markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
Hydrogen atom\begin{equation}\label{eq1}-\frac{\hbar^2}{2 \mu} \left[ \frac{1}{r^2} \frac{\partial }{\partial r} \left( r^2 \frac{ \partial \psi}{\partial r}\right) + \frac{1}{r^2 \sin \theta} \frac{\partial }{\partial \theta} \left( \sin \theta \frac{\partial \psi}{\partial \theta}\right) + \frac{1}{r^2 \sin^2 \theta} \frac{\partial^2 \psi}{\partial \phi^2} \right] - \frac{e^2}{ 4 \pi \epsilon_0 r} \psi= E \psi\end{equation}\begin{equation}\label{eqsol1}\psi_{n\ell m}(r,\vartheta,\varphi) = \sqrt {{\left ( \frac{2}{n a^*_0} \right )}^3 \frac{(n-\ell-1)!}{2n(n+\ell)!}} e^{- \rho / 2} \rho^{\ell} L_{n-\ell-1}^{2\ell+1}(\rho) Y_{\ell}^{m}(\vartheta, \varphi ) \end{equation} | import k3d
from ipywidgets import interact, FloatSlider
import numpy as np
import scipy.special
import scipy.misc
r = lambda x,y,z: np.sqrt(x**2+y**2+z**2)
theta = lambda x,y,z: np.arccos(z/r(x,y,z))
phi = lambda x,y,z: np.arctan2(y,x)
a0 = 1.
R = lambda r,n,l: (2*r/(n*a0))**l * np.exp(-r/n/a0) * scipy.special.genlaguerre(n-l-1,2*l+1)(2*r/n/a0)
WF = lambda r,theta,phi,n,l,m: R(r,n,l) * scipy.special.sph_harm(m,l,phi,theta)
absWF = lambda r,theta,phi,n,l,m: abs(WF(r,theta,phi,n,l,m)).astype(np.float32)**2
N = 50j
a = 30.0
x,y,z = np.ogrid[-a:a:N,-a:a:N,-a:a:N]
x = x.astype(np.float32)
y = y.astype(np.float32)
z = z.astype(np.float32)
orbital = WF(r(x,y,z),theta(x,y,z),phi(x,y,z),4,1,0).real.astype(np.float32) # 4p
plt_vol = k3d.volume(orbital)
plt_label = k3d.text2d(r'n=1\; l=0\; m=0',(0.,0.))
plot = k3d.plot()
plot += plt_vol
plot += plt_label
plt_vol.opacity_function = [0. , 0. , 0.21327923, 0.98025 , 0.32439035,
0. , 0.5 , 0. , 0.67560965, 0. ,
0.74537706, 0.9915 , 1. , 0. ]
plt_vol.color_map = k3d.colormaps.paraview_color_maps.Cool_to_Warm_Extended
plt_vol.color_range = (-0.5,0.5)
plot.display() | _____no_output_____ | MIT | atomic_orbitals_wave_function.ipynb | OpenDreamKit/k3d_demo |
animation single wave function is sent at a time | E = 4
for l in range(E):
for m in range(-l,l+1):
psi2 = WF(r(x,y,z),theta(x,y,z),phi(x,y,z),E,l,m).real.astype(np.float32)
plt_vol.volume = psi2/np.max(psi2)
plt_label.text = 'n=%d \quad l=%d \quad m=%d'%(E,l,m)
| _____no_output_____ | MIT | atomic_orbitals_wave_function.ipynb | OpenDreamKit/k3d_demo |
using time series - series of volumetric data are sent to k3d, - player interpolates between | E = 4
psi_t = {}
t = 0.0
for l in range(E):
for m in range(-l,l+1):
psi2 = WF(r(x,y,z),theta(x,y,z),phi(x,y,z),E,l,m)
psi_t[str(t)] = (psi2.real/np.max(np.abs(psi2))).astype(np.float32)
t += 0.3
plt_vol.volume = psi_t
| _____no_output_____ | MIT | atomic_orbitals_wave_function.ipynb | OpenDreamKit/k3d_demo |
 Demo text-mining: Pharma caseIn this demo, I will demonstrate what are the basic steps that you will have to use in most text-mining cases. This are also some of the steps that have been used in the ResuMe app, available here: [ResuMe](https://resume.businessdecision.be/). The case that we will cover here, is a simplified version of a project that has actually been carried out by B&D, where the goal was to identify if a given paper is treating about Pharmacovigilance or not. Pharmacovigilance is a domain of study in healthcare about drug safety. Consequently, we would like to predict, based on the text of the scientific article if the article treats about Pharmacovigilance or not.For this we can use any kind of model, but in any case we will have to transform the words in numbers in some way. We'll see different methods and compare their performance. Downloading the dataset from PubMed | #import documents from PubMed
from Bio import Entrez
# Function to search for a certain number articles based on a certain keyword
def search(keyword,number=20):
Entrez.email = '[email protected]'
handle = Entrez.esearch(db='pubmed',
sort='relevance',
retmax=str(number),
retmode='xml',
term=keyword)
results = Entrez.read(handle)
return results
# Function to retrieve the results of previous search query
def fetch_details(id_list):
ids = ','.join(id_list)
Entrez.email = '[email protected]'
handle = Entrez.efetch(db='pubmed',
retmode='xml',
id=ids)
results = Entrez.read(handle)
return results
| _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Retrieving top 200 articles with Pharmacovigilance keyword | results = search('Pharmacovigilance', 200) #querying PubMed
id_list = results['IdList']
papers_pharmacov = fetch_details(id_list) #retrieving the info about the articles in nested lists & dictionary format
# checking article title for the first 10 retrieved articles
for i, paper in enumerate(papers_pharmacov['PubmedArticle'][:10]):
print("%d) %s" % (i+1,paper['MedlineCitation']['Article']['ArticleTitle'])) | 1) FarmaREL: An Italian pharmacovigilance project to monitor and evaluate adverse drug reactions in haematologic patients.
2) Feasibility and Educational Value of a Student-Run Pharmacovigilance Programme: A Prospective Cohort Study.
3) Developing a Crowdsourcing Approach and Tool for Pharmacovigilance Education Material Delivery.
4) Promoting and Protecting Public Health: How the European Union Pharmacovigilance System Works.
5) Effect of an educational intervention on knowledge and attitude regarding pharmacovigilance and consumer pharmacovigilance among community pharmacists in Lalitpur district, Nepal.
6) Pharmacovigilance and Biomedical Informatics: A Model for Future Development.
7) Pharmacovigilance in Europe: Place of the Pharmacovigilance Risk Assessment Committee (PRAC) in organisation and decisional processes.
8) Tamoxifen Pharmacovigilance: Implications for Safe Use in the Future.
9) Pharmacovigilance Skills, Knowledge and Attitudes in our Future Doctors - A Nationwide Study in the Netherlands.
10) Adverse drug reactions reporting in Calabria (Southern Italy) in the four-year period 2011-2014: impact of a regional pharmacovigilance project in light of the new European Legislation.
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Retrieving top 1.000 articles with Pharma keywordThis will be our base of comparison, we want to separate them from the others | results = search('Pharma', 1000) #querying PubMed
id_list = results['IdList']
papers_pharma = fetch_details(id_list)#retrieving the info about the articles in nested lists & dictionary format
# checking article title for the first 10 retrieved articles
for i, paper in enumerate(papers_pharma['PubmedArticle'][:10]):
print("%d) %s" % (i+1,paper['MedlineCitation']['Article']['ArticleTitle'])) | 1) Recent trends in specialty pharma business model.
2) The moderating role of absorptive capacity and the differential effects of acquisitions and alliances on Big Pharma firms' innovation performance.
3) Space-related pharma-motifs for fast search of protein binding motifs and polypharmacological targets.
4) Pharma Websites and "Professionals-Only" Information: The Implications for Patient Trust and Autonomy.
5) BRIC Health Systems and Big Pharma: A Challenge for Health Policy and Management.
6) Developing Deep Learning Applications for Life Science and Pharma Industry.
7) Exzellenz in der Bildung für eine innovative Schweiz: Die Position des Wirtschaftsdachverbandes Chemie Pharma Biotech.
8) Shaking Up Biotech/Pharma: Can Cues Be Taken from the Tech Industry?
9) Pharma-Nutritional Properties of Olive Oil Phenols. Transfer of New Findings to Human Nutrition.
10) Pharma Success in Product Development—Does Biotechnology Change the Paradigm in Product Development and Attrition.
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Saving ID's, labels and title + abstracts of the articlesWhen an article was retrieved via the Pharmacovigilance keyword, it will receive the label = 1 and = 0 else. We'll per article put the article title and article abstract together as our text data on the article. | # Save ids & label 1 = pharmacovigilance , 0 = not pharmacovigilance
# & Save title + abstract in dico
ids = []
labels = []
data = []
for i, paper in enumerate(papers_pharmacov['PubmedArticle']):
if 'Abstract' in paper['MedlineCitation']['Article'].keys(): #check that abstract info is available
ids.append(str(paper['MedlineCitation']['PMID']))
labels.append(1)
title = paper['MedlineCitation']['Article']['ArticleTitle'] #Article title
abstract = paper['MedlineCitation']['Article']['Abstract']['AbstractText'][0] #Abstract
data.append( title + abstract )
for i, paper in enumerate(papers_pharma['PubmedArticle']):
if 'Abstract' in paper['MedlineCitation']['Article'].keys(): #check that abstract info is available
ids.append(str(paper['MedlineCitation']['PMID']))
labels.append(0)
title = paper['MedlineCitation']['Article']['ArticleTitle'] #Article title
abstract = paper['MedlineCitation']['Article']['Abstract']['AbstractText'][0] #Abstract
data.append( title + abstract )
# Check result for one paper
ids[0] # ID
labels[0] # 1 = pharmacovigilance , 0 = not pharmacovigilance
data[0] # Title & abstract | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Transform to numeric attributesWe will now **transform** the **text into numeric attributes**. For this, we will convert every word to a number, but we first need to **split** the full text into **separate words**. This is done by using a ***Tokenizer***. The tokenizer will split the full text based on a certain pattern you specify. Here we'll take a very basic pattern and take any words that contain only upper- or lowercase letters and we will convert everything to lowercase. | from nltk.tokenize.regexp import RegexpTokenizer #import a tokenizer, to split the full text into separate words
def Tokenize_text_value(value):
tokenizer1 = RegexpTokenizer(r"[A-Za-z]+") # our self defined tokenizera
value = value.lower() # convert all words to lowercase
return tokenizer1.tokenize(value) # tokenize each text
# example of our tokenizer
Tokenize_text_value(data[0]) | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Using the ***bag-of-words*** method we can transform any document to a vector. Using this method you have **one column per word and one row per document** and either a binary value 1 if the word is present in a certain document, 0 if not or a count value of the number of times the word appears in the document. For instance, the following three sentences:1. Intelligent applications creates intelligent business processes2. Bots are intelligent applications3. I do business intelligenceCan be represented in the following matrix using the counts of each word as values in the matrix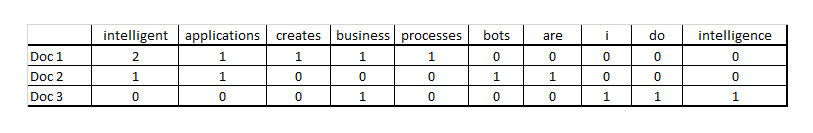 | # transform non-processed data to nummeric features:
from sklearn.feature_extraction.text import TfidfVectorizer
binary_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', binary = True,
tokenizer=Tokenize_text_value) # initialize the binary vectorizer
count_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=False,
tokenizer=Tokenize_text_value) # initialize the count vectorizer
binary_matrix = binary_vectorizer.fit_transform(data) # fit & transform
count_matrix = count_vectorizer.fit_transform(data) # fit & transform
# Check our output matrix shape: rows = documents, columns = words
binary_matrix.shape | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Check performance in a basic modelWe'll apply now a model on our 2 matrices. For this we will use the ***Naive Bayes model***, which (as the name tells) is based on the probabilistic Bayes theorem. It is used a lot in text-mining as it is really **fast** to train and apply and is able to **handle a lot of features**, which is often the case in text-mining, when you have one column per word. We will use the ***kappa*** measure to evaluate model performance. Kappa is a metric that is robust to class-imbalances in the data and varies from -1 to +1 with 0 being a random performance and +1 a perfect performance. | # apply cross validation Naive Bayes model
from sklearn.model_selection import cross_val_score
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import (cohen_kappa_score, make_scorer)
NB = MultinomialNB() # our Naive Bayes Model initialisation
scorer = make_scorer(cohen_kappa_score) # Our kappa score
scores = cross_val_score(NB,binary_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Binary matrix with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,count_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Count matrix with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var())) | Cross validation on Count matrix with a mean kappa score of 0.064193 and variance of 0.000682
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
TF-IDF transformationAn alternative to the binary and count matrix is the **tf-idf transformation**. It stands for ***Term Frequency - Inverse Document Frequency*** and is a measure that will try to find the words that are unique to each document and that characterizes the document compared to the other documents. How this achieved is by taking the term frequency (which is the same as the count that we have defined before) and multiplying it by the inverse document frequency (which is low when the term appears in all other documents and high when it appears in few other documents):*Copyright © Chris Albon, 2018* | # transform non-processed data to nummeric features:
from sklearn.feature_extraction.text import TfidfVectorizer
tfidf_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=True, smooth_idf = True ,
tokenizer=Tokenize_text_value) # initialize the tf-idf vectorizer
tfidf_matrix = tfidf_vectorizer.fit_transform(data) # fit & transform
scores = cross_val_score(NB,tfidf_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on TF-IDF matrix with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var())) | Cross validation on TF-IDF matrix with a mean kappa score of 0.320332 and variance of 0.003960
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
How to improve this score? How come the TF-IDF works the best, followed closely by the binary matrix and with the count matrix far behind? Let's have a look at the words that occur the most in the different documents: | import numpy as np
# Find words with maximum occurence for each document in the count_matrix
max_counts_per_doc = np.asarray(np.argmax(count_matrix,axis = 1)).ravel()
# Count how many times every word is the most occuring word across all documents
unique, counts = np.unique(max_counts_per_doc,return_counts=True)
# Keep only the words that are the most frequent word of at least 5 different documents
frequent = unique[counts > 5]
# Retrieve the vocabulary of our count matrix
vocab = count_vectorizer.get_feature_names()
# print out the words in frequent
for i in frequent:
print(vocab[i]) | a
and
for
in
of
the
to
with
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
As you can see those words are all words without any added value as they are mostly used to link certain words together in sentences, but have no standalone value. This is what we call ***Stop words***. So knowing that, we can find an intuition of why the tf-idf and binary transformations worked better than the count one. In the count one, we have seen that words that appear a lot, but have no value as such, get a high weight/value, whereas in binary every word gets the same weight and in tf-idf, the words that appear a lot in the other documents are automatically given a lower weight thanks to the IDF part. To avoid this problem we usually remove stop words Removing Stop words | # Remove the stop words
binary_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', binary = True,
tokenizer=Tokenize_text_value, stop_words = 'english') # initialize the binary vectorizer
count_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=False, smooth_idf=False,
tokenizer=Tokenize_text_value, stop_words = 'english') # initialize the count vectorizer
tfidf_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=True, smooth_idf=True,
tokenizer=Tokenize_text_value, stop_words = 'english') # initialize the tf-idf vectorizer
binary_matrix = binary_vectorizer.fit_transform(data) # fit & transform
count_matrix = count_vectorizer.fit_transform(data) # fit & transform
tfidf_matrix = tfidf_vectorizer.fit_transform(data) # fit & transform
scores = cross_val_score(NB,binary_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Binary matrix by removing stop-words with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,count_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Count matrix by removing stop-words with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,tfidf_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on TF-IDF matrix by removing stop-words with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var())) | Cross validation on TF-IDF matrix by removing stop-words with a mean kappa score of 0.682766 and variance of 0.011562
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
We have a big improvement in our performance when we remove the stop words. How can we go a step further? Now the following steps are mostly domain dependent. You have to think about your problem and what you would need to solve it. In this case, if we are using only the abstracts and the titles, if we had to do it ourselves, we would have a look at the most common keywords you have in the articles about Pharmacovigilance and when we have a new article to classify, we would look if we find those same keywords back. However, here we are analyzing all words (minus the stopwords) and not only the keywords. So we could try to filter out to keep only words that appear at least a certain number of times across all documents. Keeping only key-words | # keep only words that appear at least in 5% of the documents:
binary_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', binary = True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05) # initialize the binary vectorizer
count_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=False, smooth_idf=False,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05) # initialize the count vectorizer
tfidf_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=True, smooth_idf=True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05) # initialize the tf-idf vectorizer
binary_matrix = binary_vectorizer.fit_transform(data) # fit & transform
count_matrix = count_vectorizer.fit_transform(data) # fit & transform
tfidf_matrix = tfidf_vectorizer.fit_transform(data) # fit & transform
scores = cross_val_score(NB,binary_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Binary matrix by keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,count_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Count matrix by keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,tfidf_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on TF-IDF matrix by keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var())) | Cross validation on TF-IDF matrix by keeping only keywords appearing in at least 5% of the documents with a mean kappa score of 0.916734 and variance of 0.001633
| MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
Final improvementsWe've made a big improvement with this one as well. We can even go further and add some extra fine-tunings. Let's have a look at the final key-words: | tfidf_vectorizer.get_feature_names() | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
We can see that some words all refer to the same thing: *report, reported, reporting, reports* all refer to one same thing *report* and should therefore be grouped together => this can be done by ***stemming*** StemmingStemming is a technique where we try to reduce words to a common base form, this is done by chopping off the last part of the word: s's are removed, -ing is removed, -ed is removed, ... | # Define a stemmer that will preprocess the text before transforming it
from nltk.stem.porter import PorterStemmer
def preprocess(value):
stemmer = PorterStemmer()
#split in tokens
return ' '.join([stemmer.stem(i) for i in Tokenize_text_value(value) ])
# Have a look at what it gives on the first article
print(' '.join([i for i in Tokenize_text_value(data[0]) ])) # original
print('\n')
print(preprocess(data[0])) #stemmed
# Preprocess the documents by stemming the words
binary_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', binary = True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05, preprocessor = preprocess) # initialize the binary vectorizer
count_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=False, smooth_idf=False,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05, preprocessor = preprocess) # initialize the count vectorizer
tfidf_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=True, smooth_idf=True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.05, preprocessor = preprocess) # initialize the tf-idf vectorizer
binary_matrix = binary_vectorizer.fit_transform(data) # fit & transform
count_matrix = count_vectorizer.fit_transform(data) # fit & transform
tfidf_matrix = tfidf_vectorizer.fit_transform(data) # fit & transform
scores = cross_val_score(NB,binary_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Binary matrix by stemming and keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,count_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Count matrix by stemming and keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,tfidf_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on TF-IDF matrix by stemming and keeping only keywords appearing in at least 5%% of the documents with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
# Check the stemmed final vocabulary
tfidf_vectorizer.get_feature_names() | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
We can see that the performance slightly decreases with the stemming. Probably, because now when we are keeping words that appear in only 5% of the documents, we have more words than before, as before words with different endings were counted separately and now they are grouped together. So to correct for this we should increase our 5% threshold to take this effect into account. | # Preprocess the documents by stemming the words and keeping only words that appear in at least 10% of the documents:
binary_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', binary = True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.1, preprocessor = preprocess) # initialize the binary vectorizer
count_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=False, smooth_idf=False,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.1, preprocessor = preprocess) # initialize the count vectorizer
tfidf_vectorizer = TfidfVectorizer(input=u'content', analyzer=u'word', use_idf=True, smooth_idf=True,
tokenizer=Tokenize_text_value, stop_words = 'english'
, min_df = 0.1, preprocessor = preprocess) # initialize the tf-idf vectorizer
binary_matrix = binary_vectorizer.fit_transform(data) # fit & transform
count_matrix = count_vectorizer.fit_transform(data) # fit & transform
tfidf_matrix = tfidf_vectorizer.fit_transform(data) # fit & transform
scores = cross_val_score(NB,binary_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Binary matrix by stemming with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,count_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on Count matrix by stemming with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
scores = cross_val_score(NB,tfidf_matrix,labels,scoring = scorer,cv = 5 )
print('Cross validation on TF-IDF matrix by stemming with a mean kappa score of %f and variance of %f' % (scores.mean(),scores.var()))
# Check the stemmed final vocabulary
tfidf_vectorizer.get_feature_names() | _____no_output_____ | MIT | Text-Mining Hands-on.ipynb | tdekelver-bd/ResuMe |
This notebook was prepared by [Donne Martin](https://github.com/donnemartin). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges). Challenge Notebook Problem: Implement a priority queue backed by an array.* [Constraints](Constraints)* [Test Cases](Test-Cases)* [Algorithm](Algorithm)* [Code](Code)* [Unit Test](Unit-Test)* [Solution Notebook](Solution-Notebook) Constraints* Do we expect the methods to be insert, extract_min, and decrease_key? * Yes* Can we assume there aren't any duplicate keys? * Yes* Do we need to validate inputs? * No* Can we assume this fits memory? * Yes Test Cases insert* `insert` general case -> inserted node extract_min* `extract_min` from an empty list -> None* `extract_min` general case -> min node decrease_key* `decrease_key` an invalid key -> None* `decrease_key` general case -> updated node AlgorithmRefer to the [Solution Notebook](priority_queue_solution.ipynb). If you are stuck and need a hint, the solution notebook's algorithm discussion might be a good place to start. Code | class PriorityQueueNode(object):
def __init__(self, obj, key):
self.obj = obj
self.key = key
def __repr__(self):
return str(self.obj) + ': ' + str(self.key)
class PriorityQueue(object):
def __init__(self):
self.array = []
def __len__(self):
return len(self.array)
def insert(self, node):
for k in range(len(self.array)):
if self.array[k].key < node.key:
# TODO: Implement me
pass
def extract_min(self):
if not self.array:
return None
else:
# TODO: Implement me
pass
def decrease_key(self, obj, new_key):
# TODO: Implement me
pass | _____no_output_____ | Apache-2.0 | arrays_strings/priority_queue_(unsolved)/priority_queue_challenge.ipynb | zzong2006/interactive-coding-challenges |
Unit Test **The following unit test is expected to fail until you solve the challenge.** | # %load test_priority_queue.py
import unittest
class TestPriorityQueue(unittest.TestCase):
def test_priority_queue(self):
priority_queue = PriorityQueue()
self.assertEqual(priority_queue.extract_min(), None)
priority_queue.insert(PriorityQueueNode('a', 20))
priority_queue.insert(PriorityQueueNode('b', 5))
priority_queue.insert(PriorityQueueNode('c', 15))
priority_queue.insert(PriorityQueueNode('d', 22))
priority_queue.insert(PriorityQueueNode('e', 40))
priority_queue.insert(PriorityQueueNode('f', 3))
priority_queue.decrease_key('f', 2)
priority_queue.decrease_key('a', 19)
mins = []
while priority_queue.array:
mins.append(priority_queue.extract_min().key)
self.assertEqual(mins, [2, 5, 15, 19, 22, 40])
print('Success: test_min_heap')
def main():
test = TestPriorityQueue()
test.test_priority_queue()
if __name__ == '__main__':
main() | _____no_output_____ | Apache-2.0 | arrays_strings/priority_queue_(unsolved)/priority_queue_challenge.ipynb | zzong2006/interactive-coding-challenges |
pIC50 Test | import numpy as np
import torch
import seaborn as sns
import malt
import pandas as pd
import dgllife
from dgllife.utils import smiles_to_bigraph, CanonicalAtomFeaturizer, CanonicalBondFeaturizer
df = pd.read_csv('../../../data/moonshot_pIC50.csv', index_col=0)
dgllife_dataset = dgllife.data.csv_dataset.MoleculeCSVDataset(
df=df,
smiles_to_graph=smiles_to_bigraph,
node_featurizer=CanonicalAtomFeaturizer(),
edge_featurizer=CanonicalBondFeaturizer(),
smiles_column='SMILES',
task_names=[
# 'MW', 'cLogP', 'r_inhibition_at_20_uM',
# 'r_inhibition_at_50_uM', 'r_avg_IC50', 'f_inhibition_at_20_uM',
# 'f_inhibition_at_50_uM', 'f_avg_IC50', 'relative_solubility_at_20_uM',
# 'relative_solubility_at_100_uM', 'trypsin_IC50',
'f_avg_pIC50'
],
init_mask=False,
cache_file_path='../../../data/moonshot_pIC50.bin'
)
sns.displot(dgllife_dataset.labels.numpy()[dgllife_dataset.labels.numpy() > 4.005])
from malt.data.collections import _dataset_from_dgllife
data = _dataset_from_dgllife(dgllife_dataset)
# mask data if it's at the limit of detection
data_masked = data[list(np.flatnonzero(np.array(data.y) > 4.005))]
data.shuffle(seed=2666)
ds_tr, ds_vl, ds_te = data_masked[:1500].split([8, 1, 1]) | _____no_output_____ | MIT | scripts/supervised/pIC50_test.ipynb | choderalab/malt |
Make model | model_choice = 'nn' # 'nn'
if model_choice == "gp":
model = malt.models.supervised_model.GaussianProcessSupervisedModel(
representation=malt.models.representation.DGLRepresentation(
out_features=128,
),
regressor=malt.models.regressor.ExactGaussianProcessRegressor(
in_features=128, out_features=2,
),
likelihood=malt.models.likelihood.HeteroschedasticGaussianLikelihood(),
)
elif model_choice == "nn":
model = malt.models.supervised_model.SimpleSupervisedModel(
representation=malt.models.representation.DGLRepresentation(
out_features=128,
),
regressor=malt.models.regressor.NeuralNetworkRegressor(
in_features=128, out_features=1,
),
likelihood=malt.models.likelihood.HomoschedasticGaussianLikelihood(),
) | _____no_output_____ | MIT | scripts/supervised/pIC50_test.ipynb | choderalab/malt |
Train and evaluate. | trainer = malt.trainer.get_default_trainer(
without_player=True,
batch_size=32,
n_epochs=3000,
learning_rate=1e-3
)
model = trainer(model, ds_tr)
r2 = malt.metrics.supervised_metrics.R2()(model, ds_te)
print(r2)
rmse = malt.metrics.supervised_metrics.RMSE()(model, ds_te)
print(rmse)
ds_te_loader = ds_te.view(batch_size=len(ds_te))
g, y = next(iter(ds_te_loader))
y_hat = model.condition(g).mean
g = sns.jointplot(x = ds_te.y, y = y_hat.detach().numpy())
g.set_axis_labels('y', '\hat{y}')
import torch
import dgl
import malt
import argparse
parser = argparse.ArgumentParser()
parser.add_argument("--data", type=str, default="esol")
parser.add_argument("--model", type=str, default="nn")
args = parser.parse_args([])
data = getattr(malt.data.collections, args.data)()
data.shuffle(seed=2666)
ds_tr, ds_vl, ds_te = data.split([8, 1, 1])
if args.model == "gp":
model = malt.models.supervised_model.GaussianProcessSupervisedModel(
representation=malt.models.representation.DGLRepresentation(
out_features=128,
),
regressor=malt.models.regressor.ExactGaussianProcessRegressor(
in_features=128, out_features=2,
),
likelihood=malt.models.likelihood.HeteroschedasticGaussianLikelihood(),
)
elif args.model == "nn":
model = malt.models.supervised_model.SimpleSupervisedModel(
representation=malt.models.representation.DGLRepresentation(
out_features=128,
),
regressor=malt.models.regressor.NeuralNetworkRegressor(
in_features=128, out_features=1,
),
likelihood=malt.models.likelihood.HomoschedasticGaussianLikelihood(),
)
trainer = malt.trainer.get_default_trainer(without_player=True, batch_size=len(ds_tr), n_epochs=3000, learning_rate=1e-3)
model = trainer(model, ds_tr)
r2 = malt.metrics.supervised_metrics.R2()(model, ds_te)
print(r2)
rmse = malt.metrics.supervised_metrics.RMSE()(model, ds_te)
print(rmse)
ds_te_loader = ds_te.view(batch_size=len(ds_te))
g, y = next(iter(ds_te_loader))
y_hat = model.condition(g).mean
g = sns.jointplot(x = ds_te.y, y = y_hat.detach().numpy())
g.set_axis_labels('y', '\hat{y}') | _____no_output_____ | MIT | scripts/supervised/pIC50_test.ipynb | choderalab/malt |
Inference and ValidationNow that you have a trained network, you can use it for making predictions. This is typically called **inference**, a term borrowed from statistics. However, neural networks have a tendency to perform *too well* on the training data and aren't able to generalize to data that hasn't been seen before. This is called **overfitting** and it impairs inference performance. To test for overfitting while training, we measure the performance on data not in the training set called the **validation** set. We avoid overfitting through regularization such as dropout while monitoring the validation performance during training. In this notebook, I'll show you how to do this in PyTorch. As usual, let's start by loading the dataset through torchvision. You'll learn more about torchvision and loading data in a later part. This time we'll be taking advantage of the test set which you can get by setting `train=False` here:```pythontestset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)```The test set contains images just like the training set. Typically you'll see 10-20% of the original dataset held out for testing and validation with the rest being used for training. | import torch
from torchvision import datasets, transforms
# Define a transform to normalize the data
transform = transforms.Compose([transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
# Download and load the training data
trainset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=64, shuffle=True)
# Download and load the test data
testset = datasets.FashionMNIST('~/.pytorch/F_MNIST_data/', download=True, train=False, transform=transform)
testloader = torch.utils.data.DataLoader(testset, batch_size=64, shuffle=True) | _____no_output_____ | MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
Here I'll create a model like normal, using the same one from my solution for part 4. | from torch import nn, optim
import torch.nn.functional as F
class Classifier(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 256)
self.fc2 = nn.Linear(256, 128)
self.fc3 = nn.Linear(128, 64)
self.fc4 = nn.Linear(64, 10)
def forward(self, x):
# make sure input tensor is flattened
x = x.view(x.shape[0], -1)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.relu(self.fc3(x))
x = F.log_softmax(self.fc4(x), dim=1)
return x | _____no_output_____ | MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
The goal of validation is to measure the model's performance on data that isn't part of the training set. Performance here is up to the developer to define though. Typically this is just accuracy, the percentage of classes the network predicted correctly. Other options are [precision and recall](https://en.wikipedia.org/wiki/Precision_and_recallDefinition_(classification_context)) and top-5 error rate. We'll focus on accuracy here. First I'll do a forward pass with one batch from the test set. | model = Classifier()
images, labels = next(iter(testloader))
# Get the class probabilities
ps = torch.exp(model(images))
# Make sure the shape is appropriate, we should get 10 class probabilities for 64 examples
print(ps.shape) | torch.Size([64, 10])
| MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
With the probabilities, we can get the most likely class using the `ps.topk` method. This returns the $k$ highest values. Since we just want the most likely class, we can use `ps.topk(1)`. This returns a tuple of the top-$k$ values and the top-$k$ indices. If the highest value is the fifth element, we'll get back 4 as the index. | top_p, top_class = ps.topk(1, dim=1)
# Look at the most likely classes for the first 10 examples
print(top_class[:10,:]) | tensor([[0],
[3],
[0],
[0],
[0],
[0],
[0],
[0],
[0],
[0]])
| MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
Now we can check if the predicted classes match the labels. This is simple to do by equating `top_class` and `labels`, but we have to be careful of the shapes. Here `top_class` is a 2D tensor with shape `(64, 1)` while `labels` is 1D with shape `(64)`. To get the equality to work out the way we want, `top_class` and `labels` must have the same shape.If we do```pythonequals = top_class == labels````equals` will have shape `(64, 64)`, try it yourself. What it's doing is comparing the one element in each row of `top_class` with each element in `labels` which returns 64 True/False boolean values for each row. | equals = top_class == labels.view(*top_class.shape) | _____no_output_____ | MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
Now we need to calculate the percentage of correct predictions. `equals` has binary values, either 0 or 1. This means that if we just sum up all the values and divide by the number of values, we get the percentage of correct predictions. This is the same operation as taking the mean, so we can get the accuracy with a call to `torch.mean`. If only it was that simple. If you try `torch.mean(equals)`, you'll get an error```RuntimeError: mean is not implemented for type torch.ByteTensor```This happens because `equals` has type `torch.ByteTensor` but `torch.mean` isn't implement for tensors with that type. So we'll need to convert `equals` to a float tensor. Note that when we take `torch.mean` it returns a scalar tensor, to get the actual value as a float we'll need to do `accuracy.item()`. | accuracy = torch.mean(equals.type(torch.FloatTensor))
print(f'Accuracy: {accuracy.item()*100}%') | Accuracy: 20.3125%
| MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
The network is untrained so it's making random guesses and we should see an accuracy around 10%. Now let's train our network and include our validation pass so we can measure how well the network is performing on the test set. Since we're not updating our parameters in the validation pass, we can speed up our code by turning off gradients using `torch.no_grad()`:```python turn off gradientswith torch.no_grad(): validation pass here for images, labels in testloader: ...```>**Exercise:** Implement the validation loop below and print out the total accuracy after the loop. You can largely copy and paste the code from above, but I suggest typing it in because writing it out yourself is essential for building the skill. In general you'll always learn more by typing it rather than copy-pasting. You should be able to get an accuracy above 80%. | model = Classifier()
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.003)
epochs = 30
steps = 0
train_losses, test_losses = [], []
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
optimizer.zero_grad()
log_ps = model(images)
loss = criterion(log_ps, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
## TODO: Implement the validation pass and print out the validation accuracy
test_loss = 0
accuracy = 0
with torch.no_grad():
for images, labels in testloader:
log_ps = model(images)
test_loss += criterion(log_ps, labels)
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor))
train_losses.append(running_loss/len(trainloader))
test_losses.append(test_loss/len(testloader))
print("Epoch: {}/{}.. ".format(e+1, epochs),
"Training Loss: {:.3f}.. ".format(running_loss/len(trainloader)),
"Test Loss: {:.3f}.. ".format(test_loss/len(testloader)),
