA:
json_data = [data for data in json_data.values() if data]
assert(len(json_data) == 1058)
```
## Persisting the Data
Now that we have the list of JSON responses, we would like to persist them for later analysis. We will use [Json Lines](http://jsonlines.org/) as it seems like a convenient format for storing structured data that may be processed one record at a time.
```
if FETCH_JSON_DATA:
with jsonlines.open(jsonl_file, 'w') as writer:
writer.write_all(json_data)
```
Let's do a quick sanity check to make sure the file contains the expected number of records.
```
if FETCH_JSON_DATA:
json_lines = read_jsonl(jsonl_file)
assert(len(json_lines) == 1058)
```
Finally, let's compress the file to reduce its footprint.
```
if FETCH_JSON_DATA:
with open(jsonl_file, 'rb') as src, gzip.open(jsonl_file + '.gz', 'wb') as dest:
shutil.copyfileobj(src, dest)
os.remove(jsonl_file)
```
|
github_jupyter
|
# 1-1. AIとは何か?簡単なAIを設計してみよう
AIブームに伴って、様々なメディアでAIや機械学習、深層学習といった言葉が使われています。本章ではAIと機械学習(ML)、深層学習の違いを理解しましょう。
## 人工知能(AI)とは?
そもそも人工知能(AI)とは何でしょうか?
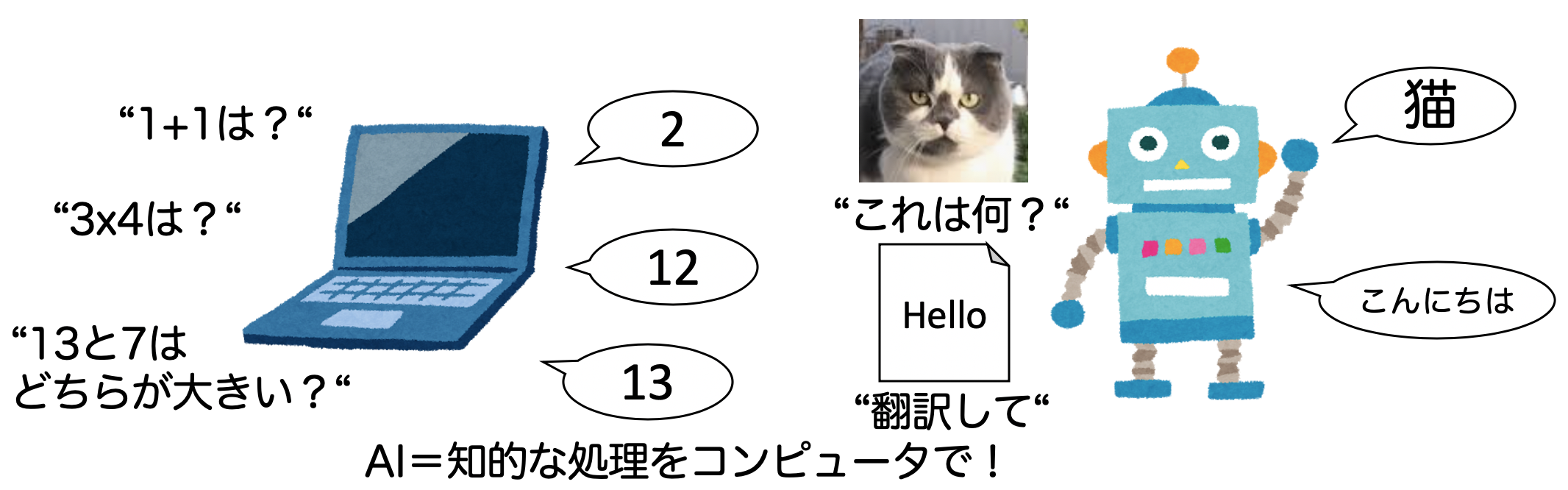
Wikipedia[1]によると、人工知能について以下のように書かれています。
人工知能(じんこうちのう、英: artificial intelligence、AI〈エーアイ〉)とは「言語の理解や推論、問題解決などの知的行動を人間に代わってコンピュータに行わせる技術」。
要は知的行動をコンピュータが行う技術のことですね。。もう少し歴史を遡ってみると、過去のコンピュータは日本語で計算機、その言葉通り「計算」をするための機械でした。今で言うと「電卓」そのものですね。つまり、電卓で行う計算
(左の図)ではできない絵の認識や翻訳(右の図)などを知的な処理とし、その処理をコンピュータ、すなわち電卓などの計算機で行うんですね。
つまり、こんな感じでしょうか。↓
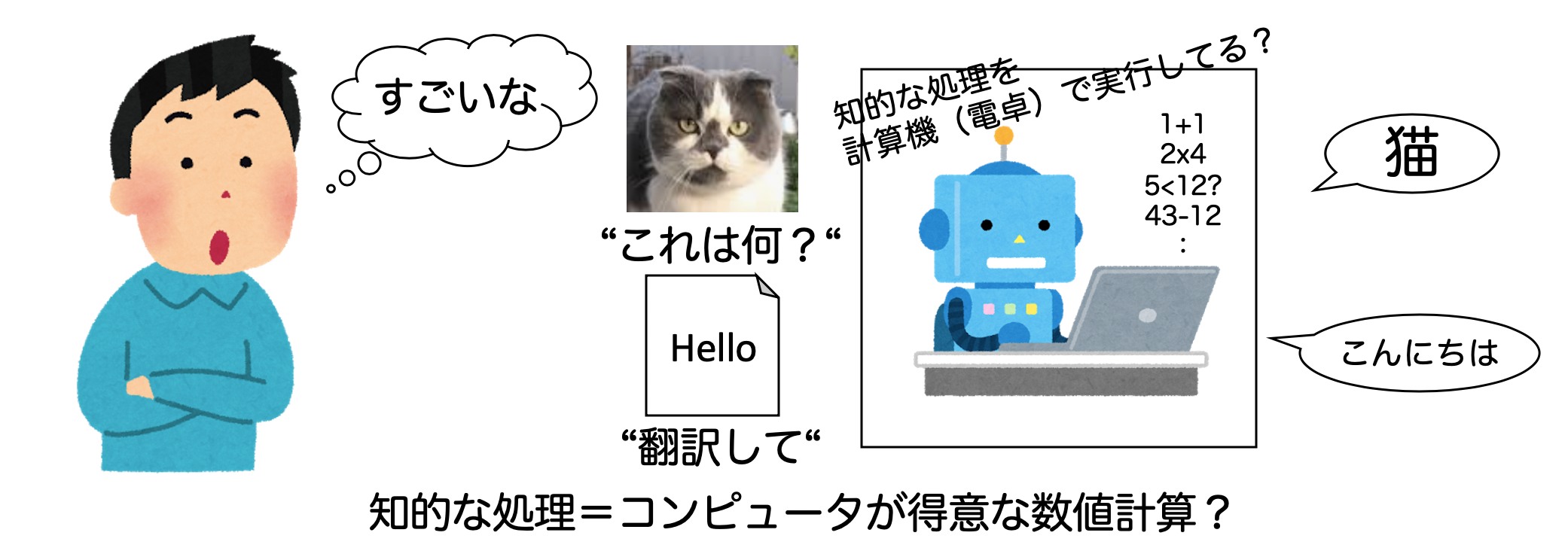
あれ?と思われるかもしれませんが、現実は皆さんの身の回りのコンピュータが認識や翻訳をやってますよね。コンピュータに脳は入っていません。つまり、脳が行っている「知的な処理」というのをコンピュータが得意な電卓で行う計算に置き換えて処理しているのです。
## 今のコンピュータでできること
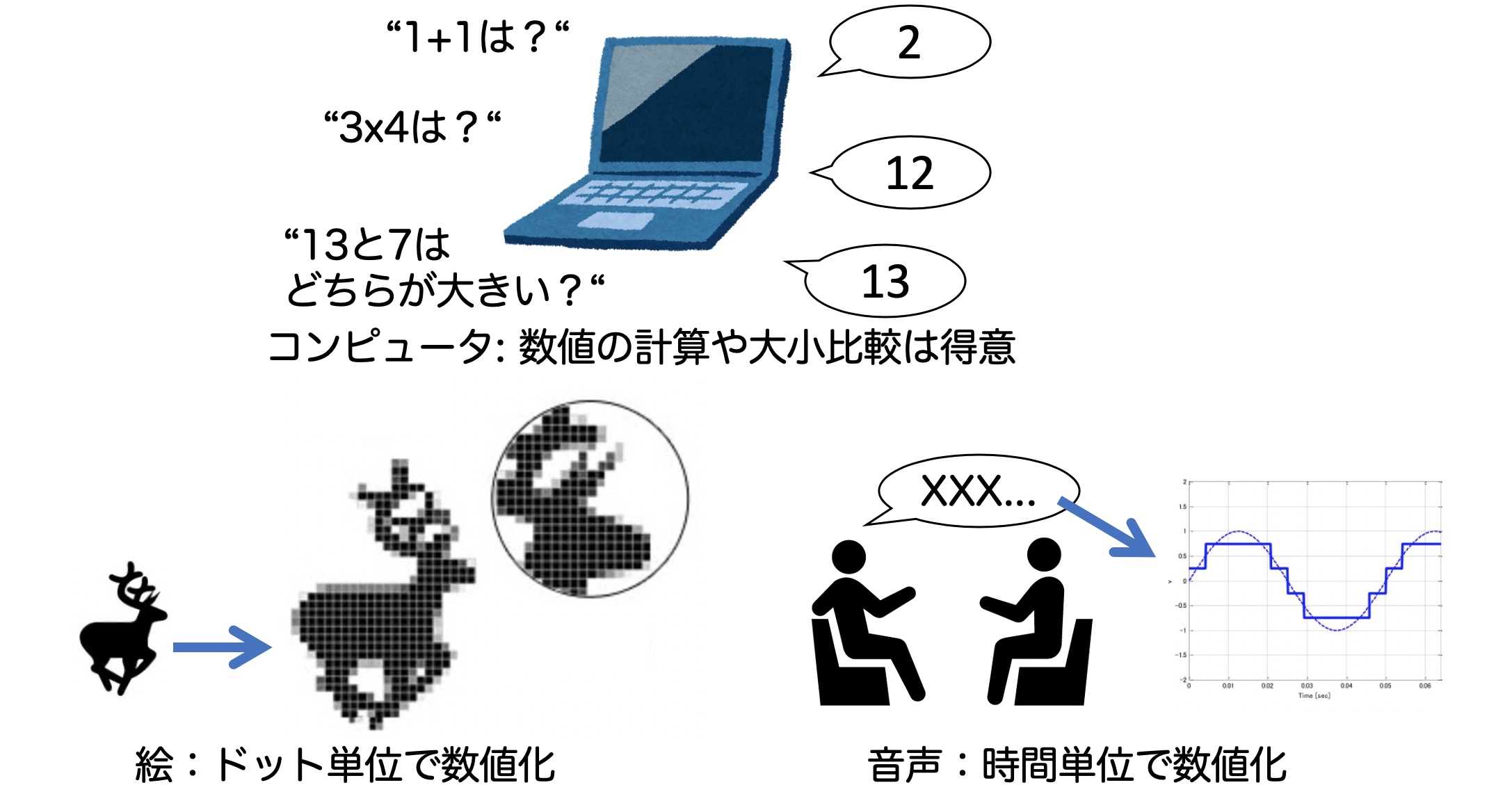
そもそもコンピュータが得意な処理とは何でしょうか?それは電卓の例でもわかるように、数値の計算です。あと、数値の大小比較も得意です。つまり、数値にしてしまえばコンピュータで色々できそうですね。実際、写真の加工や音声の合成などはそれぞれデータを数値化することでコンピュータが処理できるようにしています。AIも数値を扱う問題に変換してしまえばよさそうですね。
# 簡単なAIを作ってみよう
## ミニトマトを出荷用に収穫するかどうか判定するAI

早速AIを作ってみてどんなものか体験してみましょう。ここではミニトマト農家になったつもりで、収穫するかどうかを自動で見分けてくれるAIを作ることにしましょう。
## コンピュータが処理できる数値の計算・比較処理に直す
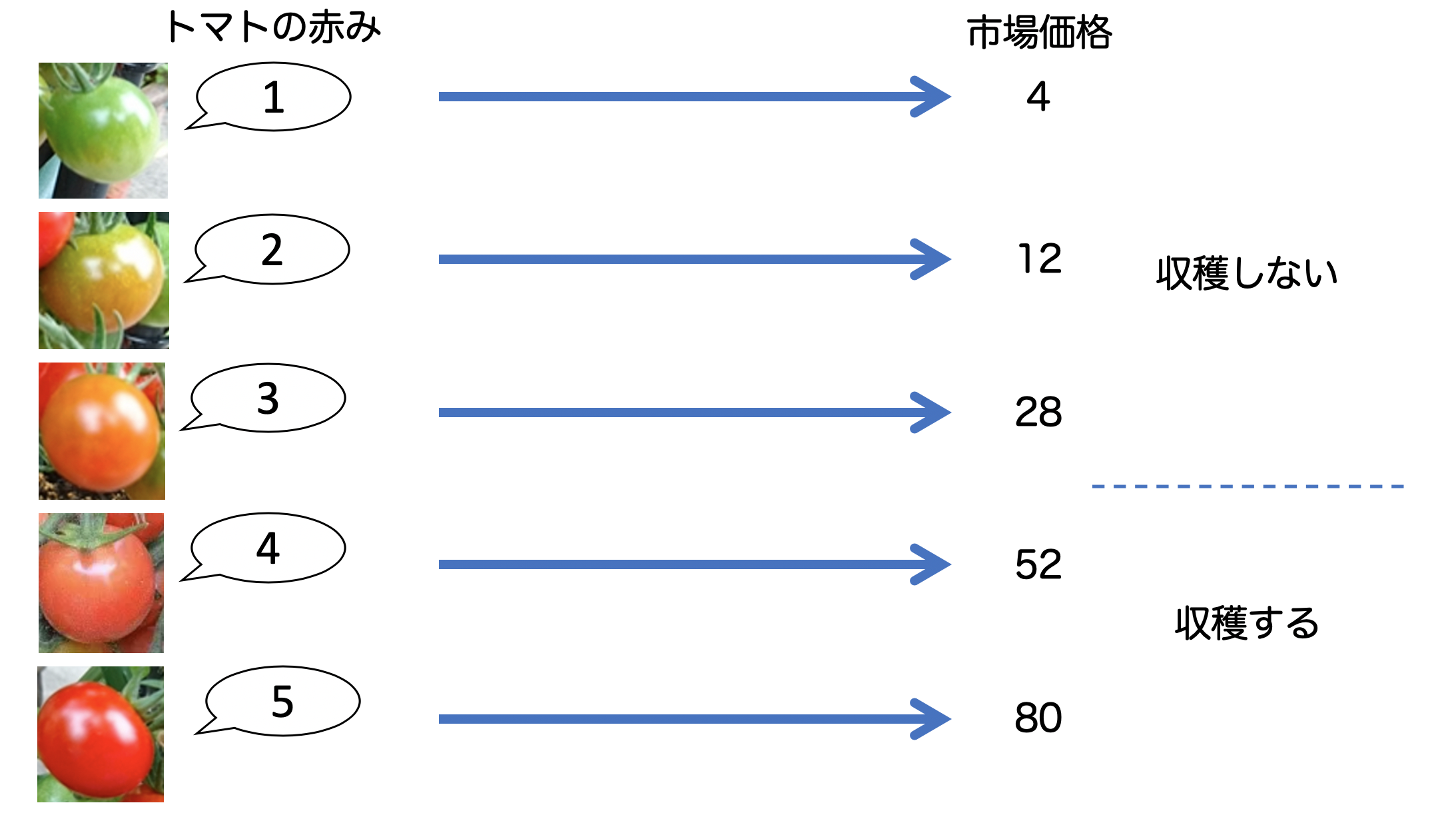
コンピュータは数値の計算と比較が得意なので、数値に直しましょう。例えばトマトの赤みを数値化することは画像処理(後半の章でOpenCVというライブラリを説明します)で比較的簡単にできます。市場価格はスーパーとかでトマトのパックを買って1個当たりの値段を算出すればわりと正確な値段が出ると思います。ここではあくまで私が適当に付けた値段ですが。。
最後に、市場価格から収穫する/しないかどうかを決めます。これは予想した市場価格に対してあらかじめ決めておいた値との比較でできますね。
```
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
x = np.array([1,2,3,4,5])
y = np.array([4,12,28,52,80])
plt.plot(x,y, label="tomato", marker="o")
```
この関係をグラフに書いてみましょう。そうです、この関係を正しく推定できるとミニトマトの収穫を判断するAIができあがるのです!
さて、どうやって推定しましょうか。。
## 直線で近似してみる
学生時代(私は中学生)に習った直線の方程式で推定してみましょう。2次元グラフは任意の2点(x1,y1)-(x2,y2)間を直線で表現できます。
直線による表現→y=ax+b でしたから、先ほどのグラフの数値(x1,y1)=(1,4), (x2,y2)=(5,80)より
傾き: a=(y2-y1)/(x2-x1)=(80-4)/(5-1)=76/4=19
切片: b=y-ax=4-19×1=-15
従って y=19x-15 という直線式を得ました。これに間の値を代入してどの程度正しく予測できたかみてみましょう。
```
a = 19
b = -15
y_est1 = a * x + b
plt.plot(x,y, label="tomato", marker="o")
plt.plot(x,y_est1, label="estimation", marker="o")
```
うーん、ダメですね。。両端は上手く予想出来ていますが、その間の誤差が大きそうです。。
## 誤差を最小に抑える
トライアンドエラーで誤差が最も小さくなる直線、すなわち傾きと切片を求めるのは大変そうです。そこで、全ての点の二乗誤差の合計を最小にするようにしましょう。二乗を使うのは正負に影響を受けないようにするためです。まず、傾きaの計算式は
a= Sum of (x-xの平均)*(y-yの平均) / Sum of (x-xの平均)^2
です。計算してみましょう。
x_mean = (1+2+3+4+5)/5=3
a = 192/10=19.2 と計算できましたね。Jupyter NotebookはPythonプログラミングもできるので、試行錯誤しながらプログラミングするのにとても向いています。
切片bは
b=yの平均 - a×xの平均
です。計算するとb=-22.4です。
```
a = 19.2
b = -22.4
y_est2 = a * x + b
plt.plot(x,y, label="tomato", marker="o")
plt.plot(x,y_est2, label="estimation (mean of squared error(MSE))", marker="o")
```
おお、今度はデータの間をちょうど通る直線が引けましたね!なんとなく実際の市場価格を推定できてそうです。
## 他の近似式では?
するどい!何も直線式で近似する必要はないです。その通りです。2次式・多項式・さらに複雑な式、、色々ありすぎて困りますね。AIの設計は近似式をどうやって決めるかがポイントですがトライアンドエラーをするしかないのが現状です。それに、近似式のいろいろな値(今回は傾きと切片)を決めるのも大変そうですね。。
最近ではクラウド上の計算機を大量に使って力技で探しています。ただ、それでも探す範囲が広すぎるので全自動化はまだ難しいのが現状です。
## これまでのステップは機械学習そのものだった
実はこれまでの一連の作業は機械学習というAIの1分野の手法を使ったのでした。具体的には
色や市場価格を準備する→データを収集する
近似式を決める→モデルを設計する
切片や傾きを求める→学習を行う(ハイパーパラメータを決める)
推定がどれくらい正確かを確認する→モデルの検定を行う
をやっていたのです。最近、機械学習・AIが紛れていましたが、なんとなく両者の関係がわかってきましたでしょうか?AIを設計する=機械学習で設計する、といっても過言でないくらい今日では機械学習が主流となっています。次回は機械学習をより詳しく説明します。
## 課題
AIを使ったアプリケーション・製品について一つ調査し、レポートを提出してください。
フォーマット:PDF形式(図、文章、参考にした文献(URL))
提出先:T2Scholar
締め切り: (講義中にアナウンスします)
## 参考文献
[1] https://ja.wikipedia.org/wiki/%E4%BA%BA%E5%B7%A5%E7%9F%A5%E8%83%BD
|
github_jupyter
|
## UBC Intro to Machine Learning
### APIs
Instructor: Socorro Dominguez
February 05, 2022
## Exercise to try in your local machine
## Motivation
For our ML class, we want to do a Classifier that differentiates images from dogs and cats.
## Problem
We need a dataset to do this. Our friends don't have enough cats and dogs.
Let's take free, open and legal data from the [Unsplash Image API](https://unsplash.com/developers).
## Caveats
Sometimes, raw data is unsuitable for machine learning algorithms. For instance, we may want:
- Only images that are landscape (i.e. width > height)
- All our images to be of the same resolution
---
## Step 1: Get cat and dog image URLs from the API
We will use the [`search/photos` GET method](https://unsplash.com/documentation#search-photos).
```
import requests
import config as cfg
# API variables
root_endpoint = 'https://api.unsplash.com/'
client_id = cfg.splash['key']
# Wrapper function for making API calls and grabbing results
def search_photos(search_term):
api_method = 'search/photos'
endpoint = root_endpoint + api_method
response = requests.get(endpoint,
params={'query': search_term, 'per_page': 30, 'client_id': client_id})
status_code, result = response.status_code, response.json()
if status_code != 200:
print(f'Bad status code: {status_code}')
image_urls = [img['urls']['small'] for img in result['results']]
return image_urls
dog_urls = search_photos('dog')
cat_urls = search_photos('cat')
cat_urls
```
---
## Step 2: Download the images from the URLs
(Step 2a: Google [how to download an image from a URL in Python](https://stackoverflow.com/a/40944159))
We'll just define the function to download an image for now. Later on, we'll use it on images one at a time (but after doing some processing).
```
from PIL import Image
def download_image(url):
image = Image.open(requests.get(url, stream=True).raw)
return image
test_img = download_image(cat_urls[0])
test_img.show()
```
---
## Step 3: Download and save images that meet our requirements
We'll need to know how to work with the [PIL Image data type](https://pillow.readthedocs.io/en/stable/reference/Image.html), which is what our `download_image(url)` function returns. Namely, we need to be able to a) get it's resolution and b) resize it.
```
import os
def is_landscape(image):
return image.width > image.height
def save_category_images(urls, category_name, resolution=(256, 256)):
save_folder = f'saved_images/{category_name}'
if not os.path.exists(save_folder):
os.mkdir(save_folder)
for i, url in enumerate(urls):
image = download_image(url)
if is_landscape(image):
image = image.resize(resolution)
filename = f'{i:05d}.jpg'
image.save(os.path.join(save_folder, filename))
save_category_images(dog_urls, 'dogs')
save_category_images(cat_urls, 'cats')
```
|
github_jupyter
|
```
#12/29/20
#runnign synthetic benchmark graphs for synthetic OR datasets generated
#making benchmark images
import keras
from keras.models import Sequential, Model, load_model
from keras.layers import Dense, Dropout, Activation, Flatten, Input, Lambda
from keras.layers import Conv2D, MaxPooling2D, AveragePooling2D, Conv1D, MaxPooling1D, LSTM, ConvLSTM2D, GRU, CuDNNLSTM, CuDNNGRU, BatchNormalization, LocallyConnected2D, Permute, TimeDistributed, Bidirectional
from keras.layers import Concatenate, Reshape, Softmax, Conv2DTranspose, Embedding, Multiply
from keras.callbacks import ModelCheckpoint, EarlyStopping, Callback
from keras import regularizers
from keras import backend as K
from keras.utils.generic_utils import Progbar
from keras.layers.merge import _Merge
import keras.losses
from keras.datasets import mnist
from functools import partial
from collections import defaultdict
import tensorflow as tf
from tensorflow.python.framework import ops
import isolearn.keras as iso
import numpy as np
import tensorflow as tf
import logging
logging.getLogger('tensorflow').setLevel(logging.ERROR)
import os
import pickle
import numpy as np
import isolearn.io as isoio
import isolearn.keras as isol
import pandas as pd
import scipy.sparse as sp
import scipy.io as spio
import matplotlib.pyplot as plt
from sequence_logo_helper import dna_letter_at, plot_dna_logo
from keras.backend.tensorflow_backend import set_session
def contain_tf_gpu_mem_usage() :
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
sess = tf.Session(config=config)
set_session(sess)
contain_tf_gpu_mem_usage()
class EpochVariableCallback(Callback) :
def __init__(self, my_variable, my_func) :
self.my_variable = my_variable
self.my_func = my_func
def on_epoch_begin(self, epoch, logs={}) :
K.set_value(self.my_variable, self.my_func(K.get_value(self.my_variable), epoch))
#ONLY RUN THIS CELL ONCE
from tensorflow.python.framework import ops
#Stochastic Binarized Neuron helper functions (Tensorflow)
#ST Estimator code adopted from https://r2rt.com/beyond-binary-ternary-and-one-hot-neurons.html
#See Github https://github.com/spitis/
def st_sampled_softmax(logits):
with ops.name_scope("STSampledSoftmax") as namescope :
nt_probs = tf.nn.softmax(logits)
onehot_dim = logits.get_shape().as_list()[1]
sampled_onehot = tf.one_hot(tf.squeeze(tf.multinomial(logits, 1), 1), onehot_dim, 1.0, 0.0)
with tf.get_default_graph().gradient_override_map({'Ceil': 'Identity', 'Mul': 'STMul'}):
return tf.ceil(sampled_onehot * nt_probs)
def st_hardmax_softmax(logits):
with ops.name_scope("STHardmaxSoftmax") as namescope :
nt_probs = tf.nn.softmax(logits)
onehot_dim = logits.get_shape().as_list()[1]
sampled_onehot = tf.one_hot(tf.argmax(nt_probs, 1), onehot_dim, 1.0, 0.0)
with tf.get_default_graph().gradient_override_map({'Ceil': 'Identity', 'Mul': 'STMul'}):
return tf.ceil(sampled_onehot * nt_probs)
@ops.RegisterGradient("STMul")
def st_mul(op, grad):
return [grad, grad]
#Gumbel Distribution Sampler
def gumbel_softmax(logits, temperature=0.5) :
gumbel_dist = tf.contrib.distributions.RelaxedOneHotCategorical(temperature, logits=logits)
batch_dim = logits.get_shape().as_list()[0]
onehot_dim = logits.get_shape().as_list()[1]
return gumbel_dist.sample()
#PWM Masking and Sampling helper functions
def mask_pwm(inputs) :
pwm, onehot_template, onehot_mask = inputs
return pwm * onehot_mask + onehot_template
def sample_pwm_st(pwm_logits) :
n_sequences = K.shape(pwm_logits)[0]
seq_length = K.shape(pwm_logits)[2]
flat_pwm = K.reshape(pwm_logits, (n_sequences * seq_length, 4))
sampled_pwm = st_sampled_softmax(flat_pwm)
return K.reshape(sampled_pwm, (n_sequences, 1, seq_length, 4))
def sample_pwm_gumbel(pwm_logits) :
n_sequences = K.shape(pwm_logits)[0]
seq_length = K.shape(pwm_logits)[2]
flat_pwm = K.reshape(pwm_logits, (n_sequences * seq_length, 4))
sampled_pwm = gumbel_softmax(flat_pwm, temperature=0.5)
return K.reshape(sampled_pwm, (n_sequences, 1, seq_length, 4))
#Generator helper functions
def initialize_sequence_templates(generator, sequence_templates, background_matrices) :
embedding_templates = []
embedding_masks = []
embedding_backgrounds = []
for k in range(len(sequence_templates)) :
sequence_template = sequence_templates[k]
onehot_template = iso.OneHotEncoder(seq_length=len(sequence_template))(sequence_template).reshape((1, len(sequence_template), 4))
for j in range(len(sequence_template)) :
if sequence_template[j] not in ['N', 'X'] :
nt_ix = np.argmax(onehot_template[0, j, :])
onehot_template[:, j, :] = -4.0
onehot_template[:, j, nt_ix] = 10.0
elif sequence_template[j] == 'X' :
onehot_template[:, j, :] = -1.0
onehot_mask = np.zeros((1, len(sequence_template), 4))
for j in range(len(sequence_template)) :
if sequence_template[j] == 'N' :
onehot_mask[:, j, :] = 1.0
embedding_templates.append(onehot_template.reshape(1, -1))
embedding_masks.append(onehot_mask.reshape(1, -1))
embedding_backgrounds.append(background_matrices[k].reshape(1, -1))
embedding_templates = np.concatenate(embedding_templates, axis=0)
embedding_masks = np.concatenate(embedding_masks, axis=0)
embedding_backgrounds = np.concatenate(embedding_backgrounds, axis=0)
generator.get_layer('template_dense').set_weights([embedding_templates])
generator.get_layer('template_dense').trainable = False
generator.get_layer('mask_dense').set_weights([embedding_masks])
generator.get_layer('mask_dense').trainable = False
generator.get_layer('background_dense').set_weights([embedding_backgrounds])
generator.get_layer('background_dense').trainable = False
#Generator construction function
def build_sampler(batch_size, seq_length, n_classes=1, n_samples=1, sample_mode='st') :
#Initialize Reshape layer
reshape_layer = Reshape((1, seq_length, 4))
#Initialize background matrix
onehot_background_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='zeros', name='background_dense')
#Initialize template and mask matrices
onehot_template_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='zeros', name='template_dense')
onehot_mask_dense = Embedding(n_classes, seq_length * 4, embeddings_initializer='ones', name='mask_dense')
#Initialize Templating and Masking Lambda layer
masking_layer = Lambda(mask_pwm, output_shape = (1, seq_length, 4), name='masking_layer')
background_layer = Lambda(lambda x: x[0] + x[1], name='background_layer')
#Initialize PWM normalization layer
pwm_layer = Softmax(axis=-1, name='pwm')
#Initialize sampling layers
sample_func = None
if sample_mode == 'st' :
sample_func = sample_pwm_st
elif sample_mode == 'gumbel' :
sample_func = sample_pwm_gumbel
upsampling_layer = Lambda(lambda x: K.tile(x, [n_samples, 1, 1, 1]), name='upsampling_layer')
sampling_layer = Lambda(sample_func, name='pwm_sampler')
permute_layer = Lambda(lambda x: K.permute_dimensions(K.reshape(x, (n_samples, batch_size, 1, seq_length, 4)), (1, 0, 2, 3, 4)), name='permute_layer')
def _sampler_func(class_input, raw_logits) :
#Get Template and Mask
onehot_background = reshape_layer(onehot_background_dense(class_input))
onehot_template = reshape_layer(onehot_template_dense(class_input))
onehot_mask = reshape_layer(onehot_mask_dense(class_input))
#Add Template and Multiply Mask
pwm_logits = masking_layer([background_layer([raw_logits, onehot_background]), onehot_template, onehot_mask])
#Compute PWM (Nucleotide-wise Softmax)
pwm = pwm_layer(pwm_logits)
#Tile each PWM to sample from and create sample axis
pwm_logits_upsampled = upsampling_layer(pwm_logits)
sampled_pwm = sampling_layer(pwm_logits_upsampled)
sampled_pwm = permute_layer(sampled_pwm)
sampled_mask = permute_layer(upsampling_layer(onehot_mask))
return pwm_logits, pwm, sampled_pwm, onehot_mask, sampled_mask
return _sampler_func
#for formulation 2 graphing
def returnXMeanLogits(e_train):
#returns x mean logits for displayign the pwm difference for the version 2 networks
#Visualize background sequence distribution
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
pseudo_count = 1.0
x_mean = (np.sum(x_train, axis=(0, 1)) + pseudo_count) / (x_train.shape[0] + 4. * pseudo_count)
x_mean_logits = np.log(x_mean / (1. - x_mean))
return x_mean_logits, x_mean
#loading testing dataset
from optimusFunctions import *
import pandas as pd
csv_to_open = "optimus5_synthetic_random_insert_if_uorf_2_start_1_stop_variable_loc_512.csv"
dataset_name = csv_to_open.replace(".csv", "")
print (dataset_name)
data_df = pd.read_csv("./" + csv_to_open) #open from scores folder
#loaded test set which is sorted by number of start/stop signals
seq_e_test = one_hot_encode(data_df, seq_len=50)
benchmarkSet_seqs = seq_e_test
x_test = np.reshape(benchmarkSet_seqs, (benchmarkSet_seqs.shape[0], 1, benchmarkSet_seqs.shape[1], benchmarkSet_seqs.shape[2]))
print (x_test.shape)
e_train = pd.read_csv("bottom5KIFuAUGTop5KIFuAUG.csv")
print ("training: ", e_train.shape[0], " testing: ", x_test.shape[0])
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_mean_logits, x_mean = returnXMeanLogits(e_train)
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
#background
#for formulation 2 graphing
def returnXMeanLogits(e_train):
#returns x mean logits for displayign the pwm difference for the version 2 networks
#Visualize background sequence distribution
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
pseudo_count = 1.0
x_mean = (np.sum(x_train, axis=(0, 1)) + pseudo_count) / (x_train.shape[0] + 4. * pseudo_count)
x_mean_logits = np.log(x_mean / (1. - x_mean))
return x_mean_logits, x_mean
e_train = pd.read_csv("bottom5KIFuAUGTop5KIFuAUG.csv")
print ("training: ", e_train.shape[0], " testing: ", x_test.shape[0])
#one hot encode with optimus encoders
seq_e_train = one_hot_encode(e_train,seq_len=50)
x_mean_logits, x_mean = returnXMeanLogits(e_train)
x_train = seq_e_train
x_train = np.reshape(x_train, (x_train.shape[0], 1, x_train.shape[1], x_train.shape[2]))
#Define sequence template for optimus
sequence_template = 'N'*50
sequence_mask = np.array([1 if sequence_template[j] == 'N' else 0 for j in range(len(sequence_template))])
#Visualize background sequence distribution
save_figs = True
plot_dna_logo(np.copy(x_mean), sequence_template=sequence_template, figsize=(14, 0.65), logo_height=1.0, plot_start=0, plot_end=50)
#Calculate mean training set conservation
entropy = np.sum(x_mean * -np.log(x_mean), axis=-1) / np.log(2.0)
conservation = 2.0 - entropy
x_mean_conservation = np.sum(conservation) / np.sum(sequence_mask)
print("Mean conservation (bits) = " + str(x_mean_conservation))
#Calculate mean training set kl-divergence against background
x_train_clipped = np.clip(np.copy(x_train[:, 0, :, :]), 1e-8, 1. - 1e-8)
kl_divs = np.sum(x_train_clipped * np.log(x_train_clipped / np.tile(np.expand_dims(x_mean, axis=0), (x_train_clipped.shape[0], 1, 1))), axis=-1) / np.log(2.0)
x_mean_kl_divs = np.sum(kl_divs * sequence_mask, axis=-1) / np.sum(sequence_mask)
x_mean_kl_div = np.mean(x_mean_kl_divs)
print("Mean KL Div against background (bits) = " + str(x_mean_kl_div))
#Initialize Encoder and Decoder networks
batch_size = 32
seq_length = 50
n_samples = 128
sample_mode = 'st'
#sample_mode = 'gumbel'
#Load sampler
sampler = build_sampler(batch_size, seq_length, n_classes=1, n_samples=n_samples, sample_mode=sample_mode)
#Load Predictor
predictor_path = 'optimusRetrainedMain.hdf5'
predictor = load_model(predictor_path)
predictor.trainable = False
predictor.compile(optimizer=keras.optimizers.SGD(lr=0.1), loss='mean_squared_error')
#Build scrambler model
dummy_class = Input(shape=(1,), name='dummy_class')
input_logits = Input(shape=(1, seq_length, 4), name='input_logits')
pwm_logits, pwm, sampled_pwm, pwm_mask, sampled_mask = sampler(dummy_class, input_logits)
scrambler_model = Model([input_logits, dummy_class], [pwm_logits, pwm, sampled_pwm, pwm_mask, sampled_mask])
#Initialize Sequence Templates and Masks
initialize_sequence_templates(scrambler_model, [sequence_template], [x_mean_logits])
scrambler_model.trainable = False
scrambler_model.compile(
optimizer=keras.optimizers.Adam(lr=0.001, beta_1=0.9, beta_2=0.999),
loss='mean_squared_error'
)
#open all score and reshape as needed
file_names = [
"l2x_" + dataset_name + "_importance_scores_test.npy",
"invase_" + dataset_name + "_conv_importance_scores_test.npy",
"l2x_" + dataset_name + "_full_data_importance_scores_test.npy",
"invase_" + dataset_name + "_conv_full_data_importance_scores_test.npy",
]
#deepexplain_optimus_utr_OR_logic_synth_1_start_2_stops_method_integrated_gradients_importance_scores_test.npy
model_names =[
"l2x",
"invase",
"l2x_full_data",
"invase_full_data",
]
model_importance_scores_test = [np.load("./" + file_name) for file_name in file_names]
for scores in model_importance_scores_test:
print (scores.shape)
for model_i in range(len(model_names)) :
if model_importance_scores_test[model_i].shape[-1] > 1 :
model_importance_scores_test[model_i] = np.sum(model_importance_scores_test[model_i], axis=-1, keepdims=True)
for scores in model_importance_scores_test:
print (scores.shape)
#reshape for mse script -> if not (3008, 1, 50, 1) make it that shape
idealShape = model_importance_scores_test[0].shape
print (idealShape)
for model_i in range(len(model_names)) :
if model_importance_scores_test[model_i].shape != idealShape:
model_importance_scores_test[model_i] = np.expand_dims(model_importance_scores_test[model_i], 1)
for scores in model_importance_scores_test:
print (scores.shape)
on_state_logit_val = 50.
