markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
OptimizersWe want to update the generator and discriminator variables separately. So we need to get the variables for each part and build optimizers for the two parts. To get all the trainable variables, we use `tf.trainable_variables()`. This creates a list of all the variables we've defined in our graph.For the generator optimizer, we only want to generator variables. Our past selves were nice and used a variable scope to start all of our generator variable names with `generator`. So, we just need to iterate through the list from `tf.trainable_variables()` and keep variables that start with `generator`. Each variable object has an attribute `name` which holds the name of the variable as a string (`var.name == 'weights_0'` for instance). We can do something similar with the discriminator. All the variables in the discriminator start with `discriminator`.Then, in the optimizer we pass the variable lists to the `var_list` keyword argument of the `minimize` method. This tells the optimizer to only update the listed variables. Something like `tf.train.AdamOptimizer().minimize(loss, var_list=var_list)` will only train the variables in `var_list`.>**Exercise: ** Below, implement the optimizers for the generator and discriminator. First you'll need to get a list of trainable variables, then split that list into two lists, one for the generator variables and another for the discriminator variables. Finally, using `AdamOptimizer`, create an optimizer for each network that update the network variables separately.
|
# Optimizers
learning_rate = 0.002
# Get the trainable_variables, split into G and D parts
t_vars = tf.trainable_variables()
g_vars = [var for var in t_vars if var.name.startswith('generator')]
d_vars = [var for var in t_vars if var.name.startswith('discriminator')]
d_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(d_loss, var_list=d_vars)
g_train_opt = tf.train.AdamOptimizer(learning_rate).minimize(g_loss, var_list=g_vars)
|
_____no_output_____
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
Training
|
batch_size = 100
epochs = 100
samples = []
losses = []
saver = tf.train.Saver(var_list = g_vars)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
for e in range(epochs):
for ii in range(mnist.train.num_examples//batch_size):
batch = mnist.train.next_batch(batch_size)
# Get images, reshape and rescale to pass to D
batch_images = batch[0].reshape((batch_size, 784))
batch_images = batch_images*2 - 1
# Sample random noise for G
batch_z = np.random.uniform(-1, 1, size=(batch_size, z_size))
# Run optimizers
_ = sess.run(d_train_opt, feed_dict={input_real: batch_images, input_z: batch_z})
_ = sess.run(g_train_opt, feed_dict={input_z: batch_z})
# At the end of each epoch, get the losses and print them out
train_loss_d = sess.run(d_loss, {input_z: batch_z, input_real: batch_images})
train_loss_g = g_loss.eval({input_z: batch_z})
print("Epoch {}/{}...".format(e+1, epochs),
"Discriminator Loss: {:.4f}...".format(train_loss_d),
"Generator Loss: {:.4f}".format(train_loss_g))
# Save losses to view after training
losses.append((train_loss_d, train_loss_g))
# Sample from generator as we're training for viewing afterwards
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, n_units=g_hidden_size, reuse=True, alpha=alpha),
feed_dict={input_z: sample_z})
samples.append(gen_samples)
saver.save(sess, './checkpoints/generator.ckpt')
# Save training generator samples
with open('train_samples.pkl', 'wb') as f:
pkl.dump(samples, f)
|
Epoch 1/100... Discriminator Loss: 0.3678... Generator Loss: 3.5897
Epoch 2/100... Discriminator Loss: 0.3645... Generator Loss: 3.6309
Epoch 3/100... Discriminator Loss: 0.4357... Generator Loss: 3.4136
Epoch 4/100... Discriminator Loss: 0.7224... Generator Loss: 6.2652
Epoch 5/100... Discriminator Loss: 0.6075... Generator Loss: 4.3486
Epoch 6/100... Discriminator Loss: 0.7519... Generator Loss: 4.1813
Epoch 7/100... Discriminator Loss: 0.9000... Generator Loss: 2.0781
Epoch 8/100... Discriminator Loss: 1.3756... Generator Loss: 1.5895
Epoch 9/100... Discriminator Loss: 0.9456... Generator Loss: 2.2028
Epoch 10/100... Discriminator Loss: 1.0948... Generator Loss: 2.3228
Epoch 11/100... Discriminator Loss: 1.0107... Generator Loss: 1.9952
Epoch 12/100... Discriminator Loss: 1.0860... Generator Loss: 1.6382
Epoch 13/100... Discriminator Loss: 1.1476... Generator Loss: 2.2547
Epoch 14/100... Discriminator Loss: 1.1930... Generator Loss: 2.0586
Epoch 15/100... Discriminator Loss: 0.7046... Generator Loss: 2.6142
Epoch 16/100... Discriminator Loss: 1.4536... Generator Loss: 1.9014
Epoch 17/100... Discriminator Loss: 0.9945... Generator Loss: 1.8042
Epoch 18/100... Discriminator Loss: 0.9138... Generator Loss: 2.2109
Epoch 19/100... Discriminator Loss: 1.9424... Generator Loss: 1.3089
Epoch 20/100... Discriminator Loss: 1.1435... Generator Loss: 2.6712
Epoch 21/100... Discriminator Loss: 1.1042... Generator Loss: 1.9536
Epoch 22/100... Discriminator Loss: 1.0478... Generator Loss: 1.8048
Epoch 23/100... Discriminator Loss: 1.0245... Generator Loss: 1.7871
Epoch 24/100... Discriminator Loss: 1.1023... Generator Loss: 1.3868
Epoch 25/100... Discriminator Loss: 1.0755... Generator Loss: 1.6908
Epoch 26/100... Discriminator Loss: 0.9878... Generator Loss: 1.5943
Epoch 27/100... Discriminator Loss: 1.0857... Generator Loss: 1.3091
Epoch 28/100... Discriminator Loss: 1.3463... Generator Loss: 1.0437
Epoch 29/100... Discriminator Loss: 1.1604... Generator Loss: 1.5717
Epoch 30/100... Discriminator Loss: 0.8951... Generator Loss: 2.0207
Epoch 31/100... Discriminator Loss: 0.7005... Generator Loss: 2.6086
Epoch 32/100... Discriminator Loss: 0.8702... Generator Loss: 3.1350
Epoch 33/100... Discriminator Loss: 0.8715... Generator Loss: 2.2553
Epoch 34/100... Discriminator Loss: 0.9684... Generator Loss: 2.1891
Epoch 35/100... Discriminator Loss: 1.1734... Generator Loss: 2.2908
Epoch 36/100... Discriminator Loss: 1.0828... Generator Loss: 1.7820
Epoch 37/100... Discriminator Loss: 1.0500... Generator Loss: 2.0008
Epoch 38/100... Discriminator Loss: 1.0380... Generator Loss: 1.8724
Epoch 39/100... Discriminator Loss: 0.9818... Generator Loss: 2.2052
Epoch 40/100... Discriminator Loss: 0.7781... Generator Loss: 2.0690
Epoch 41/100... Discriminator Loss: 0.9980... Generator Loss: 2.0535
Epoch 42/100... Discriminator Loss: 0.9168... Generator Loss: 1.9688
Epoch 43/100... Discriminator Loss: 0.8056... Generator Loss: 2.1651
Epoch 44/100... Discriminator Loss: 0.9269... Generator Loss: 2.4960
Epoch 45/100... Discriminator Loss: 0.8217... Generator Loss: 2.0567
Epoch 46/100... Discriminator Loss: 0.8549... Generator Loss: 2.3850
Epoch 47/100... Discriminator Loss: 0.7160... Generator Loss: 2.2929
Epoch 48/100... Discriminator Loss: 0.8896... Generator Loss: 2.0800
Epoch 49/100... Discriminator Loss: 1.0880... Generator Loss: 1.6579
Epoch 50/100... Discriminator Loss: 1.0534... Generator Loss: 2.2300
Epoch 51/100... Discriminator Loss: 0.9481... Generator Loss: 2.1894
Epoch 52/100... Discriminator Loss: 0.9767... Generator Loss: 2.1380
Epoch 53/100... Discriminator Loss: 1.1231... Generator Loss: 1.8526
Epoch 54/100... Discriminator Loss: 0.9081... Generator Loss: 1.9529
Epoch 55/100... Discriminator Loss: 1.0239... Generator Loss: 2.2693
Epoch 56/100... Discriminator Loss: 1.0482... Generator Loss: 1.7395
Epoch 57/100... Discriminator Loss: 0.9157... Generator Loss: 1.8313
Epoch 58/100... Discriminator Loss: 0.8839... Generator Loss: 2.2156
Epoch 59/100... Discriminator Loss: 0.9230... Generator Loss: 1.8060
Epoch 60/100... Discriminator Loss: 0.9655... Generator Loss: 2.0184
Epoch 61/100... Discriminator Loss: 0.9161... Generator Loss: 1.9261
Epoch 62/100... Discriminator Loss: 0.8266... Generator Loss: 2.0038
Epoch 63/100... Discriminator Loss: 0.8978... Generator Loss: 2.2338
Epoch 64/100... Discriminator Loss: 1.0432... Generator Loss: 1.6098
Epoch 65/100... Discriminator Loss: 1.1114... Generator Loss: 1.4504
Epoch 66/100... Discriminator Loss: 0.9215... Generator Loss: 1.7533
Epoch 67/100... Discriminator Loss: 0.9408... Generator Loss: 1.9942
Epoch 68/100... Discriminator Loss: 1.1266... Generator Loss: 1.8851
Epoch 69/100... Discriminator Loss: 1.1030... Generator Loss: 1.5994
Epoch 70/100... Discriminator Loss: 0.8640... Generator Loss: 2.1240
Epoch 71/100... Discriminator Loss: 0.9544... Generator Loss: 2.0405
Epoch 72/100... Discriminator Loss: 0.9874... Generator Loss: 1.6178
Epoch 73/100... Discriminator Loss: 0.9185... Generator Loss: 2.0491
Epoch 74/100... Discriminator Loss: 1.1316... Generator Loss: 1.4504
Epoch 75/100... Discriminator Loss: 0.9773... Generator Loss: 1.5728
Epoch 76/100... Discriminator Loss: 0.9489... Generator Loss: 1.8277
Epoch 77/100... Discriminator Loss: 1.1911... Generator Loss: 1.6216
Epoch 78/100... Discriminator Loss: 1.0081... Generator Loss: 2.2352
Epoch 79/100... Discriminator Loss: 0.9159... Generator Loss: 1.9558
Epoch 80/100... Discriminator Loss: 1.0335... Generator Loss: 1.7723
Epoch 81/100... Discriminator Loss: 1.1103... Generator Loss: 1.7102
Epoch 82/100... Discriminator Loss: 0.9083... Generator Loss: 2.1996
Epoch 83/100... Discriminator Loss: 1.0865... Generator Loss: 1.8519
Epoch 84/100... Discriminator Loss: 0.9847... Generator Loss: 1.6953
Epoch 85/100... Discriminator Loss: 0.8754... Generator Loss: 2.0061
Epoch 86/100... Discriminator Loss: 1.0773... Generator Loss: 1.7892
Epoch 87/100... Discriminator Loss: 0.8109... Generator Loss: 1.9592
Epoch 88/100... Discriminator Loss: 0.9158... Generator Loss: 2.1380
Epoch 89/100... Discriminator Loss: 0.8513... Generator Loss: 2.1318
Epoch 90/100... Discriminator Loss: 0.8617... Generator Loss: 2.1592
Epoch 91/100... Discriminator Loss: 0.9953... Generator Loss: 1.7510
Epoch 92/100... Discriminator Loss: 0.9552... Generator Loss: 2.0343
Epoch 93/100... Discriminator Loss: 0.8665... Generator Loss: 1.7532
Epoch 94/100... Discriminator Loss: 0.8872... Generator Loss: 1.5990
Epoch 95/100... Discriminator Loss: 1.1236... Generator Loss: 1.5481
Epoch 96/100... Discriminator Loss: 0.8847... Generator Loss: 2.2585
Epoch 97/100... Discriminator Loss: 0.9323... Generator Loss: 1.6943
Epoch 98/100... Discriminator Loss: 1.1218... Generator Loss: 1.4691
Epoch 99/100... Discriminator Loss: 0.9928... Generator Loss: 2.0241
Epoch 100/100... Discriminator Loss: 0.9447... Generator Loss: 2.0391
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
Training lossHere we'll check out the training losses for the generator and discriminator.
|
%matplotlib inline
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
losses = np.array(losses)
plt.plot(losses.T[0], label='Discriminator')
plt.plot(losses.T[1], label='Generator')
plt.title("Training Losses")
plt.legend()
|
_____no_output_____
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
Generator samples from trainingHere we can view samples of images from the generator. First we'll look at images taken while training.
|
def view_samples(epoch, samples):
fig, axes = plt.subplots(figsize=(7,7), nrows=4, ncols=4, sharey=True, sharex=True)
for ax, img in zip(axes.flatten(), samples[epoch]):
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
im = ax.imshow(img.reshape((28,28)), cmap='Greys_r')
return fig, axes
# Load samples from generator taken while training
with open('train_samples.pkl', 'rb') as f:
samples = pkl.load(f)
|
_____no_output_____
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
These are samples from the final training epoch. You can see the generator is able to reproduce numbers like 5, 7, 3, 0, 9. Since this is just a sample, it isn't representative of the full range of images this generator can make.
|
_ = view_samples(-1, samples)
|
_____no_output_____
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
Below I'm showing the generated images as the network was training, every 10 epochs. With bonus optical illusion!
