hexsha
stringlengths 40
40
| size
int64 6
14.9M
| ext
stringclasses 1
value | lang
stringclasses 1
value | max_stars_repo_path
stringlengths 6
260
| max_stars_repo_name
stringlengths 6
119
| max_stars_repo_head_hexsha
stringlengths 40
41
| max_stars_repo_licenses
list | max_stars_count
int64 1
191k
⌀ | max_stars_repo_stars_event_min_datetime
stringlengths 24
24
⌀ | max_stars_repo_stars_event_max_datetime
stringlengths 24
24
⌀ | max_issues_repo_path
stringlengths 6
260
| max_issues_repo_name
stringlengths 6
119
| max_issues_repo_head_hexsha
stringlengths 40
41
| max_issues_repo_licenses
list | max_issues_count
int64 1
67k
⌀ | max_issues_repo_issues_event_min_datetime
stringlengths 24
24
⌀ | max_issues_repo_issues_event_max_datetime
stringlengths 24
24
⌀ | max_forks_repo_path
stringlengths 6
260
| max_forks_repo_name
stringlengths 6
119
| max_forks_repo_head_hexsha
stringlengths 40
41
| max_forks_repo_licenses
list | max_forks_count
int64 1
105k
⌀ | max_forks_repo_forks_event_min_datetime
stringlengths 24
24
⌀ | max_forks_repo_forks_event_max_datetime
stringlengths 24
24
⌀ | avg_line_length
float64 2
1.04M
| max_line_length
int64 2
11.2M
| alphanum_fraction
float64 0
1
| cells
list | cell_types
list | cell_type_groups
list |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
ec6f7d382455327fce95024862bcda52558c3f95 | 195,015 | ipynb | Jupyter Notebook | DRLND-Navigation/Navigation.ipynb | jaredjxyz/deep-reinforcement-learning | eaf6ff16bfb0d7a684c9d296d192e61086132238 | [
"MIT"
]
| null | null | null | DRLND-Navigation/Navigation.ipynb | jaredjxyz/deep-reinforcement-learning | eaf6ff16bfb0d7a684c9d296d192e61086132238 | [
"MIT"
]
| null | null | null | DRLND-Navigation/Navigation.ipynb | jaredjxyz/deep-reinforcement-learning | eaf6ff16bfb0d7a684c9d296d192e61086132238 | [
"MIT"
]
| null | null | null | 57.458751 | 99,431 | 0.713453 | [
[
[
"# Navigation\n\n---\n\nYou are welcome to use this coding environment to train your agent for the project. Follow the instructions below to get started!\n\n### 1. Start the Environment\n\nRun the next code cell to install a few packages. This line will take a few minutes to run!",
"_____no_output_____"
]
],
[
[
"!pip -q install ./python",
"_____no_output_____"
]
],
[
[
"The environment is already saved in the Workspace and can be accessed at the file path provided below. Please run the next code cell without making any changes.",
"_____no_output_____"
]
],
[
[
"from unityagents import UnityEnvironment\nimport numpy as np\n\n# please do not modify the line below\nenv = UnityEnvironment(file_name=\"/data/Banana_Linux_NoVis/Banana.x86_64\")",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\t\nUnity brain name: BananaBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 37\n Number of stacked Vector Observation: 1\n Vector Action space type: discrete\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]",
"_____no_output_____"
]
],
[
[
"### 2. Examine the State and Action Spaces\n\nRun the code cell below to print some information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of agents in the environment\nprint('Number of agents:', len(env_info.agents))\n\n# number of actions\naction_size = brain.vector_action_space_size\nprint('Number of actions:', action_size)\n\n# examine the state space \nstate = env_info.vector_observations[0]\nprint('States look like:', state)\nstate_size = len(state)\nprint('States have length:', state_size)",
"Number of agents: 1\nNumber of actions: 4\nStates look like: [ 1. 0. 0. 0. 0.84408134 0. 0.\n 1. 0. 0.0748472 0. 1. 0. 0.\n 0.25755 1. 0. 0. 0. 0.74177343\n 0. 1. 0. 0. 0.25854847 0. 0.\n 1. 0. 0.09355672 0. 1. 0. 0.\n 0.31969345 0. 0. ]\nStates have length: 37\n"
]
],
[
[
"### 3. Take Random Actions in the Environment\n\nIn the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.\n\nNote that **in this coding environment, you will not be able to watch the agent while it is training**, and you should set `train_mode=True` to restart the environment.",
"_____no_output_____"
]
],
[
[
"env_info = env.reset(train_mode=True)[brain_name] # reset the environment\nstate = env_info.vector_observations[0] # get the current state\nscore = 0 # initialize the score\nwhile True:\n action = np.random.randint(action_size) # select an action\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = env_info.local_done[0] # see if episode has finished\n score += reward # update the score\n state = next_state # roll over the state to next time step\n if done: # exit loop if episode finished\n break\n \nprint(\"Score: {}\".format(score))",
"Score: 0.0\n"
]
],
[
[
"When finished, you can close the environment.",
"_____no_output_____"
]
],
[
[
"#env.close()",
"_____no_output_____"
]
],
[
[
"### 4. It's Your Turn!\n\nNow it's your turn to train your own agent to solve the environment! A few **important notes**:\n- When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:\n```python\nenv_info = env.reset(train_mode=True)[brain_name]\n```\n- To structure your work, you're welcome to work directly in this Jupyter notebook, or you might like to start over with a new file! You can see the list of files in the workspace by clicking on **_Jupyter_** in the top left corner of the notebook.\n- In this coding environment, you will not be able to watch the agent while it is training. However, **_after training the agent_**, you can download the saved model weights to watch the agent on your own machine! ",
"_____no_output_____"
],
[
"## Define neural network",
"_____no_output_____"
]
],
[
[
"import torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\n\n# class QNetwork(nn.Module):\n# def __init__(self, state_size, action_size, seed, *args):\n# super().__init__()\n# self.seed = torch.manual_seed(seed)\n# self.fc1 = nn.Linear(state_size, 64)\n# self.fc2 = nn.Linear(64, 64)\n# self.fc3 = nn.Linear(64, action_size)\n \n# def forward(self, state):\n# print('Forward')\n# state = F.relu(self.fc1(state))\n# state = F.relu(self.fc2(state))\n# state = self.fc3(state)\n# return state\n \nclass QNetwork(nn.Module):\n def __init__(self, state_size, action_size, seed, hidden_layer_sizes=[]):\n super().__init__()\n self.seed = torch.manual_seed(seed)\n self.fc_layers = nn.ModuleList()\n layer_sizes = [state_size] + list(hidden_layer_sizes) + [action_size]\n for size1, size2 in zip(layer_sizes[:-1], layer_sizes[1:]):\n self.fc_layers.append(nn.Linear(size1, size2))\n \n def forward(self, state):\n for layer in self.fc_layers[:-1]:\n state = F.leaky_relu(layer(state))\n state = self.fc_layers[-1](state)\n return state",
"_____no_output_____"
]
],
[
[
"## Define agent",
"_____no_output_____"
]
],
[
[
"device = torch.device(\"cuda:0\" if torch.cuda.is_available() else \"cpu\")\nprint(\"Using device\", device)\nfrom collections import namedtuple, deque\nimport random\n\n# Uses the Double q-learning method to learn\nclass DQN_Agent:\n def __init__(self, state_size, action_size, seed,\n buffer_size = int(300 * 1e5),\n batch_size = 512,\n gamma = 0.99,\n learning_rate = 5e-3,\n update_every = 20,\n tau = 2e-2,\n middle_layers = [32, 32]):\n self.state_size = state_size\n self.action_size = action_size\n self.seed = seed\n self.gamma = gamma\n self.learning_rate = learning_rate\n self.update_every = update_every\n self.tau = tau\n self.middle_layers = middle_layers\n \n self.qnetwork = QNetwork(state_size, action_size, seed, middle_layers).to(device)\n self.qnetwork_target = QNetwork(state_size, action_size, seed, middle_layers).to(device)\n self.optimizer = optim.Adam(self.qnetwork.parameters(), lr=learning_rate)\n \n self.memory = ReplayBuffer(buffer_size, batch_size, seed)\n \n self.time_step = 0\n \n def step(self, state, action, reward, next_state, done):\n self.memory.add(state, action, reward, next_state, done)\n \n self.time_step = (self.time_step + 1) % self.update_every\n if self.time_step == 0:\n if len(self.memory) > self.memory.batch_size:\n experiences = self.memory.sample()\n self.learn(experiences, self.gamma)\n \n def act(self, state, eps=0.0):\n state = torch.from_numpy(state).float().unsqueeze(0).to(device)\n self.qnetwork.eval()\n with torch.no_grad():\n action_values = self.qnetwork(state).max(1)[1]\n self.qnetwork.train()\n \n if random.random() > eps:\n return action_values.cpu().numpy()\n else:\n return random.choice(np.arange(self.action_size))\n \n def learn(self, experiences, gamma):\n states, actions, rewards, next_states, dones = experiences\n \n next_qs = self.qnetwork(next_states).detach()\n next_qs_target = self.qnetwork_target(next_states).detach()\n max_next_qs, best_next_actions = next_qs.max(1, keepdim=True)\n \n q = self.qnetwork(states).gather(1, actions)\n q_next = next_qs_target.gather(1, best_next_actions) # Double q-learning step\n \n loss = F.mse_loss(rewards + gamma * q_next * (1 - dones), q)\n self.optimizer.zero_grad()\n loss.backward()\n self.optimizer.step()\n self.soft_update(self.qnetwork, self.qnetwork_target, self.tau)\n \n # Update\n def soft_update(self, local_model, target_model, tau):\n \"\"\"Soft update model parameters.\n θ_target = τ*θ_local + (1 - τ)*θ_target\n\n Params\n ======\n local_model (PyTorch model): weights will be copied from\n target_model (PyTorch model): weights will be copied to\n tau (float): interpolation parameter \n \"\"\"\n for target_param, local_param in zip(target_model.parameters(), local_model.parameters()):\n target_param.data.copy_(tau*local_param.data + (1.0-tau)*target_param.data)\n \n def values(self, state):\n state = torch.from_numpy(state).float().unsqueeze(0).to(device)\n self.qnetwork.eval()\n with torch.no_grad():\n action_values = self.qnetwork(state).squeeze(1)\n self.qnetwork.train()\n return action_values.cpu().numpy()\n \n def save(self, location):\n torch.save(self.qnetwork_target.state_dict(), location)\n \n def load(self, location):\n self.qnetwork.load_state_dict(torch.load(location))\n self.qnetwork_target.load_state_dict(torch.load(location))\n \nclass ReplayBuffer:\n def __init__(self, buffer_size, batch_size, seed):\n self.memory = deque(maxlen=buffer_size)\n self.batch_size = batch_size\n self.experience = namedtuple(\"Experience\", field_names=['state', 'action', 'reward', 'next_state', 'done'])\n self.seed = random.seed(seed)\n \n def add(self, state, action, reward, next_state, done):\n e = self.experience(state, action, reward, next_state, done)\n self.memory.append(e)\n \n def sample(self):\n experiences = random.sample(self.memory, k=self.batch_size)\n \n states = torch.from_numpy(np.vstack([e.state for e in experiences if e is not None])).float().to(device)\n actions = torch.from_numpy(np.vstack([e.action for e in experiences if e is not None])).long().to(device)\n rewards = torch.from_numpy(np.vstack([e.reward for e in experiences if e is not None])).float().to(device)\n next_states = torch.from_numpy(np.vstack([e.next_state for e in experiences if e is not None])).float().to(device)\n dones = torch.from_numpy(np.vstack([e.done for e in experiences if e is not None])).float().to(device)\n \n return (states, actions, rewards, next_states, dones)\n \n def __len__(self):\n return len(self.memory)",
"Using device cpu\n"
]
],
[
[
"### Make the agent",
"_____no_output_____"
]
],
[
[
"agent = DQN_Agent(state_size=state_size, action_size=action_size, seed=0)",
"_____no_output_____"
]
],
[
[
"### Train the agent",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom tqdm import tqdm_notebook as tqdm\nfrom collections import deque\n\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n%matplotlib notebook\nfig = plt.figure()\nax = fig.add_subplot(111)\naction_axes = [fig.add_subplot(111) for action in range(action_size)]\nplt.ion()\n#fig.show()\nfig.canvas.draw()\n\nnum_episodes = 2000\nepisodes = []\nsmoothing_buffer = 5\nscores = deque(maxlen=smoothing_buffer)\nmeans = []\naction_values = [[] for i in range(action_size)]\nfor i in tqdm(range(num_episodes)):\n env_info = env.reset(train_mode=True)[brain_name] # reset the environment\n state = env_info.vector_observations[0] # get the current state\n initial_state = state\n score = 0 # initialize the score\n moves = 0\n explore_rate = max((1 - i / num_episodes*2), .05) # linear for half of the time, .05 for the other half\n print(explore_rate)\n \n while True:\n action = agent.act(state, explore_rate) # select an action\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = int(env_info.local_done[0]) # see if episode has finished\n agent.step(state, action, reward, next_state, done)\n values = agent.values(state)\n score += reward # update the score\n state = next_state # roll over the state to next time step\n moves += 1\n if done: # exit loop if episode finished\n break\n \n scores.append(score)\n values = agent.values(initial_state)\n if len(scores) == smoothing_buffer:\n episodes.append(i)\n means.append(np.mean(scores))\n for action_num in range(len(values)):\n action_values[action_num].append(values[action_num])\n if i % 1 == 0:\n ax.clear()\n ax.plot(episodes, means)\n# for action in range(action_size):\n# ax.plot(episodes, action_values[action])\n# ax.legend()\n \n fig.canvas.draw()\nagent.save('q_checkpoint.pt')\n ",
"_____no_output_____"
]
],
[
[
"## Discussion on future work\n\nThere are several things I wanted to try that I didn't have to in order to get this to work.\nI am curious what would happen ono a much larger neural network. Let's say a nerual net with 10 middle layers of size 128 a piece. I didn't get to try this because of time and GPU size restrictions, but it's an experiment that I think is worth trying.\n\nThe other thing I think would help this implementation quite a bit is picking experience that the agent will learn from more (prioritized experience replay). Through the current implementation, I think the last bit of data has the agent doing the same thing over and over, which is good for getting through the last push of finding a great policy after it's done some exploration, but has the drawback of looking at the same examples over and over. Choosing which of those examples to look at intelligently could potentially really help this implementation.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6f7f7f351fe4645467d6d5572304a96d63e7f9 | 28,115 | ipynb | Jupyter Notebook | notebooks/layers/recurrent/SimpleRNN.ipynb | jefffriesen/keras-js | 130cca841fc4e8fd5fc157ae57061f9576d8588f | [
"MIT"
]
| null | null | null | notebooks/layers/recurrent/SimpleRNN.ipynb | jefffriesen/keras-js | 130cca841fc4e8fd5fc157ae57061f9576d8588f | [
"MIT"
]
| null | null | null | notebooks/layers/recurrent/SimpleRNN.ipynb | jefffriesen/keras-js | 130cca841fc4e8fd5fc157ae57061f9576d8588f | [
"MIT"
]
| 1 | 2020-03-12T21:03:06.000Z | 2020-03-12T21:03:06.000Z | 43.387346 | 454 | 0.577059 | [
[
[
"import numpy as np\nfrom keras.models import Model\nfrom keras.layers import Input\nfrom keras.layers.recurrent import SimpleRNN\nfrom keras import backend as K",
"Using TensorFlow backend.\n"
],
[
"def format_decimal(arr, places=6):\n return [round(x * 10**places) / 10**places for x in arr]",
"_____no_output_____"
]
],
[
[
"### SimpleRNN",
"_____no_output_____"
],
[
"**[recurrent.SimpleRNN.0] output_dim=4, activation='tanh'**\n\nNote dropout_W and dropout_U are only applied during training phase",
"_____no_output_____"
]
],
[
[
"data_in_shape = (3, 6)\nrnn = SimpleRNN(4, activation='tanh')\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3400 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [-0.045589, 0.415186, -0.562532, 0.417194, 0.595636, 0.384863, -0.421095, 0.531931, 0.892653, -0.9421, -0.522872, -0.37874, -0.768283, -0.196357, -0.818039, -0.631257, -0.405011, -0.035917, -0.48787, 0.181399, 0.150278, -0.910744, 0.68533, 0.571771]\nU shape: (4, 4)\nU: [-0.720918, -0.952173, -0.727704, 0.156292, -0.355836, -0.862534, 0.167887, 0.9923, -0.726801, 0.346909, 0.339642, 0.91009, 0.52891, -0.857623, -0.906373, 0.492599]\nb shape: (4,)\nb: [-0.836587, 0.897901, -0.267459, -0.930645]\n\nin shape: (3, 6)\nin: [-0.409861, -0.508697, -0.23829, 0.215855, -0.570529, 0.272606, -0.304086, -0.907375, -0.030361, 0.792806, 0.0388, -0.782223, 0.098008, -0.99904, 0.356238, -0.490761, 0.905586, 0.839691]\nout shape: (4,)\nout: [-0.921578, 0.582325, 0.984568, 0.313785]\n"
]
],
[
[
"**[recurrent.SimpleRNN.1] output_dim=5, activation='sigmoid'**\n\nNote dropout_W and dropout_U are only applied during training phase",
"_____no_output_____"
]
],
[
[
"data_in_shape = (8, 5)\nrnn = SimpleRNN(5, activation='sigmoid')\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3500 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (5, 5)\nW: [0.219175, 0.170157, -0.559083, 0.254571, -0.07401, -0.665539, -0.673129, -0.205758, -0.480003, 0.337035, 0.051406, -0.81763, -0.428026, -0.092167, 0.685529, -0.5668, -0.457007, 0.928876, 0.658722, -0.336033, -0.424178, -0.833065, 0.861114, 0.468557, -0.098388]\nU shape: (5, 5)\nU: [-0.048881, 0.614161, 0.195963, 0.306777, 0.135452, -0.773353, 0.075287, 0.283726, -0.340782, -0.184696, 0.292298, 0.660226, -0.914862, -0.785209, 0.617659, 0.937712, -0.58992, 0.752688, -0.120278, -0.591822, 0.717781, 0.903103, 0.992716, -0.891614, -0.697178]\nb shape: (5,)\nb: [0.698618, 0.022064, 0.814655, 0.729225, -0.278111]\n\nin shape: (8, 5)\nin: [0.010183, -0.266283, -0.587731, 0.343666, -0.894301, 0.745644, 0.221028, -0.386505, 0.599566, 0.769919, 0.609677, -0.28979, 0.967391, 0.717751, -0.140432, -0.631171, 0.947662, -0.028251, 0.421945, 0.204412, -0.366352, -0.920305, -0.748026, -0.124691, -0.578315, -0.684603, -0.336723, -0.918999, -0.820263, -0.186095, 0.571209, 0.206034, 0.477405, -0.478543, 0.401324, -0.582631, 0.920882, 0.54878, -0.473092, -0.504899]\nout shape: (5,)\nout: [0.706439, 0.65058, 0.610492, 0.185008, 0.596629]\n"
]
],
[
[
"**[recurrent.SimpleRNN.2] output_dim=4, activation='tanh', return_sequences=True**\n\nNote dropout_W and dropout_U are only applied during training phase",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=True)\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3600 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [0.201127, 0.727021, -0.937503, -0.008165, -0.433634, 0.261543, 0.5643, 0.254178, 0.716909, -0.796969, -0.216724, -0.388474, 0.775137, -0.574555, 0.846263, 0.402263, -0.551731, -0.068827, 0.998469, -0.197222, -0.178234, -0.714859, -0.203858, -0.318714]\nU shape: (4, 4)\nU: [-0.61537, -0.160725, -0.055996, 0.737985, -0.204782, 0.97122, -0.736425, -0.935545, 0.910577, -0.988285, 0.689019, -0.336002, -0.598416, 0.842676, 0.543717, 0.66669]\nb shape: (4,)\nb: [0.555658, 0.481608, 0.539114, -0.411614]\n\nin shape: (7, 6)\nin: [-0.687216, 0.901262, -0.801191, -0.323217, -0.809709, 0.80387, -0.356349, 0.675151, 0.355832, 0.909097, 0.416688, -0.59589, -0.005416, 0.78297, 0.744056, -0.249837, 0.188897, -0.414898, -0.917833, -0.074002, -0.263692, -0.502035, 0.957567, 0.960735, -0.30952, 0.192052, -0.086556, -0.013007, -0.370762, 0.274436, -0.744553, -0.779231, 0.064843, 0.771956, 0.438702, 0.582149, -0.750711, 0.828857, 0.742574, -0.204235, -0.410779, -0.4297]\nout shape: (7, 4)\nout: [-0.458042, 0.480022, 0.55094, -0.091951, 0.943342, -0.091388, 0.986228, -0.733112, 0.861592, -0.865843, 0.828059, -0.497206, -0.174145, -0.990565, 0.989866, -0.179369, 0.948069, -0.951829, 0.951084, -0.000203, 0.908789, -0.996825, 0.995605, 0.591596, 0.725882, -0.919538, 0.991781, 0.865701]\n"
]
],
[
[
"**[recurrent.SimpleRNN.3] output_dim=4, activation='tanh', return_sequences=False, go_backwards=True**\n\nNote dropout_W and dropout_U are only applied during training phase",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=False, go_backwards=True)\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3700 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [0.705929, -0.455616, -0.247481, -0.289738, -0.172745, 0.10867, -0.245206, -0.281163, 0.153902, 0.822921, 0.411496, 0.06129, 0.725935, -0.914002, 0.057156, -0.839685, 0.078484, 0.920066, 0.853763, 0.567047, 0.417124, 0.267793, 0.388957, -0.996824]\nU shape: (4, 4)\nU: [-0.219586, 0.206946, -0.331215, 0.127084, 0.22716, -0.850342, -0.866678, -0.96958, 0.788851, 0.549344, -0.182773, 0.765206, -0.856816, -0.163875, 0.80548, -0.385654]\nb shape: (4,)\nb: [-0.679828, 0.865029, 0.861333, 0.591202]\n\nin shape: (7, 6)\nin: [0.719399, 0.281407, 0.778497, 0.881326, 0.143583, -0.039836, -0.753727, -0.868352, -0.161102, 0.474482, 0.424175, -0.308505, 0.686509, 0.049983, 0.650185, 0.484433, 0.822685, -0.522601, 0.480813, 0.102252, -0.83675, 0.66675, 0.586352, -0.71142, 0.861089, -0.674528, -0.594313, 0.465266, 0.559367, -0.546443, -0.187902, -0.661597, 0.474318, 0.32583, 0.357901, -0.232202, -0.585644, 0.103958, -0.779314, -0.743425, -0.864946, -0.640665]\nout shape: (4,)\nout: [0.670791, 0.295618, 0.839228, -0.451159]\n"
]
],
[
[
"**[recurrent.SimpleRNN.4] output_dim=4, activation='tanh', return_sequences=True, go_backwards=True**\n\nNote dropout_W and dropout_U are only applied during training phase",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=True, go_backwards=True)\n\nlayer_0 = Input(shape=data_in_shape)\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3800 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [-0.390491, -0.578366, 0.174977, -0.794259, 0.275087, 0.428053, -0.915046, -0.096302, -0.744594, 0.049007, -0.159581, 0.213776, 0.238386, 0.329687, 0.619891, 0.36543, -0.482228, 0.841286, -0.270654, 0.132222, 0.042815, 0.057958, 0.870286, -0.57147]\nU shape: (4, 4)\nU: [0.467358, -0.740084, -0.552357, -0.475388, -0.053543, -0.749294, -0.89908, -0.196198, -0.7233, 0.67584, 0.165427, -0.011422, -0.766433, -0.250531, 0.959118, -0.582048]\nb shape: (4,)\nb: [-0.217165, 0.276023, -0.853592, -0.465207]\n\nin shape: (7, 6)\nin: [0.112672, -0.828695, 0.336233, 0.388093, -0.295826, -0.480416, 0.225187, -0.364328, -0.206269, 0.961105, 0.039125, 0.391735, -0.24781, 0.492028, 0.557904, 0.264287, -0.540591, -0.777572, 0.350376, 0.03832, -0.48235, -0.424034, 0.750492, -0.167177, -0.479996, 0.359486, 0.660316, 0.152612, 0.784523, 0.748462, -0.844083, 0.33961, 0.123796, -0.800752, -0.643821, 0.431799, -0.516504, -0.732232, -0.660103, -0.264003, 0.933494, 0.275952]\nout shape: (7, 4)\nout: [-0.222866, 0.736716, -0.331714, -0.250841, 0.49324, -0.391338, -0.971192, -0.304377, 0.425853, 0.686683, -0.837114, -0.211837, 0.442765, -0.660864, -0.988027, -0.767739, 0.892751, -0.063246, -0.982079, 0.571859, 0.583632, -0.788749, 0.403633, -0.84945, 0.148118, 0.352691, -0.528363, 0.328677]\n"
]
],
[
[
"**[recurrent.SimpleRNN.5] output_dim=4, activation='tanh', return_sequences=False, go_backwards=False, stateful=True**\n\nNote dropout_W and dropout_U are only applied during training phase\n\n**To test statefulness, model.predict is run twice**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=False, go_backwards=False, stateful=True)\n\nlayer_0 = Input(batch_shape=(1, *data_in_shape))\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3800 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [-0.390491, -0.578366, 0.174977, -0.794259, 0.275087, 0.428053, -0.915046, -0.096302, -0.744594, 0.049007, -0.159581, 0.213776, 0.238386, 0.329687, 0.619891, 0.36543, -0.482228, 0.841286, -0.270654, 0.132222, 0.042815, 0.057958, 0.870286, -0.57147]\nU shape: (4, 4)\nU: [0.467358, -0.740084, -0.552357, -0.475388, -0.053543, -0.749294, -0.89908, -0.196198, -0.7233, 0.67584, 0.165427, -0.011422, -0.766433, -0.250531, 0.959118, -0.582048]\nb shape: (4,)\nb: [-0.217165, 0.276023, -0.853592, -0.465207]\n\nin shape: (7, 6)\nin: [0.112672, -0.828695, 0.336233, 0.388093, -0.295826, -0.480416, 0.225187, -0.364328, -0.206269, 0.961105, 0.039125, 0.391735, -0.24781, 0.492028, 0.557904, 0.264287, -0.540591, -0.777572, 0.350376, 0.03832, -0.48235, -0.424034, 0.750492, -0.167177, -0.479996, 0.359486, 0.660316, 0.152612, 0.784523, 0.748462, -0.844083, 0.33961, 0.123796, -0.800752, -0.643821, 0.431799, -0.516504, -0.732232, -0.660103, -0.264003, 0.933494, 0.275952]\nout shape: (4,)\nout: [0.883934, 0.439214, -0.654698, -0.106698]\n"
]
],
[
[
"**[recurrent.SimpleRNN.6] output_dim=4, activation='tanh', return_sequences=True, go_backwards=False, stateful=True**\n\nNote dropout_W and dropout_U are only applied during training phase\n\n**To test statefulness, model.predict is run twice**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=True, go_backwards=False, stateful=True)\n\nlayer_0 = Input(batch_shape=(1, *data_in_shape))\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3810 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [-0.161197, 0.910156, -0.96216, 0.204728, -0.041548, 0.558612, 0.037287, -0.169732, 0.935044, 0.8204, -0.479019, 0.291815, -0.161405, -0.395158, -0.834952, 0.685206, 0.707996, 0.048291, 0.280055, 0.300212, 0.824877, -0.858207, -0.453055, -0.057151]\nU shape: (4, 4)\nU: [-0.014444, 0.56472, -0.720144, -0.252002, 0.391629, -0.228955, -0.013911, 0.40588, -0.317771, 0.989316, 0.037567, -0.715889, 0.541733, -0.044509, 0.932983, 0.318657]\nb shape: (4,)\nb: [-0.775538, -0.82662, 0.808256, 0.069654]\n\nin shape: (7, 6)\nin: [-0.456878, 0.673716, 0.754458, 0.99948, -0.576319, -0.213528, -0.00711, 0.125772, -0.928776, 0.838621, -0.457717, -0.7566, 0.24007, -0.48667, -0.422729, -0.613233, -0.059555, 0.618784, 0.729787, 0.218327, 0.522534, -0.778783, 0.232627, 0.370789, -0.852386, -0.119443, 0.713664, -0.672227, 0.506668, 0.47509, 0.406951, -0.599353, -0.824306, -0.711984, -0.781436, -0.08695, -0.584066, 0.534988, 0.994046, 0.781186, -0.41391, -0.664649]\nout shape: (7, 4)\nout: [-0.617819, -0.609403, 0.701484, 0.601676, -0.99344, -0.647588, 0.948856, -0.144808, -0.845821, -0.755636, 0.921907, -0.812145, -0.682638, 0.774936, 0.298497, -0.821957, 0.582644, -0.892139, 0.904271, -0.198371, -0.993326, 0.279982, 0.568939, -0.95815, -0.866132, -0.021665, 0.278295, 0.244629]\n"
]
],
[
[
"**[recurrent.SimpleRNN.7] output_dim=4, activation='tanh', return_sequences=False, go_backwards=True, stateful=True**\n\nNote dropout_W and dropout_U are only applied during training phase\n\n**To test statefulness, model.predict is run twice**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=False, go_backwards=True, stateful=True)\n\nlayer_0 = Input(batch_shape=(1, *data_in_shape))\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3820 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [-0.967442, -0.753649, 0.635901, 0.084272, -0.464285, 0.37727, 0.587541, 0.603524, 0.615927, 0.196817, -0.319359, 0.719587, 0.173497, -0.151731, -0.566144, -0.290326, -0.745915, -0.862802, -0.961024, -0.672173, 0.646805, -0.664107, -0.990968, 0.056156]\nU shape: (4, 4)\nU: [0.3366, 0.31007, -0.976141, 0.926255, -0.711283, -0.530559, -0.756235, 0.439944, 0.552108, -0.871472, 0.14125, -0.962905, -0.338603, -0.300054, -0.417717, 0.450066]\nb shape: (4,)\nb: [-0.322072, 0.24445, -0.973474, -0.322077]\n\nin shape: (7, 6)\nin: [0.768144, -0.420612, -0.87321, -0.115841, 0.698756, -0.909438, 0.037401, -0.96988, -0.708589, -0.067676, 0.01788, 0.837723, -0.043675, 0.59153, -0.895703, 0.080775, 0.923127, -0.989487, 0.754247, 0.719781, -0.728688, 0.297508, -0.853478, -0.284425, 0.705101, 0.530558, -0.090936, 0.332791, -0.950894, -0.671238, -0.281409, -0.035306, 0.859823, -0.330751, -0.00469, 0.668596, -0.123336, -0.886897, -0.835349, 0.887526, 0.940587, -0.187323]\nout shape: (4,)\nout: [-0.898236, 0.423821, 0.101186, -0.917313]\n"
]
],
[
[
"**[recurrent.SimpleRNN.8] output_dim=4, activation='tanh', return_sequences=True, go_backwards=True, stateful=True**\n\nNote dropout_W and dropout_U are only applied during training phase\n\n**To test statefulness, model.predict is run twice**",
"_____no_output_____"
]
],
[
[
"data_in_shape = (7, 6)\nrnn = SimpleRNN(4, activation='tanh', return_sequences=True, go_backwards=True, stateful=True)\n\nlayer_0 = Input(batch_shape=(1, *data_in_shape))\nlayer_1 = rnn(layer_0)\nmodel = Model(input=layer_0, output=layer_1)\n\n# set weights to random (use seed for reproducibility)\nweights = []\nfor i, w in enumerate(model.get_weights()):\n np.random.seed(3830 + i)\n weights.append(2 * np.random.random(w.shape) - 1)\nmodel.set_weights(weights)\nweight_names = ['W', 'U', 'b']\nfor w_i, w_name in enumerate(weight_names):\n print('{} shape:'.format(w_name), weights[w_i].shape)\n print('{}:'.format(w_name), format_decimal(weights[w_i].ravel().tolist()))\n\ndata_in = 2 * np.random.random(data_in_shape) - 1\nprint('')\nprint('in shape:', data_in_shape)\nprint('in:', format_decimal(data_in.ravel().tolist()))\nresult = model.predict(np.array([data_in]))\nresult = model.predict(np.array([data_in]))\nprint('out shape:', result[0].shape)\nprint('out:', format_decimal(result[0].ravel().tolist()))",
"W shape: (6, 4)\nW: [0.541657, -0.064909, -0.116436, 0.256712, 0.300281, 0.896851, 0.047865, 0.850134, -0.129581, 0.39304, 0.80892, -0.384143, 0.563076, -0.119239, -0.744437, 0.402776, -0.780366, 0.468597, -0.804769, -0.799921, 0.435465, 0.450867, 0.98813, 0.159434]\nU shape: (4, 4)\nU: [0.58305, -0.918477, 0.10927, -0.3796, -0.375415, 0.123171, -0.463354, 0.442039, 0.928033, -0.848093, -0.899042, 0.294531, 0.46675, 0.785351, 0.802663, -0.89208]\nb shape: (4,)\nb: [0.110875, 0.51125, -0.445144, 0.43158]\n\nin shape: (7, 6)\nin: [0.624168, -0.518746, -0.020951, 0.465103, -0.901332, -0.915009, 0.406032, 0.172798, 0.867637, -0.348888, 0.084563, 0.343669, 0.007207, -0.192566, 0.993381, -0.764461, -0.265019, 0.13182, -0.170015, 0.069884, -0.977789, 0.466722, -0.502645, 0.495466, 0.261529, 0.428916, -0.634665, 0.791701, 0.406729, 0.372998, 0.224605, 0.412702, -0.760375, -0.216756, 0.519871, 0.375917, -0.733975, -0.868739, -0.225755, -0.08227, -0.222005, 0.881433]\nout shape: (7, 4)\nout: [0.484936, -0.81626, -0.172498, 0.157695, 0.532515, 0.62373, -0.234853, -0.04739, 0.530358, 0.567308, -0.917707, 0.835725, 0.586255, 0.817835, 0.554356, 0.482036, 0.490133, 0.221862, 0.682954, -0.304563, 0.69191, -0.024181, -0.157429, 0.523323, 0.879457, -0.756114, -0.410511, 0.133938]\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6f8e84e01cdf62f0827411920dcdc4b840c17c | 44,253 | ipynb | Jupyter Notebook | fashion_mnist.ipynb | philbasford/aws-deeplearning-lambda | e592746ea20d708d3cedcbc13d7819b198207e2a | [
"Apache-2.0"
]
| null | null | null | fashion_mnist.ipynb | philbasford/aws-deeplearning-lambda | e592746ea20d708d3cedcbc13d7819b198207e2a | [
"Apache-2.0"
]
| null | null | null | fashion_mnist.ipynb | philbasford/aws-deeplearning-lambda | e592746ea20d708d3cedcbc13d7819b198207e2a | [
"Apache-2.0"
]
| null | null | null | 70.02057 | 10,360 | 0.820351 | [
[
[
"tensorflow-2-0-with-fashion-mnist-dataset\n=======\n\nThe following notebook is based on https://medium.com/datadriveninvestor/introduction-to-tensorflow-2-0-with-fashion-mnist-dataset-2f243423c563 which in tuen is based on https://www.tensorflow.org/tutorials/quickstart/beginner\n\nload tensor flow\n-----------------",
"_____no_output_____"
],
[
"!pip install matplotlib\n\nimport tensorflow as tf\nfrom tensorflow import keras\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nimport datetime",
"_____no_output_____"
],
[
"prepare data\n-------------\nload data",
"_____no_output_____"
]
],
[
[
"mnist = keras.datasets.fashion_mnist\n(X_train, y_train), (X_test, y_test) = mnist.load_data()",
"_____no_output_____"
]
],
[
[
"check shape",
"_____no_output_____"
]
],
[
[
"X_train.shape, y_train.shape",
"_____no_output_____"
]
],
[
[
"definig class name",
"_____no_output_____"
]
],
[
[
"class_name = ['top', 'trouser', 'pullover', 'dress', 'coat', 'sandal',\n 'shirt', 'sneaker', 'bag', 'ankle boot']",
"_____no_output_____"
]
],
[
[
"plot image using colorbar",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.imshow(X_train[26])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"Preprocessing/Wrangle data\n--------------------\nchanging scale 0-255 to 0-1",
"_____no_output_____"
]
],
[
[
"X_train = X_train/255.0\nX_test = X_test/255.0",
"_____no_output_____"
]
],
[
[
"checking plot image using colorbar again",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.imshow(X_train[26])\nplt.colorbar()",
"_____no_output_____"
]
],
[
[
"building model\n-------------------\n\nusing keras to build out the model",
"_____no_output_____"
]
],
[
[
"from tensorflow.keras import Sequential\nfrom tensorflow.keras.layers import Flatten, Dense, Dropout\n\nmodel = Sequential()",
"_____no_output_____"
]
],
[
[
"we can pass an array in sequential model or just add another layer",
"_____no_output_____"
]
],
[
[
"model.add(Flatten(input_shape = (28,28)))",
"_____no_output_____"
]
],
[
[
"flatten is used to change input data in 1d",
"_____no_output_____"
]
],
[
[
"model.add(Dense(128, activation = 'relu'))\nmodel.add(Dropout(0.2))\nmodel.add(Dense(10, activation = 'softmax'))\nmodel.summary()",
"Model: \"sequential_12\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nflatten_6 (Flatten) (None, 784) 0 \n_________________________________________________________________\ndense_13 (Dense) (None, 128) 100480 \n_________________________________________________________________\ndropout_5 (Dropout) (None, 128) 0 \n_________________________________________________________________\ndense_14 (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 101,770\nTrainable params: 101,770\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"compilation of our model",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer = 'adam', loss = 'sparse_categorical_crossentropy',\n metrics = ['accuracy'])\n ",
"_____no_output_____"
]
],
[
[
"train model\n-----------------\nfit the model",
"_____no_output_____"
]
],
[
[
"model.fit(X_train, y_train, epochs=2)",
"Epoch 1/2\n1875/1875 [==============================] - 5s 3ms/step - loss: 0.5362 - accuracy: 0.8099\nEpoch 2/2\n1875/1875 [==============================] - 5s 3ms/step - loss: 0.4017 - accuracy: 0.8547\n"
]
],
[
[
"chechking test_loss, test_acc",
"_____no_output_____"
]
],
[
[
"test_loss, test_acc = model.evaluate(X_test, y_test)\ntest_loss, test_acc",
"313/313 [==============================] - 1s 2ms/step - loss: 0.3994 - accuracy: 0.8578\n"
]
],
[
[
"validated model\n-----------------\ncheck accuracy (should be around 85%) ",
"_____no_output_____"
]
],
[
[
"from sklearn.metrics import accuracy_score\ny_pred = model.predict_classes(X_test)\naccuracy_score(y_test,y_pred)",
"_____no_output_____"
]
],
[
[
"checking pred",
"_____no_output_____"
]
],
[
[
"x=16\npred = model.predict(X_test)\npred[x]",
"_____no_output_____"
]
],
[
[
"now lets make it a little more human readable and use probability",
"_____no_output_____"
]
],
[
[
"probability_model = tf.keras.Sequential([model, \n tf.keras.layers.Softmax()])\n\npred = probability_model.predict(X_test)",
"_____no_output_____"
]
],
[
[
"so now we should see for 10 classes the propability that the image is of each one (and hopefull our class is the highest)",
"_____no_output_____"
]
],
[
[
"pred[x]",
"_____no_output_____"
]
],
[
[
"look like it is, so plotting with image",
"_____no_output_____"
]
],
[
[
"plt.figure()\nplt.imshow(X_test[x])\nplt.colorbar()\nnp.argmax(pred[x])",
"_____no_output_____"
]
],
[
[
"Now to use out model later we need to save model and then zip it.",
"_____no_output_____"
]
],
[
[
"import os\nmobilenet_save_path = os.path.join('./model/', \"imageclass/2/\")\ntf.saved_model.save(probability_model, mobilenet_save_path)\n!tar -zcvf model.tar.gz model/",
"INFO:tensorflow:Assets written to: ./model/imageclass/2/assets\n"
]
],
[
[
"check decoding of prediction",
"_____no_output_____"
]
],
[
[
"encoded = np.argmax(pred[x])",
"_____no_output_____"
],
[
"class_name[encoded]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6f8f3ff38e2395a384ed4166e2429037dda510 | 2,379 | ipynb | Jupyter Notebook | visualizations/bokeh/notebooks/glyphs/.ipynb_checkpoints/line-checkpoint.ipynb | martinpeck/apryor6.github.io | 52a227cc65cf04fb7ca3162d405349b41c311a42 | [
"MIT"
]
| 1 | 2016-11-06T23:46:58.000Z | 2016-11-06T23:46:58.000Z | visualizations/bokeh/notebooks/glyphs/.ipynb_checkpoints/line-checkpoint.ipynb | martinpeck/apryor6.github.io | 52a227cc65cf04fb7ca3162d405349b41c311a42 | [
"MIT"
]
| 1 | 2019-09-06T22:32:16.000Z | 2019-09-06T22:32:16.000Z | visualizations/bokeh/notebooks/glyphs/.ipynb_checkpoints/line-checkpoint.ipynb | martinpeck/apryor6.github.io | 52a227cc65cf04fb7ca3162d405349b41c311a42 | [
"MIT"
]
| 2 | 2019-09-06T13:54:42.000Z | 2020-03-11T09:33:59.000Z | 33.507042 | 336 | 0.580076 | [
[
[
"# Bokeh Line Glyph",
"_____no_output_____"
]
],
[
[
"from bokeh.plotting import figure, output_file, show\nfrom bokeh.models import Range1d\nfrom bokeh.io import export_png\n\nfill_color = '#a6bddb'\nline_color = '#1c9099'\noutput_file(\"../../figures/line.html\")\n\np = figure(plot_width=400, plot_height=400)\np.line(x=[0, 0.5],y=[0, .5],size=100, fill_alpha=1,fill_color=fill_color,\n line_alpha=1, line_color=line_color, line_dash='dashed', line_width=5)\np.line(x=[0.25, 0.75],y=[0.25, 0.75],size=100, fill_alpha=0.8, fill_color=fill_color,\n line_alpha=1, line_color=line_color, line_dash='dotdash', line_width=8)\n# p.line(x=1,y=0,size=100, fill_alpha=0.6, fill_color = fill_color,\n# line_alpha=1, line_color=line_color, line_dash='dotted', line_width=13)\n# p.line(x=1,y=1,size=100, fill_alpha=0.4, fill_color = fill_color,\n# line_alpha=1, line_color=line_color, line_dash='solid', line_width=17)\np.x_range = Range1d(-0.5,1.5, bounds=(-1,2))\np.y_range = Range1d(-0.5,1.5, bounds=(-1,2))\nshow(p)\nexport_png(p, filename=\"../../figures/line.png\");",
"_____no_output_____"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
ec6f9733480c85ffb0dc971a3bef97d0077f48d9 | 955,198 | ipynb | Jupyter Notebook | Time_Series_Forecasting/Energy_Consumption_Exercise.ipynb | Harshit4199/ML_Sagemaker_Studies | c1ff23f5211f06f5f74f27cf8a227198c1b7f5e9 | [
"MIT"
]
| null | null | null | Time_Series_Forecasting/Energy_Consumption_Exercise.ipynb | Harshit4199/ML_Sagemaker_Studies | c1ff23f5211f06f5f74f27cf8a227198c1b7f5e9 | [
"MIT"
]
| null | null | null | Time_Series_Forecasting/Energy_Consumption_Exercise.ipynb | Harshit4199/ML_Sagemaker_Studies | c1ff23f5211f06f5f74f27cf8a227198c1b7f5e9 | [
"MIT"
]
| null | null | null | 399.163393 | 207,392 | 0.906049 | [
[
[
"# Time Series Forecasting \n\nA time series is data collected periodically, over time. Time series forecasting is the task of predicting future data points, given some historical data. It is commonly used in a variety of tasks from weather forecasting, retail and sales forecasting, stock market prediction, and in behavior prediction (such as predicting the flow of car traffic over a day). There is a lot of time series data out there, and recognizing patterns in that data is an active area of machine learning research!\n\n<img src='notebook_ims/time_series_examples.png' width=80% />\n\nIn this notebook, we'll focus on one method for finding time-based patterns: using SageMaker's supervised learning model, [DeepAR](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html).\n\n\n### DeepAR\n\nDeepAR utilizes a recurrent neural network (RNN), which is designed to accept some sequence of data points as historical input and produce a predicted sequence of points. So, how does this model learn?\n\nDuring training, you'll provide a training dataset (made of several time series) to a DeepAR estimator. The estimator looks at *all* the training time series and tries to identify similarities across them. It trains by randomly sampling **training examples** from the training time series. \n* Each training example consists of a pair of adjacent **context** and **prediction** windows of fixed, predefined lengths. \n * The `context_length` parameter controls how far in the *past* the model can see.\n * The `prediction_length` parameter controls how far in the *future* predictions can be made.\n * You can find more details, in [this documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_how-it-works.html).\n\n<img src='notebook_ims/context_prediction_windows.png' width=50% />\n\n> Since DeepAR trains on several time series, it is well suited for data that exhibit **recurring patterns**.\n\nIn any forecasting task, you should choose the context window to provide enough, **relevant** information to a model so that it can produce accurate predictions. In general, data closest to the prediction time frame will contain the information that is most influential in defining that prediction. In many forecasting applications, like forecasting sales month-to-month, the context and prediction windows will be the same size, but sometimes it will be useful to have a larger context window to notice longer-term patterns in data.\n\n### Energy Consumption Data\n\nThe data we'll be working with in this notebook is data about household electric power consumption, over the globe. The dataset is originally taken from [Kaggle](https://www.kaggle.com/uciml/electric-power-consumption-data-set), and represents power consumption collected over several years from 2006 to 2010. With such a large dataset, we can aim to predict over long periods of time, over days, weeks or months of time. Predicting energy consumption can be a useful task for a variety of reasons including determining seasonal prices for power consumption and efficiently delivering power to people, according to their predicted usage. \n\n**Interesting read**: An inversely-related project, recently done by Google and DeepMind, uses machine learning to predict the *generation* of power by wind turbines and efficiently deliver power to the grid. You can read about that research, [in this post](https://deepmind.com/blog/machine-learning-can-boost-value-wind-energy/).\n\n### Machine Learning Workflow\n\nThis notebook approaches time series forecasting in a number of steps:\n* Loading and exploring the data\n* Creating training and test sets of time series\n* Formatting data as JSON files and uploading to S3\n* Instantiating and training a DeepAR estimator\n* Deploying a model and creating a predictor\n* Evaluating the predictor \n\n---\n\nLet's start by loading in the usual resources.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"# Load and Explore the Data\n\nWe'll be loading in some data about global energy consumption, collected over a few years. The below cell downloads and unzips this data, giving you one text file of data, `household_power_consumption.txt`.",
"_____no_output_____"
]
],
[
[
"! wget https://s3.amazonaws.com/video.udacity-data.com/topher/2019/March/5c88a3f1_household-electric-power-consumption/household-electric-power-consumption.zip\n! unzip household-electric-power-consumption",
"--2020-05-13 09:31:37-- https://s3.amazonaws.com/video.udacity-data.com/topher/2019/March/5c88a3f1_household-electric-power-consumption/household-electric-power-consumption.zip\nResolving s3.amazonaws.com (s3.amazonaws.com)... 52.216.249.62\nConnecting to s3.amazonaws.com (s3.amazonaws.com)|52.216.249.62|:443... connected.\nHTTP request sent, awaiting response... 200 OK\nLength: 20805339 (20M) [application/zip]\nSaving to: ‘household-electric-power-consumption.zip’\n\nhousehold-electric- 100%[===================>] 19.84M 21.3MB/s in 0.9s \n\n2020-05-13 09:31:38 (21.3 MB/s) - ‘household-electric-power-consumption.zip’ saved [20805339/20805339]\n\nArchive: household-electric-power-consumption.zip\n inflating: household_power_consumption.txt \n"
]
],
[
[
"### Read in the `.txt` File\n\nThe next cell displays the first few lines in the text file, so we can see how it is formatted.",
"_____no_output_____"
]
],
[
[
"# display first ten lines of text data\nn_lines = 10\n\nwith open('household_power_consumption.txt') as file:\n head = [next(file) for line in range(n_lines)]\n \ndisplay(head)",
"_____no_output_____"
]
],
[
[
"## Pre-Process the Data\n\nThe 'household_power_consumption.txt' file has the following attributes:\n * Each data point has a date and time (hour:minute:second) of recording\n * The various data features are separated by semicolons (;)\n * Some values are 'nan' or '?', and we'll treat these both as `NaN` values\n\n### Managing `NaN` values\n\nThis DataFrame does include some data points that have missing values. So far, we've mainly been dropping these values, but there are other ways to handle `NaN` values, as well. One technique is to just fill the missing column values with the **mean** value from that column; this way the added value is likely to be realistic.\n\nI've provided some helper functions in `txt_preprocessing.py` that will help to load in the original text file as a DataFrame *and* fill in any `NaN` values, per column, with the mean feature value. This technique will be fine for long-term forecasting; if I wanted to do an hourly analysis and prediction, I'd consider dropping the `NaN` values or taking an average over a small, sliding window rather than an entire column of data.\n\n**Below, I'm reading the file in as a DataFrame and filling `NaN` values with feature-level averages.**",
"_____no_output_____"
]
],
[
[
"import txt_preprocessing as pprocess\n\n# create df from text file\ninitial_df = pprocess.create_df('household_power_consumption.txt', sep=';')\n\n# fill NaN column values with *average* column value\ndf = pprocess.fill_nan_with_mean(initial_df)\n\n# print some stats about the data\nprint('Data shape: ', df.shape)\ndf.head()",
"Data shape: (2075259, 7)\n"
]
],
[
[
"## Global Active Power \n\nIn this example, we'll want to predict the global active power, which is the household minute-averaged active power (kilowatt), measured across the globe. So, below, I am getting just that column of data and displaying the resultant plot.",
"_____no_output_____"
]
],
[
[
"# Select Global active power data\npower_df = df['Global_active_power'].copy()\nprint(power_df.shape)",
"(2075259,)\n"
],
[
"# display the data \nplt.figure(figsize=(12,6))\n# all data points\npower_df.plot(title='Global active power', color='blue') \nplt.show()",
"_____no_output_____"
]
],
[
[
"Since the data is recorded each minute, the above plot contains *a lot* of values. So, I'm also showing just a slice of data, below.",
"_____no_output_____"
]
],
[
[
"# can plot a slice of hourly data\nend_mins = 1440 # 1440 mins = 1 day\n\nplt.figure(figsize=(12,6))\npower_df[0:end_mins].plot(title='Global active power, over one day', color='blue') \nplt.show()",
"_____no_output_____"
]
],
[
[
"### Hourly vs Daily\n\nThere is a lot of data, collected every minute, and so I could go one of two ways with my analysis:\n1. Create many, short time series, say a week or so long, in which I record energy consumption every hour, and try to predict the energy consumption over the following hours or days.\n2. Create fewer, long time series with data recorded daily that I could use to predict usage in the following weeks or months.\n\nBoth tasks are interesting! It depends on whether you want to predict time patterns over a day/week or over a longer time period, like a month. With the amount of data I have, I think it would be interesting to see longer, *recurring* trends that happen over several months or over a year. So, I will resample the 'Global active power' values, recording **daily** data points as averages over 24-hr periods.\n\n> I can resample according to a specified frequency, by utilizing pandas [time series tools](https://pandas.pydata.org/pandas-docs/stable/user_guide/timeseries.html), which allow me to sample at points like every hour ('H') or day ('D'), etc.\n\n",
"_____no_output_____"
]
],
[
[
"# resample over day (D)\nfreq = 'D'\n# calculate the mean active power for a day\nmean_power_df = power_df.resample(freq).mean()\n\n# display the mean values\nplt.figure(figsize=(15,8))\nmean_power_df.plot(title='Global active power, mean per day', color='blue') \nplt.tight_layout()\nplt.show()",
"_____no_output_____"
]
],
[
[
"In this plot, we can see that there are some interesting trends that occur over each year. It seems that there are spikes of energy consumption around the end/beginning of each year, which correspond with heat and light usage being higher in winter months. We also see a dip in usage around August, when global temperatures are typically higher.\n\nThe data is still not very smooth, but it shows noticeable trends, and so, makes for a good use case for machine learning models that may be able to recognize these patterns.",
"_____no_output_____"
],
[
"---\n## Create Time Series \n\nMy goal will be to take full years of data, from 2007-2009, and see if I can use it to accurately predict the average Global active power usage for the next several months in 2010!\n\nNext, let's make one time series for each complete year of data. This is just a design decision, and I am deciding to use full years of data, starting in January of 2017 because there are not that many data points in 2006 and this split will make it easier to handle leap years; I could have also decided to construct time series starting at the first collected data point, just by changing `t_start` and `t_end` in the function below.\n\nThe function `make_time_series` will create pandas `Series` for each of the passed in list of years `['2007', '2008', '2009']`.\n* All of the time series will start at the same time point `t_start` (or t0). \n * When preparing data, it's important to use a consistent start point for each time series; DeepAR uses this time-point as a frame of reference, which enables it to learn recurrent patterns e.g. that weekdays behave differently from weekends or that Summer is different than Winter.\n * You can change the start and end indices to define any time series you create.\n* We should account for leap years, like 2008, in the creation of time series.\n* Generally, we create `Series` by getting the relevant global consumption data (from the DataFrame) and date indices.\n\n```\n# get global consumption data\ndata = mean_power_df[start_idx:end_idx]\n\n# create time series for the year\nindex = pd.DatetimeIndex(start=t_start, end=t_end, freq='D')\ntime_series.append(pd.Series(data=data, index=index))\n```",
"_____no_output_____"
]
],
[
[
"def make_time_series(mean_power_df, years, freq='D', start_idx=16):\n '''Creates as many time series as there are complete years. This code\n accounts for the leap year, 2008.\n :param mean_power_df: A dataframe of global power consumption, averaged by day.\n This dataframe should also be indexed by a datetime.\n :param years: A list of years to make time series out of, ex. ['2007', '2008'].\n :param freq: The frequency of data recording (D = daily)\n :param start_idx: The starting dataframe index of the first point in the first time series.\n The default, 16, points to '2017-01-01'. \n :return: A list of pd.Series(), time series data.\n '''\n \n # store time series\n time_series = []\n \n # store leap year in this dataset\n leap = '2008'\n\n # create time series for each year in years\n for i in range(len(years)):\n\n year = years[i]\n if(year == leap):\n end_idx = start_idx+366\n else:\n end_idx = start_idx+365\n\n # create start and end datetimes\n t_start = year + '-01-01' # Jan 1st of each year = t_start\n t_end = year + '-12-31' # Dec 31st = t_end\n\n # get global consumption data\n data = mean_power_df[start_idx:end_idx]\n\n # create time series for the year\n index = pd.DatetimeIndex(start=t_start, end=t_end, freq=freq)\n time_series.append(pd.Series(data=data, index=index))\n \n start_idx = end_idx\n \n # return list of time series\n return time_series\n ",
"_____no_output_____"
]
],
[
[
"## Test the results\n\nBelow, let's construct one time series for each complete year of data, and display the results.",
"_____no_output_____"
]
],
[
[
"# test out the code above\n\n# yearly time series for our three complete years\nfull_years = ['2007', '2008', '2009']\nfreq='D' # daily recordings\n\n# make time series\ntime_series = make_time_series(mean_power_df, full_years, freq=freq)",
"/home/ec2-user/anaconda3/envs/amazonei_mxnet_p36/lib/python3.6/site-packages/ipykernel/__main__.py:36: FutureWarning: Creating a DatetimeIndex by passing range endpoints is deprecated. Use `pandas.date_range` instead.\n"
],
[
"# display first time series\ntime_series_idx = 0\n\nplt.figure(figsize=(12,6))\ntime_series[time_series_idx].plot()\nplt.show()",
"_____no_output_____"
]
],
[
[
"---\n# Splitting in Time\n\nWe'll evaluate our model on a test set of data. For machine learning tasks like classification, we typically create train/test data by randomly splitting examples into different sets. For forecasting it's important to do this train/test split in **time** rather than by individual data points. \n> In general, we can create training data by taking each of our *complete* time series and leaving off the last `prediction_length` data points to create *training* time series. \n\n### EXERCISE: Create training time series\n\nComplete the `create_training_series` function, which should take in our list of complete time series data and return a list of truncated, training time series.\n\n* In this example, we want to predict about a month's worth of data, and we'll set `prediction_length` to 30 (days).\n* To create a training set of data, we'll leave out the last 30 points of *each* of the time series we just generated, so we'll use only the first part as training data. \n* The **test set contains the complete range** of each time series.\n",
"_____no_output_____"
]
],
[
[
"# create truncated, training time series\ndef create_training_series(complete_time_series, prediction_length):\n '''Given a complete list of time series data, create training time series.\n :param complete_time_series: A list of all complete time series.\n :param prediction_length: The number of points we want to predict.\n :return: A list of training time series.\n '''\n # get training series\n time_series_training = []\n\n for ts in complete_time_series:\n # truncate trailing `prediction_length` pts\n time_series_training.append(ts[:-prediction_length])\n\n return time_series_training\n ",
"_____no_output_____"
],
[
"# test your code!\n\n# set prediction length\nprediction_length = 30 # 30 days ~ a month\n\ntime_series_training = create_training_series(time_series, prediction_length)\n",
"_____no_output_____"
]
],
[
[
"### Training and Test Series\n\nWe can visualize what these series look like, by plotting the train/test series on the same axis. We should see that the test series contains all of our data in a year, and a training series contains all but the last `prediction_length` points.",
"_____no_output_____"
]
],
[
[
"# display train/test time series\ntime_series_idx = 0\n\nplt.figure(figsize=(15,8))\n# test data is the whole time series\ntime_series[time_series_idx].plot(label='test', lw=3)\n# train data is all but the last prediction pts\ntime_series_training[time_series_idx].plot(label='train', ls=':', lw=3)\n\nplt.legend()\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Convert to JSON \n\nAccording to the [DeepAR documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar.html), DeepAR expects to see input training data in a JSON format, with the following fields:\n\n* **start**: A string that defines the starting date of the time series, with the format 'YYYY-MM-DD HH:MM:SS'.\n* **target**: An array of numerical values that represent the time series.\n* **cat** (optional): A numerical array of categorical features that can be used to encode the groups that the record belongs to. This is useful for finding models per class of item, such as in retail sales, where you might have {'shoes', 'jackets', 'pants'} encoded as categories {0, 1, 2}.\n\nThe input data should be formatted with one time series per line in a JSON file. Each line looks a bit like a dictionary, for example:\n```\n{\"start\":'2007-01-01 00:00:00', \"target\": [2.54, 6.3, ...], \"cat\": [1]}\n{\"start\": \"2012-01-30 00:00:00\", \"target\": [1.0, -5.0, ...], \"cat\": [0]} \n...\n```\nIn the above example, each time series has one, associated categorical feature and one time series feature.\n\n### EXERCISE: Formatting Energy Consumption Data\n\nFor our data:\n* The starting date, \"start,\" will be the index of the first row in a time series, Jan. 1st of that year.\n* The \"target\" will be all of the energy consumption values that our time series holds.\n* We will not use the optional \"cat\" field.\n\nComplete the following utility function, which should convert `pandas.Series` objects into the appropriate JSON strings that DeepAR can consume.",
"_____no_output_____"
]
],
[
[
"def series_to_json_obj(ts):\n '''Returns a dictionary of values in DeepAR, JSON format.\n :param ts: A single time series.\n :return: A dictionary of values with \"start\" and \"target\" keys.\n '''\n json_obj = {\"start\": str(ts.index[0]), \"target\": list(ts)}\n return json_obj\n",
"_____no_output_____"
],
[
"# test out the code\nts = time_series[0]\n\njson_obj = series_to_json_obj(ts)\n\nprint(json_obj)",
"{'start': '2007-01-01 00:00:00', 'target': [1.9090305555555664, 0.8814138888888893, 0.7042041666666671, 2.263480555555549, 1.8842805555555506, 1.047484722222222, 1.6997361111111127, 1.5564999999999982, 1.2979541666666659, 1.496388888888886, 1.5661069444444446, 1.0147888888888905, 2.2130652777777766, 2.0895191771086794, 1.4921374999999986, 1.1711138888888888, 1.9775611111111107, 1.2649041666666674, 1.0280833333333352, 2.176202777777775, 2.3661541666666652, 1.5142319444444445, 1.2344722222222224, 2.07489861111111, 1.1085722222222205, 1.1235916666666654, 1.419494444444445, 2.146733065997562, 1.3768500000000001, 1.1859708333333323, 1.6414944444444453, 1.2671944444444443, 1.1581499999999982, 2.798418055555553, 2.4971805555555573, 1.1425555555555553, 0.8843611111111115, 1.6166791666666673, 1.2509819444444445, 1.1251375000000008, 1.9648111111111108, 2.4800194444444434, 1.3038958333333335, 0.9823236111111113, 1.7351888888888864, 1.3787124999999985, 1.1823111111111093, 1.4423000000000017, 2.659556944444449, 1.5184819444444428, 2.187450000000003, 1.4394958333333352, 2.3414314097729143, 0.9121388888888906, 0.4964736111111122, 0.3484305555555563, 0.3947805555555556, 0.3597999999999982, 0.3616486111111118, 0.3594194444444455, 0.358116666666667, 0.5688347222222228, 1.451875000000001, 1.8474527777777783, 0.8656249999999984, 1.6494486111111129, 1.091758333333331, 0.8837583333333331, 1.7254944444444502, 2.417108333333334, 1.354630555555556, 0.8959333333333357, 1.2453916666666665, 1.308898611111112, 1.1819819444444437, 1.4083958333333346, 1.6465597222222195, 1.4948041666666658, 1.448769444444444, 1.4875638888888894, 1.0104013888888888, 0.9090333333333334, 2.0614097222222245, 2.3175108437753487, 0.9833902777777774, 0.7674583333333332, 1.620762500000001, 1.1044486111111105, 0.9738847222222207, 2.437159722222223, 1.9346888888888931, 1.3616513888888915, 1.1408472222222201, 1.4249083333333314, 1.2806097222222221, 1.1119027777777781, 1.079113888888888, 1.1172333333333315, 0.6950833333333352, 0.5398361111111113, 0.616306944444446, 0.3734069444444451, 0.3826541666666676, 0.3849083333333345, 0.5042805555555557, 0.6613819444444443, 0.849815277777779, 0.6932263888888891, 0.7143458333333358, 0.571236111111111, 1.1622611111111096, 1.5511902777777784, 0.7323374999999995, 0.7167361111111107, 0.8778902777777773, 0.8857402777777782, 0.7599527777777767, 1.0914859283295342, 1.091615036500725, 0.9472065219004123, 1.1554569444444436, 0.6968652777777783, 0.7571097222222218, 0.6931444444444462, 1.0854527777777792, 1.4597944444444448, 1.1414333333333337, 1.242563888888889, 1.1567611111111105, 0.49158611111111133, 0.6799680555555554, 1.0010874999999977, 1.1352347222222203, 1.0192930555555566, 0.62805, 1.1756555555555552, 0.8554277777777767, 1.1330069444444428, 0.9926680555555539, 1.3668944444444437, 0.7778208333333345, 0.7867027777777772, 1.0173166666666695, 0.8016583333333344, 1.1209861111111095, 1.0501722222222214, 1.4956999999999978, 1.137130555555556, 0.7370416666666654, 1.1179624999999997, 0.6517708333333337, 0.7263955659975689, 1.030806944444443, 1.463906944444444, 0.6330680555555547, 1.0131194444444456, 1.1768955659975675, 0.6840291666666676, 1.4916986111111106, 0.7647884523521012, 0.9311194444444444, 0.5518430555555551, 0.6380319444444437, 0.6439388888888893, 0.8743638888888887, 0.6872250000000003, 1.4012236111111112, 1.352145833333335, 0.4711625000000016, 0.5864883542173611, 0.7460319444444469, 0.560379166666666, 0.39325972222222155, 0.33164305555555645, 0.49628888888888706, 0.7445847222222217, 0.6561861111111094, 1.0283513888888873, 0.8996097222222214, 0.906451121553125, 0.9273013888888878, 0.7129611111111108, 0.6715499999999989, 0.7360499999999994, 0.6968125000000016, 0.734229166666665, 0.7427249999999991, 0.9431986111111104, 0.7563055555555565, 0.8195541666666663, 0.6307805555555537, 0.5016333333333299, 0.7427652777777777, 0.5516541666666681, 0.8945624999999994, 0.7353708019063109, 0.5928291666666665, 0.4997444444444466, 0.52575138888889, 0.47075416666666736, 0.7711263888888894, 0.7362833333333343, 0.9561566771086814, 0.6882069444444442, 0.5731138888888893, 0.3281666666666694, 0.6420138888888899, 0.8350791666666663, 1.1262930555555566, 0.2229972222222228, 0.3241777777777761, 0.5641194444444436, 0.7522652192823055, 0.7079277777777766, 0.7384472222222229, 0.7951319444444429, 0.5465958333333305, 0.7567499999999994, 1.0302638888888895, 0.7815847222222178, 0.7442222222222233, 0.7138958333333341, 0.7580902777777812, 0.8624333333333364, 0.8007027777777773, 0.6536041666666672, 0.8276805555555493, 0.6656222222222172, 0.7013138888888948, 0.2416388888888889, 0.9877041666666672, 0.9297513888888894, 0.44621944444444595, 0.8401444444444471, 0.7724388888888885, 1.1509066771086796, 0.7992222222222208, 0.9131291666666661, 0.8414791666666658, 0.692363888888888, 0.5194388888888899, 0.83504027777778, 0.8887652777777755, 1.1815874999999996, 1.154491666666666, 0.7214736111111109, 1.1006555555555562, 0.8587305555555576, 0.6260236111111122, 0.9281222222222196, 1.0206611111111112, 1.1663208333333344, 0.7759555555555581, 0.6830361111111114, 0.8942805555555555, 0.796098611111113, 0.7885208333333332, 0.7606333333333342, 1.1473777777777783, 1.0147124999999988, 0.8107777777777783, 0.9018638888888884, 0.871897222222219, 0.860263888888889, 0.8167861111111107, 0.9847013888888875, 0.9406458333333346, 1.2252375000000006, 1.266977243106251, 0.9670833333333325, 1.1670125000000016, 1.5948069444444422, 1.0529791666666668, 1.1950847222222218, 1.5269888888888836, 0.8892041666666652, 1.0544847222222216, 0.9681194444444439, 1.2196111111111123, 1.4625791666666648, 0.7605847222222217, 1.1721236111111104, 0.8103972222222212, 1.2030875, 1.3295166666666667, 1.3380055555555561, 1.4314250000000006, 0.8793958333333348, 1.3641958333333348, 1.0950902777777778, 1.054534722222222, 1.448366666666666, 1.15294861111111, 1.3485694444444438, 1.147844444444447, 1.3589105764395841, 1.2955805555555562, 1.2874430555555554, 0.9983111111111123, 1.240061111111108, 0.3556652777777766, 0.37876944444444327, 0.5631999999999971, 0.8911180555555571, 0.34459444444444476, 1.002501388888888, 0.34204861111111057, 1.0317263888888877, 1.0420777777777788, 1.446095833333335, 1.2810819444444468, 1.042402777777777, 1.507795833333338, 1.8136111111111097, 1.3105027777777782, 1.4973236111111095, 1.5377763888888916, 1.4222180555555546, 1.0361027777777765, 1.6039958333333328, 1.812565277777774, 1.516568055555557, 1.1969819444444456, 1.4554555555555526, 1.2173108437753466, 1.3419291666666693, 1.751818055555553, 1.318787500000002, 0.9691652777777785, 1.0900805555555562, 1.9451472222222208, 1.1423194444444453, 1.1946677882197925, 1.6192555555555597, 2.177043055555557, 2.0459638888888865, 1.2236527777777777, 1.4358583333333348, 1.4952222222222211, 1.727731944444445, 1.2284833333333336, 1.537086111111109, 1.579572222222225, 1.2213569444444459, 1.6007666666666638, 1.1028208333333336, 1.2831569444444444, 1.3713944444444444, 1.9006083333333303, 1.9182069444444376, 1.2487205659975713, 1.8216625000000024, 1.525036111111112, 1.3539986111111106, 1.376788888888891, 1.8133777777777769, 1.6729499999999993, 1.5679499999999997, 1.8715055555555555, 1.7918611111111102, 1.7584708333333323, 2.161841666666663, 2.2909416666666647, 1.7770249999999976, 1.5392652777777778]}\n"
]
],
[
[
"### Saving Data, Locally\n\nThe next helper function will write one series to a single JSON line, using the new line character '\\n'. The data is also encoded and written to a filename that we specify.",
"_____no_output_____"
]
],
[
[
"# import json for formatting data\nimport json\nimport os # and os for saving\n\ndef write_json_dataset(time_series, filename): \n with open(filename, 'wb') as f:\n # for each of our times series, there is one JSON line\n for ts in time_series:\n json_line = json.dumps(series_to_json_obj(ts)) + '\\n'\n json_line = json_line.encode('utf-8')\n f.write(json_line)\n print(filename + ' saved.')",
"_____no_output_____"
],
[
"# save this data to a local directory\ndata_dir = 'json_energy_data'\n\n# make data dir, if it does not exist\nif not os.path.exists(data_dir):\n os.makedirs(data_dir)",
"_____no_output_____"
],
[
"# directories to save train/test data\ntrain_key = os.path.join(data_dir, 'train.json')\ntest_key = os.path.join(data_dir, 'test.json')\n\n# write train/test JSON files\nwrite_json_dataset(time_series_training, train_key) \nwrite_json_dataset(time_series, test_key)",
"json_energy_data/train.json saved.\njson_energy_data/test.json saved.\n"
]
],
[
[
"---\n## Uploading Data to S3\n\nNext, to make this data accessible to an estimator, I'll upload it to S3.\n\n### Sagemaker resources\n\nLet's start by specifying:\n* The sagemaker role and session for training a model.\n* A default S3 bucket where we can save our training, test, and model data.",
"_____no_output_____"
]
],
[
[
"import boto3\nimport sagemaker\nfrom sagemaker import get_execution_role",
"_____no_output_____"
],
[
"# session, role, bucket\nsagemaker_session = sagemaker.Session()\nrole = get_execution_role()\n\nbucket = sagemaker_session.default_bucket()\n",
"_____no_output_____"
]
],
[
[
"### EXERCISE: Upoad *both* training and test JSON files to S3\n\nSpecify *unique* train and test prefixes that define the location of that data in S3.\n* Upload training data to a location in S3, and save that location to `train_path`\n* Upload test data to a location in S3, and save that location to `test_path`",
"_____no_output_____"
]
],
[
[
"# general prefix\nprefix='deepar-energy-consumption'\n\n# *unique* train/test prefixes\ntrain_prefix = '{}/{}'.format(prefix, 'train')\ntest_prefix = '{}/{}'.format(prefix, 'test')\n\n# uploading data to S3, and saving locations\ntrain_path = sagemaker_session.upload_data(train_key, bucket=bucket, key_prefix=train_prefix)\ntest_path = sagemaker_session.upload_data(test_key, bucket=bucket, key_prefix=test_prefix)",
"_____no_output_____"
],
[
"# check locations\nprint('Training data is stored in: '+ train_path)\nprint('Test data is stored in: '+ test_path)",
"Training data is stored in: s3://sagemaker-us-east-2-771023245618/deepar-energy-consumption/train/train.json\nTest data is stored in: s3://sagemaker-us-east-2-771023245618/deepar-energy-consumption/test/test.json\n"
]
],
[
[
"---\n# Training a DeepAR Estimator\n\nSome estimators have specific, SageMaker constructors, but not all. Instead you can create a base `Estimator` and pass in the specific image (or container) that holds a specific model.\n\nNext, we configure the container image to be used for the region that we are running in.",
"_____no_output_____"
]
],
[
[
"from sagemaker.amazon.amazon_estimator import get_image_uri\n\nimage_name = get_image_uri(boto3.Session().region_name, # get the region\n 'forecasting-deepar') # specify image\n",
"_____no_output_____"
]
],
[
[
"### EXERCISE: Instantiate an Estimator \n\nYou can now define the estimator that will launch the training job. A generic Estimator will be defined by the usual constructor arguments and an `image_name`. \n> You can take a look at the [estimator source code](https://github.com/aws/sagemaker-python-sdk/blob/master/src/sagemaker/estimator.py#L595) to view specifics.\n",
"_____no_output_____"
]
],
[
[
"from sagemaker.estimator import Estimator\n\n# dir to save model artifacts\ns3_output_path = \"s3://{}/{}/output\".format(bucket, prefix)\n\n# instantiate a DeepAR estimator\nestimator = Estimator(sagemaker_session=sagemaker_session,\n image_name=image_name,\n role=role,\n train_instance_count=1,\n train_instance_type='ml.c4.xlarge',\n output_path=s3_output_path\n )",
"_____no_output_____"
]
],
[
[
"## Setting Hyperparameters\n\nNext, we need to define some DeepAR hyperparameters that define the model size and training behavior. Values for the epochs, frequency, prediction length, and context length are required.\n\n* **epochs**: The maximum number of times to pass over the data when training.\n* **time_freq**: The granularity of the time series in the dataset ('D' for daily).\n* **prediction_length**: A string; the number of time steps (based off the unit of frequency) that the model is trained to predict. \n* **context_length**: The number of time points that the model gets to see *before* making a prediction. \n\n### Context Length\n\nTypically, it is recommended that you start with a `context_length`=`prediction_length`. This is because a DeepAR model also receives \"lagged\" inputs from the target time series, which allow the model to capture long-term dependencies. For example, a daily time series can have yearly seasonality and DeepAR automatically includes a lag of one year. So, the context length can be shorter than a year, and the model will still be able to capture this seasonality. \n\nThe lag values that the model picks depend on the frequency of the time series. For example, lag values for daily frequency are the previous week, 2 weeks, 3 weeks, 4 weeks, and year. You can read more about this in the [DeepAR \"how it works\" documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_how-it-works.html).\n\n### Optional Hyperparameters\n\nYou can also configure optional hyperparameters to further tune your model. These include parameters like the number of layers in our RNN model, the number of cells per layer, the likelihood function, and the training options, such as batch size and learning rate. \n\nFor an exhaustive list of all the different DeepAR hyperparameters you can refer to the DeepAR [hyperparameter documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar_hyperparameters.html).",
"_____no_output_____"
]
],
[
[
"freq='D'\ncontext_length=30 # same as prediction_length\n\nhyperparameters = {\n \"epochs\": \"50\",\n \"time_freq\": freq,\n \"prediction_length\": str(prediction_length),\n \"context_length\": str(context_length),\n \"num_cells\": \"50\",\n \"num_layers\": \"2\",\n \"mini_batch_size\": \"128\",\n \"learning_rate\": \"0.001\",\n \"early_stopping_patience\": \"10\"\n}",
"_____no_output_____"
],
[
"# set the hyperparams\nestimator.set_hyperparameters(**hyperparameters)",
"_____no_output_____"
]
],
[
[
"## Training Job\n\nNow, we are ready to launch the training job! SageMaker will start an EC2 instance, download the data from S3, start training the model and save the trained model.\n\nIf you provide the `test` data channel, as we do in this example, DeepAR will also calculate accuracy metrics for the trained model on this test data set. This is done by predicting the last `prediction_length` points of each time series in the test set and comparing this to the *actual* value of the time series. The computed error metrics will be included as part of the log output.\n\nThe next cell may take a few minutes to complete, depending on data size, model complexity, and training options.",
"_____no_output_____"
]
],
[
[
"%%time\n# train and test channels\ndata_channels = {\n \"train\": train_path,\n \"test\": test_path\n}\n\n# fit the estimator\nestimator.fit(inputs=data_channels)",
"2020-05-13 10:26:21 Starting - Starting the training job...\n2020-05-13 10:26:23 Starting - Launching requested ML instances......\n2020-05-13 10:27:27 Starting - Preparing the instances for training...\n2020-05-13 10:28:22 Downloading - Downloading input data......\n2020-05-13 10:29:05 Training - Downloading the training image..\u001b[34mArguments: train\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Reading default configuration from /opt/amazon/lib/python2.7/site-packages/algorithm/resources/default-input.json: {u'num_dynamic_feat': u'auto', u'dropout_rate': u'0.10', u'mini_batch_size': u'128', u'test_quantiles': u'[0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]', u'_tuning_objective_metric': u'', u'_num_gpus': u'auto', u'num_eval_samples': u'100', u'learning_rate': u'0.001', u'num_cells': u'40', u'num_layers': u'2', u'embedding_dimension': u'10', u'_kvstore': u'auto', u'_num_kv_servers': u'auto', u'cardinality': u'auto', u'likelihood': u'student-t', u'early_stopping_patience': u''}\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Reading provided configuration from /opt/ml/input/config/hyperparameters.json: {u'learning_rate': u'0.001', u'num_cells': u'50', u'prediction_length': u'30', u'epochs': u'50', u'time_freq': u'D', u'context_length': u'30', u'num_layers': u'2', u'mini_batch_size': u'128', u'early_stopping_patience': u'10'}\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Final configuration: {u'dropout_rate': u'0.10', u'test_quantiles': u'[0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9]', u'_tuning_objective_metric': u'', u'num_eval_samples': u'100', u'learning_rate': u'0.001', u'num_layers': u'2', u'epochs': u'50', u'embedding_dimension': u'10', u'num_cells': u'50', u'_num_kv_servers': u'auto', u'mini_batch_size': u'128', u'likelihood': u'student-t', u'num_dynamic_feat': u'auto', u'cardinality': u'auto', u'_num_gpus': u'auto', u'prediction_length': u'30', u'time_freq': u'D', u'context_length': u'30', u'_kvstore': u'auto', u'early_stopping_patience': u'10'}\u001b[0m\n\u001b[34mProcess 1 is a worker.\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Detected entry point for worker worker\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Using early stopping with patience 10\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] [cardinality=auto] `cat` field was NOT found in the file `/opt/ml/input/data/train/train.json` and will NOT be used for training.\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] [num_dynamic_feat=auto] `dynamic_feat` field was NOT found in the file `/opt/ml/input/data/train/train.json` and will NOT be used for training.\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Training set statistics:\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Real time series\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] number of time series: 3\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] number of observations: 1006\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] mean target length: 335\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] min/mean/max target: 0.173818051815/1.05969123006/2.79841804504\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] mean abs(target): 1.05969123006\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] contains missing values: no\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Small number of time series. Doing 427 passes over dataset with prob 0.999219359875 per epoch.\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Test set statistics:\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Real time series\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] number of time series: 3\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] number of observations: 1096\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] mean target length: 365\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] min/mean/max target: 0.173818051815/1.08920513626/2.79841804504\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] mean abs(target): 1.08920513626\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] contains missing values: no\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] nvidia-smi took: 0.0252130031586 secs to identify 0 gpus\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Number of GPUs being used: 0\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Create Store: local\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"get_graph.time\": {\"count\": 1, \"max\": 141.448974609375, \"sum\": 141.448974609375, \"min\": 141.448974609375}}, \"EndTime\": 1589365768.400006, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365768.257615}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:28 INFO 140382612440896] Number of GPUs being used: 0\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"initialize.time\": {\"count\": 1, \"max\": 370.8229064941406, \"sum\": 370.8229064941406, \"min\": 370.8229064941406}}, \"EndTime\": 1589365768.628586, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365768.400084}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:29 INFO 140382612440896] Epoch[0] Batch[0] avg_epoch_loss=2.285124\u001b[0m\n\u001b[34m[05/13/2020 10:29:29 INFO 140382612440896] #quality_metric: host=algo-1, epoch=0, batch=0 train loss <loss>=2.28512430191\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] Epoch[0] Batch[5] avg_epoch_loss=1.391992\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] #quality_metric: host=algo-1, epoch=0, batch=5 train loss <loss>=1.39199231068\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] Epoch[0] Batch [5]#011Speed: 743.14 samples/sec#011loss=1.391992\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] processed a total of 1254 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"epochs\": {\"count\": 1, \"max\": 50, \"sum\": 50.0, \"min\": 50}, \"update.time\": {\"count\": 1, \"max\": 2207.641124725342, \"sum\": 2207.641124725342, \"min\": 2207.641124725342}}, \"EndTime\": 1589365770.836371, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365768.628649}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=567.998030236 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] #progress_metric: host=algo-1, completed 2 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] #quality_metric: host=algo-1, epoch=0, train loss <loss>=1.19816408157\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:30 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_808ffa15-b4a4-42cb-b5b6-048396c8e94b-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 23.999929428100586, \"sum\": 23.999929428100586, \"min\": 23.999929428100586}}, \"EndTime\": 1589365770.860982, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365770.83645}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:31 INFO 140382612440896] Epoch[1] Batch[0] avg_epoch_loss=0.853938\u001b[0m\n\u001b[34m[05/13/2020 10:29:31 INFO 140382612440896] #quality_metric: host=algo-1, epoch=1, batch=0 train loss <loss>=0.853937804699\u001b[0m\n\u001b[34m[05/13/2020 10:29:32 INFO 140382612440896] Epoch[1] Batch[5] avg_epoch_loss=0.708400\u001b[0m\n\u001b[34m[05/13/2020 10:29:32 INFO 140382612440896] #quality_metric: host=algo-1, epoch=1, batch=5 train loss <loss>=0.708399752776\u001b[0m\n\u001b[34m[05/13/2020 10:29:32 INFO 140382612440896] Epoch[1] Batch [5]#011Speed: 735.90 samples/sec#011loss=0.708400\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] Epoch[1] Batch[10] avg_epoch_loss=0.613172\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] #quality_metric: host=algo-1, epoch=1, batch=10 train loss <loss>=0.498898273706\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] Epoch[1] Batch [10]#011Speed: 754.48 samples/sec#011loss=0.498898\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] processed a total of 1294 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2288.2699966430664, \"sum\": 2288.2699966430664, \"min\": 2288.2699966430664}}, \"EndTime\": 1589365773.149376, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365770.861047}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=565.462602979 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] #progress_metric: host=algo-1, completed 4 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] #quality_metric: host=algo-1, epoch=1, train loss <loss>=0.613171807744\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_b56d83c5-fa7c-47fe-8298-47a04ffbc5c8-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 25.77805519104004, \"sum\": 25.77805519104004, \"min\": 25.77805519104004}}, \"EndTime\": 1589365773.175783, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365773.149458}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] Epoch[2] Batch[0] avg_epoch_loss=0.369981\u001b[0m\n\u001b[34m[05/13/2020 10:29:33 INFO 140382612440896] #quality_metric: host=algo-1, epoch=2, batch=0 train loss <loss>=0.369981318712\u001b[0m\n\u001b[34m[05/13/2020 10:29:34 INFO 140382612440896] Epoch[2] Batch[5] avg_epoch_loss=0.378516\u001b[0m\n\u001b[34m[05/13/2020 10:29:34 INFO 140382612440896] #quality_metric: host=algo-1, epoch=2, batch=5 train loss <loss>=0.378515988588\u001b[0m\n\u001b[34m[05/13/2020 10:29:34 INFO 140382612440896] Epoch[2] Batch [5]#011Speed: 658.69 samples/sec#011loss=0.378516\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] processed a total of 1259 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2468.4250354766846, \"sum\": 2468.4250354766846, \"min\": 2468.4250354766846}}, \"EndTime\": 1589365775.644355, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365773.175861}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=510.017633692 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] #progress_metric: host=algo-1, completed 6 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] #quality_metric: host=algo-1, epoch=2, train loss <loss>=0.377673172951\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:35 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_3f22545a-74be-47e1-99a0-8e1e497c860c-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 31.029939651489258, \"sum\": 31.029939651489258, \"min\": 31.029939651489258}}, \"EndTime\": 1589365775.675957, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365775.644433}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:36 INFO 140382612440896] Epoch[3] Batch[0] avg_epoch_loss=0.397179\u001b[0m\n\u001b[34m[05/13/2020 10:29:36 INFO 140382612440896] #quality_metric: host=algo-1, epoch=3, batch=0 train loss <loss>=0.397179305553\u001b[0m\n\n2020-05-13 10:29:25 Training - Training image download completed. Training in progress.\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] Epoch[3] Batch[5] avg_epoch_loss=0.356190\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] #quality_metric: host=algo-1, epoch=3, batch=5 train loss <loss>=0.356189568837\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] Epoch[3] Batch [5]#011Speed: 715.19 samples/sec#011loss=0.356190\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] Epoch[3] Batch[10] avg_epoch_loss=0.319859\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] #quality_metric: host=algo-1, epoch=3, batch=10 train loss <loss>=0.276262110472\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] Epoch[3] Batch [10]#011Speed: 740.43 samples/sec#011loss=0.276262\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] processed a total of 1311 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2295.116901397705, \"sum\": 2295.116901397705, \"min\": 2295.116901397705}}, \"EndTime\": 1589365777.971212, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365775.676032}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=571.182117173 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] #progress_metric: host=algo-1, completed 8 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] #quality_metric: host=algo-1, epoch=3, train loss <loss>=0.319858905944\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:37 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_ae5ff9ae-ada5-4e49-902e-6e033ad07b53-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.899866104125977, \"sum\": 22.899866104125977, \"min\": 22.899866104125977}}, \"EndTime\": 1589365777.994772, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365777.971296}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:38 INFO 140382612440896] Epoch[4] Batch[0] avg_epoch_loss=0.221055\u001b[0m\n\u001b[34m[05/13/2020 10:29:38 INFO 140382612440896] #quality_metric: host=algo-1, epoch=4, batch=0 train loss <loss>=0.221054896712\u001b[0m\n\u001b[34m[05/13/2020 10:29:39 INFO 140382612440896] Epoch[4] Batch[5] avg_epoch_loss=0.265531\u001b[0m\n\u001b[34m[05/13/2020 10:29:39 INFO 140382612440896] #quality_metric: host=algo-1, epoch=4, batch=5 train loss <loss>=0.265530814727\u001b[0m\n\u001b[34m[05/13/2020 10:29:39 INFO 140382612440896] Epoch[4] Batch [5]#011Speed: 734.48 samples/sec#011loss=0.265531\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] Epoch[4] Batch[10] avg_epoch_loss=0.250094\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] #quality_metric: host=algo-1, epoch=4, batch=10 train loss <loss>=0.23156914711\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] Epoch[4] Batch [10]#011Speed: 743.50 samples/sec#011loss=0.231569\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] processed a total of 1293 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2270.0021266937256, \"sum\": 2270.0021266937256, \"min\": 2270.0021266937256}}, \"EndTime\": 1589365780.264889, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365777.994836}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=569.57565166 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] #progress_metric: host=algo-1, completed 10 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] #quality_metric: host=algo-1, epoch=4, train loss <loss>=0.250093693083\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_29c63689-53e2-443d-9c50-16ea6ba72491-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 24.94192123413086, \"sum\": 24.94192123413086, \"min\": 24.94192123413086}}, \"EndTime\": 1589365780.290477, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365780.264966}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] Epoch[5] Batch[0] avg_epoch_loss=0.257301\u001b[0m\n\u001b[34m[05/13/2020 10:29:40 INFO 140382612440896] #quality_metric: host=algo-1, epoch=5, batch=0 train loss <loss>=0.257301181555\u001b[0m\n\u001b[34m[05/13/2020 10:29:41 INFO 140382612440896] Epoch[5] Batch[5] avg_epoch_loss=0.258020\u001b[0m\n\u001b[34m[05/13/2020 10:29:41 INFO 140382612440896] #quality_metric: host=algo-1, epoch=5, batch=5 train loss <loss>=0.258019750317\u001b[0m\n\u001b[34m[05/13/2020 10:29:41 INFO 140382612440896] Epoch[5] Batch [5]#011Speed: 722.46 samples/sec#011loss=0.258020\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] processed a total of 1248 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2100.404977798462, \"sum\": 2100.404977798462, \"min\": 2100.404977798462}}, \"EndTime\": 1589365782.391017, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365780.290548}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=594.139702797 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] #progress_metric: host=algo-1, completed 12 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] #quality_metric: host=algo-1, epoch=5, train loss <loss>=0.233308123052\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_4aac5a97-40a9-4fc3-948e-33276156dc0b-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 31.780004501342773, \"sum\": 31.780004501342773, \"min\": 31.780004501342773}}, \"EndTime\": 1589365782.4234, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365782.391091}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] Epoch[6] Batch[0] avg_epoch_loss=0.233674\u001b[0m\n\u001b[34m[05/13/2020 10:29:42 INFO 140382612440896] #quality_metric: host=algo-1, epoch=6, batch=0 train loss <loss>=0.233674049377\u001b[0m\n\u001b[34m[05/13/2020 10:29:43 INFO 140382612440896] Epoch[6] Batch[5] avg_epoch_loss=0.214551\u001b[0m\n\u001b[34m[05/13/2020 10:29:43 INFO 140382612440896] #quality_metric: host=algo-1, epoch=6, batch=5 train loss <loss>=0.214551289876\u001b[0m\n\u001b[34m[05/13/2020 10:29:43 INFO 140382612440896] Epoch[6] Batch [5]#011Speed: 748.13 samples/sec#011loss=0.214551\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] processed a total of 1225 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2198.8770961761475, \"sum\": 2198.8770961761475, \"min\": 2198.8770961761475}}, \"EndTime\": 1589365784.622408, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365782.423469}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=557.041288852 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] #progress_metric: host=algo-1, completed 14 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] #quality_metric: host=algo-1, epoch=6, train loss <loss>=0.195748349279\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:44 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_a79d4ba2-f38d-4784-9c36-6d1fd7e3cb0b-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 27.443885803222656, \"sum\": 27.443885803222656, \"min\": 27.443885803222656}}, \"EndTime\": 1589365784.650699, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365784.622612}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:45 INFO 140382612440896] Epoch[7] Batch[0] avg_epoch_loss=0.196261\u001b[0m\n\u001b[34m[05/13/2020 10:29:45 INFO 140382612440896] #quality_metric: host=algo-1, epoch=7, batch=0 train loss <loss>=0.196261182427\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] Epoch[7] Batch[5] avg_epoch_loss=0.174724\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] #quality_metric: host=algo-1, epoch=7, batch=5 train loss <loss>=0.174724444747\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] Epoch[7] Batch [5]#011Speed: 740.90 samples/sec#011loss=0.174724\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] Epoch[7] Batch[10] avg_epoch_loss=0.224538\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] #quality_metric: host=algo-1, epoch=7, batch=10 train loss <loss>=0.284315294027\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] Epoch[7] Batch [10]#011Speed: 721.80 samples/sec#011loss=0.284315\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] processed a total of 1319 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2305.7568073272705, \"sum\": 2305.7568073272705, \"min\": 2305.7568073272705}}, \"EndTime\": 1589365786.956598, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365784.650776}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=572.018119998 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] #progress_metric: host=algo-1, completed 16 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] #quality_metric: host=algo-1, epoch=7, train loss <loss>=0.224538467147\u001b[0m\n\u001b[34m[05/13/2020 10:29:46 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:29:47 INFO 140382612440896] Epoch[8] Batch[0] avg_epoch_loss=0.168205\u001b[0m\n\u001b[34m[05/13/2020 10:29:47 INFO 140382612440896] #quality_metric: host=algo-1, epoch=8, batch=0 train loss <loss>=0.168204680085\u001b[0m\n\u001b[34m[05/13/2020 10:29:48 INFO 140382612440896] Epoch[8] Batch[5] avg_epoch_loss=0.152630\u001b[0m\n\u001b[34m[05/13/2020 10:29:48 INFO 140382612440896] #quality_metric: host=algo-1, epoch=8, batch=5 train loss <loss>=0.152629859746\u001b[0m\n\u001b[34m[05/13/2020 10:29:48 INFO 140382612440896] Epoch[8] Batch [5]#011Speed: 745.60 samples/sec#011loss=0.152630\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] Epoch[8] Batch[10] avg_epoch_loss=0.152451\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=8, batch=10 train loss <loss>=0.152237153053\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] Epoch[8] Batch [10]#011Speed: 730.39 samples/sec#011loss=0.152237\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] processed a total of 1313 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2308.852195739746, \"sum\": 2308.852195739746, \"min\": 2308.852195739746}}, \"EndTime\": 1589365789.26592, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365786.956676}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=568.65376707 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] #progress_metric: host=algo-1, completed 18 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=8, train loss <loss>=0.152451356704\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_f9644baf-1feb-43f3-856c-5bd4d53dca46-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 36.219120025634766, \"sum\": 36.219120025634766, \"min\": 36.219120025634766}}, \"EndTime\": 1589365789.302672, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365789.265995}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] Epoch[9] Batch[0] avg_epoch_loss=0.116971\u001b[0m\n\u001b[34m[05/13/2020 10:29:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=9, batch=0 train loss <loss>=0.116971261799\u001b[0m\n\u001b[34m[05/13/2020 10:29:50 INFO 140382612440896] Epoch[9] Batch[5] avg_epoch_loss=0.122035\u001b[0m\n\u001b[34m[05/13/2020 10:29:50 INFO 140382612440896] #quality_metric: host=algo-1, epoch=9, batch=5 train loss <loss>=0.122034928451\u001b[0m\n\u001b[34m[05/13/2020 10:29:50 INFO 140382612440896] Epoch[9] Batch [5]#011Speed: 750.09 samples/sec#011loss=0.122035\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] Epoch[9] Batch[10] avg_epoch_loss=0.172588\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] #quality_metric: host=algo-1, epoch=9, batch=10 train loss <loss>=0.233252303302\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] Epoch[9] Batch [10]#011Speed: 688.16 samples/sec#011loss=0.233252\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] processed a total of 1301 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2368.3130741119385, \"sum\": 2368.3130741119385, \"min\": 2368.3130741119385}}, \"EndTime\": 1589365791.671118, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365789.302746}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=549.309999358 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] #progress_metric: host=algo-1, completed 20 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] #quality_metric: host=algo-1, epoch=9, train loss <loss>=0.172588280656\u001b[0m\n\u001b[34m[05/13/2020 10:29:51 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:29:52 INFO 140382612440896] Epoch[10] Batch[0] avg_epoch_loss=0.084035\u001b[0m\n\u001b[34m[05/13/2020 10:29:52 INFO 140382612440896] #quality_metric: host=algo-1, epoch=10, batch=0 train loss <loss>=0.0840349271894\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] Epoch[10] Batch[5] avg_epoch_loss=0.106007\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] #quality_metric: host=algo-1, epoch=10, batch=5 train loss <loss>=0.10600658382\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] Epoch[10] Batch [5]#011Speed: 743.54 samples/sec#011loss=0.106007\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] Epoch[10] Batch[10] avg_epoch_loss=0.130081\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] #quality_metric: host=algo-1, epoch=10, batch=10 train loss <loss>=0.158970734477\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] Epoch[10] Batch [10]#011Speed: 747.82 samples/sec#011loss=0.158971\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] processed a total of 1323 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2255.156993865967, \"sum\": 2255.156993865967, \"min\": 2255.156993865967}}, \"EndTime\": 1589365793.927056, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365791.671197}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=586.622641617 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] #progress_metric: host=algo-1, completed 22 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] #quality_metric: host=algo-1, epoch=10, train loss <loss>=0.130081197755\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:53 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_0840a242-fa3c-4441-b4d7-4dcf2f5929db-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 21.74520492553711, \"sum\": 21.74520492553711, \"min\": 21.74520492553711}}, \"EndTime\": 1589365793.94947, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365793.92714}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:54 INFO 140382612440896] Epoch[11] Batch[0] avg_epoch_loss=0.113716\u001b[0m\n\u001b[34m[05/13/2020 10:29:54 INFO 140382612440896] #quality_metric: host=algo-1, epoch=11, batch=0 train loss <loss>=0.113715589046\u001b[0m\n\u001b[34m[05/13/2020 10:29:55 INFO 140382612440896] Epoch[11] Batch[5] avg_epoch_loss=0.114304\u001b[0m\n\u001b[34m[05/13/2020 10:29:55 INFO 140382612440896] #quality_metric: host=algo-1, epoch=11, batch=5 train loss <loss>=0.114304396013\u001b[0m\n\u001b[34m[05/13/2020 10:29:55 INFO 140382612440896] Epoch[11] Batch [5]#011Speed: 751.01 samples/sec#011loss=0.114304\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] Epoch[11] Batch[10] avg_epoch_loss=0.126607\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] #quality_metric: host=algo-1, epoch=11, batch=10 train loss <loss>=0.141369990259\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] Epoch[11] Batch [10]#011Speed: 749.30 samples/sec#011loss=0.141370\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] processed a total of 1305 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2258.431911468506, \"sum\": 2258.431911468506, \"min\": 2258.431911468506}}, \"EndTime\": 1589365796.208016, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365793.949534}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=577.807960447 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] #progress_metric: host=algo-1, completed 24 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] #quality_metric: host=algo-1, epoch=11, train loss <loss>=0.126606938853\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_252fb7b2-1287-4cd9-8b7f-44b6ca63c6c5-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 34.915924072265625, \"sum\": 34.915924072265625, \"min\": 34.915924072265625}}, \"EndTime\": 1589365796.243447, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365796.208089}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] Epoch[12] Batch[0] avg_epoch_loss=0.087924\u001b[0m\n\u001b[34m[05/13/2020 10:29:56 INFO 140382612440896] #quality_metric: host=algo-1, epoch=12, batch=0 train loss <loss>=0.0879236087203\u001b[0m\n\u001b[34m[05/13/2020 10:29:57 INFO 140382612440896] Epoch[12] Batch[5] avg_epoch_loss=0.082218\u001b[0m\n\u001b[34m[05/13/2020 10:29:57 INFO 140382612440896] #quality_metric: host=algo-1, epoch=12, batch=5 train loss <loss>=0.0822178591043\u001b[0m\n\u001b[34m[05/13/2020 10:29:57 INFO 140382612440896] Epoch[12] Batch [5]#011Speed: 712.27 samples/sec#011loss=0.082218\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] processed a total of 1236 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2187.774896621704, \"sum\": 2187.774896621704, \"min\": 2187.774896621704}}, \"EndTime\": 1589365798.431373, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365796.24353}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=564.924207139 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] #progress_metric: host=algo-1, completed 26 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] #quality_metric: host=algo-1, epoch=12, train loss <loss>=0.0628971852362\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_2f9a7360-1d30-4205-af64-4e65a50fcb30-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.105932235717773, \"sum\": 22.105932235717773, \"min\": 22.105932235717773}}, \"EndTime\": 1589365798.454131, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365798.431462}\n\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] Epoch[13] Batch[0] avg_epoch_loss=0.087569\u001b[0m\n\u001b[34m[05/13/2020 10:29:58 INFO 140382612440896] #quality_metric: host=algo-1, epoch=13, batch=0 train loss <loss>=0.087568834424\u001b[0m\n\u001b[34m[05/13/2020 10:29:59 INFO 140382612440896] Epoch[13] Batch[5] avg_epoch_loss=0.059229\u001b[0m\n\u001b[34m[05/13/2020 10:29:59 INFO 140382612440896] #quality_metric: host=algo-1, epoch=13, batch=5 train loss <loss>=0.0592288697759\u001b[0m\n\u001b[34m[05/13/2020 10:29:59 INFO 140382612440896] Epoch[13] Batch [5]#011Speed: 717.04 samples/sec#011loss=0.059229\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] processed a total of 1239 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2126.3267993927, \"sum\": 2126.3267993927, \"min\": 2126.3267993927}}, \"EndTime\": 1589365800.580589, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365798.454201}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=582.666713608 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] #progress_metric: host=algo-1, completed 28 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] #quality_metric: host=algo-1, epoch=13, train loss <loss>=0.0534487454221\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:00 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_6ed524ad-c4c1-4d66-b641-c2ad7bb74538-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 21.45099639892578, \"sum\": 21.45099639892578, \"min\": 21.45099639892578}}, \"EndTime\": 1589365800.60276, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365800.580658}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:01 INFO 140382612440896] Epoch[14] Batch[0] avg_epoch_loss=0.078639\u001b[0m\n\u001b[34m[05/13/2020 10:30:01 INFO 140382612440896] #quality_metric: host=algo-1, epoch=14, batch=0 train loss <loss>=0.0786394327879\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] Epoch[14] Batch[5] avg_epoch_loss=0.042168\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] #quality_metric: host=algo-1, epoch=14, batch=5 train loss <loss>=0.0421677608974\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] Epoch[14] Batch [5]#011Speed: 717.42 samples/sec#011loss=0.042168\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] processed a total of 1230 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2105.5068969726562, \"sum\": 2105.5068969726562, \"min\": 2105.5068969726562}}, \"EndTime\": 1589365802.708387, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365800.602827}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=584.106897915 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] #progress_metric: host=algo-1, completed 30 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] #quality_metric: host=algo-1, epoch=14, train loss <loss>=0.0733171797357\u001b[0m\n\u001b[34m[05/13/2020 10:30:02 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:03 INFO 140382612440896] Epoch[15] Batch[0] avg_epoch_loss=0.040853\u001b[0m\n\u001b[34m[05/13/2020 10:30:03 INFO 140382612440896] #quality_metric: host=algo-1, epoch=15, batch=0 train loss <loss>=0.0408528186381\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] Epoch[15] Batch[5] avg_epoch_loss=0.035749\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] #quality_metric: host=algo-1, epoch=15, batch=5 train loss <loss>=0.0357490122163\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] Epoch[15] Batch [5]#011Speed: 718.54 samples/sec#011loss=0.035749\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] processed a total of 1270 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2182.194948196411, \"sum\": 2182.194948196411, \"min\": 2182.194948196411}}, \"EndTime\": 1589365804.891408, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365802.708626}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=581.955111996 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] #progress_metric: host=algo-1, completed 32 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] #quality_metric: host=algo-1, epoch=15, train loss <loss>=0.0339481100906\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:04 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_751252ea-5c2b-47d1-ace4-e4b887cc536b-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.980928421020508, \"sum\": 22.980928421020508, \"min\": 22.980928421020508}}, \"EndTime\": 1589365804.915004, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365804.891474}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:05 INFO 140382612440896] Epoch[16] Batch[0] avg_epoch_loss=0.046296\u001b[0m\n\u001b[34m[05/13/2020 10:30:05 INFO 140382612440896] #quality_metric: host=algo-1, epoch=16, batch=0 train loss <loss>=0.046296145767\u001b[0m\n\u001b[34m[05/13/2020 10:30:06 INFO 140382612440896] Epoch[16] Batch[5] avg_epoch_loss=0.017626\u001b[0m\n\u001b[34m[05/13/2020 10:30:06 INFO 140382612440896] #quality_metric: host=algo-1, epoch=16, batch=5 train loss <loss>=0.0176261936043\u001b[0m\n\u001b[34m[05/13/2020 10:30:06 INFO 140382612440896] Epoch[16] Batch [5]#011Speed: 744.96 samples/sec#011loss=0.017626\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] processed a total of 1230 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2089.8489952087402, \"sum\": 2089.8489952087402, \"min\": 2089.8489952087402}}, \"EndTime\": 1589365807.005189, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365804.91528}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=588.53362209 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] #progress_metric: host=algo-1, completed 34 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] #quality_metric: host=algo-1, epoch=16, train loss <loss>=0.02333825432\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_12cd57af-02db-49f7-9f9e-09bcc0261b6c-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.337913513183594, \"sum\": 22.337913513183594, \"min\": 22.337913513183594}}, \"EndTime\": 1589365807.028126, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365807.005244}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] Epoch[17] Batch[0] avg_epoch_loss=0.022488\u001b[0m\n\u001b[34m[05/13/2020 10:30:07 INFO 140382612440896] #quality_metric: host=algo-1, epoch=17, batch=0 train loss <loss>=0.0224878005683\u001b[0m\n\u001b[34m[05/13/2020 10:30:08 INFO 140382612440896] Epoch[17] Batch[5] avg_epoch_loss=0.022596\u001b[0m\n\u001b[34m[05/13/2020 10:30:08 INFO 140382612440896] #quality_metric: host=algo-1, epoch=17, batch=5 train loss <loss>=0.0225963490472\u001b[0m\n\u001b[34m[05/13/2020 10:30:08 INFO 140382612440896] Epoch[17] Batch [5]#011Speed: 732.27 samples/sec#011loss=0.022596\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] Epoch[17] Batch[10] avg_epoch_loss=0.070359\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] #quality_metric: host=algo-1, epoch=17, batch=10 train loss <loss>=0.127673883364\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] Epoch[17] Batch [10]#011Speed: 743.90 samples/sec#011loss=0.127674\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] processed a total of 1295 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2295.7279682159424, \"sum\": 2295.7279682159424, \"min\": 2295.7279682159424}}, \"EndTime\": 1589365809.32397, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365807.028188}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=564.065683312 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] #progress_metric: host=algo-1, completed 36 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] #quality_metric: host=algo-1, epoch=17, train loss <loss>=0.0703588646456\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] Epoch[18] Batch[0] avg_epoch_loss=0.004557\u001b[0m\n\u001b[34m[05/13/2020 10:30:09 INFO 140382612440896] #quality_metric: host=algo-1, epoch=18, batch=0 train loss <loss>=0.00455685891211\u001b[0m\n\u001b[34m[05/13/2020 10:30:10 INFO 140382612440896] Epoch[18] Batch[5] avg_epoch_loss=0.022645\u001b[0m\n\u001b[34m[05/13/2020 10:30:10 INFO 140382612440896] #quality_metric: host=algo-1, epoch=18, batch=5 train loss <loss>=0.0226447856985\u001b[0m\n\u001b[34m[05/13/2020 10:30:10 INFO 140382612440896] Epoch[18] Batch [5]#011Speed: 667.89 samples/sec#011loss=0.022645\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] processed a total of 1266 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2266.187906265259, \"sum\": 2266.187906265259, \"min\": 2266.187906265259}}, \"EndTime\": 1589365811.591033, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365809.324041}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=558.617552684 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] #progress_metric: host=algo-1, completed 38 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] #quality_metric: host=algo-1, epoch=18, train loss <loss>=0.0222950534895\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:11 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_15a0dfc8-ed63-4c9b-a49a-f96a24dd69cf-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 31.078815460205078, \"sum\": 31.078815460205078, \"min\": 31.078815460205078}}, \"EndTime\": 1589365811.622647, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365811.591106}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:12 INFO 140382612440896] Epoch[19] Batch[0] avg_epoch_loss=0.005779\u001b[0m\n\u001b[34m[05/13/2020 10:30:12 INFO 140382612440896] #quality_metric: host=algo-1, epoch=19, batch=0 train loss <loss>=0.00577916065231\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] Epoch[19] Batch[5] avg_epoch_loss=0.012726\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] #quality_metric: host=algo-1, epoch=19, batch=5 train loss <loss>=0.0127259235984\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] Epoch[19] Batch [5]#011Speed: 742.50 samples/sec#011loss=0.012726\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] processed a total of 1277 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2117.9189682006836, \"sum\": 2117.9189682006836, \"min\": 2117.9189682006836}}, \"EndTime\": 1589365813.740703, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365811.62272}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=602.9178474 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] #progress_metric: host=algo-1, completed 40 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] #quality_metric: host=algo-1, epoch=19, train loss <loss>=-0.000462670018896\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:13 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_2165c710-55f4-423e-b8d1-1843726c79d8-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 24.909019470214844, \"sum\": 24.909019470214844, \"min\": 24.909019470214844}}, \"EndTime\": 1589365813.766185, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365813.74078}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:14 INFO 140382612440896] Epoch[20] Batch[0] avg_epoch_loss=-0.006543\u001b[0m\n\u001b[34m[05/13/2020 10:30:14 INFO 140382612440896] #quality_metric: host=algo-1, epoch=20, batch=0 train loss <loss>=-0.00654338859022\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] Epoch[20] Batch[5] avg_epoch_loss=-0.009487\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] #quality_metric: host=algo-1, epoch=20, batch=5 train loss <loss>=-0.00948691088706\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] Epoch[20] Batch [5]#011Speed: 735.72 samples/sec#011loss=-0.009487\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] processed a total of 1275 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2104.543924331665, \"sum\": 2104.543924331665, \"min\": 2104.543924331665}}, \"EndTime\": 1589365815.87084, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365813.766238}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=605.797384816 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] #progress_metric: host=algo-1, completed 42 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] #quality_metric: host=algo-1, epoch=20, train loss <loss>=-0.0158227193169\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:15 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_031c3360-a29f-4187-85af-18f9c70ce76a-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 28.18608283996582, \"sum\": 28.18608283996582, \"min\": 28.18608283996582}}, \"EndTime\": 1589365815.899716, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365815.870921}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:16 INFO 140382612440896] Epoch[21] Batch[0] avg_epoch_loss=-0.021988\u001b[0m\n\u001b[34m[05/13/2020 10:30:16 INFO 140382612440896] #quality_metric: host=algo-1, epoch=21, batch=0 train loss <loss>=-0.0219877958298\u001b[0m\n\u001b[34m[05/13/2020 10:30:17 INFO 140382612440896] Epoch[21] Batch[5] avg_epoch_loss=-0.023795\u001b[0m\n\u001b[34m[05/13/2020 10:30:17 INFO 140382612440896] #quality_metric: host=algo-1, epoch=21, batch=5 train loss <loss>=-0.0237953779482\u001b[0m\n\u001b[34m[05/13/2020 10:30:17 INFO 140382612440896] Epoch[21] Batch [5]#011Speed: 716.39 samples/sec#011loss=-0.023795\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] processed a total of 1273 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2112.2090816497803, \"sum\": 2112.2090816497803, \"min\": 2112.2090816497803}}, \"EndTime\": 1589365818.012071, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365815.899793}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=602.650902828 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] #progress_metric: host=algo-1, completed 44 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] #quality_metric: host=algo-1, epoch=21, train loss <loss>=-0.0240660685231\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_a494dae2-215d-44b5-b52f-46bcf32f15f6-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 34.607887268066406, \"sum\": 34.607887268066406, \"min\": 34.607887268066406}}, \"EndTime\": 1589365818.047227, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365818.012154}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] Epoch[22] Batch[0] avg_epoch_loss=-0.015403\u001b[0m\n\u001b[34m[05/13/2020 10:30:18 INFO 140382612440896] #quality_metric: host=algo-1, epoch=22, batch=0 train loss <loss>=-0.0154030378908\u001b[0m\n\u001b[34m[05/13/2020 10:30:19 INFO 140382612440896] Epoch[22] Batch[5] avg_epoch_loss=-0.030710\u001b[0m\n\u001b[34m[05/13/2020 10:30:19 INFO 140382612440896] #quality_metric: host=algo-1, epoch=22, batch=5 train loss <loss>=-0.0307101585592\u001b[0m\n\u001b[34m[05/13/2020 10:30:19 INFO 140382612440896] Epoch[22] Batch [5]#011Speed: 741.02 samples/sec#011loss=-0.030710\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] Epoch[22] Batch[10] avg_epoch_loss=0.005170\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] #quality_metric: host=algo-1, epoch=22, batch=10 train loss <loss>=0.0482272613794\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] Epoch[22] Batch [10]#011Speed: 740.08 samples/sec#011loss=0.048227\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] processed a total of 1316 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2324.2459297180176, \"sum\": 2324.2459297180176, \"min\": 2324.2459297180176}}, \"EndTime\": 1589365820.371604, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365818.047295}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=566.178834095 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] #progress_metric: host=algo-1, completed 46 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] #quality_metric: host=algo-1, epoch=22, train loss <loss>=0.00517048686743\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] Epoch[23] Batch[0] avg_epoch_loss=-0.054595\u001b[0m\n\u001b[34m[05/13/2020 10:30:20 INFO 140382612440896] #quality_metric: host=algo-1, epoch=23, batch=0 train loss <loss>=-0.0545947477221\u001b[0m\n\u001b[34m[05/13/2020 10:30:21 INFO 140382612440896] Epoch[23] Batch[5] avg_epoch_loss=-0.035302\u001b[0m\n\u001b[34m[05/13/2020 10:30:21 INFO 140382612440896] #quality_metric: host=algo-1, epoch=23, batch=5 train loss <loss>=-0.0353024373762\u001b[0m\n\u001b[34m[05/13/2020 10:30:21 INFO 140382612440896] Epoch[23] Batch [5]#011Speed: 697.28 samples/sec#011loss=-0.035302\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] processed a total of 1263 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2174.631118774414, \"sum\": 2174.631118774414, \"min\": 2174.631118774414}}, \"EndTime\": 1589365822.54673, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365820.371675}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=580.758549581 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] #progress_metric: host=algo-1, completed 48 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] #quality_metric: host=algo-1, epoch=23, train loss <loss>=-0.0377370188478\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:22 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_2155e01d-31c4-4a51-a83e-c308b885815a-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 26.7179012298584, \"sum\": 26.7179012298584, \"min\": 26.7179012298584}}, \"EndTime\": 1589365822.573997, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365822.546802}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:23 INFO 140382612440896] Epoch[24] Batch[0] avg_epoch_loss=-0.019537\u001b[0m\n\u001b[34m[05/13/2020 10:30:23 INFO 140382612440896] #quality_metric: host=algo-1, epoch=24, batch=0 train loss <loss>=-0.019537108019\u001b[0m\n\u001b[34m[05/13/2020 10:30:23 INFO 140382612440896] Epoch[24] Batch[5] avg_epoch_loss=-0.039477\u001b[0m\n\u001b[34m[05/13/2020 10:30:23 INFO 140382612440896] #quality_metric: host=algo-1, epoch=24, batch=5 train loss <loss>=-0.0394767836357\u001b[0m\n\u001b[34m[05/13/2020 10:30:23 INFO 140382612440896] Epoch[24] Batch [5]#011Speed: 743.42 samples/sec#011loss=-0.039477\u001b[0m\n\u001b[34m[05/13/2020 10:30:24 INFO 140382612440896] processed a total of 1256 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2110.553979873657, \"sum\": 2110.553979873657, \"min\": 2110.553979873657}}, \"EndTime\": 1589365824.6847, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365822.574079}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:24 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=595.076723225 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:24 INFO 140382612440896] #progress_metric: host=algo-1, completed 50 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:24 INFO 140382612440896] #quality_metric: host=algo-1, epoch=24, train loss <loss>=-0.0312322543934\u001b[0m\n\u001b[34m[05/13/2020 10:30:24 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:25 INFO 140382612440896] Epoch[25] Batch[0] avg_epoch_loss=-0.004981\u001b[0m\n\u001b[34m[05/13/2020 10:30:25 INFO 140382612440896] #quality_metric: host=algo-1, epoch=25, batch=0 train loss <loss>=-0.00498093245551\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] Epoch[25] Batch[5] avg_epoch_loss=-0.043468\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] #quality_metric: host=algo-1, epoch=25, batch=5 train loss <loss>=-0.0434680226414\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] Epoch[25] Batch [5]#011Speed: 748.30 samples/sec#011loss=-0.043468\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] processed a total of 1261 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2080.104112625122, \"sum\": 2080.104112625122, \"min\": 2080.104112625122}}, \"EndTime\": 1589365826.765376, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365824.68476}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=606.187625744 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] #progress_metric: host=algo-1, completed 52 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] #quality_metric: host=algo-1, epoch=25, train loss <loss>=-0.0536721154582\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:26 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_da473660-f18b-4dce-80f5-3bd0d49151d4-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.900104522705078, \"sum\": 22.900104522705078, \"min\": 22.900104522705078}}, \"EndTime\": 1589365826.788914, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365826.765448}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:27 INFO 140382612440896] Epoch[26] Batch[0] avg_epoch_loss=-0.042160\u001b[0m\n\u001b[34m[05/13/2020 10:30:27 INFO 140382612440896] #quality_metric: host=algo-1, epoch=26, batch=0 train loss <loss>=-0.0421602204442\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] Epoch[26] Batch[5] avg_epoch_loss=-0.056362\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] #quality_metric: host=algo-1, epoch=26, batch=5 train loss <loss>=-0.056361855939\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] Epoch[26] Batch [5]#011Speed: 735.69 samples/sec#011loss=-0.056362\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] processed a total of 1279 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2173.762798309326, \"sum\": 2173.762798309326, \"min\": 2173.762798309326}}, \"EndTime\": 1589365828.962808, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365826.78898}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=588.351367797 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] #progress_metric: host=algo-1, completed 54 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] #quality_metric: host=algo-1, epoch=26, train loss <loss>=-0.0602754935622\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:28 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_3de82f30-4549-456c-b934-297a59548890-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.695064544677734, \"sum\": 22.695064544677734, \"min\": 22.695064544677734}}, \"EndTime\": 1589365828.986159, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365828.962878}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:29 INFO 140382612440896] Epoch[27] Batch[0] avg_epoch_loss=-0.057286\u001b[0m\n\u001b[34m[05/13/2020 10:30:29 INFO 140382612440896] #quality_metric: host=algo-1, epoch=27, batch=0 train loss <loss>=-0.0572861805558\u001b[0m\n\u001b[34m[05/13/2020 10:30:30 INFO 140382612440896] Epoch[27] Batch[5] avg_epoch_loss=-0.056217\u001b[0m\n\u001b[34m[05/13/2020 10:30:30 INFO 140382612440896] #quality_metric: host=algo-1, epoch=27, batch=5 train loss <loss>=-0.0562166906893\u001b[0m\n\u001b[34m[05/13/2020 10:30:30 INFO 140382612440896] Epoch[27] Batch [5]#011Speed: 731.19 samples/sec#011loss=-0.056217\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] Epoch[27] Batch[10] avg_epoch_loss=-0.082458\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] #quality_metric: host=algo-1, epoch=27, batch=10 train loss <loss>=-0.113946881145\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] Epoch[27] Batch [10]#011Speed: 738.54 samples/sec#011loss=-0.113947\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] processed a total of 1285 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2276.947021484375, \"sum\": 2276.947021484375, \"min\": 2276.947021484375}}, \"EndTime\": 1589365831.263238, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365828.986229}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=564.324696281 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] #progress_metric: host=algo-1, completed 56 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] #quality_metric: host=algo-1, epoch=27, train loss <loss>=-0.0824576863511\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_895c755d-b746-4d5d-83f3-6326dfa37ff3-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 30.091047286987305, \"sum\": 30.091047286987305, \"min\": 30.091047286987305}}, \"EndTime\": 1589365831.293903, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365831.263314}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] Epoch[28] Batch[0] avg_epoch_loss=-0.045311\u001b[0m\n\u001b[34m[05/13/2020 10:30:31 INFO 140382612440896] #quality_metric: host=algo-1, epoch=28, batch=0 train loss <loss>=-0.0453113541007\u001b[0m\n\u001b[34m[05/13/2020 10:30:32 INFO 140382612440896] Epoch[28] Batch[5] avg_epoch_loss=-0.042745\u001b[0m\n\u001b[34m[05/13/2020 10:30:32 INFO 140382612440896] #quality_metric: host=algo-1, epoch=28, batch=5 train loss <loss>=-0.0427454933524\u001b[0m\n\u001b[34m[05/13/2020 10:30:32 INFO 140382612440896] Epoch[28] Batch [5]#011Speed: 726.83 samples/sec#011loss=-0.042745\u001b[0m\n\u001b[34m[05/13/2020 10:30:33 INFO 140382612440896] processed a total of 1234 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2115.0379180908203, \"sum\": 2115.0379180908203, \"min\": 2115.0379180908203}}, \"EndTime\": 1589365833.409158, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365831.293981}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:33 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=583.388663702 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:33 INFO 140382612440896] #progress_metric: host=algo-1, completed 58 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:33 INFO 140382612440896] #quality_metric: host=algo-1, epoch=28, train loss <loss>=-0.0232894606888\u001b[0m\n\u001b[34m[05/13/2020 10:30:33 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:34 INFO 140382612440896] Epoch[29] Batch[0] avg_epoch_loss=-0.081790\u001b[0m\n\u001b[34m[05/13/2020 10:30:34 INFO 140382612440896] #quality_metric: host=algo-1, epoch=29, batch=0 train loss <loss>=-0.0817899703979\u001b[0m\n\u001b[34m[05/13/2020 10:30:34 INFO 140382612440896] Epoch[29] Batch[5] avg_epoch_loss=-0.075884\u001b[0m\n\u001b[34m[05/13/2020 10:30:34 INFO 140382612440896] #quality_metric: host=algo-1, epoch=29, batch=5 train loss <loss>=-0.0758844744414\u001b[0m\n\u001b[34m[05/13/2020 10:30:34 INFO 140382612440896] Epoch[29] Batch [5]#011Speed: 730.32 samples/sec#011loss=-0.075884\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] Epoch[29] Batch[10] avg_epoch_loss=-0.063318\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] #quality_metric: host=algo-1, epoch=29, batch=10 train loss <loss>=-0.0482389759272\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] Epoch[29] Batch [10]#011Speed: 731.96 samples/sec#011loss=-0.048239\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] processed a total of 1305 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2342.456102371216, \"sum\": 2342.456102371216, \"min\": 2342.456102371216}}, \"EndTime\": 1589365835.752108, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365833.409233}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=557.081419508 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] #progress_metric: host=algo-1, completed 60 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] #quality_metric: host=algo-1, epoch=29, train loss <loss>=-0.0633183387531\u001b[0m\n\u001b[34m[05/13/2020 10:30:35 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:36 INFO 140382612440896] Epoch[30] Batch[0] avg_epoch_loss=-0.062437\u001b[0m\n\u001b[34m[05/13/2020 10:30:36 INFO 140382612440896] #quality_metric: host=algo-1, epoch=30, batch=0 train loss <loss>=-0.0624365620315\u001b[0m\n\u001b[34m[05/13/2020 10:30:37 INFO 140382612440896] Epoch[30] Batch[5] avg_epoch_loss=-0.072066\u001b[0m\n\u001b[34m[05/13/2020 10:30:37 INFO 140382612440896] #quality_metric: host=algo-1, epoch=30, batch=5 train loss <loss>=-0.0720655545592\u001b[0m\n\u001b[34m[05/13/2020 10:30:37 INFO 140382612440896] Epoch[30] Batch [5]#011Speed: 725.86 samples/sec#011loss=-0.072066\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] Epoch[30] Batch[10] avg_epoch_loss=-0.067985\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] #quality_metric: host=algo-1, epoch=30, batch=10 train loss <loss>=-0.0630872502923\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] Epoch[30] Batch [10]#011Speed: 736.14 samples/sec#011loss=-0.063087\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] processed a total of 1304 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2339.876890182495, \"sum\": 2339.876890182495, \"min\": 2339.876890182495}}, \"EndTime\": 1589365838.092483, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365835.752183}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=557.267589541 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] #progress_metric: host=algo-1, completed 62 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] #quality_metric: host=algo-1, epoch=30, train loss <loss>=-0.0679845071652\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] Epoch[31] Batch[0] avg_epoch_loss=-0.038171\u001b[0m\n\u001b[34m[05/13/2020 10:30:38 INFO 140382612440896] #quality_metric: host=algo-1, epoch=31, batch=0 train loss <loss>=-0.0381709933281\u001b[0m\n\u001b[34m[05/13/2020 10:30:39 INFO 140382612440896] Epoch[31] Batch[5] avg_epoch_loss=-0.081714\u001b[0m\n\u001b[34m[05/13/2020 10:30:39 INFO 140382612440896] #quality_metric: host=algo-1, epoch=31, batch=5 train loss <loss>=-0.0817135795951\u001b[0m\n\u001b[34m[05/13/2020 10:30:39 INFO 140382612440896] Epoch[31] Batch [5]#011Speed: 716.21 samples/sec#011loss=-0.081714\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] processed a total of 1250 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2204.22101020813, \"sum\": 2204.22101020813, \"min\": 2204.22101020813}}, \"EndTime\": 1589365840.297186, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365838.092559}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=567.062671431 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] #progress_metric: host=algo-1, completed 64 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] #quality_metric: host=algo-1, epoch=31, train loss <loss>=-0.0813482768834\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] Epoch[32] Batch[0] avg_epoch_loss=-0.123286\u001b[0m\n\u001b[34m[05/13/2020 10:30:40 INFO 140382612440896] #quality_metric: host=algo-1, epoch=32, batch=0 train loss <loss>=-0.123285926878\u001b[0m\n\u001b[34m[05/13/2020 10:30:41 INFO 140382612440896] Epoch[32] Batch[5] avg_epoch_loss=-0.101575\u001b[0m\n\u001b[34m[05/13/2020 10:30:41 INFO 140382612440896] #quality_metric: host=algo-1, epoch=32, batch=5 train loss <loss>=-0.101575067888\u001b[0m\n\u001b[34m[05/13/2020 10:30:41 INFO 140382612440896] Epoch[32] Batch [5]#011Speed: 743.94 samples/sec#011loss=-0.101575\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] processed a total of 1273 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2119.1060543060303, \"sum\": 2119.1060543060303, \"min\": 2119.1060543060303}}, \"EndTime\": 1589365842.416897, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365840.297267}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=600.693243121 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] #progress_metric: host=algo-1, completed 66 