text
stringlengths 7
328k
| id
stringlengths 14
166
| metadata
dict | __index_level_0__
int64 0
459
|
|---|---|---|---|
# coding=utf-8
# Copyright 2018 the HuggingFace Inc. team.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import copy
import unittest
import numpy as np
from transformers.data.data_collator import default_data_collator
from transformers.testing_utils import require_accelerate, require_torch
from transformers.trainer_utils import RemoveColumnsCollator, find_executable_batch_size
from transformers.utils import is_torch_available
if is_torch_available():
import torch
from torch import nn
from torch.utils.data import IterableDataset
from transformers.modeling_outputs import SequenceClassifierOutput
from transformers.tokenization_utils_base import BatchEncoding
from transformers.trainer_pt_utils import (
DistributedLengthGroupedSampler,
DistributedSamplerWithLoop,
DistributedTensorGatherer,
IterableDatasetShard,
LabelSmoother,
LengthGroupedSampler,
SequentialDistributedSampler,
ShardSampler,
get_parameter_names,
numpy_pad_and_concatenate,
torch_pad_and_concatenate,
)
class TstLayer(nn.Module):
def __init__(self, hidden_size):
super().__init__()
self.linear1 = nn.Linear(hidden_size, hidden_size)
self.ln1 = nn.LayerNorm(hidden_size)
self.linear2 = nn.Linear(hidden_size, hidden_size)
self.ln2 = nn.LayerNorm(hidden_size)
self.bias = nn.Parameter(torch.zeros(hidden_size))
def forward(self, x):
h = self.ln1(nn.functional.relu(self.linear1(x)))
h = nn.functional.relu(self.linear2(x))
return self.ln2(x + h + self.bias)
class RandomIterableDataset(IterableDataset):
# For testing, an iterable dataset of random length
def __init__(self, p_stop=0.01, max_length=1000):
self.p_stop = p_stop
self.max_length = max_length
self.generator = torch.Generator()
def __iter__(self):
count = 0
stop = False
while not stop and count < self.max_length:
yield count
count += 1
number = torch.rand(1, generator=self.generator).item()
stop = number < self.p_stop
@require_torch
class TrainerUtilsTest(unittest.TestCase):
def test_distributed_tensor_gatherer(self):
# Simulate a result with a dataset of size 21, 4 processes and chunks of lengths 2, 3, 1
world_size = 4
num_samples = 21
input_indices = [
[0, 1, 6, 7, 12, 13, 18, 19],
[2, 3, 4, 8, 9, 10, 14, 15, 16, 20, 0, 1],
[5, 11, 17, 2],
]
predictions = np.random.normal(size=(num_samples, 13))
gatherer = DistributedTensorGatherer(world_size=world_size, num_samples=num_samples)
for indices in input_indices:
gatherer.add_arrays(predictions[indices])
result = gatherer.finalize()
self.assertTrue(np.array_equal(result, predictions))
# With nested tensors
gatherer = DistributedTensorGatherer(world_size=world_size, num_samples=num_samples)
for indices in input_indices:
gatherer.add_arrays([predictions[indices], [predictions[indices], predictions[indices]]])
result = gatherer.finalize()
self.assertTrue(isinstance(result, list))
self.assertEqual(len(result), 2)
self.assertTrue(isinstance(result[1], list))
self.assertEqual(len(result[1]), 2)
self.assertTrue(np.array_equal(result[0], predictions))
self.assertTrue(np.array_equal(result[1][0], predictions))
self.assertTrue(np.array_equal(result[1][1], predictions))
def test_distributed_tensor_gatherer_different_shapes(self):
# Simulate a result with a dataset of size 21, 4 processes and chunks of lengths 2, 3, 1
world_size = 4
num_samples = 21
input_indices = [
[0, 1, 6, 7, 12, 13, 18, 19],
[2, 3, 4, 8, 9, 10, 14, 15, 16, 20, 0, 1],
[5, 11, 17, 2],
]
sequence_lengths = [8, 10, 13]
predictions = np.random.normal(size=(num_samples, 13))
gatherer = DistributedTensorGatherer(world_size=world_size, num_samples=num_samples)
for indices, seq_length in zip(input_indices, sequence_lengths):
gatherer.add_arrays(predictions[indices, :seq_length])
result = gatherer.finalize()
# Remove the extra samples added at the end for a round multiple of num processes.
actual_indices = [input_indices[0], input_indices[1][:-2], input_indices[2][:-1]]
for indices, seq_length in zip(actual_indices, sequence_lengths):
self.assertTrue(np.array_equal(result[indices, :seq_length], predictions[indices, :seq_length]))
# With nested tensors
predictions = np.random.normal(size=(num_samples, 13))
gatherer = DistributedTensorGatherer(world_size=world_size, num_samples=num_samples)
for indices, seq_length in zip(input_indices, sequence_lengths):
gatherer.add_arrays([predictions[indices, :seq_length], predictions[indices]])
result = gatherer.finalize()
for indices, seq_length in zip(actual_indices, sequence_lengths):
self.assertTrue(np.array_equal(result[0][indices, :seq_length], predictions[indices, :seq_length]))
self.assertTrue(np.array_equal(result[1], predictions))
# Check if works if varying seq_length is second
gatherer = DistributedTensorGatherer(world_size=world_size, num_samples=num_samples)
for indices, seq_length in zip(input_indices, sequence_lengths):
gatherer.add_arrays([predictions[indices], predictions[indices, :seq_length]])
result = gatherer.finalize()
self.assertTrue(np.array_equal(result[0], predictions))
for indices, seq_length in zip(actual_indices, sequence_lengths):
self.assertTrue(np.array_equal(result[1][indices, :seq_length], predictions[indices, :seq_length]))
def test_label_smoothing(self):
epsilon = 0.1
num_labels = 12
random_logits = torch.randn(4, 5, num_labels)
random_labels = torch.randint(0, num_labels, (4, 5))
loss = nn.functional.cross_entropy(random_logits.view(-1, num_labels), random_labels.view(-1))
model_output = SequenceClassifierOutput(logits=random_logits)
label_smoothed_loss = LabelSmoother(0.1)(model_output, random_labels)
log_probs = -nn.functional.log_softmax(random_logits, dim=-1)
expected_loss = (1 - epsilon) * loss + epsilon * log_probs.mean()
self.assertTrue(torch.allclose(label_smoothed_loss, expected_loss))
# With a few -100 labels
random_labels[0, 1] = -100
random_labels[2, 1] = -100
random_labels[2, 3] = -100
loss = nn.functional.cross_entropy(random_logits.view(-1, num_labels), random_labels.view(-1))
model_output = SequenceClassifierOutput(logits=random_logits)
label_smoothed_loss = LabelSmoother(0.1)(model_output, random_labels)
log_probs = -nn.functional.log_softmax(random_logits, dim=-1)
# Mask the log probs with the -100 labels
log_probs[0, 1] = 0.0
log_probs[2, 1] = 0.0
log_probs[2, 3] = 0.0
expected_loss = (1 - epsilon) * loss + epsilon * log_probs.sum() / (num_labels * 17)
self.assertTrue(torch.allclose(label_smoothed_loss, expected_loss))
def test_group_by_length(self):
# Get some inputs of random lengths
lengths = torch.randint(0, 25, (100,)).tolist()
# Put one bigger than the others to check it ends up in first position
lengths[32] = 50
indices = list(LengthGroupedSampler(4, lengths=lengths))
# The biggest element should be first
self.assertEqual(lengths[indices[0]], 50)
# The indices should be a permutation of range(100)
self.assertEqual(sorted(indices), list(range(100)))
def test_group_by_length_with_dict(self):
# Get some inputs of random lengths
data = []
for _ in range(6):
input_ids = torch.randint(0, 25, (100,)).tolist()
data.append({"input_ids": input_ids})
# Put one bigger than the others to check it ends up in first position
data[3]["input_ids"] = torch.randint(0, 25, (105,)).tolist()
indices = list(LengthGroupedSampler(4, dataset=data))
# The biggest element should be first
self.assertEqual(len(data[indices[0]]["input_ids"]), 105)
# The indices should be a permutation of range(6)
self.assertEqual(sorted(indices), list(range(6)))
def test_group_by_length_with_batch_encoding(self):
# Get some inputs of random lengths
data = []
for _ in range(6):
input_ids = torch.randint(0, 25, (100,)).tolist()
data.append(BatchEncoding({"input_ids": input_ids}))
# Put one bigger than the others to check it ends up in first position
data[3]["input_ids"] = torch.randint(0, 25, (105,)).tolist()
indices = list(LengthGroupedSampler(4, dataset=data))
# The biggest element should be first
self.assertEqual(len(data[indices[0]]["input_ids"]), 105)
# The indices should be a permutation of range(6)
self.assertEqual(sorted(indices), list(range(6)))
def test_distributed_length_grouped(self):
# Get some inputs of random lengths
lengths = torch.randint(0, 25, (100,)).tolist()
# Put one bigger than the others to check it ends up in first position
lengths[32] = 50
indices_process_0 = list(DistributedLengthGroupedSampler(4, num_replicas=2, rank=0, lengths=lengths))
indices_process_1 = list(DistributedLengthGroupedSampler(4, num_replicas=2, rank=1, lengths=lengths))
# The biggest element should be first
self.assertEqual(lengths[indices_process_0[0]], 50)
# The indices should be a permutation of range(100)
self.assertEqual(sorted(indices_process_0 + indices_process_1), list(range(100)))
def test_get_parameter_names(self):
model = nn.Sequential(TstLayer(128), nn.ModuleList([TstLayer(128), TstLayer(128)]))
# fmt: off
self.assertEqual(
get_parameter_names(model, [nn.LayerNorm]),
['0.linear1.weight', '0.linear1.bias', '0.linear2.weight', '0.linear2.bias', '0.bias', '1.0.linear1.weight', '1.0.linear1.bias', '1.0.linear2.weight', '1.0.linear2.bias', '1.0.bias', '1.1.linear1.weight', '1.1.linear1.bias', '1.1.linear2.weight', '1.1.linear2.bias', '1.1.bias']
)
# fmt: on
def test_distributed_sampler_with_loop(self):
batch_size = 16
for length in [23, 64, 123]:
dataset = list(range(length))
shard1 = DistributedSamplerWithLoop(dataset, batch_size, num_replicas=2, rank=0)
shard2 = DistributedSamplerWithLoop(dataset, batch_size, num_replicas=2, rank=1)
# Set seeds
shard1.set_epoch(0)
shard2.set_epoch(0)
# Sample
samples1 = list(shard1)
samples2 = list(shard2)
self.assertTrue(len(samples1) % batch_size == 0)
self.assertTrue(len(samples2) % batch_size == 0)
total = []
for sample1, sample2 in zip(samples1, samples2):
total += [sample1, sample2]
self.assertEqual(set(total[:length]), set(dataset))
self.assertEqual(set(total[length:]), set(total[: (len(total) - length)]))
def test_sequential_distributed_sampler(self):
batch_size = 16
for length in [23, 64, 123]:
dataset = list(range(length))
shard1 = SequentialDistributedSampler(dataset, num_replicas=2, rank=0)
shard2 = SequentialDistributedSampler(dataset, num_replicas=2, rank=1)
# Sample
samples1 = list(shard1)
samples2 = list(shard2)
total = samples1 + samples2
self.assertListEqual(total[:length], dataset)
self.assertListEqual(total[length:], dataset[: (len(total) - length)])
# With a batch_size passed
shard1 = SequentialDistributedSampler(dataset, num_replicas=2, rank=0, batch_size=batch_size)
shard2 = SequentialDistributedSampler(dataset, num_replicas=2, rank=1, batch_size=batch_size)
# Sample
samples1 = list(shard1)
samples2 = list(shard2)
self.assertTrue(len(samples1) % batch_size == 0)
self.assertTrue(len(samples2) % batch_size == 0)
total = samples1 + samples2
self.assertListEqual(total[:length], dataset)
self.assertListEqual(total[length:], dataset[: (len(total) - length)])
def check_iterable_dataset_shard(self, dataset, batch_size, drop_last, num_processes=2, epoch=0):
# Set the seed for the base dataset to get the proper reference.
dataset.generator.manual_seed(epoch)
reference = list(dataset)
shards = [
IterableDatasetShard(
dataset, batch_size=batch_size, drop_last=drop_last, num_processes=num_processes, process_index=i
)
for i in range(num_processes)
]
for shard in shards:
shard.set_epoch(epoch)
shard_lists = [list(shard) for shard in shards]
for shard in shard_lists:
# All shards have a number of samples that is a round multiple of batch size
self.assertTrue(len(shard) % batch_size == 0)
# All shards have the same number of samples
self.assertEqual(len(shard), len(shard_lists[0]))
for shard in shards:
# All shards know the total number of samples
self.assertEqual(shard.num_examples, len(reference))
observed = []
for idx in range(0, len(shard_lists[0]), batch_size):
for shard in shard_lists:
observed += shard[idx : idx + batch_size]
# If drop_last is False we loop through samples at the beginning to have a size that is a round multiple of
# batch_size
if not drop_last:
while len(reference) < len(observed):
reference += reference
self.assertListEqual(observed, reference[: len(observed)])
# Check equivalence between IterableDataset and ShardSampler
dataset.generator.manual_seed(epoch)
reference = list(dataset)
sampler_shards = [
ShardSampler(
reference, batch_size=batch_size, drop_last=drop_last, num_processes=num_processes, process_index=i
)
for i in range(num_processes)
]
for shard, sampler_shard in zip(shard_lists, sampler_shards):
self.assertListEqual(shard, list(sampler_shard))
def test_iterable_dataset_shard(self):
dataset = RandomIterableDataset()
self.check_iterable_dataset_shard(dataset, 4, drop_last=True, num_processes=2, epoch=0)
self.check_iterable_dataset_shard(dataset, 4, drop_last=False, num_processes=2, epoch=0)
self.check_iterable_dataset_shard(dataset, 4, drop_last=True, num_processes=3, epoch=42)
self.check_iterable_dataset_shard(dataset, 4, drop_last=False, num_processes=3, epoch=42)
def test_iterable_dataset_shard_with_length(self):
sampler_shards = [
IterableDatasetShard(list(range(100)), batch_size=4, drop_last=True, num_processes=2, process_index=i)
for i in range(2)
]
# Build expected shards: each process will have batches of size 4 until there is not enough elements to
# form two full batches (so we stop at 96 = (100 // (4 * 2)) * 4)
expected_shards = [[], []]
current_shard = 0
for i in range(0, 96, 4):
expected_shards[current_shard].extend(list(range(i, i + 4)))
current_shard = 1 - current_shard
self.assertListEqual([list(shard) for shard in sampler_shards], expected_shards)
self.assertListEqual([len(shard) for shard in sampler_shards], [len(shard) for shard in expected_shards])
sampler_shards = [
IterableDatasetShard(list(range(100)), batch_size=4, drop_last=False, num_processes=2, process_index=i)
for i in range(2)
]
# When drop_last=False, we get two last full batches by looping back to the beginning.
expected_shards[0].extend(list(range(96, 100)))
expected_shards[1].extend(list(range(0, 4)))
self.assertListEqual([list(shard) for shard in sampler_shards], expected_shards)
self.assertListEqual([len(shard) for shard in sampler_shards], [len(shard) for shard in expected_shards])
def check_shard_sampler(self, dataset, batch_size, drop_last, num_processes=2):
shards = [
ShardSampler(
dataset, batch_size=batch_size, drop_last=drop_last, num_processes=num_processes, process_index=i
)
for i in range(num_processes)
]
shard_lists = [list(shard) for shard in shards]
for shard in shard_lists:
# All shards have a number of samples that is a round multiple of batch size
self.assertTrue(len(shard) % batch_size == 0)
# All shards have the same number of samples
self.assertEqual(len(shard), len(shard_lists[0]))
observed = []
for idx in range(0, len(shard_lists[0]), batch_size):
for shard in shard_lists:
observed += shard[idx : idx + batch_size]
# If drop_last is False we loop through samples at the beginning to have a size that is a round multiple of
# batch_size
reference = copy.copy(dataset)
if not drop_last:
while len(reference) < len(observed):
reference += reference
self.assertListEqual(observed, reference[: len(observed)])
def test_shard_sampler(self):
for n_elements in [64, 123]:
dataset = list(range(n_elements))
self.check_shard_sampler(dataset, 4, drop_last=True, num_processes=2)
self.check_shard_sampler(dataset, 4, drop_last=False, num_processes=2)
self.check_shard_sampler(dataset, 4, drop_last=True, num_processes=3)
self.check_shard_sampler(dataset, 4, drop_last=False, num_processes=3)
@require_accelerate
def test_executable_batch_size(self):
batch_sizes = []
@find_executable_batch_size(starting_batch_size=64, auto_find_batch_size=True)
def mock_training_loop_function(batch_size):
nonlocal batch_sizes
batch_sizes.append(batch_size)
if batch_size > 16:
raise RuntimeError("CUDA out of memory.")
mock_training_loop_function()
self.assertEqual(batch_sizes, [64, 32, 16])
@require_accelerate
def test_executable_batch_size_no_search(self):
batch_sizes = []
@find_executable_batch_size(starting_batch_size=64, auto_find_batch_size=False)
def mock_training_loop_function(batch_size):
nonlocal batch_sizes
batch_sizes.append(batch_size)
mock_training_loop_function()
self.assertEqual(batch_sizes, [64])
@require_accelerate
def test_executable_batch_size_with_error(self):
@find_executable_batch_size(starting_batch_size=64, auto_find_batch_size=False)
def mock_training_loop_function(batch_size):
raise RuntimeError("CUDA out of memory.")
with self.assertRaises(RuntimeError) as cm:
mock_training_loop_function()
self.assertEqual("CUDA out of memory", cm.args[0])
def test_pad_and_concatenate_with_1d(self):
"""Tests whether pad_and_concatenate works with scalars."""
array1 = 1.0
array2 = 2.0
result = numpy_pad_and_concatenate(array1, array2)
self.assertTrue(np.array_equal(np.array([1.0, 2.0]), result))
tensor1 = torch.tensor(1.0)
tensor2 = torch.tensor(2.0)
result = torch_pad_and_concatenate(tensor1, tensor2)
self.assertTrue(torch.equal(result, torch.Tensor([1.0, 2.0])))
def test_remove_columns_collator(self):
class MockLogger:
def __init__(self) -> None:
self.called = 0
def info(self, msg):
self.called += 1
self.last_msg = msg
data_batch = [
{"col1": 1, "col2": 2, "col3": 3},
{"col1": 1, "col2": 2, "col3": 3},
]
logger = MockLogger()
remove_columns_collator = RemoveColumnsCollator(
default_data_collator, ["col1", "col2"], logger, "model", "training"
)
self.assertNotIn("col3", remove_columns_collator(data_batch))
# check that the logging message is printed out only once
remove_columns_collator(data_batch)
remove_columns_collator(data_batch)
self.assertEqual(logger.called, 1)
self.assertIn("col3", logger.last_msg)
| transformers/tests/trainer/test_trainer_utils.py/0 | {
"file_path": "transformers/tests/trainer/test_trainer_utils.py",
"repo_id": "transformers",
"token_count": 9624
} | 399 |
# coding=utf-8
# Copyright 2021 HuggingFace Inc.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import os
import tempfile
import unittest
import datasets
import numpy as np
import pytest
from huggingface_hub.file_download import http_get
from requests import ConnectTimeout, ReadTimeout
from tests.pipelines.test_pipelines_document_question_answering import INVOICE_URL
from transformers import is_torch_available, is_vision_available
from transformers.image_utils import ChannelDimension, get_channel_dimension_axis, make_list_of_images
from transformers.testing_utils import is_flaky, require_torch, require_vision
if is_torch_available():
import torch
if is_vision_available():
import PIL.Image
from transformers import ImageFeatureExtractionMixin
from transformers.image_utils import get_image_size, infer_channel_dimension_format, load_image
def get_random_image(height, width):
random_array = np.random.randint(0, 256, (height, width, 3), dtype=np.uint8)
return PIL.Image.fromarray(random_array)
@require_vision
class ImageFeatureExtractionTester(unittest.TestCase):
def test_conversion_image_to_array(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
# Conversion with defaults (rescale + channel first)
array1 = feature_extractor.to_numpy_array(image)
self.assertTrue(array1.dtype, np.float32)
self.assertEqual(array1.shape, (3, 16, 32))
# Conversion with rescale and not channel first
array2 = feature_extractor.to_numpy_array(image, channel_first=False)
self.assertTrue(array2.dtype, np.float32)
self.assertEqual(array2.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array1, array2.transpose(2, 0, 1)))
# Conversion with no rescale and channel first
array3 = feature_extractor.to_numpy_array(image, rescale=False)
self.assertTrue(array3.dtype, np.uint8)
self.assertEqual(array3.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array1, array3.astype(np.float32) * (1 / 255.0)))
# Conversion with no rescale and not channel first
array4 = feature_extractor.to_numpy_array(image, rescale=False, channel_first=False)
self.assertTrue(array4.dtype, np.uint8)
self.assertEqual(array4.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array2, array4.astype(np.float32) * (1 / 255.0)))
def test_conversion_array_to_array(self):
feature_extractor = ImageFeatureExtractionMixin()
array = np.random.randint(0, 256, (16, 32, 3), dtype=np.uint8)
# By default, rescale (for an array of ints) and channel permute
array1 = feature_extractor.to_numpy_array(array)
self.assertTrue(array1.dtype, np.float32)
self.assertEqual(array1.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array1, array.transpose(2, 0, 1).astype(np.float32) * (1 / 255.0)))
# Same with no permute
array2 = feature_extractor.to_numpy_array(array, channel_first=False)
self.assertTrue(array2.dtype, np.float32)
self.assertEqual(array2.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array2, array.astype(np.float32) * (1 / 255.0)))
# Force rescale to False
array3 = feature_extractor.to_numpy_array(array, rescale=False)
self.assertTrue(array3.dtype, np.uint8)
self.assertEqual(array3.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array3, array.transpose(2, 0, 1)))
# Force rescale to False and no channel permute
array4 = feature_extractor.to_numpy_array(array, rescale=False, channel_first=False)
self.assertTrue(array4.dtype, np.uint8)
self.assertEqual(array4.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array4, array))
# Now test the default rescale for a float array (defaults to False)
array5 = feature_extractor.to_numpy_array(array2)
self.assertTrue(array5.dtype, np.float32)
self.assertEqual(array5.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array5, array1))
def test_make_list_of_images_numpy(self):
# Test a single image is converted to a list of 1 image
images = np.random.randint(0, 256, (16, 32, 3))
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 1)
self.assertTrue(np.array_equal(images_list[0], images))
self.assertIsInstance(images_list, list)
# Test a batch of images is converted to a list of images
images = np.random.randint(0, 256, (4, 16, 32, 3))
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 4)
self.assertTrue(np.array_equal(images_list[0], images[0]))
self.assertIsInstance(images_list, list)
# Test a list of images is not modified
images = [np.random.randint(0, 256, (16, 32, 3)) for _ in range(4)]
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 4)
self.assertTrue(np.array_equal(images_list[0], images[0]))
self.assertIsInstance(images_list, list)
# Test batched masks with no channel dimension are converted to a list of masks
masks = np.random.randint(0, 2, (4, 16, 32))
masks_list = make_list_of_images(masks, expected_ndims=2)
self.assertEqual(len(masks_list), 4)
self.assertTrue(np.array_equal(masks_list[0], masks[0]))
self.assertIsInstance(masks_list, list)
@require_torch
def test_make_list_of_images_torch(self):
# Test a single image is converted to a list of 1 image
images = torch.randint(0, 256, (16, 32, 3))
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 1)
self.assertTrue(np.array_equal(images_list[0], images))
self.assertIsInstance(images_list, list)
# Test a batch of images is converted to a list of images
images = torch.randint(0, 256, (4, 16, 32, 3))
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 4)
self.assertTrue(np.array_equal(images_list[0], images[0]))
self.assertIsInstance(images_list, list)
# Test a list of images is left unchanged
images = [torch.randint(0, 256, (16, 32, 3)) for _ in range(4)]
images_list = make_list_of_images(images)
self.assertEqual(len(images_list), 4)
self.assertTrue(np.array_equal(images_list[0], images[0]))
self.assertIsInstance(images_list, list)
@require_torch
def test_conversion_torch_to_array(self):
feature_extractor = ImageFeatureExtractionMixin()
tensor = torch.randint(0, 256, (16, 32, 3))
array = tensor.numpy()
# By default, rescale (for a tensor of ints) and channel permute
array1 = feature_extractor.to_numpy_array(array)
self.assertTrue(array1.dtype, np.float32)
self.assertEqual(array1.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array1, array.transpose(2, 0, 1).astype(np.float32) * (1 / 255.0)))
# Same with no permute
array2 = feature_extractor.to_numpy_array(array, channel_first=False)
self.assertTrue(array2.dtype, np.float32)
self.assertEqual(array2.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array2, array.astype(np.float32) * (1 / 255.0)))
# Force rescale to False
array3 = feature_extractor.to_numpy_array(array, rescale=False)
self.assertTrue(array3.dtype, np.uint8)
self.assertEqual(array3.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array3, array.transpose(2, 0, 1)))
# Force rescale to False and no channel permute
array4 = feature_extractor.to_numpy_array(array, rescale=False, channel_first=False)
self.assertTrue(array4.dtype, np.uint8)
self.assertEqual(array4.shape, (16, 32, 3))
self.assertTrue(np.array_equal(array4, array))
# Now test the default rescale for a float tensor (defaults to False)
array5 = feature_extractor.to_numpy_array(array2)
self.assertTrue(array5.dtype, np.float32)
self.assertEqual(array5.shape, (3, 16, 32))
self.assertTrue(np.array_equal(array5, array1))
def test_conversion_image_to_image(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
# On an image, `to_pil_image1` is a noop.
image1 = feature_extractor.to_pil_image(image)
self.assertTrue(isinstance(image, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image), np.array(image1)))
def test_conversion_array_to_image(self):
feature_extractor = ImageFeatureExtractionMixin()
array = np.random.randint(0, 256, (16, 32, 3), dtype=np.uint8)
# By default, no rescale (for an array of ints)
image1 = feature_extractor.to_pil_image(array)
self.assertTrue(isinstance(image1, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image1), array))
# If the array is channel-first, proper reordering of the channels is done.
image2 = feature_extractor.to_pil_image(array.transpose(2, 0, 1))
self.assertTrue(isinstance(image2, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image2), array))
# If the array has floating type, it's rescaled by default.
image3 = feature_extractor.to_pil_image(array.astype(np.float32) * (1 / 255.0))
self.assertTrue(isinstance(image3, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image3), array))
# You can override the default to rescale.
image4 = feature_extractor.to_pil_image(array.astype(np.float32), rescale=False)
self.assertTrue(isinstance(image4, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image4), array))
# And with floats + channel first.
image5 = feature_extractor.to_pil_image(array.transpose(2, 0, 1).astype(np.float32) * (1 / 255.0))
self.assertTrue(isinstance(image5, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image5), array))
@require_torch
def test_conversion_tensor_to_image(self):
feature_extractor = ImageFeatureExtractionMixin()
tensor = torch.randint(0, 256, (16, 32, 3))
array = tensor.numpy()
# By default, no rescale (for a tensor of ints)
image1 = feature_extractor.to_pil_image(tensor)
self.assertTrue(isinstance(image1, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image1), array))
# If the tensor is channel-first, proper reordering of the channels is done.
image2 = feature_extractor.to_pil_image(tensor.permute(2, 0, 1))
self.assertTrue(isinstance(image2, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image2), array))
# If the tensor has floating type, it's rescaled by default.
image3 = feature_extractor.to_pil_image(tensor.float() / 255.0)
self.assertTrue(isinstance(image3, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image3), array))
# You can override the default to rescale.
image4 = feature_extractor.to_pil_image(tensor.float(), rescale=False)
self.assertTrue(isinstance(image4, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image4), array))
# And with floats + channel first.
image5 = feature_extractor.to_pil_image(tensor.permute(2, 0, 1).float() * (1 / 255.0))
self.assertTrue(isinstance(image5, PIL.Image.Image))
self.assertTrue(np.array_equal(np.array(image5), array))
def test_resize_image_and_array(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
array = np.array(image)
# Size can be an int or a tuple of ints.
resized_image = feature_extractor.resize(image, 8)
self.assertTrue(isinstance(resized_image, PIL.Image.Image))
self.assertEqual(resized_image.size, (8, 8))
resized_image1 = feature_extractor.resize(image, (8, 16))
self.assertTrue(isinstance(resized_image1, PIL.Image.Image))
self.assertEqual(resized_image1.size, (8, 16))
# Passing an array converts it to a PIL Image.
resized_image2 = feature_extractor.resize(array, 8)
self.assertTrue(isinstance(resized_image2, PIL.Image.Image))
self.assertEqual(resized_image2.size, (8, 8))
self.assertTrue(np.array_equal(np.array(resized_image), np.array(resized_image2)))
resized_image3 = feature_extractor.resize(image, (8, 16))
self.assertTrue(isinstance(resized_image3, PIL.Image.Image))
self.assertEqual(resized_image3.size, (8, 16))
self.assertTrue(np.array_equal(np.array(resized_image1), np.array(resized_image3)))
def test_resize_image_and_array_non_default_to_square(self):
feature_extractor = ImageFeatureExtractionMixin()
heights_widths = [
# height, width
# square image
(28, 28),
(27, 27),
# rectangular image: h < w
(28, 34),
(29, 35),
# rectangular image: h > w
(34, 28),
(35, 29),
]
# single integer or single integer in tuple/list
sizes = [22, 27, 28, 36, [22], (27,)]
for (height, width), size in zip(heights_widths, sizes):
for max_size in (None, 37, 1000):
image = get_random_image(height, width)
array = np.array(image)
size = size[0] if isinstance(size, (list, tuple)) else size
# Size can be an int or a tuple of ints.
# If size is an int, smaller edge of the image will be matched to this number.
# i.e, if height > width, then image will be rescaled to (size * height / width, size).
if height < width:
exp_w, exp_h = (int(size * width / height), size)
if max_size is not None and max_size < exp_w:
exp_w, exp_h = max_size, int(max_size * exp_h / exp_w)
elif width < height:
exp_w, exp_h = (size, int(size * height / width))
if max_size is not None and max_size < exp_h:
exp_w, exp_h = int(max_size * exp_w / exp_h), max_size
else:
exp_w, exp_h = (size, size)
if max_size is not None and max_size < size:
exp_w, exp_h = max_size, max_size
resized_image = feature_extractor.resize(image, size=size, default_to_square=False, max_size=max_size)
self.assertTrue(isinstance(resized_image, PIL.Image.Image))
self.assertEqual(resized_image.size, (exp_w, exp_h))
# Passing an array converts it to a PIL Image.
resized_image2 = feature_extractor.resize(array, size=size, default_to_square=False, max_size=max_size)
self.assertTrue(isinstance(resized_image2, PIL.Image.Image))
self.assertEqual(resized_image2.size, (exp_w, exp_h))
self.assertTrue(np.array_equal(np.array(resized_image), np.array(resized_image2)))
@require_torch
def test_resize_tensor(self):
feature_extractor = ImageFeatureExtractionMixin()
tensor = torch.randint(0, 256, (16, 32, 3))
array = tensor.numpy()
# Size can be an int or a tuple of ints.
resized_image = feature_extractor.resize(tensor, 8)
self.assertTrue(isinstance(resized_image, PIL.Image.Image))
self.assertEqual(resized_image.size, (8, 8))
resized_image1 = feature_extractor.resize(tensor, (8, 16))
self.assertTrue(isinstance(resized_image1, PIL.Image.Image))
self.assertEqual(resized_image1.size, (8, 16))
# Check we get the same results as with NumPy arrays.
resized_image2 = feature_extractor.resize(array, 8)
self.assertTrue(np.array_equal(np.array(resized_image), np.array(resized_image2)))
resized_image3 = feature_extractor.resize(array, (8, 16))
self.assertTrue(np.array_equal(np.array(resized_image1), np.array(resized_image3)))
def test_normalize_image(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
array = np.array(image)
mean = [0.1, 0.5, 0.9]
std = [0.2, 0.4, 0.6]
# PIL Image are converted to NumPy arrays for the normalization
normalized_image = feature_extractor.normalize(image, mean, std)
self.assertTrue(isinstance(normalized_image, np.ndarray))
self.assertEqual(normalized_image.shape, (3, 16, 32))
# During the conversion rescale and channel first will be applied.
expected = array.transpose(2, 0, 1).astype(np.float32) * (1 / 255.0)
np_mean = np.array(mean).astype(np.float32)[:, None, None]
np_std = np.array(std).astype(np.float32)[:, None, None]
expected = (expected - np_mean) / np_std
self.assertTrue(np.array_equal(normalized_image, expected))
def test_normalize_array(self):
feature_extractor = ImageFeatureExtractionMixin()
array = np.random.random((16, 32, 3))
mean = [0.1, 0.5, 0.9]
std = [0.2, 0.4, 0.6]
# mean and std can be passed as lists or NumPy arrays.
expected = (array - np.array(mean)) / np.array(std)
normalized_array = feature_extractor.normalize(array, mean, std)
self.assertTrue(np.array_equal(normalized_array, expected))
normalized_array = feature_extractor.normalize(array, np.array(mean), np.array(std))
self.assertTrue(np.array_equal(normalized_array, expected))
# Normalize will detect automatically if channel first or channel last is used.
array = np.random.random((3, 16, 32))
expected = (array - np.array(mean)[:, None, None]) / np.array(std)[:, None, None]
normalized_array = feature_extractor.normalize(array, mean, std)
self.assertTrue(np.array_equal(normalized_array, expected))
normalized_array = feature_extractor.normalize(array, np.array(mean), np.array(std))
self.assertTrue(np.array_equal(normalized_array, expected))
@require_torch
def test_normalize_tensor(self):
feature_extractor = ImageFeatureExtractionMixin()
tensor = torch.rand(16, 32, 3)
mean = [0.1, 0.5, 0.9]
std = [0.2, 0.4, 0.6]
# mean and std can be passed as lists or tensors.
expected = (tensor - torch.tensor(mean)) / torch.tensor(std)
normalized_tensor = feature_extractor.normalize(tensor, mean, std)
self.assertTrue(torch.equal(normalized_tensor, expected))
normalized_tensor = feature_extractor.normalize(tensor, torch.tensor(mean), torch.tensor(std))
self.assertTrue(torch.equal(normalized_tensor, expected))
# Normalize will detect automatically if channel first or channel last is used.
tensor = torch.rand(3, 16, 32)
expected = (tensor - torch.tensor(mean)[:, None, None]) / torch.tensor(std)[:, None, None]
normalized_tensor = feature_extractor.normalize(tensor, mean, std)
self.assertTrue(torch.equal(normalized_tensor, expected))
normalized_tensor = feature_extractor.normalize(tensor, torch.tensor(mean), torch.tensor(std))
self.assertTrue(torch.equal(normalized_tensor, expected))
def test_center_crop_image(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
# Test various crop sizes: bigger on all dimensions, on one of the dimensions only and on both dimensions.
crop_sizes = [8, (8, 64), 20, (32, 64)]
for size in crop_sizes:
cropped_image = feature_extractor.center_crop(image, size)
self.assertTrue(isinstance(cropped_image, PIL.Image.Image))
# PIL Image.size is transposed compared to NumPy or PyTorch (width first instead of height first).
expected_size = (size, size) if isinstance(size, int) else (size[1], size[0])
self.assertEqual(cropped_image.size, expected_size)
def test_center_crop_array(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
array = feature_extractor.to_numpy_array(image)
# Test various crop sizes: bigger on all dimensions, on one of the dimensions only and on both dimensions.
crop_sizes = [8, (8, 64), 20, (32, 64)]
for size in crop_sizes:
cropped_array = feature_extractor.center_crop(array, size)
self.assertTrue(isinstance(cropped_array, np.ndarray))
expected_size = (size, size) if isinstance(size, int) else size
self.assertEqual(cropped_array.shape[-2:], expected_size)
# Check result is consistent with PIL.Image.crop
cropped_image = feature_extractor.center_crop(image, size)
self.assertTrue(np.array_equal(cropped_array, feature_extractor.to_numpy_array(cropped_image)))
@require_torch
def test_center_crop_tensor(self):
feature_extractor = ImageFeatureExtractionMixin()
image = get_random_image(16, 32)
array = feature_extractor.to_numpy_array(image)
tensor = torch.tensor(array)
# Test various crop sizes: bigger on all dimensions, on one of the dimensions only and on both dimensions.
crop_sizes = [8, (8, 64), 20, (32, 64)]
for size in crop_sizes:
cropped_tensor = feature_extractor.center_crop(tensor, size)
self.assertTrue(isinstance(cropped_tensor, torch.Tensor))
expected_size = (size, size) if isinstance(size, int) else size
self.assertEqual(cropped_tensor.shape[-2:], expected_size)
# Check result is consistent with PIL.Image.crop
cropped_image = feature_extractor.center_crop(image, size)
self.assertTrue(torch.equal(cropped_tensor, torch.tensor(feature_extractor.to_numpy_array(cropped_image))))
@require_vision
class LoadImageTester(unittest.TestCase):
def test_load_img_url(self):
img = load_image(INVOICE_URL)
img_arr = np.array(img)
self.assertEqual(img_arr.shape, (1061, 750, 3))
@is_flaky()
def test_load_img_url_timeout(self):
with self.assertRaises((ReadTimeout, ConnectTimeout)):
load_image(INVOICE_URL, timeout=0.001)
def test_load_img_local(self):
img = load_image("./tests/fixtures/tests_samples/COCO/000000039769.png")
img_arr = np.array(img)
self.assertEqual(
img_arr.shape,
(480, 640, 3),
)
def test_load_img_base64_prefix(self):
try:
tmp_file = tempfile.mktemp()
with open(tmp_file, "wb") as f:
http_get(
"https://huggingface.co/datasets/hf-internal-testing/dummy-base64-images/raw/main/image_0.txt", f
)
with open(tmp_file, encoding="utf-8") as b64:
img = load_image(b64.read())
img_arr = np.array(img)
finally:
os.remove(tmp_file)
self.assertEqual(img_arr.shape, (64, 32, 3))
def test_load_img_base64(self):
try:
tmp_file = tempfile.mktemp()
with open(tmp_file, "wb") as f:
http_get(
"https://huggingface.co/datasets/hf-internal-testing/dummy-base64-images/raw/main/image_1.txt", f
)
with open(tmp_file, encoding="utf-8") as b64:
img = load_image(b64.read())
img_arr = np.array(img)
finally:
os.remove(tmp_file)
self.assertEqual(img_arr.shape, (64, 32, 3))
def test_load_img_rgba(self):
# we use revision="refs/pr/1" until the PR is merged
# https://hf.co/datasets/hf-internal-testing/fixtures_image_utils/discussions/1
dataset = datasets.load_dataset("hf-internal-testing/fixtures_image_utils", split="test", revision="refs/pr/1")
img = load_image(dataset[0]["image"]) # img with mode RGBA
img_arr = np.array(img)
self.assertEqual(
img_arr.shape,
(512, 512, 3),
)
def test_load_img_la(self):
# we use revision="refs/pr/1" until the PR is merged
# https://hf.co/datasets/hf-internal-testing/fixtures_image_utils/discussions/1
dataset = datasets.load_dataset("hf-internal-testing/fixtures_image_utils", split="test", revision="refs/pr/1")
img = load_image(dataset[1]["image"]) # img with mode LA
img_arr = np.array(img)
self.assertEqual(
img_arr.shape,
(512, 768, 3),
)
def test_load_img_l(self):
# we use revision="refs/pr/1" until the PR is merged
# https://hf.co/datasets/hf-internal-testing/fixtures_image_utils/discussions/1
dataset = datasets.load_dataset("hf-internal-testing/fixtures_image_utils", split="test", revision="refs/pr/1")
img = load_image(dataset[2]["image"]) # img with mode L
img_arr = np.array(img)
self.assertEqual(
img_arr.shape,
(381, 225, 3),
)
def test_load_img_exif_transpose(self):
# we use revision="refs/pr/1" until the PR is merged
# https://hf.co/datasets/hf-internal-testing/fixtures_image_utils/discussions/1
dataset = datasets.load_dataset("hf-internal-testing/fixtures_image_utils", split="test", revision="refs/pr/1")
img_without_exif_transpose = dataset[3]["image"]
img_arr_without_exif_transpose = np.array(img_without_exif_transpose)
self.assertEqual(
img_arr_without_exif_transpose.shape,
(333, 500, 3),
)
img_with_exif_transpose = load_image(dataset[3]["image"])
img_arr_with_exif_transpose = np.array(img_with_exif_transpose)
self.assertEqual(
img_arr_with_exif_transpose.shape,
(500, 333, 3),
)
class UtilFunctionTester(unittest.TestCase):
def test_get_image_size(self):
# Test we can infer the size and channel dimension of an image.
image = np.random.randint(0, 256, (32, 64, 3))
self.assertEqual(get_image_size(image), (32, 64))
image = np.random.randint(0, 256, (3, 32, 64))
self.assertEqual(get_image_size(image), (32, 64))
# Test the channel dimension can be overriden
image = np.random.randint(0, 256, (3, 32, 64))
self.assertEqual(get_image_size(image, channel_dim=ChannelDimension.LAST), (3, 32))
def test_infer_channel_dimension(self):
# Test we fail with invalid input
with pytest.raises(ValueError):
infer_channel_dimension_format(np.random.randint(0, 256, (10, 10)))
with pytest.raises(ValueError):
infer_channel_dimension_format(np.random.randint(0, 256, (10, 10, 10, 10, 10)))
# Test we fail if neither first not last dimension is of size 3 or 1
with pytest.raises(ValueError):
infer_channel_dimension_format(np.random.randint(0, 256, (10, 1, 50)))
# But if we explicitly set one of the number of channels to 50 it works
inferred_dim = infer_channel_dimension_format(np.random.randint(0, 256, (10, 1, 50)), num_channels=50)
self.assertEqual(inferred_dim, ChannelDimension.LAST)
# Test we correctly identify the channel dimension
image = np.random.randint(0, 256, (3, 4, 5))
inferred_dim = infer_channel_dimension_format(image)
self.assertEqual(inferred_dim, ChannelDimension.FIRST)
image = np.random.randint(0, 256, (1, 4, 5))
inferred_dim = infer_channel_dimension_format(image)
self.assertEqual(inferred_dim, ChannelDimension.FIRST)
image = np.random.randint(0, 256, (4, 5, 3))
inferred_dim = infer_channel_dimension_format(image)
self.assertEqual(inferred_dim, ChannelDimension.LAST)
image = np.random.randint(0, 256, (4, 5, 1))
inferred_dim = infer_channel_dimension_format(image)
self.assertEqual(inferred_dim, ChannelDimension.LAST)
# We can take a batched array of images and find the dimension
image = np.random.randint(0, 256, (1, 3, 4, 5))
inferred_dim = infer_channel_dimension_format(image)
self.assertEqual(inferred_dim, ChannelDimension.FIRST)
def test_get_channel_dimension_axis(self):
# Test we correctly identify the channel dimension
image = np.random.randint(0, 256, (3, 4, 5))
inferred_axis = get_channel_dimension_axis(image)
self.assertEqual(inferred_axis, 0)
image = np.random.randint(0, 256, (1, 4, 5))
inferred_axis = get_channel_dimension_axis(image)
self.assertEqual(inferred_axis, 0)
image = np.random.randint(0, 256, (4, 5, 3))
inferred_axis = get_channel_dimension_axis(image)
self.assertEqual(inferred_axis, 2)
image = np.random.randint(0, 256, (4, 5, 1))
inferred_axis = get_channel_dimension_axis(image)
self.assertEqual(inferred_axis, 2)
# We can take a batched array of images and find the dimension
image = np.random.randint(0, 256, (1, 3, 4, 5))
inferred_axis = get_channel_dimension_axis(image)
self.assertEqual(inferred_axis, 1)
| transformers/tests/utils/test_image_utils.py/0 | {
"file_path": "transformers/tests/utils/test_image_utils.py",
"repo_id": "transformers",
"token_count": 13075
} | 400 |
# coding=utf-8
# Copyright 2023 The HuggingFace Inc. team.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""
This script is responsible for cleaning the list of doctests by making sure the entries all exist and are in
alphabetical order.
Usage (from the root of the repo):
Check that the doctest list is properly sorted and all files exist (used in `make repo-consistency`):
```bash
python utils/check_doctest_list.py
```
Auto-sort the doctest list if it is not properly sorted (used in `make fix-copies`):
```bash
python utils/check_doctest_list.py --fix_and_overwrite
```
"""
import argparse
import os
# All paths are set with the intent you should run this script from the root of the repo with the command
# python utils/check_doctest_list.py
REPO_PATH = "."
DOCTEST_FILE_PATHS = ["not_doctested.txt", "slow_documentation_tests.txt"]
def clean_doctest_list(doctest_file: str, overwrite: bool = False):
"""
Cleans the doctest in a given file.
Args:
doctest_file (`str`):
The path to the doctest file to check or clean.
overwrite (`bool`, *optional*, defaults to `False`):
Whether or not to fix problems. If `False`, will error when the file is not clean.
"""
non_existent_paths = []
all_paths = []
with open(doctest_file, "r", encoding="utf-8") as f:
for line in f:
line = line.strip().split(" ")[0]
path = os.path.join(REPO_PATH, line)
if not (os.path.isfile(path) or os.path.isdir(path)):
non_existent_paths.append(line)
all_paths.append(line)
if len(non_existent_paths) > 0:
non_existent_paths = "\n".join([f"- {f}" for f in non_existent_paths])
raise ValueError(f"`{doctest_file}` contains non-existent paths:\n{non_existent_paths}")
sorted_paths = sorted(all_paths)
if all_paths != sorted_paths:
if not overwrite:
raise ValueError(
f"Files in `{doctest_file}` are not in alphabetical order, run `make fix-copies` to fix "
"this automatically."
)
with open(doctest_file, "w", encoding="utf-8") as f:
f.write("\n".join(sorted_paths) + "\n")
if __name__ == "__main__":
parser = argparse.ArgumentParser()
parser.add_argument("--fix_and_overwrite", action="store_true", help="Whether to fix inconsistencies.")
args = parser.parse_args()
for doctest_file in DOCTEST_FILE_PATHS:
doctest_file = os.path.join(REPO_PATH, "utils", doctest_file)
clean_doctest_list(doctest_file, args.fix_and_overwrite)
| transformers/utils/check_doctest_list.py/0 | {
"file_path": "transformers/utils/check_doctest_list.py",
"repo_id": "transformers",
"token_count": 1179
} | 401 |
# coding=utf-8
# Copyright 2020 The HuggingFace Inc. team.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# this script reports modified .py files under the desired list of top-level sub-dirs passed as a list of arguments, e.g.:
# python ./utils/get_modified_files.py utils src tests examples
#
# it uses git to find the forking point and which files were modified - i.e. files not under git won't be considered
# since the output of this script is fed into Makefile commands it doesn't print a newline after the results
import re
import subprocess
import sys
fork_point_sha = subprocess.check_output("git merge-base main HEAD".split()).decode("utf-8")
modified_files = (
subprocess.check_output(f"git diff --diff-filter=d --name-only {fork_point_sha}".split()).decode("utf-8").split()
)
joined_dirs = "|".join(sys.argv[1:])
regex = re.compile(rf"^({joined_dirs}).*?\.py$")
relevant_modified_files = [x for x in modified_files if regex.match(x)]
print(" ".join(relevant_modified_files), end="")
| transformers/utils/get_modified_files.py/0 | {
"file_path": "transformers/utils/get_modified_files.py",
"repo_id": "transformers",
"token_count": 448
} | 402 |
import torch
from transformers import PreTrainedModel
from .custom_configuration import CustomConfig, NoSuperInitConfig
class CustomModel(PreTrainedModel):
config_class = CustomConfig
def __init__(self, config):
super().__init__(config)
self.linear = torch.nn.Linear(config.hidden_size, config.hidden_size)
def forward(self, x):
return self.linear(x)
def _init_weights(self, module):
pass
class NoSuperInitModel(PreTrainedModel):
config_class = NoSuperInitConfig
def __init__(self, config):
super().__init__(config)
self.linear = torch.nn.Linear(config.attribute, config.attribute)
def forward(self, x):
return self.linear(x)
def _init_weights(self, module):
pass
| transformers/utils/test_module/custom_modeling.py/0 | {
"file_path": "transformers/utils/test_module/custom_modeling.py",
"repo_id": "transformers",
"token_count": 289
} | 403 |
# How to contribute
## How to get started
Before you start contributing make sure you installed all the dev tools:
```bash
make dev
```
## Did you find a bug?
* Ensure the bug was not already reported by searching on GitHub under Issues.
* If you're unable to find an open issue addressing the problem, open a new one. Be sure to include a title and clear description, as much relevant information as possible, and a code sample or an executable test case demonstrating the expected behavior that is not occurring.
* Be sure to add the complete error messages.
#### Did you write a patch that fixes a bug?
* Open a new GitHub pull request with the patch.
* Ensure that your PR includes a test that fails without your patch, and pass with it.
* Ensure the PR description clearly describes the problem and solution. Include the relevant issue number if applicable.
## PR submission guidelines
* Keep each PR focused. While it's more convenient, do not combine several unrelated fixes together. Create as many branches as needing to keep each PR focused.
* Do not mix style changes/fixes with "functional" changes. It's very difficult to review such PRs and it most likely get rejected.
* Do not add/remove vertical whitespace. Preserve the original style of the file you edit as much as you can.
* Do not turn an already submitted PR into your development playground. If after you submitted PR, you discovered that more work is needed - close the PR, do the required work and then submit a new PR. Otherwise each of your commits requires attention from maintainers of the project.
* If, however, you submitted a PR and received a request for changes, you should proceed with commits inside that PR, so that the maintainer can see the incremental fixes and won't need to review the whole PR again. In the exception case where you realize it'll take many many commits to complete the requests, then it's probably best to close the PR, do the work and then submit it again. Use common sense where you'd choose one way over another.
### Before you submit a PR
First you want to make sure that all the tests pass:
```bash
make test
```
Then before submitting your PR make sure the code quality follows the standards. You can run the following command to format:
```bash
make precommit
```
Make sure to install `pre-commit` before running the command:
```bash
pip install pre-commit
```
## Do you want to contribute to the documentation?
* Docs are in the `docs/` folder and can be updated there.
| trl/CONTRIBUTING.md/0 | {
"file_path": "trl/CONTRIBUTING.md",
"repo_id": "trl",
"token_count": 579
} | 404 |
#!/bin/bash
#SBATCH --job-name=trl
#SBATCH --partition=hopper-prod
#SBATCH --gpus-per-task={{gpus_per_task}}
#SBATCH --cpus-per-gpu={{cpus_per_gpu}}
#SBATCH --ntasks={{ntasks}}
#SBATCH --output=slurm/logs/%x_%j.out
#SBATCH --array={{array}}
##SBATCH --exclude=ip-26-0-149-199
module load cuda/12.1
{{nodes}}
seeds={{seeds}}
seed=${seeds[$SLURM_ARRAY_TASK_ID % {{len_seeds}}]}
echo "Running task $SLURM_ARRAY_TASK_ID with seed: $seed"
srun {{command}} --seed $seed
| trl/benchmark/trl.slurm_template/0 | {
"file_path": "trl/benchmark/trl.slurm_template",
"repo_id": "trl",
"token_count": 217
} | 405 |
# Installation
You can install TRL either from pypi or from source:
## pypi
Install the library with pip:
```bash
pip install trl
```
### Source
You can also install the latest version from source. First clone the repo and then run the installation with `pip`:
```bash
git clone https://github.com/huggingface/trl.git
cd trl/
pip install -e .
```
If you want the development install you can replace the pip install with the following:
```bash
pip install -e ".[dev]"
``` | trl/docs/source/installation.mdx/0 | {
"file_path": "trl/docs/source/installation.mdx",
"repo_id": "trl",
"token_count": 147
} | 406 |
# Using LLaMA models with TRL
We've begun rolling out examples to use Meta's LLaMA models in `trl` (see [Meta's LLaMA release](https://ai.facebook.com/blog/large-language-model-llama-meta-ai/) for the original LLaMA model).
## Efficient training strategies
Even training the smallest LLaMA model requires an enormous amount of memory. Some quick math: in bf16, every parameter uses 2 bytes (in fp32 4 bytes) in addition to 8 bytes used, e.g., in the Adam optimizer (see the [performance docs](https://huggingface.co/docs/transformers/perf_train_gpu_one#optimizer) in Transformers for more info). So a 7B parameter model would use `(2+8)*7B=70GB` just to fit in memory and would likely need more when you compute intermediate values such as attention scores. So you couldn’t train the model even on a single 80GB A100 like that. You can use some tricks, like more efficient optimizers of half-precision training, to squeeze a bit more into memory, but you’ll run out sooner or later.
Another option is to use Parameter-Efficient Fine-Tuning (PEFT) techniques, such as the [`peft`](https://github.com/huggingface/peft) library, which can perform low-rank adaptation (LoRA) on a model loaded in 8-bit.
For more on `peft` + `trl`, see the [docs](https://huggingface.co/docs/trl/sentiment_tuning_peft).
Loading the model in 8bit reduces the memory footprint drastically since you only need one byte per parameter for the weights (e.g. 7B LlaMa is 7GB in memory).
Instead of training the original weights directly, LoRA adds small adapter layers on top of some specific layers (usually the attention layers); thus, the number of trainable parameters is drastically reduced.
In this scenario, a rule of thumb is to allocate ~1.2-1.4GB per billion parameters (depending on the batch size and sequence length) to fit the entire fine-tuning setup.
This enables fine-tuning larger models (up to 50-60B scale models on a NVIDIA A100 80GB) at low cost.
Now we can fit very large models into a single GPU, but the training might still be very slow.
The simplest strategy in this scenario is data parallelism: we replicate the same training setup into separate GPUs and pass different batches to each GPU.
With this, you can parallelize the forward/backward passes of the model and scale with the number of GPUs.
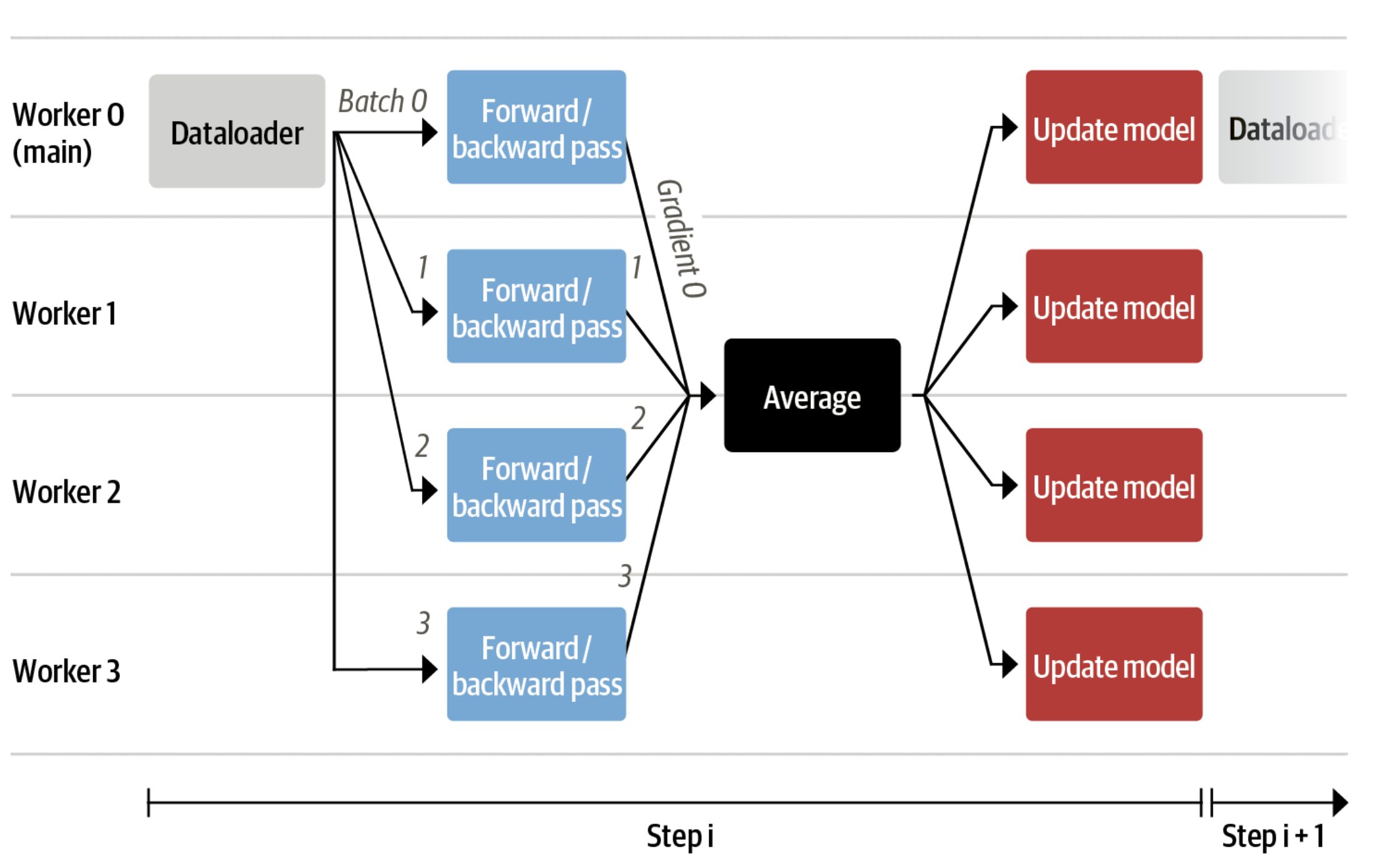
We use either the `transformers.Trainer` or `accelerate`, which both support data parallelism without any code changes, by simply passing arguments when calling the scripts with `torchrun` or `accelerate launch`. The following runs a training script with 8 GPUs on a single machine with `accelerate` and `torchrun`, respectively.
```bash
accelerate launch --multi_gpu --num_machines 1 --num_processes 8 my_accelerate_script.py
torchrun --nnodes 1 --nproc_per_node 8 my_torch_script.py
```
## Supervised fine-tuning
Before we start training reward models and tuning our model with RL, it helps if the model is already good in the domain we are interested in.
In our case, we want it to answer questions, while for other use cases, we might want it to follow instructions, in which case instruction tuning is a great idea.
The easiest way to achieve this is by continuing to train the language model with the language modeling objective on texts from the domain or task.
The [StackExchange dataset](https://huggingface.co/datasets/HuggingFaceH4/stack-exchange-preferences) is enormous (over 10 million instructions), so we can easily train the language model on a subset of it.
There is nothing special about fine-tuning the model before doing RLHF - it’s just the causal language modeling objective from pretraining that we apply here.
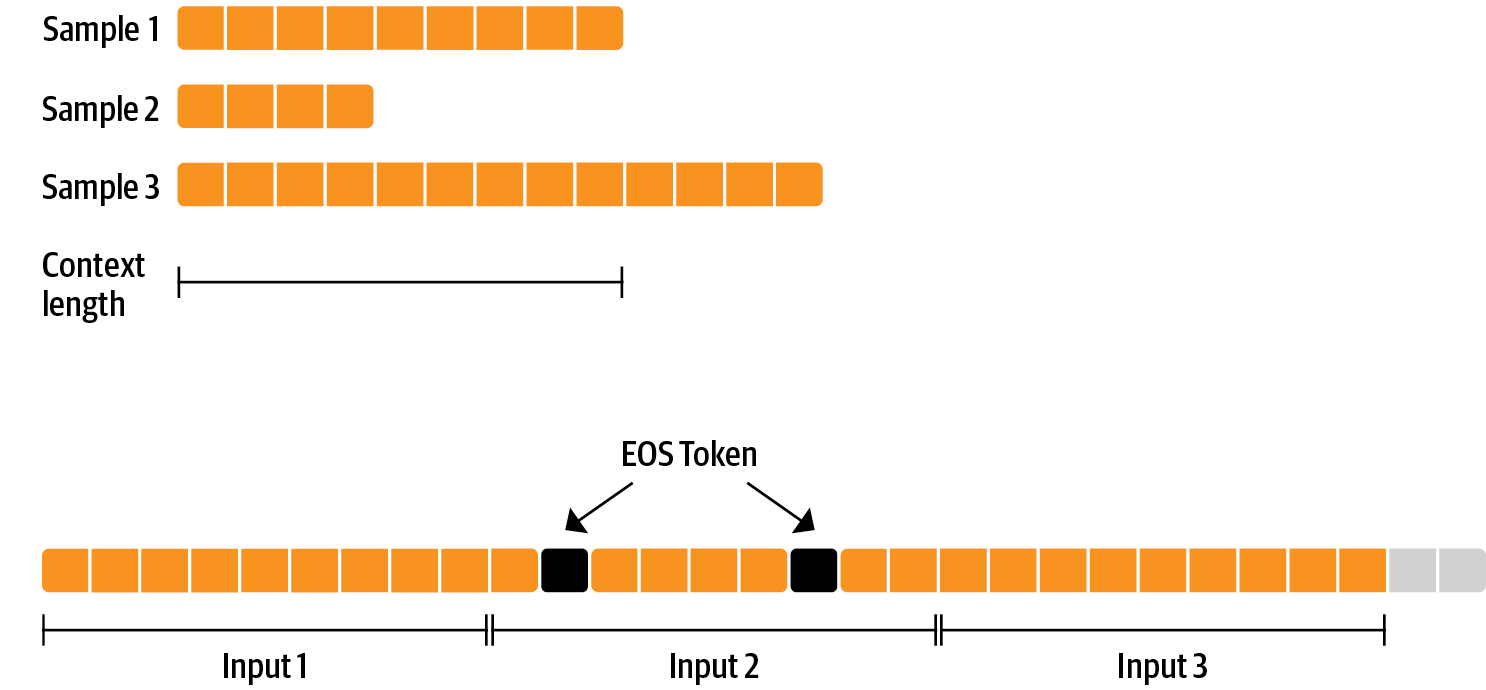
To use the data efficiently, we use a technique called packing: instead of having one text per sample in the batch and then padding to either the longest text or the maximal context of the model, we concatenate a lot of texts with a EOS token in between and cut chunks of the context size to fill the batch without any padding.
With this approach the training is much more efficient as each token that is passed through the model is also trained in contrast to padding tokens which are usually masked from the loss.
If you don't have much data and are more concerned about occasionally cutting off some tokens that are overflowing the context you can also use a classical data loader.
The packing is handled by the `ConstantLengthDataset` and we can then use the `Trainer` after loading the model with `peft`. First, we load the model in int8, prepare it for training, and then add the LoRA adapters.
```python
# load model in 8bit
model = AutoModelForCausalLM.from_pretrained(
args.model_path,
load_in_8bit=True,
device_map={"": Accelerator().local_process_index}
)
model = prepare_model_for_kbit_training(model)
# add LoRA to model
lora_config = LoraConfig(
r=16,
lora_alpha=32,
lora_dropout=0.05,
bias="none",
task_type="CAUSAL_LM",
)
model = get_peft_model(model, config)
```
We train the model for a few thousand steps with the causal language modeling objective and save the model.
Since we will tune the model again with different objectives, we merge the adapter weights with the original model weights.
**Disclaimer:** due to LLaMA's license, we release only the adapter weights for this and the model checkpoints in the following sections.
You can apply for access to the base model's weights by filling out Meta AI's [form](https://docs.google.com/forms/d/e/1FAIpQLSfqNECQnMkycAp2jP4Z9TFX0cGR4uf7b_fBxjY_OjhJILlKGA/viewform) and then converting them to the 🤗 Transformers format by running this [script](https://github.com/huggingface/transformers/blob/main/src/transformers/models/llama/convert_llama_weights_to_hf.py).
Note that you'll also need to install 🤗 Transformers from source until the `v4.28` is released.
Now that we have fine-tuned the model for the task, we are ready to train a reward model.
## Reward modeling and human preferences
In principle, we could fine-tune the model using RLHF directly with the human annotations.
However, this would require us to send some samples to humans for rating after each optimization iteration.
This is expensive and slow due to the number of training samples needed for convergence and the inherent latency of human reading and annotator speed.
A trick that works well instead of direct feedback is training a reward model on human annotations collected before the RL loop.
The goal of the reward model is to imitate how a human would rate a text. There are several possible strategies to build a reward model: the most straightforward way would be to predict the annotation (e.g. a rating score or a binary value for “good”/”bad”).
In practice, what works better is to predict the ranking of two examples, where the reward model is presented with two candidates `(y_k, y_j)` for a given prompt `x` and has to predict which one would be rated higher by a human annotator.
With the StackExchange dataset, we can infer which of the two answers was preferred by the users based on the score.
With that information and the loss defined above, we can then modify the `transformers.Trainer` by adding a custom loss function.
```python
class RewardTrainer(Trainer):
def compute_loss(self, model, inputs, return_outputs=False):
rewards_j = model(input_ids=inputs["input_ids_j"], attention_mask=inputs["attention_mask_j"])[0]
rewards_k = model(input_ids=inputs["input_ids_k"], attention_mask=inputs["attention_mask_k"])[0]
loss = -nn.functional.logsigmoid(rewards_j - rewards_k).mean()
if return_outputs:
return loss, {"rewards_j": rewards_j, "rewards_k": rewards_k}
return loss
```
We utilize a subset of a 100,000 pair of candidates and evaluate on a held-out set of 50,000. With a modest training batch size of 4, we train the Llama model using the LoRA `peft` adapter for a single epoch using the Adam optimizer with BF16 precision. Our LoRA configuration is:
```python
peft_config = LoraConfig(
task_type=TaskType.SEQ_CLS,
inference_mode=False,
r=8,
lora_alpha=32,
lora_dropout=0.1,
)
```
As detailed in the next section, the resulting adapter can be merged into the frozen model and saved for further downstream use.
## Reinforcement Learning from Human Feedback
With the fine-tuned language model and the reward model at hand, we are now ready to run the RL loop. It follows roughly three steps:
1. Generate responses from prompts,
2. Rate the responses with the reward model,
3. Run a reinforcement learning policy-optimization step with the ratings.
The Query and Response prompts are templated as follows before being tokenized and passed to the model:
```bash
Question: <Query>
Answer: <Response>
```
The same template was used for SFT, RM and RLHF stages.
Once more, we utilize `peft` for memory-efficient training, which offers an extra advantage in the RLHF context.
Here, the reference model and policy share the same base, the SFT model, which we load in 8-bit and freeze during training.
We exclusively optimize the policy's LoRA weights using PPO while sharing the base model's weights.
```python
for epoch, batch in tqdm(enumerate(ppo_trainer.dataloader)):
question_tensors = batch["input_ids"]
# sample from the policy and to generate responses
response_tensors = ppo_trainer.generate(
question_tensors,
return_prompt=False,
length_sampler=output_length_sampler,
**generation_kwargs,
)
batch["response"] = tokenizer.batch_decode(response_tensors, skip_special_tokens=True)
# Compute sentiment score
texts = [q + r for q, r in zip(batch["query"], batch["response"])]
pipe_outputs = sentiment_pipe(texts, **sent_kwargs)
rewards = [torch.tensor(output[0]["score"] - script_args.reward_baseline) for output in pipe_outputs]
# Run PPO step
stats = ppo_trainer.step(question_tensors, response_tensors, rewards)
# Log stats to Wandb
ppo_trainer.log_stats(stats, batch, rewards)
```
For the rest of the details and evaluation, please refer to our [blog post on StackLLaMA](https://huggingface.co/blog/stackllama).
| trl/docs/source/using_llama_models.mdx/0 | {
"file_path": "trl/docs/source/using_llama_models.mdx",
"repo_id": "trl",
"token_count": 2999
} | 407 |
# Research projects that use TRL
Welcome to the research projects folder! Here you can find the scripts used for some research projects that used TRL and maintained by the developers and the community (LM de-toxification, Stack-Llama, etc.). Check out the READMEs in the subfolders for more information!
- [De-detoxifying language models](https://github.com/huggingface/trl/tree/main/examples/research_projects/toxicity)
- [Stack-Llama](https://github.com/huggingface/trl/tree/main/examples/research_projects/stack_llama)
- [Stack-Llama-2](https://github.com/huggingface/trl/tree/main/examples/research_projects/stack_llama_2) | trl/examples/research_projects/README.md/0 | {
"file_path": "trl/examples/research_projects/README.md",
"repo_id": "trl",
"token_count": 189
} | 408 |
# flake8: noqa
# Copyright 2024 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from trl.commands.cli_utils import init_zero_verbose
init_zero_verbose()
import copy
import json
import os
import pwd
import re
import time
from threading import Thread
import torch
from rich.console import Console
from rich.live import Live
from rich.markdown import Markdown
from transformers import AutoModelForCausalLM, AutoTokenizer, TextIteratorStreamer
from trl.commands.cli_utils import ChatArguments, TrlParser, init_zero_verbose
from trl.trainer.utils import get_kbit_device_map, get_quantization_config
HELP_STRING = """\
**TRL CHAT INTERFACE**
The chat interface is a simple tool to try out a chat model.
Besides talking to the model there are several commands:
- **clear**: clears the current conversation and start a new one
- **example {NAME}**: load example named `{NAME}` from the config and use it as the user input
- **set {SETTING_NAME}={SETTING_VALUE};**: change the system prompt or generation settings (multiple settings are separated by a ';').
- **reset**: same as clear but also resets the generation configs to defaults if they have been changed by **set**
- **save {SAVE_NAME} (optional)**: save the current chat and settings to file by default to `./chat_history/{MODEL_NAME}/chat_{DATETIME}.yaml` or `{SAVE_NAME}` if provided
- **exit**: closes the interface
"""
SUPPORTED_GENERATION_KWARGS = [
"max_new_tokens",
"do_sample",
"num_beams",
"temperature",
"top_p",
"top_k",
"repetition_penalty",
]
SETTING_RE = r"^set\s+[A-Za-z\s_]+=[A-Za-z\d\s.!\"#$%&'()*+,-/:<=>?@\[\]^_`{|}~]+(?:;\s*[A-Za-z\s_]+=[A-Za-z\d\s.!\"#$%&'()*+,-/:<=>?@\[\]^_`{|}~]+)*$"
class RichInterface:
def __init__(self, model_name=None, user_name=None):
self._console = Console()
if model_name is None:
self.model_name = "assistant"
else:
self.model_name = model_name
if user_name is None:
self.user_name = "user"
else:
self.user_name = user_name
def stream_output(self, output_stream):
"""Stream output from a role."""
# This method is originally from the FastChat CLI: https://github.com/lm-sys/FastChat/blob/main/fastchat/serve/cli.py
# Create a Live context for updating the console output
text = ""
self._console.print(f"[bold blue]<{self.model_name}>:")
with Live(console=self._console, refresh_per_second=4) as live:
# Read lines from the stream
for i, outputs in enumerate(output_stream):
if not outputs or i == 0:
continue
text += outputs
# Render the accumulated text as Markdown
# NOTE: this is a workaround for the rendering "unstandard markdown"
# in rich. The chatbots output treat "\n" as a new line for
# better compatibility with real-world text. However, rendering
# in markdown would break the format. It is because standard markdown
# treat a single "\n" in normal text as a space.
# Our workaround is adding two spaces at the end of each line.
# This is not a perfect solution, as it would
# introduce trailing spaces (only) in code block, but it works well
# especially for console output, because in general the console does not
# care about trailing spaces.
lines = []
for line in text.splitlines():
lines.append(line)
if line.startswith("```"):
# Code block marker - do not add trailing spaces, as it would
# break the syntax highlighting
lines.append("\n")
else:
lines.append(" \n")
markdown = Markdown("".join(lines).strip(), code_theme="github-dark")
# Update the Live console output
live.update(markdown)
self._console.print()
return text
def input(self):
input = self._console.input(f"[bold red]<{self.user_name}>:\n")
self._console.print()
return input
def clear(self):
self._console.clear()
def print_user_message(self, text):
self._console.print(f"[bold red]<{self.user_name}>:[/ bold red]\n{text}")
self._console.print()
def print_green(self, text):
self._console.print(f"[bold green]{text}")
self._console.print()
def print_red(self, text):
self._console.print(f"[bold red]{text}")
self._console.print()
def print_help(self):
self._console.print(Markdown(HELP_STRING))
self._console.print()
def get_username():
return pwd.getpwuid(os.getuid())[0]
def create_default_filename(model_name):
time_str = time.strftime("%Y-%m-%d_%H-%M-%S")
return f"{model_name}/chat_{time_str}.json"
def save_chat(chat, args, filename):
output_dict = {}
output_dict["settings"] = vars(args)
output_dict["chat_history"] = chat
folder = args.save_folder
if filename is None:
filename = create_default_filename(args.model_name_or_path)
filename = os.path.join(folder, filename)
os.makedirs(os.path.dirname(filename), exist_ok=True)
with open(filename, "w") as f:
json.dump(output_dict, f, indent=4)
return os.path.abspath(filename)
def clear_chat_history(system_prompt):
if system_prompt is None:
chat = []
else:
chat = [{"role": "system", "content": system_prompt}]
return chat
def parse_settings(user_input, current_args, interface):
settings = user_input[4:].strip().split(";")
settings = [(setting.split("=")[0], setting[len(setting.split("=")[0]) + 1 :]) for setting in settings]
settings = dict(settings)
error = False
for name in settings:
if hasattr(current_args, name):
try:
if isinstance(getattr(current_args, name), bool):
if settings[name] == "True":
settings[name] = True
elif settings[name] == "False":
settings[name] = False
else:
raise ValueError
else:
settings[name] = type(getattr(current_args, name))(settings[name])
except ValueError:
interface.print_red(
f"Cannot cast setting {name} (={settings[name]}) to {type(getattr(current_args, name))}."
)
else:
interface.print_red(f"There is no '{name}' setting.")
if error:
interface.print_red("There was an issue parsing the settings. No settings have been changed.")
return current_args, False
else:
for name in settings:
setattr(current_args, name, settings[name])
interface.print_green(f"Set {name} to {settings[name]}.")
time.sleep(1.5) # so the user has time to read the changes
return current_args, True
def load_model_and_tokenizer(args):
tokenizer = AutoTokenizer.from_pretrained(args.model_name_or_path, revision=args.model_revision)
torch_dtype = args.torch_dtype if args.torch_dtype in ["auto", None] else getattr(torch, args.torch_dtype)
quantization_config = get_quantization_config(args)
model_kwargs = dict(
revision=args.model_revision,
trust_remote_code=args.trust_remote_code,
attn_implementation=args.attn_implementation,
torch_dtype=torch_dtype,
device_map=get_kbit_device_map() if quantization_config is not None else None,
quantization_config=quantization_config,
)
model = AutoModelForCausalLM.from_pretrained(args.model_name_or_path, **model_kwargs)
if getattr(model, "hf_device_map", None) is None:
model = model.to(args.device)
return model, tokenizer
def chat_cli():
parser = TrlParser(ChatArguments)
args = parser.parse_args_into_dataclasses()[0]
if args.config == "default":
args.config = os.path.join(os.path.dirname(__file__), "config/default_chat_config.yaml")
if args.config.lower() == "none":
args.config = None
args = parser.update_dataclasses_with_config([args])[0]
if args.examples is None:
args.examples = {}
current_args = copy.deepcopy(args)
if args.user is None:
user = get_username()
else:
user = args.user
model, tokenizer = load_model_and_tokenizer(args)
generation_streamer = TextIteratorStreamer(tokenizer, skip_special_tokens=True)
interface = RichInterface(model_name=args.model_name_or_path, user_name=user)
interface.clear()
chat = clear_chat_history(current_args.system_prompt)
while True:
try:
user_input = interface.input()
if user_input == "clear":
chat = clear_chat_history(current_args.system_prompt)
interface.clear()
continue
if user_input == "help":
interface.print_help()
continue
if user_input == "exit":
break
if user_input == "reset":
interface.clear()
current_args = copy.deepcopy(args)
chat = clear_chat_history(current_args.system_prompt)
continue
if user_input.startswith("save") and len(user_input.split()) < 2:
split_input = user_input.split()
if len(split_input) == 2:
filename = split_input[1]
else:
filename = None
filename = save_chat(chat, current_args, filename)
interface.print_green(f"Chat saved in {filename}!")
continue
if re.match(SETTING_RE, user_input):
current_args, success = parse_settings(user_input, current_args, interface)
if success:
chat = []
interface.clear()
continue
if user_input.startswith("example") and len(user_input.split()) == 2:
example_name = user_input.split()[1]
if example_name in current_args.examples:
interface.clear()
chat = []
interface.print_user_message(current_args.examples[example_name]["text"])
user_input = current_args.examples[example_name]["text"]
else:
interface.print_red(
f"Example {example_name} not found in list of available examples: {list(current_args.examples.keys())}."
)
continue
chat.append({"role": "user", "content": user_input})
generation_kwargs = dict(
inputs=tokenizer.apply_chat_template(chat, return_tensors="pt", add_generation_prompt=True).to(
model.device
),
streamer=generation_streamer,
max_new_tokens=current_args.max_new_tokens,
do_sample=current_args.do_sample,
num_beams=current_args.num_beams,
temperature=current_args.temperature,
top_k=current_args.top_k,
top_p=current_args.top_p,
repetition_penalty=current_args.repetition_penalty,
pad_token_id=tokenizer.pad_token_id,
eos_token_id=tokenizer.eos_token_id,
)
thread = Thread(target=model.generate, kwargs=generation_kwargs)
thread.start()
model_output = interface.stream_output(generation_streamer)
thread.join()
chat.append({"role": "assistant", "content": model_output})
except KeyboardInterrupt:
break
if __name__ == "__main__":
chat_cli()
| trl/examples/scripts/chat.py/0 | {
"file_path": "trl/examples/scripts/chat.py",
"repo_id": "trl",
"token_count": 5625
} | 409 |
# Copyright 2022 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import gc
import tempfile
import unittest
import pytest
import torch
from transformers import AutoModel, AutoModelForCausalLM, AutoModelForSeq2SeqLM
from trl import AutoModelForCausalLMWithValueHead, AutoModelForSeq2SeqLMWithValueHead, create_reference_model
ALL_CAUSAL_LM_MODELS = [
"trl-internal-testing/tiny-random-CodeGenForCausalLM",
"trl-internal-testing/tiny-random-GPTJForCausalLM",
"trl-internal-testing/tiny-random-GPTNeoForCausalLM",
"trl-internal-testing/tiny-random-GPTNeoXForCausalLM",
"trl-internal-testing/tiny-random-OPTForCausalLM",
"trl-internal-testing/tiny-random-BloomForCausalLM",
"trl-internal-testing/tiny-random-GPT2LMHeadModel",
"trl-internal-testing/tiny-random-CodeGenForCausalLM-sharded",
"trl-internal-testing/tiny-random-GPTNeoXForCausalLM-safetensors-sharded",
"trl-internal-testing/tiny-random-GPTNeoXForCausalLM-safetensors",
# "trl-internal-testing/tiny-random-LlamaForCausalLM", uncomment on the next transformers release
]
ALL_SEQ2SEQ_MODELS = [
"trl-internal-testing/tiny-random-BartForConditionalGeneration",
"trl-internal-testing/tiny-random-BigBirdPegasusForConditionalGeneration",
"trl-internal-testing/tiny-random-BlenderbotForConditionalGeneration",
"trl-internal-testing/tiny-random-BlenderbotSmallForConditionalGeneration",
"trl-internal-testing/tiny-random-FSMTForConditionalGeneration",
"trl-internal-testing/tiny-random-LEDForConditionalGeneration",
"trl-internal-testing/tiny-random-LongT5ForConditionalGeneration",
"trl-internal-testing/tiny-random-M2M100ForConditionalGeneration",
"trl-internal-testing/tiny-random-MarianMTModel",
"trl-internal-testing/tiny-random-MBartForConditionalGeneration",
"trl-internal-testing/tiny-random-MT5ForConditionalGeneration",
"trl-internal-testing/tiny-random-MvpForConditionalGeneration",
"trl-internal-testing/tiny-random-PegasusForConditionalGeneration",
"trl-internal-testing/tiny-random-PegasusXForConditionalGeneration",
"trl-internal-testing/tiny-random-PLBartForConditionalGeneration",
"trl-internal-testing/tiny-random-ProphetNetForConditionalGeneration",
"trl-internal-testing/tiny-random-SwitchTransformersForConditionalGeneration",
"trl-internal-testing/tiny-random-T5ForConditionalGeneration",
]
class VHeadModelTester:
all_model_names = None
trl_model_class = None
transformers_model_class = None
def test_value_head(self):
r"""
Test if the v-head is added to the model successfully
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
assert hasattr(model, "v_head")
def test_value_head_shape(self):
r"""
Test if the v-head has the correct shape
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
assert model.v_head.summary.weight.shape[0] == 1
def test_value_head_init_random(self):
r"""
Test if the v-head has been randomly initialized.
We can check that by making sure the bias is different
than zeros by default.
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
assert not torch.allclose(model.v_head.summary.bias, torch.zeros_like(model.v_head.summary.bias))
def test_value_head_not_str(self):
r"""
Test if the v-head is added to the model successfully, by passing a non `PretrainedModel`
as an argument to `from_pretrained`.
"""
for model_name in self.all_model_names:
pretrained_model = self.transformers_model_class.from_pretrained(model_name)
model = self.trl_model_class.from_pretrained(pretrained_model)
assert hasattr(model, "v_head")
def test_from_save_trl(self):
"""
Test if the model can be saved and loaded from a directory and get the same weights
Including the additional modules (e.g. v_head)
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
with tempfile.TemporaryDirectory() as tmp_dir:
model.save_pretrained(tmp_dir)
model_from_save = self.trl_model_class.from_pretrained(tmp_dir)
# Check if the weights are the same
for key in model_from_save.state_dict():
assert torch.allclose(model_from_save.state_dict()[key], model.state_dict()[key])
def test_from_save_trl_sharded(self):
"""
Test if the model can be saved and loaded from a directory and get the same weights - sharded case
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
with tempfile.TemporaryDirectory() as tmp_dir:
model.save_pretrained(tmp_dir)
model_from_save = self.trl_model_class.from_pretrained(tmp_dir)
# Check if the weights are the same
for key in model_from_save.state_dict():
assert torch.allclose(model_from_save.state_dict()[key], model.state_dict()[key])
def test_from_save_transformers_sharded(self):
"""
Test if the model can be saved and loaded using transformers and get the same weights - sharded case
"""
for model_name in self.all_model_names:
transformers_model = self.trl_model_class.transformers_parent_class.from_pretrained(model_name)
trl_model = self.trl_model_class.from_pretrained(model_name)
with tempfile.TemporaryDirectory() as tmp_dir:
trl_model.save_pretrained(tmp_dir, max_shard_size="1MB")
transformers_model_from_save = self.trl_model_class.transformers_parent_class.from_pretrained(tmp_dir)
# Check if the weights are the same
for key in transformers_model.state_dict():
assert torch.allclose(
transformers_model_from_save.state_dict()[key], transformers_model.state_dict()[key]
)
def test_from_save_transformers(self):
"""
Test if the model can be saved and loaded using transformers and get the same weights.
We override the test of the super class to check if the weights are the same.
