AIDO.StructureEncoder
AIDO.StructureEncoder is the encoder-only component of AIDO.StructureTokenizer for tokenization of protein structures.
Model Description
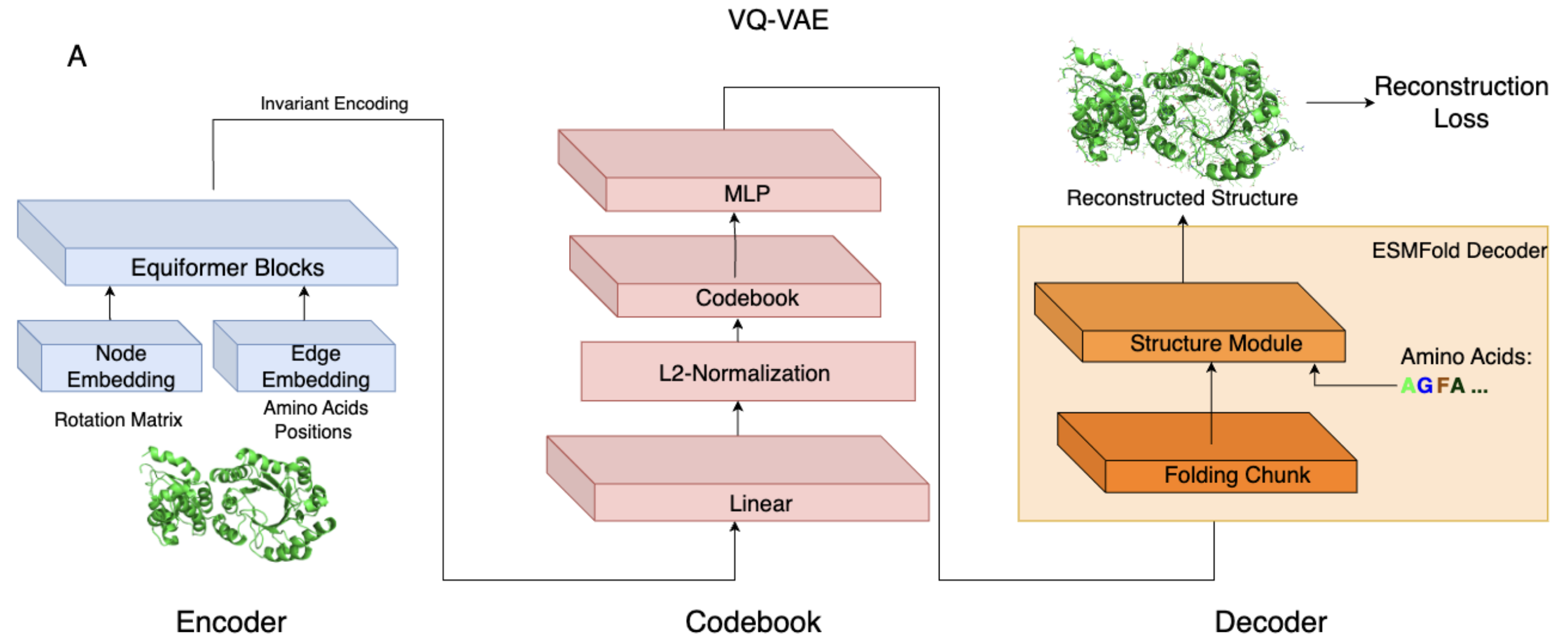
AIDO.StructureTokenizer is built on a Vector Quantized Variational Autoencoder (VQ-VAE) architecture with the following components:
- Equivariant Encoder (6M): Encodes backbone structures into a latent space that maintains rotational and translational symmetries using the Equiformer architecture.
- Discrete Codebook: Maps continuous latent vectors into 512 discrete structural tokens.
- Invariant Decoder (300M): Reconstructs full 3D structures, including side chains, from the structural tokens using an architecture adapted from ESMFold.
This model strikes a balance between reconstruction fidelity and structural locality, optimizing its suitability for downstream tasks such as structure prediction, homology detection, and multimodal protein modeling.
Key Features
- Encoding Structures into Tokens (See below)
- Decoding Tokens into Structures (See genbio-ai/AIDO.StructureDecoder)
- Reconstructing Structures (See genbio-ai/AIDO.StructureTokenizer)
- Structure Prediction (See this section in genbio-ai/AIDO.Protein2StructureToken-16B)
How to Use
Please see experiments/AIDO.StructureTokenizer in Model Generator for more details.