print (x_test.shape)
dummy_test = np.zeros((x_test.shape[0], 1))
x_test_logits = 2. * x_test - 1.
print (x_test_logits.shape)
print (dummy_test.shape)
x_test_squeezed = np.squeeze(x_test)
y_pred_ref = predictor.predict([x_test_squeezed], batch_size=32, verbose=True)[0]
_, _, _, pwm_mask, sampled_mask = scrambler_model.predict([x_test_logits, dummy_test], batch_size=batch_size)
feature_quantiles = [0.76, 0.82, 0.88]
for name in model_names:
for quantile in feature_quantiles:
totalName = name + "_" + str(quantile).replace(".","_") + "_quantile_MSE"
data_df[totalName] = None
print (data_df.columns)
feature_quantiles = [0.76, 0.82, 0.88]
#batch_size = 128
from sklearn import metrics
model_mses = []
for model_i in range(len(model_names)) :
print("Benchmarking model '" + str(model_names[model_i]) + "'...")
feature_quantile_mses = []
for feature_quantile_i, feature_quantile in enumerate(feature_quantiles) :
print("Feature quantile = " + str(feature_quantile))
if len(model_importance_scores_test[model_i].shape) >= 5 :
importance_scores_test = np.abs(model_importance_scores_test[model_i][feature_quantile_i, ...])
else :
importance_scores_test = np.abs(model_importance_scores_test[model_i])
n_to_test = importance_scores_test.shape[0] // batch_size * batch_size
importance_scores_test = importance_scores_test[:n_to_test]
importance_scores_test *= np.expand_dims(np.max(pwm_mask[:n_to_test], axis=-1), axis=-1)
quantile_vals = np.quantile(importance_scores_test, axis=(1, 2, 3), q=feature_quantile, keepdims=True)
quantile_vals = np.tile(quantile_vals, (1, importance_scores_test.shape[1], importance_scores_test.shape[2], importance_scores_test.shape[3]))
top_logits_test = np.zeros(importance_scores_test.shape)
top_logits_test[importance_scores_test > quantile_vals] = on_state_logit_val
top_logits_test = np.tile(top_logits_test, (1, 1, 1, 4)) * x_test_logits[:n_to_test]
_, _, samples_test, _, _ = scrambler_model.predict([top_logits_test, dummy_test[:n_to_test]], batch_size=batch_size)
print (samples_test.shape)
msesPerPoint = []
for data_ix in range(samples_test.shape[0]) :
#for each sample, look at kl divergence for the 128 size batch generated
#for MSE, just track the pred vs original pred
if data_ix % 1000 == 0 :
print("Processing example " + str(data_ix) + "...")
#from optimus R^2, MSE, Pearson R script
justPred = np.expand_dims(np.expand_dims(x_test[data_ix, 0, :, :], axis=0), axis=-1)
justPredReshape = np.reshape(justPred, (1,50,4))
expanded = np.expand_dims(samples_test[data_ix, :, 0, :, :], axis=-1) #batch size is 128
expandedReshape = np.reshape(expanded, (n_samples, 50,4))
y_test_hat_ref = predictor.predict(x=justPredReshape, batch_size=1)[0][0]
y_test_hat = predictor.predict(x=[expandedReshape], batch_size=32)
pwmGenerated = y_test_hat.tolist()
tempOriginals = [y_test_hat_ref]*y_test_hat.shape[0]
asArrayOrig = np.array(tempOriginals)
asArrayGen = np.array(pwmGenerated)
squeezed = np.squeeze(asArrayGen)
mse = metrics.mean_squared_error(asArrayOrig, squeezed)
#msesPerPoint.append(mse)
totalName = model_names[model_i] + "_" + str(feature_quantile).replace(".","_") + "_quantile_MSE"
data_df.at[data_ix, totalName] = mse
msesPerPoint.append(mse)
msesPerPoint = np.array(msesPerPoint)
feature_quantile_mses.append(msesPerPoint)
model_mses.append(feature_quantile_mses)
#Store benchmark results as tables
save_figs = False
mse_table = np.zeros((len(model_mses), len(model_mses[0])))
for i, model_name in enumerate(model_names) :
for j, feature_quantile in enumerate(feature_quantiles) :
mse_table[i, j] = np.mean(model_mses[i][j])
#Plot and store mse table
f = plt.figure(figsize = (4, 6))
cells = np.round(mse_table, 3).tolist()
print("--- MSEs ---")
max_len = np.max([len(model_name.upper().replace("\n", " ")) for model_name in model_names])
print(("-" * max_len) + " " + " ".join([(str(feature_quantile) + "0")[:4] for feature_quantile in feature_quantiles]))
for i in range(len(cells)) :
curr_len = len([model_name.upper().replace("\n", " ") for model_name in model_names][i])
row_str = [model_name.upper().replace("\n", " ") for model_name in model_names][i] + (" " * (max_len - curr_len))
for j in range(len(cells[i])) :
cells[i][j] = (str(cells[i][j]) + "00000")[:4]
row_str += " " + cells[i][j]
print(row_str)
print("")
table = plt.table(cellText=cells, rowLabels=[model_name.upper().replace("\n", " ") for model_name in model_names], colLabels=feature_quantiles, loc='center')
ax = plt.gca()
#f.patch.set_visible(False)
ax.axis('off')
ax.axis('tight')
plt.tight_layout()
if save_figs :
plt.savefig(dataset_name + "_l2x_and_invase_full_data" + "_mse_table.png", dpi=300, transparent=True)
plt.savefig(dataset_name + "_l2x_and_invase_full_data" + "_mse_table.eps")
plt.show()
```
|
github_jupyter
|
# Signal Autoencoder
```
import numpy as np
import scipy as sp
import scipy.stats
import itertools
import logging
import matplotlib.pyplot as plt
import pandas as pd
import torch.utils.data as utils
import math
import time
import tqdm
import torch
import torch.optim as optim
import torch.nn.functional as F
from argparse import ArgumentParser
from torch.distributions import MultivariateNormal
import torch.nn as nn
import torch.nn.init as init
import sys
sys.path.append("../new_flows")
from flows import RealNVP, Planar, MAF
from models import NormalizingFlowModel
####MAF
class VAE_NF(nn.Module):
def __init__(self, K, D):
super().__init__()
self.dim = D
self.K = K
self.encoder = nn.Sequential(
nn.Linear(16, 50),
nn.LeakyReLU(True),
nn.Linear(50, 48),
nn.LeakyReLU(True),
nn.Linear(48, D * 2)
)
self.decoder = nn.Sequential(
nn.Linear(D, 48),
nn.LeakyReLU(True),
nn.Linear(48, 50),
nn.LeakyReLU(True),
nn.Linear(50, 16)
)
flow_init = MAF(dim=D)
flows_init = [flow_init for _ in range(K)]
prior = MultivariateNormal(torch.zeros(D).cuda(), torch.eye(D).cuda())
self.flows = NormalizingFlowModel(prior, flows_init)
def forward(self, x):
# Run Encoder and get NF params
enc = self.encoder(x)
mu = enc[:, :self.dim]
log_var = enc[:, self.dim: self.dim * 2]
# Re-parametrize
sigma = (log_var * .5).exp()
z = mu + sigma * torch.randn_like(sigma)
kl_div = -0.5 * torch.sum(1 + log_var - mu.pow(2) - log_var.exp())
# Construct more expressive posterior with NF
z_k, _, sum_ladj = self.flows(z)
kl_div = kl_div / x.size(0) - sum_ladj.mean() # mean over batch
# Run Decoder
x_prime = self.decoder(z_k)
return x_prime, kl_div
prong_2 = pd.read_hdf("/data/t3home000/spark/QUASAR/preprocessing/delphes_output_5000_850_450.h5")
#prong_3 = pd.read_hdf("/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_3prong_rnd.h5")
columns = prong_2.columns
columns
dt = prong_2.values
correct = (dt[:,3]>0) &(dt[:,19]>0) & (dt[:,1]>0) & (dt[:,2]>0) &(dt[:,2]>0) & (dt[:,16]>0) & (dt[:,32]>0)
dt = dt[correct]
for i in range(13,19):
dt[:,i] = dt[:,i]/dt[:,3]
for i in range(29,35):
dt[:,i] = dt[:,i]/(dt[:,19])
correct = (dt[:,16]>0) & (dt[:,29]>=0) &(dt[:,29]<=1)&(dt[:,30]>=0) &(dt[:,30]<=1)&(dt[:,31]>=0) &(dt[:,31]<=1)&(dt[:,32]>=0) &(dt[:,32]<=1)&(dt[:,33]>=0) &(dt[:,33]<=1)&(dt[:,34]>=-0.01) &(dt[:,34]<=1)
dt = dt[correct]
#Y = dt[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
#Y = dt[:,[4,5,6,7,8,11,12,13,14,15,16,17,18,20,21,22,23,24,27,28,29,30,31,32,33,34]] # When no jet 1,2 raw mass included
#Y = dt[:,[3,4,5,6,11,12,13,14,15,16,17,18,19,20,21,22,27,28,29,30,31,32,33,34]]
#idx = dt[:,-1]
#bkg_idx = np.where(idx==0)[0]
#signal_idx = np.where((idx==1) & (dt[:,3]>300))[0]
#signal_idx = np.where((idx==1)) [0]
#dt = dt[signal_idx]
bsmlike = np.where(dt[:,16]>0.9)[0]
dt = dt[bsmlike]
dt.shape
j1sdb = dt[:,3]*dt[:,16]
j2sdb = dt[:,19]*dt[:,32]
pt = dt[:,1]
m = j1sdb[:]
m2 = j2sdb[:]
tau21 = dt[:,4]
tau32 = dt[:,5]
tau43 = dt[:,6]
tau54 = dt[:,7]
tau65 = dt[:,8]
massratio = dt[:,16]
rho = np.log((m*m)/(pt*pt))
rhoprime = np.log((m*m)/(pt*1))
tau21prime = tau21 + rhoprime * 0.088
tau32prime = tau32 + rhoprime * 0.025
tau43prime = tau43 + rhoprime * 0.01
tau54prime = tau54 + rhoprime * 0.001
j2pt = dt[:,2]
#m = j1sdb[mrange]
j2m = j2sdb[:]
j2tau21 = dt[:,20]
j2tau32 = dt[:,21]
j2tau43 = dt[:,22]
j2tau54 = dt[:,23]
j2tau65 = dt[:,24]
j2massratio = dt[:,32]
j2rho = np.log((j2m*j2m)/(j2pt*j2pt))
j2rhoprime = np.log((j2m*j2m)/(j2pt*1))
j2tau21prime = j2tau21 + j2rhoprime * 0.086
j2tau32prime = j2tau32 + j2rhoprime * 0.025
j2tau43prime = j2tau43 + j2rhoprime * 0.01
j2tau54prime = j2tau54 + j2rhoprime * 0.001
dt[:,4] = tau21prime
dt[:,5] = tau32prime
dt[:,6] = tau43prime
dt[:,7] = tau54prime
dt[:,20] = j2tau21prime
dt[:,21] = j2tau32prime
dt[:,22] = j2tau43prime
dt[:,23] = j2tau54prime
columns[19]
m1minusm2 = dt[:,3] - dt[:,19]
dt[:,19] = m1minusm2
Y = dt[:,[3,4,5,6,7,8,11,12,19,20,21,22,23,24,27,28]]
Y.shape
#if nprong == 3:
# dt = prong_3.values
# correct = (dt[:,3]>20) &(dt[:,19]>20)
# dt = dt[correct]
# for i in range(13,19):
# dt[:,i] = dt[:,i]/dt[:,3]
# for i in range(29,35):
# dt[:,i] = dt[:,i]/(dt[:,19])
# correct = (dt[:,29]>=0) &(dt[:,29]<=1)&(dt[:,30]>=0) &(dt[:,30]<=1)&(dt[:,31]>=0) &(dt[:,31]<=1)&(dt[:,32]>=0) &(dt[:,32]<=1)&(dt[:,33]>=0) &(dt[:,33]<=1)&(dt[:,34]>=-0.01) &(dt[:,34]<=1)
# dt = dt[correct]
# Y = dt[:,[4,5,6,7,8,11,12,13,14,15,16,17,18,20,21,22,23,24,27,28,29,30,31,32,33,34]]
# #Y = dt[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
# idx = dt[:,-1]
# bkg_idx = np.where(idx==0)[0]
# signal_idx = np.where((idx==1) & (dt[:,3]>400))[0]
# #signal_idx = np.where((idx==1)) [0]
# Y = Y[signal_idx]
bins = np.linspace(0,1,100)
bins.shape
column = 5
#print(f_rnd.columns[column])
plt.hist(dt[:,16],bins,alpha=0.5,color='b');
#plt.hist(sigout[:,column],bins,alpha=0.5,color='r');
#plt.hist(out2[:,column],bins,alpha=0.5,color='g');
#plt.axvline(np.mean(Y[:,column]))
Y.shape
sig_mean = []
sig_std = []
for i in range(16):
mean = np.mean(Y[:,i])
std = np.std(Y[:,i])
sig_mean.append(mean)
sig_std.append(std)
Y[:,i] = (Y[:,i]-mean)/std
sig_mean
sig_std
total_sig = torch.tensor(Y)
total_sig.shape
bins = np.linspace(-3,3,100)
bins.shape
column = 5
#print(f_rnd.columns[column])
plt.hist(Y[:,1],bins,alpha=0.5,color='b');
#plt.hist(sigout[:,column],bins,alpha=0.5,color='r');
#plt.hist(out2[:,column],bins,alpha=0.5,color='g');
#plt.axvline(np.mean(Y[:,column]))
N_EPOCHS = 30
PRINT_INTERVAL = 2000
NUM_WORKERS = 4
LR = 1e-6
#N_FLOWS = 6
#Z_DIM = 8
N_FLOWS = 10
Z_DIM = 6
n_steps = 0
sigmodel = VAE_NF(N_FLOWS, Z_DIM).cuda()
print(sigmodel)
bs = 800
sig_train_iterator = utils.DataLoader(total_sig, batch_size=bs, shuffle=True)
sig_test_iterator = utils.DataLoader(total_sig, batch_size=bs)
sigoptimizer = optim.Adam(sigmodel.parameters(), lr=1e-6)
beta = 1
def sigtrain():
global n_steps
train_loss = []
sigmodel.train()
for batch_idx, x in enumerate(sig_train_iterator):
start_time = time.time()
x = x.float().cuda()
x_tilde, kl_div = sigmodel(x)
mseloss = nn.MSELoss(size_average=False)
huberloss = nn.SmoothL1Loss(size_average=False)
#loss_recons = F.binary_cross_entropy(x_tilde, x, size_average=False) / x.size(0)
loss_recons = mseloss(x_tilde,x ) / x.size(0)
#loss_recons = huberloss(x_tilde,x ) / x.size(0)
loss = loss_recons + beta* kl_div
sigoptimizer.zero_grad()
loss.backward()
sigoptimizer.step()
train_loss.append([loss_recons.item(), kl_div.item()])
if (batch_idx + 1) % PRINT_INTERVAL == 0:
print('\tIter [{}/{} ({:.0f}%)]\tLoss: {} Time: {:5.3f} ms/batch'.format(
batch_idx * len(x), 50000,
PRINT_INTERVAL * batch_idx / 50000,
np.asarray(train_loss)[-PRINT_INTERVAL:].mean(0),
1000 * (time.time() - start_time)
))
n_steps += 1
def sigevaluate(split='valid'):
global n_steps
start_time = time.time()
val_loss = []
sigmodel.eval()
with torch.no_grad():
for batch_idx, x in enumerate(sig_test_iterator):
x = x.float().cuda()
x_tilde, kl_div = sigmodel(x)
mseloss = nn.MSELoss(size_average=False)
huberloss = nn.SmoothL1Loss(size_average=False)
#loss_recons = F.binary_cross_entropy(x_tilde, x, size_average=False) / x.size(0)
loss_recons = mseloss(x_tilde,x ) / x.size(0)
#loss_recons = huberloss(x_tilde,x ) / x.size(0)
loss = loss_recons + beta * kl_div
val_loss.append(loss.item())
#writer.add_scalar('loss/{}/ELBO'.format(split), loss.item(), n_steps)
#writer.add_scalar('loss/{}/reconstruction'.format(split), loss_recons.item(), n_steps)
#writer.add_scalar('loss/{}/KL'.format(split), kl_div.item(), n_steps)
print('\nEvaluation Completed ({})!\tLoss: {:5.4f} Time: {:5.3f} s'.format(
split,
np.asarray(val_loss).mean(0),
time.time() - start_time
))
return np.asarray(val_loss).mean(0)
ae_def = {
"type":"sig",
"trainon":"BB2refined",
"features":"tauDDTwithm1andm1minusm2",
"architecture":"MAF",
"selection":"turnoncutandj1sdbcut0p9",
"trainloss":"MSELoss",
"beta":"beta1",
"zdimnflow":"z6f10",
}
ae_def
N_EPOCHS = 10
BEST_LOSS = 99
LAST_SAVED = -1
PATIENCE_COUNT = 0
PATIENCE_LIMIT = 5
for epoch in range(1, 1000):
print("Epoch {}:".format(epoch))
sigtrain()
cur_loss = sigevaluate()
if cur_loss <= BEST_LOSS:
PATIENCE_COUNT = 0
BEST_LOSS = cur_loss
LAST_SAVED = epoch
print("Saving model!")