"Test Accuracy: {:.3f}".format(accuracy/len(testloader)))
| Epoch: 1/30.. Training Loss: 0.511.. Test Loss: 0.455.. Test Accuracy: 0.839
Epoch: 2/30.. Training Loss: 0.392.. Test Loss: 0.411.. Test Accuracy: 0.846
Epoch: 3/30.. Training Loss: 0.353.. Test Loss: 0.403.. Test Accuracy: 0.853
Epoch: 4/30.. Training Loss: 0.333.. Test Loss: 0.376.. Test Accuracy: 0.867
Epoch: 5/30.. Training Loss: 0.318.. Test Loss: 0.392.. Test Accuracy: 0.857
Epoch: 6/30.. Training Loss: 0.303.. Test Loss: 0.388.. Test Accuracy: 0.864
Epoch: 7/30.. Training Loss: 0.295.. Test Loss: 0.363.. Test Accuracy: 0.871
Epoch: 8/30.. Training Loss: 0.282.. Test Loss: 0.352.. Test Accuracy: 0.877
Epoch: 9/30.. Training Loss: 0.279.. Test Loss: 0.396.. Test Accuracy: 0.863
Epoch: 10/30.. Training Loss: 0.269.. Test Loss: 0.363.. Test Accuracy: 0.874
Epoch: 11/30.. Training Loss: 0.260.. Test Loss: 0.385.. Test Accuracy: 0.869
Epoch: 12/30.. Training Loss: 0.249.. Test Loss: 0.382.. Test Accuracy: 0.879
Epoch: 13/30.. Training Loss: 0.247.. Test Loss: 0.376.. Test Accuracy: 0.876
Epoch: 14/30.. Training Loss: 0.244.. Test Loss: 0.381.. Test Accuracy: 0.878
Epoch: 15/30.. Training Loss: 0.235.. Test Loss: 0.369.. Test Accuracy: 0.880
Epoch: 16/30.. Training Loss: 0.228.. Test Loss: 0.389.. Test Accuracy: 0.877
Epoch: 17/30.. Training Loss: 0.232.. Test Loss: 0.380.. Test Accuracy: 0.877
Epoch: 18/30.. Training Loss: 0.220.. Test Loss: 0.379.. Test Accuracy: 0.880
Epoch: 19/30.. Training Loss: 0.217.. Test Loss: 0.383.. Test Accuracy: 0.883
Epoch: 20/30.. Training Loss: 0.214.. Test Loss: 0.371.. Test Accuracy: 0.880
Epoch: 21/30.. Training Loss: 0.211.. Test Loss: 0.423.. Test Accuracy: 0.873
Epoch: 22/30.. Training Loss: 0.207.. Test Loss: 0.401.. Test Accuracy: 0.885
Epoch: 23/30.. Training Loss: 0.203.. Test Loss: 0.414.. Test Accuracy: 0.876
Epoch: 24/30.. Training Loss: 0.203.. Test Loss: 0.401.. Test Accuracy: 0.880
Epoch: 25/30.. Training Loss: 0.200.. Test Loss: 0.442.. Test Accuracy: 0.874
Epoch: 26/30.. Training Loss: 0.196.. Test Loss: 0.393.. Test Accuracy: 0.886
Epoch: 27/30.. Training Loss: 0.190.. Test Loss: 0.421.. Test Accuracy: 0.883
Epoch: 28/30.. Training Loss: 0.187.. Test Loss: 0.408.. Test Accuracy: 0.878
Epoch: 29/30.. Training Loss: 0.185.. Test Loss: 0.423.. Test Accuracy: 0.880
Epoch: 30/30.. Training Loss: 0.180.. Test Loss: 0.466.. Test Accuracy: 0.874
| MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
OverfittingIf we look at the training and validation losses as we train the network, we can see a phenomenon known as overfitting.The network learns the training set better and better, resulting in lower training losses. However, it starts having problems generalizing to data outside the training set leading to the validation loss increasing. The ultimate goal of any deep learning model is to make predictions on new data, so we should strive to get the lowest validation loss possible. One option is to use the version of the model with the lowest validation loss, here the one around 8-10 training epochs. This strategy is called *early-stopping*. In practice, you'd save the model frequently as you're training then later choose the model with the lowest validation loss.The most common method to reduce overfitting (outside of early-stopping) is *dropout*, where we randomly drop input units. This forces the network to share information between weights, increasing it's ability to generalize to new data. Adding dropout in PyTorch is straightforward using the [`nn.Dropout`](https://pytorch.org/docs/stable/nn.htmltorch.nn.Dropout) module.```pythonclass Classifier(nn.Module): def __init__(self): super().__init__() self.fc1 = nn.Linear(784, 256) self.fc2 = nn.Linear(256, 128) self.fc3 = nn.Linear(128, 64) self.fc4 = nn.Linear(64, 10) Dropout module with 0.2 drop probability self.dropout = nn.Dropout(p=0.2) def forward(self, x): make sure input tensor is flattened x = x.view(x.shape[0], -1) Now with dropout x = self.dropout(F.relu(self.fc1(x))) x = self.dropout(F.relu(self.fc2(x))) x = self.dropout(F.relu(self.fc3(x))) output so no dropout here x = F.log_softmax(self.fc4(x), dim=1) return x```During training we want to use dropout to prevent overfitting, but during inference we want to use the entire network. So, we need to turn off dropout during validation, testing, and whenever we're using the network to make predictions. To do this, you use `model.eval()`. This sets the model to evaluation mode where the dropout probability is 0. You can turn dropout back on by setting the model to train mode with `model.train()`. In general, the pattern for the validation loop will look like this, where you turn off gradients, set the model to evaluation mode, calculate the validation loss and metric, then set the model back to train mode.```python turn off gradientswith torch.no_grad(): set model to evaluation mode model.eval() validation pass here for images, labels in testloader: ... set model back to train modemodel.train()``` > **Exercise:** Add dropout to your model and train it on Fashion-MNIST again. See if you can get a lower validation loss or higher accuracy. | ## TODO: Define your model with dropout added
class Classifier(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(784, 256)
self.fc2 = nn.Linear(256, 128)
self.fc3 = nn.Linear(128, 64)
self.fc4 = nn.Linear(64, 10)
self.dropout = nn.Dropout(p=0.2)
def forward(self,x):
x = x.view(x.shape[0], -1)
# Now with dropout
x = self.dropout(F.relu(self.fc1(x)))
x = self.dropout(F.relu(self.fc2(x)))
x = self.dropout(F.relu(self.fc3(x)))
# output so no dropout here
x = F.log_softmax(self.fc4(x), dim=1)
return x
model = Classifier()
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.003)
epochs = 30
steps = 0
train_losses, test_losses = [], []
for e in range(epochs):
running_loss = 0
for images, labels in trainloader:
optimizer.zero_grad()
log_ps = model(images)
loss = criterion(log_ps, labels)
loss.backward()
optimizer.step()
running_loss += loss.item()
else:
test_loss = 0
accuracy = 0
# Turn off gradients for validation, saves memory and computations
with torch.no_grad():
model.eval()
for images, labels in testloader:
log_ps = model(images)
test_loss += criterion(log_ps, labels)
ps = torch.exp(log_ps)
top_p, top_class = ps.topk(1, dim=1)
equals = top_class == labels.view(*top_class.shape)
accuracy += torch.mean(equals.type(torch.FloatTensor))
model.train()
train_losses.append(running_loss/len(trainloader))
test_losses.append(test_loss/len(testloader))
print("Epoch: {}/{}.. ".format(e+1, epochs),
"Training Loss: {:.3f}.. ".format(running_loss/len(trainloader)),
"Test Loss: {:.3f}.. ".format(test_loss/len(testloader)),
"Test Accuracy: {:.3f}".format(accuracy/len(testloader)))
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib.pyplot as plt
plt.plot(train_losses, label='Training loss')
plt.plot(test_losses, label='Validation loss')
plt.legend(frameon=False) | _____no_output_____ | MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
InferenceNow that the model is trained, we can use it for inference. We've done this before, but now we need to remember to set the model in inference mode with `model.eval()`. You'll also want to turn off autograd with the `torch.no_grad()` context. | # Import helper module (should be in the repo)
import helper
# Test out your network!
model.eval()
dataiter = iter(testloader)
images, labels = dataiter.next()
img = images[0]
# Convert 2D image to 1D vector
img = img.view(1, 784)
# Calculate the class probabilities (softmax) for img
with torch.no_grad():
output = model.forward(img)
ps = torch.exp(output)
# Plot the image and probabilities
helper.view_classify(img.view(1, 28, 28), ps, version='Fashion') | _____no_output_____ | MIT | intro-to-pytorch/Part 5 - Inference and Validation (Exercises).ipynb | adilfaiz001/deep-learning-v2-pytorch |
Requirements | # !pip install --upgrade transformers bertviz checklist | _____no_output_____ | Apache-2.0 | notebooks/old/DeprecatedTitles_t0_gpt.ipynb | IlyaGusev/NewsCausation |
Data loading | # !rm -rf ru_news_cause_v1.tsv*
# !wget https://www.dropbox.com/s/kcxnhjzfut4guut/ru_news_cause_v1.tsv.tar.gz
# !tar -xzvf ru_news_cause_v1.tsv.tar.gz
# !cat ru_news_cause_v1.tsv | wc -l
# !head ru_news_cause_v1.tsv | _____no_output_____ | Apache-2.0 | notebooks/old/DeprecatedTitles_t0_gpt.ipynb | IlyaGusev/NewsCausation |
GPTCause | from transformers import GPT2LMHeadModel, GPT2TokenizerFast
device = 'cuda'
model_id = 'sberbank-ai/rugpt3small_based_on_gpt2'
model = GPT2LMHeadModel.from_pretrained(model_id).to(device)
tokenizer = GPT2TokenizerFast.from_pretrained(model_id)
import torch
max_length = model.config.n_positions
def gpt_assess(s1, s2):
encodings = tokenizer(f'{s1} {s2}', return_tensors='pt')
with torch.no_grad():
outputs = model(encodings.input_ids.to(device), labels=encodings.input_ids.to(device))
log_likelihood = outputs[0] * encodings.input_ids.size(1)
return log_likelihood.detach().cpu().numpy()
def gpt_assess_pair(s1,s2):
ppl1 = gpt_assess(s1,s2)
ppl2 = gpt_assess(s2,s1)
if ppl1<ppl2:
return 0, ppl1/ppl2
else:
return 1, ppl1/ppl2
print(gpt_assess('Привет!', 'Как дела?'))
print(gpt_assess('Как дела?', 'Привет!'))
print(gpt_assess_pair('Как дела?', 'Привет!'))
print(gpt_assess_pair('Привет!', 'Как дела?'))
| 11.083624
23.637806
(1, 2.1326785)
(0, 0.46889395)
| Apache-2.0 | notebooks/old/DeprecatedTitles_t0_gpt.ipynb | IlyaGusev/NewsCausation |
Scoring | import csv
labels = []
texts = []
preds = []
confs = []
with open("ru_news_cause_v1.tsv", "r", encoding='utf-8') as r:
reader = csv.reader(r, delimiter="\t")
header = next(reader)
for row in reader:
r = dict(zip(header, row))
if float(r["confidence"]) < 0.69:
continue
result = r["result"]
mapping = {
"left_right_cause": 0,
"left_right_cancel": 0,
"right_left_cause": 1,
"right_left_cancel": 1
}
if result not in mapping:
continue
r["label"] = mapping[result]
labels.append(r['label'])
texts.append( (r["left_title"], r["right_title"] ) )
p, c = gpt_assess_pair( r["left_title"], r["right_title"] )
preds.append( p )
confs.append( c )
from collections import Counter
print('labels', Counter(labels))
print('preds', Counter(preds))
import matplotlib.pyplot as plt
plt.hist(confs)
plt.show()
from sklearn.metrics import classification_report, balanced_accuracy_score, confusion_matrix
y_true = labels
y_pred = preds
print(classification_report(y_true, y_pred))
print('balanced_accuracy_score', balanced_accuracy_score(y_true, y_pred))
print('\nconfusion_matrix\n',confusion_matrix(y_true, y_pred))
import numpy as np
confidence_th = .1
mask = np.array(list(map(lambda x:abs(x-1.), confs)))>confidence_th
y_true = np.array(labels)[mask]
y_pred = np.array(preds)[mask]
print(classification_report(y_true, y_pred))
print('balanced_accuracy_score', balanced_accuracy_score(y_true, y_pred))
print('\nconfusion_matrix\n',confusion_matrix(y_true, y_pred))
from sklearn.metrics import f1_score
xs = []
ys = []
for th_idx in range(250):
th = th_idx/1000.
mask = np.array(list(map(lambda x:abs(x-1.), confs)))>th
y_true = np.array(labels)[mask]
y_pred = np.array(preds)[mask]
xs.append( th )
ys.append( f1_score(y_true, y_pred) )
import matplotlib.pyplot as plt
plt.plot(xs, ys)
plt.suptitle('f1 by conf_th')
plt.show() | _____no_output_____ | Apache-2.0 | notebooks/old/DeprecatedTitles_t0_gpt.ipynb | IlyaGusev/NewsCausation |
CS224N Assignment 1: Exploring Word Vectors (25 Points) Due 4:30pm, Tue Jan 14 Welcome to CS224n! Before you start, make sure you read the README.txt in the same directory as this notebook. You will find many provided codes in the notebook. We highly encourage you to read and understand the provided codes as part of the learning :-) | # All Import Statements Defined Here
# Note: Do not add to this list.
# ----------------
import sys
assert sys.version_info[0]==3
assert sys.version_info[1] >= 5
from gensim.models import KeyedVectors
from gensim.test.utils import datapath
import pprint
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [10, 5]
import nltk
nltk.download('reuters')
from nltk.corpus import reuters
import numpy as np
import random
import scipy as sp
from sklearn.decomposition import TruncatedSVD
from sklearn.decomposition import PCA
START_TOKEN = '<START>'
END_TOKEN = '<END>'
np.random.seed(0)
random.seed(0)
# ---------------- | [nltk_data] Downloading package reuters to
[nltk_data] /usr/local/share/nltk_data...
[nltk_data] Package reuters is already up-to-date!
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Word VectorsWord Vectors are often used as a fundamental component for downstream NLP tasks, e.g. question answering, text generation, translation, etc., so it is important to build some intuitions as to their strengths and weaknesses. Here, you will explore two types of word vectors: those derived from *co-occurrence matrices*, and those derived via *GloVe*. **Assignment Notes:** Please make sure to save the notebook as you go along. Submission Instructions are located at the bottom of the notebook.**Note on Terminology:** The terms "word vectors" and "word embeddings" are often used interchangeably. The term "embedding" refers to the fact that we are encoding aspects of a word's meaning in a lower dimensional space. As [Wikipedia](https://en.wikipedia.org/wiki/Word_embedding) states, "*conceptually it involves a mathematical embedding from a space with one dimension per word to a continuous vector space with a much lower dimension*". Part 1: Count-Based Word Vectors (10 points)Most word vector models start from the following idea:*You shall know a word by the company it keeps ([Firth, J. R. 1957:11](https://en.wikipedia.org/wiki/John_Rupert_Firth))*Many word vector implementations are driven by the idea that similar words, i.e., (near) synonyms, will be used in similar contexts. As a result, similar words will often be spoken or written along with a shared subset of words, i.e., contexts. By examining these contexts, we can try to develop embeddings for our words. With this intuition in mind, many "old school" approaches to constructing word vectors relied on word counts. Here we elaborate upon one of those strategies, *co-occurrence matrices* (for more information, see [here](http://web.stanford.edu/class/cs124/lec/vectorsemantics.video.pdf) or [here](https://medium.com/data-science-group-iitr/word-embedding-2d05d270b285)). Co-OccurrenceA co-occurrence matrix counts how often things co-occur in some environment. Given some word $w_i$ occurring in the document, we consider the *context window* surrounding $w_i$. Supposing our fixed window size is $n$, then this is the $n$ preceding and $n$ subsequent words in that document, i.e. words $w_{i-n} \dots w_{i-1}$ and $w_{i+1} \dots w_{i+n}$. We build a *co-occurrence matrix* $M$, which is a symmetric word-by-word matrix in which $M_{ij}$ is the number of times $w_j$ appears inside $w_i$'s window among all documents.**Example: Co-Occurrence with Fixed Window of n=1**:Document 1: "all that glitters is not gold"Document 2: "all is well that ends well"| * | `` | all | that | glitters | is | not | gold | well | ends | `` ||----------|-------|-----|------|----------|------|------|-------|------|------|-----|| `` | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 || all | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 || that | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 || glitters | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 || is | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 || not | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 || gold | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 || well | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 || ends | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 || `` | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |**Note:** In NLP, we often add `` and `` tokens to represent the beginning and end of sentences, paragraphs or documents. In thise case we imagine `` and `` tokens encapsulating each document, e.g., "`` All that glitters is not gold ``", and include these tokens in our co-occurrence counts.The rows (or columns) of this matrix provide one type of word vectors (those based on word-word co-occurrence), but the vectors will be large in general (linear in the number of distinct words in a corpus). Thus, our next step is to run *dimensionality reduction*. In particular, we will run *SVD (Singular Value Decomposition)*, which is a kind of generalized *PCA (Principal Components Analysis)* to select the top $k$ principal components. Here's a visualization of dimensionality reduction with SVD. In this picture our co-occurrence matrix is $A$ with $n$ rows corresponding to $n$ words. We obtain a full matrix decomposition, with the singular values ordered in the diagonal $S$ matrix, and our new, shorter length-$k$ word vectors in $U_k$.This reduced-dimensionality co-occurrence representation preserves semantic relationships between words, e.g. *doctor* and *hospital* will be closer than *doctor* and *dog*. **Notes:** If you can barely remember what an eigenvalue is, here's [a slow, friendly introduction to SVD](https://davetang.org/file/Singular_Value_Decomposition_Tutorial.pdf). If you want to learn more thoroughly about PCA or SVD, feel free to check out lectures [7](https://web.stanford.edu/class/cs168/l/l7.pdf), [8](http://theory.stanford.edu/~tim/s15/l/l8.pdf), and [9](https://web.stanford.edu/class/cs168/l/l9.pdf) of CS168. These course notes provide a great high-level treatment of these general purpose algorithms. Though, for the purpose of this class, you only need to know how to extract the k-dimensional embeddings by utilizing pre-programmed implementations of these algorithms from the numpy, scipy, or sklearn python packages. In practice, it is challenging to apply full SVD to large corpora because of the memory needed to perform PCA or SVD. However, if you only want the top $k$ vector components for relatively small $k$ — known as [Truncated SVD](https://en.wikipedia.org/wiki/Singular_value_decompositionTruncated_SVD) — then there are reasonably scalable techniques to compute those iteratively. Plotting Co-Occurrence Word EmbeddingsHere, we will be using the Reuters (business and financial news) corpus. If you haven't run the import cell at the top of this page, please run it now (click it and press SHIFT-RETURN). The corpus consists of 10,788 news documents totaling 1.3 million words. These documents span 90 categories and are split into train and test. For more details, please see https://www.nltk.org/book/ch02.html. We provide a `read_corpus` function below that pulls out only articles from the "crude" (i.e. news articles about oil, gas, etc.) category. The function also adds `` and `` tokens to each of the documents, and lowercases words. You do **not** have to perform any other kind of pre-processing. | def read_corpus(category="crude"):
""" Read files from the specified Reuter's category.