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] #quality_metric: host=algo-1, epoch=32, train loss <loss>=-0.0965427014977\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_1d3a290f-1734-4fea-9a0a-078d1c2a23d5-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.274017333984375, \"sum\": 22.274017333984375, \"min\": 22.274017333984375}}, \"EndTime\": 1589365842.439856, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365842.416968}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] Epoch[33] Batch[0] avg_epoch_loss=-0.108497\u001b[0m\n\u001b[34m[05/13/2020 10:30:42 INFO 140382612440896] #quality_metric: host=algo-1, epoch=33, batch=0 train loss <loss>=-0.108497492969\u001b[0m\n\u001b[34m[05/13/2020 10:30:43 INFO 140382612440896] Epoch[33] Batch[5] avg_epoch_loss=-0.084747\u001b[0m\n\u001b[34m[05/13/2020 10:30:43 INFO 140382612440896] #quality_metric: host=algo-1, epoch=33, batch=5 train loss <loss>=-0.0847466041644\u001b[0m\n\u001b[34m[05/13/2020 10:30:43 INFO 140382612440896] Epoch[33] Batch [5]#011Speed: 743.74 samples/sec#011loss=-0.084747\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] Epoch[33] Batch[10] avg_epoch_loss=-0.104371\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] #quality_metric: host=algo-1, epoch=33, batch=10 train loss <loss>=-0.127919696271\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] Epoch[33] Batch [10]#011Speed: 717.61 samples/sec#011loss=-0.127920\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] processed a total of 1289 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2288.723945617676, \"sum\": 2288.723945617676, \"min\": 2288.723945617676}}, \"EndTime\": 1589365844.728703, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365842.439921}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=563.168821548 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] #progress_metric: host=algo-1, completed 68 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] #quality_metric: host=algo-1, epoch=33, train loss <loss>=-0.10437073694\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:44 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_281f987f-15a1-4257-a978-1a0990c49a97-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 34.8200798034668, \"sum\": 34.8200798034668, \"min\": 34.8200798034668}}, \"EndTime\": 1589365844.764087, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365844.728778}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:45 INFO 140382612440896] Epoch[34] Batch[0] avg_epoch_loss=-0.049360\u001b[0m\n\u001b[34m[05/13/2020 10:30:45 INFO 140382612440896] #quality_metric: host=algo-1, epoch=34, batch=0 train loss <loss>=-0.0493599586189\u001b[0m\n\u001b[34m[05/13/2020 10:30:46 INFO 140382612440896] Epoch[34] Batch[5] avg_epoch_loss=-0.064794\u001b[0m\n\u001b[34m[05/13/2020 10:30:46 INFO 140382612440896] #quality_metric: host=algo-1, epoch=34, batch=5 train loss <loss>=-0.0647938493639\u001b[0m\n\u001b[34m[05/13/2020 10:30:46 INFO 140382612440896] Epoch[34] Batch [5]#011Speed: 748.09 samples/sec#011loss=-0.064794\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] Epoch[34] Batch[10] avg_epoch_loss=-0.054197\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] #quality_metric: host=algo-1, epoch=34, batch=10 train loss <loss>=-0.0414804279804\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] Epoch[34] Batch [10]#011Speed: 741.89 samples/sec#011loss=-0.041480\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] processed a total of 1309 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2312.131881713867, \"sum\": 2312.131881713867, \"min\": 2312.131881713867}}, \"EndTime\": 1589365847.076344, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365844.764153}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=566.105240517 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] #progress_metric: host=algo-1, completed 70 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] #quality_metric: host=algo-1, epoch=34, train loss <loss>=-0.0541968396442\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] Epoch[35] Batch[0] avg_epoch_loss=-0.022293\u001b[0m\n\u001b[34m[05/13/2020 10:30:47 INFO 140382612440896] #quality_metric: host=algo-1, epoch=35, batch=0 train loss <loss>=-0.0222932957113\u001b[0m\n\u001b[34m[05/13/2020 10:30:48 INFO 140382612440896] Epoch[35] Batch[5] avg_epoch_loss=-0.060317\u001b[0m\n\u001b[34m[05/13/2020 10:30:48 INFO 140382612440896] #quality_metric: host=algo-1, epoch=35, batch=5 train loss <loss>=-0.0603172015399\u001b[0m\n\u001b[34m[05/13/2020 10:30:48 INFO 140382612440896] Epoch[35] Batch [5]#011Speed: 742.07 samples/sec#011loss=-0.060317\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] Epoch[35] Batch[10] avg_epoch_loss=-0.082018\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=35, batch=10 train loss <loss>=-0.10805946216\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] Epoch[35] Batch [10]#011Speed: 728.89 samples/sec#011loss=-0.108059\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] processed a total of 1310 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2334.0070247650146, \"sum\": 2334.0070247650146, \"min\": 2334.0070247650146}}, \"EndTime\": 1589365849.410881, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365847.076457}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=561.238886511 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] #progress_metric: host=algo-1, completed 72 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=35, train loss <loss>=-0.0820182290944\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] Epoch[36] Batch[0] avg_epoch_loss=-0.005969\u001b[0m\n\u001b[34m[05/13/2020 10:30:49 INFO 140382612440896] #quality_metric: host=algo-1, epoch=36, batch=0 train loss <loss>=-0.0059692543\u001b[0m\n\u001b[34m[05/13/2020 10:30:50 INFO 140382612440896] Epoch[36] Batch[5] avg_epoch_loss=-0.071218\u001b[0m\n\u001b[34m[05/13/2020 10:30:50 INFO 140382612440896] #quality_metric: host=algo-1, epoch=36, batch=5 train loss <loss>=-0.0712176843857\u001b[0m\n\u001b[34m[05/13/2020 10:30:50 INFO 140382612440896] Epoch[36] Batch [5]#011Speed: 740.91 samples/sec#011loss=-0.071218\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] Epoch[36] Batch[10] avg_epoch_loss=-0.075411\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] #quality_metric: host=algo-1, epoch=36, batch=10 train loss <loss>=-0.0804433491081\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] Epoch[36] Batch [10]#011Speed: 698.31 samples/sec#011loss=-0.080443\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] processed a total of 1340 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2326.37095451355, \"sum\": 2326.37095451355, \"min\": 2326.37095451355}}, \"EndTime\": 1589365851.737828, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365849.410958}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=575.976696809 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] #progress_metric: host=algo-1, completed 74 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] #quality_metric: host=algo-1, epoch=36, train loss <loss>=-0.0754111683504\u001b[0m\n\u001b[34m[05/13/2020 10:30:51 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:52 INFO 140382612440896] Epoch[37] Batch[0] avg_epoch_loss=-0.042230\u001b[0m\n\u001b[34m[05/13/2020 10:30:52 INFO 140382612440896] #quality_metric: host=algo-1, epoch=37, batch=0 train loss <loss>=-0.0422302410007\u001b[0m\n\u001b[34m[05/13/2020 10:30:53 INFO 140382612440896] Epoch[37] Batch[5] avg_epoch_loss=-0.088638\u001b[0m\n\u001b[34m[05/13/2020 10:30:53 INFO 140382612440896] #quality_metric: host=algo-1, epoch=37, batch=5 train loss <loss>=-0.0886381889383\u001b[0m\n\u001b[34m[05/13/2020 10:30:53 INFO 140382612440896] Epoch[37] Batch [5]#011Speed: 740.45 samples/sec#011loss=-0.088638\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] Epoch[37] Batch[10] avg_epoch_loss=-0.079393\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] #quality_metric: host=algo-1, epoch=37, batch=10 train loss <loss>=-0.0682980820537\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] Epoch[37] Batch [10]#011Speed: 727.07 samples/sec#011loss=-0.068298\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] processed a total of 1361 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2326.1780738830566, \"sum\": 2326.1780738830566, \"min\": 2326.1780738830566}}, \"EndTime\": 1589365854.06447, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365851.737904}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=585.050262822 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] #progress_metric: host=algo-1, completed 76 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] #quality_metric: host=algo-1, epoch=37, train loss <loss>=-0.0793926858089\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] Epoch[38] Batch[0] avg_epoch_loss=-0.104801\u001b[0m\n\u001b[34m[05/13/2020 10:30:54 INFO 140382612440896] #quality_metric: host=algo-1, epoch=38, batch=0 train loss <loss>=-0.10480145365\u001b[0m\n\u001b[34m[05/13/2020 10:30:55 INFO 140382612440896] Epoch[38] Batch[5] avg_epoch_loss=-0.103159\u001b[0m\n\u001b[34m[05/13/2020 10:30:55 INFO 140382612440896] #quality_metric: host=algo-1, epoch=38, batch=5 train loss <loss>=-0.103158875679\u001b[0m\n\u001b[34m[05/13/2020 10:30:55 INFO 140382612440896] Epoch[38] Batch [5]#011Speed: 741.28 samples/sec#011loss=-0.103159\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] processed a total of 1251 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2088.9551639556885, \"sum\": 2088.9551639556885, \"min\": 2088.9551639556885}}, \"EndTime\": 1589365856.154038, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365854.064549}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=598.83479424 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] #progress_metric: host=algo-1, completed 78 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] #quality_metric: host=algo-1, epoch=38, train loss <loss>=-0.112328095362\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_04ccd820-c75d-48bc-bb4c-0406bea54690-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.050857543945312, \"sum\": 22.050857543945312, \"min\": 22.050857543945312}}, \"EndTime\": 1589365856.176787, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365856.154105}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] Epoch[39] Batch[0] avg_epoch_loss=-0.122241\u001b[0m\n\u001b[34m[05/13/2020 10:30:56 INFO 140382612440896] #quality_metric: host=algo-1, epoch=39, batch=0 train loss <loss>=-0.122240789235\u001b[0m\n\u001b[34m[05/13/2020 10:30:57 INFO 140382612440896] Epoch[39] Batch[5] avg_epoch_loss=-0.126664\u001b[0m\n\u001b[34m[05/13/2020 10:30:57 INFO 140382612440896] #quality_metric: host=algo-1, epoch=39, batch=5 train loss <loss>=-0.126664002736\u001b[0m\n\u001b[34m[05/13/2020 10:30:57 INFO 140382612440896] Epoch[39] Batch [5]#011Speed: 723.34 samples/sec#011loss=-0.126664\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] Epoch[39] Batch[10] avg_epoch_loss=-0.086272\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] #quality_metric: host=algo-1, epoch=39, batch=10 train loss <loss>=-0.0378016546369\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] Epoch[39] Batch [10]#011Speed: 738.98 samples/sec#011loss=-0.037802\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] processed a total of 1305 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2299.730062484741, \"sum\": 2299.730062484741, \"min\": 2299.730062484741}}, \"EndTime\": 1589365858.476635, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365856.17685}\n\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=567.432725546 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] #progress_metric: host=algo-1, completed 80 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] #quality_metric: host=algo-1, epoch=39, train loss <loss>=-0.0862720263275\u001b[0m\n\u001b[34m[05/13/2020 10:30:58 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:30:59 INFO 140382612440896] Epoch[40] Batch[0] avg_epoch_loss=-0.103539\u001b[0m\n\u001b[34m[05/13/2020 10:30:59 INFO 140382612440896] #quality_metric: host=algo-1, epoch=40, batch=0 train loss <loss>=-0.103538669646\u001b[0m\n\u001b[34m[05/13/2020 10:30:59 INFO 140382612440896] Epoch[40] Batch[5] avg_epoch_loss=-0.111421\u001b[0m\n\u001b[34m[05/13/2020 10:30:59 INFO 140382612440896] #quality_metric: host=algo-1, epoch=40, batch=5 train loss <loss>=-0.111421365291\u001b[0m\n\u001b[34m[05/13/2020 10:30:59 INFO 140382612440896] Epoch[40] Batch [5]#011Speed: 744.61 samples/sec#011loss=-0.111421\u001b[0m\n\u001b[34m[05/13/2020 10:31:00 INFO 140382612440896] processed a total of 1246 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2099.642038345337, \"sum\": 2099.642038345337, \"min\": 2099.642038345337}}, \"EndTime\": 1589365860.57674, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365858.476704}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:00 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=593.40423432 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:00 INFO 140382612440896] #progress_metric: host=algo-1, completed 82 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:00 INFO 140382612440896] #quality_metric: host=algo-1, epoch=40, train loss <loss>=-0.110480416566\u001b[0m\n\u001b[34m[05/13/2020 10:31:00 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:01 INFO 140382612440896] Epoch[41] Batch[0] avg_epoch_loss=-0.167133\u001b[0m\n\u001b[34m[05/13/2020 10:31:01 INFO 140382612440896] #quality_metric: host=algo-1, epoch=41, batch=0 train loss <loss>=-0.167132794857\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] Epoch[41] Batch[5] avg_epoch_loss=-0.138516\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] #quality_metric: host=algo-1, epoch=41, batch=5 train loss <loss>=-0.138516493142\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] Epoch[41] Batch [5]#011Speed: 737.43 samples/sec#011loss=-0.138516\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] Epoch[41] Batch[10] avg_epoch_loss=-0.101241\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] #quality_metric: host=algo-1, epoch=41, batch=10 train loss <loss>=-0.0565112575889\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] Epoch[41] Batch [10]#011Speed: 722.31 samples/sec#011loss=-0.056511\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] processed a total of 1295 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2341.8381214141846, \"sum\": 2341.8381214141846, \"min\": 2341.8381214141846}}, \"EndTime\": 1589365862.919197, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365860.57681}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=552.959708108 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] #progress_metric: host=algo-1, completed 84 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] #quality_metric: host=algo-1, epoch=41, train loss <loss>=-0.101241386072\u001b[0m\n\u001b[34m[05/13/2020 10:31:02 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:03 INFO 140382612440896] Epoch[42] Batch[0] avg_epoch_loss=-0.097393\u001b[0m\n\u001b[34m[05/13/2020 10:31:03 INFO 140382612440896] #quality_metric: host=algo-1, epoch=42, batch=0 train loss <loss>=-0.0973930507898\u001b[0m\n\u001b[34m[05/13/2020 10:31:04 INFO 140382612440896] Epoch[42] Batch[5] avg_epoch_loss=-0.118158\u001b[0m\n\u001b[34m[05/13/2020 10:31:04 INFO 140382612440896] #quality_metric: host=algo-1, epoch=42, batch=5 train loss <loss>=-0.118157595396\u001b[0m\n\u001b[34m[05/13/2020 10:31:04 INFO 140382612440896] Epoch[42] Batch [5]#011Speed: 722.39 samples/sec#011loss=-0.118158\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] Epoch[42] Batch[10] avg_epoch_loss=-0.096109\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] #quality_metric: host=algo-1, epoch=42, batch=10 train loss <loss>=-0.0696504056454\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] Epoch[42] Batch [10]#011Speed: 738.72 samples/sec#011loss=-0.069650\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] processed a total of 1281 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2328.733205795288, \"sum\": 2328.733205795288, \"min\": 2328.733205795288}}, \"EndTime\": 1589365865.248419, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365862.919267}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=550.044386606 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] #progress_metric: host=algo-1, completed 86 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] #quality_metric: host=algo-1, epoch=42, train loss <loss>=-0.0961088727821\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] Epoch[43] Batch[0] avg_epoch_loss=-0.101425\u001b[0m\n\u001b[34m[05/13/2020 10:31:05 INFO 140382612440896] #quality_metric: host=algo-1, epoch=43, batch=0 train loss <loss>=-0.101424530149\u001b[0m\n\u001b[34m[05/13/2020 10:31:06 INFO 140382612440896] Epoch[43] Batch[5] avg_epoch_loss=-0.120040\u001b[0m\n\u001b[34m[05/13/2020 10:31:06 INFO 140382612440896] #quality_metric: host=algo-1, epoch=43, batch=5 train loss <loss>=-0.120039508988\u001b[0m\n\u001b[34m[05/13/2020 10:31:06 INFO 140382612440896] Epoch[43] Batch [5]#011Speed: 739.64 samples/sec#011loss=-0.120040\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] processed a total of 1231 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2085.263967514038, \"sum\": 2085.263967514038, \"min\": 2085.263967514038}}, \"EndTime\": 1589365867.334328, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365865.24855}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=590.299257437 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] #progress_metric: host=algo-1, completed 88 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] #quality_metric: host=algo-1, epoch=43, train loss <loss>=-0.113350253925\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_e0fa4943-36b0-4963-9082-63da248f9637-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 32.752037048339844, \"sum\": 32.752037048339844, \"min\": 32.752037048339844}}, \"EndTime\": 1589365867.367729, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365867.334409}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] Epoch[44] Batch[0] avg_epoch_loss=-0.147835\u001b[0m\n\u001b[34m[05/13/2020 10:31:07 INFO 140382612440896] #quality_metric: host=algo-1, epoch=44, batch=0 train loss <loss>=-0.147834822536\u001b[0m\n\u001b[34m[05/13/2020 10:31:08 INFO 140382612440896] Epoch[44] Batch[5] avg_epoch_loss=-0.139445\u001b[0m\n\u001b[34m[05/13/2020 10:31:08 INFO 140382612440896] #quality_metric: host=algo-1, epoch=44, batch=5 train loss <loss>=-0.139444820583\u001b[0m\n\u001b[34m[05/13/2020 10:31:08 INFO 140382612440896] Epoch[44] Batch [5]#011Speed: 732.39 samples/sec#011loss=-0.139445\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] Epoch[44] Batch[10] avg_epoch_loss=-0.147891\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] #quality_metric: host=algo-1, epoch=44, batch=10 train loss <loss>=-0.158026784658\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] Epoch[44] Batch [10]#011Speed: 743.71 samples/sec#011loss=-0.158027\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] processed a total of 1287 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2309.7100257873535, \"sum\": 2309.7100257873535, \"min\": 2309.7100257873535}}, \"EndTime\": 1589365869.677596, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365867.367818}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=557.173982105 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] #progress_metric: host=algo-1, completed 90 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] #quality_metric: host=algo-1, epoch=44, train loss <loss>=-0.14789116789\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:31:09 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_3f6b6342-b80c-48fe-b250-2ff00168643b-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 22.49622344970703, \"sum\": 22.49622344970703, \"min\": 22.49622344970703}}, \"EndTime\": 1589365869.700772, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365869.677717}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:10 INFO 140382612440896] Epoch[45] Batch[0] avg_epoch_loss=-0.116227\u001b[0m\n\u001b[34m[05/13/2020 10:31:10 INFO 140382612440896] #quality_metric: host=algo-1, epoch=45, batch=0 train loss <loss>=-0.116227336228\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] Epoch[45] Batch[5] avg_epoch_loss=-0.152262\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] #quality_metric: host=algo-1, epoch=45, batch=5 train loss <loss>=-0.152261807273\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] Epoch[45] Batch [5]#011Speed: 740.36 samples/sec#011loss=-0.152262\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] processed a total of 1240 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2079.4689655303955, \"sum\": 2079.4689655303955, \"min\": 2079.4689655303955}}, \"EndTime\": 1589365871.78038, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365869.700849}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=596.274843271 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] #progress_metric: host=algo-1, completed 92 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] #quality_metric: host=algo-1, epoch=45, train loss <loss>=-0.153892410547\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:31:11 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_67d6bfbe-1dac-47a5-985f-c0e43dbdd446-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 28.723955154418945, \"sum\": 28.723955154418945, \"min\": 28.723955154418945}}, \"EndTime\": 1589365871.809637, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365871.780454}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:12 INFO 140382612440896] Epoch[46] Batch[0] avg_epoch_loss=-0.201754\u001b[0m\n\u001b[34m[05/13/2020 10:31:12 INFO 140382612440896] #quality_metric: host=algo-1, epoch=46, batch=0 train loss <loss>=-0.201753750443\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] Epoch[46] Batch[5] avg_epoch_loss=-0.173621\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] #quality_metric: host=algo-1, epoch=46, batch=5 train loss <loss>=-0.173620510846\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] Epoch[46] Batch [5]#011Speed: 734.58 samples/sec#011loss=-0.173621\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] processed a total of 1264 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2099.2650985717773, \"sum\": 2099.2650985717773, \"min\": 2099.2650985717773}}, \"EndTime\": 1589365873.909034, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365871.809709}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=602.067062291 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] #progress_metric: host=algo-1, completed 94 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] #quality_metric: host=algo-1, epoch=46, train loss <loss>=-0.17895764485\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] best epoch loss so far\u001b[0m\n\u001b[34m[05/13/2020 10:31:13 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/state_619a8869-d2b6-4c82-b080-805d2affd957-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"state.serialize.time\": {\"count\": 1, \"max\": 34.101009368896484, \"sum\": 34.101009368896484, \"min\": 34.101009368896484}}, \"EndTime\": 1589365873.943747, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365873.909167}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:14 INFO 140382612440896] Epoch[47] Batch[0] avg_epoch_loss=-0.150843\u001b[0m\n\u001b[34m[05/13/2020 10:31:14 INFO 140382612440896] #quality_metric: host=algo-1, epoch=47, batch=0 train loss <loss>=-0.150843143463\u001b[0m\n\u001b[34m[05/13/2020 10:31:15 INFO 140382612440896] Epoch[47] Batch[5] avg_epoch_loss=-0.159525\u001b[0m\n\u001b[34m[05/13/2020 10:31:15 INFO 140382612440896] #quality_metric: host=algo-1, epoch=47, batch=5 train loss <loss>=-0.159524808327\u001b[0m\n\u001b[34m[05/13/2020 10:31:15 INFO 140382612440896] Epoch[47] Batch [5]#011Speed: 740.72 samples/sec#011loss=-0.159525\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] Epoch[47] Batch[10] avg_epoch_loss=-0.175258\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] #quality_metric: host=algo-1, epoch=47, batch=10 train loss <loss>=-0.194137874246\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] Epoch[47] Batch [10]#011Speed: 750.05 samples/sec#011loss=-0.194138\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] processed a total of 1286 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2305.495023727417, \"sum\": 2305.495023727417, \"min\": 2305.495023727417}}, \"EndTime\": 1589365876.24936, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365873.943807}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=557.770898099 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] #progress_metric: host=algo-1, completed 96 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] #quality_metric: host=algo-1, epoch=47, train loss <loss>=-0.175258020108\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] Epoch[48] Batch[0] avg_epoch_loss=-0.060925\u001b[0m\n\u001b[34m[05/13/2020 10:31:16 INFO 140382612440896] #quality_metric: host=algo-1, epoch=48, batch=0 train loss <loss>=-0.0609254576266\u001b[0m\n\u001b[34m[05/13/2020 10:31:17 INFO 140382612440896] Epoch[48] Batch[5] avg_epoch_loss=-0.101799\u001b[0m\n\u001b[34m[05/13/2020 10:31:17 INFO 140382612440896] #quality_metric: host=algo-1, epoch=48, batch=5 train loss <loss>=-0.101798729971\u001b[0m\n\u001b[34m[05/13/2020 10:31:17 INFO 140382612440896] Epoch[48] Batch [5]#011Speed: 713.27 samples/sec#011loss=-0.101799\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] Epoch[48] Batch[10] avg_epoch_loss=-0.134969\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] #quality_metric: host=algo-1, epoch=48, batch=10 train loss <loss>=-0.174773459136\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] Epoch[48] Batch [10]#011Speed: 741.76 samples/sec#011loss=-0.174773\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] processed a total of 1286 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2318.8579082489014, \"sum\": 2318.8579082489014, \"min\": 2318.8579082489014}}, \"EndTime\": 1589365878.568682, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365876.249437}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=554.553934499 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] #progress_metric: host=algo-1, completed 98 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] #quality_metric: host=algo-1, epoch=48, train loss <loss>=-0.13496906141\u001b[0m\n\u001b[34m[05/13/2020 10:31:18 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:19 INFO 140382612440896] Epoch[49] Batch[0] avg_epoch_loss=-0.153256\u001b[0m\n\u001b[34m[05/13/2020 10:31:19 INFO 140382612440896] #quality_metric: host=algo-1, epoch=49, batch=0 train loss <loss>=-0.153255745769\u001b[0m\n\u001b[34m[05/13/2020 10:31:19 INFO 140382612440896] Epoch[49] Batch[5] avg_epoch_loss=-0.157532\u001b[0m\n\u001b[34m[05/13/2020 10:31:19 INFO 140382612440896] #quality_metric: host=algo-1, epoch=49, batch=5 train loss <loss>=-0.157532066107\u001b[0m\n\u001b[34m[05/13/2020 10:31:19 INFO 140382612440896] Epoch[49] Batch [5]#011Speed: 734.51 samples/sec#011loss=-0.157532\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] processed a total of 1257 examples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"update.time\": {\"count\": 1, \"max\": 2090.8098220825195, \"sum\": 2090.8098220825195, \"min\": 2090.8098220825195}}, \"EndTime\": 1589365880.660084, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365878.568766}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] #throughput_metric: host=algo-1, train throughput=601.171881489 records/second\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] #progress_metric: host=algo-1, completed 100 % of epochs\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] #quality_metric: host=algo-1, epoch=49, train loss <loss>=-0.173314021528\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] loss did not improve\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Final loss: -0.17895764485 (occurred at epoch 46)\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] #quality_metric: host=algo-1, train final_loss <loss>=-0.17895764485\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Worker algo-1 finished training.\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 WARNING 140382612440896] wait_for_all_workers will not sync workers since the kv store is not running distributed\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] All workers finished. Serializing model for prediction.\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"get_graph.time\": {\"count\": 1, \"max\": 184.5688819885254, \"sum\": 184.5688819885254, \"min\": 184.5688819885254}}, \"EndTime\": 1589365880.84567, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365880.660154}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Number of GPUs being used: 0\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"finalize.time\": {\"count\": 1, \"max\": 263.0009651184082, \"sum\": 263.0009651184082, \"min\": 263.0009651184082}}, \"EndTime\": 1589365880.924069, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365880.845735}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Serializing to /opt/ml/model/model_algo-1\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Saved checkpoint to \"/opt/ml/model/model_algo-1-0000.params\"\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"model.serialize.time\": {\"count\": 1, \"max\": 12.99905776977539, \"sum\": 12.99905776977539, \"min\": 12.99905776977539}}, \"EndTime\": 1589365880.93718, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365880.924132}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Successfully serialized the model for prediction.\u001b[0m\n\u001b[34m[05/13/2020 10:31:20 INFO 140382612440896] Evaluating model accuracy on testset using 100 samples\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"model.bind.time\": {\"count\": 1, \"max\": 0.04291534423828125, \"sum\": 0.04291534423828125, \"min\": 0.04291534423828125}}, \"EndTime\": 1589365880.937973, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365880.937242}\n\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"model.score.time\": {\"count\": 1, \"max\": 4174.936056137085, \"sum\": 4174.936056137085, \"min\": 4174.936056137085}}, \"EndTime\": 1589365885.112857, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365880.938032}\n\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, RMSE): 0.355022096707\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, mean_absolute_QuantileLoss): 18.671423994170297\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, mean_wQuantileLoss): 0.14619091857801914\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.1]): 0.07771903523175669\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.2]): 0.125737096078515\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.3]): 0.15330517326593907\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.4]): 0.1733076178788926\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.5]): 0.18704738450336203\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.6]): 0.18675151888145858\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.7]): 0.17284284787338977\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.8]): 0.14399965026315908\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #test_score (algo-1, wQuantileLoss[0.9]): 0.09500794322569937\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #quality_metric: host=algo-1, test mean_wQuantileLoss <loss>=0.146190918578\u001b[0m\n\u001b[34m[05/13/2020 10:31:25 INFO 140382612440896] #quality_metric: host=algo-1, test RMSE <loss>=0.355022096707\u001b[0m\n\u001b[34m#metrics {\"Metrics\": {\"totaltime\": {\"count\": 1, \"max\": 117070.307970047, \"sum\": 117070.307970047, \"min\": 117070.307970047}, \"setuptime\": {\"count\": 1, \"max\": 8.718013763427734, \"sum\": 8.718013763427734, \"min\": 8.718013763427734}}, \"EndTime\": 1589365885.133846, \"Dimensions\": {\"Host\": \"algo-1\", \"Operation\": \"training\", \"Algorithm\": \"AWS/DeepAR\"}, \"StartTime\": 1589365885.112923}\n\u001b[0m\n\n2020-05-13 10:31:34 Uploading - Uploading generated training model\n2020-05-13 10:31:34 Completed - Training job completed\nTraining seconds: 192\nBillable seconds: 192\nCPU times: user 806 ms, sys: 22.8 ms, total: 829 ms\nWall time: 5min 43s\n"
]
],
[
[
"## Deploy and Create a Predictor\n\nNow that we have trained a model, we can use it to perform predictions by deploying it to a predictor endpoint.\n\nRemember to **delete the endpoint** at the end of this notebook. A cell at the very bottom of this notebook will be provided, but it is always good to keep, front-of-mind.",
"_____no_output_____"
]
],
[
[
"%%time\n\n# create a predictor\npredictor = estimator.deploy(\n initial_instance_count=1,\n instance_type='ml.t2.medium',\n content_type=\"application/json\" # specify that it will accept/produce JSON\n)",
"---------------!CPU times: user 259 ms, sys: 4.34 ms, total: 263 ms\nWall time: 7min 31s\n"
]
],
[
[
"---\n# Generating Predictions\n\nAccording to the [inference format](https://docs.aws.amazon.com/sagemaker/latest/dg/deepar-in-formats.html) for DeepAR, the `predictor` expects to see input data in a JSON format, with the following keys:\n* **instances**: A list of JSON-formatted time series that should be forecast by the model.\n* **configuration** (optional): A dictionary of configuration information for the type of response desired by the request.\n\nWithin configuration the following keys can be configured:\n* **num_samples**: An integer specifying the number of samples that the model generates when making a probabilistic prediction.\n* **output_types**: A list specifying the type of response. We'll ask for **quantiles**, which look at the list of num_samples generated by the model, and generate [quantile estimates](https://en.wikipedia.org/wiki/Quantile) for each time point based on these values.\n* **quantiles**: A list that specified which quantiles estimates are generated and returned in the response.\n\n\nBelow is an example of what a JSON query to a DeepAR model endpoint might look like.\n\n```\n{\n \"instances\": [\n { \"start\": \"2009-11-01 00:00:00\", \"target\": [4.0, 10.0, 50.0, 100.0, 113.0] },\n { \"start\": \"1999-01-30\", \"target\": [2.0, 1.0] }\n ],\n \"configuration\": {\n \"num_samples\": 50,\n \"output_types\": [\"quantiles\"],\n \"quantiles\": [\"0.5\", \"0.9\"]\n }\n}\n```\n\n\n## JSON Prediction Request\n\nThe code below accepts a **list** of time series as input and some configuration parameters. It then formats that series into a JSON instance and converts the input into an appropriately formatted JSON_input.",
"_____no_output_____"
]
],
[
[
"def json_predictor_input(input_ts, num_samples=50, quantiles=['0.1', '0.5', '0.9']):\n '''Accepts a list of input time series and produces a formatted input.\n :input_ts: An list of input time series.\n :num_samples: Number of samples to calculate metrics with.\n :quantiles: A list of quantiles to return in the predicted output.\n :return: The JSON-formatted input.\n '''\n # request data is made of JSON objects (instances)\n # and an output configuration that details the type of data/quantiles we want\n \n instances = []\n for k in range(len(input_ts)):\n # get JSON objects for input time series\n instances.append(series_to_json_obj(input_ts[k]))\n\n # specify the output quantiles and samples\n configuration = {\"num_samples\": num_samples, \n \"output_types\": [\"quantiles\"], \n \"quantiles\": quantiles}\n\n request_data = {\"instances\": instances, \n \"configuration\": configuration}\n\n json_request = json.dumps(request_data).encode('utf-8')\n \n return json_request",
"_____no_output_____"
]
],
[
[
"### Get a Prediction\n\nWe can then use this function to get a prediction for a formatted time series!\n\nIn the next cell, I'm getting an input time series and known target, and passing the formatted input into the predictor endpoint to get a resultant prediction.",
"_____no_output_____"
]
],
[
[
"# get all input and target (test) time series\ninput_ts = time_series_training\ntarget_ts = time_series\n\n# get formatted input time series\njson_input_ts = json_predictor_input(input_ts)\n\n# get the prediction from the predictor\njson_prediction = predictor.predict(json_input_ts)\n\nprint(json_prediction)",
"b'{\"predictions\":[{\"quantiles\":{\"0.1\":[1.0026136637,1.081076622,1.3667051792,1.1711808443,1.0109467506,1.2457735538,1.3729652166,1.2885881662,1.1347281933,1.1827311516,1.0506105423,0.9972769022,1.1678456068,0.9720013738,1.151907444,0.8764561415,0.9251003265,0.8382130861,0.6767332554,0.8609979153,0.7878181338,0.5686401129,0.7179458141,0.6427162886,0.7480871677,0.5571811199,0.7568116188,0.7451438904,0.8316267133,0.5637689829],\"0.9\":[1.8645830154,1.6136842966,1.8777201176,1.5980273485,1.586761713,1.9140461683,2.0562529564,1.9208176136,1.5949819088,1.8232643604,1.6936215162,1.670081377,1.9476565123,1.8908939362,1.8992513418,1.6357431412,1.7412010431,1.8597700596,1.7427530289,1.9231746197,2.0208640099,1.8791079521,1.5770751238,1.6604226828,1.5388629436,1.3741146326,1.7850370407,1.9282718897,2.0410134792,1.5520406961],\"0.5\":[1.4265850782,1.3108716011,1.6138243675,1.4024649858,1.2970371246,1.6086105108,1.7178781033,1.6470944881,1.3511847258,1.5417058468,1.4374513626,1.3587596416,1.5214468241,1.4698103666,1.428828001,1.2291196585,1.4179273844,1.270057559,1.2146611214,1.3048044443,1.4095323086,1.3453462124,1.1249461174,1.1484526396,1.102236867,0.9823955894,1.2399948835,1.3260910511,1.1806399822,1.0872488022]}},{\"quantiles\":{\"0.1\":[1.0777281523,1.1495184898,1.0764914751,0.9647977352,1.2535099983,1.3165515661,1.0636926889,1.0921618938,1.2108998299,1.0488165617,1.0497514009,1.1651301384,1.3282637596,0.9957325459,1.0787723064,0.9363592863,1.0238156319,0.9946718216,1.1999300718,1.0442993641,0.6812716126,0.9332908988,0.8179636598,0.6549167037,0.6784488559,0.5183277726,0.4712751508,0.5651699901,0.6874364614,0.5522155166],\"0.9\":[1.6109440327,1.7669690847,1.662694335,1.6433532238,2.029613018,2.1281456947,1.5539854765,1.6947711706,1.8066313267,1.8988884687,1.9287501574,2.2598965168,2.3059532642,1.6825387478,1.6980576515,1.66350317,1.7514383793,1.7307454348,2.1252789497,2.0756313801,1.7149684429,1.7464799881,1.6410441399,1.778859973,1.6773813963,1.6165577173,1.9361001253,1.3969168663,1.5597472191,1.5975737572],\"0.5\":[1.2881650925,1.3701171875,1.3607953787,1.2264208794,1.6541969776,1.6387724876,1.3162523508,1.4893579483,1.4833191633,1.499557972,1.4416195154,1.5806183815,1.6859767437,1.4673388004,1.3673807383,1.3453086615,1.4063556194,1.3344279528,1.6525703669,1.5301046371,1.2617179155,1.2888580561,1.1437391043,1.207824707,1.1977437735,1.1131711006,1.2311378717,1.0559087992,1.0062556267,1.1630951166]}},{\"quantiles\":{\"0.1\":[1.0481187105,0.8132203221,0.7876354456,1.1233110428,1.0282701254,0.9654512405,0.9786347151,1.1780645847,0.9972060919,1.1013047695,1.317784667,1.1177604198,0.888450861,0.9546551704,1.096114397,0.9778335094,0.845166862,1.0342233181,1.0251860619,0.7783746719,0.8259630203,0.6378747821,0.8206300735,0.8874892592,0.751517415,0.7685925364,0.5855062008,0.6851963997,0.7650864124,0.7875103951],\"0.9\":[1.6698682308,1.5900255442,1.7294301987,2.1714744568,2.264452219,1.4892622232,1.6853501797,1.8920325041,1.5509400368,1.7846522331,2.1634893417,1.9543870687,1.4884784222,1.5769213438,1.7715735435,1.5906778574,1.6897567511,1.858669281,1.9781782627,1.5525105,1.6547709703,1.7690329552,1.5374096632,1.5592378378,1.8052735329,2.1049304008,1.508680582,1.6624677181,1.6968944073,1.6559429169],\"0.5\":[1.3668756485,1.2035001516,1.1846226454,1.6044806242,1.5284872055,1.2611374855,1.3843978643,1.5034894943,1.3618115187,1.3404150009,1.6989277601,1.5380866528,1.2366991043,1.3769735098,1.4040151834,1.2329754829,1.3857285976,1.5065131187,1.3298882246,1.1678228378,1.2057222128,1.2171969414,1.106808424,1.1375465393,1.2978264093,1.444720149,1.0737991333,1.254685998,1.2616313696,1.101059556]}}]}'\n"
]
],
[
[
"## Decoding Predictions\n\nThe predictor returns JSON-formatted prediction, and so we need to extract the predictions and quantile data that we want for visualizing the result. The function below, reads in a JSON-formatted prediction and produces a list of predictions in each quantile.",
"_____no_output_____"
]
],
[
[
"# helper function to decode JSON prediction\ndef decode_prediction(prediction, encoding='utf-8'):\n '''Accepts a JSON prediction and returns a list of prediction data.\n '''\n prediction_data = json.loads(prediction.decode(encoding))\n prediction_list = []\n for k in range(len(prediction_data['predictions'])):\n prediction_list.append(pd.DataFrame(data=prediction_data['predictions'][k]['quantiles']))\n return prediction_list\n",
"_____no_output_____"
],
[
"# get quantiles/predictions\nprediction_list = decode_prediction(json_prediction)\n\n# should get a list of 30 predictions \n# with corresponding quantile values\nprint(prediction_list[0])",
" 0.1 0.9 0.5\n0 1.002614 1.864583 1.426585\n1 1.081077 1.613684 1.310872\n2 1.366705 1.877720 1.613824\n3 1.171181 1.598027 1.402465\n4 1.010947 1.586762 1.297037\n5 1.245774 1.914046 1.608611\n6 1.372965 2.056253 1.717878\n7 1.288588 1.920818 1.647094\n8 1.134728 1.594982 1.351185\n9 1.182731 1.823264 1.541706\n10 1.050611 1.693622 1.437451\n11 0.997277 1.670081 1.358760\n12 1.167846 1.947657 1.521447\n13 0.972001 1.890894 1.469810\n14 1.151907 1.899251 1.428828\n15 0.876456 1.635743 1.229120\n16 0.925100 1.741201 1.417927\n17 0.838213 1.859770 1.270058\n18 0.676733 1.742753 1.214661\n19 0.860998 1.923175 1.304804\n20 0.787818 2.020864 1.409532\n21 0.568640 1.879108 1.345346\n22 0.717946 1.577075 1.124946\n23 0.642716 1.660423 1.148453\n24 0.748087 1.538863 1.102237\n25 0.557181 1.374115 0.982396\n26 0.756812 1.785037 1.239995\n27 0.745144 1.928272 1.326091\n28 0.831627 2.041013 1.180640\n29 0.563769 1.552041 1.087249\n"
]
],
[
[
"## Display the Results!\n\nThe quantile data will give us all we need to see the results of our prediction.\n* Quantiles 0.1 and 0.9 represent higher and lower bounds for the predicted values.\n* Quantile 0.5 represents the median of all sample predictions.\n",
"_____no_output_____"
]
],
[
[
"# display the prediction median against the actual data\ndef display_quantiles(prediction_list, target_ts=None):\n # show predictions for all input ts\n for k in range(len(prediction_list)):\n plt.figure(figsize=(12,6))\n # get the target month of data\n if target_ts is not None:\n target = target_ts[k][-prediction_length:]\n plt.plot(range(len(target)), target, label='target')\n # get the quantile values at 10 and 90%\n p10 = prediction_list[k]['0.1']\n p90 = prediction_list[k]['0.9']\n # fill the 80% confidence interval\n plt.fill_between(p10.index, p10, p90, color='y', alpha=0.5, label='80% confidence interval')\n # plot the median prediction line\n prediction_list[k]['0.5'].plot(label='prediction median')\n plt.legend()\n plt.show()",
"_____no_output_____"
],
[
"# display predictions\ndisplay_quantiles(prediction_list, target_ts)",
"_____no_output_____"
]
],
[
[
"## Predicting the Future\n\nRecall that we did not give our model any data about 2010, but let's see if it can predict the energy consumption given **no target**, only a known start date!\n\n### EXERCISE: Format a request for a \"future\" prediction\n\nCreate a formatted input to send to the deployed `predictor` passing in my usual parameters for \"configuration\". The \"instances\" will, in this case, just be one instance, defined by the following:\n* **start**: The start time will be time stamp that you specify. To predict the first 30 days of 2010, start on Jan. 1st, '2010-01-01'.\n* **target**: The target will be an empty list because this year has no, complete associated time series; we specifically withheld that information from our model, for testing purposes.\n```\n{\"start\": start_time, \"target\": []} # empty target\n```",
"_____no_output_____"
]
],
[
[
"# Starting my prediction at the beginning of 2010\nstart_date = '2010-01-01'\ntimestamp = '00:00:00'\n\n# formatting start_date\nstart_time = start_date +' '+ timestamp\n\n# format the request_data\n# with \"instances\" and \"configuration\"\nrequest_data = {\"instances\": [{\"start\": start_time, \"target\": []}],\n \"configuration\": {\"num_samples\": 50,\n \"output_types\": [\"quantiles\"],\n \"quantiles\": ['0.1', '0.5', '0.9']}\n }\n\n\n# create JSON input\njson_input = json.dumps(request_data).encode('utf-8')\n\nprint('Requesting prediction for '+start_time)",
"Requesting prediction for 2010-01-01 00:00:00\n"
]
],
[
[
"Then get and decode the prediction response, as usual.",
"_____no_output_____"
]
],
[
[
"# get prediction response\njson_prediction = predictor.predict(json_input)\n\nprediction_2010 = decode_prediction(json_prediction)\n",
"_____no_output_____"
]
],
[
[
"Finally, I'll compare the predictions to a known target sequence. This target will come from a time series for the 2010 data, which I'm creating below.",
"_____no_output_____"
]
],
[
[
"# create 2010 time series\nts_2010 = []\n# get global consumption data\n# index 1112 is where the 2011 data starts\ndata_2010 = mean_power_df.values[1112:]\n\nindex = pd.DatetimeIndex(start=start_date, periods=len(data_2010), freq='D')\nts_2010.append(pd.Series(data=data_2010, index=index))\n",
"/home/ec2-user/anaconda3/envs/amazonei_mxnet_p36/lib/python3.6/site-packages/ipykernel/__main__.py:7: FutureWarning: Creating a DatetimeIndex by passing range endpoints is deprecated. Use `pandas.date_range` instead.\n"
],
[
"# range of actual data to compare\nstart_idx=0 # days since Jan 1st 2010\nend_idx=start_idx+prediction_length\n\n# get target data\ntarget_2010_ts = [ts_2010[0][start_idx:end_idx]]\n\n# display predictions\ndisplay_quantiles(prediction_2010, target_2010_ts)",
"_____no_output_____"
]
],
[
[
"## Delete the Endpoint\n\nTry your code out on different time series. You may want to tweak your DeepAR hyperparameters and see if you can improve the performance of this predictor.\n\nWhen you're done with evaluating the predictor (any predictor), make sure to delete the endpoint.",
"_____no_output_____"
]
],
[
[
"## TODO: delete the endpoint\npredictor.delete_endpoint()",
"_____no_output_____"
]
],
[
[
"## Conclusion\n\nNow you've seen one complex but far-reaching method for time series forecasting. You should have the skills you need to apply the DeepAR model to data that interests you!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec6f9a1c1054dea833e966d02c614e7e169cab0d | 584,346 | ipynb | Jupyter Notebook | 02 Supervised Learning - Regression/Week 5 - Intro to Supervised Learning - Linear Regression/.ipynb_checkpoints/02 Practice Exercise - Solution-checkpoint.ipynb | senthilprabhut/DataScience | 82556b9ef1f105c0709c1eb8b13e51dddce74851 | [
"Apache-2.0"
]
| null | null | null | 02 Supervised Learning - Regression/Week 5 - Intro to Supervised Learning - Linear Regression/.ipynb_checkpoints/02 Practice Exercise - Solution-checkpoint.ipynb | senthilprabhut/DataScience | 82556b9ef1f105c0709c1eb8b13e51dddce74851 | [
"Apache-2.0"
]
| null | null | null | 02 Supervised Learning - Regression/Week 5 - Intro to Supervised Learning - Linear Regression/.ipynb_checkpoints/02 Practice Exercise - Solution-checkpoint.ipynb | senthilprabhut/DataScience | 82556b9ef1f105c0709c1eb8b13e51dddce74851 | [
"Apache-2.0"
]
| null | null | null | 316.033532 | 112,132 | 0.911054 | [
[
[
"# Practice Exercise - Linear Regression",
"_____no_output_____"
],
[
"### Problem Statement\n\nThe problem at hand is to predict the housing prices of a town or a suburb based on the features of the locality provided to us. In the process, we need to identify the most important features in the dataset. We need to employ techniques of data preprocessing and build a linear regression model that predicts the prices for us. \n\n### Data Information\n\nEach record in the database describes a Boston suburb or town. The data was drawn from the Boston Standard Metropolitan Statistical Area (SMSA) in 1970. Detailed attribute information can be found below-\n\nAttribute Information (in order):\n- CRIM: per capita crime rate by town\n- ZN: proportion of residential land zoned for lots over 25,000 sq. ft.\n- INDUS: proportion of non-retail business acres per town\n- CHAS: Charles River dummy variable (= 1 if tract bounds river; 0 otherwise)\n- NOX: nitric oxides concentration (parts per 10 million)\n- RM: average number of rooms per dwelling\n- AGE: proportion of owner-occupied units built prior to 1940\n- DIS: weighted distances to five Boston employment centers\n- RAD: index of accessibility to radial highways\n- TAX: full-value property-tax rate per 10,000 dollars\n- PTRATIO: pupil-teacher ratio by town\n- LSTAT: %lower status of the population\n- MEDV: Median value of owner-occupied homes in 1000 dollars.",
"_____no_output_____"
],
[
"### Import Necessary Libraries",
"_____no_output_____"
]
],
[
[
"# this will help in making the Python code more structured automatically (good coding practice)\n%load_ext nb_black\n\n# Libraries to help with reading and manipulating data\nimport numpy as np\nimport pandas as pd\n\n# Libraries to help with data visualization\nimport matplotlib.pyplot as plt\nimport seaborn as sns\n\n# to split the data into train and test\nfrom sklearn.model_selection import train_test_split\n\n# to build linear regression_model\nfrom sklearn.linear_model import LinearRegression\n\n# to check model performance\nfrom sklearn.metrics import mean_squared_error",
"_____no_output_____"
]
],
[
[
"### Load the dataset",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(\"boston.csv\")\ndf.head()",
"_____no_output_____"
]
],
[
[
"### Check the shape of the dataset",
"_____no_output_____"
]
],
[
[
"df.shape",
"_____no_output_____"
]
],
[
[
"### Get the info regarding column datatypes",
"_____no_output_____"
]
],
[
[
"df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 506 entries, 0 to 505\nData columns (total 13 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 CRIM 506 non-null float64\n 1 ZN 506 non-null float64\n 2 INDUS 506 non-null float64\n 3 CHAS 506 non-null int64 \n 4 NX 506 non-null float64\n 5 RM 506 non-null float64\n 6 AGE 506 non-null float64\n 7 DIS 506 non-null float64\n 8 RAD 506 non-null int64 \n 9 TAX 506 non-null int64 \n 10 PTRATIO 506 non-null float64\n 11 LSTAT 506 non-null float64\n 12 MEDV 506 non-null float64\ndtypes: float64(10), int64(3)\nmemory usage: 51.5 KB\n"
]
],
[
[
"### Get summary statistics for the numerical columns",
"_____no_output_____"
]
],
[
[
"df.describe()",
"_____no_output_____"
]
],
[
[
"### Exploratory Data Analysis",
"_____no_output_____"
],
[
"**Plot the distribution plots for all the numerical features and list your observations.**",
"_____no_output_____"
]
],
[
[
"# let's plot all the columns to look at their distributions\n\nimport seaborn as sns\n\nfor i in df.columns:\n plt.figure(figsize=(7, 4))\n sns.histplot(data=df, x=i)\n plt.show()",
"_____no_output_____"
]
],
[
[
"* CRIM and ZN have heavily skewed distributions.\n* RM and MEDV have close to normal distributions.",
"_____no_output_____"
],
[
"**Plot the scatterplots for features and the target variable `MEDV` and list your observations.**",
"_____no_output_____"
]
],
[
[
"# let's plot the scatterplots of median price with all the features\n\nfor i in df.columns:\n plt.figure(figsize=(6, 6))\n sns.scatterplot(data=df, x=i, y=\"MEDV\")\n plt.show()",
"_____no_output_____"
]
],
[
[
"* LSTAT and RM show a slightly strong linear relationship with MEDV.",
"_____no_output_____"
],
[
"**Plot the correlation heatmap and list your observations.**",
"_____no_output_____"
]
],
[
[
"plt.figure(figsize=(15, 7))\nsns.heatmap(df.corr(), annot=True, vmin=-1, vmax=1, fmt=\".2f\", cmap=\"Spectral\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"* NX and TAX show a slightly strong positive linear relationship with INDUS, while DIS shows a slightly strong negative linear relationship with INDUS.\n* NX shows a slightly strong positive linear relationship with AGE, while DIS shows a slightly strong negative linear relationship with AGE.\n* RM shows a slightly strong positive linear relationship with MEDV, while LSTAT shows a slightly strong negative linear relationship with MEDV.",
"_____no_output_____"
],
[
"### Split the dataset\n\nSplit the data into the dependent and independent variables, and further split it in a ratio of 70:30 for train and test sets.",
"_____no_output_____"
]
],
[
[
"X = df.drop(\"MEDV\", axis=1)\ny = df[\"MEDV\"]",
"_____no_output_____"
],
[
"# splitting the data in 70:30 ratio for train to test data\n\nX_train, X_test, y_train, y_test = train_test_split(\n X, y, test_size=0.30, random_state=1\n)",
"_____no_output_____"
]
],
[
[
"### Model Building (sklearn)",
"_____no_output_____"
],
[
"**Fit the model to the training set**",
"_____no_output_____"
]
],
[
[
"# intialize the model to be fit and fit the model on the train data\n\nregression_model = LinearRegression()\nregression_model.fit(X_train, y_train)",
"_____no_output_____"
]
],
[
[
"**Get the score on training set**",
"_____no_output_____"
]
],
[
[
"# get the score (R-squared) on the training set\n\nprint(\n \"The score (R-squared) on the training set is \",\n regression_model.score(X_train, y_train),\n)",
"The score (R-squared) on the training set is 0.7073732058856179\n"
]
],
[
[
"**Write your own function for the R-squared score.**",
"_____no_output_____"
]
],
[
[
"def r_squared(model, X, y):\n y_mean = y.mean()\n SST = ((y - y_mean) ** 2).sum()\n SSE = ((y - model.predict(X)) ** 2).sum()\n r_square = 1 - SSE / SST\n return SSE, SST, r_square\n\n\nSSE, SST, r_square = r_squared(regression_model, X_train, y_train)\nprint(\"SSE: \", SSE)\nprint(\"SST: \", SST)\nprint(\"R-squared: \", r_square)",
"SSE: 8410.365734587129\nSST: 28740.928389830508\nR-squared: 0.7073732058856179\n"
]
],
[
[
"**Get the score on test set**",
"_____no_output_____"
]
],
[
[
"# get the score (R-squared) on the test set\n\nprint(\n \"The score (R-squared) on the test set is \", regression_model.score(X_test, y_test)\n)",
"The score (R-squared) on the test set is 0.7721684899134138\n"
]
],
[
[
"**Get the RMSE on train set**",
"_____no_output_____"
]
],
[
[
"print(\n \"The Root Mean Square Error (RMSE) of the model for the training set is \",\n np.sqrt(mean_squared_error(y_train, regression_model.predict(X_train))),\n)",
"The Root Mean Square Error (RMSE) of the model for the training set is 4.874227661429436\n"
]
],
[
[
"**Get the RMSE on test set**",
"_____no_output_____"
]
],
[
[
"print(\n \"The Root Mean Square Error (RMSE) of the model for the test set is \",\n np.sqrt(mean_squared_error(y_test, regression_model.predict(X_test))),\n)",
"The Root Mean Square Error (RMSE) of the model for the test set is 4.569658652745821\n"
]
],
[
[
"**Get the model coefficients.**",
"_____no_output_____"
]
],
[
[
"# let's check the coefficients and intercept of the model\n\ncoef_df = pd.DataFrame(\n np.append(regression_model.coef_, regression_model.intercept_),\n index=X_train.columns.tolist() + [\"Intercept\"],\n columns=[\"Coefficients\"],\n)\n\ncoef_df",
"_____no_output_____"
]
],
[
[
"**Automate the equation of the fit**",
"_____no_output_____"
]
],
[
[
"# Let us write the equation of linear regression\n\nEquation = \"Price = \" + str(regression_model.intercept_)\nprint(Equation, end=\" \")\n\nfor i in range(len(X_train.columns)):\n if i != len(X_train.columns) - 1:\n print(\n \"+ (\",\n regression_model.coef_[i],\n \")*(\",\n X_train.columns[i],\n \")\",\n end=\" \",\n )\n else:\n print(\"+ (\", regression_model.coef_[i], \")*(\", X_train.columns[i], \")\")",
"Price = 49.88523466381757 + ( -0.11384484836914055 )*( CRIM ) + ( 0.06117026804060482 )*( ZN ) + ( 0.05410346495874324 )*( INDUS ) + ( 2.517511959122706 )*( CHAS ) + ( -22.248502345084436 )*( NX ) + ( 2.6984128200099073 )*( RM ) + ( 0.004836047284750189 )*( AGE ) + ( -1.5342953819992673 )*( DIS ) + ( 0.29883325485901513 )*( RAD ) + ( -0.011413580552025081 )*( TAX ) + ( -0.9889146257039375 )*( PTRATIO ) + ( -0.5861328508499127 )*( LSTAT )\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6fa79a2b2cbac06961e2eda3c81ea3d6fa09bb | 597,979 | ipynb | Jupyter Notebook | keras-ssd-master/.ipynb_checkpoints/ssd300_training-checkpoint.ipynb | dlsaavedra/Detector_GDXray | 1e120f8fa548819eef1b86ccfbbe306b44405b6f | [
"MIT"
]
| 3 | 2021-05-27T07:27:44.000Z | 2022-02-19T05:20:16.000Z | keras-ssd-master/.ipynb_checkpoints/ssd300_training-checkpoint.ipynb | dlsaavedra/Detector_GDXray | 1e120f8fa548819eef1b86ccfbbe306b44405b6f | [
"MIT"
]
| 8 | 2020-09-25T22:34:27.000Z | 2022-02-10T01:09:19.000Z | keras-ssd-master/.ipynb_checkpoints/ssd300_training-checkpoint.ipynb | dlsaavedra/Detector_GDXray | 1e120f8fa548819eef1b86ccfbbe306b44405b6f | [
"MIT"
]
| 3 | 2020-03-18T20:27:18.000Z | 2021-11-03T03:10:11.000Z | 780.651436 | 555,100 | 0.928337 | [
[
[
"# SSD300 Training Tutorial\n\nThis tutorial explains how to train an SSD300 on the Pascal VOC datasets. The preset parameters reproduce the training of the original SSD300 \"07+12\" model. Training SSD512 works simiarly, so there's no extra tutorial for that. The same goes for training on other datasets.\n\nYou can find a summary of a full training here to get an impression of what it should look like:\n[SSD300 \"07+12\" training summary](https://github.com/pierluigiferrari/ssd_keras/blob/master/training_summaries/ssd300_pascal_07%2B12_training_summary.md)",
"_____no_output_____"
]
],
[
[
"from keras.optimizers import Adam, SGD\nfrom keras.callbacks import ModelCheckpoint, LearningRateScheduler, TerminateOnNaN, CSVLogger\nfrom keras import backend as K\nfrom keras.models import load_model\nfrom math import ceil\nimport numpy as np\nfrom matplotlib import pyplot as plt\n\nfrom models.keras_ssd300 import ssd_300\nfrom keras_loss_function.keras_ssd_loss import SSDLoss\nfrom keras_layers.keras_layer_AnchorBoxes import AnchorBoxes\nfrom keras_layers.keras_layer_DecodeDetections import DecodeDetections\nfrom keras_layers.keras_layer_DecodeDetectionsFast import DecodeDetectionsFast\nfrom keras_layers.keras_layer_L2Normalization import L2Normalization\n\nfrom ssd_encoder_decoder.ssd_input_encoder import SSDInputEncoder\nfrom ssd_encoder_decoder.ssd_output_decoder import decode_detections, decode_detections_fast\n\nfrom data_generator.object_detection_2d_data_generator import DataGenerator\nfrom data_generator.object_detection_2d_geometric_ops import Resize\nfrom data_generator.object_detection_2d_photometric_ops import ConvertTo3Channels\nfrom data_generator.data_augmentation_chain_original_ssd import SSDDataAugmentation\nfrom data_generator.object_detection_2d_misc_utils import apply_inverse_transforms\n\n%matplotlib inline",
"/home/dlsaavedra/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:36: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"## 0. Preliminary note\n\nAll places in the code where you need to make any changes are marked `TODO` and explained accordingly. All code cells that don't contain `TODO` markers just need to be executed.",
"_____no_output_____"
],
[
"## 1. Set the model configuration parameters\n\nThis section sets the configuration parameters for the model definition. The parameters set here are being used both by the `ssd_300()` function that builds the SSD300 model as well as further down by the constructor for the `SSDInputEncoder` object that is needed to run the training. Most of these parameters are needed to define the anchor boxes.\n\nThe parameters as set below produce the original SSD300 architecture that was trained on the Pascal VOC datsets, i.e. they are all chosen to correspond exactly to their respective counterparts in the `.prototxt` file that defines the original Caffe implementation. Note that the anchor box scaling factors of the original SSD implementation vary depending on the datasets on which the models were trained. The scaling factors used for the MS COCO datasets are smaller than the scaling factors used for the Pascal VOC datasets. The reason why the list of scaling factors has 7 elements while there are only 6 predictor layers is that the last scaling factor is used for the second aspect-ratio-1 box of the last predictor layer. Refer to the documentation for details.\n\nAs mentioned above, the parameters set below are not only needed to build the model, but are also passed to the `SSDInputEncoder` constructor further down, which is responsible for matching and encoding ground truth boxes and anchor boxes during the training. In order to do that, it needs to know the anchor box parameters.",
"_____no_output_____"
]
],
[
[
"img_height = 300 # Height of the model input images\nimg_width = 300 # Width of the model input images\nimg_channels = 3 # Number of color channels of the model input images\nmean_color = [123, 117, 104] # The per-channel mean of the images in the dataset. Do not change this value if you're using any of the pre-trained weights.\nswap_channels = [2, 1, 0] # The color channel order in the original SSD is BGR, so we'll have the model reverse the color channel order of the input images.\nn_classes = 20 # Number of positive classes, e.g. 20 for Pascal VOC, 80 for MS COCO\nscales_pascal = [0.1, 0.2, 0.37, 0.54, 0.71, 0.88, 1.05] # The anchor box scaling factors used in the original SSD300 for the Pascal VOC datasets\nscales_coco = [0.07, 0.15, 0.33, 0.51, 0.69, 0.87, 1.05] # The anchor box scaling factors used in the original SSD300 for the MS COCO datasets\nscales = scales_pascal\naspect_ratios = [[1.0, 2.0, 0.5],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5, 3.0, 1.0/3.0],\n [1.0, 2.0, 0.5],\n [1.0, 2.0, 0.5]] # The anchor box aspect ratios used in the original SSD300; the order matters\ntwo_boxes_for_ar1 = True\nsteps = [8, 16, 32, 64, 100, 300] # The space between two adjacent anchor box center points for each predictor layer.\noffsets = [0.5, 0.5, 0.5, 0.5, 0.5, 0.5] # The offsets of the first anchor box center points from the top and left borders of the image as a fraction of the step size for each predictor layer.\nclip_boxes = False # Whether or not to clip the anchor boxes to lie entirely within the image boundaries\nvariances = [0.1, 0.1, 0.2, 0.2] # The variances by which the encoded target coordinates are divided as in the original implementation\nnormalize_coords = True",
"_____no_output_____"
]
],
[
[
"## 2. Build or load the model\n\nYou will want to execute either of the two code cells in the subsequent two sub-sections, not both.",
"_____no_output_____"
],
[
"### 2.1 Create a new model and load trained VGG-16 weights into it (or trained SSD weights)\n\nIf you want to create a new SSD300 model, this is the relevant section for you. If you want to load a previously saved SSD300 model, skip ahead to section 2.2.\n\nThe code cell below does the following things:\n1. It calls the function `ssd_300()` to build the model.\n2. It then loads the weights file that is found at `weights_path` into the model. You could load the trained VGG-16 weights or you could load the weights of a trained model. If you want to reproduce the original SSD training, load the pre-trained VGG-16 weights. In any case, you need to set the path to the weights file you want to load on your local machine. Download links to all the trained weights are provided in the [README](https://github.com/pierluigiferrari/ssd_keras/blob/master/README.md) of this repository.\n3. Finally, it compiles the model for the training. In order to do so, we're defining an optimizer (Adam) and a loss function (SSDLoss) to be passed to the `compile()` method.\n\nNormally, the optimizer of choice would be Adam (commented out below), but since the original implementation uses plain SGD with momentum, we'll do the same in order to reproduce the original training. Adam is generally the superior optimizer, so if your goal is not to have everything exactly as in the original training, feel free to switch to Adam. You might need to adjust the learning rate scheduler below slightly in case you use Adam.\n\nNote that the learning rate that is being set here doesn't matter, because further below we'll pass a learning rate scheduler to the training function, which will overwrite any learning rate set here, i.e. what matters are the learning rates that are defined by the learning rate scheduler.\n\n`SSDLoss` is a custom Keras loss function that implements the multi-task that consists of a log loss for classification and a smooth L1 loss for localization. `neg_pos_ratio` and `alpha` are set as in the paper.",
"_____no_output_____"
]
],
[
[
"# 1: Build the Keras model.\n\nK.clear_session() # Clear previous models from memory.\n\nmodel = ssd_300(image_size=(img_height, img_width, img_channels),\n n_classes=n_classes,\n mode='training',\n l2_regularization=0.0005,\n scales=scales,\n aspect_ratios_per_layer=aspect_ratios,\n two_boxes_for_ar1=two_boxes_for_ar1,\n steps=steps,\n offsets=offsets,\n clip_boxes=clip_boxes,\n variances=variances,\n normalize_coords=normalize_coords,\n subtract_mean=mean_color,\n swap_channels=swap_channels)\n\n# 2: Load some weights into the model.\n\n# TODO: Set the path to the weights you want to load.\nweights_path = 'path/to/VGG_ILSVRC_16_layers_fc_reduced.h5'\n\nmodel.load_weights(weights_path, by_name=True)\n\n# 3: Instantiate an optimizer and the SSD loss function and compile the model.\n# If you want to follow the original Caffe implementation, use the preset SGD\n# optimizer, otherwise I'd recommend the commented-out Adam optimizer.\n\n#adam = Adam(lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=1e-08, decay=0.0)\nsgd = SGD(lr=0.001, momentum=0.9, decay=0.0, nesterov=False)\n\nssd_loss = SSDLoss(neg_pos_ratio=3, alpha=1.0)\n\nmodel.compile(optimizer=sgd, loss=ssd_loss.compute_loss)",
"_____no_output_____"
]
],
[
[
"### 2.2 Load a previously created model\n\nIf you have previously created and saved a model and would now like to load it, execute the next code cell. The only thing you need to do here is to set the path to the saved model HDF5 file that you would like to load.\n\nThe SSD model contains custom objects: Neither the loss function nor the anchor box or L2-normalization layer types are contained in the Keras core library, so we need to provide them to the model loader.\n\nThis next code cell assumes that you want to load a model that was created in 'training' mode. If you want to load a model that was created in 'inference' or 'inference_fast' mode, you'll have to add the `DecodeDetections` or `DecodeDetectionsFast` layer type to the `custom_objects` dictionary below.",
"_____no_output_____"
]
],
[
[
"# TODO: Set the path to the `.h5` file of the model to be loaded.\nmodel_path = 'path/to/trained/model.h5'\n\n# We need to create an SSDLoss object in order to pass that to the model loader.\nssd_loss = SSDLoss(neg_pos_ratio=3, alpha=1.0)\n\nK.clear_session() # Clear previous models from memory.\n\nmodel = load_model(model_path, custom_objects={'AnchorBoxes': AnchorBoxes,\n 'L2Normalization': L2Normalization,\n 'compute_loss': ssd_loss.compute_loss})",
"_____no_output_____"
]
],
[
[
"## 3. Set up the data generators for the training\n\nThe code cells below set up the data generators for the training and validation datasets to train the model. The settings below reproduce the original SSD training on Pascal VOC 2007 `trainval` plus 2012 `trainval` and validation on Pascal VOC 2007 `test`.\n\nThe only thing you need to change here are the filepaths to the datasets on your local machine. Note that parsing the labels from the XML annotations files can take a while.\n\nNote that the generator provides two options to speed up the training. By default, it loads the individual images for a batch from disk. This has two disadvantages. First, for compressed image formats like JPG, this is a huge computational waste, because every image needs to be decompressed again and again every time it is being loaded. Second, the images on disk are likely not stored in a contiguous block of memory, which may also slow down the loading process. The first option that `DataGenerator` provides to deal with this is to load the entire dataset into memory, which reduces the access time for any image to a negligible amount, but of course this is only an option if you have enough free memory to hold the whole dataset. As a second option, `DataGenerator` provides the possibility to convert the dataset into a single HDF5 file. This HDF5 file stores the images as uncompressed arrays in a contiguous block of memory, which dramatically speeds up the loading time. It's not as good as having the images in memory, but it's a lot better than the default option of loading them from their compressed JPG state every time they are needed. Of course such an HDF5 dataset may require significantly more disk space than the compressed images (around 9 GB total for Pascal VOC 2007 `trainval` plus 2012 `trainval` and another 2.6 GB for 2007 `test`). You can later load these HDF5 datasets directly in the constructor.\n\nThe original SSD implementation uses a batch size of 32 for the training. In case you run into GPU memory issues, reduce the batch size accordingly. You need at least 7 GB of free GPU memory to train an SSD300 with 20 object classes with a batch size of 32.\n\nThe `DataGenerator` itself is fairly generic. I doesn't contain any data augmentation or bounding box encoding logic. Instead, you pass a list of image transformations and an encoder for the bounding boxes in the `transformations` and `label_encoder` arguments of the data generator's `generate()` method, and the data generator will then apply those given transformations and the encoding to the data. Everything here is preset already, but if you'd like to learn more about the data generator and its data augmentation capabilities, take a look at the detailed tutorial in [this](https://github.com/pierluigiferrari/data_generator_object_detection_2d) repository.\n\nThe data augmentation settings defined further down reproduce the data augmentation pipeline of the original SSD training. The training generator receives an object `ssd_data_augmentation`, which is a transformation object that is itself composed of a whole chain of transformations that replicate the data augmentation procedure used to train the original Caffe implementation. The validation generator receives an object `resize`, which simply resizes the input images.\n\nAn `SSDInputEncoder` object, `ssd_input_encoder`, is passed to both the training and validation generators. As explained above, it matches the ground truth labels to the model's anchor boxes and encodes the box coordinates into the format that the model needs.\n\nIn order to train the model on a dataset other than Pascal VOC, either choose `DataGenerator`'s appropriate parser method that corresponds to your data format, or, if `DataGenerator` does not provide a suitable parser for your data format, you can write an additional parser and add it. Out of the box, `DataGenerator` can handle datasets that use the Pascal VOC format (use `parse_xml()`), the MS COCO format (use `parse_json()`) and a wide range of CSV formats (use `parse_csv()`).",
"_____no_output_____"
]
],
[
[
"# 1: Instantiate two `DataGenerator` objects: One for training, one for validation.\n\n# Optional: If you have enough memory, consider loading the images into memory for the reasons explained above.\n\ntrain_dataset = DataGenerator(load_images_into_memory=False, hdf5_dataset_path=None)\nval_dataset = DataGenerator(load_images_into_memory=False, hdf5_dataset_path=None)\n\n# 2: Parse the image and label lists for the training and validation datasets. This can take a while.\n\n# TODO: Set the paths to the datasets here.\n\n# The directories that contain the images.\nVOC_2007_images_dir = '../../datasets/VOCdevkit/VOC2007/JPEGImages/'\nVOC_2012_images_dir = '../../datasets/VOCdevkit/VOC2012/JPEGImages/'\n\n# The directories that contain the annotations.\nVOC_2007_annotations_dir = '../../datasets/VOCdevkit/VOC2007/Annotations/'\nVOC_2012_annotations_dir = '../../datasets/VOCdevkit/VOC2012/Annotations/'\n\n# The paths to the image sets.\nVOC_2007_train_image_set_filename = '../../datasets/VOCdevkit/VOC2007/ImageSets/Main/train.txt'\nVOC_2012_train_image_set_filename = '../../datasets/VOCdevkit/VOC2012/ImageSets/Main/train.txt'\nVOC_2007_val_image_set_filename = '../../datasets/VOCdevkit/VOC2007/ImageSets/Main/val.txt'\nVOC_2012_val_image_set_filename = '../../datasets/VOCdevkit/VOC2012/ImageSets/Main/val.txt'\nVOC_2007_trainval_image_set_filename = '../../datasets/VOCdevkit/VOC2007/ImageSets/Main/trainval.txt'\nVOC_2012_trainval_image_set_filename = '../../datasets/VOCdevkit/VOC2012/ImageSets/Main/trainval.txt'\nVOC_2007_test_image_set_filename = '../../datasets/VOCdevkit/VOC2007/ImageSets/Main/test.txt'\n\n# The XML parser needs to now what object class names to look for and in which order to map them to integers.\nclasses = ['background',\n 'aeroplane', 'bicycle', 'bird', 'boat',\n 'bottle', 'bus', 'car', 'cat',\n 'chair', 'cow', 'diningtable', 'dog',\n 'horse', 'motorbike', 'person', 'pottedplant',\n 'sheep', 'sofa', 'train', 'tvmonitor']\n\ntrain_dataset.parse_xml(images_dirs=[VOC_2007_images_dir,\n VOC_2012_images_dir],\n image_set_filenames=[VOC_2007_trainval_image_set_filename,\n VOC_2012_trainval_image_set_filename],\n annotations_dirs=[VOC_2007_annotations_dir,\n VOC_2012_annotations_dir],\n classes=classes,\n include_classes='all',\n exclude_truncated=False,\n exclude_difficult=False,\n ret=False)\n\nval_dataset.parse_xml(images_dirs=[VOC_2007_images_dir],\n image_set_filenames=[VOC_2007_test_image_set_filename],\n annotations_dirs=[VOC_2007_annotations_dir],\n classes=classes,\n include_classes='all',\n exclude_truncated=False,\n exclude_difficult=True,\n ret=False)\n\n# Optional: Convert the dataset into an HDF5 dataset. This will require more disk space, but will\n# speed up the training. Doing this is not relevant in case you activated the `load_images_into_memory`\n# option in the constructor, because in that cas the images are in memory already anyway. If you don't\n# want to create HDF5 datasets, comment out the subsequent two function calls.\n\ntrain_dataset.create_hdf5_dataset(file_path='dataset_pascal_voc_07+12_trainval.h5',\n resize=False,\n variable_image_size=True,\n verbose=True)\n\nval_dataset.create_hdf5_dataset(file_path='dataset_pascal_voc_07_test.h5',\n resize=False,\n variable_image_size=True,\n verbose=True)",
"Processing image set 'trainval.txt': 100%|██████████| 5011/5011 [00:14<00:00, 339.96it/s]\nProcessing image set 'trainval.txt': 100%|██████████| 11540/11540 [00:30<00:00, 381.10it/s]\nLoading images into memory: 100%|██████████| 16551/16551 [01:12<00:00, 228.31it/s]\nProcessing image set 'test.txt': 100%|██████████| 4952/4952 [00:13<00:00, 367.21it/s]\nLoading images into memory: 100%|██████████| 4952/4952 [00:19<00:00, 260.61it/s]\nCreating HDF5 dataset: 100%|██████████| 16551/16551 [01:48<00:00, 152.80it/s]\nCreating HDF5 dataset: 100%|██████████| 4952/4952 [00:40<00:00, 122.75it/s]\n"
],
[
"# 3: Set the batch size.\n\nbatch_size = 32 # Change the batch size if you like, or if you run into GPU memory issues.\n\n# 4: Set the image transformations for pre-processing and data augmentation options.\n\n# For the training generator:\nssd_data_augmentation = SSDDataAugmentation(img_height=img_height,\n img_width=img_width,\n background=mean_color)\n\n# For the validation generator:\nconvert_to_3_channels = ConvertTo3Channels()\nresize = Resize(height=img_height, width=img_width)\n\n# 5: Instantiate an encoder that can encode ground truth labels into the format needed by the SSD loss function.\n\n# The encoder constructor needs the spatial dimensions of the model's predictor layers to create the anchor boxes.\npredictor_sizes = [model.get_layer('conv4_3_norm_mbox_conf').output_shape[1:3],\n model.get_layer('fc7_mbox_conf').output_shape[1:3],\n model.get_layer('conv6_2_mbox_conf').output_shape[1:3],\n model.get_layer('conv7_2_mbox_conf').output_shape[1:3],\n model.get_layer('conv8_2_mbox_conf').output_shape[1:3],\n model.get_layer('conv9_2_mbox_conf').output_shape[1:3]]\n\nssd_input_encoder = SSDInputEncoder(img_height=img_height,\n img_width=img_width,\n n_classes=n_classes,\n predictor_sizes=predictor_sizes,\n scales=scales,\n aspect_ratios_per_layer=aspect_ratios,\n two_boxes_for_ar1=two_boxes_for_ar1,\n steps=steps,\n offsets=offsets,\n clip_boxes=clip_boxes,\n variances=variances,\n matching_type='multi',\n pos_iou_threshold=0.5,\n neg_iou_limit=0.5,\n normalize_coords=normalize_coords)\n\n# 6: Create the generator handles that will be passed to Keras' `fit_generator()` function.\n\ntrain_generator = train_dataset.generate(batch_size=batch_size,\n shuffle=True,\n transformations=[ssd_data_augmentation],\n label_encoder=ssd_input_encoder,\n returns={'processed_images',\n 'encoded_labels'},\n keep_images_without_gt=False)\n\nval_generator = val_dataset.generate(batch_size=batch_size,\n shuffle=False,\n transformations=[convert_to_3_channels,\n resize],\n label_encoder=ssd_input_encoder,\n returns={'processed_images',\n 'encoded_labels'},\n keep_images_without_gt=False)\n\n# Get the number of samples in the training and validations datasets.\ntrain_dataset_size = train_dataset.get_dataset_size()\nval_dataset_size = val_dataset.get_dataset_size()\n\nprint(\"Number of images in the training dataset:\\t{:>6}\".format(train_dataset_size))\nprint(\"Number of images in the validation dataset:\\t{:>6}\".format(val_dataset_size))",
"Number of images in the training dataset:\t 16551\nNumber of images in the validation dataset:\t 4952\n"
]
],
[
[
"## 4. Set the remaining training parameters\n\nWe've already chosen an optimizer and set the batch size above, now let's set the remaining training parameters. I'll set one epoch to consist of 1,000 training steps. The next code cell defines a learning rate schedule that replicates the learning rate schedule of the original Caffe implementation for the training of the SSD300 Pascal VOC \"07+12\" model. That model was trained for 120,000 steps with a learning rate of 0.001 for the first 80,000 steps, 0.0001 for the next 20,000 steps, and 0.00001 for the last 20,000 steps. If you're training on a different dataset, define the learning rate schedule however you see fit.\n\nI'll set only a few essential Keras callbacks below, feel free to add more callbacks if you want TensorBoard summaries or whatever. We obviously need the learning rate scheduler and we want to save the best models during the training. It also makes sense to continuously stream our training history to a CSV log file after every epoch, because if we didn't do that, in case the training terminates with an exception at some point or if the kernel of this Jupyter notebook dies for some reason or anything like that happens, we would lose the entire history for the trained epochs. Finally, we'll also add a callback that makes sure that the training terminates if the loss becomes `NaN`. Depending on the optimizer you use, it can happen that the loss becomes `NaN` during the first iterations of the training. In later iterations it's less of a risk. For example, I've never seen a `NaN` loss when I trained SSD using an Adam optimizer, but I've seen a `NaN` loss a couple of times during the very first couple of hundred training steps of training a new model when I used an SGD optimizer.",
"_____no_output_____"
]
],
[
[
"# Define a learning rate schedule.\n\ndef lr_schedule(epoch):\n if epoch < 80:\n return 0.001\n elif epoch < 100:\n return 0.0001\n else:\n return 0.00001",
"_____no_output_____"
],
[
"# Define model callbacks.\n\n# TODO: Set the filepath under which you want to save the model.\nmodel_checkpoint = ModelCheckpoint(filepath='ssd300_pascal_07+12_epoch-{epoch:02d}_loss-{loss:.4f}_val_loss-{val_loss:.4f}.h5',\n monitor='val_loss',\n verbose=1,\n save_best_only=True,\n save_weights_only=False,\n mode='auto',\n period=1)\n#model_checkpoint.best = \n\ncsv_logger = CSVLogger(filename='ssd300_pascal_07+12_training_log.csv',\n separator=',',\n append=True)\n\nlearning_rate_scheduler = LearningRateScheduler(schedule=lr_schedule,\n verbose=1)\n\nterminate_on_nan = TerminateOnNaN()\n\ncallbacks = [model_checkpoint,\n csv_logger,\n learning_rate_scheduler,\n terminate_on_nan]",
"_____no_output_____"
]
],
[
[
"## 5. Train",
"_____no_output_____"
],
[
"In order to reproduce the training of the \"07+12\" model mentioned above, at 1,000 training steps per epoch you'd have to train for 120 epochs. That is going to take really long though, so you might not want to do all 120 epochs in one go and instead train only for a few epochs at a time. You can find a summary of a full training [here](https://github.com/pierluigiferrari/ssd_keras/blob/master/training_summaries/ssd300_pascal_07%2B12_training_summary.md).\n\nIn order to only run a partial training and resume smoothly later on, there are a few things you should note:\n1. Always load the full model if you can, rather than building a new model and loading previously saved weights into it. Optimizers like SGD or Adam keep running averages of past gradient moments internally. If you always save and load full models when resuming a training, then the state of the optimizer is maintained and the training picks up exactly where it left off. If you build a new model and load weights into it, the optimizer is being initialized from scratch, which, especially in the case of Adam, leads to small but unnecessary setbacks every time you resume the training with previously saved weights.\n2. In order for the learning rate scheduler callback above to work properly, `fit_generator()` needs to know which epoch we're in, otherwise it will start with epoch 0 every time you resume the training. Set `initial_epoch` to be the next epoch of your training. Note that this parameter is zero-based, i.e. the first epoch is epoch 0. If you had trained for 10 epochs previously and now you'd want to resume the training from there, you'd set `initial_epoch = 10` (since epoch 10 is the eleventh epoch). Furthermore, set `final_epoch` to the last epoch you want to run. To stick with the previous example, if you had trained for 10 epochs previously and now you'd want to train for another 10 epochs, you'd set `initial_epoch = 10` and `final_epoch = 20`.\n3. In order for the model checkpoint callback above to work correctly after a kernel restart, set `model_checkpoint.best` to the best validation loss from the previous training. If you don't do this and a new `ModelCheckpoint` object is created after a kernel restart, that object obviously won't know what the last best validation loss was, so it will always save the weights of the first epoch of your new training and record that loss as its new best loss. This isn't super-important, I just wanted to mention it.",
"_____no_output_____"
]
],
[
[
"# If you're resuming a previous training, set `initial_epoch` and `final_epoch` accordingly.\ninitial_epoch = 0\nfinal_epoch = 120\nsteps_per_epoch = 1000\n\nhistory = model.fit_generator(generator=train_generator,\n steps_per_epoch=steps_per_epoch,\n epochs=final_epoch,\n callbacks=callbacks,\n validation_data=val_generator,\n validation_steps=ceil(val_dataset_size/batch_size),\n initial_epoch=initial_epoch)",
"_____no_output_____"
]
],
[
[
"## 6. Make predictions\n\nNow let's make some predictions on the validation dataset with the trained model. For convenience we'll use the validation generator that we've already set up above. Feel free to change the batch size.\n\nYou can set the `shuffle` option to `False` if you would like to check the model's progress on the same image(s) over the course of the training.",
"_____no_output_____"
]
],
[
[
"# 1: Set the generator for the predictions.\n\npredict_generator = val_dataset.generate(batch_size=1,\n shuffle=True,\n transformations=[convert_to_3_channels,\n resize],\n label_encoder=None,\n returns={'processed_images',\n 'filenames',\n 'inverse_transform',\n 'original_images',\n 'original_labels'},\n keep_images_without_gt=False)",
"_____no_output_____"
],
[
"# 2: Generate samples.\n\nbatch_images, batch_filenames, batch_inverse_transforms, batch_original_images, batch_original_labels = next(predict_generator)\n\ni = 0 # Which batch item to look at\n\nprint(\"Image:\", batch_filenames[i])\nprint()\nprint(\"Ground truth boxes:\\n\")\nprint(np.array(batch_original_labels[i]))",
"Image: ../../datasets/VOCdevkit/VOC2007/JPEGImages/003819.jpg\n\nGround truth boxes:\n\n[[ 12 146 52 386 264]\n [ 12 69 208 322 360]\n [ 15 1 1 221 235]]\n"
],
[
"# 3: Make predictions.\n\ny_pred = model.predict(batch_images)",
"_____no_output_____"
]
],
[
[
"Now let's decode the raw predictions in `y_pred`.\n\nHad we created the model in 'inference' or 'inference_fast' mode, then the model's final layer would be a `DecodeDetections` layer and `y_pred` would already contain the decoded predictions, but since we created the model in 'training' mode, the model outputs raw predictions that still need to be decoded and filtered. This is what the `decode_detections()` function is for. It does exactly what the `DecodeDetections` layer would do, but using Numpy instead of TensorFlow (i.e. on the CPU instead of the GPU).\n\n`decode_detections()` with default argument values follows the procedure of the original SSD implementation: First, a very low confidence threshold of 0.01 is applied to filter out the majority of the predicted boxes, then greedy non-maximum suppression is performed per class with an intersection-over-union threshold of 0.45, and out of what is left after that, the top 200 highest confidence boxes are returned. Those settings are for precision-recall scoring purposes though. In order to get some usable final predictions, we'll set the confidence threshold much higher, e.g. to 0.5, since we're only interested in the very confident predictions.",
"_____no_output_____"
]
],
[
[
"# 4: Decode the raw predictions in `y_pred`.\n\ny_pred_decoded = decode_detections(y_pred,\n confidence_thresh=0.5,\n iou_threshold=0.4,\n top_k=200,\n normalize_coords=normalize_coords,\n img_height=img_height,\n img_width=img_width)",
"_____no_output_____"
]
],
[
[
"We made the predictions on the resized images, but we'd like to visualize the outcome on the original input images, so we'll convert the coordinates accordingly. Don't worry about that opaque `apply_inverse_transforms()` function below, in this simple case it just aplies `(* original_image_size / resized_image_size)` to the box coordinates.",
"_____no_output_____"
]
],
[
[
"# 5: Convert the predictions for the original image.\n\ny_pred_decoded_inv = apply_inverse_transforms(y_pred_decoded, batch_inverse_transforms)\n\nnp.set_printoptions(precision=2, suppress=True, linewidth=90)\nprint(\"Predicted boxes:\\n\")\nprint(' class conf xmin ymin xmax ymax')\nprint(y_pred_decoded_inv[i])",
"Predicted boxes:\n\n class conf xmin ymin xmax ymax\n[[ 9. 0.8 364.79 5.24 496.51 203.59]\n [ 12. 1. 115.44 50. 384.22 330.76]\n [ 12. 0.86 68.99 212.78 331.63 355.72]\n [ 15. 0.95 2.62 20.18 235.83 253.07]]\n"
]
],
[
[
"Finally, let's draw the predicted boxes onto the image. Each predicted box says its confidence next to the category name. The ground truth boxes are also drawn onto the image in green for comparison.",
"_____no_output_____"
]
],
[
[
"# 5: Draw the predicted boxes onto the image\n\n# Set the colors for the bounding boxes\ncolors = plt.cm.hsv(np.linspace(0, 1, n_classes+1)).tolist()\nclasses = ['background',\n 'aeroplane', 'bicycle', 'bird', 'boat',\n 'bottle', 'bus', 'car', 'cat',\n 'chair', 'cow', 'diningtable', 'dog',\n 'horse', 'motorbike', 'person', 'pottedplant',\n 'sheep', 'sofa', 'train', 'tvmonitor']\n\nplt.figure(figsize=(20,12))\nplt.imshow(batch_original_images[i])\n\ncurrent_axis = plt.gca()\n\nfor box in batch_original_labels[i]:\n xmin = box[1]\n ymin = box[2]\n xmax = box[3]\n ymax = box[4]\n label = '{}'.format(classes[int(box[0])])\n current_axis.add_patch(plt.Rectangle((xmin, ymin), xmax-xmin, ymax-ymin, color='green', fill=False, linewidth=2)) \n current_axis.text(xmin, ymin, label, size='x-large', color='white', bbox={'facecolor':'green', 'alpha':1.0})\n\nfor box in y_pred_decoded_inv[i]:\n xmin = box[2]\n ymin = box[3]\n xmax = box[4]\n ymax = box[5]\n color = colors[int(box[0])]\n label = '{}: {:.2f}'.format(classes[int(box[0])], box[1])\n current_axis.add_patch(plt.Rectangle((xmin, ymin), xmax-xmin, ymax-ymin, color=color, fill=False, linewidth=2)) \n current_axis.text(xmin, ymin, label, size='x-large', color='white', bbox={'facecolor':color, 'alpha':1.0})",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6fb579969612bff93c7d9c8afde971f5d5e1c0 | 7,290 | ipynb | Jupyter Notebook | serverless/dev/nmf-lda-topics.ipynb | ChenLi0830/Clevo-Emotion-Detection-Service | 2983963e01b92deac447b0b953adffe8e0455c29 | [
"Apache-2.0"
]
| null | null | null | serverless/dev/nmf-lda-topics.ipynb | ChenLi0830/Clevo-Emotion-Detection-Service | 2983963e01b92deac447b0b953adffe8e0455c29 | [
"Apache-2.0"
]
| null | null | null | serverless/dev/nmf-lda-topics.ipynb | ChenLi0830/Clevo-Emotion-Detection-Service | 2983963e01b92deac447b0b953adffe8e0455c29 | [
"Apache-2.0"
]
| null | null | null | 33.136364 | 515 | 0.550069 | [
[
[
"from __future__ import print_function\nfrom time import time\n\nfrom sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer\nfrom sklearn.decomposition import NMF, LatentDirichletAllocation\nfrom sklearn.datasets import fetch_20newsgroups\n\nimport pandas as ps\nimport numpy as np\nimport json\n\nn_features = 10000\nn_topics = 3\nn_top_words = 10\n\ndef print_top_words(model, feature_names, n_top_words):\n for topic_idx, topic in enumerate(model.components_):\n print(\"Topic #%d:\" % topic_idx)\n print(\" \".join([feature_names[i]\n for i in topic.argsort()[:-n_top_words - 1:-1]]))\n print()\n\n\nprint(\"Loading dataset...\")\nt0 = time()\ndf = ps.read_csv(\"data/transcription_table.csv\")\ndata_samples = sum(df.transcriptionText.map(lambda x: map(lambda y : y['onebest'], json.loads(x))), [])\nn_samples = len(data_samples)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\n# Use tf-idf features for NMF.\nprint(\"Extracting tf-idf features for NMF...\")\ntfidf_vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,\n max_features=n_features, ngram_range=(1,2))\nt0 = time()\ntfidf = tfidf_vectorizer.fit_transform(data_samples)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\n# Use tf (raw term count) features for LDA.\nprint(\"Extracting tf features for LDA...\")\ntf_vectorizer = CountVectorizer(max_df=0.95, min_df=2,\n max_features=n_features)\nt0 = time()\ntf = tf_vectorizer.fit_transform(data_samples)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\n# Fit the NMF model\nprint(\"Fitting the NMF model with tf-idf features, \"\n \"n_samples=%d and n_features=%d...\"\n % (n_samples, n_features))\nt0 = time()\nnmf = NMF(n_components=n_topics, random_state=1,\n alpha=.1, l1_ratio=.5)\nw_nmf = nmf.fit_transform(tfidf)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\nprint(\"\\nTopics in NMF model:\")\ntfidf_feature_names = tfidf_vectorizer.get_feature_names()\nprint_top_words(nmf, tfidf_feature_names, n_top_words)\n\nprint(\"Fitting LDA models with tf features, \"\n \"n_samples=%d and n_features=%d...\"\n % (n_samples, n_features))\nlda = LatentDirichletAllocation(n_topics=n_topics, max_iter=5,\n learning_method='online',\n learning_offset=50.,\n random_state=0)\nt0 = time()\nw_lda = lda.fit_transform(tf)\nprint(\"done in %0.3fs.\" % (time() - t0))\n\nprint(\"\\nTopics in LDA model:\")\ntf_feature_names = tf_vectorizer.get_feature_names()\nprint_top_words(lda, tf_feature_names, n_top_words)",
"Loading dataset...\ndone in 0.027s.\nExtracting tf-idf features for NMF...\ndone in 0.076s.\nExtracting tf features for LDA...\ndone in 0.046s.\nFitting the NMF model with tf-idf features, n_samples=7571 and n_features=10000...\ndone in 0.058s.\n\nTopics in NMF model:\nTopic #0:\n您好 请问有什么能帮您的吗 您好 966为您服务 966为您服务 966为您服务 请问有什么能帮您的吗 请问有什么能帮你了吗 您好 感谢耐心等待 感谢耐心等待 您好 九六为您服务 九六为您服务\nTopic #1:\n先生 还有其他能帮你的吗 先生 您好 先生 还有其他能帮你的吗 先生 我们这边是第三方支付平台 我们这边是第三方支付平台 还有其他的帮您的吗 还有其他能帮您的吗 先生 是的 先生 您是手机话费扣款了吗 先生\nTopic #2:\n您稍等 好的 好的 您稍等 嗯好的 嗯好的 您稍等 您稍等 我帮您查询 您稍等 我帮您进行查询 你好 我帮您查询 您稍等 我再帮您查询一下\n\nFitting LDA models with tf features, n_samples=7571 and n_features=10000...\n"
],
[
"np.argmax(w_nmf[1:10], axis=1)",
"_____no_output_____"
],
[
"np.argmax(w_lda[1:10], axis=1)",
"_____no_output_____"
],
[
"pwd",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
ec6fb793740751ced75a4dcb07291d19e51f6160 | 152,220 | ipynb | Jupyter Notebook | Walmart_TripType_Classification/Preprocessing - make X_train, y_train, X_test.ipynb | ds-bigwater/workspace_with_Kaggle | eb7c284f64c5d0275ef0fe35832622a7300ba5b9 | [
"MIT"
]
| null | null | null | Walmart_TripType_Classification/Preprocessing - make X_train, y_train, X_test.ipynb | ds-bigwater/workspace_with_Kaggle | eb7c284f64c5d0275ef0fe35832622a7300ba5b9 | [
"MIT"
]
| null | null | null | Walmart_TripType_Classification/Preprocessing - make X_train, y_train, X_test.ipynb | ds-bigwater/workspace_with_Kaggle | eb7c284f64c5d0275ef0fe35832622a7300ba5b9 | [
"MIT"
]
| null | null | null | 29.482859 | 269 | 0.341 | [
[
[
"# Preprocessing\n\n### part 1 : fill in missing values & refine\n\n- DepartmentDescription\n- FinelineNumber\n- Upc\n- All of DepartmentDescription, FinelineNumber, Upc is NaN\n- Divide the Upc into manufacturer(company) number and product number.\n \n=======================================================================\n### part 2 : Encode & Derivation\n\n- DepartmentDescription\n- Weekday\n- FinelineNumber\n- company Upc\n- ScanCount\n - Divide by abs_scancount for each columns\n- Refund rate\n\n=======================================================================\n### part 3 : Feature Selection\n- Feature Selection\n\n- X_train\n- y_train\n- X_test",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\nfrom tqdm import tqdm\nimport preprocessing_functions as pf\nfrom functools import partial\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"### Data load",
"_____no_output_____"
]
],
[
[
"train = pd.read_csv(\"train.csv\")\n\nprint(train.shape)\ntrain.tail()",
"(647054, 7)\n"
],
[
"train.dtypes",
"_____no_output_____"
],
[
"test = pd.read_csv(\"test.csv\")\n\nprint(test.shape)\ntest.tail()",
"(653646, 6)\n"
],
[
"test.dtypes",
"_____no_output_____"
]
],
[
[
"## part 1 : fill in missing values & refine",
"_____no_output_____"
],
[
"### DepartmentDescription\n- 각각의 VisitNumber 별 DepartmentDescription의 최빈값으로 DepartmentDescription의 빈값을 채운다.\n- 유추할 수 없는 191개의 값은 'UNKNOWN' 으로 대체한다.\n- \"MENSWEAR\" change to \"MENS WEAR\"\n- \"HEALTH AND BEAUTY AIDS\" change to \"BEAUTY\"",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train.loc[train[\"VisitNumber\"]==259, \"DepartmentDescription\"]",
"_____no_output_____"
],
[
"DD_VN_list = train[train[\"DepartmentDescription\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(DD_VN_list): # if: 특정 VisitNumber 따른 DepartmentDescription 값이 모두 비어있는 경우 제외\n if len(train[train[\"VisitNumber\"] == loc][\"DepartmentDescription\"].value_counts().index) != 0:\n train.loc[(train[\"VisitNumber\"] == loc)&(train[\"DepartmentDescription\"].isna()), \"DepartmentDescription\"] = train[train[\"VisitNumber\"] == loc][\"DepartmentDescription\"].value_counts().index[0]",
"100%|██████████| 1172/1172 [00:33<00:00, 34.52it/s]\n"
],
[
"train.loc[train[\"VisitNumber\"]==259, \"DepartmentDescription\"]",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test.loc[test[\"VisitNumber\"]==874, \"DepartmentDescription\"]",
"_____no_output_____"
],
[
"DD_VN_list_t = test[test[\"DepartmentDescription\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(DD_VN_list_t): # if: 특정 VisitNumber 따른 DepartmentDescription 값이 모두 비어있는 경우 제외\n if len(test[test[\"VisitNumber\"] == loc][\"DepartmentDescription\"].value_counts().index) != 0:\n test.loc[(test[\"VisitNumber\"] == loc)&(test[\"DepartmentDescription\"].isna()), \"DepartmentDescription\"] = test[test[\"VisitNumber\"] == loc][\"DepartmentDescription\"].value_counts().index[0]",
"100%|██████████| 1141/1141 [00:32<00:00, 35.13it/s]\n"
],
[
"test.loc[test[\"VisitNumber\"]==874, \"DepartmentDescription\"]",
"_____no_output_____"
]
],
[
[
"#### refine DepartmentDescription",
"_____no_output_____"
],
[
"#### \"MENSWEAR\" change to \"MENS WEAR\"",
"_____no_output_____"
]
],
[
[
"train.loc[train[\"DepartmentDescription\"] == \"MENSWEAR\", \"DepartmentDescription\"] = \"MENS WEAR\"\ntrain[train[\"DepartmentDescription\"] == \"MENSWEAR\"]",
"_____no_output_____"
]
],
[
[
"#### \"HEALTH AND BEAUTY AIDS\" change to \"BEAUTY\"\n- train data has only two rows of \"HEALTH AND BEAUTY AIDS\", and test data doesn't",
"_____no_output_____"
]
],
[
[
"train.loc[train[\"DepartmentDescription\"] == \"HEALTH AND BEAUTY AIDS\", \"DepartmentDescription\"] = \"BEAUTY\"\ntrain[train[\"DepartmentDescription\"] == \"HEALTH AND BEAUTY AIDS\"]",
"_____no_output_____"
]
],
[
[
"### FinelineNumber\n- DepartmentDescription이 'PHARMACY RX'일때, FinelineNumber가 빈값인 경우 DepartmentDescription이 'PHARMACY RX'일때의 FinelineNumber의 최빈값으로 채운다.\n- 각각의 VisitNumber 별 FinelineNumber의 최빈값으로 FinelineNumber의 빈값을 채운다.\n- 191개의 유추할 수 없는 값은 기존에 있던 값과 중복되지 않는 -9999 값으로 대체한다.",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train[train[\"DepartmentDescription\"] == 'PHARMACY RX'][\"FinelineNumber\"].value_counts()",
"_____no_output_____"
],
[
"Pharmacy_idx = train[train[\"DepartmentDescription\"]=='PHARMACY RX'].index\nnumber_idx = np.arange(2922)\nidx_box = zip(number_idx, Pharmacy_idx)\n\n\nfor idx, Pha_idx in tqdm(idx_box):\n if idx % 2 == 0:\n train.loc[Pha_idx, \"FinelineNumber\"] = 4822.0\n else:\n train.loc[Pha_idx, \"FinelineNumber\"] = 5615.0\n \ntrain[train[\"DepartmentDescription\"] == 'PHARMACY RX'][[\"DepartmentDescription\", \"FinelineNumber\"]].head()",
"2922it [00:09, 313.50it/s]\n"
],
[
"train.loc[train[\"VisitNumber\"]==259, \"FinelineNumber\"]",
"_____no_output_____"
],
[
"FN_VN_list = train[train[\"FinelineNumber\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(FN_VN_list): # if: 특정 VisitNumber 따른 FinelineNumber 값이 모두 비어있는 경우 제외\n if len(train[train[\"VisitNumber\"] == loc][\"FinelineNumber\"].value_counts().index) != 0:\n train.loc[(train[\"VisitNumber\"] == loc)&(train[\"FinelineNumber\"].isna()), \"FinelineNumber\"] = train[train[\"VisitNumber\"] == loc][\"FinelineNumber\"].value_counts().index[0]",
"100%|██████████| 1172/1172 [00:10<00:00, 113.46it/s]\n"
],
[
"train.loc[train[\"VisitNumber\"]==259, \"FinelineNumber\"]",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test[test[\"DepartmentDescription\"] == 'PHARMACY RX'][\"FinelineNumber\"].value_counts()",
"_____no_output_____"
],
[
"Pharmacy_idx = test[test[\"DepartmentDescription\"]=='PHARMACY RX'].index\nnumber_idx = np.arange(2784)\nidx_box = zip(number_idx, Pharmacy_idx)\n\n\nfor idx, Pha_idx in tqdm(idx_box):\n if idx % 2 == 0:\n test.loc[Pha_idx, \"FinelineNumber\"] = 4822.0\n else:\n test.loc[Pha_idx, \"FinelineNumber\"] = 5615.0\n \ntest[test[\"DepartmentDescription\"] == 'PHARMACY RX'][[\"DepartmentDescription\", \"FinelineNumber\"]].head()",
"2784it [00:09, 306.35it/s]\n"
],
[
"test.loc[test[\"VisitNumber\"]==874, \"FinelineNumber\"]",
"_____no_output_____"
],
[
"FN_VN_list_t = test[test[\"FinelineNumber\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(FN_VN_list_t): # if: 특정 VisitNumber 따른 FinelineNumber 값이 모두 비어있는 경우 제외\n if len(test[test[\"VisitNumber\"] == loc][\"FinelineNumber\"].value_counts().index) != 0:\n test.loc[(test[\"VisitNumber\"] == loc)&(test[\"FinelineNumber\"].isna()), \"FinelineNumber\"] = test[test[\"VisitNumber\"] == loc][\"FinelineNumber\"].value_counts().index[0]",
"100%|██████████| 1141/1141 [00:09<00:00, 115.74it/s]\n"
],
[
"test.loc[test[\"VisitNumber\"]==874, \"FinelineNumber\"]",
"_____no_output_____"
]
],
[
[
"### UPC\n- DepartmentDescription이 'PHARMACY RX'일때, UPC가 빈값인 경우 DepartmentDescription이 'PHARMACY RX'일때의 UPC의 최빈값으로 채운다.\n- 각각의 VisitNumber 별 UPC의 최빈값으로 UPC의 빈값을 채운다.\n- 191개의 유추할 수 없는 값은 기존에 있던 값과 중복되지 않는 '0000599996' 값으로 대체한다.",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train.loc[train[\"VisitNumber\"]==259, \"Upc\"]",
"_____no_output_____"
],
[
"Upc_VN_list = train[train[\"Upc\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(Upc_VN_list): # if: 특정 VisitNumber 따른 Upc 값이 모두 비어있는 경우 제외\n if len(train[train[\"VisitNumber\"] == loc][\"Upc\"].value_counts().index) != 0:\n train.loc[(train[\"VisitNumber\"] == loc)&(train[\"Upc\"].isna()), \"Upc\"] = train[train[\"VisitNumber\"] == loc][\"Upc\"].value_counts().index[0]",
"100%|██████████| 2754/2754 [00:15<00:00, 182.57it/s]\n"
],
[
"train.loc[train[\"VisitNumber\"]==259, \"Upc\"]",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test.loc[test[\"VisitNumber\"]==874, \"Upc\"]",
"_____no_output_____"
],
[
"Upc_VN_list_t = test[test[\"Upc\"].isna()][\"VisitNumber\"].unique()",
"_____no_output_____"
],
[
"for loc in tqdm(Upc_VN_list_t): # if: 특정 VisitNumber 따른 Upc 값이 모두 비어있는 경우 제외\n if len(test[test[\"VisitNumber\"] == loc][\"Upc\"].value_counts().index) != 0:\n test.loc[(test[\"VisitNumber\"] == loc)&(test[\"Upc\"].isna()), \"Upc\"] = test[test[\"VisitNumber\"] == loc][\"Upc\"].value_counts().index[0]",
"100%|██████████| 2706/2706 [00:14<00:00, 180.60it/s]\n"
],
[
"test.loc[test[\"VisitNumber\"]==874, \"Upc\"]",
"_____no_output_____"
]
],
[
[
"### All of DepartmentDescription, FinelineNumber, Upc is NaN\n\n- 총 191개의 다른 컬럼과의 관계로 추론 불가능한 DepartmentDescription, FinelineNumber, Upc의 값이 모두 비어있는 경우, 기존에 train, test 데이터에 없는 \"UNKNOWN\", -9999, '0000599996' 값으로 각각 채운다.\n- 이 경우에 모든 row들은 TripType이 999이다.",
"_____no_output_____"
]
],
[
[
"empty_df = train[(train[\"DepartmentDescription\"].isna())&(train[\"DepartmentDescription\"].isna())&(train[\"DepartmentDescription\"].isna())][[\"VisitNumber\", \"DepartmentDescription\", \"FinelineNumber\", \"Upc\", \"TripType\", \"Weekday\", \"ScanCount\"]]\n\nprint(empty_df.shape)\nempty_df[[\"DepartmentDescription\", \"FinelineNumber\", \"Upc\"]].head()",
"(191, 7)\n"
],
[
"print(\"191개의 빈 row들은 모두 triptype이 {}이다.\".format(empty_df[\"TripType\"].value_counts().index[0]))\nempty_df[\"TripType\"].value_counts()",
"191개의 빈 row들은 모두 triptype이 999이다.\n"
]
],
[
[
"#### train",
"_____no_output_____"
]
],
[
[
"train.loc[train[\"DepartmentDescription\"].isna(), \"DepartmentDescription\"] = \"UNKNOWN\"\ntrain[train[\"DepartmentDescription\"].isna()]",
"_____no_output_____"
],
[
"train.loc[train[\"FinelineNumber\"].isna(), \"FinelineNumber\"] = -9999.0\ntrain[train[\"FinelineNumber\"].isna()]",
"_____no_output_____"
],
[
"train.loc[train[\"Upc\"].isna(), \"Upc\"] = 0000599996.0\ntrain[train[\"Upc\"].isna()]",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test.loc[test[\"DepartmentDescription\"].isna(), \"DepartmentDescription\"] = \"UNKNOWN\"\ntest[test[\"DepartmentDescription\"].isna()]",
"_____no_output_____"
],
[
"test.loc[test[\"FinelineNumber\"].isna(), \"FinelineNumber\"] = -9999.0\ntest[test[\"FinelineNumber\"].isna()]",
"_____no_output_____"
],
[
"test.loc[test[\"Upc\"].isna(), \"Upc\"] = 0000599996.0\ntest[test[\"Upc\"].isna()]",
"_____no_output_____"
]
],
[
[
"### Divide the Upc into manufacturer(company) number and product number.\n\n- 3~12자리의 여러자기 종류의 UPC를 모두 12자리로 복원 후, 분류에 필요하다고 판단한 company_Upc와 product_Upc로 나누어 인코딩\n- preprocessing function 사용(custom function)",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train[\"Upc\"] = train[\"Upc\"].astype(str)",
"_____no_output_____"
],
[
"train[\"full_Upc\"] = train[\"Upc\"].apply(pf.upc_789101112_to_10)\ntrain[\"full_Upc\"] = train[\"full_Upc\"].apply(pf.upc_3456_to_10)\n\ntrain[\"company_Upc\"] = train[\"full_Upc\"].apply(pf.company_part_Upc)\ntrain[\"product_Upc\"] = train[\"full_Upc\"].apply(pf.product_part_Upc) \n\ntrain[[\"Upc\", \"full_Upc\", \"company_Upc\", \"product_Upc\"]].tail()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test[\"Upc\"] = test[\"Upc\"].astype(str)",
"_____no_output_____"
],
[
"test[\"full_Upc\"] = test[\"Upc\"].apply(pf.upc_789101112_to_10)\ntest[\"full_Upc\"] = test[\"full_Upc\"].apply(pf.upc_3456_to_10)\n\ntest[\"company_Upc\"] = test[\"full_Upc\"].apply(pf.company_part_Upc)\ntest[\"product_Upc\"] = test[\"full_Upc\"].apply(pf.product_part_Upc) \n\ntest[[\"Upc\", \"full_Upc\", \"company_Upc\", \"product_Upc\"]].tail()",
"_____no_output_____"
]
],
[
[
"## part 2 : Encode & Derivation",
"_____no_output_____"
],
[
"## Encode",
"_____no_output_____"
],
[
"### Count the DepartmentDescription for each VisitNumber(Encode)",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train_department = pd.pivot_table(data=train, index='VisitNumber', columns='DepartmentDescription', values='ScanCount', aggfunc='sum')\ntrain_department = train_department.fillna(0)",
"_____no_output_____"
],
[
"train_department.head()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test_department = pd.pivot_table(data=test, index='VisitNumber', columns='DepartmentDescription', values='ScanCount', aggfunc='sum')\ntest_department = test_department.fillna(0)",
"_____no_output_____"
],
[
"test_department.head()",
"_____no_output_____"
]
],
[
[
"### Count the Weekday for each VisitNumber(Encode)",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train_weekday = pd.pivot_table(data=train, index='VisitNumber', columns='Weekday', values='ScanCount', aggfunc='sum')\ntrain_weekday = train_weekday.fillna(0)",
"_____no_output_____"
],
[
"train_weekday.head()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test_weekday = pd.pivot_table(data=test, index='VisitNumber', columns='Weekday', values='ScanCount', aggfunc='sum')\ntest_weekday = test_weekday.fillna(0)",
"_____no_output_____"
],
[
"test_weekday.head()",
"_____no_output_____"
]
],
[
[
"### Count the FinelineNumber for each VisitNumber(Encode)",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train_fineline = pd.pivot_table(data=train, index='VisitNumber', columns='FinelineNumber', values='ScanCount', aggfunc='sum')\ntrain_fineline = train_fineline.fillna(0)",
"_____no_output_____"
],
[
"train_fineline.head()",
"_____no_output_____"
],
[
"test_fineline = pd.pivot_table(data=test, index='VisitNumber', columns='FinelineNumber', values='ScanCount', aggfunc='sum')\ntest_fineline = test_fineline.fillna(0)",
"_____no_output_____"
],
[
"test_fineline.head()",
"_____no_output_____"
]
],
[
[
"### Count the company_Upc for each VisitNumber(Encode)",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train[\"company_Upc\"] = train[\"company_Upc\"].astype('str')\ntrain_company_upc = pd.pivot_table(data=train, index='VisitNumber', columns='company_Upc', values='ScanCount', aggfunc='sum')\ntrain_company_upc = train_company_upc.fillna(0)",
"_____no_output_____"
],
[
"train_company_upc.head()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test[\"company_Upc\"] = test[\"company_Upc\"].astype('str')\ntest_company_upc = pd.pivot_table(data=test, index='VisitNumber', columns='company_Upc', values='ScanCount', aggfunc='sum')\ntest_company_upc = test_company_upc.fillna(0)",
"_____no_output_____"
],
[
"test_company_upc.head()",
"_____no_output_____"
]
],
[
[
"### Concat above columns",
"_____no_output_____"
]
],
[
[
"train_df = pd.concat([train_department, train_fineline, train_company_upc, train_weekday], axis=1)\n\nprint(train_df.shape)\ntrain_df.tail()",
"(95674, 10954)\n"
],
[
"test_df = pd.concat([test_department, test_fineline, test_company_upc, test_weekday], axis=1)\ntest_df = test_df.reset_index(drop=False)",
"_____no_output_____"
]
],
[
[
"## Derivation",
"_____no_output_____"
],
[
"### 파생 컬럼 생성을 위한 abs_ScanCount생성",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train[\"abs_ScanCount\"] = np.abs(train[\"ScanCount\"])",
"_____no_output_____"
],
[
"train_abs_ScanCount = train.groupby(by=\"VisitNumber\").sum().reset_index()[\"abs_ScanCount\"]\ntrain_abs_ScanCount.head()",
"_____no_output_____"
],
[
"train_ScanCount = train.groupby(by=\"VisitNumber\").sum().reset_index()[\"ScanCount\"]\ntrain_ScanCount.head()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test[\"abs_ScanCount\"] = np.abs(test[\"ScanCount\"])",
"_____no_output_____"
],
[
"test_abs_ScanCount = test.groupby(by=\"VisitNumber\").sum().reset_index()[\"abs_ScanCount\"]\ntest_abs_ScanCount.head()",
"_____no_output_____"
],
[
"test_ScanCount = test.groupby(by=\"VisitNumber\").sum().reset_index()[\"ScanCount\"]\ntest_ScanCount.head()",
"_____no_output_____"
]
],
[
[
"### Divide by abs_scancount for each columns\n- ScanCount로 각 컬럼을 나눠 산 물건 종류의 비율을 할당\n- 절대값 사용 이유는 한번 샀다가 환불한 경우도 구매했던 이력으로 간주",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train_df = train_df.reset_index(drop=False)\ntrain_df[\"abs_ScanCount\"] = train_abs_ScanCount\n\ntrain_columns = list(train_df.columns)\n\nfor col_name in tqdm(train_columns):\n train_df[col_name] = train_df[col_name] / train_df[\"abs_ScanCount\"]",
"100%|██████████| 10956/10956 [00:08<00:00, 1280.03it/s]\n"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test_df = test_df.reset_index(drop=False)\ntest_df[\"abs_ScanCount\"] = test_abs_ScanCount\ntest_columns = list(test_df.columns)\n\nfor col_name in tqdm(test_columns):\n test_df[col_name] = test_df[col_name] / test_df[\"abs_ScanCount\"]\n ",
"100%|██████████| 10986/10986 [00:10<00:00, 1057.54it/s]\n"
]
],
[
[
"### Refund rate\n- 999타입은 대부분이 환불 고객이며, 환불 비율에 따라 고객 유형 구분",
"_____no_output_____"
],
[
"#### train",
"_____no_output_____"
]
],
[
[
"train_df[\"ScanCount\"] = train_ScanCount\ntrain_df[\"refund_rate\"] = ((train_abs_ScanCount - train_ScanCount) / 2) / train_abs_ScanCount",
"_____no_output_____"
],
[
"train_df[\"refund_rate\"].head()",
"_____no_output_____"
]
],
[
[
"#### test",
"_____no_output_____"
]
],
[
[
"test_df[\"ScanCount\"] = test_ScanCount\ntest_df[\"refund_rate\"] = ((test_abs_ScanCount - test_ScanCount) / 2) / test_abs_ScanCount",
"_____no_output_____"
],
[
"test_df[\"refund_rate\"].head()",
"_____no_output_____"
]
],
[
[
"### part3 : Feature Selection\n- feature로 필요하지 않은 abs_ScanCount, VisitNumber 제거\n- train, test 데이터 중 중복되는 company_upc(5304 columns), finelinenumber(5045 columns)만 사용",
"_____no_output_____"
]
],
[
[
"del train_df[\"abs_ScanCount\"]\ndel train_df[\"VisitNumber\"]\n\ndel test_df[\"abs_ScanCount\"]\ndel test_df[\"VisitNumber\"]\n\ntrain_company_list = list(train[\"company_Upc\"].value_counts().index)\ntest_company_list = list(test[\"company_Upc\"].value_counts().index)\n\ntrain_fineline_list = list(train[\"FinelineNumber\"].value_counts().index)\ntest_fineline_list = list(test[\"FinelineNumber\"].value_counts().index)\n\ncompany_feature = list(set(train_company_list) & set(test_company_list))\nfineline_feature = list(set(train_fineline_list) & set(test_fineline_list))\n\nfeature_names = list(train_department.columns) + list(train_weekday.columns) + company_feature + fineline_feature + [\"ScanCount\", \"refund_rate\"]\nprint(len(feature_names))",
"10425\n"
]
],
[
[
"### X_train",
"_____no_output_____"
]
],
[
[
"X_train = train_df[feature_names]\n\nprint(X_train.shape)\nX_train.tail()",
"(95674, 10425)\n"
]
],
[
[
"### y_train",
"_____no_output_____"
]
],
[
[
"y_train = train.groupby(by=\"VisitNumber\").mean().reset_index()[\"TripType\"]\n\nprint(y_train.shape)\ny_train[:5]",
"(95674,)\n"
]
],
[
[
"### X_test",
"_____no_output_____"
]
],
[
[
"X_test = test_df[feature_names]\n\nprint(X_test.shape)\nX_test.tail()",
"(95674, 10425)\n"
]
],
[
[
"### Sparse matrix - csr matrix",
"_____no_output_____"
]
],
[
[
"from scipy.sparse import csr_matrix",
"_____no_output_____"
],
[
"X_train = csr_matrix(X_train)\n\nX_train",
"_____no_output_____"
],
[
"X_test = csr_matrix(X_test)\n\nX_test",
"_____no_output_____"
]
],
[
[
"### Label Encode",
"_____no_output_____"
]
],
[
[
"from sklearn.preprocessing import LabelEncoder\n\nencoder = LabelEncoder()\ny_train = encoder.fit_transform(y_train)\n\nprint(y_train.shape)\ny_train[:5]",
"(95674,)\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6fb7a3ec0817d97f976834ed2c21c12e31c158 | 74,946 | ipynb | Jupyter Notebook | HMM Tagger.ipynb | hlbowers/hmm-tagger | 0e3354b2bac97de73866767d1e0a1f70dd9587ca | [
"MIT"
]
| null | null | null | HMM Tagger.ipynb | hlbowers/hmm-tagger | 0e3354b2bac97de73866767d1e0a1f70dd9587ca | [
"MIT"
]
| null | null | null | HMM Tagger.ipynb | hlbowers/hmm-tagger | 0e3354b2bac97de73866767d1e0a1f70dd9587ca | [
"MIT"
]
| null | null | null | 58.643192 | 721 | 0.503736 | [
[
[
"# Project: Part of Speech Tagging with Hidden Markov Models \n---\n### Introduction\n\nPart of speech tagging is the process of determining the syntactic category of a word from the words in its surrounding context. It is often used to help disambiguate natural language phrases because it can be done quickly with high accuracy. Tagging can be used for many NLP tasks like determining correct pronunciation during speech synthesis (for example, _dis_-count as a noun vs dis-_count_ as a verb), for information retrieval, and for word sense disambiguation.\n\nIn this notebook, you'll use the [Pomegranate](http://pomegranate.readthedocs.io/) library to build a hidden Markov model for part of speech tagging using a \"universal\" tagset. Hidden Markov models have been able to achieve [>96% tag accuracy with larger tagsets on realistic text corpora](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf). Hidden Markov models have also been used for speech recognition and speech generation, machine translation, gene recognition for bioinformatics, and human gesture recognition for computer vision, and more. \n\n\n\nThe notebook already contains some code to get you started. You only need to add some new functionality in the areas indicated to complete the project; you will not need to modify the included code beyond what is requested. Sections that begin with **'IMPLEMENTATION'** in the header indicate that you must provide code in the block that follows. Instructions will be provided for each section, and the specifics of the implementation are marked in the code block with a 'TODO' statement. Please be sure to read the instructions carefully!",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Once you have completed all of the code implementations, you need to finalize your work by exporting the iPython Notebook as an HTML document. Before exporting the notebook to html, all of the code cells need to have been run so that reviewers can see the final implementation and output. You must then **export the notebook** by running the last cell in the notebook, or by using the menu above and navigating to **File -> Download as -> HTML (.html)** Your submissions should include both the `html` and `ipynb` files.\n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\">\n**Note:** Code and Markdown cells can be executed using the `Shift + Enter` keyboard shortcut. Markdown cells can be edited by double-clicking the cell to enter edit mode.\n</div>",
"_____no_output_____"
],
[
"### The Road Ahead\nYou must complete Steps 1-3 below to pass the project. The section on Step 4 includes references & resources you can use to further explore HMM taggers.\n\n- [Step 1](#Step-1:-Read-and-preprocess-the-dataset): Review the provided interface to load and access the text corpus\n- [Step 2](#Step-2:-Build-a-Most-Frequent-Class-tagger): Build a Most Frequent Class tagger to use as a baseline\n- [Step 3](#Step-3:-Build-an-HMM-tagger): Build an HMM Part of Speech tagger and compare to the MFC baseline\n- [Step 4](#Step-4:-[Optional]-Improving-model-performance): (Optional) Improve the HMM tagger",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-warning\">\n**Note:** Make sure you have selected a **Python 3** kernel in Workspaces or the hmm-tagger conda environment if you are running the Jupyter server on your own machine.\n</div>",
"_____no_output_____"
]
],
[
[
"# Jupyter \"magic methods\" -- only need to be run once per kernel restart\n%load_ext autoreload\n%aimport helpers, tests\n%autoreload 1",
"_____no_output_____"
],
[
"# import python modules -- this cell needs to be run again if you make changes to any of the files\nimport matplotlib.pyplot as plt\nimport numpy as np\n\nfrom IPython.core.display import HTML\nfrom itertools import chain\nfrom collections import Counter, defaultdict\nfrom helpers import show_model, Dataset\nfrom pomegranate import State, HiddenMarkovModel, DiscreteDistribution",
"_____no_output_____"
]
],
[
[
"## Step 1: Read and preprocess the dataset\n---\nWe'll start by reading in a text corpus and splitting it into a training and testing dataset. The data set is a copy of the [Brown corpus](https://en.wikipedia.org/wiki/Brown_Corpus) (originally from the [NLTK](https://www.nltk.org/) library) that has already been pre-processed to only include the [universal tagset](https://arxiv.org/pdf/1104.2086.pdf). You should expect to get slightly higher accuracy using this simplified tagset than the same model would achieve on a larger tagset like the full [Penn treebank tagset](https://www.ling.upenn.edu/courses/Fall_2003/ling001/penn_treebank_pos.html), but the process you'll follow would be the same.\n\nThe `Dataset` class provided in helpers.py will read and parse the corpus. You can generate your own datasets compatible with the reader by writing them to the following format. The dataset is stored in plaintext as a collection of words and corresponding tags. Each sentence starts with a unique identifier on the first line, followed by one tab-separated word/tag pair on each following line. Sentences are separated by a single blank line.\n\nExample from the Brown corpus. \n```\nb100-38532\nPerhaps\tADV\nit\tPRON\nwas\tVERB\nright\tADJ\n;\t.\n;\t.\n\nb100-35577\n...\n```",
"_____no_output_____"
]
],
[
[
"data = Dataset(\"tags-universal.txt\", \"brown-universal.txt\", train_test_split=0.8)\n\nprint(\"There are {} sentences in the corpus.\".format(len(data)))\nprint(\"There are {} sentences in the training set.\".format(len(data.training_set)))\nprint(\"There are {} sentences in the testing set.\".format(len(data.testing_set)))\n\nassert len(data) == len(data.training_set) + len(data.testing_set), \\\n \"The number of sentences in the training set + testing set should sum to the number of sentences in the corpus\"",
"There are 57340 sentences in the corpus.\nThere are 45872 sentences in the training set.\nThere are 11468 sentences in the testing set.\n"
]
],
[
[
"### The Dataset Interface\n\nYou can access (mostly) immutable references to the dataset through a simple interface provided through the `Dataset` class, which represents an iterable collection of sentences along with easy access to partitions of the data for training & testing. Review the reference below, then run and review the next few cells to make sure you understand the interface before moving on to the next step.\n\n```\nDataset-only Attributes:\n training_set - reference to a Subset object containing the samples for training\n testing_set - reference to a Subset object containing the samples for testing\n\nDataset & Subset Attributes:\n sentences - a dictionary with an entry {sentence_key: Sentence()} for each sentence in the corpus\n keys - an immutable ordered (not sorted) collection of the sentence_keys for the corpus\n vocab - an immutable collection of the unique words in the corpus\n tagset - an immutable collection of the unique tags in the corpus\n X - returns an array of words grouped by sentences ((w11, w12, w13, ...), (w21, w22, w23, ...), ...)\n Y - returns an array of tags grouped by sentences ((t11, t12, t13, ...), (t21, t22, t23, ...), ...)\n N - returns the number of distinct samples (individual words or tags) in the dataset\n\nMethods:\n stream() - returns an flat iterable over all (word, tag) pairs across all sentences in the corpus\n __iter__() - returns an iterable over the data as (sentence_key, Sentence()) pairs\n __len__() - returns the nubmer of sentences in the dataset\n```\n\nFor example, consider a Subset, `subset`, of the sentences `{\"s0\": Sentence((\"See\", \"Spot\", \"run\"), (\"VERB\", \"NOUN\", \"VERB\")), \"s1\": Sentence((\"Spot\", \"ran\"), (\"NOUN\", \"VERB\"))}`. The subset will have these attributes:\n\n```\nsubset.keys == {\"s1\", \"s0\"} # unordered\nsubset.vocab == {\"See\", \"run\", \"ran\", \"Spot\"} # unordered\nsubset.tagset == {\"VERB\", \"NOUN\"} # unordered\nsubset.X == ((\"Spot\", \"ran\"), (\"See\", \"Spot\", \"run\")) # order matches .keys\nsubset.Y == ((\"NOUN\", \"VERB\"), (\"VERB\", \"NOUN\", \"VERB\")) # order matches .keys\nsubset.N == 7 # there are a total of seven observations over all sentences\nlen(subset) == 2 # because there are two sentences\n```\n\n<div class=\"alert alert-block alert-info\">\n**Note:** The `Dataset` class is _convenient_, but it is **not** efficient. It is not suitable for huge datasets because it stores multiple redundant copies of the same data.\n</div>",
"_____no_output_____"
],
[
"#### Sentences\n\n`Dataset.sentences` is a dictionary of all sentences in the training corpus, each keyed to a unique sentence identifier. Each `Sentence` is itself an object with two attributes: a tuple of the words in the sentence named `words` and a tuple of the tag corresponding to each word named `tags`.",
"_____no_output_____"
]
],
[
[
"key = 'b100-38532'\nprint(\"Sentence: {}\".format(key))\nprint(\"words:\\n\\t{!s}\".format(data.sentences[key].words))\nprint(\"tags:\\n\\t{!s}\".format(data.sentences[key].tags))",
"Sentence: b100-38532\nwords:\n\t('Perhaps', 'it', 'was', 'right', ';', ';')\ntags:\n\t('ADV', 'PRON', 'VERB', 'ADJ', '.', '.')\n"
]
],
[
[
"<div class=\"alert alert-block alert-info\">\n**Note:** The underlying iterable sequence is **unordered** over the sentences in the corpus; it is not guaranteed to return the sentences in a consistent order between calls. Use `Dataset.stream()`, `Dataset.keys`, `Dataset.X`, or `Dataset.Y` attributes if you need ordered access to the data.\n</div>\n\n#### Counting Unique Elements\n\nYou can access the list of unique words (the dataset vocabulary) via `Dataset.vocab` and the unique list of tags via `Dataset.tagset`.",
"_____no_output_____"
]
],
[
[
"print(\"There are a total of {} samples of {} unique words in the corpus.\"\n .format(data.N, len(data.vocab)))\nprint(\"There are {} samples of {} unique words in the training set.\"\n .format(data.training_set.N, len(data.training_set.vocab)))\nprint(\"There are {} samples of {} unique words in the testing set.\"\n .format(data.testing_set.N, len(data.testing_set.vocab)))\nprint(\"There are {} words in the test set that are missing in the training set.\"\n .format(len(data.testing_set.vocab - data.training_set.vocab)))\n\nassert data.N == data.training_set.N + data.testing_set.N, \\\n \"The number of training + test samples should sum to the total number of samples\"",
"There are a total of 1161192 samples of 56057 unique words in the corpus.\nThere are 928458 samples of 50536 unique words in the training set.\nThere are 232734 samples of 25112 unique words in the testing set.\nThere are 5521 words in the test set that are missing in the training set.\n"
]
],
[
[
"#### Accessing word and tag Sequences\nThe `Dataset.X` and `Dataset.Y` attributes provide access to ordered collections of matching word and tag sequences for each sentence in the dataset.",
"_____no_output_____"
]
],
[
[
"# accessing words with Dataset.X and tags with Dataset.Y \nfor i in range(2): \n print(\"Sentence {}:\".format(i + 1), data.X[i])\n print()\n print(\"Labels {}:\".format(i + 1), data.Y[i])\n print()",
"Sentence 1: ('Mr.', 'Podger', 'had', 'thanked', 'him', 'gravely', ',', 'and', 'now', 'he', 'made', 'use', 'of', 'the', 'advice', '.')\n\nLabels 1: ('NOUN', 'NOUN', 'VERB', 'VERB', 'PRON', 'ADV', '.', 'CONJ', 'ADV', 'PRON', 'VERB', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\nSentence 2: ('But', 'there', 'seemed', 'to', 'be', 'some', 'difference', 'of', 'opinion', 'as', 'to', 'how', 'far', 'the', 'board', 'should', 'go', ',', 'and', 'whose', 'advice', 'it', 'should', 'follow', '.')\n\nLabels 2: ('CONJ', 'PRT', 'VERB', 'PRT', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'ADP', 'ADV', 'ADV', 'DET', 'NOUN', 'VERB', 'VERB', '.', 'CONJ', 'DET', 'NOUN', 'PRON', 'VERB', 'VERB', '.')\n\n"
]
],
[
[
"#### Accessing (word, tag) Samples\nThe `Dataset.stream()` method returns an iterator that chains together every pair of (word, tag) entries across all sentences in the entire corpus.",
"_____no_output_____"
]
],
[
[
"# use Dataset.stream() (word, tag) samples for the entire corpus\nprint(\"\\nStream (word, tag) pairs:\\n\")\nfor i, pair in enumerate(data.stream()):\n print(\"\\t\", pair)\n if i > 5: break",
"\nStream (word, tag) pairs:\n\n\t ('Mr.', 'NOUN')\n\t ('Podger', 'NOUN')\n\t ('had', 'VERB')\n\t ('thanked', 'VERB')\n\t ('him', 'PRON')\n\t ('gravely', 'ADV')\n\t (',', '.')\n"
]
],
[
[
"\nFor both our baseline tagger and the HMM model we'll build, we need to estimate the frequency of tags & words from the frequency counts of observations in the training corpus. In the next several cells you will complete functions to compute the counts of several sets of counts. ",
"_____no_output_____"
],
[
"## Step 2: Build a Most Frequent Class tagger\n---\n\nPerhaps the simplest tagger (and a good baseline for tagger performance) is to simply choose the tag most frequently assigned to each word. This \"most frequent class\" tagger inspects each observed word in the sequence and assigns it the label that was most often assigned to that word in the corpus.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Pair Counts\n\nComplete the function below that computes the joint frequency counts for two input sequences.",
"_____no_output_____"
]
],
[
[
"def pair_counts(sequences_A, sequences_B):\n \"\"\"Return a dictionary keyed to each unique value in the first sequence list\n that counts the number of occurrences of the corresponding value from the\n second sequences list.\n \n For example, if sequences_A is tags and sequences_B is the corresponding\n words, then if 1244 sequences contain the word \"time\" tagged as a NOUN, then\n you should return a dictionary such that pair_counts[NOUN][time] == 1244\n \"\"\"\n pair_counts={}\n icnt=0\n for i in sequences_A: \n for (seqA, seqB) in zip(sequences_A[icnt], sequences_B[icnt]):\n if pair_counts.get(seqA,None)== None:\n pair_counts[seqA]={}\n pair_counts[seqA][seqB]=1\n else:\n if (pair_counts[seqA].get(seqB,None) == None):\n pair_counts[seqA][seqB]=1\n else:\n pair_counts[seqA][seqB]+=1 \n icnt+=1\n return pair_counts\n\n# Calculate C(t_i, w_i)\nemission_counts = pair_counts(data.training_set.Y,data.training_set.X)\n\nassert len(emission_counts) == 12, \\\n \"Uh oh. There should be 12 tags in your dictionary.\"\nassert max(emission_counts[\"NOUN\"], key=emission_counts[\"NOUN\"].get) == 'time', \\\n \"Hmmm...'time' is expected to be the most common NOUN.\"\nHTML('<div class=\"alert alert-block alert-success\">Your emission counts look good!</div>')\n \n\n",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Most Frequent Class Tagger\n\nUse the `pair_counts()` function and the training dataset to find the most frequent class label for each word in the training data, and populate the `mfc_table` below. The table keys should be words, and the values should be the appropriate tag string.\n\nThe `MFCTagger` class is provided to mock the interface of Pomegranite HMM models so that they can be used interchangeably.",
"_____no_output_____"
]
],
[
[
"# Create a lookup table mfc_table where mfc_table[word] contains the tag label most frequently assigned to that word\nfrom collections import namedtuple\n\nFakeState = namedtuple(\"FakeState\", \"name\")\n\nclass MFCTagger:\n # NOTE: You should not need to modify this class or any of its methods\n missing = FakeState(name=\"<MISSING>\")\n \n def __init__(self, table):\n self.table = defaultdict(lambda: MFCTagger.missing)\n self.table.update({word: FakeState(name=tag) for word, tag in table.items()})\n \n def viterbi(self, seq):\n \"\"\"This method simplifies predictions by matching the Pomegranate viterbi() interface\"\"\"\n return 0., list(enumerate([\"<start>\"] + [self.table[w] for w in seq] + [\"<end>\"]))\n\n\n# TODO: calculate the frequency of each tag being assigned to each word (hint: similar, but not\n# the same as the emission probabilities) and use it to fill the mfc_table\n\nword_counts={}\nword_counts = pair_counts(data.training_set.X,data.training_set.Y)\n\nmfc_table = {}\n\nicnt=0\nfor i in word_counts:\n #print(i)\n mfc_table[i] = max(word_counts[i], key=word_counts[i].get)\n icnt+=1\n #if (icnt==10):\n # break\n \n\n# DO NOT MODIFY BELOW THIS LINE\nmfc_model = MFCTagger(mfc_table) # Create a Most Frequent Class tagger instance\n\nassert len(mfc_table) == len(data.training_set.vocab), \"\"\nassert all(k in data.training_set.vocab for k in mfc_table.keys()), \"\"\nassert sum(int(k not in mfc_table) for k in data.testing_set.vocab) == 5521, \"\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger has all the correct words!</div>')",
"_____no_output_____"
]
],
[
[
"### Making Predictions with a Model\nThe helper functions provided below interface with Pomegranate network models & the mocked MFCTagger to take advantage of the [missing value](http://pomegranate.readthedocs.io/en/latest/nan.html) functionality in Pomegranate through a simple sequence decoding function. Run these functions, then run the next cell to see some of the predictions made by the MFC tagger.",
"_____no_output_____"
]
],
[
[
"def replace_unknown(sequence):\n \"\"\"Return a copy of the input sequence where each unknown word is replaced\n by the literal string value 'nan'. Pomegranate will ignore these values\n during computation.\n \"\"\"\n return [w if w in data.training_set.vocab else 'nan' for w in sequence]\n\ndef simplify_decoding(X, model):\n \"\"\"X should be a 1-D sequence of observations for the model to predict\"\"\"\n _, state_path = model.viterbi(replace_unknown(X))\n return [state[1].name for state in state_path[1:-1]] # do not show the start/end state predictions",
