markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
ロジスティック回帰を適用した結果を表示します。 | w0, w1, w2, err_rate, result = run_logistic(train_set)
fig = plt.figure(figsize=(6, 12))
subplot = fig.add_subplot(2,1,1)
show_result(subplot, train_set, w0, w1, w2, err_rate)
subplot = fig.add_subplot(2,1,2)
draw_roc(subplot, result) | _____no_output_____ | Apache-2.0 | 05-roc_curve.ipynb | RXV06021/test_ml4se_colab |
*Python Machine Learning 2nd Edition* by [Sebastian Raschka](https://sebastianraschka.com), Packt Publishing Ltd. 2017Code Repository: https://github.com/rasbt/python-machine-learning-book-2nd-editionCode License: [MIT License](https://github.com/rasbt/python-machine-learning-book-2nd-edition/blob/master/LICENSE.txt) Python Machine Learning - Code Examples Chapter 11 - Working with Unlabeled Data – Clustering Analysis Note that the optional watermark extension is a small IPython notebook plugin that I developed to make the code reproducible. You can just skip the following line(s). | %load_ext watermark
%watermark -a "Sebastian Raschka" -u -d -v -p numpy,pandas,matplotlib,scipy,sklearn | Sebastian Raschka
last updated: 2017-08-25
CPython 3.6.1
IPython 6.1.0
numpy 1.12.1
pandas 0.20.3
matplotlib 2.0.2
scipy 0.19.1
sklearn 0.19.0
| MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
*The use of `watermark` is optional. You can install this IPython extension via "`pip install watermark`". For more information, please see: https://github.com/rasbt/watermark.* Overview - [Grouping objects by similarity using k-means](Grouping-objects-by-similarity-using-k-means) - [K-means clustering using scikit-learn](K-means-clustering-using-scikit-learn) - [A smarter way of placing the initial cluster centroids using k-means++](A-smarter-way-of-placing-the-initial-cluster-centroids-using-k-means++) - [Hard versus soft clustering](Hard-versus-soft-clustering) - [Using the elbow method to find the optimal number of clusters](Using-the-elbow-method-to-find-the-optimal-number-of-clusters) - [Quantifying the quality of clustering via silhouette plots](Quantifying-the-quality-of-clustering-via-silhouette-plots)- [Organizing clusters as a hierarchical tree](Organizing-clusters-as-a-hierarchical-tree) - [Grouping clusters in bottom-up fashion](Grouping-clusters-in-bottom-up-fashion) - [Performing hierarchical clustering on a distance matrix](Performing-hierarchical-clustering-on-a-distance-matrix) - [Attaching dendrograms to a heat map](Attaching-dendrograms-to-a-heat-map) - [Applying agglomerative clustering via scikit-learn](Applying-agglomerative-clustering-via-scikit-learn)- [Locating regions of high density via DBSCAN](Locating-regions-of-high-density-via-DBSCAN)- [Summary](Summary) | from IPython.display import Image
%matplotlib inline | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Grouping objects by similarity using k-means K-means clustering using scikit-learn | from sklearn.datasets import make_blobs
X, y = make_blobs(n_samples=150,
n_features=2,
centers=3,
cluster_std=0.5,
shuffle=True,
random_state=0)
import matplotlib.pyplot as plt
plt.scatter(X[:, 0], X[:, 1],
c='white', marker='o', edgecolor='black', s=50)
plt.grid()
plt.tight_layout()
#plt.savefig('images/11_01.png', dpi=300)
plt.show()
from sklearn.cluster import KMeans
km = KMeans(n_clusters=3,
init='random',
n_init=10,
max_iter=300,
tol=1e-04,
random_state=0)
y_km = km.fit_predict(X)
plt.scatter(X[y_km == 0, 0],
X[y_km == 0, 1],
s=50, c='lightgreen',
marker='s', edgecolor='black',
label='cluster 1')
plt.scatter(X[y_km == 1, 0],
X[y_km == 1, 1],
s=50, c='orange',
marker='o', edgecolor='black',
label='cluster 2')
plt.scatter(X[y_km == 2, 0],
X[y_km == 2, 1],
s=50, c='lightblue',
marker='v', edgecolor='black',
label='cluster 3')
plt.scatter(km.cluster_centers_[:, 0],
km.cluster_centers_[:, 1],
s=250, marker='*',
c='red', edgecolor='black',
label='centroids')
plt.legend(scatterpoints=1)
plt.grid()
plt.tight_layout()
#plt.savefig('images/11_02.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
A smarter way of placing the initial cluster centroids using k-means++ ... Hard versus soft clustering ... Using the elbow method to find the optimal number of clusters | print('Distortion: %.2f' % km.inertia_)
distortions = []
for i in range(1, 11):
km = KMeans(n_clusters=i,
init='k-means++',
n_init=10,
max_iter=300,
random_state=0)
km.fit(X)
distortions.append(km.inertia_)
plt.plot(range(1, 11), distortions, marker='o')
plt.xlabel('Number of clusters')
plt.ylabel('Distortion')
plt.tight_layout()
#plt.savefig('images/11_03.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Quantifying the quality of clustering via silhouette plots | import numpy as np
from matplotlib import cm
from sklearn.metrics import silhouette_samples
km = KMeans(n_clusters=3,
init='k-means++',
n_init=10,
max_iter=300,
tol=1e-04,
random_state=0)
y_km = km.fit_predict(X)
cluster_labels = np.unique(y_km)
n_clusters = cluster_labels.shape[0]
silhouette_vals = silhouette_samples(X, y_km, metric='euclidean')
y_ax_lower, y_ax_upper = 0, 0
yticks = []
for i, c in enumerate(cluster_labels):
c_silhouette_vals = silhouette_vals[y_km == c]
c_silhouette_vals.sort()
y_ax_upper += len(c_silhouette_vals)
color = cm.jet(float(i) / n_clusters)
plt.barh(range(y_ax_lower, y_ax_upper), c_silhouette_vals, height=1.0,
edgecolor='none', color=color)
yticks.append((y_ax_lower + y_ax_upper) / 2.)
y_ax_lower += len(c_silhouette_vals)
silhouette_avg = np.mean(silhouette_vals)
plt.axvline(silhouette_avg, color="red", linestyle="--")
plt.yticks(yticks, cluster_labels + 1)
plt.ylabel('Cluster')
plt.xlabel('Silhouette coefficient')
plt.tight_layout()
#plt.savefig('images/11_04.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Comparison to "bad" clustering: | km = KMeans(n_clusters=2,
init='k-means++',
n_init=10,
max_iter=300,
tol=1e-04,
random_state=0)
y_km = km.fit_predict(X)
plt.scatter(X[y_km == 0, 0],
X[y_km == 0, 1],
s=50,
c='lightgreen',
edgecolor='black',
marker='s',
label='cluster 1')
plt.scatter(X[y_km == 1, 0],
X[y_km == 1, 1],
s=50,
c='orange',
edgecolor='black',
marker='o',
label='cluster 2')
plt.scatter(km.cluster_centers_[:, 0], km.cluster_centers_[:, 1],
s=250, marker='*', c='red', label='centroids')
plt.legend()
plt.grid()
plt.tight_layout()
#plt.savefig('images/11_05.png', dpi=300)
plt.show()
cluster_labels = np.unique(y_km)
n_clusters = cluster_labels.shape[0]
silhouette_vals = silhouette_samples(X, y_km, metric='euclidean')
y_ax_lower, y_ax_upper = 0, 0
yticks = []
for i, c in enumerate(cluster_labels):
c_silhouette_vals = silhouette_vals[y_km == c]
c_silhouette_vals.sort()
y_ax_upper += len(c_silhouette_vals)
color = cm.jet(float(i) / n_clusters)
plt.barh(range(y_ax_lower, y_ax_upper), c_silhouette_vals, height=1.0,
edgecolor='none', color=color)
yticks.append((y_ax_lower + y_ax_upper) / 2.)
y_ax_lower += len(c_silhouette_vals)
silhouette_avg = np.mean(silhouette_vals)
plt.axvline(silhouette_avg, color="red", linestyle="--")
plt.yticks(yticks, cluster_labels + 1)
plt.ylabel('Cluster')
plt.xlabel('Silhouette coefficient')
plt.tight_layout()
#plt.savefig('images/11_06.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Organizing clusters as a hierarchical tree Grouping clusters in bottom-up fashion | Image(filename='./images/11_05.png', width=400)
import pandas as pd
import numpy as np
np.random.seed(123)
variables = ['X', 'Y', 'Z']
labels = ['ID_0', 'ID_1', 'ID_2', 'ID_3', 'ID_4']
X = np.random.random_sample([5, 3])*10
df = pd.DataFrame(X, columns=variables, index=labels)
df | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Performing hierarchical clustering on a distance matrix | from scipy.spatial.distance import pdist, squareform
row_dist = pd.DataFrame(squareform(pdist(df, metric='euclidean')),
columns=labels,
index=labels)
row_dist | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
We can either pass a condensed distance matrix (upper triangular) from the `pdist` function, or we can pass the "original" data array and define the `metric='euclidean'` argument in `linkage`. However, we should not pass the squareform distance matrix, which would yield different distance values although the overall clustering could be the same. | # 1. incorrect approach: Squareform distance matrix
from scipy.cluster.hierarchy import linkage
row_clusters = linkage(row_dist, method='complete', metric='euclidean')
pd.DataFrame(row_clusters,
columns=['row label 1', 'row label 2',
'distance', 'no. of items in clust.'],
index=['cluster %d' % (i + 1)
for i in range(row_clusters.shape[0])])
# 2. correct approach: Condensed distance matrix
row_clusters = linkage(pdist(df, metric='euclidean'), method='complete')
pd.DataFrame(row_clusters,
columns=['row label 1', 'row label 2',
'distance', 'no. of items in clust.'],
index=['cluster %d' % (i + 1)
for i in range(row_clusters.shape[0])])
# 3. correct approach: Input sample matrix
row_clusters = linkage(df.values, method='complete', metric='euclidean')
pd.DataFrame(row_clusters,
columns=['row label 1', 'row label 2',
'distance', 'no. of items in clust.'],
index=['cluster %d' % (i + 1)
for i in range(row_clusters.shape[0])])
from scipy.cluster.hierarchy import dendrogram
# make dendrogram black (part 1/2)
# from scipy.cluster.hierarchy import set_link_color_palette
# set_link_color_palette(['black'])
row_dendr = dendrogram(row_clusters,
labels=labels,
# make dendrogram black (part 2/2)
# color_threshold=np.inf
)
plt.tight_layout()
plt.ylabel('Euclidean distance')
#plt.savefig('images/11_11.png', dpi=300,
# bbox_inches='tight')
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Attaching dendrograms to a heat map | # plot row dendrogram
fig = plt.figure(figsize=(8, 8), facecolor='white')
axd = fig.add_axes([0.09, 0.1, 0.2, 0.6])
# note: for matplotlib < v1.5.1, please use orientation='right'
row_dendr = dendrogram(row_clusters, orientation='left')
# reorder data with respect to clustering
df_rowclust = df.iloc[row_dendr['leaves'][::-1]]
axd.set_xticks([])
axd.set_yticks([])
# remove axes spines from dendrogram
for i in axd.spines.values():
i.set_visible(False)
# plot heatmap
axm = fig.add_axes([0.23, 0.1, 0.6, 0.6]) # x-pos, y-pos, width, height
cax = axm.matshow(df_rowclust, interpolation='nearest', cmap='hot_r')
fig.colorbar(cax)
axm.set_xticklabels([''] + list(df_rowclust.columns))
axm.set_yticklabels([''] + list(df_rowclust.index))
#plt.savefig('images/11_12.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Applying agglomerative clustering via scikit-learn | from sklearn.cluster import AgglomerativeClustering
ac = AgglomerativeClustering(n_clusters=3,
affinity='euclidean',
linkage='complete')
labels = ac.fit_predict(X)
print('Cluster labels: %s' % labels)
ac = AgglomerativeClustering(n_clusters=2,
affinity='euclidean',
linkage='complete')
labels = ac.fit_predict(X)
print('Cluster labels: %s' % labels) | Cluster labels: [0 1 1 0 0]
| MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Locating regions of high density via DBSCAN | Image(filename='images/11_13.png', width=500)
from sklearn.datasets import make_moons
X, y = make_moons(n_samples=200, noise=0.05, random_state=0)
plt.scatter(X[:, 0], X[:, 1])
plt.tight_layout()
#plt.savefig('images/11_14.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
K-means and hierarchical clustering: | f, (ax1, ax2) = plt.subplots(1, 2, figsize=(8, 3))
km = KMeans(n_clusters=2, random_state=0)
y_km = km.fit_predict(X)
ax1.scatter(X[y_km == 0, 0], X[y_km == 0, 1],
edgecolor='black',
c='lightblue', marker='o', s=40, label='cluster 1')
ax1.scatter(X[y_km == 1, 0], X[y_km == 1, 1],
edgecolor='black',
c='red', marker='s', s=40, label='cluster 2')
ax1.set_title('K-means clustering')
ac = AgglomerativeClustering(n_clusters=2,
affinity='euclidean',
linkage='complete')
y_ac = ac.fit_predict(X)
ax2.scatter(X[y_ac == 0, 0], X[y_ac == 0, 1], c='lightblue',
edgecolor='black',
marker='o', s=40, label='cluster 1')
ax2.scatter(X[y_ac == 1, 0], X[y_ac == 1, 1], c='red',
edgecolor='black',
marker='s', s=40, label='cluster 2')
ax2.set_title('Agglomerative clustering')
plt.legend()
plt.tight_layout()
# plt.savefig('images/11_15.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Density-based clustering: | from sklearn.cluster import DBSCAN
db = DBSCAN(eps=0.2, min_samples=5, metric='euclidean')
y_db = db.fit_predict(X)
plt.scatter(X[y_db == 0, 0], X[y_db == 0, 1],
c='lightblue', marker='o', s=40,
edgecolor='black',
label='cluster 1')
plt.scatter(X[y_db == 1, 0], X[y_db == 1, 1],
c='red', marker='s', s=40,
edgecolor='black',
label='cluster 2')
plt.legend()
plt.tight_layout()
#plt.savefig('images/11_16.png', dpi=300)
plt.show() | _____no_output_____ | MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Summary ... ---Readers may ignore the next cell. | ! python ../.convert_notebook_to_script.py --input ch11.ipynb --output ch11.py | [NbConvertApp] Converting notebook ch11.ipynb to script
[NbConvertApp] Writing 14002 bytes to ch11.py
| MIT | python-machine-learning-book-2nd-edition/code/ch11/ch11.ipynb | gopala-kr/ds-notebooks |
Batch NormalizationOne way to make deep networks easier to train is to use more sophisticated optimization procedures such as SGD+momentum, RMSProp, or Adam. Another strategy is to change the architecture of the network to make it easier to train. One idea along these lines is batch normalization which was proposed by [3] in 2015.The idea is relatively straightforward. _Machine learning methods tend to work better when their input data consists of uncorrelated features with zero mean and unit variance._ When training a neural network, we can preprocess the data before feeding it to the network to explicitly decorrelate its features; this will ensure that the first layer of the network sees data that follows a nice distribution. However, even if we preprocess the input data, the activations at deeper layers of the network will likely no longer be decorrelated and will no longer have zero mean or unit variance since they are output from earlier layers in the network. Even worse, during the training process the distribution of features at each layer of the network will shift as the weights of each layer are updated.The authors of [3] hypothesize that the shifting distribution of features inside deep neural networks may make training deep networks more difficult. To overcome this problem, [3] proposes to insert batch normalization layers into the network. At training time, a batch normalization layer uses a minibatch of data to estimate the mean and standard deviation of each feature. These estimated means and standard deviations are then used to center and normalize the features of the minibatch. A running average of these means and standard deviations is kept during training, and at test time these running averages are used to center and normalize features.It is possible that this normalization strategy could reduce the representational power of the network, since it may sometimes be optimal for certain layers to have features that are not zero-mean or unit variance. To this end, the batch normalization layer includes learnable shift and scale parameters for each feature dimension.[3] [Sergey Ioffe and Christian Szegedy, "Batch Normalization: Accelerating Deep Network Training by ReducingInternal Covariate Shift", ICML 2015.](https://arxiv.org/abs/1502.03167) | # As usual, a bit of setup
import time
import numpy as np
import matplotlib.pyplot as plt
from cs231n.classifiers.fc_net import *
from cs231n.data_utils import get_CIFAR10_data
from cs231n.gradient_check import eval_numerical_gradient, eval_numerical_gradient_array
from cs231n.solver import Solver
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# for auto-reloading external modules
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
def rel_error(x, y):
""" returns relative error """
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
def print_mean_std(x,axis=0):
print(' means: ', x.mean(axis=axis))
print(' stds: ', x.std(axis=axis))
print()
# Load the (preprocessed) CIFAR10 data.
data = get_CIFAR10_data()
for k, v in data.items():
print('%s: ' % k, v.shape) | X_train: (49000, 3, 32, 32)
y_train: (49000,)
X_val: (1000, 3, 32, 32)
y_val: (1000,)
X_test: (1000, 3, 32, 32)
y_test: (1000,)
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Batch normalization: forwardIn the file `cs231n/layers.py`, implement the batch normalization forward pass in the function `batchnorm_forward`. Once you have done so, run the following to test your implementation.Referencing the paper linked to above would be helpful! | # Check the training-time forward pass by checking means and variances
# of features both before and after batch normalization
# Simulate the forward pass for a two-layer network
np.random.seed(231)
N, D1, D2, D3 = 200, 50, 60, 3
X = np.random.randn(N, D1)
W1 = np.random.randn(D1, D2)
W2 = np.random.randn(D2, D3)
a = np.maximum(0, X.dot(W1)).dot(W2)
print('Before batch normalization:')
print_mean_std(a,axis=0)
gamma = np.ones((D3,))
beta = np.zeros((D3,))
# Means should be close to zero and stds close to one
print('After batch normalization (gamma=1, beta=0)')
a_norm, _ = batchnorm_forward(a, gamma, beta, {'mode': 'train'})
print_mean_std(a_norm,axis=0)
gamma = np.asarray([1.0, 2.0, 3.0])
beta = np.asarray([11.0, 12.0, 13.0])
# Now means should be close to beta and stds close to gamma
print('After batch normalization (gamma=', gamma, ', beta=', beta, ')')
a_norm, _ = batchnorm_forward(a, gamma, beta, {'mode': 'train'})
print_mean_std(a_norm,axis=0)
# Check the test-time forward pass by running the training-time
# forward pass many times to warm up the running averages, and then
# checking the means and variances of activations after a test-time
# forward pass.
np.random.seed(231)
N, D1, D2, D3 = 200, 50, 60, 3
W1 = np.random.randn(D1, D2)
W2 = np.random.randn(D2, D3)
bn_param = {'mode': 'train'}
gamma = np.ones(D3)
beta = np.zeros(D3)
for t in range(50):
X = np.random.randn(N, D1)
a = np.maximum(0, X.dot(W1)).dot(W2)
batchnorm_forward(a, gamma, beta, bn_param)
bn_param['mode'] = 'test'
X = np.random.randn(N, D1)
a = np.maximum(0, X.dot(W1)).dot(W2)
a_norm, _ = batchnorm_forward(a, gamma, beta, bn_param)
