markdown
stringlengths 0
1.02M
| code
stringlengths 0
832k
| output
stringlengths 0
1.02M
| license
stringlengths 3
36
| path
stringlengths 6
265
| repo_name
stringlengths 6
127
|
|---|---|---|---|---|---|
**Option 3:** Draw once before loop | np.random.seed(1917)
x = np.random.normal(0,1,size=100)
print(f'var(x) = {np.var(x):.3f}')
y_ = np.random.normal(0,1,size=x.size)
for sigma in [0.5,1.0,0.5]:
y = sigma*y_
print(f'sigma = {sigma:2f}: f = {f(x,y):.4f}') | var(x) = 0.951
sigma = 0.500000: f = 0.5522
sigma = 1.000000: f = 0.0143
sigma = 0.500000: f = 0.5522
| MIT | web/06/Examples_and_overview.ipynb | Jovansam/lectures-2021 |
Image Combination Joint Single Dish and Interferometer Image Reconstruction The SDINT imaging algorithm allows joint reconstruction of wideband single dish and interferometer data. This algorithm is available in the task [sdintimaging](../api/casatasks.rstimaging) and described in [Rau, Naik & Braun (2019)](https://iopscience.iop.org/article/10.3847/1538-3881/ab1aa7/meta).Joint reconstruction of wideband single dish and interferometer data in CASA is experimental. Please use at own discretion.The usage modes that have been tested are documented below. SDINT AlgorithmInterferometer data are gridded into an image cube (and corresponding PSF). The single dish image and PSF cubes are combined with the interferometer cubes in a feathering step. The joint image and PSF cubes then form inputs to any deconvolution algorithm (in either *cube* or *mfs/mtmfs* modes). Model images from the deconvolution algorithm are translated back to model image cubes prior to subtraction from both the single dish image cube as well as the interferometer data to form a new pair of residual image cubes to be feathered in the next iteration. In the case of mosaic imaging, primary beam corrections are performed per channel of the image cube, followed by a multiplication by a common primary beam, prior to deconvolution. Therefore, for mosaic imaging, this task always implements *conjbeams=True* and *normtype='flatnoise'*.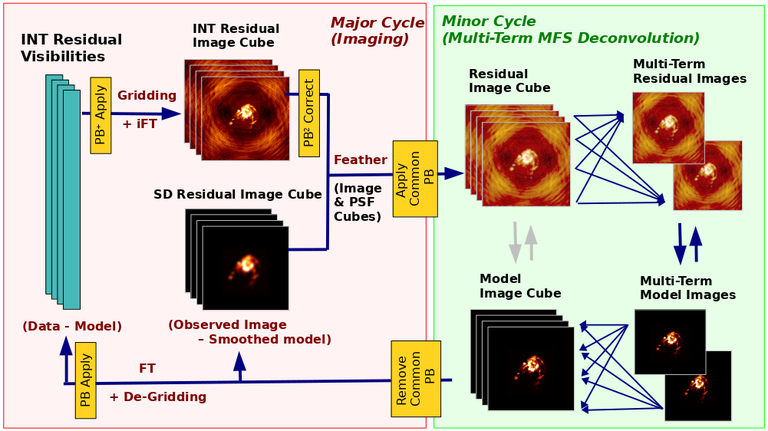{.image-inline width="674" height="378"}The input single dish data are the single dish image and psf cubes. The input interferometer data is a MeasurementSet. In addition to imaging and deconvolution parameters from interferometric imaging (task **tclean**), there are controls for a feathering step to combine interferometer and single dish cubes within the imaging iterations. Note that the above diagram shows only the \'mtmfs\' variant. Cube deconvolution proceeds directly with the cubes in the green box above, without the extra conversion back and forth to the multi-term basis. Primary beam handling is also not shown in this diagram, but full details (via pseudocode) are available in the [reference publication.](https://iopscience.iop.org/article/10.3847/1538-3881/ab1aa7)The parameters used for controlling the joint deconvolution are described on the [sdintimaging](../api/casatasks.rstimaging) task pages. Usage ModesThe task **sdintimaging** contains the algorithm for joint reconstruction of wideband single dish and interferometer data. The **sdintimaging** task shares a significant number of parameters with the **tclean** task, but also contains unique parameters. A detailed overview of these parameters, and how to use them, can be found in the CASA Docs [task pages of sdintimaging](../api/casatasks.rstimaging).As seen from the diagram above and described on the **sdintimaging** task pages, there is considerable flexibility in usage modes. One can choose between interferometer-only, singledish-only and joint interferometer-singledish imaging. Outputs are restored images and associated data products (similar to task tclean).The following usage modes are available in the (experimental) sdintimaging task. Tested modes include all 12 combinations of:- Cube Imaging : All combinations of the following options. - *specmode = 'cube'* - *deconvolver = 'multiscale', 'hogbom'* - *usedata = 'sdint', 'sd' , 'int'* - *gridder = 'standard', 'mosaic'* - *parallel = False, True*- Wideband Multi-Term Imaging : All combinations of the following options. - *specmode = 'mfs'* - *deconvolver = 'mtmfs'* ( *nterms=1* for a single-term MFS image, and *nterms>1* for multi-term MFS image. Tests use *nterms=2* ) - *usedata = 'sdint', 'sd' , 'int'* - *gridder = 'standard', 'mosaic'* - *parallel = False, True***NOTE**: When the INT and/or SD cubes have flagged (and therefore empty) channels, only those channels that have non-zero images in both the INT and SD cubes are used for the joint reconstruction.**NOTE**: Single-plane joint imaging may be run with deconvolver='mtmfs' and nterms=1.**NOTE**: All other modes allowed by the new sdintimaging task are currently untested. Tests will be added in subsequent releases. Examples/Demos Basic test resultsThe sdintimaging task was run on a pair of simulated test datasets. Both contain a flat spectrum extended emission feature plus three point sources, two of which have spectral index=-1.0 and one which is flat-spectrum (rightmost point). The scale of the top half of the extended structure was chosen to lie within the central hole in the spatial-frequency plane at the middle frequency of the band so as to generate a situation where the interferometer-only imaging is difficult.Please refer to the [publication](https://iopscience.iop.org/article/10.3847/1538-3881/ab1aa7/meta) for a more detailed analysis of the imaging quality and comparisons of images without and with SD data. Images from a run on the ALMA M100 12m+7m+TP Science Verification Data suite are also shown below.*Single Pointing Simulation :*Wideband Multi-Term Imaging ( deconvolver=\'mtmfs\', specmode=\'mfs\' )- SD + INT A joint reconstruction accurately reconstructs both intensity and spectral index for the extended emission as well as the compact sources.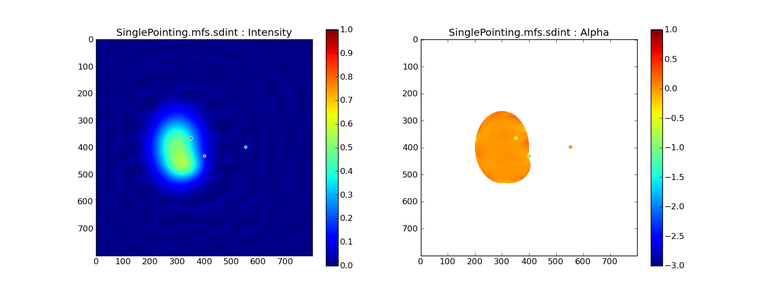- INT-only The intensity has negative bowls and the spectral index is overly steep, especially for the top half of the extended component.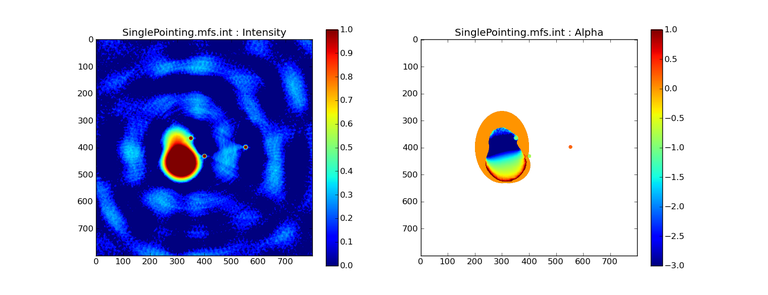- SD-only The spectral index of the extended emission is accurate (at 0.0) and the point sources are barely visible at this SD angular resolution.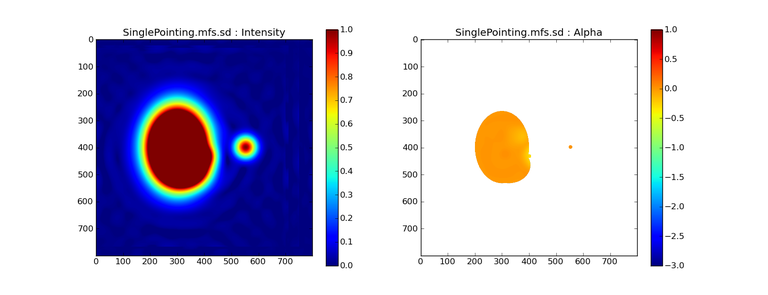Cube Imaging ( deconvolver=\'multiscale\', specmode=\'cube\' )- SD + INT A joint reconstruction has lower artifacts and more accurate intensities in all three channels, compared to the int-only reconstructions below 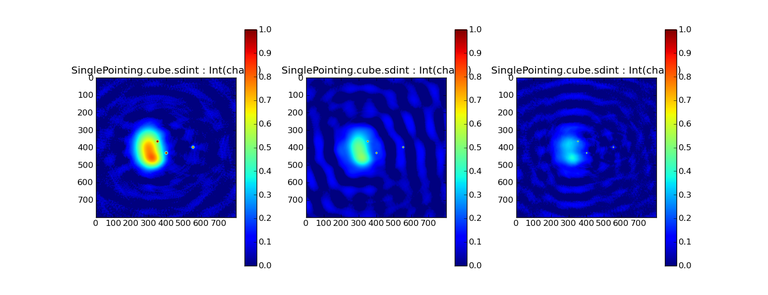- INT-only The intensity has negative bowls in the lower frequency channels and the extended emission is largely absent at the higher frequencies.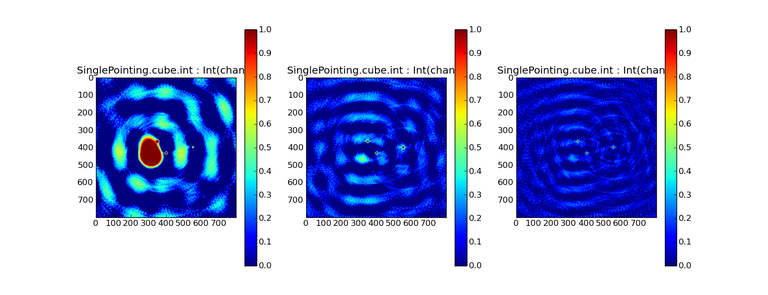- SD-only A demonstration of single-dish cube imaging with deconvolution of the SD-PSF. In this example, iterations have not been run until full convergence, which is why the sources still contain signatures of the PSF.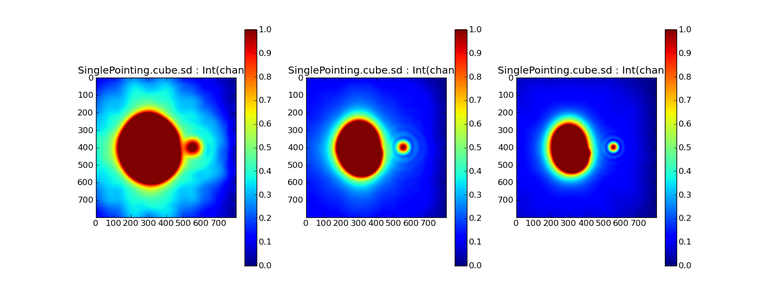*Mosaic Simulation*An observation of the same sky brightness was simulated with 25 pointings.Wideband Multi-Term Mosaic Imaging ( deconvolver=\'mtmfs\', specmode=\'mfs\' , gridder=\'mosaic\' )- SD + INT A joint reconstruction accurately reconstructs both intensity and spectral index for the extended emission as well as the compact sources. This is a demonstration of joint mosaicing along with wideband single-dish and interferometer combination.- INT-only The intensity has negative bowls and the spectral index is strongly inaccurate. Note that the errors are slightly less than the situation with the single-pointing example (where there was only one pointing's worth of uv-coverage).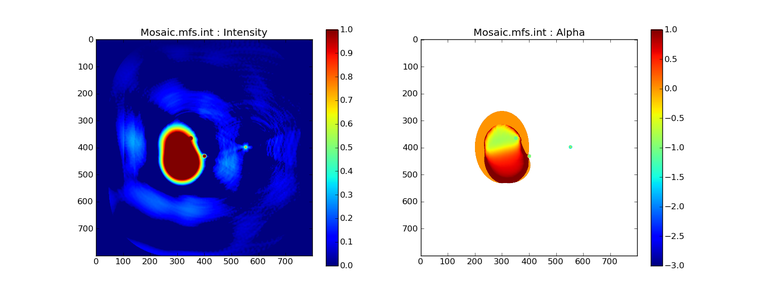Cube Mosaic Imaging ( deconvolver='multiscale', specmode='cube', gridder='mosaic' )- SD + INT A joint reconstruction produces better per-channel reconstructions compared to the INT-only situation shown below. This is a demonstration of cube mosaic imaging along with SD+INT joint reconstruction. 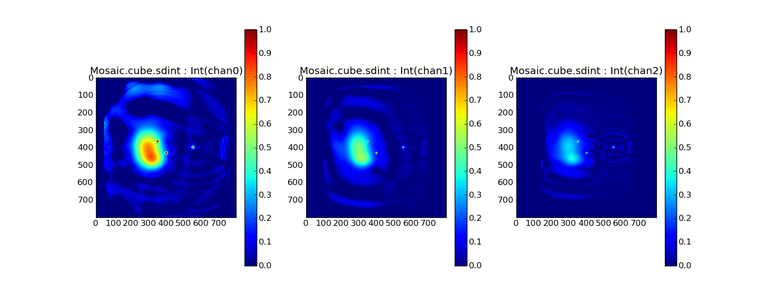- INT-only Cube mosaic imaging with only interferometer data. This clearly shows negative bowls and artifacts arising from the missing flux.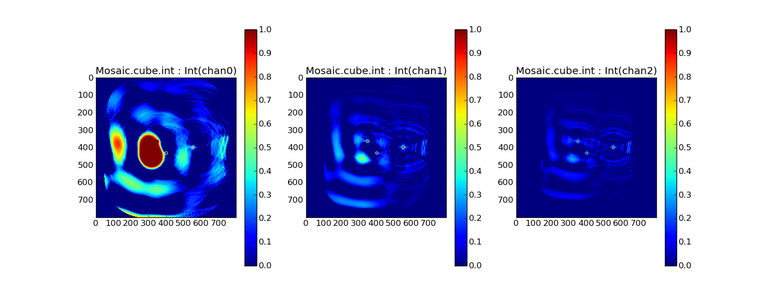 ALMA M100 Spectral Cube Imaging : 12m + 7m + TPThe sdintimaging task was run on the [ALMA M100 Science Verification Datasets](https://almascience.nrao.edu/alma-data/science-verification).\(1\) The single dish (TP) cube was pre-processed by adding per-plane restoringbeam information.\(2\) Cube specification parameters were obtained from the SD Image as follows```from sdint_helper import * sdintlib = SDINT_helper() sdintlib.setup_cube_params(sdcube='M100_TmP')Output : Shape of SD cube : [90 90 1 70\] Coordinate ordering : ['Direction', 'Direction', 'Stokes', 'Spectral']nchan = 70start = 114732899312.0Hzwidth = -1922516.74324HzFound 70 per-plane restoring beams\(For specmode='mfs' in sdintimaging, please remember to set 'reffreq' to a value within the freq range of the cube.Returned Dict : {'nchan': 70, 'start': '114732899312.0Hz', 'width': '-1922516.74324Hz'}```\(3\) Task sdintimaging was run with automatic SD-PSF generation, n-sigma stopping thresholds, a pb-based mask at the 0.3 gain level, and no other deconvolution masks (interactive=False).```sdintimaging(usedata="sdint", sdimage="../M100_TP", sdpsf="",sdgain=3.0, dishdia=12.0, vis="../M100_12m_7m", imagename="try_sdint_niter5k", imsize=1000, cell="0.5arcsec", phasecenter="J2000 12h22m54.936s +15d48m51.848s", stokes="I", specmode="cube", reffreq="", nchan=70, start="114732899312.0Hz", width="-1922516.74324Hz", outframe="LSRK", veltype="radio", restfreq="115.271201800GHz", interpolation="linear", perchanweightdensity=True, gridder="mosaic", mosweight=True, pblimit=0.2, deconvolver="multiscale", scales=[0, 5, 10, 15, 20], smallscalebias=0.0, pbcor=False, weighting="briggs", robust=0.5, niter=5000, gain=0.1, threshold=0.0, nsigma=3.0, interactive=False, usemask="user", mask="", pbmask=0.3)```**Results from two channels are show below. **LEFT : INT only (12m+7m) and RIGHT : SD+INT (12m + 7m + TP)Channel 23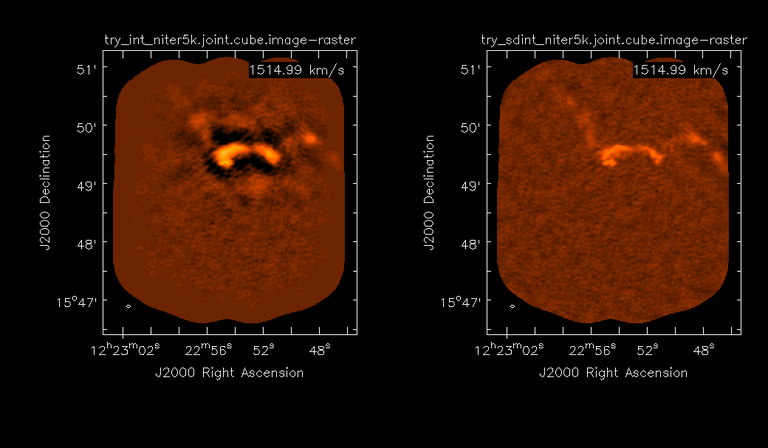Channel 43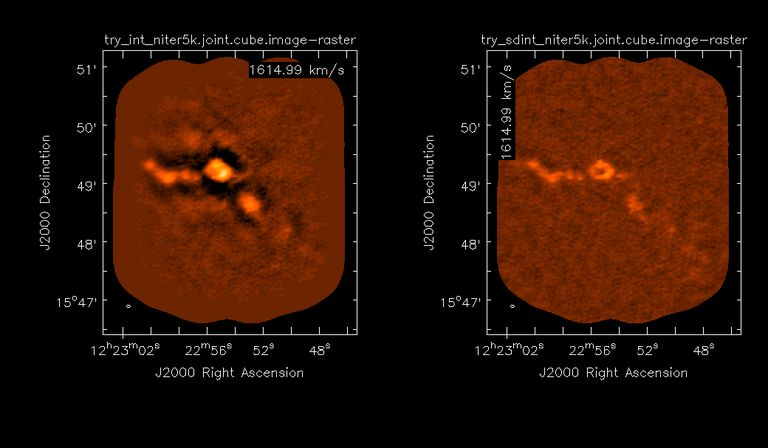 Moment 0 Maps : LEFT : INT only. MIDDLE : SD + INT with sdgain=1.0 RIGHT : SD + INT with sdgain=3.0Moment 1 Maps : LEFT : INT only. MIDDLE : SD + INT with sdgain=1.0 RIGHT : SD + INT with sdgain=3.0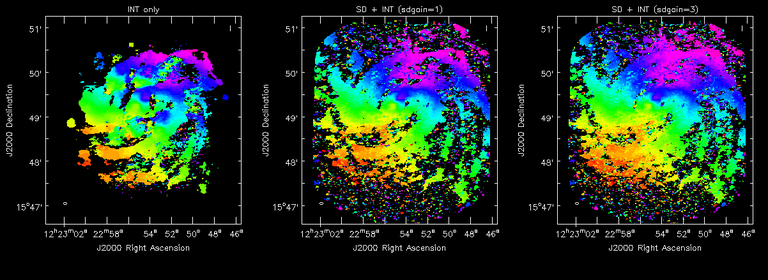A comparison (shown for one channel) with and without masking is shown below.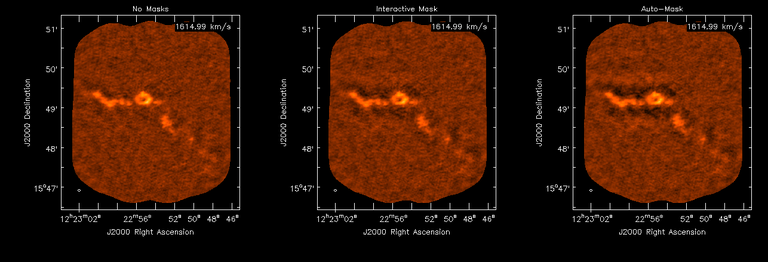 Notes : - In the reconstructed cubes, negative bowls have clearly been eliminated by using sdintimaging to combine interferometry + SD data. Residual images are close to noise-like too (not pictured above) suggesting a well-constrained and steadily converging imaging run. - The source structure is visibly different from the INT-only case, with high and low resolution structure appearing more well defined. However, the *high-resolution* peak flux in the SDINT image cube is almost a factor of 3 lower than the INT-only. While this may simply be because of deconvolution uncertainty in the ill-constrained INT-only reconstruction, it requires more investigation to evaluate absolute flux correctness. For example, it will be useful to evaluate if the INT-only reconstructed flux changes significantly with careful hand-masking. - Compare with a Feathered image : http://www.astroexplorer.org/details/apjaa60c2f1 : The reconstructed structure is consistent.- The middle and right panels compare reconstructions with different values of sdgain (1.0 and 3.0). The sdgain=3.0 run has a noticeable emphasis on the SD flux in the reconstructed moment maps, while the high resolution structures have the same are the same between sdgain=1 and 3. This is consistent with expectations from the algorithm, but requires further investigation to evaluate robustness in general.- Except for the last panel, no deconvolution masks were used (apart from a *pbmask* at the 0.3 gain level). The deconvolution quality even without masking is consistent with the expectation that when supplied with better data constraints in a joint reconstruction, the native algorithms are capable of converging on their own. In this example (same *niter* and *sdgain*), iterative cleaning with interactive and auto-masks (based mostly on interferometric peaks in the images) resulted in more artifacts compared to a run that allowed multi-scale clean to proceed on its own.- The results using sdintimaging on these ALMA data can be compared with performance results when [using feather](https://casaguides.nrao.edu/index.php?title=M100_Band3_Combine_5.4), and when [using tp2vis](https://science.nrao.edu/facilities/alma/alma-develop-old-022217/tp2vis_final_report.pdf) (ALMA study by J. Koda and P. Teuben). Fitting a new restoring beam to the Feathered PSFSince the deconvolution uses a joint SD+INT point spread function, the restoring beam is re-fitted after the feather step within the sdintimaging task. As a convenience feature, the corresponding tool method is also available to the user and may be used to invoke PSF refitting standalone, without needing an MS or any gridding of weights to make the PSF. This method will look for the imagename.psf (or imagename.psf.tt0), fit and set the new restoring beam. It is tied to the naming convention of tclean.```synu = casac.synthesisutils();synu.fitPsfBeam(imagename='qq', psfcutoff=0.3) Cubessynu.fitPsfBeam(imagename='qq', nterms=2, psfcutoff=0.3) Multi-term``` Tested Use CasesThe following is a list of use cases that have simulation-based functional verification tests within CASA.1. Wideband mulit-term imaging (SD+Int) Wideband data single field imaging by joint-reconstruction from single dish and interferometric data to obtain the high resolution of the interferometer while account for the zero spacing information. Use multi-term multi-frequency synthesis (MTMFS) algorithm to properly account for spectral information of the source.2. Wideband multi-term imaging: Int only The same as 1 except for using interferometric data only, which is useful to make a comparison with 1 (i.e. effect of missing flux). This is equivalent to running 'mtmfs' with specmode='mfs' and gridder='standard' in tclean 3. Wideband multi-term imaging: SD only The same as 1 expect for using single dish data only which is useful to make a comparison with 1 (i.e. to see how much high resolution information is missing). Also, sometimes, the SD PSF has significant sidelobes (Airy disk) and even single dish images can benefit from deconvolution. This is a use case where wideband multi-term imaging is applied to SD data alone to make images at the highest possible resolution as well as to derive spectral index information. 4. Single field cube imaging: SD+Int Spectral cube single field imaging by joint reconstruction of single dish and interferometric data to obtain single field spectral cube image. Use multi-scale clean for deconvolution 5. Single field cube imaging: Int only The same as 4 except for using the interferometric data only, which is useful to make a comparison with 4 (i.e. effect of missing flux). This is equivalent to running 'multiscale' with specmode='cube' and gridder='standard' in tclean.6. Single field cube imaging: SD only The same as 4 except for using the single dish data only, which is useful to make a comparison with 4 (i.e. to see how much high resolution information is missing) Also, it addresses the use case where SD PSF sidelobes are significant and where the SD images could benefit from multiscale (or point source) deconvolution per channel. 7. Wideband multi-term mosaic Imaging: SD+Int Wideband data mosaic imaging by joint-reconstruction from single dish and interferometric data to obtain the high resolution of the interferometer while account for the zero spacing information. Use multi-term multi-frequency synthesis (MTMFS) algorithm to properly account for spectral information of the source. Implement the concept of conjbeams (i.e. frequency dependent primary beam correction) for wideband mosaicing. 8. Wideband multi-term mosaic imaging: Int only The same as 7 except for using interferometric data only, which is useful to make a comparison with 7 (i.e. effect of missing flux). Also, this is an alternate implementation of the concept of conjbeams ( frequency dependent primary beam correction) available via tclean, and which is likely to be more robust to uv-coverage variations (and sumwt) across frequency. 9. Wideband multi-term mosaic imaging: SD only The same as 7 expect for using single dish data only which is useful to make a comparison with 7 (i.e. to see how much high resolution information is missing). This is the same situation as (3), but made on an image coordinate system that matches an interferometer mosaic mtmfs image. 