Spaces:
Running
license: apache-2.0
library_name: transformers
pipeline_tag: feature-extraction
tags:
- chemistry
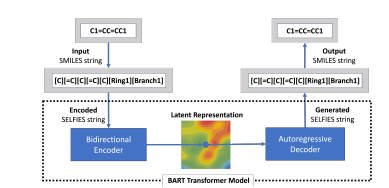
selfies-ted
selfies-ted is a project for encoding SMILES (Simplified Molecular Input Line Entry System) into SELFIES (SELF-referencing Embedded Strings) and generating embeddings for molecular representations.
Model Architecture
Configuration details
Encoder and Decoder FFN dimensions: 256 Number of attention heads: 4 Number of encoder and decoder layers: 2 Total number of hidden layers: 6 Maximum position embeddings: 128 Model dimension (d_model): 256
Pretrained Models and Training Logs
We provide checkpoints of the selfies-ted model pre-trained on a dataset of molecules curated from PubChem. The pre-trained model shows competitive performance on molecular representation tasks. For model weights: "HuggingFace link".
To install and use the pre-trained model:
Download the selfies_ted_model.pkl file from the "HuggingFace link". Add the selfies-ted selfies_ted_model.pkl to the models/ directory. The directory structure should look like the following:
models/
└── selfies_ted_model.pkl
Installation
To use this project, you'll need to install the required dependencies. We recommend using a virtual environment:
python -m venv venv
source venv/bin/activate # On Windows use `venv\Scripts\activate`
Install the required dependencies
pip install -r requirements.txt
Usage
Import
import load
Training the Model
To train the model, use the train.py script:
python train.py -f <path_to_your_data_file>
Note: The actual usage may depend on the specific implementation in load.py. Please refer to the source code for detailed functionality.
Load the model and tokenizer
load.load("path/to/checkpoint.pkl")
Encode SMILES strings
smiles_list = ["COC", "CCO"]
embeddings = load.encode(smiles_list)
Example Notebook
Example notebook of this project is selfies-ted-example.ipynb.