repo_name
stringlengths 4
136
| issue_id
stringlengths 5
10
| text
stringlengths 37
4.84M
|
|---|---|---|
fsprojects/FSharp.Formatting | 696888938 | Title: cref to members are not resolving to best possible link
Question:
username_0: In DiffSharp, consider
```
/// <summary>
/// Represents a multi-dimensional matrix containing elements of a single data type.
/// </summary>
///
/// <example>
/// A tensor can be constructed from a list or sequence using <see cref="M:DiffSharp.dsharp.tensor(System.Object)" />
///
/// <code>
/// let t = dsharp.tensor([[1.; -1.]; [1.; -1.]])
/// </code>
/// </example>
```
The `cref` is resolving to `http://localhost:8901/reference/diffsharp-dsharp.html` which is the best link for the containing type not the specific member<issue_closed>
Status: Issue closed |
brodul/najdi-si-sms | 125761579 | Title: Bug on ubuntu 15.10 "AttributeError: 'module' object has no attribute 'PROTOCOL_SSLv3'"
Question:
username_0: Traceback (most recent call last):
File "/usr/local/bin/najdisi-sms", line 9, in <module>
load_entry_point('najdisi-sms==0.3.1', 'console_scripts', 'najdisi-sms')()
File "/usr/lib/python2.7/dist-packages/pkg_resources/__init__.py", line 558, in load_entry_point
return get_distribution(dist).load_entry_point(group, name)
File "/usr/lib/python2.7/dist-packages/pkg_resources/__init__.py", line 2682, in load_entry_point
return ep.load()
File "/usr/lib/python2.7/dist-packages/pkg_resources/__init__.py", line 2355, in load
return self.resolve()
File "/usr/lib/python2.7/dist-packages/pkg_resources/__init__.py", line 2361, in resolve
module = __import__(self.module_name, fromlist=['__name__'], level=0)
File "/usr/local/lib/python2.7/dist-packages/najdisi_sms.py", line 7, in <module>
import requests
File "/usr/local/lib/python2.7/dist-packages/requests/__init__.py", line 53, in <module>
from .packages.urllib3.contrib import pyopenssl
File "/usr/local/lib/python2.7/dist-packages/requests/packages/urllib3/contrib/pyopenssl.py", line 62, in <module>
ssl.PROTOCOL_SSLv3: OpenSSL.SSL.SSLv3_METHOD,
AttributeError: 'module' object has no attribute 'PROTOCOL_SSLv3'
Answers:
username_0: Fixed with the `requests` update
Status: Issue closed
|
CATcher-org/CATcher | 710234157 | Title: [Windows 10] Unable to Open CATcher.3.3.1.exe
Question:
username_0: Hi, I was unable to start CATcher.3.3.1.exe that was downloaded from [here](https://github.com/CATcher-org/CATcher/releases/).
Actions which I have tried but did not work:
1. Followed the instructions for Windows Users from the [Usage Notes](https://github.com/CATcher-org/CATcher/blob/master/docs/usage-notes.md)
2. Tried starting on command prompt but no error messages were seen.

3. Tried running older versions of CATcher (CATcher.3.2.4.exe, CATcher.3.3.0) but none worked
I suspect it is due to some configurations on my computer and am wondering if similar issues have been encountered before. Thank you.
Answers:
username_1: Hello @username_0,
Were there any error message when you tried to start CATcher through the user interface?
username_0: Hi @username_1,
Which user interface are you referring to? If you are referring to the command line then, no, I wasn't able to see any error messages. Basically, nothing happens and the window (below) doesn't appear after I double click on the executable.

username_2: When you click on the executable, is the CATcher application visible in the list of running processes in Task Manager?
username_0: No, it is not visible. I believe it didn't even start at all, but I am not sure how to view the error messages.
username_1: Are you able to run other .exe files?
Maybe you can try downloading the "Portable" version of [ScreenToGif](https://www.screentogif.com/). Inside the zip file there is an .exe file.
Also, are you using any anti-virus software on your computer? They might flag out false-positive without any prompt.
username_0: Hi <NAME>,
The ScreenToGif executable works for me.

But may I clarify about this line in the [Usage Notes](https://github.com/CATcher-org/CATcher/blob/master/docs/usage-notes.md)?
"Windows Security" and under "App & browser control", click on "Reputation-based protection settings" and make sure CATcher is not blocked here.
For mine, the Windows Security notification showed up and I clicked on "Run Anyways", but nothing happened after that. Also, I wasn't able to find the "Reputation-based protection" settings. Perhaps I was missing this final step?
username_1: For newer version of Windows, you would probably see this:

Or are you using an older version of Windows? You might see this instead:

username_0: Yep, I am seeing the second one. It is set to "Warn" for mine.
username_3: @username_0 which OS version/edition are you using?
username_0: 
username_3: @username_1 Does CATcher produce any log file somewhere that may be useful here?
username_1: Nope, CATcher doesn't produce log files.
Maybe we could create a Windows VM that runs on same version.
username_3: I think it is better to assume the same thing might happen to some of the students and try to take some pre-emptive measures in case it does. e.g., produce some logs by default (or when run with a debug flag) We can get @username_0 's help to try out the solutions as he can repro the problem reliably.
username_2: @username_0, if you have some free time, do you think you could try building and starting CATcher from the source code?
We might be able to identify the problem, if building from source fails as well.
You'll need to have Node version 12 installed (you can use [node version manager](https://github.com/nvm-sh/nvm) to easily switch between versions)
Building and starting CATcher from source requires the following steps:
1. Clone this repository.
2. Make sure you are using Node 12 for this project.
3. Install dependencies with npm: `npm install`
4. Compile and start the application: `npm start`
username_0: @username_2 There was an error when i tried to run `npm install`
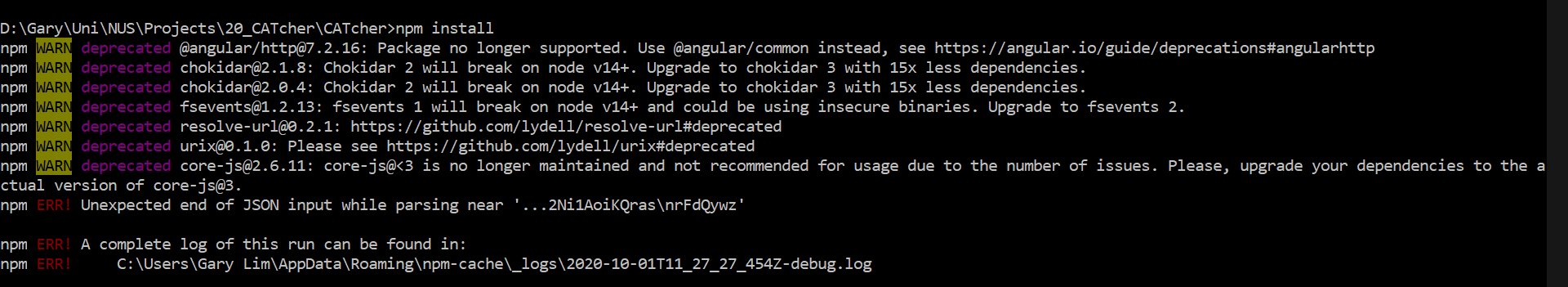
The log file is attached
[2020-10-01T11_27_27_454Z-debug.log](https://github.com/CATcher-org/CATcher/files/5311591/2020-10-01T11_27_27_454Z-debug.log)
username_0: Hi @username_2, I am able to run CATcher on my computer with `npm start`. I resolved the problem above by executing the following steps
1. Call `npm cache clean --force` based on the link [here](https://stackoverflow.com/questions/47675478/npm-install-errorunexpected-end-of-json-input-while-parsing-near-nt-webpack)
2. `npm install`
3. `npm start`
@username_3 I am able to create a new issue successfully on alpha2 with the CATcher I built on my computer.
username_3: @username_0 thanks for the update.
Dev team, does this help us find a solution? Perhaps we can ask students to build from source if a similar problem happens to others?
username_2: Thanks for trying this out @username_0.
Based on the result, I would guess that the problem lies in our executable generation step, and not the CATcher application itself.
username_4: May I ask if there is a fix to this issue?
username_2: @username_4 are you unable to launch CATcher on Windows as well?
username_4: Yes, unable to load or open anything when i click on it, or run in command prompt
username_5: Hello, I am facing the same problem as well. I am unable to load/open/run CATcher in any way.
username_1: @username_4 and @username_5, have you guys followed the [troubleshooting instructions](https://github.com/CATcher-org/CATcher/blob/master/docs/usage-notes.md#for-windows-users)?
Also please check to ensure that if you have an antivirus software, make sure it is not blocking it.
Lastly, if the above steps doesn't resolve your issue, can check whether the following file path exist? If so please provide us with the log files.
```
%USERPROFILE%\AppData\Roaming\CATcher\logs\*.log
```
username_5: @username_1 Yes I followed the troubleshooting instructions but I still cannot open the CATcher. That file path does not exist on my laptop.
username_5: @username_2 Yep, it does not exist in my computer.
username_4: same, I do not have AppData
username_2: @username_5 @username_4 thanks very much for your responses.
Could I get your OS version information ? (info similar to this [here](https://github.com/CATcher-org/CATcher/issues/476#issuecomment-700619986))
username_5: @username_2 Here is my OS information.

username_3: @username_4
You can use the web version of CATcher available at https://catcher-org.github.io/webapp/
Please give it a try to ensure you can use it. It is new and hasn't been tested as thoroughly as the desktop version. So, to be used only for cases where the student cannot use the normal desktop version.
Failing the above, you can run CATcher using the source, as explained [here](https://github.com/CATcher-org/CATcher/issues/476#issuecomment-701930310). Remember to pulls the latest code though, as there has been some updates to the code recently.
Status: Issue closed
username_2: Closing this, as the web app is now a stable alternative |
crazybilly/muadc | 154046605 | Title: Move aliases to proper functions
Question:
username_0: Currently, aliases are stored in a data/aliases.rda. They should actually be properly assigned and documented functions in R/.
Answers:
username_0: Gave this a go, but it's still not working with View. Where does View() that RStudio uses call from? |
Hallm13/portfolio | 366384234 | Title: more screens for initial concepts & testing
Question:
username_0: something to clearly show what each of the concepts looked like, what the tests showed, and why decisions were made to change them
Look at https://codydennis.design/fruition.html for examples on covering the iterations and testing<issue_closed>
Status: Issue closed |
modoboa/modoboa | 368978846 | Title: Not able to send email to external addresses
Question:
username_0: # Impacted versions
* Modoboa: 1.10.7
* installer used: Yes
* Webserver: Nginx
# Steps to reproduce
Send email to my external email such as a corporate, hotmail or gmail
# Current behavior
not able to send email to external account
# Expected behavior
not able to send email to external account
# Video/Screenshot link (optional)
1 06:13:56 whmtlmail01 postfix/qmgr[824]: F37E02226F1: from=<<EMAIL>>, size=1072, nrcpt=1 (queue active)
Oct 11 06:14:27 whmtlmail01 postfix/smtp[14127]: connect to mydomain-com.mail.protection.outlook.com[192.168.3.11]:25: Connection timed out
Oct 11 06:14:27 whmtlmail01 postfix/smtp[14127]: F37E02226F1: to=<<EMAIL>>, relay=none, delay=490, delays=460/0.08/30/0, dsn=4.4.1, status=deferred (connect to mydomain-com.mail.protection.outlook.com[172.16.31.10$
Oct 11 06:15:48 whmtlmail01 dovecot: imap-login: Login: user=<<EMAIL>>, method=PLAIN, rip=192.168.3.3, lip=172.16.1.8, mpid=14282, TLS, session=<Whnd5u1336bAqAMD>
Oct 11 06:18:15 whmtlmail01 postfix/pickup[823]: E0EEE2226FC: uid=111 from=<amavis>
Oct 11 06:18:15 whmtlmail01 postfix/cleanup[14553]: E0EEE2226FC: message-id=<<EMAIL>>
Oct 11 06:18:15 whmtlmail01 opendkim[5186]: E0EEE2226FC: no signing table match for '<EMAIL>'
Oct 11 06:18:15 whmtlmail01 opendkim[5186]: E0EEE2226FC: no signature data
Oct 11 06:18:15 whmtlmail01 postfix/qmgr[824]: E0EEE2226FC: from=<<EMAIL>>, size=760, nrcpt=1 (queue active)
Oct 11 06:18:15 whmtlmail01 postfix/local[14561]: E0EEE2226FC: to=<<EMAIL>, orig_to=<amavis>, relay=local, delay=0.54, delays=0.41/0.06/0/0.07, dsn=2.0.0, status=sent (delivered to mailbox)
Oct 11 06:18:15 whmtlmail01 postfix/qmgr[824]: E0EEE2226FC: removed
Oct 11 06:18:56 whmtlmail01 postfix/qmgr[824]: 2D2642226B1: from=<<EMAIL>>, size=1367, nrcpt=1 (queue active)
Oct 11 06:19:06 whmtlmail01 dovecot: imap(<EMAIL>): Disconnected for inactivity in=111 out=794
Oct 11 06:19:26 whmtlmail01 postfix/smtp[14592]: connect to mydomain-com.mail.protection.outlook.com[172.16.17.32]:25: Connection timed out
Oct 11 06:19:26 whmtlmail01 postfix/smtp[14592]: 2D2642226B1: to=<<EMAIL>>, relay=none, delay=2360, delays=2329/0.05/31/0, dsn=4.4.1, status=deferred (connect to mydomain-com.mail.protection.outlook.com[23.103.157$
Oct 11 06:23:56 whmtlmail01 postfix/qmgr[824]: F37E02226F1: from=<<EMAIL>>, size=1072, nrcpt=1 (queue active)
Oct 11 06:23:56 whmtlmail01 postfix/qmgr[824]: CC2E62226E7: from=<<EMAIL>>, size=3327, nrcpt=1 (queue active)
Oct 11 06:24:26 whmtlmail01 postfix/smtp[15241]: connect to msn-com.olc.protection.outlook.com[172.16.17.32]:25: Connection timed out
Answers:
username_1: Hi,
As you can see in your logs
```
Oct 11 06:14:27 whmtlmail01 postfix/smtp[14127]: connect to mydomain-com.mail.protection.outlook.com[192.168.3.11]:25: Connection timed out
Oct 11 06:19:26 whmtlmail01 postfix/smtp[14592]: connect to mydomain-com.mail.protection.outlook.com[172.16.17.32]:25: Connection timed out
```
your server can't contact the remote one. It's either a routing problem or a firewall issue in my opinion. So nothing related to modoboa.
username_1: You don't have to open specific ports for modoboa. Of course you have to open postfix and dovecot ports.
The problem here is you can't connect to a remote server. It may be an issue with your firewall filtering outgoing packets, not incoming ones. Did you try to ping the remote addresses ?
username_2: Hi @username_0
Try:
```
ping mydomain-com.mail.protection.outlook.com
traceroute mydomain-com.mail.protection.outlook.com
telnet mydomain-com.mail.protection.outlook.com 25
```
A mail server generally need ports: 25, 587, 995 and 993.
Status: Issue closed
|
zalando/patroni | 619129165 | Title: Disable creation of PGPASS if using kerberos
Question:
username_0: Hello,
It doesn't make sense to create `PGPASS` file if we are using [kerberos](https://github.com/zalando/patroni/pull/1015). Can we disable in that case.
PGPASS is being created in [here](https://github.com/zalando/patroni/blob/881bba9e1cd081f7cd4c340f9a6ad977daa92c58/patroni/postgresql/config.py#L369).
Answers:
username_1: @username_0 you are right. Feel free to create a PR implementing the necessary check.
Status: Issue closed
|
forkdelta/forkdelta.github.io | 330981957 | Title: Deposit of RiseCoinToken - RSCT fails with Bad Jump Destination error
Question:
username_0: When I tried to deposit RiseCoinToken RSCT the balance is more than 50,000 RSCT and I tried to deposit 10,000 RSCT still its not happening.
When I tried depositing it created 2 transaction 1st to RSCT contract and the other to etherdelta_2 contract.
The first transaction is approved which is to RSCT contract and RSCT approved it however etherdelta contract showed an exception.
Below are the same.
Kindly advise if this is a problem with exchange or RSCT contract because I am able to transfer RSCT to some other wallets but not on etherdelta and forkdelta. Just to tell you it is not listed on etherdelta or forkdelta. Is it because of that?
1st.
https://etherscan.io/tx/0xae7a79f815d767f5c96fd5f189ad93aea5b9d75d20d1358551eb1e605145b595
2nd.
https://etherscan.io/tx/0xb9474830bfc1d95069f3245b3c67f0e085bc4c699fd0cc759782af7e5f088eb4
Answers:
username_1: @username_0 Thank you for your report. I noticed that there are a few successful approvals for the current trading contract, but no successful transfers. I was unable to track down the exact issue, I would recommend reaching out to the token issuer for a more thorough investigation.
Status: Issue closed
|
phusion/passenger | 300891469 | Title: RuntimeError: Illegal character ‘’ in raw string ““
Question:
username_0: DEBUG [358f9ead] : #<RuntimeError: Illegal character ‘’ in raw string ““> (REXML::ParseException
DEBUG [358f9ead] )
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/text.rb:162:in `block in check’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/text.rb:154:in `scan’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/text.rb:154:in `check’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/text.rb:121:in `initialize’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/parsers/treeparser.rb:47:in `new’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/parsers/treeparser.rb:47:in `parse’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/document.rb:288:in `build’
/usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/document.rb:45:in `initialize’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:257:in `new’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:257:in `query_pool_xml’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:207:in `select_app_group_name_by_app_root_regex’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:146:in `select_app_group_name’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:46:in `run’
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/main.rb:78:in `run!'
/usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/bin/passenger-config:38:in `<top (required)>'
/usr/local/rvm/gems/ruby-2.3.4/bin/passenger-config:23:in `load’
/usr/local/rvm/gems/ruby-2.3.4/bin/passenger-config:23:in `<main>'
/usr/local/rvm/gems/ruby-2.3.4/bin/ruby_executable_hooks:15:in `eval’
/usr/local/rvm/gems/ruby-2.3.4/bin/ruby_executable_hooks:15:in `<main>'
...
Illegal character ‘’ in raw string “”
Line: 2
Position: 24953
Last 80 unconsumed characters:
</code_revision>
from /usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/parsers/treeparser.rb:21:in `parse’
from /usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/document.rb:288:in `build’
from /usr/local/rvm/rubies/ruby-2.3.4/lib/ruby/2.3.0/rexml/document.rb:45:in `initialize’
DEBUG [358f9ead] from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:257:in `new’
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:257:in `query_pool_xml’
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:207:in `select_app_group_name_by_app_root_regex’
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:146:in `select_app_group_name’
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/restart_app_command.rb:46:in `run’
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/src/ruby_supportlib/phusion_passenger/config/main.rb:78:in `run!'
from /usr/local/rvm/gems/ruby-2.3.4/gems/passenger-5.1.8/bin/passenger-config:38:in `<top (required)>'
from /usr/local/rvm/gems/ruby-2.3.4/bin/passenger-config:23:in `load’
from /usr/local/rvm/gems/ruby-2.3.4/bin/passenger-config:23:in `<main>'
from /usr/local/rvm/gems/ruby-2.3.4/bin/ruby_executable_hooks:15:in `eval’
from /usr/local/rvm/gems/ruby-2.3.4/bin/ruby_executable_hooks:15:in `<main>'
Answers:
username_1: The invalid character is coming from your app's output, perhaps check the character encoding is consistent in your app. On an unrelated note you shouldn't use 5.1.8 there have been vulnerabilities fixed in Passenger since then, so you should try and upgrade to the latest version.
username_0: @username_1 thanks for your help, i'll check them :)
Status: Issue closed
|
flutter/flutter | 1106539388 | Title: 【Flutter】I want to mute video music when I back to this page
Question:
username_0: I've recently started fluttering.
I want to mute video music when I back to this page
```
import 'package:flutter/cupertino.dart';
import 'package:flutter/material.dart';
import 'package:appinio_video_player/appinio_video_player.dart';
import 'package:flutter_riverpod/flutter_riverpod.dart';
import 'package:sample/user/user_model.dart';
import 'package:cloud_firestore/cloud_firestore.dart';
class VideoPlayerPage extends ConsumerWidget {
const VideoPlayerPage({Key? key}) : super(key: key);
@override
Widget build(BuildContext context, ScopedReader watch) {
final clickvideo = watch(clickprogramProvider).state;
return _VideoPlayerWidget(clickvideo);
}
}
Future<String> getURL(String documentName) async {
var getData = await FirebaseFirestore.instance
.collection('programs')
.doc(documentName)
.get();
String url = getData.get("url").toString();
return url;
}
Widget _VideoPlayerWidget(String name) {
late VideoPlayerController videoPlayerController;
CustomVideoPlayerController customVideoPlayerController =
CustomVideoPlayerController();
@override
void dispose() {
videoPlayerController.dispose();
customVideoPlayerController.dispose();
}
return FutureBuilder(
future: getURL(name),
builder: (context, snapshot) {
if (!snapshot.hasData) {
return const Scaffold(
backgroundColor: Colors.white,
);
}
videoPlayerController =
VideoPlayerController.network(snapshot.data.toString())
..initialize();
return CupertinoPageScaffold(
navigationBar: CupertinoNavigationBar(
middle: Text(name),
),
child: SafeArea(
child: Padding(
padding: const EdgeInsets.all(20.0),
child: ListView(
children: [
CustomVideoPlayer(
customVideoPlayerController: customVideoPlayerController,
videoPlayerController: videoPlayerController,
customVideoPlayerSettings: CustomVideoPlayerSettings(
customVideoPlayerProgressBarSettings:
const CustomVideoPlayerProgressBarSettings(),
),
),
],
),
),
),
);
});
}
```
Answers:
username_1: @username_0
Looks like you need code assistance to achieve your use case, for which, stackoverflow is the correct platform to ask this question.
Closing from here as this isn't a bug or a feature request.
Status: Issue closed
|
akhoury/ginx | 126327660 | Title: When calling `parseLine` without an options param, an exception is thrown
Question:
username_0: The docs indicate that the `options` param on `parseLine()` is optional. When you don't provide it, though, you get this error:
```
[GINX][ERROR][uncaughtException] TypeError: Cannot read property 'file' of undefined
[GINX-WARN] Exiting - No cursors were stored
/opt/pinecast-softmirror/src/tailor/node_modules/ginx/lib/ginx.js:519
throw err;
^
TypeError: Cannot read property 'file' of undefined
at Ginx.eval [as hardParseLine] (eval at generateParseLine (/opt/pinecast-softmirror/src/tailor/node_modules/ginx/lib/ginx.js:347:19), <anonymous>:60:24)
at Ginx.parseLine (/opt/pinecast-softmirror/src/tailor/node_modules/ginx/lib/ginx.js:182:10)
```
It seems like [this line](https://github.com/username_1/ginx/blob/master/lib/ginx.js#L182) should be updated to something like this:
```js
this.hardParseLine(line, callback, options || {});
```
That said, I don't know for sure.
Status: Issue closed
Answers:
username_1: i am not 100% sure how to reproduce the issue, but that should do it.
Anyways, didn't really know that anyone is still using this module, lol - it has been unmaintained for like 3 years
username_0: Thanks for the quick fix, man. I'm using ginx to parse some nginx access logs for a log aggregation service. I figured it'd be better to use it than rolling my own :)
A simple way to reproduce the issue is to just create a new Ginx object and call parseLine without an options param:
```js
var Ginx = require('ginx');
var g = new Ginx(null, {persistent: false});
g.parseLine('foo', function() {}); // no options param
```
The third line will throw the exception. The generated function code looks at `options.file`, which fails because `options` is undefined. |
ho-ansible/wireguard | 533747386 | Title: OpenVZ
Question:
username_0: May need to pre-compile wireguard-go and copy binary to host.
Answers:
username_0: To compile (only needed on one host):
debian pkg `golang-go` has the compiler, linker, and precompiled stdlib, probably enough to compile wireguard-go. The metapackage `golang` also pulls in docs and src files.
Download latest src release of wireguard-go: https://git.zx2c4.com/wireguard-go/refs/tags
Adjust `PreallocatedBuffersPerPool` if desired, to limit RAM usage.
Add `WG_I_PREFER_BUGGY_USERSPACE_TO_POLISHED_KMOD=1` to an *.env file for systemd-networkd, or an `Environment=` entry in the systemd service drop-in.
Hope that systemd-networkd knows to use the wg binary.
Thanks to [Daniel](https://d.sb/2019/07/wireguard-on-openvz-lxc) |
bethgelab/foolbox | 278499843 | Title: Training Samples
Question:
username_0: How create training samples from adversarially perturbed original training sampels?
To be simple, suppose I had 100 training images and wanted to use Deep Fool and FGSM to perturb these samples, I should now end up with 200 adversarial samples and 100 originals to train on. How to go about this in the most efficient way with this library?
Sample code **very** much appreciated! :D
Answers:
username_1: Have a look at the sample code in the [README](https://github.com/bethgelab/foolbox/blob/master/README.rst). In the case of 100 images and fast attacks like FGSM and DeepFool, basically all you need is to put a loop around the last line:
```python3
import foolbox
import keras
import numpy as np
from keras.applications.resnet50 import ResNet50
# instantiate model
keras.backend.set_learning_phase(0)
kmodel = ResNet50(weights='imagenet')
preprocessing = (np.array([104, 116, 123]), 1)
fmodel = foolbox.models.KerasModel(kmodel, bounds=(0, 255), preprocessing=preprocessing)
# get source image and label
image, label = foolbox.utils.imagenet_example()
# apply attack on source image
attack = foolbox.attacks.FGSM(fmodel)
fgsm_adversarials = []
for image, label in zip(some_training_images, corresponding_labels):
adversarial = attack(image[:,:,::-1], label)
fgsm_adversarials.append(adversarial)
```
If you want to do this for a much larger set of images or on the fly during training, _Foolbox_ might not be the right tool – performance is not it's focus. The power of _Foolbox_ is a large set of attacks that makes it easy to reliably test the robustness of models.
username_0: Makes sense. Thanks for FAST response. Once Foolbox exposes fagility of my models which I expect it (this is a FANTASTIC tool) I want to re-train on adversarial samples and re-test.
Also what is this code doing `image[:,:,::-1]` ? Why do we need to go in reverse order on the channel axis of the image? Trying to figure out intuition.
Thanks again for your help!!
username_1: That's just part of the preprocessing expected by the ResNet implementation in Keras, i.e. it expects BGR color channel ordering and channel mean subtraction (done a few lines before that).
username_1: @username_0 can this be closed?
username_0: Absolutely!! Thank you!!!
Status: Issue closed
username_0: @username_1
Can foolbox be used on a model that I trained on a different domain than imagenet? Will the Keras attack take as input any Keras mode I build?
username_1: @username_0 Foolbox can be used to attack any machine learning model, nothing is specific to ImageNet. The `foolbox.models.KerasModel` model wrapper for Keras models should be able to handle any Keras model that follows the conventions of the `keras.models.Model` [class](https://keras.io/models/model/#model-class-api).
username_0: Awesome! Thank you so much! |
npm/cli | 1173350027 | Title: [BUG] `npm run` changes user using default (1000) instead of root (0)
Question:
username_0: 1000
root@7f44813ba8d7:/app# node -v
v16.13.2
```
### Expected Behavior
When I run a script as root `process.getuid()` should return `0` even when running that script using `npm run`
### Steps To Reproduce
Described above
### Environment
- npm: 8.1.2
- Node.js: 16.13.2
- OS/System/Arch: Linux 360f89fb056b 5.13.0-35-generic #40~20.04.1-Ubuntu SMP Mon Mar 7 09:18:32 UTC 2022 x86_64 GNU/Linux
- npm config:
```ini
; "user" config from /root/.npmrc
unsafe-perm = true
user = "0"
; node bin location = /usr/local/bin/node
; cwd = /app
; HOME = /root
; Run `npm config ls -l` to show all defaults.
```
Status: Issue closed
Answers:
username_1: This is the expected behavior. See https://github.com/npm/cli/issues/4095, https://github.com/npm/cli/issues/3773, https://github.com/npm/cli/issues/3110, and https://docs.npmjs.com/cli/v7/using-npm/scripts#user |
FasterXML/jackson-module-kotlin | 616982278 | Title: Deserialization of lists is lost under certain terminal node conditions
Question:
username_0: **Describe the bug**
Deserialization of lists is lost under certain conditions. When I change the terminal nodes to have fake key names, the full structure is returned with those terminal nodes having null values. When I change the terminal nodes to reference real keys in the xml, the list is not maintained and instead returns the last entry that makes up the list with the correct terminal values.
**To Reproduce**
Small subset of xml data
```
<?xml version="1.0" encoding="utf-8" standalone="yes"?>
<feed xml:base="http://data.treasury.gov/Feed.svc/" xmlns:d="http://schemas.microsoft.com/ado/2007/08/dataservices" xmlns:m="http://schemas.microsoft.com/ado/2007/08/dataservices/metadata" xmlns="http://www.w3.org/2005/Atom">
<title type="text">DailyTreasuryYieldCurveRateData</title>
<id>http://data.treasury.gov/feed.svc/DailyTreasuryYieldCurveRateData</id>
<updated>2020-05-08T22:36:11Z</updated>
<link rel="self" title="DailyTreasuryYieldCurveRateData" href="DailyTreasuryYieldCurveRateData" />
<entry>
<id>http://data.treasury.gov/Feed.svc/DailyTreasuryYieldCurveRateData(1)</id>
<title type="text"></title>
<updated>2020-05-08T22:36:11Z</updated>
<author>
<name />
</author>
<link rel="edit" title="DailyTreasuryYieldCurveRateDatum" href="DailyTreasuryYieldCurveRateData(1)" />
<category term="TreasuryDataWarehouseModel.DailyTreasuryYieldCurveRateDatum" scheme="http://schemas.microsoft.com/ado/2007/08/dataservices/scheme" />
<content type="application/xml">
<m:properties>
<d:Id m:type="Edm.Int32">1</d:Id>
<d:NEW_DATE m:type="Edm.DateTime">1997-01-02T00:00:00</d:NEW_DATE>
<d:BC_1MONTH m:type="Edm.Double" m:null="true" />
<d:BC_2MONTH m:type="Edm.Double" m:null="true" />
<d:BC_3MONTH m:type="Edm.Double">5.190000057220459</d:BC_3MONTH>
<d:BC_6MONTH m:type="Edm.Double">5.3499999046325684</d:BC_6MONTH>
<d:BC_1YEAR m:type="Edm.Double">5.630000114440918</d:BC_1YEAR>
<d:BC_2YEAR m:type="Edm.Double">5.96999979019165</d:BC_2YEAR>
<d:BC_3YEAR m:type="Edm.Double">6.130000114440918</d:BC_3YEAR>
<d:BC_5YEAR m:type="Edm.Double">6.3000001907348633</d:BC_5YEAR>
<d:BC_7YEAR m:type="Edm.Double">6.4499998092651367</d:BC_7YEAR>
<d:BC_10YEAR m:type="Edm.Double">6.5399999618530273</d:BC_10YEAR>
<d:BC_20YEAR m:type="Edm.Double">6.8499999046325684</d:BC_20YEAR>
<d:BC_30YEAR m:type="Edm.Double">6.75</d:BC_30YEAR>
<d:BC_30YEARDISPLAY m:type="Edm.Double">0</d:BC_30YEARDISPLAY>
</m:properties>
</content>
</entry>
<entry>
<id>http://data.treasury.gov/Feed.svc/DailyTreasuryYieldCurveRateData(2)</id>
<title type="text"></title>
<updated>2020-05-08T22:36:11Z</updated>
<author>
<name />
</author>
<link rel="edit" title="DailyTreasuryYieldCurveRateDatum" href="DailyTreasuryYieldCurveRateData(2)" />
<category term="TreasuryDataWarehouseModel.DailyTreasuryYieldCurveRateDatum" scheme="http://schemas.microsoft.com/ado/2007/08/dataservices/scheme" />
<content type="application/xml">
<m:properties>
<d:Id m:type="Edm.Int32">2</d:Id>
<d:NEW_DATE m:type="Edm.DateTime">1996-12-31T00:00:00</d:NEW_DATE>
<d:BC_1MONTH m:type="Edm.Double" m:null="true" />
<d:BC_2MONTH m:type="Edm.Double" m:null="true" />
<d:BC_3MONTH m:type="Edm.Double">5.2100000381469727</d:BC_3MONTH>
<d:BC_6MONTH m:type="Edm.Double">5.3299999237060547</d:BC_6MONTH>
<d:BC_1YEAR m:type="Edm.Double">5.5100002288818359</d:BC_1YEAR>
[Truncated]
data class Properties(
@JacksonXmlProperty(localName = "NEW_DATE")
var newDate: String?,
@JacksonXmlProperty(localName = "BC_1MONTH")
var oneMonth: String?
)
```
```
Feed(entry=[Entry(id=http://data.treasury.gov/Feed.svc/DailyTreasuryYieldCurveRateData(7596), updated=2020-05-08T22:36:11Z, content=Content(properties=Properties(newDate=2020-05-08T00:00:00, oneMonth=0.1)))])
```
**Versions**
Kotlin:1.3.61
Jackson-module-kotlin:2.9.7 and 2.11.0
Jackson-databind: 2.9.7 and 2.11.0
**Additional context**
I'm not really sure if this should be posted here or in one of the core jackson libraries.
Answers:
username_1: Quick note: if it would be possible to reproduce this with java-only (by translating into similar pojo classes), it'd be easier to resolve, challenge being that XML format module is quite different from basic json handling and this problem is almost certainly specific to that module.
But it might not be related to Kotlin handling.
Practical challenge being that neither Kotlin module nor XML format module have dependency to each other (even test dependency), so reproduction as-is could not be used for automated tests. |
openfoodfacts/openfoodfacts-ios | 284795520 | Title: Check UX with different Network Conditioner profiles
Question:
username_0: With Network Conditioner set to `100% loss`, loggin in, for instance, timeouts but the user has no feedback. In this case a HUD should happear telling the user we are trying to log in to avoid him tapping the Login button again and again.
Check all use cases in the app to find if there are other UX mistakes on slow networks.
Answers:
username_1: This is a good idea. |
msiemens/tinydb | 251473263 | Title: A version attr for tinydb - Enhancement
Question:
username_0: It would be nice to have a version attr for tinydb. After my embarrassing mistake of running such an old version of tinydb, in the small app I am doing i wanted to do a version test on the lib. I still consider myself a beginner, so there may be other ways to do this that I am unaware of. But I have seen other Libs like SQLite for example have a version attr.
For backward compatibility we could use hasattr(db, 'version') before accessing it directly which if failed would tell us the maximum version it could be, if does not fail could then use the version attr.
Seems to me to this would be ok given, you typically would do this once at the star of your app.
Status: Issue closed
Answers:
username_1: That's a good idea! I'll release TinyDB v3.4.1 shortly which exposes the TinyDB version from the `tinydb` module like this:
```
import tinydb
print(tinydb.__version__)
# or
from tinydb import TinyDB, __version__ as tinydb_version
print(tinydb_version)
```
username_0: Perfect. Thanks. That will be useful for me, I hope others also |
TechnionYP5777/SmartCity-Accessibility | 200804474 | Title: Change getLocation to void
Question:
username_0: Since we want getLocation to run in the background it should be void functoin that gets a getCallback function, if I am not mistaken. But the function is already in use in some parts of the code. How should we change it?
Answers:
username_1: create a new getLocation function that works with the callback and put a deperecated label on the previous, than transfer the issue the to people using the old one so they know they need to update
username_0: Ok thanks
Status: Issue closed
|
ioBroker/AdapterRequests | 625189704 | Title: Adapter für IEC 60870-5-101/102/103/104 Protokoll
Question:
username_0: Einen Adapter der für die Fernwirkprotokolle 60870-5-... kommuniziert
https://de.wikipedia.org/wiki/IEC_60870
Evtl kann ich eine Schnitstelle/Gerät bereitstellen. Es gibt aber auch simulationssoftware im netz.
Answers:
username_1: Are there any libraries for nodejs available or such?
username_2: Eigentlich sollte IEC 60870-5-104 (Telegramme werden per TCP/IP übertragen) reichen, oder? Libraries sind vorwiegend für C / C++ verfügbar.
Beispiel:
https://sourceforge.net/projects/iec-104-source-code-library
username_0: Ich habe auch noch mal genau durchgelesen 101 ist ja ehr seriell und es geht ehr um die IP version. Daher würde das iec 60870-5-104 Reichen. Ich habe auch diverse lib gefunden allerdings auch nur für C/C++ php usw. allerdings nichts für nodejs. Kann man die irgendwie auch nutzen?
username_2: @username_0 : welchen Use Case möchtest du abbilden?
Vermutlich wird es einfacher sein, ein beispielsweise "MQTT <-> IEC 60870-5-104" Gateway in C/C++ zu implementieren.
username_0: ES geht darum eine SPS die im Freien feld sitzt und mittels IEC 60870-5-104 kommuniziert über lan an den IObroker anzubringen. Ich würde gerne kein Gateway benutzen
username_3: Die Firma Lucom bietet für ihre Linux basierenden Router ein kostenloser User Module für die Konvertierung von IEC Protokollen an.
https://www.lucom.de/user-module.html#protocol-iec101-104
Eventuell hilft das hier bei der Entwicklung weiter. Eventuell könnte ich auch dementsprechende Hardware bereit stellen. Muss ich allerdings erst abklären.
username_3: Es gibt auch einen wirklich gut funktionierenden IEC Simulator https://sourceforge.net/projects/iec-104-client-simulator/ |
andamira/forandar | 139298694 | Title: ANSI terminal fails when run through pub
Question:
username_0: When the CLI interface is run with `pub get run`, or after activating it with `pub global activate` and the ANSI terminal is used it gives the following error:
```
StdoutException: Could not get terminal size, OS Error: Inappropriate ioctl for device, errno = 25
```
It only works when using directly the `dart` command.
This issue depends on dart-lang/pub/issues/1204 |
dreamRs/apexcharter | 804088395 | Title: fill colors in bar chart
Question:
username_0: Getting a bar chart with different colored bars is harder than you think, and the end result is still not satisfying as it shows empty (NA) labels:
A natural (by `ggplot2` standards) approach is to try:
```{r}
library(apexcharter)
plotdata <- data.frame(x=c("a","b"), counts=5:6, color=1:2)
apex(
data = plotdata,
type = "column",
mapping = aes(x = x, y = counts, fill = color)
)
```
but this "drops" the b category and produces

One can complete the plotdata through
```{r}
library(tidyr)
plotdata1 <- complete(plotdata, x, color)
```
and then get two different colors for the two bars
```{r}
apex(
data = plotdata1,
type = "column",
mapping = aes(x = x, y = counts, fill = color)
)
```
but there are empty bars at each category on the x-axis (because of the NA's generated in `complete`):

Ideally, I was hoping code like
```{r}
apex(
data = plotdata,
type = "column",
mapping = aes(x = x, y = counts, fill = color)
) %>%
ax_colors(c("red","orange"))
```
would give two bars, the one over "a" in one red, the other over "b" in orange. Instead, I get:

But perhaps I don't quite understand how to get this with apexcharter.
Thanks!
Bernhard
Answers:
username_1: Hello,
Yes there's a bug somewhere with your first example, I'll investigate.
In your last example, bars are gray because colors names don't work, you have to use hex code.
To achieve what you desire, I see 2 options:
* Use special aesthetic `fillColor`:
```r
library(apexcharter)
plotdata <- data.frame(x=c("a","b"), counts=5:6, color=1:2, fillColor = c("#FE2E2E", "#FF8000"))
apex(
data = plotdata,
type = "column",
mapping = aes(x = x, y = counts, fillColor = fillColor)
)
```

* use `fill` aes like you did but with stacked bars:
```r
library(tidyr)
plotdata1 <- complete(plotdata, x, color)
apex(
data = plotdata1,
type = "column",
mapping = aes(x = x, y = counts, fill = color)
) %>%
ax_chart(stacked = TRUE) %>%
ax_colors(c("#FE2E2E", "#FF8000"))
```

Hope it helps !
Victor
username_0: Wonderful, works perfectly, thanks!
Status: Issue closed
|
malaybaku/VMagicMirror | 712006801 | Title: 手がめちゃくちゃ小さいモデル用に手指パラメータに0とかマイナスを適用したい
Question:
username_0: ## 概要
題の通りです。ユーザーフィードバックです。
## 対応方針
- 下限が0でもいいものはWPF側だけ改修して下限を0にする。
- 現状の設定でマイナス相当のパラメタが欲しいものについては、(GUIの設定値-定数)みたいな値が適用されるようにUnity側だけいじるのが良さそう
Answers:
username_0: としているのに対応する変更はUnity側では #405 の対応ついでに該当パラメータを削除します(ここのissue comment以外に書くのに適したとこが無いのでメモ程度で)。
Status: Issue closed
|
react-dropzone/react-dropzone | 466698970 | Title: TypeError: arr[Symbol.iterator] is not a function
Question:
username_0: <!-- Before creating an issue please make sure you are using the latest version of react-dropzone. -->
**Do you want to request a *feature* or report a *bug*?**
<!-- Please ask questions on StackOverflow. Questions will be closed. -->
- [x] I found a bug
- [ ] I want to propose a feature
**What is the current behavior?**
**If the current behavior is a bug, please provide the steps to reproduce.**
<!-- You can use https://codesandbox.io/s/vvw87k06k5 to get started and illustrate the issue you're experiencing. -->
1. I was just following the official 'previews' example to make a function component, and then I
got a error message -- TypeError arr[Symbol.iterator] is not a function.
This is my [code](https://codesandbox.io/s/rough-sound-nf5z2).
**What is the expected behavior?**
**If this is a feature request, what is motivation or use case for changing the behavior?**
**Please mention other relevant information such as the browser version, Operating System and react-dropzone version.**
Browser: Google Chrome Version 75.0.3770.100
OS: windows 10
react-dropzone: 10.1.5
Status: Issue closed
Answers:
username_1: @username_0 why was this closed? I've got the same issue coming up in Safari 10 (with Dropzone)
username_0: Because this is not a issue. Pay a close attention to the following code:
const {getRootProps, getInputProps, isDragActive} = useDropzone({onDrop});
If you use array destructuring rather than object destructuring, you would get this error. |
SpankChain/spankbank-web3 | 356710934 | Title: Hard coded Ledger timeout is too short
Question:
username_0: Using a Ledger Nano S, it's not possible to approve the transaction before the 7 second hard coded timeout elapses, in `spankbank.ts`:
```
transport.exchangeTimeout = 7000
```
The default timeout (30 sec) would be sufficient.
Answers:
username_1: @alexanvl please evaluate
Status: Issue closed
username_2: Thanks for the report, @username_0!
The reason for the lower timeout was so that we could more quickly detect whether or not a Ledger device is attached.
I've made the timeout configurable, here: https://github.com/SpankChain/spankbank-web3/commit/0a8ac60fcd913441a49466caf9266a0f65ed61b7
And the SpankBank will be updated as soon as this PR is accepted: https://github.com/SpankChain/spankbank-explorer_frontend/pull/3 |
cerner/terra-clinical | 290646925 | Title: Fix padding in list view of ItemCollection
Question:
username_0: # Issue Description
Currently, you are allowed to set isTablePadded to get padding on the table view of an ItemCollection. This padding does not carryover to the list view and makes everything seemed very squished.
## Issue Type
- [ ] New Feature
- [ ] Enhancement
- [x] Bug
- [ ] Other
## Expected Behavior
As there is padding on the table when isTablePadded is true, there should be padding on the list as well.
## Current Behavior
<img width="404" alt="screen shot 2018-01-22 at 4 52 59 pm" src="https://user-images.githubusercontent.com/8262756/35248952-ae265e80-ff95-11e7-96f6-9ed36bc02ba5.png">
<img width="1299" alt="screen shot 2018-01-22 at 4 52 48 pm" src="https://user-images.githubusercontent.com/8262756/35248953-ae3bca2c-ff95-11e7-894f-04dd72d57930.png">
## Steps to Reproduce
Create an ItemCollection with rows, add isTablePadded on the ItemCollection. In the large/table view, see the padding around the edges of each row. Shrink down to a list view, see how there is no padding around the edges of each row.
## Environment
* Component Version: 2.2.0
* Browser Name: Chrome
* Operating System and version (desktop or mobile): desktop
Status: Issue closed
Answers:
username_1: @avinashg1994 we are actually planning on deprecating the item collection (https://github.com/cerner/terra-clinical/issues/464). We realize there are many gaps in functionality that exist but the easiest way to manage that is for teams to create their own wrapper that switches between a list and table (which is also going under changes https://github.com/cerner/terra-core/issues/2195). That is going to be our plan going forward. |
puma/puma | 395985201 | Title: Failed to build gem native extension.
Question:
username_0: ubuntu 16.04, ruby 1.8.7
tried puma 2.16.0, 3.4.0
```
Fetching: puma-3.4.0.gem (100%)
Building native extensions. This could take a while...
ERROR: Error installing puma:
ERROR: Failed to build gem native extension.
/usr/local/rbenv/versions/1.8.7/bin/ruby extconf.rb
checking for BIO_read() in -lcrypto... yes
checking for SSL_CTX_new() in -lssl... yes
checking for openssl/bio.h... yes
creating Makefile
make
/usr/bin/gcc -I. -I. -I/usr/local/rbenv/versions/1.8.7/lib/ruby/1.8/x86_64-linux -I. -DHAVE_OPENSSL_BIO_H -I/usr/local/rbenv/versions/1.8.7/include -I/usr/local/rbenv/versions/1.8.7/include -fPIC -g -O2 -c puma_http11.c
/usr/bin/gcc -I. -I. -I/usr/local/rbenv/versions/1.8.7/lib/ruby/1.8/x86_64-linux -I. -DHAVE_OPENSSL_BIO_H -I/usr/local/rbenv/versions/1.8.7/include -I/usr/local/rbenv/versions/1.8.7/include -fPIC -g -O2 -c io_buffer.c
io_buffer.c:7:3: error: unknown type name ‘uint8_t’
uint8_t* top;
^
io_buffer.c:8:3: error: unknown type name ‘uint8_t’
uint8_t* cur;
^
io_buffer.c: In function ‘buf_append’:
io_buffer.c:49:5: error: unknown type name ‘uint8_t’
uint8_t* top;
^
io_buffer.c:50:5: error: unknown type name ‘uint8_t’
uint8_t* old;
^
io_buffer.c: In function ‘buf_append2’:
io_buffer.c:90:5: error: unknown type name ‘uint8_t’
uint8_t* top;
^
io_buffer.c:91:5: error: unknown type name ‘uint8_t’
uint8_t* old;
^
Makefile:149: recipe for target 'io_buffer.o' failed
make: *** [io_buffer.o] Error 1
Gem files will remain installed in /usr/local/rbenv/versions/1.8.7/lib/ruby/gems/1.8/gems/puma-3.4.0 for inspection.
Results logged to /usr/local/rbenv/versions/1.8.7/lib/ruby/gems/1.8/gems/puma-3.4.0/ext/puma_http11/gem_make.out
```
Answers:
username_1: I believe Ruby 1.8.7 was removed from CI testing on [2016-02-25](https://github.com/puma/puma/commit/b4a2fded22299dd8a11fd64ea88aeb3b33a0846e#diff-354f30a63fb0907d4ad57269548329e3). The Travis for that commit used [Ubuntu 12.04](https://travis-ci.org/puma/puma/jobs/111866664#L9-L13).
Someone may be able to help, but...
username_2: You have a broken complier / libc-dev setup. uint8_t is a system type.
Status: Issue closed
|
bcgov/entity | 721698642 | Title: store PAD agreement details
Question:
username_0: the pad agreement details like time of agreement ,user details etc should be captured in the DB.
Also current endpoint doesn't support supplying PAD TOS to the UI afaik.We have to support a param in the input API so that PAD TOS is avaiable to the UI.
Answers:
username_1: This ticket is to cover happy path - storing initial PAD agreement.
Status: Issue closed
|
Hannah-Sten/TeXiFy-IDEA | 508322385 | Title: java.lang.IndexOutOfBoundsException: Wrong startOffset: 9426; documentLength: 2385
Question:
username_0: ### Description
Used "Fix label name" on chapter name.
Linux Mint 19
IntelliJ IDEA 2019.2.3 (Ultimate Edition)
Build #IU-192.6817.14, built on September 24, 2019
Runtime version: 11.0.4+10-b304.69 amd64
VM: OpenJDK 64-Bit Server VM by JetBrains s.r.o
Linux 4.15.0-65-generic
GC: ParNew, ConcurrentMarkSweep
Memory: 4029M
Cores: 8
Registry:
Non-Bundled Plugins: Lua, com.jetbrains.plugins.ini4idea, mobi.hsz.idea.latex, nl.rubensten.texifyidea, Pythonid, org.jetbrains.plugins.ruby
### Stacktrace
```
java.lang.IndexOutOfBoundsException: Wrong startOffset: 9426; documentLength: 2385
at com.intellij.openapi.editor.impl.DocumentImpl.assertBounds(DocumentImpl.java:667)
at com.intellij.openapi.editor.impl.DocumentImpl.replaceString(DocumentImpl.java:612)
at com.intellij.openapi.editor.impl.DocumentImpl.replaceString(DocumentImpl.java:608)
at nl.hannahsten.texifyidea.util.DocumentsKt.replaceString(Documents.kt:44)
at nl.hannahsten.texifyidea.inspections.latex.LatexLabelConventionInspection$LabelPreFix.applyFix(LatexLabelConventionInspection.kt:138)
at nl.hannahsten.texifyidea.inspections.latex.LatexLabelConventionInspection$LabelPreFix.applyFix(LatexLabelConventionInspection.kt:108)
at com.intellij.codeInspection.ex.QuickFixWrapper.invoke(QuickFixWrapper.java:73)
at com.intellij.codeInsight.intention.impl.IntentionActionWithTextCaching$MyIntentionAction.invoke(IntentionActionWithTextCaching.java:179)
at com.intellij.codeInsight.intention.impl.ShowIntentionActionsHandler.lambda$invokeIntention$4(ShowIntentionActionsHandler.java:214)
at com.intellij.openapi.application.WriteAction.run(WriteAction.java:92)
at com.intellij.codeInsight.intention.impl.ShowIntentionActionsHandler.invokeIntention(ShowIntentionActionsHandler.java:216)
at com.intellij.codeInsight.intention.impl.ShowIntentionActionsHandler.lambda$null$2(ShowIntentionActionsHandler.java:191)
at com.intellij.openapi.application.TransactionGuardImpl.runSyncTransaction(TransactionGuardImpl.java:82)
at com.intellij.openapi.application.TransactionGuardImpl.submitTransactionAndWait(TransactionGuardImpl.java:148)
at com.intellij.codeInsight.intention.impl.ShowIntentionActionsHandler.lambda$chooseActionAndInvoke$3(ShowIntentionActionsHandler.java:190)
at com.intellij.openapi.command.impl.CoreCommandProcessor.executeCommand(CoreCommandProcessor.java:220)
at com.intellij.openapi.command.impl.CoreCommandProcessor.executeCommand(CoreCommandProcessor.java:178)
at com.intellij.openapi.command.impl.CoreCommandProcessor.executeCommand(CoreCommandProcessor.java:168)
at com.intellij.openapi.command.impl.CoreCommandProcessor.executeCommand(CoreCommandProcessor.java:154)
at com.intellij.codeInsight.intention.impl.ShowIntentionActionsHandler.chooseActionAndInvoke(ShowIntentionActionsHandler.java:189)
at com.intellij.codeInsight.intention.impl.IntentionListStep.lambda$applyAction$0(IntentionListStep.java:107)
at com.intellij.openapi.application.TransactionGuardImpl.performUserActivity(TransactionGuardImpl.java:192)
at com.intellij.ui.popup.AbstractPopup.lambda$dispose$13(AbstractPopup.java:1428)
at com.intellij.util.ui.UIUtil.invokeLaterIfNeeded(UIUtil.java:2464)
at com.intellij.ide.IdeEventQueue.ifFocusEventsInTheQueue(IdeEventQueue.java:168)
at com.intellij.ide.IdeEventQueue.executeWhenAllFocusEventsLeftTheQueue(IdeEventQueue.java:120)
at com.intellij.openapi.wm.impl.FocusManagerImpl.doWhenFocusSettlesDown(FocusManagerImpl.java:161)
at com.intellij.openapi.wm.impl.IdeFocusManagerImpl.doWhenFocusSettlesDown(IdeFocusManagerImpl.java:58)
at com.intellij.ui.popup.AbstractPopup.dispose(AbstractPopup.java:1424)
at com.intellij.ui.popup.WizardPopup.dispose(WizardPopup.java:162)
at com.intellij.ui.popup.list.ListPopupImpl.dispose(ListPopupImpl.java:318)
at com.intellij.openapi.util.Disposer$1.execute(Disposer.java:47)
at com.intellij.openapi.util.Disposer$1.execute(Disposer.java:43)
at com.intellij.openapi.util.objectTree.ObjectNode$1.execute(ObjectNode.java:135)
at com.intellij.openapi.util.objectTree.ObjectNode$1.execute(ObjectNode.java:104)
at com.intellij.openapi.util.objectTree.ObjectTree.executeActionWithRecursiveGuard(ObjectTree.java:194)
at com.intellij.openapi.util.objectTree.ObjectNode.execute(ObjectNode.java:104)
at com.intellij.openapi.util.objectTree.ObjectTree.executeAll(ObjectTree.java:142)
at com.intellij.openapi.util.Disposer.dispose(Disposer.java:136)
at com.intellij.openapi.util.Disposer.dispose(Disposer.java:132)
at com.intellij.ui.popup.WizardPopup.disposeAllParents(WizardPopup.java:263)
at com.intellij.ui.popup.list.ListPopupImpl.handleNextStep(ListPopupImpl.java:453)
at com.intellij.ui.popup.list.ListPopupImpl._handleSelect(ListPopupImpl.java:407)
at com.intellij.ui.popup.list.ListPopupImpl.handleSelect(ListPopupImpl.java:348)
at com.intellij.ui.popup.list.ListPopupImpl$1.actionPerformed(ListPopupImpl.java:261)
at com.intellij.ui.popup.WizardPopup.proceedKeyEvent(WizardPopup.java:378)
at com.intellij.ui.popup.WizardPopup.dispatch(WizardPopup.java:358)
at com.intellij.ui.popup.PopupDispatcher.dispatchKeyEvent(PopupDispatcher.java:112)
at com.intellij.ui.popup.PopupDispatcher.dispatch(PopupDispatcher.java:148)
at com.intellij.ide.IdePopupManager.dispatch(IdePopupManager.java:93)
at com.intellij.ide.IdeEventQueue._dispatchEvent(IdeEventQueue.java:714)
at com.intellij.ide.IdeEventQueue.lambda$dispatchEvent$8(IdeEventQueue.java:412)
at com.intellij.openapi.progress.impl.CoreProgressManager.computePrioritized(CoreProgressManager.java:704)
at com.intellij.ide.IdeEventQueue.dispatchEvent(IdeEventQueue.java:411)
at java.desktop/java.awt.EventDispatchThread.pumpOneEventForFilters(EventDispatchThread.java:203)
at java.desktop/java.awt.EventDispatchThread.pumpEventsForFilter(EventDispatchThread.java:124)
at java.desktop/java.awt.EventDispatchThread.pumpEventsForHierarchy(EventDispatchThread.java:113)
at java.desktop/java.awt.EventDispatchThread.pumpEvents(EventDispatchThread.java:109)
at java.desktop/java.awt.EventDispatchThread.pumpEvents(EventDispatchThread.java:101)
at java.desktop/java.awt.EventDispatchThread.run(EventDispatchThread.java:90)
```
Answers:
username_1: Thanks for the report!
This was already reported in #950 and is fixed by #953 so will be included in 0.6.7.
If you want you can install the latest alpha build which includes the fix by going to https://plugins.jetbrains.com/plugin/9473-texify-idea/versions and clicking on Alpha, then click the download button for the latest version.
You can also [subscribe to alpha releases](https://github.com/Hannah-Sten/TeXiFy-IDEA/wiki/Alpha-builds#subscribing-to-the-alpha-channel).
Status: Issue closed
username_0: Thanks for the heads up. Will check out the alpha release then. |
RyanKung/stack | 156319966 | Title: Do not overwrite my setup
Question:
username_0: The `stack init` command has downloaded and installed some dev version of pip (that I do not want).
Now if I run any `pip[2|3] --version` I get the dev version. I didn't want the dev version, why did you change it without asking me?
Now if I run `mkvirtualenv test` I am given python3.5 - I don't want python3.5 by default, why did you change it without asking me? `VIRTUALENVWRAPPER_PYTHON` is `/usr/local/bin/python` still. `/usr/local/bin/python --version` is `2.7.11`. Why are you giving me python3 now when I create a virtualenv?
Answers:
username_1: @username_0 Thanks for your response
* about `dev version pip`
`Stack` will install the `dev version` of `pip` which I forked from the origin pip. Because there is some hidden bug on `pip._vendor.distlib`. When calling a command with `os.system`, `subprocess.call` or `fabric.api.local`, it will add wrong `shabang` on the `executable` files, such as `nosetests` or `scafford`.
**And `YES`, I should ask you before change it.**
ref :
https://github.com/pypa/pip/issues/3649
https://github.com/pypa/pip/pull/3650
* about virtualenv
`Stack` won't change your virtualenv setting, It just installed latest virtualenv (15.0.1 for now), and the default setting of that venv is python3.
username_1: Hi @username_0
`pip install pip --upgrade`
You may need run this to fix your pip, because after installed `python-stack`, the default pip was replaced to the 'forked version of pip', you need to reinstall pip for fix that, I'm so sorry for this, it's a mistake.
And he new version of `stack` will give up the `forked pip`, just using the default system pip.
New stack will scan all scripts under osconfig.get_path('scripts'), and register as tasks.
It's still unstable (I should note that), and needing more issue/pullrequest.
Thanks.
Status: Issue closed
|
acowley/Frames | 316956229 | Title: Header order independent data frames
Question:
username_0: It would be nice to have a programs based on a type with two columns based on names rather than order. That is, one file with the header "h1,h2" and another file with the header "h2,h1" should both work -- the input should be name dependent, not order dependent.
Answers:
username_1: I want this, too. Off the top of my head, I think the way you could do this today is to `rcast` a record value you get (from a callee of your function or the TH stuff) to the order you want.
username_0: Unfortunately that would still require the same input file format. |
atulbhats/atulbhats.com-terminal-version | 409013032 | Title: It's lacking the license file
Question:
username_0: I want to edit this code for, but may not only personal use.
BUT!
It's lacking the license which means you have all rights reserved, for the sake of open source and our kin, please license it under an opensource license... |
JuliaArrays/StaticArrays.jl | 267104211 | Title: Restriction on sizes in matrix-matrix multiplication.
Question:
username_0: ```julia
sa99 = rand(SMatrix{9,9,Float64,9*9})
sa99'*sa99
```
all ok, but
```julia
sa98 = rand(SMatrix{9,8,Float64,9*8})
sa98'*sa98
```
gives
`ERROR: No precise constructor for SVector{8,Float64} found. Length of input was 9.`
Answers:
username_1: Oops, the medium sized non-square case wasn't covered in the tests. I think I've got a fix.
Status: Issue closed
|
goodjoblife/GoodJobShare | 314677032 | Title: GA page view 未正常搜集資訊
Question:
username_0: https://github.com/goodjoblife/GoodJobShare/commit/9fef7a86316670ce6def4d17f3778dc83b825c3a#diff-9b543e7a3cf09c571ce437f3f8b7b4f9
router 更新的時候忘記 onUpdate 要 call logPageView
Answers:
username_1: 原 PR https://github.com/goodjoblife/GoodJobShare/pull/411/files#diff-9b543e7a3cf09c571ce437f3f8b7b4f9L39
Status: Issue closed
|
openstreetmap/iD | 627722098 | Title: Add shop=trophy preset
Question:
username_0: https://wiki.openstreetmap.org/wiki/Tag:shop=trophy
Don't know if there is a suitable icon, couldn't see a trophy from a brief look at the maki icon set.
Status: Issue closed
Answers:
username_1: Maki icons are focused on traditional map features. For general objects like this we tend to have better luck with [Font Awesome](https://fontawesome.com/) (or drawing our own). |
baudren/montepython_public | 103842446 | Title: Import error of cosmohammer modules
Question:
username_0: ```
Here is the error message from montepython.
```python
$ python montepython/MontePython.py run -p input/lcdm.ini -o chains/test -m CH
run
Running Monte Python v2.1.4
with CLASS v2.4.3
Testing likelihoods for:
-> fake_planck_bluebook
Traceback (most recent call last):
File "montepython/MontePython.py", line 40, in <module>
sys.exit(run())
File "/home/osatokn/work/montepython/montepython/run.py", line 44, in run
sampler.run(cosmo, data, command_line)
File "/home/osatokn/work/montepython/montepython/sampler.py", line 49, in run
import cosmo_hammer as hammer
File "/home/osatokn/work/montepython/montepython/cosmo_hammer.py", line 25, in <module>
from cosmoHammer.likelihood.chain.LikelihoodComputationChain import (
ImportError: No module named likelihood.chain.LikelihoodComputationChain
```
Many thanks,
Ken |
miyamotok0105/html_css_sample | 565922819 | Title: 勉強
Question:
username_0: 108 Responsive & Free HTML Website Templates 2020
https://colorlib.com/wp/free-html-website-templates/
https://colorlib.com/wp/template/consulotion/
30 Helpful PSD to HTML and CSS TutorialsBy
http://streetsmash.com/blog/2013/12/17/psd-to-html-and-css-tutorials/
MD Bootstrap 5-minute Quick Start
https://mdbootstrap.com/education/bootstrap/quick-start/
HTML Templates
https://www.quackit.com/html/templates/
Learn CSS Layout
https://learnlayout.com/
XD Landing Page
https://xdresources.co/resources/landing-page
XD video(Adobe XDとPhotoshopによるランディングページのエクスペリエンスのデザイン)
https://helpx.adobe.com/jp/xd/how-to/landing-page-in-xd-photoshop.html
XD sample
https://www.behance.net/gallery/47599819/Freebie-Start-up-Landing-Page-for-Adobe-Xd
https://www.behance.net/gallery/90368513/Development-Co-website-UI
https://www.behance.net/ |
Keats/rust-bcrypt | 259671319 | Title: rustc-serialize is deprecated
Question:
username_0: The `rustc-serialize` crate is [deprecated](https://github.com/rust-lang-deprecated/rustc-serialize), and this crate should be migrated to use [`base64`](https://github.com/alicemaz/rust-base64) instead
Answers:
username_1: Uploaded 0.1.4 which uses base64 instead, thanks!
Sadly rust-crypto is not maintained too but I can't find another implementation of bcrypt
Status: Issue closed
|
rayrutjes/php-geteventstore-client | 181900512 | Title: HttpClient loads all events from a stream in memory at once
Question:
username_0: Thanks for this library! I haven't test it extensively yet but it seems very promising.
However, I did find one issue so far: Even though EventRecordCollection implements \Iterator, and is actually a decorator around an "inner" iterator, the object is created using an array which forces HttpClient to load all events from a stream into memory:
https://github.com/username_1/php-geteventstore-client/blob/master/src/Client/Http/HttpClient.php#L143-L148
This can potentially cause issues for very long streams.
I think this could be fixed by adding a fromEventStreamIterator() method on EventRecordCollection which would simply inject the EventStreamIterator and set it as the "inner" iterator:
```
public static function fromEventStreamIterator(EventStreamIterator $eventStreamIterator) {
return new self($eventStreamIterator);
}
```
Answers:
username_1: Hi @username_0 !
Thx a lot for suggesting, unfortunately it won't work because `EventStreamIterator ` is not `Countable` so can't be used as internal iterator of the `EventRecordCollection`.
When dealing with huge stream history, a workaround would be to make snapshots.
As this library is more a proof of concept than anything else, snapshotting is not possible either ;)
I'd gladly merge your PRs if you feel moving forward with this library!
username_0: You're correct, forgot about that.
I may be mistaken but aren't event store snapshots primarily used for fixing performance issues on the write side, when rebuilding an event-sourced aggregate from history?
My initial issue was primarily about event listeners/projectors in php on the read side which would require a complete replay if you found a bug or would have to make changes to the projector logic.
Maybe using Generators instead of Iterators could fix that?
username_1: @username_0 ,
An event store is an append only database. Performance is not the reason you should go with Event Sourcing. An event store ensures consistency, and holds all the aggregates mutations.
Snapshots are indeed used for the use case you described.
This lib might require some work to make it more production ready ;)
username_0: I'm not that concerned about the performance of writing to the store or even loading specific aggregate streams, but more when reading all events from `$by_category` streams to replay projectors and rebuilding the query/read models for multiple aggregate streams. I guess we're in agreement about that but some things are getting lost in translation :)
I'm planning on using this library in a prototype I'm building for an app, I can look into fixing the read issue by using Generators and submitting a PR sometime, if that's okay with you.
Do you plan on actively maintaining this library over time or was this more as a proof-of-concept / reference library with no plans to make it production ready? |
microsoft/PowerToys | 854537090 | Title: "ctrl + =" -> "prt" Not working
Question:
username_0: ### Microsoft PowerToys version
0.35.0
### Running as admin
Yes
### Area(s) with issue?
Keyboard Manager
### Steps to reproduce
"ctrl + =" -> prt
Not working
### ✔️ Expected Behavior
"ctrl + =" -> prt
working
### ❌ Actual Behavior
"ctrl + =" -> prt
Not working
### Other Software
_No response_
Answers:
username_1: @username_0
please post a screenshot of the remapping. |
bitfinexcom/bitfinex-api-py | 562351869 | Title: V2 Public Rest Endpoint Stats is not available through the library
Question:
username_0: #### Issue type
- [ ] bug
- [x] missing functionality
- [ ] performance
- [ ] feature request
#### Brief description
Public Rest Stats endpoint is not support: https://docs.bitfinex.com/reference#rest-public-stats1
#### Steps to reproduce
- Attempt to access stats endpoint
##### Additional Notes:
-
Answers:
username_1: Thanks @username_0 - we will add this to our TODO list |
status-im/nim-bncurve | 364980965 | Title: Code review
Question:
username_0: # arith.nim
1.
https://github.com/status-im/nim-bncurve/blob/ea38d7b5b8441376e6c1b170cf48d6939e6a2ef4/bncurve/arith.nim#L18-L21
It seems like randomBytes for openarray is returning the number of elements in the openarray instead of the number of bytes set, naively I would expect the number of bytes set.
2. noinit is not needed if there is no return value :
https://github.com/status-im/nim-bncurve/blob/ea38d7b5b8441376e6c1b170cf48d6939e6a2ef4/bncurve/arith.nim#L18
https://github.com/status-im/nim-bncurve/blob/ea38d7b5b8441376e6c1b170cf48d6939e6a2ef4/bncurve/arith.nim#L27
https://github.com/status-im/nim-bncurve/blob/ea38d7b5b8441376e6c1b170cf48d6939e6a2ef4/bncurve/arith.nim#L34
Furthermore following https://github.com/nim-lang/Nim/issues/8745, it shouldn't be needed anymore for ref objects, and for stack I think we always want to zero-init in crypto (and there are some GCC/LLVM flag to enforce that in C/C++).
3. https://github.com/status-im/nim-bncurve/blob/ea38d7b5b8441376e6c1b170cf48d6939e6a2ef4/bncurve/arith.nim#L41-L43
Can be:
```
proc zero*(t: typedesc[BNU256]): BNU256 {.inline.} =
## Return zero 256bit integer.
discard
```
4. setBit, to be tested but here are 2 constant-time alternatives (w.r.t. a) if needed
From:
```Nim
proc setBit*(a: var openarray[uint64], n: int,
to: bool): bool {.inline, noinit.} =
## Set bit of integer ``a`` at position ``n`` to value ``to``.
if n >= 256:
return
let part = n shr 6
let bit = n - (part shl 6)
if to:
a[part] = a[part] or (1'u64 shl bit)
else:
a[part] = a[part] and not(1'u64 shl bit)
result = true
```
to
```Nim
func setBitAlt1(a: var BNU256, n: int, to: bool): bool {.inline.}=
if n >= 256:
return
let
index = n and 63
part = n shr 6
value = to.uint64
a[part] = a[part] and not(1'u64 shl index) or (value shl index)
result = true
```
Or
```Nim
template `-`(x: uint64): uint64 =
x.not + 1
func setBitAlt2(a: var BNU256, n: int, to: bool): bool {.inline.}=
if n >= 256:
return
let
index = n and 63
part = n shr 6
value = to.uint64
a[part] = a[part] xor ((-value xor a[part]) and (1'u64 shl index))
result = true
```
5. Not sure what macDigit is, I suppose it's part of Montgomery reduction?
# fp, groups, fq2, fq6, fq12
Too unfamiliar at the moment for review.
# Final thoughs
Great job!
Answers:
username_1: Points `1, 2, 3, 4` has been fixed in https://github.com/status-im/nim-bncurve/pull/2
Status: Issue closed
|
iron-io/docs | 8003144 | Title: Combine all /worker/languages pages
Question:
username_0: All /worker/languages pages should use the same template, and we should just be plugging in words, phrases, and code samples. There's no reason to be maintaining half a dozen versions of the same exact page.
Ideally, I'd like to remove all those pages, and just include the information they had in /worker, with a language switcher--it seems like we have two "Getting Started" guides in Dev. I'd like to only have one. Thoughts on that? @frommww? @carimura? @dstaley?<issue_closed>
Status: Issue closed |
awslabs/aws-lambda-rust-runtime | 581792966 | Title: Fails when used with httpApi gateway
Question:
username_0: When using a lambda with [HTTP API Gateways](https://aws.amazon.com/blogs/compute/announcing-http-apis-for-amazon-api-gateway/) it fails to parse the incoming event. I think it's because http apis don't pass a resource id, so the deserializing fails. Maybe something like this would fix it: https://github.com/username_0/aws-lambda-rust-runtime/commit/b0091cb21fa38e9f96b91370ce711dd29854c980?
You can repro by using something like the following in a serverless.yml config:
```
...
functions:
fooFn:
handler: fooHandler
events:
- httpApi:
path: /
method: GET
```
Answers:
username_1: Yeah, I'm sorry about that this not working and for the delay in responding. The API Gateway HTTP APIs introduced a new format for HTTP requests and responses which will require some work to properly support, beyond just the resource ID change. Beyond the changing the format in serde, the multivalue headers map has gone away (https://aws.amazon.com/blogs/compute/building-better-apis-http-apis-now-generally-available/?nc1=b_rp), which makes parsing some headers harder to do correctly. I've been trying to avoid thinking too hard about it :P
username_2: I've been MIA for a while I'd be interested in picking this up again. I also noticed there's a v2 req/resp format. I can do some exploration on this
username_0: Nice! Let me know if there's anything I can do to help
username_3: Does anyone know of the best way to get around this for now?
username_2: @username_0 wanted to keep you updated. Progress is being made
https://github.com/awslabs/aws-lambda-rust-runtime/pull/217
@username_3 It depends on what version you are using. Api gateway REST API and ALB triggers are known to work. API gateway HTTP APIs are not yet but will soon be supported. See link above. In the meantime, you can always define a custom deserializable for all trigger types for a fallback.
username_4: Using master, I just define a `lambda::Handler`, receive the json value, and deserialize using `aws_lambda_events`. I then process it and serialize it back to json so it works with handler.
Works well with Http api.
Status: Issue closed
username_5: Looks like the PR that addresses this was merged in very recently. Let us know if that presents issues. Thanks! |
frontendbr/vagas | 300734840 | Title: [Sorocaba/SP] Fullstack Developer na Goomer
Question:
username_0: ## Descrição da vaga
A Goomer é uma start-up de Sorocaba que trabalha com tecnologias de auto atendimento para restaurantes. Temos 3 anos de vida e já ajudamos os restaurantes no atendimento de 6,5 milhões de pedidos. Temos a missão de melhorar a experiência de consumo dos restaurantes do Brasil.
A vaga é para desenvolvedor full stack com foco em PHP (Laravel), programação web (html, css e javascript) e Node.js
## Local
Sorocaba/SP
## Benefícios
- Vale Refeição
- Vale Transporte
- Cursos
- Horário flexível
- Stock Option (participação na empresa)
## Requisitos
**Obrigatórios:**
- PHP
- Javascript
- HTML e CSS
**Desejáveis:**
- Conhecimentos em Laravel
- Conhecimentos de Node.js
**Diferenciais:**
- AngularJS, Ractive.js ou Vue.js
## Contratação
CLT ou PJ (a combinar)
## Como se candidatar
Inscreva-se pelo link: https://goo.gl/forms/wsY6rjZ1re4IVGZY2
## Labels
- CLT
- Pleno
- Remoto - Parcial
Answers:
username_1: Boa tarde. Existe chance de remoto total?
username_0: Boa tarde @username_1 infelizmente para essa vaga ainda não. Nós gostamos de envolver todos (desde designers, desenvolvedores, galera de vendas, customer success) no negócio. Todos participam das decisões, por isso a necessidade de estar presente pelo menos em alguns dias da semana.
username_1: Que pena. Obrigada :smile:
username_2: Essa vaga já está inativa por mais dias do que o permitido. Caso ainda tenha a vaga, reabra uma nova issue.
Mais informações: https://github.com/frontendbr/vagas#importante
Status: Issue closed
|
markitosgv/JWTRefreshTokenBundle | 568276616 | Title: Auto refresh with lexik
Question:
username_0: It appear that the problem is already reported but closed here [https://github.com/markitosgv/JWTRefreshTokenBundle/issues/147](url).
But my question is less specific.
In fact I would like to split this solution in 3 cases:
1. Request with not expired token => everything is fine
2. Request with expired token but refresh_token is in headers (or cookie as you want), so refresh action is call.
3. Request with all token expired, return 401.
I would like to know if any better solution exist, or if #147 is the only way ?
Finally, I find this features very perfect because we can control jwt and stay stateless, and if jwt is not enough we can control user integrity with refresh_token in database (not stateless), so, nothing suggests such an implementation?
Thanks for your time and your mercy 🙏
Answers:
username_1: I see what you mean is "automation" but not "stateless". I don't think you need refresh token anymore cause you can just persist your jwt into database and when it become expired, just generate a new one, save it to database and return to user (so frontend just save the new jwt instead).
username_2: I think it would be better to have the client periodically explicitly refresh the JWT (by hitting the `/refresh` endpoint with a valid `refresh_token`) rather than have the server do this 'under the hood' |
uqfoundation/pathos | 438714649 | Title: AttributeError: 'int' object has no attribute 'encode'
Question:
username_0: 
In the picture , I showed you how I used multiprocessing to 'run'. The error which I get is
Exception in thread Thread-3:
Traceback (most recent call last):
File "/usr/lib/python2.7/threading.py", line 801, in __bootstrap_inner
self.run()
File "/usr/lib/python2.7/threading.py", line 754, in run
self.__target(*self.__args, **self.__kwargs)
File "/usr/lib/python2.7/multiprocessing/pool.py", line 392, in _handle_results
task = get()
File "/home/ubuntu/.local/lib/python2.7/site-packages/mysql/connector/errors.py", line 194, in __init__
'msg': self.msg.encode('utf8') if PY2 else self.msg
AttributeError: 'int' object has no attribute 'encode'
Details:
OS : ubuntu 18.04
If you need any additional details, please ask me<issue_closed>
Status: Issue closed |
redwoodjs/redwood | 1124230725 | Title: API Testing - Scenario Types
Question:
username_0: The scenario generator works great, and even has basic typescript support!
However, when functions are embedded into the scenario creation object to support relations, the redwood typescript fails. For example, I'm trying to set up the following scenario:
```
export const standard = defineScenario ({
organization: {
orgA: (): Prisma.OrganizationCreateArgs => ({
data: {
name: 'Org A',
},
}),
},
user: {
userA: (scenario): Prisma.UserCreateArgs => ({
data: {
nameFirst: 'Bob',
nameLast: 'Builder',
email: '<EMAIL>',
phone: '123-456-7890',
title: 'boss',
roles: 'abc,def',
active: true,
uid: 'aaa',
organizationId: scenario.organization.orgA.id,
},
}),
},
project: {
projectA: (): Prisma.ProjectCreateArgs => ({
data: {
name: 'Project 1',
description: 'Project 1 desc',
owner: 'Project 1 Owner',
},
}),
},
milestone: {
one: (scenario): Prisma.MilestoneCreateArgs => ({
data: {
name: 'String',
createdById: scenario.user.userA.id,
dateTargetActive: true,
dateTarget: '2021-06-07T21:02:43Z',
dateActual: '2021-06-07T21:02:43Z',
projectId: scenario.project.projectA.id,
},
}),
two: (scenario): Prisma.MilestoneCreateArgs => ({
data: {
name: 'String',
createdById: scenario.user.userA.id,
dateTargetActive: true,
dateTarget: '2021-06-07T21:02:43Z',
dateActual: '2021-06-10T21:02:43Z',
projectId: scenario.project.projectA.id,
},
}),
},
[Truncated]
export type StandardScenario = {
organization: {
orgA: Organization
}
user: {
userA: User
}
project: {
projectA: Project
}
milestone: {
one: Milestone
two: Milestone
}
}
```
If one of our typescript gurus could find an automatic way of typing the `StandardScenario` type from the `standard` definition , that would be awesome! I took a stab at it, but this goes past my current Typescript knowledge! If for some reason this proves challenging, then at a minimum we should probably update the docs regarding the typescript support.
Thanks!
Answers:
username_1: @username_0 just to confirm, in your example, the scenario is for your `Organization` model (e.g. the file's called `organization.scenarios.ts` and `defineScenario` initially had a Prisma generic passed to it, like `defineScenario<Prisma.OrganizationCreateArgs>`), but since you're creating `User`s and `Project`s in the same scenario (which is totally allowed!) the typing breaks down in two ways:
1. The relation functions aren't being appropriately typed (which is why you have to be explicit about it in your example above)
2. The exported type `StandardScenario` is wrong
username_0: @username_1, thanks for looking into this!
Yes, I think you got it. I'll clarify a bit more, numbered as per your comments:
1. By default, the scaffold generator creates a scenario using the following line:
`export const standard = defineScenario<Prisma.ModelCreateArgs>({`
As we need to create a multi-model scenario, this definition is no longer correct. It's easy enough to get rid of the type on the `defineScenario` function and type each function as shown in my example above to make sure that the entry is typed correctly. It would be great if the scaffold generator could do this by default, but this would be the lower priority of the two issues in my mind, but it may be the easier issue to change.
2. The type of the exported `StandardScenario` would be for me the higher priority. When using a multi-model scenario, I had to completely replace redwoods automatically generated `StandardScenario` with a manual one, as shown in the original post. It would be great if we could create a generic type system to create the StandardScenario type. The trick is that the resulting objects under the user property for example are no longer a `Prisma.UserCreateArgs`, but rather a prisma `User` type.
Other than for the most basic apps, I believe that multi-model scenarios are the most common use case. Note that I've also stitched together multiple model scenario files as follows:
```
import { standard as milestoneStandard } from ....
export const standard = {
...milestoneStandard,
requirement: {...}
}
```
Does that clarify things?
username_1: @username_0 yes it does thank you very much! |
ThomasCassimon/SignalProcessing | 282212545 | Title: Fix error when pushing cancel when loading data
Question:
username_0: When pressing cancel when loading data you get an error because there is no data in the dataset. (I tried fixing this, you only need to fix a default value for dataset and sampleFreq if cancel has been pressed)
Answers:
username_1: Fixed.
Status: Issue closed
|
vintasoftware/django-react-boilerplate | 560459825 | Title: Error: EISDIR: illegal operation on a directory, open 'webpack-stats.json'
Question:
username_0: **Describe the bug**
webpack-stats.json appears to be a directory, but something is attempting to open it as a file.
```
frontend_1 | Error: EISDIR: illegal operation on a directory, open 'webpack-stats.json'
frontend_1 | at Object.openSync (fs.js:457:3)
frontend_1 | at Object.writeFileSync (fs.js:1298:35)
frontend_1 | at Plugin.writeOutput (/app/node_modules/webpack-bundle-tracker/lib/index.js:124:6)
frontend_1 | at compile (/app/node_modules/webpack-bundle-tracker/lib/index.js:58:10)
frontend_1 | at SyncHook.eval (eval at create (/app/node_modules/tapable/lib/HookCodeFactory.js:19:10), <anonymous>:5:1)
frontend_1 | at SyncHook.lazyCompileHook (/app/node_modules/tapable/lib/Hook.js:154:20)
frontend_1 | at /app/node_modules/webpack/lib/Compiler.js:665:23
frontend_1 | at AsyncSeriesHook.eval [as callAsync] (eval at create (/app/node_modules/tapable/lib/HookCodeFactory.js:33:10), <anonymous>:4:1)
frontend_1 | at AsyncSeriesHook.lazyCompileHook (/app/node_modules/tapable/lib/Hook.js:154:20)
frontend_1 | at Compiler.compile (/app/node_modules/webpack/lib/Compiler.js:662:28) {
frontend_1 | errno: -21,
frontend_1 | syscall: 'open',
frontend_1 | code: 'EISDIR',
frontend_1 | path: 'webpack-stats.json'
frontend_1 | }
```
In the backend:
```
backend_1 | Internal Server Error: /
backend_1 | Traceback (most recent call last):
backend_1 | File "/usr/local/lib/python3.6/site-packages/webpack_loader/loader.py", line 26, in _load_assets
backend_1 | with open(self.config['STATS_FILE'], encoding="utf-8") as f:
backend_1 | IsADirectoryError: [Errno 21] Is a directory: '/home/user/app/../webpack-stats.json'
backend_1 |
...
```
**To Reproduce**
Steps to reproduce the behavior:
1. Install following README instructions
2. Run docker-compose up
3. Look at the logs
**Expected behavior**
There should be no errors when following the setup instructions any modification.
**Screenshots**

