hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ec6b5d838fd8f92fc7c0d89db0943c9df7be65b4 | 27,215 | ipynb | Jupyter Notebook | face-mask-detection.ipynb | charlielito/face-mask-detection | a9bf2d51680ea5eda08ff61f631a70371ddfd9ab | [
"MIT"
]
| 228 | 2020-08-19T05:09:14.000Z | 2022-03-29T13:53:14.000Z | face-mask-detection.ipynb | LaudateCorpus1/face-mask-detection-2 | dea5e027bc47a646b12984e07e2971a3f00b4ce9 | [
"MIT"
]
| 33 | 2020-08-19T08:49:00.000Z | 2022-03-24T07:24:02.000Z | face-mask-detection.ipynb | LaudateCorpus1/face-mask-detection-2 | dea5e027bc47a646b12984e07e2971a3f00b4ce9 | [
"MIT"
]
| 85 | 2020-08-28T09:02:40.000Z | 2022-03-12T19:10:56.000Z | 39.442029 | 639 | 0.621055 | [
[
[
"# Face Mask Detection using NVIDIA TLT ",
"_____no_output_____"
],
[
"The MIT License (MIT)\n\nCopyright (c) 2019-2020, NVIDIA CORPORATION.\n\nPermission is hereby granted, free of charge, to any person obtaining a copy of\nthis software and associated documentation files (the \"Software\"), to deal in\nthe Software without restriction, including without limitation the rights to\nuse, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of\nthe Software, and to permit persons to whom the Software is furnished to do so,\nsubject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS\nFOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR\nCOPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER\nIN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN\nCONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.",
"_____no_output_____"
],
[
"## DetectNet_v2 with ResNet-18 example usecase\n\nThe goal of this notebook is to utilize NVIDIA TLT to train and make Face Mask detection model deploy ready.\nWhile working on such application, this notebook will serve as an example usecase of Object Detection using DetectNet_v2 in the Transfer Learning Toolkit.\n\n0. [Set up env variables](#head-0)\n1. [Prepare dataset and pre-trained model](#head-1)\n 1. [Download dataset and convert in KITTI Format](#head-1-1)\n 1. [Prepare tfrecords from kitti format dataset](#head-1-2)\n 2. [Download pre-trained model](#head-1-3)\n2. [Provide training specification](#head-2)\n3. [Run TLT training](#head-3)\n4. [Evaluate trained models](#head-4)\n5. [Prune trained models](#head-5)\n6. [Retrain pruned models](#head-6)\n7. [Evaluate retrained model](#head-7)\n8. [Visualize inferences](#head-8)\n9. [Deploy](#head-9)\n 1. [Int8 Optimization](#head-9-1)\n 2. [Generate TensorRT engine](#head-9-2)\n10. [Verify Deployed Model](#head-10)\n 1. [Inference using TensorRT engine](#head-10-1)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"## 0. Set up env variables <a class=\"anchor\" id=\"head-0\"></a>\nWhen using the purpose-built pretrained models from NGC, please make sure to set the `$KEY` environment variable to the key as mentioned in the model overview. Failing to do so, can lead to errors when trying to load them as pretrained models.\n\n*Note: Please make sure to remove any stray artifacts/files from the `$USER_EXPERIMENT_DIR` or `$DATA_DOWNLOAD_DIR` paths as mentioned below, that may have been generated from previous experiments. Having checkpoint files etc may interfere with creating a training graph for a new experiment.*",
"_____no_output_____"
]
],
[
[
"# Setting up env variables for cleaner command line commands.\nprint(\"Update directory paths if needed\")\n%env KEY=tlt_encode\n# User directory - pre-trained/unpruned/pruned/final models will be saved here\n%env USER_EXPERIMENT_DIR=/home/detectnet_v2 \n# Download directory - tfrecords will be generated here\n%env DATA_DOWNLOAD_DIR=/home/data_fm_0916 \n# Spec Directory\n%env SPECS_DIR=/home/detectnet_v2/specs \n# Number of GPUs used for training\n%env NUM_GPUS=1",
"_____no_output_____"
]
],
[
[
"## 1. Prepare dataset and pre-trained model <a class=\"anchor\" id=\"head-1\"></a>",
"_____no_output_____"
],
[
"### A. Download dataset and convert in KITTI Format <a class=\"anchor\" id=\"head-1-1\"></a>\n\nIn this experiment we will be using 4 different datasets; \n\n1. Faces with Mask:\n - Kaggle Medical Mask Dataset [Download Link](https://www.kaggle.com/ivandanilovich/medical-masks-dataset-images-tfrecords)\n - MAFA - MAsked FAces [Download Link](https://drive.google.com/drive/folders/1nbtM1n0--iZ3VVbNGhocxbnBGhMau_OG)\n2. Faces without Mask:\n - FDDB Dataset [Download Link](http://vis-www.cs.umass.edu/fddb/)\n - WiderFace Dataset [Download Link](http://shuoyang1213.me/WIDERFACE/)",
"_____no_output_____"
],
[
"- Download the data using provided links, such that all images and label files are in one folder. We expect in structure noted in GitHub repo.\n- Convert dataset to KITTI format \n- Use KITTI format directory as \"$DATA_DOWNLOAD_DIR\"\n\n\nNote: We do not use all the images from MAFA and WiderFace. Combining we will use about 6000 faces each with and without mask",
"_____no_output_____"
],
[
"### B. Prepare tf records from kitti format dataset <a class=\"anchor\" id=\"head-1-2\"></a>\n\n* Update the tfrecords spec file to take in your kitti format dataset\n* Create the tfrecords using the tlt-dataset-convert \n\n*Note: TfRecords only need to be generated once.*",
"_____no_output_____"
]
],
[
[
"print(\"TFrecords conversion spec file for kitti training\")\n!cat $SPECS_DIR/detectnet_v2_tfrecords_kitti_trainval.txt",
"_____no_output_____"
],
[
"# Creating a new directory for the output tfrecords dump.\nprint(\"Converting Tfrecords for kitti trainval dataset\")\n!tlt-dataset-convert -d $SPECS_DIR/detectnet_v2_tfrecords_kitti_trainval.txt \\\n -o $DATA_DOWNLOAD_DIR/tfrecords/kitti_trainval/kitti_trainval",
"_____no_output_____"
],
[
"!ls -rlt $DATA_DOWNLOAD_DIR/tfrecords/kitti_trainval/",
"_____no_output_____"
]
],
[
[
"### C. Download pre-trained model <a class=\"anchor\" id=\"head-1-3\"></a>\nDownload the correct pretrained model from the NGC model registry for your experiment. Please note that for DetectNet_v2, the input is expected to be 0-1 normalized with input channels in RGB order. Therefore, for optimum results please download models with `*_detectnet_v2` in their name string. All other models expect input preprocessing with mean subtraction and input channels in BGR order. Thus, using them as pretrained weights may result in suboptimal performance. ",
"_____no_output_____"
]
],
[
[
"# List models available in the model registry.\n!ngc registry model list nvidia/tlt_pretrained_detectnet_v2:*",
"_____no_output_____"
],
[
"# Create the target destination to download the model.\n!mkdir -p $USER_EXPERIMENT_DIR/pretrained_resnet18/",
"_____no_output_____"
],
[
"# Download the pretrained model from NGC\n!ngc registry model download-version nvidia/tlt_pretrained_detectnet_v2:resnet18 \\\n --dest $USER_EXPERIMENT_DIR/pretrained_resnet18",
"_____no_output_____"
],
[
"!ls -rlt $USER_EXPERIMENT_DIR/pretrained_resnet18/tlt_pretrained_detectnet_v2_vresnet18",
"_____no_output_____"
]
],
[
[
"## 2. Provide training specification <a class=\"anchor\" id=\"head-2\"></a>\n* Tfrecords for the train datasets\n * In order to use the newly generated tfrecords, update the dataset_config parameter in the spec file at `$SPECS_DIR/detectnet_v2_train_resnet18_kitti.txt` \n * Update the fold number to use for evaluation. In case of random data split, please use fold `0` only\n * For sequence-wise split, you may use any fold generated from the dataset convert tool\n* Pre-trained models\n* Augmentation parameters for on the fly data augmentation\n* Other training (hyper-)parameters such as batch size, number of epochs, learning rate etc.",
"_____no_output_____"
]
],
[
[
"!cat $SPECS_DIR/detectnet_v2_train_resnet18_kitti.txt",
"_____no_output_____"
]
],
[
[
"## 3. Run TLT training <a class=\"anchor\" id=\"head-3\"></a>\n* Provide the sample spec file and the output directory location for models\n\n*Note: The training may take hours to complete. Also, the remaining notebook, assumes that the training was done in single-GPU mode. When run in multi-GPU mode, please expect to update the pruning and inference steps with new pruning thresholds and updated parameters in the clusterfile.json accordingly for optimum performance.*\n\n*Detectnet_v2 now supports restart from checkpoint. Incase, the training job is killed prematurely, you may resume training from the closest checkpoint by simply re-running the same command line. Please do make sure to use the same number of GPUs when restarting the training.*",
"_____no_output_____"
]
],
[
[
"!tlt-train detectnet_v2 -e $SPECS_DIR/detectnet_v2_train_resnet18_kitti.txt \\\n -r $USER_EXPERIMENT_DIR/experiment_dir_unpruned \\\n -k $KEY \\\n -n resnet18_detector \\\n --gpus $NUM_GPUS",
"_____no_output_____"
],
[
"print('Model for each epoch:')\nprint('---------------------')\n!ls -lh $USER_EXPERIMENT_DIR/experiment_dir_unpruned/weights",
"_____no_output_____"
]
],
[
[
"## 4. Evaluate the trained model <a class=\"anchor\" id=\"head-4\"></a>",
"_____no_output_____"
]
],
[
[
"!tlt-evaluate detectnet_v2 -e $SPECS_DIR/detectnet_v2_train_resnet18_kitti.txt\\\n -m $USER_EXPERIMENT_DIR/experiment_dir_unpruned/weights/resnet18_detector.tlt \\\n -k $KEY",
"_____no_output_____"
]
],
[
[
"## 5. Prune the trained model <a class=\"anchor\" id=\"head-5\"></a>\n* Specify pre-trained model\n* Equalization criterion (`Applicable for resnets and mobilenets`)\n* Threshold for pruning.\n* A key to save and load the model\n* Output directory to store the model\n\n*Usually, you just need to adjust `-pth` (threshold) for accuracy and model size trade off. Higher `pth` gives you smaller model (and thus higher inference speed) but worse accuracy. The threshold to use is depend on the dataset. A pth value `5.2e-6` is just a start point. If the retrain accuracy is good, you can increase this value to get smaller models. Otherwise, lower this value to get better accuracy.*\n\n*For some internal studies, we have noticed that a pth value of 0.01 is a good starting point for detectnet_v2 models.*",
"_____no_output_____"
]
],
[
[
"# Create an output directory if it doesn't exist.\n!mkdir -p $USER_EXPERIMENT_DIR/experiment_dir_pruned",
"_____no_output_____"
],
[
"print(\"Change Threshold (-pth) value according to you experiments\")\n\n!tlt-prune -m $USER_EXPERIMENT_DIR/experiment_dir_unpruned/weights/resnet18_detector.tlt \\\n -o $USER_EXPERIMENT_DIR/experiment_dir_pruned/resnet18_nopool_bn_detectnet_v2_pruned.tlt \\\n -eq union \\\n -pth 0.8 \\\n -k $KEY",
"_____no_output_____"
],
[
"!ls -rlt $USER_EXPERIMENT_DIR/experiment_dir_pruned/",
"_____no_output_____"
]
],
[
[
"## 6. Retrain the pruned model <a class=\"anchor\" id=\"head-6\"></a>\n* Model needs to be re-trained to bring back accuracy after pruning\n* Specify re-training specification with pretrained weights as pruned model.\n\n*Note: For retraining, please set the `load_graph` option to `true` in the model_config to load the pruned model graph. Also, if after retraining, the model shows some decrease in mAP, it could be that the originally trained model, was pruned a little too much. Please try reducing the pruning threshold, thereby reducing the pruning ratio, and use the new model to retrain.*",
"_____no_output_____"
]
],
[
[
"# Printing the retrain experiment file. \n# Note: We have updated the experiment file to include the \n# newly pruned model as a pretrained weights and, the\n# load_graph option is set to true \n!cat $SPECS_DIR/detectnet_v2_retrain_resnet18_kitti.txt",
"_____no_output_____"
],
[
"# Retraining using the pruned model as pretrained weights \n!tlt-train detectnet_v2 -e $SPECS_DIR/detectnet_v2_retrain_resnet18_kitti.txt \\\n -r $USER_EXPERIMENT_DIR/experiment_dir_retrain \\\n -k $KEY \\\n -n resnet18_detector_pruned \\\n --gpus $NUM_GPUS",
"_____no_output_____"
],
[
"# Listing the newly retrained model.\n!ls -rlt $USER_EXPERIMENT_DIR/experiment_dir_retrain/weights",
"_____no_output_____"
]
],
[
[
"## 7. Evaluate the retrained model <a class=\"anchor\" id=\"head-7\"></a>",
"_____no_output_____"
],
[
"This section evaluates the pruned and retrained model, using `tlt-evaluate`.",
"_____no_output_____"
]
],
[
[
"!tlt-evaluate detectnet_v2 -e $SPECS_DIR/detectnet_v2_retrain_resnet18_kitti.txt \\\n -m $USER_EXPERIMENT_DIR/experiment_dir_retrain/weights/resnet18_detector_pruned.tlt \\\n -k $KEY",
"_____no_output_____"
]
],
[
[
"## 8. Visualize inferences <a class=\"anchor\" id=\"head-8\"></a>\nIn this section, we run the `tlt-infer` tool to generate inferences on the trained models. To render bboxes from more classes, please edit the spec file `detectnet_v2_inference_kitti_tlt.txt` to include all the classes you would like to visualize and edit the rest of the file accordingly.",
"_____no_output_____"
],
[
"For this you will need to create `test_images` directory containing at least 8 images with masked and no-masked faces, it can be from test data or simply face captures from your own photos. ",
"_____no_output_____"
]
],
[
[
"# Running inference for detection on n images\n!tlt-infer detectnet_v2 -e $SPECS_DIR/detectnet_v2_inference_kitti_tlt.txt \\\n -o $USER_EXPERIMENT_DIR/tlt_infer_testing \\\n -i $DATA_DOWNLOAD_DIR/test_images \\\n -k $KEY",
"_____no_output_____"
]
],
[
[
"The `tlt-infer` tool produces two outputs. \n1. Overlain images in `$USER_EXPERIMENT_DIR/tlt_infer_testing/images_annotated`\n2. Frame by frame bbox labels in kitti format located in `$USER_EXPERIMENT_DIR/tlt_infer_testing/labels`\n\n*Note: To run inferences for a single image, simply replace the path to the -i flag in `tlt-infer` command with the path to the image.*",
"_____no_output_____"
]
],
[
[
"# Simple grid visualizer\nimport matplotlib.pyplot as plt\nimport os\nfrom math import ceil\nvalid_image_ext = ['.jpg', '.png', '.jpeg', '.ppm']\n\ndef visualize_images(image_dir, num_cols=4, num_images=10):\n output_path = os.path.join(os.environ['USER_EXPERIMENT_DIR'], image_dir)\n num_rows = int(ceil(float(num_images) / float(num_cols)))\n f, axarr = plt.subplots(num_rows, num_cols, figsize=[80,30])\n f.tight_layout()\n a = [os.path.join(output_path, image) for image in os.listdir(output_path) \n if os.path.splitext(image)[1].lower() in valid_image_ext]\n for idx, img_path in enumerate(a[:num_images]):\n col_id = idx % num_cols\n row_id = idx / num_cols\n img = plt.imread(img_path)\n axarr[row_id, col_id].imshow(img) ",
"_____no_output_____"
],
[
"# Visualizing the first 12 images.\nOUTPUT_PATH = 'tlt_infer_testing/images_annotated' # relative path from $USER_EXPERIMENT_DIR.\nCOLS = 4 # number of columns in the visualizer grid.\nIMAGES = 8 # number of images to visualize.\n\nvisualize_images(OUTPUT_PATH, num_cols=COLS, num_images=IMAGES)",
"_____no_output_____"
]
],
[
[
"## 9. Deploy! <a class=\"anchor\" id=\"head-9\"></a>",
"_____no_output_____"
]
],
[
[
"!mkdir -p $USER_EXPERIMENT_DIR/experiment_dir_final\n# Removing a pre-existing copy of the etlt if there has been any.\nimport os\noutput_file=os.path.join(os.environ['USER_EXPERIMENT_DIR'],\n \"experiment_dir_final/resnet18_detector.etlt\")\nif os.path.exists(output_file):\n os.system(\"rm {}\".format(output_file))\n!tlt-export detectnet_v2 \\\n -m $USER_EXPERIMENT_DIR/experiment_dir_retrain/weights/resnet18_detector_pruned.tlt \\\n -o $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.etlt \\\n -k $KEY",
"_____no_output_____"
],
[
"print('Exported model:')\nprint('------------')\n!ls -lh $USER_EXPERIMENT_DIR/experiment_dir_final",
"_____no_output_____"
]
],
[
[
"### A. Int8 Optimization <a class=\"anchor\" id=\"head-9-1\"></a>\nDetectNet_v2 model supports int8 inference mode in TRT. In order to use int8 mode, we must calibrate the model to run 8-bit inferences. This involves 2 steps\n\n* Generate calibration tensorfile from the training data using tlt-int8-tensorfile\n* Use tlt-export to generate int8 calibration table.\n\n*Note: For this example, we generate a calibration tensorfile containing 10 batches of training data.\nIdeally, it is best to use atleast 10-20% of the training data to calibrate the model. The more data provided during calibration, the closer int8 inferences are to fp32 inferences.*",
"_____no_output_____"
]
],
[
[
"!tlt-int8-tensorfile detectnet_v2 -e $SPECS_DIR/detectnet_v2_retrain_resnet18_kitti.txt \\\n -m 40 \\\n -o $USER_EXPERIMENT_DIR/experiment_dir_final/calibration.tensor",
"_____no_output_____"
],
[
"!rm -rf $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.etlt\n!rm -rf $USER_EXPERIMENT_DIR/experiment_dir_final/calibration.bin\n!tlt-export detectnet_v2 \\\n -m $USER_EXPERIMENT_DIR/experiment_dir_retrain/weights/resnet18_detector_pruned.tlt \\\n -o $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.etlt \\\n -k $KEY \\\n --cal_data_file $USER_EXPERIMENT_DIR/experiment_dir_final/calibration.tensor \\\n --data_type int8 \\\n --batches 20 \\\n --batch_size 4 \\\n --max_batch_size 4\\\n --engine_file $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.trt.int8 \\\n --cal_cache_file $USER_EXPERIMENT_DIR/experiment_dir_final/calibration.bin \\\n --verbose",
"_____no_output_____"
]
],
[
[
"### B. Generate TensorRT engine <a class=\"anchor\" id=\"head-9-2\"></a>\nVerify engine generation using the `tlt-converter` utility included with the docker.\n\nThe `tlt-converter` produces optimized tensorrt engines for the platform that it resides on. Therefore, to get maximum performance, please instantiate this docker and execute the `tlt-converter` command, with the exported `.etlt` file and calibration cache (for int8 mode) on your target device. The converter utility included in this docker only works for x86 devices, with discrete NVIDIA GPU's. \n\nFor the jetson devices, please download the converter for jetson from the dev zone link [here](https://developer.nvidia.com/tlt-converter). \n\nIf you choose to integrate your model into deepstream directly, you may do so by simply copying the exported `.etlt` file along with the calibration cache to the target device and updating the spec file that configures the `gst-nvinfer` element to point to this newly exported model. Usually this file is called `config_infer_primary.txt` for detection models and `config_infer_secondary_*.txt` for classification models.",
"_____no_output_____"
]
],
[
[
"!tlt-converter $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.etlt \\\n -k $KEY \\\n -c $USER_EXPERIMENT_DIR/experiment_dir_final/calibration.bin \\\n -o output_cov/Sigmoid,output_bbox/BiasAdd \\\n -d 3,544,960 \\\n -i nchw \\\n -m 64 \\\n -t int8 \\\n -e $USER_EXPERIMENT_DIR/experiment_dir_final/resnet18_detector.trt \\\n -b 4",
"_____no_output_____"
]
],
[
[
"## 10. Verify Deployed Model <a class=\"anchor\" id=\"head-10\"></a>\nVerify the exported model by visualizing inferences on TensorRT.\nIn addition to running inference on a `.tlt` model in [step 8](#head-8), the `tlt-infer` tool is also capable of consuming the converted `TensorRT engine` from [step 9.B](#head-9-2).\n\n*If after int-8 calibration the accuracy of the int-8 inferences seem to degrade, it could be because the there wasn't enough data in the calibration tensorfile used to calibrate thee model or, the training data is not entirely representative of your test images, and the calibration maybe incorrect. Therefore, you may either regenerate the calibration tensorfile with more batches of the training data, and recalibrate the model, or calibrate the model on a few images from the test set. This may be done using `--cal_image_dir` flag in the `tlt-export` tool. For more information, please follow the instructions in the USER GUIDE.",
"_____no_output_____"
],
[
"### A. Inference using TensorRT engine <a class=\"anchor\" id=\"head-10-1\"></a>",
"_____no_output_____"
]
],
[
[
"!tlt-infer detectnet_v2 -e $SPECS_DIR/detectnet_v2_inference_kitti_etlt.txt \\\n -o $USER_EXPERIMENT_DIR/etlt_infer_testing \\\n -i $DATA_DOWNLOAD_DIR/test_images \\\n -k $KEY",
"_____no_output_____"
],
[
"# visualize the first 12 inferenced images.\nOUTPUT_PATH = 'etlt_infer_testing/images_annotated' # relative path from $USER_EXPERIMENT_DIR.\nCOLS = 4 # number of columns in the visualizer grid.\nIMAGES = 8 # number of images to visualize.\n\nvisualize_images(OUTPUT_PATH, num_cols=COLS, num_images=IMAGES)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
ec6b6279608be60effd02732bfd40adea947d614 | 10,297 | ipynb | Jupyter Notebook | jupyter/paddlepaddle/paddle_ocr_java_zh.ipynb | bytes-lost/djl | 6dc2949d368e863172a37be969479f212569fac8 | [
"Apache-2.0"
]
| null | null | null | jupyter/paddlepaddle/paddle_ocr_java_zh.ipynb | bytes-lost/djl | 6dc2949d368e863172a37be969479f212569fac8 | [
"Apache-2.0"
]
| null | null | null | jupyter/paddlepaddle/paddle_ocr_java_zh.ipynb | bytes-lost/djl | 6dc2949d368e863172a37be969479f212569fac8 | [
"Apache-2.0"
]
| null | null | null | 31.879257 | 223 | 0.578324 | [
[
[
"# PaddleOCR在DJL 上的實現\n在這個教程裡,我們會展示利用 PaddleOCR 下載預訓練好文字處理模型並對指定的照片進行文學文字檢測 (OCR)。這個教程總共會分成三個部分:\n\n- 文字區塊檢測: 從圖片檢測出文字區塊\n- 文字角度檢測: 確認文字是否需要旋轉\n- 文字識別: 確認區塊內的文字\n\n## 導入相關環境依賴及子類別\n在這個例子中的前處理飛槳深度學習引擎需要搭配DJL混合模式進行深度學習推理,原因是引擎本身沒有包含ND數組操作,因此需要藉用其他引擎的數組操作能力來完成。這邊我們導入Pytorch來做協同的前處理工作:",
"_____no_output_____"
]
],
[
[
"// %mavenRepo snapshots https://oss.sonatype.org/content/repositories/snapshots/\n\n%maven ai.djl:api:0.11.0\n%maven ai.djl.paddlepaddle:paddlepaddle-model-zoo:0.11.0\n%maven ai.djl.paddlepaddle:paddlepaddle-native-auto:2.0.2\n%maven org.slf4j:slf4j-api:1.7.26\n%maven org.slf4j:slf4j-simple:1.7.26\n\n// second engine to do preprocessing and postprocessing\n%maven ai.djl.pytorch:pytorch-engine:0.11.0\n%maven ai.djl.pytorch:pytorch-native-auto:1.8.1",
"_____no_output_____"
],
[
"import ai.djl.*;\nimport ai.djl.inference.Predictor;\nimport ai.djl.modality.Classifications;\nimport ai.djl.modality.cv.Image;\nimport ai.djl.modality.cv.ImageFactory;\nimport ai.djl.modality.cv.output.*;\nimport ai.djl.modality.cv.util.NDImageUtils;\nimport ai.djl.ndarray.*;\nimport ai.djl.ndarray.types.DataType;\nimport ai.djl.ndarray.types.Shape;\nimport ai.djl.repository.zoo.*;\nimport ai.djl.paddlepaddle.zoo.cv.objectdetection.PpWordDetectionTranslator;\nimport ai.djl.paddlepaddle.zoo.cv.imageclassification.PpWordRotateTranslator;\nimport ai.djl.paddlepaddle.zoo.cv.wordrecognition.PpWordRecognitionTranslator;\nimport ai.djl.translate.*;\nimport java.util.concurrent.ConcurrentHashMap;",
"_____no_output_____"
]
],
[
[
"## 圖片讀取\n首先讓我們載入這次教程會用到的機票範例圖片:",
"_____no_output_____"
]
],
[
[
"String url = \"https://resources.djl.ai/images/flight_ticket.jpg\";\nImage img = ImageFactory.getInstance().fromUrl(url);\nimg.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"## 文字區塊檢測\n我們首先從 [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-detection-model-to-inference-model) 開發套件中讀取文字檢測的模型,之後我們可以生成一個DJL `Predictor` 並將其命名為 `detector`.\n",
"_____no_output_____"
]
],
[
[
"var criteria1 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, DetectedObjects.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/det_db.zip\")\n .optTranslator(new PpWordDetectionTranslator(new ConcurrentHashMap<String, String>()))\n .build();\nvar detectionModel = ModelZoo.loadModel(criteria1);\nvar detector = detectionModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"接著我們檢測出圖片中的文字區塊,這個模型的原始輸出是含有標註所有文字區域的圖算法(Bitmap),我們可以利用`PpWordDetectionTranslator` 函式將圖算法的輸出轉成長方形的方框來裁剪圖片",
"_____no_output_____"
]
],
[
[
"var detectedObj = detector.predict(img);\nImage newImage = img.duplicate(Image.Type.TYPE_INT_ARGB);\nnewImage.drawBoundingBoxes(detectedObj);\nnewImage.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"如上所示,所標註的文字區塊都非常窄,且沒有包住所有完整的文字區塊。讓我們嘗試使用`extendRect`函式來擴展文字框的長寬到需要的大小, 再利用 `getSubImage` 裁剪並擷取出文子區塊。",
"_____no_output_____"
]
],
[
[
"Image getSubImage(Image img, BoundingBox box) {\n Rectangle rect = box.getBounds();\n double[] extended = extendRect(rect.getX(), rect.getY(), rect.getWidth(), rect.getHeight());\n int width = img.getWidth();\n int height = img.getHeight();\n int[] recovered = {\n (int) (extended[0] * width),\n (int) (extended[1] * height),\n (int) (extended[2] * width),\n (int) (extended[3] * height)\n };\n return img.getSubimage(recovered[0], recovered[1], recovered[2], recovered[3]);\n}\n\ndouble[] extendRect(double xmin, double ymin, double width, double height) {\n double centerx = xmin + width / 2;\n double centery = ymin + height / 2;\n if (width > height) {\n width += height * 2.0;\n height *= 3.0;\n } else {\n height += width * 2.0;\n width *= 3.0;\n }\n double newX = centerx - width / 2 < 0 ? 0 : centerx - width / 2;\n double newY = centery - height / 2 < 0 ? 0 : centery - height / 2;\n double newWidth = newX + width > 1 ? 1 - newX : width;\n double newHeight = newY + height > 1 ? 1 - newY : height;\n return new double[] {newX, newY, newWidth, newHeight};\n}",
"_____no_output_____"
]
],
[
[
"讓我們輸出其中一個文字區塊",
"_____no_output_____"
]
],
[
[
"List<DetectedObjects.DetectedObject> boxes = detectedObj.items();\nvar sample = getSubImage(img, boxes.get(5).getBoundingBox());\nsample.getWrappedImage();",
"_____no_output_____"
]
],
[
[
"## 文字角度檢測\n我們從 [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-angle-classification-model-to-inference-model) 輸出這個模型並確認圖片及文字是否需要旋轉。以下的代碼會讀入這個模型並生成a `rotateClassifier` 子類別",
"_____no_output_____"
]
],
[
[
"var criteria2 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, Classifications.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/cls.zip\")\n .optTranslator(new PpWordRotateTranslator())\n .build();\nvar rotateModel = ModelZoo.loadModel(criteria2);\nvar rotateClassifier = rotateModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"## 文字識別\n\n我們從 [PaddleOCR](https://github.com/PaddlePaddle/PaddleOCR/blob/release/2.0/doc/doc_en/inference_en.md#convert-recognition-model-to-inference-model) 輸出這個模型並識別圖片中的文字, 我們一樣仿造上述的步驟讀取這個模型\n",
"_____no_output_____"
]
],
[
[
"var criteria3 = Criteria.builder()\n .optEngine(\"PaddlePaddle\")\n .setTypes(Image.class, String.class)\n .optModelUrls(\"https://resources.djl.ai/test-models/paddleOCR/mobile/rec_crnn.zip\")\n .optTranslator(new PpWordRecognitionTranslator())\n .build();\nvar recognitionModel = ModelZoo.loadModel(criteria3);\nvar recognizer = recognitionModel.newPredictor();",
"_____no_output_____"
]
],
[
[
"接著我們可以試著套用這兩個模型在先前剪裁好的文字區塊上",
"_____no_output_____"
]
],
[
[
"System.out.println(rotateClassifier.predict(sample));\nrecognizer.predict(sample);",
"_____no_output_____"
]
],
[
[
"最後我們把這些模型串連在一起並套用在整張圖片上看看結果會如何。DJL提供了豐富的影像工具包讓你可以從圖片中擷取出文字並且完美呈現",
"_____no_output_____"
]
],
[
[
"Image rotateImg(Image image) {\n try (NDManager manager = NDManager.newBaseManager()) {\n NDArray rotated = NDImageUtils.rotate90(image.toNDArray(manager), 1);\n return ImageFactory.getInstance().fromNDArray(rotated);\n }\n}\n\nList<String> names = new ArrayList<>();\nList<Double> prob = new ArrayList<>();\nList<BoundingBox> rect = new ArrayList<>();\n\nfor (int i = 0; i < boxes.size(); i++) {\n Image subImg = getSubImage(img, boxes.get(i).getBoundingBox());\n if (subImg.getHeight() * 1.0 / subImg.getWidth() > 1.5) {\n subImg = rotateImg(subImg);\n }\n Classifications.Classification result = rotateClassifier.predict(subImg).best();\n if (\"Rotate\".equals(result.getClassName()) && result.getProbability() > 0.8) {\n subImg = rotateImg(subImg);\n }\n String name = recognizer.predict(subImg);\n names.add(name);\n prob.add(-1.0);\n rect.add(boxes.get(i).getBoundingBox());\n}\nnewImage.drawBoundingBoxes(new DetectedObjects(names, prob, rect));\nnewImage.getWrappedImage();",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6b6366f1d7c4ce6963e2643097b30533bdf1a4 | 11,736 | ipynb | Jupyter Notebook | Convolutional Neural Networks in TensorFlow/week1 Exploring a Larger Dataset/Exercise_5_Answer.ipynb | beckswu/TensorFlow | 3b85f9748d2dff56523de98816d3d0221abfa01d | [
"MIT"
]
| null | null | null | Convolutional Neural Networks in TensorFlow/week1 Exploring a Larger Dataset/Exercise_5_Answer.ipynb | beckswu/TensorFlow | 3b85f9748d2dff56523de98816d3d0221abfa01d | [
"MIT"
]
| null | null | null | Convolutional Neural Networks in TensorFlow/week1 Exploring a Larger Dataset/Exercise_5_Answer.ipynb | beckswu/TensorFlow | 3b85f9748d2dff56523de98816d3d0221abfa01d | [
"MIT"
]
| null | null | null | 35.456193 | 134 | 0.46532 | [
[
[
"import os\nimport zipfile\nimport random\nimport tensorflow as tf\nfrom tensorflow.keras.optimizers import RMSprop\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom shutil import copyfile",
"_____no_output_____"
],
[
"# If the URL doesn't work, visit https://www.microsoft.com/en-us/download/confirmation.aspx?id=54765\n# And right click on the 'Download Manually' link to get a new URL to the dataset\n\n# Note: This is a very large dataset and will take time to download\n\n!wget --no-check-certificate \\\n \"https://download.microsoft.com/download/3/E/1/3E1C3F21-ECDB-4869-8368-6DEBA77B919F/kagglecatsanddogs_3367a.zip\" \\\n -O \"/tmp/cats-and-dogs.zip\"\n\nlocal_zip = '/tmp/cats-and-dogs.zip'\nzip_ref = zipfile.ZipFile(local_zip, 'r')\nzip_ref.extractall('/tmp')\nzip_ref.close()\n",
"_____no_output_____"
],
[
"print(len(os.listdir('/tmp/PetImages/Cat/')))\nprint(len(os.listdir('/tmp/PetImages/Dog/')))\n\n# Expected Output:\n# 12501\n# 12501",
"_____no_output_____"
],
[
"try:\n #YOUR CODE GOES HERE\n os.mkdir('/tmp/cats-v-dogs')\n os.mkdir('/tmp/cats-v-dogs/training')\n os.mkdir('/tmp/cats-v-dogs/testing')\n os.mkdir('/tmp/cats-v-dogs/training/cats')\n os.mkdir('/tmp/cats-v-dogs/training/dogs')\n os.mkdir('/tmp/cats-v-dogs/testing/cats')\n os.mkdir('/tmp/cats-v-dogs/testing/dogs')\nexcept OSError:\n pass",
"_____no_output_____"
],
[
"def split_data(SOURCE, TRAINING, TESTING, SPLIT_SIZE):\n # YOUR CODE STARTS HERE\n files = []\n for filename in os.listdir(SOURCE):\n file = SOURCE + filename\n if os.path.getsize(file) > 0:\n files.append(filename)\n else:\n print(filename + \" is zero length, so ignoring.\")\n\n training_length = int(len(files) * SPLIT_SIZE)\n testing_length = int(len(files) - training_length)\n shuffled_set = random.sample(files, len(files))\n training_set = shuffled_set[0:training_length]\n testing_set = shuffled_set[-testing_length:]\n\n for filename in training_set:\n this_file = SOURCE + filename\n destination = TRAINING + filename\n copyfile(this_file, destination)\n\n for filename in testing_set:\n this_file = SOURCE + filename\n destination = TESTING + filename\n copyfile(this_file, destination)\n # YOUR CODE END HERE\n\n\nCAT_SOURCE_DIR = \"/tmp/PetImages/Cat/\"\nTRAINING_CATS_DIR = \"/tmp/cats-v-dogs/training/cats/\"\nTESTING_CATS_DIR = \"/tmp/cats-v-dogs/testing/cats/\"\nDOG_SOURCE_DIR = \"/tmp/PetImages/Dog/\"\nTRAINING_DOGS_DIR = \"/tmp/cats-v-dogs/training/dogs/\"\nTESTING_DOGS_DIR = \"/tmp/cats-v-dogs/testing/dogs/\"\n\nsplit_size = .9\nsplit_data(CAT_SOURCE_DIR, TRAINING_CATS_DIR, TESTING_CATS_DIR, split_size)\nsplit_data(DOG_SOURCE_DIR, TRAINING_DOGS_DIR, TESTING_DOGS_DIR, split_size)\n\n# Expected output\n# 666.jpg is zero length, so ignoring\n# 11702.jpg is zero length, so ignoring",
"_____no_output_____"
],
[
"print(len(os.listdir('/tmp/cats-v-dogs/training/cats/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/training/dogs/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/testing/cats/')))\nprint(len(os.listdir('/tmp/cats-v-dogs/testing/dogs/')))\n\n# Expected output:\n# 11250\n# 11250\n# 1250\n# 1250",
"_____no_output_____"
],
[
"model = tf.keras.models.Sequential([\n # YOUR CODE STARTS HERE\n tf.keras.layers.Conv2D(16, (3, 3), activation='relu', input_shape=(150, 150, 3)),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(32, (3, 3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(64, (3, 3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(512, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid')\n])\n\nmodel.compile(optimizer=RMSprop(lr=0.001), loss='binary_crossentropy', metrics=['acc'])\n",
"_____no_output_____"
],
[
"\nTRAINING_DIR = \"/tmp/cats-v-dogs/training/\" #YOUR CODE HERE\ntrain_datagen = ImageDataGenerator(rescale=1.0/255.) #YOUR CODE HERE\ntrain_generator = train_datagen.flow_from_directory(TRAINING_DIR, #YOUR CODE HERE\n batch_size=100,\n class_mode='binary',\n target_size=(150, 150))\n\nVALIDATION_DIR = \"/tmp/cats-v-dogs/testing/\" #YOUR CODE HERE\nvalidation_datagen = ImageDataGenerator(rescale=1.0/255.) #YOUR CODE HERE\nvalidation_generator = validation_datagen.flow_from_directory(VALIDATION_DIR, #YOUR CODE HERE\n batch_size=100,\n class_mode='binary',\n target_size=(150, 150))\n\n# Expected Output:\n# Found 22498 images belonging to 2 classes.\n# Found 2500 images belonging to 2 classes.",
"_____no_output_____"
],
[
"# Note that this may take some time.\nhistory = model.fit_generator(train_generator,\n epochs=50,\n verbose=1,\n validation_data=validation_generator)",
"_____no_output_____"
],
[
"%matplotlib inline\n\nimport matplotlib.image as mpimg\nimport matplotlib.pyplot as plt\n\n#-----------------------------------------------------------\n# Retrieve a list of list results on training and test data\n# sets for each training epoch\n#-----------------------------------------------------------\nacc=history.history['acc']\nval_acc=history.history['val_acc']\nloss=history.history['loss']\nval_loss=history.history['val_loss']\n\nepochs=range(len(acc)) # Get number of epochs\n\n#------------------------------------------------\n# Plot training and validation accuracy per epoch\n#------------------------------------------------\nplt.plot(epochs, acc, 'r', \"Training Accuracy\")\nplt.plot(epochs, val_acc, 'b', \"Validation Accuracy\")\nplt.title('Training and validation accuracy')\nplt.figure()\n\n#------------------------------------------------\n# Plot training and validation loss per epoch\n#------------------------------------------------\nplt.plot(epochs, loss, 'r', \"Training Loss\")\nplt.plot(epochs, val_loss, 'b', \"Validation Loss\")\nplt.figure()\n\n\n# Desired output. Charts with training and validation metrics. No crash :)",
"_____no_output_____"
],
[
"# Here's a codeblock just for fun. You should be able to upload an image here \n# and have it classified without crashing\nimport numpy as np\nfrom google.colab import files\nfrom keras.preprocessing import image\n\nuploaded = files.upload()\n\nfor fn in uploaded.keys():\n \n # predicting images\n path = '/content/' + fn\n img = image.load_img(path, target_size=(150, 150)) #YOUR CODE HERE\n x = image.img_to_array(img)\n x = np.expand_dims(x, axis=0)\n\n images = np.vstack([x])\n classes = model.predict(images, batch_size=10)\n print(classes[0])\n if classes[0]>0.5:\n print(fn + \" is a dog\")\n else:\n print(fn + \" is a cat\")",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6b6ca41b258aa2fe712bb2de1a97e791cf12cc | 32,319 | ipynb | Jupyter Notebook | notebooks/DC3.ipynb | deeplycloudy/brawl4d | f76e4acc230ccde689b6dccabba7e7c7f8189232 | [
"BSD-2-Clause"
]
| 4 | 2015-02-27T20:49:46.000Z | 2016-04-04T23:01:51.000Z | notebooks/DC3.ipynb | deeplycloudy/brawl4d | f76e4acc230ccde689b6dccabba7e7c7f8189232 | [
"BSD-2-Clause"
]
| 1 | 2015-12-10T17:49:08.000Z | 2015-12-23T19:05:56.000Z | notebooks/DC3.ipynb | deeplycloudy/brawl4d | f76e4acc230ccde689b6dccabba7e7c7f8189232 | [
"BSD-2-Clause"
]
| 5 | 2015-03-27T17:02:49.000Z | 2016-09-06T21:57:34.000Z | 26.000805 | 536 | 0.466196 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec6b6cb92ff2af7e9f198f3b7635bd83fa5e350a | 11,910 | ipynb | Jupyter Notebook | examples/1_H_examples/weightVisualise.py.ipynb | t2mhanh/caffe_anaconda | 4f31996df63d9148de046d1b54fa1cc4e862ba83 | [
"BSD-2-Clause"
]
| 1 | 2018-11-23T08:45:35.000Z | 2018-11-23T08:45:35.000Z | examples/1_H_examples/weightVisualise.py.ipynb | t2mhanh/caffe_anaconda | 4f31996df63d9148de046d1b54fa1cc4e862ba83 | [
"BSD-2-Clause"
]
| null | null | null | examples/1_H_examples/weightVisualise.py.ipynb | t2mhanh/caffe_anaconda | 4f31996df63d9148de046d1b54fa1cc4e862ba83 | [
"BSD-2-Clause"
]
| null | null | null | 93.046875 | 2,338 | 0.679261 | [
[
[
"import sys\nprint sys.path\n#sys.path.insert(0,'/home/csunix/schtmt/NewFolder/caffe_May2017/python')\nsys.path.append('/home/csunix/schtmt/NewFolder/caffe_May2017/python')\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport caffe\ndef visualize_weights(net, layer_name, padding=4, filename=''):\n # The parameters are a list of [weights, biases]\n data = np.copy(net.params[layer_name][0].data)\n # N is the total number of convolutions\n N = data.shape[0]*data.shape[1]\n # Ensure the resulting image is square\n filters_per_row = int(np.ceil(np.sqrt(N)))\n # Assume the filters are square\n filter_size = data.shape[2]\n # Size of the result image including padding\n result_size = filters_per_row*(filter_size + padding) - padding\n # Initialize result image to all zeros\n result = np.zeros((result_size, result_size))\n \n # Tile the filters into the result image\n filter_x = 0\n filter_y = 0\n for n in range(data.shape[0]):\n for c in range(data.shape[1]):\n if filter_x == filters_per_row:\n filter_y += 1\n filter_x = 0\n for i in range(filter_size):\n for j in range(filter_size):\n result[filter_y*(filter_size + padding) + i, filter_x*(filter_size + padding) + j] = data[n, c, i, j]\n filter_x += 1\n \n # Normalize image to 0-1\n min = result.min()\n max = result.max()\n result = (result - min) / (max - min)\n \n # Plot figure\n plt.figure(figsize=(10, 10))\n plt.axis('off')\n plt.imshow(result, cmap='gray', interpolation='nearest')\n \n \n # Save plot if filename is set\n if filename != '':\n plt.savefig(filename, bbox_inches='tight', pad_inches=0)\n \n plt.show()\n\n# Load model\nnet = caffe.Net('/home/csunix/schtmt/NewFolder/caffe_May2017/examples/1_H_examples/mnist_wta_autoencoder/mnist_wta_autoencoder.prototxt',\n '/usr/not-backed-up/1_convlstm/mnist_wta_AE/mnist_wta_autoencoder_iter_6000.caffemodel',\n caffe.TEST)\n\n \n ",
"['', '/home/csunix/schtmt/.local/lib/python2.7/site-packages/cudamat-0.3-py2.7-linux-x86_64.egg', '/home/cserv1_a/soc_pg/schtmt/NewFolder/caffe_May2017/examples/1_H_examples', '/home/cserv1_a/soc_pg/schtmt/NewFolder/caffe_May2017/examples/1_H_examples/usr/not-backed-up/unsupervised-videos-master/cudamat', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python27.zip', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/plat-linux2', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/lib-tk', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/lib-old', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/lib-dynload', '/home/csunix/schtmt/.local/lib/python2.7/site-packages', '/home/cserv1_b/apps/install/anaconda/4.1.1/lib/python2.7/site-packages/Sphinx-1.4.1-py2.7.egg', '/home/cserv1_b/apps/install/anaconda/4.1.1/lib/python2.7/site-packages/setuptools-23.0.0-py2.7.egg', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/site-packages', '/home/csunix/linux/apps/install/anaconda/4.1.1/lib/python2.7/site-packages/IPython/extensions', '/home/cserv1_a/soc_pg/schtmt/.ipython', '/home/csunix/schtmt/NewFolder/caffe_May2017/python']\n"
],
[
"visualize_weights(net, 'conv1', filename='conv1.png') ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
ec6b6dee112ea558fb3eba5ba2838e9128616f06 | 186,275 | ipynb | Jupyter Notebook | dense_correspondence/evaluation/evaluation_quantitative_tutorial.ipynb | thomasweng15/pytorch-dense-correspondence | de6d57943c33b252f11c443e85bad2eed90767b1 | [
"BSD-3-Clause"
]
| null | null | null | dense_correspondence/evaluation/evaluation_quantitative_tutorial.ipynb | thomasweng15/pytorch-dense-correspondence | de6d57943c33b252f11c443e85bad2eed90767b1 | [
"BSD-3-Clause"
]
| null | null | null | dense_correspondence/evaluation/evaluation_quantitative_tutorial.ipynb | thomasweng15/pytorch-dense-correspondence | de6d57943c33b252f11c443e85bad2eed90767b1 | [
"BSD-3-Clause"
]
| 1 | 2020-05-26T12:41:52.000Z | 2020-05-26T12:41:52.000Z | 724.805447 | 53,992 | 0.928198 | [
[
[
"# Example quantitative plots\n\nHow to plot the results of the quantitative evaluation.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport os\nimport fnmatch\nimport pandas as pd\nimport sklearn.metrics as sm\nimport scipy.stats as ss\nimport matplotlib.pyplot as plt\n\nimport dense_correspondence_manipulation.utils.utils as utils\nutils.add_dense_correspondence_to_python_path()\n\nfrom dense_correspondence.evaluation.evaluation import DenseCorrespondenceEvaluationPlotter as DCEP",
"_____no_output_____"
]
],
[
[
"If you have multiple networks trained, you can add them to the `nets_list` below, and they will be plotted together.",
"_____no_output_____"
]
],
[
[
"folder_name = \"tutorials\"\npath_to_nets = os.path.join(\"code/data_volume/pdc_synthetic/trained_models\", folder_name)\npath_to_nets = utils.convert_to_absolute_path(path_to_nets)\nall_nets = sorted(os.listdir(path_to_nets))\nnets_to_plot = []\n\nnets_list = [\"wolf_3\"]\nfor net in nets_list:\n nets_to_plot.append(os.path.join(folder_name,net))",
"_____no_output_____"
]
],
[
[
"# Training \nEvaluate the network on the training scenes. Correspondences are all within scene",
"_____no_output_____"
]
],
[
[
"p = DCEP()\ndc_source_dir = utils.getDenseCorrespondenceSourceDir()\n\nnetwork_name = nets_to_plot[0]\npath_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/train/data.csv\")\nfig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, save=False)\n\nfor network_name in nets_to_plot[1:]:\n path_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/train/data.csv\")\n fig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, previous_fig_axes=fig_axes, save=False)\n\n_, axes = fig_axes\n# axes[0].set_title(\"Training Set\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Test\nEvaluate the network on the test scenes. Correspondences are all within scene",
"_____no_output_____"
]
],
[
[
"p = DCEP()\ndc_source_dir = utils.getDenseCorrespondenceSourceDir()\n\nnetwork_name = nets_to_plot[0]\npath_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/test/data.csv\")\nfig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, save=False)\n\nfor network_name in nets_to_plot[1:]:\n path_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/test/data.csv\")\n fig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, previous_fig_axes=fig_axes, save=False)\n\n_, axes = fig_axes\n# axes[0].set_title(\"Test Set\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Cross Scene Single Object\nEvaluate the network on correspondences that come from different scenes. These correspondences were manually annotated only for evaluation purposes.",
"_____no_output_____"
]
],
[
[
"p = DCEP()\ndc_source_dir = utils.getDenseCorrespondenceSourceDir()\n\nnetwork_name = nets_to_plot[0]\npath_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/cross_scene/data.csv\")\nfig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, save=False)\n\nfor network_name in nets_to_plot[1:]:\n path_to_csv = os.path.join(dc_source_dir, \"data_volume\", \"pdc_synthetic\", \"trained_models\", network_name, \"analysis/cross_scene/data.csv\")\n fig_axes = DCEP.run_on_single_dataframe(path_to_csv, label=network_name, previous_fig_axes=fig_axes, save=False)\n\n_, axes = fig_axes\n# axes[0].set_title(\"Cross Scene Set\")\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6b8b8b577f546c2ac97110262cec5adb77ffab | 7,518 | ipynb | Jupyter Notebook | examples/infer.ipynb | theSoenke/facenet-pytorch | bba6756753f3f338f6e501782b364d1e078cebd3 | [
"MIT"
]
| 2 | 2020-03-11T03:12:04.000Z | 2021-04-18T16:13:32.000Z | examples/infer.ipynb | theSoenke/facenet-pytorch | bba6756753f3f338f6e501782b364d1e078cebd3 | [
"MIT"
]
| null | null | null | examples/infer.ipynb | theSoenke/facenet-pytorch | bba6756753f3f338f6e501782b364d1e078cebd3 | [
"MIT"
]
| null | null | null | 30.560976 | 402 | 0.585262 | [
[
[
"# Face detection and recognition inference pipeline\n\nThe following example illustrates how to use the `facenet_pytorch` python package to perform face detection and recogition on an image dataset using an Inception Resnet V1 pretrained on the VGGFace2 dataset.\n\nThe following Pytorch methods are included:\n* Datasets\n* Dataloaders\n* GPU/CPU processing",
"_____no_output_____"
]
],
[
[
"from facenet_pytorch import MTCNN, InceptionResnetV1\nimport torch\nfrom torch.utils.data import DataLoader\nfrom torchvision import datasets\nimport numpy as np\nimport pandas as pd\nimport os\n\nworkers = 0 if os.name == 'nt' else 4",
"_____no_output_____"
]
],
[
[
"#### Determine if an nvidia GPU is available",
"_____no_output_____"
]
],
[
[
"device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')\nprint('Running on device: {}'.format(device))",
"Running on device: cuda:0\n"
]
],
[
[
"#### Define MTCNN module\n\nDefault params shown for illustration, but not needed. Note that, since MTCNN is a collection of neural nets and other code, the device must be passed in the following way to enable copying of objects when needed internally.\n\nSee `help(MTCNN)` for more details.",
"_____no_output_____"
]
],
[
[
"mtcnn = MTCNN(\n image_size=160, margin=0, min_face_size=20,\n thresholds=[0.6, 0.7, 0.7], factor=0.709, prewhiten=True,\n device=device\n)",
"_____no_output_____"
]
],
[
[
"#### Define Inception Resnet V1 module\n\nSet classify=True for pretrained classifier. For this example, we will use the model to output embeddings/CNN features. Note that for inference, it is important to set the model to `eval` mode.\n\nSee `help(InceptionResnetV1)` for more details.",
"_____no_output_____"
]
],
[
[
"resnet = InceptionResnetV1(pretrained='vggface2').eval().to(device)",
"_____no_output_____"
]
],
[
[
"#### Define a dataset and data loader\n\nWe add the `idx_to_class` attribute to the dataset to enable easy recoding of label indices to identity names later one.",
"_____no_output_____"
]
],
[
[
"def collate_fn(x):\n return x[0]\n\ndataset = datasets.ImageFolder('../data/test_images')\ndataset.idx_to_class = {i:c for c, i in dataset.class_to_idx.items()}\nloader = DataLoader(dataset, collate_fn=collate_fn, num_workers=workers)",
"_____no_output_____"
]
],
[
[
"#### Perfom MTCNN facial detection\n\nIterate through the DataLoader object and detect faces and associated detection probabilities for each. The `MTCNN` forward method returns images cropped to the detected face, if a face was detected. By default only a single detected face is returned - to have `MTCNN` return all detected faces, set `keep_all=True` when creating the MTCNN object above.\n\nTo obtain bounding boxes rather than cropped face images, you can instead call the lower-level `mtcnn.detect()` function. See `help(mtcnn.detect)` for details.",
"_____no_output_____"
]
],
[
[
"aligned = []\nnames = []\nfor x, y in loader:\n x_aligned, prob = mtcnn(x, return_prob=True)\n if x_aligned is not None:\n print('Face detected with probability: {:8f}'.format(prob))\n aligned.append(x_aligned)\n names.append(dataset.idx_to_class[y])",
"Face detected with probability: 0.999957\nFace detected with probability: 0.999927\nFace detected with probability: 0.999662\nFace detected with probability: 0.999873\nFace detected with probability: 0.999991\n"
]
],
[
[
"#### Calculate image embeddings\n\nMTCNN will return images of faces all the same size, enabling easy batch processing with the Resnet recognition module. Here, since we only have a few images, we build a single batch and perform inference on it. \n\nFor real datasets, code should be modified to control batch sizes being passed to the Resnet, particularly if being processed on a GPU. For repeated testing, it is best to separate face detection (using MTCNN) from embedding or classification (using InceptionResnetV1), as calculation of cropped faces or bounding boxes can then be performed a single time and detected faces saved for future use.",
"_____no_output_____"
]
],
[
[
"aligned = torch.stack(aligned).to(device)\nembeddings = resnet(aligned).detach().cpu()",
"_____no_output_____"
]
],
[
[
"#### Print distance matrix for classes",
"_____no_output_____"
]
],
[
[
"dists = [[(e1 - e2).norm().item() for e2 in embeddings] for e1 in embeddings]\nprint(pd.DataFrame(dists, columns=names, index=names))",
" angelina_jolie bradley_cooper kate_siegel paul_rudd \\\nangelina_jolie 0.000000 1.344806 0.781201 1.425579 \nbradley_cooper 1.344806 0.000000 1.256238 0.922126 \nkate_siegel 0.781201 1.256238 0.000000 1.366423 \npaul_rudd 1.425579 0.922126 1.366423 0.000000 \nshea_whigham 1.448495 0.891145 1.416447 0.985438 \n\n shea_whigham \nangelina_jolie 1.448495 \nbradley_cooper 0.891145 \nkate_siegel 1.416447 \npaul_rudd 0.985438 \nshea_whigham 0.000000 \n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6ba466808e998cfcc0e2b48909cf6350f8b5d9 | 29,166 | ipynb | Jupyter Notebook | jupyter/Teste Categorial Enginnering.ipynb | darosantos/RFNS | 2b17fcb51bd387740650690f3cdbac7bbde74378 | [
"Apache-2.0"
]
| 1 | 2019-10-01T16:10:04.000Z | 2019-10-01T16:10:04.000Z | jupyter/Teste Categorial Enginnering.ipynb | darosantos/RFNS | 2b17fcb51bd387740650690f3cdbac7bbde74378 | [
"Apache-2.0"
]
| 5 | 2019-09-17T17:29:12.000Z | 2019-10-12T13:59:21.000Z | jupyter/Teste Categorial Enginnering.ipynb | darosantos/RFNS | 2b17fcb51bd387740650690f3cdbac7bbde74378 | [
"Apache-2.0"
]
| 1 | 2019-10-31T20:39:44.000Z | 2019-10-31T20:39:44.000Z | 43.466468 | 232 | 0.500549 | [
[
[
"import sys\nsys.path.append('C:\\\\Users\\\\Danilo Santos\\\\Desktop\\\\Qualificação PPGCC\\\\abordagem\\\\RFNS')\nfrom grimoire.EnginneringForest import EnginneringForest",
"_____no_output_____"
],
[
"import pandas as pd\n\nimport numpy as np\n\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
],
[
"path = 'C:\\\\Users\\\\Danilo Santos\\\\Desktop\\\\Qualificação PPGCC\\\\abordagem\\\\RFNS'\ndf_acute = pd.read_csv(path+'\\\\datasets\\\\acute\\\\diagnosis.csv',\n engine='c', \n memory_map=True, \n low_memory=True)",
"_____no_output_____"
],
[
"X = df_acute[['temperatura', 'nausea', 'dorlombar', \n 'urinepushing', 'miccao', 'queimacao', 'inflamacao']]\n# Labels\ny = df_acute['target']\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, \n random_state=100, \n shuffle=True, \n stratify=y)",
"_____no_output_____"
],
[
"X.shape",
"_____no_output_____"
],
[
"X_train.shape",
"_____no_output_____"
],
[
"X.head()",
"_____no_output_____"
],
[
"model = EnginneringForest(3)",
"_____no_output_____"
],
[
"model.is_data_categorical = True\nmodel.encoder_enable = True\nmodel.encoder_target = True\nmodel.encoder_data = True",
"_____no_output_____"
],
[
"model.fit(X_train, y_train)",
"_____no_output_____"
],
[
"model.n_features_",
"_____no_output_____"
],
[
"model.name_features_",
"_____no_output_____"
],
[
"model.group_features_",
"_____no_output_____"
],
[
"model.encoder_feature",
"_____no_output_____"
],
[
"for c in model.encoder_feature.item():\n print(c)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6ba82af31bc98dcbd887ce2f50eaa6bfc14fda | 147,600 | ipynb | Jupyter Notebook | exercise-1/raw_code.ipynb | saiga143/geopython | 32c981a62423eb355d26176594d5e0134121b6bd | [
"MIT"
]
| null | null | null | exercise-1/raw_code.ipynb | saiga143/geopython | 32c981a62423eb355d26176594d5e0134121b6bd | [
"MIT"
]
| null | null | null | exercise-1/raw_code.ipynb | saiga143/geopython | 32c981a62423eb355d26176594d5e0134121b6bd | [
"MIT"
]
| null | null | null | 581.102362 | 138,248 | 0.941762 | [
[
[
"# Preparing the seasonal temperature anamolies data",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\n\nfp = 'data/2315676.txt' #fp means filepath\n\ndata = pd.read_csv(fp, na_values = -9999, skiprows = [1], delim_whitespace = True)\n\ndef estimates(df):\n if pd.isnull(df.TAVG):\n return (df.TMAX + df.TMIN)/2\n else:\n return df.TAVG\n \ndata['TAVG_EST'] = data.apply(estimates, axis = 'columns')\n\ndata.dropna(subset=['TAVG_EST'], inplace=True)\n\ndef fahr_to_celsius(temp_fahr):\n \"\"\"Function to convert temperatures from fahranhiet to celsius\n \n Parameters\n --------\n \n temp_fahr: int | float\n Input temperature in fahranheit (should be a number)\n \n Returns\n ------\n \n Temperature in celsius (float)\n \n \"\"\"\n return (temp_fahr -32)/(1.8)\n\ndata['TAVG_C'] = fahr_to_celsius(data['TAVG_EST'])\n\n\ndata['date_str'] = data['DATE'].astype(str)\n\ndata['YRMO'] = data['date_str'].str.slice(start=0,stop=6)\ndata['YRMO'] = data['YRMO'].astype(int)\n\ndata['year'] = data['date_str'].str.slice(start=0, stop=4)\ndata['year'] = data['year'].astype(int)\n\ndata['month'] = data['date_str'].str.slice(start=4, stop=6)\ndata['month'] = data['month'].astype(int)\n\n#For the period of 1909 - 2019\n\nwinter_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==12) | (data.month ==1) | (data.month ==2)]\nspring_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==3) | (data.month ==4) | (data.month ==5)]\nsummer_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==6) | (data.month ==7) | (data.month ==8)]\nautumn_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==9) | (data.month ==10) | (data.month ==11)]\n\n\nspring_data = spring_data.loc[(spring_data.year > 1908) & (spring_data.year < 2020)]\nspring_grouped = spring_data['TAVG_C'].groupby(spring_data['year']).mean()\n\nsummer_data = summer_data.loc[(summer_data.year > 1908) & (summer_data.year < 2020)]\nsummer_grouped = summer_data['TAVG_C'].groupby(summer_data['year']).mean()\n\nautumn_data = autumn_data.loc[(autumn_data.year > 1908) & (autumn_data.year < 2020)]\nautumn_grouped = autumn_data['TAVG_C'].groupby(autumn_data['year']).mean()\n\n\nwinter_data = winter_data.loc[winter_data.year != 2020]\ngroup1 = winter_data['TAVG_C'].groupby(winter_data['YRMO']).mean()\ngroup1 = group1.drop(labels=[190801, 190802, 201912])\ngroup2 = group1.to_frame()\n\nyear_list = []\nfor i in range(1909, 2020):\n year_list = year_list + [i,i,i]\n \ngroup2.insert(1, 'year', year_list)\nwinter_grouped = group2['TAVG_C'].groupby(group2['year']).mean()\n\n#For the reference period of 1951 - 1980\n\nref_winter_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==12) | (data.month ==1) | (data.month ==2)]\nref_spring_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==3) | (data.month ==4) | (data.month ==5)]\nref_summer_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==6) | (data.month ==7) | (data.month ==8)]\nref_autumn_data = data[['TAVG_C', 'YRMO', 'year','month']].loc[(data.month ==9) | (data.month ==10) | (data.month ==11)]\n\nref_winter_data = ref_winter_data.loc[(ref_winter_data.year > 1949) & (ref_winter_data.year < 1981)]\n\nref_spring_data = ref_spring_data.loc[(ref_spring_data.year > 1950) & (ref_spring_data.year < 1981)]\nref_spring_grouped = ref_spring_data['TAVG_C'].groupby(ref_spring_data['year']).mean()\n\nref_summer_data = ref_summer_data.loc[(ref_summer_data.year > 1950) & (ref_summer_data.year < 1981)]\nref_summer_grouped = ref_summer_data['TAVG_C'].groupby(ref_summer_data['year']).mean()\n\nref_autumn_data = ref_autumn_data.loc[(ref_autumn_data.year > 1950) & (ref_autumn_data.year < 1981)]\nref_autumn_grouped = ref_autumn_data['TAVG_C'].groupby(ref_autumn_data['year']).mean()\n\ngroup3 = ref_winter_data['TAVG_C'].groupby(ref_winter_data['YRMO']).mean()\ngroup3 = group3.drop(labels=[195001, 195002, 198012])\ngroup4 = group3.to_frame()\n\nref_year_list = []\nfor i in range(1951, 1981):\n ref_year_list = ref_year_list + [i,i,i]\n \ngroup4.insert(1, 'year', ref_year_list)\nref_winter_grouped = group4['TAVG_C'].groupby(group4['year']).mean()\n\n#Finding anamolies \nwinter_temp = ref_winter_grouped.mean()\nsummer_temp = ref_summer_grouped.mean()\nspring_temp = ref_spring_grouped.mean()\nautumn_temp = ref_autumn_grouped.mean()\n\nwinter_grouped = winter_grouped - winter_temp\nsummer_grouped = summer_grouped - summer_temp\nspring_grouped = spring_grouped - spring_temp\nautumn_grouped = autumn_grouped - autumn_temp",
"_____no_output_____"
]
],
[
[
"# Plotting the graph",
"_____no_output_____"
]
],
[
[
"# Find lower limit for y-axis\nmin_temp = min(winter_grouped.min(), spring_grouped.min(), summer_grouped.min(), autumn_grouped.min())\nmin_temp = min_temp - 5.0\n\n# Find upper limit for y-axis\nmax_temp = max(winter_grouped.max(), spring_grouped.max(), summer_grouped.max(), autumn_grouped.max())\nmax_temp = max_temp + 5.0\n\n\nfig, axs = plt.subplots(nrows=2, ncols=2, figsize=(16,10));\naxs\n\nax11 = axs[0][0]\nax12 = axs[0][1]\nax21 = axs[1][0]\nax22 = axs[1][1]\n\n# Set plot line width\nline_width = 1.5\n\n# Plot data\nwinter_grouped.plot(ax=ax11, c='blue', lw=line_width, style = 'ro-', \n ylim=[min_temp, max_temp], grid=True)\nspring_grouped.plot(ax=ax12, c='orange', lw=line_width, style = 'ro-',\n ylim=[min_temp, max_temp], grid=True)\nsummer_grouped.plot(ax=ax21, c='red', lw=line_width, style = 'ro-',\n ylim=[min_temp, max_temp], grid=True)\nautumn_grouped.plot(ax=ax22, c='brown', lw=line_width, style = 'ro-',\n ylim=[min_temp, max_temp], grid=True)\n\n# Set figure title\nfig.suptitle('1909 - 2019 Seasonal Temperature Anamolies - Sodankyala', size = 20)\n\n# Rotate the x-axis labels so they don't overlap\nplt.setp(ax11.xaxis.get_majorticklabels(), rotation=20)\nplt.setp(ax12.xaxis.get_majorticklabels(), rotation=20)\nplt.setp(ax21.xaxis.get_majorticklabels(), rotation=20)\nplt.setp(ax22.xaxis.get_majorticklabels(), rotation=20)\n\n# Axis labels\nax21.set_xlabel('Year', size = 15)\nax22.set_xlabel('Year', size = 15)\nax11.set_ylabel('Temperature Anamoly [°C]', size = 15)\nax11.set_xlabel(\"\")\nax12.set_xlabel(\"\")\nax21.set_ylabel('Temperature Anamoly[°C]', size = 15)\n\n# Season label text\nax11.text(2005, -7.5, 'Winter', size = 15)\nax12.text(2005, -7.5, 'Spring', size = 15)\nax21.text(2004, -7.5, 'Summer', size = 15)\nax22.text(2005, -7.5, 'Autumn', size = 15)\n\nfig.tight_layout()\n\n#plt.savefig('final_plot.jpeg', )",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6baa51b8139e864cdf6a14284559be5da6bbca | 679,123 | ipynb | Jupyter Notebook | Arvato_project_workbook.ipynb | IveyZ21/customer-segmentation | 0c021eb6ebf16a4d873abf710bc3bfb958b8921c | [
"zlib-acknowledgement"
]
| null | null | null | Arvato_project_workbook.ipynb | IveyZ21/customer-segmentation | 0c021eb6ebf16a4d873abf710bc3bfb958b8921c | [
"zlib-acknowledgement"
]
| null | null | null | Arvato_project_workbook.ipynb | IveyZ21/customer-segmentation | 0c021eb6ebf16a4d873abf710bc3bfb958b8921c | [
"zlib-acknowledgement"
]
| null | null | null | 63.791377 | 42,356 | 0.652974 | [
[
[
"# Capstone Project: Create a Customer Segmentation Report for Arvato Financial Services\n\nIn this project, you will analyze demographics data for customers of a mail-order sales company in Germany, comparing it against demographics information for the general population. You'll use unsupervised learning techniques to perform customer segmentation, identifying the parts of the population that best describe the core customer base of the company. Then, you'll apply what you've learned on a third dataset with demographics information for targets of a marketing campaign for the company, and use a model to predict which individuals are most likely to convert into becoming customers for the company. The data that you will use has been provided by our partners at Bertelsmann Arvato Analytics, and represents a real-life data science task.\n\nIf you completed the first term of this program, you will be familiar with the first part of this project, from the unsupervised learning project. The versions of those two datasets used in this project will include many more features and has not been pre-cleaned. You are also free to choose whatever approach you'd like to analyzing the data rather than follow pre-determined steps. In your work on this project, make sure that you carefully document your steps and decisions, since your main deliverable for this project will be a blog post reporting your findings.",
"_____no_output_____"
]
],
[
[
"# import libraries here; add more as necessary\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# magic word for producing visualizations in notebook\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## Part 0: Get to Know the Data\n\nThere are four data files associated with this project:\n\n- `Udacity_AZDIAS_052018.csv`: Demographics data for the general population of Germany; 891 211 persons (rows) x 366 features (columns).\n- `Udacity_CUSTOMERS_052018.csv`: Demographics data for customers of a mail-order company; 191 652 persons (rows) x 369 features (columns).\n- `Udacity_MAILOUT_052018_TRAIN.csv`: Demographics data for individuals who were targets of a marketing campaign; 42 982 persons (rows) x 367 (columns).\n- `Udacity_MAILOUT_052018_TEST.csv`: Demographics data for individuals who were targets of a marketing campaign; 42 833 persons (rows) x 366 (columns).\n\nEach row of the demographics files represents a single person, but also includes information outside of individuals, including information about their household, building, and neighborhood. Use the information from the first two files to figure out how customers (\"CUSTOMERS\") are similar to or differ from the general population at large (\"AZDIAS\"), then use your analysis to make predictions on the other two files (\"MAILOUT\"), predicting which recipients are most likely to become a customer for the mail-order company.\n\nThe \"CUSTOMERS\" file contains three extra columns ('CUSTOMER_GROUP', 'ONLINE_PURCHASE', and 'PRODUCT_GROUP'), which provide broad information about the customers depicted in the file. The original \"MAILOUT\" file included one additional column, \"RESPONSE\", which indicated whether or not each recipient became a customer of the company. For the \"TRAIN\" subset, this column has been retained, but in the \"TEST\" subset it has been removed; it is against that withheld column that your final predictions will be assessed in the Kaggle competition.\n\nOtherwise, all of the remaining columns are the same between the three data files. For more information about the columns depicted in the files, you can refer to two Excel spreadsheets provided in the workspace. [One of them](./DIAS Information Levels - Attributes 2017.xlsx) is a top-level list of attributes and descriptions, organized by informational category. [The other](./DIAS Attributes - Values 2017.xlsx) is a detailed mapping of data values for each feature in alphabetical order.\n\nIn the below cell, we've provided some initial code to load in the first two datasets. Note for all of the `.csv` data files in this project that they're semicolon (`;`) delimited, so an additional argument in the [`read_csv()`](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html) call has been included to read in the data properly. Also, considering the size of the datasets, it may take some time for them to load completely.\n\nYou'll notice when the data is loaded in that a warning message will immediately pop up. Before you really start digging into the modeling and analysis, you're going to need to perform some cleaning. Take some time to browse the structure of the data and look over the informational spreadsheets to understand the data values. Make some decisions on which features to keep, which features to drop, and if any revisions need to be made on data formats. It'll be a good idea to create a function with pre-processing steps, since you'll need to clean all of the datasets before you work with them.",
"_____no_output_____"
]
],
[
[
"# load in the data\nazdias = pd.read_csv('./Udacity_AZDIAS_052018.csv', sep=',', index_col=0)\ncustomers = pd.read_csv('./Udacity_CUSTOMERS_052018.csv', sep=',', index_col=0)",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/IPython/core/interactiveshell.py:3072: DtypeWarning: Columns (19,20) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"# Be sure to add in a lot more cells (both markdown and code) to document your\n# approach and findings!\nazdias.head()",
"_____no_output_____"
],
[
"pd.isna(azdias).sum().sort_values(ascending=False)[:10]",
"_____no_output_____"
],
[
"customers.head()",
"_____no_output_____"
],
[
"pd.isna(customers).sum().sort_values(ascending=False)[:10]",
"_____no_output_____"
],
[
"azdias.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 37436 entries, 0 to 37435\nColumns: 366 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(289), int64(71), object(6)\nmemory usage: 104.8+ MB\n"
],
[
"customers.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 39545 entries, 0 to 39544\nColumns: 369 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(276), int64(85), object(8)\nmemory usage: 111.6+ MB\n"
]
],
[
[
"## Data Preprocessing - Handling missing values",
"_____no_output_____"
],
[
"We notice there are mismatches in Azdias and Customers file. We need to first align the features of these two datasets.",
"_____no_output_____"
]
],
[
[
"mismatch_col = set(customers.columns) - set(azdias.columns)\nmismatch_col",
"_____no_output_____"
],
[
"customers.drop(mismatch_col, axis=1, inplace = True)\ncustomers.shape",
"_____no_output_____"
]
],
[
[
"Read in attributes files",
"_____no_output_____"
]
],
[
[
"! pip install openpyxl",
"Requirement already satisfied: openpyxl in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (3.0.6)\r\nRequirement already satisfied: jdcal in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from openpyxl) (1.4.1)\r\nRequirement already satisfied: et-xmlfile in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from openpyxl) (1.0.1)\r\n"
],
[
"import openpyxl\n\ninfo_levels = pd.read_excel(\n './DIAS Information Levels - Attributes 2017.xlsx',\n engine='openpyxl',\n index_col = 0,\n header=1\n)\ninfo_levels.index = np.arange(len(info_levels))\n\nattributes = pd.read_excel(\n './DIAS Attributes - Values 2017.xlsx',\n engine='openpyxl',\n index_col = 0,\n header = 1\n)\nattributes.index = np.arange(len(attributes))",
"_____no_output_____"
],
[
"info_levels.head()",
"_____no_output_____"
],
[
"attributes.head()",
"_____no_output_____"
],
[
"print('There are {} attributes in Azdias file not explained by the Attribute file'.format(len(set(azdias.columns.unique()) - set(attributes['Attribute'].unique()))))\nprint('There are {} attributes in Customers file not explained by the Attribute file'.format(len(set(customers.columns.unique()) - set(attributes['Attribute'].unique()))))",
"There are 94 attributes in Azdias file not explained by the Attribute file\nThere are 94 attributes in Customers file not explained by the Attribute file\n"
],
[
"attributes ",
"_____no_output_____"
]
],
[
[
"Check if there is any invalid attributes in Attribute file. <br>",
"_____no_output_____"
]
],
[
[
"attributes['Attribute'] = attributes['Attribute'].ffill()\nattributes['Attribute'].value_counts()",
"_____no_output_____"
]
],
[
[
"We notice there are few attributes with only 1 value. Let's take a look into that.",
"_____no_output_____"
]
],
[
[
"attribute_col = attributes['Attribute'].value_counts()[attributes['Attribute'].value_counts() == 1].index\nattributes.loc[attributes['Attribute'].isin(attribute_col)]",
"_____no_output_____"
]
],
[
[
"It seems fine. Next step, we need to explore the 'unkown' and 'no classification possible' values.",
"_____no_output_____"
]
],
[
[
"unkown_attribute = attributes[attributes['Meaning'].str.contains(\"unknown|no \").astype(bool)]\nunkown_attribute.head(10)",
"_____no_output_____"
],
[
"unknown_values = unkown_attribute[['Attribute', 'Value']]\nunknown_values.loc[:, 'Value'] = unknown_values.loc[:, 'Value'].astype(str)\nunknown_values = unknown_values.groupby('Attribute')['Value'].apply(', '.join).reset_index()\nunknown_values.loc[:, 'Value'] = unknown_values.loc[:, 'Value'].str.split(', ')\nunknown_values.head()",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/pandas/core/indexing.py:1781: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n self.obj[item_labels[indexer[info_axis]]] = value\n"
],
[
"# convert the unknown values in azdias to NaNs\nfor att in unknown_values['Attribute']:\n if att in azdias.columns:\n azdias_unknown = azdias.loc[:, att].isin(unknown_values.loc[unknown_values['Attribute']==att, 'Value'].iloc[0])\n azdias.loc[azdias_unknown, att] = np.nan\n else:\n continue\nazdias.head()",
"_____no_output_____"
],
[
"# convert the unknown values in customers to NaNs\nfor att in unknown_values['Attribute']:\n if att in customers.columns:\n azdias_unknown = customers.loc[:, att].isin(unknown_values.loc[unknown_values['Attribute']==att, 'Value'].iloc[0])\n customers.loc[azdias_unknown, att] = np.nan\n else:\n continue\ncustomers.head()",
"_____no_output_____"
]
],
[
[
"We noticed there are many columns with NaN, we need to explore the missing values for both datasets. <br>\nBelow are the attributes that have over 30% of NaN values",
"_____no_output_____"
]
],
[
[
"cols = azdias.columns\nazdias_na = pd.isna(azdias).sum() / len(azdias)\ncol_names_azdias = azdias_na[azdias_na > 0.3].index\ndata = [i for i in azdias_na if i >=0.3]\ny_pos = np.arange(len(col_names_azdias))\nprint(len(data), len(y_pos))\nf, ax = plt.subplots(figsize=(5,8))\nplt.barh(y_pos, data, align='center', alpha=0.5)\nplt.yticks(y_pos, col_names_azdias)\nplt.xlabel('percentage (%)')\nplt.title('Percentage of nans in each columns of azdias')\nplt.show()",
"41 41\n"
]
],
[
[
"Drop the colums with missing values over 30%",
"_____no_output_____"
]
],
[
[
"azdias.drop(col_names_azdias, axis=1, inplace = True)",
"_____no_output_____"
]
],
[
[
"We then need to look into the missing values for every row in both datasets. ",
"_____no_output_____"
]
],
[
[
"# find proportion of missing value in each row\n\nmissing_row_azdias = pd.isna(azdias).sum(axis=1) / azdias.shape[1]\nmissing_row_azdias.hist()\nplt.xlabel('Proportion of missing values')\nplt.ylabel('Counts')\nplt.title('Distribution of missing values per row in Azdias Data')",
"_____no_output_____"
]
],
[
[
"According to the above figures, the proportion of the missing values in the majority of rows is below 30%. Therefore, we can remove rows containing over 30% empty values. ",
"_____no_output_____"
]
],
[
[
"azdias.drop(missing_row_azdias[missing_row_azdias > 0.3].index, axis=0, inplace=True)",
"_____no_output_____"
],
[
"azdias.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 33461 entries, 1 to 37434\nColumns: 325 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(279), int64(40), object(6)\nmemory usage: 83.2+ MB\n"
]
],
[
[
"## Transform Data Type",
"_____no_output_____"
],
[
"From above information, there are 6 object data types, we need to transform the object to correct data type",
"_____no_output_____"
]
],
[
[
"obj_colname = []\nfor col in azdias.columns:\n if 'object' in str(azdias[col].dtype):\n print(col)\n obj_colname.append(col)",
"CAMEO_DEU_2015\nCAMEO_DEUG_2015\nCAMEO_INTL_2015\nD19_LETZTER_KAUF_BRANCHE\nEINGEFUEGT_AM\nOST_WEST_KZ\n"
],
[
"azdias[obj_colname]",
"_____no_output_____"
],
[
"azdias['CAMEO_DEUG_2015'].value_counts()",
"_____no_output_____"
],
[
"# change X to NaN\nazdias['CAMEO_DEUG_2015'].replace('X', np.nan, inplace=True)",
"_____no_output_____"
],
[
"azdias['CAMEO_INTL_2015'].value_counts()",
"_____no_output_____"
],
[
"# change XX to np.nan\nazdias['CAMEO_INTL_2015'].replace('XX', np.nan, inplace=True)",
"_____no_output_____"
],
[
"azdias[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']] = azdias[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']].astype(float)",
"_____no_output_____"
],
[
"azdias['OST_WEST_KZ'].value_counts()",
"_____no_output_____"
],
[
"attributes.loc[attributes['Attribute'] == 'OST_WEST_KZ']",
"_____no_output_____"
],
[
"# encode OST_WEST_KZ to 0 (O) and 1 (W)\nazdias['OST_WEST_KZ'].replace({'W': 1, 'O': 0}, inplace=True)",
"_____no_output_____"
],
[
"attributes.loc[attributes['Attribute'] == 'EINGEFUEGT_AM']",
"_____no_output_____"
],
[
"attributes.loc[attributes['Attribute'] == 'D19_LETZTER_KAUF_BRANCHE']",
"_____no_output_____"
],
[
"attributes.loc[attributes['Attribute'] == 'CAMEO_DEU_2015']",
"_____no_output_____"
],
[
"azdias['CAMEO_DEU_2015'].value_counts()",
"_____no_output_____"
],
[
"# encode CAMEO_DEU_2015 \nazdias['CAMEO_DEU_2015'].replace('XX', np.nan, inplace=True)\ndeu_values = attributes.loc[attributes['Attribute'] == 'CAMEO_DEU_2015', 'Value']\ndeu_values.index = np.arange(len(deu_values))\ndeu_dict = deu_values.to_dict()\ndeu_dict = dict(zip(deu_dict.values(), deu_dict.keys()))\nazdias.replace({'CAMEO_DEU_2015': deu_dict}, inplace=True)\nazdias['CAMEO_DEU_2015'].value_counts()",
"_____no_output_____"
]
],
[
[
"Since we cannot find meaning for 'EINGEFUEGT_AM' and 'D19_LETZTER_KAUF_BRANCHE' in Attribute file, we will remove these two columns ",
"_____no_output_____"
]
],
[
[
"azdias.drop(['EINGEFUEGT_AM', 'D19_LETZTER_KAUF_BRANCHE'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"### Apply the similar procedure to Customer file",
"_____no_output_____"
]
],
[
[
"customers.drop(col_names_azdias, axis=1, inplace = True)\ncustomers.drop(missing_row_azdias[missing_row_azdias > 0.3].index, axis=0, inplace=True)\ncustomers['CAMEO_DEUG_2015'].replace('X', np.nan, inplace=True)\ncustomers['CAMEO_INTL_2015'].replace('XX', np.nan, inplace=True)\ncustomers[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']] = customers[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']].astype(float)\ncustomers['OST_WEST_KZ'].replace({'W': 1, 'O': 0}, inplace=True)\n\n# encode CAMEO_DEU_2015 \ncustomers['CAMEO_DEU_2015'].replace('XX', np.nan, inplace=True)\ncustomers.replace({'CAMEO_DEU_2015': deu_dict}, inplace=True)\n\ncustomers.drop(['EINGEFUEGT_AM', 'D19_LETZTER_KAUF_BRANCHE'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"azdias.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 33461 entries, 1 to 37434\nColumns: 323 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(282), int64(41)\nmemory usage: 82.7 MB\n"
],
[
"customers.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 35570 entries, 1 to 39544\nColumns: 323 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(282), int64(41)\nmemory usage: 87.9 MB\n"
]
],
[
[
"## Imputing and scaling",
"_____no_output_____"
]
],
[
[
"from sklearn.impute import SimpleImputer\n\nimputer = SimpleImputer()\n\nazdias_col = azdias.columns\nazdias = imputer.fit_transform(azdias)\nazdias = pd.DataFrame(azdias, columns=azdias_col)\n\ncustomers_col = customers.columns\ncustomers = imputer.fit_transform(customers)\ncustomers = pd.DataFrame(customers, columns=customers_col)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import StandardScaler\n\nscaler = StandardScaler()\n\nazdias = scaler.fit_transform(azdias)\nazdias = pd.DataFrame(azdias, columns=azdias_col)\n\ncustomers = scaler.fit_transform(customers)\ncustomers = pd.DataFrame(customers, columns=customers_col)",
"_____no_output_____"
]
],
[
[
"## Part 1: Customer Segmentation Report\n\nThe main bulk of your analysis will come in this part of the project. Here, you should use unsupervised learning techniques to describe the relationship between the demographics of the company's existing customers and the general population of Germany. By the end of this part, you should be able to describe parts of the general population that are more likely to be part of the mail-order company's main customer base, and which parts of the general population are less so.",
"_____no_output_____"
],
[
"### PCA",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\n\npca_fit = PCA().fit(azdias)\npca_transform = pca_fit.transform(azdias)\n\nfeatures = np.arange(pca_fit.n_components_)\nplt.bar(features, pca_fit.explained_variance_ratio_)\nplt.xlabel('PCA features')\nplt.ylabel('variance %')\nplt.xlim(left = 0, right = 100)",
"_____no_output_____"
],
[
"explained_var = pca_fit.explained_variance_ratio_\nplt.plot(np.cumsum(explained_var))\nplt.xlabel('number of components')\nplt.ylabel('cumulative variance explained')",
"_____no_output_____"
],
[
"len(explained_var.cumsum()) - (explained_var.cumsum() > 0.90).sum()",
"_____no_output_____"
]
],
[
[
"If we have 277 components, then the explained variance will exceed 99%",
"_____no_output_____"
]
],
[
[
"n = 166\nazdias_pca_fit = PCA(n).fit(azdias)\nazdias_pca_transform = azdias_pca_fit.transform(azdias)\n\nplt.plot(np.cumsum(azdias_pca_fit.explained_variance_ratio_))\nplt.xlabel('number of components')\nplt.ylabel('cumulative variance explained')",
"_____no_output_____"
]
],
[
[
"### Clustering",
"_____no_output_____"
]
],
[
[
"customers_pca_transform = azdias_pca_fit.transform(customers)",
"_____no_output_____"
],
[
"from sklearn.cluster import KMeans\n\nscores = []\nfor i in range(1, 21):\n kmeans = KMeans(i)\n model = kmeans.fit(azdias_pca_transform)\n score = model.inertia_\n scores.append(score)",
"_____no_output_____"
],
[
"plt.plot(np.arange(20), scores, marker='o')\nplt.xlabel('Number of clusters')\nplt.ylabel('Distances')",
"_____no_output_____"
],
[
"azdias_kmeans = KMeans(8)\nazdias_model = azdias_kmeans.fit(azdias_pca_transform)\nazdias_predicted = azdias_model.predict(azdias_pca_transform)\n\nazdias_cluster = pd.Series(azdias_predicted)\nazdias_cluster = azdias_cluster.value_counts().sort_index()\nazdias_cluster_percentage = azdias_cluster / azdias_cluster.sum()\n\ncustomers_model = azdias_kmeans.fit(customers_pca_transform)\ncustomers_predicted = customers_model.predict(customers_pca_transform)\n\ncustomers_cluster = pd.Series(customers_predicted)\ncustomers_cluster = customers_cluster.value_counts().sort_index()\ncustomers_cluster_percentage = customers_cluster / customers_cluster.sum()\n\ndf = pd.DataFrame()\ndf['cluster'] = azdias_cluster_percentage\ndf['group'] = 'general_population'\n\ndf1 = pd.DataFrame()\ndf1['cluster'] = customers_cluster_percentage\ndf1['group'] = 'customers'\n\ndf_final = pd.concat([df, df1])",
"_____no_output_____"
],
[
"df_final = df_final.groupby('group')['cluster'].value_counts(normalize=True).mul(100)\ndf_final = df_final.rename('percent').reset_index()",
"_____no_output_____"
],
[
"bar1 = azdias_cluster_percentage.index\nbar2 = customers_cluster_percentage.index\nw = 0.4\nplt.bar(bar2, height = customers_cluster_percentage, width = w, label='AZDIAS')\nplt.bar(bar1 + w, height = azdias_cluster_percentage, width = w, label = 'customers')\n\nplt.xlabel(\"Cluster Number\")\nplt.ylabel(\"Percentage\")\nplt.title(\"Percentage of clusters in each population\")\nplt.legend()",
"_____no_output_____"
]
],
[
[
"## Part 2: Supervised Learning Model\n\nNow that you've found which parts of the population are more likely to be customers of the mail-order company, it's time to build a prediction model. Each of the rows in the \"MAILOUT\" data files represents an individual that was targeted for a mailout campaign. Ideally, we should be able to use the demographic information from each individual to decide whether or not it will be worth it to include that person in the campaign.\n\nThe \"MAILOUT\" data has been split into two approximately equal parts, each with almost 43 000 data rows. In this part, you can verify your model with the \"TRAIN\" partition, which includes a column, \"RESPONSE\", that states whether or not a person became a customer of the company following the campaign. In the next part, you'll need to create predictions on the \"TEST\" partition, where the \"RESPONSE\" column has been withheld.",
"_____no_output_____"
]
],
[
[
"mailout_train = pd.read_csv('./Udacity_MAILOUT_052018_TRAIN.csv', sep=',', index_col=0)",
"_____no_output_____"
],
[
"mailout_train.head()",
"_____no_output_____"
],
[
"mailout_train['RESPONSE']",
"_____no_output_____"
],
[
"plt.hist(mailout_train['RESPONSE'])\nplt.xlabel('Responses')\nplt.ylabel('Counts')\nplt.title('Responses from Mailout')",
"_____no_output_____"
]
],
[
[
"The data from Mailout-train is highly imbalanced",
"_____no_output_____"
],
[
"We need to clean data for mailout-train and mailout-test",
"_____no_output_____"
]
],
[
[
"def convert_nan(df, unknown_values):\n for att in unknown_values['Attribute']:\n if att in df.columns:\n azdias_unknown = df.loc[:, att].isin(unknown_values.loc[unknown_values['Attribute']==att, 'Value'].iloc[0])\n df.loc[azdias_unknown, att] = np.nan\n else:\n continue\n\n return df\n",
"_____no_output_____"
],
[
"def drop_missing(df, col_names_azdias):\n df.drop(col_names_azdias, axis=1, inplace = True)\n \n missing_row = pd.isna(df).sum(axis=1) / df.shape[1]\n df.drop(missing_row[missing_row > 0.3].index, axis=0, inplace=True)\n \n return df",
"_____no_output_____"
],
[
"def encode_data(df, deu_dict):\n \n df['CAMEO_DEUG_2015'].replace('X', np.nan, inplace=True)\n df['CAMEO_INTL_2015'].replace('XX', np.nan, inplace=True)\n df[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']] = df[['CAMEO_DEUG_2015', 'CAMEO_INTL_2015']].astype(float)\n df['OST_WEST_KZ'].replace({'W': 1, 'O': 0}, inplace=True)\n\n # encode CAMEO_DEU_2015 \n df['CAMEO_DEU_2015'].replace('XX', np.nan, inplace=True)\n df.replace({'CAMEO_DEU_2015': deu_dict}, inplace=True)\n\n df.drop(['EINGEFUEGT_AM', 'D19_LETZTER_KAUF_BRANCHE'], axis=1, inplace=True)\n \n return df",
"_____no_output_____"
],
[
"mailout_train = convert_nan(mailout_train, unknown_values)\nmailout_train = drop_missing(mailout_train, col_names_azdias)\nmailout_train = encode_data(mailout_train, deu_dict)",
"_____no_output_____"
],
[
"mailout_train.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 22473 entries, 0 to 27509\nColumns: 324 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(319), int64(5)\nmemory usage: 55.7 MB\n"
],
[
"X = mailout_train.drop('RESPONSE', axis=1)\ny = mailout_train['RESPONSE']",
"_____no_output_____"
],
[
"# impute NaNs \nX = pd.DataFrame(imputer.fit_transform(X), columns=X.columns)\n\n# feature scaling\nX = pd.DataFrame(scaler.fit_transform(X), columns=X.columns)",
"_____no_output_____"
],
[
"!pip install lightgbm",
"Collecting lightgbm\n Downloading lightgbm-3.3.2-py3-none-manylinux1_x86_64.whl (2.0 MB)\n |████████████████████████████████| 2.0 MB 21.4 MB/s \n\u001b[?25hRequirement already satisfied: scikit-learn!=0.22.0 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from lightgbm) (0.24.1)\nRequirement already satisfied: scipy in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from lightgbm) (1.5.3)\nRequirement already satisfied: numpy in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from lightgbm) (1.19.5)\nRequirement already satisfied: wheel in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from lightgbm) (0.36.2)\nRequirement already satisfied: joblib>=0.11 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from scikit-learn!=0.22.0->lightgbm) (1.0.1)\nRequirement already satisfied: threadpoolctl>=2.0.0 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from scikit-learn!=0.22.0->lightgbm) (2.1.0)\nInstalling collected packages: lightgbm\nSuccessfully installed lightgbm-3.3.2\n"
],
[
"from sklearn.ensemble import GradientBoostingClassifier\nfrom sklearn.ensemble import RandomForestClassifier\nimport xgboost as xgb\nfrom sklearn.model_selection import GridSearchCV\nimport lightgbm as lgb\nimport catboost\nfrom sklearn.ensemble import AdaBoostClassifier",
"_____no_output_____"
],
[
"model_CBR = catboost.CatBoostRegressor()\n",
"_____no_output_____"
],
[
"bt = xgb.XGBClassifier(use_label_encoder=False, objective='binary:logistic') # Setup xgboost model\nbt.fit(X, y, verbose=False)",
"[21:17:24] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n"
],
[
"clf = lgb.LGBMClassifier()\n",
"_____no_output_____"
],
[
"def classifier(estimator, param_grid, X=X, y=y):\n grid = GridSearchCV(estimator=estimator, param_grid=param_grid, scoring='roc_auc', cv=5)\n grid.fit(X, y)\n print('Estimator:', grid.best_estimator_)\n print('Score:', grid.best_score_)\n \n return grid.best_score_, grid.best_estimator_",
"_____no_output_____"
],
[
"estimators = [GradientBoostingClassifier(), RandomForestClassifier(), bt, model_CBR, clf, AdaBoostClassifier()]\nscores = []\nbest_paras = []\nfor estimator in estimators:\n score, paras = classifier(estimator, {})\n scores.append(score)\n best_paras.append(paras)",
"Estimator: GradientBoostingClassifier(random_state=0)\nScore: 0.7828728816159398\nEstimator: RandomForestClassifier(random_state=0)\nScore: 0.6055916377001709\n[22:04:25] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n[22:04:37] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n[22:04:50] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n[22:05:02] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n[22:05:17] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\n[22:05:29] WARNING: ../src/learner.cc:1115: Starting in XGBoost 1.3.0, the default evaluation metric used with the objective 'binary:logistic' was changed from 'error' to 'logloss'. Explicitly set eval_metric if you'd like to restore the old behavior.\nEstimator: XGBClassifier(base_score=0.5, booster='gbtree', colsample_bylevel=1,\n colsample_bynode=1, colsample_bytree=1, enable_categorical=False,\n gamma=0, gpu_id=-1, importance_type=None,\n interaction_constraints='', learning_rate=0.300000012,\n max_delta_step=0, max_depth=6, min_child_weight=1, missing=nan,\n monotone_constraints='()', n_estimators=100, n_jobs=2,\n num_parallel_tree=1, predictor='auto', random_state=0,\n reg_alpha=0, reg_lambda=1, scale_pos_weight=1, subsample=1,\n tree_method='exact', use_label_encoder=False,\n validate_parameters=1, verbosity=None)\nScore: 0.7458473768523899\nLearning rate set to 0.064629\n0:\tlearn: 0.1143586\ttotal: 15ms\tremaining: 15s\n1:\tlearn: 0.1142048\ttotal: 32.1ms\tremaining: 16s\n2:\tlearn: 0.1140291\ttotal: 48.2ms\tremaining: 16s\n"
],
[
"pd.DataFrame({\n 'classifier': ['GradientBoosting', 'RandomForest', 'XGBoosting', 'CatBoost', 'LightGBM', 'AdaBoost'],\n 'ROC_AUC': scores})",
"_____no_output_____"
],
[
"estimator = GradientBoostingClassifier()\nparam_grid = {\n 'learning_rate': [0.01, 0.1, 1],\n 'n_estimators': [10, 100, 200]\n}\nbest_estimator = classifier(estimator, param_grid)",
"Estimator: GradientBoostingClassifier(learning_rate=0.01, n_estimators=200)\nScore: 0.7846704294359511\n"
],
[
"estimator = clf\nbest_estimator = classifier(estimator, param_grid)",
"Estimator: LGBMClassifier(learning_rate=0.01, n_estimators=200)\nScore: 0.7890216713009354\n"
],
[
"best_estimator[1]",
"_____no_output_____"
]
],
[
[
"LightGBM has higher score, we choose LightGBM as our final classifier",
"_____no_output_____"
]
],
[
[
"#grid = GridSearchCV(estimator=clf, param_grid={'learning_rate': 0.01, 'n_estimators': 200}, scoring='roc_auc', cv=5)\nmodel_pred = lgb.LGBMClassifier(learning_rate=0.01, n_estimators=200, eval_metric='roc_auc')\nmodel_pred.fit(X, y)",
"_____no_output_____"
]
],
[
[
"## Part 3: Kaggle submission",
"_____no_output_____"
]
],
[
[
"mailout_test = pd.read_csv('./Udacity_MAILOUT_052018_TEST.csv', sep=',', index_col=0)",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/IPython/core/interactiveshell.py:3072: DtypeWarning: Columns (19,20) have mixed types.Specify dtype option on import or set low_memory=False.\n interactivity=interactivity, compiler=compiler, result=result)\n"
],
[
"mailout_test = convert_nan(mailout_test, unknown_values)\nmailout_test.drop(col_names_azdias, axis=1, inplace = True)\nmailout_test = encode_data(mailout_test, deu_dict)",
"_____no_output_____"
],
[
"mailout_test.info()",
"<class 'pandas.core.frame.DataFrame'>\nInt64Index: 18669 entries, 0 to 18668\nColumns: 323 entries, LNR to ALTERSKATEGORIE_GROB\ndtypes: float64(283), int64(40)\nmemory usage: 46.1 MB\n"
],
[
"mailout_pred = model_pred.predict_proba(mailout_test)",
"_____no_output_____"
],
[
"df = pd.DataFrame({'LNR': mailout_test['LNR'], 'RESPONSE': mailout_pred[:, 1]})",
"_____no_output_____"
],
[
"df.to_csv('submission.csv', index=False)",
"_____no_output_____"
],
[
"from sklearn.linear_model import LogisticRegression\nclassifier(LogisticRegression(), {})",
"/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/sklearn/linear_model/_logistic.py:765: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/sklearn/linear_model/_logistic.py:765: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/sklearn/linear_model/_logistic.py:765: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/sklearn/linear_model/_logistic.py:765: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n/home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages/sklearn/linear_model/_logistic.py:765: ConvergenceWarning: lbfgs failed to converge (status=1):\nSTOP: TOTAL NO. of ITERATIONS REACHED LIMIT.\n\nIncrease the number of iterations (max_iter) or scale the data as shown in:\n https://scikit-learn.org/stable/modules/preprocessing.html\nPlease also refer to the documentation for alternative solver options:\n https://scikit-learn.org/stable/modules/linear_model.html#logistic-regression\n extra_warning_msg=_LOGISTIC_SOLVER_CONVERGENCE_MSG)\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6be65f567f5c2d90b9e3d8157452f9ad573a57 | 493,320 | ipynb | Jupyter Notebook | iris_data_notebook.ipynb | kalyons11/notebooks | 1bf568b9d390d978419fee62f864c086d199f20f | [
"MIT"
]
| null | null | null | iris_data_notebook.ipynb | kalyons11/notebooks | 1bf568b9d390d978419fee62f864c086d199f20f | [
"MIT"
]
| null | null | null | iris_data_notebook.ipynb | kalyons11/notebooks | 1bf568b9d390d978419fee62f864c086d199f20f | [
"MIT"
]
| null | null | null | 311.046658 | 109,236 | 0.919468 | [
[
[
"# Iris Data Notebook",
"_____no_output_____"
],
[
"All code taken from Randal S. Olson's [tutorial](https://github.com/rhiever/Data-Analysis-and-Machine-Learning-Projects/blob/master/example-data-science-notebook/Example%20Machine%20Learning%20Notebook.ipynb).",
"_____no_output_____"
],
[
"## 1. Answering the question.",
"_____no_output_____"
],
[
"> Did you specify the type of data analytic question (e.g. exploration, association causality) before touching the data?\n\n> Did you define the metric for success before beginning?\n\n> Did you record the experimental design?\n\n> Did you consider whether the question could be answered with the available data?",
"_____no_output_____"
],
[
"## 2. Checking the data. ",
"_____no_output_____"
],
[
"- Is there anything wrong with the data?\n- Are there any quirks with the data?\n- Do I need to fix or remove any of the data?",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n\niris_data = pd.read_csv('data/iris-data.csv')\niris_data.head()",
"_____no_output_____"
]
],
[
[
"Need to look for missing data!!!\n\nLet's fill in the missing NA values.",
"_____no_output_____"
]
],
[
[
"iris_data = pd.read_csv('data/iris-data.csv', na_values=['NA'])",
"_____no_output_____"
]
],
[
[
"Now, we can print some stat summaries.",
"_____no_output_____"
]
],
[
[
"iris_data.describe()",
"_____no_output_____"
]
],
[
[
"Now, we know that five `petal_width_cm` entries are missing.\n\nBut, numbers don't always help that much. So, let's plot some things instead.",
"_____no_output_____"
]
],
[
[
"# Show plots inside notebook\n%matplotlib inline\n\nimport matplotlib.pyplot as plt\nimport seaborn as sb\n\n# Create scatterplot matrix - dist along diagonal, then scatters for every pair of classes\nsb.pairplot(iris_data.dropna(), hue='class')",
"_____no_output_____"
]
],
[
[
"We can already see 3 issues with our data.\n\n1. There are five classes when there should only be three, meaning there were some coding errors.\n\n2. There are some clear outliers in the measurements that may be erroneous: one `sepal_width_cm` entry for Iris-setosa falls well outside its normal range, and several `sepal_length_cm entries` for Iris-versicolor are near-zero for some reason.\n\n3. We had to drop those rows with missing values.",
"_____no_output_____"
],
[
"## 3. Tidying the data.",
"_____no_output_____"
]
],
[
[
"# Fix class type issues\niris_data.loc[iris_data['class'] == 'versicolor', 'class'] = 'Iris-versicolor'\niris_data.loc[iris_data['class'] == 'Iris-setossa', 'class'] = 'Iris-setosa'\n\niris_data['class'].unique()",
"_____no_output_____"
],
[
"# This line drops any 'Iris-setosa' rows with a separal width less than 2.5 cm\niris_data = iris_data.loc[(iris_data['class'] != 'Iris-setosa') | (iris_data['sepal_width_cm'] >= 2.5)]\niris_data.loc[iris_data['class'] == 'Iris-setosa', 'sepal_width_cm'].hist()",
"_____no_output_____"
],
[
"# Address near-zero petal lengths\niris_data.loc[(iris_data['class'] == 'Iris-versicolor') &\n (iris_data['sepal_length_cm'] < 1.0)]",
"_____no_output_____"
]
],
[
[
"Interesting - all off by 100! Probably didn't convert to cm.",
"_____no_output_____"
]
],
[
[
"iris_data.loc[(iris_data['class'] == 'Iris-versicolor') &\n (iris_data['sepal_length_cm'] < 1.0),\n 'sepal_length_cm'] *= 100.0\n\niris_data.loc[iris_data['class'] == 'Iris-versicolor', 'sepal_length_cm'].hist()",
"_____no_output_____"
]
],
[
[
"Now, we need to deal with the rows with missing values.",
"_____no_output_____"
]
],
[
[
"iris_data.loc[(iris_data['sepal_length_cm'].isnull()) |\n (iris_data['sepal_width_cm'].isnull()) |\n (iris_data['petal_length_cm'].isnull()) |\n (iris_data['petal_width_cm'].isnull())]",
"_____no_output_____"
],
[
"# Let's examine the mean\niris_data.loc[iris_data['class'] == 'Iris-setosa', 'petal_width_cm'].hist()",
"_____no_output_____"
],
[
"# Update to have the mean value\naverage_petal_width = iris_data.loc[iris_data['class'] == 'Iris-setosa', 'petal_width_cm'].mean()\n\niris_data.loc[(iris_data['class'] == 'Iris-setosa') &\n (iris_data['petal_width_cm'].isnull()),\n 'petal_width_cm'] = average_petal_width\n\niris_data.loc[(iris_data['class'] == 'Iris-setosa') &\n (iris_data['petal_width_cm'] == average_petal_width)]",
"_____no_output_____"
]
],
[
[
"Make sure our data is good now!",
"_____no_output_____"
]
],
[
[
"iris_data.loc[(iris_data['sepal_length_cm'].isnull()) |\n (iris_data['sepal_width_cm'].isnull()) |\n (iris_data['petal_length_cm'].isnull()) |\n (iris_data['petal_width_cm'].isnull())]",
"_____no_output_____"
],
[
"# Save fresh data and re-plot\niris_data.to_csv('data/iris-data-clean.csv', index=False)\niris_data_clean = pd.read_csv('data/iris-data-clean.csv')\nsb.pairplot(iris_data_clean, hue='class')",
"_____no_output_____"
]
],
[
[
"Here are some good general takeaways:\n\n- Make sure your data is encoded properly\n\n- Make sure your data falls within the expected range, and use domain knowledge whenever possible to define that expected range\n\n- Deal with missing data in one way or another: replace it if you can or drop it\n\n- Never tidy your data manually because that is not easily reproducible\n\n- Use code as a record of how you tidied your data\n\n- Plot everything you can about the data at this stage of the analysis so you can visually confirm everything looks correct",
"_____no_output_____"
],
[
"## 4. Exploratory analysis. ",
"_____no_output_____"
],
[
"- How is my data distributed?\n\n- Are there any correlations in my data?\n\n- Are there any confounding factors that explain these correlations?",
"_____no_output_____"
]
],
[
[
"sb.pairplot(iris_data_clean, hue='class')",
"_____no_output_____"
]
],
[
[
"- Correlations between petal length/width and sepal length/width.\n- Can classify well using only petal info.\n- Normal distributions.",
"_____no_output_____"
]
],
[
[
"# Make some violin plots\nplt.figure(figsize=(10, 10))\n\nfor column_index, column in enumerate(iris_data_clean.columns):\n if column == 'class':\n continue\n plt.subplot(2, 2, column_index + 1)\n sb.violinplot(x='class', y=column, data=iris_data_clean)",
"_____no_output_____"
]
],
[
[
"## 5. Classification. ",
"_____no_output_____"
]
],
[
[
"# Convert data to proper format with matrices\niris_data_clean = pd.read_csv('data/iris-data-clean.csv')\n\n# We're using all four measurements as inputs\n# Note that scikit-learn expects each entry to be a list of values, e.g.,\n# [ [val1, val2, val3],\n# [val1, val2, val3],\n# ... ]\n# such that our input data set is represented as a list of lists\n\n# We can extract the data in this format from pandas like this:\nall_inputs = iris_data_clean[['sepal_length_cm', 'sepal_width_cm',\n 'petal_length_cm', 'petal_width_cm']].values\n\n# Similarly, we can extract the classes\nall_classes = iris_data_clean['class'].values\n\n# Make sure that you don't mix up the order of the entries\n# all_inputs[5] inputs should correspond to the class in all_classes[5]\n\n# Here's what a subset of our inputs looks like:\nall_inputs[:5]",
"_____no_output_____"
],
[
"# Split data\nfrom sklearn.model_selection import train_test_split\n\n(training_inputs,\n testing_inputs,\n training_classes,\n testing_classes) = train_test_split(all_inputs, all_classes, train_size=0.75, random_state=1)",
"/Users/Kevin/Documents/mit/freelance/notebooks/env/lib/python3.6/site-packages/sklearn/model_selection/_split.py:2026: FutureWarning: From version 0.21, test_size will always complement train_size unless both are specified.\n FutureWarning)\n"
]
],
[
[
"Let's make a basic decisision tree classifier.",
"_____no_output_____"
]
],
[
[
"from sklearn.tree import DecisionTreeClassifier\n\n# Create the classifier\ndecision_tree_classifier = DecisionTreeClassifier()\n\n# Train the classifier on the training set\ndecision_tree_classifier.fit(training_inputs, training_classes)\n\n# Validate the classifier on the testing set using classification accuracy\ndecision_tree_classifier.score(testing_inputs, testing_classes)",
"_____no_output_____"
]
],
[
[
"But, there's a catch to this number... Let's train a lot more times.",
"_____no_output_____"
]
],
[
[
"model_accuracies = []\n\nfor repetition in range(1000):\n (training_inputs,\n testing_inputs,\n training_classes,\n testing_classes) = train_test_split(all_inputs, all_classes, train_size=0.75)\n \n decision_tree_classifier = DecisionTreeClassifier()\n decision_tree_classifier.fit(training_inputs, training_classes)\n classifier_accuracy = decision_tree_classifier.score(testing_inputs, testing_classes)\n model_accuracies.append(classifier_accuracy)\n \nsb.distplot(model_accuracies)",
"/Users/Kevin/Documents/mit/freelance/notebooks/env/lib/python3.6/site-packages/sklearn/model_selection/_split.py:2026: FutureWarning: From version 0.21, test_size will always complement train_size unless both are specified.\n FutureWarning)\n"
]
],
[
[
"Let's run some 10 fold cross validation (stratified to maintain class balance) to avoid this **overfitting**.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom sklearn.cross_validation import StratifiedKFold\nfrom sklearn.cross_validation import cross_val_score\n\ndecision_tree_classifier = DecisionTreeClassifier()\n\n# cross_val_score returns a list of the scores, which we can visualize\n# to get a reasonable estimate of our classifier's performance\ncv_scores = cross_val_score(decision_tree_classifier, all_inputs, all_classes, cv=10)\nsb.distplot(cv_scores)\nplt.title('Average score: {}'.format(np.mean(cv_scores)))",
"_____no_output_____"
]
],
[
[
"Tuning some parameters using a grid search.",
"_____no_output_____"
]
],
[
[
"from sklearn.model_selection import GridSearchCV\n\ndecision_tree_classifier = DecisionTreeClassifier()\n\nparameter_grid = {'max_depth': [1, 2, 3, 4, 5],\n 'max_features': [1, 2, 3, 4]}\n\ncross_validation = StratifiedKFold(all_classes, n_folds=10)\n\ngrid_search = GridSearchCV(decision_tree_classifier,\n param_grid=parameter_grid,\n cv=cross_validation)\n\ngrid_search.fit(all_inputs, all_classes)\nprint('Best score: {}'.format(grid_search.best_score_))\nprint('Best parameters: {}'.format(grid_search.best_params_))",
"Best score: 0.959731543624161\nBest parameters: {'max_depth': 3, 'max_features': 4}\n"
]
],
[
[
"We can visualize the results of the gridsearch using a heatmap.",
"_____no_output_____"
]
],
[
[
"grid_visualization = []\n\nfor grid_pair in grid_search.grid_scores_:\n grid_visualization.append(grid_pair.mean_validation_score)\n \ngrid_visualization = np.array(grid_visualization)\ngrid_visualization.shape = (5, 4)\nsb.heatmap(grid_visualization, cmap='Blues')\nplt.xticks(np.arange(4) + 0.5, grid_search.param_grid['max_features'])\nplt.yticks(np.arange(5) + 0.5, grid_search.param_grid['max_depth'][::-1])\nplt.xlabel('max_features')\nplt.ylabel('max_depth')",
"_____no_output_____"
]
],
[
[
"Let's add some more parameters and re-search.",
"_____no_output_____"
]
],
[
[
"decision_tree_classifier = DecisionTreeClassifier()\n\nparameter_grid = {'criterion': ['gini', 'entropy'],\n 'splitter': ['best', 'random'],\n 'max_depth': [1, 2, 3, 4, 5],\n 'max_features': [1, 2, 3, 4]}\n\ncross_validation = StratifiedKFold(all_classes, n_folds=10)\n\ngrid_search = GridSearchCV(decision_tree_classifier,\n param_grid=parameter_grid,\n cv=cross_validation)\n\ngrid_search.fit(all_inputs, all_classes)\nprint('Best score: {}'.format(grid_search.best_score_))\nprint('Best parameters: {}'.format(grid_search.best_params_))",
"Best score: 0.9664429530201343\nBest parameters: {'criterion': 'entropy', 'max_depth': 3, 'max_features': 3, 'splitter': 'best'}\n"
],
[
"# Take the best classifier\ndecision_tree_classifier = grid_search.best_estimator_\ndecision_tree_classifier",
"_____no_output_____"
],
[
"# Visualize with GraphViz\nimport sklearn.tree as tree\nfrom sklearn.externals.six import StringIO\n\nwith open('iris_dtc.dot', 'w') as out_file:\n out_file = tree.export_graphviz(decision_tree_classifier, out_file=out_file)",
"_____no_output_____"
]
],
[
[
"## 6. Reproducibility. ",
"_____no_output_____"
]
],
[
[
"%load_ext watermark",
"_____no_output_____"
],
[
"%watermark -a 'Kevin Lyons' -nmv --packages numpy,pandas,sklearn,matplotlib,seaborn",
"Kevin Lyons Fri Dec 29 2017 \n\nCPython 3.6.4\nIPython 6.2.1\n\nnumpy 1.13.3\npandas 0.21.1\nsklearn 0.19.1\nmatplotlib 2.1.1\nseaborn 0.8.1\n\ncompiler : GCC 4.2.1 Compatible Clang 4.0.1 (tags/RELEASE_401/final)\nsystem : Darwin\nrelease : 17.3.0\nmachine : x86_64\nprocessor : i386\nCPU cores : 4\ninterpreter: 64bit\n"
]
],
[
[
"Now, it is nice if we convert our work into a real pipeline for later use.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas as pd\nimport seaborn as sb\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.cross_validation import train_test_split\nfrom sklearn.cross_validation import cross_val_score\n\n# We can jump directly to working with the clean data because we saved our cleaned data set\niris_data_clean = pd.read_csv('data/iris-data-clean.csv')\n\n# Testing our data: Our analysis will stop here if any of these assertions are wrong\n\n# We know that we should only have three classes\nassert len(iris_data_clean['class'].unique()) == 3\n\n# We know that sepal lengths for 'Iris-versicolor' should never be below 2.5 cm\nassert iris_data_clean.loc[iris_data_clean['class'] == 'Iris-versicolor', 'sepal_length_cm'].min() >= 2.5\n\n# We know that our data set should have no missing measurements\nassert len(iris_data_clean.loc[(iris_data_clean['sepal_length_cm'].isnull()) |\n (iris_data_clean['sepal_width_cm'].isnull()) |\n (iris_data_clean['petal_length_cm'].isnull()) |\n (iris_data_clean['petal_width_cm'].isnull())]) == 0\n\nall_inputs = iris_data_clean[['sepal_length_cm', 'sepal_width_cm',\n 'petal_length_cm', 'petal_width_cm']].values\n\nall_classes = iris_data_clean['class'].values\n\n# This is the classifier that came out of Grid Search\nrandom_forest_classifier = RandomForestClassifier(bootstrap=True, class_weight=None, criterion='gini',\n max_depth=None, max_features=3, max_leaf_nodes=None,\n min_samples_leaf=1, min_samples_split=2,\n min_weight_fraction_leaf=0.0, n_estimators=5, n_jobs=1,\n oob_score=False, random_state=None, verbose=0, warm_start=True)\n\n# All that's left to do now is plot the cross-validation scores\nrf_classifier_scores = cross_val_score(random_forest_classifier, all_inputs, all_classes, cv=10)\nsb.boxplot(rf_classifier_scores)\nsb.stripplot(rf_classifier_scores, jitter=True, color='white')\n\n# ...and show some of the predictions from the classifier\n(training_inputs,\n testing_inputs,\n training_classes,\n testing_classes) = train_test_split(all_inputs, all_classes, train_size=0.75)\n\nrandom_forest_classifier.fit(training_inputs, training_classes)\n\nfor input_features, prediction, actual in zip(testing_inputs[:10],\n random_forest_classifier.predict(testing_inputs[:10]),\n testing_classes[:10]):\n print('{}\\t-->\\t{}\\t(Actual: {})'.format(input_features, prediction, actual))",
"[ 5.6 2.9 3.6 1.3]\t-->\tIris-versicolor\t(Actual: Iris-versicolor)\n[ 4.8 3.4 1.6 0.25]\t-->\tIris-setosa\t(Actual: Iris-setosa)\n[ 6.8 2.8 4.8 1.4]\t-->\tIris-versicolor\t(Actual: Iris-versicolor)\n[ 6.8 3.2 5.9 2.3]\t-->\tIris-virginica\t(Actual: Iris-virginica)\n[ 7.7 2.6 6.9 2.3]\t-->\tIris-virginica\t(Actual: Iris-virginica)\n[ 5.7 2.6 3.5 1. ]\t-->\tIris-versicolor\t(Actual: Iris-versicolor)\n[ 4.8 3. 1.4 0.1]\t-->\tIris-setosa\t(Actual: Iris-setosa)\n[ 6.3 2.5 4.9 1.5]\t-->\tIris-versicolor\t(Actual: Iris-versicolor)\n[ 4.7 3.2 1.6 0.2]\t-->\tIris-setosa\t(Actual: Iris-setosa)\n[ 5.1 3.5 1.4 0.3]\t-->\tIris-setosa\t(Actual: Iris-setosa)\n"
]
],
[
[
"And that is all!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6be7a740ded3559a3765db436529bf8251cff0 | 12,811 | ipynb | Jupyter Notebook | PyTorch Materials/Intro-to-PyTorch/Part 8 - Transfer Learning (Exercises).ipynb | Kabongosalomon/AMMI-NLP-Sg- | cdcca8b989f20976ab7d68b980d7c46fbe1fbd66 | [
"Apache-2.0"
]
| 2 | 2020-11-30T17:29:25.000Z | 2020-12-30T14:11:43.000Z | PyTorch Materials/Intro-to-PyTorch/Part 8 - Transfer Learning (Exercises).ipynb | Gedeon-m-gedus/AMMI-Ghana-2019-2020 | 090426b16b39a4c07f39c5e43e6b730a5d5bc9bc | [
"Apache-2.0"
]
| null | null | null | PyTorch Materials/Intro-to-PyTorch/Part 8 - Transfer Learning (Exercises).ipynb | Gedeon-m-gedus/AMMI-Ghana-2019-2020 | 090426b16b39a4c07f39c5e43e6b730a5d5bc9bc | [
"Apache-2.0"
]
| null | null | null | 41.060897 | 662 | 0.562563 | [
[
[
"# Transfer Learning\n\nIn this notebook, you'll learn how to use pre-trained networks to solved challenging problems in computer vision. Specifically, you'll use networks trained on [ImageNet](http://www.image-net.org/) [available from torchvision](http://pytorch.org/docs/0.3.0/torchvision/models.html). \n\nImageNet is a massive dataset with over 1 million labeled images in 1000 categories. It's used to train deep neural networks using an architecture called convolutional layers. I'm not going to get into the details of convolutional networks here, but if you want to learn more about them, please [watch this](https://www.youtube.com/watch?v=2-Ol7ZB0MmU).\n\nOnce trained, these models work astonishingly well as feature detectors for images they weren't trained on. Using a pre-trained network on images not in the training set is called transfer learning. Here we'll use transfer learning to train a network that can classify our cat and dog photos with near perfect accuracy.\n\nWith `torchvision.models` you can download these pre-trained networks and use them in your applications. We'll include `models` in our imports now.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%config InlineBackend.figure_format = 'retina'\n\nimport matplotlib.pyplot as plt\n\nimport torch\nfrom torch import nn\nfrom torch import optim\nimport torch.nn.functional as F\nfrom torchvision import datasets, transforms, models",
"_____no_output_____"
]
],
[
[
"Most of the pretrained models require the input to be 224x224 images. Also, we'll need to match the normalization used when the models were trained. Each color channel was normalized separately, the means are `[0.485, 0.456, 0.406]` and the standard deviations are `[0.229, 0.224, 0.225]`.",
"_____no_output_____"
]
],
[
[
"data_dir = 'Cat_Dog_data'\n\n# TODO: Define transforms for the training data and testing data\ntrain_transforms = transforms.Compose([transforms.RandomRotation(30),\n transforms.RandomResizedCrop(224),\n transforms.RandomHorizontalFlip(),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406],\n [0.229, 0.224, 0.225])])\n\ntest_transforms = transforms.Compose([transforms.Resize(255),\n transforms.CenterCrop(224),\n transforms.ToTensor(),\n transforms.Normalize([0.485, 0.456, 0.406],\n [0.229, 0.224, 0.225])])\n\n\n# Pass transforms in here, then run the next cell to see how the transforms look\ntrain_data = datasets.ImageFolder(data_dir + '/train', transform=train_transforms)\ntest_data = datasets.ImageFolder(data_dir + '/test', transform=test_transforms)\n\ntrainloader = torch.utils.data.DataLoader(train_data, batch_size=64, shuffle=True)\ntestloader = torch.utils.data.DataLoader(test_data, batch_size=64)",
"_____no_output_____"
]
],
[
[
"We can load in a model such as [DenseNet](http://pytorch.org/docs/0.3.0/torchvision/models.html#id5). Let's print out the model architecture so we can see what's going on.",
"_____no_output_____"
]
],
[
[
"model = models.densenet121(pretrained=True)\nmodel",
"_____no_output_____"
]
],
[
[
"This model is built out of two main parts, the features and the classifier. The features part is a stack of convolutional layers and overall works as a feature detector that can be fed into a classifier. The classifier part is a single fully-connected layer `(classifier): Linear(in_features=1024, out_features=1000)`. This layer was trained on the ImageNet dataset, so it won't work for our specific problem. That means we need to replace the classifier, but the features will work perfectly on their own. In general, I think about pre-trained networks as amazingly good feature detectors that can be used as the input for simple feed-forward classifiers.",
"_____no_output_____"
]
],
[
[
"# Freeze parameters so we don't backprop through them\nfor param in model.parameters():\n param.requires_grad = False\n\nfrom collections import OrderedDict\nclassifier = nn.Sequential(OrderedDict([\n ('fc1', nn.Linear(1024, 500)),\n ('relu', nn.ReLU()),\n ('fc2', nn.Linear(500, 2)),\n ('output', nn.LogSoftmax(dim=1))\n ]))\n \nmodel.classifier = classifier",
"_____no_output_____"
]
],
[
[
"With our model built, we need to train the classifier. However, now we're using a **really deep** neural network. If you try to train this on a CPU like normal, it will take a long, long time. Instead, we're going to use the GPU to do the calculations. The linear algebra computations are done in parallel on the GPU leading to 100x increased training speeds. It's also possible to train on multiple GPUs, further decreasing training time.\n\nPyTorch, along with pretty much every other deep learning framework, uses [CUDA](https://developer.nvidia.com/cuda-zone) to efficiently compute the forward and backwards passes on the GPU. In PyTorch, you move your model parameters and other tensors to the GPU memory using `model.to('cuda')`. You can move them back from the GPU with `model.to('cpu')` which you'll commonly do when you need to operate on the network output outside of PyTorch. As a demonstration of the increased speed, I'll compare how long it takes to perform a forward and backward pass with and without a GPU.",
"_____no_output_____"
]
],
[
[
"import time",
"_____no_output_____"
],
[
"for device in ['cpu', 'cuda']:\n\n criterion = nn.NLLLoss()\n # Only train the classifier parameters, feature parameters are frozen\n optimizer = optim.Adam(model.classifier.parameters(), lr=0.001)\n\n model.to(device)\n\n for ii, (inputs, labels) in enumerate(trainloader):\n\n # Move input and label tensors to the GPU\n inputs, labels = inputs.to(device), labels.to(device)\n\n start = time.time()\n\n outputs = model.forward(inputs)\n loss = criterion(outputs, labels)\n loss.backward()\n optimizer.step()\n\n if ii==3:\n break\n \n print(f\"Device = {device}; Time per batch: {(time.time() - start)/3:.3f} seconds\")",
"_____no_output_____"
]
],
[
[
"You can write device agnostic code which will automatically use CUDA if it's enabled like so:\n```python\n# at beginning of the script\ndevice = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\n\n...\n\n# then whenever you get a new Tensor or Module\n# this won't copy if they are already on the desired device\ninput = data.to(device)\nmodel = MyModule(...).to(device)\n```\n\nFrom here, I'll let you finish training the model. The process is the same as before except now your model is much more powerful. You should get better than 95% accuracy easily.\n\n>**Exercise:** Train a pretrained models to classify the cat and dog images. Continue with the DenseNet model, or try ResNet, it's also a good model to try out first. Make sure you are only training the classifier and the parameters for the features part are frozen.",
"_____no_output_____"
]
],
[
[
"## TODO: Use a pretrained model to classify the cat and dog images\n\n# Use GPU if it's available\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\n\nmodel = models.densenet121(pretrained=True)\n\n# Freeze parameters so we don't backprop through them\nfor param in model.parameters():\n param.requires_grad = False\n \nmodel.classifier = nn.Sequential(nn.Linear(1024, 256),\n nn.ReLU(),\n nn.Dropout(0.2),\n nn.Linear(256, 2),\n nn.LogSoftmax(dim=1))\n\ncriterion = nn.NLLLoss()\n\n# Only train the classifier parameters, feature parameters are frozen\noptimizer = optim.Adam(model.classifier.parameters(), lr=0.003)\n\nmodel.to(device)",
"_____no_output_____"
],
[
"epochs = 1\nsteps = 0\nrunning_loss = 0\nprint_every = 5\nfor epoch in range(epochs):\n for inputs, labels in trainloader:\n steps += 1\n # Move input and label tensors to the default device\n inputs, labels = inputs.to(device), labels.to(device)\n \n optimizer.zero_grad()\n \n logps = model.forward(inputs)\n loss = criterion(logps, labels)\n loss.backward()\n optimizer.step()\n\n running_loss += loss.item()\n \n if steps % print_every == 0:\n test_loss = 0\n accuracy = 0\n model.eval()\n with torch.no_grad():\n for inputs, labels in testloader:\n inputs, labels = inputs.to(device), labels.to(device)\n logps = model.forward(inputs)\n batch_loss = criterion(logps, labels)\n \n test_loss += batch_loss.item()\n \n # Calculate accuracy\n ps = torch.exp(logps)\n top_p, top_class = ps.topk(1, dim=1)\n equals = top_class == labels.view(*top_class.shape)\n accuracy += torch.mean(equals.type(torch.FloatTensor)).item()\n \n print(f\"Epoch {epoch+1}/{epochs}.. \"\n f\"Train loss: {running_loss/print_every:.3f}.. \"\n f\"Test loss: {test_loss/len(testloader):.3f}.. \"\n f\"Test accuracy: {accuracy/len(testloader):.3f}\")\n running_loss = 0\n model.train()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6bf8ed10f55ddc6b5d05091eb2930d32d7244f | 158,801 | ipynb | Jupyter Notebook | coreference_resolution.ipynb | yxw-11/coreference-resolution | 19b5e72809d6ed6516bc44660dc384886249a62d | [
"MIT"
]
| null | null | null | coreference_resolution.ipynb | yxw-11/coreference-resolution | 19b5e72809d6ed6516bc44660dc384886249a62d | [
"MIT"
]
| null | null | null | coreference_resolution.ipynb | yxw-11/coreference-resolution | 19b5e72809d6ed6516bc44660dc384886249a62d | [
"MIT"
]
| null | null | null | 34.476987 | 576 | 0.482138 | [
[
[
"#IEEE Coreference Resolution Task\n##SetSimilaritySearch + Bert-base embedding semantic similarity",
"_____no_output_____"
],
[
"##Mount Google Drive",
"_____no_output_____"
]
],
[
[
"# Mount Google Drive\nfrom google.colab import drive\ndrive.mount('/content/drive/')",
"Mounted at /content/drive/\n"
]
],
[
[
"##Get Entities from NER task",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport json\nimport pandas as pd",
"_____no_output_____"
],
[
"def get_json_list(multiclass_file, software = 'doccano'):\n with open(multiclass_file, 'r') as json_file:\n json_list = list(json_file)\n\n annotations_json = []\n for line in json_list:\n annotations_json.append(json.loads(line))\n\n if software == 'prodigy':\n annotations_json = prodigy_to_doccano(annotations_json)\n\n return annotations_json",
"_____no_output_____"
],
[
"def multiclass_to_multilabel(abstract):\n one_hot = [0] * 4\n for annotation in abstract['annotations']:\n original_label = annotation['label'] \n one_hot_copy = one_hot.copy()\n one_hot_copy[original_label] = 1\n annotation['label_one_hot'] = one_hot_copy\n\n return abstract",
"_____no_output_____"
],
[
"multiclass_file = '/content/drive/MyDrive/ieee_ner_coref/assets/data/annotations/yx_converted_abs_combine_114.jsonl' # Input from Doccano/Prodigy",
"_____no_output_____"
],
[
"# Set software to 'doccano' or 'prodigy'\nannotations_json = get_json_list(multiclass_file, software = 'doccano')",
"_____no_output_____"
],
[
"for abstract in annotations_json:\n abstract = multiclass_to_multilabel(abstract)\n abstract['id'] = abstract['meta']['paperid']\n abstract['text'] = abstract['text'].strip('®')",
"_____no_output_____"
]
],
[
[
"##Define related classes",
"_____no_output_____"
]
],
[
[
"from enum import Enum\nclass EntityClass(Enum):\n '''Represents the class of an entity, e.g. \"Method\"'''\n NONE = 0\n ORG = 1\n METHOD = 2\n PRODUCT = 3\n\nclass Entity:\n '''An entity that has been identified as part of a document\n\n Attributes:\n parent_doc_id: an identifier for the document this entity exists within\n start: offset of the first character of this entity\n in the parent doc\n text: text from the parent document with this entity\n klass: class of this entity, e.g. \"Method\"\n '''\n def __init__(self,id,start,text,klass):\n parent_doc_id: int\n start: int\n text: str\n klass: EntityClass\n self.parent_doc_id = id\n self.start = start\n self.text = text\n self.klass = klass\n\n def __str__(self):\n return self.text\n\n @property\n def end(self):\n '''Offset of the last character of this entity in the parent doc'''\n return self.start + len(self.text)\n\n @property\n def location(self):\n return (self.start, self.end)",
"_____no_output_____"
]
],
[
[
"##Construct dataframe",
"_____no_output_____"
]
],
[
[
"#The context of entities\nfor abstract in annotations_json:\n for annotation in abstract['annotations']:\n if sum(np.asarray(annotation['label_one_hot'])) > 0:\n text = abstract['text']\n start = annotation['start_offset']\n end = annotation['end_offset']\n print(text[start:end])",
"simultaneous backward geocoding\nairborne InSAR\nmulti-aspect SAR\nVoting Logic Fusion\ncoarse to fine\nHaar-like\nHOG\nCAVIAR\nfull-body detection (FBD)\nhead-shoulder detection (HSD)\nFBD\nHSD\nMIT, INRIA\nbsp-tree\nbsp-tree\nBP neural network\nBP neural network\nBP neural networks\nhold-out\n10-fold cross validation\nSIFT\nsurveillance context (scale invariant image transform (SIFT) keypoints\n geometric primitive features\nLatent Semantic Indexing\nExpectation Maximization\nNormalized Score (ENS)\nlatent semantic indexing\nhyperspectral band grouping\nhyperspectral analysis\nspatial adaptivity\nbilateral filtering\nSIFT (scale invariant feature transform)\nSIFT\nSIFT\nK-means\nK-means\nK-means\nK-means\nphase correlation\nBayesian learning\nsparse Bayesian learning (SBL)\nSBL\nBayesian-MCMC\nBayesian theorem\nreversible jump MCMC\nfull orthostereoscopic image capture and projection\nconceptual graph formalism (CGF)\nOpenNLP\nCGF\nconceptual graph (CG)\nCGs\nCG\nCG\nCG\nVerbNet\nWordNet\nSignal Detection Theory\nSensor fusion\nTransferable Belief Model\nPrincipal component analysis (PCA)\nSparse PCA\nsparse PCA\nsparse loading PCA (slPCA)\nPCA\nsvPCA\nsvPCA\nPCA\nslPCA\nsvPCA\npenalized expectation-maximization (pEM)\ntactile information processing\ntactile sensory suppression\nLight field rendering\nlight field rendering\nPSNR\nlow-level features vector quantization\nmultiple coil MRI(Magnetic Resonance Imaging) \nnon-stationary noise estimation\nmulti-coil MRI\ncoherent subspace methods\ndynamic time warping\nbigram models\nNIST\nshape-from-shading\nshape-from-shading\nAttribute-based Decision Graph\nAbDG\nAbDG\nAbDG\nAbDG\nC4.5\nMulti-Interval ID3\nanalytic hierarchy process (AHP)\nAHP\nPredictive Diagnostic Optimisation\nDiagnostic Optimisation\nLocalist Attractor Network\nTemporal Decision Tree\nNaive Bayes Classifier\nhierarchical-like particle filter\nLazy learning\nmemory learning strategy\nlazy learning\nHenon map\nsocially assistive robotics (SAR)\nparametric masking\nstimulus onset asynchrony (SOA)\nSOA\nSOA\nSOA\nmonaural separation\nfixed dimension modified sinusoidal model (FDMSM)\nSupport Vector Machine (SVM)\nRelevance Vector Machine (RVM)\nk Nearest Neighbor (k-NN)\nMulti-Layer Perceptron (MLP)\nimaging three-dimensional ladar (I3DL)\nI3DL\nmodulation transfer function (MTF)\nline spread function (LSF)\nI3DL\nMTF\nMTF\npixels binning\ngaussian mixtures\nexpectation maximization\nlocally weighted regression (LWR)\nfuzzy cerebellar model articulation controller (FCMAC)\nLWR\nfuzzy CMAC\nfragile watermarking\nsemi-fragile watermark\nfragile watermarking\nHuman Visual System (HVS)\nPKI (Public Key Infrastructure) based semi-fragile watermarking\nDCT\nwatermark embedding\nstereo rectification\nstereo rectification\nimage of the absolute conic (IAC)\neye filter\nAdaBoost\neye filter\nCAS-PEAL\nJAFFE\nBodiPod\nstereoscopic rendering\nModified Newton-Raphson\nEMT\nLBP\nXbox 360\nclassifier integration model(CIM)\nCIM\nCIM\nCIM\nKalman filter\nlinear minimum variance recursive one-shot procedure\ncontour person\ncontour person (CP)\nCP\n3D SCAPE\n2D pose estimation\npictorial-structures\n1-inch UniTouch\nconvolutional neural network (CNN)\nFG-NET\nPrincipal Component Analysis (PCA)\nNon-negative Matrix Factorization (NMF)\nPCA\nNMF\nPCA\nNMF\nPCA\nNMF\nPCA\nNMF\nReceiver Operator Characteristic curves\nSIFT-based face recognition\nSIFT\nSIFT\nExtended Yale face database B\nScale Invariant Feature Transform (or SIFT)\nSIFT-based\nDisplaced Phase Center Antenna (DPCA)\nRedundant Phase Center (RPC)\nautomated microassembly\nVision-based microassembly\nartificial potential field\nRoad segment Partitioning\n(RPat)\ndouble Markov random field\nhuman-machine-environment cyber-physical system (HME-CPS)\nC++\nProlog\nC++\nProlog\nC++\nProlog\nHME-CPS\nhuman-machine collaboration (HMC)\nHME-CPS\ntwo layer hierarchical classification\ncoarse-to-fine optical flow\nFirst Register Then Average And Subtract (FRTAAS)\nnon-local means\npower spectral density estimation\ndyadic wavelet\ncomplex wavelet\nmulti-band wavelet\nConditional Colored Petri Nets\nleast-squares\nSugeno integral\nSugeno integral\nSugeno integral\nFERET\nSugeno integral\nSugeno Measures\ninterval type-2 Sugeno integral\ntype-1 Sugeno integral\ntype-1 Sugeno integral\ninterval type-2 Sugeno integral\ninterval type-2 fuzzy logic\nmodular neural network\nNon-hierarchical k-means\nhierarchical clustering\nhierarchical clustering\nAnchor model\nanchor models\nanchor model\nYOHO\nEER\nman-machine cooperative intelligent reduction\nIRAEK\nMicro-electromechanical Systems (MEMS)\nIRAEK\nIntelligent Reduction Algorithm based on Expert Knowledge\nacoustic finger prints\nsuper-resolution mapping analyses\nsoft image classification\nHopfield neural network\nHopfield neural network\nHopfield neural network\nHopfield neural network\nsuper-resolution mapping\nincremental induction of decision trees\nonline incremental induction of decision trees\ncollaborative filtering (CF)\nCF\nminimax optimization\nregion-based flocking\nParticle swarm optimization\nNaive Bayes\nNaive Bayes\nFPGA-based Naive Bayes\nnetwork traffic classification\nnetwork traffic classification\nNaive Bayes\ncolor stereo matching\nMiddlebury1\nTNO MARS/Prescan2\ninvariant extended Kalman filter (IEKF)\nEKF-SLAM\nIEKFs\nstochastic cloning\nMR spectroscopy\ndecision trees\ndecision tree\ndecision tree\nscale-space filtered coordinate functions\nHotelling transform\neven symmetric Gabor filters\nfuzzy inference\nRBF network\nradial basis function (RBF) network\nRBF network\nRBF network\nPredictive Annotation\nPredictive Annotation\nGestalt laws\nrecurrent network\ncontextual vector quantization (CVQ)\nedge-preserving filter\ngenetic programming\nfully (self-) connected neural network\nGenetically programmed neural net (GenNet)\nGenNets\nGenNets\nGenNets\ngenetic programming\nGenNet\ngenetic programming\nDarwin machines\nblock feature motion estimation\nmixed-mode multiresolution motion estimation\nsign truncated feature (STF)\nblock matching\nSTF\nblock matching\nmean matching\nbinary phase matching\nbinary phase matching\nfull-search block matching\nfull-search block matching\ncross-correlation\nphase-correlation\nphase-correlation\ncross-correlation\nknowledge-based document-analysis\nANASTASIL\nbest-first search\nreinforcement learning\nreinforcement learning\nregion-based image coding\nlossless encoding\nfully connected recurrent network\nSD-Scicon UK Ltd\nKalman filter\nexample-based approach\n"
],
[
"entities = []",
"_____no_output_____"
],
[
"for abstract in annotations_json:\n for annotation in abstract['annotations']:\n if sum(np.asarray(annotation['label_one_hot'])) > 0:\n text = abstract['text']\n start = annotation['start_offset']\n end = annotation['end_offset']\n id = abstract['id']\n label = annotation['label']\n if label == 1:\n item = Entity(id,start,text[start:end],EntityClass.ORG)\n if label == 2:\n item = Entity(id,start,text[start:end],EntityClass.METHOD)\n if label == 3: \n item = Entity(id,start,text[start:end],EntityClass.PRODUCT)\n print(item)\n entities.append(item)",
"_____no_output_____"
],
[
"np.save(\"/content/entities.npy\", entities)",
"_____no_output_____"
]
],
[
[
"##SetSimilaritySearch Clustering",
"_____no_output_____"
]
],
[
[
"!pip install transformers",
"Collecting transformers\n Downloading transformers-4.9.2-py3-none-any.whl (2.6 MB)\n\u001b[K |████████████████████████████████| 2.6 MB 4.1 MB/s \n\u001b[?25hRequirement already satisfied: importlib-metadata in /usr/local/lib/python3.7/dist-packages (from transformers) (4.6.3)\nCollecting pyyaml>=5.1\n Downloading PyYAML-5.4.1-cp37-cp37m-manylinux1_x86_64.whl (636 kB)\n\u001b[K |████████████████████████████████| 636 kB 54.6 MB/s \n\u001b[?25hCollecting huggingface-hub==0.0.12\n Downloading huggingface_hub-0.0.12-py3-none-any.whl (37 kB)\nCollecting sacremoses\n Downloading sacremoses-0.0.45-py3-none-any.whl (895 kB)\n\u001b[K |████████████████████████████████| 895 kB 58.2 MB/s \n\u001b[?25hRequirement already satisfied: filelock in /usr/local/lib/python3.7/dist-packages (from transformers) (3.0.12)\nRequirement already satisfied: tqdm>=4.27 in /usr/local/lib/python3.7/dist-packages (from transformers) (4.62.0)\nRequirement already satisfied: regex!=2019.12.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (2019.12.20)\nCollecting tokenizers<0.11,>=0.10.1\n Downloading tokenizers-0.10.3-cp37-cp37m-manylinux_2_5_x86_64.manylinux1_x86_64.manylinux_2_12_x86_64.manylinux2010_x86_64.whl (3.3 MB)\n\u001b[K |████████████████████████████████| 3.3 MB 19.2 MB/s \n\u001b[?25hRequirement already satisfied: packaging in /usr/local/lib/python3.7/dist-packages (from transformers) (21.0)\nRequirement already satisfied: numpy>=1.17 in /usr/local/lib/python3.7/dist-packages (from transformers) (1.19.5)\nRequirement already satisfied: requests in /usr/local/lib/python3.7/dist-packages (from transformers) (2.23.0)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.7/dist-packages (from huggingface-hub==0.0.12->transformers) (3.7.4.3)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.7/dist-packages (from packaging->transformers) (2.4.7)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.7/dist-packages (from importlib-metadata->transformers) (3.5.0)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2.10)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (3.0.4)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.7/dist-packages (from requests->transformers) (2021.5.30)\nRequirement already satisfied: joblib in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.0.1)\nRequirement already satisfied: six in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (1.15.0)\nRequirement already satisfied: click in /usr/local/lib/python3.7/dist-packages (from sacremoses->transformers) (7.1.2)\nInstalling collected packages: tokenizers, sacremoses, pyyaml, huggingface-hub, transformers\n Attempting uninstall: pyyaml\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\nSuccessfully installed huggingface-hub-0.0.12 pyyaml-5.4.1 sacremoses-0.0.45 tokenizers-0.10.3 transformers-4.9.2\n"
],
[
"from transformers import (\n BertTokenizer, BertTokenizerFast, BatchEncoding,\n DataCollatorForTokenClassification, BertForTokenClassification,\n Trainer, TrainingArguments\n)",
"_____no_output_____"
],
[
"###NER model \nclass BertForTokenClassificationML(BertForTokenClassification):\n def set_label_weights(self, weights):\n self.label_weights = weights\n\n def forward(\n self,\n input_ids=None,\n attention_mask=None,\n token_type_ids=None,\n position_ids=None,\n head_mask=None,\n inputs_embeds=None,\n labels=None,\n output_attentions=None,\n output_hidden_states=None,\n return_dict=None,\n ):\n r\"\"\"\n labels (:obj:`torch.LongTensor` of shape :obj:`(batch_size, sequence_length)`, `optional`):\n Labels for computing the token classification loss. Indices should be in ``[0, ..., config.num_labels -\n 1]``.\n \"\"\"\n return_dict = return_dict if return_dict is not None else self.config.use_return_dict\n\n outputs = self.bert(\n input_ids,\n attention_mask=attention_mask,\n token_type_ids=token_type_ids,\n position_ids=position_ids,\n head_mask=head_mask,\n inputs_embeds=inputs_embeds,\n output_attentions=output_attentions,\n output_hidden_states=output_hidden_states,\n return_dict=return_dict,\n )\n\n sequence_output = outputs[0]\n\n sequence_output = self.dropout(sequence_output)\n logits = self.classifier(sequence_output)\n\n loss = None\n if labels is not None:\n loss_fct = torch.nn.BCEWithLogitsLoss()\n \n # Only keep active parts of the loss\n if attention_mask is not None:\n active_loss = attention_mask == 1\n active_logits = logits.view(-1, self.num_labels)[active_loss.view(-1), :]\n active_labels = labels[active_loss, :]\n\n loss_fct = torch.nn.BCEWithLogitsLoss()\n loss = loss_fct(active_logits, active_labels.float())\n else:\n loss = loss_fct(logits.view(-1, self.num_labels), labels.view(-1))\n\n if not return_dict:\n output = (logits,) + outputs[2:]\n return ((loss,) + output) if loss is not None else output\n\n return TokenClassifierOutput(\n loss=loss,\n logits=logits,\n hidden_states=outputs.hidden_states,\n attentions=outputs.attentions,\n )",
"_____no_output_____"
],
[
"import torch\n##Load fine-tuned model from NER task\nPATH = \"/content/drive/MyDrive/model/bert_finetune.pt\"\nmodel = torch.load(PATH,map_location=torch.device('cpu'))",
"_____no_output_____"
],
[
"from src.ieee_ner_coref import EntityClusterer\nfrom src.models import Entity\nfrom src.models import EntityClass\n\ndata = []\nfile = \"/content/entities.npy\"\nwith open(file, \"r\") as f:\n for line in f.readlines():\n line = line.strip('\\n') \n data.append(line)\ndata = np.load(file,allow_pickle=True)\ndata = data.tolist()\n###Get the results from SetSimilaritySearch Clustering\nentity_groups = EntityClusterer(method='best').cluster(data)",
"_____no_output_____"
]
],
[
[
"### Find unclustered entities",
"_____no_output_____"
]
],
[
[
"un_clustered_l = []\nfor idx in range(len(data)):\n tag = True\n for i in range(len(entity_groups)):\n for j in range(len(entity_groups[i])):\n if data[idx].text in entity_groups[i][j].text:\n tag = False\n if tag == True:\n un_clustered_l.append(data[idx])",
"_____no_output_____"
]
],
[
[
"###Save text for unclustered entities",
"_____no_output_____"
]
],
[
[
"un_cluster = []\nfor i in range(len(un_clustered_l)):\n un_cluster.append(un_clustered_l[i].text)",
"_____no_output_____"
]
],
[
[
"### Get all the entities which are clustered",
"_____no_output_____"
]
],
[
[
"cluster_entities = []\nfor i in range(len(entity_groups)):\n for j in range(len(entity_groups[i])):\n cluster_entities.append(entity_groups[i][j].text)",
"_____no_output_____"
],
[
"##Directly load saving lists\nfile1 = '/content/cl_entities.npy'\ncluster_entities = np.load(file1,allow_pickle=True)\ncluster_entities = cluster_entities.tolist()\nfile2 = '/content/un_entities.npy'\nun_cluster = np.load(file2,allow_pickle=True)\nun_cluster = un_cluster.tolist()",
"_____no_output_____"
]
],
[
[
"##BERT-based embedding measurement",
"_____no_output_____"
]
],
[
[
"### Get the dictionary for the Bert tokenizer\n!wget \"https://s3.amazonaws.com/models.huggingface.co/bert/bert-base-cased-vocab.txt\"",
"--2021-08-16 05:11:47-- https://s3.amazonaws.com/models.huggingface.co/bert/bert-base-cased-vocab.txt\nResolving s3.amazonaws.com (s3.amazonaws.com)... 52.216.131.173\nConnecting to s3.amazonaws.com (s3.amazonaws.com)|52.216.131.173|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 213450 (208K) [text/plain]\nSaving to: ‘bert-base-cased-vocab.txt’\n\nbert-base-cased-voc 100%[===================>] 208.45K 283KB/s in 0.7s \n\n2021-08-16 05:11:48 (283 KB/s) - ‘bert-base-cased-vocab.txt’ saved [213450/213450]\n\n"
],
[
"vocab = []\nfile = './bert-base-cased-vocab.txt'",
"_____no_output_____"
],
[
"with open(file, \"r\") as f:\n for line in f.readlines():\n line = line.strip('\\n') \n vocab.append(line)\nlen(vocab)",
"_____no_output_____"
],
[
"crf_w2i = {w : i for i, w in enumerate(vocab)}",
"_____no_output_____"
],
[
"def get_similarity(embeddings,\n w2i,\n term1,\n term2):\n \"\"\"\n\n \"\"\"\n ## Check Terms\n for term in term1 :\n if term not in w2i:\n raise KeyError(f\"Term `{term}` not found\")\n for term in term2 :\n if term not in w2i:\n raise KeyError(f\"Term `{term}` not found\")\n ## Get Indices\n embeddings = embeddings.cpu()\n term1_ind = [] \n term2_ind = []\n for term in term1 :\n term1_ind.append(torch.LongTensor([w2i[term]]))\n for term in term2 :\n term2_ind.append(torch.LongTensor([w2i[term]]))\n \n ## Retrieve Embeddings and Compute Cosine Similarity\n term1_embed = 0 \n term2_embed = 0\n for idx1 in term1_ind:\n term1_embed += embeddings(idx1)\n for idx2 in term2_ind:\n term2_embed += embeddings(idx2)\n term1_embed = term1_embed/(len(term1_ind))\n term2_embed = term2_embed/(len(term2_ind))\n\n distance = float(cosine_distances(term1_embed.detach(), term2_embed.detach()))\n return distance\n # print(term1, term2, distance)",
"_____no_output_____"
],
[
"### Retrieve embedding layer and the tokenizer\nembed = model.bert.embeddings.word_embeddings\ntokenizer = BertTokenizerFast.from_pretrained('bert-base-cased')",
"_____no_output_____"
],
[
"### Using cosine distance to measure similarity\nfrom sklearn.metrics.pairwise import cosine_distances",
"_____no_output_____"
],
[
"entities = []\nfile1 = './entities.txt'",
"_____no_output_____"
],
[
"with open(file1, \"r\") as f:\n for line in f.readlines():\n line = line.strip('\\n') \n entities.append(line)\nlen(entities)",
"_____no_output_____"
],
[
"### This function is for showing the most similar entities\ndef show_similarity_top(term,data,num)->dict:\n sort_dic = {}\n for item in data:\n item1 = tokenizer.tokenize(item)\n temp = get_similarity(embed, crf_w2i, term,item1)\n sort_dic[item] = temp\n sorted_dic = dict(sorted(sort_dic.items(), key = lambda kv:(kv[1], kv[0])))\n # print(sorted_dic)\n cnt = 0 \n for key, value in sorted_dic.items():\n cnt += 1\n if cnt > num:\n break\n print(\"{}:{}\".format(key, value)) ",
"_____no_output_____"
],
[
"def show_similarity(term,data)->dict:\n sort_dic = {}\n for item in data:\n item1 = tokenizer.tokenize(item)\n temp = get_similarity(embed, crf_w2i, term,item1)\n sort_dic[item] = temp\n sorted_dic = dict(sorted(sort_dic.items(), key = lambda kv:(kv[1], kv[0])))\n return sorted_dic",
"_____no_output_____"
]
],
[
[
"###Test the functions",
"_____no_output_____"
]
],
[
[
"term = tokenizer.tokenize(\"CGs\")\nshow_similarity_top(term,entities,10)",
"CGs:0.0\nCG:0.11934900283813477\nCGF:0.2058941125869751\nAbDG:0.3184809684753418\nIEKFs:0.3396766781806946\nFG-NET:0.35870277881622314\nHOG:0.37542617321014404\nconceptual graph (CG):0.3831406235694885\nC4.5:0.4044969081878662\nconceptual graph formalism (CGF):0.4098879098892212\n"
],
[
"term1 = tokenizer.tokenize('two layer hierarchical classification')\nterm2 = tokenizer.tokenize('hierarchical clustering')\nget_similarity(embed,crf_w2i,term1,term2)",
"_____no_output_____"
]
],
[
[
"Load the clustered entities and unclustered entities",
"_____no_output_____"
]
],
[
[
"file1 = '/content/cl_entities.npy'\ncluster = np.load(file1,allow_pickle=True)\ncluster = cluster.tolist()\nfile2 = '/content/un_entities.npy'\nun_cluster = np.load(file2,allow_pickle=True)\nun_cluster = un_cluster.tolist()",
"_____no_output_____"
],
[
"### Load the clustered entities (only contain text)\nfile3 = '/content/cluster_item.npy'\ncluster_item = np.load(file3,allow_pickle=True)",
"_____no_output_____"
],
[
"### Number of clusters\nlen(cluster_item)",
"_____no_output_____"
],
[
"### This function is to get the average tensor for each clusters\ndef cluster_avg_embed(cluster_item,embeddings,tokenizer,w2i):\n cluster_embed = []\n for idx in range(len(cluster_item)):\n avg = 0\n for j in range(len(cluster_item[idx])):\n embed_t = 0\n term1 = tokenizer.tokenize(cluster_item[idx][j])\n term_ind = []\n for term in term1 :\n term_ind.append(torch.LongTensor([w2i[term]]))\n for idx1 in term_ind:\n embed_t += embeddings(idx1)\n embed_t = embed_t/(len(term_ind))\n avg += embed_t\n avg = avg/(len(cluster_item[idx]))\n cluster_embed.append(avg)\n return cluster_embed",
"_____no_output_____"
],
[
"## Get the embedding representations for each cluster\ncluster_embed = cluster_avg_embed(cluster_item,embed,tokenizer,crf_w2i)",
"_____no_output_____"
],
[
"## Save embedding tensors\nval= torch.tensor([item.cpu().detach().numpy() for item in cluster_embed])\nnp.save('/content/cluster_embed.npy',val)\n## Load embedding tensors\ncluster_embed = np.load('/content/cluster_embed.npy')\ncluster_embed = cluster_embed.tolist()",
"_____no_output_____"
],
[
"###This function is to get the embedding representation of a single term\ndef get_embedding(term,tokenizer,w2i,embeddings):\n term1 = tokenizer.tokenize(term)\n temp = 0\n term_ind = []\n for t in term1 :\n term_ind.append(torch.LongTensor([w2i[t]]))\n for idx1 in term_ind:\n temp += embeddings(idx1)\n temp = temp/(len(term_ind))\n return temp",
"_____no_output_____"
],
[
"### This function is to calculate distance between embeddings\ndef embed_cos_distance(cluster_embed_,term):\n dict_r = {}\n for i in range(len(cluster_embed_)):\n distance = float(cosine_distances(cluster_embed_[i].detach(), term.detach()))\n dict_r[i] = distance\n return dict_r",
"_____no_output_____"
]
],
[
[
"TEST",
"_____no_output_____"
]
],
[
[
"term1 = tokenizer.tokenize('multi-coil MRI')\nterm2 = tokenizer.tokenize('multiple coil MRI(Magnetic Resonance Imaging) ')\nget_similarity(embed,crf_w2i,term1,term2)",
"_____no_output_____"
]
],
[
[
"###Find entities which should be included into the clusters",
"_____no_output_____"
]
],
[
[
"mod_list_ = []\n### Basically, for each unclustered entity comparing cosine distance with each cluster and decide whether it should be included. \nfor w in un_cluster:\n dict_ = {}\n w_embed = get_embedding(w,tokenizer,crf_w2i,embed)\n res = embed_cos_distance(cluster_embed,w_embed)\n if len(w)<=8:\n if res[min(res, key=res.get)]<0.28:\n dict_[w] = min(res, key=res.get)\n mod_list_.append(dict_)\n else:\n if res[min(res, key=res.get)]<0.35:\n dict_[w] = min(res, key=res.get)\n mod_list_.append(dict_)",
"_____no_output_____"
],
[
"### The entities which should be included (of course a small part of this would be misplaced)\n### The keys in the dict are entities text and values are clusters index\nmod_list_",
"_____no_output_____"
],
[
"add_list = []\nfor i in range(len(mod_list_)):\n key, = mod_list_[i].keys()\n add_list.append(key)",
"_____no_output_____"
],
[
"term1 = tokenizer.tokenize('Bayesian theorem')\nterm2 = tokenizer.tokenize('Bayesian-MCMC')\nget_similarity(embed,crf_w2i,term1,term2)",
"_____no_output_____"
],
[
"term1 = tokenizer.tokenize('hyperspectral band grouping')\nterm2 = tokenizer.tokenize('hyperspectral analysis')\nget_similarity(embed,crf_w2i,term1,term2)",
"_____no_output_____"
],
[
"## remained entities of un_clustered list after Bert-embedding process\nremain_list = list(set(un_cluster).difference(set(add_list)))",
"_____no_output_____"
],
[
"remain_list",
"_____no_output_____"
]
],
[
[
"###Add entities which should be included to the original clusters",
"_____no_output_____"
]
],
[
[
"###In order to keep the consistency of entities, construct \nfile2 = '/content/entities.npy'\nentities = np.load(file2,allow_pickle=True)",
"_____no_output_____"
],
[
"def get_Entity(text,entities):\n res = 0\n for x in entities:\n if text == x.text:\n res = x\n break\n return res",
"_____no_output_____"
],
[
"###Using following operations to add mishandled entities into clusters\nfor item in mod_list_:\n for i in range(len(entity_groups)):\n flag = 0\n for j in range(len(entity_groups[i])):\n if item==entity_groups[i][j].text:\n temp = get_Entity(item,entities)\n entity_groups[i].append(temp)\n flag = 1\n break\n if flag == 1:\n break",
"_____no_output_____"
]
],
[
[
"###DBSCAN(find new clusters)",
"_____no_output_____"
]
],
[
[
"# from sklearn import datasets\nX = remain_list.copy()\nimport numpy as np\nimport random\nimport time\ndef findNeighbor(j,X,eps,distance,embed,crf_w2i,tokenizer):\n N=[]\n for p in range(len(X)): #Find all objects in a neighbourhood\n term1 = tokenizer.tokenize(X[j])\n term2 = tokenizer.tokenize(X[p])\n temp = distance(embed,crf_w2i,term1,term2) #cosine distance\n # temp=np.sqrt(np.sum(np.square(X[j]-X[p]))) #Euclidean distance\n if(temp<=eps):\n N.append(p)\n return N\n\n\ndef dbscan(X,eps,min_Pts,distance,embed,crf_w2i,tokenizer):\n k=-1\n NeighborPts=[] #array,all items in a neighbourhood\n Ner_NeighborPts=[]\n fil=[] #visited list to be empty at start\n gama=[x for x in range(len(X))] # all the items should be un-visited at start\n cluster=[-1 for y in range(len(X))]\n while len(gama)>0:\n j=random.choice(gama)\n gama.remove(j) #remove from un-visited\n fil.append(j) #add to visited\n NeighborPts=findNeighbor(j,X,eps,distance,embed,crf_w2i,tokenizer)\n if len(NeighborPts) < min_Pts:\n cluster[j]=-1 #mark as a noise point\n else:\n k=k+1\n cluster[j]=k\n for i in NeighborPts:\n if i not in fil:\n gama.remove(i)\n fil.append(i)\n Ner_NeighborPts=findNeighbor(i,X,eps,distance,embed,crf_w2i,tokenizer)\n if len(Ner_NeighborPts) >= min_Pts:\n for a in Ner_NeighborPts:\n if a not in NeighborPts:\n NeighborPts.append(a)\n if (cluster[i]==-1):\n cluster[i]=k\n return cluster\n\n\neps=0.27\nmin_Pts=2\nbegin=time.time()\nC=dbscan(X,eps,min_Pts,get_similarity,embed,crf_w2i,tokenizer)\nend=time.time()\nprint (\"time using:\",end-begin)\n",
"time using: 13.966970920562744\n"
],
[
"##Get index for each new clusters\nfrom collections import defaultdict\nd = defaultdict(list)\nfor k,va in [(v,i) for i,v in enumerate(C)]:\n d[k].append(va)\nprint(d)",
"defaultdict(<class 'list'>, {-1: [0, 1, 2, 4, 5, 7, 8, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 23, 24, 25, 27, 28, 29, 31, 32, 33, 35, 36, 37, 38, 39, 40, 41, 42, 43, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 86, 87, 88, 89, 91, 92, 93, 94, 95, 96, 97, 98, 99, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 118, 119], 3: [3, 101], 4: [6, 117], 1: [9, 85], 5: [22, 34], 2: [26, 100], 6: [30, 57], 0: [44, 90]})\n"
],
[
"new_cluster_l = []\n# cluster_num = len(entity_groups)\ncluster_num = 80\nfor i in range(max(C)+1):\n temp = {}\n temp[cluster_num+i] = [X[idx] for idx in d[i]]\n new_cluster_l.append(temp)\nnew_cluster_l",
"_____no_output_____"
],
[
"### Add new clusters into original list\nentity_groups.append(new_cluster_l)",
"_____no_output_____"
]
],
[
[
"##SpanBERT for embedding measurement",
"_____no_output_____"
]
],
[
[
"# Import generic wrappers\nfrom transformers import AutoModel, AutoTokenizer \n\n\n# Define the model repo\nmodel_name = \"SpanBERT/spanbert-base-cased\" \n\n\n# Download pytorch model\nmodel = AutoModel.from_pretrained(model_name)\ntokenizer = AutoTokenizer.from_pretrained(model_name)",
"_____no_output_____"
],
[
"span_embed = model.embeddings.word_embeddings",
"_____no_output_____"
],
[
"###Test the feasibility of tokenizer and embedding\nterm1 = tokenizer.tokenize('two layer hierarchical classification')\nterm2 = tokenizer.tokenize('hierarchical clustering')\nget_similarity(span_embed,crf_w2i,term1,term2)",
"_____no_output_____"
],
[
"def show_similarity_top1(term,data,num)->dict:\n sort_dic = {}\n for item in data:\n item1 = tokenizer.tokenize(item)\n temp = get_similarity(span_embed, crf_w2i, term,item1)\n sort_dic[item] = temp\n sorted_dic = dict(sorted(sort_dic.items(), key = lambda kv:(kv[1], kv[0])))\n # print(sorted_dic)\n cnt = 0 \n for key, value in sorted_dic.items():\n cnt += 1\n if cnt > num:\n break\n print(\"{}:{}\".format(key, value)) ",
"_____no_output_____"
],
[
"term = tokenizer.tokenize(\"CGs\")\nshow_similarity_top1(term,entities,10)",
"CGs:4.76837158203125e-07\nCG:0.18933910131454468\nCGF:0.2702394723892212\nHME-CPS:0.2835298180580139\nCP:0.30165767669677734\nCF:0.3144184350967407\nCIM:0.32503771781921387\nhuman-machine-environment cyber-physical system (HME-CPS):0.34812843799591064\nconceptual graph (CG):0.360026478767395\nCAS-PEAL:0.36104893684387207\n"
],
[
"cluster_embed_span = cluster_avg_embed(cluster_item,span_embed,tokenizer,crf_w2i)",
"_____no_output_____"
],
[
"mod_list_span = []\nfor w in un_cluster:\n dict_ = {}\n w_embed = get_embedding(w,tokenizer,crf_w2i,span_embed)\n res = embed_cos_distance(cluster_embed_span,w_embed)\n if res[min(res, key=res.get)]<0.20:\n dict_[w] = min(res, key=res.get)\n mod_list_span.append(dict_)",
"_____no_output_____"
]
],
[
[
"Note: spanBert embedding dose not perform well in this case",
"_____no_output_____"
]
],
[
[
"mod_list_span",
"_____no_output_____"
]
],
[
[
"##Function for searching relative Entities",
"_____no_output_____"
],
[
"note: you can get the corresponding cluster information by this function",
"_____no_output_____"
]
],
[
[
"def find_crf(word,new_cluster):\n for i in range(len(new_cluster)):\n flag = False\n for j in range(len(new_cluster[i])):\n if word == new_cluster[i][j].text:\n flag = True \n if flag == True:\n print(new_cluster[i])\n # res.append(new_cluster[i])\n # return res\n\n### Example\nfind_crf('FCMAC',entity_groups)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6bfb4e7ffa537b60209742e00b798e6e4e11b6 | 277,660 | ipynb | Jupyter Notebook | Debt_Collection.ipynb | jaimemv/DebtCollectionPrediction | 74e5e835a2919cb835e7bba1b080a31ad4db3144 | [
"MIT"
]
| 1 | 2021-11-09T10:51:47.000Z | 2021-11-09T10:51:47.000Z | Debt_Collection.ipynb | jaimemv/DebtCollectionPrediction | 74e5e835a2919cb835e7bba1b080a31ad4db3144 | [
"MIT"
]
| null | null | null | Debt_Collection.ipynb | jaimemv/DebtCollectionPrediction | 74e5e835a2919cb835e7bba1b080a31ad4db3144 | [
"MIT"
]
| null | null | null | 277,660 | 277,660 | 0.935075 | [
[
[