"_____no_output_____"
]
],
[
[
"### Example Decoding Sequences with MFC Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, mfc_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"Sentence Key: b100-28144\n\nPredicted labels:\n-----------------\n['CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.']\n\nActual labels:\n--------------\n('CONJ', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'NOUN', 'NUM', '.', 'CONJ', 'NOUN', 'NUM', '.', '.', 'NOUN', '.', '.')\n\n\nSentence Key: b100-23146\n\nPredicted labels:\n-----------------\n['PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.']\n\nActual labels:\n--------------\n('PRON', 'VERB', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', 'NOUN', 'VERB', 'VERB', '.', 'ADP', 'VERB', 'DET', 'NOUN', 'ADP', 'NOUN', 'ADP', 'DET', 'NOUN', '.')\n\n\nSentence Key: b100-35462\n\nPredicted labels:\n-----------------\n['DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', '<MISSING>', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADV', 'NOUN', '.']\n\nActual labels:\n--------------\n('DET', 'ADJ', 'NOUN', 'VERB', 'VERB', 'VERB', 'ADP', 'DET', 'ADJ', 'ADJ', 'NOUN', 'ADP', 'DET', 'ADJ', 'NOUN', '.', 'ADP', 'ADJ', 'NOUN', '.', 'CONJ', 'ADP', 'DET', 'NOUN', 'ADP', 'ADJ', 'ADJ', '.', 'ADJ', '.', 'CONJ', 'ADJ', 'NOUN', 'ADP', 'ADJ', 'NOUN', '.')\n\n\n"
]
],
[
[
"### Evaluating Model Accuracy\n\nThe function below will evaluate the accuracy of the MFC tagger on the collection of all sentences from a text corpus. ",
"_____no_output_____"
]
],
[
[
"def accuracy(X, Y, model):\n \"\"\"Calculate the prediction accuracy by using the model to decode each sequence\n in the input X and comparing the prediction with the true labels in Y.\n \n The X should be an array whose first dimension is the number of sentences to test,\n and each element of the array should be an iterable of the words in the sequence.\n The arrays X and Y should have the exact same shape.\n \n X = [(\"See\", \"Spot\", \"run\"), (\"Run\", \"Spot\", \"run\", \"fast\"), ...]\n Y = [(), (), ...]\n \"\"\"\n correct = total_predictions = 0\n for observations, actual_tags in zip(X, Y):\n \n # The model.viterbi call in simplify_decoding will return None if the HMM\n # raises an error (for example, if a test sentence contains a word that\n # is out of vocabulary for the training set). Any exception counts the\n # full sentence as an error (which makes this a conservative estimate).\n try:\n most_likely_tags = simplify_decoding(observations, model)\n correct += sum(p == t for p, t in zip(most_likely_tags, actual_tags))\n except:\n pass\n total_predictions += len(observations)\n return correct / total_predictions",
"_____no_output_____"
]
],
[
[
"#### Evaluate the accuracy of the MFC tagger\nRun the next cell to evaluate the accuracy of the tagger on the training and test corpus.",
"_____no_output_____"
]
],
[
[
"mfc_training_acc = accuracy(data.training_set.X, data.training_set.Y, mfc_model)\nprint(\"training accuracy mfc_model: {:.2f}%\".format(100 * mfc_training_acc))\n\nmfc_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, mfc_model)\nprint(\"testing accuracy mfc_model: {:.2f}%\".format(100 * mfc_testing_acc))\n\nassert mfc_training_acc >= 0.955, \"Uh oh. Your MFC accuracy on the training set doesn't look right.\"\nassert mfc_testing_acc >= 0.925, \"Uh oh. Your MFC accuracy on the testing set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your MFC tagger accuracy looks correct!</div>')",
"training accuracy mfc_model: 95.72%\ntesting accuracy mfc_model: 93.01%\n"
]
],
[
[
"## Step 3: Build an HMM tagger\n---\nThe HMM tagger has one hidden state for each possible tag, and parameterized by two distributions: the emission probabilties giving the conditional probability of observing a given **word** from each hidden state, and the transition probabilities giving the conditional probability of moving between **tags** during the sequence.\n\nWe will also estimate the starting probability distribution (the probability of each **tag** being the first tag in a sequence), and the terminal probability distribution (the probability of each **tag** being the last tag in a sequence).\n\nThe maximum likelihood estimate of these distributions can be calculated from the frequency counts as described in the following sections where you'll implement functions to count the frequencies, and finally build the model. The HMM model will make predictions according to the formula:\n\n$$t_i^n = \\underset{t_i^n}{\\mathrm{argmax}} \\prod_{i=1}^n P(w_i|t_i) P(t_i|t_{i-1})$$\n\nRefer to Speech & Language Processing [Chapter 10](https://web.stanford.edu/~jurafsky/slp3/10.pdf) for more information.",
"_____no_output_____"
],
[
"### IMPLEMENTATION: Unigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each symbol over all of the input sequences. The unigram probabilities in our HMM model are estimated from the formula below, where N is the total number of samples in the input. (You only need to compute the counts for now.)\n\n$$P(tag_1) = \\frac{C(tag_1)}{N}$$",
"_____no_output_____"
]
],
[
[
"def unigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequence list that\n counts the number of occurrences of the value in the sequences list. The sequences\n collection should be a 2-dimensional array.\n \n For example, if the tag NOUN appears 275558 times over all the input sequences,\n then you should return a dictionary such that your_unigram_counts[NOUN] == 275558.\n \"\"\"\n unigram_counts={}\n icnt=0\n for i in sequences:\n for j in sequences[icnt]:\n if unigram_counts.get(j,None)== None:\n unigram_counts[j]=1\n else:\n unigram_counts[j]+=1 \n icnt+=1\n return unigram_counts \n \n \n # TODO: Finish this function!\n #raise NotImplementedError\n\n# TODO: call unigram_counts with a list of tag sequences from the training set\ntag_unigrams = unigram_counts(data.training_set.Y)\n\nassert set(tag_unigrams.keys()) == data.training_set.tagset, \\\n \"Uh oh. It looks like your tag counts doesn't include all the tags!\"\nassert min(tag_unigrams, key=tag_unigrams.get) == 'X', \\\n \"Hmmm...'X' is expected to be the least common class\"\nassert max(tag_unigrams, key=tag_unigrams.get) == 'NOUN', \\\n \"Hmmm...'NOUN' is expected to be the most common class\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag unigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Bigram Counts\n\nComplete the function below to estimate the co-occurrence frequency of each pair of symbols in each of the input sequences. These counts are used in the HMM model to estimate the bigram probability of two tags from the frequency counts according to the formula: $$P(tag_2|tag_1) = \\frac{C(tag_2|tag_1)}{C(tag_2)}$$\n",
"_____no_output_____"
]
],
[
[
"# Testing out the end conditions\ndcnt=4\nbigram_counts={}\nfor i in data.training_set.Y:\n print(len(i))\n print(i)\n len_i_p=len(i)\n if (len_i_p%2!=0):\n len_i_p=len_i_p-1\n for jcnt in range(0,len_i_p-1):\n print(i[jcnt],\" \",i[jcnt+1])\n k=i[jcnt]\n l=i[jcnt+1]\n m=(k,l)\n if bigram_counts.get(m,None)== None:\n bigram_counts[m]=1\n else:\n bigram_counts[m]+=1\n print(bigram_counts)\n dcnt=dcnt-1\n if (dcnt==1):\n break",
"36\n('ADV', 'NOUN', '.', 'ADV', '.', 'VERB', 'ADP', 'ADJ', 'NOUN', 'CONJ', 'VERB', 'ADJ', 'NOUN', '.', 'DET', 'VERB', 'ADJ', 'PRT', 'ADP', 'NUM', 'NOUN', '.', 'PRON', 'VERB', 'PRT', 'VERB', 'NOUN', '.', 'VERB', 'NOUN', 'NUM', '.', 'NUM', '.', '.', '.')\nADV NOUN\nNOUN .\n. ADV\nADV .\n. VERB\nVERB ADP\nADP ADJ\nADJ NOUN\nNOUN CONJ\nCONJ VERB\nVERB ADJ\nADJ NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 1, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 1, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 1}\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 2, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 1, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 1}\n. DET\nDET VERB\nVERB ADJ\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 2, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 1, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1}\nADJ PRT\nPRT ADP\nADP NUM\nNUM NOUN\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 3, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 1, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1}\n. PRON\nPRON VERB\nVERB PRT\nPRT VERB\nVERB NOUN\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 1, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 1}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 1}\nVERB NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2}\nNOUN NUM\nNUM .\n. NUM\nNUM .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1}\n. .\n. .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 1, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2}\n42\n('ADP', 'ADV', 'NUM', 'NOUN', '.', 'DET', 'NOUN', 'CONJ', 'DET', 'NOUN', 'VERB', 'ADP', 'NOUN', 'VERB', 'ADP', 'DET', 'NOUN', 'ADP', 'NOUN', 'CONJ', 'NOUN', '.', 'NOUN', '.', 'NOUN', '.', 'NOUN', '.', 'VERB', '.', 'VERB', 'ADP', 'NOUN', '.', 'VERB', '.', 'VERB', '.', 'VERB', '.', 'VERB', '.')\nADP ADV\nADV NUM\nNUM NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 4, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1}\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 1, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1}\n. DET\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 1, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1}\nDET NOUN\nNOUN CONJ\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 1}\nCONJ DET\nDET NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 1, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 2, ('CONJ', 'DET'): 1}\nNOUN VERB\nVERB ADP\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 2, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 2, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 1}\nADP NOUN\nNOUN VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 2, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 2, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 1}\nVERB ADP\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 2, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 1}\nADP DET\nDET NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 1, ('ADP', 'DET'): 1}\nNOUN ADP\nADP NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 2, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1}\nNOUN CONJ\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 5, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1}\nCONJ NOUN\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 6, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1}\n. NOUN\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 7, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 1}\n. NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 7, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 2}\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 8, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 2}\n. NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 8, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3}\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 9, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 2, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 9, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 3, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3}\nVERB .\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 9, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 4, ('VERB', 'ADP'): 3, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 1}\nVERB ADP\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 9, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 4, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 2, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 1}\nADP NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 9, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 4, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 1}\nNOUN .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 4, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 1}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 5, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 1}\nVERB .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 5, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 2}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 6, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 2}\nVERB .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 6, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 3}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 7, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 3}\nVERB .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 7, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 4}\n. VERB\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 8, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 4}\nVERB .\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 8, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 2, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 5}\n2\n('ADJ', 'NOUN')\nADJ NOUN\n{('ADV', 'NOUN'): 1, ('NOUN', '.'): 10, ('.', 'ADV'): 1, ('ADV', '.'): 1, ('.', 'VERB'): 8, ('VERB', 'ADP'): 4, ('ADP', 'ADJ'): 1, ('ADJ', 'NOUN'): 3, ('NOUN', 'CONJ'): 3, ('CONJ', 'VERB'): 1, ('VERB', 'ADJ'): 2, ('.', 'DET'): 2, ('DET', 'VERB'): 1, ('ADJ', 'PRT'): 1, ('PRT', 'ADP'): 1, ('ADP', 'NUM'): 1, ('NUM', 'NOUN'): 2, ('.', 'PRON'): 1, ('PRON', 'VERB'): 1, ('VERB', 'PRT'): 1, ('PRT', 'VERB'): 1, ('VERB', 'NOUN'): 2, ('NOUN', 'NUM'): 1, ('NUM', '.'): 2, ('.', 'NUM'): 1, ('.', '.'): 2, ('ADP', 'ADV'): 1, ('ADV', 'NUM'): 1, ('DET', 'NOUN'): 3, ('CONJ', 'DET'): 1, ('NOUN', 'VERB'): 2, ('ADP', 'NOUN'): 3, ('ADP', 'DET'): 1, ('NOUN', 'ADP'): 1, ('CONJ', 'NOUN'): 1, ('.', 'NOUN'): 3, ('VERB', '.'): 5}\n"
],
[
"def bigram_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique PAIR of values in the input sequences\n list that counts the number of occurrences of pair in the sequences list. The input\n should be a 2-dimensional array.\n \n For example, if the pair of tags (NOUN, VERB) appear 61582 times, then you should\n return a dictionary such that your_bigram_counts[(NOUN, VERB)] == 61582\n \"\"\"\n bigram_counts={}\n icnt=0\n for i in sequences:\n maxlen=len(i)\n if (maxlen%2 !=0):\n maxlen=maxlen-1\n for jcnt in range(0,maxlen-1):\n k=i[jcnt]\n l=i[jcnt+1]\n m=(k,l)\n if bigram_counts.get(m,None)== None:\n bigram_counts[m]=1\n else:\n bigram_counts[m]+=1\n icnt+=1\n return bigram_counts\n # TODO: Finish this function!\n #raise NotImplementedError\n\n# TODO: call bigram_counts with a list of tag sequences from the training set\ntag_bigrams = bigram_counts(data.training_set.Y)\n\nassert len(tag_bigrams) == 144, \\\n \"Uh oh. There should be 144 pairs of bigrams (12 tags x 12 tags)\"\nassert min(tag_bigrams, key=tag_bigrams.get) in [('X', 'NUM'), ('PRON', 'X')], \\\n \"Hmmm...The least common bigram should be one of ('X', 'NUM') or ('PRON', 'X').\"\nassert max(tag_bigrams, key=tag_bigrams.get) in [('DET', 'NOUN')], \\\n \"Hmmm...('DET', 'NOUN') is expected to be the most common bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your tag bigrams look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Starting Counts\nComplete the code below to estimate the bigram probabilities of a sequence starting with each tag.",
"_____no_output_____"
]
],
[
[
"def starting_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the beginning of\n a sequence.\n \n For example, if 8093 sequences start with NOUN, then you should return a\n dictionary such that your_starting_counts[NOUN] == 8093\n \"\"\"\n \n starting_counts={}\n icnt=0\n for i in sequences:\n m=i[0]\n if starting_counts.get(m,None)== None:\n starting_counts[m]=1\n else:\n starting_counts[m]+=1\n icnt+=1\n return starting_counts\n # TODO: Finish this function!\n #raise NotImplementedError\n\n# TODO: Calculate the count of each tag starting a sequence\ntag_starts = starting_counts(data.training_set.Y)\n\nassert len(tag_starts) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_starts, key=tag_starts.get) == 'X', \"Hmmm...'X' is expected to be the least common starting bigram.\"\nassert max(tag_starts, key=tag_starts.get) == 'DET', \"Hmmm...'DET' is expected to be the most common starting bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your starting tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Sequence Ending Counts\nComplete the function below to estimate the bigram probabilities of a sequence ending with each tag.",
"_____no_output_____"
]
],
[
[
"def ending_counts(sequences):\n \"\"\"Return a dictionary keyed to each unique value in the input sequences list\n that counts the number of occurrences where that value is at the end of\n a sequence.\n \n For example, if 18 sequences end with DET, then you should return a\n dictionary such that your_starting_counts[DET] == 18\n \"\"\"\n \n ending_counts={}\n icnt=0\n for i in sequences:\n m=i[len(i)-1]\n if ending_counts.get(m,None)== None:\n ending_counts[m]=1\n else:\n ending_counts[m]+=1\n icnt+=1\n return ending_counts\n # TODO: Finish this function!\n #raise NotImplementedError\n\n\n# TODO: Calculate the count of each tag ending a sequence\ntag_ends = ending_counts(data.training_set.Y)\n\nassert len(tag_ends) == 12, \"Uh oh. There should be 12 tags in your dictionary.\"\nassert min(tag_ends, key=tag_ends.get) in ['X', 'CONJ'], \"Hmmm...'X' or 'CONJ' should be the least common ending bigram.\"\nassert max(tag_ends, key=tag_ends.get) == '.', \"Hmmm...'.' is expected to be the most common ending bigram.\"\nHTML('<div class=\"alert alert-block alert-success\">Your ending tag counts look good!</div>')",
"_____no_output_____"
]
],
[
[
"### IMPLEMENTATION: Basic HMM Tagger\nUse the tag unigrams and bigrams calculated above to construct a hidden Markov tagger.\n\n- Add one state per tag\n - The emission distribution at each state should be estimated with the formula: $P(w|t) = \\frac{C(t, w)}{C(t)}$\n- Add an edge from the starting state `basic_model.start` to each tag\n - The transition probability should be estimated with the formula: $P(t|start) = \\frac{C(start, t)}{C(start)}$\n- Add an edge from each tag to the end state `basic_model.end`\n - The transition probability should be estimated with the formula: $P(end|t) = \\frac{C(t, end)}{C(t)}$\n- Add an edge between _every_ pair of tags\n - The transition probability should be estimated with the formula: $P(t_2|t_1) = \\frac{C(t_1, t_2)}{C(t_1)}$",
"_____no_output_____"
]
],
[
[
"basic_model = HiddenMarkovModel(name=\"base-hmm-tagger\")\n\n# TODO: create states with emission probability distributions P(word | tag) and add to the model\n# (Hint: you may need to loop & create/add new states)\nbasic_model.add_states()\n\n# TODO: add edges between states for the observed transition frequencies P(tag_i | tag_i-1)\n# (Hint: you may need to loop & add transitions\nbasic_model.add_transition()\n\n\n# NOTE: YOU SHOULD NOT NEED TO MODIFY ANYTHING BELOW THIS LINE\n# finalize the model\nbasic_model.bake()\n\nassert all(tag in set(s.name for s in basic_model.states) for tag in data.training_set.tagset), \\\n \"Every state in your network should use the name of the associated tag, which must be one of the training set tags.\"\nassert basic_model.edge_count() == 168, \\\n (\"Your network should have an edge from the start node to each state, one edge between every \" +\n \"pair of tags (states), and an edge from each state to the end node.\")\nHTML('<div class=\"alert alert-block alert-success\">Your HMM network topology looks good!</div>')",
"_____no_output_____"
],
[
"hmm_training_acc = accuracy(data.training_set.X, data.training_set.Y, basic_model)\nprint(\"training accuracy basic hmm model: {:.2f}%\".format(100 * hmm_training_acc))\n\nhmm_testing_acc = accuracy(data.testing_set.X, data.testing_set.Y, basic_model)\nprint(\"testing accuracy basic hmm model: {:.2f}%\".format(100 * hmm_testing_acc))\n\nassert hmm_training_acc > 0.97, \"Uh oh. Your HMM accuracy on the training set doesn't look right.\"\nassert hmm_testing_acc > 0.955, \"Uh oh. Your HMM accuracy on the testing set doesn't look right.\"\nHTML('<div class=\"alert alert-block alert-success\">Your HMM tagger accuracy looks correct! Congratulations, you\\'ve finished the project.</div>')",
"_____no_output_____"
]
],
[
[
"### Example Decoding Sequences with the HMM Tagger",
"_____no_output_____"
]
],
[
[
"for key in data.testing_set.keys[:3]:\n print(\"Sentence Key: {}\\n\".format(key))\n print(\"Predicted labels:\\n-----------------\")\n print(simplify_decoding(data.sentences[key].words, basic_model))\n print()\n print(\"Actual labels:\\n--------------\")\n print(data.sentences[key].tags)\n print(\"\\n\")",
"_____no_output_____"
]
],
[
[
"\n## Finishing the project\n---\n\n<div class=\"alert alert-block alert-info\">\n**Note:** **SAVE YOUR NOTEBOOK**, then run the next cell to generate an HTML copy. You will zip & submit both this file and the HTML copy for review.\n</div>",
"_____no_output_____"
]
],
[
[
"!!jupyter nbconvert *.ipynb",
"_____no_output_____"
]
],
[
[
"## Step 4: [Optional] Improving model performance\n---\nThere are additional enhancements that can be incorporated into your tagger that improve performance on larger tagsets where the data sparsity problem is more significant. The data sparsity problem arises because the same amount of data split over more tags means there will be fewer samples in each tag, and there will be more missing data tags that have zero occurrences in the data. The techniques in this section are optional.\n\n- [Laplace Smoothing](https://en.wikipedia.org/wiki/Additive_smoothing) (pseudocounts)\n Laplace smoothing is a technique where you add a small, non-zero value to all observed counts to offset for unobserved values.\n\n- Backoff Smoothing\n Another smoothing technique is to interpolate between n-grams for missing data. This method is more effective than Laplace smoothing at combatting the data sparsity problem. Refer to chapters 4, 9, and 10 of the [Speech & Language Processing](https://web.stanford.edu/~jurafsky/slp3/) book for more information.\n\n- Extending to Trigrams\n HMM taggers have achieved better than 96% accuracy on this dataset with the full Penn treebank tagset using an architecture described in [this](http://www.coli.uni-saarland.de/~thorsten/publications/Brants-ANLP00.pdf) paper. Altering your HMM to achieve the same performance would require implementing deleted interpolation (described in the paper), incorporating trigram probabilities in your frequency tables, and re-implementing the Viterbi algorithm to consider three consecutive states instead of two.\n\n### Obtain the Brown Corpus with a Larger Tagset\nRun the code below to download a copy of the brown corpus with the full NLTK tagset. You will need to research the available tagset information in the NLTK docs and determine the best way to extract the subset of NLTK tags you want to explore. If you write the following the format specified in Step 1, then you can reload the data using all of the code above for comparison.\n\nRefer to [Chapter 5](http://www.nltk.org/book/ch05.html) of the NLTK book for more information on the available tagsets.",
"_____no_output_____"
]
],
[
[
"import nltk\nfrom nltk import pos_tag, word_tokenize\nfrom nltk.corpus import brown\n\nnltk.download('brown')\ntraining_corpus = nltk.corpus.brown\ntraining_corpus.tagged_sents()[0]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec6fc61c29b49084baf5d446b0a9d5d113628657 | 285,787 | ipynb | Jupyter Notebook | scripts/visualize_neurons_by_decade.ipynb | ludazhao/art_history_net | 396f7b99b3c4fe4f9da752d8f7eb0f3395752f57 | [
"MIT"
]
| 15 | 2016-12-30T21:03:48.000Z | 2022-01-31T08:09:09.000Z | scripts/visualize_neurons_by_decade.ipynb | ludazhao/ArtHistoryNet | 396f7b99b3c4fe4f9da752d8f7eb0f3395752f57 | [
"MIT"
]
| null | null | null | scripts/visualize_neurons_by_decade.ipynb | ludazhao/ArtHistoryNet | 396f7b99b3c4fe4f9da752d8f7eb0f3395752f57 | [
"MIT"
]
| null | null | null | 249.813811 | 80,596 | 0.896248 | [
[
[
"import os.path\nimport re\nimport sys\nimport tarfile\nimport time\nimport multiprocessing as mp\nimport itertools\n\nimport tensorflow.python.platform\nfrom six.moves import urllib\nimport numpy as np\nimport tensorflow as tf\nimport h5py\nimport glob\nimport cPickle as pickle\nimport matplotlib as mpl\nimport matplotlib.pyplot as plt\nimport matplotlib.image as mpimg\nfrom tensorflow.python.platform import gfile\n\nfrom run_inference import predict_star, predict\n\n%matplotlib inline\n%load_ext autoreload\n%autoreload 2\n\nBOTTLENECK_TENSOR_NAME = 'pool_3/_reshape'\nBOTTLENECK_TENSOR_SIZE = 2048\nMODEL_INPUT_WIDTH = 299\nMODEL_INPUT_HEIGHT = 299\nMODEL_INPUT_DEPTH = 3\nJPEG_DATA_TENSOR_NAME = 'DecodeJpeg/contents'\nRESIZED_INPUT_TENSOR_NAME = 'ResizeBilinear'\n\ndef ensure_name_has_port(tensor_name):\n \"\"\"Makes sure that there's a port number at the end of the tensor name.\n\n Args:\n tensor_name: A string representing the name of a tensor in a graph.\n\n Returns:\n The input string with a :0 appended if no port was specified.\n \"\"\"\n if ':' not in tensor_name:\n name_with_port = tensor_name + ':0'\n else:\n name_with_port = tensor_name\n return name_with_port",
"The autoreload extension is already loaded. To reload it, use:\n %reload_ext autoreload\n"
],
[
"def create_graph(pb_file):\n \"\"\"\"Creates a graph from saved GraphDef file and returns a Graph object.\n Returns:\n Graph holding the trained Inception network.\n \"\"\"\n model_filename = pb_file\n with gfile.FastGFile(model_filename, 'rb') as f:\n graph_def = tf.GraphDef()\n graph_def.ParseFromString(f.read())\n tf.import_graph_def(graph_def, name='')",
"_____no_output_____"
],
[
"#create_graph(\"/data/retrain_manualtags1/output_graphfinal.pb\")\ncreate_graph(\"/data/classify_image_graph_def.pb\")\nsess = tf.InteractiveSession()",
"_____no_output_____"
],
[
"params = pickle.load(open(\"/data/datenet_decorations_outputs/output_params7600.pkl\", 'r')) # use 7600 for decorations; top-1 = 65.5%\nfw = params[\"final_weights\"]\nfb = params[\"final_biases\"]\nprint fb.shape\nprint fw.shape",
"(7,)\n(2048, 7)\n"
],
[
"# I currently don't know what labels correspond to each of the 7 elements; will figure out",
"_____no_output_____"
],
[
"# from http://www.socouldanyone.com/2013/03/converting-grayscale-to-rgb-with-numpy.html\ndef to_rgb(im):\n w, h = im.shape[:2]\n ret = np.empty((w, h, 3), dtype=np.uint8)\n ret[:, :, 0] = im\n ret[:, :, 1] = im\n ret[:, :, 2] = im\n return ret\n\n# from http://www.socouldanyone.com/2013/03/converting-grayscale-to-rgb-with-numpy.html\ndef to_rgbs(im):\n n, w, h = im.shape[:3]\n ret = np.empty((n, w, h, 3), dtype=np.uint8)\n ret[:, :, :, 0] = im[:,:,:,0]\n ret[:, :, :, 1] = im[:,:,:,0]\n ret[:, :, :, 2] = im[:,:,:,0]\n return ret\n\n# from https://gist.github.com/yusugomori/4462221\ndef softmax(x):\n e = np.exp(x - np.max(x)) # prevent overflow\n if e.ndim == 1:\n return e / np.sum(e, axis=0)\n else: \n return e / np.array([np.sum(e, axis=1)]).T # ndim = 2",
"_____no_output_____"
],
[
"# read in hdf5 file\nimage_hdf5 = h5py.File('/data/image_data.hdf5','r')\n(image_metadata, book_metadata, image_to_idx) = pickle.load(open(\"/data/all_metadata_1M_tags.pkl\", 'r'))",
"_____no_output_____"
],
[
"# get a list of images to look at for each date\ndate_to_images = {} # each value is a dictionary with {filename --> idx}\nfor folder in glob.glob(\"/data/decorations_by_date/*/\"):\n era = os.path.basename(folder[:-1])\n imgs = glob.glob(folder + \"/*.jpg\")\n fns = np.random.choice([os.path.basename(i)[:-4] for i in imgs], 5, False)\n idxs = [image_to_idx[fn] for fn in fns]\n date_to_images[era] = dict(zip(fns, idxs))\nprint [(x, len(date_to_images[x].keys())) for x in date_to_images]\nclass_count = len(date_to_images)",
"[('1870-1889', 5), ('1750-1799', 5), ('post-1890', 5), ('pre-1700', 5), ('1700-1749', 5), ('1800-1849', 5), ('1850-1869', 5)]\n7\n"
]
],
[
[
"## Add Final Layer\n",
"_____no_output_____"
]
],
[
[
"#Add final layers\nbottleneck_tensor = sess.graph.get_tensor_by_name(ensure_name_has_port(BOTTLENECK_TENSOR_NAME))\nlayer_weights = tf.Variable(\n tf.truncated_normal([2048, class_count], stddev=0.001),\n name='final_weights')\nlayer_biases = tf.Variable(tf.zeros([class_count]), name='final_biases')\nlogits = tf.matmul(bottleneck_tensor, layer_weights) + layer_biases\na = tf.nn.softmax(logits, name=final_tensor_name)\nground_truth_placeholder = tf.placeholder(tf.float32,\n [None, class_count],\n name=ground_truth_tensor_name)\ncross_entropy = tf.nn.softmax_cross_entropy_with_logits(\n logits, ground_truth_placeholder)\ncross_entropy_mean = tf.reduce_mean(cross_entropy)",
"_____no_output_____"
],
[
"x = [(a.name, a.values()) for a in sess.graph.get_operations()]\nfor layer, values in x[:20]:\n print layer\n #res = values[0].eval()\n \n#pool3_tensor = sess.graph.get_tensor_by_name(':0')\nimage = to_rgb(image_hdf5['Chunk5'][0][:,:,0])\nprint np.array(image).shape\nplt.imshow(image, cmap=mpl.cm.gray)\n\n# x = [(a.name, a.values()) for a in sess.graph.get_operations()]\n# for layer, values in x[:20]:\n# print layer\n\n#sess.run(pool3_tensor, {'ExpandDims:0': a})",
"DecodeJpeg/contents\nDecodeJpeg\nCast\nExpandDims/dim\nExpandDims\nResizeBilinear/size\nResizeBilinear\nSub/y\nSub\nMul/y\nMul\nconv/conv2d_params\nconv/Conv2D\nconv/batchnorm/beta\nconv/batchnorm/gamma\nconv/batchnorm/moving_mean\nconv/batchnorm/moving_variance\nconv/batchnorm\nconv/CheckNumerics\nconv/control_dependency\n(224, 224, 3)\nDecodeJpeg/contents\nDecodeJpeg\nCast\nExpandDims/dim\nExpandDims\nResizeBilinear/size\nResizeBilinear\nSub/y\nSub\nMul/y\nMul\nconv/conv2d_params\nconv/Conv2D\nconv/batchnorm/beta\nconv/batchnorm/gamma\nconv/batchnorm/moving_mean\nconv/batchnorm/moving_variance\nconv/batchnorm\nconv/CheckNumerics\nconv/control_dependency\n"
],
[
"date_to_pool3_scores = {}\ndate_to_softmax_scores = {}\npool3_tensor = sess.graph.get_tensor_by_name('pool_3:0')\n\nfor era in date_to_images:\n print era\n date_to_pool3_scores[era] = {}\n date_to_softmax_scores[era] = {}\n\n for img in date_to_images[era]:\n idx = date_to_images[era][img]\n \n chk = idx/5000\n chunk = \"Chunk\" + str(chk)\n i = idx % 5000\n if chunk not in image_hdf5: continue\n\n a = [to_rgb(image_hdf5[chunk][i][:,:,0])]\n predictions = sess.run(pool3_tensor, {'ExpandDims:0': a})[:,0,0,:]\n date_to_pool3_scores[era][img] = predictions[0]\n \n scores = np.dot(predictions, fw) + fb\n scores = np.array([softmax(scores[j]) for j in range(scores.shape[0])])\n date_to_softmax_scores[era][img] = scores[0]\n\npickle.dump((date_to_pool3_scores, date_to_softmax_scores), open(\"/data/decorations_by_date/pool3_softmax_scores.pkl\", 'w'))",
"1870-1889\n1750-1799\npost-1890\npre-1700\n1700-1749\n1800-1849\n1850-1869\n"
],
[
"date_to_pool3_scores",
"_____no_output_____"
],
[
"date_to_softmax_scores",
"_____no_output_____"
],
[
"import pandas as pd\nimport seaborn as sns",
"_____no_output_____"
],
[
"# learn the order of labels\ndate_to_softmax_arr = {}\nfor era in date_to_softmax_scores:\n date_to_softmax_arr[era] = pd.DataFrame.from_dict(date_to_softmax_scores[era]).mean(axis=1)\ndate_to_softmax_arr = pd.DataFrame.from_dict(date_to_softmax_arr)",
"_____no_output_____"
],
[
"date_to_softmax_arr",
"_____no_output_____"
],
[
"labels = [\"1850-1869\", \"1800-1849\", \"1870-1889\", \"1750-1799\", \"1700-1749\", \"pre-1700\", \"post-1890\"]\nlabels_sorted = [\"pre-1700\", \"1700-1749\", \"1750-1799\", \"1800-1849\", \"1850-1869\", \"1870-1889\", \"post-1890\"]\nlabels_idx_pos = [labels]\ndate_to_softmax_arr = date_to_softmax_arr[labels]\ndate_to_softmax_arr",
"_____no_output_____"
],
[
"sns.heatmap(date_to_softmax_arr) # we can see that the closer the dates the more likely things are to cluster",
"_____no_output_____"
],
[
"# heatmap it, rank neurons from low->high to high->low\ndate_to_pool3_arr = {}\nfor era in date_to_softmax_scores:\n date_to_pool3_arr[era] = pd.DataFrame.from_dict(date_to_pool3_scores[era]).mean(axis=1)\ndate_to_pool3_arr = pd.DataFrame.from_dict(date_to_pool3_arr)\ndate_to_pool3_arr = date_to_pool3_arr[labels_sorted]",
"_____no_output_____"
],
[
"diff = date_to_pool3_arr[\"post-1890\"] - date_to_pool3_arr[\"pre-1700\"]\ndiff.sort_values(inplace=True)\nsns.tsplot(diff)\nplt.xlabel(\"Ranked neurons\")\nplt.ylabel(\"1890 activation - 1700 activation\")",
"_____no_output_____"
],
[
"sns.heatmap(date_to_pool3_arr.iloc[diff.index[:100]])\nplt.title(\"100 filters whose activations decrease most over time\")",
"_____no_output_____"
],
[
"date_to_pool3_arr.iloc[diff.index[:5]]",
"_____no_output_____"
],
[
"sns.heatmap(date_to_pool3_arr.iloc[diff.index[-100:]])\nplt.title(\"100 filters whose activations increase most over time\")",
"_____no_output_____"
],
[
"date_to_pool3_arr.iloc[diff.index[-5:]]",
"_____no_output_____"
],
[
"sns.tsplot(date_to_pool3_arr.iloc[diff.index[-5:]].iloc[0])\nsns.tsplot(date_to_pool3_arr.iloc[diff.index[-5:]].iloc[1])\nsns.tsplot(date_to_pool3_arr.iloc[diff.index[-5:]].iloc[2])",
"_____no_output_____"
],
[
"# can do clustering so that we can see which neurons are most activated for each era\n",
"_____no_output_____"
],
[
"# what other layers are there that we could look at?",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6fc7053ee57a45fdc9ec19e5d5b3187d0bc089 | 10,548 | ipynb | Jupyter Notebook | Week_1/Wednesday/Tables.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| 3 | 2021-06-14T19:27:45.000Z | 2021-06-21T03:19:08.000Z | Week_1/Wednesday/Tables.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| null | null | null | Week_1/Wednesday/Tables.ipynb | ds-modules/BUDS-SU21-Dev | 04c8dd0973efec728b22916767a1a7396b3baece | [
"BSD-3-Clause"
]
| 1 | 2021-07-06T00:01:44.000Z | 2021-07-06T00:01:44.000Z | 28.89863 | 324 | 0.589306 | [
[
[
"# Tables: Part 1\n\n\n### Table of Contents\n\n1. <a href='#section 1'>Tables</a>\n\n a. <a href='#subsection 1a'>Attributes</a>\n\n b. <a href='#subsection 1b'>Transformations</a><br><br>\n\n2. <a href='#section 2'>Sorting Tables</a>",
"_____no_output_____"
]
],
[
[
"# dependencies: THIS CELL MUST BE RUN\nfrom datascience import *\nimport numpy as np\nimport math\nimport matplotlib.pyplot as plt\nplt.style.use('fivethirtyeight')\nimport ipywidgets as widgets\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## 1. Tables <a id='section 1'></a>\n\nThe last section covered four basic concepts of python: data, expressions, names, and functions. In this next section, we'll see just how much we can do to examine and manipulate our data with only these minimal Python skills.",
"_____no_output_____"
],
[
"**Tables** are fundamental ways of organizing and displaying data. Run the next cell to load the data.",
"_____no_output_____"
]
],
[
[
"ratings = Table.read_table(\"../data/imdb_ratings.csv\")\nratings",
"_____no_output_____"
]
],
[
[
"This table is organized into **columns**: one for each *category* of information collected:\n\nYou can also think about the table in terms of its **rows**. Each row represents all the information collected about a particular instance, which can be a person, location, action, or other unit. \n\n\n<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> What do the rows in this table represent? By default only the first ten rows are shown. Can you see how many rows there are in total?\n </div>\n",
"_____no_output_____"
],
[
"Answer here: ",
"_____no_output_____"
],
[
"### 1a. Table Attributes <a id='subsection 1a'></a>\n\nEvery table has **attributes** that give information about the table, like the number of rows and the number of columns. Table attributes are accessed using the dot method. But, since an attribute doesn't perform an operation on the table, there are no parentheses (like there would be in a call expression).\n\nAttributes you'll use frequently include `num_rows` and `num_columns`, which give the number of rows and columns in the table, respectively.",
"_____no_output_____"
]
],
[
[
"# get the number of columns\nratings.num_columns",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> Use `num_rows` to get the number of rows in our table.\n</div>",
"_____no_output_____"
]
],
[
[
"# get the number of rows in the table\n",
"_____no_output_____"
]
],
[
[
"### 1b. Table Transformation <a id='subsection 1b'></a>\n\nNot all of our columns are relevant to every question we want to ask. We can save computational resources and avoid confusion by *transforming* our table before we start work.\n\n#### Subsetting columns with `select` and `drop`\n\n\n\n### `select`\nThe `select` function is used to get a table containing only particular columns. `select` is called on a table using dot notation and takes one or more arguments: the name or names of the column or columns you want. Not this does not change the original table. To save your changes, you must assign your change a name.",
"_____no_output_____"
]
],
[
[
"# make a new table with only selected columns\nratings.select(\"Votes\", \"Title\")",
"_____no_output_____"
],
[
"# No changes made to the original table\nratings",
"_____no_output_____"
]
],
[
[
"### `drop`\n\nIf instead you need all columns except a few, the `drop` function can get rid of specified columns. `drop` works very similarly to `select`: call it on the table using dot notation, then give it the name or names of what you want to drop.",
"_____no_output_____"
]
],
[
[
"# drop a column\nratings.drop(\"Decade\")",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> Pick two columns from our table. Create a new table containing only those two columns two different ways: once using `select` and once using `drop`. \n</div>",
"_____no_output_____"
]
],
[
[
"# use select\n",
"_____no_output_____"
],
[
"# use drop\n",
"_____no_output_____"
]
],
[
[
"## 2. Sorting Tables <a id='section 2'></a>\n\nThe last section covered select and drop. In this next section, we'll test our knowledge and use what we have learned and understand how we can sort and manipulate data that are placed in tables.\n",
"_____no_output_____"
],
[
"### `show`\n\nIn a table, we can display a specific amount of rows using the `show` operation. The `show` operations allows you to enter the amount of rows you want displayed from a table.",
"_____no_output_____"
]
],
[
[
"# use show to display 20 rows\nratings.show(20)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> Create a new table and display 33 different rows, using the 'show' operation. \n</div>",
"_____no_output_____"
],
[
"\n### `sort`\n\n\nSome details about sort from the Data 8 lab 2 (Spring 2020):\n\n1. The first argument to `sort` is the name of a column to sort by.\n2. If the column has text in it, `sort` will sort alphabetically; if the column has numbers, it will sort numerically.\n3. The `descending=True` bit is called an *optional argument*. It has a default value of `False`, so when you explicitly tell the function `descending=True`, then the function will sort in descending order.\n4. The `distinct=True` bit is also an *optional argument*. When the function `distinct=True` is used, the function will delete any duplicate values based on the column_or_label value that was also passed in the `sort` function.\n4. Rows always stick together when a table is sorted. It wouldn't make sense to sort just one column and leave the other columns alone. For example, in this case, if we sorted just the `Year` column, the movies would all end up with the wrong year.\n\nThe format for sorting code is written as....\n\n\n\n`Table.sort(column_or_label, descending=False, distinct=False)`",
"_____no_output_____"
]
],
[
[
"# sort a column\nratings.sort(\"Year\")",
"_____no_output_____"
],
[
"# Sorted from most recent year to oldest year\nratings.sort(\"Year\", descending=True)",
"_____no_output_____"
]
],
[
[
"<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> Sort the table from highest ranked movie to lowest ranked.\n</div>",
"_____no_output_____"
],
[
"<div class=\"alert alert-warning\">\n<b>PRACTICE:</b> Sort the table by year in ascending order, making sure each year is distinct.\n</div>",
"_____no_output_____"
],
[
"#### References\n\n- Sections of \"Intro to Jupyter\", \"Table Transformation\" adapted from materials by Kelly Chen and Ashley Chien in [UC Berkeley Data Science Modules core resources](http://github.com/ds-modules/core-resources)\n- \"A Note on Errors\" subsection and \"error\" image adapted from materials by Chris Hench and Mariah Rogers for the Medieval Studies 250: Text Analysis for Graduate Medievalists [data science module](https://github.com/ds-modules/MEDST-250).\n- Rocket Fuel data and discussion questions adapted from materials by Zsolt Katona and Brian Bell, BerkeleyHaas Case Series",
"_____no_output_____"
],
[
"Author: Keeley Takimoto",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec6fca0c0ac26f987f92b28676eb59ac06572d47 | 463,862 | ipynb | Jupyter Notebook | setup_notes_001.ipynb | briesenberg07/jupyter | 39c7942c5344450233a1133c0fd87a7d3fc95839 | [
"CC0-1.0"
]
| null | null | null | setup_notes_001.ipynb | briesenberg07/jupyter | 39c7942c5344450233a1133c0fd87a7d3fc95839 | [
"CC0-1.0"
]
| null | null | null | setup_notes_001.ipynb | briesenberg07/jupyter | 39c7942c5344450233a1133c0fd87a7d3fc95839 | [
"CC0-1.0"
]
| null | null | null | 4,335.158879 | 182,320 | 0.963465 | [
[
[
"# Jupyter setup notes 001\n\n## If I install a Python Library using the terminal, can I use it in a notebook?\n---\n### Windows 10: Install a library using Anaconda Powershell Prompt\n\nI installed 64-bit Anaconda for Windows (see [README](https://github.com/briesenberg07/jupyter#windows-10) for a few more details), and this comes with a couple of different Anaconda terminals...one of these is the Anaconda Powershell Prompt. \n\nI used it to [install rdflib and rdflib-jsonld beforehand](https://uwnetid-my.sharepoint.com/:t:/g/personal/ries07_uw_edu/EZhwp6mSVuVBpuXOwAt5Ze0BVaFgi2xydF7aHRSIED89Nw?e=TAJ0g3)\\*, and then ran some code using these libraries without any problem:\n\n\n\n\n\\**Link access requires a UW Net ID*\n\n---\n\n### Ubuntu 20.04: Install a library using the terminal\n\nI installed Project Jupyter on Ubuntu 20.04 using the Ubuntu Software GUI tool. Not being very sure about the details of this installation, I decided to see whether I could open Jupyter using the command `jupyter notebook` from my terminal, and it worked.\n\n\n\n\\**Not pictured: The Jupyter home directory opening in my browser, etc.*\n\nGreat. This seems to indicate to me that--uh, well--whatever python packages I have available to me for running Python scripts from the terminal, I'll have available in my notebooks.\n\nSo, I've installed the rdflib library and rdflib-jsonld add-on in my terminal:\n\n\n\n\nSeems (to my very general and not particularly well-informed way of thinking) I should now be able to use these in a notebook. Let's see whether this is the case:\n\n\n\n*NOTE to self:* Where/how do I need to install my packages for use in notebooks on my Ubuntu machine, if not in the terminal??\n",
"_____no_output_____"
],
[
"# A workaround for Ubuntu (?)\nI'll use `pip` from inside a notebook to install the libraries I need to...wherever it is that they need to be so that I can use them in my notebooks:\n\n\n*Will this work?*\n\n",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
ec6fd44ce5663e85ffdd9f46e4b67621668f3055 | 126,350 | ipynb | Jupyter Notebook | Stacked_LSTM.ipynb | PSindhuri/Research-Project | 1293fad7081031e14c474f24b3551c2f40b8fed2 | [
"Artistic-1.0-cl8"
]
| 4 | 2019-05-07T04:55:57.000Z | 2021-05-29T02:45:40.000Z | Stacked_LSTM.ipynb | PSindhuri/Research-Project | 1293fad7081031e14c474f24b3551c2f40b8fed2 | [
"Artistic-1.0-cl8"
]
| 1 | 2020-09-24T08:28:01.000Z | 2020-09-24T08:28:01.000Z | Stacked_LSTM.ipynb | PSindhuri/Research-Project | 1293fad7081031e14c474f24b3551c2f40b8fed2 | [
"Artistic-1.0-cl8"
]
| 1 | 2018-06-22T01:40:33.000Z | 2018-06-22T01:40:33.000Z | 42.201069 | 10,708 | 0.565445 | [
[
[
"Best Model : 10 bin hist color stacked lstm ",
"_____no_output_____"
]
],
[
[
"\nimport numpy \nimport matplotlib.pyplot as plt\nimport pandas \nimport math\n\nfrom sklearn.preprocessing import MinMaxScaler\nfrom sklearn.metrics import mean_squared_error,mean_absolute_error\nfrom numpy import array\n\nfrom keras.layers import Input ,Dense, Dropout, Activation, LSTM\nfrom keras.layers import Convolution2D, MaxPooling2D, Flatten, Reshape,Conv2D\nfrom keras.models import Sequential\nfrom keras.layers.wrappers import TimeDistributed\n#from keras.layers.pooling import GlobalAveragePooling1D\n#from keras.optimizers import SGD\nfrom keras.utils import np_utils\n#from keras.models import Model\n\nimport numpy as np\nimport cv2\nfrom keras.preprocessing.image import img_to_array\n\n\n\n\n\n\n\n\nfrom keras import models\n\nfrom matplotlib import pyplot\nfrom numpy import array\nfrom pandas import DataFrame\nimport numpy as np\n\nfrom numpy import array\nfrom keras.models import Sequential\nfrom keras.layers import Dense\nfrom matplotlib import pyplot\n\n\n\n\n\n\n\n\nfrom keras import optimizers\n\n\n\n\n\n\n\nimport numpy as np\nimport pandas as pd\nimport matplotlib.pyplot as plt\n%matplotlib inline\nfrom keras.datasets import mnist\nfrom keras.models import Sequential\nfrom keras.layers import Dense, Dropout, Activation, Flatten\nfrom keras.optimizers import Adam\nfrom keras.layers.normalization import BatchNormalization\nfrom keras.utils import np_utils\nfrom keras.layers import Conv2D, MaxPooling2D, ZeroPadding2D, GlobalAveragePooling2D\nfrom keras.layers.advanced_activations import LeakyReLU \nfrom keras.preprocessing.image import ImageDataGenerator\n",
"/Users/SindhuPhani/anaconda3/lib/python3.6/site-packages/h5py/__init__.py:34: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`.\n from ._conv import register_converters as _register_converters\nUsing TensorFlow backend.\n"
]
],
[
[
"# Hist of pixel 16 bin Stacked LSTM ",
"_____no_output_____"
]
],
[
[
"num_bins = 16\nresults_rmse = []\nresults_mae = []",
"_____no_output_____"
],
[
"def train_optimizer_models(opt):\n np.random.seed(3)\n time_steps=19\n # load the dataset\n dataframe = pandas.read_csv('./Trainold.csv')\n dataset = dataframe.values\n\n scaler = MinMaxScaler(feature_range=(0, 1))\n\n\n #print(dataset)\n\n # we group by day so we can process a video at a time.\n grouped = dataframe.groupby(dataframe.VidName)\n\n per_vid = []\n for _, group in grouped:\n per_vid.append(group)\n\n\n\n print(len(per_vid))\n\n\n\n trainX=[]\n trainY=[]\n\n # generate sequences a vid at a time\n for i,vid in enumerate(per_vid):\n histValuesList=[]\n scoreList=[]\n # if we have less than 20 datapoints for a vid we skip over the\n # vid assuming something is missing in the raw data\n\n total = vid.iloc[:,4:20].values\n vidImPath=vid.iloc[:,0:2].values \n\n if len(total) < time_steps :\n continue\n scoreVal=vid[\"Score\"].values[0] + 1 \n max_total_for_vid = scoreVal.tolist()\n #max_total_for_vid = vid[\"Score\"].values[0].tolist()\n\n for i in range(0,time_steps):\n videoName=vidImPath[i][0]\n imgName=vidImPath[i][1]\n path=\"./IMAGES/Train/\"+videoName+\"/\"+imgName\n image = cv2.imread(path,0)\n hist = cv2.calcHist([image],[0],None,[num_bins],[0,256])\n\n hist_arr = hist.flatten()\n #img_arr = img_to_array(image)\n histValuesList.append(hist_arr)\n #scoreList.append(max_total_for_vid)\n\n trainX.append(histValuesList)\n trainY.append([max_total_for_vid])\n #trainY.append(scoreList)\n\n\n print(len(trainX[0]))\n #trainX = np.array([np.array(xi) for xi in trainX])\n\n trainX=numpy.array(trainX)\n trainY=numpy.array(trainY)\n\n print(trainX.shape,trainY.shape)\n\n\n\n\n\n time_steps=19\n # load the dataset\n dataframe = pandas.read_csv('./Test.csv')\n dataset = dataframe.values\n\n #print(dataset)\n\n # we group by day so we can process a video at a time.\n grouped = dataframe.groupby(dataframe.VidName)\n\n per_vid = []\n for _, group in grouped:\n per_vid.append(group)\n\n\n\n print(len(per_vid))\n\n\n\n testX=[]\n testY=[]\n\n # generate sequences a vid at a time\n for i,vid in enumerate(per_vid):\n histValuesList=[]\n scoreList=[]\n # if we have less than 20 datapoints for a vid we skip over the\n # vid assuming something is missing in the raw data\n\n total = vid.iloc[:,4:20].values\n\n vidImPath=vid.iloc[:,0:2].values\n\n\n if len(total)<time_steps :\n continue\n scoreVal=vid[\"Score\"].values[0] + 1 \n max_total_for_vid = scoreVal.tolist()\n #max_total_for_vid = vid[\"Score\"].values[0].tolist()\n\n\n for i in range(0,time_steps):\n #histValuesList.append(total[i])\n #print(\"Vid and Img name\")\n #print(req[i][0],req[i][1])\n videoName=vidImPath[i][0]\n imgName=vidImPath[i][1]\n path=\"./IMAGES/Test/\"+videoName+\"/\"+imgName\n image = cv2.imread(path,0)\n hist = cv2.calcHist([image],[0],None,[num_bins],[0,256])\n hist_arr = hist.flatten()\n\n histValuesList.append(hist_arr)\n #scoreList.append(max_total_for_vid)\n\n\n testX.append(histValuesList)\n testY.append([max_total_for_vid])\n #testY.append(scoreList)\n\n\n print(len(testX[0]))\n #trainX = np.array([np.array(xi) for xi in trainX])\n\n testX=numpy.array(testX)\n testY=numpy.array(testY)\n\n print(testX.shape,testY.shape)\n\n\n\n\n trainX=numpy.array(trainX)\n trainX=trainX.reshape(-1,num_bins)\n trainX=trainX.reshape(-1,19,num_bins)\n print(numpy.max(trainX))\n\n\n testX=numpy.array(testX)\n testX=testX.reshape(-1,num_bins)\n testX=testX.reshape(-1,19,num_bins)\n print(numpy.max(testX))\n\n\n\n trainX=numpy.array(trainX)\n trainX=trainX.reshape(-1,num_bins)\n\n trainX = trainX/numpy.max(trainX)\n\n trainX=trainX.reshape(-1,19,num_bins)\n print(trainX.shape,trainY.shape)\n\n\n\n\n testX=numpy.array(testX)\n testX=testX.reshape(-1,num_bins)\n\n testX = testX/numpy.max(testX)\n\n testX=testX.reshape(-1,19,num_bins)\n print(testX.shape,testY.shape)\n\n\n\n print(trainX.shape,trainY.shape)\n print(testX.shape,testY.shape)\n #print(valX.shape,valY.shape)\n\n\n\n\n\n\n print('Build model...')\n # Build Model\n model = Sequential() \n model.add(LSTM(100, input_shape=(19, num_bins),return_sequences=True))#100\n model.add(LSTM(50, input_shape=(19, num_bins),return_sequences=True)) #100\n model.add(LSTM(20, input_shape=(19, num_bins))) #100\n model.add(Dense(1))\n #model.add(Dropout(0.1))\n\n #activation=act_func, recurrent_activation=act_func,dropout=0.1,recurrent_dropout=0.1\n\n model.compile(loss='mse', optimizer=opt, metrics=['mse'])\n\n #model.compile(optimizer='adam', loss = 'mae')\n\n model.summary()\n\n history =model.fit(trainX, trainY, nb_epoch=500, batch_size=20, verbose=2,shuffle=True) #500 batch =2 \n\n\n\n # make predictions\n trainPredict = model.predict(trainX)\n\n\n trainScore = mean_squared_error(trainY, trainPredict)\n print('Train Score: %.2f MSE' % (trainScore))\n\n\n from keras import backend as K\n\n def root_mean_squared_error(y_true, y_pred):\n return K.sqrt(K.mean(K.square(y_pred - y_true), axis=-1)) \n\n pred=model.predict(testX)\n\n print(pred.shape)\n print(testY.shape)\n\n\n\n # calculate root mean squared error\n #trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:,0]))\n #print('Train Score: %.2f RMSE' % (trainScore))\n testScore = mean_squared_error(testY, pred)\n print('Test Score: %.2f MSE' % (testScore))\n\n #maeScore = root_mean_squared_error(testY, pred)\n #print('RMSE Score: %.2f MAE' % (maeScore))\n\n rmse = np.sqrt(((pred - testY) ** 2).mean(axis=0))\n print('RMSE Score: %.2f rmse' % (rmse))\n\n\n mae = mean_absolute_error(testY, pred)\n print('MAE Score: %.2f mae' % (mae))\n\n for i in range(0,len(pred)):\n\n print(testY[i],pred[i])\n \n results_rmse.append(rmse)\n results_mae.append(mae)",
"_____no_output_____"
],
[
"\n# Stacked LSTM with 16 bin hist of pixel values\n\nadam1 = optimizers.Adam(lr=0.001)\nsgd1 = optimizers.SGD(lr=0.005) \n\ntrain_optimizer_models(sgd1)",
"117\n19\n(114, 19, 16) (114, 1)\n55\n19\n(52, 19, 16) (52, 1)\n174999.0\n157653.0\n(114, 19, 16) (114, 1)\n(52, 19, 16) (52, 1)\n(114, 19, 16) (114, 1)\n(52, 19, 16) (52, 1)\nBuild model...\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nlstm_1 (LSTM) (None, 19, 100) 46800 \n_________________________________________________________________\nlstm_2 (LSTM) (None, 19, 50) 30200 \n_________________________________________________________________\nlstm_3 (LSTM) (None, 20) 5680 \n_________________________________________________________________\ndense_1 (Dense) (None, 1) 21 \n=================================================================\nTotal params: 82,701\nTrainable params: 82,701\nNon-trainable params: 0\n_________________________________________________________________\n"
]
],
[
[
"# Histogram of Color 10 bin Stacked LSTM",
"_____no_output_____"
]
],
[
[
"# Stacked LSTM with 10 bin hist of color values\nnum_bins = 10\n\n\ndef train_optimizer_models(opt):\n np.random.seed(3)\n time_steps=19\n # load the dataset\n dataframe = pandas.read_csv('./Trainold.csv')\n dataset = dataframe.values\n\n scaler = MinMaxScaler(feature_range=(0, 1))\n\n\n #print(dataset)\n\n # we group by day so we can process a video at a time.\n grouped = dataframe.groupby(dataframe.VidName)\n\n per_vid = []\n for _, group in grouped:\n per_vid.append(group)\n\n\n\n print(len(per_vid))\n\n\n\n trainX=[]\n trainY=[]\n\n # generate sequences a vid at a time\n for i,vid in enumerate(per_vid):\n histValuesList=[]\n scoreList=[]\n # if we have less than 20 datapoints for a vid we skip over the\n # vid assuming something is missing in the raw data\n\n total = vid.iloc[:,4:20].values\n vidImPath=vid.iloc[:,0:2].values \n\n if len(total) < time_steps :\n continue\n scoreVal=vid[\"Score\"].values[0] + 1 \n max_total_for_vid = scoreVal.tolist()\n #max_total_for_vid = vid[\"Score\"].values[0].tolist()\n\n for i in range(0,time_steps):\n videoName=vidImPath[i][0]\n imgName=vidImPath[i][1]\n path=\"./IMAGES/Train/\"+videoName+\"/\"+imgName\n image = cv2.imread(path)\n hist = cv2.calcHist([image], [0, 1, 2], None, [num_bins, num_bins, num_bins], [0, 256, 0, 256, 0, 256])\n hist_arr = hist.flatten()\n #img_arr = img_to_array(image)\n histValuesList.append(hist_arr)\n #scoreList.append(max_total_for_vid)\n\n trainX.append(histValuesList)\n trainY.append([max_total_for_vid])\n #trainY.append(scoreList)\n\n\n print(len(trainX[0]))\n #trainX = np.array([np.array(xi) for xi in trainX])\n\n trainX=numpy.array(trainX)\n trainY=numpy.array(trainY)\n\n print(trainX.shape,trainY.shape)\n\n\n\n\n # load the dataset\n dataframe = pandas.read_csv('./Test.csv')\n dataset = dataframe.values\n\n #print(dataset)\n\n # we group by day so we can process a video at a time.\n grouped = dataframe.groupby(dataframe.VidName)\n\n per_vid = []\n for _, group in grouped:\n per_vid.append(group)\n\n\n\n print(len(per_vid))\n\n\n\n testX=[]\n testY=[]\n\n # generate sequences a vid at a time\n for i,vid in enumerate(per_vid):\n histValuesList=[]\n scoreList=[]\n # if we have less than 20 datapoints for a vid we skip over the\n # vid assuming something is missing in the raw data\n\n total = vid.iloc[:,4:20].values\n\n vidImPath=vid.iloc[:,0:2].values\n\n\n if len(total)<time_steps :\n continue\n scoreVal=vid[\"Score\"].values[0] + 1 \n max_total_for_vid = scoreVal.tolist()\n #max_total_for_vid = vid[\"Score\"].values[0].tolist()\n\n\n for i in range(0,time_steps):\n #histValuesList.append(total[i])\n #print(\"Vid and Img name\")\n #print(req[i][0],req[i][1])\n videoName=vidImPath[i][0]\n imgName=vidImPath[i][1]\n path=\"./IMAGES/Test/\"+videoName+\"/\"+imgName\n image = cv2.imread(path)\n #img_arr = img_to_array(image)\n hist = cv2.calcHist([image], [0, 1, 2], None, [num_bins, num_bins, num_bins], [0, 256, 0, 256, 0, 256])\n hist_arr = hist.flatten()\n\n histValuesList.append(hist_arr)\n #scoreList.append(max_total_for_vid)\n\n\n testX.append(histValuesList)\n testY.append([max_total_for_vid])\n #testY.append(scoreList)\n\n\n print(len(testX[0]))\n #trainX = np.array([np.array(xi) for xi in trainX])\n\n testX=numpy.array(testX)\n testY=numpy.array(testY)\n\n print(testX.shape,testY.shape)\n\n\n\n\n trainX=numpy.array(trainX)\n trainX=trainX.reshape(-1,num_bins**3)\n\n trainX = trainX/numpy.max(trainX)\n\n trainX=trainX.reshape(-1,19,num_bins**3)\n print(trainX.shape,trainY.shape)\n\n\n\n\n testX=numpy.array(testX)\n testX=testX.reshape(-1,num_bins**3)\n\n testX = testX/numpy.max(testX)\n\n testX=testX.reshape(-1,19,num_bins**3)\n print(testX.shape,testY.shape)\n\n\n\n\n\n\n\n\n print('Build model...')\n # Build Model\n model = Sequential() \n model.add(LSTM(100, input_shape=(19, num_bins**3),return_sequences=True))#100\n model.add(LSTM(50, input_shape=(19, num_bins**3),return_sequences=True)) #100\n model.add(LSTM(20, input_shape=(19, num_bins**3))) #100\n model.add(Dense(1))\n #model.add(Dropout(0.1))\n\n #activation=act_func, recurrent_activation=act_func,dropout=0.1,recurrent_dropout=0.1\n\n model.compile(loss='mse', optimizer=opt, metrics=['mse'])\n\n #model.compile(optimizer='adam', loss = 'mae')\n\n model.summary()\n\n history =model.fit(trainX, trainY, nb_epoch=500, batch_size=20, verbose=2,shuffle=True) #500 batch =2 \n\n\n\n # make predictions\n trainPredict = model.predict(trainX)\n\n\n trainScore = mean_squared_error(trainY, trainPredict)\n print('Train Score: %.2f MSE' % (trainScore))\n\n\n from keras import backend as K\n\n def root_mean_squared_error(y_true, y_pred):\n return K.sqrt(K.mean(K.square(y_pred - y_true), axis=-1)) \n\n pred=model.predict(testX)\n\n print(pred.shape)\n print(testY.shape)\n\n\n\n # calculate root mean squared error\n #trainScore = math.sqrt(mean_squared_error(trainY[0], trainPredict[:,0]))\n #print('Train Score: %.2f RMSE' % (trainScore))\n testScore = mean_squared_error(testY, pred)\n print('Test Score: %.2f MSE' % (testScore))\n\n #maeScore = root_mean_squared_error(testY, pred)\n #print('RMSE Score: %.2f MAE' % (maeScore))\n\n rmse = np.sqrt(((pred - testY) ** 2).mean(axis=0))\n print('RMSE Score: %.2f rmse' % (rmse))\n\n\n mae = mean_absolute_error(testY, pred)\n print('MAE Score: %.2f mae' % (mae))\n\n for i in range(0,len(pred)):\n\n print(testY[i],pred[i])\n \n results_rmse.append(rmse)\n results_mae.append(mae)\n \n \n \n \n \n\n# All parameter gradients will be clipped to\n# a maximum norm of 1.\n\nadam1 = optimizers.Adam(lr=0.001)\nsgd1 = optimizers.SGD(lr=0.005) \n\ntrain_optimizer_models(sgd1)\n \n \n\n\nnewArr1=[]\nnewArr2=[]\nfor item in results_rmse:\n newArr1.append(float(item))\n \nfor item in results_mae:\n newArr2.append(float(item))\n\n\nrmse_val = tuple(newArr1)\nmae_val = tuple(newArr2) \n\n",
"117\n19\n(114, 19, 1000) (114, 1)\n55\n19\n(52, 19, 1000) (52, 1)\n(114, 19, 1000) (114, 1)\n(52, 19, 1000) (52, 1)\nBuild model...\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nlstm_4 (LSTM) (None, 19, 100) 440400 \n_________________________________________________________________\nlstm_5 (LSTM) (None, 19, 50) 30200 \n_________________________________________________________________\nlstm_6 (LSTM) (None, 20) 5680 \n_________________________________________________________________\ndense_2 (Dense) (None, 1) 21 \n=================================================================\nTotal params: 476,301\nTrainable params: 476,301\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"\n# data to plot\nn_groups = 2\n\n\n# create plot\nfig, ax = plt.subplots()\nindex = np.arange(n_groups)\nbar_width = 0.2\nopacity = 0.8\n \nrects1 = plt.bar(index, rmse_val, bar_width,alpha=0.5,color='g',label='RMSE')\n \nrects2 = plt.bar(index + bar_width, mae_val, bar_width,alpha=0.3,color='b',label='MAE')\n \n \nplt.xlabel('Optimizer')\nplt.ylabel('Error Values')\nplt.title('Error by feature set')\nplt.xticks(index + bar_width, ('16-bin pixel','10-bin color'))\nplt.legend()\n \nplt.tight_layout()\n\naxes = plt.gca()\naxes.set_ylim([0.20,1.3])\n\nplt.show()\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6fdd2c4ddd9105fcc20fbf9d37088ee7c86b20 | 13,713 | ipynb | Jupyter Notebook | keyword.ipynb | afry-south/mix-match-cv | 4718d087be7d197d1b725e24d1a2f42b7d316ea0 | [
"MIT"
]
| null | null | null | keyword.ipynb | afry-south/mix-match-cv | 4718d087be7d197d1b725e24d1a2f42b7d316ea0 | [
"MIT"
]
| 4 | 2021-06-03T08:17:00.000Z | 2021-06-03T10:40:52.000Z | keyword.ipynb | afry-south/mix-match-cv | 4718d087be7d197d1b725e24d1a2f42b7d316ea0 | [
"MIT"
]
| 1 | 2021-06-03T07:54:34.000Z | 2021-06-03T07:54:34.000Z | 47.947552 | 6,153 | 0.596004 | [
[
[
"import pandas as pd\nimport flair\nfrom bpemb import BPEmb\nfrom sklearn.feature_extraction.text import TfidfVectorizer\nfrom scipy.spatial.distance import cdist\nfrom sklearn.decomposition import TruncatedSVD\nimport regex as re\nfrom flair.data import Sentence\nfrom flair.models import SequenceTagger\nfrom flair.data import Corpus\nfrom flair.datasets import ColumnCorpus\nfrom flair.embeddings import TokenEmbeddings, WordEmbeddings, StackedEmbeddings, BytePairEmbeddings\n#import spacy",
"_____no_output_____"
],
[
"full_files_assignment = [f\"data/assignment/{x}/assignment.txt\" for x in os.listdir('data/assignment')]\nfull_files_data_assignment = [open(x).read() for x in full_files_assignment if not 'annot' in x and not '.DS' in x]\nall_text = '\\n'.join(full_files_data_assignment) # split into test/train",
"_____no_output_____"
],
[
"multi_space = re.compile(\"[_ \\t]+\")\nnon_ascii = re.compile(\"[^\\x00-\\x7FåäöÅÄÖ\\s\\n\\p{Punct}]+\")\npunct = re.compile('([\\p{Punct}\\n])+')\n#splitter = re.compile(\"[\\n\\.]\")\n#t = all_text.replace('/', ' ')\nt = non_ascii.sub('', all_text)\nt = punct.sub(' \\g<1> ', t)\nt = multi_space.sub(' ', t)\nt = t.replace('\\n', '.')\nt = t.lower()",
"_____no_output_____"
],
[
"t[:300]",
"_____no_output_____"
],
[
"keywords = {'wireshark', 'cyber', 'docker', 'kanban', 'latest technologies', 'teaching skills', 'jenkins', 'sae j2847', 'system design', 'android', 'ccc', 'measurements', 'thermal conditioning', 'product development process', 'sales', 'verification tests', 'plc', 'test automation', 'private equity', 'data handling', 'maven', 'in-vehicle software', 'project management', 'validate', 'autonomous driver', 'insurances', 'drug', 'problem solver', 'infotainment', 'sensor fusion', 'ats', 'documentation', 'integration transformation', 'test planning', 'wallet-sizing analysis', 'software requirement', 'propulsion', 'test plans', 'canalyzer', 'nvidia', 'research and development', 'team efficiency', 'modems', 'automated test', 'matlab simulink', 'spice', 'lead', 'canoe', 'stk', 'control', 'product owner', 'lin', 'signal processing', 'insurance', 'frontend', 'customer', 'mil', 'backend', 'github', 'database', 'engineering tests', 'web services', 'automotive standards', 'valuation gap', 'university degree', 'software', 'cabel harness', 'application maintenance', 'sw test', 'collaborative', 'engineering', 'pcb and manufacturing', 'documenting processes', 'planning', 'safety requirement', 'coordinate', 'microprocessor system', 'interfaces', 'javascript', 'communication skills', 'user intent algorithms', 'target setting', 'rtos', 'hil', 'devops', 'digital design', 'manufacturing', 'electrical engineering', 'fund', 'sensors', 'soc', 'front-end', '5+', 'kola', 'network', 'gerrit', 'financial', 'signals', 'recruitment', 'low-level', 'phd', '>5', 'ci/cd', 'mcu', 'design for immunity', 'bus chassis', 'integration testing', 'ad/adas', 'stakeholders', 'm.sc.', 'equity analyst', 'bts', 'gpu development', 'back-end', 'artifactory', 'verbal', 'sae j3068', 'biopharmaceutical', 'offsite', 'schematic design', 'coverage model', 'etas inca', 'standardized requirement work', 'can', 'networking', 'data-centric', 'stake holder communication', 'erp system', 'electronics', 'saber', 'logistics systems', 'telecom', 'mechatronics', 'adas', 'polyspace', 'controller', 'quality assurance', 'coaching', 'canalayzer', 'c', 'invoicing', 'machine learning', 'train management', 'system lead', 'bachelor', 'technical', 'manage', 'windows', 'cmmi', 'project manager', 'cross-functional', 'management consulting', 'warranty', 'powertrain', 'end-to-end', 'bash', 'iso 15118', 'drivetrain', 'test programs', 'systems development', 'coordinating', 'robot framework', 'hazard analysis', 'oas', 'system testing', 'sensor perception layer', 'functional safety manager', 'report', 'efficiency', 'html', 'emc certification', 'uwb', 'it', 'diagnostics', 'esd', 'designer', 'oracle', 'testweaver', 'developer', 'healthcare', 'portfolio strategy', 'technical debt', 'confluence', 'unit testing', 'architecture', 'client discussion', '7740', 'sts', 'supply chain designer', 'electric motors', 'global coordination', 'aerospace', 'academic degree', 'e2e', 'efficient', 'flexray', 'financial modelling', 'embedded software', 'certification tests', 'design', 'business experience', 'configuration management', 'php', 'mechanical engineering', 'elektra', 'cloud', 'computer engineering', 'retail', 'vehicle electronics', 'eeprom', 'drug substance', 'high speed signals', 'power company', 'industrialization', 'filter designs', 'test', 'sap', 'css', 'system control', 'implementation', 'inverters', '5-8 years', 'validation', 'ad', 'electronics designer', 'application development', 'activism defense', 'front end', 'back end', 'sil1', 'cable harness engineer', 'it solutions', 'apriso', 'r&d', 'sql', 'wso2', 'software architect', 'track record', 'cd', 'business development', 'security', 'phone as key', 'jira', 'banking', 'system simulators', 'embedded system', 'hedging', 'sil', 'embedded systems', \"master's degree\", 'r3', 'pl', 'autonomous', 'app-centric', 'team lead', 'autosar', 'ai', 'bidding', 'traction', 'safe', 'budget', 'databases', 'international', 'rest api', 'saas', 'swedish', 'order management', 'automation', 'cts', 'budgeting', 'cameras', 'ccs', 'git', 'embedded', 'analog design', 'sw', 'emc', 'quality', 'customer value', 'vehicle certification', 'e-mobility', 'team leader', 'system verification', 'product manager', 'batteries', 'driving', '> 5', 'international business', 'scm', 'financial institutions', 'advanced driver assistance system', 'carve-out', 'team work', 'b.sc.', 'battery', 'dc/dc', 'linux', 'functional development', 'research & development', 'iso26262', 'home office', 'vehicle', 'operational team management', 'workshops', 'warranties', 'optimization', 'debugging', 'carweaver', 'hardware', 'pcb-a', 'italian', 'system development', 'din 70121', 'claim management', 'software developer', 'review', 'business', 'logistics', 'asil', 'computer science', 'cyber security', 'verification', 'chargers', 'vts', 'initiative', 'keycloak', 'qnx', 'carcom', 'ultrasonic perception', 'strategic vulnerabilities', 'payments', 'design for emissions', 'c/c++', 'bid teams', 'epsm', 'simulink', 'hil testing', 'angular', 'python', 'restful', 'sensor signaling', 'financing', 'digital key', 'fintech', 'operational', 'user experience', 'leader', 'railway', 'pcb', 'polarion', 'portuguese', 'continuous integration', 'radars', 'design guidelines', 'ethernet', 'electronic designer', 'vehicle dynamics', 'mba', 'matlab', 'economy', 'testing methodology', 'capl', 'java', 'master of science', 'ram', '10+', 'automotive', 'traction batteries', 'management', 'driving license', 'inca', 'english', 'pm', 'ci', 'system architecture', 'architect', 'stk calculations', 'coordination', 'gtest', 'cybersecurity', 'sell-side', 's4hana', 'intellij', 'ul4600', 'cable harness designer', 'finance', 'agile', 'nand', 'communication', 'canalyser', 'aspice', 'sw architect', 'ad platform', 'technology', 'engineering physics', 'invoice', 'test methods', 'safety', 'construction', 'customer requirement', 'mälardalen', 'software requirements', 'robot', \"bachelor's degree\", 'nfc', 'ble', 'integrity', 'rest', 'risk assessment', 'big data', 'c++', 'supply strategy', 'project leader', 'change management', 'design alternatives', 'spring', 'scrum', 'system architect'}",
"_____no_output_____"
],
[
"tokens = t.split(' ')\ntokens[:5]",
"_____no_output_____"
],
[
"sorted([x.count(' ') for x in keywords], reverse=True)[:3]",
"_____no_output_____"
],
[
"tagged_token = []\nprev_index = 0\nfor i in range(len(tokens)):\n if i < prev_index:\n continue\n token = tokens[i]\n num_tokens = 0\n \n if i < len(tokens) - 3 and ' '.join(tokens[i:i+4]) in keywords:\n num_tokens = 4\n elif i < len(tokens) - 2 and ' '.join(tokens[i:i+3]) in keywords:\n num_tokens = 3\n elif i < len(tokens) - 1 and ' '.join(tokens[i:i+2]) in keywords:\n num_tokens = 2\n elif token in keywords:\n num_tokens = 1\n \n if num_tokens > 0:\n for j in range(num_tokens):\n if j == 0:\n tagged_token.append(f\"{tokens[i+j]} B-KEY\")\n else:\n tagged_token.append(f\"{tokens[i+j]} I-KEY\")\n prev_index = i + num_tokens\n else:\n prev_index = i\n tagged_token.append(f\"{token} 0\")",
"_____no_output_____"
],
[
"tagged_token[:20]",
"_____no_output_____"
],
[
"with open('data/keyword_ds/ds.txt', 'w') as f:\n f.write('\\n'.join(tagged_token))",
"_____no_output_____"
],
[
"columns = {0: 'text', 1: 'ner'}\ncorpus: Corpus = ColumnCorpus('data/keyword_ds', columns, train_file='ds.txt')\n# 2. what tag do we want to predict?\ntag_type = 'ner'\n\n# 3. make the tag dictionary from the corpus\ntag_dictionary = corpus.make_tag_dictionary(tag_type=tag_type)\nprint(tag_dictionary)",
"2021-06-03 18:59:52,362 Reading data from data/keyword_ds\n2021-06-03 18:59:52,363 Train: data/keyword_ds/ds.txt\n2021-06-03 18:59:52,365 Dev: None\n2021-06-03 18:59:52,365 Test: None\nDictionary with 7 tags: <unk>, O, B-KEY, 0, I-KEY, <START>, <STOP>\n"
],
[
"embedding_types = [\n\tBytePairEmbeddings('sv'),\n\tFlairEmbeddings(\"sv-forward\"),\n\tFlairEmbeddings(\"sv-backward\")\n]\n\nembeddings: StackedEmbeddings = StackedEmbeddings(embeddings=embedding_types)\n\n# 5. initialize sequence tagger\ntagger: SequenceTagger = SequenceTagger(hidden_size=256,\n\t\t\t\t\t\t\t\t\t\tembeddings=embeddings,\n\t\t\t\t\t\t\t\t\t\ttag_dictionary=tag_dictionary,\n\t\t\t\t\t\t\t\t\t\ttag_type=tag_type,\n\t\t\t\t\t\t\t\t\t\tuse_crf=True)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6fdf4c8e6371c6136f8ca92a28a9c751d57737 | 83,155 | ipynb | Jupyter Notebook | GPT_with_validation.ipynb | shun-lin/Project-Lion | 2cd9a217ad322159b58d91febd5081d4a09428d1 | [
"MIT"
]
| null | null | null | GPT_with_validation.ipynb | shun-lin/Project-Lion | 2cd9a217ad322159b58d91febd5081d4a09428d1 | [
"MIT"
]
| null | null | null | GPT_with_validation.ipynb | shun-lin/Project-Lion | 2cd9a217ad322159b58d91febd5081d4a09428d1 | [
"MIT"
]
| null | null | null | 83,155 | 83,155 | 0.815176 | [
[
[
"pip install pytorch-pretrained-bert",
"Requirement already satisfied: pytorch-pretrained-bert in /usr/local/lib/python3.6/dist-packages (0.6.2)\nRequirement already satisfied: tqdm in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (4.28.1)\nRequirement already satisfied: regex in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (2018.1.10)\nRequirement already satisfied: boto3 in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (1.9.137)\nRequirement already satisfied: torch>=0.4.1 in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (1.0.1.post2)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (2.21.0)\nRequirement already satisfied: numpy in /usr/local/lib/python3.6/dist-packages (from pytorch-pretrained-bert) (1.16.3)\nRequirement already satisfied: s3transfer<0.3.0,>=0.2.0 in /usr/local/lib/python3.6/dist-packages (from boto3->pytorch-pretrained-bert) (0.2.0)\nRequirement already satisfied: botocore<1.13.0,>=1.12.137 in /usr/local/lib/python3.6/dist-packages (from boto3->pytorch-pretrained-bert) (1.12.137)\nRequirement already satisfied: jmespath<1.0.0,>=0.7.1 in /usr/local/lib/python3.6/dist-packages (from boto3->pytorch-pretrained-bert) (0.9.4)\nRequirement already satisfied: idna<2.9,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->pytorch-pretrained-bert) (2.8)\nRequirement already satisfied: chardet<3.1.0,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->pytorch-pretrained-bert) (3.0.4)\nRequirement already satisfied: urllib3<1.25,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->pytorch-pretrained-bert) (1.24.2)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->pytorch-pretrained-bert) (2019.3.9)\nRequirement already satisfied: docutils>=0.10 in /usr/local/lib/python3.6/dist-packages (from botocore<1.13.0,>=1.12.137->boto3->pytorch-pretrained-bert) (0.14)\nRequirement already satisfied: python-dateutil<3.0.0,>=2.1; python_version >= \"2.7\" in /usr/local/lib/python3.6/dist-packages (from botocore<1.13.0,>=1.12.137->boto3->pytorch-pretrained-bert) (2.5.3)\nRequirement already satisfied: six>=1.5 in /usr/local/lib/python3.6/dist-packages (from python-dateutil<3.0.0,>=2.1; python_version >= \"2.7\"->botocore<1.13.0,>=1.12.137->boto3->pytorch-pretrained-bert) (1.12.0)\n"
],
[
"from google.colab import drive\ndrive.mount('/content/gdrive')",
"Drive already mounted at /content/gdrive; to attempt to forcibly remount, call drive.mount(\"/content/gdrive\", force_remount=True).\n"
],
[
"import numpy as np\nimport os, sys\nimport json\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport string\nimport torch\n\nfrom collections import defaultdict\nfrom random import sample \nfrom torch.nn import functional as F\nfrom pytorch_pretrained_bert import GPT2Tokenizer, GPT2LMHeadModel, BertTokenizer, BertForPreTraining, OpenAIAdam\n\nsys.path.insert(0, \"/content/gdrive/My Drive/Project-Lion/\") \nroot_folder = \"/content/gdrive/My Drive/Project-Lion/\"",
"_____no_output_____"
],
[
"def clean_data(lst, in_place=True, keep_punc=\"\"):\n if in_place:\n new_lst = lst\n else:\n new_lst = lst.copy()\n \n for i in range(len(new_lst)):\n joke = lst[i]\n joke = joke.replace(\"\\r\", \" \").replace(\"\\n\", \" \").replace(\"/\", \" or \")\n punc_to_remove = string.punctuation\n for punc in keep_punc:\n punc_to_remove = punc_to_remove.replace(punc, \"\")\n joke = joke.translate(str.maketrans(\"\", \"\", punc_to_remove)) # Remove punctuation\n joke = joke.lower() # Lowercase\n joke = joke.strip() # Remove leading and ending whitespace\n new_lst[i] = joke\n return new_lst",
"_____no_output_____"
],
[
"df = pd.read_csv(root_folder + 'data/kaggle/shortjokes.csv')\ndf = df.loc[df[\"Joke\"] != \"\"]",
"_____no_output_____"
],
[
"data = clean_data(df[\"Joke\"].tolist(), keep_punc=\"?,':.\")\nprint(\"Length of data:\", len(data))\nprint(data[0])",
"Length of data: 231657\nme narrating a documentary about narrators i can't hear what they're saying cuz i'm talking\n"
]
],
[
[
"# GPT",
"_____no_output_____"
]
],
[
[
"best_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/best_model.pt\"\nbest_train_losses_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/best_train_losses\"\nbest_val_losses_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/best_val_losses\"\nlatest_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/latest_model.pt\"\nlatest_train_losses_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/latest_train_losses\"\nlatest_val_losses_path = root_folder + \"gpt_output/gpt_finetuning_with_validation/latest_val_losses\"",
"_____no_output_____"
],
[
"tokenizer = GPT2Tokenizer.from_pretrained('gpt2')\nmodel = GPT2LMHeadModel.from_pretrained('gpt2')\nsaved_model = GPT2LMHeadModel.from_pretrained('gpt2')\noptimizer = OpenAIAdam(model.parameters(), lr=0.001)\niteration = 1\ntrain_losses = []\nval_losses = []",
"t_total value of -1 results in schedule not being applied\n"
],
[
"train_size = len(data) * 6 / 7\n\ngpt_train_data = defaultdict(list) # Map from length of data -> list of data\ngpt_val_data = defaultdict(list) \n\nfor i in range(len(data)):\n d = data[i]\n tokens = tokenizer.encode(d)\n if len(tokens) > 6 and len(tokens) < 50:\n if i < train_size:\n gpt_train_data[len(tokens)].append(tokens)\n else:\n gpt_val_data[len(tokens)].append(tokens)\n\ngpt_train_data = dict(gpt_train_data)\ngpt_train_data_lens = list(gpt_train_data.keys())\ngpt_val_data = dict(gpt_val_data)\ngpt_val_data_lens = list(gpt_val_data.keys())\n\nweighted_gpt_train_data_lens = []\nfor lens in gpt_train_data_lens:\n weighted_gpt_train_data_lens.extend([lens] * len(gpt_train_data[lens]))\n \nweighted_gpt_val_data_lens = []\nfor lens in gpt_val_data_lens:\n weighted_gpt_val_data_lens.extend([lens] * len(gpt_val_data[lens]))",
"_____no_output_____"
],
[
"print(\"Training data size:\", len(weighted_gpt_train_data_lens))\nprint(\"Validation data size:\", len(weighted_gpt_val_data_lens))",
"Training data size: 196955\nValidation data size: 32786\n"
],
[
"a_train = []\nfor lens in gpt_train_data_lens:\n a_train.append((lens, len(gpt_train_data[lens])))\n \na_val = []\nfor lens in gpt_val_data_lens:\n a_val.append((lens, len(gpt_val_data[lens])))\n \nmin_tup_train = min(a_train, key=lambda x: x[1])\nmin_tup_val = min(a_val, key=lambda x: x[1])\n \nprint(\"Training - Length with smallest batch: {0}. Length of batch: {1}\".format(min_tup_train[0], min_tup_train[1]))\nprint(\"Validation - Length with smallest batch: {0}. Length of batch: {1}\".format(min_tup_val[0], min_tup_val[1]))",
"Training - Length with smallest batch: 49. Length of batch: 232\nValidation - Length with smallest batch: 49. Length of batch: 33\n"
]
],
[
[
"## Load Previous Latest model",
"_____no_output_____"
],
[
"Run to load most recent model to continue training",
"_____no_output_____"
]
],
[
[
"checkpoint = torch.load(latest_path)\nmodel.load_state_dict(checkpoint['model_state_dict'])\noptimizer.load_state_dict(checkpoint['optimizer_state_dict'])\niteration = checkpoint['iteration'] + 1\nloss = checkpoint['loss']\ntrain_losses = np.load(latest_train_losses_path + \".npy\").tolist()\nval_losses = np.load(latest_val_losses_path + \".npy\").tolist()",
"_____no_output_____"
]
],
[
[
"## Training",
"_____no_output_____"
]
],
[
[
"train_batch_size = 32\nval_batch_size = 32\nkeep_model_trials = 5\nepochs = 20\niterations_per_epoch = 1000\ntotal_iterations = (iterations_per_epoch * epochs) + iteration - 1\n\nfor epoch in range(1, epochs + 1):\n for _ in range(iterations_per_epoch):\n\n length = np.random.choice(weighted_gpt_train_data_lens)\n batch_input = torch.tensor(sample(gpt_train_data[length], train_batch_size))\n\n loss = model(batch_input, lm_labels=batch_input)\n train_losses.append((iteration, loss.data.item()))\n\n optimizer.zero_grad()\n loss.backward()\n optimizer.step()\n \n if iteration % 10 == 0:\n print(\"Iteration {0}/{1}: Loss - {2}\".format(str(iteration), total_iterations, str(loss.data.item())))\n \n if iteration % 100 == 0:\n curr_loss, saved_loss = 0, 0\n for _ in range(keep_model_trials):\n length = np.random.choice(weighted_gpt_val_data_lens)\n batch_input = torch.tensor(sample(gpt_val_data[length], val_batch_size))\n\n checkpoint = torch.load(best_path)\n saved_model.load_state_dict(checkpoint['model_state_dict'])\n saved_model_loss = saved_model(batch_input, lm_labels=batch_input).data.item()\n model_loss = model(batch_input, lm_labels=batch_input).data.item()\n \n curr_loss, saved_loss = curr_loss + model_loss, saved_loss + saved_model_loss\n \n avg_curr_loss, avg_saved_loss = curr_loss / keep_model_trials, saved_loss / keep_model_trials\n loss_str = \"Average current model loss: {0}. Average saved model loss: {1}. \".format(avg_curr_loss, avg_saved_loss)\n \n val_losses.append((iteration, avg_curr_loss))\n \n if avg_curr_loss < avg_saved_loss:\n print(loss_str + \"Updating best model.\")\n torch.save({\n 'epoch': epoch,\n 'iteration': iteration,\n 'model_state_dict': model.state_dict(),\n 'optimizer_state_dict': optimizer.state_dict(),\n 'loss': loss\n }, best_path)\n \n np.save(best_train_losses_path, train_losses)\n np.save(best_val_losses_path, val_losses)\n \n else:\n print(loss_str + \"Keeping previous best model.\")\n \n torch.save({\n 'epoch': epoch,\n 'iteration': iteration,\n 'model_state_dict': model.state_dict(),\n 'optimizer_state_dict': optimizer.state_dict(),\n 'loss': loss\n }, latest_path)\n \n np.save(latest_train_losses_path, train_losses)\n np.save(latest_val_losses_path, val_losses)\n \n iteration += 1\n ",
"Iteration 9510/29500: Loss - 2.399461507797241\nIteration 9520/29500: Loss - 3.415865182876587\nIteration 9530/29500: Loss - 2.724057197570801\nIteration 9540/29500: Loss - 2.693467140197754\nIteration 9550/29500: Loss - 3.0270423889160156\nIteration 9560/29500: Loss - 3.5230624675750732\nIteration 9570/29500: Loss - 2.801056385040283\nIteration 9580/29500: Loss - 3.3204121589660645\nIteration 9590/29500: Loss - 2.6354641914367676\nIteration 9600/29500: Loss - 3.3820719718933105\nAverage current model loss: 3.522420597076416. Average saved model loss: 3.557790660858154. Updating best model.\nIteration 9610/29500: Loss - 3.0922203063964844\nIteration 9620/29500: Loss - 2.8020617961883545\nIteration 9630/29500: Loss - 3.086568593978882\nIteration 9640/29500: Loss - 2.2697863578796387\nIteration 9650/29500: Loss - 2.6908209323883057\nIteration 9660/29500: Loss - 2.7342865467071533\nIteration 9670/29500: Loss - 2.5063493251800537\nIteration 9680/29500: Loss - 2.4366679191589355\nIteration 9690/29500: Loss - 2.5203120708465576\nIteration 9700/29500: Loss - 2.768739938735962\nAverage current model loss: 3.9378710269927977. Average saved model loss: 3.951335096359253. Updating best model.\nIteration 9710/29500: Loss - 2.5374338626861572\nIteration 9720/29500: Loss - 3.179771661758423\nIteration 9730/29500: Loss - 2.4374892711639404\nIteration 9740/29500: Loss - 2.4816229343414307\nIteration 9750/29500: Loss - 3.7139275074005127\nIteration 9760/29500: Loss - 2.804251194000244\nIteration 9770/29500: Loss - 3.363471269607544\nIteration 9780/29500: Loss - 3.5627105236053467\nIteration 9790/29500: Loss - 3.318007230758667\nIteration 9800/29500: Loss - 3.288594961166382\nAverage current model loss: 3.851444149017334. Average saved model loss: 3.832876968383789. Keeping previous best model.\nIteration 9810/29500: Loss - 3.075065851211548\nIteration 9820/29500: Loss - 3.4898672103881836\nIteration 9830/29500: Loss - 2.629016399383545\nIteration 9840/29500: Loss - 2.4387972354888916\nIteration 9850/29500: Loss - 3.031240224838257\nIteration 9860/29500: Loss - 3.0897340774536133\nIteration 9870/29500: Loss - 2.6472647190093994\nIteration 9880/29500: Loss - 2.7046990394592285\nIteration 9890/29500: Loss - 2.6952919960021973\nIteration 9900/29500: Loss - 2.4661331176757812\nAverage current model loss: 3.7396754741668703. Average saved model loss: 3.731161642074585. Keeping previous best model.\nIteration 9910/29500: Loss - 3.164320468902588\nIteration 9920/29500: Loss - 2.6906051635742188\nIteration 9930/29500: Loss - 3.2928009033203125\nIteration 9940/29500: Loss - 3.318248987197876\nIteration 9950/29500: Loss - 3.2320821285247803\nIteration 9960/29500: Loss - 2.7054247856140137\nIteration 9970/29500: Loss - 3.0654592514038086\nIteration 9980/29500: Loss - 3.1193790435791016\nIteration 9990/29500: Loss - 2.5705206394195557\nIteration 10000/29500: Loss - 2.827897071838379\nAverage current model loss: 3.794435501098633. Average saved model loss: 3.825198793411255. Updating best model.\nIteration 10010/29500: Loss - 2.552640199661255\n"
]
],
[
[
"## Load Best Model",
"_____no_output_____"
]
],
[
[
"checkpoint = torch.load(best_path)\nsaved_model.load_state_dict(checkpoint['model_state_dict'])\nbest_train_losses = np.load(best_train_losses_path + \".npy\").tolist()\nbest_val_losses = np.load(best_val_losses_path + \".npy\").tolist()\nprint(\"Number of Iterations:\", checkpoint['iteration'])",
"Number of Iterations: 10000\n"
]
],
[
[
"## Plot",
"_____no_output_____"
]
],
[
[
"x_train = []\ny_train, y_sum = [], []\nfor x, y in best_train_losses:\n y_sum.append(y)\n if x % 100 == 0:\n x_train.append(x)\n y_train.append(sum(y_sum) / 100)\n y_sum = []\n \nx_val, y_val = [], []\nfor x, y in best_val_losses:\n x_val.append(x)\n y_val.append(y)\n \nplt.plot(x_train, y_train, label=\"Training loss\")\nplt.plot(x_val, y_val, label=\"Validation loss\")\nplt.legend(loc=\"lower left\")\nplt.xlabel(\"Iterations\")\nplt.ylabel(\"Loss\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Evaluation",
"_____no_output_____"
]
],
[
[
"batch_size = 32\nlength = np.random.choice(weighted_gpt_val_data_lens)\nbatch_input = torch.tensor(sample(gpt_val_data[length], batch_size))\nsaved_model_loss = saved_model(batch_input, lm_labels=batch_input).data.item()\nprint(saved_model_loss)",
"3.249004364013672\n"
]
],
[
[
"## Generate Text",
"_____no_output_____"
]
],
[
[
"text = tokenizer.encode(\"why did the\")\ninput, past = torch.tensor([text]), None\nfor _ in range(30):\n logits, past = saved_model(input, past=past)\n input = torch.multinomial(F.softmax(logits[:, -1]), 1)\n text.append(input.item())\n \ntokenizer.decode(text)",
"/usr/local/lib/python3.6/dist-packages/ipykernel_launcher.py:5: UserWarning: Implicit dimension choice for softmax has been deprecated. Change the call to include dim=X as an argument.\n \"\"\"\n"
],
[
"text = tokenizer.encode(\"why did the\")\n\n",
"_____no_output_____"
]
],
[
[
"# BERT",
"_____no_output_____"
]
],
[
[
"!python /content/gdrive/My\\ Drive/Project-Lion/pytorch-pretrained-BERT-master/examples/lm_finetuning/simple_lm_finetuning.py --train_corpus gdrive/My\\ Drive/Project-Lion/bert_training_data.txt --bert_model bert-base-uncased --num_train_epochs 4.0 --do_lower_case --output_dir gdrive/My\\ Drive/Project-Lion/gpt_output/ --do_train",
"Traceback (most recent call last):\n File \"/content/gdrive/My Drive/Project-Lion/pytorch-pretrained-BERT-master/examples/lm_finetuning/simple_lm_finetuning.py\", line 34, in <module>\n from pytorch_pretrained_bert.optimization import BertAdam, warmup_linear\nImportError: cannot import name 'warmup_linear'\n"
],
[
"model = BertForPreTraining.from_pretrained(root_folder + \"gpt_output/\")",
"_____no_output_____"
],
[
"bert_tokenizer = BertTokenizer.from_pretrained(\"bert-base-uncased\")\ntokenizer = bert_tokenizer.basic_tokenizer",
"_____no_output_____"
],
[
"tokens = tokenizer.tokenize(\"Why did the chicken\")",
"_____no_output_____"
],
[
"ids = bert_tokenizer.convert_tokens_to_ids(tokens)",
"_____no_output_____"
],
[
"text = []\ninput = torch.tensor([ids])\noutput1, output2 = model(input)\nprint(output1.shape)\na = torch.multinomial(F.softmax(output1[:, 3]), 1)\ntext.append(a.item())\nprint(text)\nprint(bert_tokenizer.convert_ids_to_tokens(text))",
"torch.Size([1, 4, 50257])\n[13]\n['[unused12]']\n"
]
],
[
[
"# Etc",
"_____no_output_____"
]
],
[
[
"tokenizer = GPT2Tokenizer.from_pretrained('gpt2')\nmodel = GPT2LMHeadModel.from_pretrained('gpt2')",
"_____no_output_____"
],
[
"text = tokenizer.encode(\"why did the chicken cross the road?\")\ninput, past = torch.tensor([text]), None\nfor _ in range(20):\n logits, past = model(input, past=past)\n print(logits.shape)\n input = torch.multinomial(F.softmax(logits[:, -1]), 1)\n text.append(input.item())\n \ntokenizer.decode(text)",
"torch.Size([1, 8, 50257])\ntensor([[383]])\ntorch.Size([1, 1, 50257])\ntensor([[22045]])\ntorch.Size([1, 1, 50257])\ntensor([[373]])\ntorch.Size([1, 1, 50257])\ntensor([[881]])\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec6fea721db7a3d428ca6a6b9e86390576532983 | 125,697 | ipynb | Jupyter Notebook | day2/DoFn_lifecycle_[with_solutions].ipynb | lcaggio/beam-college | d6912ee06213a59f217af099b6f3f537e9c6c828 | [
"Apache-2.0"
]
| 54 | 2021-04-06T18:39:04.000Z | 2022-03-29T11:06:48.000Z | day2/DoFn_lifecycle_[with_solutions].ipynb | lcaggio/beam-college | d6912ee06213a59f217af099b6f3f537e9c6c828 | [
"Apache-2.0"
]
| null | null | null | day2/DoFn_lifecycle_[with_solutions].ipynb | lcaggio/beam-college | d6912ee06213a59f217af099b6f3f537e9c6c828 | [
"Apache-2.0"
]
| 36 | 2021-04-02T15:49:48.000Z | 2022-01-13T18:11:09.000Z | 80.678434 | 48,238 | 0.608153 | [
[
[
"<a href=\"https://colab.research.google.com/github/griscz/beam-college/blob/main/day2/DoFn_lifecycle_%5Bwith_solutions%5D.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# DoFn lifecycle in Apache Beam\n\nLet's explore how `DoFn`s work in Apache Beam, and what are the different steps that are followed when executing a `DoFn`.",
"_____no_output_____"
],
[
"## Install Apache Beam",
"_____no_output_____"
]
],
[
[
"!pip install apache_beam[interactive]==2.28.0 --quiet",
"\u001b[K |████████████████████████████████| 9.0MB 9.4MB/s \n\u001b[K |████████████████████████████████| 61kB 6.5MB/s \n\u001b[K |████████████████████████████████| 153kB 46.5MB/s \n\u001b[K |████████████████████████████████| 61kB 6.5MB/s \n\u001b[K |████████████████████████████████| 2.2MB 49.4MB/s \n\u001b[K |████████████████████████████████| 829kB 45.9MB/s \n\u001b[K |████████████████████████████████| 17.7MB 209kB/s \n\u001b[K |████████████████████████████████| 122kB 46.6MB/s \n\u001b[K |████████████████████████████████| 788kB 50.0MB/s \n\u001b[K |████████████████████████████████| 122kB 49.6MB/s \n\u001b[K |████████████████████████████████| 112kB 47.2MB/s \n\u001b[K |████████████████████████████████| 368kB 48.2MB/s \n\u001b[?25h Building wheel for dill (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for future (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for avro-python3 (setup.py) ... \u001b[?25l\u001b[?25hdone\n Building wheel for timeloop (setup.py) ... \u001b[?25l\u001b[?25hdone\n\u001b[31mERROR: multiprocess 0.70.11.1 has requirement dill>=0.3.3, but you'll have dill 0.3.1.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: jupyter-console 5.2.0 has requirement prompt-toolkit<2.0.0,>=1.0.0, but you'll have prompt-toolkit 3.0.18 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipykernel~=4.10, but you'll have ipykernel 5.5.3 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement ipython~=5.5.0, but you'll have ipython 7.22.0 which is incompatible.\u001b[0m\n\u001b[31mERROR: google-colab 1.0.0 has requirement requests~=2.23.0, but you'll have requests 2.25.1 which is incompatible.\u001b[0m\n\u001b[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.\u001b[0m\n"
]
],
[
[
"## Some imports",
"_____no_output_____"
]
],
[
[
"import apache_beam as beam\nfrom apache_beam.runners.interactive.interactive_runner import InteractiveRunner\nimport apache_beam.runners.interactive.interactive_beam as ib\n\nfrom datetime import datetime",
"_____no_output_____"
]
],
[
[
"# The friends of `ParDo``\n\nFor many situations, we will not need to create a fully featured `DoFn`, we can use some of the \"friends\" of `ParDo`.\n\nFor instance, let's take some numbers as input, and multiply each number by 10:\n",
"_____no_output_____"
],
[
"## Solution using a normal `DoFn`\n\nWe need to create our own class for a `DoFn` and override the process method:",
"_____no_output_____"
]
],
[
[
"class MultiplyByTenDoFn(beam.DoFn):\n def process(self, element):\n yield element*10",
"_____no_output_____"
],
[
"with beam.Pipeline(InteractiveRunner()) as p:\n input = p | beam.Create([1, 2, 3, 4, 5])\n ib.show(input)\n\n output = input | beam.ParDo(MultiplyByTenDoFn())\n ib.show(output)\n",
"_____no_output_____"
]
],
[
[
"## Solution using `Map`\n\nThe same problem can be simply solved with a lambda. We don't need a fully featured `DoFn` just for this task:",
"_____no_output_____"
]
],
[
[
"with beam.Pipeline(InteractiveRunner()) as p:\n input = p | beam.Create([1, 2, 3, 4, 5])\n ib.show(input)\n\n output = input | beam.Map(lambda n: n*10)\n ib.show(output)\n",
"_____no_output_____"
]
],
[
[
"# The lifecycle of `DoFn`\n\nNow, in some situations we can benefit from controlling the lifecycle of our `DoFn`.\n\nIn this notebook, we are going to see just how this lifecycle works.\n\nThis will make much more sense when you learn about states and timers, in a later session at the Beam College.\n\n## Input data for this pipeline",
"_____no_output_____"
]
],
[
[
"p = beam.Pipeline(InteractiveRunner())\n\nlines = [\"En un lugar de la Mancha, de cuyo nombre no quiero acordarme,\",\n \"no ha mucho tiempo\",\n \"que vivía un hidalgo de los de lanza en astillero, adarga antigua,\",\n \"rocín flaco y galgo corredor.\"]\n\nwords = p | beam.Create(lines) \\\n | beam.FlatMap(lambda x: x.split(\" \"))\n\nib.show(words)",
"_____no_output_____"
]
],
[
[
"## Is a `DoFn` the same as a map function?\n\nWe may think that executing a `DoFn` is just the same as running a `map`.\n\nLet's see an example of a map application. Let's apply a function to sanitize the words in our `PCollection`.\n\nThis function is completely stateless. It accepts a word as input, and will return a new word. There is no initialization, shutdown of any kind of connection, aggregation, state, etc. It is just a stateless map: you can pass a reference to a function or a lambda. In the above cells, we showed how to use for instance a lambda with `FlatMap`.",
"_____no_output_____"
]
],
[
[
"def sanitize_word(w):\n w = w.replace(\".\", \"\").replace(\",\", \"\").replace(\";\", \"\")\n w = w.lower()\n return w",
"_____no_output_____"
],
[
"sane = words | beam.Map(sanitize_word)\n\nib.show(sane)",
"_____no_output_____"
]
],
[
[
"## The methods of a `DoFn``\n\nYou may use a `DoFn` just for a map function. For that, overwrite the `process` method and you are good to go.\n\nHowever, there are many more methods that you can override to control the lifecycle of a `DoFn`: creation of the worker, start of a new bundle, end of a new bundle, deletion of the worker, and of course, process of every element.\n\nThe `setup` and `teardown` methods will be executed only once per worker.\n\nThe `start_bundle` and `finish_bundle` will be executed once by data bundle",
"_____no_output_____"
]
],
[
[
"class ChangeWordDoFn(beam.DoFn):\n def process(self, element):\n print(\"Processing element: %s\" % element)\n yield element.upper()\n\n def start_bundle(self):\n print(\"Bundle started\")\n\n def finish_bundle(self):\n print(\"Bundle finished\")\n\n def setup(self):\n dt_string = datetime.now().strftime(\"%d/%m/%Y %H:%M:%S\")\n print(\"Worker started %s\" % dt_string)\n\n def teardown(self):\n dt_string = datetime.now().strftime(\"%d/%m/%Y %H:%M:%S\") \n print(\"Worker finished %s\" % dt_string)",
"_____no_output_____"
]
],
[
[
"Our `DoFn` is printing some messages at different stages in the lifecycle. In this case, we have only a handful of data, so there is only a bundle. But if we had much more data, we would see more bundles,",
"_____no_output_____"
]
],
[
[
"changed = sane | beam.ParDo(ChangeWordDoFn())\n\nib.show(changed)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec6fefda7f8808791ca1ded76d86c6e82fc00b98 | 3,345 | ipynb | Jupyter Notebook | .ipynb_checkpoints/summpy-checkpoint.ipynb | kikagaku/summpy | 771825fe54bc9a074b8d3a7f33c72169da26af09 | [
"MIT"
]
| 3 | 2019-07-26T08:27:09.000Z | 2022-01-17T04:31:59.000Z | .ipynb_checkpoints/summpy-checkpoint.ipynb | kikagaku/summpy | 771825fe54bc9a074b8d3a7f33c72169da26af09 | [
"MIT"
]
| null | null | null | .ipynb_checkpoints/summpy-checkpoint.ipynb | kikagaku/summpy | 771825fe54bc9a074b8d3a7f33c72169da26af09 | [
"MIT"
]
| 1 | 2020-04-07T03:38:04.000Z | 2020-04-07T03:38:04.000Z | 22.601351 | 132 | 0.534828 | [
[
[
"# Summpy",
"_____no_output_____"
]
],
[
[
"with open('kikagaku.txt') as f:\n text = ''.join(f.readlines())",
"_____no_output_____"
],
[
"text[:100]",
"_____no_output_____"
],
[
"from summpy.lexrank import summarize",
"_____no_output_____"
],
[
"sentences, debug_info = summarize(text, sent_limit=5)",
"_____no_output_____"
],
[
"sentences",
"_____no_output_____"
],
[
"for sentence in sentences:\n print(sentence)\n print('- ' * 10)",
"このように、いまではキカガクが『AI教育の会社』として紹介される場面が多くありますが、会社の方針として、教育だけではなく、機械学習のPDCAを回す工程の全体をスコープとして定めました。\n\n\n- - - - - - - - - - \nその時にできた最初のセミナーが『人工知能・機械学習 脱ブラックボックスセミナー』です。\n\n\n- - - - - - - - - - \n機械学習を専攻している自分としては最初から全自動でこのようなイベント運営をできればと思う気持ちもあったのですが、\n\n- - - - - - - - - - \n7月からはじまったDeep Learning ハンズオンセミナーが一段落して、今西くんにセミナー講義も任せられるようになり、時間に余裕ができたため、他の人からお話を頂いていたイベント登壇やセミナー協業を進めていきました。\n\n\n- - - - - - - - - - \n日経BigData – 「機械学習のデータはそもそも企業内にない、地道に整える企業が優位に立てる」、AI活用人材育成のキカガク吉崎社長(2017.10.20)\n\n- - - - - - - - - - \n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec6ff34d437226b5c38572c7ed0123f16a78aa7f | 34,312 | ipynb | Jupyter Notebook | analysis/Split-halves analysis.ipynb | cogtoolslab/curiotower_CogSci2021 | e8abe7030c0d1536c0214b138fc709455f29a8c1 | [
"MIT"
]
| null | null | null | analysis/Split-halves analysis.ipynb | cogtoolslab/curiotower_CogSci2021 | e8abe7030c0d1536c0214b138fc709455f29a8c1 | [
"MIT"
]
| null | null | null | analysis/Split-halves analysis.ipynb | cogtoolslab/curiotower_CogSci2021 | e8abe7030c0d1536c0214b138fc709455f29a8c1 | [
"MIT"
]
| null | null | null | 47.326897 | 11,696 | 0.632782 | [
[
[
"# Split Halves Analysis\nThis notebook will conduct an analysis of inter-rater reliability using split-halves analysis\n1. We first split the dataset into our two conditions: interesting/stable\n2. We then take each tower and randomly assign the rating to two groups.\n3. We calcualte the mean for each group in each tower\n4. Then take the correlation of the two group means across towers.\n\nWe run this process many times to get a sampling distribution of correlations, then compare the mean correlation (and CI) of stable to interesting using a t-test",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nfrom statistics import mean\n%precision %.2f\nimport matplotlib.pyplot as plt\n\nimport warnings\nwarnings.filterwarnings(\"ignore\")",
"_____no_output_____"
]
],
[
[
"## Read in most recet data",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv('curiotower_raw_data_run_0.csv')\nprint(df.shape)\ndf.head(2)",
"(24048, 47)\n"
]
],
[
[
"### split into two conditions",
"_____no_output_____"
]
],
[
[
"df_stable = df[df['condition'] == 'stable']\ndf_interesting = df[df['condition'] == 'interesting']",
"_____no_output_____"
]
],
[
[
"### Create dummy df of prolific IDs for randomization",
"_____no_output_____"
]
],
[
[
"df_subject = pd.DataFrame(df['prolificID'].unique(), \n columns = ['prolificID'])\ngrup_num = np.random.randint(2, size=len(df_subject['prolificID']))\ndf_subject['group_num'] = pd.Series(grup_num)\ndf_subject ",
"_____no_output_____"
],
[
"df_test = pd.merge(df, df_subject, left_on='prolificID', right_on='prolificID', how='left')\ndf_test[[\"prolificID\", 'group_num']].head()",
"_____no_output_____"
]
],
[
[
"## split-halves design",
"_____no_output_____"
]
],
[
[
"conditions = ['stable', 'interesting']\ncorr_stable = []\ncorr_interesting = []\nfor condition in conditions:\n print('sampling from:', condition)\n df_condition = df[df['condition'] == condition]\n for i in range(0,1000):\n df_subject = pd.DataFrame(df['prolificID'].unique(), \n columns = ['prolificID'])\n rand_group = np.random.randint(2, size=len(df_subject['prolificID']))\n df_subject['rand_group'] = pd.Series(rand_group)\n df_condition_rand = pd.merge(df_condition, df_subject, left_on='prolificID', right_on='prolificID', how='left')\n \n# rand_group = np.random.randint(2, size=len(df_condition['towerID']))\n# df_condition['rand_group'] = pd.Series(rand_group)\n\n out = df_condition_rand.pivot_table(index=[\"towerID\"], \n columns='rand_group', \n values='button_pressed',\n aggfunc='mean').reset_index()\n out.columns = ['towerID', 'group0', 'group1']\n sample_corr = out['group0'].corr(out['group1'])\n if condition == 'stable':\n #corr_stable.append(sample_corr)\n corr_stable.append(2*sample_corr/(1+sample_corr))\n elif condition == 'interesting':\n #corr_interesting.append(sample_corr)\n #Spearman brown correction\n corr_interesting.append(2*sample_corr/(1+sample_corr))\n\n\n\nplt.xlim([min(corr_stable + corr_interesting)-0.01, max(corr_stable + corr_interesting)+0.01])\n\nplt.hist(corr_stable, alpha=0.5, label = 'stable')\nplt.hist(corr_interesting, alpha = 0.5, color = 'orange', label = 'interesting')\nplt.title('Sample Distritbutions of Corr for Conditions')\nplt.xlabel('Corr')\nplt.ylabel('count')\nplt.legend()\nplt.show()",
"sampling from: stable\nsampling from: interesting\n"
],
[
"print(\"Mean prop for stable:\", \n round(mean(corr_stable),3),\n \"+/-\", \n round((1.96*np.std(corr_stable)/\n np.sqrt(len(corr_stable))),3))\nprint(\"Mean prop for interesting:\",\n round(mean(corr_interesting),3), \n \"+/-\", \n round((1.96*np.std(corr_interesting)/\n np.sqrt(len(corr_interesting))),10))",
"Mean prop for stable: 0.997 +/- 0.0\nMean prop for interesting: 0.996 +/- 7.48176e-05\n"
]
],
[
[
"## Now caluclate the r for each model and compute the proportion of variance explained\n",
"_____no_output_____"
]
],
[
[
"dat = df[df['condition'] == 'interesting']\ndat.columns\ndat = pd.get_dummies(dat, columns=['stability'])",
"_____no_output_____"
],
[
"dat.columns",
"_____no_output_____"
],
[
"from sklearn.linear_model import LinearRegression\nfrom sklearn.preprocessing import PolynomialFeatures\n\nregressors = ['num_blocks']\n# regressors = ['num_blocks', 'stability_high', 'stability_low', 'stability_med']\n#regressors = ['num_blocks', 'stability_high', 'stability_low', 'stability_med']\n# X = dat[regressors]\n# interaction = PolynomialFeatures(degree=3, include_bias=False, interaction_only=True)\n# X = interaction.fit_transform(X)\n\nregressors = ['num_blocks', 'stability_high', 'stability_low', 'stability_med']\nX = dat[regressors]\ny = dat['button_pressed']\n##Sklearn regression\nreg = LinearRegression().fit(X, y)\nreg.score(X,y)\n\n\n",
"_____no_output_____"
]
],
[
[
"## Mode analysis\n\nfor each condition;\n for each tower;\n calculate proportion of responses selecting mode; \n --> then average acrosss towers (and report CI)",
"_____no_output_____"
]
],
[
[
"#annoying function to get min when equal counts for mode\ndef try_min(value):\n try:\n return min(value)\n except:\n return value",
"_____no_output_____"
],
[
"df_stable_mode = df[df['condition']=='stable'].groupby(['towerID']).agg(pd.Series.mode).reset_index()[['towerID','button_pressed']]\ndf_stable_mode['mode'] = df_stable_mode['button_pressed'].apply(lambda x: try_min(x))\n",
"_____no_output_____"
],
[
"\n\nconditions = ['stable', 'interesting']\ntowers = df['towerID'].unique()\nmode_proportion_list_stable = []\nmode_proportion_list_interesting = []\nfor condition in conditions:\n print('sampling from:', condition)\n df_condition = df[df['condition'] == condition]\n df_mode = df_condition.groupby(['towerID']).agg(pd.Series.mode).reset_index()[['towerID','button_pressed']]\n df_mode['mode'] = df_mode['button_pressed'].apply(lambda x: try_min(x))\n\n for tower in towers:\n mode_response = int(df_mode[df_mode['towerID'] == tower]['mode'])\n prop = (len(df_condition.loc[(df_condition['towerID'] == tower) & (df_condition['button_pressed'] == mode_response)])/\n len(df_condition.loc[(df_condition['towerID'] == tower)]))\n if condition == 'stable':\n mode_proportion_list_stable.append(prop)\n elif condition == 'interesting':\n mode_proportion_list_interesting.append(prop)\n \n \n\nprint(\"Mean prop for stable:\", \n round(mean(mode_proportion_list_stable),3),\n \"+/-\", \n round((1.96*np.std(mode_proportion_list_stable)/\n np.sqrt(len(mode_proportion_list_stable))),3))\nprint(\"Mean prop for interesting:\",\n round(mean(mode_proportion_list_interesting),3), \n \"+/-\", \n round((1.96*np.std(mode_proportion_list_interesting)/\n np.sqrt(len(mode_proportion_list_interesting))),3))\n ",
"sampling from: stable\nsampling from: interesting\nMean prop for stable: 0.516 +/- 0.027\nMean prop for interesting: 0.542 +/- 0.031\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec6ff9216f30b8933d43d5c66fdc7759010eda8e | 185,218 | ipynb | Jupyter Notebook | Modulo2/Clase10_DiversificacionFuentesRiesgoII.ipynb | eortizt/PorInv2018-2 | 5a86eeb8fbb7be4b36f74c3414b6510ee2b652df | [
"MIT"
]
| 1 | 2018-08-27T16:54:10.000Z | 2018-08-27T16:54:10.000Z | Modulo2/Clase10_DiversificacionFuentesRiesgoII.ipynb | kitziafigueroa/PorInv2018-2 | 13192518e6e366d070c8d247a857918f3856d90c | [
"MIT"
]
| null | null | null | Modulo2/Clase10_DiversificacionFuentesRiesgoII.ipynb | kitziafigueroa/PorInv2018-2 | 13192518e6e366d070c8d247a857918f3856d90c | [
"MIT"
]
| null | null | null | 78.782646 | 37,860 | 0.722792 | [
[
[
"# Diversificación y fuentes de riesgo en un portafolio II - Una ilustración con mercados internacionales.\n\n<img style=\"float: right; margin: 0px 0px 15px 15px;\" src=\"https://upload.wikimedia.org/wikipedia/commons/5/5f/Map_International_Markets.jpg\" width=\"500px\" height=\"300px\" />\n\n> Entonces, la clase pasada vimos cómo afecta la correlación entre pares de activos en un portafolio. Dijimos que como un par de activos nunca tienen correlación perfecta, al combinarlos en un portafolio siempre conseguimos diversificación del riesgo.\n\n> Vimos también que no todo el riesgo se puede diversificar. Dos fuentes de riesgo:\n> - Sistemático: afecta de igual manera a todos los activos. No se puede diversificar.\n> - Idiosincrático: afecta a cada activo en particular por razones específicas. Se puede diversificar.\n\nEn esta clase veremos un ejemplo de diversificación en un portafolio, usando datos de mercados de activos internacionales.\n\nEn el camino, definiremos términos como *frontera media/varianza*, *portafolio de varianza mínima* y *portafolios eficientes*, los cuales son básicos para la construcción de la **teoría moderna de portafolios**.\n\nEstos portafolios los aprenderemos a obtener formalmente en el siguiente módulo. Por ahora nos bastará con agarrar intuición.",
"_____no_output_____"
],
[
"**Objetivo:**\n- Ver los beneficios de la diversificación ilustrativamente.\n- ¿Qué es la frontera de mínima varianza?\n- ¿Qué son el portafolio de varianza mínima y portafolios eficientes?\n\n*Referencia:*\n- Notas del curso \"Portfolio Selection and Risk Management\", Rice University, disponible en Coursera.\n___",
"_____no_output_____"
],
[
"## 1. Ejemplo\n\n**Los datos:** tenemos el siguiente reporte de rendimientos esperados y volatilidad (anuales) para los mercados de acciones en los países integrantes del $G5$: EU, RU, Francia, Alemania y Japón.",
"_____no_output_____"
]
],
[
[
"# Importamos pandas y numpy\nimport pandas as pd\nimport numpy as np",
"_____no_output_____"
],
[
"# Resumen en base anual de rendimientos esperados y volatilidades\nannual_ret_summ = pd.DataFrame(columns=['EU', 'RU', 'Francia', 'Alemania', 'Japon'], index=['Media', 'Volatilidad'])\nannual_ret_summ.loc['Media'] = np.array([0.1355, 0.1589, 0.1519, 0.1435, 0.1497])\nannual_ret_summ.loc['Volatilidad'] = np.array([0.1535, 0.2430, 0.2324, 0.2038, 0.2298])\n\nannual_ret_summ.round(4)",
"_____no_output_____"
]
],
[
[
"¿Qué podemos notar?\n- Los rendimientos esperados rondan todos por los mismos valores 14%-15%.\n- En cuanto al riesgo, la medida de riesgo del mercado de Estados Unidos es mucho menor respecto a las demás.",
"_____no_output_____"
],
[
"Además, tenemos el siguiente reporte de la matriz de correlación:",
"_____no_output_____"
]
],
[
[
"# Matriz de correlación\ncorr = pd.DataFrame(data= np.array([[1.0000, 0.5003, 0.4398, 0.3681, 0.2663],\n [0.5003, 1.0000, 0.5420, 0.4265, 0.3581],\n [0.4398, 0.5420, 1.0000, 0.6032, 0.3923],\n [0.3681, 0.4265, 0.6032, 1.0000, 0.3663],\n [0.2663, 0.3581, 0.3923, 0.3663, 1.0000]]),\n columns=annual_ret_summ.columns, index=annual_ret_summ.columns)\ncorr.round(4)",
"_____no_output_____"
]
],
[
[
"¿Qué se puede observar?\n- Los mercados de acciones que presentan la correlación más baja son los de EU y Japón.\n\nRecordar: correlaciones bajas significan una gran oportunidad para diversificación.",
"_____no_output_____"
],
[
"### Nos enfocaremos entonces únicamente en dos mercados: EU y Japón\n\n- ¿Cómo construiríamos un portafolio que consiste de los mercados de acciones de EU y Japón?\n- ¿Cuáles serían las posibles combinaciones?",
"_____no_output_____"
],
[
"#### 1. Supongamos que $w$ es la participación del mercado de EU en nuestro portafolio.\n- ¿Cuál es la participación del mercado de Japón entonces?: $1-w$\n\n- Luego, nuestras fórmulas de rendimiento esperado y varianza de portafolios son:\n\n$$E[r_p]=wE[r_{EU}]+(1-w)E[r_J]$$\n\n$$\\sigma_p^2=w^2\\sigma_{EU}^2+(1-w)^2\\sigma_J^2+2w(1-w)\\sigma_{EU,J}$$",
"_____no_output_____"
],
[
"#### 2. Con lo anterior...\n- podemos variar $w$ con pasos pequeños entre $0$ y $1$, y\n- calcular el rendimiento esperado y volatilidad para cada valor de $w$.",
"_____no_output_____"
]
],
[
[
"# Vector de w variando entre 0 y 1 con n pasos\nw = np.linspace(0, 1, 100)\n# Rendimientos esperados individuales\n# Activo1: EU, Activo2:Japon\nE1 = annual_ret_summ['EU']['Media']\nE2 = annual_ret_summ['Japon']['Media']\n# Volatilidades individuales\ns1 = annual_ret_summ['EU']['Volatilidad']\ns2 = annual_ret_summ['Japon']['Volatilidad']\n# Correlacion\nr12 = corr['EU']['Japon']",
"_____no_output_____"
],
[
"# Crear un DataFrame cuyas columnas sean rendimiento\n# y volatilidad del portafolio para cada una de las w\n# generadas\nportafolios2 = pd.DataFrame(index=w,columns=['Rend','Vol'])\nportafolios2.index.name = 'w'\nportafolios2.Rend = w*E1+(1-w)*E2\nportafolios2.Vol = np.sqrt((w*s1)**2+((1-w)*s2)**2+2*w*(1-w)*r12*s1*s2)\nportafolios2.round(4)",
"_____no_output_____"
]
],
[
[
"#### 3. Finalmente, \n- cada una de las combinaciones las podemos graficar en el espacio de rendimiento esperado (eje $y$) contra volatilidad (eje $x$).",
"_____no_output_____"
]
],
[
[
"# Importar matplotlib\nimport matplotlib.pyplot as plt\n%matplotlib inline",
"_____no_output_____"
],
[
"# Graficar el lugar geométrico del portafolio en el\n# espacio rendimiento esperado vs. volatilidad.\n# Especificar también los puntos relativos a los casos\n# extremos.\nplt.figure(figsize=(8,6))\nplt.plot(s1, E1, 'ro', ms = 10, label='EU')\nplt.plot(s2, E2, 'bo', ms = 10, label='Japón')\nplt.plot(portafolios2.Vol, portafolios2.Rend, 'k-', lw = 4, label='Portafolios')\nplt.xlabel('Volatilidad ($\\sigma$)')\nplt.ylabel('Rendimiento esperado ($E[r]$)')\nplt.legend(loc='best')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"#### De la gráfica,\n1. Ver casos extremos.\n2. ¿Conviene invertir 100% en el mercado de EU? ¿Porqué?\n3. ¿Porqué ocurre esto?\n4. Definición: frontera de mínima varianza. Caso particular: dos activos.\n5. Definición: portafolio de varianza mínima.\n6. Definición: portafolios eficientes.\n\n#### 1. Definición: la frontera de mínima varianza es el lugar geométrico en el espacio de rendimiento esperado vs. volatilidad de los portafolios que para un nivel de rendimiento esperado brindan la mínima varianza posible. En el caso de dos activos, la frontera de mínima varianza corresponde exactamente a todos los portafolios que se pueden generar con estos dos activos.\n\n#### 2. Definición: el portafolio de mínima varianza es el portafolio que está más a la izquierda sobre la frontera de mínima varianza.\n\n#### 3. Definición: los portafolios eficientes son los portafolios que están en la parte superior de la frontera de mínima varianza y a partir del portafolio de mínima varianza.\n___",
"_____no_output_____"
],
[
"## 2. ¿Cómo hallar el portafolio de varianza mínima?\n\nBien, esta será nuestra primera selección de portafolio. Si bien se hace de manera básica e intuitiva, nos servirá como introducción al siguiente módulo. \n\n**Comentario:** estrictamente, el portafolio que está más a la izquierda en la curva de arriba es el de *volatilidad mínima*. Sin embargo, como tanto la volatilidad es una medida siempre positiva, minimizar la volatilidad equivale a minimizar la varianza. Por lo anterior, llamamos a dicho portafolio, el portafolio de *varianza mínima*.",
"_____no_output_____"
],
[
"De modo que la búsqueda del portafolio de varianza mínima corresponde a la solución del siguiente problema de optimización:\n\n- Para un portafolio con $n$ activos ($\\boldsymbol{w}=[w_1,\\dots,w_n]^T\\in\\mathbb{R}^n$):\n\\begin{align*}\n&\\min_{\\boldsymbol{w}}\\sigma_p^2 &=\\boldsymbol{w}^T\\Sigma\\boldsymbol{w}\\\\\n&\\text{s.t.} \\qquad & \\boldsymbol{w}\\geq0,\\\\\n& & w_1+\\dots+w_n=1\n\\end{align*}\ndonde $\\Sigma$ es la matriz de varianza-covarianza de los rendimientos de los $n$ activos.",
"_____no_output_____"
],
[
"- En particular, para un portafolio con dos activos el problema anterior se reduce a:\n\\begin{align*}\n&\\min_{w_1,w_2}\\sigma_p^2=w_1^2\\sigma_1^2+w_2^2\\sigma_2^2+2w_1w_2\\rho_{12}\\sigma_1\\sigma_2\\\\\n&\\text{s.t.} \\qquad w_1,w_2\\geq0,\n\\end{align*}\ndonde $\\sigma_1,\\sigma_2$ son las volatilidades de los activos individuales y $\\rho_{12}$ es la correlación entre los activos. Equivalentemente, haciendo $w_1=w$ y $w_2=1-w$, el problema anterior se puede reescribir de la siguiente manera:\n\\begin{align*}\n&\\min_{w}\\sigma_p^2=w^2\\sigma_1^2+(1-w)^2\\sigma_2^2+2w(1-w)\\rho_{12}\\sigma_1\\sigma_2\\\\\n&\\text{s.t.} \\qquad 0\\leq w\\leq1,\n\\end{align*}",
"_____no_output_____"
],
[
"1. Los anteriores son problemas de **programación cuadrática** (función convexa sobre dominio convexo: mínimo absoluto asegurado). \n2. Existen diversos algoritmos para problemas de programación cuadrática. Por ejemplo, en la librería cvxopt. Más adelante la instalaremos y la usaremos.\n3. En scipy.optimize no hay un algoritmo dedicado a la solución de este tipo de problemas de optimización. Sin embargo, la función mínimize nos permite resolver problemas de optimización en general (es un poco limitada, pero nos sirve por ahora).",
"_____no_output_____"
],
[
"### 2.1. Antes de resolver el problema con la función minimize: resolverlo a mano en el tablero.",
"_____no_output_____"
]
],
[
[
"w_minvar = (s2**2-(r12*s1*s2))/(s1**2+s2**2-2*(r12*s1*s2))\nw_minvar",
"_____no_output_____"
]
],
[
[
"**Conclusiones:**\n- El portafolio de mínima varianza se obtiene al invertir el $75.39\\%$ de la riqueza en el mercado de EU.",
"_____no_output_____"
],
[
"### 2.2. Ahora sí, con la función scipy.optimize.minimize",
"_____no_output_____"
]
],
[
[
"# Importar el módulo optimize\nimport scipy.optimize as opt",
"_____no_output_____"
],
[
"# Función minimize\nhelp(opt.minimize)",
"Help on function minimize in module scipy.optimize._minimize:\n\nminimize(fun, x0, args=(), method=None, jac=None, hess=None, hessp=None, bounds=None, constraints=(), tol=None, callback=None, options=None)\n Minimization of scalar function of one or more variables.\n \n Parameters\n ----------\n fun : callable\n The objective function to be minimized.\n \n ``fun(x, *args) -> float``\n \n where x is an 1-D array with shape (n,) and `args`\n is a tuple of the fixed parameters needed to completely\n specify the function.\n x0 : ndarray, shape (n,)\n Initial guess. Array of real elements of size (n,),\n where 'n' is the number of independent variables.\n args : tuple, optional\n Extra arguments passed to the objective function and its\n derivatives (`fun`, `jac` and `hess` functions).\n method : str or callable, optional\n Type of solver. Should be one of\n \n - 'Nelder-Mead' :ref:`(see here) <optimize.minimize-neldermead>`\n - 'Powell' :ref:`(see here) <optimize.minimize-powell>`\n - 'CG' :ref:`(see here) <optimize.minimize-cg>`\n - 'BFGS' :ref:`(see here) <optimize.minimize-bfgs>`\n - 'Newton-CG' :ref:`(see here) <optimize.minimize-newtoncg>`\n - 'L-BFGS-B' :ref:`(see here) <optimize.minimize-lbfgsb>`\n - 'TNC' :ref:`(see here) <optimize.minimize-tnc>`\n - 'COBYLA' :ref:`(see here) <optimize.minimize-cobyla>`\n - 'SLSQP' :ref:`(see here) <optimize.minimize-slsqp>`\n - 'trust-constr':ref:`(see here) <optimize.minimize-trustconstr>`\n - 'dogleg' :ref:`(see here) <optimize.minimize-dogleg>`\n - 'trust-ncg' :ref:`(see here) <optimize.minimize-trustncg>`\n - 'trust-exact' :ref:`(see here) <optimize.minimize-trustexact>`\n - 'trust-krylov' :ref:`(see here) <optimize.minimize-trustkrylov>`\n - custom - a callable object (added in version 0.14.0),\n see below for description.\n \n If not given, chosen to be one of ``BFGS``, ``L-BFGS-B``, ``SLSQP``,\n depending if the problem has constraints or bounds.\n jac : {callable, '2-point', '3-point', 'cs', bool}, optional\n Method for computing the gradient vector. Only for CG, BFGS,\n Newton-CG, L-BFGS-B, TNC, SLSQP, dogleg, trust-ncg, trust-krylov,\n trust-exact and trust-constr. If it is a callable, it should be a\n function that returns the gradient vector:\n \n ``jac(x, *args) -> array_like, shape (n,)``\n \n where x is an array with shape (n,) and `args` is a tuple with\n the fixed parameters. Alternatively, the keywords\n {'2-point', '3-point', 'cs'} select a finite\n difference scheme for numerical estimation of the gradient. Options\n '3-point' and 'cs' are available only to 'trust-constr'.\n If `jac` is a Boolean and is True, `fun` is assumed to return the\n gradient along with the objective function. If False, the gradient\n will be estimated using '2-point' finite difference estimation.\n hess : {callable, '2-point', '3-point', 'cs', HessianUpdateStrategy}, optional\n Method for computing the Hessian matrix. Only for Newton-CG, dogleg,\n trust-ncg, trust-krylov, trust-exact and trust-constr. If it is\n callable, it should return the Hessian matrix:\n \n ``hess(x, *args) -> {LinearOperator, spmatrix, array}, (n, n)``\n \n where x is a (n,) ndarray and `args` is a tuple with the fixed\n parameters. LinearOperator and sparse matrix returns are\n allowed only for 'trust-constr' method. Alternatively, the keywords\n {'2-point', '3-point', 'cs'} select a finite difference scheme\n for numerical estimation. Or, objects implementing\n `HessianUpdateStrategy` interface can be used to approximate\n the Hessian. Available quasi-Newton methods implementing\n this interface are:\n \n - `BFGS`;\n - `SR1`.\n \n Whenever the gradient is estimated via finite-differences,\n the Hessian cannot be estimated with options\n {'2-point', '3-point', 'cs'} and needs to be\n estimated using one of the quasi-Newton strategies.\n Finite-difference options {'2-point', '3-point', 'cs'} and\n `HessianUpdateStrategy` are available only for 'trust-constr' method.\n hessp : callable, optional\n Hessian of objective function times an arbitrary vector p. Only for\n Newton-CG, trust-ncg, trust-krylov, trust-constr.\n Only one of `hessp` or `hess` needs to be given. If `hess` is\n provided, then `hessp` will be ignored. `hessp` must compute the\n Hessian times an arbitrary vector:\n \n ``hessp(x, p, *args) -> ndarray shape (n,)``\n \n where x is a (n,) ndarray, p is an arbitrary vector with\n dimension (n,) and `args` is a tuple with the fixed\n parameters.\n bounds : sequence or `Bounds`, optional\n Bounds on variables for L-BFGS-B, TNC, SLSQP and\n trust-constr methods. There are two ways to specify the bounds:\n \n 1. Instance of `Bounds` class.\n 2. Sequence of ``(min, max)`` pairs for each element in `x`. None\n is used to specify no bound.\n \n constraints : {Constraint, dict} or List of {Constraint, dict}, optional\n Constraints definition (only for COBYLA, SLSQP and trust-constr).\n Constraints for 'trust-constr' are defined as a single object or a\n list of objects specifying constraints to the optimization problem.\n Available constraints are:\n \n - `LinearConstraint`\n - `NonlinearConstraint`\n \n Constraints for COBYLA, SLSQP are defined as a list of dictionaries.\n Each dictionary with fields:\n \n type : str\n Constraint type: 'eq' for equality, 'ineq' for inequality.\n fun : callable\n The function defining the constraint.\n jac : callable, optional\n The Jacobian of `fun` (only for SLSQP).\n args : sequence, optional\n Extra arguments to be passed to the function and Jacobian.\n \n Equality constraint means that the constraint function result is to\n be zero whereas inequality means that it is to be non-negative.\n Note that COBYLA only supports inequality constraints.\n tol : float, optional\n Tolerance for termination. For detailed control, use solver-specific\n options.\n options : dict, optional\n A dictionary of solver options. All methods accept the following\n generic options:\n \n maxiter : int\n Maximum number of iterations to perform.\n disp : bool\n Set to True to print convergence messages.\n \n For method-specific options, see :func:`show_options()`.\n callback : callable, optional\n Called after each iteration. For 'trust-constr' it is a callable with\n the signature:\n \n ``callback(xk, OptimizeResult state) -> bool``\n \n where ``xk`` is the current parameter vector. and ``state``\n is an `OptimizeResult` object, with the same fields\n as the ones from the return. If callback returns True\n the algorithm execution is terminated.\n For all the other methods, the signature is:\n \n ``callback(xk)``\n \n where ``xk`` is the current parameter vector.\n \n Returns\n -------\n res : OptimizeResult\n The optimization result represented as a ``OptimizeResult`` object.\n Important attributes are: ``x`` the solution array, ``success`` a\n Boolean flag indicating if the optimizer exited successfully and\n ``message`` which describes the cause of the termination. See\n `OptimizeResult` for a description of other attributes.\n \n \n See also\n --------\n minimize_scalar : Interface to minimization algorithms for scalar\n univariate functions\n show_options : Additional options accepted by the solvers\n \n Notes\n -----\n This section describes the available solvers that can be selected by the\n 'method' parameter. The default method is *BFGS*.\n \n **Unconstrained minimization**\n \n Method :ref:`Nelder-Mead <optimize.minimize-neldermead>` uses the\n Simplex algorithm [1]_, [2]_. This algorithm is robust in many\n applications. However, if numerical computation of derivative can be\n trusted, other algorithms using the first and/or second derivatives\n information might be preferred for their better performance in\n general.\n \n Method :ref:`Powell <optimize.minimize-powell>` is a modification\n of Powell's method [3]_, [4]_ which is a conjugate direction\n method. It performs sequential one-dimensional minimizations along\n each vector of the directions set (`direc` field in `options` and\n `info`), which is updated at each iteration of the main\n minimization loop. The function need not be differentiable, and no\n derivatives are taken.\n \n Method :ref:`CG <optimize.minimize-cg>` uses a nonlinear conjugate\n gradient algorithm by Polak and Ribiere, a variant of the\n Fletcher-Reeves method described in [5]_ pp. 120-122. Only the\n first derivatives are used.\n \n Method :ref:`BFGS <optimize.minimize-bfgs>` uses the quasi-Newton\n method of Broyden, Fletcher, Goldfarb, and Shanno (BFGS) [5]_\n pp. 136. It uses the first derivatives only. BFGS has proven good\n performance even for non-smooth optimizations. This method also\n returns an approximation of the Hessian inverse, stored as\n `hess_inv` in the OptimizeResult object.\n \n Method :ref:`Newton-CG <optimize.minimize-newtoncg>` uses a\n Newton-CG algorithm [5]_ pp. 168 (also known as the truncated\n Newton method). It uses a CG method to the compute the search\n direction. See also *TNC* method for a box-constrained\n minimization with a similar algorithm. Suitable for large-scale\n problems.\n \n Method :ref:`dogleg <optimize.minimize-dogleg>` uses the dog-leg\n trust-region algorithm [5]_ for unconstrained minimization. This\n algorithm requires the gradient and Hessian; furthermore the\n Hessian is required to be positive definite.\n \n Method :ref:`trust-ncg <optimize.minimize-trustncg>` uses the\n Newton conjugate gradient trust-region algorithm [5]_ for\n unconstrained minimization. This algorithm requires the gradient\n and either the Hessian or a function that computes the product of\n the Hessian with a given vector. Suitable for large-scale problems.\n \n Method :ref:`trust-krylov <optimize.minimize-trustkrylov>` uses\n the Newton GLTR trust-region algorithm [14]_, [15]_ for unconstrained\n minimization. This algorithm requires the gradient\n and either the Hessian or a function that computes the product of\n the Hessian with a given vector. Suitable for large-scale problems.\n On indefinite problems it requires usually less iterations than the\n `trust-ncg` method and is recommended for medium and large-scale problems.\n \n Method :ref:`trust-exact <optimize.minimize-trustexact>`\n is a trust-region method for unconstrained minimization in which\n quadratic subproblems are solved almost exactly [13]_. This\n algorithm requires the gradient and the Hessian (which is\n *not* required to be positive definite). It is, in many\n situations, the Newton method to converge in fewer iteraction\n and the most recommended for small and medium-size problems.\n \n **Bound-Constrained minimization**\n \n Method :ref:`L-BFGS-B <optimize.minimize-lbfgsb>` uses the L-BFGS-B\n algorithm [6]_, [7]_ for bound constrained minimization.\n \n Method :ref:`TNC <optimize.minimize-tnc>` uses a truncated Newton\n algorithm [5]_, [8]_ to minimize a function with variables subject\n to bounds. This algorithm uses gradient information; it is also\n called Newton Conjugate-Gradient. It differs from the *Newton-CG*\n method described above as it wraps a C implementation and allows\n each variable to be given upper and lower bounds.\n \n **Constrained Minimization**\n \n Method :ref:`COBYLA <optimize.minimize-cobyla>` uses the\n Constrained Optimization BY Linear Approximation (COBYLA) method\n [9]_, [10]_, [11]_. The algorithm is based on linear\n approximations to the objective function and each constraint. The\n method wraps a FORTRAN implementation of the algorithm. The\n constraints functions 'fun' may return either a single number\n or an array or list of numbers.\n \n Method :ref:`SLSQP <optimize.minimize-slsqp>` uses Sequential\n Least SQuares Programming to minimize a function of several\n variables with any combination of bounds, equality and inequality\n constraints. The method wraps the SLSQP Optimization subroutine\n originally implemented by Dieter Kraft [12]_. Note that the\n wrapper handles infinite values in bounds by converting them into\n large floating values.\n \n Method :ref:`trust-constr <optimize.minimize-trustconstr>` is a\n trust-region algorithm for constrained optimization. It swiches\n between two implementations depending on the problem definition.\n It is the most versatile constrained minimization algorithm\n implemented in SciPy and the most appropriate for large-scale problems.\n For equality constrained problems it is an implementation of Byrd-Omojokun\n Trust-Region SQP method described in [17]_ and in [5]_, p. 549. When\n inequality constraints are imposed as well, it swiches to the trust-region\n interior point method described in [16]_. This interior point algorithm,\n in turn, solves inequality constraints by introducing slack variables\n and solving a sequence of equality-constrained barrier problems\n for progressively smaller values of the barrier parameter.\n The previously described equality constrained SQP method is\n used to solve the subproblems with increasing levels of accuracy\n as the iterate gets closer to a solution.\n \n **Finite-Difference Options**\n \n For Method :ref:`trust-constr <optimize.minimize-trustconstr>`\n the gradient and the Hessian may be approximated using\n three finite-difference schemes: {'2-point', '3-point', 'cs'}.\n The scheme 'cs' is, potentially, the most accurate but it\n requires the function to correctly handles complex inputs and to\n be differentiable in the complex plane. The scheme '3-point' is more\n accurate than '2-point' but requires twice as much operations.\n \n **Custom minimizers**\n \n It may be useful to pass a custom minimization method, for example\n when using a frontend to this method such as `scipy.optimize.basinhopping`\n or a different library. You can simply pass a callable as the ``method``\n parameter.\n \n The callable is called as ``method(fun, x0, args, **kwargs, **options)``\n where ``kwargs`` corresponds to any other parameters passed to `minimize`\n (such as `callback`, `hess`, etc.), except the `options` dict, which has\n its contents also passed as `method` parameters pair by pair. Also, if\n `jac` has been passed as a bool type, `jac` and `fun` are mangled so that\n `fun` returns just the function values and `jac` is converted to a function\n returning the Jacobian. The method shall return an ``OptimizeResult``\n object.\n \n The provided `method` callable must be able to accept (and possibly ignore)\n arbitrary parameters; the set of parameters accepted by `minimize` may\n expand in future versions and then these parameters will be passed to\n the method. You can find an example in the scipy.optimize tutorial.\n \n .. versionadded:: 0.11.0\n \n References\n ----------\n .. [1] Nelder, J A, and R Mead. 1965. A Simplex Method for Function\n Minimization. The Computer Journal 7: 308-13.\n .. [2] Wright M H. 1996. Direct search methods: Once scorned, now\n respectable, in Numerical Analysis 1995: Proceedings of the 1995\n Dundee Biennial Conference in Numerical Analysis (Eds. D F\n Griffiths and G A Watson). Addison Wesley Longman, Harlow, UK.\n 191-208.\n .. [3] Powell, M J D. 1964. An efficient method for finding the minimum of\n a function of several variables without calculating derivatives. The\n Computer Journal 7: 155-162.\n .. [4] Press W, S A Teukolsky, W T Vetterling and B P Flannery.\n Numerical Recipes (any edition), Cambridge University Press.\n .. [5] Nocedal, J, and S J Wright. 2006. Numerical Optimization.\n Springer New York.\n .. [6] Byrd, R H and P Lu and J. Nocedal. 1995. A Limited Memory\n Algorithm for Bound Constrained Optimization. SIAM Journal on\n Scientific and Statistical Computing 16 (5): 1190-1208.\n .. [7] Zhu, C and R H Byrd and J Nocedal. 1997. L-BFGS-B: Algorithm\n 778: L-BFGS-B, FORTRAN routines for large scale bound constrained\n optimization. ACM Transactions on Mathematical Software 23 (4):\n 550-560.\n .. [8] Nash, S G. Newton-Type Minimization Via the Lanczos Method.\n 1984. SIAM Journal of Numerical Analysis 21: 770-778.\n .. [9] Powell, M J D. A direct search optimization method that models\n the objective and constraint functions by linear interpolation.\n 1994. Advances in Optimization and Numerical Analysis, eds. S. Gomez\n and J-P Hennart, Kluwer Academic (Dordrecht), 51-67.\n .. [10] Powell M J D. Direct search algorithms for optimization\n calculations. 1998. Acta Numerica 7: 287-336.\n .. [11] Powell M J D. A view of algorithms for optimization without\n derivatives. 2007.Cambridge University Technical Report DAMTP\n 2007/NA03\n .. [12] Kraft, D. A software package for sequential quadratic\n programming. 1988. Tech. Rep. DFVLR-FB 88-28, DLR German Aerospace\n Center -- Institute for Flight Mechanics, Koln, Germany.\n .. [13] Conn, A. R., Gould, N. I., and Toint, P. L.\n Trust region methods. 2000. Siam. pp. 169-200.\n .. [14] F. Lenders, C. Kirches, A. Potschka: \"trlib: A vector-free\n implementation of the GLTR method for iterative solution of\n the trust region problem\", https://arxiv.org/abs/1611.04718\n .. [15] N. Gould, S. Lucidi, M. Roma, P. Toint: \"Solving the\n Trust-Region Subproblem using the Lanczos Method\",\n SIAM J. Optim., 9(2), 504--525, (1999).\n .. [16] Byrd, Richard H., Mary E. Hribar, and Jorge Nocedal. 1999.\n An interior point algorithm for large-scale nonlinear programming.\n SIAM Journal on Optimization 9.4: 877-900.\n .. [17] Lalee, Marucha, Jorge Nocedal, and Todd Plantega. 1998. On the\n implementation of an algorithm for large-scale equality constrained\n optimization. SIAM Journal on Optimization 8.3: 682-706.\n \n Examples\n --------\n Let us consider the problem of minimizing the Rosenbrock function. This\n function (and its respective derivatives) is implemented in `rosen`\n (resp. `rosen_der`, `rosen_hess`) in the `scipy.optimize`.\n \n >>> from scipy.optimize import minimize, rosen, rosen_der\n \n A simple application of the *Nelder-Mead* method is:\n \n >>> x0 = [1.3, 0.7, 0.8, 1.9, 1.2]\n >>> res = minimize(rosen, x0, method='Nelder-Mead', tol=1e-6)\n >>> res.x\n array([ 1., 1., 1., 1., 1.])\n \n Now using the *BFGS* algorithm, using the first derivative and a few\n options:\n \n >>> res = minimize(rosen, x0, method='BFGS', jac=rosen_der,\n ... options={'gtol': 1e-6, 'disp': True})\n Optimization terminated successfully.\n Current function value: 0.000000\n Iterations: 26\n Function evaluations: 31\n Gradient evaluations: 31\n >>> res.x\n array([ 1., 1., 1., 1., 1.])\n >>> print(res.message)\n Optimization terminated successfully.\n >>> res.hess_inv\n array([[ 0.00749589, 0.01255155, 0.02396251, 0.04750988, 0.09495377], # may vary\n [ 0.01255155, 0.02510441, 0.04794055, 0.09502834, 0.18996269],\n [ 0.02396251, 0.04794055, 0.09631614, 0.19092151, 0.38165151],\n [ 0.04750988, 0.09502834, 0.19092151, 0.38341252, 0.7664427 ],\n [ 0.09495377, 0.18996269, 0.38165151, 0.7664427, 1.53713523]])\n \n \n Next, consider a minimization problem with several constraints (namely\n Example 16.4 from [5]_). The objective function is:\n \n >>> fun = lambda x: (x[0] - 1)**2 + (x[1] - 2.5)**2\n \n There are three constraints defined as:\n \n >>> cons = ({'type': 'ineq', 'fun': lambda x: x[0] - 2 * x[1] + 2},\n ... {'type': 'ineq', 'fun': lambda x: -x[0] - 2 * x[1] + 6},\n ... {'type': 'ineq', 'fun': lambda x: -x[0] + 2 * x[1] + 2})\n \n And variables must be positive, hence the following bounds:\n \n >>> bnds = ((0, None), (0, None))\n \n The optimization problem is solved using the SLSQP method as:\n \n >>> res = minimize(fun, (2, 0), method='SLSQP', bounds=bnds,\n ... constraints=cons)\n \n It should converge to the theoretical solution (1.4 ,1.7).\n\n"