**Desktop (please complete the following information):**
- OS: Mac OS Catalina
- Browser [e.g. chrome, safari] Chrome
- Version [e.g. 22]
Answers:
username_1: Hi @username_0, thanks for bringing this issue. We're working on a fix now.
This happens because docker tries to bind `webpack-stats.json` on backend before this file is created by the frontend, so it ends up creating a directory. A workaround while we don't push a fix is:
1. Remove the directory created by docker: `rm -rf webpack-stats.json/`
1. Create this file manually: `echo {} >> webpack-stats.json`
1. Run `docker-compose up`
If you check `webpack-stats.json`, it should have a new content with info about the bundle.
username_0: Thanks for the response, but it doesn't seem to work:
```
$ rm -rf webpack-stats.json/
$ echo {} >> webpack-stats.json
$ docker-compose up
Starting frontend_1 ... done
Starting db_1 ... done
Starting broker_1 ... done
Starting result_1 ... done
Starting celery_1 ...
Starting backend_1 ... error
ERROR: for backend_1 Cannot start service backend: OCI runtime create failed: container_linux.go:346: starting container process caused "process_linux.go:449: container init caused \"rootfs_linux.go:58: mounting \\\"/.../webpack-stats.json\\\" to rootfs \\\"/var/lib/docker/overlay2/78fe24f7b5a9851d7b0a430a0e046a7135e9ef8e7a83700ffee535Starting celery_1 ... done
Are you trying to mount a directory onto a file (or vice-versa)? Check if the specified host path exists and is the expected type
ERROR: for backend Cannot start service backend: OCI runtime create failed: container_linux.go:346: starting container process caused "process_linux.go:449: container init caused \"rootfs_linux.go:58: mounting \\\"/.../webpack-stats.json\\\" to rootfs \\\"/var/lib/docker/overlay2/78fe24f7b5a9851d7b0a430a0e046a7135e9ef8e7a83700ffee535faf632c3d7/merged\\\" at \\\"/var/lib/docker/overlay2/78fe24f7b5a9851d7b0a430a0e046a7135e9ef8e7a83700ffee535faf632c3d7/merged/home/user/webpack-stats.json\\\" caused \\\"not a directory\\\"\"": unknown: Are you trying to mount a directory onto a file (or vice-versa)? Check if the specified host path exists and is the expected type
ERROR: Encountered errors while bringing up the project.
```
username_1: @username_0 Got it, seems like we still have something from the `webpack-stats.json` directory in this container.
Does `ls -d */ | grep webpack` return anything? If yes, the removal probably didn't work. Does it break if you try to build the frontend and backend again, then try to run them?
username_0: Thanks, building it again helped!
Status: Issue closed
|
aleksaan/diskusage | 394761799 | Title: Network Share Folders Size?
Question:
username_0: 



using "diskusage.exe" i get size ~44 GB
using "network share" from another PC I get ~20 GB
If I get properties directly on respective server I get ~15 GB
Could you please explain me , how exactly size is being calculated by diskusage.exe and why is that different from other two
Answers:
username_1: Hi, <NAME>
I suppose two possibilities:
1) symlink from your disk C:\ to another disk (I see D: on your screenshot)
2) some access rights problem in diskusage and how it works with remote disk
I'm starting to test that case.
username_0: 1. Symlink? You mean Mount ? D: C: are on my PC not on remote PC/Server for which I'm trying to get Diskusage. So, ignore D:.
My concern was How to get Diskusage of network drives ?
username_1: username_0,
can you just test your case please
Status: Issue closed
username_0: Okay sure but I'll test it on 17th January 2019 cause I'm on vacation. |
graphql-go/graphql | 368448301 | Title: Supporting Path on ResolveInfo
Question:
username_0: Hi,
I saw a line in graphql-js (of Facebook) that support `Path` in `ResolveInfo`. I am not sure if we should support this? I would love to do pull request for this.
https://github.com/graphql/graphql-js/blob/c82ff68f52722c20f10da69c9e50a030a1f218ae/src/type/definition.js#L494
Thanks a lot!
Answers:
username_1: Hi @username_0, thanks for reaching us for an feature request, which was addressed via: https://github.com/graphql-go/graphql/pull/403
Closing this one via: https://github.com/graphql-go/graphql/pull/403
Status: Issue closed
|
t00/Dapper.SqlGenerator | 730128318 | Title: Create a Select(columnSet) function
Question:
username_0: If column sets are used, it would be easier to generate a full SQL SELECT statement with to retrieve a subset of columns or computed expressions from DB. Along with a table alias feature it can be used to build a filtered select query.<issue_closed>
Status: Issue closed |
phpcr/phpcr-utils | 14709565 | Title: [command] node:touch force type when setting value
Question:
username_0: Would be nice to be able to force type when setting a property, for example, (boolean) 0, (integer) 5. At the moment all are evaluated as string.
Answers:
username_1: @username_0 is this handled by phpcr-shell now?
Status: Issue closed
username_0: yep
````
PHPCRSH > node:property:set --help
Usage:
node:property:set [--type="..."] path [value]
````
I guess I will close.
username_1: should we deprecate node:touch and drop it in 2.0? |
pytorch/pytorch | 971449754 | Title: How to build a release version libtorch1.8.1 on windows
Question:
username_0: ## Issue description
I am doing some work with libtorch on Windows 10 recently. I want to build the libtorch library since I have addedd some new features. The build work was done successfully on develop environment. However, when I copy the exe(including dependent DLLs) to another PC(running env, mentioned below), it could not run normally. I have also built the corresponding python package and it could run normally on the develop environment. Also, it shows that the official release version of 1.8.1 has more libs than mine. It seems that the official version has more torch_cuda*.dll than mine and their size is relatively larger than mine.
I just want to know how to configure the env and can build nearly the same as the official one.
Self-build release version
2021/08/12 11:22 242,176 asmjit.dll
2021/08/10 21:06 92,664 asmjit.lib
2021/08/12 11:22 417,792 c10.dll
2021/08/10 21:09 303,982 c10.lib
2021/08/12 14:02 10,382,510 c10d.lib
2021/08/12 11:25 246,784 c10_cuda.dll
2021/08/10 21:09 27,942 c10_cuda.lib
2021/08/12 14:03 20,131,328 caffe2_detectron_ops_gpu.dll
2021/08/10 22:22 34,660 caffe2_detectron_ops_gpu.lib
2021/08/12 14:02 70,144 caffe2_module_test_dynamic.dll
2021/08/10 22:21 24,130 caffe2_module_test_dynamic.lib
2021/08/12 11:25 15,872 caffe2_nvrtc.dll
2021/08/10 21:09 1,850 caffe2_nvrtc.lib
2021/08/12 11:21 18,316 clog.lib
2021/08/12 11:21 118,076 cpuinfo.lib
2021/08/12 11:25 323,147,004 dnnl.lib
2021/08/12 11:22 3,282,432 fbgemm.dll
2021/08/10 21:06 1,156,374 fbgemm.lib
2021/08/12 11:22 15,456,424 gloo.lib
2021/08/12 11:22 36,464,038 gloo_cuda.lib
2021/08/12 14:00 135,168 jitbackend_test.dll
2021/08/10 22:19 21,206 jitbackend_test.lib
2015/01/22 09:23 1,146,272 libiomp5md.dll
2021/08/12 11:20 5,220,954 libprotobuf-lite.lib
2021/08/12 11:21 36,811,506 libprotobuf.lib
2021/08/12 11:21 38,708,546 libprotoc.lib
2021/08/12 11:25 323,147,004 mkldnn.lib
2021/08/12 11:21 142,874 pthreadpool.lib
2021/08/12 13:59 9,728 torch.dll
2021/08/10 22:18 1,832 torch.lib
2021/08/12 14:00 339,456 torchbind_test.dll
2021/08/10 22:19 21,154 torchbind_test.lib
2021/08/12 12:07 201,914,368 torch_cpu.dll
2021/08/10 21:41 17,095,310 torch_cpu.lib
2021/08/12 13:59 149,539,840 torch_cuda.dll
2021/08/12 13:58 3,024,940 torch_cuda.lib
2021/08/12 11:22 9,728 torch_global_deps.dll
2020/09/18 19:02 195,072 uv.dll
2021/08/12 11:21 5,984,034 XNNPACK.lib
Official release version
2021/03/24 11:07 241,664 asmjit.dll
2021/03/24 11:07 92,664 asmjit.lib
2021/03/24 11:08 418,304 c10.dll
2021/03/24 11:08 303,982 c10.lib
2021/03/24 12:44 10,209,446 c10d.lib
2021/03/24 11:08 249,344 c10_cuda.dll
2021/03/24 11:08 27,942 c10_cuda.lib
2021/03/24 12:47 20,971,008 caffe2_detectron_ops_gpu.dll
2021/03/24 12:47 34,660 caffe2_detectron_ops_gpu.lib
2021/03/24 12:45 69,632 caffe2_module_test_dynamic.dll
2021/03/24 12:45 24,130 caffe2_module_test_dynamic.lib
2021/03/24 11:08 15,872 caffe2_nvrtc.dll
2021/03/24 11:08 1,850 caffe2_nvrtc.lib
2021/03/24 11:07 18,300 clog.lib
[Truncated]
[conda] numpy 1.17.0 py36h19fb1c0_0
[conda] numpy-base 1.17.0 py36hc3f5095_0
- PyTorch or Caffe2: Pytorch
- How you installed PyTorch (conda, pip, source): source
- Build command you used (if compiling from source):
conda create -n torch-build python=3.6
conda activate torch-build
conda install astunparse numpy ninja pyyaml mkl mkl-include setuptools cmake cffi typing_extensions future six requests dataclasses
conda install -c conda-forge libuv=1.39
mkdir build_libtorch && cd build_libtorch
python ../tools/build_libtorch.py
- OS: Windows 19042.1165
- PyTorch version: 1.8.1
- Python version: 3.6
- CUDA/cuDNN version: 11.1/ 8.0
- GPU models and configuration:
- GCC version (if compiling from source):
- CMake version: 3.21.1
- Versions of any other relevant libraries: Visual Studio 16 2019
Answers:
username_1: Well, the best way to do it is to actually use our binary packaging scripts, but for this particular case I bet it's because you aren't asking for enough SM versions for the compiled CUDA code. You'll wanna set TORCH_CUDA_ARCH_LIST to something like "6.0;6.1;7.0;7.5" or something
username_2: Official binaries are build to support all GPU architectures, while by default local build only picks architectures available locally
username_3: Feel free to continue the conversation here but going to close this issue since this is more of a question than something that's broken
Status: Issue closed
|
shakedkialy/Coderage | 925352632 | Title: Error when running the program after closing.
Question:
username_0: After installation the program works with the test examples. After closing and reopening there is a problem with the 'Results' folder. It is being created without all of the html files. That causes the bug:

The bug still occurs even after deleting the 'Results' folder and running again, while it should be ran as if it is the first time.
Answers:
username_0: Important to note that the bug accures mainly when running threw pycharm.
Status: Issue closed
username_0: the bug was solved after changing the installation to pip install -e. |
VirtualFlyBrain/VFB_reporting | 502759220 | Title: Write reporting scripts for CATMAID
Question:
username_0: @mmc46 @Robbie1977 - do you know how to get this via the API?
@Robbie1977 - do you have any existing code to share?
---
@mmc46 commented on [Wed May 15 2019](https://github.com/VirtualFlyBrain/Management/issues/190#issuecomment-492651417)
/{project_id}/annotations/query-targets
http://l1em.catmaid.virtualflybrain.org/apis/#!/annotations/annotations_query_targets_create
Try
```
project_id 1
annotated_with papers
with_annotations true
annotation_reference name
```
to get
```
curl -X POST --header 'Content-Type: application/x-www-form-urlencoded' --header 'Accept: application/json' --header 'X-CSRFToken: <KEY>' -d 'annotated_with=papers&with_annotations=true&annotation_reference=name' 'http://l1em.catmaid.virtualflybrain.org/1/annotations/query-targets'
```
and you'll see the output.
---
@jefferis commented on [Wed May 15 2019](https://github.com/VirtualFlyBrain/Management/issues/190#issuecomment-492684812)
Just a note that the CSRF token must be acquired in a separate login step in order to allow POST actions on the catmaid server. If you are happy to use the R [catmaid package](https://jefferis.github.io/rcatmaid), you can do:
```
library(catmaid)
catmaid_login(server='http://l1em.catmaid.virtualflybrain.org/')
catmaid_query_by_annotation('papers')
```
and get
```
id name type skid
1 19496665 Zwart et al. 2016 annotation NA
2 19711421 Berck, Khandelwal et al. 2016 annotation NA
3 19711583 <NAME>, Litwin-Kumar et al. 2017 annotation NA
4 19711972 Larderet, Fritsch et al. 2017 annotation NA
5 19712046 Ohyama, Schneider-Mizell et al. 2015 annotation NA
6 19712156 Jovanic, Schneider-Mizell et al. 2016 annotation NA
7 19712202 Schlegel et al. 2016 annotation NA
8 19712396 Fushiki et al. 2016 annotation NA
9 19716589 Takagi et al. 2017 annotation NA
10 19756504 Heckscher et al. 2015 annotation NA
11 19756692 Gerhard et al. 2017 annotation NA
12 19862922 Burgos et al 2018 annotation NA
13 20282757 Miroschnikow et al. 2018 annotation NA
14 20284758 "Carreira-Rosario, <NAME>, Clark et al. 2018" annotation NA
```
---
[Truncated]
[{'id': 19496665, 'name': 'Zwart et al. 2016', 'type': 'annotation'}, {'id': 19711421, 'name': 'Berck, Khandelwal et al. 2016', 'type': 'annotation'}, {'id': 19711583, 'name': '<NAME> et al. 2017', 'type': 'annotation'}, {'id': 19711972, 'name': 'Larderet, Fritsch et al. 2017', 'type': 'annotation'}, {'id': 19712046, 'name': 'Ohyama, Schneider-Mizell et al. 2015', 'type': 'annotation'}, {'id': 19712156, 'name': 'Jovanic, Schneider-Mizell et al. 2016', 'type': 'annotation'}, {'id': 19712202, 'name': 'Schlegel et al. 2016', 'type': 'annotation'}, {'id': 19712396, 'name': 'Fushiki et al. 2016', 'type': 'annotation'}, {'id': 19716589, 'name': 'Takagi et al. 2017', 'type': 'annotation'}, {'id': 19756504, 'name': 'Heckscher et al. 2015', 'type': 'annotation'}, {'id': 19756692, 'name': 'Gerhard et al. 2017', 'type': 'annotation'}, {'id': 19862922, 'name': 'Burgos et al 2018', 'type': 'annotation'}, {'id': 20282757, 'name': 'Miroschnikow et al. 2018', 'type': 'annotation'}, {'id': 20284758, 'name': '"Carreira-Rosario, <NAME>, Clark et al. 2018"', 'type': 'annotation'}]
```
```python
call_papers["annotation_reference"] = "id"
for paper in papers:
call_papers["annotated_with"] = paper["id"]
neurons = client.post("%s/%d/annotations/query-targets" % (URL,PROJECT_ID), data=call_papers, headers={"Referer": URL, "X-CSRFToken":csrftoken}).json()["entities"]
paper["neurons"] = neurons
print(papers)
```
```json
[{'id': 19496665, 'name': '<NAME>. 2016', 'type': 'annotation', 'neurons': [{'id': 4028115, 'name': 'MN5 LO1_a1l', 'type': 'neuron', 'skeleton_ids': [4028114]}, {'id': 4037552, 'name': 'A14a_a1r', 'type': 'neuron', 'skeleton_ids': [5298032]}, {'id': 4038809, 'name': 'A27k_a2l', 'type': 'neuron', 'skeleton_ids': [4038808]}, {'id': 4042316, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1l', 'type': 'neuron', 'skeleton_ids': [4042315]}, {'id': 4071033, 'name': 'A02f_a1l ', 'type': 'neuron', 'skeleton_ids': [4071032]}, {'id': 4102729, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1r ', 'type': 'neuron', 'skeleton_ids': [4102728]}, {'id': 4104477, 'name': 'T01d1_t3l', 'type': 'neuron', 'skeleton_ids': [4104476]}, {'id': 4113534, 'name': 'A01c1_a1l', 'type': 'neuron', 'skeleton_ids': [4113533]}, {'id': 4113613, 'name': 'A27k_a2r', 'type': 'neuron', 'skeleton_ids': [4113612]}, {'id': 4121536, 'name': 'MN23/24 (RF: LT3/LT4) LT4_a1r ', 'type': 'neuron', 'skeleton_ids': [4121534]}, {'id': 4124564, 'name': 'A27n_a1r', 'type': 'neuron', 'skeleton_ids': [4124563]}, {'id': 4289774, 'name': 'MN23/24 (RF: LT3/LT4) LT4_a1l', 'type': 'neuron', 'skeleton_ids': [4289773]}, {'id': 4326469, 'name': 'MN8 (RF:SBM) LT3_a1l', 'type': 'neuron', 'skeleton_ids': [4326468]}, {'id': 4535769, 'name': 'MN5 LO1_a1r', 'type': 'neuron', 'skeleton_ids': [4535767]}, {'id': 4538682, 'name': 'T03a1_t3r', 'type': 'neuron', 'skeleton_ids': [4538681]}, {'id': 4538739, 'name': 'A23a_a1l', 'type': 'neuron', 'skeleton_ids': [4538738]}, {'id': 4543282, 'name': 'A03a4_a2r', 'type': 'neuron', 'skeleton_ids': [4543281]}, {'id': 4729047, 'name': 'A23a_a1r', 'type': 'neuron', 'skeleton_ids': [4729046]}, {'id': 4881630, 'name': 'MN8 (RF:SBM) LT3_a1r ', 'type': 'neuron', 'skeleton_ids': [4881629]}, {'id': 4936150, 'name': 'A01c1_a1r', 'type': 'neuron', 'skeleton_ids': [4936149]}, {'id': 4978455, 'name': 'A19l_a1r', 'type': 'neuron', 'skeleton_ids': [4978454]}, {'id': 5078761, 'name': 'A18j_a1l', 'type': 'neuron', 'skeleton_ids': [5078760]}, {'id': 5084469, 'name': 'A01c2_ipsi_dendrite_a1l', 'type': 'neuron', 'skeleton_ids': [5084467]}, {'id': 5309933, 'name': 'A03g_a1l', 'type': 'neuron', 'skeleton_ids': [5309932]}, {'id': 5358139, 'name': 'A19l_a1l', 'type': 'neuron', 'skeleton_ids': [5358138]}, {'id': 6467482, 'name': 'MN21/22 (RF:LT1/LT2) LT2_a1l', 'type': 'neuron', 'skeleton_ids': [6467481]}, {'id': 7934160, 'name': 'A27n_a1l', 'type': 'neuron', 'skeleton_ids': [7934159]}, {'id': 8688632, 'name': 'A03a4_a2l', 'type': 'neuron', 'skeleton_ids': [8688631]}, {'id': 10090270, 'name': 'A27e(2)_a1l', 'type': 'neuron', 'skeleton_ids': [10090269]}, {'id': 10185586, 'name': 'T01d1_t3r', 'type': 'neuron', 'skeleton_ids': [10185585]}, {'id': 10855207, 'name': 'A03g_a1r', 'type': 'neuron', 'skeleton_ids': [10855206]}, {'id': 12820278, 'name': 'A14a_a1l', 'type': 'neuron', 'skeleton_ids': [12820277]}, {'id': 12953346, 'name': 'A02f_a1r', 'type': 'neuron', 'skeleton_ids': [12953345]}, {'id': 13122057, 'name': 'A00g_a1', 'type': 'neuron', 'skeleton_ids': [13122056]}, {'id': 13258643, 'name': 'A26f_a1l', 'type': 'neuron', 'skeleton_ids': [13258642]}, {'id': 14242930, 'name': 'T03a1_t3l', 'type': 'neuron', 'skeleton_ids': [14242929]}, {'id': 15290336, 'name': 'A27e(2)_a1r', 'type': 'neuron', 'skeleton_ids': [15290335]}, {'id': 16226513, 'name': 'A01c2_ipsi_dendrite_a1r', 'type': 'neuron', 'skeleton_ids': [16226512]}, {'id': 16690921, 'name': 'A18j_a1r', 'type': 'neuron', 'skeleton_ids': [16690920]}, {'id': 16713122, 'name': 'MN21/22 (RF:LT1/LT2) LT2_a1r ', 'type': 'neuron', 'skeleton_ids': [16713121]}, {'id': 17071667, 'name': 'A26f_a1r', 'type': 'neuron', 'skeleton_ids': [17071666]}]}, {'id': 19711421, 'name': '<NAME> al. 2016', 'type': 'annotation', 'neurons': [{'id': 40042, 'name': '22c ORN left', 'type': 'neuron', 'skeleton_ids': [40045]}, {'id': 2568466, 'name': 'mPN iACT B1_left', 'type': 'neuron', 'skeleton_ids': [2568465]}, {'id': 2571762, 'name': '83a ORN right', 'type': 'neuron', 'skeleton_ids': [2571761]}, {'id': 2582392, 'name': '1a ORN right', 'type': 'neuron', 'skeleton_ids': [2582391]}, {'id': 2584839, 'name': '30a ORN right', 'type': 'neuron', 'skeleton_ids': [2584838]}, {'id': 2608844, 'name': '42b ORN right', 'type': 'neuron', 'skeleton_ids': [2608843]}, {'id': 2611806, 'name': '1a ORN left', 'type': 'neuron', 'skeleton_ids': [2611805]}, {'id': 2612528, 'name': 'mPN iACT C2 left', 'type': 'neuron', 'skeleton_ids': [2612527]}, {'id': 3158544, 'name': '63a ORN left', 'type': 'neuron', 'skeleton_ids': [3158543]}, {'id': 3410507, 'name': '85c ORN left', 'type': 'neuron', 'skeleton_ids': [3410506]}, {'id': 3410513, 'name': '67b ORN left', 'type': 'neuron', 'skeleton_ids': [6791927]}, {'id': 3450443, 'name': 'mPN iACT B2 left', 'type': 'neuron', 'skeleton_ids': [3450442]}, {'id': 3464774, 'name': '42a ORN left', 'type': 'neuron', 'skeleton_ids': [3464773]}, {'id': 3468160, 'name': '74a ORN left', 'type': 'neuron', 'skeleton_ids': [7542938]}, {'id': 3480832, 'name': '42b ORN left', 'type': 'neuron', 'skeleton_ids': [3410499]}, {'id': 3486382, 'name': '42a PN left', 'type': 'neuron', 'skeleton_ids': [3486381]}, {'id': 3486951, 'name': 'vine left', 'type': 'neuron', 'skeleton_ids': [3486950]}, {'id': 3511997, 'name': '94a & 94b ORN left', 'type': 'neuron', 'skeleton_ids': [3511996]}, {'id': 3538130, 'name': 'mPN D right', 'type': 'neuron', 'skeleton_ids': [3538129]}, {'id': 3547398, 'name': '83a ORN left', 'type': 'neuron', 'skeleton_ids': [3547397]}, {'id': 3592998, 'name': '24a ORN left', 'type': 'neuron', 'skeleton_ids': [3592997]}, {'id': 3694516, 'name': '47a & 33b ORN left', 'type': 'neuron', 'skeleton_ids': [3694515]}, {'id': 3708876, 'name': '33a ORN left', 'type': 'neuron', 'skeleton_ids': [3708875]}, {'id': 3712894, 'name': 'mPN iACT bilateral LOWER left', 'type': 'neuron', 'skeleton_ids': [3712893]}, {'id': 3716798, 'name': '45a ORN left', 'type': 'neuron', 'skeleton_ids': [3716797]}, {'id': 3736501, 'name': 'ginkgo 2 left', 'type': 'neuron', 'skeleton_ids': [3736500]}, {'id': 3768026, 'name': '30a ORN left', 'type': 'neuron', 'skeleton_ids': [3768025]}, {'id': 3771795, 'name': '49a ORN left', 'type': 'neuron', 'skeleton_ids': [3771794]}, {'id': 3774569, 'name': '82a ORN left', 'type': 'neuron', 'skeleton_ids': [3774568]}, {'id': 3790005, 'name': '67b ORN right', 'type': 'neuron', 'skeleton_ids': [3790004]}, {'id': 3805642, 'name': 'ventral LN right', 'type': 'neuron', 'skeleton_ids': [3805641]}, {'id': 3806574, 'name': 'picky 2 right', 'type': 'neuron', 'skeleton_ids': [3806573]}, {'id': 3806653, 'name': 'spruce right', 'type': 'neuron', 'skeleton_ids': [3806652]}, {'id': 3813488, 'name': 'Ladder (olfactory) post.', 'type': 'neuron', 'skeleton_ids': [3813487]}, {'id': 3813724, 'name': '45b ORN right', 'type': 'neuron', 'skeleton_ids': [3813723]}, {'id': 3827212, 'name': '42a ORN right', 'type': 'neuron', 'skeleton_ids': [3827211]}, {'id': 3840346, 'name': '35a ORN right', 'type': 'neuron', 'skeleton_ids': [3840345]}, {'id': 3851373, 'name': '63a ORN right', 'type': 'neuron', 'skeleton_ids': [7719547]}, {'id': 3865953, 'name': '24a ORN right', 'type': 'neuron', 'skeleton_ids': [3865952]}, {'id': 3872654, 'name': '33a ORN right', 'type': 'neuron', 'skeleton_ids': [3872653]}, {'id': 3885216, 'name': '74a ORN right', 'type': 'neuron', 'skeleton_ids': [3885215]}, {'id': 3893196, 'name': '85c ORN right', 'type': 'neuron', 'skeleton_ids': [3893195]}, {'id': 3896630, 'name': '47a & 33b ORN right', 'type': 'neuron', 'skeleton_ids': [3896629]}, {'id': 3954015, 'name': '22c ORN right', 'type': 'neuron', 'skeleton_ids': [3954014]}, {'id': 3958235, 'name': '94a & 94b ORN right', 'type': 'neuron', 'skeleton_ids': [3958234]}, {'id': 3963892, 'name': '45a ORN right', 'type': 'neuron', 'skeleton_ids': [3963891]}, {'id': 3980316, 'name': '82a ORN right', 'type': 'neuron', 'skeleton_ids': [3980315]}, {'id': 3993292, 'name': '49a ORN right', 'type': 'neuron', 'skeleton_ids': [7658146]}, {'id': 4073354, 'name': '13a ORN right', 'type': 'neuron', 'skeleton_ids': [4073353]}, {'id': 4076390, 'name': '59a ORN right', 'type': 'neuron', 'skeleton_ids': [4076389]}, {'id': 4116175, 'name': 'lAL-1 (octopaminergic) right', 'type': 'neuron', 'skeleton_ids': [4116174]}, {'id': 4119388, 'name': 'mPN iACT A1 right', 'type': 'neuron', 'skeleton_ids': [4119387]}, {'id': 4137843, 'name': '33a PN left', 'type': 'neuron', 'skeleton_ids': [8021456]}, {'id': 4247100, 'name': 'choosy 2 right', 'type': 'neuron', 'skeleton_ids': [4247099]}, {'id': 4324899, 'name': 'mPN seahorse right', 'type': 'neuron', 'skeleton_ids': [4324898]}, {'id': 4327685, 'name': 'mPN iACT A3 right', 'type': 'neuron', 'skeleton_ids': [4327684]}, {'id': 4354934, 'name': '85c PN left', 'type': 'neuron', 'skeleton_ids': [4354933]}, {'id': 4391979, 'name': 'mPN iACT C2 right', 'type': 'neuron', 'skeleton_ids': [4391978]}, {'id': 4493200, 'name': '13a PN left', 'type': 'neuron', 'skeleton_ids': [4493199]}, {'id': 4651930, 'name': '94a & 94b PN right', 'type': 'neuron', 'skeleton_ids': [4651929]}, {'id': 4679140, 'name': 'mPN iACT A2 right', 'type': 'neuron', 'skeleton_ids': [4679139]}, {'id': 4700079, 'name': 'mPN iACT B2 right', 'type': 'neuron', 'skeleton_ids': [4700078]}, {'id': 4700183, 'name': 'vine right', 'type': 'neuron', 'skeleton_ids': [4700182]}, {'id': 4736309, 'name': 'keystone left', 'type': 'neuron', 'skeleton_ids': [5030808]}, {'id': 4955475, 'name': 'broad T2 left', 'type': 'neuron', 'skeleton_ids': [8311264]}, {'id': 5090550, 'name': '45b ORN left', 'type': 'neuron', 'skeleton_ids': [5090549]}, {'id': 5247103, 'name': 'picky 4 right', 'type': 'neuron', 'skeleton_ids': [5247102]}, {'id': 5248752, 'name': '35a ORN left', 'type': 'neuron', 'skeleton_ids': [3465659]}, {'id': 5261425, 'name': 'mPN iACT A2 left', 'type': 'neuron', 'skeleton_ids': [5261424]}, {'id': 5291793, 'name': 'picky 0 left', 'type': 'neuron', 'skeleton_ids': [5291791]}, {'id': 5478205, 'name': '74a PN left', 'type': 'neuron', 'skeleton_ids': [7843730]}, {'id': 6143931, 'name': 'CSD right', 'type': 'neuron', 'skeleton_ids': [6143930]}, {'id': 6340461, 'name': '59a ORN left', 'type': 'neuron', 'skeleton_ids': [6340439]}, {'id': 6364988, 'name': '13a ORN left', 'type': 'neuron', 'skeleton_ids': [7527710]}, {'id': 6451333, 'name': 'choosy 1 left', 'type': 'neuron', 'skeleton_ids': [7394271]}, {'id': 6557582, 'name': 'broad T3 right', 'type': 'neuron', 'skeleton_ids': [6557581]}, {'id': 6703262, 'name': '94a & 94b PN left', 'type': 'neuron', 'skeleton_ids': [8399890]}, {'id': 7001631, 'name': '49a PN left', 'type': 'neuron', 'skeleton_ids': [7515491]}, {'id': 7092418, 'name': '47a & 33b PN left', 'type': 'neuron', 'skeleton_ids': [7092417]}, {'id': 7107434, 'name': '45b PN left', 'type': 'neuron', 'skeleton_ids': [7107433]}, {'id': 7198339, 'name': '24a PN left', 'type': 'neuron', 'skeleton_ids': [7198338]}, {'id': 7503200, 'name': 'mPN iACT C1 right', 'type': 'neuron', 'skeleton_ids': [7503199]}, {'id': 7600296, 'name': 'picky 1 right', 'type': 'neuron', 'skeleton_ids': [7600311]}, {'id': 7845338, 'name': '45a PN left', 'type': 'neuron', 'skeleton_ids': [7852250]}, {'id': 7845347, 'name': '63a PN left', 'type': 'neuron', 'skeleton_ids': [7845346]}, {'id': 7851154, 'name': '82a PN left', 'type': 'neuron', 'skeleton_ids': [7851153]}, {'id': 7864824, 'name': '59a PN left', 'type': 'neuron', 'skeleton_ids': [7864823]}, {'id': 7865653, 'name': '22c PN left', 'type': 'neuron', 'skeleton_ids': [7865652]}, {'id': 7865697, 'name': '1a PN left', 'type': 'neuron', 'skeleton_ids': [7865696]}, {'id': 7865768, 'name': '83a PN left', 'type': 'neuron', 'skeleton_ids': [7865767]}, {'id': 7873136, 'name': '42b PN left', 'type': 'neuron', 'skeleton_ids': [7873135]}, {'id': 7916873, 'name': '30a PN left', 'type': 'neuron', 'skeleton_ids': [7916593]}, {'id': 7934905, 'name': '35a PN bilateral left', 'type': 'neuron', 'skeleton_ids': [7934904]}, {'id': 7935089, 'name': '35a PN bilateral right', 'type': 'neuron', 'skeleton_ids': [7935088]}, {'id': 7939892, 'name': 'broad T3 left', 'type': 'neuron', 'skeleton_ids': [7939890]}, {'id': 7939980, 'name': 'broad T1 left', 'type': 'neuron', 'skeleton_ids': [7939979]}, {'id': 7941643, 'name': 'broad D2 left', 'type': 'neuron', 'skeleton_ids': [7941642]}, {'id': 7941653, 'name': 'broad D1 left', 'type': 'neuron', 'skeleton_ids': [7941652]}, {'id': 8102936, 'name': 'picky 1 left', 'type': 'neuron', 'skeleton_ids': [8102935]}, {'id': 8140110, 'name': '74a PN right', 'type': 'neuron', 'skeleton_ids': [8140109]}, {'id': 8198239, 'name': 'broad T2 right', 'type': 'neuron', 'skeleton_ids': [8198238]}, {'id': 8198318, 'name': 'broad T1 right', 'type': 'neuron', 'skeleton_ids': [8198317]}, {'id': 8198417, 'name': 'broad D1 right', 'type': 'neuron', 'skeleton_ids': [8198416]}, {'id': 8198514, 'name': 'broad D2 right', 'type': 'neuron', 'skeleton_ids': [8198513]}, {'id': 8199673, 'name': 'picky 0 right', 'type': 'neuron', 'skeleton_ids': [8199671]}, {'id': 8231265, 'name': 'choosy 1 right', 'type': 'neuron', 'skeleton_ids': [8231264]}, {'id': 8243456, 'name': '83a PN right', 'type': 'neuron', 'skeleton_ids': [8243455]}, {'id': 8244724, 'name': '42a PN right', 'type': 'neuron', 'skeleton_ids': [8244723]}, {'id': 8244824, 'name': '13a PN right', 'type': 'neuron', 'skeleton_ids': [8244823]}, {'id': 8245015, 'name': '59a PN right', 'type': 'neuron', 'skeleton_ids': [8245014]}, {'id': 8245147, 'name': '82a PN right', 'type': 'neuron', 'skeleton_ids': [8245146]}, {'id': 8245514, 'name': '63a PN right', 'type': 'neuron', 'skeleton_ids': [8245513]}, {'id': 8245693, 'name': '42b PN right', 'type': 'neuron', 'skeleton_ids': [8245692]}, {'id': 8245871, 'name': '24a PN right', 'type': 'neuron', 'skeleton_ids': [8245869]}, {'id': 8246082, 'name': '67b PN right', 'type': 'neuron', 'skeleton_ids': [8246081]}, {'id': 8246751, 'name': '47a & 33b PN right', 'type': 'neuron', 'skeleton_ids': [8246750]}, {'id': 8247452, 'name': '45a PN right', 'type': 'neuron', 'skeleton_ids': [8247451]}, {'id': 8248632, 'name': '22c PN right', 'type': 'neuron', 'skeleton_ids': [8248631]}, {'id': 8252068, 'name': '1a PN right', 'type': 'neuron', 'skeleton_ids': [8252067]}, {'id': 8253414, 'name': '45b PN right', 'type': 'neuron', 'skeleton_ids': [8253413]}, {'id': 8256187, 'name': '30a PN right', 'type': 'neuron', 'skeleton_ids': [8256186]}, {'id': 8273370, 'name': 'choosy 2 left', 'type': 'neuron', 'skeleton_ids': [8273369]}, {'id': 8274022, 'name': 'picky 3 left', 'type': 'neuron', 'skeleton_ids': [8274021]}, {'id': 8276783, 'name': 'mPN iACT A1 left', 'type': 'neuron', 'skeleton_ids': [8276782]}, {'id': 8480222, 'name': 'mPN seahorse left', 'type': 'neuron', 'skeleton_ids': [8480221]}, {'id': 8509535, 'name': 'mPN cobra left', 'type': 'neuron', 'skeleton_ids': [8509534]}, {'id': 8704080, 'name': 'mPN iACT B3 left', 'type': 'neuron', 'skeleton_ids': [8704079]}, {'id': 8877796, 'name': 'ventral LN left', 'type': 'neuron', 'skeleton_ids': [8877795]}, {'id': 8877972, 'name': 'picky 2 left', 'type': 'neuron', 'skeleton_ids': [8877971]}, {'id': 9119559, 'name': 'mPN iACT VUM L', 'type': 'neuron', 'skeleton_ids': [9119558]}, {'id': 9589147, 'name': 'mPN iACT C1 left', 'type': 'neuron', 'skeleton_ids': [9589146]}, {'id': 9681371, 'name': 'mPN D left', 'type': 'neuron', 'skeleton_ids': [6451332]}, {'id': 10158973, 'name': 'clamp right', 'type': 'neuron', 'skeleton_ids': [10158972]}, {'id': 10167079, 'name': 'mPN iACT B1_right', 'type': 'neuron', 'skeleton_ids': [10167078]}, {'id': 10555410, 'name': 'picky 4 left', 'type': 'neuron', 'skeleton_ids': [10555409]}, {'id': 10831006, 'name': 'keystone right', 'type': 'neuron', 'skeleton_ids': [10831005]}, {'id': 10865026, 'name': 'lAL-1 (octopaminergic) left', 'type': 'neuron', 'skeleton_ids': [10865025]}, {'id': 10990804, 'name': 'mPN iACT VUM R', 'type': 'neuron', 'skeleton_ids': [10990803]}, {'id': 11051277, 'name': 'picky 3 right', 'type': 'neuron', 'skeleton_ids': [11051276]}, {'id': 11105183, 'name': 'clamp left', 'type': 'neuron', 'skeleton_ids': [11105182]}, {'id': 11279245, 'name': '49a PN right', 'type': 'neuron', 'skeleton_ids': [11279244]}, {'id': 11543213, 'name': 'mPN iACT bilateral LOWER right', 'type': 'neuron', 'skeleton_ids': [11543212]}, {'id': 12225667, 'name': 'mPN iACT A3 left', 'type': 'neuron', 'skeleton_ids': [12225666]}, {'id': 13380661, 'name': 'ginkgo 1 left', 'type': 'neuron', 'skeleton_ids': [13380660]}, {'id': 13589252, 'name': '33a PN right', 'type': 'neuron', 'skeleton_ids': [13589251]}, {'id': 13803033, 'name': 'Projection from LH 2_right', 'type': 'neuron', 'skeleton_ids': [13803032]}, {'id': 13849558, 'name': 'CSD left', 'type': 'neuron', 'skeleton_ids': [13849557]}, {'id': 14154155, 'name': 'cypress right', 'type': 'neuron', 'skeleton_ids': [14154154]}, {'id': 14437743, 'name': 'spruce left', 'type': 'neuron', 'skeleton_ids': [14437742]}, {'id': 14517692, 'name': 'ginkgo 1 right', 'type': 'neuron', 'skeleton_ids': [14517691]}, {'id': 14604029, 'name': '85c PN right', 'type': 'neuron', 'skeleton_ids': [14604028]}, {'id': 16259596, 'name': '67b PN left', 'type': 'neuron', 'skeleton_ids': [16259595]}, {'id': 16515872, 'name': 'mPN cobra right', 'type': 'neuron', 'skeleton_ids': [16515871]}, {'id': 16621865, 'name': 'ginkgo 2 right', 'type': 'neuron', 'skeleton_ids': [16621864]}, {'id': 16735587, 'name': 'mPN iACT B3 right', 'type': 'neuron', 'skeleton_ids': [16735586]}, {'id': 16795525, 'name': 'mPN iACT bilateral UPPER right', 'type': 'neuron', 'skeleton_ids': [16795524]}, {'id': 16795839, 'name': 'mPN iACT bilateral UPPER left', 'type': 'neuron', 'skeleton_ids': [16795838]}, {'id': 17068731, 'name': 'Ladder (olfactory) ant.', 'type': 'neuron', 'skeleton_ids': [17068730]}, {'id': 17545696, 'name': 'cypress left', 'type': 'neuron', 'skeleton_ids': [17545695]}, {'id': 17844901, 'name': 'Giant-step left', 'type': 'neuron', 'skeleton_ids': [17844900]}]}, {'id': 19711583, 'name': '<NAME>, Litwin-Kumar et al. 2017', 'type': 'annotation', 'neurons': [{'id': 26, 'name': 'KC #0', 'type': 'neuron', 'skeleton_ids': [29]}, {'id': 677706, 'name': 'KC1', 'type': 'neuron', 'skeleton_ids': [677717]}, {'id': 1872624, 'name': 'nc1;right', 'type': 'neuron', 'skeleton_ids': [7555968]}, {'id': 2301628, 'name': 'nc1;left', 'type': 'neuron', 'skeleton_ids': [8111135]}, {'id': 2506051, 'name': 'MBE4a right', 'type': 'neuron', 'skeleton_ids': [2506050]}, {'id': 2612528, 'name': 'mPN iACT C2 left', 'type': 'neuron', 'skeleton_ids': [2612527]}, {'id': 2862581, 'name': 'CPLd interneuron', 'type': 'neuron', 'skeleton_ids': [8598886]}, {'id': 2966609, 'name': 'MBE10a left', 'type': 'neuron', 'skeleton_ids': [8689674]}, {'id': 3234818, 'name': 'MBE12 right', 'type': 'neuron', 'skeleton_ids': [3234817]}, {'id': 3347709, 'name': 'KC2', 'type': 'neuron', 'skeleton_ids': [3347708]}, {'id': 3450752, 'name': 'mPN BAmd1-g right', 'type': 'neuron', 'skeleton_ids': [3450751]}, {'id': 3486382, 'name': '42a PN left', 'type': 'neuron', 'skeleton_ids': [3486381]}, {'id': 3497904, 'name': 'KC4', 'type': 'neuron', 'skeleton_ids': [3497901]}, {'id': 3522294, 'name': 'KC5', 'type': 'neuron', 'skeleton_ids': [3522293]}, {'id': 3523218, 'name': 'Thermo KC B_right - mbi2 - unknown', 'type': 'neuron', 'skeleton_ids': [3664102]}, {'id': 3570509, 'name': 'Handlebar PN (CB on left)', 'type': 'neuron', 'skeleton_ids': [3991518]}, {'id': 3580923, 'name': 'KC8 young', 'type': 'neuron', 'skeleton_ids': [3605630]}, {'id': 3587613, 'name': 'KC7', 'type': 'neuron', 'skeleton_ids': [3587612]}, {'id': 3679412, 'name': 'KC9 young', 'type': 'neuron', 'skeleton_ids': [3679411]}, {'id': 3691222, 'name': 'Lower Looping PN Right - AVM004 (BAla1/2)', 'type': 'neuron', 'skeleton_ids': [3690921]}, {'id': 3709978, 'name': 'backwards handlebar left (looks like 35a)', 'type': 'neuron', 'skeleton_ids': [3709977]}, {'id': 3712894, 'name': 'mPN iACT bilateral LOWER left', 'type': 'neuron', 'skeleton_ids': [3712893]}, {'id': 3756660, 'name': 'PN Only Connects To Togethers Right', 'type': 'neuron', 'skeleton_ids': [3756659]}, {'id': 3813488, 'name': 'Ladder (olfactory) post.', 'type': 'neuron', 'skeleton_ids': [3813487]}, {'id': 3819158, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [3819157]}, {'id': 3819172, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [3819171]}, {'id': 3886357, 'name': 'MBE4h left', 'type': 'neuron', 'skeleton_ids': [3886356]}, {'id': 3890029, 'name': 'MBE24a right', 'type': 'neuron', 'skeleton_ids': [3890028]}, {'id': 4022541, 'name': 'MBE20 right', 'type': 'neuron', 'skeleton_ids': [4022539]}, {'id': 4108017, 'name': 'Upper Looping PN Left - AVM004', 'type': 'neuron', 'skeleton_ids': [4108016]}, {'id': 4119388, 'name': 'mPN iACT A1 right', 'type': 'neuron', 'skeleton_ids': [4119387]}, {'id': 4134666, 'name': 'KC72', 'type': 'neuron', 'skeleton_ids': [4117305]}, {'id': 4134669, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [4134539]}, {'id': 4137843, 'name': '33a PN left', 'type': 'neuron', 'skeleton_ids': [8021456]}, {'id': 4152023, 'name': 'from upper looping PN - peptidergic right', 'type': 'neuron', 'skeleton_ids': [4152020]}, {'id': 4154422, 'name': 'Lower Looping PN Left - AVM004', 'type': 'neuron', 'skeleton_ids': [4154421]}, {'id': 4195013, 'name': 'MBE13a right', 'type': 'neuron', 'skeleton_ids': [4195012]}, {'id': 4227305, 'name': 'KC10', 'type': 'neuron', 'skeleton_ids': [4227304]}, {'id': 4227545, 'name': 'KC11', 'type': 'neuron', 'skeleton_ids': [4227544]}, {'id': 4230062, 'name': 'KC12', 'type': 'neuron', 'skeleton_ids': [4230061]}, {'id': 4230740, 'name': 'KC20', 'type': 'neuron', 'skeleton_ids': [5019924]}, {'id': 4234010, 'name': 'MBE33 right', 'type': 'neuron', 'skeleton_ids': [4234009]}, {'id': 4235140, 'name': 'AVM011f right', 'type': 'neuron', 'skeleton_ids': [4235139]}, {'id': 4241193, 'name': 'KC14', 'type': 'neuron', 'skeleton_ids': [4241192]}, {'id': 4241238, 'name': 'MBE6 right', 'type': 'neuron', 'skeleton_ids': [4241237]}, {'id': 4242746, 'name': 'MBE13b right', 'type': 'neuron', 'skeleton_ids': [4230749]}, {'id': 4274739, 'name': 'MBE15a right', 'type': 'neuron', 'skeleton_ids': [4598363]}, {'id': 4354934, 'name': '85c PN left', 'type': 'neuron', 'skeleton_ids': [4354933]}, {'id': 4381130, 'name': 'MBE19 left', 'type': 'neuron', 'skeleton_ids': [4381129]}, {'id': 4381378, 'name': 'MBE4f left', 'type': 'neuron', 'skeleton_ids': [4381377]}, {'id': 4391979, 'name': 'mPN iACT C2 right', 'type': 'neuron', 'skeleton_ids': [4391978]}, {'id': 4393050, 'name': 'non-olfactory PN right', 'type': 'neuron', 'skeleton_ids': [4393049]}, {'id': 4404664, 'name': 'PVL005 - 2 Left', 'type': 'neuron', 'skeleton_ids': [4635057]}, {'id': 4411506, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [4411505]}, {'id': 4412037, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [4412036]}, {'id': 4412316, 'name': 'KC2/24', 'type': 'neuron', 'skeleton_ids': [4412315]}, {'id': 4414164, 'name': 'MBE1d left', 'type': 'neuron', 'skeleton_ids': [4414163]}, {'id': 4414185, 'name': 'MBE1d right', 'type': 'neuron', 'skeleton_ids': [4414184]}, {'id': 4444355, 'name': 'KC17', 'type': 'neuron', 'skeleton_ids': [4444354]}, {'id': 4493200, 'name': '13a PN left', 'type': 'neuron', 'skeleton_ids': [4493199]}, {'id': 4518547, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [4518546]}, {'id': 4651930, 'name': '94a & 94b PN right', 'type': 'neuron', 'skeleton_ids': [4651929]}, {'id': 4876705, 'name': 'PN in Olfactory Bundle 1 right', 'type': 'neuron', 'skeleton_ids': [4876704]}, {'id': 4884580, 'name': 'MBE1b right', 'type': 'neuron', 'skeleton_ids': [4884579]}, {'id': 4919896, 'name': 'KC18 young', 'type': 'neuron', 'skeleton_ids': [4919895]}, {'id': 4920797, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8177991]}, {'id': 4982173, 'name': 'KC19', 'type': 'neuron', 'skeleton_ids': [4982172]}, {'id': 5049772, 'name': 'MBE5a left', 'type': 'neuron', 'skeleton_ids': [7802210]}, {'id': 5051343, 'name': 'KC21', 'type': 'neuron', 'skeleton_ids': [5051342]}, {'id': 5169588, 'name': 'MBE16 left', 'type': 'neuron', 'skeleton_ids': [8922644]}, {'id': 5395824, 'name': 'MBE15b left', 'type': 'neuron', 'skeleton_ids': [5395823]}, {'id': 5478205, 'name': '74a PN left', 'type': 'neuron', 'skeleton_ids': [7843730]}, {'id': 5669276, 'name': 'MBE16 right', 'type': 'neuron', 'skeleton_ids': [8297018]}, {'id': 5835800, 'name': 'KC22', 'type': 'neuron', 'skeleton_ids': [5835799]}, {'id': 5836036, 'name': 'KC24', 'type': 'neuron', 'skeleton_ids': [5836035]}, {'id': 5836182, 'name': 'KC26', 'type': 'neuron', 'skeleton_ids': [5836181]}, {'id': 5836207, 'name': 'KC28', 'type': 'neuron', 'skeleton_ids': [5836206]}, {'id': 5836251, 'name': 'KC40', 'type': 'neuron', 'skeleton_ids': [7891312]}, {'id': 5937085, 'name': 'KC65;_42a left', 'type': 'neuron', 'skeleton_ids': [5937084]}, {'id': 5966100, 'name': 'MBE4h right', 'type': 'neuron', 'skeleton_ids': [5966099]}, {'id': 5969256, 'name': 'MBE34 right', 'type': 'neuron', 'skeleton_ids': [4598109]}, {'id': 6611895, 'name': 'MBE1c right', 'type': 'neuron', 'skeleton_ids': [6611894]}, {'id': 6699206, 'name': 'MBE30 right', 'type': 'neuron', 'skeleton_ids': [6699205]}, {'id': 6703242, 'name': 'MBE23 right', 'type': 'neuron', 'skeleton_ids': [6703240]}, {'id': 6703262, 'name': '94a & 94b PN left', 'type': 'neuron', 'skeleton_ids': [8399890]}, {'id': 6704233, 'name': 'KC54', 'type': 'neuron', 'skeleton_ids': [6704232]}, {'id': 7001631, 'name': '49a PN left', 'type': 'neuron', 'skeleton_ids': [7515491]}, {'id': 7020345, 'name': 'handlebar-like 4 right', 'type': 'neuron', 'skeleton_ids': [7020344]}, {'id': 7055858, 'name': 'MBE6 left', 'type': 'neuron', 'skeleton_ids': [7055857]}, {'id': 7092087, 'name': 'PN in Olfactory Bundle 1 left', 'type': 'neuron', 'skeleton_ids': [7092086]}, {'id': 7092418, 'name': '47a & 33b PN left', 'type': 'neuron', 'skeleton_ids': [7092417]}, {'id': 7107434, 'name': '45b PN left', 'type': 'neuron', 'skeleton_ids': [7107433]}, {'id': 7187385, 'name': 'Thermo KC B_left - mbi2 - unknown', 'type': 'neuron', 'skeleton_ids': [7187384]}, {'id': 7196395, 'name': 'KC31', 'type': 'neuron', 'skeleton_ids': [7196392]}, {'id': 7198339, 'name': '24a PN left', 'type': 'neuron', 'skeleton_ids': [7198338]}, {'id': 7215583, 'name': 'KC103 very young', 'type': 'neuron', 'skeleton_ids': [7215582]}, {'id': 7616957, 'name': 'MBE9a right', 'type': 'neuron', 'skeleton_ids': [7616956]}, {'id': 7690946, 'name': 'KC33', 'type': 'neuron', 'skeleton_ids': [7690945]}, {'id': 7786054, 'name': 'KC34', 'type': 'neuron', 'skeleton_ids': [7786053]}, {'id': 7829847, 'name': 'KC36', 'type': 'neuron', 'skeleton_ids': [7829784]}, {'id': 7845338, 'name': '45a PN left', 'type': 'neuron', 'skeleton_ids': [7852250]}, {'id': 7845347, 'name': '63a PN left', 'type': 'neuron', 'skeleton_ids': [7845346]}, {'id': 7851154, 'name': '82a PN left', 'type': 'neuron', 'skeleton_ids': [7851153]}, {'id': 7864824, 'name': '59a PN left', 'type': 'neuron', 'skeleton_ids': [7864823]}, {'id': 7865653, 'name': '22c PN left', 'type': 'neuron', 'skeleton_ids': [7865652]}, {'id': 7865697, 'name': '1a PN left', 'type': 'neuron', 'skeleton_ids': [7865696]}, {'id': 7865768, 'name': '83a PN left', 'type': 'neuron', 'skeleton_ids': [7865767]}, {'id': 7873136, 'name': '42b PN left', 'type': 'neuron', 'skeleton_ids': [7873135]}, {'id': 7910625, 'name': 'MBE2as left', 'type': 'neuron', 'skeleton_ids': [7910624]}, {'id': 7916873, 'name': '30a PN left', 'type': 'neuron', 'skeleton_ids': [7916593]}, {'id': 7934905, 'name': '35a PN bilateral left', 'type': 'neuron', 'skeleton_ids': [7934904]}, {'id': 7935089, 'name': '35a PN bilateral right', 'type': 'neuron', 'skeleton_ids': [7935088]}, {'id': 7983900, 'name': 'MBE9a left', 'type': 'neuron', 'skeleton_ids': [7983899]}, {'id': 8012954, 'name': 'PN in Olfactory Bundle 2 left', 'type': 'neuron', 'skeleton_ids': [8012953]}, {'id': 8067289, 'name': 'KC42', 'type': 'neuron', 'skeleton_ids': [8066542]}, {'id': 8069366, 'name': 'MBE18 right', 'type': 'neuron', 'skeleton_ids': [8980589]}, {'id': 8069897, 'name': 'KC78', 'type': 'neuron', 'skeleton_ids': [9556700]}, {'id': 8070223, 'name': 'KC41 young', 'type': 'neuron', 'skeleton_ids': [8066411]}, {'id': 8094861, 'name': 'MBE11a right', 'type': 'neuron', 'skeleton_ids': [8094859]}, {'id': 8118476, 'name': 'Thermo KC A_Left - Mushroom Body Right', 'type': 'neuron', 'skeleton_ids': [8118474]}, {'id': 8138802, 'name': 'PVL005 - 2 Right (V1); BLVa34 a 11', 'type': 'neuron', 'skeleton_ids': [9275210]}, {'id': 8140110, 'name': '74a PN right', 'type': 'neuron', 'skeleton_ids': [8140109]}, {'id': 8158302, 'name': 'KC35', 'type': 'neuron', 'skeleton_ids': [7828725]}, {'id': 8160266, 'name': 'KC39', 'type': 'neuron', 'skeleton_ids': [7868195]}, {'id': 8243456, 'name': '83a PN right', 'type': 'neuron', 'skeleton_ids': [8243455]}, {'id': 8244724, 'name': '42a PN right', 'type': 'neuron', 'skeleton_ids': [8244723]}, {'id': 8244824, 'name': '13a PN right', 'type': 'neuron', 'skeleton_ids': [8244823]}, {'id': 8245015, 'name': '59a PN right', 'type': 'neuron', 'skeleton_ids': [8245014]}, {'id': 8245147, 'name': '82a PN right', 'type': 'neuron', 'skeleton_ids': [8245146]}, {'id': 8245514, 'name': '63a PN right', 'type': 'neuron', 'skeleton_ids': [8245513]}, {'id': 8245693, 'name': '42b PN right', 'type': 'neuron', 'skeleton_ids': [8245692]}, {'id': 8245871, 'name': '24a PN right', 'type': 'neuron', 'skeleton_ids': [8245869]}, {'id': 8246082, 'name': '67b PN right', 'type': 'neuron', 'skeleton_ids': [8246081]}, {'id': 8246751, 'name': '47a & 33b PN right', 'type': 'neuron', 'skeleton_ids': [8246750]}, {'id': 8247316, 'name': 'KC104 very young', 'type': 'neuron', 'skeleton_ids': [8247315]}, {'id': 8247452, 'name': '45a PN right', 'type': 'neuron', 'skeleton_ids': [8247451]}, {'id': 8248632, 'name': '22c PN right', 'type': 'neuron', 'skeleton_ids': [8248631]}, {'id': 8248707, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8248706]}, {'id': 8251399, 'name': 'KC50', 'type': 'neuron', 'skeleton_ids': [8419602]}, {'id': 8252068, 'name': '1a PN right', 'type': 'neuron', 'skeleton_ids': [8252067]}, {'id': 8253414, 'name': '45b PN right', 'type': 'neuron', 'skeleton_ids': [8253413]}, {'id': 8254896, 'name': 'PN in Olfactory Bundle 2 right', 'type': 'neuron', 'skeleton_ids': [8254894]}, {'id': 8256187, 'name': '30a PN right', 'type': 'neuron', 'skeleton_ids': [8256186]}, {'id': 8259180, 'name': 'KC43 very young', 'type': 'neuron', 'skeleton_ids': [8259179]}, {'id': 8259193, 'name': 'KC44 young', 'type': 'neuron', 'skeleton_ids': [8259192]}, {'id': 8259200, 'name': 'KC45 very young', 'type': 'neuron', 'skeleton_ids': [8259199]}, {'id': 8260012, 'name': 'KC46 young', 'type': 'neuron', 'skeleton_ids': [8260011]}, {'id': 8260936, 'name': 'KC47 very young', 'type': 'neuron', 'skeleton_ids': [8260935]}, {'id': 8262303, 'name': 'KC21', 'type': 'neuron', 'skeleton_ids': [8262302]}, {'id': 8262373, 'name': 'LON KC left', 'type': 'neuron', 'skeleton_ids': [8262372]}, {'id': 8262504, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8262503]}, {'id': 8263141, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8263140]}, {'id': 8263286, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8263285]}, {'id': 8267549, 'name': 'LON KC right', 'type': 'neuron', 'skeleton_ids': [8068173]}, {'id': 8274903, 'name': 'KC48', 'type': 'neuron', 'skeleton_ids': [8274902]}, {'id': 8318679, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [8318678]}, {'id': 8319312, 'name': 'MBE10a right', 'type': 'neuron', 'skeleton_ids': [8319311]}, {'id': 8393982, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8393981]}, {'id': 8413664, 'name': 'KC49', 'type': 'neuron', 'skeleton_ids': [8413663]}, {'id': 8427678, 'name': 'KC51', 'type': 'neuron', 'skeleton_ids': [8427677]}, {'id': 8436847, 'name': 'KC52', 'type': 'neuron', 'skeleton_ids': [8504660]}, {'id': 8509988, 'name': 'KC53', 'type': 'neuron', 'skeleton_ids': [8509987]}, {'id': 8513390, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8513389]}, {'id': 8559608, 'name': 'KC54', 'type': 'neuron', 'skeleton_ids': [8559607]}, {'id': 8723398, 'name': 'KC55 young', 'type': 'neuron', 'skeleton_ids': [8723397]}, {'id': 8761401, 'name': 'KC56 young', 'type': 'neuron', 'skeleton_ids': [8761400]}, {'id': 8764751, 'name': 'KC57 very young', 'type': 'neuron', 'skeleton_ids': [8764750]}, {'id': 8765866, 'name': 'KC58 very young', 'type': 'neuron', 'skeleton_ids': [8765865]}, {'id': 8766922, 'name': 'KC59 young', 'type': 'neuron', 'skeleton_ids': [8766921]}, {'id': 8773166, 'name': 'KC60 very young', 'type': 'neuron', 'skeleton_ids': [8773165]}, {'id': 8773717, 'name': 'KC61 very young', 'type': 'neuron', 'skeleton_ids': [8773716]}, {'id': 8773880, 'name': 'KC62 young', 'type': 'neuron', 'skeleton_ids': [8773879]}, {'id': 8792121, 'name': 'KC63 young', 'type': 'neuron', 'skeleton_ids': [8792120]}, {'id': 8798011, 'name': 'MBE13b left', 'type': 'neuron', 'skeleton_ids': [8798010]}, {'id': 8835969, 'name': 'KC64', 'type': 'neuron', 'skeleton_ids': [8835968]}, {'id': 8850803, 'name': 'KC65;_42a right', 'type': 'neuron', 'skeleton_ids': [8850802]}, {'id': 8864097, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8864096]}, {'id': 8865200, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [8865199]}, {'id': 8891316, 'name': 'KC66', 'type': 'neuron', 'skeleton_ids': [8891315]}, {'id': 8924187, 'name': 'KC67', 'type': 'neuron', 'skeleton_ids': [8924186]}, {'id': 8957326, 'name': 'KC68 young', 'type': 'neuron', 'skeleton_ids': [8957325]}, {'id': 8978118, 'name': 'KC69 young', 'type': 'neuron', 'skeleton_ids': [8978117]}, {'id': 8997282, 'name': 'KC70', 'type': 'neuron', 'skeleton_ids': [8997281]}, {'id': 9012342, 'name': 'KC71 young', 'type': 'neuron', 'skeleton_ids': [9012340]}, {'id': 9074102, 'name': 'MBE35 right', 'type': 'neuron', 'skeleton_ids': [9074101]}, {'id': 9109800, 'name': 'MBE35 left', 'type': 'neuron', 'skeleton_ids': [9109799]}, {'id': 9251784, 'name': 'MBE13a left', 'type': 'neuron', 'skeleton_ids': [8338584]}, {'id': 9462897, 'name': 'AVL011 PN Left', 'type': 'neuron', 'skeleton_ids': [4985759]}, {'id': 9469520, 'name': 'MBE11b right', 'type': 'neuron', 'skeleton_ids': [9469519]}, {'id': 9511774, 'name': 'KC74 young', 'type': 'neuron', 'skeleton_ids': [9511772]}, {'id': 9517478, 'name': 'KC75 young', 'type': 'neuron', 'skeleton_ids': [9517477]}, {'id': 9525493, 'name': 'KC76', 'type': 'neuron', 'skeleton_ids': [9525492]}, {'id': 9531723, 'name': 'KC77', 'type': 'neuron', 'skeleton_ids': [9531722]}, {'id': 9574260, 'name': 'KC79 young', 'type': 'neuron', 'skeleton_ids': [9574259]}, {'id': 9578082, 'name': 'PVL005 1 right', 'type': 'neuron', 'skeleton_ids': [7971031]}, {'id': 9578603, 'name': 'KC80 young', 'type': 'neuron', 'skeleton_ids': [9578602]}, {'id': 9606355, 'name': 'KC81 very young', 'type': 'neuron', 'skeleton_ids': [9606354]}, {'id': 9609050, 'name': 'KC82', 'type': 'neuron', 'skeleton_ids': [9609048]}, {'id': 9621721, 'name': 'AVL011 PN Right 2?', 'type': 'neuron', 'skeleton_ids': [4620453]}, {'id': 9654512, 'name': 'KC3', 'type': 'neuron', 'skeleton_ids': [3347834]}, {'id': 9654689, 'name': 'Thermo KC A_Right - Mushroom Body Left', 'type': 'neuron', 'skeleton_ids': [4229950]}, {'id': 9654697, 'name': 'KC13', 'type': 'neuron', 'skeleton_ids': [4230739]}, {'id': 9654726, 'name': 'KC15', 'type': 'neuron', 'skeleton_ids': [4414426]}, {'id': 9654744, 'name': 'KC16', 'type': 'neuron', 'skeleton_ids': [4415241]}, {'id': 9654874, 'name': 'KC27', 'type': 'neuron', 'skeleton_ids': [5836197]}, {'id': 9654888, 'name': 'KC23', 'type': 'neuron', 'skeleton_ids': [5835815]}, {'id': 9654902, 'name': 'KC29', 'type': 'neuron', 'skeleton_ids': [5836387]}, {'id': 9654922, 'name': 'KC25', 'type': 'neuron', 'skeleton_ids': [5836163]}, {'id': 9654949, 'name': 'AVL011 PN Right', 'type': 'neuron', 'skeleton_ids': [9291474]}, {'id': 9655184, 'name': 'MBE7a left', 'type': 'neuron', 'skeleton_ids': [8877158]}, {'id': 9671611, 'name': 'MBE9b right', 'type': 'neuron', 'skeleton_ids': [7840791]}, {'id': 9679236, 'name': 'MBE2a right', 'type': 'neuron', 'skeleton_ids': [3299214]}, {'id': 9679266, 'name': 'MBE2as right', 'type': 'neuron', 'skeleton_ids': [7897469]}, {'id': 9679286, 'name': 'MBE4a left', 'type': 'neuron', 'skeleton_ids': [7901791]}, {'id': 9680201, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [6702830]}, {'id': 9681014, 'name': 'KC37', 'type': 'neuron', 'skeleton_ids': [7838405]}, {'id': 9681190, 'name': 'V1: possible PVL005 - 3 Right?', 'type': 'neuron', 'skeleton_ids': [2921933]}, {'id': 9681280, 'name': 'KC6 young', 'type': 'neuron', 'skeleton_ids': [3565102]}, {'id': 9681301, 'name': 'MBE15a left', 'type': 'neuron', 'skeleton_ids': [4401350]}, {'id': 9681324, 'name': 'MBE1c left', 'type': 'neuron', 'skeleton_ids': [7057894]}, {'id': 9681389, 'name': 'KC38', 'type': 'neuron', 'skeleton_ids': [7840203]}, {'id': 9707942, 'name': 'KC83', 'type': 'neuron', 'skeleton_ids': [9707941]}, {'id': 9730874, 'name': 'KC84', 'type': 'neuron', 'skeleton_ids': [9730873]}, {'id': 9741464, 'name': 'KC85 young', 'type': 'neuron', 'skeleton_ids': [9741463]}, {'id': 9747419, 'name': 'PPLV - 2 Left (maybe PVL014c)', 'type': 'neuron', 'skeleton_ids': [9747418]}, {'id': 9747712, 'name': 'Handlebar PN (CB on Right)', 'type': 'neuron', 'skeleton_ids': [9747710]}, {'id': 9766794, 'name': 'KC86', 'type': 'neuron', 'skeleton_ids': [9766793]}, {'id': 9769560, 'name': '<NAME>', 'type': 'neuron', 'skeleton_ids': [9769559]}, {'id': 9770017, 'name': '<NAME>', 'type': 'neuron', 'skeleton_ids': [9770016]}, {'id': 9782027, 'name': '<NAME>', 'type': 'neuron', 'skeleton_ids': [9782026]}, {'id': 9796813, 'name': 'KC90', 'type': 'neuron', 'skeleton_ids': [9796812]}, {'id': 9809627, 'name': 'KC91', 'type': 'neuron', 'skeleton_ids': [9809626]}, {'id': 9813189, 'name': 'KC92', 'type': 'neuron', 'skeleton_ids': [9813188]}, {'id': 9847389, 'name': 'KC93', 'type': 'neuron', 'skeleton_ids': [9847388]}, {'id': 9889096, 'name': 'KC94', 'type': 'neuron', 'skeleton_ids': [9889095]}, {'id': 9897224, 'name': 'KC95', 'type': 'neuron', 'skeleton_ids': [9897223]}, {'id': 9983192, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [9983191]}, {'id': 10056281, 'name': 'KC96', 'type': 'neuron', 'skeleton_ids': [10056280]}, {'id': 10160251, 'name': 'mPN BAmd1-g left', 'type': 'neuron', 'skeleton_ids': [10160250]}, {'id': 10163419, 'name': 'MBE7a right', 'type': 'neuron', 'skeleton_ids': [10163418]}, {'id': 10169397, 'name': 'KC97', 'type': 'neuron', 'skeleton_ids': [10169396]}, {'id': 10246567, 'name': 'KC98 young', 'type': 'neuron', 'skeleton_ids': [10246566]}, {'id': 10259968, 'name': 'KC99', 'type': 'neuron', 'skeleton_ids': [10259967]}, {'id': 10450308, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [10450307]}, {'id': 10455235, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [10455234]}, {'id': 10673896, 'name': 'MBE4f right', 'type': 'neuron', 'skeleton_ids': [10673895]}, {'id': 11017762, 'name': 'MBE26 left', 'type': 'neuron', 'skeleton_ids': [11017761]}, {'id': 11067380, 'name': 'non-olfactory PN left', 'type': 'neuron', 'skeleton_ids': [11067379]}, {'id': 11279245, 'name': '49a PN right', 'type': 'neuron', 'skeleton_ids': [11279244]}, {'id': 11333055, 'name': 'Gustatory? PN left', 'type': 'neuron', 'skeleton_ids': [11333054]}, {'id': 11524048, 'name': 'MBE15b right', 'type': 'neuron', 'skeleton_ids': [11524047]}, {'id': 11525715, 'name': 'MBE22 left', 'type': 'neuron', 'skeleton_ids': [11525714]}, {'id': 11543213, 'name': 'mPN iACT bilateral LOWER right', 'type': 'neuron', 'skeleton_ids': [11543212]}, {'id': 11691604, 'name': 'KC17', 'type': 'neuron', 'skeleton_ids': [11691603]}, {'id': 11691641, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [11691640]}, {'id': 11725128, 'name': 'KC12', 'type': 'neuron', 'skeleton_ids': [11725127]}, {'id': 11725804, 'name': 'KC42', 'type': 'neuron', 'skeleton_ids': [11725803]}, {'id': 11785628, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [11785627]}, {'id': 11813288, 'name': 'KC100', 'type': 'neuron', 'skeleton_ids': [11813287]}, {'id': 12156010, 'name': 'handlebar-like 4 (no soma) left ', 'type': 'neuron', 'skeleton_ids': [12156009]}, {'id': 12217573, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [12217572]}, {'id': 12475433, 'name': 'MBE24a left', 'type': 'neuron', 'skeleton_ids': [12475432]}, {'id': 12726583, 'name': 'KC16', 'type': 'neuron', 'skeleton_ids': [12726582]}, {'id': 12805364, 'name': 'MBE22 right', 'type': 'neuron', 'skeleton_ids': [12805363]}, {'id': 12871994, 'name': 'MBE1b left', 'type': 'neuron', 'skeleton_ids': [12871993]}, {'id': 12948471, 'name': 'PVL005 1 left', 'type': 'neuron', 'skeleton_ids': [12948470]}, {'id': 13589252, 'name': '33a PN right', 'type': 'neuron', 'skeleton_ids': [13589251]}, {'id': 13789483, 'name': 'backwards handlebar right', 'type': 'neuron', 'skeleton_ids': [13789482]}, {'id': 14082323, 'name': 'MBE9b left', 'type': 'neuron', 'skeleton_ids': [14082322]}, {'id': 14255343, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [14255342]}, {'id': 14319961, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [14319960]}, {'id': 14460114, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [14460113]}, {'id': 14519049, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [14519048]}, {'id': 14522125, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [14522124]}, {'id': 14541928, 'name': 'AVM011f left', 'type': 'neuron', 'skeleton_ids': [14541927]}, {'id': 14547260, 'name': 'KC32', 'type': 'neuron', 'skeleton_ids': [14547259]}, {'id': 14604029, 'name': '85c PN right', 'type': 'neuron', 'skeleton_ids': [14604028]}, {'id': 14898296, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [14898295]}, {'id': 14898817, 'name': 'KC13', 'type': 'neuron', 'skeleton_ids': [14898816]}, {'id': 14981211, 'name': 'KC very young - DPLc s Left', 'type': 'neuron', 'skeleton_ids': [14981210]}, {'id': 15253922, 'name': 'MBE34 left', 'type': 'neuron', 'skeleton_ids': [15253921]}, {'id': 15398731, 'name': 'MBE23 left', 'type': 'neuron', 'skeleton_ids': [15398730]}, {'id': 15404208, 'name': 'MBE19 right', 'type': 'neuron', 'skeleton_ids': [15404207]}, {'id': 15421364, 'name': 'MBE2a left', 'type': 'neuron', 'skeleton_ids': [15421363]}, {'id': 15571195, 'name': 'MBE12 left', 'type': 'neuron', 'skeleton_ids': [15571194]}, {'id': 15592097, 'name': 'MBE4g left', 'type': 'neuron', 'skeleton_ids': [15592096]}, {'id': 15617306, 'name': 'MBE7b right', 'type': 'neuron', 'skeleton_ids': [15617305]}, {'id': 15691115, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [15691114]}, {'id': 15691119, 'name': 'KC young no claw', 'type': 'neuron', 'skeleton_ids': [15691118]}, {'id': 15691123, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15691122]}, {'id': 15721920, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [15721919]}, {'id': 15722307, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [15722306]}, {'id': 15722868, 'name': 'KC82', 'type': 'neuron', 'skeleton_ids': [15722867]}, {'id': 15734667, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15734666]}, {'id': 15734989, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [15734988]}, {'id': 15741783, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [15741782]}, {'id': 15741952, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15741951]}, {'id': 15764961, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [15764960]}, {'id': 15788420, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15788419]}, {'id': 15788432, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [15788431]}, {'id': 15810984, 'name': 'MBE30 left', 'type': 'neuron', 'skeleton_ids': [15810983]}, {'id': 15828094, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15828093]}, {'id': 15861525, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [15861524]}, {'id': 15939131, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [15939130]}, {'id': 15939180, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [15939179]}, {'id': 15995075, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [15995074]}, {'id': 16015739, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16015738]}, {'id': 16017114, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16017113]}, {'id': 16026043, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16026042]}, {'id': 16039471, 'name': 'KC14', 'type': 'neuron', 'skeleton_ids': [16039470]}, {'id': 16053651, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [16053650]}, {'id': 16053671, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16053670]}, {'id': 16054241, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [16054240]}, {'id': 16058861, 'name': 'KC100', 'type': 'neuron', 'skeleton_ids': [16058860]}, {'id': 16145457, 'name': 'KC101 very young', 'type': 'neuron', 'skeleton_ids': [16145456]}, {'id': 16178284, 'name': 'MBE26 right', 'type': 'neuron', 'skeleton_ids': [16178283]}, {'id': 16210989, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16210988]}, {'id': 16220043, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [16220042]}, {'id': 16223538, 'name': 'MBE18 left', 'type': 'neuron', 'skeleton_ids': [16223537]}, {'id': 16240570, 'name': 'MBE4g right', 'type': 'neuron', 'skeleton_ids': [16240569]}, {'id': 16259596, 'name': '67b PN left', 'type': 'neuron', 'skeleton_ids': [16259595]}, {'id': 16353833, 'name': 'MBE10b right', 'type': 'neuron', 'skeleton_ids': [16353832]}, {'id': 16435407, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16435406]}, {'id': 16435966, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16435965]}, {'id': 16438191, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16438190]}, {'id': 16440650, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16440649]}, {'id': 16440705, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16440704]}, {'id': 16443533, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16443532]}, {'id': 16443537, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16443536]}, {'id': 16443545, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16443544]}, {'id': 16443549, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [16443548]}, {'id': 16443748, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [16443747]}, {'id': 16444328, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16444327]}, {'id': 16448927, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16448926]}, {'id': 16448931, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [16448930]}, {'id': 16448935, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16448934]}, {'id': 16448955, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16448954]}, {'id': 16448968, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16448967]}, {'id': 16448972, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [16448971]}, {'id': 16448980, 'name': 'KC young no claws', 'type': 'neuron', 'skeleton_ids': [16448979]}, {'id': 16448984, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16448983]}, {'id': 16464615, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16464614]}, {'id': 16485080, 'name': 'LH bilateral axon 2 MB_right; BLVa12 b 3', 'type': 'neuron', 'skeleton_ids': [16485079]}, {'id': 16625433, 'name': 'KC30', 'type': 'neuron', 'skeleton_ids': [16625432]}, {'id': 16627774, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16627773]}, {'id': 16627839, 'name': 'KC49?', 'type': 'neuron', 'skeleton_ids': [16627838]}, {'id': 16627862, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16627861]}, {'id': 16627951, 'name': 'KC no claws', 'type': 'neuron', 'skeleton_ids': [16627950]}, {'id': 16628001, 'name': 'KC young', 'type': 'neuron', 'skeleton_ids': [16628000]}, {'id': 16629758, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16629757]}, {'id': 16629789, 'name': 'KC34', 'type': 'neuron', 'skeleton_ids': [16629788]}, {'id': 16629932, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16629931]}, {'id': 16630025, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16630024]}, {'id': 16630265, 'name': 'KC25', 'type': 'neuron', 'skeleton_ids': [16630264]}, {'id': 16630386, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16630385]}, {'id': 16630942, 'name': 'KC22', 'type': 'neuron', 'skeleton_ids': [16630941]}, {'id': 16631115, 'name': 'KC78?', 'type': 'neuron', 'skeleton_ids': [16631114]}, {'id': 16631223, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16631222]}, {'id': 16631249, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [16631248]}, {'id': 16720241, 'name': 'KC72', 'type': 'neuron', 'skeleton_ids': [16720240]}, {'id': 16782831, 'name': 'MBE11b left', 'type': 'neuron', 'skeleton_ids': [16782830]}, {'id': 16795525, 'name': 'mPN iACT bilateral UPPER right', 'type': 'neuron', 'skeleton_ids': [16795524]}, {'id': 16795839, 'name': 'mPN iACT bilateral UPPER left', 'type': 'neuron', 'skeleton_ids': [16795838]}, {'id': 16797673, 'name': 'MBE5a right', 'type': 'neuron', 'skeleton_ids': [16797672]}, {'id': 16846806, 'name': 'MBE5b left', 'type': 'neuron', 'skeleton_ids': [16846805]}, {'id': 16850889, 'name': 'KC73', 'type': 'neuron', 'skeleton_ids': [16850888]}, {'id': 16868924, 'name': 'MBE24b left', 'type': 'neuron', 'skeleton_ids': [16868923]}, {'id': 16883910, 'name': 'MBE33 left', 'type': 'neuron', 'skeleton_ids': [16883909]}, {'id': 16939376, 'name': 'KC67', 'type': 'neuron', 'skeleton_ids': [16939375]}, {'id': 16977882, 'name': 'KC90', 'type': 'neuron', 'skeleton_ids': [16977881]}, {'id': 17013074, 'name': 'MBE24b right', 'type': 'neuron', 'skeleton_ids': [17013073]}, {'id': 17016975, 'name': 'MBE20 left', 'type': 'neuron', 'skeleton_ids': [17016974]}, {'id': 17068731, 'name': 'Ladder (olfactory) ant.', 'type': 'neuron', 'skeleton_ids': [17068730]}, {'id': 17147411, 'name': 'KC no pair', 'type': 'neuron', 'skeleton_ids': [17147410]}, {'id': 17147699, 'name': 'MBE11a left', 'type': 'neuron', 'skeleton_ids': [17147698]}, {'id': 17295913, 'name': 'MBE10b left', 'type': 'neuron', 'skeleton_ids': [17295912]}, {'id': 17355758, 'name': 'MBE7b left', 'type': 'neuron', 'skeleton_ids': [17355757]}, {'id': 17434826, 'name': 'LH bilateral axon 2 MB_left; BLVa12 b 3', 'type': 'neuron', 'skeleton_ids': [17434825]}, {'id': 17454964, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17454963]}, {'id': 17476874, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476873]}, {'id': 17476917, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476916]}, {'id': 17476921, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476920]}, {'id': 17476935, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476934]}, {'id': 17476965, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476964]}, {'id': 17476976, 'name': 'KC very young', 'type': 'neuron', 'skeleton_ids': [17476975]}, {'id': 17884300, 'name': 'Gustatory PN right', 'type': 'neuron', 'skeleton_ids': [17884299]}, {'id': 18028398, 'name': 'MBE5b right', 'type': 'neuron', 'skeleton_ids': [18028397]}]}, {'id': 19711972, 'name': '<NAME>. 