"# 1. Frame the problem\nThe objective of this notebook is fitting a model that predicts the likelihood of debtors to repay their debts within the next 12 months.\n\nTo achieve this objective, three separate models will be fitted to predict the likelihood of repay within the following 3, 6 and 12 months respectively.\n\nTherefore the problem is a Classification one. Instead of training one Classifier to predict the likelihood to pay in the next 3, 6, 12 months or not pay at all, 3 independent binary classifiers will be fitted to get the likelihood of payment at each of the mentioned periods.\n\nIn terms of metrics, I consider the most important one optimise is **precision**. It is because at the time of establishing a budget allocation for the financial year, False Positives may lead the Company believe that can count with X amount of money collected back from debts.\n\nSince I do not have perfect information about the dataset and I do not know which specific outliers may be miss-type or any other kind of error, some of them will be dropped in order to not blurry the model.\n\nHowever, since debt collection is a sensitive issue, I would perform specific risk studies for each of the complex cases.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\npd.options.display.float_format = \"{:,.2f}\".format\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.impute import KNNImputer\nfrom sklearn.compose import ColumnTransformer\nfrom sklearn.pipeline import Pipeline\nfrom sklearn.metrics import accuracy_score, precision_score\nfrom xgboost import XGBClassifier\nfrom scipy import stats\nfrom scipy.stats import norm\nfrom moduleresources import path",
"_____no_output_____"
],
[
"drive_route = path\ndf = pd.read_csv(drive_route, delimiter=\";\")",
"_____no_output_____"
]
],
[
[
"# EDA",
"_____no_output_____"
]
],
[
[
"df.sample(10)",
"_____no_output_____"
]
],
[
[
"- `IS_PAID_12M`, `IS_PAID_6M`, `IS_PAID_3M` Will be the target variables.",
"_____no_output_____"
]
],
[
[
"df.SNAPSHOT_DATE.value_counts()",
"_____no_output_____"
]
],
[
[
"`SNAPSHOT_DATE` has a unique value: 2019-12-31",
"_____no_output_____"
],
[
"Define our predictors:",
"_____no_output_____"
]
],
[
[
"num_cols = ['CASE_AGE_MONTHS', 'INTEREST_RATE', 'REM_PRINCIPAL_AMT',\n 'REM_PRINCIPAL_INTEREST_AMT', 'REM_TOTAL_AMT', 'DEBTOR_AGE',\n 'PAID_AMT_12M', 'PAID_AMT_6M', 'PAID_AMT_3M', 'INCOME_TOTAL_LAST_YEAR',\n 'TOTAL_DEBT_AMT']",
"_____no_output_____"
]
],
[
[
"Let's get rid of useless features:",
"_____no_output_____"
]
],
[
[
"df.drop([\"DEBTOR_NO\", \"RAND_NO\", \"SNAPSHOT_DATE\"], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"Let's check how many null values we have to face",
"_____no_output_____"
]
],
[
[
"df.isna().sum()[df.isna().sum()!=0]",
"_____no_output_____"
],
[
"df[df['REM_PRINCIPAL_AMT'].isna()]",
"_____no_output_____"
]
],
[
[
"Let's drop the row where there 2 null values",
"_____no_output_____"
]
],
[
[
"df.drop([572], inplace=True)",
"_____no_output_____"
],
[
"def hist_box(data, title):\n\n fig, ax = plt.subplots(figsize = (20,5), ncols=2)\n\n # Histogram\n data_wo_outliers = data[data.between(data.quantile(.10), data.quantile(.99))] \n ax[0].hist(data_wo_outliers, bins=20, alpha = 0.5)\n ax[0].grid(False)\n plt.style.use(\"bmh\")\n\n # Boxplot\n try:\n ax[1].boxplot(data.dropna())\n except AttributeError:\n ax[1].boxplot(data)\n \n fig.suptitle(f\"Distribution of {title}\", fontsize=20)\n\n plt.show()",
"_____no_output_____"
],
[
"for i, col in enumerate(df[num_cols].columns):\n hist_box(df[col], col)",
"_____no_output_____"
]
],
[
[
"## Outlier Removal\nAfter exploring the data, some outliers have been found, specially on the upper part of the distributions. Then I'll be shortening the distribution from percentile 10 to percentile 99. I don't want to \"castrate\" the data and miss values such as `PAID_AMT_3M` non-zero values that are just few of them but of big utility. Then the upper bound has been chosen to be 99th percentile.",
"_____no_output_____"
]
],
[
[
"def outliers_removal(feature, feature_name, dataset):\n \n q10, q99 = np.percentile(feature, 25), np.percentile(feature, 99)\n feat_iqr = q99 - q10\n \n feat_cut_off = feat_iqr * 1.5\n feat_lower, feat_upper = q10 - feat_cut_off, q99 + feat_cut_off\n \n outliers = [x for x in feature if x < feat_lower or x > feat_upper]\n\n dataset = dataset.drop(dataset[(dataset[feature_name] > feat_upper) | (dataset[feature_name] < feat_lower)].index)\n \n return dataset\n\nfor col in df[num_cols]:\n df_clean = outliers_removal(df[col],str(col),df)",
"_____no_output_____"
]
],
[
[
"# Data Pipeline",
"_____no_output_____"
],
[
"The training data will be standarised and null values (only found on `INCOME_TOTAL_LAST_YEAR`) will be imputed with K-Nearest-Neighbors imputer.",
"_____no_output_____"
]
],
[
[
"numeric_pipeline = Pipeline(\n steps=[\n (\"scale\", StandardScaler()),\n (\"knn\", KNNImputer())\n ]\n)\n\nfull_processor = ColumnTransformer(\n transformers=[\n (\"numeric\", numeric_pipeline, num_cols)\n ]\n)\nX_processed = full_processor.fit_transform(df)",
"_____no_output_____"
]
],
[
[
"# XGBoost",
"_____no_output_____"
],
[
"XGBoost Classifier will be the model that I'll be using. It may converge fast due to the amount of data and perform well in the binary classification task.",
"_____no_output_____"
]
],
[
[
"y_list = ['IS_PAID_12M', 'IS_PAID_6M', 'IS_PAID_3M']",
"_____no_output_____"
],
[
"for y in y_list:\n y_processed = df[y].copy()\n\n X_train, X_test, y_train, y_test = train_test_split(\n X_processed, y_processed, stratify=y_processed, random_state=11091993\n )\n xgb_cl = XGBClassifier()\n xgb_cl.fit(X_train, y_train)\n # Predict\n preds = xgb_cl.predict(X_test)\n\n # Score\n accuracy_score(y_test, preds)\n print(f\"----- METRICS FOR {y} -----\\n\")\n print(f\"Confusion matrix:\")\n print(confusion_matrix(y_test, preds), \"\\n\")\n print(f\"The Accuracy score of XGBoost Classifier for {y} is: {accuracy_score(y_test, preds)*100:.2f}%\")\n print(f\"The Precision score of XGBoost Classifier for {y} is: {precision_score(y_test, preds)*100:.2f}%\")\n",
"----- METRICS FOR IS_PAID_12M -----\n\nConfusion matrix:\n[[3350 55]\n [ 523 109]] \n\nThe Accuracy score of XGBoost Classifier for IS_PAID_12M is: 85.68%\nThe Precision score of XGBoost Classifier for IS_PAID_12M is: 66.46%\n----- METRICS FOR IS_PAID_6M -----\n\nConfusion matrix:\n[[3590 32]\n [ 361 54]] \n\nThe Accuracy score of XGBoost Classifier for IS_PAID_6M is: 90.27%\nThe Precision score of XGBoost Classifier for IS_PAID_6M is: 62.79%\n----- METRICS FOR IS_PAID_3M -----\n\nConfusion matrix:\n[[3788 23]\n [ 204 22]] \n\nThe Accuracy score of XGBoost Classifier for IS_PAID_3M is: 94.38%\nThe Precision score of XGBoost Classifier for IS_PAID_3M is: 48.89%\n"
]
],
[
[
"# Conclusions\n- **Accuracy** oscillates between ~85%-94% for the three periods in advance. It is a good result.\n- **Precision** ranges from ~48-66%. For 12 and 6 months in advance the results are fair. However for the 3 months in advance the results are poor, it is even better random guess. This result can be improved by testing new models, performing a more extensive data post-processing and of course by gathering more data.\n\n# Further work\n\nThis project is a first approach to a robust debt collection Classificator. For further work I would continue by:\n\n- Fitting and testing more models may lead to better results.\n\n- The features of the data are a bit scarce so any algorithm may find hard to infere patterns from it. Adding more quality signals may bring a big improvement. Working closer to the Data Engineering team that establish the data collection criteria and the signals to be collected may be a handy solution.\n\n- In order to make the model more reliable, data should be enlarged in both: (1) number of time-periods for currently included debtors and (2) more debtors.\n\n- Talk to an expert who knows about the dataset to get a better understanding of it, potential erroneous values, etc.\n\n- Perform oversampling of the undersampled target.",
"_____no_output_____"
]
],
[
[
"username='jaimemv'\ngit_token = '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'\nrepository = 'DebtCollectionPrediction'\n\n!git remote add origin https://{git_token}@github.com/{username}/{repository}.git",
"fatal: remote origin already exists.\n"
],
[
"!git rm -rf --cached .\n!git add .\n!git commit -m \"UPDATE .gitignore\"",
"[master 1cdb938] UPDATE .gitignore\n 1 file changed, 1 insertion(+), 1 deletion(-)\n"
],
[
"!git status",
"On branch master\nChanges not staged for commit:\n (use \"git add <file>...\" to update what will be committed)\n (use \"git checkout -- <file>...\" to discard changes in working directory)\n\n\t\u001b[31mmodified: Debt_Collection.ipynb\u001b[m\n\nno changes added to commit (use \"git add\" and/or \"git commit -a\")\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec6bfc5664a510ea2d41aecd4e823f204115fab9 | 14,173 | ipynb | Jupyter Notebook | 1 Typical workflow.ipynb | MichaMucha/odsc2019-voila-jupyter-web-app | adab4a28d76eca085490282cd090cc5b5b855d38 | [
"BSD-3-Clause"
]
| 3 | 2019-11-13T08:52:51.000Z | 2019-11-22T18:54:46.000Z | 1 Typical workflow.ipynb | MichaMucha/odsc2019-voila-jupyter-web-app | adab4a28d76eca085490282cd090cc5b5b855d38 | [
"BSD-3-Clause"
]
| null | null | null | 1 Typical workflow.ipynb | MichaMucha/odsc2019-voila-jupyter-web-app | adab4a28d76eca085490282cd090cc5b5b855d38 | [
"BSD-3-Clause"
]
| 6 | 2019-11-18T14:59:14.000Z | 2019-11-20T18:19:46.000Z | 26.246296 | 138 | 0.540464 | [
[
[
"# Example financial decision:\n\n> **Whether to commit to owning a home & what does this entail**",
"_____no_output_____"
],
[
"Stakeholders - K&J\n\n## Question space\n\n> What is the wealth outcome over time? \n> What is the cashflow commitment? \n> What do the numbers mean with the QE shenanigans in play? \n> What scenarios are we looking at? \n> How does letting affect the calculation? \n> By when are we in the money, in real terms? \n> What do exit options look like over the years? ",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport datetime\n\npd.set_option('plotting.backend', 'pandas_bokeh')\npd.plotting.output_notebook()\n\nfrom bokeh.models.formatters import NumeralTickFormatter, PrintfTickFormatter\nfrom bokeh.plotting import show",
"_____no_output_____"
]
],
[
[
"## Example listing details",
"_____no_output_____"
]
],
[
[
"price, property_name, location, photo = (\n 250_000, \n 'Pretty home', \n 'Richmond, Surrey', \n 'https://unsplash.com/home.png')",
"_____no_output_____"
]
],
[
[
"### Mortgage calculation",
"_____no_output_____"
]
],
[
[
"# mortgage calculation\n# (price, downpayment, rate, term, interest-only) -> time series of debt balance, time series of accumulated payments\n\ndef mortgage(price:int, downpayment:int, rate:float, term_years:int, interest_only=False) -> pd.DataFrame:\n \n principal = price - downpayment\n rate = rate / 12\n term = term_years * 12\n periods = np.arange(term) + 1\n \n if not interest_only:\n full_payments = np.pmt(rate, term, principal) * np.ones_like(periods)\n principal_payments = np.ppmt(rate, periods, term, principal)\n interest_payments = np.ipmt(rate, periods, term, principal)\n else:\n full_payments = (principal * rate) * -np.ones_like(periods)\n principal_payments = np.zeros_like(periods)\n interest_payments = full_payments\n \n debt_balance = (principal + principal_payments.cumsum()).round(2)\n \n return pd.DataFrame(\n data=np.array([debt_balance, full_payments, principal_payments, interest_payments]).T,\n columns='debt_balance, full_payments, principal_payments, interest_payments'.split(', '),\n index=pd.date_range(start=datetime.date.today(), periods=term, freq='M')\n )\n\nmortgage(price, 0, 0.04, 30)",
"_____no_output_____"
]
],
[
[
"### Annualized mortgage cash flow",
"_____no_output_____"
]
],
[
[
"mortgage(price, 0, 0.04, 25).resample('Y')['full_payments'].sum()",
"_____no_output_____"
]
],
[
[
"### Home value appreciation",
"_____no_output_____"
]
],
[
[
"def home_value(price:int, growth_rate:float, term_years:int) -> pd.Series:\n periods = np.arange(term_years)\n value = np.fv(growth_rate, periods, 0, -price)\n return pd.Series(\n name='home_value',\n data=value,\n index=pd.date_range(start=datetime.date.today(), periods=term_years, freq='Y')\n )",
"_____no_output_____"
],
[
"# optional letting cash flow -> time series of cash payments\ndef letting_cash_flow(monthly_rate:int, term_years:int, growth_rate:float, vacancy_rate:float=2/12) -> pd.Series:\n periods = np.arange(term_years)\n value = np.fv(growth_rate, periods, 0, -1) * monthly_rate * (1-vacancy_rate)\n return pd.Series(\n name='rental_income',\n data=value,\n index=pd.date_range(start=datetime.date.today(), periods=term_years, freq='Y')\n )",
"_____no_output_____"
],
[
"home_value(price, 0.01, 15)",
"_____no_output_____"
],
[
"letting_cash_flow(1250, 25, .02)",
"_____no_output_____"
]
],
[
[
"## Step 1: Scenario - investment outcomes",
"_____no_output_____"
]
],
[
[
"# Parameters:\ntransaction_cost = 0.05 * price\ndownpayment = 0.05 * price \nyears = 25\ninflation = .018\ninflation_houseprice = 0.2\nrate_mortgage = 0.135\nrental_rate = 0\ninterest_only = False\n\nprint('## Scenario')\nprint('House price: £', price)\nprint('transaction_cost: £', transaction_cost)\nprint('downpayment: £', downpayment)\nprint('term (years)', years)\nprint(f'inflation {100*inflation:.2f}%')\nprint(f'house price appreciation: {100*inflation_houseprice:.2f}%')\nprint(f'rate_mortgage: {100*rate_mortgage:.2f}%')\nprint('monthly rental income:', 0)\nprint('interest-only:', interest_only)",
"_____no_output_____"
],
[
"# Schedule and outcomes over time\n\nloan_schedule = mortgage(price, downpayment, rate_mortgage, years, interest_only=interest_only)\ndebt_balance = loan_schedule.debt_balance.resample('Y').last()\ninterest_paid = loan_schedule.interest_payments.resample('Y').sum().cumsum()\n\n# Home value and letting income\nhome = home_value(price, inflation_houseprice, years)\nletting_income = letting_cash_flow(rental_rate, years, inflation).cumsum()\n\n# Actual cash flow\ncash_flow = (\n loan_schedule.full_payments.resample('Y').sum()\n + letting_income\n)\ncash_flow.iloc[0] -= transaction_cost\ncash_flow.iloc[0] -= downpayment\ncash_flow = cash_flow.cumsum()\n\n# Real situation\nwealth = home + letting_income - downpayment - debt_balance - transaction_cost",
"_____no_output_____"
],
[
"wealth",
"_____no_output_____"
],
[
"cash_flow",
"_____no_output_____"
],
[
"# discounted value at time T - discounting by inflation\n\ndf = pd.DataFrame(\n data = [debt_balance, interest_paid, cash_flow, wealth],\n index = 'debt_balance, interest_payments, cash_flow, wealth'.split(', ')\n).T.dropna()\n\ndiscounting_factor = np.pv(inflation, np.arange(len(df)), 0, fv=-1)\ndf = df.mul(discounting_factor, axis=0)",
"_____no_output_____"
]
],
[
[
"## Outcomes chart",
"_____no_output_____"
]
],
[
[
"p = df.plot(kind='line', \n show_figure=False,\n toolbar_location=None,\n panning=False,\n zooming=False,\n plot_data_points=True,\n plot_data_points_size=5\n )\np.hover.tooltips = [('year', '$index')] + list(\n (c, f'£@{c}'+'{0,0}')\nfor c in df.columns)\np.hover.mode='mouse'\np.yaxis[0].formatter = NumeralTickFormatter(format='£0,0')\np.xaxis.major_label_orientation = 3.14/5\np.legend.location = \"top_left\"\n# p.yaxis[0].formatter.use_scientific = False\nshow(p)",
"_____no_output_____"
]
],
[
[
"## Wealth chart",
"_____no_output_____"
]
],
[
[
"p = (df.wealth + df.cash_flow).rename('Net wealth').plot.line(\n show_figure=False,\n toolbar_location=None,\n hovertool_string=r\"\"\"<h4> Net wealth: </h4> £@{Net wealth}{0,0}\"\"\",\n panning=False,\n zooming=False\n)\np.yaxis[0].formatter = NumeralTickFormatter(format='$0,0')\np.xaxis.major_label_orientation = 3.14/5\np.legend.location = \"top_left\"\nshow(p)",
"_____no_output_____"
]
],
[
[
"## Outcomes chart in `pygal`",
"_____no_output_____"
]
],
[
[
"from IPython.display import HTML\nimport pygal",
"_____no_output_____"
],
[
"line_chart = pygal.Line(dynamic_print_values=True, value_formatter=lambda x: f'£{x:,.0f}')\nline_chart.title = 'Investment Outcomes:'\nline_chart.x_labels = map(str, range(0, years+1))\n\nline_chart = pygal.Line(dynamic_print_values=True, value_formatter=lambda x: f'£{x:,.0f}')\nfor c in df.columns:\n line_chart.add(c, df[c])\n\npygal_script = '<script type=\"text/javascript\" src=\"http://kozea.github.com/pygal.js/latest/pygal-tooltips.min.js\"></script>'\nHTML(line_chart.render(is_unicode=True)+pygal_script)",
"_____no_output_____"
]
],
[
[
"## Outcomes chart in `altair`",
"_____no_output_____"
]
],
[
[
"import altair as alt\nalt.renderers.enable('notebook')",
"_____no_output_____"
],
[
"altair_format_df = (\n df.reset_index()\n .melt('index', var_name='Factor', value_name='Amount')\n .rename(columns={'index':'Year'}))",
"_____no_output_____"
],
[
"alt.Chart(altair_format_df).mark_line(size=7, point=True).encode(\n x='Year:T',\n y='Amount:Q',\n color='Factor', \n tooltip=[\n 'Year:T', 'Factor:N', \n alt.Tooltip('Amount:Q', format='$,.2f')]\n).interactive()\n",
"_____no_output_____"
],
[
"alt.Chart(altair_format_df.query('Factor == \"wealth\"')).mark_area(opacity=0.4).encode(\n x='Year:T',\n y=alt.Y('Amount:Q', stack=None),\n color='Factor'\n)",
"_____no_output_____"
]
],
[
[
"# Step 2: Turn it into a callable\n\nOur intention is to examine multiple scenarios!",
"_____no_output_____"
]
],
[
[
"from download_listing import get_listing\nfrom mortgage import mortgage\nfrom home_value import home_value, letting_cash_flow\n\nfrom charts import altair_outcomes_chart, pygal_outcomes_chart, altair_wealth_chart, altair_format",
"_____no_output_____"
],
[
"def scenario(\n price=250_000,\n transaction_cost = 0.05,\n downpayment = 0.05,\n years = 25,\n inflation = .018,\n inflation_houseprice = 0.045,\n rate_mortgage = 0.035,\n rental_rate = 0,\n interest_only = False\n ):\n # ...\n return \"result\"",
"_____no_output_____"
],
[
"scenario(price=1_000_000, years=20, inflation=.1)",
"_____no_output_____"
]
],
[
[
"# Step 3: Make it interactive",
"_____no_output_____"
]
],
[
[
"from ipywidgets import interact",
"_____no_output_____"
],
[
"interact(scenario)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6c0d89038cd479f3d983f76675bf3ce6603f7c | 3,504 | ipynb | Jupyter Notebook | python/Generator_review .ipynb | djangojeng-e/djangoproejcts | 1efc3bc04a4a1bef039c906584cfecf0231c177f | [
"MIT"
]
| null | null | null | python/Generator_review .ipynb | djangojeng-e/djangoproejcts | 1efc3bc04a4a1bef039c906584cfecf0231c177f | [
"MIT"
]
| null | null | null | python/Generator_review .ipynb | djangojeng-e/djangoproejcts | 1efc3bc04a4a1bef039c906584cfecf0231c177f | [
"MIT"
]
| null | null | null | 21.9 | 99 | 0.478596 | [
[
[
"# Generator \n\n# Generator functions allow you to declare a function that behaves like an iterator. \n# i.e. it can be used in a for loop ",
"_____no_output_____"
],
[
"# simple function \n\ndef firstn(n):\n num = 0\n nums = []\n while num < n:\n nums.append(num)\n num += 1\n return nums\n\nsum_of_first_n = sum(firstn(1000000))\nprint(sum_of_first_n)\n\n# The code is simple but it does put the full list into the memory. \n# If the n goes to larger and larger, the memory will consume more. \n\n# Hence, alternatively, we can use the generator pattern. ",
"499999500000\n"
],
[
"# Generator Pattern \n\nclass firstn(object):\n def __init__(self, n):\n self.n = n \n self.num = 0\n \n def __iter__(self):\n return self\n \n def __next__(self):\n return self.next()\n \n def next(self):\n if self.num < self.n:\n cur, self.num = self.num, self.num+1\n return cur \n else:\n raise StopIteration()\n \n\nsum_of_first_n = sum(firstn(1000000))\nsum_of_first_n",
"_____no_output_____"
],
[
"# Instead of using the above class, \n# We can think of using generator function as a convenient shortcut to building iterators \n# Building the above iterator as a generator function as below \n\n# A generator that yields items instead of returning a list \n\ndef firstn(n):\n num = 0\n while num < n:\n yield num \n num += 1 \n \nsum_of_first_n = sum(firstn(1000000))\nsum_of_first_n",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
ec6c0f6dcb05eb7f697a712d671aa3f2948a7eba | 71,457 | ipynb | Jupyter Notebook | tfx/examples/chicago_taxi_pipeline/taxi_pipeline_interactive.ipynb | jkim1014/tfx | 5a4229639343a373fe4da9295b1ee5113912a902 | [
"Apache-2.0"
]
| 3 | 2020-07-20T18:37:16.000Z | 2021-11-17T11:24:27.000Z | tfx/examples/chicago_taxi_pipeline/taxi_pipeline_interactive.ipynb | jkim1014/tfx | 5a4229639343a373fe4da9295b1ee5113912a902 | [
"Apache-2.0"
]
| 2 | 2020-08-11T00:19:14.000Z | 2020-08-26T20:10:31.000Z | tfx/examples/chicago_taxi_pipeline/taxi_pipeline_interactive.ipynb | jkim1014/tfx | 5a4229639343a373fe4da9295b1ee5113912a902 | [
"Apache-2.0"
]
| 1 | 2020-11-06T11:44:33.000Z | 2020-11-06T11:44:33.000Z | 36.625833 | 538 | 0.554263 | [
[
[
"##### Copyright © 2019 The TensorFlow Authors.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# TFX Component Tutorial\n\n***A Component-by-Component Introduction to TensorFlow Extended (TFX)***",
"_____no_output_____"
],
[
"Note: We recommend running this tutorial in a Colab notebook, with no setup required! Just click \"Run in Google Colab\".\n\n<div class=\"devsite-table-wrapper\"><table class=\"tfo-notebook-buttons\" align=\"left\">\n<td><a target=\"_blank\" href=\"https://www.tensorflow.org/tfx/tutorials/tfx/components\">\n<img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a></td>\n<td><a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/tfx/blob/master/docs/tutorials/tfx/components.ipynb\">\n<img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\">Run in Google Colab</a></td>\n<td><a target=\"_blank\" href=\"https://github.com/tensorflow/tfx/tree/master/docs/tutorials/tfx/components.ipynb\">\n<img width=32px src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\">View source on GitHub</a></td>\n</table></div>",
"_____no_output_____"
],
[
"\nThis Colab-based tutorial will interactively walk through each built-in component of TensorFlow Extended (TFX).\n\nIt covers every step in an end-to-end machine learning pipeline, from data ingestion to pushing a model to serving.\n\nWhen you're done, the contents of this notebook can be automatically exported as TFX pipeline source code, which you can orchestrate with Apache Airflow and Apache Beam.\n\nNote: This notebook and its associated APIs are **experimental** and are\nin active development. Major changes in functionality, behavior, and\npresentation are expected.",
"_____no_output_____"
],
[
"## Background\nThis notebook demonstrates how to use TFX in a Jupyter/Colab environment. Here, we walk through the Chicago Taxi example in an interactive notebook.\n\nWorking in an interactive notebook is a useful way to become familiar with the structure of a TFX pipeline. It's also useful when doing development of your own pipelines as a lightweight development environment, but you should be aware that there are differences in the way interactive notebooks are orchestrated, and how they access metadata artifacts.\n\n### Orchestration\n\nIn a production deployment of TFX, you will use an orchestrator such as Apache Airflow, Kubeflow Pipelines, or Apache Beam to orchestrate a pre-defined pipeline graph of TFX components. In an interactive notebook, the notebook itself is the orchestrator, running each TFX component as you execute the notebook cells.\n\n### Metadata\n\nIn a production deployment of TFX, you will access metadata through the ML Metadata (MLMD) API. MLMD stores metadata properties in a database such as MySQL or SQLite, and stores the metadata payloads in a persistent store such as on your filesystem. In an interactive notebook, both properties and payloads are stored in an ephemeral SQLite database in the `/tmp` directory on the Jupyter notebook or Colab server.",
"_____no_output_____"
],
[
"## Setup\nFirst, we install and import the necessary packages, set up paths, and download data.",
"_____no_output_____"
],
[
"### Install TFX\n\n**Note: In Google Colab, because of package updates, the first time you run this cell you must restart the runtime (Runtime > Restart runtime ...).**",
"_____no_output_____"
]
],
[
[
"!pip install \"tfx>=0.21.1,<0.22\" \"tensorflow>=2.1,<2.2\" \"tensorboard>=2.1,<2.3\"",
"_____no_output_____"
]
],
[
[
"## Did you restart the runtime?\n\nIf you are using Google Colab, the first time that you run the cell above, you must restart the runtime (Runtime > Restart runtime ...). This is because of the way that Colab loads packages.",
"_____no_output_____"
],
[
"### Import packages\nWe import necessary packages, including standard TFX component classes.",
"_____no_output_____"
]
],
[
[
"import os\nimport pprint\nimport tempfile\nimport urllib\n\nimport absl\nimport tensorflow as tf\nimport tensorflow_model_analysis as tfma\ntf.get_logger().propagate = False\npp = pprint.PrettyPrinter()\n\nimport tfx\nfrom tfx.components import CsvExampleGen\nfrom tfx.components import Evaluator\nfrom tfx.components import ExampleValidator\nfrom tfx.components import Pusher\nfrom tfx.components import ResolverNode\nfrom tfx.components import SchemaGen\nfrom tfx.components import StatisticsGen\nfrom tfx.components import Trainer\nfrom tfx.components import Transform\nfrom tfx.dsl.experimental import latest_blessed_model_resolver\nfrom tfx.orchestration import metadata\nfrom tfx.orchestration import pipeline\nfrom tfx.orchestration.experimental.interactive.interactive_context import InteractiveContext\nfrom tfx.proto import pusher_pb2\nfrom tfx.proto import trainer_pb2\nfrom tfx.proto.evaluator_pb2 import SingleSlicingSpec\nfrom tfx.utils.dsl_utils import external_input\nfrom tfx.types import Channel\nfrom tfx.types.standard_artifacts import Model\nfrom tfx.types.standard_artifacts import ModelBlessing\n\n%load_ext tfx.orchestration.experimental.interactive.notebook_extensions.skip",
"_____no_output_____"
]
],
[
[
"Let's check the library versions.",
"_____no_output_____"
]
],
[
[
"print('TensorFlow version: {}'.format(tf.__version__))\nprint('TFX version: {}'.format(tfx.__version__))",
"_____no_output_____"
]
],
[
[
"### Set up pipeline paths",
"_____no_output_____"
]
],
[
[
"# This is the root directory for your TFX pip package installation.\n_tfx_root = tfx.__path__[0]\n\n# This is the directory containing the TFX Chicago Taxi Pipeline example.\n_taxi_root = os.path.join(_tfx_root, 'examples/chicago_taxi_pipeline')\n\n# This is the path where your model will be pushed for serving.\n_serving_model_dir = os.path.join(\n tempfile.mkdtemp(), 'serving_model/taxi_simple')\n\n# Set up logging.\nabsl.logging.set_verbosity(absl.logging.INFO)",
"_____no_output_____"
]
],
[
[
"### Download example data\nWe download the example dataset for use in our TFX pipeline.\n\nThe dataset we're using is the [Taxi Trips dataset](https://data.cityofchicago.org/Transportation/Taxi-Trips/wrvz-psew) released by the City of Chicago. The columns in this dataset are:\n\n<table>\n<tr><td>pickup_community_area</td><td>fare</td><td>trip_start_month</td></tr>\n<tr><td>trip_start_hour</td><td>trip_start_day</td><td>trip_start_timestamp</td></tr>\n<tr><td>pickup_latitude</td><td>pickup_longitude</td><td>dropoff_latitude</td></tr>\n<tr><td>dropoff_longitude</td><td>trip_miles</td><td>pickup_census_tract</td></tr>\n<tr><td>dropoff_census_tract</td><td>payment_type</td><td>company</td></tr>\n<tr><td>trip_seconds</td><td>dropoff_community_area</td><td>tips</td></tr>\n</table>\n\nWith this dataset, we will build a model that predicts the `tips` of a trip.",
"_____no_output_____"
]
],
[
[
"_data_root = tempfile.mkdtemp(prefix='tfx-data')\nDATA_PATH = 'https://raw.githubusercontent.com/tensorflow/tfx/master/tfx/examples/chicago_taxi_pipeline/data/simple/data.csv'\n_data_filepath = os.path.join(_data_root, \"data.csv\")\nurllib.request.urlretrieve(DATA_PATH, _data_filepath)",
"_____no_output_____"
]
],
[
[
"Take a quick look at the CSV file.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\n!head {_data_filepath}",
"_____no_output_____"
]
],
[
[
"*Disclaimer: This site provides applications using data that has been modified for use from its original source, www.cityofchicago.org, the official website of the City of Chicago. The City of Chicago makes no claims as to the content, accuracy, timeliness, or completeness of any of the data provided at this site. The data provided at this site is subject to change at any time. It is understood that the data provided at this site is being used at one’s own risk.*",
"_____no_output_____"
],
[
"### Create the InteractiveContext\nLast, we create an InteractiveContext, which will allow us to run TFX components interactively in this notebook.",
"_____no_output_____"
]
],
[
[
"# Here, we create an InteractiveContext using default parameters. This will\n# use a temporary directory with an ephemeral ML Metadata database instance.\n# To use your own pipeline root or database, the optional properties\n# `pipeline_root` and `metadata_connection_config` may be passed to\n# InteractiveContext. Calls to InteractiveContext are no-ops outside of the\n# notebook.\ncontext = InteractiveContext()",
"_____no_output_____"
]
],
[
[
"## Run TFX components interactively\nIn the cells that follow, we create TFX components one-by-one, run each of them, and visualize their output artifacts.",
"_____no_output_____"
],
[
"### ExampleGen\n\nThe `ExampleGen` component is usually at the start of a TFX pipeline. It will:\n\n1. Split data into training and evaluation sets (by default, 2/3 training + 1/3 eval)\n2. Convert data into the `tf.Example` format\n3. Copy data into the `_tfx_root` directory for other components to access\n\n`ExampleGen` takes as input the path to your data source. In our case, this is the `_data_root` path that contains the downloaded CSV.\n\nNote: In this notebook, we can instantiate components one-by-one and run them with `InteractiveContext.run()`. By contrast, in a production setting, we would specify all the components upfront in a `Pipeline` to pass to the orchestrator (see the \"Export to Pipeline\" section).",
"_____no_output_____"
]
],
[
[
"example_gen = CsvExampleGen(input=external_input(_data_root))\ncontext.run(example_gen)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `ExampleGen`. This component produces two artifacts, training examples and evaluation examples:\n\nNote: The `%%skip_for_export` cell magic will omit the contents of this cell in the exported pipeline file (see the \"Export to pipeline\" section). This is useful for notebook-specific code that you don't want to run in an orchestrated pipeline.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nartifact = example_gen.outputs['examples'].get()[0]\nprint(artifact.split_names, artifact.uri)",
"_____no_output_____"
]
],
[
[
"We can also take a look at the first three training examples:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\n# Get the URI of the output artifact representing the training examples, which is a directory\ntrain_uri = os.path.join(example_gen.outputs['examples'].get()[0].uri, 'train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"Now that `ExampleGen` has finished ingesting the data, the next step is data analysis.",
"_____no_output_____"
],
[
"### StatisticsGen\nThe `StatisticsGen` component computes statistics over your dataset for data analysis, as well as for use in downstream components. It uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`StatisticsGen` takes as input the dataset we just ingested using `ExampleGen`.",
"_____no_output_____"
]
],
[
[
"statistics_gen = StatisticsGen(\n examples=example_gen.outputs['examples'])\ncontext.run(statistics_gen)",
"_____no_output_____"
]
],
[
[
"After `StatisticsGen` finishes running, we can visualize the outputted statistics. Try playing with the different plots!",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(statistics_gen.outputs['statistics'])",
"_____no_output_____"
]
],
[
[
"### SchemaGen\n\nThe `SchemaGen` component generates a schema based on your data statistics. (A schema defines the expected bounds, types, and properties of the features in your dataset.) It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`SchemaGen` will take as input the statistics that we generated with `StatisticsGen`, looking at the training split by default.",
"_____no_output_____"
]
],
[
[
"schema_gen = SchemaGen(\n statistics=statistics_gen.outputs['statistics'],\n infer_feature_shape=False)\ncontext.run(schema_gen)",
"_____no_output_____"
]
],
[
[
"After `SchemaGen` finishes running, we can visualize the generated schema as a table.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(schema_gen.outputs['schema'])",
"_____no_output_____"
]
],
[
[
"Each feature in your dataset shows up as a row in the schema table, alongside its properties. The schema also captures all the values that a categorical feature takes on, denoted as its domain.\n\nTo learn more about schemas, see [the SchemaGen documentation](https://www.tensorflow.org/tfx/guide/schemagen).",
"_____no_output_____"
],
[
"### ExampleValidator\nThe `ExampleValidator` component detects anomalies in your data, based on the expectations defined by the schema. It also uses the [TensorFlow Data Validation](https://www.tensorflow.org/tfx/data_validation/get_started) library.\n\n`ExampleValidator` will take as input the statistics from `StatisticsGen`, and the schema from `SchemaGen`.\n\nBy default, it compares the statistics from the evaluation split to the schema from the training split.",
"_____no_output_____"
]
],
[
[
"example_validator = ExampleValidator(\n statistics=statistics_gen.outputs['statistics'],\n schema=schema_gen.outputs['schema'])\ncontext.run(example_validator)",
"_____no_output_____"
]
],
[
[
"After `ExampleValidator` finishes running, we can visualize the anomalies as a table.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(example_validator.outputs['anomalies'])",
"_____no_output_____"
]
],
[
[
"In the anomalies table, we can see that the `company` feature takes on new values that were not in the training split. This information can be used to debug model performance, understand how your data evolves over time, and identify data errors.\n\nIn our case, this `company` anomaly is innocuous, but the `payment_type` could be fixed. For now we move on to the next step of transforming the data.",
"_____no_output_____"
],
[
"### Transform\nThe `Transform` component performs feature engineering for both training and serving. It uses the [TensorFlow Transform](https://www.tensorflow.org/tfx/transform/get_started) library.\n\n`Transform` will take as input the data from `ExampleGen`, the schema from `SchemaGen`, as well as a module that contains user-defined Transform code.\n\nLet's see an example of user-defined Transform code below (for an introduction to the TensorFlow Transform APIs, [see the tutorial](https://www.tensorflow.org/tfx/tutorials/transform/simple)). First, we define a few constants for feature engineering:\n\nNote: The `%%writefile` cell magic will save the contents of the cell as a `.py` file on disk. This allows the `Transform` component to load your code as a module.\n\n",
"_____no_output_____"
]
],
[
[
"_taxi_constants_module_file = 'taxi_constants.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_constants_module_file}\n\n# Categorical features are assumed to each have a maximum value in the dataset.\nMAX_CATEGORICAL_FEATURE_VALUES = [24, 31, 12]\n\nCATEGORICAL_FEATURE_KEYS = [\n 'trip_start_hour', 'trip_start_day', 'trip_start_month',\n 'pickup_census_tract', 'dropoff_census_tract', 'pickup_community_area',\n 'dropoff_community_area'\n]\n\nDENSE_FLOAT_FEATURE_KEYS = ['trip_miles', 'fare', 'trip_seconds']\n\n# Number of buckets used by tf.transform for encoding each feature.\nFEATURE_BUCKET_COUNT = 10\n\nBUCKET_FEATURE_KEYS = [\n 'pickup_latitude', 'pickup_longitude', 'dropoff_latitude',\n 'dropoff_longitude'\n]\n\n# Number of vocabulary terms used for encoding VOCAB_FEATURES by tf.transform\nVOCAB_SIZE = 1000\n\n# Count of out-of-vocab buckets in which unrecognized VOCAB_FEATURES are hashed.\nOOV_SIZE = 10\n\nVOCAB_FEATURE_KEYS = [\n 'payment_type',\n 'company',\n]\n\n# Keys\nLABEL_KEY = 'tips'\nFARE_KEY = 'fare'\n\ndef transformed_name(key):\n return key + '_xf'",
"_____no_output_____"
]
],
[
[
"Next, we write a `preprocessing_fn` that takes in raw data as input, and returns transformed features that our model can train on:",
"_____no_output_____"
]
],
[
[
"_taxi_transform_module_file = 'taxi_transform.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_transform_module_file}\n\nimport tensorflow as tf\nimport tensorflow_transform as tft\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_FARE_KEY = taxi_constants.FARE_KEY\n_LABEL_KEY = taxi_constants.LABEL_KEY\n_transformed_name = taxi_constants.transformed_name\n\n\ndef preprocessing_fn(inputs):\n \"\"\"tf.transform's callback function for preprocessing inputs.\n Args:\n inputs: map from feature keys to raw not-yet-transformed features.\n Returns:\n Map from string feature key to transformed feature operations.\n \"\"\"\n outputs = {}\n for key in _DENSE_FLOAT_FEATURE_KEYS:\n # Preserve this feature as a dense float, setting nan's to the mean.\n outputs[_transformed_name(key)] = tft.scale_to_z_score(\n _fill_in_missing(inputs[key]))\n\n for key in _VOCAB_FEATURE_KEYS:\n # Build a vocabulary for this feature.\n outputs[_transformed_name(key)] = tft.compute_and_apply_vocabulary(\n _fill_in_missing(inputs[key]),\n top_k=_VOCAB_SIZE,\n num_oov_buckets=_OOV_SIZE)\n\n for key in _BUCKET_FEATURE_KEYS:\n outputs[_transformed_name(key)] = tft.bucketize(\n _fill_in_missing(inputs[key]), _FEATURE_BUCKET_COUNT)\n\n for key in _CATEGORICAL_FEATURE_KEYS:\n outputs[_transformed_name(key)] = _fill_in_missing(inputs[key])\n\n # Was this passenger a big tipper?\n taxi_fare = _fill_in_missing(inputs[_FARE_KEY])\n tips = _fill_in_missing(inputs[_LABEL_KEY])\n outputs[_transformed_name(_LABEL_KEY)] = tf.where(\n tf.math.is_nan(taxi_fare),\n tf.cast(tf.zeros_like(taxi_fare), tf.int64),\n # Test if the tip was > 20% of the fare.\n tf.cast(\n tf.greater(tips, tf.multiply(taxi_fare, tf.constant(0.2))), tf.int64))\n\n return outputs\n\n\ndef _fill_in_missing(x):\n \"\"\"Replace missing values in a SparseTensor.\n Fills in missing values of `x` with '' or 0, and converts to a dense tensor.\n Args:\n x: A `SparseTensor` of rank 2. Its dense shape should have size at most 1\n in the second dimension.\n Returns:\n A rank 1 tensor where missing values of `x` have been filled in.\n \"\"\"\n default_value = '' if x.dtype == tf.string else 0\n return tf.squeeze(\n tf.sparse.to_dense(\n tf.SparseTensor(x.indices, x.values, [x.dense_shape[0], 1]),\n default_value),\n axis=1)",
"_____no_output_____"
]
],
[
[
"Now, we pass in this feature engineering code to the `Transform` component and run it to transform your data.",
"_____no_output_____"
]
],
[
[
"transform = Transform(\n examples=example_gen.outputs['examples'],\n schema=schema_gen.outputs['schema'],\n module_file=os.path.abspath(_taxi_transform_module_file))\ncontext.run(transform)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Transform`. This component produces two types of outputs:\n\n* `transform_graph` is the graph that can perform the preprocessing operations (this graph will be included in the serving and evaluation models).\n* `transformed_examples` represents the preprocessed training and evaluation data.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ntransform.outputs",
"_____no_output_____"
]
],
[
[
"Take a peek at the `transform_graph` artifact. It points to a directory containing three subdirectories.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ntrain_uri = transform.outputs['transform_graph'].get()[0].uri\nos.listdir(train_uri)",
"_____no_output_____"
]
],
[
[
"The `transformed_metadata` subdirectory contains the schema of the preprocessed data. The `transform_fn` subdirectory contains the actual preprocessing graph. The `metadata` subdirectory contains the schema of the original data.\n\nWe can also take a look at the first three transformed examples:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\n# Get the URI of the output artifact representing the transformed examples, which is a directory\ntrain_uri = os.path.join(transform.outputs['transformed_examples'].get()[0].uri, 'train')\n\n# Get the list of files in this directory (all compressed TFRecord files)\ntfrecord_filenames = [os.path.join(train_uri, name)\n for name in os.listdir(train_uri)]\n\n# Create a `TFRecordDataset` to read these files\ndataset = tf.data.TFRecordDataset(tfrecord_filenames, compression_type=\"GZIP\")\n\n# Iterate over the first 3 records and decode them.\nfor tfrecord in dataset.take(3):\n serialized_example = tfrecord.numpy()\n example = tf.train.Example()\n example.ParseFromString(serialized_example)\n pp.pprint(example)",
"_____no_output_____"
]
],
[
[
"After the `Transform` component has transformed your data into features, and the next step is to train a model.",
"_____no_output_____"
],
[
"### Trainer\nThe `Trainer` component will train a model that you define in TensorFlow (either using the Estimator API or the Keras API with [`model_to_estimator`](https://www.tensorflow.org/api_docs/python/tf/keras/estimator/model_to_estimator)).\n\n`Trainer` takes as input the schema from `SchemaGen`, the transformed data and graph from `Transform`, training parameters, as well as a module that contains user-defined model code.\n\nLet's see an example of user-defined model code below (for an introduction to the TensorFlow Estimator APIs, [see the tutorial](https://www.tensorflow.org/tutorials/estimator/premade)):",
"_____no_output_____"
]
],
[
[
"_taxi_trainer_module_file = 'taxi_trainer.py'",
"_____no_output_____"
],
[
"%%skip_for_export\n%%writefile {_taxi_trainer_module_file}\n\nimport tensorflow as tf\nimport tensorflow_model_analysis as tfma\nimport tensorflow_transform as tft\nfrom tensorflow_transform.tf_metadata import schema_utils\n\nimport taxi_constants\n\n_DENSE_FLOAT_FEATURE_KEYS = taxi_constants.DENSE_FLOAT_FEATURE_KEYS\n_VOCAB_FEATURE_KEYS = taxi_constants.VOCAB_FEATURE_KEYS\n_VOCAB_SIZE = taxi_constants.VOCAB_SIZE\n_OOV_SIZE = taxi_constants.OOV_SIZE\n_FEATURE_BUCKET_COUNT = taxi_constants.FEATURE_BUCKET_COUNT\n_BUCKET_FEATURE_KEYS = taxi_constants.BUCKET_FEATURE_KEYS\n_CATEGORICAL_FEATURE_KEYS = taxi_constants.CATEGORICAL_FEATURE_KEYS\n_MAX_CATEGORICAL_FEATURE_VALUES = taxi_constants.MAX_CATEGORICAL_FEATURE_VALUES\n_LABEL_KEY = taxi_constants.LABEL_KEY\n_transformed_name = taxi_constants.transformed_name\n\n\ndef _transformed_names(keys):\n return [_transformed_name(key) for key in keys]\n\n\n# Tf.Transform considers these features as \"raw\"\ndef _get_raw_feature_spec(schema):\n return schema_utils.schema_as_feature_spec(schema).feature_spec\n\n\ndef _gzip_reader_fn(filenames):\n \"\"\"Small utility returning a record reader that can read gzip'ed files.\"\"\"\n return tf.data.TFRecordDataset(\n filenames,\n compression_type='GZIP')\n\n\ndef _build_estimator(config, hidden_units=None, warm_start_from=None):\n \"\"\"Build an estimator for predicting the tipping behavior of taxi riders.\n Args:\n config: tf.estimator.RunConfig defining the runtime environment for the\n estimator (including model_dir).\n hidden_units: [int], the layer sizes of the DNN (input layer first)\n warm_start_from: Optional directory to warm start from.\n Returns:\n A dict of the following:\n - estimator: The estimator that will be used for training and eval.\n - train_spec: Spec for training.\n - eval_spec: Spec for eval.\n - eval_input_receiver_fn: Input function for eval.\n \"\"\"\n real_valued_columns = [\n tf.feature_column.numeric_column(key, shape=())\n for key in _transformed_names(_DENSE_FLOAT_FEATURE_KEYS)\n ]\n categorical_columns = [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_VOCAB_SIZE + _OOV_SIZE, default_value=0)\n for key in _transformed_names(_VOCAB_FEATURE_KEYS)\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity(\n key, num_buckets=_FEATURE_BUCKET_COUNT, default_value=0)\n for key in _transformed_names(_BUCKET_FEATURE_KEYS)\n ]\n categorical_columns += [\n tf.feature_column.categorical_column_with_identity( # pylint: disable=g-complex-comprehension\n key,\n num_buckets=num_buckets,\n default_value=0) for key, num_buckets in zip(\n _transformed_names(_CATEGORICAL_FEATURE_KEYS),\n _MAX_CATEGORICAL_FEATURE_VALUES)\n ]\n return tf.estimator.DNNLinearCombinedClassifier(\n config=config,\n linear_feature_columns=categorical_columns,\n dnn_feature_columns=real_valued_columns,\n dnn_hidden_units=hidden_units or [100, 70, 50, 25],\n warm_start_from=warm_start_from)\n\n\ndef _example_serving_receiver_fn(tf_transform_graph, schema):\n \"\"\"Build the serving in inputs.\n Args:\n tf_transform_graph: A TFTransformOutput.\n schema: the schema of the input data.\n Returns:\n Tensorflow graph which parses examples, applying tf-transform to them.\n \"\"\"\n raw_feature_spec = _get_raw_feature_spec(schema)\n raw_feature_spec.pop(_LABEL_KEY)\n\n raw_input_fn = tf.estimator.export.build_parsing_serving_input_receiver_fn(\n raw_feature_spec, default_batch_size=None)\n serving_input_receiver = raw_input_fn()\n\n transformed_features = tf_transform_graph.transform_raw_features(\n serving_input_receiver.features)\n\n return tf.estimator.export.ServingInputReceiver(\n transformed_features, serving_input_receiver.receiver_tensors)\n\n\ndef _eval_input_receiver_fn(tf_transform_graph, schema):\n \"\"\"Build everything needed for the tf-model-analysis to run the model.\n Args:\n tf_transform_graph: A TFTransformOutput.\n schema: the schema of the input data.\n Returns:\n EvalInputReceiver function, which contains:\n - Tensorflow graph which parses raw untransformed features, applies the\n tf-transform preprocessing operators.\n - Set of raw, untransformed features.\n - Label against which predictions will be compared.\n \"\"\"\n # Notice that the inputs are raw features, not transformed features here.\n raw_feature_spec = _get_raw_feature_spec(schema)\n\n serialized_tf_example = tf.compat.v1.placeholder(\n dtype=tf.string, shape=[None], name='input_example_tensor')\n\n # Add a parse_example operator to the tensorflow graph, which will parse\n # raw, untransformed, tf examples.\n features = tf.io.parse_example(serialized_tf_example, raw_feature_spec)\n\n # Now that we have our raw examples, process them through the tf-transform\n # function computed during the preprocessing step.\n transformed_features = tf_transform_graph.transform_raw_features(\n features)\n\n # The key name MUST be 'examples'.\n receiver_tensors = {'examples': serialized_tf_example}\n\n # NOTE: Model is driven by transformed features (since training works on the\n # materialized output of TFT, but slicing will happen on raw features.\n features.update(transformed_features)\n\n return tfma.export.EvalInputReceiver(\n features=features,\n receiver_tensors=receiver_tensors,\n labels=transformed_features[_transformed_name(_LABEL_KEY)])\n\n\ndef _input_fn(filenames, tf_transform_graph, batch_size=200):\n \"\"\"Generates features and labels for training or evaluation.\n Args:\n filenames: [str] list of CSV files to read data from.\n tf_transform_graph: A TFTransformOutput.\n batch_size: int First dimension size of the Tensors returned by input_fn\n Returns:\n A (features, indices) tuple where features is a dictionary of\n Tensors, and indices is a single Tensor of label indices.\n \"\"\"\n transformed_feature_spec = (\n tf_transform_graph.transformed_feature_spec().copy())\n\n dataset = tf.data.experimental.make_batched_features_dataset(\n filenames, batch_size, transformed_feature_spec, reader=_gzip_reader_fn)\n\n transformed_features = (\n tf.compat.v1.data.make_one_shot_iterator(dataset).get_next())\n # We pop the label because we do not want to use it as a feature while we're\n # training.\n return transformed_features, transformed_features.pop(\n _transformed_name(_LABEL_KEY))\n\n\n# TFX will call this function\ndef trainer_fn(trainer_fn_args, schema):\n \"\"\"Build the estimator using the high level API.\n Args:\n trainer_fn_args: Holds args used to train the model as name/value pairs.\n schema: Holds the schema of the training examples.\n Returns:\n A dict of the following:\n - estimator: The estimator that will be used for training and eval.\n - train_spec: Spec for training.\n - eval_spec: Spec for eval.\n - eval_input_receiver_fn: Input function for eval.\n \"\"\"\n # Number of nodes in the first layer of the DNN\n first_dnn_layer_size = 100\n num_dnn_layers = 4\n dnn_decay_factor = 0.7\n\n train_batch_size = 40\n eval_batch_size = 40\n\n tf_transform_graph = tft.TFTransformOutput(trainer_fn_args.transform_output)\n\n train_input_fn = lambda: _input_fn( # pylint: disable=g-long-lambda\n trainer_fn_args.train_files,\n tf_transform_graph,\n batch_size=train_batch_size)\n\n eval_input_fn = lambda: _input_fn( # pylint: disable=g-long-lambda\n trainer_fn_args.eval_files,\n tf_transform_graph,\n batch_size=eval_batch_size)\n\n train_spec = tf.estimator.TrainSpec( # pylint: disable=g-long-lambda\n train_input_fn,\n max_steps=trainer_fn_args.train_steps)\n\n serving_receiver_fn = lambda: _example_serving_receiver_fn( # pylint: disable=g-long-lambda\n tf_transform_graph, schema)\n\n exporter = tf.estimator.FinalExporter('chicago-taxi', serving_receiver_fn)\n eval_spec = tf.estimator.EvalSpec(\n eval_input_fn,\n steps=trainer_fn_args.eval_steps,\n exporters=[exporter],\n name='chicago-taxi-eval')\n\n run_config = tf.estimator.RunConfig(\n save_checkpoints_steps=999, keep_checkpoint_max=1)\n\n run_config = run_config.replace(model_dir=trainer_fn_args.serving_model_dir)\n\n estimator = _build_estimator(\n # Construct layers sizes with exponetial decay\n hidden_units=[\n max(2, int(first_dnn_layer_size * dnn_decay_factor**i))\n for i in range(num_dnn_layers)\n ],\n config=run_config,\n warm_start_from=trainer_fn_args.base_model)\n\n # Create an input receiver for TFMA processing\n receiver_fn = lambda: _eval_input_receiver_fn( # pylint: disable=g-long-lambda\n tf_transform_graph, schema)\n\n return {\n 'estimator': estimator,\n 'train_spec': train_spec,\n 'eval_spec': eval_spec,\n 'eval_input_receiver_fn': receiver_fn\n }",
"_____no_output_____"
]
],
[
[
"Now, we pass in this model code to the `Trainer` component and run it to train the model.",
"_____no_output_____"
]
],
[
[
"trainer = Trainer(\n module_file=os.path.abspath(_taxi_trainer_module_file),\n transformed_examples=transform.outputs['transformed_examples'],\n schema=schema_gen.outputs['schema'],\n transform_graph=transform.outputs['transform_graph'],\n train_args=trainer_pb2.TrainArgs(num_steps=10000),\n eval_args=trainer_pb2.EvalArgs(num_steps=5000))\ncontext.run(trainer)",
"_____no_output_____"
]
],
[
[
"#### Analyze Training with TensorBoard\nOptionally, we can connect TensorBoard to the Trainer to analyze our model's training curves.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\n# Get the URI of the output artifact representing the training logs, which is a directory\nmodel_dir = trainer.outputs['model'].get()[0].uri\n\n%load_ext tensorboard\n%tensorboard --logdir {model_dir}",
"_____no_output_____"
]
],
[
[
"### Evaluator\nThe `Evaluator` component computes model performance metrics over the evaluation set. It uses the [TensorFlow Model Analysis](https://www.tensorflow.org/tfx/model_analysis/get_started) library. The `Evaluator` can also optionally validate that a newly trained model is better than the previous model. This is useful in a production pipeline setting where you may automatically train and validate a model every day. In this notebook, we only train one model, so the `Evaluator` automatically will label the model as \"good\". \n\n`Evaluator` will take as input the data from `ExampleGen`, the trained model from `Trainer`, and slicing configuration. The slicing configuration allows you to slice your metrics on feature values (e.g. how does your model perform on taxi trips that start at 8am versus 8pm?). See an example of this configuration below:",
"_____no_output_____"
]
],
[
[
"eval_config = tfma.EvalConfig(\n model_specs=[\n # Using signature 'eval' implies the use of an EvalSavedModel. To use\n # a serving model remove the signature to defaults to 'serving_default'\n # and add a label_key.\n tfma.ModelSpec(signature_name='eval')\n ],\n metrics_specs=[\n tfma.MetricsSpec(\n # The metrics added here are in addition to those saved with the\n # model (assuming either a keras model or EvalSavedModel is used).\n # Any metrics added into the saved model (for example using\n # model.compile(..., metrics=[...]), etc) will be computed\n # automatically.\n metrics=[\n tfma.MetricConfig(class_name='ExampleCount')\n ],\n # To add validation thresholds for metrics saved with the model,\n # add them keyed by metric name to the thresholds map.\n thresholds = {\n 'accuracy': tfma.MetricThreshold(\n value_threshold=tfma.GenericValueThreshold(\n lower_bound={'value': 0.5}),\n change_threshold=tfma.GenericChangeThreshold(\n direction=tfma.MetricDirection.HIGHER_IS_BETTER,\n absolute={'value': -1e-10}))\n }\n )\n ],\n slicing_specs=[\n # An empty slice spec means the overall slice, i.e. the whole dataset.\n tfma.SlicingSpec(),\n # Data can be sliced along a feature column. In this case, data is\n # sliced along feature column trip_start_hour.\n tfma.SlicingSpec(feature_keys=['trip_start_hour'])\n ])",
"_____no_output_____"
]
],
[
[
"Next, we give this configuration to `Evaluator` and run it.",
"_____no_output_____"
]
],
[
[
"# Use TFMA to compute a evaluation statistics over features of a model and\n# validate them against a baseline.\n\n# The model resolver is only required if performing model validation in addition\n# to evaluation. In this case we validate against the latest blessed model. If\n# no model has been blessed before (as in this case) the evaluator will make our\n# candidate the first blessed model.\nmodel_resolver = ResolverNode(\n instance_name='latest_blessed_model_resolver',\n resolver_class=latest_blessed_model_resolver.LatestBlessedModelResolver,\n model=Channel(type=Model),\n model_blessing=Channel(type=ModelBlessing))\ncontext.run(model_resolver)\n\nevaluator = Evaluator(\n examples=example_gen.outputs['examples'],\n model=trainer.outputs['model'],\n #baseline_model=model_resolver.outputs['model'],\n # Change threshold will be ignored if there is no baseline (first run).\n eval_config=eval_config)\ncontext.run(evaluator)",
"_____no_output_____"
]
],
[
[
"Now let's examine the output artifacts of `Evaluator`. ",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nevaluator.outputs",
"_____no_output_____"
]
],
[
[
"Using the `evaluation` output we can show the default visualization of global metrics on the entire evaluation set.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\ncontext.show(evaluator.outputs['evaluation'])",
"_____no_output_____"
]
],
[
[
"To see the visualization for sliced evaluation metrics, we can directly call the TensorFlow Model Analysis library.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nimport tensorflow_model_analysis as tfma\n\n# Get the TFMA output result path and load the result.\nPATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\ntfma_result = tfma.load_eval_result(PATH_TO_RESULT)\n\n# Show data sliced along feature column trip_start_hour.\ntfma.view.render_slicing_metrics(\n tfma_result, slicing_column='trip_start_hour')",
"_____no_output_____"
]
],
[
[
"This visualization shows the same metrics, but computed at every feature value of `trip_start_hour` instead of on the entire evaluation set.\n\nTensorFlow Model Analysis supports many other visualizations, such as Fairness Indicators and plotting a time series of model performance. To learn more, see [the tutorial](https://www.tensorflow.org/tfx/tutorials/model_analysis/tfma_basic).",
"_____no_output_____"
],
[
"Since we added thresholds to our config, validation output is also available. The precence of a `blessing` artifact indicates that our model passed validation. Since this is the first validation being performed the candidate is automatically blessed.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nblessing_uri = evaluator.outputs.blessing.get()[0].uri\n!ls -l {blessing_uri}",
"_____no_output_____"
]
],
[
[
"Now can also verify the success by loading the validation result record:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\nPATH_TO_RESULT = evaluator.outputs['evaluation'].get()[0].uri\nprint(tfma.load_validation_result(PATH_TO_RESULT))",
"_____no_output_____"
]
],
[
[
"### Pusher\nThe `Pusher` component is usually at the end of a TFX pipeline. It checks whether a model has passed validation, and if so, exports the model to `_serving_model_dir`.",
"_____no_output_____"
]
],
[
[
"pusher = Pusher(\n model=trainer.outputs['model'],\n model_blessing=evaluator.outputs['blessing'],\n push_destination=pusher_pb2.PushDestination(\n filesystem=pusher_pb2.PushDestination.Filesystem(\n base_directory=_serving_model_dir)))\ncontext.run(pusher)",
"_____no_output_____"
]
],
[
[
"Let's examine the output artifacts of `Pusher`. ",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\npusher.outputs",
"_____no_output_____"
]
],
[
[
"In particular, the Pusher will export your model in the SavedModel format, which looks like this:",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n\npush_uri = pusher.outputs.model_push.get()[0].uri\nlatest_version = max(os.listdir(push_uri))\nlatest_version_path = os.path.join(push_uri, latest_version)\nmodel = tf.saved_model.load(latest_version_path)\n\nfor item in model.signatures.items():\n pp.pprint(item)",
"_____no_output_____"
]
],
[
[
"We're finished our tour of built-in TFX components!\n\nAfter you're happy with experimenting with TFX components and code in this notebook, you may want to export it as a pipeline to be orchestrated with Apache Airflow or Apache Beam. See the final section.",
"_____no_output_____"
],
[
"## Export to pipeline\n\nTo export the contents of this notebook as a pipeline to be orchestrated with Airflow or Beam, follow the instructions below.\n\nIf you're using Colab, make sure to **save this notebook to Google Drive** (`File` → `Save a Copy in Drive`) before exporting.",
"_____no_output_____"
],
[
"### 1. Mount Google Drive (Colab-only)\n\nIf you're using Colab, this notebook needs to mount your Google Drive to be able to access its own `.ipynb` file.",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n#docs_infra: no_execute\n\n#@markdown Run this cell and enter the authorization code to mount Google Drive.\n\nimport sys\n\nif 'google.colab' in sys.modules:\n # Colab.\n from google.colab import drive\n\n drive.mount('/content/drive')",
"_____no_output_____"
]
],
[
[
"### 2. Select an orchestrator",
"_____no_output_____"
]
],
[
[
"_runner_type = 'beam' #@param [\"beam\", \"airflow\"]\n_pipeline_name = 'chicago_taxi_%s' % _runner_type",
"_____no_output_____"
]
],
[
[
"### 3. Set up paths for the pipeline",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n# For Colab notebooks only.\n# TODO(USER): Fill out the path to this notebook.\n_notebook_filepath = (\n '/content/drive/My Drive/Colab Notebooks/components.ipynb')\n\n# For Jupyter notebooks only.\n# _notebook_filepath = os.path.join(os.getcwd(),\n# 'taxi_pipeline_interactive.ipynb')\n\n# TODO(USER): Fill out the paths for the exported pipeline.\n_tfx_root = os.path.join(os.environ['HOME'], 'tfx')\n_taxi_root = os.path.join(os.environ['HOME'], 'taxi')\n_serving_model_dir = os.path.join(_taxi_root, 'serving_model')\n_data_root = os.path.join(_taxi_root, 'data', 'simple')\n_pipeline_root = os.path.join(_tfx_root, 'pipelines', _pipeline_name)\n_metadata_path = os.path.join(_tfx_root, 'metadata', _pipeline_name,\n 'metadata.db')",
"_____no_output_____"
]
],
[
[
"### 4. Choose components to include in the pipeline",
"_____no_output_____"
]
],
[
[
"#docs_infra: no_execute\n# TODO(USER): Specify components to be included in the exported pipeline.\ncomponents = [\n example_gen, statistics_gen, schema_gen, example_validator, transform,\n trainer, evaluator, pusher\n]",
"_____no_output_____"
]
],
[
[
"### 5. Generate pipeline files",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n#docs_infra: no_execute\n\n#@markdown Run this cell to generate the pipeline files.\n\nif get_ipython().magics_manager.auto_magic:\n print('Warning: %automagic is ON. Line magics specified without the % prefix '\n 'will not be scrubbed during export to pipeline.')\n\n_pipeline_export_filepath = 'export_%s.py' % _pipeline_name\ncontext.export_to_pipeline(notebook_filepath=_notebook_filepath,\n export_filepath=_pipeline_export_filepath,\n runner_type=_runner_type)",
"_____no_output_____"
]
],
[
[
"### 6. Download pipeline files",
"_____no_output_____"
]
],
[
[
"%%skip_for_export\n#docs_infra: no_execute\n\n#@markdown Run this cell to download the pipeline files as a `.zip`.\n\nif 'google.colab' in sys.modules:\n from google.colab import files\n import zipfile\n\n zip_export_path = os.path.join(\n tempfile.mkdtemp(), 'export.zip')\n with zipfile.ZipFile(zip_export_path, mode='w') as export_zip:\n export_zip.write(_pipeline_export_filepath)\n export_zip.write(_taxi_constants_module_file)\n export_zip.write(_taxi_transform_module_file)\n export_zip.write(_taxi_trainer_module_file)\n\n files.download(zip_export_path)",
"_____no_output_____"
]
],
[
[
"To learn how to run the orchestrated pipeline with Apache Airflow, please refer to the [TFX Orchestration Tutorial](https://www.tensorflow.org/tfx/tutorials/tfx/airflow_workshop).",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6c2f98e369c4d7a02d7ce713c6b94f7bc97a3e | 44,309 | ipynb | Jupyter Notebook | notebooks/Resource_estimator_Hannah.ipynb | Geothermal-Resource-Capacity/Power-Density | 09261ad705725e8c9213f39d96f0dfa7cb762cb4 | [
"Apache-2.0"
]
| 1 | 2021-06-08T01:42:00.000Z | 2021-06-08T01:42:00.000Z | notebooks/Resource_estimator_Hannah.ipynb | Geothermal-Resource-Capacity/Power-Density | 09261ad705725e8c9213f39d96f0dfa7cb762cb4 | [
"Apache-2.0"
]
| 4 | 2021-07-03T07:51:48.000Z | 2021-09-16T06:15:54.000Z | notebooks/Resource_estimator_Hannah-checkpoint.ipynb | Geothermal-Resource-Capacity/Power-Density | 09261ad705725e8c9213f39d96f0dfa7cb762cb4 | [
"Apache-2.0"
]
| 1 | 2021-06-01T04:38:11.000Z | 2021-06-01T04:38:11.000Z | 139.336478 | 19,448 | 0.882259 | [
[
[
"# Geothermal Power Capacity Reserve Estimation",
"_____no_output_____"
]
],
[
[
"from platform import python_version\nprint(python_version())",
"3.8.5\n"
],
[
"# Import libraries\nimport numpy as np\nimport scipy\nfrom scipy.stats import norm, lognorm\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"### Appraisal and Development: *Assuming it's there, how big is it?*",
"_____no_output_____"
]
],
[
[
"# USER INPUT REQUIRED\nTmax = 280 #startup averages temperature for P90 reserves (degrees C)\nTmin = 250 #minimum temperature for the P10 reservoir (degrees C)",
"_____no_output_____"
],
[
"# USER INPUT REQUIRED\n# Area in km2 above the minimum temperature\nArea_P90 = 1\nArea_P10 = 10",
"_____no_output_____"
],
[
"# USER INPUT REQUIRED\n# Power Density in (MWe/km2) within the P90 and P10 temperatures (Tmin and Tmax as specified by user)\nPowerDens_P90 = 10\nPowerDens_P10 = 24",
"_____no_output_____"
],
[
"def Expected_development_size(area_p90, area_p10, powerdens_p90, powerdens_p10):\n \n ''' Calculate and plot the cumulative distribution function for expected development size'''\n\n #### Calculate nu and sigma (the mean and variance in log units required for specifying lognormal distributions) ####\n \n # nu and sigma for area\n area_nu = ((np.log(area_p90)+np.log(area_p10))/2)\n area_sigma = (np.log(area_p10)-np.log(area_p90))/((norm.ppf(0.9)-(norm.ppf(0.1))))\n \n # nu and sigma for power density\n powerdens_nu = ((np.log(powerdens_p90)+np.log(powerdens_p10))/2)\n powerdens_sigma = (np.log(powerdens_p10)-np.log(powerdens_p90))/((norm.ppf(0.9)-(norm.ppf(0.1))))\n \n # nu and sigma for MWe Capacity\n capacity_nu = area_nu + powerdens_nu\n capacity_sigma = ((area_sigma**2)+(powerdens_sigma**2))**0.5\n \n \n #### Calculate cumulative confidence curve for expected development size ####\n\n prob = [0.1]\n prob_desc = []\n expected_development_size_cdf=[]\n\n # Specify probability range\n for i in range(1,100):\n prob.append(i)\n\n for j in prob:\n \n # Calculate 100-prob for plotting descending cumulative probability\n desc = 100-j\n prob_desc.append(desc) \n \n # Calculate expected development size distribution\n eds_cdf = lognorm.ppf(j/100, capacity_sigma, loc=0, scale=np.exp(capacity_nu))\n expected_development_size_cdf.append(eds_cdf)\n \n \n #### Plot expected development size cumulative distribution ####\n\n plt.plot(expected_development_size_cdf, prob_desc)\n plt.xlabel(\"Expected Development Size (MW)\")\n plt.ylabel(\"Cumulative Confidence %\")\n plt.title(\"Cumulative Confidence in Developed Reservoir Size\")\n #axs[0].set_xlim([0, 350])\n plt.ylim([10, 100]) \n",
"_____no_output_____"
],
[
"Expected_development_size(Area_P90, Area_P10, PowerDens_P90, PowerDens_P10)",
"_____no_output_____"
]
],
[
[
"### Exploration: *Is it there?*",
"_____no_output_____"
]
],
[
[
"# USER INPUT REQUIRED\nPtemp = 0.8\nPperm = 0.4\nPchem = 0.5",
"_____no_output_____"
],
[
"POSexpl = Ptemp * Pperm * Pchem\nprint(\"Probability of exploration success = {:.0f}%\".format(POSexpl*100))\n# Could potentially code in user option to adjust number of decimal places for POS",
"Probability of exploration success = 16%\n"
]
],
[
[
"#### Expected Power Capacity\n\nExpected power capacity = expected development size * POSexpl",
"_____no_output_____"
]
],
[
[
"def Expected_power_capacity(area_p90, area_p10, powerdens_p90, powerdens_p10, posexpl):\n \n ''' Calculate and plot the cumulative distribution function for expected power capacity\n \n Expected power capacity = expected development size * POSexpl\n '''\n\n #### Calculate nu and sigma (the mean and variance in log units required for specifying lognormal distributions) ####\n \n # nu and sigma for area\n area_nu = ((np.log(area_p90)+np.log(area_p10))/2)\n area_sigma = (np.log(area_p10)-np.log(area_p90))/((norm.ppf(0.9)-(norm.ppf(0.1))))\n \n # nu and sigma for power density\n powerdens_nu = ((np.log(powerdens_p90)+np.log(powerdens_p10))/2)\n powerdens_sigma = (np.log(powerdens_p10)-np.log(powerdens_p90))/((norm.ppf(0.9)-(norm.ppf(0.1))))\n \n # nu and sigma for MWe Capacity\n capacity_nu = area_nu + powerdens_nu\n capacity_sigma = ((area_sigma**2)+(powerdens_sigma**2))**0.5\n \n \n #### Calculate cumulative confidence curve for expected power capacity ####\n\n prob = [0.1]\n prob_desc = []\n expected_development_size_cdf=[]\n\n # Specify probability range\n for i in range(1,100):\n prob.append(i)\n\n for j in prob:\n \n # Calculate 100-prob for plotting descending cumulative probability\n desc = 100-j\n prob_desc.append(desc) \n \n # Calculate expected power capacity\n eds_cdf = lognorm.ppf(j/100, capacity_sigma, loc=0, scale=np.exp(capacity_nu))*posexpl\n expected_development_size_cdf.append(eds_cdf)\n \n\n # Plot power capacity cumulative distribution\n plt.plot(expected_power_capacity, prob_desc)\n plt.xlabel(\"Expected Power Capacity (MWe potential reserves)\")\n plt.ylabel(\"Cumulative Confidence %\")\n plt.title(\"Cumulative Confidence in Power Capacity\")\n #axs[0].set_xlim([0, 350])\n plt.ylim([10, 100]) \n",
"_____no_output_____"
],
[
"Expected_power_capacity(Area_P90, Area_P10, PowerDens_P90, PowerDens_P10, POSexpl)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6c4e266e1df3fdf879bd4e6b95db9d85824401 | 133,000 | ipynb | Jupyter Notebook | notebooks/artificial_neural_network.ipynb | Joshua-Robison/MLRegression | bd0058da816871a6f3f4c9893789286397459612 | [
"MIT"
]
| null | null | null | notebooks/artificial_neural_network.ipynb | Joshua-Robison/MLRegression | bd0058da816871a6f3f4c9893789286397459612 | [
"MIT"
]
| null | null | null | notebooks/artificial_neural_network.ipynb | Joshua-Robison/MLRegression | bd0058da816871a6f3f4c9893789286397459612 | [
"MIT"
]
| null | null | null | 103.421462 | 19,748 | 0.75191 | [
[
[
"# Artificial Neural Network",
"_____no_output_____"
],
[
"## Example 1: Housing Data",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
],
[
"df = pd.read_csv('../data/kc_house_data.csv')\ndf.head(n=3)",
"_____no_output_____"
],
[
"df.drop(['id', 'date'], axis=1, inplace=True)",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"selected_features = ['bedrooms', 'bathrooms', 'sqft_living', 'sqft_lot', 'sqft_basement', 'sqft_above', 'floors', 'yr_built']\n\nX = df[selected_features]\ny = df['price']",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)",
"_____no_output_____"
],
[
"from sklearn.preprocessing import MinMaxScaler\n\nsc = MinMaxScaler().fit(X_train)\nX_train_scaled = sc.transform(X_train)\nX_test_scaled = sc.transform(X_test)",
"_____no_output_____"
],
[
"from keras.models import Sequential\nfrom keras.layers import Dense",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(50, input_dim=X_train_scaled.shape[1], activation='relu'))\nmodel.add(Dense(50, activation='relu'))\nmodel.add(Dense(1, activation='linear'))",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense (Dense) (None, 50) 450 \n_________________________________________________________________\ndense_1 (Dense) (None, 50) 2550 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 51 \n=================================================================\nTotal params: 3,051\nTrainable params: 3,051\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer='Adam', loss='mean_squared_error')",
"_____no_output_____"
],
[
"epochs_hist = model.fit(X_train_scaled, y_train, epochs=100, batch_size=50, validation_split=0.2)",
"Epoch 1/100\n277/277 [==============================] - 9s 2ms/step - loss: 418794578096.8057 - val_loss: 397580730368.0000\nEpoch 2/100\n277/277 [==============================] - 0s 1ms/step - loss: 421897067011.6835 - val_loss: 394115153920.0000\nEpoch 3/100\n277/277 [==============================] - 0s 1ms/step - loss: 422035317502.1583 - val_loss: 384691503104.0000\nEpoch 4/100\n277/277 [==============================] - 0s 1ms/step - loss: 408443065881.7842 - val_loss: 367477620736.0000\nEpoch 5/100\n277/277 [==============================] - 0s 1ms/step - loss: 387613002273.1511 - val_loss: 342339289088.0000\nEpoch 6/100\n277/277 [==============================] - 0s 1ms/step - loss: 352906849861.9856 - val_loss: 310176940032.0000\nEpoch 7/100\n277/277 [==============================] - 0s 1ms/step - loss: 332582368852.7194 - val_loss: 273027186688.0000\nEpoch 8/100\n277/277 [==============================] - 0s 1ms/step - loss: 308426375050.1295 - val_loss: 233846767616.0000\nEpoch 9/100\n277/277 [==============================] - 0s 1ms/step - loss: 254238719941.0648 - val_loss: 195527606272.0000\nEpoch 10/100\n277/277 [==============================] - 0s 1ms/step - loss: 195336489092.6043 - val_loss: 161097695232.0000\nEpoch 11/100\n277/277 [==============================] - 0s 1ms/step - loss: 173928631877.9856 - val_loss: 133399379968.0000\nEpoch 12/100\n277/277 [==============================] - 0s 977us/step - loss: 152107768618.3597 - val_loss: 113280098304.0000\nEpoch 13/100\n277/277 [==============================] - 0s 1ms/step - loss: 143262091337.6691 - val_loss: 100772225024.0000\nEpoch 14/100\n277/277 [==============================] - 0s 1ms/step - loss: 123327215674.9353 - val_loss: 93967466496.0000\nEpoch 15/100\n277/277 [==============================] - 0s 1ms/step - loss: 116800803847.3669 - val_loss: 90980360192.0000\nEpoch 16/100\n277/277 [==============================] - 0s 1ms/step - loss: 117870129991.8273 - val_loss: 89677881344.0000\nEpoch 17/100\n277/277 [==============================] - 0s 977us/step - loss: 106782583447.0216 - val_loss: 89032130560.0000\nEpoch 18/100\n277/277 [==============================] - 0s 1ms/step - loss: 111039990953.4388 - val_loss: 88510537728.0000\nEpoch 19/100\n277/277 [==============================] - 0s 977us/step - loss: 101055213008.1151 - val_loss: 87978000384.0000\nEpoch 20/100\n277/277 [==============================] - 0s 1ms/step - loss: 106260034022.2158 - val_loss: 87415504896.0000\nEpoch 21/100\n277/277 [==============================] - 0s 1ms/step - loss: 100424972243.7986 - val_loss: 86822264832.0000\nEpoch 22/100\n277/277 [==============================] - 0s 1ms/step - loss: 104559066546.6475 - val_loss: 86206152704.0000\nEpoch 23/100\n277/277 [==============================] - 0s 1ms/step - loss: 113305120797.4676 - val_loss: 85560434688.0000\nEpoch 24/100\n277/277 [==============================] - 0s 1ms/step - loss: 102085765458.8777 - val_loss: 84902764544.0000\nEpoch 25/100\n277/277 [==============================] - 0s 1ms/step - loss: 109996046910.6187 - val_loss: 84254294016.0000\nEpoch 26/100\n277/277 [==============================] - 0s 1ms/step - loss: 93990862685.9281 - val_loss: 83606388736.0000\nEpoch 27/100\n277/277 [==============================] - 0s 1ms/step - loss: 107390710673.4964 - val_loss: 82907250688.0000\nEpoch 28/100\n277/277 [==============================] - 0s 1ms/step - loss: 99001186038.7914 - val_loss: 82240028672.0000\nEpoch 29/100\n277/277 [==============================] - 0s 1ms/step - loss: 102797783776.6906 - val_loss: 81545453568.0000\nEpoch 30/100\n277/277 [==============================] - 0s 1ms/step - loss: 95291582729.2086 - val_loss: 80866533376.0000\nEpoch 31/100\n277/277 [==============================] - 0s 1ms/step - loss: 93479449032.7482 - val_loss: 80178176000.0000\nEpoch 32/100\n277/277 [==============================] - 0s 1ms/step - loss: 94397746551.7122 - val_loss: 79500500992.0000\nEpoch 33/100\n277/277 [==============================] - 0s 1ms/step - loss: 98413977349.5252 - val_loss: 78778949632.0000\nEpoch 34/100\n277/277 [==============================] - 0s 1ms/step - loss: 102364481521.2662 - val_loss: 78051590144.0000\nEpoch 35/100\n277/277 [==============================] - 0s 1ms/step - loss: 100628228759.0216 - val_loss: 77361414144.0000\nEpoch 36/100\n277/277 [==============================] - 0s 1ms/step - loss: 97438898647.4820 - val_loss: 76664553472.0000\nEpoch 37/100\n277/277 [==============================] - 0s 1ms/step - loss: 88498138458.2446 - val_loss: 75974713344.0000\nEpoch 38/100\n277/277 [==============================] - 0s 1ms/step - loss: 89064461658.2446 - val_loss: 75242356736.0000\nEpoch 39/100\n277/277 [==============================] - 0s 976us/step - loss: 86249593951.7698 - val_loss: 74530586624.0000\nEpoch 40/100\n277/277 [==============================] - 0s 1ms/step - loss: 92492723428.3741 - val_loss: 73794658304.0000\nEpoch 41/100\n277/277 [==============================] - 0s 1ms/step - loss: 97259502054.2158 - val_loss: 73055420416.0000\nEpoch 42/100\n277/277 [==============================] - 0s 1ms/step - loss: 89845919478.7914 - val_loss: 72351211520.0000\nEpoch 43/100\n277/277 [==============================] - 0s 977us/step - loss: 93723005649.9568 - val_loss: 71617159168.0000\nEpoch 44/100\n277/277 [==============================] - 0s 1ms/step - loss: 84758949401.7842 - val_loss: 70899245056.0000\nEpoch 45/100\n277/277 [==============================] - 0s 1ms/step - loss: 81802216587.9712 - val_loss: 70213517312.0000\nEpoch 46/100\n277/277 [==============================] - 0s 1ms/step - loss: 86481940207.4245 - val_loss: 69435678720.0000\nEpoch 47/100\n277/277 [==============================] - 0s 1ms/step - loss: 80543670802.4173 - val_loss: 68745281536.0000\nEpoch 48/100\n277/277 [==============================] - 0s 1ms/step - loss: 81474872150.5612 - val_loss: 68005576704.0000\nEpoch 49/100\n277/277 [==============================] - 0s 1ms/step - loss: 78837802949.0647 - val_loss: 67319111680.0000\nEpoch 50/100\n277/277 [==============================] - 0s 1ms/step - loss: 89281776779.9712 - val_loss: 66612015104.0000\nEpoch 51/100\n277/277 [==============================] - 0s 1ms/step - loss: 85070273941.1799 - val_loss: 65914781696.0000\nEpoch 52/100\n277/277 [==============================] - 0s 1ms/step - loss: 76852138308.1439 - val_loss: 65275834368.0000\nEpoch 53/100\n277/277 [==============================] - 0s 1ms/step - loss: 79457289326.5036 - val_loss: 64597377024.0000\nEpoch 54/100\n277/277 [==============================] - 0s 1ms/step - loss: 85745540854.7914 - val_loss: 63934963712.0000\nEpoch 55/100\n277/277 [==============================] - 0s 1ms/step - loss: 76450373027.9137 - val_loss: 63287181312.0000\nEpoch 56/100\n277/277 [==============================] - 0s 979us/step - loss: 82610755083.0504 - val_loss: 62657269760.0000\nEpoch 57/100\n277/277 [==============================] - 0s 1ms/step - loss: 78619837535.7698 - val_loss: 62032003072.0000\nEpoch 58/100\n277/277 [==============================] - 0s 1ms/step - loss: 70788918500.3741 - val_loss: 61480300544.0000\nEpoch 59/100\n277/277 [==============================] - 0s 1ms/step - loss: 71928046407.8273 - val_loss: 60893413376.0000\nEpoch 60/100\n277/277 [==============================] - 0s 1ms/step - loss: 77752315859.7986 - val_loss: 60346826752.0000\nEpoch 61/100\n277/277 [==============================] - 0s 977us/step - loss: 71430322043.3957 - val_loss: 59837353984.0000\nEpoch 62/100\n277/277 [==============================] - 0s 976us/step - loss: 74061614661.9856 - val_loss: 59330424832.0000\nEpoch 63/100\n277/277 [==============================] - 0s 1ms/step - loss: 69222900699.1655 - val_loss: 58883407872.0000\nEpoch 64/100\n277/277 [==============================] - 0s 1ms/step - loss: 75910355474.4173 - val_loss: 58413404160.0000\nEpoch 65/100\n277/277 [==============================] - 0s 1ms/step - loss: 68521123368.5180 - val_loss: 58019889152.0000\nEpoch 66/100\n277/277 [==============================] - 0s 1ms/step - loss: 77161994726.2158 - val_loss: 57598808064.0000\nEpoch 67/100\n277/277 [==============================] - 0s 1ms/step - loss: 66369182300.0863 - val_loss: 57245855744.0000\n"
],
[
"plt.plot(epochs_hist.history['loss'])\nplt.plot(epochs_hist.history['val_loss'])\nplt.title('Training Neural Network');\nplt.xlabel('epoch');\nplt.ylabel('loss');\nplt.legend(['Training Loss', 'Validation Loss']);",
"_____no_output_____"
],
[
"predictions = model.predict(X_test_scaled)\ndelta = y_test - predictions.reshape(-1)\n\nplt.scatter(range(len(delta)), delta);\nplt.xlabel('Test Sample Index');\nplt.ylabel('Truth - Model Prediction');\nplt.title('Prediction Delta');",
"_____no_output_____"
],
[
"from sklearn.metrics import r2_score\n\nr2 = r2_score(y_test, predictions)\nprint('R2:', r2)",
"R2: 0.5131462015151431\n"
]
],
[
[
"### Ways to improve/change model:\n\n- Use dropout layers\n- Change selected features\n- Measure correlation between features and target\n- Make model larger\n- Change activations",
"_____no_output_____"
]
],
[
[
"from keras.layers import Dropout",
"_____no_output_____"
],
[
"X = df.drop(['price'], axis=1)\ny = df['price']\n\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)\n\nsc = MinMaxScaler().fit(X_train)\nX_train_scaled = sc.transform(X_train)\nX_test_scaled = sc.transform(X_test)",
"_____no_output_____"
],
[
"model = Sequential()\nmodel.add(Dense(100, input_dim=X_train_scaled.shape[1], activation='relu'))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(100, activation='relu'))\nmodel.add(Dropout(0.5))\nmodel.add(Dense(1, activation='linear'))",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_3 (Dense) (None, 100) 1900 \n_________________________________________________________________\ndropout (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_4 (Dense) (None, 100) 10100 \n_________________________________________________________________\ndropout_1 (Dropout) (None, 100) 0 \n_________________________________________________________________\ndense_5 (Dense) (None, 1) 101 \n=================================================================\nTotal params: 12,101\nTrainable params: 12,101\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer='Adam', loss='mean_squared_error')",
"_____no_output_____"
],
[
"epochs_hist = model.fit(X_train_scaled, y_train, epochs=100, batch_size=50, validation_split=0.2)",
"Epoch 1/100\n277/277 [==============================] - 2s 2ms/step - loss: 432643226940.7770 - val_loss: 394087792640.0000\nEpoch 2/100\n277/277 [==============================] - 0s 1ms/step - loss: 411740458094.5036 - val_loss: 361420652544.0000\nEpoch 3/100\n277/277 [==============================] - 0s 1ms/step - loss: 365208628157.6978 - val_loss: 286941577216.0000\nEpoch 4/100\n277/277 [==============================] - 0s 2ms/step - loss: 282173102242.0720 - val_loss: 192234536960.0000\nEpoch 5/100\n277/277 [==============================] - 0s 1ms/step - loss: 188059195730.8777 - val_loss: 119033659392.0000\nEpoch 6/100\n277/277 [==============================] - 0s 2ms/step - loss: 139902558679.4820 - val_loss: 88471429120.0000\nEpoch 7/100\n277/277 [==============================] - 0s 2ms/step - loss: 113028898432.9209 - val_loss: 81444814848.0000\nEpoch 8/100\n277/277 [==============================] - 0s 1ms/step - loss: 101925165719.0216 - val_loss: 79620104192.0000\nEpoch 9/100\n277/277 [==============================] - 0s 1ms/step - loss: 103523211558.6763 - val_loss: 78399840256.0000\nEpoch 10/100\n277/277 [==============================] - 0s 1ms/step - loss: 107622131373.1223 - val_loss: 77259808768.0000\nEpoch 11/100\n277/277 [==============================] - 0s 2ms/step - loss: 113813854856.2878 - val_loss: 76150120448.0000\nEpoch 12/100\n277/277 [==============================] - 0s 2ms/step - loss: 105370750386.6475 - val_loss: 74903584768.0000\nEpoch 13/100\n277/277 [==============================] - 0s 2ms/step - loss: 104773619535.1942 - val_loss: 73685360640.0000\nEpoch 14/100\n277/277 [==============================] - 0s 2ms/step - loss: 100014846953.8993 - val_loss: 72396931072.0000\nEpoch 15/100\n277/277 [==============================] - 0s 1ms/step - loss: 91583146750.1583 - val_loss: 71099834368.0000\nEpoch 16/100\n277/277 [==============================] - 0s 1ms/step - loss: 85380432166.6763 - val_loss: 69729222656.0000\nEpoch 17/100\n277/277 [==============================] - 0s 2ms/step - loss: 86941417516.2014 - val_loss: 68398473216.0000\nEpoch 18/100\n277/277 [==============================] - 0s 1ms/step - loss: 81853293369.0935 - val_loss: 66989903872.0000\nEpoch 19/100\n277/277 [==============================] - 0s 1ms/step - loss: 92686860346.9353 - val_loss: 65652068352.0000\nEpoch 20/100\n277/277 [==============================] - 0s 1ms/step - loss: 92548083048.9784 - val_loss: 64233185280.0000\nEpoch 21/100\n277/277 [==============================] - 0s 2ms/step - loss: 100719200233.8993 - val_loss: 62846099456.0000\nEpoch 22/100\n277/277 [==============================] - 0s 2ms/step - loss: 81026543085.5827 - val_loss: 61396885504.0000\nEpoch 23/100\n277/277 [==============================] - 0s 2ms/step - loss: 88296631907.4532 - val_loss: 60011081728.0000\nEpoch 24/100\n277/277 [==============================] - 0s 1ms/step - loss: 81325984937.4388 - val_loss: 58608238592.0000\nEpoch 25/100\n277/277 [==============================] - 0s 1ms/step - loss: 74227859956.9496 - val_loss: 57176547328.0000\nEpoch 26/100\n277/277 [==============================] - 0s 1ms/step - loss: 77398633125.7554 - val_loss: 55872581632.0000\nEpoch 27/100\n277/277 [==============================] - 0s 1ms/step - loss: 74351504376.6331 - val_loss: 54560403456.0000\nEpoch 28/100\n277/277 [==============================] - 0s 1ms/step - loss: 73126371504.8058 - val_loss: 53212205056.0000\nEpoch 29/100\n277/277 [==============================] - 0s 1ms/step - loss: 65474272543.3093 - val_loss: 51966771200.0000\nEpoch 30/100\n277/277 [==============================] - 0s 1ms/step - loss: 71724402791.1367 - val_loss: 50862837760.0000\nEpoch 31/100\n277/277 [==============================] - 0s 1ms/step - loss: 75670151757.3525 - val_loss: 49737064448.0000\nEpoch 32/100\n277/277 [==============================] - 0s 1ms/step - loss: 70840119296.0000 - val_loss: 48729313280.0000\nEpoch 33/100\n277/277 [==============================] - 0s 2ms/step - loss: 57759203062.7914 - val_loss: 47670005760.0000\nEpoch 34/100\n277/277 [==============================] - 0s 1ms/step - loss: 78810340926.6187 - val_loss: 46798852096.0000\nEpoch 35/100\n277/277 [==============================] - 0s 2ms/step - loss: 71417075137.3813 - val_loss: 45967974400.0000\nEpoch 36/100\n277/277 [==============================] - 0s 1ms/step - loss: 67628975126.1007 - val_loss: 45222563840.0000\nEpoch 37/100\n277/277 [==============================] - 0s 1ms/step - loss: 60116320580.1439 - val_loss: 44452966400.0000\nEpoch 38/100\n277/277 [==============================] - 0s 1ms/step - loss: 60947436558.7338 - val_loss: 43867996160.0000\nEpoch 39/100\n277/277 [==============================] - 0s 1ms/step - loss: 67186586484.0288 - val_loss: 43367006208.0000\nEpoch 40/100\n277/277 [==============================] - 0s 1ms/step - loss: 61018268281.5540 - val_loss: 42783870976.0000\nEpoch 41/100\n277/277 [==============================] - 0s 2ms/step - loss: 55404778135.0216 - val_loss: 42332983296.0000\nEpoch 42/100\n277/277 [==============================] - 0s 1ms/step - loss: 56188206168.4029 - val_loss: 41935405056.0000\nEpoch 43/100\n277/277 [==============================] - 0s 1ms/step - loss: 61902545205.4101 - val_loss: 41545404416.0000\nEpoch 44/100\n277/277 [==============================] - 0s 1ms/step - loss: 58184827123.1079 - val_loss: 41218605056.0000\nEpoch 45/100\n277/277 [==============================] - 0s 1ms/step - loss: 61493745472.4604 - val_loss: 40933093376.0000\nEpoch 46/100\n277/277 [==============================] - 0s 1ms/step - loss: 55670499615.3093 - val_loss: 40642043904.0000\nEpoch 47/100\n277/277 [==============================] - 0s 1ms/step - loss: 58303785785.0935 - val_loss: 40438652928.0000\nEpoch 48/100\n277/277 [==============================] - 0s 2ms/step - loss: 60517959819.9712 - val_loss: 40263348224.0000\nEpoch 49/100\n277/277 [==============================] - 0s 1ms/step - loss: 62731984601.3237 - val_loss: 40022106112.0000\nEpoch 50/100\n277/277 [==============================] - 0s 1ms/step - loss: 56070056186.4748 - val_loss: 39766130688.0000\nEpoch 51/100\n277/277 [==============================] - 0s 1ms/step - loss: 55932323810.5324 - val_loss: 39625240576.0000\nEpoch 52/100\n277/277 [==============================] - 0s 1ms/step - loss: 54165328446.6187 - val_loss: 39456489472.0000\nEpoch 53/100\n277/277 [==============================] - 0s 1ms/step - loss: 52986084241.4964 - val_loss: 39348334592.0000\nEpoch 54/100\n277/277 [==============================] - 0s 1ms/step - loss: 59811080575.0791 - val_loss: 39228551168.0000\nEpoch 55/100\n277/277 [==============================] - 0s 1ms/step - loss: 53404308089.5540 - val_loss: 39016148992.0000\nEpoch 56/100\n277/277 [==============================] - 0s 1ms/step - loss: 51767580907.7410 - val_loss: 38844506112.0000\nEpoch 57/100\n277/277 [==============================] - 0s 1ms/step - loss: 51522787372.2014 - val_loss: 38747512832.0000\nEpoch 58/100\n277/277 [==============================] - 0s 2ms/step - loss: 58750909285.2950 - val_loss: 38706524160.0000\nEpoch 59/100\n277/277 [==============================] - ETA: 0s - loss: 52794654140.235 - 0s 1ms/step - loss: 52835575962.7050 - val_loss: 38495834112.0000\nEpoch 60/100\n277/277 [==============================] - 0s 2ms/step - loss: 49206897715.5683 - val_loss: 38361628672.0000\nEpoch 61/100\n277/277 [==============================] - 0s 2ms/step - loss: 56579796977.2662 - val_loss: 38339772416.0000\nEpoch 62/100\n277/277 [==============================] - 0s 2ms/step - loss: 57682145235.7986 - val_loss: 38202933248.0000\nEpoch 63/100\n277/277 [==============================] - 0s 1ms/step - loss: 57633030939.6259 - val_loss: 38099537920.0000\nEpoch 64/100\n277/277 [==============================] - 0s 1ms/step - loss: 53424804223.0791 - val_loss: 38005768192.0000\nEpoch 65/100\n277/277 [==============================] - 0s 1ms/step - loss: 56984717518.2734 - val_loss: 37922676736.0000\nEpoch 66/100\n277/277 [==============================] - 0s 2ms/step - loss: 55795331248.8058 - val_loss: 37814755328.0000\nEpoch 67/100\n277/277 [==============================] - 0s 1ms/step - loss: 55261054371.9137 - val_loss: 37738041344.0000\n"
],
[
"plt.plot(epochs_hist.history['loss'])\nplt.plot(epochs_hist.history['val_loss'])\nplt.title('Training Neural Network');\nplt.xlabel('epoch');\nplt.ylabel('loss');\nplt.legend(['Training Loss', 'Validation Loss']);",
"_____no_output_____"
],
[
"predictions = model.predict(X_test_scaled)\ndelta = y_test - predictions.reshape(-1)\n\nplt.scatter(range(len(delta)), delta);\nplt.xlabel('Test Sample Index');\nplt.ylabel('Truth - Model Prediction');\nplt.title('Prediction Delta');",
"_____no_output_____"
],
[
"r2 = r2_score(y_test, predictions)\nprint('R2:', r2)",
"R2: 0.6835791040821051\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6c5e865d37a89b5e434357996249ab2c133ca1 | 106,362 | ipynb | Jupyter Notebook | Class Exercise/simpleregression-Ex1.ipynb | ArmaghanHJ/Machine-Learning | c243e662ecfd3920882c133db18257fb9c8c7ac9 | [
"CC0-1.0"
]
| null | null | null | Class Exercise/simpleregression-Ex1.ipynb | ArmaghanHJ/Machine-Learning | c243e662ecfd3920882c133db18257fb9c8c7ac9 | [
"CC0-1.0"
]
| null | null | null | Class Exercise/simpleregression-Ex1.ipynb | ArmaghanHJ/Machine-Learning | c243e662ecfd3920882c133db18257fb9c8c7ac9 | [
"CC0-1.0"
]
| null | null | null | 68.798189 | 32,732 | 0.733457 | [
[
[
"# Regression Week 1:\n \n \n ",
"_____no_output_____"
],
[
"  \n# Exercise 1-1\n# Fining Intercept & Slope with 2 Loop\nRSS calculates all regular pairs of ws using loops, and from them we select the w pair with the lowest RSS.\n* In this code \"SQ-FT\" is my feature\n* Because this algorithm has a high time complexity, I worked with some of this data.(N)\n* In this code I have defined two functions.(f-total-error , f-best-error)\n * f-total-error ---> RSS($ w_0 $ , $ w_1 $)\n * f-best-error ---> min(RSS)\n* $ w_0 $ and $ w_1 $ are calculated in two loops up to a range of 500.(range_number)",
"_____no_output_____"
],
[
"## Import libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom sklearn.model_selection import train_test_split",
"_____no_output_____"
]
],
[
[
"## Load data",
"_____no_output_____"
]
],
[
[
"my_data = pd.read_csv(\"data/week1_home_data.csv\")\nmy_data.head()",
"_____no_output_____"
]
],
[
[
"## Show data",
"_____no_output_____"
]
],
[
[
"fig, ax= plt.subplots(figsize=(10,6))\n\nax.scatter(x=my_data['sqft_living'],y=my_data['price'])\n\nax.set(title='Show data',\n xlabel='sqft_living',\n ylabel='price')\n\nfig.show()",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\matplotlib\\figure.py:459: UserWarning: matplotlib is currently using a non-GUI backend, so cannot show the figure\n \"matplotlib is currently using a non-GUI backend, \"\n"
]
],
[
[
"## Finding Intercept & Slope\n#### Select the number of data that we want to find Ws on it\n* All of them \n* or part of it",
"_____no_output_____"
]
],
[
[
"N = len(my_data[:100])",
"_____no_output_____"
]
],
[
[
"### Find the best line ($ \\hat W_0 $ , $ \\hat W_1 $ )\n A line that has the least error\n\n#### Define basic variables",
"_____no_output_____"
]
],
[
[
"w0 = 0\nw1 = 0\nbest_error = None",
"_____no_output_____"
]
],
[
[
"### Functions\n\n#### Function for finding --> Total Error",
"_____no_output_____"
]
],
[
[
"def f_total_error(w0,w1,N):\n total_error = 0\n for i in range(N):\n error = my_data['price'][i] - (w0+w1*my_data['sqft_living'][i])\n total_error = total_error + error**2\n return total_error",
"_____no_output_____"
]
],
[
[
"#### Function for finding \n> * Best Error and \n* $ \\hat W_0 $ \n* $ \\hat W_1 $",
"_____no_output_____"
]
],
[
[
"def f_best_error(data,N,best_error,range_number):\n best_w0 = 0\n best_w1 = 0\n for w0 in range(range_number):\n for w1 in range(range_number):\n total_error = f_total_error(w0,w1,N)\n if w0==0 and w1==0:\n best_error = total_error\n elif best_error > total_error:\n best_error = total_error\n best_w0 = w0\n best_w1 = w1\n return best_w0,best_w1,best_error",
"_____no_output_____"
]
],
[
[
"#### Run Function",
"_____no_output_____"
]
],
[
[
"w0,w1,best_error = f_best_error(my_data,N,best_error,500)\nprint('w0=%d \\nw1=%d \\nbest_error=%d'%(w0,w1,best_error))",
"w0=499 \nw1=246 \nbest_error=5314503397591\n"
]
],
[
[
"### Making a column $ \\hat Y $",
"_____no_output_____"
]
],
[
[
"my_data['y_hat'] = w0 + w1*my_data['sqft_living']\nmy_data.head()",
"_____no_output_____"
]
],
[
[
"### Show data & Estimated Line",
"_____no_output_____"
]
],
[
[
"fig, ax = plt.subplots(figsize=(10,6))\n\nax.plot(my_data['sqft_living'],my_data['price'],'.',\n my_data['sqft_living'],my_data['y_hat'],'-')\n\nfig.show()",
"C:\\ProgramData\\Anaconda3\\lib\\site-packages\\matplotlib\\figure.py:459: UserWarning: matplotlib is currently using a non-GUI backend, so cannot show the figure\n \"matplotlib is currently using a non-GUI backend, \"\n"
]
],
[
[
"## Predicting Value",
"_____no_output_____"
]
],
[
[
"print('sq_ft = %d\\nprice = %d\\nY_hat = %d\\n'%(my_data['sqft_living'][0],my_data['price'][0],my_data['y_hat'][0]))\nprint('error = %d'%(my_data['price'][0]-my_data['y_hat'][0]))",
"sq_ft = 1180\nprice = 221900\nY_hat = 290779\n\nerror = -68879\n"
]
],
[
[
" \n## Result\nFinding the best line, or in other words, finding the parameters w and z so that they have the best data coverage or the least amount of RSS, using two loops, has a high time complexity.I defined data as follow:\n* N = 100\n* Range_number = 500\n\nThe results obtained with this data are as follows:\n* $w_0$ = 499 \n* $w_1$ = 246 \n* best_error = 5314503397591\n\nIn the predict section, the test performed and the estimated number had an error of 68879. Our model is not very good because it has a high error.\n\n ",
"_____no_output_____"
],
[
"    \n# Exercise 1- 2\n# Regression Week 1: Simple Linear Regression",
"_____no_output_____"
],
[
"In this notebook we will use data on house sales in King County to predict house prices using simple (one input) linear regression. You will:\n* Use array and panda and numpy functions to compute important summary statistics\n* Write a function to compute the Simple Linear Regression weights using the closed form solution\n* Write a function to make predictions of the output given the input feature\n* Turn the regression around to predict the input given the output\n* Compare two different models for predicting house prices\n\nIn this notebook you will be provided with some already complete code as well as some code that you should complete yourself in order to answer quiz questions. The code we provide to complte is optional and is there to assist you with solving the problems but feel free to ignore the helper code and write your own.",
"_____no_output_____"
],
[
"  \n# Load house sales data\n\nDataset is from house sales in King County, the region where the city of Seattle, WA is located.",
"_____no_output_____"
]
],
[
[
"dtype_dict = {'bathrooms':float, 'waterfront':int, 'sqft_above':int, 'sqft_living15':float, 'grade':int, 'yr_renovated':int, 'price':float, 'bedrooms':float, 'zipcode':str, 'long':float, 'sqft_lot15':float, 'sqft_living':float, 'floors':str, 'condition':int, 'lat':float, 'date':str, 'sqft_basement':int, 'yr_built':int, 'id':str, 'sqft_lot':int, 'view':int}\n\nsales = pd.read_csv(\"data/week2_kc_house_data.csv\")\nsales.head()",
"_____no_output_____"
]
],
[
[
"# Split data into training and testing",
"_____no_output_____"
],
[
"We use seed=0 so that everyone running this notebook gets the same results. In practice, you may set a random seed (or let Turi Create pick a random seed for you). ",
"_____no_output_____"
]
],
[
[
"train_data,test_data = train_test_split(sales,test_size=0.2)",
"_____no_output_____"
],
[
"print(\"#tran_data = {0}\\n#test_data = {1}\".format(len(train_data),len(test_data))) ",
"#tran_data = 17290\n#test_data = 4323\n"
]
],
[
[
"# Useful functions",
"_____no_output_____"
],
[
"In order to make use of the closed form solution as well as take advantage of some functions we will review some important ones. In particular:\n* Computing the sum \n* Computing the arithmetic average (mean)\n* multiplying arrays by constants\n* multiplying arrays by other arrays",
"_____no_output_____"
]
],
[
[
"# Let's compute the mean of the House Prices in King County in 2 different ways.\nprices = sales['price'] # extract the price column of the sales SFrame -- this is now an SArray\n\n# recall that the arithmetic average (the mean) is the sum of the prices divided by the total number of houses:\nsum_prices = prices.sum()\nnum_houses = len(prices) # when prices is an SArray len() returns its length\navg_price_1 = sum_prices/num_houses\navg_price_2 = prices.mean() # if you just want the average, the .mean() function\nprint(\"average price via method 1: \" + str(avg_price_1))\nprint(\"average price via method 2: \" + str(avg_price_2))",
"average price via method 1: 540088.1417665294\naverage price via method 2: 540088.1417665294\n"
]
],
[
[
"As we see we get the same answer both ways",
"_____no_output_____"
]
],
[
[
"# if we want to multiply every price by 0.5 it's a simple as:\nhalf_prices = 0.5*prices\n\n# Let's compute the sum of squares of price. We can multiply two arrays of the same length elementwise also with *\nprices_squared = prices*prices\nsum_prices_squared = prices_squared.sum() # price_squared is an array of the squares and we want to add them up.\nprint(\"the sum of price squared is: \" + str(sum_prices_squared))",
"the sum of price squared is: 9217325138472070.0\n"
]
],
[
[
"Aside: The python notation x.xxe+yy means x.xx \\* 10^(yy). e.g 100 = 10^2 = 1*10^2 = 1e2 ",
"_____no_output_____"
],
[
"# Build a generic simple linear regression function ",
"_____no_output_____"
],
[
"Armed with these array functions we can use the closed form solution found from lecture to compute the slope and intercept for a simple linear regression on observations stored as arrays: input_feature, output.\n\nComplete the following function (or write your own) to compute the simple linear regression slope and intercept:",
"_____no_output_____"
],
[
"We can test that our function works by passing it something where we know the answer. In particular we can generate a feature and then put the output exactly on a line: output = 1 + 1\\*input_feature then we know both our slope and intercept should be 1",
"_____no_output_____"
]
],
[
[
"def simple_linear_regression(input_feature, output):\n # compute the sum of input_feature and output\n sum_x = input_feature.sum()\n sum_y = output.sum()\n \n # compute the product of the output and the input_feature and its sum\n product_x_y = input_feature * output\n sum_product_xy = product_x_y.sum()\n \n # compute the squared value of the input_feature and its sum\n x_power_2 = input_feature * input_feature\n sum_x_power_2 = x_power_2.sum()\n \n # use the formula for the slope\n N = len(input_feature)\n slope = (sum_product_xy - ((sum_y * sum_x)/N))/(sum_x_power_2 - ((sum_x*sum_x)/N))\n \n # use the formula for the intercept\n intercept = (sum_y / N) - (slope * (sum_x/N))\n \n return (intercept, slope)",
"_____no_output_____"
],
[
"# test_feature = turicreate.SArray(range(5))\ntest_feature = pd.Series(range(5))\n\n# test_output = turicreate.SArray(1 + 1*test_feature)\ntest_output = pd.Series(1 + 1*test_feature)\n\n(test_intercept, test_slope) = simple_linear_regression(test_feature, test_output)\nprint(\"Intercept: \" + str(test_intercept))\nprint(\"Slope: \" + str(test_slope))",
"Intercept: 1.0\nSlope: 1.0\n"
]
],
[
[
"Now that we know it works let's build a regression model for predicting price based on sqft_living. Rembember that we train on train_data!",
"_____no_output_____"
]
],
[
[
"sqft_intercept, sqft_slope = simple_linear_regression(train_data['sqft_living'], train_data['price'])\n\nprint(\"Intercept: \" + str(sqft_intercept))\nprint(\"Slope: \" + str(sqft_slope))",
"Intercept: -53170.19465091487\nSlope: 285.94846907460175\n"
]
],
[
[
"# Predicting Values",
"_____no_output_____"
],
[
"Now that we have the model parameters: intercept & slope we can make predictions. Using arrays it's easy to multiply an array by a constant and add a constant value. Complete the following function to return the predicted output given the input_feature, slope and intercept:",
"_____no_output_____"
]
],
[
[
"def get_regression_predictions(input_feature, intercept, slope):\n # calculate the predicted values:\n predicted_values = intercept + slope*input_feature\n \n return predicted_values",
"_____no_output_____"
]
],
[
[
"Now that we can calculate a prediction given the slope and intercept let's make a prediction. Use (or alter) the following to find out the estimated price for a house with 2650 squarefeet according to the squarefeet model we estiamted above.\n\n**Quiz Question: Using your Slope and Intercept from (4), What is the predicted price for a house with 2650 sqft?**",
"_____no_output_____"
]
],
[
[
"my_house_sqft = 2650\nestimated_price = get_regression_predictions(my_house_sqft, sqft_intercept, sqft_slope)\nprint(\"The estimated price for a house with %d squarefeet is $%.2f\" % (my_house_sqft, estimated_price))",
"The estimated price for a house with 2650 squarefeet is $704593.25\n"
]
],
[
[
"# Residual Sum of Squares",
"_____no_output_____"
],
[
"Now that we have a model and can make predictions let's evaluate our model using Residual Sum of Squares (RSS). Recall that RSS is the sum of the squares of the residuals and the residuals is just a fancy word for the difference between the predicted output and the true output. \n\nComplete the following (or write your own) function to compute the RSS of a simple linear regression model given the input_feature, output, intercept and slope:",
"_____no_output_____"
]
],
[
[
"def get_residual_sum_of_squares(input_feature, output, intercept, slope):\n # First get the predictions\n prediction_value = get_regression_predictions(input_feature,intercept,slope)\n\n # then compute the residuals (since we are squaring it doesn't matter which order you subtract)\n y_y_hat = output - prediction_value\n\n # square the residuals and add them up\n square_y_y_hat = y_y_hat * y_y_hat\n RSS = square_y_y_hat.sum()\n\n return(RSS)",
"_____no_output_____"
]
],
[
[
"Let's test our get_residual_sum_of_squares function by applying it to the test model where the data lie exactly on a line. Since they lie exactly on a line the residual sum of squares should be zero!",
"_____no_output_____"
]
],
[
[
"print(get_residual_sum_of_squares(test_feature, test_output, test_intercept, test_slope)) # should be 0.0",
"0.0\n"
]
],
[
[
"Now use your function to calculate the RSS on training data from the squarefeet model calculated above.\n\n**Quiz Question: According to this function and the slope and intercept from the squarefeet model What is the RSS for the simple linear regression using squarefeet to predict prices on TRAINING data?**",
"_____no_output_____"
]
],
[
[
"rss_prices_on_sqft = get_residual_sum_of_squares(train_data['sqft_living'], train_data['price'], sqft_intercept, sqft_slope)\nprint('The RSS of predicting Prices based on Square Feet is : ' + str(rss_prices_on_sqft))",
"The RSS of predicting Prices based on Square Feet is : 1222917739017968.5\n"
]
],
[
[
"# Predict the squarefeet given price",
"_____no_output_____"
],
[
"What if we want to predict the squarefoot given the price? Since we have an equation y = a + b\\*x we can solve the function for x. So that if we have the intercept (a) and the slope (b) and the price (y) we can solve for the estimated squarefeet (x).\n\nComplete the following function to compute the inverse regression estimate, i.e. predict the input_feature given the output.",
"_____no_output_____"
]
],
[
[
"def inverse_regression_predictions(output, intercept, slope):\n # solve output = intercept + slope*input_feature for input_feature. Use this equation to compute the inverse predictions:\n estimated_feature = (output - intercept)/slope\n\n return estimated_feature",
"_____no_output_____"
]
],
[
[
"Now that we have a function to compute the squarefeet given the price from our simple regression model let's see how big we might expect a house that costs $800,000 to be.\n\n**Quiz Question: According to this function and the regression slope and intercept from (3) what is the estimated square-feet for a house costing $800,000?**",
"_____no_output_____"
]
],
[
[
"my_house_price = 800000\nestimated_squarefeet = inverse_regression_predictions(my_house_price, sqft_intercept, sqft_slope)\nprint(\"The estimated squarefeet for a house worth $%.2f is %d\" % (my_house_price, estimated_squarefeet))",
"The estimated squarefeet for a house worth $800000.00 is 2983\n"
]
],
[
[
"# New Model: estimate prices from bedrooms",
"_____no_output_____"
],
[
"We have made one model for predicting house prices using squarefeet, but there are many other features in the sales SFrame. \nUse your simple linear regression function to estimate the regression parameters from predicting Prices based on number of bedrooms. Use the training data!",
"_____no_output_____"
]
],
[
[
"# Estimate the slope and intercept for predicting 'price' based on 'bedrooms'\nbed_intercept, bed_slope = simple_linear_regression(train_data['bedrooms'], train_data['price'])\n\nprint(\"Intercept: \" + str(bed_intercept))\nprint(\"Slope: \" + str(bed_slope))\n",
"Intercept: 100093.8959485789\nSlope: 131162.87121634884\n"
]
],
[
[
"# Test your Linear Regression Algorithm",
"_____no_output_____"
],
[
"Now we have two models for predicting the price of a house. How do we know which one is better? Calculate the RSS on the TEST data (remember this data wasn't involved in learning the model). Compute the RSS from predicting prices using bedrooms and from predicting prices using squarefeet.\n\n**Quiz Question: Which model (square feet or bedrooms) has lowest RSS on TEST data? Think about why this might be the case.**",
"_____no_output_____"
]
],
[
[
"# Compute RSS when using bedrooms on TEST data:\nprint(get_residual_sum_of_squares(test_data['bedrooms'], test_data['price'], test_intercept, test_slope))",
"1716233579121577.0\n"
],
[
"# Compute RSS when using squarefeet on TEST data:\nprint(get_residual_sum_of_squares(test_data['sqft_living'], test_data['price'], test_intercept, test_slope))",
"1704876907976277.0\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
]
]
|
ec6c6bc2cb3a84bdd4010fa4f7e6643cddc929de | 494,102 | ipynb | Jupyter Notebook | trending_event_detection.ipynb | SavannahY/Trending-Event-Detection-for-COVID-19-Twitter-chatter | f5b918a4f3bd2317884dfec03945fb9972c00202 | [
"MIT"
]
| null | null | null | trending_event_detection.ipynb | SavannahY/Trending-Event-Detection-for-COVID-19-Twitter-chatter | f5b918a4f3bd2317884dfec03945fb9972c00202 | [
"MIT"
]
| null | null | null | trending_event_detection.ipynb | SavannahY/Trending-Event-Detection-for-COVID-19-Twitter-chatter | f5b918a4f3bd2317884dfec03945fb9972c00202 | [
"MIT"
]
| null | null | null | 133.181132 | 49,240 | 0.802474 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec6c721de6f4aa4db83afe7456c3f87d286a8ad5 | 7,094 | ipynb | Jupyter Notebook | notebooks/pandas/sample_df.ipynb | codenamewei/pydata-science-playground | aa147b003aa4bd2afa2a6a5f00101cc0cb340f9f | [
"MIT"
]
| null | null | null | notebooks/pandas/sample_df.ipynb | codenamewei/pydata-science-playground | aa147b003aa4bd2afa2a6a5f00101cc0cb340f9f | [
"MIT"
]
| null | null | null | notebooks/pandas/sample_df.ipynb | codenamewei/pydata-science-playground | aa147b003aa4bd2afa2a6a5f00101cc0cb340f9f | [
"MIT"
]
| null | null | null | 24.047458 | 149 | 0.386101 | [
[
[
"import pandas as pd \r\n \r\n# assign data of lists. \r\ndata = {'date': ['20110201', '20110202', '20110203', '20110204', '20110205', '20110201', '20110202', '20110212', '20110211', '20110205'], \r\n 'count': [20, 21, 19, 18, 20, 5, 1, 0, 21, 2]} \r\n\r\n# Create DataFrame \r\ndf = pd.DataFrame(data)\r\n\r\nprint(df)\r\n",
" date count\n0 20110201 20\n1 20110202 21\n2 20110203 19\n3 20110204 18\n4 20110205 20\n5 20110201 5\n6 20110202 1\n7 20110212 0\n8 20110211 21\n9 20110205 2\n"
]
],
[
[
"# Sample by percentage\r\n\r\n- [API Reference](https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.sample.html)",
"_____no_output_____"
]
],
[
[
"sample_percentage= 0.2 # 20 percent\r\n\r\ndf.sample(frac = sample_percentage)",
"_____no_output_____"
]
],
[
[
"# Sample by number of items \r\n**Note**: sample by number of items cannot be used together with percentage",
"_____no_output_____"
]
],
[
[
"sample_num = 5\r\n\r\ndf.sample(n = sample_num)",
"_____no_output_____"
]
],
[
[
"# Sample by specific values",
"_____no_output_____"
]
],
[
[
"df.loc[df['date'] == '20110205']",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6c79892a8bf0d6648bfc4d4290ee98247ba9d1 | 154,050 | ipynb | Jupyter Notebook | python/ch13/13.5-bayesian-inference-unknown-variance.ipynb | krishnonwork/mathematical-methods-in-deep-learning | 12a7e7a9981f8639b4524b7977bd185f82c04e2d | [
"MIT"
]
| 1 | 2020-03-20T20:46:58.000Z | 2020-03-20T20:46:58.000Z | python/ch13/13.5-bayesian-inference-unknown-variance.ipynb | sthagen/mathematical-methods-in-deep-learning-ipython | 12a7e7a9981f8639b4524b7977bd185f82c04e2d | [
"MIT"
]
| null | null | null | python/ch13/13.5-bayesian-inference-unknown-variance.ipynb | sthagen/mathematical-methods-in-deep-learning-ipython | 12a7e7a9981f8639b4524b7977bd185f82c04e2d | [
"MIT"
]
| null | null | null | 540.526316 | 144,676 | 0.941136 | [
[
[
"import torch\nfrom torch.distributions import Normal, Gamma\nimport math\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"## Bayesian Inferencing of Precision of Gaussian likelihood, known Mean\n\nWe will study the case where the mean is known but the variance is unknown and expressed as a random variable. The computations become quite a bit simpler if we use precision (inverse of variance).\n\nWe will only express the variance as a random variable while the mean will be a constant.\n\nPrecision, $\\lambda$ which is related to variance $\\sigma$ as $\\lambda = \\frac{ 1 } { \\sigma^{2} }$.\n\nThe likelihood term becomes\n$$\np\\left( X \\middle\\vert \\lambda\\right) = \\frac{ \\lambda^{ \\frac{ n }{ 2 } } } { { \\sqrt {2\\pi } } } e^{ -\\frac{ \\lambda }{ 2 } \\sum_{i=1}^n \\left( {x^{ \\left( i \\right) } - \\mu } \\right)^2 }$$\n\nThe prior for the precision is a Gamma distribution\n$$\np\\left( \\lambda \\right) = \\frac{ \\beta_{0}^{ \\alpha_{0} } }{ \\Gamma\\left( \\alpha_{0} \\right) } \\lambda ^{ \\left( \\alpha_{0} - 1 \\right)} e^{ - \\beta_{0} \\lambda }\n$$\n\nThe corresponding posterior is also a Gamma distribution, such that \n\n$$p\\left( \\lambda \\middle\\vert X \\right) = \\frac{ \\beta_{n}^{ \\alpha_{n} } }{ \\Gamma\\left( \\alpha_{n} \\right) } \\lambda ^{ \\left( \\alpha_{n} - 1 \\right)} e^{ - \\beta_{n} \\lambda} $$\n\nwhere \n$$ \\alpha_{n} = \\frac{ n }{ 2 } + \\alpha_{0} \\\\\n\\beta_{n} = \\frac{ 1 }{ 2 } \\sum_{i=1}^{n} \\left( {x^{ \\left( i \\right) } - \\mu } \\right)^2 + \\beta_{0} = \\frac{ n }{ 2 } s + \\beta_{0} $$",
"_____no_output_____"
]
],
[
[
"def inference_unknown_variance(X, prior_dist):\n sigma_mle = torch.std(X)\n n = X.shape[0]\n \n # Parameters of the prior\n alpha_0 = prior_dist.concentration\n beta_0 = prior_dist.rate\n \n # Parameters of posterior\n alpha_n = n / 2 + alpha_0\n beta_n = n / 2 * sigma_mle ** 2 + beta_0\n posterior_dist = Gamma(alpha_n, beta_n)\n return posterior_dist",
"_____no_output_____"
],
[
"def plot_distribution(dists, legend, precisions, precision_legend, title, xlim=(0, 4)):\n fig, ax = plt.subplots(dpi=400)\n ax.set_title(title)\n ax.set_ylabel(\"PDF(X)\")\n ax.set_xlabel(\"X\")\n y_lim = 0\n for dist in dists:\n x = torch.linspace(0.01, 10, 1000)\n pdf = dist.log_prob(x).exp()\n ax.plot(x, pdf)\n y_lim = max(torch.max(pdf), y_lim)\n for precision in precisions:\n ax.plot([precision] * len(x) , x, '--')\n ax.set_xlim(*xlim)\n ax.set_ylim(0, y_lim+0.01)\n legend.extend(precision_legend)\n ax.legend(legend, loc='upper right')\n return ax",
"_____no_output_____"
],
[
"# Let us assume that the true distribution is a normal distribution. The true distribution corresponds \n# to a single class.\nmu_known = 20\ntrue_dist = Normal(mu_known, 5)",
"_____no_output_____"
],
[
"# Case 1\n# Let us assume our prior is a Gamma distribution with a good estimate of the variance\nprior_dist = Gamma(1, 10)\n\n# Let us set a seed for reproducability\ntorch.manual_seed(42)\n\n# Number of samples is low. \nn = 3\nX = true_dist.sample((n,))\nposterior_dist_low_n = inference_unknown_variance(X, prior_dist)\n\nprecision_mle = (1 / X.std()**2)\n\ntrue_precision = (1/ true_dist.scale**2)\n\nmax_posterior_precision = (posterior_dist_low_n.concentration-1) / posterior_dist_low_n.rate\n\n# When n is low, the posterior is dominated by the prior. Thus, a good prior can help offset the lack of data.\n# We can see this in the following case. \n\n# With a small sample (n=3), the MLE estimate of precision is 3.70, which is way off from the true value of 0.04\n# Using a good prior here helps offset it. The MAP estimate of precision, 0.14 is much better. \n\nprint(f\"True precision: {true_precision:0.2f}\")\nprint(f\"MAP precision: {max_posterior_precision:0.2f}\")\nprint(f\"MLE precision: {precision_mle:0.2f}\")",
"True precision: 0.04\nMAP precision: 0.14\nMLE precision: 3.70\n"
],
[
"dists = [prior_dist, posterior_dist_low_n]\nlegend = [\"Prior\", \"Posterior n=3\"]\n\nprecisions = [true_precision, precision_mle, max_posterior_precision]\nlegend_p = [\"True Precision\", \"MLE Precision\", \"MAP Precision\"]\n\nplot_distribution(dists, legend, precisions, legend_p, \"Low data n=3\",)\nplt.show()",
"_____no_output_____"
],
[
"# Case 2\n# Let us assume our prior is a Gamma distribution with a good estimate of the variance\nprior_dist = Gamma(1, 10)\n\n# Let us set a seed for reproducability\ntorch.manual_seed(42)\n\n# Number of samples is high. \nn = 100\nX = true_dist.sample((n,))\nposterior_dist_high_n = inference_unknown_variance(X, prior_dist)\n\nprecision_mle = (1 / X.std()**2)\n\ntrue_precision = (1/ true_dist.scale**2)\n\nmax_posterior_precision = (posterior_dist_high_n.concentration-1) / posterior_dist_high_n.rate\n\n\n# When n is high, the MLE tends to converge to the true distribution. The MAP also tends to converge to the MLE, \n# and in turn converges to the true distribution\nprint(f\"True precision: {true_precision:0.2f}\")\nprint(f\"MAP precision: {max_posterior_precision:0.2f}\")\nprint(f\"MLE precision: {precision_mle:0.2f}\")",
"True precision: 0.04\nMAP precision: 0.04\nMLE precision: 0.04\n"
]
],
[
[
"### How to use the estimated variance parameter?\n\nAs before, we will find the parameter value that maximizes the posterior density. Here we will maximize the posterior probability density function for the precision $p\\left( \\lambda \\middle\\vert X \\right) = \\lambda^{ \\left( \\alpha_{n} - 1 \\right) } e^{ -\\beta_{n} \\lambda }$ by taking the derivative and equating to zero.\n\nThis gives us $$\\lambda = \\frac{ \\alpha - 1 }{ \\beta }$$\n\nThus, our estimated distribution for the class of points corresponding to the training data is $\\mathcal{N}\\left( x ; \\;\\; \\mu, \\sigma_{n} \\right)$ where $\\frac{1}{ \\sigma_{n}^{2} } = \\left( \\frac{ \\alpha_{n} - 1 }{ \\beta_{n} } \\right)^{ -\\frac{1}{2} }$.",
"_____no_output_____"
]
],
[
[
"map_precision = (posterior_dist_high_n.concentration-1) / posterior_dist_high_n.rate\nmap_dist = Normal(mu_known, 1/ math.sqrt(map_precision))\nprint(f\"MAP distribution mu: {map_dist.mean:0.2f} std:{map_dist.scale:0.2f}\")",
"MAP distribution mu: 20.00 std:4.95\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6c7d6d1670e1c72d6be3be4c17d188c62fc324 | 47,587 | ipynb | Jupyter Notebook | practicos/p3/p3.ipynb | alejopaullier96/um-data-science | c43ef75f3d5cfea9ccf8f43281cca6ee66d948f3 | [
"MIT"
]
| null | null | null | practicos/p3/p3.ipynb | alejopaullier96/um-data-science | c43ef75f3d5cfea9ccf8f43281cca6ee66d948f3 | [
"MIT"
]
| null | null | null | practicos/p3/p3.ipynb | alejopaullier96/um-data-science | c43ef75f3d5cfea9ccf8f43281cca6ee66d948f3 | [
"MIT"
]
| null | null | null | 66.648459 | 15,616 | 0.724526 | [
[
[
"</center><img src=\"https://www3.um.edu.uy/logoum.jpg\" width=300></center>\n<h1 align=\"center\">Introducción a la Ciencia de Datos</h1>\n<h2 align=\"center\"> <font color='gray'>Práctico 3: Visualizaciones no tradicionales</font></h2>",
"_____no_output_____"
],
[
"</center><img src=\"images/charts.jpg\" width=700></center>",
"_____no_output_____"
],
[
"### <font color='289C4E'>Tabla de contenidos<font>\n- [Importe las librerías necesarias](#0)\n- [Word cloud (*)](#1)\n- [Word cloud con masks (***)](#2)\n- [Venn diagrams (**)](#3)\n- [Diagrama radial (***)](#4)\n- [Diagramas de Sankey (*)](#5)\n- [Bullet graph (**)](#6)\n- [Geographical maps (**)](#7)\n- [Stacked bars (***)](#8)\n- [Lollipop graph (**)](#9)\n- [Dot plot (**)](#10)\n- [Area graph (***)](#11)\n \nAl finalizar el práctico usted aprenderá:\n- Mapas relacionados a Natural Language Processing (Word clouds).\n- Mapas relacionados a matemática y estadística (Venn Diagrams).\n- Mapas relacionados a rendimientos (Radial Diagram).\n- Mapas relacionados a ingeniería de procesos industriales (Sankey Diagram).\n- Mapas geográficos.\n- Mapas comparativos.\n\n<font color='B60F04'>A lo largo del práctico usted notará asteriscos (*) al lado de cada ejercicio. Estos indican el nivel de dificultad. Se sugiere realizar el trabajo en conjunto y no por separado.<font>",
"_____no_output_____"
],
[
"### <font color='289C4E'>0) Importe las librerías necesarias<font> <a class='anchor' id='0'></a>",
"_____no_output_____"
]
],
[
[
"import json\nimport matplotlib.pyplot as plt\nimport numpy as np\nimport os\nimport pandas as pd\nimport plotly.express as px\nimport regex as re\nimport seaborn as sns\n\nfrom IPython.display import Image as im\nfrom math import pi\nfrom matplotlib.sankey import Sankey\nfrom matplotlib_venn import venn3, venn3_circles\nfrom PIL import Image\nfrom wordcloud import WordCloud, STOPWORDS, ImageColorGenerator",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>1) Word cloud (*)<font> <a class='anchor' id='1'></a>\n \nLos Word clouds son frecuentemente utilizados para visualizar las palabras más utilizadas en un corpus (lista de textos). Es una gran herramienta visual para analizar sentimientos, reviews y tweets (entre otros) y así obtener información vital de nuestros datos.\n \nEn el siguiente ejemplo usted analizará cuáles son las palabras más frecuentemente utilizadas por integrantes del partido republicano y democrático. Analizará aproximadamente 86k tweets y visualizará las palabras usando la librería [wordcloud](https://amueller.github.io/word_cloud/index.html).\n\nPara esto:\n- Cargue el archivo CSV que contiene los tweets y divida el dataframe en dos: uno llamado \"republicans\" que contiene los tweets republicanos y otro \"democrats\" que contiene los tweets demócratas.\n- En la celda siguiente se le proporciona un código de pre-procesamiento de lenguaje natural (un tema de este curso que usted verá más adelante). No tiene por qué entender este código, simplemente usted debe ingresar una lista que contiene los tweets y este código le devolverá una lista con las palabras utilizadas en todos los tweets del dataframe para cada partido político. La celda incluye comentarios en caso de que usted quiera comprender este pre-procesamiento.\n- Básese en el siguiente [ejemplo](https://amueller.github.io/word_cloud/auto_examples/simple.html)\n- En el ejemplo provisto el archivo llamado \"text\" equivale a nuestras listas de palabras.\n- Configure los [parámetros](https://amueller.github.io/word_cloud/generated/wordcloud.WordCloud.html#wordcloud.WordCloud) del WordCloud de tal forma que: \n 1. El fondo sea blanco.\n 2. La cantidad de palabras este limitada a 1000.\n 3. El colormap sea 'Reds' para republicanos y 'Blues' para demócratas.\n- Ajuste `figsize` a (12, 8) y añádale un título.\n\nSus gráficas deberían verse similares a las siguientes:\n<table><tr>\n<td> <img src=\"images/1a.png\" alt=\"Drawing\" style=\"width: 400px;\"/> </td>\n<td> <img src=\"images/1b.png\" alt=\"Drawing\" style=\"width: 400px;\"/> </td>\n</tr></table>",
"_____no_output_____"
]
],
[
[
"tweets = pd.read_csv('./data/tweets.csv', sep= ',')\nSTOPWORDS.update(['amp','today','day','new','thank','morning','year','will','week','bill'])",
"_____no_output_____"
],
[
"republican_tweets = # ingrese aquí la lista de tweets\nraw_string = ''.join(republican_tweets) # join tweets\nno_links = re.sub(r'http\\S+', '', raw_string) # sustituir HTTP\nno_unicode = re.sub(r\"\\\\[a-z][a-z]?[0-9]+\", '', no_links) # sustituir unicode\nno_special_characters = re.sub('[^A-Za-z ]+', '', no_unicode) # sustituir caracteres especiales\nwords = no_special_characters.split(\" \") # split \nwords = [w for w in words if len(w) > 2] # ignorar short words\nwords = [w.lower() for w in words] # hacer lowercase\nrepublican_words = [w for w in words if w not in STOPWORDS] # quitar stop words\n\ndemocrat_tweets = # ingrese aqui la lista de tweets\nraw_string = ''.join(democrat_tweets)\nno_links = re.sub(r'http\\S+', '', raw_string)\nno_unicode = re.sub(r\"\\\\[a-z][a-z]?[0-9]+\", '', no_links)\nno_special_characters = re.sub('[^A-Za-z ]+', '', no_unicode)\nwords = no_special_characters.split(\" \")\nwords = [w for w in words if len(w) > 2]\nwords = [w.lower() for w in words]\ndemocrat_words = [w for w in words if w not in STOPWORDS]",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>2) Word cloud con masks (***)<font> <a class='anchor' id='2'></a>\n\nPara hacer los WordClouds más visualmente atractivos podemos agregarles **masks**. Una mask es una imagen (puede ser negra o a color y generalmente con fondo blanco o negro) la cual le proporciona el contorno que adopta la WordCloud.\n\nEn este ejercicio, usted cargará dos imágenes: \"rep_mask.png\" y \"dem_mask.png\", las cuales contienen los logos a color de cada partido político. Estas servirán para el contorno y color de la WordCloud.\n\nPara realizar el ejercicio:\n- Cargue las masks basándose en el siguiente [ejemplo](https://amueller.github.io/word_cloud/auto_examples/masked.html). Configure `contour_width` = 3, `contour_color`='black' y que la cantidad de palabras sea 200.\n- Básese en este [ejemplo](https://amueller.github.io/word_cloud/auto_examples/colored.html) para colorear la imagen en base a los colores de la imagen.\n- Ajuste el figsize a (15,12) y añádale un título.\n\nSus gráficas deberían verse similares a las siguientes:\n<table><tr>\n<td> <img src=\"images/2i.png\" alt=\"Drawing\" style=\"width: 400px;\"/> </td>\n<td> <img src=\"images/2ii.png\" alt=\"Drawing\" style=\"width: 400px;\"/> </td>\n</tr></table>",
"_____no_output_____"
],
[
"### <font color='289C4E'>3) Venn diagrams (**)<font> <a class='anchor' id='3'></a>\n\nLos diagramas de Venn son muy usados para representar grupos que se intersectan. Nos permite visualizar niveles de jerarquía, intersecciones de conjuntos y subgrupos. \n\nReproduzca el siguiente [diagrama](https://1.bp.blogspot.com/-ju4m6PBOrgo/V-E5qz99SaI/AAAAAAAAMF0/gle0zsZz_nIBEMVg0EdZHoGJhjlnBzv1gCLcB/s1600/moz-screenshot-3-729576.png) sin tener en cuenta el círculo más externo (el que dice Data Science).\n- Instale la librería [matplotlib-venn](https://pypi.org/project/matplotlib-venn/). Hágalo mediante pip, no use easy-install.\n- En la celda siguiente se le proporciona un ejemplo sencillo.\n- Como `matplotlib-venn` solo acepta valores numéricos no interesa como esté relleno (puede rellenarlo con unos). Para que aparezcan las labels que nos importan debe usar el comando `v.get_label_by_id().set_text()`. Por ejemplo, si quisiésemos que la label \"Computer Science\" aparezca en el círculo rojo sería: `v.get_label_by_id('100').set_text('Computer Science')`.\n- Deberá determinar qué ids se mapean con qué posiciones dentro del diagrama.\n- Puede ser de utilidad mirar los ejemplos de la librería original.\n- Configure `figsize` a (15,10). Añadale un título a la gráfica.\n\nSu gráfica debería verse similar a la siguiente: \n</center><img src=\"images/3.png\" width=400></center>",
"_____no_output_____"
]
],
[
[
"v = venn3(subsets = (20, 10, 12, 10, 9, 4, 3), set_labels = ('Group A', 'Group B', 'Group C'), alpha = 0.5)\nv.get_label_by_id('10').set_text('a')",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>4) Diagrama radial (***)<font> <a class='anchor' id='4'></a>\n\nLos diagramas radiales nos permiten visualizar el área formada por datos multivariantes y medir rendimientos . Éstos gráficos son utilizados en los más diversos rubros como la psicología, el diseño de hardware, contabilidad, etc.\n\nA continuación usted deberá evaluar los gastos semestrales de una empresa, la cual se le proporcionan sus datos en la celda siguiente:\n\n- Corra la siguiente celda para cargar el dataframe.\n- Para realizar la gráfica básese fuertemente en el siguiente [ejemplo](https://python-graph-gallery.com/391-radar-chart-with-several-individuals/). Casi no deberá modificar el código provisto en el mismo.\n- Ajuste los ticks y labels para mostrar todo el rango de valores.\n- Agréguele `pad` a los ticks para que las labels con los nombres de los distintos rubros no se superpongan con la gráfica. **Tip:** use [plt.tick_params()](https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.tick_params.html).\n- Configure `figsize` = (12, 8)\n- Añádale un título a la gráfica.\n- Agréguele `pad` al título.\n\nSu gráfica debería verse similar a la siguiente: \n</center><img src=\"images/4.png\" width=400></center>",
"_____no_output_____"
]
],
[
[
"# Datos de la empresa\ndf = pd.DataFrame({\n'group': ['1er semestre','2do semestre'],\n'Ventas': [31, 28],\n'Marketing': [39, 19],\n'Administración': [18, 27],\n'Desarrollo': [7, 35],\n'Atención al cliente': [12, 43],\n'Ingeniería': [47, 40]\n})\ndf.head()",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>5) Diagramas de Sankey (*)<font> <a class='anchor' id='5'></a>\n\nEn clase ha visto los diagramas de Sankey. Éstos son utilizados para diagramar flujos tanto de dinero (empresas) como de energía (industrias). Son diagramas muy utilizados en contabilidad y termodinámica de procesos energéticos.\n\nUna empresa ha lanzado un producto. Inicialmente el 100% de la financiación proviene:\n- 70% de la inversión inicial.\n- 20% de inversores extranjeros.\n- 10% de financiación estatal.\n\nLuego se gasta dinero en el proceso de desarrollo del producto. Los porcentajes de dinero son respecto al capital inicial. A fin de año el balance es negativo:\n- 40% gasto en desarrollo.\n- 20% gasto en investigación.\n- 30% gasto en testing del producto.\n- 25% gasto en marketing y lanzamiento del producto al mercado.\n\nGrafique un diagrama de Sankey para el flujo de dinero en el desarrollo del producto:\n- Básese en el siguiente [ejemplo](https://python-graph-gallery.com/220-sankey-diagram-with-matplotlib/) para entender como realizar el diagrama.\n- Configure `figsize` = (15,8).\n- Añádale un título a la gráfica.\n- Apáguele el `axis` a la gráfica.\n- Las labels deben estar no superpuestas con el diagrama. Puede usar `.texts[].set_position(xy=(,))`.\n\nSu gráfica debería verse similar a la siguiente: \n</center><img src=\"images/5.png\" width=600></center>",
"_____no_output_____"
],
[
"### <font color='289C4E'>6) Bullet graph (**)<font> <a class='anchor' id='6'></a>\n\nEn clase ha visto los bullet graphs. Éstos son utilizados para comparar performances relativas, hacer benchmarkings y visualizar rangos de aceptabilidad de performance.\n\nReproduzca el ejemplo visto en clase. Para esto:\n- Se le proporciona a continuación en la siguiente celda una función hecha por [Chriss Moffit](https://pbpython.com/bullet-graph.html). No tiene que entender la función sino leer los comentarios en rojo para entender sus parámetros. También al final de la celda se le proporciona un ejemplo que le aclarará mejor su uso.\n- Se le proporcionan los 5 datos del gráfico que deberá reproducir.\n- Deberá graficar 5 bulletgraphs, uno para cada set de datos.\n- Use axis label para configurar las unidades.\n- Cambie el target color de manera que la barra sea negra.\n- Averigüe acerca de las **seaborn palettes**. Las mismas le servirán para que el color sea un gris dégradé.\n- Configure el parámetro limits para establecer los distintos niveles de aceptabilidad.\n\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/6.png\" width=600></center>",
"_____no_output_____"
]
],
[
[
"def bulletgraph(data=None, limits=None, labels=None, axis_label=None, title=None,\n size=(5, 3), palette=None, formatter=None, target_color=\"gray\",\n bar_color=\"black\", label_color=\"gray\"):\n \"\"\" Build out a bullet graph image\n Args:\n data = List of labels, measures and targets\n limits = list of range valules\n labels = list of descriptions of the limit ranges\n axis_label = string describing x axis\n title = string title of plot\n size = tuple for plot size\n palette = a seaborn palette\n formatter = matplotlib formatter object for x axis\n target_color = color string for the target line\n bar_color = color string for the small bar\n label_color = color string for the limit label text\n Returns:\n a matplotlib figure\n \"\"\"\n # Determine the max value for adjusting the bar height\n # Dividing by 10 seems to work pretty well\n h = limits[-1] / 10\n\n # Use the green palette as a sensible default\n if palette is None:\n palette = sns.light_palette(\"green\", len(limits), reverse=False)\n\n # Must be able to handle one or many data sets via multiple subplots\n if len(data) == 1:\n fig, ax = plt.subplots(figsize=size, sharex=True)\n else:\n fig, axarr = plt.subplots(len(data), figsize=size, sharex=True)\n\n # Add each bullet graph bar to a subplot\n for idx, item in enumerate(data):\n\n # Get the axis from the array of axes returned when the plot is created\n if len(data) > 1:\n ax = axarr[idx]\n\n # Formatting to get rid of extra marking clutter\n ax.set_aspect('equal')\n ax.set_yticklabels([item[0]])\n ax.set_yticks([1])\n ax.spines['bottom'].set_visible(False)\n ax.spines['top'].set_visible(False)\n ax.spines['right'].set_visible(False)\n ax.spines['left'].set_visible(False)\n\n prev_limit = 0\n for idx2, lim in enumerate(limits):\n # Draw the bar\n ax.barh([1], lim - prev_limit, left=prev_limit, height=h,\n color=palette[idx2])\n prev_limit = lim\n rects = ax.patches\n # The last item in the list is the value we're measuring\n # Draw the value we're measuring\n ax.barh([1], item[1], height=(h / 3), color=bar_color)\n\n # Need the ymin and max in order to make sure the target marker\n # fits\n ymin, ymax = ax.get_ylim()\n ax.vlines(\n item[2], ymin * .9, ymax * .9, linewidth=1.5, color=target_color)\n\n # Now make some labels\n if labels is not None:\n for rect, label in zip(rects, labels):\n height = rect.get_height()\n ax.text(\n rect.get_x() + rect.get_width() / 2,\n -height * .4,\n label,\n ha='center',\n va='bottom',\n color=label_color)\n if formatter:\n ax.xaxis.set_major_formatter(formatter)\n if axis_label:\n ax.set_xlabel(axis_label)\n if title:\n fig.suptitle(title, fontsize=14)\n fig.subplots_adjust(hspace=0)\n\n### Ejemplo:\n \ndata_to_plot2 = [(\"John Smith\", 105, 120),\n (\"Jane Jones\", 99, 110),\n (\"Fred Flintstone\", 109, 125),\n (\"Barney Rubble\", 135, 123),\n (\"Mr T\", 45, 105)]\nbulletgraph(data_to_plot2, limits=[20, 60, 100, 160],\n labels=[\"Poor\", \"OK\", \"Good\", \"Excellent\"], size=(8,5),\n axis_label=\"Performance Measure\", label_color=\"black\",\n bar_color=\"#252525\", target_color='#f7f7f7',\n title=\"Sales Rep Performance\")",
"_____no_output_____"
],
[
"data_1 = [(\"Revenue\", 275, 250)]\ndata_2 = [(\"Profit\", 22.5, 26)]\ndata_3 = [(\"Avg Order Size\", 310, 550)]\ndata_4 = [(\"New customers\", 1600, 2100)]\ndata_5 = [(\"Satisfaction\", 4.75, 4.6)]",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>7) Geographical maps (**)<font> <a class='anchor' id='7'></a>\n\nEn otros prácticos ya ha visto mapas geográficos. Estos sirven para representar datos por zonas geográficas.\n\nA continuación, usted recreará un mapa geográfico de los distintos estados de la India, resaltando a color las distintas poblaciones y densidades de población por estado.\n\nPara graficar mapas se utilizan archivos GEOJSON. Un formato estandarizado para representar zonas geográficas. Los archivos GEOJSON contienen un campo \"geometry\" que contiene coordenadas geográficas que forman un polígono (área) que será graficado. Además, contiene un campo \"properties\" con metadatos de esa región, como por ejemplo: ID de la región y nombre.\n\n1. Usted graficará el mapa usando [choropleth](https://plotly.com/python/choropleth-maps/):\n - Corra la siguiente celda para obtener el archivo GEOJSON y los datos a graficar.\n - Grafique de tal manera que el color quede determinado por la población de los estados.\n - El parámetro locations debe estar configurado como \"id\" para que exista un mapeo entre el dataframe y el GEOJSON.\n - Establezca un título para la gráfica.\n\n\n2. Ahora grafique la densidad de población:\n - El color debe ser determinado por el campo \"DensityScale\".\n - Los parámetros `hover` (flotar en inglés) permiten mostrar datos cuando uno pasa el cursor por encima de las regiones. \n - Configure los parámetros `hover_name` y `hover_data` de tal modo que aparezca el nombre del estado y el campo \"Density\" al pasar el cursor por encima de la gráfica.\n - Estableza un título.\n\n**Nota:** si es de su interés puede crear sus propios mapas geográficas usando [geojson.io](https://geojson.io/#map=2/20.0/0.0)\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/7.png\" width=600></center>",
"_____no_output_____"
]
],
[
[
"import plotly.io as pio\npio.renderers.default = 'browser'\nindia_states = json.load(open(\"data.geojson\", \"r\"))\ndf = pd.read_csv('india_census.csv',sep=',')",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>8) Stacked bars (***)<font> <a class='anchor' id='8'></a>\n\nEn clase ha visto las stacked columns o stacked bars. Estas son utilizadas para representar porcentajes relativos de un total.\n\nHa continuación se le proporcionan datos de ventas mensuales de tres empleados: Alejo, Victoria y Celeste. Usted es su manager y quiere ver el rendimiento relativo de los tres empleados y decide usar stacked columns.\n\nPara crear la gráfica:\n1. Genere una lista \"totals\" con la suma total de las ventas de cada mes.\n2. Genere tres listas, una para cada empleado, que contenga el porcentaje de ventas de cada empleado. Es decir, las ventas del empleado en ese mes dividido la cantidad de ventas de todo el mes.\n3. Utilize la función [plt.bar()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.bar.html) para graficar las barras. Tendrá que utilizarla tres veces, una para cada empleado.\n4. Configure los parámetros:\n - `label` = nombre del empleado.\n - `width` = 0.85\n - `edgecolor` = 'white'\n - `color` = un color que usted quiera.\n - `xticks` = use los xticks proporcionados en la celda siguiente.\n5. Deberá configurar el parámetro \"bottom\" (la cual es una lista) de la función plt.bar() de tal manera que la misma sea la suma cumulativa de los porcentajes relativos de los empleados. Es decir, si Alejo es el primer empleado que graficará entonces no precisará lista, luego si Victoria es la segunda empleada entonces el valor de \"bottom\" será los porcentajes de Alejo, y por último, si Celeste es la tercer empleada que grafica la lista \"bottom\" será la suma de los procentajes de Alejo y Victoria.\n6. Agregue los rótulos y el título de la gráfica.\n7. Utilize `plt.legend(bbox_to_anchor=(1.05, 1), loc='upper left')` para que la leyenda no quede dentro de la gráfica.\n\n\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/8.png\" width=600></center>",
"_____no_output_____"
]
],
[
[
"# Data\nxticks = [0,1,2,3,4]\nnames = ('Enero','Febrero','Marzo','Abril','Mayo')\nraw_data = {'Alejo': [14, 20, 5, 32, 19],\n 'Victoria': [10, 20, 7, 28, 26],\n 'Celeste': [17, 15, 10, 25, 17]}\ndf = pd.DataFrame(raw_data)",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>9) Lollipop graph (**)<font> <a class='anchor' id='9'></a>\n \nLos gráficos Lollipop son una buena alternativa a los gráficos de barras y columnas estándar. Son especialmente útiles cuando la visualización es abrumadora debido a su tinta masiva porque el lollipop centra la atención en el valor.\n\nGrafique el ejemplo visto en clase. Para eso:\n- Cargue los datos en la celda siguiente.\n- Haga dos subplots.\n- En la primer subplot grafique el lollipop graph usando la función [plt.stem()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.stem.html). Esta graficará los datos verticalmente por lo que deberá rotar las xlabels.\n- En la segunda subplot grafique los lollipops usando [plt.hlines()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.hlines.html) para generar el \"palito\" del lollipop y luego [plt.scatter()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.scatter.html) para generar el \"punto\" del lollipop. \n- Agregue títulos.\n\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/9i.png\" width=600></center>",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('respondents.csv',sep=',')",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>10) Dot plot (**)<font> <a class='anchor' id='10'></a>\n\nEn clase usted ha visto los dot plots. Son útiles para mostrar diferencias entre valores de una misma variable.\nReproduzca el ejemplo visto en clase para las diferencias porcentuales de logins de grandes compañías de software.\n\nPara esto:\n- Ejecute la celda siguiente para obtener el dataframe con los valores.\n- Cree dos listas: una con los xticks y otra con los xlabels.\n- Configure `figsize` = (12,8)\n- Utilize la función [plt.hlines()](https://matplotlib.org/api/_as_gen/matplotlib.pyplot.hlines.html) para generar los segmentos que uniran los puntos con los valores.\n- Utilize dos veces [plt.scatter()](https://matplotlib.org/stable/api/_as_gen/matplotlib.pyplot.scatter.html) para graficar los puntos con los extremos.\n- Agregue `xticks`, `xlabels`, `yticks`, `ylabels`.\n- Agregue rótulos, título y leyenda.\n- Agregue una grilla\n\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/10.png\" width=600></center>",
"_____no_output_____"
]
],
[
[
"# Create a dataframe\nvalue1=[0.35, 0.43, 0.06, 0.07, 0.02, 0.06]\nvalue2=[0.40, 0.45, 0.05, 0.04, 0.03, 0.05]\ncompanies = ['Google', 'Facebook', 'Twitter','Yahoo', 'LinkedIn', 'Others']\ndf = pd.DataFrame({'group':companies, 'value1':value1 , 'value2':value2 })",
"_____no_output_____"
]
],
[
[
"### <font color='289C4E'>11) Area graph (***)<font> <a class='anchor' id='11'></a>\n\nEn clase ha visto las gráficas de áreas. Son gráficos apropiados cuando se quiere mostrar tendencias que representan el 100% a lo largo del tiempo. Un gráfico de área es esencialmente un gráfico de líneas, con cada segmento apilado uno encima del otro. De esta forma, muestra partes de un todo. \n\nReproduzca el ejemplo visto en clase. Para eso:\n- Ejecute la celda siguiente para obtener los datos necesarios.\n- Configure los `xticks`, `xlabels` y colores necesarios.\n- Utilize la función [plt.stackplot()](https://matplotlib.org/3.1.1/api/_as_gen/matplotlib.pyplot.stackplot.html) para hacer el gráfico de área.\n- Haga dos for loops para anotar los textos necesarios en la gráfica. Le será de utilidad el dataframe `cumsum` que contiene la suma cumulativa de los datos del dataframe. Utilize la función [ax.annotate()](https://matplotlib.org/stable/api/_as_gen/matplotlib.axes.Axes.annotate.html) para hacer las anotaciones necesarias.\n- Remueva los bordes del gráfica usando [ax.spines()](https://matplotlib.org/stable/api/spines_api.html).\n- Configure los rótulos, la leyenda y el título.\n\nSu gráfica debería verse similar a la siguiente:\n</center><img src=\"images/11.png\" width=800></center>",
"_____no_output_____"
]
],
[
[
"y1=[26.8,24.97,25.69,24.07]\ny2=[21.74,19.58,20.7,21.09]\ny3=[13.1,12.45,12.75,10.79]\ny4=[9.38,8.18,8.79,6.75]\ny5=[12.1,10.13,10.76,8.03]\ny6=[4.33,3.73,3.78,3.75]\n\ndf = pd.DataFrame([y1,y2,y3,y4,y5,y6])\n\ncumsum = df.cumsum()\ncumsum",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6c7ff8a2d7c5e62b25b5d0d601cd73c3cabfc5 | 7,800 | ipynb | Jupyter Notebook | lessons/python/ESPIN-04- For loops and conditionals.ipynb | BCampforts/espin | 376e98eed45352af6b4f66345bec1f5792a64124 | [
"MIT"
]
| null | null | null | lessons/python/ESPIN-04- For loops and conditionals.ipynb | BCampforts/espin | 376e98eed45352af6b4f66345bec1f5792a64124 | [
"MIT"
]
| null | null | null | lessons/python/ESPIN-04- For loops and conditionals.ipynb | BCampforts/espin | 376e98eed45352af6b4f66345bec1f5792a64124 | [
"MIT"
]
| null | null | null | 29.213483 | 655 | 0.578205 | [
[
[
"<a href=\"https://csdms.colorado.edu/wiki/ESPIn2020\"><img style=\"float: center; width: 75%\" src=\"../../media/ESPIn.png\"></a>",
"_____no_output_____"
],
[
"# Programming with Python\n## For loops\n### minutes: 30\n---\n> ## Learning Objectives {.objectives}\n>\n> * Explain what a for loop does\n> * Correctly write for loops to repeat simple calculations\n> * Trace changes to a loop variable as the loop runs\n> * Trace changes to other variables as they are updated by a for loop\n\n",
"_____no_output_____"
],
[
"## For loops\n\nAutomating repetitive tasks is best accomplished with a loop. A For Loop repeats a set of actions for every item in a collection (every letter in a word, every number in some range, every name in a list) until it runs out of items:",
"_____no_output_____"
]
],
[
[
"word = 'lead'\nfor char in word:\n print( char)",
"_____no_output_____"
]
],
[
[
"This is shorter than writing individual statements for printing every letter in the word and it easy scales to longer or shorter words:",
"_____no_output_____"
]
],
[
[
"word = 'aluminium'\nfor char in word:\n print( char)",
"_____no_output_____"
],
[
"word = 'tin'\nfor char in word:\n print (char)",
"_____no_output_____"
]
],
[
[
"The general form of a for loop is:",
"_____no_output_____"
]
],
[
[
"# for item in collection:\n# do things with item",
"_____no_output_____"
]
],
[
[
"A for loop starts with the word \"for\", then the variable name that each item in the collection is going to take inside the loop, then the word \"in\", and then the collection or sequence of items to loop through.\n\nIn Python, there must be a colon at the end of the line starting the loop. The commands that are run repeatedly inside the loop are indented below that. Unlike many other languages, there is no command to end a loop (e.g. `end for`): the loop ends once the indentation moves back.\n\n### Practice your skills\nMake a for loop to count the letters in the word elephant. ",
"_____no_output_____"
],
[
"It’s worth tracing the execution of this little program step by step. Since there are eight characters in ‘elephant’, the statement inside the loop will be executed eight times. The first time around, `length` is zero (the value assigned to it on line 1) and `letter` is \"e\". The code adds 1 to the old value of `length`, producing 1, and updates `length` to refer to that new value. The next time the loop starts, `letter` is \"l\" and `length` is 1, so `length` is updated to 2. Once there are no characters left in \"elephant\" for Python to assign to `letter`, the loop finishes and the `print` statement tells us the final value of length.\n\nNote that a loop variable is just a variable that’s being used to record progress in a loop. It still exists after the loop is over (and has the last value it had inside the loop). We can re-use variables previously defined as loop variables, overwriting their value:",
"_____no_output_____"
]
],
[
[
"letter = 'z'\nfor letter in 'abc':\n print (letter)\nprint ('after the loop, letter is', letter)",
"_____no_output_____"
]
],
[
[
"## Making Choices\n\nWhen analyzing data, we’ll often want to automatically recognize differences between values and take different actions on the data depending on some conditions. Here, we’ll learn how to write code that runs only when certain conditions are true.\n\n### Conditionals\nWe can ask Python to running different commands depending on a condition with an if statement:",
"_____no_output_____"
]
],
[
[
"num = 42\n\nif num > 100:\n print ('greater')\nelse:\n print ('not greater')\n \nprint ('done')",
"_____no_output_____"
]
],
[
[
"The second line of this code uses the keyword `if` to tell Python that we want to make a choice. If the test that follows the `if` statement is true, the commands in the indented block are executed. If the test is false, the indented block beneath the else is executed instead. Only one or the other is ever executed.\n\nConditional statements don’t have to include an `else`. If there isn’t one, Python simply does nothing if the test is false:",
"_____no_output_____"
]
],
[
[
"num = 42\nprint ('before conditional...')\nif num > 100:\n print (num, 'is greater than 100')\nprint ('...after conditional')",
"_____no_output_____"
]
],
[
[
"We can also chain several tests together using `elif`, which is short for “else if”. The following Python code uses elif to print the sign of a number. We use a double equals sign `==` to test for equality between two values. The single equal sign is used for assignment:",
"_____no_output_____"
]
],
[
[
"num = -3\n\nif num > 0:\n print (num, \"is positive\")\nelif num == 0:\n print (num, \"is zero\")\nelse:\n print (num, \"is negative\")",
"_____no_output_____"
]
],
[
[
"We can also combine tests using `and` and `or`. `and` is only true if both parts are true:",
"_____no_output_____"
]
],
[
[
"if (1 > 0) and (-1 > 0):\n print ('both tests are true')\nelse:\n print ('at least one test is false')",
"_____no_output_____"
]
],
[
[
"while `or` is true if at least one part is true:",
"_____no_output_____"
]
],
[
[
"if (1 > 0) or (-1 > 0):\n print ('at least one test is true')\nelse:\n print ('neither test is true')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6c88b60b8f50877f65615cddd5c9b5769efedd | 796,962 | ipynb | Jupyter Notebook | StyleGAN2_training.ipynb | seven320/Spacecat | c9035e84e3d8928b3ac8af6cec9d3b2fd86526a4 | [