"""
for model_name in self.all_model_names:
transformers_model = self.trl_model_class.transformers_parent_class.from_pretrained(model_name)
trl_model = self.trl_model_class.from_pretrained(model_name)
with tempfile.TemporaryDirectory() as tmp_dir:
trl_model.save_pretrained(tmp_dir)
transformers_model_from_save = self.trl_model_class.transformers_parent_class.from_pretrained(tmp_dir)
# Check if the weights are the same
for key in transformers_model.state_dict():
assert torch.allclose(
transformers_model_from_save.state_dict()[key], transformers_model.state_dict()[key]
)
# Check if the trl model has the same keys as the transformers model
# except the v_head
for key in trl_model.state_dict():
if "v_head" not in key:
assert key in transformers_model.state_dict()
# check if the weights are the same
assert torch.allclose(trl_model.state_dict()[key], transformers_model.state_dict()[key])
# check if they have the same modules
assert set(transformers_model_from_save.state_dict().keys()) == set(transformers_model.state_dict().keys())
class CausalLMValueHeadModelTester(VHeadModelTester, unittest.TestCase):
"""
Testing suite for v-head models.
"""
all_model_names = ALL_CAUSAL_LM_MODELS
trl_model_class = AutoModelForCausalLMWithValueHead
transformers_model_class = AutoModelForCausalLM
def tearDown(self):
# free memory
gc.collect()
def test_inference(self):
r"""
Test if the model can be used for inference and outputs 3 values
- logits, loss, and value states
"""
EXPECTED_OUTPUT_SIZE = 3
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
outputs = model(input_ids)
# Check if the outputs are of the right size - here
# we always output 3 values - logits, loss, and value states
assert len(outputs) == EXPECTED_OUTPUT_SIZE
def test_dropout_config(self):
r"""
Test if we instantiate a model by adding `summary_drop_prob` to the config
it will be added to the v_head
"""
for model_name in self.all_model_names:
pretrained_model = self.transformers_model_class.from_pretrained(model_name)
pretrained_model.config.summary_dropout_prob = 0.5
model = self.trl_model_class.from_pretrained(pretrained_model)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == pretrained_model.config.summary_dropout_prob
def test_dropout_kwargs(self):
r"""
Test if we instantiate a model by adding `summary_drop_prob` to the config
it will be added to the v_head
"""
for model_name in self.all_model_names:
v_head_kwargs = {"summary_dropout_prob": 0.5}
model = self.trl_model_class.from_pretrained(model_name, **v_head_kwargs)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == 0.5
model = self.trl_model_class.from_pretrained(model_name, summary_dropout_prob=0.5)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == 0.5
def test_generate(self):
r"""
Test if `generate` works for every model
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
# Just check if the generation works
_ = model.generate(input_ids)
def test_raise_error_not_causallm(self):
# Test with a model without a LM head
model_id = "trl-internal-testing/tiny-random-GPT2Model"
# This should raise a ValueError
with pytest.raises(ValueError):
pretrained_model = AutoModelForCausalLM.from_pretrained(model_id)
_ = AutoModelForCausalLMWithValueHead.from_pretrained(pretrained_model.transformer)
def test_transformers_bf16_kwargs(self):
r"""
Test if the transformers kwargs are correctly passed
Here we check that loading a model in half precision works as expected, i.e. the weights of
the `pretrained_model` attribute is loaded in half precision and you can run a dummy
forward pass without any issue.
"""
for model_name in self.all_model_names:
trl_model = self.trl_model_class.from_pretrained(model_name, torch_dtype=torch.bfloat16)
lm_head_namings = self.trl_model_class.lm_head_namings
assert any(hasattr(trl_model.pretrained_model, lm_head_naming) for lm_head_naming in lm_head_namings)
for lm_head_naming in lm_head_namings:
if hasattr(trl_model.pretrained_model, lm_head_naming):
assert getattr(trl_model.pretrained_model, lm_head_naming).weight.dtype == torch.bfloat16
dummy_input = torch.LongTensor([[0, 1, 0, 1]])
# check dummy forward pass works in half precision
_ = trl_model(dummy_input)
@unittest.skip("This test needs to be run manually due to HF token issue.")
def test_push_to_hub(self):
for model_name in self.all_model_names:
model = AutoModelForCausalLMWithValueHead.from_pretrained(model_name)
if "sharded" in model_name:
model.push_to_hub(model_name + "-ppo", use_auth_token=True, max_shard_size="1MB")
else:
model.push_to_hub(model_name + "-ppo", use_auth_token=True)
model_from_pretrained = AutoModelForCausalLMWithValueHead.from_pretrained(model_name + "-ppo")
# check all keys
assert model.state_dict().keys() == model_from_pretrained.state_dict().keys()
for name, param in model.state_dict().items():
assert torch.allclose(
param, model_from_pretrained.state_dict()[name]
), f"Parameter {name} is not the same after push_to_hub and from_pretrained"
class Seq2SeqValueHeadModelTester(VHeadModelTester, unittest.TestCase):
"""
Testing suite for v-head models.
"""
all_model_names = ALL_SEQ2SEQ_MODELS
trl_model_class = AutoModelForSeq2SeqLMWithValueHead
transformers_model_class = AutoModelForSeq2SeqLM
def tearDown(self):
# free memory
gc.collect()
def test_inference(self):
r"""
Test if the model can be used for inference and outputs 3 values
- logits, loss, and value states
"""
EXPECTED_OUTPUT_SIZE = 3
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
decoder_input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
outputs = model(input_ids, decoder_input_ids=decoder_input_ids)
# Check if the outputs are of the right size - here
# we always output 3 values - logits, loss, and value states
assert len(outputs) == EXPECTED_OUTPUT_SIZE
def test_dropout_config(self):
r"""
Test if we instantiate a model by adding `summary_drop_prob` to the config
it will be added to the v_head
"""
for model_name in self.all_model_names:
pretrained_model = self.transformers_model_class.from_pretrained(model_name)
pretrained_model.config.summary_dropout_prob = 0.5
model = self.trl_model_class.from_pretrained(pretrained_model)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == pretrained_model.config.summary_dropout_prob
def test_dropout_kwargs(self):
r"""
Test if we instantiate a model by adding `summary_drop_prob` to the config
it will be added to the v_head
"""
for model_name in self.all_model_names:
v_head_kwargs = {"summary_dropout_prob": 0.5}
model = self.trl_model_class.from_pretrained(model_name, **v_head_kwargs)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == 0.5
model = self.trl_model_class.from_pretrained(model_name, summary_dropout_prob=0.5)
# Check if v head of the model has the same dropout as the config
assert model.v_head.dropout.p == 0.5
def test_generate(self):
r"""
Test if `generate` works for every model
"""
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
decoder_input_ids = torch.tensor([[1, 2, 3, 4, 5, 6, 7, 8, 9, 10]])
# Just check if the generation works
_ = model.generate(input_ids, decoder_input_ids=decoder_input_ids)
def test_raise_error_not_causallm(self):
# Test with a model without a LM head
model_id = "trl-internal-testing/tiny-random-T5Model"
# This should raise a ValueError
with pytest.raises(ValueError):
pretrained_model = AutoModel.from_pretrained(model_id)
_ = self.trl_model_class.from_pretrained(pretrained_model)
@unittest.skip("This test needs to be run manually due to HF token issue.")
def test_push_to_hub(self):
for model_name in self.all_model_names:
model = self.trl_model_class.from_pretrained(model_name)
if "sharded" in model_name:
model.push_to_hub(model_name + "-ppo", use_auth_token=True, max_shard_size="1MB")
else:
model.push_to_hub(model_name + "-ppo", use_auth_token=True)
model_from_pretrained = self.trl_model_class.from_pretrained(model_name + "-ppo")
# check all keys
assert model.state_dict().keys() == model_from_pretrained.state_dict().keys()
for name, param in model.state_dict().items():
assert torch.allclose(
param, model_from_pretrained.state_dict()[name]
), f"Parameter {name} is not the same after push_to_hub and from_pretrained"
def test_transformers_bf16_kwargs(self):
r"""
Test if the transformers kwargs are correctly passed
Here we check that loading a model in half precision works as expected, i.e. the weights of
the `pretrained_model` attribute is loaded in half precision and you can run a dummy
forward pass without any issue.
"""
for model_name in self.all_model_names:
trl_model = self.trl_model_class.from_pretrained(model_name, torch_dtype=torch.bfloat16)
lm_head_namings = self.trl_model_class.lm_head_namings
if model_name == "trl-internal-testing/tiny-random-FSMTForConditionalGeneration":
# skip the test for FSMT as it does not support mixed-prec
continue
assert any(hasattr(trl_model.pretrained_model, lm_head_naming) for lm_head_naming in lm_head_namings)
for lm_head_naming in lm_head_namings:
if hasattr(trl_model.pretrained_model, lm_head_naming):
assert getattr(trl_model.pretrained_model, lm_head_naming).weight.dtype == torch.bfloat16
dummy_input = torch.LongTensor([[0, 1, 0, 1]])
# check dummy forward pass works in half precision
_ = trl_model(input_ids=dummy_input, decoder_input_ids=dummy_input)
class ReferenceModelTest(unittest.TestCase):
def setUp(self):
self.model = AutoModelForCausalLMWithValueHead.from_pretrained(
"trl-internal-testing/tiny-random-GPT2LMHeadModel"
)
self.test_input = torch.tensor([[0, 1, 2, 3]])
self.optimizer = torch.optim.AdamW(self.model.parameters(), lr=1)
self.layer_format = "pretrained_model.transformer.h.{layer}.attn.c_attn.weight"
def test_independent_reference(self):
layer_0 = self.layer_format.format(layer=0)
layer_5 = self.layer_format.format(layer=4)
ref_model = create_reference_model(self.model)
first_layer_before = self.model.get_parameter(layer_0).data.clone()
last_layer_before = self.model.get_parameter(layer_5).data.clone()
first_ref_layer_before = ref_model.get_parameter(layer_0).data.clone()
last_ref_layer_before = ref_model.get_parameter(layer_5).data.clone()
output = self.model(input_ids=self.test_input, labels=self.test_input)
output[1].backward()
self.optimizer.step()
first_layer_after = self.model.get_parameter(layer_0).data.clone()
last_layer_after = self.model.get_parameter(layer_5).data.clone()
first_ref_layer_after = ref_model.get_parameter(layer_0).data.clone()
last_ref_layer_after = ref_model.get_parameter(layer_5).data.clone()
# before optimization ref and model are identical
assert (first_layer_before == first_ref_layer_before).all()
assert (last_layer_before == last_ref_layer_before).all()
# ref model stays identical after optimization
assert (first_ref_layer_before == first_ref_layer_after).all()
assert (last_ref_layer_before == last_ref_layer_after).all()
# optimized model changes
assert not (first_layer_before == first_layer_after).all()
assert not (last_layer_before == last_layer_after).all()
def test_shared_layers(self):
layer_0 = self.layer_format.format(layer=0)
layer_1 = self.layer_format.format(layer=1)
ref_model = create_reference_model(self.model, num_shared_layers=1)
first_layer_before = self.model.get_parameter(layer_0).data.clone()
second_layer_before = self.model.get_parameter(layer_1).data.clone()
first_ref_layer_before = ref_model.get_parameter(layer_0).data.clone()
second_ref_layer_before = ref_model.get_parameter(layer_1).data.clone()
output = self.model(input_ids=self.test_input, labels=self.test_input)
output[1].backward()
self.optimizer.step()
first_layer_after = self.model.get_parameter(layer_0).data.clone()
second_layer_after = self.model.get_parameter(layer_1).data.clone()
first_ref_layer_after = ref_model.get_parameter(layer_0).data.clone()
second_ref_layer_after = ref_model.get_parameter(layer_1).data.clone()
# before optimization ref and model are identical
assert (first_layer_before == first_ref_layer_before).all()
assert (second_layer_before == second_ref_layer_before).all()
# ref model stays identical after optimization
assert (first_ref_layer_before == first_ref_layer_after).all()
assert (second_ref_layer_before == second_ref_layer_after).all()
# first layer of optimized model stays the same
assert (first_layer_before == first_layer_after).all()
# other layers in optimized model change
assert not (second_layer_before == second_layer_after).all()
| trl/tests/test_modeling_value_head.py/0 | {
"file_path": "trl/tests/test_modeling_value_head.py",
"repo_id": "trl",
"token_count": 9527
} | 410 |
from typing import Any, Callable, List, Optional, Union
import torch
from transformers import GenerationConfig, PreTrainedTokenizer, PreTrainedTokenizerFast
from ..core import set_seed
from ..models import SUPPORTED_ARCHITECTURES, PreTrainedModelWrapper
class BestOfNSampler:
def __init__(
self,
model: PreTrainedModelWrapper,
tokenizer: Union[PreTrainedTokenizer, PreTrainedTokenizerFast],
queries_to_scores: Callable[[List[str]], List[float]],
length_sampler: Any,
sample_size: int = 4,
seed: Optional[int] = None,
n_candidates: int = 1,
generation_config: Optional[GenerationConfig] = None,
) -> None:
r"""
Initialize the sampler for best-of-n generation
Args:
model (`PreTrainedModelWrapper`):
The pretrained model to use for generation
tokenizer (`PreTrainedTokenizer` or `PreTrainedTokenizerFast`):
Tokenizer associated with the pretrained model
queries_to_scores (`Callable[[List[str]], List[float]]`):
Callable that takes a list of generated texts and returns the associated reward scores
length_sampler (`Any`):
Sampler used to sample the length of the generated text
sample_size (`int`):
Number of samples to generate for each query
seed (`int`, *optional*):
Random seed used to control generation
n_candidates (`int`):
Number of candidates to return for each query
generation_config (`GenerationConfig`, *optional*):
Generation config passed to the underlying model's `generate` method.
See `GenerationConfig` (https://huggingface.co/docs/transformers/v4.29.1/en/main_classes/text_generation#transformers.GenerationConfig) for more details
"""
if seed is not None:
set_seed(seed)
if not isinstance(tokenizer, (PreTrainedTokenizer, PreTrainedTokenizerFast)):
raise ValueError(
f"tokenizer must be a PreTrainedTokenizer or PreTrainedTokenizerFast, got {type(tokenizer)}"
)
if not isinstance(model, (SUPPORTED_ARCHITECTURES)):
raise ValueError(
f"model must be a PreTrainedModelWrapper, got {type(model)} - supported architectures are: {SUPPORTED_ARCHITECTURES}"
)
self.model = model
self.tokenizer = tokenizer
self.queries_to_scores = queries_to_scores
self.length_sampler = length_sampler
self.gen_config = generation_config
self.sample_size = sample_size
self.n_candidates = n_candidates
def generate(
self,
tokenized_query: Union[List[int], torch.Tensor, List[torch.Tensor], List[List[int]]],
skip_special_tokens: bool = True,
device: Optional[Union[str, torch.device]] = None,
**generation_kwargs,
) -> List[List[str]]:
r"""
Generate the best of n samples for input queries
Args:
tokenized_query (`List[int]` or `torch.Tensor` or `List[torch.Tensor]` or `List[int]`):
represents either a single tokenized query (a single tensor or a list of integers) or a batch of tokenized queries (a list of tensors or a list of lists of integers)
skip_special_tokens (`bool`):
Whether to remove the special tokens from the output
device (`str` or `torch.device`, *optional*):
The device on which the model will be loaded
**generation_kwargs (`dict`, *optional*):
Additional keyword arguments passed along to the underlying model's `generate` method.
This is used to override generation config
Returns:
List[List[str]]: A list of lists of generated texts
"""
queries = None
if isinstance(tokenized_query, torch.Tensor) and tokenized_query.ndim == 1:
queries = tokenized_query.unsqueeze(0)
elif isinstance(tokenized_query, List):
element_type = type(tokenized_query[0])
if element_type == int:
queries = torch.tensor(tokenized_query).unsqueeze(0)
elif element_type == torch.Tensor:
queries = [tensor.reshape((1, -1)) for tensor in tokenized_query]
else:
queries = [torch.tensor(query).reshape((1, -1)) for query in tokenized_query]
result = []
for query in queries:
queries = query.repeat((self.sample_size, 1))
output = self.model.generate(
queries.to(device),
max_new_tokens=self.length_sampler(),
generation_config=self.gen_config,
**generation_kwargs,
).squeeze()
output = self.tokenizer.batch_decode(output, skip_special_tokens=skip_special_tokens)
scores = torch.tensor(self.queries_to_scores(output))
output = [output[i] for i in scores.topk(self.n_candidates).indices]
result.append(output)
return result
| trl/trl/extras/best_of_n_sampler.py/0 | {
"file_path": "trl/trl/extras/best_of_n_sampler.py",
"repo_id": "trl",
"token_count": 2253
} | 411 |
# KTO Authors: Kawin Ethayarajh, Winnie Xu, Niklas Muennighoff, Dan Jurafsky, and Douwe Kiela
# Copyright 2024 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import inspect
import random
import warnings
from collections import defaultdict
from contextlib import nullcontext
from copy import deepcopy
from functools import wraps
from operator import itemgetter
from typing import Any, Callable, Dict, List, Literal, Optional, Tuple, Union
import numpy as np
import torch
import torch.nn as nn
import torch.nn.functional as F
from accelerate.utils import is_deepspeed_available, tqdm
from datasets import Dataset, concatenate_datasets, interleave_datasets
from torch.utils.data import DataLoader, SequentialSampler
from transformers import (
AutoModelForCausalLM,
DataCollator,
PreTrainedModel,
PreTrainedTokenizerBase,
Trainer,
TrainingArguments,
)
from transformers.trainer_callback import TrainerCallback
from transformers.trainer_utils import EvalLoopOutput, has_length
from ..import_utils import is_peft_available, is_wandb_available
from ..models import PreTrainedModelWrapper, create_reference_model
from .kto_config import KTOConfig
from .utils import (
DPODataCollatorWithPadding,
disable_dropout_in_model,
pad_to_length,
peft_module_casting_to_bf16,
trl_sanitze_kwargs_for_tagging,
)
if is_peft_available():
from peft import PeftModel, get_peft_model, prepare_model_for_kbit_training
if is_wandb_available():
import wandb
if is_deepspeed_available():
import deepspeed
class KTOTrainer(Trainer):
r"""
Initialize KTOTrainer.
Args:
model (`transformers.PreTrainedModel`):
The model to train, preferably an `AutoModelForSequenceClassification`.
ref_model (`PreTrainedModelWrapper`):
Hugging Face transformer model with a casual language modelling head. Used for implicit reward computation and loss. If no
reference model is provided, the trainer will create a reference model with the same architecture as the model to be optimized.
args (`KTOConfig`):
The arguments to use for training.
train_dataset (`datasets.Dataset`):
The dataset to use for training.
eval_dataset (`datasets.Dataset`):
The dataset to use for evaluation.
tokenizer (`transformers.PreTrainedTokenizerBase`):
The tokenizer to use for training. This argument is required if you want to use the default data collator.
data_collator (`transformers.DataCollator`, *optional*, defaults to `None`):
The data collator to use for training. If None is specified, the default data collator (`DPODataCollatorWithPadding`) will be used
which will pad the sequences to the maximum length of the sequences in the batch, given a dataset of paired sequences.
model_init (`Callable[[], transformers.PreTrainedModel]`):
The model initializer to use for training. If None is specified, the default model initializer will be used.
callbacks (`List[transformers.TrainerCallback]`):
The callbacks to use for training.
optimizers (`Tuple[torch.optim.Optimizer, torch.optim.lr_scheduler.LambdaLR]`):
The optimizer and scheduler to use for training.
preprocess_logits_for_metrics (`Callable[[torch.Tensor, torch.Tensor], torch.Tensor]`):
The function to use to preprocess the logits before computing the metrics.
peft_config (`Dict`, defaults to `None`):
The PEFT configuration to use for training. If you pass a PEFT configuration, the model will be wrapped in a PEFT model.
disable_dropout (`bool`, defaults to `True`):
Whether or not to disable dropouts in `model` and `ref_model`.
compute_metrics (`Callable[[EvalPrediction], Dict]`, *optional*):
The function to use to compute the metrics. Must take a `EvalPrediction` and return
a dictionary string to metric values.
"""
_tag_names = ["trl", "kto"]
def __init__(
self,
model: Union[PreTrainedModel, nn.Module, str] = None,
ref_model: Optional[Union[PreTrainedModel, nn.Module, str]] = None,
args: KTOConfig = None,
train_dataset: Optional[Dataset] = None,
eval_dataset: Optional[Union[Dataset, Dict[str, Dataset]]] = None,
tokenizer: Optional[PreTrainedTokenizerBase] = None,
data_collator: Optional[DataCollator] = None,
model_init: Optional[Callable[[], PreTrainedModel]] = None,
callbacks: Optional[List[TrainerCallback]] = None,
optimizers: Tuple[torch.optim.Optimizer, torch.optim.lr_scheduler.LambdaLR] = (None, None),
preprocess_logits_for_metrics: Optional[Callable[[torch.Tensor, torch.Tensor], torch.Tensor]] = None,
peft_config: Optional[Dict] = None,
compute_metrics: Optional[Callable[[EvalLoopOutput], Dict]] = None,
):
if type(args) == TrainingArguments:
raise ValueError("Please use `KTOConfig` instead TrainingArguments.")
if args.model_init_kwargs is None:
model_init_kwargs = {}
elif not isinstance(model, str):
raise ValueError("You passed model_kwargs to the KTOTrainer. But your model is already instantiated.")
else:
model_init_kwargs = args.model_init_kwargs
model_init_kwargs["torch_dtype"] = (
model_init_kwargs["torch_dtype"]
if model_init_kwargs["torch_dtype"] in ["auto", None]
else getattr(torch, model_init_kwargs["torch_dtype"])
)
if args.ref_model_init_kwargs is None:
ref_model_init_kwargs = {}
elif not isinstance(ref_model, str):
raise ValueError(
"You passed ref_model_kwargs to the KTOTrainer. But your ref_model is already instantiated."
)
else:
ref_model_init_kwargs = args.ref_model_init_kwargs
ref_model_init_kwargs["torch_dtype"] = (
ref_model_init_kwargs["torch_dtype"]
if ref_model_init_kwargs["torch_dtype"] in ["auto", None]
else getattr(torch, ref_model_init_kwargs["torch_dtype"])
)
if isinstance(model, str):
warnings.warn(
"You passed a model_id to the KTOTrainer. This will automatically create an "
"`AutoModelForCausalLM` or a `PeftModel` (if you passed a `peft_config`) for you."
)
model = AutoModelForCausalLM.from_pretrained(model, **model_init_kwargs)
if isinstance(ref_model, str):
warnings.warn(
"You passed a ref model_id to the KTOTrainer. This will automatically create an "
"`AutoModelForCausalLM`"
)
ref_model = AutoModelForCausalLM.from_pretrained(ref_model, **ref_model_init_kwargs)
# Initialize this variable to False. This helps tracking the case when `peft_module_casting_to_bf16`
# has been called in order to properly call autocast if needed.
self._peft_has_been_casted_to_bf16 = False
if not is_peft_available() and peft_config is not None:
raise ValueError(
"PEFT is not installed and you passed a `peft_config` in the trainer's kwargs, please install it with `pip install peft` to use the PEFT models"
)
elif is_peft_available() and peft_config is not None:
# if model is a peft model and we have a peft_config, we merge and unload it first
if isinstance(model, PeftModel):
model = model.merge_and_unload()
if getattr(model, "is_loaded_in_8bit", False) or getattr(model, "is_loaded_in_4bit", False):
_support_gc_kwargs = hasattr(
args, "gradient_checkpointing_kwargs"
) and "gradient_checkpointing_kwargs" in list(
inspect.signature(prepare_model_for_kbit_training).parameters
)
prepare_model_kwargs = {"use_gradient_checkpointing": args.gradient_checkpointing}
if _support_gc_kwargs:
prepare_model_kwargs["gradient_checkpointing_kwargs"] = args.gradient_checkpointing_kwargs
model = prepare_model_for_kbit_training(model, **prepare_model_kwargs)
elif getattr(args, "gradient_checkpointing", False):
# For backward compatibility with older versions of transformers
if hasattr(model, "enable_input_require_grads"):
model.enable_input_require_grads()
else:
def make_inputs_require_grad(module, input, output):
output.requires_grad_(True)
model.get_input_embeddings().register_forward_hook(make_inputs_require_grad)
# get peft model with the given config
model = get_peft_model(model, peft_config)
if args.bf16 and getattr(model, "is_loaded_in_4bit", False):
peft_module_casting_to_bf16(model)
# If args.bf16 we need to explicitly call `generate` with torch amp autocast context manager
self._peft_has_been_casted_to_bf16 = True
# For models that use gradient_checkpointing, we need to attach a hook that enables input
# to explicitly have `requires_grad=True`, otherwise training will either silently
# fail or completely fail.
elif getattr(args, "gradient_checkpointing", False):
# For backward compatibility with older versions of transformers
if hasattr(model, "enable_input_require_grads"):
model.enable_input_require_grads()
else:
def make_inputs_require_grad(module, input, output):
output.requires_grad_(True)
model.get_input_embeddings().register_forward_hook(make_inputs_require_grad)
if args.generate_during_eval and not is_wandb_available():
raise ValueError(
"`generate_during_eval=True` requires Weights and Biases to be installed."
" Please install with `pip install wandb` to resolve."
)
if model is not None:
self.is_encoder_decoder = model.config.is_encoder_decoder
elif args.is_encoder_decoder is None:
raise ValueError("When no model is provided, you need to pass the parameter is_encoder_decoder.")
else:
self.is_encoder_decoder = args.is_encoder_decoder
self.is_peft_model = is_peft_available() and isinstance(model, PeftModel)
if ref_model:
self.ref_model = ref_model
elif self.is_peft_model or args.precompute_ref_log_probs:
# The `model` with adapters turned off will be used as the reference model
self.ref_model = None
else:
self.ref_model = create_reference_model(model)
if tokenizer is None:
raise ValueError(
"max_length or a tokenizer must be specified when using the default DPODataCollatorWithPadding"
)
if args.max_length is None:
warnings.warn(
"When using DPODataCollatorWithPadding, you should set `max_length` in the KTOTrainer's init"
" it will be set to `512` by default, but you should do it yourself in the future.",
UserWarning,
)
max_length = 512
if args.max_length is not None:
max_length = args.max_length
if args.max_prompt_length is None:
warnings.warn(
"When using DPODataCollatorWithPadding, you should set `max_prompt_length` in the KTOTrainer's init"
" it will be set to `128` by default, but you should do it yourself in the future.",
UserWarning,
)
max_prompt_length = 128
if args.max_prompt_length is not None:
max_prompt_length = args.max_prompt_length
max_completion_length = None
if args.max_completion_length is None and self.is_encoder_decoder:
warnings.warn(
"When using DPODataCollatorWithPadding with an encoder decoder architecture, you should set `max_completion_length` in the KTOTrainer's init"
" it will be set to `128` by default, but you should do it yourself in the future.",
UserWarning,
)
max_completion_length = 128
if args.max_completion_length is not None and self.is_encoder_decoder:
max_completion_length = args.max_completion_length
if data_collator is None:
data_collator = DPODataCollatorWithPadding(
pad_token_id=tokenizer.pad_token_id,
label_pad_token_id=args.label_pad_token_id,
is_encoder_decoder=self.is_encoder_decoder,
)
if args.remove_unused_columns:
args.remove_unused_columns = False
# warn users
warnings.warn(
"When using DPODataCollatorWithPadding, you should set `remove_unused_columns=False` in your KTOConfig"
" we have set it for you, but you should do it yourself in the future.",
UserWarning,
)
self.use_dpo_data_collator = True
else:
self.use_dpo_data_collator = False
# disable dropout in the model and reference model
disable_dropout_in_model(model)
if self.ref_model is not None:
disable_dropout_in_model(self.ref_model)
self.max_length = max_length
self.generate_during_eval = args.generate_during_eval
self.label_pad_token_id = args.label_pad_token_id
self.padding_value = args.padding_value if args.padding_value is not None else tokenizer.pad_token_id
self.max_prompt_length = max_prompt_length
self.truncation_mode = args.truncation_mode
self.max_completion_length = max_completion_length
self.tokenizer = tokenizer
self.precompute_ref_log_probs = args.precompute_ref_log_probs
# Since ref_logs are precomputed on the first call to get_train/eval_dataloader
# keep track of first called to avoid computation of future calls
self._precomputed_train_ref_log_probs = False
self._precomputed_eval_ref_log_probs = False
# metric
self._stored_metrics = defaultdict(lambda: defaultdict(list))
# KTO parameter
self.beta = args.beta
self.desirable_weight = args.desirable_weight
self.undesirable_weight = args.undesirable_weight
# get KL datasets
total_batch_size = (
max(torch.cuda.device_count(), 1) * args.per_device_train_batch_size * args.gradient_accumulation_steps
)
if total_batch_size <= 1:
raise ValueError(
"Batch size is 1 (too small). KTO will not work properly because the KL term will be equivalent to the implied reward."
)
# note: for best results, mismatched outputs y' used to estimate the KL term for a batch should be the
# same as the matched outputs y used to estimate the rewards in that batch, just paired with different x
train_KL_dataset = train_dataset.map(self.get_KL_dataset, batched=True, batch_size=total_batch_size)
if eval_dataset is not None:
eval_KL_dataset = eval_dataset.map(self.get_KL_dataset, batched=True, batch_size=total_batch_size)
# tokenize the datasets
train_dataset = train_dataset.map(
lambda row: self.tokenize_row(row, prefix=""), remove_columns=train_dataset.column_names
)
train_KL_dataset = train_KL_dataset.map(
lambda row: self.tokenize_row(row, prefix="KL_"), remove_columns=train_KL_dataset.column_names
)
# merge the datasets
train_dataset = concatenate_datasets([train_dataset, train_KL_dataset], axis=1)
if eval_dataset is not None:
eval_dataset = eval_dataset.map(
lambda row: self.tokenize_row(row, prefix=""), remove_columns=eval_dataset.column_names
)
eval_KL_dataset = eval_KL_dataset.map(
lambda row: self.tokenize_row(row, prefix="KL_"), remove_columns=eval_KL_dataset.column_names
)
# merge the datasets
eval_dataset = concatenate_datasets([eval_dataset, eval_KL_dataset], axis=1)
desirable = train_dataset.filter(lambda x: x["label"])
undesirable = train_dataset.filter(lambda x: not x["label"])
if len(desirable) != len(undesirable):
# The lower and upper bounds come from Eq. (8) of https://arxiv.org/abs/2402.01306
des_weight_lower_bound = round((len(undesirable) * self.undesirable_weight / len(desirable)) * 1, 2)
des_weight_upper_bound = round((len(undesirable) * self.undesirable_weight / len(desirable)) * 1.33, 2)
und_weight_lower_bound = round((len(desirable) * self.desirable_weight / len(undesirable)) / 1.33, 2)
und_weight_upper_bound = round((len(desirable) * self.desirable_weight / len(undesirable)) / 1, 2)
des_weight_in_range = des_weight_lower_bound <= self.desirable_weight <= des_weight_upper_bound
und_weight_in_range = und_weight_lower_bound <= self.undesirable_weight <= und_weight_upper_bound
if not (des_weight_in_range or und_weight_in_range):
warnings.warn(
f"""
You have different amounts of desirable/positive and undesirable/negative examples but the
weights on the desirable and undesirable losses don't seem to be in an ideal range. Based
on your data, we recommend EITHER desirable_weight in [{des_weight_lower_bound}, {des_weight_upper_bound}]
or undesirable_weight in [{und_weight_lower_bound}, {und_weight_upper_bound}] (but NOT BOTH).
See the documentation on how to optimally set these weights.""",
UserWarning,
)
# split the dataset and interleave them together with equal probability of choosing chosen or rejected
interleaved_train_dataset = interleave_datasets(
[desirable, undesirable],
stopping_strategy="all_exhausted",
)
interleaved_train_dataset = interleaved_train_dataset.shuffle(seed=args.data_seed)
if eval_dataset is not None:
interleaved_eval_dataset = interleave_datasets(
[eval_dataset.filter(lambda x: x["label"]), eval_dataset.filter(lambda x: not x["label"])],
stopping_strategy="all_exhausted",
)
else:
interleaved_eval_dataset = None
super().__init__(
model=model,
args=args,
data_collator=data_collator,
train_dataset=interleaved_train_dataset,
eval_dataset=interleaved_eval_dataset,
tokenizer=tokenizer,
model_init=model_init,
compute_metrics=compute_metrics,
callbacks=callbacks,
optimizers=optimizers,
preprocess_logits_for_metrics=preprocess_logits_for_metrics,
)
# Add tags for models that have been loaded with the correct transformers version
if hasattr(self.model, "add_model_tags"):
self.model.add_model_tags(self._tag_names)
if not hasattr(self, "accelerator"):
raise AttributeError(
"Your `Trainer` does not have an `accelerator` object. Consider upgrading `transformers`."
)
# Deepspeed Zero-3 does not support precompute_ref_log_probs
if self.is_deepspeed_enabled:
if self.accelerator.state.deepspeed_plugin.zero_stage == 3 and self.precompute_ref_log_probs:
raise ValueError(
"You cannot use `precompute_ref_log_probs=True` with Deepspeed ZeRO-3. Please set `precompute_ref_log_probs=False`."
)
if self.ref_model is None:
if not (self.is_peft_model or self.precompute_ref_log_probs):
raise ValueError(
"No reference model and model is not a Peft model. Try setting `precompute_ref_log_probs=True`"
)
else:
if self.is_deepspeed_enabled:
self.ref_model = self._prepare_deepspeed(self.ref_model)
else:
self.ref_model = self.accelerator.prepare_model(self.ref_model, evaluation_mode=True)
def _prepare_deepspeed(self, model: PreTrainedModelWrapper):
# Adapted from accelerate: https://github.com/huggingface/accelerate/blob/739b135f8367becb67ffaada12fe76e3aa60fefd/src/accelerate/accelerator.py#L1473
deepspeed_plugin = self.accelerator.state.deepspeed_plugin
config_kwargs = deepcopy(deepspeed_plugin.deepspeed_config)
if model is not None:
if hasattr(model, "config"):
hidden_size = (
max(model.config.hidden_sizes)
if getattr(model.config, "hidden_sizes", None)
else getattr(model.config, "hidden_size", None)
)
if hidden_size is not None and config_kwargs["zero_optimization"]["stage"] == 3:
# Note that `stage3_prefetch_bucket_size` can produce DeepSpeed messages like: `Invalidate trace cache @ step 0: expected module 1, but got module 0`
# This is expected and is not an error, see: https://github.com/microsoft/DeepSpeed/discussions/4081
config_kwargs.update(
{
"zero_optimization.reduce_bucket_size": hidden_size * hidden_size,
"zero_optimization.stage3_param_persistence_threshold": 10 * hidden_size,
"zero_optimization.stage3_prefetch_bucket_size": 0.9 * hidden_size * hidden_size,
}
)
# If ZeRO-3 is used, we shard both the active and reference model.
# Otherwise, we assume the reference model fits in memory and is initialized on each device with ZeRO disabled (stage 0)
if config_kwargs["zero_optimization"]["stage"] != 3:
config_kwargs["zero_optimization"]["stage"] = 0
model, *_ = deepspeed.initialize(model=model, config=config_kwargs)
model.eval()
return model
def get_train_dataloader(self) -> DataLoader:
"""
Returns the training [`~torch.utils.data.DataLoader`].
Subclass of transformers.src.transformers.trainer.get_train_dataloader to precompute `ref_log_probs`.
"""
if self.precompute_ref_log_probs and not self._precomputed_train_ref_log_probs:
dataloader_params = {
"batch_size": self.args.per_device_train_batch_size,
"collate_fn": self.data_collator,
"num_workers": self.args.dataloader_num_workers,
"pin_memory": self.args.dataloader_pin_memory,
"shuffle": False,
}
# prepare dataloader
data_loader = self.accelerator.prepare(DataLoader(self.train_dataset, **dataloader_params))
reference_completion_logps = []
reference_KL_logps = []
for padded_batch in tqdm(iterable=data_loader, desc="Train dataset reference log probs"):
reference_completion_logp, reference_KL_logp = self.compute_reference_log_probs(padded_batch)
reference_completion_logp = self.accelerator.gather_for_metrics(reference_completion_logp)
reference_completion_logps.append(reference_completion_logp.cpu())
reference_KL_logp = self.accelerator.gather_for_metrics(reference_KL_logp)
reference_KL_logps.append(reference_KL_logp.cpu())
self.train_dataset = self.train_dataset.add_column(
name="reference_logps", column=torch.cat(reference_completion_logps).float().numpy()
)
self.train_dataset = self.train_dataset.add_column(
name="reference_KL_logps", column=torch.cat(reference_KL_logps).float().numpy()
)
self._precomputed_train_ref_log_probs = True
return super().get_train_dataloader()
def get_eval_dataloader(self, eval_dataset: Optional[Dataset] = None) -> DataLoader:
"""
Returns the evaluation [`~torch.utils.data.DataLoader`].
Subclass of transformers.src.transformers.trainer.get_eval_dataloader to precompute `ref_log_probs`.
Args:
eval_dataset (`torch.utils.data.Dataset`, *optional*):
If provided, will override `self.eval_dataset`. If it is a [`~datasets.Dataset`], columns not accepted
by the `model.forward()` method are automatically removed. It must implement `__len__`.
"""
if eval_dataset is None and self.eval_dataset is None:
raise ValueError("Trainer: evaluation requires an eval_dataset.")
eval_dataset = eval_dataset if eval_dataset is not None else self.eval_dataset
if self.precompute_ref_log_probs and not self._precomputed_eval_ref_log_probs:
dataloader_params = {
"batch_size": self.args.per_device_eval_batch_size,
"collate_fn": self.data_collator,
"num_workers": self.args.dataloader_num_workers,
"pin_memory": self.args.dataloader_pin_memory,
"shuffle": False,
}
# prepare dataloader
data_loader = self.accelerator.prepare(DataLoader(eval_dataset, **dataloader_params))
reference_completion_logps = []
reference_KL_logps = []
for padded_batch in tqdm(iterable=data_loader, desc="Eval dataset reference log probs"):
reference_completion_logp, reference_KL_logp = self.compute_reference_log_probs(padded_batch)
reference_completion_logp = self.accelerator.gather_for_metrics(reference_completion_logp)
reference_completion_logps.append(reference_completion_logp.cpu())
reference_KL_logp = self.accelerator.gather_for_metrics(reference_KL_logp)
reference_KL_logps.append(reference_KL_logp.cpu())
eval_dataset = eval_dataset.add_column(
name="reference_logps", column=torch.cat(reference_completion_logps).float().numpy()
)
eval_dataset = eval_dataset.add_column(
name="reference_KL_logps", column=torch.cat(reference_KL_logps).float().numpy()
)
# Save calculated reference_chosen_logps and reference_rejected_logps to the eval_dataset for subsequent runs
if self.eval_dataset is not None:
self.eval_dataset = eval_dataset
self._precomputed_eval_ref_log_probs = True
return super().get_eval_dataloader(eval_dataset=eval_dataset)
def compute_reference_log_probs(self, padded_batch: Dict) -> Dict:
"""Computes log probabilities of the reference model for a single padded batch of a KTO specific dataset."""
with torch.no_grad():
if self.ref_model is None:
with self.accelerator.unwrap_model(
self.model
).disable_adapter() if self.is_peft_model else nullcontext():
if self.is_encoder_decoder:
completion_logits = self.model(
padded_batch["prompt_input_ids"],
attention_mask=padded_batch["prompt_attention_mask"],
decoder_input_ids=padded_batch.get("completion_decoder_input_ids"),
labels=padded_batch["completion_labels"],
).logits
KL_logits = self.model(
padded_batch["KL_prompt_input_ids"],
attention_mask=padded_batch["KL_prompt_attention_mask"],
decoder_input_ids=padded_batch.get("KL_completion_decoder_input_ids"),
labels=padded_batch["KL_completion_labels"],
).logits
else:
completion_logits = self.model(
padded_batch["completion_input_ids"],
attention_mask=padded_batch["completion_attention_mask"],
).logits
KL_logits = self.model(
padded_batch["KL_completion_input_ids"],
attention_mask=padded_batch["KL_completion_attention_mask"],
).logits
else:
if self.is_encoder_decoder:
completion_logits = self.ref_model(
padded_batch["prompt_input_ids"],
attention_mask=padded_batch["prompt_attention_mask"],
decoder_input_ids=padded_batch.get("completion_decoder_input_ids"),
labels=padded_batch["completion_labels"],
).logits
KL_logits = self.ref_model(
padded_batch["KL_prompt_input_ids"],
attention_mask=padded_batch["KL_prompt_attention_mask"],
decoder_input_ids=padded_batch.get("KL_completion_decoder_input_ids"),
labels=padded_batch["KL_completion_labels"],
).logits
else:
completion_logits = self.ref_model(
padded_batch["completion_input_ids"], attention_mask=padded_batch["completion_attention_mask"]
).logits
KL_logits = self.ref_model(
padded_batch["KL_completion_input_ids"],
attention_mask=padded_batch["KL_completion_attention_mask"],
).logits
completion_logps = self.get_batch_logps(
completion_logits,
padded_batch["completion_labels"],
average_log_prob=False,
is_encoder_decoder=self.is_encoder_decoder,
label_pad_token_id=self.label_pad_token_id,
)
KL_logps = self.get_batch_logps(
KL_logits,
padded_batch["KL_completion_labels"],
average_log_prob=False,
is_encoder_decoder=self.is_encoder_decoder,
label_pad_token_id=self.label_pad_token_id,
)
return completion_logps, KL_logps
def build_tokenized_answer(self, prompt, answer):
"""
Llama tokenizer does not satisfy `enc(a + b) = enc(a) + enc(b)`.
It does ensure `enc(a + b) = enc(a) + enc(a + b)[len(enc(a)):]`.
Reference:
https://github.com/EleutherAI/lm-evaluation-harness/pull/531#issuecomment-1595586257
"""
full_tokenized = self.tokenizer(prompt + answer, add_special_tokens=False)
prompt_input_ids = self.tokenizer(prompt, add_special_tokens=False)["input_ids"]
answer_input_ids = full_tokenized["input_ids"][len(prompt_input_ids) :]
answer_attention_mask = full_tokenized["attention_mask"][len(prompt_input_ids) :]
# Concat tokens to form `enc(a) + enc(a + b)[len(enc(a)):]`
full_concat_input_ids = np.concatenate([prompt_input_ids, answer_input_ids])
# Prepare input tokens for token by token comparison
full_input_ids = np.array(full_tokenized["input_ids"])
if len(full_input_ids) != len(full_concat_input_ids):
raise ValueError("Prompt input ids and answer input ids should have the same length.")
# On some tokenizers, like Llama-2 tokenizer, there are occasions where tokens
# can be merged together when tokenizing prompt+answer. This could result
# on the last token from the prompt being different when tokenized on its own
# vs when done as prompt+answer.
response_token_ids_start_idx = len(prompt_input_ids)
# If tokenized prompt is different than both prompt+answer, then it means the
# last token has changed due to merging.
if prompt_input_ids != full_tokenized["input_ids"][:response_token_ids_start_idx]:
response_token_ids_start_idx -= 1
prompt_input_ids = full_tokenized["input_ids"][:response_token_ids_start_idx]
prompt_attention_mask = full_tokenized["attention_mask"][:response_token_ids_start_idx]
if len(prompt_input_ids) != len(prompt_attention_mask):
raise ValueError("Prompt input ids and attention mask should have the same length.")
answer_input_ids = full_tokenized["input_ids"][response_token_ids_start_idx:]
answer_attention_mask = full_tokenized["attention_mask"][response_token_ids_start_idx:]
return dict(
prompt_input_ids=prompt_input_ids,
prompt_attention_mask=prompt_attention_mask,
answer_input_ids=answer_input_ids,
answer_attention_mask=answer_attention_mask,
)
def get_KL_dataset(self, batch) -> Dict:
"""Creates mismatched pairs of prompts and completions for the KL dataset."""
batch["completion"] = batch["completion"][::-1]
return batch
def tokenize_row(self, feature, model: Union[PreTrainedModel, nn.Module] = None, prefix="") -> Dict:
"""Tokenize a single row from a KTO specific dataset.
At this stage, we don't convert to PyTorch tensors yet; we just handle the truncation
in case the prompt + completion responses is/are too long. First
we truncate the prompt; if we're still too long, we truncate the completion.
We also create the labels for the completion responses, which are of length equal to
the sum of the length of the prompt and the completion response, with
label_pad_token_id for the prompt tokens.
"""
prompt = feature["prompt"]
completion = feature["completion"]
batch = {
f"{prefix}prompt": prompt,
f"{prefix}completion": completion,
f"{prefix}label": feature["label"],
}
if not self.is_encoder_decoder:
# Check issues below for more details
# 1. https://github.com/huggingface/trl/issues/907
# 2. https://github.com/EleutherAI/lm-evaluation-harness/pull/531#issuecomment-1595586257
# 3. https://github.com/LianjiaTech/BELLE/issues/337
if not isinstance(prompt, str):
raise ValueError(f"prompt should be an str but got {type(prompt)}")
if not isinstance(completion, str):
raise ValueError(f"completion should be an str but got {type(completion)}")
# keys of format prompt_* refers to just the prompt and answer_* refers to just the answer
all_tokens = self.build_tokenized_answer(prompt, completion)
max_length = self.max_length - 2
# if combined sequence is too long (> max_length - 1 for BOS token - 1 for EOS), truncate the prompt
if len(all_tokens["prompt_input_ids"]) + len(all_tokens["answer_input_ids"]) > max_length:
for k in ["prompt_input_ids", "prompt_attention_mask"]:
if self.truncation_mode == "keep_start":
all_tokens[k] = all_tokens[k][: self.max_prompt_length]
elif self.truncation_mode == "keep_end":
all_tokens[k] = all_tokens[k][-self.max_prompt_length :]
else:
raise ValueError(f"Unknown truncation mode: {self.truncation_mode}")
# if that's still too long, truncate the response
if len(all_tokens["prompt_input_ids"]) + len(all_tokens["answer_input_ids"]) > max_length:
for k in ["answer_input_ids", "answer_attention_mask"]:
all_tokens[k] = all_tokens[k][: max_length - self.max_prompt_length]
# for legacy reasons, use the completion_* prefix to now refer to the joint sequence
batch[f"{prefix}prompt_input_ids"] = [self.tokenizer.bos_token_id] + all_tokens["prompt_input_ids"]
batch[f"{prefix}prompt_attention_mask"] = [1] + all_tokens["prompt_attention_mask"]
batch[f"{prefix}completion_input_ids"] = (
[self.tokenizer.bos_token_id]
+ all_tokens["prompt_input_ids"]
+ all_tokens["answer_input_ids"]
+ [self.tokenizer.eos_token_id]
)
batch[f"{prefix}completion_attention_mask"] = (
[1] + all_tokens["prompt_attention_mask"] + all_tokens["answer_attention_mask"] + [1]
)
batch[f"{prefix}completion_labels"] = batch[f"{prefix}completion_input_ids"][:]
batch[f"{prefix}completion_labels"][: len(batch[f"{prefix}prompt_input_ids"])] = [
self.label_pad_token_id
] * len(batch[f"{prefix}prompt_input_ids"])
else:
completion_tokens = self.tokenizer(
completion, truncation=True, max_length=self.max_completion_length, add_special_tokens=True
)
prompt_tokens = self.tokenizer(
prompt, truncation=True, max_length=self.max_prompt_length, add_special_tokens=True
)
batch[f"{prefix}prompt_input_ids"] = prompt_tokens["input_ids"]
batch[f"{prefix}prompt_attention_mask"] = prompt_tokens["attention_mask"]
batch[f"{prefix}completion_labels"] = completion_tokens["input_ids"]
batch[f"{prefix}completion_attention_mask"] = completion_tokens["attention_mask"]
if model is not None and hasattr(model, "prepare_decoder_input_ids_from_labels"):
batch[f"{prefix}completion_decoder_input_ids"] = model.prepare_decoder_input_ids_from_labels(
labels=torch.tensor(batch["completion_labels"])
)
return batch
@staticmethod
def get_batch_logps(
logits: torch.FloatTensor,
labels: torch.LongTensor,
average_log_prob: bool = False,
label_pad_token_id: int = -100,
is_encoder_decoder: bool = False,
) -> torch.FloatTensor:
"""Compute the log probabilities of the given labels under the given logits.
Args:
logits: Logits of the model (unnormalized). Shape: (batch_size, sequence_length, vocab_size)
labels: Labels for which to compute the log probabilities. Label tokens with a value of label_pad_token_id are ignored. Shape: (batch_size, sequence_length)
average_log_prob: If True, return the average log probability per (non-masked) token. Otherwise, return the sum of the log probabilities of the (non-masked) tokens.
Returns:
A tensor of shape (batch_size,) containing the average/sum log probabilities of the given labels under the given logits.
"""
if logits.shape[:-1] != labels.shape:
raise ValueError("Logits (batch and sequence length dim) and labels must have the same shape.")
if not is_encoder_decoder:
labels = labels[:, 1:].clone()
logits = logits[:, :-1, :]
else:
# Fixes end-dec RuntimeError
labels = labels.clone()
loss_mask = labels != label_pad_token_id
# dummy token; we'll ignore the losses on these tokens later
labels[labels == label_pad_token_id] = 0
per_token_logps = torch.gather(logits.log_softmax(-1), dim=2, index=labels.unsqueeze(2)).squeeze(2)
if average_log_prob:
return (per_token_logps * loss_mask).sum(-1) / loss_mask.sum(-1)
else:
return (per_token_logps * loss_mask).sum(-1)
def forward(
self, model: nn.Module, batch: Dict[str, Union[List, torch.LongTensor]]
) -> Tuple[torch.FloatTensor, torch.FloatTensor, torch.FloatTensor, torch.FloatTensor]:
if self.is_encoder_decoder:
with torch.no_grad():
KL_logits = model(
batch["KL_prompt_input_ids"],
attention_mask=batch["KL_prompt_attention_mask"],
decoder_input_ids=batch.get("KL_completion_decoder_input_ids"),
labels=batch["KL_completion_labels"],
).logits
completion_logits = model(
batch["prompt_input_ids"],
attention_mask=batch["prompt_attention_mask"],
decoder_input_ids=batch.get("completion_decoder_input_ids"),
labels=batch["completion_labels"],
).logits
else:
with torch.no_grad():
KL_logits = model(
batch["KL_completion_input_ids"],
attention_mask=batch["KL_completion_attention_mask"],
).logits
completion_logits = model(
batch["completion_input_ids"],
attention_mask=batch["completion_attention_mask"],
).logits
completion_logps = self.get_batch_logps(
completion_logits,
batch["completion_labels"],
average_log_prob=False,
is_encoder_decoder=self.is_encoder_decoder,
label_pad_token_id=self.label_pad_token_id,
)
KL_logps = self.get_batch_logps(
KL_logits,
batch["KL_completion_labels"],
average_log_prob=False,
is_encoder_decoder=self.is_encoder_decoder,
label_pad_token_id=self.label_pad_token_id,
)
if completion_logps.shape[0] != len(batch["label"]):
raise ValueError(
"There is a mismatch between the number of examples in this batch and the number of "
"examples for which an output sequence was predicted."
)
chosen_idx = [i for i in range(completion_logps.shape[0]) if batch["label"][i] is True]
rejected_idx = [i for i in range(completion_logps.shape[0]) if batch["label"][i] is False]
chosen_logps = completion_logps[chosen_idx, ...]
rejected_logps = completion_logps[rejected_idx, ...]
chosen_logits = completion_logits[chosen_idx, ...]
rejected_logits = completion_logits[rejected_idx, ...]
return (chosen_logps, rejected_logps, chosen_logits, rejected_logits, KL_logps)
def kto_loss(
self,
policy_chosen_logps: torch.FloatTensor,
policy_rejected_logps: torch.FloatTensor,
policy_KL_logps: torch.FloatTensor,
reference_chosen_logps: torch.FloatTensor,
reference_rejected_logps: torch.FloatTensor,
reference_KL_logps: torch.FloatTensor,
) -> Tuple[torch.FloatTensor, torch.FloatTensor, torch.FloatTensor, torch.FloatTensor]:
"""Compute the KTO loss for a batch of policy and reference model log probabilities.
Args:
policy_chosen_logps: Log probabilities of the policy model for the chosen responses. Shape: (batch_size,)
policy_rejected_logps: Log probabilities of the policy model for the rejected responses. Shape: (batch_size,)
policy_KL_logps: Log probabilities of the policy model for the KL responses. Shape: (batch_size,)
reference_chosen_logps: Log probabilities of the reference model for the chosen responses. Shape: (batch_size,)
reference_rejected_logps: Log probabilities of the reference model for the rejected responses. Shape: (batch_size,)
reference_KL_logps: Log probabilities of the reference model for the KL responses. Shape: (batch_size,)
Returns:
A tuple of four tensors: (losses, chosen_rewards, rejected_rewards, KL).
The losses tensor contains the KTO loss for each example in the batch.
The chosen_rewards and rejected_rewards tensors contain the rewards for the chosen and rejected responses, respectively.
The KL tensor contains the detached KL divergence estimate between the policy and reference models.
"""
KL = (policy_KL_logps - reference_KL_logps).mean().detach()
KL = self.accelerator.gather(KL).mean().clamp(min=0)
if policy_chosen_logps.shape[0] != 0 or reference_chosen_logps.shape[0] != 0:
chosen_logratios = policy_chosen_logps - reference_chosen_logps
chosen_losses = 1 - F.sigmoid(self.beta * (chosen_logratios - KL))
chosen_rewards = self.beta * chosen_logratios.detach()
else:
# lists can't be empty -- if they are, then accelerate.gather will hang
chosen_losses = torch.Tensor([torch.nan]).to(self.accelerator.device)
chosen_rewards = torch.Tensor([torch.nan]).to(self.accelerator.device)
if policy_rejected_logps.shape[0] != 0 or reference_rejected_logps.shape[0] != 0:
rejected_logratios = policy_rejected_logps - reference_rejected_logps
rejected_losses = 1 - F.sigmoid(self.beta * (KL - rejected_logratios))
rejected_rewards = self.beta * rejected_logratios.detach()
else:
# lists can't be empty -- if they are, then accelerate.gather will hang
rejected_losses = torch.Tensor([torch.nan]).to(self.accelerator.device)
rejected_rewards = torch.Tensor([torch.nan]).to(self.accelerator.device)
losses = torch.cat(
(self.desirable_weight * chosen_losses, self.undesirable_weight * rejected_losses),
0,
)
return losses, chosen_rewards, rejected_rewards, KL
def get_batch_loss_metrics(
self,
model,
batch: Dict[str, Union[List, torch.LongTensor]],
train_eval: Literal["train", "eval"] = "train",
):
"""Compute the KTO loss and other metrics for the given batch of inputs for train or test."""
metrics = {}
batch = {k: (v.to(self.accelerator.device) if isinstance(v, torch.Tensor) else v) for k, v in batch.items()}
(
policy_chosen_logps,
policy_rejected_logps,
policy_chosen_logits,
policy_rejected_logits,
policy_KL_logps,
) = self.forward(model, batch)
# if reference_logps in batch use them, otherwise use the reference model
if "reference_logps" in batch:
chosen_idx = [i for i in range(batch["reference_logps"].shape[0]) if batch["label"][i] is True]
rejected_idx = [i for i in range(batch["reference_logps"].shape[0]) if batch["label"][i] is False]
reference_chosen_logps = batch["reference_logps"][chosen_idx, ...]
reference_rejected_logps = batch["reference_logps"][rejected_idx, ...]
reference_KL_logps = batch["reference_KL_logps"]
else:
with torch.no_grad():
if self.ref_model is None:
with self.accelerator.unwrap_model(self.model).disable_adapter():
(
reference_chosen_logps,
reference_rejected_logps,
_,
_,
reference_KL_logps,
) = self.forward(self.model, batch)
else:
(
reference_chosen_logps,
reference_rejected_logps,
_,
_,
reference_KL_logps,
) = self.forward(self.ref_model, batch)
losses, chosen_rewards, rejected_rewards, kl = self.kto_loss(
policy_chosen_logps,
policy_rejected_logps,
policy_KL_logps,
reference_chosen_logps,
reference_rejected_logps,
reference_KL_logps,
)
mean_chosen_reward = chosen_rewards.nanmean().detach()
mean_rejected_reward = rejected_rewards.nanmean().detach()
mean_chosen_logps = policy_chosen_logps.nanmean().detach()
mean_rejected_logps = policy_rejected_logps.nanmean().detach()
prefix = "eval_" if train_eval == "eval" else ""
metrics[f"{prefix}rewards/chosen"] = self.accelerator.gather(mean_chosen_reward).nanmean().cpu()
metrics[f"{prefix}rewards/rejected"] = self.accelerator.gather(mean_rejected_reward).nanmean().cpu()
metrics[f"{prefix}rewards/margins"] = metrics[f"{prefix}rewards/chosen"] - metrics[f"{prefix}rewards/rejected"]
metrics[f"{prefix}kl"] = kl.item() # has already been gathered in kto_loss
metrics[f"{prefix}logps/chosen"] = self.accelerator.gather(mean_chosen_logps).nanmean().cpu()
metrics[f"{prefix}logps/rejected"] = self.accelerator.gather(mean_rejected_logps).nanmean().cpu()
loss = (
losses.mean()
if losses.shape[0] != 0
else torch.tensor(float("nan"), requires_grad=True).to(self.accelerator.device)
)
return loss, metrics
def compute_loss(
self,
model: Union[PreTrainedModel, nn.Module],
inputs: Dict[str, Union[torch.Tensor, Any]],
return_outputs=False,
) -> Union[torch.Tensor, Tuple[torch.Tensor, Dict[str, torch.Tensor]]]:
if not self.use_dpo_data_collator:
warnings.warn(
"compute_loss is only implemented for DPODataCollatorWithPadding, and you passed a datacollator that is different than "
"DPODataCollatorWithPadding - you might see unexpected behavior. Alternatively, you can implement your own prediction_step method if you are using a custom data collator"
)
loss, metrics = self.get_batch_loss_metrics(model, inputs, train_eval="train")
# force log the metrics
if self.accelerator.is_main_process:
self.store_metrics(metrics, train_eval="train")
if return_outputs:
return (loss, metrics)
return loss
def store_metrics(self, metrics: Dict[str, float], train_eval: Literal["train", "eval"] = "train") -> None:
for key, value in metrics.items():
self._stored_metrics[train_eval][key].append(value)
def _get_train_sampler(self) -> Optional[torch.utils.data.Sampler]:
# We use a sequential sampler for training as the order of the interleaved dataset is important
if self.train_dataset is None or not has_length(self.train_dataset):
return None
return SequentialSampler(self.train_dataset)
def get_batch_samples(self, model, batch: Dict[str, torch.LongTensor]) -> Tuple[str, str]:
"""Generate samples from the model and reference model for the given batch of inputs."""
# If one uses `generate_during_eval` with peft + bf16, we need to explictly call generate with
# the torch cuda amp context manager as some hidden states are silently casted to full precision.
generate_context_manager = nullcontext if not self._peft_has_been_casted_to_bf16 else torch.cuda.amp.autocast
with generate_context_manager():
policy_output = model.generate(
input_ids=batch["prompt_input_ids"],
attention_mask=batch["prompt_attention_mask"],
max_length=self.max_length,
do_sample=True,
pad_token_id=self.tokenizer.pad_token_id,
)
# if reference_output in batch use that otherwise use the reference model
if "reference_output" in batch:
reference_output = batch["reference_output"]
else:
if self.ref_model is None:
with self.null_ref_context():
reference_output = self.model.generate(
input_ids=batch["prompt_input_ids"],
attention_mask=batch["prompt_attention_mask"],
max_length=self.max_length,
do_sample=True,
pad_token_id=self.tokenizer.pad_token_id,
)
else:
reference_output = self.ref_model.generate(
input_ids=batch["prompt_input_ids"],
attention_mask=batch["prompt_attention_mask"],
max_length=self.max_length,
do_sample=True,
pad_token_id=self.tokenizer.pad_token_id,
)
policy_output = pad_to_length(policy_output, self.max_length, self.tokenizer.pad_token_id)
policy_output_decoded = self.tokenizer.batch_decode(policy_output, skip_special_tokens=True)
reference_output = pad_to_length(reference_output, self.max_length, self.tokenizer.pad_token_id)
reference_output_decoded = self.tokenizer.batch_decode(reference_output, skip_special_tokens=True)
return policy_output_decoded, reference_output_decoded
def prediction_step(
self,
model: Union[PreTrainedModel, nn.Module],
inputs: Dict[str, Union[torch.Tensor, Any]],
prediction_loss_only: bool,
ignore_keys: Optional[List[str]] = None,
):
if not self.use_dpo_data_collator:
warnings.warn(
"prediction_step is only implemented for DPODataCollatorWithPadding, and you passed a datacollator that is different than "
"DPODataCollatorWithPadding - you might see unexpected behavior. Alternatively, you can implement your own prediction_step method if you are using a custom data collator"
)
if ignore_keys is None:
if hasattr(model, "config"):
ignore_keys = getattr(model.config, "keys_to_ignore_at_inference", [])
else:
ignore_keys = []
prediction_context_manager = torch.cuda.amp.autocast if self._peft_has_been_casted_to_bf16 else nullcontext
with torch.no_grad(), prediction_context_manager():
loss, metrics = self.get_batch_loss_metrics(model, inputs, train_eval="eval")
# force log the metrics
if self.accelerator.is_main_process:
self.store_metrics(metrics, train_eval="eval")
if prediction_loss_only:
return (loss.detach(), None, None)
# logits for the chosen and rejected samples from model
logits_dict = {
"eval_logits/chosen": metrics["eval_logits/chosen"],
"eval_logits/rejected": metrics["eval_logits/rejected"],
}
logits = tuple(v.unsqueeze(dim=0) for k, v in logits_dict.items() if k not in ignore_keys)
logits = torch.stack(logits).mean(axis=1).to(self.accelerator.device)
labels = torch.zeros(logits.shape[0], device=self.accelerator.device)
return (loss.detach(), logits, labels)
def evaluation_loop(
self,
dataloader: DataLoader,
description: str,
prediction_loss_only: Optional[bool] = None,
ignore_keys: Optional[List[str]] = None,
metric_key_prefix: str = "eval",
) -> EvalLoopOutput:
"""
Overriding built-in evaluation loop to store metrics for each batch.
Prediction/evaluation loop, shared by `Trainer.evaluate()` and `Trainer.predict()`.
Works both with or without labels.
"""
# Sample and save to game log if requested (for one batch to save time)
if self.generate_during_eval:
# Generate random indices within the range of the total number of samples
num_samples = len(dataloader.dataset)
random_indices = random.sample(range(num_samples), k=self.args.eval_batch_size)
# Use dataloader.dataset.select to get the random batch without iterating over the DataLoader
random_batch_dataset = dataloader.dataset.select(random_indices)
random_batch = self.data_collator(random_batch_dataset)
random_batch = self._prepare_inputs(random_batch)
target_indicies = [i for i in range(len(random_batch["kl"])) if random_batch["kl"][i] is False]
target_batch = {
"prompt_input_ids": itemgetter(*target_indicies)(random_batch["prompt_input_ids"]),
"prompt_attention_mask": itemgetter(*target_indicies)(random_batch["prompt_attention_mask"]),
"prompt": itemgetter(*target_indicies)(random_batch["prompt"]),
}
policy_output_decoded, ref_output_decoded = self.get_batch_samples(self.model, target_batch)
self.log(
{
"game_log": wandb.Table(
columns=["Prompt", "Policy", "Ref Model"],
rows=[
[prompt, pol[len(prompt) :], ref[len(prompt) :]]
for prompt, pol, ref in zip(
target_batch["prompt"], policy_output_decoded, ref_output_decoded
)
],
)
}
)
self.state.log_history.pop()
# Base evaluation
initial_output = super().evaluation_loop(
dataloader, description, prediction_loss_only, ignore_keys, metric_key_prefix
)
return initial_output
def log(self, logs: Dict[str, float]) -> None:
"""
Log `logs` on the various objects watching training, including stored metrics.
Args:
logs (`Dict[str, float]`):
The values to log.
"""
# logs either has 'loss' or 'eval_loss'
train_eval = "train" if "loss" in logs else "eval"
# Add averaged stored metrics to logs
for key, metrics in self._stored_metrics[train_eval].items():
logs[key] = torch.tensor(metrics).mean().item()
del self._stored_metrics[train_eval]
return super().log(logs)
@wraps(Trainer.push_to_hub)
def push_to_hub(self, commit_message: Optional[str] = "End of training", blocking: bool = True, **kwargs) -> str:
"""
Overwrite the `push_to_hub` method in order to force-add the tag "sft" when pushing the
model on the Hub. Please refer to `~transformers.Trainer.push_to_hub` for more details.
"""
kwargs = trl_sanitze_kwargs_for_tagging(model=self.model, tag_names=self._tag_names, kwargs=kwargs)
return super().push_to_hub(commit_message=commit_message, blocking=blocking, **kwargs)
| trl/trl/trainer/kto_trainer.py/0 | {
"file_path": "trl/trl/trainer/kto_trainer.py",
"repo_id": "trl",
"token_count": 27065
} | 412 |
<!---
Copyright 2023 The HuggingFace Team. All rights reserved.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
-->
# Generating the documentation
To generate the documentation, you first have to build it. Several packages are necessary to build the doc,
you can install them with the following command, at the root of the code repository:
```bash
pip install -e ".[docs]"
```
Then you need to install our special tool that builds the documentation:
```bash
pip install git+https://github.com/huggingface/doc-builder
```
---
**NOTE**
You only need to generate the documentation to inspect it locally (if you're planning changes and want to
check how they look before committing for instance). You don't have to commit the built documentation.
---
## Building the documentation
Once you have setup the `doc-builder` and additional packages, you can generate the documentation by
typing the following command:
```bash
doc-builder build accelerate docs/source/ --build_dir ~/tmp/test-build
```
You can adapt the `--build_dir` to set any temporary folder that you prefer. This command will create it and generate
the MDX files that will be rendered as the documentation on the main website. You can inspect them in your favorite
Markdown editor.
## Previewing the documentation
To preview the docs, first install the `watchdog` module with:
```bash
pip install watchdog
```
Then run the following command:
```bash
doc-builder preview {package_name} {path_to_docs}
```
For example:
```bash
doc-builder preview accelerate docs/source/
```
The docs will be viewable at [http://localhost:3000](http://localhost:3000). You can also preview the docs once you have opened a PR. You will see a bot add a comment to a link where the documentation with your changes lives.
---
**NOTE**
The `preview` command only works with existing doc files. When you add a completely new file, you need to update `_toctree.yml` & restart `preview` command (`ctrl-c` to stop it & call `doc-builder preview ...` again).
---
## Adding a new element to the navigation bar
Accepted files are Markdown (.md).
Create a file with its extension and put it in the source directory. You can then link it to the toc-tree by putting
the filename without the extension in the [`_toctree.yml`](https://github.com/huggingface/accelerate/blob/main/docs/source/_toctree.yml) file.
## Renaming section headers and moving sections
It helps to keep the old links working when renaming the section header and/or moving sections from one document to another. This is because the old links are likely to be used in Issues, Forums, and Social media and it'd make for a much more superior user experience if users reading those months later could still easily navigate to the originally intended information.
Therefore, we simply keep a little map of moved sections at the end of the document where the original section was. The key is to preserve the original anchor.
So if you renamed a section from: "Section A" to "Section B", then you can add at the end of the file:
```
Sections that were moved:
[ <a href="#section-b">Section A</a><a id="section-a"></a> ]
```
and of course, if you moved it to another file, then:
```
Sections that were moved:
[ <a href="../new-file#section-b">Section A</a><a id="section-a"></a> ]
```
Use the relative style to link to the new file so that the versioned docs continue to work.
## Writing Documentation - Specification
The `huggingface/accelerate` documentation follows the
[Google documentation](https://sphinxcontrib-napoleon.readthedocs.io/en/latest/example_google.html) style for docstrings,
although we can write them directly in Markdown.
### Adding a new tutorial
Adding a new tutorial or section is done in two steps:
- Add a new file under `./source`. This file can either be ReStructuredText (.rst) or Markdown (.md).
- Link that file in `./source/_toctree.yml` on the correct toc-tree.
Make sure to put your new file under the proper section. It's unlikely to go in the first section (*Get Started*), so
depending on the intended targets (beginners, more advanced users, or researchers) it should go in sections two, three, or
four.
### Writing source documentation
Values that should be put in `code` should either be surrounded by backticks: \`like so\`. Note that argument names
and objects like True, None, or any strings should usually be put in `code`.
When mentioning a class, function, or method, it is recommended to use our syntax for internal links so that our tool
adds a link to its documentation with this syntax: \[\`XXXClass\`\] or \[\`function\`\]. This requires the class or
function to be in the main package.
If you want to create a link to some internal class or function, you need to
provide its path. For instance: \[\`utils.gather\`\]. This will be converted into a link with
`utils.gather` in the description. To get rid of the path and only keep the name of the object you are
linking to in the description, add a ~: \[\`~utils.gather\`\] will generate a link with `gather` in the description.
The same works for methods so you can either use \[\`XXXClass.method\`\] or \[~\`XXXClass.method\`\].
#### Defining arguments in a method
Arguments should be defined with the `Args:` (or `Arguments:` or `Parameters:`) prefix, followed by a line return and
an indentation. The argument should be followed by its type, with its shape if it is a tensor, a colon, and its
description:
```
Args:
n_layers (`int`): The number of layers of the model.
```
If the description is too long to fit in one line (more than 119 characters in total), another indentation is necessary
before writing the description after the argument.
Finally, to maintain uniformity if any *one* description is too long to fit on one line, the
rest of the parameters should follow suit and have an indention before their description.
Here's an example showcasing everything so far:
```
Args:
gradient_accumulation_steps (`int`, *optional*, default to 1):
The number of steps that should pass before gradients are accumulated. A number > 1 should be combined with `Accelerator.accumulate`.
cpu (`bool`, *optional*):
Whether or not to force the script to execute on CPU. Will ignore GPU available if set to `True` and force the execution on one process only.
```
For optional arguments or arguments with defaults we follow the following syntax: imagine we have a function with the
following signature:
```
def my_function(x: str = None, a: float = 1):
```
then its documentation should look like this:
```
Args:
x (`str`, *optional*):
This argument controls ... and has a description longer than 119 chars.
a (`float`, *optional*, defaults to 1):
This argument is used to ... and has a description longer than 119 chars.
```
Note that we always omit the "defaults to \`None\`" when None is the default for any argument. Also note that even
if the first line describing your argument type and its default gets long, you can't break it on several lines. You can
however write as many lines as you want in the indented description (see the example above with `input_ids`).
#### Writing a multi-line code block
Multi-line code blocks can be useful for displaying examples. They are done between two lines of three backticks as usual in Markdown:
````
```python
# first line of code
# second line
# etc
```
````
#### Writing a return block
The return block should be introduced with the `Returns:` prefix, followed by a line return and an indentation.
The first line should be the type of the return, followed by a line return. No need to indent further for the elements
building the return.
Here's an example of a single value return:
```
Returns:
`List[int]`: A list of integers in the range [0, 1] --- 1 for a special token, 0 for a sequence token.
```
Here's an example of a tuple return, comprising several objects:
```
Returns:
`tuple(torch.FloatTensor)` comprising various elements depending on the configuration ([`BertConfig`]) and inputs:
- ** loss** (*optional*, returned when `masked_lm_labels` is provided) `torch.FloatTensor` of shape `(1,)` --
Total loss is the sum of the masked language modeling loss and the next sequence prediction (classification) loss.
- **prediction_scores** (`torch.FloatTensor` of shape `(batch_size, sequence_length, config.vocab_size)`) --
Prediction scores of the language modeling head (scores for each vocabulary token before SoftMax).
```
## Styling the docstring
We have an automatic script running with the `make style` comment that will make sure that:
- the docstrings fully take advantage of the line width
- all code examples are formatted using black, like the code of the Transformers library
This script may have some weird failures if you made a syntax mistake or if you uncover a bug. Therefore, it's
recommended to commit your changes before running `make style`, so you can revert the changes done by that script
easily.
## Writing documentation examples
The syntax for Example docstrings can look as follows:
```
Example:
```python
>>> import time
>>> from accelerate import Accelerator
>>> accelerator = Accelerator()
>>> if accelerator.is_main_process:
... time.sleep(2)
>>> else:
... print("I'm waiting for the main process to finish its sleep...")
>>> accelerator.wait_for_everyone()
>>> # Should print on every process at the same time
>>> print("Everyone is here")
```
```
The docstring should give a minimal, clear example of how the respective function
is to be used in inference and also include the expected (ideally sensible)
output.
Often, readers will try out the example before even going through the function
or class definitions. Therefore, it is of utmost importance that the example
works as expected. | accelerate/docs/README.md/0 | {
"file_path": "accelerate/docs/README.md",
"repo_id": "accelerate",
"token_count": 2883
} | 0 |
<!--Copyright 2022 The HuggingFace Team. All rights reserved.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with
the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on
an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the
specific language governing permissions and limitations under the License.
⚠️ Note that this file is in Markdown but contain specific syntax for our doc-builder (similar to MDX) that may not be
rendered properly in your Markdown viewer.
-->
# Training on TPUs with 🤗 Accelerate
Training on TPUs can be slightly different from training on multi-gpu, even with 🤗 Accelerate. This guide aims to show you
where you should be careful and why, as well as the best practices in general.
## Training in a Notebook
The main carepoint when training on TPUs comes from the [`notebook_launcher`]. As mentioned in the [notebook tutorial](../usage_guides/notebook), you need to
restructure your training code into a function that can get passed to the [`notebook_launcher`] function and be careful about not declaring any tensors on the GPU.
While on a TPU that last part is not as important, a critical part to understand is that when you launch code from a notebook you do so through a process called **forking**.
When launching from the command-line, you perform **spawning**, where a python process is not currently running and you *spawn* a new process in. Since your Jupyter notebook is already
utilizing a python process, you need to *fork* a new process from it to launch your code.
Where this becomes important is in regard to declaring your model. On forked TPU processes, it is recommended that you instantiate your model *once* and pass this into your
training function. This is different than training on GPUs where you create `n` models that have their gradients synced and back-propagated at certain moments. Instead, one
model instance is shared between all the nodes and it is passed back and forth. This is important especially when training on low-resource TPUs such as those provided in Kaggle kernels or
on Google Colaboratory.
Below is an example of a training function passed to the [`notebook_launcher`] if training on CPUs or GPUs:
<Tip>
This code snippet is based off the one from the `simple_nlp_example` notebook found [here](https://github.com/huggingface/notebooks/blob/main/examples/accelerate_examples/simple_nlp_example.ipynb) with slight
modifications for the sake of simplicity
</Tip>
```python
def training_function():
# Initialize accelerator
accelerator = Accelerator()
model = AutoModelForSequenceClassification.from_pretrained("bert-base-cased", num_labels=2)
train_dataloader, eval_dataloader = create_dataloaders(
train_batch_size=hyperparameters["train_batch_size"], eval_batch_size=hyperparameters["eval_batch_size"]
)
# Instantiate optimizer
optimizer = AdamW(params=model.parameters(), lr=hyperparameters["learning_rate"])
# Prepare everything
# There is no specific order to remember, we just need to unpack the objects in the same order we gave them to the
# prepare method.
model, optimizer, train_dataloader, eval_dataloader = accelerator.prepare(
model, optimizer, train_dataloader, eval_dataloader
)
num_epochs = hyperparameters["num_epochs"]
# Now we train the model
for epoch in range(num_epochs):
model.train()
for step, batch in enumerate(train_dataloader):
outputs = model(**batch)
loss = outputs.loss
accelerator.backward(loss)
optimizer.step()
optimizer.zero_grad()
```
```python
from accelerate import notebook_launcher
notebook_launcher(training_function)
```
<Tip>
The `notebook_launcher` will default to 8 processes if 🤗 Accelerate has been configured for a TPU
</Tip>
If you use this example and declare the model *inside* the training loop, then on a low-resource system you will potentially see an error
like:
```
ProcessExitedException: process 0 terminated with signal SIGSEGV
```
This error is *extremely* cryptic but the basic explanation is you ran out of system RAM. You can avoid this entirely by reconfiguring the training function to
accept a single `model` argument, and declare it in an outside cell:
```python
# In another Jupyter cell
model = AutoModelForSequenceClassification.from_pretrained("bert-base-cased", num_labels=2)
```
```diff
+ def training_function(model):
# Initialize accelerator
accelerator = Accelerator()
- model = AutoModelForSequenceClassification.from_pretrained("bert-base-cased", num_labels=2)
train_dataloader, eval_dataloader = create_dataloaders(
train_batch_size=hyperparameters["train_batch_size"], eval_batch_size=hyperparameters["eval_batch_size"]
)
...
```
And finally calling the training function with:
```diff
from accelerate import notebook_launcher
- notebook_launcher(training_function)
+ notebook_launcher(training_function, (model,))
```
<Tip>
The above workaround is only needed when launching a TPU instance from a Jupyter Notebook on a low-resource server such as Google Colaboratory or Kaggle. If
using a script or launching on a much beefier server declaring the model beforehand is not needed.
</Tip>
## Mixed Precision and Global Variables
As mentioned in the [mixed precision tutorial](../usage_guides/mixed_precision), 🤗 Accelerate supports fp16 and bf16, both of which can be used on TPUs.
That being said, ideally `bf16` should be utilized as it is extremely efficient to use.
There are two "layers" when using `bf16` and 🤗 Accelerate on TPUs, at the base level and at the operation level.
At the base level, this is enabled when passing `mixed_precision="bf16"` to `Accelerator`, such as:
```python
accelerator = Accelerator(mixed_precision="bf16")
```
By default, this will cast `torch.float` and `torch.double` to `bfloat16` on TPUs.
The specific configuration being set is an environmental variable of `XLA_USE_BF16` is set to `1`.
There is a further configuration you can perform which is setting the `XLA_DOWNCAST_BF16` environmental variable. If set to `1`, then
`torch.float` is `bfloat16` and `torch.double` is `float32`.
This is performed in the `Accelerator` object when passing `downcast_bf16=True`:
```python
accelerator = Accelerator(mixed_precision="bf16", downcast_bf16=True)
```
Using downcasting instead of bf16 everywhere is good for when you are trying to calculate metrics, log values, and more where raw bf16 tensors would be unusable.
## Training Times on TPUs
As you launch your script, you may notice that training seems exceptionally slow at first. This is because TPUs
first run through a few batches of data to see how much memory to allocate before finally utilizing this configured
memory allocation extremely efficiently.
If you notice that your evaluation code to calculate the metrics of your model takes longer due to a larger batch size being used,
it is recommended to keep the batch size the same as the training data if it is too slow. Otherwise the memory will reallocate to this
new batch size after the first few iterations.
<Tip>
Just because the memory is allocated does not mean it will be used or that the batch size will increase when going back to your training dataloader.
</Tip>
| accelerate/docs/source/concept_guides/training_tpu.md/0 | {
"file_path": "accelerate/docs/source/concept_guides/training_tpu.md",
"repo_id": "accelerate",
"token_count": 2214
} | 1 |
<!--Copyright 2023 The HuggingFace Team. All rights reserved.
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with
the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on
an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the
specific language governing permissions and limitations under the License.
⚠️ Note that this file is in Markdown but contain specific syntax for our doc-builder (similar to MDX) that may not be
rendered properly in your Markdown viewer.
-->
# Quantization
## `bitsandbytes` Integration
🤗 Accelerate brings `bitsandbytes` quantization to your model. You can now load any pytorch model in 8-bit or 4-bit with a few lines of code.
If you want to use 🤗 Transformers models with `bitsandbytes`, you should follow this [documentation](https://huggingface.co/docs/transformers/main_classes/quantization).
To learn more about how the `bitsandbytes` quantization works, check out the blog posts on [8-bit quantization](https://huggingface.co/blog/hf-bitsandbytes-integration) and [4-bit quantization](https://huggingface.co/blog/4bit-transformers-bitsandbytes).
### Pre-Requisites
You will need to install the following requirements:
- Install `bitsandbytes` library
```bash
pip install bitsandbytes
```
- Install latest `accelerate` from source
```bash
pip install git+https://github.com/huggingface/accelerate.git
```
- Install `minGPT` and `huggingface_hub` to run examples
```bash
git clone https://github.com/karpathy/minGPT.git
pip install minGPT/
pip install huggingface_hub
```
### How it works
First, we need to initialize our model. To save memory, we can initialize an empty model using the context manager [`init_empty_weights`].
Let's take the GPT2 model from minGPT library.
```py
from accelerate import init_empty_weights
from mingpt.model import GPT
model_config = GPT.get_default_config()
model_config.model_type = 'gpt2-xl'
model_config.vocab_size = 50257
model_config.block_size = 1024
with init_empty_weights():
empty_model = GPT(model_config)
```
Then, we need to get the path to the weights of your model. The path can be the state_dict file (e.g. "pytorch_model.bin") or a folder containing the sharded checkpoints.
```py
from huggingface_hub import snapshot_download
weights_location = snapshot_download(repo_id="marcsun13/gpt2-xl-linear-sharded")
```
Finally, you need to set your quantization configuration with [`~utils.BnbQuantizationConfig`].
Here's an example for 8-bit quantization:
```py
from accelerate.utils import BnbQuantizationConfig
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True, llm_int8_threshold = 6)
```
Here's an example for 4-bit quantization:
```py
from accelerate.utils import BnbQuantizationConfig
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True, bnb_4bit_compute_dtype=torch.bfloat16, bnb_4bit_use_double_quant=True, bnb_4bit_quant_type="nf4")
```
To quantize your empty model with the selected configuration, you need to use [`~utils.load_and_quantize_model`].
```py
from accelerate.utils import load_and_quantize_model
quantized_model = load_and_quantize_model(empty_model, weights_location=weights_location, bnb_quantization_config=bnb_quantization_config, device_map = "auto")
```
### Saving and loading 8-bit model
You can save your 8-bit model with accelerate using [`~Accelerator.save_model`].
```py
from accelerate import Accelerator
accelerate = Accelerator()
new_weights_location = "path/to/save_directory"
accelerate.save_model(quantized_model, new_weights_location)
quantized_model_from_saved = load_and_quantize_model(empty_model, weights_location=new_weights_location, bnb_quantization_config=bnb_quantization_config, device_map = "auto")
```
Note that 4-bit model serialization is currently not supported.
### Offload modules to cpu and disk
You can offload some modules to cpu/disk if you don't have enough space on the GPU to store the entire model on your GPUs.
This uses big model inference under the hood. Check this [documentation](https://huggingface.co/docs/accelerate/usage_guides/big_modeling) for more details.
For 8-bit quantization, the selected modules will be converted to 8-bit precision.
For 4-bit quantization, the selected modules will be kept in `torch_dtype` that the user passed in `BnbQuantizationConfig`. We will add support to convert these offloaded modules in 4-bit when 4-bit serialization will be possible.
You just need to pass a custom `device_map` in order to offload modules on cpu/disk. The offload modules will be dispatched on the GPU when needed. Here's an example :
```py
device_map = {
"transformer.wte": 0,
"transformer.wpe": 0,
"transformer.drop": 0,
"transformer.h": "cpu",
"transformer.ln_f": "disk",
"lm_head": "disk",
}
```
### Fine-tune a quantized model
It is not possible to perform pure 8bit or 4bit training on these models. However, you can train these models by leveraging parameter efficient fine tuning methods (PEFT) and train for example adapters on top of them. Please have a look at [peft](https://github.com/huggingface/peft) library for more details.
Currently, you can't add adapters on top of any quantized model. However, with the official support of adapters with 🤗 Transformers models, you can fine-tune quantized models. If you want to finetune a 🤗 Transformers model , follow this [documentation](https://huggingface.co/docs/transformers/main_classes/quantization) instead. Check out this [demo](https://colab.research.google.com/drive/1VoYNfYDKcKRQRor98Zbf2-9VQTtGJ24k?usp=sharing) on how to fine-tune a 4-bit 🤗 Transformers model.