Params:
category (string): category name
Return:
list of lists, with words from each of the processed files
"""
files = reuters.fileids(category)
return [[START_TOKEN] + [w.lower() for w in list(reuters.words(f))] + [END_TOKEN] for f in files]
| _____no_output_____ | MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Let's have a look what these documents are like…. | reuters_corpus = read_corpus()
pprint.pprint(reuters_corpus[:3], compact=True, width=100) | [['<START>', 'japan', 'to', 'revise', 'long', '-', 'term', 'energy', 'demand', 'downwards', 'the',
'ministry', 'of', 'international', 'trade', 'and', 'industry', '(', 'miti', ')', 'will', 'revise',
'its', 'long', '-', 'term', 'energy', 'supply', '/', 'demand', 'outlook', 'by', 'august', 'to',
'meet', 'a', 'forecast', 'downtrend', 'in', 'japanese', 'energy', 'demand', ',', 'ministry',
'officials', 'said', '.', 'miti', 'is', 'expected', 'to', 'lower', 'the', 'projection', 'for',
'primary', 'energy', 'supplies', 'in', 'the', 'year', '2000', 'to', '550', 'mln', 'kilolitres',
'(', 'kl', ')', 'from', '600', 'mln', ',', 'they', 'said', '.', 'the', 'decision', 'follows',
'the', 'emergence', 'of', 'structural', 'changes', 'in', 'japanese', 'industry', 'following',
'the', 'rise', 'in', 'the', 'value', 'of', 'the', 'yen', 'and', 'a', 'decline', 'in', 'domestic',
'electric', 'power', 'demand', '.', 'miti', 'is', 'planning', 'to', 'work', 'out', 'a', 'revised',
'energy', 'supply', '/', 'demand', 'outlook', 'through', 'deliberations', 'of', 'committee',
'meetings', 'of', 'the', 'agency', 'of', 'natural', 'resources', 'and', 'energy', ',', 'the',
'officials', 'said', '.', 'they', 'said', 'miti', 'will', 'also', 'review', 'the', 'breakdown',
'of', 'energy', 'supply', 'sources', ',', 'including', 'oil', ',', 'nuclear', ',', 'coal', 'and',
'natural', 'gas', '.', 'nuclear', 'energy', 'provided', 'the', 'bulk', 'of', 'japan', "'", 's',
'electric', 'power', 'in', 'the', 'fiscal', 'year', 'ended', 'march', '31', ',', 'supplying',
'an', 'estimated', '27', 'pct', 'on', 'a', 'kilowatt', '/', 'hour', 'basis', ',', 'followed',
'by', 'oil', '(', '23', 'pct', ')', 'and', 'liquefied', 'natural', 'gas', '(', '21', 'pct', '),',
'they', 'noted', '.', '<END>'],
['<START>', 'energy', '/', 'u', '.', 's', '.', 'petrochemical', 'industry', 'cheap', 'oil',
'feedstocks', ',', 'the', 'weakened', 'u', '.', 's', '.', 'dollar', 'and', 'a', 'plant',
'utilization', 'rate', 'approaching', '90', 'pct', 'will', 'propel', 'the', 'streamlined', 'u',
'.', 's', '.', 'petrochemical', 'industry', 'to', 'record', 'profits', 'this', 'year', ',',
'with', 'growth', 'expected', 'through', 'at', 'least', '1990', ',', 'major', 'company',
'executives', 'predicted', '.', 'this', 'bullish', 'outlook', 'for', 'chemical', 'manufacturing',
'and', 'an', 'industrywide', 'move', 'to', 'shed', 'unrelated', 'businesses', 'has', 'prompted',
'gaf', 'corp', '&', 'lt', ';', 'gaf', '>,', 'privately', '-', 'held', 'cain', 'chemical', 'inc',
',', 'and', 'other', 'firms', 'to', 'aggressively', 'seek', 'acquisitions', 'of', 'petrochemical',
'plants', '.', 'oil', 'companies', 'such', 'as', 'ashland', 'oil', 'inc', '&', 'lt', ';', 'ash',
'>,', 'the', 'kentucky', '-', 'based', 'oil', 'refiner', 'and', 'marketer', ',', 'are', 'also',
'shopping', 'for', 'money', '-', 'making', 'petrochemical', 'businesses', 'to', 'buy', '.', '"',
'i', 'see', 'us', 'poised', 'at', 'the', 'threshold', 'of', 'a', 'golden', 'period', ',"', 'said',
'paul', 'oreffice', ',', 'chairman', 'of', 'giant', 'dow', 'chemical', 'co', '&', 'lt', ';',
'dow', '>,', 'adding', ',', '"', 'there', "'", 's', 'no', 'major', 'plant', 'capacity', 'being',
'added', 'around', 'the', 'world', 'now', '.', 'the', 'whole', 'game', 'is', 'bringing', 'out',
'new', 'products', 'and', 'improving', 'the', 'old', 'ones', '."', 'analysts', 'say', 'the',
'chemical', 'industry', "'", 's', 'biggest', 'customers', ',', 'automobile', 'manufacturers',
'and', 'home', 'builders', 'that', 'use', 'a', 'lot', 'of', 'paints', 'and', 'plastics', ',',
'are', 'expected', 'to', 'buy', 'quantities', 'this', 'year', '.', 'u', '.', 's', '.',
'petrochemical', 'plants', 'are', 'currently', 'operating', 'at', 'about', '90', 'pct',
'capacity', ',', 'reflecting', 'tighter', 'supply', 'that', 'could', 'hike', 'product', 'prices',
'by', '30', 'to', '40', 'pct', 'this', 'year', ',', 'said', 'john', 'dosher', ',', 'managing',
'director', 'of', 'pace', 'consultants', 'inc', 'of', 'houston', '.', 'demand', 'for', 'some',
'products', 'such', 'as', 'styrene', 'could', 'push', 'profit', 'margins', 'up', 'by', 'as',
'much', 'as', '300', 'pct', ',', 'he', 'said', '.', 'oreffice', ',', 'speaking', 'at', 'a',
'meeting', 'of', 'chemical', 'engineers', 'in', 'houston', ',', 'said', 'dow', 'would', 'easily',
'top', 'the', '741', 'mln', 'dlrs', 'it', 'earned', 'last', 'year', 'and', 'predicted', 'it',
'would', 'have', 'the', 'best', 'year', 'in', 'its', 'history', '.', 'in', '1985', ',', 'when',
'oil', 'prices', 'were', 'still', 'above', '25', 'dlrs', 'a', 'barrel', 'and', 'chemical',
'exports', 'were', 'adversely', 'affected', 'by', 'the', 'strong', 'u', '.', 's', '.', 'dollar',
',', 'dow', 'had', 'profits', 'of', '58', 'mln', 'dlrs', '.', '"', 'i', 'believe', 'the',
'entire', 'chemical', 'industry', 'is', 'headed', 'for', 'a', 'record', 'year', 'or', 'close',
'to', 'it', ',"', 'oreffice', 'said', '.', 'gaf', 'chairman', 'samuel', 'heyman', 'estimated',
'that', 'the', 'u', '.', 's', '.', 'chemical', 'industry', 'would', 'report', 'a', '20', 'pct',
'gain', 'in', 'profits', 'during', '1987', '.', 'last', 'year', ',', 'the', 'domestic',
'industry', 'earned', 'a', 'total', 'of', '13', 'billion', 'dlrs', ',', 'a', '54', 'pct', 'leap',
'from', '1985', '.', 'the', 'turn', 'in', 'the', 'fortunes', 'of', 'the', 'once', '-', 'sickly',
'chemical', 'industry', 'has', 'been', 'brought', 'about', 'by', 'a', 'combination', 'of', 'luck',
'and', 'planning', ',', 'said', 'pace', "'", 's', 'john', 'dosher', '.', 'dosher', 'said', 'last',
'year', "'", 's', 'fall', 'in', 'oil', 'prices', 'made', 'feedstocks', 'dramatically', 'cheaper',
'and', 'at', 'the', 'same', 'time', 'the', 'american', 'dollar', 'was', 'weakening', 'against',
'foreign', 'currencies', '.', 'that', 'helped', 'boost', 'u', '.', 's', '.', 'chemical',
'exports', '.', 'also', 'helping', 'to', 'bring', 'supply', 'and', 'demand', 'into', 'balance',
'has', 'been', 'the', 'gradual', 'market', 'absorption', 'of', 'the', 'extra', 'chemical',
'manufacturing', 'capacity', 'created', 'by', 'middle', 'eastern', 'oil', 'producers', 'in',
'the', 'early', '1980s', '.', 'finally', ',', 'virtually', 'all', 'major', 'u', '.', 's', '.',
'chemical', 'manufacturers', 'have', 'embarked', 'on', 'an', 'extensive', 'corporate',
'restructuring', 'program', 'to', 'mothball', 'inefficient', 'plants', ',', 'trim', 'the',
'payroll', 'and', 'eliminate', 'unrelated', 'businesses', '.', 'the', 'restructuring', 'touched',
'off', 'a', 'flurry', 'of', 'friendly', 'and', 'hostile', 'takeover', 'attempts', '.', 'gaf', ',',
'which', 'made', 'an', 'unsuccessful', 'attempt', 'in', '1985', 'to', 'acquire', 'union',
'carbide', 'corp', '&', 'lt', ';', 'uk', '>,', 'recently', 'offered', 'three', 'billion', 'dlrs',
'for', 'borg', 'warner', 'corp', '&', 'lt', ';', 'bor', '>,', 'a', 'chicago', 'manufacturer',
'of', 'plastics', 'and', 'chemicals', '.', 'another', 'industry', 'powerhouse', ',', 'w', '.',
'r', '.', 'grace', '&', 'lt', ';', 'gra', '>', 'has', 'divested', 'its', 'retailing', ',',
'restaurant', 'and', 'fertilizer', 'businesses', 'to', 'raise', 'cash', 'for', 'chemical',
'acquisitions', '.', 'but', 'some', 'experts', 'worry', 'that', 'the', 'chemical', 'industry',
'may', 'be', 'headed', 'for', 'trouble', 'if', 'companies', 'continue', 'turning', 'their',
'back', 'on', 'the', 'manufacturing', 'of', 'staple', 'petrochemical', 'commodities', ',', 'such',
'as', 'ethylene', ',', 'in', 'favor', 'of', 'more', 'profitable', 'specialty', 'chemicals',
'that', 'are', 'custom', '-', 'designed', 'for', 'a', 'small', 'group', 'of', 'buyers', '.', '"',
'companies', 'like', 'dupont', '&', 'lt', ';', 'dd', '>', 'and', 'monsanto', 'co', '&', 'lt', ';',
'mtc', '>', 'spent', 'the', 'past', 'two', 'or', 'three', 'years', 'trying', 'to', 'get', 'out',
'of', 'the', 'commodity', 'chemical', 'business', 'in', 'reaction', 'to', 'how', 'badly', 'the',
'market', 'had', 'deteriorated', ',"', 'dosher', 'said', '.', '"', 'but', 'i', 'think', 'they',
'will', 'eventually', 'kill', 'the', 'margins', 'on', 'the', 'profitable', 'chemicals', 'in',
'the', 'niche', 'market', '."', 'some', 'top', 'chemical', 'executives', 'share', 'the',
'concern', '.', '"', 'the', 'challenge', 'for', 'our', 'industry', 'is', 'to', 'keep', 'from',
'getting', 'carried', 'away', 'and', 'repeating', 'past', 'mistakes', ',"', 'gaf', "'", 's',
'heyman', 'cautioned', '.', '"', 'the', 'shift', 'from', 'commodity', 'chemicals', 'may', 'be',
'ill', '-', 'advised', '.', 'specialty', 'businesses', 'do', 'not', 'stay', 'special', 'long',
'."', 'houston', '-', 'based', 'cain', 'chemical', ',', 'created', 'this', 'month', 'by', 'the',
'sterling', 'investment', 'banking', 'group', ',', 'believes', 'it', 'can', 'generate', '700',
'mln', 'dlrs', 'in', 'annual', 'sales', 'by', 'bucking', 'the', 'industry', 'trend', '.',
'chairman', 'gordon', 'cain', ',', 'who', 'previously', 'led', 'a', 'leveraged', 'buyout', 'of',
'dupont', "'", 's', 'conoco', 'inc', "'", 's', 'chemical', 'business', ',', 'has', 'spent', '1',
'.', '1', 'billion', 'dlrs', 'since', 'january', 'to', 'buy', 'seven', 'petrochemical', 'plants',
'along', 'the', 'texas', 'gulf', 'coast', '.', 'the', 'plants', 'produce', 'only', 'basic',
'commodity', 'petrochemicals', 'that', 'are', 'the', 'building', 'blocks', 'of', 'specialty',
'products', '.', '"', 'this', 'kind', 'of', 'commodity', 'chemical', 'business', 'will', 'never',
'be', 'a', 'glamorous', ',', 'high', '-', 'margin', 'business', ',"', 'cain', 'said', ',',
'adding', 'that', 'demand', 'is', 'expected', 'to', 'grow', 'by', 'about', 'three', 'pct',
'annually', '.', 'garo', 'armen', ',', 'an', 'analyst', 'with', 'dean', 'witter', 'reynolds', ',',
'said', 'chemical', 'makers', 'have', 'also', 'benefitted', 'by', 'increasing', 'demand', 'for',
'plastics', 'as', 'prices', 'become', 'more', 'competitive', 'with', 'aluminum', ',', 'wood',
'and', 'steel', 'products', '.', 'armen', 'estimated', 'the', 'upturn', 'in', 'the', 'chemical',
'business', 'could', 'last', 'as', 'long', 'as', 'four', 'or', 'five', 'years', ',', 'provided',
'the', 'u', '.', 's', '.', 'economy', 'continues', 'its', 'modest', 'rate', 'of', 'growth', '.',
'<END>'],
['<START>', 'turkey', 'calls', 'for', 'dialogue', 'to', 'solve', 'dispute', 'turkey', 'said',
'today', 'its', 'disputes', 'with', 'greece', ',', 'including', 'rights', 'on', 'the',
'continental', 'shelf', 'in', 'the', 'aegean', 'sea', ',', 'should', 'be', 'solved', 'through',
'negotiations', '.', 'a', 'foreign', 'ministry', 'statement', 'said', 'the', 'latest', 'crisis',
'between', 'the', 'two', 'nato', 'members', 'stemmed', 'from', 'the', 'continental', 'shelf',
'dispute', 'and', 'an', 'agreement', 'on', 'this', 'issue', 'would', 'effect', 'the', 'security',
',', 'economy', 'and', 'other', 'rights', 'of', 'both', 'countries', '.', '"', 'as', 'the',
'issue', 'is', 'basicly', 'political', ',', 'a', 'solution', 'can', 'only', 'be', 'found', 'by',
'bilateral', 'negotiations', ',"', 'the', 'statement', 'said', '.', 'greece', 'has', 'repeatedly',
'said', 'the', 'issue', 'was', 'legal', 'and', 'could', 'be', 'solved', 'at', 'the',
'international', 'court', 'of', 'justice', '.', 'the', 'two', 'countries', 'approached', 'armed',
'confrontation', 'last', 'month', 'after', 'greece', 'announced', 'it', 'planned', 'oil',
'exploration', 'work', 'in', 'the', 'aegean', 'and', 'turkey', 'said', 'it', 'would', 'also',
'search', 'for', 'oil', '.', 'a', 'face', '-', 'off', 'was', 'averted', 'when', 'turkey',
'confined', 'its', 'research', 'to', 'territorrial', 'waters', '.', '"', 'the', 'latest',
'crises', 'created', 'an', 'historic', 'opportunity', 'to', 'solve', 'the', 'disputes', 'between',
'the', 'two', 'countries', ',"', 'the', 'foreign', 'ministry', 'statement', 'said', '.', 'turkey',
"'", 's', 'ambassador', 'in', 'athens', ',', 'nazmi', 'akiman', ',', 'was', 'due', 'to', 'meet',
'prime', 'minister', 'andreas', 'papandreou', 'today', 'for', 'the', 'greek', 'reply', 'to', 'a',
'message', 'sent', 'last', 'week', 'by', 'turkish', 'prime', 'minister', 'turgut', 'ozal', '.',
'the', 'contents', 'of', 'the', 'message', 'were', 'not', 'disclosed', '.', '<END>']]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Question 1.1: Implement `distinct_words` [code] (2 points)Write a method to work out the distinct words (word types) that occur in the corpus. You can do this with `for` loops, but it's more efficient to do it with Python list comprehensions. In particular, [this](https://coderwall.com/p/rcmaea/flatten-a-list-of-lists-in-one-line-in-python) may be useful to flatten a list of lists. If you're not familiar with Python list comprehensions in general, here's [more information](https://python-3-patterns-idioms-test.readthedocs.io/en/latest/Comprehensions.html).You may find it useful to use [Python sets](https://www.w3schools.com/python/python_sets.asp) to remove duplicate words. | def distinct_words(corpus):
""" Determine a list of distinct words for the corpus.
Params:
corpus (list of list of strings): corpus of documents
Return:
corpus_words (list of strings): list of distinct words across the corpus, sorted (using python 'sorted' function)
num_corpus_words (integer): number of distinct words across the corpus
"""
# ------------------
# Write your implementation here.
corpus_words = list(sorted(set([token for sentences in corpus for token in sentences])))
num_corpus_words = len(corpus_words)
# ------------------
return corpus_words, num_corpus_words
# ---------------------
# Run this sanity check
# Note that this not an exhaustive check for correctness.
# ---------------------
# Define toy corpus
test_corpus = ["{} All that glitters isn't gold {}".format(START_TOKEN, END_TOKEN).split(" "), "{} All's well that ends well {}".format(START_TOKEN, END_TOKEN).split(" ")]
test_corpus_words, num_corpus_words = distinct_words(test_corpus)
# Correct answers
ans_test_corpus_words = sorted([START_TOKEN, "All", "ends", "that", "gold", "All's", "glitters", "isn't", "well", END_TOKEN])
ans_num_corpus_words = len(ans_test_corpus_words)
# Test correct number of words
assert(num_corpus_words == ans_num_corpus_words), "Incorrect number of distinct words. Correct: {}. Yours: {}".format(ans_num_corpus_words, num_corpus_words)
# Test correct words
assert (test_corpus_words == ans_test_corpus_words), "Incorrect corpus_words.\nCorrect: {}\nYours: {}".format(str(ans_test_corpus_words), str(test_corpus_words))
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80) | --------------------------------------------------------------------------------
Passed All Tests!
--------------------------------------------------------------------------------
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Question 1.2: Implement `compute_co_occurrence_matrix` [code] (3 points)Write a method that constructs a co-occurrence matrix for a certain window-size $n$ (with a default of 4), considering words $n$ before and $n$ after the word in the center of the window. Here, we start to use `numpy (np)` to represent vectors, matrices, and tensors. If you're not familiar with NumPy, there's a NumPy tutorial in the second half of this cs231n [Python NumPy tutorial](http://cs231n.github.io/python-numpy-tutorial/). | def compute_co_occurrence_matrix(corpus, window_size=4):
""" Compute co-occurrence matrix for the given corpus and window_size (default of 4).
