code
stringlengths 38
801k
| repo_path
stringlengths 6
263
|
|---|---|
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="iXojb3z1X_fo" colab_type="text"
# **Imports**
# + id="HJ9-UNNjX_qN" colab_type="code" colab={}
import tensorflow as tf
import numpy as np
import rcwa_utils
import tensor_utils
import solver
import matplotlib.pyplot as plt
# + [markdown] id="76Zw4T37Yjsl" colab_type="text"
# **Loss Function Definition**
# + id="N52BVsa8YkBK" colab_type="code" colab={}
def loss_func():
# Global parameters dictionary.
global params
# Generate permitivitty and permeability distributions.
ER_t, UR_t = solver.generate_cylindrical_nanoposts(var_duty, params)
# Set the device layer thickness based on the length variable.
thickness_coeff = tf.clip_by_value(var_length, clip_value_min = params['length_min'], clip_value_max = params['length_max'])
thickness_coeff = tf.cast(thickness_coeff, dtype = tf.complex64)
length_shape = (1, 1, 1, 1, 1, 1)
substrate_layer = tf.ones(shape = length_shape, dtype = tf.complex64)
device_layer = thickness_coeff * tf.ones(shape = length_shape, dtype = tf.complex64)
wavelength = params['lam0'][0, 0, 0, 0, 0, 0].numpy()
params['L'] = wavelength * tf.concat([device_layer, substrate_layer], axis = 3)
# Simulate the system.
outputs = solver.simulate(ER_t, UR_t, params)
# Maximize the product of the reflectances.
ref_lambda1 = outputs['REF'][0, 0, 0]
ref_lambda2 = outputs['REF'][1, 0, 0]
return -ref_lambda1 * ref_lambda2
# + [markdown] id="zsz_XCZjYqtr" colab_type="text"
# **Setup and Initialize Variables**
# + id="trCBNNrHYq0a" colab_type="code" colab={}
# Initialize global params dictionary.
params = solver.initialize_params(wavelengths = [632.0, 530.0],
thetas = [0.0, 0.0],
phis = [0.0, 0.0],
pte = [1.0, 1.0],
ptm = [0.0, 0.0])
params['erd'] = 6.76 # Grating layer permittivity.
params['ers'] = 2.25 # Subtrate layer permittivity.
params['PQ'] = [11, 11] # Fourier Harmonics.
# Initialize grating duty cycle variable.
var_shape = (1, params['pixelsX'], params['pixelsY'])
duty_initial = 0.8 * np.ones(shape = var_shape)
var_duty = tf.Variable(duty_initial, dtype = tf.float32)
# Initialize grating thickness variable.
length_initial = 0.25
var_length = tf.Variable(length_initial, dtype = tf.float32)
# + [markdown] id="WvVK-VXMYq79" colab_type="text"
# **Optimize**
# + id="WBLtRB3gYrDG" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="c2de1613-6457-41c9-c739-6e74e46f93bb"
# Number of optimization iterations.
N = 200
# Define an optimizer and data to be stored.
opt = tf.keras.optimizers.Adam(learning_rate = 0.0005)
loss = np.zeros(N + 1)
# Compute initial loss.
loss[0] = loss_func().numpy()
# Optimize.
print('Optimizing...')
for i in range(N):
opt.minimize(loss_func, var_list = [var_duty, var_length])
loss[i + 1] = loss_func().numpy()
# + [markdown] id="jEcnCtAiYrLa" colab_type="text"
# **Display Learning Curve**
# + id="Axql3myAaZ2V" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 278} outputId="6b69cc04-a97e-431c-b65e-f842b431fb61"
plt.plot(loss)
plt.xlabel('Iterations')
plt.ylabel('Loss')
plt.xlim(0, N)
plt.show()
| examples/gratings/reflective_grating_two_wavelengths.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Seafloor Bacterial Floc Analysis
#
# This notebook shows an example of doing an analysis of water column "floc" using Pangeo. The goal of this work is to understand changes in the concentration of floc, which is bacterial material that has been flushed from the hydrothermal system into the ocean. Changes in floc are a potential indicator of changes in the hydrothermal system, possibly resulting from a magmatic event or seismic swarm.
#
# In this notebook we analyze a large number of OOI HD video camera frames to establish a proxy for the floc concentration, and then display the results using a two-dimensional multivariate histogram.
# ## Get a list of CamHD files to process
import pandas as pd
dbcamhd = pd.read_json('dbcamhd.json', orient="records", lines=True)
dbcamhd.tail()
print("Total files: %i" % len(dbcamhd))
print("Total frames: %i" % dbcamhd.frame_count.sum())
blob_urls = list(dbcamhd.blob_url[(dbcamhd.deployment == 2) & (dbcamhd.frame_count > 5000) & (dbcamhd.frame_count < 30000)])
#blob_urls = list(dbcamhd.blob_url[(dbcamhd.frame_count > 5000) & (dbcamhd.frame_count < 30000)])
blob_urls.sort()
blob_urls[0]
len(blob_urls)
# ### Define the frame numbers from each file to process
frame_numbers = [3841, 3933, 4052, 4171, 4263, 4382]
# These frame numbers correspond to times when the camera system is looking over the "shoulder" of Mushroom vent.
# ## Set up a delayed Dask array of images
import pycamhd as camhd
import numpy as np
from dask import delayed
import dask.array as dsa
delayed_frame_list = []
for blob_url in blob_urls:
delayed_moov_atom = delayed(camhd.get_moov_atom)(blob_url)
for frame_number in frame_numbers:
delayed_frame = delayed(camhd.get_frame)(blob_url, frame_number, 'gray16le', delayed_moov_atom)
delayed_frame_list.append(dsa.from_delayed(delayed_frame, (1080, 1920), np.uint16))
delayed_frame_array = dsa.stack(delayed_frame_list)
delayed_frame_array
# A dask array is in many ways like a numpy array, except in this case it holds a set of instructions for how to acquire each chunk of the array, which makes it easy to farm this array out to workers in the cloud using the [distributed](http://distributed.readthedocs.io/en/latest/#) scheduler.
# ## Show one of the images
frame = delayed_frame_array[1700*6].compute()
frame.shape
# %matplotlib inline
# %config InlineBackend.figure_format = 'svg'
import matplotlib.pyplot as plt
import matplotlib.patches as patches
plt.rc('figure', figsize=(11, 11))
fig, ax = plt.subplots()
im1 = ax.imshow(frame)
im1.set_cmap('gray')
plt.yticks(np.arange(0,1081,270))
plt.xticks(np.arange(0,1921,480))
rect = patches.Rectangle((10,10),1024,1024,linewidth=1.5,edgecolor='w',facecolor='none')
ax.add_patch(rect)
plt.show();
# ## Show the filter that will be used to filter images in the frequency domain
# To deal with variations in lighting and high-frequency noise, we filter each subimage using a Butterworth bandpass filter.
def butterworth(d1, d2, n):
x = np.arange(-1024/2+0.5,1024/2+1-0.5)
xx, yy = np.meshgrid(x, x)
d = np.sqrt(xx**2+yy**2)
bff = (1 - (1./(1 + (d/d1)**(2*n))))*(1/(1 + (d/d2)**(2*n)))
return bff
d1 = 20 # low cut wavenumber
d2 = 400 # high cut wavenumber
n = 4
bff = butterworth(d1, d2, n)
plt.rc('figure', figsize=(6, 6))
imgplot = plt.imshow(bff, cmap='gray')
# ## Define the floc proxy function
# The floc proxy is simply the number of pixels in each filtered subimage that have a value greater than 4000.
def frame_filter(frame, d1, d2, n):
if frame.ndim == 3 and frame.shape[0] == 1:
I = np.squeeze(frame[0, 0:1024, 0:1024])
else:
I = frame[0:1024, 0:1024]
bff = butterworth(d1, d2, n)
I_fft = np.fft.fft2(I)
I_fft_shift = np.fft.fftshift(I_fft)
I_fft_shift_filt = I_fft_shift*bff # filter with the Butterworth filter
I_fft_filt = np.fft.ifftshift(I_fft_shift_filt)
I_filt = np.fft.ifft2(I_fft_filt)
return I_filt
def calc_floc_proxy(frame, d1, d2, n):
I_filt = frame_filter(frame, d1, d2, n)
return np.array([(np.absolute(I_filt)>4000).sum()])
# ## Show example for one frame
I_filt = frame_filter(frame, d1, d2, n)
plt.rc('figure', figsize=(6, 6))
imgplot = plt.imshow(np.absolute(I_filt)>4000, cmap='gray')
plt.title('floc_proxy value = %i' % (np.absolute(I_filt)>4000).sum());
# ## Assemble a new Dask array including our computation using map_blocks
floc_proxy = dsa.map_blocks(calc_floc_proxy, delayed_frame_array, d1, d2, n, dtype='i8', drop_axis=[1,2])
floc_proxy
# ## Start a Dask cluster
# Use the new Dask extension!
client
# %%time
results = floc_proxy[0::10].compute()
# ## Calculate the results
print('Number of images: %i' % len(floc_proxy))
print('Size of dataset (GB): %i' % round(len(floc_proxy)*1080*1920*2/1024/1024/1024))
# %%time
results = floc_proxy.compute()
# ## Get a timestamp for each frame
import datetime, math
import matplotlib.dates as dates
frame_timestamp = []
for blob_url in blob_urls:
timestamp = dbcamhd['timestamp'][dbcamhd.blob_url == blob_url].iloc[0]
for frame_number in frame_numbers:
timestamp = timestamp + frame_number/29.97
dt = datetime.datetime.fromtimestamp(timestamp)
frame_timestamp.append(dates.date2num(dt))
# ## Plot a two-dimensional multivariate histogram of the results
plt.rc('font', size=11)
fig, ax = plt.subplots()
fig.set_size_inches(14, 6)
fig.frameon = False
hb1 = ax.hexbin(frame_timestamp[0::10], results, vmin=0.1, vmax=10, bins='log', linewidths=0.25,
gridsize=(200, 4000), mincnt=1, cmap=plt.cm.BuPu)
fig.colorbar(hb1)
ax.set_ylim([0, 8000])
ax.set_xlim([frame_timestamp[0],frame_timestamp[-1]])
ax.yaxis.grid(True)
ax.xaxis.grid(True)
months = dates.MonthLocator(interval=6) # every month
monthsFmt = dates.DateFormatter('%b %Y')
ax.xaxis.set_major_locator(months)
ax.xaxis.set_major_formatter(monthsFmt)
plt.ylabel('Floc Proxy Value');
# Starting in mid-June a large "floc event" occurs where the floc proxy values increase on average by about a factor of ten. The cause of this floc event is being investigated.
# ## References
#
# - [Pangeo](http://pangeo-data.org/)
# - [PyCamHD](https://github.com/tjcrone/pycamhd)
# - [CamHD Raw Data Archive](https://rawdata.oceanobservatories.org/files/RS03ASHS/PN03B/06-CAMHDA301)
# - [AGU Abstract](https://agu.confex.com/agu/fm16/meetingapp.cgi/Paper/192670)
# - [AGU Poster](https://drive.google.com/open?id=0B-dWW4GM434obGpTM0FZME10Nkk)
# - [Dask](http://dask.pydata.org/en/latest/)
| notebooks/camhd/floc.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# %load_ext autoreload
# %autoreload 2
import sys
sys.path.append("..")
from acse_9_irp_wafflescore import MiscHelpers as mh
from acse_9_irp_wafflescore import SOMsHelpers as sh
from acse_9_irp_wafflescore import FCMHelpers as fh
from acse_9_irp_wafflescore import dataPreprocessing as dp
import numpy as np
from timeit import default_timer as timer
import logging
import sys
logging.basicConfig(format='%(asctime)s | %(levelname)s : %(message)s',
level=logging.INFO, stream=sys.stdout)
# -
# # Tested with model with smaller anomaly (M5b)
# +
model = 'M5b'
pdir = '../data/' + model + '_xz_pos.npy'
xz_pos = np.load(pdir)
x = xz_pos[:, 0]
z = xz_pos[:, 1]
ldir = '../data/' + model + '_data_label.npy'
label = np.load(ldir)
cdir = '../data/' + model + '_clean_data.npy'
data = np.load(cdir)
ddir = '../data/' + model + '_init_data.npy'
idata = np.load(cdir)
means, stds = dp.compMeanStd(data)
norm_data = dp.normalize(data, means, stds)
col_n = ['vp', 'vs', 'dn', 'vp/vs', 'qp', 'qs', 'x', 'z']
mh.plot_fields(idata[:,:-2], x, z, titles=col_n[:-2])
# -
means, stds = dp.compMeanStd(data)
norm_data = dp.normalize(data, means, stds)
g_res = (34, 4000, 0.5, 2)
cluster_labels, avg_sils, avg_ch, dim, it, lr, sig = sh.random_search_som(norm_data, g_res)
# ### Result from random search
#
# | Dimension | Iteration | Learning Rate | Sigma | Silhouette Score | <NAME> |
# |-----------|-----------|---------------|----------|------------------|-------------------|
# | 42 | 3500 | 0.455185 | 2.034483 | 0.099022 | 1025.587367 |
# | 42 | 3500 | 0.333614 | 1.965517 | 0.052766 | 1133.190408 |
# | 39 | 3900 | 0.627987 | 2.724138 | 0.012874 | 849.064418 |
# | 34 | 3500 | 0.352648 | 2.586207 | 0.101516 | 1166.716261 |
# | 28 | 4300 | 0.411920 | 1.827586 | 0.183970 | 735.002153 |
# | 37 | 3900 | 0.649246 | 1.551724 | 0.063963 | 773.470431 |
# | 38 | 3700 | 0.656490 | 1.620690 | 0.008489 | 939.783383 |
# | 41 | 4100 | 0.460264 | 2.034483 | 0.352136 | 930.653439 |
# | 26 | 4300 | 0.394036 | 2.862069 | 0.085335 | 1247.999601 |
# | 40 | 3700 | 0.341101 | 1.896552 | 0.111925 | 1363.543003 |
# | 41 | 3500 | 0.308680 | 1.827586 | 0.120439 | 1032.707676 |
# | 38 | 3700 | 0.656490 | 1.551724 | 0.065325 | 862.401445 |
# | 28 | 4300 | 0.520021 | 1.344828 | 0.114659 | 820.655592 |
# | 42 | 3700 | 0.430617 | 3.000000 | 0.188410 | 1320.232533 |
# | 32 | 4300 | 0.491953 | 2.310345 | 0.083481 | 870.997504 |
# | 38 | 4100 | 0.301905 | 1.896552 | 0.085886 | 869.885199 |
# | 39 | 3700 | 0.475845 | 2.862069 | -0.034931 | 1001.385126 |
# | 31 | 3900 | 0.600721 | 1.068966 | -0.039465 | 884.298641 |
# | 33 | 3700 | 0.642081 | 2.034483 | 0.046745 | 1052.423670 |
# | 40 | 4100 | 0.470594 | 2.448276 | 0.150802 | 1223.052308 |
# first plot: (41, 4100, 0.460264, 2.034483)
# second plot: (40, 3700, 0.341101, 1.896552)
for c in cluster_labels:
mh.plot_e_model(c, x, z, sep_label=True)
# From observing the two final result, seems like dim, iter_cnt, lr, sigma = (40, 3700, 0.341101, 1.896552) is the best hyperparameter for model5b
# +
dim, iter_cnt, lr, sigma = (40, 3700, 0.341101, 1.896552)
som, cluster_labels = sh.run_SOMs(norm_data, dim, iter_cnt, lr, sigma)
n_map = som.neuron_map(norm_data)
u_matrix = som.distance_map().T
watershed_bins = sh.histedges_equalN(u_matrix.flatten())
ws_labels = sh.watershed_level(u_matrix, watershed_bins, plot=False)
n_map = som.neuron_map(norm_data)
cluster_labels, sils, csc, best_idx = sh.eval_ws(norm_data, ws_labels, n_map, re_all=True)
# -
# plot of best silhouette score
i = 27
print('(dim:', dim, ',iter:', iter_cnt, ',lr', lr, ',sigma:', sigma, ')')
print('sil:', sils[i], ',ch:', csc[i])
mh.plot_e_model(cluster_labels[i], x, z, sep_label=True)
| Notebooks/SOMs.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import tensorflow as tf
import numpy as np
filename = 'C:/Users/HP/Downloads/download.jpg'
from IPython.display import Image
Image(filename = 'C:/Users/HP/Downloads/download.jpg', width= 224, height= 224)
from tensorflow.keras.preprocessing import image
img = image.load_img(filename, target_size = (224,224))
import matplotlib.pyplot as plt
plt.imshow(img)
import cv2
import cv2.cv as cv
imgg = cv2.imread(filename)
plt.imshow(imgg)
imgg = cv2.resize(imgg, (224,224))
plt.imshow(imgg)
plt.imshow(cv2.cvtColor(imgg, cv2.COLOR_BGR2RGB))
from PIL import Image
im = Image.open(filename)
im = im.resize((224,224))
plt.imshow(im)
mobile = tf.keras.applications.mobilenet.MobileNet()
from tensorflow.keras.preprocessing import image
img = image.load_img(filename, target_size = (224,224))
plt.imshow(img)
resized_img = image.img_to_array(img)
final_image = np.expand_dims(resized_img,axis =0)
final_image=tf.keras.applications.mobilenet.preprocess_input(final_image)
| image loader.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Advanced Tutorial
# ### Adding visualization
#
# So far, we've built a model, run it, and analyzed some output afterwards. However, one of the advantages of agent-based models is that we can often watch them run step by step, potentially spotting unexpected patterns, behaviors or bugs, or developing new intuitions, hypotheses, or insights. Other times, watching a model run can explain it to an unfamiliar audience better than static explanations. Like many ABM frameworks, Mesa allows you to create an interactive visualization of the model. In this section we'll walk through creating a visualization using built-in components, and (for advanced users) how to create a new visualization element.
#
# First, a quick explanation of how Mesa's interactive visualization works. Visualization is done in a browser window, using JavaScript to draw the different things being visualized at each step of the model. To do this, Mesa launches a small web server, which runs the model, turns each step into a JSON object (essentially, structured plain text) and sends those steps to the browser.
#
# A visualization is built up of a few different modules: for example, a module for drawing agents on a grid, and another one for drawing a chart of some variable. Each module has a Python part, which runs on the server and turns a model state into JSON data; and a JavaScript side, which takes that JSON data and draws it in the browser window. Mesa comes with a few modules built in, and let you add your own as well.
# #### Grid Visualization
#
# To start with, let's have a visualization where we can watch the agents moving around the grid. For this, you will need to put your model code in a separate Python source file; for example, `MoneyModel.py`. Next, either in the same file or in a new one (e.g. `MoneyModel_Viz.py`) import the server class and the Canvas Grid class (so-called because it uses HTML5 canvas to draw a grid). If you're in a new file, you'll also need to import the actual model object.
# +
from mesa.visualization.modules import CanvasGrid
from mesa.visualization.ModularVisualization import ModularServer
# If MoneyModel.py is where your code is:
# from MoneyModel import MoneyModel
# -
# `CanvasGrid` works by looping over every cell in a grid, and generating a portrayal for every agent it finds. A portrayal is a dictionary (which can easily be turned into a JSON object) which tells the JavaScript side how to draw it. The only thing we need to provide is a function which takes an agent, and returns a portrayal object. Here's the simplest one: it'll draw each agent as a red, filled circle which fills half of each cell.
def agent_portrayal(agent):
portrayal = {"Shape": "circle",
"Color": "red",
"Filled": "true",
"Layer": 0,
"r": 0.5}
return portrayal
# In addition to the portrayal method, we instantiate a canvas grid with its width and height in cells, and in pixels. In this case, let's create a 10x10 grid, drawn in 500 x 500 pixels.
grid = CanvasGrid(agent_portrayal, 10, 10, 500, 500)
# Now we create and launch the actual server. We do this with the following arguments:
#
# * The model class we're running and visualizing; in this case, `MoneyModel`.
# * A list of module objects to include in the visualization; here, just `[grid]`
# * The title of the model: "Money Model"
# * Any inputs or arguments for the model itself. In this case, 100 agents, and height and width of 10.
#
# Once we create the server, we set the port for it to listen on (you can treat this as just a piece of the URL you'll open in the browser). Finally, when you're ready to run the visualization, use the server's `launch()` method.
#
# ```python
# server = ModularServer(MoneyModel,
# [grid],
# "Money Model",
# 100, 10, 10)
# server.port = 8521 # The default
# server.launch()
# ```
# The full code should now look like:
#
# ```python
# from MoneyModel import *
# from mesa.visualization.modules import CanvasGrid
# from mesa.visualization.ModularVisualization import ModularServer
#
#
# def agent_portrayal(agent):
# portrayal = {"Shape": "circle",
# "Filled": "true",
# "Layer": 0,
# "Color": "red",
# "r": 0.5}
# return portrayal
#
# grid = CanvasGrid(agent_portrayal, 10, 10, 500, 500)
# server = ModularServer(MoneyModel,
# [grid],
# "Money Model",
# 100, 10, 10)
# server.port = 8521 # The default
# server.launch()
# ```
# Now run this file; this should launch the interactive visualization server and open your web browser automatically. (If the browser doesn't open automatically, try pointing it at [http://127.0.0.1:8521](http://127.0.0.1:8521) manually. If this doesn't show you the visualization, something may have gone wrong with the server launch.)
#
# You should see something like the figure below: the model title, an empty space where the grid will be, and a control panel off to the right.
#
# 
#
# Click the 'reset' button on the control panel, and you should see the grid fill up with red circles, representing agents.
#
# 
#
# Click 'step' to advance the model by one step, and the agents will move around. Click 'run' and the agents will keep moving around, at the rate set by the 'fps' (frames per second) slider at the top. Try moving it around and see how the speed of the model changes. Pressing 'pause' will (as you'd expect) pause the model; presing 'run' again will restart it. Finally, 'reset' will start a new instantiation of the model.
#
# To stop the visualization server, go back to the terminal where you launched it, and press Control+c.
# #### Changing the agents
#
# In the visualization above, all we could see is the agents moving around -- but not how much money they had, or anything else of interest. Let's change it so that agents who are broke (wealth 0) are drawn in grey, smaller, and above agents who still have money.
#
# To do this, we go back to our `agent_portrayal` code and add some code to change the portrayal based on the agent properties.
#
# ```python
# def agent_portrayal(agent):
# portrayal = {"Shape": "circle",
# "Filled": "true",
# "r": 0.5}
#
# if agent.wealth > 0:
# portrayal["Color"] = "red"
# portrayal["Layer"] = 0
# else:
# portrayal["Color"] = "grey"
# portrayal["Layer"] = 1
# portrayal["r"] = 0.2
# return portrayal
# ```
#
# Now launch the server again - this will open a new browser window pointed at the updated visualization. Initially it looks the same, but advance the model and smaller grey circles start to appear. Note that since the zero-wealth agents have a higher layer number, they are drawn on top of the red agents.
#
# 
# #### Adding a chart
#
# Next, let's add another element to the visualization: a chart, tracking the model's Gini Coefficient. This is another built-in element that Mesa provides.
#
# ```python
# from mesa.visualization.modules import ChartModule
# ```
#
# The basic chart pulls data from the model's DataCollector, and draws it as a line graph using the [Charts.js](http://www.chartjs.org/) JavaScript libraries. We instantiate a chart element with a list of series for the chart to track. Each series is defined in a dictionary, and has a `Label` (which must match the name of a model-level variable collected by the DataCollector) and a `Color` name. We can also give the chart the name of the DataCollector object in the model.
#
# Finally, we add the chart to the list of elements in the server. The elements are added to the visualization in the order they appear, so the chart will appear underneath the grid.
#
# ```python
# chart = ChartModule([{"Label": "Gini",
# "Color": "Black"}],
# data_collector_name='datacollector')
#
# server = ModularServer(MoneyModel,
# [grid, chart],
# "Money Model",
# 100, 10, 10)
# ```
#
# Launch the visualization and start a model run, and you'll see a line chart underneath the grid. Every step of the model, the line chart updates along with the grid. Reset the model, and the chart resets too.
#
# 
#
# **Note:** You might notice that the chart line only starts after a couple of steps; this is due to a bug in Charts.js which will hopefully be fixed soon.
# ### Building your own visualization component
#
# **Note:** This section is for users who have a basic familiarity with JavaScript. If that's not you, don't worry! (If you're an advanced JavaScript coder and find things that we've done wrong or inefficiently, please [let us know](https://github.com/projectmesa/mesa/issues)!)
#
# If the visualization elements provided by Mesa aren't enough for you, you can build your own and plug them into the model server.
#
# First, you need to understand how the visualization works under the hood. Remember that each visualization module has two sides: a Python object that runs on the server and generates JSON data from the model state (the server side), and a JavaScript object that runs in the browser and turns the JSON into something it renders on the screen (the client side).
#
# Obviously, the two sides of each visualization must be designed in tandem. They result in one Python class, and one JavaScript `.js` file. The path to the JavaScript file is a property of the Python class.
#
# For this example, let's build a simple histogram visualization, which can count the number of agents with each value of wealth. We'll use the [Charts.js](http://www.chartjs.org/) JavaScript library, which is already included with Mesa. If you go and look at its documentation, you'll see that it had no histogram functionality, which means we have to build our own out of a bar chart. We'll keep the histogram as simple as possible, giving it a fixed number of integer bins. If you were designing a more general histogram to add to the Mesa repository for everyone to use across different models, obviously you'd want something more general.
# #### Client-Side Code
#
# In general, the server- and client-side are written in tandem. However, if you're like me and more comfortable with Python than JavaScript, it makes sense to figure out how to get the JavaScript working first, and then write the Python to be compatible with that.
#
# In the same directory as your model, create a new file called `HistogramModule.js`. This will store the JavaScript code for the client side of the new module.
#
# JavaScript classes can look alien to people coming from other languages -- specifically, they can look like functions. (The Mozilla [Introduction to Object-Oriented JavaScript](https://developer.mozilla.org/en-US/docs/Web/JavaScript/Introduction_to_Object-Oriented_JavaScript) is a good starting point). In `HistogramModule.js`, start by creating the class itself:
#
# ```javascript
# var HistogramModule = function(bins, canvas_width, canvas_height) {
# // The actual code will go here.
# };
# ```
#
# Note that our object is instantiated with three arguments: the number of integer bins, and the width and height (in pixels) the chart will take up in the visualization window.
#
# When the visualization object is instantiated, the first thing it needs to do is prepare to draw on the current page. To do so, it adds a [canvas](https://developer.mozilla.org/en-US/docs/Web/API/Canvas_API) tag to the page, using [JQuery's](https://jquery.com/) dollar-sign syntax (JQuery is already included with Mesa). It also gets the canvas' context, which is required for doing anything with it.
#
# ```javascript
# var HistogramModule = function(bins, canvas_width, canvas_height) {
# // Create the tag:
# var canvas_tag = "<canvas width='" + canvas_width + "' height='" + canvas_height + "' ";
# canvas_tag += "style='border:1px dotted'></canvas>";
# // Append it to #elements:
# var canvas = $(canvas_tag)[0];
# $("#elements").append(canvas);
# // Create the context and the drawing controller:
# var context = canvas.getContext("2d");
# };
# ```
#
# Look at the Charts.js [bar chart documentation](http://www.chartjs.org/docs/#bar-chart-introduction). You'll see some of the boilerplate needed to get a chart set up. Especially important is the `data` object, which includes the datasets, labels, and color options. In this case, we want just one dataset (we'll keep things simple and name it "Data"); it has `bins` for categories, and the value of each category starts out at zero. Finally, using these boilerplate objects and the canvas context we created, we can create the chart object.
#
# ```javascript
# var HistogramModule = function(bins, canvas_width, canvas_height) {
# // Create the tag:
# var canvas_tag = "<canvas width='" + canvas_width + "' height='" + canvas_height + "' ";
# canvas_tag += "style='border:1px dotted'></canvas>";
# // Append it to #elements:
# var canvas = $(canvas_tag)[0];
# $("#elements").append(canvas);
# // Create the context and the drawing controller:
# var context = canvas.getContext("2d");
#
# // Prep the chart properties and series:
# var datasets = [{
# label: "Data",
# fillColor: "rgba(151,187,205,0.5)",
# strokeColor: "rgba(151,187,205,0.8)",
# highlightFill: "rgba(151,187,205,0.75)",
# highlightStroke: "rgba(151,187,205,1)",
# data: []
# }];
#
# // Add a zero value for each bin
# for (var i in bins)
# datasets[0].data.push(0);
#
# var data = {
# labels: bins,
# datasets: datasets
# };
#
# var options = {
# scaleBeginsAtZero: true
# };
#
# // Create the chart object
# var chart = new Chart(context, {type: 'bar', data: data, options: options});
#
# // Now what?
# };
# ```
#
# There are two methods every client-side visualization class must implement to be able to work: `render(data)` to render the incoming data, and `reset()` which is called to clear the visualization when the user hits the reset button and starts a new model run.
#
# In this case, the easiest way to pass data to the histogram is as an array, one value for each bin. We can then just loop over the array and update the values in the chart's dataset.
#
# There are a few ways to reset the chart, but the easiest is probably to destroy it and create a new chart object in its place.
#
# With that in mind, we can add these two methods to the class:
#
# ```javascript
# var HistogramModule = function(bins, canvas_width, canvas_height) {
# // ...Everything from above...
# this.render = function(data) {
# datasets[0].data = data;
# chart.update();
# };
#
# this.reset = function() {
# chart.destroy();
# chart = new Chart(context, {type: 'bar', data: data, options: options});
# };
# };
# ```
#
# Note the `this`. before the method names. This makes them public and ensures that they are accessible outside of the object itself. All the other variables inside the class are only accessible inside the object itself, but not outside of it.
# #### Server-Side Code
#
# Can we get back to Python code? Please?
#
# Every JavaScript visualization element has an equal and opposite server-side Python element. The Python class needs to also have a `render` method, to get data out of the model object and into a JSON-ready format. It also needs to point towards the code where the relevant JavaScript lives, and add the JavaScript object to the model page.
#
# In a Python file (either its own, or in the same file as your visualization code), import the `VisualizationElement` class we'll inherit from, and create the new visualization class.
#
# ```python
# from mesa.visualization.ModularVisualization import VisualizationElement
#
# class HistogramModule(VisualizationElement):
# package_includes = ["Chart.min.js"]
# local_includes = ["HistogramModule.js"]
#
# def __init__(self, bins, canvas_height, canvas_width):
# self.canvas_height = canvas_height
# self.canvas_width = canvas_width
# self.bins = bins
# new_element = "new HistogramModule({}, {}, {})"
# new_element = new_element.format(bins,
# canvas_width,
# canvas_height)
# self.js_code = "elements.push(" + new_element + ");"
# ```
#
# There are a few things going on here. `package_includes` is a list of JavaScript files that are part of Mesa itself that the visualization element relies on. You can see the included files in [mesa/visualization/templates/](https://github.com/projectmesa/mesa/tree/tutorial_update/mesa/visualization/templates). Similarly, `local_includes` is a list of JavaScript files in the same directory as the class code itself. Note that both of these are class variables, not object variables -- they hold for all particular objects.
#
# Next, look at the `__init__` method. It takes three arguments: the number of bins, and the width and height for the histogram. It then uses these values to populate the `js_code` property; this is code that the server will insert into the visualization page, which will run when the page loads. In this case, it creates a new HistogramModule (the class we created in JavaScript in the step above) with the desired bins, width and height; it then appends (`push`es) this object to `elements`, the list of visualization elements that the visualization page itself maintains.
#
# Now, the last thing we need is the `render` method. If we were making a general-purpose visualization module we'd want this to be more general, but in this case we can hard-code it to our model.
#
# ```python
# import numpy as np
#
# class HistogramModule(VisualizationElement):
# # ... Everything from above...
#
# def render(self, model):
# wealth_vals = [agent.wealth for agent in model.schedule.agents]
# hist = np.histogram(wealth_vals, bins=self.bins)[0]
# return [int(x) for x in hist]
# ```
#
# Every time the render method is called (with a model object as the argument) it uses numpy to generate counts of agents with each wealth value in the bins, and then returns a list of these values. Note that the `render` method doesn't return a JSON string -- just an object that can be turned into JSON, in this case a Python list (with Python integers as the values; the `json` library doesn't like dealing with numpy's integer type).
#
# Now, you can create your new HistogramModule and add it to the server:
#
# ```python
# histogram = HistogramModule(list(range(10)), 200, 500)
# server = ModularServer(MoneyModel,
# [grid, histogram, chart],
# "Money Model",
# 100, 10, 10)
# server.launch()
# ```
#
# Run this code, and you should see your brand-new histogram added to the visualization and updating along with the model!
#
# 
#
# If you've felt comfortable with this section, it might be instructive to read the code for the [ModularServer](https://github.com/projectmesa/mesa/blob/master/mesa/visualization/ModularVisualization.py#L259) and the [modular_template](https://github.com/projectmesa/mesa/blob/master/mesa/visualization/templates/modular_template.html) to get a better idea of how all the pieces fit together.
# ### Happy Modeling!
#
# This document is a work in progress. If you see any errors, exclusions or have any problems please contact [us](https://github.com/projectmesa/mesa/issues).
| docs/tutorials/adv_tutorial.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LinearRegression, SGDRegressor
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import mean_squared_error
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import IsolationForest, RandomForestRegressor
from sklearn.feature_selection import SelectKBest, f_classif
# -
def mape(y_true, y_pred):
y_true, y_pred = np.array(y_true), np.array(y_pred)
return np.mean(np.abs((y_true - y_pred) / y_true)) * 100
X = pd.read_csv('features_data_without_ld', index_col=0)
y = pd.read_csv('target_data', index_col=0)
X.shape
X_clean = X[~X.isna_Al2O3].drop(['isna_Al2O3', 'isna_SiO2'], axis=1)
y_clean = y[~y.isna_Al2O3].drop(['isna_Al2O3', 'isna_SiO2'], axis=1)
X_clean.shape
print('Линейная регрессия')
for name in y_clean.columns:
model = LinearRegression()
x_train, x_test, y_train, y_test = train_test_split(X_clean, y_clean[name], train_size = 0.75)
lg.fit(x_train, y_train)
y_predicted = lg.predict(x_test)
print(name, 'mse:', round(mape(y_test, y_predicted),2))
print('SGDRegressor')
for name in y_clean.columns:
model = SGDRegressor()
x_train, x_test, y_train, y_test = train_test_split(X_clean, y_clean[name], train_size = 0.75)
model.fit(x_train, y_train)
y_predicted = model.predict(x_test)
print(name, 'mse:', round(mape(y_test, y_predicted),2))
parameters_grid = {
'loss' : [ 'log', 'squared_loss'],
'penalty' : ['l1', 'l2'],
'n_iter' : range(5,10),
'alpha' : np.linspace(0.0001, 0.001, num = 5),
}
model_selection = GridSearchCV(SGDRegressor(), param_grid=parameters_grid)
model_selection.fit(X_clean, y_clean)
| model_selection.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.8.5 64-bit
# metadata:
# interpreter:
# hash: 1ee38ef4a5a9feb55287fd749643f13d043cb0a7addaab2a9c224cbe137c0062
# name: python3
# ---
# # Machine Actionable Data Management Plan connections
#
# Data management plans (DMPs) are documents accompanying research proposals and project outputs. DMPs are created as textual narratives and describe the data and tools employed in scientific investigations.They are sometimes seen as an administrative exercise and not as an integral part of research practice. Machine Actionable DMPs (maDMPs) take the DMP concept further by using PIDs and PIDs services to connect all resources associated with a DMP.
#
# This notebook displays in a human-friendly way all of the connections embedded in a maDMP. By the end of this notebook, you will be able to succinctly display the essential components of the maDMP vision using persistent identifiers (PIDs): Open Researcher and Contributor IDs (ORCIDs), funders IDs, organizations Org IDs, and Dataset IDs (DOIs). To demonstrate this we use an example DMP, viz. https://doi.org/10.4124/test/.879w8. The notebook fetches all the PIDs associated with this DMP and displays it in a Tree Diagram. See below. The diagram puts the DMP at center and there are four main branches: datasets, funders, organisations, and people. Each branch gives birth to individual entities of those branches. For example, the name of all the people that contributed to the DMP.
#
#
# <img src="example_plot.png"/>
#
#
# The process of displaying the DMP visulisation is very simple. First, and after a initial setup, we fetch all the we need from the DataCite GraphQL API. Then, we transform this data into a data structure that can be use for visulisation. Finally, we take the data tranformation and supply it to a Vega visulisation specification to generate the Chart you can see above.
#
#
#
# %%capture
# Install required Python packages
# !pip install dfply altair altair_saver vega altair_viewer
# +
import json
import pandas as pd
import numpy as np
from dfply import *
import altair.vega.v5 as alt
from altair_saver import save
# alt.renderers.enable('notebook')
# +
# Prepare the GraphQL client
import requests
from gql import gql, Client
from gql.transport.requests import RequestsHTTPTransport
_transport = RequestsHTTPTransport(
url='https://api.datacite.org/graphql',
use_json=True,
)
client = Client(
transport=_transport,
fetch_schema_from_transport=True,
)
# -
from IPython.display import display, Markdown
import ipywidgets as widgets
f = widgets.Dropdown(
options=['https://doi.org/10.48321/D17G67', 'https://doi.org/10.48321/D1H59R', 'https://doi.org/10.1575/1912/bco-dmo.775500.1', 'https://doi.org/10.48321/D1G59F','https://doi.org/10.48321/D14S38','https://doi.org/10.48321/D1101N','https://doi.org/10.48321/D1W88T','https://doi.org/10.48321/D1RG6W','https://doi.org/10.48321/D1MS3M','https://doi.org/10.48321/D1H010','https://doi.org/10.48321/D1C885','https://doi.org/10.48321/D17G67','https://doi.org/10.48321/D13S3Z','https://doi.org/10.48321/D1001B','https://doi.org/10.48321/D1V88H','https://doi.org/10.48321/D1QG6K','https://doi.org/10.48321/D1KS39','https://doi.org/10.48321/D1G01P'
],
value='https://doi.org/10.48321/D17G67',
description='Choose DOI:',
disabled=False,
)
display(f)
# ## Fetching Data
#
# We obtain all the data from the DataCite GraphQL API.
#
# +
# Generate the GraphQL query to retrieve up to 100 outputs of University of Oxford, with at least 100 views each.
query_params = {
"id" : f.value,
"maxOutputs": 100,
"minViews" : 100
}
query = gql("""query getOutputs($id: ID!)
{
dataManagementPlan(id: $id) {
id
name: titles(first:1) {
title
}
datasets: citations(query:"types.resourceTypeGeneral:Dataset") {
totalCount
nodes {
id: doi
name: titles(first:1) {
title
}
}
}
publications: citations(query:"types.resourceTypeGeneral:Text") {
totalCount
nodes {
id: doi
name: titles(first:1) {
title
}
}
}
producer: contributors(contributorType: "Producer") {
id
name
contributorType
}
fundingReferences {
id: funderIdentifier
name: funderName
award: awardUri
}
creators {
id
name
type
affiliation{
id
name
}
}
pis: contributors(contributorType: "ProjectLeader") {
id
name
contributorType
affiliation{
id
name
}
}
curators: contributors(contributorType: "DataCurator") {
id
name
type
affiliation{
id
name
}
}
}
}
""")
# -
def get_data():
return client.execute(query, variable_values=json.dumps(query_params))["dataManagementPlan"]
# ## Data Transformation
#
# Simple transformations are performed to convert the graphql response into an array that can be take by Vega.
def get_affiliation(series_element):
if len(series_element) == 0:
return "None"
return series_element[0]['name']
def add_node_attributes(dataframe, parent=2):
"""Modifies each item to include attributes needed for the node visulisation
Parameters:
dataframe (dataframe): A dataframe with all the itemss
parent (int): The id of the parent node
Returns:
dataframe:Returning vthe same dataframe with new attributes
"""
if (dataframe) is None:
return pd.DataFrame()
else:
# print(dataframe)
return (dataframe >>
mutate(
id = X.id,
tooltip = X.id,
parent = parent,
))
def create_node(array=[], parent=2):
"""creates a node for the chart and formats it
Parameters:
array (array): An array with all the itemss
parent (int): The id of the parent node
Returns:
dict:Dict with all the nodes
"""
# print(array)
if len(array) == 0:
return {}
else:
# return {} if (array) is None else array
df = add_node_attributes(pd.DataFrame(array,columns=array[0].keys()), parent)
return df.to_dict(orient='records')
def merge_nodes(dmpTitle,id,dataset=[],references=[],funders=[],orgs=[],people=[]):
"""Merges all the nodes lists
Parameters:
datasets (array): dataset nodes
funders (array): funders nodes
orgs (array): orgs nodes
people (array): people nodes
Returns:
array:Array with all the nodes
"""
dataset = [] if len(dataset) == 0 else dataset
references = [] if len(references) == 0 else references
funders = [] if len(funders) == 0 else funders
orgs = [] if len(orgs) == 0 else orgs
people = [] if len(people) == 0 else people
dmp = {"id":id, "name": dmpTitle}
datasets_node = {"id":2, "name": "Datasets", "parent":id}
references_node = {"id":6, "name": "Publications", "parent":id}
funders_node = {"id":3, "name": "Funders", "parent":id}
organisations_node = {"id":4, "name": "Organisations", "parent":id}
people_node = {"id":5, "name": "People", "parent":id}
nodes_list = [dmp, datasets_node, references_node, funders_node,organisations_node,people_node] + dataset + references + funders + orgs + people,
# return np.array(nodes_list, dtype=object)
return nodes_list[0]
def get_title(series_element):
if len(series_element) == 0:
return "None"
return series_element[0]['title']
def extract_titles(list):
if len(list) == 0:
return []
return (pd.DataFrame(list) >>
mutate(
name = X.name.apply(get_title)
)).to_dict('records')
# + tags=[]
data = get_data()
datasets = create_node(extract_titles(data["datasets"]["nodes"]),2)
references = create_node(extract_titles(data["publications"]["nodes"]),6)
orgs = create_node(data["producer"],4)
people = create_node(data["creators"] + data["pis"] + data["curators"],5)
dmp_title = str('"' + data["name"][0]["title"] + '"')
funders = create_node(data["fundingReferences"],3)
id = data["id"]
nodes = merge_nodes(" ",id, datasets, references, funders, orgs, people)
# -
# ## Visulization
#
# All transofrmed data is then feeded into a Vega specification for display.
#
#
#
#
def vega_template(data):
"""Injects data into the vega specification
Parameters:
data (array): Array of nodes
Returns:
VegaSpec:Specification with data
"""
return """
{
"$schema": "https://vega.github.io/schema/vega/v5.json",
"description": "An example of a radial layout for a node-link diagram of hierarchical data.",
"width": 1024,
"height": 720,
"padding": 5,
"autosize": "none",
"signals": [
{"name": "Chart", "value": """ + dmp_title + """, "bind": {"input": "url", "size":100}},
{"name": "labels", "value": true, "bind": {"input": "checkbox"}},
{
"name": "radius",
"value": 280,
"bind": {"input": "range", "min": 20, "max": 600}
},
{
"name": "extent",
"value": 360,
"bind": {"input": "range", "min": 0, "max": 360, "step": 1}
},
{
"name": "rotate",
"value": 0,
"bind": {"input": "range", "min": 0, "max": 360, "step": 1}
},
{
"name": "layout",
"value": "cluster",
"bind": {"input": "radio", "options": ["tidy", "cluster"]}
},
{
"name": "links",
"value": "orthogonal",
"bind": {
"input": "select",
"options": ["line", "curve", "diagonal", "orthogonal"]
}
},
{"name": "originX", "update": "width / 2"},
{"name": "originY", "update": "height / 2"}
],
"data": [
{
"name": "tree",
"values": """ + data + """,
"transform": [
{"type": "stratify", "key": "id", "parentKey": "parent"},
{
"type": "tree",
"method": {"signal": "layout"},
"size": [1, {"signal": "radius"}],
"as": ["alpha", "radius", "depth", "children"]
},
{
"type": "formula",
"expr": "(rotate + extent * datum.alpha + 270) % 360",
"as": "angle"
},
{"type": "formula", "expr": "PI * datum.angle / 180", "as": "radians"},
{
"type": "formula",
"expr": "inrange(datum.angle, [90, 270])",
"as": "leftside"
},
{
"type": "formula",
"expr": "originX + datum.radius * cos(datum.radians)",
"as": "x"
},
{
"type": "formula",
"expr": "originY + datum.radius * sin(datum.radians)",
"as": "y"
}
]
},
{
"name": "links",
"source": "tree",
"transform": [
{"type": "treelinks"},
{
"type": "linkpath",
"shape": {"signal": "links"},
"orient": "radial",
"sourceX": "source.radians",
"sourceY": "source.radius",
"targetX": "target.radians",
"targetY": "target.radius"
}
]
}
],
"scales": [
{
"name": "color",
"type": "linear",
"range": {"scheme": "viridis"},
"domain": {"data": "tree", "field": "depth"},
"zero": true
}
],
"marks": [
{
"type": "path",
"from": {"data": "links"},
"encode": {
"update": {
"x": {"signal": "originX"},
"y": {"signal": "originY"},
"path": {"field": "path"},
"stroke": {"value": "#ccc"}
}
}
},
{
"type": "symbol",
"from": {"data": "tree"},
"encode": {
"enter": {
"size": {"value": 300}, "stroke": {"value": "#fff"}
},
"update": {
"x": {"field": "x"},
"y": {"field": "y"},
"fill": {"scale": "color", "field": "depth"}
}
}
},
{
"type": "text",
"from": {"data": "tree"},
"encode": {
"enter": {
"text": {"field": "name"},
"fontSize": {"value": 12},
"baseline": {"value": "middle"},
"tooltip": {"signal":
"{'Identifier': datum.tooltip, 'Affiliation': datum.affiliation, 'Contribution': datum.contributorType, 'Award': datum.award}"}
},
"update": {
"x": {"field": "x"},
"y": {"field": "y"},
"dx": {"signal": "(datum.leftside ? -1 : 1) * 12"},
"align": {"signal": "datum.leftside ? 'right' : 'left'"},
"opacity": {"signal": "labels ? 1 : 0"}
}
}
}
]
}
"""
chart = alt.vega(json.loads(vega_template(json.dumps(nodes))))
# A series of sliders and option are included to interact with the visulisation is displayed. One can remove the labels, rotate the nodes, zoom in/out, and adjust the layout.
| dmp/user-story-single-dmp-connections.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # First Programming Language Tweets, etc
#
# There is a Twitter meme that is currently circulating that was started (I think) by [@cotufa82](https://twitter.com/cotufa82) where you list programming languages by particular categories including:
#
# * first language
# * had difficulties
# * most used
# * totally hate
# * most loved
# * for beginners
#
# I thought it could be interesting to try to collect these and tabulate them...
# <a href="https://twitter.com/cotufa82/status/1179601883448655874"><img style="border: thin solid black;" width=700 src="images/tweet.png"></a>
# We can use [twint](https://github.com/twintproject/twint) to download search results for the query ["First language" AND "Most used" AND "Most loved"](https://twitter.com/search?q=%22First%20language%22%20AND%20%22Most%20used%22%20AND%20%22Most%20loved%22) from Twitter's search results.
#
# Note: it skips doing the data collection if the data file is already present...
# +
import os
import twint
config = twint.Config()
config.Search = '"First language" AND "Most used" AND "Most loved"'
config.Store_json = True
config.Output = "data/languages.json"
config.Hide_output = True
# this is needed when using twint in a Jupyter Notebook
# or else you'll see a weird error about the event loop already running
import nest_asyncio
nest_asyncio.apply()
if not os.path.isfile('data/languages.json'):
twint.run.Search(config)
# -
# Collect the text of the tweets into a simple list by reading the line oriented JSON file that twint created or the search results.
# +
import json
tweets = []
for line in open('data/languages.json'):
t = json.loads(line)
tweets.append(t['tweet'])
print(len(tweets))
# -
tweets[15]
# Now for the nasty part: parsing the text of the tweets. Unfortunately twint strips new lines from the text of tweets, so it's a bit harder that it needs to be. After identifying the categories, each category can have multiple languages that are separated by commas, "and", slashes, etc. For example:
#
# For beginners: C, Java, ML, Scheme, Haskell, JavaScript
# +
import re
def parse(s):
results = {}
parts = [p[0] for p in re.findall(r'\d\. (.+?)(?=(\d\.)|( http)|( What about you)|$)', s)]
for part in parts:
try:
k, v = [s.strip() for s in re.split(r":|-", part)[0:2]]
category = k.lower()
langs = [l.lower() for l in re.split(' *(?:/|(?:and)|\&|,) *', v)]
results[category] = langs
except ValueError as e:
return None
return results
print('tweet text: ', tweets[15])
print()
print('parsed: ', parse(tweets[15]))
# -
# Now iterate through all the tweets and count them all by category:
# +
from collections import defaultdict, Counter
counter = defaultdict(Counter)
for tweet in tweets:
results = parse(tweet)
if results is not None:
for cat, langs in results.items():
for lang in langs:
counter[cat][lang] += 1
print(counter['first language'].most_common(25))
# +
import altair
import pandas
altair.renderers.enable('notebook')
def chart(title, counts, top=25):
df = pandas.DataFrame(counts.most_common(25), columns=['language', 'tweets'])
chart = altair.Chart(df, title=title).mark_bar().encode(
altair.X('tweets:Q'),
altair.Y('language:O', sort=altair.EncodingSortField(field='tweets', order='descending'))
)
return chart
chart('First Language', counter['first language']).display()
# -
chart('Had Difficulties', counter['had difficulties']).display()
chart('Most Used', counter['most used']).display()
chart('Totally Hate', counter['totally hate']).display()
chart('Most Loved', counter['most loved']).display()
chart('For Beginners', counter['for beginners']).display()
# People also invented their own categories. Let's see what the most invented cateogries were:
# +
alt_cats = Counter()
for cat_name in counter.keys():
if cat_name not in ['first language', 'had difficulties', 'most used',
'totally hate', 'most loved', 'for beginners']:
alt_cats[cat_name] = sum(counter[cat_name].values())
for cat, count in alt_cats.most_common(100):
print('{:>10} - {}'.format(count, cat))
# -
# As you can see there a bunch of expected typos. But there were a few categories that indicated that the desire to learn a language, like:
#
# - curious about
# - want to learn
# - need to learn
# - want to learn (dabbled in)
# - would like to explore
# - most excited about lately
# - next to try
# - want to try
# - i want to learn more
#
# So I decided to merge them, and see what they looked like:
# +
learn = counter['curious about'] + counter['want to learn'] + counter['need to learn'] + \
counter['want to learn (dabbled in)'] + counter['would like to explore'] + \
counter['most excited about lately'] + counter['next to try'] + counter['want to try'] + \
counter['i want to learn more']
chart('Learning', learn).display()
# -
| Languages.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Face Generation
#
# In this project, I have defined and train a DCGAN on a dataset of faces. The goal is to get a generator network to generate *new* images of faces that look as realistic as possible!
#
# The project will be broken down into a series of tasks from **loading in data to defining and training adversarial networks**. At the end of the notebook, you'll be able to visualize the results of your trained Generator to see how it performs; your generated samples should look like fairly realistic faces with small amounts of noise.
#
# ### Get the Data
#
# You'll be using the [CelebFaces Attributes Dataset (CelebA)](http://mmlab.ie.cuhk.edu.hk/projects/CelebA.html) to train your adversarial networks.
#
# This dataset is more complex than the number datasets (like MNIST or SVHN) you've been working with, and so, you should prepare to define deeper networks and train them for a longer time to get good results. It is suggested that you utilize a GPU for training.
#
# ### Pre-processed Data
#
# Since the project's main focus is on building the GANs, we've done *some* of the pre-processing for you. Each of the CelebA images has been cropped to remove parts of the image that don't include a face, then resized down to 64x64x3 NumPy images. Some sample data is show below.
#
# <img src='assets/processed_face_data.png' width=60% />
#
# > If you are working locally, you can download this data [by clicking here](https://s3.amazonaws.com/video.udacity-data.com/topher/2018/November/5be7eb6f_processed-celeba-small/processed-celeba-small.zip)
#
# This is a zip file that you'll need to extract in the home directory of this notebook for further loading and processing. After extracting the data, you should be left with a directory of data `processed_celeba_small/`
# can comment out after executing
# !unzip processed_celeba_small.zip
# +
data_dir = 'processed_celeba_small/'
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import pickle as pkl
import matplotlib.pyplot as plt
import numpy as np
import problem_unittests as tests
#import helper
# %matplotlib inline
# -
# ## Visualize the CelebA Data
#
# The [CelebA](http://mmlab.ie.cuhk.edu.hk/projects/CelebA.html) dataset contains over 200,000 celebrity images with annotations. Since you're going to be generating faces, you won't need the annotations, you'll only need the images. Note that these are color images with [3 color channels (RGB)](https://en.wikipedia.org/wiki/Channel_(digital_image)#RGB_Images) each.
#
# ### Pre-process and Load the Data
#
# Since the project's main focus is on building the GANs, we've done *some* of the pre-processing for you. Each of the CelebA images has been cropped to remove parts of the image that don't include a face, then resized down to 64x64x3 NumPy images. This *pre-processed* dataset is a smaller subset of the very large CelebA data.
#
# > There are a few other steps that you'll need to **transform** this data and create a **DataLoader**.
#
# #### ImageFolder
#
# To create a dataset given a directory of images, it's recommended that you use PyTorch's [ImageFolder](https://pytorch.org/docs/stable/torchvision/datasets.html#imagefolder) wrapper, with a root directory `processed_celeba_small/` and data transformation passed in.
# necessary imports
import torch
from torchvision import datasets
from torchvision import transforms
from torch.utils.data import DataLoader
def get_dataloader(batch_size, image_size, data_dir='processed_celeba_small/'):
"""
Batch the neural network data using DataLoader
:param batch_size: The size of each batch; the number of images in a batch
:param img_size: The square size of the image data (x, y)
:param data_dir: Directory where image data is located
:return: DataLoader with batched data
"""
# TODO: Implement function and return a dataloader
transform = transforms.Compose([
transforms.Resize(image_size),
transforms.ToTensor()
])
dataset = datasets.ImageFolder(data_dir, transform)
loader = DataLoader(dataset, batch_size=batch_size, shuffle=True)
return loader
# ## Create a DataLoader
#
# #### Exercise: Create a DataLoader `celeba_train_loader` with appropriate hyperparameters.
#
# Call the above function and create a dataloader to view images.
# * You can decide on any reasonable `batch_size` parameter
# * Your `image_size` **must be** `32`. Resizing the data to a smaller size will make for faster training, while still creating convincing images of faces!
# +
# Define function hyperparameters
batch_size = 32
img_size = 32
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# Call your function and get a dataloader
celeba_train_loader = get_dataloader(batch_size, img_size)
# -
# Next, you can view some images! You should seen square images of somewhat-centered faces.
#
# Note: You'll need to convert the Tensor images into a NumPy type and transpose the dimensions to correctly display an image, suggested `imshow` code is below, but it may not be perfect.
# +
# helper display function
def imshow(img):
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# obtain one batch of training images
dataiter = iter(celeba_train_loader)
images, _ = dataiter.next() # _ for no labels
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(20, 4))
plot_size=20
for idx in np.arange(plot_size):
ax = fig.add_subplot(2, plot_size/2, idx+1, xticks=[], yticks=[])
imshow(images[idx])
# -
# #### Exercise: Pre-process your image data and scale it to a pixel range of -1 to 1
#
# You need to do a bit of pre-processing; you know that the output of a `tanh` activated generator will contain pixel values in a range from -1 to 1, and so, we need to rescale our training images to a range of -1 to 1. (Right now, they are in a range from 0-1.)
# TODO: Complete the scale function
def scale(x, feature_range=(-1, 1)):
''' Scale takes in an image x and returns that image, scaled
with a feature_range of pixel values from -1 to 1.
This function assumes that the input x is already scaled from 0-1.'''
# assume x is scaled to (0, 1)
# scale to feature_range and return scaled x
range_min, range_max = feature_range
return (range_max - range_min) * x + range_min
return x
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
# check scaled range
# should be close to -1 to 1
img = images[0]
scaled_img = scale(img)
print('Min: ', scaled_img.min())
print('Max: ', scaled_img.max())
# -
# ---
# # Define the Model
#
# A GAN is comprised of two adversarial networks, a discriminator and a generator.
#
# ## Discriminator
#
# Your first task will be to define the discriminator. This is a convolutional classifier like you've built before, only without any maxpooling layers. To deal with this complex data, it's suggested you use a deep network with **normalization**. You are also allowed to create any helper functions that may be useful.
#
# #### Discriminator class
# * The inputs to the discriminator are 32x32x3 tensor images
# * The output should be a single value that will indicate whether a given image is real or fake
#
import torch.nn as nn
import torch.nn.functional as F
# +
def conv(inputs, outputs, kernal=4, stride=2, padding=1, batch_norm=True):
layers=[]
conv_layer = nn.Conv2d(inputs, outputs, kernal, stride, padding, bias=False)
layers.append(conv_layer)
if batch_norm:
layers.append(nn.BatchNorm2d(outputs))
return nn.Sequential(*layers)
def de_conv(inputs, outputs, kernal=4, stride=2, padding=1, batch_norm=True):
layers=[]
deconv_layer = nn.ConvTranspose2d(inputs, outputs, kernal, stride, padding, bias=False)
layers.append(deconv_layer)
if batch_norm:
layers.append(nn.BatchNorm2d(outputs))
return nn.Sequential(*layers)
# +
class Discriminator(nn.Module):
def __init__(self, conv_dim):
"""
Initialize the Discriminator Module
:param conv_dim: The depth of the first convolutional layer
"""
super(Discriminator, self).__init__()
# complete init function
self.conv_dim = conv_dim
self.leaky_relu = 0.2
self.conv1 = conv(3, conv_dim, batch_norm=False)
self.conv2 = conv(conv_dim, conv_dim*2)
self.conv3 = conv(conv_dim*2, conv_dim*4)
self.conv4 = conv(conv_dim*4, conv_dim*8)
self.fc = nn.Linear(conv_dim*32, 1)
self.convolution = [
self.conv1, self.conv2, self.conv3, self.conv4
]
def forward(self, x):
"""
Forward propagation of the neural network
:param x: The input to the neural network
:return: Discriminator logits; the output of the neural network
"""
# define feedforward behavior
for i,layer in enumerate(self.convolution):
x = F.leaky_relu(layer(x), self.leaky_relu)
x = x.view(-1, self.conv_dim*32)
x = self.fc(x)
return x
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_discriminator(Discriminator)
# -
# ## Generator
#
# The generator should upsample an input and generate a *new* image of the same size as our training data `32x32x3`. This should be mostly transpose convolutional layers with normalization applied to the outputs.
#
# #### Generator class
# * The inputs to the generator are vectors of some length `z_size`
# * The output should be a image of shape `32x32x3`
# +
class Generator(nn.Module):
def __init__(self, z_size, conv_dim):
"""
Initialize the Generator Module
:param z_size: The length of the input latent vector, z
:param conv_dim: The depth of the inputs to the *last* transpose convolutional layer
"""
super(Generator, self).__init__()
# complete init function
self.conv_dim = conv_dim
self.de_conv1 = de_conv(conv_dim*8, conv_dim*4)
self.de_conv2 = de_conv(conv_dim*4, conv_dim*2)
self.de_conv3 = de_conv(conv_dim*2, conv_dim)
self.de_conv4 = de_conv(conv_dim, 3, batch_norm=False)
self.fc = nn.Linear(z_size, conv_dim * 32)
self.de_convolution = [
self.de_conv1, self.de_conv2, self.de_conv3, self.de_conv4
]
def forward(self, x):
"""
Forward propagation of the neural network
:param x: The input to the neural network
:return: A 32x32x3 Tensor image as output
"""
# define feedforward behavior
x = self.fc(x)
batch_size = x.shape[0]
x = x.view(batch_size, self.conv_dim*8, 2, 2)
for i, layer in enumerate(self.de_convolution):
if (i < len(self.de_convolution)-1):
x = F.relu(layer(x))
else:
x = layer(x)
x = torch.tanh(x)
return x
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
tests.test_generator(Generator)
# -
# ## Initialize the weights of your networks
#
# To help your models converge, you should initialize the weights of the convolutional and linear layers in your model. From reading the [original DCGAN paper](https://arxiv.org/pdf/1511.06434.pdf), they say:
# > All weights were initialized from a zero-centered Normal distribution with standard deviation 0.02.
#
# So, your next task will be to define a weight initialization function that does just this!
#
# You can refer back to the lesson on weight initialization or even consult existing model code, such as that from [the `networks.py` file in CycleGAN Github repository](https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix/blob/master/models/networks.py) to help you complete this function.
#
# #### weight initialization function
#
# * This should initialize only **convolutional** and **linear** layers
# * Initialize the weights to a normal distribution, centered around 0, with a standard deviation of 0.02.
# * The bias terms, if they exist, may be left alone or set to 0.
def weights_init_normal(m):
"""
Applies initial weights to certain layers in a model .
The weights are taken from a normal distribution
with mean = 0, std dev = 0.02.
:param m: A module or layer in a network
"""
# classname will be something like:
# `Conv`, `BatchNorm2d`, `Linear`, etc.
classname = m.__class__.__name__
# TODO: Apply initial weights to convolutional and linear layers
if 'Conv' in classname:
torch.nn.init.normal_(m.weight.data, 0.0, 0.02)
elif 'Linear' in classname:
torch.nn.init.normal_(m.weight.data, 0.0, 0.02)
# ## Build complete network
#
# Define your models' hyperparameters and instantiate the discriminator and generator from the classes defined above. Make sure you've passed in the correct input arguments.
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
def build_network(d_conv_dim, g_conv_dim, z_size):
# define discriminator and generator
D = Discriminator(d_conv_dim)
G = Generator(z_size=z_size, conv_dim=g_conv_dim)
# initialize model weights
D.apply(weights_init_normal)
G.apply(weights_init_normal)
print(D)
print()
print(G)
return D, G
# #### Exercise: Define model hyperparameters
# +
# Define model hyperparams
d_conv_dim = 32
g_conv_dim = 32
z_size = 100
"""
DON'T MODIFY ANYTHING IN THIS CELL THAT IS BELOW THIS LINE
"""
D, G = build_network(d_conv_dim, g_conv_dim, z_size)
# -
# ### Training on GPU
#
# Check if you can train on GPU. Here, we'll set this as a boolean variable `train_on_gpu`. Later, you'll be responsible for making sure that
# >* Models,
# * Model inputs, and
# * Loss function arguments
#
# Are moved to GPU, where appropriate.
# +
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
import torch
# Check for a GPU
train_on_gpu = torch.cuda.is_available()
if not train_on_gpu:
print('No GPU found. Please use a GPU to train your neural network.')
else:
print('Training on GPU!')
# -
# ---
# ## Discriminator and Generator Losses
#
# Now we need to calculate the losses for both types of adversarial networks.
#
# ### Discriminator Losses
#
# > * For the discriminator, the total loss is the sum of the losses for real and fake images, `d_loss = d_real_loss + d_fake_loss`.
# * Remember that we want the discriminator to output 1 for real images and 0 for fake images, so we need to set up the losses to reflect that.
#
#
# ### Generator Loss
#
# The generator loss will look similar only with flipped labels. The generator's goal is to get the discriminator to *think* its generated images are *real*.
#
# #### Real and fake loss functions
#
# **You may choose to use either cross entropy or a least squares error loss to complete the following `real_loss` and `fake_loss` functions.**
# +
def real_loss(D_out, smooth=False):
'''Calculates how close discriminator outputs are to being real.
param, D_out: discriminator logits
return: real loss'''
batch_size = D_out.shape[0]
if smooth:
labels = torch.ones(batch_size)*0.9
else:
labels = torch.ones(batch_size)
labels = labels.to(device)
criterion = nn.BCEWithLogitsLoss()
loss = criterion(D_out.squeeze(), labels)
return loss
def fake_loss(D_out):
'''Calculates how close discriminator outputs are to being fake.
param, D_out: discriminator logits
return: fake loss'''
batch_size = D_out.shape[0]
labels = torch.zeros(batch_size)
labels = labels.to(device)
criterion = nn.BCEWithLogitsLoss()
loss = criterion(D_out.squeeze(), labels)
return loss
# -
# ## Optimizers
#
# #### Optimizers for your Discriminator (D) and Generator (G)
#
# Define optimizers for your models with appropriate hyperparameters.
# +
import torch.optim as optim
# Create optimizers for the discriminator D and generator G
beta1 = 0.5
beta2 = 0.99
lr = 0.0002
d_optimizer = optim.Adam(D.parameters(), lr, betas=(beta1, beta2))
g_optimizer = optim.Adam(G.parameters(), lr, betas=(beta1, beta2))
# -
# ---
# ## Training
#
# Training will involve alternating between training the discriminator and the generator. You'll use your functions `real_loss` and `fake_loss` to help you calculate the discriminator losses.
#
# * You should train the discriminator by alternating on real and fake images
# * Then the generator, which tries to trick the discriminator and should have an opposing loss function
#
#
# #### Saving Samples
#
# You've been given some code to print out some loss statistics and save some generated "fake" samples.
# #### Training function
#
# Keep in mind that, if you've moved your models to GPU, you'll also have to move any model inputs to GPU.
def train(D, G, n_epochs, print_every=50):
'''Trains adversarial networks for some number of epochs
param, D: the discriminator network
param, G: the generator network
param, n_epochs: number of epochs to train for
param, print_every: when to print and record the models' losses
return: D and G losses'''
# move models to GPU
if train_on_gpu:
D.cuda()
G.cuda()
# keep track of loss and generated, "fake" samples
samples = []
losses = []
# Get some fixed data for sampling. These are images that are held
# constant throughout training, and allow us to inspect the model's performance
sample_size=16
fixed_z = np.random.uniform(-1, 1, size=(sample_size, z_size))
fixed_z = torch.from_numpy(fixed_z).float()
# move z to GPU if available
if train_on_gpu:
fixed_z = fixed_z.cuda()
# epoch training loop
for epoch in range(n_epochs):
# batch training loop
for batch_i, (real_images, _) in enumerate(celeba_train_loader):
batch_size = real_images.size(0)
real_images = scale(real_images)
# ===============================================
# YOUR CODE HERE: TRAIN THE NETWORKS
# ===============================================
# 1. Train the discriminator on real and fake images
#real
d_optimizer.zero_grad()
real_images = real_images.to(device)
D_real = D(real_images)
d_real_loss = real_loss(D_real)
#fake
z = np.random.uniform(-1, 1, size=(batch_size, z_size))
z = torch.from_numpy(z).float()
# move x to GPU, if available
z = z.to(device)
fake_images = G(z)
D_fake = D(fake_images)
d_fake_loss = fake_loss(D_fake)
d_loss = d_real_loss + d_fake_loss
d_loss.backward()
d_optimizer.step()
# 2. Train the generator with an adversarial loss
g_optimizer.zero_grad()
#fake
z = np.random.uniform(-1, 1, size=(batch_size, z_size))
z = torch.from_numpy(z).float()
z = z.to(device)
fake_images = G(z)
D_fake = D(fake_images)
g_loss = real_loss(D_fake, smooth=True)
g_loss.backward()
g_optimizer.step()
# ===============================================
# END OF YOUR CODE
# ===============================================
# Print some loss stats
if batch_i % print_every == 0:
# append discriminator loss and generator loss
losses.append((d_loss.item(), g_loss.item()))
# print discriminator and generator loss
print('Epoch [{:5d}/{:5d}] | d_loss: {:6.4f} | g_loss: {:6.4f}'.format(
epoch+1, n_epochs, d_loss.item(), g_loss.item()))
## AFTER EACH EPOCH##
# this code assumes your generator is named G, feel free to change the name
# generate and save sample, fake images
G.eval() # for generating samples
samples_z = G(fixed_z)
samples.append(samples_z)
G.train() # back to training mode
# Save training generator samples
with open('train_samples.pkl', 'wb') as f:
pkl.dump(samples, f)
# finally return losses
return losses
# Set your number of training epochs and train your GAN!
# set number of epochs
n_epochs = 10
device = 'cuda' if torch.cuda.is_available() else 'cpu'
"""
DON'T MODIFY ANYTHING IN THIS CELL
"""
# call training function
losses = train(D, G, n_epochs=n_epochs)
# ## Training loss
#
# Plot the training losses for the generator and discriminator, recorded after each epoch.
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator', alpha=0.5)
plt.plot(losses.T[1], label='Generator', alpha=0.5)
plt.title("Training Losses")
plt.legend()
# ## Generator samples from training
#
# View samples of images from the generator, and answer a question about the strengths and weaknesses of your trained models.
# helper function for viewing a list of passed in sample images
def view_samples(epoch, samples):
fig, axes = plt.subplots(figsize=(16,4), nrows=2, ncols=8, sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
img = img.detach().cpu().numpy()
img = np.transpose(img, (1, 2, 0))
img = ((img + 1)*255 / (2)).astype(np.uint8)
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
im = ax.imshow(img.reshape((32,32,3)))
# Load samples from generator, taken while training
with open('train_samples.pkl', 'rb') as f:
samples = pkl.load(f)
_ = view_samples(-1, samples)
# ## END
| dlnd_face_generation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (Data Science)
# language: python
# name: python3__SAGEMAKER_INTERNAL__arn:aws:sagemaker:ap-northeast-2:806072073708:image/datascience-1.0
# ---
# # Part 1 : Data Preparation, Process, and Store Features
# <a id='all-up-overview'></a>
#
# ## [Overview](./0-AutoClaimFraudDetection.ipynb)
# * [Notebook 0: Overview, Architecture and Data Exploration](./0-AutoClaimFraudDetection.ipynb)
# * **[Notebook 1: Data Preparation, Process, and Store Features](./1-data-prep-e2e.ipynb)**
# * **[Architecture](#arch)**
# * **[Getting started](#aud-getting-started)**
# * **[DataSets](#aud-datasets)**
# * **[SageMaker Feature Store](#aud-feature-store)**
# * **[Create train and test datasets](#aud-dataset)**
# * [Notebook 2: Train, Check Bias, Tune, Record Lineage, and Register a Model](./2-lineage-train-assess-bias-tune-registry-e2e.ipynb)
# * [Notebook 3: Mitigate Bias, Train New Model, Store in Registry](./3-mitigate-bias-train-model2-registry-e2e.ipynb)
# * [Notebook 4: Deploy Model, Run Predictions](./4-deploy-run-inference-e2e.ipynb)
# * [Notebook 5: Create and Run an End-to-End Pipeline to Deploy the Model](./5-pipeline-e2e.ipynb)
# 이 노트북의 목적은 ML 수명주기(lifecycle)의 데이터 준비 단계를 수행하는 것입니다. 주요 데이터 랭글링(data wrangling), 데이터 수집 및 다중 변환(multiple transformatio)이 SageMaker Studio Data Wrangler GUI를 통해 수행됩니다.
#
# 이 노트북에서는 raw 데이터에 대한 변환을 정의하는 `.flow` 파일을 가져옵니다. `.csv` 파일로 S3 버킷에 저장된 raw 데이터에 이러한 변환을 적용하는 SageMaker Processing job을 사용하여 적용합니다.
# <a id='arch'> </a>
# ## Architecture for Data Prep, Process and Store Features
# [overview](#all-up-overview)
# ___
# 
# ### Install required and/or update third-party libraries
# +
import sys
import IPython
install_needed = False
if install_needed:
print("installing deps and restarting kernel")
# !python -m pip install -Uq pip
# !python -m pip install -q awscli==1.20.25 awswrangler==2.10.0 imbalanced-learn==0.8.0 sagemaker==2.54.0 boto3==1.18.25
IPython.Application.instance().kernel.do_shutdown(True)
# -
# ### Loading stored variables
#
# 이전에 이 노트북을 실행한 경우, AWS에서 생성한 리소스를 재사용할 수 있습니다. 아래 셀을 실행하여 이전에 생성된 변수를 로드합니다. 기존 변수의 출력물이 표시되어야 합니다. 인쇄된 내용이 보이지 않으면 노트북을 처음 실행한 것일 수 있습니다.
# +
# #%store -z
# -
# %store -r
# %store
# **<font color='red'>Important</font>: StoreMagic 명령을 사용하여 변수를 검색하려면 이전 노트북을 실행해야 합니다.**
# ### Import libraries
# +
import json
import time
import boto3
import string
import sagemaker
import pandas as pd
import awswrangler as wr
from sagemaker.feature_store.feature_group import FeatureGroup
# -
# <a id='aud-getting-started'></a>
# ## Getting started: Creating Resources
#
# [overview](#all-up-overview)
# ___
#
# 이 노트북을 성공적으로 실행하려면, 몇 가지 AWS 리소스를 생성해야 합니다. 먼저 이 자습서의 모든 데이터를 저장하기 위해 S3 버킷이 생성됩니다. 생성된 후에는 IAM 콘솔을 사용하여 AWS Glue role을 생성한 다음, 이 노트북에 대한 FeatureStore 액세스를 허용하는 policy를 S3 버킷에 연결해야 합니다. 이미 이 노트북을 실행하고 중단한 부분을 선택하는 경우 아래 셀을 실행하면 추가 리소스를 생성하는 대신, 이미 생성한 리소스를 선택해야 합니다.
# #### Add FeatureStore policy to Studio's execution role
#
# 
#
# 1. 별도의 브라우저 탭에서 AWS 콘솔의 IAM 섹션으로 이동합니다.
# 2. Roles 섹션으로 이동하여 SageMaker Studio user에서 사용 중인 실행 role을 선택합니다.
# * 어떤 role을 사용하고 있는지 확실하지 않은 경우, 아래 셀을 실행하여 출력하세요.
# 3. 이 role에 <font color='green'> AmazonSageMakerFeatureStoreAccess </font> policy를 연결합니다. 연결되면 변경 사항이 즉시 적용됩니다.
print('SageMaker Role:', sagemaker.get_execution_role().split('/')[-1])
# ### Set region, boto3 and SageMaker SDK variables
#You can change this to a region of your choice
import sagemaker
region = sagemaker.Session().boto_region_name
print("Using AWS Region: {}".format(region))
# +
boto3.setup_default_session(region_name=region)
boto_session = boto3.Session(region_name=region)
s3_client = boto3.client('s3', region_name=region)
sagemaker_boto_client = boto_session.client('sagemaker')
sagemaker_session = sagemaker.session.Session(
boto_session=boto_session,
sagemaker_client=sagemaker_boto_client)
# -
# Note: SageMaker Studio 또는 SageMaker Classic 노트북에서 이 노트북을 실행하지 않는 경우, SageMakerFullAccess 및 SageMakerFeatureStoreFullAccess가 있는 AWS role로 sagemaker_execution_role_name을 인스턴스화해야 합니다.
# +
sagemaker_execution_role_name = 'AmazonSageMaker-ExecutionRole-20210107T234882'
try:
sagemaker_role = sagemaker.get_execution_role()
except ValueError:
iam = boto3.client('iam')
sagemaker_role = iam.get_role(RoleName=sagemaker_execution_role_name)['Role']['Arn']
print(f"\n instantiating sagemaker_role with supplied role name : {sagemaker_role}")
account_id = boto3.client('sts').get_caller_identity()["Account"]
# -
# ### Create a directory in the SageMaker default bucket for this tutorial
if 'bucket' not in locals():
bucket = sagemaker_session.default_bucket()
prefix = 'fraud-detect-demo'
# %store bucket
# %store prefix
print(f'Creating bucket: {bucket}...')
# 이미 존재하는 자체 S3 버킷을 사용하려면 아래 코드 셀의 주석 처리를 제거하고 다음 예제 코드를 활용하세요.
'''
try:
s3_client.create_bucket(Bucket=bucket, ACL='private', CreateBucketConfiguration={'LocationConstraint': region})
print('Create S3 bucket: SUCCESS')
except Exception as e:
if e.response['Error']['Code'] == 'BucketAlreadyOwnedByYou':
print(f'Using existing bucket: {bucket}/{prefix}')
else:
raise(e)
'''
# +
#======> Tons of output_paths
traing_job_output_path = f's3://{bucket}/{prefix}/training_jobs'
bias_report_1_output_path = f's3://{bucket}/{prefix}/clarify-bias-1'
bias_report_2_output_path = f's3://{bucket}/{prefix}/clarify-bias-2'
explainability_output_path = f's3://{bucket}/{prefix}/clarify-explainability'
train_data_uri = f's3://{bucket}/{prefix}/data/train/train.csv'
test_data_uri = f's3://{bucket}/{prefix}/data/test/test.csv'
#=======> variables used for parameterizing the notebook run
train_instance_count = 1
train_instance_type = "ml.m4.xlarge"
claify_instance_count = 1
clairfy_instance_type = 'ml.c5.xlarge'
predictor_instance_count = 1
predictor_instance_type = "ml.c5.xlarge"
# -
# ### Upload raw data to S3
#
# SageMaker Data Wrangler를 사용하여 raw 데이터를 전처리하려면, 데이터가 S3에 있어야 합니다.
s3_client.upload_file(Filename='data/claims.csv', Bucket=bucket, Key=f'{prefix}/data/raw/claims.csv')
s3_client.upload_file(Filename='data/customers.csv', Bucket=bucket, Key=f'{prefix}/data/raw/customers.csv')
# ### Update attributes within the `.flow` file
#
# DataWrangler는 `.flow` 파일을 생성합니다. 여기에는 랭글링 중에 사용된 S3 버킷에 대한 참조가 포함됩니다. 이것은 이 노트북에 기본으로 설정되어 있는 것과 다를 수 있습니다. 예를 들어 다른 사람이 랭글링을 수행한 경우, 해당 버킷에 액세스할 수 없으므로 실제로 로드할 수 있도록 자신의 S3 버킷을 가리킴으로써 `.flow` 파일을 Wrangler에 저장하거나 데이터에 액세스할 수 있습니다.
#
# 아래 셀을 실행한 후 `claim.flow` 및 `customers.flow` 파일을 열고 데이터를 S3로 내보내거나 제공된 `data/claims_preprocessed.csv` 및 `data/customers_preprocessed.csv` 파일을 사용하여 가이드를 계속할 수 있습니다.
# +
claims_flow_template_file = "claims_flow_template"
with open(claims_flow_template_file, 'r') as f:
variables = {'bucket': bucket, 'prefix': prefix}
template = string.Template(f.read())
claims_flow = template.substitute(variables)
claims_flow = json.loads(claims_flow)
with open('claims.flow', 'w') as f:
json.dump(claims_flow, f)
customers_flow_template_file = "customers_flow_template"
with open(customers_flow_template_file, 'r') as f:
variables = {'bucket': bucket, 'prefix': prefix}
template = string.Template(f.read())
customers_flow = template.substitute(variables)
customers_flow = json.loads(customers_flow)
with open('customers.flow', 'w') as f:
json.dump(customers_flow, f)
# -
# ### Load preprocessed data from Data Wrangler job
#
# `claim.flow` 및 `customers.flow` 에서 Data Wrangler job을 실행한 경우 여기에서 전처리된 데이터를 로드할 수 있습니다. Data Wrangler job을 실행하지 않은 경우에도 이 예제의 `/data` 디렉토리에서 미리 만들어진 데이터셋을 로드하여 시작할 수 있습니다.
# <a id='aud-datasets'></a>
# ## DataSets and Feature Types
# [overview](#all-up-overview)
# ___
# +
claims_dtypes = {
"policy_id": int,
"incident_severity": int,
"num_vehicles_involved": int,
"num_injuries": int,
"num_witnesses": int,
"police_report_available": int,
"injury_claim": float,
"vehicle_claim": float,
"total_claim_amount": float,
"incident_month": int,
"incident_day": int,
"incident_dow": int,
"incident_hour": int,
"fraud": int,
"driver_relationship_self": int,
"driver_relationship_na": int,
"driver_relationship_spouse": int,
"driver_relationship_child": int,
"driver_relationship_other": int,
"incident_type_collision": int,
"incident_type_breakin": int,
"incident_type_theft": int,
"collision_type_front": int,
"collision_type_rear": int,
"collision_type_side": int,
"collision_type_na": int,
"authorities_contacted_police": int,
"authorities_contacted_none": int,
"authorities_contacted_fire": int,
"authorities_contacted_ambulance": int,
"event_time": float,
}
customers_dtypes = {
"policy_id": int,
"customer_age": int,
"customer_education": int,
"months_as_customer": int,
"policy_deductable": int,
"policy_annual_premium": int,
"policy_liability": int,
"auto_year": int,
"num_claims_past_year": int,
"num_insurers_past_5_years": int,
"customer_gender_male": int,
"customer_gender_female": int,
"policy_state_ca": int,
"policy_state_wa": int,
"policy_state_az": int,
"policy_state_or": int,
"policy_state_nv": int,
"policy_state_id": int,
"event_time": float,
}
# +
#======> This is your DataFlow output path if you decide to redo the work in DataFlow on your own
flow_output_path = 'YOUR_PATH_HERE'
try:
# this will try to load the exported dataframes from the claims and customers .flow files
claims_s3_path = f'{flow_output_path}/claims_output'
customers_s3_path = f'{flow_output_path}/customers_output'
claims_preprocessed = wr.s3.read_csv(
path=claims_s3_path,
dataset=True,
index_col=0,
dtype=claims_dtypes)
customers_preprocessed = wr.s3.read_csv(
path=customers_s3_path,
dataset=True,
index_col=0,
dtype=customers_dtypes)
except:
# if the Data Wrangler job was not run, the claims and customers dataframes will be loaded from local copies
timestamp = pd.to_datetime('now').timestamp()
print('Unable to load Data Wrangler output. Loading pre-made dataframes...')
claims_preprocessed = pd.read_csv(
filepath_or_buffer='data/claims_preprocessed.csv',
dtype=claims_dtypes)
# a timestamp column is required by the feature store, so one is added with a current timestamp
claims_preprocessed['event_time'] = timestamp
customers_preprocessed = pd.read_csv(
filepath_or_buffer='data/customers_preprocessed.csv',
dtype=customers_dtypes)
customers_preprocessed['event_time'] = timestamp
print('Complete')
# -
# 이제 올바른 데이터 유형과 함께 고객 및 청구 데이터를 포함하는 Pandas 데이터프레임이 있습니다. Dat Wrangler가 피쳐를 원-핫 인코딩 피쳐로 인코딩할 때, 결과 피쳐에 대한 데이터 유형을 float로 기본 설정합니다.
#
# <font color ='red'> Note: </font> : Data Wrangler에서 생성 된 범주형 피쳐에 대한 데이터 유형을 명시적으로 변환하는 이유는, Clarify에서 범주형 변수로 처리할 수 있도록 정수 유형인지 확인하기 위한 것입니다.
# <a id='aud-feature-store'></a>
# ## SageMaker Feature Store
#
# [overview](#all-up-overview)
# ___
#
# Amazon SageMaker Feature Store는 피쳐를 저장하고 액세스할 수 있는 전용 리포지토리이므로, 팀 간에 이름을 지정하고 구성하고 재사용하기가 훨씬 쉽습니다. SageMaker Feature Store는 추가 코드를 작성하거나 피쳐를 일관되게 유지하기 위해 수동 프로세스를 생성할 필요 없이 훈련 및 실시간 추론 중 피쳐에 대한 통합 저장소를 제공합니다. SageMaker Feature Store는 저장된 피쳐의 메타데이터 (예: 피쳐 이름 또는 버전 번호)를 추적하므로 대화형 쿼리 서비스인 Amazon Athena를 사용하여 배치 또는 실시간으로 올바른 속성에 대한 피쳐를 쿼리할 수 있습니다. SageMaker Feature Store는 추론 중에 새 데이터가 생성될 때 단일 리포지토리가 업데이트되어 모델이 훈련 및 추론 중에 사용할 수 있도록 항상 새로운 피쳐를 사용할 수 있기 때문에 피쳐를 업데이트된 상태로 유지합니다.
#
# 피쳐 저장소는 S3에 저장된 오프라인 컴포넌트와 지연 시간이 짧은 데이터베이스에 저장된 온라인 컴포넌트로 구성됩니다. 온라인 데이터베이스는 선택 사항이지만 추론에서 사용할 수 있는 추가 피쳐가 필요한 경우 매우 유용합니다. 이 섹션에서는 보험 청구 및 고객 데이터셋에 대한 피쳐 그룹을 생성합니다. 보험 청구 및 고객 데이터를 각 피쳐 그룹에 삽입한 후, Athena를 사용하여 오프라인 스토어를 쿼리하여 훈련 데이터 세트를 구축해야 합니다.
#
# SageMaker 피쳐 저장소에 대한 자세한 내용은 [SageMaker Developer Guide](https://docs.aws.amazon.com/sagemaker/latest/dg/feature-store.html)를 참조해 주십시오.
# +
featurestore_runtime = boto_session.client(
service_name='sagemaker-featurestore-runtime',
region_name=region
)
feature_store_session = sagemaker.Session(
boto_session=boto_session,
sagemaker_client=sagemaker_boto_client,
sagemaker_featurestore_runtime_client=featurestore_runtime
)
# -
# ### Configure the feature groups
#
# 각 피쳐의 데이터 유형은 데이터프레임을 전달하고, 적절한 데이터 유형을 유추하여 설정됩니다. 피쳐 데이터 유형은 설정 변수(config variable)를 통해 설정할 수도 있지만, 피쳐 그룹에 수집될 때 Pandas 데이터프레임의 해당 Python 데이터 유형과 일치해야 합니다.
# +
claims_fg_name = f'{prefix}-claims'
customers_fg_name = f'{prefix}-customers'
# %store claims_fg_name
# %store customers_fg_name
claims_feature_group = FeatureGroup(
name=claims_fg_name,
sagemaker_session=feature_store_session)
customers_feature_group = FeatureGroup(
name=customers_fg_name,
sagemaker_session=feature_store_session)
claims_feature_group.load_feature_definitions(data_frame=claims_preprocessed);
customers_feature_group.load_feature_definitions(data_frame=customers_preprocessed);
# -
# ### Create the feature groups
# 데이터프레임에서 필요한 레코드 식별자 및 이벤트 타임 피쳐에 해당하는 컬럼을 피쳐 그룹에 알려야 합니다.
print(f"{customers_fg_name} -- {claims_fg_name} are the feature group names in use")
# +
record_identifier_feature_name = 'policy_id'
event_time_feature_name = 'event_time'
try:
print(f"\n Using s3://{bucket}/{prefix}")
claims_feature_group.create(
s3_uri=f"s3://{bucket}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name=event_time_feature_name,
role_arn=sagemaker_role,
enable_online_store=True
)
print(f'Create "claims" feature group: SUCCESS')
except Exception as e:
code = e.response.get('Error').get('Code')
if code == 'ResourceInUse':
print(f'Using existing feature group: {claims_fg_name}')
else:
raise(e)
try:
customers_feature_group.create(
s3_uri=f"s3://{bucket}/{prefix}",
record_identifier_name=record_identifier_feature_name,
event_time_feature_name=event_time_feature_name,
role_arn=sagemaker_role,
enable_online_store=True
)
print(f'Create "customers" feature group: SUCCESS')
except Exception as e:
code = e.response.get('Error').get('Code')
if code == 'ResourceInUse':
print(f'Using existing feature group: {customers_fg_name}')
else:
raise(e)
# -
# ### Wait until feature group creation has fully completed
# +
def wait_for_feature_group_creation_complete(feature_group):
status = feature_group.describe().get("FeatureGroupStatus")
while status == "Creating":
print("Waiting for Feature Group Creation")
time.sleep(5)
status = feature_group.describe().get("FeatureGroupStatus")
if status != "Created":
raise RuntimeError(f"Failed to create feature group {feature_group.name}")
print(f"FeatureGroup {feature_group.name} successfully created.")
wait_for_feature_group_creation_complete(feature_group=claims_feature_group)
wait_for_feature_group_creation_complete(feature_group=customers_feature_group)
# -
# ### Ingest records into the Feature Groups
#
# 피쳐 그룹이 생성된 후, PutRecord API를 사용하여 각 store에 데이터를 넣을 수 있습니다. 이 API는 높은 TPS를 처리할 수 있으며 다른 스트림에서 호출되도록 설계되었습니다. 이러한 모든 Put 요청의 데이터는 버퍼링되어 s3에 chunk로 기록됩니다. 수집 후 몇 분 이내에 파일이 오프라인 저장소에 기록됩니다.
# +
if 'claims_table' in locals():
print("You may have already ingested the data into your Feature Groups. If you'd like to do this again, you can run the ingest methods outside of the 'if/else' statement.")
else:
claims_feature_group.ingest(data_frame=claims_preprocessed, max_workers=3, wait=True);
customers_feature_group.ingest(data_frame=customers_preprocessed, max_workers=3, wait=True);
# -
# ### Wait for offline store data to become available
# 아래 코드 셀은 약 5-8분이 소요됩니다.
# +
if 'claims_table' not in locals():
claims_table = (
claims_feature_group.describe()["OfflineStoreConfig"]["DataCatalogConfig"]["TableName"]
)
if 'customers_table' not in locals():
customers_table = (
customers_feature_group.describe()["OfflineStoreConfig"]["DataCatalogConfig"]["TableName"]
)
claims_feature_group_s3_prefix = (
f"{prefix}/{account_id}/sagemaker/{region}/offline-store/{claims_table}/data"
)
customers_feature_group_s3_prefix = (
f"{prefix}/{account_id}/sagemaker/{region}/offline-store/{customers_table}/data"
)
offline_store_contents = None
while offline_store_contents is None:
objects_in_bucket = s3_client.list_objects(
Bucket=bucket, Prefix=customers_feature_group_s3_prefix
)
if "Contents" in objects_in_bucket and len(objects_in_bucket["Contents"]) > 1:
offline_store_contents = objects_in_bucket["Contents"]
else:
print("Waiting for data in offline store...")
time.sleep(60)
print("\nData available.")
# -
# <a id='aud-dataset'></a>
# ## Create train and test datasets
#
# [overview](#all-up-overview)
# ___
#
# 오프라인 스토어에서 데이터를 사용할 수있게 되면 자동으로 카탈로그화되고 Athena 테이블에 로드됩니다. (기본적으로 수행되지만 끌 수 있습니다.) 훈련 및 테스트 데이터셋을 구축하기 위해 Athena에서 생성된 Claims 및 Customers 테이블을 조인하는 SQL 쿼리를 실행합니다.