# Means should be close to zero and stds close to one, but will be
# noisier than training-time forward passes.
print('After batch normalization (test-time):')
print_mean_std(a_norm,axis=0) | After batch normalization (test-time):
means: [-0.03927353 -0.04349151 -0.10452686]
stds: [1.01531399 1.01238345 0.97819961]
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Batch normalization: backwardNow implement the backward pass for batch normalization in the function `batchnorm_backward`.To derive the backward pass you should write out the computation graph for batch normalization and backprop through each of the intermediate nodes. Some intermediates may have multiple outgoing branches; make sure to sum gradients across these branches in the backward pass.Once you have finished, run the following to numerically check your backward pass. | # Gradient check batchnorm backward pass
np.random.seed(231)
N, D = 4, 5
x = 5 * np.random.randn(N, D) + 12
gamma = np.random.randn(D)
beta = np.random.randn(D)
dout = np.random.randn(N, D)
bn_param = {'mode': 'train'}
fx = lambda x: batchnorm_forward(x, gamma, beta, bn_param)[0]
fg = lambda a: batchnorm_forward(x, a, beta, bn_param)[0]
fb = lambda b: batchnorm_forward(x, gamma, b, bn_param)[0]
dx_num = eval_numerical_gradient_array(fx, x, dout)
da_num = eval_numerical_gradient_array(fg, gamma.copy(), dout)
db_num = eval_numerical_gradient_array(fb, beta.copy(), dout)
_, cache = batchnorm_forward(x, gamma, beta, bn_param)
dx, dgamma, dbeta = batchnorm_backward(dout, cache)
#You should expect to see relative errors between 1e-13 and 1e-8
print('dx error: ', rel_error(dx_num, dx))
print('dgamma error: ', rel_error(da_num, dgamma))
print('dbeta error: ', rel_error(db_num, dbeta)) | dx error: 1.6674604875341426e-09
dgamma error: 7.417225040694815e-13
dbeta error: 2.379446949959628e-12
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Batch normalization: alternative backwardIn class we talked about two different implementations for the sigmoid backward pass. One strategy is to write out a computation graph composed of simple operations and backprop through all intermediate values. Another strategy is to work out the derivatives on paper. For example, you can derive a very simple formula for the sigmoid function's backward pass by simplifying gradients on paper.Surprisingly, it turns out that you can do a similar simplification for the batch normalization backward pass too. Given a set of inputs $X=\begin{bmatrix}x_1\\x_2\\...\\x_N\end{bmatrix}$, we first calculate the mean $\mu=\frac{1}{N}\sum_{k=1}^N x_k$ and variance $v=\frac{1}{N}\sum_{k=1}^N (x_k-\mu)^2.$ With $\mu$ and $v$ calculated, we can calculate the standard deviation $\sigma=\sqrt{v+\epsilon}$ and normalized data $Y$ with $y_i=\frac{x_i-\mu}{\sigma}.$The meat of our problem is to get $\frac{\partial L}{\partial X}$ from the upstream gradient $\frac{\partial L}{\partial Y}.$ It might be challenging to directly reason about the gradients over $X$ and $Y$ - try reasoning about it in terms of $x_i$ and $y_i$ first.You will need to come up with the derivations for $\frac{\partial L}{\partial x_i}$, by relying on the Chain Rule to first calculate the intermediate $\frac{\partial \mu}{\partial x_i}, \frac{\partial v}{\partial x_i}, \frac{\partial \sigma}{\partial x_i},$ then assemble these pieces to calculate $\frac{\partial y_i}{\partial x_i}$. You should make sure each of the intermediary steps are all as simple as possible. After doing so, implement the simplified batch normalization backward pass in the function `batchnorm_backward_alt` and compare the two implementations by running the following. Your two implementations should compute nearly identical results, but the alternative implementation should be a bit faster. | np.random.seed(231)
N, D = 100, 500
x = 5 * np.random.randn(N, D) + 12
gamma = np.random.randn(D)
beta = np.random.randn(D)
dout = np.random.randn(N, D)
bn_param = {'mode': 'train'}
out, cache = batchnorm_forward(x, gamma, beta, bn_param)
t1 = time.time()
dx1, dgamma1, dbeta1 = batchnorm_backward(dout, cache)
t2 = time.time()
dx2, dgamma2, dbeta2 = batchnorm_backward_alt(dout, cache)
t3 = time.time()
print('dx difference: ', rel_error(dx1, dx2))
print('dgamma difference: ', rel_error(dgamma1, dgamma2))
print('dbeta difference: ', rel_error(dbeta1, dbeta2))
print('speedup: %.2fx' % ((t2 - t1) / (t3 - t2))) | dx difference: 9.890497291190823e-13
dgamma difference: 0.0
dbeta difference: 0.0
speedup: 3.19x
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Fully Connected Nets with Batch NormalizationNow that you have a working implementation for batch normalization, go back to your `FullyConnectedNet` in the file `cs231n/classifiers/fc_net.py`. Modify your implementation to add batch normalization.Concretely, when the `normalization` flag is set to `"batchnorm"` in the constructor, you should insert a batch normalization layer before each ReLU nonlinearity. The outputs from the last layer of the network should not be normalized. Once you are done, run the following to gradient-check your implementation.HINT: You might find it useful to define an additional helper layer similar to those in the file `cs231n/layer_utils.py`. If you decide to do so, do it in the file `cs231n/classifiers/fc_net.py`. | np.random.seed(231)
N, D, H1, H2, C = 2, 15, 20, 30, 10
X = np.random.randn(N, D)
y = np.random.randint(C, size=(N,))
# You should expect losses between 1e-4~1e-10 for W,
# losses between 1e-08~1e-10 for b,
# and losses between 1e-08~1e-09 for beta and gammas.
for reg in [0, 3.14]:
print('Running check with reg = ', reg)
model = FullyConnectedNet([H1, H2], input_dim=D, num_classes=C,
reg=reg, weight_scale=5e-2, dtype=np.float64,
normalization='batchnorm')
loss, grads = model.loss(X, y)
print('Initial loss: ', loss)
for name in sorted(grads):
f = lambda _: model.loss(X, y)[0]
grad_num = eval_numerical_gradient(f, model.params[name], verbose=False, h=1e-5)
print('%s relative error: %.2e' % (name, rel_error(grad_num, grads[name])))
if reg == 0: print() | Running check with reg = 0
Initial loss: 2.2611955101340957
W1 relative error: 1.10e-04
W2 relative error: 3.11e-06
W3 relative error: 4.05e-10
b1 relative error: 4.44e-08
b2 relative error: 2.22e-08
b3 relative error: 1.01e-10
beta1 relative error: 7.33e-09
beta2 relative error: 1.89e-09
gamma1 relative error: 6.96e-09
gamma2 relative error: 2.41e-09
Running check with reg = 3.14
Initial loss: 5.884829928987633
W1 relative error: 1.98e-06
W2 relative error: 2.29e-06
W3 relative error: 6.29e-10
b1 relative error: 5.55e-09
b2 relative error: 2.22e-08
b3 relative error: 2.10e-10
beta1 relative error: 6.65e-09
beta2 relative error: 3.39e-09
gamma1 relative error: 6.27e-09
gamma2 relative error: 5.28e-09
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Batchnorm for deep networksRun the following to train a six-layer network on a subset of 1000 training examples both with and without batch normalization. | np.random.seed(231)
# Try training a very deep net with batchnorm
hidden_dims = [100, 100, 100, 100, 100]
num_train = 1000
small_data = {
'X_train': data['X_train'][:num_train],
'y_train': data['y_train'][:num_train],
'X_val': data['X_val'],
'y_val': data['y_val'],
}
weight_scale = 2e-2
bn_model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization='batchnorm')
model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization=None)
bn_solver = Solver(bn_model, small_data,
num_epochs=10, batch_size=50,
update_rule='adam',
optim_config={
'learning_rate': 1e-3,
},
verbose=True,print_every=20)
bn_solver.train()
solver = Solver(model, small_data,
num_epochs=10, batch_size=50,
update_rule='adam',
optim_config={
'learning_rate': 1e-3,
},
verbose=True, print_every=20)
solver.train() | (Iteration 1 / 200) loss: 2.340974
(Epoch 0 / 10) train acc: 0.107000; val_acc: 0.107000
(Epoch 1 / 10) train acc: 0.324000; val_acc: 0.264000
(Iteration 21 / 200) loss: 1.996679
(Epoch 2 / 10) train acc: 0.426000; val_acc: 0.303000
(Iteration 41 / 200) loss: 2.038482
(Epoch 3 / 10) train acc: 0.483000; val_acc: 0.313000
(Iteration 61 / 200) loss: 1.719336
(Epoch 4 / 10) train acc: 0.599000; val_acc: 0.334000
(Iteration 81 / 200) loss: 1.314874
(Epoch 5 / 10) train acc: 0.633000; val_acc: 0.305000
(Iteration 101 / 200) loss: 1.346061
(Epoch 6 / 10) train acc: 0.723000; val_acc: 0.348000
(Iteration 121 / 200) loss: 0.948622
(Epoch 7 / 10) train acc: 0.751000; val_acc: 0.348000
(Iteration 141 / 200) loss: 1.048444
(Epoch 8 / 10) train acc: 0.764000; val_acc: 0.330000
(Iteration 161 / 200) loss: 0.798128
(Epoch 9 / 10) train acc: 0.847000; val_acc: 0.347000
(Iteration 181 / 200) loss: 0.814854
(Epoch 10 / 10) train acc: 0.847000; val_acc: 0.355000
(Iteration 1 / 200) loss: 2.302332
(Epoch 0 / 10) train acc: 0.114000; val_acc: 0.122000
(Epoch 1 / 10) train acc: 0.263000; val_acc: 0.223000
(Iteration 21 / 200) loss: 2.083578
(Epoch 2 / 10) train acc: 0.305000; val_acc: 0.233000
(Iteration 41 / 200) loss: 1.856229
(Epoch 3 / 10) train acc: 0.367000; val_acc: 0.299000
(Iteration 61 / 200) loss: 1.687658
(Epoch 4 / 10) train acc: 0.410000; val_acc: 0.315000
(Iteration 81 / 200) loss: 1.566294
(Epoch 5 / 10) train acc: 0.446000; val_acc: 0.326000
(Iteration 101 / 200) loss: 1.597560
(Epoch 6 / 10) train acc: 0.500000; val_acc: 0.327000
(Iteration 121 / 200) loss: 1.445800
(Epoch 7 / 10) train acc: 0.558000; val_acc: 0.345000
(Iteration 141 / 200) loss: 1.180650
(Epoch 8 / 10) train acc: 0.601000; val_acc: 0.339000
(Iteration 161 / 200) loss: 0.985692
(Epoch 9 / 10) train acc: 0.644000; val_acc: 0.361000
(Iteration 181 / 200) loss: 0.960458
(Epoch 10 / 10) train acc: 0.711000; val_acc: 0.364000
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Run the following to visualize the results from two networks trained above. You should find that using batch normalization helps the network to converge much faster. | def plot_training_history(title, label, baseline, bn_solvers, plot_fn, bl_marker='.', bn_marker='.', labels=None):
"""utility function for plotting training history"""
plt.title(title)
plt.xlabel(label)
bn_plots = [plot_fn(bn_solver) for bn_solver in bn_solvers]
bl_plot = plot_fn(baseline)
num_bn = len(bn_plots)
for i in range(num_bn):
label='with_norm'
if labels is not None:
label += str(labels[i])
plt.plot(bn_plots[i], bn_marker, label=label)
label='baseline'
if labels is not None:
label += str(labels[0])
plt.plot(bl_plot, bl_marker, label=label)
plt.legend(loc='lower center', ncol=num_bn+1)
plt.subplot(3, 1, 1)
plot_training_history('Training loss','Iteration', solver, [bn_solver], \
lambda x: x.loss_history, bl_marker='o', bn_marker='o')
plt.subplot(3, 1, 2)
plot_training_history('Training accuracy','Epoch', solver, [bn_solver], \
lambda x: x.train_acc_history, bl_marker='-o', bn_marker='-o')
plt.subplot(3, 1, 3)
plot_training_history('Validation accuracy','Epoch', solver, [bn_solver], \
lambda x: x.val_acc_history, bl_marker='-o', bn_marker='-o')
plt.gcf().set_size_inches(15, 15)
plt.show() | _____no_output_____ | MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Batch normalization and initializationWe will now run a small experiment to study the interaction of batch normalization and weight initialization.The first cell will train 8-layer networks both with and without batch normalization using different scales for weight initialization. The second layer will plot training accuracy, validation set accuracy, and training loss as a function of the weight initialization scale. | np.random.seed(231)
# Try training a very deep net with batchnorm
hidden_dims = [50, 50, 50, 50, 50, 50, 50]
num_train = 1000
small_data = {
'X_train': data['X_train'][:num_train],
'y_train': data['y_train'][:num_train],
'X_val': data['X_val'],
'y_val': data['y_val'],
}
bn_solvers_ws = {}
solvers_ws = {}
weight_scales = np.logspace(-4, 0, num=20)
for i, weight_scale in enumerate(weight_scales):
print('Running weight scale %d / %d' % (i + 1, len(weight_scales)))
bn_model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization='batchnorm')
model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization=None)
bn_solver = Solver(bn_model, small_data,
num_epochs=10, batch_size=50,
update_rule='adam',
optim_config={
'learning_rate': 1e-3,
},
verbose=False, print_every=200)
bn_solver.train()
bn_solvers_ws[weight_scale] = bn_solver
solver = Solver(model, small_data,
num_epochs=10, batch_size=50,
update_rule='adam',
optim_config={
'learning_rate': 1e-3,
},
verbose=False, print_every=200)
solver.train()
solvers_ws[weight_scale] = solver
# Plot results of weight scale experiment
best_train_accs, bn_best_train_accs = [], []
best_val_accs, bn_best_val_accs = [], []
final_train_loss, bn_final_train_loss = [], []
for ws in weight_scales:
best_train_accs.append(max(solvers_ws[ws].train_acc_history))
bn_best_train_accs.append(max(bn_solvers_ws[ws].train_acc_history))
best_val_accs.append(max(solvers_ws[ws].val_acc_history))
bn_best_val_accs.append(max(bn_solvers_ws[ws].val_acc_history))
final_train_loss.append(np.mean(solvers_ws[ws].loss_history[-100:]))
bn_final_train_loss.append(np.mean(bn_solvers_ws[ws].loss_history[-100:]))
plt.subplot(3, 1, 1)
plt.title('Best val accuracy vs weight initialization scale')
plt.xlabel('Weight initialization scale')
plt.ylabel('Best val accuracy')
plt.semilogx(weight_scales, best_val_accs, '-o', label='baseline')
plt.semilogx(weight_scales, bn_best_val_accs, '-o', label='batchnorm')
plt.legend(ncol=2, loc='lower right')
plt.subplot(3, 1, 2)
plt.title('Best train accuracy vs weight initialization scale')
plt.xlabel('Weight initialization scale')
plt.ylabel('Best training accuracy')
plt.semilogx(weight_scales, best_train_accs, '-o', label='baseline')
plt.semilogx(weight_scales, bn_best_train_accs, '-o', label='batchnorm')
plt.legend()
plt.subplot(3, 1, 3)
plt.title('Final training loss vs weight initialization scale')
plt.xlabel('Weight initialization scale')
plt.ylabel('Final training loss')
plt.semilogx(weight_scales, final_train_loss, '-o', label='baseline')
plt.semilogx(weight_scales, bn_final_train_loss, '-o', label='batchnorm')
plt.legend()
plt.gca().set_ylim(1.0, 3.5)
plt.gcf().set_size_inches(15, 15)
plt.show() | _____no_output_____ | MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Inline Question 1:Describe the results of this experiment. How does the scale of weight initialization affect models with/without batch normalization differently, and why? Answer:Bacth norm is robust to weight initialisatio scale upto a point - after which both break. Batch normalization and batch sizeWe will now run a small experiment to study the interaction of batch normalization and batch size.The first cell will train 6-layer networks both with and without batch normalization using different batch sizes. The second layer will plot training accuracy and validation set accuracy over time. | def run_batchsize_experiments(normalization_mode):
np.random.seed(231)
# Try training a very deep net with batchnorm
hidden_dims = [100, 100, 100, 100, 100]
num_train = 1000
small_data = {
'X_train': data['X_train'][:num_train],
'y_train': data['y_train'][:num_train],
'X_val': data['X_val'],
'y_val': data['y_val'],
}
n_epochs=10
weight_scale = 2e-2
batch_sizes = [5,10,50]
lr = 10**(-3.5)
solver_bsize = batch_sizes[0]
print('No normalization: batch size = ',solver_bsize)
model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization=None)
solver = Solver(model, small_data,
num_epochs=n_epochs, batch_size=solver_bsize,
update_rule='adam',
optim_config={
'learning_rate': lr,
},
verbose=False)
solver.train()
bn_solvers = []
for i in range(len(batch_sizes)):
b_size=batch_sizes[i]
print('Normalization: batch size = ',b_size)
bn_model = FullyConnectedNet(hidden_dims, weight_scale=weight_scale, normalization=normalization_mode)
bn_solver = Solver(bn_model, small_data,
num_epochs=n_epochs, batch_size=b_size,
update_rule='adam',
optim_config={
'learning_rate': lr,
},
verbose=False)
bn_solver.train()
bn_solvers.append(bn_solver)
return bn_solvers, solver, batch_sizes
batch_sizes = [5,10,50]
bn_solvers_bsize, solver_bsize, batch_sizes = run_batchsize_experiments('batchnorm')
plt.subplot(2, 1, 1)
plot_training_history('Training accuracy (Batch Normalization)','Epoch', solver_bsize, bn_solvers_bsize, \
lambda x: x.train_acc_history, bl_marker='-^', bn_marker='-o', labels=batch_sizes)
plt.subplot(2, 1, 2)
plot_training_history('Validation accuracy (Batch Normalization)','Epoch', solver_bsize, bn_solvers_bsize, \
lambda x: x.val_acc_history, bl_marker='-^', bn_marker='-o', labels=batch_sizes)
plt.gcf().set_size_inches(15, 10)
plt.show() | _____no_output_____ | MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Inline Question 2:Describe the results of this experiment. What does this imply about the relationship between batch normalization and batch size? Why is this relationship observed? Answer:Batchnorm is sensitive to batch size because mean and variance depend on these minibatches of data. Layer NormalizationBatch normalization has proved to be effective in making networks easier to train, but the dependency on batch size makes it less useful in complex networks which have a cap on the input batch size due to hardware limitations. Several alternatives to batch normalization have been proposed to mitigate this problem; one such technique is Layer Normalization [4]. Instead of normalizing over the batch, we normalize over the features. In other words, when using Layer Normalization, each feature vector corresponding to a single datapoint is normalized based on the sum of all terms within that feature vector.[4] [Ba, Jimmy Lei, Jamie Ryan Kiros, and Geoffrey E. Hinton. "Layer Normalization." stat 1050 (2016): 21.](https://arxiv.org/pdf/1607.06450.pdf) Inline Question 3:Which of these data preprocessing steps is analogous to batch normalization, and which is analogous to layer normalization?1. Scaling each image in the dataset, so that the RGB channels for each row of pixels within an image sums up to 1.2. Scaling each image in the dataset, so that the RGB channels for all pixels within an image sums up to 1. 3. Subtracting the mean image of the dataset from each image in the dataset.4. Setting all RGB values to either 0 or 1 depending on a given threshold. Answer: Layer Normalization: ImplementationNow you'll implement layer normalization. This step should be relatively straightforward, as conceptually the implementation is almost identical to that of batch normalization. One significant difference though is that for layer normalization, we do not keep track of the moving moments, and the testing phase is identical to the training phase, where the mean and variance are directly calculated per datapoint.Here's what you need to do:* In `cs231n/layers.py`, implement the forward pass for layer normalization in the function `layernorm_backward`. Run the cell below to check your results.* In `cs231n/layers.py`, implement the backward pass for layer normalization in the function `layernorm_backward`. Run the second cell below to check your results.* Modify `cs231n/classifiers/fc_net.py` to add layer normalization to the `FullyConnectedNet`. When the `normalization` flag is set to `"layernorm"` in the constructor, you should insert a layer normalization layer before each ReLU nonlinearity. Run the third cell below to run the batch size experiment on layer normalization. | N, D = x.shape
print('X-shape :', x.shape)
mean = np.mean(x, axis = 1, keepdims = True)/D
print('mean-shape :', mean.shape)
#xmu = (x.T - mu.T).T
#print('xmu-shape :', xmu.shape)
temp = np.ones((1,D))
munew = np.matmul(mean, temp)
xmu = x - munew
print('xmu-shape :', xmu.shape)
# Check the training-time forward pass by checking means and variances
# of features both before and after layer normalization
# Simulate the forward pass for a two-layer network
np.random.seed(231)
N, D1, D2, D3 =4, 50, 60, 3
X = np.random.randn(N, D1)
W1 = np.random.randn(D1, D2)
W2 = np.random.randn(D2, D3)
a = np.maximum(0, X.dot(W1)).dot(W2)
print('Before layer normalization:')
print_mean_std(a,axis=1)
gamma = np.ones(D3)
beta = np.zeros(D3)
# Means should be close to zero and stds close to one
print('After layer normalization (gamma=1, beta=0)')
a_norm, _ = layernorm_forward(a, gamma, beta, {'mode': 'train'})
print_mean_std(a_norm,axis=1)
gamma = np.asarray([3.0,3.0,3.0])
beta = np.asarray([5.0,5.0,5.0])
# Now means should be close to beta and stds close to gamma
print('After layer normalization (gamma=', gamma, ', beta=', beta, ')')
a_norm, _ = layernorm_forward(a, gamma, beta, {'mode': 'train'})
print_mean_std(a_norm,axis=1)
# Gradient check batchnorm backward pass
np.random.seed(231)
N, D = 4, 5
x = 5 * np.random.randn(N, D) + 12
gamma = np.random.randn(D)
beta = np.random.randn(D)
dout = np.random.randn(N, D)
ln_param = {}
fx = lambda x: layernorm_forward(x, gamma, beta, ln_param)[0]
fg = lambda a: layernorm_forward(x, a, beta, ln_param)[0]
fb = lambda b: layernorm_forward(x, gamma, b, ln_param)[0]
dx_num = eval_numerical_gradient_array(fx, x, dout)
da_num = eval_numerical_gradient_array(fg, gamma.copy(), dout)
db_num = eval_numerical_gradient_array(fb, beta.copy(), dout)
_, cache = layernorm_forward(x, gamma, beta, ln_param)
dx, dgamma, dbeta = layernorm_backward(dout, cache)
#You should expect to see relative errors between 1e-12 and 1e-8
print('dx error: ', rel_error(dx_num, dx))
print('dgamma error: ', rel_error(da_num, dgamma))
print('dbeta error: ', rel_error(db_num, dbeta)) | dx error: 2.107277492956569e-09
dgamma error: 4.519489546032799e-12
dbeta error: 2.5842537629899423e-12
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
Layer Normalization and batch sizeWe will now run the previous batch size experiment with layer normalization instead of batch normalization. Compared to the previous experiment, you should see a markedly smaller influence of batch size on the training history! | ln_solvers_bsize, solver_bsize, batch_sizes = run_batchsize_experiments('layernorm')
plt.subplot(2, 1, 1)