Note: Each word in a document should be at the center of a window. Words near edges will have a smaller
number of co-occurring words.
For example, if we take the document "<START> All that glitters is not gold <END>" with window size of 4,
"All" will co-occur with "<START>", "that", "glitters", "is", and "not".
Params:
corpus (list of list of strings): corpus of documents
window_size (int): size of context window
Return:
M (a symmetric numpy matrix of shape (number of unique words in the corpus , number of unique words in the corpus)):
Co-occurence matrix of word counts.
The ordering of the words in the rows/columns should be the same as the ordering of the words given by the distinct_words function.
word2Ind (dict): dictionary that maps word to index (i.e. row/column number) for matrix M.
"""
words, num_words = distinct_words(corpus)
# ------------------
# Write your implementation here.
M = np.zeros((num_words, num_words), dtype=np.int)
co_time = {ii: [] for ii in range(num_words)}
word2Ind = dict(zip(words, range(num_words)))
for sent in corpus:
for idx, center in enumerate(sent):
center_id = word2Ind[center]
context = sent[max(0, idx - window_size):idx + window_size + 1]
context_id = [word2Ind[ii] for ii in context if ii != center]
co_time[center_id].extend(context_id)
for center, co_list in co_time.items():
unique, counts = np.unique(co_list, return_counts=True)
co_map = dict(zip(unique, counts))
for context, time in co_map.items():
M[center][context] = time
# ------------------
return M, word2Ind
# ---------------------
# Run this sanity check
# Note that this is not an exhaustive check for correctness.
# ---------------------
# Define toy corpus and get student's co-occurrence matrix
test_corpus = ["{} All that glitters isn't gold {}".format(START_TOKEN, END_TOKEN).split(" "), "{} All's well that ends well {}".format(START_TOKEN, END_TOKEN).split(" ")]
M_test, word2Ind_test = compute_co_occurrence_matrix(test_corpus, window_size=1)
# Correct M and word2Ind
M_test_ans = np.array(
[[0., 0., 0., 0., 0., 0., 1., 0., 0., 1.,],
[0., 0., 1., 1., 0., 0., 0., 0., 0., 0.,],
[0., 1., 0., 0., 0., 0., 0., 0., 1., 0.,],
[0., 1., 0., 0., 0., 0., 0., 0., 0., 1.,],
[0., 0., 0., 0., 0., 0., 0., 0., 1., 1.,],
[0., 0., 0., 0., 0., 0., 0., 1., 1., 0.,],
[1., 0., 0., 0., 0., 0., 0., 1., 0., 0.,],
[0., 0., 0., 0., 0., 1., 1., 0., 0., 0.,],
[0., 0., 1., 0., 1., 1., 0., 0., 0., 1.,],
[1., 0., 0., 1., 1., 0., 0., 0., 1., 0.,]]
)
ans_test_corpus_words = sorted([START_TOKEN, "All", "ends", "that", "gold", "All's", "glitters", "isn't", "well", END_TOKEN])
word2Ind_ans = dict(zip(ans_test_corpus_words, range(len(ans_test_corpus_words))))
# Test correct word2Ind
assert (word2Ind_ans == word2Ind_test), "Your word2Ind is incorrect:\nCorrect: {}\nYours: {}".format(word2Ind_ans, word2Ind_test)
# Test correct M shape
assert (M_test.shape == M_test_ans.shape), "M matrix has incorrect shape.\nCorrect: {}\nYours: {}".format(M_test.shape, M_test_ans.shape)
# Test correct M values
for w1 in word2Ind_ans.keys():
idx1 = word2Ind_ans[w1]
for w2 in word2Ind_ans.keys():
idx2 = word2Ind_ans[w2]
student = M_test[idx1, idx2]
correct = M_test_ans[idx1, idx2]
if student != correct:
print("Correct M:")
print(M_test_ans)
print("Your M: ")
print(M_test)
raise AssertionError("Incorrect count at index ({}, {})=({}, {}) in matrix M. Yours has {} but should have {}.".format(idx1, idx2, w1, w2, student, correct))
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80) | --------------------------------------------------------------------------------
Passed All Tests!
--------------------------------------------------------------------------------
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Question 1.3: Implement `reduce_to_k_dim` [code] (1 point)Construct a method that performs dimensionality reduction on the matrix to produce k-dimensional embeddings. Use SVD to take the top k components and produce a new matrix of k-dimensional embeddings. **Note:** All of numpy, scipy, and scikit-learn (`sklearn`) provide *some* implementation of SVD, but only scipy and sklearn provide an implementation of Truncated SVD, and only sklearn provides an efficient randomized algorithm for calculating large-scale Truncated SVD. So please use [sklearn.decomposition.TruncatedSVD](https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html). | def reduce_to_k_dim(M, k=2):
""" Reduce a co-occurence count matrix of dimensionality (num_corpus_words, num_corpus_words)
to a matrix of dimensionality (num_corpus_words, k) using the following SVD function from Scikit-Learn:
- http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.TruncatedSVD.html
Params:
M (numpy matrix of shape (number of unique words in the corpus , number of unique words in the corpus)): co-occurence matrix of word counts
k (int): embedding size of each word after dimension reduction
Return:
M_reduced (numpy matrix of shape (number of corpus words, k)): matrix of k-dimensioal word embeddings.
In terms of the SVD from math class, this actually returns U * S
"""
n_iters = 10 # Use this parameter in your call to `TruncatedSVD`
print("Running Truncated SVD over %i words..." % (M.shape[0]))
# ------------------
# Write your implementation here.
svd = TruncatedSVD(n_components=k, n_iter=n_iters)
svd.fit(M.T)
M_reduced = svd.components_.T
# ------------------
print("Done.")
return M_reduced
# ---------------------
# Run this sanity check
# Note that this is not an exhaustive check for correctness
# In fact we only check that your M_reduced has the right dimensions.
# ---------------------
# Define toy corpus and run student code
test_corpus = ["{} All that glitters isn't gold {}".format(START_TOKEN, END_TOKEN).split(" "), "{} All's well that ends well {}".format(START_TOKEN, END_TOKEN).split(" ")]
M_test, word2Ind_test = compute_co_occurrence_matrix(test_corpus, window_size=1)
M_test_reduced = reduce_to_k_dim(M_test, k=2)
# Test proper dimensions
assert (M_test_reduced.shape[0] == 10), "M_reduced has {} rows; should have {}".format(M_test_reduced.shape[0], 10)
assert (M_test_reduced.shape[1] == 2), "M_reduced has {} columns; should have {}".format(M_test_reduced.shape[1], 2)
# Print Success
print ("-" * 80)
print("Passed All Tests!")
print ("-" * 80) | Running Truncated SVD over 10 words...
Done.
--------------------------------------------------------------------------------
Passed All Tests!
--------------------------------------------------------------------------------
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Question 1.4: Implement `plot_embeddings` [code] (1 point)Here you will write a function to plot a set of 2D vectors in 2D space. For graphs, we will use Matplotlib (`plt`).For this example, you may find it useful to adapt [this code](https://www.pythonmembers.club/2018/05/08/matplotlib-scatter-plot-annotate-set-text-at-label-each-point/). In the future, a good way to make a plot is to look at [the Matplotlib gallery](https://matplotlib.org/gallery/index.html), find a plot that looks somewhat like what you want, and adapt the code they give. | def plot_embeddings(M_reduced, word2Ind, words):
""" Plot in a scatterplot the embeddings of the words specified in the list "words".
NOTE: do not plot all the words listed in M_reduced / word2Ind.
Include a label next to each point.
Params:
M_reduced (numpy matrix of shape (number of unique words in the corpus , 2)): matrix of 2-dimensioal word embeddings
word2Ind (dict): dictionary that maps word to indices for matrix M
words (list of strings): words whose embeddings we want to visualize
"""
# ------------------
# Write your implementation here.
for w in words:
x = M_reduced[word2Ind[w]][0]
y = M_reduced[word2Ind[w]][1]
plt.scatter(x, y, marker='x', color='red')
plt.text(x, y, w)
plt.show()
# ------------------
# ---------------------
# Run this sanity check
# Note that this is not an exhaustive check for correctness.
# The plot produced should look like the "test solution plot" depicted below.
# ---------------------
print ("-" * 80)
print ("Outputted Plot:")
M_reduced_plot_test = np.array([[1, 1], [-1, -1], [1, -1], [-1, 1], [0, 0]])
word2Ind_plot_test = {'test1': 0, 'test2': 1, 'test3': 2, 'test4': 3, 'test5': 4}
words = ['test1', 'test2', 'test3', 'test4', 'test5']
plot_embeddings(M_reduced_plot_test, word2Ind_plot_test, words)
print ("-" * 80) | --------------------------------------------------------------------------------
Outputted Plot:
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
**Test Plot Solution** Question 1.5: Co-Occurrence Plot Analysis [written] (3 points)Now we will put together all the parts you have written! We will compute the co-occurrence matrix with fixed window of 4 (the default window size), over the Reuters "crude" (oil) corpus. Then we will use TruncatedSVD to compute 2-dimensional embeddings of each word. TruncatedSVD returns U\*S, so we need to normalize the returned vectors, so that all the vectors will appear around the unit circle (therefore closeness is directional closeness). **Note**: The line of code below that does the normalizing uses the NumPy concept of *broadcasting*. If you don't know about broadcasting, check out[Computation on Arrays: Broadcasting by Jake VanderPlas](https://jakevdp.github.io/PythonDataScienceHandbook/02.05-computation-on-arrays-broadcasting.html).Run the below cell to produce the plot. It'll probably take a few seconds to run. What clusters together in 2-dimensional embedding space? What doesn't cluster together that you might think should have? **Note:** "bpd" stands for "barrels per day" and is a commonly used abbreviation in crude oil topic articles. | # -----------------------------
# Run This Cell to Produce Your Plot
# ------------------------------
reuters_corpus = read_corpus()
M_co_occurrence, word2Ind_co_occurrence = compute_co_occurrence_matrix(reuters_corpus)
M_reduced_co_occurrence = reduce_to_k_dim(M_co_occurrence, k=2)
# Rescale (normalize) the rows to make them each of unit-length
M_lengths = np.linalg.norm(M_reduced_co_occurrence, axis=1)
M_normalized = M_reduced_co_occurrence / M_lengths[:, np.newaxis] # broadcasting
words = ['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']
plot_embeddings(M_normalized, word2Ind_co_occurrence, words) | Running Truncated SVD over 8185 words...
Done.
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
The 'ecuador', 'energy', 'kuwait', 'oil', 'output', 'venezuela' cluster together which is intuitive.And the 'bpd', 'barrels' doesn't cluster together which should have. Part 2: Prediction-Based Word Vectors (15 points)As discussed in class, more recently prediction-based word vectors have demonstrated better performance, such as word2vec and GloVe (which also utilizes the benefit of counts). Here, we shall explore the embeddings produced by GloVe. Please revisit the class notes and lecture slides for more details on the word2vec and GloVe algorithms. If you're feeling adventurous, challenge yourself and try reading [GloVe's original paper](https://nlp.stanford.edu/pubs/glove.pdf).Then run the following cells to load the GloVe vectors into memory. **Note**: If this is your first time to run these cells, i.e. download the embedding model, it will take about 15 minutes to run. If you've run these cells before, rerunning them will load the model without redownloading it, which will take about 1 to 2 minutes. | def load_embedding_model():
""" Load GloVe Vectors
Return:
wv_from_bin: All 400000 embeddings, each lengh 200
"""
import gensim.downloader as api
wv_from_bin = api.load("glove-wiki-gigaword-200")
print("Loaded vocab size %i" % len(wv_from_bin.vocab.keys()))
return wv_from_bin
# -----------------------------------
# Run Cell to Load Word Vectors
# Note: This will take several minutes
# -----------------------------------
wv_from_bin = load_embedding_model() | Loaded vocab size 400000
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Note: If you are receiving reset by peer error, rerun the cell to restart the download. Reducing dimensionality of Word EmbeddingsLet's directly compare the GloVe embeddings to those of the co-occurrence matrix. In order to avoid running out of memory, we will work with a sample of 10000 GloVe vectors instead.Run the following cells to:1. Put 10000 Glove vectors into a matrix M2. Run reduce_to_k_dim (your Truncated SVD function) to reduce the vectors from 200-dimensional to 2-dimensional. | def get_matrix_of_vectors(wv_from_bin, required_words=['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']):
""" Put the GloVe vectors into a matrix M.
Param:
wv_from_bin: KeyedVectors object; the 400000 GloVe vectors loaded from file
Return:
M: numpy matrix shape (num words, 200) containing the vectors
word2Ind: dictionary mapping each word to its row number in M
"""
import random
words = list(wv_from_bin.vocab.keys())
print("Shuffling words ...")
random.seed(224)
random.shuffle(words)
words = words[:10000]
print("Putting %i words into word2Ind and matrix M..." % len(words))
word2Ind = {}
M = []
curInd = 0
for w in words:
try:
M.append(wv_from_bin.word_vec(w))
word2Ind[w] = curInd
curInd += 1
except KeyError:
continue
for w in required_words:
if w in words:
continue
try:
M.append(wv_from_bin.word_vec(w))
word2Ind[w] = curInd
curInd += 1
except KeyError:
continue
M = np.stack(M)
print("Done.")
return M, word2Ind
# -----------------------------------------------------------------
# Run Cell to Reduce 200-Dimensional Word Embeddings to k Dimensions
# Note: This should be quick to run
# -----------------------------------------------------------------
M, word2Ind = get_matrix_of_vectors(wv_from_bin)
M_reduced = reduce_to_k_dim(M, k=2)
# Rescale (normalize) the rows to make them each of unit-length
M_lengths = np.linalg.norm(M_reduced, axis=1)
M_reduced_normalized = M_reduced / M_lengths[:, np.newaxis] # broadcasting | Shuffling words ...
Putting 10000 words into word2Ind and matrix M...
Done.
Running Truncated SVD over 10010 words...
Done.
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
**Note: If you are receiving out of memory issues on your local machine, try closing other applications to free more memory on your device. You may want to try restarting your machine so that you can free up extra memory. Then immediately run the jupyter notebook and see if you can load the word vectors properly. If you still have problems with loading the embeddings onto your local machine after this, please follow the Piazza instructions, as how to run remotely on Stanford Farmshare machines.** Question 2.1: GloVe Plot Analysis [written] (4 points)Run the cell below to plot the 2D GloVe embeddings for `['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']`.What clusters together in 2-dimensional embedding space? What doesn't cluster together that you might think should have? How is the plot different from the one generated earlier from the co-occurrence matrix? What is a possible reason for causing the difference? | words = ['barrels', 'bpd', 'ecuador', 'energy', 'industry', 'kuwait', 'oil', 'output', 'petroleum', 'venezuela']
plot_embeddings(M_reduced_normalized, word2Ind, words) | _____no_output_____ | MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
venezuela with ecuador and industry with energy cluster together. petroleum and oil shoulf cluster together.The plot contine more semantic information than the co-occurrence matrix method.Maybe the counter of word is small in the data set. Cosine SimilarityNow that we have word vectors, we need a way to quantify the similarity between individual words, according to these vectors. One such metric is cosine-similarity. We will be using this to find words that are "close" and "far" from one another.We can think of n-dimensional vectors as points in n-dimensional space. If we take this perspective [L1](http://mathworld.wolfram.com/L1-Norm.html) and [L2](http://mathworld.wolfram.com/L2-Norm.html) Distances help quantify the amount of space "we must travel" to get between these two points. Another approach is to examine the angle between two vectors. From trigonometry we know that:Instead of computing the actual angle, we can leave the similarity in terms of $similarity = cos(\Theta)$. Formally the [Cosine Similarity](https://en.wikipedia.org/wiki/Cosine_similarity) $s$ between two vectors $p$ and $q$ is defined as:$$s = \frac{p \cdot q}{||p|| ||q||}, \textrm{ where } s \in [-1, 1] $$ Question 2.2: Words with Multiple Meanings (2 points) [code + written] Polysemes and homonyms are words that have more than one meaning (see this [wiki page](https://en.wikipedia.org/wiki/Polysemy) to learn more about the difference between polysemes and homonyms ). Find a word with at least 2 different meanings such that the top-10 most similar words (according to cosine similarity) contain related words from *both* meanings. For example, "leaves" has both "vanishes" and "stalks" in the top 10, and "scoop" has both "handed_waffle_cone" and "lowdown". You will probably need to try several polysemous or homonymic words before you find one. Please state the word you discover and the multiple meanings that occur in the top 10. Why do you think many of the polysemous or homonymic words you tried didn't work (i.e. the top-10 most similar words only contain **one** of the meanings of the words)?**Note**: You should use the `wv_from_bin.most_similar(word)` function to get the top 10 similar words. This function ranks all other words in the vocabulary with respect to their cosine similarity to the given word. For further assistance please check the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.htmlgensim.models.keyedvectors.FastTextKeyedVectors.most_similar)__. | # ------------------
# Write your implementation here.
wv_from_bin.most_similar("run")
# ------------------ | _____no_output_____ | MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
The run have running and start two meaning. Question 2.3: Synonyms & Antonyms (2 points) [code + written] When considering Cosine Similarity, it's often more convenient to think of Cosine Distance, which is simply 1 - Cosine Similarity.Find three words (w1,w2,w3) where w1 and w2 are synonyms and w1 and w3 are antonyms, but Cosine Distance(w1,w3) < Cosine Distance(w1,w2). For example, w1="happy" is closer to w3="sad" than to w2="cheerful". Once you have found your example, please give a possible explanation for why this counter-intuitive result may have happened.You should use the the `wv_from_bin.distance(w1, w2)` function here in order to compute the cosine distance between two words. Please see the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.htmlgensim.models.keyedvectors.FastTextKeyedVectors.distance)__ for further assistance. | # ------------------
# Write your implementation here.
w1 = "design"
w2 = "proposal"
w3 = "borrow"
w12_dist = wv_from_bin.distance(w1, w2)
w13_dist = wv_from_bin.distance(w1, w3)
print("Synonyms {}, {} have cosine distance: {:.2f}".format(w1, w2, w12_dist))
print("Antonyms {}, {} have cosine distance: {:.2f}".format(w1, w3, w13_dist))
# ------------------ | Synonyms design, proposal have cosine distance: 0.69
Antonyms design, borrow have cosine distance: 0.90
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Comapring proposal and design, the cosine distrance is 0.69. And the cosine distance between design and borrow is 0.9. Solving Analogies with Word VectorsWord vectors have been shown to *sometimes* exhibit the ability to solve analogies. As an example, for the analogy "man : king :: woman : x" (read: man is to king as woman is to x), what is x?In the cell below, we show you how to use word vectors to find x. The `most_similar` function finds words that are most similar to the words in the `positive` list and most dissimilar from the words in the `negative` list. The answer to the analogy will be the word ranked most similar (largest numerical value).**Note:** Further Documentation on the `most_similar` function can be found within the __[GenSim documentation](https://radimrehurek.com/gensim/models/keyedvectors.htmlgensim.models.keyedvectors.FastTextKeyedVectors.most_similar)__. | # Run this cell to answer the analogy -- man : king :: woman : x
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'king'], negative=['man'])) | [('queen', 0.6978678703308105),
('princess', 0.6081745028495789),
('monarch', 0.5889754891395569),
('throne', 0.5775108933448792),
('prince', 0.5750998854637146),
('elizabeth', 0.546359658241272),
('daughter', 0.5399125814437866),
('kingdom', 0.5318052768707275),
('mother', 0.5168544054031372),
('crown', 0.5164472460746765)]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Question 2.4: Finding Analogies [code + written] (2 Points)Find an example of analogy that holds according to these vectors (i.e. the intended word is ranked top). In your solution please state the full analogy in the form x:y :: a:b. If you believe the analogy is complicated, explain why the analogy holds in one or two sentences.**Note**: You may have to try many analogies to find one that works! | # ------------------