|
rows, cols = 10, 6
fig, axes = plt.subplots(figsize=(7,12), nrows=rows, ncols=cols, sharex=True, sharey=True)
for sample, ax_row in zip(samples[::int(len(samples)/rows)], axes):
for img, ax in zip(sample[::int(len(sample)/cols)], ax_row):
ax.imshow(img.reshape((28,28)), cmap='Greys_r')
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
|
_____no_output_____
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
It starts out as all noise. Then it learns to make only the center white and the rest black. You can start to see some number like structures appear out of the noise. Looks like 1, 9, and 8 show up first. Then, it learns 5 and 3. Sampling from the generatorWe can also get completely new images from the generator by using the checkpoint we saved after training. We just need to pass in a new latent vector $z$ and we'll get new samples!
|
saver = tf.train.Saver(var_list=g_vars)
with tf.Session() as sess:
saver.restore(sess, tf.train.latest_checkpoint('checkpoints'))
sample_z = np.random.uniform(-1, 1, size=(16, z_size))
gen_samples = sess.run(
generator(input_z, input_size, n_units=g_hidden_size, reuse=True, alpha=alpha),
feed_dict={input_z: sample_z})
view_samples(0, [gen_samples])
|
INFO:tensorflow:Restoring parameters from checkpoints\generator.ckpt
|
Apache-2.0
|
Intro_to_GANs_Exercises.ipynb
|
agoila/gan_mnist
|
Intro to Machine Learning with Classification Contents1. **Loading** iris dataset2. Splitting into **train**- and **test**-set3. Creating a **model** and training it4. **Predicting** test set5. **Evaluating** the result6. Selecting **features** This notebook will introduce you to Machine Learning and classification, using our most valued Python data science toolkit: [ScikitLearn](http://scikit-learn.org/).Classification will allow you to automatically classify data, based on the classification of previous data. The algorithm determines automatically which features it will use to classify, so the programmer does not have to think of this anymore (although it helps).First, we will transform a dataset into a set of features with labels that the algorithm can use. Then we will predict labels and validate them. Last we will select features manually and see if we can make the prediction better.Let's start with some imports.
|
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
1. Loading iris dataset We load the dataset from the datasets module in sklearn.
|
iris = datasets.load_iris()
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
This dataset contains information about iris flowers. Every entry describes a flower, more specifically its - sepal length- sepal width- petal length- petal widthSo every entry has four columns. 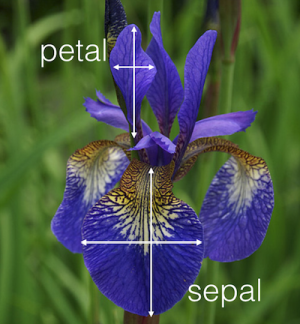 We can visualise the data with Pandas, a Python library to handle dataframes. This gives us a pretty table to see what our data looks like.We will not cover Pandas in this notebook, so don't worry about this piece of code.
|
import pandas as pd
df = pd.DataFrame(data=iris.data, columns=iris.feature_names)
df["target"] = iris.target
df.sample(n=10) # show 10 random rows
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
There are 3 different species of irises in the dataset. Every species has 50 samples, so there are 150 entries in total.We can confirm this by checking the "data"-element of the iris variable. The "data"-element is a 2D-array that contains all our entries. We can use the python function `.shape` to check its dimensions.
|
iris.data.shape
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
To get an example of the data, we can print the first ten rows:
|
print(iris.data[0:10, :]) # 0:10 gets rows 0-10, : gets all the columns
|
[[5.1 3.5 1.4 0.2]
[4.9 3. 1.4 0.2]
[4.7 3.2 1.3 0.2]
[4.6 3.1 1.5 0.2]
[5. 3.6 1.4 0.2]
[5.4 3.9 1.7 0.4]
[4.6 3.4 1.4 0.3]
[5. 3.4 1.5 0.2]
[4.4 2.9 1.4 0.2]
[4.9 3.1 1.5 0.1]]
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
The labels that we're looking for are in the "target"-element of the iris variable. This 1D-array contains the iris species for each of the entries.
|
iris.target.shape
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
Let's have a look at the target values:
|
print(iris.target)
|
[0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 2 2 2 2 2 2
2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2
2 2]
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
There are three categories so each entry will be classified as 0, 1 or 2. To get the names of the corresponding species we can print `target_names`.
|
print(iris.target_names)
|
['setosa' 'versicolor' 'virginica']
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
The iris variable is a dataset from sklearn and also contains a description of itself. We already provided the information you need to know about the data, but if you want to check, you can print the `.DESCR` method of the iris dataset.
|
print(iris.DESCR)
|
.. _iris_dataset:
Iris plants dataset
--------------------
**Data Set Characteristics:**
:Number of Instances: 150 (50 in each of three classes)
:Number of Attributes: 4 numeric, predictive attributes and the class
:Attribute Information:
- sepal length in cm
- sepal width in cm
- petal length in cm
- petal width in cm
- class:
- Iris-Setosa
- Iris-Versicolour
- Iris-Virginica
:Summary Statistics:
============== ==== ==== ======= ===== ====================
Min Max Mean SD Class Correlation
============== ==== ==== ======= ===== ====================
sepal length: 4.3 7.9 5.84 0.83 0.7826
sepal width: 2.0 4.4 3.05 0.43 -0.4194
petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)
petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)
============== ==== ==== ======= ===== ====================
:Missing Attribute Values: None
:Class Distribution: 33.3% for each of 3 classes.
:Creator: R.A. Fisher
:Donor: Michael Marshall (MARSHALL%[email protected])
:Date: July, 1988
The famous Iris database, first used by Sir R.A. Fisher. The dataset is taken
from Fisher's paper. Note that it's the same as in R, but not as in the UCI
Machine Learning Repository, which has two wrong data points.
This is perhaps the best known database to be found in the
pattern recognition literature. Fisher's paper is a classic in the field and
is referenced frequently to this day. (See Duda & Hart, for example.) The
data set contains 3 classes of 50 instances each, where each class refers to a
type of iris plant. One class is linearly separable from the other 2; the
latter are NOT linearly separable from each other.
.. topic:: References
- Fisher, R.A. "The use of multiple measurements in taxonomic problems"
Annual Eugenics, 7, Part II, 179-188 (1936); also in "Contributions to
Mathematical Statistics" (John Wiley, NY, 1950).
- Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.
(Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.
- Dasarathy, B.V. (1980) "Nosing Around the Neighborhood: A New System
Structure and Classification Rule for Recognition in Partially Exposed
Environments". IEEE Transactions on Pattern Analysis and Machine
Intelligence, Vol. PAMI-2, No. 1, 67-71.
- Gates, G.W. (1972) "The Reduced Nearest Neighbor Rule". IEEE Transactions
on Information Theory, May 1972, 431-433.
- See also: 1988 MLC Proceedings, 54-64. Cheeseman et al"s AUTOCLASS II
conceptual clustering system finds 3 classes in the data.
- Many, many more ...
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
Now we have a good idea what our data looks like.Our task now is to solve a **supervised** learning problem: Predict the species of an iris using the measurements that serve as our so-called **features**.
|
# First, we store the features we use and the labels we want to predict into two different variables
X = iris.data
y = iris.target
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
2. Splitting into train- and test-set We want to evaluate our model on data with labels that our model has not seen yet. This will give us an idea on how well the model can predict new data, and makes sure we are not [overfitting](https://en.wikipedia.org/wiki/Overfitting). If we would test and train on the same data, we would just learn this dataset really really well, but not be able to tell anything about other data.So we split our dataset into a train- and test-set. Sklearn has a function to do this: `train_test_split`. Have a look at the [documentation](http://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html) of this function and see if you can split `iris.data` and `iris.target` into train- and test-sets with a test-size of 33%.
|
from sklearn.model_selection import train_test_split
??train_test_split
X_train, X_test, y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.33, stratify=iris.target)# TODO: split iris.data and iris.target into test and train
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
We can now check the size of the resulting arrays. The shapes should be `(100, 4)`, `(100,)`, `(50, 4)` and `(50,)`.
|
print("X_train shape: {}, y_train shape: {}".format(X_train.shape, y_train.shape))
print("X_test shape: {} , y_test shape: {}".format(X_test.shape, y_test.shape))
|
X_train shape: (100, 4), y_train shape: (100,)
X_test shape: (50, 4) , y_test shape: (50,)
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
3. Creating a model and training it Now we will give the data to a model. We will use a Decision Tree Classifier model for this.This model will create a decision tree based on the X_train and y_train values and include decisions like this: 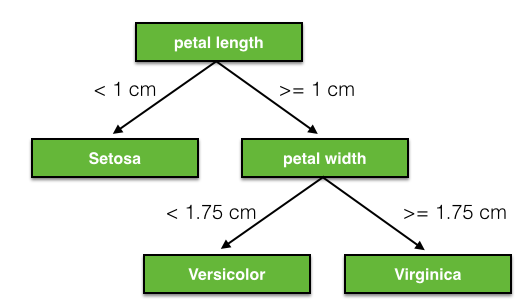 Find the Decision Tree Classifier in sklearn and call its constructor. It might be useful to set the random_state parameter to 0, otherwise a different tree will be generated each time you run the code.
|
from sklearn import tree
model = tree.DecisionTreeClassifier(random_state=0)# TODO: create a decision tree classifier
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
The model is still empty and doesn't know anything. Train (fit) it with our train-data, so that it learns things about our iris-dataset.
|
model = model.fit(X_train, y_train)# TODO: fit the train-data to the model
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
4. Predicting test set We now have a model that contains a decision tree. This decision tree knows how to turn our X_train values into y_train values. We will now let it run on our X_test values and have a look at the result.We don't want to overwrite our actual y_test values, so we store the predicted y_test values as y_pred.
|
y_pred = model.predict(X_test)# TODO: predict y_pred from X_test
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
5. Evaluating the result We now have y_test (the real values for X_test) and y_pred. We can print these values and compare them, to get an idea of how good the model predicted the data.
|
print(y_test)
print("-"*75) # print a line
print(y_pred)
|
[0 2 0 2 2 1 1 0 1 1 2 0 2 0 2 1 2 0 0 1 0 2 2 1 1 1 2 0 1 2 2 0 1 0 0 1 1
2 2 0 1 1 1 0 2 0 1 0 0 2]
---------------------------------------------------------------------------
[0 2 0 2 1 1 1 0 1 1 2 0 2 0 2 1 2 0 0 1 0 2 2 1 1 1 2 0 2 2 2 0 1 0 0 2 1
2 2 0 1 1 1 0 2 0 1 0 0 2]
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
If we look at the values closely, we can discover that all but two values are predicted correctly. However, it is bothersome to compare the numbers one by one. There are only thirty of them, but what if there were one hundred? We will need an easier method to compare our results.Luckily, this can also be found in sklearn. Google for sklearn's accuracy score and compare our y_test and y_pred. This will give us the percentage of entries that was predicted correctly.
|
from sklearn import metrics
accuracy = metrics.accuracy_score(y_test, y_pred) # TODO: calculate accuracy score of y_test and y_pred
print(accuracy)
|
0.94
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
That's pretty good, isn't it?To understand what our classifier actually did, have a look at the following picture: 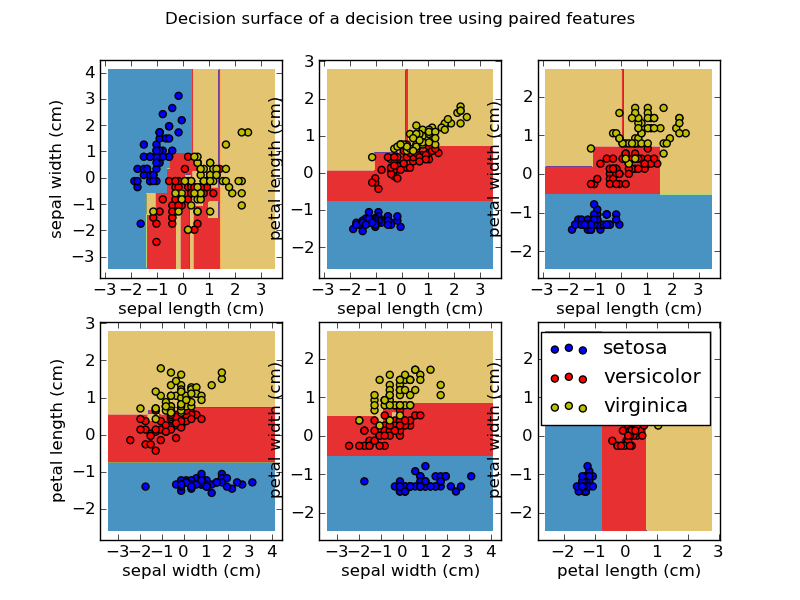 We see the distribution of all our features, compared with each other. Some have very clear distinctions between two categories, so our decision tree probably used those to make predictions about our data. 6. Selecting features In our dataset, there are four features to describe the flowers. Using these four features, we got a pretty high accuracy to predict the species. But maybe some of our features were not necessary. Maybe some did not improve our prediction, or even made it worse.It's worth a try to see if a subset of features is better at predicting the labels than all features.We still have our X_train, X_test, y_train and y_test variables. We will try removing a few columns from X_train and X_test and recalculate our accuracy.First, create a feature selector that will select the 2 features X_train that best describe y_train.(Hint: look at the imports)
|
from sklearn.feature_selection import SelectKBest, chi2
selector = SelectKBest(chi2, k=2) # TODO: create a selector for the 2 best features and fit X_train and y_train to it
X_train, X_test, y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.33, random_state=42)
selector = selector.fit(X_train, y_train)
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
We can check which features our selector selected, using the following function:
|
print(selector.get_support())
|
[False False True True]
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
It gives us an array of True and False values that represent the columns of the original X_train. The values that are marked by True are considered the most informative by the selector. Let's use the selector to select (transform) these features from the X_train values.
|
X_train_new = selector.transform(X_train) # TODO: use selector to transform X_train
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
The dimensions of X_train have now changed:
|
X_train_new.shape
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
If we want to use these values in our model, we will need to adjust X_test as well. We would get in trouble later if X_train has only 2 columns and X_test has 4. So perform the same selection on X_test.