torch.save(sigmodel.state_dict(),f"/data/t3home000/spark/QUASAR/weights/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['architecture']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}.h5")
else:
PATIENCE_COUNT += 1
print("Not saving model! Last saved: {}".format(LAST_SAVED))
if PATIENCE_COUNT > 10:
print("Patience Limit Reached")
break
sigmodel.load_state_dict(torch.load(f"/data/t3home000/spark/QUASAR/weights/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['architecture']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}.h5"))
sigout = sigmodel(torch.tensor(Y).float().cuda())[0]
sigout = sigout.data.cpu().numpy()
bins = np.linspace(-3,3,100)
bins.shape
column = 3
#print(f_rnd.columns[column]
plt.hist(Y[:,column],bins,alpha=0.5,color='b');
plt.hist(sigout[:,column],bins,alpha=0.5,color='r');
#plt.hist(out2[:,column],bins,alpha=0.5,color='g');
plt.axvline(np.mean(Y[:,column]))
inputlist = [
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_BB1_rnd.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_BB2.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_BB3.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_background.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_rnd.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_rnd.h5',
'/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_3prong_rnd.h5'
]
ae_def
outputlist_waic = [
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_bb1.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_bb2.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_bb3.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_purebkg.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_rndbkg.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_2prong.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_WAICloss_3prong.npy",
]
outputlist_justloss = [
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_bb1.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_bb2.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_bb3.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_purebkg.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_rndbkg.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_2prong.npy",
f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_3prong.npy",
]
exist_signalflag = [
False,
False,
False,
False,
True,
True,
True,
]
is_signal = [
False,
False,
False,
False,
False,
True,
True
]
nprong = [
None,
None,
None,
None,
None,
'2prong',
'3prong'
]
for in_file, out_file_waic, out_file_justloss, sigbit_flag, is_sig, n_prong in zip(inputlist,outputlist_waic,outputlist_justloss,exist_signalflag,is_signal, nprong):
f_bb = pd.read_hdf(in_file)
dt = f_bb.values
correct = (dt[:,3]>0) &(dt[:,19]>0) & (dt[:,1]>0) & (dt[:,2]>0) &(dt[:,2]>0) & (dt[:,16]>0) & (dt[:,32]>0)
dt = dt[correct]
for i in range(13,19):
dt[:,i] = dt[:,i]/dt[:,3]
for i in range(29,35):
dt[:,i] = dt[:,i]/(dt[:,19])
correct = (dt[:,16]>0) & (dt[:,29]>=0) &(dt[:,29]<=1)&(dt[:,30]>=0) &(dt[:,30]<=1)&(dt[:,31]>=0) &(dt[:,31]<=1)&(dt[:,32]>=0) &(dt[:,32]<=1)&(dt[:,33]>=0) &(dt[:,33]<=1)&(dt[:,34]>=-0.01) &(dt[:,34]<=1)
dt = dt[correct]
correct = (dt[:,3]>100)
dt = dt[correct]
correct = (dt[:,19]>20)
dt = dt[correct]
correct = (dt[:,0]>=2800)
dt = dt[correct]
bsmlike = np.where(dt[:,16]>0.9)[0]
dt = dt[bsmlike]
j1sdb = dt[:,3]*dt[:,16]
j2sdb = dt[:,19]*dt[:,32]
pt = dt[:,1]
m = j1sdb[:]
m2 = j2sdb[:]
tau21 = dt[:,4]
tau32 = dt[:,5]
tau43 = dt[:,6]
tau54 = dt[:,7]
tau65 = dt[:,8]
massratio = dt[:,16]
rho = np.log((m*m)/(pt*pt))
rhoprime = np.log((m*m)/(pt*1))
tau21prime = tau21 + rhoprime * 0.088
tau32prime = tau32 + rhoprime * 0.025
tau43prime = tau43 + rhoprime * 0.01
tau54prime = tau54 + rhoprime * 0.001
j2pt = dt[:,2]
#m = j1sdb[mrange]
j2m = j2sdb[:]
j2tau21 = dt[:,20]
j2tau32 = dt[:,21]
j2tau43 = dt[:,22]
j2tau54 = dt[:,23]
j2tau65 = dt[:,24]
j2massratio = dt[:,32]
j2rho = np.log((j2m*j2m)/(j2pt*j2pt))
j2rhoprime = np.log((j2m*j2m)/(j2pt*1))
j2tau21prime = j2tau21 + j2rhoprime * 0.086
j2tau32prime = j2tau32 + j2rhoprime * 0.025
j2tau43prime = j2tau43 + j2rhoprime * 0.01
j2tau54prime = j2tau54 + j2rhoprime * 0.001
dt[:,4] = tau21prime
dt[:,5] = tau32prime
dt[:,6] = tau43prime
dt[:,7] = tau54prime
dt[:,20] = j2tau21prime
dt[:,21] = j2tau32prime
dt[:,22] = j2tau43prime
dt[:,23] = j2tau54prime
if sigbit_flag:
idx = dt[:,-1]
sigidx = (idx == 1)
bkgidx = (idx == 0)
if is_sig:
dt = dt[sigidx]
else:
dt = dt[bkgidx]
if n_prong == '2prong':
correct = dt[:,3] > 300
dt = dt[correct]
if n_prong == '3prong':
correct = dt[:,3] > 400
dt = dt[correct]
m1minusm2 = dt[:,3] - dt[:,19]
dt[:,19] = m1minusm2
Y = dt[:,[3,4,5,6,7,8,11,12,19,20,21,22,23,24,27,28]]
#Y = dt[:,[3,4,5,6,11,12,13,14,15,16,17,18,19,20,21,22,27,28,29,30,31,32,33,34]]
#Y = dt[:,[3,4,5,6,11,12,13,14,15,16,17,18,19,20,21,22,27,28,29,30,31,32,33,34]]
#Y = dt[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
print(Y.shape)
for i in range(16):
Y[:,i] = (Y[:,i]-sig_mean[i])/sig_std[i]
total_bb_test = torch.tensor(Y)
#huberloss = nn.SmoothL1Loss(reduction='none')
sigae_bbloss = torch.mean((sigmodel(total_bb_test.float().cuda())[0]- total_bb_test.float().cuda())**2,dim=1).data.cpu().numpy()
bbvar = torch.var((sigmodel(total_bb_test.float().cuda())[0]- total_bb_test.float().cuda())**2,dim=1).data.cpu().numpy()
waic = sigae_bbloss + bbvar
#sigae_bbloss = torch.mean(huberloss(model(total_bb_test.float().cuda())[0],total_bb_test.float().cuda()),dim=1).data.cpu().numpy()
print(waic[0:10])
plt.hist(waic,bins=np.linspace(0,10,1001),density=True);
plt.xlim([0,2])
np.save(out_file_waic,waic)
np.save(out_file_justloss,sigae_bbloss)
loss_prong3 = np.load(f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_3prong.npy")
loss_prong2 = np.load(f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_2prong.npy")
loss_purebkg = np.load(f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_purebkg.npy")
loss_rndbkg = np.load(f"../data_strings/{ae_def['type']}_{ae_def['trainon']}_{ae_def['features']}_{ae_def['selection']}_{ae_def['trainloss']}_{ae_def['beta']}_{ae_def['zdimnflow']}_Justloss_rndbkg.npy")
plt.hist(loss_purebkg,bins=np.linspace(0,4,100),density=False,alpha=0.3,label='Pure Bkg');
#plt.hist(loss_rndbkg,bins=np.linspace(0,2,100),density=False,alpha=0.3,label='(rnd) bkg');
plt.hist(loss_prong2,bins=np.linspace(0,4,100),density=False,alpha=0.3,label='2prong (rnd)sig');
plt.hist(loss_prong3,bins=np.linspace(0,4,100),density=False,alpha=0.3,label='3prong (rnd)sig');
#plt.yscale('log')
plt.xlabel('Loss (SigAE trained on 2prong sig)')
plt.legend(loc='upper right')
#plt.savefig('sigae_trained_on_2prongsig.png')
ae_def
len(loss_prong2)
outputlist_waic
outputlist_justloss
sigae_bbloss
ae_def
sigae_bbloss
plt.hist(sigae_bbloss,bins=np.linspace(0,10,1001));
np.save('../data_strings/sigae_2prong_loss_bb3.npy',sigae_bbloss)
X_bkg = dt[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
X_bkg = X_bkg[bkg_idx]
for i in range(12):
X_bkg[:,i] = (X_bkg[:,i]-sig_mean[i])/sig_std[i]
total_bkg_test = torch.tensor(X_bkg)
sigae_bkgloss = torch.mean((sigmodel(total_bkg_test.float().cuda())[0]- total_bkg_test.float().cuda())**2,dim=1).data.cpu().numpy()
sigae_sigloss = torch.mean((sigmodel(total_sig.float().cuda())[0]- total_sig.float().cuda())**2,dim=1).data.cpu().numpy()
f_3prong = pd.read_hdf("/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_3prong_rnd.h5")
f_bb1 = pd.read_hdf('/data/t3home000/spark/QUASAR/preprocessing/conventional_tau_BB1_rnd.h5')
dt_bb1 = f_bb1.values
X_bb1 = dt_bb1[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
X_bb1.shape
sig_mean
sig_std
for i in range(12):
X_bb1[:,i] = (X_bb1[:,i]-sig_mean[i])/sig_std[i]
plt.hist(X_bb1[:,0],bins = np.linspace(-2,2,10))
(torch.tensor(dt[i * chunk_size:(i + 1) * chunk_size]) for i in range )
def get_loss(dt):
chunk_size=5000
total_size=1000000
i = 0
i_max = total_size // chunk_size
print(i_max)
gen = (torch.tensor(dt[i*chunk_size: (i + 1) * chunk_size]) for i in range(i_max))
with torch.no_grad():
loss = [
n
for total_in_selection in gen
for n in torch.mean((sigmodel(total_in_selection.float().cuda())[0]- total_in_selection.float().cuda())**2,dim=1).data.cpu().numpy()
]
return loss
def get_loss(dt):
def generator(dt, chunk_size=5000, total_size=1000000):
i = 0
i_max = total_size // chunk_size
print(i_max)
for i in range(i_max):
start=i * chunk_size
stop=(i + 1) * chunk_size
yield torch.tensor(dt[start:stop])
loss = []
with torch.no_grad():
for total_in_selection in generator(dt,chunk_size=5000, total_size=1000000):
loss.extend(torch.mean((sigmodel(total_in_selection.float().cuda())[0]- total_in_selection.float().cuda())**2,dim=1).data.cpu().numpy())
return loss
bb1_loss_sig = get_loss(X_bb1)
bb1_loss_sig = np.array(bb1_loss_sig,dtype=np.float)
print(bb1_loss_sig)
plt.hist(bb1_loss_sig,bins=np.linspace(0,100,1001));
np.save('../data_strings/sigaeloss_bb1.npy',bb1_loss_sig)
dt_3prong = f_3prong.values
Z = dt_3prong[:,[3,4,5,6,11,12,19,20,21,22,27,28]]
Z.shape
for i in range(12):
Z[:,i] = (Z[:,i]-sig_mean[i])/sig_std[i]
total_3prong = torch.tensor(Z)
bkgae_bkgloss = torch.mean((model(total_bkg_test.float().cuda())[0]- total_bkg_test.float().cuda())**2,dim=1).data.cpu().numpy()
bkgae_3prongloss = torch.mean((model(total_3prong.float().cuda())[0]- total_3prong.float().cuda())**2,dim=1).data.cpu().numpy()
sigae_3prongloss = torch.mean((sigmodel(total_3prong.float().cuda())[0]- total_3prong.float().cuda())**2,dim=1).data.cpu().numpy()
sigae_3prongloss.shape
bins = np.linspace(0,10,1001)
plt.hist(sigae_sigloss,bins,weights = np.ones(len(signal_idx))*10,alpha=0.4,color='r',label='2 prong signal');
plt.hist(sigae_3prongloss,bins,weights = np.ones(100000)*10,alpha=0.5,color='g',label='3 prong signal');
plt.hist(sigae_bkgloss,bins,alpha=0.4,color='b',label='background');
#plt.legend(bbox_to_anchor=(1.05, 1.0), loc='upper left')
plt.legend(loc='upper right')
plt.xlabel('Signal AE Loss',fontsize=15)
def get_tpr_fpr(sigloss,bkgloss,aetype='sig'):
bins = np.linspace(0,50,1001)
tpr = []
fpr = []
for cut in bins:
if aetype == 'sig':
tpr.append(np.where(sigloss<cut)[0].shape[0]/len(sigloss))
fpr.append(np.where(bkgloss<cut)[0].shape[0]/len(bkgloss))
if aetype == 'bkg':
tpr.append(np.where(sigloss>cut)[0].shape[0]/len(sigloss))
fpr.append(np.where(bkgloss>cut)[0].shape[0]/len(bkgloss))
return tpr,fpr
def get_precision_recall(sigloss,bkgloss,aetype='bkg'):
bins = np.linspace(0,100,1001)
tpr = []
fpr = []
precision = []
for cut in bins:
if aetype == 'sig':
tpr.append(np.where(sigloss<cut)[0].shape[0]/len(sigloss))
precision.append((np.where(sigloss<cut)[0].shape[0])/(np.where(bkgloss<cut)[0].shape[0]+np.where(sigloss<cut)[0].shape[0]))
if aetype == 'bkg':
tpr.append(np.where(sigloss>cut)[0].shape[0]/len(sigloss))
precision.append((np.where(sigloss>cut)[0].shape[0])/(np.where(bkgloss>cut)[0].shape[0]+np.where(sigloss>cut)[0].shape[0]))
return precision,tpr
tpr_2prong, fpr_2prong = get_tpr_fpr(sigae_sigloss,sigae_bkgloss,'sig')
tpr_3prong, fpr_3prong = get_tpr_fpr(sigae_3prongloss,sigae_bkgloss,'sig')
plt.plot(fpr_2prong,tpr_2prong,label='signal AE')
#plt.plot(VAE_bkg_fpr,VAE_bkg_tpr,label='Bkg VAE-Vanilla')
plt.plot(bkg_fpr4,bkg_tpr4,label='Bkg NFlowVAE-Planar')
plt.xlabel(r'$1-\epsilon_{bkg}$',fontsize=15)
plt.ylabel(r'$\epsilon_{sig}$',fontsize=15)
#plt.semilogy()
#plt.legend(bbox_to_anchor=(1.05, 1.0), loc='upper left')
plt.legend(loc='lower right')
plt.xlim([0.0,1.0])
plt.ylim([0.0,1.0])
plt.savefig('ROC_Curve_sigae.png')
precision,recall = get_precision_recall(loss_sig,loss_bkg,aetype='bkg')
np.save('NFLOWVAE_PlanarNEW_22var_sigloss.npy',loss_sig)
np.save('NFLOWVAE_PlanarNEW_22var_bkgloss.npy',loss_bkg)
np.save('NFLOWVAE_PlanarNEW_precision.npy',precision)
np.save('NFLOWVAE_PlanarNEW_recall.npy',recall)
np.save('NFLOWVAE_PlanarNEW_bkgAE_fpr.npy',bkg_fpr)
np.save('NFLOWVAE_PlanarNEW_bkgAE_tpr.npy',bkg_tpr)
np.save('NFLOWVAE_PlanarNEW_sigloss.npy',loss_sig)
np.save('NFLOWVAE_PlanarNEW_bkgloss.npy',loss_bkg)
plt.plot(recall,precision)
flows = [1,2,3,4,5,6]
zdim = [1,2,3,4,5]
for N_flows in flows:
for Z_DIM in zdim:
model = VAE_NF(N_FLOWS, Z_DIM).cuda()
optimizer = optim.Adam(model.parameters(), lr=LR)
BEST_LOSS = 99999
LAST_SAVED = -1
PATIENCE_COUNT = 0
PATIENCE_LIMIT = 5
for epoch in range(1, N_EPOCHS):
print("Epoch {}:".format(epoch))
train()
cur_loss = evaluate()
if cur_loss <= BEST_LOSS:
PATIENCE_COUNT = 0
BEST_LOSS = cur_loss
LAST_SAVED = epoch
print("Saving model!")
if mode == 'ROC':
torch.save(model.state_dict(),f"/data/t3home000/spark/QUASAR/weights/bkg_vae_NF_planar_RND_22var_z{Z_DIM}_f{N_FLOWS}.h5")
else:
torch.save(model.state_dict(), f"/data/t3home000/spark/QUASAR/weights/bkg_vae_NF_planar_PureBkg_22var_z{Z_DIM}_f{N_FLOWS}.h5")
else:
PATIENCE_COUNT += 1
print("Not saving model! Last saved: {}".format(LAST_SAVED))
if PATIENCE_COUNT > 3:
print("Patience Limit Reached")
break
loss_bkg = get_loss(dt_PureBkg[bkg_idx])
loss_sig = get_loss(dt_PureBkg[signal_idx])
np.save(f'NFLOWVAE_PlanarNEW_22var_z{Z_DIM}_f{N_flows}_sigloss.npy',loss_sig)
np.save(f'NFLOWVAE_PlanarNEW_22var_z{Z_DIM}_f{N_flows}_bkgloss.npy',loss_bkg)
```
|
github_jupyter
|
# Trabalhando com Arquivos
Tabela Modos de arquivo

# Métodos de uma lista usando biblioteca rich import inspect
```
from rich import inspect
a = open('arquivo1.txt', 'wt+')
inspect(a, methods=True)
```
# Criando Arquivo w(write) e x
# .close()
```
# cria arquivo ou abre apagando os dados de qualquer arquivo existente
a = open('arquivo1.txt', 'wt+') # w(write text) + (se não existir o arquivo crie) (t decodifica os caracteres Unicode-é default não precisa colocar)
a.close()
# cria arquivo, mas falha se o mesmo ja existir
a = open('arquivo1.txt', 'x')
a.close()
```
# .writefile()
Criar arquivo pelo Jupyter Notebook
```
%%writefile teste.txt
Olá este arquivo foi gerado pelo próprio Jupyter Notebook.
Podemos gerar quantas linhas quisermos e o Jupyter gera o arquivo final.
la...
la....
```
# Abrindo/Lendo arquivos r(read)
```
#Abre arquivo como leitura
a = open('arquivo1.txt', 'r' ,encoding="utf-8") # rt(read text) (,encoding="utf-8")
a.close()
#Abre arquivo como escrita e não apaga o anterior
a = open('arquivo1.txt', 'a') # at(append text)
a.close()
```
# .read() e encoding="utf-8"
O método read() somente funciona se abrir o arquivo como leitura ('r')
Ao abrir o arquivo com uma codificação diferente da que ele foi escrito, alguns caracteres podem apresentar erros, ou, em alguns sistemas operacionais, como no Mac OS, pode ser lançada uma exceção
Tipos de encoding:
https://docs.python.org/3/library/codecs.html#standard-encodings
```
arq4 = open("teste.txt", 'r',encoding="utf-8")
print(arq4.read())
a = open('arquivo1.txt', 'rt',encoding="utf-8") # (,encoding="utf-8") mostra as acentuações
print(a.read())
a = open('arquivo1.txt', 'rt')
print(a.read())
a.close()
a = open('arquivo1.txt', 'rt',encoding="utf-8")
print(a.read(3)) # Lendo os três primeiros caracteres
a.close()
```
# .read() e encoding="latin_1"
O método read() somente funciona se abrir o arquivo como leitura ('r')
Ao abrir o arquivo com uma codificação diferente da que ele foi escrito, alguns caracteres podem apresentar erros, ou, em alguns sistemas operacionais, como no Mac OS, pode ser lançada uma exceção
Tipos de encoding:
https://docs.python.org/3/library/codecs.html#standard-encodings
```
a = open('contatos.csv', encoding='latin_1')
print(a.read())
a.close()
```
# .readlines()
readlines ler linha por linha e coloca em uma lista
```
a = open("teste.txt", 'r',encoding="utf-8")
print(a.read())
a.seek(0)
print(a.readlines())
a.close()
```
# readline() Vs readlines()
readline() - ler somente uma linha
readlines() - coloca todas as linhas em um lista
```
# Testando readline()
from sys import getsizeof
with open('contatos.csv', 'r', encoding='latin_1') as a:
conteudo = a.readline()
print(conteudo)
print(f'conteudo = {getsizeof(conteudo)} bytes')
# Testando readlines()
from sys import getsizeof
with open('contatos.csv', 'r', encoding='latin_1') as a:
conteudo = a.readlines()
print(conteudo)
print(f'conteudo = {getsizeof(conteudo)} bytes')
```
# .seek()
```
arq4 = open("teste.txt", 'r',encoding="utf-8")
print(f'Lendo o arquivo a primeira vez:\n\n{arq4.read()}')
print(f'Tentando ler novamente e não conseguimos\n\n{arq4.read()}') # como ja lemos o arquivo ate o final temos que retornar com seek()
arq4.seek(0)
print(f'Após o uso do seek conseguimos ler novamente!\n\n{arq4.read()}')
```
# .split()
separar os caracteres
```
# separando caracteres por linhas
f = open('salarios.csv', 'r')
data = f.read()
rows = data.split('\n') # '\n' é um espaço separamos por espaço
print(rows) # cada '' é uma linha
f.close()
# separando caracteres por colunas
f = open('salarios.csv', 'r')
data = f.read()
rows = data.split('\n') # '\n' é um espaço separamos por espaço
dados = []
for row in rows:
split_row = row.split(',') # agora dentro de '' vamos separar por "," pois o arquivo é um csv
dados.append(split_row)
print(dados)
f.close()
```
# .tell()
Contar o número de caracteres
```
a = open('arquivo1.txt', 'r', encoding="utf-8")
a.read() # Se não ler o mesmo o .tell() não funciona
print(a.tell())
a.close()
```
# .flush()
Uma característica de quando a gente está trabalhando com escrita de arquivo no Python. A gente precisa fechar o arquivo para indicar que a gente não está mais trabalhando com ele. Somente após fechar é que as edições serão salvas, mas e se não podermos fechar o arquivo?
Com método **flush**, os dados vão ser escritos, porém o arquivo vai continuar aberto.
```
arquivos_contatos = open('contatos.csv', mode='a', encoding='latin_1')
novo_contato = '11,Livio,[email protected]\n'
arquivos_contatos.write(novo_contato)
arquivos_contatos.flush()
arquivos_contatos.close()
```
# <font color=#FF0000>**with open**</font>
```
with open('arquivo1.txt', mode='r', encoding="utf-8") as a:
conteudo = a.read()
print(conteudo)
```
# <font color=#FF0000>**with open - newline=''**</font>
Ao final de cada linha em um arquivo temos uma instrução de quebra de linha '\n' que significa que o texto irá para proxima linha. Este caractere é oculto, mas conseguimos ve-lo colocando **readlines()**.
Quando não usamos o newline='' o caractere é igual á '\n' (padrão Linux/unix/python) ao utilizar o newline='' o caractere muda para '\r\n' (padrão Microsoft)
# <font color=#FF0000>**diferença entre \n e \r\n e newline=''**</font>
O **\n** significa "new line" ou "line-feed", ou seja, **"nova linha"**.
O **\r** significa "carriage return", ou seja **"retorno de linha"**.
Quando a tabela ASCII foi padronizada, o \n recebeu o código 10 e \r recebeu o código 13.
_A ideia originalmente, quando as tabelas de codificação de caracteres como sequências de bits foram concebidas, é que o \n fosse interpretado como o comando para fazer o cursor se mover para baixo, e o \r o comando para ele se mover de volta até o começo da linha._
> Essa distinção era importante para as máquinas de escrever digitais que precederam os computadores, para telégrafos digitais, para teletipos e para a programação das primeiras impressoras que surgiram. De fato, isso é surpreendentemente mais antigo do que se pensa, já aparecendo no ano de 1901 junto com algumas das primeiras dessas tabelas de codificação de caracteres.
Assim sendo, em **um texto para que uma quebra-de-linha fosse inserida, fazia-se necessário utilizar-se \r\n**. Primeiro o cursor deveria se mover até o começo da linha e depois para baixo. **E foi esse o padrão de quebra-de-linha adotado muito mais tarde pela Microsoft.**
Já o Multics (e posteriormente o Unix) seguiram um caminho diferente, e decidiram implementar o **\n** como quebra-de-linha, o que já incluía um retorno ao começo da linha. Afinal de contas, não tem lá muito sentido ter uma coisa sem ter a outra junto, e **ao utilizá-los como sendo uma coisa só, garante-se que nunca serão separados**. Isso também tem a vantagem de economizar espaço ao usar um só byte para codificar a quebra-de-linha ao invés de dois, e naqueles anos aonde a memória era pequena e o processamento de baixo poder, cada byte economizado contava bastante.
Outras empresas, como a Apple e a Commodore, também seguiram um caminho semelhante ao do Unix, mas ao invés de adotarem o \n para quebras-de-linha, adotaram o \r.
Outras empresas menores adotaram outros códigos para a quebra-de-linha. Por exemplo, o QNX adotou o caractere 30 da tabela ASCII. A Atari adotou o 155. A Acorn e o RISC OS adotaram o \n\r ao invés de \r\n. A Sinclair adotou o 118.
**_Em resumo: Linux utiliza \n que representa \r(retorno ao primeiro caractere da linha) e \n(nova linha). A Apple utiliza \r que representa \r(retorno ao primeiro caractere da linha) e \n(nova linha). Já a Microsoft utiliza o padrão como \r\n. Ao usar o redline='' representamos a quebra de linha como \r\n se ocultarmos o mesmo a quebra de linha será \n._**
```
# Sem newline='' - caractere de fim de linha = '\n' padrão UNIX/Python
with open('arquivo1.txt', mode='r', encoding="utf-8") as a:
print(a.readlines())
# veja que ao final de cada linha temos o '\n'. É usado para indicar o fim de uma linha de texto.
# Com newline='' - caractere de fim de linha = '\r\n' padrão Microsoft
with open('arquivo1.txt', mode='r', encoding="utf-8", newline='') as a:
print(a.readlines())
# veja que ao final de cada linha temos o '\n'. É usado para indicar o fim de uma linha de texto..
```
# Escrevendo no arquivos a(append)
```
with open('arquivo1.txt', 'a', encoding="utf-8") as a:
a.write('\nEditando arquivo!!!') #\n é um enter, se iniciarmos com ele daremos um enter e apos isso escreveremos.
# a.read() se usar este comando ira dar erro, lembre-se que .read() somente se abrir o arquivo como leitura ('r')
with open('arquivo1.txt', 'r', encoding="utf-8") as a:
print(a.read())
```
# Trabalhando em modo b(binário) (imagens)
```
# criando uma copia da imagem python-logo.png
with open("python-logo.png", "rb") as imagem:
data = imagem.read()
with open("python-logo2.png", "wb") as imagem2:
imagem2.write(data)
```
# Lendo arquivos linha a linha e protegendo uso de memoria
```
from sys import getsizeof
with open('contatos.csv', 'r', encoding='latin_1') as a:
for numero, linha in enumerate(a):
print(f'Imprimindo linha {numero} | {getsizeof(linha)}-bytes\n {linha}', end='')
```
# Erros comuns ao tentar abrir um arquivo.
1. **FileNotFoundError** - Não encontrar o arquivo no local especificado.
1. **PermissionError** - Não tem permissão de escrita/criação no diretorio.
## try + finally
```
# Tratando erros com try:
try:
arquivo = open('contatos.csv', mode='a+', encoding='latin_1')
# Em mode='a' o arquivo abre na ultima linha, colocamos seek(0) para retornar a 1ª linha
# assim o readlines funcionar.
arquivo.seek(0)
conteudo = arquivo.readlines()
print(conteudo)
# finally será executando sempre, é comum colocarmos este tratamento para fechar o arquivo,
# apos o uso. Assim liberando o mesmo para outras pessoas.
finally:
arquivo.close()
```
## simulando FileNotFoundError
* Modificando o nome do arquivo para um arquivo que não existe.
* Abrindo em mode='r', pois em w e a se não existir o arquivo o Python cria
```
try:
arquivo = open('arquivo_nao_existe.csv', mode='r', encoding='latin_1')
arquivo.seek(0)
conteudo = arquivo.readlines()
print(conteudo)
finally:
arquivo.close()
```
### Solução com except FileNotFoundError:
**Agora nosso script não quebra caso não encontre o arquivo**
```
try:
arquivo = open('arquivo_nao_existe.csv', mode='r', encoding='latin_1')
arquivo.seek(0)
conteudo = arquivo.readlines()
print(conteudo)
except FileNotFoundError:
print('Arquivo não encontrado')
except PermissionError:
print('Sem permissão de escrita')
finally:
arquivo.close()
```
### Substituindo finally por with
* with fecha automaticamente um arquivo
* usando Lists Comprehensions simples para imprimir linha a linha
> Utilizamos o comando with para gerenciar o contexto de utilização do arquivo. Além de arquivos, podemos utilizar o with para gerenciar processos que precisam de uma pré e pós condição de execução; por exemplo: abrir e fechar o arquivo, realizar conexão com o banco de dados, sockets, entre outros.
> O objeto que está sendo manipulado pelo with precisa implementar dois métodos mágicos: \_\_enter__() e \_\_exit__().
> O método \_\_enter__() é executado logo no início da chamada da função e retorna uma representação do objeto que está sendo executada no contexto (ou context guard). Ao final, o método \_\_exit__() é invocado, e o contexto da execução, finalizado.
```
try:
with open('contatos.csv', mode='r', encoding='latin_1') as arquivo:
[print(linha, end='') for linha in arquivo]
except FileNotFoundError:
print('Arquivo não encontrado')
except PermissionError:
print('Sem permissão de escrita')
```
# De csv p/ Python
* converter um arquivo csv para um objeto list no python
* usando modulo csv
* criando uma função
## Criando uma class contatos
```
class Contato():
def __init__(self, id: int, nome: str, email: str):
self.id = id
self.nome = nome
self.email = email
```
## Criando uma função csv para list python
```
import csv
def csv_para_contatos(caminho: str, encoding: str = 'Latin_1'):
contatos: list = []
try:
with open(caminho, encoding=encoding) as a:
leitor = csv.reader(a)
for linha in leitor:
id, nome, email = linha # desencapsulando
contato = Contato(int(id), nome, email)
contatos.append(contato)
return contatos
except FileNotFoundError:
print('Arquivo não encontrado')
except PermissionError:
print('Sem permissão de escrita')
```
## Testando com arquivo contatos.csv
```
contatos = csv_para_contatos('contatos.csv')
lista = [print(f'{contato.id} - {contato.nome} - {contato.email}') for contato in contatos]
```
# De objeto Python para json
* converter um objeto python para um arquivo json
* usando modulo json
* criando uma função
## Criando uma função objeto python para json
```
import json
# escrita
def contatos_para_json(contatos, caminho: str):
try:
with open(caminho, mode='w') as a:
json.dump(contatos, a, default=__contato_para_json)
except FileNotFoundError:
print('Arquivo não encontrado')
except PermissionError:
print('Sem permissão de escrita')
def __contato_para_json(contato):
return contato.__dict__
# leitura
def json_para_contatos(caminho: str):
contatos = []
try:
with open(caminho, mode='r') as a:
contatos_json = json.load(a)
# Contato(contato['id'], contato['nome'], contato['email']) = Contato(**contato)
# assim estariamos desempacotando
[contatos.append(Contato(contato['id'], contato['nome'], contato['email']))
for contato in contatos_json]
return contatos
except FileNotFoundError:
print('Arquivo não encontrado')
except PermissionError:
print('Sem permissão de escrita')
```
## Testando de objeto python para json
```
# transformando csv em objeto python
contatos = csv_para_contatos('contatos.csv')
# transformando objeto python em json
contatos_para_json(contatos, 'contatos.json')
# transformando json em objeto python
contatos = json_para_contatos('contatos.json')
lista = [print(f'{contato.id} - {contato.nome} - {contato.email}') for contato in contatos]
import json
# json.dump = usado para gravar dados de objeto python em arquivo json
# json.dumps = usado para transformar objetos python em objetos str json
# json.load = usado para ler um arquivo json e transforma-lo em objto python
# Codificando hierarquias básicas de objetos Python:
lista = ['foo', {'bar': ('baz', None, 1.0, 2)}]
json_dump = json.dumps(lista)
print(f'{json_dump = }')
dicionario = {"c": 0, "b": 0, "a": 0}
json_dump = json.dumps(dicionario, sort_keys=True)
print(f'{json_dump = }')
# Codificação compacta:
lista = [1, 2, 3, {'4': 5, '6': 7}]
print(json.dumps(lista, separators=(',', ':')))
print(json.dumps(lista))
# Impressão bonita:
dicionario = {'4': 5, '6': 7}
print(json.dumps(dicionario, sort_keys=True, indent=4))
# Decodificando JSON:
texto = '["foo", {"bar":["baz", null, 1.0, 2]}]'
print(json.loads(texto))
import json
developer_Dict = {
"name": "Jane Doe",
"salary": 9000,
"skills": ["Python", "Machine Learning", "Web Development"],
"email": "[email protected]"
}
print(type(developer_Dict))
developer_str = json.dumps(developer_Dict)
print(developer_Dict)
print(type(developer_str))
import json
sampleDict = {
"colorList": ["Red", "Green", "Blue"],
"carTuple": ("BMW", "Audi", "range rover"),
"sampleString": "pynative.com",
"sampleInteger": 457,
"sampleFloat": 225.48,
"booleantrue": True,
"booleanfalse": False,
"nonevalue": None
}
print("Converting Python primitive types into JSON")
resultJSON = json.dumps(sampleDict)
print("Done converting Python primitive types into JSON")
print(resultJSON)
```
# <font color=#FF0000>**json**</font>
Geralmente, anexar dados a um arquivo JSON não é uma ideia muito boa porque, para cada pequena atualização, você deve ler e analisar todo o objeto de arquivo. Se o seu arquivo JSON tiver n entradas, a complexidade do tempo de execução de apenas atualizá-lo é O (n) .
**_Uma abordagem melhor seria armazenar os dados como um arquivo CSV que pode ser lido linha por linha que simplifica a análise e atualização significativamente, apenas acrescentando uma única linha ao arquivo que tem complexidade de tempo de execução constante._**
# Sintaxe de <font color=#FF0000>**json.dump()**</font> e <font color=#FF0000>**json.dumps()**</font>
>json.dump(obj, fp, *, skipkeys=False, ensure_ascii=True, check_circular=True, allow_nan=True, cls=None, indent=None, separators=None, default=None, sort_keys=False, **kw)
**É usado para gravar um objeto Python em um arquivo como dados formatados em JSON.**
>json.dumps(obj, *, skipkeys=False, ensure_ascii=True, check_circular=True, allow_nan=True, cls=None, indent=None, separators=None, default=None, sort_keys=False, **kw)
**É usado para escrever um objeto Python em uma String JSON.**
* **obj** nada mais é que um objeto serializável Python que você deseja converter em um formato JSON.
* A **fp** é um ponteiro de arquivo usado para gravar dados formatados em JSON em um arquivo. O módulo json Python sempre produz objetos de string, não objetos de bytes, portanto, fp.write()deve suportar a entrada de string.
* Se **skipkeysfor** verdadeiro (padrão: False), então as chaves de dict que não são de um tipo básico, (str, int, float, bool, None) serão ignoradas em vez de aumentar a TypeError. Por exemplo, se uma de suas chaves de dicionário for um objeto Python personalizado, essa chave será omitida durante a conversão do dicionário em JSON.
* Se **ensure_ascii** for verdadeiro (o padrão), a saída terá a garantia de ter todos os caracteres não ASCII de entrada com escape. Se ensure_asciifor falso, esses caracteres serão reproduzidos no estado em que se encontram.