2017', 'type': 'annotation', 'neurons': [{'id': 37362, 'name': 'Rh6PR-9;left', 'type': 'neuron', 'skeleton_ids': [37365]}, {'id': 581558, 'name': 'Rh5PR-5;right', 'type': 'neuron', 'skeleton_ids': [7568455]}, {'id': 671648, 'name': 'Rh6PR-4;right', 'type': 'neuron', 'skeleton_ids': [1856512]}, {'id': 765513, 'name': 'Rh5PR-2;right', 'type': 'neuron', 'skeleton_ids': [902877]}, {'id': 766235, 'name': 'Rh5PR-1;right', 'type': 'neuron', 'skeleton_ids': [766246]}, {'id': 871060, 'name': 'pOLP;right', 'type': 'neuron', 'skeleton_ids': [8124177]}, {'id': 899088, 'name': 'Rh5PR-3;right', 'type': 'neuron', 'skeleton_ids': [7485368]}, {'id': 900001, 'name': 'Rh6PR-1;right', 'type': 'neuron', 'skeleton_ids': [900032]}, {'id': 900006, 'name': 'Rh6PR-2;right', 'type': 'neuron', 'skeleton_ids': [7555204]}, {'id': 901490, 'name': 'Rh5PR-6;right', 'type': 'neuron', 'skeleton_ids': [7778722]}, {'id': 902034, 'name': 'Rh6PR-3;right', 'type': 'neuron', 'skeleton_ids': [902038]}, {'id': 945804, 'name': 'Rh6PR-5;right', 'type': 'neuron', 'skeleton_ids': [2544902]}, {'id': 945938, 'name': 'Rh6PR-10;right', 'type': 'neuron', 'skeleton_ids': [8447845]}, {'id': 946306, 'name': 'Rh5PR-4;right', 'type': 'neuron', 'skeleton_ids': [2552701]}, {'id': 946482, 'name': 'Rh6PR-8;right', 'type': 'neuron', 'skeleton_ids': [946481]}, {'id': 946589, 'name': 'Rh6PR-7;right', 'type': 'neuron', 'skeleton_ids': [7566199]}, {'id': 946760, 'name': 'Rh6PR-6;right', 'type': 'neuron', 'skeleton_ids': [2532841]}, {'id': 1807567, 'name': 'LN3;right', 'type': 'neuron', 'skeleton_ids': [2775943]}, {'id': 1816103, 'name': 'anlagen cell_right', 'type': 'neuron', 'skeleton_ids': [1816102]}, {'id': 1829226, 'name': 'LN1;right', 'type': 'neuron', 'skeleton_ids': [7874184]}, {'id': 1872624, 'name': 'nc1;right', 'type': 'neuron', 'skeleton_ids': [7555968]}, {'id': 1902900, 'name': '5thLN;right', 'type': 'neuron', 'skeleton_ids': [8717219]}, {'id': 2200029, 'name': 'Rh6PR-2;left', 'type': 'neuron', 'skeleton_ids': [2200028]}, {'id': 2200809, 'name': 'Rh5PR-1;left', 'type': 'neuron', 'skeleton_ids': [7480731]}, {'id': 2204468, 'name': 'LN2;left', 'type': 'neuron', 'skeleton_ids': [2204467]}, {'id': 2206280, 'name': 'Rh6PR-3;left', 'type': 'neuron', 'skeleton_ids': [2206279]}, {'id': 2208921, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [2208920]}, {'id': 2209092, 'name': 'Rh5PR-4;left', 'type': 'neuron', 'skeleton_ids': [2209091]}, {'id': 2211700, 'name': 'Rh5PR-3;left', 'type': 'neuron', 'skeleton_ids': [7483922]}, {'id': 2213030, 'name': 'LN3;left', 'type': 'neuron', 'skeleton_ids': [8118929]}, {'id': 2215248, 'name': 'Rh6PR-7;left', 'type': 'neuron', 'skeleton_ids': [2215247]}, {'id': 2215448, 'name': 'Rh6PR-4;left', 'type': 'neuron', 'skeleton_ids': [2215447]}, {'id': 2215623, 'name': 'Rh6PR-8;left', 'type': 'neuron', 'skeleton_ids': [2215622]}, {'id': 2216132, 'name': 'Rh6PR-6;left', 'type': 'neuron', 'skeleton_ids': [2216131]}, {'id': 2216188, 'name': 'Rh5PR-2;left', 'type': 'neuron', 'skeleton_ids': [2216187]}, {'id': 2216446, 'name': 'Rh6PR-1;left', 'type': 'neuron', 'skeleton_ids': [2216445]}, {'id': 2237150, 'name': 'GlulOLP;left', 'type': 'neuron', 'skeleton_ids': [8700125]}, {'id': 2301628, 'name': 'nc1;left', 'type': 'neuron', 'skeleton_ids': [8111135]}, {'id': 2301631, 'name': '5thLN;left', 'type': 'neuron', 'skeleton_ids': [7952981]}, {'id': 2436792, 'name': 'LONI;left', 'type': 'neuron', 'skeleton_ids': [7694589]}, {'id': 2512603, 'name': 'LN2;right', 'type': 'neuron', 'skeleton_ids': [2512602]}, {'id': 2524757, 'name': 'LONI;right', 'type': 'neuron', 'skeleton_ids': [8271034]}, {'id': 2528621, 'name': 'ChalOLP;right', 'type': 'neuron', 'skeleton_ids': [8247641]}, {'id': 2528629, 'name': 'GlulOLP;right', 'type': 'neuron', 'skeleton_ids': [7897051]}, {'id': 2528711, 'name': 'OLP4;right', 'type': 'neuron', 'skeleton_ids': [8480418]}, {'id': 2775708, 'name': 'LN4;right', 'type': 'neuron', 'skeleton_ids': [2775707]}, {'id': 7938415, 'name': 'nc2;left', 'type': 'neuron', 'skeleton_ids': [8340950]}, {'id': 7940801, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [7940800]}, {'id': 8243569, 'name': 'nc2;right', 'type': 'neuron', 'skeleton_ids': [8270683]}, {'id': 8267746, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8337623]}, {'id': 8353475, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8353473]}, {'id': 8358801, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8358800]}, {'id': 8361380, 'name': 'anlagen cell-older_left', 'type': 'neuron', 'skeleton_ids': [8361378]}, {'id': 8362222, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8362221]}, {'id': 8432029, 'name': 'anlagen cell_right', 'type': 'neuron', 'skeleton_ids': [8432028]}, {'id': 8448458, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448457]}, {'id': 8448465, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448464]}, {'id': 8448478, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448477]}, {'id': 8448569, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448568]}, {'id': 8448797, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448796]}, {'id': 8448810, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8448809]}, {'id': 8449458, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8449457]}, {'id': 8449479, 'name': 'anlagen cell_left', 'type': 'neuron', 'skeleton_ids': [8449478]}, {'id': 8470257, 'name': 'anlagen cell_right', 'type': 'neuron', 'skeleton_ids': [8470256]}, {'id': 9654958, 'name': 'PVL09 right', 'type': 'neuron', 'skeleton_ids': [9539868]}, {'id': 9681173, 'name': 'PVL09 left', 'type': 'neuron', 'skeleton_ids': [9567051]}, {'id': 9927083, 'name': 'LN1;left', 'type': 'neuron', 'skeleton_ids': [9927082]}, {'id': 9940383, 'name': 'pOLP;left', 'type': 'neuron', 'skeleton_ids': [9940382]}, {'id': 10241717, 'name': 'Rh6PR-5;left', 'type': 'neuron', 'skeleton_ids': [10241716]}, {'id': 16087800, 'name': 'LN4;left', 'type': 'neuron', 'skeleton_ids': [16087799]}, {'id': 16267293, 'name': 'Rh6PR-9;right', 'type': 'neuron', 'skeleton_ids': [16267292]}, {'id': 17414716, 'name': 'ChalOLP;left', 'type': 'neuron', 'skeleton_ids': [17414715]}]}, {'id': 19712046, 'name': '<NAME>. 2015', 'type': 'annotation', 'neurons': [{'id': 32801, 'name': 'vchA/B a1l', 'type': 'neuron', 'skeleton_ids': [7790597]}, {'id': 159277, 'name': 'lch5-2/4 a1l', 'type': 'neuron', 'skeleton_ids': [159280]}, {'id': 162290, 'name': 'lch5-2/4 a1l', 'type': 'neuron', 'skeleton_ids': [162293]}, {'id': 210250, 'name': 'A02o_a3l Wave-1', 'type': 'neuron', 'skeleton_ids': [2194374]}, {'id': 514842, 'name': 'Basin-1 a4l', 'type': 'neuron', 'skeleton_ids': [514841]}, {'id': 519625, 'name': 'A09a_a4l Basin-2', 'type': 'neuron', 'skeleton_ids': [519624]}, {'id': 587084, 'name': 'A09c_a4l Basin-4', 'type': 'neuron', 'skeleton_ids': [587083]}, {'id': 1339401, 'name': 'A00c_a3l', 'type': 'neuron', 'skeleton_ids': [1339400]}, {'id': 1398261, 'name': 'Basin-1 a4r', 'type': 'neuron', 'skeleton_ids': [1398260]}, {'id': 1398328, 'name': 'A09a_a4r Basin-2', 'type': 'neuron', 'skeleton_ids': [1398327]}, {'id': 1501517, 'name': 'A00c_a6l', 'type': 'neuron', 'skeleton_ids': [1042538]}, {'id': 1690577, 'name': 'A00c_a6r', 'type': 'neuron', 'skeleton_ids': [1690576]}, {'id': 1923550, 'name': 'A00c_a4l', 'type': 'neuron', 'skeleton_ids': [2511238]}, {'id': 2349743, 'name': 'Basin-3 a4l', 'type': 'neuron', 'skeleton_ids': [2349742]}, {'id': 2354658, 'name': 'A00c_a5r', 'type': 'neuron', 'skeleton_ids': [2619565]}, {'id': 2368378, 'name': 'lch5-2/4 a1r', 'type': 'neuron', 'skeleton_ids': [7577773]}, {'id': 2457598, 'name': 'A00c_a4r', 'type': 'neuron', 'skeleton_ids': [3093284]}, {'id': 2743339, 'name': 'hook-2_a2l', 'type': 'neuron', 'skeleton_ids': [2743404]}, {'id': 2816093, 'name': 'A00c_a5l', 'type': 'neuron', 'skeleton_ids': [2816457]}, {'id': 2890760, 'name': 'TePn04_a9l', 'type': 'neuron', 'skeleton_ids': [2891695]}, {'id': 3037784, 'name': 'A09b a1l Basin-1', 'type': 'neuron', 'skeleton_ids': [3034133]}, {'id': 3040475, 'name': 'A09c a1l Basin-4', 'type': 'neuron', 'skeleton_ids': [3040481]}, {'id': 3040478, 'name': 'A09a a1l Basin-2', 'type': 'neuron', 'skeleton_ids': [3041612]}, {'id': 3074107, 'name': 'A09b a1r Basin-1', 'type': 'neuron', 'skeleton_ids': [3282869]}, {'id': 3090350, 'name': 'A09c a1r Basin-4', 'type': 'neuron', 'skeleton_ids': [4049878]}, {'id': 3091625, 'name': 'A09a a1r Basin-2', 'type': 'neuron', 'skeleton_ids': [3074106]}, {'id': 3091944, 'name': 'A09g a1r Basin-3', 'type': 'neuron', 'skeleton_ids': [3091943]}, {'id': 3173038, 'name': 'A10j_a3l', 'type': 'neuron', 'skeleton_ids': [9354613]}, {'id': 3304284, 'name': 'A02n_a1r Pseudolooper-4', 'type': 'neuron', 'skeleton_ids': [2637812]}, {'id': 3513767, 'name': 'ddaC_a1l', 'type': 'neuron', 'skeleton_ids': [986460]}, {'id': 3546335, 'name': 'A02m_a1r Pseudolooper-3', 'type': 'neuron', 'skeleton_ids': [3546334]}, {'id': 3546836, 'name': 'A02n_a1l Pseudolooper-4', 'type': 'neuron', 'skeleton_ids': [3546835]}, {'id': 3764793, 'name': 'telegoro-1R', 'type': 'neuron', 'skeleton_ids': [3764792]}, {'id': 3792036, 'name': 'A05q_a2l', 'type': 'neuron', 'skeleton_ids': [3792035]}, {'id': 3863489, 'name': 'hook-2_a2r', 'type': 'neuron', 'skeleton_ids': [3863488]}, {'id': 3957605, 'name': 'T05t_t3r', 'type': 'neuron', 'skeleton_ids': [3957604]}, {'id': 3979182, 'name': 'ipsigoro left', 'type': 'neuron', 'skeleton_ids': [3979181]}, {'id': 4022795, 'name': 'Handle-A a1', 'type': 'neuron', 'skeleton_ids': [4022794]}, {'id': 4060629, 'name': 'A08m_a1l', 'type': 'neuron', 'skeleton_ids': [4060628]}, {'id': 4081480, 'name': 'A19c_a1l', 'type': 'neuron', 'skeleton_ids': [4081479]}, {'id': 4117791, 'name': 'A10l_a2l', 'type': 'neuron', 'skeleton_ids': [4117790]}, {'id': 4139769, 'name': 'A12m_a1r', 'type': 'neuron', 'skeleton_ids': [4139768]}, {'id': 4163235, 'name': 'A10j_a3r', 'type': 'neuron', 'skeleton_ids': [4163234]}, {'id': 4262163, 'name': 'A00c_a3r', 'type': 'neuron', 'skeleton_ids': [4262162]}, {'id': 4302563, 'name': 'A03o_a1l', 'type': 'neuron', 'skeleton_ids': [4302562]}, {'id': 4321719, 'name': 'Handle-B a1', 'type': 'neuron', 'skeleton_ids': [4321712]}, {'id': 4394778, 'name': 'A12q_a1l', 'type': 'neuron', 'skeleton_ids': [4394777]}, {'id': 4536018, 'name': 'A10f_a2r', 'type': 'neuron', 'skeleton_ids': [4536017]}, {'id': 4555764, 'name': 'A08h_a1l', 'type': 'neuron', 'skeleton_ids': [4555763]}, {'id': 4591946, 'name': 'TePn04_a9r', 'type': 'neuron', 'skeleton_ids': [4591945]}, {'id': 4740180, 'name': 'A05q_a1l', 'type': 'neuron', 'skeleton_ids': [4740178]}, {'id': 4760070, 'name': 'A12m_a1l', 'type': 'neuron', 'skeleton_ids': [1767828]}, {'id': 4766067, 'name': 'T08x_t1l', 'type': 'neuron', 'skeleton_ids': [4766066]}, {'id': 4778440, 'name': 'A02m_a1l Pseudolooper-3', 'type': 'neuron', 'skeleton_ids': [4778439]}, {'id': 4826453, 'name': 'A10l_a2r', 'type': 'neuron', 'skeleton_ids': [4826452]}, {'id': 4829814, 'name': 'A12q_a1r', 'type': 'neuron', 'skeleton_ids': [4829813]}, {'id': 4833169, 'name': 'A05q_a1r', 'type': 'neuron', 'skeleton_ids': [4833168]}, {'id': 4839571, 'name': 'A08d_a1r', 'type': 'neuron', 'skeleton_ids': [4839570]}, {'id': 4901447, 'name': 'A10f_a2l', 'type': 'neuron', 'skeleton_ids': [4901446]}, {'id': 5064748, 'name': 'A05q_a2r', 'type': 'neuron', 'skeleton_ids': [5064747]}, {'id': 5108526, 'name': 'A08h_a1r right', 'type': 'neuron', 'skeleton_ids': [5108525]}, {'id': 5121541, 'name': 'telephone-1R', 'type': 'neuron', 'skeleton_ids': [3862443]}, {'id': 5132887, 'name': 'A23g_a1l', 'type': 'neuron', 'skeleton_ids': [5132886]}, {'id': 5185730, 'name': 'A05e_a1r', 'type': 'neuron', 'skeleton_ids': [5185729]}, {'id': 5198940, 'name': 'T08x_t2r', 'type': 'neuron', 'skeleton_ids': [5198939]}, {'id': 5238495, 'name': 'A08m_a2l', 'type': 'neuron', 'skeleton_ids': [5238494]}, {'id': 5243058, 'name': 'gorogoro t2r', 'type': 'neuron', 'skeleton_ids': [5206247]}, {'id': 5268582, 'name': 'basin-related descending 1 sog_l', 'type': 'neuron', 'skeleton_ids': [5268581]}, {'id': 5336477, 'name': 'A19c_a1r', 'type': 'neuron', 'skeleton_ids': [5336476]}, {'id': 5343533, 'name': 'T05t_t3l', 'type': 'neuron', 'skeleton_ids': [5343532]}, {'id': 5343579, 'name': 'A08x_a1l', 'type': 'neuron', 'skeleton_ids': [5343578]}, {'id': 5548413, 'name': 'A08m_a2r', 'type': 'neuron', 'skeleton_ids': [5548412]}, {'id': 5690363, 'name': 'A10a_a1r Chair-1', 'type': 'neuron', 'skeleton_ids': [5690362]}, {'id': 5691935, 'name': 'telephone-1L', 'type': 'neuron', 'skeleton_ids': [5691934]}, {'id': 5754347, 'name': 'telegoro-1L', 'type': 'neuron', 'skeleton_ids': [5754346]}, {'id': 5794679, 'name': 'ipsigoro right', 'type': 'neuron', 'skeleton_ids': [5794678]}, {'id': 5997122, 'name': 'A23g_a1r', 'type': 'neuron', 'skeleton_ids': [5997121]}, {'id': 6294413, 'name': 'A08x_a1r', 'type': 'neuron', 'skeleton_ids': [7184082]}, {'id': 6450382, 'name': 'A05e_a2l', 'type': 'neuron', 'skeleton_ids': [6450381]}, {'id': 6484770, 'name': 'basin-related descending 1 sog_r', 'type': 'neuron', 'skeleton_ids': [6264270]}, {'id': 6535162, 'name': 'A10a_a1l Chair-1', 'type': 'neuron', 'skeleton_ids': [6535161]}, {'id': 6755286, 'name': 'A08m_a1r', 'type': 'neuron', 'skeleton_ids': [6755064]}, {'id': 7152432, 'name': 'A08d_a1l', 'type': 'neuron', 'skeleton_ids': [4975387]}, {'id': 7431703, 'name': 'lch5-2/4 a1r', 'type': 'neuron', 'skeleton_ids': [6276557]}, {'id': 7483663, 'name': 'lch5-3 a1r', 'type': 'neuron', 'skeleton_ids': [7686464]}, {'id': 7491399, 'name': 'lch5-1 a1r', 'type': 'neuron', 'skeleton_ids': [7732444]}, {'id': 7498915, 'name': 'lch5-5 a1r', 'type': 'neuron', 'skeleton_ids': [7694722]}, {'id': 7628069, 'name': "v'ch a1r", 'type': 'neuron', 'skeleton_ids': [7628068]}, {'id': 7631267, 'name': 'vchA/B a1r', 'type': 'neuron', 'skeleton_ids': [7746883]}, {'id': 7759340, 'name': 'lch5-5 a1l', 'type': 'neuron', 'skeleton_ids': [7763691]}, {'id': 7764786, 'name': 'lch5-1 a1l', 'type': 'neuron', 'skeleton_ids': [7764785]}, {'id': 7793417, 'name': 'vchA/B a1l', 'type': 'neuron', 'skeleton_ids': [7796832]}, {'id': 7801256, 'name': "v'ch a1l", 'type': 'neuron', 'skeleton_ids': [7801255]}, {'id': 7836571, 'name': "v'ada_a1r", 'type': 'neuron', 'skeleton_ids': [3786734]}, {'id': 8749848, 'name': 'lch5-3 a1l', 'type': 'neuron', 'skeleton_ids': [8031834]}, {'id': 9654437, 'name': 'A03o_a1r', 'type': 'neuron', 'skeleton_ids': [3180525]}, {'id': 9654573, 'name': 'gorogoro t2l', 'type': 'neuron', 'skeleton_ids': [3720037]}, {'id': 9681075, 'name': 'A09c_a4r Basin-4', 'type': 'neuron', 'skeleton_ids': [1400270]}, {'id': 9681086, 'name': 'Basin-3 a4r', 'type': 'neuron', 'skeleton_ids': [1401011]}, {'id': 9720615, 'name': 'Ipsiphone R', 'type': 'neuron', 'skeleton_ids': [9720614]}, {'id': 9771413, 'name': 'vchA/B a1r', 'type': 'neuron', 'skeleton_ids': [9771412]}, {'id': 10110271, 'name': "v'ada_a1l", 'type': 'neuron', 'skeleton_ids': [10110270]}, {'id': 10110673, 'name': 'ddaC_a1r', 'type': 'neuron', 'skeleton_ids': [10110672]}, {'id': 10179502, 'name': 'A09g a1l Basin-3', 'type': 'neuron', 'skeleton_ids': [10179501]}, {'id': 10273503, 'name': 'vdaB_a1r', 'type': 'neuron', 'skeleton_ids': [10273502]}, {'id': 10290530, 'name': 'vdaB_a1l ', 'type': 'neuron', 'skeleton_ids': [10290529]}, {'id': 10893104, 'name': 'Ipsiphone L', 'type': 'neuron', 'skeleton_ids': [10893103]}, {'id': 12946336, 'name': 'A02o_a3r Wave-1', 'type': 'neuron', 'skeleton_ids': [12946335]}]}, {'id': 19712156, 'name': '<NAME> et al. 2016', 'type': 'annotation', 'neurons': [{'id': 11001, 'name': 'Ladder-f a2', 'type': 'neuron', 'skeleton_ids': [7657358]}, {'id': 11992, 'name': 'Ladder-a a1', 'type': 'neuron', 'skeleton_ids': [11995]}, {'id': 32801, 'name': 'vchA/B a1l', 'type': 'neuron', 'skeleton_ids': [7790597]}, {'id': 159277, 'name': 'lch5-2/4 a1l', 'type': 'neuron', 'skeleton_ids': [159280]}, {'id': 162290, 'name': 'lch5-2/4 a1l', 'type': 'neuron', 'skeleton_ids': [162293]}, {'id': 471672, 'name': 'Ladder-c a1', 'type': 'neuron', 'skeleton_ids': [471720]}, {'id': 471679, 'name': 'Ladder-d a1', 'type': 'neuron', 'skeleton_ids': [471705]}, {'id': 471752, 'name': 'Ladder-b a1', 'type': 'neuron', 'skeleton_ids': [471755]}, {'id': 2368378, 'name': 'lch5-2/4 a1r', 'type': 'neuron', 'skeleton_ids': [7577773]}, {'id': 3037784, 'name': 'A09b a1l Basin-1', 'type': 'neuron', 'skeleton_ids': [3034133]}, {'id': 3040475, 'name': 'A09c a1l Basin-4', 'type': 'neuron', 'skeleton_ids': [3040481]}, {'id': 3040478, 'name': 'A09a a1l Basin-2', 'type': 'neuron', 'skeleton_ids': [3041612]}, {'id': 3074107, 'name': 'A09b a1r Basin-1', 'type': 'neuron', 'skeleton_ids': [3282869]}, {'id': 3090350, 'name': 'A09c a1r Basin-4', 'type': 'neuron', 'skeleton_ids': [4049878]}, {'id': 3091625, 'name': 'A09a a1r Basin-2', 'type': 'neuron', 'skeleton_ids': [3074106]}, {'id': 3091944, 'name': 'A09g a1r Basin-3', 'type': 'neuron', 'skeleton_ids': [3091943]}, {'id': 4022795, 'name': 'Handle-A a1', 'type': 'neuron', 'skeleton_ids': [4022794]}, {'id': 4096367, 'name': 'Drunken-1 a1r', 'type': 'neuron', 'skeleton_ids': [4096366]}, {'id': 4305391, 'name': 'Griddle-2 t3r', 'type': 'neuron', 'skeleton_ids': [4305390]}, {'id': 4321719, 'name': 'Handle-B a1', 'type': 'neuron', 'skeleton_ids': [4321712]}, {'id': 4824879, 'name': 'Griddle-1 a1r', 'type': 'neuron', 'skeleton_ids': [4824878]}, {'id': 6207233, 'name': 'Drunken-1 a1l', 'type': 'neuron', 'skeleton_ids': [6207232]}, {'id': 7195652, 'name': 'Griddle-1 a1l', 'type': 'neuron', 'skeleton_ids': [7195651]}, {'id': 7431703, 'name': 'lch5-2/4 a1r', 'type': 'neuron', 'skeleton_ids': [6276557]}, {'id': 7483663, 'name': 'lch5-3 a1r', 'type': 'neuron', 'skeleton_ids': [7686464]}, {'id': 7491399, 'name': 'lch5-1 a1r', 'type': 'neuron', 'skeleton_ids': [7732444]}, {'id': 7498915, 'name': 'lch5-5 a1r', 'type': 'neuron', 'skeleton_ids': [7694722]}, {'id': 7628069, 'name': "v'ch a1r", 'type': 'neuron', 'skeleton_ids': [7628068]}, {'id': 7631267, 'name': 'vchA/B a1r', 'type': 'neuron', 'skeleton_ids': [7746883]}, {'id': 7683402, 'name': 'Griddle-2 a1r', 'type': 'neuron', 'skeleton_ids': [7683401]}, {'id': 7759340, 'name': 'lch5-5 a1l', 'type': 'neuron', 'skeleton_ids': [7763691]}, {'id': 7764786, 'name': 'lch5-1 a1l', 'type': 'neuron', 'skeleton_ids': [7764785]}, {'id': 7793417, 'name': 'vchA/B a1l', 'type': 'neuron', 'skeleton_ids': [7796832]}, {'id': 7801256, 'name': "v'ch a1l", 'type': 'neuron', 'skeleton_ids': [7801255]}, {'id': 8045801, 'name': 'Griddle-1 t3r', 'type': 'neuron', 'skeleton_ids': [8045800]}, {'id': 8577318, 'name': 'Ladder-f a1', 'type': 'neuron', 'skeleton_ids': [8577316]}, {'id': 8614659, 'name': 'Ladder-a t3', 'type': 'neuron', 'skeleton_ids': [8614658]}, {'id': 8749848, 'name': 'lch5-3 a1l', 'type': 'neuron', 'skeleton_ids': [8031834]}, {'id': 9680932, 'name': 'Ladder-e a1', 'type': 'neuron', 'skeleton_ids': [7599503]}, {'id': 9771413, 'name': 'vchA/B a1r', 'type': 'neuron', 'skeleton_ids': [9771412]}, {'id': 9867542, 'name': 'Griddle-1 t3l', 'type': 'neuron', 'skeleton_ids': [9867541]}, {'id': 10179502, 'name': 'A09g a1l Basin-3', 'type': 'neuron', 'skeleton_ids': [10179501]}, {'id': 10804042, 'name': 'Griddle-2 a1l', 'type': 'neuron', 'skeleton_ids': [10804041]}, {'id': 16310626, 'name': 'Griddle-2 t3l', 'type': 'neuron', 'skeleton_ids': [16310625]}]}, {'id': 19712202, 'name': '<NAME>. 2016', 'type': 'annotation', 'neurons': [{'id': 40149, 'name': 'AN-L-Sens-B1-ACa-10', 'type': 'neuron', 'skeleton_ids': [40152]}, {'id': 864518, 'name': 'SOG Neuron into Thoracic - gap crossing unfinished', 'type': 'neuron', 'skeleton_ids': [864517]}, {'id': 1367172, 'name': 'AN-L-Sens-B1-ACa-04', 'type': 'neuron', 'skeleton_ids': [1367171]}, {'id': 1377587, 'name': 'AN-L-Sens-B1-ACa-07', 'type': 'neuron', 'skeleton_ids': [1377586]}, {'id': 1395972, 'name': 'AN-L-Sens-B1-ACa-11', 'type': 'neuron', 'skeleton_ids': [1395971]}, {'id': 1398254, 'name': 'AN-L-Sens-B1-ACa-05', 'type': 'neuron', 'skeleton_ids': [1398253]}, {'id': 1405494, 'name': 'AN-L-Sens-B1-ACa-09', 'type': 'neuron', 'skeleton_ids': [1405493]}, {'id': 1406464, 'name': 'AN-L-Sens-B1-ACa-14', 'type': 'neuron', 'skeleton_ids': [1406463]}, {'id': 1414207, 'name': 'AN-L-Sens-B1-AVa-16', 'type': 'neuron', 'skeleton_ids': [1414206]}, {'id': 1428272, 'name': 'AN-L-Sens-B1-AVa-22', 'type': 'neuron', 'skeleton_ids': [1428271]}, {'id': 1429970, 'name': 'AN-L-Sens-B1-AVa-20', 'type': 'neuron', 'skeleton_ids': [1429969]}, {'id': 1435467, 'name': 'AN-L-Sens-B1-AVa-21', 'type': 'neuron', 'skeleton_ids': [1435466]}, {'id': 1706717, 'name': 'AN-L-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [1706716]}, {'id': 1711077, 'name': 'A26f_a3r', 'type': 'neuron', 'skeleton_ids': [1711076]}, {'id': 2138430, 'name': 'hugin-RG left 1', 'type': 'neuron', 'skeleton_ids': [2138427]}, {'id': 2141153, 'name': 'IPC 7 right', 'type': 'neuron', 'skeleton_ids': [2355126]}, {'id': 2147244, 'name': 'IPC 2 right', 'type': 'neuron', 'skeleton_ids': [2852273]}, {'id': 2147974, 'name': 'AN-R-Sens-B1-ACa-01', 'type': 'neuron', 'skeleton_ids': [2456826]}, {'id': 2156260, 'name': 'IPC 3 right', 'type': 'neuron', 'skeleton_ids': [2283495]}, {'id': 2240256, 'name': 'hugin-PH left 2', 'type': 'neuron', 'skeleton_ids': [2606279]}, {'id': 2240356, 'name': 'hugin-PH left 1', 'type': 'neuron', 'skeleton_ids': [2679278]}, {'id': 2260756, 'name': 'DH44 1 right', 'type': 'neuron', 'skeleton_ids': [2260755]}, {'id': 2369445, 'name': 'Z: contra_left ', 'type': 'neuron', 'skeleton_ids': [2369444]}, {'id': 2438197, 'name': 'IPC 6 right', 'type': 'neuron', 'skeleton_ids': [2357110]}, {'id': 2535125, 'name': 'hugin-PH right 1', 'type': 'neuron', 'skeleton_ids': [2811369]}, {'id': 2540477, 'name': 'hugin-PH right 2', 'type': 'neuron', 'skeleton_ids': [2810998]}, {'id': 2541120, 'name': 'RG Hugin Input -> paired with #5613144? Check gap crossings (best match atm)', 'type': 'neuron', 'skeleton_ids': [7381964]}, {'id': 2597152, 'name': 'Short-presynapti-D-1R', 'type': 'neuron', 'skeleton_ids': [2597151]}, {'id': 2613533, 'name': 'hugin-VNC left 2', 'type': 'neuron', 'skeleton_ids': [2613532]}, {'id': 2613541, 'name': 'hugin-VNC left 1', 'type': 'neuron', 'skeleton_ids': [2613540]}, {'id': 2619765, 'name': 'neuron - paired with #6796532', 'type': 'neuron', 'skeleton_ids': [2619764]}, {'id': 2620300, 'name': 'Premotor for PaN + MN - paired with #10885848?', 'type': 'neuron', 'skeleton_ids': [2620299]}, {'id': 2622503, 'name': 'Projection Neuron right - Type I - paired with #8010889', 'type': 'neuron', 'skeleton_ids': [2622502]}, {'id': 2622541, 'name': 'A01?_a4r', 'type': 'neuron', 'skeleton_ids': [2622540]}, {'id': 2632169, 'name': 'neuron 2632169', 'type': 'neuron', 'skeleton_ids': [2632168]}, {'id': 2635424, 'name': 'AN-R-Sens-B1-ACa-02', 'type': 'neuron', 'skeleton_ids': [2340336]}, {'id': 2679036, 'name': 'SEZ-IN-15-right', 'type': 'neuron', 'skeleton_ids': [2679035]}, {'id': 2725744, 'name': 'DMS 1 right', 'type': 'neuron', 'skeleton_ids': [2659386]}, {'id': 2815284, 'name': 'Pan Hugin Input 1 - paired with 6', 'type': 'neuron', 'skeleton_ids': [2815283]}, {'id': 2815510, 'name': 'PaN Hugin Input 2', 'type': 'neuron', 'skeleton_ids': [3266033]}, {'id': 2815573, 'name': 'SEZ local interneuron', 'type': 'neuron', 'skeleton_ids': [2815572]}, {'id': 2896541, 'name': 'AN-L-Sens-B1-ACa-01', 'type': 'neuron', 'skeleton_ids': [2284021]}, {'id': 2935174, 'name': 'WS: from olfactory to Hugin interneuron right', 'type': 'neuron', 'skeleton_ids': [2935173]}, {'id': 2935199, 'name': 'AF3 thermo 3rd right', 'type': 'neuron', 'skeleton_ids': [2935198]}, {'id': 3046504, 'name': 'IPC 7 left', 'type': 'neuron', 'skeleton_ids': [3046503]}, {'id': 3158086, 'name': 'AF5: Short-presynaptic-D-1L', 'type': 'neuron', 'skeleton_ids': [3158085]}, {'id': 3192953, 'name': 'AN-R-Sens-B2-AVa-25', 'type': 'neuron', 'skeleton_ids': [3192952]}, {'id': 3276004, 'name': 'L1: Short-presynapticD-3-R', 'type': 'neuron', 'skeleton_ids': [3276003]}, {'id': 3281685, 'name': 'IPC 5 left', 'type': 'neuron', 'skeleton_ids': [3281684]}, {'id': 3299768, 'name': 'hugin-PC right 2', 'type': 'neuron', 'skeleton_ids': [3299767]}, {'id': 3315002, 'name': 'hugin-PC right 1', 'type': 'neuron', 'skeleton_ids': [3315001]}, {'id': 3420916, 'name': 'SOG into brain contralaterally right - paired with #3622234', 'type': 'neuron', 'skeleton_ids': [3420915]}, {'id': 3436421, 'name': 'Pre PMN R', 'type': 'neuron', 'skeleton_ids': [3436420]}, {'id': 3561725, 'name': 'IPC 5 right', 'type': 'neuron', 'skeleton_ids': [3561724]}, {'id': 3594706, 'name': 'hugin-VNC right 2', 'type': 'neuron', 'skeleton_ids': [3594705]}, {'id': 3595838, 'name': 'Brain neuron descending - gap crossings missing', 'type': 'neuron', 'skeleton_ids': [3595837]}, {'id': 3598001, 'name': 'Brain Neuron descending into VNC - right', 'type': 'neuron', 'skeleton_ids': [3598000]}, {'id': 3612559, 'name': 'SOG into Brain contralaterally left - paired with #3420915', 'type': 'neuron', 'skeleton_ids': [3622234]}, {'id': 3613277, 'name': 'Interneuron W right - Inputs from SE0 Serotonin - paired with #9787565', 'type': 'neuron', 'skeleton_ids': [3613276]}, {'id': 3722788, 'name': 'A23f2 (= Pre-goro9)_a3l', 'type': 'neuron', 'skeleton_ids': [3722787]}, {'id': 3795425, 'name': "v'td_a5l (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [3795424]}, {'id': 3915836, 'name': "v'td-2_a3l", 'type': 'neuron', 'skeleton_ids': [3915835]}, {'id': 3946433, 'name': 'WS: from olfactory to Hugin interneuron left', 'type': 'neuron', 'skeleton_ids': [3946432]}, {'id': 4179670, 'name': 'A26g_a2l', 'type': 'neuron', 'skeleton_ids': [4179669]}, {'id': 4284107, 'name': 'Holder_left', 'type': 'neuron', 'skeleton_ids': [4284106]}, {'id': 4363884, 'name': "v'td_a3l (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [4363883]}, {'id': 4364827, 'name': 'Grumpy_left', 'type': 'neuron', 'skeleton_ids': [4364826]}, {'id': 4400768, 'name': 'DH44 2 left', 'type': 'neuron', 'skeleton_ids': [4400767]}, {'id': 4453486, 'name': 'IPC 4 left', 'type': 'neuron', 'skeleton_ids': [4453485]}, {'id': 4506510, 'name': 'IPC 6 left', 'type': 'neuron', 'skeleton_ids': [4506509]}, {'id': 4523585, 'name': 'DMS 1 left', 'type': 'neuron', 'skeleton_ids': [4523584]}, {'id': 4523804, 'name': 'DMS 2 left', 'type': 'neuron', 'skeleton_ids': [4523803]}, {'id': 4595610, 'name': 'pCC a1l', 'type': 'neuron', 'skeleton_ids': [4595609]}, {'id': 4641121, 'name': 'PaN Hugin Input 6 - paired with 1 - gap crossings unfinished', 'type': 'neuron', 'skeleton_ids': [4641120]}, {'id': 4780448, 'name': 'A26f_a3l', 'type': 'neuron', 'skeleton_ids': [4780447]}, {'id': 5038704, 'name': 'hugin-RG right 1', 'type': 'neuron', 'skeleton_ids': [5038703]}, {'id': 5038921, 'name': 'MBE - Input of Hugin RG Candidates right 1 - paired with #5688091', 'type': 'neuron', 'skeleton_ids': [5038920]}, {'id': 5311300, 'name': 'T1r Ascending into PC_right', 'type': 'neuron', 'skeleton_ids': [5311299]}, {'id': 5349962, 'name': 'Happy Right ', 'type': 'neuron', 'skeleton_ids': [5349961]}, {'id': 5378684, 'name': 'T1l Ascending into PC_left', 'type': 'neuron', 'skeleton_ids': [5378683]}, {'id': 5382877, 'name': "v'td_a3r (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [5382876]}, {'id': 5404888, 'name': 'Happy Left ', 'type': 'neuron', 'skeleton_ids': [5404887]}, {'id': 5417850, 'name': "v'td_a4r (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [9503203]}, {'id': 5500959, 'name': 'Coaster (NPF) right', 'type': 'neuron', 'skeleton_ids': [5500958]}, {'id': 5539468, 'name': 'T1l descending ', 'type': 'neuron', 'skeleton_ids': [5644800]}, {'id': 5601925, 'name': 'hugin-RG right 2', 'type': 'neuron', 'skeleton_ids': [5601924]}, {'id': 5607157, 'name': 'A26x_a4r', 'type': 'neuron', 'skeleton_ids': [5607156]}, {'id': 5613145, 'name': 'Inputs to RG candidates -> paired with #7381964? Check gap crossings (best match atm)', 'type': 'neuron', 'skeleton_ids': [5613144]}, {'id': 5614979, 'name': 'T1l asc contra 01', 'type': 'neuron', 'skeleton_ids': [5614978]}, {'id': 5615962, 'name': 'hugin-RG left 2', 'type': 'neuron', 'skeleton_ids': [5615961]}, {'id': 5617166, 'name': "v'td-2_a2l", 'type': 'neuron', 'skeleton_ids': [5617164]}, {'id': 5688092, 'name': 'Brain neuron into SOG - paired with #5038920 - missing gap crossing', 'type': 'neuron', 'skeleton_ids': [5688091]}, {'id': 5860971, 'name': 'A23f2 (= Pre-goro9)_a3r', 'type': 'neuron', 'skeleton_ids': [5860970]}, {'id': 6445995, 'name': 'Pre PMN L', 'type': 'neuron', 'skeleton_ids': [6445994]}, {'id': 6795359, 'name': 'hugin-VNC right 1', 'type': 'neuron', 'skeleton_ids': [6795358]}, {'id': 6796533, 'name': 'neuron - paired with #2619764', 'type': 'neuron', 'skeleton_ids': [6796532]}, {'id': 6802554, 'name': 'T1l asc contra 02', 'type': 'neuron', 'skeleton_ids': [6802553]}, {'id': 6883832, 'name': 'AN-R-Sens-B1-AVa-16', 'type': 'neuron', 'skeleton_ids': [6883831]}, {'id': 7136255, 'name': 'Moggy_left', 'type': 'neuron', 'skeleton_ids': [7136254]}, {'id': 7165166, 'name': 'Medial Unpaired T3?', 'type': 'neuron', 'skeleton_ids': [7165165]}, {'id': 7331556, 'name': "v'td_a6r (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [7331555]}, {'id': 7338310, 'name': 'Brain into VNC_l', 'type': 'neuron', 'skeleton_ids': [7338309]}, {'id': 7340633, 'name': "v'td_a6l (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [7340629]}, {'id': 7340665, 'name': 'Midline Projection Neuron _ a3l', 'type': 'neuron', 'skeleton_ids': [7340664]}, {'id': 7557605, 'name': 'L1: Short-presynapticD-3-L', 'type': 'neuron', 'skeleton_ids': [7557604]}, {'id': 8005610, 'name': "v'td_a2l (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [8005609]}, {'id': 8008618, 'name': 'T1r descending', 'type': 'neuron', 'skeleton_ids': [8008617]}, {'id': 8010890, 'name': 'Projection Neuron left - Type I - paired with #2622502', 'type': 'neuron', 'skeleton_ids': [8010889]}, {'id': 8748453, 'name': 'A26f_a2l', 'type': 'neuron', 'skeleton_ids': [8748462]}, {'id': 9319256, 'name': 'A01?_a4l', 'type': 'neuron', 'skeleton_ids': [9319255]}, {'id': 9563255, 'name': 'neuron 9563255', 'type': 'neuron', 'skeleton_ids': [9563254]}, {'id': 9654603, 'name': 'Sweetness Right 130521_Cand1_OSN2_IN5 Bilateral', 'type': 'neuron', 'skeleton_ids': [9455820]}, {'id': 9654797, 'name': 'AF3 thermo 3rd left', 'type': 'neuron', 'skeleton_ids': [4785242]}, {'id': 9679159, 'name': 'AN-L-Sens-B1-AVa-17', 'type': 'neuron', 'skeleton_ids': [1375101]}, {'id': 9716859, 'name': 'AN-R-Sens-B1-AVa-19', 'type': 'neuron', 'skeleton_ids': [9716858]}, {'id': 9748270, 'name': 'AN-R-Sens-B1-AVa-18', 'type': 'neuron', 'skeleton_ids': [9748269]}, {'id': 9748580, 'name': 'hugin-PC right 3', 'type': 'neuron', 'skeleton_ids': [9748579]}, {'id': 9748982, 'name': 'Moggy_right', 'type': 'neuron', 'skeleton_ids': [9748981]}, {'id': 9758504, 'name': 'Holder_right', 'type': 'neuron', 'skeleton_ids': [9758503]}, {'id': 9787566, 'name': 'Interneuron W left - Inputs from SE0 Serotonin - paired with #3613276', 'type': 'neuron', 'skeleton_ids': [9787565]}, {'id': 9789131, 'name': 'hugin-PC left 1', 'type': 'neuron', 'skeleton_ids': [9789130]}, {'id': 9793490, 'name': 'BC ; Left', 'type': 'neuron', 'skeleton_ids': [9793489]}, {'id': 9796598, 'name': 'hugin-PC left 2', 'type': 'neuron', 'skeleton_ids': [9796597]}, {'id': 9805521, 'name': 'hugin-PC left 3', 'type': 'neuron', 'skeleton_ids': [9805520]}, {'id': 9805589, 'name': 'Angler_left', 'type': 'neuron', 'skeleton_ids': [9805588]}, {'id': 9813903, 'name': 'hugin-PC left 4', 'type': 'neuron', 'skeleton_ids': [9813902]}, {'id': 9835170, 'name': "v'td_a4l (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [9835169]}, {'id': 9838295, 'name': 'AN-L-Sens-B1-ACa-13', 'type': 'neuron', 'skeleton_ids': [9838294]}, {'id': 9903958, 'name': 'Sweetness Left SOG neuron', 'type': 'neuron', 'skeleton_ids': [9903957]}, {'id': 9935753, 'name': 'T1r asc contra 01', 'type': 'neuron', 'skeleton_ids': [9935752]}, {'id': 10119917, 'name': 'Z: contra_right', 'type': 'neuron', 'skeleton_ids': [10119916]}, {'id': 10329209, 'name': 'IPC 1 left', 'type': 'neuron', 'skeleton_ids': [10329208]}, {'id': 10418395, 'name': 'DH44 1 left', 'type': 'neuron', 'skeleton_ids': [10418394]}, {'id': 10459724, 'name': 'IPC 2 left', 'type': 'neuron', 'skeleton_ids': [10459723]}, {'id': 10563756, 'name': 'DH44 3 left', 'type': 'neuron', 'skeleton_ids': [10563755]}, {'id': 10716081, 'name': 'Brain descending into VNC', 'type': 'neuron', 'skeleton_ids': [10716080]}, {'id': 10728319, 'name': 'Brain into VNC', 'type': 'neuron', 'skeleton_ids': [10728318]}, {'id': 10852876, 'name': "v'td_a2r (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [10852875]}, {'id': 10868478, 'name': "v'td-2_a1r", 'type': 'neuron', 'skeleton_ids': [10868477]}, {'id': 10885849, 'name': 'T1 Local Interneuron - paired with #2620299? a27l t2?', 'type': 'neuron', 'skeleton_ids': [10885848]}, {'id': 11121241, 'name': 'A00g_a3 ?', 'type': 'neuron', 'skeleton_ids': [11121240]}, {'id': 11291345, 'name': "v'td_a5r (Abdominal Sensory)", 'type': 'neuron', 'skeleton_ids': [11291344]}, {'id': 11291635, 'name': 'Midline Projection Neuron - paired with #7340664', 'type': 'neuron', 'skeleton_ids': [11291634]}, {'id': 11517137, 'name': 'neuron 11517137', 'type': 'neuron', 'skeleton_ids': [11517136]}, {'id': 11734961, 'name': 'Medial Unpaired T2', 'type': 'neuron', 'skeleton_ids': [11734960]}, {'id': 11868261, 'name': 'Angler_right', 'type': 'neuron', 'skeleton_ids': [11868260]}, {'id': 11874131, 'name': "v'td-2_a1l", 'type': 'neuron', 'skeleton_ids': [11874130]}, {'id': 12042431, 'name': 'VNC interneuron', 'type': 'neuron', 'skeleton_ids': [12042430]}, {'id': 12634155, 'name': 'IPC 3 left', 'type': 'neuron', 'skeleton_ids': [12634154]}, {'id': 12824060, 'name': 'post B5 37', 'type': 'neuron', 'skeleton_ids': [12824059]}, {'id': 12920746, 'name': 'post B5 17', 'type': 'neuron', 'skeleton_ids': [12920745]}, {'id': 12944430, 'name': 'neuron 12944430', 'type': 'neuron', 'skeleton_ids': [12944429]}, {'id': 13429046, 'name': 'AF4: Large Spider Neuron right', 'type': 'neuron', 'skeleton_ids': [13429045]}, {'id': 13535381, 'name': 'IPC 1 right', 'type': 'neuron', 'skeleton_ids': [13535380]}, {'id': 13535388, 'name': 'DH44 3 right', 'type': 'neuron', 'skeleton_ids': [13535387]}, {'id': 13581831, 'name': 'hugin-PC right 4', 'type': 'neuron', 'skeleton_ids': [13581830]}, {'id': 13744426, 'name': 'AN-L-Sens-B1-ACa-02', 'type': 'neuron', 'skeleton_ids': [13744425]}, {'id': 13751195, 'name': 'DH44 2 right', 'type': 'neuron', 'skeleton_ids': [13751194]}, {'id': 13793213, 'name': 'DMS 2 right', 'type': 'neuron', 'skeleton_ids': [13793212]}, {'id': 14728551, 'name': 'A09e_a3r', 'type': 'neuron', 'skeleton_ids': [14728550]}, {'id': 14795311, 'name': 'A09e_a3l', 'type': 'neuron', 'skeleton_ids': [14795310]}, {'id': 15202264, 'name': 'IPC 4 right', 'type': 'neuron', 'skeleton_ids': [15202263]}, {'id': 15242800, 'name': 'BC ; Right', 'type': 'neuron', 'skeleton_ids': [15242799]}, {'id': 15506095, 'name': 'AN-R-Sens-B2-ACp-18', 'type': 'neuron', 'skeleton_ids': [15506094]}, {'id': 15532106, 'name': 'AN-R-Sens-B2-AVa-24', 'type': 'neuron', 'skeleton_ids': [15532105]}, {'id': 15541945, 'name': 'AN-R-Sens-B2-ACp-02', 'type': 'neuron', 'skeleton_ids': [15541944]}, {'id': 15541977, 'name': 'AN-R-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [15541976]}, {'id': 15543069, 'name': 'AN-R-Sens-B1-AVa-21', 'type': 'neuron', 'skeleton_ids': [15543068]}, {'id': 15562138, 'name': 'AN-R-Sens-B1-ACa-07', 'type': 'neuron', 'skeleton_ids': [15562137]}, {'id': 15564707, 'name': 'AN-R-Sens-B1-AVa-24', 'type': 'neuron', 'skeleton_ids': [15564706]}, {'id': 15564783, 'name': 'AN-R-Sens-B1-AVa-23', 'type': 'neuron', 'skeleton_ids': [15564782]}, {'id': 15564808, 'name': 'AN-R-Sens-B1-AVa-22', 'type': 'neuron', 'skeleton_ids': [15564807]}, {'id': 15572415, 'name': 'AN-R-Sens-B1-AVa-25', 'type': 'neuron', 'skeleton_ids': [15572414]}, {'id': 15573397, 'name': 'AN-R-Sens-B1-ACa-09', 'type': 'neuron', 'skeleton_ids': [15573396]}, {'id': 15573451, 'name': 'AN-R-Sens-B1-ACa-14', 'type': 'neuron', 'skeleton_ids': [15573450]}, {'id': 15574232, 'name': 'AN-R-Sens-B1-ACa-08', 'type': 'neuron', 'skeleton_ids': [15574231]}, {'id': 15574329, 'name': 'AN-R-Sens-B1-AVa-15', 'type': 'neuron', 'skeleton_ids': [15574328]}, {'id': 15575328, 'name': 'AN-R-Sens-B1-AVa-26', 'type': 'neuron', 'skeleton_ids': [15575327]}, {'id': 15769420, 'name': 'AN-L-Sens-B1-ACa-08', 'type': 'neuron', 'skeleton_ids': [15769419]}, {'id': 15769681, 'name': 'AN-L-Sens-B1-AVa-19', 'type': 'neuron', 'skeleton_ids': [15769680]}, {'id': 15770033, 'name': 'AN-L-Sens-B1-ACa-03', 'type': 'neuron', 'skeleton_ids': [15770032]}, {'id': 15770905, 'name': 'AN-L-Sens-B1-AVa-24', 'type': 'neuron', 'skeleton_ids': [15770904]}, {'id': 15995930, 'name': 'AN-L-Sens-B2-ACp-20', 'type': 'neuron', 'skeleton_ids': [15995929]}, {'id': 15997667, 'name': 'AN-L-Sens-B2-ACp-01', 'type': 'neuron', 'skeleton_ids': [15997666]}, {'id': 15998754, 'name': 'AN-L-Sens-B2-AVa-25', 'type': 'neuron', 'skeleton_ids': [15998753]}, {'id': 16442345, 'name': 'AN-R-Sens-B1-AVa-20', 'type': 'neuron', 'skeleton_ids': [16442344]}, {'id': 16575804, 'name': 'Grumpy_right', 'type': 'neuron', 'skeleton_ids': [16575803]}, {'id': 17641914, 'name': 'AF4: TOH left', 'type': 'neuron', 'skeleton_ids': [17641913]}, {'id': 18461989, 'name': 'Contralateral axon', 'type': 'neuron', 'skeleton_ids': [18461988]}, {'id': 18520223, 'name': 'A26f_a2r', 'type': 'neuron', 'skeleton_ids': [18520222]}, {'id': 19120854, 'name': 'pCC a1r', 'type': 'neuron', 'skeleton_ids': [19120853]}, {'id': 19912141, 'name': 'T1r asc contra 02', 'type': 'neuron', 'skeleton_ids': [19912140]}]}, {'id': 19712396, 'name': 'Fushiki et al. 2016', 'type': 'annotation', 'neurons': [{'id': 3365861, 'name': 'A02j Looper-10_a3l', 'type': 'neuron', 'skeleton_ids': [3365860]}, {'id': 4038809, 'name': 'A27k_a2l', 'type': 'neuron', 'skeleton_ids': [4038808]}, {'id': 4113613, 'name': 'A27k_a2r', 'type': 'neuron', 'skeleton_ids': [4113612]}, {'id': 4213418, 'name': 'A27k_a1l', 'type': 'neuron', 'skeleton_ids': [4213417]}, {'id': 4348666, 'name': 'A02j Looper-10_a3r', 'type': 'neuron', 'skeleton_ids': [4348665]}, {'id': 4732075, 'name': 'T01d4_t3r', 'type': 'neuron', 'skeleton_ids': [4732074]}, {'id': 4847162, 'name': 'A03xC_a2r pre SNC', 'type': 'neuron', 'skeleton_ids': [4847161]}, {'id': 4848301, 'name': 'A03x_a2r (EGHBAL)', 'type': 'neuron', 'skeleton_ids': [4848300]}, {'id': 4850741, 'name': 'A18f?_a3l', 'type': 'neuron', 'skeleton_ids': [4850740]}, {'id': 4990881, 'name': 'A05k_a1l', 'type': 'neuron', 'skeleton_ids': [4990880]}, {'id': 5351822, 'name': 'A05a_a1r', 'type': 'neuron', 'skeleton_ids': [5351821]}, {'id': 5458047, 'name': 'Saaghi-1_a2l (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [5458046]}, {'id': 6811350, 'name': 'A01x3_a1r', 'type': 'neuron', 'skeleton_ids': [6811349]}, {'id': 7019057, 'name': 'A01a?_a1r', 'type': 'neuron', 'skeleton_ids': [7019050]}, {'id': 7783765, 'name': 'vdaC_a1l (class II)', 'type': 'neuron', 'skeleton_ids': [7783764]}, {'id': 7808342, 'name': 'crescent a1l', 'type': 'neuron', 'skeleton_ids': [7808341]}, {'id': 7924479, 'name': 'A01b2_a1l ', 'type': 'neuron', 'skeleton_ids': [7924478]}, {'id': 7926497, 'name': 'A02j Looper-10_a2r_pre-motor', 'type': 'neuron', 'skeleton_ids': [7926496]}, {'id': 7987964, 'name': 'T03g2?_t3r ', 'type': 'neuron', 'skeleton_ids': [7987962]}, {'id': 8039618, 'name': 'A08e3_a1r', 'type': 'neuron', 'skeleton_ids': [8039617]}, {'id': 8059284, 'name': 'A08s1_a1l ', 'type': 'neuron', 'skeleton_ids': [8059283]}, {'id': 8257689, 'name': 'A27p a1r', 'type': 'neuron', 'skeleton_ids': [8257688]}, {'id': 8421330, 'name': 'A08e3_a1l ', 'type': 'neuron', 'skeleton_ids': [8421329]}, {'id': 9544993, 'name': 'A01x3_a1l', 'type': 'neuron', 'skeleton_ids': [9544992]}, {'id': 9976876, 'name': 'A27p a1l', 'type': 'neuron', 'skeleton_ids': [9976875]}, {'id': 10004244, 'name': 'A27k_a1r', 'type': 'neuron', 'skeleton_ids': [10004243]}, {'id': 10017074, 'name': 'A27k_up_a1r', 'type': 'neuron', 'skeleton_ids': [10017073]}, {'id': 10053305, 'name': 'A05k_a1r', 'type': 'neuron', 'skeleton_ids': [10053304]}, {'id': 10053735, 'name': 'vdaC_a1r (class II)', 'type': 'neuron', 'skeleton_ids': [10053734]}, {'id': 10089194, 'name': 'A03a6_a1l ', 'type': 'neuron', 'skeleton_ids': [10089193]}, {'id': 10089253, 'name': 'A03x_a2l (EGHBAL)', 'type': 'neuron', 'skeleton_ids': [10089252]}, {'id': 10089446, 'name': 'Saaghi-1_a2r (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [10089445]}, {'id': 10133526, 'name': 'A14 dude?_a1r', 'type': 'neuron', 'skeleton_ids': [10133525]}, {'id': 10223573, 'name': 'A27h_a1l', 'type': 'neuron', 'skeleton_ids': [10223572]}, {'id': 10227540, 'name': 'crescent a1r', 'type': 'neuron', 'skeleton_ids': [10227539]}, {'id': 10376513, 'name': 'A27h_a2r', 'type': 'neuron', 'skeleton_ids': [10376512]}, {'id': 10398172, 'name': 'A27h_a1r', 'type': 'neuron', 'skeleton_ids': [10398171]}, {'id': 10414726, 'name': 'A27h_a2l', 'type': 'neuron', 'skeleton_ids': [10414725]}, {'id': 10474992, 'name': 'A00g_a2', 'type': 'neuron', 'skeleton_ids': [10474991]}, {'id': 10492060, 'name': 'A31f_a1r?', 'type': 'neuron', 'skeleton_ids': [10492059]}, {'id': 10492186, 'name': 'vdaA_a1l (class II)', 'type': 'neuron', 'skeleton_ids': [10492185]}, {'id': 10492632, 'name': 'A05a_t3r', 'type': 'neuron', 'skeleton_ids': [10492631]}, {'id': 12140926, 'name': 'A01a_a1l', 'type': 'neuron', 'skeleton_ids': [12140925]}, {'id': 12643186, 'name': 'A03a4_a3l', 'type': 'neuron', 'skeleton_ids': [12643185]}, {'id': 12778087, 'name': 'A18b3_a2l', 'type': 'neuron', 'skeleton_ids': [12778086]}, {'id': 12787697, 'name': 'A01b2_a1r', 'type': 'neuron', 'skeleton_ids': [12787696]}, {'id': 12809153, 'name': 'A31d_a1l', 'type': 'neuron', 'skeleton_ids': [12809152]}, {'id': 12809875, 'name': 'T03g2?