|
X_test_new = selector.transform(X_test) # TODO: use selector to transform X_test
X_test_new.shape
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
Now we can repeat the earlier steps: create a model, fit the data to it and predict our y_test values.
|
model = tree.DecisionTreeClassifier(random_state=0) # TODO: create model as before
model = model.fit(X_train_new, y_train) # TODO: fit model as before, but use X_train_new
y_pred = model.predict(X_test_new) # TODO: predict values as before, but use X_test_new
|
_____no_output_____
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
Let's have a look at the accuracy score of our new prediction.
|
accuracy = metrics.accuracy_score(y_test, y_pred) # TODO: calculate accuracy score of y_test and y_pred
print(accuracy) # TODO: calculate accuracy score as before
|
1.0
|
MIT
|
PXL_DIGITAL_JAAR_2/Data Advanced/Bestanden/notebooks_data/Machine Learning Exercises/Own solutions/Machine_Learning_1_Classification.ipynb
|
Limoentaart/PXL_IT_JAAR_1
|
This Dataset was taken online
|
import pandas as pd
import numpy as np
dataset = pd.read_csv('url_dataset.csv')
#deleting all columns except url
dataset.drop(dataset.columns.difference(['URL']), 1, inplace=True)
dataset.head(5)
dataset.to_csv('cleaned_link_dataset.csv')
#split protocol from the other part of link
cleaned_dataset = pd.read_csv('cleaned_link_dataset.csv')
protocol = cleaned_dataset['URL'].str.split('://',expand=True)
#renaming columns
protocol.head()
protocol.rename(columns={0: 'protocol'}, inplace= True)
protocol.head()
#dividing domain name from the address of the link and create a new column with it
domain = protocol[1].str.split('/', 1,expand=True)
domain.head()
domain.rename(columns ={0:'domain', 1:'address'}, inplace= True)
domain.head()
protocol.head()
#joining datasets together in one dataframe using pd.concat
full_dataset = pd.concat([protocol['protocol'],domain],axis=1)
full_dataset.head(5)
full_dataset.to_csv('full_cleaned.csv')
|
_____no_output_____
|
MIT
|
Link Dataset Cleaning.ipynb
|
geridashja/phishing-links-detection
|
420-A52-SF - Algorithmes d'apprentissage supervisé - Hiver 2020 - Spécialisation technique en Intelligence Artificielle - Mikaël Swawola, M.Sc.**Objectif:** cette séance de travaux pratique est consacrée à la mise en oeuvre de l'ensemble des connaissances acquises jusqu'alors sur un nouveau jeu de données, *NBA*
|
%reload_ext autoreload
%autoreload 2
%matplotlib inline
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
0 - Chargement des bibliothèques
|
# Manipulation de données
import numpy as np
import pandas as pd
# Visualisation de données
import matplotlib.pyplot as plt
import seaborn as sns
# Configuration de la visualisation
sns.set(style="darkgrid", rc={'figure.figsize':(11.7,8.27)})
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
1 - Lecture du jeu de données *NBA* **Lire le fichier `NBA_train.csv`**
|
# Compléter le code ci-dessous ~ 1 ligne
NBA = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Afficher les dix premières lignes de la trame de données**
|
# Compléter le code ci-dessous ~ 1 ligne
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
Ci-dessous, la description des différentes variables explicatives du jeu de données| Variable | Description || ------------- |:-------------------------------------------------------------:|| SeasonEnd | Année de fin de la saison || Team | Nom de l'équipe || Playoffs | Indique si l'équipe est allée en playoffs || W | Nombre de victoires au cours de la saison régulière || PTS | Nombre de points obtenus (saison régulière) || oppPTS | Nombre de points obtenus pas les opposants (saison régulière) || FG | Nombre de Field Goals réussis || FGA | Nombre de tentatives de Field Goals || 2P | Nombre de 2-pointers réussis || 2PA | Nombre de tentatives de 2-pointers || 3P | Nombre de 3-pointers réussis || 3PA | Nombre de tentatives de 3-pointers || FT | Nombre de Free throws réussis || FTA | Nombre de tentatives de Free throws || ORB | Nombre de rebonds offensifs || DRB | Nombre de rebonds défensifs || AST | Nombre de passes décisives (assists) || STL | Nombre d'interceptions (steals) || BLK | Nombre de contres (blocks) || TOV | Nombre de turnovers | 1 - Régression linéaire simple Nous allons dans un premier temps effectuer la prédiction du nombre de victoires au cours de la saison régulière en fonction de la différence de points obtenus pas l'équipe et par ses opposantsNous commencons donc par un peu d'**ingénierie de données**. Une nouvelle variable explicative correspondant à la différence de points obtenus pas l'équipe et par ses opposants est crée **Créer un nouvelle variable PTSdiff, représentant la différence entre PTS et oppPTS**
|
# Compléter le code ci-dessous ~ 1 ligne
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Stocker le nombre de lignes du jeu de donnée (nombre d'exemples d'entraînement) dans la variable `m`**
|
# Compléter le code ci-dessous ~ 1 ligne
m = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Stocker le nombre de victoires au cours de la saison dans la variable `y`. Il s'agira de la variable que l'on cherche à prédire**
|
# Compléter le code ci-dessous ~ 1 ligne
y = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Créer la matrice des prédicteurs `X`.** Indice: `X` doit avoir 2 colonnes...
|
# Compléter le code ci-dessous ~ 3 lignes
X = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Vérifier la dimension de la matrice des prédicteurs `X`. Quelle est la dimension de `X` ?**
|
# Compléter le code ci-dessous ~ 1 ligne
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Créer le modèle de référence (baseline)**
|
# Compléter le code ci-dessous ~ 1 ligne
y_baseline = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**À l'aide de l'équation normale, trouver les paramètres optimaux du modèle de régression linéaire simple**
|
# Compléter le code ci-dessous ~ 1 ligne
theta = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer la somme des carrées des erreurs (SSE)**
|
# Compléter le code ci-dessous ~ 1 ligne
SSE = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer la racine carrée de l'erreur quadratique moyenne (RMSE)**
|
# Compléter le code ci-dessous ~ 1 ligne
RMSE = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer le coefficient de détermination $R^2$**
|
# Compléter le code ci-dessous ~ 1-2 lignes
R2 = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Affichage des résultats**
|
fig, ax = plt.subplots()
ax.scatter(x1, y,label="Data points")
reg_x = np.linspace(-1000,1000,50)
reg_y = theta[0] + np.linspace(-1000,1000,50)* theta[1]
ax.plot(reg_x, np.repeat(y_baseline,50), color='#777777', label="Baseline", lw=2)
ax.plot(reg_x, reg_y, color="g", lw=2, label="Modèle")
ax.set_xlabel("Différence de points", fontsize=16)
ax.set_ylabel("Nombre de victoires", fontsize=16)
ax.legend(loc='upper left', fontsize=16)
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
3 - Régression linéaire multiple Nous allons maintenant tenter de prédire le nombre de points obtenus par une équipe donnée au cours de la saison régulière en fonction des autres variables explicatives disponibles. Nous allons mettre en oeuvre plusieurs modèles de régression linéaire multiple **Stocker le nombre de points marqués au cours de la saison dans la variable `y`. Il s'agira de la varible que l'on cherche à prédire**
|
# Compléter le code ci-dessous ~ 1 ligne
y = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Créer la matrice des prédicteurs `X` à partir des variables `2PA` et `3PA`**
|
# Compléter le code ci-dessous ~ 3 lignes
X = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Vérifier la dimension de la matrice des prédicteurs `X`. Quelle est la dimension de `X` ?**
|
# Compléter le code ci-dessous ~ 1 ligne
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Créer le modèle de référence (baseline)**
|
# Compléter le code ci-dessous ~ 1 ligne
y_baseline = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**À l'aide de l'équation normale, trouver les paramètres optimaux du modèle de régression linéaire**
|
# Compléter le code ci-dessous ~ 1 ligne
theta = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer la somme des carrées des erreurs (SSE)**
|
# Compléter le code ci-dessous ~ 1 ligne
SSE = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer la racine carrée de l'erreur quadratique moyenne (RMSE)**
|
# Compléter le code ci-dessous ~ 1 ligne
RMSE = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
**Calculer le coefficient de détermination $R^2$**
|
# Compléter le code ci-dessous ~ 1-2 lignes
R2 = None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
3 - Ajouter les variables explicatives FTA et AST **Recommencer les étapes ci-dessus en incluant les variables FTA et AST**
|
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
4 - Ajouter les variables explicatives ORB et STL **Recommencer les étapes ci-dessus en incluant les variables ORB et STL**
|
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
5 - Ajouter les variables explicatives DRB et BLK **Recommencer les étapes ci-dessus en incluant les variables DRB et BLK**
|
None
|
_____no_output_____
|
MIT
|
nbs/06-metriques-et-evaluation-des-modeles-de-regression/06-TP.ipynb
|
tiombo/TP12
|
Multi-Perceptor VQGAN + CLIP (v.3.2021.11.29)by [@remi_durant](https://twitter.com/remi_durant)Lots drawn from or inspired by other colabs, chief among them is [@jbusted1](https://twitter.com/jbusted1)'s MSE regularized VQGAN + Clip, and [@RiversHaveWings](https://twitter.com/RiversHaveWings) VQGAN + Clip with Z+Quantize. Standing on the shoulders of giants. - Multi-clip mode sends the same cutouts to whatever clip models you want. If they have different perceptor resolutions, the cuts are generated at each required size, replaying the same augments across both scales- Alternate random noise generation options to use as start point (perlin, pyramid, or vqgan random z tokens)- MSE Loss doesn't apply if you have no init_image until after reaching the first epoch.- MSE epoch targets z.tensor, not z.average, to allow for more creativity- Grayscale augment added for better structure- Padding fix for perspective and affine augments to not always be black barred- Automatic disable of cudnn for A100
|
#@title First check what GPU you got and make sure it's a good one.