# +
claims_query = claims_feature_group.athena_query()
customers_query = customers_feature_group.athena_query()
claims_table = claims_query.table_name
customers_table = customers_query.table_name
database_name = customers_query.database
# %store claims_table
# %store customers_table
# %store database_name
feature_columns = list(set(claims_preprocessed.columns) ^ set(customers_preprocessed.columns))
feature_columns_string = ", ".join(f'"{c}"' for c in feature_columns)
feature_columns_string = f'"{claims_table}".policy_id as policy_id, ' + feature_columns_string
query_string = f"""
SELECT DISTINCT {feature_columns_string}
FROM "{claims_table}" LEFT JOIN "{customers_table}"
ON "{claims_table}".policy_id = "{customers_table}".policy_id
"""
# -
claims_query.run(query_string=query_string, output_location=f"s3://{bucket}/{prefix}/query_results")
claims_query.wait()
dataset = claims_query.as_dataframe()
dataset.head()
dataset.to_csv("./data/claims_customer.csv")
# +
col_order = ["fraud"] + list(dataset.drop(["fraud", "policy_id"], axis=1).columns)
# %store col_order
train = dataset.sample(frac=0.80, random_state=0)[col_order]
test = dataset.drop(train.index)[col_order]
# -
# ### Write train, test data to S3
#
# 훈련 데이터셋 및 테스트 데이터셋을 S3로 업로드합니다.
train.to_csv("data/train.csv", index=False)
test.to_csv("data/test.csv", index=False)
dataset.to_csv("data/dataset.csv", index=True)
s3_client.upload_file(Filename='data/train.csv', Bucket=bucket, Key=f'{prefix}/data/train/train.csv')
s3_client.upload_file(Filename='data/test.csv', Bucket=bucket, Key=f'{prefix}/data/test/test.csv')
# %store train_data_uri
# %store test_data_uri
train.head(5)
test.head(5)
# ___
#
# ### Next Notebook: [Train, Check Bias, Tune, Record Lineage, Register Model](./2-lineage-train-assess-bias-tune-registry-e2e.ipynb)
| 1-data-prep-e2e.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import numpy as np
from typing import List, Tuple
# + tags=["parameters"]
#### Parameters
spot_filename:str = "/tmp/index.csv"
options_filename:str = "../../data/nifty_options_eod.h5"
output_filename:str = '/tmp/output.csv' # Friendly name for output
step:int = 100
options:Tuple[Tuple[str, float]] = (
('p', 0),
('p', 0.01),
('p', 0.02),
('p', 0.03),
('c', 0.0),
('c', 0.01),
('c', 0.02),
('c', 0.03)
)
# -
index = pd.read_csv(spot_filename, parse_dates=['date'])
opt = pd.read_hdf(options_filename).sort_values(by=['date'])
rename = {
'open_price': 'open',
'high_price': 'high',
'low_price': 'low',
'close_pric': 'close'
}
opt = opt.rename(columns=rename)
opt = opt[(opt.date.dt.year == opt.expiry_date.dt.year) &\
(opt.date.dt.month == opt.expiry_date.dt.month)].sort_values(by='date')
opt['de'] = (opt['expiry_date'] - opt['date']).dt.days
opt['year'] = opt.date.dt.year
opt['month'] = opt.date.dt.month
opt['strike_pr'] = opt.contract_d.str[24:].astype(int)
expiry_dates = opt.groupby(['year', 'month']).expiry_date.max().reset_index()
opt2 = opt[opt.expiry_date.isin(expiry_dates.expiry_date.values)]
ce = opt2.query("opt_type=='CE'")
pe = opt2.query("opt_type=='PE'")
def get_result(options_data, spot_data, opt='p', itm=0, step=100):
if opt.lower() == 'p':
m = 1-itm
else:
m = 1+itm
first_dates = options_data.groupby(['year', 'month']).date.min().reset_index()
opts = options_data.merge(first_dates, on=['year', 'month', 'date'])
opts['spot'] = opts.undrlng_st.copy()
opts['strike'] = [int((x*m)/step)*step for x in opts.undrlng_st.values]
if opt.lower() == 'p':
opts['strike'] = opts['strike'] + step
opts = opts.query('strike==strike_pr')
cols = ['date', 'contract_d', 'strike', 'expiry_date']
entries = opts[cols].copy().drop_duplicates()
entries['entry_date'] = entries.date.copy()
last_dates = options_data.groupby(['year', 'month']).date.max().reset_index()
exits = entries.copy()
del exits['entry_date']
exits['date'] = last_dates.date.values
exits['exit_date'] = exits.date.copy()
frame1 = entries.merge(options_data, on=cols)
frame2 = exits.merge(options_data, on=cols)
trades = frame1.merge(frame2, on=['contract_d', 'expiry_date'])
rename = {
'date_x': 'date',
'close_x': 'entry_price',
'undrlng_st_x': 'entry_spot',
'close_y': 'exit_price',
'undrlng_st_y': 'exit_spot'
}
trades = trades.rename(columns=rename)
cols = ['date', 'contract_d', 'expiry_date', 'entry_date', 'entry_price',
'exit_date', 'exit_price', 'entry_spot', 'exit_spot']
trades['pnl'] = trades.eval('exit_price-entry_price')
trades['de'] = (trades['expiry_date'] - trades['entry_date']).dt.days
return trades[cols + ['pnl', 'de']]
collect = []
for i,(opt,strike) in enumerate(options):
data = ce if opt =='c' else pe
temp = get_result(data, index, itm=strike,step=step, opt=opt)
temp['name'] = f"opt{i}"
collect.append(temp)
res = pd.concat(collect)
del collect
res.pnl.describe()
res.groupby('name').pnl.describe()
res.to_csv(output_filename)
| options/options_backtest_monthly.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# ## Using Iris data to looking at some some Machine Learning algorithms
from sklearn.datasets import load_iris
iris= load_iris()
type(iris)
print iris.data
print iris.feature_names
print iris.target
print iris.target_names
print type(iris.data)
print type(iris.target)
print iris.data.shape
print iris.target.shape
# +
x=iris.data
y=iris.target
# -
x
y
print x.shape
print y.shape
# ## K-Nearest Neighbors
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=1) # instantiate the estimator
knn # implicitly, it fills in a lot of implicit information.
knn.fit(x, y)
knn.predict([3, 5, 4, 2])
x_new = [[3, 5, 4, 2], [5, 4, 3, 2]]
knn.predict(x_new)
# Note that we have no way of knowing the target for x_new. This motivates the need to divide up the dataset into training, and testing dataset
# ## Logistic Regression
from sklearn.linear_model import LogisticRegression
logreg=LogisticRegression() #instantiating
logreg.fit(x, y) # fitting the logistic model with the complete dataset
logreg.predict(x_new)
# ## Split the dataset into training, and testing sets
from sklearn.cross_validation import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.4)
#test_size=.4 means it should let 40% of the dataset be part of the testing set
print x_train.shape
print y_train.shape
print x_test.shape
print y_test.shape
| Sklearn/Machine Learning using Scikit-Learn.ipynb |
(* --- *)
(* jupyter: *)
(* jupytext: *)
(* text_representation: *)
(* extension: .ml *)
(* format_name: light *)
(* format_version: '1.5' *)
(* jupytext_version: 1.14.4 *)
(* kernelspec: *)
(* display_name: OCaml 4.04.2 *)
(* language: OCaml *)
(* name: ocaml-jupyter-4.04.2 *)
(* --- *)
(* # Random dataset generation *)
(* *)
(* This example shows generation of two-dimensional dataset follows bivariate Gaussian distribution. *)
(* *)
(* [Gsl_randist.bivariate_gaussian](http://mmottl.github.io/gsl-ocaml/api/Gsl_randist.html#VALbivariate_gaussian) *)
(* is a binding to [gsl_ran_bivariate_gaussian](https://www.gnu.org/software/gsl/manual/html_node/The-Bivariate-Gaussian-Distribution.html), *)
(* a function generates a two random numbers following bivariate Gaussian distribution defined as *)
(* *)
(* $$ *)
(* \newcommand{d}{\mathrm{d}} *)
(* p(x,y) \d x \d y = *)
(* \frac{1}{2 \pi \sigma_x \sigma_y \sqrt{1-\rho^2}} *)
(* \exp\left( *)
(* -\frac{x^2 / \sigma_x^2 + y^2 / \sigma_y^2 - 2 \rho x y / (\sigma_x \sigma_y)}{2(1-\rho^2)} *)
(* \right) \d x \d y *)
(* $$ *)
(* *)
(* where $\sigma_x$ and $\sigma_y$ are standard deviations of $x$ and $y$ respectively, *)
(* and $\rho \in [-1,+1]$ is a correlation coefficient between $x$ and $y$. *)
#thread ;;
#require "gsl" ;;
#require "jupyter-archimedes" ;;
(* +
let rng = Gsl.Rng.(make MT19937) ;; (* Mersenne Twister *)
(* Generate positive examples *)
let positive_xys =
Array.init 100 (fun _ -> Gsl.Randist.bivariate_gaussian rng ~sigma_x:0.4 ~sigma_y:0.9 ~rho:0.4)
|> Array.map (fun (x, y) -> (x +. 0.5, y -. 0.1))
(* Generate negative examples *)
let negative_xys =
Array.init 100 (fun _ -> Gsl.Randist.bivariate_gaussian rng ~sigma_x:0.6 ~sigma_y:1.2 ~rho:0.3)
|> Array.map (fun (x, y) -> (x -. 0.8, y +. 0.4))
(* +
let vp = A.init ["jupyter"] in
A.Axes.box vp ;
A.set_color vp A.Color.red ;
A.Array.xy_pairs vp positive_xys ;
A.set_color vp A.Color.blue ;
A.Array.xy_pairs vp negative_xys ;
A.close vp
(* -
let oc = open_out "datasets/bivariate_gaussian_2d.csv" in
let ppf = Format.formatter_of_out_channel oc in
Array.iter
(fun (x, y) -> Format.fprintf ppf "%g,%g,0@." x y)
negative_xys ;
Array.iter
(fun (x, y) -> Format.fprintf ppf "%g,%g,1@." x y)
positive_xys ;
close_out
| notebooks/random_dataset_generation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # <center>Graph Colouring with Quantum Annealing</center>
# In this notebook we have solved a graph colouring problem on a randomly generated graph of `n` nodes with randomly generated connectivity between nodes.
#
# Different solvers, inlcuding RacingBranches and DWaveSampler were used and it was concluded that ```QBSolv``` performs best.
from dwave.system import DWaveSampler, EmbeddingComposite
from neal import SimulatedAnnealingSampler
import numpy as np
import dimod
import networkx as nx
import matplotlib.pyplot as plt
import hybrid
from dwave_qbsolv import QBSolv
import matplotlib.colors as mcolors
from geopy import geocoders
# %matplotlib inline
# +
#---------------------------------------------------------------
#Change this value to change the number of nodes
n_nodes = 10
n_colors = 9
#---------------------------------------------------------------
def plot_graph(G, colors = ['beige']*n_nodes):
'''
Parameters: G: network.Graph()
colors: a list of colors for coloring the nodes
'''
plt.figure(figsize=(12, 8))
pos = nx.spring_layout(G)
default_axes = plt.axes(frameon=True)
nx.draw_networkx(G, node_color=colors, node_size=600, alpha=.8, ax=default_axes, pos=pos)
plt.show()
def generate_random_graph(n, p = 0.6):
'''
Parameters: n: number of nodes
p: probability of nodes being connected
Return: nx.Graph()
'''
return(nx.gnp_random_graph(n, p))
def graph_to_adjacency(G):
'''
Parameters: G: network.Graph()
Return: Adjacency matrix (2D-list)
'''
mat = nx.adjacency_matrix(G).todense()
return(tuple(mat.tolist()))
def graphcolour_QUBO(G, ncolours, constr=10):
#This function returns the appropriate QUBO dictionary for given parameters
Q = {}
for i in range(0,len(G)):
for j in range(0,ncolours):
for k in range(0,len(G)):
for l in range(0,ncolours):
Q[(str(i) + "_" + str(j), str(k) + "_" + str(l))] = 0
#Objective, we dont want adjacent vertices to share the colour
for i in range(0,len(G)):
for j in range(0,ncolours):
for k in range(i+1,len(G)):
if G[i][k] == 1:
Q[(str(i) + "_" + str(j), str(k) + "_" + str(j))] = 1
#Constraint: Every vertex is painted of exactly one colour
for i in range(0,len(G)):
for j in range(0,ncolours):
for k in range(0,ncolours):
if j == k:
Q[(str(i) + "_" + str(j), str(i) + "_" + str(j))] = Q[(str(i) + "_" + str(j), str(i) + "_" + str(j))] - constr
else:
Q[(str(i) + "_" + str(j), str(i) + "_" + str(k))] = Q[(str(i) + "_" + str(j), str(i) + "_" + str(k))] + constr
return (Q)
def get_colors(G, final_state):
#This function returns the colors of each node when a solution state is passed as parameter
colors = ['0'] * len(list(G.nodes))
c_map = list(set(list(mcolors.CSS4_COLORS.keys())))
c_map = c_map[0:n_colors]
for k, v in final_state.first[0].items():
if v == 1:
if len(k) == 3:
node = int(k[0])
color = int(k[-1])
colors[node] = c_map[color]
if len(k) == 4:
node = int(k[0:2])
color = int(k[-1])
colors[node] = c_map[color]
return(colors)
def verify_results(G, colors):
#Checks if the results are correct and no two adjacent nodes have the same color
connections = list(G.edges)
for connection in connections:
node_1 = connection[0]
node_2 = connection[1]
if colors[node_1] == colors[node_2]:
print("Test Failed at", connection)
'''
#Not required
def get_j(Q):
j = {}
for i in list(Q.keys()):
if i[0] == i[1]:
j[i[0]] = Q[(i[0], i[1])]
return(j)
def get_H(Q):
H = {}
for i in list(Q.keys()):
if i[0] != i[1]:
H[(i[0], i[1])] = Q[(i[0], i[1])]
return(H)
'''
#Generate random graph
G = generate_random_graph(n_nodes)
#Plot Graph
plot_graph(G)
#Get adjacency matrix
adj = graph_to_adjacency(G)
#Get Qubomax(dict(G.degree()).items(), key = lambda x: x[1])[1]
Q = graphcolour_QUBO(adj, n_colors)
'''
#Get linear and quadratic terms
j, H = get_j(Q), get_H(Q)
bqm = dimod.BinaryQuadraticModel(j, H, 0, dimod.BINARY)
# Define the workflow
iteration = hybrid.RacingBranches(
hybrid.Identity(),
hybrid.InterruptableTabuSampler(),
hybrid.EnergyImpactDecomposer(size=2)
| hybrid.QPUSubproblemAutoEmbeddingSampler()
| hybrid.SplatComposer()
) | hybrid.ArgMin()
workflow = hybrid.LoopUntilNoImprovement(iteration, convergence=3)
# Solve the problem
init_state = hybrid.State.from_problem(bqm)
final_state = workflow.run(init_state).result()
# Print results
print("Solution: sample={.samples.first}".format(final_state))
'''
#QBSolv reaches a solution much faster than RacingBranches, SimulatedAnnealing and DWaveSampler
sampler = QBSolv()
final_state = sampler.sample_qubo(Q, num_reads = 1000)
print(final_state.first)
c = get_colors(G, final_state)
plot_graph(G, c)
verify_results(G, c) #This function will print a faliure message if the solution is incorrect.
# +
locs = [
"Wausaw, WI",
"Chicago, IL",
"Detroit, MI",
"Indianapolis, IN",
"Columbus, OH",
"Chapman, PA",
"Kilington, VT",
"Corning, NY",
"New York, NY",
"Charlotte, NC",
"Roanoke, VA",
"Little Rock, AR",
"Conway, AR",
"Baton Rouge, LA",
"Jackson, MS",
"Birmingham, AL",
"Louisville, KY",
"Nashville, TN",
"Atlanta, GA",
"Acala, FL",
"Miami, FL"
]
def convert_to_coordinates(locs):
coord_lst = {}
gn = geocoders.ArcGIS()
for idx, loc in enumerate(locs):
loc_obj = gn.geocode(loc)
coords = (loc_obj.latitude, loc_obj.longitude)
coord_lst[idx] = coords
return(coord_lst)
def get_loc_graph(locs):
coord_lst = convert_to_coordinates(locs)
G = nx.Graph()
for key, val in coord_lst.items():
G.add_node(key, pos=val)
blocks = [[0,5], [5,9], [9,11], [11,14], [14,18], [18,21]]
for block in blocks:
for i in range(block[0],block[1]):
for j in range(i+1, block[1]):
G.add_edge(i, j)
return(G)
def solve_map():
G = get_loc_graph(locs)
plt.figure(figsize=(12,12))
nx.draw_networkx(G, node_size=600, pos=nx.get_node_attributes(G, 'pos'))
plt.show()
#Get adjacency matrix
adj = graph_to_adjacency(G)
#Get Qubomax(dict(G.degree()).items(), key = lambda x: x[1])[1]
Q = graphcolour_QUBO(adj, 5)
sampler = QBSolv()
final_state = sampler.sample_qubo(Q, num_reads = 1000)
print(final_state.first)
c = get_colors(G, final_state)
plt.figure(figsize=(12,12))
nx.draw_networkx(G, node_color=c, node_size=600, pos=nx.get_node_attributes(G, 'pos'))
plt.show()
# -
solve_map()
| DWave_GraphColoring.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %pylab inline
from scipy.interpolate import interpn
from multiprocessing import Pool
from constant import *
import warnings
import math
warnings.filterwarnings("ignore")
np.printoptions(precision=2)
# wealth discretization
ws = np.array([10,25,50,75,100,125,150,175,200,250,500,750,1000,1500,3000])
w_grid_size = len(ws)
# 401k amount discretization
ns = np.array([1, 5, 10, 15, 25, 50, 100, 150, 400, 1000])
n_grid_size = len(ns)
# Mortgage amount
Ms = np.array([0.01*H,0.05*H,0.1*H,0.2*H,0.3*H,0.4*H,0.5*H,0.8*H]) * pt
M_grid_size = len(Ms)
points = (ws,ns,Ms)
# +
import quantecon as qe
import timeit
mc = qe.MarkovChain(Ps)
#Vgrid = np.load("Vgrid_i.npy")
cgrid = np.load("cgrid" + str(H)+ ".npy")
bgrid = np.load("bgrid" + str(H)+ ".npy")
kgrid = np.load("kgrid" + str(H)+ ".npy")
qgrid = np.load("qgrid" + str(H)+ ".npy")
def action(t, x):
w, n, M, e, s, z = x
c = interpn(points, cgrid[:,:,:,e,s,z,t], x[:3], method = "nearest", bounds_error = False, fill_value = None)[0]
b = interpn(points, bgrid[:,:,:,e,s,z,t], x[:3], method = "nearest", bounds_error = False, fill_value = None)[0]
k = interpn(points, kgrid[:,:,:,e,s,z,t], x[:3], method = "nearest", bounds_error = False, fill_value = None)[0]
q = interpn(points, qgrid[:,:,:,e,s,z,t], x[:3], method = "nearest", bounds_error = False, fill_value = None)[0]
if q <= 0.75:
q = 0.5
else:
q = 1
return (c,b,k,q)
def transition(x, a, t, s_next):
'''
Input: state and action and time
Output: possible future states and corresponding probability
'''
w, n, M, e, s, z = x
c,b,k,q = a
# variables used to collect possible states and probabilities
x_next = []
m = M/D[T_max-t]
M_next = M*(1+rh) - m
# transition of z
if z == 1:
z_next = 1
else:
if k == 0:
z_next = 0
else:
z_next = 1
n_next = gn(t, n, x, (r_k[s_next]+r_b[int(s)])/2)
w_next = b*(1+r_b[int(s)]) + k*(1+r_k[s_next])
if t >= T_R:
return [w_next, n_next, M_next, 0, s_next, z_next]
else:
if e == 1:
for e_next in [0,1]:
x_next.append([w_next, n_next, M_next, e_next, s_next, z_next])
prob_next = [Pe[s,e], 1 - Pe[s,e]]
else:
for e_next in [0,1]:
x_next.append([w_next, n_next, M_next, e_next, s_next, z_next])
prob_next = [1-Pe[s,e], Pe[s,e]]
return x_next[np.random.choice(len(prob_next), 1, p = prob_next)[0]]
# +
'''
Start with:
Ms = H * 0.8 * 0.25
w = 20
n = 0
e = 1
s = 1
z = 0
1000 agents for 1 economy, 100 economies.
use numpy array to contain the variable change:
wealth, rFund, Mortgage, employment, sState, participation
salary, consumption, hConsumption, bond, stock, hPercentage,
life. Shape: (T_max-T_min, numAgents*numEcons)
'''
x0 = [20, 0, H * 0.8 * pt, 1, 1, 0]
numAgents = 1000
numEcons = 500
# -
import random as rd
EconStates = [mc.simulate(ts_length=T_max - T_min, init=0) for _ in range(numEcons)]
def simulation(i):
track = np.zeros((T_max - T_min,13))
econState = EconStates[i//numAgents]
alive = True
x = x0
for t in range(1,len(econState)-1):
if rd.random() > Pa[t]:
alive = False
if alive:
track[t, 0] = x[0]
track[t, 1] = x[1]
track[t, 2] = x[2]
track[t, 3] = x[3]
track[t, 4] = x[4]
track[t, 5] = x[5]
track[t, 6] = y(t,x)
a = action(t, x)
track[t, 7] = a[0]
track[t, 9] = a[1]
track[t, 10] = a[2]
track[t, 11] = a[3]
track[t, 12] = 1
# calculate housing consumption
if a[3] == 1:
Vh = (1+kappa)*H
else:
Vh = (1-kappa)*(H/2)
track[t, 8] = Vh
s_next = econState[t+1]
x = transition(x, a, t, s_next)
return track
# %%time
pool = Pool()
agentsHistory = pool.map(simulation, list(range(numAgents*numEcons)))
pool.close()
len(agentsHistory)
np.save("agents", np.array(agentsHistory))
agents = np.load("agents.npy")
#numpy array used to store
attributesNames = ['wealth', 'rFund', 'Mortgage','employment','sState', 'participation',
'salary', 'consumption', 'hConsumption', 'bond', 'stock','hPer', 'life']
attributes = [np.zeros((T_max-T_min, numAgents*numEcons)) for i in range(len(attributesNames))]
#shape the collected stats into the right format
def separateAttributes(agents):
for i in range(numAgents*numEcons):
for j in range(len(attributesNames)):
attributes[j][:,i] = agents[i][:,j]
separateAttributes(agents)
#save the data
for i in range(len(attributes)):
np.save(attributesNames[i], attributes[i])
wealth = np.load("wealth.npy")
rFund = np.load("rFund.npy")
Mortgage = np.load("Mortgage.npy")
employment = np.load("employment.npy")
sState = np.load("sState.npy")
salary = np.load("salary.npy")
consumption = np.load("consumption.npy")
hConsumption = np.load("hConsumption.npy")
bond = np.load("bond.npy")
stock = np.load("stock.npy")
hPer = np.load("hPer.npy")
life = np.load("life.npy")
participation = np.load("participation.npy")
# Population during the entire simulation period
plt.plot(np.mean(life, axis = 1))
plt.plot(Pa[:60])
# +
def quantileForPeopleWholive(attribute, quantiles = [0.25, 0.5, 0.75]):
qList = []
for i in range(T_max):
if len(np.where(life[i,:] == 1)[0]) == 0:
qList.append(np.array([0] * len(quantiles)))
else:
qList.append(np.quantile(attribute[i, np.where(life[i,:] == 1)], q = quantiles))
return np.array(qList)
def meanForPeopleWholive(attribute):
means = []
for i in range(T_max):
if len(np.where(life[i,:] == 1)[0]) == 0:
means.append(np.array([0]))
else:
means.append(np.mean(attribute[i, np.where(life[i,:] == 1)]))
return np.array(means)
# -
# plot participation ratio
plt.plot(meanForPeopleWholive(participation))
# plot the 0.25, 0.5, 0.75 quantiles of hConsumption
plt.plot(quantileForPeopleWholive(hConsumption))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(wealth))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(rFund))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(Mortgage))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(consumption))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(bond))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(stock))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.plot(quantileForPeopleWholive(hPer))
# plot the 0.25, 0.5, 0.75 quantiles of wealth
plt.figure(figsize = [14,8])
plt.plot(meanForPeopleWholive(wealth), label = "wealth")
plt.plot(meanForPeopleWholive(consumption), label = "Consumption")
plt.plot(meanForPeopleWholive(bond), label = "Bond")
plt.plot(meanForPeopleWholive(stock), label = "Stock")
# plt.plot(meanForPeopleWholive(rFund), label = "401k")
plt.legend()
plt.plot(meanForPeopleWholive(rFund), label = "401k")
| 20201026/.ipynb_checkpoints/simulationOwning-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
#### Importing all required libraries
import pandas as pd
from selenium import webdriver
from webdriver_manager.chrome import ChromeDriverManager
#### Installing chrome Driver based on current version
#### And Create a new instance of google chrome
browser = webdriver.Chrome(ChromeDriverManager().install())
base_url = 'https://www.linkedin.com/jobs/search?keywords=Data%20Science&location=Hyderabad%2C%20Telangana%2C%20India&geoId=105556991&trk=public_jobs_jobs-search-bar_search-submit&redirect=false&position=1&pageNum='
list_jobs = []
for pageNo in range(0, 40):
#### Access google chrome and open Linkedin website
browser.get(base_url+str(pageNo*25))
#### Extracting table data using class name
soup = browser.find_elements_by_xpath((".//div[@class='result-card__contents job-result-card__contents']"))
for i in range(len(soup)):
dict_jobs = {}
#### Extracting 'Title' of the job
dict_jobs["Title"] = soup[i].find_element_by_xpath((".//h3[@class='result-card__title job-result-card__title']")).text
#### Extracting Name of the 'Company'
dict_jobs["company"] = soup[i].find_element_by_xpath((".//h4[@class='result-card__subtitle job-result-card__subtitle']")).text
#### Extracting 'Location' of the jon
dict_jobs["location"] = soup[i].find_element_by_xpath((".//span[@class='job-result-card__location']")).text
#### Extracting 'date' of the job
dict_jobs["date"] = soup[i].find_element_by_tag_name('time').get_attribute('datetime')
list_jobs.append(dict_jobs)
#### Closing chrome browser
browser.close()
#### Converting all the extracted elements into dataframe
jobs_df = pd.DataFrame(list_jobs)
#### Printing top 5 rows of the data
jobs_df.head()
# -
#### Saving the data in to csv file
jobs_df.to_csv('jobs.csv')
| Selenium/Linkedin.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
import matplotlib.pyplot as plt
import h5py
# Reconstruct all neurons by looking at the neuron ids associated with the dendrite fields.
dendrites = h5py.File('dendrites.hdf5', 'r')
dendrites['dendrites']['agglomerate']
# +
segments = h5py.File('segments.hdf5', 'r')
segment_positions = np.array(segments['segments']['position'])
vc = np.array(segments['segments']['voxelCount'])
num_voxels = []
neuron_id = []
for i, agg_id in enumerate(dendrites['dendrites']['agglomerate'].keys()):
ds = dendrites['dendrites']['agglomerate'][agg_id]
num_voxels.append(vc[np.array(ds)].sum())
neuronId = np.array(dendrites['dendrites']['neuronId'])
neuronId[neuronId != 0]
# +
# Reconstruct all the neurons
import collections
d = collections.defaultdict(lambda: [])
for i, id in enumerate(neuronId):
if id > 0:
# Append the dendrite ids to the right slot.
d[id] += np.array(dendrites['dendrites']['agglomerate'][str(i + 1)]).tolist()
# +
import numpy_indexed as npi
def repaint(neuron_segments, segment_positions, voxel_pos):
cube = h5py.File('x%dy%dz%d.hdf5' % voxel_pos, 'r')
data = np.array(cube['data'])
unique_values = set(np.unique(data.ravel()))
value_map = {idx: 0 for idx in unique_values}
for k in neuron_segments.keys():
values_to_remap = set(np.array(neuron_segments[k])).intersection(unique_values)
for j in values_to_remap:
value_map[j] = k
return data, value_map
data, value_map = repaint(d, segment_positions, (0, 0, 0))
# -
# %load_ext cython
# + magic_args="-a" language="cython"
# import numpy as np
# cimport cython
#
# @cython.boundscheck(False)
# def c_remap(unsigned int[:] data, unsigned int[:] the_map):
# new_data = np.zeros(data.size, dtype=np.uint32)
# cdef unsigned int[:] new_data_view = new_data
# cdef int M = data.size
#
# for j in range(M):
# new_data_view[j] = the_map[data[j]]
# return new_data
# +
def remap(data, value_map):
value_arr = np.zeros(max(value_map.keys()) + 1, dtype=np.uint32)
for k, v in value_map.items():
value_arr[k] = v
remapped = c_remap(data.ravel(), value_arr)
return remapped.reshape(data.shape)
remapped = remap(data, value_map)
# -
f = h5py.File('x0y0z0_remapped.hdf5', 'w')
f.create_dataset('data', remapped.shape, compression="gzip", data=remapped)
f.close()
print(np.unique(remapped.ravel()))
value_map
unique_values[:9034][-1]
list(value_map.keys())[:9034][-1]
cube = h5py.File('x0y0z0.hdf5', 'r')
subd = cube['data'][:512, :512, :512]
xi, yi, zi = np.meshgrid(np.arange(512) + 1, np.arange(512) + 1, np.arange(512) + 1)
selidx = subd == 2
(zi * (selidx)).sum() / (selidx).sum()
counts = np.array(f['segments'][0])
which_ones = counts.argsort()
counts.sort()
which_ones[-3]
plt.plot(counts[-200:])
P = np.array(f['segments']['position'])
P[:, which_ones[-4]]
tinydata = cube['data'][:256, :256, :256]
f = h5py.File('x0y0z0_subset.hdf5', 'w')
f['data'] = tinydata
f.close()
C = np.array(cube['data'])
C.max()
counts = np.array([np.sum(C == x) for x in which_ones[-30:-20]])
counts
counts
| Read one neuron.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Dask Subscriber
# +
import os
from dotenv import load_dotenv
load_dotenv()
config = {'db_name': os.getenv("DB_NAME"),
'db_username': os.getenv("DB_USERNAME"),
'db_password': os.getenv("DB_PASSWORD"),
'db_system': os.getenv("DB_SYSTEM"),
'db_config_file_path': os.getenv("DB_CONFIG_PATH"),
'db_queries_file_path': os.getenv("DB_QUERIES_PATH"),
'scheduler_address': os.getenv("DASK_SCHEDULER")}
# -
# ### Instantiate a PubSub object to subscribe
# +
from src.pubsub import PubSub
subscriber = PubSub(uid='test_subscriber', config=config)
# -
# ### Register a callback function to process the incoming messages
# +
def process_msg(msg):
print('processed msg', msg)
subscriber.subscribe(topic='trade_ev', callback=process_msg)
# -
# ### Listen for incoming messages
subscriber.listen(timeout=None)
| tutorial/subscriber.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.9.10 ('conda-forge')
# language: python
# name: python3
# ---
# +
import heapq
def largestPermutation(k, arr):
'''
It seems that k can be larger than the length of arr.
It takes n-1 swaps to get to the largest permutation.
'''
k = min(k, len(arr))
# make an index lookup table
ind_dict = {v: i for i, v in enumerate(arr)}
# create a max heap
max_heap = arr.copy()
heapq._heapify_max(max_heap)
for i in range(k):
max_num = heapq._heappop_max(max_heap)
max_idx = ind_dict[max_num]
ind_dict[arr[i]] = max_idx
arr[max_idx], arr[i] = arr[i], arr[max_idx]
return arr
# -
print(largestPermutation(1, [4, 2, 3, 5, 1]))
print(largestPermutation(1, [2, 1, 3]))
print(largestPermutation(1, [2, 1]))
print(largestPermutation(1, [5, 2, 5, 5, 5])) # this input does not occur since the array is a permutation from 1 to n.
# The first implementation has an issue.
# When the max element is already in position, no swap should happen and no moves should be wasted.
def largestPermutation2(k, arr):
# make an index lookup table
ind_dict = {v: i for i, v in enumerate(arr)}
# create a max heap
max_heap = arr.copy()
heapq._heapify_max(max_heap)
for i in range(len(arr)):
max_num = heapq._heappop_max(max_heap)
max_idx = ind_dict[max_num]
if max_idx != i:
ind_dict[arr[i]] = max_idx
arr[max_idx], arr[i] = arr[i], arr[max_idx]
k -= 1
if k == 0: break
return arr
print(largestPermutation2(1, [4, 2, 3, 5, 1]))
print(largestPermutation2(1, [2, 1, 3]))
print(largestPermutation2(1, [2, 1]))
# Great. The second implementation passes!
# However, the reference solution is much more beautiful in the sense that
| hacker-rank/largest-permutation.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
## import required packages for a parameter estimation technique
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
import pandas as pd
import math
##Import Experimental Data:
rev_exp_data = pd.read_csv("data/10mVs_Reversible.csv")
current_exp=rev_exp_data['current(A)'].values
voltage_exp=rev_exp_data['voltage(mV)'].values
time_exp=rev_exp_data['time(s)'].values
## all appropriate packages and the singular experimental data file is imported now
# +
# Import the dimensionless voltammagram (V I) for reversible reactions
rev_dim_values = pd.read_csv("data/dimensionless_values_rev.csv")
rev_dim_current=rev_dim_values['dimensionless_current'].values
rev_dim_voltage=rev_dim_values['dimensionless_Voltage'].values
##We will now prompt the user to submit known parameters (THESE CAN BE CHANGED OR MADE MORE CONVENIENT)
sweep_rate= float(input("What is the Voltage sweep rate in mV/s?(10)"))
electrode_surface_area= float(input("What is the electrode surface area in cm^2?(.2)"))
concentration_initial= float(input("What is the initial concentration in mol/cm^3?(.00001)"))
Temp= float(input("What is the temperature in K?(298)"))
eq_pot= float(input("What is the equilibrium potential in V?(.10)"))
##we are inserting a diffusion coefficient to check math here, we will estimate this later:
Diff_coeff=0.00001
## Here we define constant variables, these can be made to user inputs if needed.
n=1
Faradays_const=96285
R_const=8.314
sigma=(n*Faradays_const*sweep_rate)/(R_const*Temp)
Pre=electrode_surface_area*concentration_initial*n*Faradays_const*math.sqrt(Diff_coeff*sigma)
output_voltage=(eq_pot+rev_dim_voltage/n)
output_current=Pre*rev_dim_current
plt.scatter(output_voltage,output_current)
# -
print(output_current)
print(rev_dim_current)
# +
from scipy import optimize
def test_func(rev_dim_current, D):
return electrode_surface_area*concentration_initial*n*Faradays_const*math.sqrt(D*sigma)*rev_dim_current
params, params_covariance = optimize.curve_fit(test_func, rev_dim_current, output_current,p0=None,bounds = (0,[1]))
print(params)
# -
| Pereira_CV_code_rough.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# # Collectl CPU Log Analysis
# ## Functionalities
# - Plot CPU utilization graphs.
#
# ## Input
# Log files are read from a directory in `../data`. This directory is assumed to have the following structure:
# ```
# logs/
# [node-1]/
# collectl.tar.gz
# ...
# [node-n]/
# collectl.tar.gz
# ```
# A tarball `collectl.tar.gz` contains log files. The log file extension identifies the type of resource monitored:
# - `.cpu.gz`: CPU monitoring log file.
# - `.numa.gz`: memory monitoring log file.
# - `.dsk.gz`: disk I/O monitoring log file.
# ## Notebook Configuration
# +
########## GENERAL
# Name of the directory in `../data`
EXPERIMENT_DIRNAME = "BuzzBlogBenchmark_2022-01-15-18-52-05"
# Ramp up duration (in sec)
RAMP_UP_DURATION = 720
# Ramp down duration (in sec)
RAMP_DOWN_DURATION = 360
########## CPU
# Analyzed metric (options: "user", "nice", "system", "wait", "irq", "soft",
# "steal", "idle", "total", "guest", "guest_n", "intrpt")
COLLECTL_CPU_METRIC = "total"
# Filter CPU cores
COLLECTL_CPU_CORES = None
# -
# ## Notebook Setup
# +
# %matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import sys
import warnings
warnings.filterwarnings("ignore")
sys.path.append(os.path.abspath(os.path.join("..")))
from parsers.collectl_parser import CollectlParser
from utils.utils import *
# -
# ## Log Parsing
# Get experiment time range, excluding ramping periods
experiment_start_time = get_experiment_start_time(EXPERIMENT_DIRNAME)
start_time = experiment_start_time + np.timedelta64(RAMP_UP_DURATION, 's')
experiment_end_time = get_experiment_end_time(EXPERIMENT_DIRNAME)
end_time = experiment_end_time - np.timedelta64(RAMP_DOWN_DURATION, 's')
# Build data frames
cpu = pd.concat([
pd.DataFrame.from_dict(CollectlParser(logfile, "cpu", start_time, end_time).parse()).assign(node_name=node_name)
for node_name, logfile in get_collectl_cpu_logfiles(EXPERIMENT_DIRNAME)
], ignore_index=True)
# Filter data frames
cpu = cpu[(cpu["hw_metric"] == COLLECTL_CPU_METRIC)]
if COLLECTL_CPU_CORES:
cpu = cpu[(cpu["hw_no"].isin(COLLECTL_CPU_CORES))]
cpu["hw_no"].unique()
# (Re) Build columns
cpu["timestamp"] = cpu.apply(lambda r: (r["timestamp"] - start_time).total_seconds(), axis=1)
cpu["window"] = cpu.apply(lambda r: int(r["timestamp"]), axis=1)
cpu["value"] = cpu.apply(lambda r: float(r["value"]), axis=1)
# (Re) Create index
cpu.set_index("timestamp", inplace=True)
cpu.sort_index(inplace=True)
# Get values
node_names = get_node_names(EXPERIMENT_DIRNAME)
# ## CPU Monitoring
# Plot CPU utilization (1-sec granularity)
fig = plt.figure(figsize=(24, len(node_names) * 12))
for (i, node_name) in enumerate(node_names):
df = cpu[(cpu["node_name"] == node_name)]
df = df.groupby(["window"])["value"].mean()
ax = fig.add_subplot(len(node_names), 1, i + 1)
ax.set_xlim((df.index.min(), df.index.max()))
ax.set_ylim((0, 100))
ax.grid(alpha=0.75)
df.plot(ax=ax, kind="line", title="%s - CPU Utilization" % node_name, xlabel="Time (seconds)",
ylabel="%s (%%)" % COLLECTL_CPU_METRIC, grid=True, legend=False, yticks=range(0, 101, 10))
# +
########## LOCAL CONFIG
# Minimum time (in seconds)
MIN_TIME = None
# Maximum time (in seconds)
MAX_TIME = None
# Plot CPU utilization (millisec granularity)
if MIN_TIME and MAX_TIME:
fig = plt.figure(figsize=(24, len(node_names) * 12))
for (i, node_name) in enumerate(node_names):
df = cpu[(cpu["node_name"] == node_name)]
df = df[(df.index >= MIN_TIME) & (df.index <= MAX_TIME)]
df = df.groupby(["timestamp", "hw_no"])["value"].mean()
df = df.unstack()
ax = fig.add_subplot(len(node_names), 1, i + 1)
ax.set_xlim((df.index.min(), df.index.max()))
ax.set_ylim((0, 100))
ax.grid(alpha=0.75)
df.plot(ax=ax, kind="line", title="%s - CPU Utilization" % node_name, xlabel="Time (seconds)",
ylabel="%s (%%)" % COLLECTL_CPU_METRIC, grid=True, legend=True, yticks=range(0, 101, 10))
| analysis/notebooks/CollectlCPULogAnalysis.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Reading a file with Pandas
# ==
# ---
# Pandas is a library widely used for statistics and analysis
# * Has functions which allow you to read a file directly into your script
# * Borrows many feature from R's data frames
# * Read a Comma Separate Values (CSV) data file with `pandas.read_csv`.
# * Uses the same notation as you used for bash ("./" accesses the current folder, "../" searches up to the parent folder)
# * Argument is the name of the file to be read.
# * Assign result to a variable to store the data that was read.
# ## Accessing Files
#
# We're using the gapminder data that we created yesterday. Remember that these are stored in the shell_lessons directory in a `data` sub-directory, which is why the path to the file is `../shell_lessons/data/gapminder_data/gapminder_final.txt`. If you forget to include `../shell_lessons/`, or if you include it but your copy of the file is somewhere else, you will get a [runtime error]({{ site.github.url }}/05-error-messages/) that ends with a line like this:
# ~~~
# OSError: File b'gapminder_final.txt' does not exist
# ~~~
#
# ** Don't forget to use the tab key for auto-completion **
# * Auto-complete works in Jupyter notebooks!
# First, import the pandas library
import pandas
# Then read the csv
df = pandas.read_csv("/Users/linhtran/SDC_02-23-2019/2019-02-23-WorkshopResources/python-lessons/gapminder_csv.txt")
df.drop
df.drop_duplicates #drop duplicates
# print the data frame
print(df)
# When we load a csv file with Pandas, it get's loaded into a DataFrame.
#
# DataFrame is the way Pandas represents a table, and Series is the data-structure Pandas use to represent a column. So, a data frame and series are synonomous with table and column.
#
#
# * The columns in a data frame are the observed variables, and the rows are the observations.
# * Pandas uses backslash `\` to show wrapped lines when output is too wide to fit the screen.
df.head(10)
df.tail(5)
# ---
# ## EtherPad
#
# Hypothetically, the data a project you are working on is stored in a file called `microbes.csv`, which is located in a folder called `field_data`. You are doing analysis in a notebook called `analysis.ipynb`in a sibling folder called `thesis`. You're directory structure looks like this:
# ~~~
# your_home_directory
# +-- field_data/
# | +-- microbes.csv
# +-- thesis/
# +-- analysis.ipynb
# ~~~
#
# What value(s) should you pass to `read_csv` to read `microbes.csv` in `analysis.ipynb`? Vote for your answer in EtherPad.
#
# a. "/field_data/microbes.csv"
# b. "./field_data/microbes.csv"
# c. "field_data/microbes.csv"
# d. "../field_data/microbes.csv"
#
# ---
# ## Use `DataFrame.info` to find out more about a data frame.
# Write your code here
df.info()
# ## Use `DataFrame.describe` to get summary statistics about data.
#
# DataFrame.describe() gets the summary statistics of only the columns that have numerical data.
# All other columns are ignored.
# Write your code here
df.describe()
DataFrame.info
df.columns
df[['country', 'year']]
df[['year','gdpPercap']].describe()
# can use logical operator if on your data too.
lowest_gdp_df = df['gdpPercap']=='
# ---
# ## EtherPad:
# 1. Use the python cell below to find the minimum GDP per capita of all countries in 1972?
#
# Hint: You will need to use the DataFrame.loc[] to find all entries of 1972
# +
# Write your code here
df[['country','year','gdpPercap']]
# dataframe_name.loc iterates over every row
#any time the year is == 1972, the entire row is put into a new df called df_1972
df_1972 = df.loc[df['year']==1972]
# -
df_1972.describe()
# Vote for your answer on EtherPad
#
# a. 331.0
# b. 357.0
# c. 415.0
# d. 424.
#
# ---
# ## The `DataFrame.columns` variable stores information about the data frame's columns.
#
# * Note that this is a variable, *not* a function.
# * Like `math.pi`.
# * So do not use `()` to try to call it.
# print out the data frame columns
# ## Use `index_col` to specify that a column's values should be used as row headings.
#
# * Row headings are numbers (0 and 1 in this case).
# * Really want to index by country.
# * Pass the name of the column to `read_csv` as its `index_col` parameter to do this.
# re-read in the gapminder data with the "country" column/series sa the index_col
df = pandas.read_csv("gapminder_csv.txt", index_col="country")
print(df.head())
# * This is a `DataFrame`
# * This gives us many rows with the same index value ("e.g. Afghanistan")
# * Not good practice
# * lets re-read the table without the index_cols
# Write your code here
# ## Writing to csv file
# As well as the `read_csv` function for reading data from a file, Pandas can write data frames to files with a `to_****` function.
# * Pandas can write data frames to csv, html, excel (xlsx), json, and many more.
# E.g.
# `df.to_csv("./my_data.csv")`
#
# ---
# ## EXERCISE:
# 1. With the `gapminder_final.txt` file read in as a data frame, write out a copy of the data frame as a csv to a new file called `gapminder_final.csv` in the `data` directory in the `python_lessons` directory ("./data").
#
# ---
# Write your code here
# # -- COMMIT YOUR WORK TO GITHUB --
# ---
# ## Keypoints:
# * Use the Pandas library to do statistics on tabular data.
# * Use `index_col` to specify that a column's values should be used as row headings.
# * Use `DataFrame.info` to find out more about a data frame.
# * The `DataFrame.columns` variable stores information about the data frame's columns.
# * Use `DataFrame.T` to transpose a data frame.
# * Use `DataFrame.describe` to get summary statistics about data.
| python-lessons/04 - Reading a file with Pandas.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .r
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: R
# language: R
# name: ir
# ---
# # 1806554 <NAME> (DA LAB)
# # 1
x <- c(1,2,3,4,5)
print(sqrt(x))
x <- c(11,23,12,14,4)
paste("min :",min(x))
paste("max :",max(x))
paste("sum :",sum(x))
# # 2
row_names = c("row1", "row2", "row3", "row4")
col_names = c("col1", "col2", "col3")
M = matrix(c(1:12), nrow = 4,ncol = 3, byrow = TRUE, dimnames = list(row_names, col_names))
print("Original Matrix:")
print(M)
print(M[1,])
print(M[3,])
print(M[,2])
# # 3
v1 <- c(5,9,3)
v2 <- c(10,11,12,13,14,15)
result <- array(c(v1,v2),dim = c(3,3,2))
print(result)
print(result[3,,2])
print(result[1,3,1])
print(result[,,2])
print(result[,,1]+result[,,2])
# # 4
# +
players <- data.frame(
player_no = c(1:5),
name = c("Ganesh","Messi","Michael","Rohit","Ronnie"),
age = c(17,31,37,29,48),
profession= c("Sprinter","Footballer","Basketball","Cricketer","Snooker"),
grade = c("C","A","B","A","A+")
)
result <- data.frame(players$name,players$grade)
print(result)
# -
result1 <- players[1:3,]
print(result1)
result <- players[c(2,5),c(1,3)]
print(result)
| DA Lab/eval_test.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # calculate rotation and transposition matrix
# +
import os
data_folder = r'D:\Pu\20211208-P_brain_CTP11-500_M1_DNA_RNA-seq_hybrid'
before_position_file = os.path.join(data_folder, 'positions_before_align.txt')
after_position_file = os.path.join(data_folder, 'positions_after_align.txt')
# -
import numpy as np
import os, sys
# 1. alignment for manually picked points
def align_manual_points(pos_file_before, pos_file_after,
save=True, save_folder=None, save_filename='', verbose=True):
"""Function to align two manually picked position files,
they should follow exactly the same order and of same length.
Inputs:
pos_file_before: full filename for positions file before translation
pos_file_after: full filename for positions file after translation
save: whether save rotation and translation info, bool (default: True)
save_folder: where to save rotation and translation info, None or string (default: same folder as pos_file_before)
save_filename: filename specified to save rotation and translation points
verbose: say something! bool (default: True)
Outputs:
R: rotation for positions, 2x2 array
T: traslation of positions, array of 2
Here's example for how to translate points
translated_ps_before = np.dot(ps_before, R) + t
"""
# load position_before
if os.path.isfile(pos_file_before):
ps_before = np.loadtxt(pos_file_before, delimiter=',')
# load position_after
if os.path.isfile(pos_file_after):
ps_after = np.loadtxt(pos_file_after, delimiter=',')
# do SVD decomposition to get best fit for rigid-translation
c_before = np.mean(ps_before, axis=0)
c_after = np.mean(ps_after, axis=0)
H = np.dot((ps_before - c_before).T, (ps_after - c_after))
U, _, V = np.linalg.svd(H) # do SVD
# calcluate rotation
R = np.dot(V, U.T).T
if np.linalg.det(R) < 0:
R[:, -1] = -1 * R[:, -1]
# calculate translation
t = - np.dot(c_before, R) + c_after
# here's example for how to translate points
# translated_ps_before = np.dot(ps_before, R) + t
# save
if save:
if save_folder is None:
save_folder = os.path.dirname(pos_file_before)
if not os.path.exists(save_folder):
os.makedirs(save_folder)
if len(save_filename) > 0:
save_filename += '_'
rotation_name = os.path.join(save_folder, save_filename+'rotation')
translation_name = os.path.join(
save_folder, save_filename+'translation')
np.save(rotation_name, R)
np.save(translation_name, t)
return R, t
R, T = align_manual_points(before_position_file, after_position_file, save=False)
R, T
# # transpose 60x positions
old_positions = np.loadtxt(os.path.join(data_folder, 'positions_all.txt'), delimiter=',')
new_positions = np.dot(old_positions, R) + T
print(new_positions)
save_filename = os.path.join(data_folder, 'translated_positions_all.txt')
print(save_filename)
np.savetxt(save_filename, new_positions, fmt='%.2f', delimiter=',')
# # further adjust manually
manual_shift = np.array([-28.1, -8.7])
adjusted_new_positions = new_positions + manual_shift
adj_save_filename = os.path.join(data_folder, 'adjusted_translated_positions_all.txt')
print(adj_save_filename)
np.savetxt(adj_save_filename, adjusted_new_positions, fmt='%.2f', delimiter=',')
| During_experiment/STORM6/20211208-align_10x_positions_seq_DNA_RNA_rnase_treat.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.7.6 64-bit (''.venv37'': venv)'
# name: python3
# ---
# +
import math ###distributionscategorical
import functools
import operator
def cat(c, pi):
elems = [math.pow(pi_k, c_k) for (c_k, pi_k) in zip(c,pi)] #pi_k^c_k
return functools.reduce(operator.mul, elems) #リストの掛け算
## サイコロを表現 ##
pi = [1/6, 1/6, 1/6, 1/6, 1/6, 1/6] #出目の確率が全て1/6
c = [0,1,0,0,0,0] #2の目が出るという事象の1-of-K表現
cat(c, pi) #確率を求めてみる -> 0.166666... = 1/6
# +
from scipy.stats import dirichlet ###distributionsdir
tau = [1,2,3,4,5]
for i in range(3):
pi = dirichlet(tau).rvs()[0] #ディリクレ分布からπを生成
print("π=", pi, "合計:", sum(pi)) #πの合計は1になる
# -
| section_inference/distributions.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:message_env] *
# language: python
# name: conda-env-message_env-py
# ---
# # Westeros bis - emissions introduction and analysis
#
# Copy of the scenario ``westeros_bis_energymix1.ipynb`` with the implementation of:
# - emission impact of gas and coal
# - emissions cap of 500 Mt of CO2.
#
# ### Pre-requisites
# - You have the MESSAGEix framework installed and working
# - You have run (``westeros_bis_baseline.ipynb``) scenario and solved it successfully
# - You have run (``westeros_bis_energymix1.ipynb``) scenario
# +
import pandas as pd
import ixmp
import message_ix
from message_ix.util import make_df
# %matplotlib inline
# -
mp = ixmp.Platform()
# Recalling the model
model = 'Westeros Electrified'
base = message_ix.Scenario(mp, model=model, scenario='energy mix1')
# Creating emission scenario by cloning energy mix 1
scen_em3 = base.clone(model, 'emissions3','adding emissions cap',
keep_solution=False)
scen_em3.check_out()
year_df = scen_em3.vintage_and_active_years()
vintage_years, act_years = year_df['year_vtg'], year_df['year_act']
model_horizon = scen_em3.set('year')
country = 'Westeros'
# Introducing emissions category, parameter, and value for coal and gas technology
# +
# first we introduce the emission of CO2 and the emission category GHG
scen_em3.add_set('emission', 'CO2')
scen_em3.add_cat('emission', 'GHG', 'CO2')
# we now add CO2 emissions to the coal powerplant
base_emission_factor = {
'node_loc': country,
'year_vtg': vintage_years,
'year_act': act_years,
'mode': 'standard',
'unit': 'tCO2/kWa',
}
# adding new units to the model library (needed only once)
mp.add_unit('tCO2/kWa')
mp.add_unit('MtCO2')
# +
emission_factor = { # values from https://doi.org/10.1016/B978-0-8155-2049-8.10007-5 converted to tCO2/kWa
'coal_ppl': 7.07,
'ngcc_ppl': 3.11,
'wind_ppl': 0,
}
for tec, val in emission_factor.items():
df = make_df(base_emission_factor, technology=tec, value=val, emission='CO2')
scen_em3.add_par('emission_factor', df)
# -
# Introducing an upper bound
scen_em3.add_par('bound_emission', [country, 'GHG', 'all', 'cumulative'],
value=500., unit='MtCO2')
scen_em3.par('emission_factor')
# Solving the model
scen_em3.commit(comment='introducing emissions and setting an upper bound')
scen_em3.set_as_default()
scen_em3.solve()
scen_em3.var('OBJ')['lvl']
# Plotting the results
# +
from message_ix.reporting import Reporter
from message_ix.util.tutorial import prepare_plots
rep = Reporter.from_scenario(scen_em3)
prepare_plots(rep)
# -
# Activity
rep.set_filters(t=["coal_ppl", "wind_ppl","ngcc_ppl"])
rep.get("plot activity")
# Capacity
rep.get("plot capacity")
# Electricy price
rep.set_filters(t=None, c=["light"])
rep.get("plot prices")
# Take a more detailed look into carbon emissions by decade, and equivalent emission prices
emissions = scen_em3.var('EMISS', {'node': 'Westeros'})
emissions
emission_prices = scen_em3.var('PRICE_EMISSION')
emission_prices
# # Comments on the results
#
# In ``westeros_bis_emissions_cap3.ipynb`` with respect to the scenario ``westeros_bis_energymix1.ipynb``:
# - total cost is considerably higher, 455626.65625 against 408462.5625
# - Wind technology is considerably increased in second decade and present in the third decade, even if with an activity lower than previous decade (gas plant has longer lifetime).
# - Coal technology activity stops before the first decade even ends, being replaced mainly by gas in first and third decade, and with a strong contribution by wind in the second one
# - Gas technology is overall leading the electricity supply.
# - Electricity price is approximately constant at about 4.5 ¢/kW·h.
#
# With this strict cap the overall system cost is quite higher, together with the equivalent carbon price, and coal is excluded from the energy mix.
#
# Therefore, with the method of the emission bound, the sweet spot is achieved in the previous scenario ``westeros_bis_emissions_cap2.ipynb`` with a cap of 600 MtCO2 and a carbon price of about (10, 16, 27) USD/tCO2 in the three decades.
#
# In the next scenario it will be explored a different method of limiting emission, that is a constant carbon tax, and the existence of a 'sweet spot carbon price' will be explored.
# Close connection to the database
mp.close_db()
| 5_westeros_bis_emissions_cap3.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="gG_sSCizMEX7"
# <div>
# <img src="https://drive.google.com/uc?export=view&id=1vK33e_EqaHgBHcbRV_m38hx6IkG0blK_" width="350"/>
# </div>
#
# #**Artificial Intelligence - MSc**
# This notebook is designed specially for the module
#
# ET5003 - MACHINE LEARNING APPLICATIONS
#
# Instructor: <NAME>
# ###ET5003_GaussianProcesses
#
# © All rights reserved to the author, do not share outside this module.
#
# + [markdown] id="papPpf1XMaht"
# ## Introduction
# + [markdown] id="OJs6JMjSueq-"
# The concept of [Gaussian processes](https://en.wikipedia.org/wiki/Gaussian_process) is named after <NAME> because it is based on the notion of the Gaussian distribution (normal distribution).
#
# * Gaussian processes can be seen as an infinite-dimensional generalization of multivariate normal distributions.
#
# * Gaussian processes are useful in statistical modelling, benefiting from properties inherited from the normal distribution.
#
# * For example, if a random process is modelled as a Gaussian process, the distributions of various derived quantities can be obtained explicitly.
#
# * Such quantities include the average value of the process over a range of times and the error in estimating the average using sample values at a small set of times.
#
# * While exact models often scale poorly as the amount of data increases, multiple approximation methods have been developed which often retain good accuracy while drastically reducing computation time.
# + [markdown] id="iy7SUUCjm7YF"
# **Acknowledgement**
#
# This notebook is refurbished taking source code from <NAME>'s webpage and from the libraries numpy, GPy, pylab, and pymc3.
# + [markdown] id="Wg7VCbX77eAA"
# ## Libraries
# + id="5Q08kEv9Czgz"
# Suppressing Warnings:
import warnings
warnings.filterwarnings("ignore")
# + id="5cfDHD9BXh0s" colab={"base_uri": "https://localhost:8080/"} outputId="d835b7c4-6579-4f8c-9dea-87e41cff6f4f"
# https://pypi.org/project/GPy/
# !pip install gpy
# + id="SwGh2bagw4kg"
import GPy as GPy
import numpy as np
import pylab as pb
import pymc3 as pm
import scipy as sp
from keras.models import Sequential
from keras.layers import Dense
# %matplotlib inline
# + [markdown] id="y9P-eIBjw4kj"
# ## Data generation
# Generate data from a nonlinear function and use a Gaussian Process to sample it.
# + id="w2u0eCmOmIzA"
# seed the legacy random number generator
# to replicate experiments
seed = None
#seed = 7
np.random.seed(seed)
# + id="u5-Qn922OUiU" colab={"base_uri": "https://localhost:8080/"} outputId="294265a7-c9e1-4eba-ad1d-192c1ce8c5d0"
# Gaussian Processes
# https://gpy.readthedocs.io/en/deploy/GPy.kern.html
# Radial Basis Functions
# https://scikit-learn.org/stable/auto_examples/svm/plot_rbf_parameters.html
# kernel is a function that specifies the degree of similarity
# between variables given their relative positions in parameter space
kernel = GPy.kern.RBF(input_dim=1,lengthscale=0.15,variance=0.2)
print(kernel)
# + id="ACwm0S7Aw4kp"
# number of samples
num_samples_train = 250
num_samples_test = 200
# intervals to sample
a, b, c = 0.2, 0.6, 0.8
# points evenly spaced over [0,1]
interval_1 = np.random.rand(int(num_samples_train/2))*b - c
interval_2 = np.random.rand(int(num_samples_train/2))*b + c
X_new_train = np.sort(np.hstack([interval_1,interval_2]))
X_new_test = np.linspace(-1,1,num_samples_test)
X_new_all = np.hstack([X_new_train,X_new_test]).reshape(-1,1)
# vector of the means
μ_new = np.zeros((len(X_new_all)))
# covariance matrix
C_new = kernel.K(X_new_all,X_new_all)
# noise factor
noise_new = 0.1
# generate samples path with mean μ and covariance C
TF_new = np.random.multivariate_normal(μ_new,C_new,1)[0,:]
y_new_train = TF_new[0:len(X_new_train)] + np.random.randn(len(X_new_train))*noise_new
y_new_test = TF_new[len(X_new_train):] + np.random.randn(len(X_new_test))*noise_new
TF_new = TF_new[len(X_new_train):]
# + [markdown] id="G4JSncgEw4kq"
# In this example, first generate a nonlinear functions and then generate noisy training data from that function.
#
# The constrains are:
# * Training samples $x$ belong to either interval $[-0.8,-0.2]$ or $[0.2,0.8]$.
# * There is not data training samples from the interval $[-0.2,0.2]$.
# * The goal is to evaluate the extrapolation error outside in the interval $[-0.2,0.2]$.
# + colab={"base_uri": "https://localhost:8080/", "height": 284} id="5v6-zQSUb1m6" outputId="b53209b6-aa57-4ff9-ea62-6a6dc98bd687"
# plot
pb.figure()
pb.plot(X_new_test,TF_new,c='b',label='True Function',zorder=100)
# training data
pb.scatter(X_new_train,y_new_train,c='g',label='Train Samples',alpha=0.5)
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.legend()
pb.savefig("New_data.pdf")
# + [markdown] id="PFz4z-k_w4kw"
# ## Gaussian process
#
# + [markdown] id="Fs57xTsUvGIv"
# They are equivalent to a NN with infinite hidden-units.
# + id="8_u6g-m1v_1W"
# quadratic polynomial kernel
def QPK(x_1, x_2, α, degree):
xx1 = x_1**np.arange(0,deg+1)
xx2 = x_2**np.arange(0,deg+1)
return np.linalg.multi_dot([xx1,np.diag(α),xx2.T])
# + id="uJsRmCFFw4kw"
# sample points
x = np.linspace(-1,1,50).reshape(-1,1)
# degree of the polynomial kernel
deg = 2
# parameters
α = np.ones(deg+1)
Q = QPK(x, x, α, deg)
Z = np.zeros(len(x))
# build a quadratic polynomial function
f1 = np.random.multivariate_normal(Z,Q,15)
# + colab={"base_uri": "https://localhost:8080/"} id="jTjMoS5l8y1G" outputId="2045956a-36af-486f-9d96-30d2839d6388"
# show shape
f1.shape
# + colab={"base_uri": "https://localhost:8080/", "height": 300} id="R6U2dcafwF9g" outputId="cb7c8841-347c-48ce-f675-e1ecb351fcde"
# plot function samples
pb.plot(x,f1.T,':')
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.title("15 function samples")
pb.savefig("poly2.pdf")
# + colab={"base_uri": "https://localhost:8080/", "height": 300} id="O2TIHt8bwaJU" outputId="994f5c9e-d808-4751-9eef-cedc7f3de382"
# plot prior
pb.plot(x,np.mean(f1,axis=0),label='mean')
pb.legend()
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.title("Prior")
pb.savefig("quadratic_polynomial.pdf")
# + id="fXF0SyOU8sIU"
# build a larger quadratic polynomial function
f2 = np.random.multivariate_normal(Z,Q,15000)
# + colab={"base_uri": "https://localhost:8080/"} id="x6rsicZ98tj5" outputId="c54fccad-12ae-4ca2-b4b2-502207db8c8b"
# show shape
f2.shape
# + id="_cLd3ufgw4kx"
# square exponential kernel
def SEK(x_1, x_2, lengthscale, sigma_f):
x1a = x_1**2/lengthscale**2
x1b = x_1/lengthscale**2
x2a = x_2**2/lengthscale**2
A = np.sum(x1a, 1).reshape(-1, 1)
B = np.sum(x2a, 1)
C = 2 * np.dot(x1b, x_2.T)
sqdist = A + B - C
return sigma_f**2 * np.exp(-0.5 * sqdist)
# + id="wJ6erZkX88oF"
# sample points
x3 = np.linspace(-1,1,40).reshape(-1,1)
lengthscale = np.ones(1)*0.3
# build a square exponential function
sigma_f3 = 1
Z3 = np.zeros(len(x3))
E3 = SEK(x3,x3, lengthscale, sigma_f3)
f3 = np.random.multivariate_normal(Z3, E3, 15)
# + colab={"base_uri": "https://localhost:8080/", "height": 302} id="zmhBjreO0OAz" outputId="49dd9435-8afd-4880-d71a-b6e5633ee649"
# plot family of functions
pb.plot(x3,f3.T,':')
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.title("lengthscale = "+str(lengthscale[0]) + " " + "$\sigma_f3$ = "+str(sigma_f3) )
pb.savefig("RBF_03.pdf")
# + id="-2dm_0BSw4kx" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="a63531e6-5bff-4747-dbe7-fb0d6d88fe9f"
# plot average
pb.plot(x3,np.mean(f3,axis=0),label='mean')
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.legend()
pb.title("Prior")
pb.savefig("rbf.pdf")
# + id="0-_iHLZXw4kx"
def posterior_predictive(X_s, X_train, y_train, l=1.0, sigma_f=1.0, sigma_y=1e-8, K=SEK):
'''
Compute sufficient statistics of the GP posterior predictive distribution
from m training, data X_train, y_train, and n new inputs X_s.
Args: X_s: new input locations (n x d)
X_train: training locations (m x d)
y_train: training targets (m x 1)
l: Kernel length parameter
sigma_f: Kernel vertical variation parameter
sigma_y: Noise parameter
K: Kernel [SEK]
Returns: Posterior mean vector (n x d) and covariance matrix (n x n).
'''
K0 = K(X_train, X_train, l, sigma_f) + sigma_y**2 * np.eye(len(X_train))
K_s = K(X_train, X_s, l, sigma_f)
K_ss = K(X_s, X_s, l, sigma_f) + 1e-8 * np.eye(len(X_s))
K_inv = np.linalg.inv(K0)
mu_s = K_s.T.dot(K_inv).dot(y_train)
cov_s = K_ss - K_s.T.dot(K_inv).dot(K_s)
return mu_s, cov_s
# + id="GDievjLmw4kx"
# compute posterior
xa = np.array([[0,0.3,-0.8,0.2,0.5,-0.4,-0.6]]).reshape(-1,1)
xa1 = np.linspace(-1,1,50).reshape(-1,1)
E4 = SEK(xa,xa, lengthscale, sigma_f3)
ya = np.random.multivariate_normal(np.zeros(len(xa)),E4,1).T
xb = xa[0:4,:]
yb = ya[0:4,:]
# get mean vector and covariance matrix
mu_A, cov_A = posterior_predictive(xa1, xb, yb, l=lengthscale, sigma_f=sigma_f3, sigma_y=1e-8, K=SEK)
# + [markdown] id="s02G6YEtw4kx"
# ## Posterior after i observations
# + id="xdpEN6DEw4kx" colab={"base_uri": "https://localhost:8080/", "height": 303} outputId="f8c0b2e6-2eec-4494-db82-02bf1255bff7"
# posterior plot
f_postA = np.random.multivariate_normal(mu_A[:,0], cov_A, 25)
pb.plot(xa1,f_postA.T,':')
pb.scatter(xb, yb,c='r',alpha=0.7,Zorder=100)
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.savefig("posterior.pdf")
pb.ylim([-3.75,3.75])
# + id="exNMzPt9w4ky" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="df96c896-4b9f-400d-98fc-d6ede48360d8"
# Average
f_postB = np.random.multivariate_normal(mu_A[:,0], cov_A, 14550)
pb.xlabel("x", fontsize=16)
pb.ylabel("y", fontsize=16,rotation=0)
pb.plot(xa1, np.mean(f_postB,axis=0), label='mean')
pb.scatter(xb, yb,c='r', alpha=0.7, Zorder=100)
pb.legend()
pb.title("Posterior Mean")
pb.savefig("PosteriorMean.pdf")
# + [markdown] id="aktzaEBiw4ky"
# ## 50 observations
# + colab={"base_uri": "https://localhost:8080/", "height": 287} id="v9c6iaD-PdXt" outputId="b10aed00-9c3a-4af5-f90b-ac470b212316"
#
lengthscale=0.15
np.random.seed(42)
x5 = np.linspace(-1,1,50).reshape(-1,1)
x5a = np.linspace(-1,1,50).reshape(-1,1)
y5 = np.random.multivariate_normal(np.zeros(len(x)),SEK(x5,x5, lengthscale, sigma_f3),1).T
pb.plot(x5,y5)
# + id="8IIba6S9w4ky" colab={"base_uri": "https://localhost:8080/", "height": 287} outputId="5182e157-56a2-42cc-ca96-024c6e9c50f3"
#
mu, cov = posterior_predictive(x5a, x5, y5, l=lengthscale, sigma_f=sigma_f3, sigma_y=1e-5, K=SEK)
f_post5 = np.random.multivariate_normal(mu[:,0], cov, 25)
pb.plot(x5a,f_post5.T,':')
pb.scatter(x5, y5,c='r',alpha=0.7,Zorder=100)
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.savefig("om.pdf")
# + id="9bS8FpOBw4ky"
#
np.random.seed(7)
xa_6 = np.array([[0,0.3,-0.8]]).reshape(-1,1)
xa1_6 = np.linspace(-1,1,50).reshape(-1,1)
ya_6 = np.random.multivariate_normal(np.zeros(len(xa_6)),SEK(xa_6,xa_6, lengthscale, sigma_f3),1).T
ya_6 = ya_6 + np.random.randn(ya_6.shape[0],ya_6.shape[1])*0.2
xb_6 = xa_6[0:3,:]
yb_6 = ya_6[0:3,:]
mu6, cov6 = posterior_predictive(xa1_6, xb_6, yb_6, l=lengthscale, sigma_f=sigma_f3, sigma_y=0.2, K=SEK)
# + id="_EFeAErcw4ky" colab={"base_uri": "https://localhost:8080/", "height": 300} outputId="22166e44-02ca-424c-9889-d482bcf22abe"
#
f_post6 = np.random.multivariate_normal(mu6[:,0], cov6, 14550)
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.plot(xa1_6,np.mean(f_post6,axis=0),label='mean')
pb.scatter(xb_6, yb_6,c='r',alpha=0.7,Zorder=100)
pb.legend()
pb.title("Posterior Mean")
pb.savefig("PosteriorMean.pdf")
# + id="iEbz2fPLWJsL"
#
np.random.seed(7)
xa_6 = np.array([[0,0.3,-0.8]]).reshape(-1,1)
xa1_6 = np.linspace(-1,1,50).reshape(-1,1)
ya_6 = np.random.multivariate_normal(np.zeros(len(xa_6)),SEK(xa_6,xa_6, lengthscale, sigma_f3),1).T
ya_6 = ya_6 + np.random.randn(ya_6.shape[0],ya_6.shape[1])*0.2
xb_6 = xa_6[0:3,:]
yb_6 = ya_6[0:3,:]
mu6, cov6 = posterior_predictive(xa1_6, xb_6, yb_6, l=lengthscale, sigma_f=sigma_f3, sigma_y=0.2, K=SEK)
# + id="rb3N9DoTWBSQ"
#
xa_7 = np.linspace(-1,1,50).reshape(-1,1)
xa1_7 = np.linspace(-1,1,50).reshape(-1,1)
ya_7 = np.random.multivariate_normal(np.zeros(len(xa_7)),SEK(xa_7,xa_7, lengthscale, sigma_f3),1).T
ya_7 = ya_7 + np.random.randn(ya_7.shape[0],ya_7.shape[1])*0.2
mu7, cov7 = posterior_predictive(xa1_7, xa_7, ya_7, l=lengthscale, sigma_f=sigma_f3, sigma_y=0.2)
# + id="-X7NiN9Ww4kz" colab={"base_uri": "https://localhost:8080/", "height": 393} outputId="88424771-44be-4154-b33d-0d41ccf323c0"
#
pb.figure(figsize=(10,6))
f_post7 = np.random.multivariate_normal(mu7[:,0], cov7, 500)
pb.plot(xa_7,np.mean(f_post7,axis=0),color='orange',label='mean')
pb.scatter(xa_7, ya_7,c='r',alpha=0.7,Zorder=100)
pb.xlabel("x",fontsize=16)
pb.ylabel("y",fontsize=16,rotation=0)
pb.legend()
pb.savefig("PosteriorMean7.pdf")
# + id="clivNkpPw4kz" colab={"base_uri": "https://localhost:8080/"} outputId="c822af2c-0dec-42cd-c6e0-2e8f2ddd6cdf"
print(f_post7)
| Week-5/ET5003_GaussianProcesses.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import tkinter
import translate
from translate import translate
def btn_click():
lang = str(translate(txt_1.get()))
txt_2.insert(0, lang)
# 画面作成
tki = tkinter.Tk()
tki.geometry('300x300')
tki.title('翻訳機')
# ラベル
lbl_1 = tkinter.Label(text='英文:')
lbl_1.place(x=30, y=70)
lbl_2 = tkinter.Label(text='日本文:')
lbl_2.place(x=30, y=100)
# テキストボックス
txt_1 = tkinter.Entry(width=20)
txt_1.place(x=90, y=70)
txt_2 = tkinter.Entry(width=20)
txt_2.place(x=90, y=100)
# ボタン
btn = tkinter.Button(tki, text='翻訳', command=btn_click)
btn.place(x=140, y=170)
# 画面をそのまま表示
tki.mainloop()
# -
| Untitled.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: bccp
# language: python
# name: bccp
# ---
# %pylab inline
from nbodykit.lab import *
from h5py import File
f1 = File('/global/cscratch1/sd/yfeng1/baoshift-hdf5/00000600-40-now/fof_0.6250.hdf5')
list(f1.keys())
pos = f1['FOFGroups']['Position'][:]
vel = f1['FOFGroups']['Velocity'][:]
mass = f1['FOFGroups']['Length'][:] * 27.75e10 * 0.3 * (1380 / 2048.) ** 3
posrandom = numpy.random.uniform(size=(100000, 3))
_ = hist2d(pos[:, 0], pos[:, 1], bins=80)
from nbodykit.cosmology import Planck15
Planck15.comoving_distance(0.5)
from numpy import arcsin, arctan2, pi
def pos2radec(pos, obs):
x, y, z= (pos - obs).T
r = (x**2+y**2+z**2) ** 0.5
dec = arcsin(z / r)
ra = arctan2(y, x)
return ra, dec, r
ra, dec, r = pos2radec(pos, [.5, .5, .5])
_ = hist2d(ra[r < 0.5], sin(dec[r < 0.5]), bins=80)
from scipy.interpolate import UnivariateSpline
from nbodykit.cosmology import Planck15
# +
# Planck15.comoving_distance?
# -
def r2z(cosmology, r1):
zgrid = numpy.linspace(0, 9.0, 10000)
rgrid = cosmology.comoving_distance(zgrid)
spl = UnivariateSpline(rgrid, zgrid)
return spl(r1)
z = r2z(Planck15, r * 1380.)
import healpy
import numpy
class DESIFootprint:
def __init__(self, desi_map):
map = healpy.read_map('/project/projectdirs/desi/mocks/GaussianRandomField/v0.0.4/desi-map.fits')
self.map = map
def f(self, ra, dec):
pix = healpy.ang2pix(nside=32, phi=ra, theta=numpy.pi / 2 - dec)
return self.map[pix]
ftp = DESIFootprint('/project/projectdirs/desi/mocks/GaussianRandomField/v0.0.4/desi-map.fits')
class Mock:
def __init__(self, pos, cosmo, ftp, obs):
ra, dec, r = pos2radec(pos, obs=obs)
z = r2z(cosmo, r)
f = ftp.f(ra, dec)
self.ra = ra
self.dec = dec
self.z = z
self.f = f
data = Mock(pos * 1380, Planck15, ftp, obs=[690, 690, 690])
random = Mock(posrandom * 1380, Planck15, ftp, obs=[690, 690, 690])
Planck15.comoving_distance([0, 0.6, 1.0, 1.5, 2, 2.5])
lowmass = (data.z < 0.3) & (data.f > 0.2) & (mass < 1e13)
highmass = (data.z < 0.3) & (data.f > 0.2) & (mass > 1e13)
randommask = (random.z < 0.3) & (random.f > 0.2)
_ = hist2d(data.ra[lowmass], sin(data.dec[lowmass]), bins=80)
_ = hist2d(random.ra[randommask], sin(random.dec[randommask]), bins=80)
from kdcount.sphere import AngularBinning
from kdcount.sphere import points
from kdcount.correlate import paircount
ds_lowmass = points(np.degrees(data.ra[lowmass]), np.degrees(data.dec[lowmass]))
bins = AngularBinning(linspace(0, 2.0, 20))
# +
#r = paircount(ds_lowmass, ds_lowmass, bins)
# -
data.ra[lowmass], data.dec[lowmass]
healpy.anafast()
from Corrfunc.mocks.DDtheta_mocks import DDtheta_mocks
bins = linspace(0, 10, 20)
data.dec.max()
# +
#DDtheta_mocks(1, 1, bins, np.degrees(data.ra[lowmass]+ pi), np.degrees(data.dec[lowmass]), verbose=True, )
# -
| notebooks/trunk/RADECMock.ipynb |
# + deletable=false editable=false
# Initialize Otter
import otter
grader = otter.Notebook()
# -
# # Exam
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
# ### Question 1
# Write an infinite generator of the Fibonacci sequence.
def fib():
a, b = 0, 1
yield 0
while True:
yield b
a, b = b, a + b
# ### Question 2
# Plot $f(x) = \cos 2x$ over $(0,10)$.
x = np.linspace(0, 10, 100)
y = np.cos(2 * x)
plt.plot(x, y);
# ## End Exam
# + [markdown] deletable=false editable=false
# ## Submission
#
# Make sure you have run all cells in your notebook in order before running the cell below, so that all images/graphs appear in the output. The cell below will generate a zipfile for you to submit. **Please save before exporting!**
# + deletable=false editable=false
# Save your notebook first, then run this cell to export your submission.
grader.export()
# -
#
| test/dist-correct-150/exam_7/test-exam.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="6emPPi8Zvt30"
# # Python - intro 101
#
# ## Lab 1 - Qué va a crear
#
# En este desafío, creará un contador de calorías que solicita al usuario lo siguiente:
#
# - La fecha actual (en cualquier formato)
# - Calorías ingeridas en el desayuno
# - Calorías ingeridas en la comida
# - Calorías ingeridas en la cena
# - Calorías ingeridas en tentempiés
#
# Después, el programa sumará todas las calorías y les dará formato de mensaje.
# + colab={"base_uri": "https://localhost:8080/"} id="mID9DJRxvvU5" outputId="605200fe-2f8e-4d49-f972-2d607fce4acc"
# Creamos las variables del contador
print("¿Què día es hoy?")
# + colab={"base_uri": "https://localhost:8080/"} id="GAHuwqUcwEEB" outputId="a8dd35c4-ca24-4db4-e355-f110f9a9a965"
# Para la fecha creamos una variable del tipo input()
date = input()
# + colab={"base_uri": "https://localhost:8080/", "height": 36} id="_hfmKA2iu1R9" outputId="c7b311a4-c931-4479-9d7f-e37b4d6496b8"
date
# + colab={"base_uri": "https://localhost:8080/"} id="zvc8A-XlwRxC" outputId="8380d820-94b6-4a1a-8529-6095d616b043"
print("<NAME>")
# + colab={"base_uri": "https://localhost:8080/"} id="XG9qlmvywdDi" outputId="1edf60f1-973f-4f79-f288-8531ddfce0d6"
breakfast = int(input())
# + colab={"base_uri": "https://localhost:8080/"} id="XuhvBuDwwkBC" outputId="ec20aad4-7158-4588-a417-6a6adf9ece3e"
breakfast
# + id="6N8Zj1NPwipJ"
# Tiene que recoger los demás datos y sumarlos
# AQUI VUESTRO CÓDIGO
# + id="Crw3e9Edw06d"
# Sumar
#totalCal = # sumamos las variables anteriores
# + id="vkhH1wwLw90S"
# OUTPUT
print("El total de las calorías del día : ")
# + id="DQCB4siBw7bm"
# + id="cSIznB79wwQz"
# + id="dyVHq-CKwgX7"
| 01_RampUp/week2/labs/Lab01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="jzhhIIfIXE-J"
# Mounted at google drive and change the path to Eval directory
# + colab={"base_uri": "https://localhost:8080/"} id="0qKUERXgWefG" executionInfo={"status": "ok", "timestamp": 1639021023087, "user_tz": -480, "elapsed": 3118, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="196d15ce-b0ec-4e50-d61d-0854e04d38b3"
# Mounted at google drive
from google.colab import drive
drive.mount('/content/drive')
# + id="6ZM3VICMsP_n" executionInfo={"status": "ok", "timestamp": 1639021023088, "user_tz": -480, "elapsed": 7, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}}
# Change the path to Eval directory
import os
os.chdir('/content/drive/MyDrive/SR_Code/Scripts/Data/Test/psnr_ssim')
# + [markdown] id="Zi_WmIBVsRaz"
# Part 1: evaluate the performance of the model with RRDB and resnet architecutres on HHFQ dataset
# + colab={"base_uri": "https://localhost:8080/", "height": 285} id="wKsbT160s33n" executionInfo={"status": "ok", "timestamp": 1639021023941, "user_tz": -480, "elapsed": 859, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="e55b636c-d9d1-4a2a-f759-0250247e2cec"
# Violinplot of PSNR and SSIM scores on FFHQ test set for RRDB and Resnet architectures
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="white")
import pandas as pd
import json
import numpy as np
with open('PSNR_SSIM_resnet.json','r') as f:
load_resnet_hq = json.load(f)
with open('PSNR_SSIM_rrdb.json','r') as f:
load_rrdb_hq = json.load(f)
with open('PSNR_SSIM_bicubic.json','r') as f:
load_bic_hq = json.load(f)
with open('PSNR_SSIM_srgan_hhfq.json','r') as f:
load_srgan_hq = json.load(f)
# assign data of lists.
data1 = {'PSNR score': load_resnet_hq['PSNR'], 'SSIM score':load_resnet_hq['SSIM'] }
# Create DataFrame.
df1 = pd.DataFrame(data1)
df1['model type'] = 'Resnet'
data2 = {'PSNR score': load_rrdb_hq['PSNR'], 'SSIM score':load_rrdb_hq['SSIM']}
# Create DataFrame.
df2 = pd.DataFrame(data2)
df2['model type'] = 'rrdb'
df_1 = pd.concat([df1,df2],axis=0,join='outer')
sns.violinplot(x='model type', y='PSNR score', showmeans = True, data = df_1)
plt.savefig('PSNR_score_1.png')
# + colab={"base_uri": "https://localhost:8080/", "height": 285} id="u66gm13xvxH3" executionInfo={"status": "ok", "timestamp": 1639021023942, "user_tz": -480, "elapsed": 17, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="601dc2f5-1f42-451c-e4d3-b1b3c79b0593"
sns.violinplot(x='model type', y='SSIM score', data = df_1)
plt.savefig('SSIM_score_1.png')
# + [markdown] id="O-EnHIgt-poc"
# Part 2: evaluate the performance of different models on HHFQ dataset
# + colab={"base_uri": "https://localhost:8080/", "height": 143} id="wM5KREqcyN2D" executionInfo={"status": "ok", "timestamp": 1639021023943, "user_tz": -480, "elapsed": 16, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="b82169c0-e13e-4cb3-d9ba-eb4e2b827041"
# Output average PSNR and SSIM score on FFHQ dataset
# Calculate the PSNR and SSIM score with bicubic model
data_bic = {'PSNR score': np.mean(load_bic_hq['PSNR']), 'SSIM score': np.mean(load_bic_hq['SSIM']) }
df_bic_hq = pd.DataFrame(data_bic, index = ['bicubic'])
# Calculate the PSNR and SSIM score with SRGAN model
data_srgan = {'PSNR score': np.mean(load_srgan_hq['PSNR']), 'SSIM score': np.mean(load_srgan_hq['SSIM']) }
df_srgan_hq = pd.DataFrame(data_srgan, index = ['srgan'])
# Calculate the PSNR and SSIM score with our model
data_resnet = {'PSNR score': np.mean(load_resnet_hq['PSNR']), 'SSIM score': np.mean(load_resnet_hq['SSIM']) }
df_resnet_hq = pd.DataFrame(data_resnet, index = ['mymodel'])
df_2 = pd.concat([df_bic_hq, df_srgan_hq, df_resnet_hq],axis=0,join='outer')
df_2
# + [markdown] id="QuXDW_CcYye4"
# Part 2: evaluate the performance of different models on self-collected dataset
# + id="Jn8D2yoJ1mwQ" executionInfo={"status": "ok", "timestamp": 1639021023944, "user_tz": -480, "elapsed": 16, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}}
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(style="white")
import pandas as pd
import json
import numpy as np
with open('PSNR_SSIM_sci_bic.json','r') as f:
load_bic_self = json.load(f)
with open('PSNR_SSIM_srgan_sci.json','r') as f:
load_srgan_self = json.load(f)
with open('PSNR_SSIM_sci.json','r') as f:
load_my_self = json.load(f)
# + colab={"base_uri": "https://localhost:8080/", "height": 143} id="pcFPzd-lXGn9" executionInfo={"status": "ok", "timestamp": 1639021023944, "user_tz": -480, "elapsed": 16, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="1bb5cd32-3aca-4991-abaa-fefc8f8d96d6"
# Output average PSNR and SSIM score on self-collected dataset
# Calculate the PSNR and SSIM score with bicubic model
data_bic = {'PSNR score': np.mean(load_bic_self['PSNR']), 'SSIM score': np.mean(load_bic_self['SSIM']) }
df_bic_self = pd.DataFrame(data_bic, index = ['bicubic'])
# Calculate the PSNR and SSIM score with SRGAN model
data_srgan = {'PSNR score': np.mean(load_srgan_self['PSNR']), 'SSIM score': np.mean(load_srgan_self['SSIM']) }
df_srgan_self = pd.DataFrame(data_srgan, index = ['srgan'])
# Calculate the PSNR and SSIM score with our model
data_my = {'PSNR score': np.mean(load_my_self['PSNR']), 'SSIM score': np.mean(load_my_self['SSIM']) }
df_my_self = pd.DataFrame(data_my, index = ['mymodel'])
df_3 = pd.concat([df_bic_self,df_srgan_self,df_my_self],axis=0,join='outer')
df_3
# + colab={"base_uri": "https://localhost:8080/", "height": 285} id="g7R8uIczXGrP" executionInfo={"status": "ok", "timestamp": 1639021040403, "user_tz": -480, "elapsed": 520, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="80e756bb-f80e-49e6-cc9f-1b207ca1aa52"
# Violinplot of PSNR and SSIM scores on self-collected dataset of Bicubic, SRGAN and our model
# assign data of lists.
data_bic = {'PSNR score': load_bic_self['PSNR'], 'SSIM score':load_bic_self['SSIM'] }
# Create DataFrame.
df1 = pd.DataFrame(data_bic)
df1['model type'] = 'bicubic'
data_srgan = {'PSNR score': load_srgan_self['PSNR'], 'SSIM score':load_srgan_self['SSIM']}
# Create DataFrame.
df2 = pd.DataFrame(data_srgan)
df2['model type'] = 'srgan'
data_my = {'PSNR score': load_my_self['PSNR'], 'SSIM score':load_my_self['SSIM']}
# Create DataFrame.
df3 = pd.DataFrame(data_my)
df3['model type'] = 'mymodel'
df_4 = pd.concat([df1,df2,df3],axis=0,join='outer')
sns.violinplot(x='model type', y='PSNR score', showmeans = True, data = df_4)
plt.savefig('PSNR_score_2.png')
# + colab={"base_uri": "https://localhost:8080/", "height": 285} id="NahfMJftXGxx" executionInfo={"status": "ok", "timestamp": 1639021051338, "user_tz": -480, "elapsed": 557, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a/default-user=s64", "userId": "03343040780355705652"}} outputId="62b4b248-187d-4f2b-913e-f2bf14180c88"
sns.violinplot(x='model type', y='SSIM score', data = df_4)
plt.savefig('SSIM_score_2.png')
| Eval_SR.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: 'Python 3.7.11 64-bit (''tf2'': conda)'
# name: python3
# ---
# <i>Copyright (c) Microsoft Corporation. All rights reserved.</i>
#
# <i>Licensed under the MIT License.</i>
# # Sequential Recommender Quick Start
#
# ### Example: SLi_Rec : Adaptive User Modeling with Long and Short-Term Preferences for Personailzed Recommendation
# Unlike a general recommender such as Matrix Factorization or xDeepFM (in the repo) which doesn't consider the order of the user's activities, sequential recommender systems take the sequence of the user behaviors as context and the goal is to predict the items that the user will interact in a short time (in an extreme case, the item that the user will interact next).
#
# This notebook aims to give you a quick example of how to train a sequential model based on a public Amazon dataset. Currently, we can support NextItNet \[4\], GRU4Rec \[2\], Caser \[3\], A2SVD \[1\], SLi_Rec \[1\], and SUM \[5\]. Without loss of generality, this notebook takes [SLi_Rec model](https://www.microsoft.com/en-us/research/uploads/prod/2019/07/IJCAI19-ready_v1.pdf) for example.
# SLi_Rec \[1\] is a deep learning-based model aims at capturing both long and short-term user preferences for precise recommender systems. To summarize, SLi_Rec has the following key properties:
#
# * It adopts the attentive "Asymmetric-SVD" paradigm for long-term modeling;
# * It takes both time irregularity and semantic irregularity into consideration by modifying the gating logic in LSTM.
# * It uses an attention mechanism to dynamic fuse the long-term component and short-term component.
#
# In this notebook, we test SLi_Rec on a subset of the public dataset: [Amazon_reviews](http://snap.stanford.edu/data/amazon/productGraph/categoryFiles/reviews_Movies_and_TV_5.json.gz) and [Amazon_metadata](http://snap.stanford.edu/data/amazon/productGraph/categoryFiles/meta_Movies_and_TV.json.gz)
#
# This notebook is tested under TF 2.6.
# ## 0. Global Settings and Imports
# +
import sys
import os
import logging
import papermill as pm
import scrapbook as sb
from tempfile import TemporaryDirectory
import numpy as np
import tensorflow.compat.v1 as tf
tf.get_logger().setLevel('ERROR') # only show error messages
from recommenders.utils.timer import Timer
from recommenders.utils.constants import SEED
from recommenders.models.deeprec.deeprec_utils import (
prepare_hparams
)
from recommenders.datasets.amazon_reviews import download_and_extract, data_preprocessing
from recommenders.datasets.download_utils import maybe_download
from recommenders.models.deeprec.models.sequential.sli_rec import SLI_RECModel as SeqModel
#### to use the other model, use one of the following lines:
# from recommenders.models.deeprec.models.sequential.asvd import A2SVDModel as SeqModel
# from recommenders.models.deeprec.models.sequential.caser import CaserModel as SeqModel
# from recommenders.models.deeprec.models.sequential.gru4rec import GRU4RecModel as SeqModel
# from recommenders.models.deeprec.models.sequential.sum import SUMModel as SeqModel
#from recommenders.models.deeprec.models.sequential.nextitnet import NextItNetModel
from recommenders.models.deeprec.io.sequential_iterator import SequentialIterator
#from recommenders.models.deeprec.io.nextitnet_iterator import NextItNetIterator
print("System version: {}".format(sys.version))
print("Tensorflow version: {}".format(tf.__version__))
# -
## ATTENTION: change to the corresponding config file, e.g., caser.yaml for CaserModel, sum.yaml for SUMModel
yaml_file = '../../recommenders/models/deeprec/config/sli_rec.yaml'
# #### Parameters
# + tags=["parameters"]
EPOCHS = 10
BATCH_SIZE = 400
RANDOM_SEED = SEED # Set None for non-deterministic result
data_path = os.path.join("..", "..", "tests", "resources", "deeprec", "slirec")
# -
# ## 1. Input data format
# The input data contains 8 columns, i.e., `<label> <user_id> <item_id> <category_id> <timestamp> <history_item_ids> <history_cateory_ids> <hitory_timestamp>` columns are seperated by `"\t"`. item_id and category_id denote the target item and category, which means that for this instance, we want to guess whether user user_id will interact with item_id at timestamp. `<history_*>` columns record the user behavior list up to `<timestamp>`, elements are separated by commas. `<label>` is a binary value with 1 for positive instances and 0 for negative instances. One example for an instance is:
#
# `1 A1QQ86H5M2LVW2 B0059XTU1S Movies 1377561600 B002ZG97WE,B004IK30PA,B000BNX3AU,B0017ANB08,B005LAIHW2 Movies,Movies,Movies,Movies,Movies 1304294400,1304812800,1315785600,1316304000,1356998400`
#
# In data preprocessing stage, we have a script to generate some ID mapping dictionaries, so user_id, item_id and category_id will be mapped into interager index starting from 1. And you need to tell the input iterator where is the ID mapping files are. (For example, in the next section, we have some mapping files like user_vocab, item_vocab, and cate_vocab). The data preprocessing script is at [recommenders/dataset/amazon_reviews.py](../../recommenders/dataset/amazon_reviews.py), you need to call the `_create_vocab(train_file, user_vocab, item_vocab, cate_vocab)` function. Note that ID vocabulary only creates from the train_file, so the new IDs in valid_file or test_file will be regarded as unknown IDs and assigned with a defualt 0 index.
#
# Only the SLi_Rec model is time-aware. For the other models, you can just pad some meaningless timestamp in the data files to fill up the format, the models will ignore these columns.
#
# We use Softmax to the loss function. In training and evalution stage, we group 1 positive instance with num_ngs negative instances. Pair-wise ranking can be regarded as a special case of Softmax ranking, where num_ngs is set to 1.
#
# More specifically, for training and evalation, you need to organize the data file such that each one positive instance is followd by num_ngs negative instances. Our program will take 1+num_ngs lines as a unit for Softmax calculation. num_ngs is a parameter you need to pass to the `prepare_hparams`, `fit` and `run_eval` function. `train_num_ngs` in `prepare_hparams` denotes the number of negative instances for training, where a recommended number is 4. `valid_num_ngs` and `num_ngs` in `fit` and `run_eval` denote the number in evalution. In evaluation, the model calculates metrics among the 1+num_ngs instances. For the `predict` function, since we only need to calcuate a socre for each individual instance, there is no need for num_ngs setting. More details and examples will be provided in the following sections.
#
# For training stage, if you don't want to prepare negative instances, you can just provide positive instances and set the parameter `need_sample=True, train_num_ngs=train_num_ngs` for function `prepare_hparams`, our model will dynamicly sample `train_num_ngs` instances as negative samples in each mini batch.
#
# ### Amazon dataset
# Now let's start with a public dataset containing product reviews and metadata from Amazon, which is widely used as a benchmark dataset in recommemdation systems field.