# Write your implementation here.
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'waitress'], negative=['man']))
# ------------------ | [('barmaid', 0.6116799116134644),
('bartender', 0.5877381563186646),
('receptionist', 0.5782569646835327),
('waiter', 0.5508327484130859),
('waitresses', 0.5503603219985962),
('hostess', 0.5346562266349792),
('housekeeper', 0.5310243368148804),
('homemaker', 0.5298492908477783),
('prostitute', 0.5254124402999878),
('housewife', 0.5207685232162476)]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
woman:man::waitress:waiter, through the probability of waiter isn't the maximum. Question 2.5: Incorrect Analogy [code + written] (1 point)Find an example of analogy that does *not* hold according to these vectors. In your solution, state the intended analogy in the form x:y :: a:b, and state the (incorrect) value of b according to the word vectors. | # ------------------
# Write your implementation here.
pprint.pprint(wv_from_bin.most_similar(positive=['high', 'jump'], negative=['low']))
# ------------------ | [('jumping', 0.6205310225486755),
('jumps', 0.5840020775794983),
('leap', 0.5402169823646545),
('jumper', 0.4817255735397339),
('climb', 0.4797284007072449),
('bungee', 0.464731365442276),
('championships', 0.4643418788909912),
('jumped', 0.46396756172180176),
('triple', 0.4550389349460602),
('throw', 0.4516879916191101)]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
the high:low shoudle == jump:fall, but the fall not in the top 10 probability list. Question 2.6: Guided Analysis of Bias in Word Vectors [written] (1 point)It's important to be cognizant of the biases (gender, race, sexual orientation etc.) implicit in our word embeddings. Bias can be dangerous because it can reinforce stereotypes through applications that employ these models.Run the cell below, to examine (a) which terms are most similar to "woman" and "worker" and most dissimilar to "man", and (b) which terms are most similar to "man" and "worker" and most dissimilar to "woman". Point out the difference between the list of female-associated words and the list of male-associated words, and explain how it is reflecting gender bias. | # Run this cell
# Here `positive` indicates the list of words to be similar to and `negative` indicates the list of words to be
# most dissimilar from.
pprint.pprint(wv_from_bin.most_similar(positive=['woman', 'worker'], negative=['man']))
print()
pprint.pprint(wv_from_bin.most_similar(positive=['man', 'worker'], negative=['woman'])) | [('employee', 0.6375863552093506),
('workers', 0.6068919897079468),
('nurse', 0.5837947726249695),
('pregnant', 0.5363885164260864),
('mother', 0.5321309566497803),
('employer', 0.5127025842666626),
('teacher', 0.5099576711654663),
('child', 0.5096741914749146),
('homemaker', 0.5019454956054688),
('nurses', 0.4970572590827942)]
[('workers', 0.6113258004188538),
('employee', 0.5983108282089233),
('working', 0.5615328550338745),
('laborer', 0.5442320108413696),
('unemployed', 0.5368517637252808),
('job', 0.5278826951980591),
('work', 0.5223963260650635),
('mechanic', 0.5088937282562256),
('worked', 0.505452036857605),
('factory', 0.4940453767776489)]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
The word most similar to "woman" and "worker" and most dissimilar to "man" is nurses.Most nurses are woman.The word most similar to "man" and "worker" and most dissimilar to "woman" is factory.The factory have many worker man. Question 2.7: Independent Analysis of Bias in Word Vectors [code + written] (1 point)Use the `most_similar` function to find another case where some bias is exhibited by the vectors. Please briefly explain the example of bias that you discover. | # ------------------
# Write your implementation here.
pprint.pprint(wv_from_bin.most_similar(positive=['elephant', 'skyscraper'], negative=['ant']))
print()
pprint.pprint(wv_from_bin.most_similar(positive=['motorcycle', 'car'], negative=['bicycle']))
# ------------------ | [('tower', 0.49941301345825195),
('skyscrapers', 0.48599374294281006),
('tallest', 0.46377506852149963),
('statue', 0.4558914303779602),
('towers', 0.44428494572639465),
('40-story', 0.4247894287109375),
('monument', 0.4171640872955322),
('high-rise', 0.41149571537971497),
('bust', 0.408037006855011),
('gleaming', 0.40352344512939453)]
[('cars', 0.6663373112678528),
('driver', 0.6263732314109802),
('vehicle', 0.6231670379638672),
('mercedes', 0.6017158627510071),
('truck', 0.581316351890564),
('driving', 0.5702999234199524),
('motorbike', 0.5668439865112305),
('bmw', 0.5605546236038208),
('vehicles', 0.5472753047943115),
('motorcycles', 0.5434989929199219)]
| MIT | CS224n/assignment1/exploring_word_vectors.ipynb | iofu728/Task |
Plotting File for [Redistributing the Gains From Trade Through Progressive Taxation](http://www.waugheconomics.com/uploads/2/2/5/6/22563786/lw_tax.pdf)This notebook imports the output from the MATLAB code and then plots it. Description is below. | from IPython.display import display, Image # Displays things nicely
import pandas as pd
import weightedcalcs as wc
import numpy as np
import statsmodels.api as sm
import statsmodels.formula.api as smf
import matplotlib.pyplot as plt
from scipy.io import loadmat # this is the SciPy module that loads mat-files
#fig_path = "C:\\Users\\mwaugh.NYC-STERN\\Documents\\GitHub\\tradeexposure\\figures" | C:\Program Files\Anaconda3\lib\site-packages\statsmodels\compat\pandas.py:56: FutureWarning: The pandas.core.datetools module is deprecated and will be removed in a future version. Please use the pandas.tseries module instead.
from pandas.core import datetools
| MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
--- Read in output from modelThis is the structure of the mat file and the naming conventions. Here it is assumed that the .mat files from Matlab are within the working directory. Then read it in, note the use of ``scipy`` package to get a .mat file into python. | #[params.trade_cost, trade, ls, move, output_per_hour, welfare, double(exit_flag)];
column_names = ["tau_p", "tau", "trade_volume", "ls", "migration", "output", "OPterm2", "welfare", "exitflag", "welfare_smth",
"trade_share"]
values = ["0.05","0.1", "0.2", "0.3", "0.4"]
all_df = pd.DataFrame([])
for val in values:
file_name = "results" + val + ".mat"
mat = loadmat(file_name)
df = pd.DataFrame(mat["results"])
df["9"] = val
all_df = all_df.append(df)
all_df.columns = column_names
all_df.head(10) | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
Now define some functions that we will use... | def cons_eqiv(df):
maxwel = float(df["welfare_smth"][df["tau_p"] == 0.18])
df["cons_eqiv"] = 100*(np.exp((1-0.95)*(df["welfare_smth"] - maxwel))-1)
# These are consumptione equivialents.
return df | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
Group on the trade share values.... | grp = all_df.groupby("trade_share")
grp = grp.apply(cons_eqiv)
grp = grp.groupby("trade_share") | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
--- Optimal policy in the modelThen the next two code cells will replicate Figure 3 and Figure 6 in the paper... | fig, ax = plt.subplots(figsize = (10,7))
val = "0.1"
ax.plot(grp.get_group(val).tau_p, grp.get_group(val).cons_eqiv,
linewidth = 4, label = "Imports/GDP = " + val,
color = "blue",alpha = 0.70)
index_max = grp.get_group(val).cons_eqiv.idxmax()
tau_max = grp.get_group(val).tau_p.iloc[index_max]
ax.plot(tau_max,
grp.get_group(val).cons_eqiv.iloc[index_max], 'ro',
markersize=10, linewidth = 50,
color = "red",alpha = 0.50)
ax.set_ylabel("Welfare (CE Units), Percent from Baseline", fontsize = 14)
ax.set_xlabel("Tax Progressivity", fontsize = 14)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.axvline(x = 0.18,
color='k',
linestyle='--',
lw = 2, alpha = 0.5)
#ax.legend(fontsize = 14, frameon=False)
ax.annotate(
"Optimal Progressivity \ntau_p* = " + str(tau_max),
xy=(tau_max, 0.15), # This is where we point at...
xycoords="data", # Not exactly sure about this
xytext=(0.4, 1.5), # This is about where the text is
horizontalalignment="left", # How the text is alined
arrowprops={
"arrowstyle": "-|>", # This is stuff about the arrow
"connectionstyle": "angle3,angleA=5,angleB=85",
"color": "black"
},
fontsize=14,
)
ax.annotate(
"US Data (HSV estimate) \ntau_p = 0.18",
xy=(0.18, 0.05), # This is where we point at...
xycoords="data", # Not exactly sure about this
xytext=(-0.10, 1.5), # This is about where the text is
horizontalalignment="left", # How the text is alined
arrowprops={
"arrowstyle": "-|>", # This is stuff about the arrow
"connectionstyle": "angle3,angleA=0,angleB=150",
"color": "black"
},
fontsize=14,
)
ax.set_xlim(-0.25,0.6)
ax.set_ylim(-3,1.5)
#plt.savefig(fig_path + "\\social_welfare_baseline.pdf", bbox_inches = "tight", dip = 3600)
plt.show()
fig, ax = plt.subplots(figsize = (10,7))
ax.set_prop_cycle('color',plt.cm.seismic(np.linspace(0,1.15,5)))
welfare_opt = []
tax_opt = []
flat_loss = []
oxford_data = pd.DataFrame()
for val in values[1:]:
ax.plot(grp.get_group(val).tau_p, grp.get_group(val).cons_eqiv,
linewidth = 4, label = "Imports/GDP = " + val, alpha = 0.70)
index_max = grp.get_group(val).cons_eqiv.idxmax()
ax.plot(grp.get_group(val).tau_p.iloc[index_max],
grp.get_group(val).cons_eqiv.iloc[index_max], marker ="o",
markersize=10, linewidth = 50,
color = "red",alpha = 0.50)
welfare_opt.append(grp.get_group(val).cons_eqiv.iloc[index_max])
tax_opt.append(grp.get_group(val).tau_p.iloc[index_max])
oxford_data = pd.concat([oxford_data, grp.get_group(val).cons_eqiv],axis=1)
flat_idx = grp.get_group(val).tau_p == 0
flat_loss.append(float(grp.get_group(val).cons_eqiv[flat_idx]))
ax.set_ylabel("Welfare (CE Units), Percent from Baseline", fontsize = 14)
ax.set_xlabel("Tax Progressivity", fontsize = 14)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.axvline(x = 0.18,
color='k',
linestyle='--',
lw = 2, alpha = 0.5)
ax.legend(fontsize = 14, frameon=False)
ax.set_xlim(-0.25,0.6)
ax.set_ylim(-7.5,1.5)
#plt.savefig(fig_path + "\\social_welfare_prog_diff_tau_fine.pdf", bbox_inches = "tight", dip = 3600)
plt.show()
###########################################################################################################
#oxford_data = pd.concat([oxford_data, grp.get_group(val).tau_p],axis=1)
#oxford_names = values
#oxford_names.append("tax_progressivity")
#oxford_data.columns = oxford_names[1:]
#values.remove("tax_progressivity")
#oxford_data.to_excel('oxford_fig2.xlsx')
print(welfare_opt)
print(tax_opt)
print(flat_loss)
#grp.get_group(val).head() | [0.10435983980994212, 0.34328431738519516, 0.7242843517205388, 1.3810768398272222]
[0.27, 0.32, 0.37, 0.44999999999999996]
[-0.8279921208671714, -1.3903253040046915, -1.9424125246026658, -2.628430117595859]
| MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
This finds the optimal tau... | opt_tau = []
tau = []
trade = []
for val in values:
index_max = grp.get_group(val).cons_eqiv.idxmax()
tau_star = grp.get_group(val).tau_p.iloc[index_max]
opt_tau.append(tau_star)
tau.append(grp.get_group(val).tau.iloc[index_max])
trade.append(float(val))
hold = {"opt_tau": opt_tau, "trade": trade, "tau": tau}
opt_df = pd.DataFrame(hold)
opt_df.head(10) | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
This then generates the output cost figure, Figure 8 in the paper. | def smooth_reg(df, series):
specification = series + "~ tau_p + np.square(tau_p) + np.power(tau_p,3)+ np.power(tau_p,4)"
results = smf.ols(specification , # This is the model in variable names we want to estimate
data=df[df["exitflag"]==0]).fit()
pred = results.predict(exog = df["tau_p"])
#print(results.summary())
return pred
fig, ax = plt.subplots(figsize = (10,7))
series = "output"
ax.set_prop_cycle('color',plt.cm.seismic(np.linspace(0,1.15,5)))
for val in values[1:]:
#baseline = float(grp.get_group(val)[grp.get_group(val).tau_p == 0.18][series])
ypred = smooth_reg(grp.get_group(val), series)
baseline = float(ypred[grp.get_group(val).tau_p == 0.18])
real = grp.get_group(val)[series]
index_max = grp.get_group(val).cons_eqiv.idxmax()
ax.plot(grp.get_group(val).tau_p, 100*(ypred/baseline - 1),
linewidth = 4, label = "Imports/GDP = " + val, alpha = 0.70)
index_max = grp.get_group(val).cons_eqiv.idxmax()
ax.plot(grp.get_group(val).tau_p.iloc[index_max],
100*(ypred.iloc[index_max]/baseline - 1), marker ="o",
markersize=10, linewidth = 50,
color = "red",alpha = 0.50)
####################################################################################
ax.axvline(x = 0.18,
color='k',
linestyle='--',
lw = 2, alpha = 0.5)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.set_ylabel("GDP, Percentage Points from Baseline", fontsize = 14)
ax.legend(fontsize = 14, frameon=False)
ax.set_xlim(-0.25,0.6)
#plt.savefig(fig_path + "\\output_cost.pdf", bbox_inches = "tight", dip = 3600)
plt.show() | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
This then generates the allocative efficiency (covariance term) figure, Figure 4 | fig, ax = plt.subplots(figsize = (10,7))
series = "output"
val = "0.1"
#baseline = float(grp.get_group(val)[grp.get_group(val).tau_p == 0.18][series])
ypred = smooth_reg(grp.get_group(val), series)
baseline = float(ypred[grp.get_group(val).tau_p == 0.18])
real = grp.get_group(val)[series]
index_max = grp.get_group(val).cons_eqiv.idxmax()
ax.plot(grp.get_group(val).tau_p, 100*(ypred /baseline-1),
linewidth = 4, label = "GDP",
color = 'blue', alpha = 0.75)
optimal_prog = grp.get_group(val).tau_p.iloc[index_max]
ax.plot(optimal_prog,
100*(ypred /baseline-1).iloc[index_max], 'ro',
markersize=10, linewidth = 50,
color = "red",alpha = 0.50)
####################################################################################
series = "OPterm2"
ypred = smooth_reg(grp.get_group(val), series)
ypred_output = smooth_reg(grp.get_group(val), "output")
baseline_output = float(ypred_output[grp.get_group(val).tau_p == 0.18])
baseline = float(ypred[grp.get_group(val).tau_p == 0.18])
real = grp.get_group(val)[series]
index_max = grp.get_group(val).cons_eqiv.idxmax()
ax.plot(grp.get_group(val).tau_p, 100*((ypred)/ypred_output - baseline/baseline_output),
linewidth = 4, label = "Covariance Term (Allocative Efficiency)",
alpha = 0.75, color = "red", linestyle='--')
####################################################################################
ax.axvline(x = 0.18,
color='k',
linestyle='--',
lw = 2, alpha = 0.5)
ax.annotate(
"Optimal Progressivity \ntau_p* =" + str(optimal_prog),
xy=(0.27, -0.35), # This is where we point at...