],
[
"# Función objetivo\ndef var2(w,s1,s2,s12):\n return (w*s1)**2+((1-w)*s2)**2+2*w*(1-w)*s12",
"_____no_output_____"
],
[
"# Dato inicial\nw0 = 0\n# Volatilidades individuales y covarianza\ns1 = annual_ret_summ['EU']['Volatilidad']\ns2 = annual_ret_summ['Japon']['Volatilidad']\ns12 = corr['EU']['Japon']*s1*s2\n# Cota de w\nbnd = (0, 1)",
"_____no_output_____"
],
[
"# Solución\nmin_var2 = opt.minimize(var2, w0, args=(s1,s2,s12), bounds=(bnd,))\nmin_var2",
"_____no_output_____"
],
[
"# Graficar el portafolio de varianza mínima\n# sobre el mismo gráfico realizado anteriormente\nplt.figure(figsize=(8,6))\nplt.plot(s1, E1, 'ro', ms = 10, label='EU')\nplt.plot(s2, E2, 'bo', ms = 10, label='Japón')\nplt.plot(portafolios2.Vol, portafolios2.Rend, 'k-', lw = 4, label='Portafolios')\nplt.plot(np.sqrt(min_var2.fun), min_var2.x*E1+(1-min_var2.x)*E2, '*m', ms=10, label='Port. Min. Var.')\n#plt.plot(np.sqrt(var2(min_var2.x,s1,s2,s12)), min_var2.x*E1+(1-min_var2.x)*E2, '*m', ms=10, label='Port. Min. Var.')\nplt.xlabel('Volatilidad ($\\sigma$)')\nplt.ylabel('Rendimiento esperado ($E[r]$)')\nplt.legend(loc='best')\nplt.grid()\nplt.show()",
"_____no_output_____"
]
],
[
[
"___",
"_____no_output_____"
],
[
"## 3. Ahora, para tres activos, obtengamos la frontera de mínima varianza",
"_____no_output_____"
]
],
[
[
"import scipy.optimize as opt",
"_____no_output_____"
],
[
"## Construcción de parámetros\n## Activo 1: EU, Activo 2: Japon, Activo 3: RU\n# 1. Sigma: matriz de varianza-covarianza\ns1 = annual_ret_summ['EU']['Volatilidad']\ns2 = annual_ret_summ['Japon']['Volatilidad']\ns3 = annual_ret_summ['RU']['Volatilidad']\ns12 = corr['EU']['Japon']*s1*s2\ns13 = corr['EU']['RU']*s1*s3\ns23 = corr['Japon']['RU']*s2*s3\nSigma = np.array([[s1**2, s12, s13],\n [s12, s2**2, s23],\n [s13, s23, s3**2]])\n# 2. Eind: rendimientos esperados activos individuales\nE1 = annual_ret_summ['EU']['Media']\nE2 = annual_ret_summ['Japon']['Media']\nE3 = annual_ret_summ['RU']['Media']\nEind = np.array([E1, E2, E3])\n# 3. Ereq: rendimientos requeridos para el portafolio\n# Número de portafolios\nN = 100\nEreq = np.linspace(Eind.min(), Eind.max(), N)",
"_____no_output_____"
],
[
"def varianza(w, Sigma):\n return w.dot(Sigma).dot(w)\ndef rendimiento_req(w, Eind, Ereq):\n return Eind.dot(w)-Ereq",
"_____no_output_____"
],
[
"# Dato inicial\nw0 = np.zeros(3,)\n# Cotas de las variables\nbnds = ((0,None), (0,None), (0,None))",
"_____no_output_____"
],
[
"# DataFrame de portafolios de la frontera\nportfolios3 = pd.DataFrame(index=range(N), columns=['w1', 'w2', 'w3', 'Ret', 'Vol'])\n\n# Construcción de los N portafolios de la frontera\nfor i in range(N):\n # Restricciones\n cons = ({'type': 'eq', 'fun': rendimiento_req, 'args': (Eind,Ereq[i])},\n {'type': 'eq', 'fun': lambda w: np.sum(w)-1})\n # Portafolio de mínima varianza para nivel de rendimiento esperado Ereq[i]\n min_var = opt.minimize(varianza, w0, args=(Sigma,), bounds=bnds, constraints=cons)\n # Pesos, rendimientos y volatilidades de los portafolio\n portfolios3.loc[i,['w1','w2','w3']] = min_var.x\n portfolios3['Ret'][i] = rendimiento_req(min_var.x, Eind, Ereq[i])+Ereq[i]\n portfolios3['Vol'][i] = np.sqrt(varianza(min_var.x, Sigma))",
"_____no_output_____"
],
[
"# Portafolios de la frontera\nportfolios3",
"_____no_output_____"
],
[
"# Portafolio de mínima varianza\ncons = ({'type': 'eq', 'fun': lambda w: np.sum(w)-1},)\nmin_var3 = opt.minimize(varianza, w0, args=(Sigma,), bounds=bnds, constraints=cons)\nmin_var3",
"_____no_output_____"
],
[
"# Graficamos junto a los portafolios de solo EU y Japón\nplt.figure(figsize=(8,6))\nplt.plot(s1,E1,'ro',ms=5,label='EU')\nplt.plot(s2,E2,'bo',ms=5,label='Japón')\nplt.plot(s3,E3,'co',ms=5,label='RU')\nplt.plot(portfolios3.Vol, portfolios3.Ret, 'k-', lw=2, label='Portafolios 3 act')\nplt.plot(portafolios2.Vol, portafolios2.Rend, 'g-', lw = 2, label='Portafolios 2 act')\nplt.plot(np.sqrt(min_var2.fun), min_var2.x*E1+(1-min_var2.x)*E2, '*m', ms=10, label='Port. Min. Var.2')\nplt.plot(np.sqrt(min_var3.fun), Eind.dot(min_var3.x), '*y', ms=10, label='Port. Min. Var.3')\nplt.xlabel('Volatilidad ($\\sigma$)')\nplt.ylabel('Rendimiento esperado ($E[r]$)')\nplt.grid()\nplt.axis([0.14,0.15,0.135,0.142])\nplt.legend(loc='best')",
"_____no_output_____"
]
],
[
[
"**Conclusión.** Mayor diversificación.\n___",
"_____no_output_____"
],
[
"## 4. Comentarios acerca de la Teoría Moderna de Portafolios.\n\n- Todo lo anterior es un abrebocas de lo que llamamos análisis de media-varianza, y que es la base de la teoría moderna de portafolios.\n- El análisis de media-varianza transformó el mundo de las inversiones cuando fué presentada por primera vez.\n- Claro, tiene ciertas limitaciones, pero se mantiene como una de las ideas principales en la selección óptima de portafolios.",
"_____no_output_____"
],
[
"### Historia.\n\n1. Fue presentada por primera vez por Harry Markowitz en 1950. Acá su [artículo](https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=1&ved=2ahUKEwjd0cOTx8XdAhUVo4MKHcLoBhcQFjAAegQICBAC&url=https%3A%2F%2Fwww.math.ust.hk%2F~maykwok%2Fcourses%2Fma362%2F07F%2Fmarkowitz_JF.pdf&usg=AOvVaw3d29hQoNJVqXvC8zPuixYG).\n2. Era un joven estudiante de Doctorado (como yo) en la Universidad de Chicago.\n3. Publicó su tesis doctoral en selección de portafolios en \"Journal of Finance\" en 1952.\n4. Su contribución transformó por completo la forma en la que entendemos el riesgo.\n5. Básicamente obtuvo una teoría que analiza como los inversionistas deberían escoger de manera óptima sus portafolios, en otras palabras, cómo distribuir la riqueza de manera óptima en diferentes activos.\n6. Casi 40 años después, Markowitz ganó el Premio Nobel en economía por esta idea.",
"_____no_output_____"
],
[
"- La suposición detrás del análisis media-varianza es que los rendimientos de los activos pueden ser caracterizados por completo por sus rendimientos esperados y volatilidad.\n- Por eso es que graficamos activos y sus combinaciones (portafolios) en el espacio de rendimiento esperado contra volatilidad.\n- El análisis media-varianza es básicamente acerca de la diversificación: la interacción de activos permite que las ganancias de unos compensen las pérdidas de otros.\n- La diversificación reduce el riesgo total mientras combinemos activos imperfectamente correlacionados. ",
"_____no_output_____"
],
[
"- En el siguiente módulo revisaremos cómo elegir portafolios óptimos como si los inversionistas sólo se preocuparan por medias y varianzas.\n- ¿Qué pasa si un inversionista también se preocupa por otros momentos (asimetría, curtosis...)?\n- La belleza del análisis media-varianza es que cuando combinamos activos correlacionados imperfectamente, las varianzas siempre decrecen (no sabemos que pasa con otras medidas de riesgo).\n- Si a un inversionista le preocupan otras medidas de riesgo, el análisis media-varianza no es el camino.",
"_____no_output_____"
],
[
"- Además, si eres una persona que le gusta el riesgo: quieres encontrar la próxima compañía top que apenas va arrancando (como Google en los 2000) e invertir todo en ella para generar ganancias extraordinarias; entonces la diversificación no es tampoco el camino.\n- La diversificación, por definición, elimina el riesgo idiosincrático (de cada compañía), y por tanto elimina estos rendimientos altísimos que brindaría un portafolio altamente concentrado.",
"_____no_output_____"
],
[
"# Anuncios parroquiales\n\n## 1. Tarea 4 y Tarea 3 - segunda entrega: varios no entregaron. Para ponernos de acuerdo en este punto, de ahora en adelante: si una de las entregas no se hace, la tarea tiene calificación de 0. Para estos dos casos recientes, como no nos habíamos puesto de acuerdo, abro las entregas de ambas tareas hasta las 2 pm de hoy (supondré, de buena fe, que olvidaron entregarlas y les doy una hora después de la clase para que entreguen).\n## 2. Examen módulos 1 y 2 para el miércoles 3 de Octubre.\n## 3. Recordar quiz la próxima clase.\n## 4. Revisar archivo de la Tarea 5.\n## 5. Las próximas dos clases son de repaso, sin embargo, el repaso no lo hago yo, lo hacen ustedes. Estaremos resolviendo todo tipo de dudas que ustedes planteen acerca de lo visto hasta ahora. Si no hay dudas, dedicarán el tiempo de la clase a tareas del curso.\n## 6. Tarea 4: tengo la primera entrega calificada para hoy. Segunda entrega para el lunes.\n## 7. Fin Módulo 2: revisar Clase0 para ver objetivos.",
"_____no_output_____"
],
[
"<script>\n $(document).ready(function(){\n $('div.prompt').hide();\n $('div.back-to-top').hide();\n $('nav#menubar').hide();\n $('.breadcrumb').hide();\n $('.hidden-print').hide();\n });\n</script>\n\n<footer id=\"attribution\" style=\"float:right; color:#808080; background:#fff;\">\nCreated with Jupyter by Esteban Jiménez Rodríguez.\n</footer>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec6ff9d805d919ffb6ad7b0fdcce2075b953b000 | 64,453 | ipynb | Jupyter Notebook | D78_訓練神經網路前的注意事項/Day078_CheckBeforeTrain.ipynb | jacky0405/100Days_ML_Marathon | c9eed3445e59d0df3ea50c7f819ac266491afe89 | [
"MIT"
]
| 5 | 2020-11-03T04:31:55.000Z | 2022-03-02T16:23:28.000Z | D78_訓練神經網路前的注意事項/Day078_CheckBeforeTrain.ipynb | jacky0405/100Days_ML_Marathon | c9eed3445e59d0df3ea50c7f819ac266491afe89 | [
"MIT"
]
| null | null | null | D78_訓練神經網路前的注意事項/Day078_CheckBeforeTrain.ipynb | jacky0405/100Days_ML_Marathon | c9eed3445e59d0df3ea50c7f819ac266491afe89 | [
"MIT"
]
| 2 | 2021-02-01T03:34:48.000Z | 2021-04-12T14:14:59.000Z | 131.002033 | 21,116 | 0.788885 | [
[
[
"## 範例重點\n### 學習在模型開始前檢查各個環節\n1. 是否有 GPU 資源\n2. 將前處理轉為函式,統一處理訓練、驗證與測試集\n3. 將超參數變數化,易於重複使用函式、模型等",
"_____no_output_____"
]
],
[
[
"## 確認硬體資源 (如果你是在 Linux, 若是在 Windows, 請參考 https://blog.csdn.net/idwtwt/article/details/78017565)\n!nvidia-smi",
"zsh:1: command not found: nvidia-smi\r\n"
],
[
"import os\nimport keras\n\n# 本範例不需使用 GPU, 將 GPU 設定為 \"無\"\nos.environ[\"CUDA_VISIBLE_DEVICES\"] = \"\"",
"_____no_output_____"
],
[
"# 從 Keras 的內建功能中,取得 train 與 test 資料集\ntrain, test = keras.datasets.cifar10.load_data()",
"_____no_output_____"
],
[
"## 資料前處理\ndef preproc_x(x, flatten=True):\n x = x / 255.\n if flatten:\n x = x.reshape((len(x), -1))\n return x\n\ndef preproc_y(y, num_classes=10):\n if y.shape[-1] == 1:\n y = keras.utils.to_categorical(y, num_classes)\n return y ",
"_____no_output_____"
],
[
"x_train, y_train = train\nx_test, y_test = test\n\n# 資料前處理 - X 標準化\nx_train = preproc_x(x_train)\nx_test = preproc_x(x_test)\n\n# 資料前處理 -Y 轉成 onehot\ny_train = preproc_y(y_train)\ny_test = preproc_y(y_test)",
"_____no_output_____"
],
[
"def build_mlp(input_shape, output_units=10, num_neurons=[512, 256, 128]):\n input_layer = keras.layers.Input(input_shape)\n \n for i, n_units in enumerate(num_neurons):\n if i == 0:\n x = keras.layers.Dense(units=n_units, activation=\"relu\", name=\"hidden_layer\"+str(i+1))(input_layer)\n else:\n x = keras.layers.Dense(units=n_units, activation=\"relu\", name=\"hidden_layer\"+str(i+1))(x)\n \n out = keras.layers.Dense(units=output_units, activation=\"softmax\", name=\"output\")(x)\n \n model = keras.models.Model(inputs=[input_layer], outputs=[out])\n return model",
"_____no_output_____"
],
[
"model = build_mlp(input_shape=x_train.shape[1:])\nmodel.summary()",
"Model: \"functional_1\"\n_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\ninput_1 (InputLayer) [(None, 3072)] 0 \n_________________________________________________________________\nhidden_layer1 (Dense) (None, 512) 1573376 \n_________________________________________________________________\nhidden_layer2 (Dense) (None, 256) 131328 \n_________________________________________________________________\nhidden_layer3 (Dense) (None, 128) 32896 \n_________________________________________________________________\noutput (Dense) (None, 10) 1290 \n=================================================================\nTotal params: 1,738,890\nTrainable params: 1,738,890\nNon-trainable params: 0\n_________________________________________________________________\n"
],
[
"## 超參數設定\nLEARNING_RATE = 0.001\nEPOCHS = 100\nBATCH_SIZE = 256",
"_____no_output_____"
],
[
"optimizer = keras.optimizers.Adam(lr=LEARNING_RATE)\nmodel.compile(loss=\"categorical_crossentropy\", metrics=[\"accuracy\"], optimizer=optimizer)",
"_____no_output_____"
],
[
"model.fit(x_train, y_train, \n epochs=EPOCHS, \n batch_size=BATCH_SIZE, \n validation_data=(x_test, y_test), \n shuffle=True)",
"Epoch 1/100\n196/196 [==============================] - 6s 28ms/step - loss: 1.9682 - accuracy: 0.2882 - val_loss: 1.8033 - val_accuracy: 0.3507\nEpoch 2/100\n196/196 [==============================] - 6s 32ms/step - loss: 1.7312 - accuracy: 0.3814 - val_loss: 1.6695 - val_accuracy: 0.4100\nEpoch 3/100\n196/196 [==============================] - 5s 27ms/step - loss: 1.6285 - accuracy: 0.4184 - val_loss: 1.6066 - val_accuracy: 0.4336\nEpoch 4/100\n196/196 [==============================] - 7s 33ms/step - loss: 1.5677 - accuracy: 0.4419 - val_loss: 1.5687 - val_accuracy: 0.4400\nEpoch 5/100\n196/196 [==============================] - 7s 36ms/step - loss: 1.5163 - accuracy: 0.4586 - val_loss: 1.5166 - val_accuracy: 0.4583\nEpoch 6/100\n196/196 [==============================] - 7s 37ms/step - loss: 1.4738 - accuracy: 0.4741 - val_loss: 1.4887 - val_accuracy: 0.4671\nEpoch 7/100\n196/196 [==============================] - 7s 36ms/step - loss: 1.4429 - accuracy: 0.4853 - val_loss: 1.4759 - val_accuracy: 0.4755\nEpoch 8/100\n196/196 [==============================] - 7s 34ms/step - loss: 1.4138 - accuracy: 0.4961 - val_loss: 1.4840 - val_accuracy: 0.4750\nEpoch 9/100\n196/196 [==============================] - 7s 34ms/step - loss: 1.3822 - accuracy: 0.5051 - val_loss: 1.4592 - val_accuracy: 0.4788\nEpoch 10/100\n196/196 [==============================] - 9s 44ms/step - loss: 1.3575 - accuracy: 0.5171 - val_loss: 1.4072 - val_accuracy: 0.5012\nEpoch 11/100\n196/196 [==============================] - 9s 44ms/step - loss: 1.3259 - accuracy: 0.5267 - val_loss: 1.4027 - val_accuracy: 0.5063\nEpoch 12/100\n196/196 [==============================] - 6s 30ms/step - loss: 1.3102 - accuracy: 0.5312 - val_loss: 1.4147 - val_accuracy: 0.4963\nEpoch 13/100\n196/196 [==============================] - 5s 24ms/step - loss: 1.2890 - accuracy: 0.5408 - val_loss: 1.4088 - val_accuracy: 0.4988\nEpoch 14/100\n196/196 [==============================] - 6s 31ms/step - loss: 1.2592 - accuracy: 0.5529 - val_loss: 1.3940 - val_accuracy: 0.5076\nEpoch 15/100\n196/196 [==============================] - 5s 24ms/step - loss: 1.2284 - accuracy: 0.5618 - val_loss: 1.3624 - val_accuracy: 0.5162\nEpoch 16/100\n196/196 [==============================] - 5s 26ms/step - loss: 1.2171 - accuracy: 0.5656 - val_loss: 1.4210 - val_accuracy: 0.4958\nEpoch 17/100\n196/196 [==============================] - 6s 30ms/step - loss: 1.1905 - accuracy: 0.5749 - val_loss: 1.3718 - val_accuracy: 0.5192\nEpoch 18/100\n196/196 [==============================] - 6s 29ms/step - loss: 1.1698 - accuracy: 0.5823 - val_loss: 1.3560 - val_accuracy: 0.5234\nEpoch 19/100\n196/196 [==============================] - 6s 28ms/step - loss: 1.1573 - accuracy: 0.5869 - val_loss: 1.3495 - val_accuracy: 0.5243\nEpoch 20/100\n196/196 [==============================] - 5s 28ms/step - loss: 1.1329 - accuracy: 0.5953 - val_loss: 1.3608 - val_accuracy: 0.5263\nEpoch 21/100\n196/196 [==============================] - 5s 28ms/step - loss: 1.1187 - accuracy: 0.6006 - val_loss: 1.3548 - val_accuracy: 0.5256\nEpoch 22/100\n196/196 [==============================] - 6s 30ms/step - loss: 1.0965 - accuracy: 0.6084 - val_loss: 1.3822 - val_accuracy: 0.5169\nEpoch 23/100\n196/196 [==============================] - 6s 33ms/step - loss: 1.0818 - accuracy: 0.6132 - val_loss: 1.3877 - val_accuracy: 0.5232\nEpoch 24/100\n196/196 [==============================] - 5s 27ms/step - loss: 1.0441 - accuracy: 0.6282 - val_loss: 1.4206 - val_accuracy: 0.5174\nEpoch 25/100\n196/196 [==============================] - 5s 27ms/step - loss: 1.0267 - accuracy: 0.6320 - val_loss: 1.4641 - val_accuracy: 0.5144\nEpoch 26/100\n196/196 [==============================] - 6s 28ms/step - loss: 1.0126 - accuracy: 0.6402 - val_loss: 1.3801 - val_accuracy: 0.5279\nEpoch 27/100\n196/196 [==============================] - 5s 26ms/step - loss: 0.9896 - accuracy: 0.6465 - val_loss: 1.3987 - val_accuracy: 0.5301\nEpoch 28/100\n196/196 [==============================] - 6s 31ms/step - loss: 0.9747 - accuracy: 0.6517 - val_loss: 1.4551 - val_accuracy: 0.5222\nEpoch 29/100\n196/196 [==============================] - 6s 29ms/step - loss: 0.9501 - accuracy: 0.6603 - val_loss: 1.4146 - val_accuracy: 0.5301\nEpoch 30/100\n196/196 [==============================] - 5s 26ms/step - loss: 0.9309 - accuracy: 0.6669 - val_loss: 1.4840 - val_accuracy: 0.5156\nEpoch 31/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.9170 - accuracy: 0.6714 - val_loss: 1.4525 - val_accuracy: 0.5234\nEpoch 32/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.8980 - accuracy: 0.6773 - val_loss: 1.4757 - val_accuracy: 0.5209\nEpoch 33/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.8882 - accuracy: 0.6817 - val_loss: 1.4710 - val_accuracy: 0.5280\nEpoch 34/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.8685 - accuracy: 0.6906 - val_loss: 1.4792 - val_accuracy: 0.5244\nEpoch 35/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.8445 - accuracy: 0.6983 - val_loss: 1.5495 - val_accuracy: 0.5191\nEpoch 36/100\n196/196 [==============================] - 6s 31ms/step - loss: 0.8287 - accuracy: 0.7031 - val_loss: 1.5257 - val_accuracy: 0.5279\nEpoch 37/100\n196/196 [==============================] - 5s 28ms/step - loss: 0.8133 - accuracy: 0.7083 - val_loss: 1.5771 - val_accuracy: 0.5158\nEpoch 38/100\n196/196 [==============================] - 6s 31ms/step - loss: 0.8001 - accuracy: 0.7127 - val_loss: 1.5526 - val_accuracy: 0.5211\nEpoch 39/100\n196/196 [==============================] - 5s 25ms/step - loss: 0.7892 - accuracy: 0.7184 - val_loss: 1.5938 - val_accuracy: 0.5162\nEpoch 40/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.7767 - accuracy: 0.7218 - val_loss: 1.6258 - val_accuracy: 0.5104\nEpoch 41/100\n196/196 [==============================] - 4s 23ms/step - loss: 0.7620 - accuracy: 0.7273 - val_loss: 1.5743 - val_accuracy: 0.5212\nEpoch 42/100\n196/196 [==============================] - 4s 22ms/step - loss: 0.7428 - accuracy: 0.7338 - val_loss: 1.6199 - val_accuracy: 0.5246\nEpoch 43/100\n196/196 [==============================] - 6s 29ms/step - loss: 0.7144 - accuracy: 0.7431 - val_loss: 1.6769 - val_accuracy: 0.5197\nEpoch 44/100\n196/196 [==============================] - 4s 23ms/step - loss: 0.7088 - accuracy: 0.7446 - val_loss: 1.7094 - val_accuracy: 0.5160\nEpoch 45/100\n196/196 [==============================] - 4s 23ms/step - loss: 0.6974 - accuracy: 0.7492 - val_loss: 1.6847 - val_accuracy: 0.5186\nEpoch 46/100\n196/196 [==============================] - 5s 24ms/step - loss: 0.6865 - accuracy: 0.7524 - val_loss: 1.7119 - val_accuracy: 0.5243\nEpoch 47/100\n196/196 [==============================] - 4s 23ms/step - loss: 0.6547 - accuracy: 0.7661 - val_loss: 1.7746 - val_accuracy: 0.5111\nEpoch 48/100\n196/196 [==============================] - 5s 26ms/step - loss: 0.6506 - accuracy: 0.7671 - val_loss: 1.8001 - val_accuracy: 0.5106\nEpoch 49/100\n196/196 [==============================] - 5s 24ms/step - loss: 0.6445 - accuracy: 0.7709 - val_loss: 1.7851 - val_accuracy: 0.5164\nEpoch 50/100\n196/196 [==============================] - 4s 23ms/step - loss: 0.6334 - accuracy: 0.7721 - val_loss: 1.8368 - val_accuracy: 0.5044\nEpoch 51/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.6278 - accuracy: 0.7761 - val_loss: 1.8405 - val_accuracy: 0.5098\nEpoch 52/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.6001 - accuracy: 0.7848 - val_loss: 1.8433 - val_accuracy: 0.5206\nEpoch 53/100\n196/196 [==============================] - 6s 32ms/step - loss: 0.5748 - accuracy: 0.7933 - val_loss: 2.1242 - val_accuracy: 0.4989\nEpoch 54/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.5934 - accuracy: 0.7873 - val_loss: 1.9603 - val_accuracy: 0.5150\nEpoch 55/100\n196/196 [==============================] - 5s 23ms/step - loss: 0.5721 - accuracy: 0.7941 - val_loss: 2.0021 - val_accuracy: 0.5159\nEpoch 56/100\n196/196 [==============================] - 5s 25ms/step - loss: 0.5469 - accuracy: 0.8046 - val_loss: 1.9911 - val_accuracy: 0.5117\nEpoch 57/100\n"
],
[
"# 訓練模型並檢視驗證集的結果\nimport matplotlib.pyplot as plt\n\ntrain_loss = model.history.history[\"loss\"]\nvalid_loss = model.history.history[\"val_loss\"]\n\ntrain_acc = model.history.history[\"accuracy\"]\nvalid_acc = model.history.history[\"val_accuracy\"]\n\nplt.plot(range(len(train_loss)), train_loss, label=\"train loss\")\nplt.plot(range(len(valid_loss)), valid_loss, label=\"valid loss\")\nplt.legend()\nplt.title(\"Loss\")\nplt.show()\n\nplt.plot(range(len(train_acc)), train_acc, label=\"train accuracy\")\nplt.plot(range(len(valid_acc)), valid_acc, label=\"valid accuracy\")\nplt.legend()\nplt.title(\"Accuracy\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Work\n1. 請嘗試將 preproc_x 替換成以每筆資料的 min/max 進行標準化至 -1 ~ 1 間,再進行訓練\n2. 請嘗試將 mlp 疊更深 (e.g 5~10 層),進行訓練後觀察 learning curve 的走勢\n3. (optional) 請改用 GPU 進行訓練 (如果你有 GPU 的話),比較使用 CPU 與 GPU 的訓練速度",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec6ffc58f10c1afa1b7a5e04d4d9b4dfe57b40db | 1,172 | ipynb | Jupyter Notebook | Java.ipynb | Frank17/Java | 940d80a7ed656e00438bf8a45dc72e72c45164a4 | [
"MIT"
]
| null | null | null | Java.ipynb | Frank17/Java | 940d80a7ed656e00438bf8a45dc72e72c45164a4 | [
"MIT"
]
| null | null | null | Java.ipynb | Frank17/Java | 940d80a7ed656e00438bf8a45dc72e72c45164a4 | [
"MIT"
]
| null | null | null | 22.113208 | 213 | 0.450512 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec6ffd3205dfacadffd6229845b3851d359d1aca | 52,178 | ipynb | Jupyter Notebook | site/en/guide/distributed_training.ipynb | coderkodor/docs | adae1941b3a1b3c0b781b94d96f4476892a44620 | [
"Apache-2.0"
]
| 3 | 2020-07-28T20:42:26.000Z | 2020-08-15T03:29:12.000Z | site/en/guide/distributed_training.ipynb | coderkodor/docs | adae1941b3a1b3c0b781b94d96f4476892a44620 | [
"Apache-2.0"
]
| null | null | null | site/en/guide/distributed_training.ipynb | coderkodor/docs | adae1941b3a1b3c0b781b94d96f4476892a44620 | [
"Apache-2.0"
]
| 1 | 2020-08-03T20:17:42.000Z | 2020-08-03T20:17:42.000Z | 48.447539 | 822 | 0.614359 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.\n",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Distributed training with TensorFlow",
"_____no_output_____"
],
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/guide/distributed_training\"><img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/distributed_training.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/en/guide/distributed_training.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n <td>\n <a href=\"https://storage.googleapis.com/tensorflow_docs/docs/site/en/guide/distributed_training.ipynb\"><img src=\"https://www.tensorflow.org/images/download_logo_32px.png\" />Download notebook</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Overview\n\n`tf.distribute.Strategy` is a TensorFlow API to distribute training \nacross multiple GPUs, multiple machines or TPUs. Using this API, you can distribute your existing models and training code with minimal code changes.\n\n`tf.distribute.Strategy` has been designed with these key goals in mind:\n\n* Easy to use and support multiple user segments, including researchers, ML engineers, etc.\n* Provide good performance out of the box.\n* Easy switching between strategies.\n\n`tf.distribute.Strategy` can be used with a high-level API like [Keras](https://www.tensorflow.org/guide/keras), and can also be used to distribute custom training loops (and, in general, any computation using TensorFlow).\n\nIn TensorFlow 2.x, you can execute your programs eagerly, or in a graph using [`tf.function`](function.ipynb). `tf.distribute.Strategy` intends to support both these modes of execution, but works best with `tf.function`. Eager mode is only recommended for debugging purpose and not supported for `TPUStrategy`. Although we discuss training most of the time in this guide, this API can also be used for distributing evaluation and prediction on different platforms.\n\nYou can use `tf.distribute.Strategy` with very few changes to your code, because we have changed the underlying components of TensorFlow to become strategy-aware. This includes variables, layers, models, optimizers, metrics, summaries, and checkpoints.\n\nIn this guide, we explain various types of strategies and how you can use them in different situations.\n\nNote: For a deeper understanding of the concepts, please watch [this deep-dive presentation](https://youtu.be/jKV53r9-H14). This is especially recommended if you plan to write your own training loop.\n",
"_____no_output_____"
]
],
[
[
"# Import TensorFlow\nimport tensorflow as tf",
"_____no_output_____"
]
],
[
[
"## Types of strategies\n`tf.distribute.Strategy` intends to cover a number of use cases along different axes. Some of these combinations are currently supported and others will be added in the future. Some of these axes are:\n\n* *Synchronous vs asynchronous training:* These are two common ways of distributing training with data parallelism. In sync training, all workers train over different slices of input data in sync, and aggregating gradients at each step. In async training, all workers are independently training over the input data and updating variables asynchronously. Typically sync training is supported via all-reduce and async through parameter server architecture.\n* *Hardware platform:* You may want to scale your training onto multiple GPUs on one machine, or multiple machines in a network (with 0 or more GPUs each), or on Cloud TPUs.\n\nIn order to support these use cases, there are six strategies available. In the next section we explain which of these are supported in which scenarios in TF 2.2 at this time. Here is a quick overview:\n\n| Training API \t| MirroredStrategy \t| TPUStrategy \t| MultiWorkerMirroredStrategy \t| CentralStorageStrategy \t| ParameterServerStrategy \t|\n|:-----------------------\t|:-------------------\t|:---------------------\t|:---------------------------------\t|:---------------------------------\t|:--------------------------\t|\n| **Keras API** \t| Supported \t| Supported \t| Experimental support \t| Experimental support \t| Supported planned post 2.3 \t|\n| **Custom training loop** \t| Supported \t| Supported \t| Experimental support \t| Experimental support \t| Supported planned post 2.3 \t|\n| **Estimator API** \t| Limited Support \t| Not supported \t| Limited Support \t| Limited Support \t| Limited Support \t|\n\nNote: [Experimental support](https://www.tensorflow.org/guide/versions#what_is_not_covered) means the APIs are not covered by any compatibilities guarantees.\n\nNote: Estimator support is limited. Basic training and evaluation are experimental, and advanced features—such as scaffold—are not implemented. We recommend using Keras or custom training loops if a use case is not covered.",
"_____no_output_____"
],
[
"### MirroredStrategy\n`tf.distribute.MirroredStrategy` supports synchronous distributed training on multiple GPUs on one machine. It creates one replica per GPU device. Each variable in the model is mirrored across all the replicas. Together, these variables form a single conceptual variable called `MirroredVariable`. These variables are kept in sync with each other by applying identical updates.\n\nEfficient all-reduce algorithms are used to communicate the variable updates across the devices.\nAll-reduce aggregates tensors across all the devices by adding them up, and makes them available on each device.\nIt’s a fused algorithm that is very efficient and can reduce the overhead of synchronization significantly. There are many all-reduce algorithms and implementations available, depending on the type of communication available between devices. By default, it uses NVIDIA NCCL as the all-reduce implementation. You can choose from a few other options we provide, or write your own.\n\nHere is the simplest way of creating `MirroredStrategy`:\n",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy()",
"_____no_output_____"
]
],
[
[
"This will create a `MirroredStrategy` instance which will use all the GPUs that are visible to TensorFlow, and use NCCL as the cross device communication.\n\nIf you wish to use only some of the GPUs on your machine, you can do so like this:",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy(devices=[\"/gpu:0\", \"/gpu:1\"])",
"_____no_output_____"
]
],
[
[
"If you wish to override the cross device communication, you can do so using the `cross_device_ops` argument by supplying an instance of `tf.distribute.CrossDeviceOps`. Currently, `tf.distribute.HierarchicalCopyAllReduce` and `tf.distribute.ReductionToOneDevice` are two options other than `tf.distribute.NcclAllReduce` which is the default.",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy(\n cross_device_ops=tf.distribute.HierarchicalCopyAllReduce())",
"_____no_output_____"
]
],
[
[
"### TPUStrategy\n`tf.distribute.experimental.TPUStrategy` lets you run your TensorFlow training on Tensor Processing Units (TPUs). TPUs are Google's specialized ASICs designed to dramatically accelerate machine learning workloads. They are available on Google Colab, the [TensorFlow Research Cloud](https://www.tensorflow.org/tfrc) and [Cloud TPU](https://cloud.google.com/tpu).\n\nIn terms of distributed training architecture, `TPUStrategy` is the same `MirroredStrategy` - it implements synchronous distributed training. TPUs provide their own implementation of efficient all-reduce and other collective operations across multiple TPU cores, which are used in `TPUStrategy`.\n\nHere is how you would instantiate `TPUStrategy`:\n\nNote: To run this code in Colab, you should select TPU as the Colab runtime. See [TensorFlow TPU Guide](https://www.tensorflow.org/guide/tpu).\n\n```\ncluster_resolver = tf.distribute.cluster_resolver.TPUClusterResolver(\n tpu=tpu_address)\ntf.config.experimental_connect_to_cluster(cluster_resolver)\ntf.tpu.experimental.initialize_tpu_system(cluster_resolver)\ntpu_strategy = tf.distribute.experimental.TPUStrategy(cluster_resolver)\n```\n\n\nThe `TPUClusterResolver` instance helps locate the TPUs. In Colab, you don't need to specify any arguments to it.\n\nIf you want to use this for Cloud TPUs:\n- You must specify the name of your TPU resource in the `tpu` argument.\n- You must initialize the tpu system explicitly at the *start* of the program. This is required before TPUs can be used for computation. Initializing the tpu system also wipes out the TPU memory, so it's important to complete this step first in order to avoid losing state.",
"_____no_output_____"
],
[
"### MultiWorkerMirroredStrategy\n\n`tf.distribute.experimental.MultiWorkerMirroredStrategy` is very similar to `MirroredStrategy`. It implements synchronous distributed training across multiple workers, each with potentially multiple GPUs. Similar to `MirroredStrategy`, it creates copies of all variables in the model on each device across all workers.\n\nIt uses [CollectiveOps](https://github.com/tensorflow/tensorflow/blob/master/tensorflow/python/ops/collective_ops.py) as the multi-worker all-reduce communication method used to keep variables in sync. A collective op is a single op in the TensorFlow graph which can automatically choose an all-reduce algorithm in the TensorFlow runtime according to hardware, network topology and tensor sizes.\n\nIt also implements additional performance optimizations. For example, it includes a static optimization that converts multiple all-reductions on small tensors into fewer all-reductions on larger tensors. In addition, we are designing it to have a plugin architecture - so that in the future, you will be able to plugin algorithms that are better tuned for your hardware. Note that collective ops also implement other collective operations such as broadcast and all-gather.\n\nHere is the simplest way of creating `MultiWorkerMirroredStrategy`:",
"_____no_output_____"
]
],
[
[
"multiworker_strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy()",
"_____no_output_____"
]
],
[
[
"`MultiWorkerMirroredStrategy` currently allows you to choose between two different implementations of collective ops. `CollectiveCommunication.RING` implements ring-based collectives using gRPC as the communication layer. `CollectiveCommunication.NCCL` uses [Nvidia's NCCL](https://developer.nvidia.com/nccl) to implement collectives. `CollectiveCommunication.AUTO` defers the choice to the runtime. The best choice of collective implementation depends upon the number and kind of GPUs, and the network interconnect in the cluster. You can specify them in the following way:\n",
"_____no_output_____"
]
],
[
[
"multiworker_strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy(\n tf.distribute.experimental.CollectiveCommunication.NCCL)",
"_____no_output_____"
]
],
[
[
"One of the key differences to get multi worker training going, as compared to multi-GPU training, is the multi-worker setup. The `TF_CONFIG` environment variable is the standard way in TensorFlow to specify the cluster configuration to each worker that is part of the cluster. Learn more about [setting up TF_CONFIG](#TF_CONFIG).",
"_____no_output_____"
],
[
"Note: This strategy is [`experimental`](https://www.tensorflow.org/guide/versions#what_is_not_covered) as we are currently improving it and making it work for more scenarios. As part of this, please expect the APIs to change in the future.",
"_____no_output_____"
],
[
"### CentralStorageStrategy\n`tf.distribute.experimental.CentralStorageStrategy` does synchronous training as well. Variables are not mirrored, instead they are placed on the CPU and operations are replicated across all local GPUs. If there is only one GPU, all variables and operations will be placed on that GPU.\n\nCreate an instance of `CentralStorageStrategy` by:\n",
"_____no_output_____"
]
],
[
[
"central_storage_strategy = tf.distribute.experimental.CentralStorageStrategy()",
"_____no_output_____"
]
],
[
[
"This will create a `CentralStorageStrategy` instance which will use all visible GPUs and CPU. Update to variables on replicas will be aggregated before being applied to variables.",
"_____no_output_____"
],
[
"Note: This strategy is [`experimental`](https://www.tensorflow.org/guide/versions#what_is_not_covered) as we are currently improving it and making it work for more scenarios. As part of this, please expect the APIs to change in the future.",
"_____no_output_____"
],
[
"### ParameterServerStrategy\n`tf.distribute.experimental.ParameterServerStrategy` supports parameter servers training on multiple machines. In this setup, some machines are designated as workers and some as parameter servers. Each variable of the model is placed on one parameter server. Computation is replicated across all GPUs of all the workers.\n\nIn terms of code, it looks similar to other strategies:\n```\nps_strategy = tf.distribute.experimental.ParameterServerStrategy()\n```",
"_____no_output_____"
],
[
"For multi worker training, `TF_CONFIG` needs to specify the configuration of parameter servers and workers in your cluster, which you can read more about in [TF_CONFIG below](#TF_CONFIG).\n\nNote: This strategy only works with the Estimator API.",
"_____no_output_____"
],
[
"### Other strategies\n\nIn addition to the above strategies, there are two other strategies which might be useful for prototyping and debugging when using `tf.distribute` APIs.",
"_____no_output_____"
],
[
"#### Default Strategy\n\nDefault strategy is a distribution strategy which is present when no explicit distribution strategy is in scope. It implements the `tf.distribute.Strategy` interface but is a pass-through and provides no actual distribution. For instance, `strategy.run(fn)` will simply call `fn`. Code written using this strategy should behave exactly as code written without any strategy. You can think of it as a \"no-op\" strategy.\n\nDefault strategy is a singleton - and one cannot create more instances of it. It can be obtained using `tf.distribute.get_strategy()` outside any explicit strategy's scope (the same API that can be used to get the current strategy inside an explicit strategy's scope). ",
"_____no_output_____"
]
],
[
[
"default_strategy = tf.distribute.get_strategy()",
"_____no_output_____"
]
],
[
[
"This strategy serves two main purposes:\n\n* It allows writing distribution aware library code unconditionally. For example, in optimizer, we can do `tf.distribute.get_strategy()` and use that strategy for reducing gradients - it will always return a strategy object on which we can call the reduce API.\n",
"_____no_output_____"
]
],
[
[
"# In optimizer or other library code\n# Get currently active strategy\nstrategy = tf.distribute.get_strategy()\nstrategy.reduce(\"SUM\", 1., axis=None) # reduce some values",
"_____no_output_____"
]
],
[
[
"* Similar to library code, it can be used to write end users' programs to work with and without distribution strategy, without requiring conditional logic. A sample code snippet illustrating this:",
"_____no_output_____"
]
],
[
[
"if tf.config.list_physical_devices('gpu'):\n strategy = tf.distribute.MirroredStrategy()\nelse: # use default strategy\n strategy = tf.distribute.get_strategy() \n\nwith strategy.scope():\n # do something interesting\n print(tf.Variable(1.))",
"_____no_output_____"
]
],
[
[
"#### OneDeviceStrategy\n`tf.distribute.OneDeviceStrategy` is a strategy to place all variables and computation on a single specified device. \n\n```\nstrategy = tf.distribute.OneDeviceStrategy(device=\"/gpu:0\")\n```\n\nThis strategy is distinct from the default strategy in a number of ways. In default strategy, the variable placement logic remains unchanged when compared to running TensorFlow without any distribution strategy. But when using `OneDeviceStrategy`, all variables created in its scope are explicitly placed on the specified device. Moreover, any functions called via `OneDeviceStrategy.run` will also be placed on the specified device. \n\nInput distributed through this strategy will be prefetched to the specified device. In default strategy, there is no input distribution.\n\nSimilar to the default strategy, this strategy could also be used to test your code before switching to other strategies which actually distribute to multiple devices/machines. This will exercise the distribution strategy machinery somewhat more than default strategy, but not to the full extent as using `MirroredStrategy` or `TPUStrategy` etc. If you want code that behaves as if no strategy, then use default strategy.",
"_____no_output_____"
],
[
"So far we've talked about what are the different strategies available and how you can instantiate them. In the next few sections, we will talk about the different ways in which you can use them to distribute your training. We will show short code snippets in this guide and link off to full tutorials which you can run end to end.",
"_____no_output_____"
],
[
"## Using `tf.distribute.Strategy` with `tf.keras.Model.fit`\nWe've integrated `tf.distribute.Strategy` into `tf.keras` which is TensorFlow's implementation of the\n[Keras API specification](https://keras.io). `tf.keras` is a high-level API to build and train models. By integrating into `tf.keras` backend, we've made it seamless for you to distribute your training written in the Keras training framework using `model.fit`.\n\nHere's what you need to change in your code:\n\n1. Create an instance of the appropriate `tf.distribute.Strategy`.\n2. Move the creation of Keras model, optimizer and metrics inside `strategy.scope`.\n\nWe support all types of Keras models - sequential, functional and subclassed.\n\nHere is a snippet of code to do this for a very simple Keras model with one dense layer:",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy()\n\nwith mirrored_strategy.scope():\n model = tf.keras.Sequential([tf.keras.layers.Dense(1, input_shape=(1,))])\n\nmodel.compile(loss='mse', optimizer='sgd')",
"_____no_output_____"
]
],
[
[
"In this example we used `MirroredStrategy` so we can run this on a machine with multiple GPUs. `strategy.scope()` indicates to Keras which strategy to use to distribute the training. Creating models/optimizers/metrics inside this scope allows us to create distributed variables instead of regular variables. Once this is set up, you can fit your model like you would normally. `MirroredStrategy` takes care of replicating the model's training on the available GPUs, aggregating gradients, and more.",
"_____no_output_____"
]
],
[
[
"dataset = tf.data.Dataset.from_tensors(([1.], [1.])).repeat(100).batch(10)\nmodel.fit(dataset, epochs=2)\nmodel.evaluate(dataset)",
"_____no_output_____"
]
],
[
[
"Here we used a `tf.data.Dataset` to provide the training and eval input. You can also use numpy arrays:",
"_____no_output_____"
]
],
[
[
"import numpy as np\ninputs, targets = np.ones((100, 1)), np.ones((100, 1))\nmodel.fit(inputs, targets, epochs=2, batch_size=10)",
"_____no_output_____"
]
],
[
[
"In both cases (dataset or numpy), each batch of the given input is divided equally among the multiple replicas. For instance, if using `MirroredStrategy` with 2 GPUs, each batch of size 10 will get divided among the 2 GPUs, with each receiving 5 input examples in each step. Each epoch will then train faster as you add more GPUs. Typically, you would want to increase your batch size as you add more accelerators so as to make effective use of the extra computing power. You will also need to re-tune your learning rate, depending on the model. You can use `strategy.num_replicas_in_sync` to get the number of replicas.",
"_____no_output_____"
]
],
[
[
"# Compute global batch size using number of replicas.\nBATCH_SIZE_PER_REPLICA = 5\nglobal_batch_size = (BATCH_SIZE_PER_REPLICA *\n mirrored_strategy.num_replicas_in_sync)\ndataset = tf.data.Dataset.from_tensors(([1.], [1.])).repeat(100)\ndataset = dataset.batch(global_batch_size)\n\nLEARNING_RATES_BY_BATCH_SIZE = {5: 0.1, 10: 0.15}\nlearning_rate = LEARNING_RATES_BY_BATCH_SIZE[global_batch_size]",
"_____no_output_____"
]
],
[
[
"### What's supported now?\n\n\n| Training API \t| MirroredStrategy \t| TPUStrategy \t| MultiWorkerMirroredStrategy \t| CentralStorageStrategy \t| ParameterServerStrategy \t|\n|----------------\t|---------------------\t|-----------------------\t|-----------------------------------\t|-----------------------------------\t|---------------------------\t|\n| Keras APIs \t| Supported \t| Supported \t| Experimental support \t| Experimental support \t| Support planned post 2.3 \t|\n\n### Examples and Tutorials\n\nHere is a list of tutorials and examples that illustrate the above integration end to end with Keras:\n\n1. [Tutorial](https://www.tensorflow.org/tutorials/distribute/keras) to train MNIST with `MirroredStrategy`.\n2. [Tutorial](https://www.tensorflow.org/tutorials/distribute/multi_worker_with_keras) to train MNIST using `MultiWorkerMirroredStrategy`.\n3. [Guide](https://www.tensorflow.org/guide/tpu#train_a_model_using_keras_high_level_apis) on training MNIST using `TPUStrategy`.\n4. TensorFlow Model Garden [repository](https://github.com/tensorflow/models/tree/master/official) containing collections of state-of-the-art models implemented using various strategies.",
"_____no_output_____"
],
[
"## Using `tf.distribute.Strategy` with custom training loops\nAs you've seen, using `tf.distribute.Strategy` with Keras `model.fit` requires changing only a couple lines of your code. With a little more effort, you can also use `tf.distribute.Strategy` with custom training loops.\n\nIf you need more flexibility and control over your training loops than is possible with Estimator or Keras, you can write custom training loops. For instance, when using a GAN, you may want to take a different number of generator or discriminator steps each round. Similarly, the high level frameworks are not very suitable for Reinforcement Learning training.\n\nTo support custom training loops, we provide a core set of methods through the `tf.distribute.Strategy` classes. Using these may require minor restructuring of the code initially, but once that is done, you should be able to switch between GPUs, TPUs, and multiple machines simply by changing the strategy instance.\n\nHere we will show a brief snippet illustrating this use case for a simple training example using the same Keras model as before.\n",
"_____no_output_____"
],
[
"First, we create the model and optimizer inside the strategy's scope. This ensures that any variables created with the model and optimizer are mirrored variables.",
"_____no_output_____"
]
],
[
[
"with mirrored_strategy.scope():\n model = tf.keras.Sequential([tf.keras.layers.Dense(1, input_shape=(1,))])\n optimizer = tf.keras.optimizers.SGD()",
"_____no_output_____"
]
],
[
[
"Next, we create the input dataset and call `tf.distribute.Strategy.experimental_distribute_dataset` to distribute the dataset based on the strategy.",
"_____no_output_____"
]
],
[
[
"dataset = tf.data.Dataset.from_tensors(([1.], [1.])).repeat(100).batch(\n global_batch_size)\ndist_dataset = mirrored_strategy.experimental_distribute_dataset(dataset)",
"_____no_output_____"
]
],
[
[
"Then, we define one step of the training. We will use `tf.GradientTape` to compute gradients and optimizer to apply those gradients to update our model's variables. To distribute this training step, we put in a function `train_step` and pass it to `tf.distrbute.Strategy.run` along with the dataset inputs that we get from `dist_dataset` created before:",
"_____no_output_____"
]
],
[
[
"loss_object = tf.keras.losses.BinaryCrossentropy(\n from_logits=True,\n reduction=tf.keras.losses.Reduction.NONE)\n\ndef compute_loss(labels, predictions):\n per_example_loss = loss_object(labels, predictions)\n return tf.nn.compute_average_loss(per_example_loss, global_batch_size=global_batch_size)\n\ndef train_step(inputs):\n features, labels = inputs\n\n with tf.GradientTape() as tape:\n predictions = model(features, training=True)\n loss = compute_loss(labels, predictions)\n\n gradients = tape.gradient(loss, model.trainable_variables)\n optimizer.apply_gradients(zip(gradients, model.trainable_variables))\n return loss\n\[email protected]\ndef distributed_train_step(dist_inputs):\n per_replica_losses = mirrored_strategy.run(train_step, args=(dist_inputs,))\n return mirrored_strategy.reduce(tf.distribute.ReduceOp.SUM, per_replica_losses,\n axis=None)",
"_____no_output_____"
]
],
[
[
"A few other things to note in the code above:\n\n1. We used `tf.nn.compute_average_loss` to compute the loss. `tf.nn.compute_average_loss` sums the per example loss and divide the sum by the global_batch_size. This is important because later after the gradients are calculated on each replica, they are aggregated across the replicas by **summing** them.\n2. We used the `tf.distribute.Strategy.reduce` API to aggregate the results returned by `tf.distribute.Strategy.run`. `tf.distribute.Strategy.run` returns results from each local replica in the strategy, and there are multiple ways to consume this result. You can `reduce` them to get an aggregated value. You can also do `tf.distribute.Strategy.experimental_local_results` to get the list of values contained in the result, one per local replica.\n3. When `apply_gradients` is called within a distribution strategy scope, its behavior is modified. Specifically, before applying gradients on each parallel instance during synchronous training, it performs a sum-over-all-replicas of the gradients.\n",
"_____no_output_____"
],
[
"Finally, once we have defined the training step, we can iterate over `dist_dataset` and run the training in a loop:",
"_____no_output_____"
]
],
[
[
"for dist_inputs in dist_dataset:\n print(distributed_train_step(dist_inputs))",
"_____no_output_____"
]
],
[
[
"In the example above, we iterated over the `dist_dataset` to provide input to your training. We also provide the `tf.distribute.Strategy.make_experimental_numpy_dataset` to support numpy inputs. You can use this API to create a dataset before calling `tf.distribute.Strategy.experimental_distribute_dataset`.\n\nAnother way of iterating over your data is to explicitly use iterators. You may want to do this when you want to run for a given number of steps as opposed to iterating over the entire dataset.\nThe above iteration would now be modified to first create an iterator and then explicitly call `next` on it to get the input data.\n",
"_____no_output_____"
]
],
[
[
"iterator = iter(dist_dataset)\nfor _ in range(10):\n print(distributed_train_step(next(iterator)))",
"_____no_output_____"
]
],
[
[
"This covers the simplest case of using `tf.distribute.Strategy` API to distribute custom training loops. We are in the process of improving these APIs. Since this use case requires more work to adapt your code, we will be publishing a separate detailed guide in the future.",
"_____no_output_____"
],
[
"### What's supported now?\n\n| Training API \t| MirroredStrategy \t| TPUStrategy \t| MultiWorkerMirroredStrategy \t| CentralStorageStrategy \t| ParameterServerStrategy \t| \n|:-----------------------\t|:-------------------\t|:-------------------\t|:-----------------------------\t|:------------------------\t|:-------------------------\t|\n| Custom Training Loop \t| Supported \t| Supported \t| Experimental support \t| Experimental support \t| Support planned post 2.3 \t|\n\n### Examples and Tutorials\nHere are some examples for using distribution strategy with custom training loops:\n\n1. [Tutorial](https://www.tensorflow.org/tutorials/distribute/custom_training) to train MNIST using `MirroredStrategy`.\n2. [Guide](https://www.tensorflow.org/guide/tpu#train_a_model_using_custom_training_loop) on training MNIST using `TPUStrategy`.\n3. TensorFlow Model Garden [repository](https://github.com/tensorflow/models/tree/master/official) containing collections of state-of-the-art models implemented using various strategies.\n",
"_____no_output_____"
],
[
"## Using `tf.distribute.Strategy` with Estimator (Limited support)\n`tf.estimator` is a distributed training TensorFlow API that originally supported the async parameter server approach. Like with Keras, we've integrated `tf.distribute.Strategy` into `tf.Estimator`. If you're using Estimator for your training, you can easily change to distributed training with very few changes to your code. With this, Estimator users can now do synchronous distributed training on multiple GPUs and multiple workers, as well as use TPUs. This support in Estimator is, however, limited. See [What's supported now](#estimator_support) section below for more details.\n\nThe usage of `tf.distribute.Strategy` with Estimator is slightly different than the Keras case. Instead of using `strategy.scope`, now we pass the strategy object into the [`RunConfig`](https://www.tensorflow.org/api_docs/python/tf/estimator/RunConfig) for the Estimator.\n\nHere is a snippet of code that shows this with a premade Estimator `LinearRegressor` and `MirroredStrategy`:\n",
"_____no_output_____"
]
],
[
[
"mirrored_strategy = tf.distribute.MirroredStrategy()\nconfig = tf.estimator.RunConfig(\n train_distribute=mirrored_strategy, eval_distribute=mirrored_strategy)\nregressor = tf.estimator.LinearRegressor(\n feature_columns=[tf.feature_column.numeric_column('feats')],\n optimizer='SGD',\n config=config)",
"_____no_output_____"
]
],
[
[
"We use a premade Estimator here, but the same code works with a custom Estimator as well. `train_distribute` determines how training will be distributed, and `eval_distribute` determines how evaluation will be distributed. This is another difference from Keras where we use the same strategy for both training and eval.\n\nNow we can train and evaluate this Estimator with an input function:\n",
"_____no_output_____"
]
],
[
[
"def input_fn():\n dataset = tf.data.Dataset.from_tensors(({\"feats\":[1.]}, [1.]))\n return dataset.repeat(1000).batch(10)\nregressor.train(input_fn=input_fn, steps=10)\nregressor.evaluate(input_fn=input_fn, steps=10)",
"_____no_output_____"
]
],
[
[
"Another difference to highlight here between Estimator and Keras is the input handling. In Keras, we mentioned that each batch of the dataset is split automatically across the multiple replicas. In Estimator, however, we do not do automatic splitting of batch, nor automatically shard the data across different workers. You have full control over how you want your data to be distributed across workers and devices, and you must provide an `input_fn` to specify how to distribute your data.\n\nYour `input_fn` is called once per worker, thus giving one dataset per worker. Then one batch from that dataset is fed to one replica on that worker, thereby consuming N batches for N replicas on 1 worker. In other words, the dataset returned by the `input_fn` should provide batches of size `PER_REPLICA_BATCH_SIZE`. And the global batch size for a step can be obtained as `PER_REPLICA_BATCH_SIZE * strategy.num_replicas_in_sync`.\n\nWhen doing multi worker training, you should either split your data across the workers, or shuffle with a random seed on each. You can see an example of how to do this in the [Multi-worker Training with Estimator](../tutorials/distribute/multi_worker_with_estimator.ipynb).",
"_____no_output_____"
],
[
"And similarly, you can use multi worker and parameter server strategies as well. The code remains the same, but you need to use `tf.estimator.train_and_evaluate`, and set `TF_CONFIG` environment variables for each binary running in your cluster.",
"_____no_output_____"
],
[
"<a name=\"estimator_support\"></a>\n### What's supported now?\n\nThere is limited support for training with Estimator using all strategies except `TPUStrategy`. Basic training and evaluation should work, but a number of advanced features such as scaffold do not yet work. There may also be a number of bugs in this integration. At this time, we do not plan to actively improve this support, and instead are focused on Keras and custom training loop support. If at all possible, you should prefer to use `tf.distribute` with those APIs instead.\n\n| Training API \t| MirroredStrategy \t| TPUStrategy \t| MultiWorkerMirroredStrategy \t| CentralStorageStrategy \t| ParameterServerStrategy \t|\n|:---------------\t|:------------------\t|:-------------\t|:-----------------------------\t|:------------------------\t|:-------------------------\t|\n| Estimator API \t| Limited Support \t| Not supported \t| Limited Support \t| Limited Support \t| Limited Support \t|\n\n### Examples and Tutorials\nHere are some examples that show end to end usage of various strategies with Estimator:\n\n1. [Multi-worker Training with Estimator](../tutorials/distribute/multi_worker_with_estimator.ipynb) to train MNIST with multiple workers using `MultiWorkerMirroredStrategy`.\n2. [End to end example](https://github.com/tensorflow/ecosystem/tree/master/distribution_strategy) for multi worker training in tensorflow/ecosystem using Kubernetes templates. This example starts with a Keras model and converts it to an Estimator using the `tf.keras.estimator.model_to_estimator` API.\n3. Official [ResNet50](https://github.com/tensorflow/models/blob/master/official/vision/image_classification/resnet_imagenet_main.py) model, which can be trained using either `MirroredStrategy` or `MultiWorkerMirroredStrategy`.",
"_____no_output_____"
],
[
"## Other topics\nIn this section, we will cover some topics that are relevant to multiple use cases.",
"_____no_output_____"
],
[
"<a name=\"TF_CONFIG\"></a>\n### Setting up TF\\_CONFIG environment variable\n\nFor multi-worker training, as mentioned before, you need to set `TF_CONFIG` environment variable for each\nbinary running in your cluster. The `TF_CONFIG` environment variable is a JSON string which specifies what\ntasks constitute a cluster, their addresses and each task's role in the cluster. We provide a Kubernetes template in the\n[tensorflow/ecosystem](https://github.com/tensorflow/ecosystem) repo which sets\n`TF_CONFIG` for your training tasks.\n\nThere are two components of TF_CONFIG: cluster and task. cluster provides information about the training cluster, which is a dict consisting of different types of jobs such as worker. In multi-worker training, there is usually one worker that takes on a little more responsibility like saving checkpoint and writing summary file for TensorBoard in addition to what a regular worker does. Such worker is referred to as the 'chief' worker, and it is customary that the worker with index 0 is appointed as the chief worker (in fact this is how tf.distribute.Strategy is implemented). task on the other hand provides information of the current task. The first component cluster is the same for all workers, and the second component task is different on each worker and specifies the type and index of that worker.\n\nOne example of `TF_CONFIG` is:\n```\nos.environ[\"TF_CONFIG\"] = json.dumps({\n \"cluster\": {\n \"worker\": [\"host1:port\", \"host2:port\", \"host3:port\"],\n \"ps\": [\"host4:port\", \"host5:port\"]\n },\n \"task\": {\"type\": \"worker\", \"index\": 1}\n})\n```\n",
"_____no_output_____"
],
[
"This `TF_CONFIG` specifies that there are three workers and two ps tasks in the\ncluster along with their hosts and ports. The \"task\" part specifies that the\nrole of the current task in the cluster, worker 1 (the second worker). Valid roles in a cluster is\n\"chief\", \"worker\", \"ps\" and \"evaluator\". There should be no \"ps\" job except when using `tf.distribute.experimental.ParameterServerStrategy`.",
"_____no_output_____"
],
[
"## What's next?\n\n`tf.distribute.Strategy` is actively under development. We welcome you to try it out and provide and your feedback using [GitHub issues](https://github.com/tensorflow/tensorflow/issues/new).",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec700d10dd90ff1785e3e2af9e862e3aa83f709c | 12,962 | ipynb | Jupyter Notebook | docs_jupyter/aliases.ipynb | lmfaber-dkfz/refgenie | faed6779548c4efb819ab987edc18c38033c95b8 | [
"BSD-2-Clause"
]
| 32 | 2017-02-20T08:03:44.000Z | 2020-04-19T03:02:26.000Z | docs_jupyter/aliases.ipynb | lmfaber-dkfz/refgenie | faed6779548c4efb819ab987edc18c38033c95b8 | [
"BSD-2-Clause"
]
| 160 | 2018-06-29T13:28:21.000Z | 2020-06-17T18:11:49.000Z | docs_jupyter/aliases.ipynb | lmfaber-dkfz/refgenie | faed6779548c4efb819ab987edc18c38033c95b8 | [
"BSD-2-Clause"
]
| 5 | 2017-01-31T05:10:49.000Z | 2020-06-16T22:33:20.000Z | 39.639144 | 843 | 0.628607 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec701732b36da518749e302498b5d53146c43fef | 245,052 | ipynb | Jupyter Notebook | tests/Taco_testing.ipynb | kartikgill/taco-box | 41d1e6977d0f5dc1d5bd1b508318afd086bdd1e5 | [
"Apache-2.0"
]
| 4 | 2021-11-28T15:58:54.000Z | 2022-03-19T14:30:34.000Z | tests/Taco_testing.ipynb | kartikgill/taco-box | 41d1e6977d0f5dc1d5bd1b508318afd086bdd1e5 | [
"Apache-2.0"
]
| null | null | null | tests/Taco_testing.ipynb | kartikgill/taco-box | 41d1e6977d0f5dc1d5bd1b508318afd086bdd1e5 | [
"Apache-2.0"
]
| null | null | null | 720.741176 | 33,620 | 0.955756 | [
[
[
"from tacobox import Taco\nimport cv2",
"_____no_output_____"
]
],
[
[
"## Taco Object",
"_____no_output_____"
]
],
[
[
"mytaco = Taco()",
"_____no_output_____"
]
],
[
[
"## Input Image",
"_____no_output_____"
]
],
[
[
"img = cv2.imread(\"../images/taco.png\", cv2.IMREAD_GRAYSCALE)",
"_____no_output_____"
],
[
"mytaco.visualize(img)",
"_____no_output_____"
]
],
[
[
"# Vertical TACO Examples",
"_____no_output_____"
]
],
[
[
"# Random pixels\naugmented = mytaco.apply_vertical_taco(img)\nmytaco.visualize(augmented, 'Random Vertical')\n# White pixels\naugmented = mytaco.apply_vertical_taco(img, 'white')\nmytaco.visualize(augmented, 'White Vertical')\n# Black pixels\naugmented = mytaco.apply_vertical_taco(img, 'black')\nmytaco.visualize(augmented, 'Black Vertical')\n# Mean Pixels\naugmented = mytaco.apply_vertical_taco(img, 'mean')\nmytaco.visualize(augmented, 'Mean Vertical')",
"_____no_output_____"
]
],
[
[
"# Horizontal TACO Examples",
"_____no_output_____"
]
],
[
[
"# Random pixels\naugmented = mytaco.apply_horizontal_taco(img)\nmytaco.visualize(augmented, 'Random Horizontal')\n# White pixels\naugmented = mytaco.apply_horizontal_taco(img, 'white')\nmytaco.visualize(augmented, 'White Horizontal')\n# Black pixels\naugmented = mytaco.apply_horizontal_taco(img, 'black')\nmytaco.visualize(augmented, 'Black Horizontal')\n# Mean Pixels\naugmented = mytaco.apply_horizontal_taco(img, 'mean')\nmytaco.visualize(augmented, 'Mean Horizontal')",
"_____no_output_____"
]
],
[
[
"# Both(Vertical + Horizontal) TACO ",
"_____no_output_____"
]
],
[
[
"# Random pixels\naugmented = mytaco.apply_taco(img)\nmytaco.visualize(augmented, 'Random (V+H)')\n# White pixels\naugmented = mytaco.apply_taco(img, 'white')\nmytaco.visualize(augmented, 'White (V+H)')\n# Black pixels\naugmented = mytaco.apply_taco(img, 'black')\nmytaco.visualize(augmented, 'Black (V+H)')\n# Mean Pixels\naugmented = mytaco.apply_taco(img, 'mean')\nmytaco.visualize(augmented, 'Mean (V+H)')",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec7022555808a2c43bf33c28ebfda17eaa4fdb15 | 8,299 | ipynb | Jupyter Notebook | coursera_ai/week1/a3_m1_v2_deep_feedforward_neural_networks_ex1_solution.ipynb | mbalgabayev/claimed | b595c25a752874602054443fb8785611c5e863e4 | [
"Apache-2.0"
]
| 308 | 2021-08-09T20:08:59.000Z | 2022-03-31T15:24:02.000Z | coursera_ai/week1/a3_m1_v2_deep_feedforward_neural_networks_ex1_solution.ipynb | mbalgabayev/claimed | b595c25a752874602054443fb8785611c5e863e4 | [
"Apache-2.0"
]
| 10 | 2021-09-30T16:47:17.000Z | 2022-03-25T14:25:16.000Z | coursera_ai/week1/a3_m1_v2_deep_feedforward_neural_networks_ex1_solution.ipynb | mbalgabayev/claimed | b595c25a752874602054443fb8785611c5e863e4 | [
"Apache-2.0"
]
| 615 | 2021-08-11T12:41:21.000Z | 2022-03-31T18:08:12.000Z | 4,149.5 | 8,298 | 0.591035 | [
[
[
"dp1 = {'partno': 100, 'maxtemp': 35, 'mintemp': 35, 'maxvibration': 12, 'asperity': 0.32}\ndp2 = {'partno': 101, 'maxtemp': 46, 'mintemp': 35, 'maxvibration': 21, 'asperity': 0.34}\ndp3 = {'partno': 130, 'maxtemp': 56, 'mintemp': 46, 'maxvibration': 3412, 'asperity': 12.42}\ndp4 = {'partno': 131, 'maxtemp': 58, 'mintemp': 48, 'maxvibration': 3542, 'asperity': 13.43}",
"_____no_output_____"
],
[
"def predict(dp):\n if dp['maxvibration']> 100:\n return 13\n else:\n return 0.33",
"_____no_output_____"
],
[
"predict(dp1)",
"_____no_output_____"
],
[
"predict(dp2)",
"_____no_output_____"
],
[
"predict(dp3)",
"_____no_output_____"
],
[
"predict(dp4)",
"_____no_output_____"
],
[
"w1 = 0.30\nw2 = 0\nw3 = 0\nw4 = 13/3412.0\n\ndef mlpredict(dp):\n return w1+w2*dp['maxtemp']+w3*dp['mintemp']+w4*dp['maxvibration']",
"_____no_output_____"
],
[
"mlpredict(dp4)",
"_____no_output_____"
],
[
"dp1 = {'partno': 100, 'maxtemp': 35, 'mintemp': 35, 'maxvibration': 12, 'broken': 0}\ndp2 = {'partno': 101, 'maxtemp': 46, 'mintemp': 35, 'maxvibration': 21, 'broken': 0}\ndp3 = {'partno': 130, 'maxtemp': 56, 'mintemp': 46, 'maxvibration': 3412, 'broken': 1}\ndp4 = {'partno': 131, 'maxtemp': 58, 'mintemp': 48, 'maxvibration': 3542, 'broken': 1}",
"_____no_output_____"
],
[
"def predict(dp):\n if dp['maxvibration']> 100:\n return 1\n else:\n return 0",
"_____no_output_____"
],
[
"predict(dp1)",
"_____no_output_____"
],
[
"predict(dp2)",
"_____no_output_____"
],
[
"predict(dp3)",
"_____no_output_____"
],
[
"predict(dp4)",
"_____no_output_____"
],
[
"import math\n\ndef sigmoid(x):\n return 1/(1+math.exp(-x))",
"_____no_output_____"
],
[
"w1 = 0.30\nw2 = 0\nw3 = 0\nw4 = 13/3412.0\n\ndef mlpredict(dp):\n return 1 if sigmoid(w1+w2*dp['maxtemp']+w3*dp['mintemp']+w4*dp['maxvibration']) > 0.7 else 0",
"_____no_output_____"
],
[
"mlpredict(dp4)",
"_____no_output_____"
],
[
"dp1 = {'partno': 100, 'maxtemp': 35, 'mintemp': 35, 'maxvibration': 12, 'asperity': 0.32}\ndp2 = {'partno': 101, 'maxtemp': 46, 'mintemp': 35, 'maxvibration': 21, 'asperity': 0.34}\ndp3 = {'partno': 130, 'maxtemp': 56, 'mintemp': 46, 'maxvibration': 3412, 'asperity': 12.42}\ndp4 = {'partno': 131, 'maxtemp': 58, 'mintemp': 48, 'maxvibration': 3542, 'asperity': 13.43}",
"_____no_output_____"
],
[
"dp1",
"_____no_output_____"
],
[
"x1 = np.array([1] + [v for k, v in dp1.items()] [1:-1])\nx2 = np.array([1] + [v for k, v in dp2.items()] [1:-1])\nx3 = np.array([1] + [v for k, v in dp3.items()] [1:-1])\nx4 = np.array([1] + [v for k, v in dp4.items()] [1:-1])",
"_____no_output_____"
],
[
"x1",
"_____no_output_____"
],
[
"w_layer1 = np.random.rand(4)\n\ndef neuron(x):\n return sigmoid(x.dot(w_layer1))",
"_____no_output_____"
],
[
"neuron(x1)",
"_____no_output_____"
],
[
"def sigmoid(x):\n return 1/(1+np.exp(-x))",
"_____no_output_____"
],
[
"w_layer1 = np.random.rand(4,4)\n\ndef layer1(x):\n return sigmoid(x.dot(w_layer1))",
"_____no_output_____"
],
[
"x = np.array([x1,x2,x3,x4])",
"_____no_output_____"
],
[
"x",
"_____no_output_____"
],
[
"layer1(x)",
"_____no_output_____"
],
[
"w_layer2 = np.random.rand(4,4)\n\ndef layer2(x):\n return sigmoid(x.dot(w_layer2))",
"_____no_output_____"
],
[
"layer2(layer1(x))",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec703c5ffbc6c5e4ef4a700407dd2ddc5f88fdd5 | 4,325 | ipynb | Jupyter Notebook | 30 Days of Challenge/Day 26 - Nested Logic.ipynb | sohammanjrekar/HackerRank | 1f5010133a1ac1e765e855a086053c97d9e958be | [
"MIT"
]
| null | null | null | 30 Days of Challenge/Day 26 - Nested Logic.ipynb | sohammanjrekar/HackerRank | 1f5010133a1ac1e765e855a086053c97d9e958be | [
"MIT"
]
| null | null | null | 30 Days of Challenge/Day 26 - Nested Logic.ipynb | sohammanjrekar/HackerRank | 1f5010133a1ac1e765e855a086053c97d9e958be | [
"MIT"
]
| null | null | null | 32.276119 | 193 | 0.540116 | [
[
[
"# Day 26: Nested Logic\n\n*Author: Eda AYDIN*\n\n## Objective\nToday’s challenge puts your understanding of nested conditional statements to the test.\n\n## Task\nYour local library needs your help! Given the expected and actual return dates for a library book, create a program that calculates the fine (if any). The fee structure is as follows:\n\n1. If the book is returned on or before the expected return date, no fine will be charged (i.e.: $fine = 0$.\n2. If the book is returned after the expected return day but still within the same calendar month and year as the expected return date, $fine = 15 Hackos x (the number of days late)$.\n3. If the book is returned after the expected return month but still within the same calendar year as the expected return date, the $fine = 500 Hackos x (the number of months late)$.\n4. If the book is returned after the calendar year in which it was expected, there is a fixed fine of $10000 Hackos$.\n\n## Example\n$d1, m1, y1 = 12312014$ returned date\n$d2, m2, y2 = 112015$ due date\n\nThe book is returned on time, so no fine is applied.\n\n$d1, m1, y1 = 112015$ returned date\n$d2, m2, y2 = 12312014$ due date\n\nThe book is returned in the following year, so the fine is a fixed 10000.\n\n## Input Format\nThe first line contains $3$ space-separated integers denoting the respective $day$, $month$, and $year$ on which the book was actually returned.\nThe second line contains $3$ space-separated integers denoting the respective $day$, $month$, and $year$ on which the book was expected to be returned (due date).\n\n## Constraints\n\n- &1 <= D <= 31&\n- &1 <= M <= 12&\n- &1 <= Y <= 3000&\n- &It is guaranteed that the dates will be valid Gregorian calendar dates.&\n\n## Output Format\nPrint a single integer denoting the library fine for the book received as input.\n\n## Sample Input\n\n```\nSTDIN Function\n----- --------\n9 6 2015 day = 9, month = 6, year = 2015 (date returned)\n6 6 2015 day = 6, month = 6, year = 2015 (date due)\n```\n## Sample Output\n```\n45\n```",
"_____no_output_____"
]
],
[
[
"returned_date = input()\nreturned_date = list(map(int,returned_date.split(\" \")))\n\ndue_date = input()\ndue_date = list(map(int,due_date.split(\" \")))\n\nreturned_day = returned_date[0]\nreturned_month = returned_date[1]\nreturned_year = returned_date[2]\n\ndue_day = due_date[0]\ndue_month = due_date[1]\ndue_year = due_date[2]\n\nfine = 0\n\nif returned_year > due_year:\n fine = 10000\nelif returned_year == due_year:\n if returned_month > due_month:\n fine = 500 * (returned_month - due_month)\n elif returned_month == due_month:\n if returned_day > due_day:\n fine = 15 * (returned_day - due_day)\nprint(fine)",
"45\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
ec7041231ccd24be52e413551b1fd3a08d85e2a8 | 153,975 | ipynb | Jupyter Notebook | examples/pole_zero_fitting/lemi_pole_zero_fitting_example.ipynb | simpeg/aurora | 1c81c464fb2e01da58e2c6b361b406b117100dcf | [
"MIT"
]
| 6 | 2021-05-14T00:38:20.000Z | 2022-03-31T14:00:27.000Z | examples/pole_zero_fitting/lemi_pole_zero_fitting_example.ipynb | simpeg/aurora | 1c81c464fb2e01da58e2c6b361b406b117100dcf | [
"MIT"
]
| 153 | 2021-05-29T19:21:38.000Z | 2022-03-27T22:16:12.000Z | examples/pole_zero_fitting/lemi_pole_zero_fitting_example.ipynb | simpeg/aurora | 1c81c464fb2e01da58e2c6b361b406b117100dcf | [
"MIT"
]
| null | null | null | 137.477679 | 35,536 | 0.882364 | [
[
[
"<h1>Poles/Zeros/Gain from Look-Up Tables (Prototyping)</h1>",
"_____no_output_____"
],
[
"## Problem Statement",
"_____no_output_____"
],
[
"The IRIS metadata archiving process had a strong preference for pole-zero ('zpk') representations of analog filters. However, calibrations of many MT instruments are characterized in the form of frequency sweeps, and the manufacturers calibrations are often provided in terms of tables of frequency, with amplitude and phase responses ('fap'). ",
"_____no_output_____"
],
[
"Thus we are faced with the problem of expressing a generic _fap_ instrument resposne in the _zpk_ format.",
"_____no_output_____"
],
[
"## Inverse Problem Setup\n\n$\\bf{d}$: The \"data\" in the the 'fap' table. This is simply a set of $N$ ordered pairs, $\\{(f_1, x_1), (f_2, x_2), ... (f_N, x_N)\\}$ where $f_i \\in \\mathbb{R}^+$ and $x_i \\in \\mathbb{C}$\n\n$\\bf{G}$: The function relating the data and the model can take on several parameterizations. One of the parameterizations is as a ratio of real valued polynomials:\n\n$ \\displaystyle h(s) = \\frac{a_{m}s^m + a_{m-1}s^{m-1} + ... + a_{1}s^{1} + a_{0}}{b_{n}s^{n} + b_{n-1}s^{n-1} + ... + b_{1}s^{1} + b_{0}} $\n\nWhere the Laplace variable _s_ = *j* $\\omega$\n\n\n$\\bf{m}$: Using the above parameterization $\\bf{m}$ would be the set of $m + n + 2$ coefficients ${a_0, a_1, ...a_m, b_0, b_1, ... b_n}$ with $a_i, b_i \\in \\mathbb{R}$\n\n<br>\n<br>\n\nThe coefficients in the numerator and denominator polynomials in the Laplace domain representation of the filter are real. They typically correspond to values of circuit parameters resistance, capacitance, inductance, etc. in analog EE applications. This means that these polynomials factor completely over the complex plane and moreover, the roots are either real or occur in complex-conjugate pairs.\n<br>\n<br>\n\nFactoring $h(s)$ on top and bottom yields the form:\n\n$ \\displaystyle h (s) = k \\frac{(s-z_1)(s-z_2)...(s-z_m) }{(s-p_1)(s-p_2)...(s-p_n)} $\n\nThere are well defined transformations between these representations and they are available through _scipy.signal.tf2zpk_ or _zpk2tf_.\n",
"_____no_output_____"
],
[
"### Recent Activity in PZ-fitting",
"_____no_output_____"
],
[
"Ben Murphy tried a variety of methods to solve this problem but invariably wound up encountering poles and zeros (roots) that did not obey the principle of complex-conjugate roots, and sometimes poles on the right hand side of the plane, and or k values that were complex-valued. The complex k values are particularly concerning because IRIS does not provide support for complex-valued k -- it is just one double-precision number. \n\nThe reason for these issues deserves some analysis but at a high level seems to be due to parameterizations that ultimately wound up searching complex-valued spaces for soltuions.\n\nKarl tried some variants on Ben's methods which involved some regularization functions that penalized non-conjugate root pairs, and imaginary parts of k, but these never matured so their value is questionable.\n\nIt seems like we should be able to use a numeric method to solve in the real valued form (even though the evaluation of the TF is Complex, the coefficients are real) and then extract the zpk afterwards. Following is an example of a numeric solution.\n\n\n\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\n\n# from matplotlib.gridspec import GridSpec\nfrom pathlib import Path\nfrom scipy import signal\nfrom scipy.optimize import minimize\n\nfrom mt_metadata.timeseries.filters.plotting_helpers import plot_response\nfrom aurora.sandbox.plot_helpers import plot_complex_response\n\n# from regularization_function_development import add_regularization_term \n# from regularization_function_development import get_pz_regularization\n\n# %matplotlib inline\n",
"_____no_output_____"
],
[
"DEG2RAD = np.pi/180",
"_____no_output_____"
]
],
[
[
"## Load the data from data repo",
"_____no_output_____"
]
],
[
[
"calibration_file_path = Path(\"Lemi-039_N131BxByBz\", \"Lemi-039_N131.cal\")\n\ndf = pd.read_csv(calibration_file_path, skiprows=3, delimiter='\\t')\ncolumn_labels = ['frequency', 'amplitude_x', 'phase_x', 'amplitude_y', 'phase_y', 'amplitude_z', 'phase_z']\ncolumns_map = dict(zip(df.columns, column_labels))\ndf = df.rename(columns=columns_map)\nprint(df.columns)\n",
"Index(['frequency', 'amplitude_x', 'phase_x', 'amplitude_y', 'phase_y',\n 'amplitude_z', 'phase_z'],\n dtype='object')\n"
]
],
[
[
"## Preliminary Response Curve",
"_____no_output_____"
]
],
[
[
"x_phase_radians = DEG2RAD*df['phase_x']\nx_phasor = np.exp(1.J*x_phase_radians) #(np.cos( x_phase_radians)+ 1.j*np.sin(x_phase_radians))\nx_complex_response = df['amplitude_x'] * x_phasor\nX_COMPLEX_RESPONSE = x_complex_response #make it global for availabilty in objective functions\nfrequencies = df['frequency'].to_numpy()\n\nplot_complex_response(df['frequency'], x_complex_response)",
"_____no_output_____"
]
],
[
[
"## Estimate the response amplitude in the pass band",
"_____no_output_____"
]
],
[
[
"nominal_passband_response_amplitude = df['amplitude_x'].iloc[10].mean()\nprint('nominal_passband_response_amplitude', nominal_passband_response_amplitude)",
"nominal_passband_response_amplitude 0.02847\n"
]
],
[
[
"## Express in units of deciBels and plot on semilog",
"_____no_output_____"
]
],
[
[
"amplitude_in_db = 20*np.log10(df['amplitude_x']/nominal_passband_response_amplitude)",
"_____no_output_____"
],
[
"plt.semilogx(df['frequency'], amplitude_in_db)\nplt.semilogx(df['frequency'], amplitude_in_db, 'r*')\nplt.grid(which='both')\nplt.title('Amplitude Response of Instrument in dB')\n",
"_____no_output_____"
]
],
[
[
"## Note that the decay is log-linear after about 5Hz",
"_____no_output_____"
]
],
[
[
"#The last 13 points are approximately a straight line on the log plot\nn_asymptotic_log_linear = 13\nfrq_line = df['frequency'][-n_asymptotic_log_linear:]\nampl_line = df['amplitude_x'][-n_asymptotic_log_linear:]\ndb_line = amplitude_in_db[-n_asymptotic_log_linear:]\nlog_f = np.array(np.log10(frq_line))\nlog_a = np.array(np.log10(ampl_line))\ndb = np.array(db_line)\ndb_per_decade = (db[-1]-db[0]) / (log_f[-1]-log_f[0])\n\nplt.plot(log_f, db_line)\nplt.xlabel('log$_{10}$ frequency')\nplt.ylabel('dB')\nplt.title(f\"db_per_decade: {db_per_decade:.2f}\")\nplt.show()",
"_____no_output_____"
]
],
[
[
"### looks like approximately -20dB/decade, which theoretically corresponds to a single pole. We obtain a decent fit with one pole, but a better fit with 3poles and one zero.\n\nIf we want to optimize we need to add constraints however. In particular, a regulariization that forces 1: complex poles to be conjugates, and the 'odd' poles to be pure real. This means modifying the LLSQ process.",
"_____no_output_____"
],
[
"If the roll off is ~20dB/ decade then we would expect a 1 pole filter, or at least a filter with one more pole than zeros",
"_____no_output_____"
]
],
[
[
"%%html\n<iframe src=\"https://eng.libretexts.org/Bookshelves/Electrical_Engineering/Electronics/Operational_Amplifiers_and_Linear_Integrated_Circuits_-_Theory_and_Application_(Fiore)/11%3A_Active_Filters/11.04%3A_Section_4-\" width=\"1200\" height=\"1000\"></iframe>",
"_____no_output_____"
],
[
"# from tf_helpers import Fit_ZerosPolesGain_toFrequencyResponse_LLSQ\n# n_zeros = 2; n_poles = 3;\n# #n_zeros = 0; n_poles = 1;\n\n# print(\"PARAMETERIZATION: {} zeros, {} poles\".format(n_zeros, n_poles))\n# w_obs = 2*np.pi*frequencies\n# resp_obs = x_complex_response#df['amplitude_x']\n# LEMI_LM_ZPK = Fit_ZerosPolesGain_toFrequencyResponse_LLSQ(w_obs, \n# resp_obs, \n# n_zeros, n_poles, useSKiter=False, \n# regularize=False)\n# print(LEMI_LM_ZPK)\n\n# plot_response(resp_obs=resp_obs, w_obs=w_obs, x_units='frequency', \n# zpk_pred=LEMI_LM_ZPK, w_values=w_obs)\n",
"_____no_output_____"
]
],
[
[
"### Lets try a numerical approach using scipy optimize and some regularization that penalizes deviations from the fundamental theorem of algebra solutions",
"_____no_output_____"
]
],
[
[
"W = 2*np.pi*frequencies\n\nN_ZEROS = 0; N_POLES = 1 #Not great\nN_ZEROS = 1; N_POLES = 2#Good when seeded with 1\n#N_ZEROS = 2; N_POLES = 3#Good when seeded with 3\n#N_ZEROS = 3; N_POLES = 4#not so good\nnp.random.seed(1)\nb0 = np.random.rand(N_ZEROS+1)\na0 = np.random.rand(N_POLES+1)\n\nMODEL_VECTOR_0 = np.hstack((b0, a0))\n#print('MODEL_VECTOR_0',MODEL_VECTOR_0)\n\n\ndef objective_function(model_vector, verbose=False):\n #unpack the model vector\n bb = model_vector[:N_ZEROS+1]\n aa = model_vector[N_ZEROS+1:]\n \n if verbose:\n print('numerator ', bb, N_ZEROS)\n print('denominator', aa, N_POLES)\n w, h = signal.freqs(bb, aa, worN=W)\n residual = X_COMPLEX_RESPONSE - h\n #print(residual)\n misfit = np.sqrt(np.mean(np.abs(residual**2)))\n# misfit = add_regularization_term(zeros, poles, k, misfit, aa=0.1)\n if verbose:\n print('misfit',misfit)\n return misfit\n\nobjective_function(MODEL_VECTOR_0, verbose=True)",
"numerator [ 0.417022 0.72032449] 1\ndenominator [ 1.14374817e-04 3.02332573e-01 1.46755891e-01] 2\nmisfit 2.46055330257\n"
],
[
"#method = 'nelder-mead'\nmethod = 'Powell'\n#method = 'CG'\n#method = 'Newton-CG' #needs Jacobian\nres = minimize(objective_function, MODEL_VECTOR_0, method=method, options={'xatol': 1e-10, 'disp': True, })",
"/home/kkappler/.local/lib/python3.6/site-packages/ipykernel_launcher.py:5: OptimizeWarning: Unknown solver options: xatol\n \"\"\"\n"
],
[
"res.x\nbb = res.x[:N_ZEROS+1]\naa = res.x[N_ZEROS+1:]\nzpk = signal.TransferFunction(bb, aa).to_zpk()\n#zpk = signal.tf2zpk(bb, aa)\nprint(zpk)\n#print(type(zpk))\n",
"ZerosPolesGainContinuous(\narray([-37.78764719]),\narray([-18.49867054+16.57300946j, -18.49867054-16.57300946j]),\n-0.4648829833859911,\ndt: None\n)\n"
],
[
"plot_response(resp_obs=resp_obs, w_obs=w_obs, x_units='frequency', \n zpk_pred=zpk, w_values=w_obs)",
"_____no_output_____"
]
],
[
[
"what percent differences are we looking at here?",
"_____no_output_____"
]
],
[
[
"resp_obs\nfrequencies\nw_pred, resp_pred = signal.freqresp(zpk, w=w_obs)\nnp.abs((resp_pred-resp_obs)/resp_obs)*100\n\n(1./DEG2RAD)*(np.angle(resp_pred) - np.angle(resp_obs))/np.angle(resp_obs)",
"_____no_output_____"
],
[
"frequencies[40]",
"_____no_output_____"
]
],
[
[
"## So, where are we?\n\nWe have a numeric method that works \"pretty well\" but could use some serious tuning\n1. We have analytic expressions so lets get some Jacobians and Hessians and try some of the more classical approaches in scipy.minimize\n\n2. Lets try a bunch of seed values for each optimization and explore the sum RMS misfits. \n3. Do (2) above for a collection of n_poles, n_zeros controls\n\n4. Tidy up supporting codes and make some metrics/reports to use for these fits\n5. Move on",
"_____no_output_____"
],
[
"#### Note that if we relax the requirement that the poles and zeros are real or conjugate-pairs we can get better fits, but it is a physical solution. There is some research to be done here. \n\nIf you are interested in exploring more check out Ben Murphy's ipynb in iris-mt-scratch repo",
"_____no_output_____"
]
],
[
[
"from scipy.optimize import minimize",
"_____no_output_____"
],
[
"from tf_helpers import Fit_ZerosPolesGain_toFrequencyResponse_LLSQ\nn_zeros = 2; n_poles = 3;\n#n_zeros = 1; n_poles = 2;\n#n_zeros = 0; n_poles = 1;\n\nprint(\"PARAMETERIZATION: {} zeros, {} poles\".format(n_zeros, n_poles))\nw_obs = 2*np.pi*frequencies\nresp_obs = x_complex_response#df['amplitude_x']\nLEMI_LM_ZPK = Fit_ZerosPolesGain_toFrequencyResponse_LLSQ(w_obs, \n resp_obs, \n n_zeros, n_poles, useSKiter=False, \n regularize=False)\nprint(LEMI_LM_ZPK)\n\nplot_response(resp_obs=resp_obs, w_obs=w_obs, x_units='frequency', \n zpk_pred=LEMI_LM_ZPK, w_values=w_obs)\n",
"PARAMETERIZATION: 2 zeros, 3 poles\nZerosPolesGainContinuous(\narray([-957.08603092+4.45450947j, -34.98372872-2.28826934j]),\narray([-485.28932769+107.50895551j, -17.26778927 -17.54658073j,\n -18.34527168 +15.52113528j]),\n(-0.2446806069717897+0.046680268003547215j),\ndt: None\n)\n"
],
[
"w_pred, resp_pred = signal.freqresp(LEMI_LM_ZPK, w=w_obs)\nnp.abs((resp_pred-resp_obs)/resp_obs)*100",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec7051388e76421e7d85407f167c3fee38495b95 | 13,642 | ipynb | Jupyter Notebook | notebooks/text_classification/text_classification_unified_information_explainer.ipynb | imatiach-msft/interpret-text | 7629b78f425459a84a40fe0acf370de5b5b27bd1 | [
"MIT"
]
| 277 | 2020-05-12T10:14:02.000Z | 2022-03-31T07:09:06.000Z | notebooks/text_classification/text_classification_unified_information_explainer.ipynb | ruiwang1994/interpret-text | d74efc985270aaa2b3ab779408597d9505e9ea1d | [
"MIT"
]
| 49 | 2020-04-30T18:15:30.000Z | 2022-02-27T01:03:04.000Z | notebooks/text_classification/text_classification_unified_information_explainer.ipynb | ruiwang1994/interpret-text | d74efc985270aaa2b3ab779408597d9505e9ea1d | [
"MIT"
]
| 48 | 2020-05-08T16:07:32.000Z | 2022-03-06T21:34:18.000Z | 29.656522 | 310 | 0.5972 | [
[
[
"*Licensed under the MIT License.*\n\n# Text Classification of MultiNLI Sentences using BERT",
"_____no_output_____"
],
[
"# Before You Start\n\n> **Tip**: If you want to run through the notebook quickly, you can set the **`QUICK_RUN`** flag in the cell below to **`True`**. This will run the notebook on a small subset of the data and a use a smaller number of epochs. \n\nIf you run into CUDA out-of-memory error or the jupyter kernel dies constantly, try reducing the `BATCH_SIZE` and `MAX_LEN`, but note that model performance will be compromised. ",
"_____no_output_____"
]
],
[
[
"## Set QUICK_RUN = True to run the notebook on a small subset of data and a smaller number of epochs.\nQUICK_RUN = True",
"_____no_output_____"
],
[
"import sys\nsys.path.append(\"../../\")\nimport os\nimport json\nimport pandas as pd\nimport numpy as np\nimport scrapbook as sb\nfrom sklearn.metrics import classification_report, accuracy_score\nfrom sklearn.preprocessing import LabelEncoder\nfrom sklearn.model_selection import train_test_split\nimport torch\nimport torch.nn as nn\n\nfrom interpret_text.experimental.common.utils_bert import Language, Tokenizer, BERTSequenceClassifier\nfrom interpret_text.experimental.common.timer import Timer\n\nfrom notebooks.test_utils.utils_mnli import load_mnli_pandas_df",
"_____no_output_____"
],
[
"from interpret_text.experimental.unified_information import UnifiedInformationExplainer",
"_____no_output_____"
]
],
[
[
"## Introduction\nIn this notebook, we fine-tune and evaluate a pretrained [BERT](https://arxiv.org/abs/1810.04805) model on a subset of the [MultiNLI](https://www.nyu.edu/projects/bowman/multinli/) dataset.\n\nWe use a [sequence classifier](https://github.com/microsoft/nlp/blob/master/utils_nlp/models/bert/sequence_classification.py) that wraps [Hugging Face's PyTorch implementation](https://github.com/huggingface/pytorch-pretrained-BERT) of Google's [BERT](https://github.com/google-research/bert).",
"_____no_output_____"
],
[
"### Set parameters\nHere we set some parameters that we use for our modeling task.",
"_____no_output_____"
]
],
[
[
"TRAIN_DATA_FRACTION = 1\nTEST_DATA_FRACTION = 1\nNUM_EPOCHS = 1\n\nif QUICK_RUN:\n TRAIN_DATA_FRACTION = 0.001\n TEST_DATA_FRACTION = 0.001\n NUM_EPOCHS = 1\n\nif torch.cuda.is_available():\n BATCH_SIZE = 1\nelse:\n BATCH_SIZE = 8\n\nDATA_FOLDER = \"./temp\"\nBERT_CACHE_DIR = \"./temp\"\nLANGUAGE = Language.ENGLISH\nTO_LOWER = True\nMAX_LEN = 150\nBATCH_SIZE_PRED = 512\nTRAIN_SIZE = 0.6\nLABEL_COL = \"genre\"\nTEXT_COL = \"sentence1\"",
"_____no_output_____"
]
],
[
[
"## Read Dataset\nWe start by loading a subset of the data. The following function also downloads and extracts the files, if they don't exist in the data folder.\n\nThe MultiNLI dataset is mainly used for natural language inference (NLI) tasks, where the inputs are sentence pairs and the labels are entailment indicators. The sentence pairs are also classified into *genres* that allow for more coverage and better evaluation of NLI models.\n\nFor our classification task, we use the first sentence only as the text input, and the corresponding genre as the label. We select the examples corresponding to one of the entailment labels (*neutral* in this case) to avoid duplicate rows, as the sentences are not unique, whereas the sentence pairs are.",
"_____no_output_____"
]
],
[
[
"df = load_mnli_pandas_df(DATA_FOLDER, \"train\")\ndf = df[df[\"gold_label\"]==\"neutral\"] # get unique sentences",
"_____no_output_____"
]
],
[
[
"These are the five genres in the dataset:",
"_____no_output_____"
]
],
[
[
"df[[LABEL_COL, TEXT_COL]].head()",
"_____no_output_____"
],
[
"df[LABEL_COL].value_counts()",
"_____no_output_____"
]
],
[
[
"We start by splitting the data for training and testing, and then we encode the class labels:",
"_____no_output_____"
]
],
[
[
"# split\ndf_train, df_test = train_test_split(df, train_size = TRAIN_SIZE, random_state=0)\ndf_train = df_train.reset_index(drop=True)\ndf_test = df_test.reset_index(drop=True)\n\nif QUICK_RUN:\n df_train = df_train.sample(frac=TRAIN_DATA_FRACTION).reset_index(drop=True)\n df_test = df_test.sample(frac=TEST_DATA_FRACTION).reset_index(drop=True)",
"_____no_output_____"
],
[
"# encode labels\nlabel_encoder = LabelEncoder()\nlabels_train = label_encoder.fit_transform(df_train[LABEL_COL])\nlabels_test = label_encoder.transform(df_test[LABEL_COL])\n\nnum_labels = len(np.unique(labels_train))",
"_____no_output_____"
],
[
"print(\"Number of unique labels: {}\".format(num_labels))\nprint(\"Number of training examples: {}\".format(df_train.shape[0]))\nprint(\"Number of testing examples: {}\".format(df_test.shape[0]))",
"_____no_output_____"
]
],
[
[
"## Tokenize and Preprocess",
"_____no_output_____"
],
[
"Before we start training, we tokenize the text documents and convert them to lists of tokens. The following steps instantiate a `BERT tokenizer` given the language, and tokenize the text of the training and testing sets.",
"_____no_output_____"
]
],
[
[
"tokenizer = Tokenizer(LANGUAGE, to_lower=TO_LOWER, cache_dir=BERT_CACHE_DIR)\n\ntokens_train = tokenizer.tokenize(list(df_train[TEXT_COL]))\ntokens_test = tokenizer.tokenize(list(df_test[TEXT_COL]))",
"_____no_output_____"
]
],
[
[
"In addition, we perform the following preprocessing steps in the cell below:\n- Convert the tokens into token indices corresponding to the BERT tokenizer's vocabulary\n- Add the special tokens [CLS] and [SEP] to mark the beginning and end of a sentence, respectively\n- Pad or truncate the token lists to the specified max length. In this case, `MAX_LEN = 150`\n- Return mask lists that indicate the paddings' positions\n- Return token type id lists that indicate which sentence the tokens belong to (not needed for one-sequence classification)\n\n*See the original [implementation](https://github.com/google-research/bert/blob/master/run_classifier.py) for more information on BERT's input format.*",
"_____no_output_____"
]
],
[
[
"tokens_train, mask_train, _ = tokenizer.preprocess_classification_tokens(tokens_train, MAX_LEN)\ntokens_test, mask_test, _ = tokenizer.preprocess_classification_tokens(tokens_test, MAX_LEN)",
"_____no_output_____"
]
],
[
[
"## Sequence Classifier Model\nNext, we use a sequence classifier that loads a pre-trained BERT model, given the language and number of labels.",
"_____no_output_____"
]
],
[
[
"classifier = BERTSequenceClassifier(language=LANGUAGE, num_labels=num_labels, cache_dir=BERT_CACHE_DIR)",
"_____no_output_____"
]
],
[
[
"## Train Model\nWe train the classifier using the training set. This involves fine-tuning the BERT Transformer and learning a linear classification layer on top of that:",
"_____no_output_____"
]
],
[
[
"with Timer() as t:\n classifier.fit(token_ids=tokens_train,\n input_mask=mask_train,\n labels=labels_train, \n num_epochs=NUM_EPOCHS,\n batch_size=BATCH_SIZE, \n verbose=True) \nprint(\"[Training time: {:.3f} hrs]\".format(t.interval / 3600))",
"_____no_output_____"
]
],
[
[
"## Score Model\nWe score the test set using the trained classifier:",
"_____no_output_____"
]
],
[
[
"preds = classifier.predict(token_ids=tokens_test, \n input_mask=mask_test, \n batch_size=BATCH_SIZE_PRED)",
"_____no_output_____"
]
],
[
[
"## Evaluate Model\nFinally, we compute the overall accuracy, precision, recall, and F1 metrics on the test set. We also look at the metrics for eact of the genres in the the dataset. ",
"_____no_output_____"
]
],
[
[
"report = classification_report(labels_test, preds, target_names=label_encoder.classes_, output_dict=True) \naccuracy = accuracy_score(labels_test, preds)\nprint(\"accuracy: {}\".format(accuracy))\nprint(json.dumps(report, indent=4, sort_keys=True))",
"_____no_output_____"
],
[
"# for testing\nsb.glue(\"accuracy\", accuracy)\nsb.glue(\"precision\", report[\"macro avg\"][\"precision\"])\nsb.glue(\"recall\", report[\"macro avg\"][\"recall\"])\nsb.glue(\"f1\", report[\"macro avg\"][\"f1-score\"])",
"_____no_output_____"
]
],
[
[
"## Explain Model",
"_____no_output_____"
]
],
[
[
"device = torch.device(\"cpu\" if not torch.cuda.is_available() else \"cuda\")\n\nclassifier.model.to(device)\nfor param in classifier.model.parameters():\n param.requires_grad = False\nclassifier.model.eval()",
"_____no_output_____"
],
[
"interpreter_unified = UnifiedInformationExplainer(model=classifier.model, \n train_dataset=list(df_train[TEXT_COL]), \n device=device, \n target_layer=14, \n classes=label_encoder.classes_)",
"_____no_output_____"
],
[
"idx = 7\ntext = df_test[TEXT_COL][idx]\ntrue_label = df_test[LABEL_COL][idx]\npredicted_label = label_encoder.inverse_transform([preds[idx]])\nprint(text, true_label, predicted_label)",
"_____no_output_____"
],
[
"explanation_unified = interpreter_unified.explain_local(text, true_label)",
"_____no_output_____"
]
],
[
[
"## Visualize Explanation",
"_____no_output_____"
]
],
[
[
"from interpret_text.experimental.widget import ExplanationDashboard",
"_____no_output_____"
],
[
"ExplanationDashboard(explanation_unified)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec7058822ca835d5d60c2e724473a18fab58cf3b | 95,186 | ipynb | Jupyter Notebook | Co-op/.ipynb_checkpoints/test_coop_round_vs_sector-checkpoint.ipynb | SYDE-25/Class-Profile1A-Data-Analysis | fad918b587009be7183f8c444f5284943aa41eb8 | [
"MIT"
]
| 4 | 2021-05-02T22:58:51.000Z | 2021-05-04T23:38:49.000Z | Co-op/test_coop_round_vs_sector.ipynb | NirmalHegde/Class-Profile1A-Data-Analysis | d7ffd5987336f68c983ec3be4235f93456749ef2 | [
"MIT"
]
| null | null | null | Co-op/test_coop_round_vs_sector.ipynb | NirmalHegde/Class-Profile1A-Data-Analysis | d7ffd5987336f68c983ec3be4235f93456749ef2 | [
"MIT"
]
| 1 | 2021-05-02T23:43:08.000Z | 2021-05-02T23:43:08.000Z | 56.997605 | 27,580 | 0.639558 | [
[
[
"import pandas as pd \nimport seaborn as sns\nimport matplotlib.pyplot as plt\n%matplotlib inline\nimport numpy as np\npd.options.mode.chained_assignment = None",
"_____no_output_____"
]
],
[
[
"Round 1 Co-op",
"_____no_output_____"
]
],
[
[
"round1_coop = round1_coop[round1_coop['Which round did you get your coop in?'].notna()]\nround1_coop.head()",
"_____no_output_____"
],
[
"round1_coop['Which sector was your first co-op in?'] = round1_coop['Which sector was your first co-op in?'].str.split(';')\n\nround1_coop = (round1_coop\n .set_index(['Which round did you get your coop in?'])['Which sector was your first co-op in?']\n .apply(pd.Series)\n .stack()\n .reset_index()\n .drop('level_1', axis=1)\n .rename(columns={0:'Which sector was your first co-op in?'}))\n\nround1_coop.head()",
"_____no_output_____"
],
[
"round1_coop[round1_coop['Which sector was your first co-op in?'] != 'Unhired']\nround1_coop = round1_coop[round1_coop['Which sector was your first co-op in?'] != 'Unhired']\nround1_coop.head()",
"_____no_output_____"
],
[
"# checking for sectors in the first round\nround1_coop = round1_coop[round1_coop['Which round did you get your coop in?'] == '1st Round']\nround1_coop.head()",
"_____no_output_____"
],
[
"round1_coop['Number of people'] = round1_coop.groupby(['Which sector was your first co-op in?'])['Which round did you get your coop in?'].transform('count')\n\nround1_coop = round1_coop.drop_duplicates(subset=['Which round did you get your coop in?', 'Which sector was your first co-op in?', 'Number of people'], keep='first')\n\nround1_coop",
"_____no_output_____"
]
],
[
[
"Round 2 Co-op",
"_____no_output_____"
]
],
[
[
"round2_coop = class_df[relevant_cols]\nround2_coop.head()",
"_____no_output_____"
],
[
"round2_coop = round2_coop[round2_coop['Which round did you get your coop in?'].notna()]\nround2_coop.head()",
"_____no_output_____"
],
[
"round2_coop['Which sector was your first co-op in?'] = round2_coop['Which sector was your first co-op in?'].str.split(';')\n\nround2_coop = (round2_coop\n .set_index(['Which round did you get your coop in?'])['Which sector was your first co-op in?']\n .apply(pd.Series)\n .stack()\n .reset_index()\n .drop('level_1', axis=1)\n .rename(columns={0:'Which sector was your first co-op in?'}))\n\nround2_coop.head()",
"_____no_output_____"
],
[
"round2_coop[round2_coop['Which sector was your first co-op in?'] != 'Unhired']\nround2_coop = round2_coop[round2_coop['Which sector was your first co-op in?'] != 'Unhired']\nround2_coop.head()",
"_____no_output_____"
],
[
"# checking for sectors in the second round\nround2_coop = round2_coop[round2_coop['Which round did you get your coop in?'] == '2nd Round']\nround2_coop.head()",
"_____no_output_____"
],
[
"round2_coop['Number of people'] = round2_coop.groupby(['Which sector was your first co-op in?'])['Which round did you get your coop in?'].transform('count')\n\nround2_coop = round2_coop.drop_duplicates(subset=['Which round did you get your coop in?', 'Which sector was your first co-op in?', 'Number of people'], keep='first')\n\nround2_coop",
"_____no_output_____"
]
],
[
[
"Continuous round co-op",
"_____no_output_____"
]
],
[
[
"cont_coop = class_df[relevant_cols]\ncont_coop.head()",
"_____no_output_____"
],
[
"cont_coop = cont_coop[cont_coop['Which round did you get your coop in?'].notna()]\ncont_coop.head()",
"_____no_output_____"
],
[
"cont_coop['Which sector was your first co-op in?'] = cont_coop['Which sector was your first co-op in?'].str.split(';')\n\ncont_coop = (cont_coop\n .set_index(['Which round did you get your coop in?'])['Which sector was your first co-op in?']\n .apply(pd.Series)\n .stack()\n .reset_index()\n .drop('level_1', axis=1)\n .rename(columns={0:'Which sector was your first co-op in?'}))\n\ncont_coop.head()",
"_____no_output_____"
],
[
"cont_coop[cont_coop['Which sector was your first co-op in?'] != 'Unhired']\ncont_coop = cont_coop[cont_coop['Which sector was your first co-op in?'] != 'Unhired']\ncont_coop.head()",
"_____no_output_____"
],
[
"cont_coop = cont_coop[cont_coop['Which round did you get your coop in?'] == 'Continuous']\ncont_coop.head()",
"_____no_output_____"
],
[
"cont_coop['Number of people'] = cont_coop.groupby(['Which sector was your first co-op in?'])['Which round did you get your coop in?'].transform('count')\n\ncont_coop = cont_coop.drop_duplicates(subset=['Which round did you get your coop in?', 'Which sector was your first co-op in?', 'Number of people'], keep='first')\n\ncont_coop",
"_____no_output_____"
]
],
[
[
"Direct offer round",
"_____no_output_____"
]
],
[
[
"direct_coop = class_df[relevant_cols]\ndirect_coop.head()",
"_____no_output_____"
],
[
"direct_coop = direct_coop[direct_coop['Which round did you get your coop in?'].notna()]\ndirect_coop.head()",
"_____no_output_____"
],
[
"direct_coop['Which sector was your first co-op in?'] = direct_coop['Which sector was your first co-op in?'].str.split(';')\n\ndirect_coop = (direct_coop\n .set_index(['Which round did you get your coop in?'])['Which sector was your first co-op in?']\n .apply(pd.Series)\n .stack()\n .reset_index()\n .drop('level_1', axis=1)\n .rename(columns={0:'Which sector was your first co-op in?'}))\n\ndirect_coop.head()",
"_____no_output_____"
],
[
"direct_coop[direct_coop['Which sector was your first co-op in?'] != 'Unhired']\ndirect_coop = direct_coop[direct_coop['Which sector was your first co-op in?'] != 'Unhired']\ndirect_coop.head()",
"_____no_output_____"
],
[
"direct_coop = direct_coop[direct_coop['Which round did you get your coop in?'] == 'Direct Offer']\ndirect_coop.head()",
"_____no_output_____"
],
[
"direct_coop['Number of people'] = direct_coop.groupby(['Which sector was your first co-op in?'])['Which round did you get your coop in?'].transform('count')\n\ndirect_coop = direct_coop.drop_duplicates(subset=['Which round did you get your coop in?', 'Which sector was your first co-op in?', 'Number of people'], keep='first')\n\ndirect_coop",
"_____no_output_____"
],
[
"\"\"\"\nx1 = round1_coop['Which sector was your first co-op in?']\nx2 = round2_coop['Which sector was your first co-op in?']\nx3 = cont_coop['Which sector was your first co-op in?']\nx4 = direct_coop['Which sector was your first co-op in?']\ny1 = round1_coop['Number of people']\ny2 = round2_coop['Number of people']\ny3 = cont_coop['Number of people']\ny4 = direct_coop['Number of people']\n\nsns.set_theme(palette=\"colorblind\")\nplt.figure(figsize=(15,8))\nplt.bar(x1, y1, label=\"First Round\", zorder=4)\nplt.bar(x2, y2,label=\"Second Round\", zorder=3)\nplt.bar(x3, y3, label=\"Continuous Round\", zorder=2)\nplt.bar(x4, y4, label=\"Direct Offer Round\", zorder=4)\n\nplt.title(\"CO-OP Round VS Sector\")\nplt.xlabel(\"Sector\", labelpad=15)\nplt.ylabel(\"Number of people\", labelpad=15)\nplt.legend(title=\"CO-OP Round\")\nplt.show()\n\"\"\"\n\nx1 = round1_coop['Which round did you get your coop in?']\nx2 = round2_coop['Which round did you get your coop in?']\nx3 = cont_coop['Which round did you get your coop in?']\nx4 = direct_coop['Which round did you get your coop in?']\ny1 = round1_coop['Number of people']\ny2 = round2_coop['Number of people']\ny3 = cont_coop['Number of people']\ny4 = direct_coop['Number of people']\n\nsns.set_theme(palette=\"colorblind\")\nplt.figure(figsize=(15,8))\nplt.bar(x1, y1, label=\"First Round\")\nplt.bar(x2, y2, label=\"Second Round\")\nplt.bar(x3, y3, label=\"Continuous Round\")\nplt.bar(x4, y4, label=\"Direct Offer Round\")\n\nplt.title(\"CO-OP Round VS Sector\")\nplt.xlabel(\"Round\", labelpad=15)\nplt.ylabel(\"Number of people\", labelpad=15)\nplt.legend()\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec708885008b5760c28b3d693d07a7b29641a268 | 23,569 | ipynb | Jupyter Notebook | PY0101EN-5-2-Numpy2D.ipynb | CardiffKT/Python-101---IBM | 1566b1cbb41552e671387e9ac3362dfa38b34b2f | [
"BSD-4-Clause-UC"
]
| 1 | 2020-02-25T13:17:10.000Z | 2020-02-25T13:17:10.000Z | PY0101EN-5-2-Numpy2D.ipynb | CardiffKT/Python-101---IBM | 1566b1cbb41552e671387e9ac3362dfa38b34b2f | [
"BSD-4-Clause-UC"
]
| null | null | null | PY0101EN-5-2-Numpy2D.ipynb | CardiffKT/Python-101---IBM | 1566b1cbb41552e671387e9ac3362dfa38b34b2f | [
"BSD-4-Clause-UC"
]
| 2 | 2020-03-12T20:58:56.000Z | 2020-10-12T22:18:43.000Z | 21.543876 | 716 | 0.508168 | [
[
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <a href=\"https://cocl.us/topNotebooksPython101Coursera\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/TopAd.png\" width=\"750\" align=\"center\">\n </a>\n</div>",
"_____no_output_____"
],
[
"<a href=\"https://cognitiveclass.ai/\">\n <img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/CCLog.png\" width=\"200\" align=\"center\">\n</a>",
"_____no_output_____"
],
[
"<h1>2D <code>Numpy</code> in Python</h1>",
"_____no_output_____"
],
[
"<p><strong>Welcome!</strong> This notebook will teach you about using <code>Numpy</code> in the Python Programming Language. By the end of this lab, you'll know what <code>Numpy</code> is and the <code>Numpy</code> operations.</p>",
"_____no_output_____"
],
[
"<h2>Table of Contents</h2>\n<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n <ul>\n <li><a href=\"create\">Create a 2D Numpy Array</a></li>\n <li><a href=\"access\">Accessing different elements of a Numpy Array</a></li>\n <li><a href=\"op\">Basic Operations</a></li>\n </ul>\n <p>\n Estimated time needed: <strong>20 min</strong>\n </p>\n</div>\n\n<hr>",
"_____no_output_____"
],
[
"<h2 id=\"create\">Create a 2D Numpy Array</h2>",
"_____no_output_____"
]
],
[
[
"# Import the libraries\n\nimport numpy as np \nimport matplotlib.pyplot as plt",
"_____no_output_____"
]
],
[
[
"Consider the list <code>a</code>, the list contains three nested lists **each of equal size**. ",
"_____no_output_____"
]
],
[
[
"# Create a list\n\na = [[11, 12, 13], [21, 22, 23], [31, 32, 33]]\na",
"_____no_output_____"
]
],
[
[
"We can cast the list to a Numpy Array as follow",
"_____no_output_____"
]
],
[
[
"# Convert list to Numpy Array\n# Every element is the same type\n\nA = np.array(a)\nA",
"_____no_output_____"
]
],
[
[
"We can use the attribute <code>ndim</code> to obtain the number of axes or dimensions referred to as the rank. ",
"_____no_output_____"
]
],
[
[
"# Show the numpy array dimensions\n\nA.ndim",
"_____no_output_____"
]
],
[
[
"Attribute <code>shape</code> returns a tuple corresponding to the size or number of each dimension.",
"_____no_output_____"
]
],
[
[
"# Show the numpy array shape\n\nA.shape",
"_____no_output_____"
]
],
[
[
"The total number of elements in the array is given by the attribute <code>size</code>.",
"_____no_output_____"
]
],
[
[
"# Show the numpy array size\n\nA.size",
"_____no_output_____"
]
],
[
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"access\">Accessing different elements of a Numpy Array</h2>",
"_____no_output_____"
],
[
"We can use rectangular brackets to access the different elements of the array. The correspondence between the rectangular brackets and the list and the rectangular representation is shown in the following figure for a 3x3 array: ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoEg.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can access the 2nd-row 3rd column as shown in the following figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFT.png\" width=\"400\" />",
"_____no_output_____"
],
[
" We simply use the square brackets and the indices corresponding to the element we would like:",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1, 2]",
"_____no_output_____"
]
],
[
[
" We can also use the following notation to obtain the elements: ",
"_____no_output_____"
]
],
[
[
"# Access the element on the second row and third column\n\nA[1][2]",
"_____no_output_____"
]
],
[
[
" Consider the elements shown in the following figure ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFF.png\" width=\"400\" />",
"_____no_output_____"
],
[
"We can access the element as follows ",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first column\n\nA[0][0]",
"_____no_output_____"
]
],
[
[
"We can also use slicing in numpy arrays. Consider the following figure. We would like to obtain the first two columns in the first row",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoFSF.png\" width=\"400\" />",
"_____no_output_____"
],
[
" This can be done with the following syntax ",
"_____no_output_____"
]
],
[
[
"# Access the element on the first row and first and second columns\n\nA[0][0:2]",
"_____no_output_____"
]
],
[
[
"Similarly, we can obtain the first two rows of the 3rd column as follows:",
"_____no_output_____"
]
],
[
[
"# Access the element on the first and second rows and third column\n\nA[0:2, 2]",
"_____no_output_____"
]
],
[
[
"Corresponding to the following figure: ",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoTST.png\" width=\"400\" />",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<h2 id=\"op\">Basic Operations</h2>",
"_____no_output_____"
],
[
"We can also add arrays. The process is identical to matrix addition. Matrix addition of <code>X</code> and <code>Y</code> is shown in the following figure:",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoAdd.png\" width=\"500\" />",
"_____no_output_____"
],
[
"The numpy array is given by <code>X</code> and <code>Y</code>",
"_____no_output_____"
]
],
[
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
]
],
[
[
" We can add the numpy arrays as follows.",
"_____no_output_____"
]
],
[
[
"# Add X and Y\n\nZ = X + Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplying a numpy array by a scaler is identical to multiplying a matrix by a scaler. If we multiply the matrix <code>Y</code> by the scaler 2, we simply multiply every element in the matrix by 2 as shown in the figure.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoDb.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can perform the same operation in numpy as follows ",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Multiply Y with 2\n\nZ = 2 * Y\nZ",
"_____no_output_____"
]
],
[
[
"Multiplication of two arrays corresponds to an element-wise product or Hadamard product. Consider matrix <code>X</code> and <code>Y</code>. The Hadamard product corresponds to multiplying each of the elements in the same position, i.e. multiplying elements contained in the same color boxes together. The result is a new matrix that is the same size as matrix <code>Y</code> or <code>X</code>, as shown in the following figure.",
"_____no_output_____"
],
[
"<img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Chapter%205/Images/NumTwoMul.png\" width=\"500\" />",
"_____no_output_____"
],
[
"We can perform element-wise product of the array <code>X</code> and <code>Y</code> as follows:",
"_____no_output_____"
]
],
[
[
"# Create a numpy array Y\n\nY = np.array([[2, 1], [1, 2]]) \nY",
"_____no_output_____"
],
[
"# Create a numpy array X\n\nX = np.array([[1, 0], [0, 1]]) \nX",
"_____no_output_____"
],
[
"# Multiply X with Y\n\nZ = X * Y\nZ",
"_____no_output_____"
]
],
[
[
"We can also perform matrix multiplication with the numpy arrays <code>A</code> and <code>B</code> as follows:",
"_____no_output_____"
],
[
"First, we define matrix <code>A</code> and <code>B</code>:",
"_____no_output_____"
]
],
[
[
"# Create a matrix A\n\nA = np.array([[0, 1, 1], [1, 0, 1]])\nA",
"_____no_output_____"
],
[
"# Create a matrix B\n\nB = np.array([[1, 1], [1, 1], [-1, 1]])\nB",
"_____no_output_____"
]
],
[
[
"We use the numpy function <code>dot</code> to multiply the arrays together.",
"_____no_output_____"
]
],
[
[
"# Calculate the dot product\n\nZ = np.dot(A,B)\nZ",
"_____no_output_____"
],
[
"# Calculate the sine of Z\n\nnp.sin(Z)",
"_____no_output_____"
]
],
[
[
"We use the numpy attribute <code>T</code> to calculate the transposed matrix",
"_____no_output_____"
]
],
[
[
"# Create a matrix C\n\nC = np.array([[1,1],[2,2],[3,3]])\nC",
"_____no_output_____"
],
[
"# Get the transposed of C\n\nC.T",
"_____no_output_____"
]
],
[
[
"<hr>\n<h2>The last exercise!</h2>\n<p>Congratulations, you have completed your first lesson and hands-on lab in Python. However, there is one more thing you need to do. The Data Science community encourages sharing work. The best way to share and showcase your work is to share it on GitHub. By sharing your notebook on GitHub you are not only building your reputation with fellow data scientists, but you can also show it off when applying for a job. Even though this was your first piece of work, it is never too early to start building good habits. So, please read and follow <a href=\"https://cognitiveclass.ai/blog/data-scientists-stand-out-by-sharing-your-notebooks/\" target=\"_blank\">this article</a> to learn how to share your work.\n<hr>",
"_____no_output_____"
],
[
"<div class=\"alert alert-block alert-info\" style=\"margin-top: 20px\">\n<h2>Get IBM Watson Studio free of charge!</h2>\n <p><a href=\"https://cocl.us/bottemNotebooksPython101Coursera\"><img src=\"https://s3-api.us-geo.objectstorage.softlayer.net/cf-courses-data/CognitiveClass/PY0101EN/Ad/BottomAd.png\" width=\"750\" align=\"center\"></a></p>\n</div>",
"_____no_output_____"
],
[
"<h3>About the Authors:</h3> \n<p><a href=\"https://www.linkedin.com/in/joseph-s-50398b136/\" target=\"_blank\">Joseph Santarcangelo</a> is a Data Scientist at IBM, and holds a PhD in Electrical Engineering. His research focused on using Machine Learning, Signal Processing, and Computer Vision to determine how videos impact human cognition. Joseph has been working for IBM since he completed his PhD.</p>",
"_____no_output_____"
],
[
"Other contributors: <a href=\"www.linkedin.com/in/jiahui-mavis-zhou-a4537814a\">Mavis Zhou</a>",
"_____no_output_____"
],
[
"<hr>",
"_____no_output_____"
],
[
"<p>Copyright © 2018 IBM Developer Skills Network. This notebook and its source code are released under the terms of the <a href=\"https://cognitiveclass.ai/mit-license/\">MIT License</a>.</p>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec708dd4829b5c2bdcedefd0682dcf23a446b1a1 | 8,158 | ipynb | Jupyter Notebook | Spring 2021/PF200_Report_Spring_2021.ipynb | AguaClara/PF200 | e17adec737ae0d07842f70726feccd6e094d4278 | [
"MIT"
]
| 1 | 2019-06-12T16:51:24.000Z | 2019-06-12T16:51:24.000Z | Spring 2021/PF200_Report_Spring_2021.ipynb | AguaClara/Prefabricated-Pilot-Plant | e17adec737ae0d07842f70726feccd6e094d4278 | [
"MIT"
]
| 4 | 2019-06-19T14:37:41.000Z | 2019-08-14T03:50:12.000Z | Spring 2021/PF200_Report_Spring_2021.ipynb | AguaClara/PF200 | e17adec737ae0d07842f70726feccd6e094d4278 | [
"MIT"
]
| 1 | 2019-07-08T20:31:55.000Z | 2019-07-08T20:31:55.000Z | 65.264 | 1,008 | 0.681417 | [
[
[
"<a href=\"https://colab.research.google.com/github/AguaClara/PF200/blob/master/PF200_Report_Spring_2021.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# PF200\n#### Sandra Cangelosi, Son-Jay Lake, Melissa Louie, Lynn Shin\n#### 19 March 2021",
"_____no_output_____"
],
[
"## Abstract\nThe PF200 is a small scale water treatment system designed to be easily shipped, fabricated, and implemented in small communities where a full scale plant is not economically viable. In the summer of 2019, a PF300 plant was built at the Cornell Water Treatment Plant and several failure points were observed in the design. The future goal is to redesign the PF200 system using data from the PF300 summer team as well as other past subteams. This semester, we are currently focused on redesigning the flocculator, by building off of the Flocculatorita design of the PF300 subteam. ",
"_____no_output_____"
],
[
"## Introduction\nThe PF200 is a small-scale pre-fabricated water treatment plant designed to serve a community of around 200 people with a flow rate of 0.67 L/s. The size of the plant makes it a more economically viable alternative to building a full-scale plant, especially for smaller rural communities. \n\nThere are currently several PF200 plants in use in different communities in Honduras, including Copan, Cuatro Comunidades, and Manzaragua. Improvements must be made for various parts of the water treatment process, and the Spring 2021 team has decided to focus on redesigning the flocculator. The considerations for the redesign include economic viability and fabrication complexity. For example, the current iteration of the flocculation design includes many pipes, which drive up the cost of the plant significantly. Plant operators have also provided the feedback that a longer residence time in the flocculator is needed for when heavy rain events cause turbidity to spike, which would increase the cost of pipes even further. In addition, fluid deformations in the flocculator are created by small baffles, which are circular sheets of plastic threaded onto a rod that is then inserted into the flocculator pipes. The process of putting this design together is time-consuming and laborious.\n\n<center>\n<img src=\"https://raw.githubusercontent.com/AguaClara/PF200/master/Spring%202021/Images/Current%20Baffle.34.40%20PM.png\" height=250>\n\nFigure 1. Current baffle fabrication process in the existing flocculator design.\n</center>\n\nTherefore, the Spring 2021 team seeks to redesign the flocculator in a way that will reduce the cost of materials and will be simpler to fabricate. The redesign will be modeled on the traditional flocculator of the full-scale plants, except it will all be contained in one box made out of PVC sheets. The only fabrication will consist of welding the sides of the box together and cutting slots into the sheets, which will then interlock with each other to create the framework for the flocculator.\n",
"_____no_output_____"
],
[
"## Literature Review and Previous Work\nSo far, the literature referenced for this new redesign is the Flocculatorita Report written by a Fall 2018 CEE 4520 team of Lily Falk, Nina Blahut, Marcin Sawczuk, and Yitzy Rosenberg.\n\nIn Fall 2018, the PF300 subteam created a new design for a small-scale version of the flocculator. This alternate design was characterized by increased flexibility, and was made to be more scalable than previous designs. The general makeup included a large diameter pipe divided into channels with baffles. This design would enable obstructed flow of water. The team solved for volume and retention time, and found numerical values for active cross-sectional area and necessary diameter of the pipe. One interesting conclusion they reached was that there wasn’t a clear constraint on the number of channels or baffles that would be included. This allows for versatility in fabrication, and allows the user to make a decision regarding how much turbulent flow will be present. \n\n<center>\n <img src=\"https://raw.githubusercontent.com/lilyfalk/CEE4520/master/IMG_4226.png\" height=300>\n \n\nFigure 1. Top view of Spring 2021 proposed flocculator design from Fall 2018 CEE 4520 design project.\n </center>\n\n\nOne problem with this design, acknowledged in the report, was the significant ‘dead space’ that exists on all sides of the rectangular channel/baffle configuration. Since the apparatus is located inside a circular pipe, there are 4 equally sized empty spaces on each side where no water is flowing, which means that space is wasted. In our design, we seek to rectify this issue while simultaneously building off the Flocculatorita design. Our goal is to eliminate the circular pipe as a whole and just keep the rectangular shape of the channels and baffles, enclosing it in a tall box constructed of PVC sheets. \n\n<center>\n <img src=\"https://raw.githubusercontent.com/AguaClara/PF200/master/Spring%202021/Images/Flocculatorita%20Top.png\" height=300>\n\nFigure 2. Top view of Spring 2021 proposed flocculator design.\n </center>\n\n<center>\n <img src=\"https://raw.githubusercontent.com/AguaClara/PF200/master/Spring%202021/Images/Flocculatorita%20Side.png\" height=400>\n\nFigure 3. Side view of Spring 2021 proposed flocculator design.\n </center>\n",
"_____no_output_____"
],
[
"## Future Work\nCurrently, we are designing the components of a flocculator for the PF200 plant. Our goals for the near future are to finish the design and fabrication of the flocculator. To complete this goal we will need to purchase PVC sheets and other materials which are utilized in the construction of the flocculator. With fabrication finished we will then move on to testing the flocculator and identifying any failure points in the design.\nOnce the improvements to the existing flocculator have been made, our subteam will move on to redesigning the other processes of the PF200 plant. This includes the redesign of the current filter and sedimentation tank. This is a lower priority, however, as the filter was already worked on in the past and the flocculator is easier to redesign by our subteam than the sedimentation tank. Work for the other components will be done further in the future. \n",
"_____no_output_____"
],
[
"## Bibliography\n\n1. Blahut, N., Falk, L., Sawczuk, M., & Bitzyspider, Y. (2019). (rep.). Flocculatorita. Ithaca, NY: AguaClara. \n",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec7095e6c37880f3ded493c614ecdbaf0324f1dd | 377,511 | ipynb | Jupyter Notebook | notebooks/block sampling AR signals.ipynb | davmre/sigvisa | 91a1f163b8f3a258dfb78d88a07f2a11da41bd04 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/block sampling AR signals.ipynb | davmre/sigvisa | 91a1f163b8f3a258dfb78d88a07f2a11da41bd04 | [
"BSD-3-Clause"
]
| null | null | null | notebooks/block sampling AR signals.ipynb | davmre/sigvisa | 91a1f163b8f3a258dfb78d88a07f2a11da41bd04 | [
"BSD-3-Clause"
]
| null | null | null | 321.285957 | 59,818 | 0.903044 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec70a58e550cba2b066077fa43ff77f8e9012c52 | 3,912 | ipynb | Jupyter Notebook | Figure_5/swan.ipynb | callumparr/TALON-paper-2020 | 8a089e51a4f36dbe57d154754a4bc7a00367d9e8 | [
"MIT"
]
| 1 | 2020-05-31T23:12:57.000Z | 2020-05-31T23:12:57.000Z | Figure_5/swan.ipynb | callumparr/TALON-paper-2020 | 8a089e51a4f36dbe57d154754a4bc7a00367d9e8 | [
"MIT"
]
| 3 | 2021-10-06T08:16:32.000Z | 2021-10-15T05:40:59.000Z | Figure_5/swan.ipynb | callumparr/TALON-paper-2020 | 8a089e51a4f36dbe57d154754a4bc7a00367d9e8 | [
"MIT"
]
| 2 | 2021-06-08T03:16:49.000Z | 2021-10-15T02:40:04.000Z | 24.759494 | 248 | 0.583078 | [
[
[
"import swan_vis as swan\nimport pandas as pd",
"/Users/fairliereese/miniconda3/lib/python3.7/site-packages/anndata/_core/anndata.py:21: FutureWarning: pandas.core.index is deprecated and will be removed in a future version. The public classes are available in the top-level namespace.\n from pandas.core.index import RangeIndex\n"
],
[
"sg = swan.SwanGraph()",
"_____no_output_____"
],
[
"# cortex \ngtf = 'Brain_revised_both.gtf'\nab_file = 'Brain_talon_abundance_filtered.tsv'\ncols = ['PacBio_Cortex_Rep1', 'PacBio_Cortex_Rep2']\nsg.add_dataset('Cortex', gtf, counts_file=ab_file, count_cols=cols)",
"\nAdding dataset Cortex to the SwanGraph\n"
],
[
"# hippocampus \ngtf = 'Brain_revised_both.gtf'\nab_file = 'Brain_talon_abundance_filtered.tsv'\ncols = ['PacBio_Hippocampus_Rep1', 'PacBio_Hippocampus_Rep2']\nsg.add_dataset('Hippocampus', gtf, counts_file=ab_file, count_cols=cols)",
"\nAdding dataset Hippocampus to the SwanGraph\n"
],
[
"sg.save_graph('swan')",
"Saving graph as swan.p\n"
],
[
"gname = 'Pnisr'\nsg.gen_report(gname, prefix='plots/{}'.format(gname),\n heatmap=True, \n cmap='viridis', \n browser=True,\n novelty=True)",
"\nPlotting transcripts for ENSMUSG00000028248.15\nSaving transcript path graph for ENCODEMT000409037 as plots/Pnisr_browser_ENCODEMT000409037_path.png\nSaving transcript path graph for ENSMUST00000029911.11 as plots/Pnisr_browser_ENSMUST00000029911.11_path.png\nSaving transcript path graph for ENSMUST00000108229.1 as plots/Pnisr_browser_ENSMUST00000108229.1_path.png\nSaving transcript path graph for ENSMUST00000152666.7 as plots/Pnisr_browser_ENSMUST00000152666.7_path.png\nSaving transcript path graph for ENSMUST00000138681.7 as plots/Pnisr_browser_ENSMUST00000138681.7_path.png\nGenerating report for ENSMUSG00000028248.15\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec70c3f1c713748aa3e5666bf1174ea09939f020 | 2,731 | ipynb | Jupyter Notebook | _notebooks/2020-04-30-InferenceData.ipynb | OriolAbril/test_fastpages | 2843f592b80bbf59458c2916ccf9ad55f08f5e25 | [
"Apache-2.0"
]
| 2 | 2022-01-21T00:00:55.000Z | 2022-01-21T05:43:55.000Z | _notebooks/2020-04-30-InferenceData.ipynb | OriolAbril/test_fastpages | 2843f592b80bbf59458c2916ccf9ad55f08f5e25 | [
"Apache-2.0"
]
| null | null | null | _notebooks/2020-04-30-InferenceData.ipynb | OriolAbril/test_fastpages | 2843f592b80bbf59458c2916ccf9ad55f08f5e25 | [
"Apache-2.0"
]
| null | null | null | 33.304878 | 630 | 0.635298 | [
[
[
"# \"Introduction to ArviZ: InferenceData objects\"\n> \"Why should you care about `InferenceData` if `az.xyz(pymc3_trace)` and `az.xyz(pystan_fit)` already work?\"\n\n- toc: true \n- author: Oriol Abril\n- badges: true\n- categories: [arviz, InferenceData, data]\n- image: images/InferenceDataStructure.png\n\nComments are not enabled for the blog, to inquiry further about the contents of the post, ask on [ArviZ Issues](https://github.com/arviz-devs/arviz/issues) or [PyMC Discourse](https://discourse.pymc.io/)",
"_____no_output_____"
],
[
"# About\n\nThis notebook is an introduction to `InferenceData` objects and their role in ArviZ. It aims to be as hands-on as possible, using examples to cover different use cases of the data in each group, examples on how to combine several `InferenceData` objects and so on.\n\nThe key idea behind `InferenceData` objects is to centralize and store all data relevant to a specific Bayesian inference run; from `observed_data` to `predictions` going through `prior` and `sample_stats`. The goal is therefore to both ease exploration and visualization of Bayesian inference results and to ease their sharing. Hence, ArviZ also provides several converter functions to transform results from common inference libraries such as PyMC3, PyStan or Pyro to `InferenceData`. In the future, we'll dedicate one post to each converter function, but before, we have to make sure the conversion process is worth it!\n",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown"
]
]
|
ec70c59da591d402444f9e8d0e7f5073c94b9713 | 3,889 | ipynb | Jupyter Notebook | questions/q1_game_of_names/GameOfNames.ipynb | aadhityasw/Competitive-Programs | 901a48d35f024a3a87c32a45b7f4531e8004a203 | [
"MIT"
]
| null | null | null | questions/q1_game_of_names/GameOfNames.ipynb | aadhityasw/Competitive-Programs | 901a48d35f024a3a87c32a45b7f4531e8004a203 | [
"MIT"
]
| 1 | 2021-05-15T07:56:51.000Z | 2021-05-15T07:56:51.000Z | questions/q1_game_of_names/GameOfNames.ipynb | aadhityasw/Competitive-Programs | 901a48d35f024a3a87c32a45b7f4531e8004a203 | [
"MIT"
]
| null | null | null | 40.510417 | 1,096 | 0.59604 | [
[
[
"# Game of Names\n\nOn a Sunday morning, some friends have joined to play: Game of Names. There is an N x N board. Each player is playing, turn by turn. At the turn of ith player, the player places first character of its name at one of the unfilled cells. The first player who places its character for 3 consecutive vertical, horizontal or diagonal cells is the winner.\nYou are provided a board with some filled and some unfilled cells. You have to tell the winner. If there is no winner, then you must print “ongoing”.\n### Input format:\nThe first line of input contains a single integer N (1 <= N <= 30), which denotes a board of size N x N. \nEach of the following N lines of input contains N characters. These denote N characters of a particular row. For unfilled cells are denoted by ‘.’ character and filled cells will be denoted by an uppercase letter.\n### Output format\nPrint the winning character or “ongoing”, as described in the task above.\n#### Sample Input 1:\n3<br>\nXOC<br>\nXOC<br>\nX..<br>\n#### Sample Output 1:\nX<br>\n#### Sample Input 2:\n4<br>\n....<br>\n..A.<br>\nAAB.<br>\n.B.B<br>\n#### Sample Output 2:\nOngoing",
"_____no_output_____"
]
],
[
[
"n = int(input('Enter thr size of the board : '))\narr = []\nfor i in range(n) :\n arr.append(input().split(''))\nprint(arr)",
"Enter thr size of the board : 3\n ZOC\n"
]
]
]
| [
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
]
]
|
ec70d83652877311274d78352164add9e19e2f19 | 42,771 | ipynb | Jupyter Notebook | tagging/entities/organization-augmentation.ipynb | huseinzol05/Malay-Dataset | e27b7617c74395c86bb5ed9f3f194b3cac2f66f6 | [
"Apache-2.0"
]
| 51 | 2020-05-20T13:26:18.000Z | 2021-05-13T07:21:17.000Z | tagging/entities/organization-augmentation.ipynb | huseinzol05/Malaya-Dataset | c9c1917a6b1cab823aef5a73bd10e0fab0bff42d | [
"Apache-2.0"
]
| 3 | 2019-02-05T11:34:38.000Z | 2020-03-19T03:18:38.000Z | tagging/entities/organization-augmentation.ipynb | huseinzol05/Malaya-Dataset | c9c1917a6b1cab823aef5a73bd10e0fab0bff42d | [
"Apache-2.0"
]
| 21 | 2019-02-08T05:17:24.000Z | 2020-05-05T09:28:50.000Z | 25.443783 | 137 | 0.431531 | [
[
[
"import numpy as np\nimport pandas as pd\nimport json",
"_____no_output_____"
],
[
"with open('malaysia-organization.json') as fopen:\n org = json.load(fopen)",
"_____no_output_____"
],
[
"org.keys()",
"_____no_output_____"
],
[
"import random\n\ndef generate_org(length):\n if str(length - 1) not in org:\n o = ' '.join(random.sample(org['free'], length))\n else:\n o = random.choice(org[str(length - 1)])\n return o",
"_____no_output_____"
],
[
"df = pd.read_csv('penganjur-acara.csv')['Nama Syarikat'].dropna()\nlocal_companies = df.tolist()",
"_____no_output_____"
],
[
"df = pd.read_csv('senarai-anak-syarikat-kplb.csv')['NAMA SYARIKAT'].dropna()\nlocal_companies.extend(df.tolist())",
"_____no_output_____"
],
[
"df = pd.read_csv('magiceventstartup.20191216.csv')['Startup Name'].dropna()\nlocal_companies.extend(df.tolist())",
"_____no_output_____"
],
[
"df = pd.read_csv('magiceventstartup.20180920.csv')['Startup Name'].dropna()\nlocal_companies.extend(df.tolist())",
"_____no_output_____"
],
[
"df = pd.read_csv('maklumat-syarikat-jualan-langsung.csv')['NAMA SYARIKAT'].dropna()\nlocal_companies.extend(df.tolist())\n\nlen(local_companies)",
"_____no_output_____"
],
[
"import re\n\nlocal_companies = [re.sub(r'[ ]+', ' ', l.replace('.', ' ')).strip().title() for l in local_companies]\nlocal_companies = list(set(local_companies))\nlen(local_companies)",
"_____no_output_____"
],
[
"with open('entities-data-v4.json') as fopen:\n entities = json.load(fopen)",
"_____no_output_____"
],
[
"entities['text'][10678: 10703]",
"_____no_output_____"
],
[
"results = []\ni = 0\nwhile i < len(entities['label']):\n r = []\n if entities['label'][i] == 'organization':\n while entities['label'][i] == 'organization':\n r.append(i)\n i += 1\n print(r)\n results.append(r)\n i += 1",