Note that you don’t need to pass `device_map` when loading the model for training. It will automatically load your model on your GPU. Please note that `device_map=auto` should be used for inference only.
### Example demo - running GPT2 1.5b on a Google Colab
Check out the Google Colab [demo](https://colab.research.google.com/drive/1T1pOgewAWVpR9gKpaEWw4orOrzPFb3yM?usp=sharing) for running quantized models on a GTP2 model. The GPT2-1.5B model checkpoint is in FP32 which uses 6GB of memory. After quantization, it uses 1.6GB with 8-bit modules and 1.2GB with 4-bit modules.
| accelerate/docs/source/usage_guides/quantization.md/0 | {
"file_path": "accelerate/docs/source/usage_guides/quantization.md",
"repo_id": "accelerate",
"token_count": 1962
} | 2 |
# Copyright 2023 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import argparse
import runhouse as rh
import torch
from nlp_example import training_function
from accelerate.utils import PrepareForLaunch, patch_environment
def launch_train(*args):
num_processes = torch.cuda.device_count()
print(f"Device count: {num_processes}")
with patch_environment(
world_size=num_processes, master_addr="127.0.0.1", master_port="29500", mixed_precision=args[1].mixed_precision
):
launcher = PrepareForLaunch(training_function, distributed_type="MULTI_GPU")
torch.multiprocessing.start_processes(launcher, args=args, nprocs=num_processes, start_method="spawn")
if __name__ == "__main__":
# Refer to https://runhouse-docs.readthedocs-hosted.com/en/main/rh_primitives/cluster.html#hardware-setup
# for cloud access setup instructions (if using on-demand hardware), and for API specifications.
# on-demand GPU
# gpu = rh.cluster(name='rh-cluster', instance_type='V100:1', provider='cheapest', use_spot=False) # single GPU
gpu = rh.cluster(name="rh-cluster", instance_type="V100:4", provider="cheapest", use_spot=False) # multi GPU
gpu.up_if_not()
# on-prem GPU
# gpu = rh.cluster(
# ips=["ip_addr"], ssh_creds={ssh_user:"<username>", ssh_private_key:"<key_path>"}, name="rh-cluster"
# )
# Set up remote function
reqs = [
"pip:./",
"transformers",
"datasets",
"evaluate",
"tqdm",
"scipy",
"scikit-learn",
"tensorboard",
"torch --upgrade --extra-index-url https://download.pytorch.org/whl/cu117",
]
launch_train_gpu = rh.function(fn=launch_train, system=gpu, reqs=reqs, name="train_bert_glue")
# Define train args/config, run train function
train_args = argparse.Namespace(cpu=False, mixed_precision="fp16")
config = {"lr": 2e-5, "num_epochs": 3, "seed": 42, "batch_size": 16}
launch_train_gpu(config, train_args, stream_logs=True)
# Alternatively, we can just run as instructed in the README (but only because there's already a wrapper CLI):
# gpu.install_packages(reqs)
# gpu.run(['accelerate launch --multi_gpu accelerate/examples/nlp_example.py'])
| accelerate/examples/multigpu_remote_launcher.py/0 | {
"file_path": "accelerate/examples/multigpu_remote_launcher.py",
"repo_id": "accelerate",
"token_count": 1026
} | 3 |
# Copyright 2022 The HuggingFace Team and Brian Chao. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""
This file contains utilities for handling input from the user and registering specific keys to specific functions,
based on https://github.com/bchao1/bullet
"""
from typing import List
from .keymap import KEYMAP, get_character
def mark(key: str):
"""
Mark the function with the key code so it can be handled in the register
"""
def decorator(func):
handle = getattr(func, "handle_key", [])
handle += [key]
func.handle_key = handle
return func
return decorator
def mark_multiple(*keys: List[str]):
"""
Mark the function with the key codes so it can be handled in the register
"""
def decorator(func):
handle = getattr(func, "handle_key", [])
handle += keys
func.handle_key = handle
return func
return decorator
class KeyHandler(type):
"""
Metaclass that adds the key handlers to the class
"""
def __new__(cls, name, bases, attrs):
new_cls = super().__new__(cls, name, bases, attrs)
if not hasattr(new_cls, "key_handler"):
new_cls.key_handler = {}
new_cls.handle_input = KeyHandler.handle_input
for value in attrs.values():
handled_keys = getattr(value, "handle_key", [])
for key in handled_keys:
new_cls.key_handler[key] = value
return new_cls
@staticmethod
def handle_input(cls):
"Finds and returns the selected character if it exists in the handler"
char = get_character()
if char != KEYMAP["undefined"]:
char = ord(char)
handler = cls.key_handler.get(char)
if handler:
cls.current_selection = char
return handler(cls)
else:
return None
def register(cls):
"""Adds KeyHandler metaclass to the class"""
return KeyHandler(cls.__name__, cls.__bases__, cls.__dict__.copy())
| accelerate/src/accelerate/commands/menu/input.py/0 | {
"file_path": "accelerate/src/accelerate/commands/menu/input.py",
"repo_id": "accelerate",
"token_count": 947
} | 4 |
# Copyright 2021 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import numpy as np
import torch
from torch.utils.data import DataLoader
from accelerate.utils.dataclasses import DistributedType
class RegressionDataset:
def __init__(self, a=2, b=3, length=64, seed=None):
rng = np.random.default_rng(seed)
self.length = length
self.x = rng.normal(size=(length,)).astype(np.float32)
self.y = a * self.x + b + rng.normal(scale=0.1, size=(length,)).astype(np.float32)
def __len__(self):
return self.length
def __getitem__(self, i):
return {"x": self.x[i], "y": self.y[i]}
class RegressionModel4XPU(torch.nn.Module):
def __init__(self, a=0, b=0, double_output=False):
super().__init__()
self.a = torch.nn.Parameter(torch.tensor([2, 3]).float())
self.b = torch.nn.Parameter(torch.tensor([2, 3]).float())
self.first_batch = True
def forward(self, x=None):
if self.first_batch:
print(f"Model dtype: {self.a.dtype}, {self.b.dtype}. Input dtype: {x.dtype}")
self.first_batch = False
return x * self.a[0] + self.b[0]
class RegressionModel(torch.nn.Module):
def __init__(self, a=0, b=0, double_output=False):
super().__init__()
self.a = torch.nn.Parameter(torch.tensor(a).float())
self.b = torch.nn.Parameter(torch.tensor(b).float())
self.first_batch = True
def forward(self, x=None):
if self.first_batch:
print(f"Model dtype: {self.a.dtype}, {self.b.dtype}. Input dtype: {x.dtype}")
self.first_batch = False
return x * self.a + self.b
def mocked_dataloaders(accelerator, batch_size: int = 16):
from datasets import load_dataset
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("bert-base-cased")
data_files = {"train": "tests/test_samples/MRPC/train.csv", "validation": "tests/test_samples/MRPC/dev.csv"}
datasets = load_dataset("csv", data_files=data_files)
label_list = datasets["train"].unique("label")
label_to_id = {v: i for i, v in enumerate(label_list)}
def tokenize_function(examples):
# max_length=None => use the model max length (it's actually the default)
outputs = tokenizer(
examples["sentence1"], examples["sentence2"], truncation=True, max_length=None, padding="max_length"
)
if "label" in examples:
outputs["labels"] = [label_to_id[l] for l in examples["label"]]
return outputs
# Apply the method we just defined to all the examples in all the splits of the dataset
tokenized_datasets = datasets.map(
tokenize_function,
batched=True,
remove_columns=["sentence1", "sentence2", "label"],
)
def collate_fn(examples):
# On TPU it's best to pad everything to the same length or training will be very slow.
if accelerator.distributed_type == DistributedType.XLA:
return tokenizer.pad(examples, padding="max_length", max_length=128, return_tensors="pt")
return tokenizer.pad(examples, padding="longest", return_tensors="pt")
# Instantiate dataloaders.
train_dataloader = DataLoader(tokenized_datasets["train"], shuffle=True, collate_fn=collate_fn, batch_size=2)
eval_dataloader = DataLoader(tokenized_datasets["validation"], shuffle=False, collate_fn=collate_fn, batch_size=1)
return train_dataloader, eval_dataloader
| accelerate/src/accelerate/test_utils/training.py/0 | {
"file_path": "accelerate/src/accelerate/test_utils/training.py",
"repo_id": "accelerate",
"token_count": 1572
} | 5 |
# Copyright 2022 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import collections
import os
import platform
import re
import socket
from contextlib import contextmanager
from functools import partial, reduce
from types import MethodType
from typing import OrderedDict
import torch
from packaging.version import Version
from safetensors.torch import save_file as safe_save_file
from ..commands.config.default import write_basic_config # noqa: F401
from ..logging import get_logger
from ..state import PartialState
from .constants import FSDP_PYTORCH_VERSION
from .dataclasses import DistributedType
from .imports import is_deepspeed_available, is_torch_distributed_available, is_torch_xla_available
from .modeling import id_tensor_storage
from .transformer_engine import convert_model
from .versions import is_torch_version
logger = get_logger(__name__)
if is_torch_xla_available():
import torch_xla.core.xla_model as xm
def is_compiled_module(module):
"""
Check whether the module was compiled with torch.compile()
"""
if is_torch_version("<", "2.0.0") or not hasattr(torch, "_dynamo"):
return False
return isinstance(module, torch._dynamo.eval_frame.OptimizedModule)
def extract_model_from_parallel(model, keep_fp32_wrapper: bool = True):
"""
Extract a model from its distributed containers.
Args:
model (`torch.nn.Module`):
The model to extract.
keep_fp32_wrapper (`bool`, *optional*):
Whether to remove mixed precision hooks from the model.
Returns:
`torch.nn.Module`: The extracted model.
"""
options = (torch.nn.parallel.DistributedDataParallel, torch.nn.DataParallel)
is_compiled = is_compiled_module(model)
if is_compiled:
compiled_model = model
model = model._orig_mod
if is_deepspeed_available():
from deepspeed import DeepSpeedEngine
options += (DeepSpeedEngine,)
if is_torch_version(">=", FSDP_PYTORCH_VERSION) and is_torch_distributed_available():
from torch.distributed.fsdp.fully_sharded_data_parallel import FullyShardedDataParallel as FSDP
options += (FSDP,)
while isinstance(model, options):
model = model.module
if not keep_fp32_wrapper:
forward = model.forward
original_forward = model.__dict__.pop("_original_forward", None)
if original_forward is not None:
while hasattr(forward, "__wrapped__"):
forward = forward.__wrapped__
if forward == original_forward:
break
model.forward = MethodType(forward, model)
if getattr(model, "_converted_to_transformer_engine", False):
convert_model(model, to_transformer_engine=False)
if is_compiled:
compiled_model._orig_mod = model
model = compiled_model
return model
def wait_for_everyone():
"""
Introduces a blocking point in the script, making sure all processes have reached this point before continuing.
<Tip warning={true}>
Make sure all processes will reach this instruction otherwise one of your processes will hang forever.
</Tip>
"""
PartialState().wait_for_everyone()
def clean_state_dict_for_safetensors(state_dict: dict):
"""
Cleans the state dictionary from a model and removes tensor aliasing if present.
Args:
state_dict (`dict`):
The state dictionary from a model
"""
ptrs = collections.defaultdict(list)
# When bnb serialization is used, weights in state dict can be strings
for name, tensor in state_dict.items():
if not isinstance(tensor, str):
ptrs[id_tensor_storage(tensor)].append(name)
# These are all pointers of tensors with shared memory
shared_ptrs = {ptr: names for ptr, names in ptrs.items() if len(names) > 1}
warn_names = set()
for names in shared_ptrs.values():
# When not all duplicates have been cleaned, we still remove those keys but put a clear warning.
# If the link between tensors was done at runtime then `from_pretrained` will not get
# the key back leading to random tensor. A proper warning will be shown
# during reload (if applicable), but since the file is not necessarily compatible with
# the config, better show a proper warning.
found_names = [name for name in names if name in state_dict]
warn_names.update(found_names[1:])
for name in found_names[1:]:
del state_dict[name]
if len(warn_names) > 0:
logger.warning(
f"Removed shared tensor {warn_names} while saving. This should be OK, but check by verifying that you don't receive any warning while reloading",
)
state_dict = {k: v.contiguous() if isinstance(v, torch.Tensor) else v for k, v in state_dict.items()}
return state_dict
def save(obj, f, save_on_each_node: bool = False, safe_serialization: bool = False):
"""
Save the data to disk. Use in place of `torch.save()`.
Args:
obj:
The data to save
f:
The file (or file-like object) to use to save the data
save_on_each_node (`bool`, *optional*, defaults to `False`):
Whether to only save on the global main process
safe_serialization (`bool`, *optional*, defaults to `False`):
Whether to save `obj` using `safetensors` or the traditional PyTorch way (that uses `pickle`).
"""
# When TorchXLA is enabled, it's necessary to transfer all data to the CPU before saving.
# Another issue arises with `id_tensor_storage`, which treats all XLA tensors as identical.
# If tensors remain on XLA, calling `clean_state_dict_for_safetensors` will result in only
# one XLA tensor remaining.
if PartialState().distributed_type == DistributedType.XLA:
obj = xm._maybe_convert_to_cpu(obj)
# Check if it's a model and remove duplicates
if safe_serialization:
save_func = partial(safe_save_file, metadata={"format": "pt"})
if isinstance(obj, OrderedDict):
obj = clean_state_dict_for_safetensors(obj)
else:
save_func = torch.save
if PartialState().is_main_process and not save_on_each_node:
save_func(obj, f)
elif PartialState().is_local_main_process and save_on_each_node:
save_func(obj, f)
@contextmanager
def clear_environment():
"""
A context manager that will temporarily clear environment variables.
When this context exits, the previous environment variables will be back.
Example:
```python
>>> import os
>>> from accelerate.utils import clear_environment
>>> os.environ["FOO"] = "bar"
>>> with clear_environment():
... print(os.environ)
... os.environ["FOO"] = "new_bar"
... print(os.environ["FOO"])
{}
new_bar
>>> print(os.environ["FOO"])
bar
```
"""
_old_os_environ = os.environ.copy()
os.environ.clear()
try:
yield
finally:
os.environ.clear() # clear any added keys,
os.environ.update(_old_os_environ) # then restore previous environment
@contextmanager
def patch_environment(**kwargs):
"""
A context manager that will add each keyword argument passed to `os.environ` and remove them when exiting.
Will convert the values in `kwargs` to strings and upper-case all the keys.
Example:
```python
>>> import os
>>> from accelerate.utils import patch_environment
>>> with patch_environment(FOO="bar"):
... print(os.environ["FOO"]) # prints "bar"
>>> print(os.environ["FOO"]) # raises KeyError
```
"""
existing_vars = {}
for key, value in kwargs.items():
key = key.upper()
if key in os.environ:
existing_vars[key] = os.environ[key]
os.environ[key] = str(value)
try:
yield
finally:
for key in kwargs:
key = key.upper()
if key in existing_vars:
# restore previous value
os.environ[key] = existing_vars[key]
else:
os.environ.pop(key, None)
def get_pretty_name(obj):
"""
Gets a pretty name from `obj`.
"""
if not hasattr(obj, "__qualname__") and not hasattr(obj, "__name__"):
obj = getattr(obj, "__class__", obj)
if hasattr(obj, "__qualname__"):
return obj.__qualname__
if hasattr(obj, "__name__"):
return obj.__name__
return str(obj)
def merge_dicts(source, destination):
"""
Recursively merges two dictionaries.
Args:
source (`dict`): The dictionary to merge into `destination`.
destination (`dict`): The dictionary to merge `source` into.
"""
for key, value in source.items():
if isinstance(value, dict):
node = destination.setdefault(key, {})
merge_dicts(value, node)
else:
destination[key] = value
return destination
def is_port_in_use(port: int = None) -> bool:
"""
Checks if a port is in use on `localhost`. Useful for checking if multiple `accelerate launch` commands have been
run and need to see if the port is already in use.
"""
if port is None:
port = 29500
with socket.socket(socket.AF_INET, socket.SOCK_STREAM) as s:
return s.connect_ex(("localhost", port)) == 0
def convert_bytes(size):
"Converts `size` from bytes to the largest possible unit"
for x in ["bytes", "KB", "MB", "GB", "TB"]:
if size < 1024.0:
return f"{round(size, 2)} {x}"
size /= 1024.0
return f"{round(size, 2)} PB"
def check_os_kernel():
"""Warns if the kernel version is below the recommended minimum on Linux."""
# see issue #1929
info = platform.uname()
system = info.system
if system != "Linux":
return
_, version, *_ = re.split(r"(\d+\.\d+\.\d+)", info.release)
min_version = "5.5.0"
if Version(version) < Version(min_version):
msg = (
f"Detected kernel version {version}, which is below the recommended minimum of {min_version}; this can "
"cause the process to hang. It is recommended to upgrade the kernel to the minimum version or higher."
)
logger.warning(msg, main_process_only=True)
def recursive_getattr(obj, attr: str):
"""
Recursive `getattr`.
Args:
obj:
A class instance holding the attribute.
attr (`str`):
The attribute that is to be retrieved, e.g. 'attribute1.attribute2'.
"""
def _getattr(obj, attr):
return getattr(obj, attr)
return reduce(_getattr, [obj] + attr.split("."))
| accelerate/src/accelerate/utils/other.py/0 | {
"file_path": "accelerate/src/accelerate/utils/other.py",
"repo_id": "accelerate",
"token_count": 4278
} | 6 |
compute_environment: LOCAL_MACHINE
debug: false
distributed_type: MULTI_CPU
downcast_bf16: 'no'
ipex_config:
ipex: true
machine_rank: 0
main_process_ip: 127.0.0.1
main_process_port: 29500
main_training_function: main
mixed_precision: 'no'
mpirun_config:
mpirun_ccl: '1'
mpirun_hostfile: /home/user/hostfile
num_machines: 4
num_processes: 16
rdzv_backend: static
same_network: true
tpu_env: []
tpu_use_cluster: false
tpu_use_sudo: false
use_cpu: true
| accelerate/tests/test_configs/0_28_0_mpi.yaml/0 | {
"file_path": "accelerate/tests/test_configs/0_28_0_mpi.yaml",
"repo_id": "accelerate",
"token_count": 193
} | 7 |
# Copyright 2023 The HuggingFace Team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import gc
import tempfile
import unittest
import torch
import torch.nn as nn
from accelerate import Accelerator, init_empty_weights
from accelerate.test_utils import (
require_bnb,
require_cuda,
require_huggingface_suite,
require_multi_gpu,
require_non_torch_xla,
slow,
)
from accelerate.utils.bnb import load_and_quantize_model
from accelerate.utils.dataclasses import BnbQuantizationConfig
class BitsAndBytesConfigIntegration(unittest.TestCase):
def test_BnbQuantizationConfig(self):
with self.assertRaises(ValueError):
BnbQuantizationConfig(load_in_8bit=True, load_in_4bit=True)
@require_non_torch_xla
@slow
@require_cuda
@require_bnb
@require_huggingface_suite
class MixedInt8EmptyModelTest(unittest.TestCase):
# We keep the constants inside the init function and model loading inside setUp function
# We need to test on relatively large models (aka >1b parameters otherwise the quantiztion may not work as expected)
# Therefore here we use only bloom-1b3 to test our module
model_name = "marcsun13/bloom-1b7_with_lm_head"
# Constant values
# This was obtained on a Quadro RTX 8000 so the number might slightly change
EXPECTED_RELATIVE_DIFFERENCE = 1.540025
input_text = "Hello my name is"
EXPECTED_OUTPUT = "Hello my name is John.\nI am a friend of the family.\n"
MAX_NEW_TOKENS = 10
def setUp(self):
"""
Setup quantized model from empty model
"""
from huggingface_hub import hf_hub_download
from transformers import AutoConfig, AutoModelForCausalLM, AutoTokenizer
# Models and tokenizer
self.model_fp16 = AutoModelForCausalLM.from_pretrained(
self.model_name, torch_dtype=torch.float16, device_map="auto"
)
# create model on meta device
with init_empty_weights():
self.model_8bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
self.model_8bit.tie_weights()
self.weights_location = hf_hub_download(self.model_name, "pytorch_model.bin")
self.bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
self.model_8bit = load_and_quantize_model(
self.model_8bit,
self.bnb_quantization_config,
weights_location=self.weights_location,
device_map={"": 0},
no_split_module_classes=["BloomBlock"],
)
self.tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-1b7")
self.accelerate = Accelerator()
def tearDown(self):
r"""
TearDown function needs to be called at the end of each test to free the GPU memory and cache, also to
avoid unexpected behaviors. Please see: https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/27
"""
del self.model_fp16
del self.model_8bit
gc.collect()
torch.cuda.empty_cache()
def test_memory_footprint(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
from bitsandbytes.nn import Int8Params
mem_fp16 = self.model_fp16.get_memory_footprint()
mem_8bit = self.model_8bit.get_memory_footprint()
assert round((mem_fp16 / mem_8bit) - self.EXPECTED_RELATIVE_DIFFERENCE, 7) >= 0
assert self.model_8bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
def test_linear_are_8bit(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
self.model_fp16.get_memory_footprint()
self.model_8bit.get_memory_footprint()
for name, module in self.model_8bit.named_modules():
if isinstance(module, torch.nn.Linear):
modules_not_converted = (
self.bnb_quantization_config.keep_in_fp32_modules + self.bnb_quantization_config.skip_modules
)
if name not in modules_not_converted:
assert module.weight.dtype == torch.int8
def test_llm_skip(self):
r"""
A simple test to check if `llm_int8_skip_modules` works as expected
"""
import bitsandbytes as bnb
from transformers import AutoConfig, AutoModelForCausalLM
bnb_quantization_config = BnbQuantizationConfig(
load_in_8bit=True, skip_modules=["lm_head", "transformer.word_embeddings"]
)
with init_empty_weights():
model = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model.tie_weights()
model = load_and_quantize_model(
model,
bnb_quantization_config,
weights_location=self.weights_location,
device_map="auto",
no_split_module_classes=["BloomBlock"],
)
assert model.transformer.h[1].mlp.dense_4h_to_h.weight.dtype == torch.int8
assert isinstance(model.transformer.h[1].mlp.dense_4h_to_h, bnb.nn.Linear8bitLt)
assert isinstance(model.lm_head, nn.Linear)
assert model.lm_head.weight.dtype != torch.int8
def check_inference_correctness(self, model):
r"""
Test the generation quality of the quantized model and see that we are matching the expected output.
Given that we are operating on small numbers + the testing model is relatively small, we might not get
the same output across GPUs. So we'll generate few tokens (5-10) and check their output.
"""
# Check that inference pass works on the model
encoded_input = self.tokenizer(self.input_text, return_tensors="pt")
# Check the exactness of the results
output_parallel = model.generate(input_ids=encoded_input["input_ids"].to(0), max_new_tokens=10)
# Get the generation
output_text = self.tokenizer.decode(output_parallel[0], skip_special_tokens=True)
assert output_text == self.EXPECTED_OUTPUT
def test_generate_quality(self):
self.check_inference_correctness(self.model_8bit)
def test_fp32_8bit_conversion(self):
r"""
Test whether it is possible to mix both `8bit` and `fp32` weights when using `keep_in_fp32_modules` correctly.
"""
from transformers import AutoConfig, AutoModelForCausalLM
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True, keep_in_fp32_modules=["lm_head"])
with init_empty_weights():
model = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model.tie_weights()
model = load_and_quantize_model(
model,
bnb_quantization_config,
weights_location=self.weights_location,
device_map="auto",
no_split_module_classes=["BloomBlock"],
)
assert model.lm_head.weight.dtype == torch.float32
@require_multi_gpu
def test_cpu_gpu_loading_custom_device_map(self):
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a custom `device_map`.
"""
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": 0,
"lm_head": "cpu",
"transformer.h.0": "cpu",
"transformer.h.1": "cpu",
"transformer.h.2": "cpu",
"transformer.h.3": 0,
"transformer.h.4": 0,
"transformer.h.5": 0,
"transformer.h.6": 0,
"transformer.h.7": 0,
"transformer.h.8": 0,
"transformer.h.9": 1,
"transformer.h.10": 0,
"transformer.h.11": 1,
"transformer.h.12": 0,
"transformer.h.13": 0,
"transformer.h.14": 1,
"transformer.h.15": 0,
"transformer.h.16": 0,
"transformer.h.17": 1,
"transformer.h.18": 1,
"transformer.h.19": 0,
"transformer.h.20": 1,
"transformer.h.21": 1,
"transformer.h.22": 0,
"transformer.h.23": 0,
"transformer.ln_f": 1,
}
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
with init_empty_weights():
model_8bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit.tie_weights()
model_8bit = load_and_quantize_model(
model_8bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
)
assert model_8bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert model_8bit.transformer.h[1].mlp.dense_4h_to_h.weight.__class__ == Int8Params
self.check_inference_correctness(model_8bit)
@require_multi_gpu
def test_cpu_gpu_loading_custom_device_map_offload_state_dict(self):
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a custom `device_map` and offload_state_dict=True.
"""
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": 0,
"lm_head": "cpu",
"transformer.h.0": "cpu",
"transformer.h.1": "cpu",
"transformer.h.2": "cpu",
"transformer.h.3": 0,
"transformer.h.4": 0,
"transformer.h.5": 0,
"transformer.h.6": 0,
"transformer.h.7": 0,
"transformer.h.8": 0,
"transformer.h.9": 1,
"transformer.h.10": 0,
"transformer.h.11": 1,
"transformer.h.12": 0,
"transformer.h.13": 0,
"transformer.h.14": 1,
"transformer.h.15": 0,
"transformer.h.16": 0,
"transformer.h.17": 1,
"transformer.h.18": 1,
"transformer.h.19": 0,
"transformer.h.20": 1,
"transformer.h.21": 1,
"transformer.h.22": 0,
"transformer.h.23": 0,
"transformer.ln_f": 1,
}
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
with init_empty_weights():
model_8bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit.tie_weights()
model_8bit = load_and_quantize_model(
model_8bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
offload_state_dict=True,
)
assert model_8bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert model_8bit.transformer.h[1].mlp.dense_4h_to_h.weight.__class__ == Int8Params
self.check_inference_correctness(model_8bit)
@require_multi_gpu
def test_cpu_gpu_disk_loading_custom_device_map_kwargs(self):
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a custom `device_map`.
This time we also add `disk` on the device_map - using the kwargs directly instead of the quantization config
"""
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": 0,
"lm_head": "cpu",
"transformer.h.0": "cpu",
"transformer.h.1": "cpu",
"transformer.h.2": "cpu",
"transformer.h.3": "disk",
"transformer.h.4": "disk",
"transformer.h.5": "disk",
"transformer.h.6": 0,
"transformer.h.7": 0,
"transformer.h.8": 0,
"transformer.h.9": 1,
"transformer.h.10": 0,
"transformer.h.11": 1,
"transformer.h.12": 0,
"transformer.h.13": 0,
"transformer.h.14": 1,
"transformer.h.15": 0,
"transformer.h.16": 0,
"transformer.h.17": 1,
"transformer.h.18": 1,
"transformer.h.19": 0,
"transformer.h.20": 1,
"transformer.h.21": 1,
"transformer.h.22": 0,
"transformer.h.23": 0,
"transformer.ln_f": 1,
}
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
with init_empty_weights():
model_8bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit.tie_weights()
with tempfile.TemporaryDirectory() as tmpdirname:
model_8bit = load_and_quantize_model(
model_8bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
offload_folder=tmpdirname,
offload_state_dict=True,
)
assert model_8bit.transformer.h[4].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert model_8bit.transformer.h[5].mlp.dense_4h_to_h.weight.__class__ == Int8Params
self.check_inference_correctness(model_8bit)
def test_int8_serialization(self):
r"""
Test whether it is possible to serialize a model in 8-bit.
"""
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
with tempfile.TemporaryDirectory() as tmpdirname:
# saving state dict for now but will save config and other in the future
self.accelerate.save_model(self.model_8bit, tmpdirname)
with init_empty_weights():
# let's suppose that we can get the right config
model_8bit_from_saved = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit_from_saved.tie_weights()
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
model_8bit_from_saved = load_and_quantize_model(
model_8bit_from_saved,
bnb_quantization_config,
weights_location=tmpdirname,
device_map="auto",
no_split_module_classes=["BloomBlock"],
)
assert model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert hasattr(model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight, "SCB")
assert hasattr(model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight, "CB")
self.check_inference_correctness(model_8bit_from_saved)
@require_multi_gpu
def test_int8_serialization_offload(self):
r"""
Test whether it is possible to serialize a model in 8-bit and offload weights to cpu/disk
"""
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
with tempfile.TemporaryDirectory() as tmpdirname:
# saving state dict for now but will save config and other in the future
self.accelerate.save_model(self.model_8bit, tmpdirname)
with init_empty_weights():
# let's suppose that we can get the right config
model_8bit_from_saved = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit_from_saved.tie_weights()
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": 0,
"lm_head": "cpu",
"transformer.h.0": "cpu",
"transformer.h.1": "cpu",
"transformer.h.2": "cpu",
"transformer.h.3": "disk",
"transformer.h.4": "disk",
"transformer.h.5": "disk",
"transformer.h.6": 0,
"transformer.h.7": 0,
"transformer.h.8": 0,
"transformer.h.9": 1,
"transformer.h.10": 0,
"transformer.h.11": 1,
"transformer.h.12": 0,
"transformer.h.13": 0,
"transformer.h.14": 1,
"transformer.h.15": 0,
"transformer.h.16": 0,
"transformer.h.17": 1,
"transformer.h.18": 1,
"transformer.h.19": 0,
"transformer.h.20": 1,
"transformer.h.21": 1,
"transformer.h.22": 0,
"transformer.h.23": 0,
"transformer.ln_f": 1,
}
model_8bit_from_saved = load_and_quantize_model(
model_8bit_from_saved,
bnb_quantization_config,
weights_location=tmpdirname,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
offload_folder=tmpdirname + "/tmp",
offload_state_dict=True,
)
assert model_8bit_from_saved.transformer.h[4].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert model_8bit_from_saved.transformer.h[5].mlp.dense_4h_to_h.weight.__class__ == Int8Params
self.check_inference_correctness(model_8bit_from_saved)
def test_int8_serialization_shard(self):
r"""
Test whether it is possible to serialize a model in 8-bit.
"""
from bitsandbytes.nn import Int8Params
from transformers import AutoConfig, AutoModelForCausalLM
with tempfile.TemporaryDirectory() as tmpdirname:
# saving state dict for now but will save config and other in the future
self.accelerate.save_model(self.model_8bit, tmpdirname, max_shard_size="1GB")
with init_empty_weights():
# let's suppose that we can get the right config
model_8bit_from_saved = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_8bit_from_saved.tie_weights()
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
model_8bit_from_saved = load_and_quantize_model(
model_8bit_from_saved,
bnb_quantization_config,
weights_location=tmpdirname,
device_map="auto",
no_split_module_classes=["BloomBlock"],
)
assert model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
assert hasattr(model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight, "SCB")
assert hasattr(model_8bit_from_saved.transformer.h[0].mlp.dense_4h_to_h.weight, "CB")
self.check_inference_correctness(model_8bit_from_saved)
@require_non_torch_xla
@slow
@require_cuda
@require_bnb
@require_huggingface_suite
class MixedInt8LoaddedModelTest(unittest.TestCase):
# We keep the constants inside the init function and model loading inside setUp function
# We need to test on relatively large models (aka >1b parameters otherwise the quantiztion may not work as expected)
# Therefore here we use only bloom-1b3 to test our module
model_name = "marcsun13/bloom-1b7_with_lm_head"
# Constant values
# This was obtained on a Quadro RTX 8000 so the number might slightly change
EXPECTED_RELATIVE_DIFFERENCE = 1.540025
input_text = "Hello my name is"
EXPECTED_OUTPUT = "Hello my name is John.\nI am a friend of the family.\n"
MAX_NEW_TOKENS = 10
def setUp(self):
"""
Setup quantized model from loaded model
"""
from transformers import AutoModelForCausalLM, AutoTokenizer
# Models and tokenizer
self.model_fp16 = AutoModelForCausalLM.from_pretrained(
self.model_name, torch_dtype=torch.float16, device_map="auto"
)
self.bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True)
self.model_8bit = AutoModelForCausalLM.from_pretrained(self.model_name, torch_dtype=torch.float16)
self.model_8bit = load_and_quantize_model(self.model_8bit, self.bnb_quantization_config)
self.tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-1b7")
def tearDown(self):
r"""
TearDown function needs to be called at the end of each test to free the GPU memory and cache, also to
avoid unexpected behaviors. Please see: https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/27
"""
del self.model_fp16
del self.model_8bit
gc.collect()
torch.cuda.empty_cache()
def test_memory_footprint(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
from bitsandbytes.nn import Int8Params
mem_fp16 = self.model_fp16.get_memory_footprint()
mem_8bit = self.model_8bit.get_memory_footprint()
assert round((mem_fp16 / mem_8bit) - self.EXPECTED_RELATIVE_DIFFERENCE, 7) >= 0
assert self.model_8bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Int8Params
def test_linear_are_8bit(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
self.model_fp16.get_memory_footprint()
self.model_8bit.get_memory_footprint()
for name, module in self.model_8bit.named_modules():
if isinstance(module, torch.nn.Linear):
modules_not_converted = (
self.bnb_quantization_config.keep_in_fp32_modules + self.bnb_quantization_config.skip_modules
)
if name not in modules_not_converted:
assert module.weight.dtype == torch.int8
def test_generate_quality(self):
r"""
Test the generation quality of the quantized model and see that we are matching the expected output.
Given that we are operating on small numbers + the testing model is relatively small, we might not get
the same output across GPUs. So we'll generate few tokens (5-10) and check their output.
"""
encoded_input = self.tokenizer(self.input_text, return_tensors="pt")
output_sequences = self.model_8bit.generate(
input_ids=encoded_input["input_ids"].to(self.model_8bit.device), max_new_tokens=10
)
assert self.tokenizer.decode(output_sequences[0], skip_special_tokens=True) == self.EXPECTED_OUTPUT
def test_fp32_8bit_conversion(self):
r"""
Test whether it is possible to mix both `8bit` and `fp32` weights when using `keep_in_fp32_modules` correctly.
"""
from transformers import AutoModelForCausalLM
bnb_quantization_config = BnbQuantizationConfig(load_in_8bit=True, keep_in_fp32_modules=["lm_head"])
model = AutoModelForCausalLM.from_pretrained(self.model_name, torch_dtype=torch.float16)
model = load_and_quantize_model(model, bnb_quantization_config)
assert model.lm_head.weight.dtype == torch.float32
@require_non_torch_xla
@slow
@require_cuda
@require_bnb
@require_huggingface_suite
class Bnb4BitEmptyModelTest(unittest.TestCase):
# We keep the constants inside the init function and model loading inside setUp function
# We need to test on relatively large models (aka >1b parameters otherwise the quantiztion may not work as expected)
# Therefore here we use only bloom-1b3 to test our module
model_name = "marcsun13/bloom-1b7_with_lm_head"
# Constant values
# This was obtained on a RTX Titan so the number might slightly change
EXPECTED_RELATIVE_DIFFERENCE = 2.109659552692574
input_text = "Hello my name is"
EXPECTED_OUTPUTS = set()
EXPECTED_OUTPUTS.add("Hello my name is John and I am a professional photographer. I")
EXPECTED_OUTPUTS.add("Hello my name is John.\nI am a friend of your father.\n")
MAX_NEW_TOKENS = 10
def setUp(self):
from huggingface_hub import hf_hub_download
from transformers import AutoConfig, AutoModelForCausalLM, AutoTokenizer
super().setUp()
# Models and tokenizer
self.model_fp16 = AutoModelForCausalLM.from_pretrained(
self.model_name, torch_dtype=torch.float16, device_map="auto"
)
# create model on meta device
with init_empty_weights():
self.model_4bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
self.model_4bit.tie_weights()
self.weights_location = hf_hub_download(self.model_name, "pytorch_model.bin")
self.bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True)
self.model_4bit = load_and_quantize_model(
self.model_4bit,
self.bnb_quantization_config,
weights_location=self.weights_location,
device_map={"": 0},
no_split_module_classes=["BloomBlock"],
)
self.tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-1b7")
def tearDown(self):
"""
TearDown function needs to be called at the end of each test to free the GPU memory and cache, also to
avoid unexpected behaviors. Please see: https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/27
"""
super().tearDown()
del self.model_fp16
del self.model_4bit
gc.collect()
torch.cuda.empty_cache()
def test_memory_footprint(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
from bitsandbytes.nn import Params4bit
mem_fp16 = self.model_fp16.get_memory_footprint()
mem_4bit = self.model_4bit.get_memory_footprint()
assert round((mem_fp16 / mem_4bit) - self.EXPECTED_RELATIVE_DIFFERENCE, 7) >= 0
assert self.model_4bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Params4bit
def check_inference_correctness(self, model):
r"""
Test the generation quality of the quantized model and see that we are matching the expected output.
Given that we are operating on small numbers + the testing model is relatively small, we might not get
the same output across GPUs. So we'll generate few tokens (5-10) and check their output.
"""
# Check that inference pass works on the model
encoded_input = self.tokenizer(self.input_text, return_tensors="pt")
# Check the exactness of the results
output_sequences = model.generate(input_ids=encoded_input["input_ids"].to(0), max_new_tokens=10)
assert self.tokenizer.decode(output_sequences[0], skip_special_tokens=True) in self.EXPECTED_OUTPUTS
def test_generate_quality(self):
self.check_inference_correctness(self.model_4bit)
def test_linear_are_4bit(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
self.model_fp16.get_memory_footprint()
self.model_4bit.get_memory_footprint()
for name, module in self.model_4bit.named_modules():
if isinstance(module, torch.nn.Linear):
if (
name
not in self.bnb_quantization_config.keep_in_fp32_modules
+ self.bnb_quantization_config.skip_modules
):
# 4-bit parameters are packed in uint8 variables
assert module.weight.dtype == torch.uint8
def test_fp32_4bit_conversion(self):
r"""
Test whether it is possible to mix both `4bit` and `fp32` weights when using `keep_in_fp32_modules` correctly.
"""
from transformers import AutoConfig, AutoModelForCausalLM
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True, keep_in_fp32_modules=["lm_head"])
with init_empty_weights():
model = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model.tie_weights()
model = load_and_quantize_model(
model,
bnb_quantization_config,
weights_location=self.weights_location,
device_map="auto",
no_split_module_classes=["BloomBlock"],
)
assert model.lm_head.weight.dtype == torch.float32
@require_multi_gpu
def test_cpu_gpu_loading_random_device_map(self):
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a random `device_map`.
"""
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": 0,
"lm_head": "cpu",
"transformer.h.0": 0,
"transformer.h.1": 0,
"transformer.h.2": 0,
"transformer.h.3": 0,
"transformer.h.4": 0,
"transformer.h.5": 0,
"transformer.h.6": 0,
"transformer.h.7": 0,
"transformer.h.8": 0,
"transformer.h.9": 1,
"transformer.h.10": 0,
"transformer.h.11": 1,
"transformer.h.12": 0,
"transformer.h.13": 0,
"transformer.h.14": 1,
"transformer.h.15": 0,
"transformer.h.16": 0,
"transformer.h.17": 1,
"transformer.h.18": 1,
"transformer.h.19": 0,
"transformer.h.20": 1,
"transformer.h.21": 1,
"transformer.h.22": 0,
"transformer.h.23": 0,
"transformer.ln_f": 1,
}
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True)
with init_empty_weights():
model_4bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_4bit.tie_weights()
model_4bit = load_and_quantize_model(
model_4bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
)
self.check_inference_correctness(model_4bit)
@require_multi_gpu
def test_cpu_gpu_loading_custom_device_map(self):
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a random `device_map`.
"""
device_map = {
"transformer.word_embeddings": "cpu",
"transformer.word_embeddings_layernorm": "cpu",
"lm_head": "cpu",
"transformer.h": 0,
"transformer.ln_f": 1,
}
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True)
with init_empty_weights():
model_4bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_4bit.tie_weights()
model_4bit = load_and_quantize_model(
model_4bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
)
self.check_inference_correctness(model_4bit)
@require_multi_gpu
def test_cpu_gpu_disk_loading_custom_device_map_kwargs(self):
from transformers import AutoConfig, AutoModelForCausalLM
r"""
A test to check is dispatching a model on cpu & gpu works correctly using a custom `device_map`.
This time we also add `disk` on the device_map - using the kwargs directly instead of the quantization config
"""
device_map = {
"transformer.word_embeddings": 0,
"transformer.word_embeddings_layernorm": "disk",
"lm_head": 0,
"transformer.h": 1,
"transformer.ln_f": "cpu",
}
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True)
with init_empty_weights():
model_4bit = AutoModelForCausalLM.from_config(AutoConfig.from_pretrained(self.model_name))
model_4bit.tie_weights()
with tempfile.TemporaryDirectory() as tmpdirname:
model_4bit = load_and_quantize_model(
model_4bit,
bnb_quantization_config,
weights_location=self.weights_location,
device_map=device_map,
no_split_module_classes=["BloomBlock"],
offload_folder=tmpdirname,
offload_state_dict=True,
)
self.check_inference_correctness(model_4bit)
@require_non_torch_xla
@slow
@require_cuda
@require_bnb
@require_huggingface_suite
class Bnb4BitTestLoadedModel(unittest.TestCase):
# We keep the constants inside the init function and model loading inside setUp function
# We need to test on relatively large models (aka >1b parameters otherwise the quantiztion may not work as expected)
# Therefore here we use only bloom-1b3 to test our module
model_name = "marcsun13/bloom-1b7_with_lm_head"
# Constant values
# This was obtained on a RTX Titan so the number might slightly change
EXPECTED_RELATIVE_DIFFERENCE = 2.109659552692574
input_text = "Hello my name is"
EXPECTED_OUTPUTS = set()
EXPECTED_OUTPUTS.add("Hello my name is John and I am a professional photographer. I")
EXPECTED_OUTPUTS.add("Hello my name is John.\nI am a friend of your father.\n")
MAX_NEW_TOKENS = 10
def setUp(self):
"""
Setup quantized model from loaded model
"""
from transformers import AutoModelForCausalLM, AutoTokenizer
super().setUp()
# Models and tokenizer
self.model_fp16 = AutoModelForCausalLM.from_pretrained(
self.model_name, torch_dtype=torch.float16, device_map="auto"
)
self.bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True)
self.model_4bit = AutoModelForCausalLM.from_pretrained(self.model_name, torch_dtype=torch.float16)
self.model_4bit = load_and_quantize_model(self.model_4bit, self.bnb_quantization_config)
self.tokenizer = AutoTokenizer.from_pretrained("bigscience/bloom-1b7")
def tearDown(self):
"""
TearDown function needs to be called at the end of each test to free the GPU memory and cache, also to
avoid unexpected behaviors. Please see: https://discuss.pytorch.org/t/how-can-we-release-gpu-memory-cache/14530/27
"""
super().tearDown()
del self.model_fp16
del self.model_4bit
gc.collect()
torch.cuda.empty_cache()
def test_memory_footprint(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
from bitsandbytes.nn import Params4bit
mem_fp16 = self.model_fp16.get_memory_footprint()
mem_4bit = self.model_4bit.get_memory_footprint()
assert round((mem_fp16 / mem_4bit) - self.EXPECTED_RELATIVE_DIFFERENCE, 7) >= 0
assert self.model_4bit.transformer.h[0].mlp.dense_4h_to_h.weight.__class__ == Params4bit
def test_linear_are_4bit(self):
r"""
A simple test to check if the model conversion has been done correctly by checking on the
memory footprint of the converted model and the class type of the linear layers of the converted models
"""
self.model_fp16.get_memory_footprint()
self.model_4bit.get_memory_footprint()
for name, module in self.model_4bit.named_modules():
if isinstance(module, torch.nn.Linear):
if (
name
not in self.bnb_quantization_config.keep_in_fp32_modules
+ self.bnb_quantization_config.skip_modules
):
# 4-bit parameters are packed in uint8 variables
assert module.weight.dtype == torch.uint8
def test_generate_quality(self):
r"""
Test the generation quality of the quantized model and see that we are matching the expected output.
Given that we are operating on small numbers + the testing model is relatively small, we might not get
the same output across GPUs. So we'll generate few tokens (5-10) and check their output.
"""
encoded_input = self.tokenizer(self.input_text, return_tensors="pt")
output_sequences = self.model_4bit.generate(
input_ids=encoded_input["input_ids"].to(self.model_4bit.device), max_new_tokens=10
)
assert self.tokenizer.decode(output_sequences[0], skip_special_tokens=True) in self.EXPECTED_OUTPUTS
def test_fp32_4bit_conversion(self):
r"""
Test whether it is possible to mix both `4bit` and `fp32` weights when using `keep_in_fp32_modules` correctly.
"""
from transformers import AutoModelForCausalLM
bnb_quantization_config = BnbQuantizationConfig(load_in_4bit=True, keep_in_fp32_modules=["lm_head"])
model = AutoModelForCausalLM.from_pretrained(self.model_name, torch_dtype=torch.float16)
model = load_and_quantize_model(model, bnb_quantization_config)
assert model.lm_head.weight.dtype == torch.float32
| accelerate/tests/test_quantization.py/0 | {
"file_path": "accelerate/tests/test_quantization.py",
"repo_id": "accelerate",
"token_count": 17682
} | 8 |
# Model arguments
model_name_or_path: alignment-handbook/zephyr-7b-sft-full
torch_dtype: null
# Data training arguments
dataset_mixer:
HuggingFaceH4/ultrafeedback_binarized: 1.0
dataset_splits:
- train_prefs
- test_prefs
preprocessing_num_workers: 12
# Training arguments with sensible defaults
bf16: true
beta: 0.01
loss_type: sigmoid
do_eval: true
evaluation_strategy: steps
eval_steps: 100
gradient_accumulation_steps: 2
gradient_checkpointing: true
gradient_checkpointing_kwargs:
use_reentrant: False
hub_model_id: zephyr-7b-align-scan
hub_model_revision: dpo-beta-0.01
learning_rate: 5.0e-7
logging_steps: 10
lr_scheduler_type: cosine
max_prompt_length: 512
num_train_epochs: 1
optim: adamw_torch
output_dir: data/zephyr-7b-align-scan-dpo-beta-0.01
per_device_train_batch_size: 8
per_device_eval_batch_size: 8
save_strategy: "steps"
save_steps: 100
save_total_limit: 1
seed: 42
warmup_ratio: 0.1 | alignment-handbook/recipes/pref_align_scan/dpo/config_zephyr.yaml/0 | {
"file_path": "alignment-handbook/recipes/pref_align_scan/dpo/config_zephyr.yaml",
"repo_id": "alignment-handbook",
"token_count": 359
} | 9 |
#!/usr/bin/env python
# coding=utf-8
# Copyright 2023 The HuggingFace Inc. team. All rights reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""
Supervised fine-tuning script for decoder language models.
"""
import logging
import random
import sys
import datasets
import torch
import transformers
from transformers import AutoModelForCausalLM, set_seed
from alignment import (
DataArguments,
H4ArgumentParser,
ModelArguments,
SFTConfig,
apply_chat_template,
decontaminate_humaneval,
get_checkpoint,
get_datasets,
get_kbit_device_map,
get_peft_config,
get_quantization_config,
get_tokenizer,
)
from trl import SFTTrainer, setup_chat_format
logger = logging.getLogger(__name__)
def main():
parser = H4ArgumentParser((ModelArguments, DataArguments, SFTConfig))
model_args, data_args, training_args = parser.parse()
# Set seed for reproducibility
set_seed(training_args.seed)
###############
# Setup logging
###############
logging.basicConfig(
format="%(asctime)s - %(levelname)s - %(name)s - %(message)s",
datefmt="%Y-%m-%d %H:%M:%S",
handlers=[logging.StreamHandler(sys.stdout)],
)
log_level = training_args.get_process_log_level()
logger.setLevel(log_level)
datasets.utils.logging.set_verbosity(log_level)
transformers.utils.logging.set_verbosity(log_level)
transformers.utils.logging.enable_default_handler()
transformers.utils.logging.enable_explicit_format()
# Log on each process a small summary
logger.warning(
f"Process rank: {training_args.local_rank}, device: {training_args.device}, n_gpu: {training_args.n_gpu}"
+ f" distributed training: {bool(training_args.local_rank != -1)}, 16-bits training: {training_args.fp16}"
)
logger.info(f"Model parameters {model_args}")
logger.info(f"Data parameters {data_args}")
logger.info(f"Training/evaluation parameters {training_args}")
# Check for last checkpoint
last_checkpoint = get_checkpoint(training_args)
if last_checkpoint is not None and training_args.resume_from_checkpoint is None:
logger.info(f"Checkpoint detected, resuming training at {last_checkpoint=}.")
###############
# Load datasets
###############
raw_datasets = get_datasets(data_args, splits=data_args.dataset_splits, configs=data_args.dataset_configs)
logger.info(
f"Training on the following datasets and their proportions: {[split + ' : ' + str(dset.num_rows) for split, dset in raw_datasets.items()]}"
)
column_names = list(raw_datasets["train"].features)
################
# Load tokenizer
################
tokenizer = get_tokenizer(model_args, data_args)
#######################
# Load pretrained model
#######################
logger.info("*** Load pretrained model ***")
torch_dtype = (
model_args.torch_dtype if model_args.torch_dtype in ["auto", None] else getattr(torch, model_args.torch_dtype)
)
quantization_config = get_quantization_config(model_args)
model_kwargs = dict(
revision=model_args.model_revision,
trust_remote_code=model_args.trust_remote_code,
use_flash_attention_2=model_args.use_flash_attention_2,
torch_dtype=torch_dtype,
use_cache=False if training_args.gradient_checkpointing else True,
device_map=get_kbit_device_map() if quantization_config is not None else None,
quantization_config=quantization_config,
)
model = model_args.model_name_or_path
# For ChatML we need to add special tokens and resize the embedding layer
if "<|im_start|>" in tokenizer.chat_template:
model = AutoModelForCausalLM.from_pretrained(model_args.model_name_or_path, **model_kwargs)
model, tokenizer = setup_chat_format(model, tokenizer)
model_kwargs = None
#####################
# Apply chat template
#####################
raw_datasets = raw_datasets.map(
apply_chat_template,
fn_kwargs={
"tokenizer": tokenizer,
"task": "sft",
"auto_insert_empty_system_msg": data_args.auto_insert_empty_system_msg,
},
num_proc=data_args.preprocessing_num_workers,
remove_columns=column_names,
desc="Applying chat template",
)
##########################
# Decontaminate benchmarks
##########################
num_raw_train_samples = len(raw_datasets["train"])
raw_datasets = raw_datasets.filter(decontaminate_humaneval, batched=True, batch_size=10_000, num_proc=1)
num_filtered_train_samples = num_raw_train_samples - len(raw_datasets["train"])
logger.info(
f"Decontaminated {num_filtered_train_samples} ({num_filtered_train_samples/num_raw_train_samples * 100:.2f}%) samples from the training set."
)
train_dataset = raw_datasets["train"]
eval_dataset = raw_datasets["test"]
with training_args.main_process_first(desc="Log a few random samples from the processed training set"):
for index in random.sample(range(len(raw_datasets["train"])), 3):
logger.info(f"Sample {index} of the processed training set:\n\n{raw_datasets['train'][index]['text']}")
########################
# Initialize the Trainer
########################
trainer = SFTTrainer(
model=model,
model_init_kwargs=model_kwargs,
args=training_args,
train_dataset=train_dataset,
eval_dataset=eval_dataset,
dataset_text_field="text",
max_seq_length=training_args.max_seq_length,
tokenizer=tokenizer,
packing=True,
peft_config=get_peft_config(model_args),
dataset_kwargs=training_args.dataset_kwargs,
)
###############
# Training loop
###############
logger.info("*** Train ***")
checkpoint = None
if training_args.resume_from_checkpoint is not None:
checkpoint = training_args.resume_from_checkpoint
elif last_checkpoint is not None:
checkpoint = last_checkpoint
train_result = trainer.train(resume_from_checkpoint=checkpoint)
metrics = train_result.metrics
metrics["train_samples"] = len(train_dataset)
trainer.log_metrics("train", metrics)
trainer.save_metrics("train", metrics)
trainer.save_state()
##################################
# Save model and create model card
##################################
logger.info("*** Save model ***")
trainer.save_model(training_args.output_dir)
logger.info(f"Model saved to {training_args.output_dir}")
# Save everything else on main process
kwargs = {
"finetuned_from": model_args.model_name_or_path,
"dataset": list(data_args.dataset_mixer.keys()),
"dataset_tags": list(data_args.dataset_mixer.keys()),
"tags": ["alignment-handbook"],
}
if trainer.accelerator.is_main_process:
trainer.create_model_card(**kwargs)
# Restore k,v cache for fast inference
trainer.model.config.use_cache = True
trainer.model.config.save_pretrained(training_args.output_dir)
##########
# Evaluate
##########
if training_args.do_eval:
logger.info("*** Evaluate ***")
metrics = trainer.evaluate()
metrics["eval_samples"] = len(eval_dataset)
trainer.log_metrics("eval", metrics)
trainer.save_metrics("eval", metrics)
if training_args.push_to_hub is True:
logger.info("Pushing to hub...")
trainer.push_to_hub(**kwargs)
logger.info("*** Training complete ***")
if __name__ == "__main__":
main()
| alignment-handbook/scripts/run_sft.py/0 | {
"file_path": "alignment-handbook/scripts/run_sft.py",
"repo_id": "alignment-handbook",
"token_count": 3115
} | 10 |
[build]
rustflags = ["-C", "target-cpu=native"]
[target.wasm32-unknown-unknown]
rustflags = ["-C", "target-feature=+simd128"]
[target.x86_64-apple-darwin]
rustflags = ["-C", "target-feature=-avx,-avx2"] | candle/.cargo/config.toml/0 | {
"file_path": "candle/.cargo/config.toml",
"repo_id": "candle",
"token_count": 84
} | 11 |
# Summary
[Introduction](README.md)
# User Guide
- [Installation](guide/installation.md)
- [Hello World - MNIST](guide/hello_world.md)
- [PyTorch cheatsheet](guide/cheatsheet.md)
# Reference Guide
- [Running a model](inference/inference.md)
- [Using the hub](inference/hub.md)
- [Error management](error_manage.md)
- [Training](training/training.md)
- [Simplified](training/simplified.md)
- [MNIST](training/mnist.md)
- [Fine-tuning]()
- [Serialization]()
- [Advanced Cuda usage]()
- [Writing a custom kernel]()
- [Porting a custom kernel]()
- [Using MKL]()
- [Creating apps]()
- [Creating a WASM app]()
- [Creating a REST api webserver]()
- [Creating a desktop Tauri app]()
| candle/candle-book/src/SUMMARY.md/0 | {
"file_path": "candle/candle-book/src/SUMMARY.md",
"repo_id": "candle",
"token_count": 274
} | 12 |
# Writing a custom kernel
| candle/candle-book/src/inference/cuda/writing.md/0 | {
"file_path": "candle/candle-book/src/inference/cuda/writing.md",
"repo_id": "candle",
"token_count": 6
} | 13 |
pub(crate) mod affine;
pub(crate) mod matmul;
pub(crate) mod random;
pub(crate) mod where_cond;
use candle_core::{Device, Result};
pub(crate) trait BenchDevice {
fn sync(&self) -> Result<()>;
fn bench_name<S: Into<String>>(&self, name: S) -> String;
}
impl BenchDevice for Device {
fn sync(&self) -> Result<()> {
match self {
Device::Cpu => Ok(()),
Device::Cuda(device) => {
#[cfg(feature = "cuda")]
return Ok(device.synchronize()?);
#[cfg(not(feature = "cuda"))]
panic!("Cuda device without cuda feature enabled: {:?}", device)
}
Device::Metal(device) => {
#[cfg(feature = "metal")]
return Ok(device.wait_until_completed()?);
#[cfg(not(feature = "metal"))]
panic!("Metal device without metal feature enabled: {:?}", device)
}
}
}
fn bench_name<S: Into<String>>(&self, name: S) -> String {
match self {
Device::Cpu => {
let cpu_type = if cfg!(feature = "accelerate") {
"accelerate"
} else if cfg!(feature = "mkl") {
"mkl"
} else {
"cpu"
};
format!("{}_{}", cpu_type, name.into())
}
Device::Cuda(_) => format!("cuda_{}", name.into()),
Device::Metal(_) => format!("metal_{}", name.into()),
}
}
}
struct BenchDeviceHandler {
devices: Vec<Device>,
}
impl BenchDeviceHandler {
pub fn new() -> Result<Self> {
let mut devices = Vec::new();
if cfg!(feature = "metal") {
devices.push(Device::new_metal(0)?);
} else if cfg!(feature = "cuda") {
devices.push(Device::new_cuda(0)?);
}
devices.push(Device::Cpu);
Ok(Self { devices })
}
}
| candle/candle-core/benches/benchmarks/mod.rs/0 | {
"file_path": "candle/candle-core/benches/benchmarks/mod.rs",
"repo_id": "candle",
"token_count": 1019
} | 14 |
use super::Cpu;
#[cfg(target_arch = "arm")]
use core::arch::arm::*;
#[cfg(target_arch = "aarch64")]
use core::arch::aarch64::*;
pub struct CurrentCpu {}
const STEP: usize = 16;
const EPR: usize = 4;
const ARR: usize = STEP / EPR;
impl CurrentCpu {
#[cfg(target_arch = "aarch64")]
unsafe fn reduce_one(x: float32x4_t) -> f32 {
vaddvq_f32(x)
}
#[cfg(target_arch = "arm")]
unsafe fn reduce_one(x: float32x4_t) -> f32 {
vgetq_lane_f32(x, 0) + vgetq_lane_f32(x, 1) + vgetq_lane_f32(x, 2) + vgetq_lane_f32(x, 3)
}
}
impl Cpu<ARR> for CurrentCpu {
type Unit = float32x4_t;
type Array = [float32x4_t; ARR];
const STEP: usize = STEP;
const EPR: usize = EPR;
fn n() -> usize {
ARR
}
unsafe fn zero() -> Self::Unit {
vdupq_n_f32(0.0)
}
unsafe fn from_f32(x: f32) -> Self::Unit {
vdupq_n_f32(x)
}
unsafe fn zero_array() -> Self::Array {
[Self::zero(); ARR]
}
unsafe fn load(mem_addr: *const f32) -> Self::Unit {
vld1q_f32(mem_addr)
}
unsafe fn vec_add(a: Self::Unit, b: Self::Unit) -> Self::Unit {
vaddq_f32(a, b)
}
unsafe fn vec_fma(a: Self::Unit, b: Self::Unit, c: Self::Unit) -> Self::Unit {
vfmaq_f32(a, b, c)
}
unsafe fn vec_store(mem_addr: *mut f32, a: Self::Unit) {
vst1q_f32(mem_addr, a);
}
unsafe fn vec_reduce(mut x: Self::Array, y: *mut f32) {
for i in 0..ARR / 2 {
x[2 * i] = vaddq_f32(x[2 * i], x[2 * i + 1]);
}
for i in 0..ARR / 4 {
x[4 * i] = vaddq_f32(x[4 * i], x[4 * i + 2]);
}
*y = Self::reduce_one(x[0]);
}
}
| candle/candle-core/src/cpu/neon.rs/0 | {
"file_path": "candle/candle-core/src/cpu/neon.rs",
"repo_id": "candle",
"token_count": 897
} | 15 |
//! Numpy support for tensors.
//!
//! The spec for the npy format can be found in
//! [npy-format](https://docs.scipy.org/doc/numpy-1.14.2/neps/npy-format.html).
//! The functions from this module can be used to read tensors from npy/npz files
//! or write tensors to these files. A npy file contains a single tensor (unnamed)
//! whereas a npz file can contain multiple named tensors. npz files are also compressed.
//!
//! These two formats are easy to use in Python using the numpy library.
//!
//! ```python
//! import numpy as np
//! x = np.arange(10)
//!
//! # Write a npy file.
//! np.save("test.npy", x)
//!
//! # Read a value from the npy file.
//! x = np.load("test.npy")
//!
//! # Write multiple values to a npz file.
//! values = { "x": x, "x_plus_one": x + 1 }
//! np.savez("test.npz", **values)
//!
//! # Load multiple values from a npz file.
//! values = np.loadz("test.npz")
//! ```
use crate::{DType, Device, Error, Result, Shape, Tensor};
use byteorder::{LittleEndian, ReadBytesExt};
use half::{bf16, f16, slice::HalfFloatSliceExt};
use std::collections::HashMap;
use std::fs::File;
use std::io::{BufReader, Read, Write};
use std::path::Path;
const NPY_MAGIC_STRING: &[u8] = b"\x93NUMPY";
const NPY_SUFFIX: &str = ".npy";
fn read_header<R: Read>(reader: &mut R) -> Result<String> {
let mut magic_string = vec![0u8; NPY_MAGIC_STRING.len()];
reader.read_exact(&mut magic_string)?;
if magic_string != NPY_MAGIC_STRING {
return Err(Error::Npy("magic string mismatch".to_string()));
}
let mut version = [0u8; 2];
reader.read_exact(&mut version)?;
let header_len_len = match version[0] {
1 => 2,
2 => 4,
otherwise => return Err(Error::Npy(format!("unsupported version {otherwise}"))),
};
let mut header_len = vec![0u8; header_len_len];
reader.read_exact(&mut header_len)?;
let header_len = header_len
.iter()
.rev()
.fold(0_usize, |acc, &v| 256 * acc + v as usize);
let mut header = vec![0u8; header_len];
reader.read_exact(&mut header)?;
Ok(String::from_utf8_lossy(&header).to_string())
}
#[derive(Debug, PartialEq)]
struct Header {
descr: DType,
fortran_order: bool,
shape: Vec<usize>,
}
impl Header {
fn shape(&self) -> Shape {
Shape::from(self.shape.as_slice())
}
fn to_string(&self) -> Result<String> {
let fortran_order = if self.fortran_order { "True" } else { "False" };
let mut shape = self
.shape
.iter()
.map(|x| x.to_string())
.collect::<Vec<_>>()
.join(",");
let descr = match self.descr {
DType::BF16 => Err(Error::Npy("bf16 is not supported".into()))?,
DType::F16 => "f2",
DType::F32 => "f4",
DType::F64 => "f8",
DType::I64 => "i8",
DType::U32 => "u4",
DType::U8 => "u1",
};
if !shape.is_empty() {
shape.push(',')
}
Ok(format!(
"{{'descr': '<{descr}', 'fortran_order': {fortran_order}, 'shape': ({shape}), }}"
))
}
// Hacky parser for the npy header, a typical example would be:
// {'descr': '<f8', 'fortran_order': False, 'shape': (128,), }
fn parse(header: &str) -> Result<Header> {
let header =
header.trim_matches(|c: char| c == '{' || c == '}' || c == ',' || c.is_whitespace());
let mut parts: Vec<String> = vec![];
let mut start_index = 0usize;
let mut cnt_parenthesis = 0i64;
for (index, c) in header.chars().enumerate() {
match c {
'(' => cnt_parenthesis += 1,
')' => cnt_parenthesis -= 1,
',' => {
if cnt_parenthesis == 0 {
parts.push(header[start_index..index].to_owned());
start_index = index + 1;
}
}
_ => {}
}
}
parts.push(header[start_index..].to_owned());
let mut part_map: HashMap<String, String> = HashMap::new();
for part in parts.iter() {
let part = part.trim();
if !part.is_empty() {
match part.split(':').collect::<Vec<_>>().as_slice() {
[key, value] => {
let key = key.trim_matches(|c: char| c == '\'' || c.is_whitespace());
let value = value.trim_matches(|c: char| c == '\'' || c.is_whitespace());
let _ = part_map.insert(key.to_owned(), value.to_owned());
}
_ => return Err(Error::Npy(format!("unable to parse header {header}"))),
}
}
}
let fortran_order = match part_map.get("fortran_order") {
None => false,
Some(fortran_order) => match fortran_order.as_ref() {
"False" => false,
"True" => true,
_ => return Err(Error::Npy(format!("unknown fortran_order {fortran_order}"))),
},
};
let descr = match part_map.get("descr") {
None => return Err(Error::Npy("no descr in header".to_string())),
Some(descr) => {
if descr.is_empty() {
return Err(Error::Npy("empty descr".to_string()));
}
if descr.starts_with('>') {
return Err(Error::Npy(format!("little-endian descr {descr}")));
}
// the only supported types in tensor are:
// float64, float32, float16,
// complex64, complex128,
// int64, int32, int16, int8,
// uint8, and bool.
match descr.trim_matches(|c: char| c == '=' || c == '<' || c == '|') {
"e" | "f2" => DType::F16,
"f" | "f4" => DType::F32,
"d" | "f8" => DType::F64,
// "i" | "i4" => DType::S32,
"q" | "i8" => DType::I64,
// "h" | "i2" => DType::S16,
// "b" | "i1" => DType::S8,
"B" | "u1" => DType::U8,
"I" | "u4" => DType::U32,
"?" | "b1" => DType::U8,
// "F" | "F4" => DType::C64,
// "D" | "F8" => DType::C128,
descr => return Err(Error::Npy(format!("unrecognized descr {descr}"))),
}
}
};
let shape = match part_map.get("shape") {
None => return Err(Error::Npy("no shape in header".to_string())),
Some(shape) => {
let shape = shape.trim_matches(|c: char| c == '(' || c == ')' || c == ',');
if shape.is_empty() {
vec![]
} else {
shape
.split(',')
.map(|v| v.trim().parse::<usize>())
.collect::<std::result::Result<Vec<_>, _>>()?
}
}
};
Ok(Header {
descr,
fortran_order,
shape,
})
}
}
impl Tensor {
// TODO: Add the possibility to read directly to a device?
pub(crate) fn from_reader<R: std::io::Read>(
shape: Shape,
dtype: DType,
reader: &mut R,
) -> Result<Self> {
let elem_count = shape.elem_count();
match dtype {
DType::BF16 => {
let mut data_t = vec![bf16::ZERO; elem_count];
reader.read_u16_into::<LittleEndian>(data_t.reinterpret_cast_mut())?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::F16 => {
let mut data_t = vec![f16::ZERO; elem_count];
reader.read_u16_into::<LittleEndian>(data_t.reinterpret_cast_mut())?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::F32 => {
let mut data_t = vec![0f32; elem_count];
reader.read_f32_into::<LittleEndian>(&mut data_t)?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::F64 => {
let mut data_t = vec![0f64; elem_count];
reader.read_f64_into::<LittleEndian>(&mut data_t)?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::U8 => {
let mut data_t = vec![0u8; elem_count];
reader.read_exact(&mut data_t)?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::U32 => {
let mut data_t = vec![0u32; elem_count];
reader.read_u32_into::<LittleEndian>(&mut data_t)?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
DType::I64 => {
let mut data_t = vec![0i64; elem_count];
reader.read_i64_into::<LittleEndian>(&mut data_t)?;
Tensor::from_vec(data_t, shape, &Device::Cpu)
}
}
}
/// Reads a npy file and return the stored multi-dimensional array as a tensor.
pub fn read_npy<T: AsRef<Path>>(path: T) -> Result<Self> {
let mut reader = File::open(path.as_ref())?;
let header = read_header(&mut reader)?;
let header = Header::parse(&header)?;
if header.fortran_order {
return Err(Error::Npy("fortran order not supported".to_string()));
}
Self::from_reader(header.shape(), header.descr, &mut reader)
}
/// Reads a npz file and returns the stored multi-dimensional arrays together with their names.
pub fn read_npz<T: AsRef<Path>>(path: T) -> Result<Vec<(String, Self)>> {
let zip_reader = BufReader::new(File::open(path.as_ref())?);
let mut zip = zip::ZipArchive::new(zip_reader)?;
let mut result = vec![];
for i in 0..zip.len() {
let mut reader = zip.by_index(i)?;
let name = {
let name = reader.name();
name.strip_suffix(NPY_SUFFIX).unwrap_or(name).to_owned()
};
let header = read_header(&mut reader)?;
let header = Header::parse(&header)?;
if header.fortran_order {
return Err(Error::Npy("fortran order not supported".to_string()));
}
let s = Self::from_reader(header.shape(), header.descr, &mut reader)?;
result.push((name, s))
}
Ok(result)
}
/// Reads a npz file and returns the stored multi-dimensional arrays for some specified names.
pub fn read_npz_by_name<T: AsRef<Path>>(path: T, names: &[&str]) -> Result<Vec<Self>> {
let zip_reader = BufReader::new(File::open(path.as_ref())?);
let mut zip = zip::ZipArchive::new(zip_reader)?;
let mut result = vec![];
for name in names.iter() {
let mut reader = match zip.by_name(&format!("{name}{NPY_SUFFIX}")) {
Ok(reader) => reader,
Err(_) => Err(Error::Npy(format!(
"no array for {name} in {:?}",
path.as_ref()
)))?,
};
let header = read_header(&mut reader)?;
let header = Header::parse(&header)?;
if header.fortran_order {
return Err(Error::Npy("fortran order not supported".to_string()));
}
let s = Self::from_reader(header.shape(), header.descr, &mut reader)?;
result.push(s)
}
Ok(result)
}
fn write<T: Write>(&self, f: &mut T) -> Result<()> {
f.write_all(NPY_MAGIC_STRING)?;
f.write_all(&[1u8, 0u8])?;
let header = Header {
descr: self.dtype(),
fortran_order: false,
shape: self.dims().to_vec(),
};
let mut header = header.to_string()?;
let pad = 16 - (NPY_MAGIC_STRING.len() + 5 + header.len()) % 16;
for _ in 0..pad % 16 {
header.push(' ')
}
header.push('\n');
f.write_all(&[(header.len() % 256) as u8, (header.len() / 256) as u8])?;
f.write_all(header.as_bytes())?;
self.write_bytes(f)
}
/// Writes a multi-dimensional array in the npy format.
pub fn write_npy<T: AsRef<Path>>(&self, path: T) -> Result<()> {
let mut f = File::create(path.as_ref())?;
self.write(&mut f)
}
/// Writes multiple multi-dimensional arrays using the npz format.
pub fn write_npz<S: AsRef<str>, T: AsRef<Tensor>, P: AsRef<Path>>(
ts: &[(S, T)],
path: P,
) -> Result<()> {
let mut zip = zip::ZipWriter::new(File::create(path.as_ref())?);
let options =
zip::write::FileOptions::default().compression_method(zip::CompressionMethod::Stored);
for (name, tensor) in ts.iter() {
zip.start_file(format!("{}.npy", name.as_ref()), options)?;
tensor.as_ref().write(&mut zip)?
}
Ok(())
}
}
/// Lazy tensor loader.
pub struct NpzTensors {
index_per_name: HashMap<String, usize>,
path: std::path::PathBuf,
// We do not store a zip reader as it needs mutable access to extract data. Instead we
// re-create a zip reader for each tensor.
}
impl NpzTensors {
pub fn new<T: AsRef<Path>>(path: T) -> Result<Self> {
let path = path.as_ref().to_owned();
let zip_reader = BufReader::new(File::open(&path)?);
let mut zip = zip::ZipArchive::new(zip_reader)?;
let mut index_per_name = HashMap::new();
for i in 0..zip.len() {
let file = zip.by_index(i)?;
let name = {
let name = file.name();
name.strip_suffix(NPY_SUFFIX).unwrap_or(name).to_owned()
};
index_per_name.insert(name, i);
}
Ok(Self {
index_per_name,
path,
})
}
pub fn names(&self) -> Vec<&String> {
self.index_per_name.keys().collect()
}
/// This only returns the shape and dtype for a named tensor. Compared to `get`, this avoids
/// reading the whole tensor data.
pub fn get_shape_and_dtype(&self, name: &str) -> Result<(Shape, DType)> {
let index = match self.index_per_name.get(name) {
None => crate::bail!("cannot find tensor {name}"),
Some(index) => *index,
};
let zip_reader = BufReader::new(File::open(&self.path)?);
let mut zip = zip::ZipArchive::new(zip_reader)?;
let mut reader = zip.by_index(index)?;
let header = read_header(&mut reader)?;
let header = Header::parse(&header)?;
Ok((header.shape(), header.descr))
}
pub fn get(&self, name: &str) -> Result<Option<Tensor>> {
let index = match self.index_per_name.get(name) {
None => return Ok(None),
Some(index) => *index,
};
// We hope that the file has not changed since first reading it.
let zip_reader = BufReader::new(File::open(&self.path)?);
let mut zip = zip::ZipArchive::new(zip_reader)?;
let mut reader = zip.by_index(index)?;
let header = read_header(&mut reader)?;
let header = Header::parse(&header)?;
if header.fortran_order {
return Err(Error::Npy("fortran order not supported".to_string()));
}
let tensor = Tensor::from_reader(header.shape(), header.descr, &mut reader)?;
Ok(Some(tensor))
}
}
#[cfg(test)]
mod tests {
use super::Header;
#[test]
fn parse() {
let h = "{'descr': '<f8', 'fortran_order': False, 'shape': (128,), }";
assert_eq!(
Header::parse(h).unwrap(),
Header {
descr: crate::DType::F64,
fortran_order: false,
shape: vec![128]
}
);
let h = "{'descr': '<f4', 'fortran_order': True, 'shape': (256,1,128), }";
let h = Header::parse(h).unwrap();
assert_eq!(
h,
Header {
descr: crate::DType::F32,
fortran_order: true,
shape: vec![256, 1, 128]
}
);
assert_eq!(
h.to_string().unwrap(),
"{'descr': '<f4', 'fortran_order': True, 'shape': (256,1,128,), }"
);
let h = Header {
descr: crate::DType::U32,
fortran_order: false,
shape: vec![],
};
assert_eq!(
h.to_string().unwrap(),
"{'descr': '<u4', 'fortran_order': False, 'shape': (), }"
);
}
}
| candle/candle-core/src/npy.rs/0 | {
"file_path": "candle/candle-core/src/npy.rs",
"repo_id": "candle",
"token_count": 8717
} | 16 |
use crate::{Result, Tensor, WithDType};
pub enum TensorScalar {
Tensor(Tensor),
Scalar(Tensor),
}
pub trait TensorOrScalar {
fn to_tensor_scalar(self) -> Result<TensorScalar>;
}
impl TensorOrScalar for &Tensor {
fn to_tensor_scalar(self) -> Result<TensorScalar> {
Ok(TensorScalar::Tensor(self.clone()))
}
}
impl<T: WithDType> TensorOrScalar for T {
fn to_tensor_scalar(self) -> Result<TensorScalar> {
let scalar = Tensor::new(self, &crate::Device::Cpu)?;
Ok(TensorScalar::Scalar(scalar))
}
}
| candle/candle-core/src/scalar.rs/0 | {
"file_path": "candle/candle-core/src/scalar.rs",
"repo_id": "candle",
"token_count": 261
} | 17 |
import numpy as np
x = np.arange(10)
# Write a npy file.
np.save("test.npy", x)
# Write multiple values to a npz file.
values = { "x": x, "x_plus_one": x + 1 }
np.savez("test.npz", **values)
| candle/candle-core/tests/npy.py/0 | {
"file_path": "candle/candle-core/tests/npy.py",
"repo_id": "candle",
"token_count": 83
} | 18 |
pub mod tinystories;
| candle/candle-datasets/src/nlp/mod.rs/0 | {
"file_path": "candle/candle-datasets/src/nlp/mod.rs",
"repo_id": "candle",
"token_count": 6
} | 19 |
# candle-convnext
[A ConvNet for the 2020s](https://arxiv.org/abs/2201.03545) and
[ConvNeXt V2: Co-designing and Scaling ConvNets with Masked Autoencoders](https://arxiv.org/abs/2301.00808).
This candle implementation uses a pre-trained ConvNeXt network for inference. The
classification head has been trained on the ImageNet dataset and returns the
probabilities for the top-5 classes.
## Running an example
```
$ cargo run --example convnext --release -- --image candle-examples/examples/yolo-v8/assets/bike.jpg --which tiny
loaded image Tensor[dims 3, 224, 224; f32]
model built
mountain bike, all-terrain bike, off-roader: 84.09%
bicycle-built-for-two, tandem bicycle, tandem: 4.15%
maillot : 0.74%
crash helmet : 0.54%
unicycle, monocycle : 0.44%
```
| candle/candle-examples/examples/convnext/README.md/0 | {
"file_path": "candle/candle-examples/examples/convnext/README.md",
"repo_id": "candle",
"token_count": 293
} | 20 |
#[cfg(feature = "mkl")]
extern crate intel_mkl_src;
#[cfg(feature = "accelerate")]
extern crate accelerate_src;
use anyhow::Result;
use candle::{DType, IndexOp, Tensor};
use candle_nn::VarBuilder;
use candle_transformers::models::encodec::{Config, Model};
use clap::{Parser, ValueEnum};
use hf_hub::api::sync::Api;
mod audio_io;
#[derive(Clone, Debug, Copy, PartialEq, Eq, ValueEnum)]
enum Action {
AudioToAudio,
AudioToCode,
CodeToAudio,
}
#[derive(Parser, Debug)]
#[command(author, version, about, long_about = None)]
struct Args {
/// The action to be performed, specifies the format for the input and output data.
action: Action,
/// The input file, either an audio file or some encodec tokens stored as safetensors.
in_file: String,
/// The output file, either a wave audio file or some encodec tokens stored as safetensors.
out_file: String,
/// Run on CPU rather than on GPU.
#[arg(long)]
cpu: bool,
/// The model weight file, in safetensor format.
#[arg(long)]
model: Option<String>,
}
fn main() -> Result<()> {
let args = Args::parse();
let device = candle_examples::device(args.cpu)?;
let model = match args.model {
Some(model) => std::path::PathBuf::from(model),
None => Api::new()?
.model("facebook/encodec_24khz".to_string())
.get("model.safetensors")?,
};
let vb = unsafe { VarBuilder::from_mmaped_safetensors(&[model], DType::F32, &device)? };
let config = Config::default();
let model = Model::new(&config, vb)?;
let codes = match args.action {
Action::CodeToAudio => {
let codes = candle::safetensors::load(args.in_file, &device)?;
codes.get("codes").expect("no codes in input file").clone()
}
Action::AudioToCode | Action::AudioToAudio => {
let pcm = if args.in_file == "-" {
println!(">>>> RECORDING AUDIO, PRESS ENTER ONCE DONE <<<<");
let (stream, input_audio) = audio_io::setup_input_stream()?;
let mut pcms = vec![];
let stdin = std::thread::spawn(|| {
let mut s = String::new();
std::io::stdin().read_line(&mut s)
});
while !stdin.is_finished() {
let input = input_audio.lock().unwrap().take_all();
if input.is_empty() {
std::thread::sleep(std::time::Duration::from_millis(100));
continue;
}
pcms.push(input)
}
drop(stream);
pcms.concat()
} else {
let (pcm, sample_rate) = audio_io::pcm_decode(args.in_file)?;
if sample_rate != 24_000 {
println!("WARNING: encodec uses a 24khz sample rate, input uses {sample_rate}, resampling...");
audio_io::resample(&pcm, sample_rate as usize, 24_000)?
} else {
pcm
}
};
let pcm_len = pcm.len();
let pcm = Tensor::from_vec(pcm, (1, 1, pcm_len), &device)?;
println!("input pcm shape: {:?}", pcm.shape());
model.encode(&pcm)?
}
};
println!("codes shape: {:?}", codes.shape());
match args.action {
Action::AudioToCode => {
codes.save_safetensors("codes", &args.out_file)?;
}
Action::AudioToAudio | Action::CodeToAudio => {
let pcm = model.decode(&codes)?;
println!("output pcm shape: {:?}", pcm.shape());
let pcm = pcm.i(0)?.i(0)?;
let pcm = candle_examples::audio::normalize_loudness(&pcm, 24_000, true)?;
let pcm = pcm.to_vec1::<f32>()?;
if args.out_file == "-" {
let (stream, ad) = audio_io::setup_output_stream()?;
{
let mut ad = ad.lock().unwrap();
ad.push_samples(&pcm)?;
}
loop {
let ad = ad.lock().unwrap();
if ad.is_empty() {
break;
}
// That's very weird, calling thread::sleep here triggers the stream to stop
// playing (the callback doesn't seem to be called anymore).
// std::thread::sleep(std::time::Duration::from_millis(100));
}
drop(stream)
} else {
let mut output = std::fs::File::create(&args.out_file)?;
candle_examples::wav::write_pcm_as_wav(&mut output, &pcm, 24_000)?;
}
}
}
Ok(())
}
| candle/candle-examples/examples/encodec/main.rs/0 | {
"file_path": "candle/candle-examples/examples/encodec/main.rs",
"repo_id": "candle",
"token_count": 2395
} | 21 |
#[cfg(feature = "mkl")]
extern crate intel_mkl_src;
#[cfg(feature = "accelerate")]
extern crate accelerate_src;
use candle::{IndexOp, D};
use clap::{Parser, ValueEnum};
#[derive(Clone, Copy, Debug, ValueEnum)]
enum Which {
SqueezeNet,
EfficientNet,
}
#[derive(Parser)]
struct Args {
#[arg(long)]
image: String,
#[arg(long)]
model: Option<String>,
/// The model to be used.
#[arg(value_enum, long, default_value_t = Which::SqueezeNet)]
which: Which,
}
pub fn main() -> anyhow::Result<()> {
let args = Args::parse();
let image = candle_examples::imagenet::load_image224(args.image)?;
let image = match args.which {
Which::SqueezeNet => image,
Which::EfficientNet => image.permute((1, 2, 0))?,
};
println!("loaded image {image:?}");
let model = match args.model {
Some(model) => std::path::PathBuf::from(model),
None => match args.which {
Which::SqueezeNet => hf_hub::api::sync::Api::new()?
.model("lmz/candle-onnx".into())
.get("squeezenet1.1-7.onnx")?,
Which::EfficientNet => hf_hub::api::sync::Api::new()?
.model("onnx/EfficientNet-Lite4".into())
.get("efficientnet-lite4-11.onnx")?,
},
};
let model = candle_onnx::read_file(model)?;
let graph = model.graph.as_ref().unwrap();
let mut inputs = std::collections::HashMap::new();
inputs.insert(graph.input[0].name.to_string(), image.unsqueeze(0)?);
let mut outputs = candle_onnx::simple_eval(&model, inputs)?;
let output = outputs.remove(&graph.output[0].name).unwrap();
let prs = match args.which {
Which::SqueezeNet => candle_nn::ops::softmax(&output, D::Minus1)?,
Which::EfficientNet => output,
};
let prs = prs.i(0)?.to_vec1::<f32>()?;
// Sort the predictions and take the top 5
let mut top: Vec<_> = prs.iter().enumerate().collect();
top.sort_by(|a, b| b.1.partial_cmp(a.1).unwrap());
let top = top.into_iter().take(5).collect::<Vec<_>>();
// Print the top predictions
for &(i, p) in &top {
println!(
"{:50}: {:.2}%",
candle_examples::imagenet::CLASSES[i],
p * 100.0
);
}
Ok(())
}
| candle/candle-examples/examples/onnx/main.rs/0 | {
"file_path": "candle/candle-examples/examples/onnx/main.rs",
"repo_id": "candle",
"token_count": 1042
} | 22 |
use super::gym_env::{GymEnv, Step};
use candle::{DType, Device, Error, Module, Result, Tensor};
use candle_nn::{
linear, ops::log_softmax, ops::softmax, sequential::seq, Activation, AdamW, Optimizer,
ParamsAdamW, VarBuilder, VarMap,
};
use rand::{distributions::Distribution, rngs::ThreadRng, Rng};
fn new_model(
input_shape: &[usize],
num_actions: usize,
dtype: DType,
device: &Device,
) -> Result<(impl Module, VarMap)> {
let input_size = input_shape.iter().product();
let mut varmap = VarMap::new();
let var_builder = VarBuilder::from_varmap(&varmap, dtype, device);
let model = seq()
.add(linear(input_size, 32, var_builder.pp("lin1"))?)