xycoords="data", # Not exactly sure about this
xytext=(0.4, 0.25), # This is about where the text is
horizontalalignment="left", # How the text is alined
arrowprops={
"arrowstyle": "-|>", # This is stuff about the arrow
"connectionstyle": "angle3,angleA=5,angleB=85",
"color": "black"
},
fontsize=14,
)
ax.set_ylabel("Percentage Points from Baseline", fontsize = 14)
ax.set_xlabel("Tax Progressivity", fontsize = 14)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.set_xlim(-0.25,0.6)
ax.legend(loc = "lower left", fontsize = 14, frameon=False)
#plt.savefig(fig_path + "\\output_baseline.pdf", bbox_inches = "tight", dip = 3600)
plt.show()
| _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
Then the migration figure, Figure 5 | fig, ax = plt.subplots(figsize = (10,7))
series = "migration"
val = "0.1"
baseline = float(grp.get_group(val)[grp.get_group(val).tau_p == 0.18][series])
ypred = smooth_reg(grp.get_group(val), series)
ax.plot(grp.get_group(val).tau_p, 100*(ypred /baseline-1),
linewidth = 4, label = "Migration",
color = 'blue', alpha = 0.70)
series = "ls"
val = "0.1"
baseline = float(grp.get_group(val)[grp.get_group(val).tau_p == 0.18][series])
ypred = smooth_reg(grp.get_group(val), series)
real = grp.get_group(val)[series]
ax.plot(grp.get_group(val).tau_p, 100*(ypred /baseline-1),
linewidth = 4, label = "Labor Supply",
color = 'red', alpha = 0.70, linestyle = "--")
###########################################################################################
ax.axvline(x = 0.18,
color='k',
linestyle='--',
lw = 2, alpha = 0.5)
ax.set_ylabel("Percentage Points from Baseline", fontsize = 14)
ax.set_xlabel("Tax Progressivity", fontsize = 14)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.legend(fontsize = 14, frameon=False)
ax.set_xlim(-0.25,0.6)
#plt.savefig(fig_path + "\\migration_baseline.pdf", bbox_inches = "tight", dip = 3600)
plt.show() | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
--- Marginal tax rates in the model | values_TAX = ["0.05","0.1b", "0.1", "0.2", "0.3", "0.4"]
mat = loadmat("opt_marg_rates")
marginal_rates = pd.DataFrame(mat["marg_rates"])
marginal_rates.columns = values_TAX
mat = loadmat("opt_incom_prct")
income_pct = pd.DataFrame(mat["incom_prct"])
income_pct.columns = values_TAX
def smooth_marg_rates(income_pct, marginal_rates, op_level):
df = pd.DataFrame([income_pct.T.loc[op_level], marginal_rates.T.loc[op_level]])
df = df.T
df.columns = ["inc_prct", "marg_rates"]
specification = '''marg_rates ~ np.log(inc_prct) + np.square(np.log(inc_prct)) +
np.power(np.log(inc_prct),3)+ np.power(np.log(inc_prct),4)'''
results = smf.ols(specification , # This is the model in variable names we want to estimate
data=df).fit()
pred = results.predict(exog = df["inc_prct"])
#print(results.summary())
return pred
fig, ax = plt.subplots(figsize = (10,7))
ax.set_prop_cycle('color',plt.cm.seismic(np.linspace(0,1.15,5)))
for val in values[1:]:
pred = smooth_marg_rates(income_pct, marginal_rates, val)
#ax.plot(100*(income_pct[val]), 100*pred,linewidth = 4,
# alpha = 0.70, label = "Imports/GDP = " + val)
if val == "0.1b":
print(" ")
ax.plot(100*income_pct[val], 100*pred,linewidth = 4,
alpha = 0.70, color = "black", linestyle = '--', label = "Baseline")
else:
ax.plot(100*income_pct[val], 100*pred,linewidth = 4,
alpha = 0.70, label = "Imports/GDP = " + val)
idx = (np.abs(income_pct[val]-0.90)).idxmin()
print(100*pred[idx])
##############################################################################
ax.set_ylabel("Marginal Tax Rates, Percent", fontsize = 14)
ax.set_xlabel("Pre-Tax Labor Income Percentile", fontsize = 14)
ax.set_ylim(-10,70)
ax.set_xlim(-2,91)
test = list(range(0,100,10))
#test.append(90)
ax.set_xticks(test)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.legend(loc = "lower right", fontsize = 14, frameon=False)
#plt.savefig(fig_path + "\\marginal_rates.pdf", bbox_inches = "tight", dip = 3600)
plt.show()
opt_tax = []
prctile_income = 0.10
for val in values_TAX[1:]:
pred = smooth_marg_rates(income_pct, marginal_rates, val)
#ax.plot(100*(income_pct[val]), 100*pred,linewidth = 4,
# alpha = 0.70, label = "Imports/GDP = " + val)
if val == "0.1b":
print(" ")
else:
idx = (np.abs(income_pct[val]-prctile_income)).idxmin()
opt_tax.append(100*pred[idx])
#######################################################################
elasticity = (opt_tax[-1] - opt_tax[1])/(40-10)
fig, ax = plt.subplots(figsize = (10,7))
ax.plot(values[1:], opt_tax, linewidth = 5, alpha = 0.70, color = "red", linestyle = '--')
#test.append(90)
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
ax.set_ylabel("Marginal Tax Rates for 90th Percentile", fontsize = 14)
ax.set_xlabel("Imports/ GDP", fontsize = 14)
#ax.set_ylim(35,70)
test = list(range(0,100,10))
#test.append(90)
ax.set_xticklabels(test)
plt.show()
def gains_trade(df, tax_policy):
new_df = df[df.tau_p == tax_policy]
basewel = float(new_df["welfare"][new_df["trade_share"] == "0.1"])
new_df["cons_eqiv"] = 100*(np.exp((1-0.95)*(new_df["welfare_smth"] - basewel))-1)
return new_df
gains_trade(all_df, 0.18)
#100*(-27.633091 / -(-26.18) + 1)
100*(np.exp((1-0.95)*(-26.528930- -27.633091))-1)
100*(-27.633091 / -(-26.528930) + 1)
100*(np.exp((1-0.95)*(-26.248914- -27.466803))-1)
100*(np.exp((1-0.95)*(-26.248914- -27.633091))-1)
7.16/5.67 | _____no_output_____ | MIT | matlab/plot_model_data/plot_model_results.ipynb | mwaugh0328/redistributing_gains_from_trade |
在上面的例子中,数据存储为多维Numpy数组,也称为张量(tensor)。当前流行的机器学习系统都以张量作为基本数据结构。所以Google的TensorFlow也拿张量命名。那张量是什么呢?张量是数据的容器(container)。这里的数据一般是数值型数据,所以是数字的容器。大家所熟悉的矩阵是二维(2D)张量。张量是广义的矩阵,它的某一维也称为轴(axis)。 标量(Scalar,0D 张量)只包含一个数字的张量称为标量(或者数量张量,零维张量,0D张量)。在Numpy中,一个float32或者float64位的数值称为数量张量。Numpy张量可用其ndim属性显示轴的序数,数量张量有0个轴(ndim == 0)。张量的轴的序数也称为阶(rank)。下面是Numpy标量: | import numpy as np
x = np.array(12)
x
x.ndim | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
向量(1D张量)数字的数组也称为向量,或者一维张量(1D张量)。一维张量只有一个轴。 | x = np.array([12, 3, 6, 14])
x
x.ndim | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
该向量有5项,也称为5维的向量。但是不要混淆5D向量和5D张量!一个5D向量只有一个轴,以及沿该轴有5个维数(元素);然而一个5D张量有5个轴,并且沿每个轴可以有任意个的维数。维度既能表示沿某个轴的项的数量(比如,上面的5D向量),又能表示一个张量中轴的数量(比如,上面的5D张量),时常容易混淆。对于后者,用更准确地技术术语来讲,应该称为5阶张量(张量的阶即是轴的数量),但人们更常用的表示方式是5D张量。 矩阵(2D张量)向量的数组称为矩阵,或者二维张量(2D张量)。矩阵有两个轴,也常称为行和列。你可以将数字排成的矩形网格看成矩阵,下面是一个Numpy矩阵: | x = np.array([[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]])
x.ndim | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
沿着第一个轴的项称为行,沿着第二个轴的项称为列。上面的例子中,[5, 78, 2, 34, 0]是矩阵 x 第一行,[5, 6, 7]是第一列。矩阵的数组称为三维张量(3D张量),你可以将其看成是数字排列成的立方体,下面是一个Numpy三维张量(注意该三维张量内部的三个二维张量的shape一致,均为(3,5)。维度是一个自然数,形状则是一个元组): | x = np.array([[[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]],
[[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]],
[[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]]])
x.ndim | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
题外话!若张量某维度的元素未对其,则这些元素成为list。 | x = np.array([[[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]],
[[5, 78, 2, 34, 0],
[6, 79, 3, 35, 1],
[7, 80, 4, 36, 2]],
[[5, 78, 2, 34, 0],
[7, 80, 4, 36, 2]]])
x
x.ndim | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
同理,将三维张量放进数组可以创建四维张量,其它更高维的张量亦是如此。深度学习中常用的张量是 0D 到 4D。如果处理视频数据,你会用到5D。 关键属性张量具有如下三个关键属性:1. 轴的数量(阶数,rank):一个三维张量有3个轴,矩阵有2个轴。Python Numpy中的张量维度为ndim。2. 形状(shape):它是一个整数元组,描述张量沿每个轴有多少维。例如,前面的例子中,矩阵的形状为(3,5),三维张量的形状为(3,3,5)。向量的形状只有一个元素,比如(5,),标量则是空形状,()。3. 数据类型:张量中包含的数据类型有float32,unit8,float64等等,调用Python的dtype属性获取。字符型张量是极少见的。注意,Numpy中不存在字符串张量,其它大部分库也不存在。因为张量存在于预先申请的、连续的内存分段;而字符是变长的。 下面来几个具体的例子,回看MNIST数据集。首先加载MNIST数据集: | from keras.datasets import mnist
(train_images, train_labels), (test_images, test_labels) = mnist.load_data() | e:\program_files\miniconda3\envs\dl\lib\site-packages\h5py\__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.
from ._conv import register_converters as _register_converters
Using TensorFlow backend.
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
接着,用ndim属性显示张量train_images的轴数量: | print(train_images.ndim) | 3
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
打印形状: | print(train_images.shape) | (60000, 28, 28)
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
使用dtype属性打印数据类型: | print(train_images.dtype) | uint8
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
所以train_images是一个8-bit 整数的三维张量。更确切地说,它是一个包含60,000个矩阵的数组,其中每个矩阵是28 x 8 的整数。每个矩阵是一个灰度图,其值为0到255。下面使用Python Matplotlib库显示三维张量中的第四幅数字图,见图2.2: | #Listing 2.6 Dispalying the fourth digit
digit = train_images[4]
import matplotlib.pyplot as plt
plt.imshow(digit, cmap=plt.cm.binary)
plt.show() | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
注:这里使用 matplotlib.cm, cm 表示 colormap。 binary map:https://en.wikipedia.org/wiki/Binary_image digit 是从这个三维张量取出的一个矩阵(二维数组/张量): | print(digit.ndim,",",digit.shape) | 2 , (28, 28)
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Numpy中的张量操作上面的例子中,使用了train_images[i]沿第一个轴选择指定的数字图。选择张量的指定元素称为张量分片(tensor slicing),下面看Numpy数组中的张量切片操作: 选择10到100(不包括100)的数字图,对应的张量形状为(90,28,28): | my_slice = train_images[10:100]
print(my_slice.shape) | (90, 28, 28)
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
其等效的表示方法有,沿每个轴为张量分片指定起始索引和终止索引。注意,“:”等效于选择整个轴的数据: | my_slice = train_images[10:100, 0:28, 0:28]
my_slice.shape | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
一般,你可以沿着张量每个轴任意选择两个索引之间的元素。例如,选择所有图片的右下角的14 x 14的像素: | my_slice = train_images[:, 14:, 14:] | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
你也可以用负索引。就像Python list中的负索引一样,它表示相对于当前轴末端的位置。剪切图片中间14 x 14像素,使用如下的方法: | my_slice = train_images[:, 7:-7, 7:-7] | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
数据批(data batch)的概念总的来说,你在深度学习中即将接触的所有数据张量的第一轴(axis 0,since indexing starts at 0)就叫“样本轴”(samples axis,也叫“样本维”)。简单地说,MINIST例子中的“样本(samples)”就是那些数字图片。 另外,深度学习模型不会一次处理整个数据集,而是将数据集分解为若干个小批次。具体来讲,下面就是MNIST数据集中一个大小为128的批次。 | # Listing 2.23 Slicing a tensor into batches
batch = train_images[:128]
# and here's the next batch
batch = train_images[128:256]
# and the n-th batch:
#batch = train_images[128 * n: 128 * (n + 1)]
batch.shape | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
考虑这样一个批张量,第一轴(axis 0)就称为“批次轴”(batch axis)或“批次维数”(batch dimension)。这是一个术语,当你使用Keras或其他深度学习库时你会经常接触到。 现实中data tensors的例子让我们让数据张量更具体,还有一些类似于你稍后会遇到的例子。 你将操作的数据几乎总是属于下列类别之一:1. 向量数据:2D 张量,shape 为(samples,features)2. 时间序列数据或序列数据:3D 张量,shape 为(samples,timesteps,features)3. 图像: 4D 张量,shape 为(samples,width,height,channels)或者(samples,channels,width,height)4. 视频:5D 张量,shape 为(samples,frames,width,height,channels)或者(samples,frames,channels,width,height) 向量数据 时间序列数据或序列数据 图像 视频 The gears of neural networks: tensor operations 就像计算机程序可以最终被降阶为一系列对二值输入的二值操作一样,深度神经网络中的所有变换可以被降阶至一系列对数值数据张量的“张量操作”。例如,可以对张量实施add,multiply等操作。 在我们最开始的例子中,我们通过逐个地堆叠 Dense 层搭建了神经网络。一个神经网络层看起来是这样的: | #Listing 2.24 A Keras layer
#keras.layers.Dense(512, activation='relu') | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
这个神经网络层,可以用一个函数(function)来解释,这个函数输入一个2D张量,并返回另一个2D张量,这个张量是对输入张量的新描述。具体地,该方程为:  让我们来分解它。这个方程里面有三个张量操作:输入张量和W张量的点乘(dot),得到的2D张量和向量b的加(+)操作,最后是一个relu操作。rulu(x) 就是简单的 max(x,0)。 虽然这些操作完全是线性代数计算,但你会发现这里没有任何数学符号。因为我们发现当没有相应数学背景的编程人员使用Python语句而不是数学方程式时,他们更能够掌握。所以这里我们一直使用Numpy代码。 Element-wise operations“relu”操作和加法操作都是element-wise操作(独立地对张量的每一个元素实施计算)。这意味着这些操作对大量并行运算是非常适合的(这类操作也叫“vectorized” implementations,向量化运算。)。如果你要写一个element-wise 操作的朴素Python实现,你将使用到for循环: | #Listing 2.25 A naive implemetation of an element-wise "relu" operation
def naive_relu(x):
# x is a 2D Numpy tensor
assert len(x.shape) == 2
x = x.copy() # Avoid overwrinting the input tensor
for i in range(x.shape[0]:
for j in range(x.shape[1]):
x[i, j] = max(x[i, j], 0)
return x | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
同样的,对于加法操作有: | #Listing 2.26 A naive implementation of element-wise addition
def naive_add(x, y):
# x and y are 2D Numpy tensors
assert len(x.shape) == 2
assert x.shape == y.shape
x = x.copy() # Avoid overwriting the input tensor
for i in range(x.shape[0]):
for j in range(x.shape[1]):
x[i, j] += y[i, h]
return x | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
使用相同的方法,我们可以实现element-wise multiplication,subtraction等运算。 In practice, when dealing with Numpy arrays, these operations are available as well-optimized built-in Numpy functions, which themselves delegate the heavy lifting to a BLAS implementation (Basic Linear Algebra Subprograms) if you have one installed, which you should. BLAS are low-level, highly-parallel, efficient tensor manipulation routines typically implemented in Fortran or C. 所以在Numpy中你可以这样实施element-wise,速度非常快。 | # Listing 2.27 Naive element-wise operation in Numpy
import numpy as np
# Element-wise addtion
#z = x + y
# Element-wise relu
#z = np.maximum(z, 0.) | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Broadcasting In our naive implementation of above, we only support the addition of 2D naive_add tensors with identical shapes. But in the layer introduced earlier, we were adding a Dense 2D tensor with a vector. What happens with addition when the shape of the two tensors being added differ? When possible and if there is no ambiguity, the smaller tensor will be "broadcasted" to match the shape of the larger tensor. Broadcasting consists in two steps: 1. 维度小的张量添加一个轴,以和维度大的张量的维度(ndim)适配(该操作为 broadcast axes)2. 维度小的张量沿着新轴拷贝,以和维度大的张量的形状(shape)适配。 让我们来看一个具体的例子:考虑shape为(32,10)的张量x,和shape为(10,)的张量y。 首先,首先我们给y张量添加一个第一轴,这时y的shape变成(1,10)。接着沿着新轴重复y 32次,得到shape为(32,10)的张量Y。即: | # Y[i,:] = y for i in range(0, 32) | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
In terms of implementation, no new 2D tensor would actually be created since that would be terribly inefficient, so the repetition operation would be entirely virtual, i.e. it would be happening at the algorithmic level rather than at the memory level. But thinking of the vector being repeated 10 times alongside a new axis is a helpful mental model. Here’s what a naive implementation would look like: | def naive_add_matrix_and_vector(x, y):
# x is a 2D Numpy tensor
# y is a Numpy vector
assert len(x.shape) == 2
assert len(y.shape) == 1
assert x.shape[10] == y.shape[0]
x = x.copy() # Avoid overwriting the input tensor
for i in range(x.shape[0]):
for j in range(x.shape[1]):
x[i, j] += y[j]
return x | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
With broadcasting, you can generally apply two-tensor element-wise operations if one tensor has shape and the other has shape (a, b, … n, n + 1, … m) and the other has shpae (n, n + 1, ... m). The broadcasting would then automatically happen for axes a to n -1. You can thus do:(注意下面这里扩展的轴的维度为2,而不是1!) | ### Listing 2.29 Applying the element-wise operation to two tensors of maximum different shapes via broadcastin
import numpy as np
# x is a random tensor with shape (64, 3, 32, 10)
x = np.random.random((64, 3, 32, 10))
# y is a random tensor with shape (32, 10)
y = np.random.random((32, 10))
# The output z has shape (64, 3, 32, 10) like x
z = np.maximum(x, y) | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Tensor dotdot操作, 也叫张量乘法(tensor product),不要将其和element-wise混淆。它是张量运算中最常见和最重要的。 与element-wise相反,它将输入张量中的元素组合在一起(组合有权重)。 Element-wise product is done with the * operator in Numpy, Keras, Theano and TensorFlow. uses a different syntax in TensorFlow, but in both Numpy and Keras it dot is done using the standard operator: | # Listing 2.30 Numpy operations between two tensors
import numpy as np
#z = np.dot(x, y) | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
In mathematical notation, you would note the operation with a dot . : z = x . y Mathematically, what does the dot operation do? Let’s start with the dot product of two vectors x and y. It is computed as such: | # Listing 2.31 A naive implementation of dot
def naive_vector_dot(x, y):
# x and y are Numpy vectors
assert len(x.shape) == 1
assert len(y.shape) == 1
assert x.shape[0] == y.shape[0]
z = 0.