"Apache-2.0"
]
| null | null | null | StyleGAN2_training.ipynb | seven320/Spacecat | c9035e84e3d8928b3ac8af6cec9d3b2fd86526a4 | [
"Apache-2.0"
]
| null | null | null | StyleGAN2_training.ipynb | seven320/Spacecat | c9035e84e3d8928b3ac8af6cec9d3b2fd86526a4 | [
"Apache-2.0"
]
| null | null | null | 1,253.084906 | 211,653 | 0.953419 | [
[
[
"<a href=\"https://colab.research.google.com/github/seven320/Spacecat/blob/main/StyleGAN2_training.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!nvidia-smi",
"Mon Mar 21 12:19:36 2022 \n+-----------------------------------------------------------------------------+\n| NVIDIA-SMI 460.32.03 Driver Version: 460.32.03 CUDA Version: 11.2 |\n|-------------------------------+----------------------+----------------------+\n| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |\n| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |\n| | | MIG M. |\n|===============================+======================+======================|\n| 0 Tesla P100-PCIE... Off | 00000000:00:04.0 Off | 0 |\n| N/A 37C P0 26W / 250W | 0MiB / 16280MiB | 0% Default |\n| | | N/A |\n+-------------------------------+----------------------+----------------------+\n \n+-----------------------------------------------------------------------------+\n| Processes: |\n| GPU GI CI PID Type Process name GPU Memory |\n| ID ID Usage |\n|=============================================================================|\n| No running processes found |\n+-----------------------------------------------------------------------------+\n"
],
[
"from typing import Tuple, List\nfrom tqdm import tqdm\n\nfrom PIL import Image",
"_____no_output_____"
]
],
[
[
"## train style GAN2 model for this cat doesn't exists\n\n* prepare kaggle account and kaggle.json in your google drive",
"_____no_output_____"
]
],
[
[
"import os, sys\nimport shutil\n\nCOLAB = \"google.colab\" in sys.modules\n\nif COLAB:\n from google.colab import drive\n from google.colab import output\n drive.mount(\"/content/drive\")",
"Mounted at /content/drive\n"
],
[
"if not os.path.exists(\"/root/.kaggle\"):\n os.makedirs(\"/root/.kaggle\")\n!cp drive/MyDrive/kaggle/kaggle.json /root/.kaggle/kaggle.json",
"_____no_output_____"
],
[
"!git clone https://github.com/seven320/stylegan2-pytorch.git\n!pip install kaggle > /dev/null\n!pip install Ninja",
"Cloning into 'stylegan2-pytorch'...\nremote: Enumerating objects: 403, done.\u001b[K\nremote: Counting objects: 100% (11/11), done.\u001b[K\nremote: Compressing objects: 100% (9/9), done.\u001b[K\nremote: Total 403 (delta 3), reused 7 (delta 2), pack-reused 392\u001b[K\nReceiving objects: 100% (403/403), 122.52 MiB | 14.02 MiB/s, done.\nResolving deltas: 100% (203/203), done.\nCollecting Ninja\n Downloading ninja-1.10.2.3-py2.py3-none-manylinux_2_5_x86_64.manylinux1_x86_64.whl (108 kB)\n\u001b[K |████████████████████████████████| 108 kB 4.3 MB/s \n\u001b[?25hInstalling collected packages: Ninja\nSuccessfully installed Ninja-1.10.2.3\n"
]
],
[
[
"# Prepare Dataset",
"_____no_output_____"
]
],
[
[
"!kaggle datasets download -d crawford/cat-dataset",
"Downloading cat-dataset.zip to /content\n100% 4.04G/4.04G [01:38<00:00, 84.9MB/s]\n100% 4.04G/4.04G [01:38<00:00, 44.2MB/s]\n"
],
[
"shutil.unpack_archive(\"cat-dataset.zip\")",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt # plotting\nimport numpy as np # linear algebra\n\nimport enum\nimport numpy as np\nimport cv2 \nfrom matplotlib import pyplot as plt\nfrom keras.layers import *\nimport os",
"_____no_output_____"
],
[
"def load_image(path):\n img = cv2.imread(path)\n img = cv2.cvtColor(img,cv2.COLOR_BGR2RGB)\n labels = load_labels(path)[1:]\n\n w,h = img.shape[:2]\n \n return img, labels , w , h\n\ndef load_labels(path):\n path = path + \".cat\"\n \n with open(path,'r') as f:\n coordinates = f.readline()\n coordinates = str(coordinates).split(' ')[:-1]\n \n return list(map(int,coordinates))",
"_____no_output_____"
],
[
"roots = [f\"CAT_0{i}\" for i in range(7)]\nroot = roots[0]\nfiles = [i for i in os.listdir(root) if i.endswith(\".jpg\")]\nfor i, file in enumerate(files):\n print(root, file)\n x,y,w,h = load_image(os.path.join(root,file))\n plt.imshow(x)\n plt.scatter(y[0:18:2], y[1:18:2])\n plt.show()\n if i == 3:\n break",
"CAT_00 00000441_010.jpg\n"
],
[
"def cut_cats_center(image:np.ndarray, y:List[int], h:int, w:int) -> np.ndarray:\n \"\"\"\n cut cat's image into a square with the cat's face in the center\n \"\"\"\n x_min = min(y[0:18:2])\n x_max = max(y[0:18:2])\n y_min = min(y[1:18:2])\n y_max = max(y[1:18:2])\n\n # calculate cat face position\n x_center =(x_min + x_max) /2\n y_center = (y_min + y_max) / 2\n\n half_w = min(x_center, w - x_center)\n half_h = min(y_center, h - y_center)\n half_edge = min(half_w, half_h)\n\n y_s = int(y_center - half_edge)\n y_e = int(y_center + half_edge)\n x_s = int(x_center - half_edge)\n x_e = int(x_center + half_edge)\n assert y_e-y_s == x_e-x_s\n return image[y_s:y_e, x_s:x_e]\n",
"_____no_output_____"
],
[
"if not os.path.exists(\"prepare_data/cats\"):\n os.makedirs(\"prepare_data/cats\")\n\nroots = [f\"CAT_0{i}\" for i in range(7)]\ntotal_index = 0\nfor root in tqdm(roots):\n files = [i for i in os.listdir(root) if i.endswith(\".jpg\")]\n for i, file in enumerate(files):\n if os.path.exists(f\"data/cats/{total_index:04}.jpg\"):\n total_index += 1\n continue\n image, label,w,h = load_image(os.path.join(root,file))\n c_image = cut_cats_center(image, label, w, h)\n try:\n Image.fromarray(c_image).save(f\"prepare_data/cats/{total_index:04}.jpg\")\n total_index += 1\n except:\n pass",
"100%|██████████| 7/7 [02:14<00:00, 19.20s/it]\n"
],
[
"path = \"prepare_data/cats/0000.jpg\"\nimg = cv2.imread(path)\nimg = cv2.cvtColor(img,cv2.COLOR_BGR2RGB)\n\nplt.imshow(img)",
"_____no_output_____"
],
[
"!python stylegan2-pytorch/prepare_data.py --out prepare_data --size 256 --n_worker 4 /content/prepare_data",
"Make dataset of image sizes: 256\n\r0it [00:00, ?it/s]/usr/local/lib/python3.7/dist-packages/torchvision/transforms/functional.py:405: UserWarning: Argument interpolation should be of type InterpolationMode instead of int. Please, use InterpolationMode enum.\n \"Argument interpolation should be of type InterpolationMode instead of int. \"\n/usr/local/lib/python3.7/dist-packages/torchvision/transforms/functional.py:405: UserWarning: Argument interpolation should be of type InterpolationMode instead of int. Please, use InterpolationMode enum.\n \"Argument interpolation should be of type InterpolationMode instead of int. \"\n/usr/local/lib/python3.7/dist-packages/torchvision/transforms/functional.py:405: UserWarning: Argument interpolation should be of type InterpolationMode instead of int. Please, use InterpolationMode enum.\n \"Argument interpolation should be of type InterpolationMode instead of int. \"\n/usr/local/lib/python3.7/dist-packages/torchvision/transforms/functional.py:405: UserWarning: Argument interpolation should be of type InterpolationMode instead of int. Please, use InterpolationMode enum.\n \"Argument interpolation should be of type InterpolationMode instead of int. \"\n9996it [00:43, 230.73it/s]\n"
],
[
"# !ls drive/MyDrive\ntry:\n os.makedirs(\"drive/MyDrive/cats/checkpoint\")\nexcept:\n print(\"file exists\")\ntry:\n os.makedirs(\"drive/MyDrive/cats/sample\")\nexcept:\n print(\"file exists\")",
"file exists\nfile exists\n"
],
[
"latest_ckpt = None\nfor i in range(10000, 10**6, 5000):\n if os.path.exists(f\"drive/MyDrive/cats/checkpoint/{i:06}.pt\"):\n latest_ckpt = f\"drive/MyDrive/cats/checkpoint/{i:06}.pt\"\n else:\n break\n\nprint(f\"latest_ckpt: {latest_ckpt}\") ",
"latest_ckpt: drive/MyDrive/cats/checkpoint/010000.pt\n"
]
],
[
[
"# Train StyleGAN2",
"_____no_output_____"
]
],
[
[
"if latest_ckpt is None:\n !python stylegan2-pytorch/train.py --iter 100000 --size 256 --batch 16 --n_sample 16 prepare_data --save_dir drive/MyDrive/cats \nelse:\n !python stylegan2-pytorch/train.py --iter 100000 --size 256 --batch 16 --n_sample 16 prepare_data --save_dir drive/MyDrive/cats --ckpt $latest_ckpt",
"load model: drive/MyDrive/cats/checkpoint/010000.pt\n 10% 10000/100000 [00:00<?, ?it/s]/content/stylegan2-pytorch/op/conv2d_gradfix.py:89: UserWarning: conv2d_gradfix not supported on PyTorch 1.10.0+cu111. Falling back to torch.nn.functional.conv2d().\n f\"conv2d_gradfix not supported on PyTorch {torch.__version__}. Falling back to torch.nn.functional.conv2d().\"\nd: 1.0290; g: 0.8291; r1: 0.0120; path: 0.0823; mean path: 0.0027; augment: 0.0000: 10% 10000/100000 [00:06<?, ?it/s]/usr/local/lib/python3.7/dist-packages/torchvision/utils.py:50: UserWarning: range will be deprecated, please use value_range instead.\n warnings.warn(warning)\nd: 0.9150; g: 1.1875; r1: 0.0203; path: 0.0124; mean path: 0.3241; augment: 0.0000: 26% 25741/100000 [14:35:14<68:39:50, 3.33s/it]"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6c93091484e3ae97cf7ad2770853f3c8710b1d | 80,271 | ipynb | Jupyter Notebook | S&P500/S&P-500 Buy on Dip Intro.ipynb | CruddyShad0w/StockBot | e8e321bc7c6c694afc04fa2d48690a96c49a680c | [
"MIT"
]
| null | null | null | S&P500/S&P-500 Buy on Dip Intro.ipynb | CruddyShad0w/StockBot | e8e321bc7c6c694afc04fa2d48690a96c49a680c | [
"MIT"
]
| null | null | null | S&P500/S&P-500 Buy on Dip Intro.ipynb | CruddyShad0w/StockBot | e8e321bc7c6c694afc04fa2d48690a96c49a680c | [
"MIT"
]
| 1 | 2021-03-15T02:19:32.000Z | 2021-03-15T02:19:32.000Z | 104.519531 | 22,996 | 0.735309 | [
[
[
"# A simple Buy-on-dip algo for Alpaca API\nThis is a simple algo that trades every day refreshing portfolio based on the EMA ranking. Among the universe (e.g. SP500 stocks), it ranks by daily (price - EMA) percentage as of trading time and keep positions in sync with lowest ranked stocks.\n\nThe rationale behind this: low (price - EMA) vs price ratio indicates there is a big dip in a short time. Since the universe is SP500 which means there is some fundamental strengths, the belief is that the price should be recovered to some extent. Let's get started by setting up our enviroment.",
"_____no_output_____"
],
[
"## Setup\nFirst you need to set your environment variables for the rest api. The rest api uses environment variables to when executing a call to the rest api. If you see an error referring to the rest.api() call this is most likely the origin.\n\nFirst we need to Verify packages are correct by installing from pip file in `~\\StockBot\\S&P500BOD\\`\n```\n$ pip install -p\n```\nIf you run into errors double check the Pipfile.lock for all dependency versions.\nAfter you have the dependencies and environment variables set be sure to restart the jupyter notebooks application.\n\nWere almost at the point of being able to run the code. The next step is to import our api key into the files that contain the algorithm. Open the file `~\\StockBot\\S&P500BOD\\samplealgo\\algo.py` and input your `key_id` and `secret_key` as well as the `base_url` that matches your alpaca api account. \n\nThe section of code starts near line 12 and looks like the following:\n\n`\napi = tradeapi.REST(\n key_id='',\n secret_key='',\n base_url='https://paper-api.alpaca.markets'\n)\n`\n\nNext run the following chunk of python code.\nIt will automagically set your environment variables for the rest api calls.",
"_____no_output_____"
]
],
[
[
"import os\nos.environ[\"APCA_API_BASE_URL\"] = \" \"\nos.environ[\"APCA_API_KEY_ID\"] = \" \"\nos.environ[\"APCA_API_SECRET_KEY\"] = \" \" ",
"_____no_output_____"
],
[
"print ('Base url: ', os.environ[\"APCA_API_BASE_URL\"] )\nprint ('key:', os.environ[\"APCA_API_KEY_ID\"] )\nprint ('secret:', os.environ[\"APCA_API_SECRET_KEY\"] )",
"_____no_output_____"
]
],
[
[
"## Running the code.\nFirst lets take a look at what btest is doing under the hood to get a better understanding of what's happening.\nLooking at the function Headers inside the `class Account` we can see there are `__init__(cash), balance_hist(), performance(), set_benchmark(), update(), fill_order(), `. It looks like `balance_hist() and performance() `could be use full for examining data after execution. It looks like `balance_hsit()` rturns `pd.DataFrame(data, index=series.index)` which is a pandas dataframe of the balance history for the backtest. `performance()` returns a dataframe of `df[[c for c in df.columns if c.endswith('perf')]`. It looks like this performance measure is returning the performance for each stock over time. Along with these function calls we can extrack the objects variables which include `cash, positions, trades, equities, _benchmark`. Well examine these after running the back test.\n\n\nThe btest module takes a default argument of `starting_cash` when creating the object. The object has stored values which we will be extracting from the object after running btest.py. This is where jupyter-notebooks comes in handy for running multiple code segments. Lets set up our test and execute a run. ",
"_____no_output_____"
]
],
[
[
"from samplealgo import algo, btest\nimport os\naccount = btest.simulate(500)",
"_____no_output_____"
],
[
"print(account.cash)\nprint(account.positions)",
"123.79829999999788\n{'KR': {'entry_timestamp': Timestamp('2019-03-12 00:00:00-0400', tz='America/New_York'), 'entry_price': 24.77, 'shares': 4.0}, 'NWS': {'entry_timestamp': Timestamp('2019-03-15 00:00:00-0400', tz='America/New_York'), 'entry_price': 12.82, 'shares': 7.0}, 'BWA': {'entry_timestamp': Timestamp('2019-03-15 00:00:00-0400', tz='America/New_York'), 'entry_price': 37.9, 'shares': 2.0}, 'ROST': {'entry_timestamp': Timestamp('2019-03-15 00:00:00-0400', tz='America/New_York'), 'entry_price': 89.09, 'shares': 1.0}, 'KHC': {'entry_timestamp': Timestamp('2019-03-15 00:00:00-0400', tz='America/New_York'), 'entry_price': 31.95, 'shares': 3.0}}\n"
]
],
[
[
"Looks like our ending account balance was 123.798 after running the backtest. You can also see the 'current portfolio' of the simulated portfolio. Now lets get a handle on the equities. The equities is a dictionary object with the equity amount at each time stamp.",
"_____no_output_____"
]
],
[
[
"account.equities",
"_____no_output_____"
]
],
[
[
"Wow so it looks like our bot was able to make a little bit of money on its strategy over the course of 2 years. This startegy could definitley use improvements. Lets take a look at a graph of this time series",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.plot(list(account.equities.values()))",
"_____no_output_____"
]
],
[
[
"Looks like our stock performed fairly well up to day 425 where it lost almost all of its profit. It would be interesting to see how the algorithm would recuperate given more data. it looks like the algorithm spotted a trend and found a good curve to ride but only more training can tell.\n\nTo start optimizing this algorithm you should go into algo.py and adjust indicators and guides for the algorithm. A good place to start would be in the `def calc_scores()` function. This is the guiding principle behind the buy and sell logic. \n\n```\nparam = 10\n\nema = df.close.ewm(span=param).mean()[dayindex]\nlast = df.close.values[dayindex]\ndiff = (last - ema) / last\ndiffs[symbol] = diff\n```\n\nIt looks like it is calculating the close ema mean() based off of a span of `10`. \nIt then returns a sorted dictionary of the diff's according to the diff amount. This is later used to rank the stocks for placement in a queue of whether to buy or not. We can see the logic in the code chunk from the `def get_orders()` function in the file `algo.py`\n```\nranked = calc_scores(price_df)\nto_buy = set()\nto_sell = set()\naccount = api.get_account()\n# take the top one twentieth out of ranking,\n# excluding stocks too expensive to buy a share\nfor symbol, _ in ranked[:len(ranked) // 20]:\n price = float(price_df[symbol].close.values[-1])\n if price > float(account.cash):\n continue\n to_buy.add(symbol) \n```\n\nFrom here it follows a script to decide what to sell.\n```\n# now get the current positions and see what to buy,\n # what to sell to transition to today's desired portfolio.\n positions = api.list_positions()\n logger.info(positions)\n holdings = {p.symbol: p for p in positions}\n holding_symbol = set(holdings.keys())\n to_sell = holding_symbol - to_buy\n to_buy = to_buy - holding_symbol\n orders = []\n```\n\nWe can see it sets the to_sell to any items in your current holdings that are not in the buy list already. \nThis is some simple logic that proves to work well over time. It simply states as long as we want to buy the stock that day we hold onto it, otherwise we sell it and buy the highest ranked stock not already in our buy list.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6c95d4a5a4211e4c053897a76b744ba600ec97 | 8,780 | ipynb | Jupyter Notebook | new_trainer.ipynb | Severus11/P1_Facial_Keypoints | 73ef94ac60cab1b85da1f690d0368126cd3c1794 | [
"MIT"
]
| null | null | null | new_trainer.ipynb | Severus11/P1_Facial_Keypoints | 73ef94ac60cab1b85da1f690d0368126cd3c1794 | [
"MIT"
]
| null | null | null | new_trainer.ipynb | Severus11/P1_Facial_Keypoints | 73ef94ac60cab1b85da1f690d0368126cd3c1794 | [
"MIT"
]
| null | null | null | 34.431373 | 148 | 0.60262 | [
[
[
"import numpy as np \nimport pandas as pd \nimport matplotlib.pyplot as plt \nimport matplotlib.image as mpimg\nimport cv2\nimport os\nimport tensorflow as tf\nfrom sklearn.model_selection import train_test_split \n\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom tensorflow.keras.layers import Dense, Input, Dropout, Flatten,Conv2D,BatchNormalization,Activation, MaxPooling2D\nfrom tensorflow.keras.models import Model, Sequential\nfrom tensorflow.keras.optimizers import Adam\nfrom tensorflow.keras.callbacks import ModelCheckpoint, ReduceLROnPlateau",
"_____no_output_____"
],
[
"key_points_df= pd.read_csv('data/training_frames_keypoints.csv')\n\ndef make_dataset(csv_dir, root_dir):\n \n key_points_frame = pd.read_csv(csv_dir)\n x=[]\n y=[]\n \n for i in range(len(key_points_frame)):\n image_name= os.path.join(root_dir,key_points_frame.iloc[i,0])\n image = cv2.imread(image_name)\n image= image[:,:,0:3]\n #image= cv2.cvtColor(image, cv2.COLOR_RGB2GRAY)\n h, w = image.shape[:2]\n image_resized = cv2.resize(image, (96,96),1)\n #image_resized = image_resized/255.0\n x.append(image_resized)\n \n key_points= key_points_frame.iloc[i, 1:].values\n key_points= key_points.astype('float').reshape(-1,2)\n key_points = key_points * [96/w, 96/h]\n key_points= key_points.reshape(-1)\n y.append(key_points)\n \n x = np.asarray(x, dtype=np.float32)\n y= np.asarray(y, dtype=np.float32)\n\n #sample={'image':x, 'points':y}\n return x, y\n\nx_train, y_train= make_dataset(\"data/training_frames_keypoints.csv\", \"data/training/\")\n\nX_train, X_test, Y_train, Y_test = train_test_split(x_train, y_train, test_size=0.2, random_state=42)\n",
"_____no_output_____"
],
[
"\nprint(X_train.shape)\nprint(X_test.shape)\n",
"_____no_output_____"
],
[
"model = tf.keras.models.Sequential()\n\nmodel.add(tf.keras.layers.Conv2D(filters =32, kernel_size =(3,3), padding='same', input_shape=(96,96,3)))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.Conv2D(filters =32, kernel_size =(3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.MaxPool2D(pool_size =2))\n\nmodel.add(tf.keras.layers.Dropout(0.5))\n\nmodel.add(tf.keras.layers.Conv2D(filters =64, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.Conv2D(filters =64, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.MaxPool2D(pool_size=2))\n\nmodel.add(tf.keras.layers.Dropout(0.3))\n\nmodel.add(tf.keras.layers.Conv2D(filters =128, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.Conv2D(filters =128, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.MaxPool2D(pool_size=2))\n\nmodel.add(tf.keras.layers.Conv2D(filters =256, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.Conv2D(filters =256, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.MaxPool2D(pool_size=2))\n\nmodel.add(tf.keras.layers.Dropout(0.25))\n\nmodel.add(tf.keras.layers.Conv2D(filters =512, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\nmodel.add(tf.keras.layers.Conv2D(filters =512, kernel_size= (3,3),padding='same'))\nmodel.add(tf.keras.layers.LeakyReLU(alpha = 0.1))\nmodel.add(tf.keras.layers.BatchNormalization())\n\nmodel.add(tf.keras.layers.Dropout(0.2))\n\nmodel.add(tf.keras.layers.Flatten())\nmodel.add(tf.keras.layers.Dense(units=1024, activation='relu'))\nmodel.add(tf.keras.layers.Dropout(0.1))\nmodel.add(tf.keras.layers.Dense(units=512, activation='relu'))\nmodel.add(tf.keras.layers.Dropout(0.1))\nmodel.add(tf.keras.layers.Dense(units=136))\nmodel.compile(optimizer = 'adam', loss = 'mse', metrics = ['accuracy', 'mae'])\n",
"_____no_output_____"
],
[
"checkpoint = tf.keras.callbacks.ModelCheckpoint('model_weights.h5', monitor=['val_accuracy'],save_weights_only=True, mode='max', verbose=1)\nreduce_lr= tf.keras.callbacks.ReduceLROnPlateau(monitor='val_loss', factor = 0.9, patience=5, min_delta=0.00001, mode='auto')\ntensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=\"./logs\")\ncallbacks = [tensorboard_callback,checkpoint]",
"_____no_output_____"
],
[
"model.fit(X_train, Y_train, validation_data=(X_test, Y_test), epochs = 250 ,batch_size = 128, callbacks=callbacks) \n",
"_____no_output_____"
],
[
"model_json = model.to_json()\nwith open(\"model_new_latest.json\", \"w\") as json_file:\n json_file.write(model_json)",
"_____no_output_____"
],
[
"score = model.evaluate(X_test, Y_test, verbose=0)\nprint('Test loss:', score[0])\nprint('Test accuracy:', score[1])\n",
"_____no_output_____"
],
[
"model.save_weights('model_weights_LATEST.h5')\nprint('model weights saved to disk')",
"_____no_output_____"
],
[
"from tensorflow.keras.preprocessing import image\ntest_image = image.load_img(\"data/test/Abdullah_Gul_31.jpg\", target_size=(96,96))\ntest_image1 = np.asarray(test_image, dtype=np.float32)\ntest_image1 = np.expand_dims(test_image, axis = 0)\nprint(test_image1.shape)\n\n",
"_____no_output_____"
],
[
"result = model.predict(test_image1)\nresult= result.astype('float').reshape(-1,2)\nplt.imshow(test_image)\nplt.scatter(result[:, 0], result[:, 1], s=20, marker='.', c='m')\n\nprint(result)\n",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6c9b09308320ec4c09ad7902e18ced3db35ae7 | 302,016 | ipynb | Jupyter Notebook | Jupyter_Notebooks/Exploratory_Data_Analysis_Final.ipynb | basketcase03/The_Reddit_Flair_Detection | b735924ce1eafc767382ff60482881d4eab1a498 | [
"bzip2-1.0.6"
]
| null | null | null | Jupyter_Notebooks/Exploratory_Data_Analysis_Final.ipynb | basketcase03/The_Reddit_Flair_Detection | b735924ce1eafc767382ff60482881d4eab1a498 | [
"bzip2-1.0.6"
]
| 3 | 2021-03-30T13:15:12.000Z | 2021-09-22T18:55:59.000Z | Jupyter_Notebooks/Exploratory_Data_Analysis_Final.ipynb | basketcase03/The_Reddit_Flair_Detection | b735924ce1eafc767382ff60482881d4eab1a498 | [
"bzip2-1.0.6"
]
| null | null | null | 149.217391 | 95,948 | 0.832052 | [
[
[
"Import pandas to work with dataframe.\nImport the visualisation libraries ( matplotlib and seaborn )",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Read the csv file with collected data and check the information.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv('final_db_2.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 19916 entries, 0 to 19915\nData columns (total 8 columns):\nauthor 19916 non-null object\ncomments 14686 non-null object\ndomain 19916 non-null object\nflair 19916 non-null object\nscore 19916 non-null int64\ntext 7923 non-null object\ntitle 19916 non-null object\nurl 19916 non-null object\ndtypes: int64(1), object(7)\nmemory usage: 1.2+ MB\n"
]
],
[
[
"Here we can see that there are many posts that do not have text. Therefore, it is probable that text is not a really good attribute to classify the flair on. Also, comments has a similar case.\nThe author is a proper noun and can hardly be expected to useful in contributing to detect the flairs.",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"Let us see and group our dataset by flair",
"_____no_output_____"
]
],
[
[
"df.groupby('flair').describe()",
"_____no_output_____"
]
],
[
[
"Let us add textlength and title length to the dataframe and see if we can get any information.",
"_____no_output_____"
]
],
[
[
"df['text_len']=df['text'].apply(str).apply(len)\ndf['title_len']=df['title'].apply(str).apply(len)",
"_____no_output_____"
],
[
"g = sns.PairGrid(df,hue='flair',palette='rainbow')\ng=g.map(plt.scatter,edgecolor=\"w\", s=40)\ng=g.add_legend()",
"_____no_output_____"
],
[
"df.groupby('flair').describe()",
"_____no_output_____"
],
[
"plt.figure(figsize=(13,4))\nsns.countplot(x='flair',data=df)",
"_____no_output_____"
]
],
[
[
"But here we see an imbalanced data. So to balance it, we go back to out gathering data notebook, and create a balanced \ndataset. We stored it in corrected_dataset.csv, so now we analyse our new dataset.",
"_____no_output_____"
]
],
[
[
"df=pd.read_csv('corrected_dataset.csv')",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.drop(['Unnamed: 0'],axis=1,inplace=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 22287 entries, 0 to 22286\nData columns (total 8 columns):\nauthor 22287 non-null object\ncomments 16418 non-null object\ndomain 22287 non-null object\nflair 22287 non-null object\nscore 22287 non-null int64\ntext 9125 non-null object\ntitle 22287 non-null object\nurl 22287 non-null object\ndtypes: int64(1), object(7)\nmemory usage: 1.4+ MB\n"
]
],
[
[
"Here too we can see that there are many posts that do not have text. Therefore, it is probable that text is not a really good attribute to classify the flair on. Also, comments has a similar case.\nThe author is a proper noun and can hardly be expected to useful in contributing to detect the flairs.\nBut every submission has a title and url, which can be useful.",
"_____no_output_____"
]
],
[
[
" df.describe()",
"_____no_output_____"
]
],
[
[
"Adding textlength and titlelength to dataframe",
"_____no_output_____"
]
],
[
[
"df['text_len']=df['text'].apply(str).apply(len)\ndf['title_len']=df['title'].apply(str).apply(len)",
"_____no_output_____"
]
],
[
[
"Visualising the data",
"_____no_output_____"
]
],
[
[
"g = sns.PairGrid(df,hue='flair',palette='rainbow')\ng=g.map(plt.scatter,edgecolor=\"w\", s=40)\ng=g.add_legend()",
"_____no_output_____"
]
],
[
[
"Grouping the data by flairs",
"_____no_output_____"
]
],
[
[
"df.groupby('flair').describe()",
"_____no_output_____"
],
[
"plt.figure(figsize=(16,6))\nsns.countplot(x='flair',data=df)",
"_____no_output_____"
],
[
"df['text_len']=df['text'].apply(str).apply(len)\ndf['title_len']=df['title'].apply(str).apply(len)",
"_____no_output_____"
],
[
"df['text_len'].hist()",
"_____no_output_____"
],
[
"df['title_len'].hist()",
"_____no_output_____"
]
],
[
[
"We now move to apply the ML classification algorithms to data.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec6cd1c2c5ec3cf38e7fbab8db6aa13cd6207766 | 28,848 | ipynb | Jupyter Notebook | b117.ipynb | AlaaALatif/bjorn | 3bc4c2b5b5f6b18c93513721f5df96c47ba68ec8 | [
"MIT"
]
| null | null | null | b117.ipynb | AlaaALatif/bjorn | 3bc4c2b5b5f6b18c93513721f5df96c47ba68ec8 | [
"MIT"
]
| null | null | null | b117.ipynb | AlaaALatif/bjorn | 3bc4c2b5b5f6b18c93513721f5df96c47ba68ec8 | [
"MIT"
]
| null | null | null | 26.273224 | 217 | 0.465024 | [
[
[
"import sys\nimport pandas as pd\nimport os\nfrom path import Path\nimport plotly\nimport plotly.express as px\nimport plotly.graph_objects as go\nfrom plotly.subplots import make_subplots\nfrom urllib.request import urlopen\nimport json\nimport statsmodels as sm\nfrom statsmodels.formula.api import ols\nfrom Bio import Seq, SeqIO, AlignIO, Phylo, Align\nfrom jinja2 import Environment, FileSystemLoader # html template engine\n# import cv2\nimport numpy as np\nimport skimage as sk\nimport matplotlib.pylab as plt\nimport datetime as dt",
"_____no_output_____"
],
[
"sys.path.append('../')",
"_____no_output_____"
],
[
"import bjorn_support as bs\nimport onion_trees as ot\nimport mutations as bm\nimport visualize as bv\nimport reports as br",
"_____no_output_____"
],
[
"msa_fp = '/home/al/analysis/gisaid/sequences_2021-01-25_08-14_aligned.fasta.gz'\nmeta_fp = '/home/al/analysis/gisaid/metadata_2021-01-25_09-11.tsv.gz'",
"_____no_output_____"
],
[
"cols = {'Virus name': 'strain', 'Collection date': 'date' , 'Additional host information': 'purpose_of_sequencing',\n 'Lineage': 'pangolin_lineage', 'Host': 'host'}\nold_meta_fp = meta_fp\nmeta_fp = '/home/al/analysis/gisaid/metadata_2021-01-26.tsv.gz'\nxtra_fp = '/home/al/analysis/gisaid/gisaid_hcov-19_2021_01_26_19.tsv'\ndrop_cols = ['Accession ID', 'Location', 'Passage', 'Specimen', 'Sequencing technology',\n 'Assembly method', 'Comment', 'Comment type']",
"_____no_output_____"
],
[
"# meta = bs.integrate_gisaid_meta(old_meta_fp, xtra_fp, msa_fp, cols, drop_cols)",
"_____no_output_____"
],
[
"meta_fp",
"_____no_output_____"
],
[
"# meta.to_csv(meta_fp, sep='\\t', compression='gzip', index=False)",
"_____no_output_____"
],
[
"# meta = pd.read_csv(meta_fp, sep='\\t', compression='gzip')\n# meta.loc[(meta['country'].str.contains('United States'))&(meta['pangolin_lineage']=='B.1.1.7')]",
"/home/al/anaconda3/envs/bjorn/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3146: DtypeWarning:\n\nColumns (8,28) have mixed types.Specify dtype option on import or set low_memory=False.\n\n"
],
[
"# gisaid, _ = bm.identify_replacements_per_sample(msa_fp, meta_fp, bm.GENE2POS, data_src='gisaid', test=False)\n# gisaid['mutation'] = gisaid['gene'] + ':' + gisaid['ref_aa'] + gisaid['codon_num'].astype(str) + gisaid['alt_aa']\n# gisaid.loc[gisaid['country']=='USA', 'country'] = 'United States of America'",
"_____no_output_____"
],
[
"# gisaid_fp = '/home/al/analysis/gisaid/subs_long_2021-01-15_14-55v2.csv.gz'\n# gisaid.to_csv(gisaid_fp, index=False, compression='gzip')",
"_____no_output_____"
],
[
"# gisaid[(gisaid['mutation']=='ORF1b:D1183Y')]['strain'].unique().shape",
"_____no_output_____"
],
[
"# feature = 'mutations'\n# values = ['S:S13I', 'S:W152C', 'S:L452R']\nfeature = 'pangolin_lineage'\n# S:Q677H, M:A85S, N:D377Y,\nvalues = ['B.1.1.7']#, 'S:W152C', 'S:L452R'] # 'ORF1ab:I4205V', \ninput_params = {\n 'gisaid_data_fp' : '/home/al/analysis/gisaid/subs_long_2021-01-25.csv.gz',\n 'gisaid_meta_fp': meta_fp,\n 'vocs': ['B.1.1.7', 'B.1.1.70'],\n 'strain': 'B117',\n 'date': '01/26/2021',\n 'msa_fp': Path(msa_fp),\n 'meta_fp' : Path('/home/al/code/HCoV-19-Genomics/metadata.csv'),\n 'tree_fp' : Path('/home/al/analysis/alab_mutations_01-01-2021/alab/seqs_aligned.fa.treefile'),\n 'subs_fp' : '/home/al/analysis/alab_mutations_01-01-2021/alab_substitutions_long_01-01-2021.csv',\n 'countries_fp' : '/home/al/data/geojsons/countries.geo.json',\n 'states_fp' : \"/home/al/data/geojsons/us-states.json\",\n 'counties_fp' : '/home/al/data/geojsons/us-counties.json',\n 'patient_zero' : 'NC_045512.2',\n 'b117_meta' : '/home/al/analysis/b117/nextstrain_groups_neherlab_ncov_S.N501_metadata.tsv',\n 'b117_tree': 'test_data/b117_seqs_aligned.fasta.treefile',\n 'sample_sz': 500,\n 'sampling_img_fp' : \"/home/al/analysis/b117/figs/sars-cov-2_EM_v3.png\"\n}",
"_____no_output_____"
],
[
"results = br.generate_voc_data(feature, values, input_params)",
"/home/al/anaconda3/envs/bjorn/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3338: DtypeWarning: Columns (8,28) have mixed types.Specify dtype option on import or set low_memory=False.\n if (await self.run_code(code, result, async_=asy)):\n"
],
[
"html = br.generate_voc_html(feature, values, results, template_name='voc.html')\n# br.save_html(html, f'test_data/orf1ab_i4205v_report.html')\nbr.save_html(html, f'test_data/b117_current_report.html')",
"_____no_output_____"
],
[
"plotly.offline.plot()",
"_____no_output_____"
],
[
"gisaid = pd.read_csv(input_params['gisaid_data_fp'], compression='gzip')",
"_____no_output_____"
],
[
"# res = (gisaid.groupby(['date', 'country', 'division', \n# 'purpose_of_sequencing',\n# 'location', 'pangolin_lineage', 'strain'])\n# .agg(mutations=('mutation', 'unique')).reset_index())\n# res['is_vui'] = res['mutations'].apply(bv.is_vui, args=(set(values),))",
"_____no_output_____"
],
[
"gisaid = pd.read_csv(input_params['gisaid_data_fp'], compression='gzip')",
"_____no_output_____"
],
[
"\ndef get_mutations(data: pd.DataFrame, lineage: str='B.1.1.7'):\n mutations = set(data[data['pangolin_lineage']==lineage]['mutation'].unique().tolist())\n return mutations",
"_____no_output_____"
],
[
"# gisaid = pd.read_csv(input_params['gisaid_data_fp'], compression='gzip')\n# gisaid.drop(columns=['mutation'], inplace=True)\n# gisaid['mutations'] = gisaid['gene'] + ':' + gisaid['ref_aa'] + gisaid['codon_num'].astype(str) + gisaid['alt_aa']\n# gisaid.to_csv(input_params['gisaid_data_fp'], compression='gzip')",
"/home/al/anaconda3/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3146: DtypeWarning: Columns (38,39,41,42,43,44,45,46) have mixed types.Specify dtype option on import or set low_memory=False.\n has_raised = await self.run_ast_nodes(code_ast.body, cell_name,\n"
],
[
"gisaid[gisaid['pangolin_lineage']=='B.1.1.7']['strain'].unique().shape",
"_____no_output_____"
],
[
"gisaid[gisaid['pangolin_lineage']=='B.1.1.70']['strain'].unique().shape",
"_____no_output_____"
],
[
"b117 = get_mutations(gisaid, lineage='B.1.1.7')\nb1170 = get_mutations(gisaid, lineage='B.1.1.70')\nb117.intersection(b1170)",
"_____no_output_____"
],
[
"# gisaid.columns",
"/home/al/anaconda3/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3146: DtypeWarning:\n\nColumns (37,38,40,41,42,43,44,45) have mixed types.Specify dtype option on import or set low_memory=False.\n\n"
]
],
[
[
"'England/205141731/2020', 'Lebanon/LAU-uk4/2020', 'England/204590625/2020', \n'England/204590575/2020', 'England/NORW-F6D33/2020'",
"_____no_output_____"
]
],
[
[
"g = gisaid.groupby(['pangolin_lineage', 'strain']).agg(mutations=('mutation', 'unique')).reset_index()\ndef check_mutation(x, mutation='S:N501Y'):\n if mutation in x:\n return True\n return False\n\ng['501Y'] = g['mutations'].apply(check_mutation)",
"_____no_output_____"
],
[
"m = \"S:D614G\"\ng['614G'] = g['mutations'].apply(check_mutation, args=(m,))",
"_____no_output_____"
],
[
"soi = g.loc[(g['614G']==False) \n & (g['pangolin_lineage']=='B.1.1.7'), \n 'strain'].unique().tolist() + [input_params['patient_zero']]",
"_____no_output_____"
],
[
"soi",
"_____no_output_____"
],
[
"bs.fetch_seqs(input_params['msa_fp'], 'test_data/b117_wo_N501Y.fasta', soi, is_aligned=True)",
"_____no_output_____"
],
[
"# gisaid[(gisaid['pangolin_lineage']=='B.1.1.7')]['location'].value_counts()",
"_____no_output_____"
],
[
"# df",
"_____no_output_____"
],
[
"results['county_map'] = fig",
"_____no_output_____"
],
[
"gisaid.loc[(gisaid['codon_num']==452) & (gisaid['gene']=='S') & (gisaid['alt_aa']=='R'), 'strain'].unique().shape",
"_____no_output_____"
],
[
"gisaid.loc[(gisaid['codon_num']==452) & (gisaid['gene']=='S') & (gisaid['alt_aa']=='R'), 'date'].min()",
"_____no_output_____"
],
[
"gisaid[gisaid['pangolin_lineage']=='B.1.1.70']",
"_____no_output_____"
],
[
"dists.loc[(dists['group']=='outgroup') & (dists['genetic_distance']>0.0008), 'pangolin_lineage'].value_counts()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6ce89922ab4c1df207f403c76354cea8ecfc55 | 73,942 | ipynb | Jupyter Notebook | week9_ML_svm_poly_norm/day1_logistic_regresion_confusion_matrix/exercises/1.multinominal_regression.ipynb | PabloEduardoMartinezPicazo/Bootcamp-DataScience-2021 | 0fa5288aec5fb14e3796877882e4f1ddc5ad4aea | [
"MIT"
]
| null | null | null | week9_ML_svm_poly_norm/day1_logistic_regresion_confusion_matrix/exercises/1.multinominal_regression.ipynb | PabloEduardoMartinezPicazo/Bootcamp-DataScience-2021 | 0fa5288aec5fb14e3796877882e4f1ddc5ad4aea | [
"MIT"
]
| null | null | null | week9_ML_svm_poly_norm/day1_logistic_regresion_confusion_matrix/exercises/1.multinominal_regression.ipynb | PabloEduardoMartinezPicazo/Bootcamp-DataScience-2021 | 0fa5288aec5fb14e3796877882e4f1ddc5ad4aea | [
"MIT"
]
| null | null | null | 286.596899 | 44,321 | 0.711666 | [
[
[
"### 1. \n\nEn el archivo \"logistic_regression_df_class\" hemos visto un ejemplo multiclase. Realiza un análisis con regresión linear multinominal teniendo como target la columna \"duración\". Es decir, averigua cuánto aumentan o disminuyen la duración cuando se aumenta una unidad los valores de las demás columnas.\n\n- ¿Qué acierto tiene? (score)\n- ¿Cuál es el RMSE? ¿Dirías que es un valor alto?\n- Viendo la matriz de correlación, ¿tienen coherencia los resultados de la regresión?\n- ¿Es la regresión un buen método para este dataset?",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn import linear_model\nfrom sklearn import model_selection\nfrom sklearn.metrics import classification_report\nfrom sklearn.metrics import confusion_matrix\nfrom sklearn.metrics import accuracy_score\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn import metrics",
"_____no_output_____"
],
[
"dataframe = pd.read_csv(\"../data/usuarios_win_mac_lin.csv\")\n#dataframe #clase -> 0 windwos, 1 linux y 2 mac ",
"_____no_output_____"
],
[
"X = dataframe.drop(['duracion'], axis=1)\nY = dataframe['duracion']\nprint(X.shape)\nY.shape",
"(170, 4)\n"
],
[
"\n \nX_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2,random_state=7)\n\nregresion = LinearRegression()\nregresion.fit(X_train, Y_train)\n\nprint(regresion.score(X_train, Y_train) *100)\n\nprint(regresion.score(X_test, Y_test) *100)\n\n\n \n\n\n",
"10.966235092000032\n-24.895550591347735\n"
],
[
"regresion.coef_",
"_____no_output_____"
],
[
"Y_pred = regresion.predict(X_test)",
"_____no_output_____"
],
[
"Y_pred",
"_____no_output_____"
],
[
" np.sqrt(metrics.mean_squared_error(Y_test, Y_pred)) #RMSE",
"_____no_output_____"
]
],
[
[
"Valor muy alto para los datos que tenemos noosotros, la media es de unos 100 segundos y nos da un valor de 185 segundos. ",
"_____no_output_____"
]
],
[
[
"sns.heatmap(dataframe.corr(), annot = True)",
"_____no_output_____"
]
],
[
[
"Muy mal metodo por la dispersion que da de los dtos con respecto a los datos de verdad",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6d00ee9a3b902d57e9f656418921e49b1c0745 | 39,203 | ipynb | Jupyter Notebook | same_class.ipynb | billalihaddaden/TERbigGAN | c2c24a4e99b0eb3ff3d16778b97caf19328a97bd | [
"MIT"
]
| null | null | null | same_class.ipynb | billalihaddaden/TERbigGAN | c2c24a4e99b0eb3ff3d16778b97caf19328a97bd | [
"MIT"
]
| null | null | null | same_class.ipynb | billalihaddaden/TERbigGAN | c2c24a4e99b0eb3ff3d16778b97caf19328a97bd | [
"MIT"
]
| null | null | null | 139.01773 | 27,703 | 0.847333 | [
[
[
"cd \"./same_class/\"",
"/Users/xuejiaxin/Dropbox/My Mac (Jiaxin的MacBook Pro)/Documents/GitHub/latent/pytorch-pretrained-BigGAN/same_class\n"
],
[
"%load_ext autoreload\n%autoreload 2\n\n# target image\nimport torch\nfrom pytorch_pretrained_biggan import (BigGAN, one_hot_from_names, truncated_noise_sample,\n save_as_images, display_in_terminal)\nimport logging\n\n\nlogging.basicConfig(level=logging.INFO)\n\nmodel = BigGAN.from_pretrained('biggan-deep-128')\n\ntruncation = 0.5\nclass_vector = one_hot_from_names([\"dog\"], batch_size=1)\nnoise_vector = truncated_noise_sample(truncation=truncation, batch_size=1, seed = 10)\n\nnoise_vector = torch.from_numpy(noise_vector)\nclass_vector = torch.from_numpy(class_vector)\n\n\nwith torch.no_grad():\n output = model(noise_vector, class_vector, truncation)\n\nsave_as_images(output, \"dog_target\")\n",
"INFO:pytorch_pretrained_biggan.model:loading model biggan-deep-128 from cache at /Users/xuejiaxin/.pytorch_pretrained_biggan/6371c3777477e4e75187da1b9b526561aac3134f38c7299a3438009ae560e20d.3434ebdfa74a8c17e0e56061cfd905fa163c92f110e88df77b47da6ce9910b48\nINFO:pytorch_pretrained_biggan.model:Model config {\n \"attention_layer_position\": 8,\n \"channel_width\": 128,\n \"class_embed_dim\": 128,\n \"eps\": 0.0001,\n \"layers\": [\n [\n false,\n 16,\n 16\n ],\n [\n true,\n 16,\n 16\n ],\n [\n false,\n 16,\n 16\n ],\n [\n true,\n 16,\n 8\n ],\n [\n false,\n 8,\n 8\n ],\n [\n true,\n 8,\n 4\n ],\n [\n false,\n 4,\n 4\n ],\n [\n true,\n 4,\n 2\n ],\n [\n false,\n 2,\n 2\n ],\n [\n true,\n 2,\n 1\n ]\n ],\n \"n_stats\": 51,\n \"num_classes\": 1000,\n \"output_dim\": 128,\n \"z_dim\": 128\n}\n\nINFO:pytorch_pretrained_biggan.utils:Saving image to dog_target_0.png\n"
],
[
"from torchvision.models import squeezenet1_0\nfrom tqdm import trange\n\nDEVICE = 'cpu'\nmodel = model.eval().to(DEVICE)\n\n\nsemantic_model = squeezenet1_0(pretrained=True).to(DEVICE)\nsemantic_model.classifier = torch.nn.Sequential(\n torch.nn.Flatten()\n )\nsemantic_model = semantic_model.eval()\n\ntrunction = 0.5\nnoise = truncated_noise_sample(truncation=truncation, batch_size=1, seed=9)\nnoise = torch.nn.Parameter(torch.tensor(noise, requires_grad=True).float().to(DEVICE))\nnoise_optim = torch.optim.Adam([noise], lr=0.05)\n\nclass_vector = one_hot_from_names(['dog'], batch_size=1)\nclass_vector = torch.from_numpy(class_vector)\n\nL = []\nL_pixel = []\nL_semantic = []\n\nfor iteration in trange(0, 200):\n noise_optim.zero_grad()\n\n y_hat = model(noise, class_vector, truncation)\n\n semantic_loss = ((semantic_model(y_hat) - semantic_model(output)) ** 2).mean() ** .5 #-cos_sim(semantic_model(y_hat), semantic_model(output))\n L_semantic.append(semantic_loss.item())\n\n pixel_loss = abs(y_hat - output).mean()\n L_pixel.append(pixel_loss.item())\n\n loss = semantic_loss + 30 * pixel_loss\n L.append(loss.item())\n\n loss.backward()\n noise_optim.step()\n\n if iteration % 5 == 0:\n save_as_images(y_hat, f\"dog2dog_{iteration}\")",
" 0%| | 0/200 [00:00<?, ?it/s]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_0_0.png\n 2%|▎ | 5/200 [00:18<12:09, 3.74s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_5_0.png\n 5%|▌ | 10/200 [00:37<11:03, 3.49s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_10_0.png\n 8%|▊ | 15/200 [00:51<08:38, 2.80s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_15_0.png\n 10%|█ | 20/200 [01:04<08:09, 2.72s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_20_0.png\n 12%|█▎ | 25/200 [01:24<09:42, 3.33s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_25_0.png\n 15%|█▌ | 30/200 [01:38<07:56, 2.80s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_30_0.png\n 18%|█▊ | 35/200 [01:51<07:27, 2.71s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_35_0.png\n 20%|██ | 40/200 [02:05<07:09, 2.69s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_40_0.png\n 22%|██▎ | 45/200 [02:18<06:58, 2.70s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_45_0.png\n 25%|██▌ | 50/200 [02:32<06:40, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_50_0.png\n 28%|██▊ | 55/200 [02:45<06:25, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_55_0.png\n 30%|███ | 60/200 [02:58<06:12, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_60_0.png\n 32%|███▎ | 65/200 [03:12<05:58, 2.65s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_65_0.png\n 35%|███▌ | 70/200 [03:25<05:43, 2.64s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_70_0.png\n 38%|███▊ | 75/200 [03:38<05:32, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_75_0.png\n 40%|████ | 80/200 [03:51<05:16, 2.64s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_80_0.png\n 42%|████▎ | 85/200 [04:05<05:03, 2.64s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_85_0.png\n 45%|████▌ | 90/200 [04:18<04:50, 2.64s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_90_0.png\n 48%|████▊ | 95/200 [04:31<04:34, 2.62s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_95_0.png\n 50%|█████ | 100/200 [04:44<04:25, 2.65s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_100_0.png\n 52%|█████▎ | 105/200 [04:58<04:16, 2.70s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_105_0.png\n 55%|█████▌ | 110/200 [05:11<04:01, 2.68s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_110_0.png\n 57%|█████▊ | 115/200 [05:25<03:46, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_115_0.png\n 60%|██████ | 120/200 [05:38<03:32, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_120_0.png\n 62%|██████▎ | 125/200 [05:51<03:17, 2.63s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_125_0.png\n 65%|██████▌ | 130/200 [06:04<03:05, 2.65s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_130_0.png\n 68%|██████▊ | 135/200 [06:18<02:54, 2.68s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_135_0.png\n 70%|███████ | 140/200 [06:31<02:40, 2.68s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_140_0.png\n 72%|███████▎ | 145/200 [06:45<02:26, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_145_0.png\n 75%|███████▌ | 150/200 [06:58<02:13, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_150_0.png\n 78%|███████▊ | 155/200 [07:11<02:00, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_155_0.png\n 80%|████████ | 160/200 [07:25<01:46, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_160_0.png\n 82%|████████▎ | 165/200 [07:38<01:33, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_165_0.png\n 85%|████████▌ | 170/200 [07:51<01:19, 2.66s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_170_0.png\n 88%|████████▊ | 175/200 [08:05<01:06, 2.64s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_175_0.png\n 90%|█████████ | 180/200 [08:18<00:53, 2.68s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_180_0.png\n 92%|█████████▎| 185/200 [08:31<00:39, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_185_0.png\n 95%|█████████▌| 190/200 [08:45<00:26, 2.67s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_190_0.png\n 98%|█████████▊| 195/200 [08:58<00:13, 2.65s/it]INFO:pytorch_pretrained_biggan.utils:Saving image to dog2dog_195_0.png\n100%|██████████| 200/200 [09:11<00:00, 2.76s/it]\n"
],
[
"import matplotlib.pyplot as plt\n\n\nplt.plot(L)\nplt.plot([x*30 for x in L_pixel], 'r')\nplt.plot(L_semantic, 'b')\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
ec6d08cbf6a28ccbcf221812c5295c0a37838ef9 | 31,663 | ipynb | Jupyter Notebook | compare-itczpositions.ipynb | aikovoigt/itcz-position | 1d2226baf2a49c581f3a536260614217995288e3 | [
"MIT"
]
| null | null | null | compare-itczpositions.ipynb | aikovoigt/itcz-position | 1d2226baf2a49c581f3a536260614217995288e3 | [
"MIT"
]
| null | null | null | compare-itczpositions.ipynb | aikovoigt/itcz-position | 1d2226baf2a49c581f3a536260614217995288e3 | [
"MIT"
]
| null | null | null | 88.444134 | 17,112 | 0.798313 | [
[
[
"# Compare two possibilities to define the ITCZ position",
"_____no_output_____"
],
[
"## Purpose",
"_____no_output_____"
],
[
"The ITCZ is typically defined by means of the precipitation centroid. The centroid offers a more stable metric compared to the location of the precipitation maximum, which for example is sensitive to small precipitation changes for the case of a \"double ITCZ\".\n\nYet, there are different choices what is meant by the centroid, and also within which latitude range it should be calculated (e.g., between 20N/S deg lat or 30N/S deg lat). The latter point is not studied here. The first point is investigated in the following.\n\nAt least two definitions of the centroid have been used:\n\n1. The \"Adam\" definition, documented in Eq.1a of Adam et al., 2016, J. Climate, Seasonal and Interannual Variations of the Energy Flux Equator and ITCZ. Part I: Zonally Averaged ITCZ Position, https://journals.ametsoc.org/doi/full/10.1175/JCLI-D-15-0512.1 with N=1. Note that in Adam et al., 2016, the latitude boundaries were 20N/S deg lat.\n\n2. The \"Voigt\" definition used in the Tracmip introduction paper Voigt et al., 2016, JAMES, https://agupubs.onlinelibrary.wiley.com/doi/full/10.1002/2016MS000748. Note that in Voigt et al., 2016, the latitude boundaries were 30N/S deg lat.\n\nBoth definitions have been used as illustrated by the following non-exhaustive list:\n\n * Frierson, D. and Y.-T. Hwang (2012), Extratropical Influence on ITCZ Shifts in Slab Ocean Simulations of Global Warming, Journal of Climate, DOI: 10.1175/JCLI-D-11-00116.1 (Voigt definition with 15N/S deg lat; indeed this is where the \"Voigt\" definition originates from)\n * Voigt, A. et al. (2014), The radiative impact of clouds on the shift of the Intertropical Convergence Zone, Geophys. Res. Lett., 41, 4308–4315, doi:10.1002/2014GL060354. (Voigt definition with 30N/S deg lat)\n * Voigt, A. et al. (2014), Compensation of Hemispheric Albedo Asymmetries by Shifts of the ITCZ and Tropical Clouds, Journal of Climate, https://doi.org/10.1175/JCLI-D-13-00205.1. (Voigt definition with 30N/S deg lat)\n * Donohoe, A. and A. Voigt (2017), Why Future Shifts in Tropical Precipitation Will Likely Be Small: The Location of the Tropical Rain Belt and the Hemispheric Contrast of Energy Input to the Atmosphere, Climate Extremes: Patterns and Mechanisms, AGU Book. (Voigt definition with 20N/S deg lat)\n\nIn the following, we will document to what extent the two definitions differ quantitatively. We do so by using precipitation from the TRACMIP aquaControl simulations that are described in Voigt et al., 2016, JAMES and that we access via the pangeo cloud. \n\nThe ITCZ definition problem might seem an academic problem - the precipitation is what it is, independent of how we decide to diagnose the ITCZ position. But in fact the choice of definition can be important for conceptual pictures of the ITCZ and tropical rainfall, such as those embodied in the so-called energetic framework that link the ITCZ position to cross-equatorial atmosphere energy transport. The ratio of ITCZ position and energy transport could thus be sensitive to how the ITCZ position is diagnosed.",
"_____no_output_____"
],
[
"## Background",
"_____no_output_____"
],
[
"The Adam-based definition uses the equation:\n \n\\begin{equation}\n \\varphi_\\text{Adam}= \\frac{ \\int_{-\\varphi_0}^{\\varphi_0} \\varphi P(\\varphi) \\cos\\varphi d\\varphi }{ \\int_{-\\varphi_0}^{\\varphi_0} P(\\varphi) \\cos\\varphi d\\varphi } .\n\\end{equation}\n\nThis defintion of the ITCZ position is in analogy to a lever, where the force is the product of the weight (precipition P) and the distance from the fulcrum (abs(latitude-ITCZ position)). Thus, for the Adam definition one has \n\n\\begin{equation}\n \\int_{-\\varphi_0}^{\\varphi_\\text{Adam}} abs(\\varphi-\\varphi_\\text{Adam}) P(\\varphi) \\cos\\varphi d\\varphi = \\int_{\\varphi_\\text{Adam}}^{\\varphi_0} abs(\\varphi-\\varphi_\\text{Adam}) P(\\varphi) \\cos\\varphi d\\varphi .\n\\end{equation}\n\nThe Voigt-based definition instead follows the idea that the ITCZ is given by the latitude for which the sum of the area-weighted precipitation south and north of it are the same:\n\n\\begin{equation}\n \\int_{-\\varphi_0}^{\\varphi_\\text{Voigt}} P(\\varphi) \\cos\\varphi d\\varphi = \\int_{\\varphi_\\text{Voigt}}^{\\varphi_0} P(\\varphi) \\cos\\varphi d\\varphi .\n\\end{equation}\n\nI.e., while the Adam definition weighs precip more strongly according to its distance from the \"fulcrum\", the Voigt definition just has two buckets of precip that need to have the same filling level but are at the same distance from the fulcrum.\n\nThis shows that while both definitions are closely related, the Adam definition gives more weight to latitudes further away from the diagnosed ITCZ. As a result, it will tend to put the ITCZ closer to the equator than the Voigt definition, for which it does not matter whether the precipitation falls near or far from the diagnosed ITCZ.",
"_____no_output_____"
],
[
"## Analysis",
"_____no_output_____"
],
[
"### Load Tracmip aquaControl precipitation",
"_____no_output_____"
]
],
[
[
"from intake import open_catalog\n# get whole pangeo catalogue\ncat = open_catalog(\"https://raw.githubusercontent.com/pangeo-data/pangeo-datastore/master/intake-catalogs/climate.yaml\")\n# select tracmip collection\ncol = cat.tracmip()\n# load data into dictionary\nds_dict = col.search(frequency='Amon', experiment=['aquaControl'],\n variable=['pr']).to_dataset_dict(zarr_kwargs={'consolidated': True})",
"Dataset(s): 0%| | 0/14 [00:00<?, ?it/s]"
],
[
"# uncomment the following line to check content of dataset dictionary ds_dict\n#print(ds_dict.keys())\n# lists of models\nmodels = list(col.df.model.unique())",
"_____no_output_____"
]
],
[
[
"### Calculate ITCZ positions according to Adam and Voigt definitions",
"_____no_output_____"
],
[
"Dictionary of model colors, same as in Voigt et al. 2016, JAMES, TRACMIP introduction paper",
"_____no_output_____"
]
],
[
[
"import numpy as np\ndict_color = {'AM21': np.array([1. , 0.8, 0.6]), 'CAM3':np.array([0.50196078, 0.50196078, 0.50196078]), \n 'CAM4': np.array([0.58039216, 1. , 0.70980392]), 'CAM5Nor':np.array([0.76078431, 0. , 0.53333333]), \n 'CNRM-AM5': np.array([0. , 0.2 , 0.50196078]), 'ECHAM61':np.array([0. , 0.45882353, 0.8627451 ]), \n 'ECHAM63':np.array([0.6 , 0.24705882, 0. ]), 'GISS-ModelE2':np.array([0.61568627, 0.8 , 0. ]),\n 'LMDZ5A':np.array([0.29803922, 0. , 0.36078431]), 'MetUM-CTL':np.array([0.09803922, 0.09803922, 0.09803922]),\n 'MetUM-ENT':np.array([0. , 0.36078431, 0.19215686]), 'MIROC5':np.array([0.16862745, 0.80784314, 0.28235294]), \n 'MPAS':np.array([0.56078431, 0.48627451, 0. ]), 'CALTECH': np.array([1. , 0.64313725, 0.01960784])}",
"_____no_output_____"
]
],
[
[
"Load my implementation of the ITCZ definitions",
"_____no_output_____"
]
],
[
[
"import itcz",
"_____no_output_____"
]
],
[
[
"**Calculate ITCZ positions using the zonal-mean time-mean precipitation from the last 30 years of the aquaControl simulation using the latitude boundaries of 20N/S deg lat**",
"_____no_output_____"
]
],
[
[
"for mod in models:\n ds_dict[mod+'.aquaControl.Amon']['itcz_adam'] = (\n itcz.get_itczposition_adam(ds_dict[mod+'.aquaControl.Amon']['pr'][-360:,:,:].mean(['time','lon']), \n ds_dict[mod+'.aquaControl.Amon'].lat, latboundary=20, dlat=0.1) )\n ds_dict[mod+'.aquaControl.Amon']['itcz_voigt'] = (\n itcz.get_itczposition_voigt(ds_dict[mod+'.aquaControl.Amon']['pr'][-360:,:,:].mean(['time','lon']), \n ds_dict[mod+'.aquaControl.Amon'].lat, latboundary=20, dlat=0.05) )",
"_____no_output_____"
]
],
[
[
"**Scatter plot of Adam versus Voigt definitions (left panel) and the ratio of the two (right panel):**\n\nThis shows that the two definitions agree well, but that the Voigt definition yields more poleward ITCZ position, in agreement with the considerations in the background section.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nplt.figure(figsize=(12,4))\nplt.subplot(1,2,1)\nfor mod in models:\n plt.plot(ds_dict[mod+'.aquaControl.Amon']['itcz_adam'],\n ds_dict[mod+'.aquaControl.Amon']['itcz_voigt'],'o', color=dict_color[mod])\nplt.xlabel('Adam ITCZ (deg lat)'); plt.ylabel('Voigt ITCZ (deg lat)');\n \nplt.subplot(1,2,2)\nfor mod in models:\n plt.plot(ds_dict[mod+'.aquaControl.Amon']['itcz_adam'],\n ds_dict[mod+'.aquaControl.Amon']['itcz_voigt']/ds_dict[mod+'.aquaControl.Amon']['itcz_adam'],'x', color=dict_color[mod])\nplt.xlabel('Adam ITCZ (deg lat)'); plt.ylabel('Voigt ITCZ / Adam ITCZ');",
"_____no_output_____"
]
],
[
[
"**Test that the Voigt and Adam results agree with the integral equations given in the background section.**\n\nWe use the criterion that for both definitions the l.h.s and r.h.s agree within 1%. Note that the ITCZ positions are recalculated within the test subroutines.",
"_____no_output_____"
]
],
[
[
"for mod in models:\n lhs, rhs = itcz.test_itczposition_adam(ds_dict[mod+'.aquaControl.Amon']['pr'][-360:,:,:].mean(['time','lon']),\n ds_dict[mod+'.aquaControl.Amon'].lat, 20, 0.1)\n if (np.abs(lhs-rhs)>0.01*rhs):\n print(mod, ': Adam test FAILED!')\n else:\n print(mod, ': Adam test passed!')",
"AM21 : Adam test passed!\nCAM4 : Adam test passed!\nCAM5Nor : Adam test passed!\nCNRM-AM5 : Adam test passed!\nECHAM61 : Adam test passed!\nECHAM63 : Adam test passed!\nGISS-ModelE2 : Adam test passed!\nMIROC5 : Adam test passed!\nMPAS : Adam test passed!\nMetUM-CTL : Adam test passed!\nMetUM-ENT : Adam test passed!\nCAM3 : Adam test passed!\nLMDZ5A : Adam test passed!\nCALTECH : Adam test passed!\n"
],
[
"for mod in models:\n lhs, rhs = itcz.test_itczposition_voigt(ds_dict[mod+'.aquaControl.Amon']['pr'][-360:,:,:].mean(['time','lon']),\n ds_dict[mod+'.aquaControl.Amon'].lat, 20, 0.05)\n if (np.abs(lhs-rhs)>0.01*rhs):\n print(mod, ': Voigt test FAILED!')\n else:\n print(mod, ': Voigt test passed!')",
"AM21 : Voigt test passed!\nCAM4 : Voigt test passed!\nCAM5Nor : Voigt test passed!\nCNRM-AM5 : Voigt test passed!\nECHAM61 : Voigt test passed!\nECHAM63 : Voigt test passed!\nGISS-ModelE2 : Voigt test passed!\nMIROC5 : Voigt test passed!\nMPAS : Voigt test passed!\nMetUM-CTL : Voigt test passed!\nMetUM-ENT : Voigt test passed!\nCAM3 : Voigt test passed!\nLMDZ5A : Voigt test passed!\nCALTECH : Voigt test passed!\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6d09fff9073d81c79969fdcbadc751264a9a63 | 11,683 | ipynb | Jupyter Notebook | Mission_to_Mars.ipynb | ayyray/Mission-to-Mars | 81ff4564d96a2104dcf94af982be96e9cb70b386 | [
"MIT"
]
| null | null | null | Mission_to_Mars.ipynb | ayyray/Mission-to-Mars | 81ff4564d96a2104dcf94af982be96e9cb70b386 | [
"MIT"
]
| null | null | null | Mission_to_Mars.ipynb | ayyray/Mission-to-Mars | 81ff4564d96a2104dcf94af982be96e9cb70b386 | [
"MIT"
]
| null | null | null | 26.313063 | 1,076 | 0.469828 | [
[
[
"# Article Scraping",
"_____no_output_____"
]
],
[
[
"# Import Splinter and BeautifulSoup\nfrom splinter import Browser\nfrom bs4 import BeautifulSoup as soup\nfrom webdriver_manager.chrome import ChromeDriverManager\nimport pandas as pd",
"_____no_output_____"
],
[
"# Set executable path and set up URL\nexecutable_path = {'executable_path': ChromeDriverManager().install()}\nbrowser = Browser('chrome', **executable_path, headless=False)",
"\n\n====== WebDriver manager ======\nCurrent google-chrome version is 98.0.4758\nGet LATEST chromedriver version for 98.0.4758 google-chrome\nTrying to download new driver from https://chromedriver.storage.googleapis.com/98.0.4758.102/chromedriver_mac64.zip\nDriver has been saved in cache [/Users/anthonyray/.wdm/drivers/chromedriver/mac64/98.0.4758.102]\n"
],
[
"# Assign URL and instruct the browser to visit it\nurl = 'https://redplanetscience.com'\nbrowser.visit(url)\n# Optional delay for loading the page (so we do not scrape too much and get booted)\nbrowser.is_element_present_by_css('div.list_text', wait_time=1)",
"_____no_output_____"
],
[
"# Setup the HTML parser\nhtml = browser.html\nnews_soup = soup(html, 'html.parser')\nslide_elem = news_soup.select_one('div.list_text')",
"_____no_output_____"
],
[
"# Begin scraping",
"_____no_output_____"
],
[
"# Title and summary text to variables to reference later\nslide_elem.find('div', class_='content_title')",
"_____no_output_____"
],
[
"# Use the parent element to find the first 'a' tag and save it as 'news_title'\nnews_title = slide_elem.find('div', class_='content_title').get_text()\nnews_title",
"_____no_output_____"
],
[
"# Use the parent element to find the paragraph text\nnews_p = slide_elem.find('div', class_='article_teaser_body').get_text()\nnews_p",
"_____no_output_____"
]
],
[
[
"# Image Scraping:\n\n## Featured Images",
"_____no_output_____"
]
],
[
[
"# Set up the URL\nurl = 'https://spaceimages-mars.com'\nbrowser.visit(url)",
"_____no_output_____"
],
[
"# Find and click the full image button\nfull_image_elem = browser.find_by_tag('button')[1]\nfull_image_elem.click()",
"_____no_output_____"
],
[
"# Parse the resulting html with soup\nhtml = browser.html\nimg_soup = soup(html, 'html.parser')",
"_____no_output_____"
],
[
"# Find the relative image url\nimg_url_rel = img_soup.find('img', class_='fancybox-image').get('src')\nimg_url_rel",
"_____no_output_____"
],
[
"# Use the base URL to create an absolute URL\nimg_url = f'https://spaceimages-mars.com/{img_url_rel}'\nimg_url",
"_____no_output_____"
],
[
"# Use the base URL to create an absolute URL\nimg_url = f'https://spaceimages-mars.com/{img_url_rel}'\nimg_url",
"_____no_output_____"
],
[
"df = pd.read_html('https://galaxyfacts-mars.com')[0]\ndf.columns=['description', 'Mars', 'Earth']\ndf.set_index('description', inplace=True)\ndf\n",
"_____no_output_____"
],
[
"df.to_html()",
"_____no_output_____"
],
[
"browser.quit()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6d162b4cf33c385e18d53265524aad0533f1ca | 224,635 | ipynb | Jupyter Notebook | 4.1.Basedon_Simliarity_v4_exp2.ipynb | balag752/Identifying-and-Analyzing-the-Interdisciplinary | c5399e964c451cd0fafad5367e8c3083081293a4 | [
"MIT"
]
| null | null | null | 4.1.Basedon_Simliarity_v4_exp2.ipynb | balag752/Identifying-and-Analyzing-the-Interdisciplinary | c5399e964c451cd0fafad5367e8c3083081293a4 | [
"MIT"
]
| null | null | null | 4.1.Basedon_Simliarity_v4_exp2.ipynb | balag752/Identifying-and-Analyzing-the-Interdisciplinary | c5399e964c451cd0fafad5367e8c3083081293a4 | [
"MIT"
]
| null | null | null | 201.647217 | 89,036 | 0.882053 | [
[
[
"import datetime\n\nimport pandas as pd\nimport spacy\nimport re\nimport string\nimport numpy as np\n\n\nimport seaborn as sns\nfrom matplotlib import cm\nfrom matplotlib.pyplot import figure\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom spacy.tokens import Token\nfrom tqdm import tqdm\nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\n\nimport gensim\nfrom gensim import corpora, models\nfrom gensim.models.phrases import Phrases, Phraser\nfrom gensim.utils import simple_preprocess\nfrom gensim.parsing.preprocessing import STOPWORDS\nfrom gensim.models.coherencemodel import CoherenceModel\nfrom gensim.models import HdpModel\n\nfrom sklearn.metrics.pairwise import cosine_similarity\nfrom collections import Counter\nimport itertools\n\nimport pyLDAvis.gensim\npyLDAvis.enable_notebook()\nfrom ipywidgets import interact\n\nimport nltk\nfrom nltk.corpus import stopwords\nfrom textblob import TextBlob\n\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.decomposition import PCA\nfrom sklearn.manifold import TSNE\n\nfrom sklearn.cluster import MiniBatchKMeans\nfrom sklearn.cluster import DBSCAN\nfrom sklearn.cluster import KMeans\n\nfrom sklearn.metrics import silhouette_score\n\nimport enchant\npd.set_option('display.max_rows', 500)",
"/Users/balaji/anaconda3/lib/python3.7/site-packages/nltk/decorators.py:68: DeprecationWarning: `formatargspec` is deprecated since Python 3.5. Use `signature` and the `Signature` object directly\n regargs, varargs, varkwargs, defaults, formatvalue=lambda value: \"\"\n/Users/balaji/anaconda3/lib/python3.7/site-packages/nltk/lm/counter.py:15: DeprecationWarning: Using or importing the ABCs from 'collections' instead of from 'collections.abc' is deprecated, and in 3.8 it will stop working\n from collections import Sequence, defaultdict\n"
],
[
"dict_check = enchant.Dict(\"en_US\")\n\n#### Importing the file ####\nPath=\"src/\"\nFilename='projects_Preprocessed.csv'\ndf=pd.read_csv(Path+Filename)\n\nCat_File=\"category_hier.csv\"\nCat_data=pd.read_csv(Path+Cat_File)\n\nvarcluster_file=\"variable_clusters.csv\"\nvarcluster=pd.read_csv(Path+varcluster_file)\n\nmanualtag=pd.read_csv(Path+'SamplesManualTagger.csv')\nvarcluster_info=pd.read_csv(Path+'variable_clusters_info_v2.csv')\n\ndf=df[df['Translates']!=\"The goal of the Heisenberg Program is to enable outstanding scientists who fulfill all the requirements for a long-term professorship to prepare for a scientific leadership role and to work on further research topics during this time. In pursuing this goal, it is not always necessary to choose and implement project-based procedures. For this reason, in the submission of applications and later in the preparation of final reports - unlike other support instruments - no 'summary' of project descriptions and project results is required. Thus, such information is not provided in GEPRIS.\"]\n\n## Filtering the null abstracts & short description\ndf=df[(pd.isnull(df.PreProcessedDescription)==False) & (df.PreProcessedDescription.str.strip()!='abstract available')& (df.PreProcessedDescription.str.len()>100) & (pd.isnull(df[\"SubjectArea\"])==False)]\n\n# Striping the category column\nCat_data.Category=Cat_data.Category.str.strip()\n\n## Merging the high level category information\n\ndf=df.merge(Cat_data[[\"File_Categories\",\"Category\"]], how=\"left\", left_on=\"SubjectArea\", right_on=\"File_Categories\")\n\ndbdf=pd.read_csv(Path+'Report_WEPCADBScanFindingsKMeansV3.csv')",
"_____no_output_____"
],
[
"## Only keeping noun and adjective\n\ndf.PreProcessedDescription=df.PreProcessedDescription.apply(lambda x : ' '.join([i for i,pos in TextBlob(x).tags if pos in ['NN','JJ']]))",
"_____no_output_____"
],
[
"manualstopwords=['research','group','subject','process','development','analysis','model','different','new','process','study','change','system','approach', 'phase', 'high', 'develop', 'allow','investigation','property','interaction','university', 'device', 'institute', 'researcher', 'science', 'fund', 'facility', 'coordination', 'cooperation']",
"_____no_output_____"
],
[
"varcluster_info.cluster_id=varcluster_info.cluster_id.astype('int32')\nvarclusterall=varcluster.merge(varcluster_info, how='left',left_on='Cluster', right_on='cluster_id')\nnewstopwords=varclusterall[(varclusterall.cluster_name=='General') | (varclusterall.cluster_name=='Text & Publish') ].Variable.tolist()+manualstopwords\n#.sort_values(by='RS_Ratio')",
"_____no_output_____"
],
[
"df.PreProcessedDescription=[' '.join([words for words in abstract.split() if(not words in newstopwords)]) for abstract in df.PreProcessedDescription ]",
"_____no_output_____"
]
],
[
[
"### Preprocessing ",
"_____no_output_____"
]
],
[
[
"# TF IDF Conversion\n\nvectorizer = TfidfVectorizer(max_features=1000, ngram_range=(1, 2))\nreview_vectors = vectorizer.fit_transform(df[\"PreProcessedDescription\"])\nfeatures_df = pd.DataFrame(review_vectors.toarray(), columns = vectorizer.get_feature_names())\n\nfeatures_df.reset_index(drop=True, inplace=True)\ndf.reset_index(drop=True, inplace=True)\nmerged_data=pd.concat([df,features_df], axis=1,ignore_index=False)\n\n#wordslist=merged_data.columns.tolist()[len(df.columns)+6:]",
"_____no_output_____"
],
[
"wordslist=merged_data.columns.tolist()[len(df.columns)+2:]\n\nplt.subplots(figsize=(14,8)) \ni=1\ncategories=[]\ncategory_words=[]\n\nfor cat,bucket in merged_data.groupby('Category'):\n \n plt.subplot(2,2,i)\n bucket[wordslist].sum().sort_values(ascending=False).head(20).plot(kind='bar',color='green')\n plt.title(cat)\n plt.xticks(rotation=60)\n i=i+1\n plt.tight_layout()\n categories.append(cat)\n category_words.append( ' '.join(bucket[wordslist].sum().sort_values(ascending=False).head(50).index.tolist()))\n\nCategoryImpoWords=pd.DataFrame({'Category':categories,'Words':category_words})",
"_____no_output_____"
],
[
"CategoryImpoWords.Words.tolist()",
"_____no_output_____"
],
[
"CategoryImpoWords.loc[CategoryImpoWords.Category=='Engineering Sciences', 'Words']='engineer method simulation component flow surface behavior technology tool parameter mechanical field measurement temperature production machine particle numerical optimization technique sensor element measure performance algorithm'\nCategoryImpoWords.loc[CategoryImpoWords.Category=='Humanities and Social Sciences', 'Words']='humanity social history country century political cultural language economic literature law early policy european literary knowledge religious society culture national manuscript market'\nCategoryImpoWords.loc[CategoryImpoWords.Category=='Life Sciences', 'Words']='biology life cell protein gene disease molecular receptor mouse signal patient human factor plant tumor tissue membrane immune genetic cellular animal specie dna vivo clinical infection brain therapy inflammatory therapeutic'\nCategoryImpoWords.loc[CategoryImpoWords.Category=='Natural Sciences', 'Words']='natural field surface quantum metal theory magnetic spin temperature measurement electron water complex particle energy compound molecule chemical electronic synthesis transition molecular optical climate spectroscopy resolution organic gas polymer sediment mathematic'",
"_____no_output_____"
],
[
"## Repeated words in category\n\nword=[]\nfor x in [cc.split() for cc in CategoryImpoWords.Words]:\n word.extend(x)\npd.Series(word).value_counts().sort_values(ascending=False).head(16)#.index.tolist()",
"_____no_output_____"
],
[
"CategoryImpoWords.loc[CategoryImpoWords.Category=='Humanities and Social Sciences', 'Words']='humanity social history country century political cultural language economic law early policy european literary knowledge religious society culture national manuscript market'\nCategoryImpoWords.loc[CategoryImpoWords.Category=='Natural Sciences', 'Words']='mathematic natural field surface quantum metal theory magnetic spin temperature measurement electron water complex particle energy compound molecule chemical electronic synthesis transition molecular optical climate spectroscopy resolution organic gas polymer sediment '\n\ndoc='direct method spatial statistic natural technology computer feedback orientate theory mathematic partner main emphasis stochastic young specialist progress thesis order individual knowledge interdisciplinary contact seminar lecture colloquium special'\n\nfor ip in CategoryImpoWords.iterrows():\n print(ip[1].Category, similarity_scores(doc, ip[1]['Words'],50))\n",
"_____no_output_____"
]
],
[
[
"#### 1.1 Word Embedding",
"_____no_output_____"
]
],
[
[
"## Word Embeddings Functions\n\n## Generate the tagged documents (tagging based on the category column)\ndef create_tagged_document(list_of_list_of_words):\n for i, list_of_words in enumerate(list_of_list_of_words):\n yield gensim.models.doc2vec.TaggedDocument(list_of_words, [i])\n\n## Generate the tagged documents (each record in single tag )\ndef create_tagged_document_based_on_tags(list_of_list_of_words, tags):\n for i in range(len(list_of_list_of_words)):\n yield gensim.models.doc2vec.TaggedDocument(list_of_list_of_words[i], [tags[i]])\n\ndef make_bigram(inputlist):\n bigram = Phrases(inputlist, min_count=3, threshold=1,delimiter=b' ')\n bigram_phraser = Phraser(bigram)\n new_list=[]\n for sent in inputlist:\n new_list.append(bigram_phraser[sent])\n return new_list \n \n## Generate output using the word embedding model prediction - takes long time to regenerate\ndef vec_for_learning(model, tagged_docs):\n sents = tagged_docs#.values\n targets, regressors = zip(*[(doc.tags[0], model.infer_vector(doc.words, steps=20)) for doc in sents])\n return targets, regressors",
"_____no_output_____"
],
[
"## creating a tagged document\n#DescDict=make_bigram([[x for x in str(i).split()] for i in df.PreProcessedDescription])\nDescDict=[[x for x in str(i).split()] for i in df.PreProcessedDescription]\n\n\n#tagged_value_tags = list(create_tagged_document_based_on_tags(DescDict, df.Category.tolist()))\ntagged_value = list(create_tagged_document(DescDict))",
"_____no_output_____"
],
[
"print(str(datetime.datetime.now()),'Started')\n\n# Init the Doc2Vec model\nmodel = gensim.models.Word2Vec(DescDict,size=50, min_count=1, alpha = 0.02, sg=0, seed=42, workers=4)\n\n#### Hyper parameter ####\nprint(str(datetime.datetime.now()),'Completed')",
"_____no_output_____"
],
[
"## Validating the model response for random words \n\nmodelchecked=model\ntarget_word='mathematic'\nprint('target_word: %r model: %s similar words:' % (target_word, modelchecked))\nfor i, (word, sim) in enumerate(modelchecked.wv.most_similar(target_word, topn=10), 1):\n print(' %d. %.2f %r' % (i, sim, word))",
"_____no_output_____"
],
[
"modelchecked.similarity('country','german')",
"_____no_output_____"
],
[
"###############################\n#### similarity functions #####\n###############################\n\ndef map_word_frequency(document):\n return Counter(itertools.chain(*document))\n \ndef get_sif_feature_vectors(sentence1, sentence2, word_emb_model=modelchecked):\n sentence1 = [token for token in sentence1.split() if token in word_emb_model.wv.vocab]\n sentence2 = [token for token in sentence2.split() if token in word_emb_model.wv.vocab]\n word_counts = map_word_frequency((sentence1 + sentence2))\n embedding_size = 50 # size of vectore in word embeddings\n a = 0.0001\n sentence_set=[]\n for sentence in [sentence1, sentence2]:\n vs = np.zeros(embedding_size)\n sentence_length = len(sentence)\n for word in sentence:\n a_value = a / (a + word_counts[word]) # smooth inverse frequency, SIF\n vs = np.add(vs, np.multiply(a_value, word_emb_model.wv[word])) # vs += sif * word_vector\n vs = np.divide(vs, sentence_length) # weighted average\n sentence_set.append(vs)\n return sentence_set\n\ndef get_cosine_similarity(feature_vec_1, feature_vec_2): \n return cosine_similarity(feature_vec_1.reshape(1, -1), feature_vec_2.reshape(1, -1))[0][0]\n\ndef similarity_scores(sentence1, sentence2, n=50):\n sentence1=sentence1.replace(',','')\n sentence2=sentence2.replace(',','')\n features=get_sif_feature_vectors(sentence1, sentence2)\n return get_cosine_similarity(features[0][:n],features[1][:n])",
"_____no_output_____"
]
],
[
[
"### Direct Comparision",
"_____no_output_____"
]
],
[
[
"Sims={'Natural Sciences':[], 'Humanities and Social Sciences':[],'Engineering Sciences':[], 'Life Sciences':[]}\nsize=len(df.PreProcessedDescription)\nwith tqdm(total=size) as bar:\n for i in range(len(df.PreProcessedDescription)):\n topicWord=df.loc[i].PreProcessedDescription\n for s in Sims.keys():\n CatWords=CategoryImpoWords[CategoryImpoWords.Category==s].Words.values[0]\n Sims[s].append(similarity_scores(topicWord, CatWords,50))\n bar.update(1)",
"_____no_output_____"
],
[
"df_Sim=pd.DataFrame(Sims)\n\ndef top_matchings(input):\n \n input=input.sort_values(ascending=False)\n \n PrimaryMatching=input.head(1).tail(1).index.values[0]\n PrimaryMatchingSim=input.head(1).tail(1).values[0]\n \n SecMatching=input.head(2).tail(1).index.values[0]\n SecMatchingSim=input.head(2).tail(1).values[0]\n \n ThrdMatching=input.head(3).tail(1).index.values[0]\n ThrdMatchingSim=input.head(3).tail(1).values[0]\n \n #if(PrimaryMatchingSim<.70):\n # PrimaryMatchingSim=0\n # PrimaryMatching=''\n #if(SecMatchingSim<.70):\n # SecMatchingSim=0\n # SecMatching=''\n \n return PrimaryMatching,PrimaryMatchingSim,SecMatching,SecMatchingSim,ThrdMatching,ThrdMatchingSim\n\ndf_Sim.head().apply(top_matchings,axis=1, result_type=\"expand\")\n \ndf_Sim1=df_Sim.apply(top_matchings,axis=1, result_type=\"expand\")\ndf_Sim1.columns=['PrimaryMatching','PrimaryMatchingSim','SecMatching','SecMatchingSim','ThrdMatching','ThrdMatchingSim']\n",
"_____no_output_____"
],
[
"Merged_df=pd.concat([df,df_Sim1], axis=1)\nthersold_1=.6\nthersold_2=.6\nthersold_3=.6\n\n\nMerged_df.loc[(Merged_df.PrimaryMatchingSim<thersold_1),'PrimaryMatching']=''\nMerged_df.loc[(Merged_df.PrimaryMatchingSim<thersold_1) | (Merged_df.SecMatchingSim<thersold_2),'SecMatching']=''\nMerged_df.loc[(Merged_df.PrimaryMatchingSim<thersold_1) | (Merged_df.SecMatchingSim<thersold_2) | (Merged_df.ThrdMatchingSim<thersold_3),'ThrdMatching']=''\n\nMerged_df['ID_Categories']=Merged_df.apply(lambda x: ','.join(sorted([x['PrimaryMatching'],x['SecMatching'],x['ThrdMatching']]) ).strip(','), axis=1)",
"_____no_output_____"
],
[
"Merged_df['Interdiscipilinary']=True\nMerged_df.loc[Merged_df.SecMatching=='','Interdiscipilinary']=False",
"_____no_output_____"
],
[
"Merged_df.Interdiscipilinary.value_counts()",
"_____no_output_____"
],
[
"#'Category',,'ThrdMatching'\n#Merged_df.groupby(['PrimaryMatching','SecMatching']).count()[['SubUrl']]\n\n#Merged_df.groupby(['Category','PrimaryMatching']).count()[['SubUrl']]\n#Merged_df[Merged_df.Category=='Life Sciences'].groupby(['PrimaryMatching','SecMatching']).count()[['SubUrl']]\nMerged_df[Merged_df.Interdiscipilinary].groupby('ID_Categories').count()[['SubUrl']].sort_values('SubUrl', ascending=False)",
"_____no_output_____"
],
[
"#Merged_df.loc[(Merged_df.Category=='Engineering Sciences') & (Merged_df.PrimaryMatching=='Life Sciences'),'Translates'].head(10).tolist()\n#Merged_df.loc[(Merged_df.PrimaryMatching=='Natural Sciences') & (Merged_df.SecMatching=='Engineering Sciences'),'Translates'].head(10).tolist()\n\nMerged_df.loc[(Merged_df.ID_Categories=='Engineering Sciences,Natural Sciences') ,'Translates'].head(10).tolist()",
"_____no_output_____"
],
[
"i=1\nplt.figure(figsize=(12, 8))\nfor cc,w in Merged_df[Merged_df.PrimaryMatching!=''].groupby(['PrimaryMatching']):\n plt.subplot(2,2,i)\n w['PrimaryMatchingSim'].hist( edgecolor='white', linewidth=1) \n plt.title(str(cc)+' Len :'+str(len(w)))\n i=i+1\nplt.tight_layout()",
"_____no_output_____"
],
[
"Merged_df.to_csv(Path+'IDBasedonSimliarity2.csv', index=False)",
"_____no_output_____"
],
[
"Merged_df.loc[Merged_df.PrimaryMatching=='','PrimaryMatchingSim']=np.NaN\nMerged_df.loc[Merged_df.SecMatching=='','SecMatchingSim']=np.NaN\nMerged_df.loc[Merged_df.ThrdMatching=='','ThrdMatchingSim']=np.NaN",
"_____no_output_____"
],
[
"for c1 in Merged_df.Category.unique():\n CategoryImpoWords[c1]=0\n for c2 in Merged_df.Category.unique():\n countvalues=len(Merged_df.loc[(Merged_df.ID_Categories.str.contains(c1,case=False)) & (Merged_df.ID_Categories.str.contains(c2,case=False))])\n if((c1==c2) | (countvalues==0) ):\n CategoryImpoWords.loc[CategoryImpoWords.Category==c2,c1]=0\n else:\n CategoryImpoWords.loc[CategoryImpoWords.Category==c2,c1]=np.log(countvalues)\n #print(c1,c2,countvalues)\n# pass\n\nCategoryImpoWords[Merged_df.Category.unique()]=CategoryImpoWords[Merged_df.Category.unique()].max().max()+0.01-CategoryImpoWords[Merged_df.Category.unique()]\n\nfor c1 in Merged_df.Category.unique():\n for c2 in Merged_df.Category.unique():\n if((c1==c2)):\n CategoryImpoWords.loc[CategoryImpoWords.Category==c2,c1]=0\n \n",
"_____no_output_____"
],
[
"## Generating coordinates from distance\n\n\ncoords = TSNE(n_components=2,perplexity=1, random_state=12).fit_transform(CategoryImpoWords[Merged_df.Category.unique()])\n#coords = PCA(n_components=2).fit_transform(CategoryImpoWords[[ 'Engineering Sciences','Humanities and Social Sciences', 'Life Sciences', 'Natural Sciences']])\n\ncoords=MinMaxScaler([0,1000]).fit_transform(coords)\ncoords=pd.DataFrame(coords, index=CategoryImpoWords.Category).reset_index()\np1=sns.scatterplot(\n x=0, y=1,\n hue=\"Category\",\n # palette=sns.color_palette(\"hls\", 4),\n data=coords,\n # legend=\"full\",\n alpha=1,\n size = 8,\n legend=False\n)\n\nfor line in range(0,coords.shape[0]):\n p1.text(coords[0][line]+0.01, coords[1][line], CategoryImpoWords.Category[line], horizontalalignment='left', size='medium', color='black')",
"_____no_output_____"
],
[
"cat_dict=pd.DataFrame(Merged_df[Merged_df.ID_Categories!='']['ID_Categories'].value_counts())\ncats=[]\n\n#temp=[ cats.extend([c+'_1',c+'_2',c+'_3']) for c in Merged_df.Category.unique()]\n#for t in cats:\n# cat_dict[t]=10#.00\n \n#for cx in cat_dict.iterrows():\n# i=1\n# counts=np.log(cx[1]['ID_Categories'])/len(cx[0].split(','))\n# for cc in cx[0].split(','):\n# cat_dict.loc[cx[0]][cc+'_'+str(i)]=10-counts\n# #print(cc+'_'+str(i) , '-',cx[1]['ID_Categories']/len(cx[0].split(',')), cat_dict.loc[cx[0]][cc+'_'+str(i)])\n# i=i+1\n\ncats=Merged_df.Category.unique()\n\nfor c in Merged_df.Category.unique():\n cat_dict[c]=10#.00\n\nfor cx in cat_dict.iterrows():\n catlist=cx[0].split(',')\n cxmeans=CategoryImpoWords[CategoryImpoWords.Category.isin(catlist)].mean()\n for c in Merged_df.Category.unique():\n #print(cx[0],':',c)\n cat_dict.loc[cx[0],c]=cxmeans[c]\n ",
"_____no_output_____"
],
[
"cat_dict.reset_index().sort_values(by='index')",
"_____no_output_____"
],
[
"#coords = TSNE(n_components=2,perplexity=5, random_state=12).fit_transform(cat_dict[cats])\ncoords = TSNE(n_components=2,perplexity=3.3, random_state=42, n_iter=850).fit_transform(cat_dict[cats])\n\ncoords=pd.DataFrame(coords, index=cat_dict.index).reset_index()\np1=sns.scatterplot(\n x=0, y=1,\n hue=\"index\",\n # palette=sns.color_palette(\"hls\", 4),\n data=coords,\n # legend=\"full\",\n alpha=1,\n size = 8,\n legend=False\n)\n\nfor line in range(0,coords.shape[0]):\n p1.text(coords[0][line]+0.01, coords[1][line], cat_dict.index[line], horizontalalignment='left', size='medium', color='black')",
"_____no_output_____"
],
[
"coords['size']=list(cat_dict['ID_Categories'])\n#coords.to_csv(Path+'IDSimliarityCats.csv', index=False)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6d1b3e565dfb957b192427f0bdd4aa748f1955 | 16,658 | ipynb | Jupyter Notebook | GroversAlgorithm/GroversAlgorithm.ipynb | thmhugo/QuantumKatas | 86c7a44e703187f7dafc4615e9cd5efa45f7d1e3 | [
"MIT"
]
| null | null | null | GroversAlgorithm/GroversAlgorithm.ipynb | thmhugo/QuantumKatas | 86c7a44e703187f7dafc4615e9cd5efa45f7d1e3 | [
"MIT"
]
| null | null | null | GroversAlgorithm/GroversAlgorithm.ipynb | thmhugo/QuantumKatas | 86c7a44e703187f7dafc4615e9cd5efa45f7d1e3 | [
"MIT"
]
| null | null | null | 37.100223 | 325 | 0.587646 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec6d1fcfcf1b49c0ba753b7783c26e39fdbdf304 | 33,506 | ipynb | Jupyter Notebook | ScrambleTree.ipynb | jfogarty/python-code-practice | 677861ee23ae98056b5e3c5a77bdda6c0952139d | [
"Unlicense"
]
| null | null | null | ScrambleTree.ipynb | jfogarty/python-code-practice | 677861ee23ae98056b5e3c5a77bdda6c0952139d | [
"Unlicense"
]
| null | null | null | ScrambleTree.ipynb | jfogarty/python-code-practice | 677861ee23ae98056b5e3c5a77bdda6c0952139d | [
"Unlicense"
]
| null | null | null | 30.966728 | 126 | 0.404345 | [
[
[
"### Determine whether Scrambled Trees are Equivalent\n\nGiven two trees which represent a scrambled word. Determine if they're equivalent.\n\nGiven a string s1, we may represent it as a binary tree by partitioning it to two non-empty substrings recursively.\n\nFor example, s1 = \"great\":\n\nTo scramble the string, we may choose any non-leaf node and swap its two children.\nFor example, if we choose the node \"gr\" and swap its two children, it produces a scrambled string \"rgeat\".\n\nWe say that \"rgeat\" is a scrambled string of \"great\".\nSimilarly, if we continue to swap the children of nodes \"eat\" and \"at\", it produces a scrambled string \"rgtae\".\n\nWe say that \"rgtae\" is a scrambled string of \"great\".\nGiven two strings s1 and s2 of the same length, determine if s2 is a scrambled string of s1.\n\nSee [The example problem](https://leetcode.com/problems/scramble-string/description/)",
"_____no_output_____"
]
],
[
[
"debugging = False\n#debugging = True\ndebug2 = False\n\nlogging = True\n\ndef dbg(f, *args):\n if debugging:\n print((' DBG:' + f).format(*args))\n\ndef dbg_cont(f, *args):\n if debugging:\n print((' DBG:' + f).format(*args), end='')\n\ndef dbg2(f, *args):\n if debug2:\n print((' -DBG2:' + f).format(*args))\n \ndef log(f, *args):\n if logging:\n print((f).format(*args))\n \ndef logError(f, *args):\n if logging:\n print(('*** ERROR:' + f).format(*args))\n \ndef className(instance):\n return type(instance).__name__",
"_____no_output_____"
]
],
[
[
"### The TestSet Mechanism",
"_____no_output_____"
]
],
[
[
"# %load TestHarness.py\nclass TestCase(object):\n def __init__(self, name, method, inputs, expected, catchExceptions=False):\n self.name = name\n self.method = method\n self.inputs = inputs\n self.expected = expected\n self.catchExceptions = catchExceptions\n \n def run(self):\n if self.catchExceptions:\n try:\n return self.method(*self.inputs)\n except Exception as x:\n return x\n else:\n return self.method(*self.inputs)\n\nimport time\nfrom datetime import timedelta\n\nclass TestSet(object):\n def __init__(self, cases):\n self.cases = cases\n \n def run_tests(self):\n count = 0\n errors = 0\n total_time = 0\n for case in self.cases:\n count += 1\n start_time = time.time()\n result = case.run()\n elapsed_time = time.time() - start_time\n total_time += elapsed_time\n if callable(case.expected):\n if not case.expected(result):\n errors += 1\n logError(\"Test {0} failed. Returned {1}\", case.name, result)\n elif result != case.expected:\n errors += 1\n logError('Test {0} failed. Returned \"{1}\", expected \"{2}\"', case.name, result, case.expected)\n if errors:\n logError(\"Tests passed: {0}; Failures: {1}\", count-errors, errors)\n else:\n log(\"All {0} tests passed.\", count)\n log(\"Elapsed test time: {0}\", timedelta(seconds=total_time))\n ",
"_____no_output_____"
],
[
"14 % 2",
"_____no_output_____"
],
[
"14 // 2",
"_____no_output_____"
]
],
[
[
"### The Unit Under Test",
"_____no_output_____"
]
],
[
[
"import math\n\nclass AutoList(list):\n def __setitem__(self, index, value):\n if index >= len(self):\n self.extend([None]*(index + 1 - len(self)))\n list.__setitem__(self, index, value)\n \nclass ATree(object):\n \"\"\" Array representation of binary tree \"\"\"\n val = AutoList()\n\n def __init__(self, v):\n self.text = v\n\n def sink(self, index=0):\n \"\"\" propagate tree changes down the tree \"\"\"\n if index == 0:\n self.val[1] = self.text\n self.sink(index=1)\n else:\n assert index < 100\n v = self.val[index]\n dbg(\"{0} Sinking[{1}] == '{2}'\", '-'*int(math.floor(math.log(index,2))), index, v)\n n = len(v)\n if n>1:\n offset = len(v) // 2\n vleft = v[0:offset]\n vright = v[offset:]\n self.val[index*2 + 0] = vleft\n self.val[index*2 + 1] = vright\n if len(vleft) > 0: \n self.sink(index*2 + 0)\n if len(vright) > 0: \n self.sink(index*2 + 1)\n\n def swim(self, index=0):\n \"\"\" propagate tree changes up the tree \"\"\"\n if index == 0: \n self.text = self.val[1]\n else:\n assert index < 100 \n v = self.val[index] \n vleft = self.val[index*2 + 0]\n vright = self.val[index*2 + 1]\n newv = vleft + vright\n self.val[index] = newv\n dbg(\"{0} Swimming[{1}] == '{2}'==>{3}\", '-'*int(math.floor(math.log(index,2))), index, v, newv)\n self.swim(index // 2)\n\n def exch(self, index=0):\n assert index > 0 # no exchanging at the text root\n assert index < len(self.val)\n v = self.val[index] \n if v is None or len(v) < 2:\n return # ignore exchange of leaf nodes\n vleft = self.val[index*2 + 0]\n vright = self.val[index*2 + 1]\n self.val[index*2 + 0] = vright\n self.val[index*2 + 1] = vleft\n self.swim(index)\n self.sink(index)\n\n def variants(self):\n pass\n ",
"_____no_output_____"
],
[
"class SolutionBad(object):\n def isScramble(self, s1, s2):\n \"\"\"\n :type s1: str\n :type s2: str\n :rtype: bool\n \"\"\"\n \n if len(s1) != len(s2):\n return False\n\n if len(s1) == 0:\n return True\n\n if len(s1) == 1:\n return True if s1[0] == s2[0] else False\n\n t1 = ETree(s1).load()\n t2 = ETree(s2).load()\n print(\"T1={0}\".format(t1.liststr()))\n print(\"T2={0}\".format(t2.liststr()))\n return t1.isequiv(t1, t2)\n # This solution fails because the comparison tree doesn't have the same\n # structure as the source tree. No amount of flipping will make them equivalent.",
"_____no_output_____"
],
[
"class ETree(object): \n val = None\n left = None\n right = None\n parent = None\n idv=-1 # initialized on tree load, does not reflect exchanges.\n\n def __init__(self, v, parent=None, index=1):\n self.val = v \n self.idv=index\n # Load the remainder of the tree\n if len(v) > 1:\n offset = len(v) // 2\n self.left = ETree(v[0:offset], parent=self, index=index*2)\n self.right = ETree(v[offset:], parent=self, index=index*2+1)\n self.parent = parent\n\n def formatNodeVerbose(self):\n return '[None]' if self is None else '[{0}:\"{1}\"]'.format(self.idv, self.val)\n\n def formatNode(self):\n return '' if self is None else '\"{0}\"'.format(self.val)\n \n def liststr(self):\n if not self:\n return \"(*)\"\n if self.left is None and self.right is None:\n return '\"' + self.val + '\"'\n else:\n leftstr = '' if self.left is None else \"[{0}:{1}]\".format(\n self.left.idv, self.left.liststr())\n rightstr = '' if self.right is None else \"[{0}:{1}]\".format(\n self.right.idv, self.right.liststr())\n return '\"{1}\"==>{2}, {3}'.format(self.idv, self.val, leftstr, rightstr) \n\n def indexorder(self, index):\n \"\"\" get the ordered tree following operations that reach a specific index \"\"\"\n order = []\n while index > 1:\n op = 'L' if index % 2 == 0 else 'R'\n order = [op] + order\n index = index // 2\n return order\n\n def indexed(self, index):\n node = self\n for op in self.indexorder(index):\n node = node.left if op == 'L' else node.right\n return node\n\n def root(self):\n \"\"\" Return the root node for this tree \"\"\"\n node = self\n return self if self.parent is None else self.parent.root()\n\n def swim(self):\n \"\"\" Propagate changes up to top of tree. \"\"\"\n if len(self.val) > 1:\n self.val = self.left.val + self.right.val\n if self.parent is not None:\n self.parent.swim()\n\n def exch(self, index=0):\n \"\"\" Exchange the left and right branches below this node. \n An optional index selects the node to exchange (root relative indexing)\n \"\"\"\n node = self\n if index > 0:\n node = self.root().indexed(index)\n #print(\"xxxx:\" + node.liststr()) \n left = node.left\n right = node.right\n node.left = right\n node.right = left\n #print(\"yyy:\" + node.liststr())\n node.swim()\n return node\n \n def variants(self):\n vals = []\n rootnode = self.root()\n def tryvars(node):\n nonlocal vals\n if node.left is not None or node.left is not None:\n node.exch() # swap the nodes and propagate changes.\n vals += [rootnode.val] \n if node.left is not None:\n tryvars(node.left)\n if node.right is not None:\n tryvars(node.right)\n node.exch() # undo the swap. \n vals += [rootnode.val]\n tryvars(self)\n return vals\n \n def listTree(self):\n lister = self.TreeLister()\n return lister.list(self)",
"_____no_output_____"
],
[
"import math\nimport pprint\nclass TreeLister(object):\n def __init__(self):\n pass\n\n def list(self, tree):\n layers = [] # The set of nodes in display form\n widths = [] # The average widths of each level\n\n def listLayer(node, layers, off='C', p=None, index=1):\n if node is not None:\n level = math.floor(math.log(index,2))\n if len(layers) <= level:\n layers.append([])\n f = ' ' + node.formatNode() + ' '\n c = index - 2**level\n datum = { 'parent':p, 'text':f, 'level':level, 'offset':off, 'col':c }\n layers[level].append(datum)\n listLayer(node.left, layers, 'L', node, index*2)\n listLayer(node.right, layers, 'R', node, index*2+1)\n listLayer(tree, layers)\n #pprint.pprint(layers)\n \n # Find a good maximum width for the displayed tree.\n count = 1\n maxwidth = 1\n for layer in layers:\n avg = math.floor(0.5 + sum(len(x['text']) for x in layer) / len(layer))\n lwidth = avg*count \n widths.append({ 'avg':avg, 'layerwidth':lwidth})\n count *= 2\n maxwidth = max(maxwidth, lwidth)\n dbg2(\"-- Maxwidth={0} widths={1}\",maxwidth,widths)\n \n # Format the actual text lines\n count = 1\n output = \"\"\n for layer in layers:\n line = \"\"\n arrow = \"\"\n center = maxwidth / 2\n dbg2(\"Layer: {0}\", count)\n for node in layer:\n colwidth = maxwidth / count\n text = node['text']\n colnum = node['col']\n leadingoffset = math.floor(colnum * colwidth) # Offset of cell from left.\n celloffset = math.floor(0.5 + (colwidth - len(text))/2)\n offset = leadingoffset + celloffset\n math.floor(0.5 + (colwidth * colnum - len(text)/2) / 2)\n dbg2(\"offset={1} colwidth={2} colnum={3} text={0}\", text, offset, colwidth, colnum)\n if len(line) < offset:\n fill = offset - len(line)\n line += ' '*fill\n rightarrowoffset = len(line)+1 \n line += text\n leftarrowoffset = len(line)-2\n # For non root nodes, format the link arrows.\n if count > 1:\n arrowoffset = leftarrowoffset if node['offset'] == 'L' else rightarrowoffset\n offset = arrowoffset\n dbg2(\"offset={1} arrowoffset={2} colnum={3} text={0} \", text, offset, arrowoffset, colnum)\n if len(arrow) < offset:\n fill = offset - len(arrow)\n arrow += ' '*fill\n arrow += '/' if node['offset'] == 'L' else '\\\\'\n if count>1:\n output += arrow + '\\n'\n output += line + '\\n'\n count *= 2\n return output",
"_____no_output_____"
],
[
"t = ETree(\"abeaseeeeeh\")\nX = ETree(\"xyztabC\")\nt.liststr()\nll = TreeLister()\nprint(ll.list(t))\nprint(ll.list(X))\n",
" \"abeaseeeeeh\" \n / \\\n \"abeas\" \"eeeeeh\" \n / \\ / \\\n \"ab\" \"eas\" \"eee\" \"eeh\" \n / \\ / \\ / \\ / \\\n \"a\" \"b\" \"e\" \"as\" \"e\" \"ee\" \"e\" \"eh\" \n / \\ / \\ / \\\n \"a\" \"s\" \"e\" \"e\" \"e\" \"h\" \n\n \"xyztabC\" \n / \\\n \"xyz\" \"tabC\" \n / \\ / \\\n \"x\" \"yz\" \"ta\" \"bC\" \n / \\ / \\ / \\\n \"y\" \"z\" \"t\" \"a\" \"b\" \"C\" \n\n"
],
[
"class Solution(object):\n\n def isScramble(self, s1, s2):\n \"\"\"\n :type s1: str\n :type s2: str\n :rtype: bool\n \"\"\"\n if len(s1) != len(s2):\n return False\n \n if len(s1) == 0:\n return True\n \n if len(s1) == 1:\n return True if s1[0] == s2[0] else False\n \n # Ignore strings that don't contain the same set of characters.\n x = s2\n for c in s1:\n i = x.find(c)\n if i < 0:\n return False\n x = x[0:i] + x[i+1:]\n \n # Now try all tree operations until success or failure\n t = ETree(s1)\n v = t.variants()\n return True if s2 in t.variants() else False\n",
"_____no_output_____"
],
[
"t = ETree(\"great\")\nt = ETree(\"abcd\")\nt = ETree(\"abb\")\n\nprint(t.liststr())\nt.indexed(3).liststr()\n\n#print('This is\\na\\ntest\\nof the\\nemergency broadcast\\nsystem.')",
"\"abb\"==>[2:\"a\"], [3:\"bb\"==>[6:\"b\"], [7:\"b\"]]\n"
],
[
"print(t.liststr())\nx = t.variants()\nprint(x)\nprint(t.liststr())\n\"bab\" in x",
"\"abb\"==>[2:\"a\"], [3:\"bb\"==>[6:\"b\"], [7:\"b\"]]\n['bba', 'bba', 'bba', 'abb']\n\"abb\"==>[2:\"a\"], [3:\"bb\"==>[6:\"b\"], [7:\"b\"]]\n"
],
[
"t.indexed(7).root().liststr()\nt.indexed(7).liststr()\nt.exch(7)\nt.exch()\nt.liststr()\nprint(t.listTree())",
"[]\n"
]
],
[
[
"### The Test Cases",
"_____no_output_____"
]
],
[
[
"t = ATree(\"great\")\nt.sink()\nprint(t.val)\n\nt.exch(2)\nt.exch(3)\n#t.exch(6)\n\nprint(t.val)",
" DBG: Sinking[1] == 'great'\n DBG:- Sinking[2] == 'gr'\n DBG:-- Sinking[4] == 'g'\n DBG:-- Sinking[5] == 'r'\n DBG:- Sinking[3] == 'eat'\n DBG:-- Sinking[6] == 'e'\n DBG:-- Sinking[7] == 'at'\n DBG:--- Sinking[14] == 'a'\n DBG:--- Sinking[15] == 't'\n[None, 'great', 'gr', 'eat', 'g', 'r', 'e', 'at', None, None, None, None, None, None, 'a', 't']\n DBG:- Swimming[2] == 'gr'==>rg\n DBG: Swimming[1] == 'great'==>rgeat\n DBG:- Sinking[2] == 'rg'\n DBG:-- Sinking[4] == 'r'\n DBG:-- Sinking[5] == 'g'\n DBG:- Swimming[3] == 'eat'==>ate\n DBG: Swimming[1] == 'rgeat'==>rgate\n DBG:- Sinking[3] == 'ate'\n DBG:-- Sinking[6] == 'a'\n DBG:-- Sinking[7] == 'te'\n DBG:--- Sinking[14] == 't'\n DBG:--- Sinking[15] == 'e'\n[None, 'rgate', 'rg', 'ate', 'r', 'g', 'a', 'te', None, None, None, None, None, None, 't', 'e']\n"
],
[
"Solution().isScramble(\"abb\", \"bab\")",
"T1=abb==>(a), (bb==>(b), (b))\nT2=bab==>(b), (ab==>(a), (b))\n DBG:-- isequiv('abb', 'bab') DBG:-- Recurse\n DBG:-- isequiv('a', 'b') DBG: Chars Same --> False\n DBG:-- isequiv('a', 'ab') DBG:-- Recurse\n DBG: DIFF --> False\n DBG: DIFF --> False\n DBG: Answer --> False\n DBG: Answer --> False\n"
],
[
"Solution().isScramble(\"great\", \"rgtae\")",
"T1=great==>(gr==>(g), (r)), (eat==>(e), (at==>(a), (t)))\nT2=rgtae==>(rg==>(r), (g)), (tae==>(t), (ae==>(a), (e)))\n DBG:-- isequiv('great', 'rgtae') DBG:-- Recurse\n DBG:-- isequiv('gr', 'rg') DBG:-- Recurse\n DBG:-- isequiv('g', 'r') DBG: Chars Same --> False\n DBG:-- isequiv('g', 'g') DBG: Chars Same --> True\n DBG:-- isequiv('r', 'r') DBG: Chars Same --> True\n DBG: Answer --> True\n DBG:-- isequiv('eat', 'tae') DBG:-- Recurse\n DBG:-- isequiv('e', 't') DBG: Chars Same --> False\n DBG:-- isequiv('e', 'ae') DBG:-- Recurse\n DBG: DIFF --> False\n DBG: DIFF --> False\n DBG: Answer --> False\n DBG: Answer --> False\n DBG:-- isequiv('gr', 'tae') DBG:-- Recurse\n DBG:-- isequiv('g', 't') DBG: Chars Same --> False\n DBG:-- isequiv('g', 'ae') DBG:-- Recurse\n DBG: DIFF --> False\n DBG: DIFF --> False\n DBG: Answer --> False\n DBG: Answer --> False\n DBG: Answer --> False\n"
],
[
"#simpletest = lambda A : [spiral_square(A)]\ndef simpletest(w1, w2):\n x = Solution()\n return x.isScramble(w1, w2)\n\nc1g = TestCase('one same char', simpletest, [\"a\", \"a\"], True)\nc1b = TestCase('one diff char', simpletest, [\"a\", \"b\"], False)\n\nc2g = TestCase('two same char', simpletest, [\"aa\", \"aa\"], True)\nc2f = TestCase('two flip char', simpletest, [\"ab\", \"ba\"], True)\nc2b = TestCase('two diff char', simpletest, [\"ab\", \"bc\"], False)\n\n\nc3g = TestCase('three same char', simpletest, [\"abc\", \"abc\"], True)\nc3f = TestCase('three flip char', simpletest, [\"abb\", \"bba\"], True)\nc3f2 = TestCase('three flip char', simpletest, [\"abb\", \"bab\"], True)\n\ntester = TestSet([c1g, c1b, c2g, c2f, c2b, c3g, c3f, c3f2])\n",
"_____no_output_____"
],
[
"tester.run_tests()",
"*** ERROR:Test three flip char failed. Returned \"False\", expected \"True\"\n*** ERROR:Tests passed: 7; Failures: 1\nElapsed test time: 0:00:00.000684\n"
],
[
"index=17\nmath.log(index,2)\nmath.floor(math.log(index,2))\n4.0",
"_____no_output_____"
],
[
"BoolArray3D(object):\n val = []\n \n def __init__(self, rows, cols, z):\n inner = [0]*z\n \n \n ",
"_____no_output_____"
],
[
"def isScramble(s1, s2):\n \"\"\"\n Let F(i, j, k) = whether the substring S1[i..i + k - 1] is a scramble of S2[j..j + k - 1] or not\n Since each of these substrings is a potential node in the tree, we need to check for all possible cuts.\n Let q be the length of a cut (hence, q < k), then we are in the following situation:\n \n S1 [ x1 | x2 ]\n i i + q i + k - 1\n \n here we have two possibilities:\n\n S2 [ y1 | y2 ]\n j j + q j + k - 1\n \n or \n \n S2 [ y1 | y2 ]\n j j + k - q j + k - 1\n \n which in terms of F means:\n \n F(i, j, k) = for some 1 <= q < k we have:\n (F(i, j, q) AND F(i + q, j + q, k - q)) OR (F(i, j + k - q, q) AND F(i + q, j, k - q))\n \n Base case is k = 1, where we simply need to check for S1[i] and S2[j] to be equal \n \"\"\"\n slen = len(s1) \n if slen != len(s2): return False\n #boolean [][][] F = new boolean[len][len][len + 1];\n F = [[[False]*(slen+1)]*slen]*slen\n k = 1\n while k<=slen: #for (int k = 1; k <= len; ++k)\n i = 0\n while i+k <= slen: #for (int i = 0; i + k <= len; ++i)\n j = 0\n while j+k <= slen: #for (int j = 0; j + k <= len; ++j)\n if k == 1:\n F[i][j][k] = s1[i] == s2[j]\n else:\n q = 1\n while q<k and not F[i][j][k]: #for (int q = 1; q < k && !F[i][j][k]; ++q) {\n t1 = F[i][j ][q] and F[i + q][j + q][k - q]\n t2 = F[i][j + k - q][q] and F[i + q][j ][k - q]\n F[i][j][k] = t1 or t2\n q += 1\n j += 1\n i += 1\n k += 1\n print(F)\n return F[0][0][slen]\n",
"_____no_output_____"
],
[
"isScramble(\"ab\", \"ba\")",
"[[[False, False, False], [False, False, False]], [[False, False, False], [False, False, False]]]\n"
]
],
[
[
"### Some Ad Hoc Tests",
"_____no_output_____"
]
],
[
[
"x = [[None]]*10 \nprint(x)\nx[0].append('happy trails')\nx[0] += 'WTF'\nx += 'GLORM'\nx\n#[x for x in range(1, 3)]",
"[[None], [None], [None], [None], [None], [None], [None], [None], [None], [None]]\n"
],
[
"[[[0]*4]*3]*2",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6d2466a45c57d0003e7a3cb102b4974a20a5a9 | 221,294 | ipynb | Jupyter Notebook | notebooks/draft_notebooks/JaccardIndexBinaryMasks_Bangalore.ipynb | DerkBarten/satsense | 7760464e435512edf6ccc892ee6df26c89ac154d | [
"Apache-2.0"
]
| null | null | null | notebooks/draft_notebooks/JaccardIndexBinaryMasks_Bangalore.ipynb | DerkBarten/satsense | 7760464e435512edf6ccc892ee6df26c89ac154d | [
"Apache-2.0"
]
| null | null | null | notebooks/draft_notebooks/JaccardIndexBinaryMasks_Bangalore.ipynb | DerkBarten/satsense | 7760464e435512edf6ccc892ee6df26c89ac154d | [
"Apache-2.0"
]
| 1 | 2020-05-19T02:37:33.000Z | 2020-05-19T02:37:33.000Z | 660.579104 | 24,856 | 0.938354 | [
[
[
"# Jaccard index between binary masks",
"_____no_output_____"
],
[
"This notebook illustrates the calculation of the Jaccard index between the binary masks of the truth and segmentaiton results for three classes (Slum, Built-up and Non-built-up) of 5 ROIs from a 2017 GoogleEarth image of Bangalore, India using `satsense` python library.",
"_____no_output_____"
]
],
[
[
"# Python and satsense imports\nimport os\nimport numpy as np\nfrom satsense.util.mask import load_mask_from_file\n\nfrom matplotlib.pyplot import imshow\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nfrom satsense.performance.jaccard_similarity import jaccard_index_binary_masks as jibm # Jaccard index computation",
"_____no_output_____"
],
[
"# parameters\nTRUTH_MASKS_PATH = '/home/elena/DynaSlum/Data/Bangalore/GEImages/masks/'\nRESULTS_MASKS_PATH = '/home/elena/DynaSlum/Results/Bangalore/Segmentation/'\n\next = 'tif'\n\nROIs = ['ROI1', 'ROI2','ROI3', 'ROI4', 'ROI5']\n",
"_____no_output_____"
],
[
"# Loading, visualizations and calculations\nfor roi in ROIs:\n print(\"---------------------------------------------------------------------------------------\")\n print(\"Loading \", roi)\n # Load the truth and segmentation masks from disk\n truth_slum_mask_fname = 'Bangalore_' + roi + '_slumMask' + '.' + ext\n truth_builtup_mask_fname = 'Bangalore_' + roi + '_builtupMask' + '.' + ext\n truth_nonbuiltup_mask_fname = 'Bangalore_' + roi + '_nonbuiltupMask' + '.' + ext\n\n truth_slum_mask_fullfname = os.path.join(TRUTH_MASKS_PATH, truth_slum_mask_fname)\n truth_builtup_mask_fullfname = os.path.join(TRUTH_MASKS_PATH, truth_builtup_mask_fname)\n truth_nonbuiltup_mask_fullfname = os.path.join(TRUTH_MASKS_PATH, truth_nonbuiltup_mask_fname)\n\n result_slum_mask_fname = 'Bangalore_' + roi + '_slumResult' + '.' + ext\n result_builtup_mask_fname = 'Bangalore_' + roi + '_builtupResult' + '.' + ext\n result_nonbuiltup_mask_fname = 'Bangalore_' + roi + '_nonbuiltupResult' + '.' + ext\n\n result_slum_mask_fullfname = os.path.join(RESULTS_MASKS_PATH, result_slum_mask_fname)\n result_builtup_mask_fullfname = os.path.join(RESULTS_MASKS_PATH, result_builtup_mask_fname)\n result_nonbuiltup_mask_fullfname = os.path.join(RESULTS_MASKS_PATH, result_nonbuiltup_mask_fname)\n\n \n # load the masks into numpy arrays\n truth_slum_mask = load_mask_from_file(truth_slum_mask_fullfname)\n result_slum_mask = load_mask_from_file(result_slum_mask_fullfname)\n\n truth_builtup_mask = load_mask_from_file(truth_builtup_mask_fullfname)\n result_builtup_mask = load_mask_from_file(result_builtup_mask_fullfname)\n\n truth_nonbuiltup_mask = load_mask_from_file(truth_nonbuiltup_mask_fullfname)\n result_nonbuiltup_mask = load_mask_from_file(result_nonbuiltup_mask_fullfname)\n\n #visualization\n print(\"Visualizing \", roi)\n \n f, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n ax1.imshow(truth_slum_mask, cmap='gray'), ax1.set_title('Slum mask: truth' + roi)\n ax2.imshow(result_slum_mask,cmap='gray'), ax2.set_title('Slum mask: result' + roi)\n \n f, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n ax1.imshow(truth_builtup_mask,cmap='gray'), ax1.set_title('Builtup mask: truth' + roi)\n ax2.imshow(result_builtup_mask,cmap='gray'), ax2.set_title('Builtup mask: result' + roi)\n \n f, (ax1, ax2) = plt.subplots(1, 2, sharey=True)\n ax1.imshow(truth_nonbuiltup_mask,cmap='gray'), ax1.set_title('Nonbuiltup mask: truth' + roi)\n ax2.imshow(result_nonbuiltup_mask,cmap='gray'), ax2.set_title('Nonbuiltup mask: result' + roi)\n \n # Jaccard Index computation\n print(\"Jacard Index computations for \", roi)\n \n # JSS between slum ground truth mask and segmentation result\n jss_slum = jibm(truth_slum_mask, result_slum_mask)\n print(\"JSS between slum ground truth mask and segmentation result: \",jss_slum)\n\n # JSS between built-up truth mask and segmentation result\n jss_builtup = jibm(truth_builtup_mask, result_builtup_mask)\n print(\"JSS between builtup manual truth mask and segmentation result: \",jss_builtup)\n\n # JSS between non-built-up truth mask and segmentation result\n jss_nonbuiltup = jibm(truth_nonbuiltup_mask, result_nonbuiltup_mask)\n print(\"JSS between non-built-up manual truth mask and segmentation result: \",jss_nonbuiltup)\n \n \n ",
"---------------------------------------------------------------------------------------\nLoading ROI1\nVisualizing ROI1\nJacard Index computations for ROI1\nJSS between slum ground truth mask and segmentation result: 1.13443976839\nJSS between builtup manual truth mask and segmentation result: 1.84221673512\nJSS between non-built-up manual truth mask and segmentation result: 0.311862968548\n---------------------------------------------------------------------------------------\nLoading ROI2\nVisualizing ROI2\nJacard Index computations for ROI2\nJSS between slum ground truth mask and segmentation result: 0.369944615735\nJSS between builtup manual truth mask and segmentation result: 1.03755905042\nJSS between non-built-up manual truth mask and segmentation result: 0.283259629108\n---------------------------------------------------------------------------------------\nLoading ROI3\nVisualizing ROI3\nJacard Index computations for ROI3\nJSS between slum ground truth mask and segmentation result: 0.0504021377958\nJSS between builtup manual truth mask and segmentation result: 0.94099422707\nJSS between non-built-up manual truth mask and segmentation result: 0.557391683695\n---------------------------------------------------------------------------------------\nLoading ROI4\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec6d36b9eb7e79fd600df723baee0bb4d7889e0f | 13,613 | ipynb | Jupyter Notebook | cookbook/ch_6/learning_Tic_Tac_Toe.ipynb | zlpmichelle/crackingtensorflow | 66c3517b60c3793ef06f904e5d58e4d044628182 | [
"Apache-2.0"
]
| 3 | 2017-10-19T23:41:26.000Z | 2019-10-22T08:59:35.000Z | cookbook/ch_6/learning_Tic_Tac_Toe.ipynb | zlpmichelle/crackingtensorflow | 66c3517b60c3793ef06f904e5d58e4d044628182 | [
"Apache-2.0"
]
| null | null | null | cookbook/ch_6/learning_Tic_Tac_Toe.ipynb | zlpmichelle/crackingtensorflow | 66c3517b60c3793ef06f904e5d58e4d044628182 | [
"Apache-2.0"
]
| null | null | null | 45.989865 | 1,179 | 0.551238 | [
[
[
"\"\"\"\nLearning Optimal Tic-Tac-Toe Moves via a Neural Network\n-------------------------------------------------------\nWe will build a one-hidden layer neural network\n to predict the optimal response given a set\n of tic-tac-toe boards.\n\n\"\"\"\n\nimport tensorflow as tf\nimport matplotlib.pyplot as plt\nimport csv\nimport numpy as np\nimport random\nfrom tensorflow.python.framework import ops\nops.reset_default_graph()\n\n# Definition of X's, O's, and empty spots:\n# X = 1\n# O = -1\n# empty = 0\n# response on 1-9 grid for placement of next '1'\n\n# For example, the 'test_board' is:\n#\n# O | - | -\n# -----------------\n# X | O | O\n# -----------------\n# - | - | X\n#\n# board above = [-1, 0, 0, 1, -1, -1, 0, 0, 1]\n# Optimal response would be position 6, where\n# the position numbers are:\n#\n# 0 | 1 | 2\n# -----------------\n# 3 | 4 | 5\n# -----------------\n# 6 | 7 | 8\n\n# Test board optimal response:\nresponse = 6\n# Set batch size and five different symmetries of board positions\nbatch_size = 50\nsymmetry = ['rotate180', 'rotate90', 'rotate270', 'flip_v', 'flip_h']\n\n\n# Print a board\ndef print_board(board):\n symbols = ['O', ' ', 'X']\n board_plus1 = [int(x) + 1 for x in board]\n board_line1 = ' {} | {} | {}'.format(symbols[board_plus1[0]],\n symbols[board_plus1[1]],\n symbols[board_plus1[2]])\n board_line2 = ' {} | {} | {}'.format(symbols[board_plus1[3]],\n symbols[board_plus1[4]],\n symbols[board_plus1[5]])\n board_line3 = ' {} | {} | {}'.format(symbols[board_plus1[6]],\n symbols[board_plus1[7]],\n symbols[board_plus1[8]])\n print(board_line1)\n print('___________')\n print(board_line2)\n print('___________')\n print(board_line3)\n\n\n# Given a board, a response, and a transformation, get the new board+response\ndef get_symmetry(board, play_response, transformation):\n \"\"\"\n :param board: list of integers 9 long:\n opposing mark = -1\n friendly mark = 1\n empty space = 0\n :param play_response: integer of where response is (0-8)\n :param transformation: one of five transformations on a board:\n 'rotate180', 'rotate90', 'rotate270', 'flip_v', 'flip_h'\n :return: tuple: (new_board, new_response)\n \"\"\"\n if transformation == 'rotate180':\n new_response = 8 - play_response\n return board[::-1], new_response\n elif transformation == 'rotate90':\n new_response = [6, 3, 0, 7, 4, 1, 8, 5, 2].index(play_response)\n tuple_board = list(zip(*[board[6:9], board[3:6], board[0:3]]))\n return [value for item in tuple_board for value in item], new_response\n elif transformation == 'rotate270':\n new_response = [2, 5, 8, 1, 4, 7, 0, 3, 6].index(play_response)\n tuple_board = list(zip(*[board[0:3], board[3:6], board[6:9]]))[::-1]\n return [value for item in tuple_board for value in item], new_response\n elif transformation == 'flip_v':\n new_response = [6, 7, 8, 3, 4, 5, 0, 1, 2].index(play_response)\n return board[6:9] + board[3:6] + board[0:3], new_response\n elif transformation == 'flip_h': # flip_h = rotate180, then flip_v\n new_response = [2, 1, 0, 5, 4, 3, 8, 7, 6].index(play_response)\n new_board = board[::-1]\n return new_board[6:9] + new_board[3:6] + new_board[0:3], new_response\n else:\n raise ValueError('Method not implemented.')\n\n\n# Read in board move csv file\ndef get_moves_from_csv(csv_file):\n \"\"\"\n :param csv_file: csv file location containing the boards w/ responses\n :return: moves: list of moves with index of best response\n \"\"\"\n play_moves = []\n with open(csv_file, 'rt') as csvfile:\n reader = csv.reader(csvfile, delimiter=',')\n for row in reader:\n play_moves.append(([int(x) for x in row[0:9]], int(row[9])))\n return play_moves\n\n\n# Get random board with optimal move\ndef get_rand_move(play_moves, rand_transforms=2):\n \"\"\"\n :param play_moves: list of the boards w/responses\n :param rand_transforms: how many random transforms performed on each\n :return: (board, response), board is a list of 9 integers, response is 1 int\n \"\"\"\n (board, play_response) = random.choice(play_moves)\n possible_transforms = ['rotate90', 'rotate180', 'rotate270', 'flip_v', 'flip_h']\n for _ in range(rand_transforms):\n random_transform = random.choice(possible_transforms)\n (board, play_response) = get_symmetry(board, play_response, random_transform)\n return board, play_response\n\n# Get list of optimal moves w/ responses\nmoves = get_moves_from_csv('base_tic_tac_toe_moves.csv')\n\n# Create a train set:\ntrain_length = 500\ntrain_set = []\nfor t in range(train_length):\n train_set.append(get_rand_move(moves))\n\n# To see if the network learns anything new, we will remove\n# all instances of the board [-1, 0, 0, 1, -1, -1, 0, 0, 1],\n# which the optimal response will be the index '6'. We will\n# Test this at the end.\ntest_board = [-1, 0, 0, 1, -1, -1, 0, 0, 1]\ntrain_set = [x for x in train_set if x[0] != test_board]\n\n\ndef init_weights(shape):\n return tf.Variable(tf.random_normal(shape))\n\n\ndef model(X, A1, A2, bias1, bias2):\n layer1 = tf.nn.sigmoid(tf.add(tf.matmul(X, A1), bias1))\n layer2 = tf.add(tf.matmul(layer1, A2), bias2)\n # Note: we don't take the softmax at the end because our cost function does that for us\n return layer2\n\nX = tf.placeholder(dtype=tf.float32, shape=[None, 9])\nY = tf.placeholder(dtype=tf.int32, shape=[None])\n\nA1 = init_weights([9, 81])\nbias1 = init_weights([81])\nA2 = init_weights([81, 9])\nbias2 = init_weights([9])\n\nmodel_output = model(X, A1, A2, bias1, bias2)\n\nloss = tf.reduce_mean(tf.nn.sparse_softmax_cross_entropy_with_logits(logits=model_output, labels=Y))\ntrain_step = tf.train.GradientDescentOptimizer(0.025).minimize(loss)\nprediction = tf.argmax(model_output, 1)\n\nsess = tf.Session()\ninit = tf.global_variables_initializer()\nsess.run(init)\n\nloss_vec = []\nfor i in range(10000):\n rand_indices = np.random.choice(range(len(train_set)), batch_size, replace=False)\n batch_data = [train_set[i] for i in rand_indices]\n x_input = [x[0] for x in batch_data]\n y_target = np.array([y[1] for y in batch_data])\n sess.run(train_step, feed_dict={X: x_input, Y: y_target})\n \n temp_loss = sess.run(loss, feed_dict={X: x_input, Y: y_target})\n loss_vec.append(temp_loss)\n if i % 500 == 0:\n print('Iteration: {}, Loss: {}'.format(i, temp_loss))\n\n\n# Print loss\nplt.plot(loss_vec, 'k-', label='Loss')\nplt.title('Loss (MSE) per Generation')\nplt.xlabel('Generation')\nplt.ylabel('Loss')\nplt.show()\n\n# Make Prediction:\ntest_boards = [test_board]\nfeed_dict = {X: test_boards}\nlogits = sess.run(model_output, feed_dict=feed_dict)\npredictions = sess.run(prediction, feed_dict=feed_dict)\nprint(predictions)\n\n\n# Declare function to check for win\ndef check(board):\n wins = [[0, 1, 2], [3, 4, 5], [6, 7, 8], [0, 3, 6], [1, 4, 7], [2, 5, 8], [0, 4, 8], [2, 4, 6]]\n for ix in range(len(wins)):\n if board[wins[ix][0]] == board[wins[ix][1]] == board[wins[ix][2]] == 1.:\n return 1\n elif board[wins[ix][0]] == board[wins[ix][1]] == board[wins[ix][2]] == -1.:\n return 1\n return 0\n\n# Let's play against our model\ngame_tracker = [0., 0., 0., 0., 0., 0., 0., 0., 0.]\nwin_logical = False\nnum_moves = 0\nwhile not win_logical:\n player_index = input('Input index of your move (0-8): ')\n num_moves += 1\n # Add player move to game\n game_tracker[int(player_index)] = 1.\n \n # Get model's move by first getting all the logits for each index\n [potential_moves] = sess.run(model_output, feed_dict={X: [game_tracker]})\n # Now find allowed moves (where game tracker values = 0.0)\n allowed_moves = [ix for ix, x in enumerate(game_tracker) if x == 0.0]\n # Find best move by taking argmax of logits if they are in allowed moves\n model_move = np.argmax([x if ix in allowed_moves else -999.0 for ix, x in enumerate(potential_moves)])\n \n # Add model move to game\n game_tracker[int(model_move)] = -1.\n print('Model has moved')\n print_board(game_tracker)\n # Now check for win or too many moves\n if check(game_tracker) == 1 or num_moves >= 5:\n print('Game Over!')\n win_logical = True",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
ec6d38ad19042bd9dc207e08b148f600c79fb8fe | 18,106 | ipynb | Jupyter Notebook | exercises/deep_learning/plot_digits_classification.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
]
| null | null | null | exercises/deep_learning/plot_digits_classification.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
]
| null | null | null | exercises/deep_learning/plot_digits_classification.ipynb | bobrokerson/kaggle | 96c13e85476e2fe0fdb2af74075f82510db90573 | [
"MIT"
]
| null | null | null | 57.66242 | 4,280 | 0.742074 | [
[
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"\n# Recognizing hand-written digits\n\nThis example shows how scikit-learn can be used to recognize images of\nhand-written digits, from 0-9.\n",
"_____no_output_____"
]
],
[
[
"# Author: Gael Varoquaux <gael dot varoquaux at normalesup dot org>\n# License: BSD 3 clause\n\n# Standard scientific Python imports\nimport matplotlib.pyplot as plt\n\n# Import datasets, classifiers and performance metrics\nfrom sklearn import datasets, svm, metrics\nfrom sklearn.model_selection import train_test_split\n\nfrom sklearn.metrics import ConfusionMatrixDisplay",
"_____no_output_____"
]
],
[
[
"## Digits dataset\n\nThe digits dataset consists of 8x8\npixel images of digits. The ``images`` attribute of the dataset stores\n8x8 arrays of grayscale values for each image. We will use these arrays to\nvisualize the first 4 images. The ``target`` attribute of the dataset stores\nthe digit each image represents and this is included in the title of the 4\nplots below.\n\nNote: if we were working from image files (e.g., 'png' files), we would load\nthem using :func:`matplotlib.pyplot.imread`.\n\n",
"_____no_output_____"
]
],
[
[
"digits = datasets.load_digits()\n\n_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))\nfor ax, image, label in zip(axes, digits.images, digits.target):\n ax.set_axis_off()\n ax.imshow(image, cmap=plt.cm.gray_r, interpolation=\"nearest\")\n ax.set_title(\"Training: %i\" % label)",
"_____no_output_____"
]
],
[
[
"## Classification\n\nTo apply a classifier on this data, we need to flatten the images, turning\neach 2-D array of grayscale values from shape ``(8, 8)`` into shape\n``(64,)``. Subsequently, the entire dataset will be of shape\n``(n_samples, n_features)``, where ``n_samples`` is the number of images and\n``n_features`` is the total number of pixels in each image.\n\nWe can then split the data into train and test subsets and fit a support\nvector classifier on the train samples. The fitted classifier can\nsubsequently be used to predict the value of the digit for the samples\nin the test subset.\n\n",
"_____no_output_____"
]
],
[
[
"# flatten the images\nn_samples = len(digits.images)\ndata = digits.images.reshape((n_samples, -1))\n\n# Create a classifier: a support vector classifier\nclf = svm.SVC(gamma=0.001)\n\n# Split data into 50% train and 50% test subsets\nX_train, X_test, y_train, y_test = train_test_split(\n data, digits.target, test_size=0.5, shuffle=False\n)\n\n# Learn the digits on the train subset\nclf.fit(X_train, y_train)\n\n# Predict the value of the digit on the test subset\npredicted = clf.predict(X_test)",
"_____no_output_____"
]
],
[
[
"Below we visualize the first 4 test samples and show their predicted\ndigit value in the title.\n\n",
"_____no_output_____"
]
],
[
[
"_, axes = plt.subplots(nrows=1, ncols=4, figsize=(10, 3))\nfor ax, image, prediction in zip(axes, X_test, predicted):\n ax.set_axis_off()\n image = image.reshape(8, 8)\n ax.imshow(image, cmap=plt.cm.gray_r, interpolation=\"nearest\")\n ax.set_title(f\"Prediction: {prediction}\")",
"_____no_output_____"
]
],
[
[
":func:`~sklearn.metrics.classification_report` builds a text report showing\nthe main classification metrics.\n\n",
"_____no_output_____"
]
],
[
[
"print(\n f\"Classification report for classifier {clf}:\\n\"\n f\"{metrics.classification_report(y_test, predicted)}\\n\"\n)",
"Classification report for classifier SVC(gamma=0.001):\n precision recall f1-score support\n\n 0 1.00 0.99 0.99 88\n 1 0.99 0.97 0.98 91\n 2 0.99 0.99 0.99 86\n 3 0.98 0.87 0.92 91\n 4 0.99 0.96 0.97 92\n 5 0.95 0.97 0.96 91\n 6 0.99 0.99 0.99 91\n 7 0.96 0.99 0.97 89\n 8 0.94 1.00 0.97 88\n 9 0.93 0.98 0.95 92\n\n accuracy 0.97 899\n macro avg 0.97 0.97 0.97 899\nweighted avg 0.97 0.97 0.97 899\n\n\n"
]
],
[
[
"We can also plot a `confusion matrix <confusion_matrix>` of the\ntrue digit values and the predicted digit values.\n\n",
"_____no_output_____"
]
],
[
[
"disp = metrics.ConfusionMatrixDisplay.from_predictions(y_test, predicted)\ndisp.figure_.suptitle(\"Confusion Matrix\")\nprint(f\"Confusion matrix:\\n{disp.confusion_matrix}\")\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6d45fe09107e8f4b038aad9f74a5a386752618 | 7,109 | ipynb | Jupyter Notebook | General/Regular expressions.ipynb | afcarl/Useful-python | 5d1947052fb25b2388704926e4692511cc162031 | [
"MIT"
]
| null | null | null | General/Regular expressions.ipynb | afcarl/Useful-python | 5d1947052fb25b2388704926e4692511cc162031 | [
"MIT"
]
| null | null | null | General/Regular expressions.ipynb | afcarl/Useful-python | 5d1947052fb25b2388704926e4692511cc162031 | [
"MIT"
]
| 1 | 2018-09-05T21:48:57.000Z | 2018-09-05T21:48:57.000Z | 23.696667 | 238 | 0.510058 | [
[
[
"https://www.tutorialspoint.com/python/python_reg_expressions.htm\n\nPython offers two different primitive operations based on regular expressions: match checks for a match only at the beginning of the string, while search checks for a match anywhere in the string (this is what Perl does by default).",
"_____no_output_____"
]
],
[
[
"import re",
"_____no_output_____"
]
],
[
[
"# Match function \n\nThis function attempts to match RE pattern to string with optional flags.",
"_____no_output_____"
]
],
[
[
"line = \"Cats are smarter than dogs\";\n\nmatchObj = re.match( r'(.*) are (.*?) .*', line, re.M|re.I)\n\nif matchObj:\n print(\"matchObj.group() : \", matchObj.group()) # This method returns entire match\n print(\"matchObj.group(1) : \", matchObj.group(1)) # This method returns specific subgroup num\n print(\"matchObj.group(2) : \", matchObj.group(2))\n print(\"matchObj.groups() : \", matchObj.groups()) # This method returns all matching subgroups in a tuple (empty if there weren't any)\nelse:\n print(\"Nothing found!!\")",
"matchObj.group() : Cats are smarter than dogs\nmatchObj.group(1) : Cats\nmatchObj.group(2) : smarter\nmatchObj.groups() : ('Cats', 'smarter')\n"
]
],
[
[
"# Search function \n\nThis function searches for first occurrence of RE pattern within string with optional flags.",
"_____no_output_____"
]
],
[
[
"line = \"Cats are smarter than dogs\";\n\nsearchObj = re.search( r'(.*) are (.*?) .*', line, re.M|re.I)\n\nif searchObj:\n print(\"searchObj.group() : \", searchObj.group())\n print(\"searchObj.group(1) : \", searchObj.group(1))\n print(\"searchObj.group(2) : \", searchObj.group(2))\n print(\"searchObj.groups() : \", searchObj.groups()) \nelse:\n print(\"Nothing found!!\")",
"searchObj.group() : Cats are smarter than dogs\nsearchObj.group(1) : Cats\nsearchObj.group(2) : smarter\nsearchObj.groups() : ('Cats', 'smarter')\n"
]
],
[
[
"# Comparison of search and match",
"_____no_output_____"
]
],
[
[
"line = \"Cats are smarter than dogs\";\n\nmatchObj = re.match( r'dogs', line, re.M|re.I)\nif matchObj:\n print(\"match --> matchObj.group() : \", matchObj.group())\nelse:\n print(\"No match!!\")\n\nsearchObj = re.search( r'dogs', line, re.M|re.I)\nif searchObj:\n print(\"search --> searchObj.group() : \", searchObj.group())\nelse:\n print(\"Nothing found!!\")",
"No match!!\nsearch --> searchObj.group() : dogs\n"
]
],
[
[
"# Search and replace\n\n re.sub(pattern, repl, string, max=0)\n\nThis method replaces all occurrences of the RE pattern in string with repl, substituting all occurrences unless max provided. This method returns modified string.",
"_____no_output_____"
]
],
[
[
"phone = \"2004-959-559 # This is Phone Number\"\n\n# Delete Python-style comments\nnum = re.sub(r'#.*$', \"\", phone)\nprint(\"Phone Num : \", num)\n\n# Remove anything other than digits\nnum = re.sub(r'\\D', \"\", phone) \nprint(\"Phone Num : \", num)",
"Phone Num : 2004-959-559 \nPhone Num : 2004959559\n"
]
],
[
[
"# RadOnc translations\n\nLine in file \n\n Prescribed dose [Gy]: 55.000\n\nR code \n \n dose.rx <- suppressWarnings(as.numeric(sub(\"^Prescribed dose.*: \", \"\", dose.rx, ignore.case=TRUE, perl=TRUE)))",
"_____no_output_____"
]
],
[
[
"dose_string = 'Prescribed dose [Gy]: 55.000'\ndose = float(re.sub(r\"^Prescribed dose.*: \", \"\", dose_string)) \ndose",
"_____no_output_____"
],
[
"structure_string = 'Structure: Bronchial tree_P'\nname = re.sub(\"^.*: (.+$)\", \"\\\\1\", structure_string)\nname",
"_____no_output_____"
],
[
"matchObj = re.match( r'Structure: (.*?) .*', structure_string, re.M|re.I)\nif matchObj:\n print(\"matchObj.group(1) : \", matchObj.group(1)) ",
"matchObj.group(1) : Bronchial\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec6d4ae4538e13fe5a70936f7cf5d62a246defc7 | 36,903 | ipynb | Jupyter Notebook | creditapprovals.ipynb | rhearai/Predicting-Credit-Card-Approvals | 8d6169e49c0d51c712e9ca3986c1a6de56d42c9f | [
"MIT"
]
| null | null | null | creditapprovals.ipynb | rhearai/Predicting-Credit-Card-Approvals | 8d6169e49c0d51c712e9ca3986c1a6de56d42c9f | [
"MIT"
]
| null | null | null | creditapprovals.ipynb | rhearai/Predicting-Credit-Card-Approvals | 8d6169e49c0d51c712e9ca3986c1a6de56d42c9f | [
"MIT"
]
| null | null | null | 36,903 | 36,903 | 0.609002 | [
[
[
"## 1. Credit card applications\n<p>Commercial banks receive <em>a lot</em> of applications for credit cards. Many of them get rejected for many reasons, like high loan balances, low income levels, or too many inquiries on an individual's credit report, for example. Manually analyzing these applications is mundane, error-prone, and time-consuming (and time is money!). Luckily, this task can be automated with the power of machine learning and pretty much every commercial bank does so nowadays. In this notebook, we will build an automatic credit card approval predictor using machine learning techniques, just like the real banks do.</p>\n<p><img src=\"https://assets.datacamp.com/production/project_558/img/credit_card.jpg\" alt=\"Credit card being held in hand\"></p>\n<p>We'll use the <a href=\"http://archive.ics.uci.edu/ml/datasets/credit+approval\">Credit Card Approval dataset</a> from the UCI Machine Learning Repository. The structure of this notebook is as follows:</p>\n<ul>\n<li>First, we will start off by loading and viewing the dataset.</li>\n<li>We will see that the dataset has a mixture of both numerical and non-numerical features, that it contains values from different ranges, plus that it contains a number of missing entries.</li>\n<li>We will have to preprocess the dataset to ensure the machine learning model we choose can make good predictions.</li>\n<li>After our data is in good shape, we will do some exploratory data analysis to build our intuitions.</li>\n<li>Finally, we will build a machine learning model that can predict if an individual's application for a credit card will be accepted.</li>\n</ul>\n<p>First, loading and viewing the dataset. We find that since this data is confidential, the contributor of the dataset has anonymized the feature names.</p>",
"_____no_output_____"
]
],
[
[
"import pandas as pd\n# ... YOUR CODE FOR TASK 1 ...\n\n# Load dataset\ncc_apps = pd.read_csv(\"datasets/cc_approvals.data\", header = None)\n\n# Inspect data\n# ... YOUR CODE FOR TASK 1 ...\ncc_apps.head()",
"_____no_output_____"
]
],
[
[
"## 2. Inspecting the applications\n<p>The output may appear a bit confusing at its first sight, but let's try to figure out the most important features of a credit card application. The features of this dataset have been anonymized to protect the privacy, but <a href=\"http://rstudio-pubs-static.s3.amazonaws.com/73039_9946de135c0a49daa7a0a9eda4a67a72.html\">this blog</a> gives us a pretty good overview of the probable features. The probable features in a typical credit card application are <code>Gender</code>, <code>Age</code>, <code>Debt</code>, <code>Married</code>, <code>BankCustomer</code>, <code>EducationLevel</code>, <code>Ethnicity</code>, <code>YearsEmployed</code>, <code>PriorDefault</code>, <code>Employed</code>, <code>CreditScore</code>, <code>DriversLicense</code>, <code>Citizen</code>, <code>ZipCode</code>, <code>Income</code> and finally the <code>ApprovalStatus</code>. This gives us a pretty good starting point, and we can map these features with respect to the columns in the output. </p>\n<p>As we can see from our first glance at the data, the dataset has a mixture of numerical and non-numerical features. This can be fixed with some preprocessing, but before we do that, let's learn about the dataset a bit more to see if there are other dataset issues that need to be fixed.</p>",
"_____no_output_____"
]
],
[
[
"# Print summary statistics\ncc_apps_description = cc_apps.describe()\nprint(cc_apps_description)\n\nprint(\"\\n\")\n\n# Print DataFrame information\ncc_apps_info = cc_apps.info()\nprint(cc_apps_info)\n\nprint(\"\\n\")\n\n# Inspect missing values in the dataset\n# ... YOUR CODE FOR TASK 2 ...\ncc_apps.tail(17)",
" 2 7 10 14\ncount 690.000000 690.000000 690.00000 690.000000\nmean 4.758725 2.223406 2.40000 1017.385507\nstd 4.978163 3.346513 4.86294 5210.102598\nmin 0.000000 0.000000 0.00000 0.000000\n25% 1.000000 0.165000 0.00000 0.000000\n50% 2.750000 1.000000 0.00000 5.000000\n75% 7.207500 2.625000 3.00000 395.500000\nmax 28.000000 28.500000 67.00000 100000.000000\n\n\n<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 690 entries, 0 to 689\nData columns (total 16 columns):\n0 690 non-null object\n1 690 non-null object\n2 690 non-null float64\n3 690 non-null object\n4 690 non-null object\n5 690 non-null object\n6 690 non-null object\n7 690 non-null float64\n8 690 non-null object\n9 690 non-null object\n10 690 non-null int64\n11 690 non-null object\n12 690 non-null object\n13 690 non-null object\n14 690 non-null int64\n15 690 non-null object\ndtypes: float64(2), int64(2), object(12)\nmemory usage: 86.3+ KB\nNone\n\n\n"
]
],
[
[
"## 3. Handling the missing values (part i)\n<p>We've uncovered some issues that will affect the performance of our machine learning model(s) if they go unchanged:</p>\n<ul>\n<li>Our dataset contains both numeric and non-numeric data (specifically data that are of <code>float64</code>, <code>int64</code> and <code>object</code> types). Specifically, the features 2, 7, 10 and 14 contain numeric values (of types float64, float64, int64 and int64 respectively) and all the other features contain non-numeric values.</li>\n<li>The dataset also contains values from several ranges. Some features have a value range of 0 - 28, some have a range of 2 - 67, and some have a range of 1017 - 100000. Apart from these, we can get useful statistical information (like <code>mean</code>, <code>max</code>, and <code>min</code>) about the features that have numerical values. </li>\n<li>Finally, the dataset has missing values, which we'll take care of in this task. The missing values in the dataset are labeled with '?', which can be seen in the last cell's output.</li>\n</ul>\n<p>Now, let's temporarily replace these missing value question marks with NaN.</p>",
"_____no_output_____"
]
],
[
[
"import numpy as np\n# ... YOUR CODE FOR TASK 3 ...\n\n# Inspect missing values in the dataset\nprint(cc_apps.tail(17))\n\n# Replace the '?'s with NaN\ncc_apps = cc_apps.replace(\"?\",np.NaN)\n\n# Inspect the missing values again\nprint(cc_apps.tail(17))",
" 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15\n673 ? 29.50 2.000 y p e h 2.000 f f 0 f g 00256 17 -\n674 a 37.33 2.500 u g i h 0.210 f f 0 f g 00260 246 -\n675 a 41.58 1.040 u g aa v 0.665 f f 0 f g 00240 237 -\n676 a 30.58 10.665 u g q h 0.085 f t 12 t g 00129 3 -\n677 b 19.42 7.250 u g m v 0.040 f t 1 f g 00100 1 -\n678 a 17.92 10.210 u g ff ff 0.000 f f 0 f g 00000 50 -\n679 a 20.08 1.250 u g c v 0.000 f f 0 f g 00000 0 -\n680 b 19.50 0.290 u g k v 0.290 f f 0 f g 00280 364 -\n681 b 27.83 1.000 y p d h 3.000 f f 0 f g 00176 537 -\n682 b 17.08 3.290 u g i v 0.335 f f 0 t g 00140 2 -\n683 b 36.42 0.750 y p d v 0.585 f f 0 f g 00240 3 -\n684 b 40.58 3.290 u g m v 3.500 f f 0 t s 00400 0 -\n685 b 21.08 10.085 y p e h 1.250 f f 0 f g 00260 0 -\n686 a 22.67 0.750 u g c v 2.000 f t 2 t g 00200 394 -\n687 a 25.25 13.500 y p ff ff 2.000 f t 1 t g 00200 1 -\n688 b 17.92 0.205 u g aa v 0.040 f f 0 f g 00280 750 -\n689 b 35.00 3.375 u g c h 8.290 f f 0 t g 00000 0 -\n 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15\n673 NaN 29.50 2.000 y p e h 2.000 f f 0 f g 00256 17 -\n674 a 37.33 2.500 u g i h 0.210 f f 0 f g 00260 246 -\n675 a 41.58 1.040 u g aa v 0.665 f f 0 f g 00240 237 -\n676 a 30.58 10.665 u g q h 0.085 f t 12 t g 00129 3 -\n677 b 19.42 7.250 u g m v 0.040 f t 1 f g 00100 1 -\n678 a 17.92 10.210 u g ff ff 0.000 f f 0 f g 00000 50 -\n679 a 20.08 1.250 u g c v 0.000 f f 0 f g 00000 0 -\n680 b 19.50 0.290 u g k v 0.290 f f 0 f g 00280 364 -\n681 b 27.83 1.000 y p d h 3.000 f f 0 f g 00176 537 -\n682 b 17.08 3.290 u g i v 0.335 f f 0 t g 00140 2 -\n683 b 36.42 0.750 y p d v 0.585 f f 0 f g 00240 3 -\n684 b 40.58 3.290 u g m v 3.500 f f 0 t s 00400 0 -\n685 b 21.08 10.085 y p e h 1.250 f f 0 f g 00260 0 -\n686 a 22.67 0.750 u g c v 2.000 f t 2 t g 00200 394 -\n687 a 25.25 13.500 y p ff ff 2.000 f t 1 t g 00200 1 -\n688 b 17.92 0.205 u g aa v 0.040 f f 0 f g 00280 750 -\n689 b 35.00 3.375 u g c h 8.290 f f 0 t g 00000 0 -\n"
]
],
[
[
"## 4. Handling the missing values (part ii)\n<p>We replaced all the question marks with NaNs. This is going to help us in the next missing value treatment that we are going to perform.</p>\n<p>An important question that gets raised here is <em>why are we giving so much importance to missing values</em>? Can't they be just ignored? Ignoring missing values can affect the performance of a machine learning model heavily. While ignoring the missing values our machine learning model may miss out on information about the dataset that may be useful for its training. Then, there are many models which cannot handle missing values implicitly such as LDA. </p>\n<p>So, to avoid this problem, we are going to impute the missing values with a strategy called mean imputation.</p>",
"_____no_output_____"
]
],
[
[
"# Impute the missing values with mean imputation\ncc_apps.fillna(cc_apps.mean(), inplace=True)\n\n# Count the number of NaNs in the dataset to verify\n# ... YOUR CODE FOR TASK 4 ...\nprint(cc_apps.isnull().values.sum())",
"67\n"
]
],
[
[
"## 5. Handling the missing values (part iii)\n<p>We have successfully taken care of the missing values present in the numeric columns. There are still some missing values to be imputed for columns 0, 1, 3, 4, 5, 6 and 13. All of these columns contain non-numeric data and this why the mean imputation strategy would not work here. This needs a different treatment. </p>\n<p>We are going to impute these missing values with the most frequent values as present in the respective columns. This is <a href=\"https://www.datacamp.com/community/tutorials/categorical-data\">good practice</a> when it comes to imputing missing values for categorical data in general.</p>",
"_____no_output_____"
]
],
[
[
"# Iterate over each column of cc_apps\nfor col in cc_apps.columns:\n # Check if the column is of object type\n if cc_apps[col].dtypes == 'object':\n # Impute with the most frequent value\n cc_apps[col] = cc_apps[col].fillna(cc_apps[col].value_counts().index[0])\n\n# Count the number of NaNs in the dataset and print the counts to verify\nprint(cc_apps.isnull().values.sum())",
"0\n"
]
],
[
[
"## 6. Preprocessing the data (part i)\n<p>The missing values are now successfully handled.</p>\n<p>There is still some minor but essential data preprocessing needed before we proceed towards building our machine learning model. We are going to divide these remaining preprocessing steps into three main tasks:</p>\n<ol>\n<li>Convert the non-numeric data into numeric.</li>\n<li>Split the data into train and test sets. </li>\n<li>Scale the feature values to a uniform range.</li>\n</ol>\n<p>First, we will be converting all the non-numeric values into numeric ones. We do this because not only it results in a faster computation but also many machine learning models (like XGBoost) (and especially the ones developed using scikit-learn) require the data to be in a strictly numeric format. We will do this by using a technique called <a href=\"http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.LabelEncoder.html\">label encoding</a>.</p>",
"_____no_output_____"
]
],
[
[
"# Import LabelEncoder\nfrom sklearn.preprocessing import LabelEncoder\n\n# Instantiate LabelEncoder\n# ... YOUR CODE FOR TASK 6 ...\nle = LabelEncoder()\n# Iterate over all the values of each column and extract their dtypes\nfor col in cc_apps.columns:\n # Compare if the dtype is object\n if cc_apps[col].dtype=='object':\n # Use LabelEncoder to do the numeric transformation\n cc_apps[col]=le.fit_transform(cc_apps[col])",
"_____no_output_____"
]
],
[
[
"## 7. Splitting the dataset into train and test sets\n<p>We have successfully converted all the non-numeric values to numeric ones.</p>\n<p>Now, we will split our data into train set and test set to prepare our data for two different phases of machine learning modeling: training and testing. Ideally, no information from the test data should be used to scale the training data or should be used to direct the training process of a machine learning model. Hence, we first split the data and then apply the scaling.</p>\n<p>Also, features like <code>DriversLicense</code> and <code>ZipCode</code> are not as important as the other features in the dataset for predicting credit card approvals. We should drop them to design our machine learning model with the best set of features. In Data Science literature, this is often referred to as <em>feature selection</em>. </p>",
"_____no_output_____"
]
],
[
[
"# Import train_test_split\n# ... YOUR CODE FOR TASK 7 ...\nfrom sklearn.model_selection import train_test_split\n\n# Drop the features 11 and 13 and convert the DataFrame to a NumPy array\ncc_apps = cc_apps.drop([cc_apps.columns[11], cc_apps.columns[13]], axis=1)\ncc_apps = cc_apps.values\n\n# Segregate features and labels into separate variables\nX,y = cc_apps[:,0:13] , cc_apps[:,13]\n\n# Split into train and test sets\nX_train, X_test, y_train, y_test = train_test_split(X,\n y,\n test_size=0.33,\n random_state=42)",
"_____no_output_____"
]
],
[
[
"## 8. Preprocessing the data (part ii)\n<p>The data is now split into two separate sets - train and test sets respectively. We are only left with one final preprocessing step of scaling before we can fit a machine learning model to the data. </p>\n<p>Now, let's try to understand what these scaled values mean in the real world. Let's use <code>CreditScore</code> as an example. The credit score of a person is their creditworthiness based on their credit history. The higher this number, the more financially trustworthy a person is considered to be. So, a <code>CreditScore</code> of 1 is the highest since we're rescaling all the values to the range of 0-1.</p>",
"_____no_output_____"
]
],
[
[
"# Import MinMaxScaler\n# ... YOUR CODE FOR TASK 8 ...\nfrom sklearn.preprocessing import MinMaxScaler\n# Instantiate MinMaxScaler and use it to rescale X_train and X_test\nscaler = MinMaxScaler(feature_range=(0,1))\nrescaledX_train = scaler.fit_transform(X_train)\nrescaledX_test = scaler.fit_transform(X_test)",
"_____no_output_____"
]
],
[
[
"## 9. Fitting a logistic regression model to the train set\n<p>Essentially, predicting if a credit card application will be approved or not is a <a href=\"https://en.wikipedia.org/wiki/Statistical_classification\">classification</a> task. <a href=\"http://archive.ics.uci.edu/ml/machine-learning-databases/credit-screening/crx.names\">According to UCI</a>, our dataset contains more instances that correspond to \"Denied\" status than instances corresponding to \"Approved\" status. Specifically, out of 690 instances, there are 383 (55.5%) applications that got denied and 307 (44.5%) applications that got approved. </p>\n<p>This gives us a benchmark. A good machine learning model should be able to accurately predict the status of the applications with respect to these statistics.</p>\n<p>Which model should we pick? A question to ask is: <em>are the features that affect the credit card approval decision process correlated with each other?</em> Although we can measure correlation, that is outside the scope of this notebook, so we'll rely on our intuition that they indeed are correlated for now. Because of this correlation, we'll take advantage of the fact that generalized linear models perform well in these cases. Let's start our machine learning modeling with a Logistic Regression model (a generalized linear model).</p>",
"_____no_output_____"
]
],
[
[
"# Import LogisticRegression\n# ... YOUR CODE FOR TASK 9 ...\nfrom sklearn.linear_model import LogisticRegression\n# Instantiate a LogisticRegression classifier with default parameter values\nlogreg = LogisticRegression()\n\n# Fit logreg to the train set\n# ... YOUR CODE FOR TASK 9 ...\nlogreg.fit(rescaledX_train, y_train)",
"_____no_output_____"
]
],
[
[
"## 10. Making predictions and evaluating performance\n<p>But how well does our model perform? </p>\n<p>We will now evaluate our model on the test set with respect to <a href=\"https://developers.google.com/machine-learning/crash-course/classification/accuracy\">classification accuracy</a>. But we will also take a look the model's <a href=\"http://www.dataschool.io/simple-guide-to-confusion-matrix-terminology/\">confusion matrix</a>. In the case of predicting credit card applications, it is equally important to see if our machine learning model is able to predict the approval status of the applications as denied that originally got denied. If our model is not performing well in this aspect, then it might end up approving the application that should have been approved. The confusion matrix helps us to view our model's performance from these aspects. </p>",
"_____no_output_____"
]
],
[
[
"# Import confusion_matrix\n# ... YOUR CODE FOR TASK 10 ...\nfrom sklearn.metrics import confusion_matrix\n# Use logreg to predict instances from the test set and store it\ny_pred = logreg.predict(rescaledX_test)\n\n# Get the accuracy score of logreg model and print it\nprint(\"Accuracy of logistic regression classifier: \", ...)\n\n# Print the confusion matrix of the logreg model\n# ... YOUR CODE FOR TASK 10 ...\nprint(\"Accuracy score of logreg\", logreg.score(rescaledX_test, y_test))\nconfusion_matrix(y_test,y_pred)",
"Accuracy of logistic regression classifier: Ellipsis\nAccuracy score of logreg 0.8377192982456141\n"
]
],
[
[
"## 11. Grid searching and making the model perform better\n<p>Our model was pretty good! It was able to yield an accuracy score of almost 84%.</p>\n<p>For the confusion matrix, the first element of the of the first row of the confusion matrix denotes the true negatives meaning the number of negative instances (denied applications) predicted by the model correctly. And the last element of the second row of the confusion matrix denotes the true positives meaning the number of positive instances (approved applications) predicted by the model correctly.</p>\n<p>Let's see if we can do better. We can perform a <a href=\"https://machinelearningmastery.com/how-to-tune-algorithm-parameters-with-scikit-learn/\">grid search</a> of the model parameters to improve the model's ability to predict credit card approvals.</p>\n<p><a href=\"http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html\">scikit-learn's implementation of logistic regression</a> consists of different hyperparameters but we will grid search over the following two:</p>\n<ul>\n<li>tol</li>\n<li>max_iter</li>\n</ul>",
"_____no_output_____"
]
],
[
[
"# Import GridSearchCV\n# ... YOUR CODE FOR TASK 11 ...\nfrom sklearn.model_selection import GridSearchCV\n# Define the grid of values for tol and max_iter\ntol = [0.01, 0.001, 0.0001]\nmax_iter = [100, 150, 200]\n\n# Create a dictionary where tol and max_iter are keys and the lists of their values are corresponding values\nparam_grid = dict(tol=tol, max_iter=max_iter)",
"_____no_output_____"
]
],
[
[
"## 12. Finding the best performing model\n<p>We have defined the grid of hyperparameter values and converted them into a single dictionary format which <code>GridSearchCV()</code> expects as one of its parameters. Now, we will begin the grid search to see which values perform best.</p>\n<p>We will instantiate <code>GridSearchCV()</code> with our earlier <code>logreg</code> model with all the data we have. Instead of passing train and test sets separately, we will supply <code>X</code> (scaled version) and <code>y</code>. We will also instruct <code>GridSearchCV()</code> to perform a <a href=\"https://www.dataschool.io/machine-learning-with-scikit-learn/\">cross-validation</a> of five folds.</p>\n<p>We'll end the notebook by storing the best-achieved score and the respective best parameters.</p>\n<p>While building this credit card predictor, we tackled some of the most widely-known preprocessing steps such as <strong>scaling</strong>, <strong>label encoding</strong>, and <strong>missing value imputation</strong>. We finished with some <strong>machine learning</strong> to predict if a person's application for a credit card would get approved or not given some information about that person.</p>",
"_____no_output_____"
]
],
[
[
"# Instantiate GridSearchCV with the required parameters\ngrid_model = GridSearchCV(estimator=logreg, param_grid=param_grid, cv=5)\n\n# Use scaler to rescale X and assign it to rescaledX\nrescaledX = scaler.fit_transform(X)\n\n# Fit data to grid_model\ngrid_model_result = grid_model.fit(rescaledX, y)\n\n# Summarize results\nbest_score, best_params = ...\nprint(\"Best: %f using %s\" % (..., ...))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6d571a2074409bb492b7a8308671e2e3d084a8 | 20,255 | ipynb | Jupyter Notebook | 10_0_develop_new_OD_demand_estimator_Sioux_uni_class/archive/uni-classt_traffic_assignment_MSA_backup.ipynb | jingzbu/InverseVITraffic | c0d33d91bdd3c014147d58866c1a2b99fb8a9608 | [
"MIT"
]
| null | null | null | 10_0_develop_new_OD_demand_estimator_Sioux_uni_class/archive/uni-classt_traffic_assignment_MSA_backup.ipynb | jingzbu/InverseVITraffic | c0d33d91bdd3c014147d58866c1a2b99fb8a9608 | [
"MIT"
]
| null | null | null | 10_0_develop_new_OD_demand_estimator_Sioux_uni_class/archive/uni-classt_traffic_assignment_MSA_backup.ipynb | jingzbu/InverseVITraffic | c0d33d91bdd3c014147d58866c1a2b99fb8a9608 | [
"MIT"
]
| null | null | null | 30.550528 | 1,292 | 0.449173 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec6d5da988c4e91ce8de74a037b03e98780c4b3b | 127,362 | ipynb | Jupyter Notebook | Topic Modelling/topic_modelling_v1.ipynb | e-olang/NLP | 12f965f622f2f5f950c8dbe491887cde4fcec415 | [
"MIT"
]
| null | null | null | Topic Modelling/topic_modelling_v1.ipynb | e-olang/NLP | 12f965f622f2f5f950c8dbe491887cde4fcec415 | [
"MIT"
]
| null | null | null | Topic Modelling/topic_modelling_v1.ipynb | e-olang/NLP | 12f965f622f2f5f950c8dbe491887cde4fcec415 | [
"MIT"
]
| null | null | null | 70.171901 | 51,398 | 0.668284 | [
[
[
"<a href=\"https://colab.research.google.com/github/Joe-Ol/NLP/blob/main/Twitter/topic_modelling_v1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"# *Intros*",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nimport sklearn\n\nimport re",
"_____no_output_____"
]
],
[
[
"You can find the used data as well as more info on it here: [How-ISIS-Uses-Twitter](https://www.kaggle.com/fifthtribe/how-isis-uses-twitter)",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('/content/drive/MyDrive/Colab Notebooks/NLP/How ISIS Uses Twitter/tweets.csv')",
"_____no_output_____"
],
[
"for col in df.columns:\n print(col)",
"name\nusername\ndescription\nlocation\nfollowers\nnumberstatuses\ntime\ntweets\n"
],
[
"df.drop(['description', 'followers', 'numberstatuses', 'time', 'location'], axis=1, inplace=True)",
"_____no_output_____"
]
],
[
[
"# **Exploraotry Analysis**",
"_____no_output_____"
]
],
[
[
"len(df.username.unique())",
"_____no_output_____"
],
[
"len(df.tweets.unique())",
"_____no_output_____"
],
[
"df",
"_____no_output_____"
],
[
"def find_retweeted(tweet):\n '''This function will extract the twitter handles of retweed people'''\n return re.findall('(?<=RT\\s)(@[A-Za-z]+[A-Za-z0-9-_]+)', tweet)\n\ndef find_mentioned(tweet):\n '''This function will extract the twitter handles of people mentioned in the tweet'''\n return re.findall('(?<!RT\\s)(@[A-Za-z]+[A-Za-z0-9-_]+)', tweet) \n\ndef find_hashtags(tweet):\n '''This function will extract hashtags'''\n return re.findall('(#[A-Za-z]+[A-Za-z0-9-_]+)', tweet) \n\ndef find_links(tweet):\n return re.findall('(?:(?:https?|ftp|file):\\/\\/|www\\.|ftp\\.)(?:\\([-A-Z0-9+&@#\\/%=~_|$?!:,.]*\\)|[-A-Z0-9+&@#\\/%=~_|$?!:,.])*(?:\\([-A-Z0-9+&@#\\/%=~_|$?!:,.]*\\)|[A-Z0-9+&@#\\/%=~_|$])', tweet) \n",