plot_training_history('Training accuracy (Layer Normalization)','Epoch', solver_bsize, ln_solvers_bsize, \
lambda x: x.train_acc_history, bl_marker='-^', bn_marker='-o', labels=batch_sizes)
plt.subplot(2, 1, 2)
plot_training_history('Validation accuracy (Layer Normalization)','Epoch', solver_bsize, ln_solvers_bsize, \
lambda x: x.val_acc_history, bl_marker='-^', bn_marker='-o', labels=batch_sizes)
plt.gcf().set_size_inches(15, 10)
plt.show() | No normalization: batch size = 5
Normalization: batch size = 5
Normalization: batch size = 10
Normalization: batch size = 50
| MIT | assignment2/BatchNormalization.ipynb | lalithnag/cs231n |
More Information about Synestias[When Earth and the Moon Were One](https://www.scientificamerican.com/article/when-earth-and-the-moon-were-one/)by Simon J. Lock and Sarah T. StewartScientific American, July 2019. Check your local library for anonline or print subscription.[Where did the Moon come from? A New Theory](https://www.ted.com/talks/sarah_t_stewart_where_did_the_moon_come_from_a_new_theory?language=en)Sarah T. Stewart, TED Talk | from IPython.display import YouTubeVideo
YouTubeVideo('7uRPPaYuu44', width=640, height=360) | _____no_output_____ | MIT | synestia-book/_build/jupyter_execute/docs/MoreInformation.ipynb | ststewart/synestiabook2 |
Introduction to Neural NetworksIn this notebook you will learn how to create and use a neural network to classify articles of clothing. To achieve this, we will use a sub module of TensorFlow called *keras*.*This guide is based on the following TensorFlow documentation.*https://www.tensorflow.org/tutorials/keras/classification KerasBefore we dive in and start discussing neural networks, I'd like to give a breif introduction to keras.From the keras official documentation (https://keras.io/) keras is described as follows."Keras is a high-level neural networks API, written in Python and capable of running on top of TensorFlow, CNTK, or Theano. It was developed with a focus on enabling fast experimentation. Use Keras if you need a deep learning library that:- Allows for easy and fast prototyping (through user friendliness, modularity, and extensibility).- Supports both convolutional networks and recurrent networks, as well as combinations of the two.- Runs seamlessly on CPU and GPU."Keras is a very powerful module that allows us to avoid having to build neural networks from scratch. It also hides a lot of mathematical complexity (that otherwise we would have to implement) inside of helpful packages, modules and methods.In this guide we will use keras to quickly develop neural networks. What is a Neural NetworkSo, what are these magical things that have been beating chess grandmasters, driving cars, detecting cancer cells and winning video games? A deep neural network is a layered representation of data. The term "deep" refers to the presence of multiple layers. Recall that in our core learning algorithms (like linear regression) data was not transformed or modified within the model, it simply existed in one layer. We passed some features to our model, some math was done, an answer was returned. The data was not changed or transformed throughout this process. A neural network processes our data differently. It attempts to represent our data in different ways and in different dimensions by applying specific operations to transform our data at each layer. Another way to express this is that at each layer our data is transformed in order to learn more about it. By performing these transformations, the model can better understand our data and therefore provide a better prediction. How it WorksBefore going into too much detail I will provide a very surface level explination of how neural networks work on a mathematical level. All the terms and concepts I discuss will be defined and explained in more detail below.On a lower level neural networks are simply a combination of elementry math operations and some more advanced linear algebra. Each neural network consists of a sequence of layers in which data passes through. These layers are made up on neurons and the neurons of one layer are connected to the next (see below). These connections are defined by what we call a weight (some numeric value). Each layer also has something called a bias, this is simply an extra neuron that has no connections and holds a single numeric value. Data starts at the input layer and is trasnformed as it passes through subsequent layers. The data at each subsequent neuron is defined as the following.> $Y =(\sum_{i=0}^n w_i x_i) + b$> $w$ stands for the weight of each connection to the neuron> $x$ stands for the value of the connected neuron from the previous value> $b$ stands for the bias at each layer, this is a constant> $n$ is the number of connections> $Y$ is the output of the current neuron> $\sum$ stands for sumThe equation you just read is called a weighed sum. We will take this weighted sum at each and every neuron as we pass information through the network. Then we will add what's called a bias to this sum. The bias allows us to shift the network up or down by a constant value. It is like the y-intercept of a line.But that equation is the not complete one! We forgot a crucial part, **the activation function**. This is a function that we apply to the equation seen above to add complexity and dimensionality to our network. Our new equation with the addition of an activation function $F(x)$ is seen below.> $Y =F((\sum_{i=0}^n w_i x_i) + b)$Our network will start with predefined activation functions (they may be different at each layer) but random weights and biases. As we train the network by feeding it data it will learn the correct weights and biases and adjust the network accordingly using a technqiue called **backpropagation** (explained below). Once the correct weights and biases have been learned our network will hopefully be able to give us meaningful predictions. We get these predictions by observing the values at our final layer, the output layer. Breaking Down The Neural Network!Before we dive into any code lets break down how a neural network works and what it does.*Figure 1* DataThe type of data a neural network processes varies drastically based on the problem being solved. When we build a neural network, we define what shape and kind of data it can accept. It may sometimes be neccessary to modify our dataset so that it can be passed to our neural network. Some common types of data a neural network uses are listed below.- Vector Data (2D)- Timeseries or Sequence (3D)- Image Data (4D)- Video Data (5D)There are of course many different types or data, but these are the main categories. LayersAs we mentioned earlier each neural network consists of multiple layers. At each layer a different transformation of data occurs. Our initial input data is fed through the layers and eventually arrives at the output layer where we will obtain the result.Input LayerThe input layer is the layer that our initial data is passed to. It is the first layer in our neural network.Output LayerThe output layer is the layer that we will retrive our results from. Once the data has passed through all other layers it will arrive here.Hidden Layer(s)All the other layers in our neural network are called "hidden layers". This is because they are hidden to us, we cannot observe them. Most neural networks consist of at least one hidden layer but can have an unlimited amount. Typically, the more complex the model the more hidden layers.NeuronsEach layer is made up of what are called neurons. Neurons have a few different properties that we will discuss later. The important aspect to understand now is that each neuron is responsible for generating/holding/passing ONE numeric value. This means that in the case of our input layer it will have as many neurons as we have input information. For example, say we want to pass an image that is 28x28 pixels, thats 784 pixels. We would need 784 neurons in our input layer to capture each of these pixels. This also means that our output layer will have as many neurons as we have output information. The output is a little more complicated to understand so I'll refrain from an example right now but hopefully you're getting the idea.But what about our hidden layers? Well these have as many neurons as we decide. We'll discuss how we can pick these values later but understand a hidden layer can have any number of neurons.Connected LayersSo how are all these layers connected? Well the neurons in one layer will be connected to neurons in the subsequent layer. However, the neurons can be connected in a variety of different ways. Take for example *Figure 1* (look above). Each neuron in one layer is connected to every neuron in the next layer. This is called a **dense** layer. There are many other ways of connecting layers but well discuss those as we see them. WeightsWeights are associated with each connection in our neural network. Every pair of connected nodes will have one weight that denotes the strength of the connection between them. These are vital to the inner workings of a neural network and will be tweaked as the neural network is trained. The model will try to determine what these weights should be to achieve the best result. Weights start out at a constant or random value and will change as the network sees training data. BiasesBiases are another important part of neural networks and will also be tweaked as the model is trained. A bias is simply a constant value associated with each layer. It can be thought of as an extra neuron that has no connections. The purpose of a bias is to shift an entire activation function by a constant value. This allows a lot more flexibllity when it comes to choosing an activation and training the network. There is one bias for each layer. Activation FunctionActivation functions are simply a function that is applied to the weighed sum of a neuron. They can be anything we want but are typically higher order/degree functions that aim to add a higher dimension to our data. We would want to do this to introduce more comolexity to our model. By transforming our data to a higher dimension, we can typically make better, more complex predictions.A list of some common activation functions and their graphs can be seen below.- Relu (Rectified Linear Unit)- Tanh (Hyperbolic Tangent)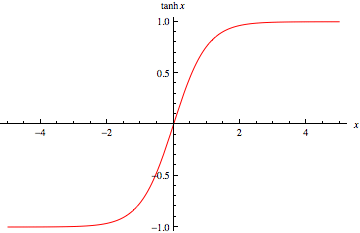- Sigmoid  BackpropagationBackpropagation is the fundemental algorithm behind training neural networks. It is what changes the weights and biases of our network. To fully explain this process, we need to start by discussing something called a cost/loss function.Loss/Cost FunctionAs we now know our neural network feeds information through the layers until it eventually reaches an output layer. This layer contains the results that we look at to determine the prediciton from our network. In the training phase it is likely that our network will make many mistakes and poor predicitions. In fact, at the start of training our network doesn't know anything (it has random weights and biases)! We need some way of evaluating if the network is doing well and how well it is doing. For our training data we have the features (input) and the labels (expected output), because of this we can compare the output from our network to the expected output. Based on the difference between these values we can determine if our network has done a good job or poor job. If the network has done a good job, we'll make minor changes to the weights and biases. If it has done a poor job our changes may be more drastic.So, this is where the cost/loss function comes in. This function is responsible for determining how well the network did. We pass it the output and the expected output, and it returns to us some value representing the cost/loss of the network. This effectively makes the networks job to optimize this cost function, trying to make it as low as possible. Some common loss/cost functions include.- Mean Squared Error- Mean Absolute Error- Hinge LossGradient DescentGradient descent and backpropagation are closely related. Gradient descent is the algorithm used to find the optimal paramaters (weights and biases) for our network, while backpropagation is the process of calculating the gradient that is used in the gradient descent step. Gradient descent requires some pretty advanced calculus and linear algebra to understand so we'll stay away from that for now. Let's just read the formal definition for now."Gradient descent is an optimization algorithm used to minimize some function by iteratively moving in the direction of steepest descent as defined by the negative of the gradient. In machine learning, we use gradient descent to update the parameters of our model." (https://ml-cheatsheet.readthedocs.io/en/latest/gradient_descent.html)And that's all we really need to know for now. I'll direct you to the video for a more in depth explination. OptimizerYou may sometimes see the term optimizer or optimization function. This is simply the function that implements the backpropagation algorithm described above. Here's a list of a few common ones.- Gradient Descent- Stochastic Gradient Descent- Mini-Batch Gradient Descent- Momentum- Nesterov Accelerated Gradient*This article explains them quite well is where I've pulled this list from.*(https://medium.com/@sdoshi579/optimizers-for-training-neural-network-59450d71caf6) Creating a Neural NetworkOkay now you have reached the exciting part of this tutorial! No more math and complex explinations. Time to get hands on and train a very basic neural network.*As stated earlier this guide is based off of the following TensorFlow tutorial.*https://www.tensorflow.org/tutorials/keras/classification Imports | %tensorflow_version 2.x # this line is not required unless you are in a notebook
# TensorFlow and tf.keras
import tensorflow as tf
from tensorflow import keras
# Helper libraries
import numpy as np
import matplotlib.pyplot as plt | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
DatasetFor this tutorial we will use the MNIST Fashion Dataset. This is a dataset that is included in keras.This dataset includes 60,000 images for training and 10,000 images for validation/testing. | fashion_mnist = keras.datasets.fashion_mnist # load dataset
(train_images, train_labels), (test_images, test_labels) = fashion_mnist.load_data() # split into tetsing and training | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Let's have a look at this data to see what we are working with. | train_images.shape | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
So we've got 60,000 images that are made up of 28x28 pixels (784 in total). | train_images[0,23,23] # let's have a look at one pixel | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Our pixel values are between 0 and 255, 0 being black and 255 being white. This means we have a grayscale image as there are no color channels. | train_labels[:10] # let's have a look at the first 10 training labels | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Our labels are integers ranging from 0 - 9. Each integer represents a specific article of clothing. We'll create an array of label names to indicate which is which. | class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot'] | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Fianlly let's look at what some of these images look like! | plt.figure()
plt.imshow(train_images[1])
plt.colorbar()
plt.grid(False)
plt.show() | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Data PreprocessingThe last step before creating our model is to *preprocess* our data. This simply means applying some prior transformations to our data before feeding it the model. In this case we will simply scale all our greyscale pixel values (0-255) to be between 0 and 1. We can do this by dividing each value in the training and testing sets by 255.0. We do this because smaller values will make it easier for the model to process our values. | train_images = train_images / 255.0
test_images = test_images / 255.0 | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Building the ModelNow it's time to build the model! We are going to use a keras *sequential* model with three different layers. This model represents a feed-forward neural network (one that passes values from left to right). We'll break down each layer and its architecture below. | model = keras.Sequential([
keras.layers.Flatten(input_shape=(28, 28)), # input layer (1)
keras.layers.Dense(128, activation='relu'), # hidden layer (2)
keras.layers.Dense(10, activation='softmax') # output layer (3)
]) | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
**Layer 1:** This is our input layer and it will conist of 784 neurons. We use the flatten layer with an input shape of (28,28) to denote that our input should come in in that shape. The flatten means that our layer will reshape the shape (28,28) array into a vector of 784 neurons so that each pixel will be associated with one neuron.**Layer 2:** This is our first and only hidden layer. The *dense* denotes that this layer will be fully connected and each neuron from the previous layer connects to each neuron of this layer. It has 128 neurons and uses the rectify linear unit activation function.**Layer 3:** This is our output later and is also a dense layer. It has 10 neurons that we will look at to determine our models output. Each neuron represnts the probabillity of a given image being one of the 10 different classes. The activation function *softmax* is used on this layer to calculate a probabillity distribution for each class. This means the value of any neuron in this layer will be between 0 and 1, where 1 represents a high probabillity of the image being that class. Compile the ModelThe last step in building the model is to define the loss function, optimizer and metrics we would like to track. I won't go into detail about why we chose each of these right now. | model.compile(optimizer='adam',
loss='sparse_categorical_crossentropy',
metrics=['accuracy']) | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Training the ModelNow it's finally time to train the model. Since we've already done all the work on our data this step is as easy as calling a single method. | model.fit(train_images, train_labels, epochs=10) # we pass the data, labels and epochs and watch the magic! | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Evaluating the ModelNow it's time to test/evaluate the model. We can do this quite easily using another builtin method from keras.The *verbose* argument is defined from the keras documentation as:"verbose: 0 or 1. Verbosity mode. 0 = silent, 1 = progress bar."(https://keras.io/models/sequential/) | test_loss, test_acc = model.evaluate(test_images, test_labels, verbose=1)
print('Test accuracy:', test_acc) | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
You'll likely notice that the accuracy here is lower than when training the model. This difference is reffered to as **overfitting**.And now we have a trained model that's ready to use to predict some values! Making PredictionsTo make predictions we simply need to pass an array of data in the form we've specified in the input layer to ```.predict()``` method. | predictions = model.predict(test_images) | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
This method returns to us an array of predictions for each image we passed it. Let's have a look at the predictions for image 1. | predictions[0] | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
If we wan't to get the value with the highest score we can use a useful function from numpy called ```argmax()```. This simply returns the index of the maximium value from a numpy array. | np.argmax(predictions[0]) | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
And we can check if this is correct by looking at the value of the cooresponding test label. | test_labels[0] | _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
Verifying PredictionsI've written a small function here to help us verify predictions with some simple visuals. | COLOR = 'white'
plt.rcParams['text.color'] = COLOR
plt.rcParams['axes.labelcolor'] = COLOR
def predict(model, image, correct_label):
class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',
'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']
prediction = model.predict(np.array([image]))
predicted_class = class_names[np.argmax(prediction)]
show_image(image, class_names[correct_label], predicted_class)
def show_image(img, label, guess):
plt.figure()
plt.imshow(img, cmap=plt.cm.binary)
plt.title("Excpected: " + label)
plt.xlabel("Guess: " + guess)
plt.colorbar()
plt.grid(False)
plt.show()
def get_number():
while True:
num = input("Pick a number: ")
if num.isdigit():
num = int(num)
if 0 <= num <= 1000:
return int(num)
else:
print("Try again...")
num = get_number()
image = test_images[num]
label = test_labels[num]
predict(model, image, label)
| _____no_output_____ | Unlicense | AI-ML/Tensorflow fcc/Instructor notebooks/Neural Networks.ipynb | f-dufour/cheat-sheets-and-snippets |
MXNet Tutorial and Hand Written Digit RecognitionIn this tutorial we will go through the basic use case of MXNet and also touch on some advanced usages. This example is based on the MNIST dataset, which contains 70,000 images of hand written characters with 28-by-28 pixel size.This tutorial covers the following topics:- network definition.- Variable naming.- Basic data loading and training with feed-forward deep neural networks.- Monitoring intermediate outputs for debuging.- Custom training loop for advanced models. First let's import the modules and setup logging: | %matplotlib inline
import mxnet as mx
import numpy as np
import cv2
import matplotlib.pyplot as plt
import logging
logger = logging.getLogger()
logger.setLevel(logging.DEBUG) | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Network DefinitionNow we can start constructing our network: | # Variables are place holders for input arrays. We give each variable a unique name.