for i in range(x.shape[0]):
z += x[i] * y[i]
return z | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
You will have noticed that the dot product between two vectors is a scalar, and that only vectors with the same number of elements are compatible for dot product. You can also take the dot product between a matrix x and a vector y, which returns a vector where coefficients are the dot products between y and the rows of x. You would implement it as such | # Listing 2.32 A naive implementation of matrix-vector dot
import numpy as np
def naive_matrix_vector_dot(x, y):
# x is a Numpy matrix
# y is a Numpy vector
assert len(x.shape) == 2
assert len(y.shape) == 1
# The 1st dimension of x must be
# the same as the 0th dimension of y!
assert x.shape[1] == y.shape[0]
# This operation returns a vector of 0s
# with the same shape as y
z = np.zeros(x.shape[0])
for i in range(x.shape[0]):
for j in range(x.shape[1]):
z[i] += x[i, j] * y[j]
return z | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
You could also be reusing the code we wrote previously, which highlights the relationship between matrix-vector product and vector product: | # Listing 2.33 Alternative naive implementation of matrix-vector dot
def naive_matrix_vector_dot(x, y):
z = np.zeros(x.shape[0])
for i in range(x.shape[0]):
z[i] = naive_vector_dot(x[i, :], y)
return z | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Note that as soon as one of the two tensors has a higher than 1, is no longer ndim dot symmetric, which is to say that is not the same as . dot(x, y) dot(y, x) Of course, dot product generalizes to tensors with arbitrary number of axes. The most common applications may be the dot product between two matrices. You can take the dot product of two matrices x and y (dot(x, y)) if and only if x.shape[1] == y.shape[0]. The result is a matrix with shape (x.shape[0], y.shape[1]) , where coefficients are the vector products between the rows of x and the columns of y. Here’s the naive implementation: | # Listing 2.34 A naive implementation of matrix-matrix dot
def naive_matrix_dot(x, y):
# x and y are Numpy matrices
assert len(x.shape) == 2
assert len(y.shape) == 2
# The 1st dimension of x must be
# the same as the 0th dimension of y!
assert x.shape[1] == y.shape[0]
# This operation returns a matrix of 0s
# with a specific shape
z = np.zeros((x.shape[0], y.shape[1]))
# We iterate over the rows of x
for i in range(x.shape[0]):
# And over the columns of y
for j in range(y.shape[1]):
row_x = x[i, :]
column_y = y[:, j]
z[i, j] = naive_vector_dot(row_x, column_y)
return z | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
To understand dot product shape compatibility, it helps to visualize the input and output tensors by aligning them in the following way: x, y and z are pictured as rectangles (literal boxes of coefficients). Because the rows and x and the columns of y must have the same size, it follows that the width of x must match the height of y. If you go on to develop new machine learning algorithms, you will likely be drawing such diagrams a lot. More generally, you can take the dot product between higher-dimensional tensors, following the same rules for shape compatibility as outlined above for the 2D case:下面这个操作在线性代数的矩阵乘法中还没有接触过,这已经超过矩阵的维度(二维)。 (a, b, c, d) . (d,) (a, b, c) (a, b, c, d) . (d, e) (a, b, c, e) Tensor reshaping第三类非常需要理解的张量运算是“tensor reshaping”. 虽然我们的第一个神经网络的例子的第一层中并没用用到该操作,但是在另一个Dense中,我们在将数据喂入网络前的数字数据预处理(pre-proccessed)上使用了reshape。 | # Listing 2.35 MNIST image tensor reshaping
#train_images = train_images.reshape((60000, 28 * 28) | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Reshaping a tensor means re-arranging its rows and columns so as to match a target shape. Naturally the reshaped tensor will have the same total number of coefficients as the initial tensor. Reshaping is best understood via simple examples:对一个张量的reshape意味着重新调整张量的行和列,以适配目标shape。所以reshape后地张量和原始的张量自然而然地拥有着相同总数的参数。reshpe可以通过下面的例子很好地理解: | # Listing 2.36 Tensor reshaping example
x = np.array([[0., 1.],
[2., 3.],
[4., 5.]])
print(x.shape)
x = x.reshape((6, 1))
x
x = x.reshape((2, 3))
x | _____no_output_____ | MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
A special case of reshaping that is commonly encountered is the transposition. "Transposing" a matrix means exchanging its rows and its columns, so that x[i, :] becomes : | # Listing 2.37 Matrix transposition
x = np.zeros((300, 20))
# Creates an all-zeros matrix of shape (300, 20)
x = np.transpose(x)
print(x.shape) | (20, 300)
| MIT | .ipynb_checkpoints/2.2&2.3 Data-representation-fo-neura-networks_cn &The-gears-of-neural-networks-tensor-operations -checkpoint.ipynb | ViolinLee/deep-learning-with-python-notebooks |
Neural Machine TranslationWelcome to your first programming assignment for this week! You will build a Neural Machine Translation (NMT) model to translate human readable dates ("25th of June, 2009") into machine readable dates ("2009-06-25"). You will do this using an attention model, one of the most sophisticated sequence to sequence models. This notebook was produced together with NVIDIA's Deep Learning Institute. Let's load all the packages you will need for this assignment. | from keras.layers import Bidirectional, Concatenate, Permute, Dot, Input, LSTM, Multiply
from keras.layers import RepeatVector, Dense, Activation, Lambda
from keras.optimizers import Adam
from keras.utils import to_categorical
from keras.models import load_model, Model
import keras.backend as K
import numpy as np
from faker import Faker
import random
from tqdm import tqdm
from babel.dates import format_date
from nmt_utils import *
import matplotlib.pyplot as plt
%matplotlib inline | Using TensorFlow backend.
| MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
1 - Translating human readable dates into machine readable datesThe model you will build here could be used to translate from one language to another, such as translating from English to Hindi. However, language translation requires massive datasets and usually takes days of training on GPUs. To give you a place to experiment with these models even without using massive datasets, we will instead use a simpler "date translation" task. The network will input a date written in a variety of possible formats (*e.g. "the 29th of August 1958", "03/30/1968", "24 JUNE 1987"*) and translate them into standardized, machine readable dates (*e.g. "1958-08-29", "1968-03-30", "1987-06-24"*). We will have the network learn to output dates in the common machine-readable format YYYY-MM-DD. <!-- Take a look at [nmt_utils.py](./nmt_utils.py) to see all the formatting. Count and figure out how the formats work, you will need this knowledge later. !--> 1.1 - DatasetWe will train the model on a dataset of 10000 human readable dates and their equivalent, standardized, machine readable dates. Let's run the following cells to load the dataset and print some examples. | m = 10000
dataset, human_vocab, machine_vocab, inv_machine_vocab = load_dataset(m)
dataset[:10] | _____no_output_____ | MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
You've loaded:- `dataset`: a list of tuples of (human readable date, machine readable date)- `human_vocab`: a python dictionary mapping all characters used in the human readable dates to an integer-valued index - `machine_vocab`: a python dictionary mapping all characters used in machine readable dates to an integer-valued index. These indices are not necessarily consistent with `human_vocab`. - `inv_machine_vocab`: the inverse dictionary of `machine_vocab`, mapping from indices back to characters. Let's preprocess the data and map the raw text data into the index values. We will also use Tx=30 (which we assume is the maximum length of the human readable date; if we get a longer input, we would have to truncate it) and Ty=10 (since "YYYY-MM-DD" is 10 characters long). | Tx = 30
Ty = 10
X, Y, Xoh, Yoh = preprocess_data(dataset, human_vocab, machine_vocab, Tx, Ty)
print("X.shape:", X.shape)
print("Y.shape:", Y.shape)
print("Xoh.shape:", Xoh.shape)
print("Yoh.shape:", Yoh.shape) | X.shape: (10000, 30)
Y.shape: (10000, 10)
Xoh.shape: (10000, 30, 37)
Yoh.shape: (10000, 10, 11)
| MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
You now have:- `X`: a processed version of the human readable dates in the training set, where each character is replaced by an index mapped to the character via `human_vocab`. Each date is further padded to $T_x$ values with a special character (). `X.shape = (m, Tx)`- `Y`: a processed version of the machine readable dates in the training set, where each character is replaced by the index it is mapped to in `machine_vocab`. You should have `Y.shape = (m, Ty)`. - `Xoh`: one-hot version of `X`, the "1" entry's index is mapped to the character thanks to `human_vocab`. `Xoh.shape = (m, Tx, len(human_vocab))`- `Yoh`: one-hot version of `Y`, the "1" entry's index is mapped to the character thanks to `machine_vocab`. `Yoh.shape = (m, Tx, len(machine_vocab))`. Here, `len(machine_vocab) = 11` since there are 11 characters ('-' as well as 0-9). Lets also look at some examples of preprocessed training examples. Feel free to play with `index` in the cell below to navigate the dataset and see how source/target dates are preprocessed. | index = 0
print("Source date:", dataset[index][0])
print("Target date:", dataset[index][1])
print()
print("Source after preprocessing (indices):", X[index])
print("Target after preprocessing (indices):", Y[index])
print()
print("Source after preprocessing (one-hot):", Xoh[index])
print("Target after preprocessing (one-hot):", Yoh[index]) | Source date: 15 october 1986
Target date: 1986-10-15
Source after preprocessing (indices): [ 4 8 0 26 15 30 26 14 17 28 0 4 12 11 9 36 36 36 36 36 36 36 36 36 36
36 36 36 36 36]
Target after preprocessing (indices): [ 2 10 9 7 0 2 1 0 2 6]
Source after preprocessing (one-hot): [[ 0. 0. 0. ..., 0. 0. 0.]
[ 0. 0. 0. ..., 0. 0. 0.]
[ 1. 0. 0. ..., 0. 0. 0.]
...,
[ 0. 0. 0. ..., 0. 0. 1.]
[ 0. 0. 0. ..., 0. 0. 1.]
[ 0. 0. 0. ..., 0. 0. 1.]]
Target after preprocessing (one-hot): [[ 0. 0. 1. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1.]
[ 0. 0. 0. 0. 0. 0. 0. 0. 0. 1. 0.]
[ 0. 0. 0. 0. 0. 0. 0. 1. 0. 0. 0.]
[ 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 0. 0. 1. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 0. 1. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 1. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 0. 0. 1. 0. 0. 0. 0. 0. 0. 0. 0.]
[ 0. 0. 0. 0. 0. 0. 1. 0. 0. 0. 0.]]
| MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
2 - Neural machine translation with attentionIf you had to translate a book's paragraph from French to English, you would not read the whole paragraph, then close the book and translate. Even during the translation process, you would read/re-read and focus on the parts of the French paragraph corresponding to the parts of the English you are writing down. The attention mechanism tells a Neural Machine Translation model where it should pay attention to at any step. 2.1 - Attention mechanismIn this part, you will implement the attention mechanism presented in the lecture videos. Here is a figure to remind you how the model works. The diagram on the left shows the attention model. The diagram on the right shows what one "Attention" step does to calculate the attention variables $\alpha^{\langle t, t' \rangle}$, which are used to compute the context variable $context^{\langle t \rangle}$ for each timestep in the output ($t=1, \ldots, T_y$). **Figure 1**: Neural machine translation with attention Here are some properties of the model that you may notice: - There are two separate LSTMs in this model (see diagram on the left). Because the one at the bottom of the picture is a Bi-directional LSTM and comes *before* the attention mechanism, we will call it *pre-attention* Bi-LSTM. The LSTM at the top of the diagram comes *after* the attention mechanism, so we will call it the *post-attention* LSTM. The pre-attention Bi-LSTM goes through $T_x$ time steps; the post-attention LSTM goes through $T_y$ time steps. - The post-attention LSTM passes $s^{\langle t \rangle}, c^{\langle t \rangle}$ from one time step to the next. In the lecture videos, we were using only a basic RNN for the post-activation sequence model, so the state captured by the RNN output activations $s^{\langle t\rangle}$. But since we are using an LSTM here, the LSTM has both the output activation $s^{\langle t\rangle}$ and the hidden cell state $c^{\langle t\rangle}$. However, unlike previous text generation examples (such as Dinosaurus in week 1), in this model the post-activation LSTM at time $t$ does will not take the specific generated $y^{\langle t-1 \rangle}$ as input; it only takes $s^{\langle t\rangle}$ and $c^{\langle t\rangle}$ as input. We have designed the model this way, because (unlike language generation where adjacent characters are highly correlated) there isn't as strong a dependency between the previous character and the next character in a YYYY-MM-DD date. - We use $a^{\langle t \rangle} = [\overrightarrow{a}^{\langle t \rangle}; \overleftarrow{a}^{\langle t \rangle}]$ to represent the concatenation of the activations of both the forward-direction and backward-directions of the pre-attention Bi-LSTM. - The diagram on the right uses a `RepeatVector` node to copy $s^{\langle t-1 \rangle}$'s value $T_x$ times, and then `Concatenation` to concatenate $s^{\langle t-1 \rangle}$ and $a^{\langle t \rangle}$ to compute $e^{\langle t, t'}$, which is then passed through a softmax to compute $\alpha^{\langle t, t' \rangle}$. We'll explain how to use `RepeatVector` and `Concatenation` in Keras below. Lets implement this model. You will start by implementing two functions: `one_step_attention()` and `model()`.**1) `one_step_attention()`**: At step $t$, given all the hidden states of the Bi-LSTM ($[a^{},a^{}, ..., a^{}]$) and the previous hidden state of the second LSTM ($s^{}$), `one_step_attention()` will compute the attention weights ($[\alpha^{},\alpha^{}, ..., \alpha^{}]$) and output the context vector (see Figure 1 (right) for details):$$context^{} = \sum_{t' = 0}^{T_x} \alpha^{}a^{}\tag{1}$$ Note that we are denoting the attention in this notebook $context^{\langle t \rangle}$. In the lecture videos, the context was denoted $c^{\langle t \rangle}$, but here we are calling it $context^{\langle t \rangle}$ to avoid confusion with the (post-attention) LSTM's internal memory cell variable, which is sometimes also denoted $c^{\langle t \rangle}$. **2) `model()`**: Implements the entire model. It first runs the input through a Bi-LSTM to get back $[a^{},a^{}, ..., a^{}]$. Then, it calls `one_step_attention()` $T_y$ times (`for` loop). At each iteration of this loop, it gives the computed context vector $c^{}$ to the second LSTM, and runs the output of the LSTM through a dense layer with softmax activation to generate a prediction $\hat{y}^{}$. **Exercise**: Implement `one_step_attention()`. The function `model()` will call the layers in `one_step_attention()` $T_y$ using a for-loop, and it is important that all $T_y$ copies have the same weights. I.e., it should not re-initiaiize the weights every time. In other words, all $T_y$ steps should have shared weights. Here's how you can implement layers with shareable weights in Keras:1. Define the layer objects (as global variables for examples).2. Call these objects when propagating the input.We have defined the layers you need as global variables. Please run the following cells to create them. Please check the Keras documentation to make sure you understand what these layers are: [RepeatVector()](https://keras.io/layers/core/repeatvector), [Concatenate()](https://keras.io/layers/merge/concatenate), [Dense()](https://keras.io/layers/core/dense), [Activation()](https://keras.io/layers/core/activation), [Dot()](https://keras.io/layers/merge/dot). | # Defined shared layers as global variables
repeator = RepeatVector(Tx)
concatenator = Concatenate(axis=-1)
densor1 = Dense(10, activation = "tanh")
densor2 = Dense(1, activation = "relu")
activator = Activation(softmax, name='attention_weights') # We are using a custom softmax(axis = 1) loaded in this notebook
dotor = Dot(axes = 1) | _____no_output_____ | MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
Now you can use these layers to implement `one_step_attention()`. In order to propagate a Keras tensor object X through one of these layers, use `layer(X)` (or `layer([X,Y])` if it requires multiple inputs.), e.g. `densor(X)` will propagate X through the `Dense(1)` layer defined above. | # GRADED FUNCTION: one_step_attention
def one_step_attention(a, s_prev):
"""
Performs one step of attention: Outputs a context vector computed as a dot product of the attention weights
"alphas" and the hidden states "a" of the Bi-LSTM.
Arguments:
a -- hidden state output of the Bi-LSTM, numpy-array of shape (m, Tx, 2*n_a)
s_prev -- previous hidden state of the (post-attention) LSTM, numpy-array of shape (m, n_s)
Returns:
context -- context vector, input of the next (post-attetion) LSTM cell
"""
### START CODE HERE ###
# Use repeator to repeat s_prev to be of shape (m, Tx, n_s) so that you can concatenate it with all hidden states "a" (≈ 1 line)
s_prev = repeator(s_prev)
# Use concatenator to concatenate a and s_prev on the last axis (≈ 1 line)
concat = concatenator([a, s_prev])
# Use densor1 to propagate concat through a small fully-connected neural network to compute the "intermediate energies" variable e. (≈1 lines)
e = densor1(concat)
# Use densor2 to propagate e through a small fully-connected neural network to compute the "energies" variable energies. (≈1 lines)
energies = densor2(e)
# Use "activator" on "energies" to compute the attention weights "alphas" (≈ 1 line)
alphas = activator(energies)
# Use dotor together with "alphas" and "a" to compute the context vector to be given to the next (post-attention) LSTM-cell (≈ 1 line)
context = dotor([alphas, a])
### END CODE HERE ###
return context | _____no_output_____ | MIT | MOOCS/Deeplearing_Specialization/Notebooks/Neural machine translation with attention-v4.ipynb | itismesam/Courses-1 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.