"_____no_output_____"
],
[
"# make new columns for retweeted usernames, mentioned usernames and hashtags\n#df['retweeted'] = df.tweets.apply(find_retweeted)\n#df['mentioned'] = df.tweets.apply(find_mentioned)\ndf['hashtags'] = df.tweets.apply(find_hashtags)\ndf['link'] = df.tweets.apply(find_links)",
"_____no_output_____"
]
],
[
[
"\n\n---\n\n",
"_____no_output_____"
],
[
"Findingthe Most Popular Hashtags in Our Dataset",
"_____no_output_____"
]
],
[
[
"# take the rows from the hashtag columns where there are actually hashtags\nhashtags_list_df = df.loc[\n df.hashtags.apply(\n lambda hashtags_list: hashtags_list !=[]\n ),['hashtags']]",
"_____no_output_____"
],
[
"# create dataframe where each use of hashtag gets its own row\nflattened_hashtags_df = pd.DataFrame(\n [hashtag for hashtags_list in hashtags_list_df.hashtags\n for hashtag in hashtags_list],\n columns=['hashtag'])",
"_____no_output_____"
],
[
"len(flattened_hashtags_df.hashtag.unique())",
"_____no_output_____"
],
[
"# count of appearances of each hashtag\npopular_hashtags = flattened_hashtags_df.groupby('hashtag').size()\\\n .reset_index(name='counts')\\\n .sort_values('counts', ascending=False)\\\n .reset_index(drop=True)",
"_____no_output_____"
],
[
"# number of times each hashtag appears\ncounts = flattened_hashtags_df.groupby(['hashtag']).size()\\\n .reset_index(name='counts')\\\n .counts\n\n# define bins for histogram \nmy_bins = np.arange(0,counts.max()+2, 5)-0.5\n\n# plot histogram of tweet counts\nplt.figure()\nplt.hist(counts, bins = my_bins)\nplt.xlabels = np.arange(1,counts.max()+1, 1)\nplt.xlabel('hashtag number of appearances')\nplt.ylabel('frequency')\nplt.yscale('log', nonposy='clip')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Finding the most common user",
"_____no_output_____"
]
],
[
[
"user_list_df = df.loc[\n df.username.apply(\n lambda username_list: username_list !=[]\n ),['username']]",
"_____no_output_____"
],
[
"popular_users = user_list_df.groupby('username').size()\\\n .reset_index(name='counts')\\\n .sort_values('counts', ascending=False)\\\n .reset_index(drop=True)",
"_____no_output_____"
],
[
"# number of times each user appears\ncounts = user_list_df.groupby(['username']).size()\\\n .reset_index(name='counts')\\\n .counts\n\n# define bins for histogram \nmy_bins_2 = np.arange(0,counts.max()+2, 5)-0.5\n\n# plot histogram of tweet counts\nplt.figure()\nplt.hist(counts, bins = my_bins_2)\nplt.xlabels = np.arange(1,counts.max()+1, 1)\nplt.xlabel('Username number of appearances')\nplt.ylabel('frequency')\nplt.yscale('log', nonposy='clip')\nplt.show()",
"_____no_output_____"
]
],
[
[
"Hashtags to Vectors",
"_____no_output_____"
]
],
[
[
"# take hashtags which appear at least this amount of times\nmin_appearance = 110\n# find popular hashtags - make into python set for efficiency\npopular_hashtags_set = set(popular_hashtags[\n popular_hashtags.counts>=min_appearance\n ]['hashtag'])",
"_____no_output_____"
],
[
"len(popular_hashtags_set)",
"_____no_output_____"
],
[
"# make a new column with only the popular hashtags\nhashtags_list_df['popular_hashtags'] = hashtags_list_df.hashtags.apply(\n lambda hashtag_list: [hashtag for hashtag in hashtag_list\n if hashtag in popular_hashtags_set])\n# drop rows without popular hashtag\npopular_hashtags_list_df = hashtags_list_df.loc[\n hashtags_list_df.popular_hashtags.apply(lambda hashtag_list: hashtag_list !=[])]\n",
"_____no_output_____"
],
[
"# make new dataframe\nhashtag_vector_df = popular_hashtags_list_df.loc[:, ['popular_hashtags']]\n\nfor hashtag in popular_hashtags_set:\n # make columns to encode presence of hashtags\n hashtag_vector_df['{}'.format(hashtag)] = hashtag_vector_df.popular_hashtags.apply(\n lambda hashtag_list: int(hashtag in hashtag_list))",
"_____no_output_____"
],
[
" hashtag_vector_df",
"_____no_output_____"
],
[
"hashtag_matrix = hashtag_vector_df.drop('popular_hashtags', axis=1)",
"_____no_output_____"
],
[
"# calculate the correlation matrix\ncorrelations = hashtag_matrix.corr()",
"_____no_output_____"
],
[
"# plot the correlation matrix\nplt.figure(figsize=(10,10))\nsns.heatmap(correlations,\n cmap='RdBu',\n vmin=-1,\n vmax=1,\n square = True,\n cbar_kws={'label':'correlation'})\nplt.show()",
"_____no_output_____"
]
],
[
[
"# **Topic Modelling**",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk.tokenize import RegexpTokenizer\nfrom nltk.corpus import stopwords\nnltk.download('stopwords')",
"[nltk_data] Downloading package stopwords to /root/nltk_data...\n[nltk_data] Unzipping corpora/stopwords.zip.\n"
],
[
"def remove_links(tweet):\n '''Takes a string and removes web links from it'''\n tweet = re.sub(r'http\\S+', '', tweet) # remove http links\n tweet = re.sub(r'bit.ly/\\S+', '', tweet) # rempve bitly links\n tweet = tweet.strip('[link]') # remove [links]\n return tweet\n\ndef remove_users(tweet):\n '''Takes a string and removes retweet and @user information'''\n tweet = re.sub('(RT\\s@[A-Za-z]+[A-Za-z0-9-_]+)', '', tweet) # remove retweet\n tweet = re.sub('(@[A-Za-z]+[A-Za-z0-9-_]+)', '', tweet) # remove tweeted at\n return tweet",
"_____no_output_____"
],
[
"my_stopwords = nltk.corpus.stopwords.words('english')\nword_rooter = nltk.stem.snowball.PorterStemmer(ignore_stopwords=False).stem\nmy_punctuation = '!\"$%&\\'()*+,-./:;<=>?[\\\\]^_`{|}~•@'\n\n# cleaning master function\ndef clean_tweet(tweet, bigrams=False):\n tweet = remove_users(tweet)\n tweet = remove_links(tweet)\n tweet = tweet.lower() # lower case\n tweet = re.sub('['+my_punctuation + ']+', ' ', tweet) # strip punctuation\n tweet = re.sub('\\s+', ' ', tweet) #remove double spacing\n tweet = re.sub('([0-9]+)', '', tweet) # remove numbers\n tweet_token_list = [word for word in tweet.split(' ')\n if word not in my_stopwords] # remove stopwords\n\n tweet_token_list = [word_rooter(word) if '#' not in word else word\n for word in tweet_token_list] # apply word rooter\n if bigrams:\n tweet_token_list = tweet_token_list+[tweet_token_list[i]+'_'+tweet_token_list[i+1]\n for i in range(len(tweet_token_list)-1)]\n tweet = ' '.join(tweet_token_list)\n return tweet",
"_____no_output_____"
],
[
"df['clean_tweet'] = df.tweets.apply(clean_tweet)",
"_____no_output_____"
],
[
"max_df = 0.9\nmin_df = 25",
"_____no_output_____"
],
[
"from sklearn.feature_extraction.text import CountVectorizer\n\n# the vectorizer object will be used to transform text to vector form\nvectorizer = CountVectorizer(max_df=0.9, min_df=25, token_pattern='\\w+|\\$[\\d\\.]+|\\S+')\n\n# apply transformation\ntf = vectorizer.fit_transform(df['clean_tweet']).toarray()\n\n# tf_feature_names tells us what word each column in the matric represents\ntf_feature_names = vectorizer.get_feature_names()",
"_____no_output_____"
],
[
"tf.shape",
"_____no_output_____"
],
[
"hashtag_vector_df.shape",
"_____no_output_____"
],
[
"# tf_feature_names",
"_____no_output_____"
]
],
[
[
"\n\n---\n\n",
"_____no_output_____"
],
[
"Using LDA - *Latent Dirichlet Allocation* Algorithm",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import LatentDirichletAllocation\n\nnumber_of_topics = 10\n\nmodel = LatentDirichletAllocation(n_components=number_of_topics, random_state=0)",
"_____no_output_____"
],
[
"model.fit(tf)",
"_____no_output_____"
],
[
"def display_topics(model, feature_names, no_top_words):\n topic_dict = {}\n for topic_idx, topic in enumerate(model.components_):\n topic_dict[\"Topic %d words\" % (topic_idx)]= ['{}'.format(feature_names[i])\n for i in topic.argsort()[:-no_top_words - 1:-1]]\n topic_dict[\"Topic %d weights\" % (topic_idx)]= ['{:.1f}'.format(topic[i])\n for i in topic.argsort()[:-no_top_words - 1:-1]]\n return pd.DataFrame(topic_dict)",
"_____no_output_____"
],
[
"no_top_words = 10\ndisplay_topics(model, tf_feature_names, no_top_words)",
"_____no_output_____"
]
],
[
[
"Note: If a word apeear in most topics with a higher weight, it may be a good idea to add it to our stopword list since it may not be adding too much meaning to our task.",
"_____no_output_____"
],
[
"\n\n---\n\n",
"_____no_output_____"
],
[
"Using LDA - *Non-Negative Matrix Factorisation* Algorithm",
"_____no_output_____"
]
],
[
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6d6a85cce1a8f86cad2741b9ccf30ba32c8f32 | 3,705 | ipynb | Jupyter Notebook | Hmwk 5.ipynb | rustymuffin99/Hmwk-5 | 12befd977b004db13af5a467d599ab701e2350ca | [
"MIT"
]
| null | null | null | Hmwk 5.ipynb | rustymuffin99/Hmwk-5 | 12befd977b004db13af5a467d599ab701e2350ca | [
"MIT"
]
| 1 | 2020-11-09T06:40:28.000Z | 2020-11-09T23:57:06.000Z | Hmwk 5.ipynb | rustymuffin99/Hmwk-5 | 12befd977b004db13af5a467d599ab701e2350ca | [
"MIT"
]
| null | null | null | 21.794118 | 75 | 0.407018 | [
[
[
"%matplotlib inline\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def func(x):\n a = np.exp(-2*x)\n b = np.cos(10*x)\n return a*b",
"_____no_output_____"
],
[
"def trapezoid_core(f,x,h):\n return 0.5*h*(f(x+h)+f(x))",
"_____no_output_____"
],
[
"def trapezoid_method(f,a,b,N):\n \n x = np.linspace(a,b,N)\n h = x[1]-x[0]\n \n Fint = 0.0\n \n for i in range(0,len(x)-1,1):\n Fint += trapezoid_core(f,x[i],h)\n \n return Fint",
"_____no_output_____"
],
[
"def simpson_core(f,x,h):\n return h*(f(x) + 4*f(x+h) + f(x+2*h))/3",
"_____no_output_____"
],
[
"def simpsons_method(f,a,b,N):\n \n x = np.linspace(a,b,N)\n h = x[1]-x[0]\n \n Fint = 0\n \n for i in range(0,len(x)-2,2):\n Fint += simpson_core(f,x[-2],0.5*h)\n \n return Fint",
"_____no_output_____"
],
[
"def romberg_core(f,a,b,i):\n \n h = b-a\n \n dh = h/2.**(i)\n \n K = h/2.**(i+1)\n \n M = 0.0\n for j in range(2**i):\n M += f(a + 0.5*dh + j*dh)\n \n return K*M",
"_____no_output_____"
],
[
"def romberg_integration(f,a,b,tol):\n \n i = 0\n \n imax = 1000\n \n delta = 100.0*np.fabs(tol)\n \n I = np.zeros(imax,dtype=float)\n \n I[0] = 0.5*(b-a)*(f(a)+f(b))\n \n i += 1\n \n while(delta>tol):\n \n I[i] = 0.5*I[i-1] + romberg_core(f,a,b,i)\n \n delta = np.fabs((I[i]-I[i-1])/I[i])\n \n print(i,I[i],I[i-1],delta)\n \n if(delta>tol):\n i += 1\n \n if(i>imax):\n print(\"max iterations reached\")\n raise StopIteration('stopping iterations after',i)\n \n return I[i]",
"_____no_output_____"
],
[
"print(\"Trapezoid\")\nprint(trapezoid_method(func,0,np.pi,50))\nprint(\"simpsons method\")\nprint(simpsons_method(func,0,np.pi,50))\nprint(\"romberg\")\ntolerance = 1.0e-6\nRI = romberg_integration(func,0,np.pi,tolerance)\nprint(RI, (RI-answer)/answer,tolerance)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6d6bc12b3b366f6c8313db0ba08ef78fea237a | 2,832 | ipynb | Jupyter Notebook | playbooks/vti_playbook.ipynb | CptOfEvilMinions/Vault-Jupyter-Notebooks | 348711513cfd5f5b22fc96026246bfeb70f51c21 | [
"MIT"
]
| null | null | null | playbooks/vti_playbook.ipynb | CptOfEvilMinions/Vault-Jupyter-Notebooks | 348711513cfd5f5b22fc96026246bfeb70f51c21 | [
"MIT"
]
| null | null | null | playbooks/vti_playbook.ipynb | CptOfEvilMinions/Vault-Jupyter-Notebooks | 348711513cfd5f5b22fc96026246bfeb70f51c21 | [
"MIT"
]
| null | null | null | 34.120482 | 144 | 0.540254 | [
[
[
"from prettytable import PrettyTable\nfrom helpers import vault\nfrom helpers import vti\nimport getpass \nimport os\n\n# Obtain the necessary values\nVAULT_ADDR = os.getenv('VAULT_ADDR') or getpass.getpass(\"Enter Vault URL: \")\nVAULT_SECRET_PATH = \"/secrets/data/incident-response/jupyter-notebooks\"\nVAULT_TOKEN = open(os.path.expanduser('~/.vault-token')).read() or os.getenv('VAULT_TOKEN') or getpass.getpass(\"Enter Vault token: \")\n\n# Request secrets for incident responders\nvault_secrets = vault.GetVaultSecrets(VAULT_ADDR, VAULT_SECRET_PATH, VAULT_TOKEN)\n\n# Request user to enter a SHA256 file hash\nsha256_file_hash = input(\"Enter SHA256 file hash: \")\n\n# Request results from Virustotal\nvti_results = vti.GetVTIresults(vault_secrets['vti-api-key'], sha256_file_hash)\n\n# Print the VTI malicious score to user\nx = PrettyTable()\nx.field_names = ['Sha256 file hash', 'Malicious score']\nmalicious = vti_results['data']['attributes']['last_analysis_stats']['malicious']\nundetected = vti_results['data']['attributes']['last_analysis_stats']['undetected']\nx.add_row([sha256_file_hash, f\"{malicious}/{malicious + undetected}\"])\nprint (x)\n\n",
"https://vault.hackinglab.local/v1/secrets/data/incident-response/jupyter-notebooks\nEnter SHA256 file hash: 423a0799efe41b28a8b765fa505699183c8278d5a7bf07658b3bd507bfa5346f\n+------------------------------------------------------------------+-----------------+\n| Sha256 file hash | Malicious score |\n+------------------------------------------------------------------+-----------------+\n| 423a0799efe41b28a8b765fa505699183c8278d5a7bf07658b3bd507bfa5346f | 53/70 |\n+------------------------------------------------------------------+-----------------+\n"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec6d6f2b658e8f57e829ae6a56042e9dbe95617e | 9,123 | ipynb | Jupyter Notebook | others/hbase_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
]
| 13 | 2018-03-19T18:16:03.000Z | 2022-03-22T03:44:13.000Z | others/hbase_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
]
| null | null | null | others/hbase_command_note.ipynb | johnnychiuchiu/Machine-Learning | 0fd6fd2c08025134cf7d20b245c39f82d5453e14 | [
"MIT"
]
| 7 | 2018-01-11T04:03:11.000Z | 2021-01-22T07:56:42.000Z | 21.315421 | 493 | 0.543242 | [
[
[
"# Hbase Command Note",
"_____no_output_____"
],
[
"**Outline**\n\n* [Reference](#refer)",
"_____no_output_____"
],
[
"> **run shell script using hbase**",
"_____no_output_____"
],
[
"[cloudera doc](http://archive.cloudera.com/cdh5/cdh/5/hbase-0.98.6-cdh5.2.0/book/ch04s04.html)",
"_____no_output_____"
],
[
"Pass the path to the command file as the only argument to the hbase shell command. Each command is executed and its output is shown. If you do not include the exit command in your script, you are returned to the HBase shell prompt. There is no way to programmatically check each individual command for success or failure. Also, though you see the output for each command, the commands themselves are not echoed to the screen so it can be difficult to line up the command with its output.",
"_____no_output_____"
]
],
[
[
"# both should work\n/usr/bin/hbase shell ./sample_commands.txt\n/usr/bin/hbase shell ./sample_commands.sh",
"_____no_output_____"
],
[
"# save output as txt file\n/usr/bin/hbase shell ./sample_commands.sh > output.txt",
"_____no_output_____"
]
],
[
[
"> **getting into hbase shell**",
"_____no_output_____"
]
],
[
[
" /usr/bin/hbase shell",
"_____no_output_____"
]
],
[
[
"> **listing a Table using HBase Shell**",
"_____no_output_____"
]
],
[
[
"list",
"_____no_output_____"
]
],
[
[
"> **create a table**",
"_____no_output_____"
]
],
[
[
"# table name: railway\n# column family: locomotive, cars, cargo\ncreate 'railway_johnny', {NAME => 'locomotive'}, {NAME => 'cars'}, {NAME => 'cargo'}",
"_____no_output_____"
]
],
[
[
"> **drop/delete a table**",
"_____no_output_____"
]
],
[
[
"disable 'railway_johnny'\ndrop 'railway_johnny'",
"_____no_output_____"
]
],
[
[
"> **inserting data using HBase Shell**",
"_____no_output_____"
],
[
"[tutorialspoint](https://www.tutorialspoint.com/hbase/hbase_create_data.htm)",
"_____no_output_____"
]
],
[
[
"# format\nput '<table name>','row1','<colfamily:colname>','<value>'\n\n# example\nput 'enron_johnny', '1', 'name:employee', 'mCarson'",
"_____no_output_____"
]
],
[
[
"> **Reading Data using HBase Shell**",
"_____no_output_____"
],
[
"[tutorialspoint](https://www.tutorialspoint.com/hbase/hbase_read_data.htm)",
"_____no_output_____"
]
],
[
[
"# format\nget '<table name>','row1'",
"_____no_output_____"
]
],
[
[
"> **view the hbase table using scan**",
"_____no_output_____"
],
[
"[tutorialspoint](https://www.tutorialspoint.com/hbase/hbase_scan.htm)",
"_____no_output_____"
]
],
[
[
"scan '<table name>'",
"_____no_output_____"
]
],
[
[
"> **delete all cells in a row**",
"_____no_output_____"
],
[
"[tutorialspoint](https://www.tutorialspoint.com/hbase/hbase_delete_data.htm)",
"_____no_output_____"
]
],
[
[
"deleteall '<table name>', '<row>'",
"_____no_output_____"
]
],
[
[
"> **delete all the data in a hbase table / clear a table in hbase**",
"_____no_output_____"
],
[
"[stackoverflow](https://stackoverflow.com/questions/9902353/how-to-clear-a-table-in-hbase)",
"_____no_output_____"
]
],
[
[
"# equivalent to: disable -> drop -> create\ntruncate 'yourTableName'",
"_____no_output_____"
]
],
[
[
"> **select: reading a specific column**",
"_____no_output_____"
],
[
"[tutorialspoint](https://www.tutorialspoint.com/hbase/hbase_read_data.htm)",
"_____no_output_____"
]
],
[
[
"# format\nget 'table name', 'rowid', {COLUMN => 'column family:column name'}\n\n# example\nget 'emp', 'row1', {COLUMN => 'personal:name'}",
"_____no_output_____"
]
],
[
[
"> **filter/where in hbase**",
"_____no_output_____"
],
[
"* [hbase-shell-commands](https://learnhbase.wordpress.com/2013/03/02/hbase-shell-commands/)\n* [cloudera filter doc](https://www.cloudera.com/documentation/enterprise/5-5-x/topics/admin_hbase_filtering.html)\n* [hbase apache doc](http://hbase.apache.org/0.94/book/thrift.html)\n* [USEFUL: Different Types of Filters in HBase Shell](https://acadgild.com/blog/different-types-of-filters-in-hbase-shell)",
"_____no_output_____"
],
[
"The following example are copied from [here](https://acadgild.com/blog/different-types-of-filters-in-hbase-shell)",
"_____no_output_____"
],
[
"<img src=\"pic/bulktable.jpg\" style=\"width: 600px;height: 200px;\"/>",
"_____no_output_____"
]
],
[
[
"# Show all the filters in hbase.\nshow_filters",
"_____no_output_____"
]
],
[
[
"**ValueFilter**: \n* output only the select cells from the hbase table\n* to select a column and output only a particular value of the selected column",
"_____no_output_____"
],
[
"<img src=\"pic/valueFilter.png\" style=\"width: 600px;height: 150px;\"/>",
"_____no_output_____"
],
[
"**SingleColumnValueFilter**: filter out rows matching the value of some column value",
"_____no_output_____"
],
[
"<img src=\"pic/singleColumnValueFilter.png\" style=\"width: 600px;height: 180px;\"/>",
"_____no_output_____"
],
[
"**ColumnPrefixFilter**: to select a particular column name \n\n**PrefixFilter**: to select a particular row name",
"_____no_output_____"
],
[
"<img src=\"pic/columnPrefixFilter.jpg\" style=\"width: 400px;height: 180px;\"/>",
"_____no_output_____"
],
[
"# <a id='refer'>Reference</a>",
"_____no_output_____"
],
[
"**HBase Stargate (REST API) client wrapper for Python.**\n* [Starbase](https://pypi.org/project/starbase/)\n* [Happybase](https://happybase.readthedocs.io/en/latest/)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec6d7b70f76d7c9e3b03f81671b198b2f0870918 | 10,841 | ipynb | Jupyter Notebook | intro_exercises1_solutions_2020.ipynb | andersx/python-intro | 8409c89da7dd9cea21e3702a0f0f47aae816eb58 | [
"CC0-1.0"
]
| 11 | 2020-05-03T11:59:01.000Z | 2021-11-15T12:33:39.000Z | intro_exercises1_solutions_2020.ipynb | andersx/python-intro | 8409c89da7dd9cea21e3702a0f0f47aae816eb58 | [
"CC0-1.0"
]
| null | null | null | intro_exercises1_solutions_2020.ipynb | andersx/python-intro | 8409c89da7dd9cea21e3702a0f0f47aae816eb58 | [
"CC0-1.0"
]
| 7 | 2020-05-10T21:15:15.000Z | 2021-12-05T15:13:54.000Z | 25.036952 | 127 | 0.42865 | [
[
[
"# Solutions to Exercises to Lecture 1: Introduction to Python 3.6 \nBy Dr. Anders S. Christensen\n`anders.christensen @ unibas.ch`\n\n\n",
"_____no_output_____"
],
[
"## Exercise 1.1: Basic math\n\nIn the lecture we saw how Python code can be used to evaluate mathmatical expressions, for example $x = 42 \\cdot 5$:",
"_____no_output_____"
]
],
[
[
"x = 42 * 5\nprint(x)",
"_____no_output_____"
]
],
[
[
"### Question 1.1.1:\nUsing Python code (similarly to the example above), what is the result of $x = 3.5^6$?",
"_____no_output_____"
]
],
[
[
"x = 3.5**6 # <-- change this line yourself\nprint(x)",
"1838.265625\n"
]
],
[
[
"## Exercise 1.2: For loop\n\nIn this exercise we use `for`-loops to carry out summations.\n\nFor example the following sum\n\\begin{equation}\nx = \\sum_{n=0}^{25} n\n\\end{equation}\n\ncan be be calculated using the code below",
"_____no_output_____"
]
],
[
[
"x = 0\n\n# Remember how range(i) goes from 0 to i-1!\nfor n in range(26):\n x = x + n\n \nprint(\"x is\", x)",
"_____no_output_____"
]
],
[
[
"\n### Question 1.2.1:\nIn this exercise, write your own code below and to calculate the answer to the following summation:\n\n\\begin{equation}\nx = \\sum_{n=0}^{100} n^2\n\\end{equation}\n",
"_____no_output_____"
]
],
[
[
"x = 0\n\nfor n in range(101):\n\n x = x + n**2\n\nprint(\"x is\", x)\n",
"x is 338350\n"
]
],
[
[
"## Exercise 1.3: Debugging\nSomeone wrote some bad code for you to print the square root of all numbers from $0$ to $9$.\n### Question 1.3.1:\nSeveral things are wrong in the code block below. Pay attention to the error message and fix the errors.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Print the square root of all numbers up to 9\nfor i in range(10):\n print(np.sqrt(i))",
"0.0\n1.0\n1.4142135623730951\n1.7320508075688772\n2.0\n2.23606797749979\n2.449489742783178\n2.6457513110645907\n2.8284271247461903\n3.0\n"
]
],
[
[
"# Exercise 1.4: NumPy functions\n\nIn the lecture, we used the `np.exp()` function from NumPy to calculate the exponential function.\n\nFor example, we can print the result of $\\exp\\left(2.0\\right)$ using this function:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\nx = np.exp(2.0)\n\nprint(x)",
"_____no_output_____"
]
],
[
[
"\n\nNumPy has a most other mathmatical function you can think of. \nYou can find the documentation for many of these math routines here:\n\nhttps://docs.scipy.org/doc/numpy/reference/routines.math.html\n\nBefore you answer the next question, take a look at the manual page in the link to see what is there.",
"_____no_output_____"
],
[
"### Question 1.4.1:\nMost problems in programming are solved by Googling or looking in the manual.\n\nFind a routine from NumPy, and use it to calculate and print the value of $x = \\sin(2.0)$.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\n# Calculate sin of 2.0\nx = np.sin(2.0) #<-- change this line yourself\n\nprint(x)",
"0.9092974268256817\n"
]
],
[
[
"# Exercise 1.5: Double loop? (harder exercise)",
"_____no_output_____"
],
[
"In the lecture we had—not only single `for`-loops—but also double `for`-loops.\n\nFor example a double sum of products\n\\begin{equation}\nn = \\sum_{x=0}^{5} \\sum_{y=0}^{5} \\left(x \\cdot y\\right)\n\\end{equation}\nCan be calculated using nested `for`-loops as demonstrated below\n\n",
"_____no_output_____"
]
],
[
[
"n = 0\n\nfor x in range(6): # <--- NOTE: SHOULD SAY range(6), not range(5)\n for y in range(6): # <--- NOTE: SHOULD SAY range(6), not range(5)\n n = n + x * y\n\nprint(\"n is\", n)",
"n is 100\n"
]
],
[
[
"### Question 1.5.1:\nIn the code-box below, evaluate the following double sum and print the answer\n\n\\begin{equation}\nz = \\sum_{x=0}^{3} \\sum_{y=0}^{10} y^{x}\n\\end{equation}",
"_____no_output_____"
]
],
[
[
"# Do-it-yourself!\n\nz = 0\n\nfor x in range(3):\n for y in range(10):\n\n z = z + y**x\n\nprint(z)\n\n# Note, technically 0^0 is not defined, so not a real number.\n# Python implementes this as 1\nprint(0**0)",
"340\n1\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6d7b725205adf654a5c6bb3520a2cbdd2a0dc8 | 8,901 | ipynb | Jupyter Notebook | numpy/zeros_and_ones.ipynb | Mr-Umidjon/Hackerrank | d11a1f6ae9051ff8c5c01b44ae1a1ac4adcddffc | [
"MIT"
]
| null | null | null | numpy/zeros_and_ones.ipynb | Mr-Umidjon/Hackerrank | d11a1f6ae9051ff8c5c01b44ae1a1ac4adcddffc | [
"MIT"
]
| null | null | null | numpy/zeros_and_ones.ipynb | Mr-Umidjon/Hackerrank | d11a1f6ae9051ff8c5c01b44ae1a1ac4adcddffc | [
"MIT"
]
| null | null | null | 23.672872 | 258 | 0.270082 | [
[
[
"### You are given the shape of the array in the form of space-separated integers, each integer representing the size of different dimensions, your task is to print an array of the given shape and integer type using the tools numpy.zeros and numpy.ones.",
"_____no_output_____"
]
],
[
[
"import numpy as np",
"_____no_output_____"
],
[
"n = tuple(map(int, input().split()))\nprint(np.zeros(n, int))\nprint(np.ones(n, int))",
" 2 3 4 5 5\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6d7cee140a9216245b382b07d72821f68f0da6 | 2,850 | ipynb | Jupyter Notebook | experiment_4_learning_rate_low.ipynb | b09/Deep-Learning-101 | 050f173b05b75553ba27cf3555ad529d7fde6680 | [
"MIT"
]
| null | null | null | experiment_4_learning_rate_low.ipynb | b09/Deep-Learning-101 | 050f173b05b75553ba27cf3555ad529d7fde6680 | [
"MIT"
]
| null | null | null | experiment_4_learning_rate_low.ipynb | b09/Deep-Learning-101 | 050f173b05b75553ba27cf3555ad529d7fde6680 | [
"MIT"
]
| 1 | 2021-06-30T22:50:37.000Z | 2021-06-30T22:50:37.000Z | 34.337349 | 87 | 0.616842 | [
[
[
"from __future__ import division, print_function, absolute_import\n\nimport tflearn\nfrom tflearn.data_utils import shuffle, to_categorical\nfrom tflearn.layers.core import input_data, dropout, fully_connected\nfrom tflearn.layers.conv import conv_2d, max_pool_2d\nfrom tflearn.layers.estimator import regression\nfrom tflearn.data_preprocessing import ImagePreprocessing\nfrom tflearn.data_augmentation import ImageAugmentation\nfrom load_data import load_data\n\n# Data loading and preprocessing\n(X, Y), (X_test, Y_test) = load_data()\nX, Y = shuffle(X, Y)\nY = to_categorical(Y, 10)\nY_test = to_categorical(Y_test, 10)\n\n# Real-time data preprocessing\nimg_prep = ImagePreprocessing()\nimg_prep.add_featurewise_zero_center()\nimg_prep.add_featurewise_stdnorm()\n\n# Real-time data augmentation\nimg_aug = ImageAugmentation()\nimg_aug.add_random_flip_leftright()\nimg_aug.add_random_rotation(max_angle=25.)\n\n# Convolutional network building\nnetwork = input_data(shape=[None, 32, 32, 3],\n data_preprocessing=img_prep,\n data_augmentation=img_aug)\nnetwork = conv_2d(network, 32, 3, activation='relu')\nnetwork = max_pool_2d(network, 2)\nnetwork = conv_2d(network, 64, 3, activation='relu')\nnetwork = conv_2d(network, 64, 3, activation='relu')\nnetwork = max_pool_2d(network, 2)\nnetwork = fully_connected(network, 512, activation='relu')\nnetwork = dropout(network, 0.5)\nnetwork = fully_connected(network, 10, activation='softmax')\nnetwork = regression(network, optimizer='adam',\n loss='categorical_crossentropy',\n learning_rate=0.0001)\n\n# Train using classifier\nmodel = tflearn.DNN(network, tensorboard_verbose=0, tensorboard_dir='/output')\nmodel.fit(X, Y, n_epoch=50, shuffle=True, validation_set=(X_test, Y_test),\n show_metric=True, batch_size=96, run_id='cifar_learning_rate_low')",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec6d8a7195eaef9cffc3071cf655f94cdfa6be4a | 2,330 | ipynb | Jupyter Notebook | Python Notebooks/LOOPS_ WHILE, BREAK _ CONTINUE.ipynb | Isaquehg/Scripts | d6d94e2d32171262b8286bae82ccd83b3baf30a5 | [
"MIT"
]
| null | null | null | Python Notebooks/LOOPS_ WHILE, BREAK _ CONTINUE.ipynb | Isaquehg/Scripts | d6d94e2d32171262b8286bae82ccd83b3baf30a5 | [
"MIT"
]
| null | null | null | Python Notebooks/LOOPS_ WHILE, BREAK _ CONTINUE.ipynb | Isaquehg/Scripts | d6d94e2d32171262b8286bae82ccd83b3baf30a5 | [
"MIT"
]
| null | null | null | 2,330 | 2,330 | 0.648069 | [
[
[
"#LOOP\r\nn = 5\r\nwhile n > 0:\r\n print (n)\r\n n = n - 1\r\nprint(\"Blastoff!\")\r\nprint(n)",
"5\n4\n3\n2\n1\nBlastoff!\n0\n"
],
[
"#LOOPS W/ BREAK.(Go out of the Loop)\r\nwhile True:\r\n line = input(\"What`s next?\")\r\n if line == (\"done\"):\r\n break\r\n print(line)\r\nprint(\"Finished\")",
"What`s next?hello\nhello\nWhat`s next?blabla\nblabla\nWhat`s next?done\nFinish\n"
],
[
"#LOOPS W/ CONTINUE.(Go to the Top of the Loop)\r\nwhile True:\r\n line = input(\"What`s next?\")\r\n if line == \"#\":\r\n continue\r\n elif line == \"done\":\r\n break\r\n else:\r\n continue\r\n print(line)\r\nprint(\"Finished.\")",
"What`s next?#\nWhat`s next?hello\nWhat`s next?done\nFinished.\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec6d8a769eaed40b29e6899f8a9b1d1f41175f1e | 25,779 | ipynb | Jupyter Notebook | LAB-01/144_01_01.ipynb | rajvaghasiya6/144_RajVaghasiya | fc7b7a369242dfecae343b74dafb96eaef1bfaeb | [
"MIT"
]
| null | null | null | LAB-01/144_01_01.ipynb | rajvaghasiya6/144_RajVaghasiya | fc7b7a369242dfecae343b74dafb96eaef1bfaeb | [
"MIT"
]
| null | null | null | LAB-01/144_01_01.ipynb | rajvaghasiya6/144_RajVaghasiya | fc7b7a369242dfecae343b74dafb96eaef1bfaeb | [
"MIT"
]
| null | null | null | 25,779 | 25,779 | 0.936615 | [
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"1)Draw Scatter Plot between age and salary for \"Data_for_Transformation.csv\" file",
"_____no_output_____"
]
],
[
[
"data=pd.read_csv('Data_for_Transformation.csv')\nplt.scatter(data['Age'],data['Salary']) \nplt.show()",
"_____no_output_____"
]
],
[
[
"2) Draw Histogram of column/feature \"Salary\"",
"_____no_output_____"
]
],
[
[
"plt.hist(data['Salary'],bins=5) \nplt.show()",
"_____no_output_____"
]
],
[
[
"3) Plot bar chart for column/feature \"Country\"",
"_____no_output_____"
]
],
[
[
"df=pd.DataFrame(data,columns=['Country'])\n\nval = df['Country'].value_counts()\n\nlabels = val.keys()\n\nfig = plt.figure(figsize = (6, 5))\nplt.bar(labels, val,width = 0.4)\nplt.xlabel(\"Country\")\nplt.ylabel(\"No. of Records\")\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6dbb7051d65cf7dbfe09030e866769c34ea596 | 15,315 | ipynb | Jupyter Notebook | Documentation.ipynb | bruxy70/ComAp-API | b167f04accb9859329179ca6ee16e2fc31097db6 | [
"MIT"
]
| 1 | 2019-09-17T17:36:28.000Z | 2019-09-17T17:36:28.000Z | Documentation.ipynb | bruxy70/ComAp-API | b167f04accb9859329179ca6ee16e2fc31097db6 | [
"MIT"
]
| null | null | null | Documentation.ipynb | bruxy70/ComAp-API | b167f04accb9859329179ca6ee16e2fc31097db6 | [
"MIT"
]
| 1 | 2019-09-17T18:14:09.000Z | 2019-09-17T18:14:09.000Z | 31.128049 | 382 | 0.573294 | [
[
[
"# Installing the ComAp package\n1. You need to have `python` 3.6 or newer installed\n2. Download the latest release from the `ComAp-API` github repository\n3. Run `setup.bat`. It will install the comap API package and lead you to generate the API secrets (see further)",
"_____no_output_____"
],
[
"# Authentication - API secrets\nTo link the API to your account, and for security reasons, each API call needs to have two parameters: `ComAp-Key` and `Token`. \nTo get the ComAp-Key, login to https://portal.websupervisor.net/developer (using your WSV Pro login credentials). There are two keys available - Primary and Secondary - pick one of them - it does not matter which one.\n\n*(The idea is, that you use one of them in production, and the second for testig. After the testing, you can Regenerate the key, to make sure it stays secret. And this will not impact the other key that is used in production.)*\n\n### Generating token\nThe `Token` is generated by calling API `Authenticate`. As part of this package, there is a script `get-token.py`, that will ask you for the `ComAp-Key`, your WSV username and password, generare the `Token` and store it in the config file. This script is called from the `setup.bat` script. But you can call it any time by running `python get-token` from the command line. \nPlease do not share this configuration file, it contains your login information. \nThe `Token` automatically expires after 1 month if it is not used. The `ComAp-Key` can be regenerated in case it gets leaked.\n\n### Storing the API secrets \nThe API key should not be hard-coded in the script. There are number of ways for storing the keys securely, that are beyond the scope of this document. For the purpose of this repository, we are storing the key in file `config.py`, with following content:\n``` python\nKEY = '<YOUR KEY GOES HERE>'\nTOKEN = '<THE TOKEN GOES HERE>'\n```\nThe individual scripts then get the stored secrets by calling:",
"_____no_output_____"
]
],
[
[
"from config import KEY, TOKEN ",
"_____no_output_____"
]
],
[
[
"# Calling WebSupervisor\nAll ComAp WebSupervisor APIs are methods of `wsv` class. To use them, you need to authenticate to an instance:",
"_____no_output_____"
]
],
[
[
"from comap.api import wsv\n\nwsv = wsv(KEY, TOKEN)",
"_____no_output_____"
]
],
[
[
"Then you can use individual functions.\n\n## Units\nTo get the list of units (gensets) under your WSV account, call method `wsv.units`.\n\nIt returns a list of units, each with the following attributes: `name`, `unitGuid`, `url`.",
"_____no_output_____"
]
],
[
[
"for unit in wsv.units():\n print(unit[\"name\"])",
"Hybrid Barcelona\nCGT Polyglass\nIL-NT\nID FLX lite\nCogeneration unit ABB Prague\nWestport \nID-Lite\nIA-NT STD\nIC-NT Mint\nCogeneration unit Hilton London\nID-EM\nHarvester Goa\nHabana hospital\nAIO GAS\nNew marine\nIL9\nController Parallel\nController Solo\nHybrid\ntest33\nsifrovani IL3\nGlobal\nC07 - Hybrid Master\nIG200_VAJL\nEnel SIL\n"
]
],
[
[
"## Info\nThis is to get the information about an unit. For this method (and all the following ones, we need to specify the unit by its `unitGuid`. You can get this information from calling units, or from the WebSupervisor web unit detail url.\n\nFor this example, let's use unitGuid `genset98a2e2828bde4ddb8d48fe5a39cade27`",
"_____no_output_____"
]
],
[
[
"unitGuid = 'genset98a2e2828bde4ddb8d48fe5a39cade27'\ninfo = wsv.info(unitGuid)\nvalues = [('Name', info['name']),\n ('Owner', info['ownerLoginId']),\n ('Timezone', info['timezone']),\n ('Position', f'{info[\"position\"][\"latitude\"]},{info[\"position\"][\"longitude\"]}')\n]\nfor desc, value in values:\n print(f'{desc:10}{value}')",
"Name ID-EM\nOwner storage\nTimezone Europe/Prague\nPosition 50.1069711208162,14.4529473779403\n"
]
],
[
[
"## Values\nWIth this you can read controller values. You can read all values (not recommended) or list of specific values. To specify the values to get, you need to include comma separated list of their `valueGuid`s. \n\nThe list of available value depends on the type of controller. You can get the list of available `valueGuid`s by calling it without any `valueGuid`s.\n\nThe list of common `valueGuid`s is available from constant `VALUE_GUID`. Let's check it:",
"_____no_output_____"
]
],
[
[
"from comap.constants import VALUE_GUID\nfor value in VALUE_GUID:\n print(f'{value:18} {VALUE_GUID[value]}')",
"genset_name 7b2ae258-65a8-40dd-bb42-5455753679f9\nserial_number 6253525D-CF01-4a15-A34B-6C232496E7B4\nlast_update 0C9117DA-495A-11DF-85EB-428556D89593\ncomm_state F0219C1C-1860-4b4d-8E6E-3CEC96279D6F\nunit_state 48EC10D1-7AF8-4ce9-B5B3-A5315993FEB3\nunit_changed 52C8C105-49E6-477B-B43D-33FD46F54640\ncontroller B9FF3CE7-0A81-4D80-8029-99C24A0E764F\nalarm_list 7EDE9575-FEB5-4540-9153-A3B9CF60DE9F\nalarm_list_ext 5E36C1AA-A2B7-4264-96B5-BB975E48B547\nmode 6a12aed6-3be0-44c4-9110-9ea33cfe3ccc\nnominal_power 72D0295A-3E65-11DF-892C-D6A856D89593\nactual_power EE83E7E9-7453-4e64-A5CB-C8C093FB2A2F\nengine_state BB2D1ADE-740E-488d-853B-6BA970D52E27\nbreaker_state B7E5B9B1-9F5E-45c1-82F4-3A336F1F97FA\nGcb_mode b0f910f3-14e4-4bd1-8db1-739e26680b9c\nMcb_mode 5bbc94e2-2497-4d2b-a3e6-1c798d0b6430\nimport_kWh 4C214D52-3D75-11DF-8AA2-529056D89593\nU_L1 4a2a2694-3049-4a8e-ba5f-cc6feeda9d59\nU_L2 e0f2f0ff-0627-4d78-bc78-8973ae8b7cea\nU_L3 201b5a65-e4b8-49e6-8f47-9144607eaf58\nmodes_available 49424e2b-1009-471c-b8e3-3e84e9c16159\nU_battery e707bcd1-2735-4ab7-8a8c-d75fd589052c\nRPM 6629f84f-4b79-43c2-ab33-1f27f1f8c42b\nnominal_RPM DFBA9743-A4E9-4759-89A9-845E861AD9FF\nfrequency_gen 1a3767fd-72e9-429b-a088-95318804ce8e\nbase_load D23D6768-AAD0-11DF-8C33-2A13DFD72085\nP_mains d61be5b5-56fa-4cd8-9fb7-00d2c01627ad\nQ_mains 441ef9cc-fdec-4064-8939-825be28a97b0\nObject_P 87a22d69-b0b7-48ba-bf5f-c3979b462b50\nfuel_level 0dc2739f-0fc9-4a17-bc91-26e5dda19ed8\nservice_time_Sd 4ef4bd3d-d8dd-45ff-93a0-b82320fddef2\nservice_time_1 57d26063-6237-489c-9e59-091c0ed13c47\nservice_time_3 b5719e22-2620-414b-b36b-54975efe3dd0\nrun_hours 74200D25-93F2-4328-B1D6-1A08923AA499\nnumber_of_starts 80042f38-a6d4-4767-a730-b573e7a0dae0\nservice_info 416E416A-CA43-4064-948E-479247B12411\n"
]
],
[
[
"Now let's read few values:",
"_____no_output_____"
]
],
[
[
"values = wsv.values(\n unitGuid,\n f'{VALUE_GUID[\"genset_name\"]},'\n f'{VALUE_GUID[\"mode\"]},'\n f'{VALUE_GUID[\"engine_state\"]}'\n)\nfor value in values:\n print(f'{value[\"name\"]:<16} : {value[\"value\"]}')",
"Motor Name : ID-EM 1.5\nControllerMode : MAN\nMotor State : Not ready\n"
]
],
[
[
"## History\nTo get log of the values for given time period (do not confuse with the controller history)",
"_____no_output_____"
]
],
[
[
"history = wsv.history(\n unitGuid,\n '09/19/2019',\n '09/20/2019',\n VALUE_GUID[\"engine_state\"]\n)\nfor event in history[0][\"history\"]:\n print(f'{event[\"value\"]:<8} valid to {event[\"validTo\"]}')",
" valid to 2019-09-19 01:00:00\n valid to 2019-09-19 04:00:00\n valid to 2019-09-19 06:00:00\n valid to 2019-09-19 12:00:00\n valid to 2019-09-19 15:00:00\n valid to 2019-09-19 20:00:00\n"
]
],
[
[
"## Files\nList the files (reports and controller history) stored on the controller.",
"_____no_output_____"
]
],
[
[
"for file in wsv.files(unitGuid):\n print(f'{file[\"generated\"]} {file[\"fileType\"]} [{file[\"fileName\"]}]')",
"2019-09-19 18:45:42 unitHistory [2019-09-19_20-44_ID-EM.csv]\n2019-09-19 18:47:27 unitHistory [2019-09-19_20-45_ID-EM.csv]\n2019-09-25 11:02:08 unitHistory [2019-09-25_13-01_ID-EM.csv]\n2019-10-10 11:08:06 unitHistory [2019-10-10_13-07_ID-EM.csv]\n"
]
],
[
[
"## Comments\nGet the comments entered on WSV Application. This can be used to automate the intereaction with the application users. ",
"_____no_output_____"
]
],
[
[
"for comment in wsv.comments(unitGuid):\n print(f'{comment[\"author\"]:<18} {comment[\"date\"]} {comment[\"text\"]}')",
"Vaclav Chaloupka 2019-09-19 17:08:15 Ciao\n"
]
],
[
[
"## Download\nThis can be used to download a file",
"_____no_output_____"
]
],
[
[
"FILENAME = 'filename.csv'\n\nif wsv.download(unitGuid,FILENAME):\n print('SUCCESS')\nelse:\n print('FAILED')",
"SUCCESS\n"
]
],
[
[
"## Command\nControls the genset. The available commands are `start`,`stop`,`faultReset`,`changeMcb` (toggle mains circuit breaker), `changeGcb` (toggle genset circuit breaker) and `changeMode`.\n\nFor changeMode enter the mode parameter e.g. to `man` or `aut`",
"_____no_output_____"
]
],
[
[
"if wsv.command(unitGuid,'stop'):\n print('Genset stop succesful')\nelse:\n print('Genset stop failed')",
"Genset stop succesful\n"
],
[
"if wsv.command(unitGuid,'changeMode','aut'):\n print('Change mode succesful')\nelse:\n print('Change mode failed')",
"Change mode succesful\n"
]
],
[
[
"# Asynchronous package\nThe previous examples are using synchronous Python package `requests`. Whilst simple for understanding, the disadvantage of this package is, that it blocks computer resources while waiting for the server response.\n\nThere is therefore also Asynchronous module available. It has the same methods as described above, but they start with a word `_async`. Explaining the `aiohttp` package is beyond the scope of this document, if you intend using the async package, I assume you are familiar with the concept.\n\nHere is an example of usage - doing the same as `wsv.units()`",
"_____no_output_____"
]
],
[
[
"# import aiohttp\n# import asyncio\n# from comap.api_async import wsv_async\n\n# async def test():\n# session = aiohttp.ClientSession(raise_for_status=True)\n# wsv = wsv_async(session, KEY, TOKEN)\n# for unit in await wsv.async_units():\n# print(f'{unit[\"name\"]}')\n# await session.close()\n\n# asyncio.run(test())",
"_____no_output_____"
]
],
[
[
"*You can also refer to the [Jupyter asyncio tests](https://nbviewer.jupyter.org/github/bruxy70/ComAp-API/blob/development/Asyncio%20test.ipynb)*",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6dd601c4979da4d36fe7218ff1d9d5f0ad1a89 | 5,528 | ipynb | Jupyter Notebook | avaliacao.ipynb | silverioleitejr/avaliacao-python | 29f04be2f039bd2488e87e21bf2183a896a108f7 | [
"BSD-2-Clause"
]
| null | null | null | avaliacao.ipynb | silverioleitejr/avaliacao-python | 29f04be2f039bd2488e87e21bf2183a896a108f7 | [
"BSD-2-Clause"
]
| null | null | null | avaliacao.ipynb | silverioleitejr/avaliacao-python | 29f04be2f039bd2488e87e21bf2183a896a108f7 | [
"BSD-2-Clause"
]
| null | null | null | 36.853333 | 1,838 | 0.508864 | [
[
[
"# connectar Dataframe\n#Bibliotecas\nfrom pandas import pandas\n\n#definir variais globais \nimport pandas as pd\npath = \"https://raw.githubusercontent.com/silverioleitejr/avaliacao-python/main/data/dataset.csv\"\nvendas_df = pd.read_csv(path)\n",
"_____no_output_____"
],
[
"def isNaN(num):\n return num != num\n",
"_____no_output_____"
],
[
"# Esta função retorna um DataFrameGroupBy por Marca\ndef agruparMediaMarca():\n\n grouped_df = vendas_df.groupby(['car_make'])\n mean_df = grouped_df.mean()\n\n return mean_df\n\n",
"_____no_output_____"
],
[
" agruparMediaMarca()\n ",
"_____no_output_____"
],
[
"def calcularMedia( fabricante = ''):\n agrupamento_df = agruparMediaMarca()\n filtro_query = 'car_make == ' '\"' + fabricante + '\"'\n filtro_df = agrupamento_df.query( filtro_query )\n\n media = filtro_df['car_value']\n\n return media\n",
"_____no_output_____"
],
[
"def criarSparkDataFrame():\n import pyspark\n from pyspark.sql import SparkSession\n import pandas as pd\n\n # Criar sessão spark\n spark = SparkSession.builder.getOrCreate()\n\n # Ler o dataframe de vendas \n spark_df = spark.createDataFrame(vendas_df)\n\n # incluir o spark df no cataloga Spark\n spark_df.createOrReplaceTempView('spark_df')\n\n",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6dfb82539c73e5bfc59a8386bfd59c5774e4fb | 1,500 | ipynb | Jupyter Notebook | index.ipynb | bwolfson97/reflection_prompts | 8ccdadf3f18e7fdd46baf9a2e01674d12b33c804 | [
"Apache-2.0"
]
| 1 | 2021-11-22T22:36:44.000Z | 2021-11-22T22:36:44.000Z | index.ipynb | bwolfson97/reflection_prompts | 8ccdadf3f18e7fdd46baf9a2e01674d12b33c804 | [
"Apache-2.0"
]
| null | null | null | index.ipynb | bwolfson97/reflection_prompts | 8ccdadf3f18e7fdd46baf9a2e01674d12b33c804 | [
"Apache-2.0"
]
| null | null | null | 15.789474 | 81 | 0.48 | [
[
[
"#hide\nfrom reflection_prompts.core import *",
"_____no_output_____"
]
],
[
[
"# Project name here\n\n> Summary description here.",
"_____no_output_____"
],
[
"This file will become your README and also the index of your documentation.",
"_____no_output_____"
],
[
"## Install",
"_____no_output_____"
],
[
"`pip install your_project_name`",
"_____no_output_____"
],
[
"## How to use",
"_____no_output_____"
],
[
"Fill me in please! Don't forget code examples:",
"_____no_output_____"
]
],
[
[
"1+1",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6dff7508b856174bf7d601eb5076cf2f949ab5 | 6,246 | ipynb | Jupyter Notebook | nb/Binary Heap.ipynb | BaiqiangGit/Data-Structure-and-Algorithms---Python3 | 964a41f210356b8d7b192bf54a2a1eb9a754b19e | [
"Apache-2.0"
]
| 2 | 2021-01-14T08:58:05.000Z | 2021-11-25T21:25:16.000Z | nb/Binary Heap.ipynb | BaiqiangGit/Data-Structure-and-Algorithms---Python3 | 964a41f210356b8d7b192bf54a2a1eb9a754b19e | [
"Apache-2.0"
]
| null | null | null | nb/Binary Heap.ipynb | BaiqiangGit/Data-Structure-and-Algorithms---Python3 | 964a41f210356b8d7b192bf54a2a1eb9a754b19e | [
"Apache-2.0"
]
| 1 | 2021-01-14T08:58:06.000Z | 2021-01-14T08:58:06.000Z | 26.133891 | 100 | 0.476945 | [
[
[
"### Binary Heap\nsemi-ordered tree based data structure\nhttps://en.wikipedia.org/wiki/Binary_heap\n* order exist hierachically, top-to-bottem is fixed\n* no order laterally, left-to-right order is arbitrary\n\n#### Definition\n* Shape property (same to complete binary tree)\n * all layers exceppt last should be full\n * last layer filled from left, no gap between nodes\n * that is why swarping the root and last first to delete, to maintain this property\n * as it's complete, binary heap can be implemented as array\n * 'abstract' implementation of ADT binary tree \n* Heap property: \n * max heap: parent.key ≥ child.key, largest key on top\n * min heap: parent.key ≤ child.key, smallest key on top\n \n#### Properties\n* make root the second item of array for convenience, then for the k-th element of array:\n * its left child is at index 2*k\n * its right child is at index (2*k + 1)\n * its parent is at index k//2 \n \n#### Applications\n* heapsort O ~ nlog(n)\n* Priority Queue\n* others:\n * K’th Largest Element in an array\n * Sort an almost sorted array\n * Merge K Sorted Arrays\n \n### Implementation\n",
"_____no_output_____"
]
],
[
[
"## add path\nimport sys\nsys.path.insert(0, '../ds/')\nfrom minBinaryHeap import *",
"_____no_output_____"
]
],
[
[
"### min binary heap",
"_____no_output_____"
]
],
[
[
"minhp = minBinaryHeap()\nminhp.heapFromList([12, 20, 20, 25, 25, 30, 30])\nminhp.insert(12)\nminhp.insert(20)\nminhp.insert(30)\nminhp.insert(25)\nprint(minhp.heap)\nprint('deleted:', minhp.delete())\nprint(minhp.heap)\nprint('deleted:', minhp.delete())\nprint(minhp.heap)",
"[0, 12, 20, 20, 12, 25, 30, 30, 25, 20, 30, 25]\ndeleted: 12\n[0, 20, 20, 25, 12, 25, 30, 30, 25, 20, 30]\ndeleted: 20\n[0, 20, 12, 25, 20, 25, 30, 30, 25, 30]\n"
]
],
[
[
"### max binary heap",
"_____no_output_____"
]
],
[
[
"from maxBinaryHeap import *\nmaxhp = maxBinaryHeap()\nmaxhp.heapFromList([0, 25, 20, 25, 12, 20])\nmaxhp.insert(12)\nmaxhp.insert(20)\nmaxhp.insert(30)\nmaxhp.insert(25)\nmaxhp.insert(20)\nmaxhp.insert(40)\nmaxhp.insert(25)\nprint(maxhp.heap)\nprint('deleted:', maxhp.delete())\nprint(maxhp.heap)\nprint('deleted:', maxhp.delete())\nprint(maxhp.heap)",
"[0, 40, 25, 30, 25, 25, 25, 12, 0, 20, 12, 20, 20, 20]\ndeleted: 40\n[0, 30, 25, 25, 25, 25, 20, 12, 0, 20, 12, 20, 20]\ndeleted: 30\n[0, 25, 25, 20, 25, 25, 20, 12, 0, 20, 12, 20]\n"
]
],
[
[
"### min heap sort",
"_____no_output_____"
]
],
[
[
"def minHeapSort(ls):\n \n ## helper functions\n def minChild(ls, index):\n if index * 2 + 1 <= len(ls) - 1 and ls[index * 2 + 1] < ls[index * 2]:\n return index *2 + 1\n else:\n return index * 2 \n \n def swimDown(ls, index):\n left_child_index = index * 2 \n while left_child_index <= len(ls) - 1:\n min_child_index = minChild(ls, index)\n if ls[min_child_index] < ls[index]:\n ls[min_child_index], ls[index] = ls[index], ls[min_child_index]\n index = min_child_index\n left_child_index = index * 2 \n else:\n return\n\n ## heap function \n def heapify(ls):\n start = len(ls)//2\n while start > 0:\n swimDown(ls, start)\n start -= 1\n \n ## load data, get size\n ls = [0] + ls\n ls_length = len(ls)\n \n ## inplace heapify\n heapify(ls)\n \n ## heap sort, pop the last\n while len(ls) > 1:\n ls[1], ls[-1] = ls[-1], ls[1]\n yield ls.pop()\n swimDown(ls,1)",
"_____no_output_____"
],
[
"ls = [12, 2, 4, 5, 2, 3, 13, 40, 45]\n[f for f in minHeapSort(ls)]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6e0cf861106b7d0e86be28f2d51cbf00909030 | 18,852 | ipynb | Jupyter Notebook | introduction_to_amazon_algorithms/factorization_machines_mnist/factorization_machines_mnist.ipynb | duytinvo/sagemaker_examples | ee5e66f159076112fa4e21597bc14caeaf7ac919 | [
"Apache-2.0"
]
| 1 | 2018-06-07T22:29:04.000Z | 2018-06-07T22:29:04.000Z | introduction_to_amazon_algorithms/factorization_machines_mnist/factorization_machines_mnist.ipynb | mlaicode/amazon-sagemaker-examples | eeadb596192c467eb3684edc6a361d5cc85ef1d0 | [
"Apache-2.0"
]
| null | null | null | introduction_to_amazon_algorithms/factorization_machines_mnist/factorization_machines_mnist.ipynb | mlaicode/amazon-sagemaker-examples | eeadb596192c467eb3684edc6a361d5cc85ef1d0 | [
"Apache-2.0"
]
| 1 | 2019-10-06T09:17:00.000Z | 2019-10-06T09:17:00.000Z | 44.779097 | 725 | 0.654042 | [
[
[
"# An Introduction to Factorization Machines with MNIST\n_**Making a Binary Prediction of Whether a Handwritten Digit is a 0**_\n\n1. [Introduction](#Introduction)\n2. [Prerequisites and Preprocessing](#Prequisites-and-Preprocessing)\n 1. [Permissions and environment variables](#Permissions-and-environment-variables)\n 2. [Data ingestion](#Data-ingestion)\n 3. [Data inspection](#Data-inspection)\n 4. [Data conversion](#Data-conversion)\n3. [Training the FM model](#Training-the-FM-model)\n4. [Set up hosting for the model](#Set-up-hosting-for-the-model)\n 1. [Import model into hosting](#Import-model-into-hosting)\n 2. [Create endpoint configuration](#Create-endpoint-configuration)\n 3. [Create endpoint](#Create-endpoint)\n5. [Validate the model for use](#Validate-the-model-for-use)\n",
"_____no_output_____"
],
[
"## Introduction\n\nWelcome to our example introducing Amazon SageMaker's Factorization Machines Algorithm! Today, we're analyzing the [MNIST](https://en.wikipedia.org/wiki/MNIST_database) dataset which consists of images of handwritten digits, from zero to nine. We'll use the individual pixel values from each 28 x 28 grayscale image to predict a yes or no label of whether the digit is a 0 or some other digit (1, 2, 3, ... 9).\n\nThe method that we'll use is a factorization machine binary classifier. A factorization machine is a general-purpose supervised learning algorithm that you which can use for both classification and regression tasks. It is an extension of a linear model that is designed to parsimoniously capture interactions between features in high dimensional sparse datasets. For example, in a click prediction system, the factorization machine model can capture click rate patterns observed when ads from a certain ad-category are placed on pages from a certain page-category. Factorization machines are a good choice for tasks dealing with high dimensional sparse datasets, such as click prediction and item recommendation.\n\nAmazon SageMaker's Factorization Machine algorithm provides a robust, highly scalable implementation of this algorithm, which has become extremely popular in ad click prediction and recommender systems. The main purpose of this notebook is to quickly show the basics of implementing Amazon SageMaker Factorization Machines, even if the use case of predicting a digit from an image is not where factorization machines shine.\n\nTo get started, we need to set up the environment with a few prerequisite steps, for permissions, configurations, and so on.",
"_____no_output_____"
],
[
"## Prequisites and Preprocessing\n\n### Permissions and environment variables\n\n_This notebook was created and tested on an ml.m4.xlarge notebook instance._\n\nLet's start by specifying:\n\n- The S3 bucket and prefix that you want to use for training and model data. This should be within the same region as the Notebook Instance, training, and hosting.\n- The IAM role arn used to give training and hosting access to your data. See the documentation for how to create these. Note, if more than one role is required for notebook instances, training, and/or hosting, please replace the boto regexp with a the appropriate full IAM role arn string(s).",
"_____no_output_____"
]
],
[
[
"bucket = '<your_s3_bucket_name_here>'\nprefix = 'sagemaker/DEMO-fm-mnist'\n \n# Define IAM role\nimport boto3\nimport re\nfrom sagemaker import get_execution_role\n\nrole = get_execution_role()",
"_____no_output_____"
]
],
[
[
"### Data ingestion\n\nNext, we read the dataset from an online URL into memory, for preprocessing prior to training. This processing could be done *in situ* by Amazon Athena, Apache Spark in Amazon EMR, Amazon Redshift, etc., assuming the dataset is present in the appropriate location. Then, the next step would be to transfer the data to S3 for use in training. For small datasets, such as this one, reading into memory isn't onerous, though it would be for larger datasets.",
"_____no_output_____"
]
],
[
[
"%%time\nimport pickle, gzip, numpy, urllib.request, json\n\n# Load the dataset\nurllib.request.urlretrieve(\"http://deeplearning.net/data/mnist/mnist.pkl.gz\", \"mnist.pkl.gz\")\nwith gzip.open('mnist.pkl.gz', 'rb') as f:\n train_set, valid_set, test_set = pickle.load(f, encoding='latin1')",
"_____no_output_____"
]
],
[
[
"### Data inspection\n\nOnce the dataset is imported, it's typical as part of the machine learning process to inspect the data, understand the distributions, and determine what type(s) of preprocessing might be needed. You can perform those tasks right here in the notebook. As an example, let's go ahead and look at one of the digits that is part of the dataset.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport matplotlib.pyplot as plt\nplt.rcParams[\"figure.figsize\"] = (2,10)\n\n\ndef show_digit(img, caption='', subplot=None):\n if subplot==None:\n _,(subplot)=plt.subplots(1,1)\n imgr=img.reshape((28,28))\n subplot.axis('off')\n subplot.imshow(imgr, cmap='gray')\n plt.title(caption)\n\nshow_digit(train_set[0][30], 'This is a {}'.format(train_set[1][30]))",
"_____no_output_____"
]
],
[
[
"### Data conversion\n\nSince algorithms have particular input and output requirements, converting the dataset is also part of the process that a data scientist goes through prior to initiating training. In this particular case, the Amazon SageMaker implementation of Factorization Machines takes recordIO-wrapped protobuf, where the data we have today is a pickle-ized numpy array on disk.\n\nMost of the conversion effort is handled by the Amazon SageMaker Python SDK, imported as `sagemaker` below.\n\n_Notice, despite the fact that most use cases for factorization machines will utilize spare input, we are writing our data out as dense tensors. This will be fine since the MNIST dataset is not particularly large or high dimensional._",
"_____no_output_____"
]
],
[
[
"import io\nimport numpy as np\nimport sagemaker.amazon.common as smac\n\nvectors = np.array([t.tolist() for t in train_set[0]]).astype('float32')\nlabels = np.where(np.array([t.tolist() for t in train_set[1]]) == 0, 1.0, 0.0).astype('float32')\n\nbuf = io.BytesIO()\nsmac.write_numpy_to_dense_tensor(buf, vectors, labels)\nbuf.seek(0)",
"_____no_output_____"
]
],
[
[
"## Upload training data\nNow that we've created our recordIO-wrapped protobuf, we'll need to upload it to S3, so that Amazon SageMaker training can use it.",
"_____no_output_____"
]
],
[
[
"import boto3\nimport os\n\nkey = 'recordio-pb-data'\nboto3.resource('s3').Bucket(bucket).Object(os.path.join(prefix, 'train', key)).upload_fileobj(buf)\ns3_train_data = 's3://{}/{}/train/{}'.format(bucket, prefix, key)\nprint('uploaded training data location: {}'.format(s3_train_data))",
"_____no_output_____"
]
],
[
[
"Let's also setup an output S3 location for the model artifact that will be output as the result of training with the algorithm.",
"_____no_output_____"
]
],
[
[
"output_location = 's3://{}/{}/output'.format(bucket, prefix)\nprint('training artifacts will be uploaded to: {}'.format(output_location))",
"_____no_output_____"
]
],
[
[
"## Training the factorization machine model\n\nOnce we have the data preprocessed and available in the correct format for training, the next step is to actually train the model using the data. Since this data is relatively small, it isn't meant to show off the performance of the Amazon SageMaker's Factorization Machines in training, although we have tested it on multi-terabyte datasets.\n\nAgain, we'll use the Amazon SageMaker Python SDK to kick off training and monitor status until it is completed. In this example that takes between 7 and 11 minutes. Despite the dataset being small, provisioning hardware and loading the algorithm container take time upfront.\n\nFirst, let's specify our containers. Since we want this notebook to run in all 4 of Amazon SageMaker's regions, we'll create a small lookup. More details on algorithm containers can be found in [AWS documentation](https://docs-aws.amazon.com/sagemaker/latest/dg/sagemaker-algo-docker-registry-paths.html).",
"_____no_output_____"
]
],
[
[
"containers = {'us-west-2': '174872318107.dkr.ecr.us-west-2.amazonaws.com/factorization-machines:latest',\n 'us-east-1': '382416733822.dkr.ecr.us-east-1.amazonaws.com/factorization-machines:latest',\n 'us-east-2': '404615174143.dkr.ecr.us-east-2.amazonaws.com/factorization-machines:latest',\n 'eu-west-1': '438346466558.dkr.ecr.eu-west-1.amazonaws.com/factorization-machines:latest'}",
"_____no_output_____"
]
],
[
[
"Next we'll kick off the base estimator, making sure to pass in the necessary hyperparameters. Notice:\n- `feature_dim` is set to 784, which is the number of pixels in each 28 x 28 image.\n- `predictor_type` is set to 'binary_classifier' since we are trying to predict whether the image is or is not a 0.\n- `mini_batch_size` is set to 200. This value can be tuned for relatively minor improvements in fit and speed, but selecting a reasonable value relative to the dataset is appropriate in most cases.\n- `num_factors` is set to 10. As mentioned initially, factorization machines find a lower dimensional representation of the interactions for all features. Making this value smaller provides a more parsimonious model, closer to a linear model, but may sacrifice information about interactions. Making it larger provides a higher-dimensional representation of feature interactions, but adds computational complexity and can lead to overfitting. In a practical application, time should be invested to tune this parameter to the appropriate value.",
"_____no_output_____"
]
],
[
[
"import boto3\nimport sagemaker\n\nsess = sagemaker.Session()\n\nfm = sagemaker.estimator.Estimator(containers[boto3.Session().region_name],\n role, \n train_instance_count=1, \n train_instance_type='ml.c4.xlarge',\n output_path=output_location,\n sagemaker_session=sess)\nfm.set_hyperparameters(feature_dim=784,\n predictor_type='binary_classifier',\n mini_batch_size=200,\n num_factors=10)\n\nfm.fit({'train': s3_train_data})",
"_____no_output_____"
]
],
[
[
"## Set up hosting for the model\nNow that we've trained our model, we can deploy it behind an Amazon SageMaker real-time hosted endpoint. This will allow out to make predictions (or inference) from the model dyanamically.\n\n_Note, Amazon SageMaker allows you the flexibility of importing models trained elsewhere, as well as the choice of not importing models if the target of model creation is AWS Lambda, AWS Greengrass, Amazon Redshift, Amazon Athena, or other deployment target._",
"_____no_output_____"
]
],
[
[
"fm_predictor = fm.deploy(initial_instance_count=1,\n instance_type='ml.m4.xlarge')",
"_____no_output_____"
]
],
[
[
"## Validate the model for use\nFinally, we can now validate the model for use. We can pass HTTP POST requests to the endpoint to get back predictions. To make this easier, we'll again use the Amazon SageMaker Python SDK and specify how to serialize requests and deserialize responses that are specific to the algorithm.\n\nSince factorization machines are so frequently used with sparse data, making inference requests with a CSV format (as is done in other algorithm examples) can be massively inefficient. Rather than waste space and time generating all of those zeros, to pad the row to the correct dimensionality, JSON can be used more efficiently. Since we trained the model using dense data, this is a bit of a moot point, as we'll have to pass all the 0s in anyway.\n\nNevertheless, we'll write our own small function to serialize our inference request in the JSON format that Amazon SageMaker Factorization Machines expects.",
"_____no_output_____"
]
],
[
[
"import json\nfrom sagemaker.predictor import json_deserializer\n\ndef fm_serializer(data):\n js = {'instances': []}\n for row in data:\n js['instances'].append({'features': row.tolist()})\n return json.dumps(js)\n\nfm_predictor.content_type = 'application/json'\nfm_predictor.serializer = fm_serializer\nfm_predictor.deserializer = json_deserializer",
"_____no_output_____"
]
],
[
[
"Now let's try getting a prediction for a single record.",
"_____no_output_____"
]
],
[
[
"result = fm_predictor.predict(train_set[0][30:31])\nprint(result)",
"_____no_output_____"
]
],
[
[
"OK, a single prediction works. We see that for one record our endpoint returned some JSON which contains `predictions`, including the `score` and `predicted_label`. In this case, `score` will be a continuous value between [0, 1] representing the probability we think the digit is a 0 or not. `predicted_label` will take a value of either `0` or `1` where (somewhat counterintuitively) `1` denotes that we predict the image is a 0, while `0` denotes that we are predicting the image is not of a 0.\n\nLet's do a whole batch of images and evaluate our predictive accuracy.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\npredictions = []\nfor array in np.array_split(test_set[0], 100):\n result = fm_predictor.predict(array)\n predictions += [r['predicted_label'] for r in result['predictions']]\n\npredictions = np.array(predictions)",
"_____no_output_____"
],
[
"import pandas as pd\n\npd.crosstab(np.where(test_set[1] == 0, 1, 0), predictions, rownames=['actuals'], colnames=['predictions'])",
"_____no_output_____"
]
],
[
[
"As we can see from the confusion matrix above, we predict 951 images of the digit 0 correctly (confusingly this is class 1). Meanwhile we predict 165 images as the digit 0 when in actuality they aren't, and we miss predicting 29 images of the digit 0 that we should have.\n\n*Note: Due to some differences in parameter initialization, your results may differ from those listed above, but should remain reasonably consistent.*",
"_____no_output_____"
],
[
"### (Optional) Delete the Endpoint\n\nIf you're ready to be done with this notebook, please run the delete_endpoint line in the cell below. This will remove the hosted endpoint you created and avoid any charges from a stray instance being left on.",
"_____no_output_____"
]
],
[
[
"import sagemaker\n\nsagemaker.Session().delete_endpoint(fm_predictor.endpoint)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6e331d822a0f8b851660de31774d7552000b10 | 30,092 | ipynb | Jupyter Notebook | site/id/tutorials/load_data/csv.ipynb | KarimaTouati/docs-l10n | 583c00a412cefd168a19975bf2f4cef2e2fc9dca | [
"Apache-2.0"
]
| null | null | null | site/id/tutorials/load_data/csv.ipynb | KarimaTouati/docs-l10n | 583c00a412cefd168a19975bf2f4cef2e2fc9dca | [
"Apache-2.0"
]
| null | null | null | site/id/tutorials/load_data/csv.ipynb | KarimaTouati/docs-l10n | 583c00a412cefd168a19975bf2f4cef2e2fc9dca | [
"Apache-2.0"
]
| null | null | null | 28.879079 | 332 | 0.509803 | [
[
[
"##### Copyright 2019 The TensorFlow Authors.\n\n",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Muat data CSV",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/tutorials/load_data/csv\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />Lihat di TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs-l10n/blob/master/site/id/tutorials/load_data/csv.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Jalankan di Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs-l10n/blob/master/site/id/tutorials/load_data/csv.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />Lihat kode di GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs-l10n/site/id/tutorials/load_data/csv.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Unduh notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"Note: Komunitas TensorFlow kami telah menerjemahkan dokumen-dokumen ini. Tidak ada jaminan bahwa translasi ini akurat, dan translasi terbaru dari [Official Dokumentasi - Bahasa Inggris](https://www.tensorflow.org/?hl=en) karena komunitas translasi ini adalah usaha terbaik dari komunitas translasi.\nJika Anda memiliki saran untuk meningkatkan terjemahan ini, silakan kirim pull request ke [tensorflow/docs](https://github.com/tensorflow/docs) repositori GitHub.\nUntuk menjadi sukarelawan untuk menulis atau memeriksa terjemahan komunitas, hubungi\n[daftar [email protected]](https://groups.google.com/a/tensorflow.org/forum/#!forum/docs).",
"_____no_output_____"
],
[
"Tutorial ini memberikan contoh cara memuat data CSV dari file ke `tf.data.Dataset`.\n\nData yang digunakan dalam tutorial ini diambil dari daftar penumpang Titanic. Model akan memprediksi kemungkinan penumpang selamat berdasarkan karakteristik seperti usia, jenis kelamin, kelas tiket, dan apakah orang tersebut bepergian sendirian.",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
]
],
[
[
"try:\n # %tensorflow_version hanya ada di Colab.\n %tensorflow_version 2.x\nexcept Exception:\n pass",
"_____no_output_____"
],
[
"from __future__ import absolute_import, division, print_function, unicode_literals\nimport functools\n\nimport numpy as np\nimport tensorflow as tf",
"_____no_output_____"
],
[
"TRAIN_DATA_URL = \"https://storage.googleapis.com/tf-datasets/titanic/train.csv\"\nTEST_DATA_URL = \"https://storage.googleapis.com/tf-datasets/titanic/eval.csv\"\n\ntrain_file_path = tf.keras.utils.get_file(\"train.csv\", TRAIN_DATA_URL)\ntest_file_path = tf.keras.utils.get_file(\"eval.csv\", TEST_DATA_URL)",
"_____no_output_____"
],
[
"# Membuat nilai numpy agar lebih mudah dibaca.\nnp.set_printoptions(precision=3, suppress=True)",
"_____no_output_____"
]
],
[
[
"## Memuat data\n\nUntuk memulai, mari kita lihat bagian atas file CSV untuk melihat formatnya.\n",
"_____no_output_____"
]
],
[
[
"!head {train_file_path}",
"_____no_output_____"
]
],
[
[
"Anda dapat [memuat ini menggunakan panda] (pandas.ipynb), dan meneruskan array NumPy ke TensorFlow. Jika Anda perlu meningkatkan ke file dengan skala besar, atau membutuhkan loader yang terintegrasi dengan [TensorFlow dan tf.data] (../../panduan/data.ipynb) kemudian gunakan fungsi `tf.data.experimental.make_csv_dataset`:",
"_____no_output_____"
],
[
"Satu-satunya kolom yang perlu Anda identifikasi secara eksplisit adalah kolom dengan nilai yang dimaksudkan untuk diprediksi oleh model.",
"_____no_output_____"
]
],
[
[
"LABEL_COLUMN = 'survived'\nLABELS = [0, 1]",
"_____no_output_____"
]
],
[
[
"Sekarang baca data CSV dari file dan buat sebuah dataset.\n\n(Untuk dokumentasi lengkap, lihat `tf.data.experimental.make_csv_dataset`)\n",
"_____no_output_____"
]
],
[
[
"def get_dataset(file_path, **kwargs):\n dataset = tf.data.experimental.make_csv_dataset(\n file_path,\n batch_size=5, # Artifisial kecil untuk membuat contoh lebih mudah ditampilkan.\n label_name=LABEL_COLUMN,\n na_value=\"?\",\n num_epochs=1,\n ignore_errors=True, \n **kwargs)\n return dataset\n\nraw_train_data = get_dataset(train_file_path)\nraw_test_data = get_dataset(test_file_path)",
"_____no_output_____"
],
[
"def show_batch(dataset):\n for batch, label in dataset.take(1):\n for key, value in batch.items():\n print(\"{:20s}: {}\".format(key,value.numpy()))",
"_____no_output_____"
]
],
[
[
"Setiap item dalam dataset adalah *batch*, direpresentasikan sebagai *tuple* dari (*banyak contoh*, *banyak label*). Data dari contoh-contoh tersebut disusun dalam tensor berbasis kolom (bukan tensor berbasis baris), masing-masing dengan elemen sebanyak ukuran batch (dalam kasus ini 5).\n\nDengan melihat sendiri, mungkin akan membantu Anda untuk memahami.",
"_____no_output_____"
]
],
[
[
"show_batch(raw_train_data)",
"_____no_output_____"
]
],
[
[
"Seperti yang Anda lihat, kolom dalam file CSV diberi nama. Konstruktor dataset akan mengambil nama-nama ini secara otomatis. Jika file yang sedang Anda kerjakan tidak mengandung nama kolom di baris pertama, berikan mereka dalam daftar string ke argumen `column_names` dalam fungsi` make_csv_dataset`.",
"_____no_output_____"
]
],
[
[
"CSV_COLUMNS = ['survived', 'sex', 'age', 'n_siblings_spouses', 'parch', 'fare', 'class', 'deck', 'embark_town', 'alone']\n\ntemp_dataset = get_dataset(train_file_path, column_names=CSV_COLUMNS)\n\nshow_batch(temp_dataset)",
"_____no_output_____"
]
],
[
[
"Contoh ini akan menggunakan semua kolom yang tersedia. Jika Anda perlu menghilangkan beberapa kolom dari dataset, buat daftar hanya kolom yang Anda rencanakan untuk digunakan, dan kirimkan ke argumen `select_columns` (opsional) dari konstruktor.\n",
"_____no_output_____"
]
],
[
[
"SELECT_COLUMNS = ['survived', 'age', 'n_siblings_spouses', 'class', 'deck', 'alone']\n\ntemp_dataset = get_dataset(train_file_path, select_columns=SELECT_COLUMNS)\n\nshow_batch(temp_dataset)",
"_____no_output_____"
]
],
[
[
"## Data pre-processing\n\nFile CSV dapat berisi berbagai tipe data. Biasanya Anda ingin mengonversi dari berbagai tipe ke vektor dengan panjang tetap sebelum memasukkan data ke dalam model Anda.\n\nTensorFlow memiliki sistem bawaan untuk menjelaskan konversi input umum: `tf.feature_column`, lihat [tutorial ini](../keras/feature_columns) untuk detailnya.\n\nAnda dapat memproses data Anda menggunakan alat apa pun yang Anda suka (seperti [nltk](https://www.nltk.org/) atau [sklearn](https://scikit-learn.org/stable/)), dan kemudian memberikan output yang telah diproses ke TensorFlow.\n\nKeuntungan utama melakukan preprocessing di dalam model Anda adalah ketika Anda mengekspor model itu termasuk dengan proses preprocessing. Dengan cara ini Anda bisa mengirimkan data mentah langsung ke model Anda.",
"_____no_output_____"
],
[
"### Continuous data",
"_____no_output_____"
],
[
"Jika data Anda sudah dalam format numerik yang sesuai, Anda bisa mengemas data ke dalam vektor sebelum meneruskannya ke model:",
"_____no_output_____"
]
],
[
[
"SELECT_COLUMNS = ['survived', 'age', 'n_siblings_spouses', 'parch', 'fare']\nDEFAULTS = [0, 0.0, 0.0, 0.0, 0.0]\ntemp_dataset = get_dataset(train_file_path, \n select_columns=SELECT_COLUMNS,\n column_defaults = DEFAULTS)\n\nshow_batch(temp_dataset)",
"_____no_output_____"
],
[
"example_batch, labels_batch = next(iter(temp_dataset)) ",
"_____no_output_____"
]
],
[
[
"Berikut adalah fungsi sederhana yang akan menyatukan semua\n\n---\n\nkolom:",
"_____no_output_____"
]
],
[
[
"def pack(features, label):\n return tf.stack(list(features.values()), axis=-1), label",
"_____no_output_____"
]
],
[
[
"Terapkan ini ke setiap elemen dataset:",
"_____no_output_____"
]
],
[
[
"packed_dataset = temp_dataset.map(pack)\n\nfor features, labels in packed_dataset.take(1):\n print(features.numpy())\n print()\n print(labels.numpy())",
"_____no_output_____"
]
],
[
[
"Jika Anda memiliki data dengan berbagai tipe, Anda mungkin ingin memisahkan data simple-numeric fields. Api `tf.feature_column` dapat menanganinya, tetapi hal ini menimbulkan beberapa overhead dan harus dihindari kecuali benar-benar diperlukan. Kembali ke kumpulan data campuran:",
"_____no_output_____"
]
],
[
[
"show_batch(raw_train_data)",
"_____no_output_____"
],
[
"example_batch, labels_batch = next(iter(temp_dataset)) ",
"_____no_output_____"
]
],
[
[
"Jadi tentukan preprosesor yang lebih umum yang memilih daftar fitur numerik dan mengemasnya ke dalam satu kolom:",
"_____no_output_____"
]
],
[
[
"class PackNumericFeatures(object):\n def __init__(self, names):\n self.names = names\n\n def __call__(self, features, labels):\n numeric_freatures = [features.pop(name) for name in self.names]\n numeric_features = [tf.cast(feat, tf.float32) for feat in numeric_freatures]\n numeric_features = tf.stack(numeric_features, axis=-1)\n features['numeric'] = numeric_features\n\n return features, labels",
"_____no_output_____"
],
[
"NUMERIC_FEATURES = ['age','n_siblings_spouses','parch', 'fare']\n\npacked_train_data = raw_train_data.map(\n PackNumericFeatures(NUMERIC_FEATURES))\n\npacked_test_data = raw_test_data.map(\n PackNumericFeatures(NUMERIC_FEATURES))",
"_____no_output_____"
],
[
"show_batch(packed_train_data)",
"_____no_output_____"
],
[
"example_batch, labels_batch = next(iter(packed_train_data)) ",
"_____no_output_____"
]
],
[
[
"#### Normalisasi Data\n\nData kontinu (continues data) harus selalu dinormalisasi.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ndesc = pd.read_csv(train_file_path)[NUMERIC_FEATURES].describe()\ndesc",
"_____no_output_____"
],
[
"MEAN = np.array(desc.T['mean'])\nSTD = np.array(desc.T['std'])",
"_____no_output_____"
],
[
"def normalize_numeric_data(data, mean, std):\n # Center the data\n return (data-mean)/std",
"_____no_output_____"
]
],
[
[
"Sekarang buat kolom angka. API `tf.feature_columns.numeric_column` menerima argumen `normalizer_fn`, yang akan dijalankan pada setiap batch.\n\nBundlekan `MEAN` dan `STD` ke normalizer fn menggunakan [`functools.partial`](https://docs.python.org/3/library/functools.html#functools.partial).",
"_____no_output_____"
]
],
[
[
"# Lihat apa yang baru saja Anda buat.\nnormalizer = functools.partial(normalize_numeric_data, mean=MEAN, std=STD)\n\nnumeric_column = tf.feature_column.numeric_column('numeric', normalizer_fn=normalizer, shape=[len(NUMERIC_FEATURES)])\nnumeric_columns = [numeric_column]\nnumeric_column",
"_____no_output_____"
]
],
[
[
"Saat Anda melatih model, sertakan kolom fitur ini untuk memilih dan memusatkan blok data numerik ini:",
"_____no_output_____"
]
],
[
[
"example_batch['numeric']",
"_____no_output_____"
],
[
"numeric_layer = tf.keras.layers.DenseFeatures(numeric_columns)\nnumeric_layer(example_batch).numpy()",
"_____no_output_____"
]
],
[
[
"Normalisasi berdasarkan rata-rata yang digunakan di sini mewajibkan kita untuk mengetahui rata-rata setiap kolom sebelumnya.",
"_____no_output_____"
],
[
"### Kategori data\n\nBeberapa kolom dalam data CSV adalah kolom kategorikal. Artinya, kontennya harus menjadi salah satu dari opsi yang ada.\n\nGunakan API `tf.feature_column` untuk membuat koleksi dengan `tf.feature_column.indicator_column` untuk setiap kolom kategorikal.",
"_____no_output_____"
]
],
[
[
"CATEGORIES = {\n 'sex': ['male', 'female'],\n 'class' : ['First', 'Second', 'Third'],\n 'deck' : ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I', 'J'],\n 'embark_town' : ['Cherbourg', 'Southhampton', 'Queenstown'],\n 'alone' : ['y', 'n']\n}",
"_____no_output_____"
],
[
"categorical_columns = []\nfor feature, vocab in CATEGORIES.items():\n cat_col = tf.feature_column.categorical_column_with_vocabulary_list(\n key=feature, vocabulary_list=vocab)\n categorical_columns.append(tf.feature_column.indicator_column(cat_col))",
"_____no_output_____"
],
[
"# See what you just created.\ncategorical_columns",
"_____no_output_____"
],
[
"categorical_layer = tf.keras.layers.DenseFeatures(categorical_columns)\nprint(categorical_layer(example_batch).numpy()[0])",
"_____no_output_____"
]
],
[
[
"Ini akan menjadi bagian dari pemrosesan input data ketika Anda membangun model.",
"_____no_output_____"
],
[
"### Layer preprocessing gabungan",
"_____no_output_____"
],
[
"Tambahkan dua koleksi kolom fitur dan teruskan ke `tf.keras.layers.DenseFeatures` untuk membuat lapisan input yang akan mengekstraksi dan memproses kedua jenis input:",
"_____no_output_____"
]
],
[
[
"preprocessing_layer = tf.keras.layers.DenseFeatures(categorical_columns+numeric_columns)",
"_____no_output_____"
],
[
"print(preprocessing_layer(example_batch).numpy()[0])",
"_____no_output_____"
]
],
[
[
"## Membangun Model",
"_____no_output_____"
],
[
"Jalankan `tf.keras.Sequential`, mulai dari `preprocessing_layer`.",
"_____no_output_____"
]
],
[
[
"model = tf.keras.Sequential([\n preprocessing_layer,\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(128, activation='relu'),\n tf.keras.layers.Dense(1, activation='sigmoid'),\n])\n\nmodel.compile(\n loss='binary_crossentropy',\n optimizer='adam',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## Latih, evaluasi, dan prediksi",
"_____no_output_____"
],
[
"Sekarang model dapat dipakai dan dilatih.",
"_____no_output_____"
]
],
[
[
"train_data = packed_train_data.shuffle(500)\ntest_data = packed_test_data",
"_____no_output_____"
],
[
"model.fit(train_data, epochs=20)",
"_____no_output_____"
]
],
[
[
"Setelah model dilatih, Anda dapat memeriksa akurasinya pada set `test_data`.",
"_____no_output_____"
]
],
[
[
"test_loss, test_accuracy = model.evaluate(test_data)\n\nprint('\\n\\nTest Loss {}, Test Accuracy {}'.format(test_loss, test_accuracy))",
"_____no_output_____"
]
],
[
[
"Gunakan `tf.keras.Model.predict` untuk menyimpulkan pada label batch atau dataset batch.\n",
"_____no_output_____"
]
],
[
[
"predictions = model.predict(test_data)\n\n# Tampilkan beberapa hasil\nfor prediction, survived in zip(predictions[:10], list(test_data)[0][1][:10]):\n print(\"Predicted survival: {:.2%}\".format(prediction[0]),\n \" | Actual outcome: \",\n (\"SURVIVED\" if bool(survived) else \"DIED\"))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6e340f800723257993b608922fa27eaddb85ad | 41,388 | ipynb | Jupyter Notebook | ee046746_tut_11_3D_deep_learning.ipynb | antonagafonov/ee046746-computer-vision | c39d485e6887d774c9a47b5f46f268b900fff9f4 | [
"MIT"
]
| null | null | null | ee046746_tut_11_3D_deep_learning.ipynb | antonagafonov/ee046746-computer-vision | c39d485e6887d774c9a47b5f46f268b900fff9f4 | [
"MIT"
]
| null | null | null | ee046746_tut_11_3D_deep_learning.ipynb | antonagafonov/ee046746-computer-vision | c39d485e6887d774c9a47b5f46f268b900fff9f4 | [
"MIT"
]
| null | null | null | 30.979042 | 278 | 0.561274 | [
[
[
"# <img src=\"https://img.icons8.com/bubbles/100/000000/3d-glasses.png\" style=\"height:50px;display:inline\"> EE 046746 - Technion - Computer Vision\n\n\n## Tutorial 11 - Introduction to 3D Deep Learning\n---\n<img src=\"./assets/tut_09_teaser0.gif\" style=\"width:800px\">\n\n* <a href=\"https://towardsdatascience.com/the-future-of-3d-point-clouds-a-new-perspective-125b35b558b9\">Image source</a>",
"_____no_output_____"
],
[
"* Cool video - Unreal Engine Point Clouds Feature:\n<a href=\"https://www.youtube.com/watch?v=R-ZXdAEGbiw&feature=youtu.be\">LiDAR point cloud support | Feature Highlight | Unreal Engine\n</a>\n",
"_____no_output_____"
],
[
"## <img src=\"https://img.icons8.com/bubbles/50/000000/checklist.png\" style=\"height:50px;display:inline\"> Agenda\n---\n* [Depth Cameras](#-Depth-Cameras)\n * [Stereo Imaging](#-Stereo-Cameras)\n * [Time of Flight](#-Time-of-Flight-Cameras)\n* [3D Deep Learning](#-3D-Deep-Learning)\n * [Voxels](#-Voxalization)\n * [Multi-View](#-Multi-View-Approach)\n * [Point Clouds](#-Apply-Deep-Learning-Directly-on-3D-Point-Clouds)\n* [3D Applications](#-3D-Deep-Learning-Applications)\n* [Recommended Tools](#-Recommended-Tools)\n* [Recommended Videos](#-Recommended-Videos)\n* [Credits](#-Credits)\n",
"_____no_output_____"
],
[
"<img src=\"./assets/tut_09_teaser.JPG\" style=\"width:800px\">\n\n<a href=\"https://cseweb.ucsd.edu/~haosu/\">Image Source</a>",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/office/80/000000/depth.png\" style=\"height:50px;display:inline\"> Depth Cameras\n---\n* **Stereo Cameras - Last week**\n* Time of flight",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/doodle/48/000000/3d-glasses.png\" style=\"height:50px;display:inline\"> Stereo Cameras\n---\n\n<img src=\"./assets/stereo_match1.jpg\" width=\"800\">",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/office/80/000000/depth.png\" style=\"height:50px;display:inline\"> Depth Cameras\n---\n* Stereo Cameras - Lecture with Anat\n* **Time of flight**",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n* Light travels at approximately a constant speed $c = 3\\times 10^8$ (meters per second).\n* Measuring the time it takes for light to travel over a distance one can infer distance.\n* Can be categorized into two types:\n 1. Direct TOF - switch laser on and off rapidly.\n 2. Indirect TOF - send out modulated light, then measure phase difference to infer depth.",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n##### 1. Direct - TOF\n* **Li**ght **D**etection **A**nd **R**anging (LiDAR) probably best example in computer vision and robotics.\n* High-energy light pulses limit influence of background illumination.\n* However, difficulty to generate short light pulses with fast rise and fall times.\n* High-accuracy time measurement required.\n* Prone to motion blur.\n* Sparser as objects grow in distance.\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n\n<img src=\"./assets/tut_09_LiDAR.GIF\" style=\"width:300px\">\n\n<a href=\"https://en.wikipedia.org/wiki/Lidar\">Gif source - Wikipedia</a>\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n<img src=\"./assets/tut_09_sydney.png\" style=\"width:800px\">\n<a href=\"http://www.acfr.usyd.edu.au/papers/SydneyUrbanObjectsDataset.shtml\">Sydney Dataset</a>\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n###### Autonomous Car - LiDAR\n<img src=\"./assets/tut_09_cam1.JPG\" style=\"width:800px\">\n\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n##### SLAM + LIDAT - Zebedee\n<img src=\"./assets/tut_09_cam2.JPG\" style=\"width:800px\">\n<a href=\"https://research.csiro.au/robotics/zebedee/\">zebedee</a>\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/android/48/000000/time.png\" style=\"height:50px;display:inline\"> Time of Flight Cameras\n---\n###### 2. Indirect - TOF\n* Continuous light waves instead of short light pulses.\n* Modulation in terms of frequency of sinusoidal waves.\n* Detected wave after reflection has shifted phase.\n* Phase shift proportional to distance from reflecting surface.\n\n<img src=\"./assets/tut_09_cam3.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"<img src=\"./assets/tut_09_cam4.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/fluent/48/000000/3d.png\" style=\"height:50px;display:inline\"> 3D Deep Learning\n---\n* Representation\n* Architectures",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n<img src=\"./assets/tut_o9_pn1.JPG\" style=\"width:800px\">\n\n* <a href=\"https://arxiv.org/abs/1612.00593\">Image Source</a>",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n\n* Calssification\n* Semantic segmentation\n* Part segmentation\n * Each point belongs to a specific part of the object\n* ...",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n##### <img src=\"https://img.icons8.com/bubbles/50/000000/question-mark.png\" style=\"height:50px;display:inline\"> Questions\n* What are the differences between 2D image and a point cloud?\n* Why it might be hard to feed a point cloud as neural network input?\n* What are the benefits of using a point cloud?",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n#### <img src=\"https://img.icons8.com/plasticine/100/000000/not-applicable.png\" style=\"height:50px;display:inline\"> Point Clouds Problems\n\n* Point Clouds Vary in Size (not constant)\n* Unordered Input\n * Data is unstructured (no grid)\n * Data is invariant to point ordering (permutations)",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n#### <img src=\"https://img.icons8.com/plasticine/100/000000/not-applicable.png\" style=\"height:50px;display:inline\"> Point Clouds Problems\n\n* Missing data\n* Noise\n* Rotations",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n##### Problem - Point Clouds Vary in Size\n\n<img src=\"./assets/tut_09_pn2.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n<img src=\"./assets/tut_09_pn3.JPG\" style=\"width:800px\">\n\n* Different point clouds represent the same object",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n#### Problem - Unordered Input\n\nPoint cloud: $N$ **orderless** points, each represented by a $D$ dim vector\n\n<img src=\"./assets/tut_09_pn4.JPG\" style=\"width:800px\">\n\n* How many semi-equal representations?\n * **Model needs to be invariant to $N!$ permutations**",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n#### <img src=\"https://img.icons8.com/carbon-copy/100/000000/switch-camera.png\" style=\"height:50px;display:inline\">Alternative 3D Representations\n\nSolution:\n* Convert the raw point clouds into Voxels or multiple 2D RGB(D) images\n\n<img src=\"./assets/tut_09_pn5.JPG\" style=\"width:800px\">\n\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/nolan/64/cloud.png\" style=\"height:50px;display:inline\"> 3D Deep Learning: Point Clouds?\n---\n\n* Other 3D representation (not in this course):\n\n<img src=\"./assets/tut_09_other.JPG\" style=\"width:800px\">\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/carbon-copy/100/000000/sugar-cubes.png\" style=\"height:50px;display:inline\"> Voxelization\n---\nIdea: generalize 2D convolutions to regular 3D grids\n\n* The straightforward approach: transform the point clouds into a voxel grid by rasterizing and use 3D CNNs\n\n<img src=\"./assets/tut_09_vox1.jpg\" style=\"width:300px\">\n\nVoxel grid is a 3D grid of equal size volumes (voxels), can be occupied by:\n* Binary 0/1 - Is there any point within the voxel?\n* Weighted - The amount of point located within each voxel\n\nUsually we use binary occupancy\n\n\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/carbon-copy/100/000000/sugar-cubes.png\" style=\"height:50px;display:inline\"> Voxelization\n---\n3D convolution uses 4D kernels\n\n<img src=\"./assets/tut_09_vox2.JPG\" style=\"width:600px\">\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/carbon-copy/100/000000/sugar-cubes.png\" style=\"height:50px;display:inline\"> Voxelization\n---\n<img src=\"./assets/tut_09_voxnet.png\" style=\"width:500px\">\n\n* <a href=\"https://www.ri.cmu.edu/pub_files/2015/9/voxnet_maturana_scherer_iros15.pdf\">Image Source</a>",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/carbon-copy/100/000000/sugar-cubes.png\" style=\"height:50px;display:inline\"> Voxelization\n---\n#### <img src=\"https://img.icons8.com/plasticine/100/000000/not-applicable.png\" style=\"height:50px;display:inline\"> Voxelization Problems\n\n* Large memory cost\n* Slow processing time\n* Limited spatial resolution\n* Quantization artifacts\n\n<img src=\"./assets/tut_09_vox3.JPG\" style=\"width:600px\">\n\n<a href=\"https://cseweb.ucsd.edu/~haosu/\">Image Source</a>",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/dusk/64/000000/multiple-cameras.png\" style=\"height:50px;display:inline\"> Multi-View Approach\n---\nIdea: Transfrom the problem into a well known domain (3D$\\rightarrow$2D)\n\n* The multi-view approach: project multiple views to 2D and use CNN to process\n * How many views do we need? (Another hyper parameter)\n\n<img src=\"./assets/tut_09_pn7.png\" style=\"width:600px\">\n\n* CNN$_1$ - We can use pre-trained networks to extract features followed by fine tuned layers\n* <a href=\"https://www.cv-foundation.org/openaccess/content_iccv_2015/papers/Su_Multi-View_Convolutional_Neural_ICCV_2015_paper.pdf\">Image Source</a>",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/doodle/48/000000/direction-sign.png\" style=\"height:50px;display:inline\"> Apply Deep Learning Directly on 3D Point Clouds\n---\nIdea: Most of the raw 3D data are point clouds - Solve the problems!\n\nNote: Point Clouds Problems:\n* Point Clouds Vary in Size (not constant)\n* Unordered Input\n * Data is unstructured (no grid)\n * Data is invariant to point ordering (permutations)\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/cute-clipart/64/000000/machine-learning.png\" style=\"height:50px;display:inline\"> PointNet\n---\n#### Permutation Invariance: Symmetric Function\n$$ f(x_1,x_2,\\dots,x_n) \\equiv f(x_{\\pi_1},x_{\\pi_2},\\dots,x_{\\pi_n}), \\ x_i \\in R^D $$\n\n$\\pi$ is a different permutation",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/cute-clipart/64/000000/machine-learning.png\" style=\"height:50px;display:inline\"> PointNet\n---\nExample:\n$$ f(x_1,x_2,\\dots,x_n) = max\\{x_1,x_2,\\dots,x_n\\}$$\n$$ f(x_1,x_2,\\dots,x_n) = x_1+x_2+\\dots+x_n$$\n",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/cute-clipart/64/000000/machine-learning.png\" style=\"height:50px;display:inline\"> PointNet\n---\n* How can we construct a family of symmetric functions by neural networks?",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/cute-clipart/64/000000/machine-learning.png\" style=\"height:50px;display:inline\"> PointNet\n---\nObserve:\n$$ f(x_1,x_2,\\dots,x_n) = \\gamma \\circ g(h(x_1),\\dots,h(x_n))$$\nis symmetric if $g$ is symmetric",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/cute-clipart/64/000000/machine-learning.png\" style=\"height:50px;display:inline\"> PointNet\n---\n\n<img src=\"./assets/tut_09_pn8.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/wired/64/000000/diversity.png\" style=\"height:50px;display:inline\"> Basic PointNet Architecture\n---\nEmpirically, we use multi-layer perceptron (MLP) and max pooling:\n\n<img src=\"./assets/tut_09_pn9.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/wired/64/000000/diversity.png\" style=\"height:50px;display:inline\"> Basic PointNet Architecture\n---\nInput MLP:\n$$h(x_i): R^{3} \\rightarrow{} R^{D}$$\nWe can look at it as $D$ functions $\\{h_k\\}_{k=1}^D$ operate on each point where $$h_k(x_i): R^{3} \\rightarrow{} R^{1}$$\n\nPooling layer:\n$$g(h(x_1),\\dots,h(x_n)): R^{N\\times D} \\rightarrow{} R^{D}$$\nWe apply the pooling over all points for each function $h_k$.\n$$g(h_k(x_1),\\dots,h_k(x_n)): R^{N\\times 1} \\rightarrow{} R^{1}$$\n\nClassification MLP:\n$$\\gamma \\circ g(h(x_1),\\dots,h(x_n)): R^{D} \\rightarrow{} R^{\\text{Num Classes}}$$",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/wired/64/000000/diversity.png\" style=\"height:50px;display:inline\"> Basic PointNet Architecture\n---\nShared mlp implementation \"trick\":\n* Use conv layers : Number of filters $C_{out}$, each filter size is ${ 1 \\times {C_{in}}} $.\n* Input: $R^{ N\\times {C_{in}}}$\n* Output: $R^{N\\times {C_{out}}}$\n* Main idea is to cast to 1D convolutions:\n * shift invariance in the $N$ dimension takes care of weight sharing.\n\n\n\n\nMLP:\n$$h: R^{C_{in}}\\rightarrow{} R^{C_1}\\rightarrow{} \\dots \\rightarrow{} R^{C_{out}}$$",
"_____no_output_____"
],
[
"#### PointNet Classification Network\n\n<img src=\"./assets/tut_09_pn10.JPG\" style=\"width:800px\">\n\n",
"_____no_output_____"
],
[
"<img src=\"./assets/tut_09_pn20.gif\" style=\"width:800px\">\n\n<a href=\"https://towardsdatascience.com/deep-learning-on-point-clouds-implementing-pointnet-in-google-colab-1fd65cd3a263\">Image source</a>",
"_____no_output_____"
],
[
"##### Transformation Invariance\n\n<img src=\"./assets/tut_09_pn21.JPG\" style=\"width:400px\">\n\n* Learn transformation matrix to improve task performance: \n * We want our network to be invariant to rigid transformations of the object.\n * Practically, more augmentation over the training dataset also solved the transformation invariance.\n\n<a href=\"https://medium.com/@luis_gonzales/an-in-depth-look-at-pointnet-111d7efdaa1a\">Image source</a>",
"_____no_output_____"
],
[
"## <img src=\"https://img.icons8.com/color/96/000000/code.png\" style=\"height:50px;display:inline\"> Code Example\n* Code based on example by <a href=\"https://github.com/nikitakaraevv/pointnet\">Nikita Karaev</a>",
"_____no_output_____"
]
],
[
[
"# imports for the tutorial\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport time\n\n# pytorch imports\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nfrom torch.utils.data import DataLoader, Dataset, ConcatDataset",
"_____no_output_____"
],
[
"class Tnet(nn.Module):\n def __init__(self, k=3):\n super().__init__()\n self.k=k\n self.conv1 = nn.Conv1d(k,64,1)\n self.conv2 = nn.Conv1d(64,128,1)\n self.conv3 = nn.Conv1d(128,1024,1)\n self.fc1 = nn.Linear(1024,512)\n self.fc2 = nn.Linear(512,256)\n self.fc3 = nn.Linear(256,k*k)\n\n self.bn1 = nn.BatchNorm1d(64)\n self.bn2 = nn.BatchNorm1d(128)\n self.bn3 = nn.BatchNorm1d(1024)\n self.bn4 = nn.BatchNorm1d(512)\n self.bn5 = nn.BatchNorm1d(256)\n\n\n def forward(self, input):\n # input.shape == (bs,n,3)\n bs = input.size(0)\n xb = F.relu(self.bn1(self.conv1(input)))\n xb = F.relu(self.bn2(self.conv2(xb)))\n xb = F.relu(self.bn3(self.conv3(xb)))\n pool = nn.MaxPool1d(xb.size(-1))(xb)\n flat = nn.Flatten(1)(pool)\n xb = F.relu(self.bn4(self.fc1(flat)))\n xb = F.relu(self.bn5(self.fc2(xb)))\n\n #initialize as identity\n init = torch.eye(self.k, requires_grad=True).repeat(bs,1,1)\n if xb.is_cuda:\n init=init.cuda()\n matrix = self.fc3(xb).view(-1,self.k,self.k) + init\n return matrix",
"_____no_output_____"
],
[
"class Transform(nn.Module):\n def __init__(self):\n super().__init__()\n self.input_transform = Tnet(k=3)\n self.feature_transform = Tnet(k=64)\n self.conv1 = nn.Conv1d(3,64,1)\n self.conv2 = nn.Conv1d(64,128,1)\n self.conv3 = nn.Conv1d(128,1024,1)\n self.bn1 = nn.BatchNorm1d(64)\n self.bn2 = nn.BatchNorm1d(128)\n self.bn3 = nn.BatchNorm1d(1024)\n\n def forward(self, input):\n \n matrix3x3 = self.input_transform(input)\n xb = torch.bmm(torch.transpose(input,1,2), matrix3x3).transpose(1,2) # batch matrix multiplication\n xb = F.relu(self.bn1(self.conv1(xb)))\n matrix64x64 = self.feature_transform(xb)\n xb = torch.bmm(torch.transpose(xb,1,2), matrix64x64).transpose(1,2)\n\n xb = F.relu(self.bn2(self.conv2(xb)))\n xb = self.bn3(self.conv3(xb))\n xb = nn.MaxPool1d(xb.size(-1))(xb)\n output = nn.Flatten(1)(xb)\n return output, matrix3x3, matrix64x64",
"_____no_output_____"
],
[
"class PointNet(nn.Module):\n def __init__(self, classes = 10):\n super().__init__()\n self.transform = Transform()\n self.fc1 = nn.Linear(1024, 512)\n self.fc2 = nn.Linear(512, 256)\n self.fc3 = nn.Linear(256, classes)\n\n\n self.bn1 = nn.BatchNorm1d(512)\n self.bn2 = nn.BatchNorm1d(256)\n self.dropout = nn.Dropout(p=0.3)\n self.logsoftmax = nn.LogSoftmax(dim=1)\n\n def forward(self, input):\n xb, matrix3x3, matrix64x64 = self.transform(input)\n xb = F.relu(self.bn1(self.fc1(xb)))\n xb = F.relu(self.bn2(self.dropout(self.fc2(xb))))\n output = self.fc3(xb)\n return self.logsoftmax(output), matrix3x3, matrix64x64",
"_____no_output_____"
]
],
[
[
"##### Results - Classification\n\n<img src=\"./assets/tut_09_pn12.JPG\" style=\"width:800px\">",
"_____no_output_____"
],
[
"#### PointNet Segmentation Network\n\n<img src=\"./assets/tut_09_pn11.JPG\" style=\"width:800px\">\n",
"_____no_output_____"
],
[
"* Extract local features - Describes each point separately\n* Extract global feature - Describes the entire point cloud\n* Concatenate the local and global features and feed them into a shared mlp:\n * The mlp learns to process the point feature according to a condition. \n * The condition is described by the global feature vector.\n",
"_____no_output_____"
],
[
"##### Semantic Scene Parsing\n\n<img src=\"./assets/tut_09_pn13.JPG\" style=\"width:800px\">\n\n<a href=\"https://arxiv.org/abs/1612.00593\">Image Source</a>",
"_____no_output_____"
],
[
"* Colab notebooks for:\n * <a href=\"https://github.com/nikitakaraevv/pointnet/blob/master/nbs/PointNetClass.ipynb\">PointNet Classification</a>\n * <a href=\"https://github.com/nikitakaraevv/pointnet/blob/master/nbs/PointNetSeg.ipynb\">PointNet Segmentation</a>",
"_____no_output_____"
],
[
"##### Results - Robustness to Missing Data (Classification example)\n* Why is PointNet so robust to missing data?\n\n<img src=\"./assets/tut_09_pn14.JPG\" style=\"width:500px\">\n\n<a href=\"https://arxiv.org/abs/1612.00593\">Image Source</a>",
"_____no_output_____"
],
[
"##### Visualizing Global Point Cloud Features\n\n<img src=\"./assets/tut_09_pn16.JPG\" style=\"width:800px\">\n",
"_____no_output_____"
],
[
"##### Visualize What is Learned by Reconstruction\n* Salient points are discovered!\n\n<img src=\"./assets/tut_09_pn15.png\" style=\"width:800px\">\n\n* The \"critical points\" are those who influenced the global feature vector, a.k.a the pooling layer. \n * The \"critical\" object's geometry is preserved.",
"_____no_output_____"
],
[
"##### Point function visualization\nFor each per-point function $h$ (mlp), calculate the values of $h(p)$ for all the points $p$ in the cube.\n\nRandom 15 functions out of the 1024 learned functions:\n\n<img src=\"./assets/tut_09_pn19.JPG\" style=\"width:800px\">\n\n* Semi-equivalent to filter responses in CNNs\n",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/plasticine/100/000000/not-applicable.png\" style=\"height:50px;display:inline\"> Limitations of PointNet\n<img src=\"./assets/tut_09_pn17.JPG\" style=\"width:800px\">\n\n* No local context for each point\n* Global feature depends on **absolute** coordinate. Hard to generalize to unseen scene configurations\n",
"_____no_output_____"
],
[
"#### Points in Metric Space\n* Learn “kernels” in 3D space and conduct convolution\n* Kernels have compact spatial support\n* For convolution, we need to find neighboring points\n* Possible strategies for range query\n * Ball query (results in more stable features)\n * k-NN query (faster)",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/plasticine/100/000000/approve-and-update.png\" style=\"height:50px;display:inline\"> PointNet v2.0 (PointNet++): Multi-Scale PointNet\n\n<img src=\"./assets/tut_09_pn18.png\" style=\"width:800px\">\n\nRepeated layers:\n* Sample anchor points\n* Find neighborhood of anchor points\n* Apply PointNet in each neighborhood to mimic convolution\n\n* <a href=\"https://arxiv.org/abs/1706.02413\">Image Source</a>",
"_____no_output_____"
],
[
"### More Point Clouds DL solutions:\n* 3DmFV\n* Dynamic Graph CNN\n* PCNN\n* PointCNN\n* KPConv",
"_____no_output_____"
],
[
"## <img src=\"https://img.icons8.com/bubbles/50/000000/list.png\" style=\"height:50px;display:inline\"> 3D Deep Learning Applications\n---",
"_____no_output_____"
],
[
"* Classification (V)\n* Semantic segmentation (V)\n* Part segmentation\n* Registration (Upcoming)\n* Generation (Upcoming)\n* Object detection (Upcoming)\n* Reconstruction\n* Sampling - Downsampling, Upsampling\n* SLAM\n* Normal Estimation\n* and many more...",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/bubbles/50/000000/list.png\" style=\"height:50px;display:inline\"> Registration:\nProblem statment: Find the rotation and translation transformation between objects\n\n<img src=\"./assets/tut_09_pnlk1.JPG\" style=\"width:800px\">\n\n* PointNetLK (blue) - Deep Learning, based on Lucas–Kanade method (Tracking lecture)\n * Comparing 2 point clouds using PointNet features\n* ICP (orange) - Classic registration method\n\n",
"_____no_output_____"
],
[
"<img src=\"./assets/tut_09_pnlk2.JPG\" style=\"width:800px\">\n\nBoth inputs (target and source) are being processed by PointNet architecture\n\n<a href=\"https://arxiv.org/abs/1903.05711\">Image Source</a>",
"_____no_output_____"
],
[
"#### <img src=\"https://img.icons8.com/bubbles/50/000000/list.png\" style=\"height:50px;display:inline\"> Detection:\n* Generate object proposals from a view (e.g., using SSD)\n* Recognize using PointNet\n\n<img src=\"./assets/tut_09_pndet1.JPG\" style=\"width:800px\">\n\n* <a href=\"https://arxiv.org/abs/1711.08488\">Image Source</a>",
"_____no_output_____"
],
[
"##### <img src=\"https://img.icons8.com/bubbles/50/000000/question-mark.png\" style=\"height:50px;display:inline\"> Questions\n* What are the differences between 2D image and a point cloud?\n * Unstructured\n * Varying number of points\n * Unordered",
"_____no_output_____"
],
[
"##### <img src=\"https://img.icons8.com/bubbles/50/000000/question-mark.png\" style=\"height:50px;display:inline\"> Questions\n* Why it might be hard to feed a point cloud as input to a neural network (NN)?\n * Does not rely on a grid\n * Does not have a fixed size\n * Different permutation represent the same point cloud\n\nAll three diffrences influence directly the ability of using NNs!",
"_____no_output_____"
],
[
"##### <img src=\"https://img.icons8.com/bubbles/50/000000/question-mark.png\" style=\"height:50px;display:inline\"> Questions\n* What are the benefits of using a point cloud?\n * Most sensors raw outputs are point clouds (LiDAR)\n * Very efficient representation of 3D data (no empty voxels)\n * Reserve geometric details (no quantization)",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/clouds/100/000000/hand-tools.png\" style=\"height:50px;display:inline\"> Recommended Tools\n---\nPython:\n* Open3D\n* trimesh\n* Ipyvolume - Visualization for Notebooks\n\nDeep Learning:\n* Pytorch3D\n* Kaolin (Pytorch)\n* TensorFlow Graphics",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/clouds/100/000000/hand-tools.png\" style=\"height:50px;display:inline\"> Recommended Tools\n---\nVisualize Tools (drop and view):\n* CloudCompare\n* MeshLab\n\nDatasets:\n* ModelNet\n* ShapeNet\n* PartNet\n* Sydney Urban Opject DAtaset\n* Stanford 3D\n* KITTI\n* ...",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/clouds/100/000000/hand-tools.png\" style=\"height:50px;display:inline\"> Recommended Tools\n---\nFor more 3D deep learnig frameworks and datasets:\n* <a href=\"https://github.com/Yochengliu/awesome-point-cloud-analysis\">awesome-point-cloud-analysis</a>\n* <a href=\"https://github.com/timzhang642/3D-Machine-Learning#datasets\">3D-Machine-Learning#datasets</a> ",
"_____no_output_____"
],
[
"### <img src=\"https://img.icons8.com/bubbles/50/000000/video-playlist.png\" style=\"height:50px;display:inline\"> Recommended Videos\n---\n#### <img src=\"https://img.icons8.com/cute-clipart/64/000000/warning-shield.png\" style=\"height:30px;display:inline\"> Warning!\n* These videos do not replace the lectures and tutorials.\n* Please use these to get a better understanding of the material, and not as an alternative to the written material.\n\n#### Video By Subject\n* 3D Deep Learning\n * General (Both highly recommended):\n * <a href=\"https://www.youtube.com/watch?time_continue=6&v=vfL6uJYFrp4&feature=emb_logo\">3D Deep Learning Tutorial from SU lab at UCSD</a> - Hao Su\n * <a href=\"https://www.youtube.com/watch?v=wLU4YsC_4NY\no\">Geometric deep learning</a> - Michael Bronstein\n * <a href=\"https://www.youtube.com/watch?v=Cge-hot0Oc0&t=24s\">PointNet</a> \n * <a href=\"https://www.youtube.com/watch?v=HIUGOKSLTcE\">3DmFV</a>\n ",
"_____no_output_____"
],
[
"## <img src=\"https://img.icons8.com/dusk/64/000000/prize.png\" style=\"height:50px;display:inline\"> Credits\n----\n* EE 046746 Spring 2021 - <a href=\"https://eliasnehme.github.io\">Elias Nehme</a>\n* EE 046746 Spring 2020 - <a href=\"https://il.linkedin.com/in/dahlia-urbach-97a816123\">Dahlia Urbach</a>\n* Slides - <a href=\"http://www.itzikbs.com/category/research-blog\">Yizhak (Itzik) Ben-Shabat</a>, <a href=\"https://ci2cv.net/people/simon-lucey/\">Simon Lucey (CMU)</a>, <a href=\"https://cseweb.ucsd.edu/~haosu/\">Hao Su, Jiayuan Gu and Minghua Liu(UCSanDiego) </a>\n* Multiple View Geometry in Computer Vision - Hartley and Zisserman - Sections 9,10\n* <a href=\"https://www.springer.com/gp/book/9781848829343\">Computer Vision: Algorithms and Applications</a> - Richard Szeliski - Sections 11,12\n* Research Papers:\n * <a href=\"https://arxiv.org/abs/1612.00593\">PointNet</a> - Qi et al. CVPR 17\n * <a href=\"https://arxiv.org/abs/1706.02413\">PointNet++</a> - Qi et al. NeuroIPS 17\n * <a href=\"https://arxiv.org/abs/1707.02392\">Generative Models for 3D Point Clouds</a> - Qi et al. ICML 18\n * <a href=\"https://arxiv.org/abs/1711.08488\">Frustum PointNets</a> - Qi et al. CVPR 18\n * <a href=\"https://arxiv.org/abs/1903.05711\">PointNetLTK</a> - Aoki et al. CVPR 19\n * etc.\n \n* Icons from <a href=\"https://icons8.com/\">Icon8.com</a> - https://icons8.com\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec6e38abee56d0be0bdc71663ea3ce4dc7b96e4d | 156,932 | ipynb | Jupyter Notebook | udacity_tensorflow/fashion_mnist.ipynb | aidiary/deep-learning-notebooks | 95783c6ba2688e15d362a1fcfec6215861d65a9a | [
"MIT"
]
| 1 | 2019-09-22T14:09:48.000Z | 2019-09-22T14:09:48.000Z | udacity_tensorflow/fashion_mnist.ipynb | aidiary/deep-learning-notebooks | 95783c6ba2688e15d362a1fcfec6215861d65a9a | [
"MIT"
]
| null | null | null | udacity_tensorflow/fashion_mnist.ipynb | aidiary/deep-learning-notebooks | 95783c6ba2688e15d362a1fcfec6215861d65a9a | [
"MIT"
]
| null | null | null | 179.146119 | 58,736 | 0.89816 | [
[
[
"!pip install -U tensorflow_datasets",
"_____no_output_____"
],
[
"import tensorflow as tf\nimport tensorflow_datasets as tfds\n\nimport math\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"import logging\nlogger = tf.get_logger()\nlogger.setLevel(logging.ERROR)",
"_____no_output_____"
],
[
"dataset, metadata = tfds.load('fashion_mnist', as_supervised=True, with_info=True)",
"_____no_output_____"
],
[
"class_names = ['T-shirt/top', 'Trouser', 'Pullover', 'Dress', 'Coat',\n 'Sandal', 'Shirt', 'Sneaker', 'Bag', 'Ankle boot']",
"_____no_output_____"
],
[
"dataset",
"_____no_output_____"
],
[
"metadata",
"_____no_output_____"
],
[
"train_dataset, test_dataset = dataset['train'], dataset['test']",
"_____no_output_____"
],
[
"num_train_examples = metadata.splits['train'].num_examples\nnum_test_examples = metadata.splits['test'].num_examples",
"_____no_output_____"
],
[
"num_train_examples",
"_____no_output_____"
],
[
"num_test_examples",
"_____no_output_____"
],
[
"train_dataset",
"_____no_output_____"
],
[
"def normalize(images, labels):\n images = tf.cast(images, tf.float32)\n images /= 255\n return images, labels",
"_____no_output_____"
],
[
"train_dataset = train_dataset.map(normalize)\ntest_dataset = test_dataset.map(normalize)",
"_____no_output_____"
],
[
"for image, label in test_dataset.take(1):\n break\nprint(type(image), type(label))\nprint(label)\nimage = image.numpy().reshape((28, 28))",
"<class 'tensorflow.python.framework.ops.EagerTensor'> <class 'tensorflow.python.framework.ops.EagerTensor'>\ntf.Tensor(6, shape=(), dtype=int64)\n"
],
[
"image.shape",
"_____no_output_____"
],
[
"plt.figure()\nplt.imshow(image, cmap=plt.cm.binary)\nplt.colorbar()\nplt.grid(False)\nplt.show()",
"_____no_output_____"
],
[
"plt.figure(figsize=(10, 10))\ni = 0\nfor (image, label) in test_dataset.take(25):\n image = image.numpy().reshape((28, 28))\n plt.subplot(5, 5, i + 1)\n plt.xticks([])\n plt.yticks([])\n plt.grid(False)\n plt.imshow(image, cmap=plt.cm.binary)\n plt.xlabel(class_names[label])\n i += 1\nplt.show()",
"_____no_output_____"
],
[
"model = tf.keras.Sequential([\n tf.keras.layers.Flatten(input_shape=(28, 28, 1)),\n tf.keras.layers.Dense(128, activation=tf.nn.relu),\n tf.keras.layers.Dense(10, activation=tf.nn.softmax)\n])",
"_____no_output_____"
],
[
"model.summary()",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten (Flatten) (None, 784) 0 \n_________________________________________________________________\ndense (Dense) (None, 128) 100480 \n_________________________________________________________________\ndense_1 (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 101,770\nTrainable params: 101,770\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"model.compile(optimizer='adam',\n loss='sparse_categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
],
[
"num = 0\nfor image, label in test_dataset:\n num += 1\nprint(num)",
"10000\n"
]
],
[
[
"- PyTorchのDataLoaderの機能をDatasetのメソッドで行う",
"_____no_output_____"
]
],
[
[
"BATCH_SIZE = 32\ntrain_dataset = train_dataset.repeat().shuffle(num_train_examples).batch(BATCH_SIZE)\ntest_dataset = test_dataset.batch(BATCH_SIZE)",
"_____no_output_____"
]
],
[
[
"- TensorFlowはPyTorchと違ってchannel-lastなので注意",
"_____no_output_____"
]
],
[
[
"for image, label in train_dataset:\n print(image.shape, label.shape)\n break",
"(32, 28, 28, 1) (32,)\n"
],
[
"# 1epochに何batchあるか?",
"_____no_output_____"
],
[
"steps_per_epoch = math.ceil(num_train_examples / BATCH_SIZE)\nsteps_per_epoch",
"_____no_output_____"
],
[
"model.fit(train_dataset, epochs=5, steps_per_epoch=steps_per_epoch)",
"Train for 1875 steps\nEpoch 1/5\n1875/1875 [==============================] - 17s 9ms/step - loss: 0.4951 - accuracy: 0.8249\nEpoch 2/5\n1875/1875 [==============================] - 10s 6ms/step - loss: 0.3721 - accuracy: 0.8660\nEpoch 3/5\n1875/1875 [==============================] - 11s 6ms/step - loss: 0.3358 - accuracy: 0.8761\nEpoch 4/5\n1875/1875 [==============================] - 10s 6ms/step - loss: 0.3120 - accuracy: 0.8856\nEpoch 5/5\n1875/1875 [==============================] - 10s 6ms/step - loss: 0.2949 - accuracy: 0.8903\n"
],
[
"test_loss, test_accuracy = model.evaluate(\n test_dataset,\n steps=math.ceil(num_test_examples / 32))",
"313/313 [==============================] - 1s 4ms/step - loss: 0.3587 - accuracy: 0.8710\n"
],
[
"test_accuracy",
"_____no_output_____"
],
[
"for test_images, test_labels in test_dataset.take(1):\n test_images = test_images.numpy()\n test_labels = test_labels.numpy()\n predictions = model.predict(test_images)",
"_____no_output_____"
],
[
"predictions.shape",
"_____no_output_____"
],
[
"predictions[0]",
"_____no_output_____"
],
[
"np.argmax(predictions[0])",
"_____no_output_____"
],
[
"test_labels[0]",
"_____no_output_____"
],
[
"def plot_image(i, predictions_array, true_labels, images):\n predictions_array, true_label, img = predictions_array[i], true_labels[i], images[i]\n plt.grid(False)\n plt.xticks([])\n plt.yticks([])\n\n plt.imshow(img[...,0], cmap=plt.cm.binary)\n\n predicted_label = np.argmax(predictions_array)\n if predicted_label == true_label:\n color = 'blue'\n else:\n color = 'red'\n \n plt.xlabel(\"{} {:2.0f}% ({})\".format(class_names[predicted_label],\n 100 * np.max(predictions_array),\n class_names[true_label]),\n color=color)\n\ndef plot_value_array(i, predictions_array, true_label):\n predictions_array, true_label = predictions_array[i], true_label[i]\n plt.grid(False)\n plt.xticks([])\n plt.yticks([])\n thisplot = plt.bar(range(10), predictions_array, color=\"#777777\")\n plt.ylim([0, 1]) \n predicted_label = np.argmax(predictions_array)\n \n thisplot[predicted_label].set_color('red')\n thisplot[true_label].set_color('blue')",
"_____no_output_____"
],
[
"i = 0\nplt.figure(figsize=(6,3))\nplt.subplot(1,2,1)\nplot_image(i, predictions, test_labels, test_images)\nplt.subplot(1,2,2)\nplot_value_array(i, predictions, test_labels)",
"_____no_output_____"
],
[
"i = 12\nplt.figure(figsize=(6,3))\nplt.subplot(1,2,1)\nplot_image(i, predictions, test_labels, test_images)\nplt.subplot(1,2,2)\nplot_value_array(i, predictions, test_labels)",
"_____no_output_____"
],
[
"# Plot the first X test images, their predicted label, and the true label\n# Color correct predictions in blue, incorrect predictions in red\nnum_rows = 5\nnum_cols = 3\nnum_images = num_rows*num_cols\nplt.figure(figsize=(2*2*num_cols, 2*num_rows))\nfor i in range(num_images):\n plt.subplot(num_rows, 2*num_cols, 2*i+1)\n plot_image(i, predictions, test_labels, test_images)\n plt.subplot(num_rows, 2*num_cols, 2*i+2)\n plot_value_array(i, predictions, test_labels)",
"_____no_output_____"
],
[
"# Grab an image from the test dataset\nimg = test_images[0]\nprint(img.shape)",
"(28, 28, 1)\n"
],
[
"img = np.array([img])\nprint(img.shape)",
"(1, 28, 28, 1)\n"
],
[
"predictions_single = model.predict(img)\npredictions_single",
"_____no_output_____"
],
[
"np.argmax(predictions_single[0])",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6e4931e5c065022cbb01ed49672aeca7483138 | 675,301 | ipynb | Jupyter Notebook | material_aula/lecture06.ipynb | nilodna/python-basico | 38cd76007347d0f1f77189862cf33546a8e60a7d | [
"Apache-2.0"
]
| 6 | 2020-10-30T23:09:37.000Z | 2021-11-26T00:35:32.000Z | material_aula/lecture06.ipynb | nilodna/python-basico | 38cd76007347d0f1f77189862cf33546a8e60a7d | [
"Apache-2.0"
]
| null | null | null | material_aula/lecture06.ipynb | nilodna/python-basico | 38cd76007347d0f1f77189862cf33546a8e60a7d | [
"Apache-2.0"
]
| 3 | 2021-07-15T13:11:37.000Z | 2021-11-26T13:40:50.000Z | 338.327154 | 129,932 | 0.922568 | [
[
[
"### Boa noite,\n\ncomeçaremos em breve.\n\n https://forms.gle/EhjkK8RFvVe6vLUU9",
"_____no_output_____"
],
[
"**Manchas de óleo (2019) - IBAMA**",
"_____no_output_____"
]
],
[
[
"import warnings\nwarnings.filterwarnings('ignore')\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\n\nfrom windrose import WindroseAxes\nimport matplotlib.cm as cm\n\n%matplotlib inline\n\nurl = \"http://www.ibama.gov.br/phocadownload/emergenciasambientais/2019/manchasdeoleo/2019-12-02_LOCALIDADES_AFETADAS.xlsx\"\ndf = pd.read_excel(\n url,\n parse_dates=[\"Data_Avist\", \"Data_Revis\"],\n)\n\n# Calculando o tempo de permanência da mancha em cada localidade\ndf[\"dias\"] = (df[\"Data_Revis\"] - df[\"Data_Avist\"]).dt.days\n\n# tratando as colunas de tempo\ndf['Data_Avist'] = df['Data_Avist'].dt.strftime('%Y-%m-%d').str.replace('NaT', 'na')\ndf[\"Data_Revis\"] = df[\"Data_Revis\"].dt.strftime(\"%Y-%m-%d\").str.replace(\"NaT\", \"na\")\ndf[\"dias\"] = df[\"dias\"].astype(float)\n\ndf.dropna(inplace=True)",
"_____no_output_____"
],
[
"# um pouco de limpeza de dados (data cleaning)\n\n# removendo linhas onde o status == nao observado\nfiltr = (df['Status'] == 'Oleo Nao Observado')\ndf = df[~filtr].copy()\n\n# removendo registros onde o dias == 0\nfiltr = (df['dias'] == 0)\ndf = df[~filtr].copy()\n\ndf.head()",
"_____no_output_____"
],
[
"# Estado mais afetado\ndf.groupby('sigla_uf').count()['estado'].plot(kind='barh')\n",
"_____no_output_____"
],
[
"# Município mais afetado\ndf.groupby('municipio').count()\\\n .sort_values(by='Data_Revis', \n ascending=False)\\\n .head(10)",
"_____no_output_____"
],
[
"# primeiros e ultimos afetados\n# filtr = (df['Data_Avist'] == df.min()['Data_Avist'])\n# df[filtr].head(5)\n\ndf[df['Data_Revis'] == df.max()['Data_Revis']].sort_values(by='dias', ascending=False).head(5)\n",
"_____no_output_____"
],
[
"# afetados por mais tempo\ndf.groupby(by='localidade').sum().sort_values('dias', ascending=False).head(5)",
"_____no_output_____"
],
[
"# funcao: será utilizada no date_parser\ndef dateparse(x):\n return pd.datetime.strptime(x, '%Y %m %d %H %M')\ndf = pd.read_csv('../dados/pnboia_vitoria.csv',\n parse_dates={'datetime': ['Year', 'Month', 'Day', 'Hour', 'Minute']},date_parser=dateparse, decimal=',')\n# agora podemos converter nossa coluna datetime em index\ndf.set_index('datetime', inplace=True)\n# replace numeros invalidos\ndf = df.replace(-9999.0, np.nan)\nws,wd = df['Wspd'], df['Wdir']\nax = WindroseAxes.from_ax()\nax.bar(wd, ws, normed=True, opening=0.8, edgecolor='white')\nax.set_legend()",
"_____no_output_____"
]
],
[
[
"# Aula 6 - Mapas\n\n",
"_____no_output_____"
],
[
"**materiais de apoio**\n\n- Documentação cartopy:\n- NaturalEarth: conjunto de informações geográficas de acesso público [[link]](https://www.naturalearthdata.com/downloads/) ou [[link]](https://github.com/nilodna/natural-earth-vector/tree/master/packages/Natural_Earth_quick_start)\n- OpenStreetMap: iniciativa colaborativa para acesso livre à informações geográficas [[link]](https://download.geofabrik.de/south-america/brazil.html)\n- ",
"_____no_output_____"
],
[
"### O que é cartopy?\n\n- processamento de dados geoespaciais\n- mapas com alta qualidade\n- jornais mundo afora\n- controle dos elementos cartográficos",
"_____no_output_____"
],
[
"**Criando geofiguras**\n\n- criar ```GeoAxes```\n- integra matplotlib e cartopy\n- utilizando uma projeção\n- Duas formas:\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n# padrão de importação: crs é o core do pacote\nimport cartopy.crs as ccrs\n\n%matplotlib inline\n\n# modo 1:\nfig = plt.figure()\nax = fig.add_subplot(projection=ccrs.PlateCarree())",
"_____no_output_____"
],
[
"fig,ax = plt.subplots(subplot_kw={'projection': ccrs.PlateCarree()})",
"_____no_output_____"
]
],
[
[
"**Principais métodos**\n\n\n- **ax.coastlines():** inserir linha de costa\n- **ax.set_global():** ajusta limites geográficos para o globo\n- **ax.set_extent():** ajusta os limites geográficos para uma região\n- **ax.stock_img():** adiciona uma imagem padrão ao mapa\n- **ax.imshow():** adiciona imagem personalizada ao mapa\n- **ax.add_geometries():** adiciona geometrias do shapely (pacote de leitura de shapefiles)\n- **ax.gridlines():** adiciona linhas de grade associadas às coordenadas",
"_____no_output_____"
]
],
[
[
"# apenas pq podemos, vamos criar uma função para retornar um mapa\n\ndef create_map(projection):\n fig,ax = plt.subplots(subplot_kw={'projection': projection})\n \n return fig,ax",
"_____no_output_____"
],
[
"proj = ccrs.PlateCarree()\n\nfig,ax = create_map(proj)\n\n# adicionando linha de costa\nax.coastlines()\n\n# adicionando gridline\nax.gridlines()\n\n# adicionando imagem padrão\nax.stock_img()",
"_____no_output_____"
]
],
[
[
"**Projeção de mapas: problemas**\n\n- Terra não é plana\n- projetar esfera em plano: distorção\n\n",
"_____no_output_____"
]
],
[
[
"# Robinson, Mercator, Orthographic, InterruptedGoodeHomolosine\n\nfig,ax = create_map(ccrs.PlateCarree())\nax.coastlines()\nax.stock_img()\nax.gridlines()",
"_____no_output_____"
]
],
[
[
"**Criando um mapa regional**\n\n- área de estudo\n- mapa da PCSE\n- Unidade de Conservação do Arquipélago de Alcatrazes (ICMBio)\n",
"_____no_output_____"
]
],
[
[
"# importando diversos pacotes novos\nimport matplotlib.pyplot as plt\nimport matplotlib.patches as mpatches\nimport os\n# pacotes relacionados às funcionalidades do cartopy\nimport cartopy.crs as ccrs\nimport cartopy.feature as cfeature\nimport cartopy.io.shapereader as shpreader\nimport shapely.geometry as sgeom",
"_____no_output_____"
]
],
[
[
"**Primeiros passos**\n\n- determinar projeção\n- limites geográficos\n- google maps para auxiliar\n - canto inferior esquerdo\n - canto superior direito",
"_____no_output_____"
]
],
[
[
"# projecao\nproj = ccrs.PlateCarree()\n\n# posicao do canto inferior esquerdo\nlower_lon = -50\nlower_lat = -30\n\n# posicao do canto superior direito\nupper_lon = -41\nupper_lat = -22",
"_____no_output_____"
]
],
[
[
"**Linha de costa**\n\n- padrão: NaturalEarth (1cm:10m)\n- download feito automaticamente\n- demora, mas depois fica rápido\n- para mais conjuntos, visite: [Natural Earth](https://www.naturalearthdata.com/)",
"_____no_output_____"
]
],
[
[
"# montando uma lista para facilitar enviar para o GeoAxes os limites geográficos. \nextent = [lower_lon, upper_lon, lower_lat, upper_lat]\n# criando mapa com a função \nfig,ax = create_map(ccrs.PlateCarree())\n# alterando para os limites criados acima\nax.set_extent(extent)\n# inserindo linha de costa com resolução de 1:110, teste com 10m e 50m\nax.coastlines('10m')",
"_____no_output_____"
]
],
[
[
"Dentre os conjuntos do **NaturalEarth**, podemos ainda utilizar:",
"_____no_output_____"
]
],
[
[
"import cartopy.feature as cfeature\n\nfig,ax = create_map(ccrs.PlateCarree())\nax.set_extent(extent)\nax.coastlines('10m')\n\ncoastline_10m = cfeature.NaturalEarthFeature(\n category='physical',name='coastline',scale='10m',\n facecolor=cfeature.COLORS['land'])\n\nbathy_0m = cfeature.NaturalEarthFeature(category='physical',name='bathymetry_L_0',scale='10m')\n\n# inserting information over the map\nax.add_feature(coastline_10m, edgecolor='black', linewidth=.05)\nax.add_feature(bathy_0m, facecolor='lightblue', alpha=.3)",
"_____no_output_____"
]
],
[
[
"criando uma função para auxiliar:",
"_____no_output_____"
]
],
[
[
"def make_map(fig=None, ax=None, extent=[-50,-41,-30,-22], projection=ccrs.PlateCarree()):\n \"\"\" \n documente-a como exercício para entender a \n ação de cada linha\n \"\"\"\n ax.set_extent(extent, crs=ccrs.PlateCarree())\n\n coastline_10m = cfeature.NaturalEarthFeature(\n category='physical',name='coastline',scale='10m',\n facecolor=cfeature.COLORS['land'])\n\n bathy_0m = cfeature.NaturalEarthFeature(\n category='physical',name='bathymetry_L_0',scale='10m')\n\n # inserting information over the map\n ax.add_feature(coastline_10m, \n edgecolor='black',\n linewidth=.1)\n ax.add_feature(bathy_0m,\n facecolor='lightblue',\n alpha=.3)\n \n return ax",
"_____no_output_____"
]
],
[
[
"**Meridianos e Paralelos como grids**\n\n- ```.gridlines()```\n- limites geográficos como referência\n- customizar: grossura, cor, transparencia, etc ...",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\nax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\n\n# criando as gridlines\ngl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,\n linewidth=.4, color='gray', \n alpha=0.5, linestyle='--', \n xlocs=np.linspace(extent[0], extent[1], 5),\n ylocs=np.linspace(extent[3], extent[2], 5))",
"_____no_output_____"
]
],
[
[
"Rótulos aparecem ao redor de todo o mapa, mas podemos customizar isso também!",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\nax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\n\n# criando as gridlines\ngl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,\n linewidth=.4, color='gray', \n alpha=0.5, linestyle='--', \n xlocs=np.linspace(extent[0], extent[1], 5),\n ylocs=np.linspace(extent[3], extent[2], 5))\n\ngl.top_labels = False\ngl.right_labels = False\n\n# dicionario com configurações dos rótulos\ndict_style_labels = {'color': 'black', 'fontsize': 8}\n\ngl.ylabel_style = dict_style_labels\ngl.xlabel_style = dict_style_labels",
"_____no_output_____"
]
],
[
[
"Vamos criar uma segunda função, para configurar o mapa em termo de meridianos e paralelos:",
"_____no_output_____"
]
],
[
[
"def configuring_map(ax, extent):\n # criando as gridlines \n gl = ax.gridlines(crs=ccrs.PlateCarree(), draw_labels=True,\n linewidth=.4, color='gray', \n alpha=0.5, linestyle='--', \n xlocs=np.linspace(extent[0], extent[1], 5),\n ylocs=np.linspace(extent[3], extent[2], 5))\n\n gl.top_labels = False\n gl.right_labels = False\n\n # dicionario com configurações dos rótulos\n dict_style_labels = {\n 'color': 'black',\n 'fontsize': 8\n }\n\n gl.ylabel_style = dict_style_labels",
"_____no_output_____"
]
],
[
[
"**Notem que:**\n\n- duas funções capazes de criar um mapa para qualquer lugar no globo\n- podemos começar a pensar em criar nossas próprias coleções de funçõs, para não nos repetirmos!\n\n",
"_____no_output_____"
]
],
[
[
"# mapa Espirito Santo (centrado em Vitória)\nlower = [-40.99, -21.177]\nupper = [-39.62, -19.579]\nextent2 = [lower[0], upper[0], lower[1], upper[1]]\n\nfig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\n\nax = make_map(fig=fig, ax=ax, extent=extent2, projection=ccrs.PlateCarree())\nconfiguring_map(ax, extent=extent2)",
"_____no_output_____"
]
],
[
[
"**Unidade de Conservação**\n\n- estabelecer limites geográficos para criar minimapa\n- inserir polígono indicando aonde iremos dar zoom no mapa",
"_____no_output_____"
]
],
[
[
"# coordenadas geográficas do limite do minimapa\nlower = [-24.3, -45.949]\nupper = [-23.8, -45.4]\nextent_UC = [lower[1], upper[1], lower[0], upper[0]]\n# usando um método novo do matplotlib, criamos um polígono\n\nextent_box = sgeom.box(extent_UC[0], extent_UC[2], extent_UC[1], extent_UC[3])\n\n# plotando mapa\nfig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\nax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\nconfiguring_map(ax, extent)\n_ = ax.add_geometries([extent_box], ccrs.PlateCarree(), \n facecolor='coral', # cor do polígono\n edgecolor='red', # cor das arestas\n alpha=.3) # transparência",
"_____no_output_____"
]
],
[
[
"**Inserindo minimapa**",
"_____no_output_____"
]
],
[
[
"# coordenadas geográficas do limite do minimapa\nlower,upper = [-24.3, -45.949], [-23.74, -45.32]\nextent_UC = [lower[1], upper[1], lower[0], upper[0]]\n# usando um método novo do matplotlib, criamos um polígono\nextent_box = sgeom.box(extent_UC[0], extent_UC[2], extent_UC[1], extent_UC[3])\n# plotando mapa\nfig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\nax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\nconfiguring_map(ax, extent)\n_ = ax.add_geometries([extent_box], ccrs.PlateCarree(), \n facecolor='coral', # cor do polígono\n edgecolor='red', # cor das arestas\n alpha=.3) # transparência\n# setting axes position and size\nleft,bottom = 0.465, 0.25\nwidth, height = 0.55, 0.20\nrect = [left,bottom,width,height]\n\n# creating axes 2 for minimap\nax2 = plt.axes(rect, projection=ccrs.PlateCarree())\nax2 = make_map(fig=fig, ax=ax2, extent=extent_UC)\n\nplt.savefig('tmp_map.png', dpi=150)\n",
"_____no_output_____"
]
],
[
[
"**Lendo shapefiles com cartopy.shapereader**\n\n- requisitos: shapely",
"_____no_output_____"
]
],
[
[
"def criando_mapa(extent):\n # coordenadas geográficas do limite do minimapa\n lower = [-24.3, -45.949]\n upper = [-23.74, -45.32]\n extent_UC = [lower[1], upper[1], lower[0], upper[0]]\n # usando um método novo do matplotlib, criamos um polígono\n extent_box = sgeom.box(extent_UC[0], extent_UC[2], extent_UC[1], extent_UC[3])\n \n # plotando mapa\n fig = plt.figure(figsize=(15/2.54,20/2.54))\n ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\n ax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\n configuring_map(ax, extent)\n _ = ax.add_geometries([extent_box], ccrs.PlateCarree(), \n facecolor='coral', # cor do polígono\n edgecolor='red', # cor das arestas\n alpha=.3) # transparência\n\n # setting axes position and size\n left,bottom = 0.465, 0.25\n width, height = 0.55, 0.20\n rect = [left,bottom,width,height]\n\n # creating axes 2 for minimap\n ax2 = plt.axes(rect, projection=ccrs.PlateCarree())\n ax2 = make_map(fig=fig, ax=ax2, extent=extent_UC)\n \n return fig,ax,ax2",
"_____no_output_____"
],
[
"# criei uma função temporaria para facilitar a visualização\nfig,ax,ax2 = criando_mapa(extent)\n\n# adicionando linha de costa do OpenStreetMap\nline = shpreader.Reader('../dados/OSM/OSM_BRA_coastline/lines.shp')\nax2.add_geometries(line.geometries(), ccrs.PlateCarree(), facecolor='none',edgecolor='black',linewidth=.05)\n\n# adicionando continente do OpenStreetMap\nland = shpreader.Reader('../dados/OSM/OSM_BRA_land/land.shp')\nax2.add_geometries(land.geometries(), ccrs.PlateCarree(),facecolor=cfeature.COLORS['land'],edgecolor='black',linewidth=0.05)\n\n# adicionando área da unidade de conservação (terra e água)\nucs = shpreader.Reader('../dados/alcatrazes/alcatrazes.shp')\nax2.add_geometries(ucs.geometries(), ccrs.PlateCarree(),facecolor='coral',alpha=.3,edgecolor='coral')\n\nplt.savefig('tmp_map.png', dpi=150)",
"_____no_output_____"
]
],
[
[
"Como temos tudo elaborado, transformei o bloco de códigos para inserir o minimapa de Alcatrazes em uma função que chamei de:\n```create_alcatrazes(ax)``` para facilitar.",
"_____no_output_____"
]
],
[
[
"def create_alcatrazes(ax):\n\n # coordenadas geográficas do limite do minimapa\n lower_lat = -24.3\n lower_lon = -45.949\n upper_lat = -23.74\n upper_lon = -45.32\n\n extent_UC = [lower_lon, upper_lon, lower_lat, upper_lat]\n\n # usando um método novo do matplotlib, criamos um polígono\n extent_box = sgeom.box(extent_UC[0], extent_UC[2], \n extent_UC[1], extent_UC[3])\n\n # adicionando polígono no mapa\n ax.add_geometries([extent_box], ccrs.PlateCarree(), \n facecolor='coral', # cor do polígono\n edgecolor='red', # cor das arestas\n alpha=.3) # transparência\n\n # setting axes position and size\n left = 0.465\n bottom = 0.25\n width = 0.55\n height = 0.20\n\n rect = [left,bottom,width,height]\n\n # creating axes 2 for minimap\n ax2 = plt.axes(rect, projection=ccrs.PlateCarree())\n ax2 = make_map(fig=fig, ax=ax2, extent=extent_UC)\n\n # adicionando linha de costa do OpenStreetMap\n line = shpreader.Reader('../dados/OSM/OSM_BRA_coastline/lines.shp')\n ax2.add_geometries(line.geometries(), ccrs.PlateCarree(),facecolor='none',edgecolor='black',linewidth=.05)\n\n # adicionando continente do OpenStreetMap\n land = shpreader.Reader('../dados/OSM/OSM_BRA_land/land.shp')\n ax2.add_geometries(land.geometries(), ccrs.PlateCarree(),facecolor=cfeature.COLORS['land'],edgecolor='black',linewidth=0.05)\n\n # adicionando área da unidade de conservação (terra e água)\n ucs = shpreader.Reader('../dados/alcatrazes/alcatrazes.shp')\n ax2.add_geometries(ucs.geometries(), ccrs.PlateCarree(),facecolor='coral',alpha=.3,edgecolor='coral')\n \n return ax2",
"_____no_output_____"
]
],
[
[
"**Inserindo localizações com scatter**\n\n- dicionário com informações para plotar\n- estações meteorológicas, fundeios, etc\n",
"_____no_output_____"
]
],
[
[
"# criando dicionário com os locais e metadados\n\ninsitu = {\n 'simcosta-css': {\n 'coords': (-23.831, -45.423), # lat,lon\n 'marker': 's',\n 'color': 'purple',\n 'label': 'SiMCosta - SP',\n 'minimap': True # plotar no minimapa?\n },\n \n 'simcosta-rj': {\n 'coords': (-22.967, -43.131),\n 'marker': 's',\n 'color': 'green',\n 'label': 'SiMCosta - RJ',\n 'minimap': False\n },\n \n 'base-ubatuba': {\n 'coords': (-23.499, -45.119),\n 'marker': 'o',\n 'color': 'k',\n 'label': 'Base Ubatuba (IOUSP)',\n 'minimap': False\n \n },\n 'base-cananeia': {\n 'coords': (-25.02, -47.925),\n 'marker': 'o',\n 'color': 'r',\n 'label': u'Base Cananéia (IOUSP)',\n 'minimap': False\n }\n}",
"_____no_output_____"
],
[
"# código completo\n# projecao\nproj = ccrs.PlateCarree()\nlower_lon, lower_lat = -50, -30\nupper_lon, upper_lat = -41, -22\nextent = [lower_lon, upper_lon, lower_lat, upper_lat]\n# criando a base do mapa:\nfig = plt.figure(figsize=(15/2.54,20/2.54))\nax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())\nax = make_map(fig=fig, ax=ax, extent=extent, projection=ccrs.PlateCarree())\nconfiguring_map(ax, extent)\n# inserindo o minimapa completo\nax2 = create_alcatrazes(ax)\n\n# inserindo marcadores de pontos de observação\nfor local,metadados in insitu.items():\n # extraindo metadados\n lat,lon = metadados['coords']\n marker = metadados['marker']\n color = metadados['color']\n label = metadados['label']\n \n # varia o ax_plot de acordo com o mapa que deve exibir a informação\n if metadados['minimap']:\n ax_plot = ax2\n else:\n ax_plot = ax\n \n ax_plot.scatter(lon, lat, s=40, marker=marker, color=color, label=label, zorder=50)\n\nplt.savefig('mapa_alcatrazes.png', dpi=150)",
"_____no_output_____"
]
],
[
[
"**Exercícios**\n\nSabendo como elaborar um mapa, utilize o conjunto de informações da última aula para reproduzir alguns mapas.\n\n1) Faça um mapa indicando os Estados mais afetados, utilizando como marker='o' (uma bola) e essa bola deve variar de tamanho segundo a quantidade de regiões atingidas apenas.\n\n2) Selecione uma das regiões acima e reproduza um mapa mais detalhado, indicando as praias que foram atingidas. Tente variar as cores dos marcadores segundo a evolução temporal: primeira afetada com uma cor e varia até a cor da última afetada.\n\n3) Utilize a coluna \"dias\" do dataframe e refaça o mapa 1), mas agora varie o tamanho dos marcadores segundo o tempo de permanência somado dos Estados. Para isso use o agrupamento do pandas com o método ```.sum()```.\n\n**Dicas**:\n\n- edite as funções que criamos hoje, para se adequar aos casos",
"_____no_output_____"
],
[
"Para auxiliar, é necessário converter o formato de latitude e longitude presente no dataframe. Para isso, use a função abaixo, obtida com o [Filipe Fernandes](ttps://gist.github.com/ocefpaf/60bec6fbd252107de11073376bad7925) e insira essa função usando o método ```.apply()``` do pandas para converter.\n\n```python\ndf['new_lon'] = df['Longitude'].apply(fix_pos)\n```",
"_____no_output_____"
]
],
[
[
"def fix_pos(pos):\n # source: https://gist.github.com/ocefpaf/60bec6fbd252107de11073376bad7925\n deg, rest = pos.split(\"°\")\n mi, rest = rest.strip().split(\"'\")\n sec, hem = rest.strip().split('\"')\n hem = hem.strip()\n\n deg, mi, sec = map(float, (deg, mi, sec))\n if hem in [\"S\", \"W\"]:\n sign = -1\n elif hem in [\"N\", \"E\"]:\n sign = +1\n else:\n raise ValueError(f\"Unrecognized sign {sign}, expected 'S', 'W', 'E', or 'N'\")\n\n return sign * (deg + mi/60 + sec/60/60)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6e6349e99fe3c4bb66744d7c462dcd2d84aaa6 | 236,813 | ipynb | Jupyter Notebook | sandbox/Transient-01-Simulations.ipynb | gnizq64/DESI-HighSchool | f9a8430b536acda8d6720792e982d603dc289a58 | [
"BSD-3-Clause"
]
| null | null | null | sandbox/Transient-01-Simulations.ipynb | gnizq64/DESI-HighSchool | f9a8430b536acda8d6720792e982d603dc289a58 | [
"BSD-3-Clause"
]
| null | null | null | sandbox/Transient-01-Simulations.ipynb | gnizq64/DESI-HighSchool | f9a8430b536acda8d6720792e982d603dc289a58 | [
"BSD-3-Clause"
]
| null | null | null | 990.849372 | 138,112 | 0.953723 | [
[
[
"# Searching for Supernova Spectra (and other Transients) in DESI\n\nDESI is designed to study dark energy by carefully measuring the 3D positions of millions of galaxies. During the survey, by chance DESI will catch a fraction of the observed galaxies containing powerful astrophysical explosions such as supernovae, kilonovae, tidal disruption events, and other energetic events. If we train our software to specifically look for the signatures of these explosions, we can potentially discover and classify thousands of them each year.\n\nFirst, we'll focus on supernova and how we tell them apart.\n\n### Supernova Types: Core-Collapse\n\nIn most cases, supernova are the final stage of the life cycle of very massive stars (>10 times the mass of the Sun). When the core of a massive star burns through its available nuclear fuel, it stops producing enough energy to hold up the outer envelope of the star. The star then collapses under its own weight in a fraction of a second and explodes. The remnant left behind will be a neutron star, or if the progenitor star is massive enough, a black hole. This is called a **core collapse** supernova for obvious reasons.\n\n<img src=\"figures/ccsn.png\" width=\"700px\"/>\n\n### Supernova Types: Binary System\n\nNot all supernova occur due to a core collapse. In binary star systems made up of a compact White Dwarf and an aging main sequence star, the White Dwarf can gravitationally attract material from the envelope of its companion star. If enough material accumulates on the surface of the White Dwarf so that its total mass exceeds 1.44 solar masses (a.k.a. the [*Chandrasekhar Limit*](https://en.wikipedia.org/wiki/Chandrasekhar_limit)) then a runaway nuclear reaction occurs that blows up the White Dwarf and produces an intensely bright explosion. These are called Type Ia supernovae, and they are very important for measuring distances in cosmology.\n\n<img src=\"figures/sn_ia.png\" width=\"700px\"/>\n\n### Telling Supernovae Types Apart\n\nSupernova types can be distinguished by looking at how their brightness changes over time (their [light curves](https://en.wikipedia.org/wiki/Light_curve)), but the most reliable method is to look at the intensity of the light as a function of wavelength -- the [spectrum](https://en.wikipedia.org/wiki/Astronomical_spectroscopy) of the supernova. Supernova spectra have distinct **very broad emission and absorption lines** produced by hot gas moving away from the explosion at high velocity. These lines provide distinct \"fingerprints\" that distinguish the various types. The figure below, from [a paper published in 1997](https://ned.ipac.caltech.edu/level5/March03/Filippenko/paper.pdf), shows the signature spectra of several observed supernovae.\n\n<img src=\"figures/sne_filippenko.png\" width=\"600px\"/>\n\nThe flow chart below, taken from the August 2004 issue of [*Astronomy Magazine*](https://astronomy.com/magazine/2004/08/know-your-supernovae), shows how the classifications are made.\n\n<img src=\"figures/supernova-matrix.GIF\" width=\"600px\"/>",
"_____no_output_____"
],
[
"## Example Spectra in the Bright Galaxy Survey\n\nThis notebook shows how supernova will appear when DESI catches them in one of the 4 million galaxies in the Brigth Galaxy Survey.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\n\nfrom astropy.io import fits\n\nmpl.rc('font', size=14)",
"_____no_output_____"
]
],
[
[
"### Spectrum Plots\n\nHere is a file containing simulated spectra from the DESI Bright Galaxy Survey (BGS). The first spectrum shows what a normal galaxy looks like. The rest show galaxies \"contaminated\" with light from a luminous supernova. Several types of supernova are simulated. Take a close look at the features in the spectrum and see if you can identify the difference.",
"_____no_output_____"
]
],
[
[
"# Open the data file and show its contents.\nhdus = fits.open('data/spectrum-examples.fits')\nhdus.info()",
"Filename: data/spectrum-examples.fits\nNo. Name Ver Type Cards Dimensions Format\n 0 WAVE 1 PrimaryHDU 8 (6265,) float64 \n 1 FLUX 1 ImageHDU 8 (6265, 6) float64 \n 2 IVAR 1 ImageHDU 8 (6265, 6) float64 \n 3 MASK 1 ImageHDU 10 (6265, 6) int32 (rescales to uint32) \n 4 SPECTYPE 1 BinTableHDU 11 6R x 1C [10A] \n"
]
],
[
[
"#### Raw Spectra\n\nThese are fluxes as they would be observed in a fiber, after we subtract known backgrounds from the night sky. They show simulated spectra from:\n1. An ordinary BGS galaxy.\n2. A BGS galaxy contaminated with light from a Type Ia supernova.\n3. A galaxy with light from a Type Ib supernova.\n4. A galaxy with light from a Type Ic supernova.\n5. A galaxy with light from a Type IIn supernova.\n6. A galaxy with light from a Type IIP supernova.\n\nThe spectra look different, but it's hard to see all the features.",
"_____no_output_____"
]
],
[
[
"# Plot all the spectra on the same wavelength grid.\nfig, axes = plt.subplots(2,3, figsize=(15,8), sharex=True, sharey=True, tight_layout=True)\n\nfor i, ax in enumerate(axes.flatten()):\n wave = hdus['WAVE'].data\n flux = hdus['FLUX'].data[i]\n ivar = hdus['IVAR'].data[i]\n spec = hdus['SPECTYPE'].data[i][0]\n \n ax.plot(wave, flux, alpha=0.5)\n ax.set(xlim=(3600, 9800), ylim=(-10,50),\n title=spec)\n\naxes[0,0].set(ylabel=r'flux [$10^{-17}$ erg cm$^{-2}$ s$^{-1}$ $\\AA^{-1}$]');\nfor i in range(0,3):\n axes[1,i].set(xlabel=r'observed wavelength [$\\AA$]')",
"_____no_output_____"
]
],
[
[
"#### Rebinned Spectra\n\nRebinning means that we take the measured intensity at several wavelengths and average them together to make one new data point at a new central wavelength. This smooths out the noisy spectra and makes the broad line features of the supernovae much easier to see.",
"_____no_output_____"
]
],
[
[
"from desispec.interpolation import resample_flux\n\n# Plot all the spectra on the same wavelength grid.\nfig, axes = plt.subplots(2,3, figsize=(15,8), sharex=True, sharey=True, tight_layout=True)\n\n# Resample the flux on a new wavelength grid with only 100 bins.\nnewwave = np.linspace(3600, 9800, 101)\n\nfor i, ax in enumerate(axes.flatten()):\n wave = hdus['WAVE'].data\n flux = hdus['FLUX'].data[i]\n ivar = hdus['IVAR'].data[i]\n spec = hdus['SPECTYPE'].data[i][0]\n \n ax.plot(wave, flux, alpha=0.5)\n \n fl, iv = resample_flux(newwave, wave, flux, ivar)\n dfl = 1./np.sqrt(iv)\n ax.plot(newwave, fl, 'k-', alpha=0.2)\n ax.errorbar(newwave, fl, yerr=dfl, fmt='k_')\n \n ax.set(xlim=(3600, 9800), ylim=(-10,50),\n title=spec)\n\naxes[0,0].set(ylabel=r'flux [$10^{-17}$ erg cm$^{-2}$ s$^{-1}$ $\\AA^{-1}$]');\nfor i in range(0,3):\n axes[1,i].set(xlabel=r'observed wavelength [$\\AA$]')",
"_____no_output_____"
]
],
[
[
"## Automatically Finding Supernovae in DESI Spectra\n\nDESI will observe so many galaxies that it will not be possible to identify supernovae or other interesting astrophysical transients such as kilonovae and tidal disruption events via manual inspection. We must do it automatically.\n\nIn a companion notebook, we will show how you can train machine-learning algorithms developed for image classification on the World Wide Web to hunt for transients in galaxy spectra.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6e64c173fa2ac8d2865d4bff8d8cdbf11e2eb6 | 1,003 | ipynb | Jupyter Notebook | Python Basics/While Loops.ipynb | Kunal-Attri/DS-1Stop-AI-learn | 42f615e16819b34a642594b412f612a5bfd7f901 | [
"MIT"
]
| null | null | null | Python Basics/While Loops.ipynb | Kunal-Attri/DS-1Stop-AI-learn | 42f615e16819b34a642594b412f612a5bfd7f901 | [
"MIT"
]
| null | null | null | Python Basics/While Loops.ipynb | Kunal-Attri/DS-1Stop-AI-learn | 42f615e16819b34a642594b412f612a5bfd7f901 | [
"MIT"
]
| null | null | null | 17 | 42 | 0.449651 | [
[
[
"x = 0\nwhile x < 5:\n print('x is :', x)\n x = x + 1",
"x is : 0\nx is : 1\nx is : 2\nx is : 3\nx is : 4\n"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec6e722ab441abadbbb3924caef4a1a3e5c8f5e5 | 88,090 | ipynb | Jupyter Notebook | ARWU University Ranking.ipynb | kaiicheng/Python-Crawler | a635df595b89dd7bd8e6b19684cde999cc1ccb97 | [
"MIT"
]
| null | null | null | ARWU University Ranking.ipynb | kaiicheng/Python-Crawler | a635df595b89dd7bd8e6b19684cde999cc1ccb97 | [
"MIT"
]
| null | null | null | ARWU University Ranking.ipynb | kaiicheng/Python-Crawler | a635df595b89dd7bd8e6b19684cde999cc1ccb97 | [
"MIT"
]
| null | null | null | 38.585195 | 112 | 0.390373 | [
[
[
"import requests\nfrom pyquery import PyQuery as pq\nfrom pandas.core.frame import DataFrame\n\nhomeRes = requests.get(\"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/index.html#\")\nhomeDoc = pq(homeRes.text) # No need to use ()\n\ndataList = []\ncateList = homeDoc(\"#rankingarea > div:nth-child(10) > a:nth-child(2)\")\n\nad = \"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/\"\nad = ad + cateList.attr(\"href\")\n\ncateRes = requests.get(ad) # request\"s\ncateDoc = pq(cateRes.text) \nnextPgDoc = cateDoc\n\nsubject = cateDoc(\"#rankingarea > span\")\n\nsubject = str(cateList)\nsubject = str(subject)\nsubject = subject.split(\">\")\nsubject = subject[1]\nsubject = subject.split(\"<\")\nsubject = subject[0]\n\nsub = cateDoc(\"#rankingnav > form:nth-child(2) > select:nth-child(2)\")\nfor item in sub.items():\n dataDict={}\n dataDict[\"title\"] = ''\n dataDict['country'] = ''\n dataDict['rank'] = ''\n dataList.append(dataDict)\n\nfor i in range(1): # while True:\n item = cateDoc(\"#UniversityRanking > tr\")\n for eachItemDoc in item.items():\n dataDict={}\n if eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")!= \"United States\":\n pass\n else:\n dataDict[\"title\"] = eachItemDoc(\".left\").text()\n dataDict[\"country\"] = eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")\n dataDict[\"rank\"] = eachItemDoc(\"tr>td:nth-child(1) \").text() \n dataList.append(dataDict)\n if len(item) == 0:\n break\n\nprint(subject)\ndata=DataFrame(dataList)\n\n#display(data)\ndata[1:50]",
"Statistics\n"
],
[
"import requests\nfrom pyquery import PyQuery as pq\nfrom pandas.core.frame import DataFrame\n\nhomeRes = requests.get(\"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/index.html#\")\nhomeDoc = pq(homeRes.text) # No need to use ()\n\ndataList = []\ncateList = homeDoc(\"#rankingarea > div:nth-child(10) > a:nth-child(1)\")\n\nad = \"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/\"\nad = ad + cateList.attr(\"href\")\n\ncateRes = requests.get(ad) # request\"s\ncateDoc = pq(cateRes.text) \nnextPgDoc = cateDoc\n\nsubject = cateDoc(\"#rankingarea > span\")\n\nsubject = str(cateList)\nsubject = str(subject)\nsubject = subject.split(\">\")\nsubject = subject[1]\nsubject = subject.split(\"<\")\nsubject = subject[0]\n\nsub = cateDoc(\"#rankingnav > form:nth-child(2) > select:nth-child(2)\")\nfor item in sub.items():\n dataDict={}\n dataDict[\"title\"] = ''\n dataDict['country'] = ''\n dataDict['rank'] = ''\n dataList.append(dataDict)\n\nfor i in range(1): # while True:\n item = cateDoc(\"#UniversityRanking > tr\")\n for eachItemDoc in item.items():\n dataDict={}\n if eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")!= \"United States\":\n pass\n else:\n dataDict[\"title\"] = eachItemDoc(\".left\").text()\n dataDict[\"country\"] = eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")\n dataDict[\"rank\"] = eachItemDoc(\"tr>td:nth-child(1) \").text() \n dataList.append(dataDict)\n if len(item) == 0:\n break\n\nprint(subject)\ndata=DataFrame(dataList)\n\n#display(data)\ndata[1:50]",
"Economics\n"
],
[
"import requests\nfrom pyquery import PyQuery as pq\nfrom pandas.core.frame import DataFrame\n\nhomeRes = requests.get(\"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/index.html#\")\nhomeDoc = pq(homeRes.text) # No need to use ()\n\ndataList = []\ncateList = homeDoc(\"#rankingarea > div:nth-child(10) > a:nth-child(9)\")\n\nad = \"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/\"\nad = ad + cateList.attr(\"href\")\n\ncateRes = requests.get(ad) # request\"s\ncateDoc = pq(cateRes.text) \nnextPgDoc = cateDoc\n\nsubject = cateDoc(\"#rankingarea > span\")\n\nsubject = str(cateList)\nsubject = str(subject)\nsubject = subject.split(\">\")\nsubject = subject[1]\nsubject = subject.split(\"<\")\nsubject = subject[0]\n\nsub = cateDoc(\"#rankingnav > form:nth-child(2) > select:nth-child(2)\")\nfor item in sub.items():\n dataDict={}\n dataDict[\"title\"] = ''\n dataDict['country'] = ''\n dataDict['rank'] = ''\n dataList.append(dataDict)\n\nfor i in range(1): # while True:\n item = cateDoc(\"#UniversityRanking > tr\")\n for eachItemDoc in item.items():\n dataDict={}\n if eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")!= \"United States\":\n pass\n else:\n dataDict[\"title\"] = eachItemDoc(\".left\").text()\n dataDict[\"country\"] = eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")\n dataDict[\"rank\"] = eachItemDoc(\"tr>td:nth-child(1) \").text() \n dataList.append(dataDict)\n if len(item) == 0:\n break\n\nprint(subject)\ndata=DataFrame(dataList)\n\n#display(data)\ndata[1:50]",
"Business Administration\n"
],
[
"import requests\nfrom pyquery import PyQuery as pq\nfrom pandas.core.frame import DataFrame\n\nhomeRes = requests.get(\"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/index.html#\")\nhomeDoc = pq(homeRes.text) # No need to use ()\n\ndataList = []\ncateList = homeDoc(\"#rankingarea > div:nth-child(10) > a:nth-child(10)\")\n\nad = \"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/\"\nad = ad + cateList.attr(\"href\")\n\ncateRes = requests.get(ad) # request\"s\ncateDoc = pq(cateRes.text) \nnextPgDoc = cateDoc\n\nsubject = cateDoc(\"#rankingarea > span\")\n\nsubject = str(cateList)\nsubject = str(subject)\nsubject = subject.split(\">\")\nsubject = subject[1]\nsubject = subject.split(\"<\")\nsubject = subject[0]\n\nsub = cateDoc(\"#rankingnav > form:nth-child(2) > select:nth-child(2)\")\nfor item in sub.items():\n dataDict={}\n dataDict[\"title\"] = ''\n dataDict['country'] = ''\n dataDict['rank'] = ''\n dataList.append(dataDict)\n\nfor i in range(1): # while True:\n item = cateDoc(\"#UniversityRanking > tr\")\n for eachItemDoc in item.items():\n dataDict={}\n if eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")!= \"United States\":\n pass\n else:\n dataDict[\"title\"] = eachItemDoc(\".left\").text()\n dataDict[\"country\"] = eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")\n dataDict[\"rank\"] = eachItemDoc(\"tr>td:nth-child(1) \").text() \n dataList.append(dataDict)\n if len(item) == 0:\n break\n\nprint(subject)\ndata=DataFrame(dataList)\n\n#display(data)\ndata[1:50]",
"Finance\n"
],
[
"import requests\nfrom pyquery import PyQuery as pq\nfrom pandas.core.frame import DataFrame\n\nhomeRes = requests.get(\"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/index.html#\")\nhomeDoc = pq(homeRes.text) # No need to use ()\n\ndataList = []\ncateList = homeDoc(\"#rankingarea > div:nth-child(10) > a:nth-child(11)\")\n\nad = \"http://www.shanghairanking.com/Shanghairanking-Subject-Rankings/\"\nad = ad + cateList.attr(\"href\")\n\ncateRes = requests.get(ad) # request\"s\ncateDoc = pq(cateRes.text) \nnextPgDoc = cateDoc\n\nsubject = cateDoc(\"#rankingarea > span\")\n\nsubject = str(cateList)\nsubject = str(subject)\nsubject = subject.split(\">\")\nsubject = subject[1]\nsubject = subject.split(\"<\")\nsubject = subject[0]\n\nsub = cateDoc(\"#rankingnav > form:nth-child(2) > select:nth-child(2)\")\nfor item in sub.items():\n dataDict={}\n dataDict[\"title\"] = ''\n dataDict['country'] = ''\n dataDict['rank'] = ''\n dataList.append(dataDict)\n\nfor i in range(1): # while True:\n item = cateDoc(\"#UniversityRanking > tr\")\n for eachItemDoc in item.items():\n dataDict={}\n if eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")!= \"United States\":\n pass\n else:\n dataDict[\"title\"] = eachItemDoc(\".left\").text()\n dataDict[\"country\"] = eachItemDoc((\"td:nth-child(3) > img\")).attr(\"title\")\n dataDict[\"rank\"] = eachItemDoc(\"tr>td:nth-child(1) \").text() \n dataList.append(dataDict)\n if len(item) == 0:\n break\n\nprint(subject)\ndata=DataFrame(dataList)\n\n#display(data)\ndata[1:50]",