# +
# for test
train_file = os.path.join(data_path, r'train_data')
valid_file = os.path.join(data_path, r'valid_data')
test_file = os.path.join(data_path, r'test_data')
user_vocab = os.path.join(data_path, r'user_vocab.pkl')
item_vocab = os.path.join(data_path, r'item_vocab.pkl')
cate_vocab = os.path.join(data_path, r'category_vocab.pkl')
output_file = os.path.join(data_path, r'output.txt')
reviews_name = 'reviews_Movies_and_TV_5.json'
meta_name = 'meta_Movies_and_TV.json'
reviews_file = os.path.join(data_path, reviews_name)
meta_file = os.path.join(data_path, meta_name)
train_num_ngs = 4 # number of negative instances with a positive instance for training
valid_num_ngs = 4 # number of negative instances with a positive instance for validation
test_num_ngs = 9 # number of negative instances with a positive instance for testing
sample_rate = 0.01 # sample a small item set for training and testing here for fast example
input_files = [reviews_file, meta_file, train_file, valid_file, test_file, user_vocab, item_vocab, cate_vocab]
if not os.path.exists(train_file):
download_and_extract(reviews_name, reviews_file)
download_and_extract(meta_name, meta_file)
data_preprocessing(*input_files, sample_rate=sample_rate, valid_num_ngs=valid_num_ngs, test_num_ngs=test_num_ngs)
#### uncomment this for the NextItNet model, because it does not need to unfold the user history
# data_preprocessing(*input_files, sample_rate=sample_rate, valid_num_ngs=valid_num_ngs, test_num_ngs=test_num_ngs, is_history_expanding=False)
# -
# #### 1.1 Prepare hyper-parameters
# prepare_hparams() will create a full set of hyper-parameters for model training, such as learning rate, feature number, and dropout ratio. We can put those parameters in a yaml file (a complete list of parameters can be found under our config folder) , or pass parameters as the function's parameters (which will overwrite yaml settings).
#
# Parameters hints: <br>
# `need_sample` controls whether to perform dynamic negative sampling in mini-batch.
# `train_num_ngs` indicates how many negative instances followed by one positive instances. <br>
# Examples: <br>
# (1) `need_sample=True and train_num_ngs=4`: There are only positive instances in your training file. Our model will dynamically sample 4 negative instances for each positive instances in mini-batch. Note that if need_sample is set to True, train_num_ngs should be greater than zero. <br>
# (2) `need_sample=False and train_num_ngs=4`: In your training file, each one positive line is followed by 4 negative lines. Note that if need_sample is set to False, you must provide a traiing file with negative instances, and train_num_ngs should match the number of negative number in your training file.
### NOTE:
### remember to use `_create_vocab(train_file, user_vocab, item_vocab, cate_vocab)` to generate the user_vocab, item_vocab and cate_vocab files, if you are using your own dataset rather than using our demo Amazon dataset.
hparams = prepare_hparams(yaml_file,
embed_l2=0.,
layer_l2=0.,
learning_rate=0.001, # set to 0.01 if batch normalization is disable
epochs=EPOCHS,
batch_size=BATCH_SIZE,
show_step=20,
MODEL_DIR=os.path.join(data_path, "model/"),
SUMMARIES_DIR=os.path.join(data_path, "summary/"),
user_vocab=user_vocab,
item_vocab=item_vocab,
cate_vocab=cate_vocab,
need_sample=True,
train_num_ngs=train_num_ngs, # provides the number of negative instances for each positive instance for loss computation.
)
# #### 1.2 Create data loader
# Designate a data iterator for the model. All our sequential models use SequentialIterator.
# data format is introduced aboved.
#
# <br>Validation and testing data are files after negative sampling offline with the number of `<num_ngs>` and `<test_num_ngs>`.
input_creator = SequentialIterator
#### uncomment this for the NextItNet model, because it needs a special data iterator for training
#input_creator = NextItNetIterator
# ## 2. Create model
# When both hyper-parameters and data iterator are ready, we can create a model:
# +
model = SeqModel(hparams, input_creator, seed=RANDOM_SEED)
## sometimes we don't want to train a model from scratch
## then we can load a pre-trained model like this:
#model.load_model(r'your_model_path')
# -
# Now let's see what is the model's performance at this point (without starting training):
# test_num_ngs is the number of negative lines after each positive line in your test_file
print(model.run_eval(test_file, num_ngs=test_num_ngs))
# AUC=0.5 is a state of random guess. We can see that before training, the model behaves like random guessing.
#
# #### 2.1 Train model
# Next we want to train the model on a training set, and check the performance on a validation dataset. Training the model is as simple as a function call:
# +
with Timer() as train_time:
model = model.fit(train_file, valid_file, valid_num_ngs=valid_num_ngs)
# valid_num_ngs is the number of negative lines after each positive line in your valid_file
# we will evaluate the performance of model on valid_file every epoch
print('Time cost for training is {0:.2f} mins'.format(train_time.interval/60.0))
# -
# #### 2.2 Evaluate model
#
# Again, let's see what is the model's performance now (after training):
res_syn = model.run_eval(test_file, num_ngs=test_num_ngs)
print(res_syn)
sb.glue("res_syn", res_syn)
# If we want to get the full prediction scores rather than evaluation metrics, we can do this:
model = model.predict(test_file, output_file)
# +
# The data was downloaded in tmpdir folder. You can delete them manually if you do not need them any more.
# -
# #### 2.3 Running models with large dataset
# Here are performances using the whole amazon dataset among popular sequential models with 1,697,533 positive instances.
# <br>Settings for reproducing the results:
# <br>`learning_rate=0.001, dropout=0.3, item_embedding_dim=32, cate_embedding_dim=8, l2_norm=0, batch_size=400,
# train_num_ngs=4, valid_num_ngs=4, test_num_ngs=49`
#
#
# We compare the running time with CPU only and with GPU on the larger dataset. It appears that GPU can significantly accelerate the training. Hardware specification for running the large dataset:
# <br>GPU: Tesla P100-PCIE-16GB
# <br>CPU: 6 cores Intel(R) Xeon(R) CPU E5-2690 v4 @ 2.60GHz
#
# | Models | AUC | g-AUC | NDCG@2 | NDCG@10 | seconds per epoch on GPU | seconds per epoch on CPU| config |
# | :------| :------: | :------: | :------: | :------: | :------: | :------: | :------ |
# | A2SVD | 0.8251 | 0.8178 | 0.2922 | 0.4264 | 249.5 | 440.0 | N/A |
# | GRU4Rec | 0.8411 | 0.8332 | 0.3213 | 0.4547 | 439.0 | 4285.0 | max_seq_length=50, hidden_size=40|
# | Caser | 0.8244 | 0.8171 | 0.283 | 0.4194 | 314.3 | 5369.9 | T=1, n_v=128, n_h=128, L=3, min_seq_length=5|
# | SLi_Rec | 0.8631 | 0.8519 | 0.3491 | 0.4842 | 549.6 | 5014.0 | attention_size=40, max_seq_length=50, hidden_size=40|
# | NextItNet* | 0.6793 | 0.6769 | 0.0602 | 0.1733 | 112.0 | 214.5 | min_seq_length=3, dilations=\[1,2,4,1,2,4\], kernel_size=3 |
# | SUM | 0.8481 | 0.8406 | 0.3394 | 0.4774 | 1005.0 | 9427.0 | hidden_size=40, slots=4, dropout=0|
#
# Note 1: The five models are grid searched with a coarse granularity and the results are for reference only.
# <br>Note 2: NextItNet model requires a dataset with strong sequence property, but the Amazon dataset used in this notebook does not meet that requirement, so NextItNet Model may not performance good. If you wish to use other datasets with strong sequence property, NextItNet is recommended.
# <br>Note 3: Time cost of NextItNet Model is significantly shorter than other models because it doesn't need a history expanding of training data.
# ## 3. Loading Trained Models
# In this section, we provide a simple example to illustrate how we can use the trained model to serve for production demand.
#
# Suppose we are in a new session. First let's load a previous trained model:
model_best_trained = SeqModel(hparams, input_creator, seed=RANDOM_SEED)
path_best_trained = os.path.join(hparams.MODEL_DIR, "best_model")
print('loading saved model in {0}'.format(path_best_trained))
model_best_trained.load_model(path_best_trained)
# Let's see if we load the model correctly. The testing metrics should be close to the numbers we have in the training stage.
model_best_trained.run_eval(test_file, num_ngs=test_num_ngs)
# And we make predictions using this model. In the next step, we will make predictions using a serving model. Then we can check if the two result files are consistent.
model_best_trained.predict(test_file, output_file)
# ## References
# \[1\] <NAME>, <NAME>, <NAME>, <NAME>, <NAME>. Adaptive User Modeling with Long and Short-Term Preferences for Personailzed Recommendation. In Proceedings of the 28th International Joint Conferences on Artificial Intelligence, IJCAI’19, Pages 4213-4219. AAAI Press, 2019.
#
# \[2\] <NAME>, <NAME>, <NAME>, <NAME>. Session-based Recommendations with Recurrent Neural Networks. ICLR (Poster) 2016
#
# \[3\] <NAME>, and <NAME>. Personalized top-n sequential recommendation via convolutional sequence embedding. Proceedings of the Eleventh ACM International Conference on Web Search and Data Mining. ACM, 2018.
#
# \[4\] <NAME>., <NAME>., <NAME>., <NAME>., & <NAME>. A Simple Convolutional Generative Network for Next Item Recommendation. WSDM, 2019
#
# \[5\] <NAME>., <NAME>., <NAME>., <NAME>., <NAME>., <NAME>., & <NAME>. Multi-Interest-Aware User Modeling for Large-Scale Sequential Recommendations. (2021) arXiv preprint arXiv:2102.09211.
| examples/00_quick_start/sequential_recsys_amazondataset.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ### Preprocessing sequencing reads (FASTQs) with Cassiopeia
#
# Cassiopeia provides an end-to-end pipeline to preprocess raw sequencing reads in FASTQ format into "Character Matrices" ready to pass into a phylogeny-inference algorithm. Given a set of FASTQs that contain RNA-seq reads from a target site construct (and NOT any endogenously expressed RNA), the preprocessing pipeline consists of the following steps.
#
# 1. **convert**: Convert the FASTQs into an unmapped BAM, while parsing any barcode and/or UMI sequences into BAM tags.
# 2. **filter_bam**: Filter reads with low-quality barcode and/or UMI sequences from the unmapped bam.
# 3. **error_correct_cellbcs_to_whitelist**: For sequencing chemistries that have a predefined (cell) barcode whitelist, this steps perform correction of sequencing errors using this whitelist.
# 4. **collapse**: Collapse reads into UMIs by constructing one or more consensus sequences for each UMI using the set of reads with that UMI.
# 5. **resolve**: Resolve a single sequence for each UMI by choosing the most likely sequencing read to represent each UMI in a cell.
# 6. **align**: Align sequences to the reference target site using the Smith-Waterman local alignment algorithm.
# 7. **call_alleles**: Call alleles with respect to the reference target site and the alignment of a sequence, thereby reporting the set of mutations that a target site sequence contains.
# 8. **error_correct_intbcs_to_whitelist**: For experimental designs for which each target site vector molecule has a unique barcode ("intBC"), and the set of intBCs present in the sequenced sample are known beforehand, this step performs sequencing error correction of these intBCs to the provided whitelist.
# 9. **error_correct_umis**: Error-correct UMIs whose mutation data is identical and whose UMI barcode sequences are similar enough.
# 10. **filter_molecule_table**: Filter UMIs that have conflicting allele information, too few reads, or do not meet other quality control criteria.
# 11. **call_lineages**: Split up cells into clonal populations, based on their shared set of integration barcodes (intBCs).
#
# The final output of this pipeline is an "AlleleTable" which stores the mutation data and clonal population identity for each cell. This data structure can then be broken up into character matrices for phylogenetic inference.
#
#
# ## Pipeline API
# All of the key modules of the preprocessing pipeline can be invoked by a call from `cassiopeia.pp`. Assuming the user would like to begin at the beginning of the pipeline, we'll start with the `convert` stage. You can find all documentation on our [main site](https://cassiopeia-lineage.readthedocs.io/en/latest/).
#
# An alternative to running the pipeline interactively is to take advantage of the command line tool `cassiopeia-preprocess`, which takes in a configuration file (for example in Cassiopeia/data/preprocess.cfg) and runs the pipeline end-to-end. For example, if you have a config called `example_config.cfg`, this can be invoked from the command line with:
#
# ```bash
# cassiopeia-preprocess example_config.cfg
# ```
#
# In this brief tutorial, we will preprocess a sample prepared with the 10X Genomics 3' version 3 chemistry and an intBC whitelist (that define the target site intBCs we know are present in the sample, obtained via DNA sequencing).
# +
import pandas as pd
import cassiopeia as cas
# +
# The raw FASTQs
input_files = [
"R1.fastq.gz", "R2.fastq.gz"
]
# The sample name, used for naming output files
name = 'test_sample'
# Directory to output results
output_directory = "test_preprocess_pipeline"
# Path to the target site reference sequence in FASTA format
reference_filepath = "../data/PCT48.ref.fasta"
# Number of threads to use, whenever parallelization is possible
n_threads = 8
# Whether to allow a single intBC to have multiple allele states
# For chemistries for which barcode == cell, this should be `False`.
allow_allele_conflicts = False
# Verbosity of logging
verbose = True
cassiopeia.pp.setup(output_dir, verbose=verbose)
# -
# ### convert
#
# Since we used the 10X Genomics 3' version 3 chemistry to prepare our samples, we provide `chemistry=10xv3`. Other supported chemistries are the following.
#
# * `dropseq`: Droplet-based scRNA-seq chemistry described in Macosco et al. 2015
#
# * `10xv2`: 10x Genomics 3' version 2
#
# * `10xv3`: 10x Genomics 3' version 3
#
# * `indropsv3`: inDrops version 3 by Zilionis et al. 2017
#
# * `slideseq2`: Slide-seq version 2
bam_fp = cas.pp.convert_fastqs_to_unmapped_bam(
input_files,
chemistry='10xv3',
output_directory=output_directory,
name=name,
n_threads=n_threads
)
# ### filter_bam
#
# The `quality_threshold` parameter controls the minimum PHRED sequencing quality the barcode and UMI sequence must have for a read to pass filtering.
bam_fp = cas.pp.filter_bam(
bam_fp,
output_directory=output_directory,
quality_threshold=10,
n_threads=n_threads,
)
# ### error_correct_cellbcs_to_whitelist
#
# The 10X Genomics 3' version 3 chemistry has a predefined barcode whitelist, to which we will correct our barcode sequences. For chemistries that do not have such a whitelist (such as Drop-seq), this step should be skipped.
#
# The `whitelist` argument may be a path to the whitelist plaintext file (with one barcode per line) or a Python list containing the whitelisted barcode sequences. Here, we downloaded the appropriate whitelist file from [here](https://github.com/10XGenomics/cellranger/raw/master/lib/python/cellranger/barcodes/3M-february-2018.txt.gz) and uncompressed it.
bam_fp = cas.pp.error_correct_cellbcs_to_whitelist(
bam_fp,
whitelist='3M-february-2018.txt',
output_directory=output_directory,
n_threads=n_threads,
)
# ### collapse
#
# The `max_hq_mismatches` and `max_indels` arguments control the threshold with which to decide whether to assign a read to a proposed consensus sequence. The defaults (`3` and `2` respectively) should work best in most cases.
#
# The `method` argument may take one of two values: `cutoff` and `likelihood`. The former uses a hard PHRED qualtiy cutoff of 30 (and any mismatches below this quality are ignored). Consensus sequences are proposed by selecting the msot common base at each position (with quality at least 30). The latter is a likelihood-based approach that selects the consensus sequences based on what is the most probable base at each position.
umi_table = cas.pp.collapse_umis(
bam_fp,
output_directory=output_directory,
max_hq_mismatches=3,
max_indels=2,
method='likelihood',
n_threads=n_threads,
)
# ### resolve
#
# The `min_umi_per_cell` and `min_avg_reads_per_umi` specify filtering thresholds to filter cells. The former is the minimum number of UMIs a cell must have to pass filtering, and the latter is the minimum average number of reads per UMI a cell must have to pass filtering.
umi_table = cas.pp.resolve_umi_sequence(
umi_table,
output_directory=output_directory,
min_umi_per_cell=10,
min_avg_reads_per_umi=2.0,
plot=True,
)
# ### align
#
# The reference target site sequence must be provided as a FASTA file to the `ref_filepath` argument or as a string to the `ref` argument. The `gap_open_penalty` and `gap_extend_penalty` specify the gap open and extend penalties to use when aligning sequences. The provided defaults should work well for most cases.
umi_table = cas.pp.align_sequences(
umi_table,
ref_filepath=reference_filepath,
gap_open_penalty=20,
gap_extend_penalty=1,
n_threads=n_threads,
)
# ### call_alleles
#
# Same as with the "align" step, the reference target site sequence must be provided with either the `ref_filepath` or `ref` arguments. The following additional arguments must be provided.
#
# * `barcode_interval`: The start and end positions for the intBC, which is the barcode that uniquely identifies each target site molecule. The interval is represented as a tuple of the form `(start, end)`, using 0-indexing and start-inclusive/end-exclusive.
#
# * `cutsite_locations`: The (center) location of each cutsite, represented as a list of indices, one element for each cutsite.
#
# * `cutsite_width`: The number of nucleotides to the left and right of the cutsite location that indels can appear in.
#
# * `context`: Whether or not to use the nucleotides surrounding the indels to identify the indels.
#
# * `context_size`: The number of bases to the left and right to include as the context.
#
# For the target sites we used for this experiment, we have the following locations.
#
# * intBC located in the interval `(20, 34)`
#
# * cutsites at `[112, 166, 120]`
umi_table = cas.pp.call_alleles(
umi_table,
ref_filepath=reference_filepath,
barcode_interval=(20, 34),
cutsite_locations=[112, 166, 220],
cutsite_width=12,
context=True,
context_size=5,
)
# ### error_correct_intbcs_to_whitelist
#
# For experiments in which the intBC sequences that are present in the sample are not known beforehand, this step should be skipped.
#
# In our case, we do have an intBC whitelist, obtained from DNA sequencing. The `intbc_dist_thresh` specifies the maximum Levenshtein (edit) distance between the intBC sequence and whitelist to be correct.
intbc_whitelist = [
'ATGATTTAACTACT', 'CGATTGGTCACTTA', 'CGTGAGTCTCTGAT', 'GAACCCACAATTCC',
'GAGTATATACCCTT', 'GCGTTTAGAATATT', 'GCCTTCAATTCCAA', 'TAACCAAGCCTACA',
'TTTCGTCGCTCTTC', 'CGCTATGGGGGGAA', 'CGATATCTTCAAGC', 'TCAGTGGGGTATTG',
'ACAATGCGTGTGGC',
]
umi_table = cas.pp.error_correct_intbcs_to_whitelist(
umi_table,
whitelist=intbc_whitelist,
intbc_dist_thresh=1
)
# ### error_correct_umis
#
# The `max_umi_distance` specifies the maximum Hamming distance between two UMIs for one to be corrected to another.
umi_table = cas.pp.error_correct_umis(
umi_table,
max_umi_distance=2,
allow_allele_conflicts=allow_allele_conflicts,
n_threads=n_threads,
)
# ### filter_molecule_table
#
# The `min_umi_per_cell` and `min_avg_reads_per_umi` behave the same as the "resolve" step.
#
# See the [documentation](https://cassiopeia-lineage.readthedocs.io/en/latest/api/reference/cassiopeia.pp.filter_molecule_table.html#cassiopeia.pp.filter_molecule_table) for more details.
umi_table = cas.pp.filter_molecule_table(
umi_table,
output_directory=output_directory,
min_umi_per_cell=10,
min_avg_reads_per_umi=2.0,
min_reads_per_umi=-1,
intbc_prop_thresh=0.5,
intbc_umi_thresh=10,
intbc_dist_thresh=1,
doublet_threshold=0.35,
allow_allele_conflicts=allow_allele_conflicts,
plot=True,
)
# ### call_lineage_groups
#
# The `min_umi_per_cell` and `min_avg_reads_per_umi` behave the same as the "resolve" step.
#
# See the [documentation](https://cassiopeia-lineage.readthedocs.io/en/latest/api/reference/cassiopeia.pp.call_lineage_groups.html#cassiopeia.pp.call_lineage_groups) for more details.
allele_table = cas.pp.call_lineage_groups(
umi_table,
output_directory=output_directory,
min_umi_per_cell=10,
min_avg_reads_per_umi=2.0,
min_cluster_prop=0.005,
min_intbc_thresh=0.05,
inter_doublet_threshold=0.35,
kinship_thresh=0.25,
plot=True,
)
allele_table.head(5)
| notebooks/preprocess.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# + [markdown] id="guJN9oUUf4uR"
# # **MicNet pipeline**
# Micnet is a package developed for the analysis of microbial abundance table. It proposes an analysis pipeline which consists of three modules:
# 1. Visualization with UMAP and HDBSCAN
# 2. Estimation of the co-ocurrence network between species or OTUS with an enhanced version of SparCC.
# 3. Estimation of several graph theory metrics to describe the topology of the resulting network.
#
# In this notebook we show an implementation of the analysis pipeline for the Komnbucha dataset described in [Arikan et al 2020](https://ift.onlinelibrary.wiley.com/doi/full/10.1111/1750-3841.14992).
#
# For more information about the Micnet package: [Favila, Madrigal-Trejo et al 2021](https://www.biorxiv.org/content/10.1101/2021.11.11.468289v1.full.pdf).
#
# If you prefer to use a GUI to implement most of the code presented here you can try our web app at: [web dashboard](http://micnetapplb-1212130533.us-east-1.elb.amazonaws.com/). Or you can run this web app in your computer deploying the GUI locally with the code specified in our [github repository](https://github.com/Labevo/MicNetToolbox).
# + [markdown] id="A3UKxaZJjwdL"
# ## **Packages and dependancies**
# ### If you have not install the micnet package yet, please run the following code and remember to always create and activate teh MicNet-env first:
# + id="s-Ea40QPjNbS"
pip install micnet==1.0.0
# + [markdown] id="eLcTmupskekB"
# ### Now we can import the packages we need to run the complete analysis pipeline
# + id="FN8iQmsKlbxQ"
import hdbscan
import micnet as mc
import pandas as pd
from pathlib import Path
import matplotlib.pyplot as plt
import seaborn as sns
from bokeh.plotting import show
from bokeh.io import output_notebook
# Call once to configure Bokeh to display plots inline in the notebook.
output_notebook()
# + [markdown] id="P4q-0FU_k-uZ"
# ## **1.** **Load Kombucha data**
#
# Now we will load and inspect the Kombucha data from the MicNet package. We can see that the data consist of 179 OTUs in the rows and in the columns we have the ASV id, the taxa classification and the abundance of 11 samples.
# + colab={"base_uri": "https://localhost:8080/", "height": 287} id="FIf2Gh3yk9cT" outputId="07b4a281-3d9a-436f-9d6b-9109e7483412"
data = mc.load_kombucha()
print(data.shape)
data.head()
# + [markdown] id="7lJslHE1pOOs"
# ## **1.1 Pre-processing the data**
# We have the option of filtering out singletons and low-abundace OTUs.In this case we will do it to have a more reliable dataset to work with.
# + colab={"base_uri": "https://localhost:8080/", "height": 1000} id="hfH1-DS6pifx" outputId="7e72ecb6-3b43-417d-d03b-ad0d139d21ad"
# the filtering functions needs you to specify if the input data contains any taxa information
from micnet.utils import filter_otus
X,Taxa,Text = filter_otus(data,taxa=True,low_abundance=True)
print(X.shape)
X
# + [markdown] id="ekIgUIqbmSOY"
# ## **2. Visualize the data with UMAP and HDBSCAN**
#
# To visualize our abundance table using dimension reduction technique UMAP and HDBSCAN for clustering we need to create a class called **Embedding_Ouput**. This is where we have to decide on the parameters of umap and hdbscan. For the Kombucha example we will set them as in the paper by Favila et al (2021).
# + id="heXTwWoUmAiz"
#Set parameter values
#The metrisc for umap and hdbscan can be picked from the following:
METRIC_UMAP=['euclidean','manhattan','canberra','braycurtis', 'cosine','correlation','hellinger']
METRIC_HDB=['euclidean','manhattan','canberra','braycurtis']
n_neighbors = 2
min_dist = 1
n_components = 2
metric_umap = METRIC_UMAP[0]
metric_hdb = METRIC_HDB[0]
min_cluster_size = 2
min_sample = 3
embedding_outliers=mc.Embedding_Output(n_neighbors=n_neighbors,min_dist=min_dist, n_components=n_components,
metric_umap=metric_umap,metric_hdb=metric_hdb,min_cluster_size=min_cluster_size,
min_sample=min_sample,output=True)
# + [markdown] id="_OXT4_NzvvYr"
# Now we the object created we can obtained the two dimensions from the UMAP reduction analysis and then we proceed to plot it in a ciruclar arrangement.
# + colab={"base_uri": "https://localhost:8080/"} id="JZ8IFav0sETE" outputId="3bb12ee7-a5da-4835-f019-bda8809ec4fd"
embedding,o,l=embedding_outliers.fit(X)
# + colab={"base_uri": "https://localhost:8080/", "height": 817} id="l_IqjYuZKW2m" outputId="c95f435f-4ebe-4b84-a8fe-0b792cf1cd8b"
mc.plot_umap(embedding,l,Text,Taxa)
# + [markdown] id="BRBKH4ui2kE5"
# To have all the data of which OTU belongs to which cluster and if it is an outlier or not we can put all teh data together in a dataframe:
# + colab={"base_uri": "https://localhost:8080/", "height": 206} id="ZvvCPNZU2jVR" outputId="f253816a-5fb4-481d-c605-7e244b53be0e"
DF=pd.DataFrame()
if len(Taxa)>1:
DF['Taxa']= Text.iloc[:,1]
DF['Outliers']=o
DF['Cluster']=l
DF.head()
# + [markdown] id="Bpn7-td9-HZe"
# ## **3. SparCC: calculating co-occurrence network**
#
# To run SparCC on our abundace table we first need to instatiate the class SparCC_MicNet with the values of the parameters that we wish Sparcc is run with as follows:
#
# + colab={"base_uri": "https://localhost:8080/", "height": 223} id="q_GwJ7VtF2Qw" outputId="1910484e-53c7-4833-d049-509afb69a8fc"
#SparCC is run wihtout any ASV or taxa so we first clean our dataset
dataSparcc = data.iloc[:,2:]
print(dataSparcc.shape)
dataSparcc.head()
# + id="rp6BUQO6-YCd"
# set parameters for SparCC
n_iteractions=3
x_iteractions=3
low_abundance=True
threshold=0.1
normalization='dirichlet'
log_transform=True
num_simulate_data=5
type_pvalues='one_sided'
#Create Sparcc object
SparCC_MN = mc.SparCC_MicNet(n_iteractions=n_iteractions,
x_iteractions=x_iteractions,
low_abundance=low_abundance,
threshold=threshold,
normalization=normalization,
log_transform=log_transform,
num_simulate_data=num_simulate_data,
type_pvalues=type_pvalues,
)
# + [markdown] id="T3FFjLjDD34O"
# Then we actually run the sparcc algorithm with the method run_all
# + id="kW2aVnS1EnTw"
SparCC_MN.run_all(data_input=dataSparcc)
# + [markdown] id="ha_0HdtrEVp1"
# sparcc will compute the correlations and the pvalues separately:
# + id="fV-tXsfyEU2g"
DF_SparCC=pd.read_csv(Path(SparCC_MN.save_corr_file).resolve(),index_col=0)
DF_PValues=pd.read_csv(Path(SparCC_MN.outfile_pvals).resolve(),index_col=0)
# + [markdown] id="JWWsFn_VEqHl"
# So we can obtain the final significant correlations found by filtering them out by their p-value:
# + colab={"base_uri": "https://localhost:8080/", "height": 287} id="BB5vDXvFEbk0" outputId="b7fc1a3a-f81a-4c5e-e5a4-7006344062f5"
sparcc_corr=DF_SparCC[DF_PValues<0.05].fillna(0)
print(f'The resulting corellation matrix is of size {sparcc_corr.shape}')
sparcc_corr.head()
# + [markdown] id="ZGya4BvyGDAp"
# ## **4.Network analysis**
#
# The final step on the proposed analysis pipeline of MicNet is to obtain large scale metrics of the network and subgroups based on their relationships.
#
# To do this we begin by building the graph of the matrix obtained from SparCC to be able to get large-scale metrics. Note that we can do this in two ways, by normalizing the correlation values to a range of (0,1) or we can leave the values as they are, this is your decision, but note that some graph theory analysis only work with normalized values.
# + id="gjrcU0S_GFox"
M = mc.build_network(sparcc_corr)
Mnorm = mc.build_normalize_network(sparcc_corr)
# + [markdown] id="ImAHCZXxKOI8"
# Now we can create a network micnet obejct which we will use to obtain most of our descriptors and analysis of the network
# + id="0VUgT8xkKWIr"
NetM=mc.NetWork_MicNet()
# -
# Les start by looking at some of the basic properties of the network, such as number of nodes, number of interactions, diameter, etc..
NetM.basic_description(corr=sparcc_corr)
# + [markdown] id="ozuSeRRARiwg"
# Now we might be interested in how many triads including different types (such as + + + or - + -) of interactions are present ir our network. We can do this by calling the structural balance method:
# + id="Kum3gui-Rh1x"
NetM.structural_balance(M)
# -
# Remember that structural balance anaylsis needs the raw data (ranging from -1 to 1), not the normalized ones, that is why in this analysis we used the network M.
# Now we can obtain the communties found in the network based on the Louvain method (which finds clusters based on increasing intragroup interactions and minimizing intergroup interactions)
Communities=NetM.community_analysis(Mnorm)
# Within the communities object we can extract the number of comminities and a table with a summary of properties of each community found as we show below:
print(Communities['Number'])
print(Communities['Community_topology'])
# Finally, it is also possible to extract the assignement of each node to the different communities found:
print(Communities['Data'].head())
# Now we obtain the centrlities of the nodes:
# + id="SsyJeezxRksO"
Centrality=NetM.key_otus(Mnorm)
# + [markdown] id="VNNEwbOPRoDx"
# For easier display we can put everything in a single database, including the info that we previously obtained from hdbscan clustering:
# + id="3lLS8G3LRqqT"
NetDF=pd.DataFrame({'OTUS':Centrality['NUM_OTUS'],
'Degree_Centrality':Centrality['Degree centrality'],
'Betweeness_Centrality':Centrality['Betweeness centrality'],
'Closeness_Centrality':Centrality['Closeness centrality'],
'PageRank':Centrality['PageRank'],
'HDBSCAN':DF['Cluster'],
'Community':Communities['Data'].values.ravel()})
# + [markdown] id="BRgfOEj2RrLP"
# We now plot the network, coloring by communities:
# + id="NnWkeHhoRx7U"
pl = mc.plot_bokeh(graph=M,frame=NetDF,
nodes = M.number_of_nodes(),
max = sparcc_corr.max().max(),
min = sparcc_corr.min().min(),
kind_network='spring',
kind='Community')
show(pl)
# -
# We could also colored the nodes according to the groups found with HDBSCAN and in a circular layout:
pl2 = mc.plot_bokeh(graph=M,frame=NetDF,
nodes = M.number_of_nodes(),
max = sparcc_corr.max().max(),
min = sparcc_corr.min().min(),
kind_network='circular',
kind='HDBSCAN')
show(pl2)
# ### 4.1 Topology comparison
# MicNet includes the computation of the distribution of several large-scale metrics under the assumption that the underlying topology is: 1) a random Erdos-Renyi network, built using function nx.erdos_renyi_graph, 2) a small world Watts-Strogatz built using nx.watts_strogatz_graph function, or 3) a scale-free Barabási-Albert network built using nx.barabasi_albert_graph function.
#
# The topology_boostrap function included in the micnet package takes a correlation matrix as input and returns three dataframes with the distribution for several large-scale metrics under the assumption that the network with the same density and average degree but with the defined topologies mentioned.
#
# Now lest run the boostrap with the Kombucha correlation data:
#
#Run boostrap
df_rand, df_small, df_scale = mc.topology_boostrap(sparcc_corr, n_boot=20)
# Note that the larger the values of n_boot (number of simuations for the boostraping), the longer it will take to run.
#
# Now, we plot as an example the distribution of the average shortest path for networks of the same size as the Kombucha example but with a topology that is random (in red), small-world (in green) or scale-free (in blue) and we see where our Kombucha data lies (black line):
# +
#Plotting average shortest path under the asumption of a random, small-world and scale-free topology.
plt.figure(figsize=(10,6))
sns.kdeplot(df_small['Average shortest path'], color='g', shade=True)
sns.kdeplot(df_rand['Average shortest path'], color='r', shade=True)
sns.kdeplot(df_scale['Average shortest path'], color='b', shade=True)
plt.axvline(1.554078, color = 'black',lw=3, ls = '--')
# -
# Degree distributions can also be used to discriminate between network topologies. Thus, we have included in the MicNet toolbox a function that plots the Complementary Cumulative Distribution Function (CCDF) of the degrees of the given network and compares it with the CCDF of a simulated comparable random, scale-free and small-word network on a log-log scale.
#
# To perform the comparison use the following code:
ccdf = mc.degree_comparison(sparcc_corr, topology ='random', bins=20)
# the degree_comparison function returns a dataframe with the bins and CCDF for the corr matrix, and the bins and CCDF for an equivalent network with defined topology (which can be specified in the topology parameter as 'random', 'small_world' or scale_free).
#
# We suggest to plot the results as follows:
plt.figure(figsize=(7.5,6))
sns.lineplot(x=ccdf.Data_bins,y=ccdf.Data_CCDF, color = 'black',lw= 3)
sns.lineplot(x=ccdf.Simulated_bins,y=ccdf.Simulated_CCDF, color = 'r',lw= 3)
plt.xlabel('Degree')
plt.ylabel('CCDF')
# ### 4.2 Percolation analysis
# The percolation analysis consists of removing nodes and their corresponding edges and analyzing how much the network's properties are disrupted. The percolation simulation consists of n iterations; in each iteration a percentage of the nodes (with default value of 0.1, but this can be specified by the user) is removed along with all of their edges. We have provided two percolations functions to run percolation either by a type of centrality or by defined groups (such as clusters or taxa groups).
#
# Lets begin by running a percolation by degree centrality. The resulting data will tell us how removing the nodes affected several metrics of the network, such as the number of communities, the modularity, the network's density, amongst others.
#Run percolation removing nodes by degree centrality
percolation = mc.percolation_sim(sparcc_corr, prem =0.1, per_type='deg_centrality')
percolation
# Note tha you can change **prem** (percentage of removal) to modify what percentange of nodes get removed in each iteration, and to modify the type of nodes that are removed first, change **per_type** which can take the values 'random', 'deg_centrality', 'clos_centrality', 'bet_centrality'.
# It is also possible to remove the nodes by a list of ids, for example, removing by HDBSCAN groups.
#Run percolation removing nodes by group
grouplist = list(DF['Cluster'])
percolation = mc.percolation_by_group(sparcc_corr, prem=0.1, groups=grouplist)
# Note that this analysis will return one table per group and will show how removing each group (in steps of 10%) found in **grouplist** affects the networks properties. So for example if we would want to know what effect would have on the network removing all the elements from group 1 identified by HDBSCAN we would look at the following table
# +
#We first look at all the tables present in the percolation object
print(percolation.keys())
#Now we only see the percolation results from group 1
percolation['Percolation_group_1']
| notebooks/Kombucha_demo.ipynb |
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .ps1
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: PowerShell
# name: powershell
# ---
# + [markdown] azdata_cell_guid="0b04834e-defa-4af2-9d95-a8aaa27e5f7b"
# # SQL Assessment API Tutorial
# You can use this tutorial to understand how to assess your SQL Server configuration for best practices. In this tutorial, you will learn:
#
# 1. How to install PowerShell SqlServer module that includes SQL Assessment API cmdlets.
# 2. How to assess your SQL Server and databases
# 3. How to save results in a sql table and graph over results
# 4. How to customize rules by disabling some rules, adding new ones, and changing thresholds
#
# Supported products and platforms: SQL Server 2012 and up, both on Windows and Linux. Azure SQL DB Managed Instance. More products to come.
#
# Microsoft ruleset ([ruleset.json](https://github.com/microsoft/sql-server-samples/blob/master/samples/manage/sql-assessment-api/ruleset.json)) is published on SQL Assessment API GitHub repo and continuously improved.
#
# Useful links at the bottom of the tutorial.
# + [markdown] azdata_cell_guid="424b5be9-3f95-4fa0-b630-b5507a0e5db2"
# ### Quick primer on cmdlets
#
# There are two cmdlets:
#
# 1. **Get-SqlAssessmentItem** shows a list of available rules for a given object (5 kinds of objects is currently supported as input: Server, RegisteredServer, Database, AvailabilityGroup, Filegroup; the default ruleset contains rules for Server/RegisteredServer and Database only). Every rule has a target that describes what kind of SQL objects this rule applies to: Object Type, Object Name, SQL Server version, SQL Server platform, SQL Server engine edition. So by the availability of a rule, we mean that when you run Get-SqlAssessmentItem or Invoke-SqlAssessment, the API first verifies what rules apply for the given object.
#
# 2. **Invoke-SqlAssessment** performs an assessment of a passed object and provides the results. It's worthwhile to mention that assessment is invoked for a passed object only, so if you want to assess a SQL Server instance and all its databases, run the cmdlet with the instance object as input and then run it with the databases as input. We'll show you different ways of doing this below.
# + [markdown] azdata_cell_guid="9575ad0b-daca-48ac-84f1-59431afd1387"
# ### 1. Setup
# You need to install PowerShell SqlServer module using the following command. It is a good practice to run Import-Module at the beginning of your session as well. Get-Module will show you the version you have installed. The minimum version you want is 21.1.18206 — it is the version of SqlServer module containing SQL Assessment API GA. You can [read more](https://docs.microsoft.com/sql/powershell/download-sql-server-ps-module) about installing and updating the SqlServer module on docs.
# + azdata_cell_guid="bf6a34ad-a006-4b24-858d-5d08b760b231"
# Uncomment and run Install-Module only the first time
# Install-Module -Name SqlServer -AllowClobber -Force
Import-Module -Name SqlServer
Get-Module
# + [markdown] azdata_cell_guid="a34fa4e1-bf4e-44c9-9274-3fca29a6c535"
# ### 2. Invoke an assessment for SQL Server instance
# There are various ways to run the assessment cmdlet. The following statements give recommendations for a local default instance. Pick whatever style works for your script.
#
# Server and RegisteredServer objects are interchangeable, so you can pass any to the SQL Assessment cmdlets to assess a SQL Server instance.
# + azdata_cell_guid="07bf073e-9268-4e6b-b142-9c9fa8dc78ca"
# Option 1
Get-SqlInstance -ServerInstance 'localhost' | Invoke-SqlAssessment
# + azdata_cell_guid="5f30b889-22cb-4f15-9729-adc555fafd8f"
# Option 2
$serverInstance = Get-SqlInstance -ServerInstance 'localhost'
Invoke-SqlAssessment $serverInstance
# + azdata_cell_guid="5e69dad3-0501-4501-bb7d-656fbac1d90e"
# Option 3
Get-Item SQLSERVER:\SQL\localhost\default | Invoke-SqlAssessment
# + azdata_cell_guid="f9241df5-8945-453b-90e1-5e724c76cf89"
# Option 4
Invoke-SqlAssessment SQLSERVER:\SQL\localhost\default
# + azdata_cell_guid="e0e5d1bc-c5ec-4952-a658-e01d9d51f5d6"
# Option 5
cd SQLSERVER:\SQL\localhost\default
Invoke-SqlAssessment -Verbose
# + azdata_cell_guid="4a5cbed3-b75c-411b-89bf-9cbd06656dd4"
# Option 6
cd SQLSERVER:\SQL\localhost
Get-Item default | Invoke-SqlAssessment
# + azdata_cell_guid="dc8b69d6-6d20-4115-a68a-f6abd39d6c4a"
# Option 7 - passing registered servers to the cmdlet
Get-ChildItem 'SQLSERVER:\SQLRegistration\Database Engine Server Group' | WHERE { $_.Mode -ne 'D'} | Invoke-SqlAssessment
# + [markdown] azdata_cell_guid="d5253a77-1c77-45d4-9d96-552024662463"
# ### 3. Invoke an assessment for SQL Server database
# You need to run Invoke-SqlAssessment against a database object to get database specific recommendations. Again, there are various ways of accomplishing this. Below are some examples.
# + azdata_cell_guid="50d4d023-fbda-4b9d-8263-9aeab26759e7"
# Option 1
$database = Get-SqlDatabase -ServerInstance 'localhost' -Name master
Invoke-SqlAssessment $database -Verbose
# + azdata_cell_guid="ad2f5c1b-3d94-4939-b2bb-f51e6b140924"
# Option 2
Invoke-SqlAssessment SQLSERVER:\SQL\localhost\default\Databases\master -Verbose
# + azdata_cell_guid="4677c0f0-8d75-404f-8add-b484394d16a4"
# Option 3
cd SQLSERVER:\SQL\localhost\default\Databases\master
Invoke-SqlAssessment
# + azdata_cell_guid="b374c2ec-8d2e-4c35-a48a-c427dec9c21c" tags=[]
# Get recommendations for all databases on local instance:
Get-SqlDatabase -ServerInstance 'localhost' | Invoke-SqlAssessment
# + [markdown] azdata_cell_guid="97414396-2557-44c4-80e2-5afa6a5a5516"
# ### 4. Browse applicable rules
#
# The full Microsoft ruleset is in [ruleset.json](https://github.com/microsoft/sql-server-samples/blob/master/samples/manage/sql-assessment-api/ruleset.json) in the GitHub repo. If you want to list the rules that apply to a particular instance or database, you can use Get-SqlAssessmentItem cmdlet. Below are some different ways of listing the rules.
# + azdata_cell_guid="56b4a035-658b-4bc4-8870-90823b8c2549"
# Get all rules available for an object:
$serverInstance = Get-SqlInstance -ServerInstance 'localhost'
Get-SqlAssessmentItem $serverInstance | Select Id, Description
# + azdata_cell_guid="b1274ddb-b9f7-4bb0-bb69-4b8c403f0b6a"
# Get all rules by a specific tag
$serverInstance = Get-SqlInstance -ServerInstance 'localhost'
Get-SqlAssessmentItem $serverInstance -Check TraceFlag
# + [markdown] azdata_cell_guid="0267d880-c35e-4fbf-90db-dd9120194da9"
# ### 5. Run a specific rule
# If you want to check a particular rule (maybe after you fixed it), you can run it by its name. You can also specify several rules in the -Check parameter, just delimit them by commas.
#
# Every rule in the default ruleset has tags to group them into logical sets. In the example below, we look for backup related issues only. Backup value used for the -Check parameter is a tag. You can use both rule names and tags at the same time in a comma delimited list.
# + azdata_cell_guid="02638a85-ca44-4e34-88fa-e8eea85f4304"
# Run a rule by its name
$serverInstance = Get-SqlInstance -ServerInstance 'localhost'
Invoke-SqlAssessment $serverInstance -Check TF634
# + azdata_cell_guid="09c8991d-1b92-4630-9a87-5361f04e2c36"
# Run a group of rules using their tag
$database = Get-SqlDatabase -ServerInstance 'localhost'
Invoke-SqlAssessment $database -Check Backup
# + [markdown] azdata_cell_guid="289f9e4a-29c4-4da3-a52d-71b95ddaf3df"
# ### 6. Store results in a table
# You probably want to save the results of an assessment to analyze and process later on. You can pipe the results of Invoke-SqlAssessment cmdlet into a table using Write-SqlTableData cmdlet. If the table doesn't exist, it creates the table and then inserts the results. If the table exists (subsequent runs), it appends the results to the table. Just remember to use -FlattenOutput parameter as it makes the Invoke-SqlAssessment output sutiable for Write-SqlTableData.
# + azdata_cell_guid="695f924d-0729-4ebb-ba83-bba43ed8a9c7"
Get-SqlInstance -ServerInstance 'localhost' | Invoke-SqlAssessment -FlattenOutput |
Write-SqlTableData -ServerInstance 'localhost' -DatabaseName SQLAssessmentDemo -SchemaName Assessment -TableName Results -Force
# + [markdown] azdata_cell_guid="0ad9518f-e770-4819-b4d1-7f5608c793b3"
# ### 7. Scaling up your Checks
# Running checks across multiple machines and writng their results back to the same table in SQL Server. You can keep you list of SQL Servers to check anywhere you want, in a text file, in an Excel spreadsheet, in a table you maintain yourself, the options are endless.
#
# Two options are shown below show features availabile that can be managed from SSMS or Azure Data Studio, and can accessed via the `SQLSERVER:\` Provider, which is part of the `SqlServer` module.
#
# 1. Registered Servers is feature of SSMS and stores a list of SQL Server instances in a local XML file.
# 2. Central Management Server relise on a SQL Server to maintain the list of SQL Server instances (instead of a _local XML file_) and is available in both SSMS & Azure Data Studio.
# + azdata_cell_guid="3a0c510d-07c7-464b-8b0a-2152df24e237"
# This approach leverages the Registered Servers feature of SSMS to obtain a list of SQL Servers, and run checks against them.
Get-ChildItem 'SQLSERVER:\SQLRegistration\Database Engine Server Group' |
Where-Object { $_.mode -ne 'd'} |
foreach
{
Get-SqlInstance -ServerInstance $_.Name |
Invoke-SqlAssessment -FlattenOutput |
Write-SqlTableData -ServerInstance localhost -DatabaseName SQLAssessmentDemo -SchemaName Assessment -TableName Results -Force
}
# + azdata_cell_guid="c6ca3196-b661-46b4-9818-502c779742ab"
<# This approach leverages the Central Management Server feature to obtain a list of SQL Servers #>
Get-ChildItem 'SQLSERVER:\SQLRegistration\Central Management Server Group' -Recurse |
Where-Object { $_.mode -ne 'd'} |
Invoke-SqlAssessment -FlattenOutput |
Write-SqlTableData -ServerInstance localhost -DatabaseName SQLAssessmentDemo -SchemaName Assessment -TableName Results -Force
# + [markdown] azdata_cell_guid="3cfbe027-21f0-40be-b6f0-b871360aff5d"
# ## Customization
# In this section you will learn how to customize existing rules and create new ones.
#
# As a prerequisite, make sure to grab the JSON files in the CustomizationSamples folder and place them in an accessible path and then edit the first script below to point at the right path and server instance for your environment. By default, we use in this notebook the following parameters:
# - SQL Instance to assess is "localhost"
# - JSON samples and DLLs are available by path "C:\SQLAsmnt\CustomizationSamples\"
#
# The final code block in this notebook has its own prerequisites, please complete them prior to running it:
# - There are 2 dlls in CustomizationSamples folder. They both should be unblocked: https://stackoverflow.com/questions/34400546/could-not-load-file-or-assembly-operation-is-not-supported-exception-from-hres/45221477
# - Then open CustomRuleCLRProbe.json and make sure that assembly key contains the right path to TestsProbeLibrary.dll, double backslashes are required.
#
# We encourage you to look into every JSON sample so you can understand better the making of customizations for SQL Assessment.
#
# + [markdown] azdata_cell_guid="88318608-c678-4692-8315-c205933331cf"
# ### Disabling/Enabling rules
# + azdata_cell_guid="5df878e7-8204-45c8-8bb2-4861131f3588" tags=[]
#Setup three parameters that are used in all the customization examples below
#$samplesPath='<replace this with the path to customization json files, e.g. "C:\SQLAsmnt\CustomizationSamples">'
$samplesPath='C:\SQLAsmnt\CustomizationSamples'
$samplesPath
#$serverInstance = Get-SqlInstance -ServerInstance 'localhost'
$serverInstance = Get-SqlInstance -ServerInstance '.\sql2017express'
$serverInstance
$sqlDbMaster = $serverInstance | Get-SqlDatabase -Name master
$sqlDbMaster
# + azdata_cell_guid="7a2a90f0-8341-4cd8-81a3-1fdedbd1adb6"
# Disable a single rule using its ID (TF634)
# To see this in action, make sure you have trace flag 634 turned on in the instance you are testing. Otherwise this rule will not fire even when enabled.
# You will see that TF634 is not enabled (On=False)
Get-SqlAssessmentItem $serverInstance -Configuration $(join-path $samplesPath "DisableTF634.json")
# + azdata_cell_guid="35ee6731-70dd-42b9-b041-f3b09ac02a4d" tags=[]
# Disable all Trace Flag rules using a tag
# You will see that all TF rules are set to False (disabled)
Get-SqlAssessmentItem $serverInstance -Configuration $(join-path $samplesPath "DisableAllTF.json")
# + azdata_cell_guid="4f79d729-4a3b-47a6-8cb3-d4a5b0c2db05"
# Combine configurations
# This example disables all trace flag rules except for performance-related ones using tags.
# The order of json files is important. First we disable all TF rules, then enable performance rules which re-enables performance-related TF rules.
Get-SqlAssessmentItem $serverInstance -Configuration $(join-path $samplesPath "DisableAllTF.json"), $(join-path $samplesPath "EnablePerformance.json")
# + [markdown] azdata_cell_guid="0d79fec1-d64d-4580-abfa-0c367a6f20f8"
# ### Creating a new rule
# The rules are defined in json files. In this example, we are creating a rule that checks for available database space. Go ahead and examine CustomRuleTSQLProbe.json.
#
# A rule has many components such as which ruleset it belongs to, what type of objects, editions, versions, platforms it targets as well as more obvious components such as id, name, description, etc.
#
# Condition is what gets evaluated. When an expression in Condition returns false, it means that the rule is violated and the user gets a recommendation from this rule.
#
# Probe is what gets the data to be evaluated in the condition. Probes can be SQL or CLR. SQL probe is a T-SQL query to pull the required data right out of SQL Server.
# CLR probe is a reference to a .NET or Core assembly with a call to a method inside the library.
# + azdata_cell_guid="1df3379f-e5fe-47ea-a43e-920265f3d0e2" tags=[]
# Create a new rule with TSQL probe
# This rule applies to databases and uses a TSQL statement to get the data for the rule.
Invoke-SqlAssessment $sqlDbMaster -configuration $(join-path $samplesPath "CustomRuleTSQLProbe.json")
# + azdata_cell_guid="953cd87b-4311-4553-9170-e219c231642a"
# Override threshold parameter
# CustomRuleThresholdChange.json defines a new threshold value for DBSpaceAvailable rule created above
Invoke-SqlAssessment $sqlDbMaster -configuration $(join-path $samplesPath "CustomRuleTSQLProbe.json"),$(join-path $samplesPath "CustomRuleThresholdChange.json")
# + [markdown] azdata_cell_guid="86aed978-b91a-47f0-8a78-f8254c5ec6f1"
# ## Probe types
# ### CmdShell
# Create a new rule with CmdShell probe. CmdShell probe executes a CMD.EXE shell command and returns lines of text in variable @stdout. Use 'CMDSHELL' instead of 'SQL' in probe definition to load a .cmd file. Use Regex parser transformation to extract data from @stdout
#
# + azdata_cell_guid="a75ebf02-e27e-4d19-b306-ce4e04481acd"
#Create new rule with cmd probe type. It runs 'dir' cmd command and checks that resulted list is'n empty.
#Make sure that xp_cmdshell is enabled
Invoke-SqlAssessment $serverInstance -configuration $(join-path $samplesPath "CustomRuleCmdShellProbe.json")
# + [markdown] azdata_cell_guid="4450f005-b008-4464-8b83-059b5d3fa8f6"
# ### PowerShell
#
# PowerShell probe executes a command in PowerShell on target machine and returns pipeline output in @Output variable.
# Use $ (dollar) sign to access probe parameters passed from checks.
# Use . (dot) to access properties of the output object. For example, if a returned object is string, then @Output.Length returns its length.
# + azdata_cell_guid="0be17782-0051-4d49-931e-a29aaca34d02"
#Create new rule with Powershell probe type.
#It runs query to get major PS version
#Make sure that xp_cmdshell is enabled and PS execution policy is RemoteSigned or Unrestricted.
Invoke-SqlAssessment $serverInstance -configuration $(join-path $samplesPath "CustomRulePowerShellProbe.json")
# + [markdown] azdata_cell_guid="9277b49b-7f19-4d9b-8e37-d3592962c03f"
# ### Registry
#
# Registry probe obtains data from target machine's registry. The key name will be returned in @RegistryKeyName. Use * (asterisk) symbol to enumerate all keys.
# + azdata_cell_guid="6a100496-4356-4e15-b857-4cdf44177010" tags=[]
#Create new rule with Registry probe
#Make sure that xp_cmdshell is enabled
Invoke-SqlAssessment $serverInstance -configuration $(join-path $samplesPath "CustomRuleRegistryProbe.json")
# + [markdown] azdata_cell_guid="3a03fb80-361d-4b41-9a52-a2eedd1e85a0"
# ### WMI
#
# WMI probe runs a WMI query and returns results in @Output variable in the same way as a PowerShell probe does.
# Use $ (dollar) sign to access probe parameters passed from checks.
# + azdata_cell_guid="143eea26-62d1-40e8-a97a-9f1ed8ac2532"
#Create new rule with WMI probe
#Make sure that xp_cmdshell is enabled.
Invoke-SqlAssessment $serverInstance -configuration $(join-path $samplesPath "CustomRuleWmiProbe.json")
# + [markdown] azdata_cell_guid="815584d6-ceac-4160-ba9a-020c59e14221"
# ### Managed code probe
#
# For CLR probe use "External" probe type.
# + azdata_cell_guid="bcff0e69-65e8-4b44-90b8-83f1241589bd"
# Create a new rule with CLR probe. CustomRuleCLRProbe.json, in addition to a check with a CLR probe, contains an ovveride to disable all the rules of the DefaultRuleset.
# !!! Complete the prerequisites below before running this block.
# !!! There are 2 dlls in CustomizationSamples folder. Make sure that they both are not blocked: https://stackoverflow.com/questions/34400546/could-not-load-file-or-assembly-operation-is-not-supported-exception-from-hres/45221477
# !!! Then open CustomRuleCLRProbe.json in the same folder and make sure that assembly key contains the right path to TestsProbeLibrary.dll, double slashes are required.
# !!! You're all set. Run this block
Invoke-SqlAssessment $serverInstance -configuration $(join-path $samplesPath "CustomRuleCLRProbe.json")
# + [markdown] azdata_cell_guid="2c572e58-0281-432f-b400-70b55b719b7e"
# ## Useful links about SQL Assessment API
#
# - [Docs online page](https://docs.microsoft.com/sql/sql-assessment-api/sql-assessment-api-overview)
# - [GitHub repo](http://aka.ms/sql-assessment-api)
# - [SQL Server blog with release announcements and other useful information](https://techcommunity.microsoft.com/t5/SQL-Server/bg-p/SQLServer)
| samples/manage/sql-assessment-api/notebooks/SQLAssessmentAPITutorialNotebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: SageMath 9.4
# language: sage
# name: sagemath-9.4
# ---
# # plot-hrf
#
# (Last updated: 2022-02-05)
#
# Modified from [**Nilearn**](https://nilearn.github.io): Statistics for NeuroImaging in Python.<br> 9.5.1 _Example of hemodynamic response functions_<br>
# https://nilearn.github.io/auto_examples/04_glm_first_level/plot_hrf.html#sphx-glr-download-auto-examples-04-glm-first-level-plot-hrf-py
#
# See also: http://nipy.org/nipy/api/generated/nipy.modalities.fmri.hrf.html
#
# Can be run from [SageMath](https://www.sagemath.org) (Kernel: `SageMath 9.4`) if `nilearn` is installed, i.e.
#
# > sage --pip install nilearn
# %matplotlib inline
# # Example of hemodynamic response functions.
#
# Within this example we are going to plot the hemodynamic response function
# (hrf) model in SPM together with the hrf shape proposed by G.Glover, as well as
# their time and dispersion derivatives.
#
# - Glover, <NAME>. Deconvolution of impulse response in event-related BOLD fMRI. Neuroimage 1999;9(4):416-429 [[link](https://pubmed.ncbi.nlm.nih.gov/10191170)] [[pdf](https://web.mit.edu/swg/ImagingPubs/experimental-design/Glover.NeuroImage.1999.pdf)]
#
#
# This example requires matplotlib.
#
# The hrf is the filter that couples neural responses to the metabolic-related
# changes in the MRI signal. hrf models are simply phenomenological.
#
# In current analysis frameworks, the choice of hrf model is essentially left to
# the user. Fortunately, using the spm or Glover model does not make a huge
# difference. Adding derivatives should be considered whenever timing
# information has some degree of uncertainty. It is actually useful to detect
# timing issues.
#
# ## Set up some parameters for model display
#
# To get an impulse response, we simulate a single event occurring at time t=0,
# with duration 1s.
#
#
import numpy as np
frame_times = np.linspace(0, 30, 61)
onset, amplitude, duration = 0., 1., 1.
exp_condition = np.array((onset, duration, amplitude)).reshape(3, 1)
# We sample this on a fris for display
#
#
stim = np.zeros_like(frame_times)
stim[(frame_times > onset) * (frame_times <= onset + duration)] = amplitude
# Now we have to define the candidate hrf models.
#
#
hrf_models = [None, 'glover + derivative', 'glover + derivative + dispersion']
# ## Sample the hrf
#
#
# +
from nilearn.glm.first_level import compute_regressor
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(9, 4))
for i, hrf_model in enumerate(hrf_models):
# obtain the signal of interest by convolution
signal, name = compute_regressor(
exp_condition, hrf_model, frame_times, con_id='main',
oversampling=16)
# plot this
plt.subplot(1, 3, i + 1)
plt.fill(frame_times, stim, 'k', alpha=.5, label='stimulus')
for j in range(signal.shape[1]):
plt.plot(frame_times, signal.T[j], label=name[j])
plt.xlabel('time (s)')
plt.legend(loc=1)
plt.title(hrf_model)
# adjust the plot
plt.subplots_adjust(bottom=.12)
plt.show()
# -
# ### Papers of relevance
# <img src="assets/rs-hrf.png" alt="drawing" width="400"/>
#
# <img src="assets/bold_deconvolved_hrf.png" alt="drawing" width="700"/>
#
# **Resting State Hemodynamic Response Function Retrieval and Deconvolution (RS-HRF) Images** (https://www.nitrc.org/projects/rshrf)
# - <NAME>; <NAME>; <NAME>, Sofie; <NAME>, et al. rsHRF: A toolbox for resting-state HRF estimation and deconvolution. Neoroimage 2021;244:11859 [[link](https://www.sciencedirect.com/science/article/pii/S1053811921008648)] [[pdf](https://www.sciencedirect.com/science/article/pii/S1053811921008648/pdfft?isDTMRedir=true&download=true)]
# [[code](https://www.nitrc.org/projects/rshrf)] [[github](https://github.com/compneuro-da/rsHRF)] [[bids-app](http://bids-apps.neuroimaging.io/rsHRF)]
# > The hemodynamic response function (HRF) greatly influences the intra- and inter-subject variability of brain activation and connectivity, and might confound the estimation of temporal precedence in connectivity analyses, making its estimation necessary for a correct interpretation of neuroimaging studies. Additionally, the HRF shape itself is a useful local measure. However, most algorithms for HRF estimation are specific for task-related fMRI data, and only a few can be directly applied to resting-state protocols. Here we introduce rsHRF, a Matlab and Python toolbox that implements HRF estimation and deconvolution from the resting-state BOLD signal.
# >
# > This toolbox is aimed to retrieve the onsets of pseudo-events triggering an hemodynamic response from resting state fMRI BOLD signal. It is based on point process theory, and fits a model to retrieve the optimal lag between the events and the HRF onset, as well as the HRF shape, using different shape parameters or combinations of basis functions.
# >
# Once that the HRF has been retrieved for each voxel/vertex, it can be deconvolved from the time series (for example to improve lag-based connectivity estimates), or one can map the shape parameters everywhere in the brain (including white matter), and use it as a pathophysiological indicator.
# >
# Input can be 2D GIfTI, 3D or 4D NIfTI images, but also on time series matrices/vectors.
# The output are three HRF shape parameters for each voxel/vertex, plus the deconvolved time series, and the number of retrieved pseudo-events. All can be written back to GIfTI or NIfTI images.
#
#
# - <NAME>, <NAME>, <NAME>. Estimated hemodynamic response function parameters obtained from resting state BOLD fMRI signals in subjects with autism spectrum disorder and matched healthy subjects. Data Brief 2018;19:1305-1309.
# > Most of the time the HRF is associated with task-based fMRI protocols, in which its onset is explicitly included in the design matrix. On the other hand, the HRF also mediates the relationship between spontaneous neural activity and the BOLD signal in resting-state protocols, in which no explicit stimulus is taken into account. It has been shown that resting-state brain dynamics can be characterized by looking at sparse BOLD events, which can be retrieved by point process analysis. These events can be then used to retrieve the HRF at rest.
#
# - <NAME>, <NAME>, <NAME>, M.J. Kaufman, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>. Reduction in BOLD fMRI response to primary visual stimulation following alcohol ingestion. Psychiatry Res 1998;82(3):135-146
#
#
# - <NAME>, <NAME>, <NAME>. Effect of hemodynamic variability on Granger causality analysis of fMRI Neuroimage 2010;52(3):884-896.
#
#
# - <NAME>, <NAME>, <NAME>. Attenuation of brain BOLD response following lipid ingestion. Hum Brain Mapp 2003;20:116-121. [[link](https://onlinelibrary.wiley.com/doi/epdf/10.1002/hbm.10131)]
# > We sought to assess whether controllable physiologic modulators, such as dietary factors, could influence the outcome of fMRI data. A high fat diet, for example, prior to a fMRI scan could change microvascular blood rheologic factors and potentially alter brain blood oxygen-level dependent (BOLD) signal patterns.
#
#
# - <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>, <NAME>. Posterior cingulate cortex-related co-activation patterns: a resting state fMRI study in propofol-induced loss of consciousness. PLoS One 2014;9(6) [[link](https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0100012)]
# > Lately, point process analysis applied on functional data has revealed that much of the information regarding brain connectivity is contained in a fraction of critical time points of a resting state dataset. In the present study we want to extend this methodology for the investigation of resting state fMRI spatial pattern changes during propofol-induced modulation of consciousness, with the aim of extracting new insights on brain networks consciousness-dependent fluctuations.
#
#
| code/HemodynamicResponseModeling/02-plot-hrf-nilearn.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
## loading packages
import pandas as pd
import numpy as np
from rdkit import Chem
from rdkit.Chem import Descriptors
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
from mpl_toolkits.mplot3d import Axes3D
from sklearn.model_selection import train_test_split
from matplotlib import pyplot
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import KFold
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
from sklearn.preprocessing import MinMaxScaler
import joblib
import sys
import import_ipynb
from feature_generation import *
import IPython
import tensorflow as tf
from tensorflow import keras
import kerastuner as kt
import kerastuner as kt
from tensorflow.keras import backend as K
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import InputLayer, Input,Activation
from tensorflow.keras.layers import Dense,LeakyReLU
from tensorflow.keras.callbacks import TensorBoard
from tensorflow.keras.optimizers import Adam,SGD,Nadam
from tensorflow.keras.models import load_model
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras import losses
from tensorflow.keras.callbacks import ModelCheckpoint
import skopt
from skopt import gp_minimize, forest_minimize
from skopt.space import Real, Categorical, Integer
from skopt.plots import plot_convergence
from skopt.plots import plot_objective, plot_evaluations
from skopt.plots import plot_objective
from skopt.utils import use_named_args
import pickle
# +
###################################loading all the data#########################
ds = pd.read_csv('final_converted2.txt',sep="\t",index_col=None,header=None)
ds=ds.rename(columns=ds.iloc[0],copy=False).iloc[1:].reset_index(drop=True)
# +
#########combine all the data
frames = [ds]
dn = pd.concat(frames)
print(dn.columns)
print(dn.info())
# +
###drop these string columns to convert rest of the columns to numerical
df = dn.drop(['Cation_Name', 'Cation_smiles', 'Anion_Name', 'Anion_smiles'],axis=1)
df=df.apply(pd.to_numeric)
#######save the cation name, smiles, anion name, anion smiles to a dataframe
dname = dn.iloc[:,0:4]
###save the cation descriptors
dcation = df.iloc[:,0:196]
###save the anion descriptors
####anion feature selection
danion = df.iloc[:,196:392]
# +
###########cation correlation
###drop any columns with na
dcation =dcation.dropna(axis='columns')
p= (dcation.columns[dcation.sum() == 0 ]) ##find out what columns have all 0 values
dcation =dcation.drop(columns=p) ##drop that
corr = dcation.corr()
columns = np.full((corr.shape[0],), True, dtype=bool)
for i in range(corr.shape[0]):
for j in range(i+1, corr.shape[0]):
if corr.iloc[i,j] >= abs(0.9):
if columns[j]:
columns[j] = False
selected_cation_columns = dcation.columns[columns]
dcation = dcation[selected_cation_columns]
#########anion correlation
danion =danion.dropna(axis='columns')
p= (danion.columns[danion.sum() == 0 ]) ##find out what columns have nan
danion =danion.drop(columns=p)
corr = danion.corr()
columns = np.full((corr.shape[0],), True, dtype=bool)
for i in range(corr.shape[0]):
for j in range(i+1, corr.shape[0]):
if corr.iloc[i,j] >= abs(0.9):
if columns[j]:
columns[j] = False
selected_anion_columns = danion.columns[columns]
danion = danion[selected_anion_columns]
cation_column = dcation.columns.tolist()
anion_column = danion.columns.tolist()
#########saving the cation descriptor and anion descriptor final column that is required to run the model
np.savetxt('cation_column.txt', cation_column, delimiter=";", fmt="%s")
np.savetxt('anion_column.txt', anion_column, delimiter=";", fmt="%s")
# +
###property column
dp = df[['Delta(prev)','Pressure/kPa','Temperature/K','Electrical_conductivity[Liquid]/S/m']]
###combine the name column, descriptor column and the property column
do = pd.concat([dname,dcation,danion,dp],axis=1,sort=False)
do =do.dropna(axis='rows')
# +
####drop duplicates with the lowest error value
do= do.sort_values('Delta(prev)', ascending=True).drop_duplicates(['Cation_smiles','Anion_smiles','Temperature/K','Pressure/kPa'],keep='last').sort_index()
###drop the error value
do = do.drop(['Delta(prev)'],axis=1)
# +
######add property filter
do = do.reset_index(drop=True)
do = do[do['Pressure/kPa'] == 101][do["Temperature/K"] < 475][do["Temperature/K"] > 275][
do['Electrical_conductivity[Liquid]/S/m']>0]
do.to_csv('processed.csv')
# +
###separate out X and y
len_row=do.shape[0]
len_coul=do.shape[1]
X= do.iloc[:,4:len_coul-1].values
y = do.iloc[:,len_coul-1].values
print(X)
print(y)
# +
########data normalization
sc =MinMaxScaler()
sy = MinMaxScaler()
X = sc.fit_transform(X)
y = np.log10(y)
y= y.reshape(-1,1)
y = sy.fit_transform(y)
joblib.dump(sc, 'scalerx_svm.gz')
joblib.dump(sy, 'scaler2x_svm.gz')
####scale the validation data
# -
X_train,X_test,y_train,y_test= train_test_split(X,y,test_size=0.1,random_state=50)
# # splitting the data into test and train##################
# +
from sklearn.svm import SVR
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
# defining parameter range
regressor = GridSearchCV(
estimator=SVR(kernel='rbf'),
param_grid={
'C': [0.1, 1,10,25,50,100,110,120,130,140,150,160,170,180,190,200,250,350,500],
'epsilon': [0.0001,0.0002,0.0003,0.0004,0.0005, 0.001,0.002,0.003,0.004,0.005],
'gamma': [0.0001,0.0002,0.0003,0.0004,0.0005,0.0006,0.0007,0.001, 0.005,0.006,0.007,0.008,0.1,0.2,0.3,0.4,1, 3, 5],
},
cv=10, scoring='neg_mean_squared_error', verbose=2, n_jobs=-1)
regressor.fit(X_train,y_train)
print(regressor.best_params_)
print(regressor.best_estimator_)
# +
from sklearn.svm import SVR
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
best_svr = SVR(kernel='rbf', C=250, epsilon=0.001, gamma=3,
max_iter=-1)
# +
best_svr.fit(X_train,y_train)
y_ptrain = best_svr.predict(X_train)
y_ptrain=y_ptrain.reshape(-1,1)
ys=y_train.reshape(-1,1)
ypn = sy.inverse_transform(y_ptrain)
ytn = sy.inverse_transform(ys)
ypn = 10**(ypn) ####model
ytn = 10**(ytn) ####experiment
print('r2 of train set is',r2_score(ytn,ypn))
print('mae of train set is',mean_absolute_error(ytn,ypn))
print('rmsd of train set is',mean_squared_error(ytn,ypn,squared=False))
pyplot.scatter(ytn,ypn,label='Training Set')
plt.xlabel('Experiment Ionic Conductivity S/m')
plt.ylabel('Predicted Ionic Conductivity S/m')
pyplot.legend()
ypn = np.concatenate(ypn,axis=0)
ytn = np.concatenate(ytn,axis=0)
ypn = ypn.tolist()
ytn = ytn.tolist()
####################################################################################
make = np.column_stack((ypn,ytn))
df = pd.DataFrame(make,columns=['Model_S/m','Exp_S/m'])
df.to_csv('svm_model_train_set.csv',sep=';',index=False)
y_tra = best_svr.predict(X_test)
y_ptrain=y_tra.reshape(-1,1)
ys=y_test.reshape(-1,1)
ypn = sy.inverse_transform(y_ptrain)
ytn = sy.inverse_transform(ys)
ypn = 10**(ypn) ####model
ytn = 10**(ytn) ####experiment
print('r2 of test set is',r2_score(ytn,ypn))
print('mae of test set is',mean_absolute_error(ytn,ypn))
print('rmsd of test set is',mean_squared_error(ytn,ypn,squared=False))
pyplot.scatter(ytn,ypn,label='Test Set')
plt.xlabel('Experiment Ionic Conductivity S/m')
plt.ylabel('Predicted Ionic Conductivity S/m')
pyplot.legend()
plt.savefig('train.png')
ypn = np.concatenate(ypn,axis=0)
ytn = np.concatenate(ytn,axis=0)
ypn = ypn.tolist()
ytn = ytn.tolist()
####################################################################################
make = np.column_stack((ypn,ytn))
df = pd.DataFrame(make,columns=['Model_S/m','Exp_S/m'])
df.to_csv('svm_model_test_set.csv',sep=';',index=False)
y_tra = best_svr.predict(X)
y_ptrain=y_tra.reshape(-1,1)
ys=y.reshape(-1,1)
ypn = sy.inverse_transform(y_ptrain)
ytn = sy.inverse_transform(ys)
ypn = 10**(ypn) ####model
ytn = 10**(ytn) ####experiment
print('r2 of entire set is',r2_score(ytn,ypn))
print('mae of entire set is',mean_absolute_error(ytn,ypn))
print('rmsd of test set is',mean_squared_error(ytn,ypn,squared=False))
# +
y_ptrain = best_svr.predict(X_train)
y_ptrain=y_ptrain.reshape(-1,1)
ys=y_train.reshape(-1,1)
ypn = sy.inverse_transform(y_ptrain)
ytn = sy.inverse_transform(ys)
ypn = 10**(ypn) ####model
ytn = 10**(ytn) ####experiment
print('r2 of train set is',r2_score(ytn,ypn))
print('mse of train set is',mean_absolute_error(ytn,ypn))
print('rmsd of train set is',mean_squared_error(ytn,ypn,squared=False))
pyplot.scatter(ytn,ypn,label='Training Set')
plt.xlabel('Experiment Ionic Conductivity S/m')
plt.ylabel('Predicted Ionic Conductivity S/m')
pyplot.legend()
ypn2 = np.concatenate(ypn,axis=0)
ytn2 = np.concatenate(ytn,axis=0)
ypn2 = ypn2.tolist()
ytn2 = ytn2.tolist()
####################################################################################
make = np.column_stack((ypn2,ytn2))
df = pd.DataFrame(make,columns=['Model_S/m','Exp_S/m'])
df.to_csv('results/svm_model_test_set.csv',sep=';',index=False)
filename = 'best_model.sav'
pickle.dump(best_svr, open(filename, 'wb'))
# +
y_ptrain = regressor.predict(X_train)
y_ptrain=y_ptrain.reshape(-1,1)
y_train=y_train.reshape(-1,1)
ypn = sy.inverse_transform(y_ptrain)
ytn = sy.inverse_transform(y_train)
ypn = 10**(ypn) ####model
ytn = 10**(ytn) ####experiment
print('r2 of train set is',r2_score(ytn,ypn))
print('mse of train set is',mean_squared_error(ytn,ypn))
print('rmsd of train set is',mean_squared_error(ytn,ypn,squared=False))
################################################################################
make = np.column_stack((ypn,ytn))
df = pd.DataFrame(make,columns=['Model_S/m','Exp_S/m'])
df.to_csv('results/svm_model_training_set.csv',sep=';',index=False)
y_p = regressor.predict(X_test)
y_p=y_p.reshape(-1,1)
y_test=y_test.reshape(-1,1)
ypn2 = sy.inverse_transform(y_p)
ytn2 = sy.inverse_transform(y_test)
ypn2 = 10**(ypn2) ####model
ytn2 = 10**(ytn2) ####experiment
####################################################################################
print('r2 of test set is',r2_score(ytn2,ypn2))
print('mse of test set is',mean_squared_error(ytn2,ypn2))
print('rmsd of test set is',mean_squared_error(ytn2,ypn2,squared=False))
ypn2 = np.concatenate(ypn2,axis=0)
ytn2 = np.concatenate(ytn2,axis=0)
ypn2 = ypn2.tolist()
ytn2 = ytn2.tolist()
####################################################################################
make = np.column_stack((ypn2,ytn2))
df = pd.DataFrame(make,columns=['Model_S/m','Exp_S/m'])
df.to_csv('results/svm_model_test_set.csv',sep=';',index=False)
######################################################################################
pyplot.scatter(ytn,ypn,label='Training Set')
pyplot.scatter(ytn2,ypn2,label='Test Set')
plt.xlabel('Experiment Ionic Conductivity S/m')
plt.ylabel('Predicted Ionic Conductivity S/m')
pyplot.legend()
| model/svr/model_development-version3-svm_kfold.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # LeetCode #566. Reshape the Matrix
#
# ## Question
#
# https://leetcode.com/problems/reshape-the-matrix/
#
# In MATLAB, there is a very useful function called 'reshape', which can reshape a matrix into a new one with different size but keep its original data.
#
# You're given a matrix represented by a two-dimensional array, and two positive integers r and c representing the row number and column number of the wanted reshaped matrix, respectively.
#
# The reshaped matrix need to be filled with all the elements of the original matrix in the same row-traversing order as they were.
#
# If the 'reshape' operation with given parameters is possible and legal, output the new reshaped matrix; Otherwise, output the original matrix.
#
# Example 1:
#
# Input:
# nums = [[1,2],
# [3,4]]
# r = 1, c = 4
#
# Output:
# [[1,2,3,4]]
#
# Explanation:
# The row-traversing of nums is [1,2,3,4]. The new reshaped matrix is a 1 * 4 matrix, fill it row by row by using the previous list.
#
# Example 2:
#
# Input:
# nums = [[1,2],
# [3,4]]
# r = 2, c = 4
#
# Output:
# [[1,2],
# [3,4]]
#
# Explanation:
# There is no way to reshape a 2 * 2 matrix to a 2 * 4 matrix. So output the original matrix.
# ## My Solution
def matrixReshape(nums, r, c):
import numpy as np
n = np.array(nums)
n = n.ravel()
if len(n) != r * c:
return nums
n = n.reshape(r, c)
return n
# test code
nums = [[1,2],
[3,4]]
r = 1
c = 4
matrixReshape(nums, r, c)
# numpy의 ravel을 사용해서 펴준 뒤 reshape로 가로 세로 길이를 맞춥니다.
# ravel로 폈을 때 길이가 r * c가 되지 않으면 처음에 입력받은 nums를 반환합니다.
# ## My Result
#
# __Runtime__ : 144 ms, faster than 18.11% of Python online submissions for Reshape the Matrix.
#
# __Memory Usage__ : 22 MB, less than 5.88% of Python online submissions for Reshape the Matrix.
# ## @StefanPochmann's Solution
import numpy as np
def matrixReshape(nums, r, c):
try:
return np.reshape(nums, (r, c)).tolist()
except:
return nums
# test code
nums = [[1,2],
[3,4]]
r = 1
c = 4
matrixReshape(nums, r, c)
# 사실 ravel을 쓸 필요가 없었습니다. 넘파이의 reshape와 tolist로 한 번에 처리했습니다.
# 길이가 다를 경우는 try, except로 처리했습니다.
# ## @StefanPochmann's Result
#
# __Runtime__ : 92 ms, faster than 46.11% of Python online submissions for Reshape the Matrix.
#
# __Memory Usage__ : 22.4 MB, less than 5.43% of Python online submissions for Reshape the Matrix.
# ## @yifan.li.1112's Solution
def matrixReshape(nums, r, c):
nrows = len(nums)
ncols = len(nums[0])
if nrows * ncols == r * c:
onedArray = []
reshaped = [[0] * c for i in range(r)]
for x in nums:
onedArray += x
for index, item in enumerate(onedArray):
placeRow = int(index / c)
placeCol = int(index % c)
reshaped[placeRow][placeCol] = item
return reshaped
else:
return nums
# test code
nums = [[1,2],
[3,4]]
r = 1
c = 4
matrixReshape(nums, r, c)
# ### About yifan.li.1112's Solution
# 1) `onedArray`
#
# onedArray = []
# nums = [[1, 2], [3, 4]]
#
# for x in nums:
# onedArray += x
#
# 위 코드는 np.ravel()과 같은 역할을 합니다.
# [] + [1, 2] + [3, 4] 과정을 통해 onedArray = [1, 2, 3, 4]가 됩니다.
#
# 2) `reshaped = [[0] * c for i in range(r)]`
#
# c와 r에 들어가는 숫자에 따라 다음 결과를 반환합니다.
#
# >>> [[0] * 1 for i in range(4)]
# [[0], [0], [0], [0]]
#
# >>> [[0] * 2 for i in range(2)]
# [[0, 0], [0, 0]]
#
# >>> [[0] * 4 for i in range(1)]
# [[0, 0, 0, 0]]
# 3)
onedArray = [1, 2, 3, 4]
for index, item in enumerate(onedArray):
## c = 4
placeRow = int(index / c) ## index를 4로 나눈 몫을 Row로 할당합니다.
placeCol = int(index % c) ## index를 4로 나눈 나머지를 Col으로 할당합니다.
print('index:', index, ' item:', item, ' c:', c, ' placeRow:', placeRow, ' placeCol:', placeCol)
# ## @yifan.li.1112's Result
#
# __Runtime__ : 80 ms, faster than 96.11% of Python online submissions for Reshape the Matrix.
#
# __Memory Usage__ : 12.8 MB, less than 81.90% of Python online submissions for Reshape the Matrix.
| LeetCode/LeetCode_566ReshapeTheMatrix.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="ZrwVQsM9TiUw"
# ##### Copyright 2019 The TensorFlow Probability Authors.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# + cellView="form" id="CpDUTVKYTowI"
#@title Licensed under the Apache License, Version 2.0 (the "License"); { display-mode: "form" }
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] id="ltPJCG6pAUoc"
# # TFP Probabilistic Layers: Regression
#
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/probability/examples/Probabilistic_Layers_Regression"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/probability/blob/main/tensorflow_probability/examples/jupyter_notebooks/Probabilistic_Layers_Regression.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/probability/blob/main/tensorflow_probability/examples/jupyter_notebooks/Probabilistic_Layers_Regression.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/probability/tensorflow_probability/examples/jupyter_notebooks/Probabilistic_Layers_Regression.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# </table>
# + [markdown] id="WRVR-tGTR31S"
# In this example we show how to fit regression models using TFP's "probabilistic layers."
# + [markdown] id="uiR4-VOt9NFX"
# ### Dependencies & Prerequisites
#
# + id="kZ0MdF1j8WJf"
#@title Import { display-mode: "form" }
from pprint import pprint
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import tensorflow.compat.v2 as tf
tf.enable_v2_behavior()
import tensorflow_probability as tfp
sns.reset_defaults()
#sns.set_style('whitegrid')
#sns.set_context('talk')
sns.set_context(context='talk',font_scale=0.7)
# %matplotlib inline
tfd = tfp.distributions
# + [markdown] id="7nnwjUdVoWN2"
# ### Make things Fast!
# + [markdown] id="2CK9RaDcoYPG"
# Before we dive in, let's make sure we're using a GPU for this demo.
#
# To do this, select "Runtime" -> "Change runtime type" -> "Hardware accelerator" -> "GPU".
#
# The following snippet will verify that we have access to a GPU.
# + id="qP_4Xr8vpA42"
if tf.test.gpu_device_name() != '/device:GPU:0':
print('WARNING: GPU device not found.')
else:
print('SUCCESS: Found GPU: {}'.format(tf.test.gpu_device_name()))
# + [markdown] id="FJRBc_S0ppfE"
# Note: if for some reason you cannot access a GPU, this colab will still work. (Training will just take longer.)
# + [markdown] id="xuqxMmryiduM"
# ## Motivation
# + [markdown] id="RtBLNF-tin2L"
# Wouldn't it be great if we could use TFP to specify a probabilistic model then simply minimize the negative log-likelihood, i.e.,
# + id="3PFfNeJzifo7"
negloglik = lambda y, rv_y: -rv_y.log_prob(y)
# + [markdown] id="cN4IP8n_jIvT"
# Well not only is it possible, but this colab shows how! (In context of linear regression problems.)
# + cellView="form" id="5zCEYpzu7bDX"
#@title Synthesize dataset.
w0 = 0.125
b0 = 5.
x_range = [-20, 60]
def load_dataset(n=150, n_tst=150):
np.random.seed(43)
def s(x):
g = (x - x_range[0]) / (x_range[1] - x_range[0])
return 3 * (0.25 + g**2.)
x = (x_range[1] - x_range[0]) * np.random.rand(n) + x_range[0]
eps = np.random.randn(n) * s(x)
y = (w0 * x * (1. + np.sin(x)) + b0) + eps
x = x[..., np.newaxis]
x_tst = np.linspace(*x_range, num=n_tst).astype(np.float32)
x_tst = x_tst[..., np.newaxis]
return y, x, x_tst
y, x, x_tst = load_dataset()
# + [markdown] id="N8Shtn_e99XC"
# ### Case 1: No Uncertainty
# + id="RxKJ_RPI0K4N"
# Build model.
model = tf.keras.Sequential([
tf.keras.layers.Dense(1),
tfp.layers.DistributionLambda(lambda t: tfd.Normal(loc=t, scale=1)),
])
# Do inference.
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01), loss=negloglik)
model.fit(x, y, epochs=1000, verbose=False);
# Profit.
[print(np.squeeze(w.numpy())) for w in model.weights];
yhat = model(x_tst)
assert isinstance(yhat, tfd.Distribution)
# + cellView="form" id="1AE9ElaKI6Er"
#@title Figure 1: No uncertainty.
w = np.squeeze(model.layers[-2].kernel.numpy())
b = np.squeeze(model.layers[-2].bias.numpy())
plt.figure(figsize=[6, 1.5]) # inches
#plt.figure(figsize=[8, 5]) # inches
plt.plot(x, y, 'b.', label='observed');
plt.plot(x_tst, yhat.mean(),'r', label='mean', linewidth=4);
plt.ylim(-0.,17);
plt.yticks(np.linspace(0, 15, 4)[1:]);
plt.xticks(np.linspace(*x_range, num=9));
ax=plt.gca();
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['left'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
#ax.spines['left'].set_smart_bounds(True)
#ax.spines['bottom'].set_smart_bounds(True)
plt.legend(loc='center left', fancybox=True, framealpha=0., bbox_to_anchor=(1.05, 0.5))
plt.savefig('/tmp/fig1.png', bbox_inches='tight', dpi=300)
# + [markdown] id="91kwRqs4O5Yv"
# ### Case 2: Aleatoric Uncertainty
# + id="TLZ97_V4PP-f"
# Build model.
model = tf.keras.Sequential([
tf.keras.layers.Dense(1 + 1),
tfp.layers.DistributionLambda(
lambda t: tfd.Normal(loc=t[..., :1],
scale=1e-3 + tf.math.softplus(0.05 * t[...,1:]))),
])
# Do inference.
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01), loss=negloglik)
model.fit(x, y, epochs=1000, verbose=False);
# Profit.
[print(np.squeeze(w.numpy())) for w in model.weights];
yhat = model(x_tst)
assert isinstance(yhat, tfd.Distribution)
# + cellView="form" id="JSSWw2-FPCiG"
#@title Figure 2: Aleatoric Uncertainty
plt.figure(figsize=[6, 1.5]) # inches
plt.plot(x, y, 'b.', label='observed');
m = yhat.mean()
s = yhat.stddev()
plt.plot(x_tst, m, 'r', linewidth=4, label='mean');
plt.plot(x_tst, m + 2 * s, 'g', linewidth=2, label=r'mean + 2 stddev');
plt.plot(x_tst, m - 2 * s, 'g', linewidth=2, label=r'mean - 2 stddev');
plt.ylim(-0.,17);
plt.yticks(np.linspace(0, 15, 4)[1:]);
plt.xticks(np.linspace(*x_range, num=9));
ax=plt.gca();
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['left'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
#ax.spines['left'].set_smart_bounds(True)
#ax.spines['bottom'].set_smart_bounds(True)
plt.legend(loc='center left', fancybox=True, framealpha=0., bbox_to_anchor=(1.05, 0.5))
plt.savefig('/tmp/fig2.png', bbox_inches='tight', dpi=300)
# + [markdown] id="xEvTd7ZJYvDx"
# ### Case 3: Epistemic Uncertainty
# + cellView="both" id="VwzbWw3_CQ2z"
# Specify the surrogate posterior over `keras.layers.Dense` `kernel` and `bias`.
def posterior_mean_field(kernel_size, bias_size=0, dtype=None):
n = kernel_size + bias_size
c = np.log(np.expm1(1.))
return tf.keras.Sequential([
tfp.layers.VariableLayer(2 * n, dtype=dtype),
tfp.layers.DistributionLambda(lambda t: tfd.Independent(
tfd.Normal(loc=t[..., :n],
scale=1e-5 + tf.nn.softplus(c + t[..., n:])),
reinterpreted_batch_ndims=1)),
])
# + cellView="both" id="aAQhyK9Y_lm1"
# Specify the prior over `keras.layers.Dense` `kernel` and `bias`.
def prior_trainable(kernel_size, bias_size=0, dtype=None):
n = kernel_size + bias_size
return tf.keras.Sequential([
tfp.layers.VariableLayer(n, dtype=dtype),
tfp.layers.DistributionLambda(lambda t: tfd.Independent(
tfd.Normal(loc=t, scale=1),
reinterpreted_batch_ndims=1)),
])
# + id="XI7ZCFzSnrWN"
# Build model.
model = tf.keras.Sequential([
tfp.layers.DenseVariational(1, posterior_mean_field, prior_trainable, kl_weight=1/x.shape[0]),
tfp.layers.DistributionLambda(lambda t: tfd.Normal(loc=t, scale=1)),
])
# Do inference.
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01), loss=negloglik)
model.fit(x, y, epochs=1000, verbose=False);
# Profit.
[print(np.squeeze(w.numpy())) for w in model.weights];
yhat = model(x_tst)
assert isinstance(yhat, tfd.Distribution)
# + cellView="form" id="Y4Bypix9UvTO"
#@title Figure 3: Epistemic Uncertainty
plt.figure(figsize=[6, 1.5]) # inches
plt.clf();
plt.plot(x, y, 'b.', label='observed');
yhats = [model(x_tst) for _ in range(100)]
avgm = np.zeros_like(x_tst[..., 0])
for i, yhat in enumerate(yhats):
m = np.squeeze(yhat.mean())
s = np.squeeze(yhat.stddev())
if i < 25:
plt.plot(x_tst, m, 'r', label='ensemble means' if i == 0 else None, linewidth=0.5)
avgm += m
plt.plot(x_tst, avgm/len(yhats), 'r', label='overall mean', linewidth=4)
plt.ylim(-0.,17);
plt.yticks(np.linspace(0, 15, 4)[1:]);
plt.xticks(np.linspace(*x_range, num=9));
ax=plt.gca();
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['left'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
#ax.spines['left'].set_smart_bounds(True)
#ax.spines['bottom'].set_smart_bounds(True)
plt.legend(loc='center left', fancybox=True, framealpha=0., bbox_to_anchor=(1.05, 0.5))
plt.savefig('/tmp/fig3.png', bbox_inches='tight', dpi=300)
# + [markdown] id="H_3At7s2fel0"
# ### Case 4: Aleatoric & Epistemic Uncertainty
# + id="GcRC3uwcft6l"
# Build model.
model = tf.keras.Sequential([
tfp.layers.DenseVariational(1 + 1, posterior_mean_field, prior_trainable, kl_weight=1/x.shape[0]),
tfp.layers.DistributionLambda(
lambda t: tfd.Normal(loc=t[..., :1],
scale=1e-3 + tf.math.softplus(0.01 * t[...,1:]))),
])
# Do inference.
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01), loss=negloglik)
model.fit(x, y, epochs=1000, verbose=False);
# Profit.
[print(np.squeeze(w.numpy())) for w in model.weights];
yhat = model(x_tst)
assert isinstance(yhat, tfd.Distribution)
# + cellView="form" id="cWhfYYzcgFak"
#@title Figure 4: Both Aleatoric & Epistemic Uncertainty
plt.figure(figsize=[6, 1.5]) # inches
plt.plot(x, y, 'b.', label='observed');
yhats = [model(x_tst) for _ in range(100)]
avgm = np.zeros_like(x_tst[..., 0])
for i, yhat in enumerate(yhats):
m = np.squeeze(yhat.mean())
s = np.squeeze(yhat.stddev())
if i < 15:
plt.plot(x_tst, m, 'r', label='ensemble means' if i == 0 else None, linewidth=1.)
plt.plot(x_tst, m + 2 * s, 'g', linewidth=0.5, label='ensemble means + 2 ensemble stdev' if i == 0 else None);
plt.plot(x_tst, m - 2 * s, 'g', linewidth=0.5, label='ensemble means - 2 ensemble stdev' if i == 0 else None);
avgm += m
plt.plot(x_tst, avgm/len(yhats), 'r', label='overall mean', linewidth=4)
plt.ylim(-0.,17);
plt.yticks(np.linspace(0, 15, 4)[1:]);
plt.xticks(np.linspace(*x_range, num=9));
ax=plt.gca();
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['left'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
#ax.spines['left'].set_smart_bounds(True)
#ax.spines['bottom'].set_smart_bounds(True)
plt.legend(loc='center left', fancybox=True, framealpha=0., bbox_to_anchor=(1.05, 0.5))
plt.savefig('/tmp/fig4.png', bbox_inches='tight', dpi=300)
# + [markdown] id="qmgmcmMKzOH7"
# ### Case 5: Functional Uncertainty
# + cellView="form" id="qtXVxLRdzHBn"
#@title Custom PSD Kernel
class RBFKernelFn(tf.keras.layers.Layer):
def __init__(self, **kwargs):
super(RBFKernelFn, self).__init__(**kwargs)
dtype = kwargs.get('dtype', None)
self._amplitude = self.add_variable(
initializer=tf.constant_initializer(0),
dtype=dtype,
name='amplitude')
self._length_scale = self.add_variable(
initializer=tf.constant_initializer(0),
dtype=dtype,
name='length_scale')
def call(self, x):
# Never called -- this is just a layer so it can hold variables
# in a way Keras understands.
return x
@property
def kernel(self):
return tfp.math.psd_kernels.ExponentiatedQuadratic(
amplitude=tf.nn.softplus(0.1 * self._amplitude),
length_scale=tf.nn.softplus(5. * self._length_scale)
)
# + id="_gJJtPMzzDyo"
# For numeric stability, set the default floating-point dtype to float64
tf.keras.backend.set_floatx('float64')
# Build model.
num_inducing_points = 40
model = tf.keras.Sequential([
tf.keras.layers.InputLayer(input_shape=[1]),
tf.keras.layers.Dense(1, kernel_initializer='ones', use_bias=False),
tfp.layers.VariationalGaussianProcess(
num_inducing_points=num_inducing_points,
kernel_provider=RBFKernelFn(),
event_shape=[1],
inducing_index_points_initializer=tf.constant_initializer(
np.linspace(*x_range, num=num_inducing_points,
dtype=x.dtype)[..., np.newaxis]),
unconstrained_observation_noise_variance_initializer=(
tf.constant_initializer(np.array(0.54).astype(x.dtype))),
),
])
# Do inference.
batch_size = 32
loss = lambda y, rv_y: rv_y.variational_loss(
y, kl_weight=np.array(batch_size, x.dtype) / x.shape[0])
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01), loss=loss)
model.fit(x, y, batch_size=batch_size, epochs=1000, verbose=False)
# Profit.
yhat = model(x_tst)
assert isinstance(yhat, tfd.Distribution)
# + cellView="form" id="Fp4qEWSRzc8m"
#@title Figure 5: Functional Uncertainty
y, x, _ = load_dataset()
plt.figure(figsize=[6, 1.5]) # inches
plt.plot(x, y, 'b.', label='observed');
num_samples = 7
for i in range(num_samples):
sample_ = yhat.sample().numpy()
plt.plot(x_tst,
sample_[..., 0].T,
'r',
linewidth=0.9,
label='ensemble means' if i == 0 else None);
plt.ylim(-0.,17);
plt.yticks(np.linspace(0, 15, 4)[1:]);
plt.xticks(np.linspace(*x_range, num=9));
ax=plt.gca();
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
ax.spines['left'].set_position(('data', 0))
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
#ax.spines['left'].set_smart_bounds(True)
#ax.spines['bottom'].set_smart_bounds(True)
plt.legend(loc='center left', fancybox=True, framealpha=0., bbox_to_anchor=(1.05, 0.5))
plt.savefig('/tmp/fig5.png', bbox_inches='tight', dpi=300)
| site/en-snapshot/probability/examples/Probabilistic_Layers_Regression.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # MNIST - Lightning ⚡️ Syft Duet - Data Owner 🎸
# ## PART 1: Launch a Duet Server and Connect
#
# As a Data Owner, you want to allow someone else to perform data science on data that you own and likely want to protect.