.add(Activation::Relu)
.add(linear(32, num_actions, var_builder.pp("lin2"))?);
Ok((model, varmap))
}
fn accumulate_rewards(steps: &[Step<i64>]) -> Vec<f64> {
let mut rewards: Vec<f64> = steps.iter().map(|s| s.reward).collect();
let mut acc_reward = 0f64;
for (i, reward) in rewards.iter_mut().enumerate().rev() {
if steps[i].terminated {
acc_reward = 0.0;
}
acc_reward += *reward;
*reward = acc_reward;
}
rewards
}
fn weighted_sample(probs: Vec<f32>, rng: &mut ThreadRng) -> Result<usize> {
let distribution = rand::distributions::WeightedIndex::new(probs).map_err(Error::wrap)?;
let mut rng = rng;
Ok(distribution.sample(&mut rng))
}
pub fn run() -> Result<()> {
let env = GymEnv::new("CartPole-v1")?;
println!("action space: {:?}", env.action_space());
println!("observation space: {:?}", env.observation_space());
let (model, varmap) = new_model(
env.observation_space(),
env.action_space(),
DType::F32,
&Device::Cpu,
)?;
let optimizer_params = ParamsAdamW {
lr: 0.01,
weight_decay: 0.01,
..Default::default()
};
let mut optimizer = AdamW::new(varmap.all_vars(), optimizer_params)?;
let mut rng = rand::thread_rng();
for epoch_idx in 0..100 {
let mut state = env.reset(rng.gen::<u64>())?;
let mut steps: Vec<Step<i64>> = vec![];
loop {
let action = {
let action_probs: Vec<f32> =
softmax(&model.forward(&state.detach().unsqueeze(0)?)?, 1)?
.squeeze(0)?
.to_vec1()?;
weighted_sample(action_probs, &mut rng)? as i64
};
let step = env.step(action)?;
steps.push(step.copy_with_obs(&state));
if step.terminated || step.truncated {
state = env.reset(rng.gen::<u64>())?;
if steps.len() > 5000 {
break;
}
} else {
state = step.state;
}
}
let total_reward: f64 = steps.iter().map(|s| s.reward).sum();
let episodes: i64 = steps
.iter()
.map(|s| (s.terminated || s.truncated) as i64)
.sum();
println!(
"epoch: {:<3} episodes: {:<5} avg reward per episode: {:.2}",
epoch_idx,
episodes,
total_reward / episodes as f64
);
let batch_size = steps.len();
let rewards = Tensor::from_vec(accumulate_rewards(&steps), batch_size, &Device::Cpu)?
.to_dtype(DType::F32)?
.detach();
let actions_mask = {
let actions: Vec<i64> = steps.iter().map(|s| s.action).collect();
let actions_mask: Vec<Tensor> = actions
.iter()
.map(|&action| {
// One-hot encoding
let mut action_mask = vec![0.0; env.action_space()];
action_mask[action as usize] = 1.0;
Tensor::from_vec(action_mask, env.action_space(), &Device::Cpu)
.unwrap()
.to_dtype(DType::F32)
.unwrap()
})
.collect();
Tensor::stack(&actions_mask, 0)?.detach()
};
let states = {
let states: Vec<Tensor> = steps.into_iter().map(|s| s.state).collect();
Tensor::stack(&states, 0)?.detach()
};
let log_probs = actions_mask
.mul(&log_softmax(&model.forward(&states)?, 1)?)?
.sum(1)?;
let loss = rewards.mul(&log_probs)?.neg()?.mean_all()?;
optimizer.backward_step(&loss)?;
}
Ok(())
}
| candle/candle-examples/examples/reinforcement-learning/policy_gradient.rs/0 | {
"file_path": "candle/candle-examples/examples/reinforcement-learning/policy_gradient.rs",
"repo_id": "candle",
"token_count": 2333
} | 23 |
## VGG Model Implementation
This example demonstrates the implementation of VGG models (VGG13, VGG16, VGG19) using the Candle library.
The VGG models are defined in `candle-transformers/src/models/vgg.rs`. The main function in `candle-examples/examples/vgg/main.rs` loads an image, selects the VGG model based on the provided argument, and applies the model to the loaded image.
You can run the example with the following command:
```bash
cargo run --example vgg --release -- --image ../yolo-v8/assets/bike.jpg --which vgg13
```
In the command above, `--image` specifies the path to the image file and `--which` specifies the VGG model to use (vgg13, vgg16, or vgg19).
| candle/candle-examples/examples/vgg/README.md/0 | {
"file_path": "candle/candle-examples/examples/vgg/README.md",
"repo_id": "candle",
"token_count": 200
} | 24 |
pub mod audio;
pub mod bs1770;
pub mod coco_classes;
pub mod imagenet;
pub mod token_output_stream;
pub mod wav;
use candle::utils::{cuda_is_available, metal_is_available};
use candle::{Device, Result, Tensor};
pub fn device(cpu: bool) -> Result<Device> {
if cpu {
Ok(Device::Cpu)
} else if cuda_is_available() {
Ok(Device::new_cuda(0)?)
} else if metal_is_available() {
Ok(Device::new_metal(0)?)
} else {
#[cfg(all(target_os = "macos", target_arch = "aarch64"))]
{
println!(
"Running on CPU, to run on GPU(metal), build this example with `--features metal`"
);
}
#[cfg(not(all(target_os = "macos", target_arch = "aarch64")))]
{
println!("Running on CPU, to run on GPU, build this example with `--features cuda`");
}
Ok(Device::Cpu)
}
}
pub fn load_image<P: AsRef<std::path::Path>>(
p: P,
resize_longest: Option<usize>,
) -> Result<(Tensor, usize, usize)> {
let img = image::io::Reader::open(p)?
.decode()
.map_err(candle::Error::wrap)?;
let (initial_h, initial_w) = (img.height() as usize, img.width() as usize);
let img = match resize_longest {
None => img,
Some(resize_longest) => {
let (height, width) = (img.height(), img.width());
let resize_longest = resize_longest as u32;
let (height, width) = if height < width {
let h = (resize_longest * height) / width;
(h, resize_longest)
} else {
let w = (resize_longest * width) / height;
(resize_longest, w)
};
img.resize_exact(width, height, image::imageops::FilterType::CatmullRom)
}
};
let (height, width) = (img.height() as usize, img.width() as usize);
let img = img.to_rgb8();
let data = img.into_raw();
let data = Tensor::from_vec(data, (height, width, 3), &Device::Cpu)?.permute((2, 0, 1))?;
Ok((data, initial_h, initial_w))
}
pub fn load_image_and_resize<P: AsRef<std::path::Path>>(
p: P,
width: usize,
height: usize,
) -> Result<Tensor> {
let img = image::io::Reader::open(p)?
.decode()
.map_err(candle::Error::wrap)?
.resize_to_fill(
width as u32,
height as u32,
image::imageops::FilterType::Triangle,
);
let img = img.to_rgb8();
let data = img.into_raw();
Tensor::from_vec(data, (width, height, 3), &Device::Cpu)?.permute((2, 0, 1))
}
/// Saves an image to disk using the image crate, this expects an input with shape
/// (c, height, width).
pub fn save_image<P: AsRef<std::path::Path>>(img: &Tensor, p: P) -> Result<()> {
let p = p.as_ref();
let (channel, height, width) = img.dims3()?;
if channel != 3 {
candle::bail!("save_image expects an input of shape (3, height, width)")
}
let img = img.permute((1, 2, 0))?.flatten_all()?;
let pixels = img.to_vec1::<u8>()?;
let image: image::ImageBuffer<image::Rgb<u8>, Vec<u8>> =
match image::ImageBuffer::from_raw(width as u32, height as u32, pixels) {
Some(image) => image,
None => candle::bail!("error saving image {p:?}"),
};
image.save(p).map_err(candle::Error::wrap)?;
Ok(())
}
pub fn save_image_resize<P: AsRef<std::path::Path>>(
img: &Tensor,
p: P,
h: usize,
w: usize,
) -> Result<()> {
let p = p.as_ref();
let (channel, height, width) = img.dims3()?;
if channel != 3 {
candle::bail!("save_image expects an input of shape (3, height, width)")
}
let img = img.permute((1, 2, 0))?.flatten_all()?;
let pixels = img.to_vec1::<u8>()?;
let image: image::ImageBuffer<image::Rgb<u8>, Vec<u8>> =
match image::ImageBuffer::from_raw(width as u32, height as u32, pixels) {
Some(image) => image,
None => candle::bail!("error saving image {p:?}"),
};
let image = image::DynamicImage::from(image);
let image = image.resize_to_fill(w as u32, h as u32, image::imageops::FilterType::CatmullRom);
image.save(p).map_err(candle::Error::wrap)?;
Ok(())
}
/// Loads the safetensors files for a model from the hub based on a json index file.
pub fn hub_load_safetensors(
repo: &hf_hub::api::sync::ApiRepo,
json_file: &str,
) -> Result<Vec<std::path::PathBuf>> {
let json_file = repo.get(json_file).map_err(candle::Error::wrap)?;
let json_file = std::fs::File::open(json_file)?;
let json: serde_json::Value =
serde_json::from_reader(&json_file).map_err(candle::Error::wrap)?;
let weight_map = match json.get("weight_map") {
None => candle::bail!("no weight map in {json_file:?}"),
Some(serde_json::Value::Object(map)) => map,
Some(_) => candle::bail!("weight map in {json_file:?} is not a map"),
};
let mut safetensors_files = std::collections::HashSet::new();
for value in weight_map.values() {
if let Some(file) = value.as_str() {
safetensors_files.insert(file.to_string());
}
}
let safetensors_files = safetensors_files
.iter()
.map(|v| repo.get(v).map_err(candle::Error::wrap))
.collect::<Result<Vec<_>>>()?;
Ok(safetensors_files)
}
| candle/candle-examples/src/lib.rs/0 | {
"file_path": "candle/candle-examples/src/lib.rs",
"repo_id": "candle",
"token_count": 2455
} | 25 |
// Inspired by
// https://github.com/NVIDIA/DALI/blob/main/include/dali/core/static_switch.h
// and https://github.com/pytorch/pytorch/blob/master/aten/src/ATen/Dispatch.h
#pragma once
/// @param COND - a boolean expression to switch by
/// @param CONST_NAME - a name given for the constexpr bool variable.
/// @param ... - code to execute for true and false
///
/// Usage:
/// ```
/// BOOL_SWITCH(flag, BoolConst, [&] {
/// some_function<BoolConst>(...);
/// });
/// ```
#define BOOL_SWITCH(COND, CONST_NAME, ...) \
[&] { \
if (COND) { \
constexpr static bool CONST_NAME = true; \
return __VA_ARGS__(); \
} else { \
constexpr static bool CONST_NAME = false; \
return __VA_ARGS__(); \
} \
}()
#define FP16_SWITCH(COND, ...) \
[&] { \
if (COND) { \
using elem_type = cutlass::half_t; \
return __VA_ARGS__(); \
} else { \
using elem_type = cutlass::bfloat16_t; \
return __VA_ARGS__(); \
} \
}()
#define FWD_HEADDIM_SWITCH(HEADDIM, ...) \
[&] { \
if (HEADDIM <= 32) { \
constexpr static int kHeadDim = 32; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 64) { \
constexpr static int kHeadDim = 64; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 96) { \
constexpr static int kHeadDim = 96; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 128) { \
constexpr static int kHeadDim = 128; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 160) { \
constexpr static int kHeadDim = 160; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 192) { \
constexpr static int kHeadDim = 192; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 224) { \
constexpr static int kHeadDim = 224; \
return __VA_ARGS__(); \
} else if (HEADDIM <= 256) { \
constexpr static int kHeadDim = 256; \
return __VA_ARGS__(); \
} \
}()
| candle/candle-flash-attn/kernels/static_switch.h/0 | {
"file_path": "candle/candle-flash-attn/kernels/static_switch.h",
"repo_id": "candle",
"token_count": 1516
} | 26 |
// WARNING: THIS IS ONLY VALID ASSUMING THAT inp IS CONTIGUOUS!
// TODO: proper error reporting when ids are larger than v_size.
#include "cuda_utils.cuh"
#include<stdint.h>
template<typename T, typename I>
__device__ void index_select(
const size_t numel,
const size_t num_dims,
const size_t *info,
const I *ids,
const T *inp,
T *out,
const size_t left_size,
const size_t src_dim_size,
const size_t ids_dim_size,
const size_t right_size
) {
const size_t *dims = info;
const size_t *strides = info + num_dims;
bool b = is_contiguous(num_dims, dims, strides);
for (unsigned int dst_i = blockIdx.x * blockDim.x + threadIdx.x; dst_i < numel; dst_i += blockDim.x * gridDim.x) {
unsigned int left_i = dst_i / (ids_dim_size * right_size);
unsigned int id_i = dst_i / right_size % ids_dim_size;
unsigned int right_i = dst_i % right_size;
unsigned int src_i = left_i * (src_dim_size * right_size) + ids[id_i] * right_size + right_i;
unsigned strided_i = b ? src_i : get_strided_index(src_i, num_dims, dims, strides);
out[dst_i] = inp[strided_i];
}
}
#define IS_OP(TYPENAME, INDEX_TYPENAME, FN_NAME) \
extern "C" __global__ void FN_NAME( \
const size_t numel, \
const size_t num_dims, \
const size_t *info, \
const INDEX_TYPENAME *ids, \
const TYPENAME *inp, \
TYPENAME *out, \
const size_t left_size, \
const size_t src_dim_size, \
const size_t ids_dim_size, \
const size_t right_size \
) { index_select(numel, num_dims, info, ids, inp, out, left_size, src_dim_size, ids_dim_size, right_size); } \
template<typename T, typename I>
__device__ void gather(
const size_t numel,
const I *ids,
const T *inp,
T *out,
const size_t left_size,
const size_t src_dim_size,
const size_t ids_dim_size,
const size_t right_size
) {
for (unsigned int i = blockIdx.x * blockDim.x + threadIdx.x; i < numel; i += blockDim.x * gridDim.x) {
size_t post = i % right_size;
size_t idx = ids[i];
size_t pre = i / (right_size * ids_dim_size);
size_t src_i = (pre * src_dim_size + idx) * right_size + post;
out[i] = inp[src_i];
}
}
#define GATHER_OP(TYPENAME, INDEX_TYPENAME, FN_NAME) \
extern "C" __global__ void FN_NAME( \
const size_t numel, \
const INDEX_TYPENAME *ids, \
const TYPENAME *inp, \
TYPENAME *out, \
const size_t left_size, \
const size_t src_dim_size, \
const size_t ids_dim_size, \
const size_t right_size \
) { gather(numel, ids, inp, out, left_size, src_dim_size, ids_dim_size, right_size); } \
template<typename T, typename I>
__device__ void index_add(
const I *ids,
const size_t ids_dim_size,
const T *inp,
T *out,
const size_t left_size,
const size_t src_dim_size,
const size_t dst_dim_size,
const size_t right_size
) {
const size_t numel = left_size * right_size;
for (unsigned int i = blockIdx.x * blockDim.x + threadIdx.x; i < numel; i += blockDim.x * gridDim.x) {
const size_t pre = i / right_size;
const size_t post = i % right_size;
for (unsigned int j = 0; j < ids_dim_size; ++j) {
const size_t idx = ids[j];
const size_t src_i = (pre * ids_dim_size + j) * right_size + post;
const size_t dst_i = (pre * dst_dim_size + idx) * right_size + post;
out[dst_i] += inp[src_i];
}
}
}
#define IA_OP(TYPENAME, INDEX_TYPENAME, FN_NAME) \
extern "C" __global__ void FN_NAME( \
const INDEX_TYPENAME *ids, \
const size_t ids_dim_size, \
const TYPENAME *inp, \
TYPENAME *out, \
const size_t left_size, \
const size_t src_dim_size, \
const size_t dst_dim_size, \
const size_t right_size \
) { index_add(ids, ids_dim_size, inp, out, left_size, src_dim_size, dst_dim_size, right_size); } \
template<typename T, typename I>
__device__ void scatter_add(
const I *ids,
const T *inp,
T *out,
const size_t left_size,
const size_t src_dim_size,
const size_t dst_dim_size,
const size_t right_size
) {
const size_t numel = left_size * right_size;
for (unsigned int i = blockIdx.x * blockDim.x + threadIdx.x; i < numel; i += blockDim.x * gridDim.x) {
const size_t pre = i / right_size;
const size_t post = i % right_size;
for (unsigned int j = 0; j < src_dim_size; ++j) {
const size_t src_i = (pre * src_dim_size + j) * right_size + post;
const size_t idx = ids[src_i];
const size_t dst_i = (pre * dst_dim_size + idx) * right_size + post;
out[dst_i] += inp[src_i];
}
}
}
#define SA_OP(TYPENAME, INDEX_TYPENAME, FN_NAME) \
extern "C" __global__ void FN_NAME( \
const INDEX_TYPENAME *ids, \
const TYPENAME *inp, \
TYPENAME *out, \
const size_t left_size, \
const size_t src_dim_size, \
const size_t dst_dim_size, \
const size_t right_size \
) { scatter_add(ids, inp, out, left_size, src_dim_size, dst_dim_size, right_size); } \
#if __CUDA_ARCH__ >= 800
IS_OP(__nv_bfloat16, int64_t, is_i64_bf16)
IS_OP(__nv_bfloat16, uint32_t, is_u32_bf16)
IS_OP(__nv_bfloat16, uint8_t, is_u8_bf16)
GATHER_OP(__nv_bfloat16, int64_t, gather_i64_bf16)
GATHER_OP(__nv_bfloat16, uint32_t, gather_u32_bf16)
GATHER_OP(__nv_bfloat16, uint8_t, gather_u8_bf16)
IA_OP(__nv_bfloat16, int64_t, ia_i64_bf16)
IA_OP(__nv_bfloat16, uint32_t, ia_u32_bf16)
IA_OP(__nv_bfloat16, uint8_t, ia_u8_bf16)
SA_OP(__nv_bfloat16, int64_t, sa_i64_bf16)
SA_OP(__nv_bfloat16, uint32_t, sa_u32_bf16)
SA_OP(__nv_bfloat16, uint8_t, sa_u8_bf16)
#endif
#if __CUDA_ARCH__ >= 530
IS_OP(__half, int64_t, is_i64_f16)
IS_OP(__half, uint32_t, is_u32_f16)
IS_OP(__half, uint8_t, is_u8_f16)
GATHER_OP(__half, int64_t, gather_i64_f16)
GATHER_OP(__half, uint32_t, gather_u32_f16)
GATHER_OP(__half, uint8_t, gather_u8_f16)
IA_OP(__half, uint32_t, ia_u32_f16)
IA_OP(__half, uint8_t, ia_u8_f16)
SA_OP(__half, uint32_t, sa_u32_f16)
SA_OP(__half, uint8_t, sa_u8_f16)
#endif
IS_OP(float, int64_t, is_i64_f32)
IS_OP(double, int64_t, is_i64_f64)
IS_OP(uint8_t, int64_t, is_i64_u8)
IS_OP(uint32_t, int64_t, is_i64_u32)
IS_OP(int64_t, int64_t, is_i64_i64)
IS_OP(float, uint32_t, is_u32_f32)
IS_OP(double, uint32_t, is_u32_f64)
IS_OP(uint8_t, uint32_t, is_u32_u8)
IS_OP(int64_t, uint32_t, is_u32_i64)
IS_OP(uint32_t, uint32_t, is_u32_u32)
IS_OP(float, uint8_t, is_u8_f32)
IS_OP(double, uint8_t, is_u8_f64)
IS_OP(uint8_t, uint8_t, is_u8_u8)
IS_OP(uint32_t, uint8_t, is_u8_u32)
IS_OP(int64_t, uint8_t, is_u8_i64)
GATHER_OP(float, int64_t, gather_i64_f32)
GATHER_OP(double, int64_t, gather_i64_f64)
GATHER_OP(uint8_t, int64_t, gather_i64_u8)
GATHER_OP(uint32_t, int64_t, gather_i64_u32)
GATHER_OP(int64_t, int64_t, gather_i64_i64)
GATHER_OP(float, uint32_t, gather_u32_f32)
GATHER_OP(double, uint32_t, gather_u32_f64)
GATHER_OP(uint8_t, uint32_t, gather_u32_u8)
GATHER_OP(int64_t, uint32_t, gather_u32_i64)
GATHER_OP(uint32_t, uint32_t, gather_u32_u32)
GATHER_OP(float, uint8_t, gather_u8_f32)
GATHER_OP(double, uint8_t, gather_u8_f64)
GATHER_OP(uint8_t, uint8_t, gather_u8_u8)
GATHER_OP(uint32_t, uint8_t, gather_u8_u32)
GATHER_OP(int64_t, uint8_t, gather_u8_i64)
IA_OP(float, int64_t, ia_i64_f32)
IA_OP(double, int64_t, ia_i64_f64)
IA_OP(uint8_t, int64_t, ia_i64_u8)
IA_OP(int64_t, int64_t, ia_i64_i64)
IA_OP(uint32_t, int64_t, ia_i64_u32)
IA_OP(float, uint32_t, ia_u32_f32)
IA_OP(double, uint32_t, ia_u32_f64)
IA_OP(uint8_t, uint32_t, ia_u32_u8)
IA_OP(int64_t, uint32_t, ia_u32_i64)
IA_OP(uint32_t, uint32_t, ia_u32_u32)
IA_OP(float, uint8_t, ia_u8_f32)
IA_OP(double, uint8_t, ia_u8_f64)
IA_OP(uint8_t, uint8_t, ia_u8_u8)
IA_OP(uint32_t, uint8_t, ia_u8_u32)
IA_OP(int64_t, uint8_t, ia_u8_i64)
SA_OP(float, int64_t, sa_i64_f32)
SA_OP(double, int64_t, sa_i64_f64)
SA_OP(uint8_t, int64_t, sa_i64_u8)
SA_OP(int64_t, int64_t, sa_i64_i64)
SA_OP(uint32_t, int64_t, sa_i64_u32)
SA_OP(float, uint32_t, sa_u32_f32)
SA_OP(double, uint32_t, sa_u32_f64)
SA_OP(uint8_t, uint32_t, sa_u32_u8)
SA_OP(int64_t, uint32_t, sa_u32_i64)
SA_OP(uint32_t, uint32_t, sa_u32_u32)
SA_OP(float, uint8_t, sa_u8_f32)
SA_OP(double, uint8_t, sa_u8_f64)
SA_OP(uint8_t, uint8_t, sa_u8_u8)
SA_OP(uint32_t, uint8_t, sa_u8_u32)
SA_OP(int64_t, uint8_t, sa_u8_i64)
| candle/candle-kernels/src/indexing.cu/0 | {
"file_path": "candle/candle-kernels/src/indexing.cu",
"repo_id": "candle",
"token_count": 4314
} | 27 |
#include <metal_stdlib>
#include <metal_integer>
#include <metal_atomic>
using namespace metal;
// Constants
// 2^32 and 1/2^32. Useful for converting between float and uint.
static constexpr constant ulong UNIF01_NORM32 = 4294967296;
static constexpr constant float UNIF01_INV32 = 2.328306436538696289e-10;
// 2 * pi
static constexpr constant float TWO_PI = 2.0 * M_PI_F;
static constexpr constant int3 S1 = {13, 19, 12};
static constexpr constant int3 S2 = {2, 25, 4};
static constexpr constant int3 S3 = {3, 11, 17};
// Used to prevent bad seeds.
static constexpr constant uint64_t PHI[16] = {
0x9E3779B97F4A7C15,
0xF39CC0605CEDC834,
0x1082276BF3A27251,
0xF86C6A11D0C18E95,
0x2767F0B153D27B7F,
0x0347045B5BF1827F,
0x01886F0928403002,
0xC1D64BA40F335E36,
0xF06AD7AE9717877E,
0x85839D6EFFBD7DC6,
0x64D325D1C5371682,
0xCADD0CCCFDFFBBE1,
0x626E33B8D04B4331,
0xBBF73C790D94F79D,
0x471C4AB3ED3D82A5,
0xFEC507705E4AE6E5,
};
// Combined Tausworthe and LCG Random Number Generator.
// https://developer.nvidia.com/gpugems/gpugems3/part-vi-gpu-computing/chapter-37-efficient-random-number-generation-and-application
// https://indico.cern.ch/event/93877/contributions/2118070/attachments/1104200/1575343/acat3_revised_final.pdf
struct HybridTaus {
float state;
HybridTaus() thread = default;
HybridTaus() threadgroup = default;
HybridTaus() device = default;
HybridTaus() constant = default;
// Generate seeds for each thread.
METAL_FUNC static uint4 seed_per_thread(const ulong4 seeds) {
return uint4(ulong4(seeds) * ulong4(PHI[0], PHI[1], PHI[2], PHI[3]) * ulong4(1099087573UL));
}
// Tausworthe generator.
METAL_FUNC static uint taus(const uint z, const int3 s, const uint M) {
uint b = (((z << s.x) ^ z) >> s.y);
return (((z & M) << s.z) ^ b);
}
// LCG generator.
METAL_FUNC static uint lcg(const uint z) {
return (1664525 * z + 1013904223UL);
}
// Initialize the RNG state.
METAL_FUNC static HybridTaus init(const ulong4 seeds) {
uint4 seed = seed_per_thread(seeds);
// Seed #1
uint z1 = taus(seed.x, S1, 4294967294UL);
uint z2 = taus(seed.y, S2, 4294967288UL);
uint z3 = taus(seed.z, S3, 4294967280UL);
uint z4 = lcg(seed.x);
// Seed #2
uint r1 = (z1^z2^z3^z4^seed.y);
z1 = taus(r1, S1, 429496729UL);
z2 = taus(r1, S2, 4294967288UL);
z3 = taus(r1, S3, 429496280UL);
z4 = lcg(r1);
// Seed #3
r1 = (z1^z2^z3^z4^seed.z);
z1 = taus(r1, S1, 429496729UL);
z2 = taus(r1, S2, 4294967288UL);
z3 = taus(r1, S3, 429496280UL);
z4 = lcg(r1);
// Seed #4
r1 = (z1^z2^z3^z4^seed.w);
z1 = taus(r1, S1, 429496729UL);
z2 = taus(r1, S2, 4294967288UL);
z3 = taus(r1, S3, 429496280UL);
z4 = lcg(r1);
HybridTaus rng;
rng.state = (z1^z2^z3^z4) * UNIF01_INV32;
return rng;
}
METAL_FUNC float rand() {
uint seed = this->state * UNIF01_NORM32;
uint z1 = taus(seed, S1, 429496729UL);
uint z2 = taus(seed, S2, 4294967288UL);
uint z3 = taus(seed, S3, 429496280UL);
uint z4 = lcg(seed);
thread float result = this->state;
this->state = (z1^z2^z3^z4) * UNIF01_INV32;
return result;
}
};
template<typename T> METAL_FUNC void rand_uniform(
constant size_t &size,
constant float &min,
constant float &max,
device atomic_uint *seed,
device T *out,
uint tid [[thread_position_in_grid]]
) {
if (tid >= size) {
return;
}
// Evenly sized vectors need an offset when writing the mirror element.
uint off = 1 - size % 2;
float diff = abs(min - max);
uint s = atomic_load_explicit(seed, memory_order_relaxed);
HybridTaus rng = HybridTaus::init({ulong(s), tid, 1, 1});
out[tid] = static_cast<T>(rng.rand() * diff + min);
if (tid == 0) {
atomic_store_explicit(seed, uint(rng.rand() * UNIF01_NORM32), memory_order_relaxed);
// Return early if tid == 0 && off == 0, otherwise we will write to out[size].
if (off == 0)
return;
}
// Use symmetry to fill the other half of the array.
out[size - off - tid] = static_cast<T>(rng.rand() * diff + min);
}
// Create Gaussian normal distribution using Box-Muller transform:
// https://en.wikipedia.org/wiki/Box–Muller_transform
template<typename T> METAL_FUNC void normal(
constant size_t &size,
constant float &mean,
constant float &stddev,
device atomic_uint *seed,
device T *out,
uint tid [[thread_position_in_grid]]
) {
if (tid >= size) {
return;
}
// Evenly sized vectors need an offset when writing the mirror element.
uint off = 1 - size % 2;
uint s = atomic_load_explicit(seed, memory_order_relaxed);
HybridTaus rng = HybridTaus::init({ulong(s), tid, 1, 1});
float u1 = rng.rand();
float u2 = rng.rand();
float cosval;
float sinval = sincos(TWO_PI * u2, cosval);
float mag = stddev * sqrt(-2.0 * log(u1));
float z0 = mag * cosval + mean;
float z1 = mag * sinval + mean;
out[tid] = static_cast<T>(z0);
if (tid == 0) {
atomic_store_explicit(seed, uint(rng.rand() * UNIF01_NORM32), memory_order_relaxed);
// Return early if tid == 0 && off == 0, otherwise we will write to out[size].
if (off == 0)
return;
}
// Use symmetry to fill the other half of the array.
out[size - off - tid] = static_cast<T>(z1);
}
#define UNIFORM_OP(NAME, T) \
kernel void rand_uniform_##NAME( \
constant size_t &size, \
constant float &min, \
constant float &max, \
device atomic_uint *seed, \
device T *out, \
uint tid [[thread_position_in_grid]] \
) { \
rand_uniform<T>(size, min, max, seed, out, tid); \
} \
#define NORMAL_OP(NAME, T) \
kernel void rand_normal_##NAME( \
constant size_t &size, \
constant float &mean, \
constant float &stddev, \
device atomic_uint *seed, \
device T *out, \
uint tid [[thread_position_in_grid]] \
) { \
normal<T>(size, mean, stddev, seed, out, tid); \
} \
#define RANDOM_OPS(NAME, T) \
UNIFORM_OP(NAME, T) \
NORMAL_OP(NAME, T) \
RANDOM_OPS(f32, float)
RANDOM_OPS(f16, half)
#if __METAL_VERSION__ >= 310
RANDOM_OPS(bf16, bfloat)
#endif
| candle/candle-metal-kernels/src/random.metal/0 | {
"file_path": "candle/candle-metal-kernels/src/random.metal",
"repo_id": "candle",
"token_count": 3671
} | 28 |
//! Embedding Layer.
use candle::{Result, Tensor};
#[derive(Clone, Debug)]
pub struct Embedding {
embeddings: Tensor,
hidden_size: usize,
}
impl Embedding {
pub fn new(embeddings: Tensor, hidden_size: usize) -> Self {
Self {
embeddings,
hidden_size,
}
}
pub fn embeddings(&self) -> &Tensor {
&self.embeddings
}
/// Get the hidden size of the embedding matrix
pub fn hidden_size(&self) -> usize {
self.hidden_size
}
}
impl crate::Module for Embedding {
fn forward(&self, indexes: &Tensor) -> Result<Tensor> {
let mut final_dims = indexes.dims().to_vec();
final_dims.push(self.hidden_size);
let indexes = indexes.flatten_all()?;
let values = self.embeddings.index_select(&indexes, 0)?;
let values = values.reshape(final_dims)?;
Ok(values)
}
}
pub fn embedding(in_size: usize, out_size: usize, vb: crate::VarBuilder) -> Result<Embedding> {
let embeddings = vb.get_with_hints(
(in_size, out_size),
"weight",
crate::Init::Randn {
mean: 0.,
stdev: 1.,
},
)?;
Ok(Embedding::new(embeddings, out_size))
}
| candle/candle-nn/src/embedding.rs/0 | {
"file_path": "candle/candle-nn/src/embedding.rs",
"repo_id": "candle",
"token_count": 571
} | 29 |
/* Equivalent PyTorch code.
import torch
from torch.nn.functional import group_norm
t = torch.tensor(
[[[-0.3034, 0.2726, -0.9659],
[-1.1845, -1.3236, 0.0172],
[ 1.9507, 1.2554, -0.8625],
[ 1.0682, 0.3604, 0.3985],
[-0.4957, -0.4461, -0.9721],
[ 1.5157, -0.1546, -0.5596]],
[[-1.6698, -0.4040, -0.7927],
[ 0.3736, -0.0975, -0.1351],
[-0.9461, 0.5461, -0.6334],
[-1.0919, -0.1158, 0.1213],
[-0.9535, 0.1281, 0.4372],
[-0.2845, 0.3488, 0.5641]]])
print(group_norm(t, num_groups=2))
print(group_norm(t, num_groups=3))
*/
#[cfg(feature = "mkl")]
extern crate intel_mkl_src;
#[cfg(feature = "accelerate")]
extern crate accelerate_src;
use anyhow::Result;
use candle::test_utils::to_vec3_round;
use candle::{Device, Tensor};
use candle_nn::{GroupNorm, Module};
#[test]
fn group_norm() -> Result<()> {
let device = &Device::Cpu;
let w = Tensor::from_vec(vec![1f32; 6], 6, device)?;
let b = Tensor::from_vec(vec![0f32; 6], 6, device)?;
let gn2 = GroupNorm::new(w.clone(), b.clone(), 6, 2, 1e-5)?;
let gn3 = GroupNorm::new(w, b, 6, 3, 1e-5)?;
let input = Tensor::new(
&[
[
[-0.3034f32, 0.2726, -0.9659],
[-1.1845, -1.3236, 0.0172],
[1.9507, 1.2554, -0.8625],
[1.0682, 0.3604, 0.3985],
[-0.4957, -0.4461, -0.9721],
[1.5157, -0.1546, -0.5596],
],
[
[-1.6698, -0.4040, -0.7927],
[0.3736, -0.0975, -0.1351],
[-0.9461, 0.5461, -0.6334],
[-1.0919, -0.1158, 0.1213],
[-0.9535, 0.1281, 0.4372],
[-0.2845, 0.3488, 0.5641],
],
],
device,
)?;
assert_eq!(
to_vec3_round(&gn2.forward(&input)?, 4)?,
&[
[
[-0.1653, 0.3748, -0.7866],
[-0.9916, -1.1220, 0.1353],
[1.9485, 1.2965, -0.6896],
[1.2769, 0.3628, 0.4120],
[-0.7427, -0.6786, -1.3578],
[1.8547, -0.3022, -0.8252]
],
[
[-1.9342, 0.0211, -0.5793],
[1.2223, 0.4945, 0.4365],
[-0.8163, 1.4887, -0.3333],
[-1.7960, -0.0392, 0.3875],
[-1.5469, 0.3998, 0.9561],
[-0.3428, 0.7970, 1.1845]
]
]
);
assert_eq!(
to_vec3_round(&gn3.forward(&input)?, 4)?,
&[
[
[0.4560, 1.4014, -0.6313],
[-0.9901, -1.2184, 0.9822],
[1.4254, 0.6360, -1.7682],
[0.4235, -0.3800, -0.3367],
[-0.3890, -0.3268, -0.9862],
[2.1325, 0.0386, -0.4691]
],
[
[-1.8797, 0.0777, -0.5234],
[1.2802, 0.5517, 0.4935],
[-1.0102, 1.5327, -0.4773],
[-1.2587, 0.4047, 0.8088],
[-1.9074, 0.1691, 0.7625],
[-0.6230, 0.5928, 1.0061]
]
]
);
Ok(())
}
| candle/candle-nn/tests/group_norm.rs/0 | {
"file_path": "candle/candle-nn/tests/group_norm.rs",
"repo_id": "candle",
"token_count": 2154
} | 30 |
## Installation
From the `candle-pyo3` directory, enable a virtual env where you will want the
candle package to be installed then run.
```bash
maturin develop -r
python test.py
```
## Generating Stub Files for Type Hinting
For type hinting support, the `candle-pyo3` package requires `*.pyi` files. You can automatically generate these files using the `stub.py` script.
### Steps:
1. Install the package using `maturin`.
2. Generate the stub files by running:
```
python stub.py
```
### Validation:
To ensure that the stub files match the current implementation, execute:
```
python stub.py --check
```
| candle/candle-pyo3/README.md/0 | {
"file_path": "candle/candle-pyo3/README.md",
"repo_id": "candle",
"token_count": 190
} | 31 |
from .module import Module
from typing import Optional, Tuple, Any
from candle import Tensor
import candle
class Embedding(Module):
"""A simple lookup table that stores embeddings of a fixed dictionary and size.
This module is often used to store word embeddings and retrieve them using indices.
The input to the module is a list of indices, and the output is the corresponding
word embeddings.
Args:
num_embeddings (int): size of the dictionary of embeddings
embedding_dim (int): the size of each embedding vector
Attributes:
weight (Tensor): the learnable weights of the module of shape (num_embeddings, embedding_dim)
initialized from :math:`\mathcal{N}(0, 1)`
Shape:
- Input: :math:`(*)`, IntTensor or LongTensor of arbitrary shape containing the indices to extract
- Output: :math:`(*, H)`, where `*` is the input shape and :math:`H=\text{embedding\_dim}`
"""
def __init__(self, num_embeddings: int, embedding_dim: int, device=None) -> None:
factory_kwargs = {"device": device}
super().__init__()
self.num_embeddings = num_embeddings
self.embedding_dim = embedding_dim
self.weight = candle.randn((num_embeddings, embedding_dim), **factory_kwargs)
def forward(self, indexes: Tensor) -> Tensor:
final_dims = list(indexes.shape)
final_dims.append(self.embedding_dim)
indexes = indexes.flatten_all()
values = self.weight.index_select(indexes, 0)
return values.reshape(final_dims)
| candle/candle-pyo3/py_src/candle/nn/sparse.py/0 | {
"file_path": "candle/candle-pyo3/py_src/candle/nn/sparse.py",
"repo_id": "candle",
"token_count": 590
} | 32 |
use candle::Result;
use candle_nn::{batch_norm, Conv2dConfig, Module, VarBuilder};
#[allow(clippy::many_single_char_names)]
fn conv2d_same(
i: usize,
o: usize,
k: usize,
c: Conv2dConfig,
vb: VarBuilder,
) -> Result<impl Module> {
let conv2d = candle_nn::conv2d(i, o, k, c, vb)?;
let s = c.stride;
let module = candle_nn::func(move |xs| {
let ih = xs.dim(2)?;
let iw = xs.dim(3)?;
let oh = (ih + s - 1) / s;
let ow = (iw + s - 1) / s;
let pad_h = usize::max((oh - 1) * s + k - ih, 0);
let pad_w = usize::max((ow - 1) * s + k - iw, 0);
if pad_h > 0 || pad_w > 0 {
xs.pad_with_zeros(3, pad_w / 2, pad_w - pad_w / 2)?
.pad_with_zeros(2, pad_h / 2, pad_h - pad_h / 2)?
.apply(&conv2d)
} else {
xs.apply(&conv2d)
}
});
Ok(module)
}
fn block(dim: usize, kernel_size: usize, vb: VarBuilder) -> Result<impl Module> {
let conv2d_cfg = Conv2dConfig {
groups: dim,
..Default::default()
};
let vb_fn = vb.pp(0).pp("fn");
let conv1 = conv2d_same(dim, dim, kernel_size, conv2d_cfg, vb_fn.pp(0))?;
let bn1 = batch_norm(dim, 1e-5, vb_fn.pp(2))?;
let conv2 = candle_nn::conv2d(dim, dim, 1, Default::default(), vb.pp(1))?;
let bn2 = batch_norm(dim, 1e-5, vb.pp(3))?;
Ok(candle_nn::func(move |xs| {
let ys = xs.apply(&conv1)?.gelu_erf()?.apply_t(&bn1, false)?;
(xs + ys)?.apply(&conv2)?.gelu_erf()?.apply_t(&bn2, false)
}))
}
fn convmixer(
nclasses: usize,
dim: usize,
depth: usize,
kernel_size: usize,
patch_size: usize,
vb: VarBuilder,
) -> Result<candle_nn::Func<'static>> {
let conv2d_cfg = Conv2dConfig {
stride: patch_size,
..Default::default()
};
let conv1 = candle_nn::conv2d(3, dim, patch_size, conv2d_cfg, vb.pp(0))?;
let bn1 = batch_norm(dim, 1e-5, vb.pp(2))?;
let blocks: Vec<_> = (0..depth)
.map(|index| block(dim, kernel_size, vb.pp(3 + index)))
.collect::<Result<Vec<_>>>()?;
let fc = candle_nn::linear(dim, nclasses, vb.pp(25))?;
Ok(candle_nn::func(move |xs| {
let mut xs = xs.apply(&conv1)?.gelu_erf()?.apply_t(&bn1, false)?;
for block in blocks.iter() {
xs = xs.apply(block)?
}
// This performs the adaptive average pooling with a target size of (1, 1).
xs.mean(3)?.mean(2)?.apply(&fc)
}))
}
pub fn c1536_20(nclasses: usize, vb: VarBuilder) -> Result<candle_nn::Func<'static>> {
convmixer(nclasses, 1536, 20, 9, 7, vb)
}
pub fn c1024_20(nclasses: usize, vb: VarBuilder) -> Result<candle_nn::Func<'static>> {
convmixer(nclasses, 1024, 20, 9, 14, vb)
}
| candle/candle-transformers/src/models/convmixer.rs/0 | {
"file_path": "candle/candle-transformers/src/models/convmixer.rs",
"repo_id": "candle",
"token_count": 1413
} | 33 |
use crate::{
quantized_nn::{layer_norm, linear_no_bias as linear, Embedding, Linear},
quantized_var_builder::VarBuilder,
};
use candle::{IndexOp, Result, Tensor};
use candle_nn::{GroupNorm, LayerNorm, Module};
pub use crate::models::rwkv_v5::{Config, State, Tokenizer};
#[derive(Debug, Clone)]
struct SelfAttention {
key: Linear,
receptance: Linear,
value: Linear,
gate: Linear,
output: Linear,
ln_x: candle_nn::GroupNorm,
time_mix_key: Tensor,
time_mix_value: Tensor,
time_mix_receptance: Tensor,
time_decay: Tensor,
time_faaaa: Tensor,
time_mix_gate: Tensor,
layer_id: usize,
n_attn_heads: usize,
}
impl SelfAttention {
fn new(layer_id: usize, cfg: &Config, vb: VarBuilder) -> Result<Self> {
let hidden_size = cfg.hidden_size;
let attn_hidden_size = cfg.attention_hidden_size;
let key = linear(hidden_size, attn_hidden_size, vb.pp("key"))?;
let receptance = linear(hidden_size, attn_hidden_size, vb.pp("receptance"))?;
let value = linear(hidden_size, attn_hidden_size, vb.pp("value"))?;
let gate = linear(hidden_size, attn_hidden_size, vb.pp("gate"))?;
let output = linear(attn_hidden_size, hidden_size, vb.pp("output"))?;
let vb_x = vb.pp("ln_x");
let ln_x_weight = vb_x.get(hidden_size, "weight")?.dequantize(vb.device())?;
let ln_x_bias = vb_x.get(hidden_size, "bias")?.dequantize(vb.device())?;
let ln_x = GroupNorm::new(
ln_x_weight,
ln_x_bias,
hidden_size,
hidden_size / cfg.head_size,
1e-5,
)?;
let time_mix_key = vb
.get((1, 1, cfg.hidden_size), "time_mix_key")?
.dequantize(vb.device())?;
let time_mix_value = vb
.get((1, 1, cfg.hidden_size), "time_mix_value")?
.dequantize(vb.device())?;
let time_mix_receptance = vb
.get((1, 1, cfg.hidden_size), "time_mix_receptance")?
.dequantize(vb.device())?;
let n_attn_heads = cfg.hidden_size / cfg.head_size;
let time_decay = vb
.get((n_attn_heads, cfg.head_size), "time_decay")?
.dequantize(vb.device())?;
let time_faaaa = vb
.get((n_attn_heads, cfg.head_size), "time_faaaa")?
.dequantize(vb.device())?;
let time_mix_gate = vb
.get((1, 1, cfg.hidden_size), "time_mix_gate")?
.dequantize(vb.device())?;
Ok(Self {
key,
value,
receptance,
gate,
output,
ln_x,
time_mix_key,
time_mix_value,
time_mix_receptance,
time_decay,
time_faaaa,
time_mix_gate,
layer_id,
n_attn_heads,
})
}
pub fn forward(&self, xs: &Tensor, state: &mut State) -> Result<Tensor> {
let h = self.time_decay.dim(0)?;
let (b, t, s) = xs.dims3()?;
let s = s / h;
let (receptance, key, value, gate) = {
// extract key-value
let shifted = state.per_layer[self.layer_id].extract_key_value.clone();
let shifted = if shifted.rank() == 2 {
shifted.unsqueeze(1)?
} else {
shifted
};
let key = ((xs * &self.time_mix_key)? + &shifted * (1.0 - &self.time_mix_key)?)?;
let value = ((xs * &self.time_mix_value)? + &shifted * (1.0 - &self.time_mix_value)?)?;
let receptance = ((xs * &self.time_mix_receptance)?
+ &shifted * (1.0 - &self.time_mix_receptance)?)?;
let gate = ((xs * &self.time_mix_gate)? + &shifted * (1.0 - &self.time_mix_gate)?)?;
let key = self.key.forward(&key)?;
let value = self.value.forward(&value)?;
let receptance = self.receptance.forward(&receptance)?;
let gate = candle_nn::ops::silu(&self.gate.forward(&gate)?)?;
state.per_layer[self.layer_id].extract_key_value = xs.i((.., t - 1))?;
(receptance, key, value, gate)
};
// linear attention
let mut state_ = state.per_layer[self.layer_id].linear_attention.clone();
let key = key.reshape((b, t, h, s))?.permute((0, 2, 3, 1))?;
let value = value.reshape((b, t, h, s))?.transpose(1, 2)?;
let receptance = receptance.reshape((b, t, h, s))?.transpose(1, 2)?;
let time_decay = self
.time_decay
.exp()?
.neg()?
.exp()?
.reshape(((), 1, 1))?
.reshape((self.n_attn_heads, (), 1))?;
let time_faaaa =
self.time_faaaa
.reshape(((), 1, 1))?
.reshape((self.n_attn_heads, (), 1))?;
let mut out: Vec<Tensor> = Vec::with_capacity(t);
for t_ in 0..t {
let rt = receptance.i((.., .., t_..t_ + 1))?.contiguous()?;
let kt = key.i((.., .., .., t_..t_ + 1))?.contiguous()?;
let vt = value.i((.., .., t_..t_ + 1))?.contiguous()?;
let at = kt.matmul(&vt)?;
let rhs = (time_faaaa.broadcast_mul(&at)? + &state_)?;
let out_ = rt.matmul(&rhs)?.squeeze(2)?;
state_ = (&at + time_decay.broadcast_mul(&state_))?;
out.push(out_)
}
let out = Tensor::cat(&out, 1)?.reshape((b * t, h * s, 1))?;
let out = out.apply(&self.ln_x)?.reshape((b, t, h * s))?;
let out = (out * gate)?.apply(&self.output)?;
state.per_layer[self.layer_id].linear_attention = state_;
Ok(out)
}
}
#[derive(Debug, Clone)]
struct FeedForward {
time_mix_key: Tensor,
time_mix_receptance: Tensor,
key: Linear,
receptance: Linear,
value: Linear,
layer_id: usize,
}
impl FeedForward {
fn new(layer_id: usize, cfg: &Config, vb: VarBuilder) -> Result<Self> {
let int_size = cfg
.intermediate_size
.unwrap_or(((cfg.hidden_size as f64 * 3.5) as usize) / 32 * 32);
let key = linear(cfg.hidden_size, int_size, vb.pp("key"))?;
let receptance = linear(cfg.hidden_size, cfg.hidden_size, vb.pp("receptance"))?;
let value = linear(int_size, cfg.hidden_size, vb.pp("value"))?;
let time_mix_key = vb
.get((1, 1, cfg.hidden_size), "time_mix_key")?
.dequantize(vb.device())?;
let time_mix_receptance = vb
.get((1, 1, cfg.hidden_size), "time_mix_receptance")?
.dequantize(vb.device())?;
Ok(Self {
key,
receptance,
value,
time_mix_key,
time_mix_receptance,
layer_id,
})
}
fn forward(&self, xs: &Tensor, state: &mut State) -> Result<Tensor> {
let shifted = &state.per_layer[self.layer_id].feed_forward;
let key = (xs.broadcast_mul(&self.time_mix_key)?
+ shifted.broadcast_mul(&(1.0 - &self.time_mix_key)?)?)?;
let receptance = (xs.broadcast_mul(&self.time_mix_receptance)?
+ shifted.broadcast_mul(&(1.0 - &self.time_mix_receptance)?)?)?;
let key = key.apply(&self.key)?.relu()?.sqr()?;
let value = key.apply(&self.value)?;
let receptance = candle_nn::ops::sigmoid(&receptance.apply(&self.receptance)?)?;
state.per_layer[self.layer_id].feed_forward = xs.i((.., xs.dim(1)? - 1))?;
let xs = (receptance * value)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
struct Block {
pre_ln: Option<LayerNorm>,
ln1: LayerNorm,
ln2: LayerNorm,
attention: SelfAttention,
feed_forward: FeedForward,
}
impl Block {
fn new(layer_id: usize, cfg: &Config, vb: VarBuilder) -> Result<Self> {
let ln1 = layer_norm(cfg.hidden_size, cfg.layer_norm_epsilon, vb.pp("ln1"))?;
let ln2 = layer_norm(cfg.hidden_size, cfg.layer_norm_epsilon, vb.pp("ln2"))?;
let pre_ln = if layer_id == 0 {
let ln = layer_norm(cfg.hidden_size, cfg.layer_norm_epsilon, vb.pp("pre_ln"))?;
Some(ln)
} else {
None
};
let attention = SelfAttention::new(layer_id, cfg, vb.pp("attention"))?;
let feed_forward = FeedForward::new(layer_id, cfg, vb.pp("feed_forward"))?;
Ok(Self {
pre_ln,
ln1,
ln2,
attention,
feed_forward,
})
}
fn forward(&self, xs: &Tensor, state: &mut State) -> Result<Tensor> {
let xs = match self.pre_ln.as_ref() {
None => xs.clone(),
Some(pre_ln) => xs.apply(pre_ln)?,
};
let attention = self.attention.forward(&xs.apply(&self.ln1)?, state)?;
let xs = (xs + attention)?;
let feed_forward = self.feed_forward.forward(&xs.apply(&self.ln2)?, state)?;
let xs = (xs + feed_forward)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
pub struct Model {
embeddings: Embedding,
blocks: Vec<Block>,
ln_out: LayerNorm,
head: Linear,
rescale_every: usize,
layers_are_rescaled: bool,
}
impl Model {
pub fn new(cfg: &Config, vb: VarBuilder) -> Result<Self> {
let vb_m = vb.pp("rwkv");
let embeddings = Embedding::new(cfg.vocab_size, cfg.hidden_size, vb_m.pp("embeddings"))?;
let mut blocks = Vec::with_capacity(cfg.num_hidden_layers);
let vb_b = vb_m.pp("blocks");
for block_index in 0..cfg.num_hidden_layers {
let block = Block::new(block_index, cfg, vb_b.pp(block_index))?;
blocks.push(block)
}
let ln_out = layer_norm(cfg.hidden_size, 1e-5, vb_m.pp("ln_out"))?;
let head = linear(cfg.hidden_size, cfg.vocab_size, vb.pp("head"))?;
Ok(Self {
embeddings,
blocks,
ln_out,
head,
rescale_every: cfg.rescale_every,
layers_are_rescaled: false, // This seem to only happen for the f16/bf16 dtypes.
})
}
pub fn forward(&self, xs: &Tensor, state: &mut State) -> Result<Tensor> {
let (_b_size, _seq_len) = xs.dims2()?;
let mut xs = xs.apply(&self.embeddings)?;
for (block_idx, block) in self.blocks.iter().enumerate() {
xs = block.forward(&xs, state)?;
if self.layers_are_rescaled && (block_idx + 1) % self.rescale_every == 0 {
xs = (xs / 2.)?
}
}
let xs = xs.apply(&self.ln_out)?.apply(&self.head)?;
state.pos += 1;
Ok(xs)
}
}
| candle/candle-transformers/src/models/quantized_rwkv_v5.rs/0 | {
"file_path": "candle/candle-transformers/src/models/quantized_rwkv_v5.rs",
"repo_id": "candle",
"token_count": 5518
} | 34 |
use candle::{Result, Tensor};
use candle_nn::{layer_norm, LayerNorm, Linear, Module, VarBuilder};
#[derive(Debug)]
struct Attention {
q_proj: Linear,
k_proj: Linear,
v_proj: Linear,
out_proj: Linear,
num_heads: usize,
}
impl Attention {
fn new(
embedding_dim: usize,
num_heads: usize,
downsample_rate: usize,
vb: VarBuilder,
) -> Result<Self> {
let internal_dim = embedding_dim / downsample_rate;
let q_proj = candle_nn::linear(embedding_dim, internal_dim, vb.pp("q_proj"))?;
let k_proj = candle_nn::linear(embedding_dim, internal_dim, vb.pp("k_proj"))?;
let v_proj = candle_nn::linear(embedding_dim, internal_dim, vb.pp("v_proj"))?;
let out_proj = candle_nn::linear(internal_dim, embedding_dim, vb.pp("out_proj"))?;
Ok(Self {
q_proj,
k_proj,
v_proj,
out_proj,
num_heads,
})
}
fn separate_heads(&self, x: &Tensor) -> Result<Tensor> {
let (b, n, c) = x.dims3()?;
x.reshape((b, n, self.num_heads, c / self.num_heads))?
.transpose(1, 2)?
.contiguous()
}
fn recombine_heads(&self, x: &Tensor) -> Result<Tensor> {
let (b, n_heads, n_tokens, c_per_head) = x.dims4()?;
x.transpose(1, 2)?
.reshape((b, n_tokens, n_heads * c_per_head))
}
fn forward(&self, q: &Tensor, k: &Tensor, v: &Tensor) -> Result<Tensor> {
let q = self.q_proj.forward(&q.contiguous()?)?;
let k = self.k_proj.forward(&k.contiguous()?)?;
let v = self.v_proj.forward(&v.contiguous()?)?;
let q = self.separate_heads(&q)?;
let k = self.separate_heads(&k)?;
let v = self.separate_heads(&v)?;
let (_, _, _, c_per_head) = q.dims4()?;
let attn = (q.matmul(&k.t()?)? / (c_per_head as f64).sqrt())?;
let attn = candle_nn::ops::softmax_last_dim(&attn)?;
let out = attn.matmul(&v)?;
self.recombine_heads(&out)?.apply(&self.out_proj)
}
}
#[derive(Debug)]
struct TwoWayAttentionBlock {
self_attn: Attention,
norm1: LayerNorm,
cross_attn_token_to_image: Attention,
norm2: LayerNorm,
mlp: super::MlpBlock,
norm3: LayerNorm,
norm4: LayerNorm,
cross_attn_image_to_token: Attention,
skip_first_layer_pe: bool,
}
impl TwoWayAttentionBlock {
fn new(
embedding_dim: usize,
num_heads: usize,
mlp_dim: usize,
skip_first_layer_pe: bool,
vb: VarBuilder,
) -> Result<Self> {
let norm1 = layer_norm(embedding_dim, 1e-5, vb.pp("norm1"))?;
let norm2 = layer_norm(embedding_dim, 1e-5, vb.pp("norm2"))?;
let norm3 = layer_norm(embedding_dim, 1e-5, vb.pp("norm3"))?;
let norm4 = layer_norm(embedding_dim, 1e-5, vb.pp("norm4"))?;
let self_attn = Attention::new(embedding_dim, num_heads, 1, vb.pp("self_attn"))?;
let cross_attn_token_to_image = Attention::new(
embedding_dim,
num_heads,
2,
vb.pp("cross_attn_token_to_image"),
)?;
let cross_attn_image_to_token = Attention::new(
embedding_dim,
num_heads,
2,
vb.pp("cross_attn_image_to_token"),
)?;
let mlp = super::MlpBlock::new(
embedding_dim,
mlp_dim,
candle_nn::Activation::Relu,
vb.pp("mlp"),
)?;
Ok(Self {
self_attn,
norm1,
cross_attn_image_to_token,
norm2,
mlp,
norm3,
norm4,
cross_attn_token_to_image,
skip_first_layer_pe,
})
}
fn forward(
&self,
queries: &Tensor,
keys: &Tensor,
query_pe: &Tensor,
key_pe: &Tensor,
) -> Result<(Tensor, Tensor)> {
// Self attention block
let queries = if self.skip_first_layer_pe {
self.self_attn.forward(queries, queries, queries)?
} else {
let q = (queries + query_pe)?;
let attn_out = self.self_attn.forward(&q, &q, queries)?;
(queries + attn_out)?
};
let queries = self.norm1.forward(&queries)?;
// Cross attention block, tokens attending to image embedding
let q = (&queries + query_pe)?;
let k = (keys + key_pe)?;
let attn_out = self.cross_attn_token_to_image.forward(&q, &k, keys)?;
let queries = (&queries + attn_out)?;
let queries = self.norm2.forward(&queries)?;
// MLP block
let mlp_out = self.mlp.forward(&queries);
let queries = (queries + mlp_out)?;
let queries = self.norm3.forward(&queries)?;
// Cross attention block, image embedding attending to tokens
let q = (&queries + query_pe)?;
let k = (keys + key_pe)?;
let attn_out = self.cross_attn_image_to_token.forward(&k, &q, &queries)?;
let keys = (keys + attn_out)?;
let keys = self.norm4.forward(&keys)?;
Ok((queries, keys))
}
}
#[derive(Debug)]
pub struct TwoWayTransformer {
layers: Vec<TwoWayAttentionBlock>,
final_attn_token_to_image: Attention,
norm_final_attn: LayerNorm,
}
impl TwoWayTransformer {
pub fn new(
depth: usize,
embedding_dim: usize,
num_heads: usize,
mlp_dim: usize,
vb: VarBuilder,
) -> Result<Self> {
let vb_l = vb.pp("layers");
let mut layers = Vec::with_capacity(depth);
for i in 0..depth {
let layer =
TwoWayAttentionBlock::new(embedding_dim, num_heads, mlp_dim, i == 0, vb_l.pp(i))?;
layers.push(layer)
}
let final_attn_token_to_image = Attention::new(
embedding_dim,
num_heads,
2,
vb.pp("final_attn_token_to_image"),
)?;
let norm_final_attn = layer_norm(embedding_dim, 1e-5, vb.pp("norm_final_attn"))?;
Ok(Self {
layers,
final_attn_token_to_image,
norm_final_attn,
})
}
pub fn forward(
&self,
image_embedding: &Tensor,
image_pe: &Tensor,
point_embedding: &Tensor,
) -> Result<(Tensor, Tensor)> {
let image_embedding = image_embedding.flatten_from(2)?.permute((0, 2, 1))?;
let image_pe = image_pe.flatten_from(2)?.permute((0, 2, 1))?;
let mut queries = point_embedding.clone();
let mut keys = image_embedding;
for layer in self.layers.iter() {
(queries, keys) = layer.forward(&queries, &keys, point_embedding, &image_pe)?
}
let q = (&queries + point_embedding)?;
let k = (&keys + image_pe)?;
let attn_out = self.final_attn_token_to_image.forward(&q, &k, &keys)?;
let queries = (queries + attn_out)?.apply(&self.norm_final_attn)?;
Ok((queries, keys))
}
}
| candle/candle-transformers/src/models/segment_anything/transformer.rs/0 | {
"file_path": "candle/candle-transformers/src/models/segment_anything/transformer.rs",
"repo_id": "candle",
"token_count": 3597
} | 35 |
// T5 Text Model
// https://github.com/huggingface/transformers/blob/main/src/transformers/models/t5/modeling_t5.py
use crate::models::with_tracing::{linear_no_bias, Embedding, Linear};
use candle::{DType, Device, Module, Result, Tensor, D};
use candle_nn::{Activation, VarBuilder};
use serde::Deserialize;
use std::sync::Arc;
fn default_relative_attention_max_distance() -> usize {
128
}
fn default_is_decoder() -> bool {
false
}
fn default_use_cache() -> bool {
true
}
fn default_tie_word_embeddings() -> bool {
true
}
fn get_mask(size: usize, device: &Device) -> Result<Tensor> {
let mask: Vec<_> = (0..size)
.flat_map(|i| (0..size).map(move |j| u8::from(j > i)))
.collect();
Tensor::from_slice(&mask, (size, size), device)
}
fn masked_fill(on_false: &Tensor, mask: &Tensor, on_true: f32) -> Result<Tensor> {
let shape = mask.shape();
let on_true = Tensor::new(on_true, on_false.device())?.broadcast_as(shape.dims())?;
let m = mask.where_cond(&on_true, on_false)?;
Ok(m)
}
#[derive(Debug, Deserialize, Default, Clone, PartialEq)]
pub struct ActivationWithOptionalGating {
pub gated: bool,
pub activation: candle_nn::Activation,
}
pub fn deserialize_feed_forward_proj_activation<'de, D>(
deserializer: D,
) -> std::result::Result<ActivationWithOptionalGating, D::Error>
where
D: serde::de::Deserializer<'de>,
{
match String::deserialize(deserializer)?.as_str() {
"gated-gelu" => Ok(ActivationWithOptionalGating {
gated: true,
activation: candle_nn::Activation::NewGelu,
}),
"gated-silu" => Ok(ActivationWithOptionalGating {
gated: true,
activation: candle_nn::Activation::Silu,
}),
buf => {
let activation = serde_plain::from_str(buf).map_err(serde::de::Error::custom)?;
Ok(ActivationWithOptionalGating {
gated: false,
activation,
})
}
}
}
#[derive(Debug, Clone, PartialEq, Deserialize)]
pub struct Config {
vocab_size: usize,
d_model: usize,
d_kv: usize,
d_ff: usize,
num_layers: usize,
num_decoder_layers: Option<usize>,
num_heads: usize,
relative_attention_num_buckets: usize,
#[serde(default = "default_relative_attention_max_distance")]
relative_attention_max_distance: usize,
dropout_rate: f64,
layer_norm_epsilon: f64,
initializer_factor: f64,
#[serde(default, deserialize_with = "deserialize_feed_forward_proj_activation")]
feed_forward_proj: ActivationWithOptionalGating,
#[serde(default = "default_tie_word_embeddings")]
tie_word_embeddings: bool,
#[serde(default = "default_is_decoder")]
is_decoder: bool,
is_encoder_decoder: bool,
#[serde(default = "default_use_cache")]
pub use_cache: bool,
pub pad_token_id: usize,
pub eos_token_id: usize,
pub decoder_start_token_id: Option<usize>,
}
impl Default for Config {
fn default() -> Self {
Self {
vocab_size: 32128,
d_model: 512,
d_kv: 64,
d_ff: 2048,
num_layers: 6,
num_decoder_layers: None,
num_heads: 8,
relative_attention_num_buckets: 32,
relative_attention_max_distance: 128,
dropout_rate: 0.1,
layer_norm_epsilon: 1e-6,
initializer_factor: 1.0,
feed_forward_proj: ActivationWithOptionalGating {
gated: false,
activation: Activation::Relu,
},
tie_word_embeddings: true,
is_decoder: false,
is_encoder_decoder: true,
use_cache: true,
pad_token_id: 0,
eos_token_id: 1,
decoder_start_token_id: Some(0),
}
}
}
impl Config {
// https://huggingface.co/facebook/musicgen-small/blob/495da4ad086b3416a27c6187f9239f9fd96f3962/config.json#L184
pub fn musicgen_small() -> Self {
Self {
d_ff: 3072,
d_kv: 64,
d_model: 768,
dropout_rate: 0.1,
eos_token_id: 1,
feed_forward_proj: ActivationWithOptionalGating {
gated: false,
activation: Activation::Relu,
},
tie_word_embeddings: true,
initializer_factor: 1.0,
is_decoder: false,
is_encoder_decoder: true,
layer_norm_epsilon: 1e-6,
num_decoder_layers: Some(12),
num_heads: 12,
num_layers: 12,
pad_token_id: 0,
decoder_start_token_id: Some(0),
relative_attention_max_distance: 128,
relative_attention_num_buckets: 32,
use_cache: true,
vocab_size: 32128,
}
}
}
#[derive(Debug, Clone)]
struct T5LayerNorm {
weight: Tensor,
variance_epsilon: f64,
span: tracing::Span,
}
impl T5LayerNorm {
fn load(h: usize, eps: f64, vb: VarBuilder) -> Result<Self> {
let weight = vb.get(h, "weight")?;
Ok(Self {
weight,
variance_epsilon: eps,
span: tracing::span!(tracing::Level::TRACE, "layer-norm"),
})
}
}
impl Module for T5LayerNorm {
fn forward(&self, xs: &Tensor) -> Result<Tensor> {
let _enter = self.span.enter();
let dtype = xs.dtype();
let xs_f32 = xs.to_dtype(DType::F32)?;
// variance = hidden_states.to(torch.float32).pow(2).mean(-1, keepdim=True)
let variance = xs_f32.sqr()?.mean_keepdim(D::Minus1)?;
let xs = xs.broadcast_div(&(variance + self.variance_epsilon)?.sqrt()?)?;
let xs = xs.to_dtype(dtype)?;
let xs = xs.broadcast_mul(&self.weight)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
struct T5DenseActDense {
wi: Linear,
wo: Linear,
act: Activation,
span: tracing::Span,
}
impl T5DenseActDense {
fn load(vb: VarBuilder, cfg: &Config) -> Result<Self> {
let wi = linear_no_bias(cfg.d_model, cfg.d_ff, vb.pp("wi"))?;
let wo = linear_no_bias(cfg.d_ff, cfg.d_model, vb.pp("wo"))?;
Ok(Self {
wi,
wo,
act: Activation::Relu,
span: tracing::span!(tracing::Level::TRACE, "dense-act-dense"),
})
}
}
impl Module for T5DenseActDense {
fn forward(&self, xs: &Tensor) -> Result<Tensor> {
let _enter = self.span.enter();
let xs = self.wi.forward(xs)?;
let xs = self.act.forward(&xs)?;
let xs = self.wo.forward(&xs)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
struct T5DenseGatedActDense {
wi_0: Linear,
wi_1: Linear,
wo: Linear,
act: Activation,
span: tracing::Span,
}
impl T5DenseGatedActDense {
fn load(vb: VarBuilder, cfg: &Config) -> Result<Self> {
let wi_0 = linear_no_bias(cfg.d_model, cfg.d_ff, vb.pp("wi_0"))?;
let wi_1 = linear_no_bias(cfg.d_model, cfg.d_ff, vb.pp("wi_1"))?;
let wo = linear_no_bias(cfg.d_ff, cfg.d_model, vb.pp("wo"))?;
Ok(Self {
wi_0,
wi_1,
wo,
act: cfg.feed_forward_proj.activation,
span: tracing::span!(tracing::Level::TRACE, "dense-gated-act-dense"),
})
}
}
impl Module for T5DenseGatedActDense {
fn forward(&self, xs: &Tensor) -> Result<Tensor> {
let _enter = self.span.enter();
let hidden_gelu = self.act.forward(&self.wi_0.forward(xs)?)?;
let hidden_linear = self.wi_1.forward(xs)?;
let xs = hidden_gelu.broadcast_mul(&hidden_linear)?;
let xs = self.wo.forward(&xs)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
struct T5LayerFF {
dense_act: Option<T5DenseActDense>,
gated_dense_act: Option<T5DenseGatedActDense>,
layer_norm: T5LayerNorm,
span: tracing::Span,
}
impl T5LayerFF {
fn load(vb: VarBuilder, cfg: &Config) -> Result<Self> {
let layer_norm =
T5LayerNorm::load(cfg.d_model, cfg.layer_norm_epsilon, vb.pp("layer_norm"))?;
let (dense_act, gated_dense_act) = if cfg.feed_forward_proj.gated {
(
None,
Some(T5DenseGatedActDense::load(vb.pp("DenseReluDense"), cfg)?),
)
} else {
(
Some(T5DenseActDense::load(vb.pp("DenseReluDense"), cfg)?),
None,
)
};
Ok(Self {
dense_act,
gated_dense_act,
layer_norm,
span: tracing::span!(tracing::Level::TRACE, "layer-ff"),
})
}
}
impl Module for T5LayerFF {
fn forward(&self, xs: &Tensor) -> Result<Tensor> {
let _enter = self.span.enter();
let ys = self.layer_norm.forward(xs)?;
let ys = match &self.dense_act {
Some(dense_act) => dense_act.forward(&ys)?,
None => self.gated_dense_act.as_ref().unwrap().forward(&ys)?,
};
let xs = (xs + ys)?;
Ok(xs)
}
}
#[derive(Debug, Clone)]
struct T5Attention {
q: Linear,
k: Linear,
v: Linear,
o: Linear,
n_heads: usize,
d_kv: usize,
relative_attention_bias: Option<Embedding>,
relative_attention_num_buckets: usize,
relative_attention_max_distance: usize,
inner_dim: usize,
use_cache: bool,
kv_cache: Option<(Tensor, Tensor)>,
span: tracing::Span,
span_cache: tracing::Span,
span_mm: tracing::Span,
span_sm: tracing::Span,
}
impl T5Attention {
fn load(
has_relative_attention_bias: bool,
decoder: bool,
vb: VarBuilder,
cfg: &Config,
) -> Result<Self> {
let inner_dim = cfg.num_heads * cfg.d_kv;
let q = linear_no_bias(cfg.d_model, inner_dim, vb.pp("q"))?;
let k = linear_no_bias(cfg.d_model, inner_dim, vb.pp("k"))?;
let v = linear_no_bias(cfg.d_model, inner_dim, vb.pp("v"))?;
let o = linear_no_bias(inner_dim, cfg.d_model, vb.pp("o"))?;
let relative_attention_bias = if has_relative_attention_bias {
let emb = Embedding::new(
cfg.relative_attention_num_buckets,
cfg.num_heads,
vb.pp("relative_attention_bias"),
)?;
Some(emb)
} else {
None
};
Ok(Self {
q,
k,
v,
o,
n_heads: cfg.num_heads,
d_kv: cfg.d_kv,
relative_attention_bias,
relative_attention_num_buckets: cfg.relative_attention_num_buckets,
relative_attention_max_distance: cfg.relative_attention_max_distance,
inner_dim,
use_cache: cfg.use_cache && decoder,
kv_cache: None,
span: tracing::span!(tracing::Level::TRACE, "attention"),
span_cache: tracing::span!(tracing::Level::TRACE, "attention-cache"),
span_mm: tracing::span!(tracing::Level::TRACE, "attention-mm"),
span_sm: tracing::span!(tracing::Level::TRACE, "attention-sm"),
})
}
fn forward(
&mut self,
xs: &Tensor,
position_bias: Option<&Tensor>,
key_value_states: Option<&Tensor>,
mask: Option<&Tensor>,
) -> Result<(Tensor, Option<Tensor>)> {
// Performs Self-attention (if key_value_states is None) or attention
// over source sentence (provided by key_value_states).
let _enter = self.span.enter();
let kv_input = match key_value_states {
None => xs,
Some(key_value_states) => key_value_states,
};
let (b_sz, q_len) = (xs.dim(0)?, xs.dim(1)?);
let kv_len = kv_input.dim(1)?;
let q = self.q.forward(xs)?;
let k = self.k.forward(kv_input)?;
let v = self.v.forward(kv_input)?;
let q = q
.reshape((b_sz, q_len, self.n_heads, self.d_kv))?
.transpose(1, 2)?
.contiguous()?;
let mut k = k
.reshape((b_sz, kv_len, self.n_heads, self.d_kv))?
.transpose(1, 2)?;
let mut v = v
.reshape((b_sz, kv_len, self.n_heads, self.d_kv))?
.transpose(1, 2)?;
if self.use_cache && key_value_states.is_none() {
let _enter = self.span_cache.enter();
if let Some((kv_cache_k, kv_cache_v)) = &self.kv_cache {
k = Tensor::cat(&[kv_cache_k, &k], 2)?;
v = Tensor::cat(&[kv_cache_v, &v], 2)?;
};
self.kv_cache = Some((k.clone(), v.clone()));
};
let k = k.contiguous()?;
let v = v.contiguous()?;
// TODO: Use flash_attn.
let scores = {
let _enter = self.span_mm.enter();
q.matmul(&k.t()?)?
};
let scores = match mask {
None => scores,
Some(mask) => masked_fill(
&scores,
&mask
.unsqueeze(0)?
.unsqueeze(0)?
.repeat((b_sz, self.n_heads))?,
f32::NEG_INFINITY,
)?,
};
let (scores, position_bias) = match position_bias {
Some(position_bias) => (
scores.broadcast_add(position_bias)?,
Some(position_bias.clone()),
),
None => match &self.relative_attention_bias {
None => (scores, None),
Some(relative_attention_bias) => {
// This only handles the bidirectional case.
let kv_len = k.dim(2)?;
let (q_start, q_end) = match self.use_cache {
true => ((kv_len - q_len) as u32, kv_len as u32),
false => (0_u32, kv_len as u32),
};
let num_buckets = self.relative_attention_num_buckets as u32 / 2;
let max_exact = num_buckets / 2;
let relative_position = (q_start..q_end)
.map(|i| {
(0..kv_len as u32)
.map(|j| {
if i < j {
if j - i < max_exact {
j - i + num_buckets
} else {
let b = f32::log(
(j - i) as f32 / max_exact as f32,
self.relative_attention_max_distance as f32
/ max_exact as f32,
) * (num_buckets - max_exact) as f32;
u32::min(
max_exact + num_buckets + b as u32,
self.relative_attention_num_buckets as u32 - 1,
)
}
} else if i - j < max_exact {
i - j
} else {
let b = f32::log(
(i - j) as f32 / max_exact as f32,
self.relative_attention_max_distance as f32
/ max_exact as f32,
) * (num_buckets - max_exact) as f32;
u32::min(max_exact + b as u32, num_buckets - 1)
}
})
.collect::<Vec<u32>>()
})
.collect::<Vec<Vec<_>>>();
let relative_buckets = Tensor::new(relative_position, q.device())?;
let position_bias = relative_attention_bias
.forward(&relative_buckets)?
.permute((2, 0, 1))?
.unsqueeze(0)?;
(scores.broadcast_add(&position_bias)?, Some(position_bias))
// TODO: position_bias_masked?
}
},
};
let attn_weights = {
let _enter = self.span_sm.enter();
candle_nn::ops::softmax_last_dim(&scores)?
};
let attn_output = attn_weights.matmul(&v)?;
let attn_output = attn_output
.transpose(1, 2)?
.reshape((b_sz, q_len, self.inner_dim))?;
let attn_output = self.o.forward(&attn_output)?;
Ok((attn_output, position_bias))
}
fn clear_kv_cache(&mut self) {
self.kv_cache = None
}
}
#[derive(Debug, Clone)]
struct T5LayerSelfAttention {
self_attention: T5Attention,
layer_norm: T5LayerNorm,
span: tracing::Span,
}
impl T5LayerSelfAttention {
fn load(h: bool, d: bool, vb: VarBuilder, cfg: &Config) -> Result<Self> {
let self_attention = T5Attention::load(h, d, vb.pp("SelfAttention"), cfg)?;
let layer_norm =
T5LayerNorm::load(cfg.d_model, cfg.layer_norm_epsilon, vb.pp("layer_norm"))?;
Ok(Self {
self_attention,
layer_norm,
span: tracing::span!(tracing::Level::TRACE, "self-attn"),
})
}
fn forward(
&mut self,
xs: &Tensor,
position_bias: Option<&Tensor>,
mask: Option<&Tensor>,
) -> Result<(Tensor, Option<Tensor>)> {
let _enter = self.span.enter();
let normed_xs = self.layer_norm.forward(xs)?;
let (ys, position_bias) =
self.self_attention
.forward(&normed_xs, position_bias, None, mask)?;
let ys = (xs + ys)?;
Ok((ys, position_bias))
}
fn clear_kv_cache(&mut self) {
self.self_attention.clear_kv_cache()
}
}
#[derive(Debug, Clone)]
struct T5LayerCrossAttention {
cross_attention: T5Attention,
layer_norm: T5LayerNorm,
span: tracing::Span,
}
impl T5LayerCrossAttention {
fn load(decoder: bool, vb: VarBuilder, cfg: &Config) -> Result<Self> {
let cross_attention = T5Attention::load(false, decoder, vb.pp("EncDecAttention"), cfg)?;
let layer_norm =
T5LayerNorm::load(cfg.d_model, cfg.layer_norm_epsilon, vb.pp("layer_norm"))?;
Ok(Self {
cross_attention,
layer_norm,
span: tracing::span!(tracing::Level::TRACE, "cross-attn"),
})
}
fn forward(
&mut self,
hidden_states: &Tensor,
position_bias: Option<&Tensor>,
key_value_states: &Tensor,
) -> Result<(Tensor, Option<Tensor>)> {
let _enter = self.span.enter();
let normed_hidden_states = self.layer_norm.forward(hidden_states)?;
let (ys, position_bias) = self.cross_attention.forward(
&normed_hidden_states,
position_bias,
Some(key_value_states),
None,
)?;
let ys = (hidden_states + ys)?;
Ok((ys, position_bias))
}
fn clear_kv_cache(&mut self) {
self.cross_attention.clear_kv_cache()
}
}
#[derive(Debug, Clone)]
struct T5Block {
self_attn: T5LayerSelfAttention,
cross_attn: Option<T5LayerCrossAttention>,
ff: T5LayerFF,
span: tracing::Span,
}
impl T5Block {
fn load(
has_relative_attention_bias: bool,
decoder: bool,
vb: VarBuilder,
cfg: &Config,
) -> Result<Self> {
let vb = vb.pp("layer");
let self_attn =
T5LayerSelfAttention::load(has_relative_attention_bias, decoder, vb.pp("0"), cfg)?;
let cross_attn = if cfg.is_decoder {
Some(T5LayerCrossAttention::load(decoder, vb.pp("1"), cfg)?)
} else {
None
};
let ff_i = if cross_attn.is_some() { 2 } else { 1 };
let ff = T5LayerFF::load(vb.pp(&ff_i.to_string()), cfg)?;
Ok(Self {
self_attn,
cross_attn,
ff,
span: tracing::span!(tracing::Level::TRACE, "block"),
})
}
fn forward(
&mut self,
xs: &Tensor,
position_bias: Option<&Tensor>,
encoder_hidden_states: Option<&Tensor>,
) -> Result<(Tensor, Option<Tensor>)> {
let _enter = self.span.enter();
// TODO: Cache masks
let mask = match self.cross_attn.is_some() {
true => {
let mask_len = xs.dim(1)?;
// If the input seq length is 1, no need for a mask, this is also helpful to avoid shape
// issues when using the KV cache in the decoder.
if mask_len <= 1 {
None
} else {
Some(get_mask(mask_len, xs.device())?)
}
}
false => None,
};
let (mut xs, position_bias) = self.self_attn.forward(xs, position_bias, mask.as_ref())?;
// TODO: clamp for f16?
if let Some(cross_attn) = &mut self.cross_attn {
(xs, _) = cross_attn.forward(&xs, None, encoder_hidden_states.unwrap())?;
// TODO: clamp for f16?
}
let xs = self.ff.forward(&xs)?;
// TODO: clamp for f16?
Ok((xs, position_bias))
}
fn clear_kv_cache(&mut self) {
self.self_attn.clear_kv_cache();
self.cross_attn.iter_mut().for_each(|c| c.clear_kv_cache());
}
}
#[derive(Debug, Clone)]
struct T5Stack {
block: Vec<T5Block>,
shared: Arc<Embedding>,
final_layer_norm: T5LayerNorm,
span: tracing::Span,
}
impl T5Stack {
fn load(decoder: bool, vb: VarBuilder, shared: &Arc<Embedding>, cfg: &Config) -> Result<Self> {
let block = (0..cfg.num_layers)
.map(|i| T5Block::load(i == 0, decoder, vb.pp(&format!("block.{i}")), cfg))
.collect::<Result<Vec<_>>>()?;
let final_layer_norm = T5LayerNorm::load(
cfg.d_model,
cfg.layer_norm_epsilon,
vb.pp("final_layer_norm"),
)?;
Ok(Self {
block,
shared: shared.clone(),
final_layer_norm,
span: tracing::span!(tracing::Level::TRACE, "stack"),
})
}
fn forward(
&mut self,
input_ids: &Tensor,
encoder_hidden_states: Option<&Tensor>,
) -> Result<Tensor> {
let _enter = self.span.enter();
let input_embeds = self.shared.as_ref().forward(input_ids)?;
let mut hidden_states = input_embeds;
let mut position_bias = None;
for block in self.block.iter_mut() {
(hidden_states, position_bias) = block.forward(
&hidden_states,
position_bias.as_ref(),
encoder_hidden_states,
)?
}
self.final_layer_norm.forward(&hidden_states)
}
fn clear_kv_cache(&mut self) {
self.block.iter_mut().for_each(|b| b.clear_kv_cache())
}
}
#[derive(Debug, Clone)]
pub struct T5EncoderModel {
encoder: T5Stack,
device: Device,
span: tracing::Span,
}
impl T5EncoderModel {
pub fn load(vb: VarBuilder, cfg: &Config) -> Result<Self> {
let shared_vb = if vb.contains_tensor("shared.weight") {
vb.pp("shared")
} else {
vb.pp("decoder").pp("embed_tokens")
};
let shared = Embedding::new(cfg.vocab_size, cfg.d_model, shared_vb)?;
let shared = Arc::new(shared);
let encoder = T5Stack::load(false, vb.pp("encoder"), &shared, cfg)?;
Ok(Self {
encoder,
device: vb.device().clone(),
span: tracing::span!(tracing::Level::TRACE, "encoder"),
})
}
pub fn forward(&mut self, input_ids: &Tensor) -> Result<Tensor> {
let _enter = self.span.enter();
self.encoder.forward(input_ids, None)
}
pub fn device(&self) -> &Device {
&self.device
}
pub fn clear_kv_cache(&mut self) {
self.encoder.clear_kv_cache()
}
}
#[derive(Debug, Clone)]
pub struct T5ForConditionalGeneration {
encoder: T5Stack,
decoder: T5Stack,
d_model: usize,
tie_word_embeddings: bool,
lm_head: Option<Linear>,
shared: Arc<Embedding>,
device: Device,
span_decode: tracing::Span,
span_decode_head: tracing::Span,
}
impl T5ForConditionalGeneration {
pub fn load(vb: VarBuilder, cfg: &Config) -> Result<Self> {
assert!(cfg.is_encoder_decoder);
let d_model = cfg.d_model;
let shared_vb = if vb.contains_tensor("shared.weight") {
vb.pp("shared")
} else {
vb.pp("decoder").pp("embed_tokens")
};
let shared = Embedding::new(cfg.vocab_size, cfg.d_model, shared_vb)?;
let shared = Arc::new(shared);
let mut encoder_cfg = cfg.clone();
encoder_cfg.is_decoder = false;
encoder_cfg.use_cache = false;
encoder_cfg.is_encoder_decoder = false;
let encoder = T5Stack::load(false, vb.pp("encoder"), &shared, &encoder_cfg)?;
let mut decoder_cfg = cfg.clone();
decoder_cfg.is_decoder = true;
decoder_cfg.is_encoder_decoder = false;
decoder_cfg.num_layers = cfg.num_decoder_layers.unwrap_or(cfg.num_layers);
let decoder = T5Stack::load(true, vb.pp("decoder"), &shared, &decoder_cfg)?;
let tie_word_embeddings = cfg.tie_word_embeddings;
let lm_head = if tie_word_embeddings {
None
} else {
Some(linear_no_bias(
cfg.d_model,
cfg.vocab_size,
vb.pp("lm_head"),
)?)