* **allow_nan** é True por padrão, então seus equivalentes JavaScript (NaN, Infinity, -Infinity) serão usados. Se for False, será um ValueError para serializar valores flutuantes fora do intervalo (nan, inf, -inf).
* Um **indent** argumento é usado para imprimir JSON para torná-lo mais legível. O padrão é (', ', ': '). Para obter a representação JSON mais compacta, você deve usar (',', ':') para eliminar os espaços em branco.
*Se **sort_keys** for verdadeiro (padrão: Falso), a saída dos dicionários será classificada por chave
# <font color=#FF0000>**json.load()**</font> - Lendo um arquivo json formatado e transformando em dict
```json
{
"permissões": {
"1": {"nome": "Desenvolvedor", "descrição": "Tem acesso full ao sistema"},
"2": {"nome": "Administrador Master", "descrição": "Tem acesso full as funcionalidades do sistema e não pode ser apagado"},
"3": {"nome": "Administrador", "descrição": "Tem acesso full as funcionalidades do sistema e pode ser apagado"},
"4": {"nome": "Escrita", "descrição": "Tem acesso para inserção de dados no sistema e pode se bloquear telas do mesmo"},
"5": {"nome": "Leitura", "descrição": "Tem acesso para leitura de dados no sistema e pode se bloquear telas do mesmo"}
},
"bloqueio_tela": {
"2": {"tela_bloqueadas": []},
"3": {"tela_bloqueadas": []},
"4": {"tela_bloqueadas": []},
"5": {"tela_bloqueadas": []}
},
"telas": {},
"menu_config": {
"0": [{"icon_left": "account", "texto": "_users", "icon_right": "chevron-right", "status_icon_right": "True", "func_icon_right": "config_user", "cor": "False"},
{"icon_left": "tools", "texto": "_project", "icon_right": "chevron-right", "status_icon_right": "True", "func_icon_right": "config_project", "cor": "False"}]
}
}
```
Abrindo arquivo json de varios niveis e transformando o mesmo em objeto dict em python. Por fim manipulando o dict.
```
import json
with open('config_app.json', mode='r', encoding='utf-8') as a:
json_obj = json.load(a)
print(f'Type = {type(json_obj)}')
for key, data in json_obj.items():
print(f'\n{key} - {data}')
print('-'*100)
print(f'\n Imprimindo nivel 2:\n{json_obj["permissões"]["5"]}')
print(f'\n Imprimindo nivel 3:\n{json_obj["permissões"]["5"]["nome"]}')
```
# <font color=#FF0000>**json.load()**</font> - Trabalhando com json.load()
transformando obj json em dict **_(json.loads)_** python e obj json **_(json.dumps)_**
```
import json
json_string = '{"first_name": "Guido", "last_name":"Rossum"}'
print(f'{json_string = }')
print(f'{type(json_string) = }')
# A mesma pode ser analisado assim:
parsed_json = json.loads(json_string)
print(f'\n{parsed_json = }')
print(f'{type(parsed_json) = }')
# e agora pode ser usado como um dicionário normal:
print(f'\n{parsed_json["first_name"] = }')
# convertendo novamente para json
json_obj = json.dumps(parsed_json)
print(f'\n{json_obj = }')
print(f'{type(json_obj) = }')
```
# <font color=#FF0000>**json.dumps()**</font> para converter tipos primitivos Python em equivalentes JSON
Existem vários cenários em que você precisa usar dados JSON serializados em seu programa. Se você precisar desses dados JSON serializados em seu aplicativo de processamento adicional, poderá convertê-los em um **str objeto Python** nativo em vez de gravá-los em um arquivo.
Por exemplo, você recebe uma solicitação HTTP para enviar detalhes do desenvolvedor. você buscou dados de desenvolvedor da tabela de banco de dados e os armazenou em um dicionário Python ou qualquer objeto Python, agora você precisa enviar esses dados de volta para o aplicativo solicitado, então você precisa converter o objeto de dicionário Python em uma string formatada em JSON para enviar como um resposta na string JSON. Para fazer isso, você precisa usar json.dumps().
O json.dumps() retorna a representação de string JSON do Python dict.
## converter o dicionário Python em uma string formatada em JSON
```
import json
def SendJsonResponse(resultDict):
print("Convert Python dictionary into JSON formatted String")
developer_str = json.dumps(resultDict)
print(developer_str)
# sample developer dict
dicionario = {
"name": "Jane Doe",
"salary": 9000,
"skills": ["Python", "Machine Learning", "Web Development"],
"email": "[email protected]"
}
print(f'Type dicionario = {type(dicionario)}')
print(f'{dicionario = }')
string_json = json.dumps(dicionario)
print(f'\nType string_json= {type(string_json)}')
print(f'{string_json = }')
```
# <font color=#FF0000>**json.dumps()**</font> - Mapeamento entre entidades JSON e Python durante a codificação
Para codificar objetos Python no módulo JSON equivalente a JSON, usa-se a seguinte tabela de conversão. A json.dump() e json.dumps() executa o método as traduções quando codificam.
Agora vamos ver como converter todos os tipos primitivos Python, tais como dict, list, set, tuple, str, números em JSON dados formatados. Consulte a tabela a seguir para saber o mapeamento entre os tipos de dados JSON e Python.
Python | Json
:---: | :---:
dict | object
list, tuple | array
str | string
int, float, int & float-derived Enums | number
True | true
False | false
None | null
```
import json
dicionario = {
"colorList": ["Red", "Green", "Blue"],
"carTuple": ("BMW", "Audi", "range rover"),
"sampleString": "pynative.com",
"sampleInteger": 457,
"sampleFloat": 225.48,
"booleantrue": True,
"booleanfalse": False,
"nonevalue": None
}
print(f'Type dicionario = {type(dicionario)}')
print(f'{dicionario = }')
string_json = json.dumps(dicionario)
print(f'\nType string_json= {type(string_json)}')
print(f'{string_json = }')
from json import dumps
#! dict para obj json
carros_dict = {'marca': 'Toyota', 'modelo': 'Corolla', 'cor': 'chumbo'}
print(carros_dict)
print(type(carros_dict))
# transformando em objeto json
carros_json = dumps(carros_dict)
print(f'\n{carros_json}')
print(type(carros_json))
from json import dumps
#! tuple() to array json []
carros_tuple = ('Toyota', 'VW', 'Honda', 'BMW')
print(carros_tuple)
print(type(carros_tuple))
# transformando em objeto json
carros_json = dumps(carros_tuple)
print(f'\n{carros_json}')
print(type(carros_json))
from json import dumps
#! list[] to array json[]
carros_list = ['Toyota', 'VW', 'Honda', 'BMW']
print(carros_list)
print(type(carros_list))
# transformando em objeto json
carros_json = dumps(carros_list)
print(f'\n{carros_json}')
print(type(carros_json))
```
# <font color=#FF0000>**json.dump()**</font> - Para codificar e gravar dados JSON em um arquivo
Para gravar a resposta JSON em um arquivo: Na maioria das vezes, ao executar uma solicitação GET, você recebe uma resposta no formato JSON e pode armazenar a resposta JSON em um arquivo para uso futuro ou para uso de um sistema subjacente.
Por exemplo, você tem dados em uma lista ou dicionário ou qualquer objeto Python e deseja codificá-los e armazená-los em um arquivo na forma de JSON.
Vamos converter o dicionário Python em um formato JSON e gravá-lo em um arquivo, sendo:
1. **SEM FORMATAÇÃO NO ARQUIVO JSON**. (file_json_sem_formatar.json)
```json
{"bloqueio_tela": {"5": {"tela_bloqueadas": []}, "3": {"tela_bloqueadas": []}, "1": {"tela_bloqueadas": []}, "2": {"tela_bloqueadas": []}}}
```
2. **RECUADOS E FORMATADOS**. (file_json_formatado.json)
* indent=4 --> _4 espaços de indentação_
* separators=(', ', ': ') --> _formato com espaço apos "," e apos ":"_
* sort_keys=True --> _as chavas são gravadas em ordem crescente_
```json
{
"bloqueio_tela": {
"1": {
"tela_bloqueadas": []
},
"2": {
"tela_bloqueadas": []
},
"3": {
"tela_bloqueadas": []
},
"5": {
"tela_bloqueadas": []
}
}
}
```
3. **CODIFICAÇÃO COMPACTA PARA ECONOMIZAR ESPAÇO**. (file_json_compacto.json)
* separators=(',', ':') --> _eliminando os espaços e formatação_
```json
{"bloqueio_tela":{"1":{"tela_bloqueadas":[]},"2":{"tela_bloqueadas":[]},"3":{"tela_bloqueadas":[]},"5":{"tela_bloqueadas":[]}}}
```
```
import json
dicionario = {"bloqueio_tela": {"5": {"tela_bloqueadas": []}, "3": {"tela_bloqueadas": []}, "1": {"tela_bloqueadas": []}, "2": {"tela_bloqueadas": []}}}
# criando um arquivo sem formatar:
with open('file_json_sem_formatar.json', mode='w', encoding='utf-8') as write_file:
json.dump(dicionario, write_file)
# criando um arquivo json formatado, com recuo, espaços apos "," e ":" e em orden crescente de chaves:
with open('file_json_formatado.json', mode='w', encoding='utf-8') as write_file:
json.dump(dicionario, write_file, indent=4, separators=(', ', ': '), sort_keys=True)
# criando um arquivo json sem formatação e sem espaço para economizar tamanho com "," e ":":
with open('file_json_compacto.json', mode='w', encoding='utf-8') as write_file:
json.dump(dicionario, write_file, separators=(',', ':'), sort_keys=True)
# podemos tambem trocar o sinal que divide key e chave com separators
print(json.dumps(dicionario, separators=(',', '='), sort_keys=True))
```
# <font color=#FF0000>**json.dump()**</font> - Pule os tipos não básicos ao gravar JSON em um arquivo usando o parâmetro skipkeys
O módulo json integrado do Python só pode lidar com tipos primitivos Python que tenham um equivalente JSON direto (por exemplo, dicionário, listas, strings, ints, Nenhum, etc.).
Se o dicionário Python contiver um objeto Python personalizado como uma das chaves e se tentarmos convertê-lo em um formato JSON, você obterá um TypeError, isto é <font color=#FF0000>**_Object of type "Your Class" is not JSON serializable_**</font>,.
Se este objeto personalizado não for necessário em dados JSON, você pode ignorá-lo usando um **_skipkeys=true_** argumento do json.dump() método.
Se **_skipkeys=true_** for True, então as dict chaves que não são de um tipo básico (str, int, float, bool, None) serão ignoradas em vez de gerar um TypeError.
```json
{"salario": 9000, "skills": ["Python", "Machine Learning", "Web Development"], "email": "[email protected]"}
```
Obs.: Sem o DadosPessoais: usuario
Artigo para transformar tipos não basicos em json:
<https://pynative.com/make-python-class-json-serializable/>
```
import json
class DadosPessoais():
def __init__(self, name: str, age: int):
self.name = name
self.age = age
def showInfo(self):
print("Nome é " + self.name, "Idade é ", self.age)
# instanciando um objeto
usuario = DadosPessoais("João", 36)
dicionario = {
DadosPessoais: usuario,
"salario": 9000,
"skills": ["Python", "Machine Learning", "Web Development"],
"email": "[email protected]"
}
# criando arquivo json sem tipos não basicos (obj DadosPessoais)
with open("file_json_sem_tipos_nao_basicos.json", mode='w', encoding='utf-8') as write_file:
json.dump(dicionario, write_file, skipkeys=True)
```
# <font color=#FF0000>**json.dumps()**</font> - Lidar com caracteres não ASCII de dados JSON ao gravá-los em um arquivo
O json.dump() método possui ensure_ascii parâmetro. O ensure_ascii é verdadeiro por padrão. A saída tem a garantia de ter todos os caracteres não ASCII de entrada com escape. Se ensure_ascii for falso, esses caracteres serão reproduzidos no estado em que se encontram. Se você deseja armazenar caracteres não ASCII, no estado em que se encontra, use o código a seguir.
Obs.: Se usar o **ensure_ascii=False** como parametro do json.dump o mesmo irá salvar palavras com acentuação no arquivo json. Uma boa pratica ao abrir estes arquivos é usar o encoding utf-8
~~~
# boa pratica se salvar um json com ensure_ascii=False é abrir o mesmo com encoding utf-8
with open(caminho, mode='r', encoding='utf-8') as read_file:
~~~
```
import json
# encoding in UTF-8
unicode_data= {
"string1": "明彦",
"string2": u"\u00f8"}
print(f'{unicode_data = }')
# dumps com ensure_ascii=False
encoded_unicode = json.dumps(unicode_data, ensure_ascii=False)
print(f'{encoded_unicode = }')
encoded_unicode = json.dumps(unicode_data, ensure_ascii=True)
print(f'{encoded_unicode = }')
# dumps com ensure_ascii=True (default)
print(json.loads(encoded_unicode))
```
# <font color=#FF0000>**CSV (Comma Separated Values)**</font> - Trabalhando com arquivo CSV
# <font color=#FF0000>**csv.reader**</font> - Leia CSV com delimitador de vírgula
csv.reader função no modo padrão para arquivos CSV com delimitador de vírgula.
csv.reader(file) ou csv.reader(file, delimiter=',') é a mesma coisa, uma vez que o delimiter default é = ','
```
import csv
with open('contatos.csv', mode='r', encoding='utf8', newline='') as file:
# csv.reader(file) ou csv.reader(file, delimiter=',') é a mesma coisa, uma vez que o delimiter default é = ','
csv_reader = csv.reader(file)
for row in csv_reader:
print(row)
```
# <font color=#FF0000>**csv.reader - delimiter='\t'**</font> - Leia CSV com delimitador diferente
Por padrão, uma vírgula é usada como delimitador em um arquivo CSV. No entanto, alguns arquivos CSV podem usar outros delimitadores além de vírgulas. Os populares | e \t(tab).
```
import csv
with open('contatos_com_delimitador_tab.csv', mode='r', encoding='utf8', newline='') as file:
# csv.reader(file) ou csv.reader(file, delimiter=',') é a mesma coisa, uma vez que o delimiter default é = ','
csv_reader = csv.reader(file)
for row in csv_reader:
print(row)
print('-' * 70)
# utilizando seek(0) para retornar ao inicio do arquivo para ler novamente
file.seek(0)
# agora usando o delimeter '\t'(\t = tab)
csv_reader = csv.reader(file, delimiter='\t')
for row in csv_reader:
print(row)
```
# <font color=#FF0000>**csv.reader - skipinitialspace=True**</font> - Leia arquivos CSV com espaços iniciais
Isso permite que o reader objeto saiba que as entradas possuem um espaço em branco inicial. Como resultado, os espaços iniciais que estavam presentes após um delimitador são removidos.
```
import csv
with open('contatos_com_espaços.csv', mode='r', encoding='utf8', newline='') as file:
csv_reader = csv.reader(file)
for row in csv_reader:
print(row)
print('-' * 70)
# utilizando seek(0) para retornar ao inicio do arquivo para ler novamente
file.seek(0)
# agora usando o skipinitialspace=True para eliminar os espaços
csv_reader = csv.reader(file, skipinitialspace=True)
for row in csv_reader:
print(row)
```
# <font color=#FF0000>**csv.reader - quoting=csv.QUOTE_ALL, skipinitialspace=True**</font> - Ler arquivos CSV com aspas
Como você pode ver, passamos csv.QUOTE_ALL para o quoting parâmetro. É uma constante definida pelo csv módulo.
csv.QUOTE_ALL especifica o objeto leitor que todos os valores no arquivo CSV estão presentes entre aspas.
Existem 3 outras constantes predefinidas que você pode passar para o quoting parâmetro:
* csv.QUOTE_MINIMAL- Especifica o reader objeto que o arquivo CSV tem aspas em torno das entradas que contêm caracteres especiais, como delimitador , quotechar ou qualquer um dos caracteres no determinador de linha .
* csv.QUOTE_NONNUMERIC- Especifica o reader objeto que o arquivo CSV tem aspas em torno das entradas não numéricas.
* csv.QUOTE_NONE - Especifica o objeto leitor que nenhuma das entradas tem aspas ao redor.
```
import csv
with open('arquivo_csv_com_aspas.csv', mode='r', encoding='utf8', newline='') as file:
csv_reader = csv.reader(file)
for row in csv_reader:
print(row)
print('-' * 70)
# utilizando seek(0) para retornar ao inicio do arquivo para ler novamente
file.seek(0)
# agora usando o quoting=csv.QUOTE_ALL, skipinitialspace=True para eliminar as aspas e espaços
csv_reader = csv.reader(file, quoting=csv.QUOTE_ALL, skipinitialspace=True)
for row in csv_reader:
print(row)
```
# <font color=#FF0000>**csv.reader - dialect='myDialect'**</font> - Ler arquivos CSV usando dialeto
Passamos vários parâmetros ( delimiter, quotinge, skipinitialspace) para a csv.reader()função.
Essa prática é aceitável ao lidar com um ou dois arquivos. Mas isso tornará o código mais redundante e feio quando começarmos a trabalhar com vários arquivos CSV com formatos semelhantes. Como solução para isso, o csv módulo oferece dialect como parâmetro opcional.
Dialeto ajuda a agrupar muitos padrões de formatação específicas, como delimiter, skipinitialspace, quoting, escapecharem um único nome dialeto.
Ele pode então ser passado como um parâmetro para várias writer ou reader instâncias.
```
import csv
with open('arquivo_csv_uso_dialetos.csv', mode='r', encoding='utf8', newline='') as file:
csv_reader = csv.reader(file)
for row in csv_reader:
print(row)
print('-' * 70)
# utilizando seek(0) para retornar ao inicio do arquivo para ler novamente
file.seek(0)
# registrando um dialeto
csv.register_dialect('myDialect', delimiter='|', skipinitialspace=True, quoting=csv.QUOTE_ALL)
# agora usando o dialect='myDialect'
csv_reader = csv.reader(file, dialect='myDialect')
for row in csv_reader:
print(row)
"""A vantagem de usar dialect é que torna o programa mais modular. Observe que podemos reutilizar
'myDialect' para abrir outros arquivos sem ter que especificar novamente o formato CSV."""
```
# <font color=#FF0000>**csv.DictReader**</font>
Entradas da primeira linha são as chaves do dicionário. E as entradas nas outras linhas são os valores do dicionário.
```
import csv
with open('contatos.csv', mode='r', encoding='utf8', newline='') as file:
csv_file = csv.DictReader(file)
for row in csv_file:
print(row) # python >= 3.8 print(dict(row)) python < 3.8
# Entradas da primeira linha são as chaves do dicionário. E as entradas nas outras linhas são os valores do dicionário.
```
# <font color=#FF0000>**csv.writer writerow**</font> - Gravando linha por linha com writerow
A csv.writer()função retorna um writer objeto que converte os dados do usuário em uma string delimitada. Esta string pode ser usada posteriormente para gravar em arquivos CSV usando a writerow()função. Vamos dar um exemplo.
```
import csv
# Gravando linha por linha com writerow
with open('arquivo_csv_writer.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file)
writer.writerow(["SN", "Movie", "Protagonist"])
writer.writerow([1, "Lord of the Rings", "Frodo Baggins"])
writer.writerow([2, "Harry Potter", "Harry Potter"])
```
# <font color=#FF0000>**csv.writer writerows**</font> - Gravando várias linhas com writerows
```
import csv
# Gravando varias linhas com writerows
lista = [["SN", "Movie", "Protagonist"], [1, "Lord of the Rings", "Frodo Baggins"], [2, "Harry Potter", "Harry Potter"]]
with open('arquivo_csv_writer_rows.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file)
writer.writerows(lista)
```
# <font color=#FF0000>**csv.writer - delimiter**</font> - Gravando em um arquivo CSV com delimitador
```
cod nome email
1 Joao [email protected]
2 Amanda [email protected]
3 Arthur [email protected]
4 Matheus [email protected]
5 Gustavo [email protected]
6 Renato [email protected]
```
```
import csv
lista = [['cod', 'nome', 'email'], ['1', 'Joao', '[email protected]'], ['2', 'Amanda', '[email protected]'],
['3', 'Arthur', '[email protected]'], ['4', 'Matheus', '[email protected]'], ['5', 'Gustavo', '[email protected]'],
['6', 'Renato', '[email protected]']]
with open('contatos_com_delimitador_tab.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file, delimiter='\t')
writer.writerows(lista)
```
# <font color=#FF0000>**csv.writer - quoting=csv.QUOTE_NONNUMERIC**</font> - Gravando em um arquivo CSV com aspas
* _csv.QUOTE_NONNUMERIC_ Especifica o writer objeto que as aspas devem ser adicionadas às entradas **não numéricas**.
* _csv.QUOTE_ALL_ Especifica o writer objeto para gravar o arquivo CSV com aspas em torno de **todas as entradas**.
* _csv.QUOTE_MINIMAL_ Especifica o writer objeto para citar apenas os campos que contêm caracteres especiais (delimitador , quotechar ou quaisquer caracteres no determinador de linha)
* _csv.QUOTE_NONE_ Especifica o writer objeto que nenhuma das entradas deve ser citada. **É o valor padrão**.
```
"cod","nome","email"
1,"Joao","[email protected]"
2,"Amanda","[email protected]"
3,"Arthur","[email protected]"
4,"Matheus","[email protected]"
5,"Gustavo","[email protected]"
6,"Renato","[email protected]"
```
```
import csv
lista = [["cod", "nome", "email"], [1, 'Joao', '[email protected]'], [2, 'Amanda', "[email protected]"],
[3, 'Arthur', '[email protected]'], [4, "Matheus", '[email protected]'], [5, 'Gustavo', '[email protected]'],
[6, "Renato", '[email protected]']]
with open('arquivo_csv_com_aspas.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file, quoting=csv.QUOTE_NONNUMERIC)
writer.writerows(lista)
```
# <font color=#FF0000>**csv.writer - quoting=csv.QUOTE_NONNUMERIC e quotechar='*'**</font> - Gravando arquivos CSV com caractere de citação personalizado
```
*cod*,*nome*,*email*
*1*,*Joao*,*[email protected]*
*2*,*Amanda*,*[email protected]*
*3*,*Arthur*,*[email protected]*
*4*,*Matheus*,*[email protected]*
*5*,*Gustavo*,*[email protected]*
*6*,*Renato*,*[email protected]*
```
```
import csv
lista = [['cod', 'nome', 'email'], ['1', 'Joao', '[email protected]'], ['2', 'Amanda', '[email protected]'],
['3', 'Arthur', '[email protected]'], ['4', 'Matheus', '[email protected]'], ['5', 'Gustavo', '[email protected]'],
['6', 'Renato', '[email protected]']]
with open('arquivo_csv_com_quotechar.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file, quoting=csv.QUOTE_NONNUMERIC, quotechar='*')
writer.writerows(lista)
```
# <font color=#FF0000>**csv.writer - dialect='myDialect'**</font> - Gravando arquivos CSV usando dialeto
A vantagem de usar dialect é que torna o programa mais modular. Observe que podemos reutilizar myDialect para gravar outros arquivos CSV sem ter que especificar novamente o formato CSV.
```
*cod*|*nome*|*email*
*1*|*Joao*|*[email protected]*
*2*|*Amanda*|*[email protected]*
*3*|*Arthur*|*[email protected]*
*4*|*Matheus*|*[email protected]*
*5*|*Gustavo*|*[email protected]*
*6*|*Renato*|*[email protected]*
```
```
import csv
lista = [['cod', 'nome', 'email'], ['1', 'Joao', '[email protected]'], ['2', 'Amanda', '[email protected]'],
['3', 'Arthur', '[email protected]'], ['4', 'Matheus', '[email protected]'], ['5', 'Gustavo', '[email protected]'],
['6', 'Renato', '[email protected]']]
csv.register_dialect('myDialect', delimiter='|', quoting=csv.QUOTE_NONNUMERIC, quotechar='*')
with open('arquivo_csv_uso_dialetos.csv', mode='w', encoding='utf8', newline='') as file:
writer = csv.writer(file, dialect='myDialect')
writer.writerows(lista)
```
# <font color=#FF0000>**csv.DictWriter**</font> - Gravando arquivos CSV atraves de uma lista de dicionarios
```
cod,nome,email
1,Joao,[email protected]
2,Amanda,[email protected]
3,Arthur,[email protected]
4,Matheus,[email protected]
5,Gustavo,[email protected]
6,Renato,[email protected]
```
```
import csv
lista = [{'cod': 1, 'nome': 'Joao', 'email': '[email protected]'}, {'cod': 2, 'nome': 'Amanda', 'email': '[email protected]'},
{'cod': 3, 'nome': 'Arthur', 'email': '[email protected]'}, {'cod': 4, 'nome': 'Matheus', 'email': '[email protected]'},
{'cod': 5, 'nome': 'Gustavo', 'email': '[email protected]'}, {'cod': 6, 'nome': 'Renato', 'email': '[email protected]'}]
with open('arquivo_csv_dictWriter.csv', mode='w', encoding='utf8', newline='') as file:
fieldnames = ['cod', 'nome', 'email']
writer = csv.DictWriter(file, fieldnames=fieldnames)
writer.writeheader()
writer.writerows(lista)
```
# <font color=#FF0000>**csv to Excel com openpyxl**</font> - Transformando arquivos CSV em Excel
```
import csv
from openpyxl import Workbook
import os
wb = Workbook()
ws = wb.active
with open('salarios.csv') as f:
reader = csv.reader(f, delimiter=',')
for row in reader:
ws.append(row)
wb.save('salarios.xlsx')
os.startfile('salarios.xlsx')
# os.system("start EXCEL.EXE salarios.xlsx")
# os.system("open -a 'path/Microsoft Excel.app' 'path/file.xlsx'")
```
|
github_jupyter
|
```
import autograd.numpy as np
import autograd.numpy.random as npr
npr.seed(0)
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
sns.set_style("white")
sns.set_context("talk")
color_names = ["windows blue",
"red",
"amber",
"faded green",
"dusty purple",
"orange",
"clay",
"pink",
"greyish",
"mint",
"light cyan",
"steel blue",
"forest green",
"pastel purple",
"salmon",
"dark brown"]
colors = sns.xkcd_palette(color_names)
import ssm
from ssm.variational import SLDSMeanFieldVariationalPosterior, SLDSTriDiagVariationalPosterior
from ssm.util import random_rotation, find_permutation
```
## Ring attractor
https://www.sciencedirect.com/science/article/pii/S0896627318303258
```
###Ring attractor (multi-attractor) model
Neu = 100 #number of cells
taum = 0.01 #membrain time constant
k = 0.1 #gain
gm = 100 #conductance
Wa = -40/gm #average weight
Wd = 33/gm #tuning-dependent
###synaptic weights
Wij = np.zeros((Neu,Neu)) #connectivity matrix
deg2rad = np.pi/180
ths = np.linspace(-90,90,Neu)*deg2rad #preferred tuning direction
for ii in range(Neu):
for jj in range(Neu):
Wij[ii,jj] = Wa + Wd/Neu*np.cos(ths[ii]-ths[jj])
#np.exp( (np.cos(ths[ii]-ths[jj])-1)/lsyn**2 )
Wij = (Wij-np.mean(Wij))*1 ##??
plt.plot(ths,Wij);
plt.xlabel('angle (rad)')
plt.ylabel('weight')
plt.figure()
plt.imshow(Wij)
plt.xlabel('i')
plt.ylabel('j')
###stimulus
T = 100 #sec
dt = 0.01 #10ms
time = np.arange(0,T,dt) #time series
b = 2
c = 0.5
Am = 0.1
def the2h(ti,tstim):
return b + c*(1-Am+Am*np.cos(ti-tstim))
#b + c*Am*np.exp( (np.cos(ti-tstim)-1)/lstim**2 )
taun = 10 #noise correlation
noise = 10
h = np.zeros(len(time))
for tt in range(0,len(time)-1):
h[tt+1] = h[tt] + (ths[int(len(ths)/2)]-h[tt])*dt/taun + np.sqrt(taun*dt)*np.random.randn()*noise
# if h[tt+1]>180:
# h[tt+1] = h[tt+1]-180
# if h[tt+1]<0:
# h[tt+1] = 180+h[tt+1]
#h = h*deg2rad
# smoothed = 200
# temp = np.convolve(np.random.randn(len(time))*180,np.exp(-np.arange(1,smoothed,1)/smoothed),'same')
h = np.mod(h,180)*deg2rad - np.pi/2
plt.plot(time,h,'o')
plt.xlabel('time (s)')
plt.ylabel('head angle (rad)')
###neural dynamics
Vr = 0
V = np.zeros((Neu,len(time))) #neurons by time
V[:,0] = Vr + np.random.randn(Neu) #initialization
r = np.zeros((Neu,len(time)))
r[:,0] = gm*np.tanh(k*V[:,0]) #k*(np.max((V[:,0]-V0)))**nn
for tt in range(0,len(time)-1):
ht = np.array([the2h(hh,h[tt]) for hh in ths]) #input bump
V[:,tt+1] = V[:,tt] + dt*(-V[:,tt] + ht + np.dot(Wij,r[:,tt]))/taum + np.sqrt(dt*taum)*np.random.randn(Neu)*1
temp = V[:,tt+1].copy()
temp[temp<0] = 0
r[:,tt+1] = gm*np.tanh(k*temp) #k*(np.max((V[:,tt+1]-V0)))**nn
extent = [0,T,ths[0],ths[-1]]
plt.imshow(r, aspect="auto",extent=extent)
#plt.plot(V.T);
plt.plot(r[:,2:].T);
###PCA test
X = r[:,2:].copy()
C = np.cov(X)
u,s,v = np.linalg.svd(C)
PCs = np.dot(u[:,:3].T,X)
from mpl_toolkits.mplot3d import Axes3D
ax = plt.axes(projection='3d')
ax.plot3D(PCs[1,:], PCs[2,:], PCs[0,:])
```
## SLDS fitting
```
XX = np.array(X[np.random.choice(np.arange(0,Neu,1),10),:])
XX = XX[:,np.arange(0,len(time),10)]
#XX = X.copy()
plt.imshow(XX, aspect="auto")
XX.shape
# Set the parameters of the HMM
T = XX.shape[1] # number of time bins
K = 3 # number of discrete states
D = 2 # number of latent dimensions
N = XX.shape[0] # number of observed dimensions
# Make an SLDS with the true parameters
true_slds = ssm.SLDS(N, K, D, transitions="recurrent_only", emissions="gaussian_orthog")
for k in range(K):
true_slds.dynamics.As[k] = .95 * random_rotation(D, theta=(k+1) * np.pi/20)
z, x, y = true_slds.sample(T)
# Mask off some data
y = XX.T.copy()
mask = npr.rand(T, N) < 0.9
y_masked = y * mask
print("Fitting SLDS with SVI")
# Create the model and initialize its parameters
slds = ssm.SLDS(N, K, D, emissions="gaussian_orthog")
slds.initialize(y_masked, masks=mask)
# Create a variational posterior
q_mf = SLDSMeanFieldVariationalPosterior(slds, y_masked, masks=mask)
q_mf_elbos = slds.fit(q_mf, y_masked, masks=mask, num_iters=1000, initialize=False)
# Get the posterior mean of the continuous states
q_mf_x = q_mf.mean[0]
# Find the permutation that matches the true and inferred states
slds.permute(find_permutation(z, slds.most_likely_states(q_mf_x, y)))
q_mf_z = slds.most_likely_states(q_mf_x, y)
# Smooth the data under the variational posterior
q_mf_y = slds.smooth(q_mf_x, y)
print("Fitting SLDS with SVI using structured variational posterior")
slds = ssm.SLDS(N, K, D, emissions="gaussian_orthog")
slds.initialize(y_masked, masks=mask)
q_struct = SLDSTriDiagVariationalPosterior(slds, y_masked, masks=mask)
q_struct_elbos = slds.fit(q_struct, y_masked, masks=mask, num_iters=1000, initialize=False)
# Get the posterior mean of the continuous states
q_struct_x = q_struct.mean[0]
# Find the permutation that matches the true and inferred states
slds.permute(find_permutation(z, slds.most_likely_states(q_struct_x, y)))
q_struct_z = slds.most_likely_states(q_struct_x, y)
# Smooth the data under the variational posterior
q_struct_y = slds.smooth(q_struct_x, y)
###try with switching!