#@markdown - Tier List: (K80 < T4 < P100 < V100 < A100)
from subprocess import getoutput
!nvidia-smi --query-gpu=name,memory.total,memory.free --format=csv,noheader
|
_____no_output_____
|
MIT
|
Multi_Perceptor_VQGAN_+_CLIP_[Public].ipynb
|
keirwilliamsxyz/keirxyz
|
Setup
|
#@title memory footprint support libraries/code
!ln -sf /opt/bin/nvidia-smi /usr/bin/nvidia-smi
!pip install gputil
!pip install psutil
!pip install humanize
import psutil
import humanize
import os
import GPUtil as GPU
#@title Print GPU details
!nvidia-smi
GPUs = GPU.getGPUs()
# XXX: only one GPU on Colab and isn’t guaranteed
gpu = GPUs[0]
def printm():
process = psutil.Process(os.getpid())
print("Gen RAM Free: " + humanize.naturalsize(psutil.virtual_memory().available), " | Proc size: " + humanize.naturalsize(process.memory_info().rss))
print("GPU RAM Free: {0:.0f}MB | Used: {1:.0f}MB | Util {2:3.0f}% | Total {3:.0f}MB".format(gpu.memoryFree, gpu.memoryUsed, gpu.memoryUtil*100, gpu.memoryTotal))
printm()
#@title Install Dependencies
# Fix for A100 issues
!pip install tensorflow==1.15.2
# Install normal dependencies
!git clone https://github.com/openai/CLIP
!git clone https://github.com/CompVis/taming-transformers
!pip install ftfy regex tqdm omegaconf pytorch-lightning
!pip install kornia
!pip install einops
!pip install transformers
#@title Load libraries and variables
import argparse
import math
from pathlib import Path
import sys
sys.path.append('./taming-transformers')
from IPython import display
from omegaconf import OmegaConf
from PIL import Image
from taming.models import cond_transformer, vqgan
import torch
from torch import nn, optim
from torch.nn import functional as F
from torchvision import transforms
from torchvision.transforms import functional as TF
from tqdm.notebook import tqdm
import numpy as np
import os.path
from os import path
from urllib.request import Request, urlopen
from CLIP import clip
import kornia
import kornia.augmentation as K
from torch.utils.checkpoint import checkpoint
from matplotlib import pyplot as plt
from fastprogress.fastprogress import master_bar, progress_bar
import random
import gc
import re
from datetime import datetime
from base64 import b64encode
import warnings
warnings.filterwarnings('ignore')
torch.set_printoptions( sci_mode=False )
def noise_gen(shape, octaves=5):
n, c, h, w = shape
noise = torch.zeros([n, c, 1, 1])
max_octaves = min(octaves, math.log(h)/math.log(2), math.log(w)/math.log(2))
for i in reversed(range(max_octaves)):
h_cur, w_cur = h // 2**i, w // 2**i
noise = F.interpolate(noise, (h_cur, w_cur), mode='bicubic', align_corners=False)
noise += torch.randn([n, c, h_cur, w_cur]) / 5
return noise
def sinc(x):
return torch.where(x != 0, torch.sin(math.pi * x) / (math.pi * x), x.new_ones([]))
def lanczos(x, a):
cond = torch.logical_and(-a < x, x < a)
out = torch.where(cond, sinc(x) * sinc(x/a), x.new_zeros([]))
return out / out.sum()
def ramp(ratio, width):
n = math.ceil(width / ratio + 1)
out = torch.empty([n])
cur = 0
for i in range(out.shape[0]):
out[i] = cur
cur += ratio
return torch.cat([-out[1:].flip([0]), out])[1:-1]
def resample(input, size, align_corners=True):
n, c, h, w = input.shape
dh, dw = size
input = input.view([n * c, 1, h, w])
if dh < h:
kernel_h = lanczos(ramp(dh / h, 2), 2).to(input.device, input.dtype)
pad_h = (kernel_h.shape[0] - 1) // 2
input = F.pad(input, (0, 0, pad_h, pad_h), 'reflect')
input = F.conv2d(input, kernel_h[None, None, :, None])
if dw < w:
kernel_w = lanczos(ramp(dw / w, 2), 2).to(input.device, input.dtype)
pad_w = (kernel_w.shape[0] - 1) // 2
input = F.pad(input, (pad_w, pad_w, 0, 0), 'reflect')
input = F.conv2d(input, kernel_w[None, None, None, :])
input = input.view([n, c, h, w])
return F.interpolate(input, size, mode='bicubic', align_corners=align_corners)
# def replace_grad(fake, real):
# return fake.detach() - real.detach() + real
class ReplaceGrad(torch.autograd.Function):
@staticmethod
def forward(ctx, x_forward, x_backward):
ctx.shape = x_backward.shape
return x_forward
@staticmethod
def backward(ctx, grad_in):
return None, grad_in.sum_to_size(ctx.shape)
class ClampWithGrad(torch.autograd.Function):
@staticmethod
def forward(ctx, input, min, max):
ctx.min = min
ctx.max = max
ctx.save_for_backward(input)
return input.clamp(min, max)
@staticmethod
def backward(ctx, grad_in):
input, = ctx.saved_tensors
return grad_in * (grad_in * (input - input.clamp(ctx.min, ctx.max)) >= 0), None, None
replace_grad = ReplaceGrad.apply
clamp_with_grad = ClampWithGrad.apply
# clamp_with_grad = torch.clamp
def vector_quantize(x, codebook):
d = x.pow(2).sum(dim=-1, keepdim=True) + codebook.pow(2).sum(dim=1) - 2 * x @ codebook.T
indices = d.argmin(-1)
x_q = F.one_hot(indices, codebook.shape[0]).to(d.dtype) @ codebook
return replace_grad(x_q, x)
class Prompt(nn.Module):
def __init__(self, embed, weight=1., stop=float('-inf')):
super().__init__()
self.register_buffer('embed', embed)
self.register_buffer('weight', torch.as_tensor(weight))
self.register_buffer('stop', torch.as_tensor(stop))
def forward(self, input):
input_normed = F.normalize(input.unsqueeze(1), dim=2)#(input / input.norm(dim=-1, keepdim=True)).unsqueeze(1)#
embed_normed = F.normalize((self.embed).unsqueeze(0), dim=2)#(self.embed / self.embed.norm(dim=-1, keepdim=True)).unsqueeze(0)#
dists = input_normed.sub(embed_normed).norm(dim=2).div(2).arcsin().pow(2).mul(2)
dists = dists * self.weight.sign()
return self.weight.abs() * replace_grad(dists, torch.maximum(dists, self.stop)).mean()
def parse_prompt(prompt):
vals = prompt.rsplit(':', 2)
vals = vals + ['', '1', '-inf'][len(vals):]
return vals[0], float(vals[1]), float(vals[2])
def one_sided_clip_loss(input, target, labels=None, logit_scale=100):
input_normed = F.normalize(input, dim=-1)
target_normed = F.normalize(target, dim=-1)
logits = input_normed @ target_normed.T * logit_scale
if labels is None:
labels = torch.arange(len(input), device=logits.device)
return F.cross_entropy(logits, labels)
class MakeCutouts(nn.Module):
def __init__(self, cut_size, cutn, cut_pow=1.):
super().__init__()
self.cut_size = cut_size
self.cutn = cutn
self.cut_pow = cut_pow
self.av_pool = nn.AdaptiveAvgPool2d((self.cut_size, self.cut_size))
self.max_pool = nn.AdaptiveMaxPool2d((self.cut_size, self.cut_size))
def set_cut_pow(self, cut_pow):
self.cut_pow = cut_pow
def forward(self, input):
sideY, sideX = input.shape[2:4]
max_size = min(sideX, sideY)
min_size = min(sideX, sideY, self.cut_size)
cutouts = []
cutouts_full = []
min_size_width = min(sideX, sideY)
lower_bound = float(self.cut_size/min_size_width)
for ii in range(self.cutn):
size = int(min_size_width*torch.zeros(1,).normal_(mean=.8, std=.3).clip(lower_bound, 1.)) # replace .5 with a result for 224 the default large size is .95
offsetx = torch.randint(0, sideX - size + 1, ())
offsety = torch.randint(0, sideY - size + 1, ())
cutout = input[:, :, offsety:offsety + size, offsetx:offsetx + size]
cutouts.append(resample(cutout, (self.cut_size, self.cut_size)))
cutouts = torch.cat(cutouts, dim=0)
return clamp_with_grad(cutouts, 0, 1)
def load_vqgan_model(config_path, checkpoint_path):
config = OmegaConf.load(config_path)
if config.model.target == 'taming.models.vqgan.VQModel':
model = vqgan.VQModel(**config.model.params)
model.eval().requires_grad_(False)
model.init_from_ckpt(checkpoint_path)
elif config.model.target == 'taming.models.cond_transformer.Net2NetTransformer':
parent_model = cond_transformer.Net2NetTransformer(**config.model.params)
parent_model.eval().requires_grad_(False)
parent_model.init_from_ckpt(checkpoint_path)
model = parent_model.first_stage_model
elif config.model.target == 'taming.models.vqgan.GumbelVQ':
model = vqgan.GumbelVQ(**config.model.params)
model.eval().requires_grad_(False)
model.init_from_ckpt(checkpoint_path)
else:
raise ValueError(f'unknown model type: {config.model.target}')
del model.loss
return model
def resize_image(image, out_size):
ratio = image.size[0] / image.size[1]
area = min(image.size[0] * image.size[1], out_size[0] * out_size[1])
size = round((area * ratio)**0.5), round((area / ratio)**0.5)
return image.resize(size, Image.LANCZOS)
class GaussianBlur2d(nn.Module):
def __init__(self, sigma, window=0, mode='reflect', value=0):
super().__init__()
self.mode = mode
self.value = value
if not window:
window = max(math.ceil((sigma * 6 + 1) / 2) * 2 - 1, 3)
if sigma:
kernel = torch.exp(-(torch.arange(window) - window // 2)**2 / 2 / sigma**2)
kernel /= kernel.sum()
else:
kernel = torch.ones([1])
self.register_buffer('kernel', kernel)
def forward(self, input):
n, c, h, w = input.shape
input = input.view([n * c, 1, h, w])
start_pad = (self.kernel.shape[0] - 1) // 2
end_pad = self.kernel.shape[0] // 2
input = F.pad(input, (start_pad, end_pad, start_pad, end_pad), self.mode, self.value)
input = F.conv2d(input, self.kernel[None, None, None, :])
input = F.conv2d(input, self.kernel[None, None, :, None])
return input.view([n, c, h, w])
class EMATensor(nn.Module):
"""implmeneted by Katherine Crowson"""
def __init__(self, tensor, decay):
super().__init__()
self.tensor = nn.Parameter(tensor)
self.register_buffer('biased', torch.zeros_like(tensor))
self.register_buffer('average', torch.zeros_like(tensor))
self.decay = decay
self.register_buffer('accum', torch.tensor(1.))
self.update()
@torch.no_grad()
def update(self):
if not self.training:
raise RuntimeError('update() should only be called during training')
self.accum *= self.decay
self.biased.mul_(self.decay)
self.biased.add_((1 - self.decay) * self.tensor)
self.average.copy_(self.biased)
self.average.div_(1 - self.accum)
def forward(self):
if self.training:
return self.tensor
return self.average
import io
import base64
def image_to_data_url(img, ext):
img_byte_arr = io.BytesIO()
img.save(img_byte_arr, format=ext)
img_byte_arr = img_byte_arr.getvalue()
# ext = filename.split('.')[-1]
prefix = f'data:image/{ext};base64,'
return prefix + base64.b64encode(img_byte_arr).decode('utf-8')
def update_random( seed, purpose ):
if seed == -1:
seed = random.seed()
seed = random.randrange(1,99999)
print( f'Using seed {seed} for {purpose}')
random.seed(seed)
torch.manual_seed(seed)
np.random.seed(seed)
return seed
def clear_memory():
gc.collect()
torch.cuda.empty_cache()
#@title Setup for A100
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
if gpu.name.startswith('A100'):
torch.backends.cudnn.enabled = False
print('Finished setup for A100')
#@title Loss Module Definitions
from typing import cast, Dict, Optional
from kornia.augmentation.base import IntensityAugmentationBase2D
class FixPadding(nn.Module):
def __init__(self, module=None, threshold=1e-12, noise_frac=0.00 ):
super().__init__()
self.threshold = threshold
self.noise_frac = noise_frac
self.module = module
def forward(self,input):
dims = input.shape
if self.module is not None:
input = self.module(input + self.threshold)
light = input.new_empty(dims[0],1,1,1).uniform_(0.,2.)
mixed = input.view(*dims[:2],-1).sum(dim=1,keepdim=True)
black = mixed < self.threshold
black = black.view(-1,1,*dims[2:4]).type(torch.float)
black = kornia.filters.box_blur( black, (5,5) ).clip(0,0.1)/0.1
mean = input.view(*dims[:2],-1).sum(dim=2) / mixed.count_nonzero(dim=2)
mean = ( mean[:,:,None,None] * light ).clip(0,1)
fill = mean.expand(*dims)
if 0 < self.noise_frac:
rng = torch.get_rng_state()
fill = fill + torch.randn_like(mean) * self.noise_frac
torch.set_rng_state(rng)
if self.module is not None:
input = input - self.threshold
return torch.lerp(input,fill,black)
class MyRandomNoise(IntensityAugmentationBase2D):
def __init__(
self,
frac: float = 0.1,
return_transform: bool = False,
same_on_batch: bool = False,
p: float = 0.5,
) -> None:
super().__init__(p=p, return_transform=return_transform, same_on_batch=same_on_batch, p_batch=1.0)
self.frac = frac
def __repr__(self) -> str:
return self.__class__.__name__ + f"({super().__repr__()})"
def generate_parameters(self, shape: torch.Size) -> Dict[str, torch.Tensor]:
noise = torch.FloatTensor(1).uniform_(0,self.frac)
# generate pixel data without throwing off determinism of augs
rng = torch.get_rng_state()
noise = noise * torch.randn(shape)
torch.set_rng_state(rng)
return dict(noise=noise)
def apply_transform(
self, input: torch.Tensor, params: Dict[str, torch.Tensor], transform: Optional[torch.Tensor] = None
) -> torch.Tensor:
return input + params['noise'].to(input.device)
class MakeCutouts2(nn.Module):
def __init__(self, cut_size, cutn):
super().__init__()
self.cut_size = cut_size
self.cutn = cutn
def forward(self, input):
sideY, sideX = input.shape[2:4]
max_size = min(sideX, sideY)
min_size = min(sideX, sideY, self.cut_size)
cutouts = []
cutouts_full = []
min_size_width = min(sideX, sideY)
lower_bound = float(self.cut_size/min_size_width)
for ii in range(self.cutn):
size = int(min_size_width*torch.zeros(1,).normal_(mean=.8, std=.3).clip(lower_bound, 1.)) # replace .5 with a result for 224 the default large size is .95
offsetx = torch.randint(0, sideX - size + 1, ())
offsety = torch.randint(0, sideY - size + 1, ())
cutout = input[:, :, offsety:offsety + size, offsetx:offsetx + size]
cutouts.append(cutout)
return cutouts
class MultiClipLoss(nn.Module):
def __init__(self, clip_models, text_prompt, normalize_prompt_weights, cutn, cut_pow=1., clip_weight=1., use_old_augs=False, simulate_old_cuts=False ):
super().__init__()
self.use_old_augs = use_old_augs
self.simulate_old_cuts = simulate_old_cuts
# Load Clip
self.perceptors = []
for cm in clip_models:
c = clip.load(cm[0], jit=False)[0].eval().requires_grad_(False).to(device)
self.perceptors.append( { 'res': c.visual.input_resolution, 'perceptor': c, 'weight': cm[1], 'prompts':[] } )
self.perceptors.sort(key=lambda e: e['res'], reverse=True)
# Make Cutouts
self.cut_sizes = list(set([p['res'] for p in self.perceptors]))
self.cut_sizes.sort( reverse=True )
self.make_cuts = MakeCutouts2(self.cut_sizes[-1], cutn)
# Get Prompt Embedings
texts = [phrase.strip() for phrase in text_prompt.split("|")]
if text_prompt == ['']:
texts = []
self.pMs = []
prompts_weight_sum = 0
parsed_prompts = []
for prompt in texts:
txt, weight, stop = parse_prompt(prompt)
parsed_prompts.append( [txt,weight,stop] )
prompts_weight_sum += max( weight, 0 )
for prompt in parsed_prompts:
txt, weight, stop = prompt
clip_token = clip.tokenize(txt).to(device)
if normalize_prompt_weights and 0 < prompts_weight_sum:
weight /= prompts_weight_sum
for p in self.perceptors:
embed = p['perceptor'].encode_text(clip_token).float()
embed_normed = F.normalize(embed.unsqueeze(0), dim=2)
p['prompts'].append({'embed_normed':embed_normed,'weight':torch.as_tensor(weight, device=device),'stop':torch.as_tensor(stop, device=device)})
# Prep Augments
self.noise_fac = 0.1
self.normalize = transforms.Normalize(mean=[0.48145466, 0.4578275, 0.40821073],
std=[0.26862954, 0.26130258, 0.27577711])
self.augs = nn.Sequential(
K.RandomHorizontalFlip(p=0.5),
K.RandomSharpness(0.3,p=0.1),
FixPadding( nn.Sequential(
K.RandomAffine(degrees=30, translate=0.1, p=0.8, padding_mode='zeros'), # padding_mode=2
K.RandomPerspective(0.2,p=0.4, ),
)),
K.ColorJitter(hue=0.01, saturation=0.01, p=0.7),
K.RandomGrayscale(p=0.15),
MyRandomNoise(frac=self.noise_fac,p=1.),
)
self.clip_weight = clip_weight
def prepare_cuts(self,img):
cutouts = self.make_cuts(img)
cutouts_out = []
rng = torch.get_rng_state()
for sz in self.cut_sizes:
cuts = [resample(c, (sz,sz)) for c in cutouts]
cuts = torch.cat(cuts, dim=0)
cuts = clamp_with_grad(cuts,0,1)
torch.set_rng_state(rng)
cuts = self.augs(cuts)
cuts = self.normalize(cuts)
cutouts_out.append(cuts)
return cutouts_out
def forward( self, i, img ):
cutouts = self.prepare_cuts( img )
loss = []
current_cuts = None
currentres = 0
for p in self.perceptors:
if currentres != p['res']:
currentres = p['res']
current_cuts = cutouts[self.cut_sizes.index( currentres )]
iii = p['perceptor'].encode_image(current_cuts).float()
input_normed = F.normalize(iii.unsqueeze(1), dim=2)
for prompt in p['prompts']:
dists = input_normed.sub(prompt['embed_normed']).norm(dim=2).div(2).arcsin().pow(2).mul(2)
dists = dists * prompt['weight'].sign()
l = prompt['weight'].abs() * replace_grad(dists, torch.maximum(dists, prompt['stop'])).mean()
loss.append(l * p['weight'])
return loss
class MSEDecayLoss(nn.Module):
def __init__(self, init_weight, mse_decay_rate, mse_epoches, mse_quantize ):
super().__init__()
self.init_weight = init_weight
self.has_init_image = False
self.mse_decay = init_weight / mse_epoches if init_weight else 0
self.mse_decay_rate = mse_decay_rate
self.mse_weight = init_weight
self.mse_epoches = mse_epoches
self.mse_quantize = mse_quantize
@torch.no_grad()
def set_target( self, z_tensor, model ):
z_tensor = z_tensor.detach().clone()
if self.mse_quantize:
z_tensor = vector_quantize(z_tensor.movedim(1, 3), model.quantize.embedding.weight).movedim(3, 1)#z.average
self.z_orig = z_tensor
def forward( self, i, z ):
if self.is_active(i):
return F.mse_loss(z, self.z_orig) * self.mse_weight / 2
return 0
def is_active(self, i):
if not self.init_weight:
return False
if i <= self.mse_decay_rate and not self.has_init_image:
return False
return True
@torch.no_grad()
def step( self, i ):
if i % self.mse_decay_rate == 0 and i != 0 and i < self.mse_decay_rate * self.mse_epoches:
if self.mse_weight - self.mse_decay > 0 and self.mse_weight - self.mse_decay >= self.mse_decay:
self.mse_weight -= self.mse_decay
else:
self.mse_weight = 0
print(f"updated mse weight: {self.mse_weight}")
return True
return False
class TVLoss(nn.Module):
def forward(self, input):
input = F.pad(input, (0, 1, 0, 1), 'replicate')
x_diff = input[..., :-1, 1:] - input[..., :-1, :-1]
y_diff = input[..., 1:, :-1] - input[..., :-1, :-1]
diff = x_diff**2 + y_diff**2 + 1e-8
return diff.mean(dim=1).sqrt().mean()
#@title Random Inits
import torch
import math
def rand_perlin_2d(shape, res, fade = lambda t: 6*t**5 - 15*t**4 + 10*t**3):
delta = (res[0] / shape[0], res[1] / shape[1])
d = (shape[0] // res[0], shape[1] // res[1])
grid = torch.stack(torch.meshgrid(torch.arange(0, res[0], delta[0]), torch.arange(0, res[1], delta[1])), dim = -1) % 1
angles = 2*math.pi*torch.rand(res[0]+1, res[1]+1)
gradients = torch.stack((torch.cos(angles), torch.sin(angles)), dim = -1)
tile_grads = lambda slice1, slice2: gradients[slice1[0]:slice1[1], slice2[0]:slice2[1]].repeat_interleave(d[0], 0).repeat_interleave(d[1], 1)
dot = lambda grad, shift: (torch.stack((grid[:shape[0],:shape[1],0] + shift[0], grid[:shape[0],:shape[1], 1] + shift[1] ), dim = -1) * grad[:shape[0], :shape[1]]).sum(dim = -1)
n00 = dot(tile_grads([0, -1], [0, -1]), [0, 0])
n10 = dot(tile_grads([1, None], [0, -1]), [-1, 0])
n01 = dot(tile_grads([0, -1],[1, None]), [0, -1])
n11 = dot(tile_grads([1, None], [1, None]), [-1,-1])
t = fade(grid[:shape[0], :shape[1]])
return math.sqrt(2) * torch.lerp(torch.lerp(n00, n10, t[..., 0]), torch.lerp(n01, n11, t[..., 0]), t[..., 1])
def rand_perlin_2d_octaves( desired_shape, octaves=1, persistence=0.5):
shape = torch.tensor(desired_shape)
shape = 2 ** torch.ceil( torch.log2( shape ) )
shape = shape.type(torch.int)
max_octaves = int(min(octaves,math.log(shape[0])/math.log(2), math.log(shape[1])/math.log(2)))
res = torch.floor( shape / 2 ** max_octaves).type(torch.int)
noise = torch.zeros(list(shape))
frequency = 1
amplitude = 1
for _ in range(max_octaves):
noise += amplitude * rand_perlin_2d(shape, (frequency*res[0], frequency*res[1]))
frequency *= 2
amplitude *= persistence
return noise[:desired_shape[0],:desired_shape[1]]
def rand_perlin_rgb( desired_shape, amp=0.1, octaves=6 ):
r = rand_perlin_2d_octaves( desired_shape, octaves )
g = rand_perlin_2d_octaves( desired_shape, octaves )
b = rand_perlin_2d_octaves( desired_shape, octaves )
rgb = ( torch.stack((r,g,b)) * amp + 1 ) * 0.5
return rgb.unsqueeze(0).clip(0,1).to(device)
def pyramid_noise_gen(shape, octaves=5, decay=1.):
n, c, h, w = shape
noise = torch.zeros([n, c, 1, 1])
max_octaves = int(min(math.log(h)/math.log(2), math.log(w)/math.log(2)))
if octaves is not None and 0 < octaves:
max_octaves = min(octaves,max_octaves)
for i in reversed(range(max_octaves)):
h_cur, w_cur = h // 2**i, w // 2**i
noise = F.interpolate(noise, (h_cur, w_cur), mode='bicubic', align_corners=False)
noise += ( torch.randn([n, c, h_cur, w_cur]) / max_octaves ) * decay**( max_octaves - (i+1) )
return noise
def rand_z(model, toksX, toksY):
e_dim = model.quantize.e_dim
n_toks = model.quantize.n_e
z_min = model.quantize.embedding.weight.min(dim=0).values[None, :, None, None]
z_max = model.quantize.embedding.weight.max(dim=0).values[None, :, None, None]
one_hot = F.one_hot(torch.randint(n_toks, [toksY * toksX], device=device), n_toks).float()
z = one_hot @ model.quantize.embedding.weight
z = z.view([-1, toksY, toksX, e_dim]).permute(0, 3, 1, 2)
return z
def make_rand_init( mode, model, perlin_octaves, perlin_weight, pyramid_octaves, pyramid_decay, toksX, toksY, f ):
if mode == 'VQGAN ZRand':
return rand_z(model, toksX, toksY)
elif mode == 'Perlin Noise':
rand_init = rand_perlin_rgb((toksY * f, toksX * f), perlin_weight, perlin_octaves )
z, *_ = model.encode(rand_init * 2 - 1)
return z
elif mode == 'Pyramid Noise':
rand_init = pyramid_noise_gen( (1,3,toksY * f, toksX * f), pyramid_octaves, pyramid_decay).to(device)
rand_init = ( rand_init * 0.5 + 0.5 ).clip(0,1)
z, *_ = model.encode(rand_init * 2 - 1)
return z
|
_____no_output_____
|
MIT
|
Multi_Perceptor_VQGAN_+_CLIP_[Public].ipynb
|
keirwilliamsxyz/keirxyz
|
Make some Art!
|
#@title Set VQGAN Model Save Location
#@markdown It's a lot faster to load model files from google drive than to download them every time you want to use this notebook.
save_vqgan_models_to_drive = True #@param {type: 'boolean'}
download_all = False
vqgan_path_on_google_drive = "/content/drive/MyDrive/Art/Models/VQGAN/" #@param {type: 'string'}
vqgan_path_on_google_drive += "/" if not vqgan_path_on_google_drive.endswith('/') else ""
#@markdown Should all the images during the run be saved to google drive?
save_output_to_drive = True #@param {type:'boolean'}
output_path_on_google_drive = "/content/drive/MyDrive/Art/" #@param {type: 'string'}
output_path_on_google_drive += "/" if not output_path_on_google_drive.endswith('/') else ""
#@markdown When saving the images, how much should be included in the name?
include_full_prompt_in_filename = False #@param {type:'boolean'}
shortname_limit = 50 #@param {type: 'number'}
filename_limit = 250
if save_vqgan_models_to_drive or save_output_to_drive:
from google.colab import drive
drive.mount('/content/drive')
vqgan_model_path = "/content/"
if save_vqgan_models_to_drive:
vqgan_model_path = vqgan_path_on_google_drive
!mkdir -p "$vqgan_path_on_google_drive"
save_output_path = "/content/art/"
if save_output_to_drive:
save_output_path = output_path_on_google_drive
!mkdir -p "$save_output_path"
model_download={
"vqgan_imagenet_f16_1024":
[["vqgan_imagenet_f16_1024.yaml", "https://heibox.uni-heidelberg.de/d/8088892a516d4e3baf92/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1"],
["vqgan_imagenet_f16_1024.ckpt", "https://heibox.uni-heidelberg.de/d/8088892a516d4e3baf92/files/?p=%2Fckpts%2Flast.ckpt&dl=1"]],
"vqgan_imagenet_f16_16384":
[["vqgan_imagenet_f16_16384.yaml", "https://heibox.uni-heidelberg.de/d/a7530b09fed84f80a887/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1"],
["vqgan_imagenet_f16_16384.ckpt", "https://heibox.uni-heidelberg.de/d/a7530b09fed84f80a887/files/?p=%2Fckpts%2Flast.ckpt&dl=1"]],
"vqgan_openimages_f8_8192":
[["vqgan_openimages_f8_8192.yaml", "https://heibox.uni-heidelberg.de/d/2e5662443a6b4307b470/files/?p=%2Fconfigs%2Fmodel.yaml&dl=1"],
["vqgan_openimages_f8_8192.ckpt", "https://heibox.uni-heidelberg.de/d/2e5662443a6b4307b470/files/?p=%2Fckpts%2Flast.ckpt&dl=1"]],
"coco":
[["coco_first_stage.yaml", "http://batbot.tv/ai/models/vqgan/coco_first_stage.yaml"],
["coco_first_stage.ckpt", "http://batbot.tv/ai/models/vqgan/coco_first_stage.ckpt"]],
"faceshq":
[["faceshq.yaml", "https://drive.google.com/uc?export=download&id=1fHwGx_hnBtC8nsq7hesJvs-Klv-P0gzT"],
["faceshq.ckpt", "https://app.koofr.net/content/links/a04deec9-0c59-4673-8b37-3d696fe63a5d/files/get/last.ckpt?path=%2F2020-11-13T21-41-45_faceshq_transformer%2Fcheckpoints%2Flast.ckpt"]],
"wikiart_1024":
[["wikiart_1024.yaml", "http://batbot.tv/ai/models/vqgan/WikiArt_augmented_Steps_7mil_finetuned_1mil.yaml"],
["wikiart_1024.ckpt", "http://batbot.tv/ai/models/vqgan/WikiArt_augmented_Steps_7mil_finetuned_1mil.ckpt"]],
"wikiart_16384":
[["wikiart_16384.yaml", "http://eaidata.bmk.sh/data/Wikiart_16384/wikiart_f16_16384_8145600.yaml"],
["wikiart_16384.ckpt", "http://eaidata.bmk.sh/data/Wikiart_16384/wikiart_f16_16384_8145600.ckpt"]],
"sflckr":
[["sflckr.yaml", "https://heibox.uni-heidelberg.de/d/73487ab6e5314cb5adba/files/?p=%2Fconfigs%2F2020-11-09T13-31-51-project.yaml&dl=1"],
["sflckr.ckpt", "https://heibox.uni-heidelberg.de/d/73487ab6e5314cb5adba/files/?p=%2Fcheckpoints%2Flast.ckpt&dl=1"]],
}
loaded_model = None
loaded_model_name = None
def dl_vqgan_model(image_model):
for curl_opt in model_download[image_model]:
modelpath = f'{vqgan_model_path}{curl_opt[0]}'
if not path.exists(modelpath):
print(f'downloading {curl_opt[0]} to {modelpath}')
!curl -L -o {modelpath} '{curl_opt[1]}'
else:
print(f'found existing {curl_opt[0]}')
def get_vqgan_model(image_model):
global loaded_model
global loaded_model_name
if loaded_model is None or loaded_model_name != image_model:
dl_vqgan_model(image_model)
print(f'loading {image_model} vqgan checkpoint')
vqgan_config= vqgan_model_path + model_download[image_model][0][0]
vqgan_checkpoint= vqgan_model_path + model_download[image_model][1][0]
print('vqgan_config',vqgan_config)
print('vqgan_checkpoint',vqgan_checkpoint)
model = load_vqgan_model(vqgan_config, vqgan_checkpoint).to(device)
if image_model == 'vqgan_openimages_f8_8192':
model.quantize.e_dim = 256
model.quantize.n_e = model.quantize.n_embed
model.quantize.embedding = model.quantize.embed
loaded_model = model
loaded_model_name = image_model
return loaded_model
def slugify(value):
value = str(value)
value = re.sub(r':([-\d.]+)', ' [\\1]', value)
value = re.sub(r'[|]','; ',value)
value = re.sub(r'[<>:"/\\|?*]', ' ', value)
return value
def get_filename(text, seed, i, ext):
if ( not include_full_prompt_in_filename ):
text = re.split(r'[|:;]',text, 1)[0][:shortname_limit]
text = slugify(text)
now = datetime.now()
t = now.strftime("%y%m%d%H%M")
if i is not None:
data = f'; r{seed} i{i} {t}{ext}'
else:
data = f'; r{seed} {t}{ext}'
return text[:filename_limit-len(data)] + data
def save_output(pil, text, seed, i):
fname = get_filename(text,seed,i,'.png')
pil.save(save_output_path + fname)
if save_vqgan_models_to_drive and download_all:
for model in model_download.keys():
dl_vqgan_model(model)
#@title Set Display Rate
#@markdown If `use_automatic_display_schedule` is enabled, the image will be output frequently at first, and then more spread out as time goes on. Turn this off if you want to specify the display rate yourself.