};
Ok(Self {
encoder,
decoder,
d_model,
tie_word_embeddings,
lm_head,
shared,
device: vb.device().clone(),
span_decode: tracing::span!(tracing::Level::TRACE, "decode"),
span_decode_head: tracing::span!(tracing::Level::TRACE, "decode-head"),
})
}
pub fn encode(&mut self, input_ids: &Tensor) -> Result<Tensor> {
self.encoder.forward(input_ids, None)
}
pub fn decode(
&mut self,
decoder_input_ids: &Tensor,
encoder_output: &Tensor,
) -> Result<Tensor> {
let _enter = self.span_decode.enter();
let decoder_output = self
.decoder
.forward(decoder_input_ids, Some(encoder_output))?;
let scaling_factor = if self.tie_word_embeddings {
// Rescale output before projecting on vocab
// See https://github.com/tensorflow/mesh/blob/fa19d69eafc9a482aff0b59ddd96b025c0cb207d/mesh_tensorflow/transformer/transformer.py#L586
(self.d_model as f64).sqrt()
} else {
1.0
};
let sequence_output = ((decoder_output
.narrow(1, decoder_output.dim(1)? - 1, 1)?
.squeeze(1)?)
* scaling_factor)?;
let output = {
let _enter = self.span_decode_head.enter();
match self.lm_head {
None => sequence_output.matmul(&self.shared.embeddings().t()?)?,
Some(ref lm_head) => lm_head.forward(&sequence_output)?,
}
};
Ok(output)
}
pub fn forward(&mut self, input_ids: &Tensor, decoder_input_ids: &Tensor) -> Result<Tensor> {
let encoder_output = self.encode(input_ids)?;
self.decode(decoder_input_ids, &encoder_output)
}
pub fn device(&self) -> &Device {
&self.device
}
pub fn clear_kv_cache(&mut self) {
self.encoder.clear_kv_cache();
self.decoder.clear_kv_cache();
}
}
| candle/candle-transformers/src/models/t5.rs/0 | {
"file_path": "candle/candle-transformers/src/models/t5.rs",
"repo_id": "candle",
"token_count": 15016
} | 36 |
use candle::{Device, Tensor};
use candle_transformers::generation::LogitsProcessor;
use candle_wasm_example_llama2::worker::{Model as M, ModelData};
use wasm_bindgen::prelude::*;
#[wasm_bindgen]
pub struct Model {
inner: M,
logits_processor: LogitsProcessor,
tokens: Vec<u32>,
repeat_penalty: f32,
}
impl Model {
fn process(&mut self, tokens: &[u32]) -> candle::Result<String> {
const REPEAT_LAST_N: usize = 64;
let dev = Device::Cpu;
let input = Tensor::new(tokens, &dev)?.unsqueeze(0)?;
let logits = self.inner.llama.forward(&input, tokens.len())?;
let logits = logits.squeeze(0)?;
let logits = if self.repeat_penalty == 1. || tokens.is_empty() {
logits
} else {
let start_at = self.tokens.len().saturating_sub(REPEAT_LAST_N);
candle_transformers::utils::apply_repeat_penalty(
&logits,
self.repeat_penalty,
&self.tokens[start_at..],
)?
};
let next_token = self.logits_processor.sample(&logits)?;
self.tokens.push(next_token);
let text = match self.inner.tokenizer.id_to_token(next_token) {
Some(text) => text.replace('▁', " ").replace("<0x0A>", "\n"),
None => "".to_string(),
};
Ok(text)
}
}
#[wasm_bindgen]
impl Model {
#[wasm_bindgen(constructor)]
pub fn new(weights: Vec<u8>, tokenizer: Vec<u8>) -> Result<Model, JsError> {
let model = M::load(ModelData {
tokenizer,
model: weights,
});
let logits_processor = LogitsProcessor::new(299792458, None, None);
match model {
Ok(inner) => Ok(Self {
inner,
logits_processor,
tokens: vec![],
repeat_penalty: 1.,
}),
Err(e) => Err(JsError::new(&e.to_string())),
}
}
#[wasm_bindgen]
pub fn get_seq_len(&mut self) -> usize {
self.inner.config.seq_len
}
#[wasm_bindgen]
pub fn init_with_prompt(
&mut self,
prompt: String,
temp: f64,
top_p: f64,
repeat_penalty: f32,
seed: u64,
) -> Result<String, JsError> {
// First reset the cache.
{
let mut cache = self.inner.cache.kvs.lock().unwrap();
for elem in cache.iter_mut() {
*elem = None
}
}
let temp = if temp <= 0. { None } else { Some(temp) };
let top_p = if top_p <= 0. || top_p >= 1. {
None
} else {
Some(top_p)
};
self.logits_processor = LogitsProcessor::new(seed, temp, top_p);
self.repeat_penalty = repeat_penalty;
self.tokens.clear();
let tokens = self
.inner
.tokenizer
.encode(prompt, true)
.map_err(|m| JsError::new(&m.to_string()))?
.get_ids()
.to_vec();
let text = self
.process(&tokens)
.map_err(|m| JsError::new(&m.to_string()))?;
Ok(text)
}
#[wasm_bindgen]
pub fn next_token(&mut self) -> Result<String, JsError> {
let last_token = *self.tokens.last().unwrap();
let text = self
.process(&[last_token])
.map_err(|m| JsError::new(&m.to_string()))?;
Ok(text)
}
}
fn main() {}
| candle/candle-wasm-examples/llama2-c/src/bin/m.rs/0 | {
"file_path": "candle/candle-wasm-examples/llama2-c/src/bin/m.rs",
"repo_id": "candle",
"token_count": 1807
} | 37 |
//load the candle SAM Model wasm module
import init, { Model } from "./build/m.js";
async function fetchArrayBuffer(url, cacheModel = true) {
if (!cacheModel)
return new Uint8Array(await (await fetch(url)).arrayBuffer());
const cacheName = "sam-candle-cache";
const cache = await caches.open(cacheName);
const cachedResponse = await cache.match(url);
if (cachedResponse) {
const data = await cachedResponse.arrayBuffer();
return new Uint8Array(data);
}
const res = await fetch(url, { cache: "force-cache" });
cache.put(url, res.clone());
return new Uint8Array(await res.arrayBuffer());
}
class SAMModel {
static instance = {};
// keep current image embeddings state
static imageArrayHash = {};
// Add a new property to hold the current modelID
static currentModelID = null;
static async getInstance(modelURL, modelID) {
if (!this.instance[modelID]) {
await init();
self.postMessage({
status: "loading",
message: `Loading Model ${modelID}`,
});
const weightsArrayU8 = await fetchArrayBuffer(modelURL);
this.instance[modelID] = new Model(
weightsArrayU8,
/tiny|mobile/.test(modelID)
);
} else {
self.postMessage({ status: "loading", message: "Model Already Loaded" });
}
// Set the current modelID to the modelID that was passed in
this.currentModelID = modelID;
return this.instance[modelID];
}
// Remove the modelID parameter from setImageEmbeddings
static setImageEmbeddings(imageArrayU8) {
// check if image embeddings are already set for this image and model
const imageArrayHash = this.getSimpleHash(imageArrayU8);
if (
this.imageArrayHash[this.currentModelID] === imageArrayHash &&
this.instance[this.currentModelID]
) {
self.postMessage({
status: "embedding",
message: "Embeddings Already Set",
});
return;
}
this.imageArrayHash[this.currentModelID] = imageArrayHash;
this.instance[this.currentModelID].set_image_embeddings(imageArrayU8);
self.postMessage({ status: "embedding", message: "Embeddings Set" });
}
static getSimpleHash(imageArrayU8) {
// get simple hash of imageArrayU8
let imageArrayHash = 0;
for (let i = 0; i < imageArrayU8.length; i += 100) {
imageArrayHash ^= imageArrayU8[i];
}
return imageArrayHash.toString(16);
}
}
async function createImageCanvas(
{ mask_shape, mask_data }, // mask
{ original_width, original_height, width, height } // original image
) {
const [_, __, shape_width, shape_height] = mask_shape;
const maskCanvas = new OffscreenCanvas(shape_width, shape_height); // canvas for mask
const maskCtx = maskCanvas.getContext("2d");
const canvas = new OffscreenCanvas(original_width, original_height); // canvas for creating mask with original image size
const ctx = canvas.getContext("2d");
const imageData = maskCtx.createImageData(
maskCanvas.width,
maskCanvas.height
);
const data = imageData.data;
for (let p = 0; p < data.length; p += 4) {
data[p] = 0;
data[p + 1] = 0;
data[p + 2] = 0;
data[p + 3] = mask_data[p / 4] * 255;
}
maskCtx.putImageData(imageData, 0, 0);
let sx, sy;
if (original_height < original_width) {
sy = original_height / original_width;
sx = 1;
} else {
sy = 1;
sx = original_width / original_height;
}
ctx.drawImage(
maskCanvas,
0,
0,
maskCanvas.width * sx,
maskCanvas.height * sy,
0,
0,
original_width,
original_height
);
const blob = await canvas.convertToBlob();
return URL.createObjectURL(blob);
}
self.addEventListener("message", async (event) => {
const { modelURL, modelID, imageURL, points } = event.data;
try {
self.postMessage({ status: "loading", message: "Starting SAM" });
const sam = await SAMModel.getInstance(modelURL, modelID);
self.postMessage({ status: "loading", message: "Loading Image" });
const imageArrayU8 = await fetchArrayBuffer(imageURL, false);
self.postMessage({ status: "embedding", message: "Creating Embeddings" });
SAMModel.setImageEmbeddings(imageArrayU8);
if (!points) {
// no points only do the embeddings
self.postMessage({
status: "complete-embedding",
message: "Embeddings Complete",
});
return;
}
self.postMessage({ status: "segmenting", message: "Segmenting" });
const { mask, image } = sam.mask_for_point({ points });
const maskDataURL = await createImageCanvas(mask, image);
// Send the segment back to the main thread as JSON
self.postMessage({
status: "complete",
message: "Segmentation Complete",
output: { maskURL: maskDataURL },
});
} catch (e) {
self.postMessage({ error: e });
}
});
| candle/candle-wasm-examples/segment-anything/samWorker.js/0 | {
"file_path": "candle/candle-wasm-examples/segment-anything/samWorker.js",
"repo_id": "candle",
"token_count": 1747
} | 38 |
<!DOCTYPE html>
<html lang="en">
<head>
<meta charset="utf-8" />
<title>Welcome to Candle!</title>
<link data-trunk rel="copy-file" href="mel_filters.safetensors" />
<!-- samples -->
<link data-trunk rel="copy-dir" href="audios" />
<!-- tiny.en -->
<link data-trunk rel="copy-dir" href="whisper-tiny.en" />
<!-- tiny -->
<link data-trunk rel="copy-dir" href="whisper-tiny" />
<!-- quantized -->
<link data-trunk rel="copy-dir" href="quantized" />
<link
data-trunk
rel="rust"
href="Cargo.toml"
data-bin="app"
data-type="main" />
<link
data-trunk
rel="rust"
href="Cargo.toml"
data-bin="worker"
data-type="worker" />
<link
rel="stylesheet"
href="https://fonts.googleapis.com/css?family=Roboto:300,300italic,700,700italic" />
<link
rel="stylesheet"
href="https://cdnjs.cloudflare.com/ajax/libs/normalize/8.0.1/normalize.css" />
<link
rel="stylesheet"
href="https://cdnjs.cloudflare.com/ajax/libs/milligram/1.4.1/milligram.css" />
</head>
<body></body>
</html>
| candle/candle-wasm-examples/whisper/index.html/0 | {
"file_path": "candle/candle-wasm-examples/whisper/index.html",
"repo_id": "candle",
"token_count": 523
} | 39 |
<html>
<head>
<meta content="text/html;charset=utf-8" http-equiv="Content-Type" />
<title>Candle YOLOv8 Rust/WASM</title>
</head>
<body></body>
</html>
<!DOCTYPE html>
<html>
<head>
<meta charset="UTF-8" />
<meta name="viewport" content="width=device-width, initial-scale=1.0" />
<style>
@import url("https://fonts.googleapis.com/css2?family=Source+Code+Pro:wght@200;300;400&family=Source+Sans+3:wght@100;200;300;400;500;600;700;800;900&display=swap");
html,
body {
font-family: "Source Sans 3", sans-serif;
}
code,
output,
select,
pre {
font-family: "Source Code Pro", monospace;
}
</style>
<script src="https://cdn.tailwindcss.com"></script>
<script
src="https://cdn.jsdelivr.net/gh/huggingface/hub-js-utils/share-canvas.js"
type="module"
></script>
<script type="module">
const MODEL_BASEURL =
"https://huggingface.co/lmz/candle-yolo-v8/resolve/main/";
const MODELS = {
yolov8n: {
model_size: "n",
url: "yolov8n.safetensors",
},
yolov8s: {
model_size: "s",
url: "yolov8s.safetensors",
},
yolov8m: {
model_size: "m",
url: "yolov8m.safetensors",
},
yolov8l: {
model_size: "l",
url: "yolov8l.safetensors",
},
yolov8x: {
model_size: "x",
url: "yolov8x.safetensors",
},
yolov8n_pose: {
model_size: "n",
url: "yolov8n-pose.safetensors",
},
yolov8s_pose: {
model_size: "s",
url: "yolov8s-pose.safetensors",
},
yolov8m_pose: {
model_size: "m",
url: "yolov8m-pose.safetensors",
},
yolov8l_pose: {
model_size: "l",
url: "yolov8l-pose.safetensors",
},
yolov8x_pose: {
model_size: "x",
url: "yolov8x-pose.safetensors",
},
};
const COCO_PERSON_SKELETON = [
[4, 0], // head
[3, 0],
[16, 14], // left lower leg
[14, 12], // left upper leg
[6, 12], // left torso
[6, 5], // top torso
[6, 8], // upper arm
[8, 10], // lower arm
[1, 2], // head
[1, 3], // right head
[2, 4], // left head
[3, 5], // right neck
[4, 6], // left neck
[5, 7], // right upper arm
[7, 9], // right lower arm
[5, 11], // right torso
[11, 12], // bottom torso
[11, 13], // right upper leg
[13, 15], // right lower leg
];
// init web worker
const yoloWorker = new Worker("./yoloWorker.js", { type: "module" });
let hasImage = false;
//add event listener to image examples
document.querySelector("#image-select").addEventListener("click", (e) => {
const target = e.target;
if (target.nodeName === "IMG") {
const href = target.src;
drawImageCanvas(href);
}
});
//add event listener to file input
document.querySelector("#file-upload").addEventListener("change", (e) => {
const target = e.target;
if (target.files.length > 0) {
const href = URL.createObjectURL(target.files[0]);
drawImageCanvas(href);
}
});
// add event listener to drop-area
const dropArea = document.querySelector("#drop-area");
dropArea.addEventListener("dragenter", (e) => {
e.preventDefault();
dropArea.classList.add("border-blue-700");
});
dropArea.addEventListener("dragleave", (e) => {
e.preventDefault();
dropArea.classList.remove("border-blue-700");
});
dropArea.addEventListener("dragover", (e) => {
e.preventDefault();
});
dropArea.addEventListener("drop", (e) => {
e.preventDefault();
dropArea.classList.remove("border-blue-700");
const url = e.dataTransfer.getData("text/uri-list");
const files = e.dataTransfer.files;
if (files.length > 0) {
const href = URL.createObjectURL(files[0]);
drawImageCanvas(href);
} else if (url) {
drawImageCanvas(url);
}
});
document.querySelector("#clear-btn").addEventListener("click", () => {
drawImageCanvas();
});
function drawImageCanvas(imgURL) {
const canvas = document.querySelector("#canvas");
const canvasResult = document.querySelector("#canvas-result");
canvasResult
.getContext("2d")
.clearRect(0, 0, canvas.width, canvas.height);
const ctx = canvas.getContext("2d");
ctx.clearRect(0, 0, canvas.width, canvas.height);
document.querySelector("#share-btn").classList.add("invisible");
document.querySelector("#clear-btn").classList.add("invisible");
document.querySelector("#detect").disabled = true;
hasImage = false;
canvas.parentElement.style.height = "auto";
if (imgURL && imgURL !== "") {
const img = new Image();
img.crossOrigin = "anonymous";
img.onload = () => {
canvas.width = img.width;
canvas.height = img.height;
ctx.drawImage(img, 0, 0);
canvas.parentElement.style.height = canvas.offsetHeight + "px";
hasImage = true;
document.querySelector("#detect").disabled = false;
document.querySelector("#clear-btn").classList.remove("invisible");
};
img.src = imgURL;
}
}
async function classifyImage(
imageURL, // URL of image to classify
modelID, // ID of model to use
modelURL, // URL to model file
modelSize, // size of model
confidence, // confidence threshold
iou_threshold, // IoU threshold
updateStatus // function receives status updates
) {
return new Promise((resolve, reject) => {
yoloWorker.postMessage({
imageURL,
modelID,
modelURL,
modelSize,
confidence,
iou_threshold,
});
function handleMessage(event) {
console.log("message", event.data);
if ("status" in event.data) {
updateStatus(event.data.status);
}
if ("error" in event.data) {
yoloWorker.removeEventListener("message", handleMessage);
reject(new Error(event.data.error));
}
if (event.data.status === "complete") {
yoloWorker.removeEventListener("message", handleMessage);
resolve(event.data);
}
}
yoloWorker.addEventListener("message", handleMessage);
});
}
// add event listener to detect button
document.querySelector("#detect").addEventListener("click", async () => {
if (!hasImage) {
return;
}
const modelID = document.querySelector("#model").value;
const modelURL = MODEL_BASEURL + MODELS[modelID].url;
const modelSize = MODELS[modelID].model_size;
const confidence = parseFloat(
document.querySelector("#confidence").value
);
const iou_threshold = parseFloat(
document.querySelector("#iou_threshold").value
);
const canvasInput = document.querySelector("#canvas");
const canvas = document.querySelector("#canvas-result");
canvas.width = canvasInput.width;
canvas.height = canvasInput.height;
const scale = canvas.width / canvas.offsetWidth;
const ctx = canvas.getContext("2d");
ctx.drawImage(canvasInput, 0, 0);
const imageURL = canvas.toDataURL();
const results = await await classifyImage(
imageURL,
modelID,
modelURL,
modelSize,
confidence,
iou_threshold,
updateStatus
);
const { output } = results;
ctx.lineWidth = 1 + 2 * scale;
ctx.strokeStyle = "#3c8566";
ctx.fillStyle = "#0dff9a";
const fontSize = 14 * scale;
ctx.font = `${fontSize}px sans-serif`;
for (const detection of output) {
// check keypoint for pose model data
let xmin, xmax, ymin, ymax, label, confidence, keypoints;
if ("keypoints" in detection) {
xmin = detection.xmin;
xmax = detection.xmax;
ymin = detection.ymin;
ymax = detection.ymax;
confidence = detection.confidence;
keypoints = detection.keypoints;
} else {
const [_label, bbox] = detection;
label = _label;
xmin = bbox.xmin;
xmax = bbox.xmax;
ymin = bbox.ymin;
ymax = bbox.ymax;
confidence = bbox.confidence;
}
const [x, y, w, h] = [xmin, ymin, xmax - xmin, ymax - ymin];
const text = `${label ? label + " " : ""}${confidence.toFixed(2)}`;
const width = ctx.measureText(text).width;
ctx.fillStyle = "#3c8566";
ctx.fillRect(x - 2, y - fontSize, width + 4, fontSize);
ctx.fillStyle = "#e3fff3";
ctx.strokeRect(x, y, w, h);
ctx.fillText(text, x, y - 2);
if (keypoints) {
ctx.save();
ctx.fillStyle = "magenta";
ctx.strokeStyle = "yellow";
for (const keypoint of keypoints) {
const { x, y } = keypoint;
ctx.beginPath();
ctx.arc(x, y, 3, 0, 2 * Math.PI);
ctx.fill();
}
ctx.beginPath();
for (const [xid, yid] of COCO_PERSON_SKELETON) {
//draw line between skeleton keypoitns
if (keypoints[xid] && keypoints[yid]) {
ctx.moveTo(keypoints[xid].x, keypoints[xid].y);
ctx.lineTo(keypoints[yid].x, keypoints[yid].y);
}
}
ctx.stroke();
ctx.restore();
}
}
});
function updateStatus(statusMessage) {
const button = document.querySelector("#detect");
if (statusMessage === "detecting") {
button.disabled = true;
button.classList.add("bg-blue-700");
button.classList.remove("bg-blue-950");
button.textContent = "Predicting...";
} else if (statusMessage === "complete") {
button.disabled = false;
button.classList.add("bg-blue-950");
button.classList.remove("bg-blue-700");
button.textContent = "Predict";
document.querySelector("#share-btn").classList.remove("invisible");
}
}
document.querySelector("#share-btn").addEventListener("click", () => {
shareToCommunity(
"lmz/candle-yolo",
"Candle + YOLOv8",
"YOLOv8 with [Candle](https://github.com/huggingface/candle)",
"canvas-result",
"share-btn"
);
});
</script>
</head>
<body class="container max-w-4xl mx-auto p-4">
<main class="grid grid-cols-1 gap-8 relative">
<span class="absolute text-5xl -ml-[1em]"> 🕯️ </span>
<div>
<h1 class="text-5xl font-bold">Candle YOLOv8</h1>
<h2 class="text-2xl font-bold">Rust/WASM Demo</h2>
<p class="max-w-lg">
This demo showcases object detection and pose estimation models in
your browser using Rust/WASM. It utilizes
<a
href="https://huggingface.co/lmz/candle-yolo-v8"
target="_blank"
class="underline hover:text-blue-500 hover:no-underline"
>
safetensor's YOLOv8 models
</a>
and a WASM runtime built with
<a
href="https://github.com/huggingface/candle/"
target="_blank"
class="underline hover:text-blue-500 hover:no-underline"
>Candle </a
>.
</p>
<p>
To run pose estimation, select a yolo pose model from the dropdown
</p>
</div>
<div>
<label for="model" class="font-medium">Models Options: </label>
<select
id="model"
class="border-2 border-gray-500 rounded-md font-light"
>
<option value="yolov8n" selected>yolov8n (6.37 MB)</option>
<option value="yolov8s">yolov8s (22.4 MB)</option>
<option value="yolov8m">yolov8m (51.9 MB)</option>
<option value="yolov8l">yolov8l (87.5 MB)</option>
<option value="yolov8x">yolov8x (137 MB)</option>
<!-- Pose models -->
<option value="yolov8n_pose">yolov8n_pose (6.65 MB)</option>
<option value="yolov8s_pose">yolov8s_pose (23.3 MB)</option>
<option value="yolov8m_pose">yolov8m_pose (53 MB)</option>
<option value="yolov8l_pose">yolov8l_pose (89.1 MB)</option>
<option value="yolov8x_pose">yolov8x_pose (139 MB)</option>
</select>
</div>
<div>
<button
id="detect"
disabled
class="bg-gray-700 hover:bg-gray-800 text-white font-normal py-2 px-4 rounded disabled:bg-gray-300 disabled:cursor-not-allowed"
>
Predict
</button>
</div>
<!-- drag and drop area -->
<div class="relative max-w-lg">
<div class="py-1">
<button
id="clear-btn"
class="text-xs bg-white rounded-md disabled:opacity-50 flex gap-1 items-center ml-auto invisible"
>
<svg
class=""
xmlns="http://www.w3.org/2000/svg"
viewBox="0 0 13 12"
height="1em"
>
<path
d="M1.6.7 12 11.1M12 .7 1.6 11.1"
stroke="#2E3036"
stroke-width="2"
/>
</svg>
Clear image
</button>
</div>
<div
id="drop-area"
class="flex flex-col items-center justify-center border-2 border-gray-300 border-dashed rounded-xl relative aspect-video w-full overflow-hidden"
>
<div
class="flex flex-col items-center justify-center space-y-1 text-center"
>
<svg
width="25"
height="25"
viewBox="0 0 25 25"
fill="none"
xmlns="http://www.w3.org/2000/svg"
>
<path
d="M3.5 24.3a3 3 0 0 1-1.9-.8c-.5-.5-.8-1.2-.8-1.9V2.9c0-.7.3-1.3.8-1.9.6-.5 1.2-.7 2-.7h18.6c.7 0 1.3.2 1.9.7.5.6.7 1.2.7 2v18.6c0 .7-.2 1.4-.7 1.9a3 3 0 0 1-2 .8H3.6Zm0-2.7h18.7V2.9H3.5v18.7Zm2.7-2.7h13.3c.3 0 .5 0 .6-.3v-.7l-3.7-5a.6.6 0 0 0-.6-.2c-.2 0-.4 0-.5.3l-3.5 4.6-2.4-3.3a.6.6 0 0 0-.6-.3c-.2 0-.4.1-.5.3l-2.7 3.6c-.1.2-.2.4 0 .7.1.2.3.3.6.3Z"
fill="#000"
/>
</svg>
<div class="flex text-sm text-gray-600">
<label
for="file-upload"
class="relative cursor-pointer bg-white rounded-md font-medium text-blue-950 hover:text-blue-700"
>
<span>Drag and drop your image here</span>
<span class="block text-xs">or</span>
<span class="block text-xs">Click to upload</span>
</label>
</div>
<input
id="file-upload"
name="file-upload"
type="file"
class="sr-only"
/>
</div>
<canvas
id="canvas"
class="absolute pointer-events-none w-full"
></canvas>
<canvas
id="canvas-result"
class="absolute pointer-events-none w-full"
></canvas>
</div>
<div class="text-right py-2">
<button
id="share-btn"
class="bg-white rounded-md hover:outline outline-orange-200 disabled:opacity-50 invisible"
>
<img
src="https://huggingface.co/datasets/huggingface/badges/raw/main/share-to-community-sm.svg"
/>
</button>
</div>
</div>
<div>
<div
class="flex gap-3 items-center overflow-x-scroll"
id="image-select"
>
<h3 class="font-medium">Examples:</h3>
<img
src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/candle/examples/sf.jpg"
class="cursor-pointer w-24 h-24 object-cover"
/>
<img
src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/candle/examples/bike.jpeg"
class="cursor-pointer w-24 h-24 object-cover"
/>
<img
src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/candle/examples/000000000077.jpg"
class="cursor-pointer w-24 h-24 object-cover"
/>
</div>
</div>
<div>
<div class="grid grid-cols-3 max-w-md items-center gap-3">
<label class="text-sm font-medium" for="confidence"
>Confidence Threshold</label
>
<input
type="range"
id="confidence"
name="confidence"
min="0"
max="1"
step="0.01"
value="0.25"
oninput="this.nextElementSibling.value = Number(this.value).toFixed(2)"
/>
<output
class="text-xs font-light px-1 py-1 border border-gray-700 rounded-md w-min"
>0.25</output
>
<label class="text-sm font-medium" for="iou_threshold"
>IoU Threshold</label
>
<input
type="range"
id="iou_threshold"
name="iou_threshold"
min="0"
max="1"
step="0.01"
value="0.45"
oninput="this.nextElementSibling.value = Number(this.value).toFixed(2)"
/>
<output
class="font-extralight text-xs px-1 py-1 border border-gray-700 rounded-md w-min"
>0.45</output
>
</div>
</div>
</main>
</body>
</html>
| candle/candle-wasm-examples/yolo/lib-example.html/0 | {
"file_path": "candle/candle-wasm-examples/yolo/lib-example.html",
"repo_id": "candle",
"token_count": 9649
} | 40 |
Dockerfile
.vscode/
.idea
.gitignore
LICENSE
README.md
node_modules/
.svelte-kit/
.env*
!.env
!.env.local | chat-ui/.dockerignore/0 | {
"file_path": "chat-ui/.dockerignore",
"repo_id": "chat-ui",
"token_count": 51
} | 41 |
<script lang="ts">
import CarbonContinue from "~icons/carbon/continue";
export let classNames = "";
</script>
<button
type="button"
on:click
class="btn flex h-8 rounded-lg border bg-white px-3 py-1 text-gray-500 shadow-sm transition-all hover:bg-gray-100 dark:border-gray-600 dark:bg-gray-700 dark:text-gray-300 dark:hover:bg-gray-600 {classNames}"
>
<CarbonContinue class="mr-2 text-xs " /> Continue
</button>
| chat-ui/src/lib/components/ContinueBtn.svelte/0 | {
"file_path": "chat-ui/src/lib/components/ContinueBtn.svelte",
"repo_id": "chat-ui",
"token_count": 149
} | 42 |
<script lang="ts">
import CarbonStopFilledAlt from "~icons/carbon/stop-filled-alt";
export let classNames = "";
</script>
<button
type="button"
on:click
class="btn flex h-8 rounded-lg border bg-white px-3 py-1 shadow-sm transition-all hover:bg-gray-100 dark:border-gray-600 dark:bg-gray-700 dark:hover:bg-gray-600 {classNames}"
>
<CarbonStopFilledAlt class="-ml-1 mr-1 h-[1.25rem] w-[1.1875rem] text-gray-300" /> Stop generating
</button>
| chat-ui/src/lib/components/StopGeneratingBtn.svelte/0 | {
"file_path": "chat-ui/src/lib/components/StopGeneratingBtn.svelte",
"repo_id": "chat-ui",
"token_count": 170
} | 43 |
<script lang="ts">
export let classNames = "";
</script>
<svg
class={classNames}
xmlns="http://www.w3.org/2000/svg"
aria-hidden="true"
focusable="false"
role="img"
width="1em"
height="1em"
fill="currentColor"
preserveAspectRatio="xMidYMid meet"
viewBox="0 0 20 20"
>
><path
fill-rule="evenodd"
d="M1.5 10a8.5 8.5 0 1 0 17 0a8.5 8.5 0 0 0-17 0m16 0a7.5 7.5 0 1 1-15 0a7.5 7.5 0 0 1 15 0"
clip-rule="evenodd"
/><path
fill-rule="evenodd"
d="M6.5 10c0 4.396 1.442 8 3.5 8s3.5-3.604 3.5-8s-1.442-8-3.5-8s-3.5 3.604-3.5 8m6 0c0 3.889-1.245 7-2.5 7s-2.5-3.111-2.5-7S8.745 3 10 3s2.5 3.111 2.5 7"
clip-rule="evenodd"
/><path
d="m3.735 5.312l.67-.742c.107.096.221.19.343.281c1.318.988 3.398 1.59 5.665 1.59c1.933 0 3.737-.437 5.055-1.19a5.59 5.59 0 0 0 .857-.597l.65.76c-.298.255-.636.49-1.01.704c-1.477.845-3.452 1.323-5.552 1.323c-2.47 0-4.762-.663-6.265-1.79a5.81 5.81 0 0 1-.413-.34m0 9.389l.67.74c.107-.096.221-.19.343-.28c1.318-.988 3.398-1.59 5.665-1.59c1.933 0 3.737.436 5.055 1.19c.321.184.608.384.857.596l.65-.76a6.583 6.583 0 0 0-1.01-.704c-1.477-.844-3.452-1.322-5.552-1.322c-2.47 0-4.762.663-6.265 1.789c-.146.11-.284.223-.413.34M2 10.5v-1h16v1z"
/></svg
>
| chat-ui/src/lib/components/icons/IconInternet.svelte/0 | {
"file_path": "chat-ui/src/lib/components/icons/IconInternet.svelte",
"repo_id": "chat-ui",
"token_count": 691
} | 44 |
import { z } from "zod";
import type { EmbeddingEndpoint, Embedding } from "../embeddingEndpoints";
import { chunk } from "$lib/utils/chunk";
export const embeddingEndpointTeiParametersSchema = z.object({
weight: z.number().int().positive().default(1),
model: z.any(),
type: z.literal("tei"),
url: z.string().url(),
authorization: z.string().optional(),
});
const getModelInfoByUrl = async (url: string, authorization?: string) => {
const { origin } = new URL(url);
const response = await fetch(`${origin}/info`, {
headers: {
Accept: "application/json",
"Content-Type": "application/json",
...(authorization ? { Authorization: authorization } : {}),
},
});
const json = await response.json();
return json;
};
export async function embeddingEndpointTei(
input: z.input<typeof embeddingEndpointTeiParametersSchema>
): Promise<EmbeddingEndpoint> {
const { url, model, authorization } = embeddingEndpointTeiParametersSchema.parse(input);
const { max_client_batch_size, max_batch_tokens } = await getModelInfoByUrl(url);
const maxBatchSize = Math.min(
max_client_batch_size,
Math.floor(max_batch_tokens / model.chunkCharLength)
);
return async ({ inputs }) => {
const { origin } = new URL(url);
const batchesInputs = chunk(inputs, maxBatchSize);
const batchesResults = await Promise.all(
batchesInputs.map(async (batchInputs) => {
const response = await fetch(`${origin}/embed`, {
method: "POST",
headers: {
Accept: "application/json",
"Content-Type": "application/json",
...(authorization ? { Authorization: authorization } : {}),
},
body: JSON.stringify({ inputs: batchInputs, normalize: true, truncate: true }),
});
const embeddings: Embedding[] = await response.json();
return embeddings;
})
);
const flatAllEmbeddings = batchesResults.flat();
return flatAllEmbeddings;
};
}
| chat-ui/src/lib/server/embeddingEndpoints/tei/embeddingEndpoints.ts/0 | {
"file_path": "chat-ui/src/lib/server/embeddingEndpoints/tei/embeddingEndpoints.ts",
"repo_id": "chat-ui",
"token_count": 664
} | 45 |
import { Address6, Address4 } from "ip-address";
import dns from "node:dns";
export async function isURLLocal(URL: URL): Promise<boolean> {
const isLocal = new Promise<boolean>((resolve, reject) => {
dns.lookup(URL.hostname, (err, address, family) => {
if (err) {
reject(err);
}
if (family === 4) {
const addr = new Address4(address);
resolve(addr.isInSubnet(new Address4("127.0.0.0/8")));
} else if (family === 6) {
const addr = new Address6(address);
resolve(
addr.isLoopback() || addr.isInSubnet(new Address6("::1/128")) || addr.isLinkLocal()
);
} else {
reject(new Error("Unknown IP family"));
}
});
});
return isLocal;
}
| chat-ui/src/lib/server/isURLLocal.ts/0 | {
"file_path": "chat-ui/src/lib/server/isURLLocal.ts",
"repo_id": "chat-ui",
"token_count": 290
} | 46 |
import { writable } from "svelte/store";
export const pendingMessage = writable<
| {
content: string;
files: File[];
}
| undefined
>();
| chat-ui/src/lib/stores/pendingMessage.ts/0 | {
"file_path": "chat-ui/src/lib/stores/pendingMessage.ts",
"repo_id": "chat-ui",
"token_count": 56
} | 47 |
import type { Timestamps } from "./Timestamps";
export interface Semaphore extends Timestamps {
key: string;
}
| chat-ui/src/lib/types/Semaphore.ts/0 | {
"file_path": "chat-ui/src/lib/types/Semaphore.ts",
"repo_id": "chat-ui",
"token_count": 35
} | 48 |
export function formatUserCount(userCount: number): string {
const userCountRanges: { min: number; max: number; label: string }[] = [
{ min: 0, max: 1, label: "1" },
{ min: 2, max: 9, label: "1-10" },
{ min: 10, max: 49, label: "10+" },
{ min: 50, max: 99, label: "50+" },
{ min: 100, max: 299, label: "100+" },
{ min: 300, max: 499, label: "300+" },
{ min: 500, max: 999, label: "500+" },
{ min: 1_000, max: 2_999, label: "1k+" },
{ min: 3_000, max: 4_999, label: "3k+" },
{ min: 5_000, max: 9_999, label: "5k+" },
{ min: 10_000, max: Infinity, label: "10k+" },
];
const range = userCountRanges.find(({ min, max }) => userCount >= min && userCount <= max);
return range?.label ?? "";
}
| chat-ui/src/lib/utils/formatUserCount.ts/0 | {
"file_path": "chat-ui/src/lib/utils/formatUserCount.ts",
"repo_id": "chat-ui",
"token_count": 308
} | 49 |
export const timeout = <T>(prom: Promise<T>, time: number): Promise<T> => {
let timer: NodeJS.Timeout;
return Promise.race([prom, new Promise<T>((_r, rej) => (timer = setTimeout(rej, time)))]).finally(
() => clearTimeout(timer)
);
};
| chat-ui/src/lib/utils/timeout.ts/0 | {
"file_path": "chat-ui/src/lib/utils/timeout.ts",
"repo_id": "chat-ui",
"token_count": 87
} | 50 |
import { PARQUET_EXPORT_DATASET, PARQUET_EXPORT_HF_TOKEN } from "$env/static/private";
import { collections } from "$lib/server/database";
import type { Message } from "$lib/types/Message";
import { error } from "@sveltejs/kit";
import { pathToFileURL } from "node:url";
import { unlink } from "node:fs/promises";
import { uploadFile } from "@huggingface/hub";
import parquet from "parquetjs";
import { z } from "zod";
// Triger like this:
// curl -X POST "http://localhost:5173/chat/admin/export" -H "Authorization: Bearer <ADMIN_API_SECRET>" -H "Content-Type: application/json" -d '{"model": "OpenAssistant/oasst-sft-6-llama-30b-xor"}'
export async function POST({ request }) {
if (!PARQUET_EXPORT_DATASET || !PARQUET_EXPORT_HF_TOKEN) {
throw error(500, "Parquet export is not configured.");
}
const { model } = z
.object({
model: z.string(),
})
.parse(await request.json());
const schema = new parquet.ParquetSchema({
title: { type: "UTF8" },
created_at: { type: "TIMESTAMP_MILLIS" },
updated_at: { type: "TIMESTAMP_MILLIS" },
messages: {
repeated: true,
fields: {
from: { type: "UTF8" },
content: { type: "UTF8" },
score: { type: "INT_8", optional: true },
},
},
});
const fileName = `/tmp/conversations-${new Date().toJSON().slice(0, 10)}-${Date.now()}.parquet`;
const writer = await parquet.ParquetWriter.openFile(schema, fileName);
let count = 0;
console.log("Exporting conversations for model", model);
for await (const conversation of collections.settings.aggregate<{
title: string;
created_at: Date;
updated_at: Date;
messages: Message[];
}>([
{
$match: {
shareConversationsWithModelAuthors: true,
sessionId: { $exists: true },
userId: { $exists: false },
},
},
{
$lookup: {
from: "conversations",
localField: "sessionId",
foreignField: "sessionId",
as: "conversations",
pipeline: [{ $match: { model, userId: { $exists: false } } }],
},
},
{ $unwind: "$conversations" },
{
$project: {
title: "$conversations.title",
created_at: "$conversations.createdAt",
updated_at: "$conversations.updatedAt",
messages: "$conversations.messages",
},
},
])) {
await writer.appendRow({
title: conversation.title,
created_at: conversation.created_at,
updated_at: conversation.updated_at,
messages: conversation.messages.map((message: Message) => ({
from: message.from,
content: message.content,
...(message.score ? { score: message.score } : undefined),
})),
});
++count;
if (count % 1_000 === 0) {
console.log("Exported", count, "conversations");
}
}
console.log("exporting convos with userId");
for await (const conversation of collections.settings.aggregate<{
title: string;
created_at: Date;
updated_at: Date;
messages: Message[];
}>([
{ $match: { shareConversationsWithModelAuthors: true, userId: { $exists: true } } },
{
$lookup: {
from: "conversations",
localField: "userId",
foreignField: "userId",
as: "conversations",
pipeline: [{ $match: { model } }],
},
},
{ $unwind: "$conversations" },
{
$project: {
title: "$conversations.title",
created_at: "$conversations.createdAt",
updated_at: "$conversations.updatedAt",
messages: "$conversations.messages",
},
},
])) {
await writer.appendRow({
title: conversation.title,
created_at: conversation.created_at,
updated_at: conversation.updated_at,
messages: conversation.messages.map((message: Message) => ({
from: message.from,
content: message.content,
...(message.score ? { score: message.score } : undefined),
})),
});
++count;
if (count % 1_000 === 0) {
console.log("Exported", count, "conversations");
}
}
await writer.close();
console.log("Uploading", fileName, "to Hugging Face Hub");
await uploadFile({
file: pathToFileURL(fileName) as URL,
credentials: { accessToken: PARQUET_EXPORT_HF_TOKEN },
repo: {
type: "dataset",
name: PARQUET_EXPORT_DATASET,
},
});
console.log("Upload done");
await unlink(fileName);
return new Response();
}
| chat-ui/src/routes/admin/export/+server.ts/0 | {
"file_path": "chat-ui/src/routes/admin/export/+server.ts",
"repo_id": "chat-ui",
"token_count": 1653
} | 51 |
import { buildPrompt } from "$lib/buildPrompt";
import { authCondition } from "$lib/server/auth";
import { collections } from "$lib/server/database";
import { models } from "$lib/server/models";
import { buildSubtree } from "$lib/utils/tree/buildSubtree";
import { isMessageId } from "$lib/utils/tree/isMessageId";
import { error } from "@sveltejs/kit";
import { ObjectId } from "mongodb";
export async function GET({ params, locals }) {
const conv =
params.id.length === 7
? await collections.sharedConversations.findOne({
_id: params.id,
})
: await collections.conversations.findOne({
_id: new ObjectId(params.id),
...authCondition(locals),
});
if (conv === null) {
throw error(404, "Conversation not found");
}
const messageId = params.messageId;
const messageIndex = conv.messages.findIndex((msg) => msg.id === messageId);
if (!isMessageId(messageId) || messageIndex === -1) {
throw error(404, "Message not found");
}
const model = models.find((m) => m.id === conv.model);
if (!model) {
throw error(404, "Conversation model not found");
}
const messagesUpTo = buildSubtree(conv, messageId);
const prompt = await buildPrompt({
preprompt: conv.preprompt,
messages: messagesUpTo,
model,
});
return new Response(
JSON.stringify(
{
note: "This is a preview of the prompt that will be sent to the model when retrying the message. It may differ from what was sent in the past if the parameters have been updated since",
prompt,
model: model.name,
parameters: {
...model.parameters,
return_full_text: false,
},
},
null,
2
),
{ headers: { "Content-Type": "application/json" } }
);
}
| chat-ui/src/routes/conversation/[id]/message/[messageId]/prompt/+server.ts/0 | {
"file_path": "chat-ui/src/routes/conversation/[id]/message/[messageId]/prompt/+server.ts",
"repo_id": "chat-ui",
"token_count": 601
} | 52 |
<script lang="ts">
import { marked } from "marked";
import privacy from "../../../PRIVACY.md?raw";
</script>
<div class="overflow-auto p-6">
<div class="prose mx-auto px-4 pb-24 pt-6 dark:prose-invert md:pt-12">
<!-- eslint-disable-next-line svelte/no-at-html-tags -->
{@html marked(privacy, { gfm: true })}
</div>
</div>
| chat-ui/src/routes/privacy/+page.svelte/0 | {
"file_path": "chat-ui/src/routes/privacy/+page.svelte",
"repo_id": "chat-ui",
"token_count": 141
} | 53 |
import { collections } from "$lib/server/database";
import type { LayoutServerLoad } from "./$types";
import type { Report } from "$lib/types/Report";
export const load = (async ({ locals, parent }) => {
const { assistants } = await parent();
let reportsByUser: string[] = [];
const createdBy = locals.user?._id ?? locals.sessionId;
if (createdBy) {
const reports = await collections.reports
.find<Pick<Report, "assistantId">>({ createdBy }, { projection: { _id: 0, assistantId: 1 } })
.toArray();
reportsByUser = reports.map((r) => r.assistantId.toString());
}
return {
assistants: assistants.map((el) => ({
...el,
reported: reportsByUser.includes(el._id),
})),
};
}) satisfies LayoutServerLoad;
| chat-ui/src/routes/settings/+layout.server.ts/0 | {
"file_path": "chat-ui/src/routes/settings/+layout.server.ts",
"repo_id": "chat-ui",
"token_count": 241
} | 54 |
import json
import os
from dataclasses import dataclass
import numpy as np
import pyarrow as pa
import datasets
from utils import get_duration
SPEED_TEST_N_EXAMPLES = 100_000_000_000
SPEED_TEST_CHUNK_SIZE = 10_000
RESULTS_BASEPATH, RESULTS_FILENAME = os.path.split(__file__)
RESULTS_FILE_PATH = os.path.join(RESULTS_BASEPATH, "results", RESULTS_FILENAME.replace(".py", ".json"))
def generate_100B_dataset(num_examples: int, chunk_size: int) -> datasets.Dataset:
table = pa.Table.from_pydict({"col": [0] * chunk_size})
table = pa.concat_tables([table] * (num_examples // chunk_size))
return datasets.Dataset(table, fingerprint="table_100B")
@dataclass
class RandIter:
low: int
high: int
size: int
seed: int
def __post_init__(self):
rng = np.random.default_rng(self.seed)
self._sampled_values = rng.integers(low=self.low, high=self.high, size=self.size).tolist()
def __iter__(self):
return iter(self._sampled_values)
def __len__(self):
return self.size
@get_duration
def get_first_row(dataset: datasets.Dataset):
_ = dataset[0]
@get_duration
def get_last_row(dataset: datasets.Dataset):
_ = dataset[-1]
@get_duration
def get_batch_of_1024_rows(dataset: datasets.Dataset):
_ = dataset[range(len(dataset) // 2, len(dataset) // 2 + 1024)]
@get_duration
def get_batch_of_1024_random_rows(dataset: datasets.Dataset):
_ = dataset[RandIter(0, len(dataset), 1024, seed=42)]
def benchmark_table_100B():
times = {"num examples": SPEED_TEST_N_EXAMPLES}
functions = (get_first_row, get_last_row, get_batch_of_1024_rows, get_batch_of_1024_random_rows)
print("generating dataset")
dataset = generate_100B_dataset(num_examples=SPEED_TEST_N_EXAMPLES, chunk_size=SPEED_TEST_CHUNK_SIZE)
print("Functions")
for func in functions:
print(func.__name__)
times[func.__name__] = func(dataset)
with open(RESULTS_FILE_PATH, "wb") as f:
f.write(json.dumps(times).encode("utf-8"))
if __name__ == "__main__": # useful to run the profiler
benchmark_table_100B()
| datasets/benchmarks/benchmark_getitem_100B.py/0 | {
"file_path": "datasets/benchmarks/benchmark_getitem_100B.py",
"repo_id": "datasets",
"token_count": 867
} | 55 |
# Datasets 🤝 Arrow
## What is Arrow?
[Arrow](https://arrow.apache.org/) enables large amounts of data to be processed and moved quickly. It is a specific data format that stores data in a columnar memory layout. This provides several significant advantages:
* Arrow's standard format allows [zero-copy reads](https://en.wikipedia.org/wiki/Zero-copy) which removes virtually all serialization overhead.
* Arrow is language-agnostic so it supports different programming languages.
* Arrow is column-oriented so it is faster at querying and processing slices or columns of data.
* Arrow allows for copy-free hand-offs to standard machine learning tools such as NumPy, Pandas, PyTorch, and TensorFlow.
* Arrow supports many, possibly nested, column types.
## Memory-mapping
🤗 Datasets uses Arrow for its local caching system. It allows datasets to be backed by an on-disk cache, which is memory-mapped for fast lookup.
This architecture allows for large datasets to be used on machines with relatively small device memory.
For example, loading the full English Wikipedia dataset only takes a few MB of RAM:
```python
>>> import os; import psutil; import timeit
>>> from datasets import load_dataset
# Process.memory_info is expressed in bytes, so convert to megabytes
>>> mem_before = psutil.Process(os.getpid()).memory_info().rss / (1024 * 1024)
>>> wiki = load_dataset("wikipedia", "20220301.en", split="train")
>>> mem_after = psutil.Process(os.getpid()).memory_info().rss / (1024 * 1024)
>>> print(f"RAM memory used: {(mem_after - mem_before)} MB")
RAM memory used: 50 MB
```
This is possible because the Arrow data is actually memory-mapped from disk, and not loaded in memory.
Memory-mapping allows access to data on disk, and leverages virtual memory capabilities for fast lookups.
## Performance
Iterating over a memory-mapped dataset using Arrow is fast. Iterating over Wikipedia on a laptop gives you speeds of 1-3 Gbit/s:
```python
>>> s = """batch_size = 1000
... for batch in wiki.iter(batch_size):
... ...
... """
>>> elapsed_time = timeit.timeit(stmt=s, number=1, globals=globals())
>>> print(f"Time to iterate over the {wiki.dataset_size >> 30} GB dataset: {elapsed_time:.1f} sec, "
... f"ie. {float(wiki.dataset_size >> 27)/elapsed_time:.1f} Gb/s")
Time to iterate over the 18 GB dataset: 31.8 sec, ie. 4.8 Gb/s
```
| datasets/docs/source/about_arrow.md/0 | {
"file_path": "datasets/docs/source/about_arrow.md",
"repo_id": "datasets",
"token_count": 682
} | 56 |
# Depth estimation
Depth estimation datasets are used to train a model to approximate the relative distance of every pixel in an
image from the camera, also known as depth. The applications enabled by these datasets primarily lie in areas like visual machine
perception and perception in robotics. Example applications include mapping streets for self-driving cars. This guide will show you how to apply transformations
to a depth estimation dataset.
Before you start, make sure you have up-to-date versions of `albumentations` installed:
```bash
pip install -U albumentations
```
[Albumentations](https://albumentations.ai/) is a Python library for performing data augmentation
for computer vision. It supports various computer vision tasks such as image classification, object
detection, segmentation, and keypoint estimation.
This guide uses the [NYU Depth V2](https://huggingface.co/datasets/sayakpaul/nyu_depth_v2) dataset which is
comprised of video sequences from various indoor scenes, recorded by RGB and depth cameras. The dataset consists of scenes from 3 cities and provides images along with
their depth maps as labels.
Load the `train` split of the dataset and take a look at an example:
```py
>>> from datasets import load_dataset
>>> train_dataset = load_dataset("sayakpaul/nyu_depth_v2", split="train")
>>> index = 17
>>> example = train_dataset[index]
>>> example
{'image': <PIL.PngImagePlugin.PngImageFile image mode=RGB size=640x480>,
'depth_map': <PIL.TiffImagePlugin.TiffImageFile image mode=F size=640x480>}
```
The dataset has two fields:
* `image`: a PIL PNG image object with `uint8` data type.
* `depth_map`: a PIL Tiff image object with `float32` data type which is the depth map of the image.
It is mention-worthy that JPEG/PNG format can only store `uint8` or `uint16` data. As the depth map is `float32` data, it can't be stored using PNG/JPEG. However, we can save the depth map using TIFF format as it supports a wider range of data types, including `float32` data.
Next, check out an image with:
```py
>>> example["image"]
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_sample.png">
</div>
Before we look at the depth map, we need to first convert its data type to `uint8` using `.convert('RGB')` as PIL can't display `float32` images. Now take a look at its corresponding depth map:
```py
>>> example["depth_map"].convert("RGB")
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_target.png">
</div>
It's all black! You'll need to add some color to the depth map to visualize it properly. To do that, either we can apply color automatically during display using `plt.imshow()` or create a colored depth map using `plt.cm` and then display it. In this example, we have used the latter one, as we can save/write the colored depth map later. (the utility below is taken from the [FastDepth repository](https://github.com/dwofk/fast-depth/blob/master/utils.py)).
```py
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> cmap = plt.cm.viridis
>>> def colored_depthmap(depth, d_min=None, d_max=None):
... if d_min is None:
... d_min = np.min(depth)
... if d_max is None:
... d_max = np.max(depth)
... depth_relative = (depth - d_min) / (d_max - d_min)
... return 255 * cmap(depth_relative)[:,:,:3]
>>> def show_depthmap(depth_map):
... if not isinstance(depth_map, np.ndarray):
... depth_map = np.array(depth_map)
... if depth_map.ndim == 3:
... depth_map = depth_map.squeeze()
... d_min = np.min(depth_map)
... d_max = np.max(depth_map)
... depth_map = colored_depthmap(depth_map, d_min, d_max)
... plt.imshow(depth_map.astype("uint8"))
... plt.axis("off")
... plt.show()
>>> show_depthmap(example["depth_map"])
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_target_viz.png">
</div>
You can also visualize several different images and their corresponding depth maps.
```py
>>> def merge_into_row(input_image, depth_target):
... if not isinstance(input_image, np.ndarray):
... input_image = np.array(input_image)
...
... d_min = np.min(depth_target)
... d_max = np.max(depth_target)
... depth_target_col = colored_depthmap(depth_target, d_min, d_max)
... img_merge = np.hstack([input_image, depth_target_col])
...
... return img_merge
>>> random_indices = np.random.choice(len(train_dataset), 9).tolist()
>>> plt.figure(figsize=(15, 6))
>>> for i, idx in enumerate(random_indices):
... example = train_dataset[idx]
... ax = plt.subplot(3, 3, i + 1)
... image_viz = merge_into_row(
... example["image"], example["depth_map"]
... )
... plt.imshow(image_viz.astype("uint8"))
... plt.axis("off")
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_collage.png">
</div>
Now apply some augmentations with `albumentations`. The augmentation transformations include:
* Random horizontal flipping
* Random cropping
* Random brightness and contrast
* Random gamma correction
* Random hue saturation
```py
>>> import albumentations as A
>>> crop_size = (448, 576)
>>> transforms = [
... A.HorizontalFlip(p=0.5),
... A.RandomCrop(crop_size[0], crop_size[1]),
... A.RandomBrightnessContrast(),
... A.RandomGamma(),
... A.HueSaturationValue()
... ]
```
Additionally, define a mapping to better reflect the target key name.
```py
>>> additional_targets = {"depth": "mask"}
>>> aug = A.Compose(transforms=transforms, additional_targets=additional_targets)
```
With `additional_targets` defined, you can pass the target depth maps to the `depth` argument of `aug` instead of `mask`. You'll notice this change
in the `apply_transforms()` function defined below.
Create a function to apply the transformation to the images as well as their depth maps:
```py
>>> def apply_transforms(examples):
... transformed_images, transformed_maps = [], []
... for image, depth_map in zip(examples["image"], examples["depth_map"]):
... image, depth_map = np.array(image), np.array(depth_map)
... transformed = aug(image=image, depth=depth_map)
... transformed_images.append(transformed["image"])
... transformed_maps.append(transformed["depth"])
...
... examples["pixel_values"] = transformed_images
... examples["labels"] = transformed_maps
... return examples
```
Use the [`~Dataset.set_transform`] function to apply the transformation on-the-fly to batches of the dataset to consume less disk space:
```py
>>> train_dataset.set_transform(apply_transforms)
```
You can verify the transformation worked by indexing into the `pixel_values` and `labels` of an example image:
```py
>>> example = train_dataset[index]
>>> plt.imshow(example["pixel_values"])
>>> plt.axis("off")
>>> plt.show()
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_sample_aug.png">
</div>
Visualize the same transformation on the image's corresponding depth map:
```py
>>> show_depthmap(example["labels"])
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_target_aug.png">
</div>
You can also visualize multiple training samples reusing the previous `random_indices`:
```py
>>> plt.figure(figsize=(15, 6))
>>> for i, idx in enumerate(random_indices):
... ax = plt.subplot(3, 3, i + 1)
... example = train_dataset[idx]
... image_viz = merge_into_row(
... example["pixel_values"], example["labels"]
... )
... plt.imshow(image_viz.astype("uint8"))
... plt.axis("off")
```
<div class="flex justify-center">
<img src="https://huggingface.co/datasets/huggingface/documentation-images/resolve/main/datasets/depth_est_aug_collage.png">
</div> | datasets/docs/source/depth_estimation.mdx/0 | {
"file_path": "datasets/docs/source/depth_estimation.mdx",
"repo_id": "datasets",
"token_count": 2848
} | 57 |
# Load text data
This guide shows you how to load text datasets. To learn how to load any type of dataset, take a look at the <a class="underline decoration-sky-400 decoration-2 font-semibold" href="./loading">general loading guide</a>.
Text files are one of the most common file types for storing a dataset. By default, 🤗 Datasets samples a text file line by line to build the dataset.
```py
>>> from datasets import load_dataset
>>> dataset = load_dataset("text", data_files={"train": ["my_text_1.txt", "my_text_2.txt"], "test": "my_test_file.txt"})
# Load from a directory
>>> dataset = load_dataset("text", data_dir="path/to/text/dataset")
```
To sample a text file by paragraph or even an entire document, use the `sample_by` parameter:
```py
# Sample by paragraph
>>> dataset = load_dataset("text", data_files={"train": "my_train_file.txt", "test": "my_test_file.txt"}, sample_by="paragraph")
# Sample by document
>>> dataset = load_dataset("text", data_files={"train": "my_train_file.txt", "test": "my_test_file.txt"}, sample_by="document")
```
You can also use grep patterns to load specific files:
```py
>>> from datasets import load_dataset
>>> c4_subset = load_dataset("allenai/c4", data_files="en/c4-train.0000*-of-01024.json.gz")
```
To load remote text files via HTTP, pass the URLs instead:
```py
>>> dataset = load_dataset("text", data_files="https://huggingface.co/datasets/lhoestq/test/resolve/main/some_text.txt")
``` | datasets/docs/source/nlp_load.mdx/0 | {
"file_path": "datasets/docs/source/nlp_load.mdx",
"repo_id": "datasets",
"token_count": 482
} | 58 |
# Troubleshooting
This guide aims to provide you the tools and knowledge required to navigate some common issues. If the suggestions listed
in this guide do not cover your such situation, please refer to the [Asking for Help](#asking-for-help) section to learn where to
find help with your specific issue.
## Issues when uploading datasets with `push_to_hub`
### Authentication issues
If you are experiencing authentication issues when sharing a dataset on 🤗 Hub using [`Dataset.push_to_hub`] and a Hugging Face
access token:
* Make sure that the Hugging Face token you're using to authenticate yourself is a token with **write** permission.
* On OSX, it may help to clean up all the huggingface.co passwords on your keychain access, as well as reconfigure `git config --global credential.helper osxkeychain`, before using `huggingface-cli login`.
Alternatively, you can use SSH keys to authenticate yourself - read more in the [🤗 Hub documentation](https://huggingface.co/docs/hub/security-git-ssh).
### Lost connection on large dataset upload
When uploading large datasets to Hub, if the number of dataset shards is large, it can create too many commits for the Hub in a
short period. This will result in a connection error.
The connection error can also be caused by a HTTP 500 error returned by AWS S3 bucket that Hub uses internally.
In either situation, you can re-run [`Dataset.push_to_hub`] to proceed with the dataset upload. Hub will check the SHAs
of already uploaded shards to avoid reuploading them.
We are working on making upload process more robust to transient errors, so updating to the latest library version is
always a good idea.
### `Too Many Requests`
Uploading large datasets via `push_to_hub()` can result in an error:
```bash
HfHubHTTPError: 429 Client Error: Too Many Requests for url: ...
You have exceeded our hourly quotas for action: commit. We invite you to retry later.
```
If you encounter this issue, you need to upgrade the `datasets` library to the latest version (or at least `2.15.0`).
## Issues when creating datasets from custom data
### Loading images and audio from a folder
When creating a dataset from a folder, one of the most common issues is that the file structure does not follow the
expected format, or there's an issue with the metadata file.
Learn more about required folder structure in corresponding documentation pages:
* [AudioFolder](https://huggingface.co/docs/datasets/audio_dataset#audiofolder)
* [ImageFolder](https://huggingface.co/docs/datasets/image_dataset#imagefolder)
### Pickling issues
#### Pickling issues when using `Dataset.from_generator`
When creating a dataset, [`IterableDataset.from_generator`] and [`Dataset.from_generator`] expect a "picklable" generator function.
This is required to hash the function using [`pickle`](https://docs.python.org/3/library/pickle.html) to be able to cache the dataset on disk.
While generator functions are generally "picklable", note that generator objects are not. So if you're using a generator object,
you will encounter a `TypeError` like this:
```bash
TypeError: cannot pickle 'generator' object
```
This error can also occur when using a generator function that uses a global object that is not "picklable", such as a
DB connection, for example. If that's the case, you can initialize such object directly inside the generator function to
avoid this error.
#### Pickling issues with `Dataset.map`
Pickling errors can also happen in the multiprocess [`Dataset.map`] - objects are pickled to be passed to child processes.
If the objects used in the transformation are not picklable, it's not possible to cache the result of `map`, which leads to an error being raised.
Here are some ways to address this issue:
* A universal solution to pickle issues is to make sure the objects (or generator classes) are pickable manually by implementing `__getstate__` / `__setstate__` / `__reduce__`.
* You can also provide your own unique hash in `map` with the `new_fingerprint` argument.
* You can also disable caching by calling `datasets.disable_caching()`, however, this is undesirable - [read more about importance of cache](cache)
## Asking for help
If the above troubleshooting advice did not help you resolve your issue, reach out for help to the community and the team.
### Forums
Ask for help on the Hugging Face forums - post your question in the [🤗Datasets category](https://discuss.huggingface.co/c/datasets/10)
Make sure to write a descriptive post with relevant context about your setup and reproducible code to maximize the likelihood that your problem is solved!
### Discord
Post a question on [Discord](http://hf.co/join/discord), and let the team and the community help you.
### Community Discussions on 🤗 Hub
If you are facing issues creating a custom dataset with a script on Hub, you can ask the Hugging Face team for help by opening
a discussion in the Community tab of your dataset with this message:
```text
# Dataset rewiew request for <Dataset name>
## Description
<brief description of the dataset>
## Files to review
- file1
- file2
- ...
cc @lhoestq @polinaeterna @mariosasko @albertvillanova
```
### GitHub Issues
Finally, if you suspect to have found a bug related to the library itself, create an Issue on the 🤗 Datasets
[GitHub repository](https://github.com/huggingface/datasets/issues). Include context regarding the bug: code snippet to reproduce,
details about your environment and data, etc. to help us figure out what's wrong and how we can fix it.
| datasets/docs/source/troubleshoot.mdx/0 | {
"file_path": "datasets/docs/source/troubleshoot.mdx",
"repo_id": "datasets",
"token_count": 1470
} | 59 |
# Metric Card for CER
## Metric description
Character error rate (CER) is a common metric of the performance of an automatic speech recognition (ASR) system. CER is similar to Word Error Rate (WER), but operates on character instead of word.
Character error rate can be computed as:
`CER = (S + D + I) / N = (S + D + I) / (S + D + C)`
where
`S` is the number of substitutions,
`D` is the number of deletions,
`I` is the number of insertions,
`C` is the number of correct characters,
`N` is the number of characters in the reference (`N=S+D+C`).
## How to use
The metric takes two inputs: references (a list of references for each speech input) and predictions (a list of transcriptions to score).
```python
from datasets import load_metric
cer = load_metric("cer")
cer_score = cer.compute(predictions=predictions, references=references)
```
## Output values
This metric outputs a float representing the character error rate.
```
print(cer_score)
0.34146341463414637
```
The **lower** the CER value, the **better** the performance of the ASR system, with a CER of 0 being a perfect score.
However, CER's output is not always a number between 0 and 1, in particular when there is a high number of insertions (see [Examples](#Examples) below).
### Values from popular papers
This metric is highly dependent on the content and quality of the dataset, and therefore users can expect very different values for the same model but on different datasets.
Multilingual datasets such as [Common Voice](https://huggingface.co/datasets/common_voice) report different CERs depending on the language, ranging from 0.02-0.03 for languages such as French and Italian, to 0.05-0.07 for English (see [here](https://github.com/speechbrain/speechbrain/tree/develop/recipes/CommonVoice/ASR/CTC) for more values).
## Examples
Perfect match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello world", "good night moon"]
references = ["hello world", "good night moon"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
0.0
```
Partial match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["this is the prediction", "there is an other sample"]
references = ["this is the reference", "there is another one"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
0.34146341463414637
```
No match between prediction and reference:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello"]
references = ["gracias"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
1.0
```
CER above 1 due to insertion errors:
```python
from datasets import load_metric
cer = load_metric("cer")
predictions = ["hello world"]
references = ["hello"]
cer_score = cer.compute(predictions=predictions, references=references)
print(cer_score)
1.2
```
## Limitations and bias
CER is useful for comparing different models for tasks such as automatic speech recognition (ASR) and optic character recognition (OCR), especially for multilingual datasets where WER is not suitable given the diversity of languages. However, CER provides no details on the nature of translation errors and further work is therefore required to identify the main source(s) of error and to focus any research effort.
Also, in some cases, instead of reporting the raw CER, a normalized CER is reported where the number of mistakes is divided by the sum of the number of edit operations (`I` + `S` + `D`) and `C` (the number of correct characters), which results in CER values that fall within the range of 0–100%.
## Citation
```bibtex
@inproceedings{morris2004,
author = {Morris, Andrew and Maier, Viktoria and Green, Phil},
year = {2004},
month = {01},
pages = {},
title = {From WER and RIL to MER and WIL: improved evaluation measures for connected speech recognition.}
}
```
## Further References
- [Hugging Face Tasks -- Automatic Speech Recognition](https://huggingface.co/tasks/automatic-speech-recognition)
| datasets/metrics/cer/README.md/0 | {
"file_path": "datasets/metrics/cer/README.md",
"repo_id": "datasets",
"token_count": 1192
} | 60 |
"""Official evaluation script for CUAD dataset."""
import argparse
import json
import re
import string
import sys
import numpy as np
IOU_THRESH = 0.5
def get_jaccard(prediction, ground_truth):
remove_tokens = [".", ",", ";", ":"]
for token in remove_tokens:
ground_truth = ground_truth.replace(token, "")
prediction = prediction.replace(token, "")
ground_truth, prediction = ground_truth.lower(), prediction.lower()
ground_truth, prediction = ground_truth.replace("/", " "), prediction.replace("/", " ")
ground_truth, prediction = set(ground_truth.split(" ")), set(prediction.split(" "))
intersection = ground_truth.intersection(prediction)
union = ground_truth.union(prediction)
jaccard = len(intersection) / len(union)
return jaccard
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r"\b(a|an|the)\b", " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def compute_precision_recall(predictions, ground_truths, qa_id):
tp, fp, fn = 0, 0, 0
substr_ok = "Parties" in qa_id
# first check if ground truth is empty
if len(ground_truths) == 0:
if len(predictions) > 0:
fp += len(predictions) # false positive for each one
else:
for ground_truth in ground_truths:
assert len(ground_truth) > 0
# check if there is a match
match_found = False
for pred in predictions:
if substr_ok:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH or ground_truth in pred
else:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH
if is_match:
match_found = True
if match_found:
tp += 1
else:
fn += 1
# now also get any fps by looping through preds
for pred in predictions:
# Check if there's a match. if so, don't count (don't want to double count based on the above)
# but if there's no match, then this is a false positive.
# (Note: we get the true positives in the above loop instead of this loop so that we don't double count
# multiple predictions that are matched with the same answer.)
match_found = False
for ground_truth in ground_truths:
assert len(ground_truth) > 0
if substr_ok:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH or ground_truth in pred
else:
is_match = get_jaccard(pred, ground_truth) >= IOU_THRESH
if is_match:
match_found = True
if not match_found:
fp += 1
precision = tp / (tp + fp) if tp + fp > 0 else np.nan
recall = tp / (tp + fn) if tp + fn > 0 else np.nan
return precision, recall
def process_precisions(precisions):
"""
Processes precisions to ensure that precision and recall don't both get worse.
Assumes the list precision is sorted in order of recalls
"""
precision_best = precisions[::-1]
for i in range(1, len(precision_best)):
precision_best[i] = max(precision_best[i - 1], precision_best[i])
precisions = precision_best[::-1]
return precisions
def get_aupr(precisions, recalls):
processed_precisions = process_precisions(precisions)
aupr = np.trapz(processed_precisions, recalls)
if np.isnan(aupr):
return 0
return aupr
def get_prec_at_recall(precisions, recalls, recall_thresh):
"""Assumes recalls are sorted in increasing order"""
processed_precisions = process_precisions(precisions)
prec_at_recall = 0
for prec, recall in zip(processed_precisions, recalls):
if recall >= recall_thresh:
prec_at_recall = prec
break
return prec_at_recall
def exact_match_score(prediction, ground_truth):
return normalize_answer(prediction) == normalize_answer(ground_truth)
def metric_max_over_ground_truths(metric_fn, predictions, ground_truths):
score = 0
for pred in predictions:
for ground_truth in ground_truths:
score = metric_fn(pred, ground_truth)
if score == 1: # break the loop when one prediction matches the ground truth
break
if score == 1:
break
return score
def evaluate(dataset, predictions):
f1 = exact_match = total = 0
precisions = []
recalls = []
for article in dataset:
for paragraph in article["paragraphs"]:
for qa in paragraph["qas"]:
total += 1
if qa["id"] not in predictions:
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
precision, recall = compute_precision_recall(prediction, ground_truths, qa["id"])
precisions.append(precision)
recalls.append(recall)
if precision == 0 and recall == 0:
f1 += 0
else:
f1 += 2 * (precision * recall) / (precision + recall)
exact_match += metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
precisions = [x for _, x in sorted(zip(recalls, precisions))]
recalls.sort()
f1 = 100.0 * f1 / total
exact_match = 100.0 * exact_match / total
aupr = get_aupr(precisions, recalls)
prec_at_90_recall = get_prec_at_recall(precisions, recalls, recall_thresh=0.9)
prec_at_80_recall = get_prec_at_recall(precisions, recalls, recall_thresh=0.8)
return {
"exact_match": exact_match,
"f1": f1,
"aupr": aupr,
"prec_at_80_recall": prec_at_80_recall,
"prec_at_90_recall": prec_at_90_recall,
}
if __name__ == "__main__":
parser = argparse.ArgumentParser(description="Evaluation for CUAD")
parser.add_argument("dataset_file", help="Dataset file")
parser.add_argument("prediction_file", help="Prediction File")
args = parser.parse_args()
with open(args.dataset_file) as dataset_file:
dataset_json = json.load(dataset_file)
dataset = dataset_json["data"]
with open(args.prediction_file) as prediction_file:
predictions = json.load(prediction_file)
print(json.dumps(evaluate(dataset, predictions)))
| datasets/metrics/cuad/evaluate.py/0 | {
"file_path": "datasets/metrics/cuad/evaluate.py",
"repo_id": "datasets",
"token_count": 3035
} | 61 |
# Copyright 2021 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Mahalanobis metric."""
import numpy as np
import datasets
_DESCRIPTION = """
Compute the Mahalanobis Distance
Mahalonobis distance is the distance between a point and a distribution.
And not between two distinct points. It is effectively a multivariate equivalent of the Euclidean distance.
It was introduced by Prof. P. C. Mahalanobis in 1936
and has been used in various statistical applications ever since
[source: https://www.machinelearningplus.com/statistics/mahalanobis-distance/]
"""
_CITATION = """\
@article{de2000mahalanobis,
title={The mahalanobis distance},
author={De Maesschalck, Roy and Jouan-Rimbaud, Delphine and Massart, D{\'e}sir{\'e} L},
journal={Chemometrics and intelligent laboratory systems},
volume={50},
number={1},
pages={1--18},
year={2000},
publisher={Elsevier}
}
"""
_KWARGS_DESCRIPTION = """
Args:
X: List of datapoints to be compared with the `reference_distribution`.
reference_distribution: List of datapoints from the reference distribution we want to compare to.
Returns:
mahalanobis: The Mahalonobis distance for each datapoint in `X`.
Examples:
>>> mahalanobis_metric = datasets.load_metric("mahalanobis")
>>> results = mahalanobis_metric.compute(reference_distribution=[[0, 1], [1, 0]], X=[[0, 1]])
>>> print(results)
{'mahalanobis': array([0.5])}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Mahalanobis(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"X": datasets.Sequence(datasets.Value("float", id="sequence"), id="X"),
}
),
)
def _compute(self, X, reference_distribution):
# convert to numpy arrays
X = np.array(X)
reference_distribution = np.array(reference_distribution)
# Assert that arrays are 2D
if len(X.shape) != 2:
raise ValueError("Expected `X` to be a 2D vector")
if len(reference_distribution.shape) != 2:
raise ValueError("Expected `reference_distribution` to be a 2D vector")
if reference_distribution.shape[0] < 2:
raise ValueError(
"Expected `reference_distribution` to be a 2D vector with more than one element in the first dimension"
)
# Get mahalanobis distance for each prediction
X_minus_mu = X - np.mean(reference_distribution)
cov = np.cov(reference_distribution.T)
try:
inv_covmat = np.linalg.inv(cov)
except np.linalg.LinAlgError:
inv_covmat = np.linalg.pinv(cov)
left_term = np.dot(X_minus_mu, inv_covmat)
mahal_dist = np.dot(left_term, X_minus_mu.T).diagonal()
return {"mahalanobis": mahal_dist}
| datasets/metrics/mahalanobis/mahalanobis.py/0 | {
"file_path": "datasets/metrics/mahalanobis/mahalanobis.py",
"repo_id": "datasets",
"token_count": 1363
} | 62 |
# Copyright 2020 The HuggingFace Datasets Authors and the current dataset script contributor.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Precision metric."""
from sklearn.metrics import precision_score
import datasets
_DESCRIPTION = """
Precision is the fraction of correctly labeled positive examples out of all of the examples that were labeled as positive. It is computed via the equation:
Precision = TP / (TP + FP)
where TP is the True positives (i.e. the examples correctly labeled as positive) and FP is the False positive examples (i.e. the examples incorrectly labeled as positive).
"""
_KWARGS_DESCRIPTION = """
Args:
predictions (`list` of `int`): Predicted class labels.
references (`list` of `int`): Actual class labels.
labels (`list` of `int`): The set of labels to include when `average` is not set to `'binary'`. If `average` is `None`, it should be the label order. Labels present in the data can be excluded, for example to calculate a multiclass average ignoring a majority negative class. Labels not present in the data will result in 0 components in a macro average. For multilabel targets, labels are column indices. By default, all labels in `predictions` and `references` are used in sorted order. Defaults to None.
pos_label (`int`): The class to be considered the positive class, in the case where `average` is set to `binary`. Defaults to 1.
average (`string`): This parameter is required for multiclass/multilabel targets. If set to `None`, the scores for each class are returned. Otherwise, this determines the type of averaging performed on the data. Defaults to `'binary'`.
- 'binary': Only report results for the class specified by `pos_label`. This is applicable only if the classes found in `predictions` and `references` are binary.
- 'micro': Calculate metrics globally by counting the total true positives, false negatives and false positives.
- 'macro': Calculate metrics for each label, and find their unweighted mean. This does not take label imbalance into account.
- 'weighted': Calculate metrics for each label, and find their average weighted by support (the number of true instances for each label). This alters `'macro'` to account for label imbalance. This option can result in an F-score that is not between precision and recall.
- 'samples': Calculate metrics for each instance, and find their average (only meaningful for multilabel classification).
sample_weight (`list` of `float`): Sample weights Defaults to None.
zero_division (`int` or `string`): Sets the value to return when there is a zero division. Defaults to 'warn'.
- 0: Returns 0 when there is a zero division.
- 1: Returns 1 when there is a zero division.
- 'warn': Raises warnings and then returns 0 when there is a zero division.
Returns:
precision (`float` or `array` of `float`): Precision score or list of precision scores, depending on the value passed to `average`. Minimum possible value is 0. Maximum possible value is 1. Higher values indicate that fewer negative examples were incorrectly labeled as positive, which means that, generally, higher scores are better.
Examples:
Example 1-A simple binary example
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0])
>>> print(results)
{'precision': 0.5}
Example 2-The same simple binary example as in Example 1, but with `pos_label` set to `0`.
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], pos_label=0)
>>> print(round(results['precision'], 2))
0.67
Example 3-The same simple binary example as in Example 1, but with `sample_weight` included.
>>> precision_metric = datasets.load_metric("precision")
>>> results = precision_metric.compute(references=[0, 1, 0, 1, 0], predictions=[0, 0, 1, 1, 0], sample_weight=[0.9, 0.5, 3.9, 1.2, 0.3])
>>> print(results)
{'precision': 0.23529411764705882}
Example 4-A multiclass example, with different values for the `average` input.
>>> predictions = [0, 2, 1, 0, 0, 1]
>>> references = [0, 1, 2, 0, 1, 2]
>>> results = precision_metric.compute(predictions=predictions, references=references, average='macro')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='micro')
>>> print(results)
{'precision': 0.3333333333333333}
>>> results = precision_metric.compute(predictions=predictions, references=references, average='weighted')
>>> print(results)
{'precision': 0.2222222222222222}
>>> results = precision_metric.compute(predictions=predictions, references=references, average=None)
>>> print([round(res, 2) for res in results['precision']])
[0.67, 0.0, 0.0]
"""
_CITATION = """
@article{scikit-learn,
title={Scikit-learn: Machine Learning in {P}ython},
author={Pedregosa, F. and Varoquaux, G. and Gramfort, A. and Michel, V.
and Thirion, B. and Grisel, O. and Blondel, M. and Prettenhofer, P.
and Weiss, R. and Dubourg, V. and Vanderplas, J. and Passos, A. and
Cournapeau, D. and Brucher, M. and Perrot, M. and Duchesnay, E.},
journal={Journal of Machine Learning Research},
volume={12},
pages={2825--2830},
year={2011}
}
"""
@datasets.utils.file_utils.add_start_docstrings(_DESCRIPTION, _KWARGS_DESCRIPTION)
class Precision(datasets.Metric):
def _info(self):
return datasets.MetricInfo(
description=_DESCRIPTION,
citation=_CITATION,
inputs_description=_KWARGS_DESCRIPTION,
features=datasets.Features(
{
"predictions": datasets.Sequence(datasets.Value("int32")),
"references": datasets.Sequence(datasets.Value("int32")),
}
if self.config_name == "multilabel"
else {
"predictions": datasets.Value("int32"),
"references": datasets.Value("int32"),
}
),
reference_urls=["https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_score.html"],
)
def _compute(
self,
predictions,
references,
labels=None,
pos_label=1,
average="binary",
sample_weight=None,
zero_division="warn",
):
score = precision_score(
references,
predictions,
labels=labels,
pos_label=pos_label,
average=average,
sample_weight=sample_weight,
zero_division=zero_division,
)
return {"precision": float(score) if score.size == 1 else score}
| datasets/metrics/precision/precision.py/0 | {
"file_path": "datasets/metrics/precision/precision.py",
"repo_id": "datasets",
"token_count": 2663
} | 63 |
"""Official evaluation script for v1.1 of the SQuAD dataset."""
import argparse
import json
import re
import string
import sys
from collections import Counter
def normalize_answer(s):
"""Lower text and remove punctuation, articles and extra whitespace."""
def remove_articles(text):
return re.sub(r"\b(a|an|the)\b", " ", text)
def white_space_fix(text):
return " ".join(text.split())
def remove_punc(text):
exclude = set(string.punctuation)
return "".join(ch for ch in text if ch not in exclude)
def lower(text):
return text.lower()
return white_space_fix(remove_articles(remove_punc(lower(s))))
def f1_score(prediction, ground_truth):
prediction_tokens = normalize_answer(prediction).split()
ground_truth_tokens = normalize_answer(ground_truth).split()
common = Counter(prediction_tokens) & Counter(ground_truth_tokens)
num_same = sum(common.values())
if num_same == 0:
return 0
precision = 1.0 * num_same / len(prediction_tokens)
recall = 1.0 * num_same / len(ground_truth_tokens)
f1 = (2 * precision * recall) / (precision + recall)
return f1
def exact_match_score(prediction, ground_truth):
return normalize_answer(prediction) == normalize_answer(ground_truth)
def metric_max_over_ground_truths(metric_fn, prediction, ground_truths):
scores_for_ground_truths = []
for ground_truth in ground_truths:
score = metric_fn(prediction, ground_truth)
scores_for_ground_truths.append(score)
return max(scores_for_ground_truths)
def evaluate(dataset, predictions):
f1 = exact_match = total = 0
for article in dataset:
for paragraph in article["paragraphs"]:
for qa in paragraph["qas"]:
total += 1
if qa["id"] not in predictions:
message = "Unanswered question " + qa["id"] + " will receive score 0."
print(message, file=sys.stderr)
continue
ground_truths = [x["text"] for x in qa["answers"]]
prediction = predictions[qa["id"]]
exact_match += metric_max_over_ground_truths(exact_match_score, prediction, ground_truths)
f1 += metric_max_over_ground_truths(f1_score, prediction, ground_truths)
exact_match = 100.0 * exact_match / total
f1 = 100.0 * f1 / total
return {"exact_match": exact_match, "f1": f1}
if __name__ == "__main__":
expected_version = "1.1"
parser = argparse.ArgumentParser(description="Evaluation for SQuAD " + expected_version)
parser.add_argument("dataset_file", help="Dataset file")
parser.add_argument("prediction_file", help="Prediction File")
args = parser.parse_args()
with open(args.dataset_file) as dataset_file:
dataset_json = json.load(dataset_file)
if dataset_json["version"] != expected_version:
print(
"Evaluation expects v-" + expected_version + ", but got dataset with v-" + dataset_json["version"],
file=sys.stderr,
)
dataset = dataset_json["data"]
with open(args.prediction_file) as prediction_file:
predictions = json.load(prediction_file)
print(json.dumps(evaluate(dataset, predictions)))
| datasets/metrics/squad/evaluate.py/0 | {
"file_path": "datasets/metrics/squad/evaluate.py",
"repo_id": "datasets",
"token_count": 1337
} | 64 |
# Metric Card for XTREME-S
## Metric Description
The XTREME-S metric aims to evaluate model performance on the Cross-lingual TRansfer Evaluation of Multilingual Encoders for Speech (XTREME-S) benchmark.
This benchmark was designed to evaluate speech representations across languages, tasks, domains and data regimes. It covers 102 languages from 10+ language families, 3 different domains and 4 task families: speech recognition, translation, classification and retrieval.
## How to Use
There are two steps: (1) loading the XTREME-S metric relevant to the subset of the benchmark being used for evaluation; and (2) calculating the metric.
1. **Loading the relevant XTREME-S metric** : the subsets of XTREME-S are the following: `mls`, `voxpopuli`, `covost2`, `fleurs-asr`, `fleurs-lang_id`, `minds14` and `babel`. More information about the different subsets can be found on the [XTREME-S benchmark page](https://huggingface.co/datasets/google/xtreme_s).
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'mls')
```
2. **Calculating the metric**: the metric takes two inputs :
- `predictions`: a list of predictions to score, with each prediction a `str`.
- `references`: a list of lists of references for each translation, with each reference a `str`.
```python
>>> references = ["it is sunny here", "paper and pen are essentials"]
>>> predictions = ["it's sunny", "paper pen are essential"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
```
It also has two optional arguments:
- `bleu_kwargs`: a `dict` of keywords to be passed when computing the `bleu` metric for the `covost2` subset. Keywords can be one of `smooth_method`, `smooth_value`, `force`, `lowercase`, `tokenize`, `use_effective_order`.
- `wer_kwargs`: optional dict of keywords to be passed when computing `wer` and `cer`, which are computed for the `mls`, `fleurs-asr`, `voxpopuli`, and `babel` subsets. Keywords are `concatenate_texts`.
## Output values
The output of the metric depends on the XTREME-S subset chosen, consisting of a dictionary that contains one or several of the following metrics:
- `accuracy`: the proportion of correct predictions among the total number of cases processed, with a range between 0 and 1 (see [accuracy](https://huggingface.co/metrics/accuracy) for more information). This is returned for the `fleurs-lang_id` and `minds14` subsets.
- `f1`: the harmonic mean of the precision and recall (see [F1 score](https://huggingface.co/metrics/f1) for more information). Its range is 0-1 -- its lowest possible value is 0, if either the precision or the recall is 0, and its highest possible value is 1.0, which means perfect precision and recall. It is returned for the `minds14` subset.