"Management\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6e756d278a9682922c37b53c638637d0242e9d | 119,522 | ipynb | Jupyter Notebook | JupyterNotebooks from Medium/Book Recommendation System.ipynb | patiljeevanr/Recommendation | eee6e9c0b07315c98a304458ed0ee1cde9c9c295 | [
"MIT"
]
| null | null | null | JupyterNotebooks from Medium/Book Recommendation System.ipynb | patiljeevanr/Recommendation | eee6e9c0b07315c98a304458ed0ee1cde9c9c295 | [
"MIT"
]
| null | null | null | JupyterNotebooks from Medium/Book Recommendation System.ipynb | patiljeevanr/Recommendation | eee6e9c0b07315c98a304458ed0ee1cde9c9c295 | [
"MIT"
]
| null | null | null | 36.719508 | 10,934 | 0.488412 | [
[
[
"**About Book Crossing Dataset**<br>\n\nThis dataset has been compiled by Cai-Nicolas Ziegler in 2004, and it comprises of three tables for users, books and ratings. Explicit ratings are expressed on a scale from 1-10 (higher values denoting higher appreciation) and implicit rating is expressed by 0",
"_____no_output_____"
],
[
"Link to dataset files<br>\nhttp://www2.informatik.uni-freiburg.de/~cziegler/BX/ ",
"_____no_output_____"
],
[
"**About this Project**\n\nThis project entails building a Book Recommender System for users based on user-based and item-based collaborative filtering approaches",
"_____no_output_____"
]
],
[
[
"#Making necesarry imports\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport sklearn.metrics as metrics\nimport numpy as np\nfrom sklearn.neighbors import NearestNeighbors\nfrom scipy.spatial.distance import correlation\nfrom sklearn.metrics.pairwise import pairwise_distances\nimport ipywidgets as widgets\nfrom IPython.display import display, clear_output\nfrom contextlib import contextmanager\nimport warnings\nwarnings.filterwarnings('ignore')\nimport numpy as np\nimport os, sys\nimport re\nimport seaborn as sns",
"_____no_output_____"
],
[
"#Setting the current working directory\nos.chdir('D:\\Data Science\\Projects\\Book Crossing Dataset - Recommender System')",
"_____no_output_____"
],
[
"from IPython.display import HTML\nHTML('''<script>\ncode_show_err=false; \nfunction code_toggle_err() {\n if (code_show_err){\n $('div.output_stderr').hide();\n } else {\n $('div.output_stderr').show();\n }\n code_show_err = !code_show_err\n} \n$( document ).ready(code_toggle_err);\n</script>\nTo toggle on/off output_stderr, click <a href=\"javascript:code_toggle_err()\">here</a>.''')",
"_____no_output_____"
],
[
"#Loading data\nbooks = pd.read_csv('books.csv', sep=';', error_bad_lines=False, encoding=\"latin-1\")\nbooks.columns = ['ISBN', 'bookTitle', 'bookAuthor', 'yearOfPublication', 'publisher', 'imageUrlS', 'imageUrlM', 'imageUrlL']\nusers = pd.read_csv('users.csv', sep=';', error_bad_lines=False, encoding=\"latin-1\")\nusers.columns = ['userID', 'Location', 'Age']\nratings = pd.read_csv('ratings.csv', sep=';', error_bad_lines=False, encoding=\"latin-1\")\nratings.columns = ['userID', 'ISBN', 'bookRating']",
"Skipping line 6452: expected 8 fields, saw 9\nSkipping line 43667: expected 8 fields, saw 10\nSkipping line 51751: expected 8 fields, saw 9\n\nSkipping line 92038: expected 8 fields, saw 9\nSkipping line 104319: expected 8 fields, saw 9\nSkipping line 121768: expected 8 fields, saw 9\n\nSkipping line 144058: expected 8 fields, saw 9\nSkipping line 150789: expected 8 fields, saw 9\nSkipping line 157128: expected 8 fields, saw 9\nSkipping line 180189: expected 8 fields, saw 9\nSkipping line 185738: expected 8 fields, saw 9\n\nSkipping line 209388: expected 8 fields, saw 9\nSkipping line 220626: expected 8 fields, saw 9\nSkipping line 227933: expected 8 fields, saw 11\nSkipping line 228957: expected 8 fields, saw 10\nSkipping line 245933: expected 8 fields, saw 9\nSkipping line 251296: expected 8 fields, saw 9\nSkipping line 259941: expected 8 fields, saw 9\nSkipping line 261529: expected 8 fields, saw 9\n\n"
],
[
"#checking shapes of the datasets\nprint books.shape\nprint users.shape\nprint ratings.shape",
"(271360, 8)\n(278858, 3)\n(1149780, 3)\n"
],
[
"#Exploring books dataset\nbooks.head()",
"_____no_output_____"
],
[
"#dropping last three columns containing image URLs which will not be required for analysis\nbooks.drop(['imageUrlS', 'imageUrlM', 'imageUrlL'],axis=1,inplace=True)",
"_____no_output_____"
],
[
"#Now the books datasets looks like....\nbooks.head()",
"_____no_output_____"
],
[
"#checking data types of columns\nbooks.dtypes",
"_____no_output_____"
],
[
"#making this setting to display full text in columns\npd.set_option('display.max_colwidth', -1)",
"_____no_output_____"
]
],
[
[
"**yearOfPublication**",
"_____no_output_____"
]
],
[
[
"#yearOfPublication should be set as having dtype as int\n#checking the unique values of yearOfPublication\nbooks.yearOfPublication.unique()\n\n#as it can be seen from below that there are some incorrect entries in this field. It looks like Publisher names \n#'DK Publishing Inc' and 'Gallimard' have been incorrectly loaded as yearOfPublication in dataset due to some errors in csv file\n#Also some of the entries are strings and same years have been entered as numbers in some places",
"_____no_output_____"
],
[
"#investigating the rows having 'DK Publishing Inc' as yearOfPublication\nbooks.loc[books.yearOfPublication == 'DK Publishing Inc',:]",
"_____no_output_____"
],
[
"#From above, it is seen that bookAuthor is incorrectly loaded with bookTitle, hence making required corrections\n#ISBN '0789466953'\nbooks.loc[books.ISBN == '0789466953','yearOfPublication'] = 2000\nbooks.loc[books.ISBN == '0789466953','bookAuthor'] = \"James Buckley\"\nbooks.loc[books.ISBN == '0789466953','publisher'] = \"DK Publishing Inc\"\nbooks.loc[books.ISBN == '0789466953','bookTitle'] = \"DK Readers: Creating the X-Men, How Comic Books Come to Life (Level 4: Proficient Readers)\"",
"_____no_output_____"
],
[
"#ISBN '078946697X'\nbooks.loc[books.ISBN == '078946697X','yearOfPublication'] = 2000\nbooks.loc[books.ISBN == '078946697X','bookAuthor'] = \"Michael Teitelbaum\"\nbooks.loc[books.ISBN == '078946697X','publisher'] = \"DK Publishing Inc\"\nbooks.loc[books.ISBN == '078946697X','bookTitle'] = \"DK Readers: Creating the X-Men, How It All Began (Level 4: Proficient Readers)\"",
"_____no_output_____"
],
[
"#rechecking\nbooks.loc[(books.ISBN == '0789466953') | (books.ISBN == '078946697X'),:]\n#corrections done",
"_____no_output_____"
],
[
"#investigating the rows having 'Gallimard' as yearOfPublication\nbooks.loc[books.yearOfPublication == 'Gallimard',:]",
"_____no_output_____"
],
[
"#making required corrections as above, keeping other fields intact\nbooks.loc[books.ISBN == '2070426769','yearOfPublication'] = 2003\nbooks.loc[books.ISBN == '2070426769','bookAuthor'] = \"Jean-Marie Gustave Le Cl�©zio\"\nbooks.loc[books.ISBN == '2070426769','publisher'] = \"Gallimard\"\nbooks.loc[books.ISBN == '2070426769','bookTitle'] = \"Peuple du ciel, suivi de 'Les Bergers\"",
"_____no_output_____"
],
[
"#rechecking\nbooks.loc[books.ISBN == '2070426769',:]\n#corrections done",
"_____no_output_____"
],
[
"#Correcting the dtypes of yearOfPublication\nbooks.yearOfPublication=pd.to_numeric(books.yearOfPublication, errors='coerce')",
"_____no_output_____"
],
[
"print sorted(books['yearOfPublication'].unique())\n#Now it can be seen that yearOfPublication has all values as integers",
"[0, 1376, 1378, 1806, 1897, 1900, 1901, 1902, 1904, 1906, 1908, 1909, 1910, 1911, 1914, 1917, 1919, 1920, 1921, 1922, 1923, 1924, 1925, 1926, 1927, 1928, 1929, 1930, 1931, 1932, 1933, 1934, 1935, 1936, 1937, 1938, 1939, 1940, 1941, 1942, 1943, 1944, 1945, 1946, 1947, 1948, 1949, 1950, 1951, 1952, 1953, 1954, 1955, 1956, 1957, 1958, 1959, 1960, 1961, 1962, 1963, 1964, 1965, 1966, 1967, 1968, 1969, 1970, 1971, 1972, 1973, 1974, 1975, 1976, 1977, 1978, 1979, 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989, 1990, 1991, 1992, 1993, 1994, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002, 2003, 2004, 2005, 2006, 2008, 2010, 2011, 2012, 2020, 2021, 2024, 2026, 2030, 2037, 2038, 2050]\n"
],
[
"#However, the value 0 is invalid and as this dataset was published in 2004, I have assumed the the years after 2006 to be \n#invalid keeping some margin in case dataset was updated thereafer\n#setting invalid years as NaN\nbooks.loc[(books.yearOfPublication > 2006) | (books.yearOfPublication == 0),'yearOfPublication'] = np.NAN",
"_____no_output_____"
],
[
"#replacing NaNs with mean value of yearOfPublication\nbooks.yearOfPublication.fillna(round(books.yearOfPublication.mean()), inplace=True)",
"_____no_output_____"
],
[
"#rechecking\nbooks.yearOfPublication.isnull().sum()\n#No NaNs",
"_____no_output_____"
],
[
"#resetting the dtype as int32\nbooks.yearOfPublication = books.yearOfPublication.astype(np.int32)",
"_____no_output_____"
]
],
[
[
"**publisher**",
"_____no_output_____"
]
],
[
[
"#exploring 'publisher' column\nbooks.loc[books.publisher.isnull(),:]\n#two NaNs",
"_____no_output_____"
],
[
"#investigating rows having NaNs\n#Checking with rows having bookTitle as Tyrant Moon to see if we can get any clues\nbooks.loc[(books.bookTitle == 'Tyrant Moon'),:]\n#no clues",
"_____no_output_____"
],
[
"#Checking with rows having bookTitle as Finder Keepers to see if we can get any clues\nbooks.loc[(books.bookTitle == 'Finders Keepers'),:]\n#all rows with different publisher and bookAuthor",
"_____no_output_____"
],
[
"#checking by bookAuthor to find patterns\nbooks.loc[(books.bookAuthor == 'Elaine Corvidae'),:]\n#all having different publisher...no clues here",
"_____no_output_____"
],
[
"#checking by bookAuthor to find patterns\nbooks.loc[(books.bookAuthor == 'Linnea Sinclair'),:]",
"_____no_output_____"
],
[
"#since there is nothing in common to infer publisher for NaNs, replacing these with 'other\nbooks.loc[(books.ISBN == '193169656X'),'publisher'] = 'other'\nbooks.loc[(books.ISBN == '1931696993'),'publisher'] = 'other'",
"_____no_output_____"
]
],
[
[
"**Users**",
"_____no_output_____"
]
],
[
[
"print users.shape\nusers.head()",
"(278858, 3)\n"
],
[
"users.dtypes",
"_____no_output_____"
]
],
[
[
"**userID**",
"_____no_output_____"
]
],
[
[
"users.userID.values\n#it can be seen that these are unique",
"_____no_output_____"
]
],
[
[
"**Age**",
"_____no_output_____"
]
],
[
[
"print sorted(users.Age.unique())\n#Age column has some invalid entries like nan, 0 and very high values like 100 and above",
"[nan, 0.0, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, 13.0, 14.0, 15.0, 16.0, 17.0, 18.0, 19.0, 20.0, 21.0, 22.0, 23.0, 24.0, 25.0, 26.0, 27.0, 28.0, 29.0, 30.0, 31.0, 32.0, 33.0, 34.0, 35.0, 36.0, 37.0, 38.0, 39.0, 40.0, 41.0, 42.0, 43.0, 44.0, 45.0, 46.0, 47.0, 48.0, 49.0, 50.0, 51.0, 52.0, 53.0, 54.0, 55.0, 56.0, 57.0, 58.0, 59.0, 60.0, 61.0, 62.0, 63.0, 64.0, 65.0, 66.0, 67.0, 68.0, 69.0, 70.0, 71.0, 72.0, 73.0, 74.0, 75.0, 76.0, 77.0, 78.0, 79.0, 80.0, 81.0, 82.0, 83.0, 84.0, 85.0, 86.0, 87.0, 88.0, 89.0, 90.0, 91.0, 92.0, 93.0, 94.0, 95.0, 96.0, 97.0, 98.0, 99.0, 100.0, 101.0, 102.0, 103.0, 104.0, 105.0, 106.0, 107.0, 108.0, 109.0, 110.0, 111.0, 113.0, 114.0, 115.0, 116.0, 118.0, 119.0, 123.0, 124.0, 127.0, 128.0, 132.0, 133.0, 136.0, 137.0, 138.0, 140.0, 141.0, 143.0, 146.0, 147.0, 148.0, 151.0, 152.0, 156.0, 157.0, 159.0, 162.0, 168.0, 172.0, 175.0, 183.0, 186.0, 189.0, 199.0, 200.0, 201.0, 204.0, 207.0, 208.0, 209.0, 210.0, 212.0, 219.0, 220.0, 223.0, 226.0, 228.0, 229.0, 230.0, 231.0, 237.0, 239.0, 244.0]\n"
],
[
"#In my view values below 5 and above 90 do not make much sense for our book rating case...hence replacing these by NaNs\nusers.loc[(users.Age > 90) | (users.Age < 5), 'Age'] = np.nan",
"_____no_output_____"
],
[
"#replacing NaNs with mean\nusers.Age = users.Age.fillna(users.Age.mean())",
"_____no_output_____"
],
[
"#setting the data type as int\nusers.Age = users.Age.astype(np.int32)",
"_____no_output_____"
],
[
"#rechecking\nprint sorted(users.Age.unique())\n#looks good now",
"[5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90]\n"
]
],
[
[
"**Ratings Dataset**",
"_____no_output_____"
]
],
[
[
"#checking shape\nratings.shape",
"_____no_output_____"
],
[
"#ratings dataset will have n_users*n_books entries if every user rated every item, this shows that the dataset is very sparse\nn_users = users.shape[0]\nn_books = books.shape[0]\nprint n_users * n_books",
"75670906880\n"
],
[
"#checking first few rows...\nratings.head(5)",
"_____no_output_____"
],
[
"ratings.bookRating.unique()",
"_____no_output_____"
],
[
"#ratings dataset should have books only which exist in our books dataset, unless new books are added to books dataset\nratings_new = ratings[ratings.ISBN.isin(books.ISBN)]",
"_____no_output_____"
],
[
"print ratings.shape\nprint ratings_new.shape\n#it can be seen that many rows having book ISBN not part of books dataset got dropped off",
"(1149780, 3)\n(1031136, 3)\n"
],
[
"#ratings dataset should have ratings from users which exist in users dataset, unless new users are added to users dataset\nratings = ratings[ratings.userID.isin(users.userID)]",
"_____no_output_____"
],
[
"print ratings.shape\nprint ratings_new.shape\n#no new users added, hence we will go with above dataset ratings_new (1031136, 3)",
"(1149780, 3)\n(1031136, 3)\n"
],
[
"print \"number of users: \" + str(n_users)\nprint \"number of books: \" + str(n_books)",
"number of users: 278858\nnumber of books: 271360\n"
],
[
"#Sparsity of dataset in %\nsparsity=1.0-len(ratings_new)/float(n_users*n_books)\nprint 'The sparsity level of Book Crossing dataset is ' + str(sparsity*100) + ' %'",
"The sparsity level of Book Crossing dataset is 99.9986373416 %\n"
],
[
"#As quoted in the description of the dataset -\n#BX-Book-Ratings contains the book rating information. Ratings are either explicit, expressed on a scale from 1-10 \n#higher values denoting higher appreciation, or implicit, expressed by 0\nratings.bookRating.unique()",
"_____no_output_____"
],
[
"#Hence segragating implicit and explict ratings datasets\nratings_explicit = ratings_new[ratings_new.bookRating != 0]\nratings_implicit = ratings_new[ratings_new.bookRating == 0]",
"_____no_output_____"
],
[
"#checking shapes\nprint ratings_new.shape\nprint ratings_explicit.shape\nprint ratings_implicit.shape",
"(1031136, 3)\n(383842, 3)\n(647294, 3)\n"
],
[
"#plotting count of bookRating\nsns.countplot(data=ratings_explicit , x='bookRating')\nplt.show()\n#It can be seen that higher ratings are more common amongst users and rating 8 has been rated highest number of times",
"_____no_output_____"
]
],
[
[
"**Simple Popularity Based Recommendation System**",
"_____no_output_____"
]
],
[
[
"#At this point , a simple popularity based recommendation system can be built based on count of user ratings for different books\nratings_count = pd.DataFrame(ratings_explicit.groupby(['ISBN'])['bookRating'].sum())\ntop10 = ratings_count.sort_values('bookRating', ascending = False).head(10)\nprint \"Following books are recommended\"\ntop10.merge(books, left_index = True, right_on = 'ISBN')\n\n#Given below are top 10 recommendations based on popularity. It is evident that books authored by J.K. Rowling are most popular",
"Following books are recommended\n"
],
[
"#Similarly segregating users who have given explicit ratings from 1-10 and those whose implicit behavior was tracked\nusers_exp_ratings = users[users.userID.isin(ratings_explicit.userID)]\nusers_imp_ratings = users[users.userID.isin(ratings_implicit.userID)]",
"_____no_output_____"
],
[
"#checking shapes\nprint users.shape\nprint users_exp_ratings.shape\nprint users_imp_ratings.shape",
"(278858, 3)\n(68091, 3)\n(52451, 3)\n"
]
],
[
[
"**Collaborative Filtering Based Recommendation Systems**",
"_____no_output_____"
]
],
[
[
"#To cope up with computing power I have and to reduce the dataset size, I am considering users who have rated atleast 100 books\n#and books which have atleast 100 ratings\ncounts1 = ratings_explicit['userID'].value_counts()\nratings_explicit = ratings_explicit[ratings_explicit['userID'].isin(counts1[counts1 >= 100].index)]\ncounts = ratings_explicit['bookRating'].value_counts()\nratings_explicit = ratings_explicit[ratings_explicit['bookRating'].isin(counts[counts >= 100].index)]",
"_____no_output_____"
],
[
"#Generating ratings matrix from explicit ratings table\nratings_matrix = ratings_explicit.pivot(index='userID', columns='ISBN', values='bookRating')\nuserID = ratings_matrix.index\nISBN = ratings_matrix.columns\nprint(ratings_matrix.shape)\nratings_matrix.head()\n#Notice that most of the values are NaN (undefined) implying absence of ratings",
"(449, 66574)\n"
],
[
"n_users = ratings_matrix.shape[0] #considering only those users who gave explicit ratings\nn_books = ratings_matrix.shape[1]\nprint n_users, n_books",
"449 66574\n"
],
[
"#since NaNs cannot be handled by training algorithms, replacing these by 0, which indicates absence of ratings\n#setting data type\nratings_matrix.fillna(0, inplace = True)\nratings_matrix = ratings_matrix.astype(np.int32)",
"_____no_output_____"
],
[
"#checking first few rows\nratings_matrix.head(5)",
"_____no_output_____"
],
[
"#rechecking the sparsity\nsparsity=1.0-len(ratings_explicit)/float(users_exp_ratings.shape[0]*n_books)\nprint 'The sparsity level of Book Crossing dataset is ' + str(sparsity*100) + ' %'",
"The sparsity level of Book Crossing dataset is 99.9977218411 %\n"
]
],
[
[
"**Training our recommendation system**",
"_____no_output_____"
]
],
[
[
"#setting global variables\nglobal metric,k\nk=10\nmetric='cosine'",
"_____no_output_____"
]
],
[
[
"**User-based Recommendation System**",
"_____no_output_____"
]
],
[
[
"#This function finds k similar users given the user_id and ratings matrix \n#These similarities are same as obtained via using pairwise_distances\ndef findksimilarusers(user_id, ratings, metric = metric, k=k):\n similarities=[]\n indices=[]\n model_knn = NearestNeighbors(metric = metric, algorithm = 'brute') \n model_knn.fit(ratings)\n loc = ratings.index.get_loc(user_id)\n distances, indices = model_knn.kneighbors(ratings.iloc[loc, :].values.reshape(1, -1), n_neighbors = k+1)\n similarities = 1-distances.flatten()\n \n return similarities,indices",
"_____no_output_____"
],
[
"#This function predicts rating for specified user-item combination based on user-based approach\ndef predict_userbased(user_id, item_id, ratings, metric = metric, k=k):\n prediction=0\n user_loc = ratings.index.get_loc(user_id)\n item_loc = ratings.columns.get_loc(item_id)\n similarities, indices=findksimilarusers(user_id, ratings,metric, k) #similar users based on cosine similarity\n mean_rating = ratings.iloc[user_loc,:].mean() #to adjust for zero based indexing\n sum_wt = np.sum(similarities)-1\n product=1\n wtd_sum = 0 \n \n for i in range(0, len(indices.flatten())):\n if indices.flatten()[i] == user_loc:\n continue;\n else: \n ratings_diff = ratings.iloc[indices.flatten()[i],item_loc]-np.mean(ratings.iloc[indices.flatten()[i],:])\n product = ratings_diff * (similarities[i])\n wtd_sum = wtd_sum + product\n \n #in case of very sparse datasets, using correlation metric for collaborative based approach may give negative ratings\n #which are handled here as below\n if prediction <= 0:\n prediction = 1 \n elif prediction >10:\n prediction = 10\n \n prediction = int(round(mean_rating + (wtd_sum/sum_wt)))\n print '\\nPredicted rating for user {0} -> item {1}: {2}'.format(user_id,item_id,prediction)\n\n return prediction",
"_____no_output_____"
],
[
"predict_userbased(11676,'0001056107',ratings_matrix);",
"\nPredicted rating for user 11676 -> item 0001056107: 2\n"
]
],
[
[
"**Item-based Recommendation Systems**",
"_____no_output_____"
]
],
[
[
"#This function finds k similar items given the item_id and ratings matrix\n\ndef findksimilaritems(item_id, ratings, metric=metric, k=k):\n similarities=[]\n indices=[]\n ratings=ratings.T\n loc = ratings.index.get_loc(item_id)\n model_knn = NearestNeighbors(metric = metric, algorithm = 'brute')\n model_knn.fit(ratings)\n \n distances, indices = model_knn.kneighbors(ratings.iloc[loc, :].values.reshape(1, -1), n_neighbors = k+1)\n similarities = 1-distances.flatten()\n\n return similarities,indices",
"_____no_output_____"
],
[
"similarities,indices=findksimilaritems('0001056107',ratings_matrix)",
"_____no_output_____"
],
[
"#This function predicts the rating for specified user-item combination based on item-based approach\ndef predict_itembased(user_id, item_id, ratings, metric = metric, k=k):\n prediction= wtd_sum =0\n user_loc = ratings.index.get_loc(user_id)\n item_loc = ratings.columns.get_loc(item_id)\n similarities, indices=findksimilaritems(item_id, ratings) #similar users based on correlation coefficients\n sum_wt = np.sum(similarities)-1\n product=1\n for i in range(0, len(indices.flatten())):\n if indices.flatten()[i] == item_loc:\n continue;\n else:\n product = ratings.iloc[user_loc,indices.flatten()[i]] * (similarities[i])\n wtd_sum = wtd_sum + product \n prediction = int(round(wtd_sum/sum_wt))\n \n #in case of very sparse datasets, using correlation metric for collaborative based approach may give negative ratings\n #which are handled here as below //code has been validated without the code snippet below, below snippet is to avoid negative\n #predictions which might arise in case of very sparse datasets when using correlation metric\n if prediction <= 0:\n prediction = 1 \n elif prediction >10:\n prediction = 10\n\n print '\\nPredicted rating for user {0} -> item {1}: {2}'.format(user_id,item_id,prediction) \n \n return prediction",
"_____no_output_____"
],
[
"prediction = predict_itembased(11676,'0001056107',ratings_matrix)",
"\nPredicted rating for user 11676 -> item 0001056107: 1\n"
],
[
"@contextmanager\ndef suppress_stdout():\n with open(os.devnull, \"w\") as devnull:\n old_stdout = sys.stdout\n sys.stdout = devnull\n try: \n yield\n finally:\n sys.stdout = old_stdout",
"_____no_output_____"
],
[
"#This function utilizes above functions to recommend items for item/user based approach and cosine/correlation. \n#Recommendations are made if the predicted rating for an item is >= to 6,and the items have not been rated already\ndef recommendItem(user_id, ratings, metric=metric): \n if (user_id not in ratings.index.values) or type(user_id) is not int:\n print \"User id should be a valid integer from this list :\\n\\n {} \".format(re.sub('[\\[\\]]', '', np.array_str(ratings_matrix.index.values)))\n else: \n ids = ['Item-based (correlation)','Item-based (cosine)','User-based (correlation)','User-based (cosine)']\n select = widgets.Dropdown(options=ids, value=ids[0],description='Select approach', width='1000px')\n def on_change(change):\n clear_output(wait=True)\n prediction = [] \n if change['type'] == 'change' and change['name'] == 'value': \n if (select.value == 'Item-based (correlation)') | (select.value == 'User-based (correlation)') :\n metric = 'correlation'\n else: \n metric = 'cosine' \n with suppress_stdout():\n if (select.value == 'Item-based (correlation)') | (select.value == 'Item-based (cosine)'):\n for i in range(ratings.shape[1]):\n if (ratings[str(ratings.columns[i])][user_id] !=0): #not rated already\n prediction.append(predict_itembased(user_id, str(ratings.columns[i]) ,ratings, metric))\n else: \n prediction.append(-1) #for already rated items\n else:\n for i in range(ratings.shape[1]):\n if (ratings[str(ratings.columns[i])][user_id] !=0): #not rated already\n prediction.append(predict_userbased(user_id, str(ratings.columns[i]) ,ratings, metric))\n else: \n prediction.append(-1) #for already rated items\n prediction = pd.Series(prediction)\n prediction = prediction.sort_values(ascending=False)\n recommended = prediction[:10]\n print \"As per {0} approach....Following books are recommended...\".format(select.value)\n for i in range(len(recommended)):\n print \"{0}. {1}\".format(i+1,books.bookTitle[recommended.index[i]].encode('utf-8')) \n select.observe(on_change)\n display(select)",
"_____no_output_____"
],
[
"#checking for incorrect entries\nrecommendItem(999999,ratings_matrix)",
"User id should be a valid integer from this list :\n\n 2033 2110 2276 4017 4385 5582 6242 6251 6543 6575\n 7286 7346 8067 8245 8681 8890 10560 11676 11993 12538\n 12824 12982 13552 13850 14422 15408 15418 16634 16795 16966\n 17950 19085 21014 23768 23872 23902 25409 25601 25981 26535\n 26544 26583 28591 28634 29259 30276 30511 30711 30735 30810\n 31315 31556 31826 32773 33145 35433 35836 35857 35859 36299\n 36554 36606 36609 36836 36907 37644 37712 37950 38023 38273\n 38281 39281 39467 40889 40943 43246 43910 46398 47316 48025\n 48494 49144 49889 51883 52199 52350 52584 52614 52917 53220\n 55187 55490 55492 56271 56399 56447 56554 56959 59172 60244\n 60337 60707 63714 63956 65258 66942 67840 68555 69078 69389\n 69697 70415 70594 70666 72352 73681 75591 75819 76151 76223\n 76499 76626 78553 78783 78834 78973 79441 81492 81560 83287\n 83637 83671 85526 85656 86189 86947 87141 87555 88283 88677\n 88693 88733 89602 91113 92652 92810 93047 93363 93629 94242\n 94347 94853 94951 95010 95359 95902 95932 96448 97754 97874\n 98391 98758 100459 100906 101209 101606 101851 102359 102647 102702\n 102967 104399 104636 105028 105517 105979 106007 107784 107951 109574\n 109901 109955 110483 110912 110934 110973 112001 113270 113519 114368\n 114868 114988 115002 115003 116599 117384 120565 122429 122793 123094\n 123608 123883 123981 125519 125774 126492 126736 127200 127359 128835\n 129074 129716 129851 130554 130571 132492 132836 133747 134434 135149\n 135265 136010 136139 136348 136382 138578 138844 140000 140358 141902\n 142524 143175 143253 143415 145449 146113 146348 147847 148199 148258\n 148744 148966 149907 149908 150979 153662 156150 156269 156300 156467\n 157247 157273 158226 158295 158433 159506 160295 162052 162639 162738\n 163759 163761 163804 163973 164096 164323 164533 164828 164905 165308\n 165319 165758 166123 166596 168047 168245 169682 170513 170634 171118\n 172030 172742 172888 173291 173415 174304 174892 177072 177432 177458\n 178522 179718 179978 180378 180651 181176 182085 182086 182993 183958\n 183995 184299 184532 185233 185384 187145 187256 187517 189139 189334\n 189835 189973 190708 190925 193458 193560 193898 194600 196077 196160\n 196502 197659 199416 200226 201290 203240 204864 205735 205943 206534\n 207782 208406 208671 209516 210485 211426 211919 212965 214786 216012\n 216444 216683 217106 217318 217740 218552 218608 219546 219683 222204\n 222296 223087 223501 224349 224525 224646 224764 225087 225199 225232\n 225595 225763 226965 227250 227447 227520 227705 229011 229329 229551\n 229741 230522 231210 232131 232945 233911 234359 234828 235105 235282\n 235935 236058 236283 236340 236757 236948 239584 239594 240144 240403\n 240543 240567 240568 241198 241666 241980 242006 242083 242409 242465\n 244627 244685 245410 245827 246311 247429 247447 248718 249894 250405\n 250709 251394 251843 251844 252695 252820 254206 254465 254899 255489\n 257204 258152 258185 258534 261105 261829 262998 264031 264082 264321\n 264525 265115 265313 265889 266056 266226 268110 268300 268932 269566\n 270713 271448 271705 273113 274061 274301 275970 277427 278418 \n"
],
[
"recommendItem(4385, ratings_matrix)",
"As per Item-based (cosine) approach....Following books are recommended...\n1. My Wicked Wicked Ways\n2. Fair Peril\n3. Wolfpointe\n4. A Nest of Ninnies\n5. A Bitter Legacy\n6. A Hymn Before Battle\n7. Thomas the Rhymer\n8. Gatherer of Clouds (Initiate Brother Duology)\n9. Wege zum Ruhm: 13 Hilfestellungen für junge Künstler und 1 Warnung\n10. Love In Bloom's\n"
],
[
"recommendItem(4385, ratings_matrix)",
"As per User-based (correlation) approach....Following books are recommended...\n1. The Gift\n2. A Close Run Thing : A Novel of Wellington's Army of 1815\n3. The Romantic: A Novel\n4. Mazurka for Two Dead Men\n5. The Titanic Conspiracy: Cover-Ups and Mysteries of the World's Most Famous Sea Disaster\n6. And Never Let Her Go : Thomas Capano: The Deadly Seducer\n7. Chop Wood, Carry Water: A Guide to Finding Spiritual Fulfillment in Everyday Life\n8. WHO NEEDS GOD\n9. Lords of the White Castle\n10. Prince Charming Isn't Coming: How Women Get Smart About Money\n"
]
],
[
[
"**Thanks for reading this notebook**",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec6e77fa7008fa50d482fb15de8790b9ffe710ab | 2,410 | ipynb | Jupyter Notebook | worded_questions/answers/is-multithreading-in-python-a-good-idea.ipynb | lhayhurst/interview-with-python | 8acf397cdd96cbf00334a567a837d6209507e41c | [
"MIT"
]
| 201 | 2016-08-29T23:02:43.000Z | 2022-03-28T14:15:29.000Z | worded_questions/answers/is-multithreading-in-python-a-good-idea.ipynb | inovizz/interview-with-python | 8acf397cdd96cbf00334a567a837d6209507e41c | [
"MIT"
]
| 5 | 2016-09-03T05:57:13.000Z | 2018-03-10T06:09:30.000Z | worded_questions/answers/is-multithreading-in-python-a-good-idea.ipynb | inovizz/interview-with-python | 8acf397cdd96cbf00334a567a837d6209507e41c | [
"MIT"
]
| 85 | 2016-09-01T14:39:13.000Z | 2022-03-17T08:16:54.000Z | 50.208333 | 775 | 0.691701 | [
[
[
"## Python and multi-threading. Is it a good idea? List some ways to get some Python code to run in a parallel way.\n",
"_____no_output_____"
],
[
"Python doesn't allow multi-threading in the truest sense of the word. It has a multi-threading package but if you want to multi-thread to speed your code up, then it's usually not a good idea to use it. Python has a construct called the Global Interpreter Lock (GIL). The GIL makes sure that only one of your 'threads' can execute at any one time. A thread acquires the GIL, does a little work, then passes the GIL onto the next thread. This happens very quickly so to the human eye it may seem like your threads are executing in parallel, but they are really just taking turns using the same CPU core. All this GIL passing adds overhead to execution. This means that if you want to make your code run faster then using the threading package often isn't a good idea.\n\nThere are reasons to use Python's threading package. If you want to run some things simultaneously, and efficiency is not a concern, then it's totally fine and convenient. Or if you are running code that needs to wait for something (like some IO) then it could make a lot of sense. But the threading library wont let you use extra CPU cores.\n\nMulti-threading can be outsourced to the operating system (by doing multi-processing), some external application that calls your Python code (eg, Spark or Hadoop), or some code that your Python code calls (eg: you could have your Python code call a C function that does the expensive multi-threaded stuff).\n\n### Why is this Important?\n\nBecause the GIL is an A-hole. Lots of people spend a lot of time trying to find bottlenecks in their fancy Python multi-threaded code before they learn what the GIL is.",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
ec6e7a3d223fc5e28a3ce1b6caa214f5f8edd926 | 70,246 | ipynb | Jupyter Notebook | Exercises/Exercise 8 - Multiclass with Signs/Exercise 8 - Answer.ipynb | Mathipe98/Tensorflow-Practice | 1f438fa203988ebe56c69c1bfdb08bed62dba5d4 | [
"MIT"
]
| null | null | null | Exercises/Exercise 8 - Multiclass with Signs/Exercise 8 - Answer.ipynb | Mathipe98/Tensorflow-Practice | 1f438fa203988ebe56c69c1bfdb08bed62dba5d4 | [
"MIT"
]
| null | null | null | Exercises/Exercise 8 - Multiclass with Signs/Exercise 8 - Answer.ipynb | Mathipe98/Tensorflow-Practice | 1f438fa203988ebe56c69c1bfdb08bed62dba5d4 | [
"MIT"
]
| null | null | null | 190.88587 | 32,008 | 0.875794 | [
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
],
[
"import csv\nimport numpy as np\nimport tensorflow as tf\nfrom tensorflow.keras.preprocessing.image import ImageDataGenerator\nfrom google.colab import files",
"_____no_output_____"
]
],
[
[
"The data for this exercise is available at: https://www.kaggle.com/datamunge/sign-language-mnist/home\n\nSign up and download to find 2 CSV files: sign_mnist_test.csv and sign_mnist_train.csv -- You will upload both of them using this button before you can continue.\n",
"_____no_output_____"
]
],
[
[
"uploaded=files.upload()",
"_____no_output_____"
],
[
"def get_data(filename):\n with open(filename) as training_file:\n csv_reader = csv.reader(training_file, delimiter=',')\n first_line = True\n temp_images = []\n temp_labels = []\n for row in csv_reader:\n if first_line:\n # print(\"Ignoring first line\")\n first_line = False\n else:\n temp_labels.append(row[0])\n image_data = row[1:785]\n image_data_as_array = np.array_split(image_data, 28)\n temp_images.append(image_data_as_array)\n images = np.array(temp_images).astype('float')\n labels = np.array(temp_labels).astype('float')\n return images, labels\n\n\ntraining_images, training_labels = get_data('sign_mnist_train.csv')\ntesting_images, testing_labels = get_data('sign_mnist_test.csv')\n\nprint(training_images.shape)\nprint(training_labels.shape)\nprint(testing_images.shape)\nprint(testing_labels.shape)\n",
"(27455, 28, 28)\n(27455,)\n(7172, 28, 28)\n(7172,)\n"
],
[
"training_images = np.expand_dims(training_images, axis=3)\ntesting_images = np.expand_dims(testing_images, axis=3)\n\ntrain_datagen = ImageDataGenerator(\n rescale=1. / 255,\n rotation_range=40,\n width_shift_range=0.2,\n height_shift_range=0.2,\n shear_range=0.2,\n zoom_range=0.2,\n horizontal_flip=True,\n fill_mode='nearest')\n\nvalidation_datagen = ImageDataGenerator(\n rescale=1. / 255)\n\nprint(training_images.shape)\nprint(testing_images.shape)",
"(27455, 28, 28, 1)\n(7172, 28, 28, 1)\n"
],
[
"model = tf.keras.models.Sequential([\n tf.keras.layers.Conv2D(64, (3, 3), activation='relu', input_shape=(28, 28, 1)),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Conv2D(64, (3, 3), activation='relu'),\n tf.keras.layers.MaxPooling2D(2, 2),\n tf.keras.layers.Flatten(),\n tf.keras.layers.Dense(128, activation=tf.nn.relu),\n tf.keras.layers.Dense(26, activation=tf.nn.softmax)])\n\nmodel.compile(optimizer = tf.optimizers.Adam(),\n loss = 'sparse_categorical_crossentropy',\n metrics=['accuracy'])\n\nhistory = model.fit(train_datagen.flow(training_images, training_labels, batch_size=32),\n steps_per_epoch=len(training_images) / 32,\n epochs=15,\n validation_data=validation_datagen.flow(testing_images, testing_labels, batch_size=32),\n validation_steps=len(testing_images) / 32)\n\nmodel.evaluate(testing_images, testing_labels)\n",
"Epoch 1/15\n857/857 [==============================] - 21s 24ms/step - loss: 2.8540 - acc: 0.1562 - val_loss: 1.9644 - val_acc: 0.3100\nEpoch 2/15\n857/857 [==============================] - 46s 54ms/step - loss: 2.1131 - acc: 0.2188 - val_loss: 1.5558 - val_acc: 0.4855\nEpoch 3/15\n857/857 [==============================] - 40s 47ms/step - loss: 1.7423 - acc: 0.3438 - val_loss: 1.1643 - val_acc: 0.6115\nEpoch 4/15\n857/857 [==============================] - 30s 35ms/step - loss: 1.4933 - acc: 0.4688 - val_loss: 1.1572 - val_acc: 0.6064\nEpoch 5/15\n857/857 [==============================] - 22s 26ms/step - loss: 1.3521 - acc: 0.7500 - val_loss: 0.8973 - val_acc: 0.7167\nEpoch 6/15\n857/857 [==============================] - 21s 24ms/step - loss: 1.2332 - acc: 0.5625 - val_loss: 0.8082 - val_acc: 0.7200\nEpoch 7/15\n857/857 [==============================] - 44s 51ms/step - loss: 1.1631 - acc: 0.5938 - val_loss: 0.8671 - val_acc: 0.7352\nEpoch 8/15\n857/857 [==============================] - 58s 67ms/step - loss: 1.0857 - acc: 0.7188 - val_loss: 0.7608 - val_acc: 0.7949\nEpoch 9/15\n857/857 [==============================] - 23s 26ms/step - loss: 1.0197 - acc: 0.6562 - val_loss: 0.6978 - val_acc: 0.7674\nEpoch 10/15\n857/857 [==============================] - 28s 33ms/step - loss: 0.9711 - acc: 0.6875 - val_loss: 0.7027 - val_acc: 0.7984\nEpoch 11/15\n857/857 [==============================] - 53s 61ms/step - loss: 0.9076 - acc: 0.5625 - val_loss: 0.5784 - val_acc: 0.8238\nEpoch 12/15\n857/857 [==============================] - 64s 75ms/step - loss: 0.8764 - acc: 0.5938 - val_loss: 0.6079 - val_acc: 0.8133\nEpoch 13/15\n857/857 [==============================] - 28s 33ms/step - loss: 0.8410 - acc: 0.7500 - val_loss: 0.4547 - val_acc: 0.8182\nEpoch 14/15\n857/857 [==============================] - 22s 26ms/step - loss: 0.8045 - acc: 0.5312 - val_loss: 0.2415 - val_acc: 0.8496\nEpoch 15/15\n857/857 [==============================] - 47s 55ms/step - loss: 0.7836 - acc: 0.7188 - val_loss: 0.3857 - val_acc: 0.8425\n7172/7172 [==============================] - 4s 596us/step - loss: 6.9243 - acc: 0.5661\n"
],
[
"import matplotlib.pyplot as plt\nacc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(len(acc))\n\nplt.plot(epochs, acc, 'r', label='Training accuracy')\nplt.plot(epochs, val_acc, 'b', label='Validation accuracy')\nplt.title('Training and validation accuracy')\nplt.legend()\nplt.figure()\n\nplt.plot(epochs, loss, 'r', label='Training Loss')\nplt.plot(epochs, val_loss, 'b', label='Validation Loss')\nplt.title('Training and validation loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6e824533cbc0d7622ee156166d5dcdc15b8855 | 34,830 | ipynb | Jupyter Notebook | Introduction/02-Libraries.ipynb | HSE-LAMBDA/MLatMisis-2019 | d04bec16858edf507131fd56d4564ae56909a1c6 | [
"MIT"
]
| null | null | null | Introduction/02-Libraries.ipynb | HSE-LAMBDA/MLatMisis-2019 | d04bec16858edf507131fd56d4564ae56909a1c6 | [
"MIT"
]
| null | null | null | Introduction/02-Libraries.ipynb | HSE-LAMBDA/MLatMisis-2019 | d04bec16858edf507131fd56d4564ae56909a1c6 | [
"MIT"
]
| null | null | null | 26.567506 | 249 | 0.449756 | [
[
[
"<a href=\"https://colab.research.google.com/github/HSE-LAMBDA/MLatMisis-2019/blob/master/Introduction/02-Libraries.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Titanic: Machine Learning from Disaster",
"_____no_output_____"
],
[
"\n\nThis notebook's gonna teach you to use the basic data science stack for python: jupyter, numpy, pandas and matplotlib.",
"_____no_output_____"
],
[
"## Part I: Jupyter notebooks recap",
"_____no_output_____"
],
[
"**The most important feature** of jupyter notebooks for this course: \n* if you're typing something, press `Tab` to see automatic suggestions, use arrow keys + enter to pick one.\n* if you move your cursor inside some function and press `Tab`, you'll get a help window.",
"_____no_output_____"
]
],
[
[
"# run this first\nimport math",
"_____no_output_____"
],
[
"# Place your cursor at the end of the unfinished line below and press tab to\n# find a function that computes arctangent from two parameters (should\n# have 2 in it's name).\n# Once you chose it, put an opening bracket character and press tab to\n# see the docs.\n\nmath.a # <---",
"_____no_output_____"
]
],
[
[
"## Part II: Loading data with Pandas",
"_____no_output_____"
],
[
"Pandas is a library that helps you load the data, prepare it and perform some lightweight analysis. The god object here is the `pandas.DataFrame` - a 2d table with batteries included. \n\nIn the cell below we use it to read the data on the infamous titanic shipwreck.\n\n__please keep running all the code cells as you read__",
"_____no_output_____"
]
],
[
[
"!wget https://github.com/HSE-LAMBDA/MLatMisis-2019/raw/master/Introduction/train.csv",
"_____no_output_____"
],
[
"import pandas as pd\ndata = pd.read_csv(\"train.csv\", index_col='PassengerId') # this yields a pandas.DataFrame",
"_____no_output_____"
],
[
"# Selecting rows\nhead = data[:10]\n\nhead #if you leave an expression at the end of a cell, jupyter will \"display\" it automatically",
"_____no_output_____"
]
],
[
[
"#### About the data\nHere's some of the columns\n* Name - a string with person's full name\n* Survived - 1 if a person survived the shipwreck, 0 otherwise.\n* Pclass - passenger class. Pclass == 3 is cheap'n'cheerful, Pclass == 1 is for moneybags.\n* Sex - a person's gender\n* Age - age in years, if available\n* Sibsp - number of siblings on a ship\n* Parch - number of parents on a ship\n* Fare - ticket cost\n* Embarked - port where the passenger embarked\n * C = Cherbourg; Q = Queenstown; S = Southampton",
"_____no_output_____"
]
],
[
[
"# table dimensions\nprint(\"len(data) = \", len(data))\nprint(\"data.shape = \", data.shape)",
"_____no_output_____"
],
[
"# select a single row\nprint(data.loc[4])",
"_____no_output_____"
],
[
"# select a single column.\nages = data[\"Age\"]\nprint(ages[:10]) # alternatively: data.Age",
"_____no_output_____"
],
[
"# select several columns and rows at once\ndata.loc[5:10, (\"Fare\", \"Pclass\")] # alternatively: data[[\"Fare\",\"Pclass\"]].loc[5:10]",
"_____no_output_____"
]
],
[
[
"### `loc` vs `iloc`",
"_____no_output_____"
],
[
"There are two ways of indexing the rows in pandas:\n * by index column values (`PassengerId` in our case) – use `data.loc` for that\n * by positional index - use `data.iloc` for that",
"_____no_output_____"
],
[
"Note that index column starts from 1, so positional index 0 will correspond to index column value 1, positional 1 to index column value 2, and so on:",
"_____no_output_____"
]
],
[
[
"print(data.index)\nprint('------')\nprint(\"data.iloc[0]:\")\nprint(data.iloc[0])\nprint('------')\nprint(\"data.loc[1]:\")\nprint(data.loc[1])",
"_____no_output_____"
]
],
[
[
"### Your turn:\n",
"_____no_output_____"
]
],
[
[
"# select passengers number 13 and 666 - did they survive?\n\n<YOUR CODE>",
"_____no_output_____"
],
[
"# compute the overall survival rate (what fraction of passengers survived the shipwreck)\n\n<YOUR CODE>",
"_____no_output_____"
]
],
[
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n",
"_____no_output_____"
],
[
"Pandas also has some basic data analysis tools. For one, you can quickly display statistical aggregates for each column using `.describe()`",
"_____no_output_____"
]
],
[
[
"data.describe()",
"_____no_output_____"
]
],
[
[
"Some columns contain __NaN__ values - this means that there is no data there. For example, passenger `#5` has unknown age. To simplify the future data analysis, we'll replace NaN values by using pandas `fillna` function.\n\n_Note: we do this so easily because it's a tutorial. In general, you think twice before you modify data like this._",
"_____no_output_____"
]
],
[
[
"data.iloc[5]",
"_____no_output_____"
],
[
"data['Age'] = data['Age'].fillna(value=data['Age'].mean())\ndata['Fare'] = data['Fare'].fillna(value=data['Fare'].mean())",
"_____no_output_____"
],
[
"data.iloc[5]",
"_____no_output_____"
]
],
[
[
"More pandas: \n* A neat [tutorial](http://pandas.pydata.org/) from pydata\n* Official [tutorials](https://pandas.pydata.org/pandas-docs/stable/tutorials.html), including this [10 minutes to pandas](https://pandas.pydata.org/pandas-docs/stable/10min.html#min)\n* Bunch of cheat sheets awaits just one google query away from you (e.g. [basics](http://blog.yhat.com/static/img/datacamp-cheat.png), [combining datasets](https://pbs.twimg.com/media/C65MaMpVwAA3v0A.jpg) and so on). ",
"_____no_output_____"
],
[
"## Part III: Numpy and vectorized computing",
"_____no_output_____"
],
[
"Almost any machine learning model requires some computational heavy lifting usually involving linear algebra problems. Unfortunately, raw python is terrible at this because each operation is interpreted at runtime. \n\nSo instead, we'll use `numpy` - a library that lets you run blazing fast computation with vectors, matrices and other tensors. Again, the god oject here is `numpy.ndarray`:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\na = np.array([1,2,3,4,5])\nb = np.array([5,4,3,2,1])\nprint(\"a = \", a)\nprint(\"b = \", b)\n\n# math and boolean operations can applied to each element of an array\nprint(\"a + 1 =\", a + 1)\nprint(\"a * 2 =\", a * 2)\nprint(\"a == 2\", a == 2)\n# ... or corresponding elements of two (or more) arrays\nprint(\"a + b =\", a + b)\nprint(\"a * b =\", a * b)",
"_____no_output_____"
],
[
"# Your turn: compute half-products of a and b elements (halves of products)\n<YOUR CODE>",
"_____no_output_____"
],
[
"# compute elementwise quotient between squared a and (b plus 1)\n<YOUR CODE>",
"_____no_output_____"
]
],
[
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```",
"_____no_output_____"
],
[
"There's a number of functions to create arrays of zeros, ones, ascending/descending numbers etc.:",
"_____no_output_____"
]
],
[
[
"np.zeros(shape=(3, 4))",
"_____no_output_____"
],
[
"np.ones(shape=(2, 5), dtype=np.bool)",
"_____no_output_____"
],
[
"np.arange(3, 15, 2) # start, stop, step",
"_____no_output_____"
],
[
"np.linspace(0, 10, 11) # divide [0, 10] interval into 11 points",
"_____no_output_____"
],
[
"np.logspace(1, 10, 10, base=2, dtype=np.int64)",
"_____no_output_____"
]
],
[
[
"You can easily reshape arrays:",
"_____no_output_____"
]
],
[
[
"np.arange(24).reshape(2, 3, 4)",
"_____no_output_____"
]
],
[
[
"or add dimensions of size 1:",
"_____no_output_____"
]
],
[
[
"print(np.arange(3)[:, np.newaxis])\nprint('---')\nprint(np.arange(3)[np.newaxis, :])",
"_____no_output_____"
]
],
[
[
"Such dimensions are automatically broadcast when doing mathematical operations:",
"_____no_output_____"
]
],
[
[
"np.arange(3)[:, np.newaxis] + np.arange(3)[np.newaxis, :]",
"_____no_output_____"
]
],
[
[
"There is also a number of ways to stack arrays together:",
"_____no_output_____"
]
],
[
[
"matrix1 = np.arange(50).reshape(10, 5)\nmatrix2 = -np.arange(20).reshape(10, 2)\n\nnp.concatenate([matrix1, matrix2], axis=1)",
"_____no_output_____"
],
[
"np.stack([matrix1[:,0], matrix2[:,0]], axis=1)",
"_____no_output_____"
]
],
[
[
"Any matrix can be transposed easily:",
"_____no_output_____"
]
],
[
[
"matrix2.T",
"_____no_output_____"
],
[
"# Your turn: make a (7 x 5) matrix with e_ij = i\n# (i - row number, j - column number)\n\n<YOUR CODE>",
"_____no_output_____"
]
],
[
[
"### How fast is it, harry?",
"_____no_output_____"
],
[
"\n\nLet's compare computation time for python and numpy\n* Two arrays of 10^6 elements\n * first - from 0 to 1 000 000\n * second - from 99 to 1 000 099\n \n* Computing:\n * elemwise sum\n * elemwise product\n * square root of first array\n * sum of all elements in the first array\n ",
"_____no_output_____"
]
],
[
[
"%%time \n# ^-- this \"magic\" measures and prints cell computation time\n\n# Option I: pure python\narr_1 = range(1000000)\narr_2 = range(99,1000099)\n\n\na_sum = []\na_prod = []\nsqrt_a1 = []\nfor i in range(len(arr_1)):\n a_sum.append(arr_1[i]+arr_2[i])\n a_prod.append(arr_1[i]*arr_2[i])\n a_sum.append(arr_1[i]**0.5)\n \narr_1_sum = sum(arr_1)\n",
"_____no_output_____"
],
[
"%%time\n\n# Option II: start from python, convert to numpy\narr_1 = range(1000000)\narr_2 = range(99,1000099)\n\narr_1, arr_2 = np.array(arr_1) , np.array(arr_2)\n\n\na_sum = arr_1 + arr_2\na_prod = arr_1 * arr_2\nsqrt_a1 = arr_1 ** .5\narr_1_sum = arr_1.sum()\n",
"_____no_output_____"
],
[
"%%time\n\n# Option III: pure numpy\narr_1 = np.arange(1000000)\narr_2 = np.arange(99,1000099)\n\na_sum = arr_1 + arr_2\na_prod = arr_1 * arr_2\nsqrt_a1 = arr_1 ** .5\narr_1_sum = arr_1.sum()\n",
"_____no_output_____"
]
],
[
[
"If you want more serious benchmarks, take a look at [this](http://brilliantlywrong.blogspot.ru/2015/01/benchmarks-of-speed-numpy-vs-all.html).",
"_____no_output_____"
],
[
"```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```\n\n```",
"_____no_output_____"
],
[
"### Other numpy functions and features",
"_____no_output_____"
],
[
"There's also a bunch of pre-implemented operations including logarithms, trigonometry, vector/matrix products and aggregations.",
"_____no_output_____"
]
],
[
[
"a = np.array([1,2,3,4,5])\nb = np.array([5,4,3,2,1])\nprint(\"numpy.sum(a) = \", np.sum(a))\nprint(\"numpy.mean(a) = \", np.mean(a))\nprint(\"numpy.min(a) = \", np.min(a))\nprint(\"numpy.argmin(b) = \", np.argmin(b)) # index of minimal element\nprint(\"numpy.dot(a,b) = \", np.dot(a, b)) # dot product. Also used for matrix/tensor multiplication\nprint(\"numpy.unique(['male','male','female','female','male']) = \", np.unique(['male','male','female','female','male']))\n\n# and tons of other stuff. see http://bit.ly/2u5q430 .",
"_____no_output_____"
]
],
[
[
"The important part: all this functionality works with dataframes, as you can get their numpy representation with `.values` (most numpy functions will even work on pure pandas objects):",
"_____no_output_____"
]
],
[
[
"# calling np.max on a pure pandas column:\nprint(\"Max ticket price: \", np.max(data[\"Fare\"]))\n\n# calling np.argmax on a numpy representation of a pandas column\n# to get its positional index:\nprint(\"\\nThe guy who paid the most:\\n\", data.iloc[np.argmax(data[\"Fare\"].values)])",
"_____no_output_____"
],
[
"# your code: compute mean passenger age and the oldest guy on the ship\n<YOUR CODE>",
"_____no_output_____"
],
[
"print(\"Boolean operations\")\n\nprint('a = ', a)\nprint('b = ', b)\nprint(\"a > 2\", a > 2)\nprint(\"numpy.logical_not(a>2) = \", np.logical_not(a>2))\nprint(\"numpy.logical_and(a>2,b>2) = \", np.logical_and(a > 2,b > 2))\nprint(\"numpy.logical_or(a>2,b<3) = \", np.logical_or(a > 2, b < 3))\n\nprint(\"\\n shortcuts\")\nprint(\"~(a > 2) = \", ~(a > 2)) #logical_not(a > 2)\nprint(\"(a > 2) & (b > 2) = \", (a > 2) & (b > 2)) #logical_and\nprint(\"(a > 2) | (b < 3) = \", (a > 2) | (b < 3)) #logical_or",
"_____no_output_____"
]
],
[
[
"Another numpy feature we'll need is indexing: selecting elements from an array. \nAside from python indexes and slices (e.g. a[1:4]), numpy also allows you to select several elements at once.",
"_____no_output_____"
]
],
[
[
"a = np.array([0, 1, 4, 9, 16, 25])\nix = np.array([1,2,5])\nprint(\"a = \", a)\nprint(\"Select by element index\")\nprint(\"a[[1,2,5]] = \", a[ix])\n\nprint(\"\\nSelect by boolean mask\")\nprint(\"a[a > 5] = \", a[a > 5]) # select all elements in a that are greater than 5\nprint(\"(a % 2 == 0) =\", a % 2 == 0) # True for even, False for odd\nprint(\"a[a % 2 == 0] =\", a[a % 2 == 0]) # select all elements in a that are even\n\n\nprint(\"data[(data['Age'] < 18) & (data['Sex'] == 'male')] = (below)\") # select male children\ndata[(data['Age'] < 18) & (data['Sex'] == 'male')]",
"_____no_output_____"
]
],
[
[
"### Your turn\n\nUse numpy and pandas to answer a few questions about data",
"_____no_output_____"
]
],
[
[
"# who on average paid more for their ticket, men or women?\n\nmean_fare_men = <YOUR CODE>\nmean_fare_women = <YOUR CODE>\n\nprint(mean_fare_men, mean_fare_women)",
"_____no_output_____"
],
[
"# who is more likely to survive: a child (<18 yo) or an adult?\n\nchild_survival_rate = <YOUR CODE>\nadult_survival_rate = <YOUR CODE>\n\nprint(child_survival_rate, adult_survival_rate)",
"_____no_output_____"
]
],
[
[
"## Part IV: plots and matplotlib",
"_____no_output_____"
],
[
"Using python to visualize the data is covered by yet another library: `matplotlib`.\n\nJust like python itself, matplotlib has an awesome tendency of keeping simple things simple while still allowing you to write complicated stuff with convenience (e.g. super-detailed plots or custom animations).",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n%matplotlib inline \n# ^-- this \"magic\" selects specific matplotlib backend suitable for\n# jupyter notebooks. For more info see:\n# https://ipython.readthedocs.io/en/stable/interactive/plotting.html#id1\n\n# line plot\nplt.plot([0,1,2,3,4,5],[0,1,4,9,16,25]);",
"_____no_output_____"
],
[
"#scatter-plot\nx = np.arange(5)\nprint(\"x =\", x)\nprint(\"x**2 =\", x**2)\nprint(\"plotting x**2 vs x:\")\nplt.scatter(x, x**2)\n\nplt.show() # show the first plot and begin drawing next one\nplt.plot(x, x**2);",
"_____no_output_____"
],
[
"# draw a scatter plot with custom markers and colors\nplt.scatter([1, 1, 2, 3, 4, 4.5], [3, 2, 2, 5, 15, 24],\n c=[\"red\", \"blue\", \"orange\", \"green\", \"cyan\", \"gray\"],\n marker=\"x\")\n\n# without plt.show(), several plots will be drawn on top of one another\nplt.plot([0, 1, 2, 3, 4, 5], [0, 1, 4, 9, 16, 25], c=\"black\")\n\n# adding more sugar\nplt.title(\"Conspiracy theory proven!!!\")\nplt.xlabel(\"Per capita alcohol consumption\")\nplt.ylabel(\"# Layers in state of the art image classifier\");\n\n# fun with correlations: http://bit.ly/1FcNnWF",
"_____no_output_____"
],
[
"# histogram - showing data density\nplt.hist([0,1,1,1,2,2,3,3,3,3,3,4,4,5,5,5,6,7,7,8,9,10])\nplt.show()\n\nplt.hist([0,1,1,1,2,2,3,3,3,3,3,4,4,5,5,5,6,7,7,8,9,10], bins=5);",
"_____no_output_____"
],
[
"# plot a histogram of age and a histogram of ticket fares on separate plots\n\n<YOUR CODE>\n\n#bonus: use tab to see if there is a way to draw a 2D histogram of age vs fare.",
"_____no_output_____"
],
[
"# make a scatter plot of passenger age vs ticket fare\n\n<YOUR CODE>\n\n# kudos if you add separate colors for men and women",
"_____no_output_____"
]
],
[
[
"* Extended [tutorial](https://matplotlib.org/2.0.2/users/pyplot_tutorial.html)\n* A [cheat sheet](http://bit.ly/2koHxNF)\n* Other libraries for more sophisticated stuff: [Seaborn](https://seaborn.pydata.org/), [Plotly](https://plot.ly/python/), and [Bokeh](https://bokeh.pydata.org/en/latest/)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec6e88d5715d11e1f05b9922f0948378345a1594 | 9,434 | ipynb | Jupyter Notebook | mission_to_mars.ipynb | dylhowe/Web-Scraping-and-Document-Databases | f2e5489c19e348168d0d1017dfbb2bc4ff87f660 | [
"ADSL"
]
| null | null | null | mission_to_mars.ipynb | dylhowe/Web-Scraping-and-Document-Databases | f2e5489c19e348168d0d1017dfbb2bc4ff87f660 | [
"ADSL"
]
| null | null | null | mission_to_mars.ipynb | dylhowe/Web-Scraping-and-Document-Databases | f2e5489c19e348168d0d1017dfbb2bc4ff87f660 | [
"ADSL"
]
| null | null | null | 32.986014 | 1,073 | 0.56625 | [
[
[
"from splinter import Browser\nfrom bs4 import BeautifulSoup\nimport pandas as pd",
"_____no_output_____"
],
[
"executable_path = {'executable_path': 'chromedriver.exe'}\nbrowser = Browser('chrome', **executable_path, headless=True)",
"_____no_output_____"
],
[
"#get Mars News article title and teaser\nurl = 'https://mars.nasa.gov/news/'\nbrowser.visit(url)\nhtml = browser.html\nsoup = BeautifulSoup(html, 'html.parser')\n\n#find the title and teaser text for the first article\narticle_title = soup.find('div', class_='content_title').text\narticle_teaser = soup.find('div', class_='article_teaser_body').text\nprint(article_title)\nprint(article_teaser)",
"Digital Creators: Apply for NASA Mars Landing Event\nUp to 30 multimedia participants will be selected for a special two-day event at JPL culminating in the InSight spacecraft's landing on Mars. Apply by Sept. 3.\n"
],
[
"#get url for fullsize featured image\nurl = 'https://www.jpl.nasa.gov/spaceimages/?search=&category=Mars'\nbrowser.visit(url)\nhtml = browser.html\nsoup = BeautifulSoup(html, 'html.parser')\n\n#click to the featured image\nbrowser.click_link_by_partial_text('FULL IMAGE')\n\n#find html for image\nimg_full_str = str(soup.find('article', class_='carousel_item')[\"style\"])\nprint(img_full_str)\n\n#slice string to get the url extension\nbegin= img_full_str.find(\"/\")\nend=img_full_str.find(\"');\")\nprint(begin)\nprint(end)\nimg_url_ext = img_full_str[begin:end]\nprint(img_url_ext)\n\n#append url extension to base url\nimg_url=\"https://www.jpl.nasa.gov\" + img_url_ext\nprint(img_url)",
"background-image: url('/spaceimages/images/wallpaper/PIA19324-1920x1200.jpg');\n23\n75\n/spaceimages/images/wallpaper/PIA19324-1920x1200.jpg\nhttps://www.jpl.nasa.gov/spaceimages/images/wallpaper/PIA19324-1920x1200.jpg\n"
],
[
"#Get the most recent Mars weather update from twitter\nurl = 'https://twitter.com/marswxreport?lang=en'\nbrowser.visit(url)\nhtml = browser.html\nsoup = BeautifulSoup(html, 'html.parser')\n\n#find text from the most recent tweet\nmars_weather = soup.find('p', class_=\"TweetTextSize TweetTextSize--normal js-tweet-text tweet-text\").text\n\n\nprint(mars_weather)",
"Sol 2147 (2018-08-21), high -15C/5F, low -68C/-90F, pressure at 8.70 hPa, daylight 05:30-17:44\n"
],
[
"#Get the table of Mars Facts\nurl = 'https://space-facts.com/mars/'\nbrowser.visit(url)\nhtml = browser.html\nsoup = BeautifulSoup(html, 'html.parser')\n\n#read the table to a dataframe, set index, and write to HTML\nfacts_df = pd.read_html(url, encoding=\"utf-8-sig\")[0].set_index(0)\n\nfacts_html = facts_df.to_html()\nfacts_html",
"_____no_output_____"
],
[
"#Get Mars hemisphere titles and images\nurl = 'https://astrogeology.usgs.gov/search/results?q=hemisphere+enhanced&k1=target&v1=Mars'\nbrowser.visit(url)\nhtml = browser.html\nsoup = BeautifulSoup(html, 'html.parser')\n\n#create list for dicts\ntitles_urls = []\n\n#get titles and links\ntitlereturn = soup.find_all('h3')\n\nfor title in titlereturn:\n \n html = browser.html\n soup = BeautifulSoup(html, 'html.parser')\n \n #save title text\n titletext=title.text\n print(titletext)\n \n #click to full image\n browser.click_link_by_partial_text(titletext)\n \n #save full image URL\n html = browser.html\n soup = BeautifulSoup(html, 'html.parser')\n \n #append url extension to base url\n img_end=str(soup.find_all(\"img\", class_=\"wide-image\")[0][\"src\"])\n imgurl = \"https://astrogeology.usgs.gov\" + img_end\n print(imgurl)\n \n #go back to homepage\n browser.click_link_by_partial_text(\"Back\")\n \n #save title and url pair to list of dicts\n titles_urls.append({\"title\":titletext, \"img_url\":imgurl})\n \ntitles_urls\n\n ",
"Cerberus Hemisphere Enhanced\nhttps://astrogeology.usgs.gov/cache/images/cfa62af2557222a02478f1fcd781d445_cerberus_enhanced.tif_full.jpg\nSchiaparelli Hemisphere Enhanced\nhttps://astrogeology.usgs.gov/cache/images/3cdd1cbf5e0813bba925c9030d13b62e_schiaparelli_enhanced.tif_full.jpg\nSyrtis Major Hemisphere Enhanced\nhttps://astrogeology.usgs.gov/cache/images/ae209b4e408bb6c3e67b6af38168cf28_syrtis_major_enhanced.tif_full.jpg\nValles Marineris Hemisphere Enhanced\nhttps://astrogeology.usgs.gov/cache/images/7cf2da4bf549ed01c17f206327be4db7_valles_marineris_enhanced.tif_full.jpg\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6e89c0394aa7fce284515edad30ed7667340a3 | 44,020 | ipynb | Jupyter Notebook | AzureNotebooks/01-turbofan_regression.ipynb | devlead/IoTEdgeAndMlSample | 624e933cc6c1f13e84e246077ed9df6519b5c7d6 | [
"MIT"
]
| null | null | null | AzureNotebooks/01-turbofan_regression.ipynb | devlead/IoTEdgeAndMlSample | 624e933cc6c1f13e84e246077ed9df6519b5c7d6 | [
"MIT"
]
| 1 | 2020-04-09T09:36:46.000Z | 2020-04-16T22:11:42.000Z | AzureNotebooks/01-turbofan_regression.ipynb | devlead/IoTEdgeAndMlSample | 624e933cc6c1f13e84e246077ed9df6519b5c7d6 | [
"MIT"
]
| null | null | null | 73.366667 | 3,707 | 0.640595 | [
[
[
"# Introduction\n\nIn this notebook, we demonstrate the steps needed create a model for predicting Remaining Useful Life for turbofan engines based on data collected by devices and routed into storage via the IoT Hub. The notebook assumes that you have complete the device data generation steps from the [IoT Edge for Machine Learning](aka.ms/IoTEdgeMLPaper). The data generated from the devices needs to be in an Azure Storage account blob container in the same Azure Subscription as you will use to create the Azure Machine Learning service workspace using this notebook. \n\nThe steps we will complete in this notebooks are:\n 1. Create a Machine Learning service workspace for managing the experiments, compute, and models for this sample\n 1. Load training data from Azure Storage\n 1. Prepare the data for training the model\n 1. Explore the data \n 1. Remotely train the model\n 1. Test the model using test data\n\nThe intent is not to provide an extensive coverage of machine learning in Azure as that is covered in much depth elsewhere [here for example](https://github.com/Azure/MachineLearningNotebooks), but to demonstrate how machine learning can be used with IoT Edge.",
"_____no_output_____"
],
[
"# Setup notebook\n\nPlease ensure that you are running the notebook under the Python 3.6 kernal. Intall fastavro and setup interactive shell to display output nicely.\n\n>You may see a warning about Matplotlib building the font cache. You may ignore the warning as it is benign.",
"_____no_output_____"
]
],
[
[
"!pip install fastavro\n\nfrom IPython.core.interactiveshell import InteractiveShell\nInteractiveShell.ast_node_interactivity = \"all\"\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### List versions of azureml modules.",
"_____no_output_____"
]
],
[
[
"!pip list | grep azureml-sdk\n\n!pip list | grep azureml-train",
"_____no_output_____"
]
],
[
[
"### azureml-sdk version check\n\nazureml-sdk has introduced breaking changes in version 1.0.76. Upgrading to azureml-train-automl from versions <=v1.0.76 can cause partial installations, causing some auto import to fail. This also affects other libraries used in this sample that are incompatible with v1.0.79. If your current version is <=1.0.79, please run the following 3 cells to resolve issues with breaking changes.\n\nNote: There is no harm in running this even if you are on later versions. This just fixes the issues for older versions.",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade tqdm\n\n!pip install --ignore-installed entrypoints",
"_____no_output_____"
],
[
"!pip install --upgrade azureml-train-automl\n\n!pip install --ignore-installed azureml-train-automl-client",
"_____no_output_____"
],
[
"!pip uninstall --yes azureml-contrib-notebook\n\n!pip install azureml-contrib-notebook\n\n!pip install --upgrade azureml-explain-model",
"_____no_output_____"
]
],
[
[
"### Install latest version of azureml-sdk and automl",
"_____no_output_____"
]
],
[
[
"!pip install --upgrade azureml-sdk[automl]",
"_____no_output_____"
]
],
[
[
"## Move data files to data directory",
"_____no_output_____"
]
],
[
[
"import os, glob, shutil\n\nif not os.path.exists('./data'):\n os.mkdir('./data')\n\nfor f in glob.glob('./*.txt') + glob.glob('./*.csv'):\n shutil.move(f, './data/')",
"_____no_output_____"
]
],
[
[
"# Set global properties\n\nThese properties will be used throughout the notebook.\n * `AZURE_SUBSCRIPTION_ID` - the Azure subscription containing the storage account where device data has been uploaded. We will create the Machine Learning service workspace (ml workspace) in this subscription.\n * `ML_WORKSPACE_NAME` name to give the ml workspace\n * `AZURE_IOT_HUB_NAME` - name of the Azure IoT Hub used in creating the device data using the DeviceHarness. See [IoT Edge for Machine Learning](aka.ms/IoTEdgeMLPaper) for details.\n * `RESOURCE_GROUP_NAME` - name of the resource group where the IoT Hub exists\n * `LOCATION` - the Azure location of the IoT Hub\n * `STORAGE_ACCOUNT_NAME` - name of the Azure Storage account where device data was routed via IoT Hub.\n * `STORAGE_ACCOUNT_KEY` - access key for the Azure Storage account\n * `STORAGE_ACCOUNT_CONTAINER` - name of Azure Storage blob container where device data was routed via IoT Hub.",
"_____no_output_____"
]
],
[
[
"AZURE_SUBSCRIPTION_ID = ''\nML_WORKSPACE_NAME = 'turbofanDemo'\nAZURE_IOT_HUB_NAME = ''\nRESOURCE_GROUP_NAME = ''\nLOCATION = ''\nSTORAGE_ACCOUNT_NAME = ''\nSTORAGE_ACCOUNT_KEY = ''\nSTORAGE_ACCOUNT_CONTAINER = 'devicedata'\n\nif (AZURE_SUBSCRIPTION_ID == ''\n or ML_WORKSPACE_NAME == ''\n or AZURE_IOT_HUB_NAME == ''\n or RESOURCE_GROUP_NAME == ''\n or LOCATION == ''\n or STORAGE_ACCOUNT_NAME == ''\n or STORAGE_ACCOUNT_KEY == ''\n or STORAGE_ACCOUNT_CONTAINER == ''):\n raise ValueError('All values must be filled in') ",
"_____no_output_____"
]
],
[
[
"# Create a workspace\n\n## What is an Azure ML Workspace and Why Do I Need One?\n\nAn Azure ML workspace is an Azure resource that organizes and coordinates the actions of many other Azure resources to assist in executing and sharing machine learning workflows. In particular, an Azure ML workspace coordinates storage, databases, and compute resources providing added functionality for machine learning experimentation, operationalization, and the monitoring of operationalized models.\n\nIn addition to creating the workspace, the cell below writes a file, config.json, to a ./.azureml/config.json, which allows the Workspace object to be reloaded later.\n\n\n\n\n><font color=gray>_Note: currently Workspaces are supported in the following regions: eastus2, eastus,westcentralus, southeastasia, westeurope, australiaeast, westus2, southcentralus_</font>\n\nYou may need to authenticate with Azure when running this cell. If so you will see a message like: \n\n```To sign in, use a web browser to open the page https://microsoft.com/devicelogin and enter the code XXXXXXXXX to authenticate.```\n\nIf you are logged in with an AAD account you will instead be prompted to allow access to Azure.\n\nOnce you authenticate, the cell will finish creating the Workspace.\n\n>To facilitate rerunning the notebook with the same Workspace, the cell first checks for the presence of a config. If it finds the config, it loads the Workspace from the config instead of creating it.",
"_____no_output_____"
]
],
[
[
"from azureml.core import Workspace\nworkspace_name = ML_WORKSPACE_NAME\nsubscription_id = AZURE_SUBSCRIPTION_ID\nresource_group = RESOURCE_GROUP_NAME\nlocation = LOCATION\n\nif not os.path.exists('./aml_config'):\n os.mkdir('./aml_config')\n\n#check to see if the workspace has already been created and persisted\nif (os.path.exists('./aml_config/.azureml/config.json')):\n ws = Workspace.from_config(path='./aml_config')\nelse:\n ws = Workspace.create(name=workspace_name,\n subscription_id=subscription_id,\n resource_group=resource_group,\n create_resource_group=True,\n location=location\n )\n\n ws.write_config(path='./aml_config')",
"_____no_output_____"
]
],
[
[
"## Workspace details\n\nPrint the Workspace details.",
"_____no_output_____"
]
],
[
[
"import azureml.core\nimport pandas as pd\nfrom azureml.core import Workspace\n\noutput = {}\noutput['SDK version'] = azureml.core.VERSION\noutput['Subscription ID'] = ws.subscription_id\noutput['Workspace'] = ws.name\noutput['Resource Group'] = ws.resource_group\noutput['Location'] = ws.location\npd.set_option('display.max_colwidth', -1)\npd.DataFrame(data=output, index=['']).T",
"_____no_output_____"
]
],
[
[
"# Download data from storage\n\nThe first step toward creating a model for RUL is to explore the data and understand its shape. We will download the data for this purpose, realizing that in the case of larger data sets only a sample of the data would be used at this step.",
"_____no_output_____"
],
[
"## Register storage account\n\nThe Datastore is a convenient construct associated the Workspace to upload/download data, and interact with it from remote compute targets. Register the Azure Storage account and container where device data was routed by IoT Hub using the information about the storage container provided at the beginning of the notebook.\n",
"_____no_output_____"
]
],
[
[
"from azureml.core import Datastore\n\nds = Datastore.register_azure_blob_container(workspace=ws,\n datastore_name='turbofan',\n container_name=STORAGE_ACCOUNT_CONTAINER,\n account_name=STORAGE_ACCOUNT_NAME,\n account_key=STORAGE_ACCOUNT_KEY,\n create_if_not_exists=False)",
"_____no_output_____"
]
],
[
[
"## Use Datastore to download data\n\nUse the Datastore to download the files to the local machine. The prefix is the top level path to download, which should be the name of the IoT Hub. ",
"_____no_output_____"
]
],
[
[
"ds.download(target_path=\"./data/download\", prefix=AZURE_IOT_HUB_NAME)",
"_____no_output_____"
]
],
[
[
"# Load train data\n\nThe data we just downloaded represent a series of messages sent by each device stored in [Apache Avro](https://avro.apache.org/docs/current/)(avro) format. We will use the fastavro package to deserialize the records from the avro files.\nHere is an example deserialized record from the avro file. \n\n```json \n{\n \"EnqueuedTimeUtc\": \"2018-12-01T01: 16: 22.0950000Z\",\n \"Properties\": {},\n \"SystemProperties\": {\n \"connectionDeviceId\": \"Client_3\",\n \"connectionAuthMethod\": {\n \"scope\": \"device\",\n \"type\": \"sas\",\n \"issuer\": \"iothub\",\n \"acceptingIpFilterRule\": null\n },\n \"connectionDeviceGenerationId\": \"636791290544434625\",\n \"contentType\": \"application/json\",\n \"contentEncoding\": \"utf-8\",\n \"enqueuedTime\": \"2018-12-01T01: 16: 22.0950000Z\"\n },\n \"Body\": b'{\n \"CycleTime\": 1,\n \"OperationalSetting1\": -0.0001,\n \"OperationalSetting2\": 0.0001,\n \"OperationalSetting3\": 100.0,\n \"Sensor1\": 518.67,\n \"Sensor2\": 642.03,\n //Sensor 3-19 ommitted for brevity\n \"Sensor20\": 38.99,\n \"Sensor21\": 23.296\n }\n}```\n\nTaken together the messages represent a time series of data for multiple engines. Each engine is operating normally at the start of each time series, and develops a fault at some point during the series. The fault grows in magnitude until system failure (i.e. the failure point for the engine is the final cycle in the set). The remaining useful life (RUL) is therefore expressed as: \n\n$$RUL_{current} = Cycle_{max} - Cycle_{current}$$\n",
"_____no_output_____"
],
[
"## Create utils for loading data from avro files\n\nDefine a set of utility methods for loading the data from the avro files. We use thes utilities to load the locally downloaded data. Later in the notebook, these same utilities will form the basis of data processing for remote training (see **Train regression using Azure AutoMl and remote execution** below)",
"_____no_output_____"
]
],
[
[
"%%writefile ./utils.py\n\nimport glob\nimport json\nimport pandas as pd\n\nfrom fastavro import reader\nfrom os.path import isfile\nfrom multiprocessing.dummy import Pool as ThreadPool \n\n# parse connectionDeviceId and return the int part\n# (e.g. Client_1 becomes 1)\ndef get_unit_num (unit_record):\n unit = unit_record[\"connectionDeviceId\"]\n return int(unit.split('_')[1])\n\n# create data row from avro file record\ndef load_cycle_row(record):\n json_body = record[\"Body\"].decode()\n row = json.loads(json_body)\n row.update({'Unit': get_unit_num(record[\"SystemProperties\"])})\n row.update({'QueueTime': pd.to_datetime(record[\"EnqueuedTimeUtc\"])})\n return row\n\n# add row to data frame\ndef append_df(base_df, append_df):\n if(base_df is None):\n base_df = pd.DataFrame(append_df)\n else:\n base_df = base_df.append(append_df, ignore_index=True)\n return base_df\n\n# sort rows and columns in dataframe\ndef sort_and_index(index_data):\n #sort rows and reset index\n index_data.sort_values(by=['Unit', 'CycleTime'], inplace=True)\n index_data.reset_index(drop=True, inplace=True)\n \n #fix up column sorting for convenience in notebook\n sorted_cols = ([\"Unit\",\"CycleTime\", \"QueueTime\"] \n + [\"OperationalSetting\"+str(i) for i in range(1,4)] \n + [\"Sensor\"+str(i) for i in range(1,22)])\n\n return index_data[sorted_cols]\n\n# load data from an avro file and return a dataframe\ndef load_avro_file(avro_file_name):\n with open(avro_file_name, 'rb') as fo:\n file_df = None\n avro_reader = reader(fo)\n print (\"load records from file: %s\" % avro_file_name)\n for record in avro_reader:\n row = load_cycle_row(record)\n file_df = append_df(base_df=file_df, append_df=[row])\n return file_df\n\n# load data from all avro files in given dir \ndef load_avro_directory(avro_dir_name):\n lst = glob.iglob(avro_dir_name, recursive=True)\n files = [x for x in lst if isfile(x)]\n pool = ThreadPool(4)\n results = pool.map(load_avro_file, files)\n pool.close()\n pool.join()\n\n dir_df = None\n for df in results:\n dir_df = append_df(base_df=dir_df, append_df=df)\n print(\"loaded %d records\" % dir_df.shape[0])\n return sort_and_index(dir_df)\n\n# add max cycle to each row in the data\ndef add_maxcycle(data_frame):\n # cleanup column if it already exists\n if 'MaxCycle' in data_frame.columns:\n data_frame.drop('MaxCycle', axis=1, inplace=True)\n\n total_cycles = data_frame.groupby(['Unit']).agg({'CycleTime' : 'max'}).reset_index()\n total_cycles.rename(columns = {'CycleTime' : 'MaxCycle'}, inplace = True)\n return data_frame.merge(total_cycles, how = 'left', left_on = 'Unit', right_on = 'Unit')\n\n# return a remaining useful life class based on RUL\ndef classify_rul(rul):\n if (rul <= 25):\n return 'F25'\n elif (rul <= 75):\n return 'F75'\n elif (rul <= 150):\n return 'F150'\n else:\n return 'Full'\n \n# add remaining useful life and remaing useful life class\n# to each row in the data\ndef add_rul(data_frame):\n data_frame = add_maxcycle(data_frame)\n \n if 'RUL' in data_frame.columns:\n data_frame.drop('RUL', axis=1, inplace=True)\n data_frame['RUL'] = data_frame.apply(lambda r: int(r['MaxCycle'] - r['CycleTime']), axis = 1)\n\n if 'RulClass' in data_frame.columns:\n data_frame.drop('RulClass', axis=1, inplace=True)\n data_frame['RulClass'] = data_frame.apply(lambda r: classify_rul(r['RUL']), axis = 1)\n \n return data_frame\n\n",
"_____no_output_____"
]
],
[
[
"## Use utils to load data from download directory\n\nThis step will take several minutes.",
"_____no_output_____"
]
],
[
[
"import utils\nimport pandas as pd\n\n# check to see if this step has run before and if it has load the data rather than recreating it\nif (os.path.exists('./data/WebServiceTrain.csv')):\n train_pd = pd.read_csv('./data/WebServiceTrain.csv')\nelse:\n train_pd = utils.load_avro_directory('./data/download/**/*')\n\ntrain_pd.head(5)",
"_____no_output_____"
]
],
[
[
"## Calculate remaining useful life and RUL class labels\n\nAdd RUL for regression training and RulClass for classification",
"_____no_output_____"
]
],
[
[
"train_pd = utils.add_rul(train_pd)\n\ncols = ['Unit', 'CycleTime', 'MaxCycle', 'RUL', 'RulClass']\n#show first 5 rows\ntrain_pd[cols].head(5)\n\n#show last 5 rows for engine 3\ntrain_pd[train_pd['Unit'] == 3][cols].tail(5)",
"_____no_output_____"
]
],
[
[
"## Display train data",
"_____no_output_____"
]
],
[
[
"#persist data so we can recover if kernel dies\ntrain_pd.to_csv('./data/WebServiceTrain.csv')\n\n#show the first five rows\ntrain_pd.head(5)",
"_____no_output_____"
]
],
[
[
"# Explore the data\n\nVisualize the data to start to get a sense of how features like sensor measurements and operations settings relate to remaining useful life (RUL)",
"_____no_output_____"
],
[
"## Sensor readings and RUL\n\nCreate a scatterplot for each sensor measurement vs. RUL. Notice that some measurements (e.g. sensor 2) seem to be correlated strongly to RUL whereas other measurements (e.g. sensor 1) stay constant throughout the life of the engine.\n\n \n><font color=gray>_Note: the data is limited to the first 10 engine units for speed of rendering_</font>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\n\n#select the data to plot\nplotData = train_pd.query('Unit < 10');\n\nsns.set()\ng = sns.PairGrid(data=plotData,\n x_vars = ['RUL'],\n y_vars = [\"Sensor\"+str(i) for i in range(1,22)],\n hue=\"Unit\",\n height=3,\n aspect=2.5,\n palette=\"Paired\")\ng = g.map(plt.scatter, alpha=0.3)\ng = g.set(xlim=(300,0))\ng = g.add_legend()\n",
"_____no_output_____"
]
],
[
[
"## Operational settings and RUL\n\nCreate a scatterplot for each operation setting vs. RUL. Operational settings do not seem to correlate with RUL.\n \n><font color=gray>_Note: the data is limited to the first 10 engine units for speed of rendering_</font>",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport seaborn as sns\nimport utils\n\nplotData = train_pd.query('Unit < 10');\nsns.set()\ng = sns.PairGrid(data=plotData,\n x_vars = ['RUL'],\n y_vars = [\"OperationalSetting\"+str(i) for i in range(1,4)],\n hue=\"Unit\",\n height=3,\n aspect=2.5,\n palette=\"Paired\")\ng = g.map(plt.scatter, alpha=0.3)\ng = g.set(xlim=(300,0))\ng = g.add_legend()\n",
"_____no_output_____"
]
],
[
[
"# Train model using Azure AutoMl and remote execution\n\nIn this section, we will use the Azure Machine Learning service to build a model to predict remaining useful life.",
"_____no_output_____"
],
[
"## Create remote compute target\n\nAzure ML Managed Compute is a managed service that enables data scientists to train machine learning models on clusters of Azure virtual machines, including VMs with GPU support. This code creates an Azure Managed Compute cluster if it does not already exist in your workspace. \n\n **Creation of the cluster takes approximately 5 minutes.** If the cluster is already in the workspace this code uses it and skips the creation process.",
"_____no_output_____"
]
],
[
[
"from azureml.core.compute import AmlCompute\nfrom azureml.core.compute import ComputeTarget\nimport os\n\nCLUSTER_NAME = \"mlturbo\"\n\n# choose a name for your cluster\nbatchai_cluster_name = CLUSTER_NAME + \"gpu\"\ncluster_min_nodes = 0\ncluster_max_nodes = 3\nvm_size = \"STANDARD_NC6\" #NC6 is GPU-enabled\n \ncts = ws.compute_targets\nif batchai_cluster_name in cts:\n found = True\n print('Found existing compute target...%s' % batchai_cluster_name)\n compute_target = cts[batchai_cluster_name]\nelse:\n print('creating a new compute target...')\n provisioning_config = AmlCompute.provisioning_configuration(vm_size = vm_size, \n # vm_priority = 'lowpriority', #optional\n min_nodes = cluster_min_nodes, \n max_nodes = cluster_max_nodes)\n\n # create the cluster\n compute_target = ComputeTarget.create(ws, batchai_cluster_name, provisioning_config)\n \n # can poll for a minimum number of nodes and for a specific timeout. \n # if no min node count is provided it will use the scale settings for the cluster\n compute_target.wait_for_completion(show_output=True, min_node_count=None, timeout_in_minutes=20)\n \n # For a more detailed view of current BatchAI cluster status, use the 'status' property \n compute_target.status.serialize()",
"_____no_output_____"
]
],
[
[
"# Create a regression model",
"_____no_output_____"
],
[
"## Configure run settings\n\nCreate a DataReferenceConfiguration object to inform the system what data folder to download to the compute target. The path_on_compute should be an absolute path to ensure that the data files are downloaded only once. The get_data method should use the same path to access the data files.",
"_____no_output_____"
],
[
"### Setup DataReference",
"_____no_output_____"
]
],
[
[
"from azureml.train.automl import AutoMLConfig\nfrom azureml.core.runconfig import DataReferenceConfiguration\n\ndr = DataReferenceConfiguration(datastore_name=ds.name, \n path_on_datastore=AZURE_IOT_HUB_NAME, \n path_on_compute='/tmp/azureml_runs',\n mode='download', # download files from datastore to compute target\n overwrite=False)",
"_____no_output_____"
]
],
[
[
"### Update run settings",
"_____no_output_____"
]
],
[
[
"from azureml.core.runconfig import RunConfiguration\nfrom azureml.core.conda_dependencies import CondaDependencies\n\n# create a new RunConfig object\nconda_run_config = RunConfiguration(framework=\"python\")\n\n# Set compute target to the Azure ML managed compute\nconda_run_config.target = compute_target\n\n# set the data reference of the run coonfiguration\nconda_run_config.data_references = {ds.name: dr}\n\n#specify package dependencies needed to load data and train the model\ncd = CondaDependencies.create(pip_packages=['azureml-sdk[automl]', 'fastavro'], conda_packages=['numpy'])\nconda_run_config.environment.python.conda_dependencies = cd",
"_____no_output_____"
]
],
[
[
"## Create data retrieval script\n\nRemote execution requires a .py file containing a get_data() function that will be used to retrieve data from the mounted storage. We will create the file for retrieving data by copying the utils.py file to our script folder as get_data.py. Then we will append a get_data(), which uses the utility methods for data loading, into the newly created get_data.py. ",
"_____no_output_____"
],
[
"### Create a directory ",
"_____no_output_____"
]
],
[
[
"import os\nscript_folder = './turbofan-regression'\nos.makedirs(script_folder, exist_ok=True)",
"_____no_output_____"
]
],
[
[
"### Create get data script",
"_____no_output_____"
]
],
[
[
"# create the script by copyting utils.py to the script_folder\nimport shutil\nshutil.copyfile('utils.py', script_folder + '/get_data.py')",
"_____no_output_____"
]
],
[
[
"### Append the get_data method to the newly created get_data.py\n",
"_____no_output_____"
]
],
[
[
"%%writefile -a $script_folder/get_data.py\n\ndef get_data():\n #for the sake of simplicity use all sensors as training features for the model\n features = [\"Sensor\"+str(i) for i in range(1,22)]\n train_pd = load_avro_directory('/tmp/azureml_runs/**/*')\n train_pd = add_rul(train_pd)\n y_train = train_pd['RUL'].values\n X_train = train_pd[features].values\n return { \"X\" : X_train, \"y\" : y_train}\n\n",
"_____no_output_____"
]
],
[
[
"## Run the experiment on Azure ML compute \n### Instantiate AutoML\n\nIn the interest of time, the cell below uses a short iteration timeout, **3 min**, and a small number of iterations, **10**. Longer iteration timeouts and a greater number of iterations will yield better results",
"_____no_output_____"
]
],
[
[
"import logging\nfrom azureml.train.automl import AutoMLConfig\n\n#name project folder and experiment\nexperiment_name = 'turbofan-regression-remote'\n\nautoml_settings = {\n \"iteration_timeout_minutes\": 3,\n \"iterations\": 10,\n \"n_cross_validations\": 10,\n \"primary_metric\": 'spearman_correlation',\n \"max_cores_per_iteration\": -1,\n \"enable_ensembling\": True,\n \"ensemble_iterations\": 5,\n \"verbosity\": logging.INFO,\n \"preprocess\": True,\n \"enable_tf\": True,\n \"auto_blacklist\": True\n}\n\nAutoml_config = AutoMLConfig(task = 'regression',\n debug_log = 'auto-regress.log',\n path=script_folder,\n run_configuration=conda_run_config,\n data_script = script_folder + \"/get_data.py\",\n **automl_settings\n )",
"_____no_output_____"
]
],
[
[
"### Run the experiment\n\nRun the experiment on the remote compute target and show results as the runs execute. Assuming you have kept the auto_ml settings set in the notebook this step will take several minutes. If you have increased the number of iterations of the iteration timeout it will take longer.\n\n>Note: unlike other cells, this one is not finished until the \"Status\" in the output below shows \"Completed\". If it shows a failure, you can check the status in the Azure portal (link will be at the bottom of the output) to learn more.",
"_____no_output_____"
]
],
[
[
"from azureml.core.experiment import Experiment\nfrom azureml.widgets import RunDetails\n\nexperiment=Experiment(ws, experiment_name)\nregression_run = experiment.submit(Automl_config, show_output=True)\nRunDetails(regression_run).show()",
"_____no_output_____"
]
],
[
[
"# Explore the results\n\nExplore the results of the automatic training using the run details.",
"_____no_output_____"
],
[
"## Reconstitute a run\nGiven the long running nature of running the experiment the notebook may have been closed or timed out. In that case, to retrieve the run from the run id set the value of `run_id` to the run_id of the experiment. We use `AutoMLRun` from `azureml.train.automl.run`",
"_____no_output_____"
]
],
[
[
"from azureml.train.automl.run import AutoMLRun\nfrom azureml.core.experiment import Experiment\nfrom azureml.core import Workspace\n\nrun_id = 'AutoML_xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxxx'\n\nif 'regression_run' not in globals():\n ws = Workspace.from_config(path='./aml_config')\n experiment_name = 'turbofan-regression-remote'\n experiment=Experiment(ws, experiment_name)\n regression_run = AutoMLRun(experiment = experiment, \n run_id = run_id)\n\nregression_run.id",
"_____no_output_____"
]
],
[
[
"## Retrieve all iterations\n\nView the experiment history and see individual metrics for each iteration run.",
"_____no_output_____"
]
],
[
[
"children = list(regression_run.get_children())\nmetricslist = {}\nfor run in children:\n properties = run.get_properties()\n metrics = {k: v for k, v in run.get_metrics().items() if isinstance(v, float)}\n metricslist[int(properties['iteration'])] = metrics\n\nimport pandas as pd\nrundata = pd.DataFrame(metricslist).sort_index(1)\nrundata",
"_____no_output_____"
]
],
[
[
"## Register the best model \n\nUse the `regression_run` object to get the best model and register it into the workspace. ",
"_____no_output_____"
]
],
[
[
"best_run, fitted_model = regression_run.get_output()\n\n# register model in workspace\ndescription = 'Aml Model ' + regression_run.id[7:15]\ntags = None\nregression_run.register_model(description=description, tags=tags)\nregression_run.model_id # Use this id to deploy the model as a web service in Azure",
"_____no_output_____"
]
],
[
[
"### Save model information for deployment\n\nPersist the information that we will need to deploy the model in the [turbofan deploy model](./turbofan_deploy_model.ipynb)",
"_____no_output_____"
]
],
[
[
"import json\nimport os\n\nmodel_information = {'regressionRunId': regression_run.id, 'modelId': regression_run.model_id, 'experimentName': experiment.name}\nwith open('./aml_config/model_config.json', 'w') as fo:\n json.dump(model_information, fo)",
"_____no_output_____"
]
],
[
[
"# Load test data\n\nIn the test set, the time series ends some time prior to system failure. The actual\nremaining useful life (RUL) are given in the RUL_*.txt files. The data in the RUL files is a single vector where the index corresponds to the unit number of the engine and the value corresponds to the actual RUL at the end of the test.\n\nThe RUL for a given cycle in the training set is given by adding the RUL at test end (from the RUL vector file) to the maximum cycle in the test data and then subtracting the current cycle:\n\n$$RUL_{current} = RUL_{TestEnd} + Cycle_{max} - Cycle_{current}$$\n\nTaking unit number 1 as an example:\n * Taking the first value from RUL_FD003.txt gives: $RUL_{TestEnd} = 44$\n * The final(max) cycle value from test_FD003.txt gives: $Cycle_{max} = 233$\n * The values for the first 5 cycles for engine 1 are:\n\n|Unit|Cycle|Max Cycle|Test End RUL|Remaining Life|\n|-----|-----|-----|-----|-----|\n|1|1|233|44|276|\n|1|2|233|44|275|\n|1|3|233|44|274|\n|1|4|233|44|273|\n|1|5|233|44|272|\n",
"_____no_output_____"
],
[
"\n## Define some methods for loading from text files",
"_____no_output_____"
]
],
[
[
"import json\nimport utils\nimport pandas as pd\nfrom os.path import isfile\n\ndef add_column_names(data_frame):\n data_frame.columns = ([\"Unit\",\"CycleTime\"]\n + [\"OperationalSetting\"+str(i) for i in range(1,4)]\n + [\"Sensor\"+str(i) for i in range(1,22)])\n\ndef read_data_file(full_file_name):\n data = pd.read_csv(full_file_name, sep = ' ', header = None)\n data.dropna(axis='columns', inplace=True)\n return data\n\ndef load_rul_data(full_file_name):\n rul_data = read_data_file(full_file_name)\n\n # add a column for the unit and fill with numbers 1..n where\n # n = number of rows of RUL data\n rul_data['Unit'] = list(range(1, len(rul_data) + 1))\n rul_data.rename(columns = {0 : 'TestEndRUL'}, inplace = True)\n return rul_data\n\n\ndef load_test_data(test_full_file_name, rul_full_file_name):\n data = read_data_file(test_full_file_name)\n add_column_names(data)\n data = utils.add_maxcycle(data)\n \n rul_data = load_rul_data(rul_full_file_name)\n data = data.merge(rul_data, how = 'left', left_on = 'Unit', right_on = 'Unit')\n data['RUL'] = data.apply(lambda r: int(r['MaxCycle'] + r['TestEndRUL'] - r['CycleTime']), axis = 1)\n data['RulClass'] = data.apply(lambda r: utils.classify_rul(r['RUL']), axis = 1)\n\n return data\n",
"_____no_output_____"
]
],
[
[
"## Read and process the test data",
"_____no_output_____"
]
],
[
[
"dataset = \"FD003\" \nrul_file_name = 'data/RUL_' + dataset + '.txt'\ntest_file_name = 'data/test_' + dataset + '.txt'\n\ntest_pd = load_test_data(test_file_name, rul_file_name)\ntest_pd.head(5)",
"_____no_output_____"
]
],
[
[
"### Serialize test data\n\nSave off the data so that we can use it when we test the web service in the [turbofan deploy model](./turbofan_deploy_model.ipynb) notebook.",
"_____no_output_____"
]
],
[
[
"test_pd.to_csv('./data/WebServiceTest.csv')",
"_____no_output_____"
]
],
[
[
"# Test regression model\n\npredict on training and test set and calculate residual values",
"_____no_output_____"
]
],
[
[
"selected_features = [\"Sensor\"+str(i) for i in range(1,22)]\n\n#reload data in case the kernel died at some point\nif 'train_pd' not in globals():\n train_pd = pd.read_csv(\"data/WebServiceTrain.csv\")\n\n#load the values used to train the model\nX_train = train_pd[selected_features].values\ny_train = train_pd['RUL'].values\n\n#predict and calculate residual values for train\ny_pred_train = fitted_model.predict(X_train)\ny_residual_train = y_train - y_pred_train\n\ntrain_pd['predicted'] = y_pred_train; \ntrain_pd['residual'] = y_residual_train\n\n#load the values from the test set\nX_test = test_pd[selected_features].values\ny_test = test_pd['RUL'].values\n\n#predict and calculate residual values for test\ny_pred_test = fitted_model.predict(X_test)\ny_residual_test = y_test - y_pred_test\n\ntest_pd['predicted'] = y_pred_test;\ntest_pd['residual'] = y_residual_test",
"_____no_output_____"
],
[
"train_pd[['Unit', 'RUL', 'predicted', 'residual']].head(5)\ntest_pd[['Unit', 'RUL', 'predicted', 'residual']].head(5)",
"_____no_output_____"
]
],
[
[
"## Predicted vs. actual\n\nPlot the predicted RUL against the actual RUL. The dashed line represents the ideal model.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\nfrom sklearn.metrics import mean_squared_error, r2_score\n\nfig, (ax1,ax2) = plt.subplots(nrows=2, sharex=True)\nfig.set_size_inches(16, 16)\n\nfont_size = 14\n\ng = sns.regplot(y='predicted', x='RUL', data=train_pd, fit_reg=False, ax=ax1)\nlim_set = g.set(ylim=(0, 500), xlim=(0, 500))\nplot = g.axes.plot([0, 500], [0, 500], c=\".3\", ls=\"--\");\n\nrmse = ax1.text(16,450,'RMSE = {0:.2f}'.format(np.sqrt(mean_squared_error(y_train, y_pred_train))), fontsize = font_size)\nr2 = ax1.text(16,425,'R2 Score = {0:.2f}'.format(r2_score(y_train, y_pred_train)), fontsize = font_size)\n\ng2 = sns.regplot(y='predicted', x='RUL', data=test_pd, fit_reg=False, ax=ax2)\nlim_set = g2.set(ylim=(0, 500), xlim=(0, 500))\nplot = g2.axes.plot([0, 500], [0, 500], c=\".3\", ls=\"--\");\n\nrmse = ax2.text(16,450,'RMSE = {0:.2f}'.format(np.sqrt(mean_squared_error(y_test, y_pred_test))), fontsize = font_size)\nr2 = ax2.text(16,425,'R2 Score = {0:.2f}'.format(r2_score(y_test, y_pred_test)), fontsize = font_size)\n\nptitle = ax1.set_title('Train data', size=font_size)\nxlabel = ax1.set_xlabel('Actual RUL', size=font_size)\nylabel = ax1.set_ylabel('Predicted RUL', size=font_size)\n\nptitle = ax2.set_title(\"Test data\", size=font_size)\nxlabel = ax2.set_xlabel('Actual RUL', size=font_size)\nylabel = ax2.set_ylabel('Predicted RUL', size=font_size)",
"_____no_output_____"
]
],
[
[
"## Predicted vs. residual",
"_____no_output_____"
]
],
[
[
"fig, (ax1,ax2) = plt.subplots(nrows=2, sharex=True)\nfig.set_size_inches(16, 16)\n\nfont_size = 14\n\ng = sns.regplot(y='residual', x='predicted', data=train_pd, fit_reg=False, ax=ax1)\nlim_set = g.set(ylim=(-350, 350), xlim=(0, 350))\nplot = g.axes.plot([0, 350], [0, 0], c=\".3\", ls=\"--\");\n\ng2 = sns.regplot(y='residual', x='predicted', data=test_pd, fit_reg=False, ax=ax2)\nlim_set = g2.set(ylim=(-350, 350), xlim=(0, 350))\nplot = g2.axes.plot([0, 350], [0, 0], c=\".3\", ls=\"--\");\n\nptitle = ax1.set_title('Train data', size=font_size)\nxlabel = ax1.set_xlabel('Predicted RUL', size=font_size)\nylabel = ax1.set_ylabel('Residual', size=font_size)\n\nptitle = ax2.set_title(\"Test data\", size=font_size)\nxlabel = ax2.set_xlabel('Predicted RUL', size=font_size)\nylabel = ax2.set_ylabel('Residual', size=font_size)",
"_____no_output_____"
]
],
[
[
"## Residual distribution\n\nPlot histogram and Q-Q plot for test and train data to check for normal distibution of residuals",
"_____no_output_____"
]
],
[
[
"import statsmodels.api as sm\nimport scipy.stats as stats\n\nfig, (ax1,ax2) = plt.subplots(nrows=2, ncols= 2)\nfig.set_size_inches(16, 16)\n\ng = sns.distplot(train_pd['residual'], ax=ax1[0], kde=False)\ng = stats.probplot(train_pd['residual'], plot=ax1[1])\n\ng2 = sns.distplot(test_pd['residual'], ax=ax2[0], kde=False)\ng2 = stats.probplot(test_pd['residual'], plot=ax2[1])\n\n\nptitle = ax1[0].set_title('Residual Histogram Train', size=font_size)\nxlabel = ax1[0].set_xlabel('Residuals', size=font_size)\nptitle = ax1[1].set_title('Q-Q Plot Train Residuals', size=font_size)\n\nptitle = ax2[0].set_title('Residual Histogram Test', size=font_size)\nxlabel = ax2[0].set_xlabel('Residuals', size=font_size)\nptitle = ax2[1].set_title('Q-Q Plot Test Residuals', size=font_size)\n\n",
"_____no_output_____"
]
],
[
[
"# Next Steps\n\nNow that we have a working model we want to deploy it as an Azure IoT Edge module. The [turbofan deploy model](./02-turbofan_deploy_model.ipynb) walks through the steps to create and Edge module.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6e8e9a027963f6e0c73600b10b20e4d68db4c5 | 19,562 | ipynb | Jupyter Notebook | Lab3.ipynb | chaitanya2334/cse5243-lab3 | 96fcd5525a524558e9e74928eeb8035622c1425a | [
"MIT"
]
| null | null | null | Lab3.ipynb | chaitanya2334/cse5243-lab3 | 96fcd5525a524558e9e74928eeb8035622c1425a | [
"MIT"
]
| null | null | null | Lab3.ipynb | chaitanya2334/cse5243-lab3 | 96fcd5525a524558e9e74928eeb8035622c1425a | [
"MIT"
]
| 1 | 2020-05-03T23:56:36.000Z | 2020-05-03T23:56:36.000Z | 37.190114 | 427 | 0.544934 | [
[
[
"\n\n<hr size=\"6\" style=\"color:#d65828;background-color:#d65828;border:none;\">\n\n# Lab3: Clustering and Association Mining\n\nThis laboratory assignment was designed by Chaitanya Kulkarni and Raghu Machiraju.\n\nDue Date: April 9, 2020, 23:59:59. No late submissions allowed unless the Night King from Game of Thrones arises again and invades our campus. Please consult instructor if you are falling behind.\n\nSome salient points:\n* The total number of points is 100 (10% of total grade).\n* The distribution for each part and question is listed below.\n* Teams of two/three enrolled students will complete this project.\n* Discussion across teams is only permitted on piazza.\n* Teams are forbidden to exchange code.\n* If any code is found and used, it should be mentioned in the notebook.\n* Submission will be through Carmen and we will accept only Jupyter notebooks.\n* It is required that the notebooks are zipped up and deposited. \n* The zip file should be named in the following way lastname1-lastame2-labN. (1<=N<=4).\n* Please please do not include the data with the .zip file.\n* It is very important that the class follows the honor code of academic conduct to maximize learning in an open manner.\n\n>### _Objective of this Lab:_\n>* Look at simple k-means implementation and understand how to best visualize and evaluate k-mean clusters.\n>* Association Mining.\n",
"_____no_output_____"
],
[
"## Dataset\n\nMNIST PreProcessed Dataset https://www.kaggle.com/oddrationale/mnist-in-csv. \n\nUse the `mnist_train.csv` (60k) only to identify the clusters. Please note that there exist 10 clusters for digits 0-9. The file `mnist_test.csv` wil not be used for this exercise.\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\ntrain_df = pd.read_csv(\"data/mnist-in-csv/mnist_train.csv\")\nprint(train_df.shape[0])\ntrain_df.head()\n\nX = train_df.iloc[:,1:].values\ntrue_labels = train_df.iloc[:, 0].values\n",
"_____no_output_____"
]
],
[
[
"## Question 1: K-Means Clustering",
"_____no_output_____"
],
[
"**To Learn :** You will need to find the requisite number of clusters from N image points in a 784 dimensional space (note that each is image 28x28 or 784 pixels). Each pixel will take a value ranging from 0-255.\n\n**Target Classes to Predict :** There should be 10 clusters and if your realization of k-means works wells, it should find 10 clusters. Further, you should be able to identify and label each of the clusters. We describe how this can be done. \n",
"_____no_output_____"
],
[
"Boilerplate starter code will be provided, as before you only have to fill in the empty code blocks. \n\nFor the k-means clustering, you will build a templated code which will allow you to change the distance metric and value of k. \n",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom numpy.linalg import norm\nfrom tqdm import tqdm\n\n\nclass MyKMeans:\n '''Implementing Kmeans algorithm.'''\n\n def __init__(self, k, max_iter=100):\n self.k = k\n self.max_iter = max_iter\n \n # Be sure to store the centroids that you \n # learn in self.fit() here.\n self.centroids = None \n\n def init_centroids(self, X):\n \"\"\"\n Initialize centroids. \n Randomly pick any k entries/rows in X.\n \n :param X: 2d input array X of shape (60000, 784) where \n dim=0 represents 60000 input images and dim=1 represents \n the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)] \n \n :return centroids: randomly initialized centroids. It will be a\n 2d array of shape (k, 784) \n where dim=0 is cluster id, and dim=1 is the individual pixel intensities.\n ie., centroids[k, :] = \\\n [0, 3, 2, 255, ... (784 values)] <- These must come from some random row in X.\n \"\"\"\n # code here\n return centroids\n \n def distance(self, X, centroids):\n \"\"\"\n Calculate the euclidean distance between every point in `X` to \n every centroid in `centroids`.\n \n :param X: 2d input array X of shape (60000, 784) where \n dim=0 represents 60000 input images and dim=1 represents \n the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)] \n :param centroids: 2d array centroids of shape (k, 784) \n where dim=0 is cluster id, and dim=1 is the individual pixel intensities.\n ie., centroids[k, :] = [0, 3, 2, 1, ... (784 values)]\n \n :return dist: return a 2d array dist of shape (60000, k) where,\n dim=0 represents a datapoint, and dim=1 represents the cluster. \n eg., dist[i, :] = [0.2, 1.5, 4.9] for 3 clusters\n \n \"\"\"\n # code here\n return distance\n\n def calc_centroids(self, X, y):\n \"\"\"\n Calculate new centroids using `y`.\n \n :param X: 2d input array X of shape (60000, 784) where \n dim=0 represents 60000 input images and dim=1 represents \n the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n :param y: 1d array where y[i] = cluster id of X[i]\n \n :return centroids: return 2d array centroids of shape (k, 784) \n where dim=0 is cluster id, and dim=1 is the individual pixel intensities.\n ie., centroids[k, :] = [0, 3, 2, 1, ... (784 values)]\n \"\"\"\n # code here\n return centroids\n\n def calc_sse(self, X, y, centroids):\n \"\"\"\n Using the input X, and the predicted cluster ids `y` \n and the exact `centroids` points calculate Squared Sum of Errors.\n \n :param X: 2d input array X of shape (60000, 784) where \n dim=0 represents 60000 input images and dim=1 represents \n the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n :param y: 1d array where y[i] = cluster id of X[i]\n :param centroids: 2d array centroids of shape (k, 784) \n where dim=0 is cluster id, and dim=1 is the individual pixel intensities.\n ie., centroids[k, :] = [0, 3, 2, 1, ... (784 values)]\n \n :return sse: Return the SSE value for the given centroids \n and predicted cluster ids.\n \"\"\"\n # code here\n return sse\n \n def fit(self, X):\n \"\"\"\n Implement K means algorithm here. The pseudo code is provided to you.\n Remember to store the learned centroids in `self.centroids`\n \n :param X: 2d input array X of shape (60000, 784) where \n dim=0 represents 60000 input images and dim=1 represents \n the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n \n :return None: this function doesnt return anything. You must however update\n self.centroids as you train your k means model.\n \"\"\"\n # step 1: init centroids\n \n # step 2: for every iteration do\n \n # step 2.1: calculate distance for each point with every centroid\n # step 2.2: Assign cluster to each point based on closest centroid\n # step 2.3: recalculate new centroids using assigned clusters\n # step 2.4: (optional) if centroids dont change, end early\n \n self.centroids = self.init_centroids(X)\n \n t = tqdm(range(self.max_iter))\n for i in t:\n old_centroids = self.centroids\n \n # code here #\n # ....\n # ....\n # ....\n # ....\n # code here #\n \n if np.all(old_centroids == self.centroids):\n break\n t.set_description(\"sse = {0:.2f}\".format(self.calc_sse(X, y, self.centroids)))\n\n \n def predict(self, X):\n \"\"\"\n Once the model is trained using fit, to visualize the cluster you will\n need to know which point was assigned with cluster. Return a \n label/cluster id for each datapoint using trained centroids.\n \n :param X: 2d input array that you which to cluster using trained centroids. \n X is of shape (n_images, 784) where dim=0 represents n_images input \n images and dim=1 represents the 784 pixel intensities.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n \n :return y: predicted label for each input image. \n 1d array where y[i] = predicted cluster id of X[i]\n \"\"\"\n # code here\n return y",
"_____no_output_____"
]
],
[
[
"## Get k=10 clusters",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import classification_report\n\nk_means = MyKMeans(k=10, max_iter=10)\n# learn clusters\nk_means.fit(X)\n\n# predict labels\ny = k_means.predict(X)",
"_____no_output_____"
]
],
[
[
"## Visualization\n\n#### First perform Dimensionality reduction : PCA from 784 dimensions to 2 principle components. ",
"_____no_output_____"
]
],
[
[
"from sklearn.decomposition import PCA\n\nX_2 = PCA(n_components=2).fit_transform(X)",
"_____no_output_____"
]
],
[
[
"\n#### Then, visualize the predicted clusters on some subset of randomly selected points. \n\nUse colors for predicted labels and shapes for true/golden labels. Mark centroids too. Centroids will be marked with a special symbol. Color should match the predicted label’s color.",
"_____no_output_____"
]
],
[
[
"# code here\ndef visualize(X, model):\n \"\"\"\n Create a scatter plot where,\n 1. the colors indicate predicted clusters and centroid (marked with special shape)\n 2. the shapes indicate true clusters\n \n :param X: 2d input array. X[i, j] where \n i is the datapoint index and j is the pixel intensity index.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n :param model: trained k_means model to visualize.\n \"\"\"\n y = model.predict(X)\n # code here",
"_____no_output_____"
]
],
[
[
"## Bonus: Assign predicted clusters to true labels\n\nMap the predicted cluster labels to the most related true label. We can do this naively through permutation by simply randomly assigning a set of true labels to the predicted labels and checking the accuracy of the prediction. Permute through all possible assignments $O(K!)$ and return the assignment that resulted in the best accuracy. \n\nA more polynomial approach is to use the Hungarian algorithm. Scikit-learn provides you with the implementation of hungarian algorithm under `sklearn.utils.linear_assignment_.linear_assignment`. This function works on a **cost matrix (cost of aligning any two labels)** and uses the least cost to assign labels. You need to hence convert your confusion matrix into a cost matrix before running the hungarian algorithm.\n",
"_____no_output_____"
]
],
[
[
"from sklearn.utils.linear_assignment_ import linear_assignment\ndef assign_labels(y, true_labels):\n conf_mat = # create confusion matrix\n cm = # convert confusion matrix into a cost matrix. pseudo code: \n # cost_matrix = max - confusion_matrix.\n assignemnt = linear_assignment(cm)\n print_assignment(assignment)\n new_conf_mat = # create a new confusion matrix that is now matches \n # the true labels. A quick sanity check is to \n # ensure the diagonal has the max number in any row.\n return new_conf_mat \n\ndef print_assignment(assignment):\n # print the assignment mapping\n # eg:\n # pred -> true\n # (0) -> (4)\n # (1) -> (3)\n # ...\n pass\n\ndef accuracy(y, true_labels):\n conf_mat = assign_labels(y, true_labels)\n \n nb_correct = np.trace(conf_mat)\n nb_total = np.sum(conf_mat)\n accuracy = nb_correct / nb_total\n return accuracy",
"_____no_output_____"
]
],
[
[
"### Now visualize this information\n\nThe difference between the `visualize` function you implemented before and this one will be the fact that now you have the most likely label your predicted cluster must correspond to. And so, you would like to visualize this additional information to get a sense of how well your k means model did compared to the true labels.",
"_____no_output_____"
]
],
[
[
"def visualize_and_label(X, model):\n \"\"\"\n Create a scatter plot where,\n 1. the colors indicate predicted clusters and centroid (marked with assigned label id)\n 2. the shapes indicate true clusters\n \n Also, add a legend for the colors, that maps the assigned predicted label id for each color. \n \n :param X: 2d input array. X[i, j] where \n i is the datapoint index and j is the pixel intensity index.\n ie., X[i, :] = [0, 3, 2, 1, ... (784 pixel intensities)]\n :param model: trained k_means model to visualize.\n \"\"\"\n y = model.predict(X)\n # code here",
"_____no_output_____"
]
],
[
[
"## Evaluations",
"_____no_output_____"
],
[
"### 1. Find Best Initial Centroids by Randoming\n\nAlthough the best strategy is k-means++, here we will make use of the most simplest strategy -- random. ",
"_____no_output_____"
]
],
[
[
"# code here",
"_____no_output_____"
]
],
[
[
"Comment on how effective (or ineffective) this strategy really is. Compare your k-means with `sklearn` implementation, initialized using kmeans++.",
"_____no_output_____"
]
],
[
[
"from sklearn.cluster import KMeans\n\nsklearn_k_means = KMeans(n_clusters=10, \n random_state=0, \n init=\"k-means++\", \n max_iter=3).fit(X)\n\nyour_k_means = MyKMeans(k=10, max_iter=3).fit(X)\n\n# code to compare here",
"_____no_output_____"
],
[
"# your comments here #",
"_____no_output_____"
]
],
[
[
"## 2. Find Best K\n\n### 2.1. Use Elbow analysis. \n\nTypically, as k increases, sum-of-squared-errors (SSE) decreases. The idea here is to find a specific k that drastically decreases SSE relative to other k values. This can be done by plotting k on the x-axis and SSE on the y-axis. If the plot looks like an arm, then the elbow on the arm (or knee if it is a leg instead) is the optimal k. \n",
"_____no_output_____"
]
],
[
[
"# code here",
"_____no_output_____"
]
],
[
[
"### 2.2. Silhouette Measure. \n\nInstead of SSE (used in Elbow analysis) lets make use of silhouette measure. This measure captures the density of or strength of a cluster vs. its separation from other clusters. Again plot k on the x-axis and plot the silhouette measure on the y-axis. The optimal k is where the silhouette value is maximum.\n",
"_____no_output_____"
]
],
[
[
"# code here",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6e8e9faa4ef1095dd4964da8e0e0639780651f | 191,772 | ipynb | Jupyter Notebook | notebooks/benchmarks/single_example/001_p-gnn2D_benchmark.ipynb | FordyceLab/tessellate | a3f0c38b4392027a7503828f48d65ec02eb24698 | [
"MIT"
]
| null | null | null | notebooks/benchmarks/single_example/001_p-gnn2D_benchmark.ipynb | FordyceLab/tessellate | a3f0c38b4392027a7503828f48d65ec02eb24698 | [
"MIT"
]
| 7 | 2021-03-31T19:41:46.000Z | 2022-01-13T02:39:45.000Z | notebooks/benchmarks/single_example/001_p-gnn2D_benchmark.ipynb | FordyceLab/tessellate | a3f0c38b4392027a7503828f48d65ec02eb24698 | [
"MIT"
]
| null | null | null | 125.014342 | 48,076 | 0.820375 | [
[
[
"# Benchmark position-aware graph neural network/2D CNN architecture\n\nThis notebook contains all of the code to overfit a P-GNN/2D CNN to four contact channels of a single structure (6E6O).\n\n## Setup\n\n",
"_____no_output_____"
],
[
"### Dataloader code ",
"_____no_output_____"
]
],
[
[
"import torch\nfrom torch.utils.data import Dataset\nimport numpy as np\nimport periodictable as pt\nimport pandas as pd\nimport os\nimport matplotlib.pyplot as plt\n\ndef read_files(acc, model, graph_dir, contacts_dir):\n \"\"\"\n Read graph and contacts files.\n \n Args:\n - acc (str) - String of the PDB ID (lowercese).\n - model (int) - Model number of the desired bioassembly.\n - graph_dir (str) - Directory containing the nodes, edges,\n and mask files.\n - contacts_dir (str) - Directory containing the .contacts\n files from get_contacts.py.\n \n Returns:\n - Dictionary of DataFrames and lists corresponding to\n graph nodes, edges, mask, and contacts. \n \"\"\"\n\n # Get the file names for the graph files\n node_file = os.path.join(graph_dir, '{}-{}_nodes.csv'.format(acc, model))\n edge_file = os.path.join(graph_dir, '{}-{}_edges.csv'.format(acc, model))\n mask_file = os.path.join(graph_dir, '{}-{}_mask.csv'.format(acc, model))\n\n # Get the contacts file\n contacts_file = os.path.join(contacts_dir, '{}-{}.contacts'.format(acc, model))\n\n # Read the nodes and edges\n nodes = pd.read_csv(node_file)\n edges = pd.read_csv(edge_file)\n\n # Check if the mask is empty\n if os.path.getsize(mask_file) > 0:\n with open(mask_file) as f:\n mask = f.read().split('\\n')\n else:\n mask = []\n\n # Read the contacts\n contacts = pd.read_table(contacts_file, sep='\\t',\n header=None, names=['type', 'start', 'end'])\n\n # Return the data\n data = {\n 'nodes': nodes,\n 'edges': edges,\n 'mask': mask,\n 'contacts': contacts\n }\n\n return data\n \n \ndef process_res_data(data):\n \"\"\"\n Process residue-level data from atom-level data.\n \n Args:\n - data (dict) - Dictionary of graph data output from `read_files`.\n \n Returns:\n - Dictionary of atom and residue graph and contact data.\n \"\"\"\n\n # Extract data form dict\n nodes = data['nodes']\n edges = data['edges']\n mask = data['mask']\n contacts = data['contacts']\n\n # Get residue nodes\n res_nodes = pd.DataFrame()\n res_nodes['res'] = [':'.join(atom.split(':')[:3]) for atom in nodes['atom']]\n res_nodes = res_nodes.drop_duplicates().reset_index(drop=True)\n\n # Get residue edges\n res_edges = edges.copy()\n res_edges['start'] = [':'.join(atom.split(':')[:3]) for atom in res_edges['start']]\n res_edges['end'] = [':'.join(atom.split(':')[:3]) for atom in res_edges['end']]\n res_edges = res_edges[res_edges['start'] != res_edges['end']].drop_duplicates().reset_index(drop=True)\n\n # Get residue contacts\n res_contacts = contacts.copy()\n res_contacts['start'] = [':'.join(atom.split(':')[:3]) for atom in res_contacts['start']]\n res_contacts['end'] = [':'.join(atom.split(':')[:3]) for atom in res_contacts['end']]\n res_contacts = res_contacts[res_contacts['start'] != res_contacts['end']].drop_duplicates().reset_index(drop=True)\n\n # Get residue mask\n res_mask = list(set([':'.join(atom.split(':')[:3]) for atom in mask]))\n\n # Return data dict\n data = {\n 'atom_nodes': nodes,\n 'atom_edges': edges,\n 'atom_contact': contacts,\n 'atom_mask': mask,\n 'res_nodes': res_nodes,\n 'res_edges': res_edges,\n 'res_contact': res_contacts,\n 'res_mask': res_mask\n }\n\n return data\n\n\ndef get_map_dicts(entity_list):\n \"\"\"\n Map identifiers to indices and vice versa.\n \n Args:\n - entity_list (list) - List of entities (atoms, residues, etc.)\n to index.\n \n Returns:\n - Tuple of the entity to index and index to entity dicts, respectively.\n \"\"\"\n \n # Create the entity:index dictionary\n ent2idx_dict = {entity: idx for idx, entity in enumerate(entity_list)}\n\n # Create the index:entity dictionary\n idx2ent_dict = {idx: entity for entity, idx in ent2idx_dict.items()}\n \n return (ent2idx_dict, idx2ent_dict)\n\n\ndef create_adj_mat(data, dict_map, mat_type):\n \"\"\"\n Creates an adjacency matrix.\n \n Args:\n - data (DataFrame) - Dataframe with 'start' and 'end' column\n for each interaction. For atom-level adjacency, 'order' \n column is also required. For atom or residue conatcts,\n 'type' column is also required.\n \n Returns:\n - Coordinate format matrix (numpy). For atom adjacency, third column\n corresponds to bond order. For contacts, third column\n corresponds to channel.\n \n Channel mappings (shorthand from get_contacts.py source):\n\n 0:\n hp hydrophobic interactions\n 1:\n hb hydrogen bonds\n lhb ligand hydrogen bonds\n hbbb backbone-backbone hydrogen bonds\n hbsb backbone-sidechain hydrogen bonds\n hbss sidechain-sidechain hydrogen bonds\n hbls ligand-sidechain residue hydrogen bonds\n hblb ligand-backbone residue hydrogen bonds\n 2:\n vdw van der Waals\n 3:\n wb water bridges\n wb2 extended water bridges\n lwb ligand water bridges\n lwb2 extended ligand water bridges\n 4:\n sb salt bridges\n 5:\n ps pi-stacking\n 6:\n pc pi-cation\n 7:\n ts t-stacking\n \"\"\"\n \n # Initialize the coordinate list\n coord_mat = []\n\n # Map channel names to numeric channels\n channel = {\n # Hydrophobic interactions in first channel\n 'hp': 0,\n\n # Hydrogen bonds in second channel\n 'hb': 1,\n 'lhb': 1, \n 'hbbb': 1,\n 'hbsb': 1,\n 'hbss': 1,\n 'hbls': 1,\n 'hblb': 1,\n\n # VdW in third channel\n 'vdw': 2,\n\n # Water bridges\n 'wb': 3, \n 'wb2': 3,\n 'lwb': 3,\n 'lwb2': 3,\n\n # Salt bridges\n 'sb': 4,\n\n # Other interactions\n 'ps': 5,\n 'pc': 6,\n 'ts': 7,\n }\n\n # Assemble the contacts\n for idx, row in data.iterrows():\n\n entry = [dict_map[row['start']], dict_map[row['end']]]\n\n # Add order or type if necessary\n if mat_type == 'atom_graph':\n entry.append(row['order'])\n elif mat_type == 'atom_contact':\n entry.append(channel[row['type']])\n elif mat_type == 'res_contact':\n entry.append(channel[row['type']])\n\n coord_mat.append(entry)\n\n return(np.array(coord_mat))\n\n\ndef create_mem_mat(atom_dict, res_dict):\n \"\"\"\n Create a membership matrix mapping atoms to residues.\n \n Args:\n - atom_dict (dict) - Dictionary mapping atoms to indices.\n - res_dict (dict) - Dictionary mapping residues to indices.\n \n Returns:\n - Coordinate format membership matrix (numpy) with first\n row being residue number and the second column being\n atom number.\n \"\"\"\n \n # Initialize the coordinate list\n mem_coord = []\n \n # Map atoms to residues\n for atom, atom_idx in atom_dict.items():\n res_idx = res_dict[':'.join(atom.split(':')[:3])]\n \n mem_coord.append([res_idx, atom_idx])\n \n mem_coord = np.array(mem_coord)\n \n return mem_coord\n\n\ndef create_idx_list(id_list, dict_map):\n \"\"\"\n Create list of indices.\n \n Args:\n - id_list (list) - List of masked atom or residue identifiers.\n - dict_map (dict) - Dictionary mapping entities to indices.\n \n Returns:\n - A numpy array of the masked indices.\n \"\"\"\n \n # Generate the numpy index array\n idx_array = np.array([dict_map[iden] for iden in id_list])\n \n return idx_array\n\n\nclass TesselateDataset(Dataset):\n \"\"\"\n Dataset class for structural data.\n \n Args:\n - accession_list (str) - File path from which to read PDB IDs for dataset.\n - graph_dir (str) - Directory containing the nodes, edges, and mask files.\n - contacts_dir (str) - Directory containing the .contacts files from\n get_contacts.py.\n - return_data (list) - List of datasets to return. Value must be 'all' or\n a subset of the following list:\n - pdb_id\n - model\n - atom_nodes\n - atom_adj\n - atom_contact\n - atom_mask\n - res_adj\n - res_contact\n - res_mask\n - mem_mat\n - idx2atom_dict\n - idx2res_dict \n \"\"\"\n \n def __init__(self, accession_list, graph_dir, contacts_dir, add_covalent=False, return_data='all', in_memory=False):\n \n if return_data == 'all':\n self.return_data = [\n 'pdb_id',\n 'model',\n 'atom_nodes',\n 'atom_adj',\n 'atom_contact',\n 'atom_mask',\n 'res_adj',\n 'res_contact',\n 'res_mask',\n 'mem_mat',\n 'idx2atom_dict',\n 'idx2res_dict'\n ]\n \n # Store reference to accession list file\n self.accession_list = accession_list\n \n # Store references to the necessary directories\n self.graph_dir = graph_dir\n self.contacts_dir = contacts_dir\n \n # Whether to add covalent bonds to prediction task and\n # remove sequence non-deterministic covalent bonds from the adjacency matrix\n self.add_covalent=add_covalent\n \n # Read in and store a list of accession IDs\n with open(accession_list, 'r') as handle:\n self.accessions = np.array([acc.strip().lower().split() for acc in handle.readlines()])\n \n self.data = {}\n\n \n def __len__(self):\n \"\"\"\n Return the length of the dataset.\n \n Returns:\n - Integer count of number of examples.\n \"\"\"\n return len(self.accessions)\n \n \n def __getitem__(self, idx):\n \"\"\"\n Get an item with a particular index value.\n \n Args:\n - idx (int) - Index of desired sample.\n \n Returns:\n - Dictionary of dataset example. All tensors are sparse when possible.\n \"\"\"\n if idx in self.data:\n return self.data[idx]\n \n # initialize the return dictionary\n return_dict = {}\n \n acc_entry = self.accessions[idx]\n \n # Get the PDB ID\n acc = acc_entry[0]\n\n # Get the model number if one exists\n if len(acc_entry) == 1:\n model = 1\n else:\n model = acc_entry[1]\n \n # Read and process the files\n data = read_files(acc, model, self.graph_dir, self.contacts_dir)\n data = process_res_data(data)\n \n # Generate the mapping dictionaries\n atom2idx_dict, idx2atom_dict = get_map_dicts(data['atom_nodes']['atom'].unique())\n res2idx_dict, idx2res_dict = get_map_dicts(data['res_nodes']['res'].unique())\n \n # Get numbers of atoms and residues per sample\n n_atoms = len(atom2idx_dict)\n n_res = len(res2idx_dict)\n \n # Handle all of the possible returned datasets\n if 'pdb_id' in self.return_data:\n return_dict['pdb_id'] = acc\n \n if 'model' in self.return_data:\n return_dict['model'] = model\n \n if 'atom_nodes' in self.return_data:\n ele_nums = [pt.elements.symbol(element).number for element in data['atom_nodes']['element']]\n return_dict['atom_nodes'] = torch.LongTensor(ele_nums)\n \n if 'atom_adj' in self.return_data:\n adj = create_adj_mat(data['atom_edges'], atom2idx_dict, mat_type='atom_graph').T\n \n x = torch.LongTensor(adj[0, :]).squeeze()\n y = torch.LongTensor(adj[1, :]).squeeze()\n val = torch.FloatTensor(adj[2, :]).squeeze()\n \n atom_adj = torch.zeros([n_atoms, n_atoms]).index_put_((x, y), val, accumulate=False)\n \n atom_adj = atom_adj.index_put_((y, x), val, accumulate=False)\n \n atom_adj[range(n_atoms), range(n_atoms)] = 1\n \n atom_adj = (atom_adj > 0).float()\n \n return_dict['atom_adj'] = atom_adj\n \n if 'atom_contact' in self.return_data:\n atom_contact = create_adj_mat(data['atom_contact'], atom2idx_dict, mat_type='atom_contact').T\n \n x = torch.LongTensor(atom_contact[0, :]).squeeze()\n y = torch.LongTensor(atom_contact[1, :]).squeeze()\n z = torch.LongTensor(atom_contact[2, :]).squeeze()\n\n atom_contact = torch.zeros([n_atoms, n_atoms, 8]).index_put_((x, y, z),\n torch.ones(len(x)))\n atom_contact = atom_contact.index_put_((y, x, z), \n torch.ones(len(x)))\n \n return_dict['atom_contact'] = atom_contact\n \n if 'atom_mask' in self.return_data:\n atom_mask = create_idx_list(data['atom_mask'], atom2idx_dict)\n \n masked_pos = torch.from_numpy(atom_mask)\n \n if self.add_covalent:\n channels = 9\n else:\n channels = 8\n \n mask = torch.ones([n_atoms, n_atoms, channels])\n mask[masked_pos, :, :] = 0\n mask[:, masked_pos, :] = 0\n \n return_dict['atom_mask'] = mask\n \n if 'res_adj' in self.return_data: \n adj = create_adj_mat(data['res_edges'], res2idx_dict, mat_type='res_graph').T\n \n x = torch.LongTensor(adj[0, :]).squeeze()\n y = torch.LongTensor(adj[1, :]).squeeze()\n \n res_adj = torch.zeros([n_res, n_res]).index_put_((x, y), torch.ones(len(x)))\n \n res_adj = res_adj.index_put_((y, x), torch.ones(len(x)))\n \n res_adj[range(n_res), range(n_res)] = 1\n \n return_dict['res_adj'] = res_adj\n \n if 'res_contact' in self.return_data:\n res_contact = create_adj_mat(data['res_contact'], res2idx_dict, mat_type='res_contact').T\n \n x = torch.LongTensor(res_contact[0, :]).squeeze()\n y = torch.LongTensor(res_contact[1, :]).squeeze()\n z = torch.LongTensor(res_contact[2, :]).squeeze()\n\n res_contact = torch.zeros([n_res, n_res, 8]).index_put_((x, y, z),\n torch.ones(len(x)))\n \n res_contact = res_contact.index_put_((y, x, z),\n torch.ones(len(x)))\n \n return_dict['res_contact'] = res_contact\n \n if 'res_mask' in self.return_data:\n res_mask = create_idx_list(data['res_mask'], res2idx_dict)\n \n masked_pos = torch.from_numpy(res_mask)\n \n if self.add_covalent:\n channels = 9\n else:\n channels = 8\n \n mask = torch.ones([n_res, n_res, channels])\n mask[masked_pos, :, :] = 0\n mask[:, masked_pos, :] = 0\n \n return_dict['res_mask'] = mask\n \n if 'mem_mat' in self.return_data:\n mem_mat = create_mem_mat(atom2idx_dict, res2idx_dict).T\n \n x = torch.LongTensor(mem_mat[0, :]).squeeze()\n y = torch.LongTensor(mem_mat[1, :]).squeeze()\n \n mem_mat = torch.zeros([n_res, n_atoms]).index_put_((x, y),\n torch.ones(len(x)))\n \n return_dict['mem_mat'] = mem_mat\n \n if 'idx2atom_dict' in self.return_data:\n return_dict['idx2atom_dict'] = idx2atom_dict\n \n if 'idx2res_dict' in self.return_data:\n return_dict['idx2res_dict'] = idx2res_dict\n \n self.data[idx] = return_dict\n \n # Return the processed data\n return return_dict",
"_____no_output_____"
]
],
[
[
"### Modules",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\nimport numpy as np\n\n\n####################\n# Embedding layers #\n####################\n\nclass AtomOneHotEmbed(nn.Module):\n \"\"\"\n Create one-hot embeddings for atom identities.\n \"\"\"\n def __init__(self):\n super(AtomOneHotEmbed, self).__init__()\n \n self.idx_map = {\n 6: 0,\n 7: 1,\n 8: 2,\n 15: 3,\n 16: 4\n }\n \n def forward(self, atomic_numbers):\n \"\"\"\n Return the embeddings for each atom in the graph.\n \n Args:\n - atoms (torch.LongTensor) - Tensor (n_atoms) containing atomic numbers.\n \n Returns:\n - torch.FloatTensor of dimension (n_atoms, n_features) containing\n the embedding vectors.\n \"\"\"\n \n embedded_atoms = torch.zeros((len(atomic_numbers), 6),\n device=atomic_numbers.device)\n for i, j in enumerate(atomic_numbers):\n j = int(j)\n if j in self.idx_map:\n embedded_atoms[i, self.idx_map[j]] = 1\n else:\n embedded_atoms[i, 5] = 1\n \n return embedded_atoms\n\n\n#########################\n# Position-aware layers #\n#########################\n\n# # PGNN layer, only pick closest node for message passing\nclass PGNN_layer(nn.Module):\n def __init__(self, input_dim, output_dim,dist_trainable=False):\n super(PGNN_layer, self).__init__()\n self.input_dim = input_dim\n self.dist_trainable = dist_trainable\n\n if self.dist_trainable:\n self.dist_compute = Nonlinear(1, output_dim, 1)\n\n self.linear_hidden = nn.Linear(input_dim*2, output_dim)\n self.linear_out_position = nn.Linear(output_dim,1)\n self.act = nn.ReLU()\n\n for m in self.modules():\n if isinstance(m, nn.Linear):\n m.weight.data = nn.init.xavier_uniform_(m.weight.data, gain=nn.init.calculate_gain('relu'))\n if m.bias is not None:\n m.bias.data = nn.init.constant_(m.bias.data, 0.0)\n\n def forward(self, feature, dists_max, dists_argmax):\n if self.dist_trainable:\n dists_max = self.dist_compute(dists_max.unsqueeze(-1)).squeeze()\n\n subset_features = feature[dists_argmax.flatten(), :]\n subset_features = subset_features.reshape((dists_argmax.shape[0], dists_argmax.shape[1],\n feature.shape[1]))\n\n messages = subset_features * dists_max.unsqueeze(-1)\n\n self_feature = feature.unsqueeze(1).repeat(1, dists_max.shape[1], 1)\n messages = torch.cat((messages, self_feature), dim=-1)\n\n messages = self.linear_hidden(messages).squeeze()\n messages = self.act(messages) # n*m*d\n\n out_position = self.linear_out_position(messages).squeeze(-1) # n*m_out\n out_structure = torch.mean(messages, dim=1) # n*d\n\n return out_position, out_structure\n\n\n### Non linearity\nclass Nonlinear(nn.Module):\n def __init__(self, input_dim, hidden_dim, output_dim):\n super(Nonlinear, self).__init__()\n\n self.linear1 = nn.Linear(input_dim, hidden_dim)\n self.linear2 = nn.Linear(hidden_dim, output_dim)\n\n self.act = nn.ReLU()\n\n for m in self.modules():\n if isinstance(m, nn.Linear):\n m.weight.data = nn.init.xavier_uniform_(m.weight.data, gain=nn.init.calculate_gain('relu'))\n if m.bias is not None:\n m.bias.data = nn.init.constant_(m.bias.data, 0.0)\n\n def forward(self, x):\n x = self.linear1(x)\n x = self.act(x)\n x = self.linear2(x)\n return x\n\n\n#################\n# Custom losses #\n#################\n\nclass FocalLoss(nn.Module):\n \n def __init__(self, gamma, reduction='mean'):\n super(FocalLoss, self).__init__()\n self.gamma = gamma\n self.reduction = reduction\n \n def forward(self, preds, target):\n \n target_colsums = torch.sum(target, dim=0)\n alpha = target_colsums / target.shape[0]\n \n pos_mask = target == 1\n neg_mask = target == 0\n alpha_mat = torch.zeros(*target.shape, device=target.device)\n alpha_mat += (pos_mask * alpha.repeat(target.shape[0], 1))\n alpha_mat += (neg_mask * (1 - alpha).repeat(target.shape[0], 1))\n \n BCE_loss = F.binary_cross_entropy_with_logits(preds, target,\n reduction='none')\n pt = torch.exp(-BCE_loss)\n focal_loss = alpha_mat * (1-pt) ** self.gamma * BCE_loss\n \n if self.reduction == 'mean':\n focal_loss = focal_loss.mean()\n elif self.reduction == 'sum':\n focal_loss = focal_loss.sum()\n \n return focal_loss",
"_____no_output_____"
]
],
[
[
"# Functions",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport torch\n\n########################################\n# Pairwise matrix generation functions #\n########################################\n\ndef pairwise_mat(nodes, method='mean'):\n \"\"\"\n Generate matrix for pairwise determination of interactions.\n \n Args:\n - nodes (torch.FloatTensor) - Tensor of node (n_nodes, n_features) features.\n - method (str) - One of 'sum' or 'mean' for combination startegy for\n pairwise combination matrix (default = 'mean').\n \n Returns:\n - torch.FloatTensor of shape (n_pairwise, n_nodes) than can be used used to\n combine feature vectors. Values are 1 if method == \"sum\" and 0.5 if\n method == \"mean\".\n \"\"\" \n\n # Get the upper triangle indices\n triu = np.vstack(np.triu_indices(nodes.shape[0]))\n \n # Loop through all indices and add to list with \n idxs = torch.from_numpy(triu).T\n \n # Convert to tensor\n combos = torch.zeros([idxs.shape[0], nodes.shape[0]]).scatter(1, idxs, 1)\n \n # Set to 0.5 if method is 'mean'\n if method == 'mean':\n combos *= 0.5\n \n return combos\n\n\ndef pairwise_3d(nodes):\n # Get the upper triangle indices\n repeated_nodes = nodes.unsqueeze(0).expand(nodes.shape[0], -1, -1)\n repeated_nodes2 = repeated_nodes.permute(1, 0, 2)\n \n return torch.cat((repeated_nodes, repeated_nodes2), dim=-1)\n\n\n############################\n# Upper triangle functions #\n############################\n\ndef triu_condense(input_tensor):\n \"\"\"\n Condense the upper triangle of a tensor into a 2d dense representation.\n \n Args:\n - input_tensor (torch.Tensor) - Tensor of shape (n, n, m).\n \n Returns:\n - Tensor of shape (n(n+1)/2, m) where elements along the third dimension in\n the original tensor are packed row-wise according to the upper\n triangular indices.\n \"\"\"\n \n # Get upper triangle index info\n row_idx, col_idx = np.triu_indices(input_tensor.shape[0])\n row_idx = torch.LongTensor(row_idx)\n col_idx = torch.LongTensor(col_idx)\n \n # Return the packed matrix\n output = input_tensor[row_idx, col_idx, :]\n \n return output\n\n\ndef triu_expand(input_matrix):\n \"\"\"\n Expand a dense representation of the upper triangle of a tensor into \n a 3D squareform representation.\n \n Args:\n - input_matrix (torch.Tensor) - Tensor of shape (n(n+1)/2, m).\n \n Returns:\n - Tensor of shape (n, n, m) where elements along the third dimension in the\n original tensor are packed row-wise according to the upper triangular\n indices.\n \"\"\"\n # Get the edge size n of the tensor\n n_elements = input_matrix.shape[0]\n n_chan = input_matrix.shape[1]\n n_res = int((-1 + np.sqrt(1 + 4 * 2 * (n_elements))) / 2)\n \n # Get upper triangle index info\n row_idx, col_idx = np.triu_indices(n_res)\n row_idx = torch.LongTensor(row_idx)\n col_idx = torch.LongTensor(col_idx)\n \n # Generate the output tensor\n output = torch.zeros((n_res, n_res, n_chan))\n \n # Input the triu values\n for i in range(n_chan):\n i_tens = torch.full((len(row_idx),), i, dtype=torch.long)\n output.index_put_((row_idx, col_idx, i_tens), input_matrix[:, i])\n \n # Input the tril values\n for i in range(n_chan):\n i_tens = torch.full((len(row_idx),), i, dtype=torch.long)\n output.index_put_((col_idx, row_idx, i_tens), input_matrix[:, i])\n \n return output\n\n\n###################\n# P-GNN functions #\n###################\n\ndef generate_dists(adj_mat):\n adj_mask = adj_mat == 0\n \n dist = adj_mat - torch.eye(adj_mat.shape[0], device=adj_mat.device)\n dist = 1 / (dist + 1)\n dist[adj_mask] = 0\n \n return dist.squeeze()\n\n\ndef get_dist_max(anchorset_id, dist):\n dist_max = torch.zeros((dist.shape[0],len(anchorset_id)),\n device=dist.device)\n dist_argmax = torch.zeros((dist.shape[0],len(anchorset_id)),\n device=dist.device).long()\n for i in range(len(anchorset_id)):\n temp_id = anchorset_id[i]\n dist_temp = dist[:, temp_id]\n dist_max_temp, dist_argmax_temp = torch.max(dist_temp, dim=-1)\n dist_max[:,i] = dist_max_temp\n dist_argmax[:,i] = dist_argmax_temp\n return dist_max, dist_argmax\n\n\ndef get_random_anchorset(n,c=0.5):\n m = int(np.log2(n))\n copy = int(c*m)\n anchorset_id = []\n for i in range(m):\n anchor_size = int(n/np.exp2(i + 1))\n for j in range(copy):\n anchorset_id.append(np.random.choice(n,size=anchor_size,replace=False))\n return anchorset_id\n\n\ndef preselect_anchor(n_nodes, dists):\n anchorset_id = get_random_anchorset(n_nodes, c=1)\n return get_dist_max(anchorset_id, dists)",
"_____no_output_____"
]
],
[
[
"### Model",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nfrom torch.nn import functional as F\nimport pytorch_lightning as pl\nfrom torch.utils.data import DataLoader\n\n\nclass PGNN2D(pl.LightningModule):\n def __init__(self, input_dim, hidden_dim, output_dim,\n anchorset_n, n_contact_channels, \n layer_num=2, train_data=None,\n val_data=None, test_data=None):\n super(PGNN2D, self).__init__()\n \n # Parameters\n self.n_contact_channels = n_contact_channels\n self.layer_num = layer_num\n \n # Datasets\n self.train_data = train_data\n self.val_data = val_data\n self.test_data = test_data\n \n # Embedding\n self.embed = AtomOneHotEmbed()\n \n # First P-GNN layer\n self.conv_atom_first = PGNN_layer(input_dim, hidden_dim)\n\n # All other P-GNN layers\n if layer_num>1:\n self.conv_atom_hidden = nn.ModuleList([PGNN_layer(hidden_dim,\n hidden_dim)\n for i in range(layer_num - 1)])\n \n # 2D convolutional layers\n self.conv1 = nn.Conv2d(anchorset_n * 2,\n 25, 3, stride=1, padding=1)\n self.conv2 = nn.Conv2d(25,\n 25, 3, stride=1, padding=1)\n self.conv3 = nn.Conv2d(25,\n 25, 3, stride=1, padding=1)\n \n # Final 2D CNN\n self.conv_final = nn.Conv2d(25, n_contact_channels, 3, stride=1, padding=1)\n \n # Focal loss\n self.loss = FocalLoss(3)\n \n self.activations = {}\n\n\n def forward(self, data):\n \n data['atom_dist'] = generate_dists(data['atom_adj'])\n\n data['atom_dist_max'], data['atom_dist_argmax'] = preselect_anchor(data['atom_adj'].squeeze().shape[0], data['atom_dist'])\n\n x = self.embed(data['atom_nodes'].squeeze())\n \n atom_embed = x.detach().cpu().numpy()\n \n x_position, x = self.conv_atom_first(x, data['atom_dist_max'], data['atom_dist_argmax'])\n x = F.relu(x)\n \n for i in range(self.layer_num-1):\n \n data['atom_dist'] = generate_dists(data['atom_adj'])\n\n data['atom_dist_max'], data['atom_dist_argmax'] = preselect_anchor(data['atom_adj'].squeeze().shape[0], data['atom_dist'])\n \n x_position, x = self.conv_atom_hidden[i](x, data['atom_dist_max'], data['atom_dist_argmax'])\n x = F.relu(x)\n \n atom_embed_update = x_position.detach().cpu().numpy()\n \n x_position = data['mem_mat'].squeeze().matmul(x_position)\n \n res_embed = x_position.detach().cpu().numpy()\n res_embed.shape\n \n pairwise = pairwise_3d(x_position).permute(2, 0, 1).unsqueeze(0)\n \n conv1_out = F.relu(self.conv1(pairwise))\n conv2_out = F.relu(self.conv2(conv1_out))\n conv3_out = F.relu(self.conv3(conv2_out))\n square_preds = self.conv_final(conv3_out)\n \n square_preds = square_preds + square_preds.permute(0, 1, 3, 2)\n \n preds = triu_condense(square_preds.squeeze().permute(1, 2, 0))\n \n self.activations = {\n 'atom_embed': atom_embed,\n 'atom_embed_update': atom_embed_update,\n 'res_embed': res_embed,\n 'combined': pairwise.detach().cpu().numpy(),\n 'preds': preds.detach().cpu().numpy()\n }\n\n return preds\n \n def training_step(self, batch, batch_nb):\n y_hat = self.forward(batch)\n y = triu_condense(batch['res_contact'].squeeze())\n loss = self.loss(y_hat, y)\n \n return {'loss': loss}\n\n def configure_optimizers(self): \n parameters = filter(lambda p: p.requires_grad, self.parameters())\n return torch.optim.SGD(parameters, lr=0.001, momentum=0.9)\n \n @pl.data_loader\n def train_dataloader(self):\n return DataLoader(self.train_data, shuffle=True, num_workers=10, pin_memory=True)",
"/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:541: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint8 = np.dtype([(\"qint8\", np.int8, 1)])\n/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:542: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint8 = np.dtype([(\"quint8\", np.uint8, 1)])\n/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:543: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint16 = np.dtype([(\"qint16\", np.int16, 1)])\n/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:544: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_quint16 = np.dtype([(\"quint16\", np.uint16, 1)])\n/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:545: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n _np_qint32 = np.dtype([(\"qint32\", np.int32, 1)])\n/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/tensorboard/compat/tensorflow_stub/dtypes.py:550: FutureWarning: Passing (type, 1) or '1type' as a synonym of type is deprecated; in a future version of numpy, it will be understood as (type, (1,)) / '(1,)type'.\n np_resource = np.dtype([(\"resource\", np.ubyte, 1)])\n"
]
],
[
[
"### Analysis functions",
"_____no_output_____"
]
],
[
[
"def plot_channels(values):\n fig, ax = plt.subplots(nrows=2, ncols=4, figsize=(20, 10))\n \n ax = ax.flatten()\n \n channel_names = [\n 'Hydrophobic',\n 'Hydrogen bond',\n 'Van der Waals',\n 'Water bridges',\n 'Salt bridges',\n 'Pi-stacking',\n 'Pi-cation',\n 'T-stacking'\n ]\n \n for channel in range(preds.shape[-1]):\n ax[channel].imshow(values[:, :, channel].squeeze(), vmin=0, vmax=1)\n ax[channel].set(title=channel_names[channel], xlabel='Residue #', ylabel='Residue #')\n \n fig.show()\n \n\n######################\n# ROC and PRC curves #\n######################\nfrom sklearn.metrics import precision_recall_curve, roc_curve, auc\n\ndef calc_metric_curve(preds, target, curve_type, squareform=False):\n \"\"\"\n Calculate ROC or PRC curves and area for the predicted contact channels.\n \n Args:\n - preds (np.ndarray) - Numpy array of model predictions either of form\n (n_res, n_res, n_chan) or (n_res * [n_res - 1] / 2, n_chan).\n - target (np.ndarray) - Numpy array of target values either of form\n (n_res, n_res, n_chan) or (n_res * [n_res - 1] / 2, n_chan),\n must match form of preds.\n - curve_type (str) - One of 'ROC' or 'PRC' to denote type of curve.\n - squareform (bool) - True if tensors are of shape (n_res, n_res, n_chan),\n False if they are of shape (n_res * [n_res - 1] / 2, n_chan)\n (default = True).\n \n Returns:\n - Tuple of x, y, and AUC values to be used for plotting the curves\n using plot_curve metric.\n \"\"\"\n \n # Get correct curve function\n if curve_type.upper() == 'ROC':\n curve_func = roc_curve\n elif curve_type.upper() == 'PRC':\n curve_func = precision_recall_curve\n \n # Generate dicts to hold outputs from curve generation functions\n x = dict()\n y = dict()\n auc_ = dict()\n \n # Handle case of squareform matrix (only get non-redundant triu indices)\n if squareform:\n indices = np.triu_indices(target.shape[0])\n\n # For each channel\n for i in range(target.shape[-1]):\n \n # Handle case of squareform\n if squareform:\n var1, var2, _ = curve_func(target[:, :, i][indices],\n preds[:, :, i][indices])\n \n # Handle case of pairwise\n else:\n var1, var2, _ = curve_func(target[:, i], preds[:, i])\n \n # Assign outputs to correct dict for plotting\n if curve_type.upper() == 'ROC':\n x[i] = var1\n y[i] = var2\n elif curve_type.upper() == 'PRC':\n x[i] = var2\n y[i] = var1\n \n # Calc AUC\n auc_[i] = auc(x[i], y[i])\n \n return (x, y, auc_)\n\n\ndef plot_curve_metric(x, y, auc, curve_type, title=None, labels=None):\n \"\"\"\n Plot ROC or PRC curves per output channel.\n \n Args:\n - x (dict) - Dict of numpy arrays for values to plot on x axis.\n - y (dict) - Dict of numpy arrays for values to plot on x axis.\n - auc (dict) - Dict of numpy arrays for areas under each curve.\n - curve_type (str) - One of 'ROC' or 'PRC' to denote type of curve.\n - title\n - labels\n \n Returns:\n - pyplot object of curves. \n \"\"\"\n \n # Generate figure\n plt.figure()\n \n # Linetype spec\n lw = 2\n curve_type = curve_type.upper()\n \n # Get the number of channels being plotted\n n_chan = len(x)\n \n # Make labels numeric if not provided\n if labels is None:\n labels = list(range(n_chan))\n \n # Check to make sure the labels are the right length\n if len(labels) != n_chan:\n raise ValueError('Number of labels ({}) does not match number of prediction channels ({}).'.format(len(labels), n_chan))\n \n # Get a lit of colors for all the channels\n color_list = plt.cm.Set1(np.linspace(0, 1, n_chan))\n \n # Plot each line\n for i, color in enumerate(color_list):\n plt.plot(x[i], y[i], color=color,\n lw=lw, label='{} (area = {:0.2f})'.format(labels[i], auc[i]))\n \n # Add labels and diagonal line for ROC\n if curve_type == 'ROC':\n xlab = 'FPR'\n ylab = 'TPR'\n plt.plot([0, 1], [0, 1], color='navy', lw=lw, linestyle='--')\n plt.legend(loc=\"lower right\")\n\n # Add labels for PRC \n elif curve_type == 'PRC':\n xlab = 'Recall'\n ylab = 'Precision'\n plt.legend(loc=\"lower left\")\n \n # Extend limits, add labels and title\n plt.xlim([-0.05, 1.05])\n plt.ylim([-0.05, 1.05])\n plt.xlabel(xlab)\n plt.ylabel(ylab)\n \n if title is not None:\n plt.title('{} for {}'.format(curve_type, title))\n else:\n plt.title('{}'.format(curve_type))\n \n plt.show()\n\ndef plot_curve(preds, target, curve_type, title=None, labels=None,\n squareform=False):\n \"\"\"\n Wrapper to directly plot curves from model output and target.\n \n Args:\n - preds (np array-like) - Array or tensor of predicted values output by\n model.\n - target (np array-like) - Array or tensor of target values.\n - curve_type (str) - One of 'ROC' or 'PRC'.\n - title (str) - Title of plot (default = None).\n - labels (list) - List of labels for each channel on the plot\n (default = None).\n - squareform (bool) - Whether the predictions and targets are in square form\n (default = False).\n \"\"\"\n x, y, auc_ = calc_metric_curve(preds, target, curve_type, squareform)\n return plot_curve_metric(x, y, auc_, curve_type, title, labels)",
"_____no_output_____"
]
],
[
[
"## Training\n\n### Instantiate dataloader and model",
"_____no_output_____"
]
],
[
[
"data_base = '/home/tshimko/tesselate/'\n\ntorch.manual_seed(0)\n\n\ntrain_data = TesselateDataset(data_base + 'test2.txt', graph_dir=data_base + 'data/graphs',\n contacts_dir=data_base + 'data/contacts',\n return_data='all', in_memory=True)\n\nmodel = PGNN2D(input_dim=6, hidden_dim=10, output_dim=10,\n anchorset_n=int(np.log2(303))**2, n_contact_channels=8,\n layer_num=4,\n train_data=train_data,\n val_data=None, test_data=None)",
"_____no_output_____"
],
[
"from pytorch_lightning import Trainer\n\ntrainer = Trainer(max_nb_epochs=15000)\ntrainer.fit(model)",
" 0%| | 0/1 [00:00<?, ?it/s]"
]
],
[
[
"# Analysis",
"_____no_output_____"
]
],
[
[
"acts = model.activations\nx = train_data[0]",
"_____no_output_____"
],
[
"# Plot the predictions\npreds = triu_expand(torch.from_numpy(acts['preds'])).sigmoid()\nplot_channels(preds)",
"_____no_output_____"
],
[
"# Plot the true values\nplot_channels(x['res_contact'])",
"_____no_output_____"
]
],
[
[
"## ROC",
"_____no_output_____"
]
],
[
[
"plot_curve(torch.from_numpy(acts['preds']).sigmoid(), triu_condense(x['res_contact'].squeeze()), 'ROC')",
"/home/tshimko/.virtualenvs/tesselate/lib/python3.6/site-packages/sklearn/metrics/ranking.py:659: UndefinedMetricWarning: No positive samples in y_true, true positive value should be meaningless\n UndefinedMetricWarning)\n"
]
],
[
[
"## PRC",
"_____no_output_____"
]
],
[
[
"plot_curve(torch.from_numpy(acts['preds']).sigmoid(), triu_condense(x['res_contact'].squeeze()), 'PRC')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6e96478067d81ba9032b63888d8954cdca4af7 | 11,318 | ipynb | Jupyter Notebook | examples/Example3_CorrelationCoefficient.ipynb | GriffithsLab/fooof-unit | 8c0144010b27d68eb1bf044dc5d21db00d9a664b | [
"MIT"
]
| null | null | null | examples/Example3_CorrelationCoefficient.ipynb | GriffithsLab/fooof-unit | 8c0144010b27d68eb1bf044dc5d21db00d9a664b | [
"MIT"
]
| 11 | 2020-08-11T15:07:50.000Z | 2020-09-14T18:21:34.000Z | examples/Example3_CorrelationCoefficient.ipynb | GriffithsLab/fooof-unit | 8c0144010b27d68eb1bf044dc5d21db00d9a664b | [
"MIT"
]
| null | null | null | 5,659 | 11,317 | 0.674854 | [
[
[
"# **Example 3: Correlation2Spectrum Test**\n\nCorrelation2Spectrum test compares neural power spectrum models. This is done with the computation of the correlation coefficient. The results is a Correlation Matrix",
"_____no_output_____"
],
[
"**Installation of SciUnit and FOOOF**",
"_____no_output_____"
]
],
[
[
"# If running in colab, uncomment\n#!pip install -q sciunit\n#!pip install fooof",
"_____no_output_____"
]
],
[
[
"**Import libraries and modules**",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport sciunit\nimport sys\nfrom fooofunit import capabilities, models, scores, tests\nfrom fooof import FOOOF\nimport os,glob, numpy as np, pandas as pd\nfrom matplotlib import pyplot as plt\nfrom scipy.io import loadmat",
"_____no_output_____"
]
],
[
[
"**Import capability, model and test of interest**",
"_____no_output_____"
]
],
[
[
"from fooofunit.capabilities import ProducesPowerSpectrum\nfrom fooofunit.models import NeuralPowerSpectra\nfrom fooofunit.tests import Correlation2Spectrum, Correlation2SpectrumFloat",
"_____no_output_____"
]
],
[
[
"**Generate Simulated Power Spectra**",
"_____no_output_____"
],
[
"The example is demonstrated on five different simulated neural power spectra. As in the previous examples, they are generated with FOOOF. The frequency range, noise, aperiodic and periodic parameters are defined. ",
"_____no_output_____"
]
],
[
[
"class GeneratePowerSpectrum:\n \n def __init__(self, freq_range, aperiodic_params, periodic_params, nlv=0):\n from fooof.sim.gen import gen_power_spectrum\n self.freqs, self.spectrum = gen_power_spectrum(freq_range, aperiodic_params, periodic_params,nlv)\n self.freq_range = freq_range",
"_____no_output_____"
],
[
"SimulatedPowerSpectra1 = GeneratePowerSpectrum([3, 40] ,[1, 1], [10, 0.3, 1])\nSimulatedPowerSpectra2 = GeneratePowerSpectrum([3, 40], [1, 500, 2], [9, 0.4, 1, 24, 0.2, 3], 0.01)\nSimulatedPowerSpectra3 = GeneratePowerSpectrum([3, 40], [1, 1], [[10, 0.2, 1.25], [30, 0.15, 2]])\nSimulatedPowerSpectra4 = GeneratePowerSpectrum([3, 40] ,[1, 2], [20, 0.4, 1])\nSimulatedPowerSpectra5 = GeneratePowerSpectrum([3, 40] ,[1, 1], [12, 0.1, 1])",
"_____no_output_____"
]
],
[
[
"**NeuralPowerSpectra class**",
"_____no_output_____"
],
[
"Each simulated power spectrum are an instance of the model class NeuralPowerSpectra.",
"_____no_output_____"
]
],
[
[
"PowerSpectra1 = NeuralPowerSpectra(SimulatedPowerSpectra1.freqs,SimulatedPowerSpectra1.spectrum, SimulatedPowerSpectra1.freq_range, name = \"Simulated Power Spectra 1\")\nPowerSpectra2 = NeuralPowerSpectra(SimulatedPowerSpectra2.freqs,SimulatedPowerSpectra2.spectrum, SimulatedPowerSpectra2.freq_range, name = \"Simulated Power Spectra 2\")\nPowerSpectra3 = NeuralPowerSpectra(SimulatedPowerSpectra3.freqs,SimulatedPowerSpectra3.spectrum, SimulatedPowerSpectra3.freq_range, name = \"Simulated Power Spectra 3\")\nPowerSpectra4 = NeuralPowerSpectra(SimulatedPowerSpectra4.freqs,SimulatedPowerSpectra4.spectrum, SimulatedPowerSpectra4.freq_range, name = \"Simulated Power Spectra 4\")\nPowerSpectra5 = NeuralPowerSpectra(SimulatedPowerSpectra5.freqs,SimulatedPowerSpectra5.spectrum, SimulatedPowerSpectra5.freq_range, name = \"Simulated Power Spectra 5\")\n",
"_____no_output_____"
]
],
[
[
"**Judge**",
"_____no_output_____"
],
[
"The judge method executes the test and returns the correlation coefficient. Only_lower_triangle is set to True in order to only compute the lower triangle and take advantage of the symmetry observed to save computational time.",
"_____no_output_____"
]
],
[
[
"CorrelationTest = Correlation2Spectrum()\nCorrelation_Coeff = CorrelationTest.judge([PowerSpectra1, PowerSpectra2, PowerSpectra3, PowerSpectra4, PowerSpectra5], only_lower_triangle=True)",
"\nFOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.50 <= 0.50\n\tLower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).\n\tToo low a limit may lead to overfitting noise as small bandwidth peaks.\n\tWe recommend a lower bound of approximately 2x the frequency resolution.\n\n\nFOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.50 <= 0.50\n\tLower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).\n\tToo low a limit may lead to overfitting noise as small bandwidth peaks.\n\tWe recommend a lower bound of approximately 2x the frequency resolution.\n\n\nFOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.50 <= 0.50\n\tLower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).\n\tToo low a limit may lead to overfitting noise as small bandwidth peaks.\n\tWe recommend a lower bound of approximately 2x the frequency resolution.\n\n\nFOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.50 <= 0.50\n\tLower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).\n\tToo low a limit may lead to overfitting noise as small bandwidth peaks.\n\tWe recommend a lower bound of approximately 2x the frequency resolution.\n\n\nFOOOF WARNING: Lower-bound peak width limit is < or ~= the frequency resolution: 0.50 <= 0.50\n\tLower bounds below frequency-resolution have no effect (effective lower bound is the frequency resolution).\n\tToo low a limit may lead to overfitting noise as small bandwidth peaks.\n\tWe recommend a lower bound of approximately 2x the frequency resolution.\n\n"