"[3, 4, 5]\n[14, 15]\n[41]\n[61]\n[63]\n[93]\n[99, 100, 101, 102]\n[197]\n[200, 201, 202]\n[228]\n[235]\n[328, 329, 330]\n[363, 364]\n[418, 419, 420, 421, 422, 423, 424, 425]\n[533, 534]\n[568]\n[589, 590]\n[603, 604, 605, 606, 607]\n[610]\n[692]\n[748]\n[768, 769]\n[774]\n[796]\n[908, 909, 910, 911]\n[952]\n[961]\n[981]\n[1047, 1048]\n[1118, 1119, 1120, 1121, 1122]\n[1158, 1159]\n[1172, 1173, 1174, 1175]\n[1214, 1215]\n[1244]\n[1301, 1302]\n[1307]\n[1327, 1328, 1329]\n[1384, 1385, 1386]\n[1399, 1400, 1401, 1402, 1403, 1404, 1405]\n[1412]\n[1421, 1422, 1423]\n[1446]\n[1474]\n[1497]\n[1540]\n[1542]\n[1545]\n[1579, 1580, 1581]\n[1583, 1584, 1585, 1586]\n[1596]\n[1620, 1621, 1622, 1623, 1624, 1625, 1626]\n[1641]\n[1658]\n[1728, 1729, 1730]\n[1735, 1736]\n[1739, 1740, 1741, 1742]\n[1746, 1747]\n[1767, 1768, 1769]\n[1797, 1798]\n[1812]\n[1829]\n[1845, 1846, 1847]\n[1855, 1856]\n[1945]\n[2005, 2006]\n[2039]\n[2107, 2108, 2109, 2110]\n[2133]\n[2163]\n[2176]\n[2207, 2208]\n[2343]\n[2400]\n[2417, 2418, 2419]\n[2439, 2440, 2441]\n[2447, 2448, 2449, 2450]\n[2459, 2460]\n[2463]\n[2472]\n[2482]\n[2493]\n[2503]\n[2515]\n[2533]\n[2538, 2539, 2540]\n[2546]\n[2563]\n[2565, 2566]\n[2574, 2575]\n[2580]\n[2584]\n[2588]\n[2599]\n[2603]\n[2644, 2645, 2646, 2647]\n[2663]\n[2680]\n[2731]\n[2755]\n[2766]\n[2772]\n[2787]\n[2804, 2805]\n[2832]\n[2893, 2894, 2895, 2896, 2897]\n[2950]\n[2967, 2968, 2969, 2970]\n[2972]\n[2992]\n[2994]\n[3073, 3074, 3075, 3076, 3077]\n[3095, 3096, 3097, 3098]\n[3164, 3165]\n[3169]\n[3188]\n[3234]\n[3242, 3243, 3244, 3245, 3246]\n[3303]\n[3345]\n[3364]\n[3375, 3376]\n[3396]\n[3408]\n[3420, 3421]\n[3611, 3612, 3613, 3614, 3615, 3616]\n[3621, 3622, 3623, 3624, 3625]\n[3663, 3664]\n[3667, 3668]\n[3709, 3710]\n[3743]\n[3752, 3753, 3754]\n[3786, 3787, 3788]\n[3816]\n[3828, 3829]\n[3833]\n[3941, 3942]\n[3964]\n[3977]\n[4139, 4140, 4141, 4142]\n[4167]\n[4174]\n[4208, 4209, 4210, 4211]\n[4230]\n[4242]\n[4282]\n[4307, 4308, 4309]\n[4339]\n[4342, 4343, 4344, 4345]\n[4349]\n[4386]\n[4395]\n[4404]\n[4433]\n[4443]\n[4467]\n[4637]\n[4639]\n[4641]\n[4643]\n[4645]\n[4648]\n[4707, 4708]\n[4726]\n[4803, 4804]\n[4838]\n[4849]\n[4879, 4880]\n[4892, 4893]\n[4895, 4896]\n[4932]\n[4934]\n[4965]\n[4968, 4969]\n[5007]\n[5011, 5012]\n[5024, 5025, 5026, 5027]\n[5050]\n[5089]\n[5124]\n[5155]\n[5199]\n[5204]\n[5222, 5223]\n[5319, 5320]\n[5429, 5430]\n[5437]\n[5446]\n[5459]\n[5490]\n[5521, 5522]\n[5568]\n[5589, 5590, 5591, 5592]\n[5600]\n[5621]\n[5632]\n[5649]\n[5655]\n[5685]\n[5711, 5712, 5713, 5714]\n[5722]\n[5728]\n[5743]\n[5753]\n[5775]\n[5790, 5791, 5792]\n[5814]\n[5834]\n[5887, 5888]\n[5912, 5913, 5914, 5915, 5916, 5917]\n[6085, 6086]\n[6091, 6092, 6093]\n[6109]\n[6112]\n[6125]\n[6148, 6149]\n[6172]\n[6203, 6204]\n[6217]\n[6224, 6225, 6226, 6227, 6228]\n[6244]\n[6249]\n[6254]\n[6257]\n[6267, 6268, 6269, 6270]\n[6296]\n[6310, 6311]\n[6350]\n[6357]\n[6374, 6375, 6376]\n[6388, 6389]\n[6460, 6461, 6462, 6463, 6464]\n[6521]\n[6547]\n[6605, 6606]\n[6635, 6636]\n[6643, 6644, 6645, 6646]\n[6714, 6715, 6716]\n[6939, 6940, 6941, 6942]\n[6970]\n[7006]\n[7052]\n[7140, 7141]\n[7273, 7274]\n[7292]\n[7368, 7369]\n[7413]\n[7432]\n[7452, 7453, 7454, 7455, 7456]\n[7471, 7472]\n[7514]\n[7571, 7572]\n[7602, 7603]\n[7635]\n[7692, 7693]\n[7785, 7786, 7787]\n[7856]\n[7875, 7876]\n[7884]\n[7900]\n[7932, 7933, 7934, 7935]\n[7967]\n[8081, 8082]\n[8087]\n[8097]\n[8125]\n[8168]\n[8176]\n[8181]\n[8194]\n[8204]\n[8218, 8219, 8220, 8221]\n[8223, 8224, 8225]\n[8227, 8228, 8229]\n[8232, 8233, 8234]\n[8250, 8251, 8252, 8253]\n[8277, 8278]\n[8281]\n[8287, 8288, 8289, 8290, 8291]\n[8305]\n[8307]\n[8319, 8320]\n[8327]\n[8331, 8332]\n[8337]\n[8357]\n[8372]\n[8400]\n[8413, 8414, 8415]\n[8432]\n[8437]\n[8444]\n[8454, 8455]\n[8457, 8458]\n[8461, 8462]\n[8464, 8465, 8466, 8467]\n[8476]\n[8490]\n[8497]\n[8499, 8500]\n[8505, 8506]\n[8510, 8511]\n[8532, 8533]\n[8539, 8540]\n[8548, 8549]\n[8570]\n[8578, 8579]\n[8613, 8614, 8615, 8616, 8617, 8618, 8619]\n[8674]\n[8689]\n[8711]\n[8768, 8769, 8770]\n[8788, 8789, 8790]\n[8801, 8802, 8803]\n[8833, 8834, 8835, 8836]\n[8891]\n[8954, 8955, 8956, 8957]\n[9167, 9168, 9169, 9170, 9171, 9172]\n[9236, 9237, 9238, 9239, 9240]\n[9282, 9283, 9284]\n[9290]\n[9368, 9369]\n[9387, 9388, 9389]\n[9394, 9395]\n[9539, 9540]\n[9743]\n[9867, 9868]\n[9876, 9877, 9878, 9879]\n[9989]\n[10029]\n[10058]\n[10062]\n[10265, 10266, 10267, 10268]\n[10418, 10419, 10420, 10421]\n[10515, 10516, 10517]\n[10587, 10588]\n[10756, 10757, 10758, 10759]\n[10772]\n[10789, 10790, 10791, 10792]\n[10879, 10880, 10881]\n[10895]\n[11163, 11164, 11165]\n[11167, 11168, 11169]\n[11171, 11172, 11173, 11174, 11175]\n[11177, 11178]\n[11180, 11181, 11182]\n[11184, 11185]\n[11187, 11188]\n[11190, 11191, 11192]\n[11194, 11195, 11196]\n[11199, 11200, 11201, 11202]\n[11272, 11273]\n[11324, 11325, 11326]\n[11413, 11414, 11415, 11416]\n[11418, 11419]\n[11433, 11434]\n[11456, 11457]\n[11473]\n[11501, 11502]\n[11524, 11525]\n[11535, 11536]\n[11549, 11550]\n[11802]\n[11821]\n[11830]\n[11921]\n[11938]\n[12002]\n[12496]\n[12636]\n[13248, 13249, 13250, 13251, 13252, 13253]\n[13319]\n[13658, 13659, 13660, 13661]\n[13744, 13745, 13746, 13747, 13748, 13749]\n[13790]\n[13804]\n[13865]\n[13933]\n[13957]\n[14047, 14048]\n[14052, 14053]\n[14066, 14067]\n[14108, 14109]\n[14146, 14147]\n[14157, 14158]\n[14161, 14162]\n[14171, 14172]\n[14188, 14189]\n[14212, 14213]\n[14258, 14259]\n[14294, 14295]\n[14325, 14326]\n[14329, 14330]\n[14343, 14344]\n[14357, 14358]\n[14438]\n[14597]\n[14649, 14650]\n[14656]\n[14675]\n[14975, 14976, 14977, 14978, 14979, 14980]\n[15044, 15045]\n[15191]\n[15206]\n[15241]\n[15692]\n[16388]\n[16487]\n[16506, 16507, 16508, 16509, 16510, 16511]\n[16519]\n[16556]\n[16604]\n[16634]\n[16660]\n[16696]\n[16766]\n[16797]\n[16845, 16846, 16847, 16848, 16849]\n[16861, 16862, 16863]\n[16913]\n[17009]\n[17013]\n[17077, 17078]\n[17294, 17295, 17296]\n[17908, 17909, 17910]\n[18251, 18252, 18253, 18254, 18255, 18256, 18257, 18258, 18259]\n[18280]\n[18319]\n[18354]\n[18689, 18690, 18691, 18692, 18693, 18694, 18695]\n[18697, 18698, 18699]\n[18915, 18916, 18917, 18918, 18919, 18920, 18921]\n[19214]\n[19288, 19289, 19290, 19291, 19292, 19293]\n[19321]\n[19400]\n[19474]\n[19495]\n[19579, 19580]\n[19700]\n[19759]\n[19765, 19766, 19767, 19768, 19769, 19770]\n[19930, 19931, 19932, 19933, 19934, 19935, 19936]\n[20212]\n[20293]\n[20423]\n[20475]\n[20496, 20497, 20498, 20499, 20500]\n[20529]\n[20534]\n[21043, 21044, 21045, 21046, 21047]\n[21069]\n[21132]\n[21269]\n[21287, 21288]\n[21336]\n[21339]\n[21354]\n[21374]\n[21410]\n[21459]\n[21471]\n[21537]\n[21614]\n[21623, 21624, 21625, 21626, 21627]\n[21637, 21638, 21639]\n[21777]\n[21795, 21796]\n[21848, 21849, 21850, 21851, 21852, 21853, 21854, 21855, 21856, 21857, 21858]\n[21862]\n[21977, 21978]\n[22118, 22119, 22120, 22121, 22122]\n[22144]\n[22207]\n[22344]\n[22426]\n[22480, 22481]\n[23661, 23662, 23663]\n[23685]\n[23777]\n[23893, 23894, 23895, 23896, 23897, 23898]\n[23964, 23965]\n[23978, 23979]\n[24012]\n[24017]\n[24067, 24068, 24069]\n[24107]\n[24118, 24119, 24120]\n[24123, 24124]\n[24134, 24135]\n[24158, 24159]\n[24165, 24166]\n[24175, 24176, 24177, 24178]\n[24181]\n[24206]\n[24228]\n[24248]\n[24301, 24302]\n[24396]\n[24421]\n[24898, 24899, 24900, 24901]\n[25119, 25120, 25121, 25122, 25123, 25124, 25125]\n[25149]\n[25205]\n[25352]\n[25365]\n[25390]\n[25412]\n[25435]\n[25550, 25551, 25552]\n[25646]\n[25922]\n[26071]\n[26122]\n[26209, 26210, 26211, 26212, 26213, 26214]\n[26299, 26300, 26301, 26302, 26303, 26304]\n[26348]\n[26353]\n[26393]\n[26397]\n[26402]\n[26461]\n[26600]\n[26771]\n[26773]\n[27198, 27199, 27200, 27201, 27202, 27203, 27204]\n[27479, 27480, 27481, 27482]\n[27542, 27543, 27544]\n[27558, 27559, 27560, 27561, 27562, 27563, 27564]\n[27806, 27807, 27808]\n[27814, 27815, 27816, 27817, 27818, 27819, 27820]\n[27845]\n[27889, 27890, 27891, 27892]\n[28345, 28346, 28347, 28348, 28349, 28350, 28351]\n[28414, 28415]\n[28490]\n[28492, 28493]\n[28557, 28558, 28559]\n[28685, 28686]\n[28821, 28822, 28823]\n[28835]\n[28850]\n[28863, 28864, 28865, 28866]\n[28886, 28887]\n[28889]\n[28891]\n[28915, 28916, 28917, 28918]\n[28967]\n[29574]\n[29740, 29741]\n[29777, 29778]\n[29803]\n[29831]\n[29881]\n[29895]\n[29956]\n[30020]\n[30074]\n[30144]\n[30212, 30213, 30214]\n[30489]\n[30526]\n[30636]\n[30915]\n[30923, 30924]\n[30963, 30964]\n[30989, 30990]\n[31050, 31051, 31052, 31053, 31054]\n[31133, 31134]\n[31143, 31144, 31145, 31146]\n[31184, 31185]\n[31228, 31229]\n[31250, 31251, 31252]\n[31341]\n[31858]\n[31875, 31876, 31877, 31878]\n[31894]\n[31907]\n[31913]\n[31952]\n[31957, 31958]\n[31960]\n[32135]\n[32137, 32138]\n[32187]\n[32257, 32258]\n[32424, 32425]\n[32656]\n[32678]\n[32705]\n[32753]\n[33052, 33053, 33054]\n[33241]\n[33277]\n[33440, 33441]\n[33572, 33573, 33574, 33575, 33576, 33577, 33578]\n[33600]\n[33626]\n[33628]\n[33632]\n[33671]\n[33716]\n[33809]\n[34004]\n[34033, 34034, 34035]\n[34038]\n[34067]\n[34106, 34107, 34108]\n[34110]\n[34165, 34166]\n[34195]\n[34355]\n[34487, 34488, 34489]\n[34503, 34504, 34505]\n[34542, 34543]\n[34606, 34607]\n[34637, 34638]\n[34647, 34648, 34649, 34650, 34651, 34652]\n[34675]\n[34683, 34684]\n[34688, 34689]\n[34713]\n[35157, 35158, 35159]\n[35469, 35470, 35471, 35472]\n[35491]\n[35495]\n[35521]\n[35710, 35711, 35712, 35713]\n[35743, 35744, 35745]\n[35838, 35839, 35840, 35841]\n[36008, 36009, 36010]\n[36079, 36080, 36081]\n[36183, 36184, 36185, 36186]\n[36219]\n[36362, 36363, 36364, 36365]\n[36370, 36371, 36372, 36373, 36374]\n[36400, 36401]\n[36404, 36405]\n[36410, 36411, 36412]\n[36417, 36418, 36419, 36420]\n[36563]\n[36612]\n[36630, 36631, 36632]\n[36689, 36690, 36691, 36692]\n[36702, 36703, 36704]\n[36722, 36723, 36724]\n[36753]\n[36771, 36772, 36773, 36774, 36775, 36776]\n[36816, 36817]\n[36845, 36846]\n[36848]\n[36850]\n[36859, 36860]\n[36862, 36863, 36864, 36865]\n[36890]\n[36895]\n[36901]\n[36905]\n[36923]\n[36935]\n[36941]\n[36951]\n[36994]\n[37000, 37001, 37002]\n[37006, 37007, 37008]\n[37053, 37054, 37055]\n[37074, 37075]\n[37080, 37081, 37082]\n[37104]\n[37131]\n[37483, 37484, 37485]\n[37540]\n[37706, 37707, 37708, 37709]\n[37727]\n[37739]\n[37768]\n[37788]\n[37795]\n[37808]\n[37816]\n[37823, 37824]\n[37833, 37834]\n[37846, 37847]\n[37857, 37858]\n[37860, 37861, 37862, 37863]\n[37875]\n[37894]\n[37896]\n[38011, 38012]\n[38052, 38053, 38054, 38055]\n[38297, 38298, 38299, 38300]\n[38371]\n[38483]\n[38485]\n[38488]\n[38524]\n[38527, 38528]\n[38541]\n[38543]\n[38657, 38658, 38659]\n[38663, 38664]\n[38676]\n[38718, 38719]\n[38723]\n[38739]\n[38751]\n[38816]\n[38976, 38977]\n[39076, 39077]\n[39301, 39302]\n[39814, 39815, 39816, 39817]\n[39908, 39909]\n[40000, 40001, 40002]\n[40022]\n[40040, 40041, 40042]\n[40147]\n[40159, 40160]\n[40187, 40188]\n[40214, 40215]\n[40274, 40275]\n[40309]\n[40399, 40400]\n[40415, 40416]\n[40532, 40533]\n[40613, 40614]\n[40699, 40700, 40701]\n[40724, 40725, 40726, 40727]\n[40878, 40879, 40880]\n[40892, 40893]\n[40905]\n[40942, 40943]\n[40962, 40963]\n[40985, 40986]\n[41039, 41040]\n[41048, 41049]\n[41096, 41097]\n[41114, 41115]\n[41218, 41219]\n[41234, 41235]\n[41244, 41245]\n[41272, 41273]\n[41280, 41281, 41282, 41283]\n[41541, 41542]\n[41612, 41613]\n[41651, 41652]\n[41721, 41722]\n[41783]\n[41883, 41884]\n[42019, 42020]\n[42025, 42026]\n[42042, 42043]\n[42121, 42122]\n[42201, 42202]\n[42272, 42273]\n[42294, 42295]\n[42329, 42330, 42331]\n[42334, 42335, 42336]\n[42407, 42408]\n[42574, 42575]\n[42625, 42626]\n[43019, 43020]\n[43054, 43055]\n[43063, 43064]\n[43165, 43166]\n[43194, 43195]\n[43202, 43203]\n[43219, 43220]\n[43242, 43243]\n[43249, 43250]\n[43587, 43588]\n[43733, 43734, 43735]\n[43912, 43913]\n[44024, 44025]\n[44082, 44083]\n[44165, 44166]\n[44168]\n[44241, 44242]\n[44244]\n[44287, 44288]\n[44315]\n[44370, 44371]\n[44435, 44436, 44437, 44438]\n[44720, 44721]\n[44871, 44872, 44873, 44874]\n[44934, 44935, 44936, 44937]\n[45023, 45024, 45025, 45026, 45027]\n[45289, 45290]\n[45296, 45297, 45298, 45299]\n[45454, 45455]\n[45577, 45578]\n[45592, 45593, 45594, 45595]\n[45598, 45599, 45600]\n[45816, 45817]\n[45854, 45855, 45856, 45857, 45858]\n[45867, 45868]\n[45981, 45982]\n[45995, 45996]\n[46039, 46040]\n[46061]\n[46077, 46078, 46079]\n[46148]\n[46193]\n[46241]\n[46277]\n[46359, 46360, 46361, 46362, 46363, 46364]\n[46463, 46464, 46465, 46466]\n[46630, 46631]\n[46752, 46753]\n[46790, 46791]\n[46852, 46853]\n[46967, 46968]\n[47052, 47053]\n[47068, 47069]\n[47094]\n[47122]\n[47126]\n[47134]\n[47144]\n[47173, 47174, 47175]\n[47182, 47183, 47184, 47185, 47186]\n[47281]\n[47447, 47448, 47449]\n[47562]\n[47570]\n[47576]\n[47591]\n[47636]\n[47657]\n[47712]\n[47810, 47811]\n[47815]\n[47882]\n[47887, 47888, 47889, 47890]\n[47909, 47910, 47911, 47912, 47913, 47914]\n[47992]\n[48015, 48016]\n[48020]\n[48064]\n[48066]\n[48074, 48075]\n[48123, 48124, 48125, 48126]\n[48162]\n[48246, 48247]\n[48284]\n[48355]\n[48420, 48421, 48422, 48423]\n[48460, 48461, 48462, 48463]\n[48515, 48516]\n[48617]\n[48632]\n[48648, 48649]\n[48668]\n[48725]\n[48731, 48732]\n[48755, 48756, 48757, 48758, 48759]\n[48770]\n[48780]\n[48790]\n[48808]\n[48815, 48816]\n[48898]\n[48900]\n[48948]\n[49055, 49056]\n[49081, 49082, 49083, 49084, 49085, 49086]\n[49119]\n[49129]\n[49168]\n[49217]\n[49248]\n[49253, 49254]\n[49283]\n[49306]\n[49331]\n[49343, 49344, 49345, 49346, 49347]\n[49376]\n[49381, 49382]\n[49424, 49425]\n[49438, 49439]\n[49458]\n[49496, 49497, 49498, 49499]\n[49501]\n[49513]\n[49544]\n[49776]\n[49820, 49821, 49822, 49823]\n[49852]\n[49864, 49865]\n[49892]\n[49907]\n[49922, 49923]\n[49940]\n[49974, 49975, 49976]\n[50011, 50012, 50013, 50014]\n[50016, 50017]\n[50040, 50041, 50042, 50043, 50044]\n[50066, 50067]\n[50130]\n[50134]\n[50182]\n[50371, 50372]\n[50422, 50423, 50424, 50425, 50426]\n[50508, 50509]\n[50572, 50573]\n[50593]\n[51018, 51019, 51020, 51021]\n[51157, 51158]\n[51311, 51312, 51313]\n[51317, 51318, 51319, 51320]\n[51351]\n[51386]\n[51433, 51434, 51435, 51436, 51437, 51438]\n[51458, 51459, 51460]\n[51516]\n[51671, 51672]\n[51921, 51922]\n[51949, 51950, 51951, 51952]\n[52028, 52029, 52030, 52031]\n[52035]\n[52223, 52224, 52225]\n[52247, 52248]\n[52260, 52261]\n[52352]\n[52390, 52391, 52392, 52393]\n[52396, 52397, 52398, 52399, 52400]\n[52425, 52426]\n[52443, 52444]\n[52516]\n[52549, 52550]\n[52606, 52607]\n[52716, 52717, 52718, 52719]\n[52768, 52769, 52770]\n[52788, 52789]\n[52841]\n[52944, 52945]\n[53091]\n[53131, 53132, 53133, 53134, 53135, 53136]\n[53143, 53144]\n[53166, 53167]\n[53230]\n[53235, 53236, 53237]\n[53248, 53249]\n[53266]\n[53293, 53294, 53295, 53296]\n[53319]\n[53339, 53340]\n[53357, 53358]\n[53410]\n[53422, 53423, 53424, 53425, 53426]\n[53448]\n[53578]\n[53638]\n[53653, 53654, 53655]\n[53679, 53680, 53681, 53682]\n[53704]\n[53943, 53944, 53945, 53946, 53947]\n[54092, 54093]\n[54106, 54107]\n[54202, 54203, 54204, 54205, 54206, 54207, 54208]\n[54210, 54211, 54212]\n[54282, 54283, 54284, 54285, 54286, 54287, 54288, 54289, 54290, 54291, 54292, 54293, 54294, 54295, 54296]\n[54298, 54299, 54300, 54301, 54302]\n[54306]\n[54315]\n[54328, 54329, 54330]\n[54343, 54344]\n[54349, 54350, 54351, 54352]\n[54388, 54389, 54390]\n[54430]\n[54483, 54484, 54485, 54486, 54487, 54488]\n[54519]\n[54550]\n[54558, 54559]\n[54683, 54684, 54685]\n[54819]\n[54825]\n[54846]\n[54864]\n[54882]\n[54890]\n[54894]\n[54906]\n[54924]\n[54952]\n[54981, 54982, 54983]\n[55020, 55021]\n[55050, 55051, 55052]\n[55074, 55075, 55076]\n[55226, 55227]\n[55284, 55285]\n[55307, 55308, 55309]\n[55323, 55324]\n[55602, 55603, 55604, 55605]\n[55718, 55719, 55720, 55721, 55722]\n[55754, 55755, 55756, 55757, 55758]\n[55805, 55806, 55807, 55808, 55809]\n[55823, 55824, 55825, 55826, 55827, 55828, 55829]\n[55849]\n[55852, 55853, 55854, 55855]\n[55914]\n[55919, 55920]\n[55971, 55972, 55973, 55974, 55975]\n[55985]\n[56100, 56101, 56102, 56103]\n[56128, 56129]\n[56218, 56219]\n[56236, 56237]\n[56292]\n[56310]\n[56324, 56325, 56326, 56327]\n[56334]\n[56384]\n[56403]\n[56443]\n[56467]\n[56514]\n[56571]\n[56592]\n[56600]\n[56618, 56619, 56620]\n[56632, 56633, 56634]\n[56653, 56654]\n[56656, 56657, 56658]\n[56682]\n[56707]\n[56709]\n[56720, 56721, 56722]\n[56928, 56929, 56930, 56931, 56932]\n[57100]\n[57133, 57134, 57135, 57136]\n[57204]\n[57239]\n[57306, 57307]\n[57364]\n[57413, 57414]\n[57435]\n[57465]\n[57489]\n[57497]\n[57507]\n[57519]\n[57538]\n[57550]\n[57561]\n[57569]\n[57586]\n[57592, 57593]\n[57596, 57597, 57598, 57599]\n[57603, 57604, 57605, 57606, 57607]\n[57624]\n[57627, 57628, 57629, 57630]\n[57646]\n[57654, 57655, 57656, 57657, 57658]\n[57663, 57664, 57665, 57666]\n[57680, 57681, 57682]\n[57693, 57694, 57695]\n[57699, 57700]\n[57702, 57703]\n[57910, 57911, 57912, 57913, 57914, 57915]\n[57922, 57923]\n[57959, 57960, 57961, 57962]\n[57975, 57976]\n[58003, 58004]\n[58065, 58066]\n[58110, 58111]\n[58129, 58130]\n[58138, 58139]\n[58204, 58205, 58206, 58207, 58208]\n[58260]\n[58287]\n[58296]\n[58382, 58383, 58384]\n[58451, 58452, 58453]\n[58508, 58509, 58510]\n[58512, 58513, 58514, 58515]\n[58520]\n[58531]\n[58540]\n[58544, 58545, 58546]\n[58548, 58549, 58550]\n[58561, 58562, 58563]\n[58565, 58566, 58567, 58568]\n[58667, 58668]\n[58681, 58682]\n[58688]\n[58722, 58723]\n[58746, 58747, 58748]\n[58755]\n[58804, 58805, 58806, 58807]\n[58854]\n[58856, 58857]\n[58961, 58962]\n[59057]\n[59119]\n[59202]\n[59225]\n[59237]\n[59240]\n[59258]\n[59281, 59282, 59283, 59284, 59285, 59286]\n[59305, 59306, 59307, 59308, 59309]\n[59330, 59331]\n[59366]\n[59381]\n[59394]\n[59470, 59471, 59472, 59473]\n[59493, 59494, 59495, 59496, 59497, 59498]\n[59502]\n[59508, 59509]\n[59514, 59515, 59516, 59517, 59518, 59519, 59520, 59521]\n[59608, 59609]\n[59626, 59627]\n[59720, 59721]\n[59799, 59800, 59801]\n[59825]\n[59847]\n[59862, 59863, 59864, 59865, 59866, 59867, 59868, 59869, 59870, 59871, 59872]\n[59875, 59876, 59877, 59878, 59879]\n[59905, 59906, 59907, 59908]\n[59951]\n[59976]\n[59988, 59989]\n[59991]\n[59996, 59997, 59998, 59999, 60000]\n[60027, 60028, 60029, 60030]\n[60057]\n[60099, 60100, 60101, 60102, 60103]\n[60233, 60234, 60235, 60236, 60237, 60238, 60239]\n[60274]\n[60286]\n[60328]\n[60339, 60340, 60341, 60342, 60343]\n[60374]\n[60378]\n[60388]\n[60408]\n[60773, 60774, 60775]\n[60956, 60957, 60958, 60959]\n[60966, 60967]\n[60973]\n[61091, 61092, 61093]\n[61172, 61173]\n[61235, 61236, 61237, 61238, 61239, 61240]\n[61365, 61366, 61367]\n[61369, 61370, 61371, 61372]\n[61388, 61389, 61390]\n[61392, 61393, 61394]\n[61401]\n[61404, 61405, 61406]\n[61423, 61424]\n[61437]\n[61501, 61502, 61503, 61504, 61505, 61506, 61507, 61508]\n[61512, 61513]\n[61525, 61526, 61527, 61528, 61529, 61530]\n[61550, 61551, 61552, 61553, 61554, 61555, 61556]\n[61563]\n[61596, 61597, 61598]\n[61600, 61601, 61602, 61603]\n[61609, 61610]\n[61617]\n[61638, 61639, 61640, 61641, 61642]\n[61644]\n[61686]\n[61696, 61697, 61698]\n[61741, 61742, 61743, 61744, 61745]\n[61766]\n[61775]\n[61780, 61781, 61782]\n[61824]\n[61864, 61865, 61866, 61867, 61868, 61869, 61870]\n[61910, 61911]\n[61928]\n[61942]\n[61984]\n[62096]\n[62117, 62118]\n[62153]\n[62155, 62156, 62157, 62158, 62159, 62160, 62161, 62162, 62163, 62164, 62165, 62166, 62167, 62168, 62169, 62170, 62171, 62172]\n[62324, 62325, 62326]\n[62448, 62449, 62450, 62451]\n[62533, 62534]\n[62546]\n[62594, 62595]\n[62597, 62598, 62599, 62600]\n[62644]\n[62815, 62816, 62817]\n[62889]\n[62973, 62974, 62975, 62976]\n[63141, 63142, 63143, 63144, 63145, 63146]\n[63176, 63177]\n[63179, 63180, 63181, 63182, 63183, 63184]\n[63186]\n[63196]\n[63243]\n[63321]\n[63383]\n[63410]\n[63442, 63443, 63444, 63445, 63446]\n[63624, 63625, 63626]\n[63759, 63760]\n[63803]\n[63814, 63815, 63816]\n[63909, 63910]\n[64028]\n[64219]\n[64259, 64260, 64261, 64262, 64263]\n[64308, 64309]\n[64332, 64333, 64334, 64335]\n[64337, 64338, 64339]\n[64386, 64387, 64388, 64389]\n[64407, 64408]\n[64412]\n[64427, 64428]\n[64443, 64444, 64445, 64446, 64447, 64448]\n[64460, 64461, 64462, 64463, 64464]\n[64481]\n[64495]\n[64504, 64505]\n[64561, 64562, 64563, 64564, 64565, 64566, 64567]\n[64569, 64570]\n[64648, 64649]\n[64871, 64872]\n[64879, 64880]\n[64910]\n[64940, 64941]\n[65034, 65035, 65036, 65037]\n[65067, 65068, 65069, 65070]\n[65087, 65088, 65089, 65090]\n[65200, 65201]\n[65245, 65246, 65247]\n[65270]\n[65343]\n[65357]\n[65374]\n[65383]\n[65507]\n[65532]\n[65589]\n[65630]\n[65640]\n[65664, 65665, 65666, 65667, 65668, 65669]\n"
],
[
"import math\n\ndef generate_index(l, name, texts, labels, length):\n cp, indices = [], []\n b = length - len(l)\n left = math.ceil(b / 2)\n right = b - left\n minus = l[0] - left\n if minus < 0:\n absolute = np.abs(minus)\n right += absolute\n left -= absolute\n\n for i in range(l[0] - left, l[0]):\n cp.append(texts[i])\n indices.append(labels[i])\n\n cp.extend(name)\n indices.extend([labels[l[0]] for _ in range(len(name))])\n try:\n for i in range(l[-1] + 1, l[-1] + right + 1):\n cp.append(texts[i])\n indices.append(labels[i])\n except Exception as e:\n print(e)\n pass\n \n return cp, indices",
"_____no_output_____"
],
[
"org.keys()",
"_____no_output_____"
],
[
"for i in range(8):\n local_companies.extend(org[str(i)])\n \nlocal_companies = list(set(local_companies))\nlen(local_companies)",
"_____no_output_____"
],
[
"from sklearn.model_selection import train_test_split\nimport random\n\ntrain_results, test_results = train_test_split(results, test_size = 0.2)\ntrain_local_companies, test_local_companies = train_test_split(local_companies, test_size = 0.2)",
"_____no_output_____"
],
[
"train_X, train_Y = [], []\n\nfor t in train_local_companies:\n x, y = generate_index(train_results[random.randint(0, len(train_results) - 1)], \n t.split(), entities['text'], entities['label'], 25)\n if len(x) != len(y):\n print('len not same')\n continue\n train_X.append(x)\n train_Y.append(y)",
"_____no_output_____"
],
[
"for r in train_results:\n for _ in range(3):\n x, y = generate_index(r, generate_org(len(r)).split(), entities['text'], entities['label'], 25)\n if len(x) != len(y):\n print('len not same')\n continue\n train_X.append(x)\n train_Y.append(y)",
"_____no_output_____"
],
[
"test_X, test_Y = [], []\n\nfor t in test_local_companies:\n x, y = generate_index(test_results[random.randint(0, len(test_results) - 1)], \n t.split(), entities['text'], entities['label'], 25)\n if len(x) != len(y):\n print('len not same')\n continue\n test_X.append(x)\n test_Y.append(y)",
"_____no_output_____"
],
[
"for r in test_results:\n for _ in range(3):\n x, y = generate_index(r, generate_org(len(r)).split(), entities['text'], entities['label'], 25)\n if len(x) != len(y):\n print('len not same')\n continue\n test_X.append(x)\n test_Y.append(y)",
"_____no_output_____"
],
[
"len(train_X), len(test_X)",
"_____no_output_____"
],
[
"with open('org-augmentation.json', 'w') as fopen:\n json.dump({'train_X': train_X, 'train_Y': train_Y, 'test_X': test_X, 'test_Y': test_Y}, fopen)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec70eae76e573fb4041c12d28fcfc89dee2ebd95 | 22,944 | ipynb | Jupyter Notebook | Python/Market Basket/Market Basket Example.ipynb | jbochenek/jbochenek.github.io | d436c079f1545133c257f51e091439647d3ecfcb | [
"Unlicense"
]
| null | null | null | Python/Market Basket/Market Basket Example.ipynb | jbochenek/jbochenek.github.io | d436c079f1545133c257f51e091439647d3ecfcb | [
"Unlicense"
]
| null | null | null | Python/Market Basket/Market Basket Example.ipynb | jbochenek/jbochenek.github.io | d436c079f1545133c257f51e091439647d3ecfcb | [
"Unlicense"
]
| null | null | null | 41.415162 | 2,098 | 0.518 | [
[
[
"# Market Basket",
"_____no_output_____"
],
[
"## An ingredient-based recommender system\nThis is an exmample of a recommender system using some recipes data and the Apriori algorithm. These data can be obtained from Kaggle:\n\n- https://www.kaggle.com/kaggle/recipe-ingredients-dataset\n\nand are packaged with the assignment in the following directory:\n\n- `./data/train.json`",
"_____no_output_____"
],
[
"To start, load the recipe data from `json` format and print the first 5 recipes.",
"_____no_output_____"
]
],
[
[
"import json\nfrom pprint import pprint\n\nwith open('./data/train.json', 'r') as train:\n recipe = json.load(train)\n\npprint(recipe [:4])",
"[{'cuisine': 'greek',\n 'id': 10259,\n 'ingredients': ['romaine lettuce',\n 'black olives',\n 'grape tomatoes',\n 'garlic',\n 'pepper',\n 'purple onion',\n 'seasoning',\n 'garbanzo beans',\n 'feta cheese crumbles']},\n {'cuisine': 'southern_us',\n 'id': 25693,\n 'ingredients': ['plain flour',\n 'ground pepper',\n 'salt',\n 'tomatoes',\n 'ground black pepper',\n 'thyme',\n 'eggs',\n 'green tomatoes',\n 'yellow corn meal',\n 'milk',\n 'vegetable oil']},\n {'cuisine': 'filipino',\n 'id': 20130,\n 'ingredients': ['eggs',\n 'pepper',\n 'salt',\n 'mayonaise',\n 'cooking oil',\n 'green chilies',\n 'grilled chicken breasts',\n 'garlic powder',\n 'yellow onion',\n 'soy sauce',\n 'butter',\n 'chicken livers']},\n {'cuisine': 'indian',\n 'id': 22213,\n 'ingredients': ['water', 'vegetable oil', 'wheat', 'salt']}]\n"
]
],
[
[
"Next, I wrote a function called `count_items(recipes)` that counts up the number of recipes that include each `ingredient`, storing each in the counter as a single-element tuple (for downstream convienience), i.e., incrementing like `counts[tuple([ingredient])] +=1`. \n\nWhen complete, exhibit this functions utility in application to the `recipes` and prints the number of 'candidates' in the output.",
"_____no_output_____"
]
],
[
[
"from collections import Counter\n\ndef count_items(recipes):\n counts = Counter()\n for r in recipes: \n for ingredient in r['ingredients']:\n counts[tuple([ingredient])] +=1\n return counts",
"_____no_output_____"
],
[
"i = count_items(recipe)\npprint(i.most_common(10))\n",
"[(('salt',), 18049),\n (('onions',), 7972),\n (('olive oil',), 7972),\n (('water',), 7457),\n (('garlic',), 7380),\n (('sugar',), 6434),\n (('garlic cloves',), 6237),\n (('butter',), 4848),\n (('ground black pepper',), 4785),\n (('all-purpose flour',), 4632)]\n"
]
],
[
[
"Then, I wrote a function called `store_frequent(candidates, threshold = 25)`, which accepts a `Counter` of `candidates`, i.e., item or itemset counts, and stores only those with count above the determined `threshold` value in a separate counter called `frequent`, which is `return`ed at the end of the function. ",
"_____no_output_____"
]
],
[
[
"def store_frequent(candidates, threshold = 25):\n frequent=[]\n for c, k in candidates.items():\n if k >= threshold:\n frequent.append([c,k])\n frequent = Counter(dict(frequent)) \n '''coercing the list i made back into a counter as frequent works best as counter not as a dict'''\n return frequent",
"_____no_output_____"
],
[
"frequent = store_frequent(count_items(recipe),25) \n\npprint(frequent.most_common(5)) # the top 5 ingredients to check that it worked \npprint(len(frequent)) #how many ingredients are higher than 25 in count",
"[(('salt',), 18049),\n (('onions',), 7972),\n (('olive oil',), 7972),\n (('water',), 7457),\n (('garlic',), 7380)]\n1487\n"
]
],
[
[
"Then I wrote a function called `get_next(recipes, frequent, threshold = 25)` that accepts the `frequent` items output from the `store_frequent()` function. With these inputs, your function should:\n\n1. create a new `Counter` called `next_candidates`\n2. compute the `size` of the itemsets for `next_candidates` from a single key in `frequent`\n3. `for` any `recipe` with _at least_ as many ingredients as `size`:\n 1. loop over all itemsets of size `size` (see combinations note below)\n 2. utilize the apriori principle and subsets of itemsets to count up potentially-frequent candidate itemsets in `next_candidates`\n4. `return(next_candidates)` \n",
"_____no_output_____"
],
[
"This is generating fewer candidates but more frequent as it increases the `size`. This follows the image below: \n\n\nAs this gets larger, it has to make combinations of ingredients from the recipes and then it has to add in all other combinations to the item set before the items less than the threshold is cut off. ",
"_____no_output_____"
]
],
[
[
"from itertools import combinations\nimport random\n\ndef get_next(recipe, frequent, threshold = 25):\n next_candidate=Counter()\n nonviable = set()\n size = len(list(frequent)[0])+1\n for re in recipe:\n if len(re['ingredients']) >= size:\n items = re['ingredients']\n for candidate in combinations(items, size):\n if candidate in next_candidate:\n next_candidate[candidate] += 1\n elif candidate not in nonviable:\n for subitemset in combinations(candidate, size-1):\n if subitemset not in frequent:\n nonviable.add(candidate)\n break\n else:\n next_candidate[candidate] += 1 \n return next_candidate\n ",
"_____no_output_____"
],
[
"next_candidate2 = get_next(recipe, frequent, threshold=25)\nprint(next_candidate2.most_common(50))",
"[(('salt', 'onions'), 2641), (('olive oil', 'salt'), 2636), (('water', 'salt'), 2503), (('pepper', 'salt'), 2427), (('sugar', 'salt'), 1915), (('salt', 'garlic'), 1907), (('garlic', 'salt'), 1842), (('onions', 'salt'), 1751), (('ground black pepper', 'salt'), 1720), (('garlic', 'onions'), 1690), (('salt', 'garlic cloves'), 1605), (('all-purpose flour', 'salt'), 1567), (('salt', 'olive oil'), 1544), (('salt', 'all-purpose flour'), 1512), (('butter', 'salt'), 1497), (('salt', 'water'), 1457), (('salt', 'pepper'), 1417), (('garlic cloves', 'salt'), 1393), (('olive oil', 'onions'), 1366), (('olive oil', 'garlic cloves'), 1331), (('olive oil', 'garlic'), 1312), (('salt', 'butter'), 1280), (('water', 'onions'), 1245), (('eggs', 'salt'), 1198), (('vegetable oil', 'salt'), 1157), (('salt', 'sugar'), 1146), (('black pepper', 'salt'), 1125), (('large eggs', 'salt'), 1073), (('tomatoes', 'salt'), 1040), (('salt', 'ground black pepper'), 1017), (('water', 'garlic'), 1014), (('salt', 'ground cumin'), 971), (('onions', 'garlic'), 969), (('salt', 'vegetable oil'), 944), (('pepper', 'onions'), 910), (('unsalted butter', 'salt'), 896), (('garlic cloves', 'onions'), 894), (('milk', 'salt'), 842), (('onions', 'olive oil'), 841), (('water', 'garlic cloves'), 837), (('pepper', 'garlic'), 836), (('sugar', 'water'), 816), (('salt', 'carrots'), 811), (('butter', 'all-purpose flour'), 803), (('ground black pepper', 'garlic'), 796), (('olive oil', 'ground black pepper'), 783), (('sugar', 'all-purpose flour'), 781), (('garlic cloves', 'olive oil'), 772), (('garlic', 'olive oil'), 768), (('water', 'sugar'), 760)]\n"
],
[
"next_candidate3 = get_next(recipe, next_candidate2, threshold=25)\nprint(next_candidate3.most_common(50))",
"[(('pepper', 'salt', 'onions'), 397), (('garlic', 'onions', 'salt'), 394), (('water', 'salt', 'onions'), 370), (('salt', 'garlic', 'onions'), 369), (('olive oil', 'salt', 'onions'), 360), (('salt', 'onions', 'garlic'), 339), (('olive oil', 'salt', 'garlic cloves'), 293), (('salt', 'pepper', 'garlic'), 289), (('salt', 'olive oil', 'garlic'), 276), (('garlic', 'salt', 'onions'), 269), (('pepper', 'olive oil', 'salt'), 261), (('salt', 'butter', 'all-purpose flour'), 260), (('olive oil', 'garlic', 'onions'), 255), (('garlic', 'pepper', 'salt'), 253), (('olive oil', 'garlic', 'salt'), 248), (('garlic', 'olive oil', 'salt'), 245), (('olive oil', 'salt', 'pepper'), 244), (('salt', 'sugar', 'all-purpose flour'), 243), (('pepper', 'salt', 'garlic'), 243), (('salt', 'baking powder', 'all-purpose flour'), 243), (('garlic cloves', 'olive oil', 'salt'), 240), (('pepper', 'onions', 'salt'), 239), (('olive oil', 'pepper', 'salt'), 234), (('water', 'garlic', 'onions'), 230), (('olive oil', 'salt', 'garlic'), 229), (('ground black pepper', 'salt', 'onions'), 226), (('all-purpose flour', 'sugar', 'salt'), 225), (('garlic', 'water', 'salt'), 224), (('salt', 'water', 'garlic'), 222), (('tomatoes', 'salt', 'onions'), 222), (('onions', 'pepper', 'salt'), 221), (('olive oil', 'ground black pepper', 'salt'), 220), (('olive oil', 'salt', 'ground black pepper'), 219), (('pepper', 'garlic', 'salt'), 218), (('pepper', 'salt', 'olive oil'), 217), (('salt', 'water', 'onions'), 211), (('all-purpose flour', 'baking powder', 'salt'), 208), (('onions', 'olive oil', 'salt'), 208), (('olive oil', 'garlic cloves', 'salt'), 207), (('water', 'onions', 'salt'), 207), (('salt', 'pepper', 'onions'), 202), (('onions', 'water', 'salt'), 199), (('salt', 'olive oil', 'garlic cloves'), 198), (('salt', 'large eggs', 'all-purpose flour'), 197), (('salt', 'unsalted butter', 'all-purpose flour'), 195), (('pepper', 'garlic', 'onions'), 195), (('salt', 'olive oil', 'onions'), 193), (('sugar', 'large eggs', 'salt'), 192), (('salt', 'onions', 'pepper'), 190), (('butter', 'all-purpose flour', 'salt'), 189)]\n"
]
],
[
[
"With this I have the pieces to run Apriori/collect frequent itemsets, collecting all frequent itemsets up to a particular `size`. To do this, I wrote a function called `train(recipes, size = 4)`, which:\n\n1. initializes two empty dictionaries, `candidates`, and `frequent`;\n2. runs the `count_items` and `store_frequent` function, storing output in the `candidates`, and `frequent` dictionaries using the integer `1` as a key;\n3. loops over sizes: 2, 3, .., `size` to compute and store the subsequent sizes candidates and frequent itemsets in the same structure as (2), but now utilizing the `get_next` function, instead of `count_items`; and\n4. `return`s the `candidates` and `frequent` itemsets.",
"_____no_output_____"
]
],
[
[
"def train(recipes, size = 4):\n candidates=dict()\n frequent=dict()\n i = 1\n candidates[i] = count_items(recipes)\n frequent[i] = store_frequent(candidates[i], threshold=25)\n while i <= (size-1):\n i=i+1\n candidates[i]= get_next(recipes, candidates[i-1], threshold = 25)\n frequent[i] = store_frequent(candidates[i], threshold=25) #these return the same thing...\n return candidates, frequent",
"_____no_output_____"
],
[
"candidates, frequent=train(random.sample(recipe,8000),size=4)",
"_____no_output_____"
],
[
"candidates, frequent=train(recipe,size=4)",
"_____no_output_____"
],
[
"pprint(candidates[3].most_common(30))\npprint(frequent[3].most_common(30))",
"[(('pepper', 'salt', 'onions'), 397),\n (('garlic', 'onions', 'salt'), 394),\n (('water', 'salt', 'onions'), 370),\n (('salt', 'garlic', 'onions'), 369),\n (('olive oil', 'salt', 'onions'), 360),\n (('salt', 'onions', 'garlic'), 339),\n (('olive oil', 'salt', 'garlic cloves'), 293),\n (('salt', 'pepper', 'garlic'), 289),\n (('salt', 'olive oil', 'garlic'), 276),\n (('garlic', 'salt', 'onions'), 269),\n (('pepper', 'olive oil', 'salt'), 261),\n (('salt', 'butter', 'all-purpose flour'), 260),\n (('olive oil', 'garlic', 'onions'), 255),\n (('garlic', 'pepper', 'salt'), 253),\n (('olive oil', 'garlic', 'salt'), 248),\n (('garlic', 'olive oil', 'salt'), 245),\n (('olive oil', 'salt', 'pepper'), 244),\n (('salt', 'sugar', 'all-purpose flour'), 243),\n (('pepper', 'salt', 'garlic'), 243),\n (('salt', 'baking powder', 'all-purpose flour'), 243),\n (('garlic cloves', 'olive oil', 'salt'), 240),\n (('pepper', 'onions', 'salt'), 239),\n (('olive oil', 'pepper', 'salt'), 234),\n (('water', 'garlic', 'onions'), 230),\n (('olive oil', 'salt', 'garlic'), 229),\n (('ground black pepper', 'salt', 'onions'), 226),\n (('all-purpose flour', 'sugar', 'salt'), 225),\n (('garlic', 'water', 'salt'), 224),\n (('salt', 'water', 'garlic'), 222),\n (('tomatoes', 'salt', 'onions'), 222)]\n[(('pepper', 'salt', 'onions'), 397),\n (('garlic', 'onions', 'salt'), 394),\n (('water', 'salt', 'onions'), 370),\n (('salt', 'garlic', 'onions'), 369),\n (('olive oil', 'salt', 'onions'), 360),\n (('salt', 'onions', 'garlic'), 339),\n (('olive oil', 'salt', 'garlic cloves'), 293),\n (('salt', 'pepper', 'garlic'), 289),\n (('salt', 'olive oil', 'garlic'), 276),\n (('garlic', 'salt', 'onions'), 269),\n (('pepper', 'olive oil', 'salt'), 261),\n (('salt', 'butter', 'all-purpose flour'), 260),\n (('olive oil', 'garlic', 'onions'), 255),\n (('garlic', 'pepper', 'salt'), 253),\n (('olive oil', 'garlic', 'salt'), 248),\n (('garlic', 'olive oil', 'salt'), 245),\n (('olive oil', 'salt', 'pepper'), 244),\n (('salt', 'sugar', 'all-purpose flour'), 243),\n (('pepper', 'salt', 'garlic'), 243),\n (('salt', 'baking powder', 'all-purpose flour'), 243),\n (('garlic cloves', 'olive oil', 'salt'), 240),\n (('pepper', 'onions', 'salt'), 239),\n (('olive oil', 'pepper', 'salt'), 234),\n (('water', 'garlic', 'onions'), 230),\n (('olive oil', 'salt', 'garlic'), 229),\n (('ground black pepper', 'salt', 'onions'), 226),\n (('all-purpose flour', 'sugar', 'salt'), 225),\n (('garlic', 'water', 'salt'), 224),\n (('salt', 'water', 'garlic'), 222),\n (('tomatoes', 'salt', 'onions'), 222)]\n"
]
],
[
[
"With the `frequent` itemsets up to `size`, I can utilize them to recommend missing ingredients from ingredient 'baskets' of at most `size - 1`. To do this, I wrote a function called `recommend(basket, frequent)` that does the following: \n\n1. initializes an empty `recommendations` list\n2. loops over all frequent `itemset`s of `size 1 greater than the `basket`\n - if there's one item left from the `itemset` when the `basket` removed, append the remaining item to the `recommendations` list in a tuple, with the number of ocurrences of the itemset in the second position\n4. `return` `recommendations`, but sorted from high to low by itemset ocurrence.\n\nWith this code complete, I have reported the top 10 recommended items to buy for recipe flexibility in the following scenarios:\n\n- `basket = tuple(['butter', 'flour'])`\n- `basket = tuple(['soy sauce', 'green onions'])`\n- `basket = tuple(['avocado', 'garlic', 'salt'])`",
"_____no_output_____"
],
[
"The function works well in pointing out additional items to add, however due to the fact that the trained data is sensitive to order in the items it comes up with the same items multiple times with different counts. This is because they used to be [(flour, butter, salt), ###] and [(butter, flour, salt), ###] and the result shows just salt with different numbers. However, it is performing within the specified limits of the question. \n\nThe data does not have a suggestion for something that goes with avocado, garlic and salt, but appropriately suggest other goods needed for baking when supplied with butter and flour, which are hallmarks of baking recipes. ",
"_____no_output_____"
]
],
[
[
"import operator\ndef recommend(basket, frequent):\n recommendations = []\n size = len(frequent)\n if (len(basket)-1) < size:\n for i in frequent:\n if (len(basket)+1) == i:\n\n for l,v in frequent[i].items():\n toup = set(basket + l)\n \n if len(toup) == i:\n\n ple = [s for s in l if s not in basket]\n recommendations.append(tuple([ple, v]))\n else:\n print(\"Cannot compute recommended next item in basket, trained itemset not large enough\")\n recommendations.sort(key = operator.itemgetter(1), reverse = True)\n return recommendations",
"_____no_output_____"
],
[
"basket = tuple(['butter', 'flour'])\n\nrecommendations = recommend(basket, frequent)\n\nprint(recommendations[:9])\n#there is some overlap that occurs with the items as the combinations stored in the first D5 code is sensitive to order, so (salt,butter,flour) and (butter,flour,salt) are separate items and have different counts.\n",
"[(['salt'], 67), (['salt'], 61), (['salt'], 56), (['salt'], 48), (['eggs'], 48), (['salt'], 39), (['milk'], 36), (['salt'], 35), (['sugar'], 35)]\n"
],
[
"\nbasket = tuple(['soy sauce', 'green onions'])\nrecommendations = recommend(basket, frequent)\n\nprint(recommendations[:9])",
"[(['sesame oil'], 93), (['garlic'], 91), (['sesame oil'], 88), (['sesame oil'], 80), (['salt'], 74), (['garlic'], 66), (['garlic'], 65), (['sesame oil'], 60), (['garlic'], 59)]\n"
],
[
"basket = tuple(['avocado', 'garlic', 'salt'])\nrecommendations = recommend(basket, frequent)\n\nprint(recommendations[:9])",
"[]\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
]
]
|
ec70f41a49d4758ab3ca9eb551709e35d4f11393 | 2,619 | ipynb | Jupyter Notebook | examples/gallery/demos/bokeh/quiver_demo.ipynb | stuarteberg/holoviews | 65136173014124b41cee00f5a0fee82acdc78f7f | [
"BSD-3-Clause"
]
| null | null | null | examples/gallery/demos/bokeh/quiver_demo.ipynb | stuarteberg/holoviews | 65136173014124b41cee00f5a0fee82acdc78f7f | [
"BSD-3-Clause"
]
| null | null | null | examples/gallery/demos/bokeh/quiver_demo.ipynb | stuarteberg/holoviews | 65136173014124b41cee00f5a0fee82acdc78f7f | [
"BSD-3-Clause"
]
| null | null | null | 23.383929 | 98 | 0.515082 | [
[
[
"URL: http://matplotlib.org/examples/pylab_examples/quiver_demo.html\n\nMost examples work across multiple plotting backends, this example is also available for:\n\n* [Matplotlib - quiver_demo](../matplotlib/quiver_demo.ipynb)",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport holoviews as hv\nfrom holoviews import opts\nhv.extension('bokeh')",
"_____no_output_____"
]
],
[
[
"## Define data",
"_____no_output_____"
]
],
[
[
"xs, ys = np.arange(0, 2 * np.pi, .2), np.arange(0, 2 * np.pi, .2)\nX, Y = np.meshgrid(xs, ys)\nU = np.cos(X)\nV = np.sin(Y)\n\n# Convert to magnitude and angle\nmag = np.sqrt(U**2 + V**2)\nangle = (np.pi/2.) - np.arctan2(U/mag, V/mag)",
"_____no_output_____"
]
],
[
[
"## Plot",
"_____no_output_____"
]
],
[
[
"label = 'Arrows scale with plot width, not view'\n\nopts.defaults(opts.VectorField(height=400, width=500, padding=0.1))\n\nvectorfield = hv.VectorField((xs, ys, angle, mag))\nvectorfield.relabel(label)",
"_____no_output_____"
],
[
"label = \"pivot='mid'; every third arrow\"\n\nvf_mid = hv.VectorField((xs[::3], ys[::3], angle[::3, ::3], mag[::3, ::3], ))\npoints = hv.Points((X[::3, ::3].flat, Y[::3, ::3].flat))\n\nopts.defaults(opts.Points(color='red'))\n\n(vf_mid * points).relabel(label)",
"_____no_output_____"
],
[
"label = \"pivot='tip'; scales with x view\"\n\nvectorfield = hv.VectorField((xs, ys, angle, mag))\npoints = hv.Points((X.flat, Y.flat))\n\n(points * vectorfield).opts(\n opts.VectorField(magnitude='Magnitude', color='Magnitude',\n pivot='tip', line_width=2, title=label))",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec70f8f2130490d44bf5520016ceb4d18a8d74a5 | 13,993 | ipynb | Jupyter Notebook | site/en/guide/_tpu.ipynb | truongsinh/tensorflow-docs | e62749e757b0b9fd124f195e0463cfb0c913a835 | [
"Apache-2.0"
]
| 1 | 2020-09-04T05:34:14.000Z | 2020-09-04T05:34:14.000Z | site/en/guide/_tpu.ipynb | MartinSamanArata2018/docs | e62749e757b0b9fd124f195e0463cfb0c913a835 | [
"Apache-2.0"
]
| null | null | null | site/en/guide/_tpu.ipynb | MartinSamanArata2018/docs | e62749e757b0b9fd124f195e0463cfb0c913a835 | [
"Apache-2.0"
]
| 1 | 2019-09-17T05:17:18.000Z | 2019-09-17T05:17:18.000Z | 36.727034 | 465 | 0.538126 | [
[
[
"##### Copyright 2018 The TensorFlow Authors.\n\nLicensed under the Apache License, Version 2.0 (the \"License\");",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\"); { display-mode: \"form\" }\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://www.tensorflow.org/{PATH}\">\n <img src=\"https://www.tensorflow.org/images/tf_logo_32px.png\" />\n View on TensorFlow.org</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/tensorflow/docs/blob/master/site/{PATH}.ipynb\">\n <img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />\n Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/tensorflow/docs/blob/master/site/{PATH}.ipynb\">\n <img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />\n View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"# Using TPUs\n\nTensor Processing Units (TPUs) are Google's specialized ASICs designed to dramatically accelerate machine learning workloads. They are available on Google Colab, the TensorFlow Research Cloud and Google Compute Engine. \n\nIn this notebook, you can try training a convolutional neural network against the Fashion MNIST dataset on Cloud TPUs using tf.keras and Distribution Strategy.\n",
"_____no_output_____"
],
[
"## Learning Objectives\n\nIn this Colab, you will learn how to:\n\n* Write a standard 4-layer conv-net with drop-out and batch normalization in Keras.\n* Use TPUs and Distribution Strategy to train the model.\n* Run a prediction to see how well the model can predict fashion categories and output the result.",
"_____no_output_____"
],
[
"## Instructions\n\nTo use TPUs in Colab:\n\n1. On the main menu, click Runtime and select **Change runtime type**. Set \"TPU\" as the hardware accelerator.\n1. Click Runtime again and select **Runtime > Run All**. You can also run the cells manually with Shift-ENTER. ",
"_____no_output_____"
],
[
"## Data, Model, and Training\n\n### Download the Data\n\nBegin by downloading the fashion MNIST dataset using `tf.keras.datasets`, as shown below. We will also need to convert the data to `float32` format, as the data types supported by TPUs are limited right now.\n\nTPUs currently do not support Eager Execution, so we disable that with `disable_eager_execution()`.",
"_____no_output_____"
]
],
[
[
"from __future__ import absolute_import, division, print_function, unicode_literals\nimport numpy as np",
"_____no_output_____"
],
[
"from __future__ import absolute_import, division, print_function\n\ntry:\n # %tensorflow_version only exists in Colab.\n import tensorflow.compat.v2 as tf\nexcept Exception:\n pass\ntf.enable_v2_behavior()\ntf.compat.v1.disable_eager_execution()\n\nimport numpy as np\n\n(x_train, y_train), (x_test, y_test) = tf.keras.datasets.fashion_mnist.load_data()\n\n# add empty color dimension\nx_train = np.expand_dims(x_train, -1)\nx_test = np.expand_dims(x_test, -1)\n\n# convert types to float32\nx_train = x_train.astype(np.float32)\nx_test = x_test.astype(np.float32)\ny_train = y_train.astype(np.float32)\ny_test = y_test.astype(np.float32)",
"_____no_output_____"
]
],
[
[
"### Initialize TPUStrategy\n\nWe first initialize the TPUStrategy object before creating the model, so that Keras knows that we are creating a model for TPUs. \n\nTo do this, we are first creating a TPUClusterResolver using the IP address of the TPU, and then creating a TPUStrategy object from the Cluster Resolver.",
"_____no_output_____"
]
],
[
[
"import os\n\nresolver = tf.distribute.cluster_resolver.TPUClusterResolver()\ntf.tpu.experimental.initialize_tpu_system(resolver)\nstrategy = tf.distribute.experimental.TPUStrategy(resolver)",
"_____no_output_____"
]
],
[
[
"### Define the Model\n\nThe following example uses a standard conv-net that has 4 layers with drop-out and batch normalization between each layer. Note that we are creating the model within a `strategy.scope`.\n",
"_____no_output_____"
]
],
[
[
"with strategy.scope():\n model = tf.keras.models.Sequential()\n model.add(tf.keras.layers.BatchNormalization(input_shape=x_train.shape[1:]))\n model.add(tf.keras.layers.Conv2D(64, (5, 5), padding='same', activation='elu'))\n model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))\n model.add(tf.keras.layers.Dropout(0.25))\n\n model.add(tf.keras.layers.BatchNormalization())\n model.add(tf.keras.layers.Conv2D(128, (5, 5), padding='same', activation='elu'))\n model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2)))\n model.add(tf.keras.layers.Dropout(0.25))\n\n model.add(tf.keras.layers.BatchNormalization())\n model.add(tf.keras.layers.Conv2D(256, (5, 5), padding='same', activation='elu'))\n model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))\n model.add(tf.keras.layers.Dropout(0.25))\n\n model.add(tf.keras.layers.BatchNormalization())\n model.add(tf.keras.layers.Conv2D(512, (5, 5), padding='same', activation='elu'))\n model.add(tf.keras.layers.MaxPooling2D(pool_size=(2, 2), strides=(2,2)))\n model.add(tf.keras.layers.Dropout(0.25))\n\n model.add(tf.keras.layers.Flatten())\n model.add(tf.keras.layers.Dense(256))\n model.add(tf.keras.layers.Activation('elu'))\n model.add(tf.keras.layers.Dropout(0.5))\n model.add(tf.keras.layers.Dense(10))\n model.add(tf.keras.layers.Activation('softmax'))\n model.summary()",
"_____no_output_____"
]
],
[
[
"### Train on the TPU\n\nTo train on the TPU, we can simply call `model.compile` under the strategy scope, and then call `model.fit` to start training. In this case, we are training for 5 epochs with 60 steps per epoch, and running evaluation at the end of 5 epochs.\n\nIt may take a while for the training to start, as the data and model has to be transferred to the TPU and compiled before training can start.",
"_____no_output_____"
]
],
[
[
"with strategy.scope():\n model.compile(\n optimizer=tf.train.AdamOptimizer(learning_rate=1e-3),\n loss=tf.keras.losses.sparse_categorical_crossentropy,\n metrics=['sparse_categorical_accuracy']\n )\n\nmodel.fit(\n (x_train, y_train),\n epochs=5,\n steps_per_epoch=60,\n validation_data=(x_test, y_test),\n validation_freq=5,\n)",
"_____no_output_____"
]
],
[
[
"### Check our results with Inference\n\nNow that we are done training, we can see how well the model can predict fashion categories:",
"_____no_output_____"
]
],
[
[
"LABEL_NAMES = ['t_shirt', 'trouser', 'pullover', 'dress', 'coat', 'sandal', 'shirt', 'sneaker', 'bag', 'ankle_boots']\n\nfrom matplotlib import pyplot\n%matplotlib inline\n\ndef plot_predictions(images, predictions):\n n = images.shape[0]\n nc = int(np.ceil(n / 4))\n f, axes = pyplot.subplots(nc, 4)\n for i in range(nc * 4):\n y = i // 4\n x = i % 4\n axes[x, y].axis('off')\n \n label = LABEL_NAMES[np.argmax(predictions[i])]\n confidence = np.max(predictions[i])\n if i > n:\n continue\n axes[x, y].imshow(images[i])\n axes[x, y].text(0.5, -1.5, label + ': %.3f' % confidence, fontsize=12)\n\n pyplot.gcf().set_size_inches(8, 8) \n\nplot_predictions(np.squeeze(x_test[:16]), \n model.predict(x_test[:16]))",
"_____no_output_____"
]
],
[
[
"### What's next\n\n* Learn about [Cloud TPUs](https://cloud.google.com/tpu/docs) that Google designed and optimized specifically to speed up and scale up ML workloads for training and inference and to enable ML engineers and researchers to iterate more quickly.\n* Explore the range of [Cloud TPU tutorials and Colabs](https://cloud.google.com/tpu/docs/tutorials) to find other examples that can be used when implementing your ML project.\n\nOn Google Cloud Platform, in addition to GPUs and TPUs available on pre-configured [deep learning VMs](https://cloud.google.com/deep-learning-vm/), you will find [AutoML](https://cloud.google.com/automl/)*(beta)* for training custom models without writing code and [Cloud ML Engine](https://cloud.google.com/ml-engine/docs/) which will allows you to run parallel trainings and hyperparameter tuning of your custom models on powerful distributed hardware.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec70f915f0cba9edb7f790e2f3273807ba42aa37 | 122,937 | ipynb | Jupyter Notebook | examples/introduction_lab_widget_comm.ipynb | gideon116/clustergrammer_not_organized | 14b622ea91e236bee08c47b91caf12178f97aa8b | [
"BSD-3-Clause"
]
| 98 | 2018-11-18T09:51:15.000Z | 2022-03-15T05:39:01.000Z | examples/introduction_lab_widget_comm.ipynb | gideon116/clustergrammer_not_organized | 14b622ea91e236bee08c47b91caf12178f97aa8b | [
"BSD-3-Clause"
]
| 78 | 2019-03-14T19:18:55.000Z | 2021-11-03T13:59:19.000Z | examples/introduction_lab_widget_comm.ipynb | gideon116/clustergrammer_not_organized | 14b622ea91e236bee08c47b91caf12178f97aa8b | [
"BSD-3-Clause"
]
| 19 | 2019-07-10T20:02:01.000Z | 2022-03-10T06:08:32.000Z | 160.492167 | 36,379 | 0.643907 | [
[
[
"# Introduction \nDemonstrating widget two-way communication.",
"_____no_output_____"
]
],
[
[
"import ipywidgets as widgets\nfrom clustergrammer2 import Network, CGM2\nimport json\nimport pandas as pd\ndf = {}\nnet = {}",
">> clustergrammer2 backend version 0.13.2\n"
]
],
[
[
"\n### Load Data",
"_____no_output_____"
]
],
[
[
"df['clean'] = pd.read_csv('rc_two_cat_clean.csv', index_col=0)\ndf['meta_col'] = pd.read_csv('meta_col.csv', index_col=0)\ndf['clean'].shape",
"_____no_output_____"
],
[
"df['meta_cat_col'] = pd.DataFrame()\ndf['meta_cat_col'].loc['Cat', 'color'] = 'red'\ndf['meta_cat_col'].loc['Dog', 'color'] = 'yellow'\ndf['meta_cat_col'].loc['Shark', 'color'] = 'black'\ndf['meta_cat_col'].loc['Snake', 'color'] = 'blue'\ndf['meta_cat_col'].loc['Lizard', 'color'] = 'green'\ndf['meta_cat_col']",
"_____no_output_____"
],
[
"viz_cats = ['Category', 'Gender']",
"_____no_output_____"
]
],
[
[
"### Heatmap 1",
"_____no_output_____"
]
],
[
[
"n1 = Network(CGM2)\nn1.load_df(df['clean'], meta_col=df['meta_col'], col_cats=viz_cats)\nn1.normalize(axis='row', norm_type='zscore')\nn1.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nn1.widget()",
"_____no_output_____"
]
],
[
[
"### Heatmap 2",
"_____no_output_____"
]
],
[
[
"n2 = Network(CGM2)\nn2.load_df(df['clean'], meta_col=df['meta_col'], col_cats=viz_cats)\nn2.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nn2.widget(link_net=n1)",
"_____no_output_____"
]
],
[
[
"### Heatmap 3",
"_____no_output_____"
]
],
[
[
"n3 = Network(CGM2)\nn3.load_df(df['clean'], meta_col=df['meta_col'], col_cats=viz_cats)\nn3.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nn3.widget(link_net=n2)",
"_____no_output_____"
],
[
"df['meta_col']['Category'].value_counts()",
"_____no_output_____"
],
[
"# net1 = Network(Clustergrammer2)\n# net2 = Network(Clustergrammer2)",
"_____no_output_____"
],
[
"# sliders1, slider2 = widgets.IntSlider(description='Slider 1'),\\\n# widgets.IntSlider(description='Slider 2')\n# l = widgets.link((sliders1, 'value'), (slider2, 'value'))\n# display(sliders1, slider2)",
"_____no_output_____"
],
[
"# def update_value(new_value):\n# w1.value = new_value",
"_____no_output_____"
],
[
"# w1.observe(update_value, )",
"_____no_output_____"
],
[
"# net2.load_df(df['clean'], meta_col=df['meta_col'], col_cats=viz_cats)\nnet2.load_df(df['clean'], meta_col=net1.meta_col, col_cats=viz_cats)\nnet2.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nw2 = net2.widget()\nw2",
"_____no_output_____"
]
],
[
[
"# Downsample Example",
"_____no_output_____"
]
],
[
[
"nds = Network(CGM2)",
"_____no_output_____"
],
[
"nds.load_df(df['clean'], meta_col=df['meta_col'], col_cats=['Gender', 'Category'])\nnds.downsample(axis='col', num_samples=5, random_state=99, ds_name='DS-Clusters', ds_cluster_name='default')\nnds.set_manual_category(col='Gender')\nnds.widget()",
"_____no_output_____"
],
[
"df['meta_ds_col'] = net.meta_ds_col\ndf['meta_ds_col']",
"_____no_output_____"
],
[
"df['meta_col']",
"_____no_output_____"
],
[
"df['ds'] = net.export_df()\ndf['ds'].head()",
"_____no_output_____"
]
],
[
[
"# Start with Downsampled Data",
"_____no_output_____"
]
],
[
[
"net.load_df(df['ds'], meta_col=df['meta_col'], \n is_downsampled=True, meta_ds_col=df['meta_ds_col'], \n col_cats=['Category', 'Gender', 'number in clust'])\n\nnet.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nnet.widget()",
"_____no_output_____"
],
[
"net.get_manual_category()",
"_____no_output_____"
],
[
"df['meta_ds_col']",
"_____no_output_____"
],
[
"df['meta_col']",
"_____no_output_____"
]
],
[
[
"### Load Subset of Cells to Relabel",
"_____no_output_____"
]
],
[
[
"keep_clusters = df['meta_ds_col'][df['meta_ds_col']['Category'] == 'New Category'].index.tolist()\nkeep_clusters",
"_____no_output_____"
],
[
"net.load_df(df['ds'][keep_clusters], meta_col=df['meta_col'],\n is_downsampled=True, meta_ds_col=df['meta_ds_col'], \n col_cats=['Category', 'Gender', 'number in clust'])\n\nnet.set_manual_category(col='Category', preferred_cats=df['meta_cat_col'])\nnet.widget()",
"_____no_output_____"
],
[
"net.get_manual_category()",
"_____no_output_____"
],
[
"df['meta_ds_col']",
"_____no_output_____"
],
[
"df['meta_col']",
"_____no_output_____"
],
[
"# df['meta_col']['Granular Cell Type'] = df['meta_col']['Category']",
"_____no_output_____"
],
[
"# df['meta_col'].to_csv('meta_col_v2.csv')",
"_____no_output_____"
],
[
"# # net.load_df(df['clean'], meta_col=df['meta_col'], meta_row=df['meta_row'], col_cats=['Gender', 'Category'])\n# net.load_df(df['ini'])\n# # net.downsample(axis='col', num_samples=5, random_state=99)\n# # net.set_manual_category(col='Gender')\n# # net.widget()",
"_____no_output_____"
],
[
"# new_tuples = [x for x in cols]\n# new_tuples",
"_____no_output_____"
],
[
"# a = widgets.Text()\n# display(a)\n# mylink = widgets.jslink((a, 'value'), (net.widget_instance, 'value'))",
"_____no_output_____"
],
[
"# net.load_df(df['ini'])\n# net.cluster(sim_mat=True)\n# net.widget('sim_col')",
"_____no_output_____"
],
[
"# # destroy-viz \n# net.widget_instance.value = 'destroy-viz'",
"_____no_output_____"
]
],
[
[
"### Random Matrix Example",
"_____no_output_____"
]
],
[
[
"# import numpy as np\n# import pandas as pd\n\n# # generate random matrix\n# num_rows = 100\n# num_cols = 100\n# np.random.seed(seed=100)\n# mat = np.random.rand(num_rows, num_cols)\n\n# # make row and col labels\n# rows = range(num_rows)\n# cols = range(num_cols)\n# rows = [str(i) for i in rows]\n# cols = [str(i) for i in cols]\n\n# # make dataframe \n# df['rand'] = pd.DataFrame(data=mat, columns=cols, index=rows)",
"_____no_output_____"
],
[
"# net.load_df(df['rand'])\n# net.widget()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec70fdc326bc113c8d6d68531e97c6300e88c053 | 45,475 | ipynb | Jupyter Notebook | 3- Mathematics and Linear Algebra/Linear Algebra for Data Scientists.ipynb | gvvynplaine/10-steps-to-become-a-data-scientist | e79ec5047eedecea54c89c413257b204859e4e06 | [
"Apache-2.0"
]
| 2 | 2020-05-17T12:49:18.000Z | 2020-05-27T15:08:13.000Z | 3- Mathematics and Linear Algebra/Linear Algebra for Data Scientists.ipynb | hongzhengjiang/10-steps-to-become-a-data-scientist | dc2884668cfa3830a9665d6e7e36244744919761 | [
"Apache-2.0"
]
| null | null | null | 3- Mathematics and Linear Algebra/Linear Algebra for Data Scientists.ipynb | hongzhengjiang/10-steps-to-become-a-data-scientist | dc2884668cfa3830a9665d6e7e36244744919761 | [
"Apache-2.0"
]
| 1 | 2022-02-20T10:54:37.000Z | 2022-02-20T10:54:37.000Z | 27.004157 | 573 | 0.569962 | [
[
[
" # <div style=\"text-align: center\">Linear Algebra for Data Scientists \n<div style=\"text-align: center\">\nHaving a basic knowledge of linear algebra is one of the requirements for any data scientist. In this tutorial we will try to cover all the necessary concepts related to linear algebra\nand also this is the third step of the <a href=\"https://www.kaggle.com/mjbahmani/10-steps-to-become-a-data-scientist\">10 Steps to Become a Data Scientist</a>. and you can learn all of the thing you need for being a data scientist with Linear Algabra.</div> \n\n<div style=\"text-align:center\">last update: <b>03/01/2019</b></div>\n\n>###### You may be interested have a look at 10 Steps to Become a Data Scientist: \n\n1. [Leren Python](https://www.kaggle.com/mjbahmani/the-data-scientist-s-toolbox-tutorial-1)\n2. [Python Packages](https://www.kaggle.com/mjbahmani/the-data-scientist-s-toolbox-tutorial-2)\n3. <font color=\"red\">You are in 3th step</font>\n4. [Programming & Analysis Tools](https://www.kaggle.com/mjbahmani/20-ml-algorithms-15-plot-for-beginners)\n5. [Big Data](https://www.kaggle.com/mjbahmani/a-data-science-framework-for-quora)\n6. [Data visualization](https://www.kaggle.com/mjbahmani/top-5-data-visualization-libraries-tutorial)\n7. [Data Cleaning](https://www.kaggle.com/mjbahmani/machine-learning-workflow-for-house-prices)\n8. [How to solve a Problem?](https://www.kaggle.com/mjbahmani/the-data-scientist-s-toolbox-tutorial-2)\n9. [Machine Learning](https://www.kaggle.com/mjbahmani/a-comprehensive-ml-workflow-with-python)\n10. [Deep Learning](https://www.kaggle.com/mjbahmani/top-5-deep-learning-frameworks-tutorial)\n\n\n\n\n\nYou can Fork code and Follow me on:\n\n> ###### [ GitHub](https://github.com/mjbahmani/10-steps-to-become-a-data-scientist)\n> ###### [Kaggle](https://www.kaggle.com/mjbahmani/)\n-------------------------------------------------------------------------------------------------------------\n <b>I hope you find this kernel helpful and some <font color='blue'>UPVOTES</font> would be very much appreciated.</b>\n \n -----------",
"_____no_output_____"
],
[
" <a id=\"top\"></a> <br>\n## Notebook Content\n1. [Introduction](#1)\n1. [What is Linear Algebra?](#2)\n1. [Notation ](#2)\n1. [Matrix Multiplication](#3)\n 1. [Vector-Vector Products](#31)\n 1. [Outer Product of Two Vectors](#32)\n 1. [Matrix-Vector Products](#33)\n 1. [Matrix-Matrix Products](#34)\n1. [Identity Matrix](#4)\n1. [Diagonal Matrix](#5)\n1. [Transpose of a Matrix](#6)\n1. [Symmetric Metrices](#7)\n1. [The Trace](#8)\n1. [Norms](#9)\n1. [Linear Independence and Rank](#10)\n1. [Subtraction and Addition of Metrices](#11)\n 1. [Inverse](#111)\n1. [Orthogonal Matrices](#12)\n1. [Range and Nullspace of a Matrix](#13)\n1. [Determinant](#14)\n1. [Tensors](#16)\n1. [Hyperplane](#17)\n1. [Eigenvalues and Eigenvectors](#18)\n1. [Exercise](#19)\n1. [Conclusion](#21)\n1. [References](#22)",
"_____no_output_____"
],
[
"<a id=\"1\"></a> <br>\n# 1-Introduction\nThis is the third step of the [10 Steps to Become a Data Scientist](https://www.kaggle.com/mjbahmani/10-steps-to-become-a-data-scientist).\nwe will cover following topic:\n1. notation\n1. Matrix Multiplication\n1. Identity Matrix\n1. Diagonal Matrix\n1. Transpose of a Matrix\n1. The Trace\n1. Norms\n1. Tensors\n1. Hyperplane\n1. Eigenvalues and Eigenvectors\n## What is linear algebra?\n**Linear algebra** is the branch of mathematics that deals with **vector spaces**. good understanding of Linear Algebra is intrinsic to analyze Machine Learning algorithms, especially for **Deep Learning** where so much happens behind the curtain.you have my word that I will try to keep mathematical formulas & derivations out of this completely mathematical topic and I try to cover all of subject that you need as data scientist.[https://medium.com/@neuralnets](https://medium.com/@neuralnets/linear-algebra-for-data-science-revisiting-high-school-9a6bbeba19c6)\n<img src='https://camo.githubusercontent.com/e42ea0e40062cc1e339a6b90054bfbe62be64402/68747470733a2f2f63646e2e646973636f72646170702e636f6d2f6174746163686d656e74732f3339313937313830393536333530383733382f3434323635393336333534333331383532382f7363616c61722d766563746f722d6d61747269782d74656e736f722e706e67' height=200 width=700>\n[image credit: https://hadrienj.github.io/posts/Deep-Learning-Book-Series-2.1-Scalars-Vectors-Matrices-and-Tensors/ ](https://hadrienj.github.io/posts/Deep-Learning-Book-Series-2.1-Scalars-Vectors-Matrices-and-Tensors/)\n <a id=\"top\"></a> <br>",
"_____no_output_____"
],
[
"<a id=\"11\"></a> <br>\n## 1-1 Import",
"_____no_output_____"
]
],
[
[
"import matplotlib.patches as patch\nimport matplotlib.pyplot as plt\nfrom scipy.stats import norm\nfrom scipy import linalg\nfrom numpy import poly1d\nfrom sklearn import svm\nimport tensorflow as tf\nimport pandas as pd\nimport numpy as np\nimport glob\nimport sys\nimport os",
"_____no_output_____"
]
],
[
[
"<a id=\"12\"></a> <br>\n## 1-2 Setup",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\n%precision 4\nplt.style.use('ggplot')\nnp.set_printoptions(suppress=True)",
"_____no_output_____"
]
],
[
[
"<a id=\"2\"></a> <br>\n# 2- What is Linear Algebra?\nLinear algebra is the branch of mathematics concerning linear equations such as\n<img src='https://wikimedia.org/api/rest_v1/media/math/render/svg/f4f0f2986d54c01f3bccf464d266dfac923c80f3'>\nLinear algebra is central to almost all areas of mathematics. [6]\n<img src='https://upload.wikimedia.org/wikipedia/commons/thumb/2/2f/Linear_subspaces_with_shading.svg/800px-Linear_subspaces_with_shading.svg.png' height=400 width=400>\n[wikipedia](https://en.wikipedia.org/wiki/Linear_algebra#/media/File:Linear_subspaces_with_shading.svg)\n",
"_____no_output_____"
]
],
[
[
"# let see how to create a multi dimentional Array with Numpy\na = np.zeros((2, 3, 4))\n#l = [[[ 0., 0., 0., 0.],\n # [ 0., 0., 0., 0.],\n # [ 0., 0., 0., 0.]],\n # [[ 0., 0., 0., 0.],\n # [ 0., 0., 0., 0.],\n # [ 0., 0., 0., 0.]]]\nprint(a)\nprint(a.shape)",
"_____no_output_____"
],
[
"\n# Declaring Vectors\n\nx = [1, 2, 3]\ny = [4, 5, 6]\n\nprint(type(x))\n\n# This does'nt give the vector addition.\nprint(x + y)\n\n# Vector addition using Numpy\n\nz = np.add(x, y)\nprint(z)\nprint(type(z))\n\n# Vector Cross Product\nmul = np.cross(x, y)\nprint(mul)",
"_____no_output_____"
]
],
[
[
"## 2-1 What is Vectorization?\nIn mathematics, especially in linear algebra and matrix theory, the vectorization of a matrix is a linear transformation which converts the matrix into a column vector. Specifically, the vectorization of an m × n matrix A, denoted vec(A), is the mn × 1 column vector obtained by stacking the columns of the matrix A on top of one another:\n<img src='https://wikimedia.org/api/rest_v1/media/math/render/svg/30ca6a8b796fd3a260ba3001d9875e990baad5ab'>\n[wikipedia](https://en.wikipedia.org/wiki/Vectorization_(mathematics) )",
"_____no_output_____"
],
[
"Vectors of the length $n$ could be treated like points in $n$-dimensional space. One can calculate the distance between such points using measures like [Euclidean Distance](https://en.wikipedia.org/wiki/Euclidean_distance). The similarity of vectors could also be calculated using [Cosine Similarity](https://en.wikipedia.org/wiki/Cosine_similarity).\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"3\"></a> <br>\n## 3- Notation\n<img src='http://s8.picofile.com/file/8349058626/la.png'>\n[linear.ups.edu](http://linear.ups.edu/html/notation.html)",
"_____no_output_____"
],
[
"<a id=\"4\"></a> <br>\n## 4- Matrix Multiplication\n<img src='https://www.mathsisfun.com/algebra/images/matrix-multiply-constant.gif'>\n\n[mathsisfun](https://www.mathsisfun.com/algebra/matrix-multiplying.html)",
"_____no_output_____"
],
[
"The result of the multiplication of two matrixes $A \\in \\mathbb{R}^{m \\times n}$ and $B \\in \\mathbb{R}^{n \\times p}$ is the matrix:",
"_____no_output_____"
]
],
[
[
"# initializing matrices \nx = np.array([[1, 2], [4, 5]]) \ny = np.array([[7, 8], [9, 10]])",
"_____no_output_____"
]
],
[
[
"$C = AB \\in \\mathbb{R}^{m \\times n}$",
"_____no_output_____"
],
[
"That is, we are multiplying the columns of $A$ with the rows of $B$:",
"_____no_output_____"
],
[
"$C_{ij}=\\sum_{k=1}^n{A_{ij}B_{kj}}$\n<img src='https://cdn.britannica.com/06/77706-004-31EE92F3.jpg'>\n[reference](https://cdn.britannica.com/06/77706-004-31EE92F3.jpg)",
"_____no_output_____"
],
[
"The number of columns in $A$ must be equal to the number of rows in $B$.\n\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"# using add() to add matrices \nprint (\"The element wise addition of matrix is : \") \nprint (np.add(x,y)) ",
"_____no_output_____"
],
[
"# using subtract() to subtract matrices \nprint (\"The element wise subtraction of matrix is : \") \nprint (np.subtract(x,y)) ",
"_____no_output_____"
],
[
"# using divide() to divide matrices \nprint (\"The element wise division of matrix is : \") \nprint (np.divide(x,y)) ",
"_____no_output_____"
],
[
"# using multiply() to multiply matrices element wise \nprint (\"The element wise multiplication of matrix is : \") \nprint (np.multiply(x,y))",
"_____no_output_____"
]
],
[
[
"<a id=\"41\"></a> <br>\n## 4-1 Vector-Vector Products\n\nnumpy.cross(a, b, axisa=-1, axisb=-1, axisc=-1, axis=None)[source]\nReturn the cross product of two (arrays of) vectors.[scipy](https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.cross.html)\n<img src='http://gamedevelopertips.com/wp-content/uploads/2017/11/image8.png'>\n[image-gredits](http://gamedevelopertips.com)",
"_____no_output_____"
]
],
[
[
"x = [1, 2, 3]\ny = [4, 5, 6]\nnp.cross(x, y)",
"_____no_output_____"
]
],
[
[
"We define the vectors $x$ and $y$ using *numpy*:",
"_____no_output_____"
]
],
[
[
"x = np.array([1, 2, 3, 4])\ny = np.array([5, 6, 7, 8])\nprint(\"x:\", x)\nprint(\"y:\", y)",
"_____no_output_____"
]
],
[
[
"We can now calculate the $dot$ or $inner product$ using the *dot* function of *numpy*:",
"_____no_output_____"
]
],
[
[
"np.dot(x, y)",
"_____no_output_____"
]
],
[
[
"The order of the arguments is irrelevant:",
"_____no_output_____"
]
],
[
[
"np.dot(y, x)",
"_____no_output_____"
]
],
[
[
"Note that both vectors are actually **row vectors** in the above code. We can transpose them to column vectors by using the *shape* property:",
"_____no_output_____"
]
],
[
[
"print(\"x:\", x)\nx.shape = (4, 1)\nprint(\"xT:\", x)\nprint(\"y:\", y)\ny.shape = (4, 1)\nprint(\"yT:\", y)",
"_____no_output_____"
]
],
[
[
"In fact, in our understanding of Linear Algebra, we take the arrays above to represent **row vectors**. *Numpy* treates them differently.",
"_____no_output_____"
],
[
"We see the issues when we try to transform the array objects. Usually, we can transform a row vector into a column vector in *numpy* by using the *T* method on vector or matrix objects:\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"x = np.array([1, 2, 3, 4])\ny = np.array([5, 6, 7, 8])\nprint(\"x:\", x)\nprint(\"y:\", y)\nprint(\"xT:\", x.T)\nprint(\"yT:\", y.T)",
"_____no_output_____"
]
],
[
[
"The problem here is that this does not do, what we expect it to do. It only works, if we declare the variables not to be arrays of numbers, but in fact a matrix:",
"_____no_output_____"
]
],
[
[
"x = np.array([[1, 2, 3, 4]])\ny = np.array([[5, 6, 7, 8]])\nprint(\"x:\", x)\nprint(\"y:\", y)\nprint(\"xT:\", x.T)\nprint(\"yT:\", y.T)\n",
"_____no_output_____"
]
],
[
[
"Note that the *numpy* functions *dot* and *outer* are not affected by this distinction. We can compute the dot product using the mathematical equation above in *numpy* using the new $x$ and $y$ row vectors:\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"print(\"x:\", x)\nprint(\"y:\", y.T)\nnp.dot(x, y.T)",
"_____no_output_____"
]
],
[
[
"Or by reverting to:",
"_____no_output_____"
]
],
[
[
"print(\"x:\", x.T)\nprint(\"y:\", y)\nnp.dot(y, x.T)",
"_____no_output_____"
]
],
[
[
"To read the result from this array of arrays, we would need to access the value this way:",
"_____no_output_____"
]
],
[
[
"np.dot(y, x.T)[0][0]",
"_____no_output_____"
]
],
[
[
"<a id=\"42\"></a> <br>\n## 4-2 Outer Product of Two Vectors\nCompute the outer product of two vectors.",
"_____no_output_____"
]
],
[
[
"x = np.array([[1, 2, 3, 4]])\nprint(\"x:\", x)\nprint(\"xT:\", np.reshape(x, (4, 1)))\nprint(\"xT:\", x.T)\nprint(\"xT:\", x.transpose())",
"_____no_output_____"
]
],
[
[
"Example\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"We can now compute the **outer product** by multiplying the column vector $x$ with the row vector $y$:",
"_____no_output_____"
]
],
[
[
"x = np.array([[1, 2, 3, 4]])\ny = np.array([[5, 6, 7, 8]])\nx.T * y",
"_____no_output_____"
]
],
[
[
"*Numpy* provides an *outer* function that does all that:",
"_____no_output_____"
]
],
[
[
"np.outer(x, y)",
"_____no_output_____"
]
],
[
[
"Note, in this simple case using the simple arrays for the data structures of the vectors does not affect the result of the *outer* function:",
"_____no_output_____"
]
],
[
[
"x = np.array([1, 2, 3, 4])\ny = np.array([5, 6, 7, 8])\nnp.outer(x, y)",
"_____no_output_____"
]
],
[
[
"<a id=\"43\"></a> <br>\n## 4-3 Matrix-Vector Products\nUse numpy.dot or a.dot(b). See the documentation [here](http://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html).",
"_____no_output_____"
]
],
[
[
"a = np.array([[ 5, 1 ,3], [ 1, 1 ,1], [ 1, 2 ,1]])\nb = np.array([1, 2, 3])\nprint (a.dot(b))",
"_____no_output_____"
]
],
[
[
"Using *numpy* we can compute $Ax$:",
"_____no_output_____"
]
],
[
[
"A = np.array([[4, 5, 6],\n [7, 8, 9]])\nx = np.array([1, 2, 3])\nA.dot(x)",
"_____no_output_____"
]
],
[
[
"<a id=\"44\"></a> <br>\n## 4-4 Matrix-Matrix Products",
"_____no_output_____"
]
],
[
[
"a = [[1, 0], [0, 1]]\nb = [[4, 1], [2, 2]]\nnp.matmul(a, b)",
"_____no_output_____"
],
[
"matrix1 = np.matrix(a)\nmatrix2 = np.matrix(b)",
"_____no_output_____"
],
[
"matrix1 + matrix2",
"_____no_output_____"
],
[
"matrix1 - matrix2",
"_____no_output_____"
]
],
[
[
"<a id=\"441\"></a> <br>\n### 4-4-1 Multiplication",
"_____no_output_____"
]
],
[
[
"np.dot(matrix1, matrix2)",
"_____no_output_____"
],
[
"\nmatrix1 * matrix2",
"_____no_output_____"
]
],
[
[
"<a id=\"5\"></a> <br>\n## 5- Identity Matrix",
"_____no_output_____"
],
[
"numpy.identity(n, dtype=None)\n\nReturn the identity array.\n[source](https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.identity.html)",
"_____no_output_____"
]
],
[
[
"np.identity(3)",
"_____no_output_____"
]
],
[
[
"How to create *identity matrix* in *numpy* ",
"_____no_output_____"
]
],
[
[
"identy = np.array([[21, 5, 7],[9, 8, 16]])\nprint(\"identy:\", identy)",
"_____no_output_____"
],
[
"identy.shape",
"_____no_output_____"
],
[
"np.identity(identy.shape[1], dtype=\"int\")",
"_____no_output_____"
],
[
"np.identity(identy.shape[0], dtype=\"int\")",
"_____no_output_____"
]
],
[
[
"<a id=\"51\"></a> <br>\n### 5-1 Inverse Matrices",
"_____no_output_____"
]
],
[
[
"inverse = np.linalg.inv(matrix1)\nprint(inverse)",
"_____no_output_____"
]
],
[
[
"<a id=\"6\"></a> <br>\n## 6- Diagonal Matrix",
"_____no_output_____"
],
[
"In *numpy* we can create a *diagonal matrix* from any given matrix using the *diag* function:",
"_____no_output_____"
]
],
[
[
"import numpy as np\nA = np.array([[0, 1, 2, 3],\n [4, 5, 6, 7],\n [8, 9, 10, 11],\n [12, 13, 14, 15]])\nnp.diag(A)",
"_____no_output_____"
],
[
"np.diag(A, k=1)",
"_____no_output_____"
],
[
"np.diag(A, k=-1)",
"_____no_output_____"
]
],
[
[
"<a id=\"7\"></a> <br>\n## 7- Transpose of a Matrix\nFor reading about Transpose of a Matrix, you can visit [this link](https://py.checkio.org/en/mission/matrix-transpose/)",
"_____no_output_____"
]
],
[
[
"a = np.array([[1, 2], [3, 4]])\na",
"_____no_output_____"
],
[
"a.transpose()",
"_____no_output_____"
]
],
[
[
"<a id=\"8\"></a> <br>\n## 8- Symmetric Matrices\nIn linear algebra, a symmetric matrix is a square matrix that is equal to its transpose. Formally,\n<img src='https://wikimedia.org/api/rest_v1/media/math/render/svg/ad8a5a3a4c95de6f7f50b0a6fb592d115fe0e95f'>\n\n[wikipedia](https://en.wikipedia.org/wiki/Symmetric_matrix)",
"_____no_output_____"
]
],
[
[
"N = 100\nb = np.random.random_integers(-2000,2000,size=(N,N))\nb_symm = (b + b.T)/2",
"_____no_output_____"
]
],
[
[
"<a id=\"9\"></a> <br>\n## 9-The Trace\nReturn the sum along diagonals of the array.",
"_____no_output_____"
]
],
[
[
"np.trace(np.eye(3))",
"_____no_output_____"
],
[
"print(np.trace(matrix1))",
"_____no_output_____"
],
[
"det = np.linalg.det(matrix1)\nprint(det)",
"_____no_output_____"
]
],
[
[
"<a id=\"10\"></a> <br>\n# 10- Norms\nnumpy.linalg.norm\nThis function is able to return one of eight different matrix norms, or one of an infinite number of vector norms (described below), depending on the value of the ord parameter. [scipy](https://docs.scipy.org/doc/numpy-1.13.0/reference/generated/numpy.linalg.norm.html)\n\n <a id=\"top\"></a> <br>",
"_____no_output_____"
]
],
[
[
"v = np.array([1,2,3,4])\nnorm.median(v)",
"_____no_output_____"
]
],
[
[
"<a id=\"11\"></a> <br>\n# 11- Linear Independence and Rank\nHow to identify the linearly independent rows from a matrix?",
"_____no_output_____"
]
],
[
[
"#How to find linearly independent rows from a matrix\nmatrix = np.array(\n [\n [0, 1 ,0 ,0],\n [0, 0, 1, 0],\n [0, 1, 1, 0],\n [1, 0, 0, 1]\n ])\n\nlambdas, V = np.linalg.eig(matrix.T)\n# The linearly dependent row vectors \nprint (matrix[lambdas == 0,:])",
"_____no_output_____"
]
],
[
[
"<a id=\"12\"></a> <br>\n# 12- Subtraction and Addition of Metrices",
"_____no_output_____"
]
],
[
[
"import numpy as np\nprint(\"np.arange(9):\", np.arange(9))\nprint(\"np.arange(9, 18):\", np.arange(9, 18))\nA = np.arange(9, 18).reshape((3, 3))\nB = np.arange(9).reshape((3, 3))\nprint(\"A:\", A)\nprint(\"B:\", B)",
"_____no_output_____"
]
],
[
[
"We can now add and subtract the two matrices $A$ and $B$:",
"_____no_output_____"
]
],
[
[
"A + B",
"_____no_output_____"
],
[
"A - B",
"_____no_output_____"
]
],
[
[
"<a id=\"121\"></a> <br>\n## 12-1 Inverse\nWe use numpy.linalg.inv() function to calculate the inverse of a matrix. The inverse of a matrix is such that if it is multiplied by the original matrix, it results in identity matrix.[tutorialspoint](https://www.tutorialspoint.com/numpy/numpy_inv.htm)",
"_____no_output_____"
]
],
[
[
"x = np.array([[1,2],[3,4]]) \ny = np.linalg.inv(x) \nprint (x )\nprint (y )\nprint (np.dot(x,y))",
"_____no_output_____"
]
],
[
[
"<a id=\"13\"></a> <br>\n## 13- Orthogonal Matrices\nHow to create random orthonormal matrix in python numpy",
"_____no_output_____"
]
],
[
[
"## based on https://stackoverflow.com/questions/38426349/how-to-create-random-orthonormal-matrix-in-python-numpy\ndef rvs(dim=3):\n random_state = np.random\n H = np.eye(dim)\n D = np.ones((dim,))\n for n in range(1, dim):\n x = random_state.normal(size=(dim-n+1,))\n D[n-1] = np.sign(x[0])\n x[0] -= D[n-1]*np.sqrt((x*x).sum())\n # Householder transformation\n Hx = (np.eye(dim-n+1) - 2.*np.outer(x, x)/(x*x).sum())\n mat = np.eye(dim)\n mat[n-1:, n-1:] = Hx\n H = np.dot(H, mat)\n # Fix the last sign such that the determinant is 1\n D[-1] = (-1)**(1-(dim % 2))*D.prod()\n # Equivalent to np.dot(np.diag(D), H) but faster, apparently\n H = (D*H.T).T\n return H",
"_____no_output_____"
]
],
[
[
"<a id=\"14\"></a> <br>\n## 14- Range and Nullspace of a Matrix",
"_____no_output_____"
]
],
[
[
"from scipy.linalg import null_space\nA = np.array([[1, 1], [1, 1]])\nns = null_space(A)\nns * np.sign(ns[0,0]) # Remove the sign ambiguity of the vector",
"_____no_output_____"
]
],
[
[
"<a id=\"15\"></a> <br>\n# 15- Determinant\nCompute the determinant of an array",
"_____no_output_____"
]
],
[
[
"a = np.array([[1, 2], [3, 4]])\nnp.linalg.det(a)",
"_____no_output_____"
]
],
[
[
"<a id=\"16\"></a> <br>\n# 16- Tensors",
"_____no_output_____"
],
[
"A [**tensor**](https://en.wikipedia.org/wiki/Tensor) could be thought of as an organized multidimensional array of numerical values. A vector could be assumed to be a sub-class of a tensor. Rows of tensors extend alone the y-axis, columns along the x-axis. The **rank** of a scalar is 0, the rank of a **vector** is 1, the rank of a **matrix** is 2, the rank of a **tensor** is 3 or higher.\n\n###### [Go to top](#top)",
"_____no_output_____"
]
],
[
[
"# credits: https://www.tensorflow.org/api_docs/python/tf/Variable\nA = tf.Variable(np.zeros((5, 5), dtype=np.float32), trainable=False)\nnew_part = tf.ones((2,3))\nupdate_A = A[2:4,2:5].assign(new_part)\nsess = tf.InteractiveSession()\ntf.global_variables_initializer().run()\nprint(update_A.eval())",
"_____no_output_____"
]
],
[
[
"<a id=\"25\"></a> <br>\n# 17- Hyperplane",
"_____no_output_____"
],
[
"The **hyperplane** is a sub-space in the ambient space with one dimension less. In a two-dimensional space the hyperplane is a line, in a three-dimensional space it is a two-dimensional plane, etc.",
"_____no_output_____"
],
[
"Hyperplanes divide an $n$-dimensional space into sub-spaces that might represent clases in a machine learning algorithm.",
"_____no_output_____"
]
],
[
[
"##based on this address: https://stackoverflow.com/questions/46511017/plot-hyperplane-linear-svm-python\nnp.random.seed(0)\nX = np.r_[np.random.randn(20, 2) - [2, 2], np.random.randn(20, 2) + [2, 2]]\nY = [0] * 20 + [1] * 20\n\nfig, ax = plt.subplots()\nclf2 = svm.LinearSVC(C=1).fit(X, Y)\n\n# get the separating hyperplane\nw = clf2.coef_[0]\na = -w[0] / w[1]\nxx = np.linspace(-5, 5)\nyy = a * xx - (clf2.intercept_[0]) / w[1]\n\n# create a mesh to plot in\nx_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1\ny_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1\nxx2, yy2 = np.meshgrid(np.arange(x_min, x_max, .2),\n np.arange(y_min, y_max, .2))\nZ = clf2.predict(np.c_[xx2.ravel(), yy2.ravel()])\n\nZ = Z.reshape(xx2.shape)\nax.contourf(xx2, yy2, Z, cmap=plt.cm.coolwarm, alpha=0.3)\nax.scatter(X[:, 0], X[:, 1], c=Y, cmap=plt.cm.coolwarm, s=25)\nax.plot(xx,yy)\n\nax.axis([x_min, x_max,y_min, y_max])\nplt.show()",
"_____no_output_____"
]
],
[
[
"<a id=\"31\"></a> <br>\n## 20- Exercises\nlet's do some exercise.",
"_____no_output_____"
],
[
"### 20-1 Create a dense meshgrid",
"_____no_output_____"
]
],
[
[
"np.mgrid[0:5,0:5]",
"_____no_output_____"
]
],
[
[
"### 20-2 Permute array dimensions",
"_____no_output_____"
]
],
[
[
"a=np.array([1,2,3])\nb=np.array([(1+5j,2j,3j), (4j,5j,6j)])\nc=np.array([[(1.5,2,3), (4,5,6)], [(3,2,1), (4,5,6)]])",
"_____no_output_____"
],
[
"np.transpose(b)",
"_____no_output_____"
],
[
"b.flatten()",
"_____no_output_____"
],
[
"np.hsplit(c,2)",
"_____no_output_____"
]
],
[
[
"## 20-3 Polynomials",
"_____no_output_____"
]
],
[
[
"p=poly1d([3,4,5])\np",
"_____no_output_____"
]
],
[
[
"## SciPy Cheat Sheet: Linear Algebra in Python\nThis Python cheat sheet is a handy reference with code samples for doing linear algebra with SciPy and interacting with NumPy.\n\n[DataCamp](https://www.datacamp.com/community/blog/python-scipy-cheat-sheet)",
"_____no_output_____"
],
[
"<a id=\"21\"></a> <br>\n# 21-Conclusion\nIf you have made this far – give yourself a pat at the back. We have covered different aspects of **Linear algebra** in this Kernel.\n###### [Go to top](#top)",
"_____no_output_____"
],
[
"<a id=\"22\"></a> <br>\n# 22-References & Credits\n1. [Linear Algbra1](https://github.com/dcavar/python-tutorial-for-ipython)\n1. [Linear Algbra2](https://www.oreilly.com/library/view/data-science-from/9781491901410/ch04.html)\n1. [datacamp](https://www.datacamp.com/community/blog/python-scipy-cheat-sheet)\n1. [damir.cavar](http://damir.cavar.me/)\n1. [Linear_algebra](https://en.wikipedia.org/wiki/Linear_algebra)\n1. [http://linear.ups.edu/html/fcla.html](http://linear.ups.edu/html/fcla.html)\n1. [mathsisfun](https://www.mathsisfun.com/algebra/matrix-multiplying.html)\n1. [scipy](https://docs.scipy.org/doc/numpy-1.15.0/reference/generated/numpy.outer.html)\n1. [tutorialspoint](https://www.tutorialspoint.com/numpy/numpy_inv.htm)\n1. [machinelearningmastery](https://machinelearningmastery.com/introduction-to-tensors-for-machine-learning/)\n1. [gamedevelopertips](http://gamedevelopertips.com/vector-in-game-development/)\n1. [Scalars-Vectors-Matrices-and-Tensors](https://hadrienj.github.io/posts/Deep-Learning-Book-Series-2.1-Scalars-Vectors-Matrices-and-Tensors/)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec70fde23a22e69384a17f87633ba03eb981a77a | 1,768 | ipynb | Jupyter Notebook | docs/ipynb/other.ipynb | stibus/ubermagtable | 17b9a97e532c975edb3abb890a46d64eda07b307 | [
"BSD-3-Clause"
]
| 20 | 2019-06-14T07:09:44.000Z | 2022-03-02T17:37:29.000Z | docs/ipynb/other.ipynb | stibus/ubermagtable | 17b9a97e532c975edb3abb890a46d64eda07b307 | [
"BSD-3-Clause"
]
| 50 | 2019-06-13T13:41:57.000Z | 2022-03-28T09:14:33.000Z | docs/ipynb/other.ipynb | stibus/ubermagtable | 17b9a97e532c975edb3abb890a46d64eda07b307 | [
"BSD-3-Clause"
]
| 7 | 2019-08-28T14:16:10.000Z | 2021-12-13T21:06:06.000Z | 30.482759 | 358 | 0.614819 | [
[
[
"# Support, License, and Acknowledgements\n\n## Support\n\nIf you require support, have questions, want to report a bug, or want to suggest an improvement, please raise an issue in [ubermag/help](https://github.com/ubermag/help) repository.\n\n## Contributions\n\nAll contributions are welcome, however small they are. If you would like to contribute, please fork the repository and create a pull request. If you are not sure how to contribute, please contact us by raising an issue in [ubermag/help](https://github.com/ubermag/help) repository, and we are going to help you get started and assist you on the way.\n\n## License\n\nLicensed under the BSD 3-Clause \"New\" or \"Revised\" License. For details, please refer to the LICENSE file.\n\n## Acknowledgements\n\n- [OpenDreamKit](http://opendreamkit.org/) – Horizon 2020 European Research Infrastructure project (676541)\n\n- EPSRC Programme Grant on [Skyrmionics](http://www.skyrmions.ac.uk) (EP/N032128/1)",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown"
]
]
|
ec71057a6a2d62cb992aa8fc698326a6fb1797ca | 13,212 | ipynb | Jupyter Notebook | utilities/video-analysis/notebooks/common/create_azure_vm.ipynb | fvneerden/live-video-analytics | 3e67d4c411fb59ab7e5279f307ff6867925f5d0d | [
"MIT"
]
| 69 | 2020-06-01T14:36:42.000Z | 2021-11-02T03:22:54.000Z | utilities/video-analysis/notebooks/common/create_azure_vm.ipynb | fvneerden/live-video-analytics | 3e67d4c411fb59ab7e5279f307ff6867925f5d0d | [
"MIT"
]
| 104 | 2020-05-18T18:17:36.000Z | 2021-07-02T01:49:48.000Z | utilities/video-analysis/notebooks/common/create_azure_vm.ipynb | fvneerden/live-video-analytics | 3e67d4c411fb59ab7e5279f307ff6867925f5d0d | [
"MIT"
]
| 95 | 2020-05-18T18:07:10.000Z | 2021-11-02T12:12:07.000Z | 42.619355 | 424 | 0.431502 | [
[
[
"# Create Azure Virtual Machine\nRecall that in [the previous section](create_azure_services.ipynb), we created an IoT Edge device identity in Azure IoT Hub. In this section, we will be creating and configuring a virtual machine (VM) to act as our IoT Edge device.",
"_____no_output_____"
],
[
"## Get Global Variables",
"_____no_output_____"
]
],
[
[
"from env_variables import *\nresourceTags = \"Owner={} Project=lva\".format(userName)",
"_____no_output_____"
]
],
[
[
"## Check VM Quota\n\nVerify that your subscription has enough quota to create your desired VM.",
"_____no_output_____"
]
],
[
[
"output = !az vm list-sizes --subscription $azureSubscriptionId --location $resourceLocation --output json\nsizes = ''.join(output)\n\nimport json\njsonDetails = json.loads(sizes)\n\nfor n in jsonDetails:\n if vm_type == n['name']:\n neededSpace = n['numberOfCores']\n print('\\n' + n['name'])\n print(\"Number of Cores Required to Deploy: %s vCPUs\" % neededSpace)",
"_____no_output_____"
],
[
"!az account set --subscription $azureSubscriptionId\noutput = !az vm list-usage --location $resourceLocation --output json\noutput = ''.join(output)",
"_____no_output_____"
],
[
"import json\njsonDetails = json.loads(output)\n\nneeded_size = vm_type.split('_')\nneeded_size = needed_size[1]\nneeded_size = ''.join([i for i in needed_size if not i.isdigit()])\n\nquota_remaining = 0\nfor n in jsonDetails:\n if (needed_size in n['localName']):\n quota_remaining = int(n['limit']) - int(n['currentValue'])\n print('\\n' + n['localName'])\n print(\"Quota Remaining: %s vCPUs\" % quota_remaining)",
"_____no_output_____"
]
],
[
[
"Check if the quota remaining is greater than the size of the VM you wish to create.\n\nIf you do not enough quota, you can try any of the following:\n\n* [Delete VMs](https://docs.microsoft.com/en-us/powershell/module/azurerm.compute/remove-azurermvm?view=azurermps-6.13.0) that might not be in use to free up quota.\n* [Increase quota](https://docs.microsoft.com/en-us/azure/azure-portal/supportability/per-vm-quota-requests) limits.",
"_____no_output_____"
],
[
"## Create the Virtual Machine\nThe code snippets below will execute a shell script to create an Ubuntu VM in your Azure subscription. \n\nBefore starting, we will be setting the parameters to be used for creating the VM. For more details on the process of creating a VM using Azure, see the tutorial on creating a [Linux VM on Azure CLI](https://docs.microsoft.com/en-us/azure/virtual-machines/linux/quick-create-cli) or the tutorial on creating a [Windows VM on Azure CLI](https://docs.microsoft.com/en-us/azure/virtual-machines/windows/quick-create-cli).",
"_____no_output_____"
]
],
[
[
"dns_name = iotDeviceId\npublic_ip_name = iotDeviceId + 'publicip'\nvnet_name=iotDeviceId + 'vnet'\nsubnet_name=iotDeviceId + 'subnet'\nvnet_prefix=\"192.168.0.0/16\"\nsubnet_name=\"FrontEnd\"\nsubnet_prefix=\"192.168.1.0/24\"\nnsg_name=iotDeviceId + 'nsg'\nnic_name=iotDeviceId + 'nic'\n\n# Static DNS name of the VM\nvm_dns_name= iotDeviceId + \".\" + resourceLocation + \".cloudapp.azure.com\"",
"_____no_output_____"
],
[
"# Create resource group\n!az group create \\\n --name $resourceGroupName \\\n --location $resourceLocation \\\n --tag $resourceTags\n\n# Create a public IP address resource with a static IP address\n!az network public-ip create \\\n --name $public_ip_name \\\n --resource-group $resourceGroupName \\\n --location $resourceLocation \\\n --allocation-method Static \\\n --dns-name $dns_name \\\n --tag $resourceTags \n\n# Create a virtual network with one subnet\n!az network vnet create \\\n --name $vnet_name \\\n --resource-group $resourceGroupName \\\n --location $resourceLocation \\\n --address-prefix $vnet_prefix \\\n --subnet-name $subnet_name \\\n --subnet-prefix $subnet_prefix \\\n --tag $resourceTags \n\n\n!az network nsg create \\\n --name $nsg_name \\\n --resource-group $resourceGroupName \\\n --tag $resourceTags \n\n# Open SSH port\n!az network nsg rule create \\\n --resource-group $resourceGroupName \\\n --nsg-name $nsg_name \\\n --name \"Default SSH\" \\\n --destination-port-ranges 22 \\\n --protocol Tcp \\\n --access Allow \\\n --priority 1020 \n\n# Create a network interface connected to the VNet with a static private IP address \n# and associate the public IP address resource to the NIC.\n!az network nic create \\\n --name $nic_name \\\n --resource-group $resourceGroupName \\\n --location $resourceLocation \\\n --subnet $subnet_name \\\n --vnet-name $vnet_name \\\n --public-ip-address $public_ip_name \\\n --network-security-group $nsg_name \\\n --tag $resourceTags \n",
"_____no_output_____"
],
[
"# Create the VM\n# For image param: Ubuntu 18.04-LTS image -> https://docs.microsoft.com/en-us/azure/virtual-machines/linux/cli-ps-findimage\n# Instead of Password login, we are uploading Private SSH key. Update the path accrodingly if needed\n!az vm create \\\n --name $iotDeviceId \\\n --resource-group $resourceGroupName \\\n --location $resourceLocation \\\n --storage-sku Standard_LRS \\\n --os-disk-name $iotDeviceId\"_osdisk\" \\\n --image \"Canonical:UbuntuServer:18.04-LTS:latest\" \\\n --size $vm_type \\\n --nics $nic_name \\\n --admin-username $userName \\\n --authentication-type ssh \\\n --tag $resourceTags \\\n --generate-ssh-keys ",
"_____no_output_____"
]
],
[
[
"## Optional: Remove Previously Trusted Host\n\nIf you previously created a VM with the same name, you may run into issues connecting the VM through SSH. As such, you can first remove this VM from your list of known hosts. ",
"_____no_output_____"
]
],
[
[
"!ssh-keygen -f ~/.ssh/known_hosts -R $vm_dns_name ",
"_____no_output_____"
]
],
[
[
"If the above command was unable to run, you can try running the following command in your terminal:\n\n`ssh-keygen -R \"<vm_dns_name>\"`\n\nwhere vm_dns_name is the variable defined above. ",
"_____no_output_____"
],
[
"## Print the SSH Connection String for the VM",
"_____no_output_____"
]
],
[
[
"# If needed, use the output command to SSH into the VM\nsshstring = \"ssh -i ~/.ssh/id_rsa {0}@{1}\".format(userName, vm_dns_name)",
"_____no_output_____"
],
[
"# save the ssh conn string into .env file\nset_key(envPath, \"SSH_STRING\", sshstring.strip())",
"_____no_output_____"
]
],
[
[
"> <span>[!NOTE]</span>\n> If you need the SSH connection string anytime in the future to connect to your IoT Edge device VM, you can find the string in the [.env file](.env) as `SSH_STRING`. ",
"_____no_output_____"
],
[
"## Next Steps\nIf you experience unresolved issues, you can always [delete your resources](https://docs.microsoft.com/en-us/azure/azure-resource-manager/management/delete-resource-group?tabs=azure-powershell) and re-run this section.\n\nIf all the code cells above have successfully finished running, return to the Readme page to continue. ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
]
]
|
ec711fc138c9a60d3905488050b9b35c8845c2fa | 74,656 | ipynb | Jupyter Notebook | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | deep0892/pandas_practice | f3c1581d2aa167416c057b6bb140c3c29677ab20 | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | deep0892/pandas_practice | f3c1581d2aa167416c057b6bb140c3c29677ab20 | [
"BSD-3-Clause"
]
| null | null | null | 02_Filtering_&_Sorting/Euro12/Exercises_with_Solutions.ipynb | deep0892/pandas_practice | f3c1581d2aa167416c057b6bb140c3c29677ab20 | [
"BSD-3-Clause"
]
| null | null | null | 34.105071 | 185 | 0.258546 | [
[
[
"# Ex2 - Filtering and Sorting Data\nCheck out [Euro 12 Exercises Video Tutorial](https://youtu.be/iqk5d48Qisg) to watch a data scientist go through the exercises",
"_____no_output_____"
],
[
"This time we are going to pull data directly from the internet.\n\n### Step 1. Import the necessary libraries",
"_____no_output_____"
]
],
[
[
"import pandas as pd",
"_____no_output_____"
]
],
[
[
"### Step 2. Import the dataset from this [address](https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv). ",
"_____no_output_____"
],
[
"### Step 3. Assign it to a variable called euro12.",
"_____no_output_____"
]
],
[
[
"euro12 = pd.read_csv('https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/02_Filtering_%26_Sorting/Euro12/Euro_2012_stats_TEAM.csv', sep=',')\neuro12",
"_____no_output_____"
]
],
[
[
"### Step 4. Select only the Goal column.",
"_____no_output_____"
]
],
[
[
"euro12.Goals",
"_____no_output_____"
]
],
[
[
"### Step 5. How many team participated in the Euro2012?",
"_____no_output_____"
]
],
[
[
"euro12.shape[0]",
"_____no_output_____"
]
],
[
[
"### Step 6. What is the number of columns in the dataset?",
"_____no_output_____"
]
],
[
[
"euro12.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 16 entries, 0 to 15\nData columns (total 35 columns):\nTeam 16 non-null object\nGoals 16 non-null int64\nShots on target 16 non-null int64\nShots off target 16 non-null int64\nShooting Accuracy 16 non-null object\n% Goals-to-shots 16 non-null object\nTotal shots (inc. Blocked) 16 non-null int64\nHit Woodwork 16 non-null int64\nPenalty goals 16 non-null int64\nPenalties not scored 16 non-null int64\nHeaded goals 16 non-null int64\nPasses 16 non-null int64\nPasses completed 16 non-null int64\nPassing Accuracy 16 non-null object\nTouches 16 non-null int64\nCrosses 16 non-null int64\nDribbles 16 non-null int64\nCorners Taken 16 non-null int64\nTackles 16 non-null int64\nClearances 16 non-null int64\nInterceptions 16 non-null int64\nClearances off line 15 non-null float64\nClean Sheets 16 non-null int64\nBlocks 16 non-null int64\nGoals conceded 16 non-null int64\nSaves made 16 non-null int64\nSaves-to-shots ratio 16 non-null object\nFouls Won 16 non-null int64\nFouls Conceded 16 non-null int64\nOffsides 16 non-null int64\nYellow Cards 16 non-null int64\nRed Cards 16 non-null int64\nSubs on 16 non-null int64\nSubs off 16 non-null int64\nPlayers Used 16 non-null int64\ndtypes: float64(1), int64(29), object(5)\nmemory usage: 4.4+ KB\n"
]
],
[
[
"### Step 7. View only the columns Team, Yellow Cards and Red Cards and assign them to a dataframe called discipline",
"_____no_output_____"
]
],
[
[
"# filter only giving the column names\n\ndiscipline = euro12[['Team', 'Yellow Cards', 'Red Cards']]\ndiscipline",
"_____no_output_____"
]
],
[
[
"### Step 8. Sort the teams by Red Cards, then to Yellow Cards",
"_____no_output_____"
]
],
[
[
"discipline.sort_values(['Red Cards', 'Yellow Cards'], ascending = False)",
"_____no_output_____"
]
],
[
[
"### Step 9. Calculate the mean Yellow Cards given per Team",
"_____no_output_____"
]
],
[
[
"round(discipline['Yellow Cards'].mean())",
"_____no_output_____"
]
],
[
[
"### Step 10. Filter teams that scored more than 6 goals",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Goals > 6]",
"_____no_output_____"
]
],
[
[
"### Step 11. Select the teams that start with G",
"_____no_output_____"
]
],
[
[
"euro12[euro12.Team.str.startswith('G')]",
"_____no_output_____"
]
],
[
[
"### Step 12. Select the first 7 columns",
"_____no_output_____"
]
],
[
[
"# use .iloc to slices via the position of the passed integers\n# : means all, 0:7 means from 0 to 7\n\neuro12.iloc[: , 0:7]",
"_____no_output_____"
]
],
[
[
"### Step 13. Select all columns except the last 3.",
"_____no_output_____"
]
],
[
[
"# use negative to exclude the last 3 columns\n\neuro12.iloc[: , :-3]",
"_____no_output_____"
]
],
[
[
"### Step 14. Present only the Shooting Accuracy from England, Italy and Russia",
"_____no_output_____"
]
],
[
[
"# .loc is another way to slice, using the labels of the columns and indexes\n\neuro12.loc[euro12.Team.isin(['England', 'Italy', 'Russia']), ['Team','Shooting Accuracy']]",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec712637e63bbcd72bf927f0b9bb0dad6fff371e | 6,556 | ipynb | Jupyter Notebook | docs/tutorials/ranking/dirichlet.ipynb | pmla/scikit-network | 6e74a2338f53f1e54a25e7e19ab9fcf56371c275 | [
"BSD-3-Clause"
]
| 457 | 2018-07-24T12:42:14.000Z | 2022-03-31T08:30:39.000Z | docs/tutorials/ranking/dirichlet.ipynb | pmla/scikit-network | 6e74a2338f53f1e54a25e7e19ab9fcf56371c275 | [
"BSD-3-Clause"
]
| 281 | 2018-07-13T05:01:19.000Z | 2022-03-31T14:13:43.000Z | docs/tutorials/ranking/dirichlet.ipynb | pmla/scikit-network | 6e74a2338f53f1e54a25e7e19ab9fcf56371c275 | [
"BSD-3-Clause"
]
| 58 | 2019-04-22T09:04:32.000Z | 2022-03-30T12:43:08.000Z | 21.565789 | 216 | 0.552471 | [
[
[
"# Dirichlet",
"_____no_output_____"
],
[
"This notebook illustrates the ranking of the nodes of a graph through the [Dirichlet problem](https://scikit-network.readthedocs.io/en/latest/reference/ranking.html#dirichlet) (heat diffusion with constraints).",
"_____no_output_____"
]
],
[
[
"from IPython.display import SVG",
"_____no_output_____"
],
[
"import numpy as np",
"_____no_output_____"
],
[
"from sknetwork.data import karate_club, painters, movie_actor\nfrom sknetwork.ranking import Dirichlet\nfrom sknetwork.visualization import svg_graph, svg_digraph, svg_bigraph",
"_____no_output_____"
]
],
[
[
"## Graphs",
"_____no_output_____"
]
],
[
[
"graph = karate_club(metadata=True)\nadjacency = graph.adjacency\nposition = graph.position\nlabels_true = graph.labels",
"_____no_output_____"
],
[
"# heat diffusion\ndirichlet = Dirichlet()\nseeds = {0: 0, 33: 1}\nscores = dirichlet.fit_transform(adjacency, seeds)",
"_____no_output_____"
],
[
"image = svg_graph(adjacency, position, scores=scores, seeds=seeds)\nSVG(image)",
"_____no_output_____"
]
],
[
[
"## Directed graphs",
"_____no_output_____"
]
],
[
[
"graph = painters(metadata=True)\nadjacency = graph.adjacency\nposition = graph.position\nnames = graph.names",
"_____no_output_____"
],
[
"picasso = 0\nmonet = 1",
"_____no_output_____"
],
[
"dirichlet = Dirichlet()\nseeds = {picasso: 0, monet: 1}\nscores = dirichlet.fit_transform(adjacency, seeds)",
"_____no_output_____"
],
[
"image = svg_digraph(adjacency, position, names, scores=scores, seeds=seeds)\nSVG(image)",
"_____no_output_____"
]
],
[
[
"## Bipartite graphs",
"_____no_output_____"
]
],
[
[
"graph = movie_actor(metadata=True)\nbiadjacency = graph.biadjacency\nnames_row = graph.names_row\nnames_col = graph.names_col",
"_____no_output_____"
],
[
"dirichlet = Dirichlet()",
"_____no_output_____"
],
[
"drive = 3\naviator = 9",
"_____no_output_____"
],
[
"seeds_row = {drive: 0, aviator: 1}\ndirichlet.fit(biadjacency, seeds_row)\nscores_row = dirichlet.scores_row_\nscores_col = dirichlet.scores_col_",
"_____no_output_____"
],
[
"image = svg_bigraph(biadjacency, names_row, names_col, scores_row=scores_row, scores_col=scores_col, \n seeds_row=seeds_row)\nSVG(image)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec7131b38285960a114e3da8a080310c3ca5ceec | 10,472 | ipynb | Jupyter Notebook | 深度学习/d2l-zh-1.1/chapter_convolutional-neural-networks/alexnet.ipynb | Blankwhiter/LearningNotes | 83e570bf386a8e2b5aa699c3d38b83e5dcdd9cb0 | [
"MIT"
]
| null | null | null | 深度学习/d2l-zh-1.1/chapter_convolutional-neural-networks/alexnet.ipynb | Blankwhiter/LearningNotes | 83e570bf386a8e2b5aa699c3d38b83e5dcdd9cb0 | [
"MIT"
]
| 3 | 2020-08-14T07:50:27.000Z | 2020-08-14T08:51:06.000Z | 深度学习/d2l-zh-1.1/chapter_convolutional-neural-networks/alexnet.ipynb | Blankwhiter/LearningNotes | 83e570bf386a8e2b5aa699c3d38b83e5dcdd9cb0 | [
"MIT"
]
| 2 | 2021-03-14T05:58:45.000Z | 2021-08-29T17:25:52.000Z | 35.14094 | 345 | 0.62395 | [
[
[
"# 深度卷积神经网络(AlexNet)\n\n在LeNet提出后的将近20年里,神经网络一度被其他机器学习方法超越,如支持向量机。虽然LeNet可以在早期的小数据集上取得好的成绩,但是在更大的真实数据集上的表现并不尽如人意。一方面,神经网络计算复杂。虽然20世纪90年代也有过一些针对神经网络的加速硬件,但并没有像之后GPU那样大量普及。因此,训练一个多通道、多层和有大量参数的卷积神经网络在当年很难完成。另一方面,当年研究者还没有大量深入研究参数初始化和非凸优化算法等诸多领域,导致复杂的神经网络的训练通常较困难。\n\n我们在上一节看到,神经网络可以直接基于图像的原始像素进行分类。这种称为端到端(end-to-end)的方法节省了很多中间步骤。然而,在很长一段时间里更流行的是研究者通过勤劳与智慧所设计并生成的手工特征。这类图像分类研究的主要流程是:\n\n1. 获取图像数据集;\n2. 使用已有的特征提取函数生成图像的特征;\n3. 使用机器学习模型对图像的特征分类。\n\n当时认为的机器学习部分仅限最后这一步。如果那时候跟机器学习研究者交谈,他们会认为机器学习既重要又优美。优雅的定理证明了许多分类器的性质。机器学习领域生机勃勃、严谨而且极其有用。然而,如果跟计算机视觉研究者交谈,则是另外一幅景象。他们会告诉你图像识别里“不可告人”的现实是:计算机视觉流程中真正重要的是数据和特征。也就是说,使用较干净的数据集和较有效的特征甚至比机器学习模型的选择对图像分类结果的影响更大。\n\n\n## 学习特征表示\n\n既然特征如此重要,它该如何表示呢?\n\n我们已经提到,在相当长的时间里,特征都是基于各式各样手工设计的函数从数据中提取的。事实上,不少研究者通过提出新的特征提取函数不断改进图像分类结果。这一度为计算机视觉的发展做出了重要贡献。\n\n然而,另一些研究者则持异议。他们认为特征本身也应该由学习得来。他们还相信,为了表征足够复杂的输入,特征本身应该分级表示。持这一想法的研究者相信,多层神经网络可能可以学得数据的多级表征,并逐级表示越来越抽象的概念或模式。以图像分类为例,并回忆[“二维卷积层”](conv-layer.ipynb)一节中物体边缘检测的例子。在多层神经网络中,图像的第一级的表示可以是在特定的位置和⻆度是否出现边缘;而第二级的表示说不定能够将这些边缘组合出有趣的模式,如花纹;在第三级的表示中,也许上一级的花纹能进一步汇合成对应物体特定部位的模式。这样逐级表示下去,最终,模型能够较容易根据最后一级的表示完成分类任务。需要强调的是,输入的逐级表示由多层模型中的参数决定,而这些参数都是学出来的。\n\n尽管一直有一群执着的研究者不断钻研,试图学习视觉数据的逐级表征,然而很长一段时间里这些野心都未能实现。这其中有诸多因素值得我们一一分析。\n\n\n### 缺失要素一:数据\n\n包含许多特征的深度模型需要大量的有标签的数据才能表现得比其他经典方法更好。限于早期计算机有限的存储和90年代有限的研究预算,大部分研究只基于小的公开数据集。例如,不少研究论文基于加州大学欧文分校(UCI)提供的若干个公开数据集,其中许多数据集只有几百至几千张图像。这一状况在2010年前后兴起的大数据浪潮中得到改善。特别是,2009年诞生的ImageNet数据集包含了1,000大类物体,每类有多达数千张不同的图像。这一规模是当时其他公开数据集无法与之相提并论的。ImageNet数据集同时推动计算机视觉和机器学习研究进入新的阶段,使此前的传统方法不再有优势。\n\n\n### 缺失要素二:硬件\n\n深度学习对计算资源要求很高。早期的硬件计算能力有限,这使训练较复杂的神经网络变得很困难。然而,通用GPU的到来改变了这一格局。很久以来,GPU都是为图像处理和计算机游戏设计的,尤其是针对大吞吐量的矩阵和向量乘法从而服务于基本的图形变换。值得庆幸的是,这其中的数学表达与深度网络中的卷积层的表达类似。通用GPU这个概念在2001年开始兴起,涌现出诸如OpenCL和CUDA之类的编程框架。这使得GPU也在2010年前后开始被机器学习社区使用。\n\n\n## AlexNet\n\n2012年,AlexNet横空出世。这个模型的名字来源于论文第一作者的姓名Alex Krizhevsky [1]。AlexNet使用了8层卷积神经网络,并以很大的优势赢得了ImageNet 2012图像识别挑战赛。它首次证明了学习到的特征可以超越手工设计的特征,从而一举打破计算机视觉研究的前状。\n\nAlexNet与LeNet的设计理念非常相似,但也有显著的区别。\n\n第一,与相对较小的LeNet相比,AlexNet包含8层变换,其中有5层卷积和2层全连接隐藏层,以及1个全连接输出层。下面我们来详细描述这些层的设计。\n\nAlexNet第一层中的卷积窗口形状是$11\\times11$。因为ImageNet中绝大多数图像的高和宽均比MNIST图像的高和宽大10倍以上,ImageNet图像的物体占用更多的像素,所以需要更大的卷积窗口来捕获物体。第二层中的卷积窗口形状减小到$5\\times5$,之后全采用$3\\times3$。此外,第一、第二和第五个卷积层之后都使用了窗口形状为$3\\times3$、步幅为2的最大池化层。而且,AlexNet使用的卷积通道数也大于LeNet中的卷积通道数数十倍。\n\n紧接着最后一个卷积层的是两个输出个数为4,096的全连接层。这两个巨大的全连接层带来将近1 GB的模型参数。由于早期显存的限制,最早的AlexNet使用双数据流的设计使一块GPU只需要处理一半模型。幸运的是,显存在过去几年得到了长足的发展,因此通常我们不再需要这样的特别设计了。\n\n第二,AlexNet将sigmoid激活函数改成了更加简单的ReLU激活函数。一方面,ReLU激活函数的计算更简单,例如它并没有sigmoid激活函数中的求幂运算。另一方面,ReLU激活函数在不同的参数初始化方法下使模型更容易训练。这是由于当sigmoid激活函数输出极接近0或1时,这些区域的梯度几乎为0,从而造成反向传播无法继续更新部分模型参数;而ReLU激活函数在正区间的梯度恒为1。因此,若模型参数初始化不当,sigmoid函数可能在正区间得到几乎为0的梯度,从而令模型无法得到有效训练。\n\n第三,AlexNet通过丢弃法(参见[“丢弃法”](../chapter_deep-learning-basics/dropout.ipynb)一节)来控制全连接层的模型复杂度。而LeNet并没有使用丢弃法。\n\n第四,AlexNet引入了大量的图像增广,如翻转、裁剪和颜色变化,从而进一步扩大数据集来缓解过拟合。我们将在后面的[“图像增广”](../chapter_computer-vision/image-augmentation.ipynb)一节详细介绍这种方法。\n\n下面我们实现稍微简化过的AlexNet。",
"_____no_output_____"
]
],
[
[
"import d2lzh as d2l\nfrom mxnet import gluon, init, nd\nfrom mxnet.gluon import data as gdata, nn\nimport os\nimport sys\n\nnet = nn.Sequential()\n# 使用较大的11 x 11窗口来捕获物体。同时使用步幅4来较大幅度减小输出高和宽。这里使用的输出通\n# 道数比LeNet中的也要大很多\nnet.add(nn.Conv2D(96, kernel_size=11, strides=4, activation='relu'),\n nn.MaxPool2D(pool_size=3, strides=2),\n # 减小卷积窗口,使用填充为2来使得输入与输出的高和宽一致,且增大输出通道数\n nn.Conv2D(256, kernel_size=5, padding=2, activation='relu'),\n nn.MaxPool2D(pool_size=3, strides=2),\n # 连续3个卷积层,且使用更小的卷积窗口。除了最后的卷积层外,进一步增大了输出通道数。\n # 前两个卷积层后不使用池化层来减小输入的高和宽\n nn.Conv2D(384, kernel_size=3, padding=1, activation='relu'),\n nn.Conv2D(384, kernel_size=3, padding=1, activation='relu'),\n nn.Conv2D(256, kernel_size=3, padding=1, activation='relu'),\n nn.MaxPool2D(pool_size=3, strides=2),\n # 这里全连接层的输出个数比LeNet中的大数倍。使用丢弃层来缓解过拟合\n nn.Dense(4096, activation=\"relu\"), nn.Dropout(0.5),\n nn.Dense(4096, activation=\"relu\"), nn.Dropout(0.5),\n # 输出层。由于这里使用Fashion-MNIST,所以用类别数为10,而非论文中的1000\n nn.Dense(10))",
"_____no_output_____"
]
],
[
[
"我们构造一个高和宽均为224的单通道数据样本来观察每一层的输出形状。",
"_____no_output_____"
]
],
[
[
"X = nd.random.uniform(shape=(1, 1, 224, 224))\nnet.initialize()\nfor layer in net:\n X = layer(X)\n print(layer.name, 'output shape:\\t', X.shape)",
"conv0 output shape:\t (1, 96, 54, 54)\npool0 output shape:\t (1, 96, 26, 26)\nconv1 output shape:\t (1, 256, 26, 26)\npool1 output shape:\t (1, 256, 12, 12)\nconv2 output shape:\t (1, 384, 12, 12)\nconv3 output shape:\t (1, 384, 12, 12)\nconv4 output shape:\t (1, 256, 12, 12)\npool2 output shape:\t (1, 256, 5, 5)\ndense0 output shape:\t (1, 4096)\ndropout0 output shape:\t (1, 4096)\ndense1 output shape:\t (1, 4096)\ndropout1 output shape:\t (1, 4096)\ndense2 output shape:\t (1, 10)\n"
]
],
[
[
"## 读取数据集\n\n虽然论文中AlexNet使用ImageNet数据集,但因为ImageNet数据集训练时间较长,我们仍用前面的Fashion-MNIST数据集来演示AlexNet。读取数据的时候我们额外做了一步将图像高和宽扩大到AlexNet使用的图像高和宽224。这个可以通过`Resize`实例来实现。也就是说,我们在`ToTensor`实例前使用`Resize`实例,然后使用`Compose`实例来将这两个变换串联以方便调用。",
"_____no_output_____"
]
],
[
[
"# 本函数已保存在d2lzh包中方便以后使用\ndef load_data_fashion_mnist(batch_size, resize=None, root=os.path.join(\n '~', '.mxnet', 'datasets', 'fashion-mnist')):\n root = os.path.expanduser(root) # 展开用户路径'~'\n transformer = []\n if resize:\n transformer += [gdata.vision.transforms.Resize(resize)]\n transformer += [gdata.vision.transforms.ToTensor()]\n transformer = gdata.vision.transforms.Compose(transformer)\n mnist_train = gdata.vision.FashionMNIST(root=root, train=True)\n mnist_test = gdata.vision.FashionMNIST(root=root, train=False)\n num_workers = 0 if sys.platform.startswith('win32') else 4\n train_iter = gdata.DataLoader(\n mnist_train.transform_first(transformer), batch_size, shuffle=True,\n num_workers=num_workers)\n test_iter = gdata.DataLoader(\n mnist_test.transform_first(transformer), batch_size, shuffle=False,\n num_workers=num_workers)\n return train_iter, test_iter\n\nbatch_size = 128\n# 如出现“out of memory”的报错信息,可减小batch_size或resize\ntrain_iter, test_iter = load_data_fashion_mnist(batch_size, resize=224)",
"_____no_output_____"
]
],
[
[
"## 训练\n\n这时候我们可以开始训练AlexNet了。相对于上一节的LeNet,这里的主要改动是使用了更小的学习率。",
"_____no_output_____"
]
],
[
[
"lr, num_epochs, ctx = 0.01, 5, d2l.try_gpu()\nnet.initialize(force_reinit=True, ctx=ctx, init=init.Xavier())\ntrainer = gluon.Trainer(net.collect_params(), 'sgd', {'learning_rate': lr})\nd2l.train_ch5(net, train_iter, test_iter, batch_size, trainer, ctx, num_epochs)",
"training on gpu(0)\n"
]
],
[
[
"## 小结\n\n* AlexNet与LeNet结构类似,但使用了更多的卷积层和更大的参数空间来拟合大规模数据集ImageNet。它是浅层神经网络和深度神经网络的分界线。\n* 虽然看上去AlexNet的实现比LeNet的实现也就多了几行代码而已,但这个观念上的转变和真正优秀实验结果的产生令学术界付出了很多年。\n\n## 练习\n\n* 尝试增加迭代周期。跟LeNet的结果相比,AlexNet的结果有什么区别?为什么?\n* AlexNet对Fashion-MNIST数据集来说可能过于复杂。试着简化模型来使训练更快,同时保证准确率不明显下降。\n* 修改批量大小,观察准确率和内存或显存的变化。\n\n\n\n\n## 参考文献\n\n[1] Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems (pp. 1097-1105).\n\n## 扫码直达[讨论区](https://discuss.gluon.ai/t/topic/1258)\n\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec713de4846826be2eb5bb6476bed18841d20e82 | 55,930 | ipynb | Jupyter Notebook | Session08_Scopes and Closures/notebooks/Session08_ClassRoom.ipynb | abdksyed/EPAi | 68bdf04fe5861b4f77a838adddd371b6fb1660c3 | [
"MIT"
]
| 3 | 2020-08-07T16:01:19.000Z | 2021-01-29T11:22:06.000Z | Session08_Scopes and Closures/notebooks/Session08_ClassRoom.ipynb | abdksyed/EPAi | 68bdf04fe5861b4f77a838adddd371b6fb1660c3 | [
"MIT"
]
| null | null | null | Session08_Scopes and Closures/notebooks/Session08_ClassRoom.ipynb | abdksyed/EPAi | 68bdf04fe5861b4f77a838adddd371b6fb1660c3 | [
"MIT"
]
| 2 | 2020-08-08T06:27:13.000Z | 2020-08-09T17:02:43.000Z | 19.882688 | 882 | 0.48836 | [
[
[
"a = 10",
"_____no_output_____"
],
[
"def my_func(n):\n c = n ** 2\n return c ",
"_____no_output_____"
],
[
"def my_func(n):\n print('global a:', a) \n c = a ** n\n return c",
"_____no_output_____"
],
[
"my_func(2)",
"global a: 10\n"
],
[
"a =10\ndef my_func(n):\n a = 20\n c = a ** n\n return c\nmy_func(2)",
"_____no_output_____"
],
[
"a",
"_____no_output_____"
],
[
"def my_func(n):\n global var\n var = 20\n c = var ** n\n return c",
"_____no_output_____"
],
[
"my_func(2)",
"_____no_output_____"
],
[
"var",
"_____no_output_____"
],
[
"var\n",
"_____no_output_____"
],
[
"def my_func(n):\n global vor\n vor = 20\n c = vor ** n\n return c",
"_____no_output_____"
],
[
"vor",
"_____no_output_____"
],
[
"my_func(2)",
"_____no_output_____"
],
[
"vor",
"_____no_output_____"
],
[
"a = 10\n\ndef my_func():\n global a\n a = 'hello'\n print('global a:', a)\nprint(a)\nmy_func()",
"10\nglobal a: hello\n"
],
[
"a",
"_____no_output_____"
],
[
"def my_func(n):\n global var\n c = var ** n\n return c\nvar = 2\nvar",
"_____no_output_____"
],
[
"my_func(2)",
"_____no_output_____"
],
[
"def my_func():\n print('global a:', a)\nmy_func()",
"global a: hello\n"
],
[
"def my_func():\n print('global a:', a)\n a = 'hello world'\n print(a)",
"_____no_output_____"
],
[
"my_func()",
"_____no_output_____"
],
[
"print(b)\nb = 1",
"_____no_output_____"
],
[
"a = 10\nf = lambda n: print(a ** n)\nf(2)",
"_____no_output_____"
],
[
"f = lambda n: print(b ** n)\nf(2)",
"_____no_output_____"
],
[
"print(True) ",
"_____no_output_____"
],
[
"def print(x):\n return 'hello {0}'.format(x)",
"_____no_output_____"
],
[
"print('world')",
"_____no_output_____"
],
[
"print('world', '!')",
"_____no_output_____"
],
[
"del print\nprint('world', '!')",
"_____no_output_____"
],
[
"# Java or C++\nfor (int i = 0; i < 10; i++){\n int x = 2 * i\n}\nsystem.out.println(x) <<< This throws error",
"_____no_output_____"
],
[
"for i in range(10):\n x = 2 * i\nprint(x)",
"_____no_output_____"
],
[
"if (True):\n y = 100\nprint(y)",
"_____no_output_____"
],
[
"if (False):\n yy = 100\nprint(yy)",
"_____no_output_____"
],
[
"def my_func():\n if(True):\n yyyy = 100\n print(yyyy)\nmy_func()\nprint(yyyy)",
"_____no_output_____"
],
[
"z = 100\ndef outer_func():\n x = 'hello'\n def inner_func():\n y = 'red'\n print(x)\n inner_func()\n\nouter_func()",
"_____no_output_____"
],
[
"z = 100\ndef outer_func():\n global z\n print(z)\n def inner_func():\n global z\n print(z)\n inner_func()\n\nouter_func()",
"_____no_output_____"
],
[
"def outer_func():\n x = 'hello'\n def inner():\n x = 'tsai'\n print('inner x:', x)\n inner()\n print('outer x:', x)",
"_____no_output_____"
],
[
"outer_func()",
"_____no_output_____"
],
[
"def outer():\n x = 'hello'\n def inner1():\n def inner2():\n nonlocal x\n x = 'python'\n inner2()\n inner1()\n print(x)\nouter()",
"_____no_output_____"
],
[
"def outer():\n x = 'hello'\n def inner1():\n nonlocal x\n x = 'inner1'\n def inner2():\n nonlocal x\n x = 'inner2'\n inner2()\n inner1()\n print(x)\nouter()",
"_____no_output_____"
],
[
"x = 'global'\n\ndef outer():\n global x\n x = 'outer'\n def inner():\n nonlocal x \n x = 'inner'\n inner()\n print(x)",
"_____no_output_____"
],
[
"del x\ndef outer():\n global x\n x = 'outer'\n def inner():\n global x \n x = 'inner'\n inner()\n print(x)\nouter()\nprint(x)",
"_____no_output_____"
],
[
"x = 0\ndef fn1():\n def fn2():\n def fn3():\n nonlocal x\n print(x)\n fn3()\n fn2()\nfn1()",
"_____no_output_____"
],
[
"def counter():\n count = 0\n def inc():\n print(count)\n count += 1\n return count\n inc()\ncounter()",
"_____no_output_____"
],
[
"def outer():\n x = 'python'\n def inner():\n print(x)\n return inner",
"_____no_output_____"
],
[
"fn = outer()",
"_____no_output_____"
],
[
"fn.__code__.co_freevars",
"_____no_output_____"
],
[
"fn.__closure__",
"_____no_output_____"
],
[
"id('python')",
"_____no_output_____"
],
[
"def outer():\n x = 123\n print(hex(id(x)))\n def inner():\n x = 123\n print(hex(id(x)))\n return inner",
"_____no_output_____"
],
[
"fn = outer()",
"_____no_output_____"
],
[
"fn()",
"_____no_output_____"
],
[
"def outer():\n x = 'tsai'\n print(hex(id(x)))\n def inner():\n y = x\n print(hex(id(y)))\n return inner\nfn = outer()",
"_____no_output_____"
],
[
"fn()",
"_____no_output_____"
],
[
"fn.__closure__",
"_____no_output_____"
],
[
"def outer():\n count = 0\n def inner():\n nonlocal count\n count += 1\n return count\n return inner\nfn = outer()\nfn.__code__.co_freevars",
"_____no_output_____"
],
[
"fn.__closure__",
"_____no_output_____"
],
[
"hex(id(0))",
"_____no_output_____"
],
[
"fn()",
"_____no_output_____"
],
[
"fn.__closure__",
"_____no_output_____"
],
[
"hex(id(1))",
"_____no_output_____"
],
[
"fn(), fn.__closure__, hex(id(2))\n",
"_____no_output_____"
],
[
"def outer():\n count = 0\n def inc1():\n nonlocal count\n count += 1\n return count\n\n def inc2():\n nonlocal count\n count += 1\n return count\n return inc1, inc2\nfn1, fn2 = outer()",
"_____no_output_____"
],
[
"fn1.__code__.co_freevars, fn2.__code__.co_freevars",
"_____no_output_____"
],
[
"fn1.__closure__, fn2.__closure__",
"_____no_output_____"
],
[
"fn1()",
"_____no_output_____"
],
[
"fn1.__closure__, fn2.__closure__",
"_____no_output_____"
],
[
"fn2()",
"_____no_output_____"
],
[
"def pow(n):\n def inner(x):\n return x ** n\n return inner",
"_____no_output_____"
],
[
"square = pow(2)",
"_____no_output_____"
],
[
"square.__closure__",
"_____no_output_____"
],
[
"hex(id(2))",
"_____no_output_____"
],
[
"square(9)",
"_____no_output_____"
],
[
"square",
"_____no_output_____"
],
[
"cube = pow(3)",
"_____no_output_____"
],
[
"cube.__closure__",
"_____no_output_____"
],
[
"cube(5)",
"_____no_output_____"
],
[
"def adder(n):\n def inner(x):\n return x + n\n return inner",
"_____no_output_____"
],
[
"add_1 = adder(1)\nadd_2 = adder(2)\nadd_3 = adder(3)",
"_____no_output_____"
],
[
"add_1.__closure__, add_2.__closure__, add_3.__closure__",
"_____no_output_____"
],
[
"add_1(10)",
"_____no_output_____"
],
[
"add_2(10)",
"_____no_output_____"
],
[
"add_3(10)",
"_____no_output_____"
],
[
"adders = []\n\nfor n in range(1, 4):\n adders.append(lambda x: x + n)",
"_____no_output_____"
],
[
"adders",
"_____no_output_____"
],
[
"n",
"_____no_output_____"
],
[
"adders[0](10)",
"_____no_output_____"
],
[
"def create_adders():\n adders = []\n for n in range(1, 4):\n adders.append(lambda x: x + n)\n return adders",
"_____no_output_____"
],
[
"adders = create_adders()",
"_____no_output_____"
],
[
"adders",
"_____no_output_____"
],
[
"adders[0].__closure__",
"_____no_output_____"
],
[
"adders[1].__closure__",
"_____no_output_____"
],
[
"def create_adders():\n adders = []\n for n in range(1, 4):\n adders.append(lambda x, y=n: x + y)\n return adders",
"_____no_output_____"
],
[
"adders = create_adders()",
"_____no_output_____"
],
[
"adders[0](10), adders[1](10), adders[2](10)",
"_____no_output_____"
],
[
"class Averager:\n def __init__(self):\n self.numbers = []\n\n def add(self, number):\n self.numbers.append(number)\n total = sum(self.numbers)\n count = len(self.numbers)\n return total/count",
"_____no_output_____"
],
[
"a = Averager()",
"_____no_output_____"
],
[
"a.add(10)",
"_____no_output_____"
],
[
"a.add(20)\na.add(45)",
"_____no_output_____"
],
[
"def averager():\n numbers = []\n def add(number):\n numbers.append(number)\n total = sum(numbers)\n count = len(numbers)\n return total/count\n return add",
"_____no_output_____"
],
[
"a = averager()\na(10)\na(20)\na(45)",
"_____no_output_____"
],
[
"b = averager()\nb(10)\nb(30)",
"_____no_output_____"
],
[
"a(25)",
"_____no_output_____"
],
[
"a.__closure__, b.__closure__",
"_____no_output_____"
],
[
"def averager():\n total = 0\n count = 0\n def add(number):\n nonlocal total\n nonlocal count\n total = total + number\n count = count + 1\n return total/count\n return add",
"_____no_output_____"
],
[
"a = averager()\na(10)",
"_____no_output_____"
],
[
"a(20)\na(45)",
"_____no_output_____"
],
[
"from time import perf_counter",
"_____no_output_____"
],
[
"perf_counter()",
"_____no_output_____"
],
[
"perf_counter()",
"_____no_output_____"
],
[
"class Timer:\n def __init__(self):\n self.start = perf_counter()\n\n def poll(self):\n return perf_counter() - self.start",
"_____no_output_____"
],
[
"t1 = Timer()\nt1.poll()",
"_____no_output_____"
],
[
"t1.poll()",
"_____no_output_____"
],
[
"class Timer:\n def __init__(self):\n self.start = perf_counter()\n\n def __call__(self):\n return perf_counter() - self.start",
"_____no_output_____"
],
[
"t1 = Timer()",
"_____no_output_____"
],
[
"t1()",
"_____no_output_____"
],
[
"t1()",
"_____no_output_____"
],
[
"# let's use a closure now\n\ndef timer():\n start = perf_counter()\n def poll():\n return perf_counter() - start\n return poll",
"_____no_output_____"
],
[
"t2 = timer()",
"_____no_output_____"
],
[
"t2()",
"_____no_output_____"
],
[
"t2()",
"_____no_output_____"
],
[
"def counter(initial_value = 0):\n def inc(increment = 1):\n nonlocal initial_value\n initial_value += increment\n return initial_value\n return inc",
"_____no_output_____"
],
[
"counter1 = counter()",
"_____no_output_____"
],
[
"counter1()",
"_____no_output_____"
],
[
"counter1()",
"_____no_output_____"
],
[
"counter1()\ncounter1()\ncounter1()\ncounter1()\n",
"_____no_output_____"
],
[
"counter1(5)",
"_____no_output_____"
],
[
"def counter(fn):\n cnt = 0\n def inner(*args, **kwargs):\n nonlocal cnt\n cnt += 1\n print('{0} has been called {1} times'.format(fn.__name__, cnt))\n return fn(*args, **kwargs)\n return inner",
"_____no_output_____"
],
[
"def add(a, b):\n return a + b\ndef mul(a, b):\n return a*b",
"_____no_output_____"
],
[
"counter_add = counter(add)",
"_____no_output_____"
],
[
"counter_add.__closure__",
"_____no_output_____"
],
[
"counter_add(1, 2)",
"add has been called 1 times\n"
],
[
"counter_add(10, 20)",
"add has been called 2 times\n"
],
[
"counter_mult = counter(mul)\ncounter_mult(2, 5)\ncounter_mult(2, 5)\ncounter_mult(2, 5)\ncounter_mult(2, 5)\ncounter_mult(2, 5)",
"mul has been called 1 times\nmul has been called 2 times\nmul has been called 3 times\nmul has been called 4 times\nmul has been called 5 times\n"
],
[
"counters = dict()\ncounters['india'] = 1\ncounters['china'] = 0\ncounters",
"_____no_output_____"
],
[
"counters['india'] +=1\ncounters",
"_____no_output_____"
],
[
"times_dict >> add: 13, mil: 1, div; 12",
"_____no_output_____"
],
[
"x = 0\ndef fn1():\n def fn2():\n x = 20\n print(f'x in fn2 is {x}')\n def fn3():\n nonlocal x\n x = 50\n print(x)\n \n fn3()\n fn2()\nfn1()",
"x in fn2 is 20\n50\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec7141e12cf4e629c7e5146fee6fc0619ad6f2e1 | 48,956 | ipynb | Jupyter Notebook | CallystoAndDataScience/types-of-correlations.ipynb | lfunderburk/online-courses | d186a4f5667c7d4265b59fb94e9d8ac2668b8866 | [
"CC-BY-4.0"
]
| null | null | null | CallystoAndDataScience/types-of-correlations.ipynb | lfunderburk/online-courses | d186a4f5667c7d4265b59fb94e9d8ac2668b8866 | [
"CC-BY-4.0"
]
| 3 | 2020-09-25T21:00:56.000Z | 2020-10-17T01:31:17.000Z | CallystoAndDataScience/types-of-correlations.ipynb | lfunderburk/online-courses | d186a4f5667c7d4265b59fb94e9d8ac2668b8866 | [
"CC-BY-4.0"
]
| 4 | 2020-05-08T20:01:12.000Z | 2020-09-25T17:46:58.000Z | 317.896104 | 12,544 | 0.934839 | [
[
[
"\n\n<a href=\"https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&subPath=folder/notebook-name.ipynb&depth=1\" target=\"_parent\"><img src=\"https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true\" width=\"123\" height=\"24\" alt=\"Open in Callysto\"/></a>",
"_____no_output_____"
],
[
"# Types of Correlations\n## Activity for Module 5 Unit 2 ",
"_____no_output_____"
]
],
[
[
"# linear\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\nx = np.linspace(1,100,100)\ny = x + 3\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
],
[
"# linear with negative slope\ny = -x + 3\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
],
[
"# exponential\ny = x ** 2\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
],
[
"# inverse\ny = 1 / x\nplt.plot(x, y)\nplt.show()",
"_____no_output_____"
]
],
[
[
"[](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec714223d30710ca79da0e87eabe35cbda75c850 | 6,328 | ipynb | Jupyter Notebook | Week5/Guided projects/Projects.ipynb | Love2612/loveCSC102 | a76f291d071629cef7155fdacede5478ba5a2a78 | [
"MIT"
]
| null | null | null | Week5/Guided projects/Projects.ipynb | Love2612/loveCSC102 | a76f291d071629cef7155fdacede5478ba5a2a78 | [
"MIT"
]
| null | null | null | Week5/Guided projects/Projects.ipynb | Love2612/loveCSC102 | a76f291d071629cef7155fdacede5478ba5a2a78 | [
"MIT"
]
| null | null | null | 27.876652 | 89 | 0.453382 | [
[
[
"#PROJECT1\ndef avoids():\n count = 0\n \n f = input(\"Enter a forbidden letter: \")\n a = input(\"Enter a word: \")\n if f not in a:\n z = count + 1\n print(\"There is no forbidden letter in this word.\")\n else:\n z = 0\n print(\"There is a forbidden letter in this word.\")\n j = input(\"Enter a forbidden letter: \")\n b = input(\"Enter a word: \")\n if j not in b:\n x = count + 1\n print(\"There is no forbidden letter in this word.\")\n else:\n x = 0\n print(\"There is a forbidden letter in this word.\")\n k = input(\"Enter a forbidden letter: \")\n c = input(\"Enter a word: \")\n if k not in c:\n v = count + 1\n print(\"There is no forbidden letter in this word.\")\n else:\n v = 0\n print(\"There is a forbidden letter in this word.\")\n l = input(\"Enter a forbidden letter: \")\n d = input(\"Enter a word: \")\n if l not in d:\n n = count + 1\n print(\"There is no forbidden letter in this word.\")\n else:\n n = 0\n print(\"There is a forbidden letter in this word.\")\n m = input(\"Enter a forbidden letter: \")\n e = input(\"Enter a word: \")\n if m not in e:\n q = count + 1\n print(\"There is no forbidden letter in this word.\")\n else:\n q = 0\n print(\"There is a forbidden letter in this word.\")\n\n p = z + x + v + n + q \n print(\"The total number of words that do not contain forbidded words: \",p)\navoids()",
"Enter a forbidden letter: i\nEnter a word: ink\nThere is a forbidden letter in this word.\nEnter a forbidden letter: g\nEnter a word: get\nThere is a forbidden letter in this word.\nEnter a forbidden letter: y\nEnter a word: our\nThere is no forbidden letter in this word.\nEnter a forbidden letter: t\nEnter a word: tad\nThere is a forbidden letter in this word.\nEnter a forbidden letter: w\nEnter a word: ask\nThere is no forbidden letter in this word.\nThe total number of words that do not contain forbidded words: 2\n"
],
[
"#PROJECT2\ndef uses_all():\n string= [\"a\",\"e\",\"i\",\"o\",\"u\",\"y\"]\n s=list(input(\"Enter the required letters: \"))\n word=input(\"Enter a word: \")\n k=[]\n for g in s:\n if g in word:\n continue\n else:\n k.append(g)\n if len(k)>0:\n print(False)\n print(\"The\",word,\"does not contain the required letters\",k)\n else:\n print(True)\n f=True\n for j in string:\n if j in word:\n continue\n else:\n f=False\n if f:\n print(\"The word contains all required letters.\")\n \nuses_all()",
"Enter the required letters: aeiouy\nEnter a word: red\nFalse\nThe red does not contain the required letters ['a', 'i', 'o', 'u', 'y']\n"
],
[
"#PROJECT3\nJane=int(input(\"Pick a number Jane : \"))\nJack=int(input(\"Pick a number Jack: \"))\njg=input(\"Is your number EVEN or ODD Jane? \")\njag=input(\"Is your number EVEN or ODD Jack? \")\nTot=0\nif (Jack+Jane)%2==0:\n Tot=1\nelse:\n Tot=2\nif Tot==1:\n if jg==\"EVEN\":\n print(\"Jane wins.\")\n elif jag==\"EVEN\":\n print(\"Jack wins.\")\n elif jg==jag==\"EVEN\":\n print(\"Both Jack and Jane wins.\")\n else:\n print(\"Both players lose.\")\n \nelif Tot==2:\n if jg==\"ODD\":\n print(\"Jane wins.\")\n elif jag==\"ODD\":\n print(\"Jack wins.\")\n elif jg==jag==\"ODD\":\n print(\"Both Jack and Jane wins.\")\n else:\n print(\"Both players lose.\")",
"Pick a number Jane : 43\nPick a number Jack: 42\nIs your number EVEN or ODD Jane? ODD\nIs your number EVEN or ODD Jack? EVEN\nJane wins.\n"
],
[
"#PROJECT4\nD=50\nT=2\nspeed=(D/T)\nprint(\"The speed of the car is \",speed)",
"The speed of the car is 25.0\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code"
]
]
|
ec71550c68df384814f08ff80178b0d1ebf3a149 | 11,935 | ipynb | Jupyter Notebook | tripyview/shapefiles/convert_mpasregion2shp.ipynb | patrickscholz/tripyview | 14c232d840db2e879d79b3e3b3286a6962211c47 | [
"MIT"
]
| null | null | null | tripyview/shapefiles/convert_mpasregion2shp.ipynb | patrickscholz/tripyview | 14c232d840db2e879d79b3e3b3286a6962211c47 | [
"MIT"
]
| null | null | null | tripyview/shapefiles/convert_mpasregion2shp.ipynb | patrickscholz/tripyview | 14c232d840db2e879d79b3e3b3286a6962211c47 | [
"MIT"
]
| null | null | null | 117.009804 | 8,976 | 0.867784 | [
[
[
"import sys\nsys.path.append(\"./src/\")\nfrom sub_mesh import * \nfrom sub_data import * \nfrom sub_plot import * \nfrom sub_climatology import *\nfrom sub_index import *\nfrom shapely.geometry import Point, Polygon, MultiPolygon, shape\nfrom shapely.vectorized import contains\nimport shapefile as shp\nimport json\nimport geopandas as gpd\n\n\n# region = 'Labrador_Sea'\n# region = 'Greenland_Sea'\n# region = 'Norwegian_Sea'\n# region = 'Irminger_Sea'\n# region = 'Weddell_Sea'\n# region = 'Chukchi_Sea'\n# region = 'Bering_Sea'\n# region = 'Beaufort_Sea'\n# region = 'Barents_Sea'\n# region = 'Indian_Ocean'\n# region = 'Canada_Basin'\n# region = 'Kara_Sea'\n# region = 'Laptev_Sea'\n# region = 'Southern_Ocean'\n# region = 'Global_Ocean_15S_to_15N'\n# region = 'North_Pacific_Ocean'\n# region = 'South_Pacific_Ocean'\n# region = 'North_Atlantic_Ocean'\n# region = 'South_Atlantic_Ocean'\n# region = 'Sea_of_Okhotsk'\n# region = 'Japan_Sea'\n# region = 'Philippine_Sea'#\n# region ='North_Sea'\n# region = 'Amundsen_Sea'\n# region = 'North_Sea'\n# region = 'Java_Sea'\n# region = 'Nino_3.4'\n# region = 'South_Atlantic_Ocean'\n# region = 'South_China_Sea'\n# region = 'Weddell_Sea_Deep'\n# region = 'Weddell_Sea_Shelf'\n# region = 'Western_Ross_Sea'\n# region = 'Western_Ross_Sea_Deep'\n# region = 'Western_Ross_Sea_Shelf'\nregion = 'Western_Weddell_Sea'\nregion = 'Western_Weddell_Sea_Deep'\nregion = 'Western_Weddell_Sea_Shelf'\ngeojsonfilepath = '/home/ollie/pscholz/software/geometric_features/geometric_data/ocean/region/'+region+'/region.geojson'\nshppath = '/home/ollie/pscholz/tripyview_github/src/shapefiles/mpas_region/'\nconvert_geojson2shp(geojsonfilepath, shppath, do_plot=True)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec7198a0a5f5b7bcecea8cbbc3da566ffc4835b0 | 17,610 | ipynb | Jupyter Notebook | principal-component-disaster.ipynb | MLStruckmann/principal-component-disaster | 4d499f447f65e74d4f97db382c60274cadac742d | [
"MIT"
]
| null | null | null | principal-component-disaster.ipynb | MLStruckmann/principal-component-disaster | 4d499f447f65e74d4f97db382c60274cadac742d | [
"MIT"
]
| null | null | null | principal-component-disaster.ipynb | MLStruckmann/principal-component-disaster | 4d499f447f65e74d4f97db382c60274cadac742d | [
"MIT"
]
| null | null | null | 30.679443 | 190 | 0.50494 | [
[
[
"# Principal component disaster",
"_____no_output_____"
],
[
"*Creator: Magnus Struckmann*\n## Table of contents\n\n- Notebook description\n- Create data sets\n- Visualize blob data\n- Visualize elbow plots of explained variance from PCA\n- Visualize PCA transformation fitted on training data\n- Visualize PCA transformation applied on testing data\n- Visualize decision boundaries and training data\n- Visualize decision boundaries and test data\n- Visualize surface probability and test data",
"_____no_output_____"
],
[
"### Notebook description\n\nTo analyze the consequences of changing the distance between blob data centers 30 different data sets are created.\nOnly the distance between the centers of the Gaussian distributed blobs is changed while the standard deviation and the random initialization parameter are kept constant.\n\nEach data set contains 1000 data points that are equally distributed among the four data classes.\nA principal component analysis is performed on each data set and a logistic regression classifier fitted to its output.\nAll results are saved to the *df_collection* dictionary.\n\nThe different data sets, PCA results and logistic regression predicitions are visualized with Plotly to conclude on the consequences of changing the distance between blob data centers.",
"_____no_output_____"
]
],
[
[
"# Import packages\nimport pandas as pd\nimport numpy as np\nimport plotly\nimport plotly.graph_objects as go\nfrom sklearn import datasets\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.preprocessing import StandardScaler\nfrom sklearn.decomposition import PCA\nfrom sklearn.linear_model import LogisticRegression",
"_____no_output_____"
]
],
[
[
"### Create data sets",
"_____no_output_____"
]
],
[
[
"df_collection = []\nfor step in np.arange(0, 3, 0.1): # Step size for distance between blob centers\n \n # Create blob data\n distance = step\n centers = [\n [distance,-distance,-distance],\n [-distance,distance,-distance],\n [-distance,distance,distance],\n [distance,-distance,distance]]\n \n X, y = datasets.make_blobs(n_samples=1000,\n random_state=42,\n n_features=3,\n cluster_std=1,\n centers=centers)\n \n data = np.column_stack((X, y))\n df = pd.DataFrame(data, columns = ['Feature 1','Feature 2','Feature 3','Cluster'])\n\n # Split data\n X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, stratify=y, random_state=0)\n \n # Standardize data\n sc = StandardScaler()\n X_train_std = sc.fit_transform(X_train)\n X_test_std = sc.transform(X_test) \n \n # Principal component analysis\n pca = PCA(n_components=3)\n pca.fit(X_train_std)\n pca_var = pca.explained_variance_ratio_\n pca = PCA(n_components=2)\n pca_X_train = pca.fit_transform(X_train_std)\n pca_X_test = pca.transform(X_test_std)\n pca_X = np.append(pca_X_train, pca_X_test, axis=0)\n \n # Fit logistic regression model\n clf = LogisticRegression()\n clf.fit(pca_X_train, y_train)\n\n # Make predictions over complete PCA feature space (min and max of training and test data)\n h = .02 # step size in the mesh\n x_min, x_max = pca_X[:, 0].min() - 1, pca_X[:, 0].max() + 1\n y_min, y_max = pca_X[:, 1].min() - 1, pca_X[:, 1].max() + 1\n xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))\n y_ = np.arange(y_min, y_max, h)\n Z_class_prediction = clf.predict(np.c_[xx.ravel(), yy.ravel()])\n Z_class_prediction = Z_class_prediction.reshape(xx.shape)\n Z_probability_label1 = clf.predict_proba(np.c_[xx.ravel(), yy.ravel()])[:,0] # probability for class 1\n Z_probability_label1 = Z_probability_label1.reshape(xx.shape)\n \n step_dict = dict(distance=round(step,1),\n data=df,\n pca_var=pca_var,\n pca_X_train=pca_X_train,\n pca_X_test=pca_X_test,\n y_train=y_train,\n y_test=y_test,\n xx_meshgrid=xx,\n y_meshgrid=y_,\n Z_class_prediction=Z_class_prediction,\n Z_probability_label1=Z_probability_label1)\n \n df_collection.append(step_dict)",
"_____no_output_____"
]
],
[
[
"### Visualize blob data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n df = step_dict['data']\n\n trace = go.Scatter3d(x=df['Feature 1'], y=df['Feature 2'], z=df['Feature 3'],\n mode='markers',\n marker=dict(size=12,color=df['Cluster']),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[10].visible = True",
"_____no_output_____"
],
[