data = mx.symbol.Variable('data')
# The input is fed to a fully connected layer that computes Y=WX+b.
# This is the main computation module in the network.
# Each layer also needs an unique name. We'll talk more about naming in the next section.
fc1 = mx.symbol.FullyConnected(data = data, name='fc1', num_hidden=128)
# Activation layers apply a non-linear function on the previous layer's output.
# Here we use Rectified Linear Unit (ReLU) that computes Y = max(X, 0).
act1 = mx.symbol.Activation(data = fc1, name='relu1', act_type="relu")
fc2 = mx.symbol.FullyConnected(data = act1, name = 'fc2', num_hidden = 64)
act2 = mx.symbol.Activation(data = fc2, name='relu2', act_type="relu")
fc3 = mx.symbol.FullyConnected(data = act2, name='fc3', num_hidden=10)
# Finally we have a loss layer that compares the network's output with label and generates gradient signals.
mlp = mx.symbol.SoftmaxOutput(data = fc3, name = 'softmax') | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
We can visualize the network we just defined with MXNet's visualization module: | mx.viz.plot_network(mlp) | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Variable NamingMXNet requires variable names to follow certain conventions:- All input arrays have a name. This includes inputs (data & label) and model parameters (weight, bias, etc).- Arrays can be renamed by creating named variable. Otherwise, a default name is given as 'SymbolName_ArrayName'. For example, FullyConnected symbol fc1's weight array is named as 'fc1_weight'.- Although you can also rename weight arrays with variables, weight array's name should always end with '_weight' and bias array '_bias'. MXNet relies on the suffixes of array names to correctly initialize & update them.Call list_arguments method on a symbol to get the names of all its inputs: | mlp.list_arguments() | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Data LoadingWe fetch and load the MNIST dataset and partition it into two sets: 60000 examples for training and 10000 examples for testing. We also visualize a few examples to get an idea of what the dataset looks like. | from sklearn.datasets import fetch_mldata
mnist = fetch_mldata('MNIST original')
np.random.seed(1234) # set seed for deterministic ordering
p = np.random.permutation(mnist.data.shape[0])
X = mnist.data[p]
Y = mnist.target[p]
for i in range(10):
plt.subplot(1,10,i+1)
plt.imshow(X[i].reshape((28,28)), cmap='Greys_r')
plt.axis('off')
plt.show()
X = X.astype(np.float32)/255
X_train = X[:60000]
X_test = X[60000:]
Y_train = Y[:60000]
Y_test = Y[60000:] | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Now we can create data iterators from our MNIST data. A data iterator returns a batch of data examples each time for the network to process. MXNet provide a suite of basic DataIters for parsing different data format. Here we use NDArrayIter, which wraps around a numpy array and each time slice a chunk from it along the first dimension. | batch_size = 100
train_iter = mx.io.NDArrayIter(X_train, Y_train, batch_size=batch_size)
test_iter = mx.io.NDArrayIter(X_test, Y_test, batch_size=batch_size) | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
TrainingWith the network and data source defined, we can finally start to train our model. We do this with MXNet's convenience wrapper for feed forward neural networks (it can also be made to handle RNNs with explicit unrolling). | model = mx.model.FeedForward(
ctx = mx.gpu(0), # Run on GPU 0
symbol = mlp, # Use the network we just defined
num_epoch = 10, # Train for 10 epochs
learning_rate = 0.1, # Learning rate
momentum = 0.9, # Momentum for SGD with momentum
wd = 0.00001) # Weight decay for regularization
model.fit(
X=train_iter, # Training data set
eval_data=test_iter, # Testing data set. MXNet computes scores on test set every epoch
batch_end_callback = mx.callback.Speedometer(batch_size, 200)) # Logging module to print out progress | INFO:root:Start training with [gpu(0)]
INFO:root:Epoch[0] Batch [200] Speed: 70941.64 samples/sec Train-accuracy=0.389050
INFO:root:Epoch[0] Batch [400] Speed: 97857.94 samples/sec Train-accuracy=0.646450
INFO:root:Epoch[0] Batch [600] Speed: 70507.97 samples/sec Train-accuracy=0.743333
INFO:root:Epoch[0] Resetting Data Iterator
INFO:root:Epoch[0] Train-accuracy=0.743333
INFO:root:Epoch[0] Time cost=1.069
INFO:root:Epoch[0] Validation-accuracy=0.950800
INFO:root:Epoch[1] Batch [200] Speed: 79912.66 samples/sec Train-accuracy=0.947300
INFO:root:Epoch[1] Batch [400] Speed: 58822.31 samples/sec Train-accuracy=0.954425
INFO:root:Epoch[1] Batch [600] Speed: 67124.87 samples/sec Train-accuracy=0.957733
INFO:root:Epoch[1] Resetting Data Iterator
INFO:root:Epoch[1] Train-accuracy=0.957733
INFO:root:Epoch[1] Time cost=0.893
INFO:root:Epoch[1] Validation-accuracy=0.959400
INFO:root:Epoch[2] Batch [200] Speed: 87015.23 samples/sec Train-accuracy=0.964450
INFO:root:Epoch[2] Batch [400] Speed: 91101.30 samples/sec Train-accuracy=0.968875
INFO:root:Epoch[2] Batch [600] Speed: 88963.21 samples/sec Train-accuracy=0.970017
INFO:root:Epoch[2] Resetting Data Iterator
INFO:root:Epoch[2] Train-accuracy=0.970017
INFO:root:Epoch[2] Time cost=0.678
INFO:root:Epoch[2] Validation-accuracy=0.963000
INFO:root:Epoch[3] Batch [200] Speed: 66986.68 samples/sec Train-accuracy=0.973750
INFO:root:Epoch[3] Batch [400] Speed: 65680.34 samples/sec Train-accuracy=0.976575
INFO:root:Epoch[3] Batch [600] Speed: 91931.16 samples/sec Train-accuracy=0.977050
INFO:root:Epoch[3] Resetting Data Iterator
INFO:root:Epoch[3] Train-accuracy=0.977050
INFO:root:Epoch[3] Time cost=0.825
INFO:root:Epoch[3] Validation-accuracy=0.968000
INFO:root:Epoch[4] Batch [200] Speed: 73709.59 samples/sec Train-accuracy=0.978950
INFO:root:Epoch[4] Batch [400] Speed: 85750.82 samples/sec Train-accuracy=0.980425
INFO:root:Epoch[4] Batch [600] Speed: 87061.38 samples/sec Train-accuracy=0.981183
INFO:root:Epoch[4] Resetting Data Iterator
INFO:root:Epoch[4] Train-accuracy=0.981183
INFO:root:Epoch[4] Time cost=0.739
INFO:root:Epoch[4] Validation-accuracy=0.967600
INFO:root:Epoch[5] Batch [200] Speed: 85031.11 samples/sec Train-accuracy=0.981950
INFO:root:Epoch[5] Batch [400] Speed: 94063.25 samples/sec Train-accuracy=0.983475
INFO:root:Epoch[5] Batch [600] Speed: 97417.46 samples/sec Train-accuracy=0.984183
INFO:root:Epoch[5] Resetting Data Iterator
INFO:root:Epoch[5] Train-accuracy=0.984183
INFO:root:Epoch[5] Time cost=0.657
INFO:root:Epoch[5] Validation-accuracy=0.972000
INFO:root:Epoch[6] Batch [200] Speed: 96185.84 samples/sec Train-accuracy=0.984650
INFO:root:Epoch[6] Batch [400] Speed: 95023.61 samples/sec Train-accuracy=0.985850
INFO:root:Epoch[6] Batch [600] Speed: 97022.32 samples/sec Train-accuracy=0.986683
INFO:root:Epoch[6] Resetting Data Iterator
INFO:root:Epoch[6] Train-accuracy=0.986683
INFO:root:Epoch[6] Time cost=0.628
INFO:root:Epoch[6] Validation-accuracy=0.971900
INFO:root:Epoch[7] Batch [200] Speed: 84764.84 samples/sec Train-accuracy=0.986350
INFO:root:Epoch[7] Batch [400] Speed: 87358.40 samples/sec Train-accuracy=0.986425
INFO:root:Epoch[7] Batch [600] Speed: 74520.63 samples/sec Train-accuracy=0.986517
INFO:root:Epoch[7] Resetting Data Iterator
INFO:root:Epoch[7] Train-accuracy=0.986517
INFO:root:Epoch[7] Time cost=0.737
INFO:root:Epoch[7] Validation-accuracy=0.973700
INFO:root:Epoch[8] Batch [200] Speed: 91634.21 samples/sec Train-accuracy=0.987450
INFO:root:Epoch[8] Batch [400] Speed: 94328.96 samples/sec Train-accuracy=0.987250
INFO:root:Epoch[8] Batch [600] Speed: 91991.24 samples/sec Train-accuracy=0.987850
INFO:root:Epoch[8] Resetting Data Iterator
INFO:root:Epoch[8] Train-accuracy=0.987850
INFO:root:Epoch[8] Time cost=0.652
INFO:root:Epoch[8] Validation-accuracy=0.976800
INFO:root:Epoch[9] Batch [200] Speed: 66583.86 samples/sec Train-accuracy=0.986800
INFO:root:Epoch[9] Batch [400] Speed: 67393.86 samples/sec Train-accuracy=0.987500
INFO:root:Epoch[9] Batch [600] Speed: 65748.40 samples/sec Train-accuracy=0.987900
INFO:root:Epoch[9] Resetting Data Iterator
INFO:root:Epoch[9] Train-accuracy=0.987900
INFO:root:Epoch[9] Time cost=0.906
INFO:root:Epoch[9] Validation-accuracy=0.973800
| Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
EvaluationAfter the model is trained, we can evaluate it on a held out test set.First, lets classity a sample image: | plt.imshow((X_test[0].reshape((28,28))*255).astype(np.uint8), cmap='Greys_r')
plt.show()
print 'Result:', model.predict(X_test[0:1])[0].argmax() | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
We can also evaluate the model's accuracy on the entire test set: | print 'Accuracy:', model.score(test_iter)*100, '%' | Accuracy: 97.38 %
| Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Now, try if your model recognizes your own hand writing.Write a digit from 0 to 9 in the box below. Try to put your digit in the middle of the box. | # run hand drawing test
from IPython.display import HTML
def classify(img):
img = img[len('data:image/png;base64,'):].decode('base64')
img = cv2.imdecode(np.fromstring(img, np.uint8), -1)
img = cv2.resize(img[:,:,3], (28,28))
img = img.astype(np.float32).reshape((1, 784))/255.0
return model.predict(img)[0].argmax()
html = """<style type="text/css">canvas { border: 1px solid black; }</style><div id="board"><canvas id="myCanvas" width="100px" height="100px">Sorry, your browser doesn't support canvas technology.</canvas><p><button id="classify" onclick="classify()">Classify</button><button id="clear" onclick="myClear()">Clear</button>Result: <input type="text" id="result_output" size="5" value=""></p></div>"""
script = """<script type="text/JavaScript" src="https://ajax.googleapis.com/ajax/libs/jquery/1.4.2/jquery.min.js?ver=1.4.2"></script><script type="text/javascript">function init() {var myCanvas = document.getElementById("myCanvas");var curColor = $('#selectColor option:selected').val();if(myCanvas){var isDown = false;var ctx = myCanvas.getContext("2d");var canvasX, canvasY;ctx.lineWidth = 8;$(myCanvas).mousedown(function(e){isDown = true;ctx.beginPath();var parentOffset = $(this).parent().offset(); canvasX = e.pageX - parentOffset.left;canvasY = e.pageY - parentOffset.top;ctx.moveTo(canvasX, canvasY);}).mousemove(function(e){if(isDown != false) {var parentOffset = $(this).parent().offset(); canvasX = e.pageX - parentOffset.left;canvasY = e.pageY - parentOffset.top;ctx.lineTo(canvasX, canvasY);ctx.strokeStyle = curColor;ctx.stroke();}}).mouseup(function(e){isDown = false;ctx.closePath();});}$('#selectColor').change(function () {curColor = $('#selectColor option:selected').val();});}init();function handle_output(out) {document.getElementById("result_output").value = out.content.data["text/plain"];}function classify() {var kernel = IPython.notebook.kernel;var myCanvas = document.getElementById("myCanvas");data = myCanvas.toDataURL('image/png');document.getElementById("result_output").value = "";kernel.execute("classify('" + data +"')", { 'iopub' : {'output' : handle_output}}, {silent:false});}function myClear() {var myCanvas = document.getElementById("myCanvas");myCanvas.getContext("2d").clearRect(0, 0, myCanvas.width, myCanvas.height);}</script>"""
HTML(html+script) | _____no_output_____ | Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
DebuggingDNNs can perform poorly for a lot of reasons, like learning rate too big/small, initialization too big/small, network structure not reasonable, etc. When this happens it's often helpful to print out the weights and intermediate outputs to understand what's going on. MXNet provides a monitor utility that does this: | def norm_stat(d):
"""The statistics you want to see.
We compute the L2 norm here but you can change it to anything you like."""
return mx.nd.norm(d)/np.sqrt(d.size)
mon = mx.mon.Monitor(
100, # Print every 100 batches
norm_stat, # The statistics function defined above
pattern='.*weight', # A regular expression. Only arrays with name matching this pattern will be included.
sort=True) # Sort output by name
model = mx.model.FeedForward(ctx = mx.gpu(0), symbol = mlp, num_epoch = 1,
learning_rate = 0.1, momentum = 0.9, wd = 0.00001)
model.fit(X=train_iter, eval_data=test_iter, monitor=mon, # Set the monitor here
batch_end_callback = mx.callback.Speedometer(100, 100)) | INFO:root:Start training with [gpu(0)]
INFO:root:Batch: 1 fc1_backward_weight 0.000519617
INFO:root:Batch: 1 fc1_weight 0.00577777
INFO:root:Batch: 1 fc2_backward_weight 0.00164324
INFO:root:Batch: 1 fc2_weight 0.00577121
INFO:root:Batch: 1 fc3_backward_weight 0.00490826
INFO:root:Batch: 1 fc3_weight 0.00581168
INFO:root:Epoch[0] Batch [100] Speed: 56125.81 samples/sec Train-accuracy=0.141400
INFO:root:Batch: 101 fc1_backward_weight 0.170696
INFO:root:Batch: 101 fc1_weight 0.0077417
INFO:root:Batch: 101 fc2_backward_weight 0.300237
INFO:root:Batch: 101 fc2_weight 0.0188219
INFO:root:Batch: 101 fc3_backward_weight 1.26234
INFO:root:Batch: 101 fc3_weight 0.0678799
INFO:root:Epoch[0] Batch [200] Speed: 76573.19 samples/sec Train-accuracy=0.419000
INFO:root:Batch: 201 fc1_backward_weight 0.224993
INFO:root:Batch: 201 fc1_weight 0.0224456
INFO:root:Batch: 201 fc2_backward_weight 0.574649
INFO:root:Batch: 201 fc2_weight 0.0481841
INFO:root:Batch: 201 fc3_backward_weight 1.50356
INFO:root:Batch: 201 fc3_weight 0.223626
INFO:root:Epoch[0] Batch [300] Speed: 82821.98 samples/sec Train-accuracy=0.574900
INFO:root:Batch: 301 fc1_backward_weight 0.128922
INFO:root:Batch: 301 fc1_weight 0.0297723
INFO:root:Batch: 301 fc2_backward_weight 0.25938
INFO:root:Batch: 301 fc2_weight 0.0623646
INFO:root:Batch: 301 fc3_backward_weight 0.623773
INFO:root:Batch: 301 fc3_weight 0.243092
INFO:root:Epoch[0] Batch [400] Speed: 81133.86 samples/sec Train-accuracy=0.662375
INFO:root:Batch: 401 fc1_backward_weight 0.244692
INFO:root:Batch: 401 fc1_weight 0.0343876
INFO:root:Batch: 401 fc2_backward_weight 0.42573
INFO:root:Batch: 401 fc2_weight 0.0708167
INFO:root:Batch: 401 fc3_backward_weight 0.813565
INFO:root:Batch: 401 fc3_weight 0.252606
INFO:root:Epoch[0] Batch [500] Speed: 79695.23 samples/sec Train-accuracy=0.716540
INFO:root:Batch: 501 fc1_backward_weight 0.208892
INFO:root:Batch: 501 fc1_weight 0.0385131
INFO:root:Batch: 501 fc2_backward_weight 0.475372
INFO:root:Batch: 501 fc2_weight 0.0783694
INFO:root:Batch: 501 fc3_backward_weight 0.984594
INFO:root:Batch: 501 fc3_weight 0.2605
INFO:root:Epoch[0] Batch [600] Speed: 78154.25 samples/sec Train-accuracy=0.754600
INFO:root:Epoch[0] Resetting Data Iterator
INFO:root:Epoch[0] Train-accuracy=0.754600
INFO:root:Epoch[0] Time cost=0.831
INFO:root:Epoch[0] Validation-accuracy=0.953200
| Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Under the hood: Custom Training Loop`mx.model.FeedForward` is a convenience wrapper for training standard feed forward networks. What if the model you are working with is more complicated? With MXNet, you can easily control every aspect of training by writing your own training loop.Neural network training typically has 3 steps: forward, backward (gradient), and update. With custom training loop, you can control the details in each step as while as insert complicated computations in between. You can also connect multiple networks together. | # ==================Binding=====================
# The symbol we created is only a graph description.
# To run it, we first need to allocate memory and create an executor by 'binding' it.
# In order to bind a symbol, we need at least two pieces of information: context and input shapes.
# Context specifies which device the executor runs on, e.g. cpu, GPU0, GPU1, etc.
# Input shapes define the executor's input array dimensions.
# MXNet then run automatic shape inference to determine the dimensions of intermediate and output arrays.
# data iterators defines shapes of its output with provide_data and provide_label property.
input_shapes = dict(train_iter.provide_data+train_iter.provide_label)
print 'input_shapes', input_shapes
# We use simple_bind to let MXNet allocate memory for us.
# You can also allocate memory youself and use bind to pass it to MXNet.
exe = mlp.simple_bind(ctx=mx.gpu(0), **input_shapes)
# ===============Initialization=================
# First we get handle to input arrays
arg_arrays = dict(zip(mlp.list_arguments(), exe.arg_arrays))
data = arg_arrays[train_iter.provide_data[0][0]]
label = arg_arrays[train_iter.provide_label[0][0]]
# We initialize the weights with uniform distribution on (-0.01, 0.01).
init = mx.init.Uniform(scale=0.01)
for name, arr in arg_arrays.items():
if name not in input_shapes:
init(name, arr)
# We also need to create an optimizer for updating weights
opt = mx.optimizer.SGD(
learning_rate=0.1,
momentum=0.9,
wd=0.00001,
rescale_grad=1.0/train_iter.batch_size)
updater = mx.optimizer.get_updater(opt)
# Finally we need a metric to print out training progress
metric = mx.metric.Accuracy()
# Training loop begines
for epoch in range(10):
train_iter.reset()
metric.reset()
t = 0
for batch in train_iter:
# Copy data to executor input. Note the [:].
data[:] = batch.data[0]
label[:] = batch.label[0]
# Forward
exe.forward(is_train=True)