10. Cube mosaic imaging: SD+Int Spectral cube mosaic imaging by joint reconstruction of single dish and interferometric data. Use multi-scale clean for deconvolution. 11. Cube mosaic imaging: Int only The same as 10 except for using the intererometric data only, which is useful to make a comparison with 10 (i.e. effect of missing flux). This is the same use case as gridder='mosaic' and deconvolver='multiscale' in tclean for specmode='cube'.12. Cube mosaic imaging: SD only The same as 10 except for using the single dish data only, which is useful to make a comparison with 10 (i.e. to see how much high resolution information is missing). This is the same situation as (6), but made on an image coordinate system that matches an interferometer mosaic cube image. 13. Wideband MTMFS SD+INT with channel 2 flagged in INT The same as 1, but with partially flagged data in the cubes. This is a practical reality with real data where the INT and SD data are likely to have gaps in the data due to radio frequency interferenece or other weight variations. 14. Cube SD+INT with channel 2 flagged The same as 4, but with partially flagged data in the cubes. This is a practical reality with real data where the INT and SD data are likely to have gaps in the data due to radio frequency interferenece or other weight variations. 15. Wideband MTMFS SD+INT with sdpsf="" The same as 1, but with an unspecified sdpsf. This triggers the auto-calculation of the SD PSF cube using restoring beam information from the regridded input sdimage. 16. INT-only cube comparison between tclean and sdintimaging Compare cube imaging results for a functionally equivalent run.17. INT-only mtmfs comparison between tclean and sdintimaging Compare mtmfs imaging results for a functionally equivalent run. Note that the sdintimaging task implements wideband primary beam correction in the image domain on the cube residual image, whereas tclean uses the 'conjbeams' parameter to apply an approximation of this correction during the gridding step.Note : Serial and Parallel Runs for an ALMA test dataset have been shown to be consistent to a 1e+6 dynamic range, consistent with differences measured for our current implementation of cube parallelization. References[Urvashi Rau, Nikhil Naik, and Timothy Braun 2019 AJ 158, 1](https://iopscience.iop.org/article/10.3847/1538-3881/ab1aa7/meta)https://github.com/urvashirau/WidebandSDINT*** Feather & CASAfeather Feathering is a technique used to combine a Single Dish (SD) image with an interferometric image of the same field.The goal of this process is to reconstruct the source emission on all spatial scales, ranging from the small spatial scales measured by the interferometer to the large-scale structure measured by the single dish. To do this, feather combines the images in Fourier space, weighting them by the spatial frequency response of each image. This technique assumes that the spatial frequencies of the single dish and interferometric data partially overlap. The subject of interferometric and single dish data combination has a long history. See the introduction of Koda et al 2011 (and references therein) [\[1\]](Bibliography) for a concise review, and Vogel et al 1984 [\[2\]](Bibliography), Stanimirovic et al 1999 [\[3\]](Bibliography), Stanimirovic 2002 [\[4\]](Bibliography), Helfer et al 2003 [\[5\]](Bibliography), and Weiss et al 2001 [\[6\]](Bibliography), among other referenced papers, for other methods and discussions concerning the combination of single dish and interferometric data.The feathering algorithm implemented in CASA is as follows: 1. Regrid the single dish image to match the coordinate system, image shape, and pixel size of the high resolution image. 2. Transform each image onto uniformly gridded spatial-frequency axes.3. Scale the Fourier-transformed low-resolution image by the ratio of the volumes of the two \'clean beams\' (high-res/low-res) to convert the single dish intensity (in Jy/beam) to that corresponding to the high resolution intensity (in Jy/beam). The volume of the beam is calculated as the volume under a two dimensional Gaussian with peak 1 and major and minor axes of the beam corresponding to the major and minor axes of the Gaussian. 4. Add the Fourier-transformed data from the high-resolution image, scaled by $(1-wt)$ where $wt$ is the Fourier transform of the \'clean beam\' defined in the low-resolution image, to the scaled low resolution image from step 3.5. Transform back to the image plane.The input images for feather must have the following characteristics:1. Both input images must have a well-defined beam shape for this task to work, which will be a \'clean beam\' for interferometric images and a \'primary-beam\' for a single-dish image. The beam for each image should be specified in the image header. If a beam is not defined in the header or feather cannot guess the beam based on the telescope parameter in the header, then you will need to add the beam size to the header using **imhead**. 2. Both input images must have the same flux density normalization scale. If necessary, the SD image should be converted from temperature units to Jy/beam. Since measuring absolute flux levels is difficult with single dishes, the single dish data is likely to be the one with the most uncertain flux calibration. The SD image flux can be scaled using the parameter *sdfactor* to place it on the same scale as the interferometer data. The casafeather task (see below) can be used to investigate the relative flux scales of the images.Feather attemps to regrid the single dish image to the interferometric image. Given that the single dish image frequently originates from other data reduction packages, CASA may have trouble performing the necessary regridding steps. If that happens, one may try to regrid the single dish image manually to the interferometric image. CASA has a few tasks to perform individual steps, including **imregrid** for coordinate transformations, **imtrans** to swap and reverse coordinate axes, the tool **ia.adddegaxes()** for adding degenerate axes (e.g. a single Stokes axis). See the \"[Image Analysis](image_analysis.ipynbimage-analysis)\" chapter for additional options. If you have trouble changing image projections, you can try the [montage package](http://montage.ipac.caltech.edu/), which also has an [associated python wrapper](http://www.astropy.org/montage-wrapper/).If you are feathering large images together, set the numbers of pixels along the X and Y axes to composite (non-prime) numbers in order to improve the algorithm speed. In general, FFTs work much faster on even and composite numbers. Then use the subimage task or tool to trim the number of pixels to something desirable. Inputs for task featherThe inputs for **feather** are: ```feather :: Combine two images using their Fourier transformsimagename = '' Name of output feathered imagehighres = '' Name of high resolution (interferometer) imagelowres = '' Name of low resolution (single dish) imagesdfactor = 1.0 Scale factor to apply to Single Dish imageeffdishdiam = -1.0 New effective SingleDish diameter to use in mlowpassfiltersd = False Filter out the high spatial frequencies of the SD image```The SD data cube is specified by the *lowres* parameter and the interferometric data cube by the *highres* parameter. The combined, feathered output cube name is given by the *imagename* parameter. The parameter *sdfactor* can be used to scale the flux calibration of the SD cube. The parameter *effdishdiam* can be used to change the weighting of the single dish image.The weighting functions for the data are usually the Fourier transform of the Single Dish beam FFT(PB~SD~) for the Single dish data, and the inverse, 1-FFT(PB~SD~), for the interferometric data. It is possible, however, to change the weighting functions by pretending that the SD is smaller in size via the *effdishdiam* parameter. This tapers the high spatial frequencies of the SD data and adds more weight to the interferometric data. The *lowpassfiltersd* can take out non-physical artifacts at very high spatial frequencies that are often present in SD data.Note that the only inputs are for images; **feather** will attempt to regrid the images to a common shape, i.e. pixel size, pixel numbers, and spectral channels. If you are having issues with the regridding inside feather, you may consider regridding using the **imregrid** and **specsmooth** tasks.The **feather** task does not perform any deconvolution but combines the single dish image with a presumably deconvolved interferometric image. The short spacings of the interferometric image that are extrapolated by the deconvolution process will be those that are down-weighted the most when combined with the single dish data. The single dish image must have a well-defined beam shape and the correct flux units for a model image (Jy/beam instead of Jy/pixel). Use the tasks **imhead** and **immath** first to convert if needed.Starting with a cleaned synthesis image and a low resolution image from a single dish telescope, the following example shows how they can be feathered: ```feather(imagename ='feather.im', Create an image called feather.im highres ='synth.im', The synthesis image is called synth.im lowres ='single_dish.im') The SD image is called single_dish.im``` Visual Interface for feather (casafeather)CASA also provides a visual interface to the **feather** task. The interface is run from a command line *outside* CASA by typing casafeather in a shell. An example of the interface is shown below. To start, one needs to specify a high and a low resolution image, typically an interferometric and a single dish map. Note that the single dish map needs to be in units of Jy/beam. The output image name can be specified. The non-deconvolved (dirty) interferometric image can also be specified to use as diagnostic of the relative flux scaling of the single dish and interferometer images. See below for more details. At the top of the display, the parameters *effdshdiameter* and *sdfactor* can be provided in the "Effective Dish Diameter" and "Low Resolution Scale Factor" input boxes. One you have specified the images and parameters, press the "Feather" button in the center of the GUI window to start the feathering process. The feathering process here includes regridding the low resolution image to the high resolution image.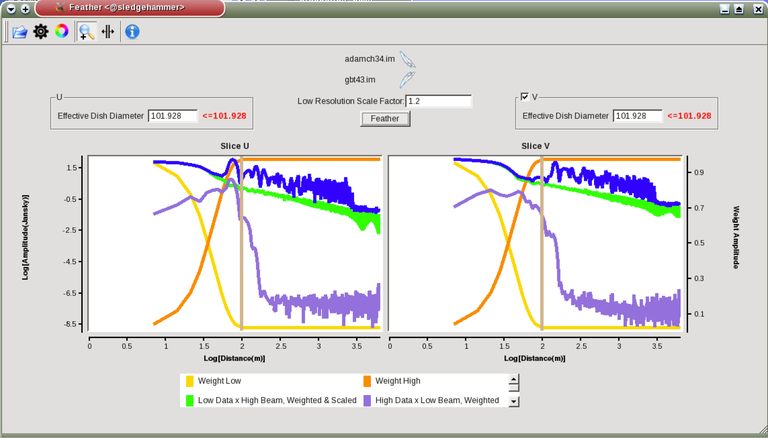>Figure 1: The panel shows the "Original Data Slice", which are cuts through the u and v directions of the Fourier-transformed input images. Green is the single dish data (low resolution) and purple the interferometric data (high resolution). To bring them on the same flux scale, the low data were convolved to the high resolution beam and vice versa (selectable in color preferences). In addition, a single dish scaling of 1.2 was applied to adjust calibration differences. The weight functions are shown in yellow (for the low resolution data) and orange (for the high resolution data). The weighting functions were also applied to the green and purple slices. Image slices of the combined, feathered output image are shown in blue. The displays also show the location of the effective dish diameter by the vertical line. This value is kept at the original single dish diameter that is taken from the respective image header. The initial casafeather display shows two rows of plots. The panel shows the "Original Data Slice", which are either cuts through the u and v directions of the Fourier-transformed input images or a radial average. A vertical line shows the location of the effective dish diameter(s). The blue lines are the combined, feathered slices.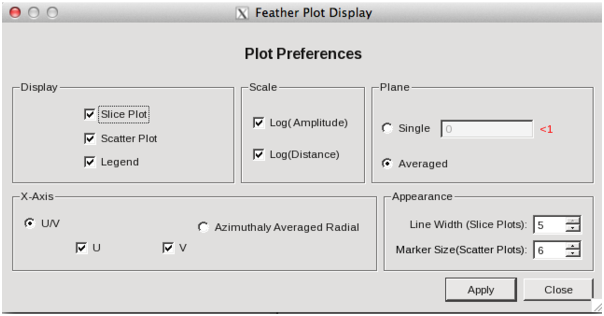>Figure 2: The casafeather "customize" window.The \'Customize\' button (gear icon on the top menu page) allows one to set the display parameters. Options are to show the slice plot, the scatter plot, or the legend. One can also select between logarithmic and linear axes; a good option is usually to make both axes logarithmic. You can also select whether the x-axis for the slices are in the u, or v, or both directions, or, alternatively a radial average in the uv-plane. For data cubes, one can also select a particular velocity plane, or to average the data across all velocity channels. The scatter plot can display any two data sets on the two axes, selected from the \'Color Preferences\' menu. The data can be the unmodified, original data, or data that have been convolved with the high or low resolution beams. One can also select to display data that were weighted and scaled by the functions discussed above.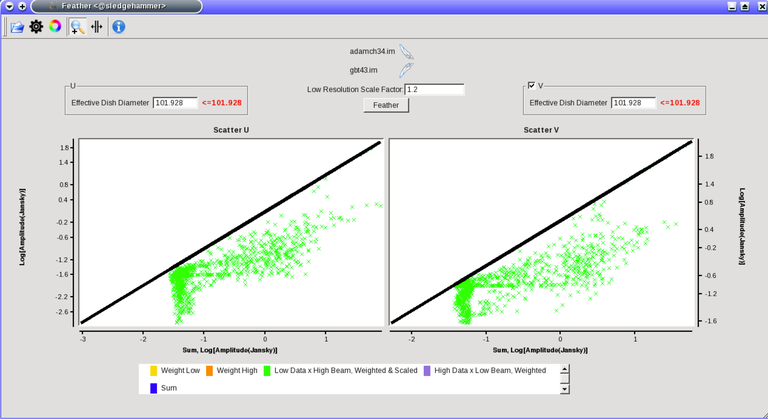>Figure 3: The scatter plot in casafeather. The low data, convolved with high beam, weighted and scaled is still somewhat below the equality line (plotted against high data, convolved with low beam, weighted). In this case one can try to adjust the \"low resolution scale factor\" to bring the values closer to the line of equality, ie. to adjust the calibration scales. Plotting the data as a scatter plot is a useful diagnostic tool for checking for differences in flux scaling between the high and low resolution data sets.The dirty interferometer image contains the actual flux measurements made by the telescope. Therefore, if the single dish scaling is correct, the flux in the dirty image convolved with the low resolution beam and with the appropriate weighting applied should be the same as the flux of the low-resolution data convolved with the high resolution beam once weighted and scaled. If not, the *sdfactor* parameter can be adjusted until they are the same. One may also use the cleaned high resolution image instead of the dirty image, if the latter is not available. However, note that the cleaned high resolution image already contains extrapolations to larger spatial scales that may bias the comparison.*** Bibliography1. Koda et al 2011 (http://adsabs.harvard.edu/abs/2011ApJS..193...19K)2. Vogel et al 1984 (http://adsabs.harvard.edu/abs/1984ApJ...283..655V)3. Stanimirovic et al 1999 (http://adsabs.harvard.edu/abs/1999MNRAS.302..417S)4. Stanimirovic et al 2002 (http://adsabs.harvard.edu/abs/2002ASPC..278..375S)5. Helfer et al 2003 (http://adsabs.harvard.edu/abs/2003ApJS..145..259H)6. Weiss et al 2001 (http://adsabs.harvard.edu/abs/2001A%26A...365..571W) | _____no_output_____ | Apache-2.0 | docs/notebooks/image_combination.ipynb | yohei99/casadocs |
|
IPL Dataset Analysis Problem StatementWe want to know as to what happens during an IPL match which raises several questions in our mind with our limited knowledge about the game called cricket on which it is based. This analysis is done to know as which factors led one of the team to win and how does it matter. About the Dataset :The Indian Premier League (IPL) is a professional T20 cricket league in India contested during April-May of every year by teams representing Indian cities. It is the most-attended cricket league in the world and ranks sixth among all the sports leagues. It has teams with players from around the world and is very competitive and entertaining with a lot of close matches between teams.The IPL and other cricket related datasets are available at [cricsheet.org](https://cricsheet.org/%c2%a0(data). Feel free to visit the website and explore the data by yourself as exploring new sources of data is one of the interesting activities a data scientist gets to do. About the dataset:Snapshot of the data you will be working on:The dataset 1452 data points and 23 features|Features|Description||-----|-----||match_code|Code pertaining to individual match||date|Date of the match played||city|Location where the match was played||team1|team1||team2|team2||toss_winner|Who won the toss out of two teams||toss_decision|toss decision taken by toss winner||winner|Winner of that match between two teams||win_type|How did the team won(by wickets or runs etc.)||win_margin|difference with which the team won| |inning|inning type(1st or 2nd)||delivery|ball delivery||batting_team|current team on batting||batsman|current batsman on strike||non_striker|batsman on non-strike||bowler|Current bowler||runs|runs scored||extras|extra run scored||total|total run scored on that delivery including runs and extras||extras_type|extra run scored by wides or no ball or legby||player_out|player that got out||wicket_kind|How did the player got out||wicket_fielders|Fielder who caught out the player by catch| Analysing data using numpy module Read the data using numpy module. | import numpy as np
# Not every data format will be in csv there are other file formats also.
# This exercise will help you deal with other file formats and how toa read it.
path = './ipl_matches_small.csv'
data_ipl = np.genfromtxt(path, delimiter=',', skip_header=1, dtype=str)
print(data_ipl) | [['392203' '2009-05-01' 'East London' ... '' '' '']
['392203' '2009-05-01' 'East London' ... '' '' '']
['392203' '2009-05-01' 'East London' ... '' '' '']
...