- `wer`: Word error rate (WER) is a common metric of the performance of an automatic speech recognition system. The lower the value, the better the performance of the ASR system, with a WER of 0 being a perfect score (see [WER score](https://huggingface.co/metrics/wer) for more information). It is returned for the `mls`, `fleurs-asr`, `voxpopuli` and `babel` subsets of the benchmark.
- `cer`: Character error rate (CER) is similar to WER, but operates on character instead of word. The lower the CER value, the better the performance of the ASR system, with a CER of 0 being a perfect score (see [CER score](https://huggingface.co/metrics/cer) for more information). It is returned for the `mls`, `fleurs-asr`, `voxpopuli` and `babel` subsets of the benchmark.
- `bleu`: the BLEU score, calculated according to the SacreBLEU metric approach. It can take any value between 0.0 and 100.0, inclusive, with higher values being better (see [SacreBLEU](https://huggingface.co/metrics/sacrebleu) for more details). This is returned for the `covost2` subset.
### Values from popular papers
The [original XTREME-S paper](https://arxiv.org/pdf/2203.10752.pdf) reported average WERs ranging from 9.2 to 14.6, a BLEU score of 20.6, an accuracy of 73.3 and F1 score of 86.9, depending on the subsets of the dataset tested on.
## Examples
For the `mls` subset (which outputs `wer` and `cer`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'mls')
>>> references = ["it is sunny here", "paper and pen are essentials"]
>>> predictions = ["it's sunny", "paper pen are essential"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'wer': 0.56, 'cer': 0.27}
```
For the `covost2` subset (which outputs `bleu`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'covost2')
>>> references = ["bonjour paris", "il est necessaire de faire du sport de temps en temp"]
>>> predictions = ["bonjour paris", "il est important de faire du sport souvent"]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'bleu': 31.65}
```
For the `fleurs-lang_id` subset (which outputs `accuracy`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'fleurs-lang_id')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'accuracy': 0.6}
```
For the `minds14` subset (which outputs `f1` and `accuracy`):
```python
>>> from datasets import load_metric
>>> xtreme_s_metric = datasets.load_metric('xtreme_s', 'minds14')
>>> references = [0, 1, 0, 0, 1]
>>> predictions = [0, 1, 1, 0, 0]
>>> results = xtreme_s_metric.compute(predictions=predictions, references=references)
>>> print({k: round(v, 2) for k, v in results.items()})
{'f1': 0.58, 'accuracy': 0.6}
```
## Limitations and bias
This metric works only with datasets that have the same format as the [XTREME-S dataset](https://huggingface.co/datasets/google/xtreme_s).
While the XTREME-S dataset is meant to represent a variety of languages and tasks, it has inherent biases: it is missing many languages that are important and under-represented in NLP datasets.
It also has a particular focus on read-speech because common evaluation benchmarks like CoVoST-2 or LibriSpeech evaluate on this type of speech, which results in a mismatch between performance obtained in a read-speech setting and a more noisy setting (in production or live deployment, for instance).
## Citation
```bibtex
@article{conneau2022xtreme,
title={XTREME-S: Evaluating Cross-lingual Speech Representations},
author={Conneau, Alexis and Bapna, Ankur and Zhang, Yu and Ma, Min and von Platen, Patrick and Lozhkov, Anton and Cherry, Colin and Jia, Ye and Rivera, Clara and Kale, Mihir and others},
journal={arXiv preprint arXiv:2203.10752},
year={2022}
}
```
## Further References
- [XTREME-S dataset](https://huggingface.co/datasets/google/xtreme_s)
- [XTREME-S github repository](https://github.com/google-research/xtreme)
| datasets/metrics/xtreme_s/README.md/0 | {
"file_path": "datasets/metrics/xtreme_s/README.md",
"repo_id": "datasets",
"token_count": 2218
} | 65 |
import platform
from argparse import ArgumentParser
import fsspec
import huggingface_hub
import pandas
import pyarrow
from datasets import __version__ as version
from datasets.commands import BaseDatasetsCLICommand
def info_command_factory(_):
return EnvironmentCommand()
class EnvironmentCommand(BaseDatasetsCLICommand):
@staticmethod
def register_subcommand(parser: ArgumentParser):
download_parser = parser.add_parser("env", help="Print relevant system environment info.")
download_parser.set_defaults(func=info_command_factory)
def run(self):
info = {
"`datasets` version": version,
"Platform": platform.platform(),
"Python version": platform.python_version(),
"`huggingface_hub` version": huggingface_hub.__version__,
"PyArrow version": pyarrow.__version__,
"Pandas version": pandas.__version__,
"`fsspec` version": fsspec.__version__,
}
print("\nCopy-and-paste the text below in your GitHub issue.\n")
print(self.format_dict(info))
return info
@staticmethod
def format_dict(d):
return "\n".join([f"- {prop}: {val}" for prop, val in d.items()]) + "\n"
| datasets/src/datasets/commands/env.py/0 | {
"file_path": "datasets/src/datasets/commands/env.py",
"repo_id": "datasets",
"token_count": 476
} | 66 |
import os
import sys
import warnings
from dataclasses import dataclass, field
from io import BytesIO
from typing import TYPE_CHECKING, Any, ClassVar, Dict, List, Optional, Union
import numpy as np
import pyarrow as pa
from .. import config
from ..download.download_config import DownloadConfig
from ..download.streaming_download_manager import xopen
from ..table import array_cast
from ..utils.file_utils import is_local_path
from ..utils.py_utils import first_non_null_value, no_op_if_value_is_null, string_to_dict
if TYPE_CHECKING:
import PIL.Image
from .features import FeatureType
_IMAGE_COMPRESSION_FORMATS: Optional[List[str]] = None
_NATIVE_BYTEORDER = "<" if sys.byteorder == "little" else ">"
# Origin: https://github.com/python-pillow/Pillow/blob/698951e19e19972aeed56df686868f1329981c12/src/PIL/Image.py#L3126 minus "|i1" which values are not preserved correctly when saving and loading an image
_VALID_IMAGE_ARRAY_DTPYES = [
np.dtype("|b1"),
np.dtype("|u1"),
np.dtype("<u2"),
np.dtype(">u2"),
np.dtype("<i2"),
np.dtype(">i2"),
np.dtype("<u4"),
np.dtype(">u4"),
np.dtype("<i4"),
np.dtype(">i4"),
np.dtype("<f4"),
np.dtype(">f4"),
np.dtype("<f8"),
np.dtype(">f8"),
]
@dataclass
class Image:
"""Image [`Feature`] to read image data from an image file.
Input: The Image feature accepts as input:
- A `str`: Absolute path to the image file (i.e. random access is allowed).
- A `dict` with the keys:
- `path`: String with relative path of the image file to the archive file.
- `bytes`: Bytes of the image file.
This is useful for archived files with sequential access.
- An `np.ndarray`: NumPy array representing an image.
- A `PIL.Image.Image`: PIL image object.
Args:
mode (`str`, *optional*):
The mode to convert the image to. If `None`, the native mode of the image is used.
decode (`bool`, defaults to `True`):
Whether to decode the image data. If `False`,
returns the underlying dictionary in the format `{"path": image_path, "bytes": image_bytes}`.
Examples:
```py
>>> from datasets import load_dataset, Image
>>> ds = load_dataset("beans", split="train")
>>> ds.features["image"]
Image(decode=True, id=None)
>>> ds[0]["image"]
<PIL.JpegImagePlugin.JpegImageFile image mode=RGB size=500x500 at 0x15E52E7F0>
>>> ds = ds.cast_column('image', Image(decode=False))
{'bytes': None,
'path': '/root/.cache/huggingface/datasets/downloads/extracted/b0a21163f78769a2cf11f58dfc767fb458fc7cea5c05dccc0144a2c0f0bc1292/train/healthy/healthy_train.85.jpg'}
```
"""
mode: Optional[str] = None
decode: bool = True
id: Optional[str] = None
# Automatically constructed
dtype: ClassVar[str] = "PIL.Image.Image"
pa_type: ClassVar[Any] = pa.struct({"bytes": pa.binary(), "path": pa.string()})
_type: str = field(default="Image", init=False, repr=False)
def __call__(self):
return self.pa_type
def encode_example(self, value: Union[str, bytes, dict, np.ndarray, "PIL.Image.Image"]) -> dict:
"""Encode example into a format for Arrow.
Args:
value (`str`, `np.ndarray`, `PIL.Image.Image` or `dict`):
Data passed as input to Image feature.
Returns:
`dict` with "path" and "bytes" fields
"""
if config.PIL_AVAILABLE:
import PIL.Image
else:
raise ImportError("To support encoding images, please install 'Pillow'.")
if isinstance(value, list):
value = np.array(value)
if isinstance(value, str):
return {"path": value, "bytes": None}
elif isinstance(value, bytes):
return {"path": None, "bytes": value}
elif isinstance(value, np.ndarray):
# convert the image array to PNG/TIFF bytes
return encode_np_array(value)
elif isinstance(value, PIL.Image.Image):
# convert the PIL image to bytes (default format is PNG/TIFF)
return encode_pil_image(value)
elif value.get("path") is not None and os.path.isfile(value["path"]):
# we set "bytes": None to not duplicate the data if they're already available locally
return {"bytes": None, "path": value.get("path")}
elif value.get("bytes") is not None or value.get("path") is not None:
# store the image bytes, and path is used to infer the image format using the file extension
return {"bytes": value.get("bytes"), "path": value.get("path")}
else:
raise ValueError(
f"An image sample should have one of 'path' or 'bytes' but they are missing or None in {value}."
)
def decode_example(self, value: dict, token_per_repo_id=None) -> "PIL.Image.Image":
"""Decode example image file into image data.
Args:
value (`str` or `dict`):
A string with the absolute image file path, a dictionary with
keys:
- `path`: String with absolute or relative image file path.
- `bytes`: The bytes of the image file.
token_per_repo_id (`dict`, *optional*):
To access and decode
image files from private repositories on the Hub, you can pass
a dictionary repo_id (`str`) -> token (`bool` or `str`).
Returns:
`PIL.Image.Image`
"""
if not self.decode:
raise RuntimeError("Decoding is disabled for this feature. Please use Image(decode=True) instead.")
if config.PIL_AVAILABLE:
import PIL.Image
import PIL.ImageOps
else:
raise ImportError("To support decoding images, please install 'Pillow'.")
if token_per_repo_id is None:
token_per_repo_id = {}
path, bytes_ = value["path"], value["bytes"]
if bytes_ is None:
if path is None:
raise ValueError(f"An image should have one of 'path' or 'bytes' but both are None in {value}.")
else:
if is_local_path(path):
image = PIL.Image.open(path)
else:
source_url = path.split("::")[-1]
pattern = (
config.HUB_DATASETS_URL
if source_url.startswith(config.HF_ENDPOINT)
else config.HUB_DATASETS_HFFS_URL
)
try:
repo_id = string_to_dict(source_url, pattern)["repo_id"]
token = token_per_repo_id.get(repo_id)
except ValueError:
token = None
download_config = DownloadConfig(token=token)
with xopen(path, "rb", download_config=download_config) as f:
bytes_ = BytesIO(f.read())
image = PIL.Image.open(bytes_)
else:
image = PIL.Image.open(BytesIO(bytes_))
image.load() # to avoid "Too many open files" errors
if image.getexif().get(PIL.Image.ExifTags.Base.Orientation) is not None:
image = PIL.ImageOps.exif_transpose(image)
if self.mode and self.mode != image.mode:
image = image.convert(self.mode)
return image
def flatten(self) -> Union["FeatureType", Dict[str, "FeatureType"]]:
"""If in the decodable state, return the feature itself, otherwise flatten the feature into a dictionary."""
from .features import Value
return (
self
if self.decode
else {
"bytes": Value("binary"),
"path": Value("string"),
}
)
def cast_storage(self, storage: Union[pa.StringArray, pa.StructArray, pa.ListArray]) -> pa.StructArray:
"""Cast an Arrow array to the Image arrow storage type.
The Arrow types that can be converted to the Image pyarrow storage type are:
- `pa.string()` - it must contain the "path" data
- `pa.binary()` - it must contain the image bytes
- `pa.struct({"bytes": pa.binary()})`
- `pa.struct({"path": pa.string()})`
- `pa.struct({"bytes": pa.binary(), "path": pa.string()})` - order doesn't matter
- `pa.list(*)` - it must contain the image array data
Args:
storage (`Union[pa.StringArray, pa.StructArray, pa.ListArray]`):
PyArrow array to cast.
Returns:
`pa.StructArray`: Array in the Image arrow storage type, that is
`pa.struct({"bytes": pa.binary(), "path": pa.string()})`.
"""
if pa.types.is_string(storage.type):
bytes_array = pa.array([None] * len(storage), type=pa.binary())
storage = pa.StructArray.from_arrays([bytes_array, storage], ["bytes", "path"], mask=storage.is_null())
elif pa.types.is_binary(storage.type):
path_array = pa.array([None] * len(storage), type=pa.string())
storage = pa.StructArray.from_arrays([storage, path_array], ["bytes", "path"], mask=storage.is_null())
elif pa.types.is_struct(storage.type):
if storage.type.get_field_index("bytes") >= 0:
bytes_array = storage.field("bytes")
else:
bytes_array = pa.array([None] * len(storage), type=pa.binary())
if storage.type.get_field_index("path") >= 0:
path_array = storage.field("path")
else:
path_array = pa.array([None] * len(storage), type=pa.string())
storage = pa.StructArray.from_arrays([bytes_array, path_array], ["bytes", "path"], mask=storage.is_null())
elif pa.types.is_list(storage.type):
bytes_array = pa.array(
[encode_np_array(np.array(arr))["bytes"] if arr is not None else None for arr in storage.to_pylist()],
type=pa.binary(),
)
path_array = pa.array([None] * len(storage), type=pa.string())
storage = pa.StructArray.from_arrays(
[bytes_array, path_array], ["bytes", "path"], mask=bytes_array.is_null()
)
return array_cast(storage, self.pa_type)
def embed_storage(self, storage: pa.StructArray) -> pa.StructArray:
"""Embed image files into the Arrow array.
Args:
storage (`pa.StructArray`):
PyArrow array to embed.
Returns:
`pa.StructArray`: Array in the Image arrow storage type, that is
`pa.struct({"bytes": pa.binary(), "path": pa.string()})`.
"""
@no_op_if_value_is_null
def path_to_bytes(path):
with xopen(path, "rb") as f:
bytes_ = f.read()
return bytes_
bytes_array = pa.array(
[
(path_to_bytes(x["path"]) if x["bytes"] is None else x["bytes"]) if x is not None else None
for x in storage.to_pylist()
],
type=pa.binary(),
)
path_array = pa.array(
[os.path.basename(path) if path is not None else None for path in storage.field("path").to_pylist()],
type=pa.string(),
)
storage = pa.StructArray.from_arrays([bytes_array, path_array], ["bytes", "path"], mask=bytes_array.is_null())
return array_cast(storage, self.pa_type)
def list_image_compression_formats() -> List[str]:
if config.PIL_AVAILABLE:
import PIL.Image
else:
raise ImportError("To support encoding images, please install 'Pillow'.")
global _IMAGE_COMPRESSION_FORMATS
if _IMAGE_COMPRESSION_FORMATS is None:
PIL.Image.init()
_IMAGE_COMPRESSION_FORMATS = list(set(PIL.Image.OPEN.keys()) & set(PIL.Image.SAVE.keys()))
return _IMAGE_COMPRESSION_FORMATS
def image_to_bytes(image: "PIL.Image.Image") -> bytes:
"""Convert a PIL Image object to bytes using native compression if possible, otherwise use PNG/TIFF compression."""
buffer = BytesIO()
if image.format in list_image_compression_formats():
format = image.format
else:
format = "PNG" if image.mode in ["1", "L", "LA", "RGB", "RGBA"] else "TIFF"
image.save(buffer, format=format)
return buffer.getvalue()
def encode_pil_image(image: "PIL.Image.Image") -> dict:
if hasattr(image, "filename") and image.filename != "":
return {"path": image.filename, "bytes": None}
else:
return {"path": None, "bytes": image_to_bytes(image)}
def encode_np_array(array: np.ndarray) -> dict:
if config.PIL_AVAILABLE:
import PIL.Image
else:
raise ImportError("To support encoding images, please install 'Pillow'.")
dtype = array.dtype
dtype_byteorder = dtype.byteorder if dtype.byteorder != "=" else _NATIVE_BYTEORDER
dtype_kind = dtype.kind
dtype_itemsize = dtype.itemsize
dest_dtype = None
# Multi-channel array case (only np.dtype("|u1") is allowed)
if array.shape[2:]:
if dtype_kind not in ["u", "i"]:
raise TypeError(
f"Unsupported array dtype {dtype} for image encoding. Only {dest_dtype} is supported for multi-channel arrays."
)
dest_dtype = np.dtype("|u1")
if dtype != dest_dtype:
warnings.warn(f"Downcasting array dtype {dtype} to {dest_dtype} to be compatible with 'Pillow'")
# Exact match
elif dtype in _VALID_IMAGE_ARRAY_DTPYES:
dest_dtype = dtype
else: # Downcast the type within the kind (np.can_cast(from_type, to_type, casting="same_kind") doesn't behave as expected, so do it manually)
while dtype_itemsize >= 1:
dtype_str = dtype_byteorder + dtype_kind + str(dtype_itemsize)
if np.dtype(dtype_str) in _VALID_IMAGE_ARRAY_DTPYES:
dest_dtype = np.dtype(dtype_str)
warnings.warn(f"Downcasting array dtype {dtype} to {dest_dtype} to be compatible with 'Pillow'")
break
else:
dtype_itemsize //= 2
if dest_dtype is None:
raise TypeError(
f"Cannot downcast dtype {dtype} to a valid image dtype. Valid image dtypes: {_VALID_IMAGE_ARRAY_DTPYES}"
)
image = PIL.Image.fromarray(array.astype(dest_dtype))
return {"path": None, "bytes": image_to_bytes(image)}
def objects_to_list_of_image_dicts(
objs: Union[List[str], List[dict], List[np.ndarray], List["PIL.Image.Image"]],
) -> List[dict]:
"""Encode a list of objects into a format suitable for creating an extension array of type `ImageExtensionType`."""
if config.PIL_AVAILABLE:
import PIL.Image
else:
raise ImportError("To support encoding images, please install 'Pillow'.")
if objs:
_, obj = first_non_null_value(objs)
if isinstance(obj, str):
return [{"path": obj, "bytes": None} if obj is not None else None for obj in objs]
if isinstance(obj, np.ndarray):
obj_to_image_dict_func = no_op_if_value_is_null(encode_np_array)
return [obj_to_image_dict_func(obj) for obj in objs]
elif isinstance(obj, PIL.Image.Image):
obj_to_image_dict_func = no_op_if_value_is_null(encode_pil_image)
return [obj_to_image_dict_func(obj) for obj in objs]
else:
return objs
else:
return objs
| datasets/src/datasets/features/image.py/0 | {
"file_path": "datasets/src/datasets/features/image.py",
"repo_id": "datasets",
"token_count": 6990
} | 67 |
from abc import ABC, abstractmethod
from typing import Optional, Union
from .. import Dataset, DatasetDict, Features, IterableDataset, IterableDatasetDict, NamedSplit
from ..utils.typing import NestedDataStructureLike, PathLike
class AbstractDatasetReader(ABC):
def __init__(
self,
path_or_paths: Optional[NestedDataStructureLike[PathLike]] = None,
split: Optional[NamedSplit] = None,
features: Optional[Features] = None,
cache_dir: str = None,
keep_in_memory: bool = False,
streaming: bool = False,
num_proc: Optional[int] = None,
**kwargs,
):
self.path_or_paths = path_or_paths
self.split = split if split or isinstance(path_or_paths, dict) else "train"
self.features = features
self.cache_dir = cache_dir
self.keep_in_memory = keep_in_memory
self.streaming = streaming
self.num_proc = num_proc
self.kwargs = kwargs
@abstractmethod
def read(self) -> Union[Dataset, DatasetDict, IterableDataset, IterableDatasetDict]:
pass
class AbstractDatasetInputStream(ABC):
def __init__(
self,
features: Optional[Features] = None,
cache_dir: str = None,
keep_in_memory: bool = False,
streaming: bool = False,
num_proc: Optional[int] = None,
**kwargs,
):
self.features = features
self.cache_dir = cache_dir
self.keep_in_memory = keep_in_memory
self.streaming = streaming
self.num_proc = num_proc
self.kwargs = kwargs
@abstractmethod
def read(self) -> Union[Dataset, IterableDataset]:
pass
| datasets/src/datasets/io/abc.py/0 | {
"file_path": "datasets/src/datasets/io/abc.py",
"repo_id": "datasets",
"token_count": 721
} | 68 |
import copy
import os
from functools import partial
from itertools import groupby
from typing import TYPE_CHECKING, Callable, Iterator, List, Optional, Tuple, TypeVar, Union
import numpy as np
import pyarrow as pa
import pyarrow.compute as pc
import pyarrow.types
from . import config
from .utils.logging import get_logger
if TYPE_CHECKING:
from .features.features import Features, FeatureType
logger = get_logger(__name__)
def inject_arrow_table_documentation(arrow_table_method):
def wrapper(fn):
fn.__doc__ = arrow_table_method.__doc__ + (fn.__doc__ if fn.__doc__ is not None else "")
fn.__doc__ = fn.__doc__.replace("pyarrow.Table", "Table")
if hasattr(arrow_table_method, "__annotations__"):
fn.__annotations__ = arrow_table_method.__annotations__
return fn
return wrapper
def _in_memory_arrow_table_from_file(filename: str) -> pa.Table:
in_memory_stream = pa.input_stream(filename)
opened_stream = pa.ipc.open_stream(in_memory_stream)
pa_table = opened_stream.read_all()
return pa_table
def _in_memory_arrow_table_from_buffer(buffer: pa.Buffer) -> pa.Table:
stream = pa.BufferReader(buffer)
opened_stream = pa.ipc.open_stream(stream)
table = opened_stream.read_all()
return table
def _memory_mapped_record_batch_reader_from_file(filename: str) -> pa.RecordBatchStreamReader:
memory_mapped_stream = pa.memory_map(filename)
return pa.ipc.open_stream(memory_mapped_stream)
def read_schema_from_file(filename: str) -> pa.Schema:
"""
Infer arrow table schema from file without loading whole file into memory.
Usefull especially while having very big files.
"""
with pa.memory_map(filename) as memory_mapped_stream:
schema = pa.ipc.open_stream(memory_mapped_stream).schema
return schema
def _memory_mapped_arrow_table_from_file(filename: str) -> pa.Table:
opened_stream = _memory_mapped_record_batch_reader_from_file(filename)
pa_table = opened_stream.read_all()
return pa_table
def _deepcopy(x, memo: dict):
"""deepcopy a regular class instance"""
cls = x.__class__
result = cls.__new__(cls)
memo[id(x)] = result
for k, v in x.__dict__.items():
setattr(result, k, copy.deepcopy(v, memo))
return result
def _interpolation_search(arr: List[int], x: int) -> int:
"""
Return the position i of a sorted array so that arr[i] <= x < arr[i+1]
Args:
arr (`List[int]`): non-empty sorted list of integers
x (`int`): query
Returns:
`int`: the position i so that arr[i] <= x < arr[i+1]
Raises:
`IndexError`: if the array is empty or if the query is outside the array values
"""
i, j = 0, len(arr) - 1
while i < j and arr[i] <= x < arr[j]:
k = i + ((j - i) * (x - arr[i]) // (arr[j] - arr[i]))
if arr[k] <= x < arr[k + 1]:
return k
elif arr[k] < x:
i, j = k + 1, j
else:
i, j = i, k
raise IndexError(f"Invalid query '{x}' for size {arr[-1] if len(arr) else 'none'}.")
class IndexedTableMixin:
def __init__(self, table: pa.Table):
self._schema: pa.Schema = table.schema
self._batches: List[pa.RecordBatch] = [
recordbatch for recordbatch in table.to_batches() if len(recordbatch) > 0
]
self._offsets: np.ndarray = np.cumsum([0] + [len(b) for b in self._batches], dtype=np.int64)
def fast_gather(self, indices: Union[List[int], np.ndarray]) -> pa.Table:
"""
Create a pa.Table by gathering the records at the records at the specified indices. Should be faster
than pa.concat_tables(table.fast_slice(int(i) % table.num_rows, 1) for i in indices) since NumPy can compute
the binary searches in parallel, highly optimized C
"""
if not len(indices):
raise ValueError("Indices must be non-empty")
batch_indices = np.searchsorted(self._offsets, indices, side="right") - 1
return pa.Table.from_batches(
[
self._batches[batch_idx].slice(i - self._offsets[batch_idx], 1)
for batch_idx, i in zip(batch_indices, indices)
],
schema=self._schema,
)
def fast_slice(self, offset=0, length=None) -> pa.Table:
"""
Slice the Table using interpolation search.
The behavior is the same as `pyarrow.Table.slice` but it's significantly faster.
Interpolation search is used to find the start and end indexes of the batches we want to keep.
The batches to keep are then concatenated to form the sliced Table.
"""
if offset < 0:
raise IndexError("Offset must be non-negative")
elif offset >= self._offsets[-1] or (length is not None and length <= 0):
return pa.Table.from_batches([], schema=self._schema)
i = _interpolation_search(self._offsets, offset)
if length is None or length + offset >= self._offsets[-1]:
batches = self._batches[i:]
batches[0] = batches[0].slice(offset - self._offsets[i])
else:
j = _interpolation_search(self._offsets, offset + length - 1)
batches = self._batches[i : j + 1]
batches[-1] = batches[-1].slice(0, offset + length - self._offsets[j])
batches[0] = batches[0].slice(offset - self._offsets[i])
return pa.Table.from_batches(batches, schema=self._schema)
class Table(IndexedTableMixin):
"""
Wraps a pyarrow Table by using composition.
This is the base class for `InMemoryTable`, `MemoryMappedTable` and `ConcatenationTable`.
It implements all the basic attributes/methods of the pyarrow Table class except
the Table transforms: `slice, filter, flatten, combine_chunks, cast, add_column,
append_column, remove_column, set_column, rename_columns` and `drop`.
The implementation of these methods differs for the subclasses.
"""
def __init__(self, table: pa.Table):
super().__init__(table)
self.table = table
def __deepcopy__(self, memo: dict):
# arrow tables are immutable, so there's no need to copy self.table
# moreover calling deepcopy on a pyarrow table seems to make pa.total_allocated_bytes() decrease for some reason
# by adding it to the memo, self.table won't be copied
memo[id(self.table)] = self.table
# same for the recordbatches used by the index
memo[id(self._batches)] = list(self._batches)
return _deepcopy(self, memo)
def validate(self, *args, **kwargs):
"""
Perform validation checks. An exception is raised if validation fails.
By default only cheap validation checks are run. Pass `full=True`
for thorough validation checks (potentially `O(n)`).
Args:
full (`bool`, defaults to `False`):
If `True`, run expensive checks, otherwise cheap checks only.
Raises:
`pa.lib.ArrowInvalid`: if validation fails
"""
return self.table.validate(*args, **kwargs)
def equals(self, *args, **kwargs):
"""
Check if contents of two tables are equal.
Args:
other ([`~datasets.table.Table`]):
Table to compare against.
check_metadata `bool`, defaults to `False`):
Whether schema metadata equality should be checked as well.
Returns:
`bool`
"""
args = tuple(arg.table if isinstance(arg, Table) else arg for arg in args)
kwargs = {k: v.table if isinstance(v, Table) else v for k, v in kwargs}
return self.table.equals(*args, **kwargs)
def to_batches(self, *args, **kwargs):
"""
Convert Table to list of (contiguous) `RecordBatch` objects.
Args:
max_chunksize (`int`, defaults to `None`):
Maximum size for `RecordBatch` chunks. Individual chunks may be
smaller depending on the chunk layout of individual columns.
Returns:
`List[pyarrow.RecordBatch]`
"""
return self.table.to_batches(*args, **kwargs)
def to_pydict(self, *args, **kwargs):
"""
Convert the Table to a `dict` or `OrderedDict`.
Returns:
`dict`
"""
return self.table.to_pydict(*args, **kwargs)
def to_pylist(self, *args, **kwargs):
"""
Convert the Table to a list
Returns:
`list`
"""
return self.table.to_pylist(*args, **kwargs)
def to_pandas(self, *args, **kwargs):
"""
Convert to a pandas-compatible NumPy array or DataFrame, as appropriate.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
Arrow MemoryPool to use for allocations. Uses the default memory
pool is not passed.
strings_to_categorical (`bool`, defaults to `False`):
Encode string (UTF8) and binary types to `pandas.Categorical`.
categories (`list`, defaults to `empty`):
List of fields that should be returned as `pandas.Categorical`. Only
applies to table-like data structures.
zero_copy_only (`bool`, defaults to `False`):
Raise an `ArrowException` if this function call would require copying
the underlying data.
integer_object_nulls (`bool`, defaults to `False`):
Cast integers with nulls to objects.
date_as_object (`bool`, defaults to `True`):
Cast dates to objects. If `False`, convert to `datetime64[ns]` dtype.
timestamp_as_object (`bool`, defaults to `False`):
Cast non-nanosecond timestamps (`np.datetime64`) to objects. This is
useful if you have timestamps that don't fit in the normal date
range of nanosecond timestamps (1678 CE-2262 CE).
If `False`, all timestamps are converted to `datetime64[ns]` dtype.
use_threads (`bool`, defaults to `True`):
Whether to parallelize the conversion using multiple threads.
deduplicate_objects (`bool`, defaults to `False`):
Do not create multiple copies Python objects when created, to save
on memory use. Conversion will be slower.
ignore_metadata (`bool`, defaults to `False`):
If `True`, do not use the 'pandas' metadata to reconstruct the
DataFrame index, if present.
safe (`bool`, defaults to `True`):
For certain data types, a cast is needed in order to store the
data in a pandas DataFrame or Series (e.g. timestamps are always
stored as nanoseconds in pandas). This option controls whether it
is a safe cast or not.
split_blocks (`bool`, defaults to `False`):
If `True`, generate one internal "block" for each column when
creating a pandas.DataFrame from a `RecordBatch` or `Table`. While this
can temporarily reduce memory note that various pandas operations
can trigger "consolidation" which may balloon memory use.
self_destruct (`bool`, defaults to `False`):
EXPERIMENTAL: If `True`, attempt to deallocate the originating Arrow
memory while converting the Arrow object to pandas. If you use the
object after calling `to_pandas` with this option it will crash your
program.
types_mapper (`function`, defaults to `None`):
A function mapping a pyarrow DataType to a pandas `ExtensionDtype`.
This can be used to override the default pandas type for conversion
of built-in pyarrow types or in absence of `pandas_metadata` in the
Table schema. The function receives a pyarrow DataType and is
expected to return a pandas `ExtensionDtype` or `None` if the
default conversion should be used for that type. If you have
a dictionary mapping, you can pass `dict.get` as function.
Returns:
`pandas.Series` or `pandas.DataFrame`: `pandas.Series` or `pandas.DataFrame` depending on type of object
"""
return self.table.to_pandas(*args, **kwargs)
def to_string(self, *args, **kwargs):
return self.table.to_string(*args, **kwargs)
def to_reader(self, max_chunksize: Optional[int] = None):
"""
Convert the Table to a RecordBatchReader.
Note that this method is zero-copy, it merely exposes the same data under a different API.
Args:
max_chunksize (`int`, defaults to `None`)
Maximum size for RecordBatch chunks. Individual chunks may be smaller depending
on the chunk layout of individual columns.
Returns:
`pyarrow.RecordBatchReader`
"""
return self.table.to_reader(max_chunksize=max_chunksize)
def field(self, *args, **kwargs):
"""
Select a schema field by its column name or numeric index.
Args:
i (`Union[int, str]`):
The index or name of the field to retrieve.
Returns:
`pyarrow.Field`
"""
return self.table.field(*args, **kwargs)
def column(self, *args, **kwargs):
"""
Select a column by its column name, or numeric index.
Args:
i (`Union[int, str]`):
The index or name of the column to retrieve.
Returns:
`pyarrow.ChunkedArray`
"""
return self.table.column(*args, **kwargs)
def itercolumns(self, *args, **kwargs):
"""
Iterator over all columns in their numerical order.
Yields:
`pyarrow.ChunkedArray`
"""
return self.table.itercolumns(*args, **kwargs)
@property
def schema(self):
"""
Schema of the table and its columns.
Returns:
`pyarrow.Schema`
"""
return self.table.schema
@property
def columns(self):
"""
List of all columns in numerical order.
Returns:
`List[pa.ChunkedArray]`
"""
return self.table.columns
@property
def num_columns(self):
"""
Number of columns in this table.
Returns:
int
"""
return self.table.num_columns
@property
def num_rows(self):
"""
Number of rows in this table.
Due to the definition of a table, all columns have the same number of
rows.
Returns:
int
"""
return self.table.num_rows
@property
def shape(self):
"""
Dimensions of the table: (#rows, #columns).
Returns:
`(int, int)`: Number of rows and number of columns.
"""
return self.table.shape
@property
def nbytes(self):
"""
Total number of bytes consumed by the elements of the table.
"""
return self.table.nbytes
@property
def column_names(self):
"""
Names of the table's columns.
"""
return self.table.column_names
def __eq__(self, other):
return self.equals(other)
def __getitem__(self, i):
return self.table[i]
def __len__(self):
return len(self.table)
def __repr__(self):
return self.table.__repr__().replace("pyarrow.Table", self.__class__.__name__)
def __str__(self):
return self.table.__str__().replace("pyarrow.Table", self.__class__.__name__)
def slice(self, *args, **kwargs):
"""
Compute zero-copy slice of this Table.
Args:
offset (`int`, defaults to `0`):
Offset from start of table to slice.
length (`int`, defaults to `None`):
Length of slice (default is until end of table starting from
offset).
Returns:
`datasets.table.Table`
"""
raise NotImplementedError()
def filter(self, *args, **kwargs):
"""
Select records from a Table. See `pyarrow.compute.filter` for full usage.
"""
raise NotImplementedError()
def flatten(self, *args, **kwargs):
"""
Flatten this Table. Each column with a struct type is flattened
into one column per struct field. Other columns are left unchanged.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
raise NotImplementedError()
def combine_chunks(self, *args, **kwargs):
"""
Make a new table by combining the chunks this table has.
All the underlying chunks in the `ChunkedArray` of each column are
concatenated into zero or one chunk.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
raise NotImplementedError()
def cast(self, *args, **kwargs):
"""
Cast table values to another schema.
Args:
target_schema (`Schema`):
Schema to cast to, the names and order of fields must match.
safe (`bool`, defaults to `True`):
Check for overflows or other unsafe conversions.
Returns:
`datasets.table.Table`
"""
raise NotImplementedError()
def replace_schema_metadata(self, *args, **kwargs):
"""
EXPERIMENTAL: Create shallow copy of table by replacing schema
key-value metadata with the indicated new metadata (which may be None,
which deletes any existing metadata
Args:
metadata (`dict`, defaults to `None`):
Returns:
`datasets.table.Table`: shallow_copy
"""
raise NotImplementedError()
def add_column(self, *args, **kwargs):
"""
Add column to Table at position.
A new table is returned with the column added, the original table
object is left unchanged.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column added.
"""
raise NotImplementedError()
def append_column(self, *args, **kwargs):
"""
Append column at end of columns.
Args:
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column added.
"""
raise NotImplementedError()
def remove_column(self, *args, **kwargs):
"""
Create new Table with the indicated column removed.
Args:
i (`int`):
Index of column to remove.
Returns:
`datasets.table.Table`: New table without the column.
"""
raise NotImplementedError()
def set_column(self, *args, **kwargs):
"""
Replace column in Table at position.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column set.
"""
raise NotImplementedError()
def rename_columns(self, *args, **kwargs):
"""
Create new table with columns renamed to provided names.
"""
raise NotImplementedError()
def drop(self, *args, **kwargs):
"""
Drop one or more columns and return a new table.
Args:
columns (`List[str]`):
List of field names referencing existing columns.
Raises:
`KeyError` : if any of the passed columns name are not existing.
Returns:
`datasets.table.Table`: New table without the columns.
"""
raise NotImplementedError()
def select(self, *args, **kwargs):
"""
Select columns of the table.
Returns a new table with the specified columns, and metadata preserved.
Args:
columns (:obj:`Union[List[str], List[int]]`):
The column names or integer indices to select.
Returns:
`datasets.table.Table`: table with only a subset of the columns
"""
raise NotImplementedError()
class TableBlock(Table):
"""
`TableBlock` is the allowed class inside a `ConcanetationTable`.
Only `MemoryMappedTable` and `InMemoryTable` are `TableBlock`.
This is because we don't want a `ConcanetationTable` made out of other `ConcanetationTables`.
"""
pass
class InMemoryTable(TableBlock):
"""
The table is said in-memory when it is loaded into the user's RAM.
Pickling it does copy all the data using memory.
Its implementation is simple and uses the underlying pyarrow Table methods directly.
This is different from the `MemoryMapped` table, for which pickling doesn't copy all the
data in memory. For a `MemoryMapped`, unpickling instead reloads the table from the disk.
`InMemoryTable` must be used when data fit in memory, while `MemoryMapped` are reserved for
data bigger than memory or when you want the memory footprint of your application to
stay low.
"""
@classmethod
def from_file(cls, filename: str):
table = _in_memory_arrow_table_from_file(filename)
return cls(table)
@classmethod
def from_buffer(cls, buffer: pa.Buffer):
table = _in_memory_arrow_table_from_buffer(buffer)
return cls(table)
@classmethod
def from_pandas(cls, *args, **kwargs):
"""
Convert pandas.DataFrame to an Arrow Table.
The column types in the resulting Arrow Table are inferred from the
dtypes of the pandas.Series in the DataFrame. In the case of non-object
Series, the NumPy dtype is translated to its Arrow equivalent. In the
case of `object`, we need to guess the datatype by looking at the
Python objects in this Series.
Be aware that Series of the `object` dtype don't carry enough
information to always lead to a meaningful Arrow type. In the case that
we cannot infer a type, e.g. because the DataFrame is of length 0 or
the Series only contains `None/nan` objects, the type is set to
null. This behavior can be avoided by constructing an explicit schema
and passing it to this function.
Args:
df (`pandas.DataFrame`):
schema (`pyarrow.Schema`, *optional*):
The expected schema of the Arrow Table. This can be used to
indicate the type of columns if we cannot infer it automatically.
If passed, the output will have exactly this schema. Columns
specified in the schema that are not found in the DataFrame columns
or its index will raise an error. Additional columns or index
levels in the DataFrame which are not specified in the schema will
be ignored.
preserve_index (`bool`, *optional*):
Whether to store the index as an additional column in the resulting
`Table`. The default of None will store the index as a column,
except for RangeIndex which is stored as metadata only. Use
`preserve_index=True` to force it to be stored as a column.
nthreads (`int`, defaults to `None` (may use up to system CPU count threads))
If greater than 1, convert columns to Arrow in parallel using
indicated number of threads.
columns (`List[str]`, *optional*):
List of column to be converted. If `None`, use all columns.
safe (`bool`, defaults to `True`):
Check for overflows or other unsafe conversions,
Returns:
`datasets.table.Table`:
Examples:
```python
>>> import pandas as pd
>>> import pyarrow as pa
>>> df = pd.DataFrame({
... 'int': [1, 2],
... 'str': ['a', 'b']
... })
>>> pa.Table.from_pandas(df)
<pyarrow.lib.Table object at 0x7f05d1fb1b40>
```
"""
return cls(pa.Table.from_pandas(*args, **kwargs))
@classmethod
def from_arrays(cls, *args, **kwargs):
"""
Construct a Table from Arrow arrays.
Args:
arrays (`List[Union[pyarrow.Array, pyarrow.ChunkedArray]]`):
Equal-length arrays that should form the table.
names (`List[str]`, *optional*):
Names for the table columns. If not passed, schema must be passed.
schema (`Schema`, defaults to `None`):
Schema for the created table. If not passed, names must be passed.
metadata (`Union[dict, Mapping]`, defaults to `None`):
Optional metadata for the schema (if inferred).
Returns:
`datasets.table.Table`
"""
return cls(pa.Table.from_arrays(*args, **kwargs))
@classmethod
def from_pydict(cls, *args, **kwargs):
"""
Construct a Table from Arrow arrays or columns.
Args:
mapping (`Union[dict, Mapping]`):
A mapping of strings to Arrays or Python lists.
schema (`Schema`, defaults to `None`):
If not passed, will be inferred from the Mapping values
metadata (`Union[dict, Mapping]`, defaults to `None`):
Optional metadata for the schema (if inferred).
Returns:
`datasets.table.Table`
"""
return cls(pa.Table.from_pydict(*args, **kwargs))
@classmethod
def from_pylist(cls, mapping, *args, **kwargs):
"""
Construct a Table from list of rows / dictionaries.
Args:
mapping (`List[dict]`):
A mapping of strings to row values.
schema (`Schema`, defaults to `None`):
If not passed, will be inferred from the Mapping values
metadata (`Union[dict, Mapping]`, defaults to `None`):
Optional metadata for the schema (if inferred).
Returns:
`datasets.table.Table`
"""
return cls(pa.Table.from_pylist(mapping, *args, **kwargs))
@classmethod
def from_batches(cls, *args, **kwargs):
"""
Construct a Table from a sequence or iterator of Arrow `RecordBatches`.
Args:
batches (`Union[Sequence[pyarrow.RecordBatch], Iterator[pyarrow.RecordBatch]]`):
Sequence of `RecordBatch` to be converted, all schemas must be equal.
schema (`Schema`, defaults to `None`):
If not passed, will be inferred from the first `RecordBatch`.
Returns:
`datasets.table.Table`:
"""
return cls(pa.Table.from_batches(*args, **kwargs))
def slice(self, offset=0, length=None):
"""
Compute zero-copy slice of this Table.
Args:
offset (`int`, defaults to `0`):
Offset from start of table to slice.
length (`int`, defaults to `None`):
Length of slice (default is until end of table starting from
offset).
Returns:
`datasets.table.Table`
"""
# Use fast slicing here
return InMemoryTable(self.fast_slice(offset=offset, length=length))
def filter(self, *args, **kwargs):
"""
Select records from a Table. See `pyarrow.compute.filter` for full usage.
"""
return InMemoryTable(self.table.filter(*args, **kwargs))
def flatten(self, *args, **kwargs):
"""
Flatten this Table. Each column with a struct type is flattened
into one column per struct field. Other columns are left unchanged.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
return InMemoryTable(table_flatten(self.table, *args, **kwargs))
def combine_chunks(self, *args, **kwargs):
"""
Make a new table by combining the chunks this table has.
All the underlying chunks in the `ChunkedArray` of each column are
concatenated into zero or one chunk.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
return InMemoryTable(self.table.combine_chunks(*args, **kwargs))
def cast(self, *args, **kwargs):
"""
Cast table values to another schema.
Args:
target_schema (`Schema`):
Schema to cast to, the names and order of fields must match.
safe (`bool`, defaults to `True`):
Check for overflows or other unsafe conversions.
Returns:
`datasets.table.Table`
"""
return InMemoryTable(table_cast(self.table, *args, **kwargs))
def replace_schema_metadata(self, *args, **kwargs):
"""
EXPERIMENTAL: Create shallow copy of table by replacing schema
key-value metadata with the indicated new metadata (which may be `None`,
which deletes any existing metadata).
Args:
metadata (`dict`, defaults to `None`):
Returns:
`datasets.table.Table`: shallow_copy
"""
return InMemoryTable(self.table.replace_schema_metadata(*args, **kwargs))
def add_column(self, *args, **kwargs):
"""
Add column to Table at position.
A new table is returned with the column added, the original table
object is left unchanged.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column added.
"""
return InMemoryTable(self.table.add_column(*args, **kwargs))
def append_column(self, *args, **kwargs):
"""
Append column at end of columns.
Args:
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column added.
"""
return InMemoryTable(self.table.append_column(*args, **kwargs))
def remove_column(self, *args, **kwargs):
"""
Create new Table with the indicated column removed.
Args:
i (`int`):
Index of column to remove.
Returns:
`datasets.table.Table`:
New table without the column.
"""
return InMemoryTable(self.table.remove_column(*args, **kwargs))
def set_column(self, *args, **kwargs):
"""
Replace column in Table at position.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column set.
"""
return InMemoryTable(self.table.set_column(*args, **kwargs))
def rename_columns(self, *args, **kwargs):
"""
Create new table with columns renamed to provided names.
"""
return InMemoryTable(self.table.rename_columns(*args, **kwargs))
def drop(self, *args, **kwargs):
"""
Drop one or more columns and return a new table.
Args:
columns (`List[str]`):
List of field names referencing existing columns.
Raises:
`KeyError` : if any of the passed columns name are not existing.
Returns:
`datasets.table.Table`:
New table without the columns.
"""
return InMemoryTable(self.table.drop(*args, **kwargs))
def select(self, *args, **kwargs):
"""
Select columns of the table.
Returns a new table with the specified columns, and metadata preserved.
Args:
columns (:obj:`Union[List[str], List[int]]`):
The column names or integer indices to select.
Returns:
:class:`datasets.table.Table`: New table with the specified columns, and metadata preserved.
"""
return InMemoryTable(self.table.select(*args, **kwargs))
# The MemoryMappedTable needs replays to properly reload tables from the disk
Replay = Tuple[str, tuple, dict]
class MemoryMappedTable(TableBlock):
"""
The table is said memory mapped when it doesn't use the user's RAM but loads the data
from the disk instead.
Pickling it doesn't copy the data into memory.
Instead, only the path to the memory mapped arrow file is pickled, as well as the list
of transforms to "replay" when reloading the table from the disk.
Its implementation requires to store an history of all the transforms that were applied
to the underlying pyarrow Table, so that they can be "replayed" when reloading the Table
from the disk.
This is different from the `InMemoryTable` table, for which pickling does copy all the
data in memory.
`InMemoryTable` must be used when data fit in memory, while `MemoryMapped` are reserved for
data bigger than memory or when you want the memory footprint of your application to
stay low.
"""
def __init__(self, table: pa.Table, path: str, replays: Optional[List[Replay]] = None):
super().__init__(table)
self.path = os.path.abspath(path)
self.replays: List[Replay] = replays if replays is not None else []
@classmethod
def from_file(cls, filename: str, replays=None):
table = _memory_mapped_arrow_table_from_file(filename)
table = cls._apply_replays(table, replays)
return cls(table, filename, replays)
def __getstate__(self):
return {"path": self.path, "replays": self.replays}
def __setstate__(self, state):
path = state["path"]
replays = state["replays"]
table = _memory_mapped_arrow_table_from_file(path)
table = self._apply_replays(table, replays)
MemoryMappedTable.__init__(self, table, path=path, replays=replays)
@staticmethod
def _apply_replays(table: pa.Table, replays: Optional[List[Replay]] = None) -> pa.Table:
if replays is not None:
for name, args, kwargs in replays:
if name == "cast":
table = table_cast(table, *args, **kwargs)
elif name == "flatten":
table = table_flatten(table, *args, **kwargs)
else:
table = getattr(table, name)(*args, **kwargs)
return table
def _append_replay(self, replay: Replay) -> List[Replay]:
replays = copy.deepcopy(self.replays)
replays.append(replay)
return replays
def slice(self, offset=0, length=None):
"""
Compute zero-copy slice of this Table.
Args:
offset (`int`, defaults to `0`):
Offset from start of table to slice.
length (`int`, defaults to `None`):
Length of slice (default is until end of table starting from
offset).
Returns:
`datasets.table.Table`
"""
replay = ("slice", (offset, length), {})
replays = self._append_replay(replay)
# Use fast slicing here
return MemoryMappedTable(self.fast_slice(offset=offset, length=length), self.path, replays)
def filter(self, *args, **kwargs):
"""
Select records from a Table. See `pyarrow.compute.filter` for full usage.
"""
replay = ("filter", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.filter(*args, **kwargs), self.path, replays)
def flatten(self, *args, **kwargs):
"""
Flatten this Table. Each column with a struct type is flattened
into one column per struct field. Other columns are left unchanged.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
replay = ("flatten", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(table_flatten(self.table, *args, **kwargs), self.path, replays)
def combine_chunks(self, *args, **kwargs):
"""
Make a new table by combining the chunks this table has.
All the underlying chunks in the ChunkedArray of each column are
concatenated into zero or one chunk.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
replay = ("combine_chunks", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.combine_chunks(*args, **kwargs), self.path, replays)
def cast(self, *args, **kwargs):
"""
Cast table values to another schema
Args:
target_schema (`Schema`):
Schema to cast to, the names and order of fields must match.
safe (`bool`, defaults to `True`):
Check for overflows or other unsafe conversions.
Returns:
`datasets.table.Table`
"""
replay = ("cast", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(table_cast(self.table, *args, **kwargs), self.path, replays)
def replace_schema_metadata(self, *args, **kwargs):
"""
EXPERIMENTAL: Create shallow copy of table by replacing schema
key-value metadata with the indicated new metadata (which may be None,
which deletes any existing metadata.
Args:
metadata (`dict`, defaults to `None`):
Returns:
`datasets.table.Table`: shallow_copy
"""
replay = ("replace_schema_metadata", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.replace_schema_metadata(*args, **kwargs), self.path, replays)
def add_column(self, *args, **kwargs):
"""
Add column to Table at position.
A new table is returned with the column added, the original table
object is left unchanged.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column added.
"""
replay = ("add_column", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.add_column(*args, **kwargs), self.path, replays)
def append_column(self, *args, **kwargs):
"""
Append column at end of columns.
Args:
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column added.
"""
replay = ("append_column", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.append_column(*args, **kwargs), self.path, replays)
def remove_column(self, *args, **kwargs):
"""
Create new Table with the indicated column removed.
Args:
i (`int`):
Index of column to remove.
Returns:
`datasets.table.Table`:
New table without the column.
"""
replay = ("remove_column", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.remove_column(*args, **kwargs), self.path, replays)
def set_column(self, *args, **kwargs):
"""
Replace column in Table at position.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column set.
"""
replay = ("set_column", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.set_column(*args, **kwargs), self.path, replays)
def rename_columns(self, *args, **kwargs):
"""
Create new table with columns renamed to provided names.
"""
replay = ("rename_columns", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.rename_columns(*args, **kwargs), self.path, replays)
def drop(self, *args, **kwargs):
"""
Drop one or more columns and return a new table.
Args:
columns (`List[str]`):
List of field names referencing existing columns.
Raises:
`KeyError` : if any of the passed columns name are not existing.
Returns:
`datasets.table.Table`:
New table without the columns.
"""
replay = ("drop", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.drop(*args, **kwargs), self.path, replays)
def select(self, *args, **kwargs):
"""
Select columns of the table.
Returns a new table with the specified columns, and metadata preserved.
Args:
columns (:obj:`Union[List[str], List[int]]`):
The column names or integer indices to select.
Returns:
:class:`datasets.table.Table`: New table with the specified columns, and metadata preserved.
"""
replay = ("select", copy.deepcopy(args), copy.deepcopy(kwargs))
replays = self._append_replay(replay)
return MemoryMappedTable(self.table.select(*args, **kwargs), self.path, replays)
# A ConcatenationTable is the concatenation of several tables.
# The ``blocks`` attributes stores a list of list of blocks.
# The first axis concatenates the tables along the axis 0 (it appends rows),
# while the second axis concatenates tables along the axis 1 (it appends columns).
TableBlockContainer = TypeVar("TableBlockContainer", TableBlock, List[TableBlock], List[List[TableBlock]])
class ConcatenationTable(Table):
"""
The table comes from the concatenation of several tables called blocks.
It enables concatenation on both axis 0 (append rows) and axis 1 (append columns).
The underlying tables are called "blocks" and can be either `InMemoryTable`
or `MemoryMappedTable` objects.
This allows to combine tables that come from memory or that are memory mapped.
When a `ConcatenationTable` is pickled, then each block is pickled:
- the `InMemoryTable` objects are pickled by copying all the data in memory.
- the MemoryMappedTable objects are pickled without copying the data into memory.
Instead, only the path to the memory mapped arrow file is pickled, as well as the list
of transforms to "replays" when reloading the table from the disk.
Its implementation requires to store each block separately.
The `blocks` attributes stores a list of list of blocks.
The first axis concatenates the tables along the axis 0 (it appends rows),
while the second axis concatenates tables along the axis 1 (it appends columns).
If some columns are missing when concatenating on axis 0, they are filled with null values.
This is done using `pyarrow.concat_tables(tables, promote=True)`.
You can access the fully combined table by accessing the `ConcatenationTable.table` attribute,
and the blocks by accessing the `ConcatenationTable.blocks` attribute.
"""
def __init__(self, table: pa.Table, blocks: List[List[TableBlock]]):
super().__init__(table)
self.blocks = blocks
# Check that all the blocks have the right type.
# Only InMemoryTable and MemoryMappedTable are allowed.
for subtables in blocks:
for subtable in subtables:
if not isinstance(subtable, TableBlock):
raise TypeError(
"The blocks of a ConcatenationTable must be InMemoryTable or MemoryMappedTable objects"
f", but got {subtable}."
)
def __getstate__(self):
return {"blocks": self.blocks, "schema": self.table.schema}
def __setstate__(self, state):
blocks = state["blocks"]
schema = state["schema"]
table = self._concat_blocks_horizontally_and_vertically(blocks)
if schema is not None and table.schema != schema:
# We fix the columns by concatenating with an empty table with the right columns
empty_table = pa.Table.from_batches([], schema=schema)
# we set promote=True to fill missing columns with null values
if config.PYARROW_VERSION.major < 14:
table = pa.concat_tables([table, empty_table], promote=True)
else:
table = pa.concat_tables([table, empty_table], promote_options="default")
ConcatenationTable.__init__(self, table, blocks=blocks)
@staticmethod
def _concat_blocks(blocks: List[Union[TableBlock, pa.Table]], axis: int = 0) -> pa.Table:
pa_tables = [table.table if hasattr(table, "table") else table for table in blocks]
if axis == 0:
# we set promote=True to fill missing columns with null values
if config.PYARROW_VERSION.major < 14:
return pa.concat_tables(pa_tables, promote=True)
else:
return pa.concat_tables(pa_tables, promote_options="default")
elif axis == 1:
for i, table in enumerate(pa_tables):
if i == 0:
pa_table = table
else:
for name, col in zip(table.column_names, table.columns):
pa_table = pa_table.append_column(name, col)
return pa_table
else:
raise ValueError("'axis' must be either 0 or 1")
@classmethod
def _concat_blocks_horizontally_and_vertically(cls, blocks: List[List[TableBlock]]) -> pa.Table:
pa_tables_to_concat_vertically = []
for i, tables in enumerate(blocks):
if not tables:
continue
pa_table_horizontally_concatenated = cls._concat_blocks(tables, axis=1)
pa_tables_to_concat_vertically.append(pa_table_horizontally_concatenated)
return cls._concat_blocks(pa_tables_to_concat_vertically, axis=0)
@classmethod
def _merge_blocks(cls, blocks: TableBlockContainer, axis: Optional[int] = None) -> TableBlockContainer:
if axis is not None:
merged_blocks = []
for is_in_memory, block_group in groupby(blocks, key=lambda x: isinstance(x, InMemoryTable)):
if is_in_memory:
block_group = [InMemoryTable(cls._concat_blocks(list(block_group), axis=axis))]
merged_blocks += list(block_group)
else: # both
merged_blocks = [cls._merge_blocks(row_block, axis=1) for row_block in blocks]
if all(len(row_block) == 1 for row_block in merged_blocks):
merged_blocks = cls._merge_blocks(
[block for row_block in merged_blocks for block in row_block], axis=0
)
return merged_blocks
@classmethod
def _consolidate_blocks(cls, blocks: TableBlockContainer) -> TableBlockContainer:
if isinstance(blocks, TableBlock):
return blocks
elif isinstance(blocks[0], TableBlock):
return cls._merge_blocks(blocks, axis=0)
else:
return cls._merge_blocks(blocks)
@classmethod
def from_blocks(cls, blocks: TableBlockContainer) -> "ConcatenationTable":
blocks = cls._consolidate_blocks(blocks)
if isinstance(blocks, TableBlock):
table = blocks
return cls(table.table, [[table]])
elif isinstance(blocks[0], TableBlock):
table = cls._concat_blocks(blocks, axis=0)
blocks = [[t] for t in blocks]
return cls(table, blocks)
else:
table = cls._concat_blocks_horizontally_and_vertically(blocks)
return cls(table, blocks)
@classmethod
def from_tables(cls, tables: List[Union[pa.Table, Table]], axis: int = 0) -> "ConcatenationTable":
"""Create `ConcatenationTable` from list of tables.
Args:
tables (list of `Table` or list of `pyarrow.Table`):
List of tables.
axis (`{0, 1}`, defaults to `0`, meaning over rows):
Axis to concatenate over, where `0` means over rows (vertically) and `1` means over columns
(horizontally).
<Added version="1.6.0"/>
"""
def to_blocks(table: Union[pa.Table, Table]) -> List[List[TableBlock]]:
if isinstance(table, pa.Table):
return [[InMemoryTable(table)]]
elif isinstance(table, ConcatenationTable):
return copy.deepcopy(table.blocks)
else:
return [[table]]
def _slice_row_block(row_block: List[TableBlock], length: int) -> Tuple[List[TableBlock], List[TableBlock]]:
sliced = [table.slice(0, length) for table in row_block]
remainder = [table.slice(length, len(row_block[0]) - length) for table in row_block]
return sliced, remainder
def _split_both_like(
result: List[List[TableBlock]], blocks: List[List[TableBlock]]
) -> Tuple[List[List[TableBlock]], List[List[TableBlock]]]:
"""
Make sure each row_block contain the same num_rows to be able to concatenate them on axis=1.
To do so, we modify both blocks sets to have the same row_blocks boundaries.
For example, if `result` has 2 row_blocks of 3 rows and `blocks` has 3 row_blocks of 2 rows,
we modify both to have 4 row_blocks of size 2, 1, 1 and 2:
[ x x x | x x x ]
+ [ y y | y y | y y ]
-----------------------------
= [ x x | x | x | x x ]
[ y y | y | y | y y ]
"""
result, blocks = list(result), list(blocks)
new_result, new_blocks = [], []
while result and blocks:
# we slice the longest row block to save two row blocks of same length
# and we replace the long row block by its remainder if necessary
if len(result[0][0]) > len(blocks[0][0]):
new_blocks.append(blocks[0])
sliced, result[0] = _slice_row_block(result[0], len(blocks.pop(0)[0]))
new_result.append(sliced)
elif len(result[0][0]) < len(blocks[0][0]):
new_result.append(result[0])
sliced, blocks[0] = _slice_row_block(blocks[0], len(result.pop(0)[0]))
new_blocks.append(sliced)
else:
new_result.append(result.pop(0))
new_blocks.append(blocks.pop(0))
if result or blocks:
raise ValueError("Failed to concatenate on axis=1 because tables don't have the same number of rows")
return new_result, new_blocks
def _extend_blocks(
result: List[List[TableBlock]], blocks: List[List[TableBlock]], axis: int = 0
) -> List[List[TableBlock]]:
if axis == 0:
result.extend(blocks)
elif axis == 1:
# We make sure each row_block have the same num_rows
result, blocks = _split_both_like(result, blocks)
for i, row_block in enumerate(blocks):
result[i].extend(row_block)
return result
blocks = to_blocks(tables[0])
for table in tables[1:]:
table_blocks = to_blocks(table)
blocks = _extend_blocks(blocks, table_blocks, axis=axis)
return cls.from_blocks(blocks)
@property
def _slices(self):
offset = 0
for tables in self.blocks:
length = len(tables[0])
yield (offset, length)
offset += length
def slice(self, offset=0, length=None):
"""
Compute zero-copy slice of this Table.
Args:
offset (`int`, defaults to `0`):
Offset from start of table to slice.
length (`int`, defaults to `None`):
Length of slice (default is until end of table starting from
offset).
Returns:
`datasets.table.Table`
"""
table = self.table.slice(offset, length=length)
length = length if length is not None else self.num_rows - offset
blocks = []
for tables in self.blocks:
n_rows = len(tables[0])
if length == 0:
break
elif n_rows <= offset:
offset = offset - n_rows
elif n_rows <= offset + length:
blocks.append([t.slice(offset) for t in tables])
length, offset = length + offset - n_rows, 0
else:
blocks.append([t.slice(offset, length) for t in tables])
length, offset = 0, 0
return ConcatenationTable(table, blocks)
def filter(self, mask, *args, **kwargs):
"""
Select records from a Table. See `pyarrow.compute.filter` for full usage.
"""
table = self.table.filter(mask, *args, **kwargs)
blocks = []
for (offset, length), tables in zip(self._slices, self.blocks):
submask = mask.slice(offset, length)
blocks.append([t.filter(submask, *args, **kwargs) for t in tables])
return ConcatenationTable(table, blocks)
def flatten(self, *args, **kwargs):
"""
Flatten this Table. Each column with a struct type is flattened
into one column per struct field. Other columns are left unchanged.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
table = table_flatten(self.table, *args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.flatten(*args, **kwargs) for t in tables])
return ConcatenationTable(table, blocks)
def combine_chunks(self, *args, **kwargs):
"""
Make a new table by combining the chunks this table has.
All the underlying chunks in the `ChunkedArray` of each column are
concatenated into zero or one chunk.
Args:
memory_pool (`MemoryPool`, defaults to `None`):
For memory allocations, if required, otherwise use default pool.
Returns:
`datasets.table.Table`
"""
table = self.table.combine_chunks(*args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.combine_chunks(*args, **kwargs) for t in tables])
return ConcatenationTable(table, blocks)
def cast(self, target_schema, *args, **kwargs):
"""
Cast table values to another schema.
Args:
target_schema (`Schema`):
Schema to cast to, the names and order of fields must match.
safe (`bool`, defaults to `True`):
Check for overflows or other unsafe conversions.
Returns:
`datasets.table.Table`
"""
from .features import Features
table = table_cast(self.table, target_schema, *args, **kwargs)
target_features = Features.from_arrow_schema(target_schema)
blocks = []
for subtables in self.blocks:
new_tables = []
fields = list(target_schema)
for subtable in subtables:
subfields = []
for name in subtable.column_names:
subfields.append(fields.pop(next(i for i, field in enumerate(fields) if field.name == name)))
subfeatures = Features({subfield.name: target_features[subfield.name] for subfield in subfields})
subschema = subfeatures.arrow_schema
new_tables.append(subtable.cast(subschema, *args, **kwargs))
blocks.append(new_tables)
return ConcatenationTable(table, blocks)
def replace_schema_metadata(self, *args, **kwargs):
"""
EXPERIMENTAL: Create shallow copy of table by replacing schema
key-value metadata with the indicated new metadata (which may be `None`,
which deletes any existing metadata).
Args:
metadata (`dict`, defaults to `None`):
Returns:
`datasets.table.Table`: shallow_copy
"""
table = self.table.replace_schema_metadata(*args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.replace_schema_metadata(*args, **kwargs) for t in tables])
return ConcatenationTable(table, self.blocks)
def add_column(self, *args, **kwargs):
"""
Add column to Table at position.
A new table is returned with the column added, the original table
object is left unchanged.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`: New table with the passed column added.
"""
raise NotImplementedError()
def append_column(self, *args, **kwargs):
"""
Append column at end of columns.
Args:
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column added.
"""
raise NotImplementedError()
def remove_column(self, i, *args, **kwargs):
"""
Create new Table with the indicated column removed.
Args:
i (`int`):
Index of column to remove.
Returns:
`datasets.table.Table`:
New table without the column.
"""
table = self.table.remove_column(i, *args, **kwargs)
name = self.table.column_names[i]
blocks = []
for tables in self.blocks:
blocks.append(
[
t.remove_column(t.column_names.index(name), *args, **kwargs) if name in t.column_names else t
for t in tables
]
)
return ConcatenationTable(table, blocks)
def set_column(self, *args, **kwargs):
"""
Replace column in Table at position.
Args:
i (`int`):
Index to place the column at.
field_ (`Union[str, pyarrow.Field]`):
If a string is passed then the type is deduced from the column
data.
column (`Union[pyarrow.Array, List[pyarrow.Array]]`):
Column data.
Returns:
`datasets.table.Table`:
New table with the passed column set.
"""
raise NotImplementedError()
def rename_columns(self, names, *args, **kwargs):
"""
Create new table with columns renamed to provided names.
"""
table = self.table.rename_columns(names, *args, **kwargs)
names = dict(zip(self.table.column_names, names))
blocks = []
for tables in self.blocks:
blocks.append(
[t.rename_columns([names[name] for name in t.column_names], *args, **kwargs) for t in tables]
)
return ConcatenationTable(table, blocks)
def drop(self, columns, *args, **kwargs):
"""
Drop one or more columns and return a new table.
Args:
columns (`List[str]`):
List of field names referencing existing columns.
Raises:
`KeyError` : if any of the passed columns name are not existing.
Returns:
`datasets.table.Table`:
New table without the columns.
"""
table = self.table.drop(columns, *args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.drop([c for c in columns if c in t.column_names], *args, **kwargs) for t in tables])
return ConcatenationTable(table, blocks)
def select(self, columns, *args, **kwargs):
"""
Select columns of the table.
Returns a new table with the specified columns, and metadata preserved.
Args:
columns (:obj:`Union[List[str], List[int]]`):
The column names or integer indices to select.
Returns:
:class:`datasets.table.Table`: New table with the specified columns, and metadata preserved.
"""
table = self.table.select(columns, *args, **kwargs)
blocks = []
for tables in self.blocks:
blocks.append([t.select([c for c in columns if c in t.column_names], *args, **kwargs) for t in tables])
return ConcatenationTable(table, blocks)
def concat_tables(tables: List[Table], axis: int = 0) -> Table:
"""
Concatenate tables.
Args:
tables (list of `Table`):
List of tables to be concatenated.
axis (`{0, 1}`, defaults to `0`, meaning over rows):
Axis to concatenate over, where `0` means over rows (vertically) and `1` means over columns
(horizontally).
<Added version="1.6.0"/>
Returns:
`datasets.table.Table`:
If the number of input tables is > 1, then the returned table is a `datasets.table.ConcatenationTable`.
Otherwise if there's only one table, it is returned as is.
"""
tables = list(tables)
if len(tables) == 1:
return tables[0]
return ConcatenationTable.from_tables(tables, axis=axis)
def list_table_cache_files(table: Table) -> List[str]:
"""
Get the cache files that are loaded by the table.
Cache file are used when parts of the table come from the disk via memory mapping.
Returns:
`List[str]`:
A list of paths to the cache files loaded by the table.
"""
if isinstance(table, ConcatenationTable):
cache_files = []
for subtables in table.blocks:
for subtable in subtables:
cache_files += list_table_cache_files(subtable)
return cache_files
elif isinstance(table, MemoryMappedTable):
return [table.path]
else:
return []
def _wrap_for_chunked_arrays(func):
"""Apply the function on each chunk of a `pyarrow.ChunkedArray`, or on the array directly"""
def wrapper(array, *args, **kwargs):
if isinstance(array, pa.ChunkedArray):
return pa.chunked_array([func(chunk, *args, **kwargs) for chunk in array.chunks])
else:
return func(array, *args, **kwargs)
return wrapper
def _are_list_values_of_length(array: pa.ListArray, length: int) -> bool:
"""Check if all the sub-lists of a `pa.ListArray` have the specified length."""
return pc.all(pc.equal(array.value_lengths(), length)).as_py() or array.null_count == len(array)
def _combine_list_array_offsets_with_mask(array: pa.ListArray) -> pa.Array:
"""Add the null bitmap to the offsets of a `pa.ListArray`."""
offsets = array.offsets
if array.null_count > 0:
offsets = pa.concat_arrays(
[
pc.replace_with_mask(offsets[:-1], array.is_null(), pa.nulls(len(array), pa.int32())),
offsets[-1:],
]
)
return offsets
def _storage_type(type: pa.DataType) -> pa.DataType:
"""Convert a (possibly nested) `pa.ExtensionType` to its storage type."""
if isinstance(type, pa.ExtensionType):
return _storage_type(type.storage_type)
elif isinstance(type, pa.StructType):
return pa.struct([pa.field(field.name, _storage_type(field.type)) for field in type])
elif isinstance(type, pa.ListType):
return pa.list_(_storage_type(type.value_type))
elif isinstance(type, pa.FixedSizeListType):
return pa.list_(_storage_type(type.value_type), type.list_size)
return type
@_wrap_for_chunked_arrays
def array_cast(array: pa.Array, pa_type: pa.DataType, allow_number_to_str=True):
"""Improved version of `pa.Array.cast`
It supports casting `pa.StructArray` objects to re-order the fields.
It also let you control certain aspects of the casting, e.g. whether
to disable numbers (`floats` or `ints`) to strings.
Args:
array (`pa.Array`):
PyArrow array to cast
pa_type (`pa.DataType`):
Target PyArrow type
allow_number_to_str (`bool`, defaults to `True`):
Whether to allow casting numbers to strings.
Defaults to `True`.
Raises:
`pa.ArrowInvalidError`: if the arrow data casting fails
`TypeError`: if the target type is not supported according, e.g.
- if a field is missing
- if casting from numbers to strings and `allow_number_to_str` is `False`
Returns:
`List[pyarrow.Array]`: the casted array
"""
_c = partial(array_cast, allow_number_to_str=allow_number_to_str)
if isinstance(array, pa.ExtensionArray):
array = array.storage
if isinstance(pa_type, pa.ExtensionType):
return pa_type.wrap_array(_c(array, pa_type.storage_type))
elif array.type == pa_type:
return array
elif pa.types.is_struct(array.type):
if pa.types.is_struct(pa_type) and ({field.name for field in pa_type} == {field.name for field in array.type}):
if array.type.num_fields == 0:
return array
arrays = [_c(array.field(field.name), field.type) for field in pa_type]
return pa.StructArray.from_arrays(arrays, fields=list(pa_type), mask=array.is_null())
elif pa.types.is_list(array.type):
if pa.types.is_fixed_size_list(pa_type):
if _are_list_values_of_length(array, pa_type.list_size):
if array.null_count > 0:
# Ensure each null value in the array translates to [null] * pa_type.list_size in the array's values array
array_type = array.type
storage_type = _storage_type(array_type)
if array_type != storage_type:
# Temporarily convert to the storage type to support extension types in the slice operation
array = _c(array, storage_type)
array = pc.list_slice(array, 0, pa_type.list_size, return_fixed_size_list=True)
array = _c(array, array_type)
else:
array = pc.list_slice(array, 0, pa_type.list_size, return_fixed_size_list=True)
array_values = array.values
if config.PYARROW_VERSION.major < 15:
return pa.Array.from_buffers(
pa_type,
len(array),
[array.is_valid().buffers()[1]],
children=[_c(array_values, pa_type.value_type)],
)
else:
return pa.FixedSizeListArray.from_arrays(
_c(array_values, pa_type.value_type), pa_type.list_size, mask=array.is_null()
)
else:
array_values = array.values[
array.offset * pa_type.length : (array.offset + len(array)) * pa_type.length
]
return pa.FixedSizeListArray.from_arrays(_c(array_values, pa_type.value_type), pa_type.list_size)
elif pa.types.is_list(pa_type):
# Merge offsets with the null bitmap to avoid the "Null bitmap with offsets slice not supported" ArrowNotImplementedError
array_offsets = _combine_list_array_offsets_with_mask(array)
return pa.ListArray.from_arrays(array_offsets, _c(array.values, pa_type.value_type))
elif pa.types.is_fixed_size_list(array.type):
if pa.types.is_fixed_size_list(pa_type):
if pa_type.list_size == array.type.list_size:
array_values = array.values[
array.offset * array.type.list_size : (array.offset + len(array)) * array.type.list_size
]
if config.PYARROW_VERSION.major < 15:
return pa.Array.from_buffers(
pa_type,
len(array),
[array.is_valid().buffers()[1]],
children=[_c(array_values, pa_type.value_type)],
)
else:
return pa.FixedSizeListArray.from_arrays(
_c(array_values, pa_type.value_type), pa_type.list_size, mask=array.is_null()
)
elif pa.types.is_list(pa_type):
array_offsets = (np.arange(len(array) + 1) + array.offset) * array.type.list_size
return pa.ListArray.from_arrays(array_offsets, _c(array.values, pa_type.value_type), mask=array.is_null())
else:
if (
not allow_number_to_str
and pa.types.is_string(pa_type)
and (pa.types.is_floating(array.type) or pa.types.is_integer(array.type))
):
raise TypeError(
f"Couldn't cast array of type {array.type} to {pa_type} since allow_number_to_str is set to {allow_number_to_str}"
)
if pa.types.is_null(pa_type) and not pa.types.is_null(array.type):
raise TypeError(f"Couldn't cast array of type {array.type} to {pa_type}")
return array.cast(pa_type)
raise TypeError(f"Couldn't cast array of type\n{array.type}\nto\n{pa_type}")
@_wrap_for_chunked_arrays
def cast_array_to_feature(array: pa.Array, feature: "FeatureType", allow_number_to_str=True):
"""Cast an array to the arrow type that corresponds to the requested feature type.
For custom features like [`Audio`] or [`Image`], it takes into account the "cast_storage" methods
they defined to enable casting from other arrow types.
Args:
array (`pa.Array`):
The PyArrow array to cast.
feature (`datasets.features.FeatureType`):
The target feature type.
allow_number_to_str (`bool`, defaults to `True`):
Whether to allow casting numbers to strings.
Defaults to `True`.
Raises:
`pa.ArrowInvalidError`: if the arrow data casting fails
`TypeError`: if the target type is not supported according, e.g.
- if a field is missing
- if casting from numbers to strings and `allow_number_to_str` is `False`
Returns:
array (`pyarrow.Array`): the casted array
"""
from .features.features import Sequence, get_nested_type
_c = partial(cast_array_to_feature, allow_number_to_str=allow_number_to_str)
if isinstance(array, pa.ExtensionArray):
array = array.storage
if hasattr(feature, "cast_storage"):
return feature.cast_storage(array)
elif pa.types.is_struct(array.type):
# feature must be a dict or Sequence(subfeatures_dict)
if isinstance(feature, Sequence) and isinstance(feature.feature, dict):
feature = {
name: Sequence(subfeature, length=feature.length) for name, subfeature in feature.feature.items()
}
if isinstance(feature, dict) and {field.name for field in array.type} == set(feature):
if array.type.num_fields == 0:
return array
arrays = [_c(array.field(name), subfeature) for name, subfeature in feature.items()]
return pa.StructArray.from_arrays(arrays, names=list(feature), mask=array.is_null())
elif pa.types.is_list(array.type):
# feature must be either [subfeature] or Sequence(subfeature)
if isinstance(feature, list):
casted_array_values = _c(array.values, feature[0])
if casted_array_values.type == array.values.type:
return array
else:
# Merge offsets with the null bitmap to avoid the "Null bitmap with offsets slice not supported" ArrowNotImplementedError
array_offsets = _combine_list_array_offsets_with_mask(array)
return pa.ListArray.from_arrays(array_offsets, casted_array_values)
elif isinstance(feature, Sequence):
if feature.length > -1:
if _are_list_values_of_length(array, feature.length):
if array.null_count > 0:
# Ensure each null value in the array translates to [null] * pa_type.list_size in the array's values array
array_type = array.type
storage_type = _storage_type(array_type)
if array_type != storage_type:
# Temporarily convert to the storage type to support extension types in the slice operation
array = array_cast(array, storage_type, allow_number_to_str=allow_number_to_str)
array = pc.list_slice(array, 0, feature.length, return_fixed_size_list=True)
array = array_cast(array, array_type, allow_number_to_str=allow_number_to_str)
else:
array = pc.list_slice(array, 0, feature.length, return_fixed_size_list=True)
array_values = array.values
casted_array_values = _c(array_values, feature.feature)
if config.PYARROW_VERSION.major < 15:
return pa.Array.from_buffers(
pa.list_(casted_array_values.type, feature.length),
len(array),
[array.is_valid().buffers()[1]],
children=[casted_array_values],
)
else:
return pa.FixedSizeListArray.from_arrays(
casted_array_values, feature.length, mask=array.is_null()
)
else:
array_values = array.values[
array.offset * feature.length : (array.offset + len(array)) * feature.length
]
return pa.FixedSizeListArray.from_arrays(_c(array_values, feature.feature), feature.length)
else:
casted_array_values = _c(array.values, feature.feature)
if casted_array_values.type == array.values.type:
return array
else:
# Merge offsets with the null bitmap to avoid the "Null bitmap with offsets slice not supported" ArrowNotImplementedError
array_offsets = _combine_list_array_offsets_with_mask(array)
return pa.ListArray.from_arrays(array_offsets, casted_array_values)
elif pa.types.is_fixed_size_list(array.type):
# feature must be either [subfeature] or Sequence(subfeature)
if isinstance(feature, list):
array_offsets = (np.arange(len(array) + 1) + array.offset) * array.type.list_size
return pa.ListArray.from_arrays(array_offsets, _c(array.values, feature[0]), mask=array.is_null())
elif isinstance(feature, Sequence):
if feature.length > -1:
if feature.length == array.type.list_size:
array_values = array.values[
array.offset * array.type.list_size : (array.offset + len(array)) * array.type.list_size
]
casted_array_values = _c(array_values, feature.feature)
if config.PYARROW_VERSION.major < 15:
return pa.Array.from_buffers(
pa.list_(casted_array_values.type, feature.length),
len(array),
[array.is_valid().buffers()[1]],
children=[casted_array_values],
)
else:
return pa.FixedSizeListArray.from_arrays(
casted_array_values, feature.length, mask=array.is_null()
)
else:
array_offsets = (np.arange(len(array) + 1) + array.offset) * array.type.list_size
return pa.ListArray.from_arrays(array_offsets, _c(array.values, feature.feature), mask=array.is_null())
if pa.types.is_null(array.type):
return array_cast(array, get_nested_type(feature), allow_number_to_str=allow_number_to_str)
elif not isinstance(feature, (Sequence, dict, list, tuple)):
return array_cast(array, feature(), allow_number_to_str=allow_number_to_str)
raise TypeError(f"Couldn't cast array of type\n{array.type}\nto\n{feature}")
@_wrap_for_chunked_arrays
def embed_array_storage(array: pa.Array, feature: "FeatureType"):
"""Embed data into an arrays's storage.
For custom features like Audio or Image, it takes into account the "embed_storage" methods
they define to embed external data (e.g. an image file) into an array.
<Added version="2.4.0"/>
Args:
array (`pa.Array`):
The PyArrow array in which to embed data.
feature (`datasets.features.FeatureType`):
Array features.
Raises:
`TypeError`: if the target type is not supported according, e.g.
- if a field is missing
Returns:
array (`pyarrow.Array`): the casted array
"""
from .features import Sequence
_e = embed_array_storage
if isinstance(array, pa.ExtensionArray):
array = array.storage
if hasattr(feature, "embed_storage"):
return feature.embed_storage(array)
elif pa.types.is_struct(array.type):
# feature must be a dict or Sequence(subfeatures_dict)
if isinstance(feature, Sequence) and isinstance(feature.feature, dict):
feature = {
name: Sequence(subfeature, length=feature.length) for name, subfeature in feature.feature.items()
}
if isinstance(feature, dict):
arrays = [_e(array.field(name), subfeature) for name, subfeature in feature.items()]
return pa.StructArray.from_arrays(arrays, names=list(feature), mask=array.is_null())
elif pa.types.is_list(array.type):
# feature must be either [subfeature] or Sequence(subfeature)
# Merge offsets with the null bitmap to avoid the "Null bitmap with offsets slice not supported" ArrowNotImplementedError
array_offsets = _combine_list_array_offsets_with_mask(array)
if isinstance(feature, list):
return pa.ListArray.from_arrays(array_offsets, _e(array.values, feature[0]))
if isinstance(feature, Sequence) and feature.length == -1:
return pa.ListArray.from_arrays(array_offsets, _e(array.values, feature.feature))
elif pa.types.is_fixed_size_list(array.type):
# feature must be Sequence(subfeature)
if isinstance(feature, Sequence) and feature.length > -1:
array_values = array.values[
array.offset * array.type.list_size : (array.offset + len(array)) * array.type.list_size
]
embedded_array_values = _e(array_values, feature.feature)
if config.PYARROW_VERSION.major < 15:
return pa.Array.from_buffers(
pa.list_(array_values.type, feature.length),
len(array),
[array.is_valid().buffers()[1]],
children=[embedded_array_values],
)
else:
return pa.FixedSizeListArray.from_arrays(embedded_array_values, feature.length, mask=array.is_null())
if not isinstance(feature, (Sequence, dict, list, tuple)):
return array
raise TypeError(f"Couldn't embed array of type\n{array.type}\nwith\n{feature}")
class CastError(ValueError):
"""When it's not possible to cast an Arrow table to a specific schema or set of features"""
def __init__(self, *args, table_column_names: List[str], requested_column_names: List[str]) -> None:
super().__init__(*args)
self.table_column_names = table_column_names
self.requested_column_names = requested_column_names
def __reduce__(self):
# Fix unpickling: TypeError: __init__() missing 2 required keyword-only arguments: 'table_column_names' and 'requested_column_names'
return partial(
CastError, table_column_names=self.table_column_names, requested_column_names=self.requested_column_names
), ()
def details(self):
new_columns = set(self.table_column_names) - set(self.requested_column_names)
missing_columns = set(self.requested_column_names) - set(self.table_column_names)
if new_columns and missing_columns:
return f"there are {len(new_columns)} new columns ({', '.join(new_columns)}) and {len(missing_columns)} missing columns ({', '.join(missing_columns)})."
elif new_columns:
return f"there are {len(new_columns)} new columns ({new_columns})"
else:
return f"there are {len(missing_columns)} missing columns ({missing_columns})"
def cast_table_to_features(table: pa.Table, features: "Features"):
"""Cast a table to the arrow schema that corresponds to the requested features.
Args:
table (`pyarrow.Table`):
PyArrow table to cast.
features ([`Features`]):
Target features.
Returns:
table (`pyarrow.Table`): the casted table
"""
if sorted(table.column_names) != sorted(features):
raise CastError(
f"Couldn't cast\n{table.schema}\nto\n{features}\nbecause column names don't match",
table_column_names=table.column_names,
requested_column_names=list(features),
)
arrays = [cast_array_to_feature(table[name], feature) for name, feature in features.items()]
return pa.Table.from_arrays(arrays, schema=features.arrow_schema)
def cast_table_to_schema(table: pa.Table, schema: pa.Schema):
"""Cast a table to the arrow schema. Different from `cast_table_to_features`, this method can preserve nullability.
Args:
table (`pa.Table`):
PyArrow table to cast.
features ([`Features`]):
Target features.
Returns:
`pa.Table`: the casted table
"""
from .features import Features
features = Features.from_arrow_schema(schema)
if sorted(table.column_names) != sorted(features):
raise CastError(
f"Couldn't cast\n{table.schema}\nto\n{features}\nbecause column names don't match",
table_column_names=table.column_names,
requested_column_names=list(features),
)
arrays = [cast_array_to_feature(table[name], feature) for name, feature in features.items()]
return pa.Table.from_arrays(arrays, schema=schema)
def embed_table_storage(table: pa.Table):
"""Embed external data into a table's storage.
<Added version="2.4.0"/>
Args:
table (`pyarrow.Table`):
PyArrow table in which to embed data.