"# Create and add slider\ndef create_sliders(df_collection):\n steps = []\n for i,step_dict in enumerate(df_collection):\n step = dict(\n method=\"restyle\",\n args=[\"visible\", [False] * len(df_collection)],\n label=str(step_dict['distance']),\n )\n step[\"args\"][1][i] = True # Toggle i'th trace to \"visible\"\n steps.append(step)\n\n sliders = [dict(\n active=10,\n currentvalue={\"prefix\": \"Distance: \"},\n pad={\"t\": 50},\n steps=steps\n )]\n return sliders",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout(\n sliders=create_sliders(df_collection),\n scene = dict(xaxis_title='Feature 1',\n yaxis_title='Feature 2',\n zaxis_title='Feature 3'),\n margin=dict(r=20, b=10, l=10, t=10))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize elbow plots of explained variance from PCA",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n pca_var = step_dict['pca_var']\n \n components=['Component 1', 'Component 2', 'Component 3']\n trace = go.Bar(x=components, \n y=pca_var,\n visible=False)\n \n fig.add_trace(trace)\n\n# Make 10th trace visible\nfig.data[10].visible = True",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout(\n sliders=create_sliders(df_collection),\n yaxis=dict(title='Explained variance ratio'))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize PCA transformation fitted on training data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n pca_X_train = step_dict['pca_X_train']\n y_train = step_dict['y_train']\n\n trace = go.Scatter(x=pca_X_train[:,0], y=pca_X_train[:,1],\n mode='markers',\n marker=dict(size=12,color=y_train),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[10].visible = True",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout(\n sliders=create_sliders(df_collection),\n xaxis=dict(title='Component 1'),\n yaxis=dict(title='Component 2'))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize PCA transformation applied on testing data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n pca_X_test = step_dict['pca_X_test']\n y_test = step_dict['y_test']\n\n trace = go.Scatter(x=pca_X_test[:,0], y=pca_X_test[:,1],\n mode='markers',\n marker=dict(size=12,color=y_test),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[10].visible = True",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout(\n sliders=create_sliders(df_collection),\n xaxis=dict(title='Component 1'),\n yaxis=dict(title='Component 2'))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize decision boundaries and training data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n X = step_dict['pca_X_train']\n y = step_dict['y_train']\n xx = step_dict['xx_meshgrid'] \n y_ = step_dict['y_meshgrid']\n Z = step_dict['Z_class_prediction']\n \n trace = go.Heatmap(x=xx[0], y=y_, z=Z,\n showscale=True,\n visible=False)\n \n fig.add_trace(trace)\n\n trace = go.Scatter(x=X[:, 0], y=X[:, 1], \n mode='markers',\n showlegend=False,\n marker=dict(size=10,color=y,line=dict(color='black', width=1)),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[19].visible = True # (because we add two traces per plot we need to multiply by two and make both visible)\nfig.data[20].visible = True",
"_____no_output_____"
],
[
"# Create and add slider (two traces per plot)\ndef create_sliders_2traces(df_collection):\n steps = []\n for i,step_dict in enumerate(df_collection):\n step = dict(\n method=\"restyle\",\n args=[\"visible\", [False] * len(df_collection)*2],\n label=str(step_dict['distance']),\n )\n step[\"args\"][1][i*2] = True # Toggle i'th trace to \"visible\"\n step[\"args\"][1][i*2+1] = True\n steps.append(step)\n\n sliders = [dict(\n active=10,\n currentvalue={\"prefix\": \"Distance: \"},\n pad={\"t\": 50},\n steps=steps\n )]\n return sliders",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout( \n sliders=create_sliders_2traces(df_collection),\n xaxis=dict(title='Component 1'),\n yaxis=dict(title='Component 2'))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize decision boundaries and test data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n X = step_dict['pca_X_test']\n y = step_dict['y_test']\n xx = step_dict['xx_meshgrid'] \n y_ = step_dict['y_meshgrid']\n Z = step_dict['Z_class_prediction']\n \n trace = go.Heatmap(x=xx[0], y=y_, z=Z,\n showscale=True,\n visible=False)\n \n fig.add_trace(trace)\n\n trace = go.Scatter(x=X[:, 0], y=X[:, 1], \n mode='markers',\n showlegend=False,\n marker=dict(size=10,color=y,line=dict(color='black', width=1)),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[19].visible = True # (because we add two traces per plot we need to multiply by two and make both visible)\nfig.data[20].visible = True",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout( \n sliders=create_sliders_2traces(df_collection),\n xaxis=dict(title='Component 1'),\n yaxis=dict(title='Component 2'))\n\nfig.show()",
"_____no_output_____"
]
],
[
[
"### Visualize surface probability and test data",
"_____no_output_____"
]
],
[
[
"fig = go.Figure()\n\n# Add traces, one for each slider step\nfor step_dict in df_collection:\n X = step_dict['pca_X_test']\n y = step_dict['y_test']\n xx = step_dict['xx_meshgrid'] \n y_ = step_dict['y_meshgrid']\n Z = step_dict['Z_probability_label1']\n \n trace = go.Heatmap(x=xx[0], y=y_, z=Z,\n showscale=True,\n visible=False)\n \n fig.add_trace(trace)\n\n trace = go.Scatter(x=X[:, 0], y=X[:, 1], \n mode='markers',\n showlegend=False,\n marker=dict(size=10,color=y,line=dict(color='black', width=1)),\n visible=False)\n\n fig.add_trace(trace)\n \n# Make 10th trace visible\nfig.data[19].visible = True # (because we add two traces per plot we need to multiply by two and make both visible)\nfig.data[20].visible = True",
"_____no_output_____"
],
[
"# Show plot\nfig.update_layout( \n sliders=create_sliders_2traces(df_collection),\n xaxis=dict(title='Component 1'),\n yaxis=dict(title='Component 2'))\n\nfig.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec719eb6d76d513ba4c42bc2b5ad470644725682 | 517,821 | ipynb | Jupyter Notebook | examples/regression2.ipynb | lukewarm/orbit | c1d65082728e627d1bd048978a3a1abb85cbbee6 | [
"Apache-2.0"
]
| 1,332 | 2020-09-23T15:15:26.000Z | 2022-03-30T00:01:18.000Z | examples/regression2.ipynb | ChakChak1234/orbit | b329326b8fd9382310645927846315714386de50 | [
"Apache-2.0"
]
| 294 | 2020-09-23T22:23:26.000Z | 2022-03-25T18:58:56.000Z | examples/regression2.ipynb | ChakChak1234/orbit | b329326b8fd9382310645927846315714386de50 | [
"Apache-2.0"
]
| 96 | 2020-09-29T23:13:37.000Z | 2022-03-30T09:20:06.000Z | 427.95124 | 125,064 | 0.931895 | [
[
[
"# Regression with Orbit",
"_____no_output_____"
],
[
"In this notebook, we want to demonstartate how to use the different arguments in DLT model to realize different setups for the regressors. Those could be very useful in practice when tuning the models. Here includes two examples, one with the icliams dataset and one for the simulation data.",
"_____no_output_____"
]
],
[
[
"import pandas as pd\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom pylab import rcParams\n\nfrom orbit.models import DLT\nfrom orbit.diagnostics.plot import plot_predicted_data, plot_predicted_components\nfrom orbit.utils.simulation import make_regression\nfrom orbit.constants.palette import OrbitPalette\nfrom orbit.utils.dataset import load_iclaims\nfrom orbit.diagnostics.metrics import smape\n\nfrom sklearn.linear_model import LinearRegression\nfrom sklearn.preprocessing import minmax_scale\n\nrcParams['figure.figsize'] = 14, 8\nfrom orbit.utils.plot import get_orbit_style\nplt.style.use(get_orbit_style())\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"## iclaims Data",
"_____no_output_____"
],
[
"*iclaims_example* is a dataset containing the weekly initial claims for US unemployment benefits against a few related google trend queries (unemploy, filling and job) from Jan 2010 - June 2018. This aims to mimick the dataset from the paper [Predicting the Present with Bayesian Structural Time Series](https://people.ischool.berkeley.edu/~hal/Papers/2013/pred-present-with-bsts.pdf) by SCOTT and VARIAN (2014).\n\nNumber of claims are obtained from [Federal Reserve Bank of St. Louis](https://fred.stlouisfed.org/series/ICNSA) while google queries are obtained through [Google Trends API](https://trends.google.com/trends/?geo=US).\n\nIn order to use this data to nowcast the US unemployment claims during COVID-19 period, we extended the dataset to Jan 2021 and added the [S&P 500 (^GSPC)](https://finance.yahoo.com/quote/%5EGSPC/history?period1=1264032000&period2=1611187200&interval=1wk&filter=history&frequency=1wk&includeAdjustedClose=true) and [VIX](https://finance.yahoo.com/quote/%5EVIX/history?p=%5EVIX) Index historical data for the same period.\n\nThe data is standardized and log-transformed for the model fitting purpose.",
"_____no_output_____"
]
],
[
[
"# load data\ndf = load_iclaims(end_date='2021-01-03')\n\ndf.dtypes",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
]
],
[
[
"We can see form the plot below, there are seasonlity trend and as well as a huge changpoint due the impact of COVID-19.",
"_____no_output_____"
]
],
[
[
"fig, axs = plt.subplots(2, 2,figsize=(20,8))\naxs[0, 0].plot(df['week'], df['claims'])\naxs[0, 0].set_title('Unemployment Claims')\naxs[0, 1].plot(df['week'], df['trend.unemploy'], 'tab:orange')\naxs[0, 1].set_title('Google trend - unemploy')\naxs[1, 0].plot(df['week'], df['vix'], 'tab:green')\naxs[1, 0].set_title('VIX')\naxs[1, 1].plot(df['week'], df['sp500'], 'tab:red')\naxs[1, 1].set_title('S&P500');",
"_____no_output_____"
],
[
"# split the dataset\n# only use the data after 2018 as we observed from above chart that it has the more clear relationship between trend.unemploy and claims\ndf = df[df['week'] > '2018-01-01'].reset_index(drop=True)\n\ntest_size = 26\n\ntrain_df = df[:-test_size]\ntest_df = df[-test_size:]",
"_____no_output_____"
]
],
[
[
"### Without Regression",
"_____no_output_____"
],
[
"We will use the DLT models here to compare the model performance without vs. with regression.",
"_____no_output_____"
]
],
[
[
"DATE_COL = 'week'\nRESPONSE_COL = 'claims'",
"_____no_output_____"
],
[
"np.random.seed(8888)",
"_____no_output_____"
],
[
"dlt = DLT(response_col=RESPONSE_COL,\n date_col=DATE_COL,\n seasonality=52,\n seed=8888,\n num_warmup=4000,\n n_bootstrap_draws=4000)",
"_____no_output_____"
],
[
"%%time\ndlt.fit(df=train_df)",
"WARNING:pystan:n_eff / iter below 0.001 indicates that the effective sample size has likely been overestimated\nWARNING:pystan:Rhat above 1.1 or below 0.9 indicates that the chains very likely have not mixed\n"
],
[
"predicted_df = dlt.predict(df=test_df)",
"_____no_output_____"
],
[
"_ = plot_predicted_data(training_actual_df=train_df,\n predicted_df=predicted_df, \n date_col=DATE_COL,\n actual_col=RESPONSE_COL, \n test_actual_df=test_df,\n title='Prediction without Regression - SMAPE:{:.3f}'.format(\n smape(predicted_df['prediction'].values, test_df['claims'].values)\n )\n )",
"_____no_output_____"
]
],
[
[
"### With Regression",
"_____no_output_____"
],
[
"The regressor columns can be supplied via argument `regressor_col`. ",
"_____no_output_____"
],
[
"#### Regressor with the sign",
"_____no_output_____"
]
],
[
[
"REGRESSOR_COL = ['trend.unemploy', 'trend.filling', 'trend.job', 'sp500', 'vix']\n\ndlt_reg = DLT(response_col=RESPONSE_COL, \n date_col=DATE_COL,\n regressor_col=REGRESSOR_COL,\n seasonality=52,\n seed=8888,\n num_warmup=4000,\n n_bootstrap_draws=4000)\n\n\ndlt_reg.fit(df=train_df)",
"WARNING:pystan:n_eff / iter below 0.001 indicates that the effective sample size has likely been overestimated\nWARNING:pystan:Rhat above 1.1 or below 0.9 indicates that the chains very likely have not mixed\n"
]
],
[
[
"The estimated regressor coefficients can be retrieved via `.get_regression_coefs()`.",
"_____no_output_____"
]
],
[
[
"dlt_reg.get_regression_coefs()",
"_____no_output_____"
],
[
"predicted_df_reg = dlt_reg.predict(test_df)",
"_____no_output_____"
],
[
"_ = plot_predicted_data(training_actual_df=train_df,\n predicted_df=predicted_df_reg, \n date_col=DATE_COL,\n actual_col=RESPONSE_COL, \n test_actual_df=test_df,\n title='Prediction with Regression - SMAPE:{:.4f}'.format(\n smape(predicted_df_reg['prediction'].values, test_df['claims'].values)\n )\n )",
"_____no_output_____"
]
],
[
[
"After adding the regressor, prediction results are more closer to the real response compared to without using regression. ",
"_____no_output_____"
],
[
"#### Adjust pirors for regressor beta and standard deviation",
"_____no_output_____"
],
[
"In the model, it is assumed $$\\beta \\sim Gaussian(\\beta_{prior}, \\sigma_{prior})$$\n\nThe default values for $\\beta_{prior}$ and $\\sigma_{prior}$ are 0 and 1, respectively.\n\nAssuming users obtain further knowledge on each regressor, they could adjust prior knowledge on the coefficients via arguments `regressor_beta_prior` and `regressor_sigma_prior`. These two lists should be of the same lenght as `regressor_col`.\n\nAlso, assuming users have some prior knowledge on direction of impact by each regressor, users can specifiy them via `regressor_sign`, with a list of values consist '=' (regular, no restriction), '+' (positive) or '-' (negative). Such list should be of the same length. The default values of `regressor_sign` is all '='.",
"_____no_output_____"
]
],
[
[
"dlt_reg_adjust = DLT(response_col=RESPONSE_COL,\n date_col=DATE_COL,\n regressor_col=REGRESSOR_COL,\n regressor_sign=[\"+\", '+', '=', '-', '+'], \n regressor_beta_prior=[0.5, 0.25, 0.07, -0.3, 0.03],\n regressor_sigma_prior=[0.05] * 5,\n seasonality=52,\n seed=8888,\n num_warmup=4000,\n n_bootstrap_draws=4000)\n\n\ndlt_reg_adjust.fit(df=train_df)",
"WARNING:pystan:n_eff / iter below 0.001 indicates that the effective sample size has likely been overestimated\nWARNING:pystan:Rhat above 1.1 or below 0.9 indicates that the chains very likely have not mixed\n"
],
[
"dlt_reg_adjust.get_regression_coefs()",
"_____no_output_____"
],
[
"predicted_df_reg_adjust = dlt_reg_adjust.predict(test_df)",
"_____no_output_____"
],
[
"_ = plot_predicted_data(training_actual_df=train_df,\n predicted_df=predicted_df_reg_adjust, \n date_col=DATE_COL,\n actual_col=RESPONSE_COL, \n test_actual_df=test_df,\n title='Prediction with Regression using adjust priors - SMAPE:{:.4f}'.format(\n smape(predicted_df_reg_adjust['prediction'].values, test_df['claims'].values))\n )",
"_____no_output_____"
]
],
[
[
"## Regression on Simulated Dataset",
"_____no_output_____"
],
[
"Leverage the simulation function to generate a dateset with regression terms",
"_____no_output_____"
]
],
[
[
"# To scale regressor values in a nicer way\nSEED = 2020\nNUM_OF_REGRESSORS = 5\nCOEFS = np.random.default_rng(SEED).normal(.03, .1, NUM_OF_REGRESSORS)\n\nCOEFS",
"_____no_output_____"
],
[
"x, y, coefs = make_regression(200, COEFS, seed=SEED)",
"_____no_output_____"
],
[
"df = pd.DataFrame(x)\nregressor_cols = [f\"regressor_{x}\" for x in range(1, NUM_OF_REGRESSORS + 1)]\ndf.columns = regressor_cols\n#df[regressor_cols] = df[regressor_cols]/REG_BASE\n#df[regressor_cols] = df[regressor_cols].apply(np.log1p)\n\n# min max scale to avoid negetive values\nresponse_col = \"response\"\ndf[response_col] = y \ndf[response_col] = minmax_scale(df[response_col])\n\n# add the date column\ndf['date'] = pd.date_range(start='2016-01-04', periods=200, freq=\"1W\")",
"_____no_output_____"
],
[
"df.head()",
"_____no_output_____"
],
[
"plt.plot(df['response']);",
"_____no_output_____"
]
],
[
[
"### Use data-driven sigma for each coefficients",
"_____no_output_____"
],
[
"Instead of using fixed standard deviations for regressors, a hyperprior can be assigned to them, i.e.\n$$\\sigma_\\beta \\sim \\text{Half-Cauchy}(0, \\text{ridge_scale})$$\n\nThis can be done by setting `regression_penalty=\"auto_ridge\"`. Notice there is a hyperprior `auto_ridge_scale` for tuning with a default of `0.5`. We can also supply stan config such as `adapt_delta` to reduce divergence see [here](https://mc-stan.org/rstanarm/reference/adapt_delta.html) for details.",
"_____no_output_____"
]
],
[
[
"mod_auto_ridge = DLT(response_col=\"response\",\n date_col=\"date\",\n regressor_col=regressor_cols,\n seasonality=52,\n seed=SEED,\n regression_penalty='auto_ridge',\n num_warmup=4000,\n num_sample=1000,\n stan_mcmc_control={'adapt_delta':0.9})\n\n\nmod_auto_ridge.fit(df=df)",
"WARNING:pystan:Maximum (flat) parameter count (1000) exceeded: skipping diagnostic tests for n_eff and Rhat.\nTo run all diagnostics call pystan.check_hmc_diagnostics(fit)\nWARNING:pystan:22 of 1000 iterations ended with a divergence (2.2 %).\nWARNING:pystan:Try running with adapt_delta larger than 0.9 to remove the divergences.\n"
],
[
"coef_auto_ridge = np.median(mod_auto_ridge._posterior_samples['beta'], axis=0)",
"_____no_output_____"
],
[
"lw = 3\nplt.figure(figsize=(20, 8))\nplt.title(\"Copmparsion of the Coefficients from Different Models\")\nplt.plot(coef_auto_ridge, color=OrbitPalette.GREEN.value, linewidth=lw, label=\"Auto Ridge\", alpha=0.5, linestyle='--')\nplt.plot(coefs, color=\"black\", linewidth=lw, label=\"Ground truth\")\nplt.legend()\nplt.grid()",
"_____no_output_____"
]
],
[
[
"The result looks reasonable comparing to the true coefficients.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec71a9263b29a7fe3f36f919a2e5ba9de2d2f908 | 4,782 | ipynb | Jupyter Notebook | nb/092620_qualitative_type_model.ipynb | myutman/contracode | f2a589e1efd2788874fd0468d1ecc30d6a14c396 | [
"Apache-2.0"
]
| 115 | 2020-07-10T02:01:34.000Z | 2022-03-27T06:21:43.000Z | nb/092620_qualitative_type_model.ipynb | myutman/contracode | f2a589e1efd2788874fd0468d1ecc30d6a14c396 | [
"Apache-2.0"
]
| 7 | 2020-08-05T00:25:17.000Z | 2021-12-26T17:06:31.000Z | nb/092620_qualitative_type_model.ipynb | myutman/contracode | f2a589e1efd2788874fd0468d1ecc30d6a14c396 | [
"Apache-2.0"
]
| 21 | 2020-07-14T11:31:47.000Z | 2022-03-25T06:10:18.000Z | 41.224138 | 1,165 | 0.635508 | [
[
[
"%cd ~/contracode\nimport numpy as np\nimport pandas as pd\nimport pickle\nimport gzip\nfrom tqdm.auto import tqdm\nfrom pathlib import Path",
"/home/ajay/contracode\n"
],
[
"import os\nimport random\n\nimport fire\nimport numpy as np\nimport sentencepiece as spm\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport tqdm\nimport wandb\nfrom loguru import logger\n\nimport sys\nprint(sys.path)\n# sys.path.append(\"/home/ajay/contracode\")\n# sys.path.append(\"/home/ajay/contracode/representjs\")\n# sys.path.append(\"..\")\n\nfrom models.typetransformer import TypeTransformer\nfrom data.deeptyper_dataset import DeepTyperDataset, load_type_vocab, get_collate_fn\nfrom data.util import Timer\nfrom representjs import RUN_DIR\nfrom utils import count_parameters, get_linear_schedule_with_warmup",
"['/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/git/ext/gitdb', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/wandb/vendor/graphql-core-1.1', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/wandb/vendor/gql-0.2.0', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/wandb/vendor', '/home/ajay/contracode/nb', '/home/ajay/miniconda3/envs/paras/lib/python38.zip', '/home/ajay/miniconda3/envs/paras/lib/python3.8', '/home/ajay/miniconda3/envs/paras/lib/python3.8/lib-dynload', '', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages', '/home/ajay/contracode', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/IPython/extensions', '/home/ajay/.ipython', '/home/ajay/miniconda3/envs/paras/lib/python3.8/site-packages/gitdb/ext/smmap']\n"
],
[
"eval_dataset = DeepTyperDataset(\n eval_filepath,\n type_vocab_filepath,\n spm_filepath,\n max_length=max_eval_seq_len,\n subword_regularization_alpha=subword_regularization_alpha,\n split_source_targets_by_tab=eval_filepath.endswith(\".json\")\n)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec71be82d2262b96e91f72646387bfae3e096d3f | 10,265 | ipynb | Jupyter Notebook | Conclusion.ipynb | linogaliana/Python_2A | 41a628a5ebb74b59d7e77249c372298adb658711 | [
"Apache-2.0"
]
| null | null | null | Conclusion.ipynb | linogaliana/Python_2A | 41a628a5ebb74b59d7e77249c372298adb658711 | [
"Apache-2.0"
]
| null | null | null | Conclusion.ipynb | linogaliana/Python_2A | 41a628a5ebb74b59d7e77249c372298adb658711 | [
"Apache-2.0"
]
| null | null | null | 33.327922 | 515 | 0.499367 | [
[
[
"# 1 - Préparation du traitement des données",
"_____no_output_____"
]
],
[
[
"import os\nimport time\n\nimport pandas as pd\nimport seaborn as sns\nfrom sklearn.pipeline import make_pipeline \nfrom sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer\nfrom sklearn.linear_model import LogisticRegression",
"_____no_output_____"
],
[
"NB_DATA_140 = 100000",
"_____no_output_____"
]
],
[
[
"On a enregistré plusieurs versions des deux jeux de données préprocessées de manières différentes avec des combinaisons différentes des arguments *stop_words*, *lemmatization* et *negation*. On va créer une fonction *data* qui, en fonction des paramètres de préprocessing choisis par l'utilisateur, chargera les deux dataframes (sentiment140 et tweets scrapés) correspondants.",
"_____no_output_____"
]
],
[
[
"def data(stop_words, lemmatization, negation):\n file = \"train\"\n if stop_words:\n file += \"_stop\"\n if lemmatization:\n file += \"_lemm\"\n if negation:\n file += \"_neg\"\n df_140 = pd.read_pickle(os.path.join(\"data\", \"sentiment140\", file + \".bz2\")).sample(NB_DATA_140, random_state=1234).reset_index(drop=True)\n\n file = \"web\"\n if stop_words:\n file += \"_stop\"\n if lemmatization:\n file += \"_lemm\"\n if negation:\n file += \"_neg\"\n df_web = pd.read_pickle(os.path.join(\"data\", \"web\", file + \".bz2\"))\n\n X_140 = df_140.text.to_list()\n y_140 = df_140.sentiment.to_list()\n\n X_web = df_web.Text.to_list()\n \n return X_140, y_140, X_web, df_web",
"_____no_output_____"
]
],
[
[
"On va sélectionner le modèle pertinent ainsi que la méthode de feature extraction.",
"_____no_output_____"
]
],
[
[
"model = make_pipeline(TfidfVectorizer(max_features=6000, ngram_range=(1,2)), LogisticRegression(max_iter=500))",
"_____no_output_____"
]
],
[
[
"# 2 - Application du modèle",
"_____no_output_____"
],
[
"On choisit nos arguments de préprocessing et on appelle la base pertinente à l’aide de la fonction data",
"_____no_output_____"
]
],
[
[
"STOP_WORDS = False\nLEMMATIZATION = True\nNEGATION = False\nX_140, y_140, X_web, df_web = data(STOP_WORDS, LEMMATIZATION, NEGATION)",
"_____no_output_____"
]
],
[
[
"Application du modèle pour labelliser les tweets scrapés",
"_____no_output_____"
]
],
[
[
"y_web = model.predict(X_web)",
"_____no_output_____"
]
],
[
[
"Ajout de la prédiction au dataframe des tweets",
"_____no_output_____"
]
],
[
[
"df_web['sentiment'] = resul",
"_____no_output_____"
]
],
[
[
"# 3 - Visualisation",
"_____no_output_____"
]
],
[
[
"sns.countplot(x='sentiment', data=df_web).sett(title='Sentiments exprimés (total)')\nplt.show()",
"_____no_output_____"
],
[
"sns.histplot(x='Film', hue='sentiment', data=df_web).set(title=\"Sentiments exprimés concernant chaque film\")\nplt.show()",
"_____no_output_____"
],
[
"df_web['Time Range'] = df_web['Time Range'].replace([\"[2021-10-08, 2021-10-09, 2021-10-10, 2021-10-11, 2021-10-12, 2021-10-13, 2021-10-14, 2021-10-15, 2021-10-16, 2021-10-17, 2021-10-18, 2021-10-19, 2021-10-20, 2021-10-21, 2021-10-22, 2021-10-23, 2021-10-24, 2021-10-25, 2021-10-26, 2021-10-27, 2021-10-28, 2021-10-29, 2021-10-30, 2021-10-31, 2021-11-01, 2021-11-02, 2021-11-03, 2021-11-04, 2021-11-05, 2021-11-06, 2021-11-07, 2021-11-08, 2021-11-09, 2021-11-10, 2021-11-11, 2021-11-12, 2021-11-13]\",\n \"[2021-07-03, 2021-07-04, 2021-07-05, 2021-07-06, 2021-07-07, 2021-07-08, 2021-07-09, 2021-07-10, 2021-07-11, 2021-07-12, 2021-07-13, 2021-07-14, 2021-07-15, 2021-07-16, 2021-07-17, 2021-07-18, 2021-07-19, 2021-07-20, 2021-07-21, 2021-07-22, 2021-07-23, 2021-07-24, 2021-07-25, 2021-07-26, 2021-07-27, 2021-07-28, 2021-07-29, 2021-07-30, 2021-07-31, 2021-08-01, 2021-08-02, 2021-08-03, 2021-08-04, 2021-08-05, 2021-08-06, 2021-08-07, 2021-08-08]\",\n \"[2020-09-09, 2020-09-10, 2020-09-11, 2020-09-12, 2020-09-13, 2020-09-14, 2020-09-15, 2020-09-16, 2020-09-17, 2020-09-18, 2020-09-19, 2020-09-20, 2020-09-21, 2020-09-22, 2020-09-23]\",\n \"[2021-07-22, 2021-07-23, 2021-07-24, 2021-07-25, 2021-07-26, 2021-07-27, 2021-07-28, 2021-07-29, 2021-07-30, 2021-07-31, 2021-08-01, 2021-08-02, 2021-08-03, 2021-08-04, 2021-08-05]\",\n \"[2021-04-03, 2021-04-04, 2021-04-05, 2021-04-06, 2021-04-07, 2021-04-08, 2021-04-09, 2021-04-10, 2021-04-11, 2021-04-12, 2021-04-13, 2021-04-14, 2021-04-15, 2021-04-16, 2021-04-17]\",\n \"[2021-06-09, 2021-06-10, 2021-06-11, 2021-06-12, 2021-06-13, 2021-06-14, 2021-06-15, 2021-06-16, 2021-06-17, 2021-06-18, 2021-06-19, 2021-06-20, 2021-06-21, 2021-06-22, 2021-06-23]\"],\n [\"Sortie Dune\", \"Sortie Space Jam\", \"Premier trailer Dune\", \"Second trailer Dune\",\"Premier trailer Space Jam\", \"Second trailer Space Jam\"])",
"_____no_output_____"
],
[
"sns.histplot(x='Time Range', hue='sentiment', data=df_web.loc[df_web['Film'] == 'dune']).set(title=\"Évolution des sentiments exprimés sur le film Dune à chaque période\")",
"_____no_output_____"
],
[
"sns.histplot(x='Time Range', hue='sentiment', data=df_web.loc[df_web['Film'] == 'space jam']).set(title=\"Évolution des sentiments exprimés sur le film Space Jam 2 à chaque période\")",
"_____no_output_____"
],
[
"df_web['day'] = [time.day for time in df_web['Datetime']]",
"_____no_output_____"
],
[
"sns.histplot(x='day', hue='sentiment', data=df_web.loc[df_web['Film'] == 'dune'].loc[df_web['Time Range'] == \"Sortie Dune\"]).set(title=\"Évolution des sentiments exprimés sur le film Dune\")",
"_____no_output_____"
],
[
"sns.histplot(x='day', hue='sentiment', data=df_web.loc[df_web['Film'] == 'space jam'].loc[df_web['Time Range'] == \"Sortie Space Jam\"]).set(title=\"Évolution des sentiments exprimés sur le film Space Jam\")",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec71cb6345588201b7b30f794b49b7fd2f94c6b0 | 46,206 | ipynb | Jupyter Notebook | Encrypted_Training.ipynb | strongrunner/Secure-Machine-Learning | a6dda3bc48bb886b6ebf39fff2b2d3f35fe62afd | [
"MIT"
]
| 2 | 2021-01-14T16:33:33.000Z | 2021-05-29T14:10:00.000Z | Encrypted_Training.ipynb | strongrunner/Secure-Machine-Learning | a6dda3bc48bb886b6ebf39fff2b2d3f35fe62afd | [
"MIT"
]
| null | null | null | Encrypted_Training.ipynb | strongrunner/Secure-Machine-Learning | a6dda3bc48bb886b6ebf39fff2b2d3f35fe62afd | [
"MIT"
]
| null | null | null | 62.950954 | 310 | 0.581375 | [
[
[
"<a href=\"https://colab.research.google.com/github/strongrunner/Secure-Machine-Learning/blob/main/Encrypted_Training.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
]
],
[
[
"!pip install tf-encrypted\n!pip install syft\n! URL=\"https://github.com/openmined/PySyft.git\" && FOLDER=\"PySyft\" && if [ ! -d $FOLDER ]; then git clone -b ryffel/4P --single-branch $URL; else (cd $FOLDER && git pull $URL && cd ..); fi;\n\n!cd PySyft; python setup.py install > /dev/null\n\nimport os\nimport sys\nmodule_path = os.path.abspath(os.path.join('./PySyft'))\nif module_path not in sys.path:\n sys.path.append(module_path)\n \n!pip install --upgrade --force-reinstall lz4\n!pip install --upgrade --force-reinstall websocket\n!pip install --upgrade --force-reinstall websockets",
"Collecting tf-encrypted\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/15/be/a4c0af9fdc5e5cee28495460538acf2766382bd572e01d4847abc7608dba/tf_encrypted-0.5.9-py3-none-manylinux1_x86_64.whl (2.7MB)\n\u001b[K |████████████████████████████████| 2.7MB 5.8MB/s \n\u001b[?25hRequirement already satisfied: numpy>=1.14.0 in /usr/local/lib/python3.6/dist-packages (from tf-encrypted) (1.18.5)\nCollecting pyyaml>=5.1\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/64/c2/b80047c7ac2478f9501676c988a5411ed5572f35d1beff9cae07d321512c/PyYAML-5.3.1.tar.gz (269kB)\n\u001b[K |████████████████████████████████| 276kB 43.2MB/s \n\u001b[?25hCollecting tensorflow<2,>=1.12.0\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/8e/64/7a19837dd54d3f53b1ce5ae346ab401dde9678e8f233220317000bfdb3e2/tensorflow-1.15.4-cp36-cp36m-manylinux2010_x86_64.whl (110.5MB)\n\u001b[K |████████████████████████████████| 110.5MB 78kB/s \n\u001b[?25hCollecting gast==0.2.2\n Downloading https://files.pythonhosted.org/packages/4e/35/11749bf99b2d4e3cceb4d55ca22590b0d7c2c62b9de38ac4a4a7f4687421/gast-0.2.2.tar.gz\nCollecting tensorflow-estimator==1.15.1\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/de/62/2ee9cd74c9fa2fa450877847ba560b260f5d0fb70ee0595203082dafcc9d/tensorflow_estimator-1.15.1-py2.py3-none-any.whl (503kB)\n\u001b[K |████████████████████████████████| 512kB 45.8MB/s \n\u001b[?25hRequirement already satisfied: grpcio>=1.8.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (1.33.2)\nRequirement already satisfied: absl-py>=0.7.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (0.10.0)\nRequirement already satisfied: opt-einsum>=2.3.2 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (3.3.0)\nRequirement already satisfied: wheel>=0.26; python_version >= \"3\" in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (0.35.1)\nRequirement already satisfied: six>=1.10.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (1.15.0)\nRequirement already satisfied: astor>=0.6.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (0.8.1)\nRequirement already satisfied: protobuf>=3.6.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (3.12.4)\nRequirement already satisfied: termcolor>=1.1.0 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (1.1.0)\nCollecting keras-applications>=1.0.8\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/71/e3/19762fdfc62877ae9102edf6342d71b28fbfd9dea3d2f96a882ce099b03f/Keras_Applications-1.0.8-py3-none-any.whl (50kB)\n\u001b[K |████████████████████████████████| 51kB 6.6MB/s \n\u001b[?25hCollecting tensorboard<1.16.0,>=1.15.0\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/1e/e9/d3d747a97f7188f48aa5eda486907f3b345cd409f0a0850468ba867db246/tensorboard-1.15.0-py3-none-any.whl (3.8MB)\n\u001b[K |████████████████████████████████| 3.8MB 43.8MB/s \n\u001b[?25hRequirement already satisfied: wrapt>=1.11.1 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (1.12.1)\nRequirement already satisfied: keras-preprocessing>=1.0.5 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (1.1.2)\nRequirement already satisfied: google-pasta>=0.1.6 in /usr/local/lib/python3.6/dist-packages (from tensorflow<2,>=1.12.0->tf-encrypted) (0.2.0)\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf>=3.6.1->tensorflow<2,>=1.12.0->tf-encrypted) (50.3.2)\nRequirement already satisfied: h5py in /usr/local/lib/python3.6/dist-packages (from keras-applications>=1.0.8->tensorflow<2,>=1.12.0->tf-encrypted) (2.10.0)\nRequirement already satisfied: markdown>=2.6.8 in /usr/local/lib/python3.6/dist-packages (from tensorboard<1.16.0,>=1.15.0->tensorflow<2,>=1.12.0->tf-encrypted) (3.3.3)\nRequirement already satisfied: werkzeug>=0.11.15 in /usr/local/lib/python3.6/dist-packages (from tensorboard<1.16.0,>=1.15.0->tensorflow<2,>=1.12.0->tf-encrypted) (1.0.1)\nRequirement already satisfied: importlib-metadata; python_version < \"3.8\" in /usr/local/lib/python3.6/dist-packages (from markdown>=2.6.8->tensorboard<1.16.0,>=1.15.0->tensorflow<2,>=1.12.0->tf-encrypted) (2.0.0)\nRequirement already satisfied: zipp>=0.5 in /usr/local/lib/python3.6/dist-packages (from importlib-metadata; python_version < \"3.8\"->markdown>=2.6.8->tensorboard<1.16.0,>=1.15.0->tensorflow<2,>=1.12.0->tf-encrypted) (3.4.0)\nBuilding wheels for collected packages: pyyaml, gast\n Building wheel for pyyaml (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for pyyaml: filename=PyYAML-5.3.1-cp36-cp36m-linux_x86_64.whl size=44619 sha256=d1d0eb6fc7a444f9456707ca8b89c3eced54a860d53a15d5ee845cc0659d6d90\n Stored in directory: /root/.cache/pip/wheels/a7/c1/ea/cf5bd31012e735dc1dfea3131a2d5eae7978b251083d6247bd\n Building wheel for gast (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for gast: filename=gast-0.2.2-cp36-none-any.whl size=7542 sha256=2ff49a0d93c5dc3e9a5e5a1512b1b98f52c62e31663d76fcf06d4168a891dca4\n Stored in directory: /root/.cache/pip/wheels/5c/2e/7e/a1d4d4fcebe6c381f378ce7743a3ced3699feb89bcfbdadadd\nSuccessfully built pyyaml gast\n\u001b[31mERROR: tensorflow-probability 0.11.0 has requirement gast>=0.3.2, but you'll have gast 0.2.2 which is incompatible.\u001b[0m\nInstalling collected packages: pyyaml, gast, tensorflow-estimator, keras-applications, tensorboard, tensorflow, tf-encrypted\n Found existing installation: PyYAML 3.13\n Uninstalling PyYAML-3.13:\n Successfully uninstalled PyYAML-3.13\n Found existing installation: gast 0.3.3\n Uninstalling gast-0.3.3:\n Successfully uninstalled gast-0.3.3\n Found existing installation: tensorflow-estimator 2.3.0\n Uninstalling tensorflow-estimator-2.3.0:\n Successfully uninstalled tensorflow-estimator-2.3.0\n Found existing installation: tensorboard 2.3.0\n Uninstalling tensorboard-2.3.0:\n Successfully uninstalled tensorboard-2.3.0\n Found existing installation: tensorflow 2.3.0\n Uninstalling tensorflow-2.3.0:\n Successfully uninstalled tensorflow-2.3.0\nSuccessfully installed gast-0.2.2 keras-applications-1.0.8 pyyaml-5.3.1 tensorboard-1.15.0 tensorflow-1.15.4 tensorflow-estimator-1.15.1 tf-encrypted-0.5.9\nCollecting syft\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/45/9f/e61e32149f1323f48f89bbbebe96a32425ecd461e3e580513eaebf45b04f/syft-0.3.0-py2.py3-none-any.whl (289kB)\n\u001b[K |████████████████████████████████| 296kB 4.1MB/s \n\u001b[?25hCollecting aiortc\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/d4/b3/cd4869592930125ab832d59517469eaaf7c4fabc0313fed3be01ae31a631/aiortc-1.0.0-cp36-cp36m-manylinux2010_x86_64.whl (2.0MB)\n\u001b[K |████████████████████████████████| 2.0MB 12.6MB/s \n\u001b[?25hCollecting forbiddenfruit>=0.1.3\n Downloading https://files.pythonhosted.org/packages/03/24/903765ffae7e2022f61dcf4fb6e972c48f9abf64cfa9eb074b6a2a7c78f3/forbiddenfruit-0.1.3.tar.gz\nCollecting sqlitedict\n Downloading https://files.pythonhosted.org/packages/5c/2d/b1d99e9ad157dd7de9cd0d36a8a5876b13b55e4b75f7498bc96035fb4e96/sqlitedict-1.7.0.tar.gz\nRequirement already satisfied: pandas in /usr/local/lib/python3.6/dist-packages (from syft) (1.1.4)\nRequirement already satisfied: nest-asyncio in /usr/local/lib/python3.6/dist-packages (from syft) (1.4.3)\nCollecting PyNaCl\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/9d/57/2f5e6226a674b2bcb6db531e8b383079b678df5b10cdaa610d6cf20d77ba/PyNaCl-1.4.0-cp35-abi3-manylinux1_x86_64.whl (961kB)\n\u001b[K |████████████████████████████████| 962kB 30.8MB/s \n\u001b[?25hRequirement already satisfied: typeguard in /usr/local/lib/python3.6/dist-packages (from syft) (2.7.1)\nRequirement already satisfied: packaging in /usr/local/lib/python3.6/dist-packages (from syft) (20.4)\nCollecting websockets\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/bb/d9/856af84843912e2853b1b6e898ac8b802989fcf9ecf8e8445a1da263bf3b/websockets-8.1-cp36-cp36m-manylinux2010_x86_64.whl (78kB)\n\u001b[K |████████████████████████████████| 81kB 4.3MB/s \n\u001b[?25hCollecting loguru\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/6d/48/0a7d5847e3de329f1d0134baf707b689700b53bd3066a5a8cfd94b3c9fc8/loguru-0.5.3-py3-none-any.whl (57kB)\n\u001b[K |████████████████████████████████| 61kB 8.0MB/s \n\u001b[?25hRequirement already satisfied: dataclasses in /usr/local/lib/python3.6/dist-packages (from syft) (0.8)\nRequirement already satisfied: protobuf in /usr/local/lib/python3.6/dist-packages (from syft) (3.12.4)\nRequirement already satisfied: requests in /usr/local/lib/python3.6/dist-packages (from syft) (2.23.0)\nRequirement already satisfied: flask in /usr/local/lib/python3.6/dist-packages (from syft) (1.1.2)\nRequirement already satisfied: torchvision in /usr/local/lib/python3.6/dist-packages (from syft) (0.8.1+cu101)\nRequirement already satisfied: typing-extensions in /usr/local/lib/python3.6/dist-packages (from syft) (3.7.4.3)\nRequirement already satisfied: torch>=1.5 in /usr/local/lib/python3.6/dist-packages (from syft) (1.7.0+cu101)\nCollecting dpcontracts\n Downloading https://files.pythonhosted.org/packages/aa/e2/cad64673297a634a623808045d416ed85bad1c470ccc99e0cdc7b13b9774/dpcontracts-0.6.0.tar.gz\nCollecting pyee>=6.0.0\n Downloading https://files.pythonhosted.org/packages/0d/0a/933b3931107e1da186963fd9bb9bceb9a613cff034cb0fb3b0c61003f357/pyee-8.1.0-py2.py3-none-any.whl\nRequirement already satisfied: cffi>=1.0.0 in /usr/local/lib/python3.6/dist-packages (from aiortc->syft) (1.14.3)\nCollecting av<9.0.0,>=8.0.0\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/9e/62/9a992be76f8e13ce0e3a24a838191b546805545116f9fc869bd11bd21b5f/av-8.0.2-cp36-cp36m-manylinux2010_x86_64.whl (36.9MB)\n\u001b[K |████████████████████████████████| 36.9MB 1.2MB/s \n\u001b[?25hCollecting aioice<0.7.0,>=0.6.17\n Downloading https://files.pythonhosted.org/packages/8b/86/e3cdf660b67da7a9a7013253db5db7cf786a52296cb40078db1206177698/aioice-0.6.18-py3-none-any.whl\nCollecting crc32c\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/2f/b4/e2e293d323f10d03929ed2b68de14cdddd523700381de6aecec561f7c2d1/crc32c-2.2-cp36-cp36m-manylinux2010_x86_64.whl (48kB)\n\u001b[K |████████████████████████████████| 51kB 7.1MB/s \n\u001b[?25hCollecting pylibsrtp>=0.5.6\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/cb/fd/4f8e286da01db4e52aaa6426eac496af6e4dc00419d807d4f87d07363d84/pylibsrtp-0.6.7-cp36-cp36m-manylinux2010_x86_64.whl (75kB)\n\u001b[K |████████████████████████████████| 81kB 10.5MB/s \n\u001b[?25hCollecting cryptography>=2.2\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/4c/a2/6565c5271a79e3c96d7a079053b4d8408a740d4bf365f0f5f244a807bd09/cryptography-3.2.1-cp35-abi3-manylinux2010_x86_64.whl (2.6MB)\n\u001b[K |████████████████████████████████| 2.6MB 40.0MB/s \n\u001b[?25hRequirement already satisfied: numpy>=1.15.4 in /usr/local/lib/python3.6/dist-packages (from pandas->syft) (1.18.5)\nRequirement already satisfied: pytz>=2017.2 in /usr/local/lib/python3.6/dist-packages (from pandas->syft) (2018.9)\nRequirement already satisfied: python-dateutil>=2.7.3 in /usr/local/lib/python3.6/dist-packages (from pandas->syft) (2.8.1)\nRequirement already satisfied: six in /usr/local/lib/python3.6/dist-packages (from PyNaCl->syft) (1.15.0)\nRequirement already satisfied: pyparsing>=2.0.2 in /usr/local/lib/python3.6/dist-packages (from packaging->syft) (2.4.7)\nCollecting aiocontextvars>=0.2.0; python_version < \"3.7\"\n Downloading https://files.pythonhosted.org/packages/db/c1/7a723e8d988de0a2e623927396e54b6831b68cb80dce468c945b849a9385/aiocontextvars-0.2.2-py2.py3-none-any.whl\nRequirement already satisfied: setuptools in /usr/local/lib/python3.6/dist-packages (from protobuf->syft) (50.3.2)\nRequirement already satisfied: idna<3,>=2.5 in /usr/local/lib/python3.6/dist-packages (from requests->syft) (2.10)\nRequirement already satisfied: urllib3!=1.25.0,!=1.25.1,<1.26,>=1.21.1 in /usr/local/lib/python3.6/dist-packages (from requests->syft) (1.24.3)\nRequirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.6/dist-packages (from requests->syft) (2020.11.8)\nRequirement already satisfied: chardet<4,>=3.0.2 in /usr/local/lib/python3.6/dist-packages (from requests->syft) (3.0.4)\nRequirement already satisfied: Jinja2>=2.10.1 in /usr/local/lib/python3.6/dist-packages (from flask->syft) (2.11.2)\nRequirement already satisfied: click>=5.1 in /usr/local/lib/python3.6/dist-packages (from flask->syft) (7.1.2)\nRequirement already satisfied: itsdangerous>=0.24 in /usr/local/lib/python3.6/dist-packages (from flask->syft) (1.1.0)\nRequirement already satisfied: Werkzeug>=0.15 in /usr/local/lib/python3.6/dist-packages (from flask->syft) (1.0.1)\nRequirement already satisfied: pillow>=4.1.1 in /usr/local/lib/python3.6/dist-packages (from torchvision->syft) (7.0.0)\nRequirement already satisfied: future in /usr/local/lib/python3.6/dist-packages (from torch>=1.5->syft) (0.16.0)\nRequirement already satisfied: pycparser in /usr/local/lib/python3.6/dist-packages (from cffi>=1.0.0->aiortc->syft) (2.20)\nCollecting netifaces\n Downloading https://files.pythonhosted.org/packages/0c/9b/c4c7eb09189548d45939a3d3a6b3d53979c67d124459b27a094c365c347f/netifaces-0.10.9-cp36-cp36m-manylinux1_x86_64.whl\nCollecting contextvars==2.4; python_version < \"3.7\"\n Downloading https://files.pythonhosted.org/packages/83/96/55b82d9f13763be9d672622e1b8106c85acb83edd7cc2fa5bc67cd9877e9/contextvars-2.4.tar.gz\nRequirement already satisfied: MarkupSafe>=0.23 in /usr/local/lib/python3.6/dist-packages (from Jinja2>=2.10.1->flask->syft) (1.1.1)\nCollecting immutables>=0.9\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/99/e0/ea6fd4697120327d26773b5a84853f897a68e33d3f9376b00a8ff96e4f63/immutables-0.14-cp36-cp36m-manylinux1_x86_64.whl (98kB)\n\u001b[K |████████████████████████████████| 102kB 12.0MB/s \n\u001b[?25hBuilding wheels for collected packages: forbiddenfruit, sqlitedict, dpcontracts, contextvars\n Building wheel for forbiddenfruit (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for forbiddenfruit: filename=forbiddenfruit-0.1.3-cp36-cp36m-linux_x86_64.whl size=23740 sha256=b52f68bb6390b89e73afeb2db83ae788ebcc5dda1e2759ec6407049d02f98d12\n Stored in directory: /root/.cache/pip/wheels/1f/50/a2/ae88927c08a82f487e68b1d96efa653b45bdbb5823252d5054\n Building wheel for sqlitedict (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for sqlitedict: filename=sqlitedict-1.7.0-cp36-none-any.whl size=14377 sha256=f6b07d9c56dfae42bade25a8db755dd24bdc31dc0b14e6c51d3c97932ba8bb9f\n Stored in directory: /root/.cache/pip/wheels/cf/c6/4f/2c64a43f041415eb8b8740bd80e15e92f0d46c5e464d8e4b9b\n Building wheel for dpcontracts (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for dpcontracts: filename=dpcontracts-0.6.0-cp36-none-any.whl size=13101 sha256=579e4d61c86c509ab364f3ee27093f4316ef1abbf48f1218c721d8082c72b718\n Stored in directory: /root/.cache/pip/wheels/d0/25/65/b9972c74b262ee4a2b3f338b90e48905d4408413f2140daa07\n Building wheel for contextvars (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for contextvars: filename=contextvars-2.4-cp36-none-any.whl size=7666 sha256=bfaa079024dc97e688c744c0f47f52a11fcead4a9cb532bc7c452b77b0466af4\n Stored in directory: /root/.cache/pip/wheels/a5/7d/68/1ebae2668bda2228686e3c1cf16f2c2384cea6e9334ad5f6de\nSuccessfully built forbiddenfruit sqlitedict dpcontracts contextvars\nInstalling collected packages: pyee, av, netifaces, aioice, crc32c, pylibsrtp, cryptography, aiortc, forbiddenfruit, sqlitedict, PyNaCl, websockets, immutables, contextvars, aiocontextvars, loguru, dpcontracts, syft\nSuccessfully installed PyNaCl-1.4.0 aiocontextvars-0.2.2 aioice-0.6.18 aiortc-1.0.0 av-8.0.2 contextvars-2.4 crc32c-2.2 cryptography-3.2.1 dpcontracts-0.6.0 forbiddenfruit-0.1.3 immutables-0.14 loguru-0.5.3 netifaces-0.10.9 pyee-8.1.0 pylibsrtp-0.6.7 sqlitedict-1.7.0 syft-0.3.0 websockets-8.1\nCloning into 'PySyft'...\nremote: Enumerating objects: 2, done.\u001b[K\nremote: Counting objects: 100% (2/2), done.\u001b[K\nremote: Compressing objects: 100% (2/2), done.\u001b[K\nremote: Total 35445 (delta 0), reused 2 (delta 0), pack-reused 35443\u001b[K\nReceiving objects: 100% (35445/35445), 36.76 MiB | 12.00 MiB/s, done.\nResolving deltas: 100% (23880/23880), done.\nzip_safe flag not set; analyzing archive contents...\nexamples.tutorials.advanced.websockets_mnist.__pycache__.start_websocket_servers.cpython-36: module references __file__\nexamples.tutorials.advanced.websockets_mnist_parallel.__pycache__.start_websocket_servers.cpython-36: module references __file__\nexamples.tutorials.websocket.pen_testing.steal_data_over_sockets.__pycache__.start_websocket_servers.cpython-36: module references __file__\ntest.__pycache__.run_websocket_server.cpython-36: module references __file__\ntest.scripts.__pycache__.run_websocket_server.cpython-36: module references __file__\nno previously-included directories found matching 'docs/build'\nwarning: no files found matching 'tornado/test/README'\nzip_safe flag not set; analyzing archive contents...\ntornado.__pycache__.autoreload.cpython-36: module references __file__\ntornado.__pycache__.options.cpython-36: module references __file__\ntornado.__pycache__.speedups.cpython-36: module references __file__\ntornado.__pycache__.testing.cpython-36: module references __file__\ntornado.test.__pycache__.httpserver_test.cpython-36: module references __file__\ntornado.test.__pycache__.iostream_test.cpython-36: module references __file__\ntornado.test.__pycache__.locale_test.cpython-36: module references __file__\ntornado.test.__pycache__.options_test.cpython-36: module references __file__\ntornado.test.__pycache__.template_test.cpython-36: module references __file__\ntornado.test.__pycache__.web_test.cpython-36: module references __file__\nzip_safe flag not set; analyzing archive contents...\nCollecting lz4\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/e3/06/f6501306032cd4015e220493514907864043376a02e83acc0be6bb1b98cf/lz4-3.1.1-cp36-cp36m-manylinux2010_x86_64.whl (1.8MB)\n\u001b[K |████████████████████████████████| 1.8MB 5.2MB/s \n\u001b[?25hInstalling collected packages: lz4\n Found existing installation: lz4 3.0.2\n Uninstalling lz4-3.0.2:\n Successfully uninstalled lz4-3.0.2\nSuccessfully installed lz4-3.1.1\nCollecting websocket\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/f2/6d/a60d620ea575c885510c574909d2e3ed62129b121fa2df00ca1c81024c87/websocket-0.2.1.tar.gz (195kB)\n\u001b[K |████████████████████████████████| 204kB 4.3MB/s \n\u001b[?25hCollecting gevent\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/3f/92/b80b922f08f222faca53c8d278e2e612192bc74b0e1f0db2f80a6ee46982/gevent-20.9.0-cp36-cp36m-manylinux2010_x86_64.whl (5.3MB)\n\u001b[K |████████████████████████████████| 5.3MB 14.9MB/s \n\u001b[?25hCollecting greenlet\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/80/d0/532e160c777b42f6f393f9de8c88abb8af6c892037c55e4d3a8a211324dd/greenlet-0.4.17-cp36-cp36m-manylinux1_x86_64.whl (44kB)\n\u001b[K |████████████████████████████████| 51kB 7.1MB/s \n\u001b[?25hCollecting zope.event\n Downloading https://files.pythonhosted.org/packages/9e/85/b45408c64f3b888976f1d5b37eed8d746b8d5729a66a49ec846fda27d371/zope.event-4.5.0-py2.py3-none-any.whl\nCollecting setuptools\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/6d/38/c21ef5034684ffc0412deefbb07d66678332290c14bb5269c85145fbd55e/setuptools-50.3.2-py3-none-any.whl (785kB)\n\u001b[K |████████████████████████████████| 788kB 34.7MB/s \n\u001b[?25hCollecting zope.interface\n\u001b[?25l Downloading https://files.pythonhosted.org/packages/82/b0/da8afd9b3bd50c7665ecdac062f182982af1173c9081f9af7261091c5588/zope.interface-5.2.0-cp36-cp36m-manylinux2010_x86_64.whl (236kB)\n\u001b[K |████████████████████████████████| 245kB 45.8MB/s \n\u001b[?25hBuilding wheels for collected packages: websocket\n Building wheel for websocket (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for websocket: filename=websocket-0.2.1-cp36-none-any.whl size=192134 sha256=45b141c751e17cbd573dadf6dbeb6a5a522aa50c4e86b069dc16b0e4506b243f\n Stored in directory: /root/.cache/pip/wheels/35/f7/5c/9e8243838269ea93f05295708519a6e183fa6b515d9ce3b636\nSuccessfully built websocket\n\u001b[31mERROR: datascience 0.10.6 has requirement folium==0.2.1, but you'll have folium 0.8.3 which is incompatible.\u001b[0m\nInstalling collected packages: setuptools, zope.event, greenlet, zope.interface, gevent, websocket\n Found existing installation: setuptools 50.3.2\n Uninstalling setuptools-50.3.2:\n Successfully uninstalled setuptools-50.3.2\nSuccessfully installed gevent-20.9.0 greenlet-0.4.17 setuptools-50.3.2 websocket-0.2.1 zope.event-4.5.0 zope.interface-5.2.0\n"
]
],
[
[
"## Imports and training configuration",
"_____no_output_____"
]
],
[
[
"from __future__ import print_function\nimport argparse\nimport numpy as np\nimport pandas as pd\nimport torch\nimport torch.nn as nn\nimport torch.nn.functional as F\nimport torch.optim as optim\n#from torchvision import datasets, transforms\n#import time\ntorch.__version__",
"_____no_output_____"
],
[
"class Arguments():\n def __init__(self):\n self.batch_size = 64\n self.test_batch_size = 64\n self.epochs = 10\n self.lr = 0.02\n self.seed = 1\n self.log_interval = 1 # Log info at each batch\n self.precision_fractional = 3\n\nargs = Arguments()\n\n_ = torch.manual_seed(args.seed)",
"_____no_output_____"
]
],
[
[
"Secret sharing of data in each worker",
"_____no_output_____"
]
],
[
[
"import syft as sy \nhook = sy.TorchHook(torch) \ndef connect_to_workers(n_workers):\n return [\n sy.VirtualWorker(hook, id=f\"worker{i+1}\")\n for i in range(n_workers)\n ]\ndef connect_to_crypto_provider():\n return sy.VirtualWorker(hook, id=\"crypto_provider\")\n\nworkers = connect_to_workers(n_workers=2)\ncrypto_provider = connect_to_crypto_provider()",
"_____no_output_____"
],
[
"\nn_train_items = 640\nn_test_items = 640\n\ndef get_private_data_loaders(precision_fractional, workers, crypto_provider):\n \n def one_hot_of(index_tensor):\n \n onehot_tensor = torch.zeros(*index_tensor.shape, 10) \n onehot_tensor = onehot_tensor.scatter(1, index_tensor.view(-1, 1), 1)\n return onehot_tensor\n \n def secret_share(tensor):\n \n return (\n tensor\n .fix_precision(precision_fractional=precision_fractional)\n .share(*workers, crypto_provider=crypto_provider, requires_grad=True)\n )\n \n transformation = transforms.Compose([\n transforms.ToTensor(),\n transforms.Normalize((0.1307,), (0.3081,))\n ])\n \n train_loader = torch.utils.data.DataLoader(\n datasets.MNIST('../data', train=True, download=True, transform=transformation),\n batch_size=args.batch_size\n )\n \n private_train_loader = [\n (secret_share(data), secret_share(one_hot_of(target)))\n for i, (data, target) in enumerate(train_loader)\n if i < n_train_items / args.batch_size\n ]\n \n test_loader = torch.utils.data.DataLoader(\n datasets.MNIST('../data', train=False, download=True, transform=transformation),\n batch_size=args.test_batch_size\n )\n \n private_test_loader = [\n (secret_share(data), secret_share(target.float()))\n for i, (data, target) in enumerate(test_loader)\n if i < n_test_items / args.test_batch_size\n ]\n \n return private_train_loader, private_test_loader\n \n \nprivate_train_loader, private_test_loader = get_private_data_loaders(\n precision_fractional=args.precision_fractional,\n workers=workers,\n crypto_provider=crypto_provider\n)",
"_____no_output_____"
],
[
"class Net(nn.Module):\n def __init__(self):\n super(Net, self).__init__()\n self.fc1 = nn.Linear(28 * 28, 128)\n self.fc2 = nn.Linear(128, 64)\n self.fc3 = nn.Linear(64, 10)\n\n def forward(self, x):\n x = x.view(-1, 28 * 28)\n x = F.relu(self.fc1(x))\n x = F.relu(self.fc2(x))\n x = self.fc3(x)\n return x",
"_____no_output_____"
],
[
"def train(args, model, private_train_loader, optimizer, epoch):\n model.train()\n for batch_idx, (data, target) in enumerate(private_train_loader): # <-- now it is a private dataset\n start_time = time.time()\n \n optimizer.zero_grad()\n \n output = model(data)\n \n # loss = F.nll_loss(output, target) <-- not possible here\n batch_size = output.shape[0]\n loss = ((output - target)**2).sum().refresh()/batch_size\n \n loss.backward()\n \n optimizer.step()\n\n if batch_idx % args.log_interval == 0:\n loss = loss.get().float_precision()\n # print('Train Epoch: {} [{}/{} ({:.0f}%)]\\tLoss: {:.6f}\\tTime: {:.3f}s'.format(\n # epoch, batch_idx * args.batch_size, len(private_train_loader) * args.batch_size,\n # 100. * batch_idx / len(private_train_loader), loss.item(), time.time() - start_time))\n ",
"_____no_output_____"
],
[
"def test(args, model, private_test_loader):\n model.eval()\n test_loss = 0\n correct = 0\n with torch.no_grad():\n for data, target in private_test_loader:\n start_time = time.time()\n \n output = model(data)\n pred = output.argmax(dim=1)\n correct += pred.eq(target.view_as(pred)).sum()\n\n correct = correct.get().float_precision()\n # print('\\nTest set: Accuracy: {}/{} ({:.0f}%)\\n'.format(\n # correct.item(), len(private_test_loader)* args.test_batch_size,\n # 100. * correct.item() / (len(private_test_loader) * args.test_batch_size)))",
"_____no_output_____"
],
[
"model = Net()\nmodel = model.fix_precision().share(*workers, crypto_provider=crypto_provider, requires_grad=True)\n\noptimizer = optim.SGD(model.parameters(), lr=args.lr)\noptimizer = optimizer.fix_precision() \n\nfor epoch in range(1, args.epochs + 1):\n train(args, model, private_train_loader, optimizer, epoch)\n test(args, model, private_test_loader)",
"Train Epoch: 1 [0/640 (0%)]\tLoss: 1.128000\tTime: 6.301s\nTrain Epoch: 1 [64/640 (10%)]\tLoss: 1.011000\tTime: 4.547s\nTrain Epoch: 1 [128/640 (20%)]\tLoss: 0.990000\tTime: 4.401s\nTrain Epoch: 1 [192/640 (30%)]\tLoss: 0.902000\tTime: 5.100s\nTrain Epoch: 1 [256/640 (40%)]\tLoss: 0.887000\tTime: 4.699s\nTrain Epoch: 1 [320/640 (50%)]\tLoss: 0.875000\tTime: 4.630s\nTrain Epoch: 1 [384/640 (60%)]\tLoss: 0.853000\tTime: 4.254s\nTrain Epoch: 1 [448/640 (70%)]\tLoss: 0.849000\tTime: 4.282s\nTrain Epoch: 1 [512/640 (80%)]\tLoss: 0.830000\tTime: 4.617s\nTrain Epoch: 1 [576/640 (90%)]\tLoss: 0.839000\tTime: 4.684s\n\nTest set: Accuracy: 221.0/640 (35%)\n\nTrain Epoch: 2 [0/640 (0%)]\tLoss: 0.782000\tTime: 4.239s\nTrain Epoch: 2 [64/640 (10%)]\tLoss: 0.732000\tTime: 4.516s\nTrain Epoch: 2 [128/640 (20%)]\tLoss: 0.794000\tTime: 4.403s\nTrain Epoch: 2 [192/640 (30%)]\tLoss: 0.717000\tTime: 4.787s\nTrain Epoch: 2 [256/640 (40%)]\tLoss: 0.705000\tTime: 4.560s\nTrain Epoch: 2 [320/640 (50%)]\tLoss: 0.707000\tTime: 4.364s\nTrain Epoch: 2 [384/640 (60%)]\tLoss: 0.703000\tTime: 4.512s\nTrain Epoch: 2 [448/640 (70%)]\tLoss: 0.720000\tTime: 4.189s\nTrain Epoch: 2 [512/640 (80%)]\tLoss: 0.711000\tTime: 4.408s\nTrain Epoch: 2 [576/640 (90%)]\tLoss: 0.745000\tTime: 4.511s\n\nTest set: Accuracy: 362.0/640 (57%)\n\nTrain Epoch: 3 [0/640 (0%)]\tLoss: 0.668000\tTime: 4.564s\nTrain Epoch: 3 [64/640 (10%)]\tLoss: 0.599000\tTime: 5.153s\nTrain Epoch: 3 [128/640 (20%)]\tLoss: 0.699000\tTime: 5.253s\nTrain Epoch: 3 [192/640 (30%)]\tLoss: 0.601000\tTime: 4.417s\nTrain Epoch: 3 [256/640 (40%)]\tLoss: 0.591000\tTime: 6.257s\nTrain Epoch: 3 [320/640 (50%)]\tLoss: 0.592000\tTime: 4.235s\nTrain Epoch: 3 [384/640 (60%)]\tLoss: 0.603000\tTime: 4.387s\nTrain Epoch: 3 [448/640 (70%)]\tLoss: 0.629000\tTime: 4.460s\nTrain Epoch: 3 [512/640 (80%)]\tLoss: 0.625000\tTime: 4.927s\nTrain Epoch: 3 [576/640 (90%)]\tLoss: 0.669000\tTime: 8.410s\n\nTest set: Accuracy: 400.0/640 (62%)\n\nTrain Epoch: 4 [0/640 (0%)]\tLoss: 0.582000\tTime: 4.425s\nTrain Epoch: 4 [64/640 (10%)]\tLoss: 0.503000\tTime: 4.328s\nTrain Epoch: 4 [128/640 (20%)]\tLoss: 0.624000\tTime: 5.263s\nTrain Epoch: 4 [192/640 (30%)]\tLoss: 0.516000\tTime: 4.542s\nTrain Epoch: 4 [256/640 (40%)]\tLoss: 0.514000\tTime: 4.775s\nTrain Epoch: 4 [320/640 (50%)]\tLoss: 0.511000\tTime: 4.206s\nTrain Epoch: 4 [384/640 (60%)]\tLoss: 0.532000\tTime: 4.116s\nTrain Epoch: 4 [448/640 (70%)]\tLoss: 0.564000\tTime: 4.073s\nTrain Epoch: 4 [512/640 (80%)]\tLoss: 0.559000\tTime: 4.096s\nTrain Epoch: 4 [576/640 (90%)]\tLoss: 0.618000\tTime: 4.170s\n\nTest set: Accuracy: 429.0/640 (67%)\n\nTrain Epoch: 5 [0/640 (0%)]\tLoss: 0.525000\tTime: 4.059s\nTrain Epoch: 5 [64/640 (10%)]\tLoss: 0.441000\tTime: 4.149s\nTrain Epoch: 5 [128/640 (20%)]\tLoss: 0.571000\tTime: 4.120s\nTrain Epoch: 5 [192/640 (30%)]\tLoss: 0.458000\tTime: 4.285s\nTrain Epoch: 5 [256/640 (40%)]\tLoss: 0.460000\tTime: 4.232s\nTrain Epoch: 5 [320/640 (50%)]\tLoss: 0.453000\tTime: 4.091s\nTrain Epoch: 5 [384/640 (60%)]\tLoss: 0.480000\tTime: 4.099s\nTrain Epoch: 5 [448/640 (70%)]\tLoss: 0.515000\tTime: 4.068s\nTrain Epoch: 5 [512/640 (80%)]\tLoss: 0.507000\tTime: 4.088s\nTrain Epoch: 5 [576/640 (90%)]\tLoss: 0.576000\tTime: 4.101s\n\nTest set: Accuracy: 445.0/640 (70%)\n\nTrain Epoch: 6 [0/640 (0%)]\tLoss: 0.475000\tTime: 4.521s\nTrain Epoch: 6 [64/640 (10%)]\tLoss: 0.392000\tTime: 4.854s\nTrain Epoch: 6 [128/640 (20%)]\tLoss: 0.528000\tTime: 4.318s\nTrain Epoch: 6 [192/640 (30%)]\tLoss: 0.410000\tTime: 4.344s\nTrain Epoch: 6 [256/640 (40%)]\tLoss: 0.414000\tTime: 4.291s\nTrain Epoch: 6 [320/640 (50%)]\tLoss: 0.408000\tTime: 4.323s\nTrain Epoch: 6 [384/640 (60%)]\tLoss: 0.438000\tTime: 4.357s\nTrain Epoch: 6 [448/640 (70%)]\tLoss: 0.474000\tTime: 4.124s\nTrain Epoch: 6 [512/640 (80%)]\tLoss: 0.466000\tTime: 4.115s\nTrain Epoch: 6 [576/640 (90%)]\tLoss: 0.543000\tTime: 4.112s\n\nTest set: Accuracy: 461.0/640 (72%)\n\nTrain Epoch: 7 [0/640 (0%)]\tLoss: 0.437000\tTime: 4.110s\nTrain Epoch: 7 [64/640 (10%)]\tLoss: 0.354000\tTime: 4.129s\nTrain Epoch: 7 [128/640 (20%)]\tLoss: 0.489000\tTime: 4.140s\nTrain Epoch: 7 [192/640 (30%)]\tLoss: 0.374000\tTime: 4.015s\nTrain Epoch: 7 [256/640 (40%)]\tLoss: 0.379000\tTime: 4.109s\nTrain Epoch: 7 [320/640 (50%)]\tLoss: 0.368000\tTime: 4.098s\nTrain Epoch: 7 [384/640 (60%)]\tLoss: 0.403000\tTime: 4.031s\nTrain Epoch: 7 [448/640 (70%)]\tLoss: 0.442000\tTime: 4.039s\nTrain Epoch: 7 [512/640 (80%)]\tLoss: 0.432000\tTime: 4.104s\nTrain Epoch: 7 [576/640 (90%)]\tLoss: 0.508000\tTime: 4.101s\n\nTest set: Accuracy: 473.0/640 (74%)\n\nTrain Epoch: 8 [0/640 (0%)]\tLoss: 0.405000\tTime: 4.445s\nTrain Epoch: 8 [64/640 (10%)]\tLoss: 0.324000\tTime: 4.074s\nTrain Epoch: 8 [128/640 (20%)]\tLoss: 0.458000\tTime: 4.056s\nTrain Epoch: 8 [192/640 (30%)]\tLoss: 0.347000\tTime: 4.056s\nTrain Epoch: 8 [256/640 (40%)]\tLoss: 0.352000\tTime: 4.081s\nTrain Epoch: 8 [320/640 (50%)]\tLoss: 0.341000\tTime: 4.026s\nTrain Epoch: 8 [384/640 (60%)]\tLoss: 0.374000\tTime: 4.076s\nTrain Epoch: 8 [448/640 (70%)]\tLoss: 0.416000\tTime: 4.137s\nTrain Epoch: 8 [512/640 (80%)]\tLoss: 0.402000\tTime: 4.091s\nTrain Epoch: 8 [576/640 (90%)]\tLoss: 0.483000\tTime: 4.138s\n\nTest set: Accuracy: 478.0/640 (75%)\n\nTrain Epoch: 9 [0/640 (0%)]\tLoss: 0.382000\tTime: 4.141s\nTrain Epoch: 9 [64/640 (10%)]\tLoss: 0.303000\tTime: 4.132s\nTrain Epoch: 9 [128/640 (20%)]\tLoss: 0.433000\tTime: 4.088s\nTrain Epoch: 9 [192/640 (30%)]\tLoss: 0.323000\tTime: 4.030s\nTrain Epoch: 9 [256/640 (40%)]\tLoss: 0.332000\tTime: 4.099s\nTrain Epoch: 9 [320/640 (50%)]\tLoss: 0.317000\tTime: 4.766s\nTrain Epoch: 9 [384/640 (60%)]\tLoss: 0.349000\tTime: 4.829s\nTrain Epoch: 9 [448/640 (70%)]\tLoss: 0.396000\tTime: 4.006s\nTrain Epoch: 9 [512/640 (80%)]\tLoss: 0.377000\tTime: 3.978s\nTrain Epoch: 9 [576/640 (90%)]\tLoss: 0.463000\tTime: 4.009s\n\nTest set: Accuracy: 482.0/640 (75%)\n\nTrain Epoch: 10 [0/640 (0%)]\tLoss: 0.359000\tTime: 4.027s\nTrain Epoch: 10 [64/640 (10%)]\tLoss: 0.281000\tTime: 4.035s\nTrain Epoch: 10 [128/640 (20%)]\tLoss: 0.406000\tTime: 4.028s\nTrain Epoch: 10 [192/640 (30%)]\tLoss: 0.306000\tTime: 4.013s\nTrain Epoch: 10 [256/640 (40%)]\tLoss: 0.311000\tTime: 4.000s\nTrain Epoch: 10 [320/640 (50%)]\tLoss: 0.298000\tTime: 3.987s\nTrain Epoch: 10 [384/640 (60%)]\tLoss: 0.326000\tTime: 4.038s\nTrain Epoch: 10 [448/640 (70%)]\tLoss: 0.377000\tTime: 4.028s\nTrain Epoch: 10 [512/640 (80%)]\tLoss: 0.357000\tTime: 4.048s\nTrain Epoch: 10 [576/640 (90%)]\tLoss: 0.442000\tTime: 4.036s\n\nTest set: Accuracy: 483.0/640 (75%)\n\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec71e2dcc9e95813624f160aa424acddd32dc7b7 | 167,827 | ipynb | Jupyter Notebook | log_analyzer/appender_metrics.ipynb | ABorovtsov/log4net | 020cc68cc4dd779457ac51437875e2fc125ea556 | [
"MIT"
]
| null | null | null | log_analyzer/appender_metrics.ipynb | ABorovtsov/log4net | 020cc68cc4dd779457ac51437875e2fc125ea556 | [
"MIT"
]
| null | null | null | log_analyzer/appender_metrics.ipynb | ABorovtsov/log4net | 020cc68cc4dd779457ac51437875e2fc125ea556 | [
"MIT"
]
| null | null | null | 927.220994 | 81,516 | 0.95114 | [
[
[
"import pandas as pd\r\n\r\n\r\npd.set_option('display.max_rows', None)\r\ninput_file_name = '.\\input\\data_5w.csv'\r\ninput_df = pd.read_csv(input_file_name)\r\ninput_df.DateTime = pd.to_datetime(input_df.DateTime, format='%Y-%m-%dT%H:%M:%S')\r\nmindate = input_df.DateTime.min()\r\ninput_df['total_seconds'] = (input_df.DateTime-mindate).dt.total_seconds()\r\ninput_df.head()\r\n",
"_____no_output_____"
],
[
"input_df.groupby(by=['total_seconds','CallerName']).size().unstack(level=-1).plot(figsize=(10,5), kind=\"bar\", title='RPS')",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\r\nfig, ax = plt.subplots()\r\n\r\nax = input_df.loc[(input_df.CallerName == 'DoAppend') & (input_df.LatencyUs <= input_df.LatencyUs.quantile(0.95))].LatencyUs.plot(figsize=(10,5), title='Latency: Enqueue VS Dequeue')\r\ninput_df.loc[(input_df.CallerName == 'Dequeue') & (input_df.LatencyUs <= input_df.LatencyUs.quantile(0.95))].LatencyUs.plot()\r\nax.legend([\"Enqueue\", \"Dequeue\"])",
"_____no_output_____"
],
[
"input_df.BufferSize.plot(figsize=(10,5), title='BufferSize')",
"_____no_output_____"
],
[
"input_df.AllocatedBytes.plot(figsize=(10,5), title='AllocatedBytes')",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code"
]
]
|
ec71eaffb6e738dc5147fda73b04aae6a9166573 | 10,552 | ipynb | Jupyter Notebook | docs/Getting_started.ipynb | gitter-badger/argopy | b549514dcf179193fd38ac3d66f8452ae1d6431a | [
"Apache-2.0"
]
| null | null | null | docs/Getting_started.ipynb | gitter-badger/argopy | b549514dcf179193fd38ac3d66f8452ae1d6431a | [
"Apache-2.0"
]
| null | null | null | docs/Getting_started.ipynb | gitter-badger/argopy | b549514dcf179193fd38ac3d66f8452ae1d6431a | [
"Apache-2.0"
]
| null | null | null | 24.257471 | 272 | 0.572688 | [
[
[
"#  Argo data python library\n\n``argopy`` is a python library that aims to ease Argo data access, visualisation and manipulation for regular users as well as Argo experts and operators.\n\nSeveral python packages exist: we are currently in the process of analysing how to merge these libraries toward a single powerfull tool. [List your tool here !](https://github.com/euroargodev/argopy/issues/3)\n\n## Install\n\nSince this is a library in active development, use direct install from this repo to benefit from the last version:\n\n```bash\npip install git+http://github.com/euroargodev/argopy.git@master\n```\n\nThe ``argopy`` library should work under all OS (Linux, Mac and Windows) and with python versions 3.6, 3.7 and 3.8.\n\n**The libray is already installed to run this notebook**.",
"_____no_output_____"
],
[
"# Import",
"_____no_output_____"
]
],
[
[
"import os\nimport numpy as np\nimport xarray as xr\n\nimport argopy\nfrom argopy import DataFetcher as ArgoDataFetcher",
"_____no_output_____"
]
],
[
[
"## Fetching Argo Data\nNote that the primary data model used to manipulate Argo data is [xarray](https://github.com/pydata/xarray).\n\nInit the default data fetcher like:\n",
"_____no_output_____"
]
],
[
[
"argo_loader = ArgoDataFetcher()\nargo_loader",
"_____no_output_____"
]
],
[
[
"#### Space/time domain",
"_____no_output_____"
]
],
[
[
"ds = argo_loader.region([-85,-45,10.,20.,0,10.]).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"you can also specify a date range:",
"_____no_output_____"
]
],
[
[
"ds = argo_loader.region([-85,-45,10.,20.,0,100.,'2012-01','2014-12']).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"#### Profiles of a given float\n\nThis fetch data for the float ID 6902746 and cycle number 34:",
"_____no_output_____"
]
],
[
[
"ds = argo_loader.profile(6902746, 34).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"You can also retrieve a list of cycles:",
"_____no_output_____"
]
],
[
[
"ds = argo_loader.profile(6902746, np.arange(12,45)).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"#### One or more floats",
"_____no_output_____"
]
],
[
[
"ds = argo_loader.float(6902746).to_xarray()\nds",
"_____no_output_____"
],
[
"ds = argo_loader.float([6902746, 6902747, 6902757, 6902766]).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"## More advanced data fetching\n\nTwo Argo data fetchers are available.",
"_____no_output_____"
],
[
"#### The Ifremer erddap\nThis is the recommended and default fetching backend.\n\nNot that it requires an internet connection.",
"_____no_output_____"
]
],
[
[
"argo_loader = ArgoDataFetcher(backend='erddap')\nds = argo_loader.profile(6902746, 34).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"#### A local copy of the GDAC ftp\n\n**If you don't know what GDAC means, you should not go down this line...**\n\nThis is a more complex fetching backend since it requires you to have a local copy of files from the GDAC ftp server.\n\nNote that Argo provides monthly snapshots of the entire datasets (eg: [2020/02](https://www.seanoe.org/data/00311/42182/#70590) and [full list here](http://www.argodatamgt.org/Access-to-data/Argo-DOI-Digital-Object-Identifier)). This is great for reproducibility.\n\nYou can also managed a [synchronised copy of the GDAC using rsync](http://www.argodatamgt.org/Access-to-data/Argo-GDAC-synchronization-service).\n\nThis data fetching method is usefull for Argo experts and operators, but also because it provides an offline access to the dataset.\n\nTo initiate the fetcher, you need to use the ``backend`` option and more importantly to specify the path toward the local copy of the ftp dataset (more specifically the path where all DAC are listed).\n\n**For demonstration purposes**, we will rely here on a dummy sample of GDAC ftp files provides by the ``argopy`` tutorial module:",
"_____no_output_____"
]
],
[
[
"ftproot, flist = argopy.tutorial.open_dataset('localftp')\nprint(ftproot)",
"_____no_output_____"
]
],
[
[
"Under this path, we have downloaded some Argo netcdf files. For instance:",
"_____no_output_____"
]
],
[
[
"flist[14]",
"_____no_output_____"
],
[
"argo_loader = ArgoDataFetcher(backend='localftp', path_ftp=os.path.join(ftproot, 'dac'))\nds = argo_loader.float(3902131).to_xarray()\nds",
"_____no_output_____"
]
],
[
[
"Note that to avoid specifying on each request the local ftp path copy, you can use the handy options setter:",
"_____no_output_____"
]
],
[
[
"argopy.set_options(local_ftp=os.path.join(ftproot, 'dac'))",
"_____no_output_____"
]
],
[
[
"Options are valid only throughout a python session, so you have to set this option at the beginning of your script.\n\nSo now, we don't have to specify the path anymore:",
"_____no_output_____"
]
],
[
[
"ArgoDataFetcher(backend='localftp').float(3902131).to_xarray()",
"_____no_output_____"
]
],
[
[
"## Fetched data manipulation",
"_____no_output_____"
],
[
"#### Point vs profile\nAs you probably have noticed now, data are returned as a collection of measurements along the dimension ``index``.\n\nIf you want to convert them into a collection of profiles, you can use methods from the xarray accessor named ``argo``:",
"_____no_output_____"
]
],
[
[
"ds_as_points = ArgoDataFetcher().float(5903248).to_xarray()\nds_as_points",
"_____no_output_____"
]
],
[
[
"Now to convert this dataset into a collection of profiles:",
"_____no_output_____"
]
],
[
[
"ds_as_profiles = ds_as_points.argo.point2profile()\nds_as_profiles",
"_____no_output_____"
]
],
[
[
"And finaly you can reverse that operation with the companion method:",
"_____no_output_____"
]
],
[
[
"ds_as_profiles.argo.profile2point()",
"_____no_output_____"
]
],
[
[
"#### Other formats",
"_____no_output_____"
],
[
"By default fetched data are returned in memory as [xarray.DataSet](http://xarray.pydata.org/en/stable/data-structures.html#dataset). \nFrom there, it is easy to convert them to other formats like a [Pandas dataframe](https://pandas.pydata.org/pandas-docs/stable/getting_started/dsintro.html#dataframe):",
"_____no_output_____"
]
],
[
[
"ds = ArgoDataFetcher().profile(6902746, 34).to_xarray()\ndf = ds.to_dataframe()\ndf",
"_____no_output_____"
]
],
[
[
"## Export data to files",
"_____no_output_____"
]
],
[
[
"ds = ArgoDataFetcher().region([-85,-45,10.,20.,0,100.,'2012-01','2012-12']).to_xarray()\nds",
"_____no_output_____"
],
[
"ds.to_netcdf('my_selection.nc')\n!ls -l my_selection.nc",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec7201caa9a5ddd071a88e37f2558aee23bb1ea6 | 22,909 | ipynb | Jupyter Notebook | notebooks/Session3 - More on Functions.ipynb | chaitu210/python-for-devops | b6507470f31dab1effc5ee88736881b4e8f07a31 | [
"Apache-2.0"
]
| 15 | 2020-05-19T09:46:48.000Z | 2022-03-14T21:37:56.000Z | notebooks/4_more_on_functions.ipynb | d9thc/python-for-devops | 526c87b5c457ee5c32cb5c6bd22e0e19940a9db1 | [
"Apache-2.0"
]
| null | null | null | notebooks/4_more_on_functions.ipynb | d9thc/python-for-devops | 526c87b5c457ee5c32cb5c6bd22e0e19940a9db1 | [
"Apache-2.0"
]
| 11 | 2020-05-21T08:49:48.000Z | 2022-02-26T04:41:55.000Z | 31.862309 | 2,062 | 0.561177 | [
[
[
"More on Functions\n===",
"_____no_output_____"
],
[
"Earlier we learned the most bare-boned versions of functions. In this section we will learn more general concepts about functions, such as how to use functions to return values, and how to pass different kinds of data structures between functions.",
"_____no_output_____"
],
[
"<a name='default_values'></a>Default argument values\n===",
"_____no_output_____"
]
],
[
[
"def hello_there(name):\n print(\"\\n Hello {}, hope you are finding this training useful!\".format(name))\n \nhello_there('Santosh')\nhello_there('Sunil')\nhello_there('Mahi')",
"\n Hello Santosh, hope you are finding this training useful!\n\n Hello Sunil, hope you are finding this training useful!\n\n Hello Mahi, hope you are finding this training useful!\n"
],
[
"last_name = \"Balchandani\"\na = \"My name is Sanchit {}\".format(last_name)\nprint(a)",
"My name is Sanchit Balchandani\n"
],
[
"print(f\"My Name is Sanchit {last_name}\")",
"My Name is Sanchit Balchandani\n"
],
[
"# same function fails if you don't pass the value\ndef hello_there(name):\n if not name:\n print(\"lkjdljfl\")\n print(\"\\n Hello {}, hope you are finding this training useful!\".format(name))\n \nhello_there('Santosh')\nhello_there('Sunil')\nhello_there()",
"\n Hello Santosh, hope you are finding this training useful!\n\n Hello Sunil, hope you are finding this training useful!\n"
]
],
[
[
"That makes sense; the function needs to have a name in order to do its work, so without a name it is stuck.\n\nIf you want your function to do something by default, even if no information is passed to it, you can do so by giving your arguments default values. You do this by specifying the default values when you define the function:",
"_____no_output_____"
]
],
[
[
"def hello_there(name=\"EveryOne\"):\n print(\"\\n Hello {}, hope you are finding this training useful!\".format(name))\n \nhello_there('Santosh')\nhello_there('Sunil')\nhello_there()",
"\n Hello Santosh, hope you are finding this training useful!\n\n Hello Sunil, hope you are finding this training useful!\n\n Hello EveryOne, hope you are finding this training useful!\n"
]
],
[
[
"This is particularly useful when you have a number of arguments in your function, and some of those arguments almost always have the same value. This allows people who use the function to only specify the values that are unique to their use of the function.",
"_____no_output_____"
],
[
"<a name=\"positional_arguments\"></a>Positional Arguments\n===",
"_____no_output_____"
],
[
"Much of what you will have to learn about using functions involves how to pass values from your calling statement to the function itself. The example we just looked at is pretty simple, in that the function only needed one argument in order to do its work. Let's take a look at a function that requires two arguments to do its work.",
"_____no_output_____"
]
],
[
[
"def describe_money_heist_character(nick_name, first_name, last_name, age):\n print(\"Nick name: {}\".format(nick_name.title()))\n print(\"First name: {}\".format(first_name.title()))\n print(\"Last name: {}\".format(last_name.title()))\n print(\"Age: {}\".format(age))\n print(\"\\n\")\n\ndescribe_money_heist_character('Professor', 'Álvaro', 'Morte', 45)\ndescribe_money_heist_character('Tokyo', 'Úrsula', 'Corberó', 30)\ndescribe_money_heist_character('Berlin', 'Pedro González', 'Alonso', 48)",
"Nick name: Professor\nFirst name: Álvaro\nLast name: Morte\nAge: 45\n\n\nNick name: Tokyo\nFirst name: Úrsula\nLast name: Corberó\nAge: 30\n\n\nNick name: Berlin\nFirst name: Pedro González\nLast name: Alonso\nAge: 48\n\n\n"
]
],
[
[
"The arguments in this function are `nick_name`, `first_name`, `last_name`, and `age`. These are called *positional arguments* because Python knows which value to assign to each by the order in which you give the function values. In the calling line\n\n describe_character('Professor', 'Álvaro', 'Morte', 45)\n\nwe send the values to the function. Python matches the first value with the first argument `nick_name`. It matches the second value with the second argument `first_name`. Finally it matches the last value with the third argument `age`.\n\nThis is pretty straightforward, but it means we have to make sure to get the arguments in the right order.",
"_____no_output_____"
],
[
"If we mess up the order, we get nonsense results or an error:",
"_____no_output_____"
]
],
[
[
"def describe_money_heist_character(nick_name, first_name, last_name, age):\n \"\"\"\n nick_name: str\n first_name: str\n last_name: str\n \n return:Dict/Str/Int\n \n \"\"\"\n print(\"Nick name: {}\".format(nick_name.title()))\n print(\"First name: {}\".format(first_name.title()))\n print(\"Last name: {}\".format(last_name.title()))\n print(\"Age: {}\".format(age))\n print(\"\\n\")\n\ndescribe_money_heist_character('Professor', 'Álvaro', 'Morte', 45)\ndescribe_money_heist_character('Tokyo', 'Úrsula', 'Corberó', 30)\ndescribe_money_heist_character(48, 'Berlin', 'Pedro González', 'Alonso')",
"Nick name: Professor\nFirst name: Álvaro\nLast name: Morte\nAge: 45\n\n\nNick name: Tokyo\nFirst name: Úrsula\nLast name: Corberó\nAge: 30\n\n\n"
]
],
[
[
"<a name='keyword_arguments'></a>Keyword arguments\n===\nPython allows us to use a syntax called *keyword arguments*. In this case, we can give the arguments in any order when we call the function, as long as we use the name of the arguments in our calling statement. Here is how the previous code can be made to work using keyword arguments:",
"_____no_output_____"
]
],
[
[
"def describe_money_heist_character(nick_name, first_name, last_name, age=None, died=None):\n print(\"Nick name: {}\".format(nick_name.title()))\n print(\"First name: {}\".format(first_name.title()))\n print(\"Last name: {}\".format(last_name.title()))\n if age and isinstance(age, int):\n print(\"Age: {}\".format(age))\n else:\n print(\"This guy is immortal\")\n if died:\n print(\"This character has died :(\")\n print(\"\\n\")\n\ndescribe_money_heist_character('Professor', 'Álvaro', 'Morte', age=\"Sanchit\")\ndescribe_money_heist_character('Professor', 'Álvaro', 'Morte')\ndescribe_money_heist_character('Tokyo', 'Úrsula', 'Corberó', age=30)\ndescribe_money_heist_character('Berlin', 'Pedro González', 'Alonso', died=True)",
"Nick name: Professor\nFirst name: Álvaro\nLast name: Morte\nThis guy is immortal\n\n\nNick name: Professor\nFirst name: Álvaro\nLast name: Morte\nThis guy is immortal\n\n\nNick name: Tokyo\nFirst name: Úrsula\nLast name: Corberó\nAge: 30\n\n\nNick name: Berlin\nFirst name: Pedro González\nLast name: Alonso\nThis guy is immortal\nThis character has died :(\n\n\n"
]
],
[
[
"<a name='arbitrary_arguments'></a>Accepting an arbitrary number of arguments\n===",
"_____no_output_____"
]
],
[
[
"def add(num_1, num_2):\n sum = num_1 + num_2\n print(\"The sum of your numbers is {}.\".format(sum))\n \n# Let's add some numbers.\nadd(1, 2)\nadd(-1, 2)\nadd(1, -2)",
"The sum of your numbers is 3.\nThe sum of your numbers is 1.\nThe sum of your numbers is -1.\n"
]
],
[
[
"This function appears to work well. But what if we pass it three numbers, which is a perfectly reasonable thing to do mathematically?",
"_____no_output_____"
]
],
[
[
"def add(num_1, num_2):\n sum = num_1 + num_2\n print(\"The sum of your numbers is {}.\".format(sum))\n \n# Let's add some numbers.\nadd(1, 2, 3)",
"_____no_output_____"
],
[
"#*args , **kwargs\ndef example_function(arg_1, arg_2, *arg_3):\n # Let's look at the argument values.\n print('\\narg_1:', arg_1)\n print('arg_2:', arg_2)\n for value in arg_3:\n print('arg_3 value:', value)\n\nexample_function(1, 2)\nexample_function(1, 2, 3)\nexample_function(1, 2, 3, 4)\nexample_function(1, 2, 3, 4, 5,6,7,8,9)",
"\narg_1: 1\narg_2: 2\n\narg_1: 1\narg_2: 2\narg_3 value: 3\n\narg_1: 1\narg_2: 2\narg_3 value: 3\narg_3 value: 4\n\narg_1: 1\narg_2: 2\narg_3 value: 3\narg_3 value: 4\narg_3 value: 5\narg_3 value: 6\narg_3 value: 7\narg_3 value: 8\narg_3 value: 9\n"
]
],
[
[
"We can now rewrite the add() function to accept two or more arguments, and print the sum of those numbers:",
"_____no_output_____"
]
],
[
[
"def add(*nums):\n \"\"\"This function adds the given numbers together and prints the sum.\"\"\"\n # Print the results.\n print(\"The sum of your numbers is {}.\".format(sum(nums)))\n \n# Let's add some numbers.\nadd(1, 2, 3)",
"The sum of your numbers is 6.\n"
],
[
"def add(num_1, num_2, *nums):\n sum = num_1 + num_2\n \n for num in nums:\n sum = sum + num\n \n print(f\"The sum of your numbers is {sum}.\")\n\nadd(1, 2)\nadd(1, 2, 3)\nadd(1, 2, 3, 4)\nadd(1, 2, 3, 4, 5)",
"The sum of your numbers is 3.\nThe sum of your numbers is 6.\nThe sum of your numbers is 10.\nThe sum of your numbers is 15.\n"
]
],
[
[
"<a name='arbitrary_keyword_arguments'></a>Accepting an arbitrary number of keyword arguments\n---\nPython also provides a syntax for accepting an arbitrary number of keyword arguments. The syntax looks like this:",
"_____no_output_____"
]
],
[
[
"def example_function(*args, **kwargs):\n print(*args)\n print(kwargs)\n for k, v in kwargs.items():\n print(k,':', v)\n \nexample_function(1, 2, 4, 5)\nexample_function(1, 3, value=1, name=5)\n",
"1 2 4 5\n{}\n1 3\n{'value': 1, 'name': 5}\nvalue : 1\nname : 5\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec72034285fc1d0ac0cda6a7195091534397cac4 | 41,958 | ipynb | Jupyter Notebook | scripts/visualisation.ipynb | smart-facility/smart-faware | a4764c78e427a98eeee5d6f73e4da639c2a21d38 | [
"MIT"
]
| null | null | null | scripts/visualisation.ipynb | smart-facility/smart-faware | a4764c78e427a98eeee5d6f73e4da639c2a21d38 | [
"MIT"
]
| null | null | null | scripts/visualisation.ipynb | smart-facility/smart-faware | a4764c78e427a98eeee5d6f73e4da639c2a21d38 | [
"MIT"
]
| null | null | null | 114.953425 | 3,940 | 0.576505 | [
[
[
"import altair as alt, pandas as pd\nfrom altair import datum\nfrom sqlalchemy import create_engine\nalt.data_transformers.enable('json')",
"_____no_output_____"
],
[
"db = create_engine('postgres://floodaware:1234@localhost:5432')",
"_____no_output_____"
],
[
"#data = pd.read_sql('SELECT * FROM experiment_data WHERE index = 1', db)\ndata1 = pd.read_sql('SELECT timestep, catchment, rain_in, flow, index FROM experiment_data WHERE index = 11', db)\ndata2 = pd.read_sql('SELECT timestep, catchment, rain_in, flow, index FROM experiment_data WHERE index = 8', db)",
"_____no_output_____"
],
[
"select = alt.selection_single(fields=['catchment'], empty='none')\ncharts = []\nfor dat in [data1, data2]:\n charts.append(alt.Chart(dat).mark_line().encode(x='timestep', y='flow', color=alt.condition(select, alt.value('darkred'), 'catchment:O'), opacity=alt.condition(select, alt.value(0.9), alt.value(0.3)), tooltip=['catchment','timestep','flow','index']).add_selection(select))\ncharts[0] | charts[1]",
"_____no_output_____"
],
[
"chart0 = alt.Chart(data1).transform_pivot('catchment', groupby=['timestep'], value='rain_in').mark_line(color='red').encode(x='timestep', y='23:Q')\nchart1 = alt.Chart(data2).transform_pivot('catchment', groupby=['timestep'], value='rain_in').mark_line(color='green').encode(x='timestep', y='23:Q')\nchart0 | chart1",
"_____no_output_____"
],
[
"sensor_data = pd.read_sql(\"SELECT id, stamp, (ahd*1000 - level)/1000 as level FROM sensor_levels JOIN sensors using(id) WHERE stamp BETWEEN '20200207' AND '20200209'\", db)",
"_____no_output_____"
],
[
"flow2height = pd.read_sql(\"SELECT catchment, timestep, regr[1] + flow*regr[2] + flow^2*regr[3] + flow^3*regr[4] as level FROM experiment_data INNER JOIN transects using(catchment) WHERE index = 8 AND catchment in (23, 18, 14) AND timestep BETWEEN '20200207' AND '20200209'\", db)",
"_____no_output_____"
],
[
"alt.Chart(sensor_data).mark_point().encode(x=\"stamp\", y=\"level\", color=\"id:N\", tooltip=\"id\") + alt.Chart(flow2height).mark_line().encode(x=\"timestep\", y=\"level\", color=\"catchment:N\", tooltip=\"catchment\")",
"_____no_output_____"
],
[
"daily_rain = pd.read_sql(\"SELECT id, sum(val) AS rainfall, stamp AS date FROM rainfall WHERE stamp::date BETWEEN '20200701' AND '20200901' GROUP BY id, date\", db)",
"_____no_output_____"
],
[
"alt.Chart(daily_rain).mark_line().encode(x=\"date\", y=\"rainfall\", color=\"id:N\", tooltip=\"date\").interactive()",
"_____no_output_____"
],
[
"rain = pd.read_sql(\"SELECT id, sum(val) AS level, stamp::date AS date FROM rainfall WHERE stamp BETWEEN '20200101' AND '20210101' GROUP BY id, date\", db)",
"_____no_output_____"
],
[
"alt.Chart(rain).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\").interactive()",
"_____no_output_____"
],
[
"sensors = pd.read_sql(\"SELECT id, AVG((ahd*1000 - level)/1000) as level, stamp::date AS date FROM sensor_levels JOIN sensors using(id) WHERE stamp BETWEEN '20200101' AND '20210101' GROUP BY id, date\", db)",
"_____no_output_____"
],
[
"alt.Chart(sensors).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\").interactive()",
"_____no_output_____"
],
[
"rainjuly = pd.read_sql(\"SELECT id, val AS level, stamp AS date FROM rainfall WHERE stamp BETWEEN '20200725' AND '20200811'\", db)\nexpjuly = pd.read_sql(\"SELECT catchment, timestep, regr[1] + flow*regr[2] + flow^2*regr[3] + flow^3*regr[4] as level FROM experiment_data INNER JOIN transects using(catchment) WHERE index = 13 AND timestep BETWEEN '20200725' AND '20200811'\", db)\nlevjuly = pd.read_sql(\"SELECT id, (ahd*1000 - level)/1000 AS level, stamp AS date FROM sensor_levels JOIN sensors using(id) WHERE id IN (22, 21, 9, 39, 4, 11) AND level < 5000 AND stamp BETWEEN '20200725' AND '20200811'\", db)\nmhlevjuly = pd.read_sql(\"SELECT * FROM levels WHERE stamp BETWEEN '20200725' AND '20200811'\", db)",
"_____no_output_____"
],
[
"(alt.Chart(rainjuly).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\", tooltip=\"id\").properties(title=\"Daily Rain\") | alt.Chart(expjuly).mark_line().encode(x=\"timestep\", y=\"level\", color=\"catchment:N\", tooltip=\"catchment\").properties(title=\"Experimental Levels\")) & (alt.Chart(levjuly).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\", tooltip=\"id\").properties(title=\"Sensor Levels\") | alt.Chart(mhlevjuly).mark_line().encode(x=\"stamp\", y=\"val\", color=\"id:N\", tooltip=\"id\").properties(title=\"MHL Levels\"))",
"_____no_output_____"
],
[
"lev = pd.read_sql(\"SELECT id, avg((ahd*1000 - level)/1000) AS level, stamp AS date FROM sensor_levels JOIN sensors using(id) WHERE id IN (22, 21, 9, 39, 4, 11) AND level < 5000 AND stamp::date BETWEEN '20191230' AND '20200831' GROUP BY id, date\", db)\nmhlev = pd.read_sql(\"SELECT id, avg(val) AS val, stamp::date AS date FROM levels WHERE stamp::date > '20191230' GROUP BY id, date\", db)",
"_____no_output_____"
],
[
"alt.Chart(mhlev).mark_line().encode(x=\"date\", y=\"val\", color=\"id:N\", tooltip=\"id\").interactive() + alt.Chart(lev).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\", tooltip=\"id\").interactive()",
"_____no_output_____"
],
[
"alt.Chart(mhlev).mark_line().encode(x=\"date\", y=\"val\", color=\"id:N\", tooltip=\"id\").transform_filter(datum.id == 214404) | alt.Chart(lev).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\", tooltip=\"id\").transform_filter(datum.id == 22)",
"_____no_output_____"
],
[
"alt.Chart(mhlev).mark_line().encode(x=\"date\", y=\"val\", color=\"id:N\", tooltip=\"id\").transform_filter(datum.id == 214405) | alt.Chart(lev).mark_line().encode(x=\"date\", y=\"level\", color=\"id:N\", tooltip=\"id\").transform_filter(datum.id == 11)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec7210a54d7cfd288989f3fac57262d2afb2c3d6 | 891,573 | ipynb | Jupyter Notebook | docs/_build/.jupyter_cache/executed/6a2dab0270d42f0e56971b4aebba9d6a/base.ipynb | sertit/eoreader | da6b1ba4055adba6e8a266552c29003f59d0e51a | [
"Apache-2.0"
]
| 47 | 2021-04-27T06:24:51.000Z | 2022-03-29T19:23:19.000Z | docs/_build/.jupyter_cache/executed/6a2dab0270d42f0e56971b4aebba9d6a/base.ipynb | sertit/eoreader | da6b1ba4055adba6e8a266552c29003f59d0e51a | [
"Apache-2.0"
]
| 27 | 2021-07-02T15:13:27.000Z | 2022-03-31T13:45:29.000Z | docs/_build/.jupyter_cache/executed/6a2dab0270d42f0e56971b4aebba9d6a/base.ipynb | sertit/eoreader | da6b1ba4055adba6e8a266552c29003f59d0e51a | [
"Apache-2.0"
]
| 3 | 2021-05-22T07:09:28.000Z | 2022-01-25T05:46:09.000Z | 540.019988 | 666,360 | 0.932313 | [
[
[
"import os\n\n# First of all, we need some VHR data, let's use some COSMO-SkyMed data\npath = os.path.join(\"/home\", \"data\", \"DATA\", \"PRODS\", \"COSMO\", \"1st_GEN\", \"1001512-735097\")",
"_____no_output_____"
],
[
"# Create logger\nimport logging\n\nlogger = logging.getLogger(\"eoreader\")\nlogger.setLevel(logging.INFO)\n\n# create console handler and set level to debug\nch = logging.StreamHandler()\nch.setLevel(logging.INFO)\n\n# create formatter\nformatter = logging.Formatter('%(message)s')\n\n# add formatter to ch\nch.setFormatter(formatter)\n\n# add ch to logger\nlogger.addHandler(ch)",
"_____no_output_____"
],
[
"from eoreader.reader import Reader\n\n# Create the reader\neoreader = Reader()",
"_____no_output_____"
],
[
"# Open your product\nprod = eoreader.open(path, remove_tmp=True)\nprint(f\"Acquisition datetime: {prod.datetime}\")\nprint(f\"Condensed name: {prod.condensed_name}\")\n\n# Please be aware that EOReader will orthorectify your SAR data with SNAP\n# Be sure to have your GPT executable in your path",
"Acquisition datetime: 2020-10-08 22:40:18.446381\nCondensed name: 20201008T224018_CSK_HI_DGM\n"
],
[
"# Open here some more interesting geographical data: extent and footprint\nbase = prod.extent.plot(color='cyan', edgecolor='black')\nprod.footprint.plot(ax=base, color='blue', edgecolor='black', alpha=0.5)",
"Executing processing graph\n"
],
[
"from eoreader.bands import *\nfrom eoreader.env_vars import DEM_PATH\n\n# Set the DEM\nos.environ[DEM_PATH] = os.path.join(\"/home\", \"data\", \"DS2\", \"BASES_DE_DONNEES\", \"GLOBAL\", \"COPDEM_30m\",\n \"COPDEM_30m.vrt\")\n\n# Select the bands you want to load\nbands = [VV, HH, VV_DSPK, HH_DSPK, HILLSHADE, SLOPE]\n\n# Be sure they exist for COSMO-SkyMed sensor:\nok_bands = [band for band in bands if prod.has_band(band)]\nprint(to_str(ok_bands)) # This product does not have VV band and HILLSHADE band cannot be computed from SAR band",
"['HH', 'HH_DSPK', 'SLOPE']\n"
],
[
"# Load those bands as a dict of xarray.DataArray, with a 20m resolution\nband_dict = prod.load(ok_bands, resolution=20.)\nband_dict[HH]",
"Executing processing graph\n"
],
[
"# Plot a subsampled version\nband_dict[SLOPE][:, ::10, ::10].plot()",
"_____no_output_____"
],
[
"# You can also stack those bands\nstack = prod.stack(ok_bands)\nstack",
"_____no_output_____"
],
[
"# Plot a subsampled version\nimport matplotlib.pyplot as plt\n\nnrows = len(stack)\nfig, axes = plt.subplots(nrows=nrows, figsize=(3 * nrows, 6 * nrows), subplot_kw={\"box_aspect\": 1})\nfor i in range(nrows):\n stack[i, ::10, ::10].plot(x=\"x\", y=\"y\", ax=axes[i])",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec72143756eeb9e2a9410a3ad7fac777077d225f | 215,309 | ipynb | Jupyter Notebook | studies/perturbation_methods/PMI-ConvectiveMesoscaleDomainAssessmentExample.ipynb | lassman135/assessment | 450f16b08e182a1aee1ab5c69dfe91215ef2fcba | [
"Apache-2.0"
]
| 2 | 2019-10-25T17:10:11.000Z | 2020-03-26T17:35:56.000Z | studies/perturbation_methods/PMI-ConvectiveMesoscaleDomainAssessmentExample.ipynb | lassman135/assessment | 450f16b08e182a1aee1ab5c69dfe91215ef2fcba | [
"Apache-2.0"
]
| 49 | 2019-02-25T21:24:34.000Z | 2021-11-22T21:34:07.000Z | studies/perturbation_methods/PMI-ConvectiveMesoscaleDomainAssessmentExample.ipynb | lassman135/assessment | 450f16b08e182a1aee1ab5c69dfe91215ef2fcba | [
"Apache-2.0"
]
| 10 | 2019-02-22T19:13:36.000Z | 2021-06-04T01:49:59.000Z | 301.553221 | 191,876 | 0.909312 | [
[
[
"%matplotlib inline\nimport os, sys\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport pandas as pd\nimport xarray as xr\nfrom netCDF4 import Dataset\nimport wrf as wrfpy",
"_____no_output_____"
]
],
[
[
"#### Load in mmctools helper functions.\nThese are from the [a2e-mmc/mmctools repository](https://github.com/a2e-mmc/mmctools) (currently only in the dev branch). ",
"_____no_output_____"
]
],
[
[
"# manually add a2e-mmc repositories to PYTHONPATH if needed\n#### If you have an 'a2e-mmc' directory under your $HOME\n#module_path = os.path.join(os.environ['HOME'],'a2e-mmc')\n#if module_path not in sys.path:\n# sys.path.append(module_path)\n#### Alternatively, use a relative path to mmctools from this notebook \n#### (which lives in assessement/studies/perturbation_methods)\n#### and assumes assessment and mmctools share some parent directory\nsys.path.append('../../../')",
"_____no_output_____"
],
[
"from mmctools.helper_functions import calc_wind, theta, model4D_calcQOIs\nfrom mmctools.wrf.utils import wrfout_seriesReader",
"_____no_output_____"
]
],
[
[
"# Example: Assessment of Mesoscale Domain Results\nModel data is the convective case from perturbations method study",
"_____no_output_____"
]
],
[
[
"TTUdata = '../../datasets/SWiFT/data/TTU_tilt_corrected_20131108-09.csv'",
"_____no_output_____"
],
[
"modeldatapath = '/glade/scratch/jsauer/A2E-MMC/PMIC1/TEST_SANITY/'\n#modeldatapath = '/scratch/equon/CBL_18Z-20Z'\nmodel_file_filter = 'wrfout_d01_2013-11-08_[12][680]:00:00'\nmodelHours = [16,18,20] #commensurate with model_file_filter",
"_____no_output_____"
],
[
"# where to save outputs (e.g., figures, assessment metrics,...)\noutputdir = modeldatapath\nmodeloutputfile = 'CBL_meso_profiles_16Z-18Z'",
"_____no_output_____"
]
],
[
[
"## Load observation data\nLoad a processed TTU dataset for demonstration purposes. The dataset can be obtained by running the notebook `process_TTU_tower.ipynb` which can be found in the [a2e-mmc/assessment repository](https://github.com/a2e-mmc/assessment/tree/dev) (currently only in the dev branch)\n\n<font color='red'>TODO: use multiindex for datetime/height</font>",
"_____no_output_____"
]
],
[
[
"df = pd.read_csv(TTUdata, parse_dates=True, index_col='datetime')\ndf.head()",
"_____no_output_____"
]
],
[
[
"## Compute derived metrics (wspd, wdir, theta) from the observation data\nDo some additional data processing to obtain windspeed, wind direction and potential temperature using helper functions. ",
"_____no_output_____"
]
],
[
[
"# Calculate wind speed and direction\ndf['wspd'], df['wdir'] = calc_wind(df)\n\n# Calculate potential temperature\ndf['theta'] = theta(df['t'],df['p'])\n\ndf.head(n=10)",
"_____no_output_____"
]
],
[
[
"## Calculate 30-minute rolling means\nUse pandas intrinsics pivot, resample, stack to find the 30-minute means of the derived metrics for the SWiFT, TTU tower observations.",
"_____no_output_____"
]
],
[
[
"df30 = df.pivot(columns='height',values=['u','v','w','wspd','wdir','theta']).resample('30min').mean().stack()\ndf30.reset_index(inplace=True)\ndf30.set_index(['datetime'],inplace=True)",
"_____no_output_____"
]
],
[
[
"## Load mesoscale modeling results data ",
"_____no_output_____"
]
],
[
[
"obs_legStrs = []\nmod_legStrs = []\ndatetimeStrs = []\nfor modelHour in modelHours:\n obs_legStrs.append('Obs-{:d}Z'.format(modelHour))\n mod_legStrs.append('Mod-{:d}Z'.format(modelHour))\n datetimeStrs.append('2013-11-08 {:2d}:00:00'.format(modelHour))\nprint(obs_legStrs)\nprint(mod_legStrs)\nprint(datetimeStrs)",
"['Obs-16Z', 'Obs-18Z', 'Obs-20Z']\n['Mod-16Z', 'Mod-18Z', 'Mod-20Z']\n['2013-11-08 16:00:00', '2013-11-08 18:00:00', '2013-11-08 20:00:00']\n"
],
[
"#%time ds = wrfout_seriesReader(modeldatapath,model_file_filter,specified_heights=df30['height'].unique()) \n%time ds = wrfout_seriesReader(modeldatapath,model_file_filter) ",
"Finished opening/concatenating datasets...\nEstablishing coordinate variables, x,y,z, zSurface...\nDestaggering data variables, u,v,w...\nExtracting data variables, p,theta...\nCalculating derived data variables, wspd, wdir...\nCPU times: user 420 ms, sys: 54.4 ms, total: 475 ms\nWall time: 542 ms\n"
],
[
"%time ds=model4D_calcQOIs(ds,'ny')",
"CPU times: user 162 ms, sys: 16.9 ms, total: 179 ms\nWall time: 184 ms\n"
],
[
"fntSize=16\nplt.rcParams['xtick.labelsize']=fntSize\nplt.rcParams['ytick.labelsize']=fntSize\nnumPlotsX = 1\nnumPlotsY = 4\ncolors = []\ncolors.append([0.25,0.25,0.75])\ncolors.append([0.35,0.45,0.75])\ncolors.append([0.75,0.45,0.35])\nfig,ax = plt.subplots(numPlotsX,numPlotsY,sharey=True,sharex=False,figsize=(24,8))\n\n# plot-windspeed\nfor it in range(len(modelHours)): \n if 'datetime' in ds['z'].dims:\n zValues = ds['z'].sel(datetime=datetimeStrs[it]).mean(axis=(1,2))\n else:\n zValues = ds['z']\n # plot-windspeed\n im = ax[0].plot(df30['wspd'].loc[datetimeStrs[it]],df30['height'].loc[datetimeStrs[it]],'*--',color=colors[it],label=obs_legStrs[it])\n im = ax[0].plot(ds['UMean'].sel(datetime=datetimeStrs[it]).mean(axis=1),\n zValues,\n 'o-',color=colors[it],markerfacecolor=\"None\",label=mod_legStrs[it])\n # plot-wind_dir.\n im = ax[1].plot(df30['wdir'].loc[datetimeStrs[it]],df30['height'].loc[datetimeStrs[it]],'*--',color=colors[it],label=obs_legStrs[it])\n im = ax[1].plot(ds['UdirMean'].sel(datetime=datetimeStrs[it]).mean(axis=1),\n zValues,\n 'o-',color=colors[it],markerfacecolor=\"None\",label=mod_legStrs[it])\n # plot-theta\n im = ax[2].plot(df30['theta'].loc[datetimeStrs[it]],df30['height'].loc[datetimeStrs[it]],'*--',color=colors[it],label=obs_legStrs[it])\n im = ax[2].plot(ds['thetaMean'].sel(datetime=datetimeStrs[it]).mean(axis=1),\n zValues,\n 'o-',color=colors[it],markerfacecolor=\"None\",label=mod_legStrs[it])\n # plot-w\n im = ax[3].plot(df30['w'].loc[datetimeStrs[it]],df30['height'].loc[datetimeStrs[it]],'*--',color=colors[it],label=obs_legStrs[it])\n im = ax[3].plot(ds['wMean'].sel(datetime=datetimeStrs[it]).mean(axis=1),\n zValues,\n 'o-',color=colors[it],markerfacecolor=\"None\",label=mod_legStrs[it])\n \nax[0].set_ylim([0,205])\nax[0].set_xlim([5,12.5])\nax[1].set_xlim([180,295])\nax[2].set_xlim([292,300])\nax[3].set_xlim([-0.25,0.25])\n\nax[0].legend(fontsize=12)\nax[0].set_xlabel(r'$U$ $[\\mathrm{ms^{-1}}]$',fontsize=fntSize)\nax[0].minorticks_on()\nax[0].grid(which='both')\n\nax[1].set_xlabel(r'$dir$ $[\\mathrm{\\circ}]$',fontsize=fntSize)\nax[1].minorticks_on()\nax[1].grid(which='both')\n\nax[2].set_xlabel(r'$\\theta$ $[\\mathrm{K}]$',fontsize=fntSize)\nax[2].minorticks_on()\nax[2].grid(which='both')\n\nax[3].set_xlabel(r'$w$ $[\\mathrm{ms^{-1}}]$',fontsize=fntSize)\nax[3].minorticks_on()\nax[3].grid(which='both')",
"_____no_output_____"
]
],
[
[
"<font color='red'>TODO: simplify this section with `mmctools.plotting.plot_profile` (update plotting library to handle xarray input)</font>",
"_____no_output_____"
],
[
"## Save mesoscale modeling results data and figure ",
"_____no_output_____"
]
],
[
[
"#Save the processed model output and share with others!\nmodelprocessedname = os.path.join(outputdir,modeloutputfile) \nprint('Writing out',modelprocessedname)\nds.to_netcdf(modelprocessedname, mode='w', format='NETCDF4', unlimited_dims='Time', compute=True)\nfigurefilename = os.path.join(outputdir, 'Profiles-PMIC1_d01_test.png')\nprint('Writing out',figurefilename)\nfig.savefig(figurefilename, dpi=200, bbox_inches='tight')",
"Writing out /glade/scratch/jsauer/A2E-MMC/PMIC1/TEST_SANITY/CBL_meso_profiles_16Z-18Z\nWriting out /glade/scratch/jsauer/A2E-MMC/PMIC1/TEST_SANITY/Profiles-PMIC1_d01_test.png\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec7252128185699d1a72c4426e18453335500f9a | 51,181 | ipynb | Jupyter Notebook | p1_navigation/Report.ipynb | toebgen/deep-reinforcement-learning | c10dbe9ad521e48152ec1d58a563f03a4400b525 | [
"MIT"
]
| null | null | null | p1_navigation/Report.ipynb | toebgen/deep-reinforcement-learning | c10dbe9ad521e48152ec1d58a563f03a4400b525 | [
"MIT"
]
| 7 | 2019-12-16T22:09:00.000Z | 2022-02-10T00:17:41.000Z | p1_navigation/Report.ipynb | toebgen/deep-reinforcement-learning | c10dbe9ad521e48152ec1d58a563f03a4400b525 | [
"MIT"
]
| null | null | null | 96.02439 | 30,928 | 0.813603 | [
[
[
"# Navigation\n\n---\n\nIn this notebook, you will learn how to use the Unity ML-Agents environment for the first project of the [Deep Reinforcement Learning Nanodegree](https://www.udacity.com/course/deep-reinforcement-learning-nanodegree--nd893).\n\n### 1. Start the Environment\n\nWe begin by importing some necessary packages. If the code cell below returns an error, please revisit the project instructions to double-check that you have installed [Unity ML-Agents](https://github.com/Unity-Technologies/ml-agents/blob/master/docs/Installation.md) and [NumPy](http://www.numpy.org/).",
"_____no_output_____"
]
],
[
[
"!pip -q install ./python",
"\u001b[31mtensorflow 1.7.1 has requirement numpy>=1.13.3, but you'll have numpy 1.12.1 which is incompatible.\u001b[0m\r\n\u001b[31mipython 6.5.0 has requirement prompt-toolkit<2.0.0,>=1.0.15, but you'll have prompt-toolkit 2.0.10 which is incompatible.\u001b[0m\r\n"
]
],
[
[
"Next, we will start the environment! **_Before running the code cell below_**, change the `file_name` parameter to match the location of the Unity environment that you downloaded.\n\n- **Mac**: `\"path/to/Banana.app\"`\n- **Windows** (x86): `\"path/to/Banana_Windows_x86/Banana.exe\"`\n- **Windows** (x86_64): `\"path/to/Banana_Windows_x86_64/Banana.exe\"`\n- **Linux** (x86): `\"path/to/Banana_Linux/Banana.x86\"`\n- **Linux** (x86_64): `\"path/to/Banana_Linux/Banana.x86_64\"`\n- **Linux** (x86, headless): `\"path/to/Banana_Linux_NoVis/Banana.x86\"`\n- **Linux** (x86_64, headless): `\"path/to/Banana_Linux_NoVis/Banana.x86_64\"`\n\nFor instance, if you are using a Mac, then you downloaded `Banana.app`. If this file is in the same folder as the notebook, then the line below should appear as follows:\n```\nenv = UnityEnvironment(file_name=\"Banana.app\")\n```",
"_____no_output_____"
]
],
[
[
"from unityagents import UnityEnvironment\nimport numpy as np\n\n# please do not modify the line below\nenv = UnityEnvironment(file_name=\"/data/Banana_Linux_NoVis/Banana.x86_64\")",
"INFO:unityagents:\n'Academy' started successfully!\nUnity Academy name: Academy\n Number of Brains: 1\n Number of External Brains : 1\n Lesson number : 0\n Reset Parameters :\n\t\t\nUnity brain name: BananaBrain\n Number of Visual Observations (per agent): 0\n Vector Observation space type: continuous\n Vector Observation space size (per agent): 37\n Number of stacked Vector Observation: 1\n Vector Action space type: discrete\n Vector Action space size (per agent): 4\n Vector Action descriptions: , , , \n"
]
],
[
[
"Environments contain **_brains_** which are responsible for deciding the actions of their associated agents. Here we check for the first brain available, and set it as the default brain we will be controlling from Python.",
"_____no_output_____"
]
],
[
[
"# get the default brain\nbrain_name = env.brain_names[0]\nbrain = env.brains[brain_name]",
"_____no_output_____"
]
],
[
[
"### 2. Examine the State and Action Spaces\n\nRun the code cell below to print some information about the environment.",
"_____no_output_____"
]
],
[
[
"# reset the environment\nenv_info = env.reset(train_mode=True)[brain_name]\n\n# number of agents in the environment\nprint('Number of agents:', len(env_info.agents))\n\n# number of actions\naction_size = brain.vector_action_space_size\nprint('Number of actions:', action_size)\n\n# examine the state space \nstate = env_info.vector_observations[0]\nprint('States look like:\\n', state)\nstate_size = len(state)\nprint('States have length:', state_size)",
"Number of agents: 1\nNumber of actions: 4\nStates look like:\n [ 1. 0. 0. 0. 0.84408134 0. 0.\n 1. 0. 0.0748472 0. 1. 0. 0.\n 0.25755 1. 0. 0. 0. 0.74177343\n 0. 1. 0. 0. 0.25854847 0. 0.\n 1. 0. 0.09355672 0. 1. 0. 0.\n 0.31969345 0. 0. ]\nStates have length: 37\n"
]
],
[
[
"### 3. Take Random Actions in the Environment\n\nIn the next code cell, you will learn how to use the Python API to control the agent and receive feedback from the environment.\n\nNote that **in this coding environment, you will not be able to watch the agent while it is training**, and you should set `train_mode=True` to restart the environment.",
"_____no_output_____"
]
],
[
[
"env_info = env.reset(train_mode=True)[brain_name] # reset the environment\nstate = env_info.vector_observations[0] # get the current state\nscore = 0 # initialize the score\nwhile True:\n action = np.random.randint(action_size) # select an action\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = env_info.local_done[0] # see if episode has finished\n score += reward # update the score\n state = next_state # roll over the state to next time step\n if done: # exit loop if episode finished\n break\n \nprint(\"Score: {}\".format(score))",
"Score: 0.0\n"
]
],
[
[
"When finished, you can close the environment.",
"_____no_output_____"
]
],
[
[
"#env.close()",
"_____no_output_____"
]
],
[
[
"### 4. It's Your Turn!\n\nNow it's your turn to train your own agent to solve the environment! A few **important notes**:\n- When training the environment, set `train_mode=True`, so that the line for resetting the environment looks like the following:\n```python\nenv_info = env.reset(train_mode=True)[brain_name]\n```\n- To structure your work, you're welcome to work directly in this Jupyter notebook, or you might like to start over with a new file! You can see the list of files in the workspace by clicking on **_Jupyter_** in the top left corner of the notebook.\n- In this coding environment, you will not be able to watch the agent while it is training. However, **_after training the agent_**, you can download the saved model weights to watch the agent on your own machine! ",
"_____no_output_____"
],
[
"### 5. Submission\n\n> NOTE: Following code is heavily based on `Lunar Lander` exercise from class.\n\nImport all necessary libraries, and especially import the classes `Agent` and `QNetwork`, which contain the main parts of code for the DQN algorithm implementation.",
"_____no_output_____"
]
],
[
[
"import random\nimport torch\nimport numpy as np\nimport matplotlib.pyplot as plt\nfrom collections import deque\nimport pickle\n\n# Use the 2 files dqn_agent.py and model.py\nfrom dqn_agent import Agent",
"_____no_output_____"
]
],
[
[
"#### Deep Q-Network algorithm\n\nThe Deep Q-Learning (DQN) algorithm is a technique to learn the action value function $Q(s,a)$.\nIt is basically an online version of a neural fitted-value iteration [paper](https://link.springer.com/chapter/10.1007/11564096_32) of 2005 (Riedmiller):\nTraining of a Q-value function represented by a Multi-Layered Perceptron.\nIn sloppy terms, this function can be referred to as the *Cheat Sheet Q*, which tells the agent which action $a$ will yield the highest reward $R$ in a given situation $s$.\n\nIn *Q-Learning* (which is the pioneer technique for DQN) the $Q$-table is supposed to provide actions for the whole state space $S$ and action space $A$, i.e. it will ultimately know the value of each possible action in each possible situation.\nThis can easily become too large in terms of memory and computational requirements.\nThis is where *Deep Q-Learning* comes in:\nInstead of trying to learn the complete $Q$-table, a deep network is used to approximate $Q(s,a)$.\n\nThe useful additions of the Deep Q-Network compared to plain Q-Learning algorithm are:\n- Use of rolling history of past data: *Experience Replay*\n- Use of a target network to represent the old Q-Function, which is used to calculate the loss of every action during training: *Fixed Q-Targets*\n\nThis helps smoothing out the behaviors, avoiding oscillations, and prevents the value estimates to get out of control.\nOverall, this allows RL-agents to converge more reliably during training.\n\nThe algorithm can be sketched out with following steps:\n1. *Initialization* of Replay memory $D$, Action value function $\\hat{q}$ and Target action-value weights $\\vec{w}^{-} \\leftarrow \\, \\vec{w}$\n1. *Sample* an action using an $\\epsilon$-greedy policy, observe reward and next state, and store the new experience tuple in replay memory\n1. *Learn* (i.e. update the weights) periodically after several experiences (minibatches), not after every new experience\n1. Repeat 2. and 3. for number of episodes\n",
"_____no_output_____"
],
[
"#### Agent\n\nThe class `Agent` is taken from class exercise code.\nIt provides necessary methods to interact with the environment, following the Deep Q-Learning algorithm with Fixed Q-Targets and Experience Replay extensions (as discussed in class).\n\nThe class most importantly holds a `ReplayBuffer` with 1000 entries to store experience tuples, that also supports random sampling, and two `QNetwork`s, in order to decouple targets from network parameters.\n\nFurther algorithm parameters are:\n- Minibatch size: `64`\n- Discount factor (gamma): `0.99`\n- Softy update factor of target parameters (tau): `0.003`\n- Learning rate (alpha): `0.0005`\n- Network update every 4 steps\n\n#### Model\n\nThe class `QNetwork` implements the model, using PyTorch.\n3 fully connected layers map the 37 inputs onto 64 hidden, another 64 hidden, and finally 4 output units.\nThe state vector is thereby mapped onto action values for all available agent actions.\nReLU activation functions are used on the first two layers.",
"_____no_output_____"
]
],
[
[
"def dqn(n_episodes=2000, max_t=1000, eps_start=1.0, eps_end=0.01, eps_decay=0.995, stop_score=13.0):\n \"\"\"Deep Q-Learning.\n \n Params\n ======\n n_episodes (int): maximum number of training episodes\n max_t (int): maximum number of timesteps per episode\n eps_start (float): starting value of epsilon, for epsilon-greedy action selection\n eps_end (float): minimum value of epsilon\n eps_decay (float): multiplicative factor (per episode) for decreasing epsilon\n \"\"\"\n scores = [] # list containing scores from each episode\n scores_window = deque(maxlen=100) # last 100 scores\n eps = eps_start # initialize epsilon\n \n for i_episode in range(1, n_episodes+1):\n #state = env.reset()\n env_info = env.reset(train_mode=True)[brain_name] # reset the environment\n state = env_info.vector_observations[0] # get the current state\n \n score = 0\n \n for t in range(max_t):\n action = agent.act(state, eps)\n \n #next_state, reward, done, _ = env.step(action)\n env_info = env.step(action)[brain_name] # send the action to the environment\n next_state = env_info.vector_observations[0] # get the next state\n reward = env_info.rewards[0] # get the reward\n done = env_info.local_done[0] # see if episode has finished\n \n agent.step(state, action, reward, next_state, done)\n state = next_state\n score += reward\n \n if done:\n break \n \n scores_window.append(score) # save most recent score\n scores.append(score) # save most recent score\n eps = max(eps_end, eps_decay*eps) # decrease epsilon\n \n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)), end=\"\")\n if i_episode % 100 == 0:\n print('\\rEpisode {}\\tAverage Score: {:.2f}'.format(i_episode, np.mean(scores_window)))\n if np.mean(scores_window)>=stop_score:\n print('\\nEnvironment solved in {:d} episodes!\\tAverage Score: {:.2f}'.format(i_episode-100, np.mean(scores_window)))\n\n torch.save(agent.qnetwork_local.state_dict(), 'model.pt')\n with open('scores.pkl', 'wb') as f:\n pickle.dump(scores, f)\n \n break\n \n return scores",
"_____no_output_____"
],
[
"agent = Agent(state_size=state_size, action_size=action_size, seed=0)\n%time scores = dqn()",
"Episode 100\tAverage Score: 1.03\nEpisode 200\tAverage Score: 3.72\nEpisode 300\tAverage Score: 7.43\nEpisode 400\tAverage Score: 10.67\nEpisode 500\tAverage Score: 12.56\nEpisode 564\tAverage Score: 13.03\nEnvironment solved in 464 episodes!\tAverage Score: 13.03\nCPU times: user 4min 4s, sys: 20.5 s, total: 4min 25s\nWall time: 6min 18s\n"
],
[
"# plot the scores\nfig = plt.figure()\nax = fig.add_subplot(111)\nplt.plot(np.arange(len(scores)), scores)\nplt.ylabel('Score')\nplt.xlabel('Episode #')\nplt.show()\n\n# with open('scores.pkl', 'rb') as f:\n# scores_from_file = pickle.load(f)",
"_____no_output_____"
],
[
"agent = Agent(state_size=state_size, action_size=action_size, seed=0)\n%time scores = dqn(stop_score=20.)",
"Episode 100\tAverage Score: 0.75\nEpisode 200\tAverage Score: 3.12\nEpisode 300\tAverage Score: 6.96\nEpisode 400\tAverage Score: 9.73\nEpisode 500\tAverage Score: 11.96\nEpisode 600\tAverage Score: 11.88\nEpisode 700\tAverage Score: 13.37\nEpisode 800\tAverage Score: 15.31\nEpisode 900\tAverage Score: 14.19\nEpisode 1000\tAverage Score: 13.61\nEpisode 1100\tAverage Score: 13.52\nEpisode 1200\tAverage Score: 13.80\nEpisode 1300\tAverage Score: 14.84\nEpisode 1400\tAverage Score: 13.48\nEpisode 1500\tAverage Score: 14.80\nEpisode 1600\tAverage Score: 14.45\nEpisode 1700\tAverage Score: 14.84\nEpisode 1800\tAverage Score: 13.69\nEpisode 1900\tAverage Score: 14.23\nEpisode 2000\tAverage Score: 15.94\nCPU times: user 14min 53s, sys: 1min 14s, total: 16min 8s\nWall time: 23min 2s\n"
],
[
"env.close()",
"_____no_output_____"
]
],
[
[
"#### Future work\n\nThe current algorithm achieves satisfactory scores of +13 after less than 500 learning episodes.\nIt achieves almost +16 after 2000 episodes.\nNevertheless, with further improvements the algorithm might perform even better.\n\nPossible ideas for future work are:\n- Investigate the model architecture in more depth, find a more optimal neural network.\n- Use an hyperparameter optimization library such as `Scikit-Optimize` or `Optuna`, in order to figure out the optimal hyperparameters for learning.\n- Use different optimizations described in the DQN literature, e.g.:\n - Prioritized Experience Replay\n - Distributional DQN\n - Noisy DQN",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec7261c90a2f6273d1d093394c204496a07fa1f1 | 17,296 | ipynb | Jupyter Notebook | self-paced-labs/tensorflow-2.x/synthetic_features_and_outliers.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
]
| 6,140 | 2016-05-23T16:09:35.000Z | 2022-03-30T19:00:46.000Z | self-paced-labs/tensorflow-2.x/synthetic_features_and_outliers.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
]
| 1,384 | 2016-07-08T22:26:41.000Z | 2022-03-24T16:39:43.000Z | self-paced-labs/tensorflow-2.x/synthetic_features_and_outliers.ipynb | KayvanShah1/training-data-analyst | 3f778a57b8e6d2446af40ca6063b2fd9c1b4bc88 | [
"Apache-2.0"
]
| 5,110 | 2016-05-27T13:45:18.000Z | 2022-03-31T18:40:42.000Z | 29.365025 | 247 | 0.57019 | [
[
[
"# Synthetic Features and Outliers",
"_____no_output_____"
],
[
"**Learning Objectives:**\n * Create a synthetic feature that is the ratio of two other features\n * Use this new feature as an input to a linear regression model\n * Improve the effectiveness of the model by identifying and clipping (removing) outliers out of the input data",
"_____no_output_____"
],
[
"## Setup",
"_____no_output_____"
],
[
"Install latest `2.x.x` release for tensorflow",
"_____no_output_____"
]
],
[
[
"!pip install tensorflow==2.0.0-beta1",
"_____no_output_____"
],
[
"from matplotlib import cm\nfrom matplotlib import gridspec\nfrom matplotlib import pyplot as plt\nimport numpy as np\nimport pandas as pd\nimport logging\nfrom packaging import version\nfrom IPython.display import display",
"_____no_output_____"
],
[
"pd.options.display.max_rows = 10\npd.options.display.float_format = '{:.1f}'.format\nlogging.getLogger('tensorflow').disabled = True",
"_____no_output_____"
],
[
"import tensorflow as tf\n\n%load_ext tensorboard",
"_____no_output_____"
]
],
[
[
"First, we'll import the California housing data into a *pandas* `DataFrame`:",
"_____no_output_____"
]
],
[
[
"from datetime import datetime\nimport io\nlogging.getLogger('tensorboard').disabled = True\n\ncalifornia_housing_dataframe = pd.read_csv(\"https://download.mlcc.google.com/mledu-datasets/california_housing_train.csv\", sep=\",\")\n\ncalifornia_housing_dataframe = california_housing_dataframe.reindex(\n np.random.permutation(california_housing_dataframe.index))\ncalifornia_housing_dataframe[\"median_house_value\"] /= 1000.0\ncalifornia_housing_dataframe",
"_____no_output_____"
]
],
[
[
"We'll set up our `plot_to_image` function to convert the matplotlib plot specified by figure to a PNG image",
"_____no_output_____"
]
],
[
[
"def plot_to_image(figure):\n \"\"\"Converts the matplotlib plot specified by 'figure' to a PNG image and\n returns it. The supplied figure is closed and inaccessible after this call.\"\"\"\n # Save the plot to a PNG in memory.\n buf = io.BytesIO()\n plt.savefig(buf, format='png')\n # Closing the figure prevents it from being displayed directly inside\n # the notebook.\n plt.close(figure)\n buf.seek(0)\n # Convert PNG buffer to TF image\n image = tf.image.decode_png(buf.getvalue(), channels=4)\n # Add the batch dimension\n image = tf.expand_dims(image, 0)\n return image",
"_____no_output_____"
]
],
[
[
"Next, we'll define the function for model training",
"_____no_output_____"
]
],
[
[
"def fit_model(learning_rate,\n steps_per_epoch,\n batch_size,\n input_feature):\n \"\"\"Trains a linear regression model of one feature.\n \n Args:\n learning_rate: A `float`, the learning rate.\n steps_per_epoch: A non-zero `int`, the total number of training steps. A training step\n consists of a forward and backward pass using a single batch.\n batch_size: A non-zero `int`, the batch size.\n input_feature: A `string` specifying a column from `california_housing_dataframe`\n to use as input feature.\n Returns:\n A Pandas `DataFrame` containing targets and the corresponding predictions done\n after training the model.\n \"\"\"\n \n epochs = 10\n features = california_housing_dataframe[[input_feature]].values\n label = \"median_house_value\"\n labels = california_housing_dataframe[label].values\n\n model = tf.keras.models.Sequential([\n tf.keras.layers.Dense(1, activation='linear', kernel_initializer='zeros')\n ])\n model.compile(optimizer=tf.keras.optimizers.SGD(learning_rate=learning_rate, clipnorm=5.0),\n loss='mse',\n metrics=[tf.keras.metrics.RootMeanSquaredError()])\n \n sample = california_housing_dataframe.sample(n=300)\n logdir = \"logs/synthetic_features_and_outliers/plots\" + datetime.now().strftime(\"%Y%m%d-%H%M%S\")\n scalars_logdir = \"logs/synthetic_features_and_outliers/scalars\" + datetime.now().strftime(\"%Y%m%d-%H%M%S\")\n file_writer = tf.summary.create_file_writer(logdir)\n \n # Set up to plot the state of our model's line each epoch.\n def create_plt_params(feature, label, epochs=10):\n colors = [cm.coolwarm(x) for x in np.linspace(-1, 1, epochs)]\n return (colors,\n (sample[feature].min(), sample[feature].max()),\n (0, sample[label].max()))\n \n def create_figure(feature, label, epochs=10):\n figure = plt.figure(figsize=(15, 6))\n plt.title(\"Learned Line by Epoch\")\n plt.ylabel(label)\n plt.xlabel(feature)\n plt.scatter(sample[feature], sample[label])\n return figure\n\n colors, x_min_max, y_min_max = create_plt_params(input_feature, label, epochs)\n\n def log(epoch, logs):\n root_mean_squared_error = logs[\"root_mean_squared_error\"]\n print(\" epoch %02d : %0.2f\" % (epoch, root_mean_squared_error))\n\n weight, bias = [x.flatten()[0] for x in model.layers[0].get_weights()]\n\n # Apply some math to ensure that the data and line are plotted neatly.\n y_extents = np.array(y_min_max)\n x_extents = (y_extents - bias) / weight\n x_extents = np.maximum(np.minimum(x_extents,\n x_min_max[1]),\n x_min_max[0])\n y_extents = weight * x_extents + bias\n figure = create_figure(input_feature, label, epochs)\n plt.plot(x_extents, y_extents, color=colors[epoch]) \n with file_writer.as_default():\n tf.summary.image(\"Learned Line by Epoch\",\n plot_to_image(figure),\n step=epoch)\n \n model_callback = tf.keras.callbacks.LambdaCallback(\n on_epoch_end=lambda epoch, logs: log(epoch, logs))\n tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=scalars_logdir,\n update_freq='epoch')\n \n print(\"Train model...\")\n print(\"RMSE (on training data):\")\n history = model.fit(features,\n labels,\n epochs=epochs,\n steps_per_epoch=steps_per_epoch,\n batch_size=batch_size,\n callbacks=[model_callback, tensorboard_callback],\n verbose=0).history\n print(\"Model training finished.\")\n\n calibration_data = pd.DataFrame()\n calibration_data[\"predictions\"] = model.predict_on_batch(features).flatten()\n calibration_data[\"targets\"] = pd.Series(labels)\n display(calibration_data.describe())\n root_mean_squared_error = history[\"root_mean_squared_error\"][9]\n print(\"Final RMSE (on training data): %0.2f\" % root_mean_squared_error)\n \n return calibration_data",
"_____no_output_____"
]
],
[
[
"## Task 1: Try a Synthetic Feature\n\nBoth the `total_rooms` and `population` features count totals for a given city block.\n\nBut what if one city block were more densely populated than another? We can explore how block density relates to median house value by creating a synthetic feature that's a ratio of `total_rooms` and `population`.",
"_____no_output_____"
]
],
[
[
"!rm -rf logs/synthetic_features_and_outliers",
"_____no_output_____"
]
],
[
[
"In the cell below, create a feature called `rooms_per_person`, and use that as the `input_feature` to `fit_model()`.",
"_____no_output_____"
]
],
[
[
"#\n# YOUR CODE HERE\n#\ncalifornia_housing_dataframe[\"rooms_per_person\"] = \n\ncalibration_data = fit_model(\n learning_rate=0.00005,\n steps_per_epoch=500,\n batch_size=5,\n input_feature=\"rooms_per_person\"\n)",
"_____no_output_____"
]
],
[
[
"What's the best performance you can get with this single feature by tweaking the learning rate? (The better the performance, the better your regression line should fit the data, and the lower\nthe final RMSE should be.)",
"_____no_output_____"
]
],
[
[
"from google.datalab.ml import TensorBoard\nTensorBoard().start('logs/synthetic_features_and_outliers')",
"_____no_output_____"
]
],
[
[
"## Task 2: Identify Outliers\n\nWe can visualize the performance of our model by creating a scatter plot of predictions vs. target values. Ideally, these would lie on a perfectly correlated diagonal line.\n\nUse Pyplot's [`scatter()`](https://matplotlib.org/gallery/shapes_and_collections/scatter.html) to create a scatter plot of predictions vs. targets, using the rooms-per-person model you trained in Task 1.\n\nDo you see any oddities? Trace these back to the source data by looking at the distribution of values in `rooms_per_person`.",
"_____no_output_____"
]
],
[
[
"logdir = \"logs/synthetic_features_and_outliers/plots\"\nfile_writer = tf.summary.create_file_writer(logdir + datetime.now().strftime(\"%Y%m%d-%H%M%S\"))",
"_____no_output_____"
]
],
[
[
"__#TODO:__ Plot a scatter graph to show the scatter points. ",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE",
"_____no_output_____"
]
],
[
[
"The calibration data shows most scatter points aligned to a line. The line is almost vertical, but we'll come back to that later. Right now let's focus on the ones that deviate from the line. We notice that they are relatively few in number.",
"_____no_output_____"
],
[
"If we plot a histogram of `rooms_per_person`, we find that we have a few outliers in our input data:",
"_____no_output_____"
]
],
[
[
"figure = plt.figure()\nplt.subplot(1, 2, 2)\n_ = california_housing_dataframe[\"rooms_per_person\"].hist()\nwith file_writer.as_default():\n tf.summary.image(\"Rooms per person\",\n plot_to_image(figure),\n step=0)",
"_____no_output_____"
],
[
"TensorBoard().start('logs/synthetic_features_and_outliers')",
"_____no_output_____"
]
],
[
[
"## Task 3: Clip Outliers\n\nSee if you can further improve the model fit by setting the outlier values of `rooms_per_person` to some reasonable minimum or maximum.\n\nFor reference, here's a quick example of how to apply a function to a Pandas `Series`:\n\n clipped_feature = my_dataframe[\"my_feature_name\"].apply(lambda x: max(x, 0))\n\nThe above `clipped_feature` will have no values less than `0`.",
"_____no_output_____"
],
[
"The histogram we created in Task 2 shows that the majority of values are less than `5`.",
"_____no_output_____"
],
[
"__#TODO:__ Let's clip rooms_per_person to 5, and plot a histogram to double-check the results.",
"_____no_output_____"
]
],
[
[
"# YOUR CODE HERE",
"_____no_output_____"
],
[
"TensorBoard().start('logs/synthetic_features_and_outliers')",
"_____no_output_____"
]
],
[
[
"To verify that clipping worked, let's train again and print the calibration data once more:",
"_____no_output_____"
]
],
[
[
"calibration_data = fit_model(\n learning_rate=0.05,\n steps_per_epoch=1000,\n batch_size=5,\n input_feature=\"rooms_per_person\")",
"_____no_output_____"
],
[
"file_writer = tf.summary.create_file_writer(logdir + datetime.now().strftime(\"%Y%m%d-%H%M%S\"))\nfigure = plt.figure()\n_ = plt.scatter(calibration_data[\"predictions\"], calibration_data[\"targets\"])\nwith file_writer.as_default():\n tf.summary.image(\"Predictions vs Targets\",\n plot_to_image(figure),\n step=0)",
"_____no_output_____"
],
[
"TensorBoard().start('logs/synthetic_features_and_outliers')",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec72780db15270fbb3024e022938a66be091f946 | 138,178 | ipynb | Jupyter Notebook | Section-06-Categorical-Encoding/06.07-Probability-Ratio-Encoding.ipynb | mpofukelvintafadzwa/feature-engineering-for-machine-learning | 3b4e778f5d6cda2dccff3961b26009a2ea7c5486 | [
"BSD-3-Clause"
]
| 2 | 2021-05-28T11:03:22.000Z | 2021-05-28T11:03:29.000Z | Section-06-Categorical-Encoding/06.07-Probability-Ratio-Encoding.ipynb | SudheeshSukumar/feature-engineering-for-machine-learning | 8b592d99ee879f1d0a368326661051574dce19a7 | [
"BSD-3-Clause"
]
| null | null | null | Section-06-Categorical-Encoding/06.07-Probability-Ratio-Encoding.ipynb | SudheeshSukumar/feature-engineering-for-machine-learning | 8b592d99ee879f1d0a368326661051574dce19a7 | [
"BSD-3-Clause"
]
| 1 | 2021-07-06T09:42:14.000Z | 2021-07-06T09:42:14.000Z | 104.521936 | 18,928 | 0.834735 | [
[
[
"## Target guided encodings\n\nIn the previous lectures in this section, we learned how to convert a label into a number, by using one hot encoding, replacing by a digit or replacing by frequency or counts of observations. These methods are simple, make (almost) no assumptions and work generally well in different scenarios.\n\nThere are however methods that allow us to capture information while pre-processing the labels of categorical variables. These methods include:\n\n- Ordering the labels according to the target\n- Replacing labels by the target mean (mean encoding / target encoding)\n- Replacing the labels by the probability ratio of the target being 1 or 0\n- Weight of evidence.\n\nAll of the above methods have something in common:\n\n- the encoding is **guided by the target**, and\n- they create a **monotonic relationship** between the variable and the target.\n\n\n### Monotonicity\n\nA monotonic relationship is a relationship that does one of the following:\n\n- (1) as the value of one variable increases, so does the value of the other variable; or\n- (2) as the value of one variable increases, the value of the other variable decreases.\n\nIn this case, as the value of the independent variable (predictor) increases, so does the target, or conversely, as the value of the variable increases, the target value decreases.\n\n\n\n### Advantages of target guided encodings\n\n- Capture information within the category, therefore creating more predictive features\n- Create a monotonic relationship between the variable and the target, therefore suitable for linear models\n- Do not expand the feature space\n\n\n### Limitations\n\n- Prone to cause over-fitting\n- Difficult to cross-validate with current libraries\n\n\n### Note\n\nThe methods discussed in this and the coming 3 lectures can be also used on numerical variables, after discretisation. This creates a monotonic relationship between the numerical variable and the target, and therefore improves the performance of linear models. I will discuss this in more detail in the section \"Discretisation\".\n\n===============================================================================\n\n## Probability Ratio Encoding\n\nThese encoding is suitable for classification problems only, where the target is binary.\n\nFor each category, we calculate the mean of target=1, that is the probability of the target being 1 ( P(1) ), and the probability of the target=0 ( P(0) ). And then, we calculate the ratio P(1)/P(0), and replace the categories by that ratio.\n\n\n## In this demo:\n\nWe will see how to perform one hot encoding with:\n- pandas\n- Feature-Engine\n\nAnd the advantages and limitations of each implementation using the Titanic dataset.",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\n\nimport matplotlib.pyplot as plt\n\n# to split the datasets\nfrom sklearn.model_selection import train_test_split\n\n# for encoding with feature-engine\nfrom feature_engine.encoding import PRatioEncoder",
"_____no_output_____"
],
[
"# load dataset\n\ndata = pd.read_csv(\n '../titanic.csv',\n usecols=['cabin', 'sex', 'embarked', 'survived'])\n\ndata.head()",
"_____no_output_____"
],
[
"# let's remove obserrvations with na in embarked\n\ndata.dropna(subset=['embarked'], inplace=True)\ndata.shape",
"_____no_output_____"
],
[
"# Now we extract the first letter of the cabin\n# to create a simpler variable for the demo\n\ndata['cabin'] = data['cabin'].astype(str).str[0]",
"_____no_output_____"
],
[
"# and we remove the observations where cabin = T\n# because they are too few\n\ndata = data[data['cabin']!= 'T']\ndata.shape",
"_____no_output_____"
],
[
"# let's have a look at how many labels each variable has\n\nfor col in data.columns:\n print(col, ': ', len(data[col].unique()), ' labels')",
"survived : 2 labels\nsex : 2 labels\ncabin : 8 labels\nembarked : 3 labels\n"
],
[
"# let's explore the unique categories\ndata['cabin'].unique()",
"_____no_output_____"
],
[
"data['sex'].unique()",
"_____no_output_____"
],
[
"data['embarked'].unique()",
"_____no_output_____"
]
],
[
[
"### Encoding important\n\nWe calculate the ratio P(1)/P(0) using the train set, and then use those mappings in the test set.\n\nNote that to implement this in pandas, we need to keep the target in the training set.",
"_____no_output_____"
]
],
[
[
"# let's separate into training and testing set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[['cabin', 'sex', 'embarked', 'survived']], # this time we keep the target!!\n data['survived'], # target\n test_size=0.3, # percentage of obs in test set\n random_state=0) # seed to ensure reproducibility\n\nX_train.shape, X_test.shape",
"_____no_output_____"
]
],
[
[
"### Explore original relationship between categorical variables and target",
"_____no_output_____"
]
],
[
[
"# let's explore the relationship of the categories with the target\n\nfor var in ['cabin', 'sex', 'embarked']:\n \n fig = plt.figure()\n fig = X_train.groupby([var])['survived'].mean().plot()\n fig.set_title('Relationship between {} and Survival'.format(var))\n fig.set_ylabel('Mean Survival')\n plt.show()",
"_____no_output_____"
]
],
[
[
"You can see that the relationship between the target and cabin and embarked goes up and down, depending on the category.\n\n\n## Probability ratio encoding with pandas\n\n\n### Advantages\n\n- quick\n- returns pandas dataframe\n\n### Limitations of pandas:\n\n- it does not preserve information from train data to propagate to test data",
"_____no_output_____"
]
],
[
[
"# let's calculate the probability of survived = 1 per category\n\nprob_df = X_train.groupby(['cabin'])['survived'].mean()\n\n# and capture it into a dataframe\nprob_df = pd.DataFrame(prob_df)\nprob_df",
"_____no_output_____"
],
[
"# and now the probability of survived = 0\n\nprob_df['died'] = 1 - prob_df['survived']\n\nprob_df",
"_____no_output_____"
],
[
"# and now the ratio\n\nprob_df['ratio'] = prob_df['survived'] / prob_df['died']\n\nprob_df",
"_____no_output_____"
],
[
"# and now let's capture the ratio in a dictionary\n\nordered_labels = prob_df['ratio'].to_dict()\n\nordered_labels",
"_____no_output_____"
],
[
"# now, we replace the labels with the ratios\n\nX_train['cabin'] = X_train['cabin'].map(ordered_labels)\nX_test['cabin'] = X_test['cabin'].map(ordered_labels)",
"_____no_output_____"
],
[
"# let's explore the result\n\nX_train['cabin'].head(10)",
"_____no_output_____"
],
[
"# we can turn the previous commands into 2 functions\n\n\ndef find_category_mappings(df, variable, target):\n\n tmp = pd.DataFrame(df.groupby([variable])[target].mean())\n \n tmp['non-target'] = 1 - tmp[target]\n \n tmp['ratio'] = tmp[target] / tmp['non-target']\n\n return tmp['ratio'].to_dict()\n\n\ndef integer_encode(train, test, variable, ordinal_mapping):\n\n train[variable] = train[variable].map(ordinal_mapping)\n test[variable] = test[variable].map(ordinal_mapping)",
"_____no_output_____"
],
[
"# and now we run a loop over the remaining categorical variables\n\nfor variable in ['sex', 'embarked']:\n \n mappings = find_category_mappings(X_train, variable, 'survived')\n \n integer_encode(X_train, X_test, variable, mappings)",
"_____no_output_____"
],
[
"# let's see the result\n\nX_train.head()",
"_____no_output_____"
],
[
"# let's inspect the newly created monotonic relationship\n# between the categorical variables and the target\n\nfor var in ['cabin', 'sex', 'embarked']:\n \n fig = plt.figure()\n fig = X_train.groupby([var])['survived'].mean().plot()\n fig.set_title('Monotonic relationship between {} and Survival'.format(var))\n fig.set_ylabel('Mean Survived')\n plt.show()",
"_____no_output_____"
]
],
[
[
"Note the monotonic relationships between the mean target and the categories.\n\n### Note\n\nReplacing categorical labels with this code and method will generate missing values for categories present in the test set that were not seen in the training set. Therefore it is extremely important to handle rare labels before-hand. I will explain how to do this, in a later notebook.\n\n**In addition, it will create NA or Inf if the probability of target = 0 is zero, as the division by zero is not defined.**",
"_____no_output_____"
],
[
"## Probability Ratio Encoding with Feature-Engine\n\nIf using Feature-Engine, instead of pandas, we do not need to keep the target variable in the training dataset.",
"_____no_output_____"
]
],
[
[
"# let's separate into training and testing set\n\nX_train, X_test, y_train, y_test = train_test_split(\n data[['cabin', 'sex', 'embarked']], # predictors\n data['survived'], # target\n test_size=0.3, # percentage of obs in test set\n random_state=0) # seed to ensure reproducibility\n\nX_train.shape, X_test.shape",