#
# In order to do this, we must load our data into a locally running server within this notebook. We call this server a "Duet".
#
# To begin, you must launch Duet and help your Duet "partner" (a Data Scientist) connect to this server.
#
# You do this by running the code below and sending the code snippet containing your unique Server ID to your partner and following the instructions it gives!
# TorchVision hotfix https://github.com/pytorch/vision/issues/3549
from syft.util import get_root_data_path
from torchvision import datasets
datasets.MNIST.resources = [
(
"https://ossci-datasets.s3.amazonaws.com/mnist/train-images-idx3-ubyte.gz",
"f68b3c2dcbeaaa9fbdd348bbdeb94873",
),
(
"https://ossci-datasets.s3.amazonaws.com/mnist/train-labels-idx1-ubyte.gz",
"d53e105ee54ea40749a09fcbcd1e9432",
),
(
"https://ossci-datasets.s3.amazonaws.com/mnist/t10k-images-idx3-ubyte.gz",
"9fb629c4189551a2d022fa330f9573f3",
),
(
"https://ossci-datasets.s3.amazonaws.com/mnist/t10k-labels-idx1-ubyte.gz",
"ec29112dd5afa0611ce80d1b7f02629c",
),
]
datasets.MNIST(get_root_data_path(), train=True, download=True)
datasets.MNIST(get_root_data_path(), train=False, download=True)
import syft as sy
duet = sy.launch_duet(loopback=True)
sy.load("pytorch_lightning")
# If you are in Jupyter Notebook (not Colab) the ☝🏾DUET LIVE STATUS above will animate
# ## PART 2: Add Request Handlers
# The MNIST Data Science Notebook makes a number of requests to access data. You can view these requests with:
# ```
# duet.requests.pandas
# ```
# Then manually accept or deny them with:
# ```
# duet.requests[0].accept()
# ```
# However for training loops this can be slow and tedious so the below code will create request handlers which will automatically respond with `accept` or `deny` depending on the name of the request.
duet.requests.pandas
duet.requests.add_handler(
action="accept"
)
duet.store.pandas
| packages/syft/examples/duet/mnist_lightning/MNIST_Lightning_Syft_Data_Owner.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] papermill={"duration": 0.017418, "end_time": "2020-03-16T21:29:51.912227", "exception": false, "start_time": "2020-03-16T21:29:51.894809", "status": "completed"} tags=[]
# # Essential: Core concepts
#
#
# ## Ploomber's core: Tasks, Products, DAG and Clients
#
# To get started with ploomber you only have to learn four concepts:
#
# 1. Task. A unit of work that takes some input and produces a persistent change
# 2. Product. A persistent change *produced* by a Task (e.g. a file in the local filesystem, a table in a remote database)
# 3. DAG. A collection of Tasks, used to specify dependencies among them (use output from Task A as input for Task B)
# 4. Client. An object that communicates with an external system (e.g. a database)
#
# There is a standard [Task API](../api.rst#ploomber.tasks.Task) defined by an abstract class, this is also true for [Products](../api.rst#ploomber.products.Product) and [Clients](../api.rst#ploomber.clients.Client). Which means you only have to learn the concept once and all concrete classes will have the same API.
#
#
# ## The DAG lifecycle: Declare, render, build
#
# A DAG goes through three steps before being executed:
#
# 1. Declaration. A DAG is created and Tasks are added to it
# 2. Rendering. Placeholders are resolved and validation is performed on Task inputs
# 3. Building. All *outdated* Tasks are executed in the appropriate order (run upstream task dependencies first)
#
# ### Declaration
# + nbsphinx="hidden"
import tempfile
import os
tmp_dir = tempfile.mkdtemp()
os.chdir(tmp_dir)
# + papermill={"duration": 2.158019, "end_time": "2020-03-16T21:29:54.082263", "exception": false, "start_time": "2020-03-16T21:29:51.924244", "status": "completed"} tags=[]
from pathlib import Path
import pandas as pd
from ploomber import DAG
from ploomber.tasks import PythonCallable, SQLUpload, SQLScript
from ploomber.clients import SQLAlchemyClient
from ploomber.products import File, SQLiteRelation
from ploomber.executors import Serial
# + [markdown] papermill={"duration": 0.010662, "end_time": "2020-03-16T21:29:54.103401", "exception": false, "start_time": "2020-03-16T21:29:54.092739", "status": "completed"} tags=[]
# The simplest Task is `PythonCallable`, which takes a callable (e.g. a function) as its first argument. The only requirement for the functions is to have a `product`
# argument, if the task has dependencies, it must have an upstream argument as well.
# + papermill={"duration": 0.024542, "end_time": "2020-03-16T21:29:54.138847", "exception": false, "start_time": "2020-03-16T21:29:54.114305", "status": "completed"} tags=[]
def _one_task(product):
pd.DataFrame({'one_column': [1, 2, 3]}).to_csv(str(product))
def _another_task(upstream, product):
df = pd.read_csv(str(upstream['one']))
df['another_column'] = df['one_column'] + 1
df.to_csv(str(product))
# + papermill={"duration": 0.033715, "end_time": "2020-03-16T21:29:54.183125", "exception": false, "start_time": "2020-03-16T21:29:54.149410", "status": "completed"} tags=[]
# instantate our DAG
# NOTE: passing the executor parameter is only required for testing purposes, can be removed
dag = DAG(executor=Serial(build_in_subprocess=False))
# instantiate two tasks and add them to the DAG
one_task = PythonCallable(_one_task, File('one_file.csv'), dag, name='one')
another_task = PythonCallable(_another_task, File('another_file.csv'), dag, name='another')
# declare dependencies: another_task depends on one_task
one_task >> another_task
# + [markdown] papermill={"duration": 0.010175, "end_time": "2020-03-16T21:29:54.203928", "exception": false, "start_time": "2020-03-16T21:29:54.193753", "status": "completed"} tags=[]
# Note that in the previous function definitions we use `str(product)`, since products are custom objects, they will not work directly when used as parameters to the `DataFrame.to_csv()` function, since our products are `File` objects, using `str` will return a the path as a string. Other products implement different logic, for example a `SQLRelation` returns a `schema.name` string.
# + papermill={"duration": 0.022911, "end_time": "2020-03-16T21:29:54.237697", "exception": false, "start_time": "2020-03-16T21:29:54.214786", "status": "completed"} tags=[]
f = File('/path/to/some/file')
print('* {} to str: "{}"'.format(repr(f), str(f)))
# SQLiteRelation takes a ('schema', 'name', 'kind') or a ('name', 'kind') tuple where kind is 'table' or 'view'
t = SQLiteRelation(('my_table', 'table'))
print('* {} to str: "{}"'.format(repr(t), str(t)))
# + [markdown] papermill={"duration": 0.010264, "end_time": "2020-03-16T21:29:54.258940", "exception": false, "start_time": "2020-03-16T21:29:54.248676", "status": "completed"} tags=[]
# ### Rendering
#
# To generate a Product, Tasks use a combination of inputs and a `source`. The kind of source depends on the kind of Task, `PythonCallable` uses a Python function as source, `SQLScript` uses a string with SQL code as source, `SQLUpload` uses a string to a file as source. Rendering is the process where any necessary preparation and validation to the source take place.
#
# One use case for this is to avoid redudant code. If a Task is declared to have an upstream dependency, it means that it will take the upstream Product as input, instead of declaring the Product twice, we can refer to it in the downstream task using a placeholder. Let's see an example using `SQLUpload`:
# + papermill={"duration": 0.025986, "end_time": "2020-03-16T21:29:54.295264", "exception": false, "start_time": "2020-03-16T21:29:54.269278", "status": "completed"} tags=[]
# Clients are objects that communicate with external systems, such as databases
client = SQLAlchemyClient('sqlite:///my_db.db')
# Tasks that use clients have a client argument, but you can also define DAG-level clients
dag.clients[SQLUpload] = client
dag.clients[SQLiteRelation] = client
dag.clients[SQLScript] = client
# Source is defined as a placeholder: take the product from the upstream task
# named "another" and use it as source
my_table = SQLUpload(source='{{upstream["another"]}}',
product=SQLiteRelation(('my_table', 'table')),
dag=dag,
name='my_table')
another_task >> my_table
# + papermill={"duration": 0.056516, "end_time": "2020-03-16T21:29:54.362916", "exception": false, "start_time": "2020-03-16T21:29:54.306400", "status": "completed"} tags=[]
# resolve placeholders by rendering
dag.render()
# let's see the rendered value:
print('my_table.source as string: "{}"'.format(str(my_table.source)))
# + [markdown] papermill={"duration": 0.011417, "end_time": "2020-03-16T21:29:54.386212", "exception": false, "start_time": "2020-03-16T21:29:54.374795", "status": "completed"} tags=[]
# Another important use case for placeholders are parametrized SQL queries. `SQLScript` runs SQL code in a database that creates a table or a view. Since ploomber requires sources (SQL code) and products (a table/view) to be declared separately we can use placeholders to only declare the product once:
# + papermill={"duration": 0.026163, "end_time": "2020-03-16T21:29:54.424318", "exception": false, "start_time": "2020-03-16T21:29:54.398155", "status": "completed"} tags=[]
source = """
-- product is a placeholder
DROP TABLE IF EXISTS {{product}};
CREATE TABLE {{product}}
AS SELECT * FROM {{upstream["my_table"]}}
WHERE one_column = 1
"""
# instead of declaring "second_table" twice, we declare it in product and refer to it in source
second_table = SQLScript(source=source,
product=SQLiteRelation(('second_table', 'table')),
dag=dag,
name='second_table')
my_table >> second_table
# + papermill={"duration": 0.06229, "end_time": "2020-03-16T21:29:54.500115", "exception": false, "start_time": "2020-03-16T21:29:54.437825", "status": "completed"} tags=[]
dag.render()
# + papermill={"duration": 0.02242, "end_time": "2020-03-16T21:29:54.536101", "exception": false, "start_time": "2020-03-16T21:29:54.513681", "status": "completed"} tags=[]
print('second_table.source:\n{}'.format(str(second_table.source)))
# + [markdown] papermill={"duration": 0.014349, "end_time": "2020-03-16T21:29:54.564523", "exception": false, "start_time": "2020-03-16T21:29:54.550174", "status": "completed"} tags=[]
# ploomber uses [jinja2](https://jinja.palletsprojects.com/en/2.11.x/api/) for rendering, which opens a wide range of possibilities rendering SQL source code. Note that this time, we didn't use the `str` operator explicitely as we did for PythonCallable, this is because jinja automatically casts objects to strings.
#
# Before building our dag, let's take a look at the current status:
# + papermill={"duration": 0.172147, "end_time": "2020-03-16T21:29:54.750764", "exception": false, "start_time": "2020-03-16T21:29:54.578617", "status": "completed"} tags=[]
dag.status()
# + [markdown] papermill={"duration": 0.015344, "end_time": "2020-03-16T21:29:54.782942", "exception": false, "start_time": "2020-03-16T21:29:54.767598", "status": "completed"} tags=[]
# ### Build
#
# Once rendering is done, we can build our DAG.
# + papermill={"duration": 0.373424, "end_time": "2020-03-16T21:29:55.173157", "exception": false, "start_time": "2020-03-16T21:29:54.799733", "status": "completed"} tags=[]
dag.build()
# + [markdown] papermill={"duration": 0.017851, "end_time": "2020-03-16T21:29:55.208361", "exception": false, "start_time": "2020-03-16T21:29:55.190510", "status": "completed"} tags=[]
# The first time we run our pipeline, all Tasks are executed, but the real power of ploomber is running builds over and over again. Ploomber keeps track of each Task's status and only executed outdated ones, since we just built our pipeline, nothing will run:
# + papermill={"duration": 0.569816, "end_time": "2020-03-16T21:29:55.795469", "exception": false, "start_time": "2020-03-16T21:29:55.225653", "status": "completed"} tags=[]
dag.build()
# + [markdown] papermill={"duration": 0.018654, "end_time": "2020-03-16T21:29:55.833352", "exception": false, "start_time": "2020-03-16T21:29:55.814698", "status": "completed"} tags=[]
# ### Task status
#
# Upon sucessful execution, a Task will save metadata along with the Product, to keep track of status in subsequent builds. Once a DAG is built (even if some tasks fail), another call to `dag.build()` will only trigger execution on outdated tasks. A task is run if any of the following conditions is true:
#
# 1. The Products doesn't exist (e.g. when a Task is run for the first time)
# 2. No metadata (e.g. when a Task crashes)
# 3. Any upstream source changed (e.g. an upstream SQL script changed)
# 4. Task's own source changed
#
# These rules enable the following use cases:
#
# 1. Fast incremental builds: Modify any Task source, next build will only run outdated Tasks
# 2. Crash recovery: If a DAG crashes, the next run will start where it was interrupted
#
#
# ### Task parameters
#
# There is one last remaining Task argument to explain: `params`, they are optional parameters whose effect varies depending on the kind of Task. `PythonCallable` just passes them when calling the underlying function, Tasks that take SQL code as source, pass them directly to the source (they are available as placeholders), `NotebookRunner` (which runs Jupyter notebooks), passes them as parameters using [papermill](https://github.com/nteract/papermill).
#
# Let's take a look at the `params` of our previous DAG:
# + papermill={"duration": 0.02861, "end_time": "2020-03-16T21:29:55.880701", "exception": false, "start_time": "2020-03-16T21:29:55.852091", "status": "completed"} tags=[]
print('one_task params:\n\t', one_task.params)
print('another_task params:\n\t', another_task.params)
# + [markdown] papermill={"duration": 0.019293, "end_time": "2020-03-16T21:29:55.920068", "exception": false, "start_time": "2020-03-16T21:29:55.900775", "status": "completed"} tags=[]
# Even though we didn't pass any `param` to the tasks, `product` and `upstream` are automateically added after doing `DAG.render()`, that's why we see those parameters.
#
#
# As a general advice, it is best to keep `params` short, their primary use case is for creating dynamic DAGs (whose number of Tasks is determined using control structures). Dynamic DAGs are covered in a more advanced tutorial.
# + nbsphinx="hidden"
import shutil
shutil.rmtree(tmp_dir)
| python-api-examples/guide/core.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: cugraph_dev
# language: python
# name: cugraph_dev
# ---
# # Core Number
#
#
# In this notebook, we will use cuGraph to compute the core number of every vertex in our test graph
#
# Notebook Credits
# * Original Authors: <NAME>
# * Created: 10/28/2019
# * Last Edit: 10/28/2019
#
# RAPIDS Versions: 0.10.0
#
# Test Hardware
# * GV100 32G, CUDA 10.0
#
#
#
# ## Introduction
#
# Core Number computes the core number for every vertex of a graph G. A k-core of a graph is a maximal subgraph that contains nodes of degree k or more. A node has a core number of k if it belongs to a k-core but not to k+1-core. This call does not support a graph with self-loops and parallel edges.
#
# For a detailed description of the algorithm see: https://en.wikipedia.org/wiki/Degeneracy_(graph_theory)
#
# It takes as input a cugraph.Graph object and returns as output a
# cudf.Dataframe object
#
#
# To compute the K-Core Number cluster in cuGraph use: <br>
# * __df = cugraph.core_number(G)__
# * G: A cugraph.Graph object
#
# Returns:
# * __df : cudf.DataFrame__
# * df['vertex'] - vertex ID
# * df['core_number'] - core number of that vertex
#
#
#
# ## cuGraph Notice
# The current version of cuGraph has some limitations:
#
# * Vertex IDs need to be 32-bit integers.
# * Vertex IDs are expected to be contiguous integers starting from 0.
#
# cuGraph provides the renumber function to mitigate this problem. Input vertex IDs for the renumber function can be either 32-bit or 64-bit integers, can be non-contiguous, and can start from an arbitrary number. The renumber function maps the provided input vertex IDs to 32-bit contiguous integers starting from 0. cuGraph still requires the renumbered vertex IDs to be representable in 32-bit integers. These limitations are being addressed and will be fixed soon.
# ### Test Data
# We will be using the Zachary Karate club dataset
# *<NAME>, An information flow model for conflict and fission in small groups, Journal of
# Anthropological Research 33, 452-473 (1977).*
#
#
# 
#
# ### Prep
# Import needed libraries
import cugraph
import cudf
# ### Read data using cuDF
# Test file
datafile='../data//karate-data.csv'
# read the data using cuDF
gdf = cudf.read_csv(datafile, delimiter='\t', names=['src', 'dst'], dtype=['int32', 'int32'] )
# create a Graph
G = cugraph.Graph()
G.add_edge_list(gdf["src"], gdf["dst"])
# ### Now compute the Core Number
# Call k-cores on the graph
df = cugraph.core_number(G)
df
# ___
# Copyright (c) 2019, NVIDIA CORPORATION.
#
# Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
# ___
| cugraph/cores/core-number.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Filtering out the warnings
import warnings
warnings.filterwarnings('ignore')
# +
# Importing the required libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# -
# # <font color = blue> IMDb Movie Assignment </font>
#
# You have the data for the 100 top-rated movies from the past decade along with various pieces of information about the movie, its actors, and the voters who have rated these movies online. In this assignment, you will try to find some interesting insights into these movies and their voters, using Python.
# ## Task 1: Reading the data
# - ### Subtask 1.1: Read the Movies Data.
#
# Read the movies data file provided and store it in a dataframe `movies`.
# Read the csv file using 'read_csv'. Please write your dataset location here.
movies = pd.read_csv(r'F:\Upgrad Notes\IMDB Assignment\Movie+Assignment+Data.csv')
movies.head()
# - ### Subtask 1.2: Inspect the Dataframe
#
# Inspect the dataframe for dimensions, null-values, and summary of different numeric columns.
# Check the number of rows and columns in the dataframe
movies.shape
# Check the column-wise info of the dataframe
movies.info()
# Check the summary for the numeric columns
movies.describe()
# ## Task 2: Data Analysis
#
# Now that we have loaded the dataset and inspected it, we see that most of the data is in place. As of now, no data cleaning is required, so let's start with some data manipulation, analysis, and visualisation to get various insights about the data.
# - ### Subtask 2.1: Reduce those Digits!
#
# These numbers in the `budget` and `gross` are too big, compromising its readability. Let's convert the unit of the `budget` and `gross` columns from `$` to `million $` first.
# Divide the 'gross' and 'budget' columns by 1000000 to convert '$' to 'million $'
def convert_million(val):
return(val/1000000)
movies.Gross=movies.Gross.apply(convert_million)
movies.budget=movies.budget.apply(convert_million)
movies.head()
# - ### Subtask 2.2: Let's Talk Profit!
#
# 1. Create a new column called `profit` which contains the difference of the two columns: `gross` and `budget`.
# 2. Sort the dataframe using the `profit` column as reference.
# 3. Extract the top ten profiting movies in descending order and store them in a new dataframe - `top10`.
# 4. Plot a scatter or a joint plot between the columns `budget` and `profit` and write a few words on what you observed.
# 5. Extract the movies with a negative profit and store them in a new dataframe - `neg_profit`
# Create the new column named 'profit' by subtracting the 'budget' column from the 'gross' column
movies['Profit']=movies['Gross']-movies['budget']
movies[['Profit']]
movies.head()
# Sort the dataframe with the 'profit' column as reference using the 'sort_values' function. Make sure to set the argument
#'ascending' to 'False'
movies= movies.sort_values(by='Profit', ascending=False)
movies.head()
# Get the top 10 profitable movies by using position based indexing. Specify the rows till 10 (0-9)
top10= movies.iloc[0:10, :]
top10
#Plot profit vs budget
sns.jointplot(movies.budget, movies.Profit, kind='scatter')
plt.show()
# The dataset contains the 100 best performing movies from the year 2010 to 2016. However scatter plot tells a different story. You can notice that there are some movies with negative profit. Although good movies do incur losses, but there appear to be quite a few movie with losses. What can be the reason behind this? Lets have a closer look at this by finding the movies with negative profit.
#Find the movies with negative profit
neg_profit= movies[movies['Profit']<0]
neg_profit
# **`Checkpoint 1:`** Can you spot the movie `Tangled` in the dataset? You may be aware of the movie 'Tangled'. Although its one of the highest grossing movies of all time, it has negative profit as per this result. If you cross check the gross values of this movie (link: https://www.imdb.com/title/tt0398286/), you can see that the gross in the dataset accounts only for the domestic gross and not the worldwide gross. This is true for may other movies also in the list.
# - ### Subtask 2.3: The General Audience and the Critics
#
# You might have noticed the column `MetaCritic` in this dataset. This is a very popular website where an average score is determined through the scores given by the top-rated critics. Second, you also have another column `IMDb_rating` which tells you the IMDb rating of a movie. This rating is determined by taking the average of hundred-thousands of ratings from the general audience.
#
# As a part of this subtask, you are required to find out the highest rated movies which have been liked by critics and audiences alike.
# 1. Firstly you will notice that the `MetaCritic` score is on a scale of `100` whereas the `IMDb_rating` is on a scale of 10. First convert the `MetaCritic` column to a scale of 10.
# 2. Now, to find out the movies which have been liked by both critics and audiences alike and also have a high rating overall, you need to -
# - Create a new column `Avg_rating` which will have the average of the `MetaCritic` and `Rating` columns
# - Retain only the movies in which the absolute difference(using abs() function) between the `IMDb_rating` and `Metacritic` columns is less than 0.5. Refer to this link to know how abs() funtion works - https://www.geeksforgeeks.org/abs-in-python/ .
# - Sort these values in a descending order of `Avg_rating` and retain only the movies with a rating equal to higher than `8` and store these movies in a new dataframe `UniversalAcclaim`.
#
movies[['MetaCritic']].head()
# Change the scale of MetaCritic
movies['MetaCritic']=movies['MetaCritic']/10
movies[['MetaCritic']].head()
movies[['IMDb_rating']].head()
# Find the average ratings
movies['Avg_rating'] = movies[['MetaCritic', 'IMDb_rating']].mean(axis=1)
x= movies[(abs(movies['IMDb_rating']- movies['MetaCritic'])<0.5)]
x
#Sort in descending order of average rating
x= x.sort_values(by='Avg_rating', ascending=False)
x.head()
# Find the movies with metacritic-rating < 0.5 and also with the average rating of >8
UniversalAcclaim=x[(movies['Avg_rating']>8)]
UniversalAcclaim
# **`Checkpoint 2:`** Can you spot a `Star Wars` movie in your final dataset?
# - ### Subtask 2.4: Find the Most Popular Trios - I
#
# You're a producer looking to make a blockbuster movie. There will primarily be three lead roles in your movie and you wish to cast the most popular actors for it. Now, since you don't want to take a risk, you will cast a trio which has already acted in together in a movie before. The metric that you've chosen to check the popularity is the Facebook likes of each of these actors.
#
# The dataframe has three columns to help you out for the same, viz. `actor_1_facebook_likes`, `actor_2_facebook_likes`, and `actor_3_facebook_likes`. Your objective is to find the trios which has the most number of Facebook likes combined. That is, the sum of `actor_1_facebook_likes`, `actor_2_facebook_likes` and `actor_3_facebook_likes` should be maximum.
# Find out the top 5 popular trios, and output their names in a list.
#
# Write your code here
movies['Total_Likes']=movies[['actor_1_facebook_likes', 'actor_2_facebook_likes', 'actor_3_facebook_likes']].sum(axis=1)
top5_likes = movies.sort_values('Total_Likes', ascending=False).head(5)
top5_likes[['actor_1_name', 'actor_2_name', 'actor_3_name']].values.tolist()
# - ### Subtask 2.5: Find the Most Popular Trios - II
#
# In the previous subtask you found the popular trio based on the total number of facebook likes. Let's add a small condition to it and make sure that all three actors are popular. The condition is **none of the three actors' Facebook likes should be less than half of the other two**. For example, the following is a valid combo:
# - actor_1_facebook_likes: 70000
# - actor_2_facebook_likes: 40000
# - actor_3_facebook_likes: 50000
#
# But the below one is not:
# - actor_1_facebook_likes: 70000
# - actor_2_facebook_likes: 40000
# - actor_3_facebook_likes: 30000
#
# since in this case, `actor_3_facebook_likes` is 30000, which is less than half of `actor_1_facebook_likes`.
#
# Having this condition ensures that you aren't getting any unpopular actor in your trio (since the total likes calculated in the previous question doesn't tell anything about the individual popularities of each actor in the trio.).
#
# You can do a manual inspection of the top 5 popular trios you have found in the previous subtask and check how many of those trios satisfy this condition. Also, which is the most popular trio after applying the condition above?
# **Write your answers below.**
#
# - **`No. of trios that satisfy the above condition:`**
#
# - **`Most popular trio after applying the condition:`**
# **`Optional:`** Even though you are finding this out by a natural inspection of the dataframe, can you also achieve this through some *if-else* statements to incorporate this. You can try this out on your own time after you are done with the assignment.
# +
# Your answer here (optional)
# -
# - ### Subtask 2.6: Runtime Analysis
#
# There is a column named `Runtime` in the dataframe which primarily shows the length of the movie. It might be intersting to see how this variable this distributed. Plot a `histogram` or `distplot` of seaborn to find the `Runtime` range most of the movies fall into.
# Runtime histogram/density plot
sns.distplot(movies.Runtime)
plt.show()
# **`Checkpoint 3:`** Most of the movies appear to be sharply 2 hour-long.
# - ### Subtask 2.7: R-Rated Movies
#
# Although R rated movies are restricted movies for the under 18 age group, still there are vote counts from that age group. Among all the R rated movies that have been voted by the under-18 age group, find the top 10 movies that have the highest number of votes i.e.`CVotesU18` from the `movies` dataframe. Store these in a dataframe named `PopularR`.
# Write your code here
PopularR= movies[(movies['content_rating']=='R')]
PopularR= PopularR.sort_values('CVotesU18', ascending=False).head(10)
PopularR
# **`Checkpoint 4:`** Are these kids watching `Deadpool` a lot?
#
# ## Task 3 : Demographic analysis
#
# If you take a look at the last columns in the dataframe, most of these are related to demographics of the voters (in the last subtask, i.e., 2.8, you made use one of these columns - CVotesU18). We also have three genre columns indicating the genres of a particular movie. We will extensively use these columns for the third and the final stage of our assignment wherein we will analyse the voters across all demographics and also see how these vary across various genres. So without further ado, let's get started with `demographic analysis`.
# - ### Subtask 3.1 Combine the Dataframe by Genres
#
# There are 3 columns in the dataframe - `genre_1`, `genre_2`, and `genre_3`. As a part of this subtask, you need to aggregate a few values over these 3 columns.
# 1. First create a new dataframe `df_by_genre` that contains `genre_1`, `genre_2`, and `genre_3` and all the columns related to **CVotes/Votes** from the `movies` data frame. There are 47 columns to be extracted in total.
# 2. Now, Add a column called `cnt` to the dataframe `df_by_genre` and initialize it to one. You will realise the use of this column by the end of this subtask.
# 3. First group the dataframe `df_by_genre` by `genre_1` and find the sum of all the numeric columns such as `cnt`, columns related to CVotes and Votes columns and store it in a dataframe `df_by_g1`.
# 4. Perform the same operation for `genre_2` and `genre_3` and store it dataframes `df_by_g2` and `df_by_g3` respectively.
# 5. Now that you have 3 dataframes performed by grouping over `genre_1`, `genre_2`, and `genre_3` separately, it's time to combine them. For this, add the three dataframes and store it in a new dataframe `df_add`, so that the corresponding values of Votes/CVotes get added for each genre.There is a function called `add()` in pandas which lets you do this. You can refer to this link to see how this function works. https://pandas.pydata.org/pandas-docs/version/0.23.4/generated/pandas.DataFrame.add.html
# 6. The column `cnt` on aggregation has basically kept the track of the number of occurences of each genre.Subset the genres that have atleast 10 movies into a new dataframe `genre_top10` based on the `cnt` column value.
# 7. Now, take the mean of all the numeric columns by dividing them with the column value `cnt` and store it back to the same dataframe. We will be using this dataframe for further analysis in this task unless it is explicitly mentioned to use the dataframe `movies`.
# 8. Since the number of votes can't be a fraction, type cast all the CVotes related columns to integers. Also, round off all the Votes related columns upto two digits after the decimal point.
#
# Create the dataframe df_by_genre
df_by_genre=movies.drop(columns=['Title', 'title_year', 'budget', 'Gross', 'actor_1_name', 'actor_2_name', 'actor_3_name', 'actor_1_facebook_likes', 'actor_2_facebook_likes', 'actor_3_facebook_likes', 'IMDb_rating', 'MetaCritic', 'Runtime', 'content_rating', 'Country', 'Profit', 'Avg_rating', 'Total_Likes'], axis=1)
df_by_genre.head()
df_by_genre.shape
# Create a column cnt and initialize it to 1
df_by_genre=df_by_genre.assign(cnt=1)
df_by_genre.head()
# +
# Group the movies by individual genres
gp1= df_by_genre.groupby('genre_1')
df_by_g1= gp1.sum()
gp2= df_by_genre.groupby('genre_2')
df_by_g2= gp2.sum()
gp3= df_by_genre.groupby('genre_3')
df_by_g3= gp3.sum()
# -
df_by_g1
df_by_g2
df_by_g3
# +
# Add the grouped data frames and store it in a new data frame
x=df_by_g1.add(df_by_g2, fill_value=0)
df_add= x.add(df_by_g3, fill_value=0)
df_add
# -
#reset column index
df_add= df_add.reset_index()
#rename column name
df_add= df_add.rename(columns={'index': 'genre'})
# Extract genres with atleast 10 occurences
genre_top10=df_add[df_add['cnt']>=10]
genre_top10
# Take the mean for every column by dividing with cnt
genre_top10.loc[:,"CVotes10":"VotesnUS"]= genre_top10.loc[:,"CVotes10":"VotesnUS"].divide(genre_top10["cnt"], axis=0)
genre_top10
# Rounding off the columns of Votes to two decimals
genre_top10.loc[:,"VotesM":"VotesnUS"]= np.round(genre_top10.loc[:,"VotesM":"VotesnUS"], decimals=2)
genre_top10
# Converting CVotes to int type
genre_top10.loc[:,"CVotes10":"CVotesnUS"]= genre_top10.loc[:,"CVotes10":"CVotesnUS"].astype('int32')
genre_top10
# If you take a look at the final dataframe that you have gotten, you will see that you now have the complete information about all the demographic (Votes- and CVotes-related) columns across the top 10 genres. We can use this dataset to extract exciting insights about the voters!
# - ### Subtask 3.2: Genre Counts!
#
# Now let's derive some insights from this data frame. Make a bar chart plotting different genres vs cnt using seaborn.
# Countplot for genres
plt.figure(figsize=[20, 5])
sns.barplot(data=df_add, x='genre', y='cnt')
plt.show()
# **`Checkpoint 5:`** Is the bar for `Drama` the tallest?
# - ### Subtask 3.3: Gender and Genre
#
# If you have closely looked at the Votes- and CVotes-related columns, you might have noticed the suffixes `F` and `M` indicating Female and Male. Since we have the vote counts for both males and females, across various age groups, let's now see how the popularity of genres vary between the two genders in the dataframe.
#
# 1. Make the first heatmap to see how the average number of votes of males is varying across the genres. Use seaborn heatmap for this analysis. The X-axis should contain the four age-groups for males, i.e., `CVotesU18M`,`CVotes1829M`, `CVotes3044M`, and `CVotes45AM`. The Y-axis will have the genres and the annotation in the heatmap tell the average number of votes for that age-male group.
#
# 2. Make the second heatmap to see how the average number of votes of females is varying across the genres. Use seaborn heatmap for this analysis. The X-axis should contain the four age-groups for females, i.e., `CVotesU18F`,`CVotes1829F`, `CVotes3044F`, and `CVotes45AF`. The Y-axis will have the genres and the annotation in the heatmap tell the average number of votes for that age-female group.
#
# 3. Make sure that you plot these heatmaps side by side using `subplots` so that you can easily compare the two genders and derive insights.
#
# 4. Write your any three inferences from this plot. You can make use of the previous bar plot also here for better insights.
# Refer to this link- https://seaborn.pydata.org/generated/seaborn.heatmap.html. You might have to plot something similar to the fifth chart in this page (You have to plot two such heatmaps side by side).
#
# 5. Repeat subtasks 1 to 4, but now instead of taking the CVotes-related columns, you need to do the same process for the Votes-related columns. These heatmaps will show you how the two genders have rated movies across various genres.
#
# You might need the below link for formatting your heatmap.
# https://stackoverflow.com/questions/56942670/matplotlib-seaborn-first-and-last-row-cut-in-half-of-heatmap-plot
#
# - Note : Use `genre_top10` dataframe for this subtask
# +
# 1st set of heat maps for CVotes-related columns
x= pd.pivot_table(genre_top10, index='genre', values=['CVotesU18M', 'CVotes1829M', 'CVotes3044M', 'CVotes45AM'])
y= pd.pivot_table(genre_top10, index='genre', values=['CVotesU18F', 'CVotes1829F', 'CVotes3044F', 'CVotes45AF'])
fig, ax =plt.subplots(1,2 ,figsize=(18, 6))
sns.heatmap(x, cmap='Greens', annot=True, ax=ax[0])
sns.heatmap(y, cmap='Greens', annot=True, ax=ax[1])
plt.show()
# -
# **`Inferences:`** A few inferences that can be seen from the heatmap above is that males have voted more than females, and Sci-Fi appears to be most popular among the 18-29 age group irrespective of their gender. What more can you infer from the two heatmaps that you have plotted? Write your three inferences/observations below:
# - Inference 1: Thriller is least voted in male votes category under age 18.
# - Inference 2: Crime is least voted in female votes category under age18.
# - Inference 3: Most of the votes are provided by the age group of 18-29 in their respective male, female categories.
# +
# 2nd set of heat maps for Votes-related columns
x= pd.pivot_table(genre_top10, index='genre', values=['VotesU18M', 'Votes1829M', 'Votes3044M', 'Votes45AM'])
y= pd.pivot_table(genre_top10, index='genre', values=['VotesU18F', 'Votes1829F', 'Votes3044F', 'Votes45AF'])
fig, ax =plt.subplots(1,2 ,figsize=(18,6))
sns.heatmap(x, cmap='Greens', annot=True, ax=ax[0])
sns.heatmap(y, cmap='Greens', annot=True, ax=ax[1])
plt.show()
# -
# **`Inferences:`** Sci-Fi appears to be the highest rated genre in the age group of U18 for both males and females. Also, females in this age group have rated it a bit higher than the males in the same age group. What more can you infer from the two heatmaps that you have plotted? Write your three inferences/observations below:
# - Inference 1: Romance appears to be the lowest rated genre in the age group of 45A for both males and females.
# - Inference 2: There is almost same vote among all the genre in Male category of 18-29 age group.
# - Inference 3: Most of the ratings are provided by the age group of U18 in their male, female categories.
# - ### Subtask 3.4: US vs non-US Cross Analysis
#
# The dataset contains both the US and non-US movies. Let's analyse how both the US and the non-US voters have responded to the US and the non-US movies.
#
# 1. Create a column `IFUS` in the dataframe `movies`. The column `IFUS` should contain the value "USA" if the `Country` of the movie is "USA". For all other countries other than the USA, `IFUS` should contain the value `non-USA`.
#
#
# 2. Now make a boxplot that shows how the number of votes from the US people i.e. `CVotesUS` is varying for the US and non-US movies. Make use of the column `IFUS` to make this plot. Similarly, make another subplot that shows how non US voters have voted for the US and non-US movies by plotting `CVotesnUS` for both the US and non-US movies. Write any of your two inferences/observations from these plots.
#
#
# 3. Again do a similar analysis but with the ratings. Make a boxplot that shows how the ratings from the US people i.e. `VotesUS` is varying for the US and non-US movies. Similarly, make another subplot that shows how `VotesnUS` is varying for the US and non-US movies. Write any of your two inferences/observations from these plots.
#
# Note : Use `movies` dataframe for this subtask. Make use of this documention to format your boxplot - https://seaborn.pydata.org/generated/seaborn.boxplot.html
# Creating IFUS column
movies['IFUS']= movies['Country'].apply(lambda x: x if x=='USA' else 'non-USA')
# Box plot - 1: CVotesUS(y) vs IFUS(x)
fig, ax =plt.subplots(1,2 ,figsize=(20, 8))
sns.boxplot(movies['IFUS'], movies['CVotesUS'], ax=ax[0])
sns.boxplot(movies['IFUS'], movies['CVotesnUS'], ax=ax[1])
plt.show()
# **`Inferences:`** Write your two inferences/observations below:
# - Inference 1: Median value in CVotesUS for USA movies lies around 5000 whereas for non-USA median lies <5000.
# - Inference 2: In CVotenUS, median value for for USA and non-USA is almost equal.
# Box plot - 2: VotesUS(y) vs IFUS(x)
fig, ax =plt.subplots(1,2 ,figsize=(20, 8))
sns.boxplot(movies['IFUS'], movies['VotesUS'], ax=ax[0])
sns.boxplot(movies['IFUS'], movies['VotesnUS'], ax=ax[1])
plt.show()
# **`Inferences:`** Write your two inferences/observations below:
# - Inference 1: Median votes in VotesUS for USA movies lies around 8.0 whereas for non-USA median lies around 7.9.
# - Inference 2: In VotenUS, median value for for USA is around 7.8 whereas for non-USA it is almost 7.5.
# - ### Subtask 3.5: Top 1000 Voters Vs Genres
#
# You might have also observed the column `CVotes1000`. This column represents the top 1000 voters on IMDb and gives the count for the number of these voters who have voted for a particular movie. Let's see how these top 1000 voters have voted across the genres.
#
# 1. Sort the dataframe genre_top10 based on the value of `CVotes1000`in a descending order.
#
# 2. Make a seaborn barplot for `genre` vs `CVotes1000`.
#
# 3. Write your inferences. You can also try to relate it with the heatmaps you did in the previous subtasks.
#
#
#
# Sorting by CVotes1000
genre_top10=genre_top10.sort_values('CVotes1000', ascending=False)
genre_top10[['genre','CVotes1000']]
# Bar plot
plt.figure(figsize=[10,5])
sns.barplot(data=genre_top10, x='genre', y='CVotes1000')
plt.show()
# **`Inferences:`** It infers that top 1000 voters have voted mostly for Sci-Fi movies.
# **`Checkpoint 6:`** The genre `Romance` seems to be most unpopular among the top 1000 voters.
# With the above subtask, your assignment is over. In your free time, do explore the dataset further on your own and see what kind of other insights you can get across various other columns.
| IMDb+Movie+Assignment.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Détecteur d'ours dangereux
# Si un ours vient frapper à votre porte, utilisez d'abord ce détecteur d'ours dangereux !
#
# Il est entrainé à distinguer les grizzlis, ours noirs et ours en peluche avec un taux de succès de 97%.
# +
from fastai2.vision.all import *
from fastai2.vision.widgets import *
learn_inf = load_learner(Path()/'bears.pkl')
btn_upload = widgets.FileUpload()
out_pl = widgets.Output()
lbl_pred = widgets.Label()
btn_run = widgets.Button(description='Classify')
def on_click_classify(change):
img = PILImage.create(btn_upload.data[-1])
out_pl.clear_output()
with out_pl: display(img.to_thumb(128,128))
pred,pred_idx,probs = learn_inf.predict(img)
lbl_pred.value = f'Prédiction: {pred}; Probabilité: {probs[pred_idx]:.04f}'
btn_run.on_click(on_click_classify)
VBox([widgets.Label("Sélectionnez une image d'ours :"),
btn_upload, btn_run, out_pl, lbl_pred])
# -
# Licence GPL v3, copyright <NAME> et <NAME> 2020.
| notebooks/bearsUI.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="Xlcokkb0bnYn" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="0f6b04d0-c1e7-466b-92f9-5bf5f32062c0" executionInfo={"status": "ok", "timestamp": 1581461201967, "user_tz": -60, "elapsed": 823, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "00941150960329998400"}}
print("Hello Github")
| HelloGithub.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ___
#
# <a href='http://www.pieriandata.com'> <img src='../Pierian_Data_Logo.png' /></a>
# ___
# # Choropleth Maps
# ## Offline Plotly Usage
# Get imports and set everything up to be working offline.
import plotly.plotly as py
import plotly.graph_objs as go
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
# Now set up everything so that the figures show up in the notebook:
init_notebook_mode(connected=True)
# More info on other options for Offline Plotly usage can be found [here](https://plot.ly/python/offline/).
# ## Choropleth US Maps
#
# Plotly's mapping can be a bit hard to get used to at first, remember to reference the cheat sheet in the data visualization folder, or [find it online here](https://images.plot.ly/plotly-documentation/images/python_cheat_sheet.pdf).
import pandas as pd
# Now we need to begin to build our data dictionary. Easiest way to do this is to use the **dict()** function of the general form:
#
# * type = 'choropleth',
# * locations = list of states
# * locationmode = 'USA-states'
# * colorscale=
#
# Either a predefined string:
#
# 'pairs' | 'Greys' | 'Greens' | 'Bluered' | 'Hot' | 'Picnic' | 'Portland' | 'Jet' | 'RdBu' | 'Blackbody' | 'Earth' | 'Electric' | 'YIOrRd' | 'YIGnBu'
#
# or create a [custom colorscale](https://plot.ly/python/heatmap-and-contour-colorscales/)
#
# * text= list or array of text to display per point
# * z= array of values on z axis (color of state)
# * colorbar = {'title':'Colorbar Title'})
#
# Here is a simple example:
data = dict(type = 'choropleth',
locations = ['AZ','CA','NY'],
locationmode = 'USA-states',
colorscale= 'Portland',
text= ['text1','text2','text3'],
z=[1.0,2.0,3.0],
colorbar = {'title':'Colorbar Title'})
# Then we create the layout nested dictionary:
layout = dict(geo = {'scope':'usa'})
# Then we use:
#
# go.Figure(data = [data],layout = layout)
#
# to set up the object that finally gets passed into iplot()
choromap = go.Figure(data = [data],layout = layout)
iplot(choromap)
# ### Real Data US Map Choropleth
#
# Now let's show an example with some real data as well as some other options we can add to the dictionaries in data and layout.
df = pd.read_csv('2011_US_AGRI_Exports')
df.head()
# Now out data dictionary with some extra marker and colorbar arguments:
data = dict(type='choropleth',
colorscale = 'YIOrRd',
locations = df['code'],
z = df['total exports'],
locationmode = 'USA-states',
text = df['text'],
marker = dict(line = dict(color = 'rgb(255,255,255)',width = 2)),
colorbar = {'title':"Millions USD"}
)
# And our layout dictionary with some more arguments:
layout = dict(title = '2011 US Agriculture Exports by State',
geo = dict(scope='usa',
showlakes = True,
lakecolor = 'rgb(85,173,240)')
)
choromap = go.Figure(data = [data],layout = layout)
iplot(choromap)
# # World Choropleth Map
#
# Now let's see an example with a World Map:
df = pd.read_csv('2014_World_GDP')
df.head()
data = dict(
type = 'choropleth',
locations = df['CODE'],
z = df['GDP (BILLIONS)'],
text = df['COUNTRY'],
colorbar = {'title' : 'GDP Billions US'},
)
layout = dict(
title = '2014 Global GDP',
geo = dict(
showframe = False,
projection = {'type':'Mercator'}
)
)
choromap = go.Figure(data = [data],layout = layout)
iplot(choromap)
# # Great Job!
| NASA Meteorites/.ipynb_checkpoints/01-Choropleth Maps-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Tutorial 02: Exchange energy term
#
# > Interactive online tutorial:
# > [](https://mybinder.org/v2/gh/ubermag/oommfc/master?filepath=docs%2Fipynb%2Findex.ipynb)
#
# Exchange energy density is computed as
#
# $$w_\text{e} = A(\nabla\mathbf{m})^{2}$$
#
# where $\mathbf{m}$ is the normalised ($|\mathbf{m}|=1$) magnetisation, and $A$ is the exchange energy constant. Exchange energy term tends to align all magnetic moments parallel to each other. Direction in which they are going to point is not defined via exchange energy.
#
# In `oommfc`, $\mathbf{m}$ is a part of the magnetisation field `system.m`. Therefore, only exchange energy constant $A$ should be provided as an input parameter to uniquely define the Exchange energy term. $A$ can be constant in space or spatially varying.
#
# ## Spatially constant $A$
#
# Let us start by assembling a simple simple simulation where $A$ does not vary in space. The sample is a "one-dimensional" chain of magnetic moments.
# +
import oommfc as oc
import discretisedfield as df
import micromagneticmodel as mm
p1 = (-10e-9, 0, 0)
p2 = (10e-9, 1e-9, 1e-9)
cell = (1e-9, 1e-9, 1e-9)
region = df.Region(p1=p1, p2=p2)
mesh = df.Mesh(region=region, cell=cell)
# -
# The system has a Hamiltonian, which consists of only exchange energy term.
A = 1e-12 # exchange energy constant (J/m)
system = mm.System(name='exchange_constant_A')
system.energy = mm.Exchange(A=A)
# We are going to minimise the system's energy using `oommfc.MinDriver` later. Therefore, we do not have to define the system's dynamics equation. Finally, we need to define the system's magnetisation (`system.m`). We are going to make it random with $M_\text{s}=8\times10^{5} \,\text{Am}^{-1}$
# +
import random
import discretisedfield as df
Ms = 8e5 # saturation magnetisation (A/m)
def m_fun(pos):
return [2*random.random()-1 for i in range(3)]
system.m = df.Field(mesh, dim=3, value=m_fun, norm=Ms)
# -
# The magnetisation, we set is
system.m.k3d_vector(color_field=system.m.z)
# Now, we can minimise the system's energy by using `oommfc.MinDriver`.
md = oc.MinDriver()
md.drive(system)
# We expect that now all magnetic moments are aligned parallel to each other.
system.m.k3d_vector(color_field=system.m.z)
# Finally, we can delete the files created by `oommfc`.
# ## Spatially varying $A$
#
# There are two different ways how a parameter can be made spatially varying, by using:
# 1. Dictionary
# 2. `discretisedfield.Field`
#
# ### Dictionary
#
# In order to define a parameter using a dictionary, regions must be defined in the mesh. Regions are defined as a dictionary, whose keys are the strings and values are `discretisedfield.Region` objects, which take two corner points of the region as input parameters.
p1 = (-10e-9, 0, 0)
p2 = (10e-9, 1e-9, 1e-9)
cell = (1e-9, 1e-9, 1e-9)
subregions = {'region1': df.Region(p1=(-10e-9, 0, 0), p2=(0, 1e-9, 1e-9)),
'region2': df.Region(p1=(0, 0, 0), p2=(10e-9, 1e-9, 1e-9))}
region = df.Region(p1=p1, p2=p2)
mesh = df.Mesh(region=region, cell=cell, subregions=subregions)
# The regions we defined are:
mesh.k3d_subregions()
# Let us say there is no exchange energy ($A=0$) in region 1, whereas in region 2 $A=10^{-12} \,\text{Jm}^{-1}$. Unlike Zeeman and anisotropy energy terms, exchange energy constant is defined between cells. Therefore, it is necessary also to define the value of $A$ between the two regions. This is achieved by adding another item to the dictionary with key `'region1:region2'`. `A` is now defined as a dictionary:
A = {'region1': 0, 'region2': 1e-12, 'region1:region2': 0.5e-12}
# The system object is
system = mm.System(name='exchange_dict_A')
system.energy = mm.Exchange(A=A)
system.m = df.Field(mesh, dim=3, value=m_fun, norm=Ms)
# Its magnetisation is
system.m.k3d_vector(color_field=system.m.z)
# After we minimise the energy
md.drive(system)
# The magnetisation is as we expected. The magnetisation remains random in region 1, and it is alligned in region 2.
system.m.k3d_vector(color_field=system.m.z)
# ### `discretisedfield.Field`
#
# Let us now define the exchange energy in the similar way as in the previous example, but this time using `discretisedfield.Field`:
#
# $\mathbf{A}(x, y, z) = \left\{
# \begin{array}{ll}
# 10^{-12} & x \le 0 \\
# 10^{-20} & x > 0 \\
# \end{array}
# \right. $
#
# This time, it is not possible to define the exchange energy constant between cells, but only in cells. Therefore, the exchange energy constant is then computed as the average between two discretisation cells.
def A_fun(pos):
x, y, z = pos
if x <= 0:
return 1e-12
else:
return 1e-20
# The exchange energy constant is
A = df.Field(mesh, dim=1, value=A_fun)
# The system is
system = mm.System(name='exchange_field_A')
system.energy = mm.Exchange(A=A)
system.m = df.Field(mesh, dim=3, value=m_fun, norm=Ms)
# and its magnetisation is
system.m.k3d_vector(color_field=system.m.z)
# After the energy minimisation, the magnetisation is:
md.drive(system)
system.m.k3d_vector(color_field=system.m.z)
| docs/ipynb/02-tutorial-exchange.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Methodology
# ## Sampling Procedure
#
# - Mark corner of site : position from landmark
# - Collect temperature and weather readings
# - Tracking, capture, mark, and mark position of trail.
# - Read: size, number of prey and type, size of prey
# - Benthic samples: position from corner of site marker.
# - Record: sand, ripple, water, slope
# - Final temperature
# - Other experimental: untracked snails / marking
# ## Habitat Quality Rankings
#
# ### Sand Quality
#
# 0 fine mud
# 1 fine sandy mud
# 2 muddy fine sand
# 3 clean fine sand
# 4 medium grain sand
# 5 coarse grain sand
#
# ### Ripple Quality
#
# 0 flat
# 1 very slight occasional ripples
# 2 regular slight ripples <= 0.25 cms
# 3 medium ripples <= 0.5 cms
# 4 high ripples <= 1.5 cms
# 5 gigantic ripples > 1.5 cms
#
# ### Water Quality
#
# 0 dry
# 1 moist
# 2 thin surface layer
# 3 pooled water
# 4 surface runoff
#
# ### Slope
#
# 0 flat
# 1 <= 2 degrees
# 2 <= 5 degrees
# 3 <= 10 degrees
# 4 > 10 degrees
#
# ### Track Events
# A+ definite attack prey seen to escape
# A possible attack prey seen close by
# A? possible attack prey not seen
# PC prey capture
# PIT prey in tow
# FOT following old trail (pred sp)
# OTE old trail encountred
# ## Predator Attack on Prey
#
# The experimental procedure to determine the effect of predator attack on prey item.
#
# One capturing a prey item the predator will remain montionless for a period ranging from 15 seconds to 2 minutes. This behaviour raises the question of what is the predator doing. Possible reasons are:
# - Evaluation of prey quality i.e. recognition that captured item is a suitable item, or size of item
# - The predator is immobilising the item in some manner
#
# The following hypothesis may be tested:
#
# - H1: prey evaluation time is independent of prey size or predator size
# - H2: prey are not immobilised in some manner
#
# ### Technique
# 1. Collect prey items and sort into three size groups
# 2. Split eac group into three sub-groups:
# - SGroup 1: control handled
# - SGroup 2: captured and cleaned
# - SGroup 3: captured and uncleaned
# 3. Determine recovery time for each group
#
#
# ## Predator Prey Detection
#
# ### Predator
# - Probability of detection of prey that has not moved
# - Probability of detection of prey that has initiated escape given distance from predator
# - Subsequent reaction by predator given a prey detection (dependent on detection strength??)
#
# ### Prey
# - Probability that prey detects approach of predator and initiates movement
# - Distance at which movement is initiated
#
# ### Outcome events
# Summation of above events will lead to the following outcome events:
# - Prey not detected:
# a) no movement
# b) movement by prey
# - Prey detected and captured: a) b) as above
# - Prey detected and not captured - given escape by prey:
# a) without attack and ARS
# b) with attack and ARS
#
# ### Data available from tracks
# - Frequency of capture without prey movement
# - Distances at which prey initiate escape behaviour and directions and distances moved
# - Given a movement by prey and distance from predator whether this caused change in track parameters
# - Frequency of A1 -> A3, P and conditions, and subsequent bahaviour of the predator
# ## Track Analysis
#
# If absolute values of turns cover a range less than 180 degrees the data
# does not have to be considered circular (Batschelet, 1965).
#
# ### Mean Linear Displacement
#
# [1] Linear displacement per step for each individual averaged across
# similar trials to calculate mean linear displacement. Compared with
# ANOVA using Tukey post hoc comparisions (Sokal and Rohlf, 1981),
# performed at selected steps (5, 10, 15, 20). Not performed among steps
# due to high degree of autocorrelation.
#
# [4] Compared beline distances of before and after capture
#
# ### Mean Step Length
#
# [1] Determined an average step length for each individual and averaged
# values for each experimental trial. Step lengths compared with ANOVA
# using Tukey post hoc comparisions (Sokal and Rohlf, 1981)
#
#
# ### Throughness
#
# [1] Sensitive to change in linear displacement and may be affected by
# exceptionally long steps. Measured for a large number of steps (20).
# Provide a good relative comparason of average search efficiency among
# trials. Indices compared with z-test (Sokal and Rohlf, 1981)
#
# ### Straightness
#
# [1] Indices compared with z-test (Sokal and Rohlf, 1981)
#
# ### Turning Angle Frequency Distribution
#
# [1] Comparisons between empirical distributions used Kolmogorov-Smirnov
# two saple test for large unequal samples sizes (Batschelet, 1981; Sokal
# and Rohlf, 1981). Comparisons between empirical and theoretical
# distributions done using Kolmogorov-Smirnov goodnes of fit test for
# intrinsic hypotheses (Sokal and Rohlf, 1981)
#
# ### Turning Direction
#
# [1] Assigned 1 to right and -1 to left and analysed with runs test
# (Brown and Downhower 1988)
#
# [3] Number of turns approx equal, showed no obvious patten.
#
# [4] Data for each sample and pooled data arranged in 2 x 2 contingency
# tables in couplets of successive turns (nth vs (n+1)th direction) right
# right, right left, left right, left left), and G (log likeihood ratio
# statistic calculated.
#
# ### Rate of Change of Direction
#
# [3] Change in direction in degrees per minute. Basal level of 300
# increasing to 1300 after feeding. Pooled results of replicates and
# plotted mean of first, second ... values of r.c.d. Return to basal level
# took about 5 minutes. Hassel & Murdie, 1973 found 30 seconds with
# houseflies. Banks, 1957 found coccinellids returned within 1 minute.
#
# ### Meander Ratio
#
# [4] Length of search path : beeline distance moved. Comparisons between
# trials used Wilcoxon's two-sample test (one-tailed test)
#
# ### References
#
# 1. Winkelman, D.L. et. al., 1991
# 2. Kareiva and Shigesada, 1983
# 3. Evans
# 4. <NAME> Falls, 1976
#
| src/ipython/15 Methodology.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# ---
# %config Application.log_level='WORKAROUND'
import logging
logging.getLogger('workspace').setLevel(logging.DEBUG)
import os
os.chdir('python/tests/azure/msi')
# +
import mlbriefcase
ws = mlbriefcase.Workspace()
# -
ws['workspacetest1'].get_url()
# +
from msrestazure.azure_active_directory import MSIAuthentication
msi_auth = MSIAuthentication('https://management.core.windows.net/')
msi_auth.set_token()
| tests/azure/msi/notebook.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Map extraction
#
# This notebook shows how we extract polygons from raw maps (gifs or pdf). We do it for a single instance and we automate this process with a script for all the directories.
# +
from pylab import contour
import matplotlib.pyplot as plt
from PIL import ImageFilter, Image, ImageDraw
from datetime import date, timedelta
import numpy as np
from PIL import Image
import cv2
from skimage import measure
import os
import pandas as pd
from scipy.spatial import distance
# %matplotlib inline
# -
# ## Extract contours
# +
def dates_between(start, stop):
while start <= stop:
yield start
start += timedelta(days=1)
def hydrological_year(at_date):
if at_date.month > 6:
return at_date.year + 1
else:
return at_date.year
# colors definitions RGB alpha
black = np.array([0, 0, 0])
white = np.array([255, 255, 255])
green = np.array([204, 255, 102])
yellow = np.array([255, 255, 0])
orange = np.array([255, 153, 0])
red = np.array([255, 0, 0])
raw_red = np.array([255, 0, 0])
raw_green = np.array([0, 255, 0])
raw_blue = np.array([0, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_pink = np.array([255, 0, 255])
raw_cyan = np.array([0, 255, 255])
raw_yellow = np.array([255, 255, 0])
# +
def keep_colors(img, colors, replace_with=white):
"""return a new image with only the `colors` selected, other pixel are `replace_with`"""
keep = np.zeros(img.shape[:2], dtype=bool)
for c in colors:
keep = keep | (c == img).all(axis=-1)
new_img = img.copy()
new_img[~keep] = replace_with
return new_img
def numpify(o):
if not isinstance(o, np.ndarray):
o = np.array(o)
return o
# remove contours areas that have more than 30% of white
WHITE_RATIO_THRESHOLD = .3
def color_contours(img, color):
img = numpify(img)
color = numpify(color)
mask = (img == color[:3]).all(axis=-1)
monocholor = img.copy()
monocholor[~mask] = 255
contours = measure.find_contours(mask, 0.5)
# heuristic filter for contours
filter_contours = []
for c in contours:
region = Image.new("L", [img.shape[1], img.shape[0]], 0)
ImageDraw.Draw(region).polygon(list(map(lambda t: (t[1],t[0]), c)), fill=1)
region = np.array(region).astype(bool)
white_ratio = (monocholor == 255).all(axis=-1)[region].mean()
if white_ratio <= WHITE_RATIO_THRESHOLD:
filter_contours.append(c)
return filter_contours
# +
def replace_color(img, color_map):
"""return a new image replacing the image colors which will be mapped to their corresponding colors in `color_map` (df)"""
new_img = img.copy()
for _, (source, target) in color_map.iterrows():
new_img[(img == source).all(axis=-1)] = target
return new_img
def build_color_map(img_arr, image_shades):
"""return colormap as dataframe"""
im_df = pd.DataFrame([img_arr[i,j,:] for i,j in np.ndindex(img_arr.shape[0],img_arr.shape[1])])
im_df = im_df.drop_duplicates()
image_colors = im_df.as_matrix()
colors = np.zeros(image_colors.shape)
dist = distance.cdist(image_colors, image_shades, 'sqeuclidean')
for j in range(dist.shape[0]):
distances = dist[j,:]
colors[j, :] = image_shades[distances.argmin()]
color_map = pd.DataFrame(
{'source': image_colors.tolist(),
'target': colors.tolist()
})
return color_map
# -
def remove_grey(img):
mask = np.std(img, axis=-1) < 10
new_img = img.copy()
new_img[mask] = 255
return new_img
# +
# path = '../data/slf/2003/nbk/de/gif/20021230_nbk_de_c.gif'
# path = '../data/slf/2001/nbk/de/gif/20001230_nbk_de_c.gif'
# path = '../data/slf/2001/nbk/de/gif/20010307_nbk_de_c.gif'
# path = '../data/slf/2002/nbk/de/gif/20011122_nbk_de_c.gif'
path = '../data/slf/2002/nbk/de/gif/20020308_nbk_de_c.gif'
img = Image.open(path)
img = img.convert('RGB')
img_arr = np.array(img)
plt.imshow(img_arr)
# -
img_no_gray = remove_grey(img_arr)
plt.imshow(img_no_gray)
# img_no_gray
color_scale = [white, green, yellow, orange, red]
color_map = build_color_map(img_no_gray, color_scale)
img_projected = replace_color(img_no_gray, color_map)
plt.imshow(img_projected)
# +
# load mask of this size
leman_west = (6.148131, 46.206042)
quatre_canton_north = (8.435177, 47.082150)
majeur_east = (8.856851, 46.151857)
east_end = (10.472221, 46.544303)
constance_nw = (9.035247, 47.812716)
landmarks_colors = {
leman_west: raw_red,
quatre_canton_north: raw_green,
majeur_east: raw_blue,
constance_nw: raw_pink,
east_end: raw_yellow,
}
def coord_color(img, color):
return np.array(list(zip(*(img == color).all(-1).nonzero())))
def open_mask(height, width):
masks_path = '../map-masks/'
mask_name = '{}x{}.gif'.format(height, width)
mask_path = os.path.join(masks_path, mask_name)
mask = Image.open(mask_path)
mask = mask.convert('RGB')
mask = np.array(mask)
landmarks_pix = {
geo_point: (width, height)
for geo_point, color in landmarks_colors.items()
for height, width in coord_color(mask, color)
}
binary_mask = (mask != 255).any(-1) # different of white
return binary_mask, landmarks_pix
binary_mask, landmarks_pix = open_mask(*img_arr.shape[:2])
fix, ax = plt.subplots()
ax.imshow(binary_mask);
widths, heights = list(zip(*landmarks_pix.values()))
ax.scatter(widths, heights);
ax.set_title('Mask informations');
# +
fig, axes = plt.subplots(2, 2, figsize=(14,10))
img_arr = img_projected
# original image
axes[0][0].imshow(img_arr);
axes[0][0].set_title('Original image');
# keep useful colors
regions_only = keep_colors(img_arr, [green, yellow, orange, red])
axes[0][1].imshow(regions_only);
axes[0][1].set_title('Keep only danger colors');
# clip the binary mask to remove color key
regions_only[~binary_mask] = 255
regions_only = Image.fromarray(regions_only).convert('RGB')
smoothed = regions_only.filter(ImageFilter.MedianFilter(7))
axes[1][0].imshow(smoothed);
axes[1][0].set_title('Smoothed with median filter');
# extract contours
axes[1][1].set_xlim([0, img_arr.shape[1]])
axes[1][1].set_ylim([0, img_arr.shape[0]])
axes[1][1].invert_yaxis()
axes[1][1].set_title('Regions contours')
for color in [green, yellow, orange, red]:
contours = color_contours(smoothed, color)
for contour in contours:
axes[1][1].plot(contour[:, 1], contour[:, 0], linewidth=2, c=[x / 255 for x in color])
# -
# ## Contours to map polygon
#
# Once we have contours we want to transform it into geographic coordinates and simplify the polygons.
#
# To do this transformation, we use 5 points on the map to learn a transformation matrix $T$ that maps a pixel of the image to a geolocation. We could use only 3 points to have a valid transformation, but to dicrease the error we use 5 and solve a leastquare problem.
#
# | Location | Color |
# | -------- | ----- |
# | <NAME> | red |
# | Quatre canton N | green |
# | Lac majeur E | blue |
# | Lac Constance NW | pink |
# | Swiss E | yellow |
landmarks_pix
# +
pix = np.array(list(map(numpify, landmarks_pix.values())))
coord = np.array(list(map(numpify, landmarks_pix.keys())))
# add 1 bias raw
pix_ext = np.vstack([np.ones((1,pix.shape[0])), pix.T])
coord_ext = np.vstack([np.ones((1,pix.shape[0])), coord.T])
# T = np.linalg.solve(
T = np.linalg.lstsq(pix_ext.T, coord_ext.T)[0]
def transform_pix2map(points):
"""n x 2 array"""
points_ext = np.hstack([np.ones((points.shape[0], 1)), points])
points_map = points_ext.dot(T)
return points_map[:, 1:]
# -
# We smooth the polygon to remove some useless points and make thme a bit lighter.
# +
import json
import visvalingamwyatt as vw
SMOOTHING_THRESHOLD = 0.0001
geo_json = {
"type": "FeatureCollection",
"features": []
}
for danger_level, color in enumerate([green, yellow, orange, red]):
for contour in color_contours(smoothed, color):
contour_right = contour.copy()
contour_right[:,0] = contour[:,1]
contour_right[:,1] = contour[:,0]
contour_right = transform_pix2map(contour_right)
simplifier = vw.Simplifier(contour_right)
contour_right = simplifier.simplify(threshold=SMOOTHING_THRESHOLD)
geo_json['features'].append({
"type": "Feature",
"properties": {
"date": "TODO",
"danger_level": danger_level + 1
},
"geometry": {
"type": "Polygon",
"coordinates": [ list(reversed(contour_right.tolist())) ]
}
})
# +
import folium
switzerland = (46.875893, 8.289321)
tiles = 'https://server.arcgisonline.com/ArcGIS/rest/services/World_Topo_Map/MapServer/tile/{z}/{y}/{x}'
attr = 'Tiles © Esri — Esri, DeLorme, NAVTEQ, TomTom, Intermap, iPC, USGS, FAO, NPS, NRCAN, GeoBase, Kadaster NL, Ordnance Survey, Esri Japan, METI, Esri China (Hong Kong), and the GIS User Community'
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
colors = ['#ccff66', '#ffff00', '#ff9900', '#ff0000']
def style_function(risk_region):
level = risk_region['properties']['danger_level']
color = colors[level - 1]
return {
'fillOpacity': .5,
'weight': 0,
'fillColor': color,
'color': 'white',
}
folium.GeoJson(
geo_json,
name='geojson',
style_function=style_function
).add_to(m)
m
# -
# Use `src/map_extractor.py` script to extract in batch GeoJSON information from the gifs. Then you can visualize some of them here.
# +
m = folium.Map(location=switzerland, zoom_start=8, tiles=tiles, attr=attr)
folium.GeoJson(
json.load(open(r'../json-maps/20001230_nbk_de_c.json')),
name='geojson',
style_function=style_function
).add_to(m)
m
# -
| notebooks/map_extraction.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import numpy as np
from tqdm import tqdm
import matplotlib.pyplot as plt
from scipy.signal import medfilt
import pickle
import glob
# ### Functions
# +
def read_annotation(annotation_path):
lines = []
with open(annotation_path) as f:
lines.append(f.read().splitlines() )
f.close()
#lines = np.sort(lines)
lines = np.hstack(lines)
return lines
def generate_train_test_list(lists):
train_list = []
test_list = []
for i in range(len(lists)):
lines = read_annotation(lists[i])
for line in lines:
file_name, flag = line.split(' ')
if flag == '1':
train_list.append(file_name.split('.')[0])
elif flag == '2':
test_list.append(file_name.split('.')[0])
return train_list,test_list
import scipy.io
def read_pose(path):
mat = scipy.io.loadmat(path)
poses = np.round(mat['pos_world'],3).swapaxes(0,2)
return poses
def generate_pose_label(pose_list,train_list,test_list):
train = {}
train['pose'] = []
train['label'] = []
test = {}
test['pose'] = []
test['label'] = []
for i in range(len(pose_list)):
label = pose_list[i].split('/')[-2]
pose_path = pose_list[i]+'/joint_positions.mat'
pose = read_pose(pose_path)
file = pose_list[i].split('/')[-1]
if file in train_list:
train['label'].append(label)
train['pose'].append(pose)
elif file in test_list:
test['label'].append(label)
test['pose'].append(pose)
return train,test
# -
# ### Config settings
class Config():
def __init__(self):
self.frame_l = 32 # the length of frames
self.joint_n = 15 # the number of joints
self.joint_d = 2 # the dimension of classes
self.data_dir = '../jhmdb_data_preprocessing/'#'/mnt/nasbi/no-backups/datasets/pose_action/JHMDB/'
self.save_dir = '../data/JHMDB/'#'/mnt/nasbi/homes/fan/projects/action/skeleton/data/JHMDB/'
C = Config()
# ### There are 3 ways of splitting the ground-truth pose data
GT_split_lists = glob.glob(C.data_dir + 'GT_splits/*.txt')
print()
GT_pose_list = glob.glob(C.data_dir + 'GT_joint_positions/*/*')
GT_lists_1 = []
GT_lists_2 = []
GT_lists_3 = []
for file in GT_split_lists:
if file.split('/')[-1].split('.')[0].split('_')[-1] == 'split1':
GT_lists_1.append(file)
elif file.split('/')[-1].split('.')[0].split('_')[-1] == 'split2':
GT_lists_2.append(file)
elif file.split('/')[-1].split('.')[0].split('_')[-1] == 'split3':
GT_lists_3.append(file)
# ## Processing each spliting method
# +
GT_train_list_1,GT_test_list_1 = generate_train_test_list(GT_lists_1)
GT_train_1,GT_test_1 = generate_pose_label(GT_pose_list,GT_train_list_1,GT_test_list_1)
print(GT_train_1)
pickle.dump(GT_train_1, open(C.save_dir+"GT_train_1.pkl", "wb"))
pickle.dump(GT_test_1, open(C.save_dir+"GT_test_1.pkl", "wb"))
# +
GT_train_list_2,GT_test_list_2 = generate_train_test_list(GT_lists_2)
GT_train_2,GT_test_2 = generate_pose_label(GT_pose_list,GT_train_list_2,GT_test_list_2)
pickle.dump(GT_train_2, open(C.save_dir+"GT_train_2.pkl", "wb"))
pickle.dump(GT_test_2, open(C.save_dir+"GT_test_2.pkl", "wb"))
# +
GT_train_list_3,GT_test_list_3 = generate_train_test_list(GT_lists_3)
GT_train_3,GT_test_3 = generate_pose_label(GT_pose_list,GT_train_list_3,GT_test_list_3)
pickle.dump(GT_train_3, open(C.save_dir+"GT_train_3.pkl", "wb"))
pickle.dump(GT_test_3, open(C.save_dir+"GT_test_3.pkl", "wb"))
# -
| JHMDB/jhmdb_data_preprocessing.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] id="AmONFY6K2e6E"
# # Plot Saliency Map of the Inputs for Pretrained Models
# + id="c_xgxu_v2rRR"
from google.colab import drive
drive.mount('/content/drive')
# + id="l-82ETg23PLk" executionInfo={"status": "ok", "timestamp": 1613376320189, "user_tz": -210, "elapsed": 24536, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}}
import os
os.chdir('drive/My Drive/Pytorch/Dataset/Saliency Map')
# + [markdown] id="65JP9aUn3qmx"
# **import necessary packages**
# + id="AviIJ4va3poJ" executionInfo={"status": "ok", "timestamp": 1613376327044, "user_tz": -210, "elapsed": 4197, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "05364799661830805697"}}
import torch
import torchvision
from torchsummary import summary
import torchvision.transforms as T
import numpy as np
from PIL import Image
import matplotlib.pyplot as plt
import requests
# + [markdown] id="tnQ6On7W4NVg"
# **load pretrained model**
# + id="I_uamRGV4Q1G"
device= 'cuda:0' if torch.cuda.is_available() else 'cpu'
model = torchvision.models.vgg19(pretrained= True)
model.to(device)
# + id="99L-rspG4l4J"
def get(url, fname):
response = requests.get(url)
with open(fname, 'wb') as f :
f.write(response.content)
# + id="64oKcGKO6W3k"
get("https://specials-images.forbesimg.com/imageserve/5db4c7b464b49a0007e9dfac/960x0.jpg?fit=scale", 'dog.jpg')
get("https://s3.amazonaws.com/cdn-origin-etr.akc.org/wp-content/uploads/2019/12/03202400/Yellow-Labrador-Retriever.jpg", 'dog_big.jpg')
# + id="UMMlZ5bu5my8"
Image.open('dog.jpg')
# + id="jz5Qw8YZMiei"
Image.open('dogs_and_cats.jpeg')
# + [markdown] id="ZG2ync8VmzxX"
# **preprocess the input image**
# + id="Ob8DYnlsm3fM" executionInfo={"status": "ok", "timestamp": 1613376468008, "user_tz": -210, "elapsed": 625, "user": {"displayName": "<NAME>", "photoUrl": "", "userId": "05364799661830805697"}}
def preprocess(image, size= 224):
transform = T.Compose([
T.Resize((size, size)),
T.ToTensor(),
T.Normalize(mean = [0.485, 0.456, 0.406]
, std= [0.229, 0.225, 0.224])])
return transform(image)
def deprocess(tensor):
transform = T.Compose([
T.Normalize(mean= [0, 0, 0]
, std = [4.3668, 4.4444, 4.4642]) ,
T.Normalize(mean= [-0.485, -0.456, -0.406]
, std = [1, 1, 1]),
T.ToPILImage()])
return transform(tensor)
def show_image(image_pil, cmap):
plt.imshow(np.array(image_pil), cmap= cmap)
plt.axis('off')
# + id="yyHt7apEogRN"
img = Image.open('dog.jpg')
t = preprocess(img)
# + id="JmmVQUZ3MoPx" executionInfo={"status": "ok", "timestamp": 1613376900939, "user_tz": -210, "elapsed": 2181, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}}
img = Image.open('dog-cat-1.jpg')
t = preprocess(img)
# + id="0DYgkg_gqrub" executionInfo={"status": "ok", "timestamp": 1613376905567, "user_tz": -210, "elapsed": 897, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}}
d = deprocess(t)
# + id="uXRh3ONOquvY"
plt.imshow(np.array(d))
# + [markdown] id="hGyEMtoWrV7u"
# **Get the Saliency Map....**
# + id="1rB0HROxrZXi" executionInfo={"status": "ok", "timestamp": 1613376910793, "user_tz": -210, "elapsed": 876, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}}
X = preprocess(img)
X = X.unsqueeze(0)
model.eval()
X.requires_grad_()
scores = model(X.to(device))
#max_index = torch.argmax(scores)
#max_value = scores[0, max_index]
#max_value.backward()
# + id="zY3E_hQ9vrBi" executionInfo={"status": "ok", "timestamp": 1613376911915, "user_tz": -210, "elapsed": 847, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}}
max_value, max_index = scores.topk(2, largest= False)
max_value[0][1].backward()
# + colab={"base_uri": "https://localhost:8080/"} id="DRFa57HyskXM" executionInfo={"status": "ok", "timestamp": 1613376912760, "user_tz": -210, "elapsed": 665, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}} outputId="ef2780f4-8c4a-40dd-ab63-b01add4b07ff"
val, idx = torch.max(X.grad.abs(), dim= 1)
val.size()
# + colab={"base_uri": "https://localhost:8080/", "height": 323} id="8wbtDzFCtxY4" executionInfo={"status": "ok", "timestamp": 1613376914780, "user_tz": -210, "elapsed": 1997, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}} outputId="6f68ff7c-56e1-49be-d97b-e39a00e8a50a"
fig, axs = plt.subplots(nrows= 1, ncols= 2, figsize= (10,10))
axs[0].imshow(img.resize(size= (224, 224)))
axs[1].imshow(np.array(val[0]), cmap= plt.cm.hot)
# + colab={"base_uri": "https://localhost:8080/", "height": 323} id="X6VIEtoSMs-V" executionInfo={"status": "ok", "timestamp": 1610800619038, "user_tz": -210, "elapsed": 2361, "user": {"displayName": "gpu keshvari", "photoUrl": "", "userId": "05364799661830805697"}} outputId="4a5fd041-a8ef-4e25-a0db-c1b34f5055f6"
fig, axs = plt.subplots(nrows= 1, ncols= 2, figsize= (10,10))
axs[0].imshow(img.resize(size= (224, 224)))
axs[1].imshow(np.array(val[0]), cmap= plt.cm.hot)
| Saliency_Map.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# Copyright 2021 NVIDIA Corporation. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
# -
# <img src="http://developer.download.nvidia.com/compute/machine-learning/frameworks/nvidia_logo.png" style="width: 90px; float: right;">
#
# # BERT Question Answering Inference with Mixed Precision
#
# ## 1. Overview
#
# Bidirectional Embedding Representations from Transformers (BERT), is a method of pre-training language representations which obtains state-of-the-art results on a wide array of Natural Language Processing (NLP) tasks.
#
# The original paper can be found here: https://arxiv.org/abs/1810.04805.
#
# NVIDIA's BERT 19.10 is an optimized version of Google's official implementation, leveraging mixed precision arithmetic and tensor cores on V100 GPUS for faster training times while maintaining target accuracy.
# ### 1.a Learning objectives
#
# This notebook demonstrates:
# - Inference on QA task with BERT Large model
# - The use/download of fine-tuned NVIDIA BERT models
# - Use of Mixed Precision for Inference
# ## 2. Requirements
#
# Please refer to the ReadMe file
# ## 3. BERT Inference: Question Answering
#
# We can run inference on a fine-tuned BERT model for tasks like Question Answering.
#
# Here we use a BERT model fine-tuned on a [SQuaD 2.0 Dataset](https://rajpurkar.github.io/SQuAD-explorer/) which contains 100,000+ question-answer pairs on 500+ articles combined with over 50,000 new, unanswerable questions.
# ### 3.a Paragraph and Queries
#
# In this example we will ask our BERT model questions related to the following paragraph:
#
# **The Apollo Program**
# _"The Apollo program, also known as Project Apollo, was the third United States human spaceflight program carried out by the National Aeronautics and Space Administration (NASA), which accomplished landing the first humans on the Moon from 1969 to 1972. First conceived during Dwight D. Eisenhower's administration as a three-man spacecraft to follow the one-man Project Mercury which put the first Americans in space, Apollo was later dedicated to President <NAME>'s national goal of landing a man on the Moon and returning him safely to the Earth by the end of the 1960s, which he proposed in a May 25, 1961, address to Congress. Project Mercury was followed by the two-man Project Gemini. The first manned flight of Apollo was in 1968. Apollo ran from 1961 to 1972, and was supported by the two-man Gemini program which ran concurrently with it from 1962 to 1966. Gemini missions developed some of the space travel techniques that were necessary for the success of the Apollo missions. Apollo used Saturn family rockets as launch vehicles. Apollo/Saturn vehicles were also used for an Apollo Applications Program, which consisted of Skylab, a space station that supported three manned missions in 1973-74, and the Apollo-Soyuz Test Project, a joint Earth orbit mission with the Soviet Union in 1975."_
#
# The questions and relative answers expected are shown below:
#
# - **Q1:** "What project put the first Americans into space?"
# - **A1:** "Project Mercury"
# - **Q2:** "What program was created to carry out these projects and missions?"
# - **A2:** "The Apollo program"
# - **Q3:** "What year did the first manned Apollo flight occur?"
# - **A3:** "1968"
# - **Q4:** "What President is credited with the original notion of putting Americans in space?"
# - **A4:** "<NAME>"
# - **Q5:** "Who did the U.S. collaborate with on an Earth orbit mission in 1975?"
# - **A5:** "Soviet Union"
# - **Q6:** "How long did Project Apollo run?"
# - **A6:** "1961 to 1972"
# - **Q7:** "What program helped develop space travel techniques that Project Apollo used?"
# - **A7:** "Gemini Mission"
# - **Q8:** "What space station supported three manned missions in 1973-1974?"
# - **A8:** "Skylab"
#
# ---
#
# The paragraph and the questions can be easily customized by changing the code below:
#
# ---
# %%writefile input.json
{"data":
[
{"title": "Project Apollo",
"paragraphs": [
{"context":"The Apollo program, also known as Project Apollo, was the third United States human spaceflight program carried out by the National Aeronautics and Space Administration (NASA), which accomplished landing the first humans on the Moon from 1969 to 1972. First conceived during Dwight D. Eisenhower's administration as a three-man spacecraft to follow the one-man Project Mercury which put the first Americans in space, Apollo was later dedicated to President <NAME>'s national goal of landing a man on the Moon and returning him safely to the Earth by the end of the 1960s, which he proposed in a May 25, 1961, address to Congress. Project Mercury was followed by the two-man Project Gemini. The first manned flight of Apollo was in 1968. Apollo ran from 1961 to 1972, and was supported by the two man Gemini program which ran concurrently with it from 1962 to 1966. Gemini missions developed some of the space travel techniques that were necessary for the success of the Apollo missions. Apollo used Saturn family rockets as launch vehicles. Apollo/Saturn vehicles were also used for an Apollo Applications Program, which consisted of Skylab, a space station that supported three manned missions in 1973-74, and the Apollo-Soyuz Test Project, a joint Earth orbit mission with the Soviet Union in 1975.",
"qas": [
{ "question": "What project put the first Americans into space?",
"id": "Q1"
},
{ "question": "What program was created to carry out these projects and missions?",
"id": "Q2"
},
{ "question": "What year did the first manned Apollo flight occur?",
"id": "Q3"
},
{ "question": "What President is credited with the original notion of putting Americans in space?",
"id": "Q4"
},
{ "question": "Who did the U.S. collaborate with on an Earth orbit mission in 1975?",
"id": "Q5"
},
{ "question": "How long did Project Apollo run?",
"id": "Q6"
},
{ "question": "What program helped develop space travel techniques that Project Apollo used?",
"id": "Q7"
},
{"question": "What space station supported three manned missions in 1973-1974?",
"id": "Q8"
}
]}]}]}
# +
import os
import sys
notebooks_dir = '../notebooks'
data_dir = '../data/download'
working_dir = '../'
if working_dir not in sys.path:
sys.path.append(working_dir)
# -
input_file = os.path.join(notebooks_dir, 'input.json')
# ### 3.b Mixed Precision
#
# Mixed precision training offers significant computational speedup by performing operations in half-precision format, while storing minimal information in single-precision to retain as much information as possible in critical parts of the network. Since the introduction of tensor cores in the Volta and Turing architectures, significant training speedups are experienced by switching to mixed precision -- up to 3x overall speedup on the most arithmetically intense model architectures.
#
# For information about:
# - How to train using mixed precision, see the [Mixed Precision Training](https://arxiv.org/abs/1710.03740) paper and [Training With Mixed Precision](https://docs.nvidia.com/deeplearning/sdk/mixed-precision-training/index.html) documentation.
# - How to access and enable AMP for TensorFlow, see [Using TF-AMP](https://docs.nvidia.com/deeplearning/dgx/tensorflow-user-guide/index.html#tfamp) from the TensorFlow User Guide.
# - Techniques used for mixed precision training, see the [Mixed-Precision Training of Deep Neural Networks](https://devblogs.nvidia.com/mixed-precision-training-deep-neural-networks/) blog.
# In this notebook we control mixed precision execution with the environmental variable:
import os
os.environ["TF_ENABLE_AUTO_MIXED_PRECISION"] = "1"
# To effectively evaluate the speedup of mixed precision try a bigger workload by uncommenting the following line:
# +
#input_file = '../data/download/squad/v2.0/dev-v2.0.json'
# -
# ## 4. Fine-Tuned NVIDIA BERT TF Models
#
# Based on the model size, we have the following two default configurations of BERT.
#
# | **Model** | **Hidden layers** | **Hidden unit size** | **Attention heads** | **Feedforward filter size** | **Max sequence length** | **Parameters** |
# |:---------:|:----------:|:----:|:---:|:--------:|:---:|:----:|
# |BERTBASE |12 encoder| 768| 12|4 x 768|512|110M|
# |BERTLARGE|24 encoder|1024| 16|4 x 1024|512|330M|
#
# We will take advantage of the fine-tuned models available on NGC (NVIDIA GPU Cluster, https://ngc.nvidia.com).
# Among the many configurations available we will download these two:
#
# - **bert_tf_ckpt_large_qa_squad2_amp_384**
#
# Which are trained on the SQuaD 2.0 Dataset.
# +
# bert_tf_ckpt_large_qa_squad2_amp_384
DATA_DIR_FT = data_dir + '/finetuned_large_model_SQUAD2.0'
# !mkdir -p $DATA_DIR_FT
# !wget --content-disposition -O $DATA_DIR_FT/bert_tf_ckpt_large_qa_squad2_amp_384_19.03.1.zip \
# https://api.ngc.nvidia.com/v2/models/nvidia/bert_tf_ckpt_large_qa_squad2_amp_384/versions/19.03.1/zip \
# && unzip -n -d $DATA_DIR_FT/ $DATA_DIR_FT/bert_tf_ckpt_large_qa_squad2_amp_384_19.03.1.zip \
# && rm -rf $DATA_DIR_FT/bert_tf_ckpt_large_qa_squad2_amp_384_19.03.1.zip
# -
# In the code that follows we will refer to these models.
# ## 5. Running QA task inference
#
# In order to run QA inference we will follow step-by-step the flow implemented in run_squad.py.
#
# Configuration:
# +
import run_squad
import json
import tensorflow as tf
import modeling
import tokenization
import time
import random
tf.logging.set_verbosity(tf.logging.INFO)
# Create the output directory where all the results are saved.
output_dir = os.path.join(working_dir, 'results')
tf.gfile.MakeDirs(output_dir)
# The config json file corresponding to the pre-trained BERT model.
# This specifies the model architecture.
bert_config_file = os.path.join(data_dir, 'finetuned_large_model_SQUAD2.0/bert_config.json')
# The vocabulary file that the BERT model was trained on.
vocab_file = os.path.join(data_dir, 'finetuned_large_model_SQUAD2.0/vocab.txt')
# Initiate checkpoint to the fine-tuned BERT Large model
init_checkpoint = os.path.join(data_dir, 'finetuned_large_model_SQUAD2.0/model.ckpt')
# Whether to lower case the input text.
# Should be True for uncased models and False for cased models.
do_lower_case = True
# Total batch size for predictions
predict_batch_size = 1
params = dict([('batch_size', predict_batch_size)])
# The maximum total input sequence length after WordPiece tokenization.
# Sequences longer than this will be truncated, and sequences shorter than this will be padded.
max_seq_length = 384
# When splitting up a long document into chunks, how much stride to take between chunks.
doc_stride = 128
# The maximum number of tokens for the question.
# Questions longer than this will be truncated to this length.
max_query_length = 64
# This is a WA to use flags from here:
flags = tf.flags
if 'f' not in tf.flags.FLAGS:
tf.app.flags.DEFINE_string('f', '', 'kernel')
FLAGS = flags.FLAGS
verbose_logging = True
# Set to True if the dataset has samples with no answers. For SQuAD 1.1, this is set to False
version_2_with_negative = False
# The total number of n-best predictions to generate in the nbest_predictions.json output file.
n_best_size = 20
# The maximum length of an answer that can be generated.
# This is needed because the start and end predictions are not conditioned on one another.
max_answer_length = 30
# -
# Let's define the tokenizer and create the model for the estimator:
# +
# Validate the casing config consistency with the checkpoint name.
tokenization.validate_case_matches_checkpoint(do_lower_case, init_checkpoint)
# Create the tokenizer.
tokenizer = tokenization.FullTokenizer(vocab_file=vocab_file, do_lower_case=do_lower_case)
# Load the configuration from file
bert_config = modeling.BertConfig.from_json_file(bert_config_file)
def model_fn(features, labels, mode, params): # pylint: disable=unused-argument
unique_ids = features["unique_ids"]
input_ids = features["input_ids"]
input_mask = features["input_mask"]
segment_ids = features["segment_ids"]
(start_logits, end_logits) = run_squad.create_model(
bert_config=bert_config,
is_training=False,
input_ids=input_ids,
input_mask=input_mask,
segment_ids=segment_ids,
use_one_hot_embeddings=False)
tvars = tf.trainable_variables()
initialized_variable_names = {}
(assignment_map, initialized_variable_names) = modeling.get_assignment_map_from_checkpoint(tvars, init_checkpoint)
tf.train.init_from_checkpoint(init_checkpoint, assignment_map)
output_spec = None
predictions = {"unique_ids": unique_ids,
"start_logits": start_logits,
"end_logits": end_logits}
output_spec = tf.estimator.EstimatorSpec(mode=mode, predictions=predictions)
return output_spec
config = tf.ConfigProto(log_device_placement=True)
run_config = tf.estimator.RunConfig(
model_dir=None,
session_config=config,
save_checkpoints_steps=1000,
keep_checkpoint_max=1)
estimator = tf.estimator.Estimator(
model_fn=model_fn,
config=run_config,
params=params)
# -
# ### 5.a Inference
# +
eval_examples = run_squad.read_squad_examples(
input_file=input_file, is_training=False)
eval_writer = run_squad.FeatureWriter(
filename=os.path.join(output_dir, "eval.tf_record"),
is_training=False)
eval_features = []
def append_feature(feature):
eval_features.append(feature)
eval_writer.process_feature(feature)
# Loads a data file into a list of InputBatch's
run_squad.convert_examples_to_features(
examples=eval_examples,
tokenizer=tokenizer,
max_seq_length=max_seq_length,
doc_stride=doc_stride,
max_query_length=max_query_length,
is_training=False,
output_fn=append_feature)
eval_writer.close()
tf.logging.info("***** Running predictions *****")
tf.logging.info(" Num orig examples = %d", len(eval_examples))
tf.logging.info(" Num split examples = %d", len(eval_features))
tf.logging.info(" Batch size = %d", predict_batch_size)
predict_input_fn = run_squad.input_fn_builder(
input_file=eval_writer.filename,
batch_size=predict_batch_size,
seq_length=max_seq_length,
is_training=False,
drop_remainder=False)
all_results = []
eval_hooks = [run_squad.LogEvalRunHook(predict_batch_size)]
eval_start_time = time.time()
for result in estimator.predict(
predict_input_fn, yield_single_examples=True, hooks=eval_hooks, checkpoint_path=init_checkpoint):
unique_id = int(result["unique_ids"])
start_logits = [float(x) for x in result["start_logits"].flat]
end_logits = [float(x) for x in result["end_logits"].flat]
all_results.append(
run_squad.RawResult(
unique_id=unique_id,
start_logits=start_logits,
end_logits=end_logits))
eval_time_elapsed = time.time() - eval_start_time
time_list = eval_hooks[-1].time_list
time_list.sort()
eval_time_wo_startup = sum(time_list[:int(len(time_list) * 0.99)])
num_sentences = eval_hooks[-1].count * predict_batch_size
avg_sentences_per_second = num_sentences * 1.0 / eval_time_wo_startup
tf.logging.info("-----------------------------")
tf.logging.info("Total Inference Time = %0.2f Inference Time W/O start up overhead = %0.2f "
"Sentences processed = %d", eval_time_elapsed, eval_time_wo_startup,
num_sentences)
tf.logging.info("Inference Performance = %0.4f sentences/sec", avg_sentences_per_second)
tf.logging.info("-----------------------------")
output_prediction_file = os.path.join(output_dir, "predictions.json")
output_nbest_file = os.path.join(output_dir, "nbest_predictions.json")
output_null_log_odds_file = os.path.join(output_dir, "null_odds.json")
run_squad.write_predictions(eval_examples, eval_features, all_results,
n_best_size, max_answer_length,
do_lower_case, output_prediction_file,
output_nbest_file, output_null_log_odds_file,
version_2_with_negative, verbose_logging)
tf.logging.info("Inference Results:")
# Here we show only the prediction results, nbest prediction is also available in the output directory
results = ""
with open(output_prediction_file, 'r') as json_file:
data = json.load(json_file)
for question in eval_examples:
results += "<tr><td>{}</td><td>{}</td><td>{}</td></tr>".format(question.qas_id, question.question_text, data[question.qas_id])
from IPython.display import display, HTML
display(HTML("<table><tr><th>Id</th><th>Question</th><th>Answer</th></tr>{}</table>".format(results)))
# -
# ## 6. What's next
# Now that you are familiar with running QA Inference on BERT, using mixed precision, you may want to try
# your own paragraphs and queries.
#
# You may also want to take a look to the notebook __bert_squad_tf_finetuning.ipynb__ on how to run fine-tuning on BERT, available in the same directory.
| DeepLearningExamples/TensorFlow/LanguageModeling/BERT/notebooks/bert_squad_tf_inference.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, LSTM, Dropout
from sklearn.preprocessing import MinMaxScaler
import matplotlib.pyplot as plt
import theano
import os
# +
##
def create_dataset( dataset, look_back = 1 ) :
dataX, dataY = [], []
for i in range(len(dataset)-look_back):
dataX.append( dataset[ i : ( i + look_back ), 0 ])
dataY.append( dataset[ i + look_back, 0 ] )
return np.array( dataX ), np.array( dataY )
# +
path_train = 'Data_RNN/train'
file_list_train = [ os.path.join( path_train, file ) for file in os.listdir( path_train ) if not file.startswith('.') ]
n_train = len( file_list_train )
for n in range( n_train ) :
print( 'Dataset : ', str( n + 1 ), '/', str( n_train ) )
F = open( file_list_train[ n ], 'r' )
dataset_train = []
for Fline in F.readlines() :
try :
A = str.split( Fline, " " )
dataset_train = np.append( dataset_train, np.log10( float( A[ 6 ] ) ) )
except ValueError :
pass
F.close()
dataset_train = dataset_train[ : , None ]
look_back = 24
trainX, trainY = create_dataset( dataset_train, look_back )
trainX = np.reshape( trainX, ( trainX.shape[ 0 ], trainX.shape[ 1 ], 1 ) )
theano.config.compute_test_value = "ignore"
batch_size = 1
model = Sequential()
# model.add( LSTM( 32, input_shape = ( None, 1 ) ), stateful = True )
model.add( LSTM( 32, batch_input_shape = ( batch_size, look_back, 1 ), stateful = True ) )
model.add( Dropout( 0.3 ) )
model.add( Dense( 1, activation = 'relu' ) )
model.compile( loss = 'mean_squared_error', optimizer = 'adam' )
model.fit( trainX, trainY, epochs=5, batch_size = batch_size, verbose = 2 )
model.reset_states()
trainScore = model.evaluate( trainX, trainY, batch_size = batch_size, verbose = 0 )
print( 'Train Score : ', trainScore )
# +
G = open( 'Data_RNN/test/201601.txt', 'r' )
dataset_test = []
for Gline in G.readlines() :
try :
A = str.split( Gline, " " )
dataset_test = np.append( dataset_test, np.log10( float( A[ 6 ] ) ) )
except ValueError :
pass
batch_size = 1
dataset_test = dataset_test[ : , None ]
look_back = 24
testX, testY = create_dataset( dataset_test, look_back )
testX = np.reshape( testX, ( testX.shape[ 0 ], testX.shape[ 1 ], 1 ) )
testScore = model.evaluate( testX[ : 240 ], testY[ : 240 ], batch_size = batch_size, verbose = 0 )
print( 'Test Score : ', testScore )
# +
look_ahead = 240
testPredict = [ np.vstack( [ testX[ -1 ][ 1 : ], testY[ -1 ] ] ) ]
predictions = np.zeros( ( look_ahead , 1 ) )
for i in range( look_ahead ) :
prediction = model.predict( np.array( [ testPredict[ -1 ] ] ), batch_size = batch_size )
predictions[ i ] = prediction
testPredict.append( np.vstack( [ testPredict[ -1 ][ 1 : ], prediction ] ) )
plt.figure(figsize=(12,5))
plt.plot( np.arange( look_ahead ), predictions, 'r' , label = "prediction" )
plt.plot( np.arange( look_ahead ), dataset_test[ 0 : look_ahead ], label = "test function" )
plt.legend()
plt.show()
| _writing/LSTM.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Instruções
#
# Esse documento é um [Jupyter notebook](http://jupyter.org/), um documento interativo que mistura texto (como esse), código (como abaixo), e o resultado de executar o código (números, texto, figuras, videos, etc).