use_automatic_display_schedule = False #@param {type:'boolean'}
display_every = 5 #@param {type:'number'}
def should_checkin(i):
if i == max_iter:
return True
if not use_automatic_display_schedule:
return i % display_every == 0
schedule = [[100,25],[500,50],[1000,100],[2000,200]]
for s in schedule:
if i <= s[0]:
return i % s[1] == 0
return i % 500 == 0
|
_____no_output_____
|
MIT
|
Multi_Perceptor_VQGAN_+_CLIP_[Public].ipynb
|
keirwilliamsxyz/keirxyz
|
Before generating, the rest of the setup steps must first be executed by pressing **`Runtime > Run All`**. This only needs to be done once.
|
#@title Do the Run
#@markdown What do you want to see?
text_prompt = 'made of buildings:200 | Ethiopian flags:40 | pollution:30 | 4k:20 | Unreal engine:20 | V-ray:20 | Cryengine:20 | Ray tracing:20 | Photorealistic:20 | Hyper-realistic:20'#@param {type:'string'}
gen_seed = -1#@param {type:'number'}
#@markdown - If you want to keep starting from the same point, set `gen_seed` to a positive number. `-1` will make it random every time.
init_image = '/content/d.png'#@param {type:'string'}
width = 300#@param {type:'number'}
height = 300#@param {type:'number'}
max_iter = 2000#@param {type:'number'}
#@markdown There are different ways of generating the random starting point, when not using an init image. These influence how the image turns out. The default VQGAN ZRand is good, but some models and subjects may do better with perlin or pyramid noise.
rand_init_mode = 'VQGAN ZRand'#@param [ "VQGAN ZRand", "Perlin Noise", "Pyramid Noise"]
perlin_octaves = 7#@param {type:"slider", min:1, max:8, step:1}
perlin_weight = 0.22#@param {type:"slider", min:0, max:1, step:0.01}
pyramid_octaves = 5#@param {type:"slider", min:1, max:8, step:1}
pyramid_decay = 0.99#@param {type:"slider", min:0, max:1, step:0.01}
ema_val = 0.99
#@markdown How many slices of the image should be sent to CLIP each iteration to score? Higher numbers are better, but cost more memory. If you are running into memory issues try lowering this value.
cut_n = 64 #@param {type:'number'}
#@markdown One clip model is good. Two is better? You may need to reduce the number of cuts to support having more than one CLIP model. CLIP is what scores the image against your prompt and each model has slightly different ideas of what things are.
#@markdown - `ViT-B/32` is fast and good and what most people use to begin with
clip_model = 'ViT-B/32' #@param ["ViT-B/16", "ViT-B/32", "RN50x16", "RN50x4"]
clip_model2 ='ViT-B/16' #@param ["None","ViT-B/16", "ViT-B/32", "RN50x16", "RN50x4"]
if clip_model2 == "None":
clip_model2 = None
clip1_weight = 0.5 #@param {type:"slider", min:0, max:1, step:0.01}
#@markdown Picking a different VQGAN model will impact how an image generates. Think of this as giving the generator a different set of brushes and paints to work with. CLIP is still the "eyes" and is judging the image against your prompt but using different brushes will make a different image.
#@markdown - `vqgan_imagenet_f16_16384` is the default and what most people use
vqgan_model = 'vqgan_imagenet_f16_16384'#@param [ "vqgan_imagenet_f16_1024", "vqgan_imagenet_f16_16384", "vqgan_openimages_f8_8192", "coco", "faceshq","wikiart_1024", "wikiart_16384", "sflckr"]
#@markdown Learning rates greatly impact how quickly an image can generate, or if an image can generate at all. The first learning rate is only for the first 50 iterations. The epoch rate is what is used after reaching the first mse epoch.
#@markdown You can try lowering the epoch rate while raising the initial learning rate and see what happens
learning_rate = 0.9#@param {type:'number'}
learning_rate_epoch = 0.2#@param {type:'number'}
#@markdown How much should we try to match the init image, or if no init image how much should we resist change after reaching the first epoch?
mse_weight = 1.8#@param {type:'number'}
#@markdown Adding some TV may make the image blurrier but also helps to get rid of noise. A good value to try might be 0.1.
tv_weight = 0.0 #@param {type:'number'}
#@markdown Should the total weight of the text prompts stay in the same range, relative to other loss functions?
normalize_prompt_weights = True #@param {type:'boolean'}
#@markdown Enabling the EMA tensor will cause the image to be slower to generate but may help it be more cohesive.
#@markdown This can also help keep the final image closer to the init image, if you are providing one.
use_ema_tensor = False #@param {type:'boolean'}
#@markdown If you want to generate a video of the run, you need to save the frames as you go. The more frequently you save, the longer the video but the slower it will take to generate.
save_art_output = True #@param {type:'boolean'}
save_frames_for_video = False #@param {type:'boolean'}
save_frequency_for_video = 3 #@param {type:'number'}
#@markdown ----
#@markdown I'd love to see what you can make with my notebook. Tweet me your art [@remi_durant](https://twitter.com/remi_durant)!
output_as_png = True
print('Using device:', device)
print('using prompts: ', text_prompt)
clear_memory()
!rm -r steps
!mkdir -p steps
model = get_vqgan_model( vqgan_model )
if clip_model2:
clip_models = [[clip_model, clip1_weight], [clip_model2, 1. - clip1_weight]]
else:
clip_models = [[clip_model, 1.0]]
print(clip_models)
clip_loss = MultiClipLoss( clip_models, text_prompt, normalize_prompt_weights=normalize_prompt_weights, cutn=cut_n)
seed = update_random( gen_seed, 'image generation')
# Make Z Init
z = 0
f = 2**(model.decoder.num_resolutions - 1)
toksX, toksY = math.ceil( width / f), math.ceil( height / f)
print(f'Outputing size: [{toksX*f}x{toksY*f}]')
has_init_image = (init_image != "")
if has_init_image:
if 'http' in init_image:
req = Request(init_image, headers={'User-Agent': 'Mozilla/5.0'})
img = Image.open(urlopen(req))
else:
img = Image.open(init_image)
pil_image = img.convert('RGB')
pil_image = pil_image.resize((toksX * f, toksY * f), Image.LANCZOS)
pil_image = TF.to_tensor(pil_image)
#if args.use_noise:
# pil_image = pil_image + args.use_noise * torch.randn_like(pil_image)
z, *_ = model.encode(pil_image.to(device).unsqueeze(0) * 2 - 1)
del pil_image
del img
else:
z = make_rand_init( rand_init_mode, model, perlin_octaves, perlin_weight, pyramid_octaves, pyramid_decay, toksX, toksY, f )
z = EMATensor(z, ema_val)
opt = optim.Adam( z.parameters(), lr=learning_rate, weight_decay=0.00000000)
mse_loss = MSEDecayLoss( mse_weight, mse_decay_rate=50, mse_epoches=5, mse_quantize=True )
mse_loss.set_target( z.tensor, model )
mse_loss.has_init_image = has_init_image
tv_loss = TVLoss()
losses = []
mb = master_bar(range(1))
gnames = ['losses']
mb.names=gnames
mb.graph_fig, axs = plt.subplots(1, 1) # For custom display
mb.graph_ax = axs
mb.graph_out = display.display(mb.graph_fig, display_id=True)
## optimizer loop
def synth(z, quantize=True, scramble=True):
z_q = 0
if quantize:
z_q = vector_quantize(z.movedim(1, 3), model.quantize.embedding.weight).movedim(3, 1)
else:
z_q = z.model
out = clamp_with_grad(model.decode(z_q).add(1).div(2), 0, 1)
return out
@torch.no_grad()
def checkin(i, z, out_pil, losses):
losses_str = ', '.join(f'{loss.item():g}' for loss in losses)
tqdm.write(f'i: {i}, loss: {sum(losses).item():g}, losses: {losses_str}')
display_format='png' if output_as_png else 'jpg'
pil_data = image_to_data_url(out_pil, display_format)
display.display(display.HTML(f'<img src="{pil_data}" />'))
def should_save_for_video(i):
return save_frames_for_video and i % save_frequency_for_video
def train(i):
global opt
global z
opt.zero_grad( set_to_none = True )
out = checkpoint( synth, z.tensor )
lossAll = []
lossAll += clip_loss( i,out )
if 0 < mse_weight:
msel = mse_loss(i,z.tensor)
if 0 < msel:
lossAll.append(msel)
if 0 < tv_weight:
lossAll.append(tv_loss(out)*tv_weight)
loss = sum(lossAll)
loss.backward()
if should_checkin(i) or should_save_for_video(i):
with torch.no_grad():
if use_ema_tensor:
out = synth( z.average )
pil = TF.to_pil_image(out[0].cpu())
if should_checkin(i):
checkin(i, z, pil, lossAll)
if save_art_output:
save_output(pil, text_prompt, seed, i)
if should_save_for_video(i):
pil.save(f'steps/step{i//save_frequency_for_video:04}.png')
# update graph
losses.append(loss)
x = range(len(losses))
mb.update_graph( [[x,losses]] )
opt.step()
if use_ema_tensor:
z.update()
i = 0
try:
with tqdm() as pbar:
while True and i <= max_iter:
if i % 200 == 0:
clear_memory()
train(i)
with torch.no_grad():
if mse_loss.step(i):
print('Reseting optimizer at mse epoch')
if mse_loss.has_init_image and use_ema_tensor:
mse_loss.set_target(z.average,model)
else:
mse_loss.set_target(z.tensor,model)
# Make sure not to spike loss when mse_loss turns on
if not mse_loss.is_active(i):
z.tensor = nn.Parameter(mse_loss.z_orig.clone())
z.tensor.requires_grad = True
if use_ema_tensor:
z = EMATensor(z.average, ema_val)
else:
z = EMATensor(z.tensor, ema_val)
opt = optim.Adam(z.parameters(), lr=learning_rate_epoch, weight_decay=0.00000000)
i += 1
pbar.update()
except KeyboardInterrupt:
pass
#@title Make a Video of Your Last Run!
#@markdown If you want to make a video, you must first enable `save_frames_for_video` during the run. Setting a higher frequency will make a longer video, and a higher framerate will make a shorter video.
fps = 24 #@param{type:'number'}
!mkdir -p "/content/video/"
vname = "/content/video/"+get_filename(text_prompt,seed,None,'.mp4')
!ffmpeg -y -v 1 -framerate $fps -i steps/step%04d.png -r $fps -vcodec libx264 -crf 32 -pix_fmt yuv420p "$vname"
if save_output_to_drive:
!cp "$vname" "$output_path_on_google_drive"
mp4 = open(vname,'rb').read()
data_url = "data:video/mp4;base64," + b64encode(mp4).decode()
display.display( display.HTML(f'<video controls><source src="{data_url}" type="video/mp4"></video>') )
|
_____no_output_____
|
MIT
|
Multi_Perceptor_VQGAN_+_CLIP_[Public].ipynb
|
keirwilliamsxyz/keirxyz
|
Vertex Pipelines: Lightweight Python function-based components, and component I/O Run in Colab View on GitHub Open in Google Cloud Notebooks OverviewThis notebooks shows how to use [the Kubeflow Pipelines (KFP) SDK](https://www.kubeflow.org/docs/components/pipelines/) to build [Vertex Pipelines](https://cloud.google.com/vertex-ai/docs/pipelines) that use lightweight Python function based components, as well as supporting component I/O using the KFP SDK. ObjectiveIn this tutorial, you use the KFP SDK to build lightweight Python function-based components.The steps performed include:- Build Python function-based components.- Pass *Artifacts* and *parameters* between components, both by path reference and by value.- Use the `kfp.dsl.importer` method. KFP Python function-based componentsA Kubeflow pipeline component is a self-contained set of code that performs one step in your ML workflow. A pipeline component is composed of:* The component code, which implements the logic needed to perform a step in your ML workflow.* A component specification, which defines the following: * The component’s metadata, its name and description. * The component’s interface, the component’s inputs and outputs.* The component’s implementation, the Docker container image to run, how to pass inputs to your component code, and how to get the component’s outputs.Lightweight Python function-based components make it easier to iterate quickly by letting you build your component code as a Python function and generating the component specification for you. This notebook shows how to create Python function-based components for use in [Vertex Pipelines](https://cloud.google.com/vertex-ai/docs/pipelines).Python function-based components use the Kubeflow Pipelines SDK to handle the complexity of passing inputs into your component and passing your function’s outputs back to your pipeline.There are two categories of inputs/outputs supported in Python function-based components: *artifacts* and *parameters*.* Parameters are passed to your component by value and typically contain `int`, `float`, `bool`, or small `string` values.* Artifacts are passed to your component as a *reference* to a path, to which you can write a file or a subdirectory structure. In addition to the artifact’s data, you can also read and write the artifact’s metadata. This lets you record arbitrary key-value pairs for an artifact such as the accuracy of a trained model, and use metadata in downstream components – for example, you could use metadata to decide if a model is accurate enough to deploy for predictions. CostsThis tutorial uses billable components of Google Cloud:* Vertex AI* Cloud StorageLearn about [Vertex AIpricing](https://cloud.google.com/vertex-ai/pricing) and [Cloud Storagepricing](https://cloud.google.com/storage/pricing), and use the [PricingCalculator](https://cloud.google.com/products/calculator/)to generate a cost estimate based on your projected usage. Set up your local development environmentIf you are using Colab or Google Cloud Notebook, your environment already meets all the requirements to run this notebook. You can skip this step.Otherwise, make sure your environment meets this notebook's requirements. You need the following:- The Cloud Storage SDK- Git- Python 3- virtualenv- Jupyter notebook running in a virtual environment with Python 3The Cloud Storage guide to [Setting up a Python development environment](https://cloud.google.com/python/setup) and the [Jupyter installation guide](https://jupyter.org/install) provide detailed instructions for meeting these requirements. The following steps provide a condensed set of instructions:1. [Install and initialize the SDK](https://cloud.google.com/sdk/docs/).2. [Install Python 3](https://cloud.google.com/python/setupinstalling_python).3. [Install virtualenv](Ihttps://cloud.google.com/python/setupinstalling_and_using_virtualenv) and create a virtual environment that uses Python 3.4. Activate that environment and run `pip3 install Jupyter` in a terminal shell to install Jupyter.5. Run `jupyter notebook` on the command line in a terminal shell to launch Jupyter.6. Open this notebook in the Jupyter Notebook Dashboard. InstallationInstall the latest version of Vertex SDK for Python.