['335987' '2008-04-21' 'Jaipur' ... '' '' '']
['335987' '2008-04-21' 'Jaipur' ... '' '' '']
['335987' '2008-04-21' 'Jaipur' ... '' '' '']]
| MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
Calculate the unique no. of matches in the provided dataset ? | # How many matches were held in total we need to know so that we can analyze further statistics keeping that in mind.im
import numpy as np
unique_match_code=np.unique(data_ipl[:,0])
print(unique_match_code) | ['335987' '392197' '392203' '392212' '501226' '729297']
| MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
Find the set of all unique teams that played in the matches in the data set. | # this exercise deals with you getting to know that which are all those six teams that played in the tournament.
import numpy as np
unique_match_team3=np.unique(data_ipl[:,3])
print(unique_match_team3)
unique_match_team4=np.unique(data_ipl[:,4])
print(unique_match_team4)
union=np.union1d(unique_match_team3,unique_match_team4)
print(union)
unique=np.unique(union)
print(unique)
| _____no_output_____ | MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
Find sum of all extras in all deliveries in all matches in the dataset | # An exercise to make you familiar with indexing and slicing up within data.
import numpy as np
extras=data_ipl[:,17]
data=extras.astype(np.int)
print(sum(data))
| 88
| MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
Get the array of all delivery numbers when a given player got out. Also mention the wicket type. | import numpy as np
deliveries=[]
wicket_type=[]
for i in data_ipl:
if(i[20]!=""):
a=i[11]
b=i[21]
deliveries.append(a)
wicket_type.append(b)
print(deliveries)
print(wicket_type)
| _____no_output_____ | MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
How many matches the team `Mumbai Indians` has won the toss? | data_arr=[]
for i in data_ipl:
if(i[5]=="Mumbai Indians"):
data_arr.append(i[0])
unique_match_id=np.unique(data_arr)
print(unique_match_id)
print(len(unique_match_id))
| ['392197' '392203']
2
| MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
Create a filter that filters only those records where the batsman scored 6 runs. Also who has scored the maximum no. of sixes overall ? | # An exercise to know who is the most aggresive player or maybe the scoring player
import numpy as np
counter=0
run_dict={}
arr=[]
for i in data_ipl:
#print(i[13])
#current_run = i[16]
#prev_run = run_dict[batsman_nm]
#batsman_nm = i[13]
#if prev_run == None:
#run_dict[batsman_nm] = current_run
#else:
#run_dict[batsman_nm] = run_dict[batsman_nm]current_run
if i[13] in run_dict:
run_dict[i[13]]=run_dict[i[13]]+int(i[16])
else:
run_dict[i[13]]=int(i[16])
print(run_dict)
| _____no_output_____ | MIT | Manipulating_Data_with_NumPy_Code_Along.ipynb | vidSanas/greyatom-python-for-data-science |
读取数据 | import torchvision.transforms as T
img_shape = (3, 224, 224)
def read_raw_img(path, resize, L=False):
img = Image.open(path)
if resize:
img = img.resize(resize)
if L:
img = img.convert('L')
return np.asarray(img)
class DogCat(data.Dataset):
def __init__(self,path, img_shape):
# self.batch_size = batch_size
self.img_shape = img_shape
imgs = os.listdir(path)
random.shuffle(imgs)
self.imgs = [os.path.join(path, img) for img in imgs]
# normalize = T.Normalize(mean = [0.485, 0.456, 0.406],
# std = [0.229, 0.224, 0.225])
# self.transforms = T.Compose([T.Resize(224),
# T.CenterCrop(224),
# T.ToTensor(),
# normalize])
def __getitem__(self, index):
# start = index * self.batch_size
# end = min(start + self.batch_size, len(self.imgs))
# size = end - start
# assert size > 0
# img_paths = self.imgs[start:end]
# a = t.zeros((size,) + self.img_shape, requires_grad=True)
# b = t.zeros((size, 1))
# for i in range(size):
img = read_raw_img(self.imgs[index], self.img_shape[1:], L=False).transpose((2,1,0))
# img = Image.open(img_paths[i])
x = t.from_numpy(img)
# a[i] = self.transforms(img)
y = 1 if 'dog' in self.imgs[index].split('/')[-1].split('.')[0] else 0
return x, y
def __len__(self):
return len(self.imgs)
train = DogCat(path+'train', img_shape) | _____no_output_____ | Apache-2.0 | Pytorch/Task4.ipynb | asd55667/DateWhale |
构建模型 | import math
class Vgg16(nn.Module):
def __init__(self, features, num_classes=1, init_weights=True):
super(Vgg16, self).__init__()
self.features = features
self.classifier = nn.Sequential(
nn.Linear(512 * 7 * 7, 4096),
nn.ReLU(True),
nn.Dropout(),
nn.Linear(4096, 4096),
nn.ReLU(True),
nn.Dropout(),
nn.Linear(4096, num_classes),
)
if init_weights:
self._initialize_weights()
def forward(self, x):
x = self.features(x)
x = x.view(x.size(0), -1)
x = self.classifier(x)
x = t.sigmoid(x)
return x
def _initialize_weights(self):
for m in self.modules():
if isinstance(m, nn.Conv2d):
n = m.kernel_size[0] * m.kernel_size[1] * m.out_channels
m.weight.data.normal_(0, math.sqrt(2. / n))
if m.bias is not None:
m.bias.data.zero_()
elif isinstance(m, nn.BatchNorm2d):
m.weight.data.fill_(1)
m.bias.data.zero_()
elif isinstance(m, nn.Linear):
m.weight.data.normal_(0, 0.01)
m.bias.data.zero_()
def make_layers(cfg, mode, batch_norm=False):
layers = []
if mode == 'RGB':
in_channels = 3
elif mode == 'L':
in_channels = 1
else:
print('only RGB or L mode')
for v in cfg:
if v == 'M':
layers += [nn.MaxPool2d(kernel_size=2, stride=2)]
else:
conv2d = nn.Conv2d(in_channels, v, kernel_size=3, padding=1)
if batch_norm:
layers += [conv2d, nn.BatchNorm2d(v), nn.ReLU(inplace=True)]
else:
layers += [conv2d, nn.ReLU(inplace=True)]
in_channels = v
return nn.Sequential(*layers)
cfg = [64, 64, 'M', 128, 128, 'M', 256, 256, 256, 'M', 512, 512, 512, 'M', 512, 512, 512, 'M'] | _____no_output_____ | Apache-2.0 | Pytorch/Task4.ipynb | asd55667/DateWhale |
损失函数与优化器 | vgg = Vgg16(make_layers(cfg, 'RGB')).cuda()
print(vgg)
criterion = nn.BCELoss()
optimizer = t.optim.Adam(vgg.parameters(),lr=0.001) | Vgg16(
(features): Sequential(
(0): Conv2d(3, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(1): ReLU(inplace)
(2): Conv2d(64, 64, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(3): ReLU(inplace)
(4): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(5): Conv2d(64, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(6): ReLU(inplace)
(7): Conv2d(128, 128, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(8): ReLU(inplace)
(9): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(10): Conv2d(128, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(11): ReLU(inplace)
(12): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(13): ReLU(inplace)
(14): Conv2d(256, 256, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(15): ReLU(inplace)
(16): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(17): Conv2d(256, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(18): ReLU(inplace)
(19): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(20): ReLU(inplace)
(21): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(22): ReLU(inplace)
(23): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(24): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(25): ReLU(inplace)
(26): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(27): ReLU(inplace)
(28): Conv2d(512, 512, kernel_size=(3, 3), stride=(1, 1), padding=(1, 1))
(29): ReLU(inplace)
(30): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
)
(classifier): Sequential(
(0): Linear(in_features=25088, out_features=4096, bias=True)
(1): ReLU(inplace)
(2): Dropout(p=0.5)
(3): Linear(in_features=4096, out_features=4096, bias=True)
(4): ReLU(inplace)
(5): Dropout(p=0.5)
(6): Linear(in_features=4096, out_features=1, bias=True)
)
)
| Apache-2.0 | Pytorch/Task4.ipynb | asd55667/DateWhale |
模型训练 | use_cuda = t.cuda.is_available()
device = t.device("cuda:0" if use_cuda else "cpu")
# cudnn.benchmark = True
# Parameters
params = {'batch_size': 64,
'shuffle': True,
'num_workers': 6}
max_epochs = 1
# train = DogCat(path+'train', img_shape)
training_generator = data.DataLoader(train, **params)
for epoch in range(max_epochs):
for x, y_ in training_generator:
x, y_ = x.float().to(device), y_.float().to(device)
y = vgg(x)
loss = criterion(y, y_)
loss.backward()
optimizer.step()
| /home/wcw/anaconda3/envs/tf/lib/python3.6/site-packages/torch/nn/functional.py:2016: UserWarning: Using a target size (torch.Size([64])) that is different to the input size (torch.Size([64, 1])) is deprecated. Please ensure they have the same size.
"Please ensure they have the same size.".format(target.size(), input.size()))
/home/wcw/anaconda3/envs/tf/lib/python3.6/site-packages/torch/nn/functional.py:2016: UserWarning: Using a target size (torch.Size([28])) that is different to the input size (torch.Size([28, 1])) is deprecated. Please ensure they have the same size.
"Please ensure they have the same size.".format(target.size(), input.size()))
| Apache-2.0 | Pytorch/Task4.ipynb | asd55667/DateWhale |
模型评估 | accs = []
test = DogCat(path+'test', img_shape=img_shape)
test_loader = data.DataLoader(test, **params)
with t.set_grad_enabled(False):
for x, y_ in test_loader:
x, y_ = x.float().to(device), y_.float().to(device)
y = vgg(x)
acc = y.eq(y_).sum().item()/y.shape[0]
# acc = t.max(y, 1)[1].eq(t.max(y_, 1)[1]).sum().item()/y.shape[0]
accs.append(acc)
np.mean(accs) | _____no_output_____ | Apache-2.0 | Pytorch/Task4.ipynb | asd55667/DateWhale |
<imgsrc="https://colab.research.google.com/assets/colab-badge.svg" alt="Open In Colab"><imgsrc="https://img.shields.io/badge/GitHub-100000?logo=github&logoColor=white" alt="GitHub"> Text Annotation Import* This notebook will provide examples of each supported annotation type for text assets. It will cover the following: * Model-assisted labeling - used to provide pre-annotated data for your labelers. This will enable a reduction in the total amount of time to properly label your assets. Model-assisted labeling does not submit the labels automatically, and will need to be reviewed by a labeler for submission. * Label Import - used to provide ground truth labels. These can in turn be used and compared against prediction labels, or used as benchmarks to see how your labelers are doing. * For information on what types of annotations are supported per data type, refer to this documentation: * https://docs.labelbox.com/docs/model-assisted-labelingoption-1-import-via-python-annotation-types-recommended * Notes: * Wait until the import job is complete before opening the Editor to make sure all annotations are imported properly. Installs | !pip install -q 'labelbox[data]' | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Imports | from labelbox.schema.ontology import OntologyBuilder, Tool, Classification, Option
from labelbox import Client, LabelingFrontend, LabelImport, MALPredictionImport
from labelbox.data.annotation_types import (
Label, TextData, Checklist, Radio, ObjectAnnotation, TextEntity,
ClassificationAnnotation, ClassificationAnswer
)
from labelbox.data.serialization import NDJsonConverter
import uuid
import json
import numpy as np | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
API Key and ClientProvide a valid api key below in order to properly connect to the Labelbox Client. | # Add your api key
API_KEY = None
client = Client(api_key=API_KEY) | INFO:labelbox.client:Initializing Labelbox client at 'https://api.labelbox.com/graphql'
| Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
---- Steps1. Make sure project is setup2. Collect annotations3. Upload Project setup We will be creating two projects, one for model-assisted labeling, and one for label imports | ontology_builder = OntologyBuilder(
tools=[
Tool(tool=Tool.Type.NER, name="named_entity")
],
classifications=[
Classification(class_type=Classification.Type.CHECKLIST, instructions="checklist", options=[
Option(value="first_checklist_answer"),
Option(value="second_checklist_answer")
]),
Classification(class_type=Classification.Type.RADIO, instructions="radio", options=[
Option(value="first_radio_answer"),
Option(value="second_radio_answer")
])])
mal_project = client.create_project(name="text_mal_project")
li_project = client.create_project(name="text_label_import_project")
dataset = client.create_dataset(name="text_annotation_import_demo_dataset")
test_txt_url = "https://storage.googleapis.com/labelbox-sample-datasets/nlp/lorem-ipsum.txt"
data_row = dataset.create_data_row(row_data=test_txt_url)
editor = next(client.get_labeling_frontends(where=LabelingFrontend.name == "Editor"))
mal_project.setup(editor, ontology_builder.asdict())
mal_project.datasets.connect(dataset)
li_project.setup(editor, ontology_builder.asdict())
li_project.datasets.connect(dataset) | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Create Label using Annotation Type Objects* It is recommended to use the Python SDK's annotation types for importing into Labelbox. Object Annotations | def create_objects():
named_enity = TextEntity(start=10,end=20)
named_enity_annotation = ObjectAnnotation(value=named_enity, name="named_entity")
return named_enity_annotation | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Classification Annotations | def create_classifications():
checklist = Checklist(answer=[ClassificationAnswer(name="first_checklist_answer"),ClassificationAnswer(name="second_checklist_answer")])
checklist_annotation = ClassificationAnnotation(value=checklist, name="checklist")
radio = Radio(answer = ClassificationAnswer(name = "second_radio_answer"))
radio_annotation = ClassificationAnnotation(value=radio, name="radio")
return checklist_annotation, radio_annotation | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Create a Label object with all of our annotations | image_data = TextData(uid=data_row.uid)
named_enity_annotation = create_objects()
checklist_annotation, radio_annotation = create_classifications()
label = Label(
data=image_data,
annotations = [
named_enity_annotation, checklist_annotation, radio_annotation
]
)
label.__dict__ | _____no_output_____ | Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Model Assisted Labeling To do model-assisted labeling, we need to convert a Label object into an NDJSON. This is easily done with using the NDJSONConverter classWe will create a Label called mal_label which has the same original structure as the label aboveNotes:* Each label requires a valid feature schema id. We will assign it using our built in `assign_feature_schema_ids` method* the NDJsonConverter takes in a list of labels | mal_label = Label(
data=image_data,
annotations = [
named_enity_annotation, checklist_annotation, radio_annotation
]
)
mal_label.assign_feature_schema_ids(ontology_builder.from_project(mal_project))
ndjson_labels = list(NDJsonConverter.serialize([mal_label]))
ndjson_labels
upload_job = MALPredictionImport.create_from_objects(
client = client,
project_id = mal_project.uid,
name="upload_label_import_job",
predictions=ndjson_labels)
# Errors will appear for each annotation that failed.
# Empty list means that there were no errors
# This will provide information only after the upload_job is complete, so we do not need to worry about having to rerun
print("Errors:", upload_job.errors) | INFO:labelbox.schema.annotation_import:Sleeping for 10 seconds...
| Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Label Import Label import is very similar to model-assisted labeling. We will need to re-assign the feature schema before continuing, but we can continue to use our NDJSonConverterWe will create a Label called li_label which has the same original structure as the label above | #for the purpose of this notebook, we will need to reset the schema ids of our checklist and radio answers
image_data = TextData(uid=data_row.uid)
named_enity_annotation = create_objects()
checklist_annotation, radio_annotation = create_classifications()
li_label = Label(
data=image_data,
annotations = [
named_enity_annotation, checklist_annotation, radio_annotation
]
)
li_label.assign_feature_schema_ids(ontology_builder.from_project(li_project))
ndjson_labels = list(NDJsonConverter.serialize([li_label]))
ndjson_labels, li_project.ontology().normalized
upload_job = LabelImport.create_from_objects(
client = client,
project_id = li_project.uid,
name="upload_label_import_job",
labels=ndjson_labels)
print("Errors:", upload_job.errors) | INFO:labelbox.schema.annotation_import:Sleeping for 10 seconds...
| Apache-2.0 | examples/model_assisted_labeling/ner_mal.ipynb | Cyniikal/labelbox-python |
Hough Lines Import resources and display the image | import numpy as np
import matplotlib.pyplot as plt
import cv2
%matplotlib inline
# Read in the image
image = cv2.imread('images/phone.jpg')
# Change color to RGB (from BGR)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
plt.imshow(image) | _____no_output_____ | MIT | 1_2_Convolutional_Filters_Edge_Detection/.ipynb_checkpoints/6_1. Hough lines-checkpoint.ipynb | sxtien/CVND_Exercises |
Perform edge detection | # Convert image to grayscale
gray = cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)
# Define our parameters for Canny
low_threshold = 50
high_threshold = 100
edges = cv2.Canny(gray, low_threshold, high_threshold)
plt.imshow(edges, cmap='gray') | _____no_output_____ | MIT | 1_2_Convolutional_Filters_Edge_Detection/.ipynb_checkpoints/6_1. Hough lines-checkpoint.ipynb | sxtien/CVND_Exercises |
Find lines using a Hough transform | # Define the Hough transform parameters
# Make a blank the same size as our image to draw on
rho = 1
theta = np.pi/180
threshold = 60
min_line_length = 50
max_line_gap = 5
line_image = np.copy(image) #creating an image copy to draw lines on
# Run Hough on the edge-detected image
lines = cv2.HoughLinesP(edges, rho, theta, threshold, np.array([]),
min_line_length, max_line_gap)
# Iterate over the output "lines" and draw lines on the image copy
for line in lines:
for x1,y1,x2,y2 in line:
cv2.line(line_image,(x1,y1),(x2,y2),(255,0,0),5)
plt.imshow(line_image) | _____no_output_____ | MIT | 1_2_Convolutional_Filters_Edge_Detection/.ipynb_checkpoints/6_1. Hough lines-checkpoint.ipynb | sxtien/CVND_Exercises |
import torch
x = torch.arange(18).view(3,2,3)
print(x)
print(x[0,0,0])
print(x[1,0,0])
print(x[1,1,1])
x[1,0:2,0:2] | _____no_output_____ | MIT | Chapter3_Slicing_3D_Tensors.ipynb | SokichiFujita/PyTorch-for-Deep-Learning-and-Computer-Vision |
|
Using the same code as before, please solve the following exercises 1. Change the number of observations to 100,000 and see what happens. 2. Play around with the learning rate. Values like 0.0001, 0.001, 0.1, 1 are all interesting to observe. 3. Change the loss function. An alternative loss for regressions is the Huber loss. The Huber loss is more appropriate than the L2-norm when we have outliers, as it is less sensitive to them (in our example we don't have outliers, but you will surely stumble upon a dataset with outliers in the future). The L2-norm loss puts all differences *to the square*, so outliers have a lot of influence on the outcome. The proper syntax of the Huber loss is 'huber_loss' Useful tip: When you change something, don't forget to RERUN all cells. This can be done easily by clicking:Kernel -> Restart & Run AllIf you don't do that, your algorithm will keep the OLD values of all parameters.You can either use this file for all the exercises, or check the solutions of EACH ONE of them in the separate files we have provided. All other files are solutions of each problem. If you feel confident enough, you can simply change values in this file. Please note that it will be nice, if you return the file to starting position after you have solved a problem, so you can use the lecture as a basis for comparison. Import the relevant libraries | # We must always import the relevant libraries for our problem at hand. NumPy and TensorFlow are required for this example.
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf | _____no_output_____ | Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Data generationWe generate data using the exact same logic and code as the example from the previous notebook. The only difference now is that we save it to an npz file. Npz is numpy's file type which allows you to save numpy arrays into a single .npz file. We introduce this change because in machine learning most often: * you are given some data (csv, database, etc.)* you preprocess it into a desired format (later on we will see methods for preprocesing)* you save it into npz files (if you're working in Python) to access laterNothing to worry about - this is literally saving your NumPy arrays into a file that you can later access, nothing more. | # First, we should declare a variable containing the size of the training set we want to generate.
observations = 1000
# We will work with two variables as inputs. You can think about them as x1 and x2 in our previous examples.
# We have picked x and z, since it is easier to differentiate them.
# We generate them randomly, drawing from an uniform distribution. There are 3 arguments of this method (low, high, size).
# The size of xs and zs is observations x 1. In this case: 1000 x 1.
xs = np.random.uniform(low=-10, high=10, size=(observations,1))
zs = np.random.uniform(-10, 10, (observations,1))
# Combine the two dimensions of the input into one input matrix.
# This is the X matrix from the linear model y = x*w + b.
# column_stack is a Numpy method, which combines two matrices (vectors) into one.
generated_inputs = np.column_stack((xs,zs))
# We add a random small noise to the function i.e. f(x,z) = 2x - 3z + 5 + <small noise>
noise = np.random.uniform(-1, 1, (observations,1))