]
],
[
[
"**Score**",
"_____no_output_____"
]
],
[
[
"Correlation_Coeff",
"_____no_output_____"
]
],
[
[
"The results show how correlated all the models are to each other. They are an indicator of similarity between the two. In this case, we can conclude that Simulated Power Spectra 1, 3 and 5 are the most similar (as the result is greather than 0.9).",
"_____no_output_____"
],
[
"# **Testing**",
"_____no_output_____"
]
],
[
[
"coeff_result1 = pd.Series(np.diag(Correlation_Coeff.score))\nexpected_result1 = pd.Series([1.0, 1.0, 1.0, 1.0, 1.0])\nassert expected_result1.equals(coeff_result1)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6e98ba2c46c3249feaf826a50012b3aeadf18d | 6,283 | ipynb | Jupyter Notebook | notebooks/workshop/3 - Silly example.ipynb | lingruiluo/jp_doodle | b3935208821898f22ab504c2b26dd4d37f08f0e4 | [
"BSD-2-Clause"
]
| 43 | 2018-10-10T08:38:07.000Z | 2022-03-19T22:44:42.000Z | notebooks/workshop/3 - Silly example.ipynb | lingruiluo/jp_doodle | b3935208821898f22ab504c2b26dd4d37f08f0e4 | [
"BSD-2-Clause"
]
| 8 | 2018-09-17T19:49:45.000Z | 2020-08-24T15:51:16.000Z | notebooks/workshop/3 - Silly example.ipynb | lingruiluo/jp_doodle | b3935208821898f22ab504c2b26dd4d37f08f0e4 | [
"BSD-2-Clause"
]
| 5 | 2019-06-13T15:53:55.000Z | 2020-11-13T01:22:56.000Z | 29.497653 | 113 | 0.573293 | [
[
[
"# A silly example to warm up\n\n- Create a canvas.\n- Make it float as a dialog.\n- Put some colored rectangles in it.\n- Add a text element.\n- Make the text element name the color of the rectangle when the rectangle is clicked.",
"_____no_output_____"
]
],
[
[
"from jp_doodle import dual_canvas\nfrom IPython.display import display\n\n# Create an empty canvas and display it\ndemo = dual_canvas.DualCanvasWidget(width=320, height=220)\ndisplay(demo)",
"_____no_output_____"
],
[
"# Pop the canvas out into a dialog so it won't scroll with the page.\ndemo.in_dialog()",
"_____no_output_____"
],
[
"# Put some colored rectangles on the canvas\n# Name them so they can respond to events keep them in a list.\ncolors = \"yellow green blue magenta red brown\".split()\n\nrectangles = []\nx = 0\nfor color in colors:\n rectangle = demo.rect(x=x, y=0, w=30, h=30, color=color, name=True)\n rectangles.append(rectangle)\n x = x + 40",
"_____no_output_____"
],
[
"# Center the rectangles in the canvas.\ndemo.fit()",
"_____no_output_____"
],
[
"# Add a changable text area to the canvas.\ntext = demo.text(x=20, y=-20, text=\"Here is some text\", color=\"white\", background=\"black\", name=True)",
"_____no_output_____"
],
[
"# Attach mouse click handlers to the rectangles which change the text.\n\ndef click_handler(event):\n color = event[\"object_info\"][\"color\"]\n text.change(background=color, text=color)\n \nfor rectangle in rectangles:\n rectangle.on(\"click\", click_handler)",
"_____no_output_____"
]
],
[
[
"# Making scatter plots instead of rectangles\n\nThe dual canvas framework is designed for making scientific visualizations\nand a scatter plot a more realistic approximation of a scientific visualization\nthan a rectangle.\n\nTo make it easy to create a lot of scatter plots we will\nuse reference frames.\n\nThe following function creates a small scatter plot on a reference frame\nusing randomly generated points.",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef random_scatter(frame, n=7, color=\"green\", circle_mouseover=None):\n points = np.random.random(n*2).reshape((n, 2))\n frame.lower_left_axes(max_tick_count=4, min_x=0, max_x=1, min_y=0, max_y=1)\n for (x,y) in points:\n c = frame.frame_circle(x=x, y=y, r=0.05, color=color, name=True)\n if circle_mouseover:\n c.on(\"mouseover\", circle_mouseover)\n\n# To test the function we create a canvas, a frame in the canvas, and draw a plot:\ntest_scatter = dual_canvas.DualCanvasWidget(width=320, height=220)\n# Create a frame which scales 1 unit to 100 pixels in both dimensions.\ntest_frame = test_scatter.rframe(scale_x=100, scale_y=100)\nrandom_scatter(test_frame, color=\"salmon\")\n\n# add some niceties:\ntest_frame.text(x=0.5, y=-0.5, text=\"Just a test...\", align=\"center\")\ntest_scatter.fit()\ndisplay(test_scatter)",
"_____no_output_____"
]
],
[
[
"# Using multiple frames with multiple plots\n\nTo create several scatter plots we create several frames offset in different\npositions and use the `random_scatter` function with those frames.",
"_____no_output_____"
]
],
[
[
"# Create the canvas.\nmultiple_scatter = dual_canvas.DualCanvasWidget(width=820, height=220)\n\n# Add a feedback text area for mouse event feedback.\nfeedback = multiple_scatter.text(\n x=20, y=-80, text=\"Mouse over a circle to see it's coordinates.\", \n color=\"#ccc\", background=\"black\", name=True, font=\"normal 30px Arial\"\n)\n\n# Define an event handler.\ndef circle_mouseover(event):\n \"When the mouse is over a circle, show the color and position of the circle.\"\n info = event[\"object_info\"]\n color = info[\"color\"]\n newtext = \"%s : %4.2f, %4.2f\" % (color, info[\"x\"], info[\"y\"])\n feedback.change(background=color, text=newtext)\n\n# Draw a scatter plot for each color.\nx_offset = 0\nfor c in colors:\n frame = multiple_scatter.rframe(scale_x=100, scale_y=100, translate_x=x_offset)\n random_scatter(frame, color=c, circle_mouseover=circle_mouseover)\n x_offset += 150\n \nmultiple_scatter.fit()\ndisplay(multiple_scatter)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6e9c0d83d102f85f64b9757edac52869770eb8 | 131,123 | ipynb | Jupyter Notebook | Data_Science_2-4 _ Assignment_Week_2.ipynb | trinhl/devC-coderSchool-dataScience | 6de7e6db8fd44570b32e471fd16171f785e4d6f7 | [
"MIT"
]
| null | null | null | Data_Science_2-4 _ Assignment_Week_2.ipynb | trinhl/devC-coderSchool-dataScience | 6de7e6db8fd44570b32e471fd16171f785e4d6f7 | [
"MIT"
]
| null | null | null | Data_Science_2-4 _ Assignment_Week_2.ipynb | trinhl/devC-coderSchool-dataScience | 6de7e6db8fd44570b32e471fd16171f785e4d6f7 | [
"MIT"
]
| null | null | null | 81.544154 | 28,002 | 0.746376 | [
[
[
"\n\n# SF Salaries Exercise \n\nExplore San Francisco city employee salary data.\n\n## Overview\n\nOne way to understand how a city government works is by looking at who it employs and how its employees are compensated. This data contains the names, job title, and compensation for San Francisco city employees on an annual basis from 2011 to 2014.\n\n\n\nJust follow along and complete the tasks outlined in bold below. The tasks will get harder and harder as you go along.\n\n## Resourses\n\n[Pandas API Reference](https://pandas.pydata.org/pandas-docs/stable/api.html)\n\n[NumPy Reference](https://docs.scipy.org/doc/numpy/reference/)\n\n[Visualization with Seaborn](https://jakevdp.github.io/PythonDataScienceHandbook/04.14-visualization-with-seaborn.html)\n",
"_____no_output_____"
],
[
"**Import libraries**",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport seaborn as sns",
"_____no_output_____"
]
],
[
[
"**Read `Salaries.csv` as a dataframe called `sal`.**",
"_____no_output_____"
]
],
[
[
"link = \"https://s3-ap-southeast-1.amazonaws.com/intro-to-ml-minhdh/Salaries.csv\"\nsal = pd.read_csv(link)",
"_____no_output_____"
]
],
[
[
"**Check the head of the DataFrame.**",
"_____no_output_____"
]
],
[
[
"sal.head()",
"_____no_output_____"
]
],
[
[
"**Use the .info() method to find out how many entries there are.**",
"_____no_output_____"
]
],
[
[
"sal.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 148654 entries, 0 to 148653\nData columns (total 13 columns):\nId 148654 non-null int64\nEmployeeName 148654 non-null object\nJobTitle 148654 non-null object\nBasePay 148045 non-null float64\nOvertimePay 148650 non-null float64\nOtherPay 148650 non-null float64\nBenefits 112491 non-null float64\nTotalPay 148654 non-null float64\nTotalPayBenefits 148654 non-null float64\nYear 148654 non-null int64\nNotes 0 non-null float64\nAgency 148654 non-null object\nStatus 0 non-null float64\ndtypes: float64(8), int64(2), object(3)\nmemory usage: 14.7+ MB\n"
]
],
[
[
"**What is the average BasePay ?**\n\nKeyword: [mean](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.mean.html)",
"_____no_output_____"
]
],
[
[
"sal.describe()",
"_____no_output_____"
],
[
"sal.describe().loc['mean', 'BasePay']",
"_____no_output_____"
]
],
[
[
"**What is the lowest and highest amount of BasePay?**\n\nKeyword: \n[min](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.min.html) , \n[max](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.max.html)",
"_____no_output_____"
]
],
[
[
"sal.describe().loc['min', 'BasePay']",
"_____no_output_____"
],
[
"sal.describe().loc['max', 'BasePay']",
"_____no_output_____"
]
],
[
[
"**How about OvertimePay in the dataset? What is the average, lowest, highest amount?**",
"_____no_output_____"
]
],
[
[
"# lowest OvertimePay\nsal['OvertimePay'].min()\n",
"_____no_output_____"
],
[
"# highest OvertimePay\nsal['OvertimePay'].max()",
"_____no_output_____"
],
[
"# average OvertimePay\nsal['OvertimePay'].mean()",
"_____no_output_____"
]
],
[
[
"**What is the job title of JOSEPH DRISCOLL ? Note: Use all caps, otherwise you may get an answer that doesn't match up (there is also a lowercase Joseph Driscoll).**",
"_____no_output_____"
]
],
[
[
"sal[sal['EmployeeName'] == \"JOSEPH DRISCOLL\"]['JobTitle']",
"_____no_output_____"
],
[
"# Now try to select Employees who have name \"GARY JIMENEZ\"\n# Your code here\nsal[sal['EmployeeName'] == \"GARY JIMENEZ\"]['JobTitle']",
"_____no_output_____"
]
],
[
[
"**How much does JOSEPH DRISCOLL make (including benefits)?**",
"_____no_output_____"
]
],
[
[
"sal[sal['EmployeeName'] == \"JOSEPH DRISCOLL\"]['TotalPayBenefits']",
"_____no_output_____"
]
],
[
[
"**What is the name of highest paid person (including benefits)?**",
"_____no_output_____"
]
],
[
[
"sal.sort_values(by='TotalPayBenefits')['EmployeeName'].tail(1)",
"_____no_output_____"
]
],
[
[
"**What is the name of lowest paid person (including benefits)? Do you notice something strange about how much he or she is paid?**",
"_____no_output_____"
]
],
[
[
"sal.sort_values(by='TotalPayBenefits')[['EmployeeName', 'TotalPayBenefits']].head(1)",
"_____no_output_____"
]
],
[
[
"**What was the average (mean) BasePay of all employees per year? (2011-2014) ?**\nKeyword: _groupby_",
"_____no_output_____"
]
],
[
[
"pd.options.display.float_format = '{:,}'.format",
"_____no_output_____"
],
[
"# print(sal['Year'].unique())\nsal.groupby('Year').mean()['BasePay']\n",
"_____no_output_____"
]
],
[
[
"**How many unique job titles are there?**\n\nKeyword: _unique, nunique_",
"_____no_output_____"
]
],
[
[
"sal['JobTitle'].nunique()",
"_____no_output_____"
]
],
[
[
"**What are the top 5 most common jobs?**\n\nKeyword: *value_counts*",
"_____no_output_____"
]
],
[
[
"sal['JobTitle'].value_counts().head(5)",
"_____no_output_____"
]
],
[
[
"**How many Job Titles were represented by only one person in 2013? (e.g. Job Titles with only one occurence in 2013?)**",
"_____no_output_____"
]
],
[
[
"sum(sal[sal['Year'] == 2013].groupby('JobTitle')['JobTitle'].count() == 1)",
"_____no_output_____"
]
],
[
[
"**How many people have the word Chief in their job title?**",
"_____no_output_____"
]
],
[
[
"sal['JobTitle'].str.contains('Chief|CHIEF', regex=True).sum()",
"_____no_output_____"
]
],
[
[
"## Data Visualization\n\n**Implement seaborn's countplot with x='Year'**",
"_____no_output_____"
]
],
[
[
"sns.countplot(data=sal, x='Year')",
"_____no_output_____"
]
],
[
[
"**Implement seaborn's distplot for BasePay of Year 2011**",
"_____no_output_____"
]
],
[
[
"sns.distplot(sal[sal['Year'] == 2011]['BasePay'], bins=10)",
"_____no_output_____"
]
],
[
[
"**How about other Year**",
"_____no_output_____"
]
],
[
[
"sns.distplot(sal[sal['Year'] == 2012]['BasePay'], bins=10);\nsns.distplot(sal[sal['Year'] == 2013]['BasePay'], bins=10);\nsns.distplot(sal[sal['Year'] == 2014]['BasePay'], bins=10);",
"/usr/local/lib/python3.6/dist-packages/numpy/lib/histograms.py:824: RuntimeWarning: invalid value encountered in greater_equal\n keep = (tmp_a >= first_edge)\n/usr/local/lib/python3.6/dist-packages/numpy/lib/histograms.py:825: RuntimeWarning: invalid value encountered in less_equal\n keep &= (tmp_a <= last_edge)\n/usr/local/lib/python3.6/dist-packages/statsmodels/nonparametric/kde.py:447: RuntimeWarning: invalid value encountered in greater\n X = X[np.logical_and(X > clip[0], X < clip[1])] # won't work for two columns.\n/usr/local/lib/python3.6/dist-packages/statsmodels/nonparametric/kde.py:447: RuntimeWarning: invalid value encountered in less\n X = X[np.logical_and(X > clip[0], X < clip[1])] # won't work for two columns.\n"
]
],
[
[
"## Bonus\n\n**Visualize top 5 Jobs with BasePay, Benefits and OvertimePay**\n\nKeyword: *index, isin, groupby, mean, plot*",
"_____no_output_____"
]
],
[
[
"# sal.groupby('JobTitle').mean().sort_values(by=['BasePay','Benefits', 'OvertimePay'], ascending=False)[['BasePay','Benefits', 'OvertimePay']].head(5)\nsal.groupby('JobTitle').sum().sort_values(by=['BasePay','Benefits', 'OvertimePay'])[['BasePay','Benefits', 'OvertimePay']].tail(5).plot()",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
],
[
[
"**Fun Fact: Is there a correlation between length of the Job Title string and Salary?**\n\n*Hint: corr()*",
"_____no_output_____"
]
],
[
[
"sal['title_len'] = sal['JobTitle'].apply(len)",
"_____no_output_____"
],
[
"sal[['TotalPayBenefits','title_len']].corr()",
"_____no_output_____"
]
],
[
[
"# Great Job!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
]
]
|
ec6eabb668253c47b9d8ef0df2c1a0b151a6a8d2 | 284,862 | ipynb | Jupyter Notebook | jupyter_examples/example_11_composite_model.ipynb | colinator/spikeflow | 109aea58c3d2c7b2da8680ddd98fa99bdaf20579 | [
"MIT"
]
| 32 | 2019-01-31T16:50:57.000Z | 2022-01-18T09:55:22.000Z | jupyter_examples/example_11_composite_model.ipynb | colinator/spikeflow | 109aea58c3d2c7b2da8680ddd98fa99bdaf20579 | [
"MIT"
]
| null | null | null | jupyter_examples/example_11_composite_model.ipynb | colinator/spikeflow | 109aea58c3d2c7b2da8680ddd98fa99bdaf20579 | [
"MIT"
]
| 6 | 2019-05-26T14:55:05.000Z | 2022-01-14T08:54:27.000Z | 1,157.97561 | 276,632 | 0.941698 | [
[
[
"# composite models: two 'columns', each containing two neuron layers",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom spikeflow import BPNNModel, CompositeLayer, IzhikevichNeuronLayer\nfrom spikeflow import Synapse, ComplexSynapseLayer\nfrom spikeflow import weights_connecting_from_to, weights_from_synapses, delays_for_weights\nfrom spikeflow import identical_sampler, normal_sampler, normal_ints_sampler\nfrom spikeflow.drawing_utils.trace_renderers import *",
"/Applications/Anaconda/anaconda/envs/mlbook/lib/python3.6/site-packages/h5py/__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\n"
]
],
[
[
"# Create a model with two 'groups' ('columns'?) of neuron layers - each group has one recurrently-connected neuron layer, connecting to one 'decoder' layer. First group connects to second group.",
"_____no_output_____"
]
],
[
[
"model_input_shape = (3,)\n\ndef create_composite_layer(name):\n \n neuron_layer_0 = IzhikevichNeuronLayer.layer_from_tuples('nl0', [\n IzhikevichNeuronLayer.C(a=0.030, b=0.2, c=-65.0, d=6.0, t=30.0, v0=0.0),\n IzhikevichNeuronLayer.C(a=0.035, b=0.2, c=-65.0, d=6.0, t=30.0, v0=0.0),\n IzhikevichNeuronLayer.C(a=0.020, b=0.2, c=-65.0, d=6.0, t=30.0, v0=0.0),\n ])\n\n neuron_layer_1 = IzhikevichNeuronLayer.layer_from_tuples('nl1', [\n IzhikevichNeuronLayer.C(a=0.010, b=0.2, c=-65.0, d=6.0, t=30.0, v0=0.0),\n IzhikevichNeuronLayer.C(a=0.015, b=0.2, c=-65.0, d=6.0, t=30.0, v0=0.0)\n ])\n\n recurrent_weights = weights_connecting_from_to(\n neuron_layer_0, \n neuron_layer_0, \n 1.0, \n normal_sampler(2.0, 1.0))\n\n recurrent_connection = ComplexSynapseLayer(\n name = 'rec_syn',\n from_layer = neuron_layer_0, \n to_layer = neuron_layer_0, \n decay = 0.95, \n weights = recurrent_weights,\n delay = delays_for_weights(recurrent_weights, normal_ints_sampler(10, 5))\n )\n \n forward_connection = ComplexSynapseLayer(\n name = 'forward_syn',\n from_layer = neuron_layer_0, \n to_layer = neuron_layer_1, \n decay = 0.95, \n delay = np.array([[5, 10], [8, 4], [3, 7]]),\n weights = weights_from_synapses(neuron_layer_0, neuron_layer_1, [\n Synapse(0, 0, 5.0),\n Synapse(1, 0, 10.0),\n Synapse(2, 0, 15.0),\n Synapse(0, 1, 8.0),\n Synapse(1, 1, 10.0),\n Synapse(2, 1, 12.0)\n ])\n )\n\n return CompositeLayer(name, \n [neuron_layer_0, neuron_layer_1], \n [recurrent_connection, forward_connection])\n\ncl0 = create_composite_layer('cl0')\ncl1 = create_composite_layer('cl1')\n\ncl0_to_cl1 = ComplexSynapseLayer(\n name = 'cl0_to_cl1',\n from_layer = cl0, \n to_layer = cl1, \n decay = 0.95, \n delay = 50,\n weights = weights_from_synapses(cl0, cl1, [\n Synapse(0, 0, 5.0),\n Synapse(0, 1, 10.0),\n Synapse(0, 2, 15.0),\n Synapse(1, 0, 8.0),\n Synapse(1, 1, 10.0),\n Synapse(1, 2, 12.0)\n ]))\n\nmodel = BPNNModel.compiled_model(model_input_shape, [cl0, cl1], [cl0_to_cl1])\n\nprint(model.ops_format())",
"{'cl0': {'nl0': ['input', 'v', 'u', 'fired'], 'nl1': ['input', 'v', 'u', 'fired'], 'rec_syn': ['input', 'output'], 'forward_syn': ['input', 'output']}, 'cl1': {'nl0': ['input', 'v', 'u', 'fired'], 'nl1': ['input', 'v', 'u', 'fired'], 'rec_syn': ['input', 'output'], 'forward_syn': ['input', 'output']}, 'cl0_to_cl1': ['input', 'output']}\n"
]
],
[
[
"# Run the model for 2000 timesteps",
"_____no_output_____"
]
],
[
[
"traces = []\ndef end_time_step_callback(i, graph, sess, results):\n traces.append(results)\n \ndef data_generator():\n for i in range(0, 2000):\n yield np.ones(3,)*(7 if (i > 1200 and i < 1800) else 0)\n \nmodel.run_time(data_generator(), end_time_step_callback)",
"_____no_output_____"
]
],
[
[
"# Extract the data we want and display",
"_____no_output_____"
]
],
[
[
"neuron_layer_0_traces = np.array([r['cl0']['nl0'] for r in traces])\nneuron_layer_1_traces = np.array([r['cl0']['nl1'] for r in traces])\nneuron_layer_2_traces = np.array([r['cl1']['nl0'] for r in traces])\nneuron_layer_3_traces = np.array([r['cl1']['nl1'] for r in traces])\n\nrender_figure([IzhikevichNeuronTraceRenderer(neuron_layer_0_traces, 'Composite 0, Layer 0 Neuron'),\n NeuronFiringsRenderer(neuron_layer_0_traces[:,3,:], 30, 'Composite 0, Layer 0 Firings'),\n IzhikevichNeuronTraceRenderer(neuron_layer_1_traces, 'Composite 0, Layer 1 Neuron'), \n NeuronFiringsRenderer(neuron_layer_1_traces[:,3,:], 30,'Composite 0, Layer 1 Firings'),\n IzhikevichNeuronTraceRenderer(neuron_layer_2_traces, 'Composite 1, Layer 0 Neuron'), \n NeuronFiringsRenderer(neuron_layer_2_traces[:,3,:], 30,'Composite 1, Layer 0 Firings'),\n IzhikevichNeuronTraceRenderer(neuron_layer_3_traces, 'Composite 1, Layer 1 Neuron'), \n NeuronFiringsRenderer(neuron_layer_3_traces[:,3,:], 30,'Composite 1, Layer 1 Firings')\n ],\n 1000, 2000, 100)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6ed254e3a482e0575e7864f52625747955e5b2 | 580,301 | ipynb | Jupyter Notebook | examples/matbench_dielectric_eda.ipynb | janosh/pymatviz | 17742d161151db1f9e1d615e6f19a3eac678be27 | [
"MIT"
]
| null | null | null | examples/matbench_dielectric_eda.ipynb | janosh/pymatviz | 17742d161151db1f9e1d615e6f19a3eac678be27 | [
"MIT"
]
| 8 | 2022-03-01T21:11:01.000Z | 2022-03-20T16:46:46.000Z | examples/matbench_dielectric_eda.ipynb | janosh/pymatviz | 17742d161151db1f9e1d615e6f19a3eac678be27 | [
"MIT"
]
| null | null | null | 2,991.242268 | 223,251 | 0.779651 | [
[
[
"import matplotlib.pyplot as plt\nimport plotly.express as px\nimport plotly.io as pio\nfrom matminer.datasets import load_dataset\nfrom tqdm import tqdm\n\nfrom pymatviz import ptable_heatmap, spacegroup_hist, spacegroup_sunburst\nfrom pymatviz.utils import get_crystal_sys\n\n\npio.templates.default = \"plotly_white\"\n# Comment out next line to get interactive plotly figures. Only used here since interactive\n# figures don't show up at all in notebooks on GitHub. https://github.com/plotly/plotly.py/issues/931\npio.renderers.default = \"svg\"\n\nplt.rc(\"font\", size=14)\nplt.rc(\"savefig\", bbox=\"tight\", dpi=200)\nplt.rc(\"axes\", titlesize=18, titleweight=\"bold\")\nplt.rcParams[\"figure.constrained_layout.use\"] = True",
"_____no_output_____"
],
[
"df_diel = load_dataset(\"matbench_dielectric\")\ndf_diel[[\"spg_symbol\", \"spg_num\"]] = [\n struct.get_space_group_info() for struct in tqdm(df_diel.structure)\n]\ndf_diel[\"crys_sys\"] = df_diel.spg_num.map(get_crystal_sys)",
"100%|██████████| 4764/4764 [00:05<00:00, 881.24it/s] \n"
],
[
"df_diel[\"volume\"] = df_diel.structure.apply(lambda cryst: cryst.volume)\ndf_diel[\"formula\"] = df_diel.structure.apply(lambda cryst: cryst.formula)\n\nptable_heatmap(df_diel.formula, log=True)\nplt.title(\"Elements in Matbench Dielectric\")",
"_____no_output_____"
],
[
"_ = df_diel.hist(bins=80, log=True, figsize=(20, 4), layout=(1, 3))",
"_____no_output_____"
],
[
"ax = spacegroup_hist(df_diel.spg_num)\n_ = ax.set_title(\"Space group histogram\", y=1.2)",
"_____no_output_____"
],
[
"fig = spacegroup_sunburst(df_diel.spg_num, show_counts=\"percent\")\nfig.update_layout(title=\"<b>Space group sunburst<b>\", title_x=0.5)\nfig.show()",
"_____no_output_____"
],
[
"labels = {\"crys_sys\": \"Crystal system\", \"n\": \"Refractive index n\"}\n\nfig = px.violin(df_diel, color=\"crys_sys\", x=\"crys_sys\", y=\"n\", labels=labels)\nfig.update_layout(\n title=\"Refractive index distribution by crystal system\",\n title_x=0.5,\n margin=dict(b=10, l=10, r=10, t=50),\n showlegend=False,\n)\nfig.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6edfea3b56d4a48b268cec351ddfbeef8f6df7 | 128,226 | ipynb | Jupyter Notebook | docs/notebooks/Carver.ipynb | rindPHI/fuzzingbook | 39e3359621aeafe915d1a28e1536cad2cd7465ce | [
"MIT"
]
| null | null | null | docs/notebooks/Carver.ipynb | rindPHI/fuzzingbook | 39e3359621aeafe915d1a28e1536cad2cd7465ce | [
"MIT"
]
| null | null | null | docs/notebooks/Carver.ipynb | rindPHI/fuzzingbook | 39e3359621aeafe915d1a28e1536cad2cd7465ce | [
"MIT"
]
| null | null | null | 29.606557 | 1,932 | 0.565658 | [
[
[
"# Carving Unit Tests\n\nSo far, we have always generated _system input_, i.e. data that the program as a whole obtains via its input channels. If we are interested in testing only a small set of functions, having to go through the system can be very inefficient. This chapter introduces a technique known as _carving_, which, given a system test, automatically extracts a set of _unit tests_ that replicate the calls seen during the unit test. The key idea is to _record_ such calls such that we can _replay_ them later – as a whole or selectively. On top, we also explore how to synthesize API grammars from carved unit tests; this means that we can _synthesize API tests without having to write a grammar at all._",
"_____no_output_____"
],
[
"**Prerequisites**\n\n* Carving makes use of dynamic traces of function calls and variables, as introduced in the [chapter on configuration fuzzing](ConfigurationFuzzer.ipynb).\n* Using grammars to test units was introduced in the [chapter on API fuzzing](APIFuzzer.ipynb).",
"_____no_output_____"
]
],
[
[
"import bookutils",
"_____no_output_____"
],
[
"import APIFuzzer",
"_____no_output_____"
]
],
[
[
"## Synopsis\n<!-- Automatically generated. Do not edit. -->\n\nTo [use the code provided in this chapter](Importing.ipynb), write\n\n```python\n>>> from fuzzingbook.Carver import <identifier>\n```\n\nand then make use of the following features.\n\n\nThis chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such \"carving\" mechanisms have the potential to run tests much faster.\n\n### Recording Calls\n\nThe `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:\n\n```python\n>>> with CallCarver() as carver:\n>>> y = my_sqrt(2)\n>>> y = my_sqrt(4)\n```\nAfter execution, `called_functions()` lists the names of functions encountered:\n\n```python\n>>> carver.called_functions()\n['my_sqrt', '__exit__']\n```\nThe `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).\n\n```python\n>>> carver.arguments('my_sqrt')\n[[('x', 2)], [('x', 4)]]\n```\nComplex arguments are properly serialized, such that they can be easily restored.\n\n### Synthesizing Calls\n\nWhile such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.\n\nThe `CallGrammarMiner` class turns a list of carved executions into a grammar.\n\n```python\n>>> my_sqrt_miner = CallGrammarMiner(carver)\n>>> my_sqrt_grammar = my_sqrt_miner.mine_call_grammar()\n>>> my_sqrt_grammar\n{'<start>': ['<call>'],\n '<call>': ['<my_sqrt>'],\n '<my_sqrt-x>': ['4', '2'],\n '<my_sqrt>': ['my_sqrt(<my_sqrt-x>)']}\n```\nThis grammar can be used to synthesize calls.\n\n```python\n>>> fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)\n>>> fuzzer.fuzz()\n'my_sqrt(2)'\n```\nThese calls can be executed in isolation, effectively extracting unit tests from system tests:\n\n```python\n>>> eval(fuzzer.fuzz())\n2.0\n```\n",
"_____no_output_____"
],
[
"## System Tests vs Unit Tests\n\nRemember the URL grammar introduced for [grammar fuzzing](Grammars.ipynb)? With such a grammar, we can happily test a Web browser again and again, checking how it reacts to arbitrary page requests.\n\nLet us define a very simple \"web browser\" that goes and downloads the content given by the URL.",
"_____no_output_____"
]
],
[
[
"import urllib.parse",
"_____no_output_____"
],
[
"def webbrowser(url):\n \"\"\"Download the http/https resource given by the URL\"\"\"\n import requests # Only import if needed\n\n r = requests.get(url)\n return r.text",
"_____no_output_____"
]
],
[
[
"Let us apply this on [fuzzingbook.org](https://www.fuzzingbook.org/) and measure the time, using the [Timer class](Timer.ipynb):",
"_____no_output_____"
]
],
[
[
"from Timer import Timer",
"_____no_output_____"
],
[
"with Timer() as webbrowser_timer:\n fuzzingbook_contents = webbrowser(\n \"http://www.fuzzingbook.org/html/Fuzzer.html\")\n\nprint(\"Downloaded %d bytes in %.2f seconds\" %\n (len(fuzzingbook_contents), webbrowser_timer.elapsed_time()))",
"Downloaded 457512 bytes in 1.95 seconds\n"
],
[
"fuzzingbook_contents[:100]",
"_____no_output_____"
]
],
[
[
"A full webbrowser, of course, would also render the HTML content. We can achieve this using these commands (but we don't, as we do not want to replicate the entire Web page here):\n\n\n```python\nfrom IPython.display import HTML, display\nHTML(fuzzingbook_contents)\n```",
"_____no_output_____"
],
[
"Having to start a whole browser (or having it render a Web page) again and again means lots of overhead, though – in particular if we want to test only a subset of its functionality. In particular, after a change in the code, we would prefer to test only the subset of functions that is affected by the change, rather than running the well-tested functions again and again.",
"_____no_output_____"
],
[
"Let us assume we change the function that takes care of parsing the given URL and decomposing it into the individual elements – the scheme (\"http\"), the network location (`\"www.fuzzingbook.com\"`), or the path (`\"/html/Fuzzer.html\"`). This function is named `urlparse()`:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import urlparse",
"_____no_output_____"
],
[
"urlparse('https://www.fuzzingbook.com/html/Carver.html')",
"_____no_output_____"
]
],
[
[
"You see how the individual elements of the URL – the _scheme_ (`\"http\"`), the _network location_ (`\"www.fuzzingbook.com\"`), or the path (`\"//html/Carver.html\"`) are all properly identified. Other elements (like `params`, `query`, or `fragment`) are empty, because they were not part of our input.",
"_____no_output_____"
],
[
"The interesting thing is that executing only `urlparse()` is orders of magnitude faster than running all of `webbrowser()`. Let us measure the factor:",
"_____no_output_____"
]
],
[
[
"runs = 1000\nwith Timer() as urlparse_timer:\n for i in range(runs):\n urlparse('https://www.fuzzingbook.com/html/Carver.html')\n\navg_urlparse_time = urlparse_timer.elapsed_time() / 1000\navg_urlparse_time",
"_____no_output_____"
]
],
[
[
"Compare this to the time required by the webbrowser",
"_____no_output_____"
]
],
[
[
"webbrowser_timer.elapsed_time()",
"_____no_output_____"
]
],
[
[
"The difference in time is huge:",
"_____no_output_____"
]
],
[
[
"webbrowser_timer.elapsed_time() / avg_urlparse_time",
"_____no_output_____"
]
],
[
[
"Hence, in the time it takes to run `webbrowser()` once, we can have _tens of thousands_ of executions of `urlparse()` – and this does not even take into account the time it takes the browser to render the downloaded HTML, to run the included scripts, and whatever else happens when a Web page is loaded. Hence, strategies that allow us to test at the _unit_ level are very promising as they can save lots of overhead.",
"_____no_output_____"
],
[
"## Carving Unit Tests\n\nTesting methods and functions at the unit level requires a very good understanding of the individual units to be tested as well as their interplay with other units. Setting up an appropriate infrastructure and writing unit tests by hand thus is demanding, yet rewarding. There is, however, an interesting alternative to writing unit tests by hand. The technique of _carving_ automatically _converts system tests into unit tests_ by means of recording and replaying function calls:\n\n1. During a system test (given or generated), we _record_ all calls into a function, including all arguments and other variables the function reads.\n2. From these, we synthesize a self-contained _unit test_ that reconstructs the function call with all arguments.\n3. This unit test can be executed (replayed) at any time with high efficiency.\n\nIn the remainder of this chapter, let us explore these steps.",
"_____no_output_____"
],
[
"## Recording Calls\n\nOur first challenge is to record function calls together with their arguments. (In the interest of simplicity, we restrict ourself to arguments, ignoring any global variables or other non-arguments that are read by the function.) To record calls and arguments, we use the mechanism [we introduced for coverage](Coverage.ipynb): By setting up a tracer function, we track all calls into individual functions, also saving their arguments. Just like `Coverage` objects, we want to use `Carver` objects to be able to be used in conjunction with the `with` statement, such that we can trace a particular code block:\n\n```python\nwith Carver() as carver:\n function_to_be_traced()\nc = carver.calls()\n```\n\nThe initial definition supports this construct:",
"_____no_output_____"
],
[
"\\todo{Get tracker from [dynamic invariants](DynamicInvariants.ipynb)}",
"_____no_output_____"
]
],
[
[
"import sys",
"_____no_output_____"
],
[
"class Carver:\n def __init__(self, log=False):\n self._log = log\n self.reset()\n\n def reset(self):\n self._calls = {}\n\n # Start of `with` block\n def __enter__(self):\n self.original_trace_function = sys.gettrace()\n sys.settrace(self.traceit)\n return self\n\n # End of `with` block\n def __exit__(self, exc_type, exc_value, tb):\n sys.settrace(self.original_trace_function)",
"_____no_output_____"
]
],
[
[
"The actual work takes place in the `traceit()` method, which records all calls in the `_calls` attribute. First, we define two helper functions:",
"_____no_output_____"
]
],
[
[
"import inspect",
"_____no_output_____"
],
[
"def get_qualified_name(code):\n \"\"\"Return the fully qualified name of the current function\"\"\"\n name = code.co_name\n module = inspect.getmodule(code)\n if module is not None:\n name = module.__name__ + \".\" + name\n return name",
"_____no_output_____"
],
[
"def get_arguments(frame):\n \"\"\"Return call arguments in the given frame\"\"\"\n # When called, all arguments are local variables\n local_variables = frame.f_locals.copy()\n arguments = [(var, frame.f_locals[var])\n for var in local_variables]\n arguments.reverse() # Want same order as call\n return arguments",
"_____no_output_____"
],
[
"class CallCarver(Carver):\n def add_call(self, function_name, arguments):\n \"\"\"Add given call to list of calls\"\"\"\n if function_name not in self._calls:\n self._calls[function_name] = []\n self._calls[function_name].append(arguments)\n\n # Tracking function: Record all calls and all args\n def traceit(self, frame, event, arg):\n if event != \"call\":\n return None\n\n code = frame.f_code\n function_name = code.co_name\n qualified_name = get_qualified_name(code)\n arguments = get_arguments(frame)\n\n self.add_call(function_name, arguments)\n if qualified_name != function_name:\n self.add_call(qualified_name, arguments)\n\n if self._log:\n print(simple_call_string(function_name, arguments))\n\n return None",
"_____no_output_____"
]
],
[
[
"Finally, we need some convenience functions to access the calls:",
"_____no_output_____"
]
],
[
[
"class CallCarver(CallCarver):\n def calls(self):\n \"\"\"Return a dictionary of all calls traced.\"\"\"\n return self._calls\n\n def arguments(self, function_name):\n \"\"\"Return a list of all arguments of the given function\n as (VAR, VALUE) pairs.\n Raises an exception if the function was not traced.\"\"\"\n return self._calls[function_name]\n\n def called_functions(self, qualified=False):\n \"\"\"Return all functions called.\"\"\"\n if qualified:\n return [function_name for function_name in self._calls.keys()\n if function_name.find('.') >= 0]\n else:\n return [function_name for function_name in self._calls.keys()\n if function_name.find('.') < 0]",
"_____no_output_____"
]
],
[
[
"### Recording my_sqrt()",
"_____no_output_____"
],
[
"Let's try out our new `Carver` class – first on a very simple function:",
"_____no_output_____"
]
],
[
[
"from Intro_Testing import my_sqrt",
"_____no_output_____"
],
[
"with CallCarver() as sqrt_carver:\n my_sqrt(2)\n my_sqrt(4)",
"_____no_output_____"
]
],
[
[
"We can retrieve all calls seen...",
"_____no_output_____"
]
],
[
[
"sqrt_carver.calls()",
"_____no_output_____"
],
[
"sqrt_carver.called_functions()",
"_____no_output_____"
]
],
[
[
"... as well as the arguments of a particular function:",
"_____no_output_____"
]
],
[
[
"sqrt_carver.arguments(\"my_sqrt\")",
"_____no_output_____"
]
],
[
[
"We define a convenience function for nicer printing of these lists:",
"_____no_output_____"
]
],
[
[
"def simple_call_string(function_name, argument_list):\n \"\"\"Return function_name(arg[0], arg[1], ...) as a string\"\"\"\n return function_name + \"(\" + \\\n \", \".join([var + \"=\" + repr(value)\n for (var, value) in argument_list]) + \")\"",
"_____no_output_____"
],
[
"for function_name in sqrt_carver.called_functions():\n for argument_list in sqrt_carver.arguments(function_name):\n print(simple_call_string(function_name, argument_list))",
"my_sqrt(x=2)\nmy_sqrt(x=4)\n__exit__(tb=None, exc_value=None, exc_type=None, self=<__main__.CallCarver object at 0x10eea5fd0>)\n"
]
],
[
[
"This is a syntax we can directly use to invoke `my_sqrt()` again:",
"_____no_output_____"
]
],
[
[
"eval(\"my_sqrt(x=2)\")",
"_____no_output_____"
]
],
[
[
"### Carving urlparse()",
"_____no_output_____"
],
[
"What happens if we apply this to `webbrowser()`?",
"_____no_output_____"
]
],
[
[
"with CallCarver() as webbrowser_carver:\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
]
],
[
[
"We see that retrieving a URL from the Web requires quite some functionality:",
"_____no_output_____"
]
],
[
[
"function_list = webbrowser_carver.called_functions(qualified=True)\nlen(function_list)",
"_____no_output_____"
],
[
"print(function_list[:50])",
"['requests.api.get', 'requests.api.request', 'requests.sessions.__init__', 'requests.utils.default_headers', 'requests.utils.default_user_agent', 'requests.structures.__init__', 'collections.abc.update', 'abc.__instancecheck__', 'requests.structures.__setitem__', 'requests.hooks.default_hooks', 'requests.hooks.<dictcomp>', 'requests.cookies.cookiejar_from_dict', 'http.cookiejar.__init__', 'threading.RLock', 'http.cookiejar.__iter__', 'requests.cookies.<listcomp>', 'http.cookiejar.deepvalues', 'http.cookiejar.vals_sorted_by_key', 'requests.adapters.__init__', 'urllib3.util.retry.__init__', 'urllib3.util.retry.<listcomp>', 'requests.adapters.init_poolmanager', 'urllib3.poolmanager.__init__', 'urllib3.request.__init__', 'urllib3._collections.__init__', 'requests.sessions.mount', 'requests.sessions.<listcomp>', 'requests.sessions.__enter__', 'requests.sessions.request', 'requests.models.__init__', 'requests.sessions.prepare_request', 'requests.cookies.merge_cookies', 'requests.cookies.update', 'requests.utils.get_netrc_auth', 'collections.abc.get', 'os.__getitem__', 'os.encode', 'requests.utils.<genexpr>', 'posixpath.expanduser', 'posixpath._get_sep', 'collections.abc.__contains__', 'os.decode', 'genericpath.exists', 'urllib.parse.urlparse', 'urllib.parse._coerce_args', 'urllib.parse.urlsplit', 'urllib.parse._splitnetloc', 'urllib.parse._checknetloc', 'urllib.parse._noop', 'netrc.__init__']\n"
]
],
[
[
"Among several other functions, we also have a call to `urlparse()`:",
"_____no_output_____"
]
],
[
[
"urlparse_argument_list = webbrowser_carver.arguments(\"urllib.parse.urlparse\")\nurlparse_argument_list",
"_____no_output_____"
]
],
[
[
"Again, we can convert this into a well-formatted call:",
"_____no_output_____"
]
],
[
[
"urlparse_call = simple_call_string(\"urlparse\", urlparse_argument_list[0])\nurlparse_call",
"_____no_output_____"
]
],
[
[
"Again, we can re-execute this call:",
"_____no_output_____"
]
],
[
[
"eval(urlparse_call)",
"_____no_output_____"
]
],
[
[
"We now have successfully carved the call to `urlparse()` out of the `webbrowser()` execution.",
"_____no_output_____"
],
[
"## Replaying Calls",
"_____no_output_____"
],
[
"Replaying calls in their entirety and in all generality is tricky, as there are several challenges to be addressed. These include:\n\n1. We need to be able to _access_ individual functions. If we access a function by name, the name must be in scope. If the name is not visible (for instance, because it is a name internal to the module), we must make it visible.\n\n2. Any _resources_ accessed outside of arguments must be recorded and reconstructed for replay as well. This can be difficult if variables refer to external resources such as files or network resources.\n\n3. _Complex objects_ must be reconstructed as well.",
"_____no_output_____"
],
[
"These constraints make carving hard or even impossible if the function to be tested interacts heavily with its environment. To illustrate these issues, consider the `email.parser.parse()` method that is invoked in `webbrowser()`:",
"_____no_output_____"
]
],
[
[
"email_parse_argument_list = webbrowser_carver.arguments(\"email.parser.parse\")",
"_____no_output_____"
]
],
[
[
"Calls to this method look like this:",
"_____no_output_____"
]
],
[
[
"email_parse_call = simple_call_string(\n \"email.parser.Parser.parse\",\n email_parse_argument_list[0])\nemail_parse_call",
"_____no_output_____"
]
],
[
[
"We see that `email.parser.Parser.parse()` is part of a `email.parser.Parser` object (`self`) and it gets a `StringIO` object (`fp`). Both are non-primitive values. How could we possibly reconstruct them?",
"_____no_output_____"
],
[
"### Serializing Objects\n\nThe answer to the problem of complex objects lies in creating a _persistent_ representation that can be _reconstructed_ at later points in time. This process is known as _serialization_; in Python, it is also known as _pickling_. The `pickle` module provides means to create a serialized representation of an object. Let us apply this on the `email.parser.Parser` object we just found:",
"_____no_output_____"
]
],
[
[
"import pickle",
"_____no_output_____"
],
[
"email_parse_argument_list",
"_____no_output_____"
],
[
"parser_object = email_parse_argument_list[0][2][1]\nparser_object",
"_____no_output_____"
],
[
"pickled = pickle.dumps(parser_object)\npickled",
"_____no_output_____"
]
],
[
[
"From this string representing the serialized `email.parser.Parser` object, we can recreate the Parser object at any time:",
"_____no_output_____"
]
],
[
[
"unpickled_parser_object = pickle.loads(pickled)\nunpickled_parser_object",
"_____no_output_____"
]
],
[
[
"The serialization mechanism allows us to produce a representation for all objects passed as parameters (assuming they can be pickled, that is). We can now extend the `simple_call_string()` function such that it automatically pickles objects. Additionally, we set it up such that if the first parameter is named `self` (i.e., it is a class method), we make it a method of the `self` object.",
"_____no_output_____"
]
],
[
[
"def call_value(value):\n value_as_string = repr(value)\n if value_as_string.find('<') >= 0:\n # Complex object\n value_as_string = \"pickle.loads(\" + repr(pickle.dumps(value)) + \")\"\n return value_as_string",
"_____no_output_____"
],
[
"def call_string(function_name, argument_list):\n \"\"\"Return function_name(arg[0], arg[1], ...) as a string, pickling complex objects\"\"\"\n if len(argument_list) > 0:\n (first_var, first_value) = argument_list[0]\n if first_var == \"self\":\n # Make this a method call\n method_name = function_name.split(\".\")[-1]\n function_name = call_value(first_value) + \".\" + method_name\n argument_list = argument_list[1:]\n\n return function_name + \"(\" + \\\n \", \".join([var + \"=\" + call_value(value)\n for (var, value) in argument_list]) + \")\"",
"_____no_output_____"
]
],
[
[
"Let us apply the extended `call_string()` method to create a call for `email.parser.parse()`, including pickled objects:",
"_____no_output_____"
]
],
[
[
"call = call_string(\"email.parser.Parser.parse\", email_parse_argument_list[0])\nprint(call)",
"email.parser.Parser.parse(headersonly=False, fp=pickle.loads(b'\\x80\\x04\\x95\\x8e\\x01\\x00\\x00\\x00\\x00\\x00\\x00\\x8c\\x03_io\\x94\\x8c\\x08StringIO\\x94\\x93\\x94)\\x81\\x94(Xe\\x01\\x00\\x00Content-Encoding: gzip\\r\\nAccept-Ranges: bytes\\r\\nAge: 422658\\r\\nCache-Control: max-age=604800\\r\\nContent-Type: text/html; charset=UTF-8\\r\\nDate: Tue, 11 Jan 2022 09:24:47 GMT\\r\\nEtag: \"3147526947\"\\r\\nExpires: Tue, 18 Jan 2022 09:24:47 GMT\\r\\nLast-Modified: Thu, 17 Oct 2019 07:18:26 GMT\\r\\nServer: ECS (dcb/7FA3)\\r\\nVary: Accept-Encoding\\r\\nX-Cache: HIT\\r\\nContent-Length: 648\\r\\n\\r\\n\\x94\\x8c\\x01\\n\\x94Me\\x01Nt\\x94b.'), self=pickle.loads(b'\\x80\\x04\\x95w\\x00\\x00\\x00\\x00\\x00\\x00\\x00\\x8c\\x0cemail.parser\\x94\\x8c\\x06Parser\\x94\\x93\\x94)\\x81\\x94}\\x94(\\x8c\\x06_class\\x94\\x8c\\x0bhttp.client\\x94\\x8c\\x0bHTTPMessage\\x94\\x93\\x94\\x8c\\x06policy\\x94\\x8c\\x11email._policybase\\x94\\x8c\\x08Compat32\\x94\\x93\\x94)\\x81\\x94ub.'))\n"
]
],
[
[
"With this call involving the pickled object, we can now re-run the original call and obtain a valid result:",
"_____no_output_____"
]
],
[
[
"import email",
"_____no_output_____"
],
[
"eval(call)",
"_____no_output_____"
]
],
[
[
"### All Calls\n\nSo far, we have seen only one call of `webbrowser()`. How many of the calls within `webbrowser()` can we actually carve and replay? Let us try this out and compute the numbers.",
"_____no_output_____"
]
],
[
[
"import traceback",
"_____no_output_____"
],
[
"import enum\nimport socket",
"_____no_output_____"
],
[
"all_functions = set(webbrowser_carver.called_functions(qualified=True))\ncall_success = set()\nrun_success = set()",
"_____no_output_____"
],
[
"exceptions_seen = set()\n\nfor function_name in webbrowser_carver.called_functions(qualified=True):\n for argument_list in webbrowser_carver.arguments(function_name):\n try:\n call = call_string(function_name, argument_list)\n call_success.add(function_name)\n\n result = eval(call)\n run_success.add(function_name)\n\n except Exception as exc:\n exceptions_seen.add(repr(exc))\n # print(\"->\", call, file=sys.stderr)\n # traceback.print_exc()\n # print(\"\", file=sys.stderr)\n continue",
"_____no_output_____"
],
[
"print(\"%d/%d calls (%.2f%%) successfully created and %d/%d calls (%.2f%%) successfully ran\" % (\n len(call_success), len(all_functions), len(\n call_success) * 100 / len(all_functions),\n len(run_success), len(all_functions), len(run_success) * 100 / len(all_functions)))",
"264/325 calls (81.23%) successfully created and 54/325 calls (16.62%) successfully ran\n"
]
],
[
[
"About a quarter of the calls succeed. Let us take a look into some of the error messages we get:",
"_____no_output_____"
]
],
[
[
"for i in range(10):\n print(list(exceptions_seen)[i])",
"TypeError(\"_coerce_args() got an unexpected keyword argument 'args'\")\nNameError(\"name 'HTTPHeaderDict' is not defined\")\nResponseNotReady('Idle')\nNameError(\"name 'ParseResult' is not defined\")\nAttributeError(\"module 'email.message' has no attribute 'get_all'\")\nSyntaxError('invalid syntax', ('<string>', 1, 1138, 'pickle.loads(b\\'\\\\x80\\\\x04\\\\x95b\\\\x02\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x8c\\\\x0bhttp.client\\\\x94\\\\x8c\\\\x0bHTTPMessage\\\\x94\\\\x93\\\\x94)\\\\x81\\\\x94}\\\\x94(\\\\x8c\\\\x06policy\\\\x94\\\\x8c\\\\x11email._policybase\\\\x94\\\\x8c\\\\x08Compat32\\\\x94\\\\x93\\\\x94)\\\\x81\\\\x94\\\\x8c\\\\x08_headers\\\\x94]\\\\x94(\\\\x8c\\\\x10Content-Encoding\\\\x94\\\\x8c\\\\x04gzip\\\\x94\\\\x86\\\\x94\\\\x8c\\\\rAccept-Ranges\\\\x94\\\\x8c\\\\x05bytes\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x03Age\\\\x94\\\\x8c\\\\x06422658\\\\x94\\\\x86\\\\x94\\\\x8c\\\\rCache-Control\\\\x94\\\\x8c\\\\x0emax-age=604800\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x0cContent-Type\\\\x94\\\\x8c\\\\x18text/html; charset=UTF-8\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x04Date\\\\x94\\\\x8c\\\\x1dTue, 11 Jan 2022 09:24:47 GMT\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x04Etag\\\\x94\\\\x8c\\\\x0c\"3147526947\"\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x07Expires\\\\x94\\\\x8c\\\\x1dTue, 18 Jan 2022 09:24:47 GMT\\\\x94\\\\x86\\\\x94\\\\x8c\\\\rLast-Modified\\\\x94\\\\x8c\\\\x1dThu, 17 Oct 2019 07:18:26 GMT\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x06Server\\\\x94\\\\x8c\\\\x0eECS (dcb/7FA3)\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x04Vary\\\\x94\\\\x8c\\\\x0fAccept-Encoding\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x07X-Cache\\\\x94\\\\x8c\\\\x03HIT\\\\x94\\\\x86\\\\x94\\\\x8c\\\\x0eContent-Length\\\\x94\\\\x8c\\\\x03648\\\\x94\\\\x86\\\\x94e\\\\x8c\\\\t_unixfrom\\\\x94N\\\\x8c\\\\x08_payload\\\\x94\\\\x8c\\\\x00\\\\x94\\\\x8c\\\\x08_charset\\\\x94N\\\\x8c\\\\x08preamble\\\\x94N\\\\x8c\\\\x08epilogue\\\\x94N\\\\x8c\\\\x07defects\\\\x94]\\\\x94\\\\x8c\\\\r_default_type\\\\x94\\\\x8c\\\\ntext/plain\\\\x94ub.\\').<listcomp>(.0=pickle.loads(b\\'\\\\x80\\\\x04\\\\x95\\\\x1b\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x8c\\\\x08builtins\\\\x94\\\\x8c\\\\x04iter\\\\x94\\\\x93\\\\x94]\\\\x94\\\\x85\\\\x94R\\\\x94.\\'))'))\nSyntaxError('invalid syntax', ('<string>', 1, 16, \"requests.utils.<genexpr>(f='.netrc', .0=pickle.loads(b'\\\\x80\\\\x04\\\\x950\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x00\\\\x8c\\\\x08builtins\\\\x94\\\\x8c\\\\x04iter\\\\x94\\\\x93\\\\x94\\\\x8c\\\\x06.netrc\\\\x94\\\\x8c\\\\x06_netrc\\\\x94\\\\x86\\\\x94\\\\x85\\\\x94R\\\\x94K\\\\x01b.'))\"))\nNameError(\"name 'Compat32' is not defined\")\nNameError(\"name 'codecs' is not defined\")\nValueError('I/O operation on closed socket.')\n"
]
],
[
[
"We see that:\n\n* **A large majority of calls could be converted into call strings.** If this is not the case, this is mostly due to having unserialized objects being passed.\n* **About a quarter of the calls could be executed.** The error messages for the failing runs are varied; the most frequent being that some internal name is invoked that is not in scope.",
"_____no_output_____"
],
[
"Our carving mechanism should be taken with a grain of salt: We still do not cover the situation where external variables and values (such as global variables) are being accessed, and the serialization mechanism cannot recreate external resources. Still, if the function of interest falls among those that _can_ be carved and replayed, we can very effectively re-run its calls with their original arguments.",
"_____no_output_____"
],
[
"## Mining API Grammars from Carved Calls\n\nSo far, we have used carved calls to replay exactly the same invocations as originally encountered. However, we can also _mutate_ carved calls to effectively fuzz APIs with previously recorded arguments.\n\nThe general idea is as follows:\n\n1. First, we record all calls of a specific function from a given execution of the program.\n2. Second, we create a grammar that incorporates all these calls, with separate rules for each argument and alternatives for each value found; this allows us to produce calls that arbitrarily _recombine_ these arguments.\n\nLet us explore these steps in the following sections.",
"_____no_output_____"
],
[
"### From Calls to Grammars\n\nLet us start with an example. The `power(x, y)` function returns $x^y$; it is but a wrapper around the equivalent `math.pow()` function. (Since `power()` is defined in Python, we can trace it – in contrast to `math.pow()`, which is implemented in C.)",
"_____no_output_____"
]
],
[
[
"import math",
"_____no_output_____"
],
[
"def power(x, y):\n return math.pow(x, y)",
"_____no_output_____"
]
],
[
[
"Let us invoke `power()` while recording its arguments:",
"_____no_output_____"
]
],
[
[
"with CallCarver() as power_carver:\n z = power(1, 2)\n z = power(3, 4)",
"_____no_output_____"
],
[
"power_carver.arguments(\"power\")",
"_____no_output_____"
]
],
[
[
"From this list of recorded arguments, we could now create a grammar for the `power()` call, with `x` and `y` expanding into the values seen:",
"_____no_output_____"
]
],
[
[
"from Grammars import START_SYMBOL, is_valid_grammar, new_symbol\nfrom Grammars import extend_grammar, Grammar",
"_____no_output_____"
],
[
"POWER_GRAMMAR: Grammar = {\n \"<start>\": [\"power(<x>, <y>)\"],\n \"<x>\": [\"1\", \"3\"],\n \"<y>\": [\"2\", \"4\"]\n}\n\nassert is_valid_grammar(POWER_GRAMMAR)",
"_____no_output_____"
]
],
[
[
"When fuzzing with this grammar, we then get arbitrary combinations of `x` and `y`; aiming for coverage will ensure that all values are actually tested at least once:",
"_____no_output_____"
]
],
[
[
"from GrammarCoverageFuzzer import GrammarCoverageFuzzer",
"_____no_output_____"
],
[
"power_fuzzer = GrammarCoverageFuzzer(POWER_GRAMMAR)\n[power_fuzzer.fuzz() for i in range(5)]",
"_____no_output_____"
]
],
[
[
"What we need is a method to automatically convert the arguments as seen in `power_carver` to the grammar as seen in `POWER_GRAMMAR`. This is what we define in the next section.",
"_____no_output_____"
],
[
"### A Grammar Miner for Calls\n\nWe introduce a class `CallGrammarMiner`, which, given a `Carver`, automatically produces a grammar from the calls seen. To initialize, we pass the carver object:",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner:\n def __init__(self, carver, log=False):\n self.carver = carver\n self.log = log",
"_____no_output_____"
]
],
[
[
"#### Initial Grammar\n\nThe initial grammar produces a single call. The possible `<call>` expansions are to be constructed later:",
"_____no_output_____"
]
],
[
[
"import copy ",
"_____no_output_____"
],
[
"class CallGrammarMiner(CallGrammarMiner):\n CALL_SYMBOL = \"<call>\"\n\n def initial_grammar(self):\n return extend_grammar(\n {START_SYMBOL: [self.CALL_SYMBOL],\n self.CALL_SYMBOL: []\n })",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\ninitial_grammar = m.initial_grammar()\ninitial_grammar",
"_____no_output_____"
]
],
[
[
"#### A Grammar from Arguments\n\nLet us start by creating a grammar from a list of arguments. The method `mine_arguments_grammar()` creates a grammar for the arguments seen during carving, such as these:",
"_____no_output_____"
]
],
[
[
"arguments = power_carver.arguments(\"power\")\narguments",
"_____no_output_____"
]
],
[
[
"The `mine_arguments_grammar()` method iterates through the variables seen and creates a mapping `variables` of variable names to a set of values seen (as strings, going through `call_value()`). In a second step, it then creates a grammar with a rule for each variable name, expanding into the values seen.",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner(CallGrammarMiner):\n def var_symbol(self, function_name, var, grammar):\n return new_symbol(grammar, \"<\" + function_name + \"-\" + var + \">\")\n\n def mine_arguments_grammar(self, function_name, arguments, grammar):\n var_grammar = {}\n\n variables = {}\n for argument_list in arguments:\n for (var, value) in argument_list:\n value_string = call_value(value)\n if self.log:\n print(var, \"=\", value_string)\n\n if value_string.find(\"<\") >= 0:\n var_grammar[\"<langle>\"] = [\"<\"]\n value_string = value_string.replace(\"<\", \"<langle>\")\n\n if var not in variables:\n variables[var] = set()\n variables[var].add(value_string)\n\n var_symbols = []\n for var in variables:\n var_symbol = self.var_symbol(function_name, var, grammar)\n var_symbols.append(var_symbol)\n var_grammar[var_symbol] = list(variables[var])\n\n return var_grammar, var_symbols",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\nvar_grammar, var_symbols = m.mine_arguments_grammar(\n \"power\", arguments, initial_grammar)",
"_____no_output_____"
],
[
"var_grammar",
"_____no_output_____"
]
],
[
[
"The additional return value `var_symbols` is a list of argument symbols in the call:",
"_____no_output_____"
]
],
[
[
"var_symbols",
"_____no_output_____"
]
],
[
[
"#### A Grammar from Calls\n\nTo get the grammar for a single function (`mine_function_grammar()`), we add a call to the function:",
"_____no_output_____"
]
],
[
[
"class CallGrammarMiner(CallGrammarMiner):\n def function_symbol(self, function_name, grammar):\n return new_symbol(grammar, \"<\" + function_name + \">\")\n\n def mine_function_grammar(self, function_name, grammar):\n arguments = self.carver.arguments(function_name)\n\n if self.log:\n print(function_name, arguments)\n\n var_grammar, var_symbols = self.mine_arguments_grammar(\n function_name, arguments, grammar)\n\n function_grammar = var_grammar\n function_symbol = self.function_symbol(function_name, grammar)\n\n if len(var_symbols) > 0 and var_symbols[0].find(\"-self\") >= 0:\n # Method call\n function_grammar[function_symbol] = [\n var_symbols[0] + \".\" + function_name + \"(\" + \", \".join(var_symbols[1:]) + \")\"]\n else:\n function_grammar[function_symbol] = [\n function_name + \"(\" + \", \".join(var_symbols) + \")\"]\n\n if self.log:\n print(function_symbol, \"::=\", function_grammar[function_symbol])\n\n return function_grammar, function_symbol",
"_____no_output_____"
],
[
"m = CallGrammarMiner(power_carver)\nfunction_grammar, function_symbol = m.mine_function_grammar(\n \"power\", initial_grammar)\nfunction_grammar",
"_____no_output_____"
]
],
[
[
"The additionally returned `function_symbol` holds the name of the function call just added:",
"_____no_output_____"
]
],
[
[
"function_symbol",
"_____no_output_____"
]
],
[
[
"#### A Grammar from all Calls\n\nLet us now repeat the above for all function calls seen during carving. To this end, we simply iterate over all function calls seen:",
"_____no_output_____"
]
],
[
[
"power_carver.called_functions()",
"_____no_output_____"
],
[
"class CallGrammarMiner(CallGrammarMiner):\n def mine_call_grammar(self, function_list=None, qualified=False):\n grammar = self.initial_grammar()\n fn_list = function_list\n if function_list is None:\n fn_list = self.carver.called_functions(qualified=qualified)\n\n for function_name in fn_list:\n if function_list is None and (function_name.startswith(\"_\") or function_name.startswith(\"<\")):\n continue # Internal function\n\n # Ignore errors with mined functions\n try:\n function_grammar, function_symbol = self.mine_function_grammar(\n function_name, grammar)\n except:\n if function_list is not None:\n raise\n\n if function_symbol not in grammar[self.CALL_SYMBOL]:\n grammar[self.CALL_SYMBOL].append(function_symbol)\n grammar.update(function_grammar)\n\n assert is_valid_grammar(grammar)\n return grammar",
"_____no_output_____"
]
],
[
[
"The method `mine_call_grammar()` is the one that clients can and should use – first for mining...",
"_____no_output_____"
]
],
[
[
"m = CallGrammarMiner(power_carver)\npower_grammar = m.mine_call_grammar()\npower_grammar",
"_____no_output_____"
]
],
[
[
"...and then for fuzzing:",
"_____no_output_____"
]
],
[
[
"power_fuzzer = GrammarCoverageFuzzer(power_grammar)\n[power_fuzzer.fuzz() for i in range(5)]",
"_____no_output_____"
]
],
[
[
"With this, we have successfully extracted a grammar from a recorded execution; in contrast to \"simple\" carving, our grammar allows us to _recombine_ arguments and thus to fuzz at the API level.",
"_____no_output_____"
],
[
"## Fuzzing Web Functions\n\nLet us now apply our grammar miner on a larger API – the `urlparse()` function we already encountered during carving.",
"_____no_output_____"
]
],
[
[
"with CallCarver() as webbrowser_carver:\n webbrowser(\"https://www.fuzzingbook.org\")\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
]
],
[
[
"We can mine a grammar from the calls encountered:",
"_____no_output_____"
]
],
[
[
"m = CallGrammarMiner(webbrowser_carver)\nwebbrowser_grammar = m.mine_call_grammar()",
"_____no_output_____"
]
],
[
[
"This is a rather large grammar:",
"_____no_output_____"
]
],
[
[
"call_list = webbrowser_grammar['<call>']\nlen(call_list)",
"_____no_output_____"
],
[
"print(call_list[:20])",
"['<webbrowser>', '<default_headers>', '<default_user_agent>', '<update>', '<default_hooks>', '<cookiejar_from_dict>', '<RLock>', '<deepvalues>', '<vals_sorted_by_key>', '<init_poolmanager>', '<mount>', '<prepare_request>', '<merge_cookies>', '<get_netrc_auth>', '<encode>', '<expanduser>', '<decode>', '<exists>', '<urlparse>', '<urlsplit>']\n"
]
],
[
[
"Here's the rule for the `urlsplit()` function:",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit>\"]",
"_____no_output_____"
]
],
[
[
"Here are the arguments. Note that although we only passed `http://www.fuzzingbook.org` as a parameter, we also see the `https:` variant. That is because opening the `http:` URL automatically redirects to the `https:` URL, which is then also processed by `urlsplit()`.",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit-url>\"]",
"_____no_output_____"
]
],
[
[
"There also is some variation in the `scheme` argument:",
"_____no_output_____"
]
],
[
[
"webbrowser_grammar[\"<urlsplit-scheme>\"]",
"_____no_output_____"
]
],
[
[
"If we now apply a fuzzer on these rules, we systematically cover all variations of arguments seen, including, of course, combinations not seen during carving. Again, we are fuzzing at the API level here.",
"_____no_output_____"
]
],
[
[
"urlsplit_fuzzer = GrammarCoverageFuzzer(\n webbrowser_grammar, start_symbol=\"<urlsplit>\")\nfor i in range(5):\n print(urlsplit_fuzzer.fuzz())",
"urlsplit(True, '', 'https://www.fuzzingbook.org')\nurlsplit(True, '', 'https://www.fuzzingbook.org/')\nurlsplit(True, '', 'http://www.example.com')\nurlsplit(True, '', 'http://www.example.com/')\nurlsplit(True, '', 'http://www.example.com')\n"
]
],
[
[
"Just as seen with carving, running tests at the API level is orders of magnitude faster than executing system tests. Hence, this calls for means to fuzz at the method level:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import urlsplit",
"_____no_output_____"
],
[
"from Timer import Timer",
"_____no_output_____"
],
[
"with Timer() as urlsplit_timer:\n urlsplit('http://www.fuzzingbook.org/', 'http', True)\nurlsplit_timer.elapsed_time()",
"_____no_output_____"
],
[
"with Timer() as webbrowser_timer:\n webbrowser(\"http://www.fuzzingbook.org\")\nwebbrowser_timer.elapsed_time()",
"_____no_output_____"
],
[
"webbrowser_timer.elapsed_time() / urlsplit_timer.elapsed_time()",
"_____no_output_____"
]
],
[
[
"But then again, the caveats encountered during carving apply, notably the requirement to recreate the original function environment. If we also alter or recombine arguments, we get the additional risk of _violating an implicit precondition_ – that is, invoking a function with arguments the function was never designed for. Such _false alarms_, resulting from incorrect invocations rather than incorrect implementations, must then be identified (typically manually) and wed out (for instance, by altering or constraining the grammar). The huge speed gains at the API level, however, may well justify this additional investment.",
"_____no_output_____"
],
[
"## Synopsis\n\nThis chapter provides means to _record and replay function calls_ during a system test. Since individual function calls are much faster than a whole system run, such \"carving\" mechanisms have the potential to run tests much faster.",
"_____no_output_____"
],
[
"### Recording Calls\n\nThe `CallCarver` class records all calls occurring while it is active. It is used in conjunction with a `with` clause:",
"_____no_output_____"
]
],
[
[
"with CallCarver() as carver:\n y = my_sqrt(2)\n y = my_sqrt(4)",
"_____no_output_____"
]
],
[
[
"After execution, `called_functions()` lists the names of functions encountered:",
"_____no_output_____"
]
],
[
[
"carver.called_functions()",
"_____no_output_____"
]
],
[
[
"The `arguments()` method lists the arguments recorded for a function. This is a mapping of the function name to a list of lists of arguments; each argument is a pair (parameter name, value).",
"_____no_output_____"
]
],
[
[
"carver.arguments('my_sqrt')",
"_____no_output_____"
]
],
[
[
"Complex arguments are properly serialized, such that they can be easily restored.",
"_____no_output_____"
],
[
"### Synthesizing Calls\n\nWhile such recorded arguments already could be turned into arguments and calls, a much nicer alternative is to create a _grammar_ for recorded calls. This allows to synthesize arbitrary _combinations_ of arguments, and also offers a base for further customization of calls.",
"_____no_output_____"
],
[
"The `CallGrammarMiner` class turns a list of carved executions into a grammar.",
"_____no_output_____"
]
],
[
[
"my_sqrt_miner = CallGrammarMiner(carver)\nmy_sqrt_grammar = my_sqrt_miner.mine_call_grammar()\nmy_sqrt_grammar",
"_____no_output_____"
]
],
[
[
"This grammar can be used to synthesize calls.",
"_____no_output_____"
]
],
[
[
"fuzzer = GrammarCoverageFuzzer(my_sqrt_grammar)\nfuzzer.fuzz()",
"_____no_output_____"
]
],
[
[
"These calls can be executed in isolation, effectively extracting unit tests from system tests:",
"_____no_output_____"
]
],
[
[
"eval(fuzzer.fuzz())",
"_____no_output_____"
]
],
[
[
"## Lessons Learned\n\n* _Carving_ allows for effective replay of function calls recorded during a system test.\n* A function call can be _orders of magnitude faster_ than a system invocation.\n* _Serialization_ allows to create persistent representations of complex objects.\n* Functions that heavily interact with their environment and/or access external resources are difficult to carve.\n* From carved calls, one can produce API grammars that arbitrarily combine carved arguments.",
"_____no_output_____"
],
[
"## Next Steps\n\nIn the next chapter, we will discuss [how to reduce failure-inducing inputs](Reducer.ipynb).",
"_____no_output_____"
],
[
"## Background\n\nCarving was invented by Elbaum et al. \\cite{Elbaum2006} and originally implemented for Java. In this chapter, we follow several of their design choices (including recording and serializing method arguments only).\n\nThe combination of carving and fuzzing at the API level is described in \\cite{Kampmann2018}.",
"_____no_output_____"
],
[
"## Exercises\n\n### Exercise 1: Carving for Regression Testing\n\nSo far, during carving, we only have looked into reproducing _calls_, but not into actually checking the _results_ of these calls. This is important for _regression testing_ – i.e. checking whether a change to code does not impede existing functionality. We can build this by recording not only _calls_, but also _return values_ – and then later compare whether the same calls result in the same values. This may not work on all occasions; values that depend on time, randomness, or other external factors may be different. Still, for functionality that abstracts from these details, checking that nothing has changed is an important part of testing.",
"_____no_output_____"
],
[
"Our aim is to design a class `ResultCarver` that extends `CallCarver` by recording both calls and return values.\n\nIn a first step, create a `traceit()` method that also tracks return values by extending the `traceit()` method. The `traceit()` event type is `\"return\"` and the `arg` parameter is the returned value. Here is a prototype that only prints out the returned values:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(CallCarver):\n def traceit(self, frame, event, arg):\n if event == \"return\":\n if self._log:\n print(\"Result:\", arg)\n\n super().traceit(frame, event, arg)\n # Need to return traceit function such that it is invoked for return\n # events\n return self.traceit",
"_____no_output_____"
],
[
"with ResultCarver(log=True) as result_carver:\n my_sqrt(2)",
"my_sqrt(x=2)\nResult: 1.414213562373095\n__exit__(tb=None, exc_value=None, exc_type=None, self=<__main__.ResultCarver object at 0x12a4938b0>)\n"
]
],
[
[
"#### Part 1: Store function results\n\nExtend the above code such that results are _stored_ in a way that associates them with the currently returning function (or method). To this end, you need to keep track of the _current stack of called functions_.",
"_____no_output_____"
],
[
"**Solution.** Here's a solution, building on the above:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(CallCarver):\n def reset(self):\n super().reset()\n self._call_stack = []\n self._results = {}\n\n def add_result(self, function_name, arguments, result):\n key = simple_call_string(function_name, arguments)\n self._results[key] = result\n\n def traceit(self, frame, event, arg):\n if event == \"call\":\n code = frame.f_code\n function_name = code.co_name\n qualified_name = get_qualified_name(code)\n self._call_stack.append(\n (function_name, qualified_name, get_arguments(frame)))\n\n if event == \"return\":\n result = arg\n (function_name, qualified_name, arguments) = self._call_stack.pop()\n self.add_result(function_name, arguments, result)\n if function_name != qualified_name:\n self.add_result(qualified_name, arguments, result)\n if self._log:\n print(\n simple_call_string(\n function_name,\n arguments),\n \"=\",\n result)\n\n # Keep on processing current calls\n super().traceit(frame, event, arg)\n\n # Need to return traceit function such that it is invoked for return\n # events\n return self.traceit",
"_____no_output_____"
],
[
"with ResultCarver(log=True) as result_carver:\n my_sqrt(2)\nresult_carver._results",
"my_sqrt(x=2)\nmy_sqrt(x=2) = 1.414213562373095\n__exit__(tb=None, exc_value=None, exc_type=None, self=<__main__.ResultCarver object at 0x12a4639d0>)\n"
]
],
[
[
"#### Part 2: Access results\n\n Give it a method `result()` that returns the value recorded for that particular function name and result:\n\n```python\nclass ResultCarver(CallCarver):\n def result(self, function_name, argument):\n \"\"\"Returns the result recorded for function_name(argument\"\"\"\n```",
"_____no_output_____"
],
[
"**Solution.** This is mostly done in the code for part 1:",
"_____no_output_____"
]
],
[
[
"class ResultCarver(ResultCarver):\n def result(self, function_name, argument):\n key = simple_call_string(function_name, arguments)\n return self._results[key]",
"_____no_output_____"
]
],
[
[
"#### Part 3: Produce assertions\n\nFor the functions called during `webbrowser()` execution, create a set of _assertions_ that check whether the result returned is still the same. Test this for `urllib.parse.urlparse()` and `urllib.parse.urlsplit()`.",
"_____no_output_____"
],
[
"**Solution.** Not too hard now:",
"_____no_output_____"
]
],
[
[
"with ResultCarver() as webbrowser_result_carver:\n webbrowser(\"http://www.example.com\")",
"_____no_output_____"
],
[
"for function_name in [\"urllib.parse.urlparse\", \"urllib.parse.urlsplit\"]:\n for arguments in webbrowser_result_carver.arguments(function_name):\n try:\n call = call_string(function_name, arguments)\n result = webbrowser_result_carver.result(function_name, arguments)\n print(\"assert\", call, \"==\", call_value(result))\n except Exception:\n continue",
"assert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com') == ParseResult(scheme='http', netloc='www.example.com', path='', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlparse(allow_fragments=True, scheme='', url='http://www.example.com/') == ParseResult(scheme='http', netloc='www.example.com', path='/', params='', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com') == SplitResult(scheme='http', netloc='www.example.com', path='', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\nassert urllib.parse.urlsplit(allow_fragments=True, scheme='', url='http://www.example.com/') == SplitResult(scheme='http', netloc='www.example.com', path='/', query='', fragment='')\n"
]
],
[
[
"We can run these assertions:",
"_____no_output_____"
]
],
[
[
"from urllib.parse import SplitResult, ParseResult, urlparse, urlsplit",
"_____no_output_____"
],
[
"assert urlparse(\n url='http://www.example.com',\n scheme='',\n allow_fragments=True) == ParseResult(\n scheme='http',\n netloc='www.example.com',\n path='',\n params='',\n query='',\n fragment='')\nassert urlsplit(\n url='http://www.example.com',\n scheme='',\n allow_fragments=True) == SplitResult(\n scheme='http',\n netloc='www.example.com',\n path='',\n query='',\n fragment='')",
"_____no_output_____"
]
],
[
[
"We can now add these carved tests to a _regression test suite_ which would be run after every change to ensure that the functionality of `urlparse()` and `urlsplit()` is not changed.",
"_____no_output_____"
],
[
"### Exercise 2: Abstracting Arguments\n\nWhen mining an API grammar from executions, set up an abstraction scheme to widen the range of arguments to be used during testing. If the values for an argument, all conform to some type `T`. abstract it into `<T>`. For instance, if calls to `foo(1)`, `foo(2)`, `foo(3)` have been seen, the grammar should abstract its calls into `foo(<int>)`, with `<int>` being appropriately defined.\n\nDo this for a number of common types: integers, positive numbers, floating-point numbers, host names, URLs, mail addresses, and more.",
"_____no_output_____"
],
[
"**Solution.** Left to the reader.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec6ee7b9a820f5599d13bc52207e5ce8ea703f44 | 28,843 | ipynb | Jupyter Notebook | notebooks/11_Evaluation_walk_through_V1.0.ipynb | surabhid0223/ADS_COVID19_Surabhid0223 | a8fbbb1c49ffa788d364da2cdbb87e1fc71d4df4 | [
"FTL"
]
| null | null | null | notebooks/11_Evaluation_walk_through_V1.0.ipynb | surabhid0223/ADS_COVID19_Surabhid0223 | a8fbbb1c49ffa788d364da2cdbb87e1fc71d4df4 | [
"FTL"
]
| null | null | null | notebooks/11_Evaluation_walk_through_V1.0.ipynb | surabhid0223/ADS_COVID19_Surabhid0223 | a8fbbb1c49ffa788d364da2cdbb87e1fc71d4df4 | [
"FTL"
]
| null | null | null | 37.073265 | 226 | 0.495788 | [
[
[
"# One Run full walktrhough",
"_____no_output_____"
],
[
"- Do the full walk through on the large data set\n- Refactor the source code and bring it to individual scripts\n- Ensure a full run with one click",
"_____no_output_____"
]
],
[
[
"## check some parameters\n## depending where you launch your notebook, the relative path might not work\n## you should start the notebook server from your base path\n## when opening the notebook, typically your path will be ../ads_covid-19/notebooks\nimport os\nif os.path.split(os.getcwd())[-1]=='notebooks':\n os.chdir(\"../\")\n\n'Your base path is at: '+os.path.split(os.getcwd())[-1]",
"_____no_output_____"
]
],
[
[
"# 1. Update all data",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\data\\get_data.py\nimport subprocess\nimport os\nfrom subprocess import Popen, PIPE\n\nimport pandas as pd\nimport numpy as np\n\nfrom datetime import datetime\n\nimport requests\nimport json\n\ndef get_johns_hopkins():\n ''' Get data by a git pull request, the source code has to be pulled first\n Result is stored in the predifined csv structure\n '''\n git_pull = subprocess.Popen( \"git pull\" ,\n cwd = os.path.dirname( r'C:\\Users\\SurabhiD\\ads_covid_19\\data\\raw\\new-covid' ),\n #cwd = os.path.dirname( '../Users/SurabhiD/ads_covid_19/data/raw/new-covid' ),\n shell = True,\n stdout = subprocess.PIPE,\n stderr = subprocess.PIPE )\n (out, error) = git_pull.communicate()\n\n\n print(\"Error : \" + str(error))\n print(\"out : \" + str(out))\n\n\ndef get_current_data_germany():\n ''' Get current data from germany, attention API endpoint not too stable\n Result data frame is stored as pd.DataFrame\n\n '''\n # 16 states\n #data=requests.get('https://services7.arcgis.com/mOBPykOjAyBO2ZKk/arcgis/rest/services/Coronaf%C3%A4lle_in_den_Bundesl%C3%A4ndern/FeatureServer/0/query?where=1%3D1&outFields=*&outSR=4326&f=json')\n\n # 400 regions / Landkreise\n data=requests.get('https://services7.arcgis.com/mOBPykOjAyBO2ZKk/arcgis/rest/services/RKI_Landkreisdaten/FeatureServer/0/query?where=1%3D1&outFields=*&outSR=4326&f=json')\n\n json_object=json.loads(data.content)\n full_list=[]\n for pos,each_dict in enumerate (json_object['features'][:]):\n full_list.append(each_dict['attributes'])\n\n pd_full_list=pd.DataFrame(full_list)\n pd_full_list.to_csv(r'C:\\Users\\SurabhiD\\ads_covid_19\\data\\raw\\NPGEO\\GER_state_data.csv',sep=';')\n print('Number of regions rows: '+str(pd_full_list.shape[0]))\n\nif __name__ == '__main__':\n get_johns_hopkins()\n get_current_data_germany()",
"Error : b'fatal: not a git repository (or any of the parent directories): .git\\n'\nout : b''\nNumber of regions rows: 412\n"
]
],
[
[
"# 2. Process Pipeline",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\data\\process_JH_data.py\nimport pandas as pd\nimport numpy as np\n\nfrom datetime import datetime\n\n\ndef store_relational_JH_data():\n ''' Transformes the COVID data in a relational data set\n\n '''\n\n data_path= r'C:\\Users\\SurabhiD\\ads_covid_19\\data\\raw\\COVID-19\\csse_covid_19_data\\csse_covid_19_time_series\\time_series_covid19_confirmed_global.csv'\n pd_raw=pd.read_csv(data_path)\n\n pd_data_base=pd_raw.rename(columns={'Country/Region':'country',\n 'Province/State':'state'})\n\n pd_data_base['state']=pd_data_base['state'].fillna('no')\n\n pd_data_base=pd_data_base.drop(['Lat','Long'],axis=1)\n test_pd = pd_data_base.set_index(['state','country']).T\n test_pd.stack(level=[0,1])\n pd_relational = test_pd.stack(level=[0,1]).reset_index()\n pd_relational = pd_relational.rename(columns={'level_0': 'date',0:'confirmed'})\n\n # pd_relational_model=pd_data_base.set_index(['state','country']) \\\n # .T \\\n # .stack(level=[0,1]) \\\n # .reset_index() \\\n # .rename(columns={'level_0':'date',\n # 0:'confirmed'},\n # )\n pd_relational['date'] = pd.to_datetime(pd_relational['date'])\n #pd_relational_model['date']=pd_relational_model.date.astype('datetime64[ns]')\n\n pd_relational.to_csv(r'C:\\Users\\SurabhiD\\ads_covid_19\\data\\processed\\COVID_relational_confirmed.csv',sep=';',index=False)\n print(' Number of rows stored: '+str(pd_relational.shape[0]))\n\nif __name__ == '__main__':\n\n store_relational_JH_data()\n",
" Number of rows stored: 46816\n"
]
],
[
[
"# 3. Filter and Doubling Rate Calculation",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\features\\build_features.py\nimport numpy as np\nimport pandas as pd\nfrom sklearn import linear_model\nreg = linear_model.LinearRegression(fit_intercept = True)\nfrom scipy import signal\n\ndef get_doubling_time_via_regression(in_array):\n\n y = np.array(in_array)\n X = np.arange(-1,2).reshape(-1,1)\n\n assert len(in_array) == 3\n reg.fit(X, y)\n intercept = reg.intercept_\n slope = reg.coef_\n\n return intercept/slope\n\ndef savgol_filter(df_input,column='confirmed',window=5):\n ''' Savgol Filter which can be used in groupby apply function\n it ensures that the data structure is kept'''\n window=5,\n degree=1\n df_result=df_input\n\n filter_in=df_input[column].fillna(0) # attention with the neutral element here\n\n result=signal.savgol_filter(np.array(filter_in),\n 5, # window size used for filtering\n 1)\n df_result[column+'_filtered']=result\n return df_result\n\ndef rolling_reg(df_input,col='confirmed'):\n ''' input has to be a data frame'''\n ''' return is single series (mandatory for group by apply)'''\n days_back=3\n result=df_input[col].rolling(\n window=days_back,\n min_periods=days_back).apply(get_doubling_time_via_regression)\n return result\n\ndef calc_filtered_data(df_input, filter_on = 'confirmed'):\n '''\n Calculates savgol filter and returns merged DataFrame\n '''\n must_contain = set(['state', 'country', filter_on])\n assert must_contain.issubset(set(df_input.columns))\n\n pd_filtered_result = df_input[['state','country',filter_on]].groupby(['state','country']).apply(savgol_filter).reset_index()\n df_output = pd.merge(df_input, pd_filtered_result[['index', filter_on +'_filtered']], on = ['index'], how = 'left')\n\n return df_output\n\ndef calc_doubling_rate(df_input, filter_on = 'confirmed'):\n '''\n Calculates approximated doubling rate and returns merged DataFrame\n '''\n must_contain = set(['state', 'country', filter_on])\n assert must_contain.issubset(set(df_input.columns))\n\n pd_DR_result = df_input[['state','country', filter_on]].groupby(['state','country']).apply(rolling_reg, filter_on).reset_index()\n pd_DR_result = pd_DR_result.rename(columns = {'level_2':'index', filter_on : filter_on+'_DR'})\n\n df_output = pd.merge(df_input, pd_DR_result[['index', filter_on+'_DR']], on = ['index'], how = 'left')\n return df_output\n\n\nif __name__ == '__main__':\n test_data = np.array([2,4,6])\n result = get_doubling_time_via_regression(test_data)\n print('The test slope is: '+str(result))\n\n pd_JH_data=pd.read_csv(r'C:\\Users\\SurabhiD\\ads_covid_19\\data\\processed\\COVID_relational_confirmed.csv',sep=';',parse_dates=[0])\n pd_JH_data = pd_JH_data.sort_values('date',ascending=True).reset_index().copy()\n\n pd_result_large = calc_filtered_data(pd_JH_data)\n pd_result_large = calc_doubling_rate(pd_result_large)\n pd_result_large = calc_doubling_rate(pd_result_large, 'confirmed_filtered')\n print(pd_result_large.head())",
"The test slope is: [2.]\n index date state country confirmed confirmed_filtered \\\n0 0 2020-01-22 Alberta Canada 0.0 0.0 \n1 169 2020-01-22 no Korea, South 1.0 0.0 \n2 170 2020-01-22 no Kosovo 0.0 -4.8 \n3 171 2020-01-22 no Kuwait 0.0 0.0 \n4 172 2020-01-22 no Kyrgyzstan 0.0 0.0 \n\n confirmed_DR confirmed_filtered_DR \n0 NaN NaN \n1 NaN NaN \n2 NaN NaN \n3 NaN NaN \n4 NaN NaN \n"
]
],
[
[
"# 4. Visual Board",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\visualization\\visualize.py\nimport pandas as pd\nimport numpy as np\n\nimport dash\ndash.__version__\nimport dash_core_components as dcc\nimport dash_html_components as html\nfrom dash.dependencies import Input, Output,State\n\nimport plotly.graph_objects as go\n\nimport os\nprint(os.getcwd())\ndf_input_large=pd.read_csv('data/processed/COVID_final_set.csv',sep=';')\n\n\nfig = go.Figure()\n\napp = dash.Dash()\napp.layout = html.Div([\n\n dcc.Markdown('''\n # Applied Data Science on COVID-19 data\n Goal of the project is to teach data science by applying a cross industry standard process,\n it covers the full walkthrough of: automated data gathering, data transformations,\n filtering and machine learning to approximating the doubling time, and\n (static) deployment of responsive dashboard.\n '''),\n\n dcc.Markdown('''\n ## Multi-Select Country for visualization\n '''),\n\n\n dcc.Dropdown(\n id='country_drop_down',\n options=[ {'label': each,'value':each} for each in df_input_large['country'].unique()],\n value=['US', 'Germany','Italy'], # which are pre-selected\n multi=True\n ),\n\n dcc.Markdown('''\n ## Select Timeline of confirmed COVID-19 cases or the approximated doubling time\n '''),\n\n\n dcc.Dropdown(\n id='doubling_time',\n options=[\n {'label': 'Timeline Confirmed ', 'value': 'confirmed'},\n {'label': 'Timeline Confirmed Filtered', 'value': 'confirmed_filtered'},\n {'label': 'Timeline Doubling Rate', 'value': 'confirmed_DR'},\n {'label': 'Timeline Doubling Rate Filtered', 'value': 'confirmed_filtered_DR'},\n ],\n value='confirmed',\n multi=False\n ),\n\n dcc.Graph(figure=fig, id='main_window_slope')\n])\n\n\n\[email protected](\n Output('main_window_slope', 'figure'),\n [Input('country_drop_down', 'value'),\n Input('doubling_time', 'value')])\ndef update_figure(country_list,show_doubling):\n\n\n if 'doubling_rate' in show_doubling:\n my_yaxis={'type':\"log\",\n 'title':'Approximated doubling rate over 3 days (larger numbers are better #stayathome)'\n }\n else:\n my_yaxis={'type':\"log\",\n 'title':'Confirmed infected people (source johns hopkins csse, log-scale)'\n }\n\n\n traces = []\n for each in country_list:\n\n df_plot=df_input_large[df_input_large['country']==each]\n\n if show_doubling=='doubling_rate_filtered':\n df_plot=df_plot[['state','country','confirmed','confirmed_filtered','confirmed_DR','confirmed_filtered_DR','date']].groupby(['country','date']).agg(np.mean).reset_index()\n else:\n df_plot=df_plot[['state','country','confirmed','confirmed_filtered','confirmed_DR','confirmed_filtered_DR','date']].groupby(['country','date']).agg(np.sum).reset_index()\n #print(show_doubling)\n\n\n traces.append(dict(x=df_plot.date,\n y=df_plot[show_doubling],\n mode='markers+lines',\n opacity=0.9,\n name=each\n )\n )\n\n return {\n 'data': traces,\n 'layout': dict (\n width=1280,\n height=720,\n\n xaxis={'title':'Timeline',\n 'tickangle':-45,\n 'nticks':20,\n 'tickfont':dict(size=14,color=\"#7f7f7f\"),\n },\n\n yaxis=my_yaxis\n )\n }\n\nif __name__ == '__main__':\n\n app.run_server(debug=True, use_reloader=False)",
"C:\\Users\\SurabhiD\\ads_covid_19\nDash is running on http://127.0.0.1:8050/\n\n Warning: This is a development server. Do not use app.run_server\n in production, use a production WSGI server like gunicorn instead.\n\n * Serving Flask app \"__main__\" (lazy loading)\n * Environment: production\n WARNING: This is a development server. Do not use it in a production deployment.\n Use a production WSGI server instead.\n * Debug mode: on\n"
]
],
[
[
"# 5. SIR Calculation",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\models\\SIR_Calculation.py\nimport pandas as pd\nimport numpy as np\nfrom datetime import datetime\nfrom scipy import optimize\nfrom scipy import integrate\n\ndef SIR_model_t(SIR, t, beta, gamma):\n S,I,R=SIR\n dS_dt = -beta*I*S/N0\n dI_dt = beta*I*S/N0 - gamma*I\n dR_dt = gamma*I\n\n return dS_dt, dI_dt, dR_dt\n\ndef fit_odeint(x, beta, gamma):\n return integrate.odeint(SIR_model_t, (S0, I0, R0), x, args=(beta, gamma))[:,1]\n\n\nif __name__ == '__main__':\n\n df_analyse=pd.read_csv('../data/raw/COVID-19/csse_covid_19_data/SIR_raw.csv',sep=';')\n\n df_analyse.sort_values('Date',ascending=True)\n N0 = 1000000\n df_data = df_analyse[35:] ## We will consider data from 35th day, which is 26th Feb 2020\n t = np.arange(df_data.shape[0])\n\n for country in df_data.columns[1:]:\n ydata = np.array(df_data[df_data[country]>0][country])\n t = np.arange(len(ydata))\n I0=ydata[0]\n S0=N0-I0\n R0=0\n popt=[0.4,0.1]\n fit_odeint(t, *popt)\n popt, pcov = optimize.curve_fit(fit_odeint, t, ydata, maxfev=5000)\n perr = np.sqrt(np.diag(pcov))\n fitted=fit_odeint(t, *popt)\n f_padded = np.concatenate((np.zeros(df_data.shape[0]-len(fitted)) ,fitted)) #to make dimentions equal\n df_data[country + '_fitted'] = f_padded\n df_data.to_csv(\"../data/processed/SIR_calculated.csv\", sep = ';', index=False)",
"C:\\Users\\SurabhiD\\anaconda3\\lib\\site-packages\\scipy\\integrate\\odepack.py:247: ODEintWarning: Excess work done on this call (perhaps wrong Dfun type). Run with full_output = 1 to get quantitative information.\n warnings.warn(warning_msg, ODEintWarning)\n<ipython-input-1-4f7b256a7d10>:10: RuntimeWarning: overflow encountered in double_scalars\n dS_dt = -beta*I*S/N0\n<ipython-input-1-4f7b256a7d10>:11: RuntimeWarning: overflow encountered in double_scalars\n dI_dt = beta*I*S/N0 - gamma*I\nC:\\Users\\SurabhiD\\anaconda3\\lib\\site-packages\\scipy\\integrate\\odepack.py:247: ODEintWarning: Illegal input detected (internal error). Run with full_output = 1 to get quantitative information.\n warnings.warn(warning_msg, ODEintWarning)\n<ipython-input-1-4f7b256a7d10>:41: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy\n df_data[country + '_fitted'] = f_padded\n<ipython-input-1-4f7b256a7d10>:11: RuntimeWarning: invalid value encountered in double_scalars\n dI_dt = beta*I*S/N0 - gamma*I\n<ipython-input-1-4f7b256a7d10>:12: RuntimeWarning: overflow encountered in double_scalars\n dR_dt = gamma*I\n"
]
],
[
[
"# 6. SIR Visual Dashboard",
"_____no_output_____"
]
],
[
[
"# %load C:\\Users\\SurabhiD\\ads_covid_19\\src\\models\\SIR_visualize_Board.py\nimport numpy as np\nimport pandas as pd\nimport dash\ndash.__version__\nimport dash_core_components as dcc\nimport dash_html_components as html\nfrom dash.dependencies import Input, Output,State\nimport random\nimport plotly.graph_objects as go\n\n\ndf_dash = pd.read_csv('../data/processed/SIR_calculated.csv',sep=';')\n\ncolor_list = []\n\nfor i in range(int((df_dash.shape[1]-1)/2)):\n random_color = '#%02x%02x%02x' % (random.randint(0, 255),random.randint(0, 255), random.randint(0, 255))\n color_list.append(random_color)\n\ncolors = {\n 'background': '#def2ff',\n 'text': '#000000'\n}\n\nfig = go.Figure()\nexternal_stylesheets = ['https://codepen.io/chriddyp/pen/bWLwgP.css']\napp = dash.Dash(external_stylesheets=external_stylesheets)\n\napp.layout = html.Div(style={'backgroundColor': colors['background'],}, children = [\n\n dcc.Markdown('''\n # Enterprise Data Science Task 2: SIR Dashboard\n ''',\n style={\n \"border\":\"2px silver solid\",\n 'textAlign': 'center',\n 'color': colors['text']\n }),\n\n dcc.Markdown('''\n ## Plot shows actual number of infected people and simulated number of infected people\\\n derived from SIR model for different countries.\n ''',\n style = {\n \"border\":\"2px silver solid\",\n 'backgroundColor': colors['background'],\n 'position' : 'fixed',\n 'left' : 7,\n 'top' : 83,\n 'width' : 500,\n 'height' : 1200,\n 'textAlign': 'left',\n 'border':'2px silver solid',\n 'color': colors['text']\n\n }),\n\n dcc.Markdown('''\n ### Select the country below:\n ''',\n style={\n 'textAlign': 'left',\n 'color': colors['text'],\n 'position':'fixed',\n 'top':350,\n 'left': 7,\n 'width' :500,\n }),\n\n\n dcc.Dropdown(\n id='country_drop_down',\n options=[ {'label': each,'value':each} for each in df_dash.columns[1:187]],\n value=['Germany'], # Which is pre-selected\n multi=True,\n style={\n 'position': 'fixed',\n 'left' : 7,\n 'top' : 425,\n 'textAlign': 'left',\n 'color': '#000000',\n 'background-color': '#e1f0fa',\n 'font-size' : 'large',\n 'height': 100,\n 'width': 500,\n }),\n\n\n dcc.Graph(\n figure=fig,\n id='SIR',\n style = {\n \"border\":\"2px silver solid\",\n 'backgroundColor': colors['background'],\n 'height' : 1200,\n 'textAlign': 'center',\n 'align' : 'right',\n 'position' : 'fixed',\n 'left' : 507,\n 'top' : 83,\n 'width' : '80%',\n 'color': colors['text']\n\n })\n])\n\n\n\[email protected](\n Output('SIR', 'figure'),\n [Input('country_drop_down', 'value')])\ndef update_figure(country_list):\n\n\n traces = []\n for pos, each in enumerate(country_list):\n\n traces.append(dict(x=df_dash.Date,\n y=df_dash[each],\n mode='lines',\n opacity=0.9,\n name=each,\n line = dict(color = color_list[pos])\n )\n )\n traces.append(dict(x=df_dash.Date,\n y=df_dash[each+'_fitted'],\n mode='markers+lines',\n opacity=0.9,\n name=each+'_simulated',\n line = dict(color = color_list[pos])\n )\n )\n\n return {\n 'data': traces,\n 'layout': dict (\n width=1800,\n height=1200,\n plot_bgcolor = colors['text'],\n paper_bgcolor = colors['background'],\n xaxis={'title':'Timeline',\n 'tickangle':-25,\n 'nticks':20,\n 'tickfont':dict(size=18,color=colors['text']),\n 'titlefont': dict(size=22, color=colors['text']),\n },\n\n yaxis={'type':\"log\", 'title':'Number of infected people (log-scale)',\n 'tickfont':dict(size=18,color=colors['text']),\n 'titlefont':dict(size=22, color=colors['text']),\n },\n title={'text': \"Real and Simulated Number of Infected People\",\n 'y':0.95,\n 'x':0.5,\n 'xanchor': 'center',\n 'yanchor': 'top',\n 'font': dict(size=25, color=colors['text'])\n }\n\n )\n\n}\n\nif __name__ == '__main__':\n\n app.run_server(debug=True, use_reloader=False)",
"Dash is running on http://127.0.0.1:8050/\n\n Warning: This is a development server. Do not use app.run_server\n in production, use a production WSGI server like gunicorn instead.\n\n * Serving Flask app \"__main__\" (lazy loading)\n * Environment: production\n WARNING: This is a development server. Do not use it in a production deployment.\n Use a production WSGI server instead.\n * Debug mode: on\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6eee98faafa739ac5e5ac40ec9b29296066eee | 930,851 | ipynb | Jupyter Notebook | cs109b_hw6.ipynb | ccc255/2020-CS109B | 7a337930e21370451c45e935e2ca42a167a8b6bb | [
"MIT"
]
| null | null | null | cs109b_hw6.ipynb | ccc255/2020-CS109B | 7a337930e21370451c45e935e2ca42a167a8b6bb | [
"MIT"
]
| null | null | null | cs109b_hw6.ipynb | ccc255/2020-CS109B | 7a337930e21370451c45e935e2ca42a167a8b6bb | [
"MIT"
]
| null | null | null | 353.935741 | 209,776 | 0.918547 | [
[
[
"# <img style=\"float: left; padding-right: 10px; width: 45px\" src=\"https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/iacs.png\"> CS109B Data Science 2: Advanced Topics in Data Science \n## Homework 6 - RNNs\n\n\n\n**Harvard University**<br/>\n**Fall 2020**<br/>\n**Instructors**: Mark Glickman, Pavlos Protopapas, & Chris Tanner\n\n\n<hr style=\"height:2pt\">",
"_____no_output_____"
]
],
[
[
"#RUN THIS CELL \nimport requests\nfrom IPython.core.display import HTML\nstyles = requests.get(\"https://raw.githubusercontent.com/Harvard-IACS/2018-CS109A/master/content/styles/cs109.css\").text\nHTML(styles)",
"_____no_output_____"
]
],
[
[
"### INSTRUCTIONS\n\n<span style=\"color:red\">**Model training can be very slow; start doing this HW early**</span>\n\n- To submit your assignment follow the instructions given in Canvas.\n\n- This homework can be submitted in pairs.\n\n- If you submit individually but you have worked with someone, please include the name of your **one** partner below.\n- Please restart the kernel and run the entire notebook again before you submit.\n\n**Names of person you have worked with goes here:**\n<br><BR>",
"_____no_output_____"
],
[
"<div class=\"theme\"> Overview: Named Entity Recognition Challenge</div>",
"_____no_output_____"
],
[
"Named entity recognition (NER) seeks to locate and classify named entities present in unstructured text into predefined categories such as organizations, locations, expressions of times, names of persons, etc. This technique is often used in real use cases such as classifying content for news providers, efficient search algorithms over large corpora and content-based recommendation systems. \n\nThis represents an interesting \"many-to-many\" problem, allowing us to experiment with recurrent architectures and compare their performances against other models.\n",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\n\nfrom sklearn.metrics import f1_score, confusion_matrix\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.decomposition import PCA\n\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nplt.style.use(\"ggplot\")",
"_____no_output_____"
],
[
"import tensorflow as tf\n\nfrom tensorflow.keras.preprocessing.sequence import pad_sequences\nfrom tensorflow.keras.utils import to_categorical\nfrom tensorflow.keras import backend\n\nfrom tensorflow.keras import Model, Sequential\nfrom tensorflow.keras.models import model_from_json\nfrom tensorflow.keras.layers import Input, SimpleRNN, Embedding, Dense, TimeDistributed, GRU, \\\n Dropout, Bidirectional, Conv1D, BatchNormalization\n\nprint(tf.keras.__version__)\nprint(tf.__version__)",
"2.2.4-tf\n2.0.0\n"
],
[
"# Set seed for repeatable results\nnp.random.seed(123)\ntf.random.set_seed(456)",
"_____no_output_____"
]
],
[
[
"<div class=\"theme\"> Part 1: Data </div>\nRead `HW6_data.csv` into a pandas dataframe using the provided code below.",
"_____no_output_____"
]
],
[
[
"# Given code\npath_dataset = './data/HW6_data.csv'\ndata = pd.read_csv(path_dataset,\n encoding=\"latin1\")\ndata = data.fillna(method=\"ffill\")\ndata.head(15)",
"_____no_output_____"
],
[
"data['Sentence'] = data['Sentence #'].apply(lambda x: int(x.split(': ')[1])-1) # add a column for the sentence as numeric\n",
"_____no_output_____"
]
],
[
[
"As you can see, we have a dataset with sentences (```Sentence #``` column), each composed of words (```Word``` column) with part-of-speech tagging (```POS``` tagging) and inside–outside–beginning (IOB) named entity tags (```Tag``` column) attached. ```POS``` will not be used for this homework. We will predict ```Tag``` using only the words themselves.\n\nEssential info about entities:\n* geo = Geographical Entity\n* org = Organization\n* per = Person\n* gpe = Geopolitical Entity\n* tim = Time indicator\n* art = Artifact\n* eve = Event\n* nat = Natural Phenomenon\n\nIOB prefix:\n* B: beginning of named entity\n* I: inside of named entity\n* O: outside of named entity\n",
"_____no_output_____"
],
[
"<div class='exercise'><b> Question 1: Data [20 points total]</b></div>\n\n**1.1** Create a list of unique words found in the 'Word' column and sort it in alphabetic order. Then append the special word \"ENDPAD\" to the end of the list, and assign it to the variable ```words```. Store the length of this list as ```n_words```. **Print your results for `n_words`**\n\n**1.2** Create a list of unique tags and sort it in alphabetic order. Then append the special word \"PAD\" to the end of the list, and assign it to the variable ```tags```. Store the length of this list as ```n_tags```. **Print your results for `n_tags`**\n\n**1.3** Process the data into a list of sentences where each sentence is a list of (word, tag) tuples. Here is an example of how the first sentence in the list should look:\n\n[('Thousands', 'O'),\n ('of', 'O'),\n ('demonstrators', 'O'),\n ('have', 'O'),\n ('marched', 'O'),\n ('through', 'O'),\n ('London', 'B-geo'),\n ('to', 'O'),\n ('protest', 'O'),\n ('the', 'O'),\n ('war', 'O'),\n ('in', 'O'),\n ('Iraq', 'B-geo'),\n ('and', 'O'),\n ('demand', 'O'),\n ('the', 'O'),\n ('withdrawal', 'O'),\n ('of', 'O'),\n ('British', 'B-gpe'),\n ('troops', 'O'),\n ('from', 'O'),\n ('that', 'O'),\n ('country', 'O'),\n ('.', 'O')]\n \n**1.4** Find out the number of words in the longest sentence, and store it to variable ```max_len```. **Print your results for `max_len`.**\n\n**1.5** It's now time to convert the sentences data in a suitable format for the RNNs training/evaluation procedures. Create a ```word2idx``` dictionary mapping distinct words from the dataset into distinct integers. Also create a ```idx2word``` dictionary.\n\n**1.6** Prepare the predictors matrix ```X```, as a list of lists, where each inner list is a sequence of words mapped into integers accordly to the ```word2idx``` dictionary. \n\n**1.7** Apply the keras ```pad_sequences``` function to standardize the predictors. You should retrieve a matrix with all padded sentences and length equal to ```max_len``` previously computed. The dimensionality should therefore be equal to ```[# of sentences, max_len]```. Run the provided cell to print your results. Your ```X[i]``` now should be something similar to this:\n\n`[ 8193 27727 31033 33289 22577 33464 23723 16665 33464 31142 31319 28267\n 27700 33246 28646 16052 21 16915 17349 7924 32879 32985 18238 23555\n 24 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178]`\n \n**1.8** Create a ```tag2idx``` dictionary mapping distinct named entity tags from the dataset into distinct integers. Also create a ```idx2tag``` dictionary.\n\n**1.9** Prepare targets matrix ```Y```, as a list of lists,where each inner list is a sequence of tags mapped into integers accordly to the ```tag2idx``` dictionary.\n\n**1.10** apply the keras ```pad_sequences``` function to standardize the targets. Inject the ```PAD``` tag for the padding words. You should retrieve a matrix with all padded sentences'tags and length equal to ```max_length``` previously computed. \n\n**1.11** Use the ```to_categorical``` keras function to one-hot encode the tags. Now your ```Y``` should have dimension ```[# of sentences, max_len, n_tags]```. Run the provided cell to print your results.\n\n**1.12** Split the dataset into train and test sets (test 10%).",
"_____no_output_____"
],
[
"## Answers",
"_____no_output_____"
],
[
"**1.1** Create a list of unique words found in the 'Word' column and sort it in alphabetic order. Then append the special word \"ENDPAD\" to the end of the list, and assign it to the variable ```words```. Store the length of this list as ```n_words```. **Print your results for `n_words`**",
"_____no_output_____"
]
],
[
[
"words = sorted(data.Word.unique())\nwords.append('ENDPAD')\nn_words = len(words)",
"_____no_output_____"
],
[
"# Run this cell to show your results for n_words\nprint(n_words)",
"35179\n"
]
],
[
[
"**1.2** Create a list of unique tags and sort it in alphabetic order. Then append the special word \"PAD\" to the end of the list, and assign it to the variable ```tags```. Store the length of this list as ```n_tags```. **Print your results for `n_tags`**",
"_____no_output_____"
]
],
[
[
"tags = sorted(data.Tag.unique())\ntags.append('PAD')\nn_tags = len(tags)\n",
"_____no_output_____"
],
[
"# Run this cell to show your results for n_tags\nprint(n_tags)",
"18\n"
]
],
[
[
"**1.3** Process the data into a list of sentences where each sentence is a list of (word, tag) tuples. Here is an example of how the first sentence in the list should look:\n\n[('Thousands', 'O'),\n ('of', 'O'),\n ('demonstrators', 'O'),\n ('have', 'O'),\n ('marched', 'O'),\n ('through', 'O'),\n ('London', 'B-geo'),\n ('to', 'O'),\n ('protest', 'O'),\n ('the', 'O'),\n ('war', 'O'),\n ('in', 'O'),\n ('Iraq', 'B-geo'),\n ('and', 'O'),\n ('demand', 'O'),\n ('the', 'O'),\n ('withdrawal', 'O'),\n ('of', 'O'),\n ('British', 'B-gpe'),\n ('troops', 'O'),\n ('from', 'O'),\n ('that', 'O'),\n ('country', 'O'),\n ('.', 'O')]\n ",
"_____no_output_____"
]
],
[
[
"sentence_tuples = [list(zip(sentence[1]['Word'], sentence[1]['Tag'])) for sentence in data.groupby('Sentence')]\nprint(sentence_tuples[0])",
"[('Thousands', 'O'), ('of', 'O'), ('demonstrators', 'O'), ('have', 'O'), ('marched', 'O'), ('through', 'O'), ('London', 'B-geo'), ('to', 'O'), ('protest', 'O'), ('the', 'O'), ('war', 'O'), ('in', 'O'), ('Iraq', 'B-geo'), ('and', 'O'), ('demand', 'O'), ('the', 'O'), ('withdrawal', 'O'), ('of', 'O'), ('British', 'B-gpe'), ('troops', 'O'), ('from', 'O'), ('that', 'O'), ('country', 'O'), ('.', 'O')]\n"
]
],
[
[
"**1.4** Find out the number of words in the longest sentence, and store it to variable ```max_len```. **Print your results for `max_len`.**",
"_____no_output_____"
]
],
[
[
"max_len = data.groupby('Sentence #').size().sort_values().iloc[-1]",
"_____no_output_____"
],
[
"# Run this cell to show your results for max_len\nprint(max_len)",
"104\n"
]
],
[
[
"**1.5** It's now time to convert the sentences data in a suitable format for the RNNs training/evaluation procedures. Create a ```word2idx``` dictionary mapping distinct words from the dataset into distinct integers. Also create a ```idx2word``` dictionary.",
"_____no_output_____"
]
],
[
[
"word2idx = dict(zip(words, np.arange(len(words))))\nidx2word = {a:b for b,a in word2idx.items()}\n",
"_____no_output_____"
]
],
[
[
"**1.6** Prepare the predictors matrix ```X```, as a list of lists, where each inner list is a sequence of words mapped into integers accordly to the ```word2idx``` dictionary. ",
"_____no_output_____"
]
],
[
[
"X = [[word2idx[word[0]] for word in sentence_tuple] for sentence_tuple in sentence_tuples]",
"_____no_output_____"
]
],
[
[
"**1.7** Apply the keras ```pad_sequences``` function to standardize the predictors. You should retrieve a matrix with all padded sentences and length equal to ```max_len``` previously computed. The dimensionality should therefore be equal to ```[# of sentences, max_len]```. Run the provided cell to print your results. Your ```X[i]``` now should be something similar to this:\n\n`[ 8193 27727 31033 33289 22577 33464 23723 16665 33464 31142 31319 28267\n 27700 33246 28646 16052 21 16915 17349 7924 32879 32985 18238 23555\n 24 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178]`",
"_____no_output_____"
]
],
[
[
"Z = [[1,2], [2,1,1,2]]\npad_sequences(Z, padding = 'post', value = 14)",
"_____no_output_____"
],
[
"X = pad_sequences(X, padding = 'post', value = word2idx['ENDPAD'])",
"_____no_output_____"
],
[
"# Run this cell to show your results #\nprint(\"The index of word 'Harvard' is: {}\\n\".format(word2idx[\"Harvard\"]))\nprint(\"Sentence 1: {}\\n\".format(X[1]))\nprint(X.shape)",
"The index of word 'Harvard' is: 7506\n\nSentence 1: [ 6283 27700 31967 25619 24853 33246 19981 25517 33246 29399 34878 19044\n 18095 34971 32712 31830 17742 1 4114 11464 11631 14985 1 17364\n 1 14484 33246 3881 24 1 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178 35178\n 35178 35178 35178 35178 35178 35178 35178 35178]\n\n(47959, 104)\n"
]
],
[
[
"**1.8** Create a ```tag2idx``` dictionary mapping distinct named entity tags from the dataset into distinct integers. Also create a ```idx2tag``` dictionary.",
"_____no_output_____"
]
],
[
[
"tag2idx = dict(zip(tags, np.arange(len(tags))))\nidx2tag = {a:b for b,a in tag2idx.items()}\n",
"_____no_output_____"
]
],
[
[
"**1.9** Prepare targets matrix ```Y```, as a list of lists,where each inner list is a sequence of tags mapped into integers accordly to the ```tag2idx``` dictionary.",
"_____no_output_____"
]
],
[
[
"Y = [[tag2idx[tag[1]] for tag in sentence_tuple] for sentence_tuple in sentence_tuples]",
"_____no_output_____"
]
],
[
[
"**1.10** apply the keras ```pad_sequences``` function to standardize the targets. Inject the ```PAD``` tag for the padding words. You should retrieve a matrix with all padded sentences'tags and length equal to ```max_length``` previously computed. ",
"_____no_output_____"
]
],
[
[
"Y = pad_sequences(Y, padding = 'post', value = tag2idx['PAD'])",
"_____no_output_____"
]
],
[
[
"**1.11** Use the ```to_categorical``` keras function to one-hot encode the tags. Now your ```Y``` should have dimension ```[# of sentences, max_len, n_tags]```. Run the provided cell to print your results.",
"_____no_output_____"
]
],
[
[
"Y = to_categorical(Y)",
"_____no_output_____"
],
[
"# Run this cell to show your results #\nprint(\"The index of tag 'B-gpe' is: {}\\n\".format(tag2idx[\"B-gpe\"]))\nprint(\"The tag of the last word in Sentence 1: {}\\n\".format(Y[0][-1]))\nprint(np.array(Y).shape)",
"The index of tag 'B-gpe' is: 3\n\nThe tag of the last word in Sentence 1: [0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 0. 1.]\n\n(47959, 104, 18)\n"
]
],
[
[
"**1.12** Split the dataset into train and test sets (test 10%).",
"_____no_output_____"
]
],
[
[
"np.random.seed(109)\nn_sentences = len(data.Sentence.unique())\ntest_indices = np.sort(np.random.choice(np.arange(n_sentences), size = n_sentences//10, replace = False))\ntrain_indices = np.sort([i for i in np.arange(n_sentences) if i not in test_indices])",
"_____no_output_____"
],
[
"X_tr = X[train_indices]\nX_te = X[test_indices]\ny_tr = Y[train_indices]\ny_te = Y[test_indices]",
"_____no_output_____"
]
],
[
[
"## Part 2: Modelling\n\nAfter preparing the train and test sets, we are ready to build five models: \n* frequency-based baseline \n* vanilla feedforward neural network\n* recurrent neural network\n* gated recurrent neural network\n* bidirectional gated recurrent neural network\n\nMore details are given about architecture in each model's section. The input/output dimensionalities will be the same for all models:\n* input: ```[# of sentences, max_len]```\n* output: ```[# of sentences, max_len, n_tags]```\n\nFollow the information in each model's section to set up the architecture of each model. And the end of each training, use the given ```store_model``` function to store the weights and architectures in the ```./models``` path for later testing;```load_keras_model()``` is also provided to you\n\nA further ```plot_training_history``` helper function is given in case you need to check the training history.\n",
"_____no_output_____"
]
],
[
[
"# Store model\ndef store_keras_model(model, model_name):\n model_json = model.to_json() # serialize model to JSON\n with open(\"./models/{}.json\".format(model_name), \"w\") as json_file:\n json_file.write(model_json)\n model.save_weights(\"./models/{}.h5\".format(model_name)) # serialize weights to HDF5\n print(\"Saved model to disk\")\n \n# Plot history\ndef plot_training_history(history):\n loss = history.history['loss']\n val_loss = history.history['val_loss']\n epochs = range(1,len(loss)+1)\n\n plt.figure()\n plt.plot(epochs, loss, 'bo', label='Training loss')\n plt.plot(epochs, val_loss, 'b', label='Validation loss')\n plt.title('Training and validation loss')\n plt.xlabel('epoch')\n plt.legend()\n plt.show()",
"_____no_output_____"
],
[
"# Load model \ndef load_keras_model(model_name):\n # Load json and create model\n json_file = open('./models/{}.json'.format(model_name), 'r')\n loaded_model_json = json_file.read()\n json_file.close()\n model = tf.keras.models.model_from_json(loaded_model_json)\n # Load weights into new model\n model.load_weights(\"./models/{}.h5\".format(model_name))\n return model",
"_____no_output_____"
]
],
[
[
"<div class='exercise'><b>Question 2: Models [40 points total]</b></div>\n\n**2.1** **Model 1: Baseline Model**\n\nPredict the tag of a word simply with the most frequently-seen named entity tag of this word from the training set.\n\ne.g. word \"Apple\" appears 10 times in the training set; 7 times it was tagged as \"Corporate\" and 3 times it was tagged as \"Fruit\". If we encounter the word \"Apple\" in the test set, we predict it as \"Corporate\".\n\n**Create an np.array ```baseline``` of length [n_words]**\nwhere the ith element ```baseline[i]``` is the index of the most commonly seen named entity tag of word i summarised from training set. (e.g. [16, 16, 16, ..., 0, 16, 16])\n\n\n**2.2** **Model 2: Vanilla Feed Forward Neural Network**\n\nThis model is provided for you. Please pay attention to the architecture of this neural network, especially the input/output dimensionalities and the Embedding layer.\n\n\n**2.2a** Explain what the embedding layer is and why we need it here.\n\n**2.2b** Explain why the Param # of Embedding layer is 1758950 (as shown in `print(model.summary())`).\n\n**2.3** **Model 3: RNN**\n\nSet up a simple RNN model by stacking the following layers in sequence:\n\n an input layer\n a simple Embedding layer transforming integer words into vectors\n a dropout layer to regularize the model\n a SimpleRNN layer\n a TimeDistributed layer with an inner Dense layer which output dimensionality is equal to n_tag\n \n*(For hyperparameters, use those provided in Model 2)*\n\n**2.3a** Define, compile, and train an RNN model. Use the provided code to save the model and plot the training history.\n\n**2.3b** Visualize outputs from the SimpleRNN layer, one subplot for B-tags and one subplot for I-tags. Comment on the patterns you observed.\n\n**2.4** **Model 4: GRU**\n\n**2.4a** Briefly explain what a GRU is and how it's different from a simple RNN.\n\n**2.4b** Define, compile, and train a GRU architecture by replacing the SimpleRNN cell with a GRU one. Use the provided code to save the model and plot the training history.\n\n**2.4c** Visualize outputs from GRU layer, one subplot for **B-tags** and one subplot for **I-tags**. Comment on the patterns you observed.\n\n**2.5** **Model 5: Bidirectional GRU**\n\n**2.5a** Explain how a Bidirectional GRU differs from GRU model above.\n\n**2.5b** Define, compile, and train a bidirectional GRU by wrapping your GRU layer in a Bidirectional one. Use the provided code to save the model and plot the training history.\n\n**2.5c** Visualize outputs from bidirectional GRU layer, one subplot for **B-tags** and one subplot for **I-tags**. Comment on the patterns you observed.",
"_____no_output_____"
],
[
"## Answers",
"_____no_output_____"
],
[
"**2.1** **Model 1: Baseline Model**\n\nPredict the tag of a word simply with the most frequently-seen named entity tag of this word from the training set.\n\ne.g. word \"Apple\" appears 10 times in the training set; 7 times it was tagged as \"Corporate\" and 3 times it was tagged as \"Fruit\". If we encounter the word \"Apple\" in the test set, we predict it as \"Corporate\".\n\n**Create an np.array ```baseline``` of length [n_words]**\nwhere the ith element ```baseline[i]``` is the index of the most commonly seen named entity tag of word i summarised from training set. (e.g. [16, 16, 16, ..., 0, 16, 16])\n",
"_____no_output_____"
]
],
[
[
"sentence2idx = dict(zip(data['Sentence #'].unique(), np.arange(n_sentences)))\ndata['sentence_idx'] = data['Sentence #'].apply(lambda x: sentence2idx[x]) # assign an index to each sentence\ndata_train = data[data['sentence_idx'].isin(train_indices)]\ndata_test = data[data['sentence_idx'].isin(test_indices)]\ntrain_words = data_train['Word'].unique() ",
"_____no_output_____"
],
[
"word_tag_counts = data_train.groupby('Word')['Tag'].value_counts()\nbaseline_tag = [word_tag_counts[word].index[0] if word in train_words else 'O' for word in words] # most frequent tag by word\nbaseline = np.array([tag2idx[i] for i in baseline_tag])\n",
"_____no_output_____"
],
[
"# Run this cell to show your results #\nprint(baseline[X].shape,'\\n')\nprint('Sentence:\\n {}\\n'.format([idx2word[w] for w in X[0]]))\nprint('Predicted Tags:\\n {}'.format([idx2tag[i] for i in baseline[X[0]]]))",
"(47959, 104) \n\nSentence:\n ['Thousands', 'of', 'demonstrators', 'have', 'marched', 'through', 'London', 'to', 'protest', 'the', 'war', 'in', 'Iraq', 'and', 'demand', 'the', 'withdrawal', 'of', 'British', 'troops', 'from', 'that', 'country', '.', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD', 'ENDPAD']\n\nPredicted Tags:\n ['O', 'O', 'O', 'O', 'O', 'O', 'B-geo', 'O', 'O', 'O', 'O', 'O', 'B-geo', 'O', 'O', 'O', 'O', 'O', 'B-gpe', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O', 'O']\n"
]
],
[
[
"**2.2** **Model 2: Vanilla Feed Forward Neural Network**\n\nThis model is provided for you. Please pay attention to the architecture of this neural network, especially the input/output dimensionalities and the Embedding layer.\n",
"_____no_output_____"
],
[
"### Use these hyperparameters for all NN models",
"_____no_output_____"
]
],
[
[
"n_units = 100\ndrop_rate = .1\ndim_embed = 50\n\noptimizer = \"rmsprop\"\nloss = \"categorical_crossentropy\"\nmetrics = [\"accuracy\"]\n\nbatch_size = 32\nepochs = 10\nvalidation_split = 0.1\nverbose = 1",
"_____no_output_____"
],
[
"# Define model\nmodel = tf.keras.Sequential()\nmodel.add(tf.keras.layers.Embedding(input_dim=n_words, output_dim=dim_embed, input_length=max_len))\nmodel.add(tf.keras.layers.Dropout(drop_rate))\nmodel.add(tf.keras.layers.Dense(n_tags, activation=\"softmax\"))\n\n# Compile model\nmodel.compile(optimizer=optimizer, loss=loss, metrics=metrics)",
"_____no_output_____"
],
[
"print(model.summary())",
"Model: \"sequential\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding (Embedding) (None, 104, 50) 1758950 \n_________________________________________________________________\ndropout (Dropout) (None, 104, 50) 0 \n_________________________________________________________________\ndense (Dense) (None, 104, 18) 918 \n=================================================================\nTotal params: 1,759,868\nTrainable params: 1,759,868\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
],
[
"# Train model\nhistory = model.fit(X_tr, y_tr, batch_size=batch_size, epochs=epochs, \n validation_split=validation_split, verbose=verbose)",
"Train on 38847 samples, validate on 4317 samples\nEpoch 1/10\n38847/38847 [==============================] - 18s 459us/sample - loss: 0.3147 - accuracy: 0.9788 - val_loss: 0.0598 - val_accuracy: 0.9858\nEpoch 2/10\n38847/38847 [==============================] - 15s 398us/sample - loss: 0.0513 - accuracy: 0.9864 - val_loss: 0.0482 - val_accuracy: 0.9869\nEpoch 3/10\n38847/38847 [==============================] - 16s 406us/sample - loss: 0.0448 - accuracy: 0.9873 - val_loss: 0.0454 - val_accuracy: 0.9875\nEpoch 4/10\n38847/38847 [==============================] - 16s 411us/sample - loss: 0.0425 - accuracy: 0.9878 - val_loss: 0.0442 - val_accuracy: 0.9877\nEpoch 5/10\n38847/38847 [==============================] - 16s 405us/sample - loss: 0.0412 - accuracy: 0.9882 - val_loss: 0.0434 - val_accuracy: 0.9879\nEpoch 6/10\n38847/38847 [==============================] - 16s 409us/sample - loss: 0.0404 - accuracy: 0.9884 - val_loss: 0.0431 - val_accuracy: 0.9880\nEpoch 7/10\n38847/38847 [==============================] - 16s 404us/sample - loss: 0.0397 - accuracy: 0.9886 - val_loss: 0.0429 - val_accuracy: 0.9881\nEpoch 8/10\n38847/38847 [==============================] - 16s 409us/sample - loss: 0.0393 - accuracy: 0.9888 - val_loss: 0.0426 - val_accuracy: 0.9881\nEpoch 9/10\n38847/38847 [==============================] - 16s 405us/sample - loss: 0.0390 - accuracy: 0.9888 - val_loss: 0.0425 - val_accuracy: 0.9882\nEpoch 10/10\n38847/38847 [==============================] - 15s 399us/sample - loss: 0.0389 - accuracy: 0.9889 - val_loss: 0.0423 - val_accuracy: 0.9882\n"
],
[
"store_keras_model(model, 'model_FFNN')",
"Saved model to disk\n"
],
[
"plot_training_history(history)",
"_____no_output_____"
]
],
[
[
"**2.2a** Explain what the embedding layer is and why we need it here.",
"_____no_output_____"
],
[
"We need an embedding layer is because it allows the model to take input data in a form that conveys similarity between words that have similar sentiments. If the input data were only one hot encoded, then the network would be very sparse (mostly 0s in the encoding and just one 1 for each word) and this requires a large amount of memory during training as well as does not minimize the number of parameters in the model.",
"_____no_output_____"
],
[
"**2.2b** Explain why the Param # of Embedding layer is 1758950 (as shown in `print(model.summary())`).",
"_____no_output_____"
]
],
[
[
"n_words*dim_embed",
"_____no_output_____"
]
],
[
[
"The parameter number of the embedding layer is equal to the number of words times the embedding dimension, as shown above. This is because each word is represented at a vector of length `n_words` which we represent as a dense vector of length `dim_embed`. To do this we multiply the one-hot vector by a matrix of size `n_words` $\\times$ `dim_embed`, which explains the number of dimensions",
"_____no_output_____"
],
[
"### Viewing Hidden Layers\nIn addition to the final result, we also want to see the intermediate results from hidden layers. Below is an example showing how to get outputs from a hidden layer, and visualize them on the reduced dimension of 2D by PCA. (**Please note that this code and the parameters cannot be simply copied and pasted for other questions; some adjustments need to be made**) ",
"_____no_output_____"
]
],
[
[
"FFNN = load_keras_model(\"model_FFNN\")\ndef create_truncated_model_FFNN(trained_model):\n model = tf.keras.Sequential()\n model.add(tf.keras.layers.Embedding(input_dim=n_words, output_dim=dim_embed, input_length=max_len))\n model.add(tf.keras.layers.Dropout(drop_rate))\n # set weights of first few layers using the weights of trained model\n for i, layer in enumerate(model.layers):\n layer.set_weights(trained_model.layers[i].get_weights())\n model.compile(optimizer=optimizer, loss=loss, metrics=metrics)\n return model\ntruncated_model = create_truncated_model_FFNN(FFNN)\nhidden_features = truncated_model.predict(X_te)\n\n# flatten data\nhidden_features = hidden_features.reshape(-1,50)\n \n# find first two PCA components\npca = PCA(n_components=2)\npca_result = pca.fit_transform(hidden_features)\nprint('Variance explained by PCA: {}'.format(np.sum(pca.explained_variance_ratio_)))",
"Variance explained by PCA: 0.9407047017321251\n"
],
[
"# visualize hidden featurs on first two PCA components\n# this plot only shows B-tags\ndef visualize_hidden_features(pca_result):\n color=['r', 'C1', 'y', 'C3', 'b', 'g', 'm', 'orange']\n category = np.argmax(y_te.reshape(-1,18), axis=1)\n fig, ax = plt.subplots()\n fig.set_size_inches(6,6) \n for cat in range(8):\n indices_B = np.where(category==cat)[0]\n #length=min(1000,len(indices_B))\n #indices_B=indices_B[:length]\n ax.scatter(pca_result[indices_B,0], pca_result[indices_B, 1], label=idx2tag[cat],s=2,color=color[cat],alpha=0.5)\n legend=ax.legend(markerscale=3)\n legend.get_frame().set_facecolor('w') \n plt.show()",
"_____no_output_____"
],
[
"visualize_hidden_features(pca_result)",
"_____no_output_____"
]
],
[
[
"### Full function for other questions ###",
"_____no_output_____"
]
],
[
[
"def get_hidden_output_PCA(model,X_te,y_te,layer_index,out_dimension):\n output = tf.keras.backend.function([model.layers[0].input],[model.layers[layer_index].output])\n hidden_feature=np.array(output([X_te]))\n hidden_feature=hidden_feature.reshape(-1,out_dimension)\n \n pca = PCA(n_components=2)\n pca_result = pca.fit_transform(hidden_feature)\n print('Variance explained by PCA: {}'.format(np.sum(pca.explained_variance_ratio_)))\n return pca_result",
"_____no_output_____"
],
[
"def visualize_B_I(pca_result):\n color = ['r', 'C1', 'y', 'C3', 'b', 'g', 'm', 'orange']\n category = np.argmax(y_te.reshape(-1,18), axis=1)\n fig, ax = plt.subplots(1,2) \n fig.set_size_inches(12,6)\n for i in range(2):\n for cat in range(8*i,8*(i+1)):\n indices = np.where(category==cat)[0]\n ax[i].scatter(pca_result[indices,0], pca_result[indices, 1], label=idx2tag[cat],s=2,color=color[cat-8*i],alpha=0.5)\n legend = ax[i].legend(markerscale=3)\n legend.get_frame().set_facecolor('w') \n ax[i].set_xlabel(\"first dimension\")\n ax[i].set_ylabel(\"second dimension\")\n fig.suptitle(\"visualization of hidden feature on reduced dimension by PCA\")\n \n plt.show()",
"_____no_output_____"
],
[
"h = get_hidden_output_PCA(FFNN,X_te,y_te,1,50)\nvisualize_B_I(h)",
"Variance explained by PCA: 0.9407051658905141\n"
]
],
[
[
"**2.3** **Model 3: RNN**\n\nSet up a simple RNN model by stacking the following layers in sequence:\n\n an input layer\n a simple Embedding layer transforming integer words into vectors\n a dropout layer to regularize the model\n a SimpleRNN layer\n a TimeDistributed layer with an inner Dense layer which output dimensionality is equal to n_tag\n \n*(For hyperparameters, use those provided in Model 2)*\n\n**2.3a** Define, compile, and train an RNN model. Use the provided code to save the model and plot the training history.",
"_____no_output_____"
]
],
[
[
"RNN = Sequential()\nRNN.add(Embedding(n_words, dim_embed, input_length = max_len))\nRNN.add(Dropout(drop_rate))\nRNN.add(SimpleRNN(n_units, return_sequences = 1))\nRNN.add(TimeDistributed(Dense(n_tags, activation = 'softmax')))\nRNN.compile(optimizer = optimizer, loss = loss)\n\nhistory = RNN.fit(X_tr, y_tr, batch_size=batch_size, epochs=epochs,\n validation_split=validation_split, verbose=verbose)",
"Train on 38847 samples, validate on 4317 samples\nEpoch 1/10\n38847/38847 [==============================] - 84s 2ms/sample - loss: 0.0921 - val_loss: 0.0329\nEpoch 2/10\n38847/38847 [==============================] - 81s 2ms/sample - loss: 0.0294 - val_loss: 0.0288\nEpoch 3/10\n38847/38847 [==============================] - 83s 2ms/sample - loss: 0.0261 - val_loss: 0.0279\nEpoch 4/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0244 - val_loss: 0.0275\nEpoch 5/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0233 - val_loss: 0.0274\nEpoch 6/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0223 - val_loss: 0.0271\nEpoch 7/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0214 - val_loss: 0.0271\nEpoch 8/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0206 - val_loss: 0.0271\nEpoch 9/10\n38847/38847 [==============================] - 83s 2ms/sample - loss: 0.0198 - val_loss: 0.0276\nEpoch 10/10\n38847/38847 [==============================] - 82s 2ms/sample - loss: 0.0193 - val_loss: 0.0275\n"
],
[
"# save your mode #\nstore_keras_model(RNN, 'model_RNN')\nRNN = load_keras_model(\"model_RNN\")",
"Saved model to disk\n"
],
[
"RNN.summary()",
"Model: \"sequential_2\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_2 (Embedding) (None, 104, 50) 1758950 \n_________________________________________________________________\ndropout_2 (Dropout) (None, 104, 50) 0 \n_________________________________________________________________\nsimple_rnn (SimpleRNN) (None, 104, 100) 15100 \n_________________________________________________________________\ntime_distributed (TimeDistri (None, 104, 18) 1818 \n=================================================================\nTotal params: 1,775,868\nTrainable params: 1,775,868\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"# run this cell to show your results #\nplot_training_history(history)",
"_____no_output_____"
]
],
[
[
"**2.3b** Visualize outputs from the SimpleRNN layer, one subplot for B-tags and one subplot for I-tags. Comment on the patterns you observed.",
"_____no_output_____"
]
],
[
[
"h = get_hidden_output_PCA(RNN,X_te,y_te,2,n_units)\nvisualize_B_I(h)",
"Variance explained by PCA: 0.8272746161397989\n"
]
],
[
[
"The clusters formed after PCA on the B-tags and I-tags show generally similar groupings. For example, the B-per tag data generally fall in the space with the first PC about 6 and the second PC about 1, while the I-per tag data generally fall in the space with the first PC about 5.5 and the second PC about 1. The B-tim data generally fall in the space with the first PC about 8 and the second PC about 3, while the I-tim data are centered around the space with first PC about 8 and the second PC about 2. The similar locations of clusters along the axes of the first two PCs indicates that clustering the data based on the B versus I tags does not materially change the identification of the essential info about the named entity (i.e., tag). However, the I-tags have more readily distinguishable clusters compared with the B-tags.",
"_____no_output_____"
],
[
"<div class='explication'> </div>",
"_____no_output_____"
],
[
"**2.4** **Model 4: GRU**\n\n**2.4a** Briefly explain what a GRU is and how it's different from a simple RNN.",
"_____no_output_____"
],
[
"A GRU is a deviation of the vanilla RNN that attempts to remember information from the distant past by avoiding vanishing and exploding gradients. It uses two gates—an *update gate* and a *forget gate*. The update gate incorporates information from the present time and the most recent hidden state. The forget gate decides what information should remain from the previous state. The GRU then combines the output of these two gates to create a new hidden state. Unlike the simple RNN, the GRU updates updates the hidden state with addition, rather than multiplication, resulting in a more stable gradient. ",
"_____no_output_____"
],
[
"**2.4b** Define, compile, and train a GRU architecture by replacing the SimpleRNN cell with a GRU one. Use the provided code to save the model and plot the training history.",
"_____no_output_____"
]
],
[
[
"GRUmod = Sequential()\nGRUmod.add(Embedding(n_words, dim_embed, input_length = max_len))\nGRUmod.add(Dropout(drop_rate))\nGRUmod.add(GRU(n_units, return_sequences = 1))\nGRUmod.add(TimeDistributed(Dense(n_tags, activation = 'softmax')))\nGRUmod.compile(optimizer = optimizer, loss = loss)\n\nhistory = GRUmod.fit(X_tr, y_tr, batch_size=batch_size, epochs=epochs,\n validation_split=validation_split, verbose=verbose)",
"Train on 38847 samples, validate on 4317 samples\nEpoch 1/10\n38847/38847 [==============================] - 44s 1ms/sample - loss: 0.0893 - val_loss: 0.0321\nEpoch 2/10\n38847/38847 [==============================] - 39s 993us/sample - loss: 0.0289 - val_loss: 0.0285\nEpoch 3/10\n38847/38847 [==============================] - 38s 990us/sample - loss: 0.0258 - val_loss: 0.0272\nEpoch 4/10\n38847/38847 [==============================] - 39s 1ms/sample - loss: 0.0240 - val_loss: 0.0268\nEpoch 5/10\n38847/38847 [==============================] - 39s 994us/sample - loss: 0.0229 - val_loss: 0.0267\nEpoch 6/10\n38847/38847 [==============================] - 39s 1ms/sample - loss: 0.0219 - val_loss: 0.0264\nEpoch 7/10\n38847/38847 [==============================] - 39s 1000us/sample - loss: 0.0210 - val_loss: 0.0265\nEpoch 8/10\n38847/38847 [==============================] - 39s 1ms/sample - loss: 0.0202 - val_loss: 0.0264\nEpoch 9/10\n38847/38847 [==============================] - 39s 1ms/sample - loss: 0.0195 - val_loss: 0.0266\nEpoch 10/10\n38847/38847 [==============================] - 39s 1ms/sample - loss: 0.0189 - val_loss: 0.0266\n"
],
[
"# save your mode #\nstore_keras_model(GRUmod, 'model_GRU')\nGRUmod = load_keras_model(\"model_GRU\")",
"Saved model to disk\n"
],
[
"# run this cell to show your results #\nplot_training_history(history)",
"_____no_output_____"
],
[
"# run this cell to show your results #\nprint(GRUmod.summary())",
"Model: \"sequential_3\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_3 (Embedding) (None, 104, 50) 1758950 \n_________________________________________________________________\ndropout_3 (Dropout) (None, 104, 50) 0 \n_________________________________________________________________\ngru (GRU) (None, 104, 100) 45600 \n_________________________________________________________________\ntime_distributed_1 (TimeDist (None, 104, 18) 1818 \n=================================================================\nTotal params: 1,806,368\nTrainable params: 1,806,368\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
]
],
[
[
"**2.4c** Visualize outputs from GRU layer, one subplot for **B-tags** and one subplot for **I-tags**. Comment on the patterns you observed.",
"_____no_output_____"
]
],
[
[
"h = get_hidden_output_PCA(GRUmod,X_te,y_te,2,n_units)\nvisualize_B_I(h)",
"Variance explained by PCA: 0.7733814424623531\n"
]
],
[
[
"Just as in the PCA plots from the RNN generated previously, we observe that the I-tags (relative to the B-tags) are more readily distinguished from each other using the first two principal components. However, in the PCA plots generated from the hidden features from the GRU, we observe that the clusters are overall more clearly defined as they were in the PCA plots from the RNN. This indicates that the hidden features for the GRU layers capture more of the information for the tags in just the first two PCs. Since PCA is a linear dimensionality reduction method, the GRU's hidden features may also be capturing more of the linear relations in the tags since the PCA plot has more readily distinguished clusters.",
"_____no_output_____"
],
[
"**2.5** **Model 5: Bidirectional GRU**\n\n**2.5a** Explain how a Bidirectional GRU differs from GRU model above.\n\n",
"_____no_output_____"
],
[
"A Bidirectional GRU is essentially a combination of two GRUs, one of which works forward and one of which works backward. The forward GRU processes the start of the first word of the sentence, feeds the hidden state to the second, and repeats until the end. The backward GRU does the same but in the reverse direction. For each word, the bidirectional GRU concatenates the hidden layer from the forward-GRU for that word and the hidden layer from the backward-GRU for that word, and passes that concatenation to the output layer. This differs from a single-directional GRU model in that it is able to incorporate information about a word using words *later* in the sentence, rather than just words that came before. ",
"_____no_output_____"
],
[
"**2.5b** Define, compile, and train a bidirectional GRU by wrapping your GRU layer in a Bidirectional one. Use the provided code to save the model and plot the training history.\n",
"_____no_output_____"
]
],
[
[
"BiGRUmod = Sequential()\nBiGRUmod.add(Embedding(n_words, dim_embed, input_length = max_len))\nBiGRUmod.add(Dropout(drop_rate))\nBiGRUmod.add(Bidirectional(GRU(n_units, return_sequences = 1)))\nBiGRUmod.add(TimeDistributed(Dense(n_tags, activation = 'softmax')))\nBiGRUmod.compile(optimizer = optimizer, loss = loss)\n\nhistory = BiGRUmod.fit(X_tr, y_tr, batch_size=batch_size, epochs=epochs,\n validation_split=validation_split, verbose=verbose)",
"Train on 38847 samples, validate on 4317 samples\nEpoch 1/10\n38847/38847 [==============================] - 75s 2ms/sample - loss: 0.0728 - val_loss: 0.0269\nEpoch 2/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0241 - val_loss: 0.0236\nEpoch 3/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0208 - val_loss: 0.0224\nEpoch 4/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0189 - val_loss: 0.0222\nEpoch 5/10\n38847/38847 [==============================] - 71s 2ms/sample - loss: 0.0177 - val_loss: 0.0221\nEpoch 6/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0166 - val_loss: 0.0217\nEpoch 7/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0156 - val_loss: 0.0221\nEpoch 8/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0146 - val_loss: 0.0222\nEpoch 9/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0137 - val_loss: 0.0227\nEpoch 10/10\n38847/38847 [==============================] - 70s 2ms/sample - loss: 0.0129 - val_loss: 0.0231\n"
],
[
"# save your model #\nstore_keras_model(BiGRUmod, 'model_BiGRU')",
"Saved model to disk\n"
],
[
"# run this cell to show your results #\nprint(BiGRUmod.summary())",
"Model: \"sequential_4\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_4 (Embedding) (None, 104, 50) 1758950 \n_________________________________________________________________\ndropout_4 (Dropout) (None, 104, 50) 0 \n_________________________________________________________________\nbidirectional (Bidirectional (None, 104, 200) 91200 \n_________________________________________________________________\ntime_distributed_2 (TimeDist (None, 104, 18) 3618 \n=================================================================\nTotal params: 1,853,768\nTrainable params: 1,853,768\nNon-trainable params: 0\n_________________________________________________________________\nNone\n"
],
[
"# run this cell to show your results #\nplot_training_history(history)",
"_____no_output_____"
]
],
[
[
"**2.5c** Visualize outputs from bidirectional GRU layer, one subplot for **B-tags** and one subplot for **I-tags**. Comment on the patterns you observed.",
"_____no_output_____"
]
],
[
[
"h = get_hidden_output_PCA(BiGRUmod,X_te,y_te,2,2*n_units)\nvisualize_B_I(h)",
"Variance explained by PCA: 0.7604033777222045\n"
]
],
[
[
"According to PCA of the BiGRU, the data are more readily distinguished when using the I-tags relative to using the B-tags. This is apparent by the PCA plots of the hidden features on the axes of the first two PCs, where the colored data are not as admixed on the right panel as they are on the left panel. However, both the B-tags and I-tags distinguish the data overall since there are distinctly colored clusters in both plots. The classes (e.g., I-per and B-per) are located in similar regions of space on the axes of the first two PCs for the B-tag and I-tag plots from the BiGRU shown above, as well as for the B-tag and I-tag plots from the RNN and GRU described previously. The data are overall more distinctly clustered by tag for the hidden features from the BiGRU compared with either the RNN or GRU, indicating that the BiGRU may be the most well-fit model of the ones studied here.",
"_____no_output_____"
],
[
"<div class='exercise'><b> Question 3: Analysis [40pt]</b></div>\n",
"_____no_output_____"
],
[
"**3.1** For each model, iteratively:\n\n- Load the model using the given function ```load_keras_model```\n\n- Apply the model to the test dataset\n\n- Compute an F1 score for each ```Tag``` and store it \n\n**3.2** Plot the F1 score per Tag and per model making use of a grouped barplot.\n\n**3.3** Briefly discuss the performance of each model\n\n\n**3.4** Which tags have the lowest f1 score? For instance, you may find from the plot above that the test accuracy on \"B-art\", and \"I-art\" are very low (just an example, your case maybe different). Here is an example when models failed to predict these tags right\n\n<img src=\"data/B_art.png\" alt=\"drawing\" width=\"600\"/>\n\n**3.5** Write functions to output another example in which the tags of the lowest accuracy was predicted wrong in a sentence (include both \"B-xxx\" and \"I-xxx\" tags). Store the results in a DataFrame (same format as the above example) and use styling functions below to print out your df.\n\n**3.6** Choose one of the most promising models you have built, improve this model to achieve an f1 score higher than 0.8 for as many tags as possible (you have lots of options here, e.g. data balancing, hyperparameter tuning, changing the structure of NN, a different optimizer, etc.)\n\n**3.7** Explain why you chose to change certain elements of the model and how effective these adjustments were.\n",
"_____no_output_____"
],
[
"## Answers",
"_____no_output_____"
],
[
"**3.1** For each model, iteratively:\n\n- Load the model using the given function ```load_keras_model```\n\n- Apply the model to the test dataset\n\n- Compute an F1 score for each ```Tag``` and store it ",
"_____no_output_____"
]
],
[
[
"models = ['FFNN','RNN','GRU','BiGRU']\nf1_scores = pd.DataFrame(columns = ['model','tag','score'])\n\nfor i, model_name in enumerate(models):\n model = load_keras_model('model_' + model_name)\n y_pred = to_categorical(model.predict_classes(X_te))\n for tag in range(n_tags):\n score = f1_score(y_te[:,:,tag].reshape(-1), y_pred[:,:,tag].reshape(-1))\n f1_scores.loc[len(f1_scores)] = [model_name,tag,score]",
"/usr/share/anaconda3/lib/python3.7/site-packages/sklearn/metrics/classification.py:1437: UndefinedMetricWarning: F-score is ill-defined and being set to 0.0 due to no predicted samples.\n 'precision', 'predicted', average, warn_for)\n"
]
],
[
[
"**3.2** Plot the F1 score per Tag and per model making use of a grouped barplot.",
"_____no_output_____"
]
],
[
[
"sns.set_style('whitegrid')\nfig, ax = plt.subplots(figsize = (10,7))\nsns.barplot(f1_scores['tag'], f1_scores['score'], hue = f1_scores['model'], palette = 'mako')\nax.legend(bbox_to_anchor = (1,1))\nax.set(xlabel = 'Tag', ylabel = 'F1 Score', title = 'F1 Score by Tag and Model');\n",
"_____no_output_____"
]
],
[
[
"**3.3** Briefly discuss the performance of each model",
"_____no_output_____"
],
[
"A higher f1 score corresponds to a higher quality model as it indicates a better combination of precision and recall. In general, we observe that the BiGRU typically outperforms the GRU across tags, which typically outperforms the RNN across tags, which outperforms the FFNN across tags. Although the tags show similar relative performance across models, some tags allow all models to perform much better. For instance, all models have an f1 score above 0.8 for tags 2, 3, 16, 17 while none score above 0.2 for tags 0 and 8. We also see that the discrepency between these models varies by tag. For instance, all models perform quite similarly on tags 3, 16, and 17, but the BiGRU vastly outperforms other models on tag 1. ",
"_____no_output_____"
],
[
"**3.4** Which tags have the lowest f1 score? For instance, you may find from the plot above that the test accuracy on \"B-art\", and \"I-art\" are very low (just an example, your case maybe different). Here is an example when models failed to predict these tags right\n\n<img src=\"data/B_art.png\" alt=\"drawing\" width=\"600\"/>",
"_____no_output_____"
]
],
[
[
"[idx2tag[tag] for tag in [0,8]]",
"_____no_output_____"
]
],
[
[
"Tags 0 and 8, which correspond to \"beginning-art\" and \"inside-art\" have the lowest f1 scores. ",
"_____no_output_____"
],
[
"**3.5** Write functions to output another example in which the tags of the lowest accuracy were predicted wrong in a sentence (include both \"B-xxx\" and \"I-xxx\" tags). Store the results in a DataFrame (same format as the above example) and use styling functions below to print out your df.",
"_____no_output_____"
]
],
[
[
"# Find a sentence that all models miss\nmissed_by_all = set(np.arange(len(y_te)))\nfor i, model_name in enumerate(models):\n # load model\n model = load_keras_model('model_' + model_name)\n # make predictions\n y_pred = to_categorical(model.predict_classes(X_te))\n # see where it missed tag 8\n missed_sentences_8 = np.argwhere( (y_te[:,:,8] == 1) & (y_pred[:,:,8] == 0))[:,0]\n # see where it missed tag 0\n missed_sentences_0 = np.argwhere( (y_te[:,:,0] == 1) & (y_pred[:,:,0] == 0))[:,0]\n # get intersection (within model)\n missed_sentence_both = set(missed_sentences_8).intersection(missed_sentences_0) # sentences missed by both\n # get intersection (across models)\n missed_by_all = missed_by_all.intersection(missed_sentence_both)\n \nmissed_by_all_index = list(missed_by_all)[0] # index \nmissed_by_all_tokens = X_te[missed_by_all_index].reshape(1,-1) # X_te for that row",
"_____no_output_____"
],
[
"# Combine that sentence into a DF \ndata_test_sentence_idx = data_test.sentence_idx.unique()[missed_by_all_index]\ndata_test_missed_by_all = data_test[data_test['sentence_idx'] == data_test_sentence_idx]\n\n\ncolumns = ['Word','y_true','baseline','FFNN','RNN','GRU','BiGRU']\nmissed_df = pd.DataFrame(index = data_test_missed_by_all.index, columns = columns)\nmissed_df['Word'] = data_test_missed_by_all['Word']\nmissed_df['y_true'] = data_test_missed_by_all['Tag']\nmissed_df['baseline'] = [idx2tag[tag] for tag in baseline[np.array([words.index(word) for word in missed_df.Word])]]\n\nfor i, model_name in enumerate(models):\n # load model\n model = load_keras_model('model_' + model_name)\n # make prediction\n y_pred = model.predict_classes(missed_by_all_tokens).reshape(-1)\n y_pred_tags = [idx2tag[tag] for tag in y_pred]\n # add in \n missed_df[model_name] = y_pred_tags[:len(missed_df)]\n",
"_____no_output_____"
],
[
"# style it\ndef correct_classification(s):\n y_true = s['y_true']\n is_true = s == y_true\n is_true[0] = True\n return ['color: red' if not v else '' for v in is_true]\nmissed_df.style.apply(axis = 1, func = correct_classification)",
"_____no_output_____"
]
],
[
[
"**3.6** Choose one of the most promising models you have built, improve this model to achieve an f1 score higher than 0.8 for as many tags as possible (you have lots of options here, e.g. data balancing, hyperparameter tuning, changing the structure of NN, a different optimizer, etc.)",
"_____no_output_____"
]
],
[
[
"mod = Sequential()\nmod.add(Embedding(n_words, dim_embed, input_length = max_len))\nmod.add(BatchNormalization(axis=1, momentum=0.2))\nmod.add(Dropout(.1))\nmod.add(Bidirectional(GRU(n_units, return_sequences = 1)))\nmod.add(TimeDistributed(Dense(n_tags, activation = 'softmax')))\nmod.compile(optimizer = 'rmsprop', loss = loss)\n\nhistory = mod.fit(X_tr, y_tr, batch_size=64, epochs=4, validation_split=validation_split, verbose=verbose)\nstore_keras_model(mod, 'bestmod')",
"Train on 38847 samples, validate on 4317 samples\nEpoch 1/4\n38847/38847 [==============================] - 44s 1ms/sample - loss: 0.0522 - val_loss: 0.0240\nEpoch 2/4\n38847/38847 [==============================] - 38s 986us/sample - loss: 0.0205 - val_loss: 0.0217\nEpoch 3/4\n38847/38847 [==============================] - 38s 986us/sample - loss: 0.0171 - val_loss: 0.0218\nEpoch 4/4\n38847/38847 [==============================] - 38s 977us/sample - loss: 0.0149 - val_loss: 0.0218\nSaved model to disk\n"
],
[
"# ATTEMPTING TO RE-WEIGHT \n# from sklearn.utils import class_weight\n# y_not_hot = data.Tag.apply(lambda x: tag2idx[x])\n# class_weights = class_weight.compute_class_weight('balanced', np.sort(y_not_hot.unique()), y_not_hot)\n# class_weights_dict = dict(zip(np.arange(len(class_weights)), class_weights))\n# class_weights_dict[17] = class_weights_dict[16] # TODO: re-weight\n\n# tags = []\n# for sentence in y_tr:\n# for word in sentence:\n# tags.append([np.where(word == 1)[0][0]])\n# tags = np.array(tags).reshape(-1)\n\n# class_weights = class_weight.compute_class_weight('balanced', np.arange(n_tags),tags)\n# class_weights_dict = dict(zip(np.arange(n_tags), class_weights))\n# plz = np.array([class_weights_dict[i] for i in tags]).reshape(len(y_tr), -1)\n\n# mod = Sequential()\n# mod.add(Embedding(n_words, dim_embed, input_length = max_len))\n# mod.add(BatchNormalization(axis=1, momentum=0.09))\n# mod.add(Dropout(.1))\n# mod.add(Bidirectional(GRU(n_units, return_sequences = 1)))\n# mod.add(TimeDistributed(Dense(n_tags, use_bias = False)))\n# mod.add(tf.keras.layers.Activation('softmax'))\n# mod.compile(optimizer = 'rmsprop', loss = loss, sample_weight_mode = 'temporal')\n# history = mod.fit(X_tr,\n# y_tr,\n# batch_size=512,\n# epochs=10,\n# validation_split = validation_split,\n# verbose=verbose,\n# sample_weight = plz)\n ",
"_____no_output_____"
],
[
"y_pred = to_categorical(mod.predict_classes(X_te))\nf1_scores = []\nfor tag in range(n_tags):\n f1_scores.append(f1_score(y_te[:,:,tag].reshape(-1), y_pred[:,:,tag].reshape(-1)))\n\nfig, ax = plt.subplots(figsize = (8,6))\nsns.barplot(np.arange(n_tags), f1_scores)\nax.set(xlabel = 'Tag', ylabel = 'F1 score', title = 'F1 Score by Tag for Improved Model');",
"/usr/share/anaconda3/lib/python3.7/site-packages/sklearn/metrics/classification.py:1437: UndefinedMetricWarning: F-score is ill-defined and being set to 0.0 due to no predicted samples.\n 'precision', 'predicted', average, warn_for)\n"
]
],
[
[
"**3.7** Explain why you chose to change certain elements of the model and how effective these adjustments were.",
"_____no_output_____"
],
[
" We chose to add batch normalization to the BiGRU because it helps to make the distributions of the activations at each layer similar to each other. This is necessary because the weights and the biases change after each layer and batch normalization serves essentially as a method of standardization during training, which can help to reduce overfitting and improve training.\n \n The modifications to the model only improve the validation loss and F1 scores slightly. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
ec6f0600e00210868dc3577f62e4614c60968719 | 25,402 | ipynb | Jupyter Notebook | src/experiments/wp_2.ipynb | harmeet773/stream | bac230bc21edbfa810a08d59c62c598f16df80da | [
"MIT"
]
| null | null | null | src/experiments/wp_2.ipynb | harmeet773/stream | bac230bc21edbfa810a08d59c62c598f16df80da | [
"MIT"
]
| null | null | null | src/experiments/wp_2.ipynb | harmeet773/stream | bac230bc21edbfa810a08d59c62c598f16df80da | [
"MIT"
]
| null | null | null | 59.489461 | 1,823 | 0.618062 | [
[
[
"import re\nimport pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom datetime import *\nimport datetime as dt\nfrom matplotlib.ticker import MaxNLocator\nimport regex\nimport emoji\nfrom seaborn import *\n# from heatmap import heatmap\n# from wordcloud import WordCloud , STOPWORDS , ImageColorGenerator\nfrom nltk import *\nfrom plotly import express as px\n\n## regex emoji seaborn heatmap nltk plotly",
"_____no_output_____"
],
[
"### Python code to extract Date from chat file \n\ndef startsWithDateAndTime(s):\n pattern = ‘^([0-9]+)(/)([0-9]+)(/)([0-9][0-9]), ([0-9]+):([0-9][0-9]) (AM|PM) -‘\n result = re.match(pattern, s)\n \n if result:\n return True\n \n return False",
"_____no_output_____"
],
[
"pip install plotly",
"Collecting plotly\n Downloading plotly-5.8.0-py2.py3-none-any.whl (15.2 MB)\n |████████████████████████████████| 15.2 MB 18 kB/s \n\u001b[?25hCollecting tenacity>=6.2.0\n Downloading tenacity-8.0.1-py3-none-any.whl (24 kB)\nInstalling collected packages: tenacity, plotly\nSuccessfully installed plotly-5.8.0 tenacity-8.0.1\nNote: you may need to restart the kernel to use updated packages.\n"
],
[
"# opening a file\nos.chdir('../data/')\n\nfilename = 'Books&Beyond.txt'\ndef read_file(file):\n '''Reads Whatsapp text file into a list of strings'''\n x = open(file,'r', encoding = 'utf-8') #Opens the text file into variable x but the variable cannot be explored yet\n y = x.read() #By now it becomes a huge chunk of string that we need to separate line by line\n content = y.splitlines() #The splitline method converts the chunk of string into a list of strings\n return content\n\nchat = read_file(filename)",
"_____no_output_____"
],
[
"### Regex pattern to extract username of Author.\n\ndef FindAuthor(s):\n patterns = [\n '([w]+):', # First Name\n '([w]+[s]+[w]+):', # First Name + Last Name\n '([w]+[s]+[w]+[s]+[w]+):', # First Name + Middle Name + Last Name\n '([+]d{2} d{5} d{5}):', # Mobile Number (India no.)\n '([+]d{2} d{3} d{3} d{4}):', # Mobile Number (US no.)\n '([w]+)[u263a-U0001f999]+:', # Name and Emoji \n ]\n pattern = '^' + '|'.join(patterns)\n result = re.match(pattern, s)\n if result:\n return True\n return False\n\nFindAuthor(chat)\n### Extracting Date, Time, Author and message from the chat file.",
"_____no_output_____"
],
[
"def getDataPoint(line): \n splitLine = line.split(' - ') \n dateTime = splitLine[0]\n date, time = dateTime.split(', ') \n message = ' '.join(splitLine[1:])\n if FindAuthor(message): \n splitMessage = message.split(': ') \n author = splitMessage[0] \n message = ' '.join(splitMessage[1:])\n else:\n author = None\n return date, time, author, message\n\n### Finally creating a dataframe and storing all data inside that dataframe.",
"_____no_output_____"
],
[
"parsedData = [] # List to keep track of data so it can be used by a Pandas dataframe\n### Uploading exported chat file\nconversationPath = 'WhatsApp Chat with TE Comp 20-21 Official.txt' # chat file\nwith open(conversationPath, encoding=\"utf-8\") as fp:\n ### Skipping first line of the file because contains information related to something about end-to-end encryption\n fp.readline() \n messageBuffer = [] \n date, time, author = None, None, None\n while True:\n line = fp.readline() \n if not line: \n break\n line = line.strip() \n if startsWithDateAndTime(line): \n if len(messageBuffer) > 0: \n parsedData.append([date, time, author, ' '.join(messageBuffer)]) \n messageBuffer.clear() \n date, time, author, message = getDataPoint(line) \n messageBuffer.append(message) \n else:\n messageBuffer.append(line)\ndf = pd.DataFrame(parsedData, columns=['Date', 'Time', 'Author', 'Message']) # Initialising a pandas Dataframe.\n### changing datatype of \"Date\" column.\ndf[\"Date\"] = pd.to_datetime(df[\"Date\"])",