# You perform operations on exe.outputs here if you need to.
# For example, you can stack a CRF on top of a neural network.
# Backward
exe.backward()
# Update
for i, pair in enumerate(zip(exe.arg_arrays, exe.grad_arrays)):
weight, grad = pair
updater(i, grad, weight)
metric.update(batch.label, exe.outputs)
t += 1
if t % 100 == 0:
print 'epoch:', epoch, 'iter:', t, 'metric:', metric.get()
| input_shapes {'softmax_label': (100,), 'data': (100, 784)}
epoch: 0 iter: 100 metric: ('accuracy', 0.1427)
epoch: 0 iter: 200 metric: ('accuracy', 0.42695)
epoch: 0 iter: 300 metric: ('accuracy', 0.5826333333333333)
epoch: 0 iter: 400 metric: ('accuracy', 0.66875)
epoch: 0 iter: 500 metric: ('accuracy', 0.72238)
epoch: 0 iter: 600 metric: ('accuracy', 0.7602166666666667)
epoch: 1 iter: 100 metric: ('accuracy', 0.9504)
epoch: 1 iter: 200 metric: ('accuracy', 0.9515)
epoch: 1 iter: 300 metric: ('accuracy', 0.9547666666666667)
epoch: 1 iter: 400 metric: ('accuracy', 0.95665)
epoch: 1 iter: 500 metric: ('accuracy', 0.95794)
epoch: 1 iter: 600 metric: ('accuracy', 0.95935)
epoch: 2 iter: 100 metric: ('accuracy', 0.9657)
epoch: 2 iter: 200 metric: ('accuracy', 0.96715)
epoch: 2 iter: 300 metric: ('accuracy', 0.9698)
epoch: 2 iter: 400 metric: ('accuracy', 0.9702)
epoch: 2 iter: 500 metric: ('accuracy', 0.97104)
epoch: 2 iter: 600 metric: ('accuracy', 0.9717)
epoch: 3 iter: 100 metric: ('accuracy', 0.976)
epoch: 3 iter: 200 metric: ('accuracy', 0.97575)
epoch: 3 iter: 300 metric: ('accuracy', 0.9772666666666666)
epoch: 3 iter: 400 metric: ('accuracy', 0.9771)
epoch: 3 iter: 500 metric: ('accuracy', 0.9771)
epoch: 3 iter: 600 metric: ('accuracy', 0.97755)
epoch: 4 iter: 100 metric: ('accuracy', 0.9805)
epoch: 4 iter: 200 metric: ('accuracy', 0.9803)
epoch: 4 iter: 300 metric: ('accuracy', 0.9814666666666667)
epoch: 4 iter: 400 metric: ('accuracy', 0.981175)
epoch: 4 iter: 500 metric: ('accuracy', 0.98132)
epoch: 4 iter: 600 metric: ('accuracy', 0.98145)
epoch: 5 iter: 100 metric: ('accuracy', 0.9837)
epoch: 5 iter: 200 metric: ('accuracy', 0.98365)
epoch: 5 iter: 300 metric: ('accuracy', 0.9835333333333334)
epoch: 5 iter: 400 metric: ('accuracy', 0.98395)
epoch: 5 iter: 500 metric: ('accuracy', 0.984)
epoch: 5 iter: 600 metric: ('accuracy', 0.9842166666666666)
epoch: 6 iter: 100 metric: ('accuracy', 0.9848)
epoch: 6 iter: 200 metric: ('accuracy', 0.98475)
epoch: 6 iter: 300 metric: ('accuracy', 0.9858333333333333)
epoch: 6 iter: 400 metric: ('accuracy', 0.98555)
epoch: 6 iter: 500 metric: ('accuracy', 0.9855)
epoch: 6 iter: 600 metric: ('accuracy', 0.9856333333333334)
epoch: 7 iter: 100 metric: ('accuracy', 0.9842)
epoch: 7 iter: 200 metric: ('accuracy', 0.98625)
epoch: 7 iter: 300 metric: ('accuracy', 0.9869)
epoch: 7 iter: 400 metric: ('accuracy', 0.9877)
epoch: 7 iter: 500 metric: ('accuracy', 0.98774)
epoch: 7 iter: 600 metric: ('accuracy', 0.9875333333333334)
epoch: 8 iter: 100 metric: ('accuracy', 0.9864)
epoch: 8 iter: 200 metric: ('accuracy', 0.9878)
epoch: 8 iter: 300 metric: ('accuracy', 0.9886666666666667)
epoch: 8 iter: 400 metric: ('accuracy', 0.98885)
epoch: 8 iter: 500 metric: ('accuracy', 0.98918)
epoch: 8 iter: 600 metric: ('accuracy', 0.9894666666666667)
epoch: 9 iter: 100 metric: ('accuracy', 0.9884)
epoch: 9 iter: 200 metric: ('accuracy', 0.98855)
epoch: 9 iter: 300 metric: ('accuracy', 0.9894666666666667)
epoch: 9 iter: 400 metric: ('accuracy', 0.98945)
epoch: 9 iter: 500 metric: ('accuracy', 0.98972)
epoch: 9 iter: 600 metric: ('accuracy', 0.9899333333333333)
| Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
New OperatorsMXNet provides a repository of common operators (or layers). However, new models often require new layers. There are several ways to [create new operators](https://mxnet.readthedocs.org/en/latest/tutorial/new_op_howto.html) with MXNet. Here we talk about the easiest way: pure python. | # Define custom softmax operator
class NumpySoftmax(mx.operator.NumpyOp):
def __init__(self):
# Call the parent class constructor.
# Because NumpySoftmax is a loss layer, it doesn't need gradient input from layers above.
super(NumpySoftmax, self).__init__(need_top_grad=False)
def list_arguments(self):
# Define the input to NumpySoftmax.
return ['data', 'label']
def list_outputs(self):
# Define the output.
return ['output']
def infer_shape(self, in_shape):
# Calculate the dimensions of the output (and missing inputs) from (some) input shapes.
data_shape = in_shape[0] # shape of first argument 'data'
label_shape = (in_shape[0][0],) # 'label' should be one dimensional and has batch_size instances.
output_shape = in_shape[0] # 'output' dimension is the same as the input.
return [data_shape, label_shape], [output_shape]
def forward(self, in_data, out_data):
x = in_data[0] # 'data'
y = out_data[0] # 'output'
# Compute softmax
y[:] = np.exp(x - x.max(axis=1).reshape((x.shape[0], 1)))
y /= y.sum(axis=1).reshape((x.shape[0], 1))
def backward(self, out_grad, in_data, out_data, in_grad):
l = in_data[1] # 'label'
l = l.reshape((l.size,)).astype(np.int) # cast to int
y = out_data[0] # 'output'
dx = in_grad[0] # gradient for 'data'
# Compute gradient
dx[:] = y
dx[np.arange(l.shape[0]), l] -= 1.0
numpy_softmax = NumpySoftmax()
data = mx.symbol.Variable('data')
fc1 = mx.symbol.FullyConnected(data = data, name='fc1', num_hidden=128)
act1 = mx.symbol.Activation(data = fc1, name='relu1', act_type="relu")
fc2 = mx.symbol.FullyConnected(data = act1, name = 'fc2', num_hidden = 64)
act2 = mx.symbol.Activation(data = fc2, name='relu2', act_type="relu")
fc3 = mx.symbol.FullyConnected(data = act2, name='fc3', num_hidden=10)
# Use the new operator we just defined instead of the standard softmax operator.
mlp = numpy_softmax(data=fc3, name = 'softmax')
model = mx.model.FeedForward(ctx = mx.gpu(0), symbol = mlp, num_epoch = 2,
learning_rate = 0.1, momentum = 0.9, wd = 0.00001)
model.fit(X=train_iter, eval_data=test_iter,
batch_end_callback = mx.callback.Speedometer(100, 100)) | INFO:root:Start training with [gpu(0)]
INFO:root:Epoch[0] Batch [100] Speed: 53975.81 samples/sec Train-accuracy=0.167800
INFO:root:Epoch[0] Batch [200] Speed: 75720.80 samples/sec Train-accuracy=0.455800
INFO:root:Epoch[0] Batch [300] Speed: 73701.82 samples/sec Train-accuracy=0.602833
INFO:root:Epoch[0] Batch [400] Speed: 65162.74 samples/sec Train-accuracy=0.684375
INFO:root:Epoch[0] Batch [500] Speed: 65920.09 samples/sec Train-accuracy=0.735120
INFO:root:Epoch[0] Batch [600] Speed: 67870.31 samples/sec Train-accuracy=0.770333
INFO:root:Epoch[0] Resetting Data Iterator
INFO:root:Epoch[0] Train-accuracy=0.770333
INFO:root:Epoch[0] Time cost=0.923
INFO:root:Epoch[0] Validation-accuracy=0.950400
INFO:root:Epoch[1] Batch [100] Speed: 54063.96 samples/sec Train-accuracy=0.946700
INFO:root:Epoch[1] Batch [200] Speed: 74701.53 samples/sec Train-accuracy=0.949700
INFO:root:Epoch[1] Batch [300] Speed: 69534.33 samples/sec Train-accuracy=0.953400
INFO:root:Epoch[1] Batch [400] Speed: 76418.05 samples/sec Train-accuracy=0.954875
INFO:root:Epoch[1] Batch [500] Speed: 68825.54 samples/sec Train-accuracy=0.956340
INFO:root:Epoch[1] Batch [600] Speed: 74324.13 samples/sec Train-accuracy=0.958083
INFO:root:Epoch[1] Resetting Data Iterator
INFO:root:Epoch[1] Train-accuracy=0.958083
INFO:root:Epoch[1] Time cost=0.879
INFO:root:Epoch[1] Validation-accuracy=0.957200
| Apache-2.0 | python/moved-from-mxnet/tutorial.ipynb | marktab/mxnet-notebooks |
Challenge 026 - Giant Squid!This challenge is taken from Advent of Code 2021 - Day 4: Giant Squid (https://adventofcode.com/2021/day/4). Problem - Part 1You're already almost 1.5km (almost a mile) below the surface of the ocean, already so deep that you can't see any sunlight. What you can see, however, is a giant squid that has attached itself to the outside of your submarine.Maybe it wants to play bingo?Bingo is played on a set of boards each consisting of a 5x5 grid of numbers. Numbers are chosen at random, and the chosen number is marked on all boards on which it appears. (Numbers may not appear on all boards.) If all numbers in any row or any column of a board are marked, that board wins. (Diagonals don't count.)The submarine has a bingo subsystem to help passengers (currently, you and the giant squid) pass the time. It automatically generates a random order in which to draw numbers and a random set of boards (your puzzle input). For example:```7,4,9,5,11,17,23,2,0,14,21,24,10,16,13,6,15,25,12,22,18,20,8,19,3,26,122 13 17 11 0 8 2 23 4 2421 9 14 16 7 6 10 3 18 5 1 12 20 15 19 3 15 0 2 22 9 18 13 17 519 8 7 25 2320 11 10 24 414 21 16 12 614 21 17 24 410 16 15 9 1918 8 23 26 2022 11 13 6 5 2 0 12 3 7```After the first five numbers are drawn (7, 4, 9, 5, and 11), there are no winners, but the boards are marked as follows (shown here adjacent to each other to save space):```22 13 17 11 0 3 15 0 2 22 14 21 17 24 4 8 2 23 4 24 9 18 13 17 5 10 16 15 9 1921 9 14 16 7 19 8 7 25 23 18 8 23 26 20 6 10 3 18 5 20 11 10 24 4 22 11 13 6 5 1 12 20 15 19 14 21 16 12 6 2 0 12 3 7```After the next six numbers are drawn (17, 23, 2, 0, 14, and 21), there are still no winners:```22 13 17 11 0 3 15 0 2 22 14 21 17 24 4 8 2 23 4 24 9 18 13 17 5 10 16 15 9 1921 9 14 16 7 19 8 7 25 23 18 8 23 26 20 6 10 3 18 5 20 11 10 24 4 22 11 13 6 5 1 12 20 15 19 14 21 16 12 6 2 0 12 3 7```Finally, 24 is drawn:```22 13 17 11 0 3 15 0 2 22 14 21 17 24 4 8 2 23 4 24 9 18 13 17 5 10 16 15 9 1921 9 14 16 7 19 8 7 25 23 18 8 23 26 20 6 10 3 18 5 20 11 10 24 4 22 11 13 6 5 1 12 20 15 19 14 21 16 12 6 2 0 12 3 7``` At this point, the third board wins because it has at least one complete row or column of marked numbers (in this case, the entire top row is marked: 14 21 17 24 4).The score of the winning board can now be calculated. Start by finding the sum of all unmarked numbers on that board; in this case, the sum is 188. Then, multiply that sum by the number that was just called when the board won, 24, to get the final score, 188 * 24 = 4512.To guarantee victory against the giant squid, figure out which board will win first. What will your final score be if you choose that board? Solution - Part 1```To run this script, you need to save input-day-04-numbers.txt and input-day-boards.txt and run it in your local machine.``` | class Board:
def __init__(self):
self.position = {}
self.bingo= {
"column": [0,0,0,0,0],
"row": [0,0,0,0,0]
}
self.playBoard = [
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
]
self.selected_number = []
def populate_board(self, playBoard):
self.playBoard = playBoard
def checkBingo(self):
return 5 in self.bingo["row"] or 5 in self.bingo["column"]
def update_board(self, value):
for idx, item in enumerate(self.playBoard):
if value not in item:
continue
self.bingo["row"][idx] = self.bingo["row"][idx] + 1
self.bingo["column"][item.index(value)] = self.bingo["column"][item.index(value)] + 1
self.selected_number.append(value)
def calculate_final_score(self):
final_score = 0
for item in self.playBoard:
for number in item:
if number in self.selected_number:
continue
final_score = final_score + number
return final_score * self.selected_number.pop()
def __str__(self):
for item in self.playBoard:
print(item)
return ""
# Get sequence of bingo number
def import_input_number():
with open ("input-day-04-numbers.txt", "r") as file:
input_data = file.read()
return input_data
# Populate Bingo playboard
def import_input_board():
with open ("input-day-04-boards.txt", "r") as file:
input_data = file.read().splitlines()
board = []
temp_board = []
item_board = []
for row, val in enumerate(input_data):
if row == len(input_data) - 1:
item_board.append(val)
temp_board.append(item_board)
elif val == '':
temp_board.append(item_board)
item_board = []
else:
item_board.append(val)
for item in temp_board:
temp_playboard = []
for row in item:
value = [ int(number) for number in row.split(" ") if number != ""]
temp_playboard.append(value)
playBoard = Board()
playBoard.populate_board(temp_playboard)
board.append(playBoard)
return board
list_number = list(map(int, import_input_number().split(',')))
list_board = import_input_board()
for number in list_number:
is_bingo = False
for idx, board in enumerate(list_board):
board.update_board(number)
is_bingo = board.checkBingo()
if is_bingo:
print("Board {} BINGO! Final score: {}".format(idx, board.calculate_final_score()))
break
if is_bingo:
break | _____no_output_____ | MIT | challenges/026-Giant_Squid/026-Day04_Giant_Squid.ipynb | jfdaniel77/interview-challenge |
Problem - Part 2On the other hand, it might be wise to try a different strategy: let the giant squid win.You aren't sure how many bingo boards a giant squid could play at once, so rather than waste time counting its arms, the safe thing to do is to figure out which board will win last and choose that one. That way, no matter which boards it picks, it will win for sure.In the above example, the second board is the last to win, which happens after 13 is eventually called and its middle column is completely marked. If you were to keep playing until this point, the second board would have a sum of unmarked numbers equal to 148 for a final score of 148 * 13 = 1924.Figure out which board will win last. Once it wins, what would its final score be? Solution - Part 2```To run this script, you need to save input-day-04-numbers.txt and input-day-boards.txt and run it in your local machine.``` | class Board:
def __init__(self):
self.position = {}
self.bingo= {
"column": [0,0,0,0,0],
"row": [0,0,0,0,0]
}
self.playBoard = [
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
[0,0,0,0,0],
]
self.selected_number = []
self.id = 0
def populate_board(self, idx, playBoard):
self.playBoard = playBoard
self.id = idx
def checkBingo(self):
return 5 in self.bingo["row"] or 5 in self.bingo["column"]
def update_board(self, value):
for idx, item in enumerate(self.playBoard):
if value not in item:
continue
self.bingo["row"][idx] = self.bingo["row"][idx] + 1
self.bingo["column"][item.index(value)] = self.bingo["column"][item.index(value)] + 1
self.selected_number.append(value)
def calculate_final_score(self):
final_score = 0
for item in self.playBoard:
for number in item:
if number in self.selected_number:
continue
final_score = final_score + number
return final_score * self.selected_number.pop()
def __str__(self):
for item in self.playBoard:
print(item)
return ""
# Get sequence of bingo number
def import_input_number():
with open ("input-day-04-numbers.txt", "r") as file:
input_data = file.read()
return input_data
# Populate Bingo playboard
def import_input_board():
with open ("input-day-04-boards.txt", "r") as file:
input_data = file.read().splitlines()
board = []
temp_board = []
item_board = []
for row, val in enumerate(input_data):
if row == len(input_data) - 1:
item_board.append(val)
temp_board.append(item_board)
elif val == '':
temp_board.append(item_board)
item_board = []
else:
item_board.append(val)
for idx, item in enumerate(temp_board):
temp_playboard = []
for row in item:
value = [ int(number) for number in row.split(" ") if number != ""]
temp_playboard.append(value)
playBoard = Board()
playBoard.populate_board(idx, temp_playboard)
board.append(playBoard)
return board
list_number = list(map(int, import_input_number().split(',')))
list_board = import_input_board()
last_board = 0
bingo_board = []
last_number = 0
count_bingo = 0
for number in list_number:
is_stop = False
for idx, board in enumerate(list_board):
if idx in bingo_board:
continue
board.update_board(number)
is_bingo = board.checkBingo()
if is_bingo:
bingo_board.append(idx)
count_bingo = count_bingo + 1
if count_bingo == len(list_board):
last_board = idx
is_stop = True
if is_stop:
last_number = number
break
print("Board {} BINGO! Final score: {}".format(last_board, list_board[last_board].calculate_final_score())) | _____no_output_____ | MIT | challenges/026-Giant_Squid/026-Day04_Giant_Squid.ipynb | jfdaniel77/interview-challenge |
Parallel Processing with Dask====<img src="http://dask.readthedocs.io/en/latest/_images/dask_horizontal.svg" width="30%" align=right alt="Dask logo"> Learning Objectives* get acquanted with the Python Dask Library* learn how to execute basic operations on large arrays which cannot fit in RAM* learn about the concepts of lazy evaluation and task scheduling graphs* learn how to work with Dask Arrays* learn how to work with Dask Delayed MotivationResearchers are overloaded with data which their traditional processing workflows are incapable to handle. Usually they are faced with two possible options:* move the processing to large machines/clusters* modify their methods to access the data only pieces at a time.They also like to test out things on their laptops, and later move to clusters, without having to modify their code a lot.[Dask](https://dask.org/) is a Python Library which makes this possible:* can perform computations on data which cannot fit into RAM* has interface similar to `numpy` and `scipy`, and lives under the hood of `xarray`* the same code used on your laptop can be run on a distributed cluster _Note: Pieces of this notebook comes from the following sources:_- https://github.com/rabernat/research_computing- https://github.com/dask/dask-examples Start a Dask distributed cluster and a Client for DashboardWe can imitate starting a cluster on a local machine. Thus we can use the same code on a cluster. |
from dask.distributed import Client
client = Client(processes=False)
client
| _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
The distributed scheduler provides nice diagnostic tools which are useful to gain insight on the computation. They can reveal processing bottlenecks and are useful when running a scalable cluster (like kubernetes) and monitoring nodes. | # If we have set up a Kubernetes cluster we can start it in the following way:
#from dask_kubernetes import KubeCluster
#cluster = KubeCluster()
#cluster.scale(4)
#cluster | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Dask ArraysA dask array looks and feels a lot like a numpy array.However, a dask array doesn't directly hold any data.Instead, it symbolically represents the computations needed to generate the data.Nothing is actually computed until the actual numerical values are needed.This mode of operation is called "lazy"; it allows one to build up complex, large calculations symbolically before turning them over the scheduler for execution.If we want to create a numpy array of all ones, we do it like this: | import numpy as np