rslds = ssm.SLDS(N, K, D,
transitions="recurrent_only",
dynamics="diagonal_gaussian",
emissions="gaussian_orthog",
single_subspace=True)
rslds.initialize(y)
q = SLDSTriDiagVariationalPosterior(rslds, y)
elbos = rslds.fit(q, y, num_iters=1000, initialize=False)
xhat = q.mean[0]
# Find the permutation that matches the true and inferred states
rslds.permute(find_permutation(z, rslds.most_likely_states(xhat, y)))
zhat = rslds.most_likely_states(xhat, y)
plt.figure()
plt.plot(elbos)
plt.xlabel("Iteration")
plt.ylabel("ELBO")
plt.plot(xhat[:,0],xhat[:,1])
# Plot the ELBOs
plt.plot(q_mf_elbos, label="MF")
plt.plot(q_struct_elbos, label="LDS")
plt.xlabel("Iteration")
plt.ylabel("ELBO")
plt.legend()
###discrete state vs. head direction
plt.subplot(211)
plt.imshow(np.row_stack((q_struct_z, q_mf_z)), aspect="auto")
plt.yticks([0, 1], ["$z_{\\mathrm{struct}}$", "$z_{\\mathrm{mf}}$"])
plt.subplot(212)
plt.plot(h)
plt.xlim(0,len(h))
plt.xlabel('time')
plt.ylabel('angle (rad)')
for kk in range(K):
pos = np.where(q_struct_z==kk)[0]
plt.plot(q_struct_x[pos,0],q_struct_x[pos,1])
plt.plot(q_mf_x[:,0],q_mf_x[:,1])
# Plot the true and inferred states
# xlim = (0, 500)
# plt.figure(figsize=(8,4))
# plt.imshow(np.row_stack((z, q_struct_z, q_mf_z)), aspect="auto")
# plt.plot(xlim, [0.5, 0.5], '-k', lw=2)
# plt.yticks([0, 1, 2], ["$z_{\\mathrm{true}}$", "$z_{\\mathrm{struct}}$", "$z_{\\mathrm{mf}}$"])
# plt.xlim(xlim)
plt.figure(figsize=(8,4))
plt.plot(x + 4 * np.arange(D), '-k')
for d in range(D):
plt.plot(q_mf_x[:,d] + 4 * d, '-', color=colors[0], label="MF" if d==0 else None)
plt.plot(q_struct_x[:,d] + 4 * d, '-', color=colors[1], label="Struct" if d==0 else None)
plt.ylabel("$x$")
#plt.xlim(xlim)
plt.legend()
# Plot the smoothed observations
plt.figure(figsize=(8,4))
for n in range(N):
plt.plot(y[:, n] + 4 * n, '-k', label="True" if n == 0 else None)
plt.plot(q_mf_y[:, n] + 4 * n, '--', color=colors[0], label="MF" if n == 0 else None)
plt.plot(q_struct_y[:, n] + 4 * n, ':', color=colors[1], label="Struct" if n == 0 else None)
plt.legend()
plt.xlabel("time")
#plt.xlim(xlim)
```
|
github_jupyter
|
# Assignment 2: Implementation of Selection Sort
## Deliverables:
We will again generate random data for this assignment.
1) Please set up five data arrays of length 5,000, 10,000, 15,000, 20,000, and 25,000 of uniformly distributed random numbers (you may use either integers or floating point).
Ensure that a common random number seed is used to generate each of the arrays.
2) Execute the base algorithm (Selection Sort) for each of the random number arrays, noting the execution time with each execution.
Use one of the timing methods we learned in class.
3) Just as in the last assignment, please organize the results of the study into a table showing the size of data array and the time taken to sort the array.
Discuss the differences in timing and how they relate to data type and length of array.
4) Use Python matpl otlib or Seaborn to generate a measure of the size of the data set on the horizontal axis and with execution time in milliseconds on the vertical axis.
The plot should show execution time against problem size for each form of the algorithm being tested.
### Prepare an exec summary of your results, referring to the table and figures you have generated. Explain how your results relate to big O notation. Describe your results in language that management can understand. This summary should be included as text paragraphs in the Jupyter notebook. Explain how the algorithm works and why it is a useful to data engineers.
# Discussion
### The selection sort algorithm as implemented below uses a nested for loop. The inner loop indentifies the smallest componenent of an array and it's index while the outer loop manipulates the arrays (adds the smallest element to the new array and removes the element from the parent array). Since we have these two for loops the algorithm grows at a rate of approximately n*n. There are two operations first we identify the smallest element, then we place it in the new array. In big O notation, this is denoted O(n^2). Figure 1 below shows the sort times as a function of the length of the array. It is apparent that the lowest point demonstrates the non-linear scaling of this algorithm which is confirmed by taking the square root of the time. Figure 2 shows the square root of time as a function of the length of the array and is approximately linear.
### In some data retrieval systems items are required to be indexed sequentially, so we need methodologies to sort them, selection sort provides this methodology in an easy to implement fashion, however it is not very efficient due to the nested operations. Below are the two functions, required for the sort:
1) FindSmallest will start at the first index of an array and set it to an object 'smallest' which will be used in a repetative logical evaluation.
As we progress through the length of the array, each time the next value is smaller than smallest, smallest is replaced and it's index also is captured in smallest index.
This continues until the entire array is processed.
2) SelectionSort will find use FindSmallest to search through a given array using FindSmallest in a nested fashion to find the smallest value ('small') in the given array and append it to a new array.
The found value is removed from the original array (via it's returned index in FindSmallest; 'smallest_index') and the algorightm continues until the are no elements in the original array. The new array is returned along with the elapsed time to complete the sort in milliseconds.
```
import numpy as np
import pandas as pd
from datetime import datetime
import seaborn as sns
import time
#FindSmallest will start at the first index of an array and set it to an object 'smallest' which will be used in a repetative logical evaluation. As we progress through the length of the array, each time the next value is smaller than smallest, smallest is replaced and it's index also is captured in smallest index. This continues until the entire array is processed.
def FindSmallest(arr):
smallest = arr[0]
smallest_index=0
for i in range(1, len(arr)):
if arr[i] < smallest:
smallest = arr[i]
smallest_index = i
return smallest_index, smallest
# SelectionSort will find use FindSmallest to search through a given array using FindSmallest in a nested fashion to find the smallest value ('small') in the given array and append it to a new array. The found value is removed from the original array (via it's returned index in FindSmallest; 'smallest_index') and the algorightm continues until the are no elements in the original array. The new array is returned along with the elapsed time to complete the sort in milliseconds.
def SelectionSort(arr):
newArr = []
start = time.perf_counter()
for i in range(len(arr)):
smallest =FindSmallest(arr)[1]
smallest_index = FindSmallest(arr)[0]
newArr.append(smallest) #adds smallest element to new array.
arr = np.delete(arr, smallest_index) # removes smallest element from parent array by index.
end = time.perf_counter()
return newArr , (end-start)*1E3
```
# A. Generate arrays with a common random seed
```
#Sets the Random Seed
RANDOM_SEED = 123
np.random.seed(RANDOM_SEED)
arr5E4 = np.random.randint(low=1, high= 1000001, size=5000)#5,000 elements, 1-1E6 (inclusive)
np.random.seed(RANDOM_SEED)
arr10E4 = np.random.randint(low=1, high= 1000001, size=10000)#10,000 elements, 1-1E6 (inclusive)
np.random.seed(RANDOM_SEED)
arr15E4 = np.random.randint(low=1, high= 1000001, size=15000)#15,000 elements, 1-1E6 (inclusive)
np.random.seed(RANDOM_SEED)
arr20E4 = np.random.randint(low=1, high= 1000001, size=20000)#20,000 elements, 1-1E6 (inclusive)
np.random.seed(RANDOM_SEED)
arr25E4 = np.random.randint(low=1, high= 1000001, size=25000)#25,000 elements, 1-1E6 (inclusive)
```
# B. Sort using SelectionSort function
```
sorted_5E4 = SelectionSort(arr5E4)
sorted_10E4 = SelectionSort(arr10E4)
sorted_15E4 = SelectionSort(arr15E4)
sorted_20E4 = SelectionSort(arr20E4)
sorted_25E4 = SelectionSort(arr25E4)
Summary = {
'NumberOfElements': [ len(sorted_5E4[0]), len(sorted_10E4[0]), len(sorted_15E4[0]),len(sorted_20E4[0]), len(sorted_25E4[0])],
'Time(ms)': [ sorted_5E4[1], sorted_10E4[1], sorted_15E4[1], sorted_20E4[1], sorted_25E4[1]]}
df = pd.DataFrame.from_dict(Summary)
df['rt(Time)'] = np.sqrt(df['Time(ms)'])
display(df)
```
## Fig 1. Sort times in milliseconds as a function of the number of elements.
```
sns.scatterplot(x=df['NumberOfElements'], y=df['Time(ms)'])
```
## Fig 2. Square root of sort times in milliseconds as a function of the number of elements.
```
sns.scatterplot(x=df['NumberOfElements'], y=df['rt(Time)'])
```
# ------------------------ END ------------------------
code graveyard
```
### This code is for testing
#np.random.seed(123)
#arr7_39 = np.random.randint(low=7, high= 39, size=12)
#print("the array is",arr7_39)
#small = FindSmallest(arr7_39)
#print('the smallest index is at', small[0], 'and has value of', small[1])
#testing = SelectionSort(arr7_39)
#print('the array sorted is:', testing[0])
#print('execution time was: ', testing[1], 'ms')
```
|
github_jupyter
|
# Demos: Lecture 17
## Demo 1: bit flip errors
```
import pennylane as qml
from pennylane import numpy as np
import matplotlib.pyplot as plt
from lecture17_helpers import *
from scipy.stats import unitary_group
dev = qml.device("default.mixed", wires=1)
@qml.qnode(dev)
def prepare_state(U, p):
qml.QubitUnitary(U, wires=0)
qml.BitFlip(p, wires=0)
#qml.DepolarizingChannel(p, wires=0)
return qml.state()
n_samples = 500
original_states = []
flipped_states = []
for _ in range(n_samples):
U = unitary_group.rvs(2)
original_state = prepare_state(U, 0)
flipped_state = prepare_state(U, 0.3)
original_states.append(convert_to_bloch_vector(original_state))
flipped_states.append(convert_to_bloch_vector(flipped_state))
plot_bloch_sphere(original_states)
plot_bloch_sphere(flipped_states)
```
## Demo 2: depolarizing noise
## Demo 3: fidelity and trace distance
$$
F(\rho, \sigma) = \left( \hbox{Tr} \sqrt{\sqrt{\rho}\sigma\sqrt{\rho}} \right)^2
$$
```
from scipy.linalg import sqrtm
def fidelity(rho, sigma):
sqrt_rho = sqrtm(rho)
inner_thing = np.linalg.multi_dot([sqrt_rho, sigma, sqrt_rho])
return np.trace(sqrtm(inner_thing)) ** 2
proj_0 = np.array([[1, 0], [0, 0]])
proj_1 = np.array([[0, 0], [0, 1]])
fidelity(proj_0, proj_0)
fidelity(proj_0, proj_1)
```
$$
T(\rho, \sigma) = \frac{1}{2} \hbox{Tr} \left( \sqrt{(\rho - \sigma)^\dagger (\rho - \sigma)} \right)
$$
```
def trace_distance(rho, sigma):
rms = rho - sigma
inner_thing = np.dot(rms.conj().T, rms)
return 0.5 * np.trace(sqrtm(inner_thing))
U = unitary_group.rvs(2)
p_vals = np.linspace(0, 1, 10)
fids = []
tr_ds = []
for p in p_vals:
original_state = prepare_state(U, 0)
error_state = prepare_state(U, p)
fids.append(fidelity(original_state, error_state))
tr_ds.append(trace_distance(original_state, error_state))
plt.scatter(p_vals, fids)
plt.scatter(p_vals, tr_ds)
```
## Demo 4: VQE for $H_2$ molecule
```
bond_length = 1.3228
symbols = ["H", "H"]
coordinates = np.array([0.0, 0.0, -bond_length/2, 0.0, 0.0, bond_length/2])
H, n_qubits = qml.qchem.molecular_hamiltonian(symbols, coordinates)
print(H)
```
Ground state of $H_2$ looks like:
$$
|\psi_g(\theta)\rangle = \cos(\theta/2) |1100\rangle - \sin(\theta/2) |0011\rangle
$$
```
dev = qml.device("default.qubit", wires=4)
def prepare_ground_state(theta):
qml.PauliX(wires=0)
qml.PauliX(wires=1)
qml.DoubleExcitation(theta, wires=range(4))
return qml.expval(H)
opt = qml.GradientDescentOptimizer(stepsize=0.5)
ideal_qnode = qml.QNode(prepare_ground_state, dev)
theta = np.array(0.0, requires_grad=True)
energies = []
for _ in range(30):
theta, _energy = opt.step_and_cost(ideal_qnode, theta)
energies.append(_energy)
plt.plot(energies)
energies[-1]
theta
```
## Demo 5: VQE on a noisy device
```
from qiskit.test.mock import FakeSantiago
from qiskit.providers.aer import QasmSimulator
from qiskit.providers.aer.noise import NoiseModel
device = QasmSimulator.from_backend(FakeSantiago())
noise_model = NoiseModel.from_backend(device, readout_error=False)
noisy_dev = qml.device(
"qiskit.aer", backend='qasm_simulator', wires=4, shots=10000, noise_model=noise_model
)
noisy_qnode = qml.QNode(prepare_ground_state, noisy_dev)
noisy_qnode(theta)
opt = qml.GradientDescentOptimizer(stepsize=0.5)
theta = np.array(0.0, requires_grad=True)
noisy_energies = []
for it in range(30):
if it % 5 == 0:
print(f"it = {it}")
theta, _energy = opt.step_and_cost(noisy_qnode, theta)
noisy_energies.append(_energy)
plt.scatter(range(30), energies)
plt.scatter(range(30), noisy_energies)
```
## Demo 6: zero-noise extrapolation
|
github_jupyter
|
# Overfitting y Regularización
El **overfitting** o sobreajuste es otro problema común al entrenar un modelo de aprendizaje automático. Consiste en entrenar modelos que aprenden a la perfección los datos de entrenamiento, perdiendo de esta forma generalidad. De modo, que si al modelo se le pasan datos nuevos que jamás ha visto, no será capaz de realizar una buena predicción.
Existe un problema opuesto al overfitting conocido como **underfitting** o subajuste, en el que el modelo no logra realizar una predicción ni siquiera cercana a los datos de entrenamiento y esta lejos de hacer una generalización.

Para evitar el underfitting y el overfitting se pueden utilizar curvas de **loss**, **f1_score** o **accuracy** utilizando los datos de entrenamiento y validación. Haciendo un análisis sobre estas curvas se logra identificar estos problemas.
# Ejercicio
Utilizar el dataset [MNIST](http://yann.lecun.com/exdb/mnist/) para identificar los problemas de **underfitting** y **overfitting**, utilizando una ANN de capas lineales.
```
#-- Descomprimimos el dataset
# !rm -r mnist
# !unzip mnist.zip
#--- Buscamos las direcciones de cada archivo de imagen
from glob import glob
train_files = glob('./mnist/train/*/*.png')
valid_files = glob('./mnist/valid/*/*.png')
test_files = glob('./mnist/test/*/*.png')
train_files[0]
#--- Ordenamos los datos de forma aleatoria para evitar sesgos
import numpy as np
np.random.shuffle(train_files)
np.random.shuffle(valid_files)
np.random.shuffle(test_files)
len(train_files), len(valid_files), len(test_files)
#--- Cargamos los datos de entrenamiento en listas
from PIL import Image
N_train = len(train_files)
X_train = []
Y_train = []
for i, train_file in enumerate(train_files):
Y_train.append( int(train_file.split('/')[3]) )
X_train.append(np.array(Image.open(train_file)))
#--- Cargamos los datos de validación en listas
N_valid = len(valid_files)
X_valid = []
Y_valid = []
for i, valid_file in enumerate(valid_files):
Y_valid.append( int(valid_file.split('/')[3]) )
X_valid.append( np.array(Image.open(valid_file)) )
#--- Cargamos los datos de testeo en listas
N_test = len(test_files)
X_test = []
Y_test = []
for i, test_file in enumerate(test_files):
Y_test.append( int(test_file.split('/')[3]) )
X_test.append( np.array(Image.open(test_file)) )
#--- Visualizamos el tamaño de cada subset
len(X_train), len(X_valid), len(X_test)
#--- Visualizamos la distribución de clases en cada subset
from PIL import Image
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(15,5))
plt.subplot(1,3,1)
plt.hist(np.sort(Y_train))
plt.xlabel('class')
plt.ylabel('counts')
plt.title('Train set')
plt.subplot(1,3,2)
plt.hist(np.sort(Y_valid))
plt.xlabel('class')
plt.ylabel('counts')
plt.title('Valid set')
plt.subplot(1,3,3)
plt.hist(np.sort(Y_test))
plt.xlabel('class')
plt.ylabel('counts')
plt.title('Test set')
plt.show()
#-- Visualizamos los datos
fig = plt.figure(figsize=(8,8))
for i in range(4):
plt.subplot(2,2,i+1)
plt.imshow(X_test[i*15])
plt.title(Y_test[i*15])
plt.axis(False)
plt.show()
#--- Convetimos las listas con los datos a tensores de torch
import torch
from torch.autograd import Variable
X_train = Variable(torch.from_numpy(np.array(X_train))).float()
Y_train = Variable(torch.from_numpy(np.array(Y_train))).long()
X_valid = Variable(torch.from_numpy(np.array(X_valid))).float()
Y_valid = Variable(torch.from_numpy(np.array(Y_valid))).long()
X_test = Variable(torch.from_numpy(np.array(X_test))).float()
Y_test = Variable(torch.from_numpy(np.array(Y_test))).long()
X_train.data.size()
#--- Definimos una función que nos permita entrenar diferentes modelos de ANN
from sklearn.metrics import f1_score
def train_valid(model, n_epoch, optimizer, criterion):
loss_train = []
f1_train = []
acc_train = []
loss_valid = []
f1_valid = []
acc_valid = []
for epoch in range(n_epoch):
model.train()
Xtr = X_train.view(X_train.size(0), -1)
Y_pred = model(Xtr)
loss = criterion(Y_pred,Y_train)
loss_train.append(loss.item())
Y_pred = torch.argmax(Y_pred, 1)
f1_train.append( f1_score(Y_train,Y_pred, average='macro') )
acc = sum(Y_train == Y_pred)/len(Y_pred)
acc_train.append(acc)
optimizer.zero_grad()
loss.backward()
optimizer.step()
print( 'Epoch [{}/{}], loss: {}. f1:{} acc: {} '.format(epoch+1,n_epoch,loss_train[-1], f1_train[-1], acc_train[-1]) )
model.eval()
Xvl = X_valid.view(X_valid.size(0), -1)
Y_pred = model(Xvl)
loss = criterion(Y_pred,Y_valid)
loss_valid.append(loss.item())
Y_pred = torch.argmax(Y_pred, 1)
f1_valid.append( f1_score(Y_valid, Y_pred, average='macro') )
acc = sum(Y_valid == Y_pred)/len(Y_pred)
acc_valid.append(acc)
fig = plt.figure(figsize=(15,5))
plt.subplot(1,3,1)
plt.plot(range(n_epoch), loss_train, label='train')
plt.plot(range(n_epoch), loss_valid, label='valid')
plt.xlabel('n_epoch')
plt.ylabel('loss')
plt.legend()
plt.grid()
plt.subplot(1,3,2)
plt.plot(range(n_epoch), f1_train, label='train')
plt.plot(range(n_epoch), f1_valid, label='valid')
plt.xlabel('n_epoch')
plt.ylabel('f1_score')
plt.legend()
plt.grid()
plt.subplot(1,3,3)
plt.plot(range(n_epoch), acc_train, label='train')
plt.plot(range(n_epoch), acc_valid, label='valid')
plt.xlabel('n_epoch')
plt.ylabel('accuracy')
plt.legend()
plt.grid()
```
## Underfitting
El **underfitting** o sub ajuste se puede presentar en las siguientes situaciones:
* **Finalización temprana**: Cuando el modelo se entrena hasta una época temprana a pesar de que la tendencia indica una posible obtención de mejores resultados.
* **Modelo Simple**: Cuando el modelo es tan básico que no es capaz de extraer ningún tipo de patrón efectivo que le permita hacer una generalización de los datos.
```
#--- Definimos una ANN simple para identificar un error de underfitting
input_dim = 28*28
out_dim = 10
model = torch.nn.Sequential(
torch.nn.Linear(input_dim, out_dim)
)
optimizer = torch.optim.Adam(model.parameters())
criterion = torch.nn.CrossEntropyLoss()
train_valid(model,30,optimizer,criterion)
#-- Evaluamos el modelo entrenado con el set de testeo
model.eval()
Xts = X_test.view(X_test.size(0), -1)
Y_pred = model(Xts)
loss = criterion(Y_pred,Y_test)
Y_pred = torch.argmax(Y_pred, 1)
f1 = f1_score(Y_test, Y_pred, average='macro')
acc = sum(Y_test == Y_pred)/len(Y_pred)
print('loss: {}, f1: {}, acc: {}'.format(loss.item(), f1, acc))
```
## Overfitting
El **overfitting** o sobreajuste es el caso opuesto al subajuste y se puede presentar en la siguiente situación:
una obtención de mejores resultados.
* **Modelo Complejo**: El modelo es tan complejo que aprendió perfectamente los datos de entrenamiento, perdiendo generalidad. Cuando el modelo vea datos nuevos, diferentes a los del entrenamiento, su predicción será errónea.
```
input_dim = 28*28
out_dim = 10
hidden = 60
model = torch.nn.Sequential(
torch.nn.Linear(input_dim, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, out_dim)
)
optimizer = torch.optim.Adam(model.parameters())
criterion = torch.nn.CrossEntropyLoss()
train_valid(model,100,optimizer,criterion)
#-- Evaluamos el modelo entrenado con el set de testeo
model.eval()
Xts = X_test.view(X_test.size(0), -1)
Y_pred = model(Xts)
loss = criterion(Y_pred,Y_test)
Y_pred = torch.argmax(Y_pred, 1)
f1 = f1_score(Y_test, Y_pred, average='macro')
acc = sum(Y_test == Y_pred)/len(Y_pred)
print('loss: {}, f1: {}, acc: {}'.format(loss.item(), f1, acc))
```
## Regularización
Un mecanismo que permite evitar el sobreajuste es conocido como **regularización**. La cantidad de regularización afectará el rendimiento de validación del modelo. Muy poca regularización no resolverá el problema de sobreajuste. Demasiada regularización hará que el modelo sea mucho menos efectivo. La regularización actúa como una restricción sobre el conjunto de posibles funciones aprendibles.
<br>
Según [Ian Goodfellow](https://en.wikipedia.org/wiki/Ian_Goodfellow), "*La regularización es cualquier modificación que hacemos a un algoritmo de aprendizaje que tiene como objetivo reducir su error de generalización pero no su error de entrenamiento.*"
<br>
**Regularización de caída de peso**
La pérdida de peso es la técnica de regularización más común (implementada en Pytorch). En PyTorch, la caída de peso se proporciona como un parámetro para el optimizador *decay_weight*. En [este](https://pytorch.org/docs/stable/optim.html) enlace se muestran otros parámetros que pueden ser usados en los optimizadores.
A la caída de peso también se le llama:
* L2
* Ridge
Para la disminución de peso, agregamos un término de penalización en la actualización de los pesos:
$w(x) = w(x) − \eta \nabla x - \alpha \eta x$
Este nuevo término en la actualización lleva los parámetros $w$ ligeramente hacia cero, agregando algo de **decaimiento** en los pesos con cada actualización.
```
input_dim = 28*28
out_dim = 10
hidden = 60
model = torch.nn.Sequential(
torch.nn.Linear(input_dim, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, hidden),
torch.nn.ReLU(),
torch.nn.Linear(hidden, out_dim)
)
optimizer = torch.optim.Adam(model.parameters(), weight_decay=0.01)
criterion = torch.nn.CrossEntropyLoss()
train_valid(model,100,optimizer,criterion)
#-- Evaluamos el modelo entrenado con el set de testeo
model.eval()
Xts = X_test.view(X_test.size(0), -1)
Y_pred = model(Xts)
loss = criterion(Y_pred,Y_test)
Y_pred = torch.argmax(Y_pred, 1)
f1 = f1_score(Y_test, Y_pred, average='macro')
acc = sum(Y_test == Y_pred)/len(Y_pred)
print('loss: {}, f1: {}, acc: {}'.format(loss.item(), f1, acc))
```
|
github_jupyter
|
# 概率潜在语义分析
概率潜在语义分析(probabilistic latent semantic analysis, PLSA),也称概率潜在语义索引(probabilistic latent semantic indexing, PLSI),是一种利用概率生成模型对文本集合进行话题分析的无监督学习方法。
模型最大特点是用隐变量表示话题,整个模型表示文本生成话题,话题生成单词,从而得到单词-文本共现数据的过程;假设每个文本由一个话题分布决定,每个话题由一个单词分布决定。
### **18.1.2 生成模型**
假设有单词集合 $W = $ {$w_{1}, w_{2}, ..., w_{M}$}, 其中M是单词个数;文本(指标)集合$D = $ {$d_{1}, d_{2}, ..., d_{N}$}, 其中N是文本个数;话题集合$Z = $ {$z_{1}, z_{2}, ..., z_{K}$},其中$K$是预先设定的话题个数。随机变量 $w$ 取值于单词集合;随机变量 $d$ 取值于文本集合,随机变量 $z$ 取值于话题集合。概率分布 $P(d)$、条件概率分布 $P(z|d)$、条件概率分布 $P(w|z)$ 皆属于多项分布,其中 $P(d)$ 表示生成文本 $d$ 的概率,$P(z|d)$ 表示文本 $d$ 生成话题 $z$ 的概率,$P(w|z)$ 表示话题 $z$ 生成单词 $w$ 的概率。
每个文本 $d$ 拥有自己的话题概率分布 $P(z|d)$,每个话题 $z$ 拥有自己的单词概率分布 $P(w|z)$;也就是说**一个文本的内容由其相关话题决定,一个话题的内容由其相关单词决定**。
生成模型通过以下步骤生成文本·单词共现数据:
(1)依据概率分布 $P(d)$,从文本(指标)集合中随机选取一个文本 $d$ , 共生成 $N$ 个文本;针对每个文本,执行以下操作;
(2)在文本$d$ 给定条件下,依据条件概率分布 $P(z|d)$, 从话题集合随机选取一个话题 $z$, 共生成 $L$ 个话题,这里 $L$ 是文本长度;
(3)在话题 $z$ 给定条件下,依据条件概率分布 $P(w|z)$ , 从单词集合中随机选取一个单词 $w$.