_t3l', 'type': 'neuron', 'skeleton_ids': [12809874]}, {'id': 12820086, 'name': 'A14b a1l', 'type': 'neuron', 'skeleton_ids': [12820085]}, {'id': 12834796, 'name': 'A14b a1r', 'type': 'neuron', 'skeleton_ids': [12834795]}, {'id': 12873723, 'name': 'A31d_a1r', 'type': 'neuron', 'skeleton_ids': [12873722]}, {'id': 12873915, 'name': 'A27e_a1l', 'type': 'neuron', 'skeleton_ids': [12873914]}, {'id': 12900706, 'name': 'A03ax_a2l', 'type': 'neuron', 'skeleton_ids': [12900705]}, {'id': 12901481, 'name': 'A03xC_a2l pre SNC', 'type': 'neuron', 'skeleton_ids': [12901480]}, {'id': 12946255, 'name': 'A03 12946255', 'type': 'neuron', 'skeleton_ids': [12946254]}, {'id': 12949241, 'name': 'T01d4_t3l', 'type': 'neuron', 'skeleton_ids': [12949240]}, {'id': 13095127, 'name': 'A31f_a1l?', 'type': 'neuron', 'skeleton_ids': [13095126]}, {'id': 13121968, 'name': 'A18b3_a2r', 'type': 'neuron', 'skeleton_ids': [13121967]}, {'id': 13122057, 'name': 'A00g_a1', 'type': 'neuron', 'skeleton_ids': [13122056]}, {'id': 13124816, 'name': 'vdaA_a1r (class II)', 'type': 'neuron', 'skeleton_ids': [13124815]}, {'id': 13302530, 'name': 'A18f?_a3r', 'type': 'neuron', 'skeleton_ids': [13302529]}, {'id': 13522679, 'name': 'A05a_a1L', 'type': 'neuron', 'skeleton_ids': [13522678]}, {'id': 13525685, 'name': 'A03a4_a3r', 'type': 'neuron', 'skeleton_ids': [13525684]}, {'id': 13545405, 'name': 'neuron 13545405', 'type': 'neuron', 'skeleton_ids': [13545404]}, {'id': 14046246, 'name': 'A27a_a1r', 'type': 'neuron', 'skeleton_ids': [14046245]}, {'id': 14046280, 'name': 'A02d_a1l ', 'type': 'neuron', 'skeleton_ids': [14046279]}, {'id': 14046295, 'name': 'A02d_a1r', 'type': 'neuron', 'skeleton_ids': [14046294]}, {'id': 14046465, 'name': 'A12b?_a1l', 'type': 'neuron', 'skeleton_ids': [14046464]}, {'id': 14046473, 'name': 'A12b?_a1r', 'type': 'neuron', 'skeleton_ids': [14046472]}, {'id': 14913094, 'name': 'A08s1_a1r', 'type': 'neuron', 'skeleton_ids': [14913093]}, {'id': 15504370, 'name': 'A27j2_a1l_GABA', 'type': 'neuron', 'skeleton_ids': [15504369]}, {'id': 15504377, 'name': 'A27j2_a1r_GABA', 'type': 'neuron', 'skeleton_ids': [15504376]}, {'id': 15565588, 'name': 'neuron 15565588', 'type': 'neuron', 'skeleton_ids': [15565587]}, {'id': 16066986, 'name': 'A27a_a1l', 'type': 'neuron', 'skeleton_ids': [16066985]}, {'id': 16257963, 'name': 'A03a6_a1r', 'type': 'neuron', 'skeleton_ids': [16257962]}, {'id': 16274374, 'name': 'A02j Looper-10_a2l_pre-motor', 'type': 'neuron', 'skeleton_ids': [16274373]}, {'id': 17077343, 'name': 'A27e_a1r', 'type': 'neuron', 'skeleton_ids': [17077342]}, {'id': 17114294, 'name': 'A27k_up_a1l', 'type': 'neuron', 'skeleton_ids': [17114293]}, {'id': 17435372, 'name': 'Saaghi-1_a1r (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [17435371]}, {'id': 18418513, 'name': 'Saaghi-1_a1l (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [18418512]}, {'id': 18427864, 'name': 't05a_t3l', 'type': 'neuron', 'skeleton_ids': [18427863]}, {'id': 18507413, 'name': 'Neuron 18507413 with a soma', 'type': 'neuron', 'skeleton_ids': [18507412]}]}, {'id': 19716589, 'name': 'Takagi et al. 2017', 'type': 'annotation', 'neurons': [{'id': 20208, 'name': 'A02o_a1l Wave-1', 'type': 'neuron', 'skeleton_ids': [918889]}, {'id': 360669, 'name': 'putative MD III/IV_T1r', 'type': 'neuron', 'skeleton_ids': [360668]}, {'id': 360862, 'name': 'Pre-goro6 right', 'type': 'neuron', 'skeleton_ids': [360861]}, {'id': 1931982, 'name': 'T19v_t1r', 'type': 'neuron', 'skeleton_ids': [1931981]}, {'id': 3931435, 'name': 'MN1 aCC_a3l', 'type': 'neuron', 'skeleton_ids': [3931434]}, {'id': 3942270, 'name': 'MN1 aCC_a3r', 'type': 'neuron', 'skeleton_ids': [3942269]}, {'id': 3952744, 'name': 'putative md III/IV_t1l', 'type': 'neuron', 'skeleton_ids': [3952743]}, {'id': 4514712, 'name': 'putative md class III/IV_t2l', 'type': 'neuron', 'skeleton_ids': [4514711]}, {'id': 4791652, 'name': 'MN6/7 RP3_a2r', 'type': 'neuron', 'skeleton_ids': [4791651]}, {'id': 5243058, 'name': 'gorogoro t2r', 'type': 'neuron', 'skeleton_ids': [5206247]}, {'id': 5904834, 'name': 'A03a5_a1l', 'type': 'neuron', 'skeleton_ids': [5904833]}, {'id': 6730711, 'name': 'goro-like t1r', 'type': 'neuron', 'skeleton_ids': [6724973]}, {'id': 8342583, 'name': 'swallowtail_t2r', 'type': 'neuron', 'skeleton_ids': [8342582]}, {'id': 9654573, 'name': 'gorogoro t2l', 'type': 'neuron', 'skeleton_ids': [3720037]}, {'id': 9654821, 'name': 'A03a5_a1r', 'type': 'neuron', 'skeleton_ids': [5297998]}, {'id': 13874214, 'name': 'MN30 RP4_a2l', 'type': 'neuron', 'skeleton_ids': [13874213]}, {'id': 13875212, 'name': 'MN2? ISN MN (U3?-MN2- NB7-1?)_a2r', 'type': 'neuron', 'skeleton_ids': [13875211]}, {'id': 13880944, 'name': 'MN RP4_a2r', 'type': 'neuron', 'skeleton_ids': [13880943]}, {'id': 13885024, 'name': 'MN ISN MN_a2l (U4/U5)', 'type': 'neuron', 'skeleton_ids': [13885023]}, {'id': 14087108, 'name': 'MN30 RP4_a1r', 'type': 'neuron', 'skeleton_ids': [14087107]}, {'id': 14194235, 'name': 'MN30 RP4_a1l', 'type': 'neuron', 'skeleton_ids': [14194234]}, {'id': 14194704, 'name': 'MN13 ISN MN-VL2_a1r', 'type': 'neuron', 'skeleton_ids': [14194703]}, {'id': 14472973, 'name': 'T07u_t2r', 'type': 'neuron', 'skeleton_ids': [14472972]}, {'id': 15892038, 'name': 'T07u_t2l', 'type': 'neuron', 'skeleton_ids': [15892037]}, {'id': 16100982, 'name': 'Pre-swallowtail1 right', 'type': 'neuron', 'skeleton_ids': [16100981]}, {'id': 16264803, 'name': 'MN6/7 RP3_a2l', 'type': 'neuron', 'skeleton_ids': [16264802]}, {'id': 16306344, 'name': 'md class IV? t1l', 'type': 'neuron', 'skeleton_ids': [16306343]}, {'id': 16418095, 'name': 'goro-like t1l', 'type': 'neuron', 'skeleton_ids': [16418094]}, {'id': 16605708, 'name': 'T19v_t1l', 'type': 'neuron', 'skeleton_ids': [16605707]}, {'id': 18181777, 'name': 'md class IV? t1r', 'type': 'neuron', 'skeleton_ids': [18181776]}, {'id': 18189978, 'name': 'swallowtail t2l', 'type': 'neuron', 'skeleton_ids': [18189977]}, {'id': 18202033, 'name': 'md class IV? t1r', 'type': 'neuron', 'skeleton_ids': [18202032]}, {'id': 18303101, 'name': 'ddac t2r', 'type': 'neuron', 'skeleton_ids': [18303100]}, {'id': 18339059, 'name': 'md class IV? t1l', 'type': 'neuron', 'skeleton_ids': [18339058]}, {'id': 18684929, 'name': 'A02o_a1r Wave-1', 'type': 'neuron', 'skeleton_ids': [18684928]}, {'id': 18694052, 'name': 'Pre-goro6 left', 'type': 'neuron', 'skeleton_ids': [18694051]}, {'id': 18695580, 'name': 'Pre-swallowtail1 left', 'type': 'neuron', 'skeleton_ids': [18695579]}, {'id': 19111931, 'name': 'MN13 ISN MN-VL2_a1l ', 'type': 'neuron', 'skeleton_ids': [19111930]}]}, {'id': 19756504, 'name': 'Heckscher et al. 2015', 'type': 'annotation', 'neurons': [{'id': 3641569, 'name': 'SN motor neuron a1r ventral dendrite', 'type': 'neuron', 'skeleton_ids': [3641568]}, {'id': 3943465, 'name': 'MN1 aCC_a1r', 'type': 'neuron', 'skeleton_ids': [4488976]}, {'id': 4028115, 'name': 'MN5 LO1_a1l', 'type': 'neuron', 'skeleton_ids': [4028114]}, {'id': 4042316, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1l', 'type': 'neuron', 'skeleton_ids': [4042315]}, {'id': 4050399, 'name': 'SN motor neuron a1l', 'type': 'neuron', 'skeleton_ids': [4050398]}, {'id': 4101977, 'name': 'MN27 (SN MN VA2)_a1r', 'type': 'neuron', 'skeleton_ids': [4101976]}, {'id': 4102729, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1r ', 'type': 'neuron', 'skeleton_ids': [4102728]}, {'id': 4121536, 'name': 'MN23/24 (RF: LT3/LT4) LT4_a1r ', 'type': 'neuron', 'skeleton_ids': [4121534]}, {'id': 4271520, 'name': 'SN motor neuron a1l ventral dendrite', 'type': 'neuron', 'skeleton_ids': [4271519]}, {'id': 4274366, 'name': 'MN29 (SN MN VA3)_a1l', 'type': 'neuron', 'skeleton_ids': [4274364]}, {'id': 4289746, 'name': 'SN motor neuron a1l posterior dendrite', 'type': 'neuron', 'skeleton_ids': [4289745]}, {'id': 4289774, 'name': 'MN23/24 (RF: LT3/LT4) LT4_a1l', 'type': 'neuron', 'skeleton_ids': [4289773]}, {'id': 4326469, 'name': 'MN8 (RF:SBM) LT3_a1l', 'type': 'neuron', 'skeleton_ids': [4326468]}, {'id': 4326521, 'name': 'MN27 (SN MN VA2)_a1l', 'type': 'neuron', 'skeleton_ids': [4326520]}, {'id': 4382896, 'name': 'MN29 (SN MN VA3)_a1r', 'type': 'neuron', 'skeleton_ids': [4382895]}, {'id': 4397289, 'name': 'MN26 (SN MN VA1)_a1r', 'type': 'neuron', 'skeleton_ids': [4397287]}, {'id': 4483082, 'name': 'SN motor neuron a1l bilateral', 'type': 'neuron', 'skeleton_ids': [4483081]}, {'id': 4524872, 'name': 'MN26 (SN MN VA1)_a1l', 'type': 'neuron', 'skeleton_ids': [4524871]}, {'id': 4535769, 'name': 'MN5 LO1_a1r', 'type': 'neuron', 'skeleton_ids': [4535767]}, {'id': 4881630, 'name': 'MN8 (RF:SBM) LT3_a1r ', 'type': 'neuron', 'skeleton_ids': [4881629]}, {'id': 5085456, 'name': 'MN20 ISN_a1r', 'type': 'neuron', 'skeleton_ids': [5085455]}, {'id': 5458047, 'name': 'Saaghi-1_a2l (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [5458046]}, {'id': 5899329, 'name': 'MN28 ISN_a1l', 'type': 'neuron', 'skeleton_ids': [5899328]}, {'id': 6195522, 'name': 'dbd_a1r', 'type': 'neuron', 'skeleton_ids': [6195521]}, {'id': 6280152, 'name': 'ddaD_a1r (class I)', 'type': 'neuron', 'skeleton_ids': [6280151]}, {'id': 6467482, 'name': 'MN21/22 (RF:LT1/LT2) LT2_a1l', 'type': 'neuron', 'skeleton_ids': [6467481]}, {'id': 7651933, 'name': 'MN18 (ISN DT1)_a1r', 'type': 'neuron', 'skeleton_ids': [6970942]}, {'id': 7772438, 'name': 'lesA_a1l', 'type': 'neuron', 'skeleton_ids': [7772437]}, {'id': 7783765, 'name': 'vdaC_a1l (class II)', 'type': 'neuron', 'skeleton_ids': [7783764]}, {'id': 7876386, 'name': 'A08e1_a1l ', 'type': 'neuron', 'skeleton_ids': [7876385]}, {'id': 8026225, 'name': 'MN10 U2_a1l', 'type': 'neuron', 'skeleton_ids': [8026224]}, {'id': 8039618, 'name': 'A08e3_a1r', 'type': 'neuron', 'skeleton_ids': [8039617]}, {'id': 8095562, 'name': 'Jaam-1_a1r (HMD1 (A12p))', 'type': 'neuron', 'skeleton_ids': [8095561]}, {'id': 8170517, 'name': 'A08e2_a1r ', 'type': 'neuron', 'skeleton_ids': [8234099]}, {'id': 8196193, 'name': 'Jaam-1_a1l (HMD1 (A12p))', 'type': 'neuron', 'skeleton_ids': [8196192]}, {'id': 8198958, 'name': 'A08e1_a1r', 'type': 'neuron', 'skeleton_ids': [8198957]}, {'id': 8234537, 'name': 'vpda classI md?_t3r', 'type': 'neuron', 'skeleton_ids': [8234536]}, {'id': 8374408, 'name': 'A08e2_a2l', 'type': 'neuron', 'skeleton_ids': [8374407]}, {'id': 8419066, 'name': 'vbd_a1l', 'type': 'neuron', 'skeleton_ids': [8419064]}, {'id': 8421330, 'name': 'A08e3_a1l ', 'type': 'neuron', 'skeleton_ids': [8421329]}, {'id': 8602633, 'name': 'A08e1_a2l', 'type': 'neuron', 'skeleton_ids': [8602632]}, {'id': 10013403, 'name': 'MN11 (ISN NB3-2)_a1r', 'type': 'neuron', 'skeleton_ids': [10013402]}, {'id': 10051928, 'name': 'MN ISN MN_a2r [MN-VL2 (13)?]', 'type': 'neuron', 'skeleton_ids': [10051927]}, {'id': 10089446, 'name': 'Saaghi-1_a2r (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [10089445]}, {'id': 10397675, 'name': 'A08e3_a2r', 'type': 'neuron', 'skeleton_ids': [10397674]}, {'id': 10413918, 'name': 'A08e3_a2l', 'type': 'neuron', 'skeleton_ids': [10413917]}, {'id': 10474937, 'name': 'Saaghi-3_a1l (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [10474936]}, {'id': 10561899, 'name': 'ddaD_a2l', 'type': 'neuron', 'skeleton_ids': [10561898]}, {'id': 10562192, 'name': 'dbd_t3l', 'type': 'neuron', 'skeleton_ids': [10562191]}, {'id': 10649844, 'name': 'MN1 aCC_a1l', 'type': 'neuron', 'skeleton_ids': [10649843]}, {'id': 10678018, 'name': 'MN12-III (ISN VL1 V-MN NB5-2)_a1l', 'type': 'neuron', 'skeleton_ids': [10678017]}, {'id': 10929872, 'name': 'A08c_a1r', 'type': 'neuron', 'skeleton_ids': [10929871]}, {'id': 11065547, 'name': 'Saaghi-3?_t3r (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [11065546]}, {'id': 11321645, 'name': 'neuron 11321645 frag run into a1 neuron', 'type': 'neuron', 'skeleton_ids': [11321644]}, {'id': 11466248, 'name': 'Jaam-2_a1l (HMD2 (A12g))', 'type': 'neuron', 'skeleton_ids': [11466247]}, {'id': 11578923, 'name': 'vpda classI md?_t3l', 'type': 'neuron', 'skeleton_ids': [11578922]}, {'id': 11928390, 'name': 'Jaam-3_a1l (HMD3 (A12 c3))', 'type': 'neuron', 'skeleton_ids': [11928389]}, {'id': 11958585, 'name': 'Saaghi-3_a1r (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [11958584]}, {'id': 12809924, 'name': 'A08e2_a1l', 'type': 'neuron', 'skeleton_ids': [12809923]}, {'id': 12825903, 'name': 'Saaghi-3?_t3l (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [12825902]}, {'id': 12960080, 'name': 'dmd1_a1l', 'type': 'neuron', 'skeleton_ids': [12960079]}, {'id': 12960218, 'name': 'ddaD_a1l (class I)', 'type': 'neuron', 'skeleton_ids': [12960217]}, {'id': 12990251, 'name': 'Jaam-2_a1r (HMD2 (A12g))', 'type': 'neuron', 'skeleton_ids': [12990250]}, {'id': 13057995, 'name': 'MNISN-Is RP2_a1r', 'type': 'neuron', 'skeleton_ids': [13057994]}, {'id': 13058241, 'name': 'MNISN-Is RP2_a1l', 'type': 'neuron', 'skeleton_ids': [13058240]}, {'id': 13124816, 'name': 'vdaA_a1r (class II)', 'type': 'neuron', 'skeleton_ids': [13124815]}, {'id': 13671420, 'name': 'dbd_a1l', 'type': 'neuron', 'skeleton_ids': [13671419]}, {'id': 13775275, 'name': 'Jaam-3_a1r (HMD3 (A12 c3))', 'type': 'neuron', 'skeleton_ids': [13775274]}, {'id': 13873398, 'name': 'MN U1_a2r', 'type': 'neuron', 'skeleton_ids': [13873397]}, {'id': 13874214, 'name': 'MN30 RP4_a2l', 'type': 'neuron', 'skeleton_ids': [13874213]}, {'id': 13874876, 'name': 'MNISNb/d-Is RP5_a2l', 'type': 'neuron', 'skeleton_ids': [13874875]}, {'id': 13875539, 'name': 'MN10 U2_a2r', 'type': 'neuron', 'skeleton_ids': [13875538]}, {'id': 13876817, 'name': 'dbd_a2r', 'type': 'neuron', 'skeleton_ids': [13876816]}, {'id': 13877014, 'name': 'ISN MN_a2r (U4/U5)', 'type': 'neuron', 'skeleton_ids': [13877013]}, {'id': 13877403, 'name': 'MN ISN MN_a2r (U4/U5)', 'type': 'neuron', 'skeleton_ids': [13877402]}, {'id': 13877720, 'name': 'MN V_a2r (MN?)', 'type': 'neuron', 'skeleton_ids': [13877719]}, {'id': 13880944, 'name': 'MN RP4_a2r', 'type': 'neuron', 'skeleton_ids': [13880943]}, {'id': 13881375, 'name': 'MNISNb/d-Is RP5_a2r', 'type': 'neuron', 'skeleton_ids': [13881374]}, {'id': 13881811, 'name': 'MN-VO4/5_a2l (NB7-1? ISN MN-15/16 ?)', 'type': 'neuron', 'skeleton_ids': [13881810]}, {'id': 13882193, 'name': 'MN ISN MN_a2l [MN-VL2 (13)?]', 'type': 'neuron', 'skeleton_ids': [13882192]}, {'id': 13883599, 'name': 'MN10U2_a2l', 'type': 'neuron', 'skeleton_ids': [13883598]}, {'id': 13883896, 'name': 'dbd_a2l', 'type': 'neuron', 'skeleton_ids': [13883895]}, {'id': 13884217, 'name': 'MN-VO4–6 (NB7-1 was- VO6 (17)? )_a2l', 'type': 'neuron', 'skeleton_ids': [13884216]}, {'id': 13885024, 'name': 'MN ISN MN_a2l (U4/U5)', 'type': 'neuron', 'skeleton_ids': [13885023]}, {'id': 13885985, 'name': 'MN9 U1_a2l', 'type': 'neuron', 'skeleton_ids': [13885984]}, {'id': 14044615, 'name': 'MNISNb/d-Is RP5_a1l', 'type': 'neuron', 'skeleton_ids': [14044614]}, {'id': 14045017, 'name': 'MN14 RP1_a1l', 'type': 'neuron', 'skeleton_ids': [14045016]}, {'id': 14045490, 'name': 'MN15/16/17 VO4–6 (NB7-1? )_a1r', 'type': 'neuron', 'skeleton_ids': [14045489]}, {'id': 14045855, 'name': 'MN3 (ISN U4 DA3 NB7-1?)_a1r', 'type': 'neuron', 'skeleton_ids': [14045854]}, {'id': 14046721, 'name': 'MN12-III (ISN VL1 V-MN NB5-2)_a1r', 'type': 'neuron', 'skeleton_ids': [14046720]}, {'id': 14047131, 'name': 'MN2 ISN(U3-NB7-1?)_a1l', 'type': 'neuron', 'skeleton_ids': [14047130]}, {'id': 14047555, 'name': 'MN9 U1_a1r', 'type': 'neuron', 'skeleton_ids': [14047554]}, {'id': 14047761, 'name': 'MN10 U2_a1r', 'type': 'neuron', 'skeleton_ids': [14047760]}, {'id': 14047946, 'name': 'MN4 (ISN U5 NB7-1?)_a1r', 'type': 'neuron', 'skeleton_ids': [14047945]}, {'id': 14048288, 'name': 'MN2 ISN(U3-NB7-1?)_a1r', 'type': 'neuron', 'skeleton_ids': [14048287]}, {'id': 14048539, 'name': 'MN15/16 VO4/5 (NB7-1? ISN)_a1r', 'type': 'neuron', 'skeleton_ids': [14048538]}, {'id': 14048877, 'name': 'MN28 ISN_a1r', 'type': 'neuron', 'skeleton_ids': [14048876]}, {'id': 14085814, 'name': 'MN9 U1_a1l', 'type': 'neuron', 'skeleton_ids': [14085813]}, {'id': 14086310, 'name': 'MN19 (ISN NB3-2)_a1l', 'type': 'neuron', 'skeleton_ids': [14086309]}, {'id': 14087108, 'name': 'MN30 RP4_a1r', 'type': 'neuron', 'skeleton_ids': [14087107]}, {'id': 14087340, 'name': 'MN4 (ISN U5 NB7-1?)_a1l', 'type': 'neuron', 'skeleton_ids': [14087339]}, {'id': 14088150, 'name': 'MN3 (ISN U4 DA3 NB7-1?)_a1l ', 'type': 'neuron', 'skeleton_ids': [14088149]}, {'id': 14088452, 'name': 'MN11 (ISN NB3-2)_a1l', 'type': 'neuron', 'skeleton_ids': [14088451]}, {'id': 14194027, 'name': 'MN ISN MN_a1r R only ', 'type': 'neuron', 'skeleton_ids': [14194026]}, {'id': 14194235, 'name': 'MN30 RP4_a1l', 'type': 'neuron', 'skeleton_ids': [14194234]}, {'id': 14194704, 'name': 'MN13 ISN MN-VL2_a1r', 'type': 'neuron', 'skeleton_ids': [14194703]}, {'id': 14198376, 'name': 'ISN MN_a1r', 'type': 'neuron', 'skeleton_ids': [14198375]}, {'id': 14199032, 'name': 'MN18 (ISN DT1)_a1l', 'type': 'neuron', 'skeleton_ids': [14199031]}, {'id': 14199566, 'name': 'MNISNb/d-Is RP5_a1r', 'type': 'neuron', 'skeleton_ids': [14199565]}, {'id': 14201008, 'name': 'A08e1_a2r', 'type': 'neuron', 'skeleton_ids': [14201007]}, {'id': 14203726, 'name': 'MN15/16 VO4/5 (NB7-1? ISN)_a1l', 'type': 'neuron', 'skeleton_ids': [14203725]}, {'id': 14206439, 'name': 'MN14 RP1_a1r', 'type': 'neuron', 'skeleton_ids': [14206438]}, {'id': 14238854, 'name': 'MN6/7 RP3_a1r', 'type': 'neuron', 'skeleton_ids': [14238853]}, {'id': 14478518, 'name': 'MN6/7 RP3_a1l', 'type': 'neuron', 'skeleton_ids': [14478517]}, {'id': 14493980, 'name': 'vbd_a1r', 'type': 'neuron', 'skeleton_ids': [14493979]}, {'id': 14573137, 'name': 'dbd_t3r', 'type': 'neuron', 'skeleton_ids': [14573136]}, {'id': 15351531, 'name': 'ddaD_a2r', 'type': 'neuron', 'skeleton_ids': [15351530]}, {'id': 15505126, 'name': 'ddaE_t3l', 'type': 'neuron', 'skeleton_ids': [15505125]}, {'id': 15551561, 'name': 'dmd1_a1r', 'type': 'neuron', 'skeleton_ids': [15551560]}, {'id': 15625866, 'name': 'Saaghi-3_a2r (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [15625865]}, {'id': 15879189, 'name': 'A08e2_a2r', 'type': 'neuron', 'skeleton_ids': [15879188]}, {'id': 16009773, 'name': 'A08c_a1l', 'type': 'neuron', 'skeleton_ids': [16009772]}, {'id': 16042165, 'name': 'SN motor neuron a1r bilateral', 'type': 'neuron', 'skeleton_ids': [16042164]}, {'id': 16181495, 'name': 'MN19 (ISN NB3-2)_a1r', 'type': 'neuron', 'skeleton_ids': [16181494]}, {'id': 16233067, 'name': 'ddaE_t3r', 'type': 'neuron', 'skeleton_ids': [16233066]}, {'id': 16264803, 'name': 'MN6/7 RP3_a2l', 'type': 'neuron', 'skeleton_ids': [16264802]}, {'id': 16314354, 'name': 'SN motor neuron a1r posterior dendrite', 'type': 'neuron', 'skeleton_ids': [16314353]}, {'id': 16433396, 'name': 'MN20 ISN_a1l', 'type': 'neuron', 'skeleton_ids': [16433395]}, {'id': 16713122, 'name': 'MN21/22 (RF:LT1/LT2) LT2_a1r ', 'type': 'neuron', 'skeleton_ids': [16713121]}, {'id': 17435372, 'name': 'Saaghi-1_a1r (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [17435371]}, {'id': 18418513, 'name': 'Saaghi-1_a1l (AAZ1 (A06l))', 'type': 'neuron', 'skeleton_ids': [18418512]}, {'id': 18514165, 'name': 'Saaghi-3_a2l (AAZ3 (A06e))', 'type': 'neuron', 'skeleton_ids': [18514164]}, {'id': 19111931, 'name': 'MN13 ISN MN-VL2_a1l ', 'type': 'neuron', 'skeleton_ids': [19111930]}, {'id': 19139254, 'name': 'MN15/16/17 VO4–6 (NB7-1? )_a1l', 'type': 'neuron', 'skeleton_ids': [19139253]}]}, {'id': 19756692, 'name': '<NAME>', 'type': 'annotation', 'neurons': [{'id': 210250, 'name': 'A02o_a3l Wave-1', 'type': 'neuron', 'skeleton_ids': [2194374]}, {'id': 210597, 'name': 'TePn19 classIV-related projection B left', 'type': 'neuron', 'skeleton_ids': [7964684]}, {'id': 439983, 'name': 'SelN138 classIV-related descending sog left', 'type': 'neuron', 'skeleton_ids': [3661586]}, {'id': 627859, 'name': 'A09e_a4r', 'type': 'neuron', 'skeleton_ids': [627858]}, {'id': 729575, 'name': 'SelN138 classIV-related descending sog right', 'type': 'neuron', 'skeleton_ids': [8067524]}, {'id': 1365253, 'name': 'TePn05 classIV-related projection C right', 'type': 'neuron', 'skeleton_ids': [2558717]}, {'id': 1370786, 'name': 'A09e_a5l', 'type': 'neuron', 'skeleton_ids': [1370785]}, {'id': 1395101, 'name': 'A08n classIV-related projection A left', 'type': 'neuron', 'skeleton_ids': [1927577]}, {'id': 1716789, 'name': 'A09o_a3l', 'type': 'neuron', 'skeleton_ids': [1716788]}, {'id': 2370291, 'name': 'A09e_a5r', 'type': 'neuron', 'skeleton_ids': [2370290]}, {'id': 2583771, 'name': 'A10a_a2l Chair-1', 'type': 'neuron', 'skeleton_ids': [2583770]}, {'id': 2693652, 'name': 'A09o_a3r', 'type': 'neuron', 'skeleton_ids': [2693651]}, {'id': 3037784, 'name': 'A09b a1l Basin-1', 'type': 'neuron', 'skeleton_ids': [3034133]}, {'id': 3040475, 'name': 'A09c a1l Basin-4', 'type': 'neuron', 'skeleton_ids': [3040481]}, {'id': 3040478, 'name': 'A09a a1l Basin-2', 'type': 'neuron', 'skeleton_ids': [3041612]}, {'id': 3056157, 'name': 'A09a_a2l Basin-2', 'type': 'neuron', 'skeleton_ids': [3056163]}, {'id': 3074107, 'name': 'A09b a1r Basin-1', 'type': 'neuron', 'skeleton_ids': [3282869]}, {'id': 3090350, 'name': 'A09c a1r Basin-4', 'type': 'neuron', 'skeleton_ids': [4049878]}, {'id': 3091625, 'name': 'A09a a1r Basin-2', 'type': 'neuron', 'skeleton_ids': [3074106]}, {'id': 3281616, 'name': 'A09l_a2l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [3281615]}, {'id': 3304284, 'name': 'A02n_a1r Pseudolooper-4', 'type': 'neuron', 'skeleton_ids': [2637812]}, {'id': 3513767, 'name': 'ddaC_a1l', 'type': 'neuron', 'skeleton_ids': [986460]}, {'id': 3546335, 'name': 'A02m_a1r Pseudolooper-3', 'type': 'neuron', 'skeleton_ids': [3546334]}, {'id': 3546836, 'name': 'A02n_a1l Pseudolooper-4', 'type': 'neuron', 'skeleton_ids': [3546835]}, {'id': 4102845, 'name': 'A09l_a1r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [4102844]}, {'id': 4262160, 'name': 'T10a t3l Chair-1', 'type': 'neuron', 'skeleton_ids': [4262125]}, {'id': 4589093, 'name': 'A02o_a2l Wave-1', 'type': 'neuron', 'skeleton_ids': [4589092]}, {'id': 4717262, 'name': 'A02o_a2r Wave-1', 'type': 'neuron', 'skeleton_ids': [4717261]}, {'id': 4778440, 'name': 'A02m_a1l Pseudolooper-3', 'type': 'neuron', 'skeleton_ids': [4778439]}, {'id': 5114805, 'name': 'A09a_a2r Basin-2', 'type': 'neuron', 'skeleton_ids': [5114804]}, {'id': 5690363, 'name': 'A10a_a1r Chair-1', 'type': 'neuron', 'skeleton_ids': [5690362]}, {'id': 5715619, 'name': 'A09l_t3l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [7124056]}, {'id': 5731369, 'name': 'A00c_A8l?', 'type': 'neuron', 'skeleton_ids': [6660660]}, {'id': 6062424, 'name': 'T10a t3r Chair-1', 'type': 'neuron', 'skeleton_ids': [6062423]}, {'id': 6535162, 'name': 'A10a_a1l Chair-1', 'type': 'neuron', 'skeleton_ids': [6535161]}, {'id': 7040211, 'name': 'A09q_a7r classIV-related projection D right', 'type': 'neuron', 'skeleton_ids': [734837]}, {'id': 7836571, 'name': "v'ada_a1r", 'type': 'neuron', 'skeleton_ids': [3786734]}, {'id': 9271227, 'name': 'A09q_a7l classIV-related projection D left', 'type': 'neuron', 'skeleton_ids': [1362085]}, {'id': 9654307, 'name': 'A09e_a4l', 'type': 'neuron', 'skeleton_ids': [183502]}, {'id': 9654870, 'name': 'A00c_A8l?', 'type': 'neuron', 'skeleton_ids': [5731368]}, {'id': 10053643, 'name': 'A09l_t3r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [10053642]}, {'id': 10110271, 'name': "v'ada_a1l", 'type': 'neuron', 'skeleton_ids': [10110270]}, {'id': 10110673, 'name': 'ddaC_a1r', 'type': 'neuron', 'skeleton_ids': [10110672]}, {'id': 10273503, 'name': 'vdaB_a1r', 'type': 'neuron', 'skeleton_ids': [10273502]}, {'id': 10290530, 'name': 'vdaB_a1l ', 'type': 'neuron', 'skeleton_ids': [10290529]}, {'id': 12468813, 'name': 'A09l_a1l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [12468812]}, {'id': 12767924, 'name': 'A10a_a2r Chair-1', 'type': 'neuron', 'skeleton_ids': [12767923]}, {'id': 12946336, 'name': 'A02o_a3r Wave-1', 'type': 'neuron', 'skeleton_ids': [12946335]}, {'id': 14728551, 'name': 'A09e_a3r', 'type': 'neuron', 'skeleton_ids': [14728550]}, {'id': 14795008, 'name': 'TePn19 classIV-related projection B right', 'type': 'neuron', 'skeleton_ids': [14795007]}, {'id': 14795311, 'name': 'A09e_a3l', 'type': 'neuron', 'skeleton_ids': [14795310]}, {'id': 14899173, 'name': 'A08n classIV-related projection A right', 'type': 'neuron', 'skeleton_ids': [14899172]}, {'id': 17693221, 'name': 'A09l_a2r Down-and-back-1\t', 'type': 'neuron', 'skeleton_ids': [17693220]}, {'id': 18322844, 'name': 'fragment', 'type': 'neuron', 'skeleton_ids': [18322843]}, {'id': 18981221, 'name': 'TePn05 classIV-related projection C left', 'type': 'neuron', 'skeleton_ids': [18981220]}]}, {'id': 19862922, 'name': 'Burgos et al 2018', 'type': 'annotation', 'neurons': [{'id': 20208, 'name': 'A02o_a1l Wave-1', 'type': 'neuron', 'skeleton_ids': [918889]}, {'id': 210250, 'name': 'A02o_a3l Wave-1', 'type': 'neuron', 'skeleton_ids': [2194374]}, {'id': 367873, 'name': 'putative class III or es', 'type': 'neuron', 'skeleton_ids': [367872]}, {'id': 519625, 'name': 'A09a_a4l Basin-2', 'type': 'neuron', 'skeleton_ids': [519624]}, {'id': 587084, 'name': 'A09c_a4l Basin-4', 'type': 'neuron', 'skeleton_ids': [587083]}, {'id': 627859, 'name': 'A09e_a4r', 'type': 'neuron', 'skeleton_ids': [627858]}, {'id': 722936, 'name': 'ddaA a1l classs III md', 'type': 'neuron', 'skeleton_ids': [722935]}, {'id': 754775, 'name': 'A02o_a6l Wave-1', 'type': 'neuron', 'skeleton_ids': [754774]}, {'id': 1115337, 'name': 'A02o_a5l Wave-1', 'type': 'neuron', 'skeleton_ids': [3437872]}, {'id': 1121744, 'name': 'A09l_a5r_ Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [1115781]}, {'id': 1365253, 'name': 'TePn05 classIV-related projection C right', 'type': 'neuron', 'skeleton_ids': [2558717]}, {'id': 1370786, 'name': 'A09e_a5l', 'type': 'neuron', 'skeleton_ids': [1370785]}, {'id': 1398328, 'name': 'A09a_a4r Basin-2', 'type': 'neuron', 'skeleton_ids': [1398327]}, {'id': 1400918, 'name': 'A09l_a4r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [1702280]}, {'id': 1401062, 'name': 'A02o_a5r Wave-1', 'type': 'neuron', 'skeleton_ids': [1401061]}, {'id': 1690664, 'name': 'A09l_a3r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [1378190]}, {'id': 2370291, 'name': 'A09e_a5r', 'type': 'neuron', 'skeleton_ids': [2370290]}, {'id': 2745673, 'name': 'A09l_a5l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [3220573]}, {'id': 2933979, 'name': 'A09l_a3l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [2693767]}, {'id': 3040475, 'name': 'A09c a1l Basin-4', 'type': 'neuron', 'skeleton_ids': [3040481]}, {'id': 3040478, 'name': 'A09a a1l Basin-2', 'type': 'neuron', 'skeleton_ids': [3041612]}, {'id': 3090350, 'name': 'A09c a1r Basin-4', 'type': 'neuron', 'skeleton_ids': [4049878]}, {'id': 3091944, 'name': 'A09g a1r Basin-3', 'type': 'neuron', 'skeleton_ids': [3091943]}, {'id': 3281616, 'name': 'A09l_a2l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [3281615]}, {'id': 3380085, 'name': 'A09a_a3r Basin-2', 'type': 'neuron', 'skeleton_ids': [888730]}, {'id': 3513767, 'name': 'ddaC_a1l', 'type': 'neuron', 'skeleton_ids': [986460]}, {'id': 3661594, 'name': 'vdaD class III md_a1r or es?', 'type': 'neuron', 'skeleton_ids': [2777891]}, {'id': 3995977, 'name': 'A01d3_a1l', 'type': 'neuron', 'skeleton_ids': [3995976]}, {'id': 4022795, 'name': 'Handle-A a1', 'type': 'neuron', 'skeleton_ids': [4022794]}, {'id': 4042316, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1l', 'type': 'neuron', 'skeleton_ids': [4042315]}, {'id': 4102729, 'name': 'MN22/23 (RF:LT2/LT3) LT1_a1r ', 'type': 'neuron', 'skeleton_ids': [4102728]}, {'id': 4102845, 'name': 'A09l_a1r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [4102844]}, {'id': 4206756, 'name': 'A10f-like a1l', 'type': 'neuron', 'skeleton_ids': [4206755]}, {'id': 4213418, 'name': 'A27k_a1l', 'type': 'neuron', 'skeleton_ids': [4213417]}, {'id': 4223189, 'name': 'A02o_t3l Wave-1', 'type': 'neuron', 'skeleton_ids': [4223188]}, {'id': 4589093, 'name': 'A02o_a2l Wave-1', 'type': 'neuron', 'skeleton_ids': [4589092]}, {'id': 4717262, 'name': 'A02o_a2r Wave-1', 'type': 'neuron', 'skeleton_ids': [4717261]}, {'id': 4740180, 'name': 'A05q_a1l', 'type': 'neuron', 'skeleton_ids': [4740178]}, {'id': 4833169, 'name': 'A05q_a1r', 'type': 'neuron', 'skeleton_ids': [4833168]}, {'id': 4936150, 'name': 'A01c1_a1r', 'type': 'neuron', 'skeleton_ids': [4936149]}, {'id': 5084469, 'name': 'A01c2_ipsi_dendrite_a1l', 'type': 'neuron', 'skeleton_ids': [5084467]}, {'id': 5114805, 'name': 'A09a_a2r Basin-2', 'type': 'neuron', 'skeleton_ids': [5114804]}, {'id': 5132887, 'name': 'A23g_a1l', 'type': 'neuron', 'skeleton_ids': [5132886]}, {'id': 5231558, 'name': 'A23g_a2r', 'type': 'neuron', 'skeleton_ids': [5231557]}, {'id': 5243058, 'name': 'gorogoro t2r', 'type': 'neuron', 'skeleton_ids': [5206247]}, {'id': 5309933, 'name': 'A03g_a1l', 'type': 'neuron', 'skeleton_ids': [5309932]}, {'id': 5310671, 'name': 'A02o_t3r Wave-1', 'type': 'neuron', 'skeleton_ids': [5310670]}, {'id': 5715619, 'name': 'A09l_t3l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [7124056]}, {'id': 5733619, 'name': 'A23g_a2l', 'type': 'neuron', 'skeleton_ids': [5733618]}, {'id': 5997122, 'name': 'A23g_a1r', 'type': 'neuron', 'skeleton_ids': [5997121]}, {'id': 6896581, 'name': 'A07c4?_a1r', 'type': 'neuron', 'skeleton_ids': [6896580]}, {'id': 7076836, 'name': 'ldaB a1r class III md', 'type': 'neuron', 'skeleton_ids': [7076835]}, {'id': 7124157, 'name': 'A10f-like a1r', 'type': 'neuron', 'skeleton_ids': [7124156]}, {'id': 7633930, 'name': "v'esA or v'esB or v'es2[1][2]_a1r", 'type': 'neuron', 'skeleton_ids': [7633929]}, {'id': 7636005, 'name': "v'pda a1r class III md", 'type': 'neuron', 'skeleton_ids': [7636004]}, {'id': 7772438, 'name': 'lesA_a1l', 'type': 'neuron', 'skeleton_ids': [7772437]}, {'id': 7792782, 'name': 'vdaD a1l class III md', 'type': 'neuron', 'skeleton_ids': [7792781]}, {'id': 7836571, 'name': "v'ada_a1r", 'type': 'neuron', 'skeleton_ids': [3786734]}, {'id': 7992967, 'name': 'A02e_a1l', 'type': 'neuron', 'skeleton_ids': [7992966]}, {'id': 9654307, 'name': 'A09e_a4l', 'type': 'neuron', 'skeleton_ids': [183502]}, {'id': 9654573, 'name': 'gor<NAME>', 'type': 'neuron', 'skeleton_ids': [3720037]}, {'id': 9681053, 'name': 'A09l_a4l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [1371419]}, {'id': 9681060, 'name': 'A09c_a3l Basin-4', 'type': 'neuron', 'skeleton_ids': [1815127]}, {'id': 9681086, 'name': 'Basin-3 a4r', 'type': 'neuron', 'skeleton_ids': [1401011]}, {'id': 10004244, 'name': 'A27k_a1r', 'type': 'neuron', 'skeleton_ids': [10004243]}, {'id': 10015770, 'name': 'A27j_a1r', 'type': 'neuron', 'skeleton_ids': [10015769]}, {'id': 10021176, 'name': 'A18l_a1l', 'type': 'neuron', 'skeleton_ids': [10021175]}, {'id': 10053643, 'name': 'A09l_t3r Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [10053642]}, {'id': 10053735, 'name': 'vdaC_a1r (class II)', 'type': 'neuron', 'skeleton_ids': [10053734]}, {'id': 10110271, 'name': "v'ada_a1l", 'type': 'neuron', 'skeleton_ids': [10110270]}, {'id': 10110673, 'name': 'ddaC_a1r', 'type': 'neuron', 'skeleton_ids': [10110672]}, {'id': 10152438, 'name': 'A07c4?_a1l', 'type': 'neuron', 'skeleton_ids': [10152437]}, {'id': 10179502, 'name': 'A09g a1l Basin-3', 'type': 'neuron', 'skeleton_ids': [10179501]}, {'id': 10273503, 'name': 'vdaB_a1r', 'type': 'neuron', 'skeleton_ids': [10273502]}, {'id': 10290530, 'name': 'vdaB_a1l ', 'type': 'neuron', 'skeleton_ids': [10290529]}, {'id': 10492186, 'name': 'vdaA_a1l (class II)', 'type': 'neuron', 'skeleton_ids': [10492185]}, {'id': 10855207, 'name': 'A03g_a1r', 'type': 'neuron', 'skeleton_ids': [10855206]}, {'id': 12140588, 'name': 'likely sensory class ii A1r', 'type': 'neuron', 'skeleton_ids': [12140587]}, {'id': 12292580, 'name': 'A18l_a1r', 'type': 'neuron', 'skeleton_ids': [12292579]}, {'id': 12468813, 'name': 'A09l_a1l Down-and-back-1', 'type': 'neuron', 'skeleton_ids': [12468812]}, {'id': 12946336, 'name': 'A02o_a3r Wave-1', 'type': 'neuron', 'skeleton_ids': [12946335]}, {'id': 14642309, 'name': 'ldaB class III_a1l', 'type': 'neuron', 'skeleton_ids': [14642308]}, {'id': 14858627, 'name': 'A02e_a1r', 'type': 'neuron', 'skeleton_ids': [14858626]}, {'id': 14866843, 'name': 'A02g_a1l', 'type': 'neuron', 'skeleton_ids': [14866842]}, {'id': 16311503, 'name': "v'esA or v'esB or v'es2[1][2]_a1l", 'type': 'neuron', 'skeleton_ids': [16311502]}, {'id': 16311521, 'name': "v'pda_a1l (class III)", 'type': 'neuron', 'skeleton_ids': [16311520]}, {'id': 16312926, 'name': 'ddaA a1r class III md', 'type': 'neuron', 'skeleton_ids': [16312925]}, {'id': 17693221, 'name': 'A09l_a2r Down-and-back-1\t', 'type': 'neuron', 'skeleton_ids': [17693220]}, {'id': 17872155, 'name': 'A01d3_a1r', 'type': 'neuron', 'skeleton_ids': [17872154]}, {'id': 17904628, 'name': 'A27j_a1l', 'type': 'neuron', 'skeleton_ids': [17904627]}, {'id': 18145094, 'name': 'A02g_a1r', 'type': 'neuron', 'skeleton_ids': [18145093]}, {'id': 18508516, 'name': 'A02o_a4l Wave-1', 'type': 'neuron', 'skeleton_ids': [18508515]}, {'id': 18684929, 'name': 'A02o_a1r Wave-1', 'type': 'neuron', 'skeleton_ids': [18684928]}, {'id': 18981221, 'name': 'TePn05 classIV-related projection C left', 'type': 'neuron', 'skeleton_ids': [18981220]}]}, {'id': 20282757, 'name': '<NAME>. 2018', 'type': 'annotation', 'neurons': [{'id': 38140, 'name': 'PaN-L-motor-02', 'type': 'neuron', 'skeleton_ids': [38147]}, {'id': 40149, 'name': 'AN-L-Sens-B1-ACa-10', 'type': 'neuron', 'skeleton_ids': [40152]}, {'id': 853527, 'name': 'AN-R-motor-15', 'type': 'neuron', 'skeleton_ids': [853526]}, {'id': 855720, 'name': 'AN-R-motor-14', 'type': 'neuron', 'skeleton_ids': [855719]}, {'id': 865802, 'name': 'AN-R-motor-17', 'type': 'neuron', 'skeleton_ids': [865801]}, {'id': 873149, 'name': 'AN-R-motor-13', 'type': 'neuron', 'skeleton_ids': [873148]}, {'id': 873783, 'name': 'AN-R-motor-16', 'type': 'neuron', 'skeleton_ids': [873782]}, {'id': 875614, 'name': 'AN-R-motor-09', 'type': 'neuron', 'skeleton_ids': [875613]}, {'id': 882340, 'name': 'AN-R-motor-06', 'type': 'neuron', 'skeleton_ids': [882339]}, {'id': 887906, 'name': 'SE0 R1', 'type': 'neuron', 'skeleton_ids': [887905]}, {'id': 896563, 'name': 'SE0 R2', 'type': 'neuron', 'skeleton_ids': [896562]}, {'id': 910065, 'name': 'AN-R-motor-11', 'type': 'neuron', 'skeleton_ids': [910064]}, {'id': 937333, 'name': 'AN-R-motor-18', 'type': 'neuron', 'skeleton_ids': [937332]}, {'id': 945310, 'name': 'AN-R-Sens-B1-AVa-44', 'type': 'neuron', 'skeleton_ids': [945309]}, {'id': 1008358, 'name': 'AN-L-Sens-B2-ACp-02', 'type': 'neuron', 'skeleton_ids': [1011675]}, {'id': 1306333, 'name': 'AN-R-motor-10', 'type': 'neuron', 'skeleton_ids': [3340561]}, {'id': 1347887, 'name': 'AN-L-Sens-B2-AVp-01', 'type': 'neuron', 'skeleton_ids': [1347886]}, {'id': 1351126, 'name': 'AN-L-Sens-B1-AVa-40', 'type': 'neuron', 'skeleton_ids': [1351125]}, {'id': 1367172, 'name': 'AN-L-Sens-B1-ACa-04', 'type': 'neuron', 'skeleton_ids': [1367171]}, {'id': 1377587, 'name': 'AN-L-Sens-B1-ACa-07', 'type': 'neuron', 'skeleton_ids': [1377586]}, {'id': 1391180, 'name': 'AN-L-Sens-B1-ACa-15', 'type': 'neuron', 'skeleton_ids': [1391179]}, {'id': 1392772, 'name': 'AN-L-Sens-B1-ACa-06', 'type': 'neuron', 'skeleton_ids': [1392771]}, {'id': 1395972, 'name': 'AN-L-Sens-B1-ACa-11', 'type': 'neuron', 'skeleton_ids': [1395971]}, {'id': 1398254, 'name': 'AN-L-Sens-B1-ACa-05', 'type': 'neuron', 'skeleton_ids': [1398253]}, {'id': 1400726, 'name': 'AN-L-Sens-B1-ACa-12', 'type': 'neuron', 'skeleton_ids': [1400725]}, {'id': 1405494, 'name': 'AN-L-Sens-B1-ACa-09', 'type': 'neuron', 'skeleton_ids': [1405493]}, {'id': 1406464, 'name': 'AN-L-Sens-B1-ACa-14', 'type': 'neuron', 'skeleton_ids': [1406463]}, {'id': 1414207, 'name': 'AN-L-Sens-B1-AVa-16', 'type': 'neuron', 'skeleton_ids': [1414206]}, {'id': 1415169, 'name': 'AN-L-Sens-B1-AVa-26', 'type': 'neuron', 'skeleton_ids': [1415168]}, {'id': 1419316, 'name': 'AN-L-Sens-B1-AVa-31', 'type': 'neuron', 'skeleton_ids': [1419315]}, {'id': 1427013, 'name': 'AN-L-Sens-B1-AVa-32', 'type': 'neuron', 'skeleton_ids': [1427012]}, {'id': 1428272, 'name': 'AN-L-Sens-B1-AVa-22', 'type': 'neuron', 'skeleton_ids': [1428271]}, {'id': 1429970, 'name': 'AN-L-Sens-B1-AVa-20', 'type': 'neuron', 'skeleton_ids': [1429969]}, {'id': 1431257, 'name': 'AN-L-Sens-B1-AVa-27', 'type': 'neuron', 'skeleton_ids': [1431256]}, {'id': 1432837, 'name': 'AN-L-Sens-B1-AVa-18', 'type': 'neuron', 'skeleton_ids': [1432836]}, {'id': 1435467, 'name': 'AN-L-Sens-B1-AVa-21', 'type': 'neuron', 'skeleton_ids': [1435466]}, {'id': 1706717, 'name': 'AN-L-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [1706716]}, {'id': 1709803, 'name': 'AN-L-Sens-B2-AVa-24', 'type': 'neuron', 'skeleton_ids': [1709802]}, {'id': 1715991, 'name': 'AN-L-Sens-B2-ACp-17', 'type': 'neuron', 'skeleton_ids': [1715990]}, {'id': 1742259, 'name': 'AN-R-motor-08', 'type': 'neuron', 'skeleton_ids': [1218587]}, {'id': 1872624, 'name': 'nc1;right', 'type': 'neuron', 'skeleton_ids': [7555968]}, {'id': 1939617, 'name': 'neuron 1939617--1', 'type': 'neuron', 'skeleton_ids': [1939616]}, {'id': 2037006, 'name': 'neuron 2037006', 'type': 'neuron', 'skeleton_ids': [2037005]}, {'id': 2141153, 'name': 'IPC 7 right', 'type': 'neuron', 'skeleton_ids': [2355126]}, {'id': 2142818, 'name': 'pre-dorsal-MAP presynaptic-D-2-R left', 'type': 'neuron', 'skeleton_ids': [2142817]}, {'id': 2144071, 'name': 'BC: presynaptic -medial - paired with #3801211', 'type': 'neuron', 'skeleton_ids': [2147157]}, {'id': 2147244, 'name': 'IPC 2 right', 'type': 'neuron', 'skeleton_ids': [2852273]}, {'id': 2147974, 'name': 'AN-R-Sens-B1-ACa-01', 'type': 'neuron', 'skeleton_ids': [2456826]}, {'id': 2156260, 'name': 'IPC 3 right', 'type': 'neuron', 'skeleton_ids': [2283495]}, {'id': 2189895, 'name': 'neuron 2189895--5', 'type': 'neuron', 'skeleton_ids': [2189894]}, {'id': 2200183, 'name': 'neuron 2200183--7', 'type': 'neuron', 'skeleton_ids': [2200182]}, {'id': 2228845, 'name': 'PaN-R-motor-02', 'type': 'neuron', 'skeleton_ids': [8574714]}, {'id': 2260756, 'name': 'DH44 1 right', 'type': 'neuron', 'skeleton_ids': [2260755]}, {'id': 2268282, 'name': 'PaN-R-Sens-B1-VM-02', 'type': 'neuron', 'skeleton_ids': [2268281]}, {'id': 2289487, 'name': 'neuron 2289487--11', 'type': 'neuron', 'skeleton_ids': [2289486]}, {'id': 2301628, 'name': 'nc1;left', 'type': 'neuron', 'skeleton_ids': [8111135]}, {'id': 2302817, 'name': 'PaN-R-Sens-B2-VM-01', 'type': 'neuron', 'skeleton_ids': [2302816]}, {'id': 2304500, 'name': 'PaN-R-Sens-B1-VM-01', 'type': 'neuron', 'skeleton_ids': [2305234]}, {'id': 2320455, 'name': 'neuron 2320455--12', 'type': 'neuron', 'skeleton_ids': [2320454]}, {'id': 2369445, 'name': 'Z: contra_left ', 'type': 'neuron', 'skeleton_ids': [2369444]}, {'id': 2376543, 'name': 'SEZ-IN-12-right--14', 'type': 'neuron', 'skeleton_ids': [2376542]}, {'id': 2391093, 'name': 'PaN-L-motor-01', 'type': 'neuron', 'skeleton_ids': [2391092]}, {'id': 2426398, 'name': 'Spiral_right', 'type': 'neuron', 'skeleton_ids': [2426397]}, {'id': 2432566, 'name': 'SEZ-IN-A01-right', 'type': 'neuron', 'skeleton_ids': [2432564]}, {'id': 2438197, 'name': 'IPC 6 right', 'type': 'neuron', 'skeleton_ids': [2357110]}, {'id': 2495309, 'name': 'PaN-L-Sens-B1-VM-01', 'type': 'neuron', 'skeleton_ids': [2495308]}, {'id': 2505676, 'name': 'PaN-L-Sens-B1-VM-04', 'type': 'neuron', 'skeleton_ids': [2505675]}, {'id': 2524027, 'name': 'Pan-L-Sens-B2-VM-02', 'type': 'neuron', 'skeleton_ids': [2524026]}, {'id': 2532791, 'name': 'PaN-L-Sens-B1-VM-02', 'type': 'neuron', 'skeleton_ids': [2668719]}, {'id': 2597152, 'name': 'Short-presynapti-D-1R', 'type': 'neuron', 'skeleton_ids': [2597151]}, {'id': 2610745, 'name': 'AN-L-Sens-B3-ACal-05 (Non-ORN 1)', 'type': 'neuron', 'skeleton_ids': [2610744]}, {'id': 2612528, 'name': 'mPN iACT C2 left', 'type': 'neuron', 'skeleton_ids': [2612527]}, {'id': 2635424, 'name': 'AN-R-Sens-B1-ACa-02', 'type': 'neuron', 'skeleton_ids': [2340336]}, {'id': 2669833, 'name': 'PaN-L-Sens-B1-VM-03', 'type': 'neuron', 'skeleton_ids': [2669832]}, {'id': 2679036, 'name': 'SEZ-IN-15-right', 'type': 'neuron', 'skeleton_ids': [2679035]}, {'id': 2725744, 'name': 'DMS 1 right', 'type': 'neuron', 'skeleton_ids': [2659386]}, {'id': 2798515, 'name': 'Pan-L-Sens-B2-VM-01', 'type': 'neuron', 'skeleton_ids': [2799380]}, {'id': 2862581, 'name': 'CPLd interneuron', 'type': 'neuron', 'skeleton_ids': [8598886]}, {'id': 2896541, 'name': 'AN-L-Sens-B1-ACa-01', 'type': 'neuron', 'skeleton_ids': [2284021]}, {'id': 2936858, 'name': 'neuron 2936858', 'type': 'neuron', 'skeleton_ids': [2936857]}, {'id': 3032521, 'name': 'MN-L-Sens-B3-AVp-03', 'type': 'neuron', 'skeleton_ids': [3032520]}, {'id': 3036250, 'name': 'MN-L-Sens-B3-VM-12', 'type': 'neuron', 'skeleton_ids': [3036249]}, {'id': 3044501, 'name': 'DPLc1 Descending L', 'type': 'neuron', 'skeleton_ids': [3044500]}, {'id': 3046504, 'name': 'IPC 7 left', 'type': 'neuron', 'skeleton_ids': [3046503]}, {'id': 3050646, 'name': 'MN-L-Sens-B3-VM-14', 'type': 'neuron', 'skeleton_ids': [3050645]}, {'id': 3054102, 'name': 'MN-L-Sens-B3-ACp-04', 'type': 'neuron', 'skeleton_ids': [3054101]}, {'id': 3071302, 'name': 'MN-R-Sens-B2-ACp-13', 'type': 'neuron', 'skeleton_ids': [3071301]}, {'id': 3123906, 'name': 'MN-R-Sens-B3-ACpl-07', 'type': 'neuron', 'skeleton_ids': [3123905]}, {'id': 3158086, 'name': 'AF5: Short-presynaptic-D-1L', 'type': 'neuron', 'skeleton_ids': [3158085]}, {'id': 3158124, 'name': 'presynaptic-D-2-MAP-L left', 'type': 'neuron', 'skeleton_ids': [3158123]}, {'id': 3189408, 'name': 'T2l asc contra 01', 'type': 'neuron', 'skeleton_ids': [3189407]}, {'id': 3192953, 'name': 'AN-R-Sens-B2-AVa-25', 'type': 'neuron', 'skeleton_ids': [3192952]}, {'id': 3199457, 'name': 'MN-R-Sens-B3-ACp-05', 'type': 'neuron', 'skeleton_ids': [3199456]}, {'id': 3267020, 'name': 'MN-L-Sens-B2-VM-19', 'type': 'neuron', 'skeleton_ids': [3267019]}, {'id': 3276004, 'name': 'L1: Short-presynapticD-3-R', 'type': 'neuron', 'skeleton_ids': [3276003]}, {'id': 3281685, 'name': 'IPC 5 left', 'type': 'neuron', 'skeleton_ids': [3281684]}, {'id': 3299768, 'name': 'hugin-PC right 2', 'type': 'neuron', 'skeleton_ids': [3299767]}, {'id': 3315002, 'name': 'hugin-PC right 1', 'type': 'neuron', 'skeleton_ids': [3315001]}, {'id': 3356290, 'name': 'SEZ-IN-A06-right', 'type': 'neuron', 'skeleton_ids': [3356289]}, {'id': 3411444, 'name': 'contra-vine right', 'type': 'neuron', 'skeleton_ids': [3414508]}, {'id': 3420916, 'name': 'SOG into brain contralaterally right - paired with #3622234', 'type': 'neuron', 'skeleton_ids': [3420915]}, {'id': 3436421, 'name': 'Pre PMN R', 'type': 'neuron', 'skeleton_ids': [3436420]}, {'id': 3443130, 'name': 'T3r asc contra 01', 'type': 'neuron', 'skeleton_ids': [3443129]}, {'id': 3450752, 'name': 'mPN BAmd1-g right', 'type': 'neuron', 'skeleton_ids': [3450751]}, {'id': 3477009, 'name': 'X left (Pre PMN)', 'type': 'neuron', 'skeleton_ids': [3477008]}, {'id': 3486382, 'name': '42a PN left', 'type': 'neuron', 'skeleton_ids': [3486381]}, {'id': 3498480, 'name': 'contra-vine left', 'type': 'neuron', 'skeleton_ids': [3498479]}, {'id': 3509023, 'name': 'AN-L-Sens-B3-ACal-03 (temp sensing left neuron together 1)', 'type': 'neuron', 'skeleton_ids': [3609202]}, {'id': 3556192, 'name': 'AN-R-Sens-B1-ACa-03', 'type': 'neuron', 'skeleton_ids': [3556191]}, {'id': 3561725, 'name': 'IPC 5 right', 'type': 'neuron', 'skeleton_ids': [3561724]}, {'id': 3562955, 'name': 'MN-R-Sens-B2-VM-21', 'type': 'neuron', 'skeleton_ids': [3562954]}, {'id': 3570509, 'name': 'Handlebar PN (CB on left)', 'type': 'neuron', 'skeleton_ids': [3991518]}, {'id': 3586985, 'name': 'AN-L-Sens-B3-ACal-07 (upstream of Multig. Olfactory PN 3) ', 'type': 'neuron', 'skeleton_ids': [3586984]}, {'id': 3601282, 'name': 'AN-L-Sens-B3-ACal-01 (temp sensing left neuron alone)', 'type': 'neuron', 'skeleton_ids': [3608397]}, {'id': 3609203, 'name': 'AN-L-Sens-B3-ACal-02 (temp sensing left neuron together 2)', 'type': 'neuron', 'skeleton_ids': [3639968]}, {'id': 3612559, 'name': 'SOG into Brain contralaterally left - paired with #3420915', 'type': 'neuron', 'skeleton_ids': [3622234]}, {'id': 3613277, 'name': 'Intern<NAME> - Inputs from SE0 Serotonin - paired with #9787565', 'type': 'neuron', 'skeleton_ids': [3613276]}, {'id': 3617171, 'name': 'AN-R-Sens-B3-ACal-03 (temp sensing right neuron together 1)', 'type': 'neuron', 'skeleton_ids': [3487306]}, {'id': 3633515, 'name': 'AN-L-Sens-B3-ACp-09 (gustatory pheromone neuron)', 'type': 'neuron', 'skeleton_ids': [5186461]}, {'id': 3650974, 'name': 'AN-R-Sens-B3-ACal-01 (temp sensing right neuron alone)', 'type': 'neuron', 'skeleton_ids': [3650600]}, {'id': 3652588, 'name': 'AN-L-Sens-B3-ACp-08', 'type': 'neuron', 'skeleton_ids': [3652587]}, {'id': 3691222, 'name': 'Lower Looping PN Right - AVM004 (BAla1/2)', 'type': 'neuron', 'skeleton_ids': [3690921]}, {'id': 3709978, 'name': 'backwards handlebar left (looks like 35a)', 'type': 'neuron', 'skeleton_ids': [3709977]}, {'id': 3712894, 'name': 'mPN iACT bilateral LOWER left', 'type': 'neuron', 'skeleton_ids': [3712893]}, {'id': 3728747, 'name': 'Munin right 1', 'type': 'neuron', 'skeleton_ids': [3728746]}, {'id': 3732323, 'name': 'SEZ-IN-32-right', 'type': 'neuron', 'skeleton_ids': [3732322]}, {'id': 3736501, 'name': 'ginkgo 2 left', 'type': 'neuron', 'skeleton_ids': [3736500]}, {'id': 3756660, 'name': 'PN Only Connects To Togethers Right', 'type': 'neuron', 'skeleton_ids': [3756659]}, {'id': 3762061, 'name': 'Bubsy Right', 'type': 'neuron', 'skeleton_ids': [3762060]}, {'id': 3795499, 'name': 'AN-R-Sens-B2-ACp-12', 'type': 'neuron', 'skeleton_ids': [3795498]}, {'id': 3801212, 'name': 'BC: neuron 3801212 - paired with #2147157', 'type': 'neuron', 'skeleton_ids': [3801211]}, {'id': 3813372, 'name': 'Bashful_right', 'type': 'neuron', 'skeleton_ids': [9455885]}, {'id': 3822524, 'name': 'AN-R-Sens-B3-ACp-08', 'type': 'neuron', 'skeleton_ids': [3822523]}, {'id': 3859196, 'name': 'Munin left 2', 'type': 'neuron', 'skeleton_ids': [3859195]}, {'id': 3945233, 'name': 'Sleepy Left', 'type': 'neuron', 'skeleton_ids': [3945232]}, {'id': 3945455, 'name': 'Sleepy-2 Left', 'type': 'neuron', 'skeleton_ids': [3945454]}, {'id': 3946167, 'name': 'Pre-goro3(Mugin PC Output and RG Input?) left', 'type': 'neuron', 'skeleton_ids': [3946166]}, {'id': 3946365, 'name': 'AF6: Munin Output left', 'type': 'neuron', 'skeleton_ids': [3946364]}, {'id': 3948034, 'name': 'SOG interneuron', 'type': 'neuron', 'skeleton_ids': [3948033]}, {'id': 3966684, 'name': 'AN-R-Sens-B3-ACal-07 (upstream of Multig. Olfactory PN 3) right', 'type': 'neuron', 'skeleton_ids': [3966683]}, {'id': 3985830, 'name': 'AN-R-Sens-B3-ACal-05 (Non-ORN 1)', 'type': 'neuron', 'skeleton_ids': [3985829]}, {'id': 4017090, 'name': 'AN-R-Sens-B3-ACal-06 (Degraded? Temp?)', 'type': 'neuron', 'skeleton_ids': [4017089]}, {'id': 4023882, 'name': 'AN-R-Sens-B3-ACal-04 (Non-ORN 2)', 'type': 'neuron', 'skeleton_ids': [4023881]}, {'id': 4030527, 'name': 'AN-R-Sens-B3-VM-14', 'type': 'neuron', 'skeleton_ids': [4030526]}, {'id': 4044655, 'name': 'AN-R-Sens-B3-VM-13', 'type': 'neuron', 'skeleton_ids': [4044654]}, {'id': 4059759, 'name': 'AN-R-Sens-B3-VM-15', 'type': 'neuron', 'skeleton_ids': [4059758]}, {'id': 4096704, 'name': 'AN-R-Sens-B3-VM-12', 'type': 'neuron', 'skeleton_ids': [4096703]}, {'id': 4097555, 'name': 'AN-R-Sens-B3-ACpl-10', 'type': 'neuron', 'skeleton_ids': [4097554]}, {'id': 4108017, 'name': 'Upper Looping PN Left - AVM004', 'type': 'neuron', 'skeleton_ids': [4108016]}, {'id': 4119388, 'name': 'mPN iACT A1 right', 'type': 'neuron', 'skeleton_ids': [4119387]}, {'id': 4120911, 'name': 'SEZ-IN-04-left', 'type': 'neuron', 'skeleton_ids': [4120910]}, {'id': 4137843, 'name': '33a PN left', 'type': 'neuron', 'skeleton_ids': [8021456]}, {'id': 4152023, 'name': 'from upper looping PN - peptidergic right', 'type': 'neuron', 'skeleton_ids': [4152020]}, {'id': 4154422, 'name': 'Lower Looping PN Left - AVM004', 'type': 'neuron', 'skeleton_ids': [4154421]}, {'id': 4249898, 'name': 'NPF_t3l', 'type': 'neuron', 'skeleton_ids': [4249897]}, {'id': 4284107, 'name': 'Holder_left', 'type': 'neuron', 'skeleton_ids': [4284106]}, {'id': 4338597, 'name': 'MN-L-Sens-B2-ACp-01', 'type': 'neuron', 'skeleton_ids': [4338596]}, {'id': 4354934, 'name': '85c PN left', 'type': 'neuron', 'skeleton_ids': [4354933]}, {'id': 4391979, 'name': 'mPN iACT C2 right', 'type': 'neuron', 'skeleton_ids': [4391978]}, {'id': 4393050, 'name': 'non-olfactory PN right', 'type': 'neuron', 