# Produce the targets according to our f(x,z) = 2x - 3z + 5 + noise definition.
# In this way, we are basically saying: the weights should be 2 and -3, while the bias is 5.
generated_targets = 2*xs - 3*zs + 5 + noise
# save into an npz file called "TF_intro"
np.savez('TF_intro', inputs=generated_inputs, targets=generated_targets) | _____no_output_____ | Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Solving with TensorFlowNote: This intro is just the basics of TensorFlow which has way more capabilities and depth than that. | # Load the training data from the NPZ
training_data = np.load('TF_intro.npz')
# Declare a variable where we will store the input size of our model
# It should be equal to the number of variables you have
input_size = 2
# Declare the output size of the model
# It should be equal to the number of outputs you've got (for regressions that's usually 1)
output_size = 1
# Outline the model
# We lay out the model in 'Sequential'
# Note that there are no calculations involved - we are just describing our network
model = tf.keras.Sequential([
# Each 'layer' is listed here
# The method 'Dense' indicates, our mathematical operation to be (xw + b)
tf.keras.layers.Dense(output_size,
# there are extra arguments you can include to customize your model
# in our case we are just trying to create a solution that is
# as close as possible to our NumPy model
kernel_initializer=tf.random_uniform_initializer(minval=-0.1, maxval=0.1),
bias_initializer=tf.random_uniform_initializer(minval=-0.1, maxval=0.1)
)
])
# We can also define a custom optimizer, where we can specify the learning rate
custom_optimizer = tf.keras.optimizers.SGD(learning_rate=0.02)
# Note that sometimes you may also need a custom loss function
# That's much harder to implement and won't be covered in this course though
# 'compile' is the place where you select and indicate the optimizers and the loss
model.compile(optimizer=custom_optimizer, loss='mean_squared_error')
# finally we fit the model, indicating the inputs and targets
# if they are not otherwise specified the number of epochs will be 1 (a single epoch of training),
# so the number of epochs is 'kind of' mandatory, too
# we can play around with verbose; we prefer verbose=2
model.fit(training_data['inputs'], training_data['targets'], epochs=100, verbose=2) | Epoch 1/100
1000/1000 - 0s - loss: 24.5755
Epoch 2/100
1000/1000 - 0s - loss: 1.1773
Epoch 3/100
1000/1000 - 0s - loss: 0.4253
Epoch 4/100
1000/1000 - 0s - loss: 0.3853
Epoch 5/100
1000/1000 - 0s - loss: 0.3727
Epoch 6/100
1000/1000 - 0s - loss: 0.3932
Epoch 7/100
1000/1000 - 0s - loss: 0.3817
Epoch 8/100
1000/1000 - 0s - loss: 0.3877
Epoch 9/100
1000/1000 - 0s - loss: 0.3729
Epoch 10/100
1000/1000 - 0s - loss: 0.3982
Epoch 11/100
1000/1000 - 0s - loss: 0.3809
Epoch 12/100
1000/1000 - 0s - loss: 0.3788
Epoch 13/100
1000/1000 - 0s - loss: 0.3714
Epoch 14/100
1000/1000 - 0s - loss: 0.3608
Epoch 15/100
1000/1000 - 0s - loss: 0.3507
Epoch 16/100
1000/1000 - 0s - loss: 0.3918
Epoch 17/100
1000/1000 - 0s - loss: 0.3697
Epoch 18/100
1000/1000 - 0s - loss: 0.3811
Epoch 19/100
1000/1000 - 0s - loss: 0.3781
Epoch 20/100
1000/1000 - 0s - loss: 0.3974
Epoch 21/100
1000/1000 - 0s - loss: 0.3974
Epoch 22/100
1000/1000 - 0s - loss: 0.3724
Epoch 23/100
1000/1000 - 0s - loss: 0.3561
Epoch 24/100
1000/1000 - 0s - loss: 0.3691
Epoch 25/100
1000/1000 - 0s - loss: 0.3650
Epoch 26/100
1000/1000 - 0s - loss: 0.3569
Epoch 27/100
1000/1000 - 0s - loss: 0.3707
Epoch 28/100
1000/1000 - 0s - loss: 0.4100
Epoch 29/100
1000/1000 - 0s - loss: 0.3703
Epoch 30/100
1000/1000 - 0s - loss: 0.3598
Epoch 31/100
1000/1000 - 0s - loss: 0.3775
Epoch 32/100
1000/1000 - 0s - loss: 0.3936
Epoch 33/100
1000/1000 - 0s - loss: 0.3968
Epoch 34/100
1000/1000 - 0s - loss: 0.3614
Epoch 35/100
1000/1000 - 0s - loss: 0.3588
Epoch 36/100
1000/1000 - 0s - loss: 0.3777
Epoch 37/100
1000/1000 - 0s - loss: 0.3637
Epoch 38/100
1000/1000 - 0s - loss: 0.3662
Epoch 39/100
1000/1000 - 0s - loss: 0.3655
Epoch 40/100
1000/1000 - 0s - loss: 0.3582
Epoch 41/100
1000/1000 - 0s - loss: 0.3759
Epoch 42/100
1000/1000 - 0s - loss: 0.4468
Epoch 43/100
1000/1000 - 0s - loss: 0.3613
Epoch 44/100
1000/1000 - 0s - loss: 0.3905
Epoch 45/100
1000/1000 - 0s - loss: 0.3825
Epoch 46/100
1000/1000 - 0s - loss: 0.3810
Epoch 47/100
1000/1000 - 0s - loss: 0.3546
Epoch 48/100
1000/1000 - 0s - loss: 0.3520
Epoch 49/100
1000/1000 - 0s - loss: 0.3878
Epoch 50/100
1000/1000 - 0s - loss: 0.3748
Epoch 51/100
1000/1000 - 0s - loss: 0.3978
Epoch 52/100
1000/1000 - 0s - loss: 0.3669
Epoch 53/100
1000/1000 - 0s - loss: 0.3650
Epoch 54/100
1000/1000 - 0s - loss: 0.3869
Epoch 55/100
1000/1000 - 0s - loss: 0.3952
Epoch 56/100
1000/1000 - 0s - loss: 0.3897
Epoch 57/100
1000/1000 - 0s - loss: 0.3698
Epoch 58/100
1000/1000 - 0s - loss: 0.3655
Epoch 59/100
1000/1000 - 0s - loss: 0.3717
Epoch 60/100
1000/1000 - 0s - loss: 0.3942
Epoch 61/100
1000/1000 - 0s - loss: 0.4334
Epoch 62/100
1000/1000 - 0s - loss: 0.3836
Epoch 63/100
1000/1000 - 0s - loss: 0.3631
Epoch 64/100
1000/1000 - 0s - loss: 0.3804
Epoch 65/100
1000/1000 - 0s - loss: 0.3671
Epoch 66/100
1000/1000 - 0s - loss: 0.3801
Epoch 67/100
1000/1000 - 0s - loss: 0.4032
Epoch 68/100
1000/1000 - 0s - loss: 0.3764
Epoch 69/100
1000/1000 - 0s - loss: 0.3549
Epoch 70/100
1000/1000 - 0s - loss: 0.3585
Epoch 71/100
1000/1000 - 0s - loss: 0.3747
Epoch 72/100
1000/1000 - 0s - loss: 0.3633
Epoch 73/100
1000/1000 - 0s - loss: 0.3493
Epoch 74/100
1000/1000 - 0s - loss: 0.3924
Epoch 75/100
1000/1000 - 0s - loss: 0.4246
Epoch 76/100
1000/1000 - 0s - loss: 0.3701
Epoch 77/100
1000/1000 - 0s - loss: 0.3959
Epoch 78/100
1000/1000 - 0s - loss: 0.3923
Epoch 79/100
1000/1000 - 0s - loss: 0.3587
Epoch 80/100
1000/1000 - 0s - loss: 0.3729
Epoch 81/100
1000/1000 - 0s - loss: 0.3649
Epoch 82/100
1000/1000 - 0s - loss: 0.3611
Epoch 83/100
1000/1000 - 0s - loss: 0.3701
Epoch 84/100
1000/1000 - 0s - loss: 0.3699
Epoch 85/100
1000/1000 - 0s - loss: 0.3494
Epoch 86/100
1000/1000 - 0s - loss: 0.3613
Epoch 87/100
1000/1000 - 0s - loss: 0.3933
Epoch 88/100
1000/1000 - 0s - loss: 0.4031
Epoch 89/100
1000/1000 - 0s - loss: 0.3814
Epoch 90/100
1000/1000 - 0s - loss: 0.3481
Epoch 91/100
1000/1000 - 0s - loss: 0.3664
Epoch 92/100
1000/1000 - 0s - loss: 0.3691
Epoch 93/100
1000/1000 - 0s - loss: 0.3599
Epoch 94/100
1000/1000 - 0s - loss: 0.3817
Epoch 95/100
1000/1000 - 0s - loss: 0.3572
Epoch 96/100
1000/1000 - 0s - loss: 0.3699
Epoch 97/100
1000/1000 - 0s - loss: 0.3666
Epoch 98/100
1000/1000 - 0s - loss: 0.3667
Epoch 99/100
1000/1000 - 0s - loss: 0.4198
Epoch 100/100
1000/1000 - 0s - loss: 0.3667
| Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Extract the weights and biasExtracting the weight(s) and bias(es) of a model is not an essential step for the machine learning process. In fact, usually they would not tell us much in a deep learning context. However, this simple example was set up in a way, which allows us to verify if the answers we get are correct. | # Extracting the weights and biases is achieved quite easily
model.layers[0].get_weights()
# We can save the weights and biases in separate variables for easier examination
# Note that there can be hundreds or thousands of them!
weights = model.layers[0].get_weights()[0]
weights
# We can save the weights and biases in separate variables for easier examination
# Note that there can be hundreds or thousands of them!
bias = model.layers[0].get_weights()[1]
bias | _____no_output_____ | Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Extract the outputs (make predictions)Once more, this is not an essential step, however, we usually want to be able to make predictions. | # We can predict new values in order to actually make use of the model
# Sometimes it is useful to round the values to be able to read the output
# Usually we use this method on NEW DATA, rather than our original training data
model.predict_on_batch(training_data['inputs']).round(1)
# If we display our targets (actual observed values), we can manually compare the outputs and the targets
training_data['targets'].round(1) | _____no_output_____ | Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Plotting the data | # The model is optimized, so the outputs are calculated based on the last form of the model
# We have to np.squeeze the arrays in order to fit them to what the plot function expects.
# Doesn't change anything as we cut dimensions of size 1 - just a technicality.
plt.plot(np.squeeze(model.predict_on_batch(training_data['inputs'])), np.squeeze(training_data['targets']))
plt.xlabel('outputs')
plt.ylabel('targets')
plt.show()
# Voila - what you see should be exactly the same as in the previous notebook!
# You probably don't see the point of TensorFlow now - it took us the same number of lines of code
# to achieve this simple result. However, once we go deeper in the next chapter,
# TensorFlow will save us hundreds of lines of code. | _____no_output_____ | Apache-2.0 | 17 - Deep Learning with TensorFlow 2.0/5_Introduction to TensorFlow 2/8_Exercises/TensorFlow_Minimal_example_All_exercises.ipynb | olayinka04/365-data-science-courses |
Loan Default Risk - Exploratory Data Analysis This notebook is focused on data exploration. The key objective is to familiarise myself with the data and to identify any issues. This could lead to data cleaning or feature engineering. Contents 1. Importing Relevant Libraries, Reading In Data 2. Anomly Detection and Correction 3. Data Exploration 4. Summary 5. Distribution of New Datasets 1.1 Importing Relevant Libraries | #Importing data wrangling library
import pandas as pd #Data Wrangling/Cleaning package for mixed data
import numpy as np #Data wrangling & manipulation for numerical data
import os
#Importing visulization libraries
from matplotlib import pyplot as plt #Importing visulization libraries
import seaborn as sns
#Importing Machine Learning Libraries(Preprocessing)
from sklearn.preprocessing import OneHotEncoder
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.impute import SimpleImputer
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import LabelEncoder # sklearn preprocessing for dealing with categorical variables
#Importing Machine Learning Libraries(Modelling And Evaluation)
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import cross_val_score
from sklearn.metrics import accuracy_score
os.getcwd() # Get working Directory | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
1.2 Reading In Data | rawfilepath = 'C:/Users/chara.geru/OneDrive - Avanade/DataScienceProject/HomeCreditModel/data/raw/'
filename = 'application_train.csv'
interimfilepath1 = 'C:/Users/chara.geru/OneDrive - Avanade/DataScienceProject/HomeCreditModel/data/interim/'
filename1 = 'df1.csv'
filename2 = 'df2.csv'
application_train = pd.read_csv(rawfilepath + filename)
df1 = pd.read_csv(interimfilepath1 + filename1)
df2 = pd.read_csv(interimfilepath1 + filename2)
print('Size of application_train data:', application_train.shape) #Printing shape of datasets | Size of application_train data: (307511, 122)
| MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
This dataset has: - 122 columns (features)- 307511 rows | application_train.columns.values #Printing all column names
pd.set_option('display.max_columns', None) #Display all columns
application_train.describe() #Get summary statistics for all columns
application_train.head() #View first 5 rows of the dataset | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
Generally the data looks good based on the statistics shown from the describe method. Potentional issues - Values in DAYS_BIRTH column are negative. They represent number of days a person was before they applied for a loan. For a better representation, I will convert them to positive values and convert days to years, for better representation of the data.- DAYS_EMPLOYED will be given the same treatment for the same reasons. 2.1 Anomly Detection | (application_train['DAYS_BIRTH']).describe()
(application_train['DAYS_EMPLOYED']).describe() | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
3.1 Check for Nulls | # "This function creates a table to summarize the null values"
def nulltable(df):
"""
This function creates a table to summarize the null values
"""
total = df.isnull().sum().sort_values(ascending = False)
percent = (df.isnull().sum()/df.isnull().count()*100).sort_values(ascending = False)
missing_df_data = pd.concat([total, percent], axis=1, keys=['Total', 'Percent'])
return missing_df_data.head(30)
nulltable(application_train) | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
- There are a large number of columns(features) with more than 50% NULLS.- I've deicided to drop these columns as they will not provide much information for training the model.- If a feaure has less than 50% NULLS, these maybe filled up using an appropriate calcualtion such as mean, median or mode 3.2 Data balanced or imbalanced | #Data balanced or imbalanced
temp = application_train["TARGET"].value_counts()
fig1, ax1 = plt.subplots()
ax1.pie(temp, labels=['Loan Repayed','Loan Not Repayed'], autopct='%1.1f%%',wedgeprops={'edgecolor':'black'})
ax1.axis('equal')
plt.title('Loan Repayed or Not')
plt.show() | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
Data is higly imbalanced - This emphasises the importance of assessing the precision/recall to evaluate results. For example, predicting all rows as not defaulted would lead to an accuracy of 91.9%.- Consider rebalancing the training data 3.3 Number of each type of column | # Number of each type of column
application_train.dtypes.value_counts() | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
- There are 16 object columns.- These will need to be encoded when building the model (using label encoder or one hot encoder) 3.4 Number of unique classes in each object column | # Number of unique classes in each object column
application_train.select_dtypes('object').apply(pd.Series.nunique, axis = 0) | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
- Gender has 3 values. This needs investigation and correction. 4. SummaryBased on the analysis so far we have identified the need to handle: - skewed data - removing rows with large number of NULLS - remove rows with third gender This led to the development of 2 new datasets, as we can't be sure which iterations would lead to the best model performance. This is a good opportunity for trial and error where I compare the perforomance of different permutations.Here are some visualisations to describe the new datasetsdf1- Negative DAYS_BIRTH converted to positive YEARS_BIRTH- Rows with third gender dropped- Dealt with features with large number of NULLsdf2- df2 has all the changes implemented to df1- In adition to that, in df2 the data for the skewed columns ('AMT_CREDIT', 'AMT_INCOME_TOTAL', 'AMT_GOODS_PRICE') hads been logged 5. Distrubutions of New Datasets 5.1 Plotting distribution of Datasets -I had initally plotted historgams for features like AMT_CREDIT, AMT_TOTAL_INCOME and AMT_GOOD_PRICE using df1. But I found that the distributions were skewed. So I logged these features to create df2 and plotted these features again. I show the comparions of the same below: | # Set the style of plots
plt.style.use('fivethirtyeight')
plt.figure(figsize = (10, 12))
plt.subplot(2, 1, 1)
plt.title("Distribution of AMT_CREDIT")
plt.hist(df1["AMT_CREDIT"], bins =20)
plt.xlabel("AMT_CREDIT")
plt.subplot(2, 1, 2)
plt.title(" Log Distribution of AMT_CREDIT")
plt.hist(df2["AMT_CREDIT"], bins =20)
plt.xlabel("Log_AMT_CREDIT")
plt.figure(figsize = (10, 12))
plt.subplot(2, 1, 1)
plt.title("Distribution of AMT_INCOME_TOTAL")
plt.hist(df1["AMT_INCOME_TOTAL"].dropna(), bins =25)
plt.subplot(2, 1, 2)
plt.title(" Log Distribution of AMT_INCOME_TOTAL")
plt.hist(df2["AMT_INCOME_TOTAL"].dropna(), bins =25)
plt.xlabel("Log_INCOME_TOTAL")
plt.figure(figsize = (10, 12))
plt.subplot(2, 1, 1)
plt.title("Distribution of AMT_GOODS_PRICE")
plt.hist(df1["AMT_GOODS_PRICE"].dropna(), bins = 20)
plt.subplot(2, 1, 2)
plt.title(" Log Distribution of AMT_GOODS_PRICE")
plt.hist(df2["AMT_GOODS_PRICE"].dropna(), bins = 20)
plt.xlabel("Log_GOODS_PRICE")
plt.hist(df1['YEARS_EMPLOYED'])
plt.xlabel('Years of Employment') | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
- It is not resonable to have such high years of employment (> 40-60 years).- As there are amny rows, I would try and repalce the values with average years of employment- I would plot the distribution of the reasonable values to get a clearer picture of the disrtibution.- Based on the ditribution, I would decide to take a log of the values to reduce the skew. | less_years = df1[df1.YEARS_EMPLOYED <= 80]
more_years = df1[df1.YEARS_EMPLOYED >80]
plt.hist(less_years['YEARS_EMPLOYED'])
plt.xlabel('Distribution of Lesser Years of Employment') | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
log this data and change it in df1 | less_years['YEARS_EMPLOYED'].mean()
df1['YEARS_EMPLOYED'] = np.where(df1['YEARS_EMPLOYED'] > 80, 7, df1['YEARS_EMPLOYED'])
plt.hist(df1['YEARS_EMPLOYED'])
plt.xlabel('Years of Employment')
plt.hist(df2['YEARS_EMPLOYED'])
plt.xlabel('Years of Employment')
##Defaul = df1[df1['TARGET'] == 1]
Not_defaul = df1[df1['TARGET'] == 0]
# Find correlations with the target and sort
correlations = df1.corr()['TARGET'].sort_values()
# Display correlations
print('\nMost Positive Correlations: \n ', correlations.tail(15))
print('\nMost Negative Correlations:\n', correlations.head(15))
# Find the correlation of the positive days since birth and target
df1['YEARS_BIRTH'] = abs(df1['YEARS_BIRTH'])
df1['YEARS_BIRTH'].corr(df1['TARGET'])
# Set the style of plots
plt.style.use('fivethirtyeight')
# Plot the distribution of ages in years
plt.hist(df1['YEARS_BIRTH'], edgecolor = 'k', bins = 25)
plt.title('Age of Client'); plt.xlabel('Age (years)'); plt.ylabel('Count');
# KDE plot of loans that were repaid on time
sns.kdeplot(df1.loc[df1['TARGET'] == 0, 'YEARS_BIRTH'] / 365, label = 'target == 0')
# KDE plot of loans which were not repaid on time
sns.kdeplot(df1.loc[df1['TARGET'] == 1, 'YEARS_BIRTH'] / 365, label = 'target == 1')
# Labeling of plot
plt.xlabel('Age (years)'); plt.ylabel('Density'); plt.title('Distribution of Ages');
age_data = df1[['TARGET', 'YEARS_BIRTH']]
age_data['YEARS_BIRTH'] = age_data['YEARS_BIRTH']
# Bin the age data
age_data['YEARS_BINNED'] = pd.cut(age_data['YEARS_BIRTH'], bins = np.linspace(20, 70, num = 11))
age_data.head(10)
# Group by the bin and calculate averages
age_groups = age_data.groupby('YEARS_BINNED').mean()
age_groups
plt.figure(figsize = (8, 8))
# Graph the age bins and the average of the target as a bar plot
plt.bar(age_groups.index.astype(str), 100 * age_groups['TARGET'])
# Plot labeling
plt.xticks(rotation = 75); plt.xlabel('Age Group (years)'); plt.ylabel('Failure to Repay (%)')
plt.title('Failure to Repay by Age Group');
# Extract the EXT_SOURCE variables and show correlations
ext_data = df1[['TARGET', 'EXT_SOURCE_1', 'EXT_SOURCE_2', 'EXT_SOURCE_3', 'YEARS_BIRTH']]
ext_data_corrs = ext_data.corr()
ext_data_corrs
# Extract the EXT_SOURCE variables and show correlations
ext_data = application_train[['TARGET', 'EXT_SOURCE_1', 'EXT_SOURCE_2', 'EXT_SOURCE_3', 'DAYS_BIRTH']]
ext_data_corrs = ext_data.corr()
ext_data_corrs
plt.figure(figsize = (8, 6))
# Heatmap of correlations
sns.heatmap(ext_data_corrs, cmap = plt.cm.RdYlBu_r, vmin = -0.25, annot = True, vmax = 0.6)
plt.title('Correlation Heatmap'); | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
DAYS_BIRTH is positively correlated with EXT_SOURCE_1 indicating that maybe one of the factors in this score is the client age.so try build model with EXT_SOURCE_1 and/or DAYS_BIRTH | plt.figure(figsize = (10, 12))
# iterate through the sources
for i, source in enumerate(['EXT_SOURCE_1', 'EXT_SOURCE_2', 'EXT_SOURCE_3']):
# create a new subplot for each source
#
plt.subplot(3, 1, i+1)
# plot repaid loans
sns.kdeplot(df1.loc[df1['TARGET'] == 0, source], label = 'target == 0')
# plot loans that were not repaid
sns.kdeplot(df1.loc[application_train['TARGET'] == 1, source], label = 'target == 1')
# Label the plots
plt.title('Distribution of %s by Target Value' % source)
plt.xlabel('%s' % source); plt.ylabel('Density');
plt.tight_layout(h_pad = 2.5)
# Make a new dataframe for polynomial features
poly_features = df1[['EXT_SOURCE_1', 'EXT_SOURCE_2', 'EXT_SOURCE_3', 'YEARS_BIRTH', 'TARGET']]
poly_features_test = df1[['EXT_SOURCE_1', 'EXT_SOURCE_2', 'EXT_SOURCE_3', 'YEARS_BIRTH']]
# imputer for handling missing values
from sklearn.preprocessing import Imputer
imputer = Imputer(strategy = 'median')
poly_target = poly_features['TARGET']
poly_features = poly_features.drop(columns = ['TARGET'])
# Need to impute missing values
poly_features = imputer.fit_transform(poly_features)
poly_features_test = imputer.transform(poly_features_test)
from sklearn.preprocessing import PolynomialFeatures
# Create the polynomial object with specified degree
poly_transformer = PolynomialFeatures(degree = 3) | _____no_output_____ | MIT | notebooks/ExploratoryDataAnalysis.ipynb | geracharu/DataScienceProject |
import numpy as np
A=([1,2,-1],[4,6,-2],[-1,3,3])
print(A)
print(round(np.linalg.det(A)))
| _____no_output_____ | Apache-2.0 | Determinant_of_Matrix.ipynb | jnrtnan/Linear-Algebra-58020 |
|
Copyright 2020 DeepMind Technologies Limited. All Rights Reserved. Licensed under the Apache License, Version 2.0 (the "License"); Full license text | # Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License. | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
A (very) basic GAN for MNIST in JAX/HaikuBased on a TensorFlow tutorial written by Mihaela Rosca.Original GAN paper: https://papers.nips.cc/paper/5423-generative-adversarial-nets.pdf Imports | # Uncomment the line below if running on colab.research.google.com.