Returns:
table (`pyarrow.Table`): the table with embedded data
"""
from .features.features import Features, require_storage_embed
features = Features.from_arrow_schema(table.schema)
arrays = [
embed_array_storage(table[name], feature) if require_storage_embed(feature) else table[name]
for name, feature in features.items()
]
return pa.Table.from_arrays(arrays, schema=features.arrow_schema)
def table_cast(table: pa.Table, schema: pa.Schema):
"""Improved version of `pa.Table.cast`.
It supports casting to feature types stored in the schema metadata.
Args:
table (`pyarrow.Table`):
PyArrow table to cast.
schema (`pyarrow.Schema`):
Target PyArrow schema.
Returns:
table (`pyarrow.Table`): the casted table
"""
if table.schema != schema:
return cast_table_to_schema(table, schema)
elif table.schema.metadata != schema.metadata:
return table.replace_schema_metadata(schema.metadata)
else:
return table
def table_flatten(table: pa.Table):
"""Improved version of `pa.Table.flatten`.
It behaves as `pa.Table.flatten` in a sense it does 1-step flatten of the columns with a struct type into one column per struct field,
but updates the metadata and skips decodable features unless the `decode` attribute of these features is set to False.
Args:
table (`pa.Table`):
PyArrow table to flatten.
Returns:
`Table`: the flattened table
"""
from .features import Features
features = Features.from_arrow_schema(table.schema)
if any(hasattr(subfeature, "flatten") and subfeature.flatten() == subfeature for subfeature in features.values()):
flat_arrays = []
flat_column_names = []
for field in table.schema:
array = table.column(field.name)
subfeature = features[field.name]
if pa.types.is_struct(field.type) and (
not hasattr(subfeature, "flatten") or subfeature.flatten() != subfeature
):
flat_arrays.extend(array.flatten())
flat_column_names.extend([f"{field.name}.{subfield.name}" for subfield in field.type])
else:
flat_arrays.append(array)
flat_column_names.append(field.name)
flat_table = pa.Table.from_arrays(
flat_arrays,
names=flat_column_names,
)
else:
flat_table = table.flatten()
# Preserve complex types in the metadata
flat_features = features.flatten(max_depth=2)
flat_features = Features({column_name: flat_features[column_name] for column_name in flat_table.column_names})
return flat_table.replace_schema_metadata(flat_features.arrow_schema.metadata)
def table_visitor(table: pa.Table, function: Callable[[pa.Array], None]):
"""Visit all arrays in a table and apply a function to them.
Args:
table (`pyarrow.Table`):
PyArrow table to visit.
function (`Callable[[pa.Array], None]`):
Function to apply to each array.
"""
from .features import Features, Sequence
features = Features.from_arrow_schema(table.schema)
def _visit(array, feature):
if isinstance(array, pa.ChunkedArray):
for chunk in array.chunks:
_visit(chunk, feature)
else:
if isinstance(array, pa.ExtensionArray):
array = array.storage
function(array, feature)
if pa.types.is_struct(array.type) and not hasattr(feature, "cast_storage"):
if isinstance(feature, Sequence) and isinstance(feature.feature, dict):
feature = {
name: Sequence(subfeature, length=feature.length)
for name, subfeature in feature.feature.items()
}
for name, subfeature in feature.items():
_visit(array.field(name), subfeature)
elif pa.types.is_list(array.type):
if isinstance(feature, list):
_visit(array.values, feature[0])
elif isinstance(feature, Sequence):
_visit(array.values, feature.feature)
for name, feature in features.items():
_visit(table[name], feature)
def table_iter(table: Table, batch_size: int, drop_last_batch=False) -> Iterator[pa.Table]:
"""Iterate over sub-tables of size `batch_size`.
Args:
table (`pyarrow.Table`):
PyArrow table to iterate over.
batch_size (`int`):
Size of each sub-table to yield.
drop_last_batch (`bool`, defaults to `False`):
Drop the last batch if it is smaller than `batch_size`.
"""
chunks_buffer = []
chunks_buffer_size = 0
for chunk in table.to_reader(max_chunksize=batch_size):
if len(chunk) == 0:
continue
elif chunks_buffer_size + len(chunk) < batch_size:
chunks_buffer.append(chunk)
chunks_buffer_size += len(chunk)
continue
elif chunks_buffer_size + len(chunk) == batch_size:
chunks_buffer.append(chunk)
yield pa.Table.from_batches(chunks_buffer)
chunks_buffer = []
chunks_buffer_size = 0
else:
cropped_chunk_length = batch_size - chunks_buffer_size
chunks_buffer.append(chunk.slice(0, cropped_chunk_length))
yield pa.Table.from_batches(chunks_buffer)
chunks_buffer = [chunk.slice(cropped_chunk_length, len(chunk) - cropped_chunk_length)]
chunks_buffer_size = len(chunk) - cropped_chunk_length
if not drop_last_batch and chunks_buffer:
yield pa.Table.from_batches(chunks_buffer)
| datasets/src/datasets/table.py/0 | {
"file_path": "datasets/src/datasets/table.py",
"repo_id": "datasets",
"token_count": 41071
} | 69 |
from typing import Callable
def is_documented_by(function_with_docstring: Callable):
"""Decorator to share docstrings across common functions.
Args:
function_with_docstring (`Callable`): Name of the function with the docstring.
"""
def wrapper(target_function):
target_function.__doc__ = function_with_docstring.__doc__
return target_function
return wrapper
| datasets/src/datasets/utils/doc_utils.py/0 | {
"file_path": "datasets/src/datasets/utils/doc_utils.py",
"repo_id": "datasets",
"token_count": 137
} | 70 |
{
"monolingual": "contains a single language",
"multilingual": "contains multiple languages",
"translation": "contains translated or aligned text",
"other": "other type of language distribution"
}
| datasets/src/datasets/utils/resources/multilingualities.json/0 | {
"file_path": "datasets/src/datasets/utils/resources/multilingualities.json",
"repo_id": "datasets",
"token_count": 55
} | 71 |
# isort: skip_file
# This is the module that test_patching.py uses to test patch_submodule()
import os # noqa: F401 - this is just for tests
import os as renamed_os # noqa: F401 - this is just for tests
from os import path # noqa: F401 - this is just for tests
from os import path as renamed_path # noqa: F401 - this is just for tests
from os.path import join # noqa: F401 - this is just for tests
from os.path import join as renamed_join # noqa: F401 - this is just for tests
open = open # noqa we just need to have a builtin inside this module to test it properly
| datasets/tests/_test_patching.py/0 | {
"file_path": "datasets/tests/_test_patching.py",
"repo_id": "datasets",
"token_count": 175
} | 72 |
import datetime
from typing import List, Tuple
from unittest import TestCase
from unittest.mock import patch
import numpy as np
import pandas as pd
import pyarrow as pa
import pytest
from datasets import Array2D
from datasets.arrow_dataset import Dataset
from datasets.features import Audio, ClassLabel, Features, Image, Sequence, Value
from datasets.features.features import (
_align_features,
_arrow_to_datasets_dtype,
_cast_to_python_objects,
_check_if_features_can_be_aligned,
cast_to_python_objects,
encode_nested_example,
generate_from_dict,
string_to_arrow,
)
from datasets.features.translation import Translation, TranslationVariableLanguages
from datasets.info import DatasetInfo
from datasets.utils.py_utils import asdict
from ..utils import require_jax, require_tf, require_torch
class FeaturesTest(TestCase):
def test_from_arrow_schema_simple(self):
data = {"a": [{"b": {"c": "text"}}] * 10, "foo": [1] * 10}
original_features = Features({"a": {"b": {"c": Value("string")}}, "foo": Value("int64")})
dset = Dataset.from_dict(data, features=original_features)
new_features = dset.features
new_dset = Dataset.from_dict(data, features=new_features)
self.assertEqual(original_features.type, new_features.type)
self.assertDictEqual(dset[0], new_dset[0])
self.assertDictEqual(dset[:], new_dset[:])
def test_from_arrow_schema_with_sequence(self):
data = {"a": [{"b": {"c": ["text"]}}] * 10, "foo": [1] * 10}
original_features = Features({"a": {"b": Sequence({"c": Value("string")})}, "foo": Value("int64")})
dset = Dataset.from_dict(data, features=original_features)
new_features = dset.features
new_dset = Dataset.from_dict(data, features=new_features)
self.assertEqual(original_features.type, new_features.type)
self.assertDictEqual(dset[0], new_dset[0])
self.assertDictEqual(dset[:], new_dset[:])
def test_string_to_arrow_bijection_for_primitive_types(self):
supported_pyarrow_datatypes = [
pa.time32("s"),
pa.time64("us"),
pa.timestamp("s"),
pa.timestamp("ns", tz="America/New_York"),
pa.date32(),
pa.date64(),
pa.duration("s"),
pa.decimal128(10, 2),
pa.decimal256(40, -3),
pa.string(),
pa.int32(),
pa.float64(),
pa.array([datetime.time(1, 1, 1)]).type, # arrow type: DataType(time64[us])
]
for dt in supported_pyarrow_datatypes:
self.assertEqual(dt, string_to_arrow(_arrow_to_datasets_dtype(dt)))
unsupported_pyarrow_datatypes = [pa.list_(pa.float64())]
for dt in unsupported_pyarrow_datatypes:
with self.assertRaises(ValueError):
string_to_arrow(_arrow_to_datasets_dtype(dt))
supported_datasets_dtypes = [
"time32[s]",
"timestamp[ns]",
"timestamp[ns, tz=+07:30]",
"duration[us]",
"decimal128(30, -4)",
"int32",
"float64",
]
for sdt in supported_datasets_dtypes:
self.assertEqual(sdt, _arrow_to_datasets_dtype(string_to_arrow(sdt)))
unsupported_datasets_dtypes = [
"time32[ns]",
"timestamp[blob]",
"timestamp[[ns]]",
"timestamp[ns, tz=[ns]]",
"duration[[us]]",
"decimal20(30, -4)",
"int",
]
for sdt in unsupported_datasets_dtypes:
with self.assertRaises(ValueError):
string_to_arrow(sdt)
def test_feature_named_type(self):
"""reference: issue #1110"""
features = Features({"_type": Value("string")})
ds_info = DatasetInfo(features=features)
reloaded_features = Features.from_dict(asdict(ds_info)["features"])
assert features == reloaded_features
def test_feature_named_self_as_kwarg(self):
"""reference: issue #5641"""
features = Features(self=Value("string"))
ds_info = DatasetInfo(features=features)
reloaded_features = Features.from_dict(asdict(ds_info)["features"])
assert features == reloaded_features
def test_class_label_feature_with_no_labels(self):
"""reference: issue #4681"""
features = Features({"label": ClassLabel(names=[])})
ds_info = DatasetInfo(features=features)
reloaded_features = Features.from_dict(asdict(ds_info)["features"])
assert features == reloaded_features
def test_reorder_fields_as(self):
features = Features(
{
"id": Value("string"),
"document": {
"title": Value("string"),
"url": Value("string"),
"html": Value("string"),
"tokens": Sequence({"token": Value("string"), "is_html": Value("bool")}),
},
"question": {
"text": Value("string"),
"tokens": Sequence(Value("string")),
},
"annotations": Sequence(
{
"id": Value("string"),
"long_answer": {
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
"end_byte": Value("int64"),
},
"short_answers": Sequence(
{
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
"end_byte": Value("int64"),
"text": Value("string"),
}
),
"yes_no_answer": ClassLabel(names=["NO", "YES"]),
}
),
}
)
other = Features( # same but with [] instead of sequences, and with a shuffled fields order
{
"id": Value("string"),
"document": {
"tokens": Sequence({"token": Value("string"), "is_html": Value("bool")}),
"title": Value("string"),
"url": Value("string"),
"html": Value("string"),
},
"question": {
"text": Value("string"),
"tokens": [Value("string")],
},
"annotations": {
"yes_no_answer": [ClassLabel(names=["NO", "YES"])],
"id": [Value("string")],
"long_answer": [
{
"end_byte": Value("int64"),
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
}
],
"short_answers": [
Sequence(
{
"text": Value("string"),
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
"end_byte": Value("int64"),
}
)
],
},
}
)
expected = Features(
{
"id": Value("string"),
"document": {
"tokens": Sequence({"token": Value("string"), "is_html": Value("bool")}),
"title": Value("string"),
"url": Value("string"),
"html": Value("string"),
},
"question": {
"text": Value("string"),
"tokens": Sequence(Value("string")),
},
"annotations": Sequence(
{
"yes_no_answer": ClassLabel(names=["NO", "YES"]),
"id": Value("string"),
"long_answer": {
"end_byte": Value("int64"),
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
},
"short_answers": Sequence(
{
"text": Value("string"),
"start_token": Value("int64"),
"end_token": Value("int64"),
"start_byte": Value("int64"),
"end_byte": Value("int64"),
}
),
}
),
}
)
reordered_features = features.reorder_fields_as(other)
self.assertDictEqual(reordered_features, expected)
self.assertEqual(reordered_features.type, other.type)
self.assertEqual(reordered_features.type, expected.type)
self.assertNotEqual(reordered_features.type, features.type)
def test_flatten(self):
features = Features({"foo": {"bar1": Value("int32"), "bar2": {"foobar": Value("string")}}})
_features = features.copy()
flattened_features = features.flatten()
assert flattened_features == {"foo.bar1": Value("int32"), "foo.bar2.foobar": Value("string")}
assert features == _features, "calling flatten shouldn't alter the current features"
def test_flatten_with_sequence(self):
features = Features({"foo": Sequence({"bar": {"my_value": Value("int32")}})})
_features = features.copy()
flattened_features = features.flatten()
assert flattened_features == {"foo.bar": [{"my_value": Value("int32")}]}
assert features == _features, "calling flatten shouldn't alter the current features"
def test_features_dicts_are_synced(self):
def assert_features_dicts_are_synced(features: Features):
assert (
hasattr(features, "_column_requires_decoding")
and features.keys() == features._column_requires_decoding.keys()
)
features = Features({"foo": Sequence({"bar": {"my_value": Value("int32")}})})
assert_features_dicts_are_synced(features)
features["barfoo"] = Image()
assert_features_dicts_are_synced(features)
del features["barfoo"]
assert_features_dicts_are_synced(features)
features.update({"foobar": Value("string")})
assert_features_dicts_are_synced(features)
features.pop("foobar")
assert_features_dicts_are_synced(features)
features.popitem()
assert_features_dicts_are_synced(features)
features.setdefault("xyz", Value("bool"))
assert_features_dicts_are_synced(features)
features.clear()
assert_features_dicts_are_synced(features)
def test_classlabel_init(tmp_path_factory):
names = ["negative", "positive"]
names_file = str(tmp_path_factory.mktemp("features") / "labels.txt")
with open(names_file, "w", encoding="utf-8") as f:
f.write("\n".join(names))
classlabel = ClassLabel(names=names)
assert classlabel.names == names and classlabel.num_classes == len(names)
classlabel = ClassLabel(names_file=names_file)
assert classlabel.names == names and classlabel.num_classes == len(names)
classlabel = ClassLabel(num_classes=len(names), names=names)
assert classlabel.names == names and classlabel.num_classes == len(names)
classlabel = ClassLabel(num_classes=len(names))
assert classlabel.names == [str(i) for i in range(len(names))] and classlabel.num_classes == len(names)
with pytest.raises(ValueError):
classlabel = ClassLabel(num_classes=len(names) + 1, names=names)
with pytest.raises(ValueError):
classlabel = ClassLabel(names=names, names_file=names_file)
with pytest.raises(ValueError):
classlabel = ClassLabel()
with pytest.raises(TypeError):
classlabel = ClassLabel(names=np.array(names))
def test_classlabel_str2int():
names = ["negative", "positive"]
classlabel = ClassLabel(names=names)
for label in names:
assert classlabel.str2int(label) == names.index(label)
with pytest.raises(ValueError):
classlabel.str2int("__bad_label_name__")
with pytest.raises(ValueError):
classlabel.str2int(1)
with pytest.raises(ValueError):
classlabel.str2int(None)
def test_classlabel_int2str():
names = ["negative", "positive"]
classlabel = ClassLabel(names=names)
for i in range(len(names)):
assert classlabel.int2str(i) == names[i]
with pytest.raises(ValueError):
classlabel.int2str(len(names))
with pytest.raises(ValueError):
classlabel.int2str(-1)
with pytest.raises(ValueError):
classlabel.int2str(None)
def test_classlabel_cast_storage():
names = ["negative", "positive"]
classlabel = ClassLabel(names=names)
# from integers
arr = pa.array([0, 1, -1, -100], type=pa.int64())
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == [0, 1, -1, -100]
arr = pa.array([0, 1, -1, -100], type=pa.int32())
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == [0, 1, -1, -100]
arr = pa.array([3])
with pytest.raises(ValueError):
classlabel.cast_storage(arr)
# from strings
arr = pa.array(["negative", "positive"])
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == [0, 1]
arr = pa.array(["__label_that_doesnt_exist__"])
with pytest.raises(ValueError):
classlabel.cast_storage(arr)
# from nulls
arr = pa.array([None])
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == [None]
# from empty
arr = pa.array([], pa.int64())
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == []
arr = pa.array([], pa.string())
result = classlabel.cast_storage(arr)
assert result.type == pa.int64()
assert result.to_pylist() == []
@pytest.mark.parametrize("class_label_arg", ["names", "names_file"])
def test_class_label_to_and_from_dict(class_label_arg, tmp_path_factory):
names = ["negative", "positive"]
names_file = str(tmp_path_factory.mktemp("features") / "labels.txt")
with open(names_file, "w", encoding="utf-8") as f:
f.write("\n".join(names))
if class_label_arg == "names":
class_label = ClassLabel(names=names)
elif class_label_arg == "names_file":
class_label = ClassLabel(names_file=names_file)
generated_class_label = generate_from_dict(asdict(class_label))
assert generated_class_label == class_label
@pytest.mark.parametrize("inner_type", [Value("int32"), {"subcolumn": Value("int32")}])
def test_encode_nested_example_sequence_with_none(inner_type):
schema = Sequence(inner_type)
obj = None
result = encode_nested_example(schema, obj)
assert result is None
def test_encode_batch_with_example_with_empty_first_elem():
features = Features(
{
"x": Sequence(Sequence(ClassLabel(names=["a", "b"]))),
}
)
encoded_batch = features.encode_batch(
{
"x": [
[["a"], ["b"]],
[[], ["b"]],
]
}
)
assert encoded_batch == {"x": [[[0], [1]], [[], [1]]]}
def test_encode_column_dict_with_none():
features = Features(
{
"x": {"a": ClassLabel(names=["a", "b"]), "b": Value("int32")},
}
)
encoded_column = features.encode_column([{"a": "a", "b": 1}, None], "x")
assert encoded_column == [{"a": 0, "b": 1}, None]
@pytest.mark.parametrize(
"feature",
[
Value("int32"),
ClassLabel(num_classes=2),
Translation(languages=["en", "fr"]),
TranslationVariableLanguages(languages=["en", "fr"]),
],
)
def test_dataset_feature_with_none(feature):
data = {"col": [None]}
features = Features({"col": feature})
dset = Dataset.from_dict(data, features=features)
item = dset[0]
assert item.keys() == {"col"}
assert item["col"] is None
batch = dset[:1]
assert len(batch) == 1
assert batch.keys() == {"col"}
assert isinstance(batch["col"], list) and all(item is None for item in batch["col"])
column = dset["col"]
assert len(column) == 1
assert isinstance(column, list) and all(item is None for item in column)
# nested tests
data = {"col": [[None]]}
features = Features({"col": Sequence(feature)})
dset = Dataset.from_dict(data, features=features)
item = dset[0]
assert item.keys() == {"col"}
assert all(i is None for i in item["col"])
data = {"nested": [{"col": None}]}
features = Features({"nested": {"col": feature}})
dset = Dataset.from_dict(data, features=features)
item = dset[0]
assert item.keys() == {"nested"}
assert item["nested"].keys() == {"col"}
assert item["nested"]["col"] is None
def iternumpy(key1, value1, value2):
if value1.dtype != value2.dtype: # check only for dtype
raise AssertionError(
f"dtype of '{key1}' key for casted object: {value1.dtype} and expected object: {value2.dtype} not matching"
)
def dict_diff(d1: dict, d2: dict): # check if 2 dictionaries are equal
np.testing.assert_equal(d1, d2) # sanity check if dict values are equal or not
for (k1, v1), (k2, v2) in zip(d1.items(), d2.items()): # check if their values have same dtype or not
if isinstance(v1, dict): # nested dictionary case
dict_diff(v1, v2)
elif isinstance(v1, np.ndarray): # checks if dtype and value of np.ndarray is equal
iternumpy(k1, v1, v2)
elif isinstance(v1, list):
for element1, element2 in zip(v1, v2): # iterates over all elements of list
if isinstance(element1, dict):
dict_diff(element1, element2)
elif isinstance(element1, np.ndarray):
iternumpy(k1, element1, element2)
class CastToPythonObjectsTest(TestCase):
def test_cast_to_python_objects_list(self):
obj = {"col_1": [{"vec": [1, 2, 3], "txt": "foo"}] * 3, "col_2": [[1, 2], [3, 4], [5, 6]]}
expected_obj = {"col_1": [{"vec": [1, 2, 3], "txt": "foo"}] * 3, "col_2": [[1, 2], [3, 4], [5, 6]]}
casted_obj = cast_to_python_objects(obj)
self.assertDictEqual(casted_obj, expected_obj)
def test_cast_to_python_objects_tuple(self):
obj = {"col_1": [{"vec": (1, 2, 3), "txt": "foo"}] * 3, "col_2": [(1, 2), (3, 4), (5, 6)]}
expected_obj = {"col_1": [{"vec": (1, 2, 3), "txt": "foo"}] * 3, "col_2": [(1, 2), (3, 4), (5, 6)]}
casted_obj = cast_to_python_objects(obj)
self.assertDictEqual(casted_obj, expected_obj)
def test_cast_to_python_or_numpy(self):
obj = {"col_1": [{"vec": np.arange(1, 4), "txt": "foo"}] * 3, "col_2": np.arange(1, 7).reshape(3, 2)}
expected_obj = {
"col_1": [{"vec": np.array([1, 2, 3]), "txt": "foo"}] * 3,
"col_2": np.array([[1, 2], [3, 4], [5, 6]]),
}
casted_obj = cast_to_python_objects(obj)
dict_diff(casted_obj, expected_obj)
def test_cast_to_python_objects_series(self):
obj = {
"col_1": pd.Series([{"vec": [1, 2, 3], "txt": "foo"}] * 3),
"col_2": pd.Series([[1, 2], [3, 4], [5, 6]]),
}
expected_obj = {"col_1": [{"vec": [1, 2, 3], "txt": "foo"}] * 3, "col_2": [[1, 2], [3, 4], [5, 6]]}
casted_obj = cast_to_python_objects(obj)
self.assertDictEqual(casted_obj, expected_obj)
def test_cast_to_python_objects_dataframe(self):
obj = pd.DataFrame({"col_1": [{"vec": [1, 2, 3], "txt": "foo"}] * 3, "col_2": [[1, 2], [3, 4], [5, 6]]})
expected_obj = {"col_1": [{"vec": [1, 2, 3], "txt": "foo"}] * 3, "col_2": [[1, 2], [3, 4], [5, 6]]}
casted_obj = cast_to_python_objects(obj)
self.assertDictEqual(casted_obj, expected_obj)
def test_cast_to_python_objects_pandas_timestamp(self):
obj = pd.Timestamp(2020, 1, 1)
expected_obj = obj.to_pydatetime()
casted_obj = cast_to_python_objects(obj)
self.assertEqual(casted_obj, expected_obj)
casted_obj = cast_to_python_objects(pd.Series([obj]))
self.assertListEqual(casted_obj, [expected_obj])
casted_obj = cast_to_python_objects(pd.DataFrame({"a": [obj]}))
self.assertDictEqual(casted_obj, {"a": [expected_obj]})
def test_cast_to_python_objects_pandas_timedelta(self):
obj = pd.Timedelta(seconds=1)
expected_obj = obj.to_pytimedelta()
casted_obj = cast_to_python_objects(obj)
self.assertEqual(casted_obj, expected_obj)
casted_obj = cast_to_python_objects(pd.Series([obj]))
self.assertListEqual(casted_obj, [expected_obj])
casted_obj = cast_to_python_objects(pd.DataFrame({"a": [obj]}))
self.assertDictEqual(casted_obj, {"a": [expected_obj]})
@require_torch
def test_cast_to_python_objects_torch(self):
import torch
obj = {
"col_1": [{"vec": torch.tensor(np.arange(1, 4)), "txt": "foo"}] * 3,
"col_2": torch.tensor(np.arange(1, 7).reshape(3, 2)),
}
expected_obj = {
"col_1": [{"vec": np.array([1, 2, 3]), "txt": "foo"}] * 3,
"col_2": np.array([[1, 2], [3, 4], [5, 6]]),
}
casted_obj = cast_to_python_objects(obj)
dict_diff(casted_obj, expected_obj)
@require_tf
def test_cast_to_python_objects_tf(self):
import tensorflow as tf
obj = {
"col_1": [{"vec": tf.constant(np.arange(1, 4)), "txt": "foo"}] * 3,
"col_2": tf.constant(np.arange(1, 7).reshape(3, 2)),
}
expected_obj = {
"col_1": [{"vec": np.array([1, 2, 3]), "txt": "foo"}] * 3,
"col_2": np.array([[1, 2], [3, 4], [5, 6]]),
}
casted_obj = cast_to_python_objects(obj)
dict_diff(casted_obj, expected_obj)
@require_jax
def test_cast_to_python_objects_jax(self):
import jax.numpy as jnp
obj = {
"col_1": [{"vec": jnp.array(np.arange(1, 4)), "txt": "foo"}] * 3,
"col_2": jnp.array(np.arange(1, 7).reshape(3, 2)),
}
assert obj["col_2"].dtype == jnp.int32
expected_obj = {
"col_1": [{"vec": np.array([1, 2, 3], dtype=np.int32), "txt": "foo"}] * 3,
"col_2": np.array([[1, 2], [3, 4], [5, 6]], dtype=np.int32),
}
casted_obj = cast_to_python_objects(obj)
dict_diff(casted_obj, expected_obj)
@patch("datasets.features.features._cast_to_python_objects", side_effect=_cast_to_python_objects)
def test_dont_iterate_over_each_element_in_a_list(self, mocked_cast):
obj = {"col_1": [[1, 2], [3, 4], [5, 6]]}
cast_to_python_objects(obj)
self.assertEqual(mocked_cast.call_count, 4) # 4 = depth of obj
SIMPLE_FEATURES = [
Features(),
Features({"a": Value("int32")}),
Features({"a": Value("int32", id="my feature")}),
Features({"a": Value("int32"), "b": Value("float64"), "c": Value("string")}),
]
CUSTOM_FEATURES = [
Features({"label": ClassLabel(names=["negative", "positive"])}),
Features({"array": Array2D(dtype="float32", shape=(4, 4))}),
Features({"image": Image()}),
Features({"audio": Audio()}),
Features({"image": Image(decode=False)}),
Features({"audio": Audio(decode=False)}),
Features({"translation": Translation(["en", "fr"])}),
Features({"translation": TranslationVariableLanguages(["en", "fr"])}),
]
NESTED_FEATURES = [
Features({"foo": {}}),
Features({"foo": {"bar": Value("int32")}}),
Features({"foo": {"bar1": Value("int32"), "bar2": Value("float64")}}),
Features({"foo": Sequence(Value("int32"))}),
Features({"foo": Sequence({})}),
Features({"foo": Sequence({"bar": Value("int32")})}),
Features({"foo": [Value("int32")]}),
Features({"foo": [{"bar": Value("int32")}]}),
]
NESTED_CUSTOM_FEATURES = [
Features({"foo": {"bar": ClassLabel(names=["negative", "positive"])}}),
Features({"foo": Sequence(ClassLabel(names=["negative", "positive"]))}),
Features({"foo": Sequence({"bar": ClassLabel(names=["negative", "positive"])})}),
Features({"foo": [ClassLabel(names=["negative", "positive"])]}),
Features({"foo": [{"bar": ClassLabel(names=["negative", "positive"])}]}),
]
@pytest.mark.parametrize("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
def test_features_to_dict(features: Features):
features_dict = features.to_dict()
assert isinstance(features_dict, dict)
reloaded = Features.from_dict(features_dict)
assert features == reloaded
@pytest.mark.parametrize("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
def test_features_to_yaml_list(features: Features):
features_yaml_list = features._to_yaml_list()
assert isinstance(features_yaml_list, list)
reloaded = Features._from_yaml_list(features_yaml_list)
assert features == reloaded
@pytest.mark.parametrize("features", SIMPLE_FEATURES + CUSTOM_FEATURES + NESTED_FEATURES + NESTED_CUSTOM_FEATURES)
def test_features_to_arrow_schema(features: Features):
arrow_schema = features.arrow_schema
assert isinstance(arrow_schema, pa.Schema)
reloaded = Features.from_arrow_schema(arrow_schema)
assert features == reloaded
NESTED_COMPARISON = [
[
[Features({"email": Value(dtype="string", id=None)}), Features({"email": Value(dtype="string", id=None)})],
[Features({"email": Value(dtype="string", id=None)}), Features({"email": Value(dtype="string", id=None)})],
],
[
[Features({"email": Value(dtype="string", id=None)}), Features({"email": Value(dtype="null", id=None)})],
[Features({"email": Value(dtype="string", id=None)}), Features({"email": Value(dtype="string", id=None)})],
],
[
[
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
],
[
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
],
],
[
[
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
Features({"speaker": {"email": Value(dtype="null", id=None)}}),
],
[
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
Features({"speaker": {"email": Value(dtype="string", id=None)}}),
],
],
]
@pytest.mark.parametrize("features", NESTED_COMPARISON)
def test_features_alignment(features: Tuple[List[Features], Features]):
inputs, expected = features
_check_if_features_can_be_aligned(inputs) # Check that we can align, will raise otherwise.
assert _align_features(inputs) == expected
| datasets/tests/features/test_features.py/0 | {
"file_path": "datasets/tests/features/test_features.py",
"repo_id": "datasets",
"token_count": 13250
} | 73 |
import shutil
import textwrap
import librosa
import numpy as np
import pytest
import soundfile as sf
from datasets import Audio, ClassLabel, Features, Value
from datasets.data_files import DataFilesDict, get_data_patterns
from datasets.download.streaming_download_manager import StreamingDownloadManager
from datasets.packaged_modules.audiofolder.audiofolder import AudioFolder
from ..utils import require_sndfile
@pytest.fixture
def cache_dir(tmp_path):
return str(tmp_path / "audiofolder_cache_dir")
@pytest.fixture
def data_files_with_labels_no_metadata(tmp_path, audio_file):
data_dir = tmp_path / "data_files_with_labels_no_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
subdir_class_0 = data_dir / "fr"
subdir_class_0.mkdir(parents=True, exist_ok=True)
subdir_class_1 = data_dir / "uk"
subdir_class_1.mkdir(parents=True, exist_ok=True)
audio_filename = subdir_class_0 / "audio_fr.wav"
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = subdir_class_1 / "audio_uk.wav"
shutil.copyfile(audio_file, audio_filename2)
data_files_with_labels_no_metadata = DataFilesDict.from_patterns(
get_data_patterns(str(data_dir)), data_dir.as_posix()
)
return data_files_with_labels_no_metadata
@pytest.fixture
def audio_files_with_labels_and_duplicated_label_key_in_metadata(tmp_path, audio_file):
data_dir = tmp_path / "audio_files_with_labels_and_label_key_in_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
subdir_class_0 = data_dir / "fr"
subdir_class_0.mkdir(parents=True, exist_ok=True)
subdir_class_1 = data_dir / "uk"
subdir_class_1.mkdir(parents=True, exist_ok=True)
audio_filename = subdir_class_0 / "audio_fr.wav"
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = subdir_class_1 / "audio_uk.wav"
shutil.copyfile(audio_file, audio_filename2)
audio_metadata_filename = tmp_path / data_dir / "metadata.jsonl"
audio_metadata = textwrap.dedent(
"""\
{"file_name": "fr/audio_fr.wav", "text": "Audio in French", "label": "Fr"}
{"file_name": "uk/audio_uk.wav", "text": "Audio in Ukrainian", "label": "Uk"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
return str(audio_filename), str(audio_filename2), str(audio_metadata_filename)
@pytest.fixture
def audio_file_with_metadata(tmp_path, audio_file):
audio_filename = tmp_path / "audio_file.wav"
shutil.copyfile(audio_file, audio_filename)
audio_metadata_filename = tmp_path / "metadata.jsonl"
audio_metadata = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "Audio transcription"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
return str(audio_filename), str(audio_metadata_filename)
@pytest.fixture
def audio_files_with_metadata_that_misses_one_audio(tmp_path, audio_file):
audio_filename = tmp_path / "audio_file.wav"
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = tmp_path / "audio_file2.wav"
shutil.copyfile(audio_file, audio_filename2)
audio_metadata_filename = tmp_path / "metadata.jsonl"
audio_metadata = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "Audio transcription"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
return str(audio_filename), str(audio_filename2), str(audio_metadata_filename)
@pytest.fixture
def data_files_with_one_split_and_metadata(tmp_path, audio_file):
data_dir = tmp_path / "audiofolder_data_dir_with_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
subdir = data_dir / "subdir"
subdir.mkdir(parents=True, exist_ok=True)
audio_filename = data_dir / "audio_file.wav"
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = data_dir / "audio_file2.wav"
shutil.copyfile(audio_file, audio_filename2)
audio_filename3 = subdir / "audio_file3.wav" # in subdir
shutil.copyfile(audio_file, audio_filename3)
audio_metadata_filename = data_dir / "metadata.jsonl"
audio_metadata = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "First audio transcription"}
{"file_name": "audio_file2.wav", "text": "Second audio transcription"}
{"file_name": "subdir/audio_file3.wav", "text": "Third audio transcription (in subdir)"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
data_files_with_one_split_and_metadata = DataFilesDict.from_patterns(
get_data_patterns(str(data_dir)), data_dir.as_posix()
)
assert len(data_files_with_one_split_and_metadata) == 1
assert len(data_files_with_one_split_and_metadata["train"]) == 4
return data_files_with_one_split_and_metadata
@pytest.fixture(params=["jsonl", "csv"])
def data_files_with_two_splits_and_metadata(request, tmp_path, audio_file):
data_dir = tmp_path / "audiofolder_data_dir_with_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
train_dir = data_dir / "train"
train_dir.mkdir(parents=True, exist_ok=True)
test_dir = data_dir / "test"
test_dir.mkdir(parents=True, exist_ok=True)
audio_filename = train_dir / "audio_file.wav" # train audio
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = train_dir / "audio_file2.wav" # train audio
shutil.copyfile(audio_file, audio_filename2)
audio_filename3 = test_dir / "audio_file3.wav" # test audio
shutil.copyfile(audio_file, audio_filename3)
train_audio_metadata_filename = train_dir / f"metadata.{request.param}"
audio_metadata = (
textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "First train audio transcription"}
{"file_name": "audio_file2.wav", "text": "Second train audio transcription"}
"""
)
if request.param == "jsonl"
else textwrap.dedent(
"""\
file_name,text
audio_file.wav,First train audio transcription
audio_file2.wav,Second train audio transcription
"""
)
)
with open(train_audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
test_audio_metadata_filename = test_dir / f"metadata.{request.param}"
audio_metadata = (
textwrap.dedent(
"""\
{"file_name": "audio_file3.wav", "text": "Test audio transcription"}
"""
)
if request.param == "jsonl"
else textwrap.dedent(
"""\
file_name,text
audio_file3.wav,Test audio transcription
"""
)
)
with open(test_audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
data_files_with_two_splits_and_metadata = DataFilesDict.from_patterns(
get_data_patterns(str(data_dir)), data_dir.as_posix()
)
assert len(data_files_with_two_splits_and_metadata) == 2
assert len(data_files_with_two_splits_and_metadata["train"]) == 3
assert len(data_files_with_two_splits_and_metadata["test"]) == 2
return data_files_with_two_splits_and_metadata
@pytest.fixture
def data_files_with_zip_archives(tmp_path, audio_file):
data_dir = tmp_path / "audiofolder_data_dir_with_zip_archives"
data_dir.mkdir(parents=True, exist_ok=True)
archive_dir = data_dir / "archive"
archive_dir.mkdir(parents=True, exist_ok=True)
subdir = archive_dir / "subdir"
subdir.mkdir(parents=True, exist_ok=True)
audio_filename = archive_dir / "audio_file.wav"
shutil.copyfile(audio_file, audio_filename)
audio_filename2 = subdir / "audio_file2.wav" # in subdir
# make sure they're two different audios
# Indeed we won't be able to compare the audio filenames, since the archive is not extracted in streaming mode
array, sampling_rate = librosa.load(str(audio_filename), sr=16000) # original sampling rate is 44100
sf.write(str(audio_filename2), array, samplerate=16000)
audio_metadata_filename = archive_dir / "metadata.jsonl"
audio_metadata = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "First audio transcription"}
{"file_name": "subdir/audio_file2.wav", "text": "Second audio transcription (in subdir)"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
shutil.make_archive(str(archive_dir), "zip", archive_dir)
shutil.rmtree(str(archive_dir))
data_files_with_zip_archives = DataFilesDict.from_patterns(get_data_patterns(str(data_dir)), data_dir.as_posix())
assert len(data_files_with_zip_archives) == 1
assert len(data_files_with_zip_archives["train"]) == 1
return data_files_with_zip_archives
@require_sndfile
# check that labels are inferred correctly from dir names
def test_generate_examples_with_labels(data_files_with_labels_no_metadata, cache_dir):
# there are no metadata.jsonl files in this test case
audiofolder = AudioFolder(data_files=data_files_with_labels_no_metadata, cache_dir=cache_dir, drop_labels=False)
audiofolder.download_and_prepare()
assert audiofolder.info.features == Features({"audio": Audio(), "label": ClassLabel(names=["fr", "uk"])})
dataset = list(audiofolder.as_dataset()["train"])
label_feature = audiofolder.info.features["label"]
assert dataset[0]["label"] == label_feature._str2int["fr"]
assert dataset[1]["label"] == label_feature._str2int["uk"]
@require_sndfile
@pytest.mark.parametrize("drop_metadata", [None, True, False])
@pytest.mark.parametrize("drop_labels", [None, True, False])
def test_generate_examples_duplicated_label_key(
audio_files_with_labels_and_duplicated_label_key_in_metadata, drop_metadata, drop_labels, cache_dir, caplog
):
fr_audio_file, uk_audio_file, audio_metadata_file = audio_files_with_labels_and_duplicated_label_key_in_metadata
audiofolder = AudioFolder(
drop_metadata=drop_metadata,
drop_labels=drop_labels,
data_files=[fr_audio_file, uk_audio_file, audio_metadata_file],
cache_dir=cache_dir,
)
if drop_labels is False:
# infer labels from directories even if metadata files are found
audiofolder.download_and_prepare()
warning_in_logs = any("ignoring metadata columns" in record.msg.lower() for record in caplog.records)
assert warning_in_logs if drop_metadata is not True else not warning_in_logs
dataset = audiofolder.as_dataset()["train"]
assert audiofolder.info.features["label"] == ClassLabel(names=["fr", "uk"])
assert all(example["label"] in audiofolder.info.features["label"]._str2int.values() for example in dataset)
else:
audiofolder.download_and_prepare()
dataset = audiofolder.as_dataset()["train"]
if drop_metadata is not True:
# labels are from metadata
assert audiofolder.info.features["label"] == Value("string")
assert all(example["label"] in ["Fr", "Uk"] for example in dataset)
else:
# drop both labels and metadata
assert audiofolder.info.features == Features({"audio": Audio()})
assert all(example.keys() == {"audio"} for example in dataset)
@require_sndfile
@pytest.mark.parametrize("drop_metadata", [None, True, False])
@pytest.mark.parametrize("drop_labels", [None, True, False])
def test_generate_examples_drop_labels(data_files_with_labels_no_metadata, drop_metadata, drop_labels):
audiofolder = AudioFolder(
drop_metadata=drop_metadata, drop_labels=drop_labels, data_files=data_files_with_labels_no_metadata
)
gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
# removing the labels explicitly requires drop_labels=True
assert gen_kwargs["add_labels"] is not bool(drop_labels)
assert gen_kwargs["add_metadata"] is False # metadata files is not present in this case
generator = audiofolder._generate_examples(**gen_kwargs)
if not drop_labels:
assert all(
example.keys() == {"audio", "label"} and all(val is not None for val in example.values())
for _, example in generator
)
else:
assert all(
example.keys() == {"audio"} and all(val is not None for val in example.values())
for _, example in generator
)
@require_sndfile
@pytest.mark.parametrize("drop_metadata", [None, True, False])
@pytest.mark.parametrize("drop_labels", [None, True, False])
def test_generate_examples_drop_metadata(audio_file_with_metadata, drop_metadata, drop_labels):
audio_file, audio_metadata_file = audio_file_with_metadata
audiofolder = AudioFolder(
drop_metadata=drop_metadata, drop_labels=drop_labels, data_files={"train": [audio_file, audio_metadata_file]}
)
gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
# since the dataset has metadata, removing the metadata explicitly requires drop_metadata=True
assert gen_kwargs["add_metadata"] is not bool(drop_metadata)
# since the dataset has metadata, adding the labels explicitly requires drop_labels=False
assert gen_kwargs["add_labels"] is (drop_labels is False)
generator = audiofolder._generate_examples(**gen_kwargs)
expected_columns = {"audio"}
if gen_kwargs["add_metadata"]:
expected_columns.add("text")
if gen_kwargs["add_labels"]:
expected_columns.add("label")
result = [example for _, example in generator]
assert len(result) == 1
example = result[0]
assert example.keys() == expected_columns
for column in expected_columns:
assert example[column] is not None
@require_sndfile
@pytest.mark.parametrize("drop_metadata", [None, True, False])
def test_generate_examples_with_metadata_in_wrong_location(audio_file, audio_file_with_metadata, drop_metadata):
_, audio_metadata_file = audio_file_with_metadata
audiofolder = AudioFolder(drop_metadata=drop_metadata, data_files={"train": [audio_file, audio_metadata_file]})
gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
generator = audiofolder._generate_examples(**gen_kwargs)
if not drop_metadata:
with pytest.raises(ValueError):
list(generator)
else:
assert all(
example.keys() == {"audio"} and all(val is not None for val in example.values())
for _, example in generator
)
@require_sndfile
@pytest.mark.parametrize("drop_metadata", [None, True, False])
def test_generate_examples_with_metadata_that_misses_one_audio(
audio_files_with_metadata_that_misses_one_audio, drop_metadata
):
audio_file, audio_file2, audio_metadata_file = audio_files_with_metadata_that_misses_one_audio
if not drop_metadata:
features = Features({"audio": Audio(), "text": Value("string")})
else:
features = Features({"audio": Audio()})
audiofolder = AudioFolder(
drop_metadata=drop_metadata,
features=features,
data_files={"train": [audio_file, audio_file2, audio_metadata_file]},
)
gen_kwargs = audiofolder._split_generators(StreamingDownloadManager())[0].gen_kwargs
generator = audiofolder._generate_examples(**gen_kwargs)
if not drop_metadata:
with pytest.raises(ValueError):
_ = list(generator)
else:
assert all(
example.keys() == {"audio"} and all(val is not None for val in example.values())
for _, example in generator
)
@require_sndfile
@pytest.mark.parametrize("streaming", [False, True])
def test_data_files_with_metadata_and_single_split(streaming, cache_dir, data_files_with_one_split_and_metadata):
data_files = data_files_with_one_split_and_metadata
audiofolder = AudioFolder(data_files=data_files, cache_dir=cache_dir)
audiofolder.download_and_prepare()
datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
for split, data_files in data_files.items():
expected_num_of_audios = len(data_files) - 1 # don't count the metadata file
assert split in datasets
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_audios
# make sure each sample has its own audio and metadata
assert len({example["audio"]["path"] for example in dataset}) == expected_num_of_audios
assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
@require_sndfile
@pytest.mark.parametrize("streaming", [False, True])
def test_data_files_with_metadata_and_multiple_splits(streaming, cache_dir, data_files_with_two_splits_and_metadata):
data_files = data_files_with_two_splits_and_metadata
audiofolder = AudioFolder(data_files=data_files, cache_dir=cache_dir)
audiofolder.download_and_prepare()
datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
for split, data_files in data_files.items():
expected_num_of_audios = len(data_files) - 1 # don't count the metadata file
assert split in datasets
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_audios
# make sure each sample has its own audio and metadata
assert len({example["audio"]["path"] for example in dataset}) == expected_num_of_audios
assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
@require_sndfile
@pytest.mark.parametrize("streaming", [False, True])
def test_data_files_with_metadata_and_archives(streaming, cache_dir, data_files_with_zip_archives):
audiofolder = AudioFolder(data_files=data_files_with_zip_archives, cache_dir=cache_dir)
audiofolder.download_and_prepare()
datasets = audiofolder.as_streaming_dataset() if streaming else audiofolder.as_dataset()
for split, data_files in data_files_with_zip_archives.items():
num_of_archives = len(data_files) # the metadata file is inside the archive
expected_num_of_audios = 2 * num_of_archives
assert split in datasets
dataset = list(datasets[split])
assert len(dataset) == expected_num_of_audios
# make sure each sample has its own audio (all arrays are different) and metadata
assert (
sum(np.array_equal(dataset[0]["audio"]["array"], example["audio"]["array"]) for example in dataset[1:])
== 0
)
assert len({example["text"] for example in dataset}) == expected_num_of_audios
assert all(example["text"] is not None for example in dataset)
@require_sndfile
def test_data_files_with_wrong_metadata_file_name(cache_dir, tmp_path, audio_file):
data_dir = tmp_path / "data_dir_with_bad_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
shutil.copyfile(audio_file, data_dir / "audio_file.wav")
audio_metadata_filename = data_dir / "bad_metadata.jsonl" # bad file
audio_metadata = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "Audio transcription"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
data_files_with_bad_metadata = DataFilesDict.from_patterns(get_data_patterns(str(data_dir)), data_dir.as_posix())
audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
audiofolder.download_and_prepare()
dataset = audiofolder.as_dataset(split="train")
# check that there are no metadata, since the metadata file name doesn't have the right name
assert "text" not in dataset.column_names
@require_sndfile
def test_data_files_with_wrong_audio_file_name_column_in_metadata_file(cache_dir, tmp_path, audio_file):
data_dir = tmp_path / "data_dir_with_bad_metadata"
data_dir.mkdir(parents=True, exist_ok=True)
shutil.copyfile(audio_file, data_dir / "audio_file.wav")
audio_metadata_filename = data_dir / "metadata.jsonl"
audio_metadata = textwrap.dedent( # with bad column "bad_file_name" instead of "file_name"
"""\
{"bad_file_name_column": "audio_file.wav", "text": "Audio transcription"}
"""
)
with open(audio_metadata_filename, "w", encoding="utf-8") as f:
f.write(audio_metadata)
data_files_with_bad_metadata = DataFilesDict.from_patterns(get_data_patterns(str(data_dir)), data_dir.as_posix())
audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
with pytest.raises(ValueError) as exc_info:
audiofolder.download_and_prepare()
assert "`file_name` must be present" in str(exc_info.value)
@require_sndfile
def test_data_files_with_with_metadata_in_different_formats(cache_dir, tmp_path, audio_file):
data_dir = tmp_path / "data_dir_with_metadata_in_different_format"
data_dir.mkdir(parents=True, exist_ok=True)
shutil.copyfile(audio_file, data_dir / "audio_file.wav")
audio_metadata_filename_jsonl = data_dir / "metadata.jsonl"
audio_metadata_jsonl = textwrap.dedent(
"""\
{"file_name": "audio_file.wav", "text": "Audio transcription"}
"""
)
with open(audio_metadata_filename_jsonl, "w", encoding="utf-8") as f:
f.write(audio_metadata_jsonl)
audio_metadata_filename_csv = data_dir / "metadata.csv"
audio_metadata_csv = textwrap.dedent(
"""\
file_name,text
audio_file.wav,Audio transcription
"""
)
with open(audio_metadata_filename_csv, "w", encoding="utf-8") as f:
f.write(audio_metadata_csv)
data_files_with_bad_metadata = DataFilesDict.from_patterns(get_data_patterns(str(data_dir)), data_dir.as_posix())
audiofolder = AudioFolder(data_files=data_files_with_bad_metadata, cache_dir=cache_dir)
with pytest.raises(ValueError) as exc_info:
audiofolder.download_and_prepare()
assert "metadata files with different extensions" in str(exc_info.value)
| datasets/tests/packaged_modules/test_audiofolder.py/0 | {
"file_path": "datasets/tests/packaged_modules/test_audiofolder.py",
"repo_id": "datasets",
"token_count": 8594
} | 74 |
from unittest import TestCase
from datasets import Sequence, Value
from datasets.arrow_dataset import Dataset
class DatasetListTest(TestCase):
def _create_example_records(self):
return [
{"col_1": 3, "col_2": "a"},
{"col_1": 2, "col_2": "b"},
{"col_1": 1, "col_2": "c"},
{"col_1": 0, "col_2": "d"},
]
def _create_example_dict(self):
data = {"col_1": [3, 2, 1, 0], "col_2": ["a", "b", "c", "d"]}
return Dataset.from_dict(data)
def test_create(self):
example_records = self._create_example_records()
dset = Dataset.from_list(example_records)
self.assertListEqual(dset.column_names, ["col_1", "col_2"])
for i, r in enumerate(dset):
self.assertDictEqual(r, example_records[i])
def test_list_dict_equivalent(self):
example_records = self._create_example_records()
dset = Dataset.from_list(example_records)
dset_from_dict = Dataset.from_dict({k: [r[k] for r in example_records] for k in example_records[0]})
self.assertEqual(dset.info, dset_from_dict.info)
def test_uneven_records(self): # checks what happens with missing columns
uneven_records = [{"col_1": 1}, {"col_2": "x"}]
dset = Dataset.from_list(uneven_records)
self.assertDictEqual(dset[0], {"col_1": 1})
self.assertDictEqual(dset[1], {"col_1": None}) # NB: first record is used for columns
def test_variable_list_records(self): # checks if the type can be inferred from the second record
list_records = [{"col_1": []}, {"col_1": [1, 2]}]
dset = Dataset.from_list(list_records)
self.assertEqual(dset.info.features["col_1"], Sequence(Value("int64")))
def test_create_empty(self):
dset = Dataset.from_list([])
self.assertEqual(len(dset), 0)
self.assertListEqual(dset.column_names, [])
| datasets/tests/test_dataset_list.py/0 | {
"file_path": "datasets/tests/test_dataset_list.py",
"repo_id": "datasets",
"token_count": 875
} | 75 |
import importlib
import os
import pickle
import shutil
import tempfile
import time
from hashlib import sha256
from multiprocessing import Pool
from pathlib import Path
from unittest import TestCase
from unittest.mock import patch
import dill
import pyarrow as pa
import pytest
import requests
import datasets
from datasets import config, load_dataset, load_from_disk
from datasets.arrow_dataset import Dataset
from datasets.arrow_writer import ArrowWriter
from datasets.builder import DatasetBuilder
from datasets.config import METADATA_CONFIGS_FIELD
from datasets.data_files import DataFilesDict, DataFilesPatternsDict
from datasets.dataset_dict import DatasetDict, IterableDatasetDict
from datasets.download.download_config import DownloadConfig
from datasets.exceptions import DatasetNotFoundError
from datasets.features import Features, Image, Value
from datasets.iterable_dataset import IterableDataset
from datasets.load import (
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithParquetExport,
HubDatasetModuleFactoryWithScript,
LocalDatasetModuleFactoryWithoutScript,
LocalDatasetModuleFactoryWithScript,
LocalMetricModuleFactory,
PackagedDatasetModuleFactory,
infer_module_for_data_files_list,
infer_module_for_data_files_list_in_archives,
load_dataset_builder,
resolve_trust_remote_code,
)
from datasets.packaged_modules.audiofolder.audiofolder import AudioFolder, AudioFolderConfig
from datasets.packaged_modules.imagefolder.imagefolder import ImageFolder, ImageFolderConfig
from datasets.packaged_modules.parquet.parquet import ParquetConfig
from datasets.utils import _datasets_server
from datasets.utils.logging import INFO, get_logger
from .utils import (
OfflineSimulationMode,
assert_arrow_memory_doesnt_increase,
assert_arrow_memory_increases,
offline,
require_not_windows,
require_pil,
require_sndfile,
set_current_working_directory_to_temp_dir,
)
DATASET_LOADING_SCRIPT_NAME = "__dummy_dataset1__"
DATASET_LOADING_SCRIPT_CODE = """
import os
import datasets
from datasets import DatasetInfo, Features, Split, SplitGenerator, Value
class __DummyDataset1__(datasets.GeneratorBasedBuilder):
def _info(self) -> DatasetInfo:
return DatasetInfo(features=Features({"text": Value("string")}))
def _split_generators(self, dl_manager):
return [
SplitGenerator(Split.TRAIN, gen_kwargs={"filepath": os.path.join(dl_manager.manual_dir, "train.txt")}),
SplitGenerator(Split.TEST, gen_kwargs={"filepath": os.path.join(dl_manager.manual_dir, "test.txt")}),
]
def _generate_examples(self, filepath, **kwargs):
with open(filepath, "r", encoding="utf-8") as f:
for i, line in enumerate(f):
yield i, {"text": line.strip()}
"""
SAMPLE_DATASET_IDENTIFIER = "hf-internal-testing/dataset_with_script" # has dataset script and also a parquet export
SAMPLE_DATASET_IDENTIFIER2 = "hf-internal-testing/dataset_with_data_files" # only has data files
SAMPLE_DATASET_IDENTIFIER3 = "hf-internal-testing/multi_dir_dataset" # has multiple data directories
SAMPLE_DATASET_IDENTIFIER4 = "hf-internal-testing/imagefolder_with_metadata" # imagefolder with a metadata file outside of the train/test directories
SAMPLE_DATASET_IDENTIFIER5 = "hf-internal-testing/imagefolder_with_metadata_no_splits" # imagefolder with a metadata file and no default split names in data files
SAMPLE_NOT_EXISTING_DATASET_IDENTIFIER = "hf-internal-testing/_dummy"
SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST = "_dummy"
SAMPLE_DATASET_NO_CONFIGS_IN_METADATA = "hf-internal-testing/audiofolder_no_configs_in_metadata"
SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA = "hf-internal-testing/audiofolder_single_config_in_metadata"
SAMPLE_DATASET_TWO_CONFIG_IN_METADATA = "hf-internal-testing/audiofolder_two_configs_in_metadata"
SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT = (
"hf-internal-testing/audiofolder_two_configs_in_metadata_with_default"
)
METRIC_LOADING_SCRIPT_NAME = "__dummy_metric1__"
METRIC_LOADING_SCRIPT_CODE = """
import datasets
from datasets import MetricInfo, Features, Value
class __DummyMetric1__(datasets.Metric):
def _info(self):
return MetricInfo(features=Features({"predictions": Value("int"), "references": Value("int")}))
def _compute(self, predictions, references):
return {"__dummy_metric1__": sum(int(p == r) for p, r in zip(predictions, references))}
"""
@pytest.fixture
def data_dir(tmp_path):
data_dir = tmp_path / "data_dir"
data_dir.mkdir()
with open(data_dir / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(data_dir / "test.txt", "w") as f:
f.write("bar\n" * 10)
return str(data_dir)
@pytest.fixture
def data_dir_with_arrow(tmp_path):
data_dir = tmp_path / "data_dir"
data_dir.mkdir()
output_train = os.path.join(data_dir, "train.arrow")
with ArrowWriter(path=output_train) as writer:
writer.write_table(pa.Table.from_pydict({"col_1": ["foo"] * 10}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 10
assert num_bytes > 0
output_test = os.path.join(data_dir, "test.arrow")
with ArrowWriter(path=output_test) as writer:
writer.write_table(pa.Table.from_pydict({"col_1": ["bar"] * 10}))
num_examples, num_bytes = writer.finalize()
assert num_examples == 10
assert num_bytes > 0
return str(data_dir)
@pytest.fixture
def data_dir_with_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_metadata"
data_dir.mkdir()
with open(data_dir / "train.jpg", "wb") as f:
f.write(b"train_image_bytes")
with open(data_dir / "test.jpg", "wb") as f:
f.write(b"test_image_bytes")
with open(data_dir / "metadata.jsonl", "w") as f:
f.write(
"""\
{"file_name": "train.jpg", "caption": "Cool tran image"}
{"file_name": "test.jpg", "caption": "Cool test image"}
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_single_config_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_one_default_config_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
with open(data_dir / "README.md", "w") as f:
f.write(
f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: custom
drop_labels: true
---
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_config_and_data_files(tmp_path):
data_dir = tmp_path / "data_dir_with_config_and_data_files"
cats_data_dir = data_dir / "data" / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "data" / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
with open(data_dir / "README.md", "w") as f:
f.write(
f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: custom
data_files: "data/**/*.jpg"
---
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_two_config_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_two_configs_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
with open(data_dir / "README.md", "w") as f:
f.write(
f"""\
---
{METADATA_CONFIGS_FIELD}:
- config_name: "v1"
drop_labels: true
default: true
- config_name: "v2"
drop_labels: false
---
"""
)
return str(data_dir)
@pytest.fixture
def data_dir_with_data_dir_configs_in_metadata(tmp_path):
data_dir = tmp_path / "data_dir_with_two_configs_in_metadata"
cats_data_dir = data_dir / "cats"
cats_data_dir.mkdir(parents=True)
dogs_data_dir = data_dir / "dogs"
dogs_data_dir.mkdir(parents=True)
with open(cats_data_dir / "cat.jpg", "wb") as f:
f.write(b"this_is_a_cat_image_bytes")
with open(dogs_data_dir / "dog.jpg", "wb") as f:
f.write(b"this_is_a_dog_image_bytes")
@pytest.fixture
def sub_data_dirs(tmp_path):
data_dir2 = tmp_path / "data_dir2"
relative_subdir1 = "subdir1"
sub_data_dir1 = data_dir2 / relative_subdir1
sub_data_dir1.mkdir(parents=True)
with open(sub_data_dir1 / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(sub_data_dir1 / "test.txt", "w") as f:
f.write("bar\n" * 10)
relative_subdir2 = "subdir2"
sub_data_dir2 = tmp_path / data_dir2 / relative_subdir2
sub_data_dir2.mkdir(parents=True)
with open(sub_data_dir2 / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(sub_data_dir2 / "test.txt", "w") as f:
f.write("bar\n" * 10)
return str(data_dir2), relative_subdir1
@pytest.fixture
def complex_data_dir(tmp_path):
data_dir = tmp_path / "complex_data_dir"
data_dir.mkdir()
(data_dir / "data").mkdir()
with open(data_dir / "data" / "train.txt", "w") as f:
f.write("foo\n" * 10)
with open(data_dir / "data" / "test.txt", "w") as f:
f.write("bar\n" * 10)
with open(data_dir / "README.md", "w") as f:
f.write("This is a readme")
with open(data_dir / ".dummy", "w") as f:
f.write("this is a dummy file that is not a data file")
return str(data_dir)
@pytest.fixture
def dataset_loading_script_dir(tmp_path):
script_name = DATASET_LOADING_SCRIPT_NAME
script_dir = tmp_path / script_name
script_dir.mkdir()
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(DATASET_LOADING_SCRIPT_CODE)
return str(script_dir)
@pytest.fixture
def dataset_loading_script_dir_readonly(tmp_path):
script_name = DATASET_LOADING_SCRIPT_NAME
script_dir = tmp_path / "readonly" / script_name
script_dir.mkdir(parents=True)
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(DATASET_LOADING_SCRIPT_CODE)
dataset_loading_script_dir = str(script_dir)
# Make this directory readonly
os.chmod(dataset_loading_script_dir, 0o555)
os.chmod(os.path.join(dataset_loading_script_dir, f"{script_name}.py"), 0o555)
return dataset_loading_script_dir
@pytest.fixture
def metric_loading_script_dir(tmp_path):
script_name = METRIC_LOADING_SCRIPT_NAME
script_dir = tmp_path / script_name
script_dir.mkdir()
script_path = script_dir / f"{script_name}.py"
with open(script_path, "w") as f:
f.write(METRIC_LOADING_SCRIPT_CODE)
return str(script_dir)
@pytest.mark.parametrize(
"data_files, expected_module, expected_builder_kwargs",
[
(["train.csv"], "csv", {}),
(["train.tsv"], "csv", {"sep": "\t"}),
(["train.json"], "json", {}),
(["train.jsonl"], "json", {}),
(["train.parquet"], "parquet", {}),
(["train.geoparquet"], "parquet", {}),
(["train.gpq"], "parquet", {}),
(["train.arrow"], "arrow", {}),
(["train.txt"], "text", {}),
(["uppercase.TXT"], "text", {}),
(["unsupported.ext"], None, {}),
([""], None, {}),
],
)
def test_infer_module_for_data_files(data_files, expected_module, expected_builder_kwargs):
module, builder_kwargs = infer_module_for_data_files_list(data_files)
assert module == expected_module
assert builder_kwargs == expected_builder_kwargs
@pytest.mark.parametrize(
"data_file, expected_module",
[
("zip_csv_path", "csv"),
("zip_csv_with_dir_path", "csv"),
("zip_uppercase_csv_path", "csv"),
("zip_unsupported_ext_path", None),
],
)
def test_infer_module_for_data_files_in_archives(
data_file, expected_module, zip_csv_path, zip_csv_with_dir_path, zip_uppercase_csv_path, zip_unsupported_ext_path
):
data_file_paths = {
"zip_csv_path": zip_csv_path,
"zip_csv_with_dir_path": zip_csv_with_dir_path,
"zip_uppercase_csv_path": zip_uppercase_csv_path,
"zip_unsupported_ext_path": zip_unsupported_ext_path,
}
data_files = [str(data_file_paths[data_file])]
inferred_module, _ = infer_module_for_data_files_list_in_archives(data_files)
assert inferred_module == expected_module
class ModuleFactoryTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(
self,
jsonl_path,
data_dir,
data_dir_with_metadata,
data_dir_with_single_config_in_metadata,
data_dir_with_config_and_data_files,
data_dir_with_two_config_in_metadata,
sub_data_dirs,
dataset_loading_script_dir,
metric_loading_script_dir,
):
self._jsonl_path = jsonl_path
self._data_dir = data_dir
self._data_dir_with_metadata = data_dir_with_metadata
self._data_dir_with_single_config_in_metadata = data_dir_with_single_config_in_metadata
self._data_dir_with_config_and_data_files = data_dir_with_config_and_data_files
self._data_dir_with_two_config_in_metadata = data_dir_with_two_config_in_metadata
self._data_dir2 = sub_data_dirs[0]
self._sub_data_dir = sub_data_dirs[1]
self._dataset_loading_script_dir = dataset_loading_script_dir
self._metric_loading_script_dir = metric_loading_script_dir
def setUp(self):
self.hf_modules_cache = tempfile.mkdtemp()
self.cache_dir = tempfile.mkdtemp()
self.download_config = DownloadConfig(cache_dir=self.cache_dir)
self.dynamic_modules_path = datasets.load.init_dynamic_modules(
name="test_datasets_modules_" + os.path.basename(self.hf_modules_cache),
hf_modules_cache=self.hf_modules_cache,
)
def test_HubDatasetModuleFactoryWithScript_dont_trust_remote_code(self):
# "lhoestq/test" has a dataset script
factory = HubDatasetModuleFactoryWithScript(
"lhoestq/test", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
with patch.object(config, "HF_DATASETS_TRUST_REMOTE_CODE", None): # this will be the default soon
self.assertRaises(ValueError, factory.get_module)
factory = HubDatasetModuleFactoryWithScript(
"lhoestq/test",
download_config=self.download_config,
dynamic_modules_path=self.dynamic_modules_path,
trust_remote_code=False,
)
self.assertRaises(ValueError, factory.get_module)
def test_HubDatasetModuleFactoryWithScript_with_github_dataset(self):
# "wmt_t2t" has additional imports (internal)
factory = HubDatasetModuleFactoryWithScript(
"wmt_t2t", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
def test_GithubMetricModuleFactory_with_internal_import(self):
# "squad_v2" requires additional imports (internal)
factory = GithubMetricModuleFactory(
"squad_v2", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.filterwarnings("ignore:GithubMetricModuleFactory is deprecated:FutureWarning")
def test_GithubMetricModuleFactory_with_external_import(self):
# "bleu" requires additional imports (external from github)
factory = GithubMetricModuleFactory(
"bleu", download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_LocalMetricModuleFactory(self):
path = os.path.join(self._metric_loading_script_dir, f"{METRIC_LOADING_SCRIPT_NAME}.py")
factory = LocalMetricModuleFactory(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_LocalDatasetModuleFactoryWithScript(self):
path = os.path.join(self._dataset_loading_script_dir, f"{DATASET_LOADING_SCRIPT_NAME}.py")
factory = LocalDatasetModuleFactoryWithScript(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert os.path.isdir(module_factory_result.builder_kwargs["base_path"])
def test_LocalDatasetModuleFactoryWithScript_dont_trust_remote_code(self):
path = os.path.join(self._dataset_loading_script_dir, f"{DATASET_LOADING_SCRIPT_NAME}.py")
factory = LocalDatasetModuleFactoryWithScript(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
with patch.object(config, "HF_DATASETS_TRUST_REMOTE_CODE", None): # this will be the default soon
self.assertRaises(ValueError, factory.get_module)
factory = LocalDatasetModuleFactoryWithScript(
path,
download_config=self.download_config,
dynamic_modules_path=self.dynamic_modules_path,
trust_remote_code=False,
)
self.assertRaises(ValueError, factory.get_module)
def test_LocalDatasetModuleFactoryWithoutScript(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert os.path.isdir(module_factory_result.builder_kwargs["base_path"])
def test_LocalDatasetModuleFactoryWithoutScript_with_data_dir(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir2, data_dir=self._sub_data_dir)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
builder_config = module_factory_result.builder_configs_parameters.builder_configs[0]
assert (
builder_config.data_files is not None
and len(builder_config.data_files["train"]) == 1
and len(builder_config.data_files["test"]) == 1
)
assert all(
self._sub_data_dir in Path(data_file).parts
for data_file in builder_config.data_files["train"] + builder_config.data_files["test"]
)
def test_LocalDatasetModuleFactoryWithoutScript_with_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir_with_metadata)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
builder_config = module_factory_result.builder_configs_parameters.builder_configs[0]
assert (
builder_config.data_files is not None
and len(builder_config.data_files["train"]) > 0
and len(builder_config.data_files["test"]) > 0
)
assert any(Path(data_file).name == "metadata.jsonl" for data_file in builder_config.data_files["train"])
assert any(Path(data_file).name == "metadata.jsonl" for data_file in builder_config.data_files["test"])
def test_LocalDatasetModuleFactoryWithoutScript_with_single_config_in_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(
self._data_dir_with_single_config_in_metadata,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "drop_labels" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["drop_labels"] is True
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 1
assert isinstance(module_builder_configs[0], ImageFolderConfig)
assert module_builder_configs[0].name == "custom"
assert module_builder_configs[0].data_files is not None
assert isinstance(module_builder_configs[0].data_files, DataFilesPatternsDict)
module_builder_configs[0]._resolve_data_files(self._data_dir_with_single_config_in_metadata, DownloadConfig())
assert isinstance(module_builder_configs[0].data_files, DataFilesDict)
assert len(module_builder_configs[0].data_files) == 1 # one train split
assert len(module_builder_configs[0].data_files["train"]) == 2 # two files
assert module_builder_configs[0].drop_labels is True # parameter is passed from metadata
# config named "default" is automatically considered to be a default config
assert module_factory_result.builder_configs_parameters.default_config_name == "custom"
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
def test_LocalDatasetModuleFactoryWithoutScript_with_config_and_data_files(self):
factory = LocalDatasetModuleFactoryWithoutScript(
self._data_dir_with_config_and_data_files,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
builder_kwargs = module_factory_result.builder_kwargs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "data_files" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["data_files"] == "data/**/*.jpg"
assert "data_files" not in builder_kwargs
def test_LocalDatasetModuleFactoryWithoutScript_data_dir_with_config_and_data_files(self):
factory = LocalDatasetModuleFactoryWithoutScript(self._data_dir_with_config_and_data_files, data_dir="data")
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
builder_kwargs = module_factory_result.builder_kwargs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "data_files" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["data_files"] == "data/**/*.jpg"
assert "data_files" in builder_kwargs
assert "train" in builder_kwargs["data_files"]
assert len(builder_kwargs["data_files"]["train"]) == 2
def test_LocalDatasetModuleFactoryWithoutScript_with_two_configs_in_metadata(self):
factory = LocalDatasetModuleFactoryWithoutScript(
self._data_dir_with_two_config_in_metadata,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 2
assert list(module_metadata_configs) == ["v1", "v2"]
assert "drop_labels" in module_metadata_configs["v1"]
assert module_metadata_configs["v1"]["drop_labels"] is True
assert "drop_labels" in module_metadata_configs["v2"]
assert module_metadata_configs["v2"]["drop_labels"] is False
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 2
module_builder_config_v1, module_builder_config_v2 = module_builder_configs
assert module_builder_config_v1.name == "v1"
assert module_builder_config_v2.name == "v2"
assert isinstance(module_builder_config_v1, ImageFolderConfig)
assert isinstance(module_builder_config_v2, ImageFolderConfig)
assert isinstance(module_builder_config_v1.data_files, DataFilesPatternsDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesPatternsDict)
module_builder_config_v1._resolve_data_files(self._data_dir_with_two_config_in_metadata, DownloadConfig())
module_builder_config_v2._resolve_data_files(self._data_dir_with_two_config_in_metadata, DownloadConfig())
assert isinstance(module_builder_config_v1.data_files, DataFilesDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesDict)
assert sorted(module_builder_config_v1.data_files) == ["train"]
assert len(module_builder_config_v1.data_files["train"]) == 2
assert sorted(module_builder_config_v2.data_files) == ["train"]
assert len(module_builder_config_v2.data_files["train"]) == 2
assert module_builder_config_v1.drop_labels is True # parameter is passed from metadata
assert module_builder_config_v2.drop_labels is False # parameter is passed from metadata
assert (
module_factory_result.builder_configs_parameters.default_config_name == "v1"
) # it's marked as a default one in yaml
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
def test_PackagedDatasetModuleFactory(self):
factory = PackagedDatasetModuleFactory(
"json", data_files=self._jsonl_path, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_PackagedDatasetModuleFactory_with_data_dir(self):
factory = PackagedDatasetModuleFactory("json", data_dir=self._data_dir, download_config=self.download_config)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
data_files = module_factory_result.builder_kwargs.get("data_files")
assert data_files is not None and len(data_files["train"]) > 0 and len(data_files["test"]) > 0
assert Path(data_files["train"][0]).parent.samefile(self._data_dir)
assert Path(data_files["test"][0]).parent.samefile(self._data_dir)
def test_PackagedDatasetModuleFactory_with_data_dir_and_metadata(self):
factory = PackagedDatasetModuleFactory(
"imagefolder", data_dir=self._data_dir_with_metadata, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
data_files = module_factory_result.builder_kwargs.get("data_files")
assert data_files is not None and len(data_files["train"]) > 0 and len(data_files["test"]) > 0
assert Path(data_files["train"][0]).parent.samefile(self._data_dir_with_metadata)
assert Path(data_files["test"][0]).parent.samefile(self._data_dir_with_metadata)
assert any(Path(data_file).name == "metadata.jsonl" for data_file in data_files["train"])
assert any(Path(data_file).name == "metadata.jsonl" for data_file in data_files["test"])
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER2, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_data_dir(self):
data_dir = "data2"
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER3, data_dir=data_dir, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
builder_config = module_factory_result.builder_configs_parameters.builder_configs[0]
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
builder_config.data_files is not None
and len(builder_config.data_files["train"]) == 1
and len(builder_config.data_files["test"]) == 1
)
assert all(
data_dir in Path(data_file).parts
for data_file in builder_config.data_files["train"] + builder_config.data_files["test"]
)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_metadata(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER4, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
builder_config = module_factory_result.builder_configs_parameters.builder_configs[0]
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
builder_config.data_files is not None
and len(builder_config.data_files["train"]) > 0
and len(builder_config.data_files["test"]) > 0
)
assert any(Path(data_file).name == "metadata.jsonl" for data_file in builder_config.data_files["train"])
assert any(Path(data_file).name == "metadata.jsonl" for data_file in builder_config.data_files["test"])
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_IDENTIFIER5, download_config=self.download_config
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
builder_config = module_factory_result.builder_configs_parameters.builder_configs[0]
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
assert (
builder_config.data_files is not None
and len(builder_config.data_files) == 1
and len(builder_config.data_files["train"]) > 0
)
assert any(Path(data_file).name == "metadata.jsonl" for data_file in builder_config.data_files["train"])
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_one_default_config_in_metadata(self):
factory = HubDatasetModuleFactoryWithoutScript(
SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA,
download_config=self.download_config,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 1
assert next(iter(module_metadata_configs)) == "custom"
assert "drop_labels" in next(iter(module_metadata_configs.values()))
assert next(iter(module_metadata_configs.values()))["drop_labels"] is True
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 1
assert isinstance(module_builder_configs[0], AudioFolderConfig)
assert module_builder_configs[0].name == "custom"
assert module_builder_configs[0].data_files is not None
assert isinstance(module_builder_configs[0].data_files, DataFilesPatternsDict)
module_builder_configs[0]._resolve_data_files(
module_factory_result.builder_kwargs["base_path"], DownloadConfig()
)
assert isinstance(module_builder_configs[0].data_files, DataFilesDict)
assert sorted(module_builder_configs[0].data_files) == ["test", "train"]
assert len(module_builder_configs[0].data_files["train"]) == 3
assert len(module_builder_configs[0].data_files["test"]) == 3
assert module_builder_configs[0].drop_labels is True # parameter is passed from metadata
# config named "default" is automatically considered to be a default config
assert module_factory_result.builder_configs_parameters.default_config_name == "custom"
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithoutScript_with_two_configs_in_metadata(self):
datasets_names = [SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT]
for dataset_name in datasets_names:
factory = HubDatasetModuleFactoryWithoutScript(dataset_name, download_config=self.download_config)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
module_metadata_configs = module_factory_result.builder_configs_parameters.metadata_configs
assert module_metadata_configs is not None
assert len(module_metadata_configs) == 2
assert list(module_metadata_configs) == ["v1", "v2"]
assert "drop_labels" in module_metadata_configs["v1"]
assert module_metadata_configs["v1"]["drop_labels"] is True
assert "drop_labels" in module_metadata_configs["v2"]
assert module_metadata_configs["v2"]["drop_labels"] is False
module_builder_configs = module_factory_result.builder_configs_parameters.builder_configs
assert module_builder_configs is not None
assert len(module_builder_configs) == 2
module_builder_config_v1, module_builder_config_v2 = module_builder_configs
assert module_builder_config_v1.name == "v1"
assert module_builder_config_v2.name == "v2"
assert isinstance(module_builder_config_v1, AudioFolderConfig)
assert isinstance(module_builder_config_v2, AudioFolderConfig)
assert isinstance(module_builder_config_v1.data_files, DataFilesPatternsDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesPatternsDict)
module_builder_config_v1._resolve_data_files(
module_factory_result.builder_kwargs["base_path"], DownloadConfig()
)
module_builder_config_v2._resolve_data_files(
module_factory_result.builder_kwargs["base_path"], DownloadConfig()
)
assert isinstance(module_builder_config_v1.data_files, DataFilesDict)
assert isinstance(module_builder_config_v2.data_files, DataFilesDict)
assert sorted(module_builder_config_v1.data_files) == ["test", "train"]
assert len(module_builder_config_v1.data_files["train"]) == 3
assert len(module_builder_config_v1.data_files["test"]) == 3
assert sorted(module_builder_config_v2.data_files) == ["test", "train"]
assert len(module_builder_config_v2.data_files["train"]) == 2
assert len(module_builder_config_v2.data_files["test"]) == 1
assert module_builder_config_v1.drop_labels is True # parameter is passed from metadata
assert module_builder_config_v2.drop_labels is False # parameter is passed from metadata
# we don't pass config params to builder in builder_kwargs, they are stored in builder_configs directly
assert "drop_labels" not in module_factory_result.builder_kwargs
if dataset_name == SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT:
assert module_factory_result.builder_configs_parameters.default_config_name == "v1"
else:
assert module_factory_result.builder_configs_parameters.default_config_name is None
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithScript(self):
factory = HubDatasetModuleFactoryWithScript(
SAMPLE_DATASET_IDENTIFIER,
download_config=self.download_config,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
assert module_factory_result.builder_kwargs["base_path"].startswith(config.HF_ENDPOINT)
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithParquetExport(self):
factory = HubDatasetModuleFactoryWithParquetExport(
SAMPLE_DATASET_IDENTIFIER,
download_config=self.download_config,
)
module_factory_result = factory.get_module()
assert module_factory_result.module_path == "datasets.packaged_modules.parquet.parquet"
assert module_factory_result.builder_configs_parameters.builder_configs
assert isinstance(module_factory_result.builder_configs_parameters.builder_configs[0], ParquetConfig)
module_factory_result.builder_configs_parameters.builder_configs[0]._resolve_data_files(
base_path="", download_config=self.download_config
)
assert module_factory_result.builder_configs_parameters.builder_configs[0].data_files == {
"train": [
"hf://datasets/hf-internal-testing/dataset_with_script@8f965694d611974ef8661618ada1b5aeb1072915/default/train/0000.parquet"
],
"validation": [
"hf://datasets/hf-internal-testing/dataset_with_script@8f965694d611974ef8661618ada1b5aeb1072915/default/validation/0000.parquet"
],
}
@pytest.mark.integration
def test_HubDatasetModuleFactoryWithParquetExport_errors_on_wrong_sha(self):
factory = HubDatasetModuleFactoryWithParquetExport(
SAMPLE_DATASET_IDENTIFIER,
download_config=self.download_config,
revision="1a21ac5846fc3f36ad5f128740c58932d3d7806f",
)
factory.get_module()
factory = HubDatasetModuleFactoryWithParquetExport(
SAMPLE_DATASET_IDENTIFIER,
download_config=self.download_config,
revision="wrong_sha",
)
with self.assertRaises(_datasets_server.DatasetsServerError):
factory.get_module()
@pytest.mark.integration
def test_CachedDatasetModuleFactory(self):
name = SAMPLE_DATASET_IDENTIFIER2
load_dataset_builder(name, cache_dir=self.cache_dir).download_and_prepare()
for offline_mode in OfflineSimulationMode:
with offline(offline_mode):
factory = CachedDatasetModuleFactory(
name,
cache_dir=self.cache_dir,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
def test_CachedDatasetModuleFactory_with_script(self):
path = os.path.join(self._dataset_loading_script_dir, f"{DATASET_LOADING_SCRIPT_NAME}.py")
factory = LocalDatasetModuleFactoryWithScript(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
for offline_mode in OfflineSimulationMode:
with offline(offline_mode):
factory = CachedDatasetModuleFactory(
DATASET_LOADING_SCRIPT_NAME,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.filterwarnings("ignore:LocalMetricModuleFactory is deprecated:FutureWarning")
@pytest.mark.filterwarnings("ignore:CachedMetricModuleFactory is deprecated:FutureWarning")
def test_CachedMetricModuleFactory(self):
path = os.path.join(self._metric_loading_script_dir, f"{METRIC_LOADING_SCRIPT_NAME}.py")
factory = LocalMetricModuleFactory(
path, download_config=self.download_config, dynamic_modules_path=self.dynamic_modules_path
)
module_factory_result = factory.get_module()
for offline_mode in OfflineSimulationMode:
with offline(offline_mode):
factory = CachedMetricModuleFactory(
METRIC_LOADING_SCRIPT_NAME,
dynamic_modules_path=self.dynamic_modules_path,
)
module_factory_result = factory.get_module()
assert importlib.import_module(module_factory_result.module_path) is not None
@pytest.mark.parametrize(
"factory_class",
[
CachedDatasetModuleFactory,
CachedMetricModuleFactory,
GithubMetricModuleFactory,
HubDatasetModuleFactoryWithoutScript,
HubDatasetModuleFactoryWithScript,
LocalDatasetModuleFactoryWithoutScript,
LocalDatasetModuleFactoryWithScript,
LocalMetricModuleFactory,
PackagedDatasetModuleFactory,
],
)
def test_module_factories(factory_class):
name = "dummy_name"
factory = factory_class(name)
assert factory.name == name
@pytest.mark.integration
class LoadTest(TestCase):
@pytest.fixture(autouse=True)
def inject_fixtures(self, caplog):
self._caplog = caplog
def setUp(self):
self.hf_modules_cache = tempfile.mkdtemp()
self.cache_dir = tempfile.mkdtemp()
self.dynamic_modules_path = datasets.load.init_dynamic_modules(
name="test_datasets_modules2", hf_modules_cache=self.hf_modules_cache
)
def tearDown(self):
shutil.rmtree(self.hf_modules_cache)
shutil.rmtree(self.cache_dir)
def _dummy_module_dir(self, modules_dir, dummy_module_name, dummy_code):
assert dummy_module_name.startswith("__")
module_dir = os.path.join(modules_dir, dummy_module_name)
os.makedirs(module_dir, exist_ok=True)
module_path = os.path.join(module_dir, dummy_module_name + ".py")
with open(module_path, "w") as f:
f.write(dummy_code)
return module_dir
def test_dataset_module_factory(self):
with tempfile.TemporaryDirectory() as tmp_dir:
# prepare module from directory path
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
dataset_module = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "hello there")
self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# prepare module from file path + check resolved_file_path
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name1__", dummy_code)
module_path = os.path.join(module_dir, "__dummy_module_name1__.py")
dataset_module = datasets.load.dataset_module_factory(
module_path, dynamic_modules_path=self.dynamic_modules_path
)
dummy_module = importlib.import_module(dataset_module.module_path)
self.assertEqual(dummy_module.MY_DUMMY_VARIABLE, "general kenobi")
self.assertEqual(dataset_module.hash, sha256(dummy_code.encode("utf-8")).hexdigest())
# missing module
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises(
(DatasetNotFoundError, ConnectionError, requests.exceptions.ConnectionError)
):
datasets.load.dataset_module_factory(
"__missing_dummy_module_name__", dynamic_modules_path=self.dynamic_modules_path
)
@pytest.mark.integration
def test_offline_dataset_module_factory(self):
repo_id = SAMPLE_DATASET_IDENTIFIER2
builder = load_dataset_builder(repo_id, cache_dir=self.cache_dir)
builder.download_and_prepare()
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
self._caplog.clear()
# allow provide the repo id without an explicit path to remote or local actual file
dataset_module = datasets.load.dataset_module_factory(repo_id, cache_dir=self.cache_dir)
self.assertEqual(dataset_module.module_path, "datasets.packaged_modules.cache.cache")
self.assertIn("Using the latest cached version of the dataset", self._caplog.text)
def test_offline_dataset_module_factory_with_script(self):
with tempfile.TemporaryDirectory() as tmp_dir:
dummy_code = "MY_DUMMY_VARIABLE = 'hello there'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
dataset_module_1 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
time.sleep(0.1) # make sure there's a difference in the OS update time of the python file
dummy_code = "MY_DUMMY_VARIABLE = 'general kenobi'"
module_dir = self._dummy_module_dir(tmp_dir, "__dummy_module_name2__", dummy_code)
dataset_module_2 = datasets.load.dataset_module_factory(
module_dir, dynamic_modules_path=self.dynamic_modules_path
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
self._caplog.clear()
# allow provide the module name without an explicit path to remote or local actual file
dataset_module_3 = datasets.load.dataset_module_factory(
"__dummy_module_name2__", dynamic_modules_path=self.dynamic_modules_path
)
# it loads the most recent version of the module
self.assertEqual(dataset_module_2.module_path, dataset_module_3.module_path)
self.assertNotEqual(dataset_module_1.module_path, dataset_module_3.module_path)
self.assertIn("Using the latest cached version of the module", self._caplog.text)
def test_load_dataset_from_hub(self):
with self.assertRaises(DatasetNotFoundError) as context:
datasets.load_dataset("_dummy")
self.assertIn(
"Dataset '_dummy' doesn't exist on the Hub",
str(context.exception),
)
with self.assertRaises(DatasetNotFoundError) as context:
datasets.load_dataset("_dummy", revision="0.0.0")
self.assertIn(
"Dataset '_dummy' doesn't exist on the Hub",
str(context.exception),
)
self.assertIn(
"at revision '0.0.0'",
str(context.exception),
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises(ConnectionError) as context:
datasets.load_dataset("_dummy")
if offline_simulation_mode != OfflineSimulationMode.HF_DATASETS_OFFLINE_SET_TO_1:
self.assertIn(
"Couldn't reach '_dummy' on the Hub",
str(context.exception),
)
def test_load_dataset_namespace(self):
with self.assertRaises(DatasetNotFoundError) as context:
datasets.load_dataset("hf-internal-testing/_dummy")
self.assertIn(
"hf-internal-testing/_dummy",
str(context.exception),
)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
with self.assertRaises(ConnectionError) as context:
datasets.load_dataset("hf-internal-testing/_dummy")
self.assertIn("hf-internal-testing/_dummy", str(context.exception), msg=offline_simulation_mode)
@pytest.mark.integration
def test_load_dataset_builder_with_metadata():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4)
assert isinstance(builder, ImageFolder)
assert builder.config.name == "default"
assert builder.config.data_files is not None
assert builder.config.drop_metadata is None
with pytest.raises(ValueError):
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4, "non-existing-config")
@pytest.mark.integration
def test_load_dataset_builder_config_kwargs_passed_as_arguments():
builder_default = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4)
builder_custom = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER4, drop_metadata=True)
assert builder_custom.config.drop_metadata != builder_default.config.drop_metadata
assert builder_custom.config.drop_metadata is True
@pytest.mark.integration
def test_load_dataset_builder_with_two_configs_in_metadata():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
assert isinstance(builder, AudioFolder)
assert builder.config.name == "v1"
assert builder.config.data_files is not None
with pytest.raises(ValueError):
datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA)
with pytest.raises(ValueError):
datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "non-existing-config")
@pytest.mark.parametrize("serializer", [pickle, dill])
def test_load_dataset_builder_with_metadata_configs_pickable(serializer):
builder = datasets.load_dataset_builder(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA)
builder_unpickled = serializer.loads(serializer.dumps(builder))
assert builder.BUILDER_CONFIGS == builder_unpickled.BUILDER_CONFIGS
assert list(builder_unpickled.builder_configs) == ["custom"]
assert isinstance(builder_unpickled.builder_configs["custom"], AudioFolderConfig)
builder2 = datasets.load_dataset_builder(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
builder2_unpickled = serializer.loads(serializer.dumps(builder2))
assert builder2.BUILDER_CONFIGS == builder2_unpickled.BUILDER_CONFIGS != builder_unpickled.BUILDER_CONFIGS
assert list(builder2_unpickled.builder_configs) == ["v1", "v2"]
assert isinstance(builder2_unpickled.builder_configs["v1"], AudioFolderConfig)
assert isinstance(builder2_unpickled.builder_configs["v2"], AudioFolderConfig)
def test_load_dataset_builder_for_absolute_script_dir(dataset_loading_script_dir, data_dir):
builder = datasets.load_dataset_builder(dataset_loading_script_dir, data_dir=data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_relative_script_dir(dataset_loading_script_dir, data_dir):
with set_current_working_directory_to_temp_dir():
relative_script_dir = DATASET_LOADING_SCRIPT_NAME
shutil.copytree(dataset_loading_script_dir, relative_script_dir)
builder = datasets.load_dataset_builder(relative_script_dir, data_dir=data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_script_path(dataset_loading_script_dir, data_dir):
builder = datasets.load_dataset_builder(
os.path.join(dataset_loading_script_dir, DATASET_LOADING_SCRIPT_NAME + ".py"), data_dir=data_dir
)
assert isinstance(builder, DatasetBuilder)
assert builder.name == DATASET_LOADING_SCRIPT_NAME
assert builder.dataset_name == DATASET_LOADING_SCRIPT_NAME
assert builder.info.features == Features({"text": Value("string")})
def test_load_dataset_builder_for_absolute_data_dir(complex_data_dir):
builder = datasets.load_dataset_builder(complex_data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == Path(complex_data_dir).name
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
def test_load_dataset_builder_for_relative_data_dir(complex_data_dir):
with set_current_working_directory_to_temp_dir():
relative_data_dir = "relative_data_dir"
shutil.copytree(complex_data_dir, relative_data_dir)
builder = datasets.load_dataset_builder(relative_data_dir)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == relative_data_dir
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
@pytest.mark.integration
def test_load_dataset_builder_for_community_dataset_with_script():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "parquet"
assert builder.dataset_name == SAMPLE_DATASET_IDENTIFIER.split("/")[-1]
assert builder.config.name == "default"
assert builder.info.features == Features({"text": Value("string")})
namespace = SAMPLE_DATASET_IDENTIFIER[: SAMPLE_DATASET_IDENTIFIER.index("/")]
assert builder._relative_data_dir().startswith(namespace)
assert builder.__module__.startswith("datasets.")