"_____no_output_____"
],
[
"ratio_enc = PRatioEncoder(\n encoding_method = 'ratio',\n variables=['cabin', 'sex', 'embarked'])",
"_____no_output_____"
],
[
"# when fitting the transformer, we need to pass the target as well\n# just like with any Scikit-learn predictor class\n\nratio_enc.fit(X_train, y_train)",
"_____no_output_____"
],
[
"# in the encoder dict we see the P(1)/P(0) for each\n# category for each of the indicated variables\n\nratio_enc.encoder_dict_",
"_____no_output_____"
],
[
"# this is the list of variables that the encoder will transform\n\nratio_enc.variables",
"_____no_output_____"
],
[
"X_train = ratio_enc.transform(X_train)\nX_test = ratio_enc.transform(X_test)\n\n# let's explore the result\nX_train.head()",
"_____no_output_____"
]
],
[
[
"**Note**\n\nIf the argument variables is left to None, then the encoder will automatically identify all categorical variables. Is that not sweet?\n\nThe encoder will not encode numerical variables. So if some of your numerical variables are in fact categories, you will need to re-cast them as object before using the encoder.\n\nIf there is a label in the test set that was not present in the train set, the encoder will through and error, to alert you of this behaviour.\n\nFinally, if the probability of target = 0 is zero for any category, the encoder will raise an error as the division by zero is not defined.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec727993e4fb183e1df44a3248df1644e4a14876 | 105,096 | ipynb | Jupyter Notebook | notebooks/state_mexico.ipynb | juancruz-py/covid-19-visualization | 4ca3089b14700066259f38fe14757f80bfef132c | [
"MIT"
]
| null | null | null | notebooks/state_mexico.ipynb | juancruz-py/covid-19-visualization | 4ca3089b14700066259f38fe14757f80bfef132c | [
"MIT"
]
| null | null | null | notebooks/state_mexico.ipynb | juancruz-py/covid-19-visualization | 4ca3089b14700066259f38fe14757f80bfef132c | [
"MIT"
]
| null | null | null | 98.68169 | 71,840 | 0.754168 | [
[
[
"import pandas as pd\nimport geopandas as gpd\nimport matplotlib.pyplot as plt\nimport io\nimport PIL\n",
"_____no_output_____"
],
[
"# Reading in the csv data\ndata = pd.read_csv('/Users/admin/Documents/Summer Research/sid-project/data/Casos_Diarios_Estado_Nacional_Confirmados_2020081.csv',\nencoding='latin-1')\n\n",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data=data.groupby('cv_ent').sum()\n",
"_____no_output_____"
],
[
"data=data.drop(columns=['poblacion'])\n",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"# Read Mexico map sh\n\nstates = gpd.read_file(r'/Users/admin/Documents/Summer Research/sid-project/Maps/mexico_states/destdv250k_2gw.shp')",
"_____no_output_____"
],
[
"states.head()",
"_____no_output_____"
],
[
"states['NUM_EDO']=states['NUM_EDO'].astype('int64')",
"_____no_output_____"
],
[
"states=states[['NUM_EDO','geometry']]\n",
"_____no_output_____"
],
[
"states.head()",
"_____no_output_____"
],
[
"merge = states.join(data,on='NUM_EDO',how='right')\nmerge.head()",
"_____no_output_____"
],
[
"image_frames = []\n\n\nfor dates in merge.columns.to_list()[210:211]:\n\n # Plot\n\n ax = merge.plot(column=dates,\n cmap='PuRd',\n figsize=(10, 10),\n legend=True,\n edgecolor='black',\n linewidth=0.4)\n\n # Add a title to the map\n ax.set_title('Total Confirmed Coronavirus Cases: ' +\n dates, fontdict={'fontsize': 20}, pad=12.5)\n \n \n \n\n\n # Removing the axes\n\n ax.set_axis_off()\n\n\n img = ax.get_figure()\n \n \n\n\n \n\n\n f = io.BytesIO()\n\n img.savefig(f, format='png', bbox_inches='tight')\n\n f.seek(0)\n\n image_frames.append(PIL.Image.open(f))\n \n ",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec72867ad79f6915cb93e4aca18fa84ef8e665c2 | 959,958 | ipynb | Jupyter Notebook | pynodal/notebooks/Pupil_apodizer_optimization.ipynb | patapisp/pyNodal | 30d0a8394d7203dc1a4aad57131bb80c2c0bf988 | [
"MIT"
]
| 1 | 2021-01-16T15:13:47.000Z | 2021-01-16T15:13:47.000Z | pynodal/notebooks/.ipynb_checkpoints/Pupil_apodizer_optimization-checkpoint.ipynb | patapisp/pyNodal | 30d0a8394d7203dc1a4aad57131bb80c2c0bf988 | [
"MIT"
]
| null | null | null | pynodal/notebooks/.ipynb_checkpoints/Pupil_apodizer_optimization-checkpoint.ipynb | patapisp/pyNodal | 30d0a8394d7203dc1a4aad57131bb80c2c0bf988 | [
"MIT"
]
| null | null | null | 160.743135 | 94,436 | 0.831257 | [
[
[
"# Optimized pupil phase mask for nodal region in focal plane",
"_____no_output_____"
]
],
[
[
"import numpy as np\nfrom astropy.io import fits\nimport matplotlib.pyplot as plt\nimport sys\nsys.path.append(\"../core/\")\nfrom pupil_masks import *\nfrom optim_functions import *\nimport os\nfrom skimage.transform import resize\nfrom matplotlib.colors import LogNorm \n\n",
"_____no_output_____"
],
[
"%matplotlib notebook",
"_____no_output_____"
],
[
"N = 4096\nx = np.arange(-N/2, N/2)\ny = np.arange(-N/2, N/2)\nX, Y = np.meshgrid(x,y)\nprint(X.shape)\nRHO, THETA = cart2pol(X,Y) \n\nlambdaOverD = 10\napRad = int(N/(2*lambdaOverD))\nprint(int(apRad))\n\n\nEP = make_Palomar_pupil(1024, apRad)\nEP = np.lib.pad(EP, (1536, 1536), 'constant', constant_values=(0,0))\n\nplot_im_LP(EP, apRad, 2)",
"(4096, 4096)\n204\n"
]
],
[
[
"## Vortex coronagraph",
"_____no_output_____"
]
],
[
[
"R0 = 0.015\n\nFP = myfft2(EP)\nFP_2 = ((1-R0)*np.exp(1j*2*THETA)*FP+R0*FP)\nLP = myifft2(FP_2)\nplot_im_LP(abs(LP)*EP, apRad, 1.2)",
"_____no_output_____"
],
[
"DI = np.abs(myfft2(LP*EP))**2\nplot_im_ld(DI/np.max(DI), 10, 10)",
"_____no_output_____"
]
],
[
[
"## Circular nodal area from 1.5 to 2.5 lD",
"_____no_output_____"
]
],
[
[
"a = 2048 + 2*lambdaOverD\n\nr = 2*lambdaOverD\n\ny,x = np.ogrid[-a:N-a, -a:N-a]\nmask = x*x + y*y <= r*r\n\n\n\nOPT_REG_FP = np.zeros((N,N))\nOPT_REG_FP[mask] =1\n#OPT_REG_FP[np.where(RHO+3*<3*lambdaOverD)] = 1\n#OPT_REG_FP[np.where(RHO<1.4*lambdaOverD)] = 0\n#OPT_REG_FP[np.where(Y<0)] = 0\n#OPT_REG_FP[np.where((Y<-20) & (Y>-35) & (X>20) & (X<35))] = 1\n#OPT_REG_FP[np.where((Y<-20) & (Y>-35) & (X<-20) & (X>-35))] = 1\nplot_im_ld(OPT_REG_FP, lambdaOverD, 10, n=3)",
"_____no_output_____"
],
[
"EPinit = np.ones((N,N))\noptEP= modified_GS_amplitude(EP, EPinit, OPT_REG_FP, 100)\n\n",
"100%|██████████| 100/100 [01:50<00:00, 1.10s/it]\n"
],
[
"optPupil = EP*np.abs(optEP)\n#optPupil[np.where(EP==0)] = np.nan\nplot_im_LP(optPupil, apRad, 0.8, color='viridis')\nplt.title(\"optimized pupil mask: dark hole at 3lD\")",
"_____no_output_____"
],
[
"#save mask\n\nhdu = fits.PrimaryHDU(data=np.angle(optEP))\nhdu.header[\"PUPIL\"] = \"VLT\"\nhdu.header[\"lD\"] = \"10\"\nhdu.header[\"iter\"] = \"800\"\nhdu.header[\"OPTREG\"]= \"Dark hole at 3lD with a 2FWHM diameter\"\nhdu.header[\"DATE\"] = \"19/09/17\"\nhdu.writeto(\"/Users/patapisp/Documents/DAC/Optimized_pupil_phase_apodizer/Apodizer_dark_hole-3lD_VLT_800iter.fits\")",
"_____no_output_____"
]
],
[
[
"## visualize results",
"_____no_output_____"
]
],
[
[
"#optEP = fits.getdata(\"/Users/patapisp/Documents/DAC/Optimized_pupil_phase_apodizer/Apodizer_ring-1.4-2.6lD_VLT_400iter.fits\")\n#optEP = np.exp(1j*optEP)\nPSF = np.abs(myfft2(EP))**2\nPSFopt = np.abs(myfft2(EP*optEP))**2/np.max(PSF)\nplot_im_ld(PSF/np.max(PSF), 10,10, n=None, log_scale=True, log_min=-4 )\n",
"_____no_output_____"
],
[
"plot_im_ld(PSFopt, 10, 10, n=None, log_scale=True, log_min=-4 )\nplt.title(\"Apodizer:dark dark ring 1.4-2.6 l/D\")\nprint(np.max(PSFopt))",
"0.03608117511212594\n"
],
[
"PSF = np.abs(myfft2(EP))**2\nEP2 = EP*np.exp(1j*0.06*Y)\nEPP = 0.5*EP2+EP\nplot_im_ld(np.abs(myfft2(EPP))**2/np.max(PSF), 10, 10, n=None, log_scale=True, log_min=-4 )",
"_____no_output_____"
],
[
"PSFopt = np.abs(myfft2(EPP*optEP))**2/np.max(PSF)\nplot_im_ld(PSFopt, 10, 10, log_scale=True, n=None, log_min=-4 )\nprint(np.max(PSFopt))",
"0.654211306171\n"
],
[
"LS = make_elliptical_pupil(1024,1, apRad)\nLS = np.lib.pad(LS, (1536, 1536), 'constant', constant_values=(0,0))\nLP = myifft2(myfft2(EPP)*np.exp(1j*2*THETA))\nLPopt = myifft2(myfft2(EPP*optEP)*np.exp(1j*2*THETA))\nPSF = np.abs(myfft2(LP*LS))**2\nPSFopt = np.abs(myfft2(LPopt*LS))**2\nplot_im_ld(PSF/np.max(PSF), 10, 10, n=None, log_scale=True, log_min=-4 )",
"/Users/patapisp/polybox/MasterProject/code/pupil_masks.py:45: RuntimeWarning: overflow encountered in power\n pupil = np.exp(-np.sqrt((X/a)**2 + (Y/b)**2)**1000)\n"
],
[
"plot_im_ld(PSFopt/np.max(PSFopt), 10, 10, n=None, log_scale=True, log_min=-4 )",
"_____no_output_____"
],
[
"m = fits.getdata(\"/Users/patapisp/Documents/DAC/ZIMPOL_Mask/data_simulations/ZIMPOL_PSF_STREHL=40-100_optimized.fits\")[-1]",
"_____no_output_____"
],
[
"PSF = np.abs(myfft2(EP))**2\nplot_im_ld(m/np.max(PSF), 10, 5, log_scale=True, log_min=-4)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec729d4ef1beea373c139f2cad0184e20b1ad3a5 | 1,009 | ipynb | Jupyter Notebook | Coursera/Data Analysis with Python-IBM/Week-1/Importing Datasets/Quiz/Python-Packages-for-Data-Science.ipynb | manipiradi/Online-Courses-Learning | 2a4ce7590d1f6d1dfa5cfde632660b562fcff596 | [
"MIT"
]
| 331 | 2019-10-22T09:06:28.000Z | 2022-03-27T13:36:03.000Z | Coursera/Data Analysis with Python-IBM/Week-1/Importing Datasets/Quiz/Python-Packages-for-Data-Science.ipynb | manipiradi/Online-Courses-Learning | 2a4ce7590d1f6d1dfa5cfde632660b562fcff596 | [
"MIT"
]
| 8 | 2020-04-10T07:59:06.000Z | 2022-02-06T11:36:47.000Z | Coursera/Data Analysis with Python-IBM/Week-1/Importing Datasets/Quiz/Python-Packages-for-Data-Science.ipynb | manipiradi/Online-Courses-Learning | 2a4ce7590d1f6d1dfa5cfde632660b562fcff596 | [
"MIT"
]
| 572 | 2019-07-28T23:43:35.000Z | 2022-03-27T22:40:08.000Z | 18.685185 | 122 | 0.53221 | [
[
[
"#### 1. What is a Python library?",
"_____no_output_____"
],
[
"##### Ans: a collection of functions and methods that allow you to perform lots of actions without writing your code",
"_____no_output_____"
],
[
"#### 2. What is the primary instrument or data-structure of Pandas?",
"_____no_output_____"
],
[
"##### Ans: dataframe",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec729f2ec6758c73935c0ed7b6b5c72baaa498a3 | 2,756 | ipynb | Jupyter Notebook | game_theory/Matching Pennies (Nashpy).ipynb | rdempsey/poc | f381104b3d92441d0f35eb80b8b6de0bd09a2dc8 | [
"Apache-2.0"
]
| null | null | null | game_theory/Matching Pennies (Nashpy).ipynb | rdempsey/poc | f381104b3d92441d0f35eb80b8b6de0bd09a2dc8 | [
"Apache-2.0"
]
| null | null | null | game_theory/Matching Pennies (Nashpy).ipynb | rdempsey/poc | f381104b3d92441d0f35eb80b8b6de0bd09a2dc8 | [
"Apache-2.0"
]
| null | null | null | 21.038168 | 254 | 0.498549 | [
[
[
"Matching pennies is the name for a simple game used in game theory. It is played between two players, Even and Odd. Each player has a penny and must secretly turn the penny to heads or tails. The players then reveal their choices simultaneously.\n\nRules:\n* If the pennies match (both heads or both tails), then Even keeps both pennies, so wins one from Odd (+1 for Even, −1 for Odd).\n* If the pennies do not match (one heads and one tails) Odd keeps both pennies, so receives one from Even (−1 for Even, +1 for Odd).\n\nThis can be represented by a 2x2 matrix:\n\n|Side|Heads|Tails|\n|---|---|---|\n|Heads|+1,-1|-1,+1|\n|Tails|-1,+1|+1,-1|",
"_____no_output_____"
],
[
"## Imports",
"_____no_output_____"
]
],
[
[
"import nashpy as nash\nimport numpy as np",
"_____no_output_____"
]
],
[
[
"## Create a game",
"_____no_output_____"
]
],
[
[
"A = np.array([[1, -1], [-1,1]])\nmp = nash.Game(A)\nmp",
"_____no_output_____"
]
],
[
[
"## Solving with support enumeration",
"_____no_output_____"
]
],
[
[
"equilibria = mp.support_enumeration()\nfor eq in equilibria:\n print(eq)",
"(array([0.5, 0.5]), array([0.5, 0.5]))\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec72ca88a91b88161492ab8d9e34e95de902faa1 | 30,448 | ipynb | Jupyter Notebook | notebooks/tf-distributed-training.ipynb | austindimmer/amazon-sagemaker-workshop | a9e410eb80d0f64d3f0c449928dc424251787a31 | [
"Apache-2.0"
]
| 381 | 2018-02-07T22:35:38.000Z | 2022-03-01T20:18:33.000Z | notebooks/tf-distributed-training.ipynb | austindimmer/amazon-sagemaker-workshop | a9e410eb80d0f64d3f0c449928dc424251787a31 | [
"Apache-2.0"
]
| 22 | 2019-03-04T04:18:02.000Z | 2022-03-09T00:21:36.000Z | notebooks/tf-distributed-training.ipynb | austindimmer/amazon-sagemaker-workshop | a9e410eb80d0f64d3f0c449928dc424251787a31 | [
"Apache-2.0"
]
| 189 | 2018-02-12T18:06:10.000Z | 2021-12-17T00:00:18.000Z | 54.566308 | 870 | 0.67006 | [
[
[
"# TensorFlow Distributed Training Options \n\nSometimes it makes sense to perform training on a single machine. For large datasets, however, it may be necessary to perform distributed training on a cluster of multiple machines. Cluster managment often is a pain point in the machine learning pipeline. Fortunately, Amazon SageMaker makes it easy to run distributed training without having to manage cluster setup and tear down. In this notebook, we'll examine some different options for performing distributed training with TensorFlow in Amazon SageMaker. In particular, we'll look at the following options:\n\n- **Parameter Servers**: processes that receive asynchronous updates from worker nodes and distribute updated gradients to all workers.\n\n- **Horovod**: a framework based on Ring-AllReduce wherein worker nodes synchronously exchange gradient updates only with two other workers at a time. \n\nThe model used for this notebook is a basic Convolutional Neural Network (CNN) based on [the Keras examples](https://github.com/keras-team/keras/blob/master/examples/cifar10_cnn.py), although we will be using the tf.keras implementation of Keras rather than the separate Keras reference implementation. We'll train the CNN to classify images using the [CIFAR-10 dataset](https://www.cs.toronto.edu/~kriz/cifar.html), a well-known computer vision dataset. It consists of 60,000 32x32 images belonging to 10 different classes, with 6,000 images per class. Here is a graphic of the classes in the dataset, as well as 10 random images from each:\n\n\n",
"_____no_output_____"
],
[
"## Setup \n\nWe'll begin with some necessary imports, and get an Amazon SageMaker session to help perform certain tasks, as well as an IAM role with the necessary permissions.",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport numpy as np\nimport os\nimport sagemaker\nfrom sagemaker import get_execution_role\n\nsagemaker_session = sagemaker.Session()\nrole = get_execution_role()\n\nbucket = sagemaker_session.default_bucket()\nprefix = 'sagemaker/DEMO-tf-distribution-options'\nprint('Bucket:\\n{}'.format(bucket))",
"_____no_output_____"
]
],
[
[
"Now we'll run a script that fetches the dataset and converts it to the TFRecord format, which provides several conveniences for training models in TensorFlow.",
"_____no_output_____"
]
],
[
[
"!wget -q https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/tf-distribution-options/generate_cifar10_tfrecords.py\n!python generate_cifar10_tfrecords.py --data-dir ./data",
"_____no_output_____"
]
],
[
[
"For Amazon SageMaker hosted training on a cluster separate from this notebook instance, training data must be stored in Amazon S3, so we'll upload the data to S3 now.",
"_____no_output_____"
]
],
[
[
"inputs = sagemaker_session.upload_data(path='data', key_prefix='data/tf-distribution-options')\ndisplay(inputs)",
"_____no_output_____"
]
],
[
[
"## Distributed Training with Parameter Servers\n\nA common pattern in distributed training is to use dedicated processes to collect gradients computed by “worker” processes, then aggregate them and distribute the updated gradients back to the workers. These processes are known as parameter servers. In general, they can be run either on their own machines or co-located on the same machines as the workers. In a parameter server cluster, each parameter server communicates with all workers (“all-to-all”). The Amazon SageMaker prebuilt TensorFlow container comes with a built-in option to use parameter servers for distributed training. The container runs a parameter server thread in each training instance, so there is a 1:1 ratio of parameter servers to workers. With this built-in option, gradient updates are made asynchronously (though some other versions of parameters servers use synchronous updates). \n\nScript Mode requires a training script, which in this case is the `train_ps.py` file in the */code* subdirectory of the [related distributed training example GitHub repository](https://github.com/aws-samples/amazon-sagemaker-script-mode/tree/master/tf-distribution-options). Once a training script is ready, the next step is to set up an Amazon SageMaker TensorFlow Estimator object with the details of the training job. It is very similar to an Estimator for training on a single machine, except we specify a `distributions` parameter to enable starting a parameter server on each training instance. We'll reference the GitHub repository so we can keep our training code version controlled and avoid downloading it locally.",
"_____no_output_____"
]
],
[
[
"from sagemaker.tensorflow import TensorFlow\n\ngit_config = {'repo': 'https://github.com/aws-samples/amazon-sagemaker-script-mode', \n 'branch': 'master'}\n\nps_instance_type = 'ml.p3.2xlarge'\nps_instance_count = 2\n\nmodel_dir = \"/opt/ml/model\"\n\ndistributions = {'parameter_server': {\n 'enabled': True}\n }\nhyperparameters = {'epochs': 60, 'batch-size' : 256}\n\nestimator_ps = TensorFlow(\n git_config=git_config,\n source_dir='tf-distribution-options/code',\n entry_point='train_ps.py', \n base_job_name='ps-cifar10-tf',\n role=role,\n framework_version='1.13',\n py_version='py3',\n hyperparameters=hyperparameters,\n train_instance_count=ps_instance_count, \n train_instance_type=ps_instance_type,\n model_dir=model_dir,\n tags = [{'Key' : 'Project', 'Value' : 'cifar10'},{'Key' : 'TensorBoard', 'Value' : 'dist'}],\n distributions=distributions)",
"_____no_output_____"
]
],
[
[
"Now we can call the `fit` method of the Estimator object to start training. After training completes, the tf.keras model will be saved in the SavedModel .pb format so it can be served by a TensorFlow Serving container. Note that the model is only saved by the the lead node (disregard any warnings about the model not being saved by all the processes).",
"_____no_output_____"
]
],
[
[
"remote_inputs = {'train' : inputs+'/train', 'validation' : inputs+'/validation', 'eval' : inputs+'/eval'}\nestimator_ps.fit(remote_inputs, wait=True)",
"_____no_output_____"
]
],
[
[
"After training is complete, it is always a good idea to take a look at training curves to diagnose problems, if any, during training and determine the representativeness of the training and validation datasets. We can do this with TensorBoard, and also with the Keras API: conveniently, the Keras `fit` invocation returns a data structure with the training history. In our training script, this history is saved on the lead training node, then uploaded with the model when training is complete.\n\nTo retrieve the history, we first download the model locally, then unzip it to gain access to the history data structure. We can then simply load the history as JSON:",
"_____no_output_____"
]
],
[
[
"import json \n\n!aws s3 cp {estimator_ps.model_data} ./ps_model/model.tar.gz\n!tar -xzf ./ps_model/model.tar.gz -C ./ps_model\n\nwith open('./ps_model/ps_history.p', \"r\") as f:\n ps_history = json.load(f)",
"_____no_output_____"
]
],
[
[
"Now we can plot the history with two graphs, one for accuracy and another for loss. Each graph shows the results for both the training and validation datasets. Although training is a stochastic process that can vary significantly between training jobs, overall you are likely to see that the training curves are converging smoothly and steadily to higher accuracy and lower loss, while the validation curves are more jagged. This is due to the validation dataset being relatively small and thus not as representative as the training dataset.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\ndef plot_training_curves(history): \n \n fig, axes = plt.subplots(1, 2, figsize=(12, 4), sharex=True)\n ax = axes[0]\n ax.plot(history['acc'], label='train')\n ax.plot(history['val_acc'], label='validation')\n ax.set(\n title='model accuracy',\n ylabel='accuracy',\n xlabel='epoch')\n ax.legend()\n\n ax = axes[1]\n ax.plot(history['loss'], label='train')\n ax.plot(history['val_loss'], label='validation')\n ax.set(\n title='model loss',\n ylabel='loss',\n xlabel='epoch')\n ax.legend()\n fig.tight_layout()\n \nplot_training_curves(ps_history)",
"_____no_output_____"
]
],
[
[
"Besides saving the training history, we also can save other artifacts and data from the training job. For example, we can include a callback in the training script to save each model checkpoint after each training epoch is complete. These checkpoints will be saved to the same Amazon S3 folder as the model, in a zipped file named `output.tar.gz` as shown below:",
"_____no_output_____"
]
],
[
[
"artifacts_dir = estimator_ps.model_data.replace('model.tar.gz', '')\nprint(artifacts_dir)\n!aws s3 ls --human-readable {artifacts_dir}",
"_____no_output_____"
]
],
[
[
"## Distributed Training with Horovod\n\nHorovod is an open source distributed training framework for TensorFlow, Keras, PyTorch, and MXNet. It is an alternative to the more \"traditional\" parameter servers method of performing distributed training demonstrated above. Horovod can be more performant than parameter servers in large, GPU-based clusters where large models are trained. In Amazon SageMaker, Horovod is only available with TensorFlow version 1.12 or newer. \n\nOnly a few lines of code are necessary to use Horovod for distributed training of a Keras model defined by the tf.keras API. For details, see the `train_hvd.py` script included with this notebook; the changes primarily relate to:\n\n- importing Horovod.\n- initializing Horovod.\n- configuring GPU options and setting a Keras/tf.session with those options.\n\nThe Estimator object for Horovod training is very similar to the parameter servers Estimator above, except we specify a `distributions` parameter describing Horovod attributes such as the number of process per host, which is set here to the number of GPUs per machine. Beyond these few simple parameters and the few lines of code in the training script, there is nothing else you need to do to use distributed training with Horovod; Amazon SageMaker handles the heavy lifting for you and manages the underlying cluster setup.",
"_____no_output_____"
]
],
[
[
"from sagemaker.tensorflow import TensorFlow\n\nhvd_instance_type = 'ml.p3.2xlarge'\nhvd_processes_per_host = 1\nhvd_instance_count = 2\n\ndistributions = {'mpi': {\n 'enabled': True,\n 'processes_per_host': hvd_processes_per_host,\n 'custom_mpi_options': '-verbose --NCCL_DEBUG=INFO -x OMPI_MCA_btl_vader_single_copy_mechanism=none'\n }\n }\n\nhyperparameters = {'epochs': 60, 'batch-size' : 256}\n\nestimator_hvd = TensorFlow(\n git_config=git_config,\n source_dir='tf-distribution-options/code',\n entry_point='train_hvd.py',\n base_job_name='hvd-cifar10-tf', \n role=role,\n framework_version='1.13',\n py_version='py3',\n hyperparameters=hyperparameters,\n train_instance_count=hvd_instance_count, \n train_instance_type=hvd_instance_type,\n tags = [{'Key' : 'Project', 'Value' : 'cifar10'},{'Key' : 'TensorBoard', 'Value' : 'dist'}],\n distributions=distributions)",
"_____no_output_____"
]
],
[
[
"With these changes to the Estimator, we can call its `fit` method to start training. After training completes, the tf.keras model will be saved in the SavedModel .pb format so it can be served by a TensorFlow Serving container. Note that the model is only saved by the the master, Horovod rank = 0 process (once again disregard any warnings about the model not being saved by all the processes).",
"_____no_output_____"
]
],
[
[
"remote_inputs = {'train' : inputs+'/train', 'validation' : inputs+'/validation', 'eval' : inputs+'/eval'}\nestimator_hvd.fit(remote_inputs, wait=True)",
"_____no_output_____"
]
],
[
[
"We can now plot training curves for the Horovod training job similar to the curves we plotted for the parameter servers training job:",
"_____no_output_____"
]
],
[
[
"!aws s3 cp {estimator_hvd.model_data} ./hvd_model/model.tar.gz\n!tar -xzf ./hvd_model/model.tar.gz -C ./hvd_model\n\nwith open('./hvd_model/hvd_history.p', \"r\") as f:\n hvd_history = json.load(f)\n \nplot_training_curves(hvd_history)",
"_____no_output_____"
]
],
[
[
"## Comparing Results from Parameter Servers and Horovod\n\nNow that both training jobs are complete, we can compare the results. The CIFAR-10 dataset is relatively small so the training jobs do not run long enough to draw detailed conclusions. However, it is likely that you can observe some differences even though training time was short, and training is a stochastic process with different results every time. The training time can be approximated for our purposes by looking at the \"Billable seconds\" log output at the end of each training job (at the bottom of the log output beneath the `fit` invocation code cells).\n\nThe Horovod training job tends to take a bit longer than the parameter server training job, while producing a somewhat more accurate model. The relative speed result is consistent with research showing that Horovod is more performant for larger clusters and models, while parameter servers have the edge for smaller clusters and models such as this one. Also, asynchronous model updates like those used by the parameter servers here require more epochs to converge to more accurate models, so it is not surprising if the parameter server model training completed faster for the same number of epochs as Horovord, but was less accurate. It also is likely that you can observe that the training curves for the Horovod training job are a bit smoother, reflecting the fact that synchronous gradient updates typically are less noisy than asynchronous updates.\n\nThe following table summarizes some general guidelines regarding performance for each option. These rules aren’t absolute, and ultimately, the best choice depends on the specific use case. Typically, the performance significantly depends on how long it takes to share gradient updates during training. In turn, this is affected by the model size, gradients size, GPU specifications, and network speed.\n\n| | Better CPU Performance | Better GPU Performance |\n| :--- | :--- | :--- |\n| larger number of gradients/bigger model size | Parameter Servers | Parameter Servers, OR Horovod on a single instance with multiple GPUs |\n| smaller number of gradients/lesser model size | Parameter Servers | Horovod |\n\nComplexity is another consideration. Parameter servers are straightforward to use for one GPU per instance. However, to use multi-GPU instances, you must set up multiple towers, with each tower assigned to a different GPU. A “tower” is a function for computing inference and gradients for a single model replica, which in turn is a copy of a model training on a subset of the complete dataset. Towers involve a form of data parallelism. Horovod also employs data parallelism but abstracts away the implementation details.\n\nFinally, cluster size makes a difference. Given larger clusters with many GPUs, parameter server all-to-all communication can overwhelm network bandwidth. Reduced scaling efficiency can result, among other adverse effects. In such situations, you might find Horovod a better option.",
"_____no_output_____"
],
[
"## Model Deployment with Amazon Elastic Inference\n\nAmazon SageMaker supports both real time inference and batch inference. In this notebook, we will focus on setting up an Amazon SageMaker hosted endpoint for real time inference with TensorFlow Serving (TFS). Additionally, we will discuss why and how to use Amazon Elastic Inference with the hosted endpoint.\n\n### Deploying the Model\n\nWhen considering the overall cost of a machine learning workload, inference often is the largest part, up to 90% of the total. If a GPU instance type is used for real time inference, it typically is not fully utilized because, unlike training, real time inference usually does not involve continuously sending large batches of data to the model. Elastic Inference provides GPU acceleration suited for inference, allowing you to add just the right amount of inference acceleration to a hosted endpoint for a fraction of the cost of using a full GPU instance.\n\nThe `deploy` method of the Estimator object creates an endpoint which serves prediction requests in near real time. To utilize Elastic Inference with the SageMaker TFS container, simply provide an `accelerator_type` parameter, which determines the type of accelerator that is attached to your endpoint. Refer to the **Inference Acceleration** section of the [instance types chart](https://aws.amazon.com/sagemaker/pricing/instance-types) for a listing of the supported types of accelerators. \n\nHere we'll use a general purpose CPU compute instance type along with an Elastic Inference accelerator: together they are much cheaper than the smallest P3 GPU instance type.",
"_____no_output_____"
]
],
[
[
"predictor = estimator_hvd.deploy(initial_instance_count=1,\n instance_type='ml.m5.xlarge',\n accelerator_type='ml.eia1.medium')",
"_____no_output_____"
]
],
[
[
"By using a general purpose CPU instance with an Elastic Inference accelerator instead of a GPU intance, substantial costs savings are achieved. As of Q4 2019, On-Demand pricing for those resources is \\\\$0.269 per hour (ml.m5.xlarge), plus \\\\$0.182 per hour (ml.eia1.medium), for a total of \\\\$0.451 per hour. The total cost compared to the pricing of the smallest P3 family (NVIDIA Volta V100) GPU instance is as follows:\n\n- Elastic Inference solution: \\\\$0.451 per hour\n- GPU instance ml.p3.2xlarge: \\\\$4.284 per hour\n\nTo summarize, the Elastic Inference solution cost is about 10% of the cost of using a full P3 family GPU instance. ",
"_____no_output_____"
],
[
"### Labels and Sample Data\n \nNow that we have a Predictor object wrapping a real time Amazon SageMaker hosted enpoint, we'll define the label names and look at a sample of 10 images, one from each class.",
"_____no_output_____"
]
],
[
[
"from IPython.display import Image, display\n\nos.system(\"aws s3 cp s3://sagemaker-workshop-pdx/cifar-10-module/sample-img ./sample-img --recursive --quiet\")\n\nlabels = ['airplane','automobile','bird','cat','deer','dog','frog','horse','ship','truck']\n\nimages = []\nfor entry in os.scandir('sample-img'):\n if entry.is_file() and entry.name.endswith(\"png\"):\n images.append('sample-img/' + entry.name)\n\nfor image in images:\n display(Image(image))",
"_____no_output_____"
]
],
[
[
"### Pre/post-postprocessing Script\n\nThe TFS container in Amazon SageMaker by default uses the TFS REST API to serve prediction requests. This requires the input data to be converted to JSON format. One way to do this is to create a Docker container to do the conversion, then create an overall Amazon SageMaker model that links the conversion container to the TFS container with the model. This is known as an Amazon SageMaker Inference Pipeline, as demonstrated in another [sample notebook](https://github.com/awslabs/amazon-sagemaker-examples/tree/master/sagemaker_batch_transform/working_with_tfrecords). \n\nHowever, as a more convenient alternative for many use cases, the Amazon SageMaker TFS container provides a data pre/post-processing script feature that allows you to simply supply a data transformation script. Using such a script, there is no need to build containers or directly work with Docker. The simplest form of a script must only (1) implement an `input_handler` and `output_handler` interface, as shown in the code below, (2) be named `inference.py`, and (3) be placed in a `/code` directory.",
"_____no_output_____"
]
],
[
[
"!wget -q https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/tf-distribution-options/code/inference.py\n!cat ./inference.py",
"_____no_output_____"
]
],
[
[
"On the input preprocessing side, the code takes an image read from Amazon S3 and converts it to the required TFS REST API input format. On the output postprocessing side, the script simply passes through the predictions in the standard TFS format without modifying them. Alternatively, we could have just returned a class label for the class with the highest score, or performed other postprocessing that would be helpful to the application consuming the predictions. ",
"_____no_output_____"
],
[
"### Requirements.txt\n\nBesides an `inference.py` script implementing the handler interface, it also may be necessary to supply a `requirements.txt` file to ensure any necessary dependencies are installed in the container along with the script. For this script, in addition to the Python standard libraries we used the Pillow and Numpy libraries.",
"_____no_output_____"
]
],
[
[
"!wget -q https://raw.githubusercontent.com/aws-samples/amazon-sagemaker-script-mode/master/tf-distribution-options/code/requirements.txt\n!cat ./requirements.txt",
"_____no_output_____"
]
],
[
[
"### Make Predictions\n\nNext we'll set up the Predictor object created by the `deploy` method call above. Since we are using a preprocessing script, we need to specify the Predictor's content type as `application/x-image` and override the default (JSON) serializer. We can now get predictions about the sample data displayed above simply by providing the raw .png image bytes to the Predictor. ",
"_____no_output_____"
]
],
[
[
"predictor.content_type = 'application/x-image'\npredictor.serializer = None\n\ndef get_prediction(file_path):\n \n with open(file_path, \"rb\") as image:\n f = image.read()\n b = bytearray(f)\n return labels[np.argmax(predictor.predict(b)['predictions'], axis=1)[0]]",
"_____no_output_____"
],
[
"predictions = [get_prediction(image) for image in images]\nprint(predictions)",
"_____no_output_____"
]
],
[
[
"# Extensions\n\nAlthough we did not demonstrate them in this notebook, Amazon SageMaker provides additional ways to make distributed training more efficient and cost effective for very large datasets:\n\n- **Sharding data**: in this example we used one large TFRecord file containing the entire CIFAR-10 dataset, which is relatively small. However, larger datasets might require that you shard the data into multiple files, particularly if Pipe Mode is used (see below). Sharding may be accomplished by specifying an Amazon S3 data source as a [manifest file or ShardedByS3Key](https://docs.aws.amazon.com/sagemaker/latest/dg/API_S3DataSource.html). \n\n- **VPC training**: performing Horovod training inside a VPC improves the network latency between nodes, leading to higher performance and stability of Horovod training jobs.\n\n- **Pipe Mode**: using [Pipe Mode](https://docs.aws.amazon.com/sagemaker/latest/dg/your-algorithms-training-algo.html#your-algorithms-training-algo-running-container-inputdataconfig) reduces startup and training times. Pipe Mode streams training data from S3 as a Linux FIFO directly to the algorithm, without saving to disk. For a small dataset such as CIFAR-10, Pipe Mode does not provide any advantage, but for very large datasets where training is I/O bound rather than CPU/GPU bound, Pipe Mode can substantially reduce startup and training times.\n\n- **Amazon FSx for Lustre and Amazon EFS**: performance on large datasets in File Mode may be improved in some circumstances using either Amazon FSx for Lustre or Amazon EFS. For more details, please refer to the related [blog post](https://aws.amazon.com/blogs/machine-learning/speed-up-training-on-amazon-sagemaker-using-amazon-efs-or-amazon-fsx-for-lustre-file-systems).\n\n- **Managed Spot Training**: uses Amazon EC2 Spot instances to run training jobs instead of on-demand instances. Managed spot training can optimize the cost of training models up to 90% over on-demand instances. For further details, please refer to the [documentation](https://docs.aws.amazon.com/sagemaker/latest/dg/model-managed-spot-training.html).",
"_____no_output_____"
],
[
"# Cleanup\n\nTo avoid incurring charges due to a stray endpoint, delete the Amazon SageMaker endpoint if you no longer need it:",
"_____no_output_____"
]
],
[
[
"sagemaker_session.delete_endpoint(predictor.endpoint)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
]
]
|
ec72e13e05ac878a2c4ae8c0f28023fe35d62fc9 | 4,898 | ipynb | Jupyter Notebook | atlas/notebooks/2017-04-01 votes.ipynb | novelo-io/atlas | db7eadf8edfcf5a386f37aeb493eec7e74910b60 | [
"MIT"
]
| 2 | 2017-02-28T18:51:00.000Z | 2017-07-10T03:57:48.000Z | atlas/notebooks/2017-04-01 votes.ipynb | novelo-io/atlas | db7eadf8edfcf5a386f37aeb493eec7e74910b60 | [
"MIT"
]
| 1 | 2017-05-13T00:37:55.000Z | 2017-05-15T14:15:47.000Z | atlas/notebooks/2017-04-01 votes.ipynb | novelo-io/atlas | db7eadf8edfcf5a386f37aeb493eec7e74910b60 | [
"MIT"
]
| 1 | 2017-03-28T19:57:13.000Z | 2017-03-28T19:57:13.000Z | 22.46789 | 112 | 0.542875 | [
[
[
"%matplotlib inline",
"_____no_output_____"
],
[
"import datetime",
"_____no_output_____"
],
[
"import pandas as pd",
"_____no_output_____"
],
[
"councilman = pd.read_csv('../data/sequential_id.csv')\nsecreataries = pd.read_csv('../data/secretary-councilman.csv')\n\n# sequencial dos vereadores de são paulo\nsequencial = councilman.sequential_id.tolist()\nsequencial.extend(secreataries.sequential_id.tolist())",
"_____no_output_____"
],
[
"df = pd.read_csv(\n '/home/flavio/Downloads/votacao_candidato_munzona_2016/votacao_candidato_munzona_2016_SP.txt', \n encoding='iso-8859-1', sep=';', decimal=',', thousands='.', \n dtype={'NOME_MUNICIPIO': str, 'NOME_PARTIDO': str},\n names=['DATA_GERACAO', 'HORA_GERACAO', 'ANO_ELEICAO', 'NUM_TURNO','DESCRICAO_ELEICAO',\n 'SIGLA_UF', 'SIGLA_UE', 'CODIGO_MUNICIPIO', 'NOME_MUNICIPIO', 'NUMERO_ZONA', \n 'CODIGO_CARGO', 'NUMERO_CAND', 'sequential_id', 'NOME_CANDIDATO', \n 'NOME_URNA_CANDIDATO', 'DESCRICAO_CARGO', 'COD_SIT_CAND_SUPERIOR', \n 'DESC_SIT_CAND_SUPERIOR', 'CODIGO_SIT_CANDIDATO', 'DESC_SIT_CANDIDATO', \n 'COD_SIT_CAND_TOT', 'DESC_SIT_CAND_TOT', 'NUMERO_PARTIDO', 'SIGLA_PARTIDO', 'NOME_PARTIDO', \n 'SEQUENCIAL_LEGENDA', 'NOME_COLIGACAO', 'COMPOSICAO_LEGENDA', 'TOTAL_VOTOS', 'TRANSITO'],\n usecols=['CODIGO_MUNICIPIO', 'NOME_CANDIDATO', 'sequential_id', \n 'DESCRICAO_CARGO', 'DESC_SIT_CAND_TOT', 'TOTAL_VOTOS']\n)",
"_____no_output_____"
],
[
"df = df[df['sequential_id'].isin(sequencial)]",
"_____no_output_____"
],
[
"votes = df.groupby('sequential_id')['TOTAL_VOTOS'].sum().reset_index()\nvotes = votes.sort_values(by=\"TOTAL_VOTOS\", ascending=False)\nvotes.columns",
"_____no_output_____"
],
[
"votes = votes.rename(columns={'TOTAL_VOTOS': 'votes'})",
"_____no_output_____"
],
[
"votes['election_year'] = 2016",
"_____no_output_____"
],
[
"today = datetime.date.today()\nvotes.to_csv(f\"../data/{today}-votes.csv\", index=False)",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec72e9cbbafaab66d0cb1e4a64cc0c056edfb092 | 175,497 | ipynb | Jupyter Notebook | 1_mosaic_data_attention_experiments/5_pretrained_10_cls_for_both_what_where/model1_lr2/.ipynb_checkpoints/Visualization-checkpoint.ipynb | lnpandey/DL_explore_synth_data | 0a5d8b417091897f4c7f358377d5198a155f3f24 | [
"MIT"
]
| 2 | 2019-08-24T07:20:35.000Z | 2020-03-27T08:16:59.000Z | 1_mosaic_data_attention_experiments/5_pretrained_10_cls_for_both_what_where/model1_lr2/Visualization.ipynb | lnpandey/DL_explore_synth_data | 0a5d8b417091897f4c7f358377d5198a155f3f24 | [
"MIT"
]
| null | null | null | 1_mosaic_data_attention_experiments/5_pretrained_10_cls_for_both_what_where/model1_lr2/Visualization.ipynb | lnpandey/DL_explore_synth_data | 0a5d8b417091897f4c7f358377d5198a155f3f24 | [
"MIT"
]
| 3 | 2019-06-21T09:34:32.000Z | 2019-09-19T10:43:07.000Z | 98.927283 | 34,968 | 0.712833 | [
[
[
"import torch.nn as nn\nimport torch.nn.functional as F\n\nimport pandas as pd\nimport numpy as np\n\n\nimport torch\nimport torchvision\nimport torchvision.transforms as transforms\nfrom torch.utils.data import Dataset, DataLoader\nfrom torchvision import transforms, utils\nimport torch.optim as optim\nfrom matplotlib import pyplot as plt\n\nimport copy\nimport pickle\n%matplotlib inline\n# Ignore warnings\nimport warnings\nwarnings.filterwarnings(\"ignore\")\ndevice = torch.device(\"cuda\" if torch.cuda.is_available() else \"cpu\")\nprint(device)",
"cuda\n"
],
[
"def imshow(img):\n img = img / 2 + 0.5 # unnormalize\n npimg = img#.numpy()\n plt.imshow(np.transpose(npimg, axes = (1, 2, 0)))\n plt.show()",
"_____no_output_____"
],
[
"df_train = pd.read_csv(\"weight_pretrained2/train_results.csv\")\ndf_test = pd.read_csv(\"weight_pretrained2/test_results.csv\")",
"_____no_output_____"
],
[
"df_train",
"_____no_output_____"
],
[
"columns = [\"epochs\", \"argmax > 0.5\" ,\"argmax < 0.5\", \"focus_true_pred_true\", \"focus_false_pred_true\", \"focus_true_pred_false\", \"focus_false_pred_false\" ]\n\ncol1 = df_train[columns[0]]\ncol2 = df_train[columns[1]] \ncol3 = df_train[columns[2]] \ncol4 = df_train[columns[3]] \ncol5 = df_train[columns[4]] \ncol6 = df_train[columns[5]] \ncol7 = df_train[columns[6]] \n\ncol1 = df_test[columns[0]] \ncol8 = df_test[columns[1]] \ncol9 = df_test[columns[2]] \ncol10 = df_test[columns[3]] \ncol11 = df_test[columns[4]] \ncol12 = df_test[columns[5]] \ncol13 = df_test[columns[6]] ",
"_____no_output_____"
],
[
"# plt.figure(12,12)\nplt.plot(col1,col2, label='argmax > 0.5')\nplt.plot(col1,col3, label='argmax < 0.5')\n\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"training data\")\nplt.title(\"On Training set\")\nplt.show()\n\nplt.plot(col1,col4, label =\"focus_true_pred_true \")\nplt.plot(col1,col5, label =\"focus_false_pred_true \")\nplt.plot(col1,col6, label =\"focus_true_pred_false \")\nplt.plot(col1,col7, label =\"focus_false_pred_false \")\nplt.title(\"On Training set\")\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"training data\")\nplt.show()",
"_____no_output_____"
],
[
"df_test",
"_____no_output_____"
],
[
"# plt.figure(12,12)\nplt.plot(col1,col8, label='argmax > 0.5')\nplt.plot(col1,col9, label='argmax < 0.5')\n\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"Testing data\")\nplt.title(\"On Testing set\")\nplt.show()\n\nplt.plot(col1,col10, label =\"focus_true_pred_true \")\nplt.plot(col1,col11, label =\"focus_false_pred_true \")\nplt.plot(col1,col12, label =\"focus_true_pred_false \")\nplt.plot(col1,col13, label =\"focus_false_pred_false \")\nplt.title(\"On Testing set\")\nplt.legend(loc='center left', bbox_to_anchor=(1, 0.5))\nplt.xlabel(\"epochs\")\nplt.ylabel(\"Testing data\")\nplt.show()",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec72f3e61db6261d2e690bd0689791012df41094 | 72,330 | ipynb | Jupyter Notebook | _build/jupyter_execute/curriculum-notebooks/Mathematics/MultiplicationDivisionFractions/multiplication-division-fractions.ipynb | BryceHaley/curriculum-jbook | d1246799ddfe62b0cf5c389394a18c2904383437 | [
"CC-BY-4.0"
]
| 1 | 2022-03-18T18:19:40.000Z | 2022-03-18T18:19:40.000Z | _build/jupyter_execute/curriculum-notebooks/Mathematics/MultiplicationDivisionFractions/multiplication-division-fractions.ipynb | callysto/curriculum-jbook | ffb685901e266b0ae91d1250bf63e05a87c456d9 | [
"CC-BY-4.0"
]
| null | null | null | _build/jupyter_execute/curriculum-notebooks/Mathematics/MultiplicationDivisionFractions/multiplication-division-fractions.ipynb | callysto/curriculum-jbook | ffb685901e266b0ae91d1250bf63e05a87c456d9 | [
"CC-BY-4.0"
]
| null | null | null | 54.261065 | 21,441 | 0.689797 | [
[
[
"\n\n<a href=\"https://hub.callysto.ca/jupyter/hub/user-redirect/git-pull?repo=https%3A%2F%2Fgithub.com%2Fcallysto%2Fcurriculum-notebooks&branch=master&subPath=Mathematics/MultiplicationDivisionFractions/multiplication-division-fractions.ipynb&depth=1\" target=\"_parent\"><img src=\"https://raw.githubusercontent.com/callysto/curriculum-notebooks/master/open-in-callysto-button.svg?sanitize=true\" width=\"123\" height=\"24\" alt=\"Open in Callysto\"/></a>",
"_____no_output_____"
]
],
[
[
"%%html\n\n<script>\n function code_toggle() {\n if (code_shown){\n $('div.input').hide('500');\n $('#toggleButton').val('Show Code')\n } else {\n $('div.input').show('500');\n $('#toggleButton').val('Hide Code')\n }\n code_shown = !code_shown\n }\n\n $( document ).ready(function(){\n code_shown=false;\n $('div.input').hide()\n });\n</script>\n<p> Code is hidden for ease of viewing. Click the Show/Hide button to see. </>\n<form action=\"javascript:code_toggle()\"><input type=\"submit\" id=\"toggleButton\" value=\"Show Code\"></form>",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport ipywidgets as widgets\nimport IPython",
"_____no_output_____"
]
],
[
[
"# Fractions: Multiplication and Division\n\nThis is a notebook on how to multiply and divide by a fraction. In this notebook, you will learn how to multiply and divide between a whole number and a fraction, between two fractions, and between two mixed numbers.\n\nThis notebook starts with pictures to help you see how multiplying and dividing by a fraction changes a number. Then has a few practice questions at the end. (But remember that this one notebook will not be enough, you'll need a lot more practice!)\n\nIf you don't know what a numerator or denominator is, how to reduce fractions using common factors, or change a mixed number into a fraction then back again, please review those first before moving on to this notebook.",
"_____no_output_____"
],
[
"## Multiplying a Whole Number with a fraction\n\nWe know that repeated addition can be written as multiplication with whole numbers, which are also know as integers, and we can do the same for fractions.\n$$\\begin{align*}\n\\frac15 + \\frac15 + \\frac15 & = \\frac35 \\\\\n3 \\times \\frac15 & = \\frac35\n\\end{align*}$$\n\nWhat if we want $4 \\times \\frac23$? Then we are saying we want two thirds of \"4\", or 4 copies of two thirds, which is the same as eight copies of one-thirds, which is eight thirds. Lets look at what we did, but with numbers:\n$$\\begin{align*}\n4 \\times \\frac23 \\\\\n4 \\times \\frac23 & = 4 \\times (2 \\times \\frac13) \\\\\n4 \\times (2 \\times \\frac13) & = (4 \\times 2) \\times \\frac13 \\\\\n(4 \\times 2) \\times \\frac13 & = 8 \\times \\frac13 \\\\\n8 \\times \\frac13 & = \\frac83 \\\\\n\\frac83 & = 2 \\frac23\n\\end{align*}\n$$\n\nDid you notice how we never had to find a common denominator? That's because multiplication doesn't need a common denominator!\n\n<font color='red'>Tip! To remember which fractions need the common denominator, think of this:</font>\n>**AS** = **A**ddition and **S**ubtraction => Denominator needs to be **A**ll the **S**ame\n>\n>**DM** = **D**ivision and **M**ultiplication => Denominator **D**oesn't **M**atter\n\n\n## Multiplying two Fractions\n\nMultiplying 2 fractions is taking a small piece from an already small piece. For example, we have half a bar of a candy bar and want to eat half of what we have, but we want to know how much of the whole candy bar we are going to be eating. This word problem describes $\\frac{1}{2} \\times \\frac{1}{2}$ and we'll learn how to solve this.",
"_____no_output_____"
],
[
"## Visualizing multiplication of fractions\n\nLet's say we want to know what $\\frac15 \\times \\frac12$ is equal to.\nLet's show what's going on using the rectangles below.\n\nWe cut our first rectangle vertically into 5 equal pieces, and select one of them (in blue) to show $\\frac15$. <br>\nWe cut our second rectangle horizontally into 2 equal pieces, and select one of them (in yellow) to show $\\frac12$.",
"_____no_output_____"
]
],
[
[
"fig01 = plt.figure(figsize=(5,1))\n\nt_data01 = [['','','','','']]\ncolors01 = [[\"b\",\"w\",\"w\",\"w\",\"w\"]]\ntable01 = plt.table(cellText = t_data01,cellColours=colors01, loc='center')\ntable01.scale(1, 2)\n\nplt.axis('off')\nplt.grid('off')\n\nfig02 = plt.figure(figsize=(5,1))\n\nt_data02 = [[''],['']]\ncolors02 = [[\"yellow\"],[ \"w\"]]\ntable02 = plt.table(cellText = t_data02,cellColours=colors02, loc='center')\n\nplt.axis('off')\nplt.grid('off')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"Now let's lay one rectangle on top of the other. Do you see that there's now 10 pieces, and only one is green? The green piece represents the product of this multiplication. Therefore: $$\\frac15 \\times \\frac12 = \\frac{1}{10}$$",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(5,1))\n\nt_data = (('','','','',''), ('','','','',''))\ncolors = [[\"lime\",\"yellow\",\"yellow\",\"yellow\",\"yellow\"],[ \"b\",\"w\",\"w\",\"w\",\"w\"]]\ntable = plt.table(cellText = t_data,cellColours=colors, loc='center')\n\nplt.axis('off')\nplt.grid('off')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"The next image shows that if we had done things the other way around, using a vertical cut for $\\frac12$ and horizontal cuts for $\\frac15$, the result is the same. This gives us a way of visualizing the fact that the order in which we multiply our fractions does not matter.\n\n$$\\frac15 \\times \\frac12 = \\frac12 \\times \\frac15 = \\frac{1}{10}$$",
"_____no_output_____"
]
],
[
[
"fig2 = plt.figure(figsize=(2,2))\n\nt_data2 = (('',''), ('',''),('',''),('',''),('',''))\ncolors2 = [[\"lime\",\"b\"],[ \"yellow\",\"w\"],[ \"yellow\",\"w\"],[ \"yellow\",\"w\"],[ \"yellow\",\"w\"]]\ntable2 = plt.table(cellText = t_data2,cellColours=colors2, loc='center')\n\nplt.axis('off')\nplt.grid('off')\n\nplt.show()",
"_____no_output_____"
]
],
[
[
"## Multiplying fractions\n\n### Method #1: sticking to the rules.\n\nIn this example, we will work out the product of two fractions using the rule of multiplying the numerators together, then the denominators together, then reducing the fraction if possible at the end.\n\nWhat is the product of $\\frac34\\times\\frac29$? Let's walk through this step by step:\n\n##### Step 1:\n\nMultiply both numerators together to get the answer's numerator.\n\n In this question, the numerators are 3 and 2, so we get $3\\times 2=6$ for the numerator of the product.\n\n##### Step 2:\n\nMultiply both denominators together to get the answer's denominator.\n\n In this question, the denominators are 4 and 9, so we get $4\\times 9=36$ for the denominator of the product.\n\n##### Step 3:\n\nReduce the fraction by dividing both the numerator and denominator by the greatest common factor (GCF). You already know how to do this!\n\n Using the numbers from step 1 and step 2, we get $\\frac34\\times \\frac29 = \\frac{6}{36}$. But the GCF between 6 and 36 is not 1, it's 6. So when we reduce by dividing the top and bottom by 6, we get a final answer of $\\frac16$.\n\n##### And You're Done! ",
"_____no_output_____"
],
[
"This method seems like the most straight-forward answer, but what if you need to calculate really big numbers like $\\frac{5}{24} \\times \\frac{12}{13}$? You can use method #1, but then you need to do large multiplication, which is hard, and then you need to reduce after, which is also hard. But, there’s a way to make it easier! What if you reduce the fraction before doing the multiplication? That’s where method #2 comes in.\n\n### Method #2: A little less work.\n\nThe second approach relies on the fact that order of multiplication doesn't matter, and that this is true not only for the product of the fractions, but for the products in the numerator and denominator as well! In other words, we are reducing the fraction before multiplying. Observe:\n\n$$\\hskip-36pt\\begin{align*}\n\\frac34\\times \\frac29 &= \\frac{3\\times 2}{4\\times 9} \\tag{Use the rule of Method #1}\\\\[5pt]\n&= \\frac{2\\times 3}{4\\times 9} \\tag{Change the order of multiplication in the numerator}\\\\[5pt]\n&= \\frac24\\times\\frac39 \\tag{The rule of Method #1, in reverse}\\\\[5pt]\n&= \\frac12\\times\\frac13 \\tag{Reduce fractions}\\\\[5pt]\n&= \\frac{1\\times 1}{2\\times 3}= \\frac16 \\tag{Multiplying one more time}\n\\end{align*}$$\n\nThis might seem like more work (certainly, there are more steps), but what it tells us is that we can *cancel between fractions*. Let's repeat the above calculation, using this shortcut:\n$$\\hskip-60pt\\large \\frac34\\times\\frac29 = \\frac{^{\\color{red}{1}}3\\!\\!\\!\\!\\diagup}{_{\\color{blue}{2}}\\,4\\!\\!\\!\\!\\diagup}\\times\\frac{2\\!\\!\\!\\!\\diagup^{\\,\\,\\color{blue}{1}}}{9\\!\\!\\!\\!\\!\\diagup_{\\,\\,\\color{red}{3}}} = \\frac{1\\times 1}{2\\times 3}=\\frac16.$$\n\nWhat we've done here is cancel the 3 in $\\frac34$ with a factor of 3 in the $9=3\\times 3$ in $\\frac29$, and similarly, the 2 in $\\frac29$ with the 4 in $\\frac34$. The advantage of this approach is that the numbers we need to multiply are smaller, and (usually, if you were careful) no further reduction is needed. To recap:\n\n##### Step 1:\n\nReduce fractions first, and you can reduce the numerator of one fraction with the denominator of the other by the same common factor.\n\n##### Step 2:\n\nMultiply both numerators together for the answer's numerator. Multiply both denominators together for the answer's denominator.\n\n##### Step 3:\n\nThough the fraction should already be reduced, check again in case it can be reduced again. Check to see if there's any other common factors.\n\n##### And You're Done!\n",
"_____no_output_____"
],
[
"Let's do another example. This time, we multiply an integer, a whole number, by a fraction. Remember that any integer, or whole number, can be written as a fraction with a denominator equal to 1. First, we will use Method #1.\n\n$$\\hskip-60pt\\begin{align*}\n\\frac{4}{7} \\times 7 & = \\frac{4}{7} \\times \\frac{7}{1} \\tag{Turn integer into fraction}\\\\[5pt]\n\\frac{4}{7} \\times \\frac{7}{1} & = \\frac{28}{7} \\tag{Multiply numerators and denominators}\\\\[5pt]\n\\frac{28}{7} & = 4 \\tag{Reduce the fraction}\\\\\n\\end{align*}$$",
"_____no_output_____"
],
[
"Here's the same example again, but this time, using the second method:\n\n$$\\hskip-60pt\\begin{align*}\n\\frac{4}{7} \\times 7 & = \\frac{4}{7} \\times \\frac{7}{1} \\tag{Turn integer into fraction}\\\\[5pt]\n\\frac{4}{7\\!\\!\\!\\!\\diagup} \\times \\frac{7\\!\\!\\!\\!\\diagup}{1} & = \\frac{4}{1} \\times \\frac{1}{1} \\tag{Reduce numbers}\\\\[5pt]\n\\frac{4}{1} \\times \\frac{1}{1} & = 4 \\tag{Multiply numerators and denominators}\n\\end{align*}$$",
"_____no_output_____"
],
[
"Let's try one more example:\n\n$$\\hskip-60pt\\begin{align*}\n\\frac{4}{9} \\times \\frac{3}{5}\\\\\n\\frac{4}{9\\!\\!\\!\\!\\diagup} \\times \\frac{3\\!\\!\\!\\!\\diagup}{5} & = \\frac{4}{3} \\times \\frac{1}{5} \\tag{Reduce numbers}\\\\\n\\frac{4}{3} \\times \\frac{1}{5} & = \\frac{4}{15} \\tag{Multiply numerators and denominators}\n\\end{align*}$$",
"_____no_output_____"
],
[
"#### Here's an animation showing the two methods on the same problem.\n\n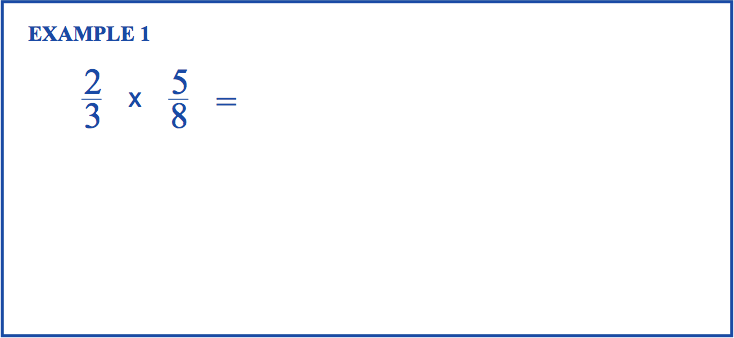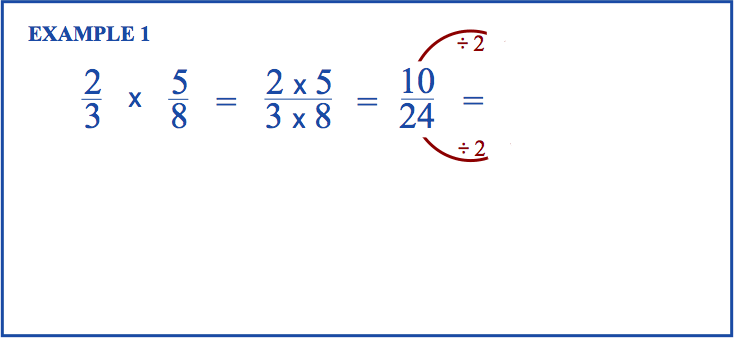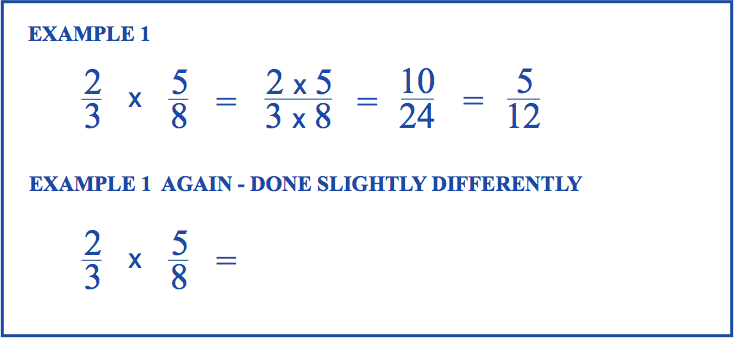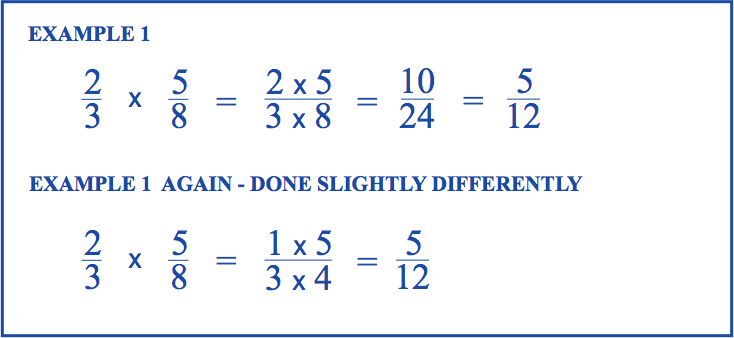",
"_____no_output_____"
],
[
"## Practice with fraction multiplication\n\nIt's time to try a few exercises. \n\nIn the following problems, fill in the missing numbers with integers. (Whole numbers.) Watch out for reductions!\n\n<br>\n$$\\hskip-60pt\\frac{2}{7} \\times \\frac{3}{5} = \\frac{6}{\\color{Red}{a}}$$",
"_____no_output_____"
]
],
[
[
"a = widgets.IntText(value=0, description='a =', disabled=False)\n\nIPython.display.display(a)\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(a)\n if a.value == 35:\n print(\"That's right! Great work!\")\n else:\n if a.value == 0:\n pass\n else:\n print(\"Sorry, try again (and it was not a reducing error)\")\n\na.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"$$\\hskip-60pt\\frac{4}{9} \\times \\frac{1}{10} = \\frac{\\color{Blue}{b}}{45}$$",
"_____no_output_____"
]
],
[
[
"b = widgets.IntText(value=0, description='b =', disabled=False)\n\nIPython.display.display(b)\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(b)\n if b.value == 2:\n print(\"That's right! Great work!\")\n else:\n if b.value == 0:\n pass\n if b.value == 4:\n print(\"Not quite, don't forget to reduce.\")\n else:\n print(\"Sorry, try again (and it was not a reducing error)\")\n\nb.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"$$\\hskip-60pt\\frac{2}{3} \\times \\frac{9}{10} = \\frac{\\color{Purple}{c}}{\\color{Green}{d}}$$",
"_____no_output_____"
]
],
[
[
"c = widgets.IntText(value=0, description='c =', disabled=False)\n\nIPython.display.display(c)\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(c)\n if c.value == 3:\n print(\"That's right! Great work!\")\n else:\n if c.value == 0:\n pass\n if c.value == 18 or c.value == 9 or c.value == 6:\n print(\"Not quite, don't forget to reduce.\")\n else:\n print(\"Sorry, try again (and it was not a reducing error)\")\n\nc.observe(check, 'value')",
"_____no_output_____"
],
[
"d = widgets.IntText(value=0, description='d =', disabled=False)\n\nIPython.display.display(d)\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(d)\n if d.value == 5:\n print(\"That's right! Great work!\")\n else:\n if d.value == 0:\n pass\n if d.value == 30 or d.value == 15 or d.value == 10:\n print(\"Not quite, don't forget to reduce.\")\n else:\n print(\"Sorry, try again (and it was not a reducing error)\")\n\nd.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"## Dividing Positive Fractions\n\n### How do we divide using fractions?\n\nIf you know how to multiply fractions, then you already know how to divide fractions! \n\nLet's go back to dividing integers,or whole numbers. We know that 10 divided by 2 is 5, because 10 can be split into 2 portions each of which has 5. Now notice that dividing 10 by 2 is the same as taking half of 10. That is,\n$$\\hskip-60pt 10\\div 2 = 10\\times \\frac12 = 5.$$\n\n\nNotice that in the equality $10 \\div 2 = 10\\times\\frac{1}{2}$, we took the reciprocal of the second number ($\\frac{2}{1} \\rightarrow \\frac{1}{2}$) then changed the division to multiplication! What is the reciprocal? It's when the old numerator becomes the new denominator and the old denominator becomes the new numerator!\n\nThat's how division with fractions works too: flip the divisor, the number after the division, and then multiply! \n\n## The golden rule of division: *division is the same as multiplying by the reciprocal*\n\nFor example, what is $10\\div \\frac{1}{3}$? If we apply the rule we just came up with, we find\n$$\\hskip-60pt 10\\div \\frac13 = 10\\times\\frac31 = 10\\times 3 = 30.$$\n\nDoes this make sense? Recall that when we compute $10\\div 2=5$, what we're working out is the number of groups of 2 we need to make 10. Similarly, the expression $10\\div \\frac13$ is asking how many $\\frac{1}{3}$ pieces can fit into 10 pieces. Since it takes 3 $\\frac{1}{3}$s to make one and 10 ones to make 10, we need $3\\times 10 = 30$ pieces of size $\\frac{1}{3}$ to make 10, therefore $10 \\div \\frac{1}{3} = 10 \\times 3 = 30$.\n\n### Here are the steps\n\n#### Step 1:\nFind the reciprocal of the divisor. (By making the numerator the denominator and the denominator the numerator of the second fraction.)\n\n#### Step 2:\nFollow the steps to multiply the fractions using either method mentioned earlier.\n\n#### And you're done!\n\n<font color='red'>Tip! To remember these steps to divide fractions, think of this:</font>\n>**KFC**: \n>\n>**K** is for **K**eep the first fraction the same\n>\n>**F** is for **F**lip the second fraction\n>\n>**C** is for **C**hange the sign from divide to multiply\n\n## But why does this work?\n\nRemember that division is the opposite of multiplication: dividing by a number \"undoes\" multiplication by that same number. This has a special name, division is the *inverse operation* of multiplication. Here, we are interested in undoing multiplication by fractions. Let's start with an integer example. Consider the multiplication of 10 by 4: $10 \\times 4 = 40$. To undo the multiplication by 4, we divide by 4: $40 \\div 4 = 10$ and we get back to the original number! Same goes for fractions: consider the product $20 \\times \\frac{1}{2} = 10$. Dividing by $\\frac12$ should undo the multiplication. Our rule for division says that to divide by $\\frac{1}{2}$, we multiply by $\\frac21 = 2$: $10 \\div \\frac{1}{2} = 10\\times 2 = 20$. Once again, we get back to the original number!\n\nIf dividing by 4 is equivalent to multiplying by $\\frac{1}{4}$, then it's true that dividing by $\\frac{1}{4}$ is equivalent to multiplying by 4. \n\nWhat if you flip the first number instead of the second? Try it with an example we've already solved. Does it change the answer or does the order not matter like multiplication?\n\n## But isn't division supposed to make the number smaller?\n\nThe short answer is, \"Yes, but only if the number we're dividing by is bigger than 1.\" Try thinking of division as repeated subtraction: $2 \\div \\frac{1}{2}$ is asking how many times can we subtract $\\frac{1}{2}$ from 2. \n\n$$\\hskip-60pt \\begin{align*}\n2 - \\frac{1}{2} &= 1\\frac{1}{2}\\tag{once}\\\\[5pt]\n1\\frac{1}{2} - \\frac{1}{2} &= 1\\tag{twice}\\\\[5pt]\n1 - \\frac{1}{2} &= \\frac{1}{2}\\tag{three times}\\\\[5pt]\n\\frac{1}{2} - \\frac{1}{2} &= 0\\tag{four times}\n\\end{align*}$$\n\nSo it took 4 subtractions of $\\frac{1}{2}$ from 2 to reach 0, therefore $2 \\div \\frac{1}{2} = 4$.\n\nThis is because we are taking smaller portions out of the original number, so we need more subtractions to get to zero.\n",
"_____no_output_____"
],
[
"Here's another example. In this example, we perform division by multiplying by the reciprocal, and we use Method #2, reducing our fractions before carrying out the multiplication.\n<br>\n$$\\hskip-36pt\\large \\frac14\\div\\frac38 = \\frac14\\times\\frac83 = \\frac{1}{_{\\color{red}{1}}\\,4\\!\\!\\!\\!\\diagup}\\times\\frac{8\\!\\!\\!\\!\\diagup^{\\,\\,\\color{red}{2}}}{3} = \\frac23.$$",
"_____no_output_____"
],
[
"Still need another explanation? Here's a video that might help:",
"_____no_output_____"
]
],
[
[
"from IPython.display import YouTubeVideo\nYouTubeVideo('f3ySpxX9oeM')",
"_____no_output_____"
]
],
[
[
"### Let's go through an example:\n\n$$\\hskip-60pt\\begin{align*}\n\\color{blue}{\\frac{4}{5} \\div \\frac{2}{7}}\\\\\n\\frac{4}{5} \\div \\frac{2}{7} & = \\frac{4}{5} \\times \\frac{7}{2} \\tag{Invert the divisor}\\\\\n\\frac{4\\!\\!\\!\\diagup}{5} \\times \\frac{7}{2\\!\\!\\!\\diagup} & = \\frac{2}{5} \\times \\frac{7}{1} \\tag{Reduce numbers}\\\\\n\\frac{2}{5} \\times \\frac{7}{1} & = \\frac{14}{5} \\tag{Multiply numerators and denominators}\\\\\n\\frac{14}{5} & = 2 \\frac{4}{5} \\tag{Write as a mixed fraction}\n\\end{align*}$$",
"_____no_output_____"
],
[
"Here's a fill in the blank question. Please read which numbers need to be filled out correctly, and then enter the correct values below.\n\n$$\\hskip-60pt\\frac{2}{3} \\div \\frac{5}{7} = \\frac{2}{3} \\times \\frac{\\color{Blue}{A}}{\\color{Green}{B}} = \\frac{\\color{Purple}{C}}{\\color{Red}{D}}$$",
"_____no_output_____"
]
],
[
[
"A = widgets.IntText(value=0, description='A =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(A)\n if A.value == 7:\n print(\"That's right! Great work!\")\n else:\n if A.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(A) \nA.observe(check, 'value')",
"_____no_output_____"
],
[
"B = widgets.IntText(value=0, description='B =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(B)\n if B.value == 5:\n print(\"That's right! Great work!\")\n else:\n if B.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(B)\nB.observe(check, 'value')",
"_____no_output_____"
],
[
"C = widgets.IntText(value=0, description='C =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(C)\n if C.value == 14:\n print(\"That's right! Great work!\")\n else:\n if C.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(C)\nC.observe(check, 'value')",
"_____no_output_____"
],
[
"D = widgets.IntText(value=0, description='D =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(D)\n if D.value == 15:\n print(\"That's right! Great work!\")\n else:\n if D.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(D)\nD.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"Here's another fill in the blank question. Please read which numbers need to be filled out correctly. <br>\nNote: there is a reduction in this one!\n\n$$\\hskip-60pt\\frac{4}{9} \\div \\frac{5}{3} = \\frac{4}{9} \\times \\frac{3}{5} = \\frac{4}{\\color{Blue}{E}} \\times \\frac{\\color{Green}{F}}{5} = \\frac{\\color{Purple}{G}}{\\color{Red}{H}}$$",
"_____no_output_____"
]
],
[
[
"E = widgets.IntText(value=0, description='E =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(E)\n if E.value == 3:\n print(\"That's right! Great work!\")\n else:\n if E.value == 0:\n pass\n if E.value == 9:\n print(\"Don't forget to reduce.\")\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(E) \nE.observe(check, 'value')",
"_____no_output_____"
],
[
"F = widgets.IntText(value=0, description='F =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(F)\n if F.value == 1:\n print(\"That's right! Great work!\")\n else:\n if F.value == 0:\n pass\n if F.value == 3:\n print(\"Don't forget to reduce.\")\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(F) \nF.observe(check, 'value')",
"_____no_output_____"
],
[
"G = widgets.IntText(value=0, description='G =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(G)\n if G.value == 4:\n print(\"That's right! Great work!\")\n else:\n if G.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(G) \nG.observe(check, 'value')",
"_____no_output_____"
],
[
"H = widgets.IntText(value=0, description='H =', disabled=False)\n\ndef check(q):\n IPython.display.clear_output(wait=False)\n IPython.display.display(H)\n if H.value == 15:\n print(\"That's right! Great work!\")\n else:\n if H.value == 0:\n pass\n else:\n print(\"Sorry, try again.\")\n \nIPython.display.display(H) \nH.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"## Multiplication and Division with Positive Mixed Fractions\n\nWith mixed numbers, we have to do a little more work, but it's only one more step:\n\n### For multiplication and division, all mixed numbers must be converted to improper fractions.\n\n**This notebook is assuming that you already know how to convert between mixed numbers and improper fractions**\n\nRemember that a mixed number is really adding an integer and a proper fraction: $3\\,\\frac14$ is really $3+\\frac14$ as one number. A proper fraction is where the numerator is less than the denominator. An improper fraction is where the numerator is bigger than or equal to the denominator.\n\nFor example, consider the product $\\frac45\\times 3\\,\\frac14$. So one way to carry out the product is to use the distributive property:\n\n$$\\hskip-60pt\\begin{align*}\n\\frac45 \\times 3\\,\\frac14 &= \\frac45(3+\\frac14)\\\\\n& = \\frac45\\times 3+\\frac45\\times\\frac14\\\\\n&= \\frac{12}{5}+\\frac15\\\\\n&= \\frac{13}{5}.\n\\end{align*}$$\n\n(Note that we've made use of two skills practised above, without writing them down: we multiplied by 3 by thinking of it as the fraction $\\frac31$, and we cancelled the fours in the product $\\frac45\\times \\frac14$.)\n\nIf it is asked for, we can do one final step, and convert back to a mixed fraction: $\\dfrac{13}{5} = 2\\,\\dfrac{3}{5}$. But that was quite a bit of work. *There must be a better way!* \n\nInstead, let's first convert our mixed number to an improper fraction:\n\n$$3\\,\\frac14 = 3+\\frac14 = 3\\times\\frac44+\\frac14 = \\frac{12}{4}+\\frac{1}{4} = \\frac{13}{4}.$$\n\nNow we can multiply using the methods we learned above:\n\n$$\\frac45\\times 3\\,\\frac14 = \\frac45\\times\\frac{13}{4} = \\frac15\\times\\frac{13}{1}=\\frac{13}{5},$$\n\nas before. If specified in the problem (or by your teacher), you can take one more step and convert the improper fraction $\\frac{13}{5}$ into the mixed number $2\\,\\frac35$.\n<br>\n\nLet's review the steps:\n\n##### Step 1:\n$\\color{red}{\\textrm{Convert all mixed numbers into improper fractions.}}$\n\n##### Step 2:\nMultiply or divide as using the methods earlier.\n\n##### Step 3: (Optional?)\nIf the result is an improper fraction, convert it back to a mixed number.\n\n##### And you're done!\n\n### Let's do some examples!\n\n1. Calculate the product $\\color{blue}{2 \\dfrac{2}{3} \\times 1 \\dfrac{3}{4}}$.<br><br>\n$$\\hskip-60pt\\begin{align*}\n2 \\frac{2}{3} \\times 1 \\frac{3}{4} & = \\frac{8}{3} \\times \\frac{7}{4} \\tag{Convert to improper fractions}\\\\[5pt]\n\\frac{8\\!\\!\\!\\diagup}{3} \\times \\frac{7}{4\\!\\!\\!\\diagup} & = \\frac{2}{3} \\times \\frac{7}{1} \\tag{Reduce numbers}\\\\[5pt]\n\\frac{2}{3} \\times \\frac{7}{1} & = \\frac{14}{3} \\tag{Multiply numerators and denomintators}\\\\[5pt]\n\\frac{14}{3} & = 4 \\frac{2}{3} \\tag{Write as a mixed fraction}\n\\end{align*}$$<br>\n2. Calculate the quotient $\\color{blue}{2 \\dfrac{2}{3} \\div 1 \\dfrac{3}{4}}$. (These are the same fractions as the first example.)<br><br>\n$$\\hskip-60pt\\begin{align*}\n2 \\frac{2}{3} \\div 1 \\frac{3}{4} & = \\frac{8}{3} \\div \\frac{7}{4} \\tag{Convert to improper fractions}\\\\[5pt]\n\\frac{8}{3} \\div \\frac{7}{4} & = \\frac{8}{3} \\times \\frac{4}{7} \\tag{Invert the divisor}\\\\[5pt]\n\\frac{8}{3} \\times \\frac{4}{7} & = \\frac{32}{21} \\tag{Multiply numerators and denomintators}\\\\[5pt]\n\\frac{32}{21} & = 1 \\frac{11}{21} \\tag{Write as a mixed fraction}\n\\end{align*}$$<br>\n3. Calculate the quotient $\\color{blue}{3 \\frac{1}{2} \\div 1 \\frac{1}{6}}$. (This time some reduction is required.)<br><br>\n$$\\hskip-60pt\\begin{align*}\n3 \\frac{1}{2} \\div 1 \\frac{1}{6} & = \\frac{7}{2} \\div \\frac{7}{6} \\tag{Convert to improper fractions}\\\\[5pt]\n\\frac{7}{2} \\div \\frac{7}{6} & = \\frac{7}{2} \\times \\frac{6}{7} \\tag{Invert the divisor}\\\\[5pt]\n\\frac{7\\!\\!\\!\\diagup}{2} \\times \\frac{6}{7\\!\\!\\!\\diagup} & = \\frac{1}{2} \\times \\frac{6}{1} \\tag{Reduce numbers}\\\\[5pt]\n\\frac{1}{2\\!\\!\\!\\diagup} \\times \\frac{6\\!\\!\\!\\diagup}{1} & = \\frac{1}{1} \\times \\frac{3}{1} \\tag{Reduce numbers again}\\\\[5pt]\n\\frac{1}{1} \\times \\frac{3}{1} & = \\frac{3}{1} \\tag{Multiply numerators and denomintators}\\\\[5pt]\n\\frac{3}{1} & = 3 \\tag{Write as an integer}\n\\end{align*}$$",
"_____no_output_____"
],
[
"### Practice Questions\n\nNow that you've seen a few examples, it's time to try some on your own. But remember: two problems worth of practice is not enough to master this skill. Make sure you take the time to work through additional problems from your teacher or textbook.\n\nGrab some extra paper and a pencil to go through the steps in this notebook to find the answer, then check to see if you're right.\n\n#### Question 1\n\n$2\\frac{1}{2} \\times 1\\frac{1}{2}$",
"_____no_output_____"
]
],
[
[
"from fractions import Fraction\n\nanswer = widgets.RadioButtons(options=['','3 3/4', '2 1/4','4','1 2/3'],\n value='', description= 'Answer')\n\ndef check(a):\n IPython.display.clear_output(wait=False)\n IPython.display.display(answer)\n if answer.value == '3 3/4':\n print(\"Correct! Great job!\")\n else:\n if answer.value == '':\n pass\n else:\n print(\"Sorry, that's not right, try again. Don't forget to convert the fraction to an improper fraction.\")\n\nIPython.display.display(answer)\nanswer.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"#### Question 2\n\n$3\\frac{1}{3} \\div 2\\frac{1}{6}$",
"_____no_output_____"
]
],
[
[
"answer2 = widgets.RadioButtons(options=['','7 2/9', '2 1/2','1 7/13','3 1/2'],\n value='', description= 'Answer')\n\ndef check(a):\n IPython.display.clear_output(wait=False)\n IPython.display.display(answer2)\n if answer2.value == '1 7/13':\n print(\"Correct! Great job!\")\n else:\n if answer.value == '':\n pass\n else:\n print(\"Sorry, that's not right, try again. Don't forget to convert the fraction to an improper fraction and invert the divisor.\")\n\nIPython.display.display(answer2)\nanswer2.observe(check, 'value')",
"_____no_output_____"
]
],
[
[
"## Summary\n\nLet's review what we've learned in this notebook.\n\n- Unlike addition, multiplying fractions *does not* need common denominators. **DM** – **D**ivision/**M**ultiplication = Denominator **D**oesn’t **M**atter \n- To multiply to fractions, multiply the numerators, then the denominators, and then reduce if necessary.\n- It is often easier if you cancel common factors before multiplying the fractions.\n- Division is the inverse of multiplication. Remember the Golden Rule: division is the same as multiplying by the reciprocal! (flipping the numerator and denominator) **KFC** - **K**eep, **F**lip, **C**hange\n- Multiplication and division involving mixed numbers is easier if you first convert any mixed numbers to improper fractions.\n\nTake the time to master your fraction multiplication and division. It may not seem like it now, but this is a much needed skill that you'll rely on for many years to come. Many careers even use fractions on a daily basis, like chefs or construction workers. As always in math, the more you practice, the easier it gets!\n",
"_____no_output_____"
],
[
"[](https://github.com/callysto/curriculum-notebooks/blob/master/LICENSE.md)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
ec72fd9cb169029d5e61b58ce6889ace0a622e75 | 8,623 | ipynb | Jupyter Notebook | notebook/integrate_azureml.ipynb | GqiqiPS/FLAML | f3fc35c438c137bb2b2f118ffbc0d890c5432da3 | [
"MIT"
]
| 1,747 | 2020-12-05T00:14:58.000Z | 2022-03-31T20:54:09.000Z | notebook/flaml_azureml.ipynb | panda4698/FLAML | 3cf8cb0761e250fb01cfecbbf8ef403e487b8c89 | [
"MIT"
]
| 229 | 2020-12-14T06:19:16.000Z | 2022-03-31T05:20:27.000Z | notebook/flaml_azureml.ipynb | sonichi/FLAML | 17b17d084ff1d1daf28090ce18039fc195f995d7 | [
"MIT"
]
| 260 | 2020-12-13T09:24:41.000Z | 2022-03-27T04:09:51.000Z | 26.289634 | 380 | 0.568016 | [
[
[
"Copyright (c) 2020-2021 Microsoft Corporation. All rights reserved. \n\nLicensed under the MIT License.\n\n# Run FLAML in AzureML\n\n\n## 1. Introduction\n\nFLAML is a Python library (https://github.com/microsoft/FLAML) designed to automatically produce accurate machine learning models \nwith low computational cost. It is fast and cheap. The simple and lightweight design makes it easy \nto use and extend, such as adding new learners. FLAML can \n- serve as an economical AutoML engine,\n- be used as a fast hyperparameter tuning tool, or \n- be embedded in self-tuning software that requires low latency & resource in repetitive\n tuning tasks.\n\nIn this notebook, we use one real data example (binary classification) to showcase how to use FLAML library together with AzureML.\n\nFLAML requires `Python>=3.6`. To run this notebook example, please install flaml with the `notebook` and `azureml` option:\n```bash\npip install flaml[notebook,azureml]\n```",
"_____no_output_____"
]
],
[
[
"!pip install flaml[notebook,azureml]",
"_____no_output_____"
]
],
[
[
"### Enable mlflow in AzureML workspace",
"_____no_output_____"
]
],
[
[
"import mlflow\nfrom azureml.core import Workspace\n\nws = Workspace.from_config()\nmlflow.set_tracking_uri(ws.get_mlflow_tracking_uri())",
"_____no_output_____"
]
],
[
[
"## 2. Classification Example\n### Load data and preprocess\n\nDownload [Airlines dataset](https://www.openml.org/d/1169) from OpenML. The task is to predict whether a given flight will be delayed, given the information of the scheduled departure.",
"_____no_output_____"
]
],
[
[
"from flaml.data import load_openml_dataset\nX_train, X_test, y_train, y_test = load_openml_dataset(dataset_id=1169, data_dir='./')",
"_____no_output_____"
]
],
[
[
"### Run FLAML\nIn the FLAML automl run configuration, users can specify the task type, time budget, error metric, learner list, whether to subsample, resampling strategy type, and so on. All these arguments have default values which will be used if users do not provide them. For example, the default ML learners of FLAML are `['lgbm', 'xgboost', 'catboost', 'rf', 'extra_tree', 'lrl1']`. ",
"_____no_output_____"
]
],
[
[
"''' import AutoML class from flaml package '''\nfrom flaml import AutoML\nautoml = AutoML()",
"_____no_output_____"
],
[
"settings = {\n \"time_budget\": 60, # total running time in seconds\n \"metric\": 'accuracy', # can be: 'r2', 'rmse', 'mae', 'mse', 'accuracy', 'roc_auc', 'roc_auc_ovr',\n # 'roc_auc_ovo', 'log_loss', 'mape', 'f1', 'ap', 'ndcg', 'micro_f1', 'macro_f1'\n \"estimator_list\": ['lgbm', 'rf', 'xgboost'], # list of ML learners\n \"task\": 'classification', # task type \n \"sample\": False, # whether to subsample training data\n \"log_file_name\": 'airlines_experiment.log', # flaml log file\n}",
"_____no_output_____"
],
[
"mlflow.set_experiment(\"flaml\")\nwith mlflow.start_run() as run:\n '''The main flaml automl API'''\n automl.fit(X_train=X_train, y_train=y_train, **settings)",
"_____no_output_____"
]
],
[
[
"### Best model and metric",
"_____no_output_____"
]
],
[
[
"''' retrieve best config and best learner'''\nprint('Best ML leaner:', automl.best_estimator)\nprint('Best hyperparmeter config:', automl.best_config)\nprint('Best accuracy on validation data: {0:.4g}'.format(1 - automl.best_loss))\nprint('Training duration of best run: {0:.4g} s'.format(automl.best_config_train_time))",
"_____no_output_____"
],
[
"automl.model",
"_____no_output_____"
],
[
"''' pickle and save the automl object '''\nimport pickle\nwith open('automl.pkl', 'wb') as f:\n pickle.dump(automl, f, pickle.HIGHEST_PROTOCOL)",
"_____no_output_____"
],
[
"''' compute predictions of testing dataset ''' \ny_pred = automl.predict(X_test)\nprint('Predicted labels', y_pred)\nprint('True labels', y_test)\ny_pred_proba = automl.predict_proba(X_test)[:,1]",
"_____no_output_____"
],
[
"''' compute different metric values on testing dataset'''\nfrom flaml.ml import sklearn_metric_loss_score\nprint('accuracy', '=', 1 - sklearn_metric_loss_score('accuracy', y_pred, y_test))\nprint('roc_auc', '=', 1 - sklearn_metric_loss_score('roc_auc', y_pred_proba, y_test))\nprint('log_loss', '=', sklearn_metric_loss_score('log_loss', y_pred_proba, y_test))",
"_____no_output_____"
]
],
[
[
"### Log history",
"_____no_output_____"
]
],
[
[
"from flaml.data import get_output_from_log\ntime_history, best_valid_loss_history, valid_loss_history, config_history, metric_history = \\\n get_output_from_log(filename = settings['log_file_name'], time_budget = 60)\n\nfor config in config_history:\n print(config)",
"_____no_output_____"
],
[
"import matplotlib.pyplot as plt\nimport numpy as np\n\nplt.title('Learning Curve')\nplt.xlabel('Wall Clock Time (s)')\nplt.ylabel('Validation Accuracy')\nplt.scatter(time_history, 1 - np.array(valid_loss_history))\nplt.step(time_history, 1 - np.array(best_valid_loss_history), where='post')\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec7303addfd9c53284637afbd1d7dd6db70f0d30 | 64,437 | ipynb | Jupyter Notebook | archives/correlation_analysis_celllevel/.ipynb_checkpoints/correlation_analysis_celllevel-generate-grand-summary-Jan8-checkpoint.ipynb | mukamel-lab/SingleCellFusion_EnhancerPaper | acbfa5184667ca57c333c04c310b0712a0e8e15e | [
"MIT"
]
| null | null | null | archives/correlation_analysis_celllevel/.ipynb_checkpoints/correlation_analysis_celllevel-generate-grand-summary-Jan8-checkpoint.ipynb | mukamel-lab/SingleCellFusion_EnhancerPaper | acbfa5184667ca57c333c04c310b0712a0e8e15e | [
"MIT"
]
| null | null | null | archives/correlation_analysis_celllevel/.ipynb_checkpoints/correlation_analysis_celllevel-generate-grand-summary-Jan8-checkpoint.ipynb | mukamel-lab/SingleCellFusion_EnhancerPaper | acbfa5184667ca57c333c04c310b0712a0e8e15e | [
"MIT"
]
| 1 | 2021-11-15T19:03:03.000Z | 2021-11-15T19:03:03.000Z | 92.982684 | 44,628 | 0.802784 | [
[
[
"import sys\nsys.path.insert(0, \"/cndd/fangming/CEMBA/snmcseq_dev\")\n\nfrom multiprocessing import Pool,cpu_count\nfrom functools import partial\n\nfrom scipy import sparse\nfrom scipy import stats\nimport importlib\nfrom __init__ import *\nfrom __init__jupyterlab import *\nimport snmcseq_utils\nimportlib.reload(snmcseq_utils)\n\nimport re\nimport pickle\nimport datetime\nfrom scipy import optimize\n\nimport tqdm\n\nimport importlib\nsys.path.insert(0, '../')\nimport enhancer_gene_utils\nimportlib.reload(enhancer_gene_utils)",
"_____no_output_____"
],
[
"today = datetime.date.today()\nprint(today)",
"2021-01-08\n"
]
],
[
[
"# Get grand summary \n### - 80% cells for 5-time downsampling",
"_____no_output_____"
]
],
[
[
"input_enh_gene_table = '/cndd2/fangming/projects/scf_enhancers/results/200521_to_evals.tsv' \noutput_res = '/cndd2/fangming/projects/scf_enhancers/results/{}_metacell_corr_res_{{}}'.format(today)\nprint(output_res)",
"/cndd2/fangming/projects/scf_enhancers/results/2021-01-08_metacell_corr_res_{}\n"
],
[
"# enhancer-gene linkage\nti = time.time()\nenhancer_gene_to_eval = (pd.read_csv(input_enh_gene_table, sep='\\t')\n [['gene', 'ens', 'dist']]\n .rename(columns={'ens': 'enh'})\n )\n\nprint(enhancer_gene_to_eval.shape)\nprint(time.time()-ti)\nenhancer_gene_to_eval.head()",
"(2589994, 3)\n6.211456298828125\n"
],
[
"# get all clusterings\ndef get_isub_clstfile(fname):\n \"\"\"Subsampling index\n \"\"\"\n pattern = '_sub[0-9]+.tsv.gz' \n isub = int(re.findall(pattern, fname)[0][len(\"_sub\"):-len('.tsv.gz')])\n return isub \n\ndef get_knn_clstfile(fname):\n \"\"\"cross-modal knn\n \"\"\"\n pattern = '_knn[0-9]+_' \n knn = int(re.findall(pattern, fname)[0][len(\"_knn\"):-len('_')])\n return knn \n\nf_pattern = (\"/cndd2/fangming/projects/miniatlas/results/\"\n \"clusterings_10x_cells_v3_mop_10x_cells_v3_snatac_gene_ka30_knn*_201130_sub*.tsv.gz\"\n )\nfnames = np.sort(glob.glob(f_pattern))\niterator_clsts = pd.DataFrame([{'i': get_isub_clstfile(fname),\n 'k': get_knn_clstfile(fname),\n 'fname': fname,\n } for fname in fnames])\nprint(iterator_clsts.shape)\n\nres_clsts = []\nfor idx, row in tqdm.tqdm(iterator_clsts.iterrows()):\n isub = row['i']\n knn = row['k']\n fname = row['fname']\n \n df = pd.read_csv(fname, index_col=0, sep='\\t')\n for col in df.columns:\n r = float(col[len('cluster_r'):])\n num_clst = len(df[col].unique())\n res_clsts.append({\n 'k': knn,\n 'i': isub,\n 'r': r,\n 'num_clst': num_clst,\n })\n# break\nres_clsts = pd.DataFrame(res_clsts)\n\nprint(res_clsts.shape)\nres_clsts.head()",
"0it [00:00, ?it/s]"
],
[
"# a little hack\nres_clsts['rM'] = (res_clsts['r']*1e6).astype(int)",
"_____no_output_____"
],
[
"fig, ax = plt.subplots()\nax.plot(res_clsts['r'].values, res_clsts['num_clst'].values, 'o', markersize=5)\nax.set_xscale('log')\nax.set_yscale('log')\nax.set_xlabel('Resolution (r)')\nax.set_ylabel('Num clusters')\nplt.show()",
"_____no_output_____"
],
[
"bins = np.linspace(-1,1,201)\nfdr_threshold = 0.2\ndistance_threshold = 1e5\nlabel = 'pairs (<100kb)'\n\ncol_orders = [\n 'num_linked_pairs',\n 'num_linked_genes',\n 'num_linked_enhs',\n 'id_linked_pairs',\n \n 'num_correlated_pairs',\n 'num_correlated_genes',\n 'num_correlated_enhs',\n 'id_correlated_pairs',\n \n 'num_total_pairs',\n 'num_total_genes',\n 'num_total_enhs',\n 'id_total_pairs',\n \n 'r_th_linked', \n 'r_th_correlated_left',\n 'r_th_correlated_right',\n ]",
"_____no_output_____"
],
[
"# get all (shared) corr analyses\ndef get_resolution(fname):\n \"\"\"Resolution\n \"\"\"\n resolution_pattern = '_cluster_r[0-9.]+_' \n r = float(re.findall(resolution_pattern, fname)[0][len(\"_cluster_r\"):-len('_')])\n return r\n \ndef get_isub(fname):\n \"\"\"Subsampling index\n \"\"\"\n pattern = '_[0-9]_cluster' \n isub = int(re.findall(pattern, fname)[0][len(\"_\"):-len('_cluster')])\n return isub \n\ndef get_knn(fname):\n \"\"\"cross-modal knn\n \"\"\"\n pattern = '_knn[0-9]+_' \n knn = int(re.findall(pattern, fname)[0][len(\"_knn\"):-len('_')])\n return knn \n\n# mC\nf_pattern = (\"/cndd2/fangming/projects/scf_enhancers/results/\"\n \"mop_10x_cells_v3_snmcseq_gene_ka30_knn*_201130\"\n \"_[0-9]_cluster_r*_spearmanr_corrs.pkl\"\n )\nfnames = np.sort(glob.glob(f_pattern))\niterator_mc = pd.DataFrame([{'r': get_resolution(fname),\n 'i': get_isub(fname),\n 'k': get_knn(fname),\n 'fname_mc': fname,\n } for fname in fnames])\nprint(iterator_mc.shape)\n\n# ATAC\nf_pattern = (\"/cndd2/fangming/projects/scf_enhancers/results/\"\n \"mop_10x_cells_v3_snatac_gene_ka30_knn*_201130\"\n \"_[0-9]_cluster_r*_spearmanr_corrs.pkl\"\n )\nfnames = np.sort(glob.glob(f_pattern))\niterator_atac = pd.DataFrame([{'r': get_resolution(fname),\n 'i': get_isub(fname),\n 'k': get_knn(fname),\n 'fname_atac': fname,\n } for fname in fnames])\nprint(iterator_atac.shape)\n\n# both\niterator_both = pd.merge(iterator_mc, iterator_atac, on=['k', 'r', 'i'])\nprint(iterator_both.shape)\niterator_both.head()",
"(750, 4)\n(750, 4)\n(750, 5)\n"
],
[
"# calc corr\niterator_both = iterator_both.sort_values(['r', 'i'])\n# ## test\n# res_corr = enhancer_gene_utils.get_corr_stats(iterator_both.iloc[[0, 1]], \n# enhancer_gene_to_eval, col_orders) \n# ## end of test\n\nres_corr = enhancer_gene_utils.get_corr_stats(iterator_both, \n enhancer_gene_to_eval, col_orders) \niterator_both = iterator_both.join(res_corr)\n# a little hack to avoid merge on float number\niterator_both['rM'] = (iterator_both['r']*1e6).astype(int)\n\n# merge with res_clsts\niterator_both = pd.merge(iterator_both, res_clsts.drop('r', axis=1), on=['rM', 'i', 'k'], how='inner')",
"25it [00:41, 2.08s/it]"
],
[
"# save this bit\noutput = output_res.format('grand_res_summary_table.tsv.gz')\nprint(output)\niterator_both.to_csv(output, sep='\\t', header=True, index=False)",
"_____no_output_____"
],
[
"iterator_both = pd.read_csv(output, sep='\\t')\nprint(iterator_both.shape)\niterator_both.head()",
"_____no_output_____"
],
[
"iterator_both.columns",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec730a280f35ef7d7cb556b9896a267a42cf2b62 | 332,669 | ipynb | Jupyter Notebook | pyddm-exploration/demo.ipynb | OmkarMehta/SSFwithNODE | 99bf8992839b3c07b6c8642a0d2b6c6f1b329d2e | [
"MIT"
]
| 1 | 2021-11-13T22:32:29.000Z | 2021-11-13T22:32:29.000Z | pyddm-exploration/demo.ipynb | OmkarMehta/SSFwithNODE | 99bf8992839b3c07b6c8642a0d2b6c6f1b329d2e | [
"MIT"
]
| null | null | null | pyddm-exploration/demo.ipynb | OmkarMehta/SSFwithNODE | 99bf8992839b3c07b6c8642a0d2b6c6f1b329d2e | [
"MIT"
]
| null | null | null | 264.653142 | 26,172 | 0.790639 | [
[
[
"System requirements\n\n* Python 3.5 or higher\n* Scipy/numpy\n* Paranoid scientist (pip install paranoid-scientist)\n* For plotting features, matplotlib\n* For parallelization support, pathos\n\n## Hello World!\n",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nfrom ddm import Model\nm = Model()\ns = m.solve() # Simulate the model using the Model.solve() method to generate a Solution object\nplt.plot(s.model.t_domain(), s.pdf_corr())\nplt.savefig(\"helloworld.png\")\nplt.show()",
"_____no_output_____"
],
[
"s.model",
"_____no_output_____"
],
[
"help(Model)",
"Help on class Model in module ddm.model:\n\nclass Model(builtins.object)\n | Model(drift=DriftConstant(drift=0), noise=NoiseConstant(noise=1), bound=BoundConstant(B=1), IC=ICPointSourceCenter(), overlay=OverlayNone(), name='', dx=0.005, dt=0.005, T_dur=2.0, fitresult=None)\n | \n | A full simulation of a single DDM-style model.\n | \n | Each model simulation depends on five key components:\n | \n | - A description of how drift rate (drift) changes throughout the simulation.\n | - A description of how variability (noise) changes throughout the simulation.\n | - A description of how the boundary changes throughout the simulation.\n | - Starting conditions for the model\n | - Specific details of a task which cause dynamic changes in the model (e.g. a stimulus intensity change)\n | \n | This class manages these, and also provides the affiliated\n | services, such as analytical or numerical simulations of the\n | resulting reaction time distribution.\n | \n | Methods defined here:\n | \n | IC(self, conditions)\n | The initial distribution at t=0.\n | \n | Returns a length N ndarray (where N is the size of x_domain())\n | which should sum to 1.\n | \n | __init__(self, drift=DriftConstant(drift=0), noise=NoiseConstant(noise=1), bound=BoundConstant(B=1), IC=ICPointSourceCenter(), overlay=OverlayNone(), name='', dx=0.005, dt=0.005, T_dur=2.0, fitresult=None)\n | Construct a Model object from the 5 key components.\n | \n | The five key components of our DDM-style models describe how\n | the drift rate (`drift`), noise (`noise`), and bounds (`bound`)\n | change over time, and the initial conditions (`IC`).\n | \n | These five components are given by the parameters `drift`,\n | `noise`, `bound`, and `IC`, respectively. They should be\n | types which inherit from the types Drift, Noise, Bound, and\n | InitialCondition, respectively. They default to constant\n | unitary values.\n | \n | Additionally, simulation parameters can be set, such as time\n | and spacial precision (`dt` and `dx`) and the simulation\n | duration `T_dur`. If not specified, they will be taken from\n | the defaults specified in the parameters file.\n | \n | If you are creating a model, `fitresult` should always be\n | None. This is provided as an optional parameter because when\n | models are output as a string (using \"str\" or \"repr\"), they\n | must save fitting information. Thus, this allows you to fit a\n | model, convert it to a string, save that string to a text\n | file, and then run \"exec\" on that file in a new script to load\n | the model.\n | \n | The `name` parameter is exclusively for convenience, and may\n | be used in plotting or in debugging.\n | \n | __repr__(self, pretty=False)\n | Return repr(self).\n | \n | __str__(self)\n | Return str(self).\n | \n | can_solve_cn(self, conditions={})\n | Check whether this model is compatible with Crank-Nicolson solver.\n | \n | All bound functions which do not depend on time are compatible.\n | \n | can_solve_explicit(self, conditions={})\n | Check explicit method stability criterion\n | \n | check_conditions_satisfied(self, conditions)\n | \n | flux(self, x, t, conditions)\n | The flux across the boundary at position `x` at time `t`.\n | \n | get_dependence(self, name)\n | Return the dependence object given by the string `name`.\n | \n | get_fit_result(self)\n | Returns a FitResult object describing how the model was fit.\n | \n | Returns the FitResult object describing the last time this\n | model was fit to data, including the loss function, fitting\n | method, and the loss function value. If the model was never\n | fit to data, this will return FitResultEmpty.\n | \n | get_model_parameter_names(self)\n | Get an ordered list of the names of all parameters in the model.\n | \n | Returns the name of each model parameter. The ordering is\n | arbitrary, but is uaranteed to be in the same order as\n | get_model_parameters() and set_model_parameters(). If multiple\n | parameters refer to the same \"Fittable\" object, then that\n | object will only be listed once, however the names of the\n | parameters will be separated by a \"/\" character.\n | \n | get_model_parameters(self)\n | Get an ordered list of all model parameters.\n | \n | Returns a list of each model parameter which can be varied\n | during a fitting procedure. The ordering is arbitrary but is\n | guaranteed to be in the same order as\n | get_model_parameter_names() and set_model_parameters(). If\n | multiple parameters refer to the same \"Fittable\" object, then\n | that object will only be listed once.\n | \n | get_model_type(self)\n | Return a dictionary which fully specifies the class of the five key model components.\n | \n | has_analytical_solution(self)\n | Is it possible to find an analytic solution for this model?\n | \n | set_model_parameters(self, params)\n | Set the parameters of the model from an ordered list.\n | \n | Takes as an argument a list of parameters in the same order as\n | those from get_model_parameters(). Sets the associated\n | parameters as a \"Fitted\" object. If multiple parameters refer\n | to the same \"Fittable\" object, then that object will only be\n | listed once.\n | \n | simulate_trial(self, conditions={}, cutoff=True, rk4=True, seed=0)\n | Simulate the decision variable for one trial.\n | \n | Given conditions `conditions`, this function will simulate the\n | decision variable for a single trial. It will cut off the\n | simulation when the decision variable crosses the boundary\n | unless `cutoff` is set to False. By default, Runge-Kutta is\n | used to simulate the trial, however if `rk4` is set to False,\n | the less efficient Euler's method is used instead. This\n | returns a trajectory of the simulated trial over time as a\n | numpy array.\n | \n | Note that this will return the same trajectory on each run\n | unless the random seed `seed` is varied.\n | \n | Also note that you shouldn't normally need to use this\n | function. To simulate an entire probability distributions,\n | call Model.solve() and the results of the simulation will be\n | in the returned Solution object. This is only useful for\n | finding individual trajectories instead of the probability\n | distribution as a whole.\n | \n | simulated_solution(self, conditions={}, size=1000, rk4=True, seed=0)\n | Simulate individual trials to obtain a distribution.\n | \n | Given conditions `conditions` and the number `size` of trials\n | to simulate, this will run the function \"simulate_trial\"\n | `size` times, and use the result to find a histogram analogous\n | to solve. Returns a Sample object.\n | \n | Note that in practice you should never need to use this\n | function. This function uses an outdated method to simulate\n | the model and should be used for comparison perposes only. To\n | produce a probability density function of boundary crosses,\n | use Model.solve(). To sample from the probability\n | distribution (e.g. for finding confidence intervals for\n | limited amounts of data), call Model.solve() and then use the\n | Solution.resample() function of the resulting Solution.\n | \n | solve(self, conditions={}, return_evolution=False)\n | Solve the model using an analytic solution if possible, and a numeric solution if not.\n | \n | Return a Solution object describing the joint PDF distribution of reaction times.\n | \n | solve_analytical(self, conditions={})\n | Solve the model with an analytic solution, if possible.\n | \n | Analytic solutions are only possible in a select number of\n | special cases; in particular, it works for simple DDM and for\n | linearly collapsing bounds and arbitrary single-point initial \n | conditions. (See Anderson (1960) for implementation details.) \n | For most reasonably complex models, the method will fail. \n | Check whether a solution is possible with has_analytic_solution().\n | \n | If successful, this returns a Solution object describing the\n | joint PDF. If unsuccessful, this will raise an exception.\n | \n | solve_numerical(self, method='cn', conditions={}, return_evolution=False)\n | Solve the DDM model numerically.\n | \n | Use `method` to solve the DDM. `method` can either be\n | \"explicit\", \"implicit\", or \"cn\" (for Crank-Nicolson). This is\n | the core DDM solver of this library.\n | \n | Crank-Nicolson is the default and works for any model with\n | constant bounds.\n | \n | Implicit is the fallback method. It should work well in most\n | cases and is generally stable.\n | \n | Normally, the explicit method should not be used. Also note\n | the stability criteria for explicit method is:\n | \n | | noise^2/2 * dt/dx^2 < 1/2\n | \n | It returns a Solution object describing the joint PDF. This\n | method should not fail for any model type.\n | \n | return_evolution(default=False) governs whether or not the function \n | returns the full evolution of the pdf as part of the Solution object. \n | This only works with methods \"explicit\" or \"implicit\", not with \"cn\".\n | \n | solve_numerical_cn(self, conditions={})\n | Solve the DDM model numerically using Crank-Nicolson.\n | \n | This uses the Crank Nicolson method to solve the DDM at each\n | timepoint. Results are then compiled together. This is the\n | core DDM solver of this library.\n | \n | It returns a Solution object describing the joint PDF.\n | \n | solve_numerical_explicit(self, conditions={}, **kwargs)\n | Solve the model using the explicit method (Forward Euler).\n | \n | solve_numerical_implicit(self, conditions={}, **kwargs)\n | Solve the model using the implicit method (Backward Euler).\n | \n | t_domain(self)\n | A list of all of the timepoints over which the joint PDF will be defined (increments of dt from 0 to T_dur).\n | \n | x_domain(self, conditions, t=None)\n | A list which spans from the lower boundary to the upper boundary by increments of dx.\n | \n | ----------------------------------------------------------------------\n | Data descriptors defined here:\n | \n | __dict__\n | dictionary for instance variables (if defined)\n | \n | __weakref__\n | list of weak references to the object (if defined)\n | \n | ----------------------------------------------------------------------\n | Data and other attributes defined here:\n | \n | __slotnames__ = []\n\n"
],
[
"from ddm import Model\nfrom ddm.models import DriftConstant, NoiseConstant, BoundConstant, OverlayNonDecision\nfrom ddm.functions import fit_adjust_model, display_model\n\nmodel = Model(name='Simple model',\n drift=DriftConstant(drift=2.2),\n noise=NoiseConstant(noise=1.5),\n bound=BoundConstant(B=1.1),\n overlay=OverlayNonDecision(nondectime=.1),\n dx=.001, dt=.01, T_dur=2)\ndisplay_model(model)\nsol = model.solve()",
"Model Simple model information:\nDrift component DriftConstant:\n constant\n Fixed parameters:\n - drift: 2.200000\nNoise component NoiseConstant:\n constant\n Fixed parameters:\n - noise: 1.500000\nBound component BoundConstant:\n constant\n Fixed parameters:\n - B: 1.100000\nIC component ICPointSourceCenter:\n point_source_center\n (No parameters)\nOverlay component OverlayNonDecision:\n Add a non-decision by shifting the histogram\n Fixed parameters:\n - nondectime: 0.100000\n\n"
],
[
"samp = sol.resample(1000)",
"_____no_output_____"
],
[
"sol.mean_decision_time()",
"_____no_output_____"
],
[
"from ddm import Fittable\nfrom ddm.models import LossRobustBIC\nfrom ddm.functions import fit_adjust_model\nmodel_fit = Model(name='Simple model (fitted)',\n drift=DriftConstant(drift=Fittable(minval=0, maxval=4)),\n noise=NoiseConstant(noise=Fittable(minval=.5, maxval=4)),\n bound=BoundConstant(B=1.1),\n overlay=OverlayNonDecision(nondectime=Fittable(minval=0, maxval=1)),\n dx=.001, dt=.01, T_dur=2)\n\nfit_adjust_model(samp, model_fit,\n fitting_method=\"differential_evolution\",\n lossfunction=LossRobustBIC, verbose=False)",
"Params [2.27693733 1.51047783 0.11324672] gave 423.26628146043083\n"
],
[
"display_model(model_fit)",
"Model Simple model (fitted) information:\nDrift component DriftConstant:\n constant\n Fitted parameters:\n - drift: 2.276937\nNoise component NoiseConstant:\n constant\n Fitted parameters:\n - noise: 1.510478\nBound component BoundConstant:\n constant\n Fixed parameters:\n - B: 1.100000\nIC component ICPointSourceCenter:\n point_source_center\n (No parameters)\nOverlay component OverlayNonDecision:\n Add a non-decision by shifting the histogram\n Fitted parameters:\n - nondectime: 0.113247\nFit information:\n Loss function: BIC\n Loss function value: 423.26628146043083\n Fitting method: differential_evolution\n Solver: auto\n Other properties:\n - nparams: 3\n - samplesize: 1000\n - mess: ''\n\n"
],
[
"samp",
"_____no_output_____"
],
[
"import ddm.plot\nimport matplotlib.pyplot as plt\nddm.plot.plot_fit_diagnostics(model=model_fit, sample=samp)\nplt.savefig(\"simple-fit.png\")\nplt.show()",
"_____no_output_____"
],
[
"print(sol.prob_correct())\nprint(sol.pdf_err())",
"0.8937015385162344\n[0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00\n 0.00000000e+00 0.00000000e+00 0.00000000e+00 2.07365499e-10\n 5.00458955e-05 2.38145936e-03 1.43848498e-02 3.90619605e-02\n 7.20389916e-02 1.07279501e-01 1.40406765e-01 1.69113545e-01\n 1.92585355e-01 2.10874478e-01 2.24459235e-01 2.33980790e-01\n 2.40101611e-01 2.43437087e-01 2.44527962e-01 2.43834117e-01\n 2.41738697e-01 2.38556580e-01 2.34544091e-01 2.29908443e-01\n 2.24816229e-01 2.19400774e-01 2.13768322e-01 2.08003174e-01\n 2.02171914e-01 1.96326847e-01 1.90508801e-01 1.84749393e-01\n 1.79072854e-01 1.73497511e-01 1.68036971e-01 1.62701084e-01\n 1.57496705e-01 1.52428321e-01 1.47498541e-01 1.42708501e-01\n 1.38058181e-01 1.33546658e-01 1.29172320e-01 1.24933024e-01\n 1.20826228e-01 1.16849097e-01 1.12998587e-01 1.09271511e-01\n 1.05664589e-01 1.02174491e-01 9.87978738e-02 9.55313982e-02\n 9.23717551e-02 8.93156782e-02 8.63599563e-02 8.35014413e-02\n 8.07370549e-02 7.80637930e-02 7.54787285e-02 7.29790134e-02\n 7.05618794e-02 6.82246385e-02 6.59646820e-02 6.37794804e-02\n 6.16665817e-02 5.96236104e-02 5.76482658e-02 5.57383200e-02\n 5.38916167e-02 5.21060687e-02 5.03796566e-02 4.87104263e-02\n 4.70964875e-02 4.55360117e-02 4.40272304e-02 4.25684328e-02\n 4.11579646e-02 3.97942261e-02 3.84756698e-02 3.72007998e-02\n 3.59681692e-02 3.47763792e-02 3.36240768e-02 3.25099542e-02\n 3.14327466e-02 3.03912310e-02 2.93842250e-02 2.84105853e-02\n 2.74692065e-02 2.65590197e-02 2.56789915e-02 2.48281226e-02\n 2.40054469e-02 2.32100303e-02 2.24409695e-02 2.16973914e-02\n 2.09784515e-02 2.02833335e-02 1.96112481e-02 1.89614321e-02\n 1.83331477e-02 1.77256813e-02 1.71383432e-02 1.65704665e-02\n 1.60214063e-02 1.54905391e-02 1.49772621e-02 1.44809925e-02\n 1.40011666e-02 1.35372398e-02 1.30886851e-02 1.26549932e-02\n 1.22356716e-02 1.18302442e-02 1.14382506e-02 1.10592456e-02\n 1.06927990e-02 1.03384944e-02 9.99592974e-03 9.66471589e-03\n 9.34447676e-03 9.03484872e-03 8.73548016e-03 8.44603115e-03\n 8.16617298e-03 7.89558788e-03 7.63396859e-03 7.38101801e-03\n 7.13644892e-03 6.89998360e-03 6.67135353e-03 6.45029908e-03\n 6.23656924e-03 6.02992132e-03 5.83012064e-03 5.63694034e-03\n 5.45016104e-03 5.26957065e-03 5.09496410e-03 4.92614312e-03\n 4.76291599e-03 4.60509738e-03 4.45250807e-03 4.30497478e-03\n 4.16232999e-03 4.02441172e-03 3.89106335e-03 3.76213346e-03\n 3.63747565e-03 3.51694836e-03 3.40041472e-03 3.28774242e-03\n 3.17880349e-03 3.07347424e-03 2.97163507e-03 2.87317032e-03\n 2.77796819e-03 2.68592056e-03 2.59692293e-03 2.51087422e-03\n 2.42767672e-03 2.34723596e-03 2.26946060e-03 2.19426231e-03\n 2.12155571e-03 2.05125823e-03 1.98329005e-03 1.91757399e-03\n 1.85403542e-03 1.79260220e-03 1.73320455e-03 1.67577504e-03\n 1.62024845e-03 1.56656172e-03 1.51465390e-03 1.46446604e-03\n 1.41594114e-03 1.36902411e-03 1.32366167e-03 1.27980231e-03\n 1.23739623e-03 1.19639526e-03 1.15675286e-03 1.11842400e-03\n 1.08136516e-03 1.04553426e-03 1.01089062e-03 9.77394888e-04\n 9.45009033e-04 9.13696280e-04 8.83421070e-04 8.54149026e-04\n 8.25846907e-04]\n"
],
[
"from ddm import Sample\nimport pandas\ndf_rt = pandas.read_csv(\"https://raw.githubusercontent.com/mwshinn/PyDDM/master/doc/downloads/roitman_rts.csv\")\ndf_rt",
"_____no_output_____"
],
[
"df_rt = df_rt[df_rt[\"monkey\"] == 1] # Only monkey 1\n \n# Remove short and long RTs, as in 10.1523/JNEUROSCI.4684-04.2005.\n# This is not strictly necessary, but is performed here for\n# compatibility with this study.\ndf_rt = df_rt[df_rt[\"rt\"] > .1] # Remove trials less than 100ms\ndf_rt = df_rt[df_rt[\"rt\"] < 1.65] # Remove trials greater than 1650ms\n \n# Create a sample object from our data. This is the standard input\n# format for fitting procedures. Since RT and correct/error are\n# both mandatory columns, their names are specified by command line\n# arguments.\nroitman_sample = Sample.from_pandas_dataframe(df_rt, rt_column_name=\"rt\", correct_column_name=\"correct\")\n",
"_____no_output_____"
],
[
"import ddm.models\nclass DriftCoherence(ddm.models.Drift):\n name = \"Drift depends linearly on coherence\"\n required_parameters = [\"driftcoh\"] # <-- Parameters we want to include in the model\n required_conditions = [\"coh\"] # <-- Task parameters (\"conditions\"). Should be the same name as in the sample.\n \n # We must always define the get_drift function, which is used to compute the instantaneous value of drift.\n def get_drift(self, conditions, **kwargs):\n return self.driftcoh * conditions['coh']",
"_____no_output_____"
],
[
"from ddm import Model, Fittable\nfrom ddm.functions import fit_adjust_model, display_model\nfrom ddm.models import NoiseConstant, BoundConstant, OverlayChain, OverlayNonDecision, OverlayPoissonMixture\nmodel_rs = Model(name='Roitman data, drift varies with coherence',\n drift=DriftCoherence(driftcoh=Fittable(minval=0, maxval=20)),\n noise=NoiseConstant(noise=1),\n bound=BoundConstant(B=Fittable(minval=.1, maxval=1.5)),\n # Since we can only have one overlay, we use\n # OverlayChain to string together multiple overlays.\n # They are applied sequentially in order. OverlayNonDecision\n # implements a non-decision time by shifting the\n # resulting distribution of response times by\n # `nondectime` seconds.\n overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fittable(minval=0, maxval=.4)),\n OverlayPoissonMixture(pmixturecoef=.02,\n rate=1)]),\n dx=.001, dt=.01, T_dur=2)\n\n# Fitting this will also be fast because PyDDM can automatically\n# determine that DriftCoherence will allow an analytical solution.\nfit_model_rs = fit_adjust_model(sample=roitman_sample, model=model_rs, verbose=False)",
"Params [10.36415557 0.74406192 0.3119136 ] gave 199.33384553433308\n"
],
[
"display_model(fit_model_rs)",
"Model Roitman data, drift varies with coherence information:\nDrift component DriftCoherence:\n Drift depends linearly on coherence\n Fitted parameters:\n - driftcoh: 10.364156\nNoise component NoiseConstant:\n constant\n Fixed parameters:\n - noise: 1.000000\nBound component BoundConstant:\n constant\n Fitted parameters:\n - B: 0.744062\nIC component ICPointSourceCenter:\n point_source_center\n (No parameters)\nOverlay component OverlayChain:\n Overlay component OverlayNonDecision:\n Add a non-decision by shifting the histogram\n Fitted parameters:\n - nondectime: 0.311914\n Overlay component OverlayPoissonMixture:\n Poisson distribution mixture model (lapse rate)\n Fixed parameters:\n - pmixturecoef: 0.020000\n - rate: 1.000000\nFit information:\n Loss function: Negative log likelihood\n Loss function value: 199.33384553433308\n Fitting method: differential_evolution\n Solver: auto\n Other properties:\n - nparams: 3\n - samplesize: 2611\n - mess: ''\n\n"
],
[
"import ddm.plot\nimport matplotlib.pyplot as plt\nddm.plot.plot_fit_diagnostics(model=fit_model_rs, sample=roitman_sample)\nplt.savefig(\"roitman-fit.png\")\nplt.show()",
"_____no_output_____"
],
[
"# ddm.plot.model_gui(model=fit_model_rs, sample=roitman_sample)",
"_____no_output_____"
],
[
"class DriftCoherenceLeak(ddm.models.Drift):\n name = \"Leaky drift depends linearly on coherence\"\n required_parameters = [\"driftcoh\", \"leak\"] # <-- Parameters we want to include in the model\n required_conditions = [\"coh\"] # <-- Task parameters (\"conditions\"). Should be the same name as in the sample.\n \n # We must always define the get_drift function, which is used to compute the instantaneous value of drift.\n def get_drift(self, x, conditions, **kwargs):\n return self.driftcoh * conditions['coh'] + self.leak * x",
"_____no_output_____"
],
[
"from ddm.models import BoundCollapsingExponential\nmodel_leak = Model(name='Roitman data, leaky drift varies with coherence',\n drift=DriftCoherenceLeak(driftcoh=Fittable(minval=0, maxval=20),\n leak=Fittable(minval=-10, maxval=10)),\n noise=NoiseConstant(noise=1),\n bound=BoundCollapsingExponential(B=Fittable(minval=0.5, maxval=3),\n tau=Fittable(minval=.0001, maxval=5)),\n # Since we can only have one overlay, we use\n # OverlayChain to string together multiple overlays.\n # They are applied sequentially in order. OverlayDelay\n # implements a non-decision time by shifting the\n # resulting distribution of response times by\n # `delaytime` seconds.\n overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fittable(minval=0, maxval=.4)),\n OverlayPoissonMixture(pmixturecoef=.02,\n rate=1)]),\n dx=.01, dt=.01, T_dur=2)",
"_____no_output_____"
],
[
"# from ddm.plot import model_gui\n# model_gui(model_leak, sample=roitman_sample)",
"_____no_output_____"
],
[
"# fit_model_leak = fit_adjust_model(sample=roitman_sample, model=model_leak)\n# ddm.plot.plot_fit_diagnostics(model=fit_model_leak, sample=roitman_sample)\n# plt.savefig(\"leak-collapse-fit.png\")",
"Model(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.542903321809481, minval=0, maxval=20), leak=Fitted(1.7326521345463863, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6897102161309165, minval=0.5, maxval=3), tau=Fitted(2.3832278058729726, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.35096334903519405, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1967.1418614641623\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.177639592668301, minval=0, maxval=20), leak=Fitted(9.110567517297387, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.142464682107733, minval=0.5, maxval=3), tau=Fitted(0.9120053245906932, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.06305226749400683, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5739.193193187177\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.211484734330153, minval=0, maxval=20), leak=Fitted(-5.222361158255124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4033542968189374, minval=0.5, maxval=3), tau=Fitted(3.554566351511098, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3999651366249339, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1957.1724192501015\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.361573370519599, minval=0, maxval=20), leak=Fitted(1.835438487253862, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.757022772156284, minval=0.5, maxval=3), tau=Fitted(4.877765735792014, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38577218494091436, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2805.2526298289877\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.537436835568695, minval=0, maxval=20), leak=Fitted(-6.39078185666055, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.551943416899259, minval=0.5, maxval=3), tau=Fitted(4.0837129155818195, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17437538882229442, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3796.138259254554\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.984442854482344, minval=0, maxval=20), leak=Fitted(0.724221849309914, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.540173801190216, minval=0.5, maxval=3), tau=Fitted(1.781842043390649, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.27575674887090934, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4121.032247360254\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.114991807937429, minval=0, maxval=20), leak=Fitted(-5.116286791213202, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.39670851796677, minval=0.5, maxval=3), tau=Fitted(1.9350929617331754, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2910116926278514, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6720.728702005034\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.3506744846401055, minval=0, maxval=20), leak=Fitted(-0.9499488780508014, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8901959220308424, minval=0.5, maxval=3), tau=Fitted(1.6978901989510478, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1481958916723394, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1936.3671952445093\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.35931026055852, minval=0, maxval=20), leak=Fitted(-3.5854375013312225, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.76999957298265, minval=0.5, maxval=3), tau=Fitted(2.659565600940745, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.37701582645469495, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8021.808762332517\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.34142114372201, minval=0, maxval=20), leak=Fitted(-9.157961677811606, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5227808245057424, minval=0.5, maxval=3), tau=Fitted(0.03123285865883485, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12236997682960665, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=110939.6746665163\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.412827887291158, minval=0, maxval=20), leak=Fitted(-4.120283854196999, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.118103761745533, minval=0.5, maxval=3), tau=Fitted(4.317386728046326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.25839703424567373, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1473.3540186071227\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.19128285503821, minval=0, maxval=20), leak=Fitted(7.080754899714408, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.918483591292807, minval=0.5, maxval=3), tau=Fitted(2.803798308945293, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32444858966643536, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1559.7075397244919\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.126986236229552, minval=0, maxval=20), leak=Fitted(-6.757327480032124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0985219833565336, minval=0.5, maxval=3), tau=Fitted(3.9734756207680704, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.24577080399663306, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2705.4085137652946\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.81024590295237, minval=0, maxval=20), leak=Fitted(4.414833168535617, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9352040071677286, minval=0.5, maxval=3), tau=Fitted(3.1794317538415537, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.0981190149127948, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5725.197278222273\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.917671444921647, minval=0, maxval=20), leak=Fitted(7.405944933563182, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.655355543598668, minval=0.5, maxval=3), tau=Fitted(2.0514875797889305, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.27138708958235086, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1579.2415886404933\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.391054383955462, minval=0, maxval=20), leak=Fitted(-2.1854671146279236, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4490639030090307, minval=0.5, maxval=3), tau=Fitted(1.4782935430434794, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21509780576878035, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5.977359880538344\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.12120117315572, minval=0, maxval=20), leak=Fitted(-2.5773791958162207, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.305815159227506, minval=0.5, maxval=3), tau=Fitted(3.9329276265909474, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14048189022126337, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8208.892193784584\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.66213550801708, minval=0, maxval=20), leak=Fitted(-4.208674746394895, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.098494814172506, minval=0.5, maxval=3), tau=Fitted(1.4316826901558302, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3934259309190975, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8326.03249439039\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.216828411447018, minval=0, maxval=20), leak=Fitted(-9.966378924477729, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9564300459216292, minval=0.5, maxval=3), tau=Fitted(0.5442469163506545, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1081117292070683, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3258.4974691630673\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.006821549469127, minval=0, maxval=20), leak=Fitted(8.296713581353528, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.525446664499696, minval=0.5, maxval=3), tau=Fitted(4.213534769524867, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10287346403935989, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=13202.694860671027\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.57059882668626, minval=0, maxval=20), leak=Fitted(6.453336787925689, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.593777457196794, minval=0.5, maxval=3), tau=Fitted(2.225800734802238, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3427598005743314, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3530.998519492379\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.711925034412583, minval=0, maxval=20), leak=Fitted(-4.524427340656998, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.048837296006622, minval=0.5, maxval=3), tau=Fitted(0.09086413073586863, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23912047849103146, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=35489.39082112373\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.4873034093977395, minval=0, maxval=20), leak=Fitted(7.862200205180187, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.658720623711777, minval=0.5, maxval=3), tau=Fitted(3.6804994246028446, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.16709734085887742, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4541.311579476601\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.1353143538374226, minval=0, maxval=20), leak=Fitted(-2.8121188665411245, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.453826207432596, minval=0.5, maxval=3), tau=Fitted(3.330241670960722, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32564344640727416, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4519.117293834119\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.370686383947106, minval=0, maxval=20), leak=Fitted(3.9982082504333527, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5796426752518755, minval=0.5, maxval=3), tau=Fitted(0.21677000693464388, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.301243260529459, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=748.6499972205659\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.0712151382052344, minval=0, maxval=20), leak=Fitted(-5.6704408999905676, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.935522069829656, minval=0.5, maxval=3), tau=Fitted(1.215142701277994, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1378720789603346, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12838.190216137778\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.8299866002008507, minval=0, maxval=20), leak=Fitted(2.665109717414569, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6043895098200496, minval=0.5, maxval=3), tau=Fitted(0.7628835045867504, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3549721335761616, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2758.9336741567736\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.999457436591813, minval=0, maxval=20), leak=Fitted(9.543538477726898, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7178716574987491, minval=0.5, maxval=3), tau=Fitted(2.307142454359793, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31306104895877596, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4641.146769079277\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.660808240049093, minval=0, maxval=20), leak=Fitted(3.0662351866574444, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1385992312550393, minval=0.5, maxval=3), tau=Fitted(4.647611917032916, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2043855464714325, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9269.37809315032\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.70982377885155, minval=0, maxval=20), leak=Fitted(-6.800419298248432, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.768239565824627, minval=0.5, maxval=3), tau=Fitted(3.434876920678647, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.02110817111991392, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11889.195042078032\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.66832697413967, minval=0, maxval=20), leak=Fitted(0.4921644982503892, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.007061826588362, minval=0.5, maxval=3), tau=Fitted(4.4207994818298415, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31615391918460084, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1368.1487255451202\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.56756363481516, minval=0, maxval=20), leak=Fitted(-0.7223831507908851, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.1706431859183954, minval=0.5, maxval=3), tau=Fitted(4.760261191611825, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.003398693824390203, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11393.171159361424\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.309148771071909, minval=0, maxval=20), leak=Fitted(1.20077183637195, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.880016048851676, minval=0.5, maxval=3), tau=Fitted(1.6489022704849163, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22556979725823584, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=227.09667567265478\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.4040191927759373, minval=0, maxval=20), leak=Fitted(-7.458202688675208, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.930354826761679, minval=0.5, maxval=3), tau=Fitted(1.147736682894549, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.36630733420361733, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=15402.661413883514\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.479588546075604, minval=0, maxval=20), leak=Fitted(-8.083588713055905, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5857379388951123, minval=0.5, maxval=3), tau=Fitted(2.741601596966736, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.007638713549017462, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3515.124593902118\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.613485659215479, minval=0, maxval=20), leak=Fitted(5.610907275252828, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.728550946841991, minval=0.5, maxval=3), tau=Fitted(4.811698419792194, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1977654856630871, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4132.9363106519\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.768243432964475, minval=0, maxval=20), leak=Fitted(6.651869885618041, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5462735049372647, minval=0.5, maxval=3), tau=Fitted(2.975245704454408, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.052871738019870135, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5674.582123415394\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.87760106313388, minval=0, maxval=20), leak=Fitted(-4.689791661264407, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2175369170718895, minval=0.5, maxval=3), tau=Fitted(4.3692084278773615, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17849619532548983, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3660.0579299619585\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.7176539080728208, minval=0, maxval=20), leak=Fitted(-6.123605808671152, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4808650722680083, minval=0.5, maxval=3), tau=Fitted(0.4024966300783519, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23239227009674812, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=16633.0704465021\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.697174232648194, minval=0, maxval=20), leak=Fitted(2.4802866112250532, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8186190795707172, minval=0.5, maxval=3), tau=Fitted(0.6306430523062001, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3396716168192412, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=199.13580478471357\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.140131462331764, minval=0, maxval=20), leak=Fitted(-3.295448348387735, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0147392628521819, minval=0.5, maxval=3), tau=Fitted(0.2727993774320825, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3040557136066886, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3918.5874695538973\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.9371837294167698, minval=0, maxval=20), leak=Fitted(8.678046234879712, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8385964901930111, minval=0.5, maxval=3), tau=Fitted(0.33899151106509073, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2558495815752162, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3019.5929609492505\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.8302846986629895, minval=0, maxval=20), leak=Fitted(-1.8397239066369875, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.994137164942317, minval=0.5, maxval=3), tau=Fitted(4.976977522418319, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.0846322352619368, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7381.92344546412\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.2345893901175797, minval=0, maxval=20), leak=Fitted(-9.598513056448347, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.3579775682328874, minval=0.5, maxval=3), tau=Fitted(3.7720005213129983, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2618198776780116, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4673.784562370676\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.351449214510761, minval=0, maxval=20), leak=Fitted(-7.640677644016273, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.9980286214857284, minval=0.5, maxval=3), tau=Fitted(3.6609646989721103, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.08977706360900047, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1430.2727961320163\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.097668447771248, minval=0, maxval=20), leak=Fitted(1.1050392579272583, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.754224624802943, minval=0.5, maxval=3), tau=Fitted(1.2761400938548721, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.033954524422048105, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=910.5143627705725\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.432397101751327, minval=0, maxval=20), leak=Fitted(7.966952743267273, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.223147186414273, minval=0.5, maxval=3), tau=Fitted(3.031993511787069, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.05412793876149302, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11595.143943251012\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.854937808756773, minval=0, maxval=20), leak=Fitted(-0.023713932836334495, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3868171233530586, minval=0.5, maxval=3), tau=Fitted(3.068387460510192, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.029135050402059298, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9796.14257814092\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.095299194503195, minval=0, maxval=20), leak=Fitted(-1.2925682982632958, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8277960615919016, minval=0.5, maxval=3), tau=Fitted(3.840091695030771, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3681091197050326, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2922.3568809585418\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.033928127058024, minval=0, maxval=20), leak=Fitted(5.3826627252158605, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7337821458473746, minval=0.5, maxval=3), tau=Fitted(3.337819224730601, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28600721699298964, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5168.037646690568\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.702433836139917, minval=0, maxval=20), leak=Fitted(8.608034815576236, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8858020748929802, minval=0.5, maxval=3), tau=Fitted(4.189471356341522, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21238957010308723, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6116.063969021941\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.247637590788464, minval=0, maxval=20), leak=Fitted(5.815824166809809, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.298298780848644, minval=0.5, maxval=3), tau=Fitted(2.55458213701347, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14955147274135394, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4866.129011550723\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.7037694419760365, minval=0, maxval=20), leak=Fitted(3.0767010714797083, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2479916281903285, minval=0.5, maxval=3), tau=Fitted(4.0099368320020465, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.02425187113048538, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10711.822437041805\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.135246248015699, minval=0, maxval=20), leak=Fitted(-3.6529078992319794, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.473513674657285, minval=0.5, maxval=3), tau=Fitted(2.5093587784326203, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2981010930285442, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4107.875190970431\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.965445486292698, minval=0, maxval=20), leak=Fitted(6.09854969896179, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8567488192353405, minval=0.5, maxval=3), tau=Fitted(1.1205908389257007, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07164077207750025, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2497.237146893574\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.4418113305347493, minval=0, maxval=20), leak=Fitted(-2.318545253277735, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6676960872636872, minval=0.5, maxval=3), tau=Fitted(0.8291335515347067, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3326860755268224, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=14051.406623579034\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.874059812656853, minval=0, maxval=20), leak=Fitted(-9.264806254942792, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