shape = (1000, 4000)
ones_np = np.ones(shape)
ones_np | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
This size of the array is: | print('%.1f MB' % (ones_np.nbytes / 1e6)) | 32.0 MB
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Now let's create the same array using dask's array interface. | import dask.array as da
ones = da.ones(shape)
ones | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
This works, but we didn't tell dask how to split up the array, so it is not optimized for distributed computation.A crucal difference with dask is that we must specify the `chunks` argument. "Chunks" describes how the array is split up over many sub-arrays._source: [Dask Array Documentation](http://dask.pydata.org/en/latest/array-overview.html)_There are [several ways to specify chunks](http://dask.pydata.org/en/latest/array-creation.htmlchunks).In this lecture, we will use a block shape. | chunk_shape = (1000, 1000)
ones = da.ones(shape, chunks=chunk_shape)
ones | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Notice that we just see a symbolic represetnation of the array, including its shape, dtype, and chunksize.No data has been generated yet.When we call `.compute()` on a dask array, the computation is trigger and the dask array becomes a numpy array. | ones.compute()
| _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Task Graphs In order to understand what happened when we called `.compute()`, we can visualize the dask _graph_, the symbolic operations that make up the array | ones.visualize() | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Our array has four chunks. To generate it, dask calls `np.ones` four times and then concatenates this together into one array.Rather than immediately loading a dask array (which puts all the data into RAM), it is more common to reduce the data somehow. For example: | sum_of_ones = ones.sum()
sum_of_ones.visualize() | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Here we see dask's strategy for finding the sum. This simple example illustrates the beauty of dask: it automatically designs an algorithm appropriate for custom operations with big data. If we make our operation more complex, the graph gets more complex. | fancy_calculation = (ones * ones[::-1, ::-1]).mean()
fancy_calculation.visualize() | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
A Bigger CalculationThe examples above were toy examples; the data (32 MB) is nowhere nearly big enough to warrant the use of dask.We can make it a lot bigger! | bigshape = (100000, 4000)
big_ones = da.ones(bigshape, chunks=chunk_shape)
big_ones
print('%.1f MB' % (big_ones.nbytes / 1e6)) | 3200.0 MB
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
This dataset is 6.4 GB, rather than 32 MB! This is probably close to or greater than the amount of available RAM than you have in your computer. Nevertheless, dask has no problem working on it._Do not try to `.visualize()` this array!_When doing a big calculation, dask also has some tools to help us understand what is happening under the hood. Let's watch the dashboard again as we do a bigger computation. | pip install bokeh
!jupyter nbextension enable --py widgetsnbextension
big_calc = (big_ones * big_ones[::-1, ::-1]).mean()
from dask.distributed import get_task_stream
with get_task_stream(filename="task-stream.html",plot=True) as ts:
big_calc.compute()
#client.profile(filename="dask-profile.html")
from bokeh.plotting import output_notebook
output_notebook()
from bokeh.plotting import show
ts.figure.plot_height=400
show(ts.figure)
#import IPython
#IPython.display.HTML(filename='task-stream.html')
#IPython.display.HTML(filename='dask-profile.html') | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Reduction All the usual numpy methods work on dask arrays.You can also apply numpy function directly to a dask array, and it will stay lazy. | big_ones_reduce = (np.cos(big_ones)**2).mean(axis=1)
big_ones_reduce
from matplotlib import pyplot as plt
%matplotlib inline
plt.rcParams['figure.figsize'] = (12,8) | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Plotting also triggers computation, since we need the actual values | plt.plot(big_ones_reduce)
| _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Dask DelayedDask.delayed is a simple and powerful way to parallelize existing code. It allows users to delay function calls into a task graph with dependencies. Dask.delayed doesn't provide any fancy parallel algorithms like Dask.dataframe, but it does give the user complete control over what they want to build.Systems like Dask.dataframe are built with Dask.delayed. If you have a problem that is paralellizable, but isn't as simple as just a big array or a big dataframe, then dask.delayed may be the right choice for you. Create simple functionsThese functions do simple operations like add two numbers together, but they sleep for a random amount of time to simulate real work. | import time
def inc(x):
time.sleep(0.1)
return x + 1
def dec(x):
time.sleep(0.1)
return x - 1
def add(x, y):
time.sleep(0.2)
return x + y | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
We can run them like normal Python functions below | %%time
x = inc(1)
y = dec(2)
z = add(x, y)
z | CPU times: user 64.8 ms, sys: 11.4 ms, total: 76.2 ms
Wall time: 401 ms
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
These ran one after the other, in sequence. Note though that the first two lines `inc(1)` and `dec(2)` don't depend on each other, we *could* have called them in parallel had we been clever. Annotate functions with Dask Delayed to make them lazyWe can call `dask.delayed` on our funtions to make them lazy. Rather than compute their results immediately, they record what we want to compute as a task into a graph that we'll run later on parallel hardware. | import dask
inc = dask.delayed(inc)
dec = dask.delayed(dec)
add = dask.delayed(add) | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Calling these lazy functions is now almost free. We're just constructing a graph | %%time
x = inc(1)
y = dec(2)
z = add(x, y)
z | CPU times: user 367 µs, sys: 0 ns, total: 367 µs
Wall time: 335 µs
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Visualize computation | z.visualize(rankdir='LR') | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Run in parallelCall `.compute()` when you want your result as a normal Python objectIf you started `Client()` above then you may want to watch the status page during computation. | %%time
z.compute() | CPU times: user 74.6 ms, sys: 7.71 ms, total: 82.3 ms
Wall time: 323 ms
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
Parallelize Normal Python codeNow we use Dask in normal for-loopy Python code. This generates graphs instead of doing computations directly, but still looks like the code we had before. Dask is a convenient way to add parallelism to existing workflows. | %%time
zs = []
for i in range(256):
x = inc(i)
y = dec(x)
z = add(x, y)
zs.append(z)
zs = dask.persist(*zs) # trigger computation in the background | CPU times: user 147 ms, sys: 16.9 ms, total: 164 ms
Wall time: 167 ms
| CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
In general `dask.delayed` is useful when the output of the individual parallel tasks are in a dask format (like dask.array) and are intended to be concatenated in one big dask object. Dask SchedulersThe Dask *Schedulers* orchestrate the tasks in the Task Graphs so that they can be run in parallel. *How* they run in parallel, though, is determined by which *Scheduler* you choose.There are 3 *local* schedulers:- **Single-Thread Local:** For debugging, profiling, and diagnosing issues- **Multi-threaded:** Using the Python built-in `threading` package (the default for all Dask operations except `Bags`)- **Multi-process:** Using the Python built-in `multiprocessing` package (the default for Dask `Bags`)and 1 *distributed* scheduler, which we will talk about later:- **Distributed:** Using the `dask.distributed` module (which uses `tornado` for TCP communication). The distributed scheduler uses a `Cluster` to manage communication between the scheduler and the "workers". This is described in the next section. Distributed Clusters (http://distributed.dask.org/)Dask can be deployed on distributed infrastructure, such as a an HPC system or a cloud computing system.- `LocalCluster` - Creates a `Cluster` that can be executed locally. Each `Cluster` includes a `Scheduler` and `Worker`s. - `Client` - Connects to and drives computation on a distributed `Cluster` Dask Jobqueue (http://jobqueue.dask.org/)- `PBSCluster`- `SlurmCluster`- `LSFCluster`- etc. Dask Kubernetes (http://kubernetes.dask.org/)- `KubeCluster` | _____no_output_____ | CC-BY-4.0 | notebooks/2_dask_colab.ipynb | oceanhackweek/Oceans19-data-science-tutorial |
|
多层感知机 --- 使用Gluon我们只需要稍微改动[多类Logistic回归](../chapter_crashcourse/softmax-regression-gluon.md)来实现多层感知机。 定义模型唯一的区别在这里,我们加了一行进来。 | from mxnet import gluon
net = gluon.nn.Sequential()
with net.name_scope():
net.add(gluon.nn.Flatten())
net.add(gluon.nn.Dense(256, activation="relu"))
net.add(gluon.nn.Dense(10))
net.initialize() | _____no_output_____ | Apache-2.0 | chapter_supervised-learning/mlp-gluon.ipynb | kyoyo/gluon_tutorials_zh_git |
读取数据并训练 | import sys
sys.path.append('..')
from mxnet import ndarray as nd
from mxnet import autograd
import utils
batch_size = 256
train_data, test_data = utils.load_data_fashion_mnist(batch_size)
softmax_cross_entropy = gluon.loss.SoftmaxCrossEntropyLoss()
trainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': 0.5})
for epoch in range(5):
train_loss = 0.
train_acc = 0.
for data, label in train_data:
with autograd.record():
output = net(data)
loss = softmax_cross_entropy(output, label)
loss.backward()
trainer.step(batch_size)
train_loss += nd.mean(loss).asscalar()
train_acc += utils.accuracy(output, label)
test_acc = utils.evaluate_accuracy(test_data, net)
print("Epoch %d. Loss: %f, Train acc %f, Test acc %f" % (
epoch, train_loss/len(train_data), train_acc/len(train_data), test_acc)) | Epoch 0. Loss: 0.694022, Train acc 0.745893, Test acc 0.817508
| Apache-2.0 | chapter_supervised-learning/mlp-gluon.ipynb | kyoyo/gluon_tutorials_zh_git |
Direct Links1. [To get top 10 by genre](best_genre)2. [To get top 10 similar users](top_ten) | # Reading the ratings data
ratings = pd.read_csv('Dataset/ratings.csv')
len(ratings)
#Just taking the required columns
ratings = ratings[['userId', 'movieId','rating']]
# Checking if the user has rated the same movie twice, in that case we just take max of them
ratings_df = ratings.groupby(['userId','movieId']).aggregate(np.max)
# In this case there are no such cases where the user has rated the same movie twice.
len(ratings_df)
# Inspecting the data
ratings.head()
ratings_df.head()
# Counting no of unique users
len(ratings['userId'].unique())
#Getting the percentage count of each rating value
count_ratings = ratings.groupby('rating').count()
count_ratings['perc_total']=round(count_ratings['userId']*100/count_ratings['userId'].sum(),1)
count_ratings
#Visualising the percentage total for each rating
count_ratings['perc_total'].plot.bar()
#reading the movies dataset
movie_list = pd.read_csv('Dataset/movies.csv')
len(movie_list)
# insepcting the movie list dataframe
movie_list.head()
# reading the tags datast
tags = pd.read_csv('Dataset/tags.csv')
# inspecting the tags data frame
tags.head()
# inspecting various genres
genres = movie_list['genres']
genres.head()
genre_list = ""
for index,row in movie_list.iterrows():
genre_list += row.genres + "|"
#split the string into a list of values
genre_list_split = genre_list.split('|')
#de-duplicate values
new_list = list(set(genre_list_split))
#remove the value that is blank
new_list.remove('')
#inspect list of genres
new_list
#Enriching the movies dataset by adding the various genres columns.
movies_with_genres = movie_list.copy()
for genre in new_list :
movies_with_genres[genre] = movies_with_genres.apply(lambda _:int(genre in _.genres), axis = 1)
movies_with_genres.head()
#Calculating the sparsity
no_of_users = len(ratings['userId'].unique())
no_of_movies = len(ratings['movieId'].unique())
sparsity = round(1.0 - len(ratings)/(1.0*(no_of_movies*no_of_users)),3)
print(sparsity)
# Counting the number of unique movies in the dataset.
len(ratings['movieId'].unique())
# Finding the average rating for movie and the number of ratings for each movie
avg_movie_rating = pd.DataFrame(ratings.groupby('movieId')['rating'].agg(['mean','count']))
#avg_movie_rating['movieId']= avg_movie_rating.index
# inspecting the average movie rating data frame
avg_movie_rating.head()
len(avg_movie_rating)
#calculate the percentile count. It gives the no of ratings at least 70% of the movies have
np.percentile(avg_movie_rating['count'],70)
#Get the average movie rating across all movies
avg_rating_all=ratings['rating'].mean()
avg_rating_all
#set a minimum threshold for number of reviews that the movie has to have
min_reviews=30
min_reviews
movie_score = avg_movie_rating.loc[avg_movie_rating['count']>min_reviews]
movie_score.head()
len(movie_score)
#create a function for weighted rating score based off count of reviews
def weighted_rating(x, m=min_reviews, C=avg_rating_all):
v = x['count']
R = x['mean']
# Calculation based on the IMDB formula
return (v/(v+m) * R) + (m/(m+v) * C)
#Calculating the weighted score for each movie
movie_score['weighted_score'] = movie_score.apply(weighted_rating, axis=1)
movie_score.head()
#join movie details to movie ratings
movies_with_genres.index.name = None
movies_with_genres = movies_with_genres.rename_axis(None)
movie_score = pd.merge(movie_score,movies_with_genres,on='movieId')
movie_score.head()
#list top scored movies over the whole range of movies
pd.DataFrame(movie_score.sort_values(['weighted_score'],ascending=False)[['title','count','mean','weighted_score','genres']][:10]) | _____no_output_____ | MIT | MovieLens_Recommendation_Notebook-Copy1.ipynb | nikita9604/Movie-Recommendation-Website-based-on-Genre |
Reading movie_score.csv directly | #movie_score.to_csv('movie_score.csv', index = False)
movie_score = pd.read_csv('movie_score.csv')
movie_score.head()
# Gives the best movies according to genre based on weighted score which is calculated using IMDB formula
def best_movies_by_genre(genre,top_n):
return pd.DataFrame(movie_score.loc[(movie_score[genre]==1)].sort_values(['weighted_score'],ascending=False)[['title','count','mean','weighted_score']][:top_n])
#run function to return top recommended movies by genre
best_movies_by_genre('Musical',10)
#run function to return top recommended movies by genre
best_movies_by_genre('Action',10)
#run function to return top recommended movies by genre
best_movies_by_genre('Children',10)
#run function to return top recommended movies by genre
best_movies_by_genre('Drama',10)
# Creating a data frame that has user ratings accross all movies in form of matrix used in matrix factorisation
ratings_df = pd.pivot_table(ratings, index='userId', columns='movieId', aggfunc=np.max)
ratings_df.head()
# Apply low rank matrix factorization to find the latent features
# U, M = matrix_factorization_utilities.low_rank_matrix_factorization(ratings_df.to_numpy(),
# num_features=5,
# regularization_amount=1.0)
ratings_df
#merging ratings and movies dataframes
ratings_movies = pd.merge(ratings,movie_list, on = 'movieId')
ratings_movies.head()
ratings_movies | _____no_output_____ | MIT | MovieLens_Recommendation_Notebook-Copy1.ipynb | nikita9604/Movie-Recommendation-Website-based-on-Genre |
Gets the other top 10 movies which are watched by the people who saw this particular movie | #ratings_movies.to_csv('ratings_movies.csv', index = False)
ratings_movies = pd.read_csv('ratings_movies.csv')
ratings_movies.head()
#Gets the other top 10 movies which are watched by the people who saw this particular movie
def get_other_movies(movie_name):
#get all users who watched a specific movie
df_movie_users_series = ratings_movies.loc[ratings_movies['title']==movie_name]['userId']
#convert to a data frame
df_movie_users = pd.DataFrame(df_movie_users_series,columns=['userId'])
#get a list of all other movies watched by these users
other_movies = pd.merge(df_movie_users,ratings_movies,on='userId')
#get a list of the most commonly watched movies by these other user
other_users_watched = pd.DataFrame(other_movies.groupby('title')['userId'].count()).sort_values('userId',ascending=False)
other_users_watched['perc_who_watched'] = round(other_users_watched['userId']*100/other_users_watched['userId'][0],1)
return other_users_watched[1:11]
# Getting other top 10 movies which are watched by the people who saw 'Gone Girl'
get_other_movies('Gone Girl (2014)')
from sklearn.neighbors import NearestNeighbors
avg_movie_rating.head()
#only include movies with more than 10 ratings
movie_plus_10_ratings = avg_movie_rating.loc[avg_movie_rating['count']>=10]
print(len(movie_plus_10_ratings))
movie_plus_10_ratings
filtered_ratings = pd.merge(movie_plus_10_ratings, ratings, on="movieId")
len(filtered_ratings)
filtered_ratings.head()
#create a matrix table with movieIds on the rows and userIds in the columns.
#replace NAN values with 0
movie_wide = filtered_ratings.pivot(index = 'movieId', columns = 'userId', values = 'rating').fillna(0)
movie_wide.head()
#specify model parameters
model_knn = NearestNeighbors(metric='cosine',algorithm='brute')
#fit model to the data set
model_knn.fit(movie_wide)
#Gets the top 10 nearest neighbours got the movie
def print_similar_movies(movie_name) :
#get the list of user ratings for a specific userId
query_index = movie_list.loc[movie_list['title']==movie_name]['movieId'].dropna().values[0]
query_index_movie_ratings = movie_wide.loc[query_index,:].values.reshape(1,-1)
#get the closest 10 movies and their distances from the movie specified
distances,indices = model_knn.kneighbors(query_index_movie_ratings,n_neighbors = 11)
#write a loop that prints the similar movies for a specified movie.