|
import os
# Google Cloud Notebook
if os.path.exists("/opt/deeplearning/metadata/env_version"):
USER_FLAG = "--user"
else:
USER_FLAG = ""
! pip3 install --upgrade google-cloud-aiplatform $USER_FLAG
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Install the latest GA version of *google-cloud-storage* library as well.
|
! pip3 install -U google-cloud-storage $USER_FLAG
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Install the latest GA version of *google-cloud-pipeline-components* library as well.
|
! pip3 install $USER kfp google-cloud-pipeline-components --upgrade
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Restart the kernelOnce you've installed the additional packages, you need to restart the notebook kernel so it can find the packages.
|
import os
if not os.getenv("IS_TESTING"):
# Automatically restart kernel after installs
import IPython
app = IPython.Application.instance()
app.kernel.do_shutdown(True)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Check the versions of the packages you installed. The KFP SDK version should be >=1.6.
|
! python3 -c "import kfp; print('KFP SDK version: {}'.format(kfp.__version__))"
! python3 -c "import google_cloud_pipeline_components; print('google_cloud_pipeline_components version: {}'.format(google_cloud_pipeline_components.__version__))"
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Before you begin GPU runtimeThis tutorial does not require a GPU runtime. Set up your Google Cloud project**The following steps are required, regardless of your notebook environment.**1. [Select or create a Google Cloud project](https://console.cloud.google.com/cloud-resource-manager). When you first create an account, you get a $300 free credit towards your compute/storage costs.2. [Make sure that billing is enabled for your project.](https://cloud.google.com/billing/docs/how-to/modify-project)3. [Enable the Vertex AI APIs, Compute Engine APIs, and Cloud Storage.](https://console.cloud.google.com/flows/enableapi?apiid=ml.googleapis.com,compute_component,storage-component.googleapis.com)4. [The Google Cloud SDK](https://cloud.google.com/sdk) is already installed in Google Cloud Notebook.5. Enter your project ID in the cell below. Then run the cell to make sure theCloud SDK uses the right project for all the commands in this notebook.**Note**: Jupyter runs lines prefixed with `!` as shell commands, and it interpolates Python variables prefixed with `$`.
|
PROJECT_ID = "[your-project-id]" # @param {type:"string"}
if PROJECT_ID == "" or PROJECT_ID is None or PROJECT_ID == "[your-project-id]":
# Get your GCP project id from gcloud
shell_output = ! gcloud config list --format 'value(core.project)' 2>/dev/null
PROJECT_ID = shell_output[0]
print("Project ID:", PROJECT_ID)
! gcloud config set project $PROJECT_ID
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
RegionYou can also change the `REGION` variable, which is used for operationsthroughout the rest of this notebook. Below are regions supported for Vertex AI. We recommend that you choose the region closest to you.- Americas: `us-central1`- Europe: `europe-west4`- Asia Pacific: `asia-east1`You may not use a multi-regional bucket for training with Vertex AI. Not all regions provide support for all Vertex AI services.Learn more about [Vertex AI regions](https://cloud.google.com/vertex-ai/docs/general/locations)
|
REGION = "us-central1" # @param {type: "string"}
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
TimestampIf you are in a live tutorial session, you might be using a shared test account or project. To avoid name collisions between users on resources created, you create a timestamp for each instance session, and append the timestamp onto the name of resources you create in this tutorial.
|
from datetime import datetime
TIMESTAMP = datetime.now().strftime("%Y%m%d%H%M%S")
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Authenticate your Google Cloud account**If you are using Google Cloud Notebook**, your environment is already authenticated. Skip this step.**If you are using Colab**, run the cell below and follow the instructions when prompted to authenticate your account via oAuth.**Otherwise**, follow these steps:In the Cloud Console, go to the [Create service account key](https://console.cloud.google.com/apis/credentials/serviceaccountkey) page.**Click Create service account**.In the **Service account name** field, enter a name, and click **Create**.In the **Grant this service account access to project** section, click the Role drop-down list. Type "Vertex" into the filter box, and select **Vertex Administrator**. Type "Storage Object Admin" into the filter box, and select **Storage Object Admin**.Click Create. A JSON file that contains your key downloads to your local environment.Enter the path to your service account key as the GOOGLE_APPLICATION_CREDENTIALS variable in the cell below and run the cell.
|
# If you are running this notebook in Colab, run this cell and follow the
# instructions to authenticate your GCP account. This provides access to your
# Cloud Storage bucket and lets you submit training jobs and prediction
# requests.
import os
import sys
# If on Google Cloud Notebook, then don't execute this code
if not os.path.exists("/opt/deeplearning/metadata/env_version"):
if "google.colab" in sys.modules:
from google.colab import auth as google_auth
google_auth.authenticate_user()
# If you are running this notebook locally, replace the string below with the
# path to your service account key and run this cell to authenticate your GCP
# account.
elif not os.getenv("IS_TESTING"):
%env GOOGLE_APPLICATION_CREDENTIALS ''
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Create a Cloud Storage bucket**The following steps are required, regardless of your notebook environment.**When you initialize the Vertex SDK for Python, you specify a Cloud Storage staging bucket. The staging bucket is where all the data associated with your dataset and model resources are retained across sessions.Set the name of your Cloud Storage bucket below. Bucket names must be globally unique across all Google Cloud projects, including those outside of your organization.
|
BUCKET_NAME = "gs://[your-bucket-name]" # @param {type:"string"}
if BUCKET_NAME == "" or BUCKET_NAME is None or BUCKET_NAME == "gs://[your-bucket-name]":
BUCKET_NAME = "gs://" + PROJECT_ID + "aip-" + TIMESTAMP
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
**Only if your bucket doesn't already exist**: Run the following cell to create your Cloud Storage bucket.
|
! gsutil mb -l $REGION $BUCKET_NAME
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Finally, validate access to your Cloud Storage bucket by examining its contents:
|
! gsutil ls -al $BUCKET_NAME
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Service Account**If you don't know your service account**, try to get your service account using `gcloud` command by executing the second cell below.
|
SERVICE_ACCOUNT = "[your-service-account]" # @param {type:"string"}
if (
SERVICE_ACCOUNT == ""
or SERVICE_ACCOUNT is None
or SERVICE_ACCOUNT == "[your-service-account]"
):
# Get your GCP project id from gcloud
shell_output = !gcloud auth list 2>/dev/null
SERVICE_ACCOUNT = shell_output[2].strip()
print("Service Account:", SERVICE_ACCOUNT)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Set service account access for Vertex PipelinesRun the following commands to grant your service account access to read and write pipeline artifacts in the bucket that you created in the previous step -- you only need to run these once per service account.
|
! gsutil iam ch serviceAccount:{SERVICE_ACCOUNT}:roles/storage.objectCreator $BUCKET_NAME
! gsutil iam ch serviceAccount:{SERVICE_ACCOUNT}:roles/storage.objectViewer $BUCKET_NAME
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Set up variablesNext, set up some variables used throughout the tutorial. Import libraries and define constants
|
import google.cloud.aiplatform as aip
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Vertex Pipelines constantsSetup up the following constants for Vertex Pipelines:
|
PIPELINE_ROOT = "{}/pipeline_root/shakespeare".format(BUCKET_NAME)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Additional imports.
|
from typing import NamedTuple
import kfp
from kfp.v2 import dsl
from kfp.v2.dsl import (Artifact, Dataset, Input, InputPath, Model, Output,
OutputPath, component)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Initialize Vertex SDK for PythonInitialize the Vertex SDK for Python for your project and corresponding bucket.
|
aip.init(project=PROJECT_ID, staging_bucket=BUCKET_NAME)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Define Python function-based pipeline componentsIn this tutorial, you define function-based components that consume parameters and produce (typed) Artifacts and parameters. Functions can produce Artifacts in three ways:* Accept an output local path using `OutputPath`* Accept an `OutputArtifact` which gives the function a handle to the output artifact's metadata* Return an `Artifact` (or `Dataset`, `Model`, `Metrics`, etc) in a `NamedTuple`These options for producing Artifacts are demonstrated. Define preprocess componentThe first component definition, `preprocess`, shows a component that outputs two `Dataset` Artifacts, as well as an output parameter. (For this example, the datasets don't reflect real data).For the parameter output, you would typically use the approach shown here, using the `OutputPath` type, for "larger" data.For "small data", like a short string, it might be more convenient to use the `NamedTuple` function output as shown in the second component instead.
|
@component
def preprocess(
# An input parameter of type string.
message: str,
# Use Output to get a metadata-rich handle to the output artifact
# of type `Dataset`.
output_dataset_one: Output[Dataset],
# A locally accessible filepath for another output artifact of type
# `Dataset`.
output_dataset_two_path: OutputPath("Dataset"),
# A locally accessible filepath for an output parameter of type string.
output_parameter_path: OutputPath(str),
):
"""'Mock' preprocessing step.
Writes out the passed in message to the output "Dataset"s and the output message.
"""
output_dataset_one.metadata["hello"] = "there"
# Use OutputArtifact.path to access a local file path for writing.
# One can also use OutputArtifact.uri to access the actual URI file path.
with open(output_dataset_one.path, "w") as f:
f.write(message)
# OutputPath is used to just pass the local file path of the output artifact
# to the function.
with open(output_dataset_two_path, "w") as f:
f.write(message)
with open(output_parameter_path, "w") as f:
f.write(message)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Define train componentThe second component definition, `train`, defines as input both an `InputPath` of type `Dataset`, and an `InputArtifact` of type `Dataset` (as well as other parameter inputs). It uses the `NamedTuple` format for function output. As shown, these outputs can be Artifacts as well as parameters.Additionally, this component writes some metrics metadata to the `model` output Artifact. This information is displayed in the Cloud Console user interface when the pipeline runs.
|
@component(
base_image="python:3.9", # Use a different base image.
)
def train(
# An input parameter of type string.
message: str,
# Use InputPath to get a locally accessible path for the input artifact
# of type `Dataset`.
dataset_one_path: InputPath("Dataset"),
# Use InputArtifact to get a metadata-rich handle to the input artifact
# of type `Dataset`.
dataset_two: Input[Dataset],
# Output artifact of type Model.
imported_dataset: Input[Dataset],
model: Output[Model],
# An input parameter of type int with a default value.
num_steps: int = 3,
# Use NamedTuple to return either artifacts or parameters.
# When returning artifacts like this, return the contents of
# the artifact. The assumption here is that this return value
# fits in memory.
) -> NamedTuple(
"Outputs",
[
("output_message", str), # Return parameter.
("generic_artifact", Artifact), # Return generic Artifact.
],
):
"""'Mock' Training step.
Combines the contents of dataset_one and dataset_two into the
output Model.
Constructs a new output_message consisting of message repeated num_steps times.