Setup
Install Model Generator
Data preparation
To reproduce the reconstruction results in the paper, we provide a preprocessed CASP15 dataset at genbio-ai/sample-structure-dataset. It could be downloaded via
huggingface-cli download genbio-ai/sample-structure-dataset --repo-type dataset --local-dir ./data/protstruct_sample_data/
This dataset is based on the CASP15 dataset, which can be referenced at:
The downloaded directory includes:
- A
registriesfolder containing a CSV file with metadata such as filenames and PDB IDs. - A
CASP15_mergedfolder containing PDB files, where domains are merged in the same way as described in Bhattacharya-Lab/CASP15.
To use customized data, you can prepare a dataset with the following structure:
- A folder containing PDB files (supported formats:
cif.gz,cif,ent.gz,pdb).
Then, you need to prepare a registry file in CSV format using the following command:
python experiments/AIDO.StructureTokenizer/register_dataset.py \
--folder_path /path/to/folder_path \
--format cif.gz \
--output_file /path/to/output_file.csv
You need to replace the folder_path and the registry_path in the following steps accordingly.
Running Encoding Task
If you use the sample dataset, you can run the encoding task using the following command:
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/encode.yaml
If you use your own dataset, you need to update the folder_path and the registry_path in the encode.yaml configuration file to point to your dataset folder and registry file. Alternatively, you can override these parameters when running the command:
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/encode.yaml \
--data.init_args.config.proteins_datasets_configs.name="your_dataset_name" \
--data.init_args.config.proteins_datasets_configs.registry_path="your_dataset_folder_path" \
--data.init_args.config.proteins_datasets_configs.folder_path="your_dataset_registry_path" \
--trainer.callbacks.dict_kwargs.output_dir="your_output_dir"
Input:
- The PDB files in the dataset folder.
- The registry file in CSV format indicating the metadata of the dataset.
Output:
- The encoded tokens will be saved in the output directory specified in the configuration file. By default it is saved in
logs/protstruct_encode/. - The encoded tokens are saved as a
.ptfile, which can be loaded using PyTorch. Inside the file, it's a dictionary that maps the name of the protein to the encoded tokens (struct_tokens) and other auxiliary information (aatype,residue_index) for reconstruction. The structure of the dictionary is as follows:{ 'T1137s5_nan': { # the nan here is the chain id and CASP15 doesn't have chain id 'struct_tokens': tensor([449, 313, 207, 129, ...]), 'aatype': tensor([ 4, 7, 5, 17, ...]), 'residue_index': tensor([ 33, 34, 35, 36, ...]), }, ... } - A codebook file (
codebook.pt) that contains the embedding of each token. The shape is(num_tokens, embedding_dim).
Notes:
- Currently, this function only supports single GPU inference due to the file saving mechanism. We plan to support multi-GPU inference in the future.
- The auxiliary information (
aatypeandresidue_index) can be substituted with placeholder values if not required.aatype: This parameter is used to reconstruct the protein sidechains. If sidechain reconstruction is not needed, you can assign dummy values (e.g., all zeros).residue_index: This parameter helps the model identify gaps in residue numbering, which can influence structural predictions. If gaps are present, the model may introduce holes in the structure. For structures without gaps, you can use a continuous sequence of integers (e.g., 0 to n-1).
- You may need to adjust the
max_nb_resparameter in the configuration file based on the maximum number of residues in your dataset. For those proteins with more residues thanmax_nb_res, the model will truncate the residues. The default value is set to 1024.
Citation
Please cite AIDO.StructureTokenizer using the following BibTex code:
@inproceedings{zhang_balancing_2024,
title = {Balancing Locality and Reconstruction in Protein Structure Tokenizer},
url = {https://www.biorxiv.org/content/10.1101/2024.12.02.626366v2},
doi = {10.1101/2024.12.02.626366},
publisher = {bioRxiv},
author = {Zhang, Jiayou and Meynard-Piganeau, Barthelemy and Gong, James and Cheng, Xingyi and Luo, Yingtao and Ly, Hugo and Song, Le and Xing, Eric},
year = {2024},
booktitle={NeurIPS 2024 Workshop on Machine Learning in Structural Biology (MLSB)},
}
- Downloads last month
- 56