"_____no_output_____"
],
[
"# Import following modules\nimport urllib.request\nimport pandas as pd \n# from pushbullet import PushBullet \n \n# Get Access Token from pushbullet.com\nAccess_token = \"Your Access Token\"\n \n# Authentication\n# pb = PushBullet(Access_token) \n \n# All pushes created by you\n# all_pushes = pb.get_pushes() \n \n# Get the latest push\n# latest_one = all_pushes[0] \n \n# Fetch the latest file URL link\n# url = latest_one['file_url'] \n \n \n# Create a new text file for storing\n# all the chats\nText_file = filename \n\n# Retrieve all the data store into\n# Text file\n# urllib.request.urlretrieve(url, Text_file)\n \n# Create an empty chat list\nchat_list = [] \n \n# Open the Text file in read mode and\n# read all the data\nwith open(Text_file, mode='r', encoding='utf8') as f:\n \n # Read all the data line-by-line\n data = f.readlines() \n \n# Excluded the first item of the list\n# first items contains some garbage\n# data\nfinal_data_set = data[1:]\n \n# Run a loop and read all the data\n# line-by-line\nfor line in final_data_set:\n # Extract the date, time, name,\n # message\n date = line.split(\",\")[0] \n tim = line.split(\"-\")[0].split(\",\")[1] \n name = line.split(\":\")[1].split(\"-\")[1] \n message = line.split(\":\")[2][:-1] \n \n # Append all the data in a List\n chat_list.append([date, tim, name, message])\n \n# Create a dataframe, for storing\n# all the data in a excel file\ndf = pd.DataFrame(chat_list,\n columns = ['Date', 'Time',\n 'Name', 'Message'])\ndf.to_excel(\"BackUp.xlsx\", index = False)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f06fe9aefd7b263c3d35ce24abab98c1cde19 | 130,792 | ipynb | Jupyter Notebook | 3.6-classifying-newswires.ipynb | Huthaifa1985/deep-learning-with-python-notebooks | bd270fb403703902126f67ba7f253b3869a925a7 | [
"MIT"
]
| null | null | null | 3.6-classifying-newswires.ipynb | Huthaifa1985/deep-learning-with-python-notebooks | bd270fb403703902126f67ba7f253b3869a925a7 | [
"MIT"
]
| null | null | null | 3.6-classifying-newswires.ipynb | Huthaifa1985/deep-learning-with-python-notebooks | bd270fb403703902126f67ba7f253b3869a925a7 | [
"MIT"
]
| null | null | null | 131.581489 | 67,450 | 0.368295 | [
[
[
"import tensorflow as tf\n\nfrom tensorflow import keras\nmnist = tf.keras.datasets.mnist",
"_____no_output_____"
]
],
[
[
"# Classifying newswires: a multi-class classification example\n\nThis notebook contains the code samples found in Chapter 3, Section 5 of [Deep Learning with Python](https://www.manning.com/books/deep-learning-with-python?a_aid=keras&a_bid=76564dff). Note that the original text features far more content, in particular further explanations and figures: in this notebook, you will only find source code and related comments.\n\n----\n\nIn the previous section we saw how to classify vector inputs into two mutually exclusive classes using a densely-connected neural network. \nBut what happens when you have more than two classes? \n\nIn this section, we will build a network to classify Reuters newswires into 46 different mutually-exclusive topics. Since we have many \nclasses, this problem is an instance of \"multi-class classification\", and since each data point should be classified into only one \ncategory, the problem is more specifically an instance of \"single-label, multi-class classification\". If each data point could have \nbelonged to multiple categories (in our case, topics) then we would be facing a \"multi-label, multi-class classification\" problem.",
"_____no_output_____"
],
[
"## The Reuters dataset\n\n\nWe will be working with the _Reuters dataset_, a set of short newswires and their topics, published by Reuters in 1986. It's a very simple, \nwidely used toy dataset for text classification. There are 46 different topics; some topics are more represented than others, but each \ntopic has at least 10 examples in the training set.\n\nLike IMDB and MNIST, the Reuters dataset comes packaged as part of Keras. Let's take a look right away:",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras.datasets import reuters\n\n(train_data, train_labels), (test_data, test_labels) = reuters.load_data(num_words=10000)",
"_____no_output_____"
]
],
[
[
"\nLike with the IMDB dataset, the argument `num_words=10000` restricts the data to the 10,000 most frequently occurring words found in the \ndata.\n\nWe have 8,982 training examples and 2,246 test examples:",
"_____no_output_____"
]
],
[
[
"len(train_data)",
"_____no_output_____"
],
[
"len(test_data)",
"_____no_output_____"
]
],
[
[
"As with the IMDB reviews, each example is a list of integers (word indices):",
"_____no_output_____"
]
],
[
[
"train_data[10]",
"_____no_output_____"
]
],
[
[
"Here's how you can decode it back to words, in case you are curious:",
"_____no_output_____"
]
],
[
[
"word_index = reuters.get_word_index()\nreverse_word_index = dict([(value, key) for (key, value) in word_index.items()])\n# Note that our indices were offset by 3\n# because 0, 1 and 2 are reserved indices for \"padding\", \"start of sequence\", and \"unknown\".\ndecoded_newswire = ' '.join([reverse_word_index.get(i - 3, '?') for i in train_data[0]])",
"_____no_output_____"
],
[
"decoded_newswire",
"_____no_output_____"
]
],
[
[
"The label associated with an example is an integer between 0 and 45: a topic index.",
"_____no_output_____"
]
],
[
[
"train_labels[10]",
"_____no_output_____"
]
],
[
[
"## Preparing the data\n\nWe can vectorize the data with the exact same code as in our previous example:",
"_____no_output_____"
]
],
[
[
"import numpy as np\n\ndef vectorize_sequences(sequences, dimension=10000):\n results = np.zeros((len(sequences), dimension))\n for i, sequence in enumerate(sequences):\n results[i, sequence] = 1.\n return results\n\n# Our vectorized training data\nx_train = vectorize_sequences(train_data)\n# Our vectorized test data\nx_test = vectorize_sequences(test_data)",
"_____no_output_____"
]
],
[
[
"\nTo vectorize the labels, there are two possibilities: we could just cast the label list as an integer tensor, or we could use a \"one-hot\" \nencoding. One-hot encoding is a widely used format for categorical data, also called \"categorical encoding\". \nFor a more detailed explanation of one-hot encoding, you can refer to Chapter 6, Section 1. \nIn our case, one-hot encoding of our labels consists in embedding each label as an all-zero vector with a 1 in the place of the label index, e.g.:",
"_____no_output_____"
]
],
[
[
"def to_one_hot(labels, dimension=46):\n results = np.zeros((len(labels), dimension))\n for i, label in enumerate(labels):\n results[i, label] = 1.\n return results\n\n# Our vectorized training labels\none_hot_train_labels = to_one_hot(train_labels)\n# Our vectorized test labels\none_hot_test_labels = to_one_hot(test_labels)",
"_____no_output_____"
]
],
[
[
"Note that there is a built-in way to do this in Keras, which you have already seen in action in our MNIST example:",
"_____no_output_____"
]
],
[
[
"\nmnist = tf.keras.datasets.mnist\nfrom tensorflow.keras.utils import to_categorical\n\none_hot_train_labels = to_categorical(train_labels)\none_hot_test_labels = to_categorical(test_labels)",
"_____no_output_____"
]
],
[
[
"## Building our network\n\n\nThis topic classification problem looks very similar to our previous movie review classification problem: in both cases, we are trying to \nclassify short snippets of text. There is however a new constraint here: the number of output classes has gone from 2 to 46, i.e. the \ndimensionality of the output space is much larger. \n\nIn a stack of `Dense` layers like what we were using, each layer can only access information present in the output of the previous layer. \nIf one layer drops some information relevant to the classification problem, this information can never be recovered by later layers: each \nlayer can potentially become an \"information bottleneck\". In our previous example, we were using 16-dimensional intermediate layers, but a \n16-dimensional space may be too limited to learn to separate 46 different classes: such small layers may act as information bottlenecks, \npermanently dropping relevant information.\n\nFor this reason we will use larger layers. Let's go with 64 units:",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras import models\nfrom tensorflow.keras import layers\n\nmodel = models.Sequential()\nmodel.add(layers.Dense(64, activation='relu', input_shape=(10000,)))\nmodel.add(layers.Dense(64, activation='relu'))\nmodel.add(layers.Dense(46, activation='softmax'))",
"_____no_output_____"
]
],
[
[
"\nThere are two other things you should note about this architecture:\n\n* We are ending the network with a `Dense` layer of size 46. This means that for each input sample, our network will output a \n46-dimensional vector. Each entry in this vector (each dimension) will encode a different output class.\n* The last layer uses a `softmax` activation. You have already seen this pattern in the MNIST example. It means that the network will \noutput a _probability distribution_ over the 46 different output classes, i.e. for every input sample, the network will produce a \n46-dimensional output vector where `output[i]` is the probability that the sample belongs to class `i`. The 46 scores will sum to 1.\n\nThe best loss function to use in this case is `categorical_crossentropy`. It measures the distance between two probability distributions: \nin our case, between the probability distribution output by our network, and the true distribution of the labels. By minimizing the \ndistance between these two distributions, we train our network to output something as close as possible to the true labels.",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='rmsprop',\n loss='categorical_crossentropy',\n metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"## Validating our approach\n\nLet's set apart 1,000 samples in our training data to use as a validation set:",
"_____no_output_____"
]
],
[
[
"x_val = x_train[:1000]\npartial_x_train = x_train[1000:]\n\ny_val = one_hot_train_labels[:1000]\npartial_y_train = one_hot_train_labels[1000:]",
"_____no_output_____"
]
],
[
[
"Now let's train our network for 20 epochs:",
"_____no_output_____"
]
],
[
[
"history = model.fit(partial_x_train,\n partial_y_train,\n epochs=20,\n batch_size=512,\n validation_data=(x_val, y_val))",
"Train on 7982 samples, validate on 1000 samples\nEpoch 1/20\n7982/7982 [==============================] - 1s 168us/sample - loss: 2.5268 - accuracy: 0.5284 - val_loss: 1.6720 - val_accuracy: 0.6430\nEpoch 2/20\n7982/7982 [==============================] - 0s 56us/sample - loss: 1.3827 - accuracy: 0.7052 - val_loss: 1.2857 - val_accuracy: 0.7230\nEpoch 3/20\n7982/7982 [==============================] - 0s 54us/sample - loss: 1.0404 - accuracy: 0.7772 - val_loss: 1.1341 - val_accuracy: 0.7650\nEpoch 4/20\n7982/7982 [==============================] - 0s 53us/sample - loss: 0.8292 - accuracy: 0.8262 - val_loss: 1.0505 - val_accuracy: 0.7680\nEpoch 5/20\n7982/7982 [==============================] - 0s 55us/sample - loss: 0.6613 - accuracy: 0.8577 - val_loss: 0.9738 - val_accuracy: 0.7920\nEpoch 6/20\n7982/7982 [==============================] - 0s 54us/sample - loss: 0.5286 - accuracy: 0.8857 - val_loss: 0.9257 - val_accuracy: 0.8070\nEpoch 7/20\n7982/7982 [==============================] - 0s 51us/sample - loss: 0.4248 - accuracy: 0.9105 - val_loss: 0.8988 - val_accuracy: 0.8150\nEpoch 8/20\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.3460 - accuracy: 0.9287 - val_loss: 0.9759 - val_accuracy: 0.7860\nEpoch 9/20\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.2816 - accuracy: 0.9400 - val_loss: 0.9086 - val_accuracy: 0.8100\nEpoch 10/20\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.2406 - accuracy: 0.9450 - val_loss: 0.8921 - val_accuracy: 0.8260\nEpoch 11/20\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.2044 - accuracy: 0.9489 - val_loss: 0.9256 - val_accuracy: 0.8200\nEpoch 12/20\n7982/7982 [==============================] - 0s 53us/sample - loss: 0.1814 - accuracy: 0.9523 - val_loss: 0.9101 - val_accuracy: 0.8230\nEpoch 13/20\n7982/7982 [==============================] - 0s 54us/sample - loss: 0.1633 - accuracy: 0.9541 - val_loss: 0.9552 - val_accuracy: 0.8070\nEpoch 14/20\n7982/7982 [==============================] - 0s 53us/sample - loss: 0.1458 - accuracy: 0.9555 - val_loss: 0.9987 - val_accuracy: 0.8040\nEpoch 15/20\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.1384 - accuracy: 0.9558 - val_loss: 1.0230 - val_accuracy: 0.8080\nEpoch 16/20\n7982/7982 [==============================] - 0s 55us/sample - loss: 0.1331 - accuracy: 0.9575 - val_loss: 1.0297 - val_accuracy: 0.8100\nEpoch 17/20\n7982/7982 [==============================] - 0s 56us/sample - loss: 0.1270 - accuracy: 0.9559 - val_loss: 1.0427 - val_accuracy: 0.8060\nEpoch 18/20\n7982/7982 [==============================] - 0s 56us/sample - loss: 0.1211 - accuracy: 0.9570 - val_loss: 1.0430 - val_accuracy: 0.8100\nEpoch 19/20\n7982/7982 [==============================] - 0s 56us/sample - loss: 0.1177 - accuracy: 0.9563 - val_loss: 1.0695 - val_accuracy: 0.8060\nEpoch 20/20\n7982/7982 [==============================] - 0s 55us/sample - loss: 0.1122 - accuracy: 0.9564 - val_loss: 1.1296 - val_accuracy: 0.7940\n"
]
],
[
[
"Let's display its loss and accuracy curves:",
"_____no_output_____"
]
],
[
[
"history_dict = history.history\nhistory_dict.keys()",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\n\nloss = history.history['loss']\nval_loss = history.history['val_loss']\n\nepochs = range(1, len(loss) + 1)\n\nplt.plot(epochs, loss, 'bo', label='Training loss')\nplt.plot(epochs, val_loss, 'b', label='Validation loss')\nplt.title('Training and validation loss')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
],
[
"plt.clf() # clear figure\n\nacc = history.history['accuracy']\nval_acc = history.history['val_accuracy']\n\nplt.plot(epochs, acc, 'bo', label='Training acc')\nplt.plot(epochs, val_acc, 'b', label='Validation acc')\nplt.title('Training and validation accuracy')\nplt.xlabel('Epochs')\nplt.ylabel('Loss')\nplt.legend()\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"It seems that the network starts overfitting after 8 epochs. Let's train a new network from scratch for 8 epochs, then let's evaluate it on \nthe test set:",
"_____no_output_____"
]
],
[
[
"model = models.Sequential()\nmodel.add(layers.Dense(64, activation='relu', input_shape=(10000,)))\nmodel.add(layers.Dense(64, activation='relu'))\nmodel.add(layers.Dense(46, activation='softmax'))\n\nmodel.compile(optimizer='rmsprop',\n loss='categorical_crossentropy',\n metrics=['accuracy'])\nmodel.fit(partial_x_train,\n partial_y_train,\n epochs=8,\n batch_size=512,\n validation_data=(x_val, y_val))\nresults = model.evaluate(x_test, one_hot_test_labels)",
"Train on 7982 samples, validate on 1000 samples\nEpoch 1/8\n7982/7982 [==============================] - 1s 145us/sample - loss: 2.5926 - accuracy: 0.5272 - val_loss: 1.6818 - val_accuracy: 0.6490\nEpoch 2/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 1.4117 - accuracy: 0.7060 - val_loss: 1.3144 - val_accuracy: 0.7140\nEpoch 3/8\n7982/7982 [==============================] - 0s 51us/sample - loss: 1.0550 - accuracy: 0.7756 - val_loss: 1.1398 - val_accuracy: 0.7520\nEpoch 4/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.8316 - accuracy: 0.8262 - val_loss: 1.0569 - val_accuracy: 0.7710\nEpoch 5/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.6631 - accuracy: 0.8608 - val_loss: 0.9863 - val_accuracy: 0.8000\nEpoch 6/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.5309 - accuracy: 0.8884 - val_loss: 0.9567 - val_accuracy: 0.7970\nEpoch 7/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.4300 - accuracy: 0.9085 - val_loss: 0.9111 - val_accuracy: 0.8150\nEpoch 8/8\n7982/7982 [==============================] - 0s 52us/sample - loss: 0.3474 - accuracy: 0.9265 - val_loss: 0.9755 - val_accuracy: 0.7860\n2246/1 [====================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================================] - 0s 66us/sample - loss: 1.8451 - accuracy: 0.7600\n"
],
[
"results",
"_____no_output_____"
]
],
[
[
"\nOur approach reaches an accuracy of ~78%. With a balanced binary classification problem, the accuracy reached by a purely random classifier \nwould be 50%, but in our case it is closer to 19%, so our results seem pretty good, at least when compared to a random baseline:",
"_____no_output_____"
]
],
[
[
"import copy\n\ntest_labels_copy = copy.copy(test_labels)\nnp.random.shuffle(test_labels_copy)\nfloat(np.sum(np.array(test_labels) == np.array(test_labels_copy))) / len(test_labels)",
"_____no_output_____"
]
],
[
[
"## Generating predictions on new data\n\nWe can verify that the `predict` method of our model instance returns a probability distribution over all 46 topics. Let's generate topic \npredictions for all of the test data:",
"_____no_output_____"
]
],
[
[
"predictions = model.predict(x_test)",
"_____no_output_____"
]
],
[
[
"Each entry in `predictions` is a vector of length 46:",
"_____no_output_____"
]
],
[
[
"predictions[0].shape",
"_____no_output_____"
]
],
[
[
"The coefficients in this vector sum to 1:",
"_____no_output_____"
]
],
[
[
"np.sum(predictions[0])",
"_____no_output_____"
]
],
[
[
"The largest entry is the predicted class, i.e. the class with the highest probability:",
"_____no_output_____"
]
],
[
[
"np.argmax(predictions[0])",
"_____no_output_____"
]
],
[
[
"## A different way to handle the labels and the loss\n\nWe mentioned earlier that another way to encode the labels would be to cast them as an integer tensor, like such:",
"_____no_output_____"
]
],
[
[
"y_train = np.array(train_labels)\ny_test = np.array(test_labels)",
"_____no_output_____"
]
],
[
[
"\nThe only thing it would change is the choice of the loss function. Our previous loss, `categorical_crossentropy`, expects the labels to \nfollow a categorical encoding. With integer labels, we should use `sparse_categorical_crossentropy`:",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='rmsprop', loss='sparse_categorical_crossentropy', metrics=['acc'])",
"_____no_output_____"
]
],
[
[
"This new loss function is still mathematically the same as `categorical_crossentropy`; it just has a different interface.",
"_____no_output_____"
],
[
"## On the importance of having sufficiently large intermediate layers\n\n\nWe mentioned earlier that since our final outputs were 46-dimensional, we should avoid intermediate layers with much less than 46 hidden \nunits. Now let's try to see what happens when we introduce an information bottleneck by having intermediate layers significantly less than \n46-dimensional, e.g. 4-dimensional.",
"_____no_output_____"
]
],
[
[
"model = models.Sequential()\nmodel.add(layers.Dense(64, activation='relu', input_shape=(10000,)))\nmodel.add(layers.Dense(4, activation='relu'))\nmodel.add(layers.Dense(46, activation='softmax'))\n\nmodel.compile(optimizer='rmsprop',\n loss='categorical_crossentropy',\n metrics=['accuracy'])\nmodel.fit(partial_x_train,\n partial_y_train,\n epochs=20,\n batch_size=128,\n validation_data=(x_val, y_val))",
"Train on 7982 samples, validate on 1000 samples\nEpoch 1/20\n7982/7982 [==============================] - 1s 167us/sample - loss: 2.9123 - accuracy: 0.3306 - val_loss: 2.1964 - val_accuracy: 0.4790\nEpoch 2/20\n7982/7982 [==============================] - 1s 77us/sample - loss: 1.8067 - accuracy: 0.5678 - val_loss: 1.6553 - val_accuracy: 0.5810\nEpoch 3/20\n7982/7982 [==============================] - 1s 78us/sample - loss: 1.4744 - accuracy: 0.5862 - val_loss: 1.5329 - val_accuracy: 0.5860\nEpoch 4/20\n7982/7982 [==============================] - 1s 83us/sample - loss: 1.3201 - accuracy: 0.6204 - val_loss: 1.4763 - val_accuracy: 0.6100\nEpoch 5/20\n7982/7982 [==============================] - 1s 75us/sample - loss: 1.2072 - accuracy: 0.6590 - val_loss: 1.4393 - val_accuracy: 0.6530\nEpoch 6/20\n7982/7982 [==============================] - 1s 75us/sample - loss: 1.1089 - accuracy: 0.7180 - val_loss: 1.4372 - val_accuracy: 0.6770\nEpoch 7/20\n7982/7982 [==============================] - 1s 85us/sample - loss: 1.0263 - accuracy: 0.7373 - val_loss: 1.4226 - val_accuracy: 0.6790\nEpoch 8/20\n7982/7982 [==============================] - 1s 86us/sample - loss: 0.9614 - accuracy: 0.7506 - val_loss: 1.4515 - val_accuracy: 0.6820\nEpoch 9/20\n7982/7982 [==============================] - 1s 81us/sample - loss: 0.9059 - accuracy: 0.7595 - val_loss: 1.4760 - val_accuracy: 0.6760\nEpoch 10/20\n1536/7982 [====>.........................] - ETA: 0s - loss: 0.8665 - accuracy: 0.7676"
]
],
[
[
"\nOur network now seems to peak at ~71% test accuracy, a 8% absolute drop. This drop is mostly due to the fact that we are now trying to \ncompress a lot of information (enough information to recover the separation hyperplanes of 46 classes) into an intermediate space that is \ntoo low-dimensional. The network is able to cram _most_ of the necessary information into these 8-dimensional representations, but not all \nof it.",
"_____no_output_____"
],
[
"## Further experiments\n\n* Try using larger or smaller layers: 32 units, 128 units...\n* We were using two hidden layers. Now try to use a single hidden layer, or three hidden layers.",
"_____no_output_____"
],
[
"## Wrapping up\n\n\nHere's what you should take away from this example:\n\n* If you are trying to classify data points between N classes, your network should end with a `Dense` layer of size N.\n* In a single-label, multi-class classification problem, your network should end with a `softmax` activation, so that it will output a \nprobability distribution over the N output classes.\n* _Categorical crossentropy_ is almost always the loss function you should use for such problems. It minimizes the distance between the \nprobability distributions output by the network, and the true distribution of the targets.\n* There are two ways to handle labels in multi-class classification:\n ** Encoding the labels via \"categorical encoding\" (also known as \"one-hot encoding\") and using `categorical_crossentropy` as your loss \nfunction.\n ** Encoding the labels as integers and using the `sparse_categorical_crossentropy` loss function.\n* If you need to classify data into a large number of categories, then you should avoid creating information bottlenecks in your network by having \nintermediate layers that are too small.",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec6f097e8851c772c15d5fb9d96f1999048ef570 | 238,199 | ipynb | Jupyter Notebook | 0621_student.ipynb | eunanomist/eunanomist.github.com | 7a484034c1355ff01f66f8d0c5b9777f7e79bd90 | [
"MIT"
]
| null | null | null | 0621_student.ipynb | eunanomist/eunanomist.github.com | 7a484034c1355ff01f66f8d0c5b9777f7e79bd90 | [
"MIT"
]
| null | null | null | 0621_student.ipynb | eunanomist/eunanomist.github.com | 7a484034c1355ff01f66f8d0c5b9777f7e79bd90 | [
"MIT"
]
| null | null | null | 238,199 | 238,199 | 0.676237 | [
[
[
"from google.colab import drive\ndrive.mount('/content/drive/')",
"Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3aietf%3awg%3aoauth%3a2.0%3aoob&response_type=code&scope=email%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdocs.test%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive%20https%3a%2f%2fwww.googleapis.com%2fauth%2fdrive.photos.readonly%20https%3a%2f%2fwww.googleapis.com%2fauth%2fpeopleapi.readonly\n\nEnter your authorization code:\n··········\nMounted at /content/drive/\n"
],
[
"import os\nos.chdir('/content/drive/My Drive/PKD')",
"_____no_output_____"
],
[
"## PKD git 가져오기\n\n#!git clone https://github.com/intersun/PKD-for-BERT-Model-Compression.git",
"_____no_output_____"
],
[
"## Torch install\n\n!conda install pytorch torchvision cudatoolkit=10.0 -c pytorch\n!pip install -r requirements.txt",
"/bin/bash: conda: command not found\nRequirement already satisfied: tqdm in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 2)) (4.41.1)\nRequirement already satisfied: boto3 in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 4)) (1.14.5)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 6)) (2.23.0)\nRequirement already satisfied: regex in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 8)) (2019.12.20)\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 10)) (1.0.5)\nRequirement already satisfied: matplotlib in /usr/local/lib/python3.6/dist-packages (from -r requirements.txt (line 11)) (3.2.2)\nRequirement already satisfied: botocore<1.18.0,>=1.17.5 in /usr/local/lib/python3.6/dist-packages (from boto3->-r requirements.txt (line 4)) (1.17.5)\nRequirement already satisfied: jmespath<1.0.0,>=0.7.1 in /usr/local/lib/python3.6/dist-packages (from boto3->-r requirements.txt (line 4)) (0.10.0)\nRequirement already satisfied: s3transfer<0.4.0,>=0.3.0 in /usr/local/lib/python3.6/dist-packages (from boto3->-r requirements.txt (line 4)) (0.3.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->-r requirements.txt (line 6)) (2020.4.5.2)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->-r requirements.txt (line 6)) (3.0.4)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->-r requirements.txt (line 6)) (2.9)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->-r requirements.txt (line 6)) (1.24.3)\nRequirement already satisfied: python-dateutil>=2.6.1 in /usr/local/lib/python3.6/dist-packages (from pandas->-r requirements.txt (line 10)) (2.8.1)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->-r requirements.txt (line 10)) (2018.9)\nRequirement already satisfied: numpy>=1.13.3 in /usr/local/lib/python3.6/dist-packages (from pandas->-r requirements.txt (line 10)) (1.18.5)\nRequirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.6/dist-packages (from matplotlib->-r requirements.txt (line 11)) (0.10.0)\nRequirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib->-r requirements.txt (line 11)) (2.4.7)\nRequirement already satisfied: kiwisolver>=1.0.1 in /usr/local/lib/python3.6/dist-packages (from matplotlib->-r requirements.txt (line 11)) (1.2.0)\nRequirement already satisfied: docutils<0.16,>=0.10 in /usr/local/lib/python3.6/dist-packages (from botocore<1.18.0,>=1.17.5->boto3->-r requirements.txt (line 4)) (0.15.2)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil>=2.6.1->pandas->-r requirements.txt (line 10)) (1.12.0)\n"
],
[
"!nvcc --version\n!python --version",
"nvcc: NVIDIA (R) Cuda compiler driver\nCopyright (c) 2005-2019 NVIDIA Corporation\nBuilt on Sun_Jul_28_19:07:16_PDT_2019\nCuda compilation tools, release 10.1, V10.1.243\nPython 3.6.9\n"
],
[
"# Apex install\n\n!git clone https://github.com/NVIDIA/apex",
"fatal: destination path 'apex' already exists and is not an empty directory.\n"
],
[
"!pip install -v --no-cache-dir --global-option=\"--cpp_ext\" --global-option=\"--cuda_ext\" ./apex",
"/usr/local/lib/python3.6/dist-packages/pip/_internal/commands/install.py:283: UserWarning: Disabling all use of wheels due to the use of --build-options / --global-options / --install-options.\n cmdoptions.check_install_build_global(options)\nCreated temporary directory: /tmp/pip-ephem-wheel-cache-wafpyosr\nCreated temporary directory: /tmp/pip-req-tracker-ef_x9t6i\nCreated requirements tracker '/tmp/pip-req-tracker-ef_x9t6i'\nCreated temporary directory: /tmp/pip-install-jblmr46c\nProcessing ./apex\n Created temporary directory: /tmp/pip-req-build-awlakocq\n Added file:///content/drive/My%20Drive/PKD/apex to build tracker '/tmp/pip-req-tracker-ef_x9t6i'\n Running setup.py (path:/tmp/pip-req-build-awlakocq/setup.py) egg_info for package from file:///content/drive/My%20Drive/PKD/apex\n Running command python setup.py egg_info\n\n\n torch.__version__ = 1.5.1+cu101\n\n\n running egg_info\n creating /tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info\n writing /tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info/PKG-INFO\n writing dependency_links to /tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info/dependency_links.txt\n writing top-level names to /tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info/top_level.txt\n writing manifest file '/tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info/SOURCES.txt'\n writing manifest file '/tmp/pip-req-build-awlakocq/pip-egg-info/apex.egg-info/SOURCES.txt'\n /tmp/pip-req-build-awlakocq/setup.py:51: UserWarning: Option --pyprof not specified. Not installing PyProf dependencies!\n warnings.warn(\"Option --pyprof not specified. Not installing PyProf dependencies!\")\n Source in /tmp/pip-req-build-awlakocq has version 0.1, which satisfies requirement apex==0.1 from file:///content/drive/My%20Drive/PKD/apex\n Removed apex==0.1 from file:///content/drive/My%20Drive/PKD/apex from build tracker '/tmp/pip-req-tracker-ef_x9t6i'\nSkipping wheel build for apex, due to binaries being disabled for it.\nInstalling collected packages: apex\n Created temporary directory: /tmp/pip-record-6u0omsn_\n Running command /usr/bin/python3 -u -c 'import sys, setuptools, tokenize; sys.argv[0] = '\"'\"'/tmp/pip-req-build-awlakocq/setup.py'\"'\"'; __file__='\"'\"'/tmp/pip-req-build-awlakocq/setup.py'\"'\"';f=getattr(tokenize, '\"'\"'open'\"'\"', open)(__file__);code=f.read().replace('\"'\"'\\r\\n'\"'\"', '\"'\"'\\n'\"'\"');f.close();exec(compile(code, __file__, '\"'\"'exec'\"'\"'))' --cpp_ext --cuda_ext install --record /tmp/pip-record-6u0omsn_/install-record.txt --single-version-externally-managed --compile\n\n\n torch.__version__ = 1.5.1+cu101\n\n\n /tmp/pip-req-build-awlakocq/setup.py:51: UserWarning: Option --pyprof not specified. Not installing PyProf dependencies!\n warnings.warn(\"Option --pyprof not specified. Not installing PyProf dependencies!\")\n\n Compiling cuda extensions with\n nvcc: NVIDIA (R) Cuda compiler driver\n Copyright (c) 2005-2019 NVIDIA Corporation\n Built on Sun_Jul_28_19:07:16_PDT_2019\n Cuda compilation tools, release 10.1, V10.1.243\n from /usr/local/cuda/bin\n\n running install\n running build\n running build_py\n creating build\n creating build/lib.linux-x86_64-3.6\n creating build/lib.linux-x86_64-3.6/apex\n copying apex/__init__.py -> build/lib.linux-x86_64-3.6/apex\n creating build/lib.linux-x86_64-3.6/apex/RNN\n copying apex/RNN/models.py -> build/lib.linux-x86_64-3.6/apex/RNN\n copying apex/RNN/RNNBackend.py -> build/lib.linux-x86_64-3.6/apex/RNN\n copying apex/RNN/__init__.py -> build/lib.linux-x86_64-3.6/apex/RNN\n copying apex/RNN/cells.py -> build/lib.linux-x86_64-3.6/apex/RNN\n creating build/lib.linux-x86_64-3.6/apex/multi_tensor_apply\n copying apex/multi_tensor_apply/multi_tensor_apply.py -> build/lib.linux-x86_64-3.6/apex/multi_tensor_apply\n copying apex/multi_tensor_apply/__init__.py -> build/lib.linux-x86_64-3.6/apex/multi_tensor_apply\n creating build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/sync_batchnorm.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/LARC.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/distributed.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/multiproc.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/optimized_sync_batchnorm_kernel.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/__init__.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/sync_batchnorm_kernel.py -> build/lib.linux-x86_64-3.6/apex/parallel\n copying apex/parallel/optimized_sync_batchnorm.py -> build/lib.linux-x86_64-3.6/apex/parallel\n creating build/lib.linux-x86_64-3.6/apex/fp16_utils\n copying apex/fp16_utils/fp16_optimizer.py -> build/lib.linux-x86_64-3.6/apex/fp16_utils\n copying apex/fp16_utils/fp16util.py -> build/lib.linux-x86_64-3.6/apex/fp16_utils\n copying apex/fp16_utils/loss_scaler.py -> build/lib.linux-x86_64-3.6/apex/fp16_utils\n copying apex/fp16_utils/__init__.py -> build/lib.linux-x86_64-3.6/apex/fp16_utils\n creating build/lib.linux-x86_64-3.6/apex/pyprof\n copying apex/pyprof/__init__.py -> build/lib.linux-x86_64-3.6/apex/pyprof\n creating build/lib.linux-x86_64-3.6/apex/normalization\n copying apex/normalization/__init__.py -> build/lib.linux-x86_64-3.6/apex/normalization\n copying apex/normalization/fused_layer_norm.py -> build/lib.linux-x86_64-3.6/apex/normalization\n creating build/lib.linux-x86_64-3.6/apex/contrib\n copying apex/contrib/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib\n creating build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/compat.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/handle.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/__version__.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/_amp_state.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/rnn_compat.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/_process_optimizer.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/__init__.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/scaler.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/_initialize.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/opt.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/amp.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/frontend.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/utils.py -> build/lib.linux-x86_64-3.6/apex/amp\n copying apex/amp/wrap.py -> build/lib.linux-x86_64-3.6/apex/amp\n creating build/lib.linux-x86_64-3.6/apex/mlp\n copying apex/mlp/mlp.py -> build/lib.linux-x86_64-3.6/apex/mlp\n copying apex/mlp/__init__.py -> build/lib.linux-x86_64-3.6/apex/mlp\n creating build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/fused_adam.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/fused_lamb.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/fused_sgd.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/fused_novograd.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/__init__.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n copying apex/optimizers/fused_adagrad.py -> build/lib.linux-x86_64-3.6/apex/optimizers\n creating build/lib.linux-x86_64-3.6/apex/reparameterization\n copying apex/reparameterization/reparameterization.py -> build/lib.linux-x86_64-3.6/apex/reparameterization\n copying apex/reparameterization/__init__.py -> build/lib.linux-x86_64-3.6/apex/reparameterization\n copying apex/reparameterization/weight_norm.py -> build/lib.linux-x86_64-3.6/apex/reparameterization\n creating build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/linear.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/index_slice_join_mutate.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/randomSample.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/base.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/loss.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/dropout.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/blas.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/embedding.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/optim.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/conv.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/misc.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/utility.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/prof.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/convert.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/data.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/usage.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/__init__.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/pooling.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/output.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/activation.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/recurrentCell.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/pointwise.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/softmax.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/normalization.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/reduction.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n copying apex/pyprof/prof/__main__.py -> build/lib.linux-x86_64-3.6/apex/pyprof/prof\n creating build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/db.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/kernel.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/parse.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/__init__.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/nvvp.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n copying apex/pyprof/parse/__main__.py -> build/lib.linux-x86_64-3.6/apex/pyprof/parse\n creating build/lib.linux-x86_64-3.6/apex/pyprof/nvtx\n copying apex/pyprof/nvtx/nvmarker.py -> build/lib.linux-x86_64-3.6/apex/pyprof/nvtx\n copying apex/pyprof/nvtx/__init__.py -> build/lib.linux-x86_64-3.6/apex/pyprof/nvtx\n creating build/lib.linux-x86_64-3.6/apex/contrib/sparsity\n copying apex/contrib/sparsity/asp.py -> build/lib.linux-x86_64-3.6/apex/contrib/sparsity\n copying apex/contrib/sparsity/sparse_masklib.py -> build/lib.linux-x86_64-3.6/apex/contrib/sparsity\n copying apex/contrib/sparsity/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib/sparsity\n creating build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/self_multihead_attn.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/fast_self_multihead_attn_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/encdec_multihead_attn_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/self_multihead_attn_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/fast_encdec_multihead_attn_norm_add_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/fast_encdec_multihead_attn_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/fast_self_multihead_attn_norm_add_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/encdec_multihead_attn.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n copying apex/contrib/multihead_attn/mask_softmax_dropout_func.py -> build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn\n creating build/lib.linux-x86_64-3.6/apex/contrib/groupbn\n copying apex/contrib/groupbn/batch_norm.py -> build/lib.linux-x86_64-3.6/apex/contrib/groupbn\n copying apex/contrib/groupbn/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib/groupbn\n creating build/lib.linux-x86_64-3.6/apex/contrib/xentropy\n copying apex/contrib/xentropy/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib/xentropy\n copying apex/contrib/xentropy/softmax_xentropy.py -> build/lib.linux-x86_64-3.6/apex/contrib/xentropy\n creating build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/fused_adam.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/fp16_optimizer.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/distributed_fused_adam.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/fused_lamb.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/distributed_fused_adam_v2.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/fused_sgd.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/__init__.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/distributed_fused_adam_v3.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n copying apex/contrib/optimizers/distributed_fused_lamb.py -> build/lib.linux-x86_64-3.6/apex/contrib/optimizers\n creating build/lib.linux-x86_64-3.6/apex/amp/lists\n copying apex/amp/lists/torch_overrides.py -> build/lib.linux-x86_64-3.6/apex/amp/lists\n copying apex/amp/lists/functional_overrides.py -> build/lib.linux-x86_64-3.6/apex/amp/lists\n copying apex/amp/lists/__init__.py -> build/lib.linux-x86_64-3.6/apex/amp/lists\n copying apex/amp/lists/tensor_overrides.py -> build/lib.linux-x86_64-3.6/apex/amp/lists\n running build_ext\n /usr/local/lib/python3.6/dist-packages/torch/utils/cpp_extension.py:305: UserWarning: Attempted to use ninja as the BuildExtension backend but we could not find ninja.. Falling back to using the slow distutils backend.\n warnings.warn(msg.format('we could not find ninja.'))\n building 'apex_C' extension\n creating build/temp.linux-x86_64-3.6\n creating build/temp.linux-x86_64-3.6/csrc\n x86_64-linux-gnu-gcc -pthread -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/include/python3.6m -c csrc/flatten_unflatten.cpp -o build/temp.linux-x86_64-3.6/csrc/flatten_unflatten.o -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=apex_C -D_GLIBCXX_USE_CXX11_ABI=0 -std=c++14\n In file included from csrc/flatten_unflatten.cpp:2:0:\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/utils/tensor_flatten.h: In member function ‘at::DeprecatedTypeProperties& torch::utils::TensorGroup::type()’:\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/utils/tensor_flatten.h:36:28: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n return tensors[0].type();\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/flatten_unflatten.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n x86_64-linux-gnu-g++ -pthread -shared -Wl,-O1 -Wl,-Bsymbolic-functions -Wl,-Bsymbolic-functions -Wl,-z,relro -Wl,-Bsymbolic-functions -Wl,-z,relro -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 build/temp.linux-x86_64-3.6/csrc/flatten_unflatten.o -L/usr/local/lib/python3.6/dist-packages/torch/lib -lc10 -ltorch -ltorch_cpu -ltorch_python -o build/lib.linux-x86_64-3.6/apex_C.cpython-36m-x86_64-linux-gnu.so\n building 'amp_C' extension\n x86_64-linux-gnu-gcc -pthread -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/amp_C_frontend.cpp -o build/temp.linux-x86_64-3.6/csrc/amp_C_frontend.o -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_sgd_kernel.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_sgd_kernel.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_scale_kernel.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_scale_kernel.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_axpby_kernel.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_axpby_kernel.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_l2norm_kernel.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_l2norm_kernel.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_lamb_stage_1.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb_stage_1.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_lamb_stage_2.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb_stage_2.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_adam.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_adam.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_adagrad.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_adagrad.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_novograd.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_novograd.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/multi_tensor_lamb.cu -o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -lineinfo -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=amp_C -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n x86_64-linux-gnu-g++ -pthread -shared -Wl,-O1 -Wl,-Bsymbolic-functions -Wl,-Bsymbolic-functions -Wl,-z,relro -Wl,-Bsymbolic-functions -Wl,-z,relro -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 build/temp.linux-x86_64-3.6/csrc/amp_C_frontend.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_sgd_kernel.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_scale_kernel.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_axpby_kernel.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_l2norm_kernel.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb_stage_1.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb_stage_2.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_adam.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_adagrad.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_novograd.o build/temp.linux-x86_64-3.6/csrc/multi_tensor_lamb.o -L/usr/local/lib/python3.6/dist-packages/torch/lib -L/usr/local/cuda/lib64 -lc10 -ltorch -ltorch_cpu -ltorch_python -lcudart -lc10_cuda -ltorch_cuda -o build/lib.linux-x86_64-3.6/amp_C.cpython-36m-x86_64-linux-gnu.so\n building 'syncbn' extension\n x86_64-linux-gnu-gcc -pthread -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/syncbn.cpp -o build/temp.linux-x86_64-3.6/csrc/syncbn.o -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=syncbn -D_GLIBCXX_USE_CXX11_ABI=0 -std=c++14\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/welford.cu -o build/temp.linux-x86_64-3.6/csrc/welford.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=syncbn -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n x86_64-linux-gnu-g++ -pthread -shared -Wl,-O1 -Wl,-Bsymbolic-functions -Wl,-Bsymbolic-functions -Wl,-z,relro -Wl,-Bsymbolic-functions -Wl,-z,relro -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 build/temp.linux-x86_64-3.6/csrc/syncbn.o build/temp.linux-x86_64-3.6/csrc/welford.o -L/usr/local/lib/python3.6/dist-packages/torch/lib -L/usr/local/cuda/lib64 -lc10 -ltorch -ltorch_cpu -ltorch_python -lcudart -lc10_cuda -ltorch_cuda -o build/lib.linux-x86_64-3.6/syncbn.cpython-36m-x86_64-linux-gnu.so\n building 'fused_layer_norm_cuda' extension\n x86_64-linux-gnu-gcc -pthread -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/layer_norm_cuda.cpp -o build/temp.linux-x86_64-3.6/csrc/layer_norm_cuda.o -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=fused_layer_norm_cuda -D_GLIBCXX_USE_CXX11_ABI=0 -std=c++14\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp: In function ‘std::vector<at::Tensor> layer_norm(at::Tensor, c10::IntArrayRef, double)’:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:129:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(input);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp: In function ‘std::vector<at::Tensor> layer_norm_affine(at::Tensor, c10::IntArrayRef, at::Tensor, at::Tensor, double)’:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:149:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(input);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:150:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(gamma);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:151:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(beta);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp: In function ‘at::Tensor layer_norm_gradient(at::Tensor, at::Tensor, at::Tensor, at::Tensor, c10::IntArrayRef, double)’:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:193:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(dout);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:194:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(mean);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:195:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(invvar);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:196:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(input);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp: In function ‘std::vector<at::Tensor> layer_norm_gradient_affine(at::Tensor, at::Tensor, at::Tensor, at::Tensor, c10::IntArrayRef, at::Tensor, at::Tensor, double)’:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:218:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(dout);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:219:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(mean);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:220:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(invvar);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:221:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(input);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:222:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(gamma);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/DeviceType.h:8:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Device.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/c10/core/Allocator.h:6,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n csrc/layer_norm_cuda.cpp:117:42: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/macros/Macros.h:141:65: note: in definition of macro ‘C10_UNLIKELY’\n #define C10_UNLIKELY(expr) (__builtin_expect(static_cast<bool>(expr), 0))\n ^~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:262:7: note: in expansion of macro ‘C10_UNLIKELY_OR_CONST’\n if (C10_UNLIKELY_OR_CONST(!(cond))) { \\\n ^~~~~~~~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/c10/util/Exception.h:273:32: note: in expansion of macro ‘TORCH_CHECK_WITH’\n #define TORCH_CHECK(cond, ...) TORCH_CHECK_WITH(Error, cond, __VA_ARGS__)\n ^~~~~~~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:117:23: note: in expansion of macro ‘TORCH_CHECK’\n #define CHECK_CUDA(x) TORCH_CHECK(x.type().is_cuda(), #x \" must be a CUDA tensor\")\n ^~~~~~~~~~~\n csrc/layer_norm_cuda.cpp:119:24: note: in expansion of macro ‘CHECK_CUDA’\n #define CHECK_INPUT(x) CHECK_CUDA(x); CHECK_CONTIGUOUS(x)\n ^~~~~~~~~~\n csrc/layer_norm_cuda.cpp:223:3: note: in expansion of macro ‘CHECK_INPUT’\n CHECK_INPUT(beta);\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/layer_norm_cuda.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/layer_norm_cuda_kernel.cu -o build/temp.linux-x86_64-3.6/csrc/layer_norm_cuda_kernel.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -maxrregcount=50 -O3 --use_fast_math -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=fused_layer_norm_cuda -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n x86_64-linux-gnu-g++ -pthread -shared -Wl,-O1 -Wl,-Bsymbolic-functions -Wl,-Bsymbolic-functions -Wl,-z,relro -Wl,-Bsymbolic-functions -Wl,-z,relro -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 build/temp.linux-x86_64-3.6/csrc/layer_norm_cuda.o build/temp.linux-x86_64-3.6/csrc/layer_norm_cuda_kernel.o -L/usr/local/lib/python3.6/dist-packages/torch/lib -L/usr/local/cuda/lib64 -lc10 -ltorch -ltorch_cpu -ltorch_python -lcudart -lc10_cuda -ltorch_cuda -o build/lib.linux-x86_64-3.6/fused_layer_norm_cuda.cpython-36m-x86_64-linux-gnu.so\n building 'mlp_cuda' extension\n x86_64-linux-gnu-gcc -pthread -DNDEBUG -g -fwrapv -O2 -Wall -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 -fPIC -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/mlp.cpp -o build/temp.linux-x86_64-3.6/csrc/mlp.o -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=mlp_cuda -D_GLIBCXX_USE_CXX11_ABI=0 -std=c++14\n csrc/mlp.cpp: In function ‘std::vector<at::Tensor> mlp_forward(int, int, std::vector<at::Tensor>)’:\n csrc/mlp.cpp:56:21: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~~~~~~~~~~\n csrc/mlp.cpp:64:77: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n auto out = at::empty({batch_size, output_features.back()}, inputs[0].type());\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n csrc/mlp.cpp:65:67: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n auto reserved_space = at::empty({reserved_size}, inputs[0].type());\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n csrc/mlp.cpp:65:68: warning: narrowing conversion of ‘reserved_size’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto reserved_space = at::empty({reserved_size}, inputs[0].type());\n ^\n csrc/mlp.cpp:65:68: warning: narrowing conversion of ‘reserved_size’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:67:54: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:129:28: note: in definition of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n const auto& the_type = TYPE; \\\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:131:56: warning: ‘c10::ScalarType detail::scalar_type(const at::DeprecatedTypeProperties&)’ is deprecated: passing at::DeprecatedTypeProperties to an AT_DISPATCH macro is deprecated, pass an at::ScalarType instead [-Wdeprecated-declarations]\n at::ScalarType _st = ::detail::scalar_type(the_type); \\\n ^\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:46:23: note: declared here\n inline at::ScalarType scalar_type(const at::DeprecatedTypeProperties &t) {\n ^~~~~~~~~~~\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:70:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp:76:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_fp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:70:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp:76:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_fp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:70:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp:76:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_fp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:67:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_forward\", [&] {\n ^\n csrc/mlp.cpp: In function ‘std::vector<at::Tensor> mlp_backward(int, int, at::Tensor, std::vector<at::Tensor>, std::vector<at::Tensor>)’:\n csrc/mlp.cpp:113:21: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~~~~~~~~~~\n csrc/mlp.cpp:119:21: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < inputs.size(); i++) {\n ~~^~~~~~~~~~~~~~~\n csrc/mlp.cpp:120:67: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n outputs.push_back(at::empty(inputs[i].sizes(), inputs[i].type())); // clone for testing now\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:123:54: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:129:28: note: in definition of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n const auto& the_type = TYPE; \\\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:131:56: warning: ‘c10::ScalarType detail::scalar_type(const at::DeprecatedTypeProperties&)’ is deprecated: passing at::DeprecatedTypeProperties to an AT_DISPATCH macro is deprecated, pass an at::ScalarType instead [-Wdeprecated-declarations]\n at::ScalarType _st = ::detail::scalar_type(the_type); \\\n ^\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:46:23: note: declared here\n inline at::ScalarType scalar_type(const at::DeprecatedTypeProperties &t) {\n ^~~~~~~~~~~\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:125:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:129:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < inputs.size(); i++) {\n ~~^~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:80: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:139:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_bp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:125:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:129:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < inputs.size(); i++) {\n ~~^~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:80: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:139:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_bp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp: In lambda function:\n csrc/mlp.cpp:125:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < num_layers; i++) {\n ~~^~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:129:23: warning: comparison between signed and unsigned integer expressions [-Wsign-compare]\n for (int i = 0; i < inputs.size(); i++) {\n ~~^~~~~~~~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:80: warning: ‘at::DeprecatedTypeProperties& at::Tensor::type() const’ is deprecated: Tensor.type() is deprecated. Instead use Tensor.options(), which in many cases (e.g. in a constructor) is a drop-in replacement. If you were using data from type(), that is now available from Tensor itself, so instead of tensor.type().scalar_type(), use tensor.scalar_type() instead and instead of tensor.type().backend() use tensor.device(). [-Wdeprecated-declarations]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Tensor.h:11:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Context.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:5,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/core/TensorBody.h:262:30: note: declared here\n DeprecatedTypeProperties & type() const {\n ^~~~\n In file included from /usr/local/lib/python3.6/dist-packages/torch/include/ATen/ATen.h:9:0,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/types.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader_options.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/base.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader/stateful.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data/dataloader.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/data.h:3,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/all.h:4,\n from /usr/local/lib/python3.6/dist-packages/torch/include/torch/extension.h:4,\n from csrc/mlp.cpp:1:\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:137:44: warning: narrowing conversion of ‘(work_size / sizeof (scalar_t))’ from ‘long unsigned int’ to ‘long int’ inside { } [-Wnarrowing]\n auto work_space = at::empty({work_size / sizeof(scalar_t)}, inputs[0].type());\n ~~~~~~~~~~^~~~~~~~~\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n csrc/mlp.cpp:139:10: warning: unused variable ‘result’ [-Wunused-variable]\n auto result = mlp_bp<scalar_t>(\n ^\n /usr/local/lib/python3.6/dist-packages/torch/include/ATen/Dispatch.h:12:12: note: in definition of macro ‘AT_PRIVATE_CASE_TYPE’\n return __VA_ARGS__(); \\\n ^~~~~~~~~~~\n csrc/mlp.cpp:123:3: note: in expansion of macro ‘AT_DISPATCH_FLOATING_TYPES_AND_HALF’\n AT_DISPATCH_FLOATING_TYPES_AND_HALF(inputs[0].type(), \"mlp_backward\", [&] {\n ^\n /usr/local/cuda/bin/nvcc -I/usr/local/lib/python3.6/dist-packages/torch/include -I/usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include -I/usr/local/lib/python3.6/dist-packages/torch/include/TH -I/usr/local/lib/python3.6/dist-packages/torch/include/THC -I/usr/local/cuda/include -I/usr/include/python3.6m -c csrc/mlp_cuda.cu -o build/temp.linux-x86_64-3.6/csrc/mlp_cuda.o -D__CUDA_NO_HALF_OPERATORS__ -D__CUDA_NO_HALF_CONVERSIONS__ -D__CUDA_NO_HALF2_OPERATORS__ --expt-relaxed-constexpr --compiler-options '-fPIC' -O3 -DVERSION_GE_1_1 -DVERSION_GE_1_3 -DVERSION_GE_1_5 -DTORCH_API_INCLUDE_EXTENSION_H -DTORCH_EXTENSION_NAME=mlp_cuda -D_GLIBCXX_USE_CXX11_ABI=0 -gencode=arch=compute_75,code=sm_75 -std=c++14\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(14): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(18): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(23): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/autograd/profiler.h(100): warning: attribute \"__visibility__\" does not apply here\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/autograd/profiler.h(115): warning: attribute \"__visibility__\" does not apply here\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(14): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(15): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(18): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(19): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(23): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/api/include/torch/nn/functional/padding.h(24): warning: integer conversion resulted in a change of sign\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/autograd/profiler.h(100): warning: attribute \"__visibility__\" does not apply here\n\n /usr/local/lib/python3.6/dist-packages/torch/include/torch/csrc/autograd/profiler.h(115): warning: attribute \"__visibility__\" does not apply here\n\n x86_64-linux-gnu-g++ -pthread -shared -Wl,-O1 -Wl,-Bsymbolic-functions -Wl,-Bsymbolic-functions -Wl,-z,relro -Wl,-Bsymbolic-functions -Wl,-z,relro -g -fstack-protector-strong -Wformat -Werror=format-security -Wdate-time -D_FORTIFY_SOURCE=2 build/temp.linux-x86_64-3.6/csrc/mlp.o build/temp.linux-x86_64-3.6/csrc/mlp_cuda.o -L/usr/local/lib/python3.6/dist-packages/torch/lib -L/usr/local/cuda/lib64 -lc10 -ltorch -ltorch_cpu -ltorch_python -lcudart -lc10_cuda -ltorch_cuda -o build/lib.linux-x86_64-3.6/mlp_cuda.cpython-36m-x86_64-linux-gnu.so\n running install_lib\n copying build/lib.linux-x86_64-3.6/apex_C.cpython-36m-x86_64-linux-gnu.so -> /usr/local/lib/python3.6/dist-packages\n creating /usr/local/lib/python3.6/dist-packages/apex\n creating /usr/local/lib/python3.6/dist-packages/apex/RNN\n copying build/lib.linux-x86_64-3.6/apex/RNN/models.py -> /usr/local/lib/python3.6/dist-packages/apex/RNN\n copying build/lib.linux-x86_64-3.6/apex/RNN/RNNBackend.py -> /usr/local/lib/python3.6/dist-packages/apex/RNN\n copying build/lib.linux-x86_64-3.6/apex/RNN/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/RNN\n copying build/lib.linux-x86_64-3.6/apex/RNN/cells.py -> /usr/local/lib/python3.6/dist-packages/apex/RNN\n creating /usr/local/lib/python3.6/dist-packages/apex/multi_tensor_apply\n copying build/lib.linux-x86_64-3.6/apex/multi_tensor_apply/multi_tensor_apply.py -> /usr/local/lib/python3.6/dist-packages/apex/multi_tensor_apply\n copying build/lib.linux-x86_64-3.6/apex/multi_tensor_apply/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/multi_tensor_apply\n creating /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/sync_batchnorm.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/LARC.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/distributed.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/multiproc.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/optimized_sync_batchnorm_kernel.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/sync_batchnorm_kernel.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n copying build/lib.linux-x86_64-3.6/apex/parallel/optimized_sync_batchnorm.py -> /usr/local/lib/python3.6/dist-packages/apex/parallel\n creating /usr/local/lib/python3.6/dist-packages/apex/fp16_utils\n copying build/lib.linux-x86_64-3.6/apex/fp16_utils/fp16_optimizer.py -> /usr/local/lib/python3.6/dist-packages/apex/fp16_utils\n copying build/lib.linux-x86_64-3.6/apex/fp16_utils/fp16util.py -> /usr/local/lib/python3.6/dist-packages/apex/fp16_utils\n copying build/lib.linux-x86_64-3.6/apex/fp16_utils/loss_scaler.py -> /usr/local/lib/python3.6/dist-packages/apex/fp16_utils\n copying build/lib.linux-x86_64-3.6/apex/fp16_utils/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/fp16_utils\n creating /usr/local/lib/python3.6/dist-packages/apex/pyprof\n creating /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/linear.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/index_slice_join_mutate.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/randomSample.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/base.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/loss.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/dropout.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/blas.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/embedding.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/optim.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/conv.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/misc.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/utility.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/prof.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/convert.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/data.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/usage.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/pooling.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/output.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/activation.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/recurrentCell.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/pointwise.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/softmax.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/normalization.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/reduction.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/prof/__main__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof\n copying build/lib.linux-x86_64-3.6/apex/pyprof/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof\n creating /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/db.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/kernel.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/parse.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/nvvp.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n copying build/lib.linux-x86_64-3.6/apex/pyprof/parse/__main__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse\n creating /usr/local/lib/python3.6/dist-packages/apex/pyprof/nvtx\n copying build/lib.linux-x86_64-3.6/apex/pyprof/nvtx/nvmarker.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/nvtx\n copying build/lib.linux-x86_64-3.6/apex/pyprof/nvtx/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/pyprof/nvtx\n copying build/lib.linux-x86_64-3.6/apex/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex\n creating /usr/local/lib/python3.6/dist-packages/apex/normalization\n copying build/lib.linux-x86_64-3.6/apex/normalization/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/normalization\n copying build/lib.linux-x86_64-3.6/apex/normalization/fused_layer_norm.py -> /usr/local/lib/python3.6/dist-packages/apex/normalization\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity\n copying build/lib.linux-x86_64-3.6/apex/contrib/sparsity/asp.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity\n copying build/lib.linux-x86_64-3.6/apex/contrib/sparsity/sparse_masklib.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity\n copying build/lib.linux-x86_64-3.6/apex/contrib/sparsity/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/self_multihead_attn.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/fast_self_multihead_attn_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/encdec_multihead_attn_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/self_multihead_attn_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/fast_encdec_multihead_attn_norm_add_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/fast_encdec_multihead_attn_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/fast_self_multihead_attn_norm_add_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/encdec_multihead_attn.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/multihead_attn/mask_softmax_dropout_func.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn\n copying build/lib.linux-x86_64-3.6/apex/contrib/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib/groupbn\n copying build/lib.linux-x86_64-3.6/apex/contrib/groupbn/batch_norm.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/groupbn\n copying build/lib.linux-x86_64-3.6/apex/contrib/groupbn/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/groupbn\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib/xentropy\n copying build/lib.linux-x86_64-3.6/apex/contrib/xentropy/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/xentropy\n copying build/lib.linux-x86_64-3.6/apex/contrib/xentropy/softmax_xentropy.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/xentropy\n creating /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/fused_adam.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/fp16_optimizer.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/distributed_fused_adam.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/fused_lamb.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/distributed_fused_adam_v2.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/fused_sgd.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/distributed_fused_adam_v3.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n copying build/lib.linux-x86_64-3.6/apex/contrib/optimizers/distributed_fused_lamb.py -> /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers\n creating /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/compat.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/handle.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/__version__.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/_amp_state.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n creating /usr/local/lib/python3.6/dist-packages/apex/amp/lists\n copying build/lib.linux-x86_64-3.6/apex/amp/lists/torch_overrides.py -> /usr/local/lib/python3.6/dist-packages/apex/amp/lists\n copying build/lib.linux-x86_64-3.6/apex/amp/lists/functional_overrides.py -> /usr/local/lib/python3.6/dist-packages/apex/amp/lists\n copying build/lib.linux-x86_64-3.6/apex/amp/lists/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/amp/lists\n copying build/lib.linux-x86_64-3.6/apex/amp/lists/tensor_overrides.py -> /usr/local/lib/python3.6/dist-packages/apex/amp/lists\n copying build/lib.linux-x86_64-3.6/apex/amp/rnn_compat.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/_process_optimizer.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/scaler.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/_initialize.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/opt.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/amp.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/frontend.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/utils.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n copying build/lib.linux-x86_64-3.6/apex/amp/wrap.py -> /usr/local/lib/python3.6/dist-packages/apex/amp\n creating /usr/local/lib/python3.6/dist-packages/apex/mlp\n copying build/lib.linux-x86_64-3.6/apex/mlp/mlp.py -> /usr/local/lib/python3.6/dist-packages/apex/mlp\n copying build/lib.linux-x86_64-3.6/apex/mlp/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/mlp\n creating /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/fused_adam.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/fused_lamb.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/fused_sgd.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/fused_novograd.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n copying build/lib.linux-x86_64-3.6/apex/optimizers/fused_adagrad.py -> /usr/local/lib/python3.6/dist-packages/apex/optimizers\n creating /usr/local/lib/python3.6/dist-packages/apex/reparameterization\n copying build/lib.linux-x86_64-3.6/apex/reparameterization/reparameterization.py -> /usr/local/lib/python3.6/dist-packages/apex/reparameterization\n copying build/lib.linux-x86_64-3.6/apex/reparameterization/__init__.py -> /usr/local/lib/python3.6/dist-packages/apex/reparameterization\n copying build/lib.linux-x86_64-3.6/apex/reparameterization/weight_norm.py -> /usr/local/lib/python3.6/dist-packages/apex/reparameterization\n copying build/lib.linux-x86_64-3.6/syncbn.cpython-36m-x86_64-linux-gnu.so -> /usr/local/lib/python3.6/dist-packages\n copying build/lib.linux-x86_64-3.6/amp_C.cpython-36m-x86_64-linux-gnu.so -> /usr/local/lib/python3.6/dist-packages\n copying build/lib.linux-x86_64-3.6/mlp_cuda.cpython-36m-x86_64-linux-gnu.so -> /usr/local/lib/python3.6/dist-packages\n copying build/lib.linux-x86_64-3.6/fused_layer_norm_cuda.cpython-36m-x86_64-linux-gnu.so -> /usr/local/lib/python3.6/dist-packages\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/RNN/models.py to models.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/RNN/RNNBackend.py to RNNBackend.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/RNN/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/RNN/cells.py to cells.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/multi_tensor_apply/multi_tensor_apply.py to multi_tensor_apply.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/multi_tensor_apply/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/sync_batchnorm.py to sync_batchnorm.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/LARC.py to LARC.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/distributed.py to distributed.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/multiproc.py to multiproc.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/optimized_sync_batchnorm_kernel.py to optimized_sync_batchnorm_kernel.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/sync_batchnorm_kernel.py to sync_batchnorm_kernel.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/parallel/optimized_sync_batchnorm.py to optimized_sync_batchnorm.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/fp16_utils/fp16_optimizer.py to fp16_optimizer.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/fp16_utils/fp16util.py to fp16util.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/fp16_utils/loss_scaler.py to loss_scaler.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/fp16_utils/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/linear.py to linear.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/index_slice_join_mutate.py to index_slice_join_mutate.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/randomSample.py to randomSample.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/base.py to base.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/loss.py to loss.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/dropout.py to dropout.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/blas.py to blas.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/embedding.py to embedding.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/optim.py to optim.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/conv.py to conv.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/misc.py to misc.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/utility.py to utility.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/prof.py to prof.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/convert.py to convert.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/data.py to data.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/usage.py to usage.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/pooling.py to pooling.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/output.py to output.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/activation.py to activation.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/recurrentCell.py to recurrentCell.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/pointwise.py to pointwise.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/softmax.py to softmax.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/normalization.py to normalization.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/reduction.py to reduction.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/prof/__main__.py to __main__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/db.py to db.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/kernel.py to kernel.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/parse.py to parse.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/nvvp.py to nvvp.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/parse/__main__.py to __main__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/nvtx/nvmarker.py to nvmarker.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/pyprof/nvtx/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/normalization/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/normalization/fused_layer_norm.py to fused_layer_norm.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity/asp.py to asp.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity/sparse_masklib.py to sparse_masklib.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/sparsity/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/self_multihead_attn.py to self_multihead_attn.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/fast_self_multihead_attn_func.py to fast_self_multihead_attn_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/encdec_multihead_attn_func.py to encdec_multihead_attn_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/self_multihead_attn_func.py to self_multihead_attn_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/fast_encdec_multihead_attn_norm_add_func.py to fast_encdec_multihead_attn_norm_add_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/fast_encdec_multihead_attn_func.py to fast_encdec_multihead_attn_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/fast_self_multihead_attn_norm_add_func.py to fast_self_multihead_attn_norm_add_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/encdec_multihead_attn.py to encdec_multihead_attn.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/multihead_attn/mask_softmax_dropout_func.py to mask_softmax_dropout_func.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/groupbn/batch_norm.py to batch_norm.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/groupbn/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/xentropy/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/xentropy/softmax_xentropy.py to softmax_xentropy.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/fused_adam.py to fused_adam.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/fp16_optimizer.py to fp16_optimizer.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/distributed_fused_adam.py to distributed_fused_adam.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/fused_lamb.py to fused_lamb.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/distributed_fused_adam_v2.py to distributed_fused_adam_v2.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/fused_sgd.py to fused_sgd.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/distributed_fused_adam_v3.py to distributed_fused_adam_v3.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/contrib/optimizers/distributed_fused_lamb.py to distributed_fused_lamb.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/compat.py to compat.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/handle.py to handle.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/__version__.py to __version__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/_amp_state.py to _amp_state.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/lists/torch_overrides.py to torch_overrides.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/lists/functional_overrides.py to functional_overrides.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/lists/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/lists/tensor_overrides.py to tensor_overrides.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/rnn_compat.py to rnn_compat.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/_process_optimizer.py to _process_optimizer.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/scaler.py to scaler.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/_initialize.py to _initialize.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/opt.py to opt.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/amp.py to amp.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/frontend.py to frontend.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/utils.py to utils.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/amp/wrap.py to wrap.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/mlp/mlp.py to mlp.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/mlp/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/fused_adam.py to fused_adam.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/fused_lamb.py to fused_lamb.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/fused_sgd.py to fused_sgd.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/fused_novograd.py to fused_novograd.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/optimizers/fused_adagrad.py to fused_adagrad.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/reparameterization/reparameterization.py to reparameterization.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/reparameterization/__init__.py to __init__.cpython-36.pyc\n byte-compiling /usr/local/lib/python3.6/dist-packages/apex/reparameterization/weight_norm.py to weight_norm.cpython-36.pyc\n running install_egg_info\n running egg_info\n creating apex.egg-info\n writing apex.egg-info/PKG-INFO\n writing dependency_links to apex.egg-info/dependency_links.txt\n writing top-level names to apex.egg-info/top_level.txt\n writing manifest file 'apex.egg-info/SOURCES.txt'\n writing manifest file 'apex.egg-info/SOURCES.txt'\n Copying apex.egg-info to /usr/local/lib/python3.6/dist-packages/apex-0.1-py3.6.egg-info\n running install_scripts\n writing list of installed files to '/tmp/pip-record-6u0omsn_/install-record.txt'\n Running setup.py install for apex ... \u001b[?25l\u001b[?25hdone\n Removing source in /tmp/pip-req-build-awlakocq\nSuccessfully installed apex-0.1\nCleaning up...\nRemoved build tracker '/tmp/pip-req-tracker-ef_x9t6i'\n"
],
[
"!sh setup.sh",
"sh: 0: Can't open setup.sh\n"
],
[
"# 204줄 from torch.optim import Adam",
"_____no_output_____"
],
[
"# Max_grad_norm 에 문제가 생김 ",
"_____no_output_____"
],
[
"",
"_____no_output_____"
],
[
"!python NLI_KD_training.py",
"06/22/2020 18:30:30 - INFO - __main__ - IN DEBUG MODE\n06/22/2020 18:30:30 - INFO - src.argument_parser - encoder checkpoint not provided, use pre-trained at /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased/pytorch_model.bin instead\n06/22/2020 18:30:31 - INFO - src.argument_parser - /content/drive/My Drive/PKD/data/outputs/KD/RTE/teacher_12layer/kd_RTE_nlayer.6_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 exist, trying next\n06/22/2020 18:30:32 - INFO - src.argument_parser - /content/drive/My Drive/PKD/data/outputs/KD/RTE/teacher_12layer/kd_RTE_nlayer.6_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-2 exist, trying next\n06/22/2020 18:30:32 - INFO - src.argument_parser - device: cuda n_gpu: 1, 16-bits training: True\n06/22/2020 18:30:32 - INFO - src.argument_parser - random seed = 97614946\n06/22/2020 18:30:32 - INFO - __main__ - actual batch size on all GPU = 32\n06/22/2020 18:30:32 - INFO - __main__ - Input Argument Information\n06/22/2020 18:30:32 - INFO - __main__ - task_name RTE\n06/22/2020 18:30:32 - INFO - __main__ - output_dir /content/drive/My Drive/PKD/data/outputs/KD/RTE/teacher_12layer/kd_RTE_nlayer.6_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-3\n06/22/2020 18:30:32 - INFO - __main__ - log_every_step 1\n06/22/2020 18:30:32 - INFO - __main__ - max_seq_length 128\n06/22/2020 18:30:32 - INFO - __main__ - seed 97614946\n06/22/2020 18:30:32 - INFO - __main__ - train_batch_size 32\n06/22/2020 18:30:32 - INFO - __main__ - eval_batch_size 32\n06/22/2020 18:30:32 - INFO - __main__ - learning_rate 2e-05\n06/22/2020 18:30:32 - INFO - __main__ - num_train_epochs 4.0\n06/22/2020 18:30:32 - INFO - __main__ - gradient_accumulation_steps 1\n06/22/2020 18:30:32 - INFO - __main__ - fp16 True\n06/22/2020 18:30:32 - INFO - __main__ - loss_scale 0\n06/22/2020 18:30:32 - INFO - __main__ - student_hidden_layers 6\n06/22/2020 18:30:32 - INFO - __main__ - teacher_prediction None\n06/22/2020 18:30:32 - INFO - __main__ - warmup_proportion 0.1\n06/22/2020 18:30:32 - INFO - __main__ - bert_model /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased\n06/22/2020 18:30:32 - INFO - __main__ - encoder_checkpoint /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased/pytorch_model.bin\n06/22/2020 18:30:32 - INFO - __main__ - cls_checkpoint None\n06/22/2020 18:30:32 - INFO - __main__ - output_all_encoded_layers False\n06/22/2020 18:30:32 - INFO - __main__ - alpha 0.0\n06/22/2020 18:30:32 - INFO - __main__ - T 10.0\n06/22/2020 18:30:32 - INFO - __main__ - beta 0.0\n06/22/2020 18:30:32 - INFO - __main__ - kd_model kd\n06/22/2020 18:30:32 - INFO - __main__ - fc_layer_idx None\n06/22/2020 18:30:32 - INFO - __main__ - weights None\n06/22/2020 18:30:32 - INFO - __main__ - normalize_patience False\n06/22/2020 18:30:32 - INFO - __main__ - do_train True\n06/22/2020 18:30:32 - INFO - __main__ - do_eval True\n06/22/2020 18:30:32 - INFO - __main__ - device cuda\n06/22/2020 18:30:32 - INFO - __main__ - n_gpu 1\n06/22/2020 18:30:32 - INFO - __main__ - raw_data_dir /content/drive/My Drive/PKD/data/data_raw/RTE\n06/22/2020 18:30:32 - INFO - __main__ - feat_data_dir /content/drive/My Drive/PKD/data/data_feat/RTE\n06/22/2020 18:30:32 - INFO - BERT.pytorch_pretrained_bert.tokenization - loading vocabulary file /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased/vocab.txt\n06/22/2020 18:30:33 - INFO - __main__ - runing simple fine-tuning because teacher's prediction is not provided\n06/22/2020 18:30:35 - INFO - src.nli_data_processing - Writing example 0 of 2490\n06/22/2020 18:30:37 - INFO - __main__ - ***** Running training *****\n06/22/2020 18:30:37 - INFO - __main__ - Num examples = 2490\n06/22/2020 18:30:37 - INFO - __main__ - Batch size = 32\n06/22/2020 18:30:37 - INFO - __main__ - Num steps = 308\n06/22/2020 18:30:37 - INFO - src.nli_data_processing - Writing example 0 of 277\n06/22/2020 18:30:37 - INFO - __main__ - ***** Running evaluation *****\n06/22/2020 18:30:37 - INFO - __main__ - Num examples = 277\n06/22/2020 18:30:37 - INFO - __main__ - Batch size = 32\n06/22/2020 18:30:38 - INFO - src.nli_data_processing - Writing example 0 of 3000\n06/22/2020 18:30:41 - INFO - __main__ - ***** Running evaluation *****\n06/22/2020 18:30:41 - INFO - __main__ - Num examples = 3000\n06/22/2020 18:30:41 - INFO - __main__ - Batch size = 32\n06/22/2020 18:30:41 - INFO - __main__ - using normal Knowledge Distillation\n06/22/2020 18:30:41 - INFO - src.nli_data_processing - predicting for RTE\n06/22/2020 18:30:41 - INFO - src.modeling - num hidden layer is set as 6\n06/22/2020 18:30:41 - INFO - src.modeling - Model config {\n \"attention_probs_dropout_prob\": 0.1,\n \"hidden_act\": \"gelu\",\n \"hidden_dropout_prob\": 0.1,\n \"hidden_size\": 768,\n \"initializer_range\": 0.02,\n \"intermediate_size\": 3072,\n \"max_position_embeddings\": 512,\n \"num_attention_heads\": 12,\n \"num_hidden_layers\": 6,\n \"type_vocab_size\": 2,\n \"vocab_size\": 30522\n}\n\n06/22/2020 18:30:43 - INFO - src.utils - loading BertForSequenceClassificationEncoder finetuned model from /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased/pytorch_model.bin\n06/22/2020 18:31:01 - INFO - src.utils - delete 98 layers, keep 103 layers\n06/22/2020 18:31:01 - INFO - src.utils - fp16 activated, now call model.half()\n06/22/2020 18:31:01 - INFO - __main__ - *****************************************************************************\n06/22/2020 18:31:01 - INFO - src.utils - no checkpoint provided for FCClassifierForSequenceClassification!\n06/22/2020 18:31:01 - INFO - src.utils - fp16 activated, now call model.half()\n06/22/2020 18:31:01 - INFO - __main__ - number of layers in student model = 6\n06/22/2020 18:31:01 - INFO - __main__ - num parameters in student model are 66955008 and 1538\n06/22/2020 18:31:01 - INFO - __main__ - FP16 activate, use apex FusedAdam\nWarning: FP16_Optimizer is deprecated and dangerous, and will be deleted soon. If it still works, you're probably getting lucky. For mixed precision, use the documented API https://nvidia.github.io/apex/amp.html, with opt_level=O1.\nFP16_Optimizer processing param group 0:\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([30522, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([512, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([2, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768, 768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([2, 768])\nFP16_Optimizer processing param group 1:\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([3072])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([768])\nFP16_Optimizer received torch.cuda.HalfTensor with torch.Size([2])\nEpoch: 0% 0/4 [00:00<?, ?it/s]\nIteration: 0% 0/78 [00:00<?, ?it/s]\u001b[AGradient overflow. Skipping step, reducing loss scale to 32768.0\n\nIteration: 1% 1/78 [00:00<00:20, 3.83it/s]\u001b[A\nIteration: 3% 2/78 [00:00<00:17, 4.37it/s]\u001b[A\nIteration: 4% 3/78 [00:00<00:14, 5.17it/s]\u001b[A\nIteration: 5% 4/78 [00:00<00:12, 5.87it/s]\u001b[A\nIteration: 6% 5/78 [00:00<00:11, 6.48it/s]\u001b[A\nIteration: 8% 6/78 [00:00<00:10, 7.00it/s]\u001b[A\nIteration: 9% 7/78 [00:00<00:09, 7.42it/s]\u001b[A\nIteration: 10% 8/78 [00:01<00:09, 7.73it/s]\u001b[A\nIteration: 12% 9/78 [00:01<00:08, 7.94it/s]\u001b[A\nIteration: 13% 10/78 [00:01<00:08, 8.17it/s]\u001b[A\nIteration: 14% 11/78 [00:01<00:08, 8.23it/s]\u001b[A\nIteration: 15% 12/78 [00:01<00:07, 8.41it/s]\u001b[A\nIteration: 17% 13/78 [00:01<00:07, 8.47it/s]\u001b[A\nIteration: 18% 14/78 [00:01<00:07, 8.49it/s]\u001b[A\nIteration: 19% 15/78 [00:01<00:07, 8.55it/s]\u001b[A\nIteration: 21% 16/78 [00:02<00:07, 8.56it/s]\u001b[A\nIteration: 22% 17/78 [00:02<00:07, 8.56it/s]\u001b[A\nIteration: 23% 18/78 [00:02<00:06, 8.60it/s]\u001b[A\nIteration: 24% 19/78 [00:02<00:06, 8.51it/s]\u001b[A\nIteration: 26% 20/78 [00:02<00:06, 8.50it/s]\u001b[A\nIteration: 27% 21/78 [00:02<00:06, 8.68it/s]\u001b[A\nIteration: 28% 22/78 [00:02<00:06, 8.64it/s]\u001b[A\nIteration: 29% 23/78 [00:02<00:06, 8.60it/s]\u001b[A\nIteration: 31% 24/78 [00:02<00:06, 8.59it/s]\u001b[A\nIteration: 32% 25/78 [00:03<00:06, 8.58it/s]\u001b[A\nIteration: 33% 26/78 [00:03<00:06, 8.53it/s]\u001b[A\nIteration: 35% 27/78 [00:03<00:05, 8.61it/s]\u001b[A\nIteration: 36% 28/78 [00:03<00:05, 8.59it/s]\u001b[A\nIteration: 37% 29/78 [00:03<00:05, 8.59it/s]\u001b[A\nIteration: 38% 30/78 [00:03<00:05, 8.61it/s]\u001b[A\nIteration: 40% 31/78 [00:03<00:05, 8.58it/s]\u001b[A\nIteration: 41% 32/78 [00:03<00:05, 8.53it/s]\u001b[A\nIteration: 42% 33/78 [00:04<00:05, 8.61it/s]\u001b[A\nIteration: 44% 34/78 [00:04<00:05, 8.59it/s]\u001b[A\nIteration: 45% 35/78 [00:04<00:05, 8.58it/s]\u001b[A\nIteration: 46% 36/78 [00:04<00:04, 8.57it/s]\u001b[A\nIteration: 47% 37/78 [00:04<00:04, 8.58it/s]\u001b[A\nIteration: 49% 38/78 [00:04<00:04, 8.59it/s]\u001b[A\nIteration: 50% 39/78 [00:04<00:04, 8.58it/s]\u001b[A\nIteration: 51% 40/78 [00:04<00:04, 8.57it/s]\u001b[A\nIteration: 53% 41/78 [00:04<00:04, 8.59it/s]\u001b[A\nIteration: 54% 42/78 [00:05<00:04, 8.60it/s]\u001b[A\nIteration: 55% 43/78 [00:05<00:04, 8.59it/s]\u001b[A\nIteration: 56% 44/78 [00:05<00:04, 8.47it/s]\u001b[A\nIteration: 58% 45/78 [00:05<00:03, 8.61it/s]\u001b[A\nIteration: 59% 46/78 [00:05<00:03, 8.50it/s]\u001b[A\nIteration: 60% 47/78 [00:05<00:03, 8.64it/s]\u001b[A\nIteration: 62% 48/78 [00:05<00:03, 8.59it/s]\u001b[A\nIteration: 63% 49/78 [00:05<00:03, 8.55it/s]\u001b[A\nIteration: 64% 50/78 [00:05<00:03, 8.61it/s]\u001b[A\nIteration: 65% 51/78 [00:06<00:03, 8.60it/s]\u001b[A\nIteration: 67% 52/78 [00:06<00:03, 8.54it/s]\u001b[A\nIteration: 68% 53/78 [00:06<00:02, 8.58it/s]\u001b[A\nIteration: 69% 54/78 [00:06<00:02, 8.59it/s]\u001b[A\nIteration: 71% 55/78 [00:06<00:02, 8.57it/s]\u001b[A\nIteration: 72% 56/78 [00:06<00:02, 8.58it/s]\u001b[A\nIteration: 73% 57/78 [00:06<00:02, 8.56it/s]\u001b[A\nIteration: 74% 58/78 [00:06<00:02, 8.58it/s]\u001b[A\nIteration: 76% 59/78 [00:07<00:02, 8.58it/s]\u001b[A\nIteration: 77% 60/78 [00:07<00:02, 8.57it/s]\u001b[A\nIteration: 78% 61/78 [00:07<00:01, 8.55it/s]\u001b[A\nIteration: 79% 62/78 [00:07<00:01, 8.59it/s]\u001b[A\nIteration: 81% 63/78 [00:07<00:01, 8.54it/s]\u001b[A\nIteration: 82% 64/78 [00:07<00:01, 8.59it/s]\u001b[A\nIteration: 83% 65/78 [00:07<00:01, 8.58it/s]\u001b[A\nIteration: 85% 66/78 [00:07<00:01, 8.50it/s]\u001b[A\nIteration: 86% 67/78 [00:07<00:01, 8.58it/s]\u001b[A\nIteration: 87% 68/78 [00:08<00:01, 8.60it/s]\u001b[A\nIteration: 88% 69/78 [00:08<00:01, 8.55it/s]\u001b[A\nIteration: 90% 70/78 [00:08<00:00, 8.61it/s]\u001b[A\nIteration: 91% 71/78 [00:08<00:00, 8.61it/s]\u001b[A\nIteration: 92% 72/78 [00:08<00:00, 8.47it/s]\u001b[A\nIteration: 94% 73/78 [00:08<00:00, 8.58it/s]\u001b[A\nIteration: 95% 74/78 [00:08<00:00, 8.59it/s]\u001b[A\nIteration: 96% 75/78 [00:08<00:00, 8.60it/s]\u001b[A\nIteration: 97% 76/78 [00:09<00:00, 8.55it/s]\u001b[A\nIteration: 99% 77/78 [00:09<00:00, 8.57it/s]\u001b[A\nIteration: 100% 78/78 [00:09<00:00, 8.43it/s]\nEpoch: 25% 1/4 [00:09<00:29, 9.97s/it]\nIteration: 0% 0/78 [00:00<?, ?it/s]\u001b[A\nIteration: 3% 2/78 [00:00<00:08, 9.48it/s]\u001b[A\nIteration: 4% 3/78 [00:00<00:08, 9.29it/s]\u001b[A\nIteration: 5% 4/78 [00:00<00:08, 9.16it/s]\u001b[A\nIteration: 6% 5/78 [00:00<00:08, 9.11it/s]\u001b[A\nIteration: 8% 6/78 [00:00<00:08, 8.98it/s]\u001b[A\nIteration: 9% 7/78 [00:00<00:07, 8.91it/s]\u001b[A\nIteration: 10% 8/78 [00:00<00:07, 8.90it/s]\u001b[A\nIteration: 12% 9/78 [00:01<00:07, 8.87it/s]\u001b[A\nIteration: 13% 10/78 [00:01<00:07, 8.87it/s]\u001b[A\nIteration: 14% 11/78 [00:01<00:07, 8.85it/s]\u001b[A\nIteration: 15% 12/78 [00:01<00:07, 8.81it/s]\u001b[A\nIteration: 17% 13/78 [00:01<00:07, 8.82it/s]\u001b[A\nIteration: 18% 14/78 [00:01<00:07, 8.82it/s]\u001b[A\nIteration: 19% 15/78 [00:01<00:07, 8.79it/s]\u001b[A\nIteration: 21% 16/78 [00:01<00:07, 8.80it/s]\u001b[A\nIteration: 22% 17/78 [00:01<00:06, 8.79it/s]\u001b[A\nIteration: 23% 18/78 [00:02<00:06, 8.81it/s]\u001b[A\nIteration: 24% 19/78 [00:02<00:06, 8.82it/s]\u001b[A\nIteration: 26% 20/78 [00:02<00:06, 8.76it/s]\u001b[A\nIteration: 27% 21/78 [00:02<00:06, 8.83it/s]\u001b[A\nIteration: 28% 22/78 [00:02<00:06, 8.80it/s]\u001b[A\nIteration: 29% 23/78 [00:02<00:06, 8.79it/s]\u001b[A\nIteration: 31% 24/78 [00:02<00:06, 8.81it/s]\u001b[A\nIteration: 32% 25/78 [00:02<00:06, 8.78it/s]\u001b[A\nIteration: 33% 26/78 [00:02<00:05, 8.76it/s]\u001b[A\nIteration: 35% 27/78 [00:03<00:05, 8.77it/s]\u001b[A\nIteration: 36% 28/78 [00:03<00:05, 8.83it/s]\u001b[A\nIteration: 37% 29/78 [00:03<00:05, 8.83it/s]\u001b[A\nIteration: 38% 30/78 [00:03<00:05, 8.78it/s]\u001b[A\nIteration: 40% 31/78 [00:03<00:05, 8.79it/s]\u001b[A\nIteration: 41% 32/78 [00:03<00:05, 8.81it/s]\u001b[A\nIteration: 42% 33/78 [00:03<00:05, 8.81it/s]\u001b[A\nIteration: 44% 34/78 [00:03<00:04, 8.84it/s]\u001b[A\nIteration: 45% 35/78 [00:03<00:04, 8.80it/s]\u001b[A\nIteration: 46% 36/78 [00:04<00:04, 8.80it/s]\u001b[A\nIteration: 47% 37/78 [00:04<00:04, 8.80it/s]\u001b[A\nIteration: 49% 38/78 [00:04<00:04, 8.77it/s]\u001b[A\nIteration: 50% 39/78 [00:04<00:04, 8.80it/s]\u001b[A\nIteration: 51% 40/78 [00:04<00:04, 8.80it/s]\u001b[A\nIteration: 53% 41/78 [00:04<00:04, 8.79it/s]\u001b[A\nIteration: 54% 42/78 [00:04<00:04, 8.78it/s]\u001b[A\nIteration: 55% 43/78 [00:04<00:03, 8.79it/s]\u001b[A\nIteration: 56% 44/78 [00:04<00:03, 8.77it/s]\u001b[A\nIteration: 58% 45/78 [00:05<00:03, 8.81it/s]\u001b[A\nIteration: 59% 46/78 [00:05<00:03, 8.83it/s]\u001b[A\nIteration: 60% 47/78 [00:05<00:03, 8.47it/s]\u001b[A\nIteration: 62% 48/78 [00:05<00:03, 8.75it/s]\u001b[A\nIteration: 63% 49/78 [00:05<00:03, 8.72it/s]\u001b[A\nIteration: 64% 50/78 [00:05<00:03, 8.86it/s]\u001b[A\nIteration: 65% 51/78 [00:05<00:03, 8.81it/s]\u001b[A\nIteration: 67% 52/78 [00:05<00:02, 8.79it/s]\u001b[A\nIteration: 68% 53/78 [00:06<00:02, 8.81it/s]\u001b[A\nIteration: 69% 54/78 [00:06<00:02, 8.74it/s]\u001b[A\nIteration: 71% 55/78 [00:06<00:02, 8.77it/s]\u001b[A\nIteration: 72% 56/78 [00:06<00:02, 8.76it/s]\u001b[A\nIteration: 73% 57/78 [00:06<00:02, 8.70it/s]\u001b[A\nIteration: 74% 58/78 [00:06<00:02, 8.82it/s]\u001b[A\nIteration: 76% 59/78 [00:06<00:02, 8.66it/s]\u001b[A\nIteration: 77% 60/78 [00:06<00:02, 8.81it/s]\u001b[A\nIteration: 78% 61/78 [00:06<00:01, 8.74it/s]\u001b[A\nIteration: 79% 62/78 [00:07<00:01, 8.86it/s]\u001b[A\nIteration: 81% 63/78 [00:07<00:01, 8.85it/s]\u001b[A\nIteration: 82% 64/78 [00:07<00:01, 8.80it/s]\u001b[A\nIteration: 83% 65/78 [00:07<00:01, 8.82it/s]\u001b[A\nIteration: 85% 66/78 [00:07<00:01, 8.66it/s]\u001b[A\nIteration: 86% 67/78 [00:07<00:01, 8.79it/s]\u001b[A\nIteration: 87% 68/78 [00:07<00:01, 8.86it/s]\u001b[A\nIteration: 88% 69/78 [00:07<00:01, 8.80it/s]\u001b[A\nIteration: 90% 70/78 [00:07<00:00, 8.83it/s]\u001b[A\nIteration: 91% 71/78 [00:08<00:00, 8.84it/s]\u001b[A\nIteration: 92% 72/78 [00:08<00:00, 8.80it/s]\u001b[A\nIteration: 94% 73/78 [00:08<00:00, 8.80it/s]\u001b[A\nIteration: 95% 74/78 [00:08<00:00, 8.73it/s]\u001b[A\nIteration: 96% 75/78 [00:08<00:00, 8.69it/s]\u001b[A\nIteration: 97% 76/78 [00:08<00:00, 8.83it/s]\u001b[A\nIteration: 100% 78/78 [00:08<00:00, 8.82it/s]\nEpoch: 50% 2/4 [00:19<00:19, 9.83s/it]\nIteration: 0% 0/78 [00:00<?, ?it/s]\u001b[A\nIteration: 1% 1/78 [00:00<00:07, 9.95it/s]\u001b[A\nIteration: 3% 2/78 [00:00<00:08, 9.43it/s]\u001b[A\nIteration: 4% 3/78 [00:00<00:08, 9.34it/s]\u001b[A\nIteration: 5% 4/78 [00:00<00:08, 9.22it/s]\u001b[A\nIteration: 6% 5/78 [00:00<00:08, 8.79it/s]\u001b[A\nIteration: 8% 6/78 [00:00<00:07, 9.09it/s]\u001b[A\nIteration: 9% 7/78 [00:00<00:07, 9.01it/s]\u001b[A\nIteration: 10% 8/78 [00:00<00:07, 8.91it/s]\u001b[A\nIteration: 12% 9/78 [00:01<00:07, 8.88it/s]\u001b[A\nIteration: 13% 10/78 [00:01<00:07, 8.85it/s]\u001b[A\nIteration: 14% 11/78 [00:01<00:07, 8.78it/s]\u001b[A\nIteration: 15% 12/78 [00:01<00:07, 8.81it/s]\u001b[A\nIteration: 17% 13/78 [00:01<00:07, 8.81it/s]\u001b[A\nIteration: 18% 14/78 [00:01<00:07, 8.80it/s]\u001b[A\nIteration: 19% 15/78 [00:01<00:07, 8.78it/s]\u001b[A\nIteration: 21% 16/78 [00:01<00:07, 8.71it/s]\u001b[A\nIteration: 22% 17/78 [00:01<00:06, 8.79it/s]\u001b[A\nIteration: 23% 18/78 [00:02<00:06, 8.76it/s]\u001b[A\nIteration: 24% 19/78 [00:02<00:06, 8.74it/s]\u001b[A\nIteration: 26% 20/78 [00:02<00:06, 8.77it/s]\u001b[A\nIteration: 27% 21/78 [00:02<00:06, 8.79it/s]\u001b[A\nIteration: 28% 22/78 [00:02<00:06, 8.76it/s]\u001b[A\nIteration: 29% 23/78 [00:02<00:06, 8.78it/s]\u001b[A\nIteration: 31% 24/78 [00:02<00:06, 8.73it/s]\u001b[A\nIteration: 32% 25/78 [00:02<00:06, 8.69it/s]\u001b[A\nIteration: 33% 26/78 [00:02<00:05, 8.75it/s]\u001b[A\nIteration: 35% 27/78 [00:03<00:05, 8.81it/s]\u001b[A\nIteration: 36% 28/78 [00:03<00:05, 8.77it/s]\u001b[A\nIteration: 37% 29/78 [00:03<00:05, 8.78it/s]\u001b[A\nIteration: 38% 30/78 [00:03<00:05, 8.76it/s]\u001b[A\nIteration: 40% 31/78 [00:03<00:05, 8.78it/s]\u001b[A\nIteration: 41% 32/78 [00:03<00:05, 8.78it/s]\u001b[A\nIteration: 42% 33/78 [00:03<00:05, 8.75it/s]\u001b[A\nIteration: 44% 34/78 [00:03<00:05, 8.80it/s]\u001b[A\nIteration: 45% 35/78 [00:03<00:04, 8.78it/s]\u001b[A\nIteration: 46% 36/78 [00:04<00:04, 8.73it/s]\u001b[A\nIteration: 47% 37/78 [00:04<00:04, 8.78it/s]\u001b[A\nIteration: 49% 38/78 [00:04<00:04, 8.75it/s]\u001b[A\nIteration: 50% 39/78 [00:04<00:04, 8.77it/s]\u001b[A\nIteration: 51% 40/78 [00:04<00:04, 8.78it/s]\u001b[A\nIteration: 53% 41/78 [00:04<00:04, 8.75it/s]\u001b[A\nIteration: 54% 42/78 [00:04<00:04, 8.79it/s]\u001b[A\nIteration: 55% 43/78 [00:04<00:03, 8.77it/s]\u001b[A\nIteration: 56% 44/78 [00:05<00:03, 8.75it/s]\u001b[A\nIteration: 58% 45/78 [00:05<00:03, 8.78it/s]\u001b[A\nIteration: 59% 46/78 [00:05<00:03, 8.69it/s]\u001b[A\nIteration: 60% 47/78 [00:05<00:03, 8.77it/s]\u001b[A\nIteration: 62% 48/78 [00:05<00:03, 8.79it/s]\u001b[A\nIteration: 63% 49/78 [00:05<00:03, 8.70it/s]\u001b[A\nIteration: 64% 50/78 [00:05<00:03, 8.78it/s]\u001b[A\nIteration: 65% 51/78 [00:05<00:03, 8.68it/s]\u001b[A\nIteration: 67% 52/78 [00:05<00:03, 8.64it/s]\u001b[A\nIteration: 68% 53/78 [00:06<00:02, 8.61it/s]\u001b[A\nIteration: 69% 54/78 [00:06<00:02, 8.69it/s]\u001b[A\nIteration: 71% 55/78 [00:06<00:02, 8.75it/s]\u001b[A\nIteration: 72% 56/78 [00:06<00:02, 8.76it/s]\u001b[A\nIteration: 73% 57/78 [00:06<00:02, 8.74it/s]\u001b[A\nIteration: 74% 58/78 [00:06<00:02, 8.74it/s]\u001b[A\nIteration: 76% 59/78 [00:06<00:02, 8.75it/s]\u001b[A\nIteration: 77% 60/78 [00:06<00:02, 8.63it/s]\u001b[A\nIteration: 78% 61/78 [00:06<00:01, 8.75it/s]\u001b[A\nIteration: 79% 62/78 [00:07<00:01, 8.78it/s]\u001b[A\nIteration: 81% 63/78 [00:07<00:01, 8.69it/s]\u001b[A\nIteration: 82% 64/78 [00:07<00:01, 8.54it/s]\u001b[A\nIteration: 83% 65/78 [00:07<00:01, 8.84it/s]\u001b[A\nIteration: 85% 66/78 [00:07<00:01, 8.86it/s]\u001b[A\nIteration: 86% 67/78 [00:07<00:01, 8.83it/s]\u001b[A\nIteration: 87% 68/78 [00:07<00:01, 8.78it/s]\u001b[A\nIteration: 88% 69/78 [00:07<00:01, 8.79it/s]\u001b[A\nIteration: 90% 70/78 [00:07<00:00, 8.78it/s]\u001b[A\nIteration: 91% 71/78 [00:08<00:00, 8.79it/s]\u001b[A\nIteration: 92% 72/78 [00:08<00:00, 8.68it/s]\u001b[A\nIteration: 94% 73/78 [00:08<00:00, 8.75it/s]\u001b[A\nIteration: 95% 74/78 [00:08<00:00, 8.78it/s]\u001b[A\nIteration: 96% 75/78 [00:08<00:00, 8.78it/s]\u001b[A\nIteration: 97% 76/78 [00:08<00:00, 8.79it/s]\u001b[A\nIteration: 100% 78/78 [00:08<00:00, 8.79it/s]\nEpoch: 75% 3/4 [00:29<00:09, 9.75s/it]\nIteration: 0% 0/78 [00:00<?, ?it/s]\u001b[A\nIteration: 1% 1/78 [00:00<00:07, 9.98it/s]\u001b[A\nIteration: 3% 2/78 [00:00<00:07, 9.51it/s]\u001b[A\nIteration: 4% 3/78 [00:00<00:08, 9.27it/s]\u001b[A\nIteration: 5% 4/78 [00:00<00:08, 9.18it/s]\u001b[A\nIteration: 6% 5/78 [00:00<00:08, 9.01it/s]\u001b[A\nIteration: 8% 6/78 [00:00<00:08, 8.97it/s]\u001b[A\nIteration: 9% 7/78 [00:00<00:08, 8.81it/s]\u001b[A\nIteration: 10% 8/78 [00:00<00:07, 8.87it/s]\u001b[A\nIteration: 12% 9/78 [00:01<00:08, 8.56it/s]\u001b[A\nIteration: 13% 10/78 [00:01<00:07, 8.87it/s]\u001b[A\nIteration: 14% 11/78 [00:01<00:07, 8.84it/s]\u001b[A\nIteration: 15% 12/78 [00:01<00:07, 8.82it/s]\u001b[A\nIteration: 17% 13/78 [00:01<00:07, 8.78it/s]\u001b[A\nIteration: 18% 14/78 [00:01<00:07, 8.79it/s]\u001b[A\nIteration: 19% 15/78 [00:01<00:07, 8.70it/s]\u001b[A\nIteration: 21% 16/78 [00:01<00:07, 8.78it/s]\u001b[A\nIteration: 22% 17/78 [00:01<00:06, 8.76it/s]\u001b[A\nIteration: 23% 18/78 [00:02<00:06, 8.76it/s]\u001b[A\nIteration: 24% 19/78 [00:02<00:06, 8.73it/s]\u001b[A\nIteration: 26% 20/78 [00:02<00:06, 8.74it/s]\u001b[A\nIteration: 27% 21/78 [00:02<00:06, 8.74it/s]\u001b[A\nIteration: 28% 22/78 [00:02<00:06, 8.69it/s]\u001b[A\nIteration: 29% 23/78 [00:02<00:06, 8.71it/s]\u001b[A\nIteration: 31% 24/78 [00:02<00:06, 8.58it/s]\u001b[A\nIteration: 32% 25/78 [00:02<00:06, 8.75it/s]\u001b[A\nIteration: 33% 26/78 [00:02<00:05, 8.78it/s]\u001b[A\nIteration: 35% 27/78 [00:03<00:05, 8.75it/s]\u001b[A\nIteration: 36% 28/78 [00:03<00:05, 8.74it/s]\u001b[A\nIteration: 37% 29/78 [00:03<00:05, 8.75it/s]\u001b[A\nIteration: 38% 30/78 [00:03<00:05, 8.75it/s]\u001b[A\nIteration: 40% 31/78 [00:03<00:05, 8.70it/s]\u001b[A\nIteration: 41% 32/78 [00:03<00:05, 8.61it/s]\u001b[A\nIteration: 42% 33/78 [00:03<00:05, 8.71it/s]\u001b[A\nIteration: 44% 34/78 [00:03<00:05, 8.73it/s]\u001b[A\nIteration: 45% 35/78 [00:03<00:04, 8.74it/s]\u001b[A\nIteration: 46% 36/78 [00:04<00:04, 8.75it/s]\u001b[A\nIteration: 47% 37/78 [00:04<00:04, 8.73it/s]\u001b[A\nIteration: 49% 38/78 [00:04<00:04, 8.73it/s]\u001b[A\nIteration: 50% 39/78 [00:04<00:04, 8.70it/s]\u001b[A\nIteration: 51% 40/78 [00:04<00:04, 8.72it/s]\u001b[A\nIteration: 53% 41/78 [00:04<00:04, 8.73it/s]\u001b[A\nIteration: 54% 42/78 [00:04<00:04, 8.72it/s]\u001b[A\nIteration: 55% 43/78 [00:04<00:04, 8.70it/s]\u001b[A\nIteration: 56% 44/78 [00:05<00:03, 8.72it/s]\u001b[A\nIteration: 58% 45/78 [00:05<00:03, 8.71it/s]\u001b[A\nIteration: 59% 46/78 [00:05<00:03, 8.66it/s]\u001b[A\nIteration: 60% 47/78 [00:05<00:03, 8.74it/s]\u001b[A\nIteration: 62% 48/78 [00:05<00:03, 8.74it/s]\u001b[A\nIteration: 63% 49/78 [00:05<00:03, 8.71it/s]\u001b[A\nIteration: 64% 50/78 [00:05<00:03, 8.70it/s]\u001b[A\nIteration: 65% 51/78 [00:05<00:03, 8.62it/s]\u001b[A\nIteration: 67% 52/78 [00:05<00:02, 8.73it/s]\u001b[A\nIteration: 68% 53/78 [00:06<00:02, 8.65it/s]\u001b[A\nIteration: 69% 54/78 [00:06<00:02, 8.74it/s]\u001b[A\nIteration: 71% 55/78 [00:06<00:02, 8.63it/s]\u001b[A\nIteration: 72% 56/78 [00:06<00:02, 8.57it/s]\u001b[A\nIteration: 73% 57/78 [00:06<00:02, 8.63it/s]\u001b[A\nIteration: 74% 58/78 [00:06<00:02, 8.78it/s]\u001b[A\nIteration: 76% 59/78 [00:06<00:02, 8.71it/s]\u001b[A\nIteration: 77% 60/78 [00:06<00:02, 8.65it/s]\u001b[A\nIteration: 78% 61/78 [00:06<00:01, 8.71it/s]\u001b[A\nIteration: 79% 62/78 [00:07<00:01, 8.70it/s]\u001b[A\nIteration: 81% 63/78 [00:07<00:01, 8.67it/s]\u001b[A\nIteration: 82% 64/78 [00:07<00:01, 8.69it/s]\u001b[A\nIteration: 83% 65/78 [00:07<00:01, 8.74it/s]\u001b[A\nIteration: 85% 66/78 [00:07<00:01, 8.60it/s]\u001b[A\nIteration: 86% 67/78 [00:07<00:01, 8.62it/s]\u001b[A\nIteration: 87% 68/78 [00:07<00:01, 8.74it/s]\u001b[A\nIteration: 88% 69/78 [00:07<00:01, 8.78it/s]\u001b[A\nIteration: 90% 70/78 [00:08<00:00, 8.78it/s]\u001b[A\nIteration: 91% 71/78 [00:08<00:00, 8.75it/s]\u001b[A\nIteration: 92% 72/78 [00:08<00:00, 8.76it/s]\u001b[A\nIteration: 94% 73/78 [00:08<00:00, 8.74it/s]\u001b[A\nIteration: 95% 74/78 [00:08<00:00, 8.69it/s]\u001b[A\nIteration: 96% 75/78 [00:08<00:00, 8.74it/s]\u001b[A\nIteration: 97% 76/78 [00:08<00:00, 8.73it/s]\u001b[A\nIteration: 100% 78/78 [00:08<00:00, 8.75it/s]\nEpoch: 100% 4/4 [00:38<00:00, 9.65s/it]\n06/22/2020 18:31:43 - INFO - __main__ - ***** Eval results *****\n06/22/2020 18:31:43 - INFO - __main__ - acc = 0.5963333333333334\n06/22/2020 18:31:43 - INFO - __main__ - eval_loss = 0.7646458402593085\n"
],
[
"!python run_glue_benchmark.py",
"06/22/2020 18:31:47 - INFO - BERT.pytorch_pretrained_bert.tokenization - loading vocabulary file /content/drive/My Drive/PKD/data/models/pretrained/bert-base-uncased/vocab.txt\n06/22/2020 18:31:47 - INFO - __main__ - sub_dir = teacher_12layer\n06/22/2020 18:31:47 - INFO - __main__ - prediction_mode = teacher\n06/22/2020 18:31:47 - INFO - __main__ - interested_set = train,dev,test\n06/22/2020 18:31:49 - INFO - __main__ - predicting for task MRPC\n06/22/2020 18:31:49 - INFO - __main__ - using model from kd_MRPC_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-3 epoch 2\n06/22/2020 18:31:49 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:31:49 - INFO - __main__ - predicting for task SST-2\n06/22/2020 18:31:49 - INFO - __main__ - using model from kd_SST-2_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 epoch 3\n06/22/2020 18:31:49 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:31:49 - INFO - __main__ - predicting for task RTE\n06/22/2020 18:31:49 - INFO - __main__ - using model from kd_RTE_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 epoch 3\n06/22/2020 18:31:49 - INFO - src.nli_data_processing - predicting for RTE\n06/22/2020 18:31:49 - INFO - src.modeling - num hidden layer is set as 12\n06/22/2020 18:31:49 - INFO - src.modeling - Model config {\n \"attention_probs_dropout_prob\": 0.1,\n \"hidden_act\": \"gelu\",\n \"hidden_dropout_prob\": 0.1,\n \"hidden_size\": 768,\n \"initializer_range\": 0.02,\n \"intermediate_size\": 3072,\n \"max_position_embeddings\": 512,\n \"num_attention_heads\": 12,\n \"num_hidden_layers\": 12,\n \"type_vocab_size\": 2,\n \"vocab_size\": 30522\n}\n\n06/22/2020 18:31:52 - INFO - src.utils - loading BertForSequenceClassificationEncoder finetuned model from /content/drive/My Drive/PKD/data/outputs/KD/RTE/teacher_12layer/kd_RTE_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1/RTE_nlayer.12_lr.2e-05_T.10.0.alpha.0.0_beta.0.0_bs.32_e.3.encoder.pkl\n06/22/2020 18:31:57 - INFO - src.utils - fp16 activated, now call model.half()\n06/22/2020 18:31:58 - INFO - src.utils - loading FCClassifierForSequenceClassification finetuned model from /content/drive/My Drive/PKD/data/outputs/KD/RTE/teacher_12layer/kd_RTE_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1/RTE_nlayer.12_lr.2e-05_T.10.0.alpha.0.0_beta.0.0_bs.32_e.3.cls.pkl\n06/22/2020 18:31:59 - INFO - src.utils - fp16 activated, now call model.half()\n06/22/2020 18:31:59 - INFO - src.nli_data_processing - Writing example 0 of 277\nIteration: 100% 9/9 [00:00<00:00, 14.09it/s]\n06/22/2020 18:31:59 - INFO - __main__ - for dev, acc = 0.6498194945848376, loss = 0.6435864169675091\n06/22/2020 18:31:59 - INFO - __main__ - debug dev acc = 0.6498194945848376\n06/22/2020 18:31:59 - INFO - src.nli_data_processing - Writing example 0 of 3000\nIteration: 100% 94/94 [00:06<00:00, 14.69it/s]\n06/22/2020 18:32:08 - INFO - __main__ - for test, acc = 0.5813333333333334, loss = 0.8422109375\n06/22/2020 18:32:08 - INFO - __main__ - debug test acc = 0.5813333333333334\n06/22/2020 18:32:08 - INFO - src.nli_data_processing - Writing example 0 of 2490\nIteration: 100% 78/78 [00:05<00:00, 14.64it/s]\n06/22/2020 18:32:16 - INFO - __main__ - for training, acc = 0.8827309236947791, loss = 0.37306570030120484\n06/22/2020 18:32:16 - INFO - __main__ - debug train acc = 0.8827309236947791\n06/22/2020 18:32:16 - INFO - __main__ - saving teacher results\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task RTE Done!\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task MNLI\n06/22/2020 18:32:16 - INFO - __main__ - using model from kd_MNLI_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-3 epoch 3\n06/22/2020 18:32:16 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task QNLI\n06/22/2020 18:32:16 - INFO - __main__ - using model from kd_QNLI_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 epoch 1\n06/22/2020 18:32:16 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task QQP\n06/22/2020 18:32:16 - INFO - __main__ - using model from kd_QQP_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 epoch 3\n06/22/2020 18:32:16 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task MNLI-mm\n06/22/2020 18:32:16 - INFO - __main__ - using model from kd_MNLI-mm_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-3 epoch 3\n06/22/2020 18:32:16 - INFO - __main__ - Skipped because not interested\n06/22/2020 18:32:16 - INFO - __main__ - predicting for task race-merge\n06/22/2020 18:32:16 - INFO - __main__ - using model from kd_race-merge_nlayer.12_lr.2e-05_T.10.0_alpha.0.0_beta.0.0_bs.32-run-1 epoch 3\n06/22/2020 18:32:16 - INFO - __main__ - Skipped because not interested\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f0a9ed0fdf49705191ed0408a90fb7f6cdc23 | 59,776 | ipynb | Jupyter Notebook | data/Python Tips # Data Merge (concat, merge, join).ipynb | goldang01/E-Commerce | 968240ba22050d9cfe6ef5aae206a6ca1695bf0d | [
"MIT"
]
| null | null | null | data/Python Tips # Data Merge (concat, merge, join).ipynb | goldang01/E-Commerce | 968240ba22050d9cfe6ef5aae206a6ca1695bf0d | [
"MIT"
]
| null | null | null | data/Python Tips # Data Merge (concat, merge, join).ipynb | goldang01/E-Commerce | 968240ba22050d9cfe6ef5aae206a6ca1695bf0d | [
"MIT"
]
| null | null | null | 26.709562 | 72 | 0.287674 | [
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"# concat으로 데이터 합치기",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({'A': [1,3,5,7,9],\n 'B': [11,12,13,14,15],\n 'C': [0,1,2,3,4],\n 'D': [101,102,103,104,105] \n},\n index = [1,2,3,4,5])\n\ndf2 = pd.DataFrame({'A': [2,4,6,8,10],\n 'B': [1,2,3,4,5],\n 'C': [30,31,32,33,34],\n 'D': [1011,1021,1031,1041,1051] \n},\n index = [6,7,8,9,10])",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"pd.concat([df1, df2])",
"_____no_output_____"
],
[
"# index가 일치하지 않아 아래와 같은 모양으로 결합됨\npd.concat([df1,df2], axis = 1)",
"_____no_output_____"
],
[
"# index 일치시키기\ndf1.reset_index(drop=True, inplace = True)\ndf2.reset_index(drop = True, inplace = True)",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"pd.concat([df1, df2], axis = 1)",
"_____no_output_____"
]
],
[
[
"# merge로 데이터 합치기",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({'id': ['X01','X02','X03','X04','X05'],\n 'A': [1,3,5,7,9],\n 'B': [11,12,13,14,15],\n 'C': [0,1,2,3,4],\n 'D': [101,102,103,104,105] \n})\n\n\ndf2 = pd.DataFrame({'id': ['X01','X02','X03','X04','X06'],\n 'E': [2,4,6,8,10],\n 'F': [1,2,3,4,5],\n 'G': [30,31,32,33,34],\n 'H': [1011,1021,1031,1041,1051] \n})",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"pd.merge(df1, df2, on = 'id')",
"_____no_output_____"
],
[
"pd.merge(df1, df2, on = 'id', how = 'inner')",
"_____no_output_____"
],
[
"pd.merge(df1, df2, on = 'id', how = 'outer')",
"_____no_output_____"
],
[
"pd.merge(df1, df2, on = 'id', how = 'left')",
"_____no_output_____"
],
[
"pd.merge(df1, df2, on = 'id', how = 'right')",
"_____no_output_____"
],
[
"# 연습을 위해 key 컬럼의 이름 변경\ndf2 = pd.DataFrame({'user_id': ['X01','X02','X03','X04','X06'],\n 'E': [2,4,6,8,10],\n 'F': [1,2,3,4,5],\n 'G': [30,31,32,33,34],\n 'H': [1011,1021,1031,1041,1051] \n})",
"_____no_output_____"
],
[
"# key 컬럼의 이름이 다른 경우\npd.merge(df1, df2, left_on = 'id', right_on='user_id')",
"_____no_output_____"
]
],
[
[
"# join으로 데이터 합치기",
"_____no_output_____"
]
],
[
[
"df1 = pd.DataFrame({'A': [1,3,5,7,9],\n 'B': [11,12,13,14,15],\n 'C': [0,1,2,3,4],\n 'D': [101,102,103,104,105] \n},\n index = [1,2,3,4,5])\n\n\ndf2 = pd.DataFrame({'E': [2,4,6,8,10],\n 'F': [1,2,3,4,5],\n 'G': [30,31,32,33,34],\n 'H': [1011,1021,1031,1041,1051] \n},\n index = [6,7,8,9,10])",
"_____no_output_____"
],
[
"df1",
"_____no_output_____"
],
[
"df2",
"_____no_output_____"
],
[
"# index가 일치하지 않아서 제대로 결합이 안됨\ndf1.join(df2)",
"_____no_output_____"
],
[
"# index 일치 시켜주기\ndf1.reset_index(drop=True, inplace=True)\ndf2.reset_index(drop=True, inplace=True)",
"_____no_output_____"
],
[
"df1.join(df2)",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f0e03a7338dd8e625ffe707031d40297950ff | 164,695 | ipynb | Jupyter Notebook | .ipynb_checkpoints/Probability and Combinatorics Exercise-checkpoint.ipynb | ivaylokanov/Math_Concepts_for_Developers | 646d4d5de48535c22b9a8fcb624973b917661c5e | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Probability and Combinatorics Exercise-checkpoint.ipynb | ivaylokanov/Math_Concepts_for_Developers | 646d4d5de48535c22b9a8fcb624973b917661c5e | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/Probability and Combinatorics Exercise-checkpoint.ipynb | ivaylokanov/Math_Concepts_for_Developers | 646d4d5de48535c22b9a8fcb624973b917661c5e | [
"MIT"
]
| null | null | null | 228.425798 | 22,322 | 0.878721 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import numpy as np\nimport matplotlib.pyplot as plt\nimport scipy.stats\nimport scipy.io.wavfile\nfrom scipy.fftpack import fft, fftfreq\n\n# Write your imports here",
"_____no_output_____"
]
],
[
[
"# Probability and Combinatorics Exercise\n## Probabilistic Events. Combinatorics and Counting. Distributions",
"_____no_output_____"
],
[
"### Problem 1. Exploring Distribution Parameters\nA good idea to visualize and explore the parameters of various distributions is just to plot them.\n\nWe can do this in either one of two ways:\n1. Draw (generate) many random variables which follow that distribution. Plot their histogram\n2. Write the distribution function directly and plot it\n\nEither of these will work but the second approach will give us better looking results. [`scipy.stats`](https://docs.scipy.org/doc/scipy-0.19.1/reference/stats.html) has a lot of built-in distributions that we can use. Each of them has its own use cases.\n\nIt's very important that we plot discrete and continuous distributions in different ways. **We must not make discrete distributions look continuous**. That is, discrete distributions are only defined for integer number of trials: $n \\in \\mathbb{N}$.\n\nLet's plot the binomial and Gaussian distributions.",
"_____no_output_____"
]
],
[
[
"def plot_binomial_distribution(x, n, p):\n \"\"\"\n Plots the binomial distribution with parameters n and p. The parameter x specifies the values\n where the function is evaluated at\n \"\"\"\n binomial = scipy.stats.binom.pmf(x, n, p)\n plt.scatter(x, binomial, color = \"blue\")\n plt.vlines(x, 0, binomial, color = \"blue\", linewidth = 5, alpha = 0.5)\n plt.show()\n \ndef plot_gaussian_distribution(mu, sigma, x):\n \"\"\"\n Plots the Gaussian distribution with parameters mu and sigma. The parameter x specifies \n the values where the function is evaluated at\n \"\"\"\n gaussian = scipy.stats.norm.pdf(x, loc = mu, scale = sigma)\n plt.plot(x, gaussian, color = \"blue\")\n plt.show()\n \nx_binomial = np.arange(1, 10)\nplot_binomial_distribution(x_binomial, 10, 0.5)\n\nx_gaussian = np.linspace(-3, 3, 1000)\nplot_gaussian_distribution(0, 1, x_gaussian)",
"_____no_output_____"
]
],
[
[
"These look similar. That's with a good reason: the Gaussian distribution is a generalization of the binomial distribution as $n \\rightarrow \\infty$.\n\nWhat do these parameters specify exactly? Let's find out. \n\nTake the binomial distribution. Keep $p = 0.5$ and change $n$. Plot several values of $n$ in the same plot, with different colors. **What values to choose?** Remember that $n$ was the number of experiments, so it should be an integer $\\ge 1$.\n\nNow keep $n$ at some reasonable value (a number between 10 and 30 should be good) and change $p$. $p$ is a probability so its values must be between 0 and 1.\n\nWhat can you conclude? How does the function shape change? When is it symmetrical and when it is not?\n\nPerform the same kind of operations on $\\mu$ and $\\sigma$ with the Gaussian distribution. What do these parameters represent?\n\nIf you get stuck, try to find what the distribution functions should look like on the Internet.",
"_____no_output_____"
]
],
[
[
"# Write your code here",
"_____no_output_____"
]
],
[
[
"### Problem 2. Central Limit Theorem\nThe **Central Limit Theorem** tells us that no matter what quirky functions we have, their sum is going to be distributed according to the normal distribution. Let's prove this.\n\nConsider the following functions:\n$$ f(x) = 1 $$\n\n$$ f(x) = 2x $$\n\n$$ f(x) = 3x^2 $$\n\n$$ f(x) = 4\\lvert x - 0,5\\rvert $$\n\n$$ f(x) = 2 - 4\\lvert x - 0,5\\rvert $$\n\nFor each of these functions `f`:\n1. Generate a big array of, say, 2000 values `x` between 0 and 1: `np.linspace(0, 1, 2000)`\n2. Generate the array f(x)\n3. Create 1000 experiments like this\n 1. Generate 25 random values $x$ between 0 and 1: `np.random.rand(25)`\n 3. Generate $y = f(x)$\n 2. Sum all 25 values $y$\n 3. Add the sum to the array of sums\n4. Plot the distribution of 1000 sums\n\nWhat do you get? Can you experiment with a combination of functions? When is the normal distribution a good approximation of the real distribution?",
"_____no_output_____"
]
],
[
[
"def plot_function(f, ax, min_x = 0, max_x = 1, values = 2000):\n x = np.linspace(min_x, max_x, values) \n y = f(x)\n ax.plot(x, y)\n\ndef perform_simulation(f, ax):\n sums = []\n for experiment in range(1001):\n random_numbers = np.random.rand(25)\n current_sum = f(random_numbers).sum()\n sums.append(current_sum)\n ax.hist(sums)\n\n \ndef plot_results(f, min_x = 0, max_x = 1, values = 2000):\n vectorized_function = np.vectorize(f)\n figure, (ax1, ax2) = plt.subplots(1,2, figsize = (12,4))\n plot_function(vectorized_function, ax1, min_x, max_x, values)\n perform_simulation(vectorized_function, ax2)\nplot_results(lambda x: 1)\nplot_results(lambda x: 2 * x)\nplot_results(lambda x: 3 * x**2)\nplot_results(lambda x: 4 * np.abs(x-0.5))\nplot_results(lambda x: 2-4 * np.abs(x-0.5))\n",
"_____no_output_____"
]
],
[
[
"### Problem 3. Birthday Paradox\nHow many people do we need to have in a room, so that the probability of two people sharing a birthday is $p(A) > 0,5$?\n\nWe suppose no leap years, so a year has 365 days. We could expect that we need about $365/2=182$ people. Well, the truth is a bit different.\n\n#### Solution\n**Random variable:** $A$: probability that two people share a birthday\nIt's sometimes easier to work with the complementary variable: $\\bar{A}$ - probability that **no people** share a birthday. \n\nLet's suppose we have $r$ people in the room. Of course, if $r = 1$, e.g. only one person, the probability is $1$ (there's no one to share a birthday with). If $r >= 365$, the probability must be 1 (by the so-called [pigeonhole principle](https://en.wikipedia.org/wiki/Pigeonhole_principle): if we have 366 people and 365 days, there's at least one day with a pair of people).\n\nOrder the people 1 to $r$. Every person's birthday is independent, so that means 365 days for the first, 365 days for the second, and so on: $365^r$ birthday possibilities in total.\n\nWe want no duplications of birthdays. The first person has 365 days to choose from, the second has 364, and so on. The $r$th person has $365-r+1$ days to choose from. Total: $365.364.363.\\cdots.(365 - r + 1)$\n\nThe probability that no people share the same birthday is the fraction of all non-shared birthdays to all possible birthdays:\n$$ p(\\bar{A})=\\frac{365.364.363.\\cdots.(365 - r + 1)}{365^r} $$\n\nWe're interested in $A$, not $\\bar{A}$ and we know that these are complementary, so their probabilities add up to 1\n$$p(A) = 1 - p(\\bar{A})$$\n\nWrite a function which plots the probability of $r$ people sharing a birthday. Remember this is a discrete distribution and should be plotted like so.",
"_____no_output_____"
]
],
[
[
"def calculate_birtday_probability(r):\n \"\"\"\n Returns the probability of r people sharing the same birthday. A year is\n supposed to have 365 days\n \"\"\"\n total_sum = []\n for year in range(365-r+1,366):\n current_sum = year/365\n total_sum.append(current_sum)\n return (1 - total_sum)",
"_____no_output_____"
],
[
"probabilities = [calculate_birtday_probability(r) for r in np.arange(2, 366)]\nplt.hist(probabilities)\nplt.show()",
"_____no_output_____"
],
[
"[print(x) for x in probabilities if x < 0.5]",
"0.0\n0.002739726027397249\n0.008204165884781345\n0.016355912466550215\n0.02713557369979347\n0.040462483649111536\n0.056235703095975365\n0.07433529235166902\n0.09462383388916673\n0.11694817771107768\n0.14114137832173312\n0.1670247888380646\n0.19441027523242926\n0.22310251200497289\n0.25290131976368635\n0.2836040052528499\n0.31500766529656055\n0.34691141787178936\n0.37911852603153673\n0.41143838358058005\n0.4436883351652058\n0.47569530766254997\n"
]
],
[
[
"At how many people do you see a transition from $p(A) < 0,5$ to $p(A) > 0,5$?\n\n**Spoiler alert:** It's 23 people.\n\nWhy so few? We're comparing everyone's birthday against everyone else's. We should **NOT** count the number of people, but the number of comparisons. In a room of 23 people, there are 252 total comparisons.\n\nIn general, we could get a 50% chance of match using $\\sqrt{n}$ people in $n$ days.",
"_____no_output_____"
],
[
"## * Breaking Cryptography: Birthday Attack\nWe already saw that if we have $n$ days in one year, it takes about $\\sqrt{n}$ people to have a 50% chance of two people sharing a birthday. This is used in cryptography for the so-called **birthday attack**.\n\nLet's first introduce **hashing functions**. A hashing function is a function which takes text (bits) of any length and **returns a fixed number of bits**. There are many such functions. Some of them are completely insecure and **ARE NOT** used in cryptography. They're useful for other purposes, such as hash tables.\n\nImportant properties of hashing functions:\n1. The output will have a fixed length, no matter whether the input is an empty string, one character, a full sentence, a full book or the entire history of mankind\n2. A concrete input will always produce the same output\n\nOne such hashing function is **MD5**. It produces 128-bit hashes (32 hexadecimal symbols). This means that it takes the space of all possible texts and converts it to $2^{128} \\approx 3.10^{38}$ possible hashes. Since the inputs are much more, by the pigeonhole principle, we can expect that many inputs will produce the same output. This is called a **hashing collision**.\n\nThe birthday paradox tells us that using $\\sqrt{n} = 2^{64} \\approx 2.10^{19}$ hashes, we have a 50% probability of collision. This is still a very large number but compare it to $3.10^{38}$ - the difference is immense.\n\nYou can see what these numbers mean in terms of CPU speed [here](https://blog.codinghorror.com/speed-hashing/).\n\nThere are other algorithms which are even faster. The fastest one returns about $2^{18}$ hashes before it finds a collision.\n\nAnother clever attack is using **rainbow tables**. These are massive dictionaries of precomputed hashes. So, for example, if the input is `password123`, its MD5 hash is `482c811da5d5b4bc6d497ffa98491e38`. Every time an algorithm sees this hash, it can convert it to its input. \n\nRainbow tables work because humans are more predictable than algorithms. When implementing any cryptography, remember that **humans are always the weakest factor of any cryptographic system**.\n\n** * Optional: ** Write a function that finds collisions in **MD5** or **SHA1**. See [this](https://www.mscs.dal.ca/~selinger/md5collision/) demo for a good example, or [this StackOverflow post](https://crypto.stackexchange.com/questions/1434/are-there-two-known-strings-which-have-the-same-md5-hash-value) for more examples.",
"_____no_output_____"
]
],
[
[
"# Write your code here",
"_____no_output_____"
]
],
[
[
"### Problem 4. Having Fun with Functions. Fourier Transform\nSometimes we can plot a **parametric curve**. We choose a parameter $t$, in this case $t \\in [0; 2\\pi]$. We then plot $x$ and $y$ as functions of $t$.\n\nPlot the function below.",
"_____no_output_____"
]
],
[
[
"t = np.linspace(0, 2 * np.pi, 2000)",
"_____no_output_____"
],
[
"x = -(721 * np.sin(t)) / 4 + 196 / 3 * np.sin(2 * t) - 86 / 3 * np.sin(3 * t) - 131 / 2 * np.sin(4 * t) + 477 / 14 * np.sin(5 * t) + 27 * np.sin(6 * t) - 29 / 2 * np.sin(7 * t) + 68 / 5 * np.sin(8 * t) + 1 / 10 * np.sin(9 * t) + 23 / 4 * np.sin(10 * t) - 19 / 2 * np.sin(12 * t) - 85 / 21 * np.sin(13 * t) + 2 / 3 * np.sin(14 * t) + 27 / 5 * np.sin(15 * t) + 7 / 4 * np.sin(16 * t) + 17 / 9 * np.sin(17 * t) - 4 * np.sin(18 * t) - 1 / 2 * np.sin(19 * t) + 1 / 6 * np.sin(20 * t) + 6 / 7 * np.sin(21 * t) - 1 / 8 * np.sin(22 * t) + 1 / 3 * np.sin(23 * t) + 3 / 2 * np.sin(24 * t) + 13 / 5 * np.sin(25 * t) + np.sin(26 * t) - 2 * np.sin(27 * t) + 3 / 5 * np.sin(28 * t) - 1 / 5 * np.sin(29 * t) + 1 / 5 * np.sin(30 * t) + (2337 * np.cos(t)\n ) / 8 - 43 / 5 * np.cos(2 * t) + 322 / 5 * np.cos(3 * t) - 117 / 5 * np.cos(4 * t) - 26 / 5 * np.cos(5 * t) - 23 / 3 * np.cos(6 * t) + 143 / 4 * np.cos(7 * t) - 11 / 4 * np.cos(8 * t) - 31 / 3 * np.cos(9 * t) - 13 / 4 * np.cos(10 * t) - 9 / 2 * np.cos(11 * t) + 41 / 20 * np.cos(12 * t) + 8 * np.cos(13 * t) + 2 / 3 * np.cos(14 * t) + 6 * np.cos(15 * t) + 17 / 4 * np.cos(16 * t) - 3 / 2 * np.cos(17 * t) - 29 / 10 * np.cos(18 * t) + 11 / 6 * np.cos(19 * t) + 12 / 5 * np.cos(20 * t) + 3 / 2 * np.cos(21 * t) + 11 / 12 * np.cos(22 * t) - 4 / 5 * np.cos(23 * t) + np.cos(24 * t) + 17 / 8 * np.cos(25 * t) - 7 / 2 * np.cos(26 * t) - 5 / 6 * np.cos(27 * t) - 11 / 10 * np.cos(28 * t) + 1 / 2 * np.cos(29 * t) - 1 / 5 * np.cos(30 * t)\ny = -(637 * np.sin(t)) / 2 - 188 / 5 * np.sin(2 * t) - 11 / 7 * np.sin(3 * t) - 12 / 5 * np.sin(4 * t) + 11 / 3 * np.sin(5 * t) - 37 / 4 * np.sin(6 * t) + 8 / 3 * np.sin(7 * t) + 65 / 6 * np.sin(8 * t) - 32 / 5 * np.sin(9 * t) - 41 / 4 * np.sin(10 * t) - 38 / 3 * np.sin(11 * t) - 47 / 8 * np.sin(12 * t) + 5 / 4 * np.sin(13 * t) - 41 / 7 * np.sin(14 * t) - 7 / 3 * np.sin(15 * t) - 13 / 7 * np.sin(16 * t) + 17 / 4 * np.sin(17 * t) - 9 / 4 * np.sin(18 * t) + 8 / 9 * np.sin(19 * t) + 3 / 5 * np.sin(20 * t) - 2 / 5 * np.sin(21 * t) + 4 / 3 * np.sin(22 * t) + 1 / 3 * np.sin(23 * t) + 3 / 5 * np.sin(24 * t) - 3 / 5 * np.sin(25 * t) + 6 / 5 * np.sin(26 * t) - 1 / 5 * np.sin(27 * t) + 10 / 9 * np.sin(28 * t) + 1 / 3 * np.sin(29 * t) - 3 / 4 * \\\n np.sin(30 * t) - (125 * np.cos(t)) / 2 - 521 / 9 * np.cos(2 * t) - 359 / 3 * np.cos(3 * t) + 47 / 3 * np.cos(4 * t) - 33 / 2 * np.cos(5 * t) - 5 / 4 * np.cos(6 * t) + 31 / 8 * np.cos(7 * t) + 9 / 10 * np.cos(8 * t) - 119 / 4 * np.cos(9 * t) - 17 / 2 * np.cos(10 * t) + 22 / 3 * np.cos(11 * t) + 15 / 4 * np.cos(12 * t) - 5 / 2 * np.cos(13 * t) + 19 / 6 * np.cos(14 * t) + \\\n 7 / 4 * np.cos(15 * t) + 31 / 4 * np.cos(16 * t) - np.cos(17 * t) + 11 / 10 * np.cos(18 * t) - 2 / 3 * np.cos(19 * t) + 13 / 3 * np.cos(20 * t) - 5 / 4 * np.cos(21 * t) + 2 / 3 * np.cos(\n 22 * t) + 1 / 4 * np.cos(23 * t) + 5 / 6 * np.cos(24 * t) + 3 / 4 * np.cos(26 * t) - 1 / 2 * np.cos(27 * t) - 1 / 10 * np.cos(28 * t) - 1 / 3 * np.cos(29 * t) - 1 / 19 * np.cos(30 * t)",
"_____no_output_____"
],
[
"plt.gca().set_aspect(\"equal\")\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Interesting... \n\nHave a closer look at the variables `x` and `y`. Note that they're linear combinations of sines and cosines. There's nothing more except sines and cosines, multiplied by coefficients. How are these able to generate the picture? Can we generate any picture?\n\nYes, we can generate pretty much anything and plot it as a parametric curve. See [this](https://www.wolframalpha.com/input/?i=Schroedinger+cat+bra-ket+curve) for example.\n\nIt turns out that **every function**, no matter what, can be represented as a linear combination of sines and cosines. This is the basis of the **Fourier transform**. We'll look at it from two different perspectives: the algebraic one and the practical one.\n\n#### Algebraic perspective: Why does this transform exist? What does it mean?\nAll functions form a **vector space**. We can see them as vectors. These vectors have infinitely many components which correspond to the infinitely many values $x \\in (-\\infty; \\infty)$. The function space has infinitely many dimensions.\n\nWe can find a basis in that space. After we've found a basis, we can express any other function as a linear combination of the basis functions. Any set of infinitely many linearly independent functions will work. But that doesn't help at all...\n\nWe know that the best kind of basis is an *orthonormal basis*. This means that all basis vectors are orthogonal and each basis vector has \"length\" 1. \n\nTwo vectors are orthogonal if their dot product is zero. Similarly, two functions are defined to be orthogonal if their product is zero, like this:\n$$ \\int_a^b f(x)g(x)dx = 0 $$\n\nIt can be shown that $1$, $\\cos(mx)$ and $\\sin(nx)$ ($m,n \\in \\mathbb{N}$) are orthogonal. So, the basis formed by them is orthogonal. They can also be made orthonormal if we divide by their norm. The norm of a function is defined by **functional analysis** - an area of mathematics which treats functions as vectors. We won't go into much more detail now. The norm for $1$ is 1, the norm for the trigonometric functions is $1/\\sqrt{2}$.\n\nThe takeaway is that ${1, \\sqrt{2}\\cos(mx), \\sqrt{2}\\sin(nx),\\ m,n \\in \\mathbb{N}}$ is an orthonormal basis in the function space. \n\nAll periodic functions with period $P$ can be described as linear combinations of these:\n$$ f(x) = \\frac{a_0}{2} + \\sum\\left(a_n\\cos\\left(\\frac{2\\pi nx}{P}\\right)+b_n\\sin\\left(\\frac{2\\pi nx}{P}\\right)\\right) $$\n\nThis definition extends to non-periodic functions as well.",
"_____no_output_____"
],
[
"#### Engineering perspective\nIn engineering, the Fourier transform **converts a function of time to a function of frequency**. The function of time is called a **signal**, and the function of frequency is the **spectrum** of that signal. There is a pair of functions - one inverts the other. We have two different options:\n1. We can inspect the spectrum\n2. We can modify the spectrum\n\nThis means that if some operation is very easy to perform in the spectrum we can perform it there using these steps:\n1. Create the spectrum from the signal - Fourier transform\n2. Perform the operation, e.g. remove a specific frequency\n3. Create the corrected signal from the corrected spectrum - inverse Fourier transform\n\nOne example usage is in audio processing. An audio signal is a 1D array of **samples** (numbers). Each audio signal has a *bitrate* which tells us how many samples are there in one second. Since audio is a function of time, we can easily get its spectrum.\n\nSome algorithms on images use the spectrum as well. The idea is exactly the same.\n\nCompare this entire process to how we created a **histogram**. Plotting a random variable $X$ as a function of the trial number is essentially plotting a function of time. To get the histogram, we counted how many times we saw each particular value. This is the same as taking the spectrum of the random variable.",
"_____no_output_____"
],
[
"### Problem 5. Working with Audio Files. Fourier Transform\nIn Python, it's easiest to work with `.wav` files. If we have other files, we can convert them first. To load audio files, we can use `scipy.io.wavfile`. Load the `c-note.wav` file. Use only one channel, e.g. the left one.",
"_____no_output_____"
]
],
[
[
"bitrate, audio = scipy.io.wavfile.read(\"c-note.wav\")\nleft_channel = audio[:, 0]\nright_channel = audio[:, 1]\nplt.plot(left_channel)\nplt.xlabel(\"Sample number\") # To get seconds, divide by the bitrate\nplt.ylabel(\"Amplitude\")\nplt.show()\n",
"_____no_output_____"
],
[
"left_fft = fft(left_channel)\n\n# fftfreq() returns the frequences in number of cycles per sample. Since we have `bitrate` samples in one second,\n# to get the frequencies in Hz, we have to multiply by the bitrate\nfrequencies = fftfreq(len(left_channel)) * bitrate\n\nplt.plot(frequencies, left_fft)\nplt.show()",
"_____no_output_____"
]
],
[
[
"Note that the signal is symmetric. This is always the case with Fourier transform. We are interested in only half the values (the ones which are $\\ge 0$).",
"_____no_output_____"
]
],
[
[
"plt.plot(frequencies, left_fft)\nplt.xlim((0, 15000))\nplt.xlabel(\"Frequency [Hz]\")\nplt.ylabel(\"Amplitude\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"We can see that some frequencies have higher intensities than others. Also, they are evenly spaced. This is because the sample is only one note: C4, which has a fundamental frequency of $261,6Hz$. Most other \"loud\" frequencies are a multiple of the fundamental frequency: these are called **obertones**. There are other frequencies as well. The combination of frequencies which one instrument emphasizes and the ones that it dampens (i.e. makes quiet) determines the specific sound, or **timbre** of that instrument.",
"_____no_output_____"
]
],
[
[
"plt.plot(frequencies, left_fft)\nplt.xlim((240, 290))\nplt.xlabel(\"Frequency [Hz]\")\nplt.ylabel(\"Amplitude\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"An interesting application of this is removal of unwanted frequencies. Look at [this video](https://www.youtube.com/watch?v=ATVbnilxIrs) for example. The highly annoying vuvuzela of 2010 World Cup turns out to produce only a single frequency which can be very easily removed from a recording.\n\nAnother interesting application of Fourier transform is filters (in audio and images).",
"_____no_output_____"
],
[
"### * Problem 6. Breaking MD5 in Different Ways\nOne relatively fast algorithm: $2^{18}$ instead of $2^{19}$ hashes for a 50% probability is described in [this](https://eprint.iacr.org/2013/170.pdf) article. It's the fastest known to date to break the MD5 function by brute force. Get familiar with it and implement the algorithm. Try to show how it works on some common (and short) plaintexts.\n\nExperiment with other types of breaking the hash, for example using rainbow tables.\n\nYou can use the following checklist:\n* What is a hash function? When are hash functions used?\n* What is a cryptographic hash function?\n* Why and how is a collision possible?\n* What is a collision attack?\n* What are the most common ways to attack hash functions?\n * Which cryptographic hash functions are no longer considered secure? Why?\n* Why do websites have to store hashes of passwords instead of plaintext?\n * How does user login work?\n* What is the \"birthday attack\"?\n * Provide an example of two plaintexts which produce the same hash\n* What is the algorithm in the article about?\n * Implement it and show the result\n* What is a rainbow table?\n * Try breaking common passwords using a rainbow table. You can generate one or use some table from the internet (e.g. English words and letters, their modifications, leaked passwords, etc.)\n * Even better, try a rainbow table first. If it doesn't work, try brute force\n* How can a website protect its database against rainbow tables?\n * What is a \"salt\"? Additionally, what is \"pepper\"?\n * Is it always secure to hash a password multiple times? What advantages and disadvantages does this provide?",
"_____no_output_____"
],
[
"### * Problem 7. Audio Filters and Equalizers\nExamine the behaviour of different filters and their influence on the input signal. The main kinds of filters are\n* High-pass / low-pass\n* Band-pass / band-stop\n\nYou can use the following checklist:\n* What is a signal? What is time domain? What is frequency domain?\n* How does the Fourier transform work?\n * Provide one (or more) examples with code\n * Optionally, show plots of some common functions (sine, step, pulse, sinc, gaussian) and their Fourier spectrums\n* What is a filter?\n* How does each type of filter work?\n * Provide examples, possibly with real data\n* How do we combine filters and why?\n* What is an equalizer?\n * Optionally, create an equalizer program or plugin that allows the user to emphasize or dampen different frequencies",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec6f18b1cacb8b81e9dfb067bb6a7549cd54677f | 319,269 | ipynb | Jupyter Notebook | discrete_fourier_transform/theorems.ipynb | spatialaudio/signals-and-systems-lecture | 93e2f3488dc8f7ae111a34732bd4d13116763c5d | [
"MIT"
]
| 243 | 2016-04-01T14:21:00.000Z | 2022-03-28T20:35:09.000Z | discrete_fourier_transform/theorems.ipynb | alirezaopmc/signals-and-systems-lecture | f819845ce82656d91f53df1f4ccd1c31b6fb09fd | [
"MIT"
]
| 6 | 2016-04-11T06:28:17.000Z | 2021-11-10T10:59:35.000Z | discrete_fourier_transform/theorems.ipynb | alirezaopmc/signals-and-systems-lecture | f819845ce82656d91f53df1f4ccd1c31b6fb09fd | [
"MIT"
]
| 63 | 2017-04-20T00:46:03.000Z | 2022-03-30T14:07:09.000Z | 63.15905 | 19,486 | 0.596578 | [
[
[
"# The Discrete Fourier Transform\n\n*This Jupyter notebook is part of a [collection of notebooks](../index.ipynb) in the bachelors module Signals and Systems, Comunications Engineering, Universität Rostock. Please direct questions and suggestions to [[email protected]](mailto:[email protected]).*",
"_____no_output_____"
],
[
"## Theorems\n\nThe theorems of the discrete Fourier transform (DFT) relate basic operations applied to discrete signals to their equivalents in the spectral domain. They are of use to transform signals composed from modified [standard signals](../discrete_signals/standard_signals.ipynb), for the computation of the response of a linear time-invariant (LTI) system and to predict the consequences of modifying a signal or system by certain operations.",
"_____no_output_____"
],
[
"### Convolution Theorem\n\nThe DFT $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$ and its inverse $x[k] = \\text{IDFT}_N \\{ X[\\mu] \\}$ are both periodic with period $N$. The linear convolution of two periodic signals is not defined. The periodic convolution introduced in the following is used instead for the convolution theorem of the DFT.",
"_____no_output_____"
],
[
"#### Periodic Convolution\n\nThe [periodic (or circular/cyclic) convolution](https://en.wikipedia.org/wiki/Circular_convolution) of two finite-length signals $x[k]$ and $h[k]$ is defined as\n\n\\begin{equation}\nx[k] \\circledast_P h[k] = \\sum_{\\kappa=0}^{P-1} \\tilde{x}_P[k - \\kappa] \\; \\tilde{h}_P[\\kappa] =\n\\sum_{\\kappa=0}^{P-1} \\tilde{x}_P[\\kappa] \\; \\tilde{h}_P[k - \\kappa]\n\\end{equation}\n\nwhere $\\circledast_P$ denotes the periodic convolution with period $P$. The periodic summations $\\tilde{x}_P[k]$ of $x[k]$ and $\\tilde{h}_P[k]$ of $h[k]$ with period $P$ are defined as\n\n\\begin{align}\n\\tilde{x}_P[k] &= \\sum_{\\nu = -\\infty}^{\\infty} x[\\nu \\cdot P + k] \\\\\n\\tilde{h}_P[k] &= \\sum_{\\nu = -\\infty}^{\\infty} h[\\nu \\cdot P + k]\n\\end{align}\n\nThe result of the circular convolution has a period of $P$. The periodic convolution of two signals is in general different to their linear convolution.\n\nFor the special case that the length of one or both of the signals $x[k]$ and $h[k]$ is smaller or equal to the period $P$, the periodic summation degenerates to a periodic continuation of the signal(s). Furthermore, the periodic continuation does only have to be performed for the shifted signal in above convolution sum. For this special case, the periodic convolution is often termed as **cyclic convolution**.",
"_____no_output_____"
],
[
"**Example - Periodic vs. linear convolution**\n\nThe periodic $y_1[k] = x[k] \\circledast_P h[k]$ and linear $y_2[k] = x[k] * h[k]$ convolution of two rectangular signals $x[k] = \\mathrm{rect}_M[k]$ and $h[k] = \\mathrm{rect}_N[k]$ is numerically evaluated. For this purpose helper functions are defined that implement the periodic summation and convolution.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\n\ndef periodic_summation(x, P):\n 'Zero-padding to length P or periodic summation with period P.'\n N = len(x)\n rows = int(np.ceil(N/P))\n\n if (N < int(P*rows)):\n x = np.pad(x, (0, int(P*rows-N)), 'constant')\n\n x = np.reshape(x, (rows, P))\n\n return np.sum(x, axis=0)\n\n\ndef periodic_convolve(x, y, P):\n 'Periodic convolution of two signals x and y with period P.'\n x = periodic_summation(x, P)\n h = periodic_summation(y, P)\n\n return np.array([np.dot(np.roll(x[::-1], k+1), h) for k in range(P)], float)",
"_____no_output_____"
]
],
[
[
"Now the signals are defined, the convolutions are computed and the signals plotted. Note, for the periodic signals $\\tilde{x}_P[k]$ and $y_1[k]$ only one period is shown.",
"_____no_output_____"
]
],
[
[
"M = 32 # length of signal x[k]\nN = 16 # length of signal h[k]\nP = 24 # period of periodic convolution\n\n\ndef rect(k, N):\n return np.where((0 <= k) & (k < N), 1.0, 0.0)\n\n\n# generate signals\nk = np.arange(M+N-1)\nx = .5 * rect(k, M)\nh = rect(k, N)\n\n# periodic convolution\ny1 = periodic_convolve(x, h, P)\n# linear convolution\ny2 = np.convolve(x, h, 'full')\n\n# plot results\nplt.figure()\nplt.stem(periodic_summation(x, P), linefmt='C0-',\n markerfmt='C0o', label=r'$\\tilde{x}_P[k]$')\nplt.stem(x, linefmt='C1--', markerfmt='C1.', label=r'$x[k]$')\nplt.xlabel(r'$k$')\nplt.xlim([0, M+N-1])\nplt.legend()\nplt.grid()\n\nplt.figure()\nplt.stem(y1, linefmt='C1-', markerfmt='C1o',\n label=r'periodic convolution $P={}$'.format(P))\nplt.stem(y2, linefmt='C0--', markerfmt='C0.', label=r'linear convolution')\nplt.xlabel(r'$k$')\nplt.xlim([0, M+N-1])\nplt.legend()\nplt.grid()",
"_____no_output_____"
]
],
[
[
"**Exercise**\n\n* Change the length $M$ of the rectangular signal $x[k]$. How does the result of the periodic convolution change?\n* Compare the result of the periodic convolution with the result of the linear convolution. For which values of $P$ are both the same?",
"_____no_output_____"
],
[
"#### Convolution Theorem\n\nThe convolution theorem states that the DFT of the cyclic convolution of two discrete signals $x[k]$ and $y[k]$ is equal to the scalar multiplication of their DFTs $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$ and $Y[\\mu] = \\text{DFT}_N \\{ y[k] \\}$\n\n\\begin{equation}\n\\text{DFT}_N \\{ x[k] \\circledast_N y[k] \\} = X[\\mu] \\cdot Y[\\mu]\n\\end{equation}\n\nfor $k, \\mu =0,1, \\dots, N-1$.\n\nThe theorem can be proven by introducing the definition of the periodic convolution into the [definition of the DFT](definition.ipynb) and changing the order of summation\n\n\\begin{align}\n\\text{DFT}_N \\{ x[k] \\circledast_N y[k] \\} &= \\sum_{k = 0}^{N-1} \\left( \\sum_{\\kappa = 0}^{N-1} \\tilde{x}_N[\\kappa] \\cdot \\tilde{y}_N[k - \\kappa] \\right) w_N^{\\mu k} \\\\\n&= \\sum_{\\kappa = 0}^{N-1} \\left( \\sum_{k = 0}^{N-1} \\tilde{y}_N[k - \\kappa] \\, w_N^{\\mu k} \\right) \\tilde{x}_N[\\kappa] \\\\\n&= Y[\\mu] \\cdot \\sum_{\\kappa = 0}^{N-1} \\tilde{x}_N[\\kappa] \\, w_N^{\\mu \\kappa} \\\\\n&= Y[\\mu] \\cdot X[\\mu]\n\\end{align}\n\nNote, $\\text{DFT}_N \\{ x[k] \\} = \\text{DFT}_N \\{ \\tilde{x}_N[k] \\}$ due to the periodicity of the DFT.\nIt can be concluded from the convolution theorem that a scalar multiplication of the two spectra results in a cyclic convolution of the corresponding signals. For a linear time-invariant (LTI) system, the output signal is given as the linear convolution of the input signal $x[k]$ with the impulse response $h[k] = \\mathcal{H} \\{ \\delta[k] \\}$. The convolution theorem cannot be applied straightforward to the computation of the output signal of an LTI system. The [fast convolution technique](fast_convolution.ipynb), introduced later, provides an efficient algorithm for the linear convolution of two signals based on the convolution theorem.",
"_____no_output_____"
],
[
"### Shift Theorem\n\nSince the convolution theorem of the DFT is given in terms of the cyclic convolution, the shift theorem of the DFT is given in terms of the periodic shift. The [periodic (circular) shift](https://en.wikipedia.org/wiki/Circular_shift) of a causal signal $x[k]$ of finite length $N$ can be expressed by a cyclic convolution with a shifted Dirac impulse\n\n\\begin{equation}\nx[k - \\kappa] = x[k] \\circledast_N \\delta[k - \\kappa]\n\\end{equation}\n\nfor $\\kappa \\in 0,1,\\dots, N-1$. This follows from the definition of the cyclic convolution in combination with the sifting property of the Dirac impulse. Applying the DFT to the left- and right-hand side and exploiting the convolution theorem yields\n\n\\begin{equation}\n\\text{DFT}_N \\{ x[k - \\kappa] \\} = X[\\mu] \\cdot e^{-j \\mu \\frac{2 \\pi}{N} \\kappa}\n\\end{equation}\n\nwhere $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$. Above relation is known as shift theorem of the DFT.\n\nExpressing the DFT $X[\\mu] = |X[\\mu]| \\cdot e^{j \\varphi[\\mu]}$ by its absolute value $|X[\\mu]|$ and phase $\\varphi[\\mu]$ results in\n\n\\begin{equation}\n\\text{DFT}_N \\{ x[k - \\kappa] \\} = |X[\\mu]| \\cdot e^{j (\\varphi[\\mu] - \\mu \\frac{2 \\pi}{N} \\kappa)}\n\\end{equation}\n\nThe periodic shift of a signal does not change the absolute value of its spectrum but subtracts the linear contribution $\\mu \\frac{2 \\pi}{N} \\kappa$ from its phase.",
"_____no_output_____"
],
[
"**Example - Shifting a signal in the spectral domain**\n\nA cosine signal $x[k] = \\cos(\\Omega_0 k)$ is shifted in the spectral domain by multiplying its spectrum $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$ with $e^{-j \\mu \\frac{2 \\pi}{N} \\kappa}$ followed by an IDFT.",
"_____no_output_____"
]
],
[
[
"from scipy.linalg import dft\n\nN = 16 # length of signals/DFT\nM = 1 # number of periods for cosine\nkappa = 2 # shift\n\n# generate signal\nW0 = M * 2*np.pi/N\nk = np.arange(N)\nx = np.cos(W0 * k)\n\n# compute DFT\nF = dft(N)\nmu = np.arange(N)\nX = np.matmul(F, x)\n\n# shift in spectral domain and IDFT\nX2 = X * np.exp(-1j * mu * 2*np.pi/N * kappa)\nIF = 1/N * np.conjugate(np.transpose(F))\nx2 = np.matmul(IF, X2)\n\n# plot signals\nplt.stem(k, x, linefmt='C0-', markerfmt='C0o', label='$x[k]$')\nplt.stem(k, np.real(x2), linefmt='C1-', markerfmt='C1o', label='$x[k - {}]$'.format(kappa))\nplt.xlabel('$k$')\nplt.legend(loc=9)",
"_____no_output_____"
]
],
[
[