注意这里为叙述方便,假设文本都是等长的,现实中不需要这个假设。
生成模型中, 单词变量 $w$ 与文本变量 $d$ 是观测变量, 话题变量 $z$ 是隐变量, 也就是说模型生成的是单词-话题-文本三元组合 ($w, z ,d$)的集合, 但观测到的单词-文本二元组 ($w, d$)的集合, 观测数据表示为单词-文本矩阵 $T$的形式,矩阵 $T$ 的行表示单词,列表示文本, 元素表示单词-文本对($w, d$)的出现次数。
从数据的生成过程可以推出,文本-单词共现数据$T$的生成概率为所有单词-文本对($w,d$)的生成概率的乘积:
$P(T) = \prod_{w,d}P(w,d)^{n(w,d)}$
这里 $n(w,d)$ 表示 ($w,d$)的出现次数,单词-文本对出现的总次数是 $N*L$。 每个单词-文本对($w,d$)的生成概率由一下公式决定:
$P(w,d) = P(d)P(w|d)$
$= P(d)\sum_{z}P(w,z|d)$
$=P(d)\sum_{z}P(z|d)P(w|z)$
### **18.1.3 共现模型**
$P(w,d) = \sum_{z\in Z}P(z)P(w|z)P(d|z)$
虽然生成模型与共现模型在概率公式意义上是等价的,但是拥有不同的性质。生成模型刻画文本-单词共现数据生成的过程,共现模型描述文本-单词共现数据拥有的模式。
如果直接定义单词与文本的共现概率 $P(w,d)$, 模型参数的个数是 $O(M*N)$, 其中 $M$ 是单词数, $N$ 是文本数。 概率潜在语义分析的生成模型和共现模型的参数个数是 $O(M*K + N*K)$, 其中 $K$ 是话题数。 现实中 $K<<M$, 所以**概率潜在语义分析通过话题对数据进行了更简洁的表示,减少了学习过程中过拟合的可能性**。
### 算法 18.1 (概率潜在语义模型参数估计的EM算法)
输入: 设单词集合为 $W = ${$w_{1}, w_{2},..., w_{M}$}, 文本集合为 $D=${$d_{1}, d_{2},..., d_{N}$}, 话题集合为 $Z=${$z_{1}, z_{2},..., z_{K}$}, 共现数据 $\left \{ n(w_{i}, d_{j}) \right \}, i = 1,2,..., M, j = 1,2,...,N;$
输出: $P(w_{i}|z_{k})$ 和 $P(z_{k}|d_{j})$.
1. 设置参数 $P(w_{i}|z_{k})$ 和 $P(z_{k}|d_{j})$ 的初始值。
2. 迭代执行以下E步,M步,直到收敛为止。
E步:
$P(z_{k}|w_{i},d_{j})=\frac{P(w_{i}|z_{k})P(z_{k}|d_{j})}{\sum_{k=1}^{K}P(w_{i}|z_{k})P(z_{k}|d_{j})}$
M步:
$P(w_{i}|z_{k})=\frac{\sum_{j=1}^{N}n(w_{i},d_{j})P(z_{k}|w_{i},d_{j})}{\sum_{m=1}^{M}\sum_{j=1}^{N}n(w_{m},d_{j})P(z_{k}|w_{m},d_{j})}$
$P(z_{k}|d_{j}) = \frac{\sum_{i=1}^{M}n(w_{i},d_{j})P(z_{k}|w_{i},d_{j})}{n(d_{j})}$
#### 习题 18.3
```
import numpy as np
X = [[0,0,1,1,0,0,0,0,0],
[0,0,0,0,0,1,0,0,1],
[0,1,0,0,0,0,0,1,0],
[0,0,0,0,0,0,1,0,1],
[1,0,0,0,0,1,0,0,0],
[1,1,1,1,1,1,1,1,1],
[1,0,1,0,0,0,0,0,0],
[0,0,0,0,0,0,1,0,1],
[0,0,0,0,0,2,0,0,1],
[1,0,1,0,0,0,0,1,0],
[0,0,0,1,1,0,0,0,0]]
X = np.asarray(X);X
X.shape
X = X.T;X
class PLSA:
def __init__(self, K, max_iter):
self.K = K
self.max_iter = max_iter
def fit(self, X):
n_d, n_w = X.shape
# P(z|w,d)
p_z_dw = np.zeros((n_d, n_w, self.K))
# P(z|d)
p_z_d = np.random.rand(n_d, self.K)
# P(w|z)
p_w_z = np.random.rand(self.K, n_w)
for i_iter in range(self.max_iter):
# E step
for di in range(n_d):
for wi in range(n_w):
sum_zk = np.zeros((self.K))
for zi in range(self.K):
sum_zk[zi] = p_z_d[di, zi] * p_w_z[zi, wi]
sum1 = np.sum(sum_zk)
if sum1 == 0:
sum1 = 1
for zi in range(self.K):
p_z_dw[di, wi, zi] = sum_zk[zi] / sum1
# M step
# update P(z|d)
for di in range(n_d):
for zi in range(self.K):
sum1 = 0.
sum2 = 0.
for wi in range(n_w):
sum1 = sum1 + X[di, wi] * p_z_dw[di, wi, zi]
sum2 = sum2 + X[di, wi]
if sum2 == 0:
sum2 = 1
p_z_d[di, zi] = sum1 / sum2
# update P(w|z)
for zi in range(self.K):
sum2 = np.zeros((n_w))
for wi in range(n_w):
for di in range(n_d):
sum2[wi] = sum2[wi] + X[di, wi] * p_z_dw[di, wi, zi]
sum1 = np.sum(sum2)
if sum1 == 0:
sum1 = 1
for wi in range(n_w):
p_w_z[zi, wi] = sum2[wi] / sum1
return p_w_z, p_z_d
# https://github.com/lipiji/PG_PLSA/blob/master/plsa.py
model = PLSA(2, 100)
p_w_z, p_z_d = model.fit(X)
p_w_z
p_z_d
```
|
github_jupyter
|
## Training a differentially private LSTM model for name classification
In this tutorial we will build a differentially-private LSTM model to classify names to their source languages, which is the same task as in the tutorial **NLP From Scratch** (https://pytorch.org/tutorials/intermediate/char_rnn_classification_tutorial.html). Since the objective of this tutorial is to demonstrate the effective use of an LSTM with privacy guarantees, we will be utilizing it in place of the bare-bones RNN model defined in the original tutorial. Specifically, we use the `DPLSTM` module from `opacus.layers.dp_lstm` to facilitate calculation of the per-example gradients, which are utilized in the addition of noise during application of differential privacy. `DPLSTM` has the same API and functionality as the `nn.LSTM`, with some restrictions (ex. we currently support single layers, the full list is given below).
## Dataset
First, let us download the dataset of names and their associated language labels as given in https://pytorch.org/tutorials/intermediate/char_rnn_classification_tutorial.html. We train our differentially-private LSTM on the same dataset as in that tutorial.
```
import os
import requests
NAMES_DATASET_URL = "https://download.pytorch.org/tutorial/data.zip"
DATA_DIR = "names"
import zipfile
import urllib
def download_and_extract(dataset_url, data_dir):
print("Downloading and extracting ...")
filename = "data.zip"
urllib.request.urlretrieve(dataset_url, filename)
with zipfile.ZipFile(filename) as zip_ref:
zip_ref.extractall(data_dir)
os.remove(filename)
print("Completed!")
download_and_extract(NAMES_DATASET_URL, DATA_DIR)
names_folder = os.path.join(DATA_DIR, 'data', 'names')
all_filenames = []
for language_file in os.listdir(names_folder):
all_filenames.append(os.path.join(names_folder, language_file))
print(os.listdir(names_folder))
import torch
import torch.nn as nn
class CharByteEncoder(nn.Module):
"""
This encoder takes a UTF-8 string and encodes its bytes into a Tensor. It can also
perform the opposite operation to check a result.
Examples:
>>> encoder = CharByteEncoder()
>>> t = encoder('Ślusàrski') # returns tensor([256, 197, 154, 108, 117, 115, 195, 160, 114, 115, 107, 105, 257])
>>> encoder.decode(t) # returns "<s>Ślusàrski</s>"
"""
def __init__(self):
super().__init__()
self.start_token = "<s>"
self.end_token = "</s>"
self.pad_token = "<pad>"
self.start_idx = 256
self.end_idx = 257
self.pad_idx = 258
def forward(self, s: str, pad_to=0) -> torch.LongTensor:
"""
Encodes a string. It will append a start token <s> (id=self.start_idx) and an end token </s>
(id=self.end_idx).
Args:
s: The string to encode.
pad_to: If not zero, pad by appending self.pad_idx until string is of length `pad_to`.
Defaults to 0.
Returns:
The encoded LongTensor of indices.
"""
encoded = s.encode()
n_pad = pad_to - len(encoded) if pad_to > len(encoded) else 0
return torch.LongTensor(
[self.start_idx]
+ [c for c in encoded] # noqa
+ [self.end_idx]
+ [self.pad_idx for _ in range(n_pad)]
)
def decode(self, char_ids_tensor: torch.LongTensor) -> str:
"""
The inverse of `forward`. Keeps the start, end and pad indices.
"""
char_ids = char_ids_tensor.cpu().detach().tolist()
out = []
buf = []
for c in char_ids:
if c < 256:
buf.append(c)
else:
if buf:
out.append(bytes(buf).decode())
buf = []
if c == self.start_idx:
out.append(self.start_token)
elif c == self.end_idx:
out.append(self.end_token)
elif c == self.pad_idx:
out.append(self.pad_token)
if buf: # in case some are left
out.append(bytes(buf).decode())
return "".join(out)
def __len__(self):
"""
The length of our encoder space. This is fixed to 256 (one byte) + 3 special chars
(start, end, pad).
Returns:
259
"""
return 259
```
## Training / Validation Set Preparation
```
from torch.nn.utils.rnn import pad_sequence
def padded_collate(batch, padding_idx=0):
x = pad_sequence(
[elem[0] for elem in batch], batch_first=True, padding_value=padding_idx
)
y = torch.stack([elem[1] for elem in batch]).long()
return x, y
from torch.utils.data import Dataset
from pathlib import Path
class NamesDataset(Dataset):
def __init__(self, root):
self.root = Path(root)
self.labels = list({langfile.stem for langfile in self.root.iterdir()})
self.labels_dict = {label: i for i, label in enumerate(self.labels)}
self.encoder = CharByteEncoder()
self.samples = self.construct_samples()
def __getitem__(self, i):
return self.samples[i]
def __len__(self):
return len(self.samples)
def construct_samples(self):
samples = []
for langfile in self.root.iterdir():
label_name = langfile.stem
label_id = self.labels_dict[label_name]
with open(langfile, "r") as fin:
for row in fin:
samples.append(
(self.encoder(row.strip()), torch.tensor(label_id).long())
)
return samples
def label_count(self):
cnt = Counter()
for _x, y in self.samples:
label = self.labels[int(y)]
cnt[label] += 1
return cnt
VOCAB_SIZE = 256 + 3 # 256 alternatives in one byte, plus 3 special characters.
```
We split the dataset into a 80-20 split for training and validation.
```
secure_rng = False
train_split = 0.8
test_every = 5
batch_size = 800
ds = NamesDataset(names_folder)
train_len = int(train_split * len(ds))
test_len = len(ds) - train_len
print(f"{train_len} samples for training, {test_len} for testing")
if secure_rng:
try:
import torchcsprng as prng
except ImportError as e:
msg = (
"To use secure RNG, you must install the torchcsprng package! "
"Check out the instructions here: https://github.com/pytorch/csprng#installation"
)
raise ImportError(msg) from e
generator = prng.create_random_device_generator("/dev/urandom")
else:
generator = None
train_ds, test_ds = torch.utils.data.random_split(
ds, [train_len, test_len], generator=generator
)
from torch.utils.data import DataLoader
from opacus.utils.uniform_sampler import UniformWithReplacementSampler
sample_rate = batch_size / len(train_ds)
train_loader = DataLoader(
train_ds,
num_workers=8,
pin_memory=True,
generator=generator,
batch_sampler=UniformWithReplacementSampler(
num_samples=len(train_ds),
sample_rate=sample_rate,
generator=generator,
),
collate_fn=padded_collate,
)
test_loader = DataLoader(
test_ds,
batch_size=2 * batch_size,
shuffle=False,
num_workers=8,
pin_memory=True,
collate_fn=padded_collate,
)
```
After splitting the dataset into a training and a validation set, we now have to convert the data into a numeric form suitable for training the LSTM model. For each name, we set a maximum sequence length of 15, and if a name is longer than the threshold, we truncate it (this rarely happens this dataset !). If a name is smaller than the threshold, we add a dummy `#` character to pad it to the desired length. We also batch the names in the dataset and set a batch size of 256 for all the experiments in this tutorial. The function `line_to_tensor()` returns a tensor of shape [15, 256] where each element is the index (in `all_letters`) of the corresponding character.
## Training/Evaluation Cycle
The training and the evaluation functions `train()` and `test()` are defined below. During the training loop, the per-example gradients are computed and the parameters are updated subsequent to gradient clipping (to bound their sensitivity) and addition of noise.
```
from statistics import mean
def train(model, criterion, optimizer, train_loader, epoch, device="cuda:0"):
accs = []
losses = []
for x, y in tqdm(train_loader):
x = x.to(device)
y = y.to(device)
logits = model(x)
loss = criterion(logits, y)
loss.backward()
optimizer.step()
optimizer.zero_grad()
preds = logits.argmax(-1)
n_correct = float(preds.eq(y).sum())
batch_accuracy = n_correct / len(y)
accs.append(batch_accuracy)
losses.append(float(loss))
printstr = (
f"\t Epoch {epoch}. Accuracy: {mean(accs):.6f} | Loss: {mean(losses):.6f}"
)
try:
privacy_engine = optimizer.privacy_engine
epsilon, best_alpha = privacy_engine.get_privacy_spent()
printstr += f" | (ε = {epsilon:.2f}, δ = {privacy_engine.target_delta}) for α = {best_alpha}"
except AttributeError:
pass
print(printstr)
return
def test(model, test_loader, privacy_engine, device="cuda:0"):
accs = []
with torch.no_grad():
for x, y in tqdm(test_loader):
x = x.to(device)
y = y.to(device)
preds = model(x).argmax(-1)
n_correct = float(preds.eq(y).sum())
batch_accuracy = n_correct / len(y)
accs.append(batch_accuracy)
printstr = "\n----------------------------\n" f"Test Accuracy: {mean(accs):.6f}"
if privacy_engine:
epsilon, best_alpha = privacy_engine.get_privacy_spent()
printstr += f" (ε = {epsilon:.2f}, δ = {privacy_engine.target_delta}) for α = {best_alpha}"
print(printstr + "\n----------------------------\n")
return
```
## Hyper-parameters
There are two sets of hyper-parameters associated with this model. The first are hyper-parameters which we would expect in any machine learning training, such as the learning rate and batch size. The second set are related to the privacy engine, where for example we define the amount of noise added to the gradients (`noise_multiplier`), and the maximum L2 norm to which the per-sample gradients are clipped (`max_grad_norm`).
```
# Training hyper-parameters
epochs = 50
learning_rate = 2.0
# Privacy engine hyper-parameters
max_per_sample_grad_norm = 1.5
delta = 8e-5
epsilon = 12.0
```
## Model
We define the name classification model in the cell below. Note that it is a simple char-LSTM classifier, where the input characters are passed through an `nn.Embedding` layer, and are subsequently input to the DPLSTM.
```
import torch
from torch import nn
from opacus.layers import DPLSTM
class CharNNClassifier(nn.Module):
def __init__(
self,
embedding_size,
hidden_size,
output_size,
num_lstm_layers=1,
bidirectional=False,
vocab_size=VOCAB_SIZE,
):
super().__init__()
self.embedding_size = embedding_size
self.hidden_size = hidden_size
self.output_size = output_size
self.vocab_size = vocab_size
self.embedding = nn.Embedding(vocab_size, embedding_size)
self.lstm = DPLSTM(
embedding_size,
hidden_size,
num_layers=num_lstm_layers,
bidirectional=bidirectional,
batch_first=True,
)
self.out_layer = nn.Linear(hidden_size, output_size)
def forward(self, x, hidden=None):
x = self.embedding(x) # -> [B, T, D]
x, _ = self.lstm(x, hidden) # -> [B, T, H]
x = x[:, -1, :] # -> [B, H]
x = self.out_layer(x) # -> [B, C]
return x
```
We now proceed to instantiate the objects (privacy engine, model and optimizer) for our differentially-private LSTM training. However, the `nn.LSTM` is replaced with a `DPLSTM` module which enables us to calculate per-example gradients.
```
# Set the device to run on a GPU
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Define classifier parameters
embedding_size = 64
hidden_size = 128 # Number of neurons in hidden layer after LSTM
n_lstm_layers = 1
bidirectional_lstm = False
model = CharNNClassifier(
embedding_size,
hidden_size,
len(ds.labels),
n_lstm_layers,
bidirectional_lstm,
).to(device)
```
## Defining the privacy engine, optimizer and loss criterion for the problem
```
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
from opacus import PrivacyEngine
privacy_engine = PrivacyEngine(
model,
sample_rate=sample_rate,
max_grad_norm=max_per_sample_grad_norm,
target_delta=delta,
target_epsilon=epsilon,
epochs=epochs,
secure_rng=secure_rng,
)
privacy_engine.attach(optimizer)
```
## Training the name classifier with privacy
Finally we can start training ! We will be training for 50 epochs iterations (where each epoch corresponds to a pass over the whole dataset). We will be reporting the privacy epsilon every `test_every` epochs. We will also benchmark this differentially-private model against a model without privacy and obtain almost identical performance. Further, the private model trained with Opacus incurs only minimal overhead in training time, with the differentially-private classifier only slightly slower (by a couple of minutes) than the non-private model.
```
from tqdm import tqdm
print("Train stats: \n")
for epoch in tqdm(range(epochs)):
train(model, criterion, optimizer, train_loader, epoch, device=device)
if test_every:
if epoch % test_every == 0:
test(model, test_loader, privacy_engine, device=device)
test(model, test_loader, privacy_engine, device=device)
```
The differentially-private name classification model obtains a test accuracy of 0.73 with an epsilon of just under 12. This shows that we can achieve a good accuracy on this task, with minimal loss of privacy.
## Training the name classifier without privacy
We also run a comparison with a non-private model to see if the performance obtained with privacy is comparable to it. To do this, we keep the parameters such as learning rate and batch size the same, and only define a different instance of the model along with a separate optimizer.
```
model_nodp = CharNNClassifier(
embedding_size,
hidden_size,
len(ds.labels),
n_lstm_layers,
bidirectional_lstm,
).to(device)
optimizer_nodp = torch.optim.SGD(model_nodp.parameters(), lr=0.5)
for epoch in tqdm(range(epochs)):
train(model_nodp, criterion, optimizer_nodp, train_loader, epoch, device=device)
if test_every:
if epoch % test_every == 0:
test(model_nodp, test_loader, None, device=device)
test(model_nodp, test_loader, None, device=device)
```
We run the training loop again, this time without privacy and for the same number of iterations.
The non-private classifier obtains a test accuracy of around 0.75 with the same parameters and number of epochs. We are effectively trading off performance on the name classification task for a lower loss of privacy.
|
github_jupyter
|
##### Copyright 2019 The TensorFlow Authors.
Licensed under the Apache License, Version 2.0 (the "License");
```
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# TFP Probabilistic Layers: Variational Auto Encoder
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/Probabilistic_Layers_VAE.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/probability/blob/master/tensorflow_probability/examples/jupyter_notebooks/Probabilistic_Layers_VAE.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
</table>
In this example we show how to fit a Variational Autoencoder using TFP's "probabilistic layers."
### Dependencies & Prerequisites
```
#@title Install { display-mode: "form" }
TF_Installation = 'TF2 Nightly (GPU)' #@param ['TF2 Nightly (GPU)', 'TF2 Stable (GPU)', 'TF1 Nightly (GPU)', 'TF1 Stable (GPU)','System']
if TF_Installation == 'TF2 Nightly (GPU)':
!pip install -q --upgrade tf-nightly-gpu-2.0-preview
print('Installation of `tf-nightly-gpu-2.0-preview` complete.')
elif TF_Installation == 'TF2 Stable (GPU)':
!pip install -q --upgrade tensorflow-gpu==2.0.0-alpha0
print('Installation of `tensorflow-gpu==2.0.0-alpha0` complete.')
elif TF_Installation == 'TF1 Nightly (GPU)':
!pip install -q --upgrade tf-nightly-gpu
print('Installation of `tf-nightly-gpu` complete.')
elif TF_Installation == 'TF1 Stable (GPU)':
!pip install -q --upgrade tensorflow-gpu
print('Installation of `tensorflow-gpu` complete.')
elif TF_Installation == 'System':
pass
else:
raise ValueError('Selection Error: Please select a valid '
'installation option.')
#@title Install { display-mode: "form" }
TFP_Installation = "Nightly" #@param ["Nightly", "Stable", "System"]
if TFP_Installation == "Nightly":
!pip install -q tfp-nightly
print("Installation of `tfp-nightly` complete.")
elif TFP_Installation == "Stable":
!pip install -q --upgrade tensorflow-probability
print("Installation of `tensorflow-probability` complete.")
elif TFP_Installation == "System":
pass
else:
raise ValueError("Selection Error: Please select a valid "
"installation option.")
#@title Import { display-mode: "form" }
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import numpy as np
import tensorflow as tf
from tensorflow.python import tf2
if not tf2.enabled():
import tensorflow.compat.v2 as tf
tf.enable_v2_behavior()
assert tf2.enabled()
import tensorflow_datasets as tfds
import tensorflow_probability as tfp
tfk = tf.keras
tfkl = tf.keras.layers
tfpl = tfp.layers
tfd = tfp.distributions
```
### Make things Fast!
Before we dive in, let's make sure we're using a GPU for this demo.
To do this, select "Runtime" -> "Change runtime type" -> "Hardware accelerator" -> "GPU".
The following snippet will verify that we have access to a GPU.
```
if tf.test.gpu_device_name() != '/device:GPU:0':
print('WARNING: GPU device not found.')
else:
print('SUCCESS: Found GPU: {}'.format(tf.test.gpu_device_name()))
```
Note: if for some reason you cannot access a GPU, this colab will still work. (Training will just take longer.)
### Load Dataset
```
datasets, datasets_info = tfds.load(name='mnist',
with_info=True,
as_supervised=False)
def _preprocess(sample):
image = tf.cast(sample['image'], tf.float32) / 255. # Scale to unit interval.
image = image < tf.random.uniform(tf.shape(image)) # Randomly binarize.
return image, image
train_dataset = (datasets['train']
.map(_preprocess)
.batch(256)
.prefetch(tf.data.experimental.AUTOTUNE)
.shuffle(int(10e3)))
eval_dataset = (datasets['test']
.map(_preprocess)
.batch(256)
.prefetch(tf.data.experimental.AUTOTUNE))
```
### VAE Code Golf
#### Specify model.
```
input_shape = datasets_info.features['image'].shape
encoded_size = 16
base_depth = 32
prior = tfd.Independent(tfd.Normal(loc=tf.zeros(encoded_size), scale=1),
reinterpreted_batch_ndims=1)
encoder = tfk.Sequential([
tfkl.InputLayer(input_shape=input_shape),
tfkl.Lambda(lambda x: tf.cast(x, tf.float32) - 0.5),
tfkl.Conv2D(base_depth, 5, strides=1,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2D(base_depth, 5, strides=2,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2D(2 * base_depth, 5, strides=1,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2D(2 * base_depth, 5, strides=2,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2D(4 * encoded_size, 7, strides=1,
padding='valid', activation=tf.nn.leaky_relu),
tfkl.Flatten(),
tfkl.Dense(tfpl.MultivariateNormalTriL.params_size(encoded_size),
activation=None),
tfpl.MultivariateNormalTriL(
encoded_size,
activity_regularizer=tfpl.KLDivergenceRegularizer(prior)),
])
decoder = tfk.Sequential([
tfkl.InputLayer(input_shape=[encoded_size]),
tfkl.Reshape([1, 1, encoded_size]),
tfkl.Conv2DTranspose(2 * base_depth, 7, strides=1,
padding='valid', activation=tf.nn.leaky_relu),
tfkl.Conv2DTranspose(2 * base_depth, 5, strides=1,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2DTranspose(2 * base_depth, 5, strides=2,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2DTranspose(base_depth, 5, strides=1,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2DTranspose(base_depth, 5, strides=2,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2DTranspose(base_depth, 5, strides=1,
padding='same', activation=tf.nn.leaky_relu),
tfkl.Conv2D(filters=1, kernel_size=5, strides=1,
padding='same', activation=None),
tfkl.Flatten(),
tfpl.IndependentBernoulli(input_shape, tfd.Bernoulli.logits),
])
vae = tfk.Model(inputs=encoder.inputs,
outputs=decoder(encoder.outputs[0]))
```
#### Do inference.
```
negloglik = lambda x, rv_x: -rv_x.log_prob(x)
vae.compile(optimizer=tf.optimizers.Adam(learning_rate=1e-3),
loss=negloglik)
vae.fit(train_dataset,
epochs=15,
validation_data=eval_dataset)
```
### Look Ma, No ~~Hands~~Tensors!
```
# We'll just examine ten random digits.
x = next(iter(eval_dataset))[0][:10]
xhat = vae(x)
assert isinstance(xhat, tfd.Distribution)
#@title Image Plot Util
import matplotlib.pyplot as plt
def display_imgs(x, y=None):
if not isinstance(x, (np.ndarray, np.generic)):
x = np.array(x)
plt.ioff()
n = x.shape[0]
fig, axs = plt.subplots(1, n, figsize=(n, 1))
if y is not None:
fig.suptitle(np.argmax(y, axis=1))
for i in xrange(n):
axs.flat[i].imshow(x[i].squeeze(), interpolation='none', cmap='gray')
axs.flat[i].axis('off')
plt.show()
plt.close()
plt.ion()
print('Originals:')
display_imgs(x)
print('Decoded Random Samples:')
display_imgs(xhat.sample())
print('Decoded Modes:')
display_imgs(xhat.mode())
print('Decoded Means:')
display_imgs(xhat.mean())
# Now, let's generate ten never-before-seen digits.
z = prior.sample(10)
xtilde = decoder(z)
assert isinstance(xtilde, tfd.Distribution)
print('Randomly Generated Samples:')
display_imgs(xtilde.sample())
print('Randomly Generated Modes:')
display_imgs(xtilde.mode())
print('Randomly Generated Means:')
display_imgs(xtilde.mean())
```
|
github_jupyter
|
# MNIST distributed training and batch transform
The SageMaker Python SDK helps you deploy your models for training and hosting in optimized, production-ready containers in SageMaker. The SageMaker Python SDK is easy to use, modular, extensible and compatible with TensorFlow and MXNet. This tutorial focuses on how to create a convolutional neural network model to train the [MNIST dataset](http://yann.lecun.com/exdb/mnist/) using TensorFlow distributed training.
## Set up the environment
First, we'll just set up a few things needed for this example
```
import sagemaker
from sagemaker import get_execution_role
from sagemaker.session import Session
sagemaker_session = sagemaker.Session()
region = sagemaker_session.boto_session.region_name
role = get_execution_role()
```
### Download the MNIST dataset
We'll now need to download the MNIST dataset, and upload it to a location in S3 after preparing for training.
```
import utils
from tensorflow.contrib.learn.python.learn.datasets import mnist
import tensorflow as tf
data_sets = mnist.read_data_sets('data', dtype=tf.uint8, reshape=False, validation_size=5000)
utils.convert_to(data_sets.train, 'train', 'data')
utils.convert_to(data_sets.validation, 'validation', 'data')
utils.convert_to(data_sets.test, 'test', 'data')
```
### Upload the data
We use the ```sagemaker.Session.upload_data``` function to upload our datasets to an S3 location. The return value inputs identifies the location -- we will use this later when we start the training job.
```
inputs = sagemaker_session.upload_data(path='data', key_prefix='data/DEMO-mnist')
```
# Construct a script for distributed training
Here is the full code for the network model:
```
!cat 'mnist.py'
```
## Create a training job
```
from sagemaker.tensorflow import TensorFlow
mnist_estimator = TensorFlow(entry_point='mnist.py',
role=role,
framework_version='1.11.0',
training_steps=1000,
evaluation_steps=100,
train_instance_count=2,
train_instance_type='ml.c4.xlarge')
mnist_estimator.fit(inputs)
```
The `fit()` method will create a training job in two ml.c4.xlarge instances. The logs above will show the instances doing training, evaluation, and incrementing the number of training steps.
In the end of the training, the training job will generate a saved model for TF serving.
## SageMaker's transformer class
After training, we use our TensorFlow estimator object to create a `Transformer` by invoking the `transformer()` method. This method takes arguments for configuring our options with the batch transform job; these do not need to be the same values as the one we used for the training job. The method also creates a SageMaker Model to be used for the batch transform jobs.
The `Transformer` class is responsible for running batch transform jobs, which will deploy the trained model to an endpoint and send requests for performing inference.
```
transformer = mnist_estimator.transformer(instance_count=1, instance_type='ml.m4.xlarge')
```
# Perform inference
Now that we've trained a model, we're going to use it to perform inference with a SageMaker batch transform job. The request handling behavior of the Endpoint deployed during the transform job is determined by the `mnist.py` script we looked at earlier.
## Run a batch transform job
For our batch transform job, we're going to use input data that contains 1000 MNIST images, located in the public SageMaker sample data S3 bucket. To create the batch transform job, we simply call `transform()` on our transformer with information about the input data.
```
input_bucket_name = 'sagemaker-sample-data-{}'.format(region)
input_file_path = 'batch-transform/mnist-1000-samples'
transformer.transform('s3://{}/{}'.format(input_bucket_name, input_file_path), content_type='text/csv')
```
Now we wait for the batch transform job to complete. We have a convenience method, `wait()`, that will block until the batch transform job has completed. We can call that here to see if the batch transform job is still running; the cell will finish running when the batch transform job has completed.
```
transformer.wait()
```
## Download the results
The batch transform job uploads its predictions to S3. Since we did not specify `output_path` when creating the Transformer, one was generated based on the batch transform job name:
```
print(transformer.output_path)
```
Now let's download the first ten results from S3:
```
import json
from six.moves.urllib import parse
import boto3
parsed_url = parse.urlparse(transformer.output_path)
bucket_name = parsed_url.netloc
prefix = parsed_url.path[1:]
s3 = boto3.resource('s3')
predictions = []
for i in range(10):
file_key = '{}/data-{}.csv.out'.format(prefix, i)
output_obj = s3.Object(bucket_name, file_key)
output = output_obj.get()["Body"].read().decode('utf-8')
predictions.extend(json.loads(output)['outputs']['classes']['int64Val'])
```
For demonstration purposes, we're also going to download the corresponding original input data so that we can see how the model did with its predictions.