#
# O notebook te fornecerá exemplos interativos que trabalham o tema abordado.
#
# As células com números ao lado, como `In [1]:`, são código [Python](https://www.python.org/). Algumas dessas células não produzem resultado e servem de preparação para os exemplos interativos. Outras, produzem gráficos interativos. **Você deve executar todas as células, uma de cada vez**, mesmo as que não produzem gráficos.
#
# **Para executar uma célula**, clique em cima dela e aperte `SHIFT+ENTER` ou presione o botão de *Run* (<button class='fa fa-play icon-play btn btn-xs btn-default'></button>) na barra de ferramentas acima. O foco (contorno verde ou cinza em torno da célula) deverá passar para a célula abaixo. Para rodá-la, aperte `SHIFT+ENTER` novamente e assim por diante. Você pode executar células de texto que não acontecerá nada.
# # Por que usar Jupyter Notebook?
# Veja o artigo [Interactive notebooks: Sharing the code](http://www.nature.com/polopoly_fs/1.16261!/menu/main/topColumns/topLeftColumn/pdf/515151a.pdf) da revista nature
# #### Autor:
# * Dr. <NAME> ([@ofmla](https://github.com/ofmla))
# # Python
#
# Python é uma linguagem de programação de alto nível, dinâmica, interpretada, modular, multiplataforma e orientada a objetos.
#
# Objetivo geral: análise de dados, desenvolvimento web, GUIs, sistemas em tempo real, etc.
#
# Atualmente a linguagem de programação que mais cresce.
#
# Em desenvolvimento ativo: Python 3.9 lançado em Outubro de 2020
#
# - Fácil de começar
# - Maior produtividade no desenvolvimento, especialmente no início do projeto
# - Da ideia ao código funcional em pouco tempo
# - Ótimo para testar ideias e experimentos rápidos
# - Biblioteca padrão abrangente: coleções, algoritmos, I/O ...
# - Python foi projetado para ser uma linguagem pequena com uma grande biblioteca padrão
# - Ecossistema de bibliotecas incrível
# - Computação numérica e científica: numpy, scipy, sympy, xarray ...
# - Visualização: matplotlib, plotly, bokeh ...
# - Tabelas e séries temporais: pandas
# - _Machine Learning_: scikit-learn, tensorflow, keras, pytorch
# - Gerenciamento de pacote padrão
# - Extremamente fácil de configurar o ambiente de desenvolvimento e instalar pacotes
# Python é uma linguagem de sintaxe relativamente simples e de fácil compreensão que tem sido cada vez mais usada por engenheiros e cientistas. Python oferece uma gama completa de recursos de linguagem de programação e há uma vasta quantidade de pacotes para cálculos científicos e de engenharia. Existem inumeros recursos na internet onde você pode ir para obter mais informações sobre programação em Python, p.ex. [ Software-Carpentry Lessons](https://software-carpentry.org/lessons/)
# ## Numpy
#
# [Numpy](https://numpy.org/) é a biblioteca central para computação científica em Python. A ideia fundamental do NumPy é o suporte a _arrays_ multidimensionais ferramentas para trabalhar com esses _arrays_.
#
# ## _Setup_
#
# Rode as células abaixo para carregar os módulos necessários para essa prática.
import sys
import numpy as np
import matplotlib.pyplot as plt
print(sys.version)
np.__version__
# %matplotlib inline
# ### _Arrays_
#
# Um numpy _array_ é uma grade de valores, todos do mesmo tipo, e é indexado por uma tupla de inteiros não negativos. O número de dimensões é o _rank_ do _array_; a forma do _array_ é uma tupla de inteiros fornecendo o tamanho do _array_ ao longo de cada dimensão.
#
# Podemos inicializar numpy _arrays_ a partir de listas Python aninhadas e acessar elementos usando colchetes:
# +
a = np.array([1, 2, 3]) # Cria um array de rank 1
print(type(a)) # Printa "<class 'numpy.ndarray'>"
print(a.shape) # Printa "(3,)"
print(a[0], a[1], a[2]) # Printa "1 2 3"
a[0] = 5 # Muda um elemento do array
print(a) # Printa "[5, 2, 3]"
b = np.array([[1,2,3],[4,5,6]]) # Cria um array de rank 2
print(b.shape) # Printa "(2, 3)"
print(b[0, 0], b[0, 1], b[1, 0]) # Printa "1 2 4"
# -
# O Numpy também oferece muitas funções para criar _arrays_:
# +
import numpy as np
a = np.zeros((2,2)) # Cria um array zerado
print(a) # Prints "[[ 0. 0.]
# [ 0. 0.]]"
b = np.ones((1,2)) # Cria um array com todos os elemntos iguais a 1
print(b) # Prints "[[ 1. 1.]]"
c = np.full((2,2), 7) # Cria um array com todos os elementos sendo um valor constate
print(c) # Prints "[[ 7. 7.]
# [ 7. 7.]]"
d = np.eye(2) # Cria uma matriz identidade 2x2
print(d) # Prints "[[ 1. 0.]
# [ 0. 1.]]"
e = np.random.random((2,2)) # Cria um array preenchido com valores aleatorios
print(e)
# -
# ## _Slicing arrays_
#
# __Fatiar__: _Arrays_ multidimensionais podem ser fatiados.
# +
# Cria o seguinte array de rank 2 e shape (3, 4)
# [[ 1 2 3 4]
# [ 5 6 7 8]
# [ 9 10 11 12]]
a = np.array([[1,2,3,4], [5,6,7,8], [9,10,11,12]])
# Use o fatiamento para obter um subarray consistendo das primeiras 2 filas
# e colunas 1 e 2; b fica da seguinte forma com shape (2, 2):
# [[2 3]
# [6 7]]
b = a[:2, 1:3]
# Uma fatia de um array é uma visualização dos mesmos dados, portanto, modificá-la
# irá modificar o array original
print(a[0, 1]) # Printa "2"
b[0, 0] = 77 # b[0, 0] são os mesmos dados que a[0, 1]
print(a[0, 1]) # Printa "77"
# +
# Cria de novo o array 'a' de rank 2 e shape (3, 4)
# [[ 1 2 3 4]
# [ 5 6 7 8]
# [ 9 10 11 12]]
a = np.array([[1,2,3,4], [5,6,7,8], [9,10,11,12]])
# Duas maneiras de acessar os dados na linha do meio do array.
# Misturar indexação de inteiro com fatias produz uma matriz de rank inferior,
# enquanto usando apenas fatias produz uma matriz do mesm rank que o
# array original:
row_r1 = a[1, :] # vista da segunda fila de a de rank 1
row_r2 = a[1:2, :] # vista da segunda fila de a de rank 2
print(row_r1, row_r1.shape) # Prints "[5 6 7 8] (4,)"
print(row_r2, row_r2.shape) # Prints "[[5 6 7 8]] (1, 4)"
# A mesma distinção pode ser feita ao acessar colunas de um array
col_r1 = a[:, 1]
col_r2 = a[:, 1:2]
print(col_r1, col_r1.shape) # Prints "[ 2 6 10] (3,)"
print(col_r2, col_r2.shape) # Prints "[[ 2]
# [ 6]
# [10]] (3, 1)"
# +
# Crie uma array novo a partir da qual selecionaremos os elementos
a = np.array([[1,2,3], [4,5,6], [7,8,9], [10, 11, 12]])
print(a) # printa "array([[ 1, 2, 3],
# [ 4, 5, 6],
# [ 7, 8, 9],
# [10, 11, 12]])"
# Crie uma série de índices
b = np.array([0, 2, 0, 1])
# Selecione um elemento de cada linha de a usando os índices em b
print(a[np.arange(4), b]) # Printa "[ 1 6 7 11]"
# Transformar um elemento de cada linha de a usando os índices em b
a[np.arange(4), b] += 10
print(a) # printa "array([[11, 2, 3],
# [ 4, 5, 16],
# [17, 8, 9],
# [10, 21, 12]])
# +
a = np.array([[1,2], [3, 4], [5, 6]])
bool_idx = (a > 2) # Encontra os elementos de a que são maiores que 2;
# isso retorna um numpy array de Booleanos da mesmo
# forma de a, onde cada elemento de bool_idx informa
# se esse elemento de a é> 2
print(bool_idx) # Printa "[[False False]
# [ True True]
# [ True True]]"
# Usamos indexação do array booleano para construir um array de rank 1
# consistindo nos elementos de a correspondente aos valores verdadeiros
# de bool_idx
# We use boolean array indexing to construct a rank 1 array
# consisting of the elements of a corresponding to the True values
# of bool_idx
print(a[bool_idx]) # Printa "[3 4 5 6]"
# Podemos fazer tudo isso em uma única declaração concisa::
print(a[a > 2]) # Printa "[3 4 5 6]"
# -
# ### Matemática em _arrays_
#
# Funções matemáticas básicas operam elemento a elemento em _arrays_ e estão disponíveis como operadores e como funções no módulo numpy:
# +
x = np.array([[1,2],[3,4]], dtype=np.float64)
y = np.array([[5,6],[7,8]], dtype=np.float64)
# Soma elemento a elemento; ambos produzem o array
# [[ 6.0 8.0]
# [10.0 12.0]]
print(x + y)
print(np.add(x, y))
# Substração elemento a elemento; ambos produzem o array
# [[-4.0 -4.0]
# [-4.0 -4.0]]
print(x - y)
print(np.subtract(x, y))
# Produto elemento a elemento; ambos produzem o array
# [[ 5.0 12.0]
# [21.0 32.0]]
print(x * y)
print(np.multiply(x, y))
# Divisão elemento a elemento; ambos produzem o array
# [[ 0.2 0.33333333]
# [ 0.42857143 0.5 ]]
print(x / y)
print(np.divide(x, y))
# Raiz quadrada elemento a elemento; ambos produzem o array
# [[ 1. 1.41421356]
# [ 1.73205081 2. ]]
print(np.sqrt(x))
# -
# A função ```dot``` é usada para calcular produtos internos de vetores, para multiplicar um vetor por uma matriz e para multiplicar matrizes. ```dot``` está disponível como uma função no módulo numpy e como um método de instância de objetos array:
# +
x = np.array([[1,2],[3,4]])
y = np.array([[5,6],[7,8]])
v = np.array([9,10])
w = np.array([11, 12])
# produto interno de vetores; ambos produzem 219
print(v.dot(w))
print(np.dot(v, w))
# produto Matriz/vetor; ambos produzem um array de rank 1 [29 67]
print(x.dot(v))
print(np.dot(x, v))
# produto Matrix/matrix ; ambos produzem array de rank 2
# [[19 22]
# [43 50]]
print(x.dot(y))
print(np.dot(x, y))
# -
# O Numpy fornece muitas funções úteis para realizar cálculos em matrizes; um dos mais úteis é a ```sum```:
# +
x = np.array([[1,2],[3,4]])
print(np.sum(x)) # Computa a soma de todos os elementos; printa "10"
print(np.sum(x, axis=0)) # Computa a soma de cada coluna; prints "[4 6]"
print(np.sum(x, axis=1)) # Computa a soma de cada fila; prints "[3 7]"
# -
# Você pode encontrar a lista completa de funções matemáticas fornecidas por numpy na [documentação](https://numpy.org/doc/stable/reference/routines.math.html).
# Além de computar funções matemáticas usando _arrays_, frequentemente precisamos mudar (_reshaping_) a forma ou manipular dados em _arrays_. O exemplo mais simples desse tipo de operação é a transposição de uma matriz; para transpor uma matriz, basta usar o atributo ```T``` de um objeto de matriz:
#
# +
x = np.array([[1,2], [3,4]])
print(x) # Printa "[[1 2]
# [3 4]]"
print(x.T) # Printa "[[1 3]
# [2 4]]"
# Observe que a transposição de um array de rank 1 não faz nada::
v = np.array([1,2,3])
print(v) # Printa "[1 2 3]"
print(v.T) # Printa "[1 2 3]"
# -
# O Numpy fornece muito mais funções para manipular _arrays_; você pode ver a lista completa na [documentação](http://docs.scipy.org/doc/numpy/reference/routines.array-manipulation.html).
# ## Matplotlib
# +
X = np.linspace(-np.pi, np.pi, 256)
C, S = np.cos(X), np.sin(X)
plt.plot(X, C)
plt.plot(X, S)
plt.show()
# -
# ### Alterar cores e larguras de linha
#
# ```
# plt.figure(figsize=(10, 6), dpi=80)
# plt.plot(X, C, color="blue", linewidth=2.5, linestyle="-")
# plt.plot(X, S, color="red", linewidth=2.5, linestyle="-")
# ```
# ### Definindo limites
# ```...
# plt.xlim(X.min() * 1.1, X.max() * 1.1)
# plt.ylim(C.min() * 1.1, C.max() * 1.1)
# ```
# ### Definindo _ticks_
# ```
# plt.xticks([-np.pi, -np.pi/2, 0, np.pi/2, np.pi])
# plt.yticks([-1, 0, +1])
# ```
# +
# Cria uma figura 8x6 polegadas, 80 pts por polegada
plt.figure(figsize=(8, 6), dpi=80)
# Cria um novo subplot 1x1
plt.subplot(1, 1, 1)
X = np.linspace(-np.pi, np.pi, 256)
C, S = np.cos(X), np.sin(X)
# Plota cosine com uma linha azul continua de espessura 1 (pixels)
plt.plot(X, C, color="blue", linewidth=1.0, linestyle="-")
# Plota sine com uma linha verde continua de espessura 1 (pixels)
plt.plot(X, S, color="green", linewidth=1.0, linestyle="-")
# Seta os limites em x
plt.xlim(-4.0, 4.0)
# Seta os ticks em x
plt.xticks(np.linspace(-4, 4, 9))
# Seta os limites em y
plt.ylim(-1.0, 1.0)
# Seta os ticks em y
plt.yticks(np.linspace(-1, 1, 5))
# Salva a figura usando 72 pts por polegada
# plt.savefig("exercise_2.png", dpi=72)
# Mostra o resultado na tela
plt.show()
# +
# Prepare o dado
x = np.linspace(0, 10, 100)
# Plote o dado
plt.plot(x, x, label='linear')
# Add a legenda
plt.legend()
# Mostre o plot
plt.show()
# -
fig = plt.figure()
ax = fig.add_subplot(111)
ax.plot([1, 2, 3, 4], [10, 20, 25, 30], color='lightblue', linewidth=3)
ax.scatter([0.3, 3.8, 1.2, 2.5], [11, 25, 9, 26], color='darkgreen', marker='^')
ax.set_xlim(0.5, 4.5)
plt.show()
plt.plot([1, 2, 3, 4], [10, 20, 25, 30], color='lightblue', linewidth=3)
plt.scatter([0.3, 3.8, 1.2, 2.5], [11, 25, 9, 26], color='darkgreen', marker='^')
plt.xlim(0.5, 4.5)
plt.show()
# +
# Inicialização do plot
fig = plt.figure(figsize=(10,5))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122)
# ou substitua as três linhas acima pela seguinte:
#fig, (ax1, ax2) = plt.subplots(1,2, figsize=(10,5))
# Plote os dados
ax1.bar([1,2,3],[3,4,5])
ax2.barh([0.5,1,2.5],[0,1,2])
# mostre o plot
plt.show()
| classes_gp_senai/intro_numpy.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] colab_type="text" id="b9R-4ezU3NH0"
# ## AutoGraph: examples of simple algorithms
#
# This notebook shows how you can use AutoGraph to compile simple algorithms and run them in TensorFlow.
#
# It requires the nightly build of TensorFlow, which is installed below.
# + colab={} colab_type="code" id="TuWj26KWz1fZ"
# !pip install -U -q tf-nightly-2.0-preview
# + colab={} colab_type="code" id="Cp7iTarmz62Y"
import tensorflow as tf
tf = tf.compat.v2
tf.enable_v2_behavior()
# + [markdown] colab_type="text" id="3kudk1elq0Gh"
# ### Fibonacci numbers
#
# https://en.wikipedia.org/wiki/Fibonacci_number
#
# + colab={"height": 187} colab_type="code" executionInfo={"elapsed": 709, "status": "ok", "timestamp": 1563825398552, "user": {"displayName": "", "photoUrl": "", "userId": ""}, "user_tz": 240} id="H7olFlMXqrHe" outputId="25243e7b-99a7-4a6d-ad00-e97c52be7d97"
@tf.function
def fib(n):
f1 = 0
f2 = 1
for i in tf.range(n):
tmp = f2
f2 = f2 + f1
f1 = tmp
tf.print(i, ': ', f2)
return f2
_ = fib(tf.constant(10))
# + [markdown] colab_type="text" id="p8zZyj-tq4K3"
# #### Generated code
# + colab={} colab_type="code" id="UeWjK8rHq6Cj"
print(tf.autograph.to_code(fib.python_function))
# + [markdown] colab_type="text" id="eIfVy6ZTrFEH"
# ### Fizz Buzz
#
# https://en.wikipedia.org/wiki/Fizz_buzz
# + colab={"height": 119} colab_type="code" executionInfo={"elapsed": 663, "status": "ok", "timestamp": 1563825401385, "user": {"displayName": "", "photoUrl": "", "userId": ""}, "user_tz": 240} id="33CAheYsrEQ7" outputId="2a88b65d-4fed-4d96-8770-0c68ffece861"
import tensorflow as tf
@tf.function(experimental_autograph_options=tf.autograph.experimental.Feature.EQUALITY_OPERATORS)
def fizzbuzz(i, n):
while i < n:
msg = ''
if i % 3 == 0:
msg += 'Fizz'
if i % 5 == 0:
msg += 'Buzz'
if msg == '':
msg = tf.as_string(i)
tf.print(msg)
i += 1
return i
_ = fizzbuzz(tf.constant(10), tf.constant(16))
# + [markdown] colab_type="text" id="Lkq3DBGOv3fA"
# #### Generated code
# + colab={} colab_type="code" id="bBhFIIaZrxvx"
print(tf.autograph.to_code(fizzbuzz.python_function))
# + [markdown] colab_type="text" id="BNRtprSvwJgk"
# ### Conway's Game of Life
#
# https://en.wikipedia.org/wiki/Conway%27s_Game_of_Life
# + [markdown] colab_type="text" id="r8_0ioEuAI-a"
# #### Testing boilerplate
# + colab={} colab_type="code" id="7moIlf8VABkl"
NUM_STEPS = 1
# + [markdown] colab_type="text" id="QlEvfIQPAYF5"
# #### Game of Life for AutoGraph
#
# Note: the code may take a while to run.
# + colab={} colab_type="code" id="5pCK2qQSAAK4"
#@test {"skip": true}
NUM_STEPS = 75
# + [markdown] colab_type="text" id="GPZANPdhMagD"
# Note: This code uses a non-vectorized algorithm, which is quite slow. For 75 steps, it will take a few minutes to run.
# + colab={"height": 309} colab_type="code" executionInfo={"elapsed": 147654, "status": "ok", "timestamp": 1563825336196, "user": {"displayName": "", "photoUrl": "", "userId": ""}, "user_tz": 240} id="hC3qMqryPDHS" outputId="56a095a3-28a3-455d-e95e-2c4c9dcd97d2"
import time
import traceback
import sys
from matplotlib import pyplot as plt
from matplotlib import animation as anim
import numpy as np
from IPython import display
@tf.autograph.experimental.do_not_convert
def render(boards):
fig = plt.figure()
ims = []
for b in boards:
im = plt.imshow(b, interpolation='none')
im.axes.get_xaxis().set_visible(False)
im.axes.get_yaxis().set_visible(False)
ims.append([im])
try:
ani = anim.ArtistAnimation(
fig, ims, interval=100, blit=True, repeat_delay=5000)
plt.close()
display.display(display.HTML(ani.to_html5_video()))
except RuntimeError:
print('Coult not render animation:')
traceback.print_exc()
return 1
return 0
def gol_episode(board):
new_board = tf.TensorArray(tf.int32, 0, dynamic_size=True)
for i in tf.range(len(board)):
for j in tf.range(len(board[i])):
num_neighbors = tf.reduce_sum(
board[tf.maximum(i-1, 0):tf.minimum(i+2, len(board)),
tf.maximum(j-1, 0):tf.minimum(j+2, len(board[i]))]
) - board[i][j]
if num_neighbors == 2:
new_cell = board[i][j]
elif num_neighbors == 3:
new_cell = 1
else:
new_cell = 0
new_board.append(new_cell)
final_board = new_board.stack()
final_board = tf.reshape(final_board, board.shape)
return final_board
@tf.function(experimental_autograph_options=(
tf.autograph.experimental.Feature.EQUALITY_OPERATORS,
tf.autograph.experimental.Feature.BUILTIN_FUNCTIONS,
tf.autograph.experimental.Feature.LISTS,
))
def gol(initial_board):
board = initial_board
boards = tf.TensorArray(tf.int32, size=0, dynamic_size=True)
i = 0
for i in tf.range(NUM_STEPS):
board = gol_episode(board)
boards.append(board)
boards = boards.stack()
tf.py_function(render, (boards,), (tf.int64,))
return i
# Gosper glider gun
# Adapted from http://www.cplusplus.com/forum/lounge/75168/
_ = 0
initial_board = tf.constant((
( _,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,1,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,1,_,1,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,1,1,_,_,_,_,_,_,1,1,_,_,_,_,_,_,_,_,_,_,_,_,1,1,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,1,_,_,_,1,_,_,_,_,1,1,_,_,_,_,_,_,_,_,_,_,_,_,1,1,_ ),
( _,1,1,_,_,_,_,_,_,_,_,1,_,_,_,_,_,1,_,_,_,1,1,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,1,1,_,_,_,_,_,_,_,_,1,_,_,_,1,_,1,1,_,_,_,_,1,_,1,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,1,_,_,_,_,_,1,_,_,_,_,_,_,_,1,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,1,_,_,_,1,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,1,1,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
( _,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_,_ ),
))
initial_board = tf.pad(initial_board, ((0, 10), (0, 5)))
_ = gol(initial_board)
# + [markdown] colab_type="text" id="7NgrSPCZxs3h"
# #### Generated code
# + colab={} colab_type="code" id="hIGYeX0Cxs3i"
print(tf.autograph.to_code(gol.python_function))
| tensorflow/contrib/autograph/examples/notebooks/algorithms.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Predictive Modeling
# ### <NAME>
#
# I will set the baseline and create the first ML model to predict song popularity.
# ## Set up Environment
# +
import pandas as pd
import numpy as np
from scipy import stats
from math import sqrt
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV, KFold
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.cluster import KMeans
from sklearn.metrics import mean_squared_error, explained_variance_score, mean_absolute_error
from sklearn.linear_model import LinearRegression, TweedieRegressor, LassoLars
from sklearn.feature_selection import RFE
from sklearn.preprocessing import PolynomialFeatures
from sklearn.ensemble import IsolationForest, RandomForestRegressor
from sklearn.svm import SVR
import sklearn.svm
import math
import itertools
import optunity
import optunity.metrics
import warnings
warnings.filterwarnings("ignore")
# -
from prepare import handle_nulls, set_index
from preprocessing import spotify_split, split_df, scale_data, encode_features
from model import get_model_features, OLS_model
# ## Acquire data
df = pd.read_csv('full-playlist.csv', index_col=0)
df.head()
df.shape
# ## Prepare data
# handle null values
df = handle_nulls(df)
# check for nulls
df.isna().sum()
# check data types
df.dtypes
# +
# set index to track_id
#df = set_index(df)
# -
# Note to self: After MVP we need to convert release_data into a Timestamp.
# ## Preprocess data
# show features
df.columns
df.head(3)
# encode features
df = encode_features(df)
df.head(3)
# chose features for MVP modeling
df = get_model_features(df)
df.head()
# split the data
X_train, y_train, X_validate, y_validate, X_test, y_test, train, validate, test = spotify_split(df, 'popularity')
train.head(3)
# scale the data
X_train_scaled, X_validate_scaled, X_test_scaled = scale_data(train, validate, test, 'popularity', 'MinMax')
X_train_scaled.head(3)
# check data types
X_train_scaled.dtypes
# ## Set the baseline
#np.median(y_train)
np.mean(y_train)
# +
#baseline = y_train.median()
baseline = y_train.mean()
baseline_rmse_train = round(sqrt(mean_squared_error(y_train, np.full(len(y_train), baseline))), 6)
print('RMSE (Root Mean Square Error) of Baseline on train data:\n', baseline_rmse_train)
baseline_rmse_validate = round(sqrt(mean_squared_error(y_validate, np.full(len(y_validate), baseline))), 6)
print('RMSE (Root Mean Square Error) of Baseline on validate data:\n', baseline_rmse_validate)
# -
# Mean performed better than median.
#
# Our baseline prediction of popularity will be ```38.46776470588235```, with an RMSE of ```22.770177``` on the train data and ```23.034868``` on the validate data.
# ## Model 1 - Ordinary Least Squares (OLS) using Linear Regression
# show available features
X_train_scaled.columns
# +
# use all features
X = X_train_scaled
y = y_train
X_v = X_validate_scaled
y_v = y_validate
lm_pred, lm_rmse, lm_pred_v, lm_rmse_v = OLS_model(X, y, X_v, y_v)
# -
# Not great results, but they did beat the baseline model.
# ## Model 2 - Support Vector Regressor using RBF Kernel
# +
# use all features
X = X_train_scaled
y = y_train
X_v = X_validate_scaled
y_v = y_validate
# most important SVR parameter is Kernel type.
# It can be linear, polynomial, or gaussian SVR.
# We have a non-linear condition so we can select polynomial or gaussian
# but here we select RBF (a gaussian type) kernel.
# create the model object
svr = SVR(kernel='rbf')
# fit the model to our training data
svr.fit(X, y)
# predict on train
svr_pred = svr.predict(X)
# compute root mean squared error
svr_rmse = sqrt(mean_squared_error(y, svr_pred))
# predict on validate
svr_pred_v = svr.predict(X_v)
# compute root mean squared error
svr_rmse_v = sqrt(mean_squared_error(y_v, svr_pred_v))
print("RMSE for SVR using RBF Kernel\n\nOn train data:\n", round(svr_rmse, 6), '\n\n',
"On validate data:\n", round(svr_rmse_v, 6))
#return svr_pred, svr_rmse, svr_pred_v, svr_rmse_v
# -
# ## Feature Selection
feature_importances = pd.DataFrame(rf.feature_importances_, index = X_train_scaled.columns,
columns=['importance']).sort_values('importance',ascending=False)
| sandbox/kwame/kwame_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Pipeline: Heterogenous data
#
# This notebook implements a pipeline for heterogeneous data.
#
#
# sources:
# Sample pipeline for text feature extraction and evaluation: https://scikit-learn.org/stable/auto_examples/model_selection/grid_search_text_feature_extraction.html
#
# Metrics and scoring: quantifying the quality of predictions: https://scikit-learn.org/stable/modules/model_evaluation.html#common-cases-predefined-values
#
#
# Demonstration of multi-metric evaluation on cross_val_score and GridSearchCV: https://scikit-learn.org/stable/auto_examples/model_selection/plot_multi_metric_evaluation.html
#
#
# ColumnTransformer for heterogeneous data:
# https://scikit-learn.org/stable/modules/compose.html#columntransformer-for-heterogeneous-data
#
#
# Column Transformer with Heterogeneous Data Sources: https://scikit-learn.org/stable/auto_examples/compose/plot_column_transformer.html
# +
import pandas as pd
import numpy as np
import glob
import os
import munge_help
from time import time
import matplotlib.pyplot as plt
# %matplotlib inline
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import GridSearchCV
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer, make_column_selector, make_column_transformer
from sklearn.preprocessing import MinMaxScaler
import utils
import xgboost as xgb
# -
# ### Load Data
X_train = utils.load_obj(path=os.path.join('data_processed', 'X_train.pkl'))
y_train = utils.load_obj(path=os.path.join('data_processed', 'y_train.pkl'))
X_train.shape
#see how many columns are numbers
X_train.select_dtypes(np.number).shape
X_train.dtypes
# +
#Control the balance of positive and negative weights, useful for unbalanced classes
#A typical value to consider:
# sum(negative instances) / sum(positive instances)
scale_pos_weight = float(np.sum(y_train == 0)) / np.sum(y_train == 1)
scale_pos_weight
# +
#define preprocessor
preprocessor = ColumnTransformer([('tfidfvect',
TfidfVectorizer(ngram_range=(1,3)),
'description')
],
remainder=MinMaxScaler(),
n_jobs=-1
)
#define pipeline
pipeline = Pipeline(steps=[('preprocessor', preprocessor),
('clf', xgb.XGBClassifier(n_estimators=100,
scale_pos_weight = scale_pos_weight,
eta=0.9,
num_boost_round=15,
)
)
])
pipeline
# +
#params for grird search
#note the double __ to get to nested elements
parameters = {
'preprocessor__tfidfvect__max_df': (0.8, 0.9),
'preprocessor__tfidfvect__min_df': (0.1, 0.15),
'preprocessor__tfidfvect__max_features': (200, 250),
'clf__max_depth': (6, 12), #Maximum depth of a tree. Increasing this value will make the model more complex and more likely to overfit
'clf__subsample': (0.5, 0.9) #take part of train data to avoid overfitting
}
#instantiate grid search
grid_search = GridSearchCV(pipeline,
parameters,
n_jobs=-1,
verbose=10, #lots of details
scoring=['roc_auc', 'f1'],
refit='roc_auc',
return_train_score=True
)
# start the timer
t0 = time()
grid_search.fit(X_train, y_train)
#time to do it
print("done in %0.3fs" % (time() - t0))
# -
print("Best score: %0.3f" % grid_search.best_score_)
print('\n')
print(20*'#')
print('\n')
print("Best parameters set:")
best_parameters = grid_search.best_estimator_.get_params()
for param_name in sorted(parameters.keys()):
print("\t%s: %r" % (param_name, best_parameters[param_name]))
utils.save_obj(obj = grid_search,
path = os.path.join('artifacts',
'grid_search_all_features_2020-11-29.pkl'))
| 04_Optimizing_Features_All.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Recreated NER twitter classifier
#
# So this is going to be a fresh start, trying to reuse the approach taken by http://didtrumptweetit.com/machine-learning-nltk/, which seems to get 98% accuracy on the same dataset. His model isn't being shared, but he's provided quite a few details. We'll see how far we get.
#
# We'll start by recreating the tfidf-logistic regression model, as although he has mentioned some issues with it, this will be far easy to create than the newer CNN one he created.
# ## Imports and Configuration
# +
# %load_ext autoreload
# %autoreload 2
# %matplotlib inline
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib
import numpy as np
matplotlib.rcParams['figure.figsize'] = [10, 6]
# -
# ## Data import and preprocessing
# We'll be training our model on trump's twitter messages in the range:
# - After his campaign anouncement (June 16, 2015)
# - Before he became president (January 20, 2017)
#
# We do this because the language use in his tweets is somewhat consistent, even though it changes throughout the campaign. We see a larger divide between his language in this period and the one before his campaign, so we therefore choose to leave those out of the training data.
# +
# Uncomment to get dataset
# # ! wget https://raw.githubusercontent.com/bpb27/political_twitter_archive/master/realdonaldtrump/realdonaldtrump.csv -P ../data
# +
twitter_df = (
pd.read_csv("../data/realdonaldtrump.csv")
.filter(["created_at", "source", "text", "is_retweet"])
.assign(created_at=lambda x: pd.to_datetime(x.created_at, format="%a %b %d %H:%M:%S +0000 %Y"))
.query('created_at > "2015-06-16"')
.query('created_at < "2017-01-20"')
.query('is_retweet == False')
.assign(text=lambda x: x.text.str.replace("&", "&"))
.drop('is_retweet', axis=1)
.sort_values("created_at")
)
twitter_df.loc[twitter_df.source != "Twitter for Android", "source"] = "Other Source"
twitter_df.describe()
# -
print(f"Twitter messages from Trump: {len(twitter_df[twitter_df.source == 'Twitter for Android'])}")
print(f"Twitter messages from other sources: {len(twitter_df[twitter_df.source != 'Twitter for Android'])}")
# ## Quick exploration of columns
# ### Sources
# +
plot_df = (
twitter_df.assign(week = lambda x: x.created_at.dt.week)
.groupby(by=["week", "source"])
.count()
.reset_index()
.filter(["week", "source", "text"])
.pivot("week", "source", "text")
)
plot_df["total"] = plot_df.sum(axis=1)
plot_df["Other Source"] = plot_df["Other Source"]/plot_df["total"]
plot_df["Twitter for Android"] = plot_df["Twitter for Android"]/plot_df["total"]
(
plot_df.filter(["Other Source", "Twitter for Android"])
.plot.area()
)
# -
# ### Twitter Messages
# +
plot_df = (
twitter_df.filter(["text", "source"])
.assign(num_words = lambda x: x.text.str.split(" ").apply(len),
num_char = lambda x: x.text.apply(len),
num_hashtags = lambda x: x.text.str.findall(r"\@[^\s\,\.\:\!\?]+").apply(len),
num_mentions = lambda x: x.text.str.findall(r"\#[^\s\,\.\:\!\?]+").apply(len))
)
sns.distplot(plot_df[plot_df["source"] == "Twitter for Android"].num_words, color="g")
sns.distplot(plot_df[plot_df["source"] == "Other Source"].num_words, color="r")
# -
sns.distplot(plot_df[plot_df["source"] == "Twitter for Android"].num_char, color="g")
sns.distplot(plot_df[plot_df["source"] == "Other Source"].num_char, color="r")
sns.distplot(plot_df[plot_df["source"] == "Twitter for Android"].num_hashtags, color="g")
sns.distplot(plot_df[plot_df["source"] == "Other Source"].num_hashtags, color="r")
sns.distplot(plot_df[plot_df["source"] == "Twitter for Android"].num_mentions, color="g")
sns.distplot(plot_df[plot_df["source"] == "Other Source"].num_mentions, color="r")
# + [markdown] toc-hr-collapsed=false
# ## Exploring NER Taggers
#
# So for this model, instead of using the actual tokens as the input for the model, we'll be using the NER tokens instead. This will give us a representation of how the sentences are structured, instead of just the word use. Essential for this is including bigrams!
#
# For creating these we'll look into using one of nltk/gensim/spacey, as they tend to represent the best/easiest implementations within python
# -
test_messages = twitter_df[twitter_df.source == "Twitter for Android"].text.values.tolist()[:10]
for msg in test_messages:
print(msg, "\n")
# ### NLTK
#
# `Didtrumptweetit.com` uses the stanford NER tagger from `nltk`, and although it probably works fine, it has an external java dependency which needs to be downloaded seperately. Since the performance difference isn't large, we'll choose for the easier options.
# + [markdown] toc-hr-collapsed=false
# ### Spacy
# -
import spacy
# We'll be using small for testing on our memory starved laptop
nlp = spacy.load('en_core_web_sm')
doc = nlp(test_messages[0])
for token in doc:
print(f"{token.text:<10} {token.tag_:>10} {spacy.explain(token.tag_)}")
doc = nlp(test_messages[1])
for token in doc:
print(f"{token.text:<10} {token.tag_:>10} {spacy.explain(token.tag_)}")
doc = nlp(test_messages[4])
for token in doc:
print(f"{token.text:<10} {token.tag_:>10} {spacy.explain(token.tag_)}")
# > Awesome, this looks like it's doing exactly what we want, we'll continue our model using this as the input
# ## Model Definition and Training
#
# We're going to package spacy's POS tagger into an sklearn transformer. We'll also need to speed it up a bit, as it's not the fastest currently.
#
# We've started with disabling the unnecessary model features, but currently the model is running in a parallel fashion either.
# ### Creating Customer Transformers
# +
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.metrics import roc_auc_score, confusion_matrix, roc_curve
# We need this for defining TfidfVectorizer such that it can be pickled
def identity(value):
return value
class NERFeatures(BaseEstimator, TransformerMixin):
def __init__(self, spacy_model):
self.spacy_model = spacy_model
self.nlp = spacy.load(spacy_model, disable=["parser", "ner"])
def fit(self, X, y=None):
return self
def transform(self, X, y=None):
return [[token.tag_ for token in doc] for doc in self.nlp.pipe(X)]
class ColumnSelector(BaseEstimator, TransformerMixin):
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
# -
def evaluate_model(X_test, y_test, clf, threshold=0.5):
print(f"Evaluating with threshold: {threshold}")
y_pred = clf.predict_proba(X_test)[:, 1] > threshold
# print Accuracy
print(f"TN: {sum(~y_test & ~y_pred)} FP: {sum(~y_test & y_pred)}")
print(f"FN: {sum(y_test & ~y_pred)} TP: {sum(y_test & y_pred)}")
# plot distributions of probabilities
probs = clf.predict_proba(X_test)[:, 0]
sns.distplot(probs[y_test == 0], color='g', norm_hist=False, label="true positive")
sns.distplot(probs[y_test == 1], color='r', norm_hist=False, label="true negative")
plt.axvline(x=threshold, color="orange")
plt.xlabel("Probability")
plt.ylabel("Density")
plt.legend()
plt.show()
# Plot ROC curve
auc = roc_auc_score(y_test, clf.predict_proba(X_test)[:,1])
fpr, tpr, threshold = roc_curve(y_test, clf.predict_proba(X_test)[:,1])
plt.plot(fpr,tpr, color="orange", label=f"AUC: {auc:.3f}")
plt.plot([0,1], [0,1], color='navy', linestyle='--')
plt.xlabel('False Positive Rate')
plt.ylabel('True Positive Rate')
plt.legend()
plt.show()
maxdiff = np.argmax(tpr - fpr)
maxdiff_val = threshold[maxdiff]
plt.plot(threshold, tpr, color="g", label="True Positive Rate")
plt.plot(threshold, fpr, color="r", label="False Positive Rate")
plt.plot(threshold, tpr - fpr, color="orange", label="TPR-FPR")
plt.xlabel("Threshold")
plt.ylabel("TP/FP Rate")
plt.xlim(0,1)
plt.legend()
plt.show()
# +
# # %%timeit -r1
# ner = NERFeatures("en_core_web_lg")
# ner.fit_transform(twitter_df.text)
# "en_core_web_sm" takes 5s for whole corpus, "lg" takes 15
# -
# ### Splitting into Train/Test
# +
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(twitter_df.drop("source", axis=1),
twitter_df.source == "Twitter for Android",
test_size = 0.1,
shuffle=False)
print(f"X_train time range: {X_train.created_at.dt.date.min()} to {X_train.created_at.dt.date.max()}")
print(f"X_train time range: {X_test.created_at.dt.date.min()} to {X_test.created_at.dt.date.max()}")
# -
# ## Model Training
# ### Model Creation and Training of Vanilla Model
# +
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import LogisticRegression
clf = Pipeline([
('selector', ColumnSelector('text')),
('ner', NERFeatures(spacy_model="en_core_web_sm")),
('tfidf', TfidfVectorizer(tokenizer=identity,
lowercase=False)),
('model', LogisticRegression(solver='lbfgs'))
])
clf.fit(X_train, y_train)
# -
evaluate_model(X_test, y_test, clf)
# ### Hyperparameter Optimization
# +
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.naive_bayes import GaussianNB
param_grid = {
'tfidf__ngram_range': [(1,2), (1,3), (1,4)],
'model__C': [1, 0.8, 0.6]
}
gs = GridSearchCV(estimator=clf,
param_grid=param_grid,
scoring='roc_auc',
cv=5,
n_jobs=-1,
verbose=True)
gs.fit(X_train, y_train)
# -
gs.best_score_
gs.best_params_
# ## Best Parameters with Large Spacy Model
# +
clf = Pipeline([
('selector', ColumnSelector('text')),
('ner', NERFeatures(spacy_model="en_core_web_sm")),
('tfidf', TfidfVectorizer(tokenizer=identity,
lowercase=False,
ngram_range=(1,3))),
('model', LogisticRegression(solver='lbfgs'))
])
clf.fit(X_train, y_train)
# -
evaluate_model(X_test, y_test, clf)
# +
import joblib
# We needed to pickle en_core_web_sm due to memory issues
joblib.dump(clf, "../models/trump_classifer_095AUC.pkl")
# -
# ## Evaluating Model Output
for msg in X_test[clf.predict(X_test)].text.tolist()[:10]:
print(msg, "\n")
for msg in X_test[~clf.predict(X_test)].text.tolist()[:10]:
print(msg, "\n")
| trump_classifier/notebooks/Recreating NER classifier.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: py2_pvfactors
# language: python
# name: py2_pvfactors
# ---
# PV Array geometry introduction
# ==========================
# In this section, we will learn how to:
#
# - create a 2D PV array geometry with PV rows at identical heights, tilt angles, and with identical widths
# - plot that PV array
# - calculate the inter-row direct shading, and get the length of the shadows on the PV rows
# Imports and settings
# +
# Import external libraries
import matplotlib.pyplot as plt
# Settings
# %matplotlib inline
# -
# ### Prepare PV array parameters
pvarray_parameters = {
'n_pvrows': 4, # number of pv rows
'pvrow_height': 1, # height of pvrows (measured at center / torque tube)
'pvrow_width': 1, # width of pvrows
'axis_azimuth': 0., # azimuth angle of rotation axis
'surface_tilt': 20., # tilt of the pv rows
'surface_azimuth': 90., # azimuth of the pv rows front surface
'solar_zenith': 40., # solar zenith angle
'solar_azimuth': 150., # solar azimuth angle
'gcr': 0.5, # ground coverage ratio
}
# ### Create a PV array and its shadows
# Import the ``OrderedPVArray`` class and create a transformed PV array object using the parameters above
# +
from pvfactors.geometry import OrderedPVArray
pvarray = OrderedPVArray.transform_from_dict_of_scalars(pvarray_parameters)
# -
# Plot the PV array
# Plot pvarray shapely geometries
f, ax = plt.subplots(figsize=(10, 3))
pvarray.plot(ax)
plt.show()
# As we can see in the plot above:
# - the blue lines represent the PV rows
# - the gray lines represent the shadows cast by the PV rows on the ground from direct light
# - the yellow lines represent the ground areas that don't get any direct shading
# - there are additional points on the ground that may seem out of place: but they are called "cut points" (or sometimes "edge points") and are necessary to calculate view factors. For instance, if you take the cut point located between the second and third shadows (counting from the left), it marks the point after which the leftmost PV row's back side is not able to see the ground anymore
# ### Situation with direct shading
# We can also create situations where direct shading happens either on the front or back surface of the PV rows.
# New configuration with direct shading
pvarray_parameters.update({'surface_tilt': 80., 'solar_zenith': 75., 'solar_azimuth': 90.})
pvarray_parameters
# Create new PV array
pvarray_w_direct_shading = OrderedPVArray.transform_from_dict_of_scalars(pvarray_parameters)
# Plot pvarray shapely geometries
f, ax = plt.subplots(figsize=(10, 3))
pvarray_w_direct_shading.plot(ax)
plt.show()
# We can now see on the plot above that some inter-row shading is happening in the PV array.
# It is also very easy to obtain the shadow length on the front surface of the shaded PV rows.
# Shaded length on first pv row (leftmost)
l = pvarray_w_direct_shading.pvrows[0].front.shaded_length
print("Shaded length on front surface of leftmost PV row: %.2f m" % l)
# Shaded length on last pv row (rightmost)
l = pvarray_w_direct_shading.pvrows[-1].front.shaded_length
print("Shaded length on front surface of rightmost PV row: %.2f m" %l)
# As we can see, the rightmost PV row is not shaded at all.
| docs/sphinx/tutorials/PVArray_introduction.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="iyEGINeSelnX" colab_type="code" colab={}
# # !pip install --upgrade tables
# # !pip install eli5
# + id="knwIQvrwd4Cf" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 172} outputId="c0894d7a-30d6-4886-8cb1-a1a79bd2212f" executionInfo={"status": "ok", "timestamp": 1583346736549, "user_tz": -60, "elapsed": 2926, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
import pandas as pd
import numpy as np
from sklearn.dummy import DummyRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import mean_absolute_error as mae
from sklearn.model_selection import cross_val_score
import eli5
from eli5.sklearn import PermutationImportance
# + id="5XkMhwN3ggwV" colab_type="code" outputId="a545515f-7c02-4886-c599-f180ddf43e1f" executionInfo={"status": "ok", "timestamp": 1583346741617, "user_tz": -60, "elapsed": 1406, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
# cd '/content/drive/My Drive/Colab Notebooks/dw_matrix/matrix_two/dw_matrix_car'
# + [markdown] id="iOTthjjrgPSW" colab_type="text"
# # Wczytywanie danych
# + id="FKWIDPr4gVYe" colab_type="code" outputId="92302b17-0994-4d02-e9a3-fa5f5f7498c5" executionInfo={"status": "ok", "timestamp": 1583346749224, "user_tz": -60, "elapsed": 4794, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
df = pd.read_hdf('data/car.h5')
df.shape
# + id="qH1EHqeAhNKF" colab_type="code" outputId="69acfb6f-f5c1-40fe-f8e8-78ac10dcd17f" executionInfo={"status": "ok", "timestamp": 1583346752932, "user_tz": -60, "elapsed": 644, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 194}
df.columns
# + id="NjskKQ8xjgP-" colab_type="code" outputId="8a7bc04e-d58f-41fe-9300-b8e91e99f4bf" executionInfo={"status": "ok", "timestamp": 1583346754930, "user_tz": -60, "elapsed": 848, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
df.select_dtypes(np.number).columns
# + [markdown] id="yvTAjccYiE-l" colab_type="text"
# # Dummy Model
# + id="12lQnxMnj9Rp" colab_type="code" outputId="636cffb3-0a56-4690-e7cd-7ac0bcfd52cd" executionInfo={"status": "ok", "timestamp": 1583346758143, "user_tz": -60, "elapsed": 731, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
df.select_dtypes(np.number).columns
# + id="cdmRFF8RiG4j" colab_type="code" outputId="ce099120-64c9-4260-b0c8-87529f92cafe" executionInfo={"status": "ok", "timestamp": 1583346760370, "user_tz": -60, "elapsed": 594, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
feats = ['car_id']
X = df[feats].values
y = df['price_value'].values
model = DummyRegressor()
model.fit(X,y)
y_pred = model.predict(X)
mae(y, y_pred)
# + id="NuaMgJ1Hi9X2" colab_type="code" outputId="b5dc271d-0e16-46a1-ce1e-2a4d86783860" executionInfo={"status": "ok", "timestamp": 1583346764585, "user_tz": -60, "elapsed": 629, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
[x for x in df.columns if 'price' in x]
# + id="SLleI9mhkTsg" colab_type="code" outputId="e6ec3900-d7ff-4482-e62a-75fbffb80d72" executionInfo={"status": "ok", "timestamp": 1583346766552, "user_tz": -60, "elapsed": 651, "user": {"displayName": "Kasia K", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 70}
df['price_currency'].value_counts()
# + id="dgNoKRIwkenh" colab_type="code" outputId="6595dfd9-4166-45d1-dfac-8c1d9af21aec" executionInfo={"status": "ok", "timestamp": 1583346769021, "user_tz": -60, "elapsed": 619, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}} colab={"base_uri": "https://localhost:8080/", "height": 34}
df = df[ df['price_currency'] != 'EUR' ]
df.shape
# + [markdown] id="7GUEE8oblJ5-" colab_type="text"
# ## Features
# + id="ImtU0Al-lLcN" colab_type="code" colab={}
SUFFIX_CAT = '__cat'
for feat in df.columns:
if isinstance(df[feat][0], list): continue
factorized_values = df[feat].factorize()[0]
if SUFFIX_CAT in feat:
df[feat] = factorized_values
else:
df[feat + SUFFIX_CAT] = factorized_values
# + id="SmHyOlb60Qrl" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="1c1ed802-2555-4a9e-df56-0fe1d1500e7f" executionInfo={"status": "ok", "timestamp": 1583346975009, "user_tz": -60, "elapsed": 857, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
cat_feats = [x for x in df.columns if SUFFIX_CAT in x]
cat_feats = [x for x in cat_feats if 'price' not in x]
len(cat_feats)
# + id="QrXkNGDy1GMH" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="b45d87c4-a4b0-4d13-d1e9-816b71cb1f18" executionInfo={"status": "ok", "timestamp": 1583347162387, "user_tz": -60, "elapsed": 4373, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
X = df[cat_feats].values
y = df['price_value'].values
model = DecisionTreeRegressor(max_depth=5)
scores = cross_val_score(model, X, y, cv=3, scoring='neg_mean_absolute_error')
np.mean(scores)
# + id="NnwAAM9T1mAL" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 383} outputId="9c4a2bb3-d6de-421f-c27f-ed8577d291ff" executionInfo={"status": "ok", "timestamp": 1583347457371, "user_tz": -60, "elapsed": 44048, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
m = DecisionTreeRegressor(max_depth=5)
m.fit(X,y)
imp = PermutationImportance(m, random_state=0).fit(X,y)
eli5.show_weights(imp, feature_names=cat_feats)
# + id="iobNJuEl3e6W" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 34} outputId="fb2d6dab-02a4-4f52-d2c4-a4168f981a41" executionInfo={"status": "ok", "timestamp": 1583347712708, "user_tz": -60, "elapsed": 1655, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
# !pwd
# + id="C4mo5QSN3kxv" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 247} outputId="4bc9abbf-0a2a-4882-eae6-20fc99af166f" executionInfo={"status": "ok", "timestamp": 1583347619323, "user_tz": -60, "elapsed": 3625, "user": {"displayName": "<NAME>", "photoUrl": "https://lh3.googleusercontent.com/a-/AOh14GjqKLyKobFs4gNUfoPNX66XsvfhkW7SFS25_UbkWQ=s64", "userId": "16687851511870375467"}}
# !git add day3_simple_model.ipynb
# !git commit -m "Add simple model"
| day3_simple_model.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# ---
# # Lista de scripts ou cli escritos em Python
#
# >Exemplos usados na aula da [semana 16](https://ericbrasiln.github.io/intro-historia-digital/mod4b/sem16.html)
# 1. Baixar vídeos do YouTube: [youtube-dl](https://github.com/ytdl-org/youtube-dl);
# 2. [resumos-anpuh-cli](https://github.com/ericbrasiln/resumos-anpuh-cli);
# 3. [Ferramentas Scielo v2](https://github.com/LABHDUFBA/ferramentas_scielo_v2);
# 4. Nuvem de palavras: [worldcloud](https://github.com/amueller/word_cloud)
#
#
#
| cclhm0069/mod4b/exemplos.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: conda_python3
# language: python
# name: conda_python3
# ---
# ## Object Detection with YOLOv5
# ### About
# ----------------
# This notebook provides a guided tour of deploying a YOLOv5 machine learning model pre-trained with MS COCO dataset using PyTorch to a Panorama appliance. More information about the model including the original model itself can be found in [this repository](https://github.com/ultralytics/yolov5) which is also included as a submodule under `3rdparty/yolov5`. More specifically, release `v3.0` of the repository was used to build this example.
#
# This example shows how to prepare a pre-trained model for deployment to a Panorama device. You can also train your own model using the resources from the aforementioned repository and deploy it to a Panorama appliance following the same steps.
#
# This is an example of inference done on an image captured from a test IP camera.
#
# 
# ### Imports & config
# ----------------
# *This notebook was tested with Torch v1.6.0 and TorchVision v0.7.0, you may need to upgrade those in case you are running older versions. If you are running this on Amazon SageMaker Notebook Instance then use `conda_python3` kernel*
# !pip install torch==1.6.0 torchvision==0.7.0
# Install extra libraries (not needed if using `conda_python3` on SageMaker Notebook Instance)
# !pip install boto3
# + jupyter={"outputs_hidden": false}
import sys
import time
import os
import random as rnd
import json
from matplotlib import pyplot as plt
from IPython.display import Image
import numpy as np
import cv2
import torch
import boto3
import utils
print(f'Using torch {torch.__version__}')
print(f'Using python {sys.version_info}')
# -
# ***Create your own AWS S3 Bucket making sure it contains `aws-panorama` in the bucket name***
# +
# Set this variable/constant value to be the full name of your bucket, for example "aws-panorama-example-xyz"
BUCKET = 'aws-panorama-<you-bucket-name-suffix>' # Bucket name must contain "aws-panorama"
MODELS_S3_PREFIX = 'models'
MODEL = 'yolov5s'
model_file = f'{MODEL}.pt'
traced_model_file = f'{MODEL}.pth'
traced_model_archive = f'{MODEL}.tar.gz'
LAMBDA = 'yolov5s'
LAMBDA_EXECUTION_ROLE_NAME = 'PanoramaYoloLambdaExecutionRole'
lambda_file = f'{LAMBDA}_lambda.py'
lambda_archive = lambda_file.replace('.py', '.zip')
TEST_IMAGE = 'test.png'
INPUT_SIZE = 640
THRESHOLD = 0.5
CLASSES = ['person', 'bicycle', 'car', 'motorcycle', 'airplane', 'bus', 'train', 'truck', 'boat', 'traffic light',
'fire hydrant', 'stop sign', 'parking meter', 'bench', 'bird', 'cat', 'dog', 'horse', 'sheep', 'cow',
'elephant', 'bear', 'zebra', 'giraffe', 'backpack', 'umbrella', 'handbag', 'tie', 'suitcase', 'frisbee',
'skis', 'snowboard', 'sports ball', 'kite', 'baseball bat', 'baseball glove', 'skateboard', 'surfboard',
'tennis racket', 'bottle', 'wine glass', 'cup', 'fork', 'knife', 'spoon', 'bowl', 'banana', 'apple',
'sandwich', 'orange', 'broccoli', 'carrot', 'hot dog', 'pizza', 'donut', 'cake', 'chair', 'couch',
'potted plant', 'bed', 'dining table', 'toilet', 'tv', 'laptop', 'mouse', 'remote', 'keyboard',
'cell phone', 'microwave', 'oven', 'toaster', 'sink', 'refrigerator', 'book', 'clock', 'vase',
'scissors', 'teddy bear', 'hair drier', 'toothbrush']
if torch.cuda.is_available():
device_type = 'GPU'
print(f'Found GPU: {torch.cuda.get_device_properties(0)}')
else:
device_type = 'CPU'
# Uncomment next like if you want to force running on CPU on a device with GPU
# device_type = 'CPU'
device = torch.device('cuda:0' if device_type == 'GPU' else 'cpu')
print(f'Using {device_type}', end='')
# -
s3_client = boto3.client("s3")
iam_client = boto3.client("iam")
lambda_client = boto3.client("lambda")
# ### The model
# ----------------
#
# Model preparation steps are completed using the `3rdparty/yolov5` submodule
#
# ##### Steps to prepare the model
# 1. Download and trace the model
# - Install YOLOv5 dependencies
# - Run a test inference. This will also download the pre-trained model as save it as `yolov5s.pt` file
# - Export the downloaded model to TorchScript format, saved as `yolov5.pth` file
# 2. Test the TorchScript model
# - Load the TorchScript model
# - Load the test image and prepare it
# - Put the test image through the model
# - Show results
# 3. Pack and upload the TorchScript model to S3
# ##### Download and trace
# !pip install -r 3rdparty/yolov5/requirements.txt
# +
device_str = 'cpu' if device_type == 'CPU' else '0'
out_dir = 'inference_results'
yolov4_dir = '3rdparty/yolov5'
# !python $yolov4_dir/detect.py --weights $model_file --img $INPUT_SIZE --conf $THRESHOLD --source $TEST_IMAGE \
# --output $out_dir --device $device_str
# !export PYTHONPATH=$yolov4_dir && python $yolov4_dir/models/export.py --weights $model_file --img-size $INPUT_SIZE
# !mv yolov5s.torchscript.pt $traced_model_file
Image(filename=f'{out_dir}/{TEST_IMAGE}', width=600)
# -
# ##### Test the traced model
traced_model = torch.jit.load(traced_model_file, map_location=device)
# Use an non-marked version of the test image
test_image = cv2.cvtColor(cv2.imread(TEST_IMAGE), cv2.COLOR_BGR2RGB)
plt.figure(figsize=(6, 6))
plt.imshow(test_image)
# Note: output of the YOLOv5 model requires further processing (Non Max Suppression) which can be done on GPU using PyTorch but on a Panorama appliance it needs to be executed on CPU (also applies to execution of model's Detector layer logic), adding significant overhead to the overall inference process.
# +
processor = utils.Processor(CLASSES, INPUT_SIZE, threshold=THRESHOLD, keep_ratio=True)
tm = time.time()
img = torch.from_numpy(processor.preprocess(test_image)).to(device)
print(f'Pre-process: {int((time.time() - tm) * 1000)} msec')
# Do a warm-up run before timing inference
traced_model(img)
tm = time.time()
results = traced_model(img)
print(f'Inference: {int((time.time() - tm) * 1000)} msec')
test_results = [r.cpu().numpy() for r in results]
tm = time.time()
_, result_image = processor.post_process(test_results, test_image.shape, test_image.copy())
print(f'Post-process: {int((time.time() - tm) * 1000)} msec')
plt.figure(figsize=(6, 6))
plt.imshow(result_image)
# -
# ##### Pack and upload the model archive to S3 bucket
#
# Take a note of an S3 location of the uploaded model archive, you'll need it during the application creation process.
# +
# !tar -czvf $traced_model_archive $traced_model_file
traced_model_key = os.path.join(MODELS_S3_PREFIX, traced_model_archive)
s3_client.upload_file(traced_model_archive, Bucket=BUCKET, Key=traced_model_key)
traced_model_s3_url = os.path.join(f's3://{BUCKET}', traced_model_key)
print(f'Uploaded model archive to {traced_model_s3_url}')
# -
# Alternatively, you can upload the model archive to S3 bucket using AWS Console or running the following AWS CLI command ***if you have AWS CLI installed and configured*** (change the cell to `Code` type before running)
# + active=""
# !aws s3 cp $traced_model_archive $traced_model_s3_url
# !aws s3 ls $traced_model_s3_url --human-readable
# -
# ### The Application
# ---------------------
# This is the script that will be deployed and executed on the Panorama Appliance as a lambda function. It can found in the same folder as this notebook along with another file `utils.py`, containing some helper scripts.
# !pygmentize $lambda_file
# #### Create and deploy lambda
#
# - If the execution of the code in this cell fails then make sure you have the rights to creates roles in AWS IAM
# - You only need to execute the next cell once. All the subsequent deployments will use the same role
lambda_execution_role_policy = {
"Version": "2012-10-17",
"Statement":[
{
"Effect": "Allow",
"Principal": {"Service": ["lambda.amazonaws.com", "events.amazonaws.com"]},
"Action": "sts:AssumeRole",
}
]
}
iam_client.create_role(
RoleName=LAMBDA_EXECUTION_ROLE_NAME,
AssumeRolePolicyDocument=json.dumps(lambda_execution_role_policy),
)
# ##### Create a new Lambda Function
#
# *Use the cell in the [OPTIONAL] cell below if you want to re-deploy lambda after the initial deployment*
#
# You can inspect the created AWS Lambda Function following the link shown after running the next cell
# !zip -o $lambda_archive $lambda_file utils.py
# +
# Uncomment and run the following if you already have a function and want to re-create it
lambda_client.delete_function(FunctionName=LAMBDA)
with open(lambda_archive, "rb") as f:
zipped_code = f.read()
lambda_execution_role = iam_client.get_role(RoleName=LAMBDA_EXECUTION_ROLE_NAME)
lambda_response = lambda_client.create_function(
FunctionName=LAMBDA,
Runtime="python3.7",
Role=lambda_execution_role["Role"]["Arn"],
Handler=lambda_file.replace('.py', '.main()'),
Code=dict(ZipFile=zipped_code),
Timeout=120,
MemorySize=2048,
Publish=True)
template = "https://console.aws.amazon.com/lambda/home?region=us-east-1#/functions/{}/versions/{}?tab=configuration"
lambda_url = template.format(lambda_response["FunctionName"], lambda_response["Version"])
print(lambda_url)
# -
# ##### [OPTIONAL] Subsequent deployments
# Convert the next cell to Code type and run the following cell if you want to re-deploy the lambda function again, e.g. if you make changes to application code and want to deploy those changes to the Panorama appliance
# + active=""
# !rm $lambda_archive
# !zip -o $lambda_archive $lambda_file utils.py
#
# with open(lambda_archive, "rb") as f:
# zipped_code = f.read()
#
# lambda_response = lambda_client.update_function_code(
# FunctionName=LAMBDA,
# ZipFile=zipped_code,
# Publish=True)
#
# template = "https://console.aws.amazon.com/lambda/home?region=us-east-1#/functions/{}/versions/{}?tab=configuration"
# lambda_url = template.format(lambda_response["FunctionName"], lambda_response["Version"])
# print(lambda_url)
# -
# ### Deploy the Application to Panorama appliance
# ---------------------
# At the time of writing this the only way to deploy the Application to the Panorama device is through the [AWS Panorama Console](https://console.aws.amazon.com/panorama). Instructions for script-based deployment will be added here after alternative means of deployment are available, e.g. via AWS CLI or Python SDK.
#
# Few things to remember when deploying the Application to your Panorama appliance in AWS Panorama Console:
#
# - when creating a new model (as part of a new Application creation process) in AWS Panorama Console:
# - use the model archive uploaded to S3 earlier in this notebook to create a new **External model** (e.g. `s3://< your bucket >/models/yolov5s.tar.gz`)
# - make sure that the **Model name** you specify matches exactly the model name used in your Application/lambda code (it is stored in the variable/constant named **MODEL** in the current version of the Application/labmda code) *
# - select `PyTorch` as *Model framework*
# - add input with **Input name** `data` and **Input shape** `1,3,640,640`
#
# - first deployment of the Application takes a bit longer due to initial conversion of the model done by AWS SageMaker Neo behind the scene. Subsequent deployments using the same model will be faster if you only change the Application code (which is usually the case)
# - to troubleshoot any issues start with looking at the logs in [AWS CloudWatch](https://console.aws.amazon.com/cloudwatch). In the AWS CloudWatch Console, click on **Log Groups** under **Logs** and select a click on a link that has a name of the lambda function corresponding to your Application (something like `/aws/greengrass/Lambda/us-east-1/<your account number>/yolov5s`)
#
# ***Note:*** *code versions may change making it out of sync with comments in this notebook, always use the latest values from the code when referred to*
# ### What's next?
# ---------------------
# This was just a taster to show you how to run a PyTorch based YOLOv5 model on Panorama appliance. Next logical step would be fine-tuning the pre-trained YOLOv5 model using your own dataset to recognise your own object types. Examples of doing that are available in the `3rdparty/yolov5` submodule.
| Object-Detection-YOLOv5/yolov5s.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: TCGA
# language: python
# name: tcga
# ---
# #%%appyter init
from appyter import magic
magic.init(lambda _=globals: _())
# # TCGA Data Loader
# ---
#
# An appyter that interfaces with the The Cancer Genome Atlas (TCGA) API to simplify the process of obtaining this dataset's RNA-seq and clinical data tables.
#
# The final output of this notebook is two dictionaries:
# 1. `cancer_rna_dfs`: maps each cancer type to a pandas DataFrame containing all RNA seq profiles obtained under that type (according to the chosen parameters).
# - For each DataFrame, the **rows** are genes and **columns** are cases (referenced by their case_id).<br>
#
#
# 2. `cancer_clinical_dfs`: maps each cancer type to a pandas DataFrame containing all clinical data obtained under that type (according to the chosen parameters).
# - For each DataFrame, the **rows** are cases (referenced by case_id) and **columns** are clinical data fields.
#
# For more information on the TCGA dataset, you can browse their [Data Dictionary](https://docs.gdc.cancer.gov/Data_Dictionary/viewer/) and [API Reference](https://docs.gdc.cancer.gov/API/Users_Guide/Getting_Started/).
#
# *Note*: it is recommended to download the notebook and run it locally, which you would have to do anyway to be able to access the produced data objects for further analysis or writing to disk.
#
# Script to acquire RNA-Seq and clinical data from TCGA
import requests
import json
import os
import re
import gzip
import shutil
import tarfile
import pathlib
import pandas as pd
import numpy as np
from maayanlab_bioinformatics.harmonization import ncbi_genes
import math
import io
from gzip import GzipFile
from IPython.display import display
from urllib.error import URLError
# +
# %%appyter hide
{% do SectionField(
name="RNASeq",
title="RNA-seq specifications",
img = "tcga-logo.png"
) %}
{% do SectionField(
name="CLINICAL",
title="Clincial data specifications",
img = "tcga-logo.png"
) %}
{% set rna_types = MultiChoiceField(
name = "RNA_types",
label = "Types of RNAs to include in the output",
choices = ['protein-coding','pseudo', 'other', 'unknown','ncRNA','tRNA','rRNA','scRNA','snoRNA','snRNA','biological-region'],
section="RNASeq",
default=[],
) %}
{% set clinical_fields = MultiChoiceField(
name = "clinical_fields",
label = "Clinical fields to load",
choices = {
'demographic': 'Demographics',
'diagnoses': 'Diagnoses',
'exposures': 'Exposures'
},
section="CLINICAL",
default=[],
) %}
# +
# %%appyter code_eval
rna_types = {{rna_types}}
map_ids = {{ BoolField(
name = "map_ids",
label = "Map ensembl IDs to Entrez gene symbols?",
description = "If a gene symbol is unavailable (as in the case of non protein-coding RNAs), it will be left as its Ensembl ID.",
default = True,
section = "RNASeq"
) }}
include_both_ids = {{BoolField(
name = "include_both_ids",
label = "Include both ensembl IDs and symbols in final output?",
default = not map_ids,
section = "RNASeq"
) }}
include_rna_types = {{BoolField(
name = "include_rna_types",
label = "Include RNA types in final output?",
default = True,
section = "RNASeq"
) }}
min_cases = {{IntField(
name = "min_documents",
label = "Minimum cases per cancer",
description = "The minimum number of cases under a single cancer type for that data to be loaded into the dataset.",
section="RNASeq",
default=150,
min=0,
max=100000
) }}
clinical_fields = {{clinical_fields}}
# +
# Endpoints
base_url = 'https://api.gdc.cancer.gov/'
files_endpt = base_url + 'files/'
genes_endpt = base_url + 'genes/'
cases_endpt = base_url + 'cases/'
data_endpt = base_url + "data/"
json_header = {"Content-Type": "application/json"}
workflow_type = "HTSeq - Counts"
# -
# ### Download RNA-seq data
# +
# Build params
filters = {
"op": "and",
"content":[
{
"op": "=",
"content":
{
"field": "files.experimental_strategy",
"value": "RNA-Seq",
}
},
{
"op": "=",
"content":
{
"field": "access",
"value": "open",
}
},
{
"op": "=",
"content":
{
"field": "files.analysis.workflow_type",
"value": workflow_type,
}
},
]
}
# +
# Get list of all cancer types returned from the search parameters
params = {
"filters": filters,
"size":"0",
"facets":"cases.diagnoses.primary_diagnosis",
}
response = requests.post(files_endpt, data = json.dumps(params), headers=json_header).json() # optionally also provide params argument
buckets = response["data"]["aggregations"]["cases.diagnoses.primary_diagnosis"]["buckets"]
print(f'{len(buckets)} total cancer types\n')
cancer_types = [ y["key"] for y in list(filter(lambda x: x["doc_count"] >= min_cases, buckets)) ]
print(f'{len(cancer_types)} cancer types with at least {min_cases} associated files')
# +
def make_cancer_params(cancer_type):
# filter results for this cancer type
cancer_filters = {
"op": "and",
"content": [
*filters["content"],
{
"op": "=",
"content":
{
"field": "cases.diagnoses.primary_diagnosis",
"value": cancer_type,
}
}
]
}
fields = "file_id,file_name,cases.case_id"
# build parameters object
return {
"fields": fields,
"filters": json.dumps(cancer_filters),
"size": 100000 # do not limit size
}
def get_uuids(cancer_type):
# get list of all files with RNA-seq results
response = requests.get(files_endpt, params = make_cancer_params(cancer_type))
data = json.loads(response.content.decode("utf-8"))
# get list of results
results = data["data"]["hits"]
# get list of file and case uuids
file_uuid_list = [ entry["file_id"] for entry in results]
case_uuid_list = [ entry["cases"][0]["case_id"] for entry in results]
# create a dictionary to map file uuids to case uuids
files_to_cases = {}
for entry in results:
files_to_cases[entry["file_id"]] = entry["cases"][0]["case_id"]
return file_uuid_list, case_uuid_list, files_to_cases
def get_files(uuid_list,files_to_cases):
params = {"ids": uuid_list}
df = pd.DataFrame({"ensembl_id": []}).set_index("ensembl_id")
# A POST is used, so the filter parameters can be passed directly as a Dict object.
response = requests.post(data_endpt, data=json.dumps(params), headers=json_header)
# filename is found in the Content-Disposition header of response
response_head_cd = response.headers["Content-Disposition"]
file_name = re.findall("filename=(.+)", response_head_cd)[0]
# write the file content (bytes) and load the tar.gz file
file_object = io.BytesIO(response.content)
tar = tarfile.open(fileobj=file_object)
# access files within the zipped file
for file in tar.getmembers():
if (file.name == "MANIFEST.txt"): continue
f=tar.extractfile(file)
bytestream=io.BytesIO(f.read())
got_text = GzipFile(None, 'rb', fileobj=bytestream).read().decode('utf-8')
# set column name to uuid of corresponding case for this file
file_name = file.name.split("/")[1].split(".")[0]
new_df = pd.read_csv(io.StringIO(got_text), sep="\t",header=None, names=["ensembl_id", file_name])
# collapse all versioned names of genes to just gene name so we can merge
new_df.ensembl_id.replace(to_replace = r'\..*$', value = "", regex=True, inplace=True)
new_df = new_df.set_index("ensembl_id")
df = pd.DataFrame.merge(df, new_df, how="outer", left_index = True, right_index = True)
tar.close()
# drop rows not corresponding to genes (i.e. metadata)
non_genes = list(filter(lambda val: not "ENSG" in val, list(df.index.values)))
df = df.drop(non_genes)
return df
def get_ncbi_df():
# Map Ensebml ids to Entrez gene symbols
ncbi = pd.DataFrame(ncbi_genes.ncbi_genes_fetch())
all_ids = ncbi.dbXrefs.values
def get_ensembl_id(ids):
ids = "".join(ids)
ensembl = re.findall("Ensembl:(.*)", ids)
if (len(ensembl) == 1):
return ensembl[0]
else:
return None
ensembl_ids = [ get_ensembl_id(ids) for ids in all_ids]
ncbi = ncbi[["dbXrefs", "Symbol", "type_of_gene"]]
ncbi["ensembl"] = ensembl_ids
ncbi = ncbi.drop(columns=["dbXrefs"])
ncbi = ncbi.set_index("ensembl")
return ncbi
def map_ncbi_data(df, ncbi, rna_types):
ensembl_to_gene_type = ncbi.to_dict()["type_of_gene"]
ensembl_to_symbol = ncbi.to_dict()["Symbol"]
data_ensembl_ids = df.index.to_list()
def id_to_type(key):
if (key in ensembl_to_gene_type):
return ensembl_to_gene_type[key]
else:
return None
# if the key is present, return it; otherwise, set the index for the corresponding row as its ensembl id
def id_to_symbol(key):
if (key in ensembl_to_symbol):
return ensembl_to_symbol[key]
else:
return key # if entrez symbol not found, keep as ensembl id
data_types = [ id_to_type(key) for key in data_ensembl_ids ]
data_symbols = [ id_to_symbol(key) for key in data_ensembl_ids ]
df["type_of_gene"] = data_types
df["symbol"] = data_symbols
df = df[df['type_of_gene'].isin(rna_types)]
df = df.reindex(columns=(['symbol','type_of_gene'] + list([a for a in df.columns if not a in ['symbol','type_of_gene']] )))
return df
# +
# %%appyter code_exec
{% if rna_types.value == [] %}
print("No RNA types were selected for data collection.")
{% else %}
# note: this will take a while!
try:
ncbi = get_ncbi_df()
cancer_rna_dfs = {}
cancer_cases = {}
for cancer in cancer_types:
file_uuid_list, case_uuid_list, files_to_cases = get_uuids(cancer)
cancer_cases[cancer] = case_uuid_list # save the case ids to retrieve for clinical data
num_files = len(file_uuid_list)
num_cases = len(case_uuid_list)
print(f"{cancer}: \n{num_files} files\n{num_cases} cases\n")
df_rna = get_files(file_uuid_list,files_to_cases)
df_rna = map_ncbi_data(df_rna, ncbi, rna_types)
if (map_ids and not include_both_ids):
df_rna = df_rna.drop("ensembl_id").set_index("symbol")
if (not include_rna_types):
df_rna = df_rna.drop("type_of_gene")
print(f"Got table for {cancer} with {df_rna.shape[0]} genes")
# display(df_rna.head())
cancer_rna_dfs[cancer] = df_rna
except URLError, e:
print e.code
print('Network error occurred. Note that these scripts run more effectively if downloaded and run locally.')
{% endif %}
# -
# ### Clinical Data
# +
# %%appyter code_exec
{% if clinical_fields.value == [] %}
print("No clinical data fields were selected.")
{% else %}
# get all clinical fields and convert to column names
cases_fields = requests.get(cases_endpt + "_mapping").json()["fields"]
def filter_field(x):
for field in clinical_fields:
if field in x: return True
return False
all_clinical_fields = list(filter(filter_field, cases_fields))
columns = list(set([ x.split(".")[1] for x in all_clinical_fields]))
print(f'{len(columns)} total clinical data columns.')
{% endif %}
# +
# %%appyter code_exec
{% if not clinical_fields.value == [] %}
# note: this will also take a while!
cancer_clinical_dfs = {}
try:
for cancer in cancer_types:
df_clinical = pd.DataFrame({}, columns=columns)
df_clinical["case_id"] = []
# get demographics and diagnosis data for each case,
# merging with pre-exisiting dataframe
for case in cancer_cases[cancer]:
fields=",".join(all_clinical_fields)
params={
"fields": fields
}
response = requests.get(cases_endpt + case, params=params).json()["data"]
all_data = {}
for field_group in clinical_fields:
data = response[field_group]
print(data)
if field_group == "diagnoses":
data = response[field_group][0]
if "treatments" in data:
del data["treatments"] # do not load treatment data
all_data = {**all_data, **data}
df_case = pd.DataFrame(all_data, index=[case])
df_case.head()
df_case["case_id"] = case
df_clinical = pd.concat([df_clinical, df_case], join="outer")
df_clinical = df_clinical.set_index("case_id")
# make first column "primary_diagnosis" for easy reference
cols = ['primary_diagnosis'] + [col for col in df_clinical.columns.values if col != 'primary_diagnosis']
df_clinical = df_clinical[cols]
cancer_clinical_dfs[cancer] = df_clinical
except URLError, e:
print e.code
print('Network error occurred. Note that these scripts run more effectively if downloaded and run locally.')
{% endif %}
# +
# utility functions to save the data
# All files are saved to the same directory, with the name "{cancer}_data.csv" and "{cancer}_clinical_data.csv"
# for RNA-seq and clinical data, respectively.
def save_rna_data(cancer_rna_dfs, path):
for cancer in cancer_types:
cancer_rna_dfs[cancer].to_csv(f"{path}/{cancer}_data.csv", encoding='utf-8')
def save_clinical_data(cancer_clinical_dfs, path):
for cancer in cancer_types:
cancer_clinical_dfs[cancer].to_csv(f"{path}/{cancer}_clinical_data.csv", encoding='utf-8')
| appyters/TCGA_Data_Loader/TCGA_Data_Loader.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="e-jf5KTgGF84" colab_type="code" colab={}
import torch
import torchvision
import torchvision.transforms as transforms
import torchvision.utils as vutils
import matplotlib.pyplot as plt
import numpy as np
import random
import math
import os
# + [markdown] id="fHNUapqlO-JF" colab_type="text"
# # Configs
# + id="1xT7wJhpO_Tp" colab_type="code" outputId="02d4836d-61cc-48d8-f4ba-5d15c8fe11a5" colab={"base_uri": "https://localhost:8080/", "height": 53}
data_PATH = os.path.join("/content/celeba/")
log_PATH = os.path.join("/gdrive/My Drive/notebooks/logs/began-128")
modelName = "BEGAN-celeba_128x128"
batch_size =16
workers = 2
h_dim = 128
gf_dim = 64
df_dim = 64
in_h = 128
in_w =128
c_dim = 3
start_step = 0
lr_update_step = 100*1000
max_step = 200 * 1000
learning_rate = 0.00008
lr_gamma = 0.5
beta1=0.5
beta2=0.9
betas = (beta1,beta2)
gamma = 0.5
lambda_k = 0.001
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
manualSeed = 3734
print("Random Seed: ",manualSeed)
random.seed(manualSeed)
torch.manual_seed(manualSeed)
# + [markdown] id="I5g40yR0YvT2" colab_type="text"
# # DataLoad
# + id="SzHzP_DgWlfQ" colab_type="code" colab={}
transform = transforms.Compose(
[
transforms.CenterCrop(160),
transforms.Resize((in_h,in_w)),
transforms.ToTensor(),
transforms.Normalize((0.5,0.5,0.5),(0.5,0.5,0.5)),
]
)
def transform_inverse (y):
t= None
if y.size()[0]==1:#1-dim
t=torchvision.transforms.Normalize((-1,),(2,))
else :#3-dim
t=torchvision.transforms.Normalize((-1,-1,-1),(2,2,2))
return t(y)
# + id="_1qx3qO0ZsYv" colab_type="code" outputId="9074cbaa-f787-43a1-dc40-c7650c017068" colab={"base_uri": "https://localhost:8080/", "height": 197}
celeba_dataset=torchvision.datasets.ImageFolder(root = data_PATH,transform=transform)
train_loader = torch.utils.data.DataLoader(celeba_dataset,batch_size=batch_size,
shuffle =True, num_workers=workers)
train_iter=iter(train_loader)
print(celeba_dataset)
# + id="q8tfW36caTwH" colab_type="code" outputId="96ec0b9a-0668-4768-b2cb-d02f274fdaa4" colab={"base_uri": "https://localhost:8080/", "height": 609}
real_batch = next(iter(train_loader))
plt.figure(figsize=(10,10))
plt.axis('off')
plt.title('Training Images')
plt.imshow(np.transpose(vutils.make_grid(real_batch[0].to(device),nrow=4,padding=2, normalize=True).cpu(),(1,2,0)))
print(real_batch[0].size())
# + [markdown] id="eijUWIipbqIC" colab_type="text"
# # Model
# + id="Lt-IQ1y9wooH" colab_type="code" colab={}
import torch.nn as nn
def conv_elu(in_dim, out_dim, kernel_size,stride,padding=0, bias=True):
return nn.Sequential(
nn.Conv2d(in_dim,out_dim, kernel_size=kernel_size, stride=stride,padding=padding,bias=bias),
nn.ELU(inplace=True)
)
def encoder_net(input_height, input_dim, df_dim,h_dim):
repeat_times = int(np.log2(input_height)) - 2
encoder_list = []
encoder_list.append( conv_elu(input_dim,df_dim,3,1,1) )
out_dim = df_dim
for idx in range(repeat_times):
prev_dim = out_dim
out_dim = df_dim * (idx + 1)
encoder_list.append(conv_elu(prev_dim,out_dim,3,1,1))
encoder_list.append(conv_elu(out_dim,out_dim,3,1,1))
if idx < repeat_times - 1:
encoder_list.append(conv_elu(out_dim,out_dim,3,2,1))
encoder_list.append(nn.Sequential(
nn.Flatten(1),
nn.Linear(8*8*out_dim,h_dim)
))
return nn.Sequential(*encoder_list)
def decoder_cnn(input_height, output_dim, gf_dim):
repeat_times = int(np.log2(input_height)) - 2
decoder_list = []
for idx in range(repeat_times):
decoder_list.append(conv_elu(gf_dim,gf_dim,3,1,1))
decoder_list.append(conv_elu(gf_dim,gf_dim,3,1,1))
if idx < repeat_times - 1:
decoder_list.append(nn.UpsamplingNearest2d(scale_factor=2))
decoder_list.append(nn.Conv2d(gf_dim, output_dim, 3,1,1))
return nn.Sequential(*decoder_list)
# + id="Pxt3wYzdcCuP" colab_type="code" colab={}
import torch.nn as nn
class Discriminator(nn.Module):
def __init__(self):
super(Discriminator,self).__init__()
self.encoder = encoder_net(in_h,c_dim,df_dim,h_dim)
self.decoder_linear = nn.Linear(h_dim,8*8*df_dim)
self.decoder_cnn = decoder_cnn(in_h,c_dim,df_dim)
def forward(self, x):
x = self.encoder(x)
x = self.decoder_linear(x).view(-1,df_dim,8,8)
x = self.decoder_cnn(x)
return x
# + id="lgCGbHPZG_vH" colab_type="code" colab={}
class Generator(nn.Module):
def __init__(self):
super(Generator,self).__init__()
self.decoder_linear = nn.Linear(h_dim,8*8*gf_dim)
self.decoder_cnn = decoder_cnn(in_h,c_dim,gf_dim)
def forward(self,x):
x = self.decoder_linear(x).view(-1,gf_dim,8,8)
x = self.decoder_cnn(x)
return x
# + [markdown] id="rMRV4pBEQ4Jy" colab_type="text"
# # Train Cofigs
# + id="eXc5NNg_Hkno" colab_type="code" colab={}
import torch.optim as optim
from torch.optim.lr_scheduler import StepLR
D = Discriminator().to(device)
G = Generator().to(device)
G_optimizer = optim.Adam(G.parameters(),lr = learning_rate, betas = betas)
D_optimizer = optim.Adam(D.parameters(),lr = learning_rate, betas = betas)
g_scheduler = StepLR(G_optimizer,lr_update_step,gamma=lr_gamma)
d_scheduler = StepLR(D_optimizer,lr_update_step,gamma=lr_gamma)
k_t = 0.0
L1_criterion = nn.L1Loss()
fixed_noise = torch.rand(64,h_dim).to(device) * 2 - 1
# + id="VLrjnSj-IeeB" colab_type="code" outputId="2831ec40-e621-4b9e-84a6-b0b7a032e2fe" colab={"base_uri": "https://localhost:8080/", "height": 1000}
def weights_init(m):
classname = m.__class__.__name__
if classname.find('Conv') != -1:
m.weight.data.normal_(0.0, 0.02)
elif classname.find('BatchNorm') != -1:
m.weight.data.normal_(1.0, 0.02)
m.bias.data.fill_(0)
G.apply(weights_init)
D.apply(weights_init)
# + id="nokFjsBdLHuK" colab_type="code" colab={}
def denorm_clipping_img(image):
return torch.clamp( ((image + 1)/2), 0, 1)
def norm_img(image):
return image/0.5 - 1.
# + id="VhDi8VQzLKVG" colab_type="code" colab={"base_uri": "https://localhost:8080/", "height": 500} outputId="0274aea5-96fb-4c7b-a5a5-9f579cdf9f83"
with torch.no_grad():
fake_batch=G(fixed_noise)
fake_batch=denorm_clipping_img(fake_batch)
plt.figure(figsize=(8,8))
plt.axis('off')
plt.title('fake Images Before learning')
plt.imshow(np.transpose(vutils.make_grid(fake_batch.to(device).view(64,c_dim,in_h,in_w),padding=2, normalize=False).cpu(),(1,2,0)))
# + id="MsK6kVMgc0vM" colab_type="code" colab={}
import pickle
img_list = []
G_losses = []
D_losses = []
real_losses = []
fake_losses = []
convergence_measure_list = []
transform_PIL=transforms.ToPILImage()
def log_list_save(l,file_name):
with open(os.path.join(log_PATH ,file_name+".logs"), "wb") as fp:
pickle.dump(l, fp)
def log_list_load(file_name):
with open(os.path.join(log_PATH ,file_name+".logs"), "rb") as fp:
return pickle.load(fp)
log_step = 100
save_step = 1000
# + [markdown] id="N029FJ4tT1h9" colab_type="text"
# # Train
# + colab_type="code" outputId="b0aa4c05-824b-4caa-c7f0-8e5e5fc84975" id="JC4DgrHgaEO7" colab={"base_uri": "https://localhost:8080/", "height": 179}
for step in range(start_step,max_step):
try:
real_data,_ = train_iter.next()
except StopIteration:
train_iter = iter(train_loader)
batch_size=real_data.shape[0]
real_data = norm_img(real_data.to(device))
z = torch.rand(batch_size,h_dim).to(device) * 2 - 1 # [-1,1] uniform
fake_data = G(z)
D.zero_grad()
d_loss_real =L1_criterion(D(real_data),real_data)
d_loss_fake =L1_criterion(D(fake_data.detach()),fake_data.detach())
d_loss = d_loss_real - k_t * d_loss_fake
d_loss.backward()
D_optimizer.step()
G.zero_grad()
g_loss = L1_criterion(D(fake_data),fake_data)
g_loss.backward()
G_optimizer.step()
balance = (gamma * d_loss_real - d_loss_fake).data
k_t = torch.clamp(k_t+lambda_k*balance, 0, 1).item()
convergence_measure = (d_loss_real + torch.abs(balance)).item()
# lr schduler
d_scheduler.step()
g_scheduler.step()
if step % log_step ==0:
print("Steps [{}/{}], convergence: {:.4f}, D_loss: {:.4f}, G_loss: {:.4f}, real_loss: {:.4f}, fake_loss: {:.4f}, k_t: {:.2f}"
.format(step,max_step, convergence_measure,d_loss.item(), g_loss.item(), d_loss_real.item(), d_loss_fake.item(), k_t))
D_losses.append(d_loss.item())
G_losses.append(g_loss.item())
real_losses.append(d_loss_real.item())
fake_losses.append(d_loss_fake.item())
convergence_measure_list.append(convergence_measure)
log_list_save(D_losses,os.path.join(log_PATH,"D_losses"))
log_list_save(G_losses,os.path.join(log_PATH,"G_losses"))
log_list_save(real_losses,os.path.join(log_PATH,"real_losses"))
log_list_save(fake_losses,os.path.join(log_PATH,"d_loss_fake"))
log_list_save(convergence_measure_list,os.path.join(log_PATH,"convergence_measure_list"))
if step % save_step == save_step - 1:
torch.save(G.state_dict(),os.path.join(log_PATH,"G_"+modelName+".pth"))
torch.save(D.state_dict(),os.path.join(log_PATH,"D_"+modelName+".pth"))
with torch.no_grad():
fake = G(fixed_noise).detach().cpu()
fake = denorm_clipping_img(fake) # instead of tanh activation
img_list.append(vutils.make_grid(torch.reshape(fake,(64,c_dim,in_h,in_w)), padding=2, normalize=False))
transform_PIL(img_list[-1]).save(os.path.join(log_PATH,str(step)+modelName+"_Last.png"))
# + [markdown] id="S1ikAplLawoR" colab_type="text"
# # Test
# + id="qIj34Rl9bKt1" colab_type="code" colab={}
with torch.no_grad():
fake_batch=G(fixed_noise)
fake_batch=denorm_clipping_img(fake_batch)
plt.figure(figsize=(8,8))
plt.axis('off')
plt.title('fake Images')
plt.imshow(np.transpose(vutils.make_grid(fake_batch.to(device).view(64,c_dim,in_h,in_w),padding=2, normalize=False).cpu(),(1,2,0)))
# + id="kg4rJm1NbPrO" colab_type="code" colab={}
for i in range(4):
if i ==0:
real_batch, _ = next(iter(train_loader))
else:
data, _ = next(iter(train_loader))
real_batch=torch.cat([real_batch, data], dim=0)
# Plot the real images
fig=plt.figure(figsize=(15,8))
plt.subplot(1,2,1)
plt.axis("off")
plt.title("Real Images")
plt.imshow(np.transpose(vutils.make_grid(real_batch.to(device), padding=5, normalize=False).cpu(),(1,2,0)))
# Plot the fake images from the last epoch
plt.subplot(1,2,2)
plt.axis("off")
plt.title("Fake Images")
plt.imshow(np.transpose(img_list[-1],(1,2,0)))
fig.savefig(os.path.join(log_PATH,"Compare_with_real"))
# + id="MHQEZN-2cjif" colab_type="code" colab={}
from PIL import Image
transform_PIL=transforms.ToPILImage()
p_img_list = [transform_PIL(p_image) for p_image in img_list]
p_img_list[0].save(os.path.join(log_PATH,modelName+'.gif'), save_all=True,append_images=p_img_list[1:], optimize=False, duration=0.5, loop=0)
p_img_list[-1].save(os.path.join(log_PATH,modelName+"_last_result.png"))
# + [markdown] id="RwSS1fgUcvyI" colab_type="text"
# # Logging
# + id="uY9mgPYqcqlI" colab_type="code" colab={}
x_len = int(max_step/log_step)
X = range(x_len)
plt.plot(X,D_losses,label="D loss")
plt.plot(X,G_losses,label="G loss")
plt.legend(loc=2)
plt.xticks(np.arange(0,x_len+1,250))
plt.ylabel("loss")
plt.xlabel("steps (0.1k)")
#plt.show()
plt.savefig(os.path.join(modelName+"_loss_figure_D,G.png"))
# + id="OnV5mGZ2hCRv" colab_type="code" colab={}
plt.plot(X,real_losses,label="Real L1")
plt.plot(X,fake_losses,label="Fake L1")
plt.legend(loc=2)
plt.xticks(np.arange(0,x_len+1,250))
plt.ylabel("loss")
plt.xlabel("steps (0.1k)")
#plt.show()
plt.savefig(os.path.join(modelName+"_loss_figure_Real,Fake.png"))
# + id="FQ7s6MEwg-ki" colab_type="code" colab={}
plt.plot(X,convergence_measure_list,label="Convergence Measure")
plt.legend(loc=2)
plt.xticks(np.arange(0,x_len+1,250))
plt.ylabel("Convergence")
plt.xlabel("steps (0.1k)")
#plt.show()
plt.savefig(os.path.join(modelName+"_loss_figure_convergence.png"))
# + id="eH5JYYFUgPTf" colab_type="code" colab={}
torch.save(G.state_dict(),os.path.join(log_PATH,"G_"+modelName+".pth"))
G.load_state_dict(torch.load(os.path.join(log_PATH,("G_"+modelName+".pth"))))
# + id="YzKpvn3xhKso" colab_type="code" colab={}
torch.save(D.state_dict(),os.path.join(log_PATH,"D_"+modelName+".pth"))
D.load_state_dict(torch.load(os.path.join(log_PATH,("D_"+modelName+".pth"))))
| notebooks/BEGAN-128x128.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
from sqlalchemy import create_engine
import pandas as pd
engine = create_engine("postgresql://zwswrynbgsidrm:3c086b43e892770d969963ca87ff3ff4a9e538affb05285e3c7b4b1d831ebaf7@ec2-35-171-250-21.compute-1.amazonaws.com:5432/d18k2ej2i6vjqm")
con = engine.connect()
final_data = pd.read_csv("finaldata.csv")
final_data.to_sql("ca_homeprice", con, if_exists = 'replace')
con.close()
| heroku_db_connect.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
在这里你的任务是创建得到一个元组,并返回一个包含三个元素(第一,第三和倒数第二的给定元组)的元组与的功能。
输入: 一个不少于三个元素的元组
输出: 一个元组.