for i in range(0,len(distances.flatten())):
#get the title of the random movie that was chosen
get_movie = movie_list.loc[movie_list['movieId']==query_index]['title']
#for the first movie in the list i.e closest print the title
if i==0:
print('Recommendations for {0}:\n'.format(get_movie))
else :
#get the indiciees for the closest movies
indices_flat = indices.flatten()[i]
#get the title of the movie
get_movie = movie_list.loc[movie_list['movieId']==movie_wide.iloc[indices_flat,:].name]['title']
#print the movie
print('{0}: {1}, with distance of {2}:'.format(i,get_movie,distances.flatten()[i]))
print_similar_movies('Godfather, The (1972)')
print_similar_movies('Toy Story (1995)')
print_similar_movies('Skyfall (2012)')
movies_with_genres.head()
#Getting the movies list with only genres like Musical and other such columns
movie_content_df_temp = movies_with_genres.copy()
movie_content_df_temp.set_index('movieId')
movie_content_df = movie_content_df_temp.drop(columns = ['movieId','title','genres'])
movie_content_df = movie_content_df.to_numpy()
movie_content_df
# Import linear_kernel
from sklearn.metrics.pairwise import linear_kernel
# Compute the cosine similarity matrix
cosine_sim = linear_kernel(movie_content_df,movie_content_df)
# Similarity of the movies based on the content
cosine_sim
cosine_sim.shape
# from numpy import savez_compressed
# savez_compressed('cosine.npz', cosine_sim)
#savetxt('cosine.csv', cosine_sim, delimiter=',')
#create a series of the movie id and title
indicies = pd.Series(movie_content_df_temp.index, movie_content_df_temp['title'])
indicies
indcs = indicies.to_dict()
rev_ind = {}
for key,val in indcs.items():
rev_ind[val] = key
rev_ind[19338]
# import pickle
# a_file = open("indicies.pkl", "wb")
# pickle.dump(indcs, a_file)
# a_file.close()
indcs['Skyfall (2012)']
# movie_content_df_temp.head()
# movie_content_df_temp.to_csv('mv_cnt_tmp.csv', index = False) | _____no_output_____ | MIT | MovieLens_Recommendation_Notebook-Copy1.ipynb | nikita9604/Movie-Recommendation-Website-based-on-Genre |
Directly getting top 10 movies based on content similarity | movie_content_df_temp = pd.read_csv('mv_cnt_tmp.csv')
movie_content_df_temp.head()
a_file = open("indicies.pkl", "rb")
inds = pickle.load(a_file)
a_file.close()
inds['Skyfall (2012)']
from numpy import load
data_dict = load('cosine.npz')
cosine_sim = data_dict['arr_0']
cosine_sim
cosine_sim.shape
#Gets the top 10 similar movies based on the content
def get_similar_movies_based_on_content(movie_name) :
movie_index = inds[movie_name]
sim_scores = list(enumerate(cosine_sim[movie_index]))
# Sort the movies based on the similarity scores
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
# Get the scores of the 10 most similar movies
sim_scores = sim_scores[0:11]
print(sim_scores)
# Get the movie indices
movie_indices = [i[0] for i in sim_scores]
if(movie_index in movie_indices):
movie_indices.remove(movie_index)
print(movie_indices)
similar_movies = pd.DataFrame(movie_content_df_temp[['title','genres']].iloc[movie_indices])
return similar_movies[:10]
#indicies["Skyfall (2012)"]
get_similar_movies_based_on_content('Skyfall (2012)')
get_similar_movies_based_on_content('Jumanji (1995)')
#get ordered list of movieIds
item_indices = pd.DataFrame(sorted(list(set(ratings['movieId']))),columns=['movieId'])
#add in data frame index value to data frame
item_indices['movie_index']=item_indices.index
#inspect data frame
item_indices.head()
#get ordered list of userIds
user_indices = pd.DataFrame(sorted(list(set(ratings['userId']))),columns=['userId'])
#add in data frame index value to data frame
user_indices['user_index']=user_indices.index
#inspect data frame
user_indices.head()
ratings.head()
#join the movie indices
df_with_index = pd.merge(ratings,item_indices,on='movieId')
#join the user indices
df_with_index=pd.merge(df_with_index,user_indices,on='userId')
#inspec the data frame
df_with_index.head()
#import train_test_split module
from sklearn.model_selection import train_test_split
#take 80% as the training set and 20% as the test set
df_train, df_test= train_test_split(df_with_index,test_size=0.2)
print(len(df_train))
print(len(df_test))
df_train.head()
df_test.head()
n_users = ratings.userId.unique().shape[0]
n_items = ratings.movieId.unique().shape[0]
print(n_users)
print(n_items) | 7120
14026
| MIT | MovieLens_Recommendation_Notebook-Copy1.ipynb | nikita9604/Movie-Recommendation-Website-based-on-Genre |
User_index is row and Movie_index is column and value is rating | #Create two user-item matrices, one for training and another for testing
train_data_matrix = np.zeros((n_users, n_items))
#for every line in the data
for line in df_train.itertuples():
#set the value in the column and row to
#line[1] is userId, line[2] is movieId and line[3] is rating, line[4] is movie_index and line[5] is user_index
train_data_matrix[line[5], line[4]] = line[3]
train_data_matrix.shape
#Create two user-item matrices, one for training and another for testing
test_data_matrix = np.zeros((n_users, n_items))
#for every line in the data
for line in df_test[:1].itertuples():
#set the value in the column and row to
#line[1] is userId, line[2] is movieId and line[3] is rating, line[4] is movie_index and line[5] is user_index
#print(line[2])
test_data_matrix[line[5], line[4]] = line[3]
#train_data_matrix[line['movieId'], line['userId']] = line['rating']
test_data_matrix.shape
pd.DataFrame(train_data_matrix).head()
df_train['rating'].max()
from sklearn.metrics import mean_squared_error
from math import sqrt
def rmse(prediction, ground_truth):
#select prediction values that are non-zero and flatten into 1 array
prediction = prediction[ground_truth.nonzero()].flatten()
#select test values that are non-zero and flatten into 1 array
ground_truth = ground_truth[ground_truth.nonzero()].flatten()
#return RMSE between values
return sqrt(mean_squared_error(prediction, ground_truth))
#Calculate the rmse sscore of SVD using different values of k (latent features)
from scipy.sparse.linalg import svds
rmse_list = []
for i in [1,2,5,20,40,60,100,200]:
#apply svd to the test data
u,s,vt = svds(train_data_matrix,k=i)
#get diagonal matrix
s_diag_matrix=np.diag(s)
#predict x with dot product of u s_diag and vt
X_pred = np.dot(np.dot(u,s_diag_matrix),vt)
#calculate rmse score of matrix factorisation predictions
rmse_score = rmse(X_pred,test_data_matrix)
rmse_list.append(rmse_score)
print("Matrix Factorisation with " + str(i) +" latent features has a RMSE of " + str(rmse_score))
#Convert predictions to a DataFrame
mf_pred = pd.DataFrame(X_pred)
mf_pred.head()
df_names = pd.merge(ratings,movie_list,on='movieId')
df_names.head()
#choose a user ID
user_id = 1
#get movies rated by this user id
users_movies = df_names.loc[df_names["userId"]==user_id]
#print how many ratings user has made
print("User ID : " + str(user_id) + " has already rated " + str(len(users_movies)) + " movies")
#list movies that have been rated
users_movies
user_index = df_train.loc[df_train["userId"]==user_id]['user_index'][:1].values[0]
#get movie ratings predicted for this user and sort by highest rating prediction
sorted_user_predictions = pd.DataFrame(mf_pred.iloc[user_index].sort_values(ascending=False))
#rename the columns
sorted_user_predictions.columns=['ratings']
#save the index values as movie id
sorted_user_predictions['movieId']=sorted_user_predictions.index
print("Top 10 predictions for User " + str(user_id))
#display the top 10 predictions for this user
pd.merge(sorted_user_predictions,movie_list, on = 'movieId')[:10]
#count number of unique users
numUsers = df_train.userId.unique().shape[0]
#count number of unitque movies
numMovies = df_train.movieId.unique().shape[0]
print(len(df_train))
print(numUsers)
print(numMovies)
#Separate out the values of the df_train data set into separate variables
Users = df_train['userId'].values
Movies = df_train['movieId'].values
Ratings = df_train['rating'].values
print(Users),print(len(Users))
print(Movies),print(len(Movies))
print(Ratings),print(len(Ratings))
#import libraries
import tensorflow as tf
from tensorflow import keras
from keras.layers import Embedding, Reshape
from keras.models import Sequential
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.utils import plot_model
# Couting no of unique users and movies
len(ratings.userId.unique()), len(ratings.movieId.unique())
# Assigning a unique value to each user and movie in range 0,no_of_users and 0,no_of_movies respectively.
ratings.userId = ratings.userId.astype('category').cat.codes.values
ratings.movieId = ratings.movieId.astype('category').cat.codes.values
# Splitting the data into train and test.
train, test = train_test_split(ratings, test_size=0.2)
train.head()
test.head()
n_users, n_movies = len(ratings.userId.unique()), len(ratings.movieId.unique())
# Returns a neural network model which performs matrix factorisation
def matrix_factorisation_model_with_n_latent_factors(n_latent_factors) :
movie_input = keras.layers.Input(shape=[1],name='Item')
movie_embedding = keras.layers.Embedding(n_movies + 1, n_latent_factors, name='Movie-Embedding')(movie_input)
movie_vec = keras.layers.Flatten(name='FlattenMovies')(movie_embedding)
user_input = keras.layers.Input(shape=[1],name='User')
user_vec = keras.layers.Flatten(name='FlattenUsers')(keras.layers.Embedding(n_users + 1, n_latent_factors,name='User-Embedding')(user_input))
prod = keras.layers.dot([movie_vec, user_vec], axes=1)
model = keras.Model([user_input, movie_input], prod)
model.compile('adam', 'mean_squared_error')
return model
model = matrix_factorisation_model_with_n_latent_factors(20)
model.summary()
#Training the model
history = model.fit([train.userId, train.movieId], train.rating, epochs=10, verbose=1)
y_hat = np.round(model.predict([test.userId, test.movieId]),0)
y_true = test.rating
from sklearn.metrics import mean_absolute_error
mean_absolute_error(y_true, y_hat)
#Getting summary of movie embeddings
movie_embedding_learnt = model.get_layer(name='Movie-Embedding').get_weights()[0]
pd.DataFrame(movie_embedding_learnt).describe()
# Getting summary of user embeddings from the model
user_embedding_learnt = model.get_layer(name='User-Embedding').get_weights()[0]
pd.DataFrame(user_embedding_learnt).describe()
from keras.constraints import non_neg
# Returns a neural network model which performs matrix factorisation with additional constraint on embeddings(that they can't be negative)
def matrix_factorisation_model_with_n_latent_factors_and_non_negative_embedding(n_latent_factors) :
movie_input = keras.layers.Input(shape=[1],name='Item')
movie_embedding = keras.layers.Embedding(n_movies + 1, n_latent_factors, name='Non-Negative-Movie-Embedding',embeddings_constraint=non_neg())(movie_input)
movie_vec = keras.layers.Flatten(name='FlattenMovies')(movie_embedding)
user_input = keras.layers.Input(shape=[1],name='User')
user_vec = keras.layers.Flatten(name='FlattenUsers')(keras.layers.Embedding(n_users + 1, n_latent_factors,name='Non-Negative-User-Embedding',embeddings_constraint=non_neg())(user_input))
prod = keras.layers.merge([movie_vec, user_vec], mode='dot',name='DotProduct')
model = keras.Model([user_input, movie_input], prod)
model.compile('adam', 'mean_squared_error')
return model
model2 = matrix_factorisation_model_with_n_latent_factors_and_non_negative_embedding(5)
model2.summary()
history_nonneg = model2.fit([train.userId, train.movieId], train.rating, epochs=50, verbose=0)
movie_embedding_learnt = model2.get_layer(name='Non-Negative-Movie-Embedding').get_weights()[0]
pd.DataFrame(movie_embedding_learnt).describe()
y_hat = np.round(model2.predict([test.userId, test.movieId]),0)
y_true = test.rating
mean_absolute_error(y_true, y_hat)
# Returns a neural network model which does recommendation
def neural_network_model(n_latent_factors_user, n_latent_factors_movie):
movie_input = keras.layers.Input(shape=[1],name='Item')
movie_embedding = keras.layers.Embedding(n_movies + 1, n_latent_factors_movie, name='Movie-Embedding')(movie_input)
movie_vec = keras.layers.Flatten(name='FlattenMovies')(movie_embedding)
movie_vec = keras.layers.Dropout(0.2)(movie_vec)
user_input = keras.layers.Input(shape=[1],name='User')
user_vec = keras.layers.Flatten(name='FlattenUsers')(keras.layers.Embedding(n_users + 1, n_latent_factors_user,name='User-Embedding')(user_input))
user_vec = keras.layers.Dropout(0.2)(user_vec)
concat = keras.layers.merge([movie_vec, user_vec], mode='concat',name='Concat')
concat_dropout = keras.layers.Dropout(0.2)(concat)
dense = keras.layers.Dense(100,name='FullyConnected')(concat)
dropout_1 = keras.layers.Dropout(0.2,name='Dropout')(dense)
dense_2 = keras.layers.Dense(50,name='FullyConnected-1')(concat)
dropout_2 = keras.layers.Dropout(0.2,name='Dropout')(dense_2)
dense_3 = keras.layers.Dense(20,name='FullyConnected-2')(dense_2)
dropout_3 = keras.layers.Dropout(0.2,name='Dropout')(dense_3)
dense_4 = keras.layers.Dense(10,name='FullyConnected-3', activation='relu')(dense_3)
result = keras.layers.Dense(1, activation='relu',name='Activation')(dense_4)
adam = Adam(lr=0.005)
model = keras.Model([user_input, movie_input], result)
model.compile(optimizer=adam,loss= 'mean_absolute_error')
return model
model3 = neural_network_model(10,13)
history_neural_network = model3.fit([train.userId, train.movieId], train.rating, epochs=50, verbose=0)
model3.summary()
y_hat = np.round(model3.predict([test.userId, test.movieId]),0)
y_true = test.rating
mean_absolute_error(y_true, y_hat) | _____no_output_____ | MIT | MovieLens_Recommendation_Notebook-Copy1.ipynb | nikita9604/Movie-Recommendation-Website-based-on-Genre |
CF Part 1 - Data loading and EDA> Collaborative Filtering on MovieLens Latest-small Part 1 - Downloading movielens latest small dataset and exploratory data analysis- toc: false- badges: true- comments: true- categories: [movie, collaborative]- image: | import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import sys
import os
from scipy.sparse import csr_matrix
from sklearn.preprocessing import LabelEncoder
!wget http://files.grouplens.org/datasets/movielens/ml-latest-small.zip
!unzip ml-latest-small.zip
DOWNLOAD_DESTINATION_DIR = "/content/ml-latest-small"
ratings_path = os.path.join(DOWNLOAD_DESTINATION_DIR, 'ratings.csv')
ratings = pd.read_csv(
ratings_path,
sep=',',
names=["userid", "itemid", "rating", "timestamp"],
skiprows=1
)
movies_path = os.path.join(DOWNLOAD_DESTINATION_DIR, 'movies.csv')
movies = pd.read_csv(
movies_path,
sep=',',
names=["itemid", "title", "genres"],
encoding='latin-1',
skiprows=1
)
ratings.head()
movies.head()
print("There are {} users and {} movies in this dataset."\
.format(ratings.userid.nunique(),
ratings.itemid.nunique()))
# histogram of ratings
ratings.groupby('rating').size().plot(kind='bar'); | _____no_output_____ | Apache-2.0 | _notebooks/2021-06-23-collaborative-filtering-movielens-latest-small-01.ipynb | recohut/notebook |
Ratings range from $0.5$ to $5.0$, with a step of $0.5$. The above histogram presents the repartition of ratings in the dataset. the two most commun ratings are $4.0$ and $3.0$ and the less common ratings are $0.5$ and $1.5$ | # average rating of movies
movie_means = ratings.join(movies['title'], on='itemid').groupby('title').rating.mean()
movie_means[:50].plot(kind='bar', grid=True, figsize=(16,6), title="mean ratings of 50 movies");
# 30 most rated movies vs. 30 less rated movies
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(16,4), sharey=True)
movie_means.nlargest(30).plot(kind='bar', ax=ax1, title="Top 30 movies in data set");
movie_means.nsmallest(30).plot(kind='bar', ax=ax2, title="Bottom 30 movies in data set");
def ids_encoder(ratings):
users = sorted(ratings['userid'].unique())
items = sorted(ratings['itemid'].unique())
# create users and items encoders
uencoder = LabelEncoder()
iencoder = LabelEncoder()
# fit users and items ids to the corresponding encoder
uencoder.fit(users)
iencoder.fit(items)
# encode userids and itemids
ratings.userid = uencoder.transform(ratings.userid.tolist())
ratings.itemid = iencoder.transform(ratings.itemid.tolist())
return ratings, uencoder, iencoder
# userids and itemids encoding
ratings, uencoder, iencoder = ids_encoder(ratings)
# transform rating dataframe to matrix
def ratings_matrix(ratings):
return csr_matrix(pd.crosstab(ratings.userid, ratings.itemid, ratings.rating, aggfunc=sum).fillna(0).values)
R = ratings_matrix(ratings)
R[:10,:10].todense()
plt.figure(figsize=(20,10))
plt.imshow(csr_matrix(R).todense(), cmap='hot', interpolation='nearest')
plt.show()
plt.figure(figsize=(5,5))
plt.imshow(csr_matrix(R[:100,:100]).todense(), cmap='hot', interpolation='nearest')
plt.show() | _____no_output_____ | Apache-2.0 | _notebooks/2021-06-23-collaborative-filtering-movielens-latest-small-01.ipynb | recohut/notebook |
multi variable에 대한 linear regression의 코드를 리뷰해보자 | import tensorflow as tf
import numpy as np
tf.set_random_seed(777) # for reproducibility
x1_data = [73., 93., 89., 96., 73.]
x2_data = [80., 88., 91., 98., 66.]
x3_data = [75., 93., 90., 100., 70.]
# Hypothesis / y hat
y_data = [152., 185., 180., 196., 142.]
x1 = tf.placeholder(tf.float32)
x2 = tf.placeholder(tf.float32)
x3 = tf.placeholder(tf.float32)
y = tf.placeholder(tf.float32)
w1 = tf.Variable(tf.random_normal([1]),name='weight_1')
w2 = tf.Variable(tf.random_normal([1]), name='weight_2')
w3 = tf.Variable(tf.random_normal([1]), name='weight_3')
b = tf.Variable(tf.random_normal([1]), name='bias')
y_hat = x1 * w1 + x2 * w2 + x3 * w3 + b
# cost/loss function
loss = tf.reduce_mean(tf.square(y_hat - y)) # sum of the squares
# optimizer
optimizer = tf.train.GradientDescentOptimizer(learning_rate=1e-5)
train = optimizer.minimize(loss)
init = tf.global_variables_initializer()
sess = tf.Session()
sess.run(init)
for step in range(2001):
loss_value, y_hat_value, _ = sess.run([loss, y_hat, train], {x1: x1_data, x2: x2_data, x3: x3_data, y: y_data})
if step % 100 == 0:
print(step, "loss: ", loss_value, "\nPrediction: ", y_hat_value)
# 대충 x1, x2, x3 값을 넣고 y 값을 prediction 해보자
x1_test = [87.]
x2_test = [82.]
x3_test = [91.]
y_hat_value = sess.run([y_hat], {x1: x1_test, x2: x2_test, x3: x3_test})
print("x1 : ", x1_test, ", x2 : ", x2_test, ", x3 : ", x3_test, "\nPrediction: ", y_hat_value[0])
| x1 : [87.0] , x2 : [82.0] , x3 : [91.0]
Prediction: [ 177.55586243]
| MIT | code/multi_variable_linear_regression_01_start.ipynb | zeran4/justdoit |
Find the missing targets | ndat = np.load('more_targets.npy', allow_pickle=True)
flare_table = Table.read('new_flares.tab', format='ascii')
lks = []
for tic in np.unique(flare_table['Target_ID']):
s = search_lightcurve('TIC {}'.format(int(tic)), author='SPOC',
exptime=120, mission='TESS')
d = s[s.year<2020][0].download_all()
lks.append(d)
tcolnames = ['ID','ra', 'dec', 'Teff', 'Tmag', 'd', 'Hmag','Jmag', 'Kmag']
gcolnames = ['teff_val', 'teff_percentile_lower', 'phot_rp_mean_mag', 'phot_bp_mean_mag',
'phot_g_mean_mag', 'bp_rp','lum_val','radius_val']
for i in range(len(lks)):
row_mgun = np.zeros(len(mgun.colnames))
row_allflares = np.zeros(len(allflares.colnames))
output_t = Catalogs.query_region(SkyCoord(lks[i][0].meta['RA_OBJ'],
lks[i][0].meta['DEC_OBJ'], unit=units.deg),
radius=0.1,
catalog='TIC')[0]
for j,k in enumerate([0,2,3,4,5,6,-3,-2,-1]):
row_mgun[k] = output_t[tcolnames[j]]
row_mgun=row_mgun.tolist()
row_mgun[1]=''
row_mgun[7]=''
mgun.add_row(row_mgun)
output_g = Catalogs.query_region(SkyCoord(lks[i][0].meta['RA_OBJ'],
lks[i][0].meta['DEC_OBJ'], unit=units.deg),
radius=0.1,
catalog='Gaia', version=2)[0]
for j,k in enumerate([0,2,3,4,5,6,-8,-7,-6]):
row_allflares[k] = output_t[tcolnames[j]]
for j,k in enumerate([19,20,21,22,31,32,24,23]):
row_allflares[k] = output_g[gcolnames[j]]
for ind in np.where(flare_table['Target_ID']==int(output_t[0]))[0]:
for j,k in enumerate(np.arange(7,13,1)):
row_allflares[k]=flare_table[ind][j]
try:
row_allflares=row_allflares.tolist()
except:
pass
row_allflares[1]=''
row_allflares[7]=''
for n in range(14,18):
row_allflares[n]='True'
allflares.add_row(row_allflares)
xmatch = np.zeros(len(mgun))
for i in range(len(tics)):
i1 = np.where(tics==tics[i])[0]
i2 = np.where(mgun['TIC_ID']==tics[i])[0]
xmatch[i2] = rots[i1]
mgun.add_column(Column(xmatch,'period_days'))
#mgun.add_column(Column(mgun_rots,'period_days'))
sub_mgun = mgun[(mgun['period_days']>0) & (mgun['N_flares']>0)]
len(sub_mgun),len(mgun)
xmatch = np.zeros(len(allflares),dtype=int)
xmatch_rots = np.zeros(len(allflares))
xmatch_time = np.zeros(len(allflares))
xmatch_lum = np.zeros(len(allflares))
xmatch_rate = np.zeros(len(allflares))
for i in range(len(sub_mgun['TIC_ID'])):
ind = np.where(allflares['TIC_ID']==sub_mgun['TIC_ID'][i])[0]
xmatch[ind] = 1
xmatch_rots[ind]=sub_mgun['period_days'][i]
xmatch_time[ind]=sub_mgun['Total_obs_time'][i]
xmatch_lum[ind]=sub_mgun['lum'][i]
xmatch_rate[ind]=sub_mgun['N_flares_per_day'][i]
try:
allflares.add_column(Column(xmatch_rots, 'Prot'))
allflares.add_column(Column(xmatch_time, 'Total_obs_time'))
allflares.add_column(Column(xmatch_rate, 'N_flares_per_day'))
except:
allflares.replace_column('Prot',xmatch_rots)
allflares.replace_column('Total_obs_time',xmatch_time)
allflares.replace_column('N_flares_per_day',xmatch_rate)
subflares = allflares[xmatch==1]
lowlim = subflares[subflares['prob']>=0.99]
medlim = subflares[(subflares['prob']>=0.9)]
upplim = subflares[(subflares['prob']>=0.5)]
mark_tab = Table()
mark_tab.add_column(Column(upplim['TIC_ID'], 'TargetID'))
mark_tab.add_column(Column(upplim['amp'], 'amp'))
mark_tab.add_column(Column(upplim['flare_energy_erg'], 'flare_energy_erg'))
mark_tab.add_column(Column(upplim['Prot'], 'Prot'))
mark_tab.add_column(Column(upplim['prob'], 'flare_probability'))
mark_tab.add_column(Column(upplim['N_flares_per_day'], 'N_flares_per_day'))
mark_tab.write('flare_outputs.csv', format='csv')
medlim.write('medlim.csv',format='csv')
lowlim.write('lowlim.csv',format='csv')
upplim.write('upplim.csv',format='csv') | WARNING: AstropyDeprecationWarning: medlim.csv already exists. Automatically overwriting ASCII files is deprecated. Use the argument 'overwrite=True' in the future. [astropy.io.ascii.ui]
WARNING: AstropyDeprecationWarning: lowlim.csv already exists. Automatically overwriting ASCII files is deprecated. Use the argument 'overwrite=True' in the future. [astropy.io.ascii.ui]
WARNING: AstropyDeprecationWarning: upplim.csv already exists. Automatically overwriting ASCII files is deprecated. Use the argument 'overwrite=True' in the future. [astropy.io.ascii.ui]
| MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Load Tables | medlim = Table.read('medlim.csv',format='csv')
lowlim = Table.read('lowlim.csv',format='csv')
upplim = Table.read('upplim.csv',format='csv') | _____no_output_____ | MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Plot the light curves | outliers = np.unique(medlim[(medlim['amp']>=1) & (medlim['Prot']<3)]['TIC_ID'])
lk = []
tic_tracker=[]
for tic in outliers:
print(tic)
d = search_lightcurve('TIC {}'.format(tic), mission='TESS',
author='SPOC').download_all().stitch()
lk.append(d)
tic_tracker.append(d.meta['TICID'])
megaflares = medlim[(medlim['amp']>2) & (medlim['Prot']<3)]
megaflares.sort('amp')
megaflares.reverse()
plt.rcParams['font.size'] = 22
megaflares
fig, axes = plt.subplots(figsize=(16,25), nrows=5)
fig.set_facecolor('w')
ax = axes.reshape(-1)
inds = [0,1,2,5,6]#,5]
for x,i in enumerate(inds):
ind = np.where(megaflares['TIC_ID'][i]==tic_tracker)[0][0]
print(ind)
for n in [megaflares['tpeak'][i]-2457000]:
left, bottom, width, height = [0.08, 0.45, 0.55, 0.5]
ax2 = ax[x].inset_axes([left, bottom, width, height])
ax2.plot(lk[ind].time.value, lk[ind].flux.value,'k.',ms=1)
ax2.vlines(n+0.01,0,100,lw=10,color=parula[100],alpha=0.4)
ax2.set_ylim(np.nanmin(lk[ind].flux.value),1.08)
if i == 0:
ax2.set_xlim(1438.2,1450.)