# !pip install dm-haiku
import functools
from typing import Any, NamedTuple
import haiku as hk
import jax
from jax.experimental import optix
import jax.numpy as jnp
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import tensorflow as tf
import tensorflow_datasets as tfds | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
Define the dataset | # Download the data once.
mnist = tfds.load("mnist")
def make_dataset(batch_size, seed=1):
def _preprocess(sample):
# Convert to floats in [0, 1].
image = tf.image.convert_image_dtype(sample["image"], tf.float32)
# Scale the data to [-1, 1] to stabilize training.
return 2.0 * image - 1.0
ds = mnist["train"]
ds = ds.map(map_func=_preprocess,
num_parallel_calls=tf.data.experimental.AUTOTUNE)
ds = ds.cache()
ds = ds.shuffle(10 * batch_size, seed=seed).repeat().batch(batch_size)
return tfds.as_numpy(ds) | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
Define the model | class Generator(hk.Module):
"""Generator network."""
def __init__(self, output_channels=(32, 1), name=None):
super().__init__(name=name)
self.output_channels = output_channels
def __call__(self, x):
"""Maps noise latents to images."""
x = hk.Linear(7 * 7 * 64)(x)
x = jnp.reshape(x, x.shape[:1] + (7, 7, 64))
for output_channels in self.output_channels:
x = jax.nn.relu(x)
x = hk.Conv2DTranspose(output_channels=output_channels,
kernel_shape=[5, 5],
stride=2,
padding="SAME")(x)
# We use a tanh to ensure that the generated samples are in the same
# range as the data.
return jnp.tanh(x)
class Discriminator(hk.Module):
"""Discriminator network."""
def __init__(self,
output_channels=(8, 16, 32, 64, 128),
strides=(2, 1, 2, 1, 2),
name=None):
super().__init__(name=name)
self.output_channels = output_channels
self.strides = strides
def __call__(self, x):
"""Classifies images as real or fake."""
for output_channels, stride in zip(self.output_channels, self.strides):
x = hk.Conv2D(output_channels=output_channels,
kernel_shape=[5, 5],
stride=stride,
padding="SAME")(x)
x = jax.nn.leaky_relu(x, negative_slope=0.2)
x = hk.Flatten()(x)
# We have two classes: 0 = input is fake, 1 = input is real.
logits = hk.Linear(2)(x)
return logits
def tree_shape(xs):
return jax.tree_map(lambda x: x.shape, xs)
def sparse_softmax_cross_entropy(logits, labels):
one_hot_labels = jax.nn.one_hot(labels, logits.shape[-1])
return -jnp.sum(one_hot_labels * jax.nn.log_softmax(logits), axis=-1)
class GANTuple(NamedTuple):
gen: Any
disc: Any
class GANState(NamedTuple):
params: GANTuple
opt_state: GANTuple
class GAN:
"""A basic GAN."""
def __init__(self, num_latents):
self.num_latents = num_latents
# Define the Haiku network transforms.
# We don't use BatchNorm so we don't use `with_state`.
self.gen_transform = hk.transform(lambda *args: Generator()(*args))
self.disc_transform = hk.transform(lambda *args: Discriminator()(*args))
# Build the optimizers.
self.optimizers = GANTuple(gen=optix.adam(1e-4, b1=0.5, b2=0.9),
disc=optix.adam(1e-4, b1=0.5, b2=0.9))
@functools.partial(jax.jit, static_argnums=0)
def initial_state(self, rng, batch):
"""Returns the initial parameters and optimize states."""
# Generate dummy latents for the generator.
dummy_latents = jnp.zeros((batch.shape[0], self.num_latents))
# Get initial network parameters.
rng_gen, rng_disc = jax.random.split(rng)
params = GANTuple(gen=self.gen_transform.init(rng_gen, dummy_latents),
disc=self.disc_transform.init(rng_disc, batch))
print("Generator: \n\n{}\n".format(tree_shape(params.gen)))
print("Discriminator: \n\n{}\n".format(tree_shape(params.disc)))
# Initialize the optimizers.
opt_state = GANTuple(gen=self.optimizers.gen.init(params.gen),
disc=self.optimizers.disc.init(params.disc))
return GANState(params=params, opt_state=opt_state)
def sample(self, rng, gen_params, num_samples):
"""Generates images from noise latents."""
latents = jax.random.normal(rng, shape=(num_samples, self.num_latents))
return self.gen_transform.apply(gen_params, latents)
def gen_loss(self, gen_params, rng, disc_params, batch):
"""Generator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# Evaluate using the discriminator. Recall class 1 is real.
fake_logits = self.disc_transform.apply(disc_params, fake_batch)
fake_probs = jax.nn.softmax(fake_logits)[:, 1]
loss = -jnp.log(fake_probs)
return jnp.mean(loss)
def disc_loss(self, disc_params, rng, gen_params, batch):
"""Discriminator loss."""
# Sample from the generator.
fake_batch = self.sample(rng, gen_params, num_samples=batch.shape[0])
# For efficiency we process both the real and fake data in one pass.
real_and_fake_batch = jnp.concatenate([batch, fake_batch], axis=0)
real_and_fake_logits = self.disc_transform.apply(disc_params,
real_and_fake_batch)
real_logits, fake_logits = jnp.split(real_and_fake_logits, 2, axis=0)
# Class 1 is real.
real_labels = jnp.ones((batch.shape[0],), dtype=jnp.int32)
real_loss = sparse_softmax_cross_entropy(real_logits, real_labels)
# Class 0 is fake.
fake_labels = jnp.zeros((batch.shape[0],), dtype=jnp.int32)
fake_loss = sparse_softmax_cross_entropy(fake_logits, fake_labels)
return jnp.mean(real_loss + fake_loss)
@functools.partial(jax.jit, static_argnums=0)
def update(self, rng, gan_state, batch):
"""Performs a parameter update."""
rng, rng_gen, rng_disc = jax.random.split(rng, 3)
# Update the discriminator.
disc_loss, disc_grads = jax.value_and_grad(self.disc_loss)(
gan_state.params.disc,
rng_disc,
gan_state.params.gen,
batch)
disc_update, disc_opt_state = self.optimizers.disc.update(
disc_grads, gan_state.opt_state.disc)
disc_params = optix.apply_updates(gan_state.params.disc, disc_update)
# Update the generator.
gen_loss, gen_grads = jax.value_and_grad(self.gen_loss)(
gan_state.params.gen,
rng_gen,
gan_state.params.disc,
batch)
gen_update, gen_opt_state = self.optimizers.gen.update(
gen_grads, gan_state.opt_state.gen)
gen_params = optix.apply_updates(gan_state.params.gen, gen_update)
params = GANTuple(gen=gen_params, disc=disc_params)
opt_state = GANTuple(gen=gen_opt_state, disc=disc_opt_state)
gan_state = GANState(params=params, opt_state=opt_state)
log = {
"gen_loss": gen_loss,
"disc_loss": disc_loss,
}
return rng, gan_state, log | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
Train the model | #@title {vertical-output: true}
num_steps = 20001
log_every = num_steps // 100
# Let's see what hardware we're working with. The training takes a few
# minutes on a GPU, a bit longer on CPU.
print(f"Number of devices: {jax.device_count()}")
print("Device:", jax.devices()[0].device_kind)
print("")
# Make the dataset.
dataset = make_dataset(batch_size=64)
# The model.
gan = GAN(num_latents=20)
# Top-level RNG.
rng = jax.random.PRNGKey(1729)
# Initialize the network and optimizer.
rng, rng1 = jax.random.split(rng)
gan_state = gan.initial_state(rng1, next(dataset))
steps = []
gen_losses = []
disc_losses = []
for step in range(num_steps):
rng, gan_state, log = gan.update(rng, gan_state, next(dataset))
# Log the losses.
if step % log_every == 0:
# It's important to call `device_get` here so we don't take up device
# memory by saving the losses.
log = jax.device_get(log)
gen_loss = log["gen_loss"]
disc_loss = log["disc_loss"]
print(f"Step {step}: "
f"gen_loss = {gen_loss:.3f}, disc_loss = {disc_loss:.3f}")
steps.append(step)
gen_losses.append(gen_loss)
disc_losses.append(disc_loss) | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
Visualize the lossesUnlike losses for classifiers or VAEs, GAN losses do not decrease steadily, instead going up and down depending on the training dynamics. | sns.set_style("whitegrid")
fig, axes = plt.subplots(1, 2, figsize=(20, 6))
# Plot the discriminator loss.
axes[0].plot(steps, disc_losses, "-")
axes[0].plot(steps, np.log(2) * np.ones_like(steps), "r--",
label="Discriminator is being fooled")
axes[0].legend(fontsize=20)
axes[0].set_title("Discriminator loss", fontsize=20)
# Plot the generator loss.
axes[1].plot(steps, gen_losses, '-')
axes[1].set_title("Generator loss", fontsize=20); | _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
Visualize samples | #@title {vertical-output: true}
def make_grid(samples, num_cols=8, rescale=True):
batch_size, height, width = samples.shape
assert batch_size % num_cols == 0
num_rows = batch_size // num_cols
# We want samples.shape == (height * num_rows, width * num_cols).
samples = samples.reshape(num_rows, num_cols, height, width)
samples = samples.swapaxes(1, 2)
samples = samples.reshape(height * num_rows, width * num_cols)
return samples
# Generate samples from the trained generator.
rng = jax.random.PRNGKey(12)
samples = gan.sample(rng, gan_state.params.gen, num_samples=64)
samples = jax.device_get(samples)
samples = samples.squeeze(axis=-1)
# Our model outputs values in [-1, 1] so scale it back to [0, 1].
samples = (samples + 1.0) / 2.0
plt.gray()
plt.axis("off")
samples_grid = make_grid(samples)
plt.imshow(samples_grid);
| _____no_output_____ | Apache-2.0 | examples/mnist_gan.ipynb | tirkarthi/dm-haiku |
¿Cómo podemos calcular con las funciones de Python los autovectores y los autovalores? | # Importamos las bibliotecas
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
# Creamos una matriz
X = np.array([[3, 2], [4, 1]])
print(X)
# Vemos la biblioteca para calcular los autovectores y autovalores de Numpy
print(np.linalg.eig(X))
# Pedimos que muestre los autovalores
autovalores, autovectores = np.linalg.eig(X)
print(autovalores)
# Pedimos cual es el autovalor asociado a cada autovector
print(autovectores[:, 0])
# Mostramos el autovector numero 1
print(autovectores[:, 1])
# Importamos nuestra función para graficar.
%run ".\\Funciones auxiliares\graficarVectores.ipynb"
# Definamos un array
v = np.array([[-1], [2]])
# Calculamos la tranformacion con el calculo del producto interno
Xv = X.dot(v)
# Y lo comparamos con el autovector anterior
v_np = autovectores[:, 1]
print(Xv)
print(v_np)
# Graficamos al que calculamos con el producto interno, el vector original y el que nos devolvió el método
graficarVectores([Xv.flatten(), v.flatten(), v_np], cols = ['green','orange','blue'])
plt.ylim(-4,2)
plt.xlim(-7,3) | _____no_output_____ | MIT | Code/4.-Cómo calcular los autovalores y autovectores.ipynb | DataEngel/Linear-algebra-applied-to-ML-with-Python |
Alert Statistics | plt.figure(figsize=(base_width, base_height), dpi=dpi)
ax1 = plt.subplot(111)
reasons = [x if x != "Poor Signalness and Localisation" else "Poor Signalness \n and Localisation" for x in non["Rejection reason"]]
reasons = [x if x != "Separation from Galactic Plane" else "Separation from \n Galactic Plane" for x in reasons]
t_min = 1803
t_max = 3303
reasons = [x for i, x in enumerate(reasons) if np.logical_and(float(non["Event"][i][2:6]) > t_min, float(non["Event"][i][2:6]) < t_max)]
labels = sorted(list(set(reasons)))
sizes = []
for l in labels:
sizes.append(list(reasons).count(l))
explode =[0.1] + [0.0 for _ in labels]
labels = ["Observed"] + labels
sizes = [len(obs)] + sizes
def absolute_value(val):
a = np.round(val/100.*np.sum(sizes), 0)
return int(a)
print(labels)
patches, texts, autotexts = ax1.pie(sizes,
explode=explode,
labels=labels,
autopct=absolute_value,
pctdistance=0.9,
textprops={'fontsize': big_fontsize}
)
[autotext.set_color('white') for autotext in autotexts]
ax1.axis('equal') # Equal aspect ratio ensures that pie is drawn as a circle.
filename = "pie.pdf"
output_path = os.path.join(output_folder, filename)
plt.savefig(os.path.join(plot_dir, filename))
plt.savefig(output_path, bbox_inches='tight', pad_inches=0) | ['Observed', 'Alert Retraction', 'Low Altitude', 'Poor Signalness \n and Localisation', 'Proximity to Sun', 'Separation from \n Galactic Plane', 'Southern Sky', 'Telescope Maintenance']
| MIT | notebooks/stats_alerts.ipynb | robertdstein/nuztfpaper |
Observed alerts (Table 1) | text = r"""
\begin{table*}
\centering
\begin{tabular}{||c | c c c c c c ||}
\hline
\textbf{Event} & \textbf{R.A. (J2000)} & \textbf{Dec (J2000)} & \textbf{90\% area} & \textbf{ZTF obs} &~ \textbf{Signalness}& \textbf{Refs}\\
& \textbf{[deg]}&\textbf{[deg]}& \textbf{[sq. deg.]}& \textbf{[sq. deg.]} &&\\
\hline
"""
tot_area = 0.
for index, row in obs.iterrows():
name = str(row["Event"].lower())
ras = json.loads(row["RA Unc (rectangle)"])
decs = json.loads(row["Dec Unc (rectangle)"])
delta_r = ras[0] - ras[1]
delta_d = decs[0] - decs[1]
area = delta_r * delta_d * np.cos(np.radians(float(row["Dec"])))
if np.isnan(float(row["Signalness"])):
s = "-"
else:
s = f'{100.*row["Signalness"]:.0f}\%'
text += f'\t {row["Event"]} & {row["RA"]} & {row["Dec"]:+.2f} & {area:.1f} & {row["Observed area (corrected for chip gaps)"]:.1f} & {s} & \cite{{{name}}} \\\\\ \n'
text += f'\t &&&&&& \cite{{{name}_ztf}} \\\\ \n'
if not isinstance(row["Additional ZTF GCN"], float):
text += f'\t &&&&&& \cite{{{name}_ztf_2}} \\\\ \n'
text += "\t \hline"
tot_area += row["Observed area (corrected for chip gaps)"]
text += f"""
\end{{tabular}}
\caption{{Summary of the {len(obs)} neutrino alerts followed up by ZTF since survey start on 2018 March 20.}}
\label{{tab:nu_alerts}}
\end{{table*}}
"""
print(text) |
\begin{table*}
\centering
\begin{tabular}{||c | c c c c c c ||}
\hline
\textbf{Event} & \textbf{R.A. (J2000)} & \textbf{Dec (J2000)} & \textbf{90\% area} & \textbf{ZTF obs} &~ \textbf{Signalness}& \textbf{Refs}\\
& \textbf{[deg]}&\textbf{[deg]}& \textbf{[sq. deg.]}& \textbf{[sq. deg.]} &&\\
\hline
IC190503A & 120.28 & +6.35 & 1.9 & 1.4 & 36\% & \cite{ic190503a} \\\
&&&&&& \cite{ic190503a_ztf} \\
\hline IC190619A & 343.26 & +10.73 & 27.2 & 21.6 & 55\% & \cite{ic190619a} \\\
&&&&&& \cite{ic190619a_ztf} \\
\hline IC190730A & 225.79 & +10.47 & 5.4 & 4.5 & 67\% & \cite{ic190730a} \\\
&&&&&& \cite{ic190730a_ztf} \\
\hline IC190922B & 5.76 & -1.57 & 4.5 & 4.1 & 51\% & \cite{ic190922b} \\\
&&&&&& \cite{ic190922b_ztf} \\
\hline IC191001A & 314.08 & +12.94 & 25.5 & 23.1 & 59\% & \cite{ic191001a} \\\
&&&&&& \cite{ic191001a_ztf} \\
\hline IC200107A & 148.18 & +35.46 & 7.6 & 6.3 & - & \cite{ic200107a} \\\
&&&&&& \cite{ic200107a_ztf} \\
\hline IC200109A & 164.49 & +11.87 & 22.5 & 22.4 & 77\% & \cite{ic200109a} \\\
&&&&&& \cite{ic200109a_ztf} \\
\hline IC200117A & 116.24 & +29.14 & 2.9 & 2.7 & 38\% & \cite{ic200117a} \\\
&&&&&& \cite{ic200117a_ztf} \\
&&&&&& \cite{ic200117a_ztf_2} \\
\hline IC200512A & 295.18 & +15.79 & 9.8 & 9.3 & 32\% & \cite{ic200512a} \\\
&&&&&& \cite{ic200512a_ztf} \\
\hline IC200530A & 255.37 & +26.61 & 25.3 & 22.0 & 59\% & \cite{ic200530a} \\\
&&&&&& \cite{ic200530a_ztf} \\
&&&&&& \cite{ic200530a_ztf_2} \\
\hline IC200620A & 162.11 & +11.95 & 1.7 & 1.2 & 32\% & \cite{ic200620a} \\\
&&&&&& \cite{ic200620a_ztf} \\
\hline IC200916A & 109.78 & +14.36 & 4.2 & 3.6 & 32\% & \cite{ic200916a} \\\
&&&&&& \cite{ic200916a_ztf} \\
&&&&&& \cite{ic200916a_ztf_2} \\
\hline IC200926A & 96.46 & -4.33 & 1.7 & 1.3 & 44\% & \cite{ic200926a} \\\
&&&&&& \cite{ic200926a_ztf} \\
\hline IC200929A & 29.53 & +3.47 & 1.1 & 0.9 & 47\% & \cite{ic200929a} \\\
&&&&&& \cite{ic200929a_ztf} \\
\hline IC201007A & 265.17 & +5.34 & 0.6 & 0.6 & 88\% & \cite{ic201007a} \\\
&&&&&& \cite{ic201007a_ztf} \\
\hline IC201021A & 260.82 & +14.55 & 6.9 & 6.3 & 30\% & \cite{ic201021a} \\\
&&&&&& \cite{ic201021a_ztf} \\
\hline IC201130A & 30.54 & -12.10 & 5.4 & 4.5 & 15\% & \cite{ic201130a} \\\
&&&&&& \cite{ic201130a_ztf} \\
\hline IC201209A & 6.86 & -9.25 & 4.7 & 3.2 & 19\% & \cite{ic201209a} \\\
&&&&&& \cite{ic201209a_ztf} \\
\hline IC201222A & 206.37 & +13.44 & 1.5 & 1.4 & 53\% & \cite{ic201222a} \\\
&&&&&& \cite{ic201222a_ztf} \\
\hline IC210210A & 206.06 & +4.78 & 2.8 & 2.1 & 65\% & \cite{ic210210a} \\\
&&&&&& \cite{ic210210a_ztf} \\
\hline IC210510A & 268.42 & +3.81 & 4.0 & 3.7 & 28\% & \cite{ic210510a} \\\
&&&&&& \cite{ic210510a_ztf} \\
\hline IC210629A & 340.75 & +12.94 & 6.0 & 4.6 & 35\% & \cite{ic210629a} \\\
&&&&&& \cite{ic210629a_ztf} \\
\hline IC210811A & 270.79 & +25.28 & 3.2 & 2.7 & 66\% & \cite{ic210811a} \\\
&&&&&& \cite{ic210811a_ztf} \\
\hline IC210922A & 60.73 & -4.18 & 1.6 & 1.2 & 92\% & \cite{ic210922a} \\\
&&&&&& \cite{ic210922a_ztf} \\
\hline
\end{tabular}
\caption{Summary of the 24 neutrino alerts followed up by ZTF since survey start on 2018 March 20.}
\label{tab:nu_alerts}
\end{table*}
| MIT | notebooks/stats_alerts.ipynb | robertdstein/nuztfpaper |
Not observed | reasons = ["Alert Retraction", "Proximity to Sun", "Low Altitude", "Southern Sky", "Separation from Galactic Plane", "Poor Signalness and Localisation", "Telescope Maintenance"]
seps = [1, 0, 0, 0, 1, 1, 1]
full_mask = np.array([float(x[2:6]) > 1802 for x in non["Event"]])
text = r"""
\begin{table*}
\centering
\begin{tabular}{||c c ||}
\hline
\textbf{Cause} & \textbf{Events} \\
\hline
"""
for i, reason in enumerate(reasons):
mask = non["Rejection reason"] == reason
names = list(non["Event"][full_mask][mask])
for j, name in enumerate(names):
names[j] = f'{name} \citep{{{name.lower()}}}'
text += f'\t {reason} & '
n_int = 2
while len(names) > n_int:
text += f'{", ".join(names[:n_int])} \\\\ \n \t & '
names = names[n_int:]
text += f'{", ".join(names)} \\\\ \n'
# if seps[i]:
if True:
text += "\t \hline \n"
text +=f"""
\end{{tabular}}
\caption{{Summary of the {np.sum(full_mask)} neutrino alerts that were not followed up by ZTF since survey start on 2018 March 20.}}
\label{{tab:nu_non_observed}}
\end{{table*}}
"""
print(text) |
\begin{table*}
\centering
\begin{tabular}{||c c ||}
\hline
\textbf{Cause} & \textbf{Events} \\
\hline
Alert Retraction & IC180423A \citep{ic180423a}, IC181031A \citep{ic181031a} \\
& IC190205A \citep{ic190205a}, IC190529A \citep{ic190529a} \\
& IC200120A \citep{ic200120a}, IC200728A \citep{ic200728a} \\
& IC201115B \citep{ic201115b}, IC210213A \citep{ic210213a} \\
& IC210322A \citep{ic210322a}, IC210519A \citep{ic210519a} \\
\hline
Proximity to Sun & IC180908A \citep{ic180908a}, IC181014A \citep{ic181014a} \\
& IC190124A \citep{ic190124a}, IC190704A \citep{ic190704a} \\
& IC190712A \citep{ic190712a}, IC190819A \citep{ic190819a} \\
& IC191119A \citep{ic191119a}, IC200227A \citep{ic200227a} \\
& IC200421A \citep{ic200421a}, IC200615A \citep{ic200615a} \\
& IC200806A \citep{ic200806a}, IC200921A \citep{ic200921a} \\
& IC200926B \citep{ic200926b}, IC201014A \citep{ic201014a} \\
& IC201115A \citep{ic201115a}, IC201221A \citep{ic201221a} \\
& IC211117A \citep{ic211117a}, IC211123A \citep{ic211123a} \\
\hline
Low Altitude & IC191215A \citep{ic191215a}, IC211023A \citep{ic211023a} \\
\hline
Southern Sky & IC190104A \citep{ic190104a}, IC190331A \citep{ic190331a} \\
& IC190504A \citep{ic190504a} \\
\hline
Separation from Galactic Plane & IC201114A \citep{ic201114a}, IC201120A \citep{ic201120a} \\
& IC210516A \citep{ic210516a}, IC210730A \citep{ic210730a} \\
\hline
Poor Signalness and Localisation & IC190221A \citep{ic190221a}, IC190629A \citep{ic190629a} \\
& IC190922A \citep{ic190922a}, IC191122A \citep{ic191122a} \\
& IC191204A \citep{ic191204a}, IC191231A \citep{ic191231a} \\
& IC200410A \citep{ic200410a}, IC200425A \citep{ic200425a} \\
& IC200523A \citep{ic200523a}, IC200614A \citep{ic200614a} \\
& IC200911A \citep{ic200911a}, IC210503A \citep{ic210503a} \\
& IC210608A \citep{ic210608a}, IC210717A \citep{ic210717a} \\
& IC211125A \citep{ic211125a} \\
\hline
Telescope Maintenance & IC181023A \citep{ic181023a}, IC211116A \citep{ic211116a} \\
& IC211208A \citep{ic211208a} \\
\hline
\end{tabular}
\caption{Summary of the 55 neutrino alerts that were not followed up by ZTF since survey start on 2018 March 20.}
\label{tab:nu_non_observed}
\end{table*}
| MIT | notebooks/stats_alerts.ipynb | robertdstein/nuztfpaper |
Full Neutrino List | text = fr"""
\begin{{longtable}}[c]{{||c c c c c c ||}}
\caption{{Summary of all {len(joint)} neutrino alerts issued since under the IceCube Realtime Program. Directions are not indicated for retracted events.}} \label{{tab:all_nu_alerts}} \\
\hline
\textbf{{Event}} & \textbf{{R.A. (J2000)}} & \textbf{{Dec (J2000)}} & \textbf{{90\% area}} &~ \textbf{{Signalness}}& \textbf{{Ref}}\\
& \textbf{{[deg]}}&\textbf{{[deg]}} & \textbf{{[sq. deg.]}} &&\\
\hline
\endfirsthead
\hline
\textbf{{Event}} & \textbf{{R.A. (J2000)}} & \textbf{{Dec (J2000)}} & \textbf{{90\% area}} &~ \textbf{{Signalness}}& \textbf{{Ref}}\\
& \textbf{{[deg]}}&\textbf{{[deg]}} & \textbf{{[sq. deg.]}} &&\\
\hline
\endhead
\hline
\endfoot
\hline
\endlastfoot
\hline%
"""
for index, row in joint.iterrows():
name = str(row["Event"].lower())
if not isinstance(row["RA Unc (rectangle)"], float):
ras = json.loads(str(row["RA Unc (rectangle)"]))
decs = json.loads(row["Dec Unc (rectangle)"])
delta_r = ras[0] - ras[1]
delta_d = decs[0] - decs[1]
area = f'{delta_r * delta_d * np.cos(np.radians(float(row["Dec"]))):.1f}'
else:
area = "-"
if np.isnan(float(row["Signalness"])):
s = "-"
else:
s = f'{100.*row["Signalness"]:.0f}\%'
if np.isnan(float(row["Dec"])):
r = "-"
d = "-"
else:
r = f'{row["RA"]}'
d = f'{row["Dec"]:+.2f}'
if name not in ["ic160731a", "ic160814a", "ic170312a"]:
c = name
else:
c = "ic_txs_mm_18"
text += f'\t {row["Event"]} & {r} & {d} & {area} & {s} & \cite{{{c}}} \\\\ \n'
text += f"""
\end{{longtable}}
"""
print(text)
# Neutrino stats
dates = [Time(f"20{x[2:4]}-{x[4:6]}-{x[6:8]}T00:00:01") for x in joint["Event"]]
plt.figure(figsize=(base_width, base_height), dpi=dpi)
ax1 = plt.subplot(111)
mjds = []
labs = []
bins = []
for year in range(2016, 2022):
for k, month in enumerate([1, 4, 7, 10]):
t = Time(f"{year}-{month}-01T00:00:00.01", format='isot', scale='utc').mjd
bins.append(t)
if (k - 1) % 2 > 0:
mjds.append(t)
labs.append(["Jan", "July"][int(k/2)] + f" {year}")
t_0 = Time(f"2016-04-01T00:00:00.01", format='isot', scale='utc').mjd
v1_t = Time(f"2019-06-17T00:00:00.01", format='isot', scale='utc').mjd
t_now = Time.now().mjd
alerts_v1 = [x.mjd for i, x in enumerate(dates) if np.logical_and(x.mjd < v1_t, not np.isnan(joint.iloc[i]["Dec"]))]
alerts_v2 = [x.mjd for i, x in enumerate(dates) if np.logical_and(
x.mjd > v1_t, not np.isnan(joint.iloc[i]["Dec"]))]
print(f'{len(alerts_v1)} V1 alerts, {len(alerts_v2)} V2 alerts')
mod = 7.