```
import os
import matplotlib.pyplot as plt
from numpy import genfromtxt
plt.rcParams['figure.figsize'] = (2,10)
def show_digit(img, caption='', subplot=None):
if subplot == None:
_,(subplot) = plt.subplots(1,1)
imgr = img.reshape((28,28))
subplot.axis('off')
subplot.imshow(imgr, cmap='gray')
plt.title(caption)
tmp_dir = '/tmp/data'
if not os.path.exists(tmp_dir):
os.makedirs(tmp_dir)
for i in range(10):
input_file_name = 'data-{}.csv'.format(i)
input_file_key = '{}/{}'.format(input_file_path, input_file_name)
s3.Bucket(input_bucket_name).download_file(input_file_key, os.path.join(tmp_dir, input_file_name))
input_data = genfromtxt(os.path.join(tmp_dir, input_file_name), delimiter=',')
show_digit(input_data)
```
Here, we can see the original labels are:
```
7, 2, 1, 0, 4, 1, 4, 9, 5, 9
```
Now let's print out the predictions to compare:
```
print(', '.join(predictions))
```
|
github_jupyter
|
##### Copyright 2018 The TensorFlow Authors.
```
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
```
# Customization basics: tensors and operations
<table class="tfo-notebook-buttons" align="left">
<td>
<a target="_blank" href="https://www.tensorflow.org/tutorials/customization/basics"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
</td>
<td>
<a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/tutorials/customization/basics.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
</td>
<td>
<a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/tutorials/customization/basics.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
</td>
<td>
<a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/tutorials/customization/basics.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
</td>
</table>
This is an introductory TensorFlow tutorial that shows how to:
* Import the required package
* Create and use tensors
* Use GPU acceleration
* Demonstrate `tf.data.Dataset`
## Import TensorFlow
To get started, import the `tensorflow` module. As of TensorFlow 2, eager execution is turned on by default. This enables a more interactive frontend to TensorFlow, the details of which we will discuss much later.
```
import tensorflow as tf
```
## Tensors
A Tensor is a multi-dimensional array. Similar to NumPy `ndarray` objects, `tf.Tensor` objects have a data type and a shape. Additionally, `tf.Tensor`s can reside in accelerator memory (like a GPU). TensorFlow offers a rich library of operations ([tf.add](https://www.tensorflow.org/api_docs/python/tf/add), [tf.matmul](https://www.tensorflow.org/api_docs/python/tf/matmul), [tf.linalg.inv](https://www.tensorflow.org/api_docs/python/tf/linalg/inv) etc.) that consume and produce `tf.Tensor`s. These operations automatically convert native Python types, for example:
```
print(tf.add(1, 2))
print(tf.add([1, 2], [3, 4]))
print(tf.square(5))
print(tf.reduce_sum([1, 2, 3]))
# Operator overloading is also supported
print(tf.square(2) + tf.square(3))
```
Each `tf.Tensor` has a shape and a datatype:
```
x = tf.matmul([[1]], [[2, 3]])
print(x)
print(x.shape)
print(x.dtype)
```
The most obvious differences between NumPy arrays and `tf.Tensor`s are:
1. Tensors can be backed by accelerator memory (like GPU, TPU).
2. Tensors are immutable.
### NumPy Compatibility
Converting between a TensorFlow `tf.Tensor`s and a NumPy `ndarray` is easy:
* TensorFlow operations automatically convert NumPy ndarrays to Tensors.
* NumPy operations automatically convert Tensors to NumPy ndarrays.
Tensors are explicitly converted to NumPy ndarrays using their `.numpy()` method. These conversions are typically cheap since the array and `tf.Tensor` share the underlying memory representation, if possible. However, sharing the underlying representation isn't always possible since the `tf.Tensor` may be hosted in GPU memory while NumPy arrays are always backed by host memory, and the conversion involves a copy from GPU to host memory.
```
import numpy as np
ndarray = np.ones([3, 3])
print("TensorFlow operations convert numpy arrays to Tensors automatically")
tensor = tf.multiply(ndarray, 42)
print(tensor)
print("And NumPy operations convert Tensors to numpy arrays automatically")
print(np.add(tensor, 1))
print("The .numpy() method explicitly converts a Tensor to a numpy array")
print(tensor.numpy())
```
## GPU acceleration
Many TensorFlow operations are accelerated using the GPU for computation. Without any annotations, TensorFlow automatically decides whether to use the GPU or CPU for an operation—copying the tensor between CPU and GPU memory, if necessary. Tensors produced by an operation are typically backed by the memory of the device on which the operation executed, for example:
```
x = tf.random.uniform([3, 3])
print("Is there a GPU available: "),
print(tf.config.experimental.list_physical_devices("GPU"))
print("Is the Tensor on GPU #0: "),
print(x.device.endswith('GPU:0'))
```
### Device Names
The `Tensor.device` property provides a fully qualified string name of the device hosting the contents of the tensor. This name encodes many details, such as an identifier of the network address of the host on which this program is executing and the device within that host. This is required for distributed execution of a TensorFlow program. The string ends with `GPU:<N>` if the tensor is placed on the `N`-th GPU on the host.
### Explicit Device Placement
In TensorFlow, *placement* refers to how individual operations are assigned (placed on) a device for execution. As mentioned, when there is no explicit guidance provided, TensorFlow automatically decides which device to execute an operation and copies tensors to that device, if needed. However, TensorFlow operations can be explicitly placed on specific devices using the `tf.device` context manager, for example:
```
import time
def time_matmul(x):
start = time.time()
for loop in range(10):
tf.matmul(x, x)
result = time.time()-start
print("10 loops: {:0.2f}ms".format(1000*result))
# Force execution on CPU
print("On CPU:")
with tf.device("CPU:0"):
x = tf.random.uniform([1000, 1000])
assert x.device.endswith("CPU:0")
time_matmul(x)
# Force execution on GPU #0 if available
if tf.config.experimental.list_physical_devices("GPU"):
print("On GPU:")
with tf.device("GPU:0"): # Or GPU:1 for the 2nd GPU, GPU:2 for the 3rd etc.
x = tf.random.uniform([1000, 1000])
assert x.device.endswith("GPU:0")
time_matmul(x)
```
## Datasets
This section uses the [`tf.data.Dataset` API](https://www.tensorflow.org/guide/datasets) to build a pipeline for feeding data to your model. The `tf.data.Dataset` API is used to build performant, complex input pipelines from simple, re-usable pieces that will feed your model's training or evaluation loops.
### Create a source `Dataset`
Create a *source* dataset using one of the factory functions like [`Dataset.from_tensors`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#from_tensors), [`Dataset.from_tensor_slices`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#from_tensor_slices), or using objects that read from files like [`TextLineDataset`](https://www.tensorflow.org/api_docs/python/tf/data/TextLineDataset) or [`TFRecordDataset`](https://www.tensorflow.org/api_docs/python/tf/data/TFRecordDataset). See the [TensorFlow Dataset guide](https://www.tensorflow.org/guide/datasets#reading_input_data) for more information.
```
ds_tensors = tf.data.Dataset.from_tensor_slices([1, 2, 3, 4, 5, 6])
# Create a CSV file
import tempfile
_, filename = tempfile.mkstemp()
with open(filename, 'w') as f:
f.write("""Line 1
Line 2
Line 3
""")
ds_file = tf.data.TextLineDataset(filename)
```
### Apply transformations
Use the transformations functions like [`map`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#map), [`batch`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#batch), and [`shuffle`](https://www.tensorflow.org/api_docs/python/tf/data/Dataset#shuffle) to apply transformations to dataset records.
```
ds_tensors = ds_tensors.map(tf.square).shuffle(2).batch(2)
ds_file = ds_file.batch(2)
```
### Iterate
`tf.data.Dataset` objects support iteration to loop over records:
```
print('Elements of ds_tensors:')
for x in ds_tensors:
print(x)
print('\nElements in ds_file:')
for x in ds_file:
print(x)
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import json
%matplotlib inline
```
### 1. Load the dataset into a data frame named loans
```
loans = pd.read_csv('../data/lending-club-data.csv')
loans.head(2)
# safe_loans = 1 => safe
# safe_loans = -1 => risky
loans['safe_loans'] = loans['bad_loans'].apply(lambda x : +1 if x==0 else -1)
#loans = loans.remove_column('bad_loans')
loans = loans.drop('bad_loans', axis=1)
features = ['grade', # grade of the loan
'term', # the term of the loan
'home_ownership', # home_ownership status: own, mortgage or rent
'emp_length', # number of years of employment
]
target = 'safe_loans'
loans = loans[features + [target]]
loans.iloc[122602]
```
## One-hot encoding
```
categorical_variables = []
for feat_name, feat_type in zip(loans.columns, loans.dtypes):
if feat_type == object:
categorical_variables.append(feat_name)
for feature in categorical_variables:
loans_one_hot_encoded = pd.get_dummies(loans[feature],prefix=feature)
#print loans_one_hot_encoded
loans = loans.drop(feature, axis=1)
for col in loans_one_hot_encoded.columns:
loans[col] = loans_one_hot_encoded[col]
print (loans.head(2) )
print (loans.columns)
with open('../data/module-5-assignment-2-train-idx.json') as train_data_file:
train_idx = json.load(train_data_file)
with open('../data/module-5-assignment-2-test-idx.json') as test_data_file:
test_idx = json.load(test_data_file)
print (train_idx[:3])
print (test_idx[:3])
print len(train_idx)
print len(test_idx)
train_data = loans.iloc[train_idx]
test_data = loans.iloc[test_idx]
print (train_data.shape)
print (test_data.shape)
```
## Decision tree implementation
## Function to count number of mistakes while predicting majority class
Recall from the lecture that prediction at an intermediate node works by predicting the majority class for all data points that belong to this node. Now, we will write a function that calculates the number of misclassified examples when predicting the majority class. This will be used to help determine which feature is the best to split on at a given node of the tree.
Note: Keep in mind that in order to compute the number of mistakes for a majority classifier, we only need the label (y values) of the data points in the node.
Steps to follow:
- Step 1: Calculate the number of safe loans and risky loans.
- Step 2: Since we are assuming majority class prediction, all the data points that are not in the majority class are considered mistakes.
- Step 3: Return the number of mistakes.
7. Now, let us write the function intermediate_node_num_mistakes which computes the number of misclassified examples of an intermediate node given the set of labels (y values) of the data points contained in the node. Your code should be analogous to
```
def intermediate_node_num_mistakes(labels_in_node):
# Corner case: If labels_in_node is empty, return 0
if len(labels_in_node) == 0:
return 0
safe_loan = (labels_in_node==1).sum()
risky_loan = (labels_in_node==-1).sum()
return min(safe_loan, risky_loan)
```
8. Because there are several steps in this assignment, we have introduced some stopping points where you can check your code and make sure it is correct before proceeding. To test your intermediate_node_num_mistakes function, run the following code until you get a Test passed!, then you should proceed. Otherwise, you should spend some time figuring out where things went wrong. Again, remember that this code is specific to SFrame, but using your software of choice, you can construct similar tests.
```
# Test case 1
example_labels = np.array([-1, -1, 1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print ('Test 1 passed!')
else:
print ('Test 1 failed... try again!')
# Test case 2
example_labels = np.array([-1, -1, 1, 1, 1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print ('Test 2 passed!')
else:
print ('Test 2 failed... try again!')
# Test case 3
example_labels = np.array([-1, -1, -1, -1, -1, 1, 1])
if intermediate_node_num_mistakes(example_labels) == 2:
print ('Test 3 passed!')
else:
print ('Test 3 failed... try again!')
```
## Function to pick best feature to split on
The function best_splitting_feature takes 3 arguments:
- The data
- The features to consider for splits (a list of strings of column names to consider for splits)
- The name of the target/label column (string)
The function will loop through the list of possible features, and consider splitting on each of them. It will calculate the classification error of each split and return the feature that had the smallest classification error when split on.
Recall that the classification error is defined as follows:
### 9. Follow these steps to implement best_splitting_feature:
- Step 1: Loop over each feature in the feature list
- Step 2: Within the loop, split the data into two groups: one group where all of the data has feature value 0 or False (we will call this the left split), and one group where all of the data has feature value 1 or True (we will call this the right split). Make sure the left split corresponds with 0 and the right split corresponds with 1 to ensure your implementation fits with our implementation of the tree building process.
- Step 3: Calculate the number of misclassified examples in both groups of data and use the above formula to compute theclassification error.
- Step 4: If the computed error is smaller than the best error found so far, store this feature and its error.
Note: Remember that since we are only dealing with binary features, we do not have to consider thresholds for real-valued features. This makes the implementation of this function much easier.
Your code should be analogous to
```
def best_splitting_feature(data, features, target):
target_values = data[target]
best_feature = None # Keep track of the best feature
best_error = 2 # Keep track of the best error so far
# Note: Since error is always <= 1, we should intialize it with something larger than 1.
# Convert to float to make sure error gets computed correctly.
num_data_points = float(len(data))
# Loop through each feature to consider splitting on that feature
for feature in features:
# The left split will have all data points where the feature value is 0
left_split = data[data[feature] == 0]
# The right split will have all data points where the feature value is 1
right_split = data[data[feature] == 1]
# Calculate the number of misclassified examples in the left split.
# Remember that we implemented a function for this! (It was called intermediate_node_num_mistakes)
left_mistakes = intermediate_node_num_mistakes(left_split[target])
# Calculate the number of misclassified examples in the right split.
right_mistakes = intermediate_node_num_mistakes(right_split[target])
# Compute the classification error of this split.
# Error = (# of mistakes (left) + # of mistakes (right)) / (# of data points)
error = (left_mistakes + right_mistakes) / num_data_points
# If this is the best error we have found so far, store the feature as best_feature and the error as best_error
if error < best_error:
best_feature = feature
best_error = error
return best_feature # Return the best feature we found
```
## Building the tree
With the above functions implemented correctly, we are now ready to build our decision tree. Each node in the decision tree is represented as a dictionary which contains the following keys and possible values:
### 10. First, we will write a function that creates a leaf node given a set of target values.
Your code should be analogous to
```
def create_leaf(target_values):
# Create a leaf node
leaf = {'splitting_feature' : None,
'left' : None,
'right' : None,
'is_leaf': True } ## YOUR CODE HERE
# Count the number of data points that are +1 and -1 in this node.
num_ones = len(target_values[target_values == +1])
num_minus_ones = len(target_values[target_values == -1])
# For the leaf node, set the prediction to be the majority class.
# Store the predicted class (1 or -1) in leaf['prediction']
if num_ones > num_minus_ones:
leaf['prediction'] = 1 ## YOUR CODE HERE
else:
leaf['prediction'] = -1 ## YOUR CODE HERE
# Return the leaf node
return leaf
```
11. Now, we will provide a Python skeleton of the learning algorithm. Note that this code is not complete; it needs to be completed by you if you are using Python. Otherwise, your code should be analogous to
1. Stopping condition 1: All data points in a node are from the same class.
1. Stopping condition 2: No more features to split on.
1. Additional stopping condition: In addition to the above two stopping conditions covered in lecture, in this assignment we will also consider a stopping condition based on the max_depth of the tree. By not letting the tree grow too deep, we will save computational effort in the learning process.
```
def decision_tree_create(data, features, target, current_depth = 0, max_depth = 10):
remaining_features = features[:] # Make a copy of the features.
target_values = data[target]
print ("--------------------------------------------------------------------")
print ("Subtree, depth = %s (%s data points)." % (current_depth, len(target_values)))
# Stopping condition 1
# (Check if there are mistakes at current node.
# Recall you wrote a function intermediate_node_num_mistakes to compute this.)
if intermediate_node_num_mistakes(target_values) == 0: ## YOUR CODE HERE
print ("No classification error in the node. Stopping for now." )
# If not mistakes at current node, make current node a leaf node
return create_leaf(target_values)
# Stopping condition 2 (check if there are remaining features to consider splitting on)
if remaining_features == []: ## YOUR CODE HERE
print ("No remaining features. Stopping for now.")
# If there are no remaining features to consider, make current node a leaf node
return create_leaf(target_values)
# Additional stopping condition (limit tree depth)
if current_depth >= max_depth: ## YOUR CODE HERE
print ("Reached maximum depth. Stopping for now.")
# If the max tree depth has been reached, make current node a leaf node
return create_leaf(target_values)
# Find the best splitting feature (recall the function best_splitting_feature implemented above)
## YOUR CODE HERE
splitting_feature = best_splitting_feature(data, remaining_features, target)
# Split on the best feature that we found.
left_split = data[data[splitting_feature] == 0]
right_split = data[data[splitting_feature] == 1] ## YOUR CODE HERE
remaining_features.remove(splitting_feature)
print ("Split on feature %s. (%s, %s)" % (\
splitting_feature, len(left_split), len(right_split)))
# Create a leaf node if the split is "perfect"
if len(left_split) == len(data):
print ("Creating leaf node.")
return create_leaf(left_split[target])
if len(right_split) == len(data):
print ("Creating leaf node.")
## YOUR CODE HERE
return create_leaf(right_split[target])
# Repeat (recurse) on left and right subtrees
left_tree = decision_tree_create(left_split, remaining_features, target, current_depth + 1, max_depth)
## YOUR CODE HERE
right_tree = decision_tree_create(right_split, remaining_features, target, current_depth + 1, max_depth)
return {'is_leaf' : False,
'prediction' : None,
'splitting_feature': splitting_feature,
'left' : left_tree,
'right' : right_tree}
```
12. Train a tree model on the train_data. Limit the depth to 6 (max_depth = 6) to make sure the algorithm doesn't run for too long. Call this tree my_decision_tree. Warning: The tree may take 1-2 minutes to learn.
```
input_features = train_data.columns
print (list(input_features))
feature_list = list(train_data.columns)
feature_list.remove('safe_loans')
my_decision_tree = decision_tree_create(train_data, feature_list, 'safe_loans', current_depth = 0, max_depth = 6)
```
#### Making predictions with a decision tree
13. As discussed in the lecture, we can make predictions from the decision tree with a simple recursive function. Write a function called classify, which takes in a learned tree and a test point x to classify. Include an option annotate that describes the prediction path when set to True. Your code should be analogous to
```
def classify(tree, x, annotate = False):
# if the node is a leaf node.
if tree['is_leaf']:
if annotate:
print ("At leaf, predicting %s" % tree['prediction'])
return tree['prediction']
else:
# split on feature.
split_feature_value = x[tree['splitting_feature']]
if annotate:
print ("Split on %s = %s" % (tree['splitting_feature'], split_feature_value))
if split_feature_value == 0:
return classify(tree['left'], x, annotate)
else:
return classify(tree['right'], x, annotate)
```
### 14. Now, let's consider the first example of the test set and see what my_decision_tree model predicts for this data point.
```
print (test_data.iloc[0])
print ('Predicted class: %s ' % classify(my_decision_tree, test_data.iloc[0]))
```
### 15. Let's add some annotations to our prediction to see what the prediction path was that lead to this predicted class:
```
classify(my_decision_tree, test_data.iloc[0], annotate=True)
```
## Quiz question:
What was the feature that my_decision_tree first split on while making the prediction for test_data[0]?
## Quiz question:
What was the first feature that lead to a right split of test_data[0]?
## Quiz question:
What was the last feature split on before reaching a leaf node for test_data[0]?
## Answer:
term_36 months
## Answer:
grade_D
## Answer:
grade_D
## Evaluating your decision tree
### 16. Now, we will write a function to evaluate a decision tree by computing the classification error of the tree on the given dataset. Write a function called evaluate_classification_error that takes in as input:
- tree (as described above)
- data (a data frame of data points)
This function should return a prediction (class label) for each row in data using the decision tree. Your code should be analogous to
```
def evaluate_classification_error(tree, data):
# Apply the classify(tree, x) to each row in your data
prediction = data.apply(lambda x: classify(tree, x), axis=1)
# Once you've made the predictions, calculate the classification error and return it
## YOUR CODE HERE
return (data['safe_loans'] != np.array(prediction)).values.sum() *1. / len(data)
```
### 17. Now, use this function to evaluate the classification error on the test set.
```
evaluate_classification_error(my_decision_tree, test_data)
```
## Quiz Question:
Rounded to 2nd decimal point, what is the classification error of my_decision_tree on the test_data?
## Answer:
0.38
## Printing out a decision stump
### 18. As discussed in the lecture, we can print out a single decision stump (printing out the entire tree is left as an exercise to the curious reader). Here we provide Python code to visualize a decision stump. If you are using different software, make sure your code is analogous to:
```
def print_stump(tree, name = 'root'):
split_name = tree['splitting_feature'] # split_name is something like 'term. 36 months'
if split_name is None:
print ("(leaf, label: %s)" % tree['prediction'])
return None
split_feature, split_value = split_name.split('_',1)
print (' %s' % name)
print( ' |---------------|----------------|')
print (' | |')
print (' | |')
print (' | |')
print (' [{0} == 0] [{0} == 1] '.format(split_name))
print (' | |')
print (' | |')
print (' | |')
print (' (%s) (%s)' \
% (('leaf, label: ' + str(tree['left']['prediction']) if tree['left']['is_leaf'] else 'subtree'),
('leaf, label: ' + str(tree['right']['prediction']) if tree['right']['is_leaf'] else 'subtree')))
```
### 19. Using this function, we can print out the root of our decision tree:
```
print_stump(my_decision_tree)
```
## Quiz Question:
What is the feature that is used for the split at the root node?
## Answer:
term_ 36 months
## Exploring the intermediate left subtree
The tree is a recursive dictionary, so we do have access to all the nodes! We can use
- my_decision_tree['left'] to go left
- my_decision_tree['right'] to go right
### 20. We can print out the left subtree by running the code
```
print_stump(my_decision_tree['left'], my_decision_tree['splitting_feature'])
print_stump(my_decision_tree['left']['left'], my_decision_tree['left']['splitting_feature'])
print_stump(my_decision_tree['right'], my_decision_tree['splitting_feature'])
print_stump(my_decision_tree['right']['right'], my_decision_tree['right']['splitting_feature'])
```
## Quiz question:
What is the path of the first 3 feature splits considered along the left-most branch of my_decision_tree?
## Quiz question:
What is the path of the first 3 feature splits considered along the right-most branch of my_decision_tree?
## Answer
- term_ 36 months
- grade_A
- grade_B
## Answer
- term_ 36 months
- grade_D
- leaf
|
github_jupyter
|
# Find the comparables: extra_features.txt
The file `extra_features.txt` contains important property information like number and quality of pools, detached garages, outbuildings, canopies, and more. Let's load this file and grab a subset with the important columns to continue our study.
```
%load_ext autoreload
%autoreload 2
from pathlib import Path
import pickle
import pandas as pd
from src.definitions import ROOT_DIR
from src.data.utils import Table, save_pickle
extra_features_fn = ROOT_DIR / 'data/external/2016/Real_building_land/extra_features.txt'
assert extra_features_fn.exists()
extra_features = Table(extra_features_fn, '2016')
extra_features.get_header()
```
# Load accounts of interest
Let's remove the account numbers that don't meet free-standing single-family home criteria that we found while processing the `building_res.txt` file.
```
skiprows = extra_features.get_skiprows()
extra_features_df = extra_features.get_df(skiprows=skiprows)
extra_features_df.head()
extra_features_df.dscr.value_counts()
```
# Grab slice of the extra features of interest
With the value counts on the extra feature description performed above we can see that the majority of the features land in the top 6 categories. Let's filter out the rests of the columns.
```
cols = extra_features_df.dscr.value_counts().head(6).index
cond0 = extra_features_df['dscr'].isin(cols)
extra_features_df = extra_features_df.loc[cond0, :]
```
# Build pivot tables for count and grade
There appear to be two important values related to each extra feature:count and grade. Let's build individual pivot tables for each and merge them before saving them out.
```
extra_features_pivot_count = extra_features_df.pivot_table(index='acct',
columns='dscr',
values='count',
fill_value=0)
extra_features_pivot_count.head()
extra_features_pivot_grade = extra_features_df.pivot_table(index='acct',
columns='dscr',
values='grade')
extra_features_pivot_grade.head()
extra_features_count_grade = extra_features_pivot_count.merge(extra_features_pivot_grade,
how='left',
left_index=True,
right_index=True,
suffixes=('_count', '_grade'),
validate='one_to_one')
extra_features_count_grade.head()
assert extra_features_count_grade.index.is_unique
```
add `acct` column to make easier the merging process ahead
```
extra_features_count_grade.reset_index(inplace=True)
```
# Export real_acct
```
save_fn = ROOT_DIR / 'data/raw/2016/extra_features_count_grade_comps.pickle'
save_pickle(extra_features_count_grade, save_fn)
```
|
github_jupyter
|
# Python good practices
## Environment setup
```
!pip install papermill
import platform
print(f"Python version: {platform.python_version()}")
assert platform.python_version_tuple() >= ("3", "6")
import os
import papermill as pm
from IPython.display import YouTubeVideo
```
## Writing pythonic code
```
import this
```
### What does "Pythonic" mean?
- Python code is considered _pythonic_ if it:
- conforms to the Python philosophy;
- takes advantage of the language's specific features.
- Pythonic code is nothing more than **idiomatic Python code** that strives to be clean, concise and readable.
### Example: swapping two variables
```
a = 3
b = 2
# Non-pythonic
tmp = a
a = b
b = tmp
# Pythonic
a, b = b, a
```
### Example: iterating on a list
```
my_list = ["a", "b", "c"]
def do_something(item):
# print(item)
pass
# Non-pythonic
i = 0
while i < len(my_list):
do_something(my_list[i])
i += 1
# Still non-pythonic
for i in range(len(my_list)):
do_something(my_list[i])
# Pythonic
for item in my_list:
do_something(item)
```
### Example: indexed traversal
```
my_list = ["a", "b", "c"]
# Non-pythonic
for i in range(len(my_list)):
print(i, "->", my_list[i])
# Pythonic
for i, item in enumerate(my_list):
print(i, "->", item)
```
### Example: searching in a list
```
fruits = ["apples", "oranges", "bananas", "grapes"]
fruit = "cherries"
# Non-pythonic
found = False
size = len(fruits)
for i in range(0, size):
if fruits[i] == fruit:
found = True
# Pythonic
found = fruit in fruits
```
### Example: generating a list
This feature is called [list comprehension](https://docs.python.org/3/tutorial/datastructures.html#list-comprehensions).
```
numbers = [1, 2, 3, 4, 5, 6]
# Non-pythonic
doubles = []
for i in range(len(numbers)):
if numbers[i] % 2 == 0:
doubles.append(numbers[i] * 2)
else:
doubles.append(numbers[i])
# Pythonic
doubles = [x * 2 if x % 2 == 0 else x for x in numbers]
```
### Code style
- [PEP8](https://www.python.org/dev/peps/pep-0008/) is the official style guide for Python:
- use 4 spaces for indentation;
- define a maximum value for line length (around 80 characters);
- organize imports at beginning of file;
- surround binary operators with a single space on each side;
- ...
- Code style should be enforced upon creation by a tool like [black](https://github.com/psf/black).
### Beyond PEP8
Focusing on style and PEP8-compliance might make you miss more fundamental code imperfections.
```
YouTubeVideo("wf-BqAjZb8M")
```
### Docstrings
A [docstring](https://www.python.org/dev/peps/pep-0257/) is a string literal that occurs as the first statement in a module, function, class, or method definition to document it.
All modules, classes, public methods and exported functions should include a docstring.
```
def complex(real=0.0, imag=0.0):
"""Form a complex number.
Keyword arguments:
real -- the real part (default 0.0)
imag -- the imaginary part (default 0.0)
"""
if imag == 0.0 and real == 0.0:
return complex_zero
```
### Code linting
- _Linting_ is the process of checking code for syntactical and stylistic problems before execution.
- It is useful to catch errors and improve code quality in dynamically typed, interpreted languages, where there is no compiler.
- Several linters exist in the Python ecosystem. The most commonly used is [pylint](https://pylint.org/).
### Type annotations
- Added in Python 3.5, [type annotations](https://www.python.org/dev/peps/pep-0484/) allow to add type hints to code entities like variables or functions, bringing a statically typed flavour to the language.
- [mypy](http://mypy-lang.org/) can automatically check the code for annotation correctness.
```
def greeting(name: str) -> str:
return "Hello " + name
# greeting('Alice') # OK
# greeting(3) # mypy error: incompatible type "int"; expected "str"
```
### Unit tests
Unit tests automate the testing of individual code elements like functions or methods, thus decreasing the risk of bugs and regressions.
They can be implemented in Python using tools like [unittest](https://docs.python.org/3/library/unittest.html) or [pytest](https://docs.pytest.org).
```
def inc(x):
return x + 1
def test_answer():
assert inc(3) == 5 # AssertionError: assert 4 == 5
```
## Packaging and dependency management
### Managing dependencies in Python
- Most Python apps depend on third-party libraries and frameworks (NumPy, Flask, Requests...).
- These tools may also have external dependencies, and so on.
- **Dependency management** is necessary to prevent version conflicts and incompatibilities. it involves two things:
- a way for the app to declare its dependencies;
- a tool to resolve these dependencies and install compatible versions.
### Semantic versioning
- Software versioning convention used in many ecosystems.
- A version number comes as a suite of three digits `X.Y.Z`.
- X = major version (potentially including breaking changes).
- Y = minor version (only non-breaking changes).
- Z = patch.
- Digits are incremented as new versions are shipped.
### pip and requirements.txt
A `requirements.txt` file is the most basic way of declaring dependencies in Python.
```text
certifi>=2020.11.0
chardet==4.0.0
click>=6.5.0, <7.1
download==0.3.5
Flask>=1.1.0
```
The [pip](https://pypi.org/project/pip/) package installer can read this file and act accordingly, downloading dependencies from [PyPI](https://pypi.org/).
```bash
pip install -r requirements.txt
```
### Virtual environments
- A **virtual environment** is an isolated Python environment where a project's dependencies are installed.
- Using them prevents the risk of mixing dependencies required by different projects on the same machine.
- Several tools exist to manage virtual environments in Python, for example [virtualenv](https://virtualenv.pypa.io) and [conda](https://docs.conda.io).
### conda and environment.yml
Installed as part of the [Anaconda](https://www.anaconda.com/) distribution, the [conda](https://docs.conda.io) package manager reads an `environment.yml` file to install the dependencies associated to a specific virtual environment.
```yaml
name: example-env
channels:
- conda-forge
- defaults
dependencies:
- python=3.7
- matplotlib
- numpy
```
### Poetry
[Poetry](https://python-poetry.org) is a recent packaging and dependency management tool for Python. It downloads packages from [PyPI](https://pypi.org/) by default.