@pytest.mark.integration
def test_load_dataset_builder_for_community_dataset_with_script_no_parquet_export():
with patch.object(config, "USE_PARQUET_EXPORT", False):
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER)
assert isinstance(builder, DatasetBuilder)
assert builder.name == SAMPLE_DATASET_IDENTIFIER.split("/")[-1]
assert builder.dataset_name == SAMPLE_DATASET_IDENTIFIER.split("/")[-1]
assert builder.config.name == "default"
assert builder.info.features == Features({"text": Value("string")})
namespace = SAMPLE_DATASET_IDENTIFIER[: SAMPLE_DATASET_IDENTIFIER.index("/")]
assert builder._relative_data_dir().startswith(namespace)
assert SAMPLE_DATASET_IDENTIFIER.replace("/", "--") in builder.__module__
@pytest.mark.integration
def test_load_dataset_builder_use_parquet_export_if_dont_trust_remote_code_keeps_features():
dataset_name = "food101"
builder = datasets.load_dataset_builder(dataset_name, trust_remote_code=False)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "parquet"
assert builder.dataset_name == dataset_name
assert builder.config.name == "default"
assert list(builder.info.features) == ["image", "label"]
assert builder.info.features["image"] == Image()
@pytest.mark.integration
def test_load_dataset_builder_for_community_dataset_without_script():
builder = datasets.load_dataset_builder(SAMPLE_DATASET_IDENTIFIER2)
assert isinstance(builder, DatasetBuilder)
assert builder.name == "text"
assert builder.dataset_name == SAMPLE_DATASET_IDENTIFIER2.split("/")[-1]
assert builder.config.name == "default"
assert isinstance(builder.config.data_files, DataFilesDict)
assert len(builder.config.data_files["train"]) > 0
assert len(builder.config.data_files["test"]) > 0
def test_load_dataset_builder_fail():
with pytest.raises(DatasetNotFoundError):
datasets.load_dataset_builder("blabla")
@pytest.mark.parametrize("keep_in_memory", [False, True])
def test_load_dataset_local_script(dataset_loading_script_dir, data_dir, keep_in_memory, caplog):
with assert_arrow_memory_increases() if keep_in_memory else assert_arrow_memory_doesnt_increase():
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, keep_in_memory=keep_in_memory)
assert isinstance(dataset, DatasetDict)
assert all(isinstance(d, Dataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
def test_load_dataset_cached_local_script(dataset_loading_script_dir, data_dir, caplog):
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir)
assert isinstance(dataset, DatasetDict)
assert all(isinstance(d, Dataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
caplog.clear()
# Load dataset from cache
dataset = datasets.load_dataset(DATASET_LOADING_SCRIPT_NAME, data_dir=data_dir)
assert len(dataset) == 2
assert "Using the latest cached version of the module" in caplog.text
assert isinstance(next(iter(dataset["train"])), dict)
with pytest.raises(DatasetNotFoundError) as exc_info:
datasets.load_dataset(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST)
assert f"Dataset '{SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST}' doesn't exist on the Hub" in str(exc_info.value)
@pytest.mark.integration
@pytest.mark.parametrize("stream_from_cache, ", [False, True])
def test_load_dataset_cached_from_hub(stream_from_cache, caplog):
dataset = load_dataset(SAMPLE_DATASET_IDENTIFIER3)
assert isinstance(dataset, DatasetDict)
assert all(isinstance(d, Dataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
for offline_simulation_mode in list(OfflineSimulationMode):
with offline(offline_simulation_mode):
caplog.clear()
# Load dataset from cache
dataset = datasets.load_dataset(SAMPLE_DATASET_IDENTIFIER3, streaming=stream_from_cache)
assert len(dataset) == 2
assert "Using the latest cached version of the dataset" in caplog.text
assert isinstance(next(iter(dataset["train"])), dict)
with pytest.raises(DatasetNotFoundError) as exc_info:
datasets.load_dataset(SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST)
assert f"Dataset '{SAMPLE_DATASET_NAME_THAT_DOESNT_EXIST}' doesn't exist on the Hub" in str(exc_info.value)
def test_load_dataset_streaming(dataset_loading_script_dir, data_dir):
dataset = load_dataset(dataset_loading_script_dir, streaming=True, data_dir=data_dir)
assert isinstance(dataset, IterableDatasetDict)
assert all(isinstance(d, IterableDataset) for d in dataset.values())
assert len(dataset) == 2
assert isinstance(next(iter(dataset["train"])), dict)
def test_load_dataset_streaming_gz_json(jsonl_gz_path):
data_files = jsonl_gz_path
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.integration
@pytest.mark.parametrize(
"path", ["sample.jsonl", "sample.jsonl.gz", "sample.tar", "sample.jsonl.xz", "sample.zip", "sample.jsonl.zst"]
)
def test_load_dataset_streaming_compressed_files(path):
repo_id = "hf-internal-testing/compressed_files"
data_files = f"https://huggingface.co/datasets/{repo_id}/resolve/main/{path}"
if data_files[-3:] in ("zip", "tar"): # we need to glob "*" inside archives
data_files = data_files[-3:] + "://*::" + data_files
return # TODO(QL, albert): support re-add support for ZIP and TAR archives streaming
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item == {
"tokens": ["Ministeri", "de", "Justícia", "d'Espanya"],
"ner_tags": [1, 2, 2, 2],
"langs": ["ca", "ca", "ca", "ca"],
"spans": ["PER: Ministeri de Justícia d'Espanya"],
}
@pytest.mark.parametrize("path_extension", ["csv", "csv.bz2"])
@pytest.mark.parametrize("streaming", [False, True])
def test_load_dataset_streaming_csv(path_extension, streaming, csv_path, bz2_csv_path):
paths = {"csv": csv_path, "csv.bz2": bz2_csv_path}
data_files = str(paths[path_extension])
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("csv", split="train", data_files=data_files, features=features, streaming=streaming)
assert isinstance(ds, IterableDataset if streaming else Dataset)
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_csv_path", "zip_csv_with_dir_path", "csv_path"])
def test_load_dataset_zip_csv(data_file, streaming, zip_csv_path, zip_csv_with_dir_path, csv_path):
data_file_paths = {
"zip_csv_path": zip_csv_path,
"zip_csv_with_dir_path": zip_csv_with_dir_path,
"csv_path": csv_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("csv", split="train", data_files=data_files, features=features, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_jsonl_path", "zip_jsonl_with_dir_path", "jsonl_path"])
def test_load_dataset_zip_jsonl(data_file, streaming, zip_jsonl_path, zip_jsonl_with_dir_path, jsonl_path):
data_file_paths = {
"zip_jsonl_path": zip_jsonl_path,
"zip_jsonl_with_dir_path": zip_jsonl_with_dir_path,
"jsonl_path": jsonl_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
features = Features({"col_1": Value("string"), "col_2": Value("int32"), "col_3": Value("float32")})
ds = load_dataset("json", split="train", data_files=data_files, features=features, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "0", "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("streaming", [False, True])
@pytest.mark.parametrize("data_file", ["zip_text_path", "zip_text_with_dir_path", "text_path"])
def test_load_dataset_zip_text(data_file, streaming, zip_text_path, zip_text_with_dir_path, text_path):
data_file_paths = {
"zip_text_path": zip_text_path,
"zip_text_with_dir_path": zip_text_with_dir_path,
"text_path": text_path,
}
data_files = str(data_file_paths[data_file])
expected_size = 8 if data_file.startswith("zip") else 4
ds = load_dataset("text", split="train", data_files=data_files, streaming=streaming)
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"text": "0"}
ds_item_counter += 1
assert ds_item_counter == expected_size
else:
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"text": "0"}
@pytest.mark.parametrize("streaming", [False, True])
def test_load_dataset_arrow(streaming, data_dir_with_arrow):
ds = load_dataset("arrow", split="train", data_dir=data_dir_with_arrow, streaming=streaming)
expected_size = 10
if streaming:
ds_item_counter = 0
for ds_item in ds:
if ds_item_counter == 0:
assert ds_item == {"col_1": "foo"}
ds_item_counter += 1
assert ds_item_counter == 10
else:
assert ds.num_rows == 10
assert ds.shape[0] == expected_size
ds_item = next(iter(ds))
assert ds_item == {"col_1": "foo"}
def test_load_dataset_text_with_unicode_new_lines(text_path_with_unicode_new_lines):
data_files = str(text_path_with_unicode_new_lines)
ds = load_dataset("text", split="train", data_files=data_files)
assert ds.num_rows == 3
def test_load_dataset_with_unsupported_extensions(text_dir_with_unsupported_extension):
data_files = str(text_dir_with_unsupported_extension)
ds = load_dataset("text", split="train", data_files=data_files)
assert ds.num_rows == 4
@pytest.mark.integration
def test_loading_from_the_datasets_hub():
with tempfile.TemporaryDirectory() as tmp_dir:
with load_dataset(SAMPLE_DATASET_IDENTIFIER, cache_dir=tmp_dir) as dataset:
assert len(dataset["train"]) == 2
assert len(dataset["validation"]) == 3
@pytest.mark.integration
def test_loading_from_the_datasets_hub_with_token():
true_request = requests.Session().request
def assert_auth(method, url, *args, headers, **kwargs):
assert headers["authorization"] == "Bearer foo"
return true_request(method, url, *args, headers=headers, **kwargs)
with patch("requests.Session.request") as mock_request:
mock_request.side_effect = assert_auth
with tempfile.TemporaryDirectory() as tmp_dir:
with offline():
with pytest.raises((ConnectionError, requests.exceptions.ConnectionError)):
load_dataset(SAMPLE_NOT_EXISTING_DATASET_IDENTIFIER, cache_dir=tmp_dir, token="foo")
mock_request.assert_called()
@pytest.mark.integration
def test_load_streaming_private_dataset(hf_token, hf_private_dataset_repo_txt_data):
ds = load_dataset(hf_private_dataset_repo_txt_data, streaming=True, token=hf_token)
assert next(iter(ds)) is not None
@pytest.mark.integration
def test_load_dataset_builder_private_dataset(hf_token, hf_private_dataset_repo_txt_data):
builder = load_dataset_builder(hf_private_dataset_repo_txt_data, token=hf_token)
assert isinstance(builder, DatasetBuilder)
@pytest.mark.integration
def test_load_streaming_private_dataset_with_zipped_data(hf_token, hf_private_dataset_repo_zipped_txt_data):
ds = load_dataset(hf_private_dataset_repo_zipped_txt_data, streaming=True, token=hf_token)
assert next(iter(ds)) is not None
@pytest.mark.integration
def test_load_dataset_config_kwargs_passed_as_arguments():
ds_default = load_dataset(SAMPLE_DATASET_IDENTIFIER4)
ds_custom = load_dataset(SAMPLE_DATASET_IDENTIFIER4, drop_metadata=True)
assert list(ds_default["train"].features) == ["image", "caption"]
assert list(ds_custom["train"].features) == ["image"]
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_single_config_in_metadata():
# load the same dataset but with no configurations (=with default parameters)
ds = load_dataset(SAMPLE_DATASET_NO_CONFIGS_IN_METADATA)
assert list(ds["train"].features) == ["audio", "label"] # assert label feature is here as expected by default
assert len(ds["train"]) == 5 and len(ds["test"]) == 4
ds2 = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA) # single config -> no need to specify it
assert list(ds2["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds2["train"]) == 3 and len(ds2["test"]) == 3
ds3 = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, "custom")
assert list(ds3["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds3["train"]) == 3 and len(ds3["test"]) == 3
with pytest.raises(ValueError):
# no config named "default"
_ = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, "default")
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_two_config_in_metadata():
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1")
assert list(ds["train"].features) == ["audio"] # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds2 = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v2")
assert list(ds2["train"].features) == [
"audio",
"label",
] # assert param `drop_labels=False` from metadata is passed
assert len(ds2["train"]) == 2 and len(ds2["test"]) == 1
with pytest.raises(ValueError):
# config is required but not specified
_ = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA)
with pytest.raises(ValueError):
# no config named "default"
_ = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "default")
ds_with_default = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA_WITH_DEFAULT)
# it's a dataset with the same data but "v1" config is marked as a default one
assert list(ds_with_default["train"].features) == list(ds["train"].features)
assert len(ds_with_default["train"]) == len(ds["train"]) and len(ds_with_default["test"]) == len(ds["test"])
@require_sndfile
@pytest.mark.integration
def test_load_hub_dataset_without_script_with_metadata_config_in_parallel():
# assert it doesn't fail (pickling of dynamically created class works)
ds = load_dataset(SAMPLE_DATASET_SINGLE_CONFIG_IN_METADATA, num_proc=2)
assert "label" not in ds["train"].features # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v1", num_proc=2)
assert "label" not in ds["train"].features # assert param `drop_labels=True` from metadata is passed
assert len(ds["train"]) == 3 and len(ds["test"]) == 3
ds = load_dataset(SAMPLE_DATASET_TWO_CONFIG_IN_METADATA, "v2", num_proc=2)
assert "label" in ds["train"].features
assert len(ds["train"]) == 2 and len(ds["test"]) == 1
@require_pil
@pytest.mark.integration
@pytest.mark.parametrize("streaming", [True])
def test_load_dataset_private_zipped_images(hf_private_dataset_repo_zipped_img_data, hf_token, streaming):
ds = load_dataset(hf_private_dataset_repo_zipped_img_data, split="train", streaming=streaming, token=hf_token)
assert isinstance(ds, IterableDataset if streaming else Dataset)
ds_items = list(ds)
assert len(ds_items) == 2
def test_load_dataset_then_move_then_reload(dataset_loading_script_dir, data_dir, tmp_path, caplog):
cache_dir1 = tmp_path / "cache1"
cache_dir2 = tmp_path / "cache2"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir1)
fingerprint1 = dataset._fingerprint
del dataset
os.rename(cache_dir1, cache_dir2)
caplog.clear()
with caplog.at_level(INFO, logger=get_logger().name):
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir2)
assert "Found cached dataset" in caplog.text
assert dataset._fingerprint == fingerprint1, "for the caching mechanism to work, fingerprint should stay the same"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="test", cache_dir=cache_dir2)
assert dataset._fingerprint != fingerprint1
def test_load_dataset_builder_then_edit_then_load_again(tmp_path: Path):
dataset_dir = tmp_path / "test_load_dataset_then_edit_then_load_again"
dataset_dir.mkdir()
with open(dataset_dir / "train.txt", "w") as f:
f.write("Hello there")
dataset_builder = load_dataset_builder(str(dataset_dir))
with open(dataset_dir / "train.txt", "w") as f:
f.write("General Kenobi !")
edited_dataset_builder = load_dataset_builder(str(dataset_dir))
assert dataset_builder.cache_dir != edited_dataset_builder.cache_dir
def test_load_dataset_readonly(dataset_loading_script_dir, dataset_loading_script_dir_readonly, data_dir, tmp_path):
cache_dir1 = tmp_path / "cache1"
cache_dir2 = tmp_path / "cache2"
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, split="train", cache_dir=cache_dir1)
fingerprint1 = dataset._fingerprint
del dataset
# Load readonly dataset and check that the fingerprint is the same.
dataset = load_dataset(dataset_loading_script_dir_readonly, data_dir=data_dir, split="train", cache_dir=cache_dir2)
assert dataset._fingerprint == fingerprint1, "Cannot load a dataset in a readonly folder."
@pytest.mark.parametrize("max_in_memory_dataset_size", ["default", 0, 50, 500])
def test_load_dataset_local_with_default_in_memory(
max_in_memory_dataset_size, dataset_loading_script_dir, data_dir, monkeypatch
):
current_dataset_size = 148
if max_in_memory_dataset_size == "default":
max_in_memory_dataset_size = 0 # default
else:
monkeypatch.setattr(datasets.config, "IN_MEMORY_MAX_SIZE", max_in_memory_dataset_size)
if max_in_memory_dataset_size:
expected_in_memory = current_dataset_size < max_in_memory_dataset_size
else:
expected_in_memory = False
with assert_arrow_memory_increases() if expected_in_memory else assert_arrow_memory_doesnt_increase():
dataset = load_dataset(dataset_loading_script_dir, data_dir=data_dir)
assert (dataset["train"].dataset_size < max_in_memory_dataset_size) is expected_in_memory
@pytest.mark.parametrize("max_in_memory_dataset_size", ["default", 0, 100, 1000])
def test_load_from_disk_with_default_in_memory(
max_in_memory_dataset_size, dataset_loading_script_dir, data_dir, tmp_path, monkeypatch
):
current_dataset_size = 512 # arrow file size = 512, in-memory dataset size = 148
if max_in_memory_dataset_size == "default":
max_in_memory_dataset_size = 0 # default
else:
monkeypatch.setattr(datasets.config, "IN_MEMORY_MAX_SIZE", max_in_memory_dataset_size)
if max_in_memory_dataset_size:
expected_in_memory = current_dataset_size < max_in_memory_dataset_size
else:
expected_in_memory = False
dset = load_dataset(dataset_loading_script_dir, data_dir=data_dir, keep_in_memory=True)
dataset_path = os.path.join(tmp_path, "saved_dataset")
dset.save_to_disk(dataset_path)
with assert_arrow_memory_increases() if expected_in_memory else assert_arrow_memory_doesnt_increase():
_ = load_from_disk(dataset_path)
@pytest.mark.integration
def test_remote_data_files():
repo_id = "hf-internal-testing/raw_jsonl"
filename = "wikiann-bn-validation.jsonl"
data_files = f"https://huggingface.co/datasets/{repo_id}/resolve/main/{filename}"
ds = load_dataset("json", split="train", data_files=data_files, streaming=True)
assert isinstance(ds, IterableDataset)
ds_item = next(iter(ds))
assert ds_item.keys() == {"langs", "ner_tags", "spans", "tokens"}
@pytest.mark.parametrize("deleted", [False, True])
def test_load_dataset_deletes_extracted_files(deleted, jsonl_gz_path, tmp_path):
data_files = jsonl_gz_path
cache_dir = tmp_path / "cache"
if deleted:
download_config = DownloadConfig(delete_extracted=True, cache_dir=cache_dir / "downloads")
ds = load_dataset(
"json", split="train", data_files=data_files, cache_dir=cache_dir, download_config=download_config
)
else: # default
ds = load_dataset("json", split="train", data_files=data_files, cache_dir=cache_dir)
assert ds[0] == {"col_1": "0", "col_2": 0, "col_3": 0.0}
assert (
[path for path in (cache_dir / "downloads" / "extracted").iterdir() if path.suffix != ".lock"] == []
) is deleted
def distributed_load_dataset(args):
data_name, tmp_dir, datafiles = args
dataset = load_dataset(data_name, cache_dir=tmp_dir, data_files=datafiles)
return dataset
def test_load_dataset_distributed(tmp_path, csv_path):
num_workers = 5
args = "csv", str(tmp_path), csv_path
with Pool(processes=num_workers) as pool: # start num_workers processes
datasets = pool.map(distributed_load_dataset, [args] * num_workers)
assert len(datasets) == num_workers
assert all(len(dataset) == len(datasets[0]) > 0 for dataset in datasets)
assert len(datasets[0].cache_files) > 0
assert all(dataset.cache_files == datasets[0].cache_files for dataset in datasets)
def distributed_load_dataset_with_script(args):
data_name, tmp_dir, download_mode = args
dataset = load_dataset(data_name, cache_dir=tmp_dir, download_mode=download_mode)
return dataset
@require_not_windows # windows doesn't support overwriting Arrow files from other processes
@pytest.mark.parametrize("download_mode", [None, "force_redownload"])
def test_load_dataset_distributed_with_script(tmp_path, download_mode):
# we need to check in the "force_redownload" case
# since in `_copy_script_and_other_resources_in_importable_dir()` we might delete the directory
# containing the .py file while the other processes use it
num_workers = 5
args = (SAMPLE_DATASET_IDENTIFIER, str(tmp_path), download_mode)
with Pool(processes=num_workers) as pool: # start num_workers processes
datasets = pool.map(distributed_load_dataset_with_script, [args] * num_workers)
assert len(datasets) == num_workers
assert all(len(dataset) == len(datasets[0]) > 0 for dataset in datasets)
assert len(datasets[0].cache_files) > 0
assert all(dataset.cache_files == datasets[0].cache_files for dataset in datasets)
def test_load_dataset_with_storage_options(mockfs):
with mockfs.open("data.txt", "w") as f:
f.write("Hello there\n")
f.write("General Kenobi !")
data_files = {"train": ["mock://data.txt"]}
ds = load_dataset("text", data_files=data_files, storage_options=mockfs.storage_options)
assert list(ds["train"]) == [{"text": "Hello there"}, {"text": "General Kenobi !"}]
@require_pil
def test_load_dataset_with_storage_options_with_decoding(mockfs, image_file):
import PIL.Image
filename = os.path.basename(image_file)
with mockfs.open(filename, "wb") as fout:
with open(image_file, "rb") as fin:
fout.write(fin.read())
data_files = {"train": ["mock://" + filename]}
ds = load_dataset("imagefolder", data_files=data_files, storage_options=mockfs.storage_options)
assert len(ds["train"]) == 1
assert isinstance(ds["train"][0]["image"], PIL.Image.Image)
def test_load_dataset_without_script_with_zip(zip_csv_path):
path = str(zip_csv_path.parent)
ds = load_dataset(path)
assert list(ds.keys()) == ["train"]
assert ds["train"].column_names == ["col_1", "col_2", "col_3"]
assert ds["train"].num_rows == 8
assert ds["train"][0] == {"col_1": 0, "col_2": 0, "col_3": 0.0}
@pytest.mark.parametrize("trust_remote_code, expected", [(False, False), (True, True), (None, True)])
def test_resolve_trust_remote_code(trust_remote_code, expected):
assert resolve_trust_remote_code(trust_remote_code, repo_id="dummy") is expected
@pytest.mark.parametrize("trust_remote_code, expected", [(False, False), (True, True), (None, ValueError)])
def test_resolve_trust_remote_code_future(trust_remote_code, expected):
with patch.object(config, "HF_DATASETS_TRUST_REMOTE_CODE", None): # this will be the default soon
if isinstance(expected, bool):
resolve_trust_remote_code(trust_remote_code, repo_id="dummy") is expected
else:
with pytest.raises(expected):
resolve_trust_remote_code(trust_remote_code, repo_id="dummy")
@pytest.mark.integration
def test_reload_old_cache_from_2_15(tmp_path: Path):
cache_dir = tmp_path / "test_reload_old_cache_from_2_15"
builder_cache_dir = (
cache_dir / "polinaeterna___audiofolder_two_configs_in_metadata/v2-374bfde4f55442bc/0.0.0/7896925d64deea5d"
)
builder_cache_dir.mkdir(parents=True)
arrow_path = builder_cache_dir / "audiofolder_two_configs_in_metadata-train.arrow"
dataset_info_path = builder_cache_dir / "dataset_info.json"
with dataset_info_path.open("w") as f:
f.write("{}")
arrow_path.touch()
builder = load_dataset_builder(
"polinaeterna/audiofolder_two_configs_in_metadata",
"v2",
data_files="v2/train/*",
cache_dir=cache_dir.as_posix(),
)
assert builder.cache_dir == builder_cache_dir.as_posix() # old cache from 2.15
builder = load_dataset_builder(
"polinaeterna/audiofolder_two_configs_in_metadata", "v2", cache_dir=cache_dir.as_posix()
)
assert (
builder.cache_dir
== (
cache_dir / "polinaeterna___audiofolder_two_configs_in_metadata" / "v2" / "0.0.0" / str(builder.hash)
).as_posix()
) # new cache
@pytest.mark.integration
def test_update_dataset_card_data_with_standalone_yaml():
# Labels defined in .huggingface.yml because they are too long to be in README.md
from datasets.utils.metadata import MetadataConfigs
with patch(
"datasets.utils.metadata.MetadataConfigs.from_dataset_card_data",
side_effect=MetadataConfigs.from_dataset_card_data,
) as card_data_read_mock:
builder = load_dataset_builder("datasets-maintainers/dataset-with-standalone-yaml")
assert card_data_read_mock.call_args.args[0]["license"] is not None # from README.md
assert card_data_read_mock.call_args.args[0]["dataset_info"] is not None # from standalone yaml
assert card_data_read_mock.call_args.args[0]["tags"] == ["test"] # standalone yaml has precedence
assert isinstance(
builder.info.features["label"], datasets.ClassLabel
) # correctly loaded from long labels list in standalone yaml
| datasets/tests/test_load.py/0 | {
"file_path": "datasets/tests/test_load.py",
"repo_id": "datasets",
"token_count": 35068
} | 76 |
import fnmatch
import gc
import os
import shutil
import tempfile
import textwrap
import time
import unittest
from io import BytesIO
from pathlib import Path
from unittest.mock import patch
import numpy as np
import pytest
from huggingface_hub import DatasetCard, HfApi
from datasets import (
Audio,
ClassLabel,
Dataset,
DatasetDict,
DownloadManager,
Features,
Image,
Value,
load_dataset,
load_dataset_builder,
)
from datasets.config import METADATA_CONFIGS_FIELD
from datasets.data_files import get_data_patterns
from datasets.packaged_modules.folder_based_builder.folder_based_builder import (
FolderBasedBuilder,
FolderBasedBuilderConfig,
)
from datasets.utils.file_utils import cached_path
from datasets.utils.hub import hf_hub_url
from tests.fixtures.hub import CI_HUB_ENDPOINT, CI_HUB_USER, CI_HUB_USER_TOKEN
from tests.utils import for_all_test_methods, require_pil, require_sndfile, xfail_if_500_502_http_error
pytestmark = pytest.mark.integration
@for_all_test_methods(xfail_if_500_502_http_error)
@pytest.mark.usefixtures("ci_hub_config", "ci_hfh_hf_hub_url")
class TestPushToHub:
_api = HfApi(endpoint=CI_HUB_ENDPOINT)
_token = CI_HUB_USER_TOKEN
def test_push_dataset_dict_to_hub_no_token(self, temporary_repo, set_ci_hub_access_token):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub_name_without_namespace(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name.split("/")[-1], token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub_datasets_with_different_features(self, cleanup_repo):
ds_train = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_test = Dataset.from_dict({"x": [True, False, True], "y": ["a", "b", "c"]})
local_ds = DatasetDict({"train": ds_train, "test": ds_test})
ds_name = f"{CI_HUB_USER}/test-{int(time.time() * 10e6)}"
try:
with pytest.raises(ValueError):
local_ds.push_to_hub(ds_name.split("/")[-1], token=self._token)
except AssertionError:
cleanup_repo(ds_name)
raise
def test_push_dataset_dict_to_hub_private(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, private=True)
hub_ds = load_dataset(ds_name, download_mode="force_redownload", token=self._token)
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub_with_pull_request(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, create_pr=True)
hub_ds = load_dataset(ds_name, revision="refs/pr/1", download_mode="force_redownload")
assert local_ds["train"].features == hub_ds["train"].features
assert list(local_ds.keys()) == list(hub_ds.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(
self._api.list_repo_files(ds_name, revision="refs/pr/1", repo_type="dataset", token=self._token)
)
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub_with_revision(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, revision="dev")
hub_ds = load_dataset(ds_name, revision="dev", download_mode="force_redownload")
assert local_ds["train"].features == hub_ds["train"].features
assert list(local_ds.keys()) == list(hub_ds.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there is a single file on the repository that has the correct name
files = sorted(self._api.list_repo_files(ds_name, revision="dev", repo_type="dataset", token=self._token))
assert files == [".gitattributes", "README.md", "data/train-00000-of-00001.parquet"]
def test_push_dataset_dict_to_hub_multiple_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
with patch("datasets.config.MAX_SHARD_SIZE", "16KB"):
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/train-00000-of-00002.parquet",
"data/train-00001-of-00002.parquet",
]
def test_push_dataset_dict_to_hub_multiple_files_with_max_shard_size(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size="16KB")
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/train-00000-of-00002.parquet",
"data/train-00001-of-00002.parquet",
]
def test_push_dataset_dict_to_hub_multiple_files_with_num_shards(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token, num_shards={"train": 2})
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/train-00000-of-00002.parquet",
"data/train-00001-of-00002.parquet",
]
def test_push_dataset_dict_to_hub_with_multiple_commits(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
local_ds = DatasetDict({"train": ds})
with temporary_repo() as ds_name:
self._api.create_repo(ds_name, token=self._token, repo_type="dataset")
num_commits_before_push = len(self._api.list_repo_commits(ds_name, repo_type="dataset", token=self._token))
with patch("datasets.config.MAX_SHARD_SIZE", "16KB"), patch(
"datasets.config.UPLOADS_MAX_NUMBER_PER_COMMIT", 1
):
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/train-00000-of-00002.parquet",
"data/train-00001-of-00002.parquet",
]
num_commits_after_push = len(self._api.list_repo_commits(ds_name, repo_type="dataset", token=self._token))
assert num_commits_after_push - num_commits_before_push > 1
def test_push_dataset_dict_to_hub_overwrite_files(self, temporary_repo):
ds = Dataset.from_dict({"x": list(range(1000)), "y": list(range(1000))})
ds2 = Dataset.from_dict({"x": list(range(100)), "y": list(range(100))})
local_ds = DatasetDict({"train": ds, "random": ds2})
# Push to hub two times, but the second time with a larger amount of files.
# Verify that the new files contain the correct dataset.
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
with tempfile.TemporaryDirectory() as tmp:
# Add a file starting with "data" to ensure it doesn't get deleted.
path = Path(tmp) / "datafile.txt"
with open(path, "w") as f:
f.write("Bogus file")
self._api.upload_file(
path_or_fileobj=str(path),
path_in_repo="datafile.txt",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size=500 << 5)
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/random-00000-of-00001.parquet",
"data/train-00000-of-00002.parquet",
"data/train-00001-of-00002.parquet",
"datafile.txt",
]
self._api.delete_file("datafile.txt", repo_id=ds_name, repo_type="dataset", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
del hub_ds
# To ensure the reference to the memory-mapped Arrow file is dropped to avoid the PermissionError on Windows
gc.collect()
# Push to hub two times, but the second time with fewer files.
# Verify that the new files contain the correct dataset and that non-necessary files have been deleted.
with temporary_repo(ds_name):
local_ds.push_to_hub(ds_name, token=self._token, max_shard_size=500 << 5)
with tempfile.TemporaryDirectory() as tmp:
# Add a file starting with "data" to ensure it doesn't get deleted.
path = Path(tmp) / "datafile.txt"
with open(path, "w") as f:
f.write("Bogus file")
self._api.upload_file(
path_or_fileobj=str(path),
path_in_repo="datafile.txt",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
local_ds.push_to_hub(ds_name, token=self._token)
# Ensure that there are two files on the repository that have the correct name
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset", token=self._token))
assert files == [
".gitattributes",
"README.md",
"data/random-00000-of-00001.parquet",
"data/train-00000-of-00001.parquet",
"datafile.txt",
]
# Keeping the "datafile.txt" breaks the load_dataset to think it's a text-based dataset
self._api.delete_file("datafile.txt", repo_id=ds_name, repo_type="dataset", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
def test_push_dataset_to_hub(self, temporary_repo):
local_ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, split="train", token=self._token)
local_ds_dict = {"train": local_ds}
hub_ds_dict = load_dataset(ds_name, download_mode="force_redownload")
assert list(local_ds_dict.keys()) == list(hub_ds_dict.keys())
for ds_split_name in local_ds_dict.keys():
local_ds = local_ds_dict[ds_split_name]
hub_ds = hub_ds_dict[ds_split_name]
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds.features.keys()) == list(hub_ds.features.keys())
assert local_ds.features == hub_ds.features
def test_push_dataset_to_hub_custom_features(self, temporary_repo):
features = Features({"x": Value("int64"), "y": ClassLabel(names=["neg", "pos"])})
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [0, 0, 1]}, features=features)
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
@require_sndfile
def test_push_dataset_to_hub_custom_features_audio(self, temporary_repo):
audio_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_audio_44100.wav")
data = {"x": [audio_path, None], "y": [0, -1]}
features = Features({"x": Audio(), "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
np.testing.assert_equal(ds[0]["x"]["array"], hub_ds[0]["x"]["array"])
assert ds[1] == hub_ds[1] # don't test hub_ds[0] since audio decoding might be slightly different
hub_ds = hub_ds.cast_column("x", Audio(decode=False))
elem = hub_ds[0]["x"]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert os.path.basename(path) == "test_audio_44100.wav"
assert bool(bytes_) == embed_external_files
@require_pil
def test_push_dataset_to_hub_custom_features_image(self, temporary_repo):
image_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_image_rgb.jpg")
data = {"x": [image_path, None], "y": [0, -1]}
features = Features({"x": Image(), "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
hub_ds = hub_ds.cast_column("x", Image(decode=False))
elem = hub_ds[0]["x"]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert bool(bytes_) == embed_external_files
@require_pil
def test_push_dataset_to_hub_custom_features_image_list(self, temporary_repo):
image_path = os.path.join(os.path.dirname(__file__), "features", "data", "test_image_rgb.jpg")
data = {"x": [[image_path], [image_path, image_path]], "y": [0, -1]}
features = Features({"x": [Image()], "y": Value("int32")})
ds = Dataset.from_dict(data, features=features)
for embed_external_files in [True, False]:
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, embed_external_files=embed_external_files, token=self._token)
hub_ds = load_dataset(ds_name, split="train", download_mode="force_redownload")
assert ds.column_names == hub_ds.column_names
assert list(ds.features.keys()) == list(hub_ds.features.keys())
assert ds.features == hub_ds.features
assert ds[:] == hub_ds[:]
hub_ds = hub_ds.cast_column("x", [Image(decode=False)])
elem = hub_ds[0]["x"][0]
path, bytes_ = elem["path"], elem["bytes"]
assert isinstance(path, str)
assert bool(bytes_) == embed_external_files
def test_push_dataset_dict_to_hub_custom_features(self, temporary_repo):
features = Features({"x": Value("int64"), "y": ClassLabel(names=["neg", "pos"])})
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [0, 0, 1]}, features=features)
local_ds = DatasetDict({"test": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["test"].features.keys()) == list(hub_ds["test"].features.keys())
assert local_ds["test"].features == hub_ds["test"].features
def test_push_dataset_to_hub_custom_splits(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, split="random", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert ds.column_names == hub_ds["random"].column_names
assert list(ds.features.keys()) == list(hub_ds["random"].features.keys())
assert ds.features == hub_ds["random"].features
def test_push_dataset_to_hub_multiple_splits_one_by_one(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
with temporary_repo() as ds_name:
ds.push_to_hub(ds_name, split="train", token=self._token)
ds.push_to_hub(ds_name, split="test", token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert sorted(hub_ds) == ["test", "train"]
assert ds.column_names == hub_ds["train"].column_names
assert list(ds.features.keys()) == list(hub_ds["train"].features.keys())
assert ds.features == hub_ds["train"].features
def test_push_dataset_dict_to_hub_custom_splits(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"random": ds})
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["random"].features.keys()) == list(hub_ds["random"].features.keys())
assert local_ds["random"].features == hub_ds["random"].features
@unittest.skip("This test cannot pass until iterable datasets have push to hub")
def test_push_streaming_dataset_dict_to_hub(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
local_ds = DatasetDict({"train": ds})
with tempfile.TemporaryDirectory() as tmp:
local_ds.save_to_disk(tmp)
local_ds = load_dataset(tmp, streaming=True)
with temporary_repo() as ds_name:
local_ds.push_to_hub(ds_name, token=self._token)
hub_ds = load_dataset(ds_name, download_mode="force_redownload")
assert local_ds.column_names == hub_ds.column_names
assert list(local_ds["train"].features.keys()) == list(hub_ds["train"].features.keys())
assert local_ds["train"].features == hub_ds["train"].features
def test_push_multiple_dataset_configs_to_hub_load_dataset_builder(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo() as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
ds_builder_default = load_dataset_builder(ds_name, download_mode="force_redownload") # default config
assert len(ds_builder_default.BUILDER_CONFIGS) == 3
assert len(ds_builder_default.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_default.config.data_files["train"][0],
"*/data/train-*",
)
ds_builder_config1 = load_dataset_builder(ds_name, "config1", download_mode="force_redownload")
assert len(ds_builder_config1.BUILDER_CONFIGS) == 3
assert len(ds_builder_config1.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_config1.config.data_files["train"][0],
"*/config1/train-*",
)
ds_builder_config2 = load_dataset_builder(ds_name, "config2", download_mode="force_redownload")
assert len(ds_builder_config2.BUILDER_CONFIGS) == 3
assert len(ds_builder_config2.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_builder_config2.config.data_files["train"][0],
"*/config2/train-*",
)
with pytest.raises(ValueError): # no config 'config3'
load_dataset_builder(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_configs_to_hub_load_dataset(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo() as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert files == [
".gitattributes",
"README.md",
"config1/train-00000-of-00001.parquet",
"config2/train-00000-of-00001.parquet",
"data/train-00000-of-00001.parquet",
]
hub_ds_default = load_dataset(ds_name, download_mode="force_redownload")
hub_ds_config1 = load_dataset(ds_name, "config1", download_mode="force_redownload")
hub_ds_config2 = load_dataset(ds_name, "config2", download_mode="force_redownload")
# only "train" split
assert len(hub_ds_default) == len(hub_ds_config1) == len(hub_ds_config2) == 1
assert ds_default.column_names == hub_ds_default["train"].column_names == ["a", "b"]
assert ds_config1.column_names == hub_ds_config1["train"].column_names == ["x", "y"]
assert ds_config2.column_names == hub_ds_config2["train"].column_names == ["foo", "bar"]
assert ds_default.features == hub_ds_default["train"].features
assert ds_config1.features == hub_ds_config1["train"].features
assert ds_config2.features == hub_ds_config2["train"].features
assert ds_default.num_rows == hub_ds_default["train"].num_rows == 1
assert ds_config1.num_rows == hub_ds_config1["train"].num_rows == 3
assert ds_config2.num_rows == hub_ds_config2["train"].num_rows == 2
with pytest.raises(ValueError): # no config 'config3'
load_dataset(ds_name, "config3", download_mode="force_redownload")
@pytest.mark.parametrize("specific_default_config_name", [False, True])
def test_push_multiple_dataset_configs_to_hub_readme_metadata_content(
self, specific_default_config_name, temporary_repo
):
ds_default = Dataset.from_dict({"a": [0], "b": [2]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
with temporary_repo() as ds_name:
if specific_default_config_name:
ds_default.push_to_hub(ds_name, config_name="config0", set_default=True, token=self._token)
else:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
# check that configs args was correctly pushed to README.md
ds_readme_path = cached_path(hf_hub_url(ds_name, "README.md"))
dataset_card_data = DatasetCard.load(ds_readme_path).data
assert METADATA_CONFIGS_FIELD in dataset_card_data
assert isinstance(dataset_card_data[METADATA_CONFIGS_FIELD], list)
assert sorted(dataset_card_data[METADATA_CONFIGS_FIELD], key=lambda x: x["config_name"]) == (
[
{
"config_name": "config0",
"data_files": [
{"split": "train", "path": "config0/train-*"},
],
"default": True,
},
]
if specific_default_config_name
else []
) + [
{
"config_name": "config1",
"data_files": [
{"split": "train", "path": "config1/train-*"},
],
},
{
"config_name": "config2",
"data_files": [
{"split": "train", "path": "config2/train-*"},
],
},
] + (
[]
if specific_default_config_name
else [
{
"config_name": "default",
"data_files": [
{"split": "train", "path": "data/train-*"},
],
},
]
)
def test_push_multiple_dataset_dict_configs_to_hub_load_dataset_builder(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"random": ds_default})
ds_config1 = DatasetDict({"random": ds_config1})
ds_config2 = DatasetDict({"random": ds_config2})
with temporary_repo() as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
ds_builder_default = load_dataset_builder(ds_name, download_mode="force_redownload") # default config
assert len(ds_builder_default.BUILDER_CONFIGS) == 3
assert len(ds_builder_default.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_default.config.data_files["random"][0],
"*/data/random-*",
)
ds_builder_config1 = load_dataset_builder(ds_name, "config1", download_mode="force_redownload")
assert len(ds_builder_config1.BUILDER_CONFIGS) == 3
assert len(ds_builder_config1.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_config1.config.data_files["random"][0],
"*/config1/random-*",
)
ds_builder_config2 = load_dataset_builder(ds_name, "config2", download_mode="force_redownload")
assert len(ds_builder_config2.BUILDER_CONFIGS) == 3
assert len(ds_builder_config2.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_builder_config2.config.data_files["random"][0],
"*/config2/random-*",
)
with pytest.raises(ValueError): # no config named 'config3'
load_dataset_builder(ds_name, "config3", download_mode="force_redownload")
def test_push_multiple_dataset_dict_configs_to_hub_load_dataset(self, temporary_repo):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"train": ds_default, "random": ds_default})
ds_config1 = DatasetDict({"train": ds_config1, "random": ds_config1})
ds_config2 = DatasetDict({"train": ds_config2, "random": ds_config2})
with temporary_repo() as ds_name:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
files = sorted(self._api.list_repo_files(ds_name, repo_type="dataset"))
assert files == [
".gitattributes",
"README.md",
"config1/random-00000-of-00001.parquet",
"config1/train-00000-of-00001.parquet",
"config2/random-00000-of-00001.parquet",
"config2/train-00000-of-00001.parquet",
"data/random-00000-of-00001.parquet",
"data/train-00000-of-00001.parquet",
]
hub_ds_default = load_dataset(ds_name, download_mode="force_redownload")
hub_ds_config1 = load_dataset(ds_name, "config1", download_mode="force_redownload")
hub_ds_config2 = load_dataset(ds_name, "config2", download_mode="force_redownload")
# two splits
expected_splits = ["random", "train"]
assert len(hub_ds_default) == len(hub_ds_config1) == len(hub_ds_config2) == 2
assert sorted(hub_ds_default) == sorted(hub_ds_config1) == sorted(hub_ds_config2) == expected_splits
for split in expected_splits:
assert ds_default[split].column_names == hub_ds_default[split].column_names == ["a", "b"]
assert ds_config1[split].column_names == hub_ds_config1[split].column_names == ["x", "y"]
assert ds_config2[split].column_names == hub_ds_config2[split].column_names == ["foo", "bar"]
assert ds_default[split].features == hub_ds_default[split].features
assert ds_config1[split].features == hub_ds_config1[split].features
assert ds_config2[split].features == hub_ds_config2["train"].features
assert ds_default[split].num_rows == hub_ds_default[split].num_rows == 1
assert ds_config1[split].num_rows == hub_ds_config1[split].num_rows == 3
assert ds_config2[split].num_rows == hub_ds_config2[split].num_rows == 2
with pytest.raises(ValueError): # no config 'config3'
load_dataset(ds_name, "config3", download_mode="force_redownload")
@pytest.mark.parametrize("specific_default_config_name", [False, True])
def test_push_multiple_dataset_dict_configs_to_hub_readme_metadata_content(
self, specific_default_config_name, temporary_repo
):
ds_default = Dataset.from_dict({"a": [0], "b": [1]})
ds_config1 = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_config2 = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
ds_default = DatasetDict({"train": ds_default, "random": ds_default})
ds_config1 = DatasetDict({"train": ds_config1, "random": ds_config1})
ds_config2 = DatasetDict({"train": ds_config2, "random": ds_config2})
with temporary_repo() as ds_name:
if specific_default_config_name:
ds_default.push_to_hub(ds_name, config_name="config0", set_default=True, token=self._token)
else:
ds_default.push_to_hub(ds_name, token=self._token)
ds_config1.push_to_hub(ds_name, "config1", token=self._token)
ds_config2.push_to_hub(ds_name, "config2", token=self._token)
# check that configs args was correctly pushed to README.md
ds_readme_path = cached_path(hf_hub_url(ds_name, "README.md"))
dataset_card_data = DatasetCard.load(ds_readme_path).data
assert METADATA_CONFIGS_FIELD in dataset_card_data
assert isinstance(dataset_card_data[METADATA_CONFIGS_FIELD], list)
assert sorted(dataset_card_data[METADATA_CONFIGS_FIELD], key=lambda x: x["config_name"]) == (
[
{
"config_name": "config0",
"data_files": [
{"split": "train", "path": "config0/train-*"},
{"split": "random", "path": "config0/random-*"},
],
"default": True,
},
]
if specific_default_config_name
else []
) + [
{
"config_name": "config1",
"data_files": [
{"split": "train", "path": "config1/train-*"},
{"split": "random", "path": "config1/random-*"},
],
},
{
"config_name": "config2",
"data_files": [
{"split": "train", "path": "config2/train-*"},
{"split": "random", "path": "config2/random-*"},
],
},
] + (
[]
if specific_default_config_name
else [
{
"config_name": "default",
"data_files": [
{"split": "train", "path": "data/train-*"},
{"split": "random", "path": "data/random-*"},
],
},
]
)
def test_push_dataset_to_hub_with_config_no_metadata_configs(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_another_config = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
parquet_buf = BytesIO()
ds.to_parquet(parquet_buf)
parquet_content = parquet_buf.getvalue()
with temporary_repo() as ds_name:
self._api.create_repo(ds_name, token=self._token, repo_type="dataset")
# old push_to_hub was uploading the parquet files only - without metadata configs
self._api.upload_file(
path_or_fileobj=parquet_content,
path_in_repo="data/train-00000-of-00001.parquet",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
ds_another_config.push_to_hub(ds_name, "another_config", token=self._token)
ds_builder = load_dataset_builder(ds_name, download_mode="force_redownload")
assert len(ds_builder.config.data_files) == 1
assert len(ds_builder.config.data_files["train"]) == 1
assert fnmatch.fnmatch(ds_builder.config.data_files["train"][0], "*/data/train-00000-of-00001.parquet")
ds_another_config_builder = load_dataset_builder(
ds_name, "another_config", download_mode="force_redownload"
)
assert len(ds_another_config_builder.config.data_files) == 1
assert len(ds_another_config_builder.config.data_files["train"]) == 1
assert fnmatch.fnmatch(
ds_another_config_builder.config.data_files["train"][0],
"*/another_config/train-00000-of-00001.parquet",
)
def test_push_dataset_dict_to_hub_with_config_no_metadata_configs(self, temporary_repo):
ds = Dataset.from_dict({"x": [1, 2, 3], "y": [4, 5, 6]})
ds_another_config = Dataset.from_dict({"foo": [1, 2], "bar": [4, 5]})
parquet_buf = BytesIO()
ds.to_parquet(parquet_buf)
parquet_content = parquet_buf.getvalue()
local_ds_another_config = DatasetDict({"random": ds_another_config})
with temporary_repo() as ds_name:
self._api.create_repo(ds_name, token=self._token, repo_type="dataset")
# old push_to_hub was uploading the parquet files only - without metadata configs
self._api.upload_file(
path_or_fileobj=parquet_content,
path_in_repo="data/random-00000-of-00001.parquet",
repo_id=ds_name,
repo_type="dataset",
token=self._token,
)
local_ds_another_config.push_to_hub(ds_name, "another_config", token=self._token)
ds_builder = load_dataset_builder(ds_name, download_mode="force_redownload")
assert len(ds_builder.config.data_files) == 1
assert len(ds_builder.config.data_files["random"]) == 1
assert fnmatch.fnmatch(ds_builder.config.data_files["random"][0], "*/data/random-00000-of-00001.parquet")
ds_another_config_builder = load_dataset_builder(
ds_name, "another_config", download_mode="force_redownload"
)
assert len(ds_another_config_builder.config.data_files) == 1
assert len(ds_another_config_builder.config.data_files["random"]) == 1
assert fnmatch.fnmatch(
ds_another_config_builder.config.data_files["random"][0],
"*/another_config/random-00000-of-00001.parquet",
)
class DummyFolderBasedBuilder(FolderBasedBuilder):
BASE_FEATURE = dict
BASE_COLUMN_NAME = "base"
BUILDER_CONFIG_CLASS = FolderBasedBuilderConfig
EXTENSIONS = [".txt"]
# CLASSIFICATION_TASK = TextClassification(text_column="base", label_column="label")
@pytest.fixture(params=[".jsonl", ".csv"])
def text_file_with_metadata(request, tmp_path, text_file):
metadata_filename_extension = request.param
data_dir = tmp_path / "data_dir"
data_dir.mkdir()
text_file_path = data_dir / "file.txt"
shutil.copyfile(text_file, text_file_path)
metadata_file_path = data_dir / f"metadata{metadata_filename_extension}"
metadata = textwrap.dedent(
"""\
{"file_name": "file.txt", "additional_feature": "Dummy file"}
"""
if metadata_filename_extension == ".jsonl"
else """\
file_name,additional_feature
file.txt,Dummy file
"""
)
with open(metadata_file_path, "w", encoding="utf-8") as f:
f.write(metadata)
return text_file_path, metadata_file_path
@for_all_test_methods(xfail_if_500_502_http_error)
@pytest.mark.usefixtures("ci_hub_config", "ci_hfh_hf_hub_url")
class TestLoadFromHub:
_api = HfApi(endpoint=CI_HUB_ENDPOINT)
_token = CI_HUB_USER_TOKEN
def test_load_dataset_with_metadata_file(self, temporary_repo, text_file_with_metadata, tmp_path):
text_file_path, metadata_file_path = text_file_with_metadata
data_dir_path = text_file_path.parent
cache_dir_path = tmp_path / ".cache"
cache_dir_path.mkdir()
with temporary_repo() as repo_id:
self._api.create_repo(repo_id, token=self._token, repo_type="dataset")
self._api.upload_folder(
folder_path=str(data_dir_path),
repo_id=repo_id,
repo_type="dataset",
token=self._token,
)
data_files = [
f"hf://datasets/{repo_id}/{text_file_path.name}",
f"hf://datasets/{repo_id}/{metadata_file_path.name}",
]
builder = DummyFolderBasedBuilder(
dataset_name=repo_id.split("/")[-1], data_files=data_files, cache_dir=str(cache_dir_path)
)
download_manager = DownloadManager()
gen_kwargs = builder._split_generators(download_manager)[0].gen_kwargs
generator = builder._generate_examples(**gen_kwargs)
result = [example for _, example in generator]
assert len(result) == 1
def test_get_data_patterns(self, temporary_repo, tmp_path):
repo_dir = tmp_path / "test_get_data_patterns"
data_dir = repo_dir / "data"
data_dir.mkdir(parents=True)
data_file = data_dir / "train-00001-of-00009.parquet"
data_file.touch()
with temporary_repo() as repo_id:
self._api.create_repo(repo_id, token=self._token, repo_type="dataset")
self._api.upload_folder(
folder_path=str(repo_dir),
repo_id=repo_id,
repo_type="dataset",
token=self._token,
)
data_file_patterns = get_data_patterns(f"hf://datasets/{repo_id}")
assert data_file_patterns == {
"train": ["data/train-[0-9][0-9][0-9][0-9][0-9]-of-[0-9][0-9][0-9][0-9][0-9]*.*"]
}
| datasets/tests/test_upstream_hub.py/0 | {
"file_path": "datasets/tests/test_upstream_hub.py",
"repo_id": "datasets",
"token_count": 22798
} | 77 |
<jupyter_start><jupyter_text>Unit 5: An Introduction to ML-Agents In this notebook, you'll learn about ML-Agents and train two agents.- The first one will learn to **shoot snowballs onto spawning targets**.- The second need to press a button to spawn a pyramid, then navigate to the pyramid, knock it over, **and move to the gold brick at the top**. To do that, it will need to explore its environment, and we will use a technique called curiosity.After that, you'll be able **to watch your agents playing directly on your browser**.For more information about the certification process, check this section 👉 https://huggingface.co/deep-rl-course/en/unit0/introductioncertification-process ⬇️ Here is an example of what **you will achieve at the end of this unit.** ⬇️ 🎮 Environments:- [Pyramids](https://github.com/Unity-Technologies/ml-agents/blob/main/docs/Learning-Environment-Examples.mdpyramids)- SnowballTarget 📚 RL-Library:- [ML-Agents](https://github.com/Unity-Technologies/ml-agents) We're constantly trying to improve our tutorials, so **if you find some issues in this notebook**, please [open an issue on the GitHub Repo](https://github.com/huggingface/deep-rl-class/issues). Objectives of this notebook 🏆At the end of the notebook, you will:- Understand how works **ML-Agents**, the environment library.- Be able to **train agents in Unity Environments**. This notebook is from the Deep Reinforcement Learning Course In this free course, you will:- 📖 Study Deep Reinforcement Learning in **theory and practice**.- 🧑💻 Learn to **use famous Deep RL libraries** such as Stable Baselines3, RL Baselines3 Zoo, CleanRL and Sample Factory 2.0.- 🤖 Train **agents in unique environments**And more check 📚 the syllabus 👉 https://huggingface.co/deep-rl-course/communication/publishing-scheduleDon’t forget to **sign up to the course** (we are collecting your email to be able to **send you the links when each Unit is published and give you information about the challenges and updates).**The best way to keep in touch is to join our discord server to exchange with the community and with us 👉🏻 https://discord.gg/ydHrjt3WP5 Prerequisites 🏗️Before diving into the notebook, you need to:🔲 📚 **Study [what is ML-Agents and how it works by reading Unit 5](https://huggingface.co/deep-rl-course/unit5/introduction)** 🤗 Let's train our agents 🚀**To validate this hands-on for the certification process, you just need to push your trained models to the Hub**. There’s no results to attain to validate this one. But if you want to get nice results you can try to attain:- For `Pyramids` : Mean Reward = 1.75- For `SnowballTarget` : Mean Reward = 15 or 30 targets hit in an episode. Set the GPU 💪- To **accelerate the agent's training, we'll use a GPU**. To do that, go to `Runtime > Change Runtime type` - `Hardware Accelerator > GPU` Clone the repository and install the dependencies 🔽<jupyter_code>%%capture
# Clone the repository
!git clone --depth 1 https://github.com/Unity-Technologies/ml-agents
# Go inside the repository and install the package
%cd ml-agents
!pip3 install -e ./ml-agents-envs
!pip3 install -e ./ml-agents<jupyter_output><empty_output><jupyter_text>SnowballTarget ⛄If you need a refresher on how this environments work check this section 👉https://huggingface.co/deep-rl-course/unit5/snowball-target Download and move the environment zip file in `./training-envs-executables/linux/`- Our environment executable is in a zip file.- We need to download it and place it to `./training-envs-executables/linux/`- We use a linux executable because we use colab, and colab machines OS is Ubuntu (linux)<jupyter_code># Here, we create training-envs-executables and linux
!mkdir ./training-envs-executables
!mkdir ./training-envs-executables/linux<jupyter_output><empty_output><jupyter_text>We downloaded the file SnowballTarget.zip from https://github.com/huggingface/Snowball-Target using `wget`<jupyter_code>!wget "https://github.com/huggingface/Snowball-Target/raw/main/SnowballTarget.zip" -O ./training-envs-executables/linux/SnowballTarget.zip<jupyter_output><empty_output><jupyter_text>We unzip the executable.zip file<jupyter_code>%%capture
!unzip -d ./training-envs-executables/linux/ ./training-envs-executables/linux/SnowballTarget.zip<jupyter_output><empty_output><jupyter_text>Make sure your file is accessible<jupyter_code>!chmod -R 755 ./training-envs-executables/linux/SnowballTarget<jupyter_output><empty_output><jupyter_text>Define the SnowballTarget config file- In ML-Agents, you define the **training hyperparameters into config.yaml files.**There are multiple hyperparameters. To know them better, you should check for each explanation with [the documentation](https://github.com/Unity-Technologies/ml-agents/blob/release_20_docs/docs/Training-Configuration-File.md)So you need to create a `SnowballTarget.yaml` config file in ./content/ml-agents/config/ppo/We'll give you here a first version of this config (to copy and paste into your `SnowballTarget.yaml file`), **but you should modify it**.```behaviors: SnowballTarget: trainer_type: ppo summary_freq: 10000 keep_checkpoints: 10 checkpoint_interval: 50000 max_steps: 200000 time_horizon: 64 threaded: true hyperparameters: learning_rate: 0.0003 learning_rate_schedule: linear batch_size: 128 buffer_size: 2048 beta: 0.005 epsilon: 0.2 lambd: 0.95 num_epoch: 3 network_settings: normalize: false hidden_units: 256 num_layers: 2 vis_encode_type: simple reward_signals: extrinsic: gamma: 0.99 strength: 1.0``` As an experimentation, you should also try to modify some other hyperparameters. Unity provides very [good documentation explaining each of them here](https://github.com/Unity-Technologies/ml-agents/blob/main/docs/Training-Configuration-File.md).Now that you've created the config file and understand what most hyperparameters do, we're ready to train our agent 🔥. Train the agentTo train our agent, we just need to **launch mlagents-learn and select the executable containing the environment.**We define four parameters:1. `mlagents-learn `: the path where the hyperparameter config file is.2. `--env`: where the environment executable is.3. `--run_id`: the name you want to give to your training run id.4. `--no-graphics`: to not launch the visualization during the training.Train the model and use the `--resume` flag to continue training in case of interruption.> It will fail first time if and when you use `--resume`, try running the block again to bypass the error. The training will take 10 to 35min depending on your config, go take a ☕️you deserve it 🤗.<jupyter_code>!mlagents-learn ./config/ppo/SnowballTarget.yaml --env=./training-envs-executables/linux/SnowballTarget/SnowballTarget --run-id="SnowballTarget1" --no-graphics<jupyter_output><empty_output><jupyter_text>Push the agent to the 🤗 Hub- Now that we trained our agent, we’re **ready to push it to the Hub to be able to visualize it playing on your browser🔥.** To be able to share your model with the community there are three more steps to follow:1️⃣ (If it's not already done) create an account to HF ➡ https://huggingface.co/join2️⃣ Sign in and then, you need to store your authentication token from the Hugging Face website.- Create a new token (https://huggingface.co/settings/tokens) **with write role**- Copy the token- Run the cell below and paste the token<jupyter_code>from huggingface_hub import notebook_login
notebook_login()<jupyter_output><empty_output><jupyter_text>If you don't want to use a Google Colab or a Jupyter Notebook, you need to use this command instead: `huggingface-cli login` Then, we simply need to run `mlagents-push-to-hf`.And we define 4 parameters:1. `--run-id`: the name of the training run id.2. `--local-dir`: where the agent was saved, it’s results/, so in my case results/First Training.3. `--repo-id`: the name of the Hugging Face repo you want to create or update. It’s always /If the repo does not exist **it will be created automatically**4. `--commit-message`: since HF repos are git repository you need to define a commit message.For instance:`!mlagents-push-to-hf --run-id="SnowballTarget1" --local-dir="./results/SnowballTarget1" --repo-id="ThomasSimonini/ppo-SnowballTarget" --commit-message="First Push"`<jupyter_code>!mlagents-push-to-hf --run-id="SnowballTarget1" --local-dir="./results/SnowballTarget1" --repo-id="ThomasSimonini/ppo-SnowballTarget" --commit-message="First Push"
!mlagents-push-to-hf --run-id= # Add your run id --local-dir= # Your local dir --repo-id= # Your repo id --commit-message= # Your commit message<jupyter_output><empty_output><jupyter_text>Else, if everything worked you should have this at the end of the process(but with a different url 😆) :```Your model is pushed to the hub. You can view your model here: https://huggingface.co/ThomasSimonini/ppo-SnowballTarget```It’s the link to your model, it contains a model card that explains how to use it, your Tensorboard and your config file. **What’s awesome is that it’s a git repository, that means you can have different commits, update your repository with a new push etc.** But now comes the best: **being able to visualize your agent online 👀.** Watch your agent playing 👀For this step it’s simple:1. Remember your repo-id2. Go here: https://huggingface.co/spaces/ThomasSimonini/ML-Agents-SnowballTarget3. Launch the game and put it in full screen by clicking on the bottom right button 1. In step 1, choose your model repository which is the model id (in my case ThomasSimonini/ppo-SnowballTarget).2. In step 2, **choose what model you want to replay**: - I have multiple one, since we saved a model every 500000 timesteps. - But if I want the more recent I choose `SnowballTarget.onnx`👉 What’s nice **is to try with different models step to see the improvement of the agent.**And don't hesitate to share the best score your agent gets on discord in rl-i-made-this channel 🔥Let's now try a harder environment called Pyramids... Pyramids 🏆 Download and move the environment zip file in `./training-envs-executables/linux/`- Our environment executable is in a zip file.- We need to download it and place it to `./training-envs-executables/linux/`- We use a linux executable because we use colab, and colab machines OS is Ubuntu (linux) We downloaded the file Pyramids.zip from from https://huggingface.co/spaces/unity/ML-Agents-Pyramids/resolve/main/Pyramids.zip using `wget`<jupyter_code>!wget "https://huggingface.co/spaces/unity/ML-Agents-Pyramids/resolve/main/Pyramids.zip" -O ./training-envs-executables/linux/Pyramids.zip<jupyter_output><empty_output><jupyter_text>We unzip the executable.zip file<jupyter_code>%%capture
!unzip -d ./training-envs-executables/linux/ ./training-envs-executables/linux/Pyramids.zip<jupyter_output><empty_output><jupyter_text>Make sure your file is accessible<jupyter_code>!chmod -R 755 ./training-envs-executables/linux/Pyramids/Pyramids<jupyter_output><empty_output><jupyter_text>Modify the PyramidsRND config file- Contrary to the first environment which was a custom one, **Pyramids was made by the Unity team**.- So the PyramidsRND config file already exists and is in ./content/ml-agents/config/ppo/PyramidsRND.yaml- You might asked why "RND" in PyramidsRND. RND stands for *random network distillation* it's a way to generate curiosity rewards. If you want to know more on that we wrote an article explaning this technique: https://medium.com/data-from-the-trenches/curiosity-driven-learning-through-random-network-distillation-488ffd8e5938For this training, we’ll modify one thing:- The total training steps hyperparameter is too high since we can hit the benchmark (mean reward = 1.75) in only 1M training steps.👉 To do that, we go to config/ppo/PyramidsRND.yaml,**and modify these to max_steps to 1000000.** As an experimentation, you should also try to modify some other hyperparameters, Unity provides a very [good documentation explaining each of them here](https://github.com/Unity-Technologies/ml-agents/blob/main/docs/Training-Configuration-File.md).We’re now ready to train our agent 🔥. Train the agentThe training will take 30 to 45min depending on your machine, go take a ☕️you deserve it 🤗.<jupyter_code>!mlagents-learn ./config/ppo/PyramidsRND.yaml --env=./training-envs-executables/linux/Pyramids/Pyramids --run-id="Pyramids Training" --no-graphics<jupyter_output><empty_output><jupyter_text>Push the agent to the 🤗 Hub- Now that we trained our agent, we’re **ready to push it to the Hub to be able to visualize it playing on your browser🔥.**<jupyter_code>!mlagents-push-to-hf --run-id= # Add your run id --local-dir= # Your local dir --repo-id= # Your repo id --commit-message= # Your commit message<jupyter_output><empty_output> | deep-rl-class/notebooks/unit5/unit5.ipynb/0 | {
"file_path": "deep-rl-class/notebooks/unit5/unit5.ipynb",
"repo_id": "deep-rl-class",
"token_count": 3877
} | 78 |
# Glossary [[glossary]]
This is a community-created glossary. Contributions are welcomed!
### Agent
An agent learns to **make decisions by trial and error, with rewards and punishments from the surroundings**.
### Environment
An environment is a simulated world **where an agent can learn by interacting with it**.
### Markov Property
It implies that the action taken by our agent is **conditional solely on the present state and independent of the past states and actions**.
### Observations/State
- **State**: Complete description of the state of the world.
- **Observation**: Partial description of the state of the environment/world.
### Actions
- **Discrete Actions**: Finite number of actions, such as left, right, up, and down.
- **Continuous Actions**: Infinite possibility of actions; for example, in the case of self-driving cars, the driving scenario has an infinite possibility of actions occurring.
### Rewards and Discounting
- **Rewards**: Fundamental factor in RL. Tells the agent whether the action taken is good/bad.
- RL algorithms are focused on maximizing the **cumulative reward**.
- **Reward Hypothesis**: RL problems can be formulated as a maximisation of (cumulative) return.
- **Discounting** is performed because rewards obtained at the start are more likely to happen as they are more predictable than long-term rewards.
### Tasks
- **Episodic**: Has a starting point and an ending point.
- **Continuous**: Has a starting point but no ending point.
### Exploration v/s Exploitation Trade-Off
- **Exploration**: It's all about exploring the environment by trying random actions and receiving feedback/returns/rewards from the environment.
- **Exploitation**: It's about exploiting what we know about the environment to gain maximum rewards.
- **Exploration-Exploitation Trade-Off**: It balances how much we want to **explore** the environment and how much we want to **exploit** what we know about the environment.
### Policy
- **Policy**: It is called the agent's brain. It tells us what action to take, given the state.
- **Optimal Policy**: Policy that **maximizes** the **expected return** when an agent acts according to it. It is learned through *training*.
### Policy-based Methods:
- An approach to solving RL problems.
- In this method, the Policy is learned directly.
- Will map each state to the best corresponding action at that state. Or a probability distribution over the set of possible actions at that state.
### Value-based Methods:
- Another approach to solving RL problems.
- Here, instead of training a policy, we train a **value function** that maps each state to the expected value of being in that state.
Contributions are welcomed 🤗
If you want to improve the course, you can [open a Pull Request.](https://github.com/huggingface/deep-rl-class/pulls)
This glossary was made possible thanks to:
- [@lucifermorningstar1305](https://github.com/lucifermorningstar1305)
- [@daspartho](https://github.com/daspartho)
- [@misza222](https://github.com/misza222)
| deep-rl-class/units/en/unit1/glossary.mdx/0 | {
"file_path": "deep-rl-class/units/en/unit1/glossary.mdx",
"repo_id": "deep-rl-class",
"token_count": 775
} | 79 |
# Mid-way Quiz [[mid-way-quiz]]
The best way to learn and [to avoid the illusion of competence](https://www.coursera.org/lecture/learning-how-to-learn/illusions-of-competence-BuFzf) **is to test yourself.** This will help you to find **where you need to reinforce your knowledge**.
### Q1: What are the two main approaches to find optimal policy?
<Question
choices={[
{
text: "Policy-based methods",
explain: "With Policy-Based methods, we train the policy directly to learn which action to take given a state.",
correct: true
},
{
text: "Random-based methods",
explain: ""
},
{
text: "Value-based methods",
explain: "With value-based methods, we train a value function to learn which state is more valuable and use this value function to take the action that leads to it.",
correct: true
},
{
text: "Evolution-strategies methods",
explain: ""
}
]}
/>
### Q2: What is the Bellman Equation?
<details>
<summary>Solution</summary>
**The Bellman equation is a recursive equation** that works like this: instead of starting for each state from the beginning and calculating the return, we can consider the value of any state as:
Rt+1 + gamma * V(St+1)
The immediate reward + the discounted value of the state that follows
</details>
### Q3: Define each part of the Bellman Equation
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/bellman4-quiz.jpg" alt="Bellman equation quiz"/>
<details>
<summary>Solution</summary>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/bellman4.jpg" alt="Bellman equation solution"/>
</details>
### Q4: What is the difference between Monte Carlo and Temporal Difference learning methods?
<Question
choices={[
{
text: "With Monte Carlo methods, we update the value function from a complete episode",
explain: "",
correct: true
},
{
text: "With Monte Carlo methods, we update the value function from a step",
explain: ""
},
{
text: "With TD learning methods, we update the value function from a complete episode",
explain: ""
},
{
text: "With TD learning methods, we update the value function from a step",
explain: "",
correct: true
},
]}
/>
### Q5: Define each part of Temporal Difference learning formula
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/td-ex.jpg" alt="TD Learning exercise"/>
<details>
<summary>Solution</summary>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/TD-1.jpg" alt="TD Exercise"/>
</details>
### Q6: Define each part of Monte Carlo learning formula
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/mc-ex.jpg" alt="MC Learning exercise"/>
<details>
<summary>Solution</summary>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit3/monte-carlo-approach.jpg" alt="MC Exercise"/>
</details>
Congrats on finishing this Quiz 🥳, if you missed some elements, take time to read again the previous sections to reinforce (😏) your knowledge.
| deep-rl-class/units/en/unit2/mid-way-quiz.mdx/0 | {
"file_path": "deep-rl-class/units/en/unit2/mid-way-quiz.mdx",
"repo_id": "deep-rl-class",
"token_count": 1100
} | 80 |
# Quiz [[quiz]]
The best way to learn and [to avoid the illusion of competence](https://www.coursera.org/lecture/learning-how-to-learn/illusions-of-competence-BuFzf) **is to test yourself.** This will help you to find **where you need to reinforce your knowledge**.
### Q1: We mentioned Q Learning is a tabular method. What are tabular methods?
<details>
<summary>Solution</summary>
*Tabular methods* is a type of problem in which the state and actions spaces are small enough to approximate value functions to be **represented as arrays and tables**. For instance, **Q-Learning is a tabular method** since we use a table to represent the state, and action value pairs.
</details>
### Q2: Why can't we use a classical Q-Learning to solve an Atari Game?
<Question
choices={[
{
text: "Atari environments are too fast for Q-Learning",
explain: ""
},
{
text: "Atari environments have a big observation space. So creating an updating the Q-Table would not be efficient",
explain: "",
correct: true
}
]}
/>
### Q3: Why do we stack four frames together when we use frames as input in Deep Q-Learning?
<details>
<summary>Solution</summary>
We stack frames together because it helps us **handle the problem of temporal limitation**: one frame is not enough to capture temporal information.
For instance, in pong, our agent **will be unable to know the ball direction if it gets only one frame**.
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit4/temporal-limitation.jpg" alt="Temporal limitation"/>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit4/temporal-limitation-2.jpg" alt="Temporal limitation"/>
</details>
### Q4: What are the two phases of Deep Q-Learning?
<Question
choices={[
{
text: "Sampling",
explain: "We perform actions and store the observed experiences tuples in a replay memory.",
correct: true,
},
{
text: "Shuffling",
explain: "",
},
{
text: "Reranking",
explain: "",
},
{
text: "Training",
explain: "We select the small batch of tuple randomly and learn from it using a gradient descent update step.",
correct: true,
}
]}
/>
### Q5: Why do we create a replay memory in Deep Q-Learning?
<details>
<summary>Solution</summary>
**1. Make more efficient use of the experiences during the training**
Usually, in online reinforcement learning, the agent interacts in the environment, gets experiences (state, action, reward, and next state), learns from them (updates the neural network), and discards them. This is not efficient.
But, with experience replay, **we create a replay buffer that saves experience samples that we can reuse during the training**.
**2. Avoid forgetting previous experiences and reduce the correlation between experiences**
The problem we get if we give sequential samples of experiences to our neural network is that it **tends to forget the previous experiences as it overwrites new experiences**. For instance, if we are in the first level and then the second, which is different, our agent can forget how to behave and play in the first level.
</details>
### Q6: How do we use Double Deep Q-Learning?
<details>
<summary>Solution</summary>
When we compute the Q target, we use two networks to decouple the action selection from the target Q value generation. We:
- Use our *DQN network* to **select the best action to take for the next state** (the action with the highest Q value).
- Use our *Target network* to calculate **the target Q value of taking that action at the next state**.
</details>
Congrats on finishing this Quiz 🥳, if you missed some elements, take time to read again the chapter to reinforce (😏) your knowledge.
| deep-rl-class/units/en/unit3/quiz.mdx/0 | {
"file_path": "deep-rl-class/units/en/unit3/quiz.mdx",
"repo_id": "deep-rl-class",
"token_count": 1099
} | 81 |
# An Introduction to Unity ML-Agents [[introduction-to-ml-agents]]
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit7/thumbnail.png" alt="thumbnail"/>
One of the challenges in Reinforcement Learning is **creating environments**. Fortunately for us, we can use game engines to do so.
These engines, such as [Unity](https://unity.com/), [Godot](https://godotengine.org/) or [Unreal Engine](https://www.unrealengine.com/), are programs made to create video games. They are perfectly suited
for creating environments: they provide physics systems, 2D/3D rendering, and more.
One of them, [Unity](https://unity.com/), created the [Unity ML-Agents Toolkit](https://github.com/Unity-Technologies/ml-agents), a plugin based on the game engine Unity that allows us **to use the Unity Game Engine as an environment builder to train agents**. In the first bonus unit, this is what we used to train Huggy to catch a stick!
<figure>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit5/example-envs.png" alt="MLAgents environments"/>
<figcaption>Source: <a href="https://github.com/Unity-Technologies/ml-agents">ML-Agents documentation</a></figcaption>
</figure>
Unity ML-Agents Toolkit provides many exceptional pre-made environments, from playing football (soccer), learning to walk, and jumping over big walls.
In this Unit, we'll learn to use ML-Agents, but **don't worry if you don't know how to use the Unity Game Engine**: you don't need to use it to train your agents.
So, today, we're going to train two agents:
- The first one will learn to **shoot snowballs onto a spawning target**.
- The second needs to **press a button to spawn a pyramid, then navigate to the pyramid, knock it over, and move to the gold brick at the top**. To do that, it will need to explore its environment, which will be done using a technique called curiosity.
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit7/envs.png" alt="Environments" />
Then, after training, **you'll push the trained agents to the Hugging Face Hub**, and you'll be able to **visualize them playing directly on your browser without having to use the Unity Editor**.
Doing this Unit will **prepare you for the next challenge: AI vs. AI where you will train agents in multi-agents environments and compete against your classmates' agents**.
Sound exciting? Let's get started!
| deep-rl-class/units/en/unit5/introduction.mdx/0 | {
"file_path": "deep-rl-class/units/en/unit5/introduction.mdx",
"repo_id": "deep-rl-class",
"token_count": 696
} | 82 |
# Designing Multi-Agents systems
For this section, you're going to watch this excellent introduction to multi-agents made by <a href="https://www.youtube.com/channel/UCq0imsn84ShAe9PBOFnoIrg"> Brian Douglas </a>.
<Youtube id="qgb0gyrpiGk" />
In this video, Brian talked about how to design multi-agent systems. He specifically took a multi-agents system of vacuum cleaners and asked: **how can can cooperate with each other**?
We have two solutions to design this multi-agent reinforcement learning system (MARL).
## Decentralized system
<figure>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit10/decentralized.png" alt="Decentralized"/>
<figcaption>
Source: <a href="https://www.youtube.com/watch?v=qgb0gyrpiGk"> Introduction to Multi-Agent Reinforcement Learning </a>
</figcaption>
</figure>
In decentralized learning, **each agent is trained independently from the others**. In the example given, each vacuum learns to clean as many places as it can **without caring about what other vacuums (agents) are doing**.
The benefit is that **since no information is shared between agents, these vacuums can be designed and trained like we train single agents**.
The idea here is that **our training agent will consider other agents as part of the environment dynamics**. Not as agents.
However, the big drawback of this technique is that it will **make the environment non-stationary** since the underlying Markov decision process changes over time as other agents are also interacting in the environment.
And this is problematic for many Reinforcement Learning algorithms **that can't reach a global optimum with a non-stationary environment**.
## Centralized approach
<figure>
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/unit10/centralized.png" alt="Centralized"/>
<figcaption>
Source: <a href="https://www.youtube.com/watch?v=qgb0gyrpiGk"> Introduction to Multi-Agent Reinforcement Learning </a>
</figcaption>
</figure>
In this architecture, **we have a high-level process that collects agents' experiences**: the experience buffer. And we'll use these experiences **to learn a common policy**.
For instance, in the vacuum cleaner example, the observation will be:
- The coverage map of the vacuums.
- The position of all the vacuums.
We use that collective experience **to train a policy that will move all three robots in the most beneficial way as a whole**. So each robot is learning from their common experience.
We now have a stationary environment since all the agents are treated as a larger entity, and they know the change of other agents' policies (since it's the same as theirs).
If we recap:
- In a *decentralized approach*, we **treat all agents independently without considering the existence of the other agents.**
- In this case, all agents **consider others agents as part of the environment**.
- **It’s a non-stationarity environment condition**, so has no guarantee of convergence.
- In a *centralized approach*:
- A **single policy is learned from all the agents**.
- Takes as input the present state of an environment and the policy outputs joint actions.
- The reward is global.
| deep-rl-class/units/en/unit7/multi-agent-setting.mdx/0 | {
"file_path": "deep-rl-class/units/en/unit7/multi-agent-setting.mdx",
"repo_id": "deep-rl-class",
"token_count": 847
} | 83 |
# Play with Huggy [[play]]
Now that you've trained Huggy and pushed it to the Hub. **You will be able to play with him ❤️**
For this step it’s simple:
- Open the Huggy game in your browser: https://huggingface.co/spaces/ThomasSimonini/Huggy
- Click on Play with my Huggy model
<img src="https://huggingface.co/datasets/huggingface-deep-rl-course/course-images/resolve/main/en/notebooks/unit-bonus1/load-huggy.jpg" alt="load-huggy" width="100%">
1. In step 1, choose your model repository which is the model id (in my case ThomasSimonini/ppo-Huggy).
2. In step 2, **choose which model you want to replay**:
- I have multiple ones, since we saved a model every 500000 timesteps.
- But if I want the most recent one I choose Huggy.onnx
👉 It's good to **try with different model checkpoints to see the improvement of the agent.**
| deep-rl-class/units/en/unitbonus1/play.mdx/0 | {
"file_path": "deep-rl-class/units/en/unitbonus1/play.mdx",
"repo_id": "deep-rl-class",
"token_count": 271
} | 84 |
# Files for typos
# Instruction: https://github.com/marketplace/actions/typos-action#getting-started
[default.extend-identifiers]
[default.extend-words]
NIN="NIN" # NIN is used in scripts/convert_ncsnpp_original_checkpoint_to_diffusers.py
nd="np" # nd may be np (numpy)
parms="parms" # parms is used in scripts/convert_original_stable_diffusion_to_diffusers.py
[files]
extend-exclude = ["_typos.toml"]
| diffusers/_typos.toml/0 | {
"file_path": "diffusers/_typos.toml",
"repo_id": "diffusers",
"token_count": 151
} | 85 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.