v1_rate = mod * float(len(alerts_v1))/(v1_t - t_0)
v2_rate = mod * float(len(alerts_v2))/(t_now - v1_t)
labels = []
for (name, rate) in [("V1", v1_rate), ("V2", v2_rate)]:
labels.append(f'{name} ({rate:.2f} per week)')
plt.xticks(mjds, labs, rotation=80)
plt.locator_params(axis="y", nbins=6)
plt.hist([alerts_v1, alerts_v2], bins=bins, stacked=True, label=labels)
plt.axvline(v1_t, linestyle=":", color="k")
plt.tick_params(axis='both', which='major', labelsize=big_fontsize)
plt.legend(fontsize=big_fontsize, loc="upper left")
plt.ylabel("Alerts (excluding retractions)", fontsize=big_fontsize)
sns.despine()
plt.ylim(0., 12.)
plt.tight_layout()
filename = "alert_hist.pdf"
output_path = os.path.join(output_folder, filename)
plt.savefig(os.path.join(plot_dir, filename))
plt.savefig(output_path, bbox_inches='tight', pad_inches=0)
plt.figure(figsize=(base_width, base_height), dpi=dpi)
ax1 = plt.subplot(111)
dates = [Time(f"20{x[2:4]}-{x[4:6]}-{x[6:8]}T00:00:01") for x in joint["Event"]]
mjds = []
labs = []
bins = []
for year in range(2016, 2022):
for k, month in enumerate([1, 4, 7, 10]):
t = Time(f"{year}-{month}-01T00:00:00.01", format='isot', scale='utc').mjd
bins.append(t)
if (k - 1) % 2 > 0:
mjds.append(t)
labs.append(["Jan", "July"][int(k/2)] + f" {year}")
t_0 = Time(f"2016-04-01T00:00:00.01", format='isot', scale='utc').mjd
v1_t = Time(f"2019-06-17T00:00:00.01", format='isot', scale='utc').mjd
t_now = Time.now().mjd
alerts_v1 = [x.mjd for i, x in enumerate(dates) if np.logical_and(x.mjd < v1_t, not np.isnan(joint.iloc[i]["Dec"]))]
alerts_v2 = [x.mjd for i, x in enumerate(dates) if np.logical_and(
x.mjd > v1_t, not np.isnan(joint.iloc[i]["Dec"]))]
print(f'{len(alerts_v1)} V1 alerts, {len(alerts_v2)} V2 alerts')
mod = 7.
v1_rate = mod * float(len(alerts_v1))/(v1_t - t_0)
v2_rate = mod * float(len(alerts_v2))/(t_now - v1_t)
labels = []
for (name, rate) in [("HESE/EHE", v1_rate), ("Gold/Bronze", v2_rate)]:
labels.append(f'{name} ({rate:.2f} per week)')
plt.xticks(mjds, labs, rotation=80)
plt.locator_params(axis="y", nbins=6)
plt.hist([alerts_v1, alerts_v2], bins=bins[:-1], stacked=True, label=labels, cumulative=True)
plt.axvline(v1_t, linestyle=":", color="k")
plt.tick_params(axis='both', which='major', labelsize=big_fontsize)
plt.legend(fontsize=big_fontsize, loc="upper left")
sns.despine()
# plt.ylim(0., 12.)
plt.ylabel("Alerts (excluding retractions)", fontsize=big_fontsize)
plt.tight_layout()
filename = "alert_cdf.pdf"
output_path = os.path.join(output_folder, filename)
plt.savefig(os.path.join(plot_dir, filename))
plt.savefig(output_path, bbox_inches='tight', pad_inches=0)
plt.figure(figsize=(base_width, base_height), dpi=dpi)
ax1 = plt.subplot(111)
dates = [Time(f"20{x[2:4]}-{x[4:6]}-{x[6:8]}T00:00:01") for x in obs["Event"]]
mjds = []
labs = []
bins = []
for year in range(2018, 2022):
for k, month in enumerate([1, 4, 7, 10]):
t = Time(f"{year}-{month}-01T00:00:00.01", format='isot', scale='utc').mjd
bins.append(t)
if (k - 1) % 2 > 0:
mjds.append(t)
labs.append(["Jan", "July"][int(k/2)] + f" {year}")
t_0 = Time(f"2018-04-01T00:00:00.01", format='isot', scale='utc').mjd
v1_t = Time(f"2019-06-17T00:00:00.01", format='isot', scale='utc').mjd
t_now = Time(f"2021-07-01T00:00:00.01", format='isot', scale='utc').mjd
t_bran_cut = Time(f"2020-02-01T00:00:00.01", format='isot', scale='utc').mjd
alerts_v1 = [x.mjd for x in dates if x.mjd < v1_t]
alerts_v2 = [x.mjd for x in dates if x.mjd > v1_t]
print(f'{len(alerts_v1)} V1 alerts, {len(alerts_v2)} V2 alerts')
mod = 7.
v1_rate = mod * float(len(alerts_v1))/(v1_t - t_0)
v2_rate = mod * float(len(alerts_v2))/(t_now - v1_t)
labels = []
for (name, rate) in [("HESE/EHE", v1_rate), ("Gold/Bronze", v2_rate)]:
labels.append(f'{name} ({rate:.2f} per week)')
plt.xticks(mjds, labs, rotation=80)
plt.locator_params(axis="y", nbins=6)
plt.hist([alerts_v1, alerts_v2], bins=bins[:-1], stacked=True, label=labels, cumulative=True)
plt.axvline(v1_t, linestyle=":", color="k")
# plt.axvline(t_bran_cut, linestyle="--", color="k")
plt.tick_params(axis='both', which='major', labelsize=big_fontsize)
plt.legend(fontsize=big_fontsize, loc="upper left")
sns.despine()
# plt.ylim(0., 12.)
plt.ylabel(r"ZTF $\nu$ follow-up campaigns", fontsize=big_fontsize)
plt.tight_layout()
filename = "ztf_cdf.pdf"
output_path = os.path.join(output_folder, filename)
plt.savefig(os.path.join(plot_dir, filename))
plt.savefig(output_path, bbox_inches='tight', pad_inches=0) | 1 V1 alerts, 23 V2 alerts
| MIT | notebooks/stats_alerts.ipynb | robertdstein/nuztfpaper |
Exploratory Data Analysis of Stringer Dataset @authors: Simone Azeglio, Chetan Dhulipalla , Khalid Saifullah Part of the code here has been taken from [Neuromatch Academy's Computational Neuroscience Course](https://compneuro.neuromatch.io/projects/neurons/README.html), and specifically from [this notebook](https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/projects/neurons/load_stringer_spontaneous.ipynb) to do list1. custom normalization: dividing by mean value per neuron1a. downsampling: convolve then downsample by 52. training validation split: withhold last 20 percent of time series for testing3. RNN for each layer: a way to capture the dynamics inside each layer instead of capturing extra dynamics from inter-layer interactions. it will be OK to compare the different RNNs. maintain same neuron count in each layer to reduce potential bias 4. layer weight regularization: L2 5. early stopping , dropout? Loading of Stringer spontaneous data | #@title Data retrieval
import os, requests
fname = "stringer_spontaneous.npy"
url = "https://osf.io/dpqaj/download"
if not os.path.isfile(fname):
try:
r = requests.get(url)
except requests.ConnectionError:
print("!!! Failed to download data !!!")
else:
if r.status_code != requests.codes.ok:
print("!!! Failed to download data !!!")
else:
with open(fname, "wb") as fid:
fid.write(r.content)
#@title Import matplotlib and set defaults
from matplotlib import rcParams
from matplotlib import pyplot as plt
rcParams['figure.figsize'] = [20, 4]
rcParams['font.size'] =15
rcParams['axes.spines.top'] = False
rcParams['axes.spines.right'] = False
rcParams['figure.autolayout'] = True | _____no_output_____ | MIT | src/NeuronBlock.ipynb | sazio/NMAs |
Exploratory Data Analysis (EDA) | #@title Data loading
import numpy as np
dat = np.load('stringer_spontaneous.npy', allow_pickle=True).item()
print(dat.keys())
# functions
def moving_avg(array, factor = 5):
"""Reducing the number of compontents by averaging of N = factor
subsequent elements of array"""
zeros_ = np.zeros((array.shape[0], 2))
array = np.hstack((array, zeros_))
array = np.reshape(array, (array.shape[0], int(array.shape[1]/factor), factor))
array = np.mean(array, axis = 2)
return array | _____no_output_____ | MIT | src/NeuronBlock.ipynb | sazio/NMAs |
Extracting Data for RNN (or LFADS)The first problem to address is that for each layer we don't have the exact same number of neurons. We'd like to have a single RNN encoding all the different layers activities, to make it easier we can take the number of neurons ($N_{neurons} = 1131$ of the least represented class (layer) and level out each remaining class. | # Extract labels from z - coordinate
from sklearn import preprocessing
x, y, z = dat['xyz']
le = preprocessing.LabelEncoder()
labels = le.fit_transform(z)
### least represented class (layer with less neurons)
n_samples = np.histogram(labels, bins=9)[0][-1]
resp = np.array(dat['sresp'])
xyz = np.array(dat['xyz'])
print(resp.shape, xyz[0].shape)
# Extracting x,y blocks
n_blocks = 9
x_range, y_range, _ = np.ptp(dat['xyz'], axis = 1)
x_block_starts = np.arange(min(x), max(x), x_range // (n_blocks ** 0.5))
y_block_starts = np.arange(min(y), max(y)+0.01, y_range // (n_blocks ** 0.5))
data_blocks = list()
labels = np.zeros(resp.shape[0])
counter = 0
for i in range(int(n_blocks ** 0.5)):
for k in range(int(n_blocks ** 0.5)):
tempx, = np.where((x >= x_block_starts[i]) & (x <= x_block_starts[i+1]))
tempy, = np.where((y >= y_block_starts[k]) & (y <= y_block_starts[k+1]))
idx = np.intersect1d(tempx, tempy)
labels[idx] = counter
counter += 1
data_blocks.append(resp[idx])
print(labels.shape)
labels = np.array(labels)
print(y_block_starts)
print(x_block_starts)
type(iunq[0])
print(iunq)
print(labels)
labels = np.int64(labels)
print(type(iunq[0]), type(labels[0]))
from matplotlib import cm
zunq, iunq = np.unique(z, return_inverse=True)
xc = np.linspace(0.0, 1.0, len(zunq))
cmap = cm.get_cmap('jet')(xc)
fig = plt.figure(figsize=(6,6))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(x[::-1],y[::-1],z[::-1], 'o', s = 4, c = cmap[labels])
ax.set(xlabel='horizontal(um)', ylabel = 'vertical(um)', zlabel='depth (um)');
for i in data_blocks:
print(i.shape)
print(x_range, y_range)
print(y_block_starts)
print(x_block_starts)
x_block_starts.shape
print(int(n_blocks ** 0.5))
for i in range(3):
print(i)
print(y_block_starts.shape)
1314 * 9 # roughly 200 neurons are lost at the boundary
### Data for LFADS / RNN
import pandas as pd
dataSet = pd.DataFrame(dat["sresp"])
dataSet["label"] = labels
# it can be done in one loop ...
data_ = []
for i in range(0, 9):
data_.append(dataSet[dataSet["label"] == i].sample(n = n_samples).iloc[:,:-1])
dataRNN = np.zeros((n_samples*9, dataSet.shape[1]-1))
for i in range(0,9):
dataRNN[n_samples*i:n_samples*(i+1), :] = data_[i]
## shuffling for training purposes
#np.random.shuffle(dataRNN)
plt.plot(dataRNN[0, :600])
print(dataRNN.shape)
#@title PCA
from sklearn.decomposition import PCA
#pca developed seperately for blocks and layers. note: the number of neurons and timepoints are constrained
block_pca = PCA(n_components = 500)
block_pca = block_pca.fit(data_blocks[0,:1131,:600].T)
compress_blocks = block_pca.transform(data_blocks[0,:1131,:1200].T)
print(compress_blocks.shape)
layer_pca = PCA(n_components = 500)
layer_pca = layer_pca.fit(dataRNN[:1131,:600].T)
compress_layers = layer_pca.transform(dataRNN[:1131,:1200].T)
print(compress_layers.shape)
var_block = np.cumsum(pca.explained_variance_ratio_)
var_layer = np.cumsum(pca2.explained_variance_ratio_)
plt.plot(var_block)
plt.title('var exp for blocks')
plt.figure()
plt.plot(var_layer)
plt.title('var exp for layers')
print(var_block[75], var_layer[75])
unshuffled = np.array(data_)
#@title Convolutions code
# convolution moving average
# kernel_length = 50
# averaging_kernel = np.ones(kernel_length) / kernel_length
# dataRNN.shape
# avgd_dataRNN = list()
# for neuron in dataRNN:
# avgd_dataRNN.append(np.convolve(neuron, averaging_kernel))
# avg_dataRNN = np.array(avgd_dataRNN)
# print(avg_dataRNN.shape)
# @title Z Score Code
# from scipy.stats import zscore
# neuron = 500
# scaled_all = zscore(avg_dataRNN)
# scaled_per_neuron = zscore(avg_dataRNN[neuron, :])
# scaled_per_layer = list()
# for layer in unshuffled:
# scaled_per_layer.append(zscore(layer))
# scaled_per_layer = np.array(scaled_per_layer)
# plt.plot(avg_dataRNN[neuron, :])
# plt.plot(avg_dataRNN[2500, :])
# plt.figure()
# plt.plot(dataRNN[neuron, :])
# plt.figure()
# plt.plot(scaled_all[neuron, :])
# plt.plot(scaled_per_neuron)
# plt.figure()
# plt.plot(scaled_per_layer[0,neuron,:])
# custom normalization
normed_dataRNN = list()
for neuron in dataRNN:
normed_dataRNN.append(neuron / neuron.mean())
normed_dataRNN = np.array(normed_dataRNN)
# downsampling and averaging
avgd_normed_dataRNN = moving_avg(normed_dataRNN, factor=5) | _____no_output_____ | MIT | src/NeuronBlock.ipynb | sazio/NMAs |
issue: does the individual scaling by layer introduce bias that may artificially increase performance of the network? Data Loader | import torch
import torch.nn as nn
import torch.nn.functional as F
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# set the seed
np.random.seed(42)
# number of neurons
NN = dataRNN.shape[0]
# let's use 270 latent components
ncomp = 10
# swapping the axes to maintain consistency with seq2seq notebook in the following code - the network takes all the neurons at a time step as input, not just one neuron
avgd_normed_dataRNN = np.swapaxes(avgd_normed_dataRNN, 0, 1)
avgd_normed_dataRNN.shape
frac = 5/6
#x1 = torch.from_numpy(dataRNN[:,:int(frac*dataRNN.shape[1])]).to(device).float().unsqueeze(0)
#x2 = torch.from_numpy(dataRNN[:,int(frac*dataRNN.shape[1]):]).to(device).float().unsqueeze(0)
x1 = torch.from_numpy(avgd_normed_dataRNN[:,:50]).to(device).float().unsqueeze(0)
x2 = torch.from_numpy(avgd_normed_dataRNN[:,:50]).to(device).float().unsqueeze(0)
NN1 = x1.shape[-1]
NN2 = x2.shape[-1]
class Net(nn.Module):
def __init__(self, ncomp, NN1, NN2, bidi=True):
super(Net, self).__init__()
# play with some of the options in the RNN!
self.rnn = nn.RNN(NN1, ncomp, num_layers = 1, dropout = 0,
bidirectional = bidi, nonlinearity = 'tanh')
self.fc = nn.Linear(ncomp, NN2)
def forward(self, x):
y = self.rnn(x)[0]
if self.rnn.bidirectional:
# if the rnn is bidirectional, it concatenates the activations from the forward and backward pass
# we want to add them instead, so as to enforce the latents to match between the forward and backward pass
q = (y[:, :, :ncomp] + y[:, :, ncomp:])/2
else:
q = y
# the softplus function is just like a relu but it's smoothed out so we can't predict 0
# if we predict 0 and there was a spike, that's an instant Inf in the Poisson log-likelihood which leads to failure
#z = F.softplus(self.fc(q), 10)
z = self.fc(q)
return z, q
# we initialize the neural network
net = Net(ncomp, NN1, NN2, bidi = True).to(device)