```bash
# Create a new poetry-compliant project
poetry new <project name>
# Initialize an already existing project for Poetry
poetry init
# Install defined dependencies
poetry install
# Add a package to project dependencies and install it
poetry add <package name>
# Update dependencies to sync them with configuration file
poetry update
```
### Poetry and virtual environments
By default, Poetry creates a virtual environment for the configured project in a user-specific folder. A standard practice is to store it in the project's folder.
```bash
# Tell Poetry to store the environment in the local project folder
poetry config virtualenvs.in-project true
# Activate the environment
poetry shell
```
### The pyproject.toml file
Poetry configuration file, soon-to-be standard for Python projects.
```toml
[tool.poetry]
name = "poetry example"
version = "0.1.0"
description = ""
[tool.poetry.dependencies]
python = ">=3.7.1,<3.10"
jupyter = "^1.0.0"
matplotlib = "^3.3.2"
sklearn = "^0.0"
pandas = "^1.1.3"
ipython = "^7.0.0"
[tool.poetry.dev-dependencies]
pytest = "^6.1.1"
```
### Caret requirements
Offers a way to precisely define dependency versions.
| Requirement | Versions allowed |
| :---------: | :--------------: |
| ^1.2.3 | >=1.2.3 <2.0.0 |
| ^1.2 | >=1.2.0 <2.0.0 |
| ~1.2.3 | >=1.2.3 <1.3.0 |
| ~1.2 | >=1.2.0 <1.3.0 |
| 1.2.3 | 1.2.3 only |
### The poetry.lock file
- The first time Poetry install dependencies, it creates a `poetry.lock` file that contains the exact versions of all installed packages.
- Subsequent installs will use these exact versions to ensure consistency.
- Removing this file and running another Poetry install will fetch the latest matching versions.
## Working with notebooks
### Advantages of Jupyter notebooks
- Standard format for mixing text, images and (executable) code.
- Open source and platform-independant.
- Useful for experimenting and prototyping.
- Growing ecosystem of [extensions](https://tljh.jupyter.org/en/latest/howto/admin/enable-extensions.html) for various purposes and cloud hosting solutions ([Colaboratory](https://colab.research.google.com/), [AI notebooks](https://www.ovhcloud.com/en/public-cloud/ai-notebook/)...).
- Integration with tools like [Visual Studio Code](https://code.visualstudio.com/docs/datascience/jupyter-notebooks).
### Drawbacks of Jupyter notebooks
- Arbitrary execution order of cells can cause confusing errors.
- Notebooks don't encourage good programming habits like modularization, linting and tests.
- Being JSON-based, their versioning is more difficult than for plain text files.
- Dependency management is also difficult, thus hindering reproducibility.
### Collaborating with notebooks
A common solution for sharing notebooks between a team is to use [Jupytext](https://jupytext.readthedocs.io). This tool can associate an `.ipynb` file with a Python file to facilitate collaboration and version control.
[](https://jupytext.readthedocs.io/en/latest/examples.html)
### Code organization
Monolithic notebooks can grow over time and become hard to understand and maintain.
Just like in a traditional software project, it is possible to split them into separate parts, thus following the [separation of concerns](https://en.wikipedia.org/wiki/Separation_of_concerns) design principle.
Code can be splitted into several sub-notebooks and/or external Python files. The latter facilitates unit testing and version control.
### Notebook workflow
Tools like [papermill](https://papermill.readthedocs.io) can orchestrate the execution of several notebooks in a row. External parameters can be passed to notebooks, and the runtime flow can depend on the execution results of each notebook.
```
# Doesn't work on Google Colaboratory. Workaround here:
# https://colab.research.google.com/github/rjdoubleu/Colab-Papermill-Patch/blob/master/Colab-Papermill-Driver.ipynb
notebook_dir = "./papermill"
result = pm.execute_notebook(
os.path.join(notebook_dir, "simple_input.ipynb"),
os.path.join(notebook_dir, "simple_output.ipynb"),
parameters={"msg": "Hello"},
)
```
|
github_jupyter
|
# Module
```
import numpy as np
import pandas as pd
import warnings
import gc
from tqdm import tqdm_notebook as tqdm
import lightgbm as lgb
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
from datetime import datetime
from sklearn.metrics import roc_auc_score
warnings.filterwarnings("ignore")
gc.enable()
pd.set_option('max_rows', 500)
pd.set_option('max_colwidth', 500)
pd.set_option('max_columns', 500)
```
# Load Data
```
train_raw = pd.read_csv('./data/train.csv')
test_raw = pd.read_csv('./data/test.csv')
train_raw.shape, test_raw.shape
del train, test, clf, data
gc.collect()
train = train_raw.copy()
test = test_raw.copy()
col_list = train.columns[2:]
train_0 = train[train.target == 0]
train_1 = train[train.target == 1]
pb_idx = np.load('./data_temp/public_LB.npy')
pv_idx = np.load('./data_temp/private_LB.npy')
test_pb = test.iloc[pb_idx].sort_index().copy()
test_pv = test.iloc[pv_idx].sort_index().copy()
test_real = test_pb.append(test_pv)
data = train.append(test_real)[['ID_code', 'target'] + col_list.tolist()]
```
# Extract Unique Value in All Data
## filter
```
# unique_df = data[['ID_code']]
con_df = data[['ID_code']]
con1_df = data[['ID_code']]
con2_df = data[['ID_code']]
con3_df = data[['ID_code']]
con4_df = data[['ID_code']]
con5_df = data[['ID_code']]
con6_df = data[['ID_code']]
con7_df = data[['ID_code']]
con8_df = data[['ID_code']]
for col in tqdm(col_list):
# unique_df[col] = data[col].map(((data[col].value_counts() == 1) * 1).to_dict())
con_df[col] = data[col].map((~(data[col].value_counts() == 1) * 1).to_dict())
con1_df[col] = data[col].map(((data[col].value_counts() == 1) * 1).to_dict())
con2_df[col] = data[col].map(((data[col].value_counts() == 2) * 1).to_dict())
con3_df[col] = data[col].map(((data[col].value_counts() == 3) * 1).to_dict())
con4_df[col] = data[col].map(((data[col].value_counts() == 4) * 1).to_dict())
con5_df[col] = data[col].map(((data[col].value_counts() == 5) * 1).to_dict())
con6_df[col] = data[col].map(((data[col].value_counts() == 6) * 1).to_dict())
con7_df[col] = data[col].map(((data[col].value_counts() == 7) * 1).to_dict())
con8_df[col] = data[col].map(((data[col].value_counts() == 8) * 1).to_dict())
order_df = data[['ID_code']]
for col in tqdm(col_list):
temp = data[col].value_counts().sort_index().to_frame()
order = [0]
for v in temp.iterrows():
order.append(order[-1] + v[1].values[0])
temp[col] = order[:-1]
temp = temp.to_dict()[col]
order_df[col] = data[col].map(temp)
```
## make data
```
for col in tqdm(col_list):
# data[col + '_unique'] = data[col] * unique_df[col]
data[col + '_con'] = data[col] * con_df[col]
data[col + '_con1'] = data[col] * con1_df[col]
data[col + '_con2'] = data[col] * con2_df[col]
data[col + '_con3'] = data[col] * con3_df[col]
data[col + '_con4'] = data[col] * con4_df[col]
data[col + '_con5'] = data[col] * con5_df[col]
data[col + '_con6'] = data[col] * con6_df[col]
data[col + '_con7'] = data[col] * con7_df[col]
data[col + '_con8'] = data[col] * con8_df[col]
for col in tqdm(col_list):
# data.loc[data[col + '_unique']==0, col + '_unique'] = np.nan
data.loc[data[col + '_con']==0, col + '_con'] = np.nan
data.loc[data[col + '_con1']==0, col + '_con1'] = np.nan
data.loc[data[col + '_con2']==0, col + '_con2'] = np.nan
data.loc[data[col + '_con3']==0, col + '_con3'] = np.nan
data.loc[data[col + '_con4']==0, col + '_con4'] = np.nan
data.loc[data[col + '_con5']==0, col + '_con5'] = np.nan
data.loc[data[col + '_con6']==0, col + '_con6'] = np.nan
data.loc[data[col + '_con7']==0, col + '_con7'] = np.nan
data.loc[data[col + '_con8']==0, col + '_con8'] = np.nan
for col in tqdm(col_list):
data[col + '_con_multi_counts'] = data[col + '_con'] * data[col].map(data[col].value_counts().to_dict())
for col in tqdm(col_list):
data[col + '_con_order'] = con_df[col] * order_df[col]
for col in tqdm(col_list):
data.loc[data[col + '_con_order']==0, col + '_con_order'] = np.nan
for col in tqdm(col_list):
data[col + '_unique_order'] = unique_df[col] * order_df[col]
for col in tqdm(col_list):
data.loc[data[col + '_unique_order']==0, col + '_unique_order'] = np.nan
data.head()
```
# Model
```
train = data[~data.target.isna()]
test = data[data.target.isna()]
target = train['target']
param = {
'bagging_freq': 5,
'bagging_fraction': 0.335,
'boost_from_average': False,
'boost': 'gbdt',
'feature_fraction_seed': 47,
'feature_fraction': 0.041,
'learning_rate': 0.01,
'max_depth': -1,
'metric':'auc',
'min_data_in_leaf': 80,
'min_sum_hessian_in_leaf': 10.0,
'num_leaves': 2,
'num_threads': 8,
'tree_learner': 'serial',
'objective': 'binary',
'verbosity': -1,
'num_threads': 8
}
```
* 0.92288
* 0.92308
```
folds = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
oof_lgb = np.zeros(len(train))
predictions_lgb = np.zeros(len(test))
feature_importance = pd.DataFrame()
train_columns = [c for c in train.columns if c not in ['ID_code', 'target']]
for fold_, (trn_idx, val_idx) in enumerate(folds.split(train, target.values)):
print("fold n°{}".format(fold_))
trn_data = lgb.Dataset(train.iloc[trn_idx][train_columns], label=target.iloc[trn_idx])
val_data = lgb.Dataset(train.iloc[val_idx][train_columns], label=target.iloc[val_idx])
num_round = 500000
clf = lgb.train(param, trn_data, num_round, valid_sets = [trn_data, val_data], verbose_eval=5000, early_stopping_rounds = 3500)
oof_lgb[val_idx] = clf.predict(train.iloc[val_idx][train_columns], num_iteration=clf.best_iteration)
predictions_lgb += clf.predict(test[train_columns], num_iteration=clf.best_iteration) / folds.n_splits
fold_importance = pd.DataFrame()
fold_importance["Feature"] = train_columns
fold_importance["importance"] = clf.feature_importance()
fold_importance["fold"] = fold_ + 1
feature_importance = pd.concat([feature_importance, fold_importance], axis=0)
print("CV score: {:<8.5f}".format(roc_auc_score(target.values[val_idx], oof_lgb[val_idx])))
print("CV score: {:<8.5f}".format(roc_auc_score(target.values, oof_lgb)))
best_features = (feature_importance[["Feature", "importance"]]
.groupby("Feature")
.mean()
.sort_values(by="importance", ascending=False)[400:])
plt.figure(figsize=(14,112))
sns.barplot(x="importance", y="Feature", data=best_features.reset_index())
plt.title('Features importance (averaged/folds)')
plt.tight_layout()
test['target'] = predictions_lgb
sub = pd.read_csv('./data/sample_submission.csv')
unchange = sub[~sub.ID_code.isin(test.ID_code)]
sub = test[['ID_code', 'target']].append(unchange).sort_index()
sample = pd.read_csv('./data/sub_lgb_5fold5aug_concategory_cv_0.9242224159538349.csv')
sample['new_target'] = sub.target
sample[sample.new_target != 0].corr()
sub.to_csv('./data/sub_lgb_noAug_cv_0.923.csv', index=False)
for col in tqdm(col_list):
data[col + '_con_category'] = np.around(data[col + '_con'], 0)
# data[col + '_unique_category'] = np.around(data[col + '_unique'], 0)
for col in tqdm(col_list):
le = LabelEncoder()
le.fit(data[col + '_con_category'].fillna(0))
data[col + '_con_category'] = le.transform(data[col + '_con_category'].fillna(0))
for col in tqdm(col_list):
le = LabelEncoder()
le.fit(data[col + '_unique_category'].fillna(0))
data[col + '_unique_category'] = le.transform(data[col + '_unique_category'].fillna(0))
for col in tqdm(col_list):
data[col + '_unique_category'] = data[col + '_unique_category'].astype('category')
data = pd.get_dummies(data, columns=[col + '_con_category' for col in col_list])
```
|
github_jupyter
|
```
import os
import random
import torch
import torchvision.transforms as standard_transforms
import scipy.io as sio
import matplotlib
import pandas as pd
import misc.transforms as own_transforms
import warnings
from torch.autograd import Variable
from torch.utils.data import DataLoader
from PIL import Image, ImageOps
from matplotlib import pyplot as plt
from tqdm import trange, tqdm
from misc.utils import *
from models.CC import CrowdCounter
from config import cfg
import CCAugmentation as cca
from datasets.SHHB.setting import cfg_data
from load_data import CustomDataset
torch.cuda.set_device(0)
torch.backends.cudnn.benchmark = True
warnings.filterwarnings('ignore')
mean_std = ([0.452016860247, 0.447249650955, 0.431981861591],[0.23242045939, 0.224925786257, 0.221840232611])
img_transform = standard_transforms.Compose([
standard_transforms.ToTensor(),
standard_transforms.Normalize(*mean_std)
])
restore = standard_transforms.Compose([
own_transforms.DeNormalize(*mean_std),
standard_transforms.ToPILImage()
])
pil_to_tensor = standard_transforms.ToTensor()
model_path = './exp/11-26_06-00_SHHB_MCNN_0.0001_[noAug]/all_ep_146_mae_23.91_mse_35.70.pth'
model_path = './exp/11-26_06-57_SHHB_MCNN_0.0001_[noAug]/all_ep_175_mae_17.92_mse_26.94.pth'
model_path = './exp/11-26_07-42_SHHB_MCNN_0.0001_[noAug]/all_ep_171_mae_18.16_mse_29.66.pth'
model_path = './exp/11-27_09-59_SHHB_MCNN_0.0001_[flipLR]/all_ep_180_mae_18.34_mse_30.49.pth'
model_path = './exp/11-27_10-44_SHHB_MCNN_0.0001_[flipLR]/all_ep_181_mae_19.11_mse_33.26.pth'
# model_path = './exp/11-27_11-30_SHHB_MCNN_0.0001_[flipLR]/all_ep_180_mae_18.16_mse_30.61.pth'
net = CrowdCounter(cfg.GPU_ID,cfg.NET)
net.load_state_dict(torch.load(model_path))
net.cuda()
net.eval()
val_pipeline = cca.Pipeline(
cca.examples.loading.SHHLoader("/dataset/ShanghaiTech", "test", "B"), []
).execute_generate()
val_loader = DataLoader(CustomDataset(val_pipeline), batch_size=cfg_data.VAL_BATCH_SIZE, num_workers=1, drop_last=False)
val_img = list(val_loader)
start = 0
N = 3
for vi, data in enumerate(val_img[start:start+N], 0):
img, gt_map = data
with torch.no_grad():
img = Variable(img).cuda()
pred_map = net.test_forward(img)
pred_map = pred_map.data.cpu().numpy()
new_img = img.data.cpu().numpy()
new_img = np.moveaxis(new_img, 1, 2)
new_img = np.moveaxis(new_img, 2, 3)
new_img = np.squeeze(new_img)[:,:,::-1]
pred_cnt = np.sum(pred_map[0])/100.0
gt_count = np.sum(gt_map.data.cpu().numpy())/100.0
fg, (ax0, ax1, ax2) = plt.subplots(1, 3, figsize=(16, 5))
plt.suptitle(' '.join([
'count_label:', str(round(gt_count, 3)),
'count_prediction:', str(round(pred_cnt, 3))
]))
ax0.imshow(np.uint8(new_img))
ax1.imshow(np.squeeze(gt_map), cmap='jet')
ax2.imshow(np.squeeze(pred_map), cmap='jet')
plt.show()
mae = np.empty(len(val_img))
mse = np.empty(len(val_img))
for vi, data in enumerate(tqdm(val_img), 0):
img, gt_map = data
with torch.no_grad():
img = Variable(img).cuda()
pred_map = net.test_forward(img)
pred_map = pred_map.data.cpu().numpy()
pred_cnt = np.sum(pred_map[0])/100.0
gt_count = np.sum(gt_map.data.cpu().numpy())/100.0
mae[vi] = np.abs(gt_count-pred_cnt)
mse[vi] = (gt_count-pred_cnt)**2
print('MAE:', round(mae.mean(),2))
print('MSE:', round(np.sqrt(mse.mean()),2))
```
|
github_jupyter
|
```
%matplotlib inline
```
# Generating an input file
This examples shows how to generate an input file in HDF5-format, which can
then be processed by the `py-fmas` library code.
This is useful when the project-specific code is separate from the `py-fmas`
library code.
.. codeauthor:: Oliver Melchert <[email protected]>
We start by importing the required `py-fmas` functionality. Since the
file-input for `py-fmas` is required to be provided in HDF5-format, we need
some python package that offers the possibility to read and write this
format. Here we opted for the python module h5py which is listed as one of
the dependencies of the `py-fmas` package.
```
import h5py
import numpy as np
import numpy.fft as nfft
```
We then define the desired propagation constant
```
def beta_fun_detuning(w):
r'''Function defining propagation constant
Implements group-velocity dispersion with expansion coefficients
listed in Tab. I of Ref. [1]. Expansion coefficients are valid for
:math:`lambda = 835\,\mathrm{nm}`, i.e. for :math:`\omega_0 \approx
2.56\,\mathrm{rad/fs}`.
References:
[1] J. M. Dudley, G. Genty, S. Coen,
Supercontinuum generation in photonic crystal fiber,
Rev. Mod. Phys. 78 (2006) 1135,
http://dx.doi.org/10.1103/RevModPhys.78.1135
Note:
A corresponding propagation constant is implemented as function
`define_beta_fun_PCF_Ranka2000` in `py-fmas` module
`propatation_constant`.
Args:
w (:obj:`numpy.ndarray`): Angular frequency detuning.
Returns:
:obj:`numpy.ndarray` Propagation constant as function of
frequency detuning.
'''
# ... EXPANSION COEFFICIENTS DISPERSION
b2 = -1.1830e-2 # (fs^2/micron)
b3 = 8.1038e-2 # (fs^3/micron)
b4 = -0.95205e-1 # (fs^4/micron)
b5 = 2.0737e-1 # (fs^5/micron)
b6 = -5.3943e-1 # (fs^6/micron)
b7 = 1.3486 # (fs^7/micron)
b8 = -2.5495 # (fs^8/micron)
b9 = 3.0524 # (fs^9/micron)
b10 = -1.7140 # (fs^10/micron)
# ... PROPAGATION CONSTANT (DEPENDING ON DETUNING)
beta_fun_detuning = np.poly1d([b10/3628800, b9/362880, b8/40320,
b7/5040, b6/720, b5/120, b4/24, b3/6, b2/2, 0., 0.])
return beta_fun_detuning(w)
```
Next, we define all parameters needed to specify a simulation run
```
# -- DEFINE SIMULATION PARAMETERS
# ... COMPUTATIONAL DOMAIN
t_max = 3500. # (fs)
t_num = 2**14 # (-)
z_max = 0.1*1e6 # (micron)
z_num = 4000 # (-)
z_skip = 20 # (-)
t = np.linspace(-t_max, t_max, t_num, endpoint=False)
w = nfft.fftfreq(t.size, d=t[1]-t[0])*2*np.pi
# ... MODEL SPECIFIC PARAMETERS
# ... PROPAGATION CONSTANT
c = 0.29979 # (fs/micron)
lam0 = 0.835 # (micron)
w0 = 2*np.pi*c/lam0 # (rad/fs)
beta_w = beta_fun_detuning(w-w0)
gam0 = 0.11e-6 # (1/W/micron)
n2 = gam0*c/w0 # (micron^2/W)
# ... PARAMETERS FOR RAMAN RESPONSE
fR = 0.18 # (-)
tau1= 12.2 # (fs)
tau2= 32.0 # (fs)
# ... INITIAL CONDITION
t0 = 28.4 # (fs)
P0 = 1e4 # (W)
E_0t_fun = lambda t: np.real(np.sqrt(P0)/np.cosh(t/t0)*np.exp(-1j*w0*t))
E_0t = E_0t_fun(t)
```
The subsequent code will store the simulation parameters defined above to the
file `input_file.h5` in the current working directory.
```
def save_data_hdf5(file_path, data_dict):
with h5py.File(file_path, 'w') as f:
for key, val in data_dict.items():
f.create_dataset(key, data=val)
data_dict = {
't_max': t_max,
't_num': t_num,
'z_min': 0.0,
'z_max': z_max,
'z_num': z_num,
'z_skip': z_skip,
'E_0t': E_0t,
'beta_w': beta_w,
'n2': n2,
'fR': fR,
'tau1': tau1,
'tau2': tau2,
'out_file_path': 'out_file.h5'
}
save_data_hdf5('input_file.h5', data_dict)
```
An example, showing how to use `py-fmas` as a black-box simulation tool that
performs a simulation run for the propagation scenario stored under the file
`input_file.h5` is available under the link below:
`sphx_glr_auto_tutorials_basics_g_app.py`
|
github_jupyter
|
# Using `bw2waterbalancer`
Notebook showing typical usage of `bw2waterbalancer`
## Generating the samples
`bw2waterbalancer` works with Brightway2. You only need set as current a project in which the database for which you want to balance water exchanges is imported.
```
import brightway2 as bw
import numpy as np
bw.projects.set_current('ei36cutoff')
```
The only Class you need is the `DatabaseWaterBalancer`:
```
from bw2waterbalancer import DatabaseWaterBalancer
```
Instantiating the DatabaseWaterBalancer will automatically identify activities that are associated with water exchanges.
```
dwb = DatabaseWaterBalancer(
ecoinvent_version="3.6", # used to identify activities with water production exchanges
database_name="ei36_cutoff", #name the LCI db in the brightway2 project
)
```
Generating presamples for the whole database is a lengthy process. Thankfully, it only ever needs to be done once per database:
```
dwb.add_samples_for_all_acts(iterations=1000)
```
The samples and associated indices are stored as attributes:
```
dwb.matrix_samples
dwb.matrix_samples.shape
dwb.matrix_indices[0:10] # First ten indices
len(dwb.matrix_indices)
```
These can directly be used to generate [`presamples`](https://presamples.readthedocs.io/):
```
presamples_id, presamples_fp = dwb.create_presamples(
name=None, #Could have specified a string as name, not passing anything will use automatically generated random name
dirpath=None, #Could have specified a directory path to save presamples somewhere specific
id_=None, #Could have specified a string as id, not passing anything will use automatically generated random id
seed='sequential', #or None, or int.
)
```
## Using the samples
The samples are formatted for use in brighway2 via the presamples package.
The following function calculates:
- Deterministic results, using `bw.LCA`
- Stochastic results, using `bw.MonteCarloLCA`
- Stochastic results using presamples, using `bw.MonteCarloLCA` and passing `presamples=[presamples_fp]`
The ratio of stochastic results to deterministic results are then plotted for Monte Carlo results with and without presamples.
Ratios for Monte Carlo with presamples are on the order of 1.
Ratios for Monte Carlo without presamples are much greater, as much (for the randomly selected activities) up to two orders of magnitude.
```
def check_presamples_act(act_key, ps_fp, lcia_method, iterations=1000):
"""Plot histrograms of Monte Carlo samples/det result for case w/ and w/o presamples"""
lca = bw.LCA({act_key:1}, method=m)
lca.lci()
lca.lcia()
mc_arr_wo = np.empty(shape=iterations)
mc = bw.MonteCarloLCA({act_key:1}, method=m)
for i in range(iterations):
mc_arr_wo[i] = next(mc)/lca.score
mc_arr_w = np.empty(shape=iterations)
mc_w = bw.MonteCarloLCA({act_key:1}, method=m, presamples=[ps_fp])
for i in range(iterations):
mc_arr_w[i] = next(mc_w)/lca.score
plt.hist(mc_arr_wo, histtype="step", color='orange', label="without presamples")
plt.hist(mc_arr_w, histtype="step", color='green', label="with presamples")
plt.legend()
```
Let's run this on a couple of random ecoinvent products with the ImpactWorld+ water scarcity LCIA method:
```
m=('IMPACTWorld+ (Default_Recommended_Midpoint 1.23)', 'Midpoint', 'Water scarcity')
import matplotlib.pyplot as plt
%matplotlib inline
act = bw.Database('ei36_cutoff').random()
print("Randomly working on ", act)
check_presamples_act(act.key, presamples_fp, m)
act = bw.Database('ei36_cutoff').random()
print("Randomly working on ", act)
check_presamples_act(act.key, presamples_fp, m)
act = bw.Database('ei36_cutoff').random()
print("Randomly working on ", act)
check_presamples_act(act.key, presamples_fp, m)
act = bw.Database('ei36_cutoff').random()
print("Randomly working on ", act)
check_presamples_act(act.key, presamples_fp, m)
act = bw.Database('ei36_cutoff').random()
print("Randomly working on ", act)
check_presamples_act(act.key, presamples_fp, m)
```
|
github_jupyter
|
```
import numpy as np
import pandas as pd
%matplotlib inline
import math
from xgboost.sklearn import XGBClassifier
from sklearn.cross_validation import cross_val_score
from sklearn import cross_validation
from sklearn.metrics import roc_auc_score
from matplotlib import pyplot
train = pd.read_csv("xtrain.csv")
target = pd.read_csv("ytrain.csv")
test = pd.read_csv("xtest.csv")
train.head()
train.describe()
target.head()
for column in train:
print column, ": ", len(train[column].unique())
cat_features = []
real_features = []
for column in train:
if len(train[column].unique()) > 21:
real_features.append(column)
else:
cat_features.append(column)
# построим гистограммы для первых 50к значений для категориальных признаков
train[cat_features].head(50000).plot.hist(bins = 100, figsize=(20, 20))
test[cat_features].head(50000).plot.hist(bins = 100, figsize=(20, 20))
# построим гистограммы для первых 50к значений для остальных признаков
train[real_features].head(50000).plot.hist(bins = 100, figsize=(20, 20))
test[real_features].head(50000).plot.hist(bins = 100, figsize=(20, 20))
#гистограммы для теста и обучающей выборки совпадают
import seaborn
seaborn.heatmap(train[real_features].corr(), square=True)
#числовые признаки не коррелируеют между собой
# в данных есть nan values в каждом столбце
train.isnull().sum()
#для категориальных признаков, nan значения заменим -1
#Для действительных признаков - заменим средним значнием
train[cat_features] = train[cat_features].fillna(-1)
for column in train[real_features]:
mean_val = train[column].mean()
train[column] = train[column].fillna(mean_val)
target.mean() #класса 0 больше чем 1
import xgboost as xgb
from sklearn.cross_validation import train_test_split
X_fit, X_eval, y_fit, y_eval= train_test_split(
train, target, test_size=0.20, random_state=1
)
clf = xgb.XGBClassifier(missing=np.nan, max_depth=3,
n_estimators=550, learning_rate=0.05, gamma =0.3, min_child_weight = 3,
subsample=0.9, colsample_bytree=0.8, seed=2000,objective= 'binary:logistic')
clf.fit(X_fit, y_fit, early_stopping_rounds=40, eval_metric="auc", eval_set=[(X_eval, y_eval)])
auc_train = roc_auc_score(y_fit.x, clf.predict(X_fit))
auc_val = roc_auc_score(y_eval.x, clf.predict(X_eval))
print 'auc_train: ', auc_train
print 'auc_val: ', auc_val
#имеет место быть переобучение
eps = 1e-5
dropped_columns = set()
C = train.columns
#Определим константные признаки
for c in C:
if train[c].var() < eps:
print '.. %-30s: too low variance ... column ignored'%(c)
dropped_columns.add(c)
#таких не обнаружено
for i, c1 in enumerate(C):
f1 = train[c1].values
for j, c2 in enumerate(C[i+1:]):
f2 = train[c2].values
if np.all(f1 == f2):
dropped_columns.add(c2)
print c2
# одинаковых полей также нет
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import ExtraTreesClassifier
import matplotlib.pyplot as plt
forest = ExtraTreesClassifier(n_estimators=150,
random_state=0)
forest.fit(train.head(100000), target.head(100000).x)
importances = forest.feature_importances_
std = np.std([tree.feature_importances_ for tree in forest.estimators_],
axis=0)
indices = np.argsort(importances)[::-1]
# Попробуем посмотреть какие признаки значимы с помощью деревьев
print("Feature ranking:")
for f in range(train.head(100000).shape[1]):
print("%d. feature %d (%f)" % (f + 1, indices[f], importances[indices[f]]))
# Построим графики
plt.figure()
plt.title("Feature importances")
plt.bar(range(train.head(100000).shape[1]), importances[indices],
color="r", yerr=std[indices], align="center")
plt.xticks(range(train.head(100000).shape[1]), indices)
plt.xlim([-1, train.head(100000).shape[1]])
plt.show()
# Явных лидеров как и аутсайдеров среди признаков не видно. Признаки анонимны,
# еще раз обучим модель с более сложными вычислительно гиперпараметрами
from sklearn.cross_validation import train_test_split
import xgboost as xgb
X_fit, X_eval, y_fit, y_eval= train_test_split(
train, target, test_size=0.20, random_state=1
)
clf = xgb.XGBClassifier(missing=np.nan, max_depth=3,
n_estimators=1200, learning_rate=0.05, gamma =0.3, min_child_weight = 3,
subsample=0.9, colsample_bytree=0.8, seed=2000,objective= 'binary:logistic')
clf.fit(X_fit, y_fit, early_stopping_rounds=40, eval_metric="auc", eval_set=[(X_eval, y_eval)])
# формирование результатов
test_target = clf.predict(test)
submission = pd.DataFrame(test_target)
submission.to_csv("test_target.csv", index=False)
```
|
github_jupyter
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.