'skeleton_ids': [4393049]}, {'id': 4400768, 'name': 'DH44 2 left', 'type': 'neuron', 'skeleton_ids': [4400767]}, {'id': 4404664, 'name': 'PVL005 - 2 Left', 'type': 'neuron', 'skeleton_ids': [4635057]}, {'id': 4453486, 'name': 'IPC 4 left', 'type': 'neuron', 'skeleton_ids': [4453485]}, {'id': 4493200, 'name': '13a PN left', 'type': 'neuron', 'skeleton_ids': [4493199]}, {'id': 4506510, 'name': 'IPC 6 left', 'type': 'neuron', 'skeleton_ids': [4506509]}, {'id': 4523585, 'name': 'DMS 1 left', 'type': 'neuron', 'skeleton_ids': [4523584]}, {'id': 4523804, 'name': 'DMS 2 left', 'type': 'neuron', 'skeleton_ids': [4523803]}, {'id': 4595610, 'name': 'pCC a1l', 'type': 'neuron', 'skeleton_ids': [4595609]}, {'id': 4603254, 'name': 'neuron 4603254', 'type': 'neuron', 'skeleton_ids': [4603253]}, {'id': 4651930, 'name': '94a & 94b PN right', 'type': 'neuron', 'skeleton_ids': [4651929]}, {'id': 4876705, 'name': 'PN in Olfactory Bundle 1 right', 'type': 'neuron', 'skeleton_ids': [4876704]}, {'id': 4992274, 'name': 'neuron 4992274', 'type': 'neuron', 'skeleton_ids': [4992273]}, {'id': 4992309, 'name': 'AN-L-Sens-B1-AVa-30', 'type': 'neuron', 'skeleton_ids': [7424178]}, {'id': 4992323, 'name': 'neuron 4992323', 'type': 'neuron', 'skeleton_ids': [4992322]}, {'id': 5057917, 'name': 'AN-L-Sens-B3-VM-11', 'type': 'neuron', 'skeleton_ids': [5057916]}, {'id': 5088294, 'name': 'AN-L-Sens-B3-ACpl-10', 'type': 'neuron', 'skeleton_ids': [5088293]}, {'id': 5093517, 'name': 'AN-L-Sens-B3-ACal-04 (Non-ORN 2)', 'type': 'neuron', 'skeleton_ids': [5093516]}, {'id': 5095270, 'name': 'AN-L-Sens-B3-VM-14', 'type': 'neuron', 'skeleton_ids': [5095269]}, {'id': 5116148, 'name': 'AN-L-Sens-B3-ACal-06 (degraded alone temperature partner neuron)', 'type': 'neuron', 'skeleton_ids': [5116147]}, {'id': 5116862, 'name': 'AN-L-Sens-B3-VM-15', 'type': 'neuron', 'skeleton_ids': [5116861]}, {'id': 5120098, 'name': 'AN-L-Sens-B3-VM-12', 'type': 'neuron', 'skeleton_ids': [5120097]}, {'id': 5120991, 'name': 'AN-L-Sens-B3-VM-13', 'type': 'neuron', 'skeleton_ids': [5120990]}, {'id': 5169588, 'name': 'MBE16 left', 'type': 'neuron', 'skeleton_ids': [8922644]}, {'id': 5311300, 'name': 'T1r Ascending into PC_right', 'type': 'neuron', 'skeleton_ids': [5311299]}, {'id': 5378684, 'name': 'T1l Ascending into PC_left', 'type': 'neuron', 'skeleton_ids': [5378683]}, {'id': 5404888, 'name': 'Happy Left ', 'type': 'neuron', 'skeleton_ids': [5404887]}, {'id': 5434747, 'name': 'MxN-R-motor-05 - MHD-B', 'type': 'neuron', 'skeleton_ids': [5434746]}, {'id': 5435751, 'name': 'MxN-R-motor-04 - MHD-A', 'type': 'neuron', 'skeleton_ids': [5435750]}, {'id': 5437705, 'name': 'MxN-R-motor-06 - MHD-C', 'type': 'neuron', 'skeleton_ids': [5437704]}, {'id': 5439603, 'name': 'MxN-R-motor-09 - MHE-C', 'type': 'neuron', 'skeleton_ids': [5439602]}, {'id': 5440617, 'name': 'MxN-R-motor-08 - MHE-B', 'type': 'neuron', 'skeleton_ids': [5440613]}, {'id': 5448623, 'name': 'MN-R-Sens-B1-ACpl-01', 'type': 'neuron', 'skeleton_ids': [5448622]}, {'id': 5450670, 'name': 'MxN-L-motor-05 - MHD-B', 'type': 'neuron', 'skeleton_ids': [5450669]}, {'id': 5452287, 'name': 'MxN-L-motor-06 - MHD-C', 'type': 'neuron', 'skeleton_ids': [5452286]}, {'id': 5453144, 'name': 'MxN-L-motor-04 - MHD-A', 'type': 'neuron', 'skeleton_ids': [5453143]}, {'id': 5478205, 'name': '74a PN left', 'type': 'neuron', 'skeleton_ids': [7843730]}, {'id': 5558725, 'name': 'BAmas d 4 neuron 5558725', 'type': 'neuron', 'skeleton_ids': [5558724]}, {'id': 5614979, 'name': 'T1l asc contra 01', 'type': 'neuron', 'skeleton_ids': [5614978]}, {'id': 5669276, 'name': 'MBE16 right', 'type': 'neuron', 'skeleton_ids': [8297018]}, {'id': 5969256, 'name': 'MBE34 right', 'type': 'neuron', 'skeleton_ids': [4598109]}, {'id': 6369545, 'name': 't2r medial soma 1', 'type': 'neuron', 'skeleton_ids': [6369544]}, {'id': 6445995, 'name': 'Pre PMN L', 'type': 'neuron', 'skeleton_ids': [6445994]}, {'id': 6703242, 'name': 'MBE23 right', 'type': 'neuron', 'skeleton_ids': [6703240]}, {'id': 6703262, 'name': '94a & 94b PN left', 'type': 'neuron', 'skeleton_ids': [8399890]}, {'id': 6794665, 'name': 'neuron 6794665', 'type': 'neuron', 'skeleton_ids': [6794664]}, {'id': 6802554, 'name': 'T1l asc contra 02', 'type': 'neuron', 'skeleton_ids': [6802553]}, {'id': 6883832, 'name': 'AN-R-Sens-B1-AVa-16', 'type': 'neuron', 'skeleton_ids': [6883831]}, {'id': 6885134, 'name': 'SEZ-IN-A05-left - paired with #18354200', 'type': 'neuron', 'skeleton_ids': [6885133]}, {'id': 7001631, 'name': '49a PN left', 'type': 'neuron', 'skeleton_ids': [7515491]}, {'id': 7020345, 'name': 'handlebar-like 4 right', 'type': 'neuron', 'skeleton_ids': [7020344]}, {'id': 7092087, 'name': 'PN in Olfactory Bundle 1 left', 'type': 'neuron', 'skeleton_ids': [7092086]}, {'id': 7092418, 'name': '47a & 33b PN left', 'type': 'neuron', 'skeleton_ids': [7092417]}, {'id': 7107434, 'name': '45b PN left', 'type': 'neuron', 'skeleton_ids': [7107433]}, {'id': 7136255, 'name': 'Moggy_left', 'type': 'neuron', 'skeleton_ids': [7136254]}, {'id': 7160161, 'name': 'AN-R-Sens-B2-AVp-01', 'type': 'neuron', 'skeleton_ids': [7160160]}, {'id': 7198339, 'name': '24a PN left', 'type': 'neuron', 'skeleton_ids': [7198338]}, {'id': 7556565, 'name': 'MN-L-Sens-B3-ACp-03', 'type': 'neuron', 'skeleton_ids': [7556564]}, {'id': 7557605, 'name': 'L1: Short-presynapticD-3-L', 'type': 'neuron', 'skeleton_ids': [7557604]}, {'id': 7571967, 'name': 'CPe3/pep asc cand Right; CP1 d 4', 'type': 'neuron', 'skeleton_ids': [7571966]}, {'id': 7845338, 'name': '45a PN left', 'type': 'neuron', 'skeleton_ids': [7852250]}, {'id': 7845347, 'name': '63a PN left', 'type': 'neuron', 'skeleton_ids': [7845346]}, {'id': 7851154, 'name': '82a PN left', 'type': 'neuron', 'skeleton_ids': [7851153]}, {'id': 7864824, 'name': '59a PN left', 'type': 'neuron', 'skeleton_ids': [7864823]}, {'id': 7865653, 'name': '22c PN left', 'type': 'neuron', 'skeleton_ids': [7865652]}, {'id': 7865697, 'name': '1a PN left', 'type': 'neuron', 'skeleton_ids': [7865696]}, {'id': 7865768, 'name': '83a PN left', 'type': 'neuron', 'skeleton_ids': [7865767]}, {'id': 7873136, 'name': '42b PN left', 'type': 'neuron', 'skeleton_ids': [7873135]}, {'id': 7916873, 'name': '30a PN left', 'type': 'neuron', 'skeleton_ids': [7916593]}, {'id': 7934905, 'name': '35a PN bilateral left', 'type': 'neuron', 'skeleton_ids': [7934904]}, {'id': 7935089, 'name': '35a PN bilateral right', 'type': 'neuron', 'skeleton_ids': [7935088]}, {'id': 8012954, 'name': 'PN in Olfactory Bundle 2 left', 'type': 'neuron', 'skeleton_ids': [8012953]}, {'id': 8055672, 'name': 'MN-L-Sens-B1-ACpl-01', 'type': 'neuron', 'skeleton_ids': [8055671]}, {'id': 8061048, 'name': 'MxN-L-motor-07 - MHE-A', 'type': 'neuron', 'skeleton_ids': [8061047]}, {'id': 8084864, 'name': 'MxN-L-motor-08 - MHE-B', 'type': 'neuron', 'skeleton_ids': [8084863]}, {'id': 8092229, 'name': 'MxN-L-motor-03 - LR-B', 'type': 'neuron', 'skeleton_ids': [8092228]}, {'id': 8094861, 'name': 'MBE11a right', 'type': 'neuron', 'skeleton_ids': [8094859]}, {'id': 8105237, 'name': 'MxN-L-motor-02 - LR-A', 'type': 'neuron', 'skeleton_ids': [8105236]}, {'id': 8134939, 'name': 'MxN-R-motor-07 - MHE-A', 'type': 'neuron', 'skeleton_ids': [8134938]}, {'id': 8138802, 'name': 'PVL005 - 2 Right (V1); BLVa34 a 11', 'type': 'neuron', 'skeleton_ids': [9275210]}, {'id': 8140110, 'name': '74a PN right', 'type': 'neuron', 'skeleton_ids': [8140109]}, {'id': 8149491, 'name': 'MxN-R-motor-01 - SGDO', 'type': 'neuron', 'skeleton_ids': [8149490]}, {'id': 8167774, 'name': 'MxN-R-motor-02 - LR-A', 'type': 'neuron', 'skeleton_ids': [8167773]}, {'id': 8169738, 'name': 'Input to MN motor neurons left - inputs to motor neurons on both sides', 'type': 'neuron', 'skeleton_ids': [8169737]}, {'id': 8177701, 'name': 'MxN-R-motor-03 - LR-B', 'type': 'neuron', 'skeleton_ids': [8177700]}, {'id': 8199013, 'name': 'MN-R-Sens-B3-?-00', 'type': 'neuron', 'skeleton_ids': [8199012]}, {'id': 8208556, 'name': 'MxN-L-motor-01 - SGDO', 'type': 'neuron', 'skeleton_ids': [8208555]}, {'id': 8213785, 'name': 'Old Fashioned Telephone Left', 'type': 'neuron', 'skeleton_ids': [8213784]}, {'id': 8238323, 'name': 'LH bilateral axon 1_left; BLVa12 b 2', 'type': 'neuron', 'skeleton_ids': [8238322]}, {'id': 8243456, 'name': '83a PN right', 'type': 'neuron', 'skeleton_ids': [8243455]}, {'id': 8244724, 'name': '42a PN right', 'type': 'neuron', 'skeleton_ids': [8244723]}, {'id': 8244824, 'name': '13a PN right', 'type': 'neuron', 'skeleton_ids': [8244823]}, {'id': 8245015, 'name': '59a PN right', 'type': 'neuron', 'skeleton_ids': [8245014]}, {'id': 8245147, 'name': '82a PN right', 'type': 'neuron', 'skeleton_ids': [8245146]}, {'id': 8245514, 'name': '63a PN right', 'type': 'neuron', 'skeleton_ids': [8245513]}, {'id': 8245693, 'name': '42b PN right', 'type': 'neuron', 'skeleton_ids': [8245692]}, {'id': 8245871, 'name': '24a PN right', 'type': 'neuron', 'skeleton_ids': [8245869]}, {'id': 8246082, 'name': '67b PN right', 'type': 'neuron', 'skeleton_ids': [8246081]}, {'id': 8246751, 'name': '47a & 33b PN right', 'type': 'neuron', 'skeleton_ids': [8246750]}, {'id': 8247452, 'name': '45a PN right', 'type': 'neuron', 'skeleton_ids': [8247451]}, {'id': 8248632, 'name': '22c PN right', 'type': 'neuron', 'skeleton_ids': [8248631]}, {'id': 8252068, 'name': '1a PN right', 'type': 'neuron', 'skeleton_ids': [8252067]}, {'id': 8253414, 'name': '45b PN right', 'type': 'neuron', 'skeleton_ids': [8253413]}, {'id': 8254896, 'name': 'PN in Olfactory Bundle 2 right', 'type': 'neuron', 'skeleton_ids': [8254894]}, {'id': 8256187, 'name': '30a PN right', 'type': 'neuron', 'skeleton_ids': [8256186]}, {'id': 8435312, 'name': 'MxN-L-motor-09 - MHE-C', 'type': 'neuron', 'skeleton_ids': [8435311]}, {'id': 8438284, 'name': 'Dopey_left', 'type': 'neuron', 'skeleton_ids': [8438283]}, {'id': 8752698, 'name': 'AN: DPMlp Lar_right', 'type': 'neuron', 'skeleton_ids': [8752697]}, {'id': 8810740, 'name': 'MN-R-Sens-B2-AVp-02', 'type': 'neuron', 'skeleton_ids': [8810739]}, {'id': 8953093, 'name': 'PaN-R-motor-01', 'type': 'neuron', 'skeleton_ids': [2220573]}, {'id': 9015746, 'name': 'Left Mid', 'type': 'neuron', 'skeleton_ids': [9015745]}, {'id': 9074102, 'name': 'MBE35 right', 'type': 'neuron', 'skeleton_ids': [9074101]}, {'id': 9109800, 'name': 'MBE35 left', 'type': 'neuron', 'skeleton_ids': [9109799]}, {'id': 9462897, 'name': 'AVL011 PN Left', 'type': 'neuron', 'skeleton_ids': [4985759]}, {'id': 9503419, 'name': 'midline projection neuron in the brain_PI ', 'type': 'neuron', 'skeleton_ids': [9503414]}, {'id': 9578082, 'name': 'PVL005 1 right', 'type': 'neuron', 'skeleton_ids': [7971031]}, {'id': 9583436, 'name': 'NI? right', 'type': 'neuron', 'skeleton_ids': [9583435]}, {'id': 9621721, 'name': 'AVL011 PN Right 2?', 'type': 'neuron', 'skeleton_ids': [4620453]}, {'id': 9654603, 'name': 'Sweetness Right 130521_Cand1_OSN2_IN5 Bilateral', 'type': 'neuron', 'skeleton_ids': [9455820]}, {'id': 9654797, 'name': 'AF3 thermo 3rd left', 'type': 'neuron', 'skeleton_ids': [4785242]}, {'id': 9654949, 'name': 'AVL011 PN Right', 'type': 'neuron', 'skeleton_ids': [9291474]}, {'id': 9655097, 'name': 'AF6 right - ASB TODO 1.1', 'type': 'neuron', 'skeleton_ids': [6133702]}, {'id': 9671611, 'name': 'MBE9b right', 'type': 'neuron', 'skeleton_ids': [7840791]}, {'id': 9679159, 'name': 'AN-L-Sens-B1-AVa-17', 'type': 'neuron', 'skeleton_ids': [1375101]}, {'id': 9679175, 'name': 'AN-L-Sens-B2-ACp-03', 'type': 'neuron', 'skeleton_ids': [7439909]}, {'id': 9680577, 'name': 'AN-L-motor-08', 'type': 'neuron', 'skeleton_ids': [407869]}, {'id': 9680663, 'name': 'BAmas12 midline neuron 1? left', 'type': 'neuron', 'skeleton_ids': [1924051]}, {'id': 9680669, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1822302]}, {'id': 9680687, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1554836]}, {'id': 9680693, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1156684]}, {'id': 9680699, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1067653]}, {'id': 9680711, 'name': 'X right (Interneuron--35 in total)', 'type': 'neuron', 'skeleton_ids': [1575789]}, {'id': 9680717, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1540458]}, {'id': 9680729, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1334471]}, {'id': 9680765, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [1149968]}, {'id': 9680801, 'name': 'Interneuron--35 in total (strong to MN motor neurons)', 'type': 'neuron', 'skeleton_ids': [768233]}, {'id': 9680813, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [692877]}, {'id': 9680831, 'name': 'SEZ-IN-10-left', 'type': 'neuron', 'skeleton_ids': [616176]}, {'id': 9680837, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [375785]}, {'id': 9680849, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [236151]}, {'id': 9680855, 'name': 'AX ; SEZ-IN-A03-left', 'type': 'neuron', 'skeleton_ids': [255222]}, {'id': 9680867, 'name': 'Interneuron--35 in total', 'type': 'neuron', 'skeleton_ids': [7439388]}, {'id': 9680873, 'name': 'interneuron missing cell bodies 5', 'type': 'neuron', 'skeleton_ids': [2321929]}, {'id': 9680885, 'name': 'SEZ-IN-A04-right', 'type': 'neuron', 'skeleton_ids': [3454765]}, {'id': 9681155, 'name': 'AN: DPMlp Lar_left', 'type': 'neuron', 'skeleton_ids': [3157405]}, {'id': 9681190, 'name': 'V1: possible PVL005 - 3 Right?', 'type': 'neuron', 'skeleton_ids': [2921933]}, {'id': 9712224, 'name': 'KS LN 1 right', 'type': 'neuron', 'skeleton_ids': [9712223]}, {'id': 9716859, 'name': 'AN-R-Sens-B1-AVa-19', 'type': 'neuron', 'skeleton_ids': [9716858]}, {'id': 9730270, 'name': 'SEZ-IN-13-right', 'type': 'neuron', 'skeleton_ids': [9730269]}, {'id': 9747419, 'name': 'PPLV - 2 Left (maybe PVL014c)', 'type': 'neuron', 'skeleton_ids': [9747418]}, {'id': 9747712, 'name': 'Handlebar PN (CB on Right)', 'type': 'neuron', 'skeleton_ids': [9747710]}, {'id': 9748270, 'name': 'AN-R-Sens-B1-AVa-18', 'type': 'neuron', 'skeleton_ids': [9748269]}, {'id': 9748580, 'name': 'hugin-PC right 3', 'type': 'neuron', 'skeleton_ids': [9748579]}, {'id': 9748982, 'name': 'Moggy_right', 'type': 'neuron', 'skeleton_ids': [9748981]}, {'id': 9787566, 'name': 'Interneuron W left - Inputs from SE0 Serotonin - paired with #3613276', 'type': 'neuron', 'skeleton_ids': [9787565]}, {'id': 9787593, 'name': 'BAmas12_ven descending 3 left', 'type': 'neuron', 'skeleton_ids': [9787592]}, {'id': 9787673, 'name': 'SEZ-IN-11-left', 'type': 'neuron', 'skeleton_ids': [9787672]}, {'id': 9789131, 'name': 'hugin-PC left 1', 'type': 'neuron', 'skeleton_ids': [9789130]}, {'id': 9795663, 'name': 'BAmas12_ven descending 4 left', 'type': 'neuron', 'skeleton_ids': [9795662]}, {'id': 9796598, 'name': 'hugin-PC left 2', 'type': 'neuron', 'skeleton_ids': [9796597]}, {'id': 9801442, 'name': 'BE ; Right', 'type': 'neuron', 'skeleton_ids': [9801441]}, {'id': 9805521, 'name': 'hugin-PC left 3', 'type': 'neuron', 'skeleton_ids': [9805520]}, {'id': 9805589, 'name': 'Angler_left', 'type': 'neuron', 'skeleton_ids': [9805588]}, {'id': 9808023, 'name': 'MN-L-Sens-B2-AVp-01', 'type': 'neuron', 'skeleton_ids': [9808022]}, {'id': 9813903, 'name': 'hugin-PC left 4', 'type': 'neuron', 'skeleton_ids': [9813902]}, {'id': 9820271, 'name': 'AN-L-Sens-B1-AVa-29', 'type': 'neuron', 'skeleton_ids': [9820270]}, {'id': 9838295, 'name': 'AN-L-Sens-B1-ACa-13', 'type': 'neuron', 'skeleton_ids': [9838294]}, {'id': 9842384, 'name': 'neuron 9842384', 'type': 'neuron', 'skeleton_ids': [9842383]}, {'id': 9853426, 'name': 'AN-R-Sens-B3-ACp-09 (gustatory pheromone neuron)', 'type': 'neuron', 'skeleton_ids': [9853425]}, {'id': 9934375, 'name': 'neuron 9934375: incomplete neuron with soma; unidentified', 'type': 'neuron', 'skeleton_ids': [9934374]}, {'id': 9935753, 'name': 'T1r asc contra 01', 'type': 'neuron', 'skeleton_ids': [9935752]}, {'id': 10119917, 'name': 'Z: contra_right', 'type': 'neuron', 'skeleton_ids': [10119916]}, {'id': 10160251, 'name': 'mPN BAmd1-g left', 'type': 'neuron', 'skeleton_ids': [10160250]}, {'id': 10263288, 'name': 'SEZ-IN-07-right', 'type': 'neuron', 'skeleton_ids': [10263287]}, {'id': 10329209, 'name': 'IPC 1 left', 'type': 'neuron', 'skeleton_ids': [10329208]}, {'id': 10418395, 'name': 'DH44 1 left', 'type': 'neuron', 'skeleton_ids': [10418394]}, {'id': 10459724, 'name': 'IPC 2 left', 'type': 'neuron', 'skeleton_ids': [10459723]}, {'id': 10563756, 'name': 'DH44 3 left', 'type': 'neuron', 'skeleton_ids': [10563755]}, {'id': 10768710, 'name': 'Bilateral near MB_right', 'type': 'neuron', 'skeleton_ids': [10768709]}, {'id': 10934439, 'name': 'MN-R-Sens-B2-ACp-18', 'type': 'neuron', 'skeleton_ids': [10934438]}, {'id': 11067380, 'name': 'non-olfactory PN left', 'type': 'neuron', 'skeleton_ids': [11067379]}, {'id': 11105183, 'name': 'clamp left', 'type': 'neuron', 'skeleton_ids': [11105182]}, {'id': 11210001, 'name': 'LH bilateral axon 1_right; BLVa12 b 2', 'type': 'neuron', 'skeleton_ids': [11210000]}, {'id': 11279245, 'name': '49a PN right', 'type': 'neuron', 'skeleton_ids': [11279244]}, {'id': 11333055, 'name': 'Gustatory? PN left', 'type': 'neuron', 'skeleton_ids': [11333054]}, {'id': 11543213, 'name': 'mPN iACT bilateral LOWER right', 'type': 'neuron', 'skeleton_ids': [11543212]}, {'id': 11637859, 'name': 'Y Neuron-right', 'type': 'neuron', 'skeleton_ids': [11637858]}, {'id': 11638512, 'name': 'Tiger_left', 'type': 'neuron', 'skeleton_ids': [11638511]}, {'id': 11639193, 'name': 'contra-vine 2 right', 'type': 'neuron', 'skeleton_ids': [11639192]}, {'id': 11642880, 'name': 'BD ; Left', 'type': 'neuron', 'skeleton_ids': [11642879]}, {'id': 11671565, 'name': 'AN-L-Sens-B2-ACp-21', 'type': 'neuron', 'skeleton_ids': [11671564]}, {'id': 11687554, 'name': 'T3l asc contra 01', 'type': 'neuron', 'skeleton_ids': [11687553]}, {'id': 11736973, 'name': 'MN-L-Sens-B2-ACp-15', 'type': 'neuron', 'skeleton_ids': [11736972]}, {'id': 11745382, 'name': 'MN-L-Sens-B2-ACp-07', 'type': 'neuron', 'skeleton_ids': [11745381]}, {'id': 11868261, 'name': 'Angler_right', 'type': 'neuron', 'skeleton_ids': [11868260]}, {'id': 11941256, 'name': 'Inter2 right', 'type': 'neuron', 'skeleton_ids': [11941255]}, {'id': 11946904, 'name': 'M1 ; Right', 'type': 'neuron', 'skeleton_ids': [11946903]}, {'id': 12073882, 'name': 'MN-L-Sens-B2-ACp-16', 'type': 'neuron', 'skeleton_ids': [12073881]}, {'id': 12156010, 'name': 'handlebar-like 4 (no soma) left ', 'type': 'neuron', 'skeleton_ids': [12156009]}, {'id': 12199927, 'name': 'SEZ-IN-10-right', 'type': 'neuron', 'skeleton_ids': [12199926]}, {'id': 12253021, 'name': 'MN-R-Sens-B2-ACp-14', 'type': 'neuron', 'skeleton_ids': [12253020]}, {'id': 12260697, 'name': 'MN-L-Sens-B2-ACp-06', 'type': 'neuron', 'skeleton_ids': [12260696]}, {'id': 12335382, 'name': 'BAmas12 midline 3 right', 'type': 'neuron', 'skeleton_ids': [12335381]}, {'id': 12634155, 'name': 'IPC 3 left', 'type': 'neuron', 'skeleton_ids': [12634154]}, {'id': 12740291, 'name': 'SEZ-IN-A01-left', 'type': 'neuron', 'skeleton_ids': [12740290]}, {'id': 12901663, 'name': 'AN-R-motor-12', 'type': 'neuron', 'skeleton_ids': [12901662]}, {'id': 12948471, 'name': 'PVL005 1 left', 'type': 'neuron', 'skeleton_ids': [12948470]}, {'id': 13142933, 'name': 'Dopey_right', 'type': 'neuron', 'skeleton_ids': [13142932]}, {'id': 13168716, 'name': 'SEZ-IN-09-left', 'type': 'neuron', 'skeleton_ids': [13168715]}, {'id': 13190772, 'name': 'fork of GPhN (right)', 'type': 'neuron', 'skeleton_ids': [13190771]}, {'id': 13429046, 'name': 'AF4: Large Spider Neuron right', 'type': 'neuron', 'skeleton_ids': [13429045]}, {'id': 13535381, 'name': 'IPC 1 right', 'type': 'neuron', 'skeleton_ids': [13535380]}, {'id': 13535388, 'name': 'DH44 3 right', 'type': 'neuron', 'skeleton_ids': [13535387]}, {'id': 13538011, 'name': 'SEZ-IN-A07-right', 'type': 'neuron', 'skeleton_ids': [13538010]}, {'id': 13581831, 'name': 'hugin-PC right 4', 'type': 'neuron', 'skeleton_ids': [13581830]}, {'id': 13589252, 'name': '33a PN right', 'type': 'neuron', 'skeleton_ids': [13589251]}, {'id': 13645611, 'name': 'DPLc1 1', 'type': 'neuron', 'skeleton_ids': [13645610]}, {'id': 13744426, 'name': 'AN-L-Sens-B1-ACa-02', 'type': 'neuron', 'skeleton_ids': [13744425]}, {'id': 13751195, 'name': 'DH44 2 right', 'type': 'neuron', 'skeleton_ids': [13751194]}, {'id': 13789483, 'name': 'backwards handlebar right', 'type': 'neuron', 'skeleton_ids': [13789482]}, {'id': 13793213, 'name': 'DMS 2 right', 'type': 'neuron', 'skeleton_ids': [13793212]}, {'id': 14035922, 'name': 'SEZ-IN-08-right', 'type': 'neuron', 'skeleton_ids': [14035921]}, {'id': 14082323, 'name': 'MBE9b left', 'type': 'neuron', 'skeleton_ids': [14082322]}, {'id': 14273874, 'name': 'neuron 14273874 alone?', 'type': 'neuron', 'skeleton_ids': [14273873]}, {'id': 14313751, 'name': 'E ; BAMD PN_left', 'type': 'neuron', 'skeleton_ids': [14313750]}, {'id': 14604029, 'name': '85c PN right', 'type': 'neuron', 'skeleton_ids': [14604028]}, {'id': 14812546, 'name': 'Bashful_left', 'type': 'neuron', 'skeleton_ids': [14812545]}, {'id': 14930785, 'name': 'SEZ-IN-12-left', 'type': 'neuron', 'skeleton_ids': [14930784]}, {'id': 14962388, 'name': 'SOG IN right', 'type': 'neuron', 'skeleton_ids': [14962387]}, {'id': 15202264, 'name': 'IPC 4 right', 'type': 'neuron', 'skeleton_ids': [15202263]}, {'id': 15225590, 'name': 'pre-dorsal-MAP presynaptic-D-2-R right', 'type': 'neuron', 'skeleton_ids': [15225589]}, {'id': 15250081, 'name': 'BAmas12_ven descending 4 right', 'type': 'neuron', 'skeleton_ids': [15250080]}, {'id': 15253922, 'name': 'MBE34 left', 'type': 'neuron', 'skeleton_ids': [15253921]}, {'id': 15398731, 'name': 'MBE23 left', 'type': 'neuron', 'skeleton_ids': [15398730]}, {'id': 15398808, 'name': 'AN-R-Sens-B2-ACp-17', 'type': 'neuron', 'skeleton_ids': [15398807]}, {'id': 15409329, 'name': 'AN-R-Sens-B2-ACp-08', 'type': 'neuron', 'skeleton_ids': [15409328]}, {'id': 15421364, 'name': 'MBE2a left', 'type': 'neuron', 'skeleton_ids': [15421363]}, {'id': 15434201, 'name': 'AN-R-Sens-B2-ACp-06', 'type': 'neuron', 'skeleton_ids': [15434200]}, {'id': 15445199, 'name': 'AN-R-Sens-B2-VM-08', 'type': 'neuron', 'skeleton_ids': [15445198]}, {'id': 15451085, 'name': 'AN-R-Sens-B2-ACp-09', 'type': 'neuron', 'skeleton_ids': [15451084]}, {'id': 15458317, 'name': 'AN-R-Sens-B2-ACp-10', 'type': 'neuron', 'skeleton_ids': [15458316]}, {'id': 15461770, 'name': 'AN-R-Sens-B2-ACp-22', 'type': 'neuron', 'skeleton_ids': [15461769]}, {'id': 15505838, 'name': 'AN-R-Sens-B2-AVp-06', 'type': 'neuron', 'skeleton_ids': [15505837]}, {'id': 15506095, 'name': 'AN-R-Sens-B2-ACp-18', 'type': 'neuron', 'skeleton_ids': [15506094]}, {'id': 15507889, 'name': 'AN-R-Sens-B2-ACp-20', 'type': 'neuron', 'skeleton_ids': [15507888]}, {'id': 15508476, 'name': 'AN-R-Sens-B2-VM-04', 'type': 'neuron', 'skeleton_ids': [15508475]}, {'id': 15508736, 'name': 'AN-R-Sens-B2-ACp-01', 'type': 'neuron', 'skeleton_ids': [15508735]}, {'id': 15522778, 'name': 'AN-R-Sens-B2-ACp-21', 'type': 'neuron', 'skeleton_ids': [15522777]}, {'id': 15525859, 'name': 'AN-R-Sens-B2-ACp-14', 'type': 'neuron', 'skeleton_ids': [15525858]}, {'id': 15530883, 'name': 'AN-R-Sens-B2-ACp-15', 'type': 'neuron', 'skeleton_ids': [15530882]}, {'id': 15531034, 'name': 'AN-R-Sens-B2-ACp-03', 'type': 'neuron', 'skeleton_ids': [15531033]}, {'id': 15531917, 'name': 'AN-R-Sens-B2-ACp-04', 'type': 'neuron', 'skeleton_ids': [15531916]}, {'id': 15532106, 'name': 'AN-R-Sens-B2-AVa-24', 'type': 'neuron', 'skeleton_ids': [15532105]}, {'id': 15533422, 'name': 'AN-R-Sens-B2-AVp-02', 'type': 'neuron', 'skeleton_ids': [15533421]}, {'id': 15541945, 'name': 'AN-R-Sens-B2-ACp-02', 'type': 'neuron', 'skeleton_ids': [15541944]}, {'id': 15541977, 'name': 'AN-R-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [15541976]}, {'id': 15542200, 'name': 'AN-R-Sens-B2-?-00', 'type': 'neuron', 'skeleton_ids': [15542199]}, {'id': 15542355, 'name': 'AN-R-Sens-B1-AVa-42', 'type': 'neuron', 'skeleton_ids': [15542354]}, {'id': 15543069, 'name': 'AN-R-Sens-B1-AVa-21', 'type': 'neuron', 'skeleton_ids': [15543068]}, {'id': 15543395, 'name': 'AN-R-Sens-B1-AVa-31', 'type': 'neuron', 'skeleton_ids': [15543394]}, {'id': 15546925, 'name': 'AN-R-Sens-B1-AVa-30', 'type': 'neuron', 'skeleton_ids': [15546924]}, {'id': 15547733, 'name': 'AN-R-Sens-B1-AVa-33', 'type': 'neuron', 'skeleton_ids': [15547732]}, {'id': 15548108, 'name': 'AN-R-Sens-B1-ACa-17', 'type': 'neuron', 'skeleton_ids': [15548107]}, {'id': 15548695, 'name': 'AN-R-Sens-B1-ACa-10', 'type': 'neuron', 'skeleton_ids': [15548694]}, {'id': 15556082, 'name': 'MN-R-Sens-B2-ACp-11', 'type': 'neuron', 'skeleton_ids': [15556081]}, {'id': 15556869, 'name': 'MN-R-Sens-B2-ACp-02', 'type': 'neuron', 'skeleton_ids': [15556868]}, {'id': 15559196, 'name': 'AN-R-Sens-B1-ACa-04', 'type': 'neuron', 'skeleton_ids': [15559195]}, {'id': 15561414, 'name': 'AN-R-Sens-B1-ACa-13', 'type': 'neuron', 'skeleton_ids': [15561413]}, {'id': 15562138, 'name': 'AN-R-Sens-B1-ACa-07', 'type': 'neuron', 'skeleton_ids': [15562137]}, {'id': 15563554, 'name': 'AN-R-Sens-B1-ACa-05', 'type': 'neuron', 'skeleton_ids': [15563553]}, {'id': 15564662, 'name': 'AN-R-Sens-B1-AVa-29', 'type': 'neuron', 'skeleton_ids': [15564661]}, {'id': 15564696, 'name': 'SE0 R3', 'type': 'neuron', 'skeleton_ids': [15564695]}, {'id': 15564707, 'name': 'AN-R-Sens-B1-AVa-24', 'type': 'neuron', 'skeleton_ids': [15564706]}, {'id': 15564783, 'name': 'AN-R-Sens-B1-AVa-23', 'type': 'neuron', 'skeleton_ids': [15564782]}, {'id': 15564808, 'name': 'AN-R-Sens-B1-AVa-22', 'type': 'neuron', 'skeleton_ids': [15564807]}, {'id': 15564883, 'name': 'AN-R-Sens-B1-AVa-37', 'type': 'neuron', 'skeleton_ids': [15564882]}, {'id': 15564912, 'name': 'AN-R-Sens-B1-AVa-27', 'type': 'neuron', 'skeleton_ids': [15564911]}, {'id': 15567385, 'name': 'AN-R-motor-07', 'type': 'neuron', 'skeleton_ids': [15567384]}, {'id': 15567467, 'name': 'AN-R-Sens-B1-AVa-41', 'type': 'neuron', 'skeleton_ids': [15567466]}, {'id': 15569562, 'name': 'AN-R-Sens-B1-AVa-28', 'type': 'neuron', 'skeleton_ids': [15569561]}, {'id': 15572415, 'name': 'AN-R-Sens-B1-AVa-25', 'type': 'neuron', 'skeleton_ids': [15572414]}, {'id': 15573284, 'name': 'AN-R-Sens-B2-ACp-13', 'type': 'neuron', 'skeleton_ids': [15573283]}, {'id': 15573332, 'name': 'AN-R-Sens-B2-ACp-16', 'type': 'neuron', 'skeleton_ids': [15573331]}, {'id': 15573382, 'name': 'AN-R-Sens-B1-ACa-12', 'type': 'neuron', 'skeleton_ids': [15573381]}, {'id': 15573397, 'name': 'AN-R-Sens-B1-ACa-09', 'type': 'neuron', 'skeleton_ids': [15573396]}, {'id': 15573429, 'name': 'SE<NAME>', 'type': 'neuron', 'skeleton_ids': [15573428]}, {'id': 15573451, 'name': 'AN-R-Sens-B1-ACa-14', 'type': 'neuron', 'skeleton_ids': [15573450]}, {'id': 15574200, 'name': 'AN-R-Sens-B1-ACa-06', 'type': 'neuron', 'skeleton_ids': [15574199]}, {'id': 15574232, 'name': 'AN-R-Sens-B1-ACa-08', 'type': 'neuron', 'skeleton_ids': [15574231]}, {'id': 15574329, 'name': 'AN-R-Sens-B1-AVa-15', 'type': 'neuron', 'skeleton_ids': [15574328]}, {'id': 15574509, 'name': 'AN-R-Sens-B1-AVa-38', 'type': 'neuron', 'skeleton_ids': [15574508]}, {'id': 15574942, 'name': 'AN-R-Sens-B1-AVa-34', 'type': 'neuron', 'skeleton_ids': [15574941]}, {'id': 15575053, 'name': 'AN-R-Sens-B1-AVa-36', 'type': 'neuron', 'skeleton_ids': [15575052]}, {'id': 15575233, 'name': 'AN-R-Sens-B1-AVa-35', 'type': 'neuron', 'skeleton_ids': [15575232]}, {'id': 15575328, 'name': 'AN-R-Sens-B1-AVa-26', 'type': 'neuron', 'skeleton_ids': [15575327]}, {'id': 15576195, 'name': 'AN-R-Sens-B1-AVa-40', 'type': 'neuron', 'skeleton_ids': [15576194]}, {'id': 15581429, 'name': 'SEZ-IN-08-left', 'type': 'neuron', 'skeleton_ids': [15581428]}, {'id': 15586537, 'name': 'AN-R-Sens-B1-ACa-11', 'type': 'neuron', 'skeleton_ids': [15586536]}, {'id': 15587167, 'name': 'AN-R-Sens-B1-AVa-39', 'type': 'neuron', 'skeleton_ids': [15587166]}, {'id': 15588692, 'name': 'MN-R-Sens-B2-ACp-12', 'type': 'neuron', 'skeleton_ids': [15588691]}, {'id': 15588801, 'name': 'MN-R-Sens-B2-ACp-20', 'type': 'neuron', 'skeleton_ids': [15588800]}, {'id': 15589363, 'name': 'MN-R-Sens-B2-ACp-06', 'type': 'neuron', 'skeleton_ids': [15589362]}, {'id': 15589412, 'name': 'MN-R-Sens-B2-ACp-21', 'type': 'neuron', 'skeleton_ids': [15589411]}, {'id': 15590762, 'name': 'AN-L-Sens-B2-ACp-07', 'type': 'neuron', 'skeleton_ids': [15590761]}, {'id': 15592697, 'name': 'AN-R-Sens-B2-ACp-11', 'type': 'neuron', 'skeleton_ids': [15592696]}, {'id': 15594039, 'name': 'AN-R-Sens-B2-VM-05', 'type': 'neuron', 'skeleton_ids': [15594038]}, {'id': 15594124, 'name': 'MN-R-Sens-B1-VM-02', 'type': 'neuron', 'skeleton_ids': [15594123]}, {'id': 15595608, 'name': 'MN-R-Sens-B2-ACp-08', 'type': 'neuron', 'skeleton_ids': [15595607]}, {'id': 15595824, 'name': 'MN-R-Sens-B2-ACp-15', 'type': 'neuron', 'skeleton_ids': [15595823]}, {'id': 15595951, 'name': 'MN-R-Sens-B2-ACpl-01 (Awesome MN sensory 2)', 'type': 'neuron', 'skeleton_ids': [15595950]}, {'id': 15599769, 'name': 'Neuron 15599769', 'type': 'neuron', 'skeleton_ids': [15599768]}, {'id': 15605658, 'name': 'AN-R-Sens-B2-AVp-03', 'type': 'neuron', 'skeleton_ids': [15605657]}, {'id': 15606919, 'name': 'MN-R-Sens-B2-VM-04', 'type': 'neuron', 'skeleton_ids': [15606918]}, {'id': 15607181, 'name': 'MN-R-Sens-B2-VM-07', 'type': 'neuron', 'skeleton_ids': [15607180]}, {'id': 15609428, 'name': 'MN-R-Sens-B2-?-00', 'type': 'neuron', 'skeleton_ids': [15609427]}, {'id': 15609886, 'name': 'MN-R-Sens-B2-ACp-17', 'type': 'neuron', 'skeleton_ids': [15609885]}, {'id': 15610388, 'name': 'MN-R-Sens-B2-VM-08', 'type': 'neuron', 'skeleton_ids': [15610387]}, {'id': 15611643, 'name': 'MN-R-Sens-B2-VM-09', 'type': 'neuron', 'skeleton_ids': [15611642]}, {'id': 15611792, 'name': 'MN-R-Sens-B2-VM-02', 'type': 'neuron', 'skeleton_ids': [15611791]}, {'id': 15612242, 'name': 'MN-R-Sens-B2-VM-19', 'type': 'neuron', 'skeleton_ids': [15612241]}, {'id': 15615043, 'name': 'MN-R-Sens-B2-ACp-09', 'type': 'neuron', 'skeleton_ids': [15615042]}, {'id': 15615082, 'name': 'MN-R-Sens-B2-ACp-16', 'type': 'neuron', 'skeleton_ids': [15615081]}, {'id': 15615306, 'name': 'MN-R-Sens-B2-VM-11', 'type': 'neuron', 'skeleton_ids': [15615305]}, {'id': 15615476, 'name': 'MN-R-Sens-B2-VM-10', 'type': 'neuron', 'skeleton_ids': [15615475]}, {'id': 15615590, 'name': 'MN-R-Sens-B2-ACp-05', 'type': 'neuron', 'skeleton_ids': [15615589]}, {'id': 15615777, 'name': 'MN-R-Sens-B2-ACpl-03', 'type': 'neuron', 'skeleton_ids': [15615776]}, {'id': 15615878, 'name': 'MN-R-Sens-B2-VM-22', 'type': 'neuron', 'skeleton_ids': [15615877]}, {'id': 15617460, 'name': 'MN-R-Sens-B2-VM-06', 'type': 'neuron', 'skeleton_ids': [15617459]}, {'id': 15618757, 'name': 'MN-R-Sens-B2-VM-20', 'type': 'neuron', 'skeleton_ids': [15618756]}, {'id': 15618977, 'name': 'MN-R-Sens-B2-VM-12', 'type': 'neuron', 'skeleton_ids': [15618976]}, {'id': 15620433, 'name': 'MN-R-Sens-B2-VM-17', 'type': 'neuron', 'skeleton_ids': [15620432]}, {'id': 15620607, 'name': 'MN-R-Sens-B2-VM-13', 'type': 'neuron', 'skeleton_ids': [15620606]}, {'id': 15629242, 'name': 'MN-R-Sens-B2-VM-15', 'type': 'neuron', 'skeleton_ids': [15629241]}, {'id': 15629356, 'name': 'MN-R-Sens-B2-VM-03', 'type': 'neuron', 'skeleton_ids': [15629355]}, {'id': 15629889, 'name': 'MN-R-Sens-B2-VM-18', 'type': 'neuron', 'skeleton_ids': [15629888]}, {'id': 15637894, 'name': 'MN-R-Sens-B3-VM-05', 'type': 'neuron', 'skeleton_ids': [15637893]}, {'id': 15638521, 'name': 'MN-R-Sens-B3-VM-06', 'type': 'neuron', 'skeleton_ids': [15638520]}, {'id': 15638743, 'name': 'MN-R-Sens-B1-VM-03', 'type': 'neuron', 'skeleton_ids': [15638742]}, {'id': 15638969, 'name': 'MN-R-Sens-B3-AVp-01', 'type': 'neuron', 'skeleton_ids': [15638968]}, {'id': 15639021, 'name': 'MN-R-Sens-B3-ACp-04', 'type': 'neuron', 'skeleton_ids': [15639020]}, {'id': 15639172, 'name': 'MN-R-Sens-B3-ACp-02', 'type': 'neuron', 'skeleton_ids': [15639171]}, {'id': 15639276, 'name': 'MN-R-Sens-B3-VM-08', 'type': 'neuron', 'skeleton_ids': [15639275]}, {'id': 15639549, 'name': 'MN-R-Sens-B3-VM-10', 'type': 'neuron', 'skeleton_ids': [15639548]}, {'id': 15640426, 'name': 'MN-R-Sens-B3-AVp-03', 'type': 'neuron', 'skeleton_ids': [15640425]}, {'id': 15640549, 'name': 'MN-R-Sens-B3-AVp-02', 'type': 'neuron', 'skeleton_ids': [15640548]}, {'id': 15640568, 'name': 'MN-R-Sens-B3-VM-13', 'type': 'neuron', 'skeleton_ids': [15640567]}, {'id': 15640725, 'name': 'MN-R-Sens-B3-VM-11', 'type': 'neuron', 'skeleton_ids': [15640724]}, {'id': 15640894, 'name': 'MN-R-Sens-B3-ACp-03', 'type': 'neuron', 'skeleton_ids': [15640893]}, {'id': 15641927, 'name': 'MN-R-Sens-B3-ACp-01', 'type': 'neuron', 'skeleton_ids': [15641926]}, {'id': 15645826, 'name': 'MN-R-Sens-B3-ACp-06', 'type': 'neuron', 'skeleton_ids': [15645825]}, {'id': 15651439, 'name': 'MN-R-Sens-B2-ACp-22', 'type': 'neuron', 'skeleton_ids': [15651438]}, {'id': 15652530, 'name': 'MN-R-Sens-B2-ACp-01', 'type': 'neuron', 'skeleton_ids': [15652529]}, {'id': 15653289, 'name': 'MN-R-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [15653288]}, {'id': 15653433, 'name': 'MN-R-Sens-B2-ACp-07', 'type': 'neuron', 'skeleton_ids': [15653432]}, {'id': 15653928, 'name': 'MN-R-Sens-B2-VM-01', 'type': 'neuron', 'skeleton_ids': [15653927]}, {'id': 15656436, 'name': 'MN-R-Sens-B2-VM-05', 'type': 'neuron', 'skeleton_ids': [15656435]}, {'id': 15658014, 'name': 'MN-R-Sens-B2-AVp-01', 'type': 'neuron', 'skeleton_ids': [15658013]}, {'id': 15671482, 'name': 'MN-R-Sens-B1-VM-05', 'type': 'neuron', 'skeleton_ids': [15671481]}, {'id': 15671849, 'name': 'MN-R-Sens-B3-VM-09', 'type': 'neuron', 'skeleton_ids': [15671848]}, {'id': 15672154, 'name': 'MN-R-Sens-B3-VM-04', 'type': 'neuron', 'skeleton_ids': [15672153]}, {'id': 15672958, 'name': 'MN-R-Sens-B3-VM-12', 'type': 'neuron', 'skeleton_ids': [15672957]}, {'id': 15673868, 'name': 'MN-L-Sens-B2-ACp-22', 'type': 'neuron', 'skeleton_ids': [15673867]}, {'id': 15673926, 'name': 'MN-L-Sens-B2-ACpl-03', 'type': 'neuron', 'skeleton_ids': [15673925]}, {'id': 15674084, 'name': 'MN-L-Sens-B2-VM-02', 'type': 'neuron', 'skeleton_ids': [15674083]}, {'id': 15674263, 'name': 'MN-L-Sens-B2-ACp-11 (Olfactory Gustatory connector 2)', 'type': 'neuron', 'skeleton_ids': [15674262]}, {'id': 15675902, 'name': 'MN-L-Sens-B2-VM-22', 'type': 'neuron', 'skeleton_ids': [15675901]}, {'id': 15677031, 'name': 'MN-L-Sens-B2-VM-05', 'type': 'neuron', 'skeleton_ids': [15677030]}, {'id': 15677324, 'name': 'MN-L-Sens-B2-ACp-18', 'type': 'neuron', 'skeleton_ids': [15677323]}, {'id': 15678442, 'name': 'MN-L-Sens-B2-VM-09', 'type': 'neuron', 'skeleton_ids': [15678441]}, {'id': 15678834, 'name': 'MN-L-Sens-B2-VM-21', 'type': 'neuron', 'skeleton_ids': [15678833]}, {'id': 15678850, 'name': 'MN-L-Sens-B2-VM-12', 'type': 'neuron', 'skeleton_ids': [15678849]}, {'id': 15679485, 'name': 'MN-L-Sens-B2-VM-08', 'type': 'neuron', 'skeleton_ids': [15679484]}, {'id': 15679578, 'name': 'MN-L-Sens-B2-ACp-10', 'type': 'neuron', 'skeleton_ids': [15679577]}, {'id': 15680094, 'name': 'MN-L-Sens-B2-ACp-19', 'type': 'neuron', 'skeleton_ids': [15680093]}, {'id': 15680711, 'name': 'MN-L-Sens-B2-VM-03', 'type': 'neuron', 'skeleton_ids': [15680710]}, {'id': 15681247, 'name': '<NAME>', 'type': 'neuron', 'skeleton_ids': [15681246]}, {'id': 15681741, 'name': 'MN-L-Sens-B2-ACp-21', 'type': 'neuron', 'skeleton_ids': [15681740]}, {'id': 15686445, 'name': 'neuron 15686445', 'type': 'neuron', 'skeleton_ids': [15686444]}, {'id': 15686513, 'name': 'MN-L-Sens-B2-ACp-14', 'type': 'neuron', 'skeleton_ids': [15686512]}, {'id': 15688359, 'name': 'MN-L-Sens-B2-ACp-20', 'type': 'neuron', 'skeleton_ids': [15688358]}, {'id': 15689374, 'name': 'MN-L-Sens-B2-ACp-12', 'type': 'neuron', 'skeleton_ids': [15689373]}, {'id': 15690094, 'name': 'MN-L-Sens-B2-ACp-09', 'type': 'neuron', 'skeleton_ids': [15690093]}, {'id': 15690393, 'name': 'MN-L-Sens-B2-VM-11', 'type': 'neuron', 'skeleton_ids': [15690392]}, {'id': 15690712, 'name': 'MN-L-Sens-B2-VM-14', 'type': 'neuron', 'skeleton_ids': [15690711]}, {'id': 15690956, 'name': 'MN-L-Sens-B2-VM-06', 'type': 'neuron', 'skeleton_ids': [15690955]}, {'id': 15698624, 'name': 'MN-L-Sens-B1-VM-02', 'type': 'neuron', 'skeleton_ids': [15698623]}, {'id': 15699716, 'name': 'MN-L-Sens-B2-VM-23', 'type': 'neuron', 'skeleton_ids': [15699715]}, {'id': 15700148, 'name': 'MN-L-Sens-B1-VM-04', 'type': 'neuron', 'skeleton_ids': [15700147]}, {'id': 15711223, 'name': 'MN-L-Sens-B2-VM-07', 'type': 'neuron', 'skeleton_ids': [15711222]}, {'id': 15711290, 'name': 'MN-L-Sens-B2-VM-16', 'type': 'neuron', 'skeleton_ids': [15711289]}, {'id': 15712025, 'name': 'MN-L-Sens-B2-VM-13', 'type': 'neuron', 'skeleton_ids': [15712024]}, {'id': 15713207, 'name': 'MN-L-Sens-B2-VM-18', 'type': 'neuron', 'skeleton_ids': [15713206]}, {'id': 15713375, 'name': 'MN-L-Sens-B2-VM-17', 'type': 'neuron', 'skeleton_ids': [15713374]}, {'id': 15714174, 'name': 'MN-L-Sens-B1-VM-05', 'type': 'neuron', 'skeleton_ids': [15714173]}, {'id': 15714331, 'name': 'MN-L-Sens-B1-VM-03', 'type': 'neuron', 'skeleton_ids': [15714330]}, {'id': 15714640, 'name': 'MN-L-Sens-B2-ACpl-01 (Awesome MN sensory 2)', 'type': 'neuron', 'skeleton_ids': [15714639]}, {'id': 15714846, 'name': 'MN-L-Sens-B2-VM-10', 'type': 'neuron', 'skeleton_ids': [15714845]}, {'id': 15715152, 'name': 'MN-L-Sens-B2-VM-01', 'type': 'neuron', 'skeleton_ids': [15715151]}, {'id': 15715644, 'name': 'MN-L-Sens-B2-ACp-04', 'type': 'neuron', 'skeleton_ids': [15715643]}, {'id': 15715797, 'name': 'MN-L-Sens-B2-VM-04', 'type': 'neuron', 'skeleton_ids': [15715796]}, {'id': 15716095, 'name': 'MN-L-Sens-B2-ACp-05', 'type': 'neuron', 'skeleton_ids': [15716094]}, {'id': 15716128, 'name': 'MN-L-Sens-B2-ACp-13', 'type': 'neuron', 'skeleton_ids': [15716127]}, {'id': 15716310, 'name': 'MN-L-Sens-B2-ACp-03', 'type': 'neuron', 'skeleton_ids': [15716309]}, {'id': 15716702, 'name': 'MN-L-Sens-B2-ACp-02', 'type': 'neuron', 'skeleton_ids': [15716701]}, {'id': 15716935, 'name': 'MN-L-Sens-B2-ACpl-02 (Awesome MN sensory 1)', 'type': 'neuron', 'skeleton_ids': [15716934]}, {'id': 15717230, 'name': 'MN-L-Sens-B2-VM-15', 'type': 'neuron', 'skeleton_ids': [15717229]}, {'id': 15717427, 'name': 'MN-L-Sens-B2-AVp-02', 'type': 'neuron', 'skeleton_ids': [15717426]}, {'id': 15717737, 'name': 'MN-L-Sens-B3-ACp-05', 'type': 'neuron', 'skeleton_ids': [15717736]}, {'id': 15717936, 'name': 'MN-L-Sens-B3-ACpl-07', 'type': 'neuron', 'skeleton_ids': [15717935]}, {'id': 15718276, 'name': 'MN-L-Sens-B3-ACp-06 (J Neuron)', 'type': 'neuron', 'skeleton_ids': [15718275]}, {'id': 15723480, 'name': 'MN-L-Sens-B3-AVp-02', 'type': 'neuron', 'skeleton_ids': [15723479]}, {'id': 15723728, 'name': 'MN-L-Sens-B3-ACp-01', 'type': 'neuron', 'skeleton_ids': [15723727]}, {'id': 15724176, 'name': 'MN-L-Sens-B3-VM-11', 'type': 'neuron', 'skeleton_ids': [15724175]}, {'id': 15724386, 'name': 'MN-L-Sens-B3-AVp-01', 'type': 'neuron', 'skeleton_ids': [15724385]}, {'id': 15724521, 'name': 'MN-L-Sens-B3-VM-06', 'type': 'neuron', 'skeleton_ids': [15724520]}, {'id': 15724637, 'name': 'MN-L-Sens-B3-VM-08', 'type': 'neuron', 'skeleton_ids': [15724636]}, {'id': 15725128, 'name': 'MN-L-Sens-B3-VM-10', 'type': 'neuron', 'skeleton_ids': [15725127]}, {'id': 15725546, 'name': 'MN-L-Sens-B3-VM-09', 'type': 'neuron', 'skeleton_ids': [15725545]}, {'id': 15725674, 'name': 'MN-L-Sens-B3-VM-07', 'type': 'neuron', 'skeleton_ids': [15725673]}, {'id': 15727305, 'name': 'MN-L-Sens-B3-VM-04', 'type': 'neuron', 'skeleton_ids': [15727304]}, {'id': 15728018, 'name': 'MN-L-Sens-B3-VM-13', 'type': 'neuron', 'skeleton_ids': [15728017]}, {'id': 15730646, 'name': 'MN-L-Sens-B3-VM-05', 'type': 'neuron', 'skeleton_ids': [15730645]}, {'id': 15730681, 'name': 'MN-L-Sens-B2-VM-20', 'type': 'neuron', 'skeleton_ids': [15730680]}, {'id': 15736148, 'name': 'MN-R-Sens-B2-ACp-03', 'type': 'neuron', 'skeleton_ids': [15736147]}, {'id': 15743906, 'name': 'MN-R-Sens-B1-VM-04', 'type': 'neuron', 'skeleton_ids': [15743905]}, {'id': 15746422, 'name': 'MN-R-Sens-B2-ACpl-02 (Awesome MN sensory 1)', 'type': 'neuron', 'skeleton_ids': [15746421]}, {'id': 15748727, 'name': 'AN-R-Sens-B1-AVa-43', 'type': 'neuron', 'skeleton_ids': [15748726]}, {'id': 15762569, 'name': 'MN-R-Sens-B2-VM-14', 'type': 'neuron', 'skeleton_ids': [15762568]}, {'id': 15767000, 'name': 'AN-L-Sens-B1-AVa-38', 'type': 'neuron', 'skeleton_ids': [15766999]}, {'id': 15769420, 'name': 'AN-L-Sens-B1-ACa-08', 'type': 'neuron', 'skeleton_ids': [15769419]}, {'id': 15769491, 'name': 'AN-L-motor-06', 'type': 'neuron', 'skeleton_ids': [15769490]}, {'id': 15769597, 'name': 'AN-L-motor-05', 'type': 'neuron', 'skeleton_ids': [15769596]}, {'id': 15769639, 'name': 'SE0 L3', 'type': 'neuron', 'skeleton_ids': [15769638]}, {'id': 15769681, 'name': 'AN-L-Sens-B1-AVa-19', 'type': 'neuron', 'skeleton_ids': [15769680]}, {'id': 15770033, 'name': 'AN-L-Sens-B1-ACa-03', 'type': 'neuron', 'skeleton_ids': [15770032]}, {'id': 15770522, 'name': 'AN-L-motor-15', 'type': 'neuron', 'skeleton_ids': [15770521]}, {'id': 15770868, 'name': 'AN-L-motor-16', 'type': 'neuron', 'skeleton_ids': [15770867]}, {'id': 15770905, 'name': 'AN-L-Sens-B1-AVa-24', 'type': 'neuron', 'skeleton_ids': [15770904]}, {'id': 15770946, 'name': 'AN-L-Sens-B1-AVa-25', 'type': 'neuron', 'skeleton_ids': [15770945]}, {'id': 15914722, 'name': 'MN-R-Sens-B3-VM-14', 'type': 'neuron', 'skeleton_ids': [15914721]}, {'id': 15938338, 'name': 'MN-L-Sens-B2-ACp-08', 'type': 'neuron', 'skeleton_ids': [15938337]}, {'id': 15982307, 'name': 'AN-L-Sens-B1-AVa-28', 'type': 'neuron', 'skeleton_ids': [15982306]}, {'id': 15982952, 'name': 'AN-L-motor-10', 'type': 'neuron', 'skeleton_ids': [15982951]}, {'id': 15983119, 'name': 'SE<NAME>', 'type': 'neuron', 'skeleton_ids': [15983118]}, {'id': 15983157, 'name': 'AN-L-Sens-B1-AVa-39', 'type': 'neuron', 'skeleton_ids': [15983156]}, {'id': 15983227, 'name': 'AN-L-Sens-B1-AVa-34', 'type': 'neuron', 'skeleton_ids': [15983225]}, {'id': 15984232, 'name': 'AN-L-Sens-B1-AVa-35', 'type': 'neuron', 'skeleton_ids': [15984231]}, {'id': 15984798, 'name': 'AN-L-Sens-B1-AVa-23', 'type': 'neuron', 'skeleton_ids': [15984797]}, {'id': 15994927, 'name': 'AN-L-Sens-B1-AVa-36', 'type': 'neuron', 'skeleton_ids': [15994926]}, {'id': 15995013, 'name': 'SE<NAME>', 'type': 'neuron', 'skeleton_ids': [15995012]}, {'id': 15995310, 'name': '<NAME>', 'type': 'neuron', 'skeleton_ids': [15995309]}, {'id': 15995328, 'name': 'AN-L-Sens-B1-AVa-37', 'type': 'neuron', 'skeleton_ids': [15995327]}, {'id': 15995361, 'name': 'AN-L-motor-09', 'type': 'neuron', 'skeleton_ids': [15995360]}, {'id': 15995369, 'name': 'AN-L-motor-17', 'type': 'neuron', 'skeleton_ids': [15995368]}, {'id': 15995395, 'name': 'AN-L-motor-14', 'type': 'neuron', 'skeleton_ids': [15995394]}, {'id': 15995405, 'name': 'AN-L-Sens-B2-ACp-22', 'type': 'neuron', 'skeleton_ids': [15995404]}, {'id': 15995862, 'name': 'AN-L-Sens-B2-VM-08', 'type': 'neuron', 'skeleton_ids': [15995861]}, {'id': 15995930, 'name': 'AN-L-Sens-B2-ACp-20', 'type': 'neuron', 'skeleton_ids': [15995929]}, {'id': 15996161, 'name': 'AN-L-Sens-B2-AVp-03', 'type': 'neuron', 'skeleton_ids': [15996160]}, {'id': 15996272, 'name': 'AN-L-Sens-B2-VM-06', 'type': 'neuron', 'skeleton_ids': [15996271]}, {'id': 15996519, 'name': 'AN-L-Sens-B2-ACp-11', 'type': 'neuron', 'skeleton_ids': [15996518]}, {'id': 15997069, 'name': 'AN-L-motor-18', 'type': 'neuron', 'skeleton_ids': [15997068]}, {'id': 15997646, 'name': 'AN-L-Sens-B2-ACp-14', 'type': 'neuron', 'skeleton_ids': [15997645]}, {'id': 15997667, 'name': 'AN-L-Sens-B2-ACp-01', 'type': 'neuron', 'skeleton_ids': [15997666]}, {'id': 15997693, 'name': 'AN-L-Sens-B2-ACp-18', 'type': 'neuron', 'skeleton_ids': [15997692]}, {'id': 15997808, 'name': 'AN-L-Sens-B2-ACp-16', 'type': 'neuron', 'skeleton_ids': [15997807]}, {'id': 15997930, 'name': 'AN-L-Sens-B2-ACp-23', 'type': 'neuron', 'skeleton_ids': [15997929]}, {'id': 15997984, 'name': 'AN-L-Sens-B2-ACp-06', 'type': 'neuron', 'skeleton_ids': [15997983]}, {'id': 15998101, 'name': 'AN-L-Sens-B2-ACp-13', 'type': 'neuron', 'skeleton_ids': [15998100]}, {'id': 15998219, 'name': 'AN-L-Sens-B2-ACp-08', 'type': 'neuron', 'skeleton_ids': [15998218]}, {'id': 15998262, 'name': 'AN-L-Sens-B2-ACp-15', 'type': 'neuron', 'skeleton_ids': [15998261]}, {'id': 15998308, 'name': 'AN-L-Sens-B2-ACp-10', 'type': 'neuron', 'skeleton_ids': [15998307]}, {'id': 15998320, 'name': 'AN-L-Sens-B2-VM-05', 'type': 'neuron', 'skeleton_ids': [15998319]}, {'id': 15998470, 'name': 'AN-L-Sens-B2-ACp-05', 'type': 'neuron', 'skeleton_ids': [15998469]}, {'id': 15998754, 'name': 'AN-L-Sens-B2-AVa-25', 'type': 'neuron', 'skeleton_ids': [15998753]}, {'id': 15998769, 'name': 'AN-L-Sens-B2-ACp-09', 'type': 'neuron', 'skeleton_ids': [15998768]}, {'id': 15998783, 'name': 'AN-L-Sens-B2-ACp-04', 'type': 'neuron', 'skeleton_ids': [15998782]}, {'id': 15998818, 'name': 'AN-L-Sens-B2-ACp-12', 'type': 'neuron', 'skeleton_ids': [15998817]}, {'id': 15998900, 'name': 'AN-L-Sens-B2-AVp-02', 'type': 'neuron', 'skeleton_ids': [15998899]}, {'id': 15998914, 'name': 'AN-L-Sens-B2-VM-04', 'type': 'neuron', 'skeleton_ids': [15998913]}, {'id': 16018124, 'name': 'T2r asc contra 01', 'type': 'neuron', 'skeleton_ids': [16018123]}, {'id': 16018441, 'name': 'SEZ to brain', 'type': 'neuron', 'skeleton_ids': [16018440]}, {'id': 16115502, 'name': 'AN-R-Sens-B3-VM-11', 'type': 'neuron', 'skeleton_ids': [16115501]}, {'id': 16123984, 'name': 'PaN-R-Sens-B1-VM-03', 'type': 'neuron', 'skeleton_ids': [16123983]}, {'id': 16124049, 'name': 'PaN-R-Sens-B1-VM-04', 'type': 'neuron', 'skeleton_ids': [16124048]}, {'id': 16124083, 'name': 'PaN-R-Sens-B1-VM-05', 'type': 'neuron', 'skeleton_ids': [16124082]}, {'id': 16223538, 'name': 'MBE18 left', 'type': 'neuron', 'skeleton_ids': [16223537]}, {'id': 16259596, 'name': '67b PN left', 'type': 'neuron', 'skeleton_ids': [16259595]}, {'id': 16288873, 'name': 'MN-L-Sens-B3-ACp-02', 'type': 'neuron', 'skeleton_ids': [16288872]}, {'id': 16317331, 'name': 'SEZ-IN-02-right', 'type': 'neuron', 'skeleton_ids': [16317330]}, {'id': 16341400, 'name': 'MN-R-Sens-B2-ACp-04', 'type': 'neuron', 'skeleton_ids': [16341399]}, {'id': 16341508, 'name': 'AN-R-Sens-B2-ACp-05', 'type': 'neuron', 'skeleton_ids': [16341507]}, {'id': 16341967, 'name': 'AN-R-Sens-B2-ACp-07', 'type': 'neuron', 'skeleton_ids': [16341966]}, {'id': 16362244, 'name': 'AN-R-Sens-B2-VM-07', 'type': 'neuron', 'skeleton_ids': [16362243]}, {'id': 16442345, 'name': 'AN-R-Sens-B1-AVa-20', 'type': 'neuron', 'skeleton_ids': [16442344]}, {'id': 16468936, 'name': 'gate1_left', 'type': 'neuron', 'skeleton_ids': [16468935]}, {'id': 16485080, 'name': 'LH bilateral axon 2 MB_right; BLVa12 b 3', 'type': 'neuron', 'skeleton_ids': [16485079]}, {'id': 16500056, 'name': 'SEZ brain projection neuron - not finished (gap)', 'type': 'neuron', 'skeleton_ids': [16500055]}, {'id': 16575804, 'name': 'Grumpy_right', 'type': 'neuron', 'skeleton_ids': [16575803]}, {'id': 16621865, 'name': 'ginkgo 2 right', 'type': 'neuron', 'skeleton_ids': [16621864]}, {'id': 16744168, 'name': 'SEZ-IN-11-right', 'type': 'neuron', 'skeleton_ids': [16744167]}, {'id': 16751956, 'name': 'Dumbo_right', 'type': 'neuron', 'skeleton_ids': [16751955]}, {'id': 16774288, 'name': 'T2r asc medial soma 02', 'type': 'neuron', 'skeleton_ids': [16774287]}, {'id': 16795525, 'name': 'mPN iACT bilateral UPPER right', 'type': 'neuron', 'skeleton_ids': [16795524]}, {'id': 16795839, 'name': 'mPN iACT bilateral UPPER left', 'type': 'neuron', 'skeleton_ids': [16795838]}, {'id': 16846806, 'name': 'MBE5b left', 'type': 'neuron', 'skeleton_ids': [16846805]}, {'id': 16868924, 'name': 'MBE24b left', 'type': 'neuron', 'skeleton_ids': [16868923]}, {'id': 16988780, 'name': 'T2l asc medial soma 02', 'type': 'neuron', 'skeleton_ids': [16988779]}, {'id': 17013074, 'name': 'MBE24b right', 'type': 'neuron', 'skeleton_ids': [17013073]}, {'id': 17096643, 'name': 'Dumbo_left', 'type': 'neuron', 'skeleton_ids': [17096642]}, {'id': 17147699, 'name': 'MBE11a left', 'type': 'neuron', 'skeleton_ids': [17147698]}, {'id': 17213785, 'name': 'Neuron 17213785', 'type': 'neuron', 'skeleton_ids': [17213784]}, {'id': 17219368, 'name': 'SEZ-IN-06-right', 'type': 'neuron', 'skeleton_ids': [17219367]}, {'id': 17220055, 'name': 'SEZ-IN-A07-left', 'type': 'neuron', 'skeleton_ids': [17220054]}, {'id': 17225494, 'name': 'SEZ IN - unfinished', 'type': 'neuron', 'skeleton_ids': [17225493]}, {'id': 17248924, 'name': 'AN-R-motor-05', 'type': 'neuron', 'skeleton_ids': [17248923]}, {'id': 17254279, 'name': 'Pre-PMN-L-08', 'type': 'neuron', 'skeleton_ids': [17254278]}, {'id': 17254291, 'name': 'Pre-PMN-R-05', 'type': 'neuron', 'skeleton_ids': [17254290]}, {'id': 17256675, 'name': 'neuron 17256675', 'type': 'neuron', 'skeleton_ids': [17256674]}, {'id': 17260594, 'name': 'neuron 17260594', 'type': 'neuron', 'skeleton_ids': [17260593]}, {'id': 17267283, 'name': 'neuron 17267283', 'type': 'neuron', 'skeleton_ids': [17267282]}, {'id': 17267289, 'name': 'SEZ-IN-A14-right', 'type': 'neuron', 'skeleton_ids': [17267288]}, {'id': 17267349, 'name': 'neuron 17267349', 'type': 'neuron', 'skeleton_ids': [17267348]}, {'id': 17271472, 'name': 'neuron 17271472', 'type': 'neuron', 'skeleton_ids': [17271471]}, {'id': 17369377, 'name': 'AN-L-motor-07', 'type': 'neuron', 'skeleton_ids': [17369376]}, {'id': 17380937, 'name': 'AN-L-Sens-B1-AVa-33', 'type': 'neuron', 'skeleton_ids': [17380936]}, {'id': 17385046, 'name': 'neuron 17385046', 'type': 'neuron', 'skeleton_ids': [17385045]}, {'id': 17399868, 'name': 'neuron 17399868', 'type': 'neuron', 'skeleton_ids': [17399867]}, {'id': 17399887, 'name': 'neuron 17399887', 'type': 'neuron', 'skeleton_ids': [17399886]}, {'id': 17400232, 'name': 'neuron 17400232', 'type': 'neuron', 'skeleton_ids': [17400231]}, {'id': 17405604, 'name': 'AN-L-motor-13', 'type': 'neuron', 'skeleton_ids': [17405603]}, {'id': 17415322, 'name': 'AN-L-motor-11', 'type': 'neuron', 'skeleton_ids': [17415321]}, {'id': 17415440, 'name': 'presynaptic-D-2-MAP right', 'type': 'neuron', 'skeleton_ids': [17415439]}, {'id': 17421903, 'name': 'neuron 17421903', 'type': 'neuron', 'skeleton_ids': [17421902]}, {'id': 17434826, 'name': 'LH bilateral axon 2 MB_left; BLVa12 b 3', 'type': 'neuron', 'skeleton_ids': [17434825]}, {'id': 17500792, 'name': 'AN-L-motor-12', 'type': 'neuron', 'skeleton_ids': [17500791]}, {'id': 17511973, 'name': 'Neuron 17511973', 'type': 'neuron', 'skeleton_ids': [17511972]}, {'id': 17587795, 'name': 'neuron 17587795', 'type': 'neuron', 'skeleton_ids': [17587794]}, {'id': 17608755, 'name': 'AN-L-Sens-B2-VM-07', 'type': 'neuron', 'skeleton_ids': [17608754]}, {'id': 17641914, 'name': 'AF4: TOH left', 'type': 'neuron', 'skeleton_ids': [17641913]}, {'id': 17651481, 'name': 'SEZ-IN-01-right', 'type': 'neuron', 'skeleton_ids': [17651480]}, {'id': 17698682, 'name': 'BAmas12 midline neuron 1? right', 'type': 'neuron', 'skeleton_ids': [17698681]}, {'id': 17706916, 'name': 'neuron 17706916', 'type': 'neuron', 'skeleton_ids': [17706915]}, {'id': 17716980, 'name': 'SEZ-IN-09-right', 'type': 'neuron', 'skeleton_ids': [17716979]}, {'id': 17717855, 'name': 'KS LN 1 left', 'type': 'neuron', 'skeleton_ids': [17717854]}, {'id': 17718178, 'name': 'AX ; SEZ-IN-A03-right', 'type': 'neuron', 'skeleton_ids': [17718177]}, {'id': 17721401, 'name': 'neuron 17721401', 'type': 'neuron', 'skeleton_ids': [17721400]}, {'id': 17816987, 'name': 'handle maxillar SEZ', 'type': 'neuron', 'skeleton_ids': [17816986]}, {'id': 17884300, 'name': 'Gustatory PN right', 'type': 'neuron', 'skeleton_ids': [17884299]}, {'id': 17951154, 'name': 'AN-R-Sens-B1-AVa-32', 'type': 'neuron', 'skeleton_ids': [17951153]}, {'id': 18028398, 'name': 'MBE5b right', 'type': 'neuron', 'skeleton_ids': [18028397]}, {'id': 18225325, 'name': 'Neuron 18225325', 'type': 'neuron', 'skeleton_ids': [18225324]}, {'id': 18232000, 'name': 'AN-R-Sens-B1-AVa-45', 'type': 'neuron', 'skeleton_ids': [18231999]}, {'id': 18239022, 'name': 'Neuron 18239022', 'type': 'neuron', 'skeleton_ids': [18239021]}, {'id': 18329063, 'name': 'SEZ-IN-A04-left', 'type': 'neuron', 'skeleton_ids': [18329062]}, {'id': 18347337, 'name': 'SEZ-IN-05-right', 'type': 'neuron', 'skeleton_ids': [18347336]}, {'id': 18354201, 'name': 'SEZ-IN-A05-right - paired with #6885133', 'type': 'neuron', 'skeleton_ids': [18354200]}, {'id': 18356794, 'name': 'SEZ-IN-04-right', 'type': 'neuron', 'skeleton_ids': [18356793]}, {'id': 18371413, 'name': 'SEZ-IN-03-left', 'type': 'neuron', 'skeleton_ids': [18371412]}, {'id': 18403156, 'name': 'SEZ-IN-18-left', 'type': 'neuron', 'skeleton_ids': [18403155]}, {'id': 18440676, 'name': 'BAmas12 midline 3 left', 'type': 'neuron', 'skeleton_ids': [18440675]}, {'id': 18620886, 'name': 'AN-R-Sens-B2-ACp-23', 'type': 'neuron', 'skeleton_ids': [18620885]}, {'id': 18625447, 'name': 'MN-R-Sens-B2-ACp-10', 'type': 'neuron', 'skeleton_ids': [18625446]}, {'id': 18630154, 'name': 'AN-R-Sens-B3-ACal-02 (temp sensing right neuron together 2)', 'type': 'neuron', 'skeleton_ids': [18630153]}, {'id': 18679890, 'name': 'MN-R-Sens-B2-VM-16', 'type': 'neuron', 'skeleton_ids': [18679889]}, {'id': 18785125, 'name': 'MN-R-Sens-B3-VM-07', 'type': 'neuron', 'skeleton_ids': [18785124]}, {'id': 18833415, 'name': 'MN-R-Sens-B2-VM-23', 'type': 'neuron', 'skeleton_ids': [18833414]}, {'id': 18934020, 'name': 'MN-L-Sens-B2-ACp-17', 'type': 'neuron', 'skeleton_ids': [18934019]}, {'id': 19042618, 'name': 'SEZ-IN-A14-left', 'type': 'neuron', 'skeleton_ids': [19042617]}, {'id': 19099537, 'name': 'neuron 19099537', 'type': 'neuron', 'skeleton_ids': [19099536]}, {'id': 19120854, 'name': 'pCC a1r', 'type': 'neuron', 'skeleton_ids': [19120853]}, {'id': 19180426, 'name': 'MN-L-Sens-B2-VM-24', 'type': 'neuron', 'skeleton_ids': [19180425]}, {'id': 19341123, 'name': 'Neuron 19341123', 'type': 'neuron', 'skeleton_ids': [19341122]}, {'id': 19431431, 'name': 'SOG IN; PN into SOG very minor AL connectivity left', 'type': 'neuron', 'skeleton_ids': [19431430]}, {'id': 19470185, 'name': 'PC-IN-left', 'type': 'neuron', 'skeleton_ids': [19470184]}, {'id': 19731865, 'name': 'Neuron 19731865', 'type': 'neuron', 'skeleton_ids': [19731864]}, {'id': 19816279, 'name': 'NPF-like 1 right', 'type': 'neuron', 'skeleton_ids': [19816278]}, {'id': 19912141, 'name': 'T1r asc contra 02', 'type': 'neuron', 'skeleton_ids': [19912140]}, {'id': 20152213, 'name': 'Neuron 20152213', 'type': 'neuron', 'skeleton_ids': [20152212]}]}, {'id': 20284758, 'name': '"Carreira-Rosario, <NAME>, Clark et al. 2018"', 'type': 'annotation', 'neurons': [{'id': 1974686, 'name': 'A27l_a3l', 'type': 'neuron', 'skeleton_ids': [2111886]}, {'id': 2072203, 'name': 'A27l_a3r', 'type': 'neuron', 'skeleton_ids': [2072202]}, {'id': 4038809, 'name': 'A27k_a2l', 'type': 'neuron', 'skeleton_ids': [4038808]}, {'id': 4113613, 'name': 'A27k_a2r', 'type': 'neuron', 'skeleton_ids': [4113612]}, {'id': 4213418, 'name': 'A27k_a1l', 'type': 'neuron', 'skeleton_ids': [4213417]}, {'id': 4542434, 'name': 'A18b_a1l', 'type': 'neuron', 'skeleton_ids': [4542433]}, {'id': 4719760, 'name': 'A27l_a2l', 'type': 'neuron', 'skeleton_ids': [4719759]}, {'id': 4727547, 'name': 'A18b_a1r', 'type': 'neuron', 'skeleton_ids': [4727546]}, {'id': 4835695, 'name': 'A27h_a4l', 'type': 'neuron', 'skeleton_ids': [4835694]}, {'id': 5308486, 'name': 'SEZ-DN1_left', 'type': 'neuron', 'skeleton_ids': [5308485]}, {'id': 5355176, 'name': 'A27l_a2r', 'type': 'neuron', 'skeleton_ids': [5355175]}, {'id': 10004244, 'name': 'A27k_a1r', 'type': 'neuron', 'skeleton_ids': [10004243]}, {'id': 10216847, 'name': 'A27h?