# special thing: we initialize the biases of the last layer in the neural network
# we set them as the mean firing rates of the neurons.
# this should make the initial predictions close to the mean, because the latents don't contribute much
net.fc.bias.data[:] = x1.mean(axis = (1,2))
# we set up the optimizer. Adjust the learning rate if the training is slow or if it explodes.
optimizer = torch.optim.Adam(net.parameters(), lr=.02)
# forward check
# net(x1) | _____no_output_____ | MIT | src/NeuronBlock.ipynb | sazio/NMAs |
Training | from tqdm.notebook import tqdm
# you can keep re-running this cell if you think the cost might decrease further
cost = nn.MSELoss()
niter = 100000
for k in tqdm(range(niter)):
# the network outputs the single-neuron prediction and the latents
z, y = net(x1)
# our cost
loss = cost(z, x2)
# train the network as usual
loss.backward()
optimizer.step()
optimizer.zero_grad()
if k % 250 == 0:
print(f' iteration {k}, cost {loss.item():.4f}')
test = net()
| _____no_output_____ | MIT | src/NeuronBlock.ipynb | sazio/NMAs |
Import CSV log | data = readfile("ver3.txt")
## normal discogan
dloss = []
gloss = []
time= []
epouch = []
for i in range(len(data)):
try:
t = data[i][data[i].find('time: ')+5: data[i].find(', [d_loss:') - len(data[i])].replace(" ","")
if len(t)<3: continue
time.append(t)
glossA = (float(data[i][data[i].find('g_loss: ')+7:len(data[i])-2].replace(" ","")))
dlossA = float(data[i][data[i].find('d_loss: ')+7: data[i].find('g_loss:') - len(data[i])-2].replace(" ","") )
if(dlossA<5):
dloss.append(100-dlossA)
else: dloss.append(95)
if (glossA<5): gloss.append(100-glossA)
else: gloss.append(95)
epouch.append(int(data[i][1:data[i].find(']')]))
except:
continue
# TO rapidmine
df = pd.DataFrame({'time':time,'epouch':epouch,'dis_loss':dloss,'gen_loss':gloss })
df.to_csv('gen2.csv', index=False)
x_max = max(gloss2)
x_min = min(gloss2)
x_maxmin = x_max - x_min
x_norm = []
x2_norm=[]
#normalize
for C in range(len(gloss)):
x2_norm.append((gloss[C]-x_min)/x_maxmin)
df = pd.DataFrame(x_norm)
df2 = pd.DataFrame(x2_norm)
plt.rcParams["figure.figsize"] = [16,9]
plt.plot(df,'r', alpha=0.3)
plt.plot(df2,'g', alpha=0.3)
plt.grid()
plt
plt.show()
df = pd.DataFrame(dloss)
plt.rcParams["figure.figsize"] = [16,9]
plt.plot(df,'r')
plt.grid()
plt
plt.show()
plt.hist(df)
plt.grid()
plt.show()
df.hist(figsize=(10,10))
plt.show() | _____no_output_____ | MIT | MylogReader.ipynb | Telexine/colorizeSketch |
L'objectif de ce notebook sera de réaliser une ébauche de solution ETL en python | #Import des packages necessaires à la réalisation du projet
import pyodbc
import pyspark
import pandas as pd
import pandasql as ps
import sqlalchemy
#creation des variables depuis le fichier de configuration qui doit être placé dans le même dossier que le notebook
from configparser import ConfigParser
#recuperation de la configuration de la connexion stg_babilou pour la donnee procare
config = ConfigParser()
config.read('config.ini')
database_procare = config['procare']['database']
sqlsUrl = config['procare']['host']
username = config['procare']['username']
password = config['procare']['password']
port = config['procare']['port']
#recuperation de la configuration de la connexion stg_babilou pour la donnee date
config = ConfigParser()
config.read('config.ini')
database_date = config['date']['database']
sqlsUrl = config['date']['host']
username = config['date']['username']
password = config['date']['password']
port = config['date']['port'] | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Utilisation de pyodbc pour la connexion à la base de donnée SQL SERVER | #String de connexion procare
connection_string_procare ='DRIVER={SQL Server Native Client 11.0};SERVER='+sqlsUrl+';DATABASE='+database_procare+';UID='+username+';PWD='+password+';Encrypt=yes;TrustServerCertificate=no;Connection Timeout=30;Authentication=ActiveDirectoryIntegrated'
#String de connexion date
connection_string_date ='DRIVER={SQL Server Native Client 11.0};SERVER='+sqlsUrl+';DATABASE='+database_date+';UID='+username+';PWD='+password+';Encrypt=yes;TrustServerCertificate=no;Connection Timeout=30;Authentication=ActiveDirectoryIntegrated'
#connexion a la base STG_BABILOU pour recuperer les donnees procare
cnxn_procare:pyodbc.Connection= pyodbc.connect(connection_string_procare)
#connexion a la base DWH_BABILOU pour recuperer les donnees de dates
cnxn_date:pyodbc.Connection= pyodbc.connect(connection_string_date)
#Requetes pour interroger les bases DW_BABILOU et STG_BABILOU
#select_query= 'Select * from STG_PROCARE_AR_SCHEDULE'
select_procare_query = '''
WITH SCHOOL AS
(SELECT
SCO.[SchoolID]
,SCO.[Code]
,SCO.[SchoolName]
,SCO.[Database]
,CHI.[ChildSchoolID]
,CHI.PersonID
FROM [dbo].[STG_PROCARE_G_SCHOOLS] SCO
INNER JOIN STG_PROCARE_AR_CHILDSCHOOL CHI
ON (SCO.SchoolID = CHI.SchoolID))
SELECT
SCH.ScheduleKeyID
,SCO.PersonID
,SCO.SchoolID
,SCH.[Database]
,(SCD.OutMinute-SCD.InMinute)/60.00 AS HoursWorked
,SCH.StartAppliesTo
,SCH.EndAppliesTo
,SCD.DayNumber
,ENR.StartDate
,ENR.EndDate
,SCH.ScheduleID
,GETDATE() AS ExtractDate
FROM STG_PROCARE_AR_SCHEDULE SCH
INNER JOIN STG_PROCARE_AR_SCHEDULEDETAIL SCD
ON (SCH.ScheduleKeyID=SCD.ScheduleKeyID
and SCH.[Database]=SCD.[Database])
INNER JOIN STG_PROCARE_G_TYPESTABLE TYP
ON (SCH.ChildSchoolID=TYP.TypeID
and SCH.[Database]=TYP.[Database])
INNER JOIN STG_PROCARE_AR_ENROLLMENT ENR
ON (SCH.ChildSchoolID=ENR.ChildSchoolID
and SCH.[Database]=ENR.[Database])
/*INNER JOIN STG_PROCARE_G_SCHOOLS SCO
ON (SCH.ChildSchoolID=SCO.SchoolID
and SCH.[Database]=SCO.[Database]) */
INNER JOIN SCHOOL SCO
ON (SCH.ChildSchoolID=SCO.ChildSchoolID)
'''
select_date_query = '''
SELECT
[DT_DATE]
,[DAY_OF_WEEK]
,[LB_DAY_NAME]
FROM [dbo].[D_TIME]
'''
# Creation du dataframe procare
data_procare = pd.read_sql(select_procare_query,cnxn_procare)
# Creation du dataframe date
data_date = pd.read_sql(select_date_query,cnxn_date)
#fermeture des connexions
cnxn_procare.close()
cnxn_date.close() | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Utilisation des packages pandas et pandasql pour respecter les règles de gestion * On cree la query qui sert à faire la jointure entre les deux dataframes precedemment crees* On fait une jointure glissante sur les dates de début et de fin de contrats* Le filtre nous permet de récupérer les données cohérentes (le premier jour de la semaine est forcément lundi) | psql_join_query ='''
SELECT * FROM data_procare pro
INNER JOIN data_date DAT
ON DAT.DT_DATE BETWEEN PRO.StartAppliesTo AND PRO.EndAppliesTo
WHERE (PRO.DayNumber = 1 AND DAT.LB_DAY_NAME ='LUNDI')
OR (PRO.DayNumber = 2 AND DAT.LB_DAY_NAME ='MARDI')
OR (PRO.DayNumber = 3 AND DAT.LB_DAY_NAME ='MERCREDI')
OR (PRO.DayNumber = 4 AND DAT.LB_DAY_NAME ='JEUDI')
OR (PRO.DayNumber = 5 AND DAT.LB_DAY_NAME ='VENDREDI')
OR (PRO.DayNumber = 6 AND DAT.LB_DAY_NAME ='SAMEDI')
OR (PRO.DayNumber = 7 AND DAT.LB_DAY_NAME ='DIMANCHE')
;
''' | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
* Execution de la query via pandasql pui affichage des 5 premières lignes | df_psql = ps.sqldf(psql_join_query)
df_psql.head(5) | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
On recupere les donnees etp dans un dataframe pour les rajouter dans le dataframe final | df_etp = pd.read_csv('etp.csv',sep=';') | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
* Conversion des donnees en decimal | df_etp_int = df_etp.astype('float64')
sql_etp_query = '''
SELECT
PRO.*,
ETP.[Correspondance ETP]
FROM df_psql PRO
INNER JOIN df_etp ETP
ON PRO.HoursWorked BETWEEN ETP.MinDailyHour AND ETP.MaxDailyHour
'''
df_join_etp = ps.sqldf(sql_etp_query)
df_join_etp.head(2) | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Creation de la table dans SQL Server via Pyodbc | #on établit une nouvelle connexion avec la base DW_BABILOU
connexion_dwh = pyodbc.connect(connection_string_date)
dwh_crusor = connexion_dwh.cursor()
#Table déjà installée
# dwh_crusor.execute('''
# CREATE TABLE [dbo].PROCARE_ETP(
# Jour DATE,
# DayNumber INT,
# [Database] VARCHAR(40),
# PersonID INT,
# SchoolID INT,
# ETP DECIMAL (3,2),
# ExtractDate DATE
# );
# ''')
#connexion_dwh.commit() #sert à confirmer les changements dans la base | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
on efface les données de la table dont l'extractdate est à la date du jour | dwh_crusor.execute('''DELETE FROM PROCARE_ETP WHERE ExtractDate = CONVERT (date, GETDATE())''')
connexion_dwh.commit() | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Insertion des donnees dans la table PROCATE_ETP nouvellement creee | # On créé un nouveau DataFrame à l'image de la table finale
df_insert_procareETP = ps.sqldf('''SELECT
date(DT_DATE) AS jour,
DayNumber,
[Database],
PersonID,
SchoolID,
[Correspondance ETP] AS ETP,
date(ExtractDate) as ExtractDate
FROM df_join_etp''')
df_insert_procareETP.head(2) | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
* quelques tests de connexion infructeux | for index,row in df_insert_procareETP.iterrows():
dwh_crusor.execute('''INSERT INTO PROCATE_ETP(
[jour],
[DayNumber],
[Database],
PersonID,
SchoolID,
ETP,
ExtractDate)
values (?,?,?,?,?,?,?)''',
row['jour'],
row['DayNumber'],
row['Database'],
row['PersonID'],
row['SchoolID'],
row['ETP'],
row['ExtractDate'])
df_insert_procareETP.to_sql('PROCARE_ETP',connexion_dwh,)
dwh_crusor.execute('''
INSERT INTO [dbo].PROCATE_ETP(
jour,
DayNumber,
[Database],
PersonID,
SchoolID,
ETP,
ExtractDate)
SELECT
CAST(DT_DATE AS DATE) AS JOUR,
DayNumber,
[Database],
PersonID,
SchoolID,
[Correspondance ETP] AS ETP,
ExtractDate
FROM df_join_etp
''')
connexion_dwh.commit() #sert à enregistrer les modifications dans la base | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Script d'insertion dans la table, pandas.to_sql et sqlachemy * On cree les informations de connexion à la table en utilisant le connexion string utilise precedemment dans pyodbc | from sqlalchemy.engine import URL,create_engine
connection_url = URL.create("mssql+pyodbc", query={"odbc_connect": connection_string_date})
engine = create_engine(connection_url,fast_executemany=True)
#https://docs.sqlalchemy.org/en/14/dialects/mssql.html#module-sqlalchemy.dialects.mssql.pyodbc
#https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.to_sql.html
#df_insert_procareETP_reduced = df_insert_procareETP.head(500)
df_insert_procareETP.to_sql('PROCARE_ETP',engine,if_exists='append',index=False,chunksize=1000)
#vérification de l'insertion des lignes
pd.read_sql('PROCARE_ETP',engine) | _____no_output_____ | CNRI-Python | Poc_ETL.ipynb | charlesLavignasse/POC-Python-ETL |
Classfication report* A Classification report is used to measure the quality of predictions from a classification algorithm. ... The report shows the main classification metrics precision, recall and f1-score on a per-class basis. The metrics are calculated by using true and false positives, true and false negatives.```sklearn.metrics.classification_report()``` | import numpy as np
from sklearn.metrics import classification_report
from matplotlib import pyplot as plt
y_true = np.array([1., 0., 1, 1, 0, 0, 1])
y_pred = np.array([1., 1., 1., 0., 0. ,1, 0]) | _____no_output_____ | MIT | 04_Evaluation_Methods/06_Classification_Report/.ipynb_checkpoints/Classification_Report-checkpoint.ipynb | CrispenGari/keras-api |
Using `scikit-learn` to generate the classification report for our predictions | labels = np.array([0., 1])
report = classification_report(y_true, y_pred, labels=labels)
print(report) | precision recall f1-score support
0.0 0.33 0.33 0.33 3
1.0 0.50 0.50 0.50 4
accuracy 0.43 7
macro avg 0.42 0.42 0.42 7
weighted avg 0.43 0.43 0.43 7
| MIT | 04_Evaluation_Methods/06_Classification_Report/.ipynb_checkpoints/Classification_Report-checkpoint.ipynb | CrispenGari/keras-api |
Ploting the ``classification_report`` | import numpy as np
import seaborn as sns
from sklearn.metrics import classification_report
import pandas as pd
report = classification_report(y_true, y_pred, labels=labels, output_dict=True)
sns.heatmap(pd.DataFrame(report).iloc[:-1, :].T, annot=True)
plt.title("Classification Report")
plt.show() | _____no_output_____ | MIT | 04_Evaluation_Methods/06_Classification_Report/.ipynb_checkpoints/Classification_Report-checkpoint.ipynb | CrispenGari/keras-api |
Синхронизация потоков Спасибо Сове Глебу, Голяр Димитрису и Николаю Васильеву за участие в написании текста Сегодня в программе:* Мьютексы MUTEX ~ MUTual EXclusion* Spinlock'и и атомики [Атомики в С на cppreference](https://ru.cppreference.com/w/c/atomic) Atomic в C и как с этим жить (раздел от Николая Васильева) Про compare_exchange_weak vs compare_exchange_strong https://stackoverflow.com/questions/4944771/stdatomic-compare-exchange-weak-vs-compare-exchange-strongThe weak compare-and-exchange operations may fail spuriously, that is, return false while leaving the contents of memory pointed to by expected before the operation is the same that same as that of the object and the same as that of expected after the operation. [ Note: This spurious failure enables implementation of compare-and-exchange on a broader class of machines, e.g., loadlocked store-conditional machines. A consequence of spurious failure is that nearly all uses of weak compare-and-exchange will be in a loop. * Condition variable (aka условные переменные)* Пример thread-safe очереди Комментарии к ДЗ[Ридинг Яковлева](https://github.com/victor-yacovlev/mipt-diht-caos/tree/master/practice/mutex-condvar-atomic) Mutex | %%cpp mutex.c
%# Санитайзер отслеживает небезопасный доступ
%# к одному и тому же участку в памяти из разных потоков
%# (а так же другие небезопасные вещи).
%# В таких задачах советую всегда использовать
%run gcc -fsanitize=thread mutex.c -lpthread -o mutex.exe # вспоминаем про санитайзеры
%run ./mutex.exe
#define _GNU_SOURCE
#include <stdio.h>
#include <stdlib.h>
#include <unistd.h>
#include <sys/types.h>
#include <sys/syscall.h>
#include <sys/time.h>
#include <pthread.h>
#include <stdatomic.h>
const char* log_prefix(const char* func, int line) {
struct timespec spec; clock_gettime(CLOCK_REALTIME, &spec); long long current_msec = spec.tv_sec * 1000L + spec.tv_nsec / 1000000;
static _Atomic long long start_msec_storage = -1; long long start_msec = -1; if (atomic_compare_exchange_strong(&start_msec_storage, &start_msec, current_msec)) start_msec = current_msec;
long long delta_msec = current_msec - start_msec;
static __thread char prefix[100]; sprintf(prefix, "%lld.%03lld %13s():%d [tid=%ld]", delta_msec / 1000, delta_msec % 1000, func, line, syscall(__NR_gettid));
return prefix;
}
#define log_printf_impl(fmt, ...) { time_t t = time(0); dprintf(2, "%s: " fmt "%s", log_prefix(__FUNCTION__, __LINE__), __VA_ARGS__); }
#define log_printf(...) log_printf_impl(__VA_ARGS__, "")
// thread-aware assert
#define ta_assert(stmt) if (stmt) {} else { log_printf("'" #stmt "' failed"); exit(EXIT_FAILURE); }
typedef enum {
VALID_STATE = 0,
INVALID_STATE = 1
} state_t;
// Инициализируем мьютекс
pthread_mutex_t mutex = PTHREAD_MUTEX_INITIALIZER; // protects: state
state_t current_state = VALID_STATE;
void thread_safe_func() {
// all function is critical section, protected by mutex
pthread_mutex_lock(&mutex); // try comment lock&unlock out and look at result
ta_assert(current_state == VALID_STATE);
current_state = INVALID_STATE; // do some work with state.
sched_yield();
current_state = VALID_STATE;
pthread_mutex_unlock(&mutex);
}
// Возвращаемое значение потока (~код возврата процесса) -- любое машинное слово.
static void* thread_func(void* arg)
{
int i = (char*)arg - (char*)NULL;
log_printf(" Thread %d started\n", i);
for (int j = 0; j < 10000; ++j) {
thread_safe_func();
}
log_printf(" Thread %d finished\n", i);
return NULL;
}
int main()
{
log_printf("Main func started\n");
const int threads_count = 2;
pthread_t threads[threads_count];
for (int i = 0; i < threads_count; ++i) {
log_printf("Creating thread %d\n", i);
ta_assert(pthread_create(&threads[i], NULL, thread_func, (char*)NULL + i) == 0);
}
for (int i = 0; i < threads_count; ++i) {
ta_assert(pthread_join(threads[i], NULL) == 0);
log_printf("Thread %d joined\n", i);
}
log_printf("Main func finished\n");
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Spinlock[spinlock в стандартной библиотеке](https://linux.die.net/man/3/pthread_spin_init) | %%cpp spinlock.c
%run gcc -fsanitize=thread -std=c11 spinlock.c -lpthread -o spinlock.exe
%run ./spinlock.exe
#define _GNU_SOURCE
#include <stdio.h>
#include <stdlib.h>
#include <unistd.h>
#include <sys/syscall.h>
#include <sys/types.h>
#include <sys/time.h>
#include <pthread.h>
#include <stdatomic.h> //! Этот заголовочный файл плохо гуглится
const char* log_prefix(const char* func, int line) {
struct timespec spec; clock_gettime(CLOCK_REALTIME, &spec); long long current_msec = spec.tv_sec * 1000L + spec.tv_nsec / 1000000;
static _Atomic long long start_msec_storage = -1; long long start_msec = -1; if (atomic_compare_exchange_strong(&start_msec_storage, &start_msec, current_msec)) start_msec = current_msec;
long long delta_msec = current_msec - start_msec;
static __thread char prefix[100]; sprintf(prefix, "%lld.%03lld %13s():%d [tid=%ld]", delta_msec / 1000, delta_msec % 1000, func, line, syscall(__NR_gettid));
return prefix;
}
#define log_printf_impl(fmt, ...) { time_t t = time(0); dprintf(2, "%s: " fmt "%s", log_prefix(__FUNCTION__, __LINE__), __VA_ARGS__); }
#define log_printf(...) log_printf_impl(__VA_ARGS__, "")
// thread-aware assert
#define ta_assert(stmt) if (stmt) {} else { log_printf("'" #stmt "' failed"); exit(EXIT_FAILURE); }
typedef enum {
VALID_STATE = 0,
INVALID_STATE = 1
} state_t;
_Atomic int lock = 0; // protects state
state_t current_state = VALID_STATE;
void sl_lock(_Atomic int* lock) {
int expected = 0;
// weak отличается от strong тем, что может выдавать иногда ложный false. Но он быстрее работает.
// atomic_compare_exchange_weak can change `expected`!
while (!atomic_compare_exchange_weak(lock, &expected, 1)) {
expected = 0;
}
}
void sl_unlock(_Atomic int* lock) {
atomic_fetch_sub(lock, 1);
}
// По сути та же функция, что и в предыдущем примере, но ипользуется spinlock вместо mutex
void thread_safe_func() {
// all function is critical section, protected by mutex
sl_lock(&lock); // try comment lock&unlock out and look at result
ta_assert(current_state == VALID_STATE);
current_state = INVALID_STATE; // do some work with state.
sched_yield(); // increase probability of fail of incorrect lock realisation
current_state = VALID_STATE;
sl_unlock(&lock);
}
// Возвращаемое значение потока (~код возврата процесса) -- любое машинное слово.
static void* thread_func(void* arg)
{
int i = (char*)arg - (char*)NULL;
log_printf(" Thread %d started\n", i);
for (int j = 0; j < 10000; ++j) {
thread_safe_func();
}
log_printf(" Thread %d finished\n", i);
return NULL;
}
int main()
{
log_printf("Main func started\n");
const int threads_count = 2;
pthread_t threads[threads_count];
for (int i = 0; i < threads_count; ++i) {
log_printf("Creating thread %d\n", i);
ta_assert(pthread_create(&threads[i], NULL, thread_func, (char*)NULL + i) == 0);
}
for (int i = 0; i < threads_count; ++i) {
ta_assert(pthread_join(threads[i], NULL) == 0);
log_printf("Thread %d joined\n", i);
}
log_printf("Main func finished\n");
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Condition variable | %%cpp condvar.c
%run gcc -fsanitize=thread condvar.c -lpthread -o condvar.exe
%run ./condvar.exe > out.txt
//%run cat out.txt
#include <stdio.h>
#include <stdlib.h>
#include <unistd.h>
#include <sys/types.h>
#include <sys/syscall.h>
#include <sys/time.h>
#include <pthread.h>
#include <stdatomic.h>
const char* log_prefix(const char* func, int line) {
struct timespec spec; clock_gettime(CLOCK_REALTIME, &spec); long long current_msec = spec.tv_sec * 1000L + spec.tv_nsec / 1000000;
static _Atomic long long start_msec_storage = -1; long long start_msec = -1; if (atomic_compare_exchange_strong(&start_msec_storage, &start_msec, current_msec)) start_msec = current_msec;
long long delta_msec = current_msec - start_msec;
static __thread char prefix[100]; sprintf(prefix, "%lld.%03lld %13s():%d [tid=%ld]", delta_msec / 1000, delta_msec % 1000, func, line, syscall(__NR_gettid));
return prefix;
}
#define log_printf_impl(fmt, ...) { time_t t = time(0); dprintf(2, "%s: " fmt "%s", log_prefix(__FUNCTION__, __LINE__), __VA_ARGS__); }
#define log_printf(...) log_printf_impl(__VA_ARGS__, "")
// thread-aware assert
#define ta_assert(stmt) if (stmt) {} else { log_printf("'" #stmt "' failed"); exit(EXIT_FAILURE); }
typedef struct {
// рекомендую порядок записи переменных:
pthread_mutex_t mutex; // мьютекс
pthread_cond_t condvar; // переменная условия (если нужна)
int value;
} promise_t;
void promise_init(promise_t* promise) {
pthread_mutex_init(&promise->mutex, NULL);
pthread_cond_init(&promise->condvar, NULL);
promise->value = -1;
}
void promise_set(promise_t* promise, int value) {
pthread_mutex_lock(&promise->mutex); // try comment lock&unlock out and look at result
promise->value = value;
pthread_mutex_unlock(&promise->mutex);
pthread_cond_signal(&promise->condvar); // notify if there was nothing and now will be elements
}
int promise_get(promise_t* promise) {
pthread_mutex_lock(&promise->mutex); // try comment lock&unlock out and look at result
while (promise->value == -1) {
// Ждем какие-либо данные, если их нет, то спим.