"### Multiplication Theorem\n\nThe transform of a multiplication of two signals $x[k] \\cdot y[k]$ is derived by introducing the signals into the definition of the DFT, expressing the signal $x[k]$ by its spectrum $X[\\mu] = \\text{IDFT}_N \\{ x[k] \\}$ and rearranging terms\n\n\\begin{align}\n\\text{DFT}_N \\{ x[k] \\cdot y[k] \\} &= \\sum_{k=0}^{N-1} x[k] \\cdot y[k] \\, w_N^{\\mu k} \\\\\n&= \\sum_{k=0}^{N-1} \\left( \\frac{1}{N} \\sum_{\\nu=0}^{N-1} X[\\nu] \\, w_N^{-\\nu k} \\right) y[k] \\, w_N^{\\mu k} \\\\\n&= \\frac{1}{N} \\sum_{\\nu=0}^{N-1} X[\\nu] \\sum_{k=0}^{N-1} y[k] \\, w_N^{(\\mu - \\nu) k} \\\\\n&= \\frac{1}{N} \\sum_{\\nu=0}^{N-1} X[\\nu] \\cdot Y[\\mu - \\nu] \\\\\n&= X[\\mu] \\circledast_N Y[\\mu]\n\\end{align}\n\nwhere $Y[\\mu] = \\text{IDFT}_N \\{ y[k] \\}$ and $k, \\mu = 0,1,\\dots,N-1$. Note, the last equality follows from the periodicity of the inverse DFT. The DFT of a multiplication of two signals $x[k] \\cdot y[k]$ is given by the cyclic convolution of their spectra $X[\\mu]$ and $Y[\\mu]$ weighted by $\\frac{1}{N}$. The cyclic convolution has a period of $N$ and it is performed with respect to the frequency index $\\mu$.\n\nApplications of the multiplication theorem include the modulation and windowing of signals. The former leads to the modulation theorem introduced in the following.",
"_____no_output_____"
],
[
"### Modulation Theorem\n\nThe complex modulation of a signal $x[k]$ is defined as $e^{j \\Omega_0 k} \\cdot x[k]$ with $\\Omega_0 = M \\frac{2 \\pi}{N}$, $M \\in \\mathbb{Z}$. The DFT of the modulated signal is derived by applying the multiplication theorem\n\n\\begin{equation}\n\\text{DFT}_N \\left\\{ e^{j M \\frac{2 \\pi}{N} k} \\cdot x[k] \\right\\} = \\delta[\\mu - M] \\circledast_N X[\\mu] = X[\\mu - M]\n\\end{equation}\n\nwhere $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$ and $X[\\mu - M]$ denotes the periodic shift of $X[\\mu]$. Above result states that the complex modulation of a signal leads to a periodic shift of its spectrum. This result is known as modulation theorem.",
"_____no_output_____"
],
[
"**Example - Decimation of a signal**\n\nAn example for the application of the modulation theorem is the [downsampling/decimation](https://en.wikipedia.org/wiki/Decimation_(signal_processing)) of a discrete signal $x[k]$. Downsampling refers to lowering the sampling rate of a signal. The example focuses on the special case of removing every second sample, hence halving the sampling rate. The downsampling is modeled by defining a signal $x_\\frac{1}{2}[k]$ where every second sample is set to zero\n\n\\begin{equation}\nx_\\frac{1}{2}[k] = \\begin{cases} \nx[k] & \\text{for even } k \\\\\n0 & \\text{for odd } k\n\\end{cases}\n\\end{equation}\n\nIn order to derive the spectrum $X_\\frac{1}{2}[\\mu] = \\text{DFT}_N \\{ x_\\frac{1}{2}[k] \\}$ for even $N$, the signal $u[k]$ is introduced where every second sample is zero\n\n\\begin{equation}\nu[k] = \\frac{1}{2} ( 1 + e^{j \\pi k} ) = \\begin{cases} 1 & \\text{for even } k \\\\\n0 & \\text{for odd } k \\end{cases}\n\\end{equation}\n\nUsing $u[k]$, the process of setting every second sample of $x[k]$ to zero can be expressed as\n\n\\begin{equation}\nx_\\frac{1}{2}[k] = u[k] \\cdot x[k]\n\\end{equation}\n\nNow the spectrum $X_\\frac{1}{2}[\\mu]$ is derived by applying the multiplication theorem and introducing the [DFT of the exponential signal](definition.ipynb#Transformation-of-the-Exponential-Signal). This results in\n\n\\begin{equation}\nX_\\frac{1}{2}[\\mu] = \\frac{1}{N} \\left( \\frac{N}{2} \\delta[\\mu] + \\frac{N}{2} \\delta[\\mu - \\frac{N}{2}] \\right) \\circledast X[\\mu] =\n\\frac{1}{2} X[\\mu] + \\frac{1}{2} X[\\mu - \\frac{N}{2}]\n\\end{equation}\n\nwhere $X[\\mu] = \\text{DFT}_N \\{ x[k] \\}$. The spectrum $X_\\frac{1}{2}[\\mu]$ consists of the spectrum of the original signal $X[\\mu]$ superimposed by the shifted spectrum $X[\\mu - \\frac{N}{2}]$ of the original signal. This may lead to overlaps that constitute aliasing. In order to avoid aliasing, the spectrum of the signal $x[k]$ has to be band-limited to $0 < \\mu < \\frac{N}{2}$ before downsampling.",
"_____no_output_____"
],
[
"The decimation of a complex exponential signal is illustrated in the following. The signal $x[k] = \\cos (\\Omega_0 k)$ is decimated by setting every second sample to zero. The DFT of the original signal and decimated signal is computed and their magnitudes are plotted for illustration.",
"_____no_output_____"
]
],
[
[
"N = 16 # length of signals/DFT\nM = 3.3 # number of periods for cosine\n\nW0 = M*2*np.pi/N\nk = np.arange(N)\nx = np.exp(1j*W0*k)\nx2 = np.copy(x)\nx2[::2] = 0\n\nF = dft(N)\nX = np.matmul(F, x)\nX2 = np.matmul(F, x2)\n\nplt.figure(figsize=(8,4))\nplt.subplot(1,2,1)\nplt.stem(abs(X))\nplt.xlabel('$\\mu$')\nplt.ylabel(r'|$X[\\mu]$|')\n\nplt.subplot(1,2,2)\nplt.stem(abs(X2))\nplt.xlabel('$\\mu$')\nplt.ylabel(r'|$X_{1/2}[\\mu]$|');",
"_____no_output_____"
]
],
[
[
"**Exercise**\n\n* What happens in the spectrum of the downsampled signal if you set $M > \\frac{N}{2}$?",
"_____no_output_____"
],
[
"### Parseval's Theorem\n\n[Parseval's theorem](https://en.wikipedia.org/wiki/Parseval's_theorem) relates the energy of a discrete signal to its spectrum. The squared absolute value of a signal $x[k]$ represents its instantaneous power. It can be expressed as\n\n\\begin{equation}\n| x[k] |^2 = x[k] \\cdot x^*[k]\n\\end{equation}\n\nwhere $x^*[k]$ denotes the complex conjugate of $x[k]$. It is assumed in the following that the signal $x[k]$ is causal and of finite length $N$ or periodic with perodid $N$. Transformation of the right-hand side and application of the multiplication theorem results in\n\n\\begin{equation}\n\\text{DFT}_N \\{ x[k] \\cdot x^*[k] \\} = \\frac{1}{N} \\cdot X[\\mu] \\circledast_{N} X^*[N -\\mu]\n\\end{equation}\n\nIntroducing the definition of the DFT and the cyclic convolution yields\n\n\\begin{equation}\n\\sum_{k = 0}^{N-1} x[k] \\cdot x^*[k] \\, w_N^{\\mu k} =\n\\frac{1}{N} \\sum_{\\nu = 0}^{N-1} \\tilde{X}[\\nu] \\cdot \\tilde{X}^*[N-\\mu-\\nu]\n\\end{equation}\n\nSetting $\\mu = 0$ followed by the substitution $\\nu = \\mu$ yields Parseval's theorem\n\n\\begin{equation}\n\\sum_{k = 0}^{N-1} | x[k] |^2 = \\frac{1}{N} \\sum_{\\nu = 0}^{N-1} |X[\\mu]|^2 \n\\end{equation}\n\nThe sum over the samples of the squared magnitude of the signal $x[k]$ is equal to the sum over its squared magnitude spectrum divided by $N$. Since the left-hand side represents the energy $E$ of the signal $x[k]$, Parseval's theorem states that the energy can be computed alternatively in the spectral domain by summing over the squared magnitude values of the spectrum.",
"_____no_output_____"
],
[
"**Copyright**\n\nThis notebook is provided as [Open Educational Resource](https://en.wikipedia.org/wiki/Open_educational_resources). Feel free to use the notebook for your own purposes. The text is licensed under [Creative Commons Attribution 4.0](https://creativecommons.org/licenses/by/4.0/), the code of the IPython examples under the [MIT license](https://opensource.org/licenses/MIT). Please attribute the work as follows: *Sascha Spors, Continuous- and Discrete-Time Signals and Systems - Theory and Computational Examples*.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec6f1e3494f1062929ef65c763bff48ba39e0a8d | 299,457 | ipynb | Jupyter Notebook | Final_regression_model.ipynb | YoungITOtaku/Yogyakarta-Food-Production-Prediction | 4b700f22065525d03f0bc6c1ee5212ff78ca3b82 | [
"MIT"
]
| null | null | null | Final_regression_model.ipynb | YoungITOtaku/Yogyakarta-Food-Production-Prediction | 4b700f22065525d03f0bc6c1ee5212ff78ca3b82 | [
"MIT"
]
| null | null | null | Final_regression_model.ipynb | YoungITOtaku/Yogyakarta-Food-Production-Prediction | 4b700f22065525d03f0bc6c1ee5212ff78ca3b82 | [
"MIT"
]
| null | null | null | 432.741329 | 48,787 | 0.771516 | [
[
[
"<h2> Importing Important Packages ",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport tensorflow as tf\nfrom tensorflow import keras\nfrom tensorflow.keras import layers",
"_____no_output_____"
]
],
[
[
"<h2>Importing Dataset that already preprocessed",
"_____no_output_____"
]
],
[
[
"path_to_file = 'data/padi.csv'\nprovince = 'DI YOGYAKARTA'\nsliding_window = 3\ntrain_test_proportion = 0.8\n\nfrom preprocessing import preprocess\n\ndata = pd.read_csv(path_to_file)\npreprocessed = preprocess(data[data['Provinsi'] == province],\\\n sliding_window)\n\npreprocessed.head()",
"_____no_output_____"
]
],
[
[
"<h2> Split Dataset into 2 gourp (train and test test)",
"_____no_output_____"
]
],
[
[
"train_data = preprocessed[:int(train_test_proportion*preprocessed.shape[0])]\ntest_data = preprocessed.drop(train_data.index)\n\ntrain_labels = train_data.pop(train_data.columns[-1])\ntest_labels = test_data.pop(test_data.columns[-1])",
"_____no_output_____"
]
],
[
[
"<h2> make Modeling function",
"_____no_output_____"
]
],
[
[
"def build_model():\n model = keras.Sequential([\n layers.Dense(2, activation=tf.nn.relu,\n input_shape=[len(train_data.keys())]),\n layers.Dense(5, activation=tf.nn.relu),\n layers.Dense(1)\n ])\n\n optimizer = tf.keras.optimizers.Adam(learning_rate=0.001)\n\n model.compile(loss='mae', optimizer='adam', metrics=['mse'])\n\n return model",
"_____no_output_____"
],
[
"tf.random.set_seed(28)\n\nmodel = build_model()\n\nmodel.summary()",
"Model: \"sequential_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ndense_3 (Dense) (None, 2) 8 \n_________________________________________________________________\ndense_4 (Dense) (None, 5) 15 \n_________________________________________________________________\ndense_5 (Dense) (None, 1) 6 \n=================================================================\nTotal params: 29\nTrainable params: 29\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"<h2> Make chackpoint to save model weights",
"_____no_output_____"
]
],
[
[
"checkpoint_path = \"model/cp_weights_model.ckpt\"\ncheckpoint_dir = os.path.dirname(checkpoint_path)\n\ncp_callback = tf.keras.callbacks.ModelCheckpoint(filepath=checkpoint_path,\n save_weights_only=True,period=10,\n verbose=1)\n\nmodel.save_weights(checkpoint_path.format(epoch=0))",
"WARNING:tensorflow:`period` argument is deprecated. Please use `save_freq` to specify the frequency in number of samples seen.\n"
]
],
[
[
"<h2> Train Regression model ",
"_____no_output_____"
]
],
[
[
"epochs = 100\nearly_stop = keras.callbacks.EarlyStopping(monitor='val_loss', patience=100)\n\nhistory = model.fit(train_data, train_labels, epochs=epochs, validation_split=0.2, verbose=1, callbacks=[early_stop])\n\nmodel.save('model')",
"Train on 12 samples, validate on 4 samples\nEpoch 1/100\n12/12 [==============================] - 0s 34ms/sample - loss: 529078.0000 - mse: 280613322752.0000 - val_loss: 666411.0625 - val_mse: 444179873792.0000\nEpoch 2/100\n12/12 [==============================] - 0s 917us/sample - loss: 524167.2500 - mse: 275437715456.0000 - val_loss: 660763.8750 - val_mse: 436681146368.0000\nEpoch 3/100\n12/12 [==============================] - 0s 916us/sample - loss: 519237.8438 - mse: 270290862080.0000 - val_loss: 655094.5000 - val_mse: 429217251328.0000\nEpoch 4/100\n12/12 [==============================] - 0s 917us/sample - loss: 514289.2500 - mse: 265173024768.0000 - val_loss: 649402.5000 - val_mse: 421788516352.0000\nEpoch 5/100\n12/12 [==============================] - 0s 833us/sample - loss: 509321.3750 - mse: 260084482048.0000 - val_loss: 643687.5000 - val_mse: 414395334656.0000\nEpoch 6/100\n12/12 [==============================] - 0s 1ms/sample - loss: 504333.7500 - mse: 255025430528.0000 - val_loss: 637949.1875 - val_mse: 407037870080.0000\nEpoch 7/100\n12/12 [==============================] - 0s 1ms/sample - loss: 499326.1250 - mse: 249996165120.0000 - val_loss: 632187.1250 - val_mse: 399716384768.0000\nEpoch 8/100\n12/12 [==============================] - 0s 1000us/sample - loss: 494298.1562 - mse: 244996915200.0000 - val_loss: 626400.8750 - val_mse: 392431239168.0000\nEpoch 9/100\n12/12 [==============================] - 0s 833us/sample - loss: 489249.4062 - mse: 240027975680.0000 - val_loss: 620590.0000 - val_mse: 385182859264.0000\nEpoch 10/100\n12/12 [==============================] - 0s 917us/sample - loss: 484179.6562 - mse: 235089625088.0000 - val_loss: 614754.2500 - val_mse: 377971474432.0000\nEpoch 11/100\n12/12 [==============================] - 0s 917us/sample - loss: 479088.4062 - mse: 230182141952.0000 - val_loss: 608893.0000 - val_mse: 370797543424.0000\nEpoch 12/100\n12/12 [==============================] - 0s 917us/sample - loss: 473975.3750 - mse: 225305870336.0000 - val_loss: 603005.8750 - val_mse: 363661328384.0000\nEpoch 13/100\n12/12 [==============================] - 0s 1ms/sample - loss: 468840.1250 - mse: 220461088768.0000 - val_loss: 597092.5000 - val_mse: 356563255296.0000\nEpoch 14/100\n12/12 [==============================] - 0s 1000us/sample - loss: 463682.3438 - mse: 215648124928.0000 - val_loss: 591152.3125 - val_mse: 349503717376.0000\nEpoch 15/100\n12/12 [==============================] - 0s 833us/sample - loss: 458501.5000 - mse: 210867322880.0000 - val_loss: 585184.8750 - val_mse: 342483173376.0000\nEpoch 16/100\n12/12 [==============================] - 0s 1ms/sample - loss: 453297.4062 - mse: 206119043072.0000 - val_loss: 579189.8750 - val_mse: 335502049280.0000\nEpoch 17/100\n12/12 [==============================] - 0s 1ms/sample - loss: 448069.5000 - mse: 201403678720.0000 - val_loss: 573166.6875 - val_mse: 328560836608.0000\nEpoch 18/100\n12/12 [==============================] - 0s 1ms/sample - loss: 442817.5000 - mse: 196721655808.0000 - val_loss: 567115.0000 - val_mse: 321660125184.0000\nEpoch 19/100\n12/12 [==============================] - 0s 1000us/sample - loss: 437541.0000 - mse: 192073351168.0000 - val_loss: 561034.3750 - val_mse: 314800373760.0000\nEpoch 20/100\n12/12 [==============================] - 0s 1ms/sample - loss: 432239.6250 - mse: 187459256320.0000 - val_loss: 554924.2500 - val_mse: 307982172160.0000\nEpoch 21/100\n12/12 [==============================] - 0s 1ms/sample - loss: 426913.0000 - mse: 182879797248.0000 - val_loss: 548784.2500 - val_mse: 301206110208.0000\nEpoch 22/100\n12/12 [==============================] - 0s 1000us/sample - loss: 421560.6562 - mse: 178335465472.0000 - val_loss: 542614.0000 - val_mse: 294472810496.0000\nEpoch 23/100\n12/12 [==============================] - 0s 917us/sample - loss: 416182.2812 - mse: 173826818048.0000 - val_loss: 536412.9375 - val_mse: 287782961152.0000\nEpoch 24/100\n12/12 [==============================] - 0s 833us/sample - loss: 410777.5312 - mse: 169354313728.0000 - val_loss: 530180.7500 - val_mse: 281137283072.0000\nEpoch 25/100\n12/12 [==============================] - 0s 1ms/sample - loss: 405345.9688 - mse: 164918542336.0000 - val_loss: 523916.8750 - val_mse: 274536398848.0000\nEpoch 26/100\n12/12 [==============================] - 0s 1000us/sample - loss: 399887.2500 - mse: 160520060928.0000 - val_loss: 517621.0312 - val_mse: 267981160448.0000\nEpoch 27/100\n12/12 [==============================] - 0s 2ms/sample - loss: 394401.0000 - mse: 156159492096.0000 - val_loss: 511292.6875 - val_mse: 261472256000.0000\nEpoch 28/100\n12/12 [==============================] - 0s 1ms/sample - loss: 388886.8438 - mse: 151837474816.0000 - val_loss: 504931.4062 - val_mse: 255010537472.0000\nEpoch 29/100\n12/12 [==============================] - 0s 1ms/sample - loss: 383344.5000 - mse: 147554631680.0000 - val_loss: 498536.9375 - val_mse: 248596922368.0000\nEpoch 30/100\n12/12 [==============================] - 0s 1ms/sample - loss: 377773.5000 - mse: 143311634432.0000 - val_loss: 492108.7500 - val_mse: 242232197120.0000\nEpoch 31/100\n12/12 [==============================] - 0s 917us/sample - loss: 372173.5938 - mse: 139109154816.0000 - val_loss: 485646.4688 - val_mse: 235917377536.0000\nEpoch 32/100\n12/12 [==============================] - 0s 916us/sample - loss: 366544.3438 - mse: 134947962880.0000 - val_loss: 479149.6562 - val_mse: 229653266432.0000\nEpoch 33/100\n12/12 [==============================] - 0s 1000us/sample - loss: 360885.3750 - mse: 130828738560.0000 - val_loss: 472617.9688 - val_mse: 223440977920.0000\nEpoch 34/100\n12/12 [==============================] - 0s 1ms/sample - loss: 355196.5000 - mse: 126752292864.0000 - val_loss: 466051.0938 - val_mse: 217281478656.0000\nEpoch 35/100\n12/12 [==============================] - 0s 1ms/sample - loss: 349477.2812 - mse: 122719444992.0000 - val_loss: 459448.5312 - val_mse: 211175833600.0000\nEpoch 36/100\n12/12 [==============================] - 0s 917us/sample - loss: 343727.4688 - mse: 118730956800.0000 - val_loss: 452809.9688 - val_mse: 205125058560.0000\nEpoch 37/100\n12/12 [==============================] - 0s 833us/sample - loss: 337946.6562 - mse: 114787680256.0000 - val_loss: 446134.9062 - val_mse: 199130267648.0000\nEpoch 38/100\n12/12 [==============================] - 0s 1ms/sample - loss: 332134.5000 - mse: 110890475520.0000 - val_loss: 439423.1250 - val_mse: 193192656896.0000\nEpoch 39/100\n12/12 [==============================] - 0s 1ms/sample - loss: 326290.7188 - mse: 107040186368.0000 - val_loss: 432674.1250 - val_mse: 187313274880.0000\nEpoch 40/100\n12/12 [==============================] - 0s 1000us/sample - loss: 320414.9688 - mse: 103237746688.0000 - val_loss: 425887.6875 - val_mse: 181493481472.0000\nEpoch 41/100\n12/12 [==============================] - 0s 917us/sample - loss: 314506.9688 - mse: 99484114944.0000 - val_loss: 419063.3125 - val_mse: 175734390784.0000\nEpoch 42/100\n12/12 [==============================] - 0s 916us/sample - loss: 308566.3438 - mse: 95780208640.0000 - val_loss: 412200.7188 - val_mse: 170037297152.0000\nEpoch 43/100\n12/12 [==============================] - 0s 917us/sample - loss: 302592.9062 - mse: 92126994432.0000 - val_loss: 405299.4375 - val_mse: 164403445760.0000\nEpoch 44/100\n12/12 [==============================] - 0s 917us/sample - loss: 296586.1250 - mse: 88525455360.0000 - val_loss: 398359.2500 - val_mse: 158834229248.0000\nEpoch 45/100\n12/12 [==============================] - 0s 1000us/sample - loss: 290545.9062 - mse: 84976656384.0000 - val_loss: 391379.7812 - val_mse: 153330958336.0000\nEpoch 46/100\n12/12 [==============================] - 0s 1ms/sample - loss: 284471.8438 - mse: 81481605120.0000 - val_loss: 384360.5625 - val_mse: 147895025664.0000\nEpoch 47/100\n12/12 [==============================] - 0s 1ms/sample - loss: 278363.6250 - mse: 78041374720.0000 - val_loss: 377301.3438 - val_mse: 142527807488.0000\nEpoch 48/100\n12/12 [==============================] - 0s 1000us/sample - loss: 272220.9375 - mse: 74657013760.0000 - val_loss: 370201.7500 - val_mse: 137230802944.0000\nEpoch 49/100\n12/12 [==============================] - 0s 1ms/sample - loss: 266043.5625 - mse: 71329701888.0000 - val_loss: 363061.4062 - val_mse: 132005421056.0000\nEpoch 50/100\n12/12 [==============================] - 0s 1000us/sample - loss: 259831.0781 - mse: 68060508160.0000 - val_loss: 355879.9375 - val_mse: 126853185536.0000\nEpoch 51/100\n12/12 [==============================] - 0s 3ms/sample - loss: 253583.2500 - mse: 64850595840.0000 - val_loss: 348657.0000 - val_mse: 121775611904.0000\nEpoch 52/100\n12/12 [==============================] - 0s 1000us/sample - loss: 247299.7031 - mse: 61701144576.0000 - val_loss: 341392.3125 - val_mse: 116774322176.0000\nEpoch 53/100\n12/12 [==============================] - 0s 1000us/sample - loss: 240980.2031 - mse: 58613362688.0000 - val_loss: 334085.4688 - val_mse: 111850848256.0000\nEpoch 54/100\n12/12 [==============================] - 0s 1ms/sample - loss: 234624.3750 - mse: 55588442112.0000 - val_loss: 326736.0312 - val_mse: 107006803968.0000\nEpoch 55/100\n12/12 [==============================] - 0s 1000us/sample - loss: 228231.9375 - mse: 52627652608.0000 - val_loss: 319343.7188 - val_mse: 102243868672.0000\nEpoch 56/100\n12/12 [==============================] - 0s 1ms/sample - loss: 221802.5781 - mse: 49732251648.0000 - val_loss: 311908.2188 - val_mse: 97563746304.0000\nEpoch 57/100\n12/12 [==============================] - 0s 917us/sample - loss: 215336.0469 - mse: 46903529472.0000 - val_loss: 304429.1250 - val_mse: 92968140800.0000\nEpoch 58/100\n12/12 [==============================] - 0s 833us/sample - loss: 208831.9219 - mse: 44142804992.0000 - val_loss: 296906.0000 - val_mse: 88458739712.0000\nEpoch 59/100\n12/12 [==============================] - 0s 1ms/sample - loss: 202289.9375 - mse: 41451409408.0000 - val_loss: 289338.5000 - val_mse: 84037378048.0000\nEpoch 60/100\n12/12 [==============================] - 0s 1000us/sample - loss: 195709.7500 - mse: 38830706688.0000 - val_loss: 281726.3438 - val_mse: 79705874432.0000\nEpoch 61/100\n12/12 [==============================] - 0s 1ms/sample - loss: 189091.0781 - mse: 36282085376.0000 - val_loss: 274069.1562 - val_mse: 75466096640.0000\nEpoch 62/100\n12/12 [==============================] - 0s 1ms/sample - loss: 182433.5781 - mse: 33806958592.0000 - val_loss: 266366.4375 - val_mse: 71319855104.0000\nEpoch 63/100\n12/12 [==============================] - 0s 917us/sample - loss: 175736.9219 - mse: 31406745600.0000 - val_loss: 258617.8750 - val_mse: 67269079040.0000\nEpoch 64/100\n12/12 [==============================] - 0s 1ms/sample - loss: 169000.7344 - mse: 29082925056.0000 - val_loss: 250823.0469 - val_mse: 63315726336.0000\nEpoch 65/100\n12/12 [==============================] - 0s 1ms/sample - loss: 162224.7500 - mse: 26836985856.0000 - val_loss: 242981.5938 - val_mse: 59461787648.0000\nEpoch 66/100\n12/12 [==============================] - 0s 1ms/sample - loss: 155408.5781 - mse: 24670437376.0000 - val_loss: 235093.1406 - val_mse: 55709265920.0000\nEpoch 67/100\n12/12 [==============================] - 0s 1ms/sample - loss: 148551.8906 - mse: 22584819712.0000 - val_loss: 227157.1719 - val_mse: 52060188672.0000\nEpoch 68/100\n12/12 [==============================] - 0s 1ms/sample - loss: 141654.2812 - mse: 20581689344.0000 - val_loss: 219173.3906 - val_mse: 48516685824.0000\nEpoch 69/100\n12/12 [==============================] - 0s 1ms/sample - loss: 134715.4844 - mse: 18662664192.0000 - val_loss: 211141.2812 - val_mse: 45080842240.0000\nEpoch 70/100\n12/12 [==============================] - 0s 1ms/sample - loss: 127735.0859 - mse: 16829363200.0000 - val_loss: 203060.5781 - val_mse: 41754882048.0000\nEpoch 71/100\n12/12 [==============================] - 0s 1ms/sample - loss: 120712.7266 - mse: 15083438080.0000 - val_loss: 194930.7188 - val_mse: 38540959744.0000\nEpoch 72/100\n12/12 [==============================] - 0s 917us/sample - loss: 113648.0234 - mse: 13426569216.0000 - val_loss: 186751.2812 - val_mse: 35441319936.0000\nEpoch 73/100\n12/12 [==============================] - 0s 833us/sample - loss: 106540.6328 - mse: 11860483072.0000 - val_loss: 178521.8906 - val_mse: 32458276864.0000\nEpoch 74/100\n12/12 [==============================] - 0s 1000us/sample - loss: 99390.1562 - mse: 10386919424.0000 - val_loss: 170241.9688 - val_mse: 29594105856.0000\nEpoch 75/100\n12/12 [==============================] - 0s 1ms/sample - loss: 92196.1250 - mse: 9007640576.0000 - val_loss: 161911.1250 - val_mse: 26851201024.0000\nEpoch 76/100\n12/12 [==============================] - 0s 1ms/sample - loss: 84958.2266 - mse: 7724476928.0000 - val_loss: 153528.8281 - val_mse: 24231962624.0000\nEpoch 77/100\n12/12 [==============================] - 0s 833us/sample - loss: 77676.0234 - mse: 6539266560.0000 - val_loss: 145094.7344 - val_mse: 21738878976.0000\nEpoch 78/100\n12/12 [==============================] - 0s 750us/sample - loss: 70349.0547 - mse: 5453880832.0000 - val_loss: 136608.2812 - val_mse: 19374436352.0000\nEpoch 79/100\n12/12 [==============================] - 0s 1000us/sample - loss: 62977.0000 - mse: 4470249472.0000 - val_loss: 128068.9375 - val_mse: 17141175296.0000\nEpoch 80/100\n12/12 [==============================] - 0s 1ms/sample - loss: 55559.3555 - mse: 3590311680.0000 - val_loss: 119476.2656 - val_mse: 15041707008.0000\nEpoch 81/100\n12/12 [==============================] - 0s 1ms/sample - loss: 48095.7227 - mse: 2816058624.0000 - val_loss: 110829.7031 - val_mse: 13078677504.0000\nEpoch 82/100\n12/12 [==============================] - 0s 1ms/sample - loss: 41136.1680 - mse: 2149514240.0000 - val_loss: 102254.2344 - val_mse: 11280004096.0000\nEpoch 83/100\n12/12 [==============================] - 0s 833us/sample - loss: 35011.3477 - mse: 1599966208.0000 - val_loss: 93869.2500 - val_mse: 9664005120.0000\nEpoch 84/100\n12/12 [==============================] - 0s 2ms/sample - loss: 30139.4785 - mse: 1169986432.0000 - val_loss: 85651.2969 - val_mse: 8217136640.0000\nEpoch 85/100\n12/12 [==============================] - 0s 1ms/sample - loss: 25364.9062 - mse: 851566592.0000 - val_loss: 77579.2344 - val_mse: 6927915520.0000\nEpoch 86/100\n12/12 [==============================] - 0s 917us/sample - loss: 20985.9434 - mse: 638037056.0000 - val_loss: 69926.6250 - val_mse: 5826454016.0000\nEpoch 87/100\n12/12 [==============================] - 0s 917us/sample - loss: 19222.6660 - mse: 526406112.0000 - val_loss: 62811.8438 - val_mse: 4907853312.0000\nEpoch 88/100\n12/12 [==============================] - 0s 1ms/sample - loss: 18274.3125 - mse: 501888768.0000 - val_loss: 56358.8594 - val_mse: 4162565376.0000\nEpoch 89/100\n12/12 [==============================] - 0s 1000us/sample - loss: 18341.1621 - mse: 545702848.0000 - val_loss: 50509.6250 - val_mse: 3559229440.0000\nEpoch 90/100\n12/12 [==============================] - 0s 1ms/sample - loss: 19238.2031 - mse: 639698240.0000 - val_loss: 45716.3906 - val_mse: 3116013056.0000\nEpoch 91/100\n12/12 [==============================] - 0s 1000us/sample - loss: 21321.5840 - mse: 755202496.0000 - val_loss: 43644.4531 - val_mse: 2794998016.0000\nEpoch 92/100\n12/12 [==============================] - 0s 917us/sample - loss: 23003.5469 - mse: 872416832.0000 - val_loss: 42369.2812 - val_mse: 2580192512.0000\nEpoch 93/100\n12/12 [==============================] - 0s 917us/sample - loss: 24602.6465 - mse: 971489728.0000 - val_loss: 41528.9375 - val_mse: 2447117568.0000\nEpoch 94/100\n12/12 [==============================] - 0s 1ms/sample - loss: 25656.8125 - mse: 1043197632.0000 - val_loss: 41081.9062 - val_mse: 2379064320.0000\nEpoch 95/100\n12/12 [==============================] - 0s 1ms/sample - loss: 26217.8496 - mse: 1083436672.0000 - val_loss: 40990.3594 - val_mse: 2365324544.0000\nEpoch 96/100\n12/12 [==============================] - 0s 1ms/sample - loss: 26333.4219 - mse: 1091910656.0000 - val_loss: 41219.3281 - val_mse: 2399677440.0000\nEpoch 97/100\n12/12 [==============================] - 0s 1ms/sample - loss: 26047.2500 - mse: 1071066688.0000 - val_loss: 41737.0156 - val_mse: 2479240704.0000\nEpoch 98/100\n12/12 [==============================] - 0s 1ms/sample - loss: 25399.2188 - mse: 1025240896.0000 - val_loss: 42514.2500 - val_mse: 2603523328.0000\nEpoch 99/100\n12/12 [==============================] - 0s 1000us/sample - loss: 24425.8223 - mse: 960006080.0000 - val_loss: 43524.4375 - val_mse: 2773702144.0000\nEpoch 100/100\n12/12 [==============================] - 0s 1ms/sample - loss: 23160.3496 - mse: 881671168.0000 - val_loss: 44743.5781 - val_mse: 2992070144.0000\nINFO:tensorflow:Assets written to: model\\assets\n"
]
],
[
[
"<h2> Plotting Training Process",
"_____no_output_____"
]
],
[
[
"hist = pd.DataFrame(history.history)\nhist['epoch'] = history.epoch\n\n#Show last Epoch matrics result\nhist.tail()",
"_____no_output_____"
],
[
"def ploting_history(history):\n\n plt.figure(figsize=(14,8))\n plt.xlabel('Epoch')\n plt.ylabel('Mean Abs Error (Target)')\n plt.title('Mean Absolute Error values change')\n plt.plot(hist['epoch'], hist['loss'], label='Train Error')\n plt.plot(hist['epoch'], hist['val_loss'], label='Val Error')\n plt.legend()\n # plt.ylim([0, 5])\n plt.savefig('plot/ABS train history.pdf') \n\n plt.figure(figsize=(14,8))\n plt.xlabel('Epoch')\n plt.ylabel('Mean Square Error (Target^2)')\n plt.title('Mean Square Error values change')\n plt.plot(hist['epoch'], hist['mse'], label='Train Error')\n plt.plot(hist['epoch'], hist['val_mse'], label='Val Error')\n plt.legend()\n # plt.ylim([0, 20])\n plt.savefig('plot/MAE train history.pdf') ",
"_____no_output_____"
],
[
"ploting_history(history)",
"_____no_output_____"
]
],
[
[
"<h2> Checking Evaluation metrics model",
"_____no_output_____"
]
],
[
[
"model.evaluate(test_data, test_labels, verbose=0)\n# print(\"Test Set Mean Abs Error : {:5.2f} Commodity in ton\".format(mae))",
"_____no_output_____"
]
],
[
[
"<h2> Predicting Test data",
"_____no_output_____"
]
],
[
[
"prediction = model.predict(test_data).flatten()\n\nresult_comparison = pd.DataFrame(list(zip(test_data.values.tolist(),prediction, test_labels.tolist())), \n columns =['Features','Prediction','Actual'])\n\nresult_comparison['error'] = (abs(result_comparison['Actual'] - result_comparison['Prediction'])/\n result_comparison['Prediction'])\nresult_comparison",
"_____no_output_____"
],
[
"print(result_comparison.error.mean())",
"0.033670833236638456\n"
],
[
"loss, mse = model.evaluate(test_data, test_labels, verbose=0)\nprint(\"Mean Absolute Error : {:5.2f} Commodity in ton\".format(loss))\nprint(\"Mean Squared Error : {:5.2f} Commodity in ton\".format(mse))\nprint(result_comparison.error.mean())",
"Mean Absolute Error : 30390.12 Commodity in ton\nMean Squared Error : 1568806912.00 Commodity in ton\n0.033670833236638456\n"
]
],
[
[
"<h2> Ploting data distribution and prediction",
"_____no_output_____"
]
],
[
[
"data_paddy = pd.read_csv('data/padi.csv')\ndata_paddy_central_java = data_paddy[data_paddy['Provinsi']==\"DI YOGYAKARTA\"]\ndata_paddy_central_java = data_paddy_central_java.drop(columns='Provinsi')\ndata_paddy_central_java = data_paddy_central_java.T\ndata_paddy_central_java = data_paddy_central_java.reset_index()\ndata_paddy_central_java = data_paddy_central_java.rename(columns={\"index\":\"Years\",13:\"Commodition_in_Ton\"})\ndata_paddy_central_java = data_paddy_central_java[['Years', 'Commodition_in_Ton']].apply(pd.to_numeric)\n\npred = data_paddy_central_java.rename(columns={\"Commodition_in_Ton\":'predict'}).iloc[-len(test_data):, :]\npred['predict'] = [round(num, 1) for num in prediction]\nfig = plt.figure(figsize=(16,9))\nplt.grid()\nplt.xlabel('Years')\nplt.ylabel('commodity values in ton')\nplt.title('Paddy Production in D.I. Yogyakarta and prediction Regression model')\nplt.plot(data_paddy_central_java['Years'], \n data_paddy_central_java['Commodition_in_Ton'], \n# kind='scatter',\n 'b-',label='Paddy Production growth')\n\nplt.plot(pred['Years'], \n pred['predict'], \n# kind='scatter',\n 'r-',label='Paddy Production prediction')\n\nplt.legend()\nplt.savefig('plot/Paddy Production growth vs prediction.pdf') \nplt.show()",
"_____no_output_____"
]
],
[
[
"<h2> Predicting paddy production for 2016",
"_____no_output_____"
]
],
[
[
"data_prediction = np.array([921824,919573,945136])\ndata_prediction = data_prediction[None, :]\n\npredict_2016 = model.predict(data_prediction).flatten()\nprint(\"Paddy Production for Year 2016 : {} Commodity in ton\".format(int(predict_2016)))",
"Paddy Production for Year 2016 : 969536 Commodity in ton\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6f2212a2d3da99733d96d7b9b9ae056c485f28 | 36,668 | ipynb | Jupyter Notebook | 2nd-ML100Days/homework/D-001/Day_001_HW.ipynb | qwerzxcv98/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
]
| 3 | 2019-08-22T15:19:11.000Z | 2019-08-24T00:54:54.000Z | 2nd-ML100Days/homework/D-001/Day_001_HW.ipynb | magikerwin1993/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
]
| null | null | null | 2nd-ML100Days/homework/D-001/Day_001_HW.ipynb | magikerwin1993/100Day-ML-Marathon | 3c86cd083b086a1e4b7a55a41b127909c7860f79 | [
"MIT"
]
| 1 | 2019-07-18T01:52:04.000Z | 2019-07-18T01:52:04.000Z | 163.696429 | 18,840 | 0.898304 | [
[
[
"# 作業1:\n\n請上 Kaggle, 在 Competitions 或 Dataset 中找一組競賽或資料並寫下:\n\n#### 1. 你選的這組資料為何重要\n https://www.kaggle.com/c/quickdraw-doodle-recognition\n 過去比較常見到的手寫資料集,大多是數字或文字\n 而\"Quick, Draw!\"這組資料集,則是收集大量的手繪圖案(動物,物品,...)\n 相較於數字與文字識別,我認為更具挑戰性與發展性\n 例如:幼兒或讀寫障礙者可以透過圖像來表達,或反過來透過生成相對應圖案來輔助理解文字\n\n#### 2. 資料從何而來 (tips: 譬如提供者是誰、以什麼方式蒐集)\n 這些資料是來自於Google開發的遊戲(https://quickdraw.withgoogle.com/)\n 該遊戲讓玩家透過塗鴉方式進行闖關,若系統判斷該塗鴉與題目夠相似合就給過關\n 過程中收集玩家所創作的50M張手繪圖案(共340類別標籤)\n\n#### 3. 蒐集而來的資料型態為何\n 手繪影像(非結構化):採用原始向量格式,並透過.csv檔保存\n 類別標籤(結構化):相同類別的手繪影像儲存在同一個.csv檔中\n\n#### 4. 這組資料想解決的問題如何評估\n 所有手繪圖案都是由玩家自由塗鴉而成,難免會有牛頭不對馬嘴的情況\n 該kaggle競賽項目希望有方法去清理問題資料\n 所以要確認分類器預測的問題資料是否真的有問題\n 屬於分類問題可使用accuracy,mAP來評估\n",
"_____no_output_____"
],
[
"# 作業2:\n\n想像你經營一個自由載客車隊,你希望能透過數據分析以提升業績,請你思考並描述你如何規劃整體的分析/解決方案:\n\n#### 1. 核心問題為何 (tips:如何定義 「提升業績 & 你的假設」)\n 如何考量所有運輸的相關成本,最小化成本,制定最佳運輸方案\n\n#### 2. 資料從何而來 (tips:哪些資料可能會對你想問的問題產生影響 & 資料如何蒐集)\n 透過駕駛與乘客手機的App來收集\n 上下車地點,行車時間,行進路線,候車時間,載客量,業績...等等。\n\n#### 3. 蒐集而來的資料型態為何\n 結構化: 行車時間,候車時間,載客量,業績,...\n 非結構化: 上下車地點,行進路線,...\n\n#### 4. 你要回答的問題,其如何評估 (tips:你的假設如何驗證)\n 先拿未優化前方案的作為基準,針對演算法給的方案進行實測實驗\n 看看是否有降低成本、增加利潤,屬於回歸問題可用MAE,RMSE評估",
"_____no_output_____"
],
[
"# 作業3:",
"_____no_output_____"
],
[
"## 練習時間\n#### 請寫一個函式用來計算 Mean Square Error\n$ MSE = \\frac{1}{n}\\sum_{i=1}^{n}{(Y_i - \\hat{Y}_i)^2} $\n\n### Hint: [如何取平方](https://googoodesign.gitbooks.io/-ezpython/unit-1.html)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def mean_absolute_error(y, yp):\n \"\"\"\n 計算 MAE\n Args:\n - y: 實際值\n - yp: 預測值\n Return:\n - mae: MAE\n \"\"\"\n mae = MAE = sum(abs(y - yp)) / len(y)\n return mae",
"_____no_output_____"
],
[
"def mean_squared_error(y, yp):\n \"\"\"\n 請完成這個 Function 後往下執行\n \"\"\"\n mse = sum((y - yp)**2) / len(y)\n return mse",
"_____no_output_____"
],
[
"w = 3\nb = 0.5\n\nx_lin = np.linspace(0, 100, 101)\n\ny = (x_lin + np.random.randn(101) * 5) * w + b\n\nplt.plot(x_lin, y, 'b.', label = 'data points')\nplt.title(\"Assume we have data points\")\nplt.legend(loc = 2)\nplt.show()",
"_____no_output_____"
],
[
"y_hat = x_lin * w + b\nplt.plot(x_lin, y, 'b.', label = 'data')\nplt.plot(x_lin, y_hat, 'r-', label = 'prediction')\nplt.title(\"Assume we have data points (And the prediction)\")\nplt.legend(loc = 2)\nplt.show()",
"_____no_output_____"
],
[
"# 執行 Function, 確認有沒有正常執行\nMSE = mean_squared_error(y, y_hat)\nMAE = mean_absolute_error(y, y_hat)\nprint(\"The Mean squared error is %.3f\" % (MSE))\nprint(\"The Mean absolute error is %.3f\" % (MAE))",
"The Mean squared error is 237.463\nThe Mean absolute error is 12.609\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f273fa8f4c1a7d113f79cf781c87788578bb4 | 12,350 | ipynb | Jupyter Notebook | tests/tf/convolutional_network_raw _new.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
]
| 1 | 2019-05-10T09:16:23.000Z | 2019-05-10T09:16:23.000Z | tests/tf/convolutional_network_raw _new.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
]
| null | null | null | tests/tf/convolutional_network_raw _new.ipynb | gopala-kr/ds-notebooks | bc35430ecdd851f2ceab8f2437eec4d77cb59423 | [
"MIT"
]
| 1 | 2019-05-10T09:17:28.000Z | 2019-05-10T09:17:28.000Z | 38.958991 | 357 | 0.575709 | [
[
[
"# Convolutional Neural Network Example\n\nBuild a convolutional neural network with TensorFlow.\n\n- Author: Aymeric Damien\n- Project: https://github.com/aymericdamien/TensorFlow-Examples/",
"_____no_output_____"
],
[
"## CNN Overview\n\n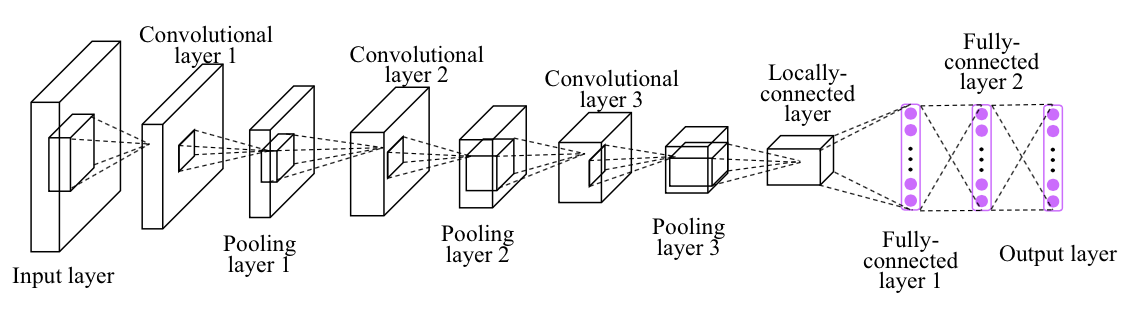\n\n## MNIST Dataset Overview\n\nThis example is using MNIST handwritten digits. The dataset contains 60,000 examples for training and 10,000 examples for testing. The digits have been size-normalized and centered in a fixed-size image (28x28 pixels) with values from 0 to 1. For simplicity, each image has been flattened and converted to a 1-D numpy array of 784 features (28*28).\n\n\n\nMore info: http://yann.lecun.com/exdb/mnist/",
"_____no_output_____"
]
],
[
[
"from __future__ import division, print_function, absolute_import\n\nimport tensorflow as tf\n\n# Import MNIST data\nfrom tensorflow.examples.tutorials.mnist import input_data\nmnist = input_data.read_data_sets(\"/tmp/data/\", one_hot=True)",
"Extracting /tmp/data/train-images-idx3-ubyte.gz\nExtracting /tmp/data/train-labels-idx1-ubyte.gz\nExtracting /tmp/data/t10k-images-idx3-ubyte.gz\nExtracting /tmp/data/t10k-labels-idx1-ubyte.gz\n"
],
[
"# Training Parameters\nlearning_rate = 0.001\nnum_steps = 500\nbatch_size = 128\ndisplay_step = 10\n\n# Network Parameters\nnum_input = 784 # MNIST data input (img shape: 28*28)\nnum_classes = 10 # MNIST total classes (0-9 digits)\ndropout = 0.75 # Dropout, probability to keep units\n\n# tf Graph input\nX = tf.placeholder(tf.float32, [None, num_input])\nY = tf.placeholder(tf.float32, [None, num_classes])\nkeep_prob = tf.placeholder(tf.float32) # dropout (keep probability)",
"_____no_output_____"
],
[
"# Create some wrappers for simplicity\ndef conv2d(x, W, b, strides=1):\n # Conv2D wrapper, with bias and relu activation\n x = tf.nn.conv2d(x, W, strides=[1, strides, strides, 1], padding='SAME')\n x = tf.nn.bias_add(x, b)\n return tf.nn.relu(x)\n\n\ndef maxpool2d(x, k=2):\n # MaxPool2D wrapper\n return tf.nn.max_pool(x, ksize=[1, k, k, 1], strides=[1, k, k, 1],\n padding='SAME')\n\n\n# Create model\ndef conv_net(x, weights, biases, dropout):\n # MNIST data input is a 1-D vector of 784 features (28*28 pixels)\n # Reshape to match picture format [Height x Width x Channel]\n # Tensor input become 4-D: [Batch Size, Height, Width, Channel]\n x = tf.reshape(x, shape=[-1, 28, 28, 1])\n\n # Convolution Layer\n conv1 = conv2d(x, weights['wc1'], biases['bc1'])\n # Max Pooling (down-sampling)\n conv1 = maxpool2d(conv1, k=2)\n\n # Convolution Layer\n conv2 = conv2d(conv1, weights['wc2'], biases['bc2'])\n # Max Pooling (down-sampling)\n conv2 = maxpool2d(conv2, k=2)\n\n # Fully connected layer\n # Reshape conv2 output to fit fully connected layer input\n fc1 = tf.reshape(conv2, [-1, weights['wd1'].get_shape().as_list()[0]])\n fc1 = tf.add(tf.matmul(fc1, weights['wd1']), biases['bd1'])\n fc1 = tf.nn.relu(fc1)\n # Apply Dropout\n fc1 = tf.nn.dropout(fc1, dropout)\n\n # Output, class prediction\n out = tf.add(tf.matmul(fc1, weights['out']), biases['out'])\n return out",
"_____no_output_____"
],
[
"# Store layers weight & bias\nweights = {\n # 5x5 conv, 1 input, 32 outputs\n 'wc1': tf.Variable(tf.random_normal([5, 5, 1, 32])),\n # 5x5 conv, 32 inputs, 64 outputs\n 'wc2': tf.Variable(tf.random_normal([5, 5, 32, 64])),\n # fully connected, 7*7*64 inputs, 1024 outputs\n 'wd1': tf.Variable(tf.random_normal([7*7*64, 1024])),\n # 1024 inputs, 10 outputs (class prediction)\n 'out': tf.Variable(tf.random_normal([1024, num_classes]))\n}\n\nbiases = {\n 'bc1': tf.Variable(tf.random_normal([32])),\n 'bc2': tf.Variable(tf.random_normal([64])),\n 'bd1': tf.Variable(tf.random_normal([1024])),\n 'out': tf.Variable(tf.random_normal([num_classes]))\n}\n\n# Construct model\nlogits = conv_net(X, weights, biases, keep_prob)\nprediction = tf.nn.softmax(logits)\n\n# Define loss and optimizer\nloss_op = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(\n logits=logits, labels=Y))\noptimizer = tf.train.AdamOptimizer(learning_rate=learning_rate)\ntrain_op = optimizer.minimize(loss_op)\n\n\n# Evaluate model\ncorrect_pred = tf.equal(tf.argmax(prediction, 1), tf.argmax(Y, 1))\naccuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))\n\n# Initialize the variables (i.e. assign their default value)\ninit = tf.global_variables_initializer()",
"_____no_output_____"
],
[
"# Start training\nwith tf.Session() as sess:\n\n # Run the initializer\n sess.run(init)\n\n for step in range(1, num_steps+1):\n batch_x, batch_y = mnist.train.next_batch(batch_size)\n # Run optimization op (backprop)\n sess.run(train_op, feed_dict={X: batch_x, Y: batch_y, keep_prob: dropout})\n if step % display_step == 0 or step == 1:\n # Calculate batch loss and accuracy\n loss, acc = sess.run([loss_op, accuracy], feed_dict={X: batch_x,\n Y: batch_y,\n keep_prob: 1.0})\n print(\"Step \" + str(step) + \", Minibatch Loss= \" + \\\n \"{:.4f}\".format(loss) + \", Training Accuracy= \" + \\\n \"{:.3f}\".format(acc))\n\n print(\"Optimization Finished!\")\n\n # Calculate accuracy for 256 MNIST test images\n print(\"Testing Accuracy:\", \\\n sess.run(accuracy, feed_dict={X: mnist.test.images[:256],\n Y: mnist.test.labels[:256],\n keep_prob: 1.0}))\n",
"Step 1, Minibatch Loss= 50639.9102, Training Accuracy= 0.086\nStep 10, Minibatch Loss= 20281.1602, Training Accuracy= 0.289\nStep 20, Minibatch Loss= 9072.8730, Training Accuracy= 0.602\nStep 30, Minibatch Loss= 5935.5635, Training Accuracy= 0.711\nStep 40, Minibatch Loss= 4546.2422, Training Accuracy= 0.766\nStep 50, Minibatch Loss= 4842.9844, Training Accuracy= 0.797\nStep 60, Minibatch Loss= 3678.5024, Training Accuracy= 0.828\nStep 70, Minibatch Loss= 2481.0059, Training Accuracy= 0.852\nStep 80, Minibatch Loss= 2640.6140, Training Accuracy= 0.875\nStep 90, Minibatch Loss= 2017.5098, Training Accuracy= 0.938\nStep 100, Minibatch Loss= 2123.2571, Training Accuracy= 0.891\nStep 110, Minibatch Loss= 2053.9116, Training Accuracy= 0.867\nStep 120, Minibatch Loss= 735.9830, Training Accuracy= 0.953\nStep 130, Minibatch Loss= 1423.4934, Training Accuracy= 0.930\nStep 140, Minibatch Loss= 1899.0032, Training Accuracy= 0.867\nStep 150, Minibatch Loss= 2214.4224, Training Accuracy= 0.852\nStep 160, Minibatch Loss= 1686.8429, Training Accuracy= 0.914\nStep 170, Minibatch Loss= 812.3309, Training Accuracy= 0.945\nStep 180, Minibatch Loss= 2307.6814, Training Accuracy= 0.875\nStep 190, Minibatch Loss= 927.6286, Training Accuracy= 0.930\nStep 200, Minibatch Loss= 654.1238, Training Accuracy= 0.938\nStep 210, Minibatch Loss= 1303.4437, Training Accuracy= 0.898\nStep 220, Minibatch Loss= 609.2120, Training Accuracy= 0.938\nStep 230, Minibatch Loss= 1332.9526, Training Accuracy= 0.938\nStep 240, Minibatch Loss= 1431.8586, Training Accuracy= 0.914\nStep 250, Minibatch Loss= 1080.5891, Training Accuracy= 0.961\nStep 260, Minibatch Loss= 706.7600, Training Accuracy= 0.961\nStep 270, Minibatch Loss= 1946.7012, Training Accuracy= 0.898\nStep 280, Minibatch Loss= 950.1468, Training Accuracy= 0.938\nStep 290, Minibatch Loss= 1305.2469, Training Accuracy= 0.914\nStep 300, Minibatch Loss= 886.6442, Training Accuracy= 0.953\nStep 310, Minibatch Loss= 1135.9377, Training Accuracy= 0.930\nStep 320, Minibatch Loss= 781.4891, Training Accuracy= 0.930\nStep 330, Minibatch Loss= 1126.2645, Training Accuracy= 0.930\nStep 340, Minibatch Loss= 708.8767, Training Accuracy= 0.953\nStep 350, Minibatch Loss= 108.6874, Training Accuracy= 0.977\nStep 360, Minibatch Loss= 347.5796, Training Accuracy= 0.969\nStep 370, Minibatch Loss= 1529.8101, Training Accuracy= 0.922\nStep 380, Minibatch Loss= 842.6948, Training Accuracy= 0.961\nStep 390, Minibatch Loss= 766.7327, Training Accuracy= 0.945\nStep 400, Minibatch Loss= 1103.9432, Training Accuracy= 0.922\nStep 410, Minibatch Loss= 428.1141, Training Accuracy= 0.969\nStep 420, Minibatch Loss= 765.8823, Training Accuracy= 0.945\nStep 430, Minibatch Loss= 278.6405, Training Accuracy= 0.953\nStep 440, Minibatch Loss= 883.9794, Training Accuracy= 0.945\nStep 450, Minibatch Loss= 894.6458, Training Accuracy= 0.906\nStep 460, Minibatch Loss= 1129.0786, Training Accuracy= 0.930\nStep 470, Minibatch Loss= 196.8833, Training Accuracy= 0.969\nStep 480, Minibatch Loss= 1986.0656, Training Accuracy= 0.906\nStep 490, Minibatch Loss= 306.7379, Training Accuracy= 0.977\nStep 500, Minibatch Loss= 897.3098, Training Accuracy= 0.938\nOptimization Finished!\nTesting Accuracy: 0.984375\n"
],
[
"test complete; Gopal",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f2b41715b27d30c6794f3225ad90b3a2b1dcd | 207,253 | ipynb | Jupyter Notebook | notebooks/coverage.ipynb | mbatchkarov/ExpLosion | 705039ec5f77c4203c98487f80d74b9d1f4fd501 | [
"BSD-3-Clause"
]
| 1 | 2015-10-21T08:53:55.000Z | 2015-10-21T08:53:55.000Z | notebooks/coverage.ipynb | mbatchkarov/ExpLosion | 705039ec5f77c4203c98487f80d74b9d1f4fd501 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/coverage.ipynb | mbatchkarov/ExpLosion | 705039ec5f77c4203c98487f80d74b9d1f4fd501 | [
"BSD-3-Clause"
]
| null | null | null | 442.848291 | 120,676 | 0.916382 | [
[
[
"%cd ~/NetBeansProjects/ExpLosion/\nfrom notebooks.common_imports import *",
"/Volumes/LocalDataHD/m/mm/mmb28/NetBeansProjects/ExpLosion\n"
],
[
"df = pd.read_csv('../thesisgenerator/coverage_stats.csv', index_col=0).convert_objects(convert_numeric=True)",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"ddf = df.query('param_composer==\"Add\" & param_algorithm==\"word2vec\" & param_unlabelled==\"wiki\"')",
"_____no_output_____"
],
[
"ddf = ddf.sort('param_unlabelled_percentage')",
"_____no_output_____"
],
[
"# plt.plot(ddf.param_unlabelled_percentage, ddf.AN_count_total, label='AN')\n# plt.plot(ddf.param_unlabelled_percentage, ddf.NN_count_total, label='NN')\nplt.plot(ddf.param_unlabelled_percentage, ddf.N_count_total, label='N')\n# plt.plot(ddf.param_unlabelled_percentage, ddf.N_count_weighted, label='NW')\n# plt.plot(ddf.param_unlabelled_percentage, ddf['1-GRAM_count_weighted'], label='1grW')\n# plt.plot(ddf.param_unlabelled_percentage, ddf['1-GRAM_count_total'], label='1gr')\nplt.plot(ddf.param_unlabelled_percentage, ddf.J_count_total, label='J')\n\nplt.plot(ddf.param_unlabelled_percentage, ddf.V_count_total, label='V')\n# plt.plot(ddf.param_unlabelled_percentage, ddf.V_count_weighted, label='VW')\nplt.axvline(15, c='k')\nplt.legend(loc='upper left')",
"_____no_output_____"
],
[
"cov = ddf['param_unlabelled_percentage N_count_total J_count_total V_count_total AN_count_total NN_count_total'.split()]\ncov.columns = 'Percent Nouns Adjs Verbs ANs NNs'.split()\ncov = pd.melt(cov, id_vars=['Percent'], value_vars='Nouns Adjs Verbs ANs NNs'.split(), \n value_name='Types', var_name='PoS')",
"_____no_output_____"
],
[
"cov['unit'] = [0]*len(cov)\ncov.head()",
"_____no_output_____"
],
[
"# convert percentages to token counts\ndef compute_token_count(row):\n corpus_sizes = {'cwiki': 525000000, 'wiki':1500000000}\n return corpus_sizes['wiki'] * (row.Percent / 100)\n\ncov['Tokens'] = cov.apply(compute_token_count, axis=1)\n\nwith sns.color_palette(\"cubehelix\", 5):\n sns.tsplot(cov, time='Tokens', condition='PoS', value='Types', unit='unit', marker='s', linewidth=4);\n plt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\n\nplt.savefig('plot-type-coverage.pdf', format='pdf', dpi=300, bbox_inches='tight', pad_inches=0.1)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f2f708bdf110b32aed02cfa71a631e066acc0 | 70,649 | ipynb | Jupyter Notebook | classifier/.ipynb_checkpoints/eyes_open-closed_inception_classifier-checkpoint.ipynb | jdpigeon/Neurodoro | 07e1bd16da78dcba648d362bbce90b0dd8b6bacc | [
"MIT"
]
| 47 | 2017-03-28T17:19:21.000Z | 2022-01-26T14:37:39.000Z | classifier/.ipynb_checkpoints/eyes_open-closed_inception_classifier-checkpoint.ipynb | jdpigeon/Neurodoro | 07e1bd16da78dcba648d362bbce90b0dd8b6bacc | [
"MIT"
]
| 28 | 2017-03-28T01:27:04.000Z | 2020-08-02T12:56:39.000Z | classifier/.ipynb_checkpoints/eyes_open-closed_inception_classifier-checkpoint.ipynb | jdpigeon/Neurodoro | 07e1bd16da78dcba648d362bbce90b0dd8b6bacc | [
"MIT"
]
| 7 | 2017-03-03T19:38:10.000Z | 2017-04-19T09:05:14.000Z | 194.090659 | 35,908 | 0.819884 | [
[
[
"# A Basic EEG classifier with Inception\n## Goals\n1. Collect Corvo data from one user, perhaps from just one long session so the electrodes are in the same position.\n2. Break data into short epochs (2s?)\n3. Convert each epoch into a spectrogram (sounds like we can just convert it into a matrix instead of an actual jpeg)\n4. Label each epoch with its associated current Corvo performance score\n5. Feed to Inception (I have a big GPU that might help here) https://www.youtube.com/watch?v=cSKfRcEDGUs\n6. Test",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport scipy.stats as stats\nimport scipy.signal as signal\nimport matplotlib.pyplot as plt\nimport sklearn as sk\nimport tensorflow as tf\nfrom tensorflow.contrib import learn\n\nEPOCH_LENGTH = 440\nVARIANCE_THRESHOLD = 550",
"_____no_output_____"
],
[
"# Data has been collected, let's import it\n\nopen_data = pd.read_csv(\"../Muse Data/DanoThursdayOpenRawEEG0.csv\", header=0, index_col=False)\nclosed_data = pd.read_csv(\"../Muse Data/DanoThursdayClosedRawEEG1.csv\", header=0, index_col=False)",
"_____no_output_____"
],
[
"# Drop difficulty, timestamp, and performance columns since we're not using them\n\nopen_data = open_data.drop(['Difficulty', 'Performance', 'Timestamp (ms)'], axis=1)\nclosed_data = closed_data.drop(['Difficulty', 'Performance', 'Timestamp (ms)'], axis=1)\n\n\n# Prune rows from tail of datasets so that they are all divisible by 440 (the desired size of our epochs)\n\nopen_overflow = open_data.shape[0] % EPOCH_LENGTH\nopen_data = open_data[0:-open_overflow]\nclosed_overflow = closed_data.shape[0] % EPOCH_LENGTH\nclosed_data = closed_data[0:-closed_overflow]",
"_____no_output_____"
],
[
"# Split DataFrames into many different dataframes 440 samples long\n\nsplit_open_data = np.stack(np.array_split(open_data, EPOCH_LENGTH), axis=1)\nsplit_closed_data = np.stack(np.array_split(closed_data, EPOCH_LENGTH), axis=1)\n\n# Transform data into a 3D pandas Panel ( n epochs x 4 channels x 440 samples )\n\nopen_panel = pd.Panel(split_open_data)\nclosed_panel = pd.Panel(split_closed_data)\n\n\nopen_panel.shape\n",
"_____no_output_____"
],
[
"# Remove epochs with too much variance\n\ndef removeNoise(panel):\n for frameIndex in panel:\n for columnIndex in panel[frameIndex]:\n if np.var(panel[frameIndex][columnIndex]) > VARIANCE_THRESHOLD:\n print('variance ', np.var(panel[frameIndex][columnIndex]), ' at electrode ', columnIndex, ' frame ', frameIndex)\n panel = panel.drop(frameIndex)\n break\n return panel\n \nclosed_panel = removeNoise(closed_panel)\nopen_panel = removeNoise(open_panel)\n",
"variance 565.2019284170628 at electrode 3 frame 72\nvariance 585.4320009092443 at electrode 3 frame 73\nvariance 596.9470221979449 at electrode 3 frame 74\nvariance 601.5649475614356 at electrode 3 frame 75\nvariance 564.6084621180363 at electrode 3 frame 76\nvariance 580.7225537057597 at electrode 3 frame 77\nvariance 581.1037362989539 at electrode 3 frame 78\nvariance 575.4028816989534 at electrode 3 frame 79\nvariance 593.2626671243911 at electrode 3 frame 80\nvariance 603.3318585880803 at electrode 3 frame 81\nvariance 607.0889083912873 at electrode 3 frame 82\nvariance 618.329252343474 at electrode 3 frame 83\nvariance 602.9789655801228 at electrode 3 frame 84\nvariance 607.3852222287647 at electrode 3 frame 85\nvariance 621.6071542852401 at electrode 3 frame 86\nvariance 627.8743596727223 at electrode 3 frame 87\nvariance 620.4504121536488 at electrode 3 frame 88\nvariance 622.3241359324637 at electrode 3 frame 89\nvariance 607.3761511117598 at electrode 3 frame 90\nvariance 590.0657343364229 at electrode 3 frame 91\nvariance 575.8104529961632 at electrode 3 frame 92\nvariance 569.5319541378939 at electrode 3 frame 133\nvariance 714.4534612214848 at electrode 3 frame 134\nvariance 916.0835804070445 at electrode 3 frame 135\nvariance 1136.7916004543645 at electrode 3 frame 136\nvariance 1392.0835266392182 at electrode 3 frame 137\nvariance 1653.0708144364266 at electrode 3 frame 138\nvariance 1818.564342976062 at electrode 3 frame 139\nvariance 1843.4597929769334 at electrode 3 frame 140\nvariance 1861.0930380873722 at electrode 3 frame 141\nvariance 1869.3656032782656 at electrode 3 frame 142\nvariance 1853.3512380693373 at electrode 3 frame 143\nvariance 1891.4706854395013 at electrode 3 frame 144\nvariance 1924.0795759878329 at electrode 3 frame 145\nvariance 1900.477060667249 at electrode 3 frame 146\nvariance 1934.0630041634647 at electrode 3 frame 147\nvariance 1964.8055648018537 at electrode 3 frame 148\nvariance 1959.9487433954341 at electrode 3 frame 149\nvariance 1948.833969420409 at electrode 3 frame 150\nvariance 1961.7047243550517 at electrode 3 frame 151\nvariance 1994.5437752789437 at electrode 3 frame 152\nvariance 1995.162008942119 at electrode 3 frame 153\nvariance 1985.7908462217063 at electrode 3 frame 154\nvariance 1970.695207656704 at electrode 3 frame 155\nvariance 1968.7063967015808 at electrode 3 frame 156\nvariance 1944.007953471432 at electrode 3 frame 157\nvariance 1922.0496444992243 at electrode 3 frame 158\nvariance 1900.3542162183069 at electrode 3 frame 159\nvariance 1890.8265522696743 at electrode 3 frame 160\nvariance 1888.3099113259416 at electrode 3 frame 161\nvariance 1871.1975076702074 at electrode 3 frame 162\nvariance 1868.9206852558796 at electrode 3 frame 163\nvariance 1868.6352875083571 at electrode 3 frame 164\nvariance 1980.0285344545682 at electrode 3 frame 165\nvariance 1964.022681279929 at electrode 3 frame 166\nvariance 867.6490569963588 at electrode 3 frame 167\nvariance 839.2241737414919 at electrode 3 frame 168\nvariance 776.6074690551703 at electrode 3 frame 169\nvariance 695.1209540925781 at electrode 3 frame 170\nvariance 637.9379742863572 at electrode 3 frame 171\nvariance 589.5089857945231 at electrode 3 frame 172\n"
],
[
"plt.figure()\nplt.subplot(2,2,1)\nplt.specgram(open_panel[20][0], NFFT=256, Fs=220, noverlap=198)\nplt.ylim(0,55)\nplt.subplot(2,2,2)\nplt.specgram(open_panel[20][1], NFFT=256, Fs=220, noverlap=198)\nplt.ylim(0,55)\nplt.subplot(2,2,3)\nplt.specgram(open_panel[20][2], NFFT=256, Fs=220, noverlap=198)\nplt.ylim(0,55)\nplt.subplot(2,2,4)\nplt.specgram(open_panel[20][3], NFFT=256, Fs=220, noverlap=198)\nplt.ylim(0,55)\nplt.show",
"/home/dano/anaconda3/lib/python3.5/site-packages/matplotlib/pyplot.py:516: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).\n max_open_warning, RuntimeWarning)\n"
],
[
"# Plot test spectograms of all 4 channels\n\ndef plotAndSave(frame, filename):\n plt.figure()\n plt.subplot(2,2,1)\n plt.specgram(frame[0], NFFT=256, Fs=220, noverlap=198)\n plt.ylim(0,55)\n plt.subplot(2,2,2)\n plt.specgram(frame[2], NFFT=256, Fs=220, noverlap=198)\n plt.ylim(0,55)\n plt.subplot(2,2,3)\n plt.specgram(frame[3], NFFT=256, Fs=220, noverlap=198)\n plt.ylim(0,55)\n plt.savefig('%s.jpg' % filename, pad_inches=0, bbox_inches='tight')\n \nfor frameIndex in open_panel:\n plotAndSave(open_panel[frameIndex], 'open%s' % frameIndex)\n\nfor frameIndex in closed_panel:\n plotAndSave(closed_panel[frameIndex], 'closed%s' % frameIndex)\n \n",
"/home/dano/anaconda3/lib/python3.5/site-packages/matplotlib/pyplot.py:516: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).\n max_open_warning, RuntimeWarning)\n"
]
],
[
[
"Questions to answer before continuing:\n- Do these spectrograms look alright?\n- Shouldn't the axis go all the way to 2 if there are 440 samples at 220hz sampling rate?\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec6f3c26a66e4007831f5d76a9eb4ef873a1b478 | 987,117 | ipynb | Jupyter Notebook | DATA-TESTER-FK.ipynb | ricardodeazambuja/IJCNN2017-2 | 51b26f81b9137b368ff93d446485ac9b85f6854c | [
"MIT"
]
| 4 | 2019-10-27T10:38:05.000Z | 2021-03-23T12:36:15.000Z | DATA-TESTER-FK.ipynb | ricardodeazambuja/IJCNN2017-2 | 51b26f81b9137b368ff93d446485ac9b85f6854c | [
"MIT"
]
| null | null | null | DATA-TESTER-FK.ipynb | ricardodeazambuja/IJCNN2017-2 | 51b26f81b9137b368ff93d446485ac9b85f6854c | [
"MIT"
]
| 1 | 2017-07-19T23:51:25.000Z | 2017-07-19T23:51:25.000Z | 2,964.315315 | 507,076 | 0.950838 | [
[
[
"import numpy\nimport time\nimport sys\nimport os\nimport save_load_file as slf # https://github.com/ricardodeazambuja/Python-UTILS\n%matplotlib inline\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"def rot_baxter(theta, alpha):\n cos = numpy.cos\n sin = numpy.sin\n l1 = numpy.array([cos(theta), -sin(theta)*cos(alpha), sin(theta)*sin(alpha)])\n l2 = numpy.array([sin(theta), cos(theta)*cos(alpha), -cos(theta)*sin(alpha)])\n l3 = numpy.array([ 0, sin(alpha), cos(alpha)])\n \n return numpy.vstack([l1,l2,l3])",
"_____no_output_____"
],
[
"def baxter_arm(s0=0,s1=0,e0=0,e1=0,w0=0,w1=0,w2=0,gripper=[0,0,147.012]):\n '''\n Returns the endpoint cartesian position\n '''\n # Corrections to match the real BAXTER\n s0=s0+numpy.pi/4\n s1=-s1\n e1=-e1+numpy.pi/2\n w1=-w1\n return numpy.array([0,0,270.35])+rot_baxter(numpy.pi/2+s0, numpy.pi/2).dot(numpy.array([69,0,0])+\\\n rot_baxter(numpy.pi/2+s1, numpy.pi/2).dot(numpy.array([0,0,364.35])+\\\n rot_baxter(0+e0, -numpy.pi/2).dot(numpy.array([-69,0,0])+\\\n rot_baxter(-numpy.pi/2+e1, numpy.pi/2).dot(numpy.array([0,0,374.29])+\\\n rot_baxter(0+w0, -numpy.pi/2).dot(numpy.array([-10,0,0])+\\\n rot_baxter(0+w1, numpy.pi/2).dot(numpy.array([0,0,229.525+gripper[2]])+\\\n rot_baxter(0+w2, 0).dot(numpy.array([gripper[0],gripper[1],0]))))))))",
"_____no_output_____"
],
[
"#\n# Controls if the results are saved to a file\n#\n\nsave2file = True",
"_____no_output_____"
],
[
"sim_set = \"experiment_0001\" # basically is the name of the folder where the data is read/saved\nbase_dir = \"simulation_data_00003\"",
"_____no_output_____"
],
[
"# %%time\n\n# number_of_trials = 100\n\n# # for percentage_of_decimated_connections in [0.01, 0.025, 0.05, 0.075, 0.1, 0.25, 0.5, 0.75, 1.0]:\n# # extra_name = \"_Monolithic_decIntConn_\"+str(percentage_of_decimated_connections)+\"_\"\n# # extra_name = \"_Modular_decIntConn_\"+str(percentage_of_decimated_connections)+\"_\"\n \n \n# # for number_of_decimated_columns in range(5):\n# # extra_name = \"_Monolithic_decColumns_\"+str(number_of_decimated_columns)+\"_\"\n# # extra_name = \"_Modular_decColumns_\"+str(number_of_decimated_columns)+\"_\"\n\n\n# # for decimated_neurons in numpy.arange(11)*6:\n# # extra_name = \"_Monolithic_decNeurons_\" + str(decimated_neurons) + \"_\"\n# # extra_name = \"_Modular_decNeurons_\" + str(decimated_neurons) + \"_\"\n\n# for decimated_neurons in numpy.arange(11)*6*5:\n# extra_name = \"_Monolithic_decNeurons_ind_col_\" + str(decimated_neurons) + \"_\"\n# # extra_name = \"_Modular_decNeurons_ind_col_\" + str(decimated_neurons) + \"_\"\n\n# simulation_type = \"parallel\"\n\n# lsm_i = \"ALL\"\n\n# trials_range = range(number_of_trials)\n\n# for trial_number in trials_range: \n# filename = \"./\"+base_dir+\"/\"+sim_set+\"/joint_angles_mean_\"+simulation_type+\"_\"+str(lsm_i)+\"_\"+str(trial_number)+extra_name+\"_LSM.npy\"\n# joint_angles_mean = numpy.load(filename)\n\n# xyz_pos = []\n# for s0, s1, e1, w1 in joint_angles_mean:\n# xyz_pos.append((baxter_arm(s0=s0,s1=s1,e1=e1,w1=w1)+numpy.array([-0.2591,-0.1362,1.054])*1E3)/1000)\n\n# filename = \"./\"+base_dir+\"/\"+sim_set+\"/baxter_xyz_joint_angles_mean_\"+simulation_type+\"_\"+str(lsm_i)+\"_\"+str(trial_number)+extra_name+\"_VREP.npy\"\n# if save2file:\n# numpy.save(filename,xyz_pos)\n\n# xyz_pos = numpy.array(xyz_pos)",
"CPU times: user 6min, sys: 1.56 s, total: 6min 1s\nWall time: 6min 4s\n"
]
],
[
[
"# Loads the values from files:",
"_____no_output_____"
]
],
[
[
"# it's for each column!!!\n# percentage_of_decimated_connections = 0.075 # [0.01, 0.025, 0.05, 0.075, 0.1, 0.25, 0.5, 0.75, 1.0]\n# extra_name = \"_Monolithic_decIntConn_\"+str(percentage_of_decimated_connections)+\"_\"\n# extra_name = \"_Modular_decIntConn_\"+str(percentage_of_decimated_connections)+\"_\"\n\n# number_of_decimated_columns = 4 # [0, 1, 2, 3, 4]\n# extra_name = \"_Monolithic_decColumns_\"+str(number_of_decimated_columns)+\"_\"\n# extra_name = \"_Modular_decColumns_\"+str(number_of_decimated_columns)+\"_\"\n\n# it's for each column!!!\n# decimated_neurons = 6 # [ 0, 6, 12, 18, 24, 30, 36, 42, 48, 54, 60]\n# extra_name = \"_Monolithic_decNeurons_\" + str(decimated_neurons) + \"_\"\n# extra_name = \"_Modular_decNeurons_\" + str(decimated_neurons) + \"_\"\n\n# the decimated neurons are located in only one column (a randomly chosen one)\ndecimated_neurons = 270 #[ 0, 30, 60, 90, 120, 150, 180, 210, 240, 270, 300]\nextra_name = \"_Monolithic_decNeurons_ind_col_\" + str(decimated_neurons) + \"_\"\n# extra_name = \"_Modular_decNeurons_ind_col_\" + str(decimated_neurons) + \"_\"\n\n\nnumber_of_trials = 100\n\n# simulation_type = \"parallel\"\n\n# lsm_i = \"ALL\"\n\nparams = {'legend.fontsize': 10,\n 'figure.figsize': (15, 5),\n 'axes.labelsize': 10,\n 'axes.titlesize':10,\n 'xtick.labelsize':10,\n 'ytick.labelsize':10}\nplt.rcParams.update(params)\n\nplt.figure(figsize =(10,10))\n\n\nfor trial_number in range(number_of_trials):\n\n filename = \"./\"+base_dir+\"/\"+sim_set+\"/baxter_xyz_joint_angles_mean_parallel_ALL_\"+str(trial_number)+extra_name+\"_VREP.npy\"\n xyz_pos = numpy.load(filename)\n\n plt.plot(xyz_pos[:,0]-xyz_pos[0,0],xyz_pos[:,1]-xyz_pos[0,1])\n \nplt.xlabel(\"x (m)\")\nplt.ylabel(\"y (m)\")\nplt.xlim([-0.05,0.2])\nplt.ylim([-0.05,0.2])\n# plt.title(\"Cartesian Movement Generated by Baxter - \"+extra_name)\n\n\nplt.axes().set_aspect('equal', 'datalim')\n\nplt.savefig(\"Results\"+extra_name+'.png', bbox_inches='tight',pad_inches=1, dpi=300)\n\nplt.show()\n",
"_____no_output_____"
],
[
"\n# the decimated neurons are located in only one column (a randomly chosen one)\ndecimated_neurons = [ 0, 30, 60, 90, 120, 150, 180, 210, 240, 270, 300]\n\n\nnumber_of_trials = 100\n\n# simulation_type = \"parallel\"\n\n# lsm_i = \"ALL\"\n\nparams = {'legend.fontsize': 10,\n 'figure.figsize': (15, 5),\n 'axes.labelsize': 10,\n 'axes.titlesize':10,\n 'xtick.labelsize':10,\n 'ytick.labelsize':10}\nplt.rcParams.update(params)\n\n\nfor di,dn in enumerate(decimated_neurons):\n \n for _extra_name,diextra in zip([\"Modular\",\"Monolithic\"],[0,len(decimated_neurons)]):\n \n plt.subplot(2,len(decimated_neurons),di+1+diextra)\n extra_name = \"_\"+_extra_name+\"_decNeurons_ind_col_\" + str(dn) + \"_\"\n\n for trial_number in range(number_of_trials):\n\n filename = \"./\"+base_dir+\"/\"+sim_set+\"/baxter_xyz_joint_angles_mean_parallel_ALL_\"+str(trial_number)+extra_name+\"_VREP.npy\"\n xyz_pos = numpy.load(filename)\n\n plt.plot(xyz_pos[:,0]-xyz_pos[0,0],xyz_pos[:,1]-xyz_pos[0,1], linewidth=0.5)\n\n if diextra:\n plt.xlabel(\"(\"+str(dn)+\")\")\n \n if di==0:\n plt.ylabel(_extra_name)\n \n plt.xlim([-0.05,0.2])\n plt.ylim([-0.05,0.2])\n plt.gca().set_aspect(1)\n plt.gca().set_xticks([])\n plt.gca().set_yticks([])\n \nplt.tight_layout(pad=0, h_pad=0, w_pad=1, rect=None)\nplt.savefig(\"Row_Results\"+extra_name+'.png', bbox_inches='tight',pad_inches=1, dpi=300)\n\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6f42c3b4a10a48af621be7b2520b9ec39ac8da | 14,349 | ipynb | Jupyter Notebook | week1_intro/crossentropy_method.ipynb | YaroslavSchubert/Practical_RL | c91c606d046ca9aaaa01d5585ac72d2db6908d19 | [
"MIT"
]
| 2 | 2019-02-13T15:47:11.000Z | 2019-02-26T19:50:11.000Z | week1_intro/crossentropy_method.ipynb | YaroslavSchubert/Practical_RL | c91c606d046ca9aaaa01d5585ac72d2db6908d19 | [
"MIT"
]
| 2 | 2019-02-18T19:33:36.000Z | 2019-02-19T15:03:35.000Z | week1_intro/crossentropy_method.ipynb | YaroslavSchubert/Practical_RL | c91c606d046ca9aaaa01d5585ac72d2db6908d19 | [
"MIT"
]
| 1 | 2019-02-14T21:12:04.000Z | 2019-02-14T21:12:04.000Z | 32.317568 | 319 | 0.55307 | [
[
[
"# Crossentropy method\n\nThis notebook will teach you to solve reinforcement learning problems with crossentropy method.",
"_____no_output_____"
]
],
[
[
"import gym\nimport numpy as np, pandas as pd\n\nenv = gym.make(\"Taxi-v2\")\nenv.reset()\nenv.render()",
"_____no_output_____"
],
[
"n_states = env.observation_space.n\nn_actions = env.action_space.n\n\nprint(\"n_states=%i, n_actions=%i\"%(n_states, n_actions))",
"_____no_output_____"
]
],
[
[
"# Create stochastic policy\n\nThis time our policy should be a probability distribution.\n\n```policy[s,a] = P(take action a | in state s)```\n\nSince we still use integer state and action representations, you can use a 2-dimensional array to represent the policy.\n\nPlease initialize policy __uniformly__, that is, probabililities of all actions should be equal.\n",
"_____no_output_____"
]
],
[
[
"policy = <your code here! Create an array to store action probabilities>",
"_____no_output_____"
],
[
"assert type(policy) in (np.ndarray,np.matrix)\nassert np.allclose(policy,1./n_actions)\nassert np.allclose(np.sum(policy,axis=1), 1)",
"_____no_output_____"
]
],
[
[
"# Play the game\n\nJust like before, but we also record all states and actions we took.",
"_____no_output_____"
]
],
[
[
"def generate_session(policy,t_max=10**4):\n \"\"\"\n Play game until end or for t_max ticks.\n :param policy: an array of shape [n_states,n_actions] with action probabilities\n :returns: list of states, list of actions and sum of rewards\n \"\"\"\n states,actions = [],[]\n total_reward = 0.\n \n s = env.reset()\n \n for t in range(t_max):\n \n a = <sample action from policy (hint: use np.random.choice)>\n \n new_s, r, done, info = env.step(a)\n \n #Record state, action and add up reward to states,actions and total_reward accordingly. \n states.append(s)\n actions.append(a)\n total_reward += r\n \n s = new_s\n if done:\n break\n return states, actions, total_reward\n ",
"_____no_output_____"
],
[
"s,a,r = generate_session(policy)\nassert type(s) == type(a) == list\nassert len(s) == len(a)\nassert type(r) in [float,np.float]",
"_____no_output_____"
],
[
"#let's see the initial reward distribution\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nsample_rewards = [generate_session(policy,t_max=1000)[-1] for _ in range(200)]\n\nplt.hist(sample_rewards,bins=20);\nplt.vlines([np.percentile(sample_rewards, 50)], [0], [100], label=\"50'th percentile\", color='green')\nplt.vlines([np.percentile(sample_rewards, 90)], [0], [100], label=\"90'th percentile\", color='red')\nplt.legend()",
"_____no_output_____"
]
],
[
[
"### Crossentropy method steps (2pts)",
"_____no_output_____"
]
],
[
[
"def select_elites(states_batch,actions_batch,rewards_batch,percentile=50):\n \"\"\"\n Select states and actions from games that have rewards >= percentile\n :param states_batch: list of lists of states, states_batch[session_i][t]\n :param actions_batch: list of lists of actions, actions_batch[session_i][t]\n :param rewards_batch: list of rewards, rewards_batch[session_i]\n \n :returns: elite_states,elite_actions, both 1D lists of states and respective actions from elite sessions\n \n Please return elite states and actions in their original order \n [i.e. sorted by session number and timestep within session]\n \n If you're confused, see examples below. Please don't assume that states are integers (they'll get different later).\n \"\"\"\n \n reward_threshold = <Compute minimum reward for elite sessions. Hint: use np.percentile>\n \n elite_states = <your code here>\n elite_actions = <your code here>\n \n return elite_states, elite_actions\n ",
"_____no_output_____"
],
[
"states_batch = [\n [1,2,3], #game1\n [4,2,0,2], #game2\n [3,1] #game3\n]\n\nactions_batch = [\n [0,2,4], #game1\n [3,2,0,1], #game2\n [3,3] #game3\n]\nrewards_batch = [\n 3, #game1\n 4, #game2\n 5, #game3\n]\n\ntest_result_0 = select_elites(states_batch, actions_batch, rewards_batch, percentile=0)\ntest_result_40 = select_elites(states_batch, actions_batch, rewards_batch, percentile=30)\ntest_result_90 = select_elites(states_batch, actions_batch, rewards_batch, percentile=90)\ntest_result_100 = select_elites(states_batch, actions_batch, rewards_batch, percentile=100)\n\nassert np.all(test_result_0[0] == [1, 2, 3, 4, 2, 0, 2, 3, 1]) \\\n and np.all(test_result_0[1] == [0, 2, 4, 3, 2, 0, 1, 3, 3]),\\\n \"For percentile 0 you should return all states and actions in chronological order\"\nassert np.all(test_result_40[0] == [4, 2, 0, 2, 3, 1]) and \\\n np.all(test_result_40[1] ==[3, 2, 0, 1, 3, 3]),\\\n \"For percentile 30 you should only select states/actions from two first\"\nassert np.all(test_result_90[0] == [3,1]) and \\\n np.all(test_result_90[1] == [3,3]),\\\n \"For percentile 90 you should only select states/actions from one game\"\nassert np.all(test_result_100[0] == [3,1]) and\\\n np.all(test_result_100[1] == [3,3]),\\\n \"Please make sure you use >=, not >. Also double-check how you compute percentile.\"\nprint(\"Ok!\")",
"_____no_output_____"
],
[
"def update_policy(elite_states,elite_actions):\n \"\"\"\n Given old policy and a list of elite states/actions from select_elites,\n return new updated policy where each action probability is proportional to\n \n policy[s_i,a_i] ~ #[occurences of si and ai in elite states/actions]\n \n Don't forget to normalize policy to get valid probabilities and handle 0/0 case.\n In case you never visited a state, set probabilities for all actions to 1./n_actions\n \n :param elite_states: 1D list of states from elite sessions\n :param elite_actions: 1D list of actions from elite sessions\n \n \"\"\"\n \n new_policy = np.zeros([n_states,n_actions])\n \n <Your code here: update probabilities for actions given elite states & actions>\n #Don't forget to set 1/n_actions for all actions in unvisited states.\n \n \n return new_policy",
"_____no_output_____"
],
[
"\nelite_states, elite_actions = ([1, 2, 3, 4, 2, 0, 2, 3, 1], [0, 2, 4, 3, 2, 0, 1, 3, 3])\n\n\nnew_policy = update_policy(elite_states,elite_actions)\n\nassert np.isfinite(new_policy).all(), \"Your new policy contains NaNs or +-inf. Make sure you don't divide by zero.\"\nassert np.all(new_policy>=0), \"Your new policy can't have negative action probabilities\"\nassert np.allclose(new_policy.sum(axis=-1),1), \"Your new policy should be a valid probability distribution over actions\"\nreference_answer = np.array([\n [ 1. , 0. , 0. , 0. , 0. ],\n [ 0.5 , 0. , 0. , 0.5 , 0. ],\n [ 0. , 0.33333333, 0.66666667, 0. , 0. ],\n [ 0. , 0. , 0. , 0.5 , 0.5 ]])\nassert np.allclose(new_policy[:4,:5],reference_answer)\nprint(\"Ok!\")",
"_____no_output_____"
]
],
[
[
"# Training loop\nGenerate sessions, select N best and fit to those.",
"_____no_output_____"
]
],
[
[
"from IPython.display import clear_output\n\ndef show_progress(batch_rewards, log, percentile, reward_range=[-990,+10]):\n \"\"\"\n A convenience function that displays training progress. \n No cool math here, just charts.\n \"\"\"\n \n mean_reward, threshold = np.mean(batch_rewards), np.percentile(batch_rewards, percentile)\n log.append([mean_reward,threshold])\n\n clear_output(True)\n print(\"mean reward = %.3f, threshold=%.3f\"%(mean_reward, threshold))\n plt.figure(figsize=[8,4])\n plt.subplot(1,2,1)\n plt.plot(list(zip(*log))[0], label='Mean rewards')\n plt.plot(list(zip(*log))[1], label='Reward thresholds')\n plt.legend()\n plt.grid()\n \n plt.subplot(1,2,2)\n plt.hist(batch_rewards,range=reward_range);\n plt.vlines([np.percentile(batch_rewards, percentile)], [0], [100], label=\"percentile\", color='red')\n plt.legend()\n plt.grid()\n\n plt.show()\n",
"_____no_output_____"
],
[
"#reset policy just in case\npolicy = np.ones([n_states, n_actions]) / n_actions ",
"_____no_output_____"
],
[
"n_sessions = 250 #sample this many sessions\npercentile = 50 #take this percent of session with highest rewards\nlearning_rate = 0.5 #add this thing to all counts for stability\n\nlog = []\n\nfor i in range(100):\n \n %time sessions = [<generate a list of n_sessions new sessions>]\n \n batch_states,batch_actions,batch_rewards = zip(*sessions)\n\n elite_states, elite_actions = <select elite states/actions>\n \n new_policy = <compute new policy>\n \n policy = learning_rate * new_policy + (1-learning_rate) * policy\n \n #display results on chart\n show_progress(batch_rewards, log, percentile)",
"_____no_output_____"
]
],
[
[
"### Reflecting on results\n\nYou may have noticed that the taxi problem quickly converges from <-1000 to a near-optimal score and then descends back into -50/-100. This is in part because the environment has some innate randomness. Namely, the starting points of passenger/driver change from episode to episode.\n\nIn case CEM failed to learn how to win from one distinct starting point, it will simply discard it because no sessions from that starting point will make it into the \"elites\".\n\nTo mitigate that problem, you can either reduce the threshold for elite sessions (duct tape way) or change the way you evaluate strategy (theoretically correct way). You can first sample an action for every possible state and then evaluate this choice of actions by running _several_ games and averaging rewards.",
"_____no_output_____"
],
[
"### Grader",
"_____no_output_____"
]
],
[
[
"sessions = [generate_session(policy) for _ in range(100)]\n_, _, session_rewards = zip(*sessions)\nsession_rewards = np.array(session_rewards)\nassert np.mean(session_rewards) > -48, \"Sorry, but you didn't pass\"\nprint(\"Congratulations!\")",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec6f4882af7fa3e4cc1872a3425d4d52a28765f7 | 10,214 | ipynb | Jupyter Notebook | convert_to_structured_data.ipynb | jamesstoneco/MyStudentData-to-csv | 8fc9760e3408c1450e26aa776c80c44cb411e972 | [
"MIT"
]
| null | null | null | convert_to_structured_data.ipynb | jamesstoneco/MyStudentData-to-csv | 8fc9760e3408c1450e26aa776c80c44cb411e972 | [
"MIT"
]
| null | null | null | convert_to_structured_data.ipynb | jamesstoneco/MyStudentData-to-csv | 8fc9760e3408c1450e26aa776c80c44cb411e972 | [
"MIT"
]
| null | null | null | 35.465278 | 1,993 | 0.498923 | [
[
[
"cols = ['Loan Type:',\n'Loan Award ID:',\n'Loan Attending School Name:',\n'Loan Date:',\n'Loan Amount:',\n'Loan Disbursed Amount:',\n'Loan Outstanding Principal Balance:',\n'Loan Outstanding Interest Balance:',\n'Loan Interest Rate Type:',\n'Loan Interest Rate:',\n'Loan Contact Name:']",
"_____no_output_____"
],
[
"delimiter = 'Loan Type:'\nloans = open('MyStudentData.txt', 'r').read().split(delimiter)",
"_____no_output_____"
],
[
"loans.pop(0) # remove header info",
"_____no_output_____"
],
[
"# devide by each loan\nproc_loans = []\nfor l in loans:\n proc_loans.append((delimiter + l).split('\\n'))",
"_____no_output_____"
],
[
"# rangle data into consistent format for pandas\ndata = []\nfor p in proc_loans:\n temp_loan = []\n for c in cols:\n for line in p:\n if line.startswith(c):\n # strip dollar signs and commas and % so they end up as floats\n temp_loan.append(line.split(':')[1].replace('$', '').replace(',', '').replace('%', ''))\n # HACK: pull off extra loan contact names and only retain the first one\n if len(temp_loan) > 11:\n for n in range(len(temp_loan) - 11):\n temp_loan.pop()\n data.append(temp_loan)\n# data",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"df = pd.DataFrame.from_records(data, columns=list(map(lambda c: c.replace(':', ''), cols)), coerce_float=True)",
"_____no_output_____"
],
[
"# convert date\ndf['Loan Date'] = df['Loan Date'].astype('datetime64')\n# convert floats\ncols_to_floats = [\n 'Loan Amount',\n 'Loan Disbursed Amount',\n 'Loan Outstanding Principal Balance',\n 'Loan Outstanding Interest Balance',\n# 'Loan Interest Rate:' # NOTE: there are blanks in my data, so would need to account for this to convert to a float\n]\nfor c in cols_to_floats:\n df[c] = df[c].astype('float')",
"_____no_output_____"
],
[
"df.describe()",
"_____no_output_____"
],
[
"df.to_csv('output.csv', index = True)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6f48c8d0a852b85d4c7b198ded9b78b446ff3f | 899,374 | ipynb | Jupyter Notebook | notebook_demos/2_VP_Redux_w_Interferon.ipynb | BrianaLane/Interferon | 11de1ef332c4ea5eed20eb01b47f200ace406d43 | [
"MIT"
]
| null | null | null | notebook_demos/2_VP_Redux_w_Interferon.ipynb | BrianaLane/Interferon | 11de1ef332c4ea5eed20eb01b47f200ace406d43 | [
"MIT"
]
| null | null | null | notebook_demos/2_VP_Redux_w_Interferon.ipynb | BrianaLane/Interferon | 11de1ef332c4ea5eed20eb01b47f200ace406d43 | [
"MIT"
]
| null | null | null | 758.965401 | 752,760 | 0.948006 | [
[
[
"import sys\nimport glob as glob\nimport numpy as np\n\nsys.path.append('../')",
"_____no_output_____"
]
],
[
[
"# Interferon\n\n- take in Remedy data products for VP and preform:\n - allow for user run custom sky subtraction\n - dither normalization from guider frames \n - flux calbiration using standard stars\n - build data cube - interpolate over dither set while conserving flux\n - spectral extraction over aperature \n - emission line fitting using Bayesian scheme\n - build emission line images \n - normalize overlapping fields ",
"_____no_output_____"
],
[
"# Define data paths",
"_____no_output_____"
]
],
[
[
"data_path = '/Volumes/B_SS/VIRUS_P/VP_reduction/20210411_test/redux'\nguider_path = '/Volumes/B_SS/VIRUS_P/VP_reduction/20210411_test/guider'\ndith_file = '../VP_config/dith_vp_6subdither.csv'\ncen_file = '../VP_config/IFUcen_VP_new_27m.csv'",
"_____no_output_____"
]
],
[
[
"# Auto build data cubes from Remedy products",
"_____no_output_____"
]
],
[
[
"from auto_VP_run import VP_run\n\nvp1 = VP_run(data_path, fits_ext=0, guider_path=guider_path, \n dith_file=dith_file, cen_file=cen_file)\n\nvp1.run_all_dithers()",
"_____no_output_____"
]
],
[
[
"# Working with individual objects \n\n 1) Guider object\n 2) VIRUS-P frame (one dither) object\n 3) Dither set object\n 4) Data cube object\n 5) Spectrum object",
"_____no_output_____"
],
[
"## Example of 1) guider object",
"_____no_output_____"
]
],
[
[
"import guider_observations as go",
"_____no_output_____"
],
[
"guid = go.guider_observations(guider_path)",
"BUILD guider observation: [GUIDER]\n"
],
[
"guid.guider_df.head(5)",
"_____no_output_____"
],
[
"guid.inspect_guider_frame(500)",
"_____no_output_____"
],
[
"ex_source_df = guid.find_guide_stars(500)\nex_source_df",
"_____no_output_____"
],
[
"ex_source_df2 = guid.measure_guide_star_params(500, ex_source_df)\nex_source_df2",
"_____no_output_____"
]
],
[
[
"## Example of 2) VIRUS-P frame object",
"_____no_output_____"
]
],
[
[
"import VP_fits_frame as vpf",
"_____no_output_____"
],
[
"file_ex = '/Volumes/B_SS/VIRUS_P/VP_reduction/20210411/redux/COOLJ0931_dither_1_20210411T030846_multi.fits'\n\nfits_ex = vpf.VP_fits_frame(file_ex, 0, cen_file=cen_file, guide_obs=guid)\n",
"BUILD VP science frame [COOLJ0931_dither_1_20210411T030846_multi.fits][EXT:0]\n"
],
[
"fits_ex.dith_num",
"_____no_output_____"
],
[
"fits_ex.build_frame_sum_spec(plot=True)",
"_____no_output_____"
]
],
[
[
"## Example of 3) dither observations object",
"_____no_output_____"
]
],
[
[
"import dither_observations as do",
"_____no_output_____"
],
[
"file_list = glob.glob(data_path+'/COOLJ0931*_multi.fits')\n\nfits_1 = vpf.VP_fits_frame(file_list[0], 0, cen_file=cen_file, guide_obs=guid)\nfits_2 = vpf.VP_fits_frame(file_list[1], 0, cen_file=cen_file, guide_obs=guid)\nfits_3 = vpf.VP_fits_frame(file_list[2], 0, cen_file=cen_file, guide_obs=guid)\nfits_4 = vpf.VP_fits_frame(file_list[3], 0, cen_file=cen_file, guide_obs=guid)\nfits_5 = vpf.VP_fits_frame(file_list[4], 0, cen_file=cen_file, guide_obs=guid)\nfits_6 = vpf.VP_fits_frame(file_list[5], 0, cen_file=cen_file, guide_obs=guid)\n\nfile_obj_lis = [fits_1, fits_2, fits_3, fits_4, fits_5, fits_6]\n\n# file_obj_lis = []\n# for f in file_list:\n# fits_ex = vpf.VP_fits_frame(f, 0, cen_file=cen_file, guide_obs=guid)\n# file_obj_lis.append(fits_ex)",
"BUILD VP science frame [COOLJ0931_dither_1_20210411T030846_multi.fits][EXT:0]\nBUILD VP science frame [COOLJ0931_dither_2_20210411T032522_multi.fits][EXT:0]\nBUILD VP science frame [COOLJ0931_dither_3_20210411T034159_multi.fits][EXT:0]\nBUILD VP science frame [COOLJ0931_dither_4_20210411T035835_multi.fits][EXT:0]\nBUILD VP science frame [COOLJ0931_dither_5_20210411T041511_multi.fits][EXT:0]\nBUILD VP science frame [COOLJ0931_dither_6_20210411T043147_multi.fits][EXT:0]\n"
],
[
"fits_1.fits_ext",
"_____no_output_____"
],
[
"dith = do.dither_observation(file_obj_lis, dither_group_id=1, dith_file=dith_file)",
"BUILD 6 dither observation: [DITHOBS:1]\n"
],
[
"dith.normalize_dithers(guid)",
" [DITHOBS:1] build normalized dithers\nGUIDE [ 111 112 113 ... 5098 5099 5100]\nOVERWRITING fits extension: [COOLJ0931_dither_1_20210411T030846_multi.fits][EXT:dithnorm]\nBUILDING new fits extension: [COOLJ0931_dither_2_20210411T032522_multi.fits][EXT:dithnorm]\nBUILDING new fits extension: [COOLJ0931_dither_3_20210411T034159_multi.fits][EXT:dithnorm]\nBUILDING new fits extension: [COOLJ0931_dither_4_20210411T035835_multi.fits][EXT:dithnorm]\nBUILDING new fits extension: [COOLJ0931_dither_5_20210411T041511_multi.fits][EXT:dithnorm]\nBUILDING new fits extension: [COOLJ0931_dither_6_20210411T043147_multi.fits][EXT:dithnorm]\n"
],
[
"for i in dith.VP_frames:\n print(i.dith_num, i.seeing, i.dithnorm, i.fits_ext)",
"1 2.414974440469334 0.9689334018819774 dithnorm\n2 2.2066830668599393 0.9835917946393833 dithnorm\n3 2.0524776279842105 1.0 dithnorm\n4 1.9797851224558247 0.9983360918629792 dithnorm\n5 2.1902447891810595 0.9633061959938931 dithnorm\n6 2.0516773838224456 0.9786423466544407 dithnorm\n"
],
[
"dith.write_data_cube()",
" [DITHOBS:1] build common wavelength solution\n [DITHOBS:1] build master dither set files\n [DITHOBS:1] build data cube and error cube\nCreating 3D WCS\n [DITHOBS:1] build fits cube files\n"
]
],
[
[
"## Example of 4) data cube object",
"_____no_output_____"
]
],
[
[
"import data_cube as dc",
"_____no_output_____"
],
[
"cube_file = data_path+'/COOLJ0931_dither_1_20210411T030846_multi_data_cube_1.fits'\ncube_err_file = data_path+'/COOLJ0931_dither_1_20210411T030846_multi_data_cube_1_err.fits'\n\ncube_1 = dc.cube(cube_file, err_cube_file=cube_err_file)",
"_____no_output_____"
],
[
"sum_frame = cube_1.collapse_frame()",
"_____no_output_____"
],
[
"cube_wcs = dith.cube_wcs\nwcs_im = WCS(dith.cube_wcs.to_header(), naxis=2)\nprint(cube_wcs.naxis, wcs_im.naxis)",
"3 2\n"
],
[
"outfile = data_path+'/COOLJ0931_dither_1_20210411T030846_multi_data_collapse_cube.png'\n\nim.plot_frame(sum_frame, wcs=wcs_im, save=True, outfile=outfile) #vmin=-0.3, vmax=2.5\n",
"_____no_output_____"
]
],
[
[
"## Example of 5) spectrum object",
"_____no_output_____"
]
],
[
[
"import IFU_spectrum as ifu_spec",
"_____no_output_____"
],
[
"fits_1.fits_ext",
"_____no_output_____"
],
[
"fits_1.filename",
"_____no_output_____"
],
[
"ex_spec = ifu_spec.spectrum(fits_1.dat[87], fits_1.wave, z=np.NaN, obj_name=fits_1.object)\nex_spec.plot_spec()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec6f4c44f2e4ea3d28f39c9a897c53588b580aa1 | 445,646 | ipynb | Jupyter Notebook | notebooks/network-spatial-autocorrelation.ipynb | anitagraser/spaghetti | c1228ae7d8c2896f6e9cc2e5b57d8f7c00192e92 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/network-spatial-autocorrelation.ipynb | anitagraser/spaghetti | c1228ae7d8c2896f6e9cc2e5b57d8f7c00192e92 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/network-spatial-autocorrelation.ipynb | anitagraser/spaghetti | c1228ae7d8c2896f6e9cc2e5b57d8f7c00192e92 | [
"BSD-3-Clause"
]
| null | null | null | 458.955716 | 171,496 | 0.940078 | [
[
[
"---------------\n\n**If any part of this notebook is used in your research, please cite with the reference found in** **[README.md](https://github.com/pysal/spaghetti#bibtex-citation).**\n\n\n----------------\n\n## Network-constrained spatial autocorrelation\n### Performing and visualizing exploratory spatial data analysis\n\n**Author: James D. Gaboardi** **<[email protected]>**\n\n**This notebook is an advanced walk-through for:**\n\n1. Demonstrating spatial autocorrelation with [pysal/esda](https://pysal.org/esda/)\n2. Calculating [Moran's *I*](https://pysal.org/esda/generated/esda.Moran.html#esda.Moran) on a segmented network\n3. Visualizing spatial autocorrelation with [pysal/splot](https://splot.readthedocs.io/en/latest/)",
"_____no_output_____"
]
],
[
[
"%load_ext watermark\n%watermark",
"2020-10-05T18:45:53+02:00\n\nCPython 3.8.5\nIPython 7.18.1\n\ncompiler : MSC v.1916 64 bit (AMD64)\nsystem : Windows\nrelease : 10\nmachine : AMD64\nprocessor : Intel64 Family 6 Model 60 Stepping 3, GenuineIntel\nCPU cores : 8\ninterpreter: 64bit\n"
],
[
"import esda\nimport libpysal\nimport matplotlib\nimport matplotlib_scalebar\nfrom matplotlib_scalebar.scalebar import ScaleBar\nimport numpy\nimport spaghetti\nimport splot\n\n%matplotlib inline\n%watermark -w\n%watermark -iv",
"watermark 2.0.2\nspaghetti 1.5.0\nmatplotlib_scalebar 0.6.2\nmatplotlib 3.3.2\nsplot 1.1.3\nnumpy 1.19.1\nesda 2.3.1\nlibpysal 4.3.0\n\n"
],
[
"try:\n from IPython.display import set_matplotlib_formats\n set_matplotlib_formats(\"retina\")\nexcept ImportError:\n pass",
"_____no_output_____"
]
],
[
[
"----------------\n\n### Instantiating a `spaghetti.Network` object and a point pattern\n#### Instantiate the network from a `.shp` file",
"_____no_output_____"
]
],
[
[
"ntw = spaghetti.Network(in_data=libpysal.examples.get_path(\"streets.shp\"))\nntw",
"_____no_output_____"
]
],
[
[
"#### Extract network arcs as a `geopandas.GeoDataFrame`",
"_____no_output_____"
]
],
[
[
"_, arc_df = spaghetti.element_as_gdf(ntw, vertices=True, arcs=True)\narc_df.head()",
"_____no_output_____"
]
],
[
[
"#### Associate the network with a point pattern",
"_____no_output_____"
]
],
[
[
"pp_name = \"crimes\"\npp_shp = libpysal.examples.get_path(\"%s.shp\" % pp_name)\nntw.snapobservations(pp_shp, pp_name, attribute=True)\nntw.pointpatterns",
"_____no_output_____"
]
],
[
[
"#### Extract the crimes point pattern as a `geopandas.GeoDataFrame`",
"_____no_output_____"
]
],
[
[
"pp_df = spaghetti.element_as_gdf(ntw, pp_name=pp_name)\npp_df.head()",
"_____no_output_____"
]
],
[
[
"--------------------------\n\n### 1. ESDA — Exploratory Spatial Data Analysis with [pysal/esda](https://esda.readthedocs.io/en/latest/)\n\n**The Moran's *I* test statistic allows for the inference of how clustered (or dispersed) a dataset is while considering both attribute values and spatial relationships. A value of closer to +1 indicates absolute clustering while a value of closer to -1 indicates absolute dispersion. Complete spatial randomness takes the value of 0. See the** [esda documentation](https://pysal.org/esda/) **for in-depth descriptions and tutorials.**",
"_____no_output_____"
]
],
[
[
"def calc_moran(net, pp_name, w):\n \"\"\"Calculate a Moran's I statistic based on network arcs.\"\"\"\n # Compute the counts\n pointpat = net.pointpatterns[pp_name]\n counts = net.count_per_link(pointpat.obs_to_arc, graph=False)\n # Build the y vector\n arcs = w.neighbors.keys()\n y = [counts[a] if a in counts.keys() else 0. for i, a in enumerate(arcs)]\n # Moran's I\n moran = esda.moran.Moran(y, w, permutations=99)\n return moran, y",
"_____no_output_____"
]
],
[
[
"#### Moran's *I* using the network representation's *W*",
"_____no_output_____"
]
],
[
[
"moran_ntwwn, yaxis_ntwwn = calc_moran(ntw, pp_name, ntw.w_network)\nmoran_ntwwn.I",
"_____no_output_____"
]
],
[
[
"#### Moran's *I* using the graph representation's *W*",
"_____no_output_____"
]
],
[
[
"moran_ntwwg, yaxis_ntwwg = calc_moran(ntw, pp_name, ntw.w_graph)\nmoran_ntwwg.I",
"_____no_output_____"
]
],
[
[
"**Interpretation:**\n\n* **Although both the network and graph representations (**``moran_ntwwn`` **and** ``moran_ntwwg``**, respectively) display minimal postive spatial autocorrelation, a slighly higher value is observed in the graph represention. This is likely due to more direct connectivity in the graph representation; a direct result of eliminating** [degree-2 vertices](https://en.wikipedia.org/wiki/Degree_(graph_theory))**. The Moran's *I* for both the network and graph representations suggest that network arcs/graph edges attributed with associated crime counts are nearly randomly distributed.**\n\n--------------------------------\n\n### 2. Moran's *I* on a segmented network\n#### Moran's *I* on a network split into 200-meter segments",
"_____no_output_____"
]
],
[
[
"n200 = ntw.split_arcs(200.0)\nn200",
"_____no_output_____"
],
[
"moran_n200, yaxis_n200 = calc_moran(n200, pp_name, n200.w_network)\nmoran_n200.I",
"_____no_output_____"
]
],
[
[
"#### Moran's *I* on a network split into 50-meter segments",
"_____no_output_____"
]
],
[
[
"n50 = ntw.split_arcs(50.0)\nn50",
"_____no_output_____"
],
[
"moran_n50, yaxis_n50 = calc_moran(n50, pp_name, n50.w_network)\nmoran_n50.I",
"_____no_output_____"
]
],
[
[
"**Interpretation:**\n\n* **Contrary to above, both the 200-meter and 50-meter segmented networks (**``moran_n200`` **and** ``moran_n50``**, respectively) display minimal negative spatial autocorrelation, with slighly lower values being observed in the 200-meter representation. However, similar to above the Moran's *I* for both the these representations suggest that network arcs attributed with associated crime counts are nearly randomly distributed.**\n\n---------------------------\n\n### 3. Visualizing ESDA with `splot`\n\n**Here we are demonstrating** [spatial lag](https://pysal.org/esda/notebooks/spatialautocorrelation.html#Attribute-Similarity)**, which refers to attribute similarity. See the** [splot documentation](https://splot.readthedocs.io/en/latest/) **for in-depth descriptions and tutorials.**",
"_____no_output_____"
]
],
[
[
"from splot.esda import moran_scatterplot, lisa_cluster, plot_moran",
"_____no_output_____"
]
],
[
[
"#### Moran scatterplot\n\n**Plotted with equal aspect**",
"_____no_output_____"
]
],
[
[
"moran_scatterplot(moran_ntwwn, aspect_equal=True);",
"_____no_output_____"
]
],
[
[
"**Plotted without equal aspect**",
"_____no_output_____"
]
],
[
[
"moran_scatterplot(moran_ntwwn, aspect_equal=False);",
"E:\\Anaconda\\envs\\spaghetti\\lib\\site-packages\\splot\\_viz_esda_mpl.py:47: MatplotlibDeprecationWarning: \nThe set_smart_bounds function was deprecated in Matplotlib 3.2 and will be removed two minor releases later.\n ax.spines['left'].set_smart_bounds(True)\nE:\\Anaconda\\envs\\spaghetti\\lib\\site-packages\\splot\\_viz_esda_mpl.py:48: MatplotlibDeprecationWarning: \nThe set_smart_bounds function was deprecated in Matplotlib 3.2 and will be removed two minor releases later.\n ax.spines['bottom'].set_smart_bounds(True)\n"
]
],
[
[
"**This scatterplot demostrates the attribute values and associated attribute similarities in space (spatial lag) for the network representation's *W* (**``moran_ntwwn``**).**\n\n#### Reference distribution and Moran scatterplot",
"_____no_output_____"
]
],
[
[
"plot_moran(moran_ntwwn, zstandard=True, figsize=(10,4));",
"_____no_output_____"
]
],
[
[
"**This figure incorporates the reference distribution of Moran's *I* values into the above scatterplot of the network representation's *W* (**``moran_ntwwn``**).**\n\n#### Local Moran's *l*\n\n**The demonstrations above considered the dataset as a whole, providing a global measure. The following demostrates the consideration of** [local spatial autocorrelation](https://pysal.org/esda/notebooks/spatialautocorrelation.html#Local-Autocorrelation:-Hot-Spots,-Cold-Spots,-and-Spatial-Outliers)**, providing a measure for each observation. This is best interpreted visually, here with another scatterplot colored to indicate** [relationship type](https://nbviewer.jupyter.org/github/pysal/splot/blob/master/notebooks/esda_morans_viz.ipynb#Visualizing-Local-Autocorrelation-with-splot---Hot-Spots,-Cold-Spots-and-Spatial-Outliers)**.**\n\n**Plotted with equal aspect**",
"_____no_output_____"
]
],
[
[
"p = 0.05\nmoran_loc_ntwwn = esda.moran.Moran_Local(yaxis_ntwwn, ntw.w_network)\nfig, ax = moran_scatterplot(moran_loc_ntwwn, p=p, aspect_equal=True)\nax.set(xlabel=\"Crimes\", ylabel=\"Spatial Lag of Crimes\");",
"_____no_output_____"
]
],
[
[
"**Plotted without equal aspect**",
"_____no_output_____"
]
],
[
[
"fig, ax = moran_scatterplot(moran_loc_ntwwn, aspect_equal=False, p=p)\nax.set(xlabel=\"Crimes\", ylabel=\"Spatial Lag of Crimes\");",
"E:\\Anaconda\\envs\\spaghetti\\lib\\site-packages\\splot\\_viz_esda_mpl.py:47: MatplotlibDeprecationWarning: \nThe set_smart_bounds function was deprecated in Matplotlib 3.2 and will be removed two minor releases later.\n ax.spines['left'].set_smart_bounds(True)\nE:\\Anaconda\\envs\\spaghetti\\lib\\site-packages\\splot\\_viz_esda_mpl.py:48: MatplotlibDeprecationWarning: \nThe set_smart_bounds function was deprecated in Matplotlib 3.2 and will be removed two minor releases later.\n ax.spines['bottom'].set_smart_bounds(True)\n"
]
],
[
[
"**Interpretation:**\n\n* **The majority of observations (network arcs) display no significant local spatial autocorrelation (shown in gray).**\n\n#### Plotting Local Indicators of Spatial Autocorrelation ([LISA](https://onlinelibrary.wiley.com/doi/epdf/10.1111/j.1538-4632.1995.tb00338.x)) ",
"_____no_output_____"
]
],
[
[
"f, ax = lisa_cluster(moran_loc_ntwwn, arc_df, p=p, figsize=(12,12), lw=5, zorder=0)\npp_df.plot(ax=ax, zorder=1, alpha=.25, color=\"g\", markersize=30)\nsuptitle = \"LISA for Crime-weighted Networks Arcs\"\nmatplotlib.pyplot.suptitle(suptitle, fontsize=20, x=.51, y=.93)\nsubtitle = \"Crimes ($n=%s$) are represented as semi-opaque green circles\"\nmatplotlib.pyplot.title(subtitle % pp_df.shape[0], fontsize=15);",
"_____no_output_____"
]
],
[
[
"-----------------",
"_____no_output_____"
]
],
[
[
"libpysal.examples.get_path(\"streets.shp\")",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6f5042bcd7256a9017279e493fa944059e7f37 | 27,713 | ipynb | Jupyter Notebook | all_systems_clustering.ipynb | guidanoli/cedae | 31195fa35da695faeb612578b0a8b3f46ef69228 | [
"MIT"
]
| 1 | 2021-11-22T18:49:03.000Z | 2021-11-22T18:49:03.000Z | all_systems_clustering.ipynb | guidanoli/inf1032 | 31195fa35da695faeb612578b0a8b3f46ef69228 | [
"MIT"
]
| null | null | null | all_systems_clustering.ipynb | guidanoli/inf1032 | 31195fa35da695faeb612578b0a8b3f46ef69228 | [
"MIT"
]
| null | null | null | 38.490278 | 321 | 0.544943 | [
[
[
"# All water supply systems (clustering)\n\nCEDAE is the coorporation that provides drinking water and wastewater services for the Rio de Janeiro State.\nThey provide plenty of data regarding the quality of the water for the press and for the population, due to laws imposed by the Ministry of Health of Brazil.\n\nHere we aim to extract, compile and analyse data from all water supply systems (that we'll be denoting by the acronym *WSS*) being monitored by CEDAE. This data is also available on their webpage. The data of every WSS is routinely gathered and anually compiled into a single report.\n\nUnfortunately, this data is not presented in high granularity, as only the mean of the measurements per month are available. Although this data has been monitored and available since 2004, we have less data samples (\\~204) than the number of daily recorded samples in a single year (\\~365).\n\nThe data available contains the following parameters:\n\n* Physical and Chemical\n * Haze (*turbidez*)\n * Aparent color\n * Residual chlorine\n* Bacteriological\n * Total coliforms (before and after recollection)\n * E. coli (before and after recollection)\n\nThe plan of this notebook is to do the following sequence of tasks:\n\n1. Download the HTML page where all links to PDFs reside\n2. Parse the HTML page and extract any link to PDFs and its metadata (year and WSS)\n3. Extract names of old reports from `input/old`\n4. Cluster similarly-named WSSs\n5. Download recent reports to `input/`\n6. Copy older reports from `input/old` to `input/`",
"_____no_output_____"
],
[
"## 1. Download the HTML page\n\nFirst we download the HTML using the `urllib.request.urlopen` method. It returns a file pointer, from which the page can be read as a stream of bytes, and decoded to UTF-8, the default string encoding for Python (and the modern internet, pretty much).",
"_____no_output_____"
]
],
[
[
"import urllib.request\n\npage_url = 'https://cedae.com.br/relatorioanual'\nwith urllib.request.urlopen(page_url) as fp:\n page = fp.read().decode() # Read from page and decode to UTF-8 string",
"_____no_output_____"
]
],
[
[
"You can check that we got indeed the HTML for the page:",
"_____no_output_____"
]
],
[
[
"print(page[:80])",
"<!DOCTYPE html>\r\n<html lang=\"pt-BR\">\r\n<head id=\"Head\">\r\n<!--*******************\n"
]
],
[
[
"## 2. Parse the HTML page\n\nFor this task, we'll be using the `HTMLParser` class from the `html.parser` module, which allows us to specify callbacks for when the parser reads the beggining tags (`<...>`), in-between text (`<a> ... </a>`), and their ending tags (`</...>`).\n\nAfter analysing the source code for the page we're parsing, we notice that all links that interest us are contained in tables, particularly inside `<td>` tags. Moreover, every table has a top row whose class is `thead` (probably for short for \"table head\") containing the year of the reports.\n\nFor building our custom parser, we inherit the `HTMLParser`.",
"_____no_output_____"
]
],
[
[
"import html.parser\nfrom datetime import datetime\n\nclass MyHTMLParser(html.parser.HTMLParser):\n \n def __init__(self):\n super().__init__()\n self.in_td = False\n self.in_thead = False\n self.year = None\n self.link = None\n self.links = {}\n \n def handle_starttag(self, tag, attrs):\n if tag == 'td':\n self.in_td = True\n elif tag == 'a':\n links = [v for k, v in attrs if k == 'href']\n if links:\n assert len(links) == 1, links\n self.link = links[0]\n elif tag == 'tr' and ('class', 'thead') in attrs:\n self.in_thead = True\n \n def handle_data(self, data):\n if self.in_thead:\n for number in [int(s) for s in data.split() if s.isdigit()]:\n # CEDAE was created in 1975\n if 1975 <= number <= datetime.now().year:\n self.year = number\n elif self.in_td and self.link is not None:\n assert self.year is not None\n data = data.strip()\n assert len(data) > 0\n if self.year not in self.links:\n self.links[self.year] = {}\n assert data not in self.links[self.year], self.links[self.year][data]\n self.links[self.year][data] = self.link\n \n def handle_endtag(self, tag):\n if tag == 'td':\n self.in_td = False\n elif tag == 'a':\n self.link = None\n elif tag == 'tr':\n self.in_thead = False\n elif tag == 'table':\n self.year = None",
"_____no_output_____"
]
],
[
[
"We now construct a parser instance and feed it with the contents of the HTML page.\n\nThe links are stored in the `links` field from the `MyHTMLParser` instance. It is a dictionary of dictionaries of strings. It is first indexed by the year of the reports, and second by the name of the water supply system, resulting in the link to the PDF of the annual report corresponding to that WSS in that year.",
"_____no_output_____"
]
],
[
[
"parser = MyHTMLParser()\nparser.feed(page)\nyearly_named_new_reports = parser.links",
"_____no_output_____"
]
],
[
[
"### 3. Extract names of old reports from `input/old`\n\nOld reports are zipped and available for download in the same page as the more recent reports.\nSo that you don't have to download them yourself, they are readily available in the `input/old` folder.\n\nIn this section, we are going to inspect the textual contents of these PDFs and guess which WSS are they related to.\n\nFirst, let's list every report located in the `input/old` folder.",
"_____no_output_____"
]
],
[
[
"import os\nimport glob\n\nyearly_old_reports = {}\nfor year in os.listdir(os.path.join('input', 'old')):\n pathname = os.path.join('input', 'old', year, '*.pdf')\n assert year.isdigit(), \"Assumed folders are numbers\"\n yearly_old_reports[int(year)] = list(glob.iglob(pathname))",
"_____no_output_____"
]
],
[
[
"Now, for each year, we'll define regular expression patterns for obtaining the name (`get_regex_specs`). This function returns the regular expression pattern to match the WSS name and the group index within that pattern that contains the name.\n\nWe also define some heuristics for PDFs we can't really parse (`get_report_name`). This function returns the name of the WSS guessed from the filename and year.",
"_____no_output_____"
]
],
[
[
"def get_regex_specs(year):\n if year < 2009:\n return [(\"([Nn]?[Oo]s? )?[Mm]unicípios? d[eo] ([^,.]*?),? \"\\\n \"(é|são|recebe|na região|e o distrito|somente o distrito|\"\\\n \"a CEDAE|no município do Rio de Janeiro)\", 2)]\n elif 2009 <= year <= 2015:\n return [(r\"SOBRE O SISTEMA (DE )?(.*?)\\s*o O MANANCIAL\", 2)]\n else:\n return None\n\ndef get_report_name(year, filename):\n if year == 2011:\n pass\n elif year in (2010, 2015, 2009):\n filename = filename.replace('_', ' ')\n elif year == 2012:\n filename = filename.split(' - ')[-1]\n elif year in (2013, 2014):\n filename = ' '.join(filename.split('_')[1:])\n else:\n return None\n return filename",
"_____no_output_____"
],
[
"import PyPDF2\nimport re\n\neol_hyphen_patt = re.compile('-\\n')\nconj_article_patt = re.compile(' [Ee](?: [OAoa])? ')\nspaces_patt = re.compile(r'\\s\\s+')\n\nyearly_named_old_reports = {}\nfor year, old_reports in yearly_old_reports.items():\n regex_spec = get_regex_specs(year)\n assert regex_spec is not None, year\n patt_strs, groups = zip(*regex_spec)\n patts = list(map(re.compile, patt_strs))\n yearly_named_old_reports[year] = {}\n for report in old_reports:\n reader = PyPDF2.PdfFileReader(report)\n text = ' '.join([page.extractText() for page in reader.pages])\n text = spaces_patt.sub(r' ', text)\n text = eol_hyphen_patt.sub('', text)\n names = set()\n for patt, group in zip(patts, groups):\n for match in patt.finditer(text):\n name = match[group]\n names.update(conj_article_patt.split(name))\n if not names:\n basename = os.path.basename(report)\n filename, fileext = os.path.splitext(basename)\n name = get_report_name(year, filename)\n assert name is not None, (year, filename)\n names = conj_article_patt.split(name)\n assert names, name\n name = '/'.join(sorted(names))\n name = name.title()\n yearly_named_old_reports[year][name] = report",
"PdfReadWarning: Xref table not zero-indexed. ID numbers for objects will be corrected. [pdf.py:1736]\n"
]
],
[
[
"For usability, we'll combine old and new reports into one dictionary...",
"_____no_output_____"
]
],
[
[
"yearly_named_reports = {**yearly_named_old_reports, **yearly_named_new_reports}",
"_____no_output_____"
]
],
[
[
"## 4. Cluster similarly-named WSSs\n\nWe know that some water supply systems are wrongly named. Let's create a list of all unique names.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nnames = set()\nfor year, named_reports in yearly_named_reports.items():\n names |= named_reports.keys()\nnames = np.asarray(sorted(names))",
"_____no_output_____"
]
],
[
[
"Then, we create an index that relates names to indices in this list.",
"_____no_output_____"
]
],
[
[
"name_index = {v: i for i, v in enumerate(names)}",
"_____no_output_____"
]
],
[
[
"Now we create a list whose item `a[i]` contains the years of reports of the `i-th` string in the list.",
"_____no_output_____"
]
],
[
[
"name_reports = [{year for year, reports in yearly_named_reports.items() if name in reports} for name in names]",
"_____no_output_____"
]
],
[
[
"Let's cluster names that don't have reports in the same year, but are very similar according to the Levenshtein distance.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom scipy.sparse import csr_matrix\nimport Levenshtein\n\nnnames = len(names)\ndmatrix = np.zeros(shape=(nnames, nnames), dtype=int) # distance matrix\n\n# Distances where user input will be requested\n# to evaluate whether two names belong in the same cluster\nmin_dist = 4\nmax_dist = 5\n\nfor i in range(nnames):\n for j in range(i):\n dist = Levenshtein.distance(names[i], names[j])\n # We evaluate first names that are substring\n if names[i] in names[j] or names[j] in names[i]:\n dist = min_dist\n dmatrix[i][j] = dmatrix[j][i] = dist\n\n# matrix indices, sorted by distance (lower first)\ndmatrix_indices = sorted(((i, j) for i in range(nnames) for j in range(i)), key=lambda k: dmatrix[k[0]][k[1]])\n\nmin_year = min(yearly_named_reports.keys())\nmax_year = max(yearly_named_reports.keys())\nnyears = max_year - min_year + 1\n\nyearmap = np.zeros(shape=(nnames, nyears), dtype=bool) # year map\n\nfor i in range(nnames):\n for year in name_reports[i]:\n yearmap[i][year - min_year] = True\n\nclusters = np.arange(nnames) # cluster ids (initially every name is in its own cluster)\n\nnot_connected = np.zeros(shape=(nnames, nnames), dtype=bool) # from input\n\nfor i, j in dmatrix_indices:\n dist = dmatrix[i][j]\n if dist > max_dist:\n break\n ci = clusters[i]\n cj = clusters[j]\n yi = np.any(yearmap[clusters == ci], axis=0)\n yj = np.any(yearmap[clusters == cj], axis=0)\n if np.any(yi & yj):\n continue # cannot merge clusters\n if np.any(not_connected[clusters == ci, clusters == cj]):\n continue # not merged before\n ci_str = \", \".join(names[clusters == ci])\n cj_str = \", \".join(names[clusters == cj])\n if dist >= min_dist:\n ok = input(\"({}) = ({})? ([Y]es/[n]o/e[x]it) \".format(ci_str, cj_str))\n if 'x' in ok:\n break\n elif 'n' in ok:\n not_connected[i][j] = not_connected[j][i] = 1\n continue # doesn't want to merge clusters\n clusters[clusters == cj] = ci",
"(Angra Dos Reis/Mangaratiba) = (Angra Dos Reis)? ([Y]es/[n]o/e[x]it) \n(Belford Roxo/Duque De Caxias/Nova Iguaçu) = (Belford Roxo)? ([Y]es/[n]o/e[x]it) \n(Bom Jesus Do Itabapoana, Bom Jesus do Itabapoana) = (Bom Jesus)? ([Y]es/[n]o/e[x]it) \n(Cachoeiras De Macacu/Tanguá) = (Cachoeiras De Macacu)? ([Y]es/[n]o/e[x]it) \n(Cardoso Moreira/Italva) = (Cardoso Moreira)? ([Y]es/[n]o/e[x]it) \n(Cordeiro) = (Cantagalo/Cordeiro)? ([Y]es/[n]o/e[x]it) \n(Cordeiro/Cantagado) = (Cantagalo/Cordeiro, Cordeiro)? ([Y]es/[n]o/e[x]it) \n(Coronel Teixeira/Batatal, Coronel Teixeirabatatal) = (Coronel Teixeira)? ([Y]es/[n]o/e[x]it) \n(Itaboraí) = (Atafona)? ([Y]es/[n]o/e[x]it) n\n(Itaguaí/Paracambi/Seropédica) = (Itaguaí, Italva)? ([Y]es/[n]o/e[x]it) \n(Japeri) = (Cacaria)? ([Y]es/[n]o/e[x]it) n\n(Japeri) = (Camorim)? ([Y]es/[n]o/e[x]it) n\n(Japeri) = (Jamapara, Jamapará)? ([Y]es/[n]o/e[x]it) n\n(Japeri/Queimados) = (Japeri)? ([Y]es/[n]o/e[x]it) \n(Japuíba) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Laranjal) = (Eta Laranjal)? ([Y]es/[n]o/e[x]it) \n(Eta Laranjal, Laranjal) = (Imunana Laranjal)? ([Y]es/[n]o/e[x]it) \n(Magé) = (Anta)? ([Y]es/[n]o/e[x]it) n\n(Magé) = (Gargau, Gargaú)? ([Y]es/[n]o/e[x]it) n\n(Mesquita) = (Mantiquira, Matiquira)? ([Y]es/[n]o/e[x]it) n\n(Mesquita/Nilópolis/Rio De Janeiro/São João Do Meriti) = (Mesquita)? ([Y]es/[n]o/e[x]it) \n(Miguel Pereira/Paty Do Alferes) = (Miguel Pereira)? ([Y]es/[n]o/e[x]it) \n(Nossa Senhora Aparecida) = (Aparecida)? ([Y]es/[n]o/e[x]it) \n(Palmas Paulo De Frontin) = (Palmas)? ([Y]es/[n]o/e[x]it) \n(Pipeiras/Palacete) = (Pipeiras)? ([Y]es/[n]o/e[x]it) \n(Rio Das Ostras) = (Barra De São João/Rio Das Ostras)? ([Y]es/[n]o/e[x]it) \n(Barra De São João/Rio Das Ostras, Rio Das Ostras) = (Casemiro De Abreu (Barra De São João)/Rio Das Ostras)? ([Y]es/[n]o/e[x]it) \n(São Francisco Do Itabapoana) = (Itabapoana)? ([Y]es/[n]o/e[x]it) \n(São Gonçalo) = (Ilha De Paquetá/São Gonçalo)? ([Y]es/[n]o/e[x]it) \n(São Sebastião dos Ferreiros) = (Ferreiros)? ([Y]es/[n]o/e[x]it) \n(Trajano De Moraes, Trajano De Morais, Trajano de Morais) = (Trajano)? ([Y]es/[n]o/e[x]it) \n(Xerem, Xerém) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Barra Do Piraí) = (Barra Do Açu, Barra Do Açú, Barra do Açu)? ([Y]es/[n]o/e[x]it) n\n(Itaboraí) = (Camorim)? ([Y]es/[n]o/e[x]it) n\n(Itaboraí) = (Ipiabas)? ([Y]es/[n]o/e[x]it) n\n(Japeri, Japeri/Queimados) = (Gargau, Gargaú)? ([Y]es/[n]o/e[x]it) n\n(Juparana, Juparanã) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Magé) = (Guandu, Guandú)? ([Y]es/[n]o/e[x]it) n\n(Manilha) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Mazomba) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Medanha, Mendanha) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Mesquita, Mesquita/Nilópolis/Rio De Janeiro/São João Do Meriti) = (Banquete)? ([Y]es/[n]o/e[x]it) n\n(Mesquita, Mesquita/Nilópolis/Rio De Janeiro/São João Do Meriti) = (Mantiquira, Matiquira)? ([Y]es/[n]o/e[x]it) n\n(Mesquita, Mesquita/Nilópolis/Rio De Janeiro/São João Do Meriti) = (Medanha, Mendanha)? ([Y]es/[n]o/e[x]it) n\n(Monnerat) = (Convento)? ([Y]es/[n]o/e[x]it) n\n(Muriqui) = (Mesquita, Mesquita/Nilópolis/Rio De Janeiro/São João Do Meriti)? ([Y]es/[n]o/e[x]it) n\n(Osório) = (Brasilio)? ([Y]es/[n]o/e[x]it) n\n(Osório) = (Camorim)? ([Y]es/[n]o/e[x]it) n\n(Osório) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Ourania, Ourânea) = (Osório)? ([Y]es/[n]o/e[x]it) n\n(Palmas, Palmas Paulo De Frontin) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Palmas, Palmas Paulo De Frontin) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Paracambi) = (Jaguarambé, Jaguarembé)? ([Y]es/[n]o/e[x]it) n\n(Paracambi) = (Marambaia)? ([Y]es/[n]o/e[x]it) n\n(Parapeuna, Parapeúna) = (Paracambi)? ([Y]es/[n]o/e[x]it) n\n(Paraíso) = (Paracambi)? ([Y]es/[n]o/e[x]it) n\n(Piabetá) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Pipeiras, Pipeiras/Palacete) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Pureza) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Rio D'Ouro, Rio De Ouro, Rio Douro) = (Rio De Janeiro)? ([Y]es/[n]o/e[x]it) n\n(Suruí) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Tachas) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Tachas) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Taylor) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Taylor) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Tingua, Tinguá) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Valença) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Varjão) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n(Varjão) = (Magé)? ([Y]es/[n]o/e[x]it) n\n(Xerem, Xerém) = (Japeri, Japeri/Queimados)? ([Y]es/[n]o/e[x]it) n\n"
]
],
[
[
"Now, we need to choose a representable sample from each cluster. We'll define a list of criterium for getting the \"best\" name. We also create a file for storing the \"codenames\" and real names of each water supply system.",
"_____no_output_____"
]
],
[
[
"import unidecode\nimport pandas as pd\nimport os\n\ndef non_ascii_char_count(s):\n return len([c for c in s if not c.isascii()])\n\ndef upper_case_char_count(s):\n return len([c for c in s if not c.isupper()])\n\ndef year_of_last_report(name):\n index = name_index[name]\n reports = name_reports[index]\n return max(reports)\n\ndef choose_name(cluster):\n cluster = sorted(cluster, key=year_of_last_report)\n cluster = sorted(cluster, key=non_ascii_char_count)\n cluster = sorted(cluster, key=upper_case_char_count)\n cluster = sorted(cluster, key=len)\n return cluster[-1]\n\ndef convert_char(c):\n if c.isalpha():\n return c.lower()\n else:\n return '_'\n\ndef format_name(name):\n name = unidecode.unidecode(name)\n g = map(convert_char, name)\n return ''.join(list(g))\n\ntry:\n os.mkdir('output')\nexcept FileExistsError:\n pass # it's ok if the output folder already exists\n\nnames_file = os.path.join('output', 'wss_codenames.csv')\nnames_df = pd.DataFrame(columns=('name',))\n\ncluster_names = {}\nfor ci in np.unique(clusters):\n ci_names = names[clusters == ci]\n name = choose_name(ci_names)\n codename = format_name(name)\n cluster_names[ci] = codename\n names_df.loc[codename] = (name,)\n\nnames_df.to_csv(names_file)",
"_____no_output_____"
]
],
[
[
"## 5. Download recent reports to `input/`\n\nNow, let's download the PDFs and store them in a nice hierarchichal directory structure composed of `input/<year>/<wss>.pdf`.",
"_____no_output_____"
]
],
[
[
"import os\nimport urllib.parse\n\npage_url_parts = urllib.parse.urlparse(page_url)\nbase_url = page_url_parts._replace(path='').geturl()\n\ndef normalize_url(url):\n if url.startswith('/'):\n url = base_url + url\n # Heuristic: if URL has %, it must be already formatted.\n # If not, format only the URL path\n if '%' in url:\n return url\n else:\n urlparts = url.split('/')\n filename = urllib.parse.quote(urlparts[-1])\n return '/'.join(urlparts[:-1] + [filename])\n\nbase_path = 'input'\nfor year, reports in yearly_named_new_reports.items():\n year_path = os.path.join(base_path, str(year))\n try:\n os.mkdir(year_path)\n except FileExistsError:\n pass # it's ok if such folder already exists\n for name, report_url in reports.items():\n index = name_index[name]\n cluster = clusters[index]\n cname = cluster_names[cluster]\n filename = cname + \".pdf\"\n filepath = os.path.join(year_path, filename)\n if os.path.exists(filepath) and os.path.isfile(filepath):\n continue # don't need to download files already local\n report_url = normalize_url(report_url)\n with urllib.request.urlopen(report_url) as webfp, open(filepath, 'wb') as localfp:\n localfp.write(webfp.read())",
"_____no_output_____"
]
],
[
[
"## 6. Copy older reports from `input/old` to `input/`\n\nFor older reports, they are already downloaded, and in the `input/old` folder. So we only need to create folders for each year in `input/<year>` and copy them with their respective \"canonical\" names to these folders.",
"_____no_output_____"
]
],
[
[
"for year, reports in yearly_named_old_reports.items():\n destination_path = os.path.join('input', str(year))\n try:\n os.mkdir(destination_path)\n except FileExistsError:\n pass # it's ok if such folder already exists\n for name, report_path in reports.items():\n index = name_index[name]\n cluster = clusters[index]\n cname = cluster_names[cluster]\n filename = cname + \".pdf\"\n filepath = os.path.join(destination_path, filename)\n if os.path.exists(filepath) and os.path.isfile(filepath):\n continue # don't need to copy files already copied\n with open(report_path, 'rb') as infp, open(filepath, 'wb') as outfp:\n outfp.write(infp.read())",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6f64a15b06704976dc6d0741313db0edcab916 | 18,942 | ipynb | Jupyter Notebook | MAIN FoodBot_Recipe_(Prolog_Code).ipynb | sadid07/AI-Prolog-Recipe-Suggestion | 1a2e303e0603f68d4f11381703c70d048c595e2f | [
"Apache-2.0"
]
| null | null | null | MAIN FoodBot_Recipe_(Prolog_Code).ipynb | sadid07/AI-Prolog-Recipe-Suggestion | 1a2e303e0603f68d4f11381703c70d048c595e2f | [
"Apache-2.0"
]
| null | null | null | MAIN FoodBot_Recipe_(Prolog_Code).ipynb | sadid07/AI-Prolog-Recipe-Suggestion | 1a2e303e0603f68d4f11381703c70d048c595e2f | [
"Apache-2.0"
]
| null | null | null | 39.544885 | 882 | 0.571323 | [
[
[
"\n# FoodBot Recipe (Prolog Code)",
"_____no_output_____"
]
],
[
[
"!apt install swi-prolog\n!pip install pyswip",
"Reading package lists... Done\nBuilding dependency tree \nReading state information... Done\nswi-prolog is already the newest version (7.6.4+dfsg-1build1).\n0 upgraded, 0 newly installed, 0 to remove and 37 not upgraded.\nRequirement already satisfied: pyswip in /usr/local/lib/python3.7/dist-packages (0.2.10)\n"
],
[
"from pyswip import Prolog\nimport requests\nimport json",
"_____no_output_____"
]
],
[
[
"### Detail on what your expert system will focus upon, and what the askables will be. You need to have a minimum of 8 askables in your expert system, regardless of the domain of your KB. [#rightproblem]\n\nWe will be using the expert system example provided in the instructions, of helping students find suitable recipes so they are incentivized to eat inside and lower risk of infection. The expert system will function on the following 8 askables: \"How much time does the student have to make the meal?\", \"How much effort in cooking do you want?\", \"What type of meal do you want? (Appetizer, Main, Dessert)\" \"Do you have any dietary goals? (Pleasure, Weight Loss, Weight Gain, etc...\", \"Which cuisine do you prefer?\", \"Which diet do you prefer?\", \"Ingredients to look for?\", \"How many calories are you aiming for?\". Some of these askables are dependent on each other. For example, if the person has a specific diet, that will influence the available options for asking what their preferred cuisine is because we may not have their diet available in all cusines.\n\n### Perform data collection for building your expert system by surveying resources available online, guided by the askables that you have chosen. [\\#evidencebased, \\#sourcequality].\nThe data will be collected from the following scraped data: https://raw.githubusercontent.com/mneedham/bbcgoodfood/master/stream_all.json. It is from a github project that scraped BBC's Good Food website for information on their recipes. While it has thousands of data points, the data needed cleaning, so the first 100 were cleaned and kept in a new JSON file presented here: https://gist.githubusercontent.com/pierre-minerva/70bccece0820fa839b53c264cc7460a3/raw/8c7afccd58cf78b42b99ceb0927025896ea108fa/recipe_data.json",
"_____no_output_____"
]
],
[
[
"recipes = []\n\n#get data\nr = requests.get(\"https://gist.githubusercontent.com/pierre-minerva/70bccece0820fa839b53c264cc7460a3/raw/8c7afccd58cf78b42b99ceb0927025896ea108fa/recipe_data.json\")\n#Save db\nwith open('recipes.json', 'wb') as f:\n f.write(r.content)\n\n#process data\nwith open('recipes.json') as f:\n for jsonObj in f:\n recipesDict = json.loads(jsonObj)\n recipes.append(recipesDict)",
"_____no_output_____"
]
],
[
[
"### Explain the logic of your expert system by showing which values of the askables lead to what specific information being provided to the end user. Any visualization that makes the logic of your expert system clear is acceptable (for example, a tree diagram or table). [#ailogic]\n\n The logic is very straightforward. \n1. First, we ask if they follow a specific diet (i.e. low-carb) in multiple choice format based on the literals from the data collection. This simply filters out all possible recipes that do not match the user preference. \n2. Then we ask if they have a dietary goal. The options are \"Weight-Loss\", \"Weight-Gain\", \"Nutrition\", and \"Pleasure\". \n3. \"Weight-Loss\" and \"Weight-Gain\" prompt a second question asking for the user's target caloric intake, which is used to filter out more results. \"Nutrition\" filters for recipes with 20g of protein or more. \"Pleasure\" doesn't filter for anything. \n4. Then we ask about total time available to prepare the meal, with options between <5m, <15m, <30m, >30m, filtering appropriately. \n5. Then we ask for the desired involvement in the cooking process, with the following choices \"Easy\", \"Some Effort\", and \"A Challenge\". \n6. Then, we ask for the type of meal, the user wants from \"Snack\", \"Main Course\", \"Dessert\", and \"Breakfast\". If they selected \"Weight-Loss\" or \"Nutrion\" earlier, the \"Dessert\" option will be left out. If \"Weight-Gain\", \"Snack\" is left out. \n7. Then we ask if the user has any ingredients they would like to use from the remaining recipes that still match the previously selected questions. \n8. Then we ask if the user has any preferred cusine from the recipes remaining that still match the previously selected questsions.",
"_____no_output_____"
],
[
"### Using this visualization to help you, code your expert system using either a native Prolog front-end, or one using the PySWIP library to interface a Python frontend. [#aicoding]",
"_____no_output_____"
]
],
[
[
"import re\n\ndef list_to_dict(lst):\n d = {}\n l = list(set(lst))\n\n for i in range(len(l)):\n d[i] = l[i]\n \n return d, l\n\ndef main():\n prolog = Prolog()\n\n #Adding of data to KB\n for recipe in recipes:\n #recipe\n title = recipe['page']['title']\n #diet\n for diet in recipe['page']['recipe']['diet_types']:\n prolog.assertz(f\"diet('{title}','{diet}')\")\n #protein\n protein = int(re.search(r'\\d+', recipes[0]['page']['recipe']['nutrition_info'][2]).group())\n prolog.assertz(f\"protein('{title}',{protein})\")\n #calories\n calories = int(re.search(r'\\d+', recipes[0]['page']['recipe']['nutrition_info'][1]).group())\n prolog.assertz(f\"calories('{title}',{calories})\")\n #time\n total_time = (recipe['page']['recipe']['cooking_time'] + recipes[0]['page']['recipe']['prep_time'])//60\n prolog.assertz(f\"time('{title}',{total_time})\")\n #effort\n skill_level = recipe['page']['recipe']['skill_level']\n prolog.assertz(f\"effort('{title}','{skill_level}')\")\n #type\n for course in recipe['page']['recipe']['courses']:\n prolog.assertz(f\"course('{title}','{course}')\")\n #ingredients\n for ing in recipe['page']['recipe']['ingredients']:\n prolog.assertz(f\"ing('{title}','{ing}')\")\n #cuisine\n cuisine = recipe['page']['recipe']['cusine']\n prolog.assertz(f\"cuisine('{title}','{cuisine}')\")\n #recipe\n prolog.assertz(f\"recipe('{title}','{diet}',{protein},{calories},{total_time},'{skill_level}','{course}','{ing}','{cuisine}' )\")\n\n prolog.assertz(\"more(X,Y) :- X @> Y\")\n\n #Q1\n prolog.assertz(\"diet(D)\")\n\n #Q2\n prolog.assertz(\"goal(G)\")\n\n #Q3\n prolog.assertz(\"protein(R) :- goal('Nutrition'); protein(R,P); more(P,20)\")\n\n #Q6\n #if loss then no dessert\n\n prolog.assertz(\"recipe2(R,D,Ci,Ti,E,Co,I,Cu)\")\n prolog.assertz(\"recipe2(R,D,Ci,Ti,E,Co,I,Cu) :- recipe(R,D,P,C,T,E,Co,I,Cu); more(T,Ti); goal('Weight-Loss'); more(Ci,C); dif(Co,'Dessert')\")\n prolog.assertz(\"recipe2(R,D,Ci,Ti,E,Co,I,Cu) :- recipe(R,D,P,C,T,E,Co,I,Cu); more(T,Ti); goal('Weight-Gain'); more(C,Ci); dif(Co,'Snack')\")\n prolog.assertz(\"recipe2(R,D,Ci,Ti,E,Co,I,Cu) :- recipe(R,D,P,C,T,E,Co,I,Cu); more(T,Ti); goal('Nutrition'); more(P,20); dif(Co,'Dessert')\")\n prolog.assertz(\"recipe2(R,D,Ci,Ti,E,Co,I,Cu) :- recipe(R,D,P,C,T,E,Co,I,Cu); more(T,Ti); goal('Pleasure')\")\n\n #Q1\n diets = []\n\n for soln in prolog.query(\"diet(R,D)\"):\n if \"_\" not in str(soln[\"D\"]):\n diets.append(str(soln[\"D\"]))\n\n diets_dict, diets_lst = list_to_dict(diets)\n print(\"Do you have any dietary preferences?\")\n print(\"Input number of choice:\")\n print(diets_dict)\n diet_choice = int(input())\n prolog.assertz(f\"diet('{diets_dict[diet_choice]}')\")\n\n #Q2\n goal_dict = {0:\"Weight-Loss\", 1: \"Weight-Gain\", 2:\"Nutrition\", 3:\"Pleasure\"}\n print(\"What is your dietary goal?\")\n print(\"Input number of choice:\")\n print(goal_dict)\n goal_choice = int(input())\n prolog.assertz(f\"goal('{goal_dict[goal_choice]}')\")\n\n #Q3\n if goal_choice == 0 or goal_choice == 1:\n print(\"What are your target calories?\")\n calories_choice = int(input())\n prolog.assertz(f\"calories({calories_choice})\")\n else:\n calories_choice = 9999\n prolog.assertz(f\"calories({calories_choice})\")\n\n #Q4\n print(\"How many minutes do you have to make the food?\")\n time_choice = int(input())\n prolog.assertz(f\"time({time_choice})\")\n\n #Q5\n effort = []\n\n for soln in prolog.query(f\"recipe2(R,'{diets_dict[diet_choice]}',{calories_choice},{time_choice},E,Co,I, Cu)\"):\n if \"_\" not in str(soln[\"E\"]):\n effort.append(str(soln[\"E\"]))\n\n effort_dict, effort_lst = list_to_dict(effort)\n print(\"How much effort do you want to put in?\")\n print(\"Input number of choice:\")\n print(effort_dict)\n effort_choice = int(input())\n prolog.assertz(f\"effort('{effort_dict[effort_choice]}')\")\n\n #Q6\n course = []\n\n for soln in prolog.query(f\"recipe2(R,'{diets_dict[diet_choice]}',{calories_choice},{time_choice},'{effort_dict[effort_choice]}',Co,I, Cu)\"):\n if str(soln[\"Co\"]) in [\"Snack\", \"Main course\", \"Dessert\", \"Breakfast\"]:\n course.append(str(soln[\"Co\"]))\n\n course_dict, course_lst = list_to_dict(course)\n print(\"What course would you like?\")\n print(\"Input number of choice:\")\n print(course_dict)\n course_choice = int(input())\n prolog.assertz(f\"course('{course_dict[course_choice]}')\")\n\n #Q7\n ing = []\n\n for soln in prolog.query(f\"recipe2(R,'{diets_dict[diet_choice]}',{calories_choice},{time_choice},'{effort_dict[effort_choice]}','{course_dict[course_choice]}',I, Cu)\"):\n if \"_\" not in str(soln[\"I\"]):\n ing.append(str(soln[\"I\"]))\n\n ing_dict, ing_lst = list_to_dict(ing)\n print(\"Which of these remaining ingredients would you like to be in your meal?\")\n print(\"Input number of choice:\")\n print(ing_dict)\n ing_choice = int(input())\n prolog.assertz(f\"ing('{ing_dict[ing_choice]}')\")\n\n #Q8\n cuisine = []\n\n for soln in prolog.query(f\"recipe2(R,'{diets_dict[diet_choice]}',{calories_choice},{time_choice},'{effort_dict[effort_choice]}','{course_dict[course_choice]}','{ing_dict[ing_choice]}', Cu)\"):\n if \"_\" not in str(soln[\"Cu\"]):\n cuisine.append(str(soln[\"Cu\"]))\n\n cuisine_dict, cuisine_lst = list_to_dict(cuisine)\n print(\"What cuisine do you want?\")\n print(\"Input number of choice:\")\n print(cuisine_dict)\n cuisine_choice = int(input())\n\n final_recipes = []\n for soln in prolog.query(f\"recipe2(R,'{diets_dict[diet_choice]}',{calories_choice},{time_choice},'{effort_dict[effort_choice]}','{course_dict[course_choice]}','{ing_dict[ing_choice]}', '{cuisine_dict[cuisine_choice]}')\"):\n if type(soln[\"R\"]) == type(\"str\"):\n final_recipes.append(soln[\"R\"])\n\n final_recipes_dict, final_recipes_lst = list_to_dict(final_recipes)\n print(\"The following recipes from BBC's Good Foods website match your needs.\")\n print(final_recipes_lst)",
"_____no_output_____"
],
[
"#Test case 1\nmain()",
"Do you have any dietary preferences?\nInput number of choice:\n{0: 'Low-salt', 1: 'Low-calorie', 2: 'Heart Healthy', 3: 'Healthy', 4: 'Low-fat', 5: 'Gluten-free', 6: 'Dairy-free', 7: 'Vegetarian', 8: 'Egg-free', 9: 'Vegan'}\n4\nWhat is your dietary goal?\nInput number of choice:\n{0: 'Weight-Loss', 1: 'Weight-Gain', 2: 'Nutrition', 3: 'Pleasure'}\n0\nWhat are your target calories?\n500\nHow many minutes do you have to make the food?\n60\nHow much effort do you want to put in?\nInput number of choice:\n{0: 'Easy'}\n0\nWhat course would you like?\nInput number of choice:\n{0: 'Breakfast', 1: 'Main course'}\n0\nWhich of these remaining ingredients would you like to be in your meal?\nInput number of choice:\n{0: 'gravadlax'}\n0\nWhat cuisine do you want?\nInput number of choice:\n{0: 'Scandinavian'}\n0\nThe following recipes from BBC's Good Foods website match your needs.\n['Potato & dill pancakes with gravadlax']\n"
],
[
"#Test case 2\nmain()",
"Do you have any dietary preferences?\nInput number of choice:\n{0: 'Low-salt', 1: 'Low-calorie', 2: 'Heart Healthy', 3: 'Healthy', 4: 'Low-fat', 5: 'Gluten-free', 6: 'Dairy-free', 7: 'Vegetarian', 8: 'Egg-free', 9: 'Vegan'}\n2\nWhat is your dietary goal?\nInput number of choice:\n{0: 'Weight-Loss', 1: 'Weight-Gain', 2: 'Nutrition', 3: 'Pleasure'}\n1\nWhat are your target calories?\n200\nHow many minutes do you have to make the food?\n60\nHow much effort do you want to put in?\nInput number of choice:\n{0: 'Easy'}\n0\nWhat course would you like?\nInput number of choice:\n{0: 'Dessert'}\n0\nWhich of these remaining ingredients would you like to be in your meal?\nInput number of choice:\n{0: 'sea salt'}\n0\nWhat cuisine do you want?\nInput number of choice:\n{0: ''}\n0\nThe following recipes from BBC's Good Foods website match your needs.\n['Chocolate, peanut butter & pretzel cookie bars']\n"
],
[
"#Test case 3\nmain()",
"Do you have any dietary preferences?\nInput number of choice:\n{0: 'Low-salt', 1: 'Low-calorie', 2: 'Heart Healthy', 3: 'Healthy', 4: 'Low-fat', 5: 'Gluten-free', 6: 'Dairy-free', 7: 'Vegetarian', 8: 'Egg-free', 9: 'Vegan'}\n0\nWhat is your dietary goal?\nInput number of choice:\n{0: 'Weight-Loss', 1: 'Weight-Gain', 2: 'Nutrition', 3: 'Pleasure'}\n3\nHow many minutes do you have to make the food?\n60\nHow much effort do you want to put in?\nInput number of choice:\n{0: 'Easy', 1: 'A challenge', 2: 'More effort'}\n0\nWhat course would you like?\nInput number of choice:\n{0: 'Snack', 1: 'Dessert'}\n0\nWhich of these remaining ingredients would you like to be in your meal?\nInput number of choice:\n{0: 'dried cranberries', 1: 'lemon'}\n1\nWhat cuisine do you want?\nInput number of choice:\n{0: 'British'}\n0\nThe following recipes from BBC's Good Foods website match your needs.\n['Rhubarb & vanilla jam']\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec6f6aa02fcd44d1224d630d52d1723108711481 | 30,294 | ipynb | Jupyter Notebook | Guided Project 4/Visualizing Earnings Based on College Majors.ipynb | AhnchBala/DataSciencePortfolio | 7fc9a4eea8718d7d1d4ede9e9e07935ab30da889 | [
"MIT"
]
| null | null | null | Guided Project 4/Visualizing Earnings Based on College Majors.ipynb | AhnchBala/DataSciencePortfolio | 7fc9a4eea8718d7d1d4ede9e9e07935ab30da889 | [
"MIT"
]
| null | null | null | Guided Project 4/Visualizing Earnings Based on College Majors.ipynb | AhnchBala/DataSciencePortfolio | 7fc9a4eea8718d7d1d4ede9e9e07935ab30da889 | [
"MIT"
]
| null | null | null | 36.498795 | 224 | 0.377236 | [
[
[
"import pandas as pd\nfrom pandas.plotting import scatter_matrix\n%matplotlib inline\nrecent_grads = pd.read_csv(\"recent-grads.csv\", encoding=\"UTF-8\")\nrecent_grads.iloc[0]\nrecent_grads.head() # acess data at top 5 rows",
"_____no_output_____"
],
[
"recent_grads.tail()",
"_____no_output_____"
],
[
"recent_grads.describe() #gives us statsitc of each column",
"_____no_output_____"
]
],
[
[
"#### Dropping NA\n\nWe can see in the .describe() that the file shows some counts to be 173 and others to be 172, so it looks like theres something wrong with the file in regards to a row having null values. We will have to drop that row.",
"_____no_output_____"
]
],
[
[
"recent_grads.isnull().sum()",
"_____no_output_____"
]
],
[
[
"In the code above we can see how there are 4 categories with one instance of null vlaue. What this means is that most likely, there is one row that has gone wrong.",
"_____no_output_____"
]
],
[
[
"recent_grads=recent_grads.dropna()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6f7588216553566aec0c2d6b922c18ea601611 | 650,771 | ipynb | Jupyter Notebook | 06_advanced_agromanagement_with_PCSE.ipynb | linghuameng/wofostmlh | 1fa20bff52af1d7adea764d22d5c69c1b1e49630 | [
"MIT"
]
| null | null | null | 06_advanced_agromanagement_with_PCSE.ipynb | linghuameng/wofostmlh | 1fa20bff52af1d7adea764d22d5c69c1b1e49630 | [
"MIT"
]
| null | null | null | 06_advanced_agromanagement_with_PCSE.ipynb | linghuameng/wofostmlh | 1fa20bff52af1d7adea764d22d5c69c1b1e49630 | [
"MIT"
]
| null | null | null | 820.644388 | 321,700 | 0.948981 | [
[
[
"<img style=\"float: right;\" src=\"data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAOIAAAAjCAYAAACJpNbGAAAABHNCSVQICAgIfAhkiAAAAAlwSFlzAAALEgAACxIB0t1+/AAAABR0RVh0Q3JlYXRpb24gVGltZQAzLzcvMTNND4u/AAAAHHRFWHRTb2Z0d2FyZQBBZG9iZSBGaXJld29ya3MgQ1M26LyyjAAACMFJREFUeJztnD1y20gWgD+6nJtzAsPhRqKL3AwqwQdYDpXDZfoEppNNTaWbmD7BUEXmI3EPMFCR2YI1UDQpdAPqBNzgvRZA/BGUZEnk9FeFIgj0z2ugX7/XP+jGer2mLv/8b6d+4Efgf/8KG0+Zn8XyXLx+bgEslqegcfzxSY3Irrx6bgEsFssBWsRGowGufwHAYtq7u+H6fUCOxTTWax4wBAbr+SRqNDKesOv3gN/133sW0yh927j1mucIaFWINl7PJ+OcvMcfW8Bol3iN44+mLIOsTCp3UJFfAETr+WRQcG8EOJpunEnTyDlYzycbeWr5xxq3jOF6PglK8ix9buv5xCsrAzBkMV1l5OwD/aJ4BXzV3+8F9z4gz/hTSbz8cxc84FuNvDc4VIsYA7+qohmGwAnycA194G22YqUYlZxv4vpN4AuwBv4oON5m8k3TVLnK4sYFcRyN86dWvCwnlCvFCeUVvwX8CkSZZ5eWs5mLJWE/VZThBMgpfirPk5J4f1SU4QsQ6LNP4+j9OkSUKdRiGlD87CWe3PcyR5PFdAhc1cz/joOziMoIeVF95GX1EGVY6bWhvsAeZQrm+kON80PDneD6PRbTi4LQpmJfsZieFaR1qXlXURh3y2BaBPyG63sspv0t6e+CKJTrf2YxHe8Qr6z8AXBdGbMoHgCTshgr4AiItfxljenPJGv5roCi+rGVw1TExTTWl99ThRsglfYHUnF7SMv+Bhjn4idxbhFLGiAu6gjXD3LuUBF5VzWi3CoAfMP1kxe7mNYZMT5DLFgf13eAXi3ZtvMOsUb3V3J5/mmqy+/66RbnTC1LFdfIu/kd8Qx2bTQeg2GBTPfiUF1TgHNE0QaIq/JDX9RKr/WBy/V8EhfEHWncWMO2EKV8S7UypYnYdE2r+o8gyj5MHXVYsZh+JnG7A+3LPQxR5g9II/UJ148ockmrybqm2+Qapo6gppwB8J7EM6jqaz8u0lhfkXgB58BKPam6rvEdh2kRARbTMa7/HXEfVqnW8hxxWwE+5+JJRTYd9CM90gxw/XFuMKMo/yTNDzUkLnbr6rCYnuH6N8igQ3CvNPJproDPuH6MKMd4Z5kMUjnrh98tn1if72/Ie729Vzq708L0YV3/HGmgB4iHsjOProhhd1lrEr4zaz/FvM4lolTnqWum/6jKmeuDmFb1jHylNg96hPQbhcU0wPVBXESvQI4W5aNshsK4jeOPhSOcOaThMVb48dhU8m2UlR+29ZHzrqyhLL0EaTROteGt67EYIsT6F1HXC/ikcvS00dl51PRwLaIwQtzCxGWRFnRMkT8v/SyAy8I+iliHJtDUsHHq7imipE42GtJanxdcB6mgQcm9MmKNs1m5F9MI13+n+cXZSEpAeV8mQgZqNkmU/HsuT7kf4PrGhXcK0h1SXv7iPKsJKCrDYvoV17+meMqhiDFlll7GEb4U3iseAf+k7mqksmU9qUoaj73E7TEtol3iZnks7Moai8WylUN3TS0WANbzyYv2rqxFtFheANYi7iGNRoPOrO2QGTQIu8vhU8vSmbWNDAHQD7vLYWfWbgFx2F3ee3FBZ9ZuIgMpTWAQdpeRXm9pPoPOrD3UMCtkQM4BRmF3ubG6ZZdxkOfCWsT9pU96CuX56KfOjeIFVC8Ar8NI0xuyOQJsVkWl8xzptQGPNY/6xFiLuL+0gIu0FVTrNESmbK7C7tLrzNpmPW0EeGF32UyFN19UnCAT4ZHGWWnYqDNrB4jViZBK/kbD9sLuMiBZSD8AVp1Z+0LD/NmZta+BIzOS3pm1xwBhd9kvkeEGUbQeqSmIdHhkXnGs5fIQRUxPV1x0Zm2zMuoq7C69rU/yBWAt4v7iAd86s/ZaDweZP+wBvwBOZ9b2SCrrmPzk+AWizA09j1QxMK4gZumcWKUWMvkdA56mfxN2l7GmHWk6V2F32Qi7yxaIsmnYHvkJ9zEQqAwBotQXwK2m0c+EN/Kk8zPTZiOkIWrp/xNTnpeOtYh7iFauN+k5W+0vXab6UsbyecAw229SxWiG3aVZ7NBCKrGHuneazy2iyBeIuxkjk9UDE1bzOtJ4IzbdwysNN0D6dnf9Rk3/iKSBWOnhUbASSWW+DbvLWM+HKreZ3O/r77gza5u842w6LxFrEfcTj+Jv3mK4q7Co63hE+fI6E94hUaT0cry+XushSuvoNZO2CdsCrlXJHDYVMUIUJso2BmhfL+wuV6rMvVR6AXnS1428XupaE7Hwnrqkg4cMGD0lr3NfpVegrUw1m2sN0+crNirEX1uTqiPbPoyI/QSKKmqA9I9aer+fcR2zxIj7GiMV+EYVIkZc3r5eH2rYI+0vnpBYIE/vGwUCdYM7s3agbqXJu58VIOwug86sfd2ZtSPNKwi7S9PHy4UnscCmXKuUZQRdsqbPwCHp2754pKYnW0akcZBO/x2df29XnvA//6iV8T3TSluBmOQlR+v5JNvaHixlDZRalRZifbZaAg3vIIrkmP6YVu6owI1M9x2r0vVIFCBGXNLS96Ph45IGY2ey6e1DY20UMaLGItUXoIhVvCv5tvDg2MWLqYNaoKBKWe6Z7gBR8OwAzZOyD4poBmtidlwt/gIxw/QHz0+oWKIoj19fRz8p3YOjoV8195F5l31ltZ5PfnluISyW+/IK6SPstRIiH/FaLHvLa2R+6F6f978AVsD7v0vf0HK4vNK9VfbVojSBceP4o/PcglgsD8GMmjaRbRCc1PEQIrbv45nlIfleIrs778XkrcWSZXMcXPZyqbvfxy7ckuyqHJPslJzH9c3We2ZRbx1O/07ziJbDI1FE2Qwp4n4DNzHJhkZF16+3bnwrCmi40U2eWoj7KZvobn7+YtKO1vPJVyyWPSZrER1kNU0TqfienpvlaWZR7oX+3tba6lxcX7MK3tNfo2RlpNc8tthsIFbAKYtpsA+TtRbLNp5/H4/EFXX0MOfbOGUxvbCKaDkEnl8Rq0jc1ayFjhFFjKwiWg6B/wNk+JCXXNBIXQAAAABJRU5ErkJggg==\">\n\n",
"_____no_output_____"
],
[
"# Advanced agromanagement with PCSE/WOFOST\n\nThis notebook will demonstrate how to implement advanced agromanagement options with PCSE/WOFOST.\n\nAllard de Wit, April 2018\n\nFor the example we will assume that data files are in the data directory within the directory where this notebook is located. This will be the case if you downloaded the notebooks from github.\n\n**Prerequisites for running this notebook**\n\nSeveral packages need to be installed for running PCSE/WOFOST:\n\n 1. PCSE and its dependencies. See the [PCSE user guide](http://pcse.readthedocs.io/en/stable/installing.html) for more information;\n 2. The `pandas` module for processing and storing WOFOST output;\n 3. The `matplotlib` module for plotting results\n\nFinally, you need a working internet connection.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport os, sys\n\nimport matplotlib\nmatplotlib.style.use(\"ggplot\")\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport yaml\n\nimport pcse\nfrom pcse.models import Wofost71_WLP_FD\nfrom pcse.fileinput import CABOFileReader, YAMLCropDataProvider\nfrom pcse.db import NASAPowerWeatherDataProvider\nfrom pcse.util import WOFOST71SiteDataProvider\nfrom pcse.base import ParameterProvider\ndata_dir = os.path.join(os.getcwd(), \"data\")\n\nprint(\"This notebook was built with:\")\nprint(\"python version: %s \" % sys.version)\nprint(\"PCSE version: %s\" % pcse.__version__)",
"This notebook was built with:\npython version: 3.7.5 (default, Oct 31 2019, 15:18:51) [MSC v.1916 64 bit (AMD64)] \nPCSE version: 5.4.2\n"
]
],
[
[
"## Input requirements\nFor running the PCSE/WOFOST (and PCSE models in general), you need three types of inputs:\n1. Model parameters that parameterize the different model components. These parameters usually\n consist of a set of crop parameters (or multiple sets in case of crop rotations), a set of soil parameters\n and a set of site parameters. The latter provide ancillary parameters that are specific for a location.\n2. Driving variables represented by weather data which can be derived from various sources.\n3. Agromanagement actions which specify the farm activities that will take place on the field that is simulated\n by PCSE.\n\n## Reading model parameters\nIn this example, we will derive the model parameters from different sources. First of all, the crop parameters will be read from my [github repository](https://github.com/ajwdewit/WOFOST_crop_parameters) using the `YAMLCropDataProvider`. Next, the soil parameters will be read from a classical CABO input file using the `CABOFileReader`. Finally, the site parameters can be defined directly using the `WOFOST71SiteDataProvider` which provides sensible defaults for site parameters. \n\nHowever, PCSE models expect a single set of parameters and therefore they need to be combined using the `ParameterProvider`:",
"_____no_output_____"
]
],
[
[
"crop = YAMLCropDataProvider()\nsoil = CABOFileReader(os.path.join(data_dir, \"soil\", \"ec3.soil\"))\nsite = WOFOST71SiteDataProvider(WAV=100,CO2=360)\nparameterprovider = ParameterProvider(soildata=soil, cropdata=crop, sitedata=site)",
"_____no_output_____"
],
[
"crop = YAMLCropDataProvider()\n",
"_____no_output_____"
]
],
[
[
"## Reading weather data\nFor reading weather data we will use the NASAPowerWeatherDataProvider. ",
"_____no_output_____"
]
],
[
[
"from pcse.fileinput import ExcelWeatherDataProvider\nweatherfile = os.path.join(data_dir, 'meteo', 'nl1.xlsx')\nweatherdataprovider = ExcelWeatherDataProvider(weatherfile)\n",
"_____no_output_____"
]
],
[
[
"## Defining agromanagement with timed events\n\nDefining agromanagement needs a bit more explanation because agromanagement is a relatively\ncomplex piece of PCSE. The agromanagement definition for PCSE is written in a format called `YAML` and for a thorough discusion have a look at the [Section on Agromanagement](https://pcse.readthedocs.io/en/stable/reference_guide.html#the-agromanager) in the PCSE manual.\nFor the current example the agromanagement definition looks like this:\n\n Version: 1.0\n AgroManagement:\n - 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: 2006-03-31\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents:\n - event_signal: irrigate\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 2006-07-10: {amount: 10, efficiency: 0.7}\n - 2006-08-05: {amount: 5, efficiency: 0.7}\n StateEvents: null\n\nThe agromanagement definition starts with `Version:` indicating the version number of the agromanagement file\nwhile the actual definition starts after the label `AgroManagement:`. Next a date must be provide which sets the\nstart date of the campaign (and the start date of the simulation). Each campaign is defined by zero or one\nCropCalendars and zero or more TimedEvents and/or StateEvents. The CropCalendar defines the crop type, date of sowing,\ndate of harvesting, etc. while the Timed/StateEvents define actions that are either connected to a date or\nto a model state.\n\nIn the current example, the campaign starts on 2006-01-01, there is a crop calendar for sugar beet starting on\n2006-03-31 with a harvest date of 2006-10-20. Next there are timed events defined for applying irrigation at 2006-07-10 and 2006-08-05. The current example has no state events. For a thorough description of all possibilities see the section on AgroManagement in the Reference Guide.\n\nLoading the agromanagement definition from a file can be done with the `YAMLAgroManagementReader`. However for this example, we can just as easily define it here and parse it directly with the YAML parser. In this case we can directly use the section after the `Agromanagement:` label.",
"_____no_output_____"
]
],
[
[
"yaml_agro = \"\"\"\n- 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: 2006-03-31\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents:\n - event_signal: irrigate\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 2006-07-10: {amount: 10, efficiency: 0.7}\n - 2006-08-05: {amount: 5, efficiency: 0.7}\n StateEvents: null\n\"\"\"\nagromanagement = yaml.load(yaml_agro)",
"_____no_output_____"
]
],
[
[
"## Starting and running the WOFOST\nWe have now all parameters, weather data and agromanagement information available to start WOFOST and make a simulation.",
"_____no_output_____"
]
],
[
[
"wofost = Wofost71_WLP_FD(parameterprovider, weatherdataprovider, agromanagement)\nwofost.run_till_terminate()",
"_____no_output_____"
]
],
[
[
"## Getting and visualizing results\n\nNext, we can easily get the output from the model using the get_output() method and turn it into a pandas DataFrame:",
"_____no_output_____"
]
],
[
[
"output = wofost.get_output()\ndf = pd.DataFrame(output).set_index(\"day\")\ndf.tail()",
"_____no_output_____"
]
],
[
[
"Finally, we can visualize the results from the pandas DataFrame with a few commands:",
"_____no_output_____"
]
],
[
[
"fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(16,8))\ndf['LAI'].plot(ax=axes[0], title=\"Leaf Area Index\")\ndf['SM'].plot(ax=axes[1], title=\"Root zone soil moisture\")\nfig.autofmt_xdate()",
"_____no_output_____"
]
],
[
[
"# Defining agromanagement with state events\n\n## Connecting events to development stages\nIt is also possible to connect irrigation events to state variables instead of dates. A logical approach is to connect an irrigation even to a development stage instead of a date, in this way changes in the sowing date will be automatically reflected in changes in irrigation events.\n\nFor this we need to change the definition of the agromanagement as below:\n\n Version: 1.0\n AgroManagement:\n - 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents: null\n StateEvents:\n - event_signal: irrigate\n event_state: DVS\n zero_condition: rising\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 0.9: {amount: 10, efficiency: 0.7}\n - 1.5: {amount: 5, efficiency: 0.7}\n - 2006-11-20: null\n \nIn this case the irrigation events are connected to the state DVS and are occurring when the simulated DVS crosses the values 0.9 and 1.5. Note that there two additional parameters: `event_state` which defines the state to which the event is connected and `zero_condition` which specifies the condition under which the state event fires, see for an explanation [here](http://pcse.readthedocs.org/en/latest/code.html#agromanagement). Finally, also note that there must be an \"empty trailing campaign\" defined which defines that the campaign that starts at 2006-01-01 ends at 2006-11-20. Otherwise PCSE cannot determine the end of the simulation period, see also the link above for an explanation.\n\nAgain, we will define the agromanagement directly on the command line and parse it with YAML.",
"_____no_output_____"
]
],
[
[
"yaml_agro = \"\"\"\n- 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: 2006-03-31\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents: null\n StateEvents:\n - event_signal: irrigate\n event_state: DVS\n zero_condition: rising\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 0.9: {amount: 10, efficiency: 0.7}\n - 1.5: {amount: 5, efficiency: 0.7}\n- 2006-11-20: null\n\"\"\"\nagromanagement = yaml.load(yaml_agro)",
"_____no_output_____"
]
],
[
[
"Again we run the model with all inputs but a changed agromanagement and plot the results",
"_____no_output_____"
]
],
[
[
"wofost2 = Wofost71_WLP_FD(parameterprovider, weatherdataprovider, agromanagement)\nwofost2.run_till_terminate()\noutput2 = wofost2.get_output()\ndf2 = pd.DataFrame(output2).set_index(\"day\")\nfig2, axes2 = plt.subplots(nrows=1, ncols=2, figsize=(16,8))\ndf2['LAI'].plot(ax=axes2[0], title=\"Leaf Area Index\")\ndf2['SM'].plot(ax=axes2[1], title=\"Root zone soil moisture\")\nfig2.autofmt_xdate()",
"_____no_output_____"
]
],
[
[
"## Connecting events to soil moisture levels\n\nThe logical approach is to connect irrigation events to stress levels that are experiences by the crop. In this case we connect the irrigation event to the state variables soil moisture (SM) and define the agromanagement like this:\n\n Version: 1.0\n AgroManagement:\n - 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: 2006-03-31\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents: null\n StateEvents:\n - event_signal: irrigate\n event_state: SM\n zero_condition: falling\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 0.2: {amount: 10, efficiency: 0.7}\n - 2006-11-20:\n \nNote that in this case the `zero_condition` is `falling` because we only want the event to trigger when the SM goes below the specified level (0.2). If we had set `zero_condition` to `either` it would trigger twice, the first time when the soil moisture gets exhausted and the second time because of the irrigation water added.",
"_____no_output_____"
]
],
[
[
"yaml_agro = \"\"\"\n- 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: 2006-03-31\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents: null\n StateEvents:\n - event_signal: irrigate\n event_state: SM\n zero_condition: falling\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 0.2: {amount: 10, efficiency: 0.7}\n- 2006-11-20: null\n\"\"\"\nagromanagement = yaml.load(yaml_agro)",
"_____no_output_____"
],
[
"wofost3 = Wofost71_WLP_FD(parameterprovider, weatherdataprovider, agromanagement)\nwofost3.run_till_terminate()\noutput3 = wofost3.get_output()\ndf3 = pd.DataFrame(output3).set_index(\"day\")\n\nfig3, axes3 = plt.subplots(nrows=1, ncols=2, figsize=(16,8))\ndf3['LAI'].plot(ax=axes3[0], title=\"Leaf Area Index\")\ndf3['SM'].plot(ax=axes3[1], title=\"Volumetric soil moisture\")\nfig3.autofmt_xdate()",
"_____no_output_____"
]
],
[
[
"Showing the differences in irrigation events\n============================================\n\nWe combine the `SM` column from the different data frames in a new dataframe and plot the results to see the effect of the differences in agromanagement.",
"_____no_output_____"
]
],
[
[
"df_all = pd.DataFrame({\"by_date\": df.SM, \n \"by_DVS\": df2.SM, \n \"by_SM\": df3.SM}, index=df.index)\nfig4, axes4 = plt.subplots(nrows=1, ncols=1, figsize=(14,12))\ndf_all.plot(ax=axes4, title=\"differences in irrigation approach.\")\naxes4.set_ylabel(\"irrigation amount [cm]\")\nfig4.autofmt_xdate()",
"_____no_output_____"
]
],
[
[
"Adjusting the sowing date with the AgroManager and making multiple runs\n==============================================\n\nThe most straightforward way of adjusting the sowing date is by editing the crop management definition in YAML format directly. Here we put a placeholder `{crop_start_date}` at the point where the crop start date is defined in the YAML format. We can then use string formatting operations to insert a new data and use `yaml.load` to load the definition in yaml directly. Note that we need double curly brackets (`{{` and `}}`) at the events table to avoid that python sees them as a placeholder.",
"_____no_output_____"
]
],
[
[
"agromanagement_yaml = \"\"\"\n- 2006-01-01:\n CropCalendar:\n crop_name: sugarbeet\n variety_name: Sugarbeet_603\n crop_start_date: {crop_start_date}\n crop_start_type: emergence\n crop_end_date: 2006-10-20\n crop_end_type: harvest\n max_duration: 300\n TimedEvents: null\n StateEvents:\n - event_signal: irrigate\n event_state: SM\n zero_condition: falling\n name: Irrigation application table\n comment: All irrigation amounts in cm\n events_table:\n - 0.2: {{amount: 10, efficiency: 0.7}}\n- 2006-11-20:\n\"\"\"",
"_____no_output_____"
]
],
[
[
"## The main loop for making several WOFOST runs",
"_____no_output_____"
]
],
[
[
"import datetime as dt\nsdate = dt.date(2006,3,1)\nstep = 10\n# Loop over six different start dates \nresults = []\nfor i in range(6):\n # get new start date\n csdate = sdate + dt.timedelta(days=i*step)\n # update agromanagement with new start date and load it with yaml.load\n tmp = agromanagement_yaml.format(crop_start_date=csdate)\n agromanagement = yaml.load(tmp)\n # run wofost and collect output\n wofost = Wofost71_WLP_FD(parameterprovider, weatherdataprovider, agromanagement)\n wofost.run_till_terminate()\n output = wofost.get_output()\n df = pd.DataFrame(output).set_index(\"day\")\n results.append(df)",
"_____no_output_____"
]
],
[
[
"## Plot the results for the different runs and variables",
"_____no_output_____"
]
],
[
[
"colors = ['k','r','g','b','m','y']\nfig5, axes5 = plt.subplots(nrows=6, ncols=2, figsize=(16,30))\nfor c, df in zip(colors, results):\n for key, axis in zip(df.columns, axes5.flatten()):\n df[key].plot(ax=axis, title=key, color=c)\nfig5.autofmt_xdate()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.