举个栗子:
easy_unpack((1, 2, 3, 4, 5, 6, 7, 9)) == (1, 3, 7)
easy_unpack((1, 1, 1, 1)) == (1, 1, 1)
easy_unpack((6, 3, 7)) == (6, 7, 3)
# +
def easy_unpack(elements: tuple) -> tuple:
"""
returns a tuple with 3 elements - first, third and second to the last
"""
# your code here
return elements[0], elements[2], elements[-2]
if __name__ == '__main__':
# These "asserts" using only for self-checking and not necessary for auto-testing
assert easy_unpack((1, 2, 3, 4, 5, 6, 7, 9)) == (1, 3, 7)
assert easy_unpack((1, 1, 1, 1)) == (1, 1, 1)
assert easy_unpack((6, 3, 7)) == (6, 7, 3)
print('Done! Go Check!')
| easy-unpack.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Recurrent Neural Networks
#
# - Sequence data
# - Natural Language
# - Speech ...
#
# ### RNN model
# 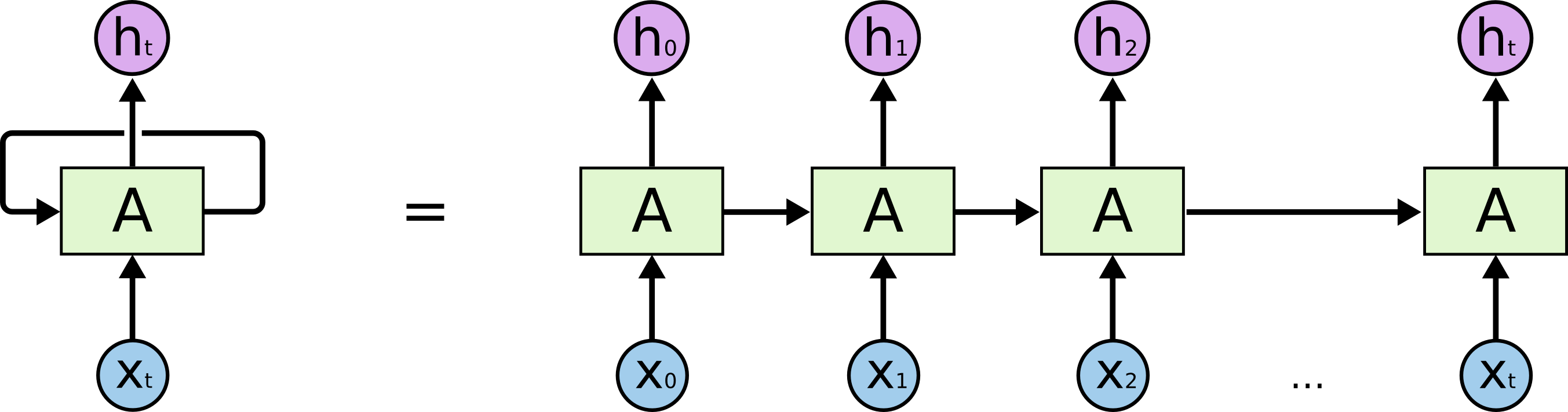
#
# ### RNN example
# 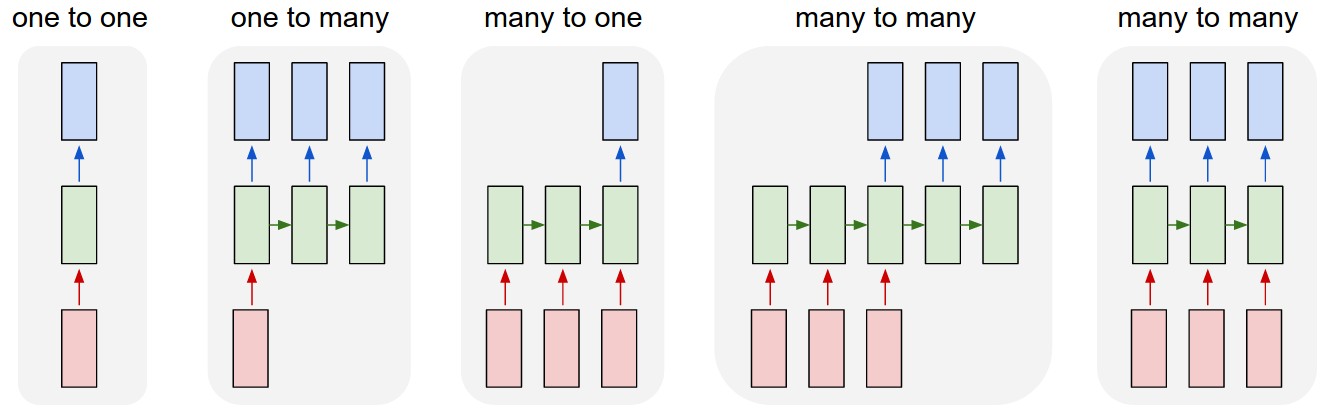
#
# ### LSTM (Long Short-Term Memory models)
# 
#
#
# ## 그래서?
#
# 1. 주가예측
# - tensorflow를 이용한 주가예측
# - keras를 이용한 주가예측
# 2. 감성분석
import numpy as np
import pandas as pd
import tensorflow as tf
from sklearn.preprocessing import MinMaxScaler
import matplotlib.pyplot as plt
import urllib
import os
# +
# S&P 500
FILE_NAME = 'all_stocks_5yr.csv'
SOURCE_URL = 'https://github.com/CNuge/kaggle-code/raw/master/stock_data/'
# YAHOO
# FILE_NAME = 'prices.csv'
#SOURCE_URL = 'https://www.kaggle.com/dgawlik/nyse/downloads/prices.csv/'
# GOOGLE
# FILE_NAME = 'trainset.csv'
# SOURCE_URL = 'https://www.kaggle.com/ptheru/googledta/downloads/trainset.csv/'
# !mkdir 'data'
filepath = './data/'+ FILE_NAME
# +
# urllib.request.urlretrieve(SOURCE_URL+'/?accessType=download', filepath)
# -
data = pd.read_csv('./data/'+FILE_NAME)
cl = data[data['Name']=='MMM']
# cl = data
cl = cl[['close']].values
def set_data(cl):
scl = MinMaxScaler()
#Scale the data
# pandas.__version__ < 0.23
# cl = cl.reshape(cl.shape[0],1)
# pandas.__version__ => 0.23
# cl = cl.as_matrix()
# cl = cl.reshape(cl.shape[0],1)
cl = scl.fit_transform(cl)
return cl
cl = set_data(cl)
def processData(data,lb):
X,Y = [],[]
for i in range(len(data)-lb-1):
X.append(data[i:(i+lb)])
Y.append(data[(i+lb),0])
return np.array(X),np.array(Y)
X, y = processData(cl,50)
y = y.reshape(y.shape[0], 1)
X_train,X_test = X[:int(X.shape[0]*0.80)],X[int(X.shape[0]*0.80):]
y_train,y_test = y[:int(y.shape[0]*0.80)],y[int(y.shape[0]*0.80):]
print(X_train.shape[0])
print(X_test.shape[0])
print(y_train.shape[0])
print(y_test.shape[0])
# train Parameters
seq_length = 50
data_dim = 1
hidden_dim = 10
output_dim = 1
learning_rate = 0.01
iterations = 500
X_test.shape
y_test.shape
# +
# input place holders
X = tf.placeholder(tf.float32, [None, seq_length, data_dim])
Y = tf.placeholder(tf.float32, [None, 1])
# build a LSTM network
#cell = tf.contrib.rnn.BasicLSTMCell(
cell = tf.contrib.rnn.BasicRNNCell(
num_units=hidden_dim, activation=tf.tanh)
outputs, _states = tf.nn.dynamic_rnn(cell, X, dtype=tf.float32)
Y_pred = tf.contrib.layers.fully_connected(
outputs[:, -1], output_dim, activation_fn=None) # We use the last cell's output
# cost/loss
loss = tf.reduce_sum(tf.square(Y_pred - Y)) # sum of the squares
# optimizer
optimizer = tf.train.AdamOptimizer(learning_rate)
train = optimizer.minimize(loss)
# RMSE
targets = tf.placeholder(tf.float32, [None, 1])
predictions = tf.placeholder(tf.float32, [None, 1])
rmse = tf.sqrt(tf.reduce_mean(tf.square(targets - predictions)))
# -
import time
start = time.time()
with tf.Session() as sess:
init = tf.global_variables_initializer()
sess.run(init)
# Training step
for i in range(iterations):
_, step_loss = sess.run([train, loss], feed_dict={
X: X_train, Y: y_train})
if i%10 == 0:
print("[step: {}] loss: {}".format(i, step_loss))
# Test step
test_predict = sess.run(Y_pred, feed_dict={X: X_test})
rmse_val = sess.run(rmse, feed_dict={
targets: y_test, predictions: test_predict})
print("RMSE: {}".format(rmse_val))
end = time.time()
print("dur : {}".format(end-start))
# Plot predictions
plt.plot(y_test, color='red')
plt.plot(test_predict, color='blue')
plt.xlabel("Time Period")
plt.ylabel("Stock Price")
plt.show()
| tfmodule/Stock_Predict_tf_RNN_1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] nbgrader={}
# # Interpolation Exercise 1
# + nbgrader={}
# %matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
# + nbgrader={}
from scipy.interpolate import interp1d
# + [markdown] nbgrader={}
# ## 2D trajectory interpolation
# + [markdown] nbgrader={}
# The file `trajectory.npz` contains 3 Numpy arrays that describe a 2d trajectory of a particle as a function of time:
#
# * `t` which has discrete values of time `t[i]`.
# * `x` which has values of the x position at those times: `x[i] = x(t[i])`.
# * `y` which has values of the y position at those times: `y[i] = y(t[i])`.
#
# Load those arrays into this notebook and save them as variables `x`, `y` and `t`:
# + deletable=false nbgrader={"checksum": "6cff4e8e53b15273846c3aecaea84a3d", "solution": true}
f=np.load('trajectory.npz')
x=np.array(f['x'])
y=np.array(f['y'])
t=np.array(f['t'])
# + deletable=false nbgrader={"checksum": "6d28ef77badc258f64b8e8844ce5329b", "grade": true, "grade_id": "interpolationex01a", "points": 4}
assert isinstance(x, np.ndarray) and len(x)==40
assert isinstance(y, np.ndarray) and len(y)==40
assert isinstance(t, np.ndarray) and len(t)==40
# + [markdown] nbgrader={}
# Use these arrays to create interpolated functions $x(t)$ and $y(t)$. Then use those functions to create the following arrays:
#
# * `newt` which has 200 points between $\{t_{min},t_{max}\}$.
# * `newx` which has the interpolated values of $x(t)$ at those times.
# * `newy` which has the interpolated values of $y(t)$ at those times.
# + deletable=false nbgrader={"checksum": "6cff4e8e53b15273846c3aecaea84a3d", "solution": true}
newt=np.linspace(t.min(),t.max(),200)
xt=interp1d(t,x,kind='cubic')
yt=interp1d(t,y,kind='cubic')
newx=xt(newt)
newy=yt(newt)
# + deletable=false nbgrader={"checksum": "50f3d594bf1df788fd1f281855440611", "grade": true, "grade_id": "interpolationex01b", "points": 4}
assert newt[0]==t.min()
assert newt[-1]==t.max()
assert len(newt)==200
assert len(newx)==200
assert len(newy)==200
# + [markdown] nbgrader={}
# Make a parametric plot of $\{x(t),y(t)\}$ that shows the interpolated values and the original points:
#
# * For the interpolated points, use a solid line.
# * For the original points, use circles of a different color and no line.
# * Customize you plot to make it effective and beautiful.
# + deletable=false nbgrader={"checksum": "6cff4e8e53b15273846c3aecaea84a3d", "solution": true}
# referenced http://goo.gl/gixqML for legend formatting
plt.plot(newx,newy,label='interpolated xy-data')
plt.plot(x,y,marker='o',linestyle='',label='original xy-data')
plt.ylim(bottom=-1.0)
plt.xlim(left=-1.0)
plt.xlabel('x-position')
plt.ylabel('y-position')
plt.legend(loc=10, bbox_to_anchor=(1.2,0.5))
plt.title('2D Trajectory');
# + deletable=false nbgrader={"checksum": "e9c74ef21797ed1bbacb14dae973b7aa", "grade": true, "grade_id": "interpolationex01c", "points": 2}
assert True # leave this to grade the trajectory plot
| assignments/assignment08/InterpolationEx01.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # NOAA extreme weather events
# The [National Oceanic and Atmospheric Administration](https://en.wikipedia.org/wiki/National_Oceanic_and_Atmospheric_Administration) has a database of extreme weather events that contains lots of detail for every year. [Link](https://www.climate.gov/maps-data/dataset/severe-storms-and-extreme-events-data-table). I'll extract the data for 2018.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
pd.set_option('display.max_columns', None) # Unlimited columns
# ## Get official list of counties
# +
import geopandas
# Import a shape file with all the counties in the US.
counties = geopandas.read_file('../data_input/1_USCounties/')
# Turn state codes from strings to integers
for col in ['STATE_FIPS', 'CNTY_FIPS', 'FIPS']:
counties[col] = counties[col].astype(int)
# Set the FIPS code as the index, and sort by it
counties = counties.set_index('FIPS').sort_index(axis=0)
# Make a list of the valid FIPS codes
official_counties = counties.index.tolist()
# -
# ## Exploring one year of NOAA data
# NOAA data comes year-by-year. Here is an exploration of that one year, which will allow me to make a function that does this for any year.
# The full table contains more than I want to use.
df1 = pd.read_csv('../data_input/4_NOAA Storms/StormEvents_details-ftp_v1.0_d2018_c20190422.csv')
print(df1.shape)
print(df1.columns)
df1.head(2)
# +
# Extract only a few useful columns
df2 = df1[['STATE','STATE_FIPS','CZ_FIPS','EVENT_TYPE']].copy()
# Create new column for complete county FIPS code
state_FIPS = [str(x) for x in df2['STATE_FIPS']]
county_FIPS = [str(x) for x in df2['CZ_FIPS']]
# Make sure the string for county FIPS is 3 digits long.
county_FIPS_2 = []
for fip in county_FIPS:
if len(fip) == 3:
fip2 = fip
elif len(fip) == 2:
fip2 = '0' + fip
elif len(fip) == 1:
fip2 = '00' + fip
county_FIPS_2.append(fip2)
# Create a full FIPS for each county
complete_FIPS = [int(x+y) for x, y in zip(state_FIPS, county_FIPS_2)]
df2['FIPS'] = complete_FIPS
# Drop entries for areas not in the list of counties that we're working
# with. This includes counties in non-state territories (eg, Puerto Rico),
# areas over lakes and oceans, and several forecast zones that don't
# correspond to particular counties.
df3 = df2[df2.FIPS.isin(official_counties)].copy()
# Drop obsolete columns
df3 = df3.drop(columns=['STATE_FIPS','CZ_FIPS'])
# -
print(df3.shape)
df3.head()
# These are all event types from the metadata, edited to remove marine events
event_types = ['Astronomical Low Tide','Avalanche','Blizzard','Coastal Flood',
'Cold/Wind Chill','Debris Flow','Dense Fog','Dense Smoke',
'Drought','Dust Devil','Dust Storm','Excessive Heat',
'Extreme Cold/Wind Chill','Flash Flood','Flood','Frost/Freeze',
'Funnel Cloud','Freezing Fog','Hail','Heat','Heavy Rain',
'Heavy Snow','High Surf','High Wind','Hurricane (Typhoon)',
'Ice Storm','Lake-Effect Snow','Lakeshore Flood','Lightning',
'Rip Current','Seiche','Sleet','Storm Surge/Tide','Strong Wind',
'Thunderstorm Wind','Tornado','Tropical Depression',
'Tropical Storm','Tsunami','Volcanic Ash','Wildfire',
'Winter Storm','Winter Weather',]
# +
df4 = df3.copy()
# Create a new column for every event type. Each row has a 1 in the column of
# its event type and a 0 for all others.
for typ in event_types:
df4[typ] = (typ == df4['EVENT_TYPE']).astype(int)
# -
df4.head()
# Group by the FIPS code, summing up all the events for that county.
# Now the dataset has FIPS code as the index and contains one column for each
# event type. Each cell is the number of events of that type for that county.
df5 = df4.groupby('FIPS').sum()
print(df5.shape)
df5.head()
# I reindex the dataframe so that it contains a row for every county in
# the official county list. Counties without events are filled with 0.
df6 = df5.reindex(official_counties, fill_value=0)
print(df6.shape)
df6.head()
# These are the county-level extreme weather events reported un 2018.
df6.sum().sort_values(ascending=False)
# ## Processing all NOAA files
# These are all event types from the metadata, edited to remove marine events
event_types = ['Astronomical Low Tide','Avalanche','Blizzard','Coastal Flood',
'Cold/Wind Chill','Debris Flow','Dense Fog','Dense Smoke',
'Drought','Dust Devil','Dust Storm','Excessive Heat',
'Extreme Cold/Wind Chill','Flash Flood','Flood','Frost/Freeze',
'Funnel Cloud','Freezing Fog','Hail','Heat','Heavy Rain',
'Heavy Snow','High Surf','High Wind','Hurricane (Typhoon)',
'Ice Storm','Lake-Effect Snow','Lakeshore Flood','Lightning',
'Rip Current','Seiche','Sleet','Storm Surge/Tide','Strong Wind',
'Thunderstorm Wind','Tornado','Tropical Depression',
'Tropical Storm','Tsunami','Volcanic Ash','Wildfire',
'Winter Storm','Winter Weather',]
def process_noaa(filepath):
"""
Process one year of NOAA Extreme weather events. Requires
the list of official counties and the list of official weather
event types.
Inputs
------
filepath (string) : file path for the CSV data file.
Outputs
-------
df6 (pandas.DataFrame) : Dataframe with the list of official FIPS
codes as index and one column for each
disaster type handled by NOAA.
"""
df1 = pd.read_csv(filepath)
# Extract only a few important columns
df2 = df1[['STATE_FIPS','CZ_FIPS','EVENT_TYPE']].copy()
# Remove any rows with nulls
df2 = df2.dropna()
# Create new column for complete county FIPS code.
# Ensure that FIPS are ints, otherwise string manipulation fails
state_FIPS = [str(x) for x in df2['STATE_FIPS'].astype(int)]
county_FIPS = [str(x) for x in df2['CZ_FIPS'].astype(int)]
# Make sure the string for county FIPS is 3 digits long,
# with trailing zeroes on the left.
county_FIPS_2 = []
for fip in county_FIPS:
if len(fip) == 3:
fip2 = fip
elif len(fip) == 2:
fip2 = '0' + fip
elif len(fip) == 1:
fip2 = '00' + fip
county_FIPS_2.append(fip2)
# Create a full FIPS for each county
complete_FIPS = [int(x+y) for x, y in zip(state_FIPS, county_FIPS_2)]
df2['FIPS'] = complete_FIPS
# Drop entries for areas not in the list of counties that we're working
# with. This includes counties in non-state territories (eg, Puerto Rico),
# areas over lakes and oceans, and several forecast zones that don't
# correspond to particular counties.
df3 = df2[df2.FIPS.isin(official_counties)].copy()
# Drop obsolete columns
df4 = df3.drop(columns=['STATE_FIPS','CZ_FIPS'])
# Create a new column for every event type. Each row has a 1 in the column of
# its event type and a 0 for all others.
for typ in event_types:
df4[typ] = (typ == df4['EVENT_TYPE']).astype(int)
# Group by the FIPS code, summing up all the events for that county.
# Now the dataset has FIPS code as the index and contains one column for each
# event type. Each cell is the number of events of that type for that county.
df5 = df4.groupby('FIPS').sum()
# I reindex the dataframe so that it contains a row for every county in
# the official county list. Counties without events are filled with 0.
df6 = df5.reindex(official_counties, fill_value=0)
return df6
# +
import glob
import os
# Read the CSV files for each year going back to 1996 (the first year
# when many of these event types started being recorded)
path = '../data_local/NOAA/'
filenames = sorted(glob.glob(os.path.join(path, '*.csv')))
years = []
layers = []
# Aggregate the dataframes in a list
for name in filenames:
year = int(name[49:53])
print(f'Processing {year}')
years.append(year)
layers.append(process_noaa(name))
# Concatenate all these dataframes into a single multi-layer dataframe
noaa = pd.concat(layers, keys=years)
# +
# Create an ordered list of the event types with highest counts
columns_by_most_events = noaa.groupby(level=0).sum().sum().sort_values(ascending=False).index
# Reorder the columns in that order
noaa = noaa[columns_by_most_events].copy()
# Create new df for comparison
noaa2 = noaa.copy()
# Rename some categories
new_names = {'Heat':'Heat old',
'Flood':'Flood old',
'Winter Weather':'Winter Weather old',
'Hurricane (Typhoon)':'Hurricane'}
noaa2 = noaa2.rename(columns=new_names)
# Create aggregate categories
noaa2['Winter Weather'] = noaa2['Winter Weather old'] +\
noaa2['Winter Storm'] +\
noaa2['Heavy Snow'] +\
noaa2['Frost/Freeze'] +\
noaa2['Freezing Fog'] +\
noaa2['Ice Storm'] +\
noaa2['Sleet'] +\
noaa2['Lake-Effect Snow'] +\
noaa2['Cold/Wind Chill'] +\
noaa2['Extreme Cold/Wind Chill'] +\
noaa2['Blizzard']
noaa2['Flood'] = noaa2['Flood old'] +\
noaa2['Flash Flood']+\
noaa2['Coastal Flood']+\
noaa2['Storm Surge/Tide']+\
noaa2['Lakeshore Flood']+\
noaa2['Debris Flow']
noaa2['Storm'] = noaa2['Thunderstorm Wind']+\
noaa2['High Wind']+\
noaa2['Funnel Cloud']+\
noaa2['Dust Storm']+\
noaa2['Strong Wind']+\
noaa2['Dust Devil']+\
noaa2['Tropical Depression']+\
noaa2['Lightning']+\
noaa2['Tropical Storm']+\
noaa2['High Surf']+\
noaa2['Heavy Rain']+\
noaa2['Hail']
noaa2['Fire'] = noaa2['Wildfire'] +\
noaa2['Dense Smoke']
noaa2['Heat'] = noaa2['Heat old'] +\
noaa2['Excessive Heat']
final_columns = ['Storm', 'Flood', 'Winter Weather', 'Tornado',
'Drought', 'Heat', 'Fire', 'Hurricane']
# Some categories are excluded because they're not important enough
deliberately_ignored = ['Dense Fog', 'Rip Current','Astronomical Low Tide','Avalanche',
'Seiche', 'Tsunami', 'Volcanic Ash']
noaa3 = noaa2[final_columns]
# -
# noaa3.groupby(level=1).sum()
noaa3.to_csv('../data_output/5__NOAA/noaa_1.csv', index_label=['year','FIPS'])
# +
# Aggregate data by county (years summed up)
noaa_county = noaa3.groupby(level=1).sum()
# Aggregate data by year (counties summed up)
noaa_year = noaa3.groupby(level=0).sum()
# Put the data by county into the geopandas file with county shapes
noaa_county_map = counties.merge(noaa_county, on='FIPS', how='left')
# -
noaa_county_log = np.log(noaa_county+1)
noaa_county_map_log = counties.merge(noaa_county_log, on='FIPS', how='left')
noaa_county_map_log.head()
print(noaa_county_map.shape)
noaa_county_map.head()
noaa_county_map_log.plot(column='Storm', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Flood', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Winter Weather', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Tornado', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Drought', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Heat', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Fire', figsize=(20,6), legend=True);
noaa_county_map_log.plot(column='Hurricane', figsize=(20,6), legend=True);
# # What about those other zones?
url = 'https://www.weather.gov/source/gis/Shapefiles/County/bp02ap19.dbx'
cols = ['STATE','ZONE','CWA','NAME','STATE_ZONE','COUNTY','FIPS',
'TIME_ZONE','FE_AREA','LAT','LON']
zones1 = pd.read_csv(url, delimiter='|', names=cols)
zones1.sort_values('FIPS')
| notebooks/archive/DMA4 - NOAA storms by county without weather zones.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Transfering knowledge through finetuning
#
# In previous chapters,
# we demonstrated how to train a neural network
# to recognize the categories corresponding to objects in images.
# We looked at toy datasets like hand-written digits,
# and thumbnail-sized pictures of animals.
# And we talked about the ImageNet dataset,
# the default academic benchmark,
# which contains 1M million images,
# 1000 each from 1000 separate classes.
#
# The ImageNet dataset categorically changed what was possible in computer vision.
# It turns out some things are possible (these days, even easy)
# on gigantic datasets, that simply aren't with smaller datasets.
# In fact, we don't know of any technique that can comparably powerful model
# on a similar photograph dataset but containing only, say, 10k images.
#
# And that's a problem.
# Because however impressive the results of CNNs on ImageNet may be,
# most people aren't interested in ImageNet itself.
# They're interested in their own problems.
# Recognize people based on pictures of their faces.
# Distinguish between photographs of $10$ different types of coral on the ocean floor.
# Usually when individuals (and not Amazon, Google, or inter-institutional *big science* initiatives)
# are interested in solving a computer vision problem,
# they come to the table with modestly sized datasets.
# A few hundred examples may be common and a few thousand examples may be as much as you can reasonably ask for.
#
# So one natural question emerges.
# Can we somehow use the powerful models trained on millions of examples for one dataset,
# and apply them to improve performance on a new problem
# with a much smaller dataset?
# This kind of problem (learning on source dataset, bringing knowledge to target dataset),
# is appropriately called *transfer learning*.
# Fortunately, we have some effective tools for solving this problem.
#
# For deep neural networks, the most popular approach is called finetuning
# and the idea is both simple and effective:
#
# * Train a neural network on the source task $S$.
# * Decapitate it, replacing it's output layer appropriate to target task $T$.
# * Initialize the weights on the new output layer randomly, keeping all other (pretrained) weights the same.
# * Begin training on the new dataset.
#
# This might be clearer if we visualize the algorithm:
#
# 
#
#
# In this section, we'll demonstrate fine-tuning,
# using the popular and compact SqueezeNet architecture.
# Since we don't want to saddle you with the burden of downloading ImageNet,
# or of training on ImageNet from scratch,
# we'll pull the weights of the pretrained Squeeze net from the internet.
# Specifically, we'll be fine-tuning a squeezenet-1.1
# that was pre-trained on imagenet-12.
# Finally, we'll fine-tune it to recognize **hotdogs**.
# 
# We'll start with the obligatory ritual of importing a bunch of stuff that you'll need later.
# %pylab inline
pylab.rcParams['figure.figsize'] = (10, 6)
# ## Settings
# We'll set a few settings up here that you can configure later to manipulate the behavior of the algorithm.
# These are mostly familiar.
# Hybrid mode, uses the just in time compiler described in [our chapter on high performance training](../chapter07_distributed-learning/hybridize.ipynb)
# to make the network much faster to train.
# Since we're not working with any crazy dynamic graphs that can't be compiled,
# there's no reason not to hybridize.
# The batch size, number of training epochs, weight decay, and learing rate should all be familiar by now.
# The positive class weight, says how much more we should upweight the importance of positive instances (photos of hot dogs) in the objective function.
# We use this to combat the extreme class imbalance (not surprisingly, most pictures do not depict hot dogs).
# +
# Demo mode uses the validation dataset for training, which is smaller and faster to train.
demo = True
log_interval = 100
# Options are imperative or hybrid. Use hybrid for better performance.
mode = 'hybrid'
# training hyperparameters
batch_size = 256
if demo:
epochs = 5
learning_rate = 0.02
wd = 0.002
else:
epochs = 40
learning_rate = 0.05
wd = 0.002
# the class weight for hotdog class to help the imbalance problem.
positive_class_weight = 5
# +
from __future__ import print_function
import logging
logging.basicConfig(level=logging.INFO)
import os
import time
from collections import OrderedDict
import skimage.io as io
import mxnet as mx
from mxnet.test_utils import download
mx.random.seed(127)
# setup the contexts; will use gpus if avaliable, otherwise cpu
gpus = mx.test_utils.list_gpus()
contexts = [mx.gpu(i) for i in gpus] if len(gpus) > 0 else [mx.cpu()]
# -
# ## Dataset
# Formally, hot dog recognition is a binary classification problem.
# We'll use $1$ to represent the hotdog class,
# and $0$ for the *not hotdog* class.
# Our hot dog dataset (the target dataset which we'll fine-tune the model to)
# contains 18,141 sample images, 2091 of which are hotdogs.
# Because the dataset is imbalanced (e.g. hotdog class is only 1% in mscoco dataset),
# sampling interesting negative samples can help to improve the performance of our algorithm.
# Thus, in the negative class in the our dataset,
# two thirds are images from food categories (e.g. pizza) other than hotdogs,
# and 30% are images from all other categories.
#
# ### Files
# We prepare the dataset in the format of MXRecord using [im2rec](http://mxnet.io/how_to/recordio.html?highlight=im2rec) tool. As of the current draft, rec files are not yet explained in the book, but if you're reading after November or December 2017 and you still see this note, [open an issue on GitHub](https://github.com/zackchase/mxnet-the-straight-dope) and let us know to stop slacking off.
#
# - not_hotdog_train.rec 641M (1882 positive, 10000 interesting negative, and 5000 random negative)
# - not_hotdog_validation.rec 49M (209 positive, 700 interesting negative, and 350 random negative)
dataset_files = {'train': ('not_hotdog_train-e6ef27b4.rec', '0aad7e1f16f5fb109b719a414a867bbee6ef27b4'),
'validation': ('not_hotdog_validation-c0201740.rec', '723ae5f8a433ed2e2bf729baec6b878ac0201740')}
# To demo the model here, we're justgoing to use the smaller validation set.
# But if you're interested in training on the full set,
# set 'demo' to False in the settings at the beginning.
# Now we're ready to download and verify the dataset.
# +
if demo:
training_dataset, training_data_hash = dataset_files['validation']
else:
training_dataset, training_data_hash = dataset_files['train']
validation_dataset, validation_data_hash = dataset_files['validation']
def verified(file_path, sha1hash):
import hashlib
sha1 = hashlib.sha1()
with open(file_path, 'rb') as f:
while True:
data = f.read(1048576)
if not data:
break
sha1.update(data)
matched = sha1.hexdigest() == sha1hash
if not matched:
logging.warn('Found hash mismatch in file {}, possibly due to incomplete download.'
.format(file_path))
return matched
url_format = 'https://apache-mxnet.s3-accelerate.amazonaws.com/gluon/dataset/{}'
if not os.path.exists(training_dataset) or not verified(training_dataset, training_data_hash):
logging.info('Downloading training dataset.')
download(url_format.format(training_dataset),
overwrite=True)
if not os.path.exists(validation_dataset) or not verified(validation_dataset, validation_data_hash):
logging.info('Downloading validation dataset.')
download(url_format.format(validation_dataset),
overwrite=True)
# -
# ### Iterators
#
# The record files can be read using [mx.io.ImageRecordIter](http://mxnet.io/api/python/io.html#mxnet.io.ImageRecordIter)
# load dataset
train_iter = mx.io.ImageRecordIter(path_imgrec=training_dataset,
min_img_size=256,
data_shape=(3, 224, 224),
rand_crop=True,
shuffle=True,
batch_size=batch_size,
max_random_scale=1.5,
min_random_scale=0.75,
rand_mirror=True)
val_iter = mx.io.ImageRecordIter(path_imgrec=validation_dataset,
min_img_size=256,
data_shape=(3, 224, 224),
batch_size=batch_size)
# ## Model
#
# The model we are finetuning is [SqueezeNet](https://arxiv.org/abs/1602.07360). Gluon module offers squeezenet v1.0 and v1.1 that are pretrained on ImageNet. This is just a convolutional neural network, with an architecture chosen to have a small number of parameters and to require a minimal amount of computation. It's especially popular for folks that need to run CNNs on low-powered devices like cell phones and other internet-of-things devices.
#
# ## Pulling the pre-trained model
# Fortunately, MXNet has a model zoo that gives us convenient access to a number of popular models,
# both their architectres and their pretrained parameters.
# Let's download SqueezeNet right now with just a few lines of code.
# +
from mxnet.gluon import nn
from mxnet.gluon.model_zoo import vision as models
# get pretrained squeezenet
net = models.squeezenet1_1(pretrained=True, prefix='deep_dog_', ctx=contexts)
# hot dog happens to be a class in imagenet.
# we can reuse the weight for that class for better performance
# here's the index for that class for later use
imagenet_hotdog_index = 713
# -
# ### DeepDog net
#
# We can now use the feature extractor part from the pretrained squeezenet to build our own network. The model zoo, even handles the decaptiation for us. All we have to do is specify the number out of output classes in our new task, which we do via the keyword argument `classes=2`.
deep_dog_net = models.squeezenet1_1(prefix='deep_dog_', classes=2)
deep_dog_net.collect_params().initialize(ctx=contexts)
deep_dog_net.features = net.features
print(deep_dog_net)
# The network can already be used for prediction. However, since it hasn't been finetuned yet, the network performance could be bad.
# +
from skimage.color import rgba2rgb
def classify_hotdog(net, url, contexts):
I = io.imread(url)
if I.shape[2] == 4:
I = rgba2rgb(I)
image = mx.nd.array(I).astype(np.uint8)
plt.subplot(1, 2, 1)
plt.imshow(image.asnumpy())
image = mx.image.resize_short(image, 256)
image, _ = mx.image.center_crop(image, (224, 224))
plt.subplot(1, 2, 2)
plt.imshow(image.asnumpy())
image = mx.image.color_normalize(image.astype(np.float32)/255,
mean=mx.nd.array([0.485, 0.456, 0.406]),
std=mx.nd.array([0.229, 0.224, 0.225]))
image = mx.nd.transpose(image.astype('float32'), (2,1,0))
image = mx.nd.expand_dims(image, axis=0)
out = mx.nd.SoftmaxActivation(net(image.as_in_context(contexts[0])))
print('Probabilities are: '+str(out[0].asnumpy()))
result = np.argmax(out.asnumpy())
outstring = ['Not hotdog!', 'Hotdog!']
print(outstring[result])
# -
classify_hotdog(deep_dog_net, '../img/real_hotdog.jpg', contexts)
# ### Reuse class weights
# As mentioned earlier, in addition to the feature extractor, we can reuse the class weights for hot dog from the pretrained model, since hot dog was already a class in the imagenet. To do that, we need to get the weight from the classifier layers of the pretrained model, find the right slice, and put it into our two-class classifier.
# let's examine the output layer and find the last conv layer
print(net.output)
# +
# the last conv layer is the second layer
pretrained_conv_params = net.output[0].params
# weights can then be found from the above parameter dict
pretrained_weight_param = pretrained_conv_params.get('weight')
pretrained_bias_param = pretrained_conv_params.get('bias')
# next, we locate the right slice that we're interested in.
hotdog_w = mx.nd.split(pretrained_weight_param.data(ctx=contexts[0]),
1000, axis=0)[imagenet_hotdog_index]
hotdog_b = mx.nd.split(pretrained_bias_param.data(ctx=contexts[0]),
1000, axis=0)[imagenet_hotdog_index]
# our classifier is for two classes. here, we reuse the hotdog class weight,
# and randomly initialize the 'not hotdog' class.
new_classifier_w = mx.nd.concat(mx.nd.random_normal(shape=hotdog_w.shape, scale=0.02, ctx=contexts[0]),
hotdog_w,
dim=0)
new_classifier_b = mx.nd.concat(mx.nd.random_normal(shape=hotdog_b.shape, scale=0.02, ctx=contexts[0]),
hotdog_b,
dim=0)
# finally, we initialize the parameter buffers and set the values.
# since classifier is a HybridSequential/Sequential, the following
# takes the zero-indexed 1-st layer of the classifier
final_conv_layer_params = deep_dog_net.output[0].params
final_conv_layer_params.get('weight').set_data(new_classifier_w)
final_conv_layer_params.get('bias').set_data(new_classifier_b)
# -
# ## Evaluation
#
# Our task is a binary classification problem with imbalanced classes. So we'll monitor performance both using accuracy and F1 score, a metric favored in settings with extreme class imbalance.
# [Note to authors: ensure that F1 score is explained earlier or explain it here in full]
# return metrics string representation
def metric_str(names, accs):
return ', '.join(['%s=%f'%(name, acc) for name, acc in zip(names, accs)])
metric = mx.metric.create(['acc', 'f1'])
# The following snippet performs inferences on evaluation dataset, and updates the metrics. Once the evaluation data iterator is exhausted, it returns the values of each of the metrics.
# +
import mxnet.gluon as gluon
from mxnet.image import color_normalize
def evaluate(net, data_iter, ctx):
data_iter.reset()
for batch in data_iter:
data = color_normalize(batch.data[0]/255,
mean=mx.nd.array([0.485, 0.456, 0.406]).reshape((1,3,1,1)),
std=mx.nd.array([0.229, 0.224, 0.225]).reshape((1,3,1,1)))
data = gluon.utils.split_and_load(data, ctx_list=ctx, batch_axis=0)
label = gluon.utils.split_and_load(batch.label[0], ctx_list=ctx, batch_axis=0)
outputs = []
for x in data:
outputs.append(net(x))
metric.update(label, outputs)
out = metric.get()
metric.reset()
return out
# -
# ## Training
# We now can train the model just as we would any supervised model.
# In this example, we set up the training loop for multi-GPU use
# as described from first principles [here](../chapter07_distributed-learning/multiple-gpus-scratch.ipynb) and in the context of gluon [here](../chapter07_distributed-learning/multiple-gpus-gluon.ipynb).
# +
import mxnet.autograd as autograd
def train(net, train_iter, val_iter, epochs, ctx):
if isinstance(ctx, mx.Context):
ctx = [ctx]
trainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': learning_rate, 'wd': wd})
loss = gluon.loss.SoftmaxCrossEntropyLoss()
best_f1 = 0
val_names, val_accs = evaluate(net, val_iter, ctx)
logging.info('[Initial] validation: %s'%(metric_str(val_names, val_accs)))
for epoch in range(epochs):
tic = time.time()
train_iter.reset()
btic = time.time()
for i, batch in enumerate(train_iter):
# the model zoo models expect normalized images
data = color_normalize(batch.data[0]/255,
mean=mx.nd.array([0.485, 0.456, 0.406]).reshape((1,3,1,1)),
std=mx.nd.array([0.229, 0.224, 0.225]).reshape((1,3,1,1)))
data = gluon.utils.split_and_load(data, ctx_list=ctx, batch_axis=0)
label = gluon.utils.split_and_load(batch.label[0], ctx_list=ctx, batch_axis=0)
outputs = []
Ls = []
with autograd.record():
for x, y in zip(data, label):
z = net(x)
# rescale the loss based on class to counter the imbalance problem
L = loss(z, y) * (1+y*positive_class_weight)/positive_class_weight
# store the loss and do backward after we have done forward
# on all GPUs for better speed on multiple GPUs.
Ls.append(L)
outputs.append(z)
for L in Ls:
L.backward()
trainer.step(batch.data[0].shape[0])
metric.update(label, outputs)
if log_interval and not (i+1)%log_interval:
names, accs = metric.get()
logging.info('[Epoch %d Batch %d] speed: %f samples/s, training: %s'%(
epoch, i, batch_size/(time.time()-btic), metric_str(names, accs)))
btic = time.time()
names, accs = metric.get()
metric.reset()
logging.info('[Epoch %d] training: %s'%(epoch, metric_str(names, accs)))
logging.info('[Epoch %d] time cost: %f'%(epoch, time.time()-tic))
val_names, val_accs = evaluate(net, val_iter, ctx)
logging.info('[Epoch %d] validation: %s'%(epoch, metric_str(val_names, val_accs)))
if val_accs[1] > best_f1:
best_f1 = val_accs[1]
logging.info('Best validation f1 found. Checkpointing...')
net.save_parameters('deep-dog-%d.params'%(epoch))
if mode == 'hybrid':
deep_dog_net.hybridize()
if epochs > 0:
deep_dog_net.collect_params().reset_ctx(contexts)
train(deep_dog_net, train_iter, val_iter, epochs, contexts)
# -
# ## Try it out!
# Once our model is trained, we can either use the `deep_dog_net` model in the notebook kernel, or load it from the best checkpoint.
# Uncomment below line and replace the file name with the last checkpoint.
# deep_dog_net.load_parameters('deep-dog-3.params', contexts)
#
# Alternatively, you can uncomment the following lines to get the model that we finetuned,
# with validation F1 score of 0.74.
download('https://apache-mxnet.s3-accelerate.amazonaws.com/gluon/models/deep-dog-5a342a6f.params',
overwrite=True)
deep_dog_net.load_parameters('deep-dog-5a342a6f.params', contexts)
classify_hotdog(deep_dog_net, '../img/real_hotdog.jpg', contexts)
classify_hotdog(deep_dog_net, '../img/leg_hotdog.jpg', contexts)
classify_hotdog(deep_dog_net, '../img/dog_hotdog.jpg', contexts)
# ## Conclusions
# As you can see, given a pretrained model, we can get a great classifier,
# even for tasks where we simply don't have enough data to train from scratch.
# That's because the representations necessary to perform both tasks have a lot in common.
# Since they both address natural images, they both require recognizing textures, shapes, edges, etc.
# Whenever you have a small enough dataset that you fear impoverishing your model,
# try thinking about what larger datasets you might be able to pre-train your model on,
# so that you can just perform fine-tuning on the task at hand.
#
#
# ## Next
#
# This section is still changing too fast to say for sure what will come next. Stay tuned!
# For whinges or inquiries, [open an issue on GitHub.](https://github.com/zackchase/mxnet-the-straight-dope)
| chapter08_computer-vision/fine-tuning.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.7
# language: python
# name: python3
# ---
# + [markdown] pycharm={"name": "#%% md\n"}
# # Modeling options and result analysis in scCODA
#
# This tutorial notebook serves as an extension to the general tutorial and presents ways to alternate the model and perform more in-depth result analysis and diagnostics.
# We will focus on:
#
# - Modifications of the model formula and reference cell type to perform different modeling tasks
# - Inference methods available in scCODA
# - Advanced interpretation and analysis of results
# - Alternative differential abundance testing using all references
#
# We will again analyze the small intestinal epithelium data of mice from [Haber et al., 2017](https://www.nature.com/articles/nature24489).
# First, we read in the data and perform the same preprocessing steps as in the general tutorial:
# + pycharm={"name": "#%%\n"}
# Setup
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import matplotlib.pyplot as plt
import arviz as az
from sccoda.util import comp_ana as mod
from sccoda.util import cell_composition_data as dat
from sccoda.util import data_visualization as viz
import sccoda.datasets as scd
# + pycharm={"name": "#%%\n"}
# Load data
cell_counts = scd.haber()
# Convert data to anndata object
data_all = dat.from_pandas(cell_counts, covariate_columns=["Mouse"])
# Extract condition from mouse name and add it as an extra column to the covariates
data_all.obs["Condition"] = data_all.obs["Mouse"].str.replace(r"_[0-9]", "", regex=True)
print(f"Entire dataset: {data_all}")
# Select control and salmonella data
data_salm = data_all[data_all.obs["Condition"].isin(["Control", "Salm"])].copy()
print(f"Salmonella dataset: {data_salm}")
viz.boxplots(data_all, feature_name="Condition")
plt.show()
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Tweaking the model formula and reference cell type
#
# First, we take a closer look at how changing the `formula` parameter of the scCODA model influences the results.
# Internally, the formula string is converted into a linear model-like design matrix via [patsy](https://patsy.readthedocs.io/en/latest/),
# which has a similar syntax to the `lm` function in the R language.
#
# ### Multi-level categories
#
# Patsy allows us to automatically handle categorical covariates, even with multiple levels.
# For example, we can model the effect of all three diseases at once:
# + pycharm={"name": "#%%\n"}
# model all three diseases at once
model_all = mod.CompositionalAnalysis(data_all, formula="Condition", reference_cell_type="Endocrine")
all_results = model_all.sample_hmc()
all_results.summary()
# + [markdown] pycharm={"name": "#%% md\n"}
# ### Different reference levels
#
# Per default, categorical variables are encoded via full-rank treatment coding. Hereby, the value of the first sample in the dataset is used as the default (control) category.
# We can select the default level by changing the model formula to `"C(<CovariateName>, Treatment('<ReferenceLevelName>'))"`:
#
# For example, we can switch the salmonella model to test diseased versus healthy samples, which switches the sign of the only credible effect (Enterocytes).
# + pycharm={"name": "#%%\n"}
# Set salmonella infection as "default" category
model_salm_switch_cond = mod.CompositionalAnalysis(data_salm, formula="C(Condition, Treatment('Salm'))", reference_cell_type="Goblet")
switch_results = model_salm_switch_cond.sample_hmc()
switch_results.summary()
# + [markdown] pycharm={"name": "#%% md\n"}
# ### Switching the reference cell type
#
# Compositional analysis generally does not allow statements on absolute abundance changes, but only in relation to a reference category,
# which is assumed to be unchanged in absolute abundance.
# The reference cell type fixes this category in scCODA. Thus, an interpretation of scCODA's effects should always be formulated like:
#
# "Using cell type xy as a reference, cell types (a, b, c) were found to credibly change in abundance"
#
# Switching the reference cell type might thus produce different results.
# For example, if we choose a different cell type as the reference (such as Enterocytes in the salmonella infection data),
# scCODA can find other credible effects on the other cell types.
# + pycharm={"name": "#%%\n"}
model_salm_ref = mod.CompositionalAnalysis(data_salm, formula="Condition", reference_cell_type="Enterocyte")
reference_results = model_salm_ref.sample_hmc()
reference_results.summary()
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Inference algorithms in scCODA
#
# Currently, scCODA performs parameter inference via Markov-chain Monte Carlo (MCMC) methods.
# There are three different MCMC sampling methods available for scCODA:
#
# - Hamiltonian Monte Carlo (HMC) sampling: `sample_hmc()`
# - HMC sampling with Dual-averaging step size adaptation ([Nesterov, 2009](https://link.springer.com/article/10.1007/s10107-007-0149-x)): `sample_hmc_da()`
# - No-U-Turn sampling ([Hoffman and Gelman, 2014](https://jmlr.org/papers/volume15/hoffman14a/hoffman14a.pdf)): `sample_nuts()`
#
# Generally, it is recommended to use the standard HMC sampling. Other methods, such as variational inference, are in consideration.
#
# For all MCMC sampling methods, properties such as the MCMC chain length and the number of burn-in samples are directly adjustable.
# + [markdown] pycharm={"name": "#%% md\n"}
# ## Result analysis and diagnostics
#
# The "getting started" tutorial explains how to do interpret the basic output of scCODA.
# To follow this up, we now take a look at how MCMC diagnostics and more advanced result analysis in scCODA can be performed.
#
# For this section, we again use the model of salmonella infection versus control group, with a reference cell type of Goblet cells.
# + pycharm={"name": "#%%\n"}
model_salm = mod.CompositionalAnalysis(data_salm, formula="Condition", reference_cell_type="Goblet")
salm_results = model_salm.sample_hmc(num_results=20000)
# + [markdown] pycharm={"name": "#%% md\n"}
# ### Extended model summary
#
# `result.summary_extended()` gives us, apart from the properties already explained in the basic tutorial, more information about the posterior inferred by the model.
# The extended summary also includes some information on the MCMC sampling procedure (chain length, burn-in, acceptance rate, duration).
#
# For both effects and intercepts, we also get the standard deviation (SD) and high density interval endpoints of the posterior density of the generated Markov chain.
#
# The effects summary also includes the spike-and-slab inclusion probability for each effect, i.e. the share of MCMC samples, for which this effect was not set to 0 by the spike-and-slab prior.
# A threshold on this value serves as the deciding factor whether an effect is considered statistically credible
#
# We can also use the summary tables from `summary_extended()` as pandas DataFrames to tweak them further.
# They are also accessible as `result.intercept_df` and `result.effect_df`, respectively.
# Furthermore, the tables a direct result of the `summary()` function in [arviz](https://arviz-devs.github.io/arviz/index.html) and support all its functionality.
# This means that we can, for example, change the credible interval:
# + pycharm={"name": "#%%\n"}
salm_results.summary_extended(hdi_prob=0.9)
# + [markdown] pycharm={"name": "#%% md\n"}
#
# ### Diagnostics and plotting
#
# Similarly to the summary dataframes being compatible with [arviz](https://arviz-devs.github.io/arviz/index.html),
# the result class itself is an extension of arviz's `Inference Data` class. This means that we can use all its MCMC diagnostic and plotting functionality.
# As an example, looking at the MCMC trace plots and kernel density estimates, we see that they are indicative of a well sampled MCMC chain:
#
# Note: Due to the spike-and-slab priors, the `beta` parameters have many values at 0, which looks like a convergence issue, but is actually not.
#
# **Caution: Trying to plot a kernel density estimate for an effect on the reference cell type results in an error, since it is constant at 0 for the entire chain.
# To avoid this, add** `coords={"cell_type": salm_results.posterior.coords["cell_type_nb"]}` **as an argument to `az.plot_trace`, which causes the plots for the reference cell type to be skipped.**
# + pycharm={"name": "#%%\n"}
az.plot_trace(
salm_results,
divergences=False,
var_names=["alpha", "beta"],
coords={"cell_type": salm_results.posterior.coords["cell_type_nb"]},
)
plt.show()
# + [markdown] pycharm={"name": "#%% md\n"}
# ### Using all cell types as reference alternatively to reference selection
#
# scCODA uses a reference cell type that is considered to be unchanged over the experiment to guarantee the unique identifiability of results.
# If no such cell type is known beforehand, setting `reference_cell_type="automatic"` will find a suited reference.
# Alternatively, it is possible to find credible effects on cell types that are mostly independent of the reference.
# By sequentially running scCODA and selecting each cell type as the reference once, we can then use a majority vote to find the cell types that were credibly changing more than half of the time.
#
# Below, an example code for this procedure on the Salmonella infection data shows that only Enterocytes were found to be credible more than half of the time.
# Indeed, they re credibly changing for every reference cell type except themselves.
# All other cell types were not found to change with any reference.
# + pycharm={"name": "#%%\n"}
# Run scCODA with each cell type as the reference
cell_types = data_salm.var.index
results_cycle = pd.DataFrame(index=cell_types, columns=["times_credible"]).fillna(0)
for ct in cell_types:
print(f"Reference: {ct}")
# Run inference
model_temp = mod.CompositionalAnalysis(data_salm, formula="Condition", reference_cell_type=ct)
temp_results = model_temp.sample_hmc(num_results=20000)
# Select credible effects
cred_eff = temp_results.credible_effects()
cred_eff.index = cred_eff.index.droplevel(level=0)
# add up credible effects
results_cycle["times_credible"] += cred_eff.astype("int")
# + pycharm={"name": "#%%\n"}
# Calculate percentages
results_cycle["pct_credible"] = results_cycle["times_credible"]/len(cell_types)
results_cycle["is_credible"] = results_cycle["pct_credible"] > 0.5
print(results_cycle)
# + pycharm={"name": "#%%\n"}
| tutorials/Modeling_options_and_result_analysis.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3.9 (tensorflow)
# language: python
# name: tensorflow
# ---
# +
import numpy as np
from keras.preprocessing import image
import matplotlib.pyplot as plt
from pathlib import Path
import tensorflow as tf
import sys
import tensorflow.keras
import pandas as pd
import sklearn as sk
print(tf.test.gpu_device_name())
print(tf.config.list_physical_devices('GPU'))
print(f"Tensor Flow Version: {tf.__version__}")
print(f"Keras Version: {tensorflow.keras.__version__}")
print()
print(f"Python {sys.version}")
print(f"Pandas {pd.__version__}")
print(f"Scikit-Learn {sk.__version__}")
gpu = len(tf.config.list_physical_devices('GPU'))>0
print("GPU is", "available" if gpu else "NOT AVAILABLE")
# +
p=Path('Train/images')
d=p.glob("*")
img_data=[]
labels=[]
for image_path in d:
a=str(image_path).split("/")[2:]
print(a)
w=[new_a.split(".") for new_a in a ]
image_number=int(w[0][0])
img=image.load_img(image_path,target_size=(40,40))
image_arr=image.img_to_array(img)
img_data.append(image_arr)
if image_number>=296 and image_number<=396:
labels.append(0) #pikachu
if image_number>=397 and image_number<=497:
labels.append(1) #bulbasaur
if image_number>=498 and image_number<=620:
labels.append(2) #charmendae
# -
print(labels)
# +
#convert to numpy
img_data=np.array(img_data,dtype='float32')/255.0
labels=np.array(labels)
print(img_data.shape,labels.shape)
# +
#randomly shuffle
import random
combined=list(zip(img_data,labels))
random.shuffle(combined)
img_data[:],labels[:]=zip(*combined)
# -
def drawImg(img):
plt.imshow(img)
plt.show()
return
for i in range(10):
drawImg(img_data[i])
# ### Convert data for ONE-VS-ONE Classification
img_data.shape
m=img_data.shape[0]
img_data=img_data.reshape(m,-1)
print(img_data.shape,labels.shape)
# +
dic={}
dic[1]=[]
dic[1].append("a,b,c,d")
dic[1].append("z,l")
dic[2]=[]
dic[2].append("o,d")
print(dic[1][1])
dic[3]={}
dic[3][2]='v'
print(dic)
# -
classes=len(np.unique(labels))
print(classes)
# +
def divide_data(x,y):
data={}
for i in range(classes):
data[i]=[]
for i in range(x.shape[0]):
data[y[i]].append(x[i])
for k in data.keys():
data[k]=np.array(data[k])
return data
# -
data=divide_data(img_data,labels)
print(data[0].shape)
print(data[1].shape)
print(data[2].shape)
# +
def combine_data(d1,d2):
samples = d1.shape[0]+d2.shape[0]
features= d1.shape[1]
l1,l2=d1.shape[0],d2.shape[0]
data_pair=np.zeros((samples,features))
data_labels=np.zeros((samples))
data_pair[:l1,:]=d1
data_pair[l1:,:]=d2
data_labels[:l1]=-1
data_labels[l1:]=+1
return data_pair,data_labels
# -
# ### SVM
class SVM:
def __init__(self,C=1.0):
self.C = C
self.W = 0
self.b = 0
def hingeLoss(self,W,b,X,Y):
loss = 0.0
loss += .5*np.dot(W,W.T)
m = X.shape[0]
for i in range(m):
ti = Y[i]*(np.dot(W,X[i].T)+b)
loss += self.C *max(0,(1-ti))
return loss[0][0]
def fit(self,X,Y,batch_size=100,learning_rate=0.001,maxItr=300):
no_of_features = X.shape[1]
no_of_samples = X.shape[0]
n = learning_rate
c = self.C
#Init the model parameters
W = np.zeros((1,no_of_features))
bias = 0
#Initial Loss
#Training from here...
# Weight and Bias update rule that we discussed!
losses = []
for i in range(maxItr):
#Training Loop
l = self.hingeLoss(W,bias,X,Y)
losses.append(l)
ids = np.arange(no_of_samples)
np.random.shuffle(ids)
#Batch Gradient Descent(Paper) with random shuffling
for batch_start in range(0,no_of_samples,batch_size):
#Assume 0 gradient for the batch
gradw = 0
gradb = 0
#Iterate over all examples in the mini batch
for j in range(batch_start,batch_start+batch_size):
if j<no_of_samples:
i = ids[j]
ti = Y[i]*(np.dot(W,X[i].T)+bias)
if ti>1:
gradw += 0
gradb += 0
else:
gradw += c*Y[i]*X[i]
gradb += c*Y[i]
#Gradient for the batch is ready! Update W,B
W = W - n*W + n*gradw
bias = bias + n*gradb
self.W = W
self.b = bias
return W,bias,losses
svm=SVM()
xp,yp=combine_data(data[0],data[1])
w,b,loss=svm.fit(xp,yp,learning_rate=0.000001,maxItr=1000)
print(loss)
plt.plot(loss)
# +
def trainSVMs(x,y):
svm_classifier={}
for i in range(classes):
svm_classifier[i]={}
for j in range(i+1,classes):
xpair,ypair=combine_data(data[i],data[j])
wts,b,loss=svm.fit(xpair,ypair,learning_rate=0.000001,maxItr=1000)
plt.plot(loss)
svm_classifier[i][j]=(wts,b)
return svm_classifier
# -
svm_classifier=trainSVMs(img_data,labels)
print(svm_classifier)
def binaryPredict(x,w,b):
z = np.dot(x,w.T) + b
if z>=0:
return 1
else:
return -1
def predict(x):
count=np.zeros((classes,))
for i in range(classes):
for j in range(i+1,classes):
w,b=svm_classifier[i][j]
#take a majority prediction
z=binaryPredict(x,w,b)
if (z==1):
count[j]+=1
else:
count[i]+=1
final_prediction=np.argmax(count)
#print(final_prediction)
return final_prediction
print(predict(img_data[10]))
print(labels[10])
def accuracy(x,y):
count=0.0
for i in range(x.shape[0]):
pred=predict(x[i])
if (pred==y[i]):
count+=1
return count/x.shape[0]
print(accuracy(img_data,labels))
| Machine Learning /SVM/Pokemon Image Classification/Pokemon Classification.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# Given the problems with incompressibility and other things, I have completely changed the strategy for the training dataset, in this notebook I will analyze the new version of the forcing data.
import xarray as xr
import holoviews as hv
import torch
import glob
from collections.abc import Mapping
from holoviews import streams
hv.extension('bokeh')
# %opts Image[colorbar=True, invert_yaxis=True, width=500, height=200] (cmap='viridis') {+framewise +axiswise}
# %opts Raster[colorbar=True, invert_yaxis=True, width=500, height=200] (cmap='viridis') {+framewise +axiswise}
# %opts QuadMesh[colorbar=True, invert_yaxis=True, width=500, height=200] (cmap='viridis') {+framewise +axiswise}
ds = xr.open_dataset("../data/training_data.nc")
ds
# Let's look at a typical grid point on the tropics (x,y) = (0,32)...
loc = ds.isel(x=0,y=32)
# %%opts Curve[width=500, height=100]
lay= hv.Raster(loc.FQT.values.T, label='FQT') + hv.Raster(loc.FSLI.values.T, label='FSLI') \
+ hv.Curve(loc.Prec.values, vdims=['Prec'])
lay.cols(1).redim(x='time', z='c', y='z')
# It looks like this new data does not have the banding artifact near the boundary layer. What about the zonal mean?
mu = ds.mean(['x', 'time'])
lay = hv.Raster(mu.FSLI.values, label='FSLI') + hv.Raster(mu.FQT.values, label='FQT')
lay.cols(1).redim(x='y', z='c', y='z')
# There still is some strange artifacts near the edge of the domain for FSLI, but that is not unexpected. The humidity forcing looks better. Maybe there are still some bugs in how I'm computing temperature.
# # Horizontal momentum tendencies
#
# These are also kind of strange. I am not sure if these are physical or not.
# +
# %%opts Raster[width=400, height=200]{+axiswise}
fields = ['U', 'V', 'FV', 'FU']
hmap = hv.HoloMap({key: hv.Raster(mu[key].values) for key in fields}).redim(z='c', x='y', y='z')
hmap.layout().cols(2)
# -
# # Comparison with debugging data
# +
id = 'deadly_becquerel'
path = f'../data/samNN/{id}/NG1_test_0000002.pkl'
def norm(x):
return np.sqrt((x**2).mean(axis=(-1,-2)))
# get first debugging point
dbg = torch.load(f'../data/samNN/{id}/NG1_test_0000001.pkl')
state, dt = dbg['args']
# get first time point
ds = xr.open_dataset("../data/training_data.nc").isel(time=0)
# -
norm(ds.W-state['W']).plot()
norm(ds['FQT']-state['FQT']).plot()
(norm(ds['FSLI']-state['FSLI'])*86400).plot()
(norm(ds['FQT']-state['FQT'])*86400).plot()
# There is still a big discrepency in FQT and FSL in the first time point
ds.FQT[5].plot()
# +
def index_like(x, y):
if isinstance(x, Mapping):
keys = set(x) & set(y.data_vars)
return xr.Dataset({key: index_like(x[key], y[key]) for key in keys})
else:
if x.shape[0] == 1:
x = x[0]
return xr.DataArray(x, dims=y.dims, coords=y.coords)
def open_debug_state_like_ds(path: str, ds: xr.Dataset) -> xr.Dataset:
"""Open SAM debugging output as xarray object
Parameters
----------
path
path to pickle saved by torch
ds
dataset to use a template
Returns
-------
state
dataset with fields from path
"""
dbg = torch.load(path)
state = dbg['args'][0]
out = dbg['out']
return index_like(state, ds)
def concat_datasets(args, name='mode'):
"""Concatenate datasets with a new named index
This function is especially useful for comparing two datasets with
shared variables with holoviews
Parameters
----------
args : list
list of (name, dataset) pairs
name : str
name of the new dimension
Returns
-------
ds : xr.Dataset
concatenated data
"""
names, vals = zip(*args)
# get list of vars
vars = set(vals[0])
for val in vals:
vars = vars & set(val)
vars = list(vars)
vals = [val[vars] for val in vals]
return xr.concat(vals, dim=pd.Index(names, name=name))
i = 4
path = glob.glob(f'../data/samNN/{id}/NG1_test_0000*.pkl')[i]
dbg = open_debug_state_like_ds(path, ds)
cds = concat_datasets([('DBG', dbg),('Train', ds)], name='source')
# -
for key in dbg:
if dbg[key].ndim > 1:
print(norm(dbg[key]-ds[key]))
# All these should be identically zero. That they are not indicates some problem.
# +
# %%opts Curve[invert_axes=True] {+framewise}
variables_to_plot = [ 'FSLI', 'FQT','SLI', 'QT', 'W', 'U']
data_to_plot = cds[variables_to_plot].to_array(dim='variable', name='value')
hv.Dataset(data_to_plot).to.curve("z", dynamic=True)\
.overlay("source")
# -
# The debugging and training data are mostly similar although not identical. However, the neural network should hopefully not be sensitive to these types of small differences. I need to check, but I suspect the problem is that the network is simply too sensitive to FQT and FSLI. Now that I have ironed out many of the issues with the training data
# there are some points with extremely different points. Here is a little interface for exploring the domain.
# +
# %%opts Curve[invert_axes=True] {+framewise}
variables_to_plot = [ 'FSLI', 'FQT','SLI', 'QT', 'W', 'U']
data_to_plot = cds[variables_to_plot].to_array(dim='variable', name='value')
def curves( x, y):
if None in [x, y]:
x, y = (0,0)
return hv.Dataset(cds['W'].sel(x=x, y=y, method='nearest')).to.curve("z").overlay("source")
w_im = hv.Image(ds.Prec)
# pointer = streams.SingleTap(transient=True, source=w_im)
pointer = streams.PointerXY(x=0,y=0, source=w_im)
dmap = hv.DynamicMap(curves, kdims=['x', 'y'], streams=[pointer]).redim.values(key=variables_to_plot)
dmap.select(key='W').redim.range(W=(-.3, .3)) + w_im
| notebooks/1.5-SAM-processed-forcings.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# %matplotlib inline
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
#Import the data
url="http://bit.ly/w-data"
data=pd.read_csv(url)
data1=data
print("The data is imported successfully")
data
data.describe()
# ##
# DATA VISUALIZATION¶
#
# Now let's plot a graph of our data so that it will give us clear idea about data.
#
# +
#Plotting the distribution of scores
data.plot(x='Hours', y='Scores', style='o')
plt.title('Hours vs Percentage')
plt.xlabel('Hours Studied')
plt.ylabel('Percentage Score')
plt.show()
# -
# # Linear Regression Model
#
# Now we prepare the data and split it in test data
#
#Splitting training and testing data
x=data.iloc[:,:-1].values
y=data.iloc[:,1].values
x_train, x_test, y_train, y_test= train_test_split(x, y,train_size=0.80,test_size=0.20,random_state=0)
# # Training the model
from sklearn.linear_model import LinearRegression
linearRegressor= LinearRegression()
linearRegressor.fit(x_train, y_train)
y_predict= linearRegressor.predict(x_train)
# # Training the Algorithm
# Now the spliting of our data into training and testing sets is done, now it's time to train our algorithm.
#
# +
regressor = LinearRegression()
regressor.fit(x_train, y_train)
print("Training complete.")
# -
# Plotting the regression line
line = regressor.coef_*x+regressor.intercept_
# Plotting for the test data
plt.scatter(x, y)
plt.plot(x, line)
plt.show()
# #### Checking the accuracy scores for training and test set
# +
print('Test Score')
print(regressor.score(x_test, y_test))
print('Training Score')
print(regressor.score(x_train, y_train))
# -
# # Now we make predictions
# +
#Let's predict the score for 9.25 hpurs
print('Score of student who studied for 9.25 hours a dat', regressor.predict([[9.25]]))
# -
# # Model Evaluation Metrics
#
| Task1.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense, Flatten, Dropout
from keras.utils import np_utils
from keras.callbacks import Callback
import json
import wandb
from wandb.keras import WandbCallback
import numpy as np
# -
# ### Logging code
# +
run = wandb.init()
config = run.config
config.epochs = 10
# -
# ### Load Data
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train[0]
X_train.shape
img_width = X_train.shape[1]
img_width
img_height = X_train.shape[2]
img_height
# ### Normalization
X_train = X_train.astype("float")
X_train /= 255.
X_test = X_test.astype("float")
X_test /= 255.
# ### One hot encoder outputs
ohe_y_train = np_utils.to_categorical(y_train)
ohe_y_train[0]
ohe_y_test = np_utils.to_categorical(y_test)
labels = range(10)
num_classes = ohe_y_train.shape[1]
num_classes
# ### Create Model
model = Sequential()
model .add(Flatten(input_shape=(img_width, img_height)))
model.add(Dense(num_classes, activation="softmax"))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
model.summary()
# ### Training
model.fit(X_train, ohe_y_train, epochs=config.epochs, validation_data=(X_test, ohe_y_test), callbacks=[WandbCallback(labels=labels, data_type="image")])
print(model.predict(X_test[:10, :, :]))
print(is_five_train[:50])
| dl-debug/debug-mnist-linear.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import sys
sys.path += ['../../lib', '../../build', '../../../build']
import numpy as np
import libry as ry
import time
C = ry.Config()
C.view()
C.addFile('../Control/minimal/scene.g')
tau=.01
# +
CS = ry.CtrlSet()
CS.add_qControlObjective(2, 1e-2*np.math.sqrt(tau), C)
CS.add_qControlObjective(1, 1e-1*np.math.sqrt(tau), C)
pos = CS.addObjective(C.feature(ry.FS.poseDiff, ["gripper", "target"], [1e0]), ry.OT.sos, .1)
CS.addObjective(C.feature(ry.FS.accumulatedCollisions, ["ALL"], [1e2]), ry.OT.eq)
# -
ctrl = ry.CtrlSolver(C, tau, 2)
# +
for t in range(0,100):
ctrl.set(CS)
ctrl.update(C)
q = ctrl.solve()
C.setJointState(q)
C.computeCollisions()
# ctrl.report();
# C.watch(false, STRING("t:" <<t));
time.sleep(.01)
# if(pos->status>AS_running) break;
#// if(CS.isConverged(ctrl.komo.pathConfig)) break;
# -
import os
os._exit(0)
import sys
sys.path += ['../../lib', '../../build', '../../../build']
import numpy as np
import libry as ry
import time
C = ry.Config()
C.addFile('../Control/minimal/pandas.g')
C.view()
tau=.01
# +
approach = ry.CtrlSet()
approach.addObjective(C.feature(ry.FS.vectorZDiff, ["object", "R_gripperCenter"], [1e1]), ry.OT.sos, .005)
approach.addObjective(C.feature(ry.FS.positionRel, ["object", "R_gripperCenter"], [1e1], [.0, 0., -.15]), ry.OT.sos, .005)
#approach.symbolicCommands.append({"openGripper", "R_gripper"});
preGrasp = ry.CtrlSet()
#immediate constraint:
preGrasp.addObjective(C.feature(ry.FS.insideBox, ["object", "R_gripperPregrasp"], [1e0]), ry.OT.ineq, -1)
#transient:
preGrasp.addObjective(C.feature(ry.FS.vectorZDiff, ["object", "R_gripperCenter"], [1e1]), ry.OT.sos, .005)
preGrasp.addObjective(C.feature(ry.FS.positionDiff, ["R_gripperCenter", "object"], [1e1]), ry.OT.sos, .002)
#preGrasp.symbolicCommands.append({"preOpenGripper", "R_gripper"});
grasp = ry.CtrlSet()
grasp.addObjective(C.feature(ry.FS.vectorZ, ["R_gripperCenter"], [], [0., 0., 1.]), ry.OT.eq, -1.)
grasp.addObjective(C.feature(ry.FS.positionDiff, ["R_gripperCenter", "object"], [1e1]), ry.OT.eq, -1.)
#grasp.symbolicCommands.append({"closeGripper", "R_gripper"});
controls = ry.CtrlSet()
controls.add_qControlObjective(2, 1e-3*np.math.sqrt(tau), C)
controls.add_qControlObjective(1, 1e-1*np.math.sqrt(tau), C)
#auto coll = ctrl.addObjective(FS_accumulatedCollisions, {}, OT_eq, {1e2});
# -
ctrl = ry.CtrlSolver(C, tau, 2)
# +
for t in range (0,100):
if grasp.isConverged(ctrl):
break;
elif grasp.canBeInitiated(ctrl):
ctrl.set(grasp)
elif preGrasp.canBeInitiated(ctrl):
ctrl.set(preGrasp)
elif approach.canBeInitiated(ctrl):
ctrl.set(approach)
ctrl.update(C)
q = ctrl.solve()
C.setJointState(q)
C.computeCollisions()
# ctrl.report();
# C.watch(false, STRING(txt <<"t:" <<t));
time.sleep(.01);
# // if(c_pos->status>AS_running) break;
# -
| test/ry/5-control.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import pandas as pd
import numpy as np
measurements = pd.DataFrame(pd.read_csv('../Resources/hawaii_measurements.csv'))
stations = pd.DataFrame(pd.read_csv('../Resources/hawaii_stations.csv'))
# **Check files info and missing values**
print(measurements.info())
print(measurements.isnull().sum())
measurements.head()
# **Check if there are any 0 values in the data**
print('0 in station:\n', measurements.loc[measurements['station'] == 0,:].count())
print()
print('0 in date:\n', measurements.loc[measurements['date'] == 0,:].count())
print()
print('0 in prcp:\n',measurements.loc[measurements['prcp'] == 0,:].count())
print()
print('0 in tobs:\n',measurements.loc[measurements['tobs'] == 0,:].count())
print(stations.isnull().sum())
print(stations.info())
stations
# **Check what stations have NaN values**
nans = measurements[pd.isnull(measurements['prcp'])]
nans['station'].value_counts()
print('% of NaN values from the whole dataset:', round((len(nans)/len(measurements)*100),2), '%')
# **Drop all null values**
clean_measure = measurements.dropna(how='any').reset_index(drop=True)
clean_measure.info()
# **Add empty index columns (weird)**
#clean_measure.insert(0, 'id', range(1, len(clean_measure)+1))
clean_stations = stations.copy()
#clean_stations.insert(0, 'id', range(1, len(clean_stations)+1))
clean_measure['id'] = ''
clean_stations['id'] = ''
# **Create csvs**
#clean_stations = stations.copy()
clean_measure.to_csv('../Resources/clean_hawaii_measurements.csv', index=False)
clean_stations.to_csv('../Resources/clean_hawaii_stations.csv', index=False)
clean_measure.head()
clean_measure['date'].max()
| Notebooks/1_data_engineering.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
# #!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Wed Jun 23 20:09:54 2021
@author: <NAME>
"""
# install the libraries
import sys
# !{sys.executable} -m pip install numpy
# !{sys.executable} -m pip install matplotlib
# !{sys.executable} -m pip install pandas
# import the libraries
import pandas as pd
from datetime import datetime
import matplotlib.pyplot as plt
# -
def parser(s):
return datetime.strptime(s, '%Y-%m-%d')
def plot_series(series1, series2):
plt.figure(figsize=(12,6))
plt.plot(series1, color='orange')
plt.plot(series2,color='green')
plt.ylabel('Windspeed', fontsize=16)
plt.legend(['Tondano', 'Winangun'], fontsize=16)
for year in range(2010, 2020):
plt.axvline(datetime(year,1,1), linestyle='--', color='k', alpha=0.5)
# read the data
dfTondano = pd.read_csv('cleaned_dataset_tondano.csv', parse_dates=[0], index_col=0, squeeze=True, date_parser=parser)
dfWinangun = pd.read_csv('cleaned_dataset_winangun.csv', parse_dates=[0], index_col=0, squeeze=True, date_parser=parser)
#print (dfTondano[pd.to_numeric(dfTondano['windspeed'], errors='coerce').isnull()])
dfTondano
dfWinangun
plot_series(dfTondano, dfWinangun)
plt.savefig('output_postprocessing/sample1.png')
# +
# Normalize
tondano_avg, tondano_dev = dfTondano.mean(), dfTondano.std()
dfTondano = (dfTondano - tondano_avg) / tondano_dev
winangun_avg, winangun_dev = dfWinangun.mean(), dfWinangun.std()
dfWinangun = (dfWinangun - winangun_avg) / winangun_dev
plot_series(dfTondano, dfWinangun)
plt.axhline(0, linestyle='--', color='k', alpha=0.3)
plt.savefig('output_postprocessing/sample2.png')
# -
# Take First Difference to Remove Trend
dfTondano = dfTondano.diff().dropna()
dfWinangun = dfWinangun.diff().dropna()
dfTondano
dfWinangun
plot_series(dfTondano, dfWinangun)
plt.axhline(0, linestyle='--', color='k', alpha=0.3)
plt.savefig('output_postprocessing/sample3.png')
tondano_annual_volatility = dfTondano.groupby(dfTondano.index.year).std()
tondano_annual_volatility
winangun_annual_volatility = dfWinangun.groupby(dfWinangun.index.year).std()
winangun_annual_volatility
tondano_annual_vol = dfTondano.index.map(lambda d: tondano_annual_volatility.loc[d.year])
tondano_annual_vol
winangun_annual_vol = dfWinangun.index.map(lambda d: winangun_annual_volatility.loc[d.year])
winangun_annual_vol
dfTondano = dfTondano / tondano_annual_vol
dfTondano
dfWinangun = dfWinangun / winangun_annual_vol
dfWinangun
plot_series(dfTondano, dfWinangun)
plt.axhline(0, linestyle='--', color='k', alpha=0.3)
plt.savefig('output_postprocessing/sample4.png')
# Remove Seasonality
tondano_month_avgs = dfTondano.groupby(dfTondano.index.month).mean()
tondano_month_avgs
winangun_month_avgs = dfWinangun.groupby(dfWinangun.index.month).mean()
winangun_month_avgs
tondano_month_avg = dfTondano.index.map(lambda d: tondano_month_avgs.loc[d.month])
tondano_month_avg
winangun_month_avg = dfWinangun.index.map(lambda d: winangun_month_avgs.loc[d.month])
winangun_month_avg
dfTondano = dfTondano - tondano_month_avg
dfTondano
dfWinangun = dfWinangun - winangun_month_avg
dfWinangun
plot_series(dfTondano, dfWinangun)
plt.axhline(0, linestyle='--', color='k', alpha=0.3)
plt.savefig('output_postprocessing/sample5.png')
# export output preprocessing to .csv file
dfTondano.to_csv(r'output_postprocessing/postprocessing_dataset_tondano.csv', index = False)
dfWinangun.to_csv(r'output_postprocessing/postprocessing_dataset_winangun.csv', index = False)
| trash/preprocessing_combine.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Getting Started with Symbulate
#
# ## Section 2. Random Variables
# <a id='contents'></a>
# <[Probability Spaces](gs_probspace.ipynb) | [Contents](index.ipynb) | [Multiple random variables and joint distributions](gs_joint.ipynb)>
# **Every time you start Symbulate**, you must first run (SHIFT-ENTER) the following commands.
from symbulate import *
# %matplotlib inline
# This section provides an introduction to the Symbulate commands for simulating and summarizing values of a random variable.
# <a id='counting_numb_heads'></a>
# ### Example 2.1: Counting the number of Heads in a sequence of coin flips
# In [Example 1.7](gs_probspace.ipynb#e2.6) we simulated the value of the number of Heads in a sequence of five coin flips. In that example, we simulated the individual coin flips (with 1 representing Heads and 0 Tails) and then used [`.apply()`](https://dlsun.github.io/symbulate/sim.html#apply) with the `sum` function to count the number of Heads. The following Symbulate commands achieve the same goal by defining an [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV), `X`, which measures the number of Heads for each outcome.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
X.sim(10000)
# The number of Heads in five coin flips is a [**random variable**](https://dlsun.github.io/symbulate/rv.html): a function that takes as an input an outcome of a probability space and returns a real number. The first argument of [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) is the [probability space](https://dlsun.github.io/symbulate/probspace.html#Probability-Spaces) on which the [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) is defined, e.g., sequences of five 1/0s. The second argument is the function which maps outcomes in the probability space to real numbers, e.g., the `sum` of the 1/0 values. Values of an [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) can be simulated with [`.sim()`](https://dlsun.github.io/symbulate/sim.html#sim).
# <a id='sum_of_two_dice'></a>
# ### Exercise 2.2: Sum of two dice
# After defining an appropriate [`BoxModel`](https://dlsun.github.io/symbulate/probspace.html#boxmodel) probability space, define an [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) `X` representing the sum of two six-sided fair dice, and simulate 10000 values of `X`.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_sum_of_two_dice)
# <a id='dist_of_five_flips'></a>
# ### Example 2.3: Summarizing simulation results with tables and plots
# In [Example 2.1](#counting_numb_heads) we defined a [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV), X, the number of Heads in a sequence of five coin flips. Simulated values of a random variable can be summarized using [`.tabulate()`](https://dlsun.github.io/symbulate/sim.html#tabulate) (with `normalize=False` (default) for frequencies (counts) or `True` for relative frequencies (proportions)).
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
sims = X.sim(10000)
sims.tabulate()
# The table above can be used to approximate the distribution of the number of Heads in five coin flips. The [*distribution*](https://dlsun.github.io/symbulate/rv.html#distribution) of a random variable specifies the possible values that the random variable can take and their relative likelihoods. The distribution of a random variable can be visualized using [`.plot()`](https://dlsun.github.io/symbulate/rv.html#Plot).
sims.plot()
# By default, [`.plot()`](https://dlsun.github.io/symbulate/rv.html#Plot) displays relative frequencies (proportions). Use [`.plot(normalize=False)`](https://dlsun.github.io/symbulate/rv.html#Plot) to display frequencies (counts).
# <a id='dist_of_sum_of_two_dice'></a>
# ### Exercise 2.4: The distribution of the sum of two dice rolls
# Continuing [Exercise 2.2](#sum_of_two_dice) summarize with a table and a plot the distribution of the sum of two rolls of a fair six-sided die.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_dist_of_sum_of_two_dice)
# <a id='prob_of_three_heads'></a>
# ### Example 2.5: Estimating probabilities from simulations
# There are several other [tools](https://dlsun.github.io/symbulate/sim.html) for summarizing simulations, like the [`count`](https://dlsun.github.io/symbulate/sim.html#count) functions. For example, the following commands approximate `P(X <= 3)` for [Example 2.1](#counting_numb_heads), the probability that in five coin flips at most three of the flips land on Heads.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
sims = X.sim(10000)
sims.count_leq(3)/10000
# <a id='prob_of_10_two_dice'></a>
# ### Exercise 2.6: Estimating probabilities for the sum of two dice rolls
# Continuing [Exercise 2.2](#sum_of_two_dice), estimate `P(X >= 10)`, the probability that the sum of two fair six-sided dice is at least 10.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_prob_of_10_two_dice)
# <a id='sim_from_binom'></a>
# ### Example 2.7: Specifying a RV by its distribution
# The plot in [Example 2.3](#dist_of_five_flips) displays the approximate distribution of the random variable `X`, the number of Heads in five flips of a fair coin. This distribution is called the [`Binomial`](https://dlsun.github.io/symbulate/common_discrete.html#Discrete-distributions) distribution with `n=5` trials (flips) and a probability that each trial (flip) results in success (1 i.e. Heads) equal to `p=0.5`.
#
# In the above examples the [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) `X` was explicitly defined on the probability space `P` - i.e. the [`BoxModel`](https://dlsun.github.io/symbulate/probspace.html#boxmodel) for the outcomes (1 or 0) of the five individual flips - via the `sum` function. This setup implied a Binomial(5, 0.5) distribution for `X`.
#
# In many situations the distribution of an [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) is assumed or specified directly, without mention of the underlying probabilty space or the function defining the random variable. For example, a problem might state "let `Y` have a Binomial distribution with `n=5` and `p=0.5`". The [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) command can also be used to define a random variable by specifying its distribution, as in the following.
Y = RV(Binomial(5, 0.5))
Y.sim(10000).plot()
# By definition, a random variable must always be a function defined on a probability space. Specifying a random variable by specifying its distribution, as in `Y = RV(Binomial(5, 0.5))`, has the effect of defining the probability space to be the distribution of the random variable and the function defined on this space to be the identity (`f(x) = x`). However, it is more appropriate to think of such a specification as defining a random variable with the given distribution on an *unspecified probability space* through an *unspecified function*.
#
#
# For example, the random variable $X$ in each of the following situations has a Binomial(5, 0.5) distribution.
# - $X$ is the number of Heads in five flips of a fair coin
# - $X$ is the number of Tails in five flips of a fair coin
# - $X$ is the number of even numbers rolled in five rolls of a fair six-sided die
# - $X$ is the number of boys in a random sample of five births
#
# Each of these situations involves a different probability space (coins, dice, births) with a random variable which counts according to different criteria (Heads, Tails, evens, boys). These examples illustrate that knowledge that a random variable has a specific distribution (e.g. Binomial(5, 0.5)) does not necessarily convey any information about the underlying observational units or variable being measured. This is why we say a specification like `X = RV(Binomial(5, 0.5))` defines a random variable `X` on an unspecified probability space via an unspecified function.
#
#
# The following code compares the two methods for definiting of a random variable with a Binomial(5, 0.5) distribution. (The `jitter=True` option offsets the vertical lines so they do not coincide.)
# +
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
X.sim(10000).plot(jitter=True)
Y = RV(Binomial(5, 0.5))
Y.sim(10000).plot(jitter=True)
# -
# In addition to [`Binomial`](https://dlsun.github.io/symbulate/common_discrete.html#binomial), many other [commonly used distributions](https://dlsun.github.io/symbulate/common_discrete.html) are built in to Symbulate.
# <a id='discrete_unif_dice'></a>
# ### Exercise 2.8: Simulating from a discrete Uniform model
# A random variable has a [`DiscreteUniform`](https://dlsun.github.io/symbulate/common_discrete.html#discrete) distribution with parameters `a` and `b` if it is equally likely to to be any of the integers between `a` and `b` (inclusive). Let `X` be the roll of a fair six-sided die. Define an `RV` `X` by specifying an appropriate `DiscreteUniform` distribution, then simulate 10000 values of `X` and summarize its approximate distribution in a plot.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_expected_discrete_unif_dice)
# <a id='numb_tails'></a>
# ### Example 2.9: Random variables versus distributions
# Continuing [Example 2.1](#counting_numb_heads), if `X` is the random variable representing number of Heads in five coin flips then `Y = 5 - X` is random variable representing the number of Tails.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
Y = 5 - X
Y.sim(10000).tabulate()
# It is important not to confuse a random variable with its distribution. Note that `X` and `Y` are two different random variables; they measure different things. For example, if the outcome of the flips is `(1, 0, 0, 1, 0)` then `X = 2` but `Y = 3`. The following code illustrates how an [`RV`](https://dlsun.github.io/symbulate/rv.html#Random-variables) can be called as a function to return its value for a particular outcome in the probability space.
outcome = (1, 0, 0, 1, 0)
X(outcome)
Y(outcome)
# In fact, in this example the values of `X` and `Y` are unequal for every outcome in the probability space . However, while `X` and `Y` are two different random variables, they do have the same distribution over many outcomes.
X.sim(10000).plot(jitter=True)
Y.sim(10000).plot(jitter=True)
# See [Example 2.7](#sim_from_binom) for further comments about the difference between random variables and distributions.
# <a id='expected_value_numb_of_heads'></a>
# ### Example 2.10: Expected value of the number of heads in five coin flips
# The expected value, or probability-weighted average value, of an [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) can be approximated by simulating many values of the random variable and finding the sample mean (i.e. average) using [`.mean()`](https://dlsun.github.io/symbulate/rv.html#Mean). Continuing [Example 2.1](#counting_numb_heads), the following code estimates the expected value of the number of Heads in five coin flips.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
X.sim(10000).mean()
# Over many sets of five coin flips, we expect that there will be on average about 2.5 Heads per set. Note that 2.5 is *not* the number of Heads we would expect in a single set of five coin flips.
# <a id='expected_value_sum_of_dice'></a>
# ### Exercise 2.11: Expected value of the sum of two dice rolls
# Continuing [Exercise 2.2](#sum_of_two_dice), approximate the expected value of the sum of two six-sided dice rolls. (Bonus: interpret the value as an appropriate long run average.)
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_expected_value_sum_of_dice)
# <a id='sd_numb_of_heads'></a>
# ### Example 2.12: Standard deviation of the number of Heads in five coin flips
# The [expected value](https://dlsun.github.io/symbulate/rv.html#Mean) of an [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) is its long run average, while the [standard deviation](https://dlsun.github.io/symbulate/rv.html#Variance) of an [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) measures the average degree to which individual values of the [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) vary from the expected value. The [standard deviation](https://dlsun.github.io/symbulate/rv.html#Variance) of an [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) can be approximated from simulated values with [`.sd()`](https://dlsun.github.io/symbulate/rv.html#Variance). Continuing [Example 2.1](#counting_numb_heads), the following code estimates the standard deviation of the number of Heads in five coin flips.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
sims = X.sim(10000)
sims.sd()
# Inspecting the plot in [Example 2.3](#dist_of_five_flips) we see there are many simulated values of 2 and 3, which are 0.5 units away from the expected value of 2.5. There are relatively fewer values of 0 and 5 which are 2.5 units away from the expected value of 2.5. Roughly, the simulated values are on average 1.1 units away from the expected value.
# *Variance* is the square of the standard deviation and can be approximated with [`.var()`](https://dlsun.github.io/symbulate/rv.html#Variance).
sims.var()
# <a id='sd_sum_of_dice'></a>
# ### Exercise 2.13: Standard deviation of the sum of two dice rolls
# Continuing [Exercise 2.2](#sum_of_two_dice), approximate the standard deviation of the sum of two six-sided dice rolls. (Bonus: interpret the value.)
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_sd_sum_of_dice)
# <a id='dist_of_normal'></a>
# ### Example 2.14: Continuous random variables
# The [`RV`](https://dlsun.github.io/symbulate/rv.html#RV)s we have seen so far have been [discrete](https://dlsun.github.io/symbulate/common_discrete.html). A discrete random variable can take at most countably many distinct values. For example, the number of Heads in five coin flips can only take values 0, 1, 2, 3, 4, 5.
#
# A [continuous](https://dlsun.github.io/symbulate/common_continuous.html) random variable can take any value in some interval of real numbers. For example, if `X` represents the height of a randomly selected U.S. adult male then `X` is a continuous random variable. Many continuous random variables are assumed to have a [Normal](https://dlsun.github.io/symbulate/common_continuous.html#normal) distribution. The following simulates values of the [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) `X` assuming it has a [Normal](https://dlsun.github.io/symbulate/common_continuous.html#normal) distribution with mean 69.1 inches and standard deviation 2.9 inches.
X = RV(Normal(mean=69.1, sd=2.9))
sims = X.sim(10000)
# The same simulation tools are available for both discrete and continuous [`RV`](https://dlsun.github.io/symbulate/rv.html#RV)s. Calling [`.plot()`](https://dlsun.github.io/symbulate/rv.html#plot) for a continuous RV produces a histogram which displays frequencies of simulated values falling in interval "bins".
sims.plot()
# The number of bins can be set using the [`bins= `](https://dlsun.github.io/symbulate/rv.html#Plot) option in [`.plot()`](https://dlsun.github.io/symbulate/rv.html#Plot)
X.sim(10000).plot(bins=60)
# It is not recommended to use [`.tabulate()`](https://dlsun.github.io/symbulate/sim.html#tabulate) with continuous [`RV`](https://dlsun.github.io/symbulate/rv.html#RV)s as almost all simulated values will only occur once.
# <a id='sim_unif'></a>
# ### Exercise 2.15: Simulating from a (continuous) uniform distribution
# The continuous analog of a [`BoxModel`](https://dlsun.github.io/symbulate/probspace.html#boxmodel) is a [`Uniform`](https://dlsun.github.io/symbulate/common_continuous.html#uniform) distribution which produces "equally likely" values in an interval with endpoints `a` and `b`. (What would you expect the plot of such a distribution to look like?)
#
# Let `X` be a random variable which has a [`Uniform`](https://dlsun.github.io/symbulate/common_continuous.html#uniform) distribution on the interval [0, 1]. Define an appropriate [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) and use simulation to display its approximate distribution. (Note that the underlying probability space is unspecified.)