// идейно convar внутри себя разблокирует mutex, чтобы другой поток мог положить в стейт то, что мы ждем
pthread_cond_wait(&promise->condvar, &promise->mutex);
// после завершения wait мьютекс снова заблокирован
}
int value = promise->value;
pthread_mutex_unlock(&promise->mutex);
return value;
}
promise_t promise_1, promise_2;
static void* thread_A_func(void* arg) {
log_printf("Func A started\n");
promise_set(&promise_1, 42);
log_printf("Func A set promise_1 with 42\n");
int value_2 = promise_get(&promise_2);
log_printf("Func A get promise_2 value = %d\n", value_2);
return NULL;
}
static void* thread_B_func(void* arg) {
log_printf("Func B started\n");
int value_1 = promise_get(&promise_1);
log_printf("Func B get promise_1 value = %d\n", value_1);
promise_set(&promise_2, value_1 * 100);
log_printf("Func B set promise_2 with %d\n", value_1 * 100)
return NULL;
}
int main()
{
promise_init(&promise_1);
promise_init(&promise_2);
log_printf("Main func started\n");
pthread_t thread_A_id;
log_printf("Creating thread A\n");
ta_assert(pthread_create(&thread_A_id, NULL, thread_A_func, NULL) == 0);
pthread_t thread_B_id;
log_printf("Creating thread B\n");
ta_assert(pthread_create(&thread_B_id, NULL, thread_B_func, NULL) == 0);
ta_assert(pthread_join(thread_A_id, NULL) == 0);
log_printf("Thread A joined\n");
ta_assert(pthread_join(thread_B_id, NULL) == 0);
log_printf("Thread B joined\n");
log_printf("Main func finished\n");
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Способ достичь успеха без боли: все изменения данных делаем под mutex. Операции с condvar тоже делаем только под заблокированным mutex. Пример thread-safe очереди | %%cpp condvar_queue.c
%run gcc -fsanitize=thread condvar_queue.c -lpthread -o condvar_queue.exe
%run (for i in $(seq 0 100000); do echo -n "$i " ; done) | ./condvar_queue.exe > out.txt
//%run cat out.txt
#include <stdio.h>
#include <stdlib.h>
#include <unistd.h>
#include <sys/types.h>
#include <sys/syscall.h>
#include <sys/time.h>
#include <pthread.h>
#include <stdatomic.h>
const char* log_prefix(const char* func, int line) {
struct timespec spec; clock_gettime(CLOCK_REALTIME, &spec); long long current_msec = spec.tv_sec * 1000L + spec.tv_nsec / 1000000;
static _Atomic long long start_msec_storage = -1; long long start_msec = -1; if (atomic_compare_exchange_strong(&start_msec_storage, &start_msec, current_msec)) start_msec = current_msec;
long long delta_msec = current_msec - start_msec;
static __thread char prefix[100]; sprintf(prefix, "%lld.%03lld %13s():%d [tid=%ld]", delta_msec / 1000, delta_msec % 1000, func, line, syscall(__NR_gettid));
return prefix;
}
#define log_printf_impl(fmt, ...) { time_t t = time(0); dprintf(2, "%s: " fmt "%s", log_prefix(__FUNCTION__, __LINE__), __VA_ARGS__); }
#define log_printf(...) log_printf_impl(__VA_ARGS__, "")
// thread-aware assert
#define ta_assert(stmt) if (stmt) {} else { log_printf("'" #stmt "' failed"); exit(EXIT_FAILURE); }
#define queue_max_size 5
struct {
// рекомендую порядок записи переменных:
pthread_mutex_t mutex; // мьютекс
pthread_cond_t condvar; // переменная условия (если нужна)
// все переменные защищаемые мьютексом
int data[queue_max_size];
int begin; // [begin, end)
int end;
} queue;
void queue_init() {
pthread_mutex_init(&queue.mutex, NULL);
pthread_cond_init(&queue.condvar, NULL);
queue.begin = queue.end = 0;
}
void queue_push(int val) {
pthread_mutex_lock(&queue.mutex); // try comment lock&unlock out and look at result
while (queue.begin + queue_max_size == queue.end) {
pthread_cond_wait(&queue.condvar, &queue.mutex); // mutex in unlocked inside this func
}
_Bool was_empty = (queue.begin == queue.end);
queue.data[queue.end++ % queue_max_size] = val;
pthread_mutex_unlock(&queue.mutex);
if (was_empty) {
pthread_cond_signal(&queue.condvar); // notify if there was nothing and now will be elements
}
}
int queue_pop() {
pthread_mutex_lock(&queue.mutex); // try comment lock&unlock out and look at result
while (queue.begin == queue.end) {
pthread_cond_wait(&queue.condvar, &queue.mutex); // mutex in unlocked inside this func
}
if (queue.end - queue.begin == queue_max_size) {
// Не важно где внутри мьютекса посылать сигнал, так как другой поток не сможет зайти в критическую секцию, пока не завершится текущая
pthread_cond_signal(&queue.condvar); // notify if buffer was full and now will have free space
}
int val = queue.data[queue.begin++ % queue_max_size];
if (queue.begin >= queue_max_size) {
queue.begin -= queue_max_size;
queue.end -= queue_max_size;
}
pthread_mutex_unlock(&queue.mutex);
return val;
}
static void* producer_func(void* arg)
{
int val;
while (scanf("%d", &val) > 0) {
queue_push(val);
//nanosleep(&(struct timespec) {.tv_nsec = 1000000}, NULL); // 1ms
}
queue_push(-1);
return NULL;
}
static void* consumer_func(void* arg)
{
int val;
while ((val = queue_pop()) >= 0) {
printf("'%d', ", val);
}
return NULL;
}
int main()
{
queue_init();
log_printf("Main func started\n");
pthread_t producer_thread;
log_printf("Creating producer thread\n");
ta_assert(pthread_create(&producer_thread, NULL, producer_func, NULL) == 0);
pthread_t consumer_thread;
log_printf("Creating producer thread\n");
ta_assert(pthread_create(&consumer_thread, NULL, consumer_func, NULL) == 0);
ta_assert(pthread_join(producer_thread, NULL) == 0);
log_printf("Producer thread joined\n");
ta_assert(pthread_join(consumer_thread, NULL) == 0);
log_printf("Consumer thread joined\n");
log_printf("Main func finished\n");
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Atomic в C и как с этим житьВ C++ атомарные переменные реализованы через `std::atomic` в силу объектной ориентированности языка. В C же к объявлению переменной приписывается _Atomic или _Atomic(). Лучше использовать второй вариант (почему, будет ниже). Ситуация усложняется отсуствием документации. Про атомарные функции с переменными можно посмотреть в ридинге Яковлева. Пример с _Atomic | %%cpp atomic_example1.c
%run gcc -fsanitize=thread atomic_example1.c -lpthread -o atomic_example1.exe
%run ./atomic_example1.exe > out.txt
%run cat out.txt
#include <stdatomic.h>
#include <stdint.h>
#include <stdio.h>
// _Atomic навешивается на `int`
_Atomic int x;
int main(int argc, char* argv[]) {
atomic_store(&x, 1);
printf("%d\n", atomic_load(&x));
int i = 2;
// изменение не пройдет, так как x = 1, а i = 2, i станет равным x
atomic_compare_exchange_strong(&x, &i, 3);
printf("%d\n", atomic_load(&x));
// тут пройдет
atomic_compare_exchange_strong(&x, &i, 3);
printf("%d\n", atomic_load(&x));
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Казалось бы все хорошо, но давайте попробуем с указателями | %%cpp atomic_example2.c
%run gcc -fsanitize=thread atomic_example2.c -lpthread -o atomic_example2.exe
%run ./atomic_example2.exe > out.txt
%run cat out.txt
#include <stdatomic.h>
#include <stdint.h>
#include <stdio.h>
#include <stdlib.h>
// ПЛОХОЙ КОД!!!
_Atomic int* x;
int main(int argc, char* argv[]) {
int data[3] = {10, 20, 30};
int* one = data + 0;
int* two = data + 1;
int* three = data + 2;
atomic_store(&x, one);
printf("%d\n", *atomic_load(&x));
int* i = two;
// изменение не пройдет, так как x = 1, а i = 2, i станет равным x
atomic_compare_exchange_strong(&x, &i, three);
printf("%d\n", *atomic_load(&x));
i = one;
// тут пройдет
atomic_compare_exchange_strong(&x, &i, three);
printf("%d\n", *atomic_load(&x));
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Получаем ад из warning/error от компилятора(все в зависимости от компилятора и платформы: `gcc 7.4.0 Ubuntu 18.04.1` - warning, `clang 11.0.0 macOS` - error).Может появиться желание написать костыль, явно прикастовав типы: | %%cpp atomic_example3.c
%run gcc -fsanitize=thread atomic_example3.c -lpthread -o atomic_example3.exe
%run ./atomic_example3.exe > out.txt
%run cat out.txt
#include <stdatomic.h>
#include <stdint.h>
#include <stdio.h>
#include <stdlib.h>
// ПЛОХОЙ КОД!!!
_Atomic int* x;
int main(int argc, char* argv[]) {
int data[3] = {10, 20, 30};
int* one = data + 0;
int* two = data + 1;
int* three = data + 2;
atomic_store(&x, (_Atomic int*) one);
printf("%d\n", *(int*)atomic_load(&x));
int* i = two;
// изменение не пройдет, так как x = 1, а i = 2, i станет равным x
atomic_compare_exchange_strong(&x, (_Atomic int**) &i, (_Atomic int*) three);
printf("%d\n", *(int*)atomic_load(&x));
i = one;
// тут пройдет
atomic_compare_exchange_strong(&x, (_Atomic int**) &i, (_Atomic int*) three);
i = (int*) atomic_load(&x);
printf("%d\n", *(int*)atomic_load(&x));
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
Теперь gcc перестает кидать warnings (в clang до сих пор error). Но код может превратиться в ад из кастов. Но! Этот код идейно полностью некорректен.Посмотрим на `_Atomic int* x;`В данном случае это работает как `(_Atomic int)* x`, а не как `_Atomic (int*) x` что легко подумать!То есть получается неатомарный указатель на атомарный `int`. Хотя задумывалось как атомарный указатель на неатомарный `int`.Поэтому лучше использовать `_Atomic (type)`.При его использовании код становится вполне читаемым и что главное - корректным. Соответственно компилируется без проблем в gcc/clang. Как надо писать | %%cpp atomic_example4.c
%run gcc -fsanitize=thread atomic_example4.c -lpthread -o atomic_example4.exe
%run ./atomic_example4.exe > out.txt
%run cat out.txt
#include <stdatomic.h>
#include <stdint.h>
#include <stdio.h>
#include <stdlib.h>
// Теперь именно атомарный указатель. Как и должно было быть.
_Atomic (int*) x;
int main(int argc, char* argv[]) {
int data[3] = {10, 20, 30};
int* one = data + 0;
int* two = data + 1;
int* three = data + 2;
atomic_store(&x, one);
printf("%d\n", *atomic_load(&x));
int* i = two;
// изменение не пройдет, так как x = 1, а i = 2, i станет равным x
atomic_compare_exchange_strong(&x, &i, three);
printf("%d\n", *atomic_load(&x));
i = one;
// тут пройдет
atomic_compare_exchange_strong(&x, &i, three);
printf("%d\n", *atomic_load(&x));
return 0;
} | _____no_output_____ | MIT | sem20-synchronizing/synchronizing.ipynb | Disadvantaged/caos_2019-2020 |
reference for calulating quartile [here](http://web.mnstate.edu/peil/MDEV102/U4/S36/S363.html:~:text=The%20third%20quartile%2C%20denoted%20by,25%25%20lie%20above%20Q3%20) | mean(case_duration_dic.values())
# quartile calculation
import statistics
def calc_third_quartile(lis):
lis.sort()
size = len(lis)
lis_upper_half = lis[size//2:-1]
third_quartile = statistics.median(lis_upper_half)
return third_quartile
case_durations = list(case_duration_dic.values())
third_quartile = calc_third_quartile(case_durations)
third_quartile | _____no_output_____ | MIT | src/dataset_div/dataset_div_bpi_12_w.ipynb | avani17101/goal-oriented-next-best-activity-recomendation |
Filter dataset for RL model | cases_gs = []
cases_gv = []
for k,v in case_duration_dic.items():
if v <= third_quartile:
cases_gs.append(k)
else:
cases_gv.append(k)
len(cases_gs), len(cases_gv)
tot = len(cases_gs)+ len(cases_gv)
percent_gs_cases = len(cases_gs) / tot
print(percent_gs_cases)
cases_train = cases_gs
cases_test = cases_gv
df.shape, len(cases_train), len(cases_test)
data_train = df.loc[df['CaseID'].isin(cases_train)]
data_test = df.loc[df['CaseID'].isin(cases_test)]
data_train
data_test | _____no_output_____ | MIT | src/dataset_div/dataset_div_bpi_12_w.ipynb | avani17101/goal-oriented-next-best-activity-recomendation |
Analysing unique events | a = get_unique_act(data_train)
len(a)
tot = get_unique_act(df)
len(tot)
lis = []
for act in tot:
if act not in a:
lis.append(act)
lis
for act in lis:
df_sub = df[df["class"] == act]
caseid_lis = list(df_sub["CaseID"])
l = len(caseid_lis)
caseid_sel = caseid_lis[:l//2]
if len(caseid_sel) == 0:
caseid_sel = caseid_lis
r = df.loc[df['CaseID'].isin(caseid_sel)]
data_train = data_train.append(r)
data_train
len(get_unique_act(data_train)), len(get_unique_act(data_test))
len(get_unique_act(df))
env_name = "bpi_12_w"
name = env_name+'_d0'
pickle.dump(data_train, open(name+"_train_RL.pkl", "wb"))
pickle.dump(data_test, open(name+"_test_RL.pkl", "wb")) | _____no_output_____ | MIT | src/dataset_div/dataset_div_bpi_12_w.ipynb | avani17101/goal-oriented-next-best-activity-recomendation |
Implementing the Gradient Descent AlgorithmIn this lab, we'll implement the basic functions of the Gradient Descent algorithm to find the boundary in a small dataset. First, we'll start with some functions that will help us plot and visualize the data. | import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
#Some helper functions for plotting and drawing lines
def plot_points(X, y):
admitted = X[np.argwhere(y==1)]
rejected = X[np.argwhere(y==0)]
plt.scatter([s[0][0] for s in rejected], [s[0][1] for s in rejected], s = 25, color = 'blue', edgecolor = 'k')
plt.scatter([s[0][0] for s in admitted], [s[0][1] for s in admitted], s = 25, color = 'red', edgecolor = 'k')
def display(m, b, color='g--'):
plt.xlim(-0.05,1.05)
plt.ylim(-0.05,1.05)
x = np.arange(-10, 10, 0.1)
plt.plot(x, m*x+b, color) | _____no_output_____ | MIT | intro-neural-networks/gradient-descent/GradientDescent.ipynb | Abhinav2604/deep-learning-v2-pytorch |
Reading and plotting the data | data = pd.read_csv('data.csv', header=None)
X = np.array(data[[0,1]])
y = np.array(data[2])
plot_points(X,y)
plt.show() | _____no_output_____ | MIT | intro-neural-networks/gradient-descent/GradientDescent.ipynb | Abhinav2604/deep-learning-v2-pytorch |
TODO: Implementing the basic functionsHere is your turn to shine. Implement the following formulas, as explained in the text.- Sigmoid activation function$$\sigma(x) = \frac{1}{1+e^{-x}}$$- Output (prediction) formula$$\hat{y} = \sigma(w_1 x_1 + w_2 x_2 + b)$$- Error function$$Error(y, \hat{y}) = - y \log(\hat{y}) - (1-y) \log(1-\hat{y})$$- The function that updates the weights$$ w_i \longrightarrow w_i + \alpha (y - \hat{y}) x_i$$$$ b \longrightarrow b + \alpha (y - \hat{y})$$ | # Implement the following functions
# Activation (sigmoid) function
def sigmoid(x):
y=1/(1+np.exp(-x))
return y
# Output (prediction) formula
def output_formula(features, weights, bias):
y_hat=sigmoid(np.dot(features,weights)+bias)
return y_hat
# Error (log-loss) formula
def error_formula(y, output):
error=-y*np.log(output)-(1-y)*np.log(1-output)
return error
# Gradient descent step
def update_weights(x, y, weights, bias, learnrate):
y_hat=output_formula(x,weights,bias)
error=y-y_hat
weights+=learnrate*error*x
bias+=learnrate*error
return weights,bias
| _____no_output_____ | MIT | intro-neural-networks/gradient-descent/GradientDescent.ipynb | Abhinav2604/deep-learning-v2-pytorch |
Training functionThis function will help us iterate the gradient descent algorithm through all the data, for a number of epochs. It will also plot the data, and some of the boundary lines obtained as we run the algorithm. | np.random.seed(44)
epochs = 100
learnrate = 0.01
def train(features, targets, epochs, learnrate, graph_lines=False):
errors = []
n_records, n_features = features.shape
last_loss = None
weights = np.random.normal(scale=1 / n_features**.5, size=n_features)
bias = 0
for e in range(epochs):
del_w = np.zeros(weights.shape)
for x, y in zip(features, targets):
output = output_formula(x, weights, bias)
error = error_formula(y, output)
weights, bias = update_weights(x, y, weights, bias, learnrate)
# Printing out the log-loss error on the training set
out = output_formula(features, weights, bias)
loss = np.mean(error_formula(targets, out))
errors.append(loss)
if e % (epochs / 10) == 0:
print("\n========== Epoch", e,"==========")
if last_loss and last_loss < loss:
print("Train loss: ", loss, " WARNING - Loss Increasing")
else:
print("Train loss: ", loss)
last_loss = loss
predictions = out > 0.5
accuracy = np.mean(predictions == targets)
print("Accuracy: ", accuracy)
if graph_lines and e % (epochs / 100) == 0:
display(-weights[0]/weights[1], -bias/weights[1])
# Plotting the solution boundary
plt.title("Solution boundary")
display(-weights[0]/weights[1], -bias/weights[1], 'black')
# Plotting the data
plot_points(features, targets)
plt.show()
# Plotting the error
plt.title("Error Plot")
plt.xlabel('Number of epochs')
plt.ylabel('Error')
plt.plot(errors)
plt.show() | _____no_output_____ | MIT | intro-neural-networks/gradient-descent/GradientDescent.ipynb | Abhinav2604/deep-learning-v2-pytorch |
Time to train the algorithm!When we run the function, we'll obtain the following:- 10 updates with the current training loss and accuracy- A plot of the data and some of the boundary lines obtained. The final one is in black. Notice how the lines get closer and closer to the best fit, as we go through more epochs.- A plot of the error function. Notice how it decreases as we go through more epochs. | train(X, y, epochs, learnrate, True) |
========== Epoch 0 ==========
Train loss: 0.7135845195381634
Accuracy: 0.4
========== Epoch 10 ==========
Train loss: 0.6225835210454962
Accuracy: 0.59
========== Epoch 20 ==========
Train loss: 0.5548744083669508
Accuracy: 0.74
========== Epoch 30 ==========
Train loss: 0.501606141872473
Accuracy: 0.84
========== Epoch 40 ==========
Train loss: 0.4593334641861401
Accuracy: 0.86
========== Epoch 50 ==========
Train loss: 0.42525543433469976
Accuracy: 0.93
========== Epoch 60 ==========
Train loss: 0.3973461571671399
Accuracy: 0.93
========== Epoch 70 ==========
Train loss: 0.3741469765239074
Accuracy: 0.93
========== Epoch 80 ==========
Train loss: 0.35459973368161973
Accuracy: 0.94
========== Epoch 90 ==========
Train loss: 0.3379273658879921
Accuracy: 0.94
| MIT | intro-neural-networks/gradient-descent/GradientDescent.ipynb | Abhinav2604/deep-learning-v2-pytorch |
ElasticNet with RobustScaler & Power Transformer This Code template is for regression analysis using the ElasticNet regressor where rescaling method used is RobustScaler and feature transformation is done via Power Transformer. Required Packages | import numpy as np
import pandas as pd
import seaborn as se
import warnings
import matplotlib.pyplot as plt
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
from sklearn.linear_model import ElasticNet
from imblearn.over_sampling import RandomOverSampler
from sklearn.preprocessing import RobustScaler, PowerTransformer
from sklearn.metrics import mean_squared_error, r2_score,mean_absolute_error
warnings.filterwarnings('ignore') | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
InitializationFilepath of CSV file | #filepath
file_path= "" | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
List of features which are required for model training. | #x_values
features=[] | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
Target feature for prediction. | #y_value
target='' | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
Data FetchingPandas is an open-source, BSD-licensed library providing high-performance, easy-to-use data manipulation and data analysis tools.We will use panda's library to read the CSV file using its storage path.And we use the head function to display the initial row or entry. | df=pd.read_csv(file_path) #reading file
df.head() | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
Feature SelectionsIt is the process of reducing the number of input variables when developing a predictive model. Used to reduce the number of input variables to both reduce the computational cost of modelling and, in some cases, to improve the performance of the model.We will assign all the required input features to X and target/outcome to Y. | X=df[features]
Y=df[target] | _____no_output_____ | Apache-2.0 | Regression/Linear Models/ElasticNet_RobustScaler_PowerTransformer.ipynb | shreepad-nade/ds-seed |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.