"""
# Directly access the passed in GCS URI as a local file (uses GCSFuse).
with open(dataset_one_path, "r") as input_file:
dataset_one_contents = input_file.read()
# dataset_two is an Artifact handle. Use dataset_two.path to get a
# local file path (uses GCSFuse).
# Alternately, use dataset_two.uri to access the GCS URI directly.
with open(dataset_two.path, "r") as input_file:
dataset_two_contents = input_file.read()
with open(model.path, "w") as f:
f.write("My Model")
with open(imported_dataset.path, "r") as f:
data = f.read()
print("Imported Dataset:", data)
# Use model.get() to get a Model artifact, which has a .metadata dictionary
# to store arbitrary metadata for the output artifact. This metadata will be
# recorded in Managed Metadata and can be queried later. It will also show up
# in the UI.
model.metadata["accuracy"] = 0.9
model.metadata["framework"] = "Tensorflow"
model.metadata["time_to_train_in_seconds"] = 257
artifact_contents = "{}\n{}".format(dataset_one_contents, dataset_two_contents)
output_message = " ".join([message for _ in range(num_steps)])
return (output_message, artifact_contents)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Define read_artifact_input componentFinally, you define a small component that takes as input the `generic_artifact` returned by the `train` component function, and reads and prints the Artifact's contents.
|
@component
def read_artifact_input(
generic: Input[Artifact],
):
with open(generic.path, "r") as input_file:
generic_contents = input_file.read()
print(f"generic contents: {generic_contents}")
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Define a pipeline that uses your components and the ImporterNext, define a pipeline that uses the components that were built in the previous section, and also shows the use of the `kfp.dsl.importer`.This example uses the `importer` to create, in this case, a `Dataset` artifact from an existing URI.Note that the `train_task` step takes as inputs three of the outputs of the `preprocess_task` step, as well as the output of the `importer` step.In the "train" inputs we refer to the `preprocess` `output_parameter`, which gives us the output string directly.The `read_task` step takes as input the `train_task` `generic_artifact` output.
|
@dsl.pipeline(
# Default pipeline root. You can override it when submitting the pipeline.
pipeline_root=PIPELINE_ROOT,
# A name for the pipeline. Use to determine the pipeline Context.
name="metadata-pipeline-v2",
)
def pipeline(message: str):
importer = kfp.dsl.importer(
artifact_uri="gs://ml-pipeline-playground/shakespeare1.txt",
artifact_class=Dataset,
reimport=False,
)
preprocess_task = preprocess(message=message)
train_task = train(
dataset_one_path=preprocess_task.outputs["output_dataset_one"],
dataset_two=preprocess_task.outputs["output_dataset_two_path"],
imported_dataset=importer.output,
message=preprocess_task.outputs["output_parameter_path"],
num_steps=5,
)
read_task = read_artifact_input( # noqa: F841
train_task.outputs["generic_artifact"]
)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Compile the pipelineNext, compile the pipeline.
|
from kfp.v2 import compiler # noqa: F811
compiler.Compiler().compile(
pipeline_func=pipeline, package_path="lightweight_pipeline.json".replace(" ", "_")
)
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Run the pipelineNext, run the pipeline.
|
DISPLAY_NAME = "shakespeare_" + TIMESTAMP
job = aip.PipelineJob(
display_name=DISPLAY_NAME,
template_path="lightweight_pipeline.json".replace(" ", "_"),
pipeline_root=PIPELINE_ROOT,
parameter_values={"message": "Hello, World"},
)
job.run()
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
Click on the generated link to see your run in the Cloud Console.<!-- It should look something like this as it is running: -->In the UI, many of the pipeline DAG nodes will expand or collapse when you click on them. Here is a partially-expanded view of the DAG (click image to see larger version). Cleaning upTo clean up all Google Cloud resources used in this project, you can [delete the Google Cloudproject](https://cloud.google.com/resource-manager/docs/creating-managing-projectsshutting_down_projects) you used for the tutorial.Otherwise, you can delete the individual resources you created in this tutorial -- *Note:* this is auto-generated and not all resources may be applicable for this tutorial:- Dataset- Pipeline- Model- Endpoint- Batch Job- Custom Job- Hyperparameter Tuning Job- Cloud Storage Bucket
|
delete_dataset = True
delete_pipeline = True
delete_model = True
delete_endpoint = True
delete_batchjob = True
delete_customjob = True
delete_hptjob = True
delete_bucket = True
try:
if delete_model and "DISPLAY_NAME" in globals():
models = aip.Model.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
model = models[0]
aip.Model.delete(model)
print("Deleted model:", model)
except Exception as e:
print(e)
try:
if delete_endpoint and "DISPLAY_NAME" in globals():
endpoints = aip.Endpoint.list(
filter=f"display_name={DISPLAY_NAME}_endpoint", order_by="create_time"
)
endpoint = endpoints[0]
endpoint.undeploy_all()
aip.Endpoint.delete(endpoint.resource_name)
print("Deleted endpoint:", endpoint)
except Exception as e:
print(e)
if delete_dataset and "DISPLAY_NAME" in globals():
if "text" == "tabular":
try:
datasets = aip.TabularDataset.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
dataset = datasets[0]
aip.TabularDataset.delete(dataset.resource_name)
print("Deleted dataset:", dataset)
except Exception as e:
print(e)
if "text" == "image":
try:
datasets = aip.ImageDataset.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
dataset = datasets[0]
aip.ImageDataset.delete(dataset.resource_name)
print("Deleted dataset:", dataset)
except Exception as e:
print(e)
if "text" == "text":
try:
datasets = aip.TextDataset.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
dataset = datasets[0]
aip.TextDataset.delete(dataset.resource_name)
print("Deleted dataset:", dataset)
except Exception as e:
print(e)
if "text" == "video":
try:
datasets = aip.VideoDataset.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
dataset = datasets[0]
aip.VideoDataset.delete(dataset.resource_name)
print("Deleted dataset:", dataset)
except Exception as e:
print(e)
try:
if delete_pipeline and "DISPLAY_NAME" in globals():
pipelines = aip.PipelineJob.list(
filter=f"display_name={DISPLAY_NAME}", order_by="create_time"
)
pipeline = pipelines[0]
aip.PipelineJob.delete(pipeline.resource_name)
print("Deleted pipeline:", pipeline)
except Exception as e:
print(e)
if delete_bucket and "BUCKET_NAME" in globals():
! gsutil rm -r $BUCKET_NAME
|
_____no_output_____
|
Apache-2.0
|
notebooks/official/pipelines/lightweight_functions_component_io_kfp.ipynb
|
diemtvu/vertex-ai-samples
|
WikiPathways and py4cytoscape Yihang Xin and Alex Pico 2020-11-10 WikiPathways is a well-known repository for biological pathways that provides unique tools to the research community for content creation, editing and utilization [@Pico2008].Python is an interpreted, high-level and general-purpose programming language.py4cytoscape leverages the WikiPathways API to communicate between Python and WikiPathways, allowing any pathway to be queried, interrogated and downloaded in both data and image formats. Queries are typically performed based on “Xrefs”, standardized identifiers for genes, proteins and metabolites. Once you can identified a pathway, you can use the WPID (WikiPathways identifier) to make additional queries.py4cytoscape leverages the CyREST API to provide a number of functions related to network visualization and analysis. InstallationThe following chunk of code installs the `py4cytoscape` module.
|
%%capture
!python3 -m pip install python-igraph requests pandas networkx
!python3 -m pip install py4cytoscape
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Prerequisites In addition to this package (py4cytoscape latest version 0.0.7), you will need:* Latest version of Cytoscape, which can be downloaded from https://cytoscape.org/download.html. Simply follow the installation instructions on screen.* Complete installation wizard* Launch CytoscapeFor this vignette, you’ll also need the WikiPathways app to access the WikiPathways database from within Cytoscape. Install the WikiPathways app from http://apps.cytoscape.org/apps/wikipathwaysInstall the filetransfer app from https://apps.cytoscape.org/apps/filetransferYou can also install app inside Python notebook by running "py4cytoscape.install_app('Your App')" Import the required package
|
import os
import sys
import requests
import pandas as pd
from lxml import etree as ET
from collections import OrderedDict
import py4cytoscape as p4c
# Check Version
p4c.cytoscape_version_info()
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Working togetherOk, with all of these components loaded and launched, you can now perform some nifty sequences. For example, search for a pathway based on a keyword search and then load it into Cytoscape.
|
def find_pathways_by_text(query, species):
base_iri = 'http://webservice.wikipathways.org/'
request_params = {'query':query, 'species':species}
response = requests.get(base_iri + 'findPathwaysByText', params=request_params)
return response
response = find_pathways_by_text("colon cancer", "Homo sapiens")
def find_pathway_dataframe(response):
data = response.text
dom = ET.fromstring(data)
pathways = []
NAMESPACES = {'ns1':'http://www.wso2.org/php/xsd','ns2':'http://www.wikipathways.org/webservice/'}
for node in dom.findall('ns1:result', NAMESPACES):
pathway_using_api_terms = {}
for child in node:
pathway_using_api_terms[ET.QName(child).localname] = child.text
pathways.append(pathway_using_api_terms)
id_list = []
score_list = []
url_list = []
name_list = []
species_list = []
revision_list = []
for p in pathways:
id_list.append(p["id"])
score_list.append(p["score"])
url_list.append(p["url"])
name_list.append(p["name"])
species_list.append(p["species"])
revision_list.append(p["revision"])
df = pd.DataFrame(list(zip(id_list,score_list,url_list,name_list,species_list,revision_list)), columns =['id', 'score','url','name','species','revision'])
return df
df = find_pathway_dataframe(response)
df.head(10)
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
We have a list of human pathways that mention “Colon Cancer”. The results include lots of information, so let’s get a unique list of just the WPIDs.
|
unique_id = list(OrderedDict.fromkeys(df["id"]))
unique_id[0]
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Let’s import the first one of these into Cytoscape!
|
cmd_list = ['wikipathways','import-as-pathway','id="',unique_id[0],'"']
cmd = " ".join(cmd_list)
p4c.commands.commands_get(cmd)
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Once in Cytoscape, you can load data, apply visual style mappings, perform analyses, and export images and data formats. See py4cytoscape package for details. From networks to pathwaysIf you are already with with networks and data in Cytoscape, you may end up focusing on one or few particular genes, proteins or metabolites, and want to query WikiPathways.For example, let’s open a sample network from Cytoscape and identify the gene with the largest number of connections, i.e., node degree.Note: this next chunk will overwrite your current session. Save if you want to keep anything.
|
p4c.session.open_session()
net_data = p4c.tables.get_table_columns(columns=['name','degree.layout','COMMON'])
max_gene = net_data[net_data["degree.layout"] == net_data["degree.layout"].max()]
max_gene
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Great. It looks like MCM1 has the larget number of connections (18) in this network. Let’s use it’s identifier (YMR043W) to query WikiPathways to learn more about the gene and its biological role, and load it into Cytoscape.Pro-tip: We need to know the datasource that provides a given identifier. In this case, it’s sort of tricky: Ensembl provides these Yeast ORF identifiers for this organism rather than they typical format. So, we’ll include the ‘En’ system code. See other vignettes for more details.
|
def find_pathways_by_xref(ids, codes):
base_iri = 'http://webservice.wikipathways.org/'
request_params = {'ids':ids, 'codes':codes}
response = requests.get(base_iri + 'findPathwaysByXref', params=request_params)
return response
response = find_pathways_by_xref('YMR043W','En')
mcm1_pathways = find_pathway_dataframe(response)
unique_id = list(OrderedDict.fromkeys(mcm1_pathways["id"]))
unique_id = "".join(unique_id)
unique_id
cmd_list = ['wikipathways','import-as-pathway','id="',unique_id,'"']
cmd = " ".join(cmd_list)
p4c.commands.commands_get(cmd)
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
And we can easily select the MCM1 node by name in the newly imported pathway to help see where exactly it plays its role.
|
p4c.network_selection.select_nodes(['Mcm1'], by_col='name')
|
_____no_output_____
|
CC0-1.0
|
for-scripters/Python/wikiPathways-and-py4cytoscape.ipynb
|
kozo2/cytoscape-automation
|
Plotting with Matplotlib What is `matplotlib`?* `matplotlib` is a 2D plotting library for Python* It provides quick way to visualize data from Python* It comes with a set plots* We can import its functions through the command```Pythonimport matplotlib.pyplot as plt```
|
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
|
_____no_output_____
|
Apache-2.0
|
02-plotting-with-matplotlib.ipynb
|
theed-ml/notebooks
|
Basic plotsYour goal is to plot the cosine and the sine functions on the same plot, using the default `matplotlib` settings. Generating the data
|
x = np.linspace(-np.pi, np.pi, 256, endpoint=True)
c, s = np.cos(x), np.sin(x)
|
_____no_output_____
|
Apache-2.0
|
02-plotting-with-matplotlib.ipynb
|
theed-ml/notebooks
|
where, * x is a vector with 256 values ranging from $-\pi$ to $\pi$ included * c and s are vectors with the cosine and the sine values of X
|
plt.plot(x, c)
plt.plot(x, s)
plt.show()
|
_____no_output_____
|
Apache-2.0
|
02-plotting-with-matplotlib.ipynb
|
theed-ml/notebooks
|
Rather than creating a plot with default size, we want to specify:* the size of the figure* the colors and type of the lines* the limits of the axes
|
# Create a figure of size 8x6 inches, 80 dots per inch
plt.figure(figsize=(8, 6), dpi=80)
# Create a new subplot from a grid of 1x1
plt.subplot(1, 1, 1)
# Plot cosine with a blue continuous line of width 1 (pixels)
plt.plot(x, c, color="blue", linewidth=2.5, linestyle="-")
# Plot sine with a green dotted line of width 1 (pixels)
plt.plot(x, s, color="green", linewidth=1.0, linestyle="dotted")
# Set x limits
plt.xlim(-4.0, 4)
# Set x ticks
plt.xticks(np.linspace(-4, 4, 9, endpoint=True))
# Set y limits
plt.ylim(-1.0, 1.0)
# Set y ticks
plt.yticks(np.linspace(-1, 1, 5, endpoint=True))
|
_____no_output_____
|
Apache-2.0
|
02-plotting-with-matplotlib.ipynb
|
theed-ml/notebooks
|
Changing colors and line widths* We can to: - make the figure more horizontal - change the color of the lines to blue and red - have slighty thicker lines
|
plt.figure(figsize=(10, 6), dpi=80)
plt.plot(x, c, color="blue", linewidth=2.5, linestyle="-")
plt.plot(x, s, color="red", linewidth=2.5, linestyle="solid")
|
_____no_output_____
|
Apache-2.0
|
02-plotting-with-matplotlib.ipynb
|
theed-ml/notebooks
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.