ax2.set_xticks(np.round(np.linspace(1438.2,1450.,4),1))
ax2.set_ylim(0.97,1.03)
ax2.set_yticks(np.round(np.linspace(0.98,1.02,3),2))
elif i == 1:
ax2.set_xlim(1328,1338)
ax2.set_xticks(np.round(np.linspace(1328,1338,4),1))
ax2.set_ylim(0.3,1.7)
elif i == 2:
ax2.set_xlim(1339.8,1353)
ax2.set_xticks(np.round(np.linspace(1339.8,1353,4),1))
elif i == 5:
ax2.set_xlim(1630.5,1639.)
ax2.set_xticks(np.round(np.linspace(1630.5,1639.,4),1))
ax2.set_ylim(0.97,1.03)
ax2.set_yticks(np.round(np.linspace(0.98,1.02,3),2))
elif i == 6:
ax2.set_xlim(1371,1381.5)
ax2.set_xticks(np.round(np.linspace(1371,1381.5,4),1))
q = ((lk[ind].time.value>=n-0.8) & (lk[ind].time.value<=n+0.2))
ax[x].plot(lk[ind].time.value[q], lk[ind].flux.value[q],c='k',lw=3)
ax[x].set_xlim(n-0.8,n+0.2)
ax[x].set_xticks(np.round(np.arange(n-0.7,n+0.3,0.2),2))
ax[x].set_xticklabels([str(e) for e in np.round(np.arange(n-0.7,n+0.3,0.2),2)])
ax[x].vlines(n+0.01,0,100,lw=60,color=parula[100],alpha=0.4)
ax[x].set_ylim(np.nanmin(lk[ind].flux.value[q])-0.08,
np.nanmax(lk[ind].flux.value[q])+0.08)
ax[x].set_rasterized(True)
sec = ax[x].secondary_yaxis('right')
sec.set_yticks([])
sec.set_ylabel('\nTIC {}'.format(megaflares['TIC_ID'][i]) +
'\n$P_{rot}$ = ' + str(np.round(megaflares['Prot'][i],2)) + ' days')
ax[-3].set_ylabel('Normalized Flux', fontsize=30)
ax[-1].set_xlabel('Time [BJD - 2457000]', fontsize=30)
plt.savefig('/Users/arcticfox/Desktop/lightcurves.pdf',dpi=300,rasterize=True,
bbox_inches='tight') | 2
4
10
14
9
| MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Rotation period plot | fig = plt.figure(figsize=(8,6))
fig.set_facecolor('w')
plt.scatter(mgun['bp']-mgun['rp'],
mgun['period_days'],
c=mgun['flare_rates'],
vmin=0, vmax=0.5,
cmap=parula_map)
plt.yscale('log')
plt.ylabel('Rotation Period [days]')
plt.xlabel('$B_p - R_p$')
plt.xlim(-1,5)
plt.colorbar(label='Flare Rate [days$^{-1}$]')
#plt.savefig('/Users/arcticfox/Desktop/rots.pdf',dpi=250,rasterize=True,
# bbox_inches='tight')
np.nanmin(mgun['bp']-mgun['rp']), np.nanmax(mgun['bp']-mgun['rp']) | _____no_output_____ | MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Fitting the Flare Frequency Distributions | def slope_fit(x, n, i=0, j=1, plot=False, init=[-1.5,-2],
bounds=((-10.0, 10.0), (-1000, 1000))):
logx = np.log10(x)
logn = np.log10(n)
q = ((np.isnan(logn) == False) & (np.isfinite(logn)==True))
if plot:
plt.plot(logx[i:j], np.log10(n[i:j]), '.', c='k')
plt.plot(logx[i:j], linear(init, logx[i:j]), '--', c='g', linewidth=3)
try:
results = minimize(linear_fit, x0=init,
args=(logx[q][i:j-1]-np.diff(logx[q][i:j])/2.,
logn[q][i:j-1],
np.sqrt(logn[q][i:j-1]) ),
bounds=bounds,
method='L-BFGS-B', tol=1e-8)
results.x[1] = 10**results.x[1]
results2 = leastsq(power_law_resid, results.x,
args=(x[q][i:j-1]-np.diff(x[q][i:j])/2.,
n[q][i:j-1],
np.sqrt(n[q][i:j-1]) ),
full_output=True)
except:
print(len(np.diff(logx[q][i:j])), len(logx[q][i:j-1]))
results = minimize(linear_fit, x0=init,
args=(logx[q][i+1:j-1]-np.diff(logx[q][i:j])/2.,
logn[q][i+1:j-1],
np.sqrt(logn[q][i+1:j-1]) ),
bounds=bounds,
method='L-BFGS-B', tol=1e-8)
results.x[1] = 10**results.x[1]
results2 = leastsq(power_law_resid, results.x,
args=(x[q][i+1:j-1]-np.diff(x[q][i:j])/2.,
n[q][i+1:j-1],
np.sqrt(n[q][i+1:j-1]) ),
full_output=True)
fit_params = results2[0]
slope_err = np.sqrt(results2[1][0][0])
model = linear([fit_params[0], np.log10(fit_params[1])], logx)
if plot:
plt.plot(logx, model, c='r')
plt.title('{} $\pm$ {}'.format(np.round(fit_params[0],2),
np.round(slope_err,2)))
plt.show()
return fit_params[0], slope_err, n, results.x[1], x, 10**model, np.log10(fit_params[1]) | _____no_output_____ | MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Amplitude Binning | bins = np.logspace(np.log10(1), np.log10(500),20)
cut = 3
outslow = []
outfast = []
for t in [medlim, upplim, lowlim]:
os = plt.hist(t[t['Prot']>=cut]['amp']*100,
bins=bins,
weights=np.full(len(t[t['Prot']>=cut]['amp']),
1.0/np.nansum(t[t['Prot']>=cut]['Total_obs_time']*
t[t['Prot']>=cut]['prob'])))
outslow.append(os)
of = plt.hist(t[t['Prot']<cut]['amp']*100,
bins=bins, color=parula[100],
weights=np.full(len(t[t['Prot']<cut]['amp']),
1.0/np.nansum(t[t['Prot']<cut]['Total_obs_time']*
t[t['Prot']<cut]['prob'])))
outfast.append(of)
plt.close()
plt.errorbar((outfast[0][1][1:]+outfast[0][1][:-1])/2,
outfast[0][0],
yerr=(outfast[1][0], outfast[2][0]), marker='o',
linestyle='')
plt.errorbar((outslow[0][1][1:]+outslow[0][1][:-1])/2,
outslow[0][0],
yerr=(outslow[1][0], outslow[2][0]), marker='o',
linestyle='')
plt.yscale('log')
plt.xscale('log')
fitslow, fitfast = [], []
for i in range(len(outslow)):
fl = slope_fit((outslow[i][1][1:]+outslow[i][1][:-1])/2,
outslow[i][0],
i=0,
j=len(outslow[i][0]),
plot=True, init=[0.988,10],
bounds=((0.988-0.0076,0.988+0.0077), (-100, 100)))
ff = slope_fit((outfast[i][1][1:]+outfast[i][1][:-1])/2,
outfast[i][0],
i=0,j=len(outfast[i][0]),
plot=True, init=[0.961,-2],
bounds=((0.961-0.0058,0.961+0.0060), (-100, 100)))
fitslow.append(fl)
fitfast.append(ff)
samples_short = np.load('/Users/arcticfox/Downloads/short_period.npy')
samples_long = np.load('/Users/arcticfox/Downloads/long_period.npy')
slow_dist = np.nanpercentile(samples_long[:,0], [5,50,95])
slow_dist[0]=slow_dist[1]-slow_dist[0]
slow_dist[2]=slow_dist[2]-slow_dist[1]
fast_dist = np.nanpercentile(samples_short[:,0], [5,50,95])
fast_dist[0]=fast_dist[1]-fast_dist[0]
fast_dist[2]=fast_dist[2]-fast_dist[1]
def pdf(a, q, astar):
norm = a0**(q-1) / mp.mpf(expint(q, a0/astar))
return norm * a**-q * np.exp(-a/astar)
a0 = np.amin(mark_tab['amp'])
asamp = np.logspace(np.log10(0.001), np.log10(1000),100)
size = 500
short_samp_pdf = np.zeros((size,len(asamp)))
long_samp_pdf = np.zeros((size,len(asamp)))
schoices = np.random.choice(np.arange(0,len(samples_short),1,dtype=int),
len(samples_short),replace=False)
lchoices = np.random.choice(np.arange(0,len(samples_long),1,dtype=int),
len(samples_long),replace=False)
for i in range(size):
short_samp_pdf[i] = pdf(asamp,
samples_short[schoices[i]][0],
10**samples_short[i][1]*100)
long_samp_pdf[i] = pdf(asamp,
samples_long[lchoices[i]][0],
10**samples_long[i][1]*100)
pdf_short = np.percentile(short_samp_pdf, [2.5,5,16,50,84,95,97.5], axis=0)
pdf_long = np.percentile(long_samp_pdf, [2.5,5,16,50,84,95,97.5], axis=0)
plt.plot(asamp, asamp*pdf_long[1],
'k', lw=3,zorder=4, label=label)
plt.fill_between(asamp,
y1=asamp*pdf_long[0],
y2=asamp*pdf_long[-1],
lw=0, color='k', alpha=0.3, zorder=4)
plt.plot(asamp, asamp*pdf_short[1],
'r', lw=3,zorder=4, label=label)
plt.fill_between(asamp,
y1=asamp*pdf_short[0],
y2=asamp*pdf_short[-1],
lw=0, color='r', alpha=0.3, zorder=4)
#plt.xlim([1e-2,3])
plt.ylim([1e-6,100])
plt.yscale('log')
plt.xscale('log')
fig, (ax2,ax3)=plt.subplots(nrows=2,figsize=(8,10),sharex=True,sharey=True)
fig.set_facecolor('w')
c1=210
c2=100
ax3.hist(medlim[medlim['Prot']>=cut]['amp']*100,
bins=bins, color=parula[c1],
weights=np.full(len(medlim[medlim['Prot']>=cut]['amp']),
1.0/np.nansum(medlim[medlim['Prot']>=cut]['Total_obs_time']*
medlim[medlim['Prot']>=cut]['prob'])),
alpha=0.6)
ax3.errorbar((outslow[0][1][1:]+outslow[0][1][:-1])/2,
outslow[0][0],
yerr=(outslow[1][0], outslow[2][0]), marker='',
linestyle='', color=parula[c1], capsize=4, lw=3,
capthick=3, zorder=3)
label = str(np.round(slow_dist[0],4)) + '$_{-'+str(np.round(slow_dist[1],4))
label += '}^{+'+str(np.round(slow_dist[2],3))+'}$'
label = r"$\alpha$' = " + label
label = r"$\alpha$' = " + str(np.round(slow_dist[1],3)) + '$\pm$' + str(np.round(slow_dist[0],3))
x = np.append(1,(outslow[0][1][1:]+outslow[0][1][:-1])/2)
x = np.append(x,1000)
N = 5
ax3.plot(asamp, pdf_long[3]/N,#*asamp,
'k', lw=3,zorder=4, label=label)
ax3.fill_between(asamp,
y1=pdf_long[0]/N,#*asamp,
y2=pdf_long[-1]/N,#*asamp,
lw=0, color='k', alpha=0.3, zorder=4)
ax3.hist(medlim[medlim['Prot']>=cut]['amp']*100,
bins=bins, color=parula[c1],
weights=np.full(len(medlim[medlim['Prot']>=cut]['amp']),
1.0/np.nansum(medlim[medlim['Prot']>=cut]['Total_obs_time']*
medlim[medlim['Prot']>=cut]['prob'])),
histtype='bar', fill=None,edgecolor=parula[c1],lw=3)
ax3.set_title('P$_{rot} \geq$'+str(cut)+ ' days')
ax2.hist(medlim[medlim['Prot']<cut]['amp']*100,
bins=bins, color=parula[c2],
weights=np.full(len(medlim[medlim['Prot']<cut]['amp']),
1.0/np.nansum(medlim[medlim['Prot']<cut]['Total_obs_time']*
medlim[medlim['Prot']<cut]['prob'])),
alpha=0.6)
ax2.hist(medlim[medlim['Prot']<cut]['amp']*100,
bins=bins, color=parula[c2],
weights=np.full(len(medlim[medlim['Prot']<cut]['amp']),
1.0/np.nansum(medlim[medlim['Prot']<cut]['Total_obs_time']*
medlim[medlim['Prot']<cut]['prob'])),
histtype='bar', fill=None, lw=3, edgecolor=parula[c2])
ax2.errorbar((outfast[0][1][1:]+outfast[0][1][:-1])/2,
outfast[0][0],
yerr=(outfast[1][0], outfast[2][0]), marker='',
linestyle='', color=parula[c2], capsize=4, lw=3,
capthick=3, zorder=3)
label = str(np.round(fast_dist[0],4)) + '$_{-'+str(np.round(fast_dist[1],4))
label += '}^{+'+str(np.round(fast_dist[2],4))+'}$'
label = r"$\alpha$' = " + label
label = r"$\alpha$' = " + str(np.round(fast_dist[1],3)) + '$\pm$' + str(np.round(fast_dist[0],3))
x = np.append(1,(outfast[0][1][1:]+outfast[0][1][:-1])/2)
x = np.append(x,1000)
N = 10
ax2.plot(asamp, pdf_short[3]/N,#*asamp,
'k', lw=3,zorder=4, label=label)
ax2.fill_between(asamp,
y1=pdf_short[0]/N,#*asamp,
y2=pdf_short[-1]/N,#*asamp,
lw=0, color='k', alpha=0.3, zorder=4)
ax2.set_title('P$_{rot} <$'+str(cut)+ ' days')
ax2.set_xscale('log')
ax2.set_yscale('log')
ax3.set_xscale('log')
ax3.set_yscale('log')
ax2.set_ylabel('Flare Rate [day$^{-1}$]')
ax3.set_ylabel('Flare Rate [day$^{-1}$]')
ax3.set_xlabel('Flare Amplitude [%]')
plt.subplots_adjust(hspace=0.3)
plt.xlim(1,500)
plt.ylim(1e-6,1e-2)
ax2.legend()
ax3.legend()
plt.savefig('/Users/arcticfox/Desktop/hist_rots.pdf',
dpi=250,rasterize=True,
bbox_inches='tight') | //anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:94: MatplotlibDeprecationWarning: savefig() got unexpected keyword argument "rasterize" which is no longer supported as of 3.3 and will become an error in 3.6
| MIT | braiding/plotting.ipynb | afeinstein20/flares_soc |
Feature Transformation with Amazon a SageMaker Processing Job and Scikit-LearnIn this notebook, we convert raw text into BERT embeddings. This will allow us to perform natural language processing tasks such as text classification.Typically a machine learning (ML) process consists of few steps. First, gathering data with various ETL jobs, then pre-processing the data, featurizing the dataset by incorporating standard techniques or prior knowledge, and finally training an ML model using an algorithm.Often, distributed data processing frameworks such as Scikit-Learn are used to pre-process data sets in order to prepare them for training. In this notebook we'll use Amazon SageMaker Processing, and leverage the power of Scikit-Learn in a managed SageMaker environment to run our processing workload. NOTE: THIS NOTEBOOK WILL TAKE A 5-10 MINUTES TO COMPLETE. PLEASE BE PATIENT.  Contents1. Setup Environment1. Setup Input Data1. Setup Output Data1. Build a Scikit-Learn container for running the processing job1. Run the Processing Job using Amazon SageMaker1. Inspect the Processed Output Data Setup EnvironmentLet's start by specifying:* The S3 bucket and prefixes that you use for training and model data. Use the default bucket specified by the Amazon SageMaker session.* The IAM role ARN used to give processing and training access to the dataset. | import sagemaker
import boto3
sess = sagemaker.Session()
role = sagemaker.get_execution_role()
bucket = sess.default_bucket()
region = boto3.Session().region_name
sm = boto3.Session().client(service_name="sagemaker", region_name=region)
s3 = boto3.Session().client(service_name="s3", region_name=region) | _____no_output_____ | Apache-2.0 | 06_prepare/02_Prepare_Dataset_BERT_Scikit_ScriptMode_FeatureStore.ipynb | MarcusFra/workshop |
Setup Input Data | %store -r s3_public_path_tsv
try:
s3_public_path_tsv
except NameError:
print("++++++++++++++++++++++++++++++++++++++++++++++++++++++++")
print("[ERROR] Please run the notebooks in the INGEST section before you continue.")
print("++++++++++++++++++++++++++++++++++++++++++++++++++++++++")
print(s3_public_path_tsv)
%store -r s3_private_path_tsv
try:
s3_private_path_tsv
except NameError:
print("++++++++++++++++++++++++++++++++++++++++++++++++++++++++")
print("[ERROR] Please run the notebooks in the INGEST section before you continue.")
print("++++++++++++++++++++++++++++++++++++++++++++++++++++++++")
print(s3_private_path_tsv)
raw_input_data_s3_uri = "s3://{}/amazon-reviews-pds/tsv/".format(bucket)
print(raw_input_data_s3_uri)
!aws s3 ls $raw_input_data_s3_uri | _____no_output_____ | Apache-2.0 | 06_prepare/02_Prepare_Dataset_BERT_Scikit_ScriptMode_FeatureStore.ipynb | MarcusFra/workshop |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.