Model description
This repo contains the model and the notebook for implementing Message Passing Neural Network (MPNN) to predict a molecular property known as blood-brain barrier permeability (BBBP). Message-passing neural network (MPNN) for molecular property prediction.
Full credits go to Alexander Kensert
Reproduced by Vu Minh Chien
Motivation: Until now, more traditional methods, such as random forests, support vector machines, etc., have been commonly used to predict molecular properties. In contrast to GNNs, these traditional approaches often operate on precomputed molecular features such as molecular weight, polarity, charge, number of carbon atoms, etc. Although these molecular features prove to be good predictors for various molecular properties, it is hypothesized that operating on these more "raw", and "low-level", features could prove even better. In this tutorial, MPNN was used to prove that GNN is a good method to predict molecular properties by using the undirected graph of molecules
Training and evaluation data
Information about the dataset can be found in A Bayesian Approach to Silico Blood-Brain Barrier Penetration Modeling and MoleculeNet: A Benchmark for Molecular Machine Learning. The dataset will be downloaded from MoleculeNet.org.
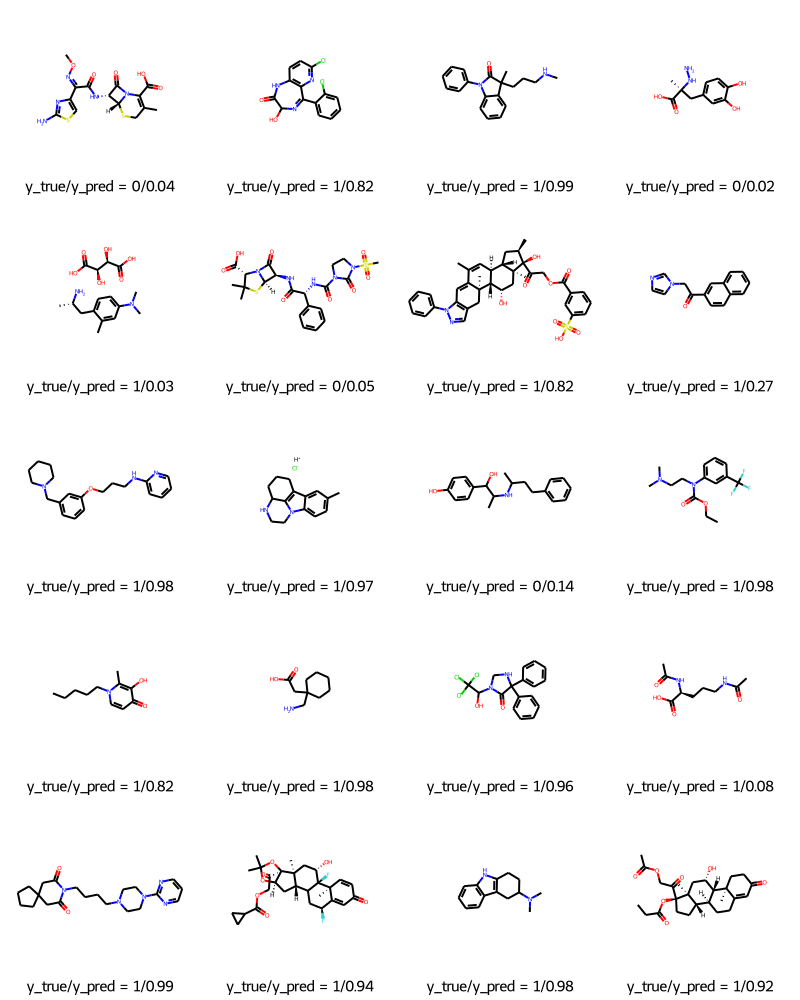
The dataset contains 2,050 molecules. Each molecule comes with a name, label, and SMILES string.
Training procedure
Training hyperparameters
The following hyperparameters were used during training:
| name | learning_rate | decay | beta_1 | beta_2 | epsilon | amsgrad | training_precision |
|---|---|---|---|---|---|---|---|
| Adam | 0.0005000000237487257 | 0.0 | 0.8999999761581421 | 0.9990000128746033 | 1e-07 | False | float32 |
Model Plot
View Model Demo
- Downloads last month
- 14