_t3l', 'type': 'neuron', 'skeleton_ids': [10216846]}, {'id': 10219171, 'name': 'A27l_a1l', 'type': 'neuron', 'skeleton_ids': [10219170]}, {'id': 10223573, 'name': 'A27h_a1l', 'type': 'neuron', 'skeleton_ids': [10223572]}, {'id': 10223755, 'name': 'DN_mx (Descending neuron_mx)_r', 'type': 'neuron', 'skeleton_ids': [10223754]}, {'id': 10499712, 'name': 'A27l_a1r', 'type': 'neuron', 'skeleton_ids': [10499711]}, {'id': 10589762, 'name': 'A18b_a2l', 'type': 'neuron', 'skeleton_ids': [10589761]}, {'id': 10728329, 'name': 'MDNb_left', 'type': 'neuron', 'skeleton_ids': [10728328]}, {'id': 10728334, 'name': 'MDNa_left', 'type': 'neuron', 'skeleton_ids': [10728333]}, {'id': 11541265, 'name': 'ThDN right', 'type': 'neuron', 'skeleton_ids': [11541264]}, {'id': 12247556, 'name': 'ThDN left', 'type': 'neuron', 'skeleton_ids': [12247555]}, {'id': 13557065, 'name': 'MDNb_right', 'type': 'neuron', 'skeleton_ids': [13557064]}, {'id': 15695113, 'name': 'A27h_a6l', 'type': 'neuron', 'skeleton_ids': [15695112]}, {'id': 15696655, 'name': 'A27h_a6r', 'type': 'neuron', 'skeleton_ids': [15696654]}, {'id': 15700464, 'name': 'A27h_a7r', 'type': 'neuron', 'skeleton_ids': [15700463]}, {'id': 15756647, 'name': 'A27h_a7l', 'type': 'neuron', 'skeleton_ids': [15756646]}, {'id': 16002927, 'name': 'A18b_a2r', 'type': 'neuron', 'skeleton_ids': [16002926]}, {'id': 17350422, 'name': 'SEZ-DN1_right', 'type': 'neuron', 'skeleton_ids': [17350421]}, {'id': 18464582, 'name': 'MDNa_right', 'type': 'neuron', 'skeleton_ids': [18464581]}, {'id': 18628920, 'name': 'A27h_a5r', 'type': 'neuron', 'skeleton_ids': [18628919]}]}]
```
---
@jefferis commented on [Thu Jun 06 2019](https://github.com/VirtualFlyBrain/Management/issues/190#issuecomment-499528701)
Don't forget that @schlegelp has https://github.com/schlegelp/pyMaid/
Answers:
username_1: duplicate of #19
Status: Issue closed
|
aurelia-contrib/aurelia-typed-observable-plugin | 311028039 | Title: installation instructions missing
Question:
username_0: Please add info on npm blah blah
cli blah blah
webpack blah blah
jspm..
(-:
and keep rocking!!!
Answers:
username_1: `observable` and `bindable` are normal function, so we use it as if we import it from aurelia. Because of that, it was kind of hard to describe how to install it like we would in a normal aurelia plugin...
username_1: Thanks, it has been added 👍
Status: Issue closed
|
grpc/grpc | 105490174 | Title: Leftover debug messages make C# output messy.
Question:
username_0: Basically all C# tests now print some debugging garbage. We should silence those.
D9/8/2015 4:36:58 PM 108345464 c:\users\username_0\github\grpc\src\core\iomgr\tcp_client_windows.c:80: on_alarm: 056F6E60
D9/8/2015 4:36:58 PM 108345464 c:\users\username_0\github\grpc\src\core\iomgr\tcp_client_windows.c:99: on_connect: 056F6E60<issue_closed>
Status: Issue closed |
dotnet/aspnetcore | 1000072769 | Title: Behavior is different to what is documented - IFormCollection.Item[String] Property
Question:
username_0: <!--
-->
### Describe the bug
When porting old .NetF code we had to make changes because Request no longer has an indexer but instead exposes sub-properties that themselves have indexers.
During a test we attempt this:
```cs
public void OnGet()
{
var a = NepHttpContext.Current;
var x = a.Request.Form["LOGON_USER"];
}
```
The code throws an exception yet it should either return the value or return `StringValues.Empty` or throw if the supplied key is null - according to the [documentation](https://docs.microsoft.com/en-us/dotnet/api/microsoft.aspnetcore.http.iformcollection.item?view=aspnetcore-5.0).
Its seems that either the documentation is wrong or this is a bug.
### To Reproduce
Just create a new empty .Net5 web app and then add the above code to the template code that's present in the project, as shown here:
```cs
namespace CoreWebApp.Pages
{
public class IndexModel : PageModel
{
private readonly ILogger<IndexModel> _logger;
public IndexModel(ILogger<IndexModel> logger)
{
_logger = logger;
}
public void OnGet()
{
var a = NepHttpContext.Current;
var x = a.Request.Form["LOGON_USER"];
}
}
}
```
The `NepHttpContext `class is our own helper class, it just gets the current request.
### Exceptions (if any)

### Further technical details
- ASP.NET Core version
- Include the output of `dotnet --info`
- The IDE (VS / VS Code/ VS4Mac) you're running on, and its version
App targets .Net5, using VS 2019, here's the donet info:

Answers:
username_1: You can read form fields in a get request. It seems you're trying to replace the indexer which merged query and form I believe. That doesn't exist in ASP.NET Core. If you want to read the query string then use .Query and if you need to read the form you can use .Form but only when a form is actually being posted.
username_0: Ahh, I see, OK so the entire scenario is invalid here, OK that makes sense this test is a bit contrived.
May I ask then, in old .NetF code we have this:
```cs
string tmpAuthIdentity = System.Web.HttpContext.Current.Request["LOGON_USER"];
```
I want to simply replace this with whatever is equivalent under ASP.NET Core, may articles talk about this being a server variable in older .NetF apps.
The above .NetF code works fine, its part of a larger library we've had for years, I'm just trying to tweak the few areas of the cod that are not compatible with .NetF.
Thanks for your prompt reply by the way!
username_0: By the way, if the access to Form["LOGON_USER"] is illegal because there is n form being posted, could the API not throw a better exception? with a message like "No form is present in this request" or something?
username_1: Seems like you're looking to read the server variables. This will only work on IIS. There's likely a modern alternative to this that doesn't require looking at this value.
You should be able to do this:
```csharp
string tmpAuthIdentity = NepHttpContext.Curent.GetServerVariable("LOGON_USER");
```
The modern alternative though is likely:
```csharp
string tmpAuthIdentity = NepHttpContext.Current.User.Identity.Name;
```
username_2: Triage: `IFormCollection.Item` API should properly document the exception case(s), and mention the sync-over-async internal implementation detail. And make the error nicer like "No form found" etc. |
sgoldenlab/simba | 635560157 | Title: Simba Create Project
Question:
username_0: Hi
I installed the Simba (pip install simba-uw-no-tf) in my local. then i launched "simba". I am trying to create a project, after filling all fields the following error came out:
Traceback (most recent call last):
File "C:\users\...\appdata\local\programspython\python37\lib\thinter\__init__.py", line 1705, in __call__
return self.func(*args)
File "C:\users\...\appdata\local\programs\python\python37\lib\site-packages\simba\simba.py", line 3056, in make_projectini
with open(no animalsPath, "r", encoding='utf8') as f:
FileNotFoundError: [errno 2] No such file or directory: 'C:\\users\\...\\pose_configurations\\no_animals\\no_animals.csv
In fact the pose configurations folder is empty.
What should I do?
Answers:
username_1: @username_0, thank you for reporting this bug. I fixed the bug and you can now download it using `pip install simba-uw-no-tf==1.1.7.
Please do not hesitate to let me know if you have anymore issue.
Thank you!
Status: Issue closed
|
nicdex/node-eventstore-client | 254335491 | Title: UUID endian issue
Question:
username_0: The eventid seems to get endian-swapped (or rather, it doesn't) on the way in & out of the eventstore.
`
%> node test.js
`
```
Appending...
Connected to eventstore at 172.18.0.5:1113
Stored event: 7dffd71b-c177-49ae-89ae-46a5998533d0
Look for it at: http://localhost:2113/web/index.html#/streams/testStream
```
`
%> curl -i -H "Accept: application/json" 'http://localhost:2113/streams/testStream/head/backward/1?embed=body'
`
```
{
"title": "Event stream 'testStream'",
"id": "http://localhost:2113/streams/testStream",
"updated": "2017-08-31T13:21:11.807372Z",
"streamId": "testStream",
"author": {
"name": "EventStore"
},
"headOfStream": true,
"selfUrl": "http://localhost:2113/streams/testStream",
"eTag": "9;-2060438500",
"links": [
{
"uri": "http://localhost:2113/streams/testStream",
"relation": "self"
},
{
"uri": "http://localhost:2113/streams/testStream/head/backward/1",
"relation": "first"
},
{
"uri": "http://localhost:2113/streams/testStream/0/forward/1",
"relation": "last"
},
{
"uri": "http://localhost:2113/streams/testStream/8/backward/1",
"relation": "next"
},
{
"uri": "http://localhost:2113/streams/testStream/10/forward/1",
"relation": "previous"
},
{
"uri": "http://localhost:2113/streams/testStream/metadata",
"relation": "metadata"
}
],
"entries": [
{
"eventId": "1bd7ff7d-77c1-ae49-89ae-46a5998533d0",
[Truncated]
buf[i+5] = buf2[i+4];
buf[i+6] = buf2[i+7];
buf[i+7] = buf2[i+6];
const bth = _byteToHex;
return bth[buf[i++]] + bth[buf[i++]] +
bth[buf[i++]] + bth[buf[i++]] + '-' +
bth[buf[i++]] + bth[buf[i++]] + '-' +
bth[buf[i++]] + bth[buf[i++]] + '-' +
bth[buf[i++]] + bth[buf[i++]] + '-' +
bth[buf[i++]] + bth[buf[i++]] +
bth[buf[i++]] + bth[buf[i++]] +
bth[buf[i++]] + bth[buf[i++]];
}
module.exports = {
parse,
unparse
};
```
Answers:
username_0: For ease of use, I made this: https://www.npmjs.com/package/uuid-parse-le
username_1: The issue is not endianess. otherwise the whole uuid would be reversed. It has to do with how Microsoft (because eventstore is in .net) stores it's uuid aka guid. So it's a guid vs uuid bytes storage issue. I will use guid instead of uuid for parsing/unparsing eventId.
See https://en.wikipedia.org/wiki/Universally_unique_identifier#Encoding (the difference in encoding is partial little-endian, so you were partly right)
username_0: Well yes, it is an endian issue, but only the first three parts differs, since the last part is a byte array where endianess is meaningless. I did of course visit that particular wiki-page during my investigation. It didn't occur to me that eventstore uses guid instead of uuid, before you mentioned it, and that the difference is basically the subtle endianess.
I came across this page, claiming that some bits hold a special meaning. Not sure if this applies in practise at all though wrt eventstore: https://www.quora.com/What-is-the-difference-between-UUID-and-GUID
Thanks for addressing this. GUID-packages on npmjs seems a bit scarce though. Perhaps I should just rename the uuid-parse-le to guid-parse? Is there anything else from a "guid-parsing" besides the endianess that we need?
username_1: Fixed in https://github.com/username_1/node-eventstore-client/commit/<PASSWORD>
Status: Issue closed
|
pytorch/pytorch | 588251397 | Title: if there is some cat operation in the model, libtorch1.4.0 can only deploy the model in cpu but not in gpu
Question:
username_0: ## 🐛 Bug
<!-- A clear and concise description of what the bug is. -->if there were some cat operations in the model, libtorch1.4.0 can only deploy the model in cpu, but not in gpu.
## To Reproduce
Steps to reproduce the behavior:
1.use pytorch1.4.0 to trace a model which has cat operations, make sure the model is GPU mode
1.use libtorch1.4.0 to deploy the traced model, it isnot ok
1.make the pytorch to cpu mode, and trace it again ,then use libtorch1.4.0 to deploy the traced model, it is ok
<!-- If you have a code sample, error messages, stack traces, please provide it here as well -->
## Expected behavior
<!-- A clear and concise description of what you expected to happen. -->
## Environment
Please copy and paste the output from our
[environment collection script](https://raw.githubusercontent.com/pytorch/pytorch/master/torch/utils/collect_env.py)
(or fill out the checklist below manually).
You can get the script and run it with:
```
wget https://raw.githubusercontent.com/pytorch/pytorch/master/torch/utils/collect_env.py
# For security purposes, please check the contents of collect_env.py before running it.
python collect_env.py
```
- PyTorch Version (e.g., 1.0):
- OS (e.g., Linux):
- How you installed PyTorch (`conda`, `pip`, source):
- Build command you used (if compiling from source):
- Python version:
- CUDA/cuDNN version:
- GPU models and configuration:
- Any other relevant information:
## Additional context
<!-- Add any other context about the problem here. -->
Answers:
username_1: What do you mean by "not ok"? Could you please elaborate a little bit?
username_1: Thanks for the update. One more question, have you copied all the DLLs from libtorch to the location of the executable?
username_0: when the program goes here:
auto CAB_output = CAB_model.forward(inputs).toTensor();
Visual Studio outputs a dialog which says:
0x00007FF839BCA839 处有未经处理的异常: Microsoft C++ 异常: std::runtime_error,位于内存位置 0x00000070A1326658 处
my libtorch version is 1.4.0.
my total deploy code is here:
#define CPU_mode false
//CAB_main
int main()
{
#if CPU_mode
std::string CAB_model_path = "./traced/traced_CAB_cpu.pt";
#else
std::string CAB_model_path = "./traced/traced_CAB_cuda.pt";
#endif
torch::jit::script::Module CAB_model;
try {
// Deserialize the ScriptModule from a file using torch::jit::load().
CAB_model = torch::jit::load(CAB_model_path);
}
catch (const c10::Error& e) {
std::cerr << "error loading the CAB model\n";
return -1;
}
at::Tensor X1_tensor = torch::zeros({ 1, 64, 48, 48 });
at::Tensor X2_tensor = torch::zeros({ 1, 64, 48, 48 });
std::vector<torch::jit::IValue> inputs;
#if CPU_mode
inputs.push_back(X1_tensor);
inputs.push_back(X2_tensor);
#else
auto X1_var = torch::autograd::make_variable(X1_tensor, false);
auto X2_var = torch::autograd::make_variable(X2_tensor, false);
inputs.push_back(X1_var.to(at::kCUDA));
inputs.push_back(X2_var.to(at::kCUDA));
#endif
auto CAB_output = CAB_model.forward(inputs).toTensor();
c10::IntList size;
size = CAB_output.sizes();
std::cout << size.at(0) << "," << size.at(1) << "," << size.at(2) << "," << size.at(3) << std::endl;
return 0;
}
username_0: No, I just copied c10.dll, c10_cuda.dll, torch.dll, libiomp5md.dll nvToolsExt64_1.dll.
thank you ,just now i copied all the DLLs from libtorch, it works
Status: Issue closed
|
umijs/umi | 323872653 | Title: 升级到新版本后PrivateRoute内Redirect会出错
Question:
username_0: 看了下新版自动生成的路由,全局`layout`跟 `index/index.js`的 `path`都是 `/`。 那如果在PrivateRoute.js文件内需要对`/`路由做鉴权的话,就会死循环Redirect
[Test](https://github.com/username_0/test)
Answers:
username_1: 你给的例子不能重现你说的这个问题,就是进了两次PrivateRoute
username_2: 是有这个问题,在 PrivateRoute 判断 props.location.path,忽略相关的路由。
Status: Issue closed
|
DefinitelyTyped/DefinitelyTyped | 451252084 | Title: react-native BUTTON interface must have an optional style type.
Question:
username_0: its nice if we have an optional stype type in button interface
https://github.com/DefinitelyTyped/DefinitelyTyped/blob/21e68259f6fd43aba37e8112e3edb792975f8266/types/react-native/index.d.ts#L7173
Status: Issue closed
Answers:
username_1: Hi thread, we're moving DefinitelyTyped to use [GitHub Discussions](https://github.com/DefinitelyTyped/DefinitelyTyped/issues/53377) for conversations the `@types` modules in DefinitelyTyped.
To help with the transition, we're closing all issues which haven't had activity in the last 6 months, which includes this issue. If you think closing this issue is a mistake, please pop into the [TypeScript Community Discord](https://discord.gg/typescript) and mention the issue in the `definitely-typed` channel. |
NYPL-discovery/discovery-designs | 181515427 | Title: Resolve user login flow for alpha
Question:
username_0: Reviewed with Kevin and Kate. Confirmed how user authentication works: oauth redirect to login page, and redirect back with user token. We need to keep in mind there may be a nypl cookie to sniff if they logged in elsewhere.
Status: Issue closed
Answers:
username_0: Reviewed with Kevin and Kate. Confirmed how user authentication works: oauth redirect to login page, and redirect back with user token. We need to keep in mind there may be a nypl cookie to sniff if they logged in elsewhere.
Status: Issue closed
|
anvc/scalar | 268461979 | Title: Security restrictions in Scalar registrations
Question:
username_0: Hello,
Is there any security restrictions that can assist stopping outside users from accessing my Scalar site?
I currently have edited the user_model.php to restrict registration to just my university's @institution.edu
[root@ace-linux11 models]# grep fordham.edu user_model.php
if (!stristr($array['email'], '[institution].edu')) throw new Exception('Only email addresses under the [institution].edu domain may register on this Scalar install');
However, Although Scalar user accounts are limited to @institution.edu email addresses, there is no validation for the @institution.edu email address. As such, anyone can register using (<EMAIL>)<EMAIL> and can be provided access to Scalar. We need confirmation that these are legitimate accounts
Answers:
username_1: Yes, see http://scalar.usc.edu/works/guide2/local-settings
I'd be careful changing Scalar system files like models as they will be overwritten if you update.
username_0: It appears our Scalar site may have been compromised. The security at the university is looking to see where these fake @university.edu accounts were created. This is a nightmare for me right now. Is there any logs to explain how these accounts got created ? Looking in /var/www/html/scalar/system/application/logs ..but NOTHING about user account are present.
My belief is that there is NO validation of email address. So even a random person can fake an @university account to register to scalar... .Goodness. Please let me know any best practices for security. Can LDAP authentication work ? How so?
username_1: Correct, there is presently no email validation in Scalar. If you wanted to write up the module and do a pull request, that'd be awesome.
You could easily set a registration key (see my previous comment).
LDAP would work perfectly. You'll see fields for that in local_settings.php, as well.
If you followed the INSTALL.txt correctly, you should be a "super admin" for your Scalar install, in which case when you head to the Dashboard you'll see extra tabs: All Users, All Books, and Tools. Between thee three you can easily delete any problem (spam) books and problem user accounts.
username_0: Can I remove the registration link from our scalar site? Can you tell me how to use ldap settings to authenticate ? Our site has been compromised. Where are the logs?
username_0: Can you let me know where the logs are for user creation ? Security is looking into the logs
username_1: Adding a registration key would block registrations.
The LDAP fields are kept in local_setting.php. The values to put there depend on how your school has set up LDAP.
If your site is compromised go to All Books, All Users, and Tools tabs in the Dashboard as a super admin and simply delete the problem books and/or users.
username_0: Hi Can you please validate my settings below. Will this work? Can I have "ou=<my OU>,dc=subdomain1,dc=subdomain2,dc=subdomain3,dc=EDU" on that line below ?
//My school LDAP settings
$config['use_ldap'] = (getenv('SCALAR_USE_LDAP') ? getenv('SCALAR_USE_LDAP') : **true**); // Default: off
$config['ldap_server'] = (getenv('SCALAR_LDAP_SERVER') ? getenv('SCALAR_LDAP_SERVER') : "**myldapderver.com**");
$config['ldap_port'] = (getenv('SCALAR_LDAP_PORT') ? getenv('SCALAR_LDAP_PORT') : **636**);
$config['ldap_basedn'] = (getenv('SCALAR_LDAP_BASEDN') ? getenv('SCALAR_LDAP_BASEDN') : "**ou=myOU,dc=domain1,dc=subdomain2,dc=subdomain3,dc=EDU**");
$config['ldap_uname_field'] = (getenv('SCALAR_LDAP_UNAME_FIELD') ? getenv('SCALAR_LDAP_UNAME_FIELD') : "**uid**");
username_1: I literally have no idea, I've never set up LDAP. Someone else committed the LDAP upgrade to our GitHub as a pull request. But I've heard other places have used it fine so I'm confidant it works correctly.
username_0: can i take down the "registration page" and "login page" from Scalar?
We want no users logging in at this point.
username_1: Is suppose you could return false in both those methods in the login_model. But then you-yourself wouldn't even be able to login.
Requisite warning about adjusting system files if you plan to update Scalar.
username_0: How does Scalar use LDAP ? Does it authenticate the user's email address? What is the field that is being looked at for LDAP authentication?
username_1: Take a look at the login_model, the LDAP function is in there.
username_0: Thanks. Where are the logs for user authentication access?
username_1: You can set the log threashhold in config.php, but I don't think the logs will contain what you're looking for.
username_0: LDAP is authenticating via email and password. Can we change this to authenticate via UID ? If so, how can we do this?
username_0: Hi. THank you for your time helping me. I learned a great deal on Scalar.
1)User creation is not validating email addresses. correct?
2)Logs for user access. Is there a way to get that? We are looking at when some possible bogus accounts may have been created. Highly important.
3)Disable registration link on scalar page (how can i do that?)
4) Support for CAS (Central Authentication Service) available?
Appreciate any help on any of these highly critical questions on securing our Scalar site.
Thank you, again.
username_1: I don't know enough about PHP's ldap functions that are kept in the user_model's get_from_ldap_by_email_and_password() method to know what this config item in local_settings.php is communicating:
$config['ldap_uname_field'] = (getenv('SCALAR_LDAP_UNAME_FIELD') ? getenv('SCALAR_LDAP_UNAME_FIELD') : "uid");
It seems to me that if the system as presently designed accepts email addresses than 'uid' would be referring to an email address. Maybe changing it to something else would accept some other user field.
username_1: No idea, never used it. But you'd need to rewrite the core to add support similar to the LDAP setup.
In all of these cases if you implemented the code in such a way that it relied on config items and ran a pull request we'd be happy to roll them into the repo.
username_0: Do you know the directory or name of file where I can find the landing home page for scalar.?
username_1: system/application/modules/book_list
You can create your own module then point to it with the $config['active_book_list'] = 'book_list'; setting.
username_0: I'm building a new Red hat Scalar server with new Scalar version 2.17a, as management has directed. Is there a way to copy all media/books and users from my old Scalar server to my new server? If so, what directories or export/import features can I perform?
Thanks.
username_0: Is there a way to display php errors? I'm testing LDAP, but no errors are showing on screen, after LDAP configuration is in place.
username_1: Change 'production' to 'development' in /codeigniter.php.
You might want to spend more time getting to know Scalar and its underlying system, a lot of these questions have well-documented answers. https://www.codeigniter.com/user_guide/general/environments.html | http://scalar.usc.edu/works.guide2
username_0: Here's my error
A PHP Error was encountered
Severity: Warning
Message: ldap_start_tls(): Unable to start TLS: Can't contact LDAP server
Filename: models/user_model.php
Line Number: 182
username_0: Can someone please show me a working ldap configuration in Scalar. Getting errors above.
username_0: What version of codeignitor is on Scalar 2.17a ?
Status: Issue closed
|
Esri/deep-learning-frameworks | 856919383 | Title: Update fails for ArcGIS API for Python after installing deep learning framework
Question:
username_0: On computers where we have installed the deep learning framework, and which are at version 1.8.3 or less of the Python API, subsequent attempts to update environments to 1.8.5 fail with an error:
`C:\Users\somebody\AppData\Local\ESRI\conda\envs\main>conda update arcgis -c esri
Fetching package metadata .............
Solving package specifications:
InvalidSpecError: Invalid spec: >=`
This happens even when using a freshly cloned Python environment in ArcGIS Pro 2.7.2.
On computers on which we have not installed the deep learning framework we have no issues updating to 1.8.5.
Answers:
username_0: After uninstalling the deep learning framework we are able to upgrade arcgis to 1.8.5 was successfully in ArcGIS Pro 2.7.2 and 2.7.3.
Status: Issue closed
|
playframework/playframework | 170716178 | Title: playUpdateSecret dies when it encounters variable substitution in a HOCON file
Question:
username_0: ```
$ sbt
[mywebapp] $ playUpdateSecret
[trace] Stack trace suppressed: run last *:playUpdateSecret for the full output.
[error] (*:playUpdateSecret) com.typesafe.config.ConfigException$NotResolved: called valueType() on value with unresolved substitutions, need to Config#resolve() first, see API docs
[error] Total time: 0 s, completed Aug 11, 2016 11:32:55 AM
[mywebapp] $ last *:playUpdateSecret
[info] Updating application secret in /var/work/training/projects/public_code/group_play/course_play_scala_intro_code/webapp1/conf/application.conf
com.typesafe.config.ConfigException$NotResolved: called valueType() on value with unresolved substitutions, need to Config#resolve() first, see API docs
at com.typesafe.config.impl.ConfigDelayedMerge.valueType(ConfigDelayedMerge.java:46)
at com.typesafe.config.impl.SimpleConfig.hasPath(SimpleConfig.java:96)
at play.sbt.ApplicationSecretGenerator$$anonfun$updateSecretTask$1.apply(ApplicationSecretGenerator.scala:47)
at play.sbt.ApplicationSecretGenerator$$anonfun$updateSecretTask$1.apply(ApplicationSecretGenerator.scala:31)
at scala.Function1$$anonfun$compose$1.apply(Function1.scala:47)
at sbt.$tilde$greater$$anonfun$$u2219$1.apply(TypeFunctions.scala:40)
at sbt.std.Transform$$anon$4.work(System.scala:63)
at sbt.Execute$$anonfun$submit$1$$anonfun$apply$1.apply(Execute.scala:228)
at sbt.Execute$$anonfun$submit$1$$anonfun$apply$1.apply(Execute.scala:228)
at sbt.ErrorHandling$.wideConvert(ErrorHandling.scala:17)
at sbt.Execute.work(Execute.scala:237)
at sbt.Execute$$anonfun$submit$1.apply(Execute.scala:228)
at sbt.Execute$$anonfun$submit$1.apply(Execute.scala:228)
at sbt.ConcurrentRestrictions$$anon$4$$anonfun$1.apply(ConcurrentRestrictions.scala:159)
at sbt.CompletionService$$anon$2.call(CompletionService.scala:28)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at java.util.concurrent.Executors$RunnableAdapter.call(Executors.java:511)
at java.util.concurrent.FutureTask.run(FutureTask.java:266)
at java.util.concurrent.ThreadPoolExecutor.runWorker(ThreadPoolExecutor.java:1142)
at java.util.concurrent.ThreadPoolExecutor$Worker.run(ThreadPoolExecutor.java:617)
at java.lang.Thread.run(Thread.java:745)
[error] (*:playUpdateSecret) com.typesafe.config.ConfigException$NotResolved: called valueType() on value with unresolved substitutions, need to Config#resolve() first, see API docs
```
`conf/application.conf` looks like this:
```
play.crypto.secret = changeme
play.crypto.secret = ${?APP_SECRET}
```
Removing the last line solves the problem, and that makes sense. I am reporting this issue in the hope that:
1. resolve() is invoked when parsing application.conf, to prevent the stack dump
1. The default value gets set and the variable substitution is not modified
1. A message is provided to the user so they know what was done (or not done)
Answers:
username_1: 👍 for the resolution |
kindredgroup/oracle-imagecopy-backup | 1054659473 | Title: how to get list of times that I can restore DB to ?!!
Question:
username_0: Is there a way to get which times I can restore DB to?
Answers:
username_1: Assuming you have archivelogging enabled and one archivelog destination is in backup area - to any point in time after the oldest existing snapshot.
username_0: I'm not able to do so, below what I did,
Restore database to time point: (yyyy-mm-dd hh24:mi:ss) 2021-11-16 14:20:00
Database unique name: FOREVER
Oracle home: /u01/app/oracle/19.0.0/db_1
Clone mount path: /backup
Target instance SID: FOR
Restore target time UTC: 2021-11-16 10:20:00+00:00
Restore target time local: 2021-11-16 14:20:00+04:00
Restored from snapshot: FOREVER-20211116T141735
Are these parameters correct? (y/n) y
and then below error appears
unable to find archived log
archived log thread=1 sequence=34
released channel: c1
RMAN-00571: ===========================================================
RMAN-00569: =============== ERROR MESSAGE STACK FOLLOWS ===============
RMAN-00571: ===========================================================
RMAN-03002: failure of recover command at 11/16/2021 14:19:40
RMAN-06054: media recovery requesting unknown archived log for thread 1 with sequence 34 and starting SCN of 2333022
username_0: I fixed it, I needed to create new clone first using ./zsnapper.py FOR create
Status: Issue closed
|
home-assistant/core | 727734920 | Title: bmw_connected_drive: authentication failed
Question:
username_0: <!-- READ THIS FIRST:
- If you need additional help with this template, please refer to https://www.home-assistant.io/help/reporting_issues/
- Make sure you are running the latest version of Home Assistant before reporting an issue: https://github.com/home-assistant/core/releases
- Do not report issues for integrations if you are using custom components or integrations.
- Provide as many details as possible. Paste logs, configuration samples and code into the backticks.
DO NOT DELETE ANY TEXT from this template! Otherwise, your issue may be closed without comment.
-->
## The problem
<!--
Describe the issue you are experiencing here to communicate to the
maintainers. Tell us what you were trying to do and what happened.
-->
BMW Connected Drive integration no longer works.
The error says: Authentication failed. Maybe your password is invalid?
Nothing has been change from my site.
Manually I can log in into BMW web site wit same credential configured in HA
## Environment
<!--
Provide details about the versions you are using, which helps us to reproduce
and find the issue quicker. Version information is found in the
Home Assistant frontend: Configuration -> Info.
-->
- Home Assistant Core release with the issue: 0.116.4
- Last working Home Assistant Core release (if known):
- Operating environment (OS/Container/Supervised/Core): HassOS 4.14
- Integration causing this issue: bmw_connectd_drive
- Link to integration documentation on our website: https://www.home-assistant.io/integrations/bmw_connected_drive/
arch | armv7l
-- | --
chassis | embedded
dev | false
docker | true
docker_version | 19.03.11
hassio | true
host_os | HassOS 4.14
installation_type | Home Assistant OS
os_name | Linux
os_version | 4.19.127-v7l
python_version | 3.8.5
supervisor | 249
timezone | Europe/Rome
version | 0.116.4
virtualenv | false
## Problem-relevant `configuration.yaml`
<!--
An example configuration that caused the problem for you. Fill this out even
if it seems unimportant to you. Please be sure to remove personal information
like passwords, private URLs and other credentials.
-->
```yaml
bmw_connected_drive:
name:
[Truncated]
File "/usr/src/homeassistant/homeassistant/setup.py", line 213, in _async_setup_component
result = await task
File "/usr/local/lib/python3.8/concurrent/futures/thread.py", line 57, in run
result = self.fn(*self.args, **self.kwargs)
File "/usr/src/homeassistant/homeassistant/components/bmw_connected_drive/__init__.py", line 52, in setup
accounts.append(setup_account(account_config, hass, name))
File "/usr/src/homeassistant/homeassistant/components/bmw_connected_drive/__init__.py", line 80, in setup_account
cd_account = BMWConnectedDriveAccount(username, password, region, name, read_only)
File "/usr/src/homeassistant/homeassistant/components/bmw_connected_drive/__init__.py", line 127, in __init__
self.account = ConnectedDriveAccount(username, password, region)
File "/usr/local/lib/python3.8/site-packages/bimmer_connected/account.py", line 61, in __init__
self._get_vehicles()
File "/usr/local/lib/python3.8/site-packages/bimmer_connected/account.py", line 226, in _get_vehicles
self._get_oauth_token()
File "/usr/local/lib/python3.8/site-packages/bimmer_connected/account.py", line 117, in _get_oauth_token
raise OSError(msg) from exception
OSError: Authentication failed. Maybe your password is invalid?
```
## Additional information
Answers:
username_0: 
username_1: Yes, this has been happening for 2 or 3 days now. I'm still on 0.115.x and it just stopped working after a reboot so I believe something has changed on BMW's end.
username_2: from @username_3 on [HA Community](https://community.home-assistant.io/t/bmw-connecteddrive-component/39569/639):
PR with the fix is made by @rikroe -> https://github.com/home-assistant/core/pull/42215 4
Hopefully it will be merged as hotfix in 0.117.
If you want to fix it in the meanwhile you can check here for instructions -> https://github.com/home-assistant/core/issues/42139 8.
username_3: This issue can be closed as it is an duplicate of #42139 which is fixed in #42215 and will be part of 0.117.
Status: Issue closed
username_4: Fixed in #42215, part of the upcoming Home Assistant 0.117 |
mne-tools/mne-bids | 687374444 | Title: Make get_matched_basenames a method of BIDSPath
Question:
username_0: As per discussion at https://github.com/mne-tools/mne-bids/pull/517#issuecomment-682042937
Answers:
username_0: Same for `get_matched_emptyroom()`, right?
username_1: Want me to just stick it into #532? Same process: i) privatize ii) call in BIDSPath and iii) do some minor update of tests.
username_0: yes, I'm currently working on it. Will file a PR shortly
Status: Issue closed
|
go-pg/pg | 681934245 | Title: EOF error when go-pg cannot parse json in row
Question:
username_0: I can only test this on v9, sorry. When there's a field tagged as json in a row but it actually contains empty string, go-pg throws an EOF error instead of reporting it as json error and closes the connection.
```
package main
import (
"fmt"
"github.com/go-pg/pg"
)
type MyParams struct {
Test int `json:"test"`
}
func main() {
// setup
DB := pg.Connect(&pg.Options{
Addr: "localhost:5432",
User: "test",
Database: "test",
})
_, err := DB.Exec("drop table test_table")
_, err = DB.Exec("create table test_table (id int, params text)")
_, err = DB.Exec("insert into test_table values (1, '{}')")
_, err = DB.Exec("insert into test_table values (2, '')")
_, err = DB.Exec(`insert into test_table values (3, '{"test":3}')`)
_, err = DB.Exec("insert into test_table values (4, 'invalid')")
// select first
var a struct {
ID int
Params MyParams
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 1")
if err != nil {
fmt.Println("1 ERROR:", err)
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 2")
if err != nil {
fmt.Println("2 ERROR:", err)
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 3")
if err != nil {
fmt.Println("3 ERROR:", err)
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 4")
if err != nil {
fmt.Println("4 ERROR:", err)
}
}
```
Produces the following:
```
2 ERROR: EOF
4 ERROR: invalid character 'i' looking for beginning of value
```
EOF is really confusing here, took me some time to figure it out. I can also see this in Postgresql logs:
```
test@test DEBUG: unexpected EOF on client connection
```
Answers:
username_1: `encoding/json` returns `EOF` on empty string:
```
r := strings.NewReader("")
var test int
err := json.NewDecoder(r).Decode(&test)
fmt.Println(test, err)
```
go-pg seems to be working as expected.
Status: Issue closed
username_0: I've checked with the pg.Connect().Conn() and it doesn't seem to close the connection.
```
package main
import (
"encoding/json"
"errors"
"fmt"
"github.com/go-pg/pg"
"io"
"strings"
)
type MyParams struct {
Test int `json:"test"`
}
func main() {
// setup
DB := pg.Connect(&pg.Options{
Addr: "localhost:5432",
User: "test",
Database: "test",
})
_, err := DB.Exec("drop table test_table")
_, err = DB.Exec("create table test_table (id int, params text)")
_, err = DB.Exec("insert into test_table values (1, '{}')")
_, err = DB.Exec("insert into test_table values (2, '')")
_, err = DB.Exec(`insert into test_table values (3, '{"test":3}')`)
_, err = DB.Exec("insert into test_table values (4, 'invalid')")
// select first
var a struct {
ID int
Params MyParams
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 1")
if err != nil {
fmt.Println("1 ERROR:", err)
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 2")
if err != nil {
fmt.Println("2 ERROR:", err, errors.Is(err, io.EOF), reflect.TypeOf(err))
}
r := strings.NewReader("")
var test int
err = json.NewDecoder(r).Decode(&test)
fmt.Println("JSON ERROR:", err, errors.Is(err, io.EOF), reflect.TypeOf(err))
_, err = DB.QueryOne(&a, "select * from test_table where id = 3")
if err != nil {
fmt.Println("3 ERROR:", err)
}
_, err = DB.QueryOne(&a, "select * from test_table where id = 4")
if err != nil {
fmt.Println("4 ERROR:", err)
}
}
```
Integrating that test shows that it's some different EOF so go-pg or something else might still be involved.
```
2 ERROR: EOF false internal.Error
JSON ERROR: EOF true *errors.errorString
```
That's probably a problem of the json library behavior, it would be much better if it returned json.SyntaxError, but alas.
username_2: The problem is that it appears go-pg is treating NULL JSONB fields as an empty string, when it shouldn't.
username_0: In my case it's just text fields, I don't use postgres jsonb type. Somewhat annoyingly, I can't just use empty strings as json placeholders, the select query will result in an error, I have to use the '{}' default.
username_2: I doubt this will be fixed, but would be nice to see if Bun has this same behavior. |
mateodelnorte/servicebus-register-handlers | 192141866 | Title: Using globbed paths
Question:
username_0: I'd like to use a glob for the path to the listeners. I was going to fork, replace this:
```
if (options.path) {
var i = 0;
var handlers = objectifyFolder({
fn: function (mod, result) {
if ( ! (mod.queueName || mod.routingKey) || ! (mod.listen || mod.subscribe)) return;
addHandler(options.pipelines, new Handler(mod));
},
path: options.path
});
```
Would that be the right place to do so? If so, I just need to make sure that for each handler that shows up in the glob is passed to addHandler as mod, correct?
I'm thinking of using something like requireDir/require-dir to get the files instead of relying on objectifyFolder.
Status: Issue closed
Answers:
username_1: The change would be in `objectify-folder`, then just update package.json here.
If you can find a direct replacement for readdirSync that brings back a list of all files, respecting globs, you may be able to essentially replace this line: https://github.com/username_1/objectify-folder/blob/master/index.js#L23
username_1: Looks like you can just do a glob.sync using https://www.npmjs.com/package/glob
username_1: Went ahead and made the updates for you.
➜ servicebus-register-handlers git:(master) npm publish
+ [email protected]
username_0: Thanks, I'll check it out. Do you have an example? Also, what's the callback in the subscribe function? What does that initiate?
username_1: Yup. Example in the tests: https://github.com/username_1/servicebus-register-handlers/blob/master/test/index.js#L74
servicebus-register-handlers works with servicebus-retry. as such, if your queues are `ack:true` when you execute the callback in listen or subscribe, it will acknowledge the message and pop it off rabbitmq. If you pass an error to either callback, it will use servicebus-retry's retry functionality until maxRetries is exceeded, then it will dead-letter the message to an error queue matching the routingKey + ".error"
username_0: thanks for the help!
username_1: Sure thing. Happy to help any time. In fact, feel free to join nycnode.com/slack and ask questions on there in real time. |
rebuy-de/aws-nuke | 309639419 | Title: Unable to remove S3 buckets if versioning is enable
Question:
username_0: As per title, when there's versioning enable on S3 bucket with files, aws-nuke won't be able to remove the old versions.
Instead it will show an error message:-
"BucketNotEmpty: The bucket you tried to delete is not empty. You must delete all versions in the bucket.
status code: 409, request id: XXXXXXXXXXXXX, host id: XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX"
Answers:
username_1: This should be already fixed in #76, but the fix was not release yet. I just did an alpha release, where deleting versioned S3 Objects should properly work: https://github.com/rebuy-de/aws-nuke/releases/tag/v2.0.0-alpha1
Status: Issue closed
|
JKISoftware/JKI-State-Machine-Objects | 1086346058 | Title: Dynamically launched SMOs from inside the Callee's Process causes intermittent errors during StopProcess call
Question:
username_0: **Describe the bug**
Dynamically launched SMOs from inside the Callee's Process causes intermittent errors during StopProcess call
**To Reproduce**
1. Launch Dynamic Processes within an SMO's process VI, after the Process:Sync has been executed (SMO started)
2. Stop SMO
**Expected behavior**
All dynamically launched SMOs should terminate before the Process that called them completes the Stop process and syncs with other components of the hierarchy.
**Screenshots**
The execution order allows the Process VI to leave memory before any dynamically launched SMOs from within that process have terminated. This does not seem to affect the dynamically launched SMOs from outside the process VI.