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_sim_unif)
# <a id='sqrt_ex'></a>
# ### Example 2.16: Transformations of random variables
# In [Example 2.9](#numb_tails) we defined a new random variable `Y = 5 - X` (the number of Tails) by transforming the `RV` `X` (the number of Heads). A [transformation of an RV](https://dlsun.github.io/symbulate/rv.html#transform) is also an [`RV`](https://dlsun.github.io/symbulate/rv.html#Random-variables). If `X` is an [`RV`](https://dlsun.github.io/symbulate/rv.html#Random-variables), define a new random variable `Y = g(X)` using `X.apply(g)`. The resulting `Y` behaves like any other [`RV`](https://dlsun.github.io/symbulate/rv.html#Random-variables).
# Note that for arithmetic operations and many common math functions (such as `exp`, `log`, `sin`) you can simply call `g(X)` rather than `X.apply(g)`.
#
# Continuing [Example 2.1](#counting_numb_heads), let $X$ represent the number of Heads in five coin flips and define the random variable $Y = \sqrt{X}$. The plot below approximates the distribution of $Y$; note that the possible values of $Y$ are 0, 1, $\sqrt{2}$, $\sqrt{3}$, 2, and $\sqrt{5}$.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
Y = X.apply(sqrt)
Y.sim(10000).plot()
# The following code uses a `g(X)` definition rather than `X.apply(g)`.
P = BoxModel([1, 0], size=5)
X = RV(P, sum)
Y = sqrt(X)
Y.sim(10000).plot()
# <a id='dif_normal'></a>
# ### Exercise 2.17 Function of a RV that has a Uniform distribution
# In [Example 2.15](#sim_unif) we encountered uniform distributions. Let $U$ be a random variable which has a Uniform distribution on the interval [0, 1]. Use simulation to display the approximate distribution of the random variable $Y = -\log(U)$.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_dif_normal)
# <a id='Numb_distinct'></a>
# ### Example 2.18: Number of switches between Heads and Tails in coin flips
# [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV)s can be defined or transformed through [user defined functions](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV). As an example, let `Y` be the number of times a sequence of five coin flips switches between Heads and Tails (not counting the first toss). For example, for the outcome (0, 1, 0, 0, 1), a switch occurs on the second third, and fifth flip so `Y = 3`. We define the random variable `Y` by first defining a function that takes as an input a list of values and returns as an output the number of times a switch from the previous value occurs in the sequence. ([Defining functions](https://docs.python.org/3/reference/compound_stmts.html#function-definitions) is one area where some familiarity with Python is helpful.)
# +
def number_switches(x):
count = 0
for i in list(range(1, len(x))):
if x[i] != x[i-1]:
count += 1
return count
number_switches((1, 1, 1, 0, 0, 1, 0, 1, 1, 1))
# -
# Now we can use the `number_switches` function to define the `RV` `Y` on the probability space corresponding to five flips of a fair coin.
# +
P = BoxModel([1, 0], size=5)
Y = RV(P, number_switches)
outcome = (0, 1, 0, 0, 1)
Y(outcome)
# -
# An [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV) defined or transformed through a user-defined function behaves like any other [`RV`](https://dlsun.github.io/symbulate/rv.html#Defining-a-random-variable-with-RV).
Y.sim(10000).plot()
# <a id='Numb_alterations'></a>
# ### Exercise 2.19: Number of distinct faces rolled in 6 rolls
# Let `X` count the number of distinct faces rolled in 6 rolls of a fair six-sided die. For example, if the result of the rolls is (3, 3, 3, 3, 3, 3) then `X = 1`; if (6, 4, 5, 4, 6, 6) then `X=3`; etc. Use the `number_distinct_values` function defined below to define the `RV` `X` on an appropriate probability space. Then simulate values of `X` and plot its approximate distribution. (The `number_distinct_values` function takes as an input a list of values and returns as an output the number of distinct values in the list. We have used the Python functions [`set`](https://docs.python.org/3/tutorial/datastructures.html#sets) and [`len`](https://docs.python.org/3/library/functions.html#len).)
# +
def number_distinct_values(x):
return len(set(x))
number_distinct_values((1, 1, 4))
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Solution](#sol_Numb_alterations)
# ## Additional Exercises
# <a id='ev_max_of_dice'></a>
# ### Exercise 2.20: Max of two dice rolls
# 1) Approximate the distribution of the max of two six-sided dice rolls.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 2) Approximate the probability that the max of two six-sided dice rolls is greater than or equal to 5.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 3) Approximate the mean and standard deviation of the max of two six-sided dice rolls.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Hint](#hint_ev_max_of_dice)
# [Solution](#sol_ev_max_of_dice)
# <a id='var_transformed_unif'></a>
# ### Exercise 2.21: Transforming a random variable
# Let $X$ have a [`Uniform`](https://dlsun.github.io/symbulate/common_continuous.html#uniform) distribution on the interval [0, 3] and let $Y = 2\cos(X)$.
#
# 1) Approximate the distribution of $Y$.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 2) Approximate the probability that the $Y$ is less than 1.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 3) Approximate the mean and standard deviation of $Y$.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Hint](#hint_var_transformed_unif)
# [Solution](#sol_var_transformed_unif)
# <a id='log_normal'></a>
# ### Exercise 2.22: Function of a random variable.
# Let $X$ be a random variable which has a Normal(0,1) distribution. Let $Y = e^X$.
# 1) Use simulation to display the approximate distribution of $Y$.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 2) Approximate the probability that the $Y$ is greater than 2.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# 3) Approximate the mean and standard deviation of $Y$.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# +
### Type your commands in this cell and then run using SHIFT-ENTER.
# -
# [Hint](#hint_log_normal)
# [Solution](#sol_log_normal)
# <a id='hints'></a>
# ## Hints for Additional Exercises
# <a id='hint_ev_max_of_dice'></a>
# ### Exercise 2.20: Hint
# In [Exercise 2.2](#sum_of_two_dice) we simulated the sum of two six-sided dice rolls. Define an [`RV`](https://dlsun.github.io/symbulate/rv.html#RV) using the `max` function to return the larger of the two rolls. In [Example 2.5](#prob_of_three_heads) we estimated the probability of a random variable taking a value. In [Example 2.10](#expected_value_numb_of_heads) we applied the [`.mean()`](https://dlsun.github.io/symbulate/rv.html#mean) funtion to return the long run expected average. In [Example 2.12](#sd_numb_of_heads) we estimated the standard deviation.
# [Back](#ev_max_of_dice)
# <a id='hint_var_transformed_unif'></a>
# ### Exercise 2.21: Hint
# [Example 2.9](#numb_tails) introduces transformations. In [Exercise 2.15](#sim_unif) we simulated an RV that had a Uniform distribution. In [Example 2.5](#prob_of_three_heads) we estimated the probabilities for a RV. In [Example 2.10](#expected_value_numb_of_heads) we applied the [`.mean()`](https://dlsun.github.io/symbulate/rv.html#mean) funtion to return the long run expected average. In [Example 2.12](#sd_numb_of_heads) we estimated the standard deviation.
# [Back](#var_transformed_unif)
# <a id='hint_log_normal'></a>
# ### Exercise 2.22: Hint
# In [Example 2.14](#dist_of_norm) we simulated an RV with a Normal distribution. In [Example 2.9](#numb_tails) we defined a random variable as a function of another random variable. In [Example 2.5](#prob_of_three_heads) we estimated the probability of a random variable taking a value. In [Example 2.10](#expected_value_numb_of_heads) we applied the [.mean()](https://dlsun.github.io/symbulate/rv.html#mean) funtion to return the long run expected average. In [Example 2.12](#sd_numb_of_heads) we estimated the standard deviation.
# [Back](#log_normal)
# ## Solutions to Exercises
# <a id='sol_sum_of_two_dice'></a>
# ## Exercise 2.2: Solution
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, sum)
X.sim(10000)
# [Back](#dist_of_five_flips)
# <a id='sol_dist_of_sum_of_two_dice'></a>
# ## Exercise 2.4: Solution
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, sum)
sims = X.sim(10000)
sims.tabulate(normalize=True)
sims.plot()
# [Back](#prob_of_three_heads)
# <a id='sol_prob_of_10_two_dice'></a>
# ## Exercise 2.6: Solution
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, sum)
sims = X.sim(10000)
sims.count_geq(10) / 10000
# [Back](#sim_from_binom)
# <a id='sol_expected_discrete_unif_dice'></a>
# ## Exercise 2.8: Solution
X = RV(DiscreteUniform(a=1, b=6))
X.sim(10000).plot(normalize=True)
# [Back](#numb_tails)
# <a id='sol_expected_value_sum_of_dice'></a>
# ## Exercise 2.11: Solution
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, sum)
X.sim(10000).mean()
# Over many pairs of rolls of fair six-sided dice, we expect that on average the sum of the two rolls will be about 7.
# [Back](#sd_numb_of_heads)
# <a id='sol_sd_sum_of_dice'></a>
# ## Exercise 2.13: Solution
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, sum)
X.sim(10000).sd()
# Over many pairs of rolls of fair six-sided dice, the values of the sum are on average roughly 2.4 units away from the expected value of 7.
# [Back](#dist_of_normal)
# <a id='sol_sim_unif'></a>
# ## Exercise 2.15: Solution
X = RV(Uniform(a=0, b=1))
X.sim(10000).plot()
# [Back](#sqrt_ex)
# <a id='sol_dif_normal'></a>
# ## Exercise 2.17: Solution
U = RV(Uniform(a=0, b=1))
Y = -log(U)
Y.sim(10000).plot()
# Note that the [`RV`](https://dlsun.github.io/symbulate/rv.html#Random-variables) has an [Exponential(1)](https://dlsun.github.io/symbulate/common_discrete.html#exponential) distribution.
# [Back](#Numb_distinct)
# <a id='sol_Numb_alterations'></a>
# ## Exercise 2.19: Solution
# +
def number_distinct_values(x):
return len(set(x))
P = BoxModel([1,2,3,4,5,6], size=6)
X = RV(P, number_distinct_values)
X.sim(10000).plot()
# -
# [Back](#ev_max_of_dice)
# <a id='sol_ev_max_of_dice'></a>
# ## Exercise 2.20: Solution
# 1) Approximate the distribution of the max of two six-sided dice rolls.
P = BoxModel([1, 2, 3, 4, 5, 6], size=2)
X = RV(P, max)
sims = X.sim(10000)
sims.plot()
# 2) Approximate the probability that the max of two six-sided dice rolls is greater than or equal to 5.
sims.count_geq(5)/10000
# 3) Approximate the mean and standard deviation of the max of two six-sided dice rolls.
sims.mean()
sims.sd()
# [Back](#var_transformed_unif)
# <a id='sol_var_transformed_unif'></a>
# ## Exercise 2.21: Solution
# 1) Approximate the distribution of $Y$.
X = RV(Uniform(0, 3))
Y = 2 * cos(X)
sims = Y.sim(10000)
sims.plot()
# Alternatively,
X = RV(Uniform(0, 3))
Y = 2 * X.apply(cos)
sims = Y.sim(10000)
sims.plot()
# 2) Approximate the probability that the Y is less than 2.
sims.count_lt(1)/10000
# 3) Approximate the mean and standard deviation of Y.
sims.mean()
sims.sd()
# [Back](#log_normal)
# <a id='sol_log_normal'></a>
# ## Exercise 2.22: Solution
# 1) Use simulation to display the approximate distribution of Y.
X = RV(Normal(0, 1))
Y = exp(X)
sims = Y.sim(10000)
sims.plot()
# 2) Approximate the probability that the Y is greater than 2.
sims.count_gt(2)/10000
# 3) Approximate the mean and standard deviation of Y.
sims.mean()
sims.sd()
# [Back](#hints)
# <[Probability Spaces](gs_probspace.ipynb) | [Contents](index.ipynb) | [Multiple random variables and joint distributions](gs_joint.ipynb)>
| tutorial/gs_rv.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # A first simple model
import pandas as pd
import pickle
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
# %matplotlib inline
# Plain Seaborn figures with matplotlib color codes mapped to the default seaborn palette
sns.set(style="white", color_codes=True)
df_merged = pd.read_csv("../data/chlamydia_cdc_census.csv")
df_merged.drop("Cases", axis=1, inplace=True)
df_merged.head()
df_zipfips= pd.read_csv("../data/ZIP_COUNTY_122014.csv", usecols={0,1})
zip2fips = dict(zip(df_zipfips["ZIP"], df_zipfips["COUNTY"]))
fips = zip2fips[10027]
target = df_merged[df_merged['FIPS']==fips]
target.shape
target_params = target.values[0]
model = pickle.load(open('../data/randomforest_params.pickle', "rb" ))
model2 = pickle.load(open('../data/gradientboosting_params.pickle', "rb" ))
Ymean = pickle.load(open('../data/Ymean.pickle', "rb"))
Ystd = pickle.load(open('../data/Ystd.pickle', "rb"))
chlamydia_rate = model.predict(target_params[1:])
# +
gender_rate = {}
gender_factor = {}
gender_number = {}
gender_rate["Male"] = 278.4e-5
gender_rate["Female"] = 627.2e-5
gender_number["Male"] = 155651602
gender_number["Female"] = 160477237
rate_average = (gender_rate["Male"]*gender_number["Male"]+gender_rate["Female"]*gender_number["Male"])/(gender_number["Male"]+gender_number["Female"])
gender_factor["Male"] = gender_rate["Male"]/rate_average
gender_factor["Female"] = gender_rate["Female"]/rate_average
gender_factor["Female"], gender_factor["Male"]
race_rate = {}
race_factor = {}
race_number = {}
race_number["Native"] = 1942876.0
race_number["Asian"] = 12721721.0
race_number["Black"] = 29489649.0
race_number["Hispanic"] = 46407173.0
race_number["Multiple"] = 5145135.0
race_number["Pacific"] = 473703.0
race_number["White"] = 161443167.0
race_rate["Native"] = 689.1e-5
race_rate["Asian"] = 115.8e-5
race_rate["Black"] = 1152.6e-5
race_rate["Hispanic"] = 376.2e-5
race_rate["Multiple"] = 116.1e-5
race_rate["Pacific"] = 641.5e-5
race_rate["White"] = 187.0e-5
US_number = race_number["Native"] + race_number["Asian"] + race_number["Black"] + race_number["Hispanic"] + race_number["Multiple"] + race_number["Pacific"] + race_number["White"]
rate_average = (race_rate["Native"]*race_number["Native"]+race_rate["Asian"]*race_number["Asian"]+race_rate["Black"]*race_number["Black"]+race_rate["Hispanic"]*race_number["Hispanic"]+race_rate["Multiple"]*race_number["Multiple"]+race_rate["Pacific"]*race_number["Multiple"]+race_rate["White"]*race_number["White"])/US_number
race_factor["Native"] = race_rate["Native"]/rate_average
race_factor["Asian"] = race_rate["Asian"]/rate_average
race_factor["Black"] = race_rate["Black"]/rate_average
race_factor["Hispanic"] = race_rate["Hispanic"]/rate_average
race_factor["Multiple"] = race_rate["Multiple"]/rate_average
race_factor["Pacific"] = race_rate["Pacific"]/rate_average
race_factor["White"] = race_rate["White"]/rate_average
age_rate = {}
age_factor = {}
age_number = {}
age_number["0-14"] = 61089123.0
age_number["15-19"] = 21158964.0
age_number["20-24"] = 22795438.0
age_number["25-29"] = 21580198.0
age_number["30-34"] = 21264389.0
age_number["35-39"] = 19603770.0
age_number["40-44"] = 20848920.0
age_number["45-54"] = 43767532.0
age_number["55-64"] = 39316431.0
age_number["65+"] = 44704074.0
age_rate["0-14"] = 20.0e-5
age_rate["15-19"] = 1804.0e-5
age_rate["20-24"] = 2484.6e-5
age_rate["25-29"] = 1176.2e-5
age_rate["30-34"] = 532.4e-5
age_rate["35-39"] = 268.0e-5
age_rate["40-44"] = 131.5e-5
age_rate["45-54"] = 56.6e-5
age_rate["55-64"] = 16.6e-5
age_rate["65+"] = 3.2e-5
US_age_number = age_number["0-14"] + age_number["15-19"] + age_number["20-24"] + age_number["25-29"] + age_number["30-34"] + age_number["35-39"] + age_number["40-44"] + age_number["45-54"] + age_number["55-64"] + age_number["65+"]
rate_average = (age_rate["0-14"]*age_number["0-14"]+age_rate["15-19"]*age_number["15-19"]+age_rate["20-24"]*age_number["20-24"]+age_rate["25-29"]*age_number["25-29"]+age_rate["30-34"]*age_number["30-34"]+age_rate["35-39"]*age_number["35-39"]+age_rate["40-44"]*age_number["40-44"]+age_rate["45-54"]*age_number["45-54"]+age_rate["55-64"]*age_number["55-64"]+age_rate["65+"]*age_number["65+"])/US_age_number
age_factor["0-14"] = age_rate["0-14"]/rate_average
age_factor["15-19"] = age_rate["15-19"]/rate_average
age_factor["20-24"] = age_rate["20-24"]/rate_average
age_factor["25-29"] = age_rate["25-29"]/rate_average
age_factor["30-34"] = age_rate["30-34"]/rate_average
age_factor["35-39"] = age_rate["35-39"]/rate_average
age_factor["40-44"] = age_rate["40-44"]/rate_average
age_factor["45-54"] = age_rate["45-54"]/rate_average
age_factor["55-64"] = age_rate["55-64"]/rate_average
age_factor["65+"] = age_rate["65+"]/rate_average
race_factor["Native"], race_factor["Asian"], race_factor["Black"], race_factor["Hispanic"], race_factor["Multiple"], race_factor["Pacific"], race_factor["White"]
age_factor["0-14"], age_factor["15-19"], age_factor["20-24"], age_factor["25-29"], age_factor["30-34"], age_factor["35-39"], age_factor["40-44"], age_factor["45-54"], age_factor["55-64"], age_factor["65+"]
# +
def calculate_rate(Zipcode, Race, Gender, Age):
fips = zip2fips[int(Zipcode)]
target = df_merged[df_merged['FIPS']==fips]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])
return chlamydia_rate*gender_factor[Gender]*race_factor[Race]*age_factor[Age]
Race = "White"
Gender = "Male"
Age = "35-39"
Zipcode = "02139"
print("Your individual chance of having Chlamydia is %.2f percent"%(calculate_rate(Zipcode, Race, Gender, Age)*100))
# -
# ## ZIP code census data base CSV file
df_zipcode = pd.read_csv("../data/census_zipcode.csv")
df_zipcode_unnormalized = pd.read_csv("../data/census_zipcode_unnormalized.csv")
df_zipcode.head()
df_zipcode_unnormalized[df_zipcode_unnormalized["geoid2"]==int(Zipcode)]
# +
def calculate_rate(Zipcode):
target = df_zipcode[df_zipcode["geoid2"]==int(Zipcode)]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
return chlamydia_rate
Race = "White"
Gender = "Male"
Age = "35-39"
Zipcode = "02139"
zipcoderate = calculate_rate(Zipcode)
target_unnormalized = df_zipcode_unnormalized[df_zipcode_unnormalized["geoid2"]==int(Zipcode)]
TOTALNR = target_unnormalized["Population"]
if Gender == "Male":
gender_table = "hd02s026"
else:
gender_table = "hd02s051"
GENDERNR = TOTALNR*target_unnormalized[gender_table]/100.0
if Race == "White":
race_table = "hd02s078"
elif Race == "Black":
race_table = "hd02s079"
elif Race == "Native":
race_table = "hd02s080"
elif Race == "Asian":
race_table = "hd02s081"
elif Race == "Pacific":
race_table = "hd02s089"
elif Race == "Multiple":
race_table = "hd02s095"
elif Race == "Hispanic":
race_table = "hd02s107"
RACENR = TOTALNR*target_unnormalized[race_table]/100.0
if Age == "0-14":
age_table = "hd02s002"
elif Age == "15-19":
age_table = "hd02s005"
elif Age == "20-24":
age_table = "hd02s006"
elif Age == "25-29":
age_table = "hd02s007"
elif Age == "30-34":
age_table = "hd02s008"
elif Age == "35-39":
age_table = "hd02s009"
elif Age == "40-44":
age_table = "hd02s010"
elif Age == "45-54":
age_table = "hd02s011"
elif Age == "55-64":
age_table = "hd02s013"
elif Age == "65+":
age_table = "hd02s015"
AGENR = TOTALNR*target_unnormalized[age_table]/100.0
the_result = 100*(zipcoderate/TOTALNR + gender_rate[Gender]/GENDERNR + race_rate[Race]/RACENR + age_rate[Age]/AGENR)/(1.0/TOTALNR+1.0/GENDERNR+1.0/RACENR+1.0/AGENR)
print("Your individual chance of having Chlamydia is %.2f percent"%(the_result))
TOTALNR, GENDERNR, RACENR, AGENR
# -
# ## Model validation with Boston Public Health Commission data
df_boston = np.genfromtxt("../data/zipcodes_boston.txt", dtype=[('Neighborhood','O'),('zip1','i8'),('zip2','i8'),('zip3','i8'),('zip4','i8'),('zip5','i8'),('zip6','i8')], delimiter=",")
df_boston, len(df_boston)
predictions = np.zeros(len(df_boston))
i = 0
for hood in df_boston:
average = 0.0
count = 0
if (hood["zip1"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip1"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip1"],round(chlamydia_rate[0]*1e5))
if (hood["zip2"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip2"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip2"],round(chlamydia_rate[0]*1e5))
if (hood["zip3"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip3"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip3"],round(chlamydia_rate[0]*1e5))
if (hood["zip4"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip4"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip4"],round(chlamydia_rate[0]*1e5))
if (hood["zip5"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip5"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip5"],round(chlamydia_rate[0]*1e5))
if (hood["zip6"]):
target = df_zipcode[df_zipcode["geoid2"]==int(hood["zip6"])]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
average += chlamydia_rate[0]*1e5
count += 1
print(hood["Neighborhood"], hood["zip6"],round(chlamydia_rate[0]*1e5))
average /= count
predictions[i] = average
i+=1
len(predictions),
df_boston_rates = np.genfromtxt("../data/rate_boston.txt", dtype=[('Neighborhood','S20'),('mean','i8'),('error','i8')], delimiter=",")
labels = df_boston["Neighborhood"]
R2 = 1.0-np.sum((predictions-df_boston_rates["mean"])**2)/np.sum((df_boston_rates["mean"]-np.mean(df_boston_rates["mean"]))**2)
R2
# +
fig = plt.figure(figsize=(10, 6))
data = np.arange(len(df_boston_rates['Neighborhood']))
ax1 = plt.scatter(data,predictions)
#plt.xlabel("Neighborhood")
plt.ylabel("Chlamydia cases [per 100,000]")
ax2 = plt.errorbar(data, df_boston_rates["mean"], yerr=df_boston_rates["error"],fmt='o', color='red')
plt.xlim([-1,15])
plt.ylim([0,1400])
x = np.arange(15)
labels = ['Hyde Park', 'West Roxbury', 'Roslindale', 'Mattapan','Jamaica Plain', 'South Dorchester', 'North Dorchester', 'Roxbury','Fenway', 'South End', 'South Boston', 'Back Bay','Charlestown', 'Allston', 'East Boston']
plt.xticks(x, labels, rotation='vertical')
plt.text(12, 1200, r'$R^2 = $%.2f'%(R2), fontsize=20)
plt.savefig('../graphics/boston_comparison.png', bbox_inches='tight', dpi=150)
# -
# # Plots for website
# +
def calculate_rate(Zipcode):
target = df_zipcode[df_zipcode["geoid2"]==int(Zipcode)]
target_params = target.values[0]
chlamydia_rate = model.predict(target_params[1:])*Ystd+Ymean
return chlamydia_rate
Race = "Black"
Gender = "Male"
Age = "30-34"
Zipcode = "02474"
target_unnormalized = df_zipcode_unnormalized[df_zipcode_unnormalized["geoid2"]==int(Zipcode)]
TOTALNR = target_unnormalized["Population"]
if Gender == "Male":
gender_table = "hd02s026"
else:
gender_table = "hd02s051"
GENDERNR = TOTALNR*target_unnormalized[gender_table]/100.0
if Race == "White":
race_table = "hd02s078"
elif Race == "Black":
race_table = "hd02s079"
elif Race == "Native":
race_table = "hd02s080"
elif Race == "Asian":
race_table = "hd02s081"
elif Race == "Pacific":
race_table = "hd02s089"
elif Race == "Multiple":
race_table = "hd02s095"
elif Race == "Hispanic":
race_table = "hd02s107"
RACENR = TOTALNR*target_unnormalized[race_table]/100.0
if Age == "0-14":
age_table = "hd02s002"
elif Age == "15-19":
age_table = "hd02s005"
elif Age == "20-24":
age_table = "hd02s006"
elif Age == "25-29":
age_table = "hd02s007"
elif Age == "30-34":
age_table = "hd02s008"
elif Age == "35-39":
age_table = "hd02s009"
elif Age == "40-44":
age_table = "hd02s010"
elif Age == "45-54":
age_table = "hd02s011"
elif Age == "55-64":
age_table = "hd02s013"
elif Age == "65+":
age_table = "hd02s015"
AGENR = TOTALNR*target_unnormalized[age_table]/100.0
zipcoderate = calculate_rate(Zipcode)*100
genderrate = gender_rate[Gender]*100
agerate = age_rate[Age]*100
racerate = race_rate[Race]*100
the_result = (zipcoderate/TOTALNR.values + genderrate/GENDERNR.values + racerate/RACENR.values + agerate/AGENR.values)/(1.0/TOTALNR.values+1.0/GENDERNR.values+1.0/RACENR.values+1.0/AGENR.values)
d = np.array([the_result[0], genderrate, agerate, racerate, zipcoderate[0]])
d_label = np.array(["You", "Your gender", "Your age group", "Your race / ethnicity", "Your location"])
d_label
# +
sns.set(style="white", context="talk")
fig, ax = plt.subplots(1, 1, figsize=(10, 6), sharex=True)
sns.barplot(d_label, d, palette="RdBu_r", ax=ax)
ax.set_ylabel("Risk", fontsize=20)
plt.title(r'Chlamydia', fontsize=20)
ax.plot([-1, len(d)], [0,0], "k-", linewidth=1.0)
sns.despine(bottom=True)
plt.setp(fig.axes, yticks=[])
plt.tight_layout(h_pad=3)
# -
| code/app_model_chlamydia.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import sys
sys.path.append('../')
# +
# # %%capture
# %run common.ipynb
# importnb allows us to load the code from other notebooks
# from importnb import Notebook
# with Notebook():
# import OfficeAutomation.common as common # will probably become OfficeAutomation.common
# -
import crestdsl.model as crest
import crestdsl.simulation as crestsim
import crestdsl.ui as ui
# + code_folding=[17, 77, 100]
@crest.dependency(source="req_electricity_out", target="switch_in")
class Lamp(ElectricalDevice, crest.Entity):
switch_in = crest.Input(res_onoff_switch, value="off") # on/off
off = current = crest.State()
on = crest.State()
need_watt = crest.Local(res_electricity, value=100)
turn_on = crest.Transition(source=off, target=on, guard=(lambda self: self.switch_in.value == "on" and self.electricity_in.value >= self.need_watt.value))
turn_off = crest.Transition(source=on, target=off, guard=(lambda self: self.switch_in.value == "off" or self.electricity_in.value < self.need_watt.value))
@crest.update(state=[on,off], target="req_electricity_out")
def update_req_on_off(self, dt):
if self.switch_in.value == "on":
return self.need_watt.value
else:
return 0
# + code_folding=[17, 77, 100]
class FragileLamp(Lamp, crest.Entity):
switch_in = crest.Input(res_onoff_switch, value="off") # on/off
spike_timeout = crest.Local(res_time, value=0.0015) # time unit = minutes hence 0.0015 is roughly .1 seconds
spike = crest.State()
broken = crest.State()
to_spike = crest.Transition(source="off", target=spike, guard=(lambda self: self.switch_in.value == "on" and self.electricity_in.value >= self.need_watt.value))
to_broken = crest.Transition(source=spike, target=broken, guard=(lambda self: self.spike_timeout.value <= 0))
@crest.update(state=spike, target=spike_timeout)
def reduce_spike_timeout(self, dt):
return self.spike_timeout.value - dt
@crest.update(state=spike, target="req_electricity_out")
def update_req_spike(self, dt):
return 1000
@crest.update(state=broken, target="req_electricity_out")
def update_req_broken(self, dt):
return 0
# crest.Validator(FragileLamp()).check_all()
# + code_folding=[]
@crest.nodependencies
class EmergencyLamp(ElectricalDevice, crest.Entity):
spike_timeout = crest.Local(res_time, value=0.0015) # time unit = minutes hence 0.0015 is roughly .1 seconds
need_watt = crest.Local(res_electricity, value=100)
off = current = crest.State()
on = crest.State()
spike = crest.State()
broken = crest.State()
turn_on = crest.Transition(source=off, target=on, guard=(lambda self: self.electricity_in.value >= self.need_watt.value))
turn_off = crest.Transition(source=on, target=off, guard=(lambda self: self.electricity_in.value < self.need_watt.value))
to_spike = crest.Transition(source=off, target=spike, guard=(lambda self: self.electricity_in.value >= self.need_watt.value))
to_broken = crest.Transition(source=spike, target=broken, guard=(lambda self: self.spike_timeout.value <= 0))
@crest.update(state=spike, target=spike_timeout)
def reduce_spike_timeout(self, dt):
return self.spike_timeout.value - dt
@crest.update(state=spike, target="req_electricity_out")
def set_spike_electricity(self, dt):
return 1000
@crest.update(state=off, target="req_electricity_out")
def update_req_off(self, dt):
return 0
@crest.update(state=on, target="req_electricity_out")
def update_req_on(self, dt):
return self.need_watt.value
# ui.plot(EmergencyLamp())
# crest.Validator(EmergencyLamp()).check_all()
| lamps.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import pandas as pd
import numpy as np
data = pd.read_csv('usedata.csv')
data.head()
# +
import category_encoders as ce
from sklearn.ensemble import RandomForestRegressor
from sklearn.impute import SimpleImputer
from sklearn.pipeline import make_pipeline
test = data[data.location == 'hawaii']
train = data[data.location != 'hawaii']
test.shape, train.shape
# +
target = 'GHI'
features = ['zen', 'temperature', 'summary','pressure','visibility','uvIndex','dewPoint']
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(),
RandomForestRegressor(n_estimators=200,min_samples_leaf=10)
)
pipeline.fit(train[features],train[target])
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
y_pred_train = pipeline.predict(train[features])
print(r2_score(train[target],y_pred_train))
print(mean_absolute_error(train[target],y_pred_train))
y_pred_test = pipeline.predict(test[features])
print(r2_score(test[target],y_pred_test))
print(mean_absolute_error(test[target],y_pred_test))
# +
pipeline = make_pipeline(
ce.OrdinalEncoder(),
SimpleImputer(),
RandomForestRegressor(n_estimators=200,min_samples_leaf=10)
)
pipeline.fit(data[features],data[target])
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error
y_pred_train = pipeline.predict(data[features])
print(r2_score(data[target],y_pred_train))
print(mean_absolute_error(data[target],y_pred_train))
# +
from joblib import dump
dump(pipeline, 'pipeline.joblib')
# -
from joblib import load
loaded_model = load('pipeline.joblib')
import requests
from pysolar.solar import *
import plotly.express as px
# +
lat, lon = 1.999140,112.940784
features = ['zen', 'temperature', 'summary','pressure','visibility','uvIndex','dewPoint']
a = requests.get('https://api.darksky.net/forecast/dc757f87dbdcb50907cdcecf02328582/' + str(lat)+ ',' + str(lon) +'?extend=hourly')
a= a.json()
a = a['hourly']['data']
df = pd.DataFrame(a)
tr = df['time'].values
def zen(time,lat,lon):
from datetime import datetime
mix = datetime.fromtimestamp(time).strftime('%Y,%m,%d,%H')
spt = mix.split(',')
import datetime
date = datetime.datetime(int(spt[0]), int(spt[1]), int(spt[2]), int(spt[3]), 0, tzinfo=datetime.timezone.utc)
return(float(90) - get_altitude(lat, lon, date))
def tim(time):
from datetime import datetime
mix = datetime.fromtimestamp(time).strftime('%Y,%m,%d,%H')
mixx = mix.split(',')
return(str(mixx[3]) + ':00')
df['date'] = df.time.apply(tim)
zenith = []
for i in tr:
zed = zen(i, lat, lon)
zenith.append(zed)
df['zen'] = zenith
res = loaded_model.predict(df[features])
df['ghi'] = res
gapminder = df.head(24)
fig = px.line(gapminder, x="date", y="ghi", title='GHI Level')
# -
fig.show()
| notebooks/makemodel.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Qiskit Runtime API
# Qiskit Runtime is a cloud service that allow you upload and run Qiskit programs in a Qiskit Runtimes near to the QPUs.
#
# In this example we are going to explore how to integrate this Qiskit Runtime in any application using only basic HTTP requests and interact with a the program running in the service.
#
#
# <div class="alert alert-block alert-info">
# <b>Note:</b> You need to have an IBM Quantum API token. You can get one by signing up at https://quantum-computing.ibm.com/
# </div>
# For this tutorial we are going to use the Python HTTP library `requests` to facilitate the API calls.
#
# You can access the online interactive runtime API documentation here: https://runtime-us-east.quantum-computing.ibm.com/openapi/
import requests
import json
import time
# ## Authenticate with the service
# +
# you need to pass your API token in the header of every call
headers = {
'Authorization': f'Bearer {Your_API_TOKEN}',
'Content-Type': 'application/json'
}
Runtime_API_URL = "https://runtime-us-east.quantum-computing.ibm.com/"
# -
# ## Get a list of program that you can invoke
# +
# Call the API passing the API Token and get the programs
response = requests.get(Runtime_API_URL + 'programs' , headers = headers)
list_of_programs = {}
if response.status_code == 200:
list_of_programs = response.json()
else:
print(f'Error:{response.status_code}')
exit()
print(f'Qiskit Runtime Programs:')
for program in list_of_programs["programs"]:
print(f'- {program["name"]}: {program["description"]} ')
# -
# ## Display information about a specific program
# +
print('Qiskit Program details:\n')
qiskit_runtime_program = json.dumps(list_of_programs["programs"][0], indent=2)
print(qiskit_runtime_program)
# -
# ## Run a Qiskit Runtime program on the Cloud
# +
Runtime_program = "sample-program"
program_input = {"iterations":2}
def run_runtime_program(program_name, program_input):
# configuing your IBM Provider data
params = json.dumps({
"program_id": Runtime_program,
"hub": "ibm-q-internal",
"group": "near-time",
"project": "qiskit-runtime",
"backend": "ibmq_montreal",
"params": program_input
})
job_ID = ''
response = requests.post(Runtime_API_URL + 'jobs', data=params, headers=headers)
if response.status_code == 200:
job_ID = response.json()['id']
return 200, job_ID
else:
return response.status_code, None
status, job_ID = run_runtime_program(Runtime_program, program_input)
print(f' status: {status}, Job: {job_ID}')
# -
# ## Get the result
# +
# You are going to get a 204 status code while the Qiskit program is still running.
final_result = False
while not final_result:
response = requests.get(Runtime_API_URL + 'jobs/'+ job_ID +'/results', headers=headers)
if response.status_code == 200:
print(f'Final Result: {response.text}')
final_result = True
elif response.status_code == 204:
print(f'Waiting for the final result')
time.sleep(2)
else:
print(f'Error:{response.status_code}')
break
| tutorials/API_direct.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# # The New Bechdel test!
# Analysis on the new Bechdel test using the Cornell Movie-Dialog Corpus
import csv
import pandas as pd
import re
import numpy as np
import plotly.plotly as py
import plotly.graph_objs as go
from unidecode import unidecode
import pickle
from sklearn.feature_extraction import DictVectorizer
from sklearn.utils import shuffle
from sklearn.tree import DecisionTreeClassifier
# Import all the datasets first
# Movie Titles Metadata.txt
mtm = 'cornell movie-dialogs corpus/movie_titles_metadata.txt'
# CSV for transforming the data
mtm_csv = 'mtm_csv.csv'
# Add header rows
with open(mtm_csv, 'wt') as csv_file:
writer = csv.DictWriter(csv_file, fieldnames = ["Movie_Number", "Movie_Name", "Year_of_Release",
"IMDB_Rating", "Number_of_Votes", "Genres"])
writer.writeheader()
# Open the .txt file and write it onto the dataframe
with open(mtm, 'r') as mtmeta:
for lines in mtmeta:
text = str(lines)
line_list = text.split('+++$+++')
with open(mtm_csv, 'a') as write_csv:
writer = csv.writer(write_csv)
writer.writerow([line_list[0].strip(), line_list[1].strip(), line_list[2].strip(),
line_list[3].strip(), line_list[4].strip(), line_list[5].strip()])
# Check the content of the csv file
mtm_df = pd.read_csv(mtm_csv, index_col = False)
print(mtm_df.shape)
mtm_df.head(5)
mtm_df.to_csv(mtm_csv, sep=',', index = False)
print("Saved to csv..")
# Movie Characters Metadata.txt
mcm = 'cornell movie-dialogs corpus/movie_characters_metadata.txt'
# CSV for transforming the data
mcm_csv = 'mcm_csv.csv'
# Add header rows
with open(mcm_csv, 'wt') as csv_file:
writer = csv.DictWriter(csv_file, fieldnames = ["Character_Id", "Character_Name", "Movie_Number",
"Movie_Title", "Gender", "Position_in_Credits"])
writer.writeheader()
# Open the second .txt file and write it onto the next dataframe
with open(mcm, 'r') as mcmeta:
for lines in mcmeta:
text = str(lines)
line_list = text.split('+++$+++')
with open(mcm_csv, 'a') as write_csv:
writer = csv.writer(write_csv)
writer.writerow([line_list[0].strip(), line_list[1].strip(), line_list[2].strip(),
line_list[3].strip(), line_list[4].strip(), line_list[5].strip()])
mcm_df = pd.read_csv(mcm_csv)
print(mcm_df.shape)
mcm_df.head(5)
mcm_df.to_csv(mcm_csv, sep=',', index = False)
print("Saved to csv..")
# Movie Characters Metadata.txt
ml = 'cornell movie-dialogs corpus/movie_lines.txt'
# CSV for transforming the data
ml_csv = 'ml_csv.csv'
# Add header rows
with open(ml_csv, 'wt') as csv_file:
writer = csv.DictWriter(csv_file, fieldnames = ["Line_Id", "Character_Id", "Movie_Number",
"Character_Name", "Dialogue"])
writer.writeheader()
# Open the second .txt file and write it onto the next dataframe
with open(ml, 'r') as mlines:
for lines in mlines:
text = str(lines)
line_list = text.split('+++$+++')
with open(ml_csv, 'a') as write_csv:
writer = csv.writer(write_csv)
writer.writerow([line_list[0].strip(), line_list[1].strip(), line_list[2].strip(),
line_list[3].strip(), line_list[4].strip()])
ml_df = pd.read_csv('ml_csv.csv')
print(ml_df.shape)
ml_df.head(5)
ml_df.to_csv(ml_csv, sep = ',', index = False)
print('Saved to csv...')
# Movie Conversations.txt
mc = 'cornell movie-dialogs corpus/movie_conversations.txt'
# CSV for transforming the data
mc_csv = 'mc_csv.csv'
# Add header rows
with open(mc_csv, 'wt') as csv_file:
writer = csv.DictWriter(csv_file, fieldnames = ['Character_Id1', 'Character_Id2', 'Movie_Number',
'List_of_Utterance'])
writer.writeheader()
# Open the second .txt file and write it onto the next dataframe
with open(mc, 'r') as mconversations:
for lines in mconversations:
line = str(lines)
line_list = line.split('+++$+++')
with open(mc_csv, 'a') as mconv:
writer = csv.writer(mconv)
writer.writerow([line_list[0].strip(), line_list[1].strip(), line_list[2].strip(),
line_list[3].strip()])
mc_df = pd.read_csv('mc_csv.csv')
print(mc_df.shape)
mc_df.head(5)
mc_df.to_csv(mc_csv, sep = ',', index = False)
print('Saved to csv...')
# # Onto Data Cleaning
# There are different aspects to this part and all the data in these csv files must be analysed in order for them to be of a consistent data
# Read all the csv files
mtm_df = pd.read_csv('mtm_csv.csv')
mcm_df = pd.read_csv('mcm_csv.csv')
mc_df = pd.read_csv('mc_csv.csv')
ml_df = pd.read_csv('ml_csv.csv')
# On the mtm dataset
# To check for any NULL values in the data
mtm_df.isnull().sum()
# Convert the years to consistent format -- elegant way
st = ""
mtm_df['Year_of_Release'] = mtm_df['Year_of_Release'].apply(lambda x: st.join(re.findall(r'[0-9]', x)))
# Convert them to Integers
mtm_df['Year_of_Release'] = mtm_df['Year_of_Release'].astype(int)
mtm_df['IMDB_Rating'] = mtm_df['IMDB_Rating'].astype(int)
mtm_df['Number_of_Votes'] = mtm_df['Number_of_Votes'].astype(int)
# Now describe the dataset
mtm_df.describe()
# More information on the dataframe
mtm_df.info()
mtm_df.to_csv(mtm_csv, sep=',', index = False)
print("Saved to csv..")
# Cleaning done on the mtm dataset
# Onto the MCM dataset
mcm_df.isnull().sum()
# The rows which has NULL values
mcm_df[mcm_df.isnull().any(axis=1)]
# Drop NULL records
print('Old size: %d' % len(mcm_df))
mcm_df = mcm_df.dropna(how = 'any', axis = 'rows')
print('New size: %d' % len(mcm_df))
mcm_df.info()
# Since, the data in the 'Gender' column has bad data, inorder to make the data consistent :-
# # A quick Gender Recognition model
# Grabbed from [nlpforhackers](https://nlpforhackers.io/introduction-machine-learning/) webpage.
# 1. Firstly convert the dataset into a numpy array to keep only gender and names
# 2. Set the feature parameters which takes in different parameters
# 3. Vectorize the parametes
# 4. Get varied train, test split and test it for validity by checking out the count of the train test split
# 5. Transform lists of feature-value mappings to vectors. (When feature values are strings, this transformer will do a binary one-hot (aka one-of-K) coding: one boolean-valued feature is constructed for each of the possible string values that the feature can take on)
# 6. Train a decision tree classifier on this and save the model as a pickle file
# +
names = pd.read_csv('gender_recognition/names_dataset.csv')
print(names.head(10))
print("%d names in dataset" % len(names))
# +
# Get the data out of the dataframe into a numpy matrix and keep only the name and gender columns
names = names.as_matrix()[:, 1:]
print(names)
# We're using 90% of the data for training
TRAIN_SPLIT = 0.90
# +
def features(name):
name = name.lower()
return {
'first-letter': name[0], # First letter
'first2-letters': name[0:2], # First 2 letters
'first3-letters': name[0:3], # First 3 letters
'last-letter': name[-1], # Last letter
'last2-letters': name[-2:], # Last 2 letters
'last3-letters': name[-3:], # Last 3 letters
}
# Feature Extraction
print(features("Alex"))
# +
# Vectorize the features function
features = np.vectorize(features)
print(features(["Anna", "Hannah", "Paul"]))
# [ array({'first2-letters': 'an', 'last-letter': 'a', 'first-letter': 'a', 'last2-letters': 'na', 'last3-letters': 'nna', 'first3-letters': 'ann'}, dtype=object)
# array({'first2-letters': 'ha', 'last-letter': 'h', 'first-letter': 'h', 'last2-letters': 'ah', 'last3-letters': 'nah', 'first3-letters': 'han'}, dtype=object)
# array({'first2-letters': 'pa', 'last-letter': 'l', 'first-letter': 'p', 'last2-letters': 'ul', 'last3-letters': 'aul', 'first3-letters': 'pau'}, dtype=object)]
# Extract the features for the whole dataset
X = features(names[:, 0]) # X contains the features
# Get the gender column
y = names[:, 1] # y contains the targets
# Test if we built the dataset correctly
print("\n\nName: %s, features=%s, gender=%s" % (names[0][0], X[0], y[0]))
# +
X, y = shuffle(X, y)
X_train, X_test = X[:int(TRAIN_SPLIT * len(X))], X[int(TRAIN_SPLIT * len(X)):]
y_train, y_test = y[:int(TRAIN_SPLIT * len(y))], y[int(TRAIN_SPLIT * len(y)):]
# Check to see if the datasets add up
print len(X_train), len(X_test), len(y_train), len(y_test)
# +
# Transforms lists of feature-value mappings to vectors.
vectorizer = DictVectorizer()
vectorizer.fit(X_train)
transformed = vectorizer.transform(features(["Mary", "John"]))
print transformed
print type(transformed) # <class 'scipy.sparse.csr.csr_matrix'>
print transformed.toarray()[0][12] # 1.0
print vectorizer.feature_names_[12] # first-letter=m
# +
clf = DecisionTreeClassifier(criterion = 'gini')
clf.fit(vectorizer.transform(X_train), y_train)
# Accuracy on training set
print clf.score(vectorizer.transform(X_train), y_train)
# Accuracy on test set
print clf.score(vectorizer.transform(X_test), y_test)
# -
# Therefore, we are getting a decent result from the names
print clf.predict(vectorizer.transform(features(["SMYSLOV", "CHASTITY", "<NAME>", "SHARON", "ALONSO", "SECONDARY OFFICER"])))
# Now since the model is built let's update the data in the dataframe
print("Number of missing genders : " , mcm_df[mcm_df.Gender == '?'].shape[0])
mcm_df.head(8)
# Let's convert all the ? with their appropriate genders by using the above trained model
for items in mcm_df.itertuples():
if items[5] == '?':
pred_val = clf.predict(vectorizer.transform(features([items[2]])))
prediction = ""
prediction = "".join(pred_val)
mcm_df.loc[items.Index, 'Gender'] = prediction.lower()
# Now check for the updated genders
print("Number of missing genders : " , mcm_df[mcm_df.Gender == '?'].shape[0])
mcm_df.head(8)
mcm_df.to_csv(mcm_csv, sep=',', index = False)
print("Saved to csv..")
# Cleaning done on the mcm dataset
# # Onto Data Exploration 🙌 🙌
# Using plotly to do visualization of all the data so that we get meaningful info out of it
# Visualization on the year of realese to get the idea of movie years we are working with
data = [go.Histogram(
x=mtm_df['Year_of_Release'],
marker = dict(color = 'rgb(17, 157, 100)'),
hoverlabel = dict(bordercolor = 'rgb(0, 0, 0)')
)]
layout = go.Layout(
title='Year of Releases',
xaxis=dict(
title='Years'
),
yaxis=dict(
title='Counts of Movie Releases'
),
bargap=0.2
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='simple-histogram1')
# +
# Scatter plot on the same data
trace = go.Scatter(
x = mtm_df['Year_of_Release'],
mode = 'markers',
)
layout = go.Layout(
title='Year of Releases',
xaxis=dict(
title='Years'
),
yaxis=dict(
title='Counts of Movie Releases'
),
)
data = [trace]
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='basic-scatter')
# -
# Visualization on the number of IMDB Ratings on these movies
data = [go.Histogram(
y=mtm_df['IMDB_Rating'],
marker = dict(color = 'rgb(17, 157, 255)'),
hoverlabel = dict(bordercolor = 'rgb(0, 0, 0)')
)]
layout = go.Layout(
title='IMDB Ratings',
xaxis=dict(
title='Number of Movies'
),
yaxis=dict(
title='Rating Scores'
),
bargap=0.2
)
fig = go.Figure(data=data, layout=layout)
py.iplot(fig, filename='simple-histogram2')
| .ipynb_checkpoints/New Bechdel-checkpoint.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python [conda env:root] *
# language: python
# name: conda-root-py
# ---
# # Continuous training with TFX and Google Cloud AI Platform
# ## Learning Objectives
#
# 1. Use the TFX CLI to build a TFX pipeline.
# 2. Deploy a new TFX pipeline version with tuning enabled to a hosted AI Platform Pipelines instance.
# 3. Create and monitor a TFX pipeline run using the TFX CLI and KFP UI.
# In this lab, you use utilize the following tools and services to deploy and run a TFX pipeline on Google Cloud that automates the development and deployment of a TensorFlow 2.3 WideDeep Classifer to predict forest cover from cartographic data:
#
# * The [**TFX CLI**](https://www.tensorflow.org/tfx/guide/cli) utility to build and deploy a TFX pipeline.
# * A hosted [**AI Platform Pipeline instance (Kubeflow Pipelines)**](https://www.tensorflow.org/tfx/guide/kubeflow) for TFX pipeline orchestration.
# * [**Dataflow**](https://cloud.google.com/dataflow) jobs for scalable, distributed data processing for TFX components.
# * A [**AI Platform Training**](https://cloud.google.com/ai-platform/) job for model training and flock management for parallel tuning trials.
# * [**AI Platform Prediction**](https://cloud.google.com/ai-platform/) as a model server destination for blessed pipeline model versions.
# * [**CloudTuner**](https://www.tensorflow.org/tfx/guide/tuner#tuning_on_google_cloud_platform_gcp) and [**AI Platform Vizier**](https://cloud.google.com/ai-platform/optimizer/docs/overview) for advanced model hyperparameter tuning using the Vizier algorithm.
#
# You will then create and monitor pipeline runs using the TFX CLI as well as the KFP UI.
# ### Setup
# #### Update lab environment PATH to include TFX CLI and skaffold
# +
import yaml
# Set `PATH` to include the directory containing TFX CLI and skaffold.
# PATH=%env PATH
# %env PATH=/home/jupyter/.local/bin:{PATH}
# -
# #### Validate lab package version installation
# !python -c "import tensorflow; print('TF version: {}'.format(tensorflow.__version__))"
# !python -c "import tfx; print('TFX version: {}'.format(tfx.__version__))"
# !python -c "import kfp; print('KFP version: {}'.format(kfp.__version__))"
# **Note**: this lab was built and tested with the following package versions:
#
# `TF version: 2.3.2`
# `TFX version: 0.25.0`
# `KFP version: 1.4.0`
# (Optional) If running the above command results in different package versions or you receive an import error, upgrade to the correct versions by running the cell below:
# %pip install --upgrade --user tensorflow==2.3.2
# %pip install --upgrade --user tfx==0.25.0
# %pip install --upgrade --user kfp==1.0.4
# Note: you may need to restart the kernel to pick up the correct package versions.
# #### Validate creation of AI Platform Pipelines cluster
#
# Navigate to [AI Platform Pipelines](https://console.cloud.google.com/ai-platform/pipelines/clusters) page in the Google Cloud Console.
#
# Note you may have already deployed an AI Pipelines instance during the Setup for the lab series. If so, you can proceed using that instance. If not:
#
# **1. Create or select an existing Kubernetes cluster (GKE) and deploy AI Platform**. Make sure to select `"Allow access to the following Cloud APIs https://www.googleapis.com/auth/cloud-platform"` to allow for programmatic access to your pipeline by the Kubeflow SDK for the rest of the lab. Also, provide an `App instance name` such as "tfx" or "mlops".
#
# Validate the deployment of your AI Platform Pipelines instance in the console before proceeding.
# ## Review: example TFX pipeline design pattern for Google Cloud
# The pipeline source code can be found in the `pipeline` folder.
# %cd pipeline
# !ls -la
# The `config.py` module configures the default values for the environment specific settings and the default values for the pipeline runtime parameters.
# The default values can be overwritten at compile time by providing the updated values in a set of environment variables. You will set custom environment variables later on this lab.
# The `pipeline.py` module contains the TFX DSL defining the workflow implemented by the pipeline.
#
# The `preprocessing.py` module implements the data preprocessing logic the `Transform` component.
#
# The `model.py` module implements the training, tuning, and model building logic for the `Trainer` and `Tuner` components.
#
# The `runner.py` module configures and executes `KubeflowDagRunner`. At compile time, the `KubeflowDagRunner.run()` method converts the TFX DSL into the pipeline package in the [argo](https://argoproj.github.io/argo/) format for execution on your hosted AI Platform Pipelines instance.
#
# The `features.py` module contains feature definitions common across `preprocessing.py` and `model.py`.
#
# ## Exercise: build your pipeline with the TFX CLI
#
# You will use TFX CLI to compile and deploy the pipeline. As explained in the previous section, the environment specific settings can be provided through a set of environment variables and embedded into the pipeline package at compile time.
# ### Configure your environment resource settings
#
# Update the below constants with the settings reflecting your lab environment.
# - `GCP_REGION` - the compute region for AI Platform Training, Vizier, and Prediction.
# - `ARTIFACT_STORE` - An existing GCS bucket. You can use any bucket or use the GCS bucket created during installation of AI Platform Pipelines. The default bucket name will contain the `kubeflowpipelines-` prefix.
# * `CUSTOM_SERVICE_ACCOUNT` - In the gcp console Click on the Navigation Menu. Navigate to `IAM & Admin`, then to `Service Accounts` and use the service account starting with prefix - `'tfx-tuner-caip-service-account'`. This enables CloudTuner and the Google Cloud AI Platform extensions Tuner component to work together and allows for distributed and parallel tuning backed by AI Platform Vizier's hyperparameter search algorithm.
# - `ENDPOINT` - set the `ENDPOINT` constant to the endpoint to your AI Platform Pipelines instance. The endpoint to the AI Platform Pipelines instance can be found on the [AI Platform Pipelines](https://console.cloud.google.com/ai-platform/pipelines/clusters) page in the Google Cloud Console. Open the *SETTINGS* for your instance and use the value of the `host` variable in the *Connect to this Kubeflow Pipelines instance from a Python client via Kubeflow Pipelines SKD* section of the *SETTINGS* window. The format is `'...pipelines.googleusercontent.com'`.
# +
PROJECT_ID = !(gcloud config get-value core/project)
PROJECT_ID = PROJECT_ID[0]
GCP_REGION = 'us-central1'
ARTIFACT_STORE_URI = f'gs://{PROJECT_ID}-kubeflowpipelines-default'
CUSTOM_SERVICE_ACCOUNT = f'<EMAIL>'
#TODO: Set your environment resource settings here for ENDPOINT.
ENDPOINT = ''
# -
# Set your resource settings as environment variables. These override the default values in pipeline/config.py.
# %env GCP_REGION={GCP_REGION}
# %env ARTIFACT_STORE_URI={ARTIFACT_STORE_URI}
# %env CUSTOM_SERVICE_ACCOUNT={CUSTOM_SERVICE_ACCOUNT}
# %env PROJECT_ID={PROJECT_ID}
# ### Set the compile time settings to first create a pipeline version without hyperparameter tuning
#
# Default pipeline runtime environment values are configured in the pipeline folder `config.py`. You will set their values directly below:
#
# * `PIPELINE_NAME` - the pipeline's globally unique name. For each pipeline update, each pipeline version uploaded to KFP will be reflected on the `Pipelines` tab in the `Pipeline name > Version name` dropdown in the format `PIPELINE_NAME_datetime.now()`.
#
# * `MODEL_NAME` - the pipeline's unique model output name for AI Platform Prediction. For multiple pipeline runs, each pushed blessed model will create a new version with the format `'v{}'.format(int(time.time()))`.
#
# * `DATA_ROOT_URI` - the URI for the raw lab dataset `gs://workshop-datasets/covertype/small`.
#
# * `CUSTOM_TFX_IMAGE` - the image name of your pipeline container build by skaffold and published by `Cloud Build` to `Cloud Container Registry` in the format `'gcr.io/{}/{}'.format(PROJECT_ID, PIPELINE_NAME)`.
#
# * `RUNTIME_VERSION` - the TensorFlow runtime version. This lab was built and tested using TensorFlow `2.3`.
#
# * `PYTHON_VERSION` - the Python runtime version. This lab was built and tested using Python `3.7`.
#
# * `USE_KFP_SA` - The pipeline can run using a security context of the GKE default node pool's service account or the service account defined in the `user-gcp-sa` secret of the Kubernetes namespace hosting Kubeflow Pipelines. If you want to use the `user-gcp-sa` service account you change the value of `USE_KFP_SA` to `True`. Note that the default AI Platform Pipelines configuration does not define the `user-gcp-sa` secret.
#
# * `ENABLE_TUNING` - boolean value indicating whether to add the `Tuner` component to the pipeline or use hyperparameter defaults. See the `model.py` and `pipeline.py` files for details on how this changes the pipeline topology across pipeline versions.
PIPELINE_NAME = 'tfx_covertype_continuous_training'
MODEL_NAME = 'tfx_covertype_classifier'
DATA_ROOT_URI = 'gs://workshop-datasets/covertype/small'
CUSTOM_TFX_IMAGE = 'gcr.io/{}/{}'.format(PROJECT_ID, PIPELINE_NAME)
RUNTIME_VERSION = '2.3'
PYTHON_VERSION = '3.7'
USE_KFP_SA=False
ENABLE_TUNING=True
# %env PIPELINE_NAME={PIPELINE_NAME}
# %env MODEL_NAME={MODEL_NAME}
# %env DATA_ROOT_URI={DATA_ROOT_URI}
# %env KUBEFLOW_TFX_IMAGE={CUSTOM_TFX_IMAGE}
# %env RUNTIME_VERSION={RUNTIME_VERSION}
# %env PYTHON_VERIONS={PYTHON_VERSION}
# %env USE_KFP_SA={USE_KFP_SA}
# %env ENABLE_TUNING={ENABLE_TUNING}
# ### Compile your pipeline code
#
# You can build and upload the pipeline to the AI Platform Pipelines instance in one step, using the `tfx pipeline create` command. The `tfx pipeline create` goes through the following steps:
# - (Optional) Builds the custom image to that provides a runtime environment for TFX components or uses the latest image of the installed TFX version
# - Compiles the pipeline code into a pipeline package
# - Uploads the pipeline package via the `ENDPOINT` to the hosted AI Platform instance.
#
# As you debug the pipeline DSL, you may prefer to first use the `tfx pipeline compile` command, which only executes the compilation step. After the DSL compiles successfully you can use `tfx pipeline create` to go through all steps.
# !tfx pipeline compile --engine kubeflow --pipeline_path runner.py
# Note: you should see a `{PIPELINE_NAME}.tar.gz` file appear in your current pipeline directory.
# ## Exercise: deploy your pipeline container to AI Platform Pipelines with TFX CLI
#
# After the pipeline code compiles without any errors you can use the `tfx pipeline create` command to perform the full build and deploy the pipeline. You will deploy your compiled pipeline container hosted on Google Container Registry e.g. `gcr.io/[PROJECT_ID]/tfx_covertype_continuous_training` to run on AI Platform Pipelines with the TFX CLI.
# +
# TODO: Your code here to use the TFX CLI to deploy your pipeline image to AI Platform Pipelines.
# !tfx pipeline create \
# --pipeline_path=runner.py \
# --endpoint={ENDPOINT} \
# --build_target_image={CUSTOM_TFX_IMAGE}
# -
# **Hint**: review the [TFX CLI documentation](https://www.tensorflow.org/tfx/guide/cli#create) on the "pipeline group" to create your pipeline. You will need to specify the `--pipeline_path` to point at the pipeline DSL and runner defined locally in `runner.py`, `--endpoint`, and `--build_target_image` arguments using the environment variables specified above.
# Note: you should see a `build.yaml` file in your pipeline folder created by skaffold. The TFX CLI compile triggers a custom container to be built with skaffold using the instructions in the `Dockerfile`.
# If you need to redeploy the pipeline you can first delete the previous version using `tfx pipeline delete` or you can update the pipeline in-place using `tfx pipeline update`.
#
# To delete the pipeline:
#
# `tfx pipeline delete --pipeline_name {PIPELINE_NAME} --endpoint {ENDPOINT}`
#
# To update the pipeline:
#
# `tfx pipeline update --pipeline_path runner.py --endpoint {ENDPOINT}`
# ### Create and monitor a pipeline run with the TFX CLI
#
# After the pipeline has been deployed, you can trigger and monitor pipeline runs using TFX CLI.
#
# *Hint*: review the [TFX CLI documentation](https://www.tensorflow.org/tfx/guide/cli#run_group) on the "run group".
# +
# TODO: your code here to trigger a pipeline run with the TFX CLI
# !tfx run create --pipeline_name={PIPELINE_NAME} --endpoint={ENDPOINT}
# -
# To view the status of existing pipeline runs:
# !tfx run list --pipeline_name {PIPELINE_NAME} --endpoint {ENDPOINT}
# To retrieve the status of a given run:
# +
RUN_ID='[YOUR RUN ID]'
# !tfx run status --pipeline_name {PIPELINE_NAME} --run_id {RUN_ID} --endpoint {ENDPOINT}
# -
# ## Important
#
# A full pipeline run with tuning enabled will take about 50 minutes to complete. You can view the run's progress using the TFX CLI commands above and in the KFP UI. While the pipeline run is in progress, there are also optional exercises below to explore your pipeline's artifacts and Google Cloud integrations while the pipeline run is in progress.
# ### Exercise (optional): review your pipeline's Dataflow jobs for data processing
#
# On the [Dataflow](https://console.cloud.google.com/dataflow) page, inspect the computational graphs parallelizing the data processing in the ExampleGen, StatisticsGen, Transform, and Evaluator pipeline components.
# ### Exercise (optional): review your pipeline's AI Platform jobs for model training and hyperparameter tuning
#
# On the [AI Platform Jobs](https://console.cloud.google.com/ai-platform/jobs) page, inspect the Training job logs. You can also launch a Tensorboard server directly from the KFP UI to monitor your training performance while the job is in progress. Click on the Trainer component in the KFP UI once it is running and navigate to the `Visualizations` tab. Scroll down to the Tensorboard widget and hit the `Open Tensorboard` button.
# ### Exercise (optional): review your pipeline's artifacts on Cloud Storage
#
# On the [Cloud Storage page](https://console.cloud.google.com/storage), review how TFX standardizes the organization of your pipeline run artifacts. You will find them organized by component and versioned in your `gs://{PROJECT_ID}-kubeflowpipelines-default` artifact storage bucket. This standardization brings reproducibility and traceability to your ML workflows and allows for easier reuse of pipeline components and artifacts across use cases.
# ## Next Steps
# In this lab, you learned how to build and deploy a TFX pipeline with the TFX CLI and then update, build and deploy a new pipeline with automatic hyperparameter tuning. You practiced triggered continuous pipeline runs using the TFX CLI as well as the Kubeflow Pipelines UI.
#
#
# In the next lab, you will construct a Cloud Build CI/CD workflow that further automates the building and deployment of the TensorFlow WideDeep Classifer pipeline code introduced in this lab.
# ## License
# <font size=-1>Licensed under the Apache License, Version 2.0 (the \"License\");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at [https://www.apache.org/licenses/LICENSE-2.0](https://www.apache.org/licenses/LICENSE-2.0)
#
# Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.</font>
| on_demand/tfx-caip/lab-02-tfx-pipeline/labs/lab-02.ipynb |
# -*- coding: utf-8 -*-
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .jl
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Julia 1.1.0
# language: julia
# name: julia-1.1
# ---
# +
# necessary packages #
#using Pkg
#Pkg.add("Distances")
using Distributions
using Random
using Distances
using LinearAlgebra
using SparseArrays
using IterativeSolvers
using ProgressMeter
using JLD2
# -
include("../../../util2.j")
# +
# unnecessary packages #
#using Pkg
#Pkg.add("UnicodePlots")
using UnicodePlots # check the structure of the sparse matrix
using BenchmarkTools
using StatsPlots
using MCMCChains
using PrettyTables
# +
#using Pkg
#Pkg.add("ProgressMeter");
# +
# Set the parameters for SLMC model #
N = 1200 # sample size
N1 = 1000; N2 = 1000;
q = 2; p = 2; K = 2
Σ = [0.4 0.15
0.15 0.3];
β = [1.0 -1.0
-5.0 2.0];
ϕ1 = 6.0; ϕ2 = 18.0; ν1 = 0.5; ν2 = 0.5; # parameter for the independent F
Λ = [1.0 1.0
0.0 1.0]; # loading matrix
# +
# Generate simulation data #
Random.seed!(1234);
coords = rand(N, 2); # random location over unit square
X = hcat(fill(1, (N,)), rand(N)); # design matrix
D = pairwise(Euclidean(), coords, dims = 1); # distance matrix
ρ1 = exp.(-ϕ1 * D); ρ2 = exp.(-ϕ2 * D); # covariance matrix
ω = [rand(MvNormal(ρ1), 1) rand(MvNormal(ρ2), 1)] * Λ; # latent process
Y = X * β + ω + transpose(rand(MvNormal(Σ), N)); # response matrix
# +
# Some data preparations #
ordx = 1:N; #sortperm(coords[1, :]);
X_ord = X[ordx, :]; Y_ord = Y[ordx, :]; ω_ord = ω[ordx, :]; # sorted data
ω_incp_obs = ω_ord + fill(1.0, (N, 1)) * transpose(β[1, :]); # latent process + intercept
coords_ord = coords[ordx, :];
S1_ind = sample(1:N, N1, replace = false, ordered = true); # observed location index for 1st response
S2_ind = sample(1:N, N2, replace = false, ordered = true); # observed location index for 2nd response
S = sort(union(S1_ind, S2_ind)); # observed index set
# -
# check the plot of the data
using RCall
@rput ω_incp_obs
@rput coords_ord
@rput S
R"""
library(MBA)
library(classInt)
library(RColorBrewer)
library(sp)
library(coda)
library(spBayes)
library(fields)
h <- 12
surf.raw1 <- mba.surf(cbind(coords_ord[S, ], ω_incp_obs[S, 1]), no.X = 300, no.Y = 300,
exten = TRUE, sp = TRUE, h = h)$xyz.est
surf.raw2 <- mba.surf(cbind(coords_ord[S, ], ω_incp_obs[S, 2]), no.X = 300, no.Y = 300,
exten = TRUE, sp = TRUE, h = h)$xyz.est
surf.brks <- classIntervals(c(surf.raw1[["z"]], surf.raw2[["z"]]), 500, 'pretty')$brks
col.pal <- colorRampPalette(brewer.pal(11,'RdBu')[11:1])
xlim <- c(0, 1.13)
zlim <- range(c(surf.raw1[["z"]], surf.raw2[["z"]]))
# size for the mapping of w
width <- 360
height <- 360
pointsize <- 16
png(paste("../../pics/sim2_r_map-w1_incp-true3.png", sep = ""),
width = width, height = height, pointsize = pointsize, family = "Courier")
par(mfrow = c(1, 1))
##Obs
i <- as.image.SpatialGridDataFrame(surf.raw1)
plot(coords_ord[S, ], typ="n", cex=0.5, xlim=xlim, axes=FALSE, ylab="y", xlab="x")
#main = "true")
axis(2, las=1)
axis(1)
image.plot(i, add=TRUE, col=rev(col.pal(length(surf.brks)-1)), zlim=zlim)
dev.off()
png(paste("../../pics/sim2_r_map-w2_incp-true3.png", sep = ""),
width = width, height = height, pointsize = pointsize, family = "Courier")
par(mfrow = c(1, 1))
##Obs
i <- as.image.SpatialGridDataFrame(surf.raw2)
plot(coords_ord[S, ], typ="n", cex=0.5, xlim=xlim, axes=FALSE, ylab="y", xlab="x")
#main = "true")
axis(2, las=1)
axis(1)
image.plot(i, add=TRUE, col=rev(col.pal(length(surf.brks)-1)), zlim=zlim)
dev.off()
"""
@save "sim2data.jld"
| sim/sim2/data/sim2_data.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
import ipywidgets as widgets
def squareit(n):
return n*n
squareit(5)
widgets.interact(squareit, n=(0, 100))
| Investment Management/Course1/lab_122.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # Gaussian Code Exercise
#
# Read through the code below and fill out the TODOs. You'll find a cell at the end of the Jupyter notebook containing unit tests. After you've run the code cell with the Gaussian class, you can run the final cell to check that your code functions as expected.
#
# This exercise includes a file called 'numbers.txt', which you can see if you click on the 'Jupyter' icon at the top of the workspace and then go into the folder titled 3.OOP_code_gaussian_class. The 'numbers.txt' file is read in by the read_data_file() method. There is also a solution in the 3.OOP_code_gaussian_class folder in a file called answer.py.
# +
import math
import matplotlib.pyplot as plt
class Gaussian():
""" Gaussian distribution class for calculating and
visualizing a Gaussian distribution.
Attributes:
mean (float) representing the mean value of the distribution
stdev (float) representing the standard deviation of the distribution
data_list (list of floats) a list of floats extracted from the data file
"""
def __init__(self, mu = 0, sigma = 1):
self.mean = mu
self.stdev = sigma
self.data = []
def calculate_mean(self):
"""Method to calculate the mean of the data set.
Args:
None
Returns:
float: mean of the data set
"""
#TODO: Calculate the mean of the data set. Remember that the data set is stored in self.data
# Change the value of the mean attribute to be the mean of the data set
# Return the mean of the data set
mean = 1.0 * (sum(self.data)/len(self.data))
self.mean = mean
return self.mean
def calculate_stdev(self, sample=True):
"""Method to calculate the standard deviation of the data set.
Args:
sample (bool): whether the data represents a sample or population
Returns:
float: standard deviation of the data set
"""
# TODO:
# Calculate the standard deviation of the data set
#
# The sample variable determines if the data set contains a sample or a population
# If sample = True, this means the data is a sample.
# Keep the value of sample in mind for calculating the standard deviation
#
# Make sure to update self.stdev and return the standard deviation as well
if sample == True:
n = len(self.data) - 1
else:
n = len(self.data)
mean = self.mean
sigma = 0
for d in self.data:
sigma += (d - mean)**2
sigma = math.sqrt(sigma / n)
self.stdev = sigma
return sigma
def read_data_file(self, file_name, sample=True):
"""Method to read in data from a txt file. The txt file should have
one number (float) per line. The numbers are stored in the data attribute.
After reading in the file, the mean and standard deviation are calculated
Args:
file_name (string): name of a file to read from
Returns:
None
"""
# This code opens a data file and appends the data to a list called data_list
with open(file_name) as file:
data_list = []
line = file.readline()
while line:
data_list.append(int(line))
line = file.readline()
file.close()
# TODO:
# Update the self.data attribute with the data_list
# Update self.mean with the mean of the data_list.
# You can use the calculate_mean() method with self.calculate_mean()
# Update self.stdev with the standard deviation of the data_list. Use the
# calcaulte_stdev() method.
self.data = data_list
self.mean = self.calculate_mean()
self.stdev = self.calculate_stdev()
def plot_histogram(self):
"""Method to output a histogram of the instance variable data using
matplotlib pyplot library.
Args:
None
Returns:
None
"""
# TODO: Plot a histogram of the data_list using the matplotlib package.
# Be sure to label the x and y axes and also give the chart a title
plt.hist(self.data)
plt.title('Histogram of Data')
plt.xlabel('Data')
plt.ylabel('Count')
def pdf(self, x):
"""Probability density function calculator for the gaussian distribution.
Args:
x (float): point for calculating the probability density function
Returns:
float: probability density function output
"""
# TODO: Calculate the probability density function of the Gaussian distribution
# at the value x. You'll need to use self.stdev and self.mean to do the calculation
return 1.0 * (1/(self.stdev * math.sqrt(2*math.pi)) * math.exp(((-1/2) * ((x - self.mean)/self.stdev)**2)))
def plot_histogram_pdf(self, n_spaces = 50):
"""Method to plot the normalized histogram of the data and a plot of the
probability density function along the same range
Args:
n_spaces (int): number of data points
Returns:
list: x values for the pdf plot
list: y values for the pdf plot
"""
#TODO: Nothing to do for this method. Try it out and see how it works.
mu = self.mean
sigma = self.stdev
min_range = min(self.data)
max_range = max(self.data)
# calculates the interval between x values
interval = 1.0 * (max_range - min_range) / n_spaces
x = []
y = []
# calculate the x values to visualize
for i in range(n_spaces):
tmp = min_range + interval*i
x.append(tmp)
y.append(self.pdf(tmp))
# make the plots
fig, axes = plt.subplots(2,sharex=True)
fig.subplots_adjust(hspace=.5)
axes[0].hist(self.data, density=True)
axes[0].set_title('Normed Histogram of Data')
axes[0].set_ylabel('Density')
axes[1].plot(x, y)
axes[1].set_title('Normal Distribution for \n Sample Mean and Sample Standard Deviation')
axes[0].set_ylabel('Density')
plt.show()
return x, y
# +
# Unit tests to check your solution
import unittest
class TestGaussianClass(unittest.TestCase):
def setUp(self):
self.gaussian = Gaussian(25, 2)
def test_initialization(self):
self.assertEqual(self.gaussian.mean, 25, 'incorrect mean')
self.assertEqual(self.gaussian.stdev, 2, 'incorrect standard deviation')
def test_pdf(self):
self.assertEqual(round(self.gaussian.pdf(25), 5), 0.19947,\
'pdf function does not give expected result')
def test_meancalculation(self):
self.gaussian.read_data_file('numbers.txt', True)
self.assertEqual(self.gaussian.calculate_mean(),\
sum(self.gaussian.data) / float(len(self.gaussian.data)), 'calculated mean not as expected')
def test_stdevcalculation(self):
self.gaussian.read_data_file('numbers.txt', True)
self.assertEqual(round(self.gaussian.stdev, 2), 92.87, 'sample standard deviation incorrect')
self.gaussian.read_data_file('numbers.txt', False)
self.assertEqual(round(self.gaussian.stdev, 2), 88.55, 'population standard deviation incorrect')
tests = TestGaussianClass()
tests_loaded = unittest.TestLoader().loadTestsFromModule(tests)
unittest.TextTestRunner().run(tests_loaded)
| Udacity_AI_Python/Gaussian Class/gaussian_code_exercise.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# <div align="center"><h1>Potential Flow Model</h1></div>
# <div align="center"><h2>log-reduction calculator</h2></div>
# +
# %reset -f
import numpy as np
import matplotlib.pyplot as plt
#import matplotlib
import ipywidgets as wd
from os import system
import os
from matplotlib.gridspec import GridSpec
#import jupypft.attachmentRateCFT as CFT
#import jupypft.plotBTC as BTC
''' GLOBAL CONSTANTS '''
PI = 3.141592
I = 10**np.linspace(-5,0,num=100)
''' FUNCTION CALCULATORS'''
def calculateAll(var_dict):
K = var_dict['K']
THETA = var_dict['n']
Qin = var_dict['Qin']/86400
f = var_dict['Qout']/var_dict['Qin']
H = var_dict['H']
r = var_dict['rs']
C0 = 1.0
decayRate = var_dict['mu']/86400
attchRate = var_dict['katt']/86400
delY,delZ = 1.35,var_dict['H']
def flowNumber(): return (4.0*K*I*H*r) / (Qin*(1+f))
def uChar(): return -(K*I*flowNumber())/(THETA*(flowNumber() + 1))
def tChar(): return -r/uChar()
def cDecay(): return C0 * np.exp(-decayRate * tChar())
def cAttach(): return C0 * np.exp(-attchRate * tChar())
def cDilut(): return (C0 * Qin) / (-uChar() * delY * delZ * THETA)
def cBoth(): return (C0 * Qin) / (-uChar() * delY * delZ * THETA) * np.exp(-decayRate * tChar())
def cTrice(): return (C0 * Qin) / (-uChar() * delY * delZ * THETA) * np.exp(-(decayRate+attchRate) * tChar())
def findSweet(): return np.argmin(np.abs(cTrice() - np.max(cTrice())))
cDec = cDecay()
cDil = cDilut()
cAtt = cAttach()
cBot = cBoth()
cAll = cTrice()
i = findSweet()
worstC = cAll[i]
worstI = I[i]
results_dict = dict(cDec=cDec,cDil=cDil,cAtt=cAtt,cBot=cBot,cAll=cAll,i=i,worstC=worstC,worstI=worstI)
return results_dict
# +
gs_Parameters = wd.GridspecLayout(8,2)
var_keys = ["<KEY>n","mu","katt"]
init_values = [0.01,20,40,0.24,2.4,0.35,0.30,0.10]
min_values = [1.0E-6,0.1,0.1,0.001,0.001,0.01,1.0E-6,1.0E-6]
max_values = [1.0E+2,1000,1000,1000,1000,0.999,1.0E+6,1.0E+6]
var_dict = {k:v for k,v in zip(var_keys,init_values)}
lbl_Parameters = [wd.Label(txt) for txt in
["Hydraulic conductivity [m/d]",
"Aquifer thickness [m]",
"Setback distance [m]",
"Contaminant influx [m3/d]",
"Extraction flow rate [m3/d]",
"Porosity [-]",
"Inactivation rate [1/d]",
"Attachment rate [1/d]"]]
val_Parameters = [wd.BoundedFloatText(value=v,min=minv,max=maxv,description=txt)
for v,minv,maxv,txt in
zip(init_values,min_values,max_values,
[r"$K$",
r"$H$",
r"$r_s$",
r"$Q_{\rm in}$",
r"$Q_{\rm out}$",
r"$n$",
r"$\mu_m$",
r"$k_{\rm att}$"])]
for i,(lbl,val) in enumerate(zip(lbl_Parameters,val_Parameters)):
gs_Parameters[i,0],gs_Parameters[i,1] = lbl,val
def update_values(_):
'''Updates var_dict with the widgets values'''
for k,v in zip(var_dict.keys(),val_Parameters):
var_dict[k] = v.value
for val_Widget in val_Parameters:
val_Widget.observe(update_values,'value')
update_values(None)
# +
layout = wd.Layout(justify_content='center',align_items='center')
output = wd.Output(layout=layout)
bbox = dict(boxstyle='round', facecolor='mintcream', alpha=0.90)
arrowprops = dict(arrowstyle="->",connectionstyle="angle,angleA=90,angleB=40,rad=5")
fontdict = dict(size=12)
@output.capture(clear_output=True)
def plot_PFM(_):
update_values(_)
cDec,cDil,cAtt,cBot,cAll,i,worstC,worstI = calculateAll(var_dict).values()
with np.errstate(divide='ignore'):
logs_cDec = -np.log10(cDec)
logs_cDil = -np.log10(cDil)
logs_cAtt = -np.log10(cAtt)
logs_cAll = -np.log10(cAll)
fig,ax = plt.subplots(figsize=(8,6))
#### log-removals
ax.plot(I,logs_cDec,label="Due decay",lw=3,ls="dashed",alpha=0.8)
ax.plot(I,logs_cDil,label="Due dilution",lw=3,ls="dashed",alpha=0.8)
ax.plot(I,logs_cAtt,label="Due attachment",lw=2,ls="dashed",alpha=0.6)
ax.plot(I,logs_cAll,label="Combined effect",lw=3,c='k',alpha=0.9)
####################################<
#Line worst case scenario
#ax.axvline(x=I[i], lw=1, ls="dashed", c="red",alpha=0.5)
ax.axhline(y=4, lw=1, ls='dashed', c='r', alpha = 0.5)
annotation = \
r"$\bf{-\log(C/C_0)} = $" + "{:.1f}".format(-np.log10(worstC)) + \
"\n@" + r" $\bf{I} = $" + "{:.1E}".format(worstI)
ax.scatter([worstI],[-np.log10(worstC)],
c='r', marker='X', s=250, zorder=2,
label="\n\nWorst case:\n" + annotation)
#####################################
ax.set(xscale="log",xlim=(1.0E-4,0.5),ylim=(-0.1,10))
ax.set_xlabel("Water table gradient\n$I$ [m/m]",fontdict=fontdict)
ax.set_ylabel("log-reductions\n$-\log(C/C_0)$ [-]",fontdict=fontdict)
ax.legend(loc="center left",bbox_to_anchor=(1.02,0.5),shadow=True)
plt.show()
# +
plotButton = wd.Button(description="Plot!",icon="fa-carrot")
plotButton.on_click(plot_PFM)
mainBox = wd.VBox([gs_Parameters,plotButton,output],layout=layout)
# -
mainBox
| PFM - Find worst case.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + id="copyright"
# Copyright 2022 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] id="title:generic,gcp"
# # E2E ML on GCP: MLOps stage 2 : experimentation: get started with Vertex Training for XGBoost
#
# <table align="left">
# <td>
# <a href="https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/main/notebooks/community/ml_ops/stage2/get_started_vertex_training_xgboost.ipynb">
# <img src="https://cloud.google.com/ml-engine/images/github-logo-32px.png" alt="GitHub logo">
# View on GitHub
# </a>
# </td>
# <td>
# <a href="https://console.cloud.google.com/ai/platform/notebooks/deploy-notebook?download_url=https://github.com/GoogleCloudPlatform/vertex-ai-samples/blob/main/notebooks/community/ml_ops/stage2/get_started_vertex_training_xgboost.ipynb">
# Open in Google Cloud Notebooks
# </a>
# </td>
# </table>
# <br/><br/><br/>
# + [markdown] id="overview:mlops"
# ## Overview
#
#
# This tutorial demonstrates how to use Vertex AI for E2E MLOps on Google Cloud in production. This tutorial covers stage 2 : experimentation: get started with Vertex Training for XGBoost.
# + [markdown] id="dataset:iris,lcn"
# ### Dataset
#
# The dataset used for this tutorial is the [Iris dataset](https://www.tensorflow.org/datasets/catalog/iris) from [TensorFlow Datasets](https://www.tensorflow.org/datasets/catalog/overview). This dataset does not require any feature engineering. The version of the dataset you will use in this tutorial is stored in a public Cloud Storage bucket. The trained model predicts the type of Iris flower species from a class of three species: setosa, virginica, or versicolor.
# + [markdown] id="objective:mlops,stage2,get_started_vertex_training_xgboost"
# ### Objective
#
# In this tutorial, you learn how to use `Vertex AI Training` for training a XGBoost custom model.
#
# This tutorial uses the following Google Cloud ML services:
#
# - `Vertex AI Training`
# - `Vertex AI Model` resource
#
# The steps performed include:
#
# - Training using a Python package.
# - Report accuracy when hyperparameter tuning.
# - Save the model artifacts to Cloud Storage using GCSFuse.
# - Create a `Vertex AI Model` resource.
# + [markdown] id="install_mlops"
# ## Installations
#
# Install *one time* the packages for executing the MLOps notebooks.
# + id="install_mlops"
ONCE_ONLY = False
if ONCE_ONLY:
# ! pip3 install -U tensorflow==2.5 $USER_FLAG
# ! pip3 install -U tensorflow-data-validation==1.2 $USER_FLAG
# ! pip3 install -U tensorflow-transform==1.2 $USER_FLAG
# ! pip3 install -U tensorflow-io==0.18 $USER_FLAG
# ! pip3 install --upgrade google-cloud-aiplatform[tensorboard] $USER_FLAG
# ! pip3 install --upgrade google-cloud-pipeline-components $USER_FLAG
# ! pip3 install --upgrade google-cloud-bigquery $USER_FLAG
# ! pip3 install --upgrade google-cloud-logging $USER_FLAG
# ! pip3 install --upgrade apache-beam[gcp] $USER_FLAG
# ! pip3 install --upgrade pyarrow $USER_FLAG
# ! pip3 install --upgrade cloudml-hypertune $USER_FLAG
# ! pip3 install --upgrade kfp $USER_FLAG
# ! pip3 install --upgrade torchvision $USER_FLAG
# ! pip3 install --upgrade rpy2 $USER_FLAG
# + [markdown] id="restart"
# ### Restart the kernel
#
# Once you've installed the additional packages, you need to restart the notebook kernel so it can find the packages.
# + id="restart"
import os
if not os.getenv("IS_TESTING"):
# Automatically restart kernel after installs
import IPython
app = IPython.Application.instance()
app.kernel.do_shutdown(True)
# + [markdown] id="project_id"
# #### Set your project ID
#
# **If you don't know your project ID**, you may be able to get your project ID using `gcloud`.
# + id="set_project_id"
PROJECT_ID = "[your-project-id]" # @param {type:"string"}
# + id="autoset_project_id"
if PROJECT_ID == "" or PROJECT_ID is None or PROJECT_ID == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = ! gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT_ID = shell_output[0]
print("Project ID:", PROJECT_ID)
# + id="set_gcloud_project_id"
# ! gcloud config set project $PROJECT_ID
# + [markdown] id="region"
# #### Region
#
# You can also change the `REGION` variable, which is used for operations
# throughout the rest of this notebook. Below are regions supported for Vertex AI. We recommend that you choose the region closest to you.
#
# - Americas: `us-central1`
# - Europe: `europe-west4`
# - Asia Pacific: `asia-east1`
#
# You may not use a multi-regional bucket for training with Vertex AI. Not all regions provide support for all Vertex AI services.
#
# Learn more about [Vertex AI regions](https://cloud.google.com/vertex-ai/docs/general/locations).
# + id="region"
REGION = "us-central1" # @param {type: "string"}
# + [markdown] id="timestamp"
# #### Timestamp
#
# If you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append the timestamp onto the name of resources you create in this tutorial.
# + id="timestamp"
from datetime import datetime
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
# + [markdown] id="bucket:mbsdk"
# ### Create a Cloud Storage bucket
#
# **The following steps are required, regardless of your notebook environment.**
#
# When you initialize the Vertex SDK for Python, you specify a Cloud Storage staging bucket. The staging bucket is where all the data associated with your dataset and model resources are retained across sessions.
#
# Set the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.
# + id="bucket"
BUCKET_NAME = "gs://[your-bucket-name]" # @param {type:"string"}
# + id="autoset_bucket"
if BUCKET_NAME == "" or BUCKET_NAME is None or BUCKET_NAME == "gs://[your-bucket-name]":
BUCKET_NAME = "gs://" + PROJECT_ID + "aip-" + TIMESTAMP
# + [markdown] id="create_bucket"
# **Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.
# + id="create_bucket"
# ! gsutil mb -l $REGION $BUCKET_NAME
# + [markdown] id="validate_bucket"
# Finally, validate access to your Cloud Storage bucket by examining its contents:
# + id="validate_bucket"
# ! gsutil ls -al $BUCKET_NAME
# + [markdown] id="setup_vars"
# ### Set up variables
#
# Next, set up some variables used throughout the tutorial.
# ### Import libraries and define constants
# + id="import_aip:mbsdk"
import google.cloud.aiplatform as aip
# + [markdown] id="init_aip:mbsdk"
# ### Initialize Vertex AI SDK for Python
#
# Initialize the Vertex AI SDK for Python for your project and corresponding bucket.
# + id="init_aip:mbsdk"
aip.init(project=PROJECT_ID, staging_bucket=BUCKET_NAME)
# + [markdown] id="accelerators:training,cpu,prediction,cpu,mbsdk"
# #### Set hardware accelerators
#
# You can set hardware accelerators for training and prediction.
#
# Set the variables `TRAIN_GPU/TRAIN_NGPU` and `DEPLOY_GPU/DEPLOY_NGPU` to use a container image supporting a GPU and the number of GPUs allocated to the virtual machine (VM) instance. For example, to use a GPU container image with 4 Nvidia Telsa K80 GPUs allocated to each VM, you would specify:
#
# (aip.AcceleratorType.NVIDIA_TESLA_K80, 4)
#
#
# Otherwise specify `(None, None)` to use a container image to run on a CPU.
#
# Learn more about [hardware accelerator support for your region](https://cloud.google.com/vertex-ai/docs/general/locations#accelerators).
#
# *Note*: TF releases before 2.3 for GPU support will fail to load the custom model in this tutorial. It is a known issue and fixed in TF 2.3. This is caused by static graph ops that are generated in the serving function. If you encounter this issue on your own custom models, use a container image for TF 2.3 with GPU support.
# + id="accelerators:training,cpu,prediction,cpu,mbsdk"
if os.getenv("IS_TESTING_TRAIN_GPU"):
TRAIN_GPU, TRAIN_NGPU = (
aip.gapic.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_TRAIN_GPU")),
)
else:
TRAIN_GPU, TRAIN_NGPU = (None, None)
if os.getenv("IS_TESTING_DEPLOY_GPU"):
DEPLOY_GPU, DEPLOY_NGPU = (
aip.gapic.AcceleratorType.NVIDIA_TESLA_K80,
int(os.getenv("IS_TESTING_DEPLOY_GPU")),
)
else:
DEPLOY_GPU, DEPLOY_NGPU = (None, None)
# + [markdown] id="container:training,prediction,xgboost"
# #### Set pre-built containers
#
# Set the pre-built Docker container image for training and prediction.
#
#
# For the latest list, see [Pre-built containers for training](https://cloud.google.com/ai-platform-unified/docs/training/pre-built-containers).
#
#
# For the latest list, see [Pre-built containers for prediction](https://cloud.google.com/ai-platform-unified/docs/predictions/pre-built-containers).
# + id="container:training,prediction,xgboost"
TRAIN_VERSION = "xgboost-cpu.1-1"
DEPLOY_VERSION = "xgboost-cpu.1-1"
TRAIN_IMAGE = "{}-docker.pkg.dev/vertex-ai/training/{}:latest".format(
REGION.split("-")[0], TRAIN_VERSION
)
DEPLOY_IMAGE = "{}-docker.pkg.dev/vertex-ai/prediction/{}:latest".format(
REGION.split("-")[0], DEPLOY_VERSION
)
# + [markdown] id="machine:training"
# #### Set machine type
#
# Next, set the machine type to use for training.
#
# - Set the variable `TRAIN_COMPUTE` to configure the compute resources for the VMs you will use for for training.
# - `machine type`
# - `n1-standard`: 3.75GB of memory per vCPU.
# - `n1-highmem`: 6.5GB of memory per vCPU
# - `n1-highcpu`: 0.9 GB of memory per vCPU
# - `vCPUs`: number of \[2, 4, 8, 16, 32, 64, 96 \]
#
# *Note: The following is not supported for training:*
#
# - `standard`: 2 vCPUs
# - `highcpu`: 2, 4 and 8 vCPUs
#
# *Note: You may also use n2 and e2 machine types for training and deployment, but they do not support GPUs*.
# + id="machine:training"
if os.getenv("IS_TESTING_TRAIN_MACHINE"):
MACHINE_TYPE = os.getenv("IS_TESTING_TRAIN_MACHINE")
else:
MACHINE_TYPE = "n1-standard"
VCPU = "4"
TRAIN_COMPUTE = MACHINE_TYPE + "-" + VCPU
print("Train machine type", TRAIN_COMPUTE)
# + [markdown] id="xgboost_intro"
# ## Introduction to XGBoost training
#
# Once you have trained a XGBoost model, you will want to save it at a Cloud Storage location, so it can subsequently be uploaded to a `Vertex AI Model` resource.
# The XGBoost package does not have support to save the model to a Cloud Storage location. Instead, you will do the following steps to save to a Cloud Storage location.
#
# 1. Save the in-memory model to the local filesystem (e.g., model.bst).
# 2. Use gsutil to copy the local copy to the specified Cloud Storage location.
#
# *Note*: You can do hyperparameter tuning with a XGBoost model.
# + [markdown] id="examine_training_package:xgboost"
# ### Examine the training package
#
# #### Package layout
#
# Before you start the training, you will look at how a Python package is assembled for a custom training job. When unarchived, the package contains the following directory/file layout.
#
# - PKG-INFO
# - README.md
# - setup.cfg
# - setup.py
# - trainer
# - \_\_init\_\_.py
# - task.py
#
# The files `setup.cfg` and `setup.py` are the instructions for installing the package into the operating environment of the Docker image.
#
# The file `trainer/task.py` is the Python script for executing the custom training job. *Note*, when we referred to it in the worker pool specification, we replace the directory slash with a dot (`trainer.task`) and dropped the file suffix (`.py`).