**Proposed Fix**
Move the Stop Dependencies before the process sync.

Answers:
username_0: Thanks @gcodetechnology and @ciozi137 for the reports and @gcodetechnology for the example code that reproduced the issue!
username_0: Unit Test created which demonstrates failure.

username_0: build 1.4.0.68 fixes this issue
Status: Issue closed
|
kivy/buildozer | 187801910 | Title: using android_new causes missing packages errors
Question:
username_0: If i try to install them with pip, it seems any of the named packages is installed
Answers:
username_1: When you install them, do you make sure you upgrade ?
like:
`sudo pip install -U colorama`
`sudo pip install -U appdirs`
`sudo pip install -U sh`
`sudo pip install -U jinja2`
username_0: Yes for my python3 installation all thoese packages are up-to-date
username_2: Had the same thing, turns out it was using anaconda's python. If you have multiple python versions make sure it's using the correct ones.
username_3: Closing as stale, buildozer handles this now.
Status: Issue closed
|
dnaeon/go-vcr | 356047144 | Title: Throw error on realHttpTransport connection
Question:
username_0: Is there a way to throw an error when an HTTP connection is made on realHttpTransport during `ModeReplay`. I imagine this is up to our own implementation of `realHttpTransport` - is there a recipe we could include in the readme.md? I find this a super helpful feature of Ruby VCR.
Thanks.
Answers:
username_0: It seems like the httpTransport will return `cassette.ErrInteractionNotFound` and it go swallowed somewhere in my stack.
Would you welcome a PR mentioning this in the readme? If not, I feel free to close.
username_1: Hey Eric,
Please feel free to submit a PR for this one.
Thanks,
Marin |
nvs-vocabs/test | 570530445 | Title: NTR: Request for S06 http://vocab.nerc.ac.uk/
Question:
username_0: <H2> Required </H2><p><b>Email Address</b><p> <EMAIL><p> <b>Term name (PrefLabel)</b><p>http://vocab.nerc.ac.uk/ <p><b>Definition</b><p> http://vocab.nerc.ac.uk/<p><H2> Optional </H2><p><b>Sources/references</b><p> http://vocab.nerc.ac.uk/<p> <b>ORCID</b><p> http://vocab.nerc.ac.uk/<p> <b>Synonym or acronym (AltLabel)</b><p> http://vocab.nerc.ac.uk/<p><b>Mapping to external terminologies</b><p> http://vocab.nerc.ac.uk/<p> |
wave-harmonic/crest | 1021796780 | Title: Graphics.CopyTexture source and destination have different master texture limits
Question:
username_0: **Versions**
2020.3.20
**Additional context**
"master texture limits" is "Project Settings > Texture Quality".
See #905 for possible fix or change _Texture Quality_ to _Full Res_.
Status: Issue closed
Answers:
username_0: Fixed for 4.14. |
bokeh/bokeh | 526457348 | Title: [FEATURE] Floodfill math functions
Question:
username_0: Hi, I'm trying to color the plot between two math functions, the area zone. I've asked about that in stackoverflow [https://stackoverflow.com/questions/58950303/coloring-the-area-between-two-curves-with-bokeh](url) .
It seems this is not avalaible with bokeh actually, and I think this feature is very important. I'm a math teacher and I'm making a web page with mathematical teory and examples. But I cannot to visualize the area about integrals without graphing the area between two or more math functions.
I like bokeh because it offers an easy way for the web readers to interact with the plots (zoom, movin' points, change colors and so on).
@bigreddot has suggested to me I make a petition about that here and it talks about a web page about to implement floodfill in html5 canvas http://www.williammalone.com/articles/html5-canvas-javascript-paint-bucket-tool/
Thanks
Answers:
username_1: Thanks for the issue @username_0 I think this would be a neat feature and also that it would be pretty self-contained and not to difficult to implement. (I could be wrong, of course...) So I am definitely interested to take a crack at it. I am not sure it will be before the upcoming 2.0 release, but hopefully fairly soon. |
MediaArea/RAWcooked | 457332480 | Title: Unsupported number 128 of slices requested
Question:
username_0: My colleague @raecasey reported an error with ffmpeg/rawcooked regarding some weird frame sizes.
We have a sequence that is 3512x2640 (you can replicate with `ffmpeg -f lavfi -i testsrc=size=3512x2640 -pix_fmt rgb48be -vframes 1 weird.dpx` ) and the rawcooked job fails due to an ffmpeg error regarding slices. If I manually specify a slice count other than 128 then all is well. I tried 256 and 16 I think. So perhaps rawcooked needs to choose a different slice count for some rasters? :
We are using an older version of rawcooked in production but I compiled from git-master on OSX El Capitan and saw the same issue.
```
$ rawcooked --file weird.dpx
License detected but decoding not supported in this version.
Using default license.
Consider to support RAWcooked: https://MediaArea.net/RAWcooked
Track 1:
weird.dpx
DPX/Raw/RGB/16bit/BE
*** This input format flavor is not supported by the current license key. ***
One or more requested features are not supported with the current license key.
Please contact <EMAIL> for a quote or a temporary key.
Ignoring the license for the moment.
ffmpeg version 4.1.1 Copyright (c) 2000-2019 the FFmpeg developers
built with Apple LLVM version 8.0.0 (clang-800.0.42.1)
configuration: --prefix=/usr/local/Cellar/ffmpeg/4.1.1 --enable-shared --enable-pthreads --enable-version3 --enable-hardcoded-tables --enable-avresample --cc=clang --host-cflags=-I/System/Library/Frameworks/JavaVM.framework/Versions/Current/Headers/ --host-ldflags= --enable-ffplay --enable-gnutls --enable-gpl --enable-libaom --enable-libbluray --enable-libmp3lame --enable-libopus --enable-librubberband --enable-libsnappy --enable-libtesseract --enable-libtheora --enable-libvorbis --enable-libvpx --enable-libx264 --enable-libx265 --enable-libxvid --enable-lzma --enable-libfontconfig --enable-libfreetype --enable-frei0r --enable-libass --enable-libopencore-amrnb --enable-libopencore-amrwb --enable-libopenjpeg --enable-librtmp --enable-libspeex --enable-videotoolbox --disable-libjack --disable-indev=jack --enable-libaom --enable-libsoxr
libavutil 56. 22.100 / 56. 22.100
libavcodec 58. 35.100 / 58. 35.100
libavformat 58. 20.100 / 58. 20.100
libavdevice 58. 5.100 / 58. 5.100
libavfilter 7. 40.101 / 7. 40.101
libavresample 4. 0. 0 / 4. 0. 0
libswscale 5. 3.100 / 5. 3.100
libswresample 3. 3.100 / 3. 3.100
libpostproc 55. 3.100 / 55. 3.100
[dpx_pipe @ 0x7fd20080c800] Stream #0: not enough frames to estimate rate; consider increasing probesize
Input #0, dpx_pipe, from 'weird.dpx':
Duration: N/A, bitrate: N/A
Stream #0:0: Video: dpx, rgb48be, 3512x2640 [SAR 1:1 DAR 439:330], 24 tbr, 24 tbn, 24 tbc
File 'weird.dpx.mkv' already exists. Overwrite ? [y/N] y
Stream mapping:
Stream #0:0 -> #0:0 (dpx (native) -> ffv1 (native))
File weird.dpx.rawcooked_reversibility_data -> Stream #0:1
Press [q] to stop, [?] for help
[ffv1 @ 0x7fd203001e00] Unsupported number 128 of slices requested, please specify a supported number with -slices (ex:4,6,9,12,16, ...)
Error initializing output stream 0:0 -- Error while opening encoder for output stream #0:0 - maybe incorrect parameters such as bit_rate, rate, width or height
Conversion failed!
```
Status: Issue closed
Answers:
username_1: @raecasey please test [latest snapshot](https://mediaarea.net/download/snapshots/binary/rawcooked/).
username_0: Many thanks! We will test ASAP. |
remote-job-boards/software-engineering | 993705974 | Title: Beat: Senior Backend Engineer
Question:
username_0: **Tags:** #golang #senior #engineer #backend
**Published on:** September 08, 2021
**Original Job Post:** https://remoteOK.io/remote-jobs/105921-remote-senior-backend-engineer-beat
<NAME> - *** Who are Beat? ***Beat is one of the most exciting companies operating in the ride-hailing space. One city at a time, all across the globe we make transportation affordable, convenient, and safe for everyone. We also help hundreds of thousands of people earn extra...<issue_closed>
Status: Issue closed |
helium/miner | 1099537041 | Title: unable to locate POC ID - failed to dial and others errors
Question:
username_0: **Hi, I have a Pisces P100 and in the dashboard I find many errors of different types.**
_2022-01-10 19:47:38.183 6 [info] <0.6412.0>@miner_onion_server:decrypt:{356,25} unable to locate POC ID <<"RgKVg_jxVZzjWwT46G9zP7L12FpyBd7oIzMDR4_jrvQ">>, dropping
2022-01-10 19:57:14.488 [error] <0.1586.0>@blockchain_state_channels_client:handle_info:{205,5} failed to dial "/p2p/11w77YQLhgUt8HUJrMtntGGr97RyXmot1ofs5Ct2ELTmbFoYsQa": timeout dropping 5 packets
2022-01-11 18:21:53.111 6 [error] <0.1483.2>@miner_onion_server:send_witness:{207,5} failed to send witness, max retry
2022-01-11 18:21:53.111 6 [info] <0.1483.2>@miner_onion_server:send_witness:{246,37} re-sending witness at RSSI: -116, Frequency: 867.3, SNR: -21.5_
**Did any of you manage to solve these problems or would you know what to do? Also it seems that my Hotspot works fine as soon as I restart and clean the blockchain but after about 20h it stops doing activity. Only if I clean the blockchain and re-sync it will it work decently.
Thanks a lot to those who can give me a hand!**
Answers:
username_1: I don't think anyone has solved anything. I have had a a bunch of different issues, and this morning I checked my logs again, I had that issue show up today as well. There is something very wrong on the p2p network, That I believe based on numbers, that on the dev team side sees nothing wrong. * say if 10000 people have the issue out of 500,000 thousand *. I think they are working more on the light hotspot role out as per the documentation it says its going to be out before the end of Q1 ... that will solve the issue since the network is getting off the p2p network. So I think they are focusing on the roll out of that and not fixing the broken issue. I could be wrong, but there is 1000s of hotspots effected by this. --- this is all speculation ---
username_2: I noticed that the ARM version is the one suffering the most, I see they put out a new AMD version yesterday which I have downloaded and finished syncing over night. I am going to try and throw my EEC card onto my AMD based laptop, with the concentrator card, And see if that resolves it.
How to run AMD version? I would like to try it in a virtual machine too.
username_1: I gave up on it, since the latest update. But I was running it on a laptop with the same basic setup instructions on the helium docs. With docker. But I had to rig up a socket in python for the i2c bus to go over USB and that won’t work on a out of the box hotspot firmware.
username_1: @username_2 Are you having an Erlang error? like a "gen_server :call/2 line 239 ??? or something similar. My original reason that I jumped over to the AMD version to see if it was calling any errors was because someone mentioned an Erlang issue with the libp2p I don't know anything about that as I mentioned I'm not a coder. But I did a quick search on google about that gen_server:call/2 line 239 error and found something on a rabbitmq forum that mentioned that the persons server was not running erlang 21 OTP, The person mentioned That they changed their server to Erlang 21 and it fixed the issue for them. I dont remember where I posted that in this forum, But I believe the list of people that I am having the Dial errrors with are running a diffrent version of Erlang on their machines, Im going to look deeper into all my failed to dial error addresses, And see if they are validators, Running the AMD version in the Cloud. I remember mentioning in that post, That who ever compiled the AMD version should compile the ARM version. because of this. Call me crazy but I think I figured this out weeks ago.
username_1: @username_2 I did a dual miner test yesterday with my setup at like 3-4am Est standard time, I’m not sure if it was a coincidence or what but my miner after 2 weeks actually synced to the helium API. I did as a test with my laptop. So it looks to interact with the API better on AMD.
@username_2 i am trying to learn this Docker buildX but I don’t find good documentation ( I don’t know docker very will) but I read in the main repo here on the build instructions , that the AMD version needs a AVX or something capable processor.
I don’t know if you got to work in VM ?? I looked to see if PI4 is AVX capable , I found in TensorFlow documentation that PI4 is AVX capable, most old miner like Nebra origonal had PI 3 which I did not see is AVX capable. Something like ARMv7 vs ARMv8 is different.
Any luck? Because I was only testing and did not want to exceed a “testing” time period I stopped the AMD version running on laptop. I’m now trying to figure out the ( learn from scratch) erlang. In the main helium miner repo, I have gone through all of the files and made some notes. I don’t think anything wrong with helium main repo and code looks fine. Issue is not there.
Today if I find some focus time I am going to go through the external repos, or I guess what they call dependency and look at the difference between the two types.
username_2: Sorry for the late reply.
Yes, I have such error messages too. (regularly)
2022-01-24 20:16:00.821 [error] <0.1655.0>@blockchain_state_channels_client:handle_info:{205,5} failed to dial "/p2p/11w77YQLhgUt8HUJrMtntGGr97RyXmot1ofs5Ct2ELTmbFoYsQa": timeout dropping 2 packets
Unfortunately, I was unable to set up the virtual machine. |
fedora-silverblue/issue-tracker | 744538508 | Title: [Little issue]: Evince icon inside thumbnails view is not visible
Question:
username_0: Related to this version:
Evince org.gnome.Evince 3.38.0 stable fedora system
I noticed that the same version on flathub doesn't have this issue.
On the upper right corner, the icon to show thumbnails of the pages isn't visible.

Answers:
username_1: @username_2 any idea? Could we miss some icons package in Evince Flatpak or Fedora runtime?
username_2: Hm, I think we are missing gdk pixbuf cache regeneration for icons that evince installs. Investigating.
username_2: This should be now fixed in https://src.fedoraproject.org/flatpaks/evince/c/3ce4e5ad231ddd6dce08bde0ec08eab71a1d2bdf?branch=master . Still waiting on the build to finish.
username_2: https://bodhi.fedoraproject.org/updates/FEDORA-FLATPAK-2020-ffc9849ee2
username_0: I've just received your update. Thanks!! :heart:
Good job!! :+1:
Status: Issue closed
|
piejanssens/premiumizer | 792541453 | Title: Permissions of files created by Docker image
Question:
username_0: I'm running premiumizer using Docker under a 'docker' user and shared group.
New files get created with **read/write** access for the **user** and only **read** access for the **group**.
Every user in the group should be able to move/rename the files.
A solution would be to allow setting a umask - https://wiki.archlinux.org/index.php/Umask
Answers:
username_1: Got exactly the same problem. Other than that, this program is awesome! :)
username_1: Got exactly the same problem. Other than that, this program is awesome! :)
username_2: no idea about docker stuff should be easy for someone to fix with the docker file ?
something like this ?
http://widerin.net/blog/change-umask-in-docker-containers/
Status: Issue closed
|
gobysec/Goby | 991653134 | Title: 不断循环请求
Question:
username_0: - 任务配置
<img width="390" alt="图片" src="https://user-images.githubusercontent.com/5812175/132604799-f3f2f369-a08b-44c0-9d0f-ad2edb98818d.png">
- 抓包结果


Answers:
username_1: bug未复现成功,请添加gobybot微信(gobyteam)详细说明 |
PaddlePaddle/Paddle | 813048276 | Title: @paddle.jit.to_static的文档是不是矛盾了?有些看不明白
Question:
username_0: https://www.paddlepaddle.org.cn/documentation/docs/zh/api/paddle/fluid/dygraph/jit/declarative_cn.html

装饰器应该返回的是函数啊?
Answers:
username_1: 1. `to_static`装饰器支持装饰普通function、类method,以及用户定义的模型Net实例(默认会在forward函数上进行装饰)。被装饰的函数会被返回一个StaticFunction对象,重写了`__call__`,以实现静态图执行
2. 被`to_static` 装饰的函数,内部会以静态图方式执行。比如 FuncA → FuncB → FuncC,如果只有中间的FuncB被装饰了,考虑到A和B依旧是动态图执行,所以被装饰后的FuncB内部以静态图运行,结果返回的是动态图Tensor,以支持“局部装饰”的功能 |
bbc/psammead | 496347530 | Title: Layout iteration - Feature 2 promo 1008+px
Question:
username_0: **Is your feature request related to a problem? Please describe.**
We should iterate the first Story Promo (Feature 2) in a section for 1008+px.
**Describe the solution you'd like**
Since the Story Promo component is getting huge and the positions of the elements of this Feature 2 promo are different, we should create a new component that matches with UX designs 1008+px.
Double check with UX if the RTL version is correct (text on the left of the image)
LTR:

RTL:

**Describe alternatives you've considered**
Reuse the current Story Promo component.
**Testing notes**
[Tester to complete]
Dev insight: Will there be any potential regression? etc
- [X] This feature is expected to need manual testing.
**Additional context**
Add any other context or screenshots about the feature request here.
Answers:
username_1: The RTL version should be a mirror of the LTR version, so on the right you should see the story headline first. Would be good to get updated designs.
username_2: Blocked until grid work is done by the Shared Components pod
Status: Issue closed
|
NationalSecurityAgency/ghidra | 1028613956 | Title: GraalVM support for headless builds
Question:
username_0: To improve the performance of the automated analysis without the GUI running, it's possible to compile the Java code using the GraalVM to the native code. It will help greatly reduce the memory consumption and increase the speed.
- https://www.graalvm.org/
- https://www.graalvm.org/docs/introduction/
- https://www.graalvm.org/reference-manual/native-image/
- https://www.graalvm.org/reference-manual/native-image/Limitations/
There are might be some limitations, but quite often they can be easily overcome by slightly modifying the source code or reducing dependencies.
Answers:
username_1: @username_0 Have you done any experimenting with the `native-image` feature and Ghidra?
"Full AOT" is known to be tricky to get working, especially in complex java applications like Ghidra. |
botman/botman | 691642541 | Title: Add 2 Redis instance/container on GitHub Actions
Question:
username_0: At the moment we are only properly testing `RedisCache` and `RedisStorage` using Travis-CI. It would be nice if we can have the same test running on GitHub Actions with the following requirements:
- Run redis on host port `6379` without password.
- Run redis on host port `6379` with password `<PASSWORD>`.
Answers:
username_1: Based on my tests I found out that the tests will fail and return `connection refused` even on my local environment (PHP 7.4.9) while connecting to redis locally, notice the `has` test which does not require an authentication.

and this is the same thing I have found on github actions.
now to run the tests with/without passwords we should split the redis group into two groups one with validation and the second without a validation, or we can create an ENV var and add it to test function like
```php
protected function isSecure()
{
return $_ENV['SECURE'];
}
/** @test */
public function valid_auth()
{
if(!$this->isSecure()) {
$this->markTestSkipped('This test needs a secure instance of redis');
}
$storage = new RedisStorage($this->getRedisHost(), $this->getAuthRedisPort(), 'secret');
$key = 'key';
$data = ['foo' => 1, 'bar' => new \DateTime()];
$storage->save($data, $key);
self::assertEquals(Collection::make($data), $storage->get($key));
}
public function get()
{
if($this->isSecure()) {
$this->markTestSkipped('This test needs an insecure instance of redis');
}
$storage = new RedisStorage($this->getRedisHost());
$key = 'key';
$data = ['foo' => 1, 'bar' => new \DateTime()];
$storage->save($data, $key);
self::assertEquals(Collection::make($data), $storage->get($key));
}
```
You can find the workflow which I've created here https://github.com/username_1/botman/blob/2.0/.github/workflows/test.yml and the results are here https://github.com/username_1/botman/actions/runs/237445099 which is always connection refused (sadly)
if anyone can test it locally at his computer and share his finding that would be helpful too.
username_1: I updated my workflow to install redis-cli to verify that redis is working and can accept connections https://github.com/username_1/botman/runs/1078750458?check_suite_focus=true, but I still got `connections refused` from the code.
username_1: @username_0 Done everything is green https://github.com/username_1/botman/actions/runs/242493405
But these are the things I have found and the recommendation we need to do:
1. The `teardown` function was missing the port which caused a connection refused whenever we finished a function.
2. $_ENV is always empty due to the `variables_order` default values in github actions, so we needed to change that in the `Setup PHP` step, or maybe we should use dotenv package for development.
3. We need to split the authenticated tests from the one without authentication **or** add a function to skip the functions based on a variable called `REDIS_SECURE` like I did in my modifications.
Please review my code and if there is any question just let me know, my solution needs a refactoring from your end for sure but now we have a green tick as a start. |
GCTC-NTGC/TalentCloud | 334216867 | Title: All Browsers - Image Requests That Don't Exist
Question:
username_0: # Description
The site is trying to load images that don't exist, which result in JavaScript 404 errors in the console.
# Steps to Reproduce
Steps to reproduce the behavior:
1. Load a page.
2. Check the console.<issue_closed>
Status: Issue closed |
mscdex/ssh2 | 179264257 | Title: Handle connection hopping to unreachable device
Question:
username_0: In your readme there's an example for connection hopping that executes the netcat command to open a connection to another device. I'm struggling to find a way to handle the case where the second device is unavailable. So for example, in your example below, how do I handle the case where `192.168.1.2` is unreachable. Right now it will just hang.
```javascript
conn1.exec('nc 192.168.1.2 22', function(err, stream) {
if (err) {
console.log('FIRST :: exec error: ' + err);
return conn1.end();
}
conn2.connect({
sock: stream,
username: 'user2',
password: '<PASSWORD>',
});
});
````
Answers:
username_1: I don't think netcat has an option for *just* initial connection timeout.
Another alternative (if the server allows it) is to use `conn1.forwardOut()` which will create an outbound connection using ssh directly. Then pass the resulting stream (if no error) to `conn2.connect()`.
username_0: Thank you for your quick response! This doesn't seem to work. I get an error after I change exec to forwardOut. This is my actual example:
```
var Client = require('ssh2').Client;
var conn1 = new Client(),
conn2 = new Client();
conn1.on('ready', function() {
console.log('FIRST :: connection ready');
conn1.forwardOut('nc 192.168.20.113 22', function(err, stream) {
if (err) {
console.log('FIRST :: exec error: ' + err);
return conn1.end();
}
conn2.connect({
sock: stream,
username: 'root',
password: '<PASSWORD>'
});
});
}).connect({
host: '10.101.139.200',
username: 'root',
password: '<PASSWORD>'
});
conn2.on('ready', function() {
console.log('SECOND :: connection ready');
conn2.exec('uptime', function(err, stream) {
if (err) {
console.log('SECOND :: exec error: ' + err);
return conn1.end();
}
stream.on('end', function() {
conn1.end(); // close parent (and this) connection
}).on('data', function(data) {
console.log(data.toString());
});
});
});
```
It gives the following error.. Does this mean the server doesn't allow it? Why would it not allow it?
```
FIRST :: connection ready
buffer.js:771
if (string.length > 0 && (length < 0 || offset < 0))
^
TypeError: Cannot read property 'length' of undefined
at Buffer.write (buffer.js:771:13)
at SSH2Stream.directTcpip (/home/slynch/WebstormProjects/testing/node_modules/ssh2-streams/lib/ssh.js:1128:7)
at openChannel (/home/slynch/WebstormProjects/testing/node_modules/ssh2/lib/client.js:1130:21)
at Client.forwardOut (/home/slynch/WebstormProjects/testing/node_modules/ssh2/lib/client.js:982:10)
at Client.<anonymous> (/home/slynch/WebstormProjects/testing/test_connection_hopping.js:12:11)
at emitNone (events.js:86:13)
at Client.emit (events.js:185:7)
at SSH2Stream.<anonymous> (/home/slynch/WebstormProjects/testing/node_modules/ssh2/lib/client.js:589:10)
at SSH2Stream.g (events.js:286:16)
at emitNone (events.js:86:13)
```
Does this mean the server doesn't allow this?
username_1: Sorry, I did not mean you just literally replace the function name. It's a different function altogether. For example:
```js
conn1.forwardOut(
// source address, can usually be any valid IP
'127.0.0.1',
// source port, this can any valid port number
12345,
// destination address, the second ssh server in your case
'127.0.0.1',
// destination port, standard ssh port unless it is something different
22, function(err, stream) {
if (err) {
console.log('FIRST :: forwardOut error: ' + err);
return conn1.end();
}
conn2.connect({
sock: stream,
username: 'root',
password: '<PASSWORD>'
});
});
```
username_0: Ah. I didn't think about how to actually use it.
Thank you. This works great.
Status: Issue closed
|
stevewithington/MuraPlugin | 96860825 | Title: Force reload for development?
Question:
username_0: Hi Steve,
Is there a way to force a reload of the plugin on each request during development, so that Mura does not need to be reloaded for each change to the eventHandler.cfc. Something similar to the config setting in the FW/1 plugins?
Thanks,
John
Status: Issue closed
Answers:
username_1: @username_0,
I've added a new setting (https://github.com/username_1/MuraPlugin/blob/65e5db18e9acc9788d295dfbf20e23a716f3de6c/plugin/settings.cfm#L19) that allows you to tell the plugin to reload on each request in the latest version.
Cheers,
Steve
username_0: @username_1 Fantastic! Thanks so much for all of your fantastic Mura work.
username_1: @username_0 hold on ... i just noticed an issue. i'll get it fixed shortly.
username_1: Hi Steve,
Is there a way to force a reload of the plugin on each request during development, so that Mura does not need to be reloaded for each change to the eventHandler.cfc. Something similar to the config setting in the FW/1 plugins?
Thanks,
John
Status: Issue closed
username_1: @username_0, ok ... should be working now.
username_0: @username_1 thanks again! |
romanz/electrs | 919859354 | Title: Bug: electrs fee estimates break Electrum Lightning trampolines
Question:
username_0: **Describe the bug**
A clear and concise description of what the bug is.
**To Reproduce**
Steps to reproduce the behavior:
1. Configure and start electrs
2. Connect with electrum client 4.1.x
3. Try opening a channel to a trampoline
4. See error as described [here](https://github.com/spesmilo/electrum/issues/7182) due to electrs estimating much lower fees than ElectrumX or whichever server the trampoline is using
**Expected behavior**
Electrs fee estimates are similar to other electrum servers
**Configuration**
Default config
**Electrum client**
Client name (if not upstream desktop Electrum) and version:
Standard electrum
Answers:
username_1: Thanks for reporting this issue!
electrs fee estimation works by inspecting the node's mempool transaction fee bands, similar to how they are shown in https://jochen-hoenicke.de/queue/ or https://mempool.space/graphs.
The main issue here is that it's possible that your node's mempool differs from the trampoline's node - since some transactions may not be broadcasted to one of the nodes. Not sure how this can be resolved automatically though :(
username_0: Yeah, it's a mystery, but I spent a good while trying to connect to all
3 trampoline nodes (many times over several days) and managed to get a
channel opened exactly once... and it was force-closed by the other side
immediately after.
This suggests there's a statistically significant difference in how fees
are/were being calculated on my end (by electrs) vs on the trampoline's
end (by ElectrumX?).
The logical next step is probably to look at how ElectrumX calculates
fees and what the differences are to electrs. If it's just a question of
changing some parameters, the obvious fix would be an "electrumx
compatibility mode fee calculations" flag in the electrs config file.
username_2: It'd be best to figure out which implementation is better and use that one consistently everywhere. |
radiasoft/sirepo | 255673602 | Title: elegant: convert string to float: (Sdelta0)
Question:
username_0: I was poking around the logs and I happened to see this (multiple times) on alpha:
```
ValueError: could not convert string to float: (Sdelta0)
[06/Sep/2017:15:43:41 +0000] "POST /run-simulation HTTP/1.1" 500 0 "https://alpha.sirepo.com/elegant" "Mozilla/5.0 (Macintosh; Intel Mac OS X 10.12; rv:55.0) Gecko/20100101 Firefox/55.0"
```
Answers:
username_1: I was causing these errors by importing an elegant example which has RPN values in bunched_beam fields. This will be fixed shortly along with ticket #716
username_0: Sorry about that. I didn't check the IP. Just happened to notice...
Status: Issue closed
|
GeoscienceAustralia/digitalearthau | 237412135 | Title: Problems with Contiguity Bit in PQ
Question:
username_0: ### Expected behaviour
Contiguous masking bit should mask data in case of no data found.
### Actual behaviour
It is not masking data even though red, green and blue bands have no data but nir band has data.
This can be tested on LS7 dataset dated 2002/12/17
### Steps to reproduce the behaviour
This is the code that can be used to check masking bit.
```
import datacube
import fiona
from datacube.api import make_mask
from datacube.utils.geometry import CRS, GeoBox, Geometry
from datacube.utils import geometry
import numpy as np
filepath = '/g/data/r78/intertidal/GA_tidal_model.shp'
mask_spec=dict()
mask_spec['flags'] = { 'contiguous': True,
'cloud_acca': 'no_cloud',
'cloud_fmask': 'no_cloud',
'cloud_shadow_acca': 'no_cloud_shadow',
'cloud_shadow_fmask': 'no_cloud_shadow',
'blue_saturated': False,
'green_saturated': False,
'red_saturated': False,
'nir_saturated': False,
'swir1_saturated': False,
'swir2_saturated': False
}
def pq_fuser(dest, src):
valid_bit = 8
valid_val = (1 << valid_bit)
no_data_dest_mask = ~(dest & valid_val).astype(bool)
np.copyto(dest, src, where=no_data_dest_mask)
both_data_mask = (valid_val & dest & src).astype(bool)
np.copyto(dest, src & dest, where=both_data_mask)
dc=datacube.Datacube(app='test')
with fiona.open(filepath) as Input:
crs = CRS(str(Input.crs_wkt))
for feature in Input:
if feature['properties']['ID'] == 136:
geom=feature['geometry']
boundary_polygon = Geometry(geom, crs)
#print (feature)
break
dt1='2002-12-17 00:00:00'
dt2='2002-12-17 23:59:00'
indexers={'geopolygon': boundary_polygon, 'group_by':'solar_day'}
pq = dc.load(product='ls7_pq_albers', time=(dt1, dt2), fuse_func=pq_fuser, **indexers)
data = dc.load(product='ls7_nbar_albers', time=(dt1, dt2), measurements=['red', 'green', 'blue', 'nir'], **indexers)
mask = make_mask(pq, **mask_spec['flags'])
data = data.where(mask['pixelquality'])
print (data.green)
print (data.nir)
```
### Environment information
* Which ``datacube --version`` are you using?
agdc-py3-prod
Answers:
username_1: An artefact demonstrated a few years ago, that was more prevalent in Landsat 7 due to the SLC-OFF. Even though nearest neighbour resampling was used, it didn't guarantee the preservation of a data value for categorical layers.
NBAR(T) are reprojected independently to PQ, but have the advantage of a defined NULL value to enforce the reprojection algorithm to preserve NULL's. PQ doesn't have this advantage, hence some differences have been observed.
username_1: PQ is only there as a prototype and not likely to persist in future collections, without being redesigned.
Status: Issue closed
|
ScottishCovidResponse/SCRCIssueTracking | 653947036 | Title: Clean up error handling
Question:
username_0: At the moment, errors are typically reported via emsg, and the code aborts, or result in uncaught exceptions which produce an ugly non-friendly error. This can sometimes make it hard to identify context.
Do we want a top-level exception handler in main()? Ideally, problems caused by bad user input or numerical issues with the simulation would be reported either with emsg with a user-friendly error message, or caught by exception handlers and turned into user-friendly errors. For problems which are bugs in the code, we probably want the full ugly exception report that appears by default, to aid in debugging (a stack trace would also be great, but I've had endless issues with getting these reliably at runtime without using a debugger). Eventually there shouldn't be any of these, anyway. Thoughts? |
json-editor/json-editor | 366984203 | Title: Json Editor used to build Json Schema
Question:
username_0: Hello.
I know this is irregular but I'm sure some time ago I saw an interesting implementation of json-editor that was used to build json-schema that could be loaded into json-editor.
I can't find this "json-schema builder" based on json-editor. Do you know if I imagined it or if it exists?
It looked like the online editor, but you could add and remove different types of fields that would in turn generate the json schema that would replicate the form afterwards.
Answers:
username_1: One of these?
https://niebert.github.io/JSON2Schema/
http://json.bootsphp.com/jsonSchemaCreator/
Status: Issue closed
username_1: Added info to Wiki.
username_0: Hi. Just got around to checking this out today.
I don't think this covers what I was looking for.
Essentially I was looking for a web that allowes the creation of the form itself, and the output was the final JSON Schema. I'm sure I used such a tool at some point, but I can't find it any more.
The two links above are nice in that the first one converts a JSON object into a probable JSON Schema one and the second one allows interactive creation by hand of the Schema.
What I was sure I had used was an interactive form that allowed the creation of the final form (by using a form that allowed the addition of fields, checkboxes, etc.) and get the final JSON Schema at the end. |
benblack86/linkclump | 695355787 | Title: Support for raw text
Question:
username_0: Would it be possible to add support for analyzing the text inside the selection box and open raw text links as well?
+ Linkclump version: 2.9.1
+ Operating system: Windows
+ Actions you have setup (screen shoot of options page is fine): not relevant in this case |
JasperFx/jasper | 217351308 | Title: Some way to use/analyze/subscribe to the built in error queues
Question:
username_0: We have the concept of a dead letter queue, but we don't do anything with it. Do some analysis and figure out something more useful to do with it. If nothing else, keep it from growing forever
Status: Issue closed
Answers:
username_0: Duplicate w/ #37 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.