#
# #### Package Assembly
#
# In the following cells, you will assemble the training package.
# + id="examine_training_package:xgboost"
# Make folder for Python training script
# ! rm -rf custom
# ! mkdir custom
# Add package information
# ! touch custom/README.md
setup_cfg = "[egg_info]\n\ntag_build =\n\ntag_date = 0"
# ! echo "$setup_cfg" > custom/setup.cfg
setup_py = "import setuptools\n\nsetuptools.setup(\n\n install_requires=[\n\n 'cloudml-hypertune',\n\n ],\n\n packages=setuptools.find_packages())"
# ! echo "$setup_py" > custom/setup.py
pkg_info = "Metadata-Version: 1.0\n\nName: Iris tabular classification\n\nVersion: 0.0.0\n\nSummary: Demostration training script\n\nHome-page: www.google.com\n\nAuthor: Google\n\nAuthor-email: <EMAIL>\n\nLicense: Public\n\nDescription: Demo\n\nPlatform: Vertex"
# ! echo "$pkg_info" > custom/PKG-INFO
# Make the training subfolder
# ! mkdir custom/trainer
# ! touch custom/trainer/__init__.py
# + [markdown] id="taskpy_contents:iris,xgboost"
# ### Create the task script for the Python training package
#
# Next, you create the `task.py` script for driving the training package. Some noteable steps include:
#
# - Command-line arguments:
# - `model-dir`: The location to save the trained model. When using Vertex AI custom training, the location will be specified in the environment variable: `AIP_MODEL_DIR`,
# - `dataset_data_url`: The location of the training data to download.
# - `dataset_labels_url`: The location of the training labels to download.
# - `boost-rounds`: Tunable hyperparameter
# - Data preprocessing (`get_data()`):
# - Download the dataset and split into training and test.
# - Training (`train_model()`):
# - Trains the model
# - Evaluation (`evaluate_model()`):
# - Evaluates the model.
# - If hyperparameter tuning, reports the metric for accuracy.
# - Model artifact saving
# - Saves the model artifacts and evaluation metrics where the Cloud Storage location specified by `model-dir`.
# + id="taskpy_contents:iris,xgboost"
# %%writefile custom/trainer/task.py
import datetime
import os
import subprocess
import sys
import pandas as pd
import xgboost as xgb
import hypertune
import argparse
import logging
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
parser = argparse.ArgumentParser()
parser.add_argument('--model-dir', dest='model_dir',
default=os.getenv('AIP_MODEL_DIR'), type=str, help='Model dir.')
parser.add_argument("--dataset-data-url", dest="dataset_data_url",
type=str, help="Download url for the training data.")
parser.add_argument("--dataset-labels-url", dest="dataset_labels_url",
type=str, help="Download url for the training data labels.")
parser.add_argument("--boost-rounds", dest="boost_rounds",
default=20, type=int, help="Number of boosted rounds")
args = parser.parse_args()
logging.getLogger().setLevel(logging.INFO)
def get_data():
logging.info("Downloading training data and labelsfrom: {}, {}".format(args.dataset_data_url, args.dataset_labels_url))
# gsutil outputs everything to stderr so we need to divert it to stdout.
subprocess.check_call(['gsutil', 'cp', args.dataset_data_url, 'data.csv'], stderr=sys.stdout)
# gsutil outputs everything to stderr so we need to divert it to stdout.
subprocess.check_call(['gsutil', 'cp', args.dataset_labels_url, 'labels.csv'], stderr=sys.stdout)
# Load data into pandas, then use `.values` to get NumPy arrays
data = pd.read_csv('data.csv').values
labels = pd.read_csv('labels.csv').values
# Convert one-column 2D array into 1D array for use with XGBoost
labels = labels.reshape((labels.size,))
train_data, test_data, train_labels, test_labels = train_test_split(data, labels, test_size=0.2, random_state=7)
# Load data into DMatrix object
dtrain = xgb.DMatrix(train_data, label=train_labels)
return dtrain, test_data, test_labels
def train_model(dtrain):
logging.info("Start training ...")
# Train XGBoost model
model = xgb.train({}, dtrain, num_boost_round=args.boost_rounds)
logging.info("Training completed")
return model
def evaluate_model(model, test_data, test_labels):
dtest = xgb.DMatrix(test_data)
pred = model.predict(dtest)
predictions = [round(value) for value in pred]
# evaluate predictions
accuracy = accuracy_score(test_labels, predictions)
logging.info(f"Evaluation completed with model accuracy: {accuracy}")
# report metric for hyperparameter tuning
hpt = hypertune.HyperTune()
hpt.report_hyperparameter_tuning_metric(
hyperparameter_metric_tag='accuracy',
metric_value=accuracy
)
return accuracy
dtrain, test_data, test_labels = get_data()
model = train_model(dtrain)
accuracy = evaluate_model(model, test_data, test_labels)
# GCSFuse conversion
gs_prefix = 'gs://'
gcsfuse_prefix = '/gcs/'
if args.model_dir.startswith(gs_prefix):
args.model_dir = args.model_dir.replace(gs_prefix, gcsfuse_prefix)
dirpath = os.path.split(args.model_dir)[0]
if not os.path.isdir(dirpath):
os.makedirs(dirpath)
# Export the classifier to a file
gcs_model_path = os.path.join(args.model_dir, 'model.bst')
logging.info("Saving model artifacts to {}". format(gcs_model_path))
model.save_model(gcs_model_path)
logging.info("Saving metrics to {}/metrics.json". format(args.model_dir))
gcs_metrics_path = os.path.join(args.model_dir, 'metrics.json')
with open(gcs_metrics_path, "w") as f:
f.write(f"{'accuracy: {accuracy}'}")
# + [markdown] id="tarball_training_script"
# #### Store training script on your Cloud Storage bucket
#
# Next, you package the training folder into a compressed tar ball, and then store it in your Cloud Storage bucket.
# + id="tarball_training_script"
# ! rm -f custom.tar custom.tar.gz
# ! tar cvf custom.tar custom
# ! gzip custom.tar
# ! gsutil cp custom.tar.gz $BUCKET_NAME/trainer_iris.tar.gz
# + [markdown] id="create_custom_pp_training_job:mbsdk"
# ### Create and run custom training job
#
#
# To train a custom model, you perform two steps: 1) create a custom training job, and 2) run the job.
#
# #### Create custom training job
#
# A custom training job is created with the `CustomTrainingJob` class, with the following parameters:
#
# - `display_name`: The human readable name for the custom training job.
# - `container_uri`: The training container image.
#
# - `python_package_gcs_uri`: The location of the Python training package as a tarball.
# - `python_module_name`: The relative path to the training script in the Python package.
# - `model_serving_container_uri`: The container image for deploying the model.
#
# *Note:* There is no requirements parameter. You specify any requirements in the `setup.py` script in your Python package.
# + id="create_custom_pp_training_job:mbsdk"
DISPLAY_NAME = "iris_" + TIMESTAMP
job = aip.CustomPythonPackageTrainingJob(
display_name=DISPLAY_NAME,
python_package_gcs_uri=f"{BUCKET_NAME}/trainer_iris.tar.gz",
python_module_name="trainer.task",
container_uri=TRAIN_IMAGE,
model_serving_container_image_uri=DEPLOY_IMAGE,
project=PROJECT_ID,
)
# + [markdown] id="prepare_custom_cmdargs:iris,xgboost"
# ### Prepare your command-line arguments
#
# Now define the command-line arguments for your custom training container:
#
# - `args`: The command-line arguments to pass to the executable that is set as the entry point into the container.
# - `--model-dir` : For our demonstrations, we use this command-line argument to specify where to store the model artifacts.
# - direct: You pass the Cloud Storage location as a command line argument to your training script (set variable `DIRECT = True`), or
# - indirect: The service passes the Cloud Storage location as the environment variable `AIP_MODEL_DIR` to your training script (set variable `DIRECT = False`). In this case, you tell the service the model artifact location in the job specification.
# - `--dataset-data-url`: The location of the training data to download.
# - `--dataset-labels-url`: The location of the training labels to download.
# - `--boost-rounds`: Tunable hyperparameter.
# + id="prepare_custom_cmdargs:iris,xgboost"
MODEL_DIR = "{}/{}".format(BUCKET_NAME, TIMESTAMP)
DATASET_DIR = "gs://cloud-samples-data/ai-platform/iris"
ROUNDS = 20
DIRECT = False
if DIRECT:
CMDARGS = [
"--dataset-data-url=" + DATASET_DIR + "/iris_data.csv",
"--dataset-labels-url=" + DATASET_DIR + "/iris_target.csv",
"--boost-rounds=" + str(ROUNDS),
"--model_dir=" + MODEL_DIR,
]
else:
CMDARGS = [
"--dataset-data-url=" + DATASET_DIR + "/iris_data.csv",
"--dataset-labels-url=" + DATASET_DIR + "/iris_target.csv",
"--boost-rounds=" + str(ROUNDS),
]
# + [markdown] id="run_custom_job:mbsdk"
# #### Run the custom training job
#
# Next, you run the custom job to start the training job by invoking the method `run`, with the following parameters:
#
# - `model_display_name`: The human readable name for the `Model` resource.
# - `args`: The command-line arguments to pass to the training script.
# - `replica_count`: The number of compute instances for training (replica_count = 1 is single node training).
# - `machine_type`: The machine type for the compute instances.
# - `accelerator_type`: The hardware accelerator type.
# - `accelerator_count`: The number of accelerators to attach to a worker replica.
# - `base_output_dir`: The Cloud Storage location to write the model artifacts to.
# - `sync`: Whether to block until completion of the job.
# + id="run_custom_job:mbsdk"
if TRAIN_GPU:
model = job.run(
model_display_name="iris_" + TIMESTAMP,
args=CMDARGS,
replica_count=1,
machine_type=TRAIN_COMPUTE,
accelerator_type=TRAIN_GPU.name,
accelerator_count=TRAIN_NGPU,
base_output_dir=MODEL_DIR,
sync=False,
)
else:
model = job.run(
model_display_name="iris_" + TIMESTAMP,
args=CMDARGS,
replica_count=1,
machine_type=TRAIN_COMPUTE,
base_output_dir=MODEL_DIR,
sync=False,
)
model_path_to_deploy = MODEL_DIR
# + [markdown] id="list_job"
# ### List a custom training job
# + id="list_job"
_job = job.list(filter=f"display_name={DISPLAY_NAME}")
print(_job)
# + [markdown] id="custom_job_wait:mbsdk"
# ### Wait for completion of custom training job
#
# Next, wait for the custom training job to complete. Alternatively, one can set the parameter `sync` to `True` in the `run()` method to block until the custom training job is completed.
# + id="custom_job_wait:mbsdk"
model.wait()
# + [markdown] id="delete_job"
# ### Delete a custom training job
#
# After a training job is completed, you can delete the training job with the method `delete()`. Prior to completion, a training job can be canceled with the method `cancel()`.
# + id="delete_job"
job.delete()
# + [markdown] id="cleanup:mbsdk"
# # Cleaning up
#
# To clean up all Google Cloud resources used in this project, you can [delete the Google Cloud
# project](https://cloud.google.com/resource-manager/docs/creating-managing-projects#shutting_down_projects) you used for the tutorial.
#
# Otherwise, you can delete the individual resources you created in this tutorial:
#
# - Dataset
# - Pipeline
# - Model
# - Endpoint
# - AutoML Training Job
# - Batch Job
# - Custom Job
# - Hyperparameter Tuning Job
# - Cloud Storage Bucket
# + id="cleanup:mbsdk"
delete_all = True
if delete_all:
# Delete the dataset using the Vertex dataset object
try:
if "dataset" in globals():
dataset.delete()
except Exception as e:
print(e)
# Delete the model using the Vertex model object
try:
if "model" in globals():
model.delete()
except Exception as e:
print(e)
# Delete the endpoint using the Vertex endpoint object
try:
if "endpoint" in globals():
endpoint.undeploy_all()
endpoint.delete()
except Exception as e:
print(e)
# Delete the AutoML or Pipeline training job
try:
if "dag" in globals():
dag.delete()
except Exception as e:
print(e)
# Delete the custom training job
try:
if "job" in globals():
job.delete()
except Exception as e:
print(e)
# Delete the batch prediction job using the Vertex batch prediction object
try:
if "batch_predict_job" in globals():
batch_predict_job.delete()
except Exception as e:
print(e)
# Delete the hyperparameter tuning job using the Vertex hyperparameter tuning object
try:
if "hpt_job" in globals():
hpt_job.delete()
except Exception as e:
print(e)
if "BUCKET_NAME" in globals():
# ! gsutil rm -r $BUCKET_NAME
| notebooks/community/ml_ops/stage2/get_started_vertex_training_xgboost.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# name: python3
# ---
# + [markdown] colab_type="text" id="-zMKQx6DkKwt"
# ##### Copyright 2019 The TensorFlow Authors.
# + cellView="form" colab={} colab_type="code" id="J307vsiDkMMW"
#@title Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# https://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# + [markdown] colab_type="text" id="vCMYwDIE9dTT"
# # The Keras Functional API in TensorFlow
# + [markdown] colab_type="text" id="lAJfkZ-K9flj"
# <table class="tfo-notebook-buttons" align="left">
# <td>
# <a target="_blank" href="https://www.tensorflow.org/beta/guide/keras/functional"><img src="https://www.tensorflow.org/images/tf_logo_32px.png" />View on TensorFlow.org</a>
# </td>
# <td>
# <a target="_blank" href="https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/r2/guide/keras/functional.ipynb"><img src="https://www.tensorflow.org/images/colab_logo_32px.png" />Run in Google Colab</a>
# </td>
# <td>
# <a target="_blank" href="https://github.com/tensorflow/docs/blob/master/site/en/r2/guide/keras/functional.ipynb"><img src="https://www.tensorflow.org/images/GitHub-Mark-32px.png" />View source on GitHub</a>
# </td>
# <td>
# <a href="https://storage.googleapis.com/tensorflow_docs/docs/site/en/r2/guide/keras/functional.ipynb"><img src="https://www.tensorflow.org/images/download_logo_32px.png" />Download notebook</a>
# </td>
# </table>
# + [markdown] colab_type="text" id="ITh3wzORxgpw"
# ## Setup
# + colab={} colab_type="code" id="HFbM9dcfxh4l"
from __future__ import absolute_import, division, print_function, unicode_literals
try:
# # %tensorflow_version only exists in Colab.
# %tensorflow_version 2.x
except Exception:
pass
import tensorflow as tf
tf.keras.backend.clear_session() # For easy reset of notebook state.
# + [markdown] colab_type="text" id="ZI47-lpfkZ5c"
#
# ## Introduction
#
# You're already familiar with the use of `keras.Sequential()` to create models.
# The Functional API is a way to create models that is more flexible than `Sequential`:
# it can handle models with non-linear topology, models with shared layers,
# and models with multiple inputs or outputs.
#
# It's based on the idea that a deep learning model
# is usually a directed acyclic graph (DAG) of layers.
# The Functional API a set of tools for **building graphs of layers**.
#
# Consider the following model:
#
# ```
# (input: 784-dimensional vectors)
# ↧
# [Dense (64 units, relu activation)]
# ↧
# [Dense (64 units, relu activation)]
# ↧
# [Dense (10 units, softmax activation)]
# ↧
# (output: probability distribution over 10 classes)
# ```
#
# It's a simple graph of 3 layers.
#
# To build this model with the functional API,
# you would start by creating an input node:
# + colab={} colab_type="code" id="Yxi0LaSHkDT-"
from tensorflow import keras
inputs = keras.Input(shape=(784,))
# + [markdown] colab_type="text" id="Mr3Z_Pxcnf-H"
# Here we just specify the shape of our data: 784-dimensional vectors.
# None that the batch size is always omitted, we only specify the shape of each sample.
# For an input meant for images of shape `(32, 32, 3)`, we would have used:
# + colab={} colab_type="code" id="0-2Q2nJNneIO"
img_inputs = keras.Input(shape=(32, 32, 3))
# + [markdown] colab_type="text" id="HoMFNu-pnkgF"
# What gets returned, `inputs`, contains information about the shape and dtype of the
# input data that you expect to feed to your model:
# + colab={} colab_type="code" id="ddIr9LPJnibj"
inputs.shape
# + colab={} colab_type="code" id="lZkLJeQonmTe"
inputs.dtype
# + [markdown] colab_type="text" id="kZnhhndTnrzC"
# You create a new node in the graph of layers by calling a layer on this `inputs` object:
# + colab={} colab_type="code" id="sMyyMTqDnpYV"
from tensorflow.keras import layers
dense = layers.Dense(64, activation='relu')
x = dense(inputs)
# + [markdown] colab_type="text" id="besm-lgFnveV"
# The "layer call" action is like drawing an arrow from "inputs" to this layer we created.
# We're "passing" the inputs to the `dense` layer, and out we get `x`.
#
# Let's add a few more layers to our graph of layers:
# + colab={} colab_type="code" id="DbF-MIO2ntf7"
x = layers.Dense(64, activation='relu')(x)
outputs = layers.Dense(10, activation='softmax')(x)
# + [markdown] colab_type="text" id="B38UlEIlnz_8"
# At this point, we can create a `Model` by specifying its inputs and outputs in the graph of layers:
# + colab={} colab_type="code" id="MrSfwvl-nx9s"
model = keras.Model(inputs=inputs, outputs=outputs)
# + [markdown] colab_type="text" id="5EeeV1xJn3jW"
# To recap, here is our full model definition process:
# + colab={} colab_type="code" id="xkz7oqj2n1-q"
inputs = keras.Input(shape=(784,), name='img')
x = layers.Dense(64, activation='relu')(inputs)
x = layers.Dense(64, activation='relu')(x)
outputs = layers.Dense(10, activation='softmax')(x)
model = keras.Model(inputs=inputs, outputs=outputs, name='mnist_model')
# + [markdown] colab_type="text" id="jJzocCbdn6qj"
# Let's check out what the model summary looks like:
# + colab={} colab_type="code" id="GirC9odQn5Ep"
model.summary()
# + [markdown] colab_type="text" id="mbNqYAlOn-vA"
# We can also plot the model as a graph:
# + colab={} colab_type="code" id="JYh2wLain8Oi"
keras.utils.plot_model(model, 'my_first_model.png')
# + [markdown] colab_type="text" id="QtgX2RoGoDZo"
# And optionally display the input and output shapes of each layer in the plotted graph:
# + colab={} colab_type="code" id="7FGesSSuoAG5"
keras.utils.plot_model(model, 'my_first_model_with_shape_info.png', show_shapes=True)
# + [markdown] colab_type="text" id="PBZ9XE6LoWvi"
#
# This figure and the code we wrote are virtually identical. In the code version,
# the connection arrows are simply replaced by the call operation.
#
# A "graph of layers" is a very intuitive mental image for a deep learning model,
# and the functional API is a way to create models that closely mirrors this mental image.
# + [markdown] colab_type="text" id="WUUHMaKLoZDn"
#
#
# ## Training, evaluation, and inference
#
# Training, evaluation, and inference work exactly in the same way for models built
# using the Functional API as for Sequential models.
#
# Here is a quick demonstration.
#
# Here we load MNIST image data, reshape it into vectors,
# fit the model on the data (while monitoring performance on a validation split),
# and finally we evaluate our model on the test data:
# + colab={} colab_type="code" id="DnHvkD22oFEY"
(x_train, y_train), (x_test, y_test) = keras.datasets.mnist.load_data()
x_train = x_train.reshape(60000, 784).astype('float32') / 255
x_test = x_test.reshape(10000, 784).astype('float32') / 255
model.compile(loss='sparse_categorical_crossentropy',
optimizer=keras.optimizers.RMSprop(),
metrics=['accuracy'])
history = model.fit(x_train, y_train,
batch_size=64,
epochs=5,
validation_split=0.2)
test_scores = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', test_scores[0])
print('Test accuracy:', test_scores[1])
# + [markdown] colab_type="text" id="c3nq2fjiLCkE"
# For a complete guide about model training and evaluation, see [Guide to Training & Evaluation](./training_and_evaluation.ipynb).
# + [markdown] colab_type="text" id="XOsL56zDorLh"
# ## Saving and serialization
#
# Saving and serialization work exactly in the same way for models built
# using the Functional API as for Sequential models.
#
# To standard way to save a Functional model is to call `model.save()` to save the whole model into a single file.
# You can later recreate the same model from this file, even if you no longer have access to the code
# that created the model.
#
# This file includes:
# - The model's architecture
# - The model's weight values (which were learned during training)
# - The model's training config (what you passed to `compile`), if any
# - The optimizer and its state, if any (this enables you to restart training where you left off)
# + colab={} colab_type="code" id="kN-AO7xvobtr"
model.save('path_to_my_model.h5')
del model
# Recreate the exact same model purely from the file:
model = keras.models.load_model('path_to_my_model.h5')
# + [markdown] colab_type="text" id="u0J0tFPHK4pb"
# For a complete guide about model saving, see [Guide to Saving and Serializing Models](./saving_and_serializing.ipynb).
# + [markdown] colab_type="text" id="lKz1WWr2LUzF"
# ## Using the same graph of layers to define multiple models
#
#
# In the functional API, models are created by specifying their inputs
# and outputs in a graph of layers. That means that a single graph of layers
# can be used to generate multiple models.
#
# In the example below, we use the same stack of layers to instantiate two models:
# an `encoder` model that turns image inputs into 16-dimensional vectors,
# and an end-to-end `autoencoder` model for training.
#
#
# + colab={} colab_type="code" id="WItZQr6LuVbF"
encoder_input = keras.Input(shape=(28, 28, 1), name='img')
x = layers.Conv2D(16, 3, activation='relu')(encoder_input)
x = layers.Conv2D(32, 3, activation='relu')(x)
x = layers.MaxPooling2D(3)(x)
x = layers.Conv2D(32, 3, activation='relu')(x)
x = layers.Conv2D(16, 3, activation='relu')(x)
encoder_output = layers.GlobalMaxPooling2D()(x)
encoder = keras.Model(encoder_input, encoder_output, name='encoder')
encoder.summary()
x = layers.Reshape((4, 4, 1))(encoder_output)
x = layers.Conv2DTranspose(16, 3, activation='relu')(x)
x = layers.Conv2DTranspose(32, 3, activation='relu')(x)
x = layers.UpSampling2D(3)(x)
x = layers.Conv2DTranspose(16, 3, activation='relu')(x)
decoder_output = layers.Conv2DTranspose(1, 3, activation='relu')(x)
autoencoder = keras.Model(encoder_input, decoder_output, name='autoencoder')
autoencoder.summary()
# + [markdown] colab_type="text" id="oNeg3WWFuYZK"
# Note that we make the decoding architecture strictly symmetrical to the encoding architecture,
# so that we get an output shape that is the same as the input shape `(28, 28, 1)`.
# The reverse of a `Conv2D` layer is a `Conv2DTranspose` layer, and the reverse of a `MaxPooling2D`
# layer is an `UpSampling2D` layer.
# + [markdown] colab_type="text" id="h1FVW4j-uc6Y"
#
# ## All models are callable, just like layers
#
# You can treat any model as if it were a layer, by calling it on an `Input` or on the output of another layer.
# Note that by calling a model you aren't just reusing the architecture of the model, you're also reusing its weights.
#
# Let's see this in action. Here's a different take on the autoencoder example that creates an encoder model, a decoder model,
# and chain them in two calls to obtain the autoencoder model:
# + colab={} colab_type="code" id="Ld7KdsQ_uZbr"
encoder_input = keras.Input(shape=(28, 28, 1), name='original_img')
x = layers.Conv2D(16, 3, activation='relu')(encoder_input)
x = layers.Conv2D(32, 3, activation='relu')(x)
x = layers.MaxPooling2D(3)(x)
x = layers.Conv2D(32, 3, activation='relu')(x)
x = layers.Conv2D(16, 3, activation='relu')(x)
encoder_output = layers.GlobalMaxPooling2D()(x)
encoder = keras.Model(encoder_input, encoder_output, name='encoder')
encoder.summary()
decoder_input = keras.Input(shape=(16,), name='encoded_img')
x = layers.Reshape((4, 4, 1))(decoder_input)
x = layers.Conv2DTranspose(16, 3, activation='relu')(x)
x = layers.Conv2DTranspose(32, 3, activation='relu')(x)
x = layers.UpSampling2D(3)(x)
x = layers.Conv2DTranspose(16, 3, activation='relu')(x)
decoder_output = layers.Conv2DTranspose(1, 3, activation='relu')(x)
decoder = keras.Model(decoder_input, decoder_output, name='decoder')
decoder.summary()
autoencoder_input = keras.Input(shape=(28, 28, 1), name='img')
encoded_img = encoder(autoencoder_input)
decoded_img = decoder(encoded_img)
autoencoder = keras.Model(autoencoder_input, decoded_img, name='autoencoder')
autoencoder.summary()
# + [markdown] colab_type="text" id="icQFny_huiXC"
# As you can see, model can be nested: a model can contain submodels (since a model is just like a layer).
#
# A common use case for model nesting is *ensembling*.
# As an example, here's how to ensemble a set of models into a single model that averages their predictions:
# + colab={} colab_type="code" id="ZBlZbRn5uk-9"
def get_model():
inputs = keras.Input(shape=(128,))
outputs = layers.Dense(1, activation='sigmoid')(inputs)
return keras.Model(inputs, outputs)
model1 = get_model()
model2 = get_model()
model3 = get_model()
inputs = keras.Input(shape=(128,))
y1 = model1(inputs)
y2 = model2(inputs)
y3 = model3(inputs)
outputs = layers.average([y1, y2, y3])
ensemble_model = keras.Model(inputs=inputs, outputs=outputs)
# + [markdown] colab_type="text" id="e1za1TZxuoId"
# ## Manipulating complex graph topologies
#
#
# ### Models with multiple inputs and outputs
#
# The functional API makes it easy to manipulate multiple inputs and outputs.
# This cannot be handled with the Sequential API.
#
# Here's a simple example.
#
# Let's say you're building a system for ranking custom issue tickets by priority and routing them to the right department.
#
# You model will have 3 inputs:
#
# - Title of the ticket (text input)
# - Text body of the ticket (text input)
# - Any tags added by the user (categorical input)
#
# It will have two outputs:
#
# - Priority score between 0 and 1 (scalar sigmoid output)
# - The department that should handle the ticket (softmax output over the set of departments)
#
# Let's built this model in a few lines with the Functional API.
# + colab={} colab_type="code" id="Gt91OtzbutJy"
num_tags = 12 # Number of unique issue tags
num_words = 10000 # Size of vocabulary obtained when preprocessing text data
num_departments = 4 # Number of departments for predictions
title_input = keras.Input(shape=(None,), name='title') # Variable-length sequence of ints
body_input = keras.Input(shape=(None,), name='body') # Variable-length sequence of ints
tags_input = keras.Input(shape=(num_tags,), name='tags') # Binary vectors of size `num_tags`
# Embed each word in the title into a 64-dimensional vector
title_features = layers.Embedding(num_words, 64)(title_input)
# Embed each word in the text into a 64-dimensional vector
body_features = layers.Embedding(num_words, 64)(body_input)
# Reduce sequence of embedded words in the title into a single 128-dimensional vector
title_features = layers.LSTM(128)(title_features)
# Reduce sequence of embedded words in the body into a single 32-dimensional vector
body_features = layers.LSTM(32)(body_features)
# Merge all available features into a single large vector via concatenation
x = layers.concatenate([title_features, body_features, tags_input])
# Stick a logistic regression for priority prediction on top of the features
priority_pred = layers.Dense(1, activation='sigmoid', name='priority')(x)
# Stick a department classifier on top of the features
department_pred = layers.Dense(num_departments, activation='softmax', name='department')(x)
# Instantiate an end-to-end model predicting both priority and department
model = keras.Model(inputs=[title_input, body_input, tags_input],
outputs=[priority_pred, department_pred])
# + [markdown] colab_type="text" id="KIS7lqW0uwh-"
# Let's plot the model:
# + colab={} colab_type="code" id="IMij4gzhuzYV"
keras.utils.plot_model(model, 'multi_input_and_output_model.png', show_shapes=True)
# + [markdown] colab_type="text" id="oOyuig2Hu00p"
# When compiling this model, we can assign different losses to each output.
# You can even assign different weights to each loss, to modulate their
# contribution to the total training loss.
# + colab={} colab_type="code" id="Crtdpi5Uu2cX"
model.compile(optimizer=keras.optimizers.RMSprop(1e-3),
loss=['binary_crossentropy', 'categorical_crossentropy'],
loss_weights=[1., 0.2])
# + [markdown] colab_type="text" id="t42Jrn0Yu5jL"
# Since we gave names to our output layers, we could also specify the loss like this:
# + colab={} colab_type="code" id="dPM0EwW_u6mV"
model.compile(optimizer=keras.optimizers.RMSprop(1e-3),
loss={'priority': 'binary_crossentropy',
'department': 'categorical_crossentropy'},
loss_weights=[1., 0.2])
# + [markdown] colab_type="text" id="bpTx2sXnu3-W"
# We can train the model by passing lists of Numpy arrays of inputs and targets:
# + colab={} colab_type="code" id="nB-upOoGu_k4"
import numpy as np
# Dummy input data
title_data = np.random.randint(num_words, size=(1280, 10))
body_data = np.random.randint(num_words, size=(1280, 100))
tags_data = np.random.randint(2, size=(1280, num_tags)).astype('float32')
# Dummy target data
priority_targets = np.random.random(size=(1280, 1))
dept_targets = np.random.randint(2, size=(1280, num_departments))
model.fit({'title': title_data, 'body': body_data, 'tags': tags_data},
{'priority': priority_targets, 'department': dept_targets},
epochs=2,
batch_size=32)
# + [markdown] colab_type="text" id="qNguhBWuvCtz"
# When calling fit with a `Dataset` object, it should yield either a
# tuple of lists like `([title_data, body_data, tags_data], [priority_targets, dept_targets])`
# or a tuple of dictionaries like
# `({'title': title_data, 'body': body_data, 'tags': tags_data}, {'priority': priority_targets, 'department': dept_targets})`.
#
# For more detailed explanation, refer to the complete guide [Guide to Training & Evaluation](./training_and_evaluation.ipynb).
# + [markdown] colab_type="text" id="tR0X5tTOvPyg"
# ### A toy resnet model
#
# In addition to models with multiple inputs and outputs,
# the Functional API makes it easy to manipulate non-linear connectivity topologies,
# that is to say, models where layers are not connected sequentially.
# This also cannot be handled with the Sequential API (as the name indicates).
#
# A common use case for this is residual connections.
#
# Let's build a toy ResNet model for CIFAR10 to demonstrate this.
# + colab={} colab_type="code" id="VzMoYrMNvXrm"
inputs = keras.Input(shape=(32, 32, 3), name='img')
x = layers.Conv2D(32, 3, activation='relu')(inputs)
x = layers.Conv2D(64, 3, activation='relu')(x)
block_1_output = layers.MaxPooling2D(3)(x)
x = layers.Conv2D(64, 3, activation='relu', padding='same')(block_1_output)
x = layers.Conv2D(64, 3, activation='relu', padding='same')(x)
block_2_output = layers.add([x, block_1_output])
x = layers.Conv2D(64, 3, activation='relu', padding='same')(block_2_output)
x = layers.Conv2D(64, 3, activation='relu', padding='same')(x)
block_3_output = layers.add([x, block_2_output])
x = layers.Conv2D(64, 3, activation='relu')(block_3_output)
x = layers.GlobalAveragePooling2D()(x)
x = layers.Dense(256, activation='relu')(x)
x = layers.Dropout(0.5)(x)
outputs = layers.Dense(10, activation='softmax')(x)
model = keras.Model(inputs, outputs, name='toy_resnet')
model.summary()
# + [markdown] colab_type="text" id="ISQX32bgrkis"
# Let's plot the model:
# + colab={} colab_type="code" id="pNFVkAd3rlCM"
keras.utils.plot_model(model, 'mini_resnet.png', show_shapes=True)
# + [markdown] colab_type="text" id="ECcG87yZrxp5"
# Let's train it:
# + colab={} colab_type="code" id="_iXGz5XEryou"
(x_train, y_train), (x_test, y_test) = keras.datasets.cifar10.load_data()
x_train = x_train.astype('float32') / 255.
x_test = x_test.astype('float32') / 255.
y_train = keras.utils.to_categorical(y_train, 10)
y_test = keras.utils.to_categorical(y_test, 10)
model.compile(optimizer=keras.optimizers.RMSprop(1e-3),
loss='categorical_crossentropy',
metrics=['acc'])
model.fit(x_train, y_train,
batch_size=64,
epochs=1,
validation_split=0.2)
# + [markdown] colab_type="text" id="XQfg0JUkr7SH"
# ## Sharing layers
#
# Another good use for the functional API are models that use shared layers. Shared layers are layer instances that get reused multiple times in a same model: they learn features that correspond to multiple paths in the graph-of-layers.
#
# Shared layers are often used to encode inputs that come from similar spaces (say, two different pieces of text that feature similar vocabulary), since they enable sharing of information across these different inputs, and they make it possible to train such a model on less data. If a given word is seen in one of the inputs, that will benefit the processing of all inputs that go through the shared layer.
#
# To share a layer in the Functional API, just call the same layer instance multiple times. For instance, here's an `Embedding` layer shared across two different text inputs:
# + colab={} colab_type="code" id="R9pAPQCnKuMR"
# Embedding for 1000 unique words mapped to 128-dimensional vectors
shared_embedding = layers.Embedding(1000, 128)
# Variable-length sequence of integers
text_input_a = keras.Input(shape=(None,), dtype='int32')
# Variable-length sequence of integers
text_input_b = keras.Input(shape=(None,), dtype='int32')
# We reuse the same layer to encode both inputs
encoded_input_a = shared_embedding(text_input_a)
encoded_input_b = shared_embedding(text_input_b)
# + [markdown] colab_type="text" id="xNEKvfUpr-Kf"
# ## Extracting and reusing nodes in the graph of layers
# + [markdown] colab_type="text" id="JHVGI6bEr-ze"
# Because the graph of layers you are manipulating in the Functional API is a static datastructure, it can be accessed and inspected. This is how we are able to plot Functional models as images, for instance.
#
# This also means that we can access the activations of intermediate layers ("nodes" in the graph) and reuse them elsewhere. This is extremely useful for feature extraction, for example!
#
# Let's look at an example. This is a VGG19 model with weights pre-trained on ImageNet:
# + colab={} colab_type="code" id="c-gl3xHBH-oX"
from tensorflow.keras.applications import VGG19
vgg19 = VGG19()
# + [markdown] colab_type="text" id="AKefin_xIGBP"
# And these are the intermediate activations of the model, obtained by querying the graph datastructure:
# + colab={} colab_type="code" id="1_Ap05fgIRgE"
features_list = [layer.output for layer in vgg19.layers]
# + [markdown] colab_type="text" id="H1zx5qM7IYu4"
# We can use these features to create a new feature-extraction model, that returns the values of the intermediate layer activations -- and we can do all of this in 3 lines.
# + colab={} colab_type="code" id="NrU82Pa8Igwo"
feat_extraction_model = keras.Model(inputs=vgg19.input, outputs=features_list)
img = np.random.random((1, 224, 224, 3)).astype('float32')
extracted_features = feat_extraction_model(img)
# + [markdown] colab_type="text" id="G-e2-jNCLIqy"
# This comes in handy when [implementing neural style transfer](https://medium.com/tensorflow/neural-style-transfer-creating-art-with-deep-learning-using-tf-keras-and-eager-execution-7d541ac31398), among other things.
# + [markdown] colab_type="text" id="t9M2Uvi3sBy0"
# ## Extending the API by writing custom layers
#
# tf.keras has a wide range of built-in layers. Here are a few examples:
#
# - Convolutional layers: `Conv1D`, `Conv2D`, `Conv3D`, `Conv2DTranspose`, etc.
# - Pooling layers: `MaxPooling1D`, `MaxPooling2D`, `MaxPooling3D`, `AveragePooling1D`, etc.
# - RNN layers: `GRU`, `LSTM`, `ConvLSTM2D`, etc.
# - `BatchNormalization`, `Dropout`, `Embedding`, etc.
#
# If you don't find what you need, it's easy to extend the API by creating your own layers.
#
# All layers subclass the `Layer` class and implement:
# - A `call` method, that specifies the computation done by the layer.
# - A `build` method, that creates the weights of the layer (note that this is just a style convention; you could create weights in `__init__` as well).
#
# To learn more about creating layers from scratch, check out the guide [Guide to writing layers and models from scratch](./custom_layers_and_models.ipynb).
#
# Here's a simple implementation of a `Dense` layer:
# + colab={} colab_type="code" id="ztAmarbgNV6V"
class CustomDense(layers.Layer):
def __init__(self, units=32):
super(CustomDense, self).__init__()
self.units = units
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], self.units),
initializer='random_normal',
trainable=True)
self.b = self.add_weight(shape=(self.units,),
initializer='random_normal',
trainable=True)
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
inputs = keras.Input((4,))
outputs = CustomDense(10)(inputs)
model = keras.Model(inputs, outputs)
# + [markdown] colab_type="text" id="NXxp_32bNWTy"
# If you want your custom layer to support serialization, you should also define a `get_config` method,
# that returns the constructor arguments of the layer instance:
# + colab={} colab_type="code" id="K3OQ4XxzNfAZ"
class CustomDense(layers.Layer):
def __init__(self, units=32):
super(CustomDense, self).__init__()
self.units = units
def build(self, input_shape):
self.w = self.add_weight(shape=(input_shape[-1], self.units),
initializer='random_normal',
trainable=True)
self.b = self.add_weight(shape=(self.units,),
initializer='random_normal',
trainable=True)
def call(self, inputs):
return tf.matmul(inputs, self.w) + self.b
def get_config(self):
return {'units': self.units}
inputs = keras.Input((4,))
outputs = CustomDense(10)(inputs)
model = keras.Model(inputs, outputs)
config = model.get_config()
new_model = keras.Model.from_config(
config, custom_objects={'CustomDense': CustomDense})
# + [markdown] colab_type="text" id="kXg6hZN_NfN8"
# Optionally, you could also implement the classmethod `from_config(cls, config)`, which is in charge of recreating a layer instance given its config dictionary. The default implementation of `from_config` is:
#
# ```python
# def from_config(cls, config):
# return cls(**config)
# ```
# + [markdown] colab_type="text" id="ifOVqn84sCNU"
# ## When to use the Functional API
#
# How to decide whether to use the Functional API to create a new model, or just subclass the `Model` class directly?
#
# In general, the Functional API is higher-level, easier & safer to use, and has a number of features that subclassed Models do not support.
#
# However, Model subclassing gives you greater flexibility when creating models that are not easily expressible as directed acyclic graphs of layers (for instance, you could not implement a Tree-RNN with the Functional API, you would have to subclass `Model` directly).
#
#
# ### Here are the strengths of the Functional API:
#
# The properties listed below are all true for Sequential models as well (which are also data structures), but they aren't true for subclassed models (which are Python bytecode, not data structures).
#
#
# #### It is less verbose.
#
# No `super(MyClass, self).__init__(...)`, no `def call(self, ...):`, etc.
#
# Compare:
#
# ```python
# inputs = keras.Input(shape=(32,))
# x = layers.Dense(64, activation='relu')(inputs)
# outputs = layers.Dense(10)(x)
# mlp = keras.Model(inputs, outputs)
# ```
#
# With the subclassed version:
#
# ```python
# class MLP(keras.Model):
#
# def __init__(self, **kwargs):
# super(MLP, self).__init__(**kwargs)
# self.dense_1 = layers.Dense(64, activation='relu')
# self.dense_2 = layers.Dense(10)
#
# def call(self, inputs):
# x = self.dense_1(inputs)
# return self.dense_2(x)
#
# # Instantiate the model.
# mlp = MLP()
# # Necessary to create the model's state.
# # The model doesn't have a state until it's called at least once.
# _ = mlp(tf.zeros((1, 32)))
# ```
#
#
# #### It validates your model while you're defining it.
#
# In the Functional API, your input specification (shape and dtype) is created in advance (via `Input`), and every time you call a layer, the layer checks that the specification passed to it matches its assumptions, and it will raise a helpful error message if not.
#
# This guarantees that any model you can build with the Functional API will run. All debugging (other than convergence-related debugging) will happen statically during the model construction, and not at execution time. This is similar to typechecking in a compiler.
#
#
# #### Your Functional model is plottable and inspectable.
#
# You can plot the model as a graph, and you can easily access intermediate nodes in this graph -- for instance, to extract and reuse the activations of intermediate layers, as we saw in a previous example:
#
# ```python
# features_list = [layer.output for layer in vgg19.layers]
# feat_extraction_model = keras.Model(inputs=vgg19.input, outputs=features_list)
# ```
#
#
# #### Your Functional model can be serialized or cloned.
#
# Because a Functional model is a data structure rather than a piece of code, it is safely serializable and can be saved as a single file that allows you to recreate the exact same model without having access to any of the original code. See our [saving and serialization guide](./saving_and_serializing.ipynb) for more details.
#
#
# ### Here are the weaknesses of the Functional API:
#
#
# #### It does not support dynamic architectures.
#
# The Functional API treats models as DAGs of layers. This is true for most deep learning architectures, but not all: for instance, recursive networks or Tree RNNs do not follow this assumption and cannot be implemented in the Functional API.
#
#
# #### Sometimes, you just need to write everything from scratch.
#
# When writing advanced achitectures, you may want to do things that are outside the scope of "defining a DAG of layers": for instance, you may want to expose multiple custom training and inference methods on your model instance. This requires subclassing.
#
#
# ---
#
#
# To dive more in-depth into the differences between the Functional API and Model subclassing, you can read [What are Symbolic and Imperative APIs in TensorFlow 2.0?](https://medium.com/tensorflow/what-are-symbolic-and-imperative-apis-in-tensorflow-2-0-dfccecb01021).
# + [markdown] colab_type="text" id="Ym1jrCqusGvj"
# ## Mix-and-matching different API styles
#
# Importantly, choosing between the Functional API or Model subclassing isn't a binary decision that restricts you to one category of models. All models in the tf.keras API can interact with each, whether they're Sequential models, Functional models, or subclassed Models/Layers written from scratch.
#
# You can always use a Functional model or Sequential model as part of a subclassed Model/Layer:
# + colab={} colab_type="code" id="9zF5YTLy_vGZ"
units = 32
timesteps = 10
input_dim = 5
# Define a Functional model
inputs = keras.Input((None, units))
x = layers.GlobalAveragePooling1D()(inputs)
outputs = layers.Dense(1, activation='sigmoid')(x)
model = keras.Model(inputs, outputs)
class CustomRNN(layers.Layer):
def __init__(self):
super(CustomRNN, self).__init__()
self.units = units
self.projection_1 = layers.Dense(units=units, activation='tanh')
self.projection_2 = layers.Dense(units=units, activation='tanh')
# Our previously-defined Functional model
self.classifier = model
def call(self, inputs):
outputs = []
state = tf.zeros(shape=(inputs.shape[0], self.units))
for t in range(inputs.shape[1]):
x = inputs[:, t, :]
h = self.projection_1(x)
y = h + self.projection_2(state)
state = y
outputs.append(y)
features = tf.stack(outputs, axis=1)
print(features.shape)
return self.classifier(features)
rnn_model = CustomRNN()
_ = rnn_model(tf.zeros((1, timesteps, input_dim)))
# + [markdown] colab_type="text" id="oxW1d0a8_ufg"
# Inversely, you can use any subclassed Layer or Model in the Functional API as long as it implements a `call` method that follows one of the following patterns:
#
# - `call(self, inputs, **kwargs)` where `inputs` is a tensor or a nested structure of tensors (e.g. a list of tensors), and where `**kwargs` are non-tensor arguments (non-inputs).
# - `call(self, inputs, training=None, **kwargs)` where `training` is a boolean indicating whether the layer should behave in training mode and inference mode.
# - `call(self, inputs, mask=None, **kwargs)` where `mask` is a boolean mask tensor (useful for RNNs, for instance).
# - `call(self, inputs, training=None, mask=None, **kwargs)` -- of course you can have both masking and training-specific behavior at the same time.
#
# In addition, if you implement the `get_config` method on your custom Layer or Model, the Functional models you create with it will still be serializable and clonable.
#
# Here's a quick example where we use a custom RNN written from scratch in a Functional model:
# + colab={} colab_type="code" id="TmTEZ6F3ArJR"
units = 32
timesteps = 10
input_dim = 5
batch_size = 16
class CustomRNN(layers.Layer):
def __init__(self):
super(CustomRNN, self).__init__()
self.units = units
self.projection_1 = layers.Dense(units=units, activation='tanh')
self.projection_2 = layers.Dense(units=units, activation='tanh')
self.classifier = layers.Dense(1, activation='sigmoid')
def call(self, inputs):
outputs = []
state = tf.zeros(shape=(inputs.shape[0], self.units))
for t in range(inputs.shape[1]):
x = inputs[:, t, :]
h = self.projection_1(x)
y = h + self.projection_2(state)
state = y
outputs.append(y)
features = tf.stack(outputs, axis=1)
return self.classifier(features)
# Note that we specify a static batch size for the inputs with the `batch_shape`
# arg, because the inner computation of `CustomRNN` requires a static batch size
# (when we create the `state` zeros tensor).
inputs = keras.Input(batch_shape=(batch_size, timesteps, input_dim))
x = layers.Conv1D(32, 3)(inputs)
outputs = CustomRNN()(x)
model = keras.Model(inputs, outputs)
rnn_model = CustomRNN()
_ = rnn_model(tf.zeros((1, 10, 5)))
# + [markdown] colab_type="text" id="6VxcYb4qArlb"
# This concludes our guide on the Functional API!
#
# Now you have at your fingertips a powerful set of tools for building deep learning models.
| site/en/r2/guide/keras/functional.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 2
# language: python
# name: python2
# ---
# +
#import the required libraries and models (estimators)
import numpy as np
from sklearn.pipeline import Pipeline
#imoort Logistic regression estimator
from sklearn.linear_model import LogisticRegression
#import Linear estimator
from sklearn.linear_model import LinearRegression
#import PCA estimator for dimensionality reduction
from sklearn.decomposition import PCA
# -
#chain the estimators together
estimator = [('dim_reduction', PCA()), ('logreg',LogisticRegression()), ('linear_model', LinearRegression())]
#put them in a pipeline object
pipeline_estimator = Pipeline(estimator)
#check the chain of estimators
pipeline_estimator
#view first step
pipeline_estimator.steps[0]
#view second step
pipeline_estimator.steps[1]
#view all the steps of the pipeline
pipeline_estimator.steps
| ML_Pipeline.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3 (ipykernel)
# language: python
# name: python3
# ---
# In this project, we will use a logistic regression model to predict whether an individual's annual income exceeds $50,000 given attributes about their educational and ethnic background, working class, and a few other features provided in [this](https://archive.ics.uci.edu/ml/datasets/adult) dataset from the UCI Machine Learning Repository.
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# First, we import the data.
cols = ['age', 'workclass', 'fnlwgt', 'education', 'education-num', \
'marital-status', 'occupation', 'relationship', 'race', 'sex', \
'capital-gain', 'capital-loss', 'hours-per-week', 'native-country', \
'income']
df_orig = pd.read_csv('adult.data', names=cols, index_col=False, sep=', ', engine='python')
df = df_orig.copy()
df.head()
# It is worth noting the missing values in some of the variables, denoted by a '?'.
print('Number of missing values for each variable:')
for col in df:
vc = df[col].value_counts()
num_missing = 0
if '?' in vc.index:
num_missing = vc['?']
print(' ', col, ':', num_missing)
# There are a number of ways of dealing with this issue. The simplest method is to drop all observations with missing values. However, this is not ideal as we are losing valuable data in the other variables associated with the dropped observations. For continuous variables, a common technique is to replace the missing values with a logical/unimpeding guess. Depending on the situation, this is usually either a zero or the mean or median of the rest of the values for that variable. A more sophisticated way of filling the missing values is to employ a technique called KNN imputation. In this technique, the observations with no missing values are used as training data to train a k-nearest neighbours algorithm, which predicts the missing values. This can be easily implemented using the `sklearn`'s `impute.KNNImputer` class. In our case, we will keep the missing values as their own unknown/other category.
# Some of the variables may require some explanation. `fnlwgt` is the final weight, which is the number of people the census believes this entry represents. For simplicity of analysis, and since it is more a quality of the population than the individual, this variable will be dropped. The variable `education-num` is an ordinal encoding of the `education` variable. The ordinal encoding will be more useful for our classification algorithm, so the education column will be removed. `relationship` represents the respondents' role in the family, which can be assessed from gender and marital status, so it, too, will be discarded. `capital-gain` and `capital-loss` represent income from sources other than wage or salary, such as investment income.
df = df.drop(['education', 'relationship', 'fnlwgt'], axis=1)
# The variable `workclass` stands for the industry in which the responding unit is employed.
df.workclass.value_counts()
# There are two small classes: `Without-pay` and `Never-worked`. I will combine these with the unknown class, `?`, in a category called `Other`. To simplify the analysis, we can group those who work for the government into a `Government` class, and both self-employed classes, incorporated and not incorporated, into a single one.
df.workclass = df.workclass.map({'?':'Other', 'Without-pay':'Other', 'Never-worked':'Other',\
'Local-gov':'Government', 'State-gov':'Government', 'Federal-gov':'Government',\
'Self-emp-not-inc':'Self-employed', 'Self-emp-inc':'Self-employed',\
'Private':'Private'})
df.workclass.value_counts()
# To investigate the distribution of the `workclass` variable and its relationship with our target variable `income`, we can plot a bar plot of the `workclass` variable, coloured by `income`.
# +
df_plot = df.groupby(['workclass', 'income']).size().reset_index().pivot(columns='income', \
index='workclass', values=0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(10,6))
plt.title('Income by Industry')
plt.ylabel('count')
annotations = df_plot.divide(df_plot.sum(axis=1), axis='index')
annotations = np.array(100*annotations).round().astype(int)
annotations = annotations.flatten(order='F')
for i, p in enumerate(ax.patches):
ax.annotate(str(annotations[i]) + '%', (p.get_x()+.18, p.get_y()+p.get_height()//2-200))
# -
# Those who are self employed have the greatest tendency of making more than $50,000 annually, while those with other or unknown employment have the lowest tendency.
# We can create a similar plot for the `education-num` variable.
# +
df_plot = df.groupby(['education-num', 'income']).size().reset_index().pivot(columns='income', \
index='education-num', values=0).fillna(0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(12,10))
plt.title('Income by Years of Education')
plt.ylabel('Count')
annotations = df_plot.divide(df_plot.sum(axis=1), axis='index')
annotations = np.array(100*annotations).round().astype(int)
annotations = annotations.flatten(order='F')
for i, p in enumerate(ax.patches):
if annotations[i] == 0:
continue
x = p.get_x()
y = max(p.get_y()+p.get_height()//2-80, 60)
if i >= len(annotations)//2:
y = max(y, 350)
ax.annotate(str(annotations[i]) + '%', (x, y))
# -
# Perhaps unsuprisingly, the proportion of people making more than $\$$50,000 annually increases with years of education. Nearly three quarters of those with doctoral degrees (16) make more than $\$$50,000 per year, while less than 10% of those with a high school education (8) or less make over $\$$50,000 annually.
df.occupation.value_counts()
# To simplify the `occupation` variable, I will group together the given categories into `White-collar`, `Professional`, `Sales`, `Service`, and `Blue-collar` categories. Because of the low number of observations with the `Armed-Forces` value, I will group them with the unknowns in a category called `Other`.
df.occupation = df.occupation.map({'Prof-specialty':'Professional', 'Craft-repair':'Blue-collar', \
'Exec-managerial':'White-collar', 'Adm-clerical':'White-collar', \
'Machine-op-inspct':'Blue-collar', 'Transport-moving':'Blue-collar', \
'Handlers-cleaners':'Blue-collar', 'Farming-fishing':'Blue-collar', \
'Other-service':'Service', 'Tech-support':'Service', 'Protective-serv':'Service', \
'Priv-house-serv':'Service', 'Sales':'Sales', \
'?':'Other', 'Armed-Forces':'Other'})
df.occupation.value_counts()
# +
df_plot = df.groupby(['occupation', 'income']).size().reset_index().pivot(columns='income', index='occupation', values=0).fillna(0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(10,6))
plt.title('Income by Occupation')
plt.ylabel('Count')
annotations = df_plot.divide(df_plot.sum(axis=1), axis='index')
annotations = np.array(100*annotations).round().astype(int)
annotations = annotations.flatten(order='F')
for i, p in enumerate(ax.patches):
if annotations[i] == 0:
continue
x = p.get_x()+.15
y = p.get_y()+p.get_height()//2-80
if i >= len(annotations)//2:
y = max(y, 350)
ax.annotate(str(annotations[i]) + '%', (x, y))
# -
# It is notable that income varies greatly across different occupations. Nearly half of those with a professional occupation make over $\$$50,000 annually, however only 13% of service workers make over $\$$50,000 per year.
# The categorical `marital-status` variable will be simplified for analysis as well.
df['marital-status'].value_counts()
# The `Married-civ-spouse`, `Married-spouse-absent`, and `Married-AF-spouse` categories will be combined into a `Married` variable.
df['marital-status'] = df['marital-status'].map({'Never-married':'Single', \
'Married-civ-spouse':'Married', 'Married-spouse-absent':'Married', 'Married-AF-spouse':'Married', \
'Divorced':'Divorced', 'Separated':'Separated', 'Widowed':'Widowed'})
df['marital-status'].value_counts()
# +
df_plot = df.groupby(['marital-status', 'income']).size().reset_index().pivot(columns='income', \
index='marital-status', values=0).fillna(0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(10,6))
plt.title('Income by Marital Status')
plt.ylabel('Count')
annotations = df_plot.divide(df_plot.sum(axis=1), axis='index')
annotations = np.array(100*annotations).round().astype(int)
annotations = annotations.flatten(order='F')
for i, p in enumerate(ax.patches):
if annotations[i] == 0:
continue
x = p.get_x()+.15
y = p.get_y()+p.get_height()//2-80
if i >= len(annotations)//2:
y = max(y, 350)
ax.annotate(str(annotations[i]) + '%', (x, y))
# -
# Almost half of married people make over $\$$50,000 annually, however, less than 10% of the rest of the respondents do.
# +
plt.figure(figsize=(10,6))
sns.histplot(df['capital-gain'])
plt.title('Histogram of Capital Gain')
plt.yscale('log')
plt.figure(figsize=(10,6))
sns.histplot(df['capital-loss'], label='capital loss')
plt.title('Histogram of Capital Loss')
plt.yscale('log')
# -
print('Proportion of zeros [capital-gain]: %.1f%%' % (100*len(df[df['capital-gain'] == 0])/len(df)))
print('Proportion of zeros [capital-loss]: %.1f%%' % (100*len(df[df['capital-loss'] == 0])/len(df)))
# As is clear from the above histograms (note the logarithmic scaling on the vertical axis) and computation, the `capital-gain` and `capital-loss` variables are both quite skewed, with a high proportion of zero values. Thus, we will exclude them from the dataset.
df_plot = df.groupby(['native-country', 'income']).size().reset_index().pivot(columns='income', \
index='native-country', values=0).fillna(0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(10,8))
plt.title('Income by Native Country')
plt.ylabel('Count')
plt.yscale('log')
# Similarly, the `native-country` variable displays high skewness as most observations are from the United States (again note the logarithmic scaling of the vertical axis on the above plot). Hence, we will exclude this variable from our model as well.
df = df.drop(['capital-gain', 'capital-loss', 'native-country'], axis=1)
# +
df_plot = df.groupby(['race', 'income']).size().reset_index().pivot(columns='income', \
index='race', values=0).fillna(0)
ax = df_plot.plot(kind='bar', stacked=True, figsize=(10,6))
plt.title('Income by Race')
plt.ylabel('Count')
annotations = df_plot.divide(df_plot.sum(axis=1), axis='index')
annotations = np.array(100*annotations).round().astype(int)
annotations = annotations.flatten(order='F')
for i, p in enumerate(ax.patches):
if annotations[i] == 0:
continue
x = p.get_x()+.15
y = max(p.get_y()+p.get_height()//2-80, 120)
if i >= len(annotations)//2:
y = max(y, 1200)
ax.annotate(str(annotations[i]) + '%', (x, y))
# -
# From the above plot, we see that the majority of respondents are white, and that white and asian-pacific islanders have the largest proportions of individuals earning more than $\$$50,000 per year.
# +
# Age hist by income
plt.figure(figsize=(10,6))
bins = np.linspace(min(df.age) - 1, max(df.age), max(df.age) - min(df.age) + 2)
plt.hist(df.age[df.income == '<=50K'], bins, alpha=0.5, label='<=50K')
plt.hist(df.age[df.income == '>50K'], bins, alpha=0.5, label='>50K')
plt.legend(loc='upper right', title='Income')
plt.xlabel('Age')
plt.ylabel('Count')
plt.title('Histograms of Age by Income')
# Age hist by gender
plt.figure(figsize=(10,6))
bins = np.linspace(min(df.age) - 1, max(df.age), max(df.age) - min(df.age) + 2)
plt.hist(df.age[df.sex == 'Male'], bins, alpha=0.5, label='Male')
plt.hist(df.age[df.sex == 'Female'], bins, alpha=0.5, label='Female')
plt.legend(loc='upper right', title='Gender')
plt.xlabel('Age')
plt.ylabel('Count')
plt.title('Histograms of Age by Gender')
# -
# Inspecting the distribution of the age variable, we see that there are significantly more observations for those who make less than $\$$50,000 annually than those who make more. Moreover, those who make more than $50,000 annually tend to be in their mid-career.
#
# Interestingly, females are underrepresented in the dataset, which could be caused by a census bias.
# +
plt.figure(figsize=(10,6))
sns.boxplot(data=df, x='hours-per-week', y='income')
plt.xlabel('Hours per week')
plt.ylabel('Income')
plt.title('Distributions of Hours per Week by Income')
# -
# Unsuprisingly, we see that those who make more than $\$$50,000 annually tend to work more hours per week than those who make less. Also, it is notable that the distribution of hours worked has a larger spread for those who make more than $\$$50,000 per year than those who don't.
# In order to perform logistic regression on the dataset, we need to numerically encode the categorical variables. For binary variables, such as our `sex` and `income` variables, we can simply map these to 0 and 1. If a logical order is present in the features, mapping the features to a range of consecutive integers is a common technique. This quantifies the data while maintaining the ordinal nature of the variable. This is what was previously done to create the `education-num` variable from the `education` variable.
#
# When a logical order is not present in the features, as is the case in our remaining categorical variables, a one-hot encoding is a common technique to quantify the categorical data. Here, each category is mapped to a vector containing a 1 or 0 to denote the presence or absence of a feature. For instance, our `marital-status` variable could represent 'Single' as $[0,1,0,0,0]$, where the vector entries correspond to 'Married', 'Single', 'Divorced', 'Separated', and 'Widowed' respectively. This method can cause issues for variables with large cardinality as it drastically increases the sparsity of the dataset, however it is a good way to quantify our data. This method can be easily implemented with the `pandas` method `get_dummies`.
df.sex = df.sex.map({'Male':0, 'Female':1})
df.income = df.income.map({'<=50K':0, '>50K':1})
df = pd.get_dummies(df, columns=['workclass', 'marital-status', 'occupation', 'race'])
df.head()
# We will preform the regression with the machine learning library PyTorch.
import torch as t
from torch import nn
from torch.optim import SGD
from torch.utils.data import Dataset, DataLoader, random_split
# The data is split into training and validation sets and the independent variables in both sets are standardized using the mean and standard deviation of the training dataset. This is best done in PyTorch by creating a class representing the data, and creating DataLoader objects to iterate over the training and validation datasets.
class Data(Dataset):
def __init__(self, df):
self.data = df.drop('income', axis=1).copy()
self.income = t.Tensor(df.income.copy()).type(t.float)
self.data = t.Tensor(self.data.to_numpy()).type(t.float)
def __len__(self):
return self.data.shape[0]
def __getitem__(self, i):
return (self.data[i], self.income[i])
# +
dataset = Data(df)
# Split the data into training and validation subsets
train_fraction = 0.75
train_len = int(train_fraction*len(dataset))
train, valid = random_split(dataset, (train_len, len(dataset) - train_len))
# Standardize the data based on the training data mean and std
train_mean = t.mean(train.dataset.data[train.indices], 0)
train_std = t.std(train.dataset.data[train.indices], 0)
train.dataset.data[train.indices] = (train.dataset.data[train.indices] - train_mean)/train_std
valid.dataset.data[valid.indices] = (valid.dataset.data[valid.indices] - train_mean)/train_std
train_loader = DataLoader(train, batch_size=len(dataset), shuffle=False)
valid_loader = DataLoader(valid, batch_size=len(dataset), shuffle=False)
# -
# We then define a simple class representing our model.
# +
class LinearRegression(nn.Module):
def __init__(self, input_size, output_size):
super(LinearRegression, self).__init__()
self.linear = nn.Linear(input_size, output_size)
def forward(self, x):
return self.linear(x)
model = LinearRegression(df.shape[1] - 1, 1)
# -
# The goal of our model is to find parameters $m_1,...,m_n,b$ such that the target variable `income`, denoted $y_{income}$, is given by $$y_{income} = \sigma(m_1x_1 + ... + m_nx_n + b)$$ where the $x_i$ are our training variables (age, workclass, etc.). Here, $\sigma$ is known as the *sigmoid function* and is given by $$\sigma:\mathbb{R} \rightarrow (0,1), x \mapsto \frac{1}{1+e^{-x}}$$
#
# For our loss function, we use the Binary Cross Entropy (BCE) loss. Of course, our model defined above does not include the sigmoid operation, it simply returns output of the linear operation. This is because, in PyTorch, it is more numerically stable to use the predefined loss function `torch.nn.BCEWithLogitsLoss`. This function applies the sigmoid operation and the loss calculation in one layer, but takes advantage of the [log-sum-exp trick](https://en.wikipedia.org/wiki/LogSumExp) for numerical stability. Thus, this is favorable over consecutively applying the sigmoid function and then the BCE calculation to increae numerical stability.
#
#
#
# +
lr = 0.03
n_epochs = 440
# This loss function combines a sigmoid function with a binary cross entropy loss function.
# It is preferred to increase numerical stability.
loss_func = nn.BCEWithLogitsLoss()
optimizer = SGD(model.parameters(), lr=lr)
# -
# We set the batch size equal to the length of the full dataset so that the entire training set is used on every parameter update. For a simple model such as this logistic regression model, it is not necessary to split the data into batches.
l = []
model.train()
for i, epoch in enumerate(range(n_epochs)):
for x, label in train_loader:
optimizer.zero_grad()
prediction = model(x)
loss = loss_func(prediction, label.unsqueeze(1))
l.append(loss.detach())
loss.backward()
optimizer.step()
if i % 20 == 0:
print('Epoch %i' % i, ' Loss %.3f' % loss)
plt.figure(figsize=(10,6))
plt.plot(range(len(l)), l)
plt.ylabel('Loss', fontsize=12)
plt.xlabel('Epoch', fontsize=12)
plt.title('Mean BCE Loss vs Training Epoch', fontsize=15)
# Now, we can test our model to see how it performs on the validation dataset. The output of the logistic regression algorithm is a probability that an individual makes more than $\$$50,000 annually. Thus, we will use a threshold of 0.5 to determine whether an individual is predicted to make more than $50,000 or not.
model.eval()
valid_data, valid_income = [*valid_loader][0]
thres = 0.5
pred_income_probabilities = nn.Sigmoid()(model(valid_data)).squeeze().detach()
pred_income = t.where(pred_income_probabilities > thres, 1, 0)
valid_income = valid_income.detach().numpy().astype(int)
pred_income = pred_income.detach().numpy().astype(int)
acc_valid = (valid_income == pred_income).sum()/len(valid_income)
print('Accuracy on validation set: %.2f%%' % (100*acc_valid))
# We achieve a fairly good accuracy for the model. To check for overfitting, we can compare the above accuracy with the accuracy on the training data.
# +
# Accuracy for train data
train_data, train_income = [*train_loader][0]
pred_train = t.where(nn.Sigmoid()(model(train_data)).squeeze() > thres, 1, 0)
pred_train = pred_train.detach().numpy().astype(int)
train_income = train_income.detach().numpy().astype(int)
acc_train = (pred_train == train_income).sum()/len(train_income)
print('Accuracy on training set: %.2f%%' % (100*acc_train))
# -
# The similar accuracy on both datasets indicates the model is not overfitting the training dataset.
#
# A confusion matrix will give us more insight into how well our model is performing.
# +
cf_matrix = np.zeros((2,2))
for real, pred in zip(valid_income, pred_income):
cf_matrix[real][pred] += 1
print('Confusion matrix:')
pd.DataFrame(cf_matrix, index=['actual_<=50k', 'actual_>50k'], \
columns=['predicted_<=50k', 'predicted_>50k']).astype(int)
# -
print('Normalized confusion matrix:')
pd.DataFrame(cf_matrix/cf_matrix.sum(), index=['actual_<=50k', 'actual_>50k'], \
columns=['predicted_<=50k', 'predicted_>50k'])
# From the confusion matrix, we can calculate a few other metrics for evaluating out model. First, we compute the misclassification rate, which is simply the fraction of predictions that were wrong.
misclass_rate = (cf_matrix[0][1]+cf_matrix[1][0])/cf_matrix.sum()
print('Misclassification rate: %.2f%%' % (100*misclass_rate))
# We can also compute the recall, precision, and $F_1$ score. Recall measures the proportion of actual positives that were identified correctly. That is, recall is defined as
#
# $$\text{Recall} = \frac{\text{True Positives}}{\text{True Positives}+\text{False Negatives}}$$
#
# Precision, on the other hand, is the proportion of positive identifications that were correct. Hence, precision is given by
#
# $$\text{Precision} = \frac{\text{True Positives}}{\text{True Positives}+\text{False Positives}}$$
#
# It is important to consider both these metrics when evaluating a classification model as it is possible to have high recall but low precision or vice versa.
#
# $F_1$ score is a single metric which combines both the precision and recall. It is given by the harmonic mean of precision and recall:
#
# $$F_1=2\cdot\frac{\text{Recall}\cdot\text{Precision}}{\text{Recall}+\text{Precision}}$$
#
# $F_1$ score is a useful metric since it is low if either precision or recall are low and allows us do sufficiently describe the effectiveness of our model in a single quantity.
# +
recall = cf_matrix[1][1]/cf_matrix[1].sum()
print('Recall: ', recall)
precision = cf_matrix[1][1]/cf_matrix.sum(axis=0)[1]
print('Precision: ', precision)
f1 = 2*precision*recall/(precision + recall)
print('F1 score: ', f1)
# -
# Our model has a greater precision than recall, and a decent $F_1$ score.
# The ROC curve is a plot of the false positive rate on the horizontal axis versus the true positive rate on the vertical axis for thresholds varying from 0 to 1. It is particularly useful for directly comparing several models, however, for a single model, the area under the ROC curve (AUC) can be used as a summary of the model's effectiveness.
from sklearn.metrics import roc_curve, roc_auc_score
fpr, tpr, thresholds = roc_curve(valid_income, pred_income_probabilities)
area = roc_auc_score(valid_income, pred_income_probabilities)
plt.figure(figsize=(10,8))
plt.plot(fpr, tpr)
plt.xlim(0,1)
plt.ylim(0,1)
plt.legend(['ROC curve (area = %.3f)' % area], loc='lower right', fontsize=14)
plt.xlabel('False Positive Rate', fontsize=14)
plt.ylabel('True Positive Rate', fontsize=14)
plt.title('ROC Curve', fontsize=18)
# Since the data is standardized before training, we can use the coefficients of our logistic regression model to determine the relative importance of each feature in predicting the class of a data sample, as well as if they contribute positively or negatively to the classification.
param = [*model.parameters()][0]
for feature, coeff in sorted(zip(df.drop(['income'], axis=1).columns, param[0]), key=lambda x: -abs(x[1])):
print(feature, ':', (26 - len(feature) + int(np.floor(coeff.item())))*' ' + '%.4f' % coeff.item())
# We see that `education-num` contributes among the most to the classification of a data sample, and does so in a positive way, whereas the `marital-status_Single` variable contributes strongly in a negative way.
import pickle
with open('income_and_predictions.pickle', 'wb') as f:
pickle.dump([valid_income, pred_income_probabilities, acc_valid, f1], f)
| Logistic Regression/IncomeViaLogisticRegression.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# +
import matplotlib.pyplot as plt
# %matplotlib inline
from sklearn import datasets
from sklearn.decomposition import PCA
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
iris = datasets.load_iris()
X = iris.data
y = iris.target
target_names = iris.target_names
X.shape
X
# +
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 0], X[y == i, 1], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 0], X[y == i, 2], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 0], X[y == i, 3], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 1], X[y == i, 2], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 1], X[y == i, 3], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X[y == i, 2], X[y == i, 3], c=c, label=target_name)
plt.legend()
plt.title('No-Transform of IRIS dataset')
# +
pca = PCA(n_components=2)
X_r = pca.fit(X).transform(X)
lda = LinearDiscriminantAnalysis(n_components=2)
X_r2 = lda.fit(X, y).transform(X)
# Percentage of variance explained for each components
print('explained variance ratio (first two components): %s'
% str(pca.explained_variance_ratio_))
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X_r[y == i, 0], X_r[y == i, 1], c=c, label=target_name)
plt.legend()
plt.title('PCA of IRIS dataset')
plt.figure()
for c, i, target_name in zip("rgb", [0, 1, 2], target_names):
plt.scatter(X_r2[y == i, 0], X_r2[y == i, 1], c=c, label=target_name)
plt.legend()
plt.title('LDA of IRIS dataset')
plt.show()
# -
| notebooks/pca_lda.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# %load_ext autoreload
# %autoreload 2
from nbdev import *
# %nbdev_default_export utils
# %nbdev_default_class_level 3
# %nbdev_export
import numpy as np
from scipy.sparse import coo_matrix
import torch
from scipy.io import loadmat
# # Utilities
# > all utilities routines
# ## Dimentions and Units
# ### Metric Prefixes
# %nbdev_export
pico = 1.e-12
nano = 1.e-9
micro = 1.e-6
milli = 1.e-3
centi = 1.e-2
deci = 1.e-1
kilo = 1.e+3
mega = 1.e+6
giga = 1.e+9
tera = 1.e+12
# ### Units
# %nbdev_export
units = {"distance":["meter", "ft", "inch"],
"mass":["kg", "gram", "pound", "Ton"],
"time":["second", "minute", "hour", "day", "year"],
"temperature":["Kelvin", "Rankine", "Celsius", "Fahrenheit"],
"volume":["litre", "gallon", "stb"],
"force":["Newton", "dyne", "lbf"],
"press":["kg", "gram", "pound", "Ton"],
"energy":["Joules", "btu", "Cal"],
"dinamicViscosity":["centiPoise", "Poise"],
"kinematicViscosity":["centiStoke", "Stoke"],
"absolutePermeability":["milliDarcy","Darcy"],
"hydraulicConductivity":["inch_second","mm_second "],
"dimensionless":["dimless"]}
# ### Distance
# %nbdev_export
meter = 1. #meters -> meters
ft = 0.3048*meter #foots -> meters
inch = 2.54*centi*meter #inches -> meters
# ### Mass
# %nbdev_export
kg = 1. #kilograms -> kilograms
gram = 1.*kg #grams -> kilograms
pound = 0.45359237*kg #pounds -> kilograms
Ton = 1000.*kg #Tons -> kilograms
# ### Time
# %nbdev_export
second = 1. #seconds -> seconds
minute = 60.*second #minutes -> seconds
hour = 60.*minute #hours -> seconds
day = 24.*hour #days -> seconds
year = 365.25*day #years -> seconds
# ### Temperature
# %nbdev_export
Kelvin = 1 #kelvin -> kelvin
Rankine = 5./9. #rankine -> kelvin
Celsius = lambda C: C+273.15 #celsius -> kelvin
Fahrenheit = lambda F: (F+459.67)*Rankine #fahrenheit -> kelvin
# ### Volume
# %nbdev_export
litre = (deci*meter)**3 #litre -> cubic meter
gallon = 231*inch**3 #US gallons -> cubic meter
stb = 42*gallon #standard barrels -> cubic meter
# ### Force
# %nbdev_export
Newton = 1. #Newtons -> Newtons
dyne = 1.e-5*Newton #dynes -> Newtons
lbf = 9.80665*pound #pound-force -> Newtons
# ### Press
# %nbdev_export
Pascal = 1. #Pascals -> Pascals
atm = 101325.*Pascal #Pascals -> Atmosphères
bar = 1e5*Pascal #Pascals -> Bars
psi = lbf/inch**2 #Pascals -> Psi
# ### Energy
# %nbdev_export
Joules = 1. #Joules -> Joules
btu = 1054.06*Joules #British thermal unit -> Joules
Cal = 4184.*Joules #Calories -> Joules
# ### Dinamic Viscosity
# %nbdev_export
centiPoise = milli*Pascal*second #centiPoise -> Pascal*second
Poise = 100*centiPoise #Poise -> Pascal*second
# ### Kinematic Viscosity
# %nbdev_export
centiStokes = 1.E-6*(meter**2)/second #centiStokes -> square-meter per second
Stokes = 1.E-4*(meter**2)/second #centipoise -> square-meter per second
# ### Absolute Permeability
# %nbdev_export
milliDarcy = 9.869233e-16*(meter**2) #milliDarcy -> square-meter
Darcy = 9.869233e-13*(meter**2) #Darcy -> square-meter
# ### Hydraulic Conductivity
# %nbdev_export
inch_second = inch*second #inches per second -> meter per second
mm_second = milli*meter*second #millimeters per second -> meter per second
# ### Dimensionless
# %nbdev_export
dimless = 1.
# ## Functions
# ### Lanczos Factorization
# %nbdev_export
def lanczos(A,xo,m,reorthog=0):
p, n = A.shape
assert p == n
assert m <= n
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
xo = xo.type(torch.float64).to(device)
A = A.type(torch.float64).to(device)
Q = torch.zeros(n,m+1, dtype=torch.float64, device=device)
T = torch.zeros(m+1,m, dtype=torch.float64, device=device)
alpha = torch.zeros(m,1, dtype=torch.float64, device=device)
beta = torch.zeros(m,1, dtype=torch.float64, device=device)
q = xo/torch.norm(xo)
Q[:,0] = q.reshape((-1))
for k in range(m):
w = torch.matmul(A,q)
alpha[k] = torch.matmul(q.T,w)
if k == 0:
w = w - alpha[0]*q
else:
w = w - alpha[k]*q - beta[k-1]*Q[:,k-1].reshape((-1,1))
if reorthog == 1:
for i in range(1,k):
h = torch.matmul(Q[:,i].reshape((-1,1)).type(torch.float64).T,w)
w = w - Q[:,i].reshape((-1,1))*h
beta[k] = torch.norm(w)
if beta[k]<1e-20:
return Q.cpu(), T.cpu()
q = w/beta[k]
Q[:,k+1] = q.reshape((-1))
#print(q)
T[:m,:] = torch.diag(beta[0:m-1].T[0],-1) + torch.diag(alpha.T[0]) + torch.diag(beta[0:m-1].T[0],1)
T[m,m-1] = beta[-1]
return Q.cpu(), T.cpu()
# ### Arnoldi Factorization
# %nbdev_export
def arnoldi(A,xo,m,reorthog=0):
p, n = A.shape
assert p == n
assert m <= n
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
xo = xo.type(torch.float64).to(device)
A = A.type(torch.float64).to(device)
Q = torch.zeros(n,m+1, dtype=torch.float64, device=device)
H = torch.zeros(m+1,m, dtype=torch.float64, device=device)
tol = n*2e-16;
q = xo/torch.norm(xo)
Q[:,0] = q.reshape((-1))
for k in range(m):
w = A@Q[:,k]
ow = torch.norm(w)
for j in range(k+1):
H[j,k] = Q[:,j].T@w
w = w - H[j,k]*Q[:,j]
if reorthog == 1:
pass
H[k+1,k] = torch.norm(w)
if H[k+1,k] <= tol*ow:
m = k
H = H[:m+1,:m]
Q = Q[:n,:m+1]
return Q.cpu(), H.cpu(), m
Q[:,k+1] = w/H[k+1,k]
return Q.cpu(), H.cpu(), m
# %nbdev_hide
notebook2script()
| 10_utils.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# # ***Introduction to Radar Using Python and MATLAB***
# ## <NAME> - Copyright (C) 2019 Artech House
# <br/>
#
# # Circular Array Antenna
# ***
# The circular array is shown in Figure 3.22. For this type of array, the radiating elements are placed on a circular path with spacing of $a \, \Delta\phi$, where $a$ is the radius, and $\Delta\phi$ is the angular difference between the elements. The normalized electric field is written as
#
# $$
# E(r, \theta, \phi) = \sum\limits_{n=1}^{N}a_n\, \frac{e^{-j k r_n}}{r_n} \hspace{0.5in} \text{(V/m)},
# $$
#
# where
#
# $$
# r_n = \sqrt{r^2 + a^2 - 2\, a\, r\, \cos \psi_n} \hspace{0.5in} \text{(m)}.
# $$
#
# For phase terms, $r_n$ is approximated by
#
# $$
# r_n \approx r - a\, \sin\theta\, \cos(\phi - \phi_n) \hspace{0.5in} \text{(m)},
# $$
#
# where $\phi_n$ is the angular position of the $n$th element, and is expressed as
#
# \begin{equation}
# \phi_n = \frac{2\pi\, n}{N} \hspace{0.5in} \text{(rad)},
# \end{equation}
#
# and for amplitude $r_n \approx r$. The coefficients, $a_n$, are represented by an amplitude and phase as
#
# $$
# a_n = A_n\, e^{j \alpha_n},
# $$
#
# where $A_n$ is the amplitude and $\alpha_n$ is the phase. The array factor for the circular array is now written as
#
# $$
# {AF}(\theta, \phi) = \sum\limits_{n=1}^{N}A_n\, e^{j [k a \sin\theta\, \cos(\phi-\phi_n) + \alpha_n]} .
# $$
# ***
# Begin by getting the library path
import lib_path
# Set the operating frequency (Hz), the number of elements, the scan angle (theta - degrees), (phi - degrees), and the radius (m)
# +
frequency = 300e6
number_of_elements = 40.0
scan_angle_theta = 30.0
scan_angle_phi = 30.0
radius = 1.1
# -
# Set up the theta and phi arrays using the `meshgrid` and `linspace` routines from `scipy`
# +
from numpy import linspace, meshgrid, radians
from scipy.constants import pi
n = 360
m = int(n / 8)
theta, phi = meshgrid(linspace(0.0 * pi, 0.5 * pi, n), linspace(0.0, 2.0 * pi, n))
# -
# Set up the keyword args
kwargs = {'number_of_elements': number_of_elements,
'scan_angle_theta': radians(scan_angle_theta),
'scan_angle_phi': radians(scan_angle_phi),
'radius': radius,
'frequency': frequency,
'theta': theta,
'phi': phi}
# Calculate the array factor using the `circular_uniform` routines
# +
from Libs.antenna.array import circular_uniform
af = circular_uniform.array_factor(**kwargs)
# -
# Create the U-V coordinates for displaying the antenna pattern
# +
from numpy import sin, cos
uu = sin(theta) * cos(phi)
vv = sin(theta) * sin(phi)
# -
# Create the color plot of the array factor using the `matplotlib` routines
# +
from matplotlib import pyplot as plt
# Set the figure size
plt.rcParams["figure.figsize"] = (15, 10)
# Set up the axes
fig, axes1 = plt.subplots()
im = axes1.pcolor(uu, vv, abs(af), cmap="jet", shading = 'auto')
cbar = fig.colorbar(im, ax=axes1, orientation='vertical')
cbar.set_label("Normalized Electric Field (V/m)", size=10)
# Set the x- and y-axis labels
axes1.set_xlabel("U (sines)", size=12)
axes1.set_ylabel("V (sines)", size=12)
# Set the plot title
plt.title('Circular Array - Array Factor', size=14)
# -
# Create the contour plot using the `matplotlib` routines
# +
# Set the figure size
plt.rcParams["figure.figsize"] = (12, 12)
plt.contour(uu, vv, abs(af), 20, cmap="jet", vmin=-0.2, vmax=1.0)
plt.grid(linestyle=':', linewidth=0.5)
# Set the x- and y-axis labels
plt.xlabel("U (sines)", size=12)
plt.ylabel("V (sines)", size=12)
# Set the plot title
plt.title('Circular Array - Array Factor', size=14)
# -
# Create the line plot using the `matplotlib` routines
# +
from numpy import degrees, log10
plt.plot(degrees(theta[0]), 20.0 * log10(abs(af[m])), '', label='E Plane')
plt.plot(degrees(theta[0]), 20.0 * log10(abs(af[0])), '--', label='H Plane')
# Set the y axis limit
plt.ylim(-60, 5)
# Set the x and y axis labels
plt.xlabel("Theta (degrees)", size=12)
plt.ylabel("Array Factor (dB)", size=12)
# Turn on the grid
plt.grid(linestyle=':', linewidth=0.5)
# Place the legend
plt.legend(loc='upper right', prop={'size': 10})
# Set the plot title
plt.title('Circular Array - Array Factor', size=14)
| jupyter/Chapter03/circular_array.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# ## Quelques exercices d'initiation en python
#
# 
#
#
# Il s'agit de progressivement s'initier à la syntaxe et aux différentes structures
#
# <u>**Ex1 :**</u> Il s'agit de générer un programme codé en python qui permette de demander votre prénom, votre année de naissance, puis qui va vous saluer en en vous donnant votre âge en 2025 comme suit :
#
# **Bonjour Mael, en 2025 vous aurez 31 ans !**
#
#
# - Vous devez utiliser les instructions **input()** et **print()**
# - Attention : la fonction input retourne toujours un caractère et non pas un nombre; de ce fait il faudra convertir l'année entrée sous forme de chaîne de caractères en entier (**int**)
# - Les variables à utiliser sont les suivantes: **prenom, annee, age, message**
prenom=input("Quel est votre prénom ? ")
annee=int(input("Quelle est votre année de naissance ? "))
age=2025-annee
print("Bonjour ",prenom,",","en 2025, vous aurez ",age, " ans")
# variante avec f-strings
print(f'Au revoir {prenom} ')
# <u>**Ex2 :**</u> Il s'agit de générer un programme codé en python qui vous demande les coordonnées de deux points A(x1,y1) et B(x2,y2), qui calcule la distance AB
#
# $$AB=\sqrt{{(x2 - x1)}^2 + {(y2 - y1)}^2}$$
#
# - Vous devez importer la fonction <i>"racine carrée"</i> de la bibliothèque(module) **math**, elle se nomme **sqrt()**
# - Les fonctions input et print seront utilisées
# - la fonction "élever au carré" doit s'écrire sous la forme suivante : 'x**2'
# - Les variables seront les suivantes : **X1, Y1, X2, Y2, AB**
# - Vous utiliserez les f-strings
# - Vous utiliserez le formatage des nombres en f-strings tel que nous ayons alignement à droite, le signe +, la largeur 10 chiffres, 2 chiffres significatifs de 2 chiffres après la virgule
#
#
from math import sqrt
X1=int(input("Entrez l'abscisse de A : "))
Y1=int(input("Entrez l'ordonnée de A : "))
X2=int(input("Entrez l'abscisse de B : "))
Y2=int(input("Entrez l'ordonnée de B : "))
AB=sqrt(((X2-X1)**2)+((Y2-Y1)**2))
print(f'La distance AB est {AB}')
print(f'La distance AB est {AB : >+.2f}')
# <u>**Ex3 :**</u> Il s'agit de générer un tracé de courbe en utilisant le module matplotlib et numpy
#
# La courbe à tracer est la suivante :
#
# $$y = e^{x}$$
#
# En abscisse, x<sub>min</sub> = 0; x<sub>max</sub> = 5
#
#
# - Vous devez importer le module <i>"pyplot"</i> de la bibliothèque **mathplotlib** et nous renommerons le tout **matplot**
# - Nous devrons également importer la bibliothèque de fonctions numpy
# - les fonctions **xlabel( ), ylabel( ), linspace( ), exp( )** seront nécessaires
# - le tracé se fera en **pointillés rouges**
#
# 
#
#
import matplotlib.pyplot as matplot
import numpy
x=numpy.linspace(0,5,20)
matplot.xlabel("l'axe des abscisse (x) ")
matplot.ylabel("l'axe des ordonnées y=f(x) ")
matplot.plot(x,numpy.exp(x), 'r--')
matplot.grid(True)
# <u>**Ex4 :**</u> Il s'agit de générer des compositions de galettes blé noir à partir d'une liste d'ingrédients situés dans un tableau. Nous travaillons sur les listes. Il faut voir une liste (tableau) comme une commode comportant un certain nombre de tiroirs contenant un élément. Nous allons étudier comment "travailler" sur ces contenus de "tiroirs" en les énumérant, en les remplaçant, en les permutant...
# 
#
# La liste des ingrédients possibles de départ dans une galette de blé noir sont les suivants :
#
# >oeuf, jambon, gruyere, champignons, tomates, andouille; cela correspond à la liste nommée **galette**
#
# Les actions à mener sont les suivantes :
#
# * **créer une liste** nommée **galette** à partir de l'énumération suivante :
#
# *'oeuf','jambon','gruyere','champignons','tomates','andouille' en utilisant la méthode **split()**
# associée au caractère ','*
#
#
# * afficher **la liste complète** des ingrédients possibles
# * afficher **le nombre total** des ingrédients possibles
# * afficher **le troisième** ingrédient
# * afficher **tous les ingrédients à partir du second**
# * afficher **tous les ingredients compris entre le second et le quatrième inclus**
# * afficher **tous les ingredients en partant du dernier**
# * **ajouter "roquefort" à la suite de gruyère** et afficher le résultat
# * **enlever "andouille"** de la liste, **le remplacer par "saumon"** et afficher le résultat
# * **trier la liste** dans l'**ordre alphabétique** et afficher le résultat
# * **rajouter** la liste suivante \['oignons','oseille'] à la liste précédente
# * afficher **le numéro d'ordre** de 'champignons'
#
#
#
# +
galette="oeuf,jambon,gruyere,champignons,tomates,andouille".split(',')
print(galette)
print(len(galette))
print(galette[2])
print(galette[1:])
print(galette[1:5])
print(galette[::-1])
galette.insert(3,'roquefort')
print(galette)
galette.remove('andouille')
print(galette)
galette.append('saumon')
print(galette)
galette=galette+['oignons','oseille']
print(galette)
print(galette.index('champignons'))
# -
# <u>**Ex5 :**</u> Il s'agit désormais de s'intéresser aux structures conditionnelles (**SI.. ALORS .. SINON) et itératives (TANT QUE ..)**
#
# 
#
# Vous devez créer un petit programme python permettant de trouver le "juste prix " d'un objet en minimisant le nombre d'essais. Ce programme devra avoir les caractéristiques suivantes :
#
# * **générer** un nombre aléatoire compris entre 1000 et 4000 (euros!!), vous utiliserez le module **random** et la fonction **randrange** pour générer ce nombre (variable **juste_prix**)
# * **afficher** " Il s'agit de donner un prix comprix entre 1000 et 4000 euros en minimisant le nombre de coups "
# * **demander** au joueur le prix estimé (variable **prix**)
# * Si le prix estimé est supérieur afficher **"c'est moins"** sinon afficher **"c'est plus**
# * **compter** le nombre d'essais (variable **nb_essais**)
# * afficher **le juste prix est (juste_prix). Vous avez réussi en (nb_essais) essais** en cas de réussite en utilisant des f-strings
#
#
from random import*
juste_prix=randrange(1000,4000) # tirage aléatoire d'un nombre entier entre 1000 et 4000
prix=0 # on initialise la reponse du joueur
nb_essais=0 # on comptabilise le nombre de réponses ou de coups
print(" Il s'agit de donner un prix comprix entre 1000 et 4000 euros en minimisant le nombre de coups ")
while juste_prix!=prix:
prix=int(input('Donner votre prix : '))
nb_essais +=1 # on incrémente le nombre de réponses
if prix > juste_prix:
print('Cest moins!')
elif prix < juste_prix:
print('Cest plus!')
else :
print('Cest gagné!')
print(f'Le juste prix est : {juste_prix} €. Vous avez réussi en {nb_essais} essais')
# <u>**Ex6 :**</u> Il s'agit désormais de s'intéresser à la structure itérative (**POUR x variant de borne 1 à borne 2 FAIRE action1** )
#
# Vous devez créer un petit programme python permettant de dresser la table des carrée et et des cubes des nombres de 10 à 30
#
# * afficher **Voici la liste des carrés et des cubes**.
# * **créer** une boucle permettant de calculer les carrés de 20 à 30 en affichant "Le carré de (nombre) est (carre) et son cube est (cube))"
# * afficher **C'est terminé !**
print('Voici la liste des carrés et des cubes')
for nombre in range(20,31):
carre=nombre**2
cube=nombre**3
print(f'Le carré de {nombre} est {carre} et son cube est {cube}')
print("C'est terminé !")
| exercices_init_cor.ipynb |
# ---
# jupyter:
# jupytext:
# text_representation:
# extension: .py
# format_name: light
# format_version: '1.5'
# jupytext_version: 1.14.4
# kernelspec:
# display_name: Python 3
# language: python
# name: python3
# ---
# + [markdown] heading_collapsed=true run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "slide"}
# # What is statistics?
# + [markdown] hidden=true run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# Probability theory computes probabilities of complex events given the underlying base probabilities.
# + [markdown] hidden=true run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# Statistics takes us in the opposite direction.
# + [markdown] hidden=true run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# We are given **data** that was generated by a **Stochastic process**
# + [markdown] hidden=true run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# We **infer** properties of the underlying base probabilities.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "slide"}
# # Example: deciding whether a coin is biased.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# In a previous video we discussed the distribution of the number of heads when flipping a fair coin many times.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# Let's turn the question around: we flip a coin 1000 times and get 570 heads.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# Can we conclude that the coin is biased (not fair) ?
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# What can we conclude if we got 507 heads?
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "slide"}
# ### The Logic of Statistical inference
# The answer uses the following logic.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Suppose that the coin is fair.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Use **probability theory** to compute the probability of getting at least 570 (or 507) heads.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * If this probability is very small, then we can **reject** <font color='red'>with confidence</font> the hypothesis that the coin is fair.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# ## Calculating the answer
# Recall the simulations we did in the video "What is probability".
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# We used $x_i=-1$ for tails and $x_i=+1$ for heads.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# We looked at the sum $S_k=\sum_{i=1}^k x_i$, here $k=1000$.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# If number of heads is $570$ then $S_{1000} = 570-430 = 140$
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# It is very unlikely that $|S_{1000}| > 4\sqrt{k} \approx 126.5$
# + run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
from math import sqrt
4*sqrt(1000)
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# It is very unlikely that the coin is unbiased.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# ### What about 507 heads?
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# 507 heads = 493 tails $ \Rightarrow S_n = 14$, $\;\;\;14 \ll 126.5$
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# We cannot conclude that coin is biased.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# ## Conclusion
# The probability that an unbiased coin would generate a sequence with 570 or more heads is extremely small. From which we can conclude, <font color='red'>with high confidence</font>, that the coin **is** biased.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# On the other hand, $\big| S_{1000} \big | \geq 507$ is quite likely. So getting 507 heads does not provide evidence that the coin is biased.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "slide"}
# # Real-World examples
# You might ask "why should I care whether a coin is biased?"
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * This is a valid critique.
# * We will give a few real-world cases in which we want to know whether a "coin" is biased or not.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# ## Case I: Polls
# * Suppose elections will take place in a few days and we want to know how people plan to vote.
# * Suppose there are just two parties: **D** and **R**.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * We could try and ask **all** potential voters.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * That would be very expensive.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Instead, we can use a poll: call up a small randomly selected set of people.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# * Call $n$ people at random and count the number of **D** votes.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Can you say <font color='red'>with confidence</font> that there are more **D** votes, or more **R** votes?
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Mathematically equivalent to flipping a biased coin and
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * asking whether you can say <font color='red'>with confidence</font> that it is biased towards "Heads" or towards "Tails"
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# ## Case 2: A/B testing
# A common practice when optimizing a web page is to perform A/B tests.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * A/B refer to two alternative designs for the page.
#
# 
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# * To see which design users prefer we randomly present design A or design B.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * We measure how long the user stayed on a page, or whether the user clicked on an advertisement.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "subslide"}
# * We want to decide, <font color='red'>with confidence</font>, which of the two designs is better.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Again: similar to making a decision <font color='red'>with confidence</font> on whether "Heads" is more probably than "Tails" or vice versa.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "slide"}
# # Summary
# Statistics is about analyzing real-world data and drawing conclusions.
#
# Examples include:
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Using polls to estimate public opinion.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * performing A/B tests to design web pages
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Estimating the rate of global warming.
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# * Deciding whether a medical procedure is effective
# + [markdown] run_control={"frozen": false, "read_only": false} slideshow={"slide_type": "fragment"}
# # The end!
| Week 1 _ Intro/Lectures/2.What-is-Statistics_.ipynb |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.