.335271652340373, minval=0.5, maxval=3), tau=Fitted(3.2414297956653177, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15639762791172174, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3084.382010808882\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.115485475179923, minval=0, maxval=20), leak=Fitted(9.76035404612401, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0387090837885962, minval=0.5, maxval=3), tau=Fitted(2.926426954353281, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09389283633411788, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11703.462227270162\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.319445216058758, minval=0, maxval=20), leak=Fitted(-7.160128053810691, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9988101569884702, minval=0.5, maxval=3), tau=Fitted(1.5500080068614421, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07952921319990024, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1918.515221275764\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.41151112341340124, minval=0, maxval=20), leak=Fitted(3.734489470969884, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5023023263287958, minval=0.5, maxval=3), tau=Fitted(3.5189911037998582, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12792306250838692, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5700.599866115208\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.492467774029389, minval=0, maxval=20), leak=Fitted(5.179148533812548, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.272088821411123, minval=0.5, maxval=3), tau=Fitted(0.4746260598418126, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.18295318033591187, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1314.1954146433582\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.131685916749026, minval=0, maxval=20), leak=Fitted(-1.6369537914847654, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8497389644488924, minval=0.5, maxval=3), tau=Fitted(2.1450754610217073, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2446236118474111, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1555.6402114185046\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.97816556884581, minval=0, maxval=20), leak=Fitted(9.218727106303186, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4186468519374333, minval=0.5, maxval=3), tau=Fitted(2.0696698736790857, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22353786432043876, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2824.1212855656267\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.130794747676992, minval=0, maxval=20), leak=Fitted(-8.378174193167938, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.120967153573368, minval=0.5, maxval=3), tau=Fitted(4.595099419092333, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.011282016195194061, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11865.280258517882\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.736428538344015, minval=0, maxval=20), leak=Fitted(4.337923085167345, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8138709836241098, minval=0.5, maxval=3), tau=Fitted(1.341157441249478, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.11262107474762145, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3716.475746666648\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.158738994504727, minval=0, maxval=20), leak=Fitted(-8.92787024034189, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.637107399032391, minval=0.5, maxval=3), tau=Fitted(4.519237281549787, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.16214827796079723, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10086.981486610955\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.07736352504598, minval=0, maxval=20), leak=Fitted(2.1692218478468472, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9308972639560213, minval=0.5, maxval=3), tau=Fitted(2.729213947363532, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3582859249792929, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1551.6500644191292\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.195784907101164, minval=0, maxval=20), leak=Fitted(-0.634128222238517, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7275366191710877, minval=0.5, maxval=3), tau=Fitted(1.0105794789631546, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38225275452393476, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4859.228062059125\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.75478051188384, minval=0, maxval=20), leak=Fitted(6.926044749350597, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.17195400042819, minval=0.5, maxval=3), tau=Fitted(0.9509342990648704, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.04395197557249128, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6787.318453629327\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.75023712195693, minval=0, maxval=20), leak=Fitted(4.761203475521604, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.3188580067266935, minval=0.5, maxval=3), tau=Fitted(1.9280731114113079, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.27854824849303894, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=148.26536686434358\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.929745510940739, minval=0, maxval=20), leak=Fitted(-8.513001799241383, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.264255810724807, minval=0.5, maxval=3), tau=Fitted(1.834733483203609, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.06882639883914962, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6116.339554106975\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.382050034722042, minval=0, maxval=20), leak=Fitted(3.403308982057378, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6876900285185692, minval=0.5, maxval=3), tau=Fitted(0.6908389365686176, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1917525800789167, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=488.8152567644936\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.058824051176941, minval=0, maxval=20), leak=Fitted(0.29084723156440395, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6130143450127488, minval=0.5, maxval=3), tau=Fitted(2.4512508631219534, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12881041223235157, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1007.8810749413897\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.612390244269072, minval=0, maxval=20), leak=Fitted(-0.3729560259449616, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7777129860097391, minval=0.5, maxval=3), tau=Fitted(4.681630673641353, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.039743950711769394, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9897.91181771521\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.555671912814596, minval=0, maxval=20), leak=Fitted(-5.949036813966995, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.602440177910261, minval=0.5, maxval=3), tau=Fitted(0.16250845781116308, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19606580553769165, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1043.7127285194201\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.633175070355268, minval=0, maxval=20), leak=Fitted(2.9426498519521216, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7912804091437411, minval=0.5, maxval=3), tau=Fitted(1.8727547291615798, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.04556059671628637, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10515.351648296488\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.177639592668301, minval=0, maxval=20), leak=Fitted(-2.7732756281299373, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6499713835665564, minval=0.5, maxval=3), tau=Fitted(0.15991237771251665, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2325819535725826, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10836.651712564677\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.245415657354524, minval=0, maxval=20), leak=Fitted(-5.222361158255124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5179466343422772, minval=0.5, maxval=3), tau=Fitted(2.2160879204509123, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28259232164498344, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=306.0458202355835\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.032351282036291, minval=0, maxval=20), leak=Fitted(-8.574228848939452, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8727569344306074, minval=0.5, maxval=3), tau=Fitted(3.5197901890770176, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38577218494091436, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6004.304481858265\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.0941031469211353, minval=0, maxval=20), leak=Fitted(-7.8887808037076335, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.036293904732351, minval=0.5, maxval=3), tau=Fitted(0.5784053833287157, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17437538882229442, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6709.259517732059\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.7487232551192484, minval=0, maxval=20), leak=Fitted(-1.1956538641787873, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6215919901851876, minval=0.5, maxval=3), tau=Fitted(1.781842043390649, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39046092600319604, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8740.40953339932\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.114991807937429, minval=0, maxval=20), leak=Fitted(1.6658105236257992, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.39670851796677, minval=0.5, maxval=3), tau=Fitted(1.9350929617331754, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17821901284133113, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=264.61209836094935\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.763244580969381, minval=0, maxval=20), leak=Fitted(8.503765112251756, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2762355399285328, minval=0.5, maxval=3), tau=Fitted(0.33657775238292054, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23466684745835298, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2113.985195490566\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.1469688107246263, minval=0, maxval=20), leak=Fitted(-6.9544019619031285, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1810652850008085, minval=0.5, maxval=3), tau=Fitted(0.05750692202537655, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.26201605481746043, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=18388.061364274854\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.150974385146232, minval=0, maxval=20), leak=Fitted(-9.157961677811606, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9666084048953891, minval=0.5, maxval=3), tau=Fitted(2.386875299912956, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.33817342276866946, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8530.345479010568\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.412827887291158, minval=0, maxval=20), leak=Fitted(-4.120283854196999, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2584286191222667, minval=0.5, maxval=3), tau=Fitted(4.317386728046326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23790484341348322, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1632.7416171856587\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.19128285503821, minval=0, maxval=20), leak=Fitted(3.042744215720403, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5229053167906894, minval=0.5, maxval=3), tau=Fitted(1.137964266248994, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.26426723151034004, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=336.31598507188727\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.126986236229552, minval=0, maxval=20), leak=Fitted(-6.757327480032124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8159379662640016, minval=0.5, maxval=3), tau=Fitted(0.16990373857938357, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.24577080399663306, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3474.9065062038835\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.923032283615042, minval=0, maxval=20), leak=Fitted(-7.0413233175597, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4838893981141563, minval=0.5, maxval=3), tau=Fitted(4.174064309618435, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1825594842728903, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1303.554042021748\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.23606223467272436, minval=0, maxval=20), leak=Fitted(0.9201856860335278, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5667322102297176, minval=0.5, maxval=3), tau=Fitted(2.2839104588929735, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3888624455039846, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3212.8750653863444\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.276289157855704, minval=0, maxval=20), leak=Fitted(-7.909247015845818, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2421829626931944, minval=0.5, maxval=3), tau=Fitted(2.261709202293513, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.028467789798590742, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3320.712876903319\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.12120117315572, minval=0, maxval=20), leak=Fitted(7.094435287704652, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.305815159227506, minval=0.5, maxval=3), tau=Fitted(1.622372597716939, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10576877554035645, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8063.1730799142215\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.66213550801708, minval=0, maxval=20), leak=Fitted(3.6809812547409204, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8313611587623053, minval=0.5, maxval=3), tau=Fitted(1.4316826901558302, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.20134870703174315, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=651.2415025082522\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.25181346188174, minval=0, maxval=20), leak=Fitted(-2.4254529177533124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.179779368859593, minval=0.5, maxval=3), tau=Fitted(0.5442469163506545, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1081117292070683, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2035.324035338101\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.45444379571422, minval=0, maxval=20), leak=Fitted(-5.997431298208199, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.038323579842466, minval=0.5, maxval=3), tau=Fitted(3.769206840061745, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10287346403935989, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2486.0113455175047\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.432686042064855, minval=0, maxval=20), leak=Fitted(6.902221678448313, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.58892915169679, minval=0.5, maxval=3), tau=Fitted(1.6273041327701616, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.019658911574563925, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12727.453033736896\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.711925034412583, minval=0, maxval=20), leak=Fitted(0.4158797298327954, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9450598847411802, minval=0.5, maxval=3), tau=Fitted(0.10380212724333093, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30055665869816695, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=966.2330726758762\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.132448184096729, minval=0, maxval=20), leak=Fitted(0.8322960621072584, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9100936017445269, minval=0.5, maxval=3), tau=Fitted(3.6804994246028446, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.16709734085887742, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7787.322583946367\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.419045041059247, minval=0, maxval=20), leak=Fitted(4.518334614202795, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0267168884367341, minval=0.5, maxval=3), tau=Fitted(3.330241670960722, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38975778682277096, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2191.868273446236\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.887199241883467, minval=0, maxval=20), leak=Fitted(3.186115209641305, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5796426752518755, minval=0.5, maxval=3), tau=Fitted(0.7783257393934342, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12028083562498915, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=86.62449676660279\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.715544457691456, minval=0, maxval=20), leak=Fitted(5.753293066971716, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5051177894204255, minval=0.5, maxval=3), tau=Fitted(1.8372109942152275, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1378720789603346, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3319.18289418731\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.8299866002008507, minval=0, maxval=20), leak=Fitted(-2.4632447891831264, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3221144107138207, minval=0.5, maxval=3), tau=Fitted(0.7628835045867504, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2709628073618001, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4475.280133770362\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.999457436591813, minval=0, maxval=20), leak=Fitted(1.2718909682488921, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2860221196567776, minval=0.5, maxval=3), tau=Fitted(2.9421079894278273, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23481982886635808, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2355.5372862732725\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.8967168080372243, minval=0, maxval=20), leak=Fitted(3.0662351866574444, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8020613941783195, minval=0.5, maxval=3), tau=Fitted(4.412389672086954, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31553451737382776, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2168.4392328219365\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.70982377885155, minval=0, maxval=20), leak=Fitted(7.863172372076321, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.768239565824627, minval=0.5, maxval=3), tau=Fitted(2.60296712130035, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14553332983088546, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9889.206847440599\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.3050947149827792, minval=0, maxval=20), leak=Fitted(-3.9992946133123395, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9241259046818622, minval=0.5, maxval=3), tau=Fitted(0.8042436980951486, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.35183867874727925, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=13191.397897380946\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.17362053727563875, minval=0, maxval=20), leak=Fitted(-0.37504863979786807, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3084109697503705, minval=0.5, maxval=3), tau=Fitted(2.361454056987139, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17358848011658626, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1650.1069363268557\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.210590482601162, minval=0, maxval=20), leak=Fitted(-4.946621452593096, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0066866739031262, minval=0.5, maxval=3), tau=Fitted(2.1654891016642623, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38490376381875374, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1364.1683940937573\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.4040191927759373, minval=0, maxval=20), leak=Fitted(2.4372515393029093, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2469716829726964, minval=0.5, maxval=3), tau=Fitted(1.147736682894549, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19960321335391282, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1152.6179211971235\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.178805860028826, minval=0, maxval=20), leak=Fitted(2.4433690356689874, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4192257320205275, minval=0.5, maxval=3), tau=Fitted(2.741601596966736, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2795035824647591, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=784.460201307262\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.644984028494372, minval=0, maxval=20), leak=Fitted(4.975384120125437, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3478785686910941, minval=0.5, maxval=3), tau=Fitted(0.47927329974438715, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1458072969537393, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2623.342867497741\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.768243432964475, minval=0, maxval=20), leak=Fitted(6.651869885618041, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8625502523091897, minval=0.5, maxval=3), tau=Fitted(2.113218538902111, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1354818474465036, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3609.6352514531645\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.78948364557884, minval=0, maxval=20), leak=Fitted(-4.227290510243346, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5840070528386396, minval=0.5, maxval=3), tau=Fitted(3.8173983558565743, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.20355892249128, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9044.758282498413\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.323689585606194, minval=0, maxval=20), leak=Fitted(4.868079403726466, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4808650722680083, minval=0.5, maxval=3), tau=Fitted(0.4024966300783519, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.04362629469565876, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2838.4973274770637\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.113888772857074, minval=0, maxval=20), leak=Fitted(-1.2292033130883118, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8186190795707172, minval=0.5, maxval=3), tau=Fitted(0.809721576748591, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3396716168192412, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=178.38812351346144\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.140131462331764, minval=0, maxval=20), leak=Fitted(-6.878237969200105, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6768390742704717, minval=0.5, maxval=3), tau=Fitted(3.3859283162430316, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.24157138203891004, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4428.963490281947\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.7884663626077275, minval=0, maxval=20), leak=Fitted(-9.064242244883317, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.46536724380734, minval=0.5, maxval=3), tau=Fitted(2.8117962359382513, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.036573727108380694, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=467.1475751700994\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.457409125010907, minval=0, maxval=20), leak=Fitted(-1.8397239066369875, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4895889059291028, minval=0.5, maxval=3), tau=Fitted(4.681969284699878, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.36056019217123914, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2218.4438701127733\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.2345893901175797, minval=0, maxval=20), leak=Fitted(-9.598513056448347, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8821662168820863, minval=0.5, maxval=3), tau=Fitted(3.2638565474120957, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14353502703096754, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1909.8689139792361\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.351449214510761, minval=0, maxval=20), leak=Fitted(-7.640677644016273, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.772845994532273, minval=0.5, maxval=3), tau=Fitted(2.9815972476291326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.34757531087397553, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9247.35327296847\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.378862618576637, minval=0, maxval=20), leak=Fitted(1.1050392579272583, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7342139240249221, minval=0.5, maxval=3), tau=Fitted(1.2761400938548721, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.02690500499091747, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9069.04346940431\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.432397101751327, minval=0, maxval=20), leak=Fitted(-2.093112017516301, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.223147186414273, minval=0.5, maxval=3), tau=Fitted(3.8942413834439513, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.06467513908524367, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10889.164182201774\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.210371543960207, minval=0, maxval=20), leak=Fitted(1.6088979030929895, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3868171233530586, minval=0.5, maxval=3), tau=Fitted(0.6255873307037183, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.02055958932280086, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4762.827143861732\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.118718129151924, minval=0, maxval=20), leak=Fitted(-0.8744984182011528, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3936153536061715, minval=0.5, maxval=3), tau=Fitted(2.9467736148429857, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2803183880992397, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=435.57359287873123\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.35997854333647, minval=0, maxval=20), leak=Fitted(5.3826627252158605, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4392695340523893, minval=0.5, maxval=3), tau=Fitted(3.337819224730601, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39432053335096107, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1952.0756768588285\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.702433836139917, minval=0, maxval=20), leak=Fitted(-6.601916508458457, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6230036788327551, minval=0.5, maxval=3), tau=Fitted(0.8042452921230834, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.29556760018728573, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-91.16602467745601\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.247637590788464, minval=0, maxval=20), leak=Fitted(5.815824166809809, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8298275145056786, minval=0.5, maxval=3), tau=Fitted(3.9298873186842167, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22796457388424743, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9183.294368740324\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.7037694419760365, minval=0, maxval=20), leak=Fitted(-5.257025637118092, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7134299318005117, minval=0.5, maxval=3), tau=Fitted(4.0099368320020465, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28289024440727784, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4576.613768769817\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.047673689467132, minval=0, maxval=20), leak=Fitted(-2.376881454347597, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3289518889167964, minval=0.5, maxval=3), tau=Fitted(2.5093587784326203, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3692288665673356, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=795.3321870925315\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.884631267090228, minval=0, maxval=20), leak=Fitted(-8.639414901543098, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.134163360744393, minval=0.5, maxval=3), tau=Fitted(1.1205908389257007, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30108452878111325, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3142.285711277733\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.1191675152272, minval=0, maxval=20), leak=Fitted(-9.231628038818018, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7982783028145426, minval=0.5, maxval=3), tau=Fitted(0.8291335515347067, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3692020026097622, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=14295.351253770223\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.874059812656853, minval=0, maxval=20), leak=Fitted(-8.708506735702748, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9501158182205072, minval=0.5, maxval=3), tau=Fitted(1.363037761970713, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15639762791172174, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5694.4129521542745\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.000439873018905, minval=0, maxval=20), leak=Fitted(9.76035404612401, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0387090837885962, minval=0.5, maxval=3), tau=Fitted(4.458257223009967, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09389283633411788, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12575.642860723083\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.156781805632846, minval=0, maxval=20), leak=Fitted(-7.410838495356792, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2258206703309056, minval=0.5, maxval=3), tau=Fitted(1.5028229891592235, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07952921319990024, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=836.3518613785488\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.332026524335845, minval=0, maxval=20), leak=Fitted(-6.133569210564125, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6686291175016184, minval=0.5, maxval=3), tau=Fitted(1.9737701886022059, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12792306250838692, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3529.3457909724098\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.702958057805624, minval=0, maxval=20), leak=Fitted(-2.616328518352562, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.272088821411123, minval=0.5, maxval=3), tau=Fitted(0.4746260598418126, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09435071555248889, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1746.0909515440467\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.131685916749026, minval=0, maxval=20), leak=Fitted(-1.6369537914847654, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7835603215521176, minval=0.5, maxval=3), tau=Fitted(3.5941899369816017, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.11114974927028504, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8975.90130604527\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.384778478284051, minval=0, maxval=20), leak=Fitted(-6.646858487787703, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7721811113748243, minval=0.5, maxval=3), tau=Fitted(2.098684183083653, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32372002234465147, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=94.76850742220148\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.592401684721094, minval=0, maxval=20), leak=Fitted(-4.225363153193975, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.51652534294875, minval=0.5, maxval=3), tau=Fitted(4.595099419092333, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15228958383701233, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2078.0996248586325\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.386615661016739, minval=0, maxval=20), leak=Fitted(-1.4901864982594581, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8138709836241098, minval=0.5, maxval=3), tau=Fitted(1.341157441249478, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23019299511637212, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1114.7192782168793\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.461270624188918, minval=0, maxval=20), leak=Fitted(5.5375423663932395, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.637107399032391, minval=0.5, maxval=3), tau=Fitted(2.83900182168418, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.16214827796079723, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9765.810378400778\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.951326492757003, minval=0, maxval=20), leak=Fitted(2.1692218478468472, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5679721953221508, minval=0.5, maxval=3), tau=Fitted(1.6528594282791316, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31416272644592996, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-19.414363111234806\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.195784907101164, minval=0, maxval=20), leak=Fitted(6.607599693202837, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6864947623364621, minval=0.5, maxval=3), tau=Fitted(1.0105794789631546, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09592604328207895, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4053.039952199267\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.187469561124146, minval=0, maxval=20), leak=Fitted(-6.2411035964954635, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.1989454524484024, minval=0.5, maxval=3), tau=Fitted(1.7052426618293945, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.04395197557249128, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1270.8959588532298\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.75023712195693, minval=0, maxval=20), leak=Fitted(5.9300778261232985, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.3188580067266935, minval=0.5, maxval=3), tau=Fitted(1.5774667644892326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32033169289912145, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=516.3505971889762\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.609377346203757, minval=0, maxval=20), leak=Fitted(-8.513001799241383, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9796893145778325, minval=0.5, maxval=3), tau=Fitted(1.834733483203609, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.06882639883914962, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1018.1227610403744\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.7016170593905393, minval=0, maxval=20), leak=Fitted(3.403308982057378, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0340857544204647, minval=0.5, maxval=3), tau=Fitted(0.4990604132664336, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.11548981814212166, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1951.6512719778734\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.620220040271752, minval=0, maxval=20), leak=Fitted(-4.136953304472781, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6130143450127488, minval=0.5, maxval=3), tau=Fitted(1.9688778558635178, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.12881041223235157, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4105.766662542889\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.325590611714693, minval=0, maxval=20), leak=Fitted(-0.42557548268072454, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7777129860097391, minval=0.5, maxval=3), tau=Fitted(1.3181006127984902, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.039743950711769394, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=237.55902460073858\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.555671912814596, minval=0, maxval=20), leak=Fitted(2.0074725854691655, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6710413482680693, minval=0.5, maxval=3), tau=Fitted(0.34335094382133224, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1521315616713835, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3033.0874096901657\ndifferential_evolution step 1: f(x)= -91.166\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.807675717383388, minval=0, maxval=20), leak=Fitted(-3.2258372989432305, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3801486091772526, minval=0.5, maxval=3), tau=Fitted(0.3826404086965032, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.36893219553202905, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6620.393668414414\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.177639592668301, minval=0, maxval=20), leak=Fitted(-6.80636931896222, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8946713693029849, minval=0.5, maxval=3), tau=Fitted(0.9120053245906932, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15173367516227643, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=408.1219225846625\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.245415657354524, minval=0, maxval=20), leak=Fitted(0.6387962040955131, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7736264822889152, minval=0.5, maxval=3), tau=Fitted(2.2160879204509123, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39220198009293594, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1758.9539695520748\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.361573370519599, minval=0, maxval=20), leak=Fitted(-6.2911432037299075, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8196311353216672, minval=0.5, maxval=3), tau=Fitted(0.26362927289349436, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38577218494091436, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=27671.817854595734\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.191469883082515, minval=0, maxval=20), leak=Fitted(-6.39078185666055, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6357722280210201, minval=0.5, maxval=3), tau=Fitted(1.4557438216504326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17437538882229442, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3089.9251953312264\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.984442854482344, minval=0, maxval=20), leak=Fitted(-6.662477212280839, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0978481340942319, minval=0.5, maxval=3), tau=Fitted(1.781842043390649, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2805107102610981, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=161.4188134739154\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.737293679747017, minval=0, maxval=20), leak=Fitted(1.6658105236257992, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1372052438267743, minval=0.5, maxval=3), tau=Fitted(1.9350929617331754, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.007581892830709924, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8040.964478985707\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.313762102968306, minval=0, maxval=20), leak=Fitted(-4.642822048350636, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5306168774443036, minval=0.5, maxval=3), tau=Fitted(1.633708339858989, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30674298476313894, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2360.021967545753\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.569414197064123, minval=0, maxval=20), leak=Fitted(-2.402239803360593, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6587687957479658, minval=0.5, maxval=3), tau=Fitted(4.319866844664604, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2774300050079683, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7473.437068772452\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.264118705577928, minval=0, maxval=20), leak=Fitted(-6.316407557184234, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8020434805605563, minval=0.5, maxval=3), tau=Fitted(0.4356128613056276, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28797742800454107, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=890.828929855362\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.412827887291158, minval=0, maxval=20), leak=Fitted(-5.750598764527986, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.118103761745533, minval=0.5, maxval=3), tau=Fitted(1.0521041954356007, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2451869275358008, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8018.509194951059\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.612207661232725, minval=0, maxval=20), leak=Fitted(3.042744215720403, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.199426289587592, minval=0.5, maxval=3), tau=Fitted(0.6022704039599724, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.26426723151034004, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=432.373182879041\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.126986236229552, minval=0, maxval=20), leak=Fitted(-6.757327480032124, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5239200179825685, minval=0.5, maxval=3), tau=Fitted(0.15613051720244542, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32745312604946075, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=477.18494404984324\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.923032283615042, minval=0, maxval=20), leak=Fitted(-7.0413233175597, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8382949201880083, minval=0.5, maxval=3), tau=Fitted(0.014659630423075942, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1825594842728903, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4965.856935744479\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.936148423286612, minval=0, maxval=20), leak=Fitted(-7.952600854597529, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9175599640780747, minval=0.5, maxval=3), tau=Fitted(0.6065019990760161, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.18614099362621328, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=15805.725487109037\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.542903321809481, minval=0, maxval=20), leak=Fitted(7.421099807916098, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4176756726406126, minval=0.5, maxval=3), tau=Fitted(1.3717231199471893, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.16995851911916793, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2498.762279438366\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.945225949223005, minval=0, maxval=20), leak=Fitted(7.094435287704652, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9978774360625113, minval=0.5, maxval=3), tau=Fitted(4.567259962818717, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1889075378807287, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9899.541377375446\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.66213550801708, minval=0, maxval=20), leak=Fitted(0.07555605266217347, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4744485592092103, minval=0.5, maxval=3), tau=Fitted(2.684207961786023, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.20134870703174315, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=805.635909355803\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.38174424966538, minval=0, maxval=20), leak=Fitted(-4.813586768384934, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9532656179500085, minval=0.5, maxval=3), tau=Fitted(1.1407958489206806, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2661168061694297, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-52.849197387711186\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.613055353013042, minval=0, maxval=20), leak=Fitted(-0.05968290653174391, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.038323579842466, minval=0.5, maxval=3), tau=Fitted(3.769206840061745, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.18732361299831035, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2664.4782324252474\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.815857908182101, minval=0, maxval=20), leak=Fitted(-4.696678769156801, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7462490009654288, minval=0.5, maxval=3), tau=Fitted(2.225800734802238, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3962077145816184, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=985.6075681669201\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.711925034412583, minval=0, maxval=20), leak=Fitted(0.4158797298327954, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5117685551494706, minval=0.5, maxval=3), tau=Fitted(4.277080827345146, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30055665869816695, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1948.5649887803552\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.4873034093977395, minval=0, maxval=20), leak=Fitted(-7.833204919706887, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8511759092206391, minval=0.5, maxval=3), tau=Fitted(2.876984868057877, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2735714554597759, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1424.565157262353\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.184964138112921, minval=0, maxval=20), leak=Fitted(-2.4496823496522815, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0267168884367341, minval=0.5, maxval=3), tau=Fitted(0.9803977862385389, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38975778682277096, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1495.014500985827\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.250725830098407, minval=0, maxval=20), leak=Fitted(3.186115209641305, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6874382325890571, minval=0.5, maxval=3), tau=Fitted(0.7783257393934342, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3082700707588369, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1714.6316080906054\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.511864495657418, minval=0, maxval=20), leak=Fitted(2.504069944969949, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5937956985530339, minval=0.5, maxval=3), tau=Fitted(2.2864991862169464, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1378720789603346, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10086.105927629827\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.96922150387807, minval=0, maxval=20), leak=Fitted(-0.9981605509224978, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.037135071844141, minval=0.5, maxval=3), tau=Fitted(0.3437863936857295, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2872239717323508, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=716.0128289363071\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.775232623926154, minval=0, maxval=20), leak=Fitted(-8.6336982638672, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1553729125436747, minval=0.5, maxval=3), tau=Fitted(2.1070589787268985, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3975935343620683, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3454.3323185148406\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.802718160891565, minval=0, maxval=20), leak=Fitted(1.9900028082231014, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8020613941783195, minval=0.5, maxval=3), tau=Fitted(1.9861169166735386, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31543816374609124, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1263.6930862050976\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.942398478296917, minval=0, maxval=20), leak=Fitted(-4.638397255146485, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3155270632104954, minval=0.5, maxval=3), tau=Fitted(2.60296712130035, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.25692160732463964, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-8.295937676361461\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.373716194220535, minval=0, maxval=20), leak=Fitted(-5.300170004508667, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6356941506537335, minval=0.5, maxval=3), tau=Fitted(0.8444697256206934, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28602997883898096, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=658.2406822212733\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.858842938022082, minval=0, maxval=20), leak=Fitted(-9.821972686694462, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7553920790484199, minval=0.5, maxval=3), tau=Fitted(0.2280871145762564, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.009073746844886932, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=33186.159703961675\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.309148771071909, minval=0, maxval=20), leak=Fitted(-4.692546363916991, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.880016048851676, minval=0.5, maxval=3), tau=Fitted(1.6489022704849163, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3558244740834831, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=11691.468854416957\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.870955937648644, minval=0, maxval=20), leak=Fitted(-8.691957114246435, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2469716829726964, minval=0.5, maxval=3), tau=Fitted(1.3171025496133335, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.357176476705058, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12648.379090319964\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.85609589118386, minval=0, maxval=20), leak=Fitted(2.4433690356689874, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8870559223851224, minval=0.5, maxval=3), tau=Fitted(2.741601596966736, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3433843289784165, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1605.3117907571332\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.533911734631191, minval=0, maxval=20), leak=Fitted(-4.511875902670476, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.505160788925693, minval=0.5, maxval=3), tau=Fitted(0.47927329974438715, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23395872366951345, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=20562.697536947246\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.203443701449665, minval=0, maxval=20), leak=Fitted(-3.6454162513666266, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8625502523091897, minval=0.5, maxval=3), tau=Fitted(2.4137873381271775, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31880654169082046, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3314.1261574356377\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.142577966386654, minval=0, maxval=20), leak=Fitted(-7.293494777730656, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2175369170718895, minval=0.5, maxval=3), tau=Fitted(0.9009581024449269, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3239486749356342, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=13866.405806380151\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.323689585606194, minval=0, maxval=20), leak=Fitted(-5.80101647339287, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.083404340461965, minval=0.5, maxval=3), tau=Fitted(2.0884802311567774, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3087955017811231, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=542.0543902297841\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.524990279153355, minval=0, maxval=20), leak=Fitted(-6.696462185682215, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8883082588192235, minval=0.5, maxval=3), tau=Fitted(0.809721576748591, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.18174829199538417, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=778.6313536311411\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.81009781212803, minval=0, maxval=20), leak=Fitted(-4.020577006890669, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.437695062949069, minval=0.5, maxval=3), tau=Fitted(3.0772361241936816, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2508198300728628, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1108.6233556218035\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.7884663626077275, minval=0, maxval=20), leak=Fitted(-0.6716496488972412, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4983252732247805, minval=0.5, maxval=3), tau=Fitted(4.703946624625609, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.036573727108380694, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10545.282712015289\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.265201616721322, minval=0, maxval=20), leak=Fitted(-5.90435543837909, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5650132470708875, minval=0.5, maxval=3), tau=Fitted(0.8411841005965459, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.009800181905823713, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5284.968058790039\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.2345893901175797, minval=0, maxval=20), leak=Fitted(-5.075892369726588, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7303670289631872, minval=0.5, maxval=3), tau=Fitted(3.6276104982702235, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2113530600989414, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1591.6463468303455\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.246289677751424, minval=0, maxval=20), leak=Fitted(-3.3388799706816297, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5108682052095024, minval=0.5, maxval=3), tau=Fitted(1.0239243940904779, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3214436779640755, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=952.4489575520859\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.097668447771248, minval=0, maxval=20), leak=Fitted(-8.71625543293783, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7618808583298122, minval=0.5, maxval=3), tau=Fitted(1.2761400938548721, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21085024786869916, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10842.492756711861\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.495348545274613, minval=0, maxval=20), leak=Fitted(-0.38610949518934845, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4198131258737807, minval=0.5, maxval=3), tau=Fitted(0.9009211924302423, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3211639572277174, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1005.5798166111044\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.552394602947988, minval=0, maxval=20), leak=Fitted(-3.0165270853552775, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3868171233530586, minval=0.5, maxval=3), tau=Fitted(0.27162861322170473, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15467825073709768, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6832.306743124194\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.871380456382944, minval=0, maxval=20), leak=Fitted(-4.159206519991374, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3936153536061715, minval=0.5, maxval=3), tau=Fitted(0.3807786153935866, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2510278875899232, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5662.576060812771\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.54710204754227, minval=0, maxval=20), leak=Fitted(5.3826627252158605, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.849929475030783, minval=0.5, maxval=3), tau=Fitted(0.06915585845883054, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39432053335096107, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=887.8043464826212\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.268500390130344, minval=0, maxval=20), leak=Fitted(-2.1854671146279236, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.1048985127885924, minval=0.5, maxval=3), tau=Fitted(0.7213841974989055, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.296679619216214, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7893.491343146927\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.247637590788464, minval=0, maxval=20), leak=Fitted(4.108273882731628, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4887389466496501, minval=0.5, maxval=3), tau=Fitted(1.9295911606114486, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1978399603872159, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3133.801806766403\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.816827327387458, minval=0, maxval=20), leak=Fitted(-5.785882034815931, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7134299318005117, minval=0.5, maxval=3), tau=Fitted(4.0099368320020465, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2967471347430104, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5247.749878455137\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.749688733403623, minval=0, maxval=20), leak=Fitted(-4.751763253920657, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.691866749259694, minval=0.5, maxval=3), tau=Fitted(1.5566820927457452, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2393519164630783, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1312.999678994953\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.48871748143484, minval=0, maxval=20), leak=Fitted(6.09854969896179, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8567488192353405, minval=0.5, maxval=3), tau=Fitted(1.1205908389257007, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07164077207750025, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4975.835953166501\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.312539066304247, minval=0, maxval=20), leak=Fitted(-2.318545253277735, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6961716764965258, minval=0.5, maxval=3), tau=Fitted(1.6099684355921615, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3326860755268224, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=827.4556711567323\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.874059812656853, minval=0, maxval=20), leak=Fitted(-4.663149641847756, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4371658784144272, minval=0.5, maxval=3), tau=Fitted(0.9358506982231534, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2817670803648292, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2809.456064972828\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.966568505774024, minval=0, maxval=20), leak=Fitted(-6.196705115070151, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0278493511340387, minval=0.5, maxval=3), tau=Fitted(1.0564954139474645, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.34002052480857264, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1688.3098111791119\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.156781805632846, minval=0, maxval=20), leak=Fitted(-7.410838495356792, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.435009851031758, minval=0.5, maxval=3), tau=Fitted(0.7529496179750508, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3119120700506244, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=16882.46377987358\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.26855937867761, minval=0, maxval=20), leak=Fitted(-1.836248159750733, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5190494714831524, minval=0.5, maxval=3), tau=Fitted(0.7745625979413273, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.37369234559345277, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=876.0461106916182\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.131937439342071, minval=0, maxval=20), leak=Fitted(5.179148533812548, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1675208855574835, minval=0.5, maxval=3), tau=Fitted(1.3489781604298947, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2756122832370753, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=897.2123132766308\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.131685916749026, minval=0, maxval=20), leak=Fitted(0.03792959617773706, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.901154879997871, minval=0.5, maxval=3), tau=Fitted(0.952134110682185, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3279081757400355, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4999.466130230696\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.276314032650651, minval=0, maxval=20), leak=Fitted(-2.2412717537718896, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5735525533452783, minval=0.5, maxval=3), tau=Fitted(2.735267151186283, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.20779569109484847, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8165.292334277054\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.028906515305897, minval=0, maxval=20), leak=Fitted(-8.13988138105323, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8947505381852, minval=0.5, maxval=3), tau=Fitted(4.595099419092333, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15228958383701233, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9581.980721057642\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.22622358434618, minval=0, maxval=20), leak=Fitted(-1.4901864982594581, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8173976784836563, minval=0.5, maxval=3), tau=Fitted(0.5493786476777154, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23995127187652698, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3449.5731798069455\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.232108104126144, minval=0, maxval=20), leak=Fitted(-2.2099931341416035, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6735978810078918, minval=0.5, maxval=3), tau=Fitted(1.0629625642506813, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.37260852337002204, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=359.55161413707873\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.930207889536693, minval=0, maxval=20), leak=Fitted(-8.333473220224896, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9838283060498263, minval=0.5, maxval=3), tau=Fitted(0.8140898567895898, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1573647441047577, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1531.117340265183\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.39370635131715, minval=0, maxval=20), leak=Fitted(6.607599693202837, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.7746636581258572, minval=0.5, maxval=3), tau=Fitted(0.4249281722742517, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.381527806315987, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2148.857549634509\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.392003548575682, minval=0, maxval=20), leak=Fitted(-6.2411035964954635, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.673074676066789, minval=0.5, maxval=3), tau=Fitted(1.7052426618293945, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3097950857754352, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10972.269480650664\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.220579461668004, minval=0, maxval=20), leak=Fitted(-8.077229664616665, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7814759865835035, minval=0.5, maxval=3), tau=Fitted(1.9280731114113079, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23522460758599478, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=534.3255169675433\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.204529989243834, minval=0, maxval=20), leak=Fitted(-8.331261266592072, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6937799112245167, minval=0.5, maxval=3), tau=Fitted(1.0489335853286614, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21223209845196722, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=991.5710102268968\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.382050034722042, minval=0, maxval=20), leak=Fitted(-7.748866420702028, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9262823279274437, minval=0.5, maxval=3), tau=Fitted(0.612887694553713, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3640138448464663, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=17408.82822962782\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.058824051176941, minval=0, maxval=20), leak=Fitted(-4.022253393271376, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2402104148054405, minval=0.5, maxval=3), tau=Fitted(2.4512508631219534, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2458741319485097, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1065.9804209506112\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.930581634831778, minval=0, maxval=20), leak=Fitted(-6.684912306205328, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7777129860097391, minval=0.5, maxval=3), tau=Fitted(0.004413167062450096, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.33593496720322796, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=40235.251756107864\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.39005174144685, minval=0, maxval=20), leak=Fitted(-4.437961229467975, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.602440177910261, minval=0.5, maxval=3), tau=Fitted(0.16250845781116308, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09608085341534377, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3225.500621004607\ndifferential_evolution step 2: f(x)= -91.166\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.030740419302083, minval=0, maxval=20), leak=Fitted(-6.178308711268689, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2791877241001148, minval=0.5, maxval=3), tau=Fitted(2.205949073074729, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.29677390434152756, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1498.7385000449854\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.955723560368078, minval=0, maxval=20), leak=Fitted(-6.80636931896222, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3042212280661594, minval=0.5, maxval=3), tau=Fitted(2.244470194033217, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15173367516227643, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1308.9991691628559\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.074995309418691, minval=0, maxval=20), leak=Fitted(-1.0976207645620284, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7399334162723434, minval=0.5, maxval=3), tau=Fitted(0.532957937110714, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30585233745658535, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=419.95278516823225\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.241405238747138, minval=0, maxval=20), leak=Fitted(-6.904647400079766, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4514564270125794, minval=0.5, maxval=3), tau=Fitted(0.6643320873065768, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3054182757914606, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7982.49230831595\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.588017723682675, minval=0, maxval=20), leak=Fitted(-6.39078185666055, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7577311122820275, minval=0.5, maxval=3), tau=Fitted(0.4386771064198167, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2362798618262316, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=740.2384103040022\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.984442854482344, minval=0, maxval=20), leak=Fitted(-1.4606408509844115, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0928689412501233, minval=0.5, maxval=3), tau=Fitted(4.368595514286731, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3167735629547485, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2642.237597025166\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.466911067998478, minval=0, maxval=20), leak=Fitted(-9.166573909670353, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5147128129478116, minval=0.5, maxval=3), tau=Fitted(0.0573516626184527, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17821901284133113, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3184.4593217268102\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.031527200274425, minval=0, maxval=20), leak=Fitted(2.990709559304918, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2966694554787088, minval=0.5, maxval=3), tau=Fitted(1.0684457587381322, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14705457313387135, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3669.6535663363725\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.293055896695583, minval=0, maxval=20), leak=Fitted(-7.725921805978897, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0161136166956886, minval=0.5, maxval=3), tau=Fitted(2.3350328069307995, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2917807784465858, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-23.386824274163416\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.176450073623965, minval=0, maxval=20), leak=Fitted(-6.923518619664244, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5634925366643895, minval=0.5, maxval=3), tau=Fitted(0.4356128613056276, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28797742800454107, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=30492.818147863552\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.374957780008687, minval=0, maxval=20), leak=Fitted(-4.120283854196999, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5382348673140247, minval=0.5, maxval=3), tau=Fitted(2.567510985915665, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2971105532986154, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4943.881831077261\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.19128285503821, minval=0, maxval=20), leak=Fitted(3.042744215720403, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5229053167906894, minval=0.5, maxval=3), tau=Fitted(1.0599575307236972, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31643335851802745, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-19.99963151685442\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.89700685216299, minval=0, maxval=20), leak=Fitted(-2.6560141871222154, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5239200179825685, minval=0.5, maxval=3), tau=Fitted(1.5446155355180173, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2483232396230553, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3511.1473883802323\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.159067654654427, minval=0, maxval=20), leak=Fitted(-9.665469886482983, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4838893981141563, minval=0.5, maxval=3), tau=Fitted(0.8664624599980519, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.25490257982403325, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=16238.503331431028\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.0670341987749, minval=0, maxval=20), leak=Fitted(-3.6084512836142526, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.655355543598668, minval=0.5, maxval=3), tau=Fitted(0.7820509401811577, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19094593285965186, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2986.3772398968545\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.18904969247762, minval=0, maxval=20), leak=Fitted(1.7326521345463863, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.016362784624726, minval=0.5, maxval=3), tau=Fitted(0.9712597506065079, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.32412417024533086, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1114.8263213218959\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.08182665953621, minval=0, maxval=20), leak=Fitted(-7.2502786577504015, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.619823130171536, minval=0.5, maxval=3), tau=Fitted(3.5042999454313213, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30807435707883923, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4916.7288886886445\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.672603519389291, minval=0, maxval=20), leak=Fitted(-6.560605929397315, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8313611587623053, minval=0.5, maxval=3), tau=Fitted(1.4316826901558302, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22470423904118744, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12430.064143692538\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.38174424966538, minval=0, maxval=20), leak=Fitted(-5.989648056791019, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9532656179500085, minval=0.5, maxval=3), tau=Fitted(1.1407958489206806, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23071388739771814, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=96.22996823038771\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.286985316235417, minval=0, maxval=20), leak=Fitted(4.015412126359679, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9395629206833962, minval=0.5, maxval=3), tau=Fitted(3.769206840061745, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28020559401412076, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1926.9076740171356\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.708468771998088, minval=0, maxval=20), leak=Fitted(-4.125782982717615, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8703227685536883, minval=0.5, maxval=3), tau=Fitted(1.586532720607829, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2583048778122204, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-81.67357229336588\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.84057835524255, minval=0, maxval=20), leak=Fitted(0.15560621647957573, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6270578811465459, minval=0.5, maxval=3), tau=Fitted(4.828233939521642, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30055665869816695, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7119.543166050579\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.680266254350405, minval=0, maxval=20), leak=Fitted(-7.833204919706887, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.844798727998535, minval=0.5, maxval=3), tau=Fitted(0.8074806512032924, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3680008850840164, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=13670.680591455386\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.458917661480693, minval=0, maxval=20), leak=Fitted(-2.4496823496522815, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.1833904052739894, minval=0.5, maxval=3), tau=Fitted(2.077414010476643, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19583787642101533, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=646.790349738794\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.887199241883467, minval=0, maxval=20), leak=Fitted(-2.712984202424187, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.5796426752518755, minval=0.5, maxval=3), tau=Fitted(0.6846872561961617, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.29717210241616693, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=13341.743968092895\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.871425490750944, minval=0, maxval=20), leak=Fitted(-2.492807125888399, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5051177894204255, minval=0.5, maxval=3), tau=Fitted(3.18000510562985, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09911599567328624, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5248.660833423043\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.99143190013196, minval=0, maxval=20), leak=Fitted(-2.4225830160589035, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5966528351825326, minval=0.5, maxval=3), tau=Fitted(1.243222883436207, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.24211754003541427, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1262.7124942615483\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.886889199395973, minval=0, maxval=20), leak=Fitted(-2.815683400230329, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.210093934375747, minval=0.5, maxval=3), tau=Fitted(0.5865949163762647, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23481982886635808, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=12906.179428003952\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.802718160891565, minval=0, maxval=20), leak=Fitted(-4.938015109896019, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8020613941783195, minval=0.5, maxval=3), tau=Fitted(1.9861169166735386, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09862080282466908, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2158.8654019611095\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.942398478296917, minval=0, maxval=20), leak=Fitted(-9.637588449809133, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3155270632104954, minval=0.5, maxval=3), tau=Fitted(3.547159978944094, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3079741566136504, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1395.2845065365736\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.409956474194372, minval=0, maxval=20), leak=Fitted(-5.300170004508667, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8015657866763264, minval=0.5, maxval=3), tau=Fitted(0.8444697256206934, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28602997883898096, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9982.945720252035\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.122821848038093, minval=0, maxval=20), leak=Fitted(-0.37504863979786807, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3084109697503705, minval=0.5, maxval=3), tau=Fitted(0.5072668553051465, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3249087727543247, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=823.5043470040655\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.947774559229579, minval=0, maxval=20), leak=Fitted(-8.17571272033274, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5679870063944434, minval=0.5, maxval=3), tau=Fitted(1.6489022704849163, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2816564188545346, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1843.2936884818869\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.473255197282874, minval=0, maxval=20), leak=Fitted(-2.5410912143469377, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2469716829726964, minval=0.5, maxval=3), tau=Fitted(0.7759186177430133, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19960321335391282, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5121.5701670908875\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(4.3940334029324735, minval=0, maxval=20), leak=Fitted(-2.1656632144893107, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8069683522252675, minval=0.5, maxval=3), tau=Fitted(2.741601596966736, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3087417570609954, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1690.765325667755\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.96304611906063, minval=0, maxval=20), leak=Fitted(-8.29813014469433, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3478785686910941, minval=0.5, maxval=3), tau=Fitted(2.0879923954542803, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19003966368368494, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=81.56449642678815\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.634199765552256, minval=0, maxval=20), leak=Fitted(-3.6454162513666266, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2992857700838554, minval=0.5, maxval=3), tau=Fitted(1.0060408180374776, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19617993941116085, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2262.6278506978733\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.87760106313388, minval=0, maxval=20), leak=Fitted(-7.284177337151888, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2175369170718895, minval=0.5, maxval=3), tau=Fitted(3.1439899126734625, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.08385585073365297, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1947.6044005366707\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.288995491979646, minval=0, maxval=20), leak=Fitted(-5.80101647339287, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.9891872633610252, minval=0.5, maxval=3), tau=Fitted(1.8583306371796615, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23069711043592595, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4145.917879779354\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.854960891979727, minval=0, maxval=20), leak=Fitted(-9.533577729339896, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8186190795707172, minval=0.5, maxval=3), tau=Fitted(2.6736417236521692, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3396716168192412, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=897.0803008215316\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.059908925231062, minval=0, maxval=20), leak=Fitted(-4.020577006890669, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7394815292461265, minval=0.5, maxval=3), tau=Fitted(0.33967137992341856, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3926583682729577, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1457.766996609064\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.330109447997533, minval=0, maxval=20), leak=Fitted(-9.064242244883317, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3931189331365954, minval=0.5, maxval=3), tau=Fitted(3.6837668501181056, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2861626589771369, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=704.4635528971965\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.896504556421752, minval=0, maxval=20), leak=Fitted(-1.8397239066369875, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4895889059291028, minval=0.5, maxval=3), tau=Fitted(3.458263260530998, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19473937042016998, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2959.2824891011705\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.368507234032409, minval=0, maxval=20), leak=Fitted(1.1032912204809442, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7303670289631872, minval=0.5, maxval=3), tau=Fitted(1.5848608783322775, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23598372507426651, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-237.46384201502656\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.246289677751424, minval=0, maxval=20), leak=Fitted(-3.3388799706816297, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4325778327285916, minval=0.5, maxval=3), tau=Fitted(1.2039529915356595, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21796031458248724, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9419.264504546332\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.097668447771248, minval=0, maxval=20), leak=Fitted(0.7224774995266126, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.754224624802943, minval=0.5, maxval=3), tau=Fitted(2.177901875408141, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1873286981912907, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=621.8354872978528\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.882484609170413, minval=0, maxval=20), leak=Fitted(-0.38610949518934845, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6643209509979948, minval=0.5, maxval=3), tau=Fitted(0.5381431740721603, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2353090105422827, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1895.616343367472\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.4143266547570992, minval=0, maxval=20), leak=Fitted(6.383662727657722, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8462289304048458, minval=0.5, maxval=3), tau=Fitted(0.6255873307037183, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.37540074327119655, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2337.5038045556853\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.118718129151924, minval=0, maxval=20), leak=Fitted(-5.325075472938767, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.611192316897464, minval=0.5, maxval=3), tau=Fitted(0.8665224390026167, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.36827210429937635, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=375.3985746618949\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.527595583505505, minval=0, maxval=20), leak=Fitted(5.3826627252158605, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.3123662910256013, minval=0.5, maxval=3), tau=Fitted(0.75548775898942, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39432053335096107, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1313.986591338055\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.179228915126888, minval=0, maxval=20), leak=Fitted(-1.018934138771217, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4659257996270787, minval=0.5, maxval=3), tau=Fitted(2.1565259854245276, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.11152264702801579, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=733.9903903737566\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(0.7319353724040933, minval=0, maxval=20), leak=Fitted(4.108273882731628, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9277959805331317, minval=0.5, maxval=3), tau=Fitted(2.168128440305765, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2713651436381712, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2457.5183288416133\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.7037694419760365, minval=0, maxval=20), leak=Fitted(-5.257025637118092, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7134299318005117, minval=0.5, maxval=3), tau=Fitted(3.585386365698042, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17831738943284722, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7374.997006884357\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.531448333564251, minval=0, maxval=20), leak=Fitted(-5.003658769859109, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9681395935279186, minval=0.5, maxval=3), tau=Fitted(2.5093587784326203, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21298248399299738, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1251.8945998339605\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.9151453634545526, minval=0, maxval=20), leak=Fitted(1.5711268592670824, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8567488192353405, minval=0.5, maxval=3), tau=Fitted(1.7868402989198273, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3191669228633267, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3894.4376529046203\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.688657557048714, minval=0, maxval=20), leak=Fitted(-2.2386209421176173, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4593286497631857, minval=0.5, maxval=3), tau=Fitted(1.6099684355921615, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.26897124431883523, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2933.6935695989814\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.751914468859542, minval=0, maxval=20), leak=Fitted(-4.663149641847756, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8599687266739378, minval=0.5, maxval=3), tau=Fitted(1.0040973143851322, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.20230357014682865, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7037.824272424863\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.966568505774024, minval=0, maxval=20), leak=Fitted(-0.6066190377395064, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2242783362520897, minval=0.5, maxval=3), tau=Fitted(1.0804099814124268, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.34002052480857264, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=465.72408461881565\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.37690227164366, minval=0, maxval=20), leak=Fitted(4.606791796006031, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2258206703309056, minval=0.5, maxval=3), tau=Fitted(3.874419155619654, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07952921319990024, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=10958.891197591503\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.7237330991124207, minval=0, maxval=20), leak=Fitted(-1.836248159750733, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6823322595166401, minval=0.5, maxval=3), tau=Fitted(3.1474947013481063, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2026014341315845, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=802.7964437083696\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.419697415633751, minval=0, maxval=20), leak=Fitted(-4.515927496190761, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.518482407424149, minval=0.5, maxval=3), tau=Fitted(2.6248451511196507, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10179359449219337, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=251.8165536465698\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(12.810675685786714, minval=0, maxval=20), leak=Fitted(-0.3520839407039611, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.932992281645003, minval=0.5, maxval=3), tau=Fitted(1.3132604306613782, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19732601771010475, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=40.13831095689673\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.384778478284051, minval=0, maxval=20), leak=Fitted(2.594492430893851, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.3404701367534466, minval=0.5, maxval=3), tau=Fitted(2.098684183083653, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19392281852178458, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=139.4474578583364\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.592401684721094, minval=0, maxval=20), leak=Fitted(-5.176784555235431, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.560815257047657, minval=0.5, maxval=3), tau=Fitted(2.846420398910258, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.18732711479601907, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=124.89205399869883\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.386615661016739, minval=0, maxval=20), leak=Fitted(7.540890083185346, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8138709836241098, minval=0.5, maxval=3), tau=Fitted(3.332770324873945, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1863165488416356, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5059.09170320434\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.169377354776884, minval=0, maxval=20), leak=Fitted(3.2419274265548337, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6735978810078918, minval=0.5, maxval=3), tau=Fitted(1.7787970699948061, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.26053052930641185, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4689.631297620253\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.951326492757003, minval=0, maxval=20), leak=Fitted(-0.11443786188160221, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.476459188410702, minval=0.5, maxval=3), tau=Fitted(1.0256740608813442, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3056268002012777, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=281.7208159254675\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.575141330555129, minval=0, maxval=20), leak=Fitted(-0.5169585904638285, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.176639985594168, minval=0.5, maxval=3), tau=Fitted(0.4249281722742517, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15591013398471026, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3531.4693878456173\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.14552482773687, minval=0, maxval=20), leak=Fitted(-2.9784631591904764, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.1989454524484024, minval=0.5, maxval=3), tau=Fitted(1.7052426618293945, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14669270009796065, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=815.2817486747299\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.26304532598127, minval=0, maxval=20), leak=Fitted(-2.2409670114010916, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.336017649572189, minval=0.5, maxval=3), tau=Fitted(1.9280731114113079, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28671530790934896, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3799.125794255725\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.204529989243834, minval=0, maxval=20), leak=Fitted(0.5750611280553097, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9979171472483667, minval=0.5, maxval=3), tau=Fitted(0.7922221230263415, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3094839939448326, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=102.1622442896563\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(10.131284648657122, minval=0, maxval=20), leak=Fitted(3.403308982057378, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6876900285185692, minval=0.5, maxval=3), tau=Fitted(0.43275458382207255, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38251563237158664, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2007.2692959657309\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.061865671318964, minval=0, maxval=20), leak=Fitted(-5.089231622472198, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.263028209214143, minval=0.5, maxval=3), tau=Fitted(2.7872502763642024, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.07768125410159053, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=25.602997110738272\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.75046456324491, minval=0, maxval=20), leak=Fitted(1.1910055382320195, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8652195512276095, minval=0.5, maxval=3), tau=Fitted(1.1417626716929976, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2508716071182817, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-101.199820661297\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.20384157597561, minval=0, maxval=20), leak=Fitted(-5.949036813966995, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.593863482835305, minval=0.5, maxval=3), tau=Fitted(2.3227258050909447, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2310071230523422, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6216.129383892307\ndifferential_evolution step 3: f(x)= -237.464\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.368507234032409, minval=0, maxval=20), leak=Fitted(3.843383358584438, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4176685718352917, minval=0.5, maxval=3), tau=Fitted(0.8023505376555375, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28467557200635235, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-45.254652279653456\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.202376696281668, minval=0, maxval=20), leak=Fitted(-1.1152472701129568, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.754671786186057, minval=0.5, maxval=3), tau=Fitted(1.1886708758132913, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15173367516227643, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4401.601967574494\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.536722298950931, minval=0, maxval=20), leak=Fitted(4.261726862322623, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6828419639130898, minval=0.5, maxval=3), tau=Fitted(0.75363626582225, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2897891687529234, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1507.4435460225254\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.408202907955278, minval=0, maxval=20), leak=Fitted(4.048644737900576, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6282260160991293, minval=0.5, maxval=3), tau=Fitted(0.07592865183760011, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.38577218494091436, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=901.1861062380931\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.588017723682675, minval=0, maxval=20), leak=Fitted(-2.591926286742564, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.0080211314743015, minval=0.5, maxval=3), tau=Fitted(3.217372023617042, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.29021166248759295, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=977.7842404443923\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.338454820550954, minval=0, maxval=20), leak=Fitted(-3.672295794220888, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7649875475682029, minval=0.5, maxval=3), tau=Fitted(2.338006599743412, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22593933321949852, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-156.9515029829631\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.046864897896324, minval=0, maxval=20), leak=Fitted(4.388720596279077, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.71618100975974, minval=0.5, maxval=3), tau=Fitted(0.4870401018240913, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2839307141802278, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=142.645234836083\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.586005550532338, minval=0, maxval=20), leak=Fitted(-0.9499488780508014, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.7930560922549192, minval=0.5, maxval=3), tau=Fitted(1.6978901989510478, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.0981607914621298, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=988.230528448673\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.293055896695583, minval=0, maxval=20), leak=Fitted(2.679917587550189, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7116392867204253, minval=0.5, maxval=3), tau=Fitted(1.1671493730259135, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3077432134435296, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1183.818576201569\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.580308972485844, minval=0, maxval=20), leak=Fitted(1.5691424613369298, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8020434805605563, minval=0.5, maxval=3), tau=Fitted(0.4356128613056276, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2200449199786344, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=677.5662113232465\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.474107319706377, minval=0, maxval=20), leak=Fitted(3.2491869395401785, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.4465257916543584, minval=0.5, maxval=3), tau=Fitted(4.317386728046326, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.14037065165845358, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=6525.686585551668\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.567607954356669, minval=0, maxval=20), leak=Fitted(-0.09912182167870709, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.5311705006702563, minval=0.5, maxval=3), tau=Fitted(0.7137481257298848, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.31643335851802745, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1446.3964958021384\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(2.643159853445222, minval=0, maxval=20), leak=Fitted(0.3232496638842397, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8760109352124414, minval=0.5, maxval=3), tau=Fitted(0.4968428697796403, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3153502364564092, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5884.899110525365\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.923032283615042, minval=0, maxval=20), leak=Fitted(-7.0413233175597, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6396353221092779, minval=0.5, maxval=3), tau=Fitted(1.451660237588177, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.1825594842728903, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4628.704389758083\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.917671444921647, minval=0, maxval=20), leak=Fitted(4.629221465111851, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.1533903511529964, minval=0.5, maxval=3), tau=Fitted(2.0169610761568455, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.284333050519989, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=252.428825144831\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.127315283218138, minval=0, maxval=20), leak=Fitted(-1.2808219489090311, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.016362784624726, minval=0.5, maxval=3), tau=Fitted(3.0481927475556763, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2739597965092133, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=676.4869837202277\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.08182665953621, minval=0, maxval=20), leak=Fitted(-0.7560108344204419, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.619823130171536, minval=0.5, maxval=3), tau=Fitted(3.5042999454313213, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.30807435707883923, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1506.1360429257818\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.7638556908023375, minval=0, maxval=20), leak=Fitted(0.8210419406106539, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.646021890484472, minval=0.5, maxval=3), tau=Fitted(3.5713413268529157, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22599584499929667, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=538.9202178007708\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(15.38174424966538, minval=0, maxval=20), leak=Fitted(-0.5920600261751785, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.880293261698849, minval=0.5, maxval=3), tau=Fitted(0.6354737360908789, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.13170750110792706, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4167.008357195672\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.042646977060684, minval=0, maxval=20), leak=Fitted(4.015412126359679, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6245294738682132, minval=0.5, maxval=3), tau=Fitted(1.2210716182404833, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.39529064263864805, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1590.905155317061\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.414632011627237, minval=0, maxval=20), leak=Fitted(-4.125782982717615, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.8703227685536883, minval=0.5, maxval=3), tau=Fitted(1.586532720607829, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.23267486665691195, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=798.9285454640684\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.420014871244291, minval=0, maxval=20), leak=Fitted(-1.3869642755262945, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.3254170555362608, minval=0.5, maxval=3), tau=Fitted(1.6357966461405167, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10858363771386753, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=169.77035454434937\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.39728994991881, minval=0, maxval=20), leak=Fitted(-0.3654873058964947, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.6790215583994366, minval=0.5, maxval=3), tau=Fitted(1.7729901971042414, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2735714554597759, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-78.79058510885596\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.504885616767893, minval=0, maxval=20), leak=Fitted(6.682557312266695, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2568711022820251, minval=0.5, maxval=3), tau=Fitted(0.9681671434350654, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.13066594873663, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3900.9868386047365\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.87012129279534, minval=0, maxval=20), leak=Fitted(1.4643557049233857, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.6733052820758325, minval=0.5, maxval=3), tau=Fitted(3.640930222919366, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2118737723335134, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=595.7204043054701\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.314814440565318, minval=0, maxval=20), leak=Fitted(5.753293066971716, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5051177894204255, minval=0.5, maxval=3), tau=Fitted(2.545690026204432, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.13059343849769756, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4472.1119652426005\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.302346365143286, minval=0, maxval=20), leak=Fitted(0.2573957133639304, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4751978940006012, minval=0.5, maxval=3), tau=Fitted(0.6104743596784257, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3381052950118263, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1146.1560486800363\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(11.607575369098907, minval=0, maxval=20), leak=Fitted(-2.228813337316975, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.6063264999550313, minval=0.5, maxval=3), tau=Fitted(2.9421079894278273, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.27702814834283945, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=5179.698207561015\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.95245439067884, minval=0, maxval=20), leak=Fitted(1.9900028082231014, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.8020613941783195, minval=0.5, maxval=3), tau=Fitted(2.1726218540084434, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.0783002713072742, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=546.1002124392357\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.091424336309498, minval=0, maxval=20), leak=Fitted(6.885885346832914, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.855919760245072, minval=0.5, maxval=3), tau=Fitted(2.60296712130035, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.15856782662542068, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=7944.929267934156\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(17.373716194220535, minval=0, maxval=20), leak=Fitted(-0.3762817965157028, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.218846675968207, minval=0.5, maxval=3), tau=Fitted(0.8444697256206934, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28602997883898096, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2776.3119496235518\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(3.087970325718709, minval=0, maxval=20), leak=Fitted(-0.37504863979786807, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.178423201095526, minval=0.5, maxval=3), tau=Fitted(0.9233662063156427, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3249087727543247, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1811.420153182869\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.074631182171164, minval=0, maxval=20), leak=Fitted(1.20077183637195, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5549002875625753, minval=0.5, maxval=3), tau=Fitted(1.3599957330431733, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19136602903948619, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=-42.837973635549076\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(16.731364877640075, minval=0, maxval=20), leak=Fitted(-7.987100935806387, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.7793133833462098, minval=0.5, maxval=3), tau=Fitted(2.4448282365502823, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.08301173154829905, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=517.7624774127937\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.0786388400247304, minval=0, maxval=20), leak=Fitted(2.00423591364451, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.0553503557991277, minval=0.5, maxval=3), tau=Fitted(2.741601596966736, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.28842127270967477, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1995.1531066021698\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(18.229132518562764, minval=0, maxval=20), leak=Fitted(-8.29813014469433, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.5547727593195229, minval=0.5, maxval=3), tau=Fitted(0.8701843590146547, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2896093144078377, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=8601.755635360376\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(13.0399294506544, minval=0, maxval=20), leak=Fitted(-5.1315483580845935, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2992857700838554, minval=0.5, maxval=3), tau=Fitted(1.0060408180374776, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.29361050238487485, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=2798.5445771602153\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.833548137405348, minval=0, maxval=20), leak=Fitted(2.8079623014274757, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.2175369170718895, minval=0.5, maxval=3), tau=Fitted(2.6100785813179233, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3182377665445639, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1875.467605812454\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.7638556908023375, minval=0, maxval=20), leak=Fitted(0.8210419406106539, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.646021890484472, minval=0.5, maxval=3), tau=Fitted(3.5713413268529157, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3087955017811231, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1587.6600752281897\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.329848467798351, minval=0, maxval=20), leak=Fitted(4.527634141625785, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.395585135879394, minval=0.5, maxval=3), tau=Fitted(1.6175883937935178, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22184968071061095, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=140.8838989051459\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.2191912323699, minval=0, maxval=20), leak=Fitted(-0.20627343054229308, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.9044057734034636, minval=0.5, maxval=3), tau=Fitted(4.10864549901708, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2508198300728628, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=775.3001696033166\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(1.1329537552466302, minval=0, maxval=20), leak=Fitted(-1.674696922854365, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.2101045561643131, minval=0.5, maxval=3), tau=Fitted(2.2987996738238228, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.19399082261501208, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1143.2900525818598\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(9.457409125010907, minval=0, maxval=20), leak=Fitted(-1.1907935144608928, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4895889059291028, minval=0.5, maxval=3), tau=Fitted(0.003322245405681379, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.22766809223226236, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=4500.597542008969\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(6.574679473443434, minval=0, maxval=20), leak=Fitted(-1.2140745631605399, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.8276026388456788, minval=0.5, maxval=3), tau=Fitted(2.948792940924128, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.24455678134621084, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=245.22236026596633\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.451110625087529, minval=0, maxval=20), leak=Fitted(-3.3388799706816297, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4223284152443, minval=0.5, maxval=3), tau=Fitted(2.6791444818677568, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.21356320324400313, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=106.40212825915685\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(14.75386126500457, minval=0, maxval=20), leak=Fitted(0.7224774995266126, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.7845442091359919, minval=0.5, maxval=3), tau=Fitted(2.177901875408141, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.2687604515414155, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3122.089591423941\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(7.9887756822755875, minval=0, maxval=20), leak=Fitted(-3.487295275882528, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4198131258737807, minval=0.5, maxval=3), tau=Fitted(0.9009211924302423, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.10188000621434763, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1247.5470528026472\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(19.775949613930184, minval=0, maxval=20), leak=Fitted(6.383662727657722, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(1.4058671942184882, minval=0.5, maxval=3), tau=Fitted(1.8377755403369176, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.3747807125064965, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=1272.444425191437\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(8.236727079676239, minval=0, maxval=20), leak=Fitted(-5.325075472938767, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(2.461104755166251, minval=0.5, maxval=3), tau=Fitted(1.4157902438328742, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.17071828277900847, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=9098.139880995068\nModel(name='Roitman data, leaky drift varies with coherence', drift=DriftCoherenceLeak(driftcoh=Fitted(5.100853022622461, minval=0, maxval=20), leak=Fitted(5.3826627252158605, minval=-10, maxval=10)), noise=NoiseConstant(noise=1), bound=BoundCollapsingExponential(B=Fitted(0.9462701971571568, minval=0.5, maxval=3), tau=Fitted(0.06915585845883054, minval=0.0001, maxval=5)), IC=ICPointSourceCenter(), overlay=OverlayChain(overlays=[OverlayNonDecision(nondectime=Fitted(0.09330760266519345, minval=0, maxval=0.4)), OverlayPoissonMixture(pmixturecoef=0.02, rate=1)]), dx=0.01, dt=0.01, T_dur=2) loss=3139.6131767938973\n"
]
],
[
[
"PyDDM can simulate models and generate artificial data, or it can fit them to data. Below are high-level overviews for how to accomplish each.\n\nTo simulate models and generate artificial data:\n\nOptionally, define unique components of your model. Models are modular, and allow specifying a dynamic drift rate, noise level, diffusion bounds, starting position of the integrator, or post-simulation modifications to the RT histogram. Many common models for these are included by default, but for advance functionality you may need to subclass Drift, Noise, Bound, InitialCondition, or Overlay. These model components may depend on “conditions”, i.e. prespecified values associated with the behavioral task which change from trial to trial (e.g. stimulus coherence), or “parameters”, i.e. values which apply to all trials and should be fit to the subject.\nDefine a model. Models are represented by creating an instance of the Model class, and specifying the model components to use for it. These model component can either be the model components included in PyDDM or ones you created in step 1. Values must be specified for all parameters required by model components.\nSimulate the model using the Model.solve() method to generate a Solution object. If you have multiple conditions, you must run Model.solve() separately for each set of conditions and generate separate Solution objects.\nRun the Solution.resample() method of the Solution object to generate a Sample. If you have multiple Solution objects (for multiple task conditions), you will need to generate multiple Sample objects as well. These can be added together with the “+” operator to form one single Sample object.",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec73337c096290e5b2a7a681416d2d18a2a95540 | 54,326 | ipynb | Jupyter Notebook | tutorials/W0D5_Statistics/student/W0D5_Tutorial1.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 2,294 | 2020-05-11T12:05:35.000Z | 2022-03-28T21:23:34.000Z | tutorials/W0D5_Statistics/student/W0D5_Tutorial1.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 629 | 2020-05-11T15:42:26.000Z | 2022-03-29T12:23:35.000Z | tutorials/W0D5_Statistics/student/W0D5_Tutorial1.ipynb | janeite/course-content | 2a3ba168c5bfb5fd5e8305fe3ae79465b0add52c | [
"CC-BY-4.0",
"BSD-3-Clause"
]
| 917 | 2020-05-11T12:47:53.000Z | 2022-03-31T12:14:41.000Z | 40.877351 | 576 | 0.611125 | [
[
[
"<a href=\"https://colab.research.google.com/github/NeuromatchAcademy/course-content/blob/master/tutorials/W0D5_Statistics/student/W0D5_Tutorial1.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"# Tutorial 1: Probability Distributions\n**Week 0, Day 5: Probability & Statistics**\n\n**By Neuromatch Academy**\n\n__Content creators:__ Ulrik Beierholm\n\n__Content reviewers:__ Natalie Schaworonkow, Keith van Antwerp, Anoop Kulkarni, Pooya Pakarian, Hyosub Kim\n\n__Production editors:__ Ethan Cheng, Ella Batty \n",
"_____no_output_____"
],
[
"**Our 2021 Sponsors, including Presenting Sponsor Facebook Reality Labs**\n\n<p align='center'><img src='https://github.com/NeuromatchAcademy/widgets/blob/master/sponsors.png?raw=True'/></p>",
"_____no_output_____"
],
[
"---\n# Tutorial Objectives\n\nWe will cover the basic ideas from probability and statistics, as a reminder of what you have hopefully previously learned. These ideas will be important for almost every one of the following topics covered in the course. \n\nThere are many additional topics within probability and statistics that we will not cover as they are not central to the main course. We also do not have time to get into a lot of details, but this should help you recall material you have previously encountered.\n\n\nBy completing the exercises in this tutorial, you should:\n* get some intuition about how stochastic randomly generated data can be\n* understand how to model data using simple probability distributions\n* understand the difference between discrete and continuous probability distributions\n* be able to plot a Gaussian distribution\n",
"_____no_output_____"
],
[
"---\n# Setup\n",
"_____no_output_____"
]
],
[
[
"# Imports\n\nimport numpy as np\nimport matplotlib.pyplot as plt\nimport scipy as sp\nfrom scipy.stats import norm # the normal probability distribution",
"_____no_output_____"
],
[
"#@title Figure settings\nimport ipywidgets as widgets # interactive display\nfrom ipywidgets import interact, fixed, HBox, Layout, VBox, interactive, Label, interact_manual\n%config InlineBackend.figure_format = 'retina'\nplt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/nma.mplstyle\")\n#plt.style.use(\"https://raw.githubusercontent.com/NeuromatchAcademy/course-content/NMA2020/nma.mplstyle\")",
"_____no_output_____"
],
[
"#@title Plotting Functions\n\ndef plot_random_sample(x, y, figtitle = None):\n \"\"\" Plot the random sample between 0 and 1 for both the x and y axes.\n\n Args:\n x (ndarray): array of x coordinate values across the random sample\n y (ndarray): array of y coordinate values across the random sample\n figtitle (str): title of histogram plot (default is no title)\n\n Returns:\n Nothing.\n \"\"\"\n fig, ax = plt.subplots()\n ax.set_xlabel('x')\n ax.set_ylabel('y')\n plt.xlim([-0.25, 1.25]) # set x and y axis range to be a bit less than 0 and greater than 1\n plt.ylim([-0.25, 1.25])\n plt.scatter(dataX, dataY)\n if figtitle is not None:\n fig.suptitle(figtitle, size=16)\n plt.show()\n\ndef plot_random_walk(x, y, figtitle = None):\n \"\"\" Plots the random walk within the range 0 to 1 for both the x and y axes.\n\n Args:\n x (ndarray): array of steps in x direction\n y (ndarray): array of steps in y direction\n figtitle (str): title of histogram plot (default is no title)\n\n Returns:\n Nothing.\n \"\"\"\n fig, ax = plt.subplots()\n plt.plot(x,y,'b-o', alpha = 0.5)\n plt.xlim(-0.1,1.1)\n plt.ylim(-0.1,1.1)\n ax.set_xlabel('x location')\n ax.set_ylabel('y location')\n plt.plot(x[0], y[0], 'go')\n plt.plot(x[-1], y[-1], 'ro')\n\n if figtitle is not None:\n fig.suptitle(figtitle, size=16)\n plt.show()\n\ndef plot_hist(data, xlabel, figtitle = None, num_bins = None):\n \"\"\" Plot the given data as a histogram.\n\n Args:\n data (ndarray): array with data to plot as histogram\n xlabel (str): label of x-axis\n figtitle (str): title of histogram plot (default is no title)\n num_bins (int): number of bins for histogram (default is 10)\n\n Returns:\n count (ndarray): number of samples in each histogram bin\n bins (ndarray): center of each histogram bin\n \"\"\"\n fig, ax = plt.subplots()\n ax.set_xlabel(xlabel)\n ax.set_ylabel('Count')\n if num_bins is not None:\n count, bins, _ = plt.hist(data, bins = num_bins)\n else:\n count, bins, _ = plt.hist(data, bins = np.arange(np.min(data)-.5, np.max(data)+.6)) # 10 bins default\n if figtitle is not None:\n fig.suptitle(figtitle, size=16)\n plt.show()\n return count, bins\n\ndef my_plot_single(x, px):\n \"\"\"\n Plots normalized Gaussian distribution\n\n Args:\n x (numpy array of floats): points at which the likelihood has been evaluated\n px (numpy array of floats): normalized probabilities for prior evaluated at each `x`\n\n Returns:\n Nothing.\n \"\"\"\n if px is None:\n px = np.zeros_like(x)\n\n fig, ax = plt.subplots()\n ax.plot(x, px, '-', color='C2', LineWidth=2, label='Prior')\n ax.legend()\n ax.set_ylabel('Probability')\n ax.set_xlabel('Orientation (Degrees)')\n\ndef plot_gaussian_samples_true(samples, xspace, mu, sigma, xlabel, ylabel):\n \"\"\" Plot a histogram of the data samples on the same plot as the gaussian\n distribution specified by the give mu and sigma values.\n\n Args:\n samples (ndarray): data samples for gaussian distribution\n xspace (ndarray): x values to sample from normal distribution\n mu (scalar): mean parameter of normal distribution\n sigma (scalar): variance parameter of normal distribution\n xlabel (str): the label of the x-axis of the histogram\n ylabel (str): the label of the y-axis of the histogram\n\n Returns:\n Nothing.\n \"\"\"\n fig, ax = plt.subplots()\n ax.set_xlabel(xlabel)\n ax.set_ylabel(ylabel)\n # num_samples = samples.shape[0]\n\n count, bins, _ = plt.hist(samples, density=True)\n plt.plot(xspace, norm.pdf(xspace, mu, sigma),'r-')\n plt.show()",
"_____no_output_____"
]
],
[
[
"---\n\n# Section 1: Stochasticity and randomness",
"_____no_output_____"
],
[
"## Section 1.1: Intro to Randomness\n",
"_____no_output_____"
]
],
[
[
"# @title Video 1: Stochastic World\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV1sU4y1G7Qt\", width=854, height=480, fs=1)\n print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"-QwTPDp7-a8\", width=854, height=480, fs=1, rel=0)\n print('Video available at https://youtube.com/watch?v=' + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"\nBefore trying out different probability distributions, let's start with the simple uniform distribution, U(a,b), which assigns equal probability to any value between a and b.\n\nTo show that we are drawing a random number $x$ from a uniform distribution with lower and upper bounds $a$ and $b$ we will use this notation:\n$x \\sim U(a,b)$. Alternatively, we can say that all the potential values of $x$ are distributed as a uniform distribution between $a$ and $b$. $x$ here is a random variable: a variable whose value depends on the outcome of a random process.",
"_____no_output_____"
],
[
"### Coding Exercise 1.1: Create randomness\n\nNumpy has many functions and capabilities related to randomness. We can draw random numbers from various probability distributions. For example, to draw 5 uniform numbers between 0 and 100, you would use `np.random.uniform(0, 100, size = (5,))`. \n\n We will use `np.random.seed` to set a specific seed for the random number generator. For example, `np.random.seed(0)` sets the seed as 0. By including this, we are actually making the random numbers reproducible, which may seem odd at first. Basically if we do the below code without that 0, we would get different random numbers every time we run it. By setting the seed to 0, we ensure we will get the same random numbers. There are lots of reasons we may want randomness to be reproducible. In NMA-world, it's so your plots will match the solution plots exactly! \n\n```\nnp.random.seed(0)\nrandom_nums = np.random.uniform(0, 100, size = (5,))\n```\n\nBelow, you will complete a function `generate_random_sample` that randomly generates `num_points` $x$ and $y$ coordinate values, all within the range 0 to 1. You will then generate 10 points and visualize.\n\n\n\n",
"_____no_output_____"
]
],
[
[
"def generate_random_sample(num_points):\n \"\"\" Generate a random sample containing a desired number of points (num_points)\n in the range [0, 1] using a random number generator object.\n\n Args:\n num_points (int): number of points desired in random sample\n\n Returns:\n dataX, dataY (ndarray, ndarray): arrays of size (num_points,) containing x\n and y coordinates of sampled points\n\n \"\"\"\n\n ###################################################################\n ## TODO for students: Draw the uniform numbers\n ## Fill out the following then remove\n raise NotImplementedError(\"Student exercise: need to complete generate_random_sample\")\n ###################################################################\n\n # Generate desired number of points uniformly between 0 and 1 (using uniform) for\n # both x and y\n dataX = ...\n dataY = ...\n\n return dataX, dataY\n\n# Set a seed\nnp.random.seed(0)\n\n# Set number of points to draw\nnum_points = 10\n\n# Draw random points\ndataX, dataY = generate_random_sample(num_points)\n\n# Visualize\nplot_random_sample(dataX, dataY, \"Random sample of 10 points\")",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_0e972635.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=1120.0 height=845.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial1_Solution_0e972635_0.png>\n\n",
"_____no_output_____"
],
[
"### Interactive Demo 1.1: Random Sample Generation from Uniform Distribution\nIn practice this may not look very uniform, although that is of course part of the randomness! Uniform randomness does not mean smoothly uniform. When we have very little data it can be hard to see the distribution.\n\nBelow, you can adjust the number of points sampled with a slider. Does it look more uniform now? Try increasingly large numbers of sampled points.",
"_____no_output_____"
]
],
[
[
"#@markdown Make sure you execute this cell to enable the widget!\n\ndef generate_random_sample(num_points):\n \"\"\" Generate a random sample containing a desired number of points (num_points)\n in the range [0, 1] using a random number generator object.\n\n Args:\n num_points (int): number of points desired in random sample\n\n Returns:\n dataX, dataY (ndarray, ndarray): arrays of size (num_points,) containing x\n and y coordinates of sampled points\n\n \"\"\"\n\n # Generate desired number of points uniformly between 0 and 1 (using uniform) for\n # both x and y\n dataX = np.random.uniform(0, 1, size = (num_points,))\n dataY = np.random.uniform(0, 1, size = (num_points,))\n\n return dataX, dataY\n\[email protected]\ndef gen_and_plot_random_sample(num_points = widgets.SelectionSlider(options=[(\"%g\"%i,i) for i in np.arange(0, 500, 10)])):\n\n dataX, dataY = generate_random_sample(num_points)\n fig, ax = plt.subplots()\n ax.set_xlabel('x')\n ax.set_ylabel('y')\n plt.xlim([-0.25, 1.25])\n plt.ylim([-0.25, 1.25])\n plt.scatter(dataX, dataY)\n fig.suptitle(\"Random sample of \" + str(num_points) + \" points\", size=16)\n plt.show()",
"_____no_output_____"
]
],
[
[
"## Section 1.2: Random walk\n\n",
"_____no_output_____"
]
],
[
[
"# @title Video 2: Random walk\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV11U4y1G7Bu\", width=854, height=480, fs=1)\n print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"Tz9gjHcqj5k\", width=854, height=480, fs=1, rel=0)\n print('Video available at https://youtube.com/watch?v=' + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"Stochastic models can be used to create models of behaviour. As an example, imagine that a rat is placed inside a novel environment, a box. We could try and model its exploration behaviour by assuming that for each time step it takes a random uniformly sampled step in any direction (simultaneous random step in x direction and random step in y direction)\n",
"_____no_output_____"
],
[
"### Coding Exercise 1.2: Modeling a random walk\n\n\nUse the `generate_random_sample` function from above to obtain the random steps the rat takes at each time step and complete the generate_random_walk function below. For plotting, the box will be represented graphically as the unit square enclosed by the points (0, 0) and (1, 1).",
"_____no_output_____"
]
],
[
[
"def generate_random_walk(num_steps, step_size):\n \"\"\" Generate the points of a random walk within a 1 X 1 box.\n\n Args:\n num_steps (int): number of steps in the random walk\n step_size (float): how much each random step size is weighted\n\n Returns:\n x, y (ndarray, ndarray): the (x, y) locations reached at each time step of the walk\n\n \"\"\"\n x = np.zeros(num_steps + 1)\n y = np.zeros(num_steps + 1)\n\n ###################################################################\n ## TODO for students: Collect random step values with function from before\n ## Fill out the following then remove\n raise NotImplementedError(\"Student exercise: need to complete generate_random_walk\")\n ###################################################################\n\n # Generate the uniformly random x, y steps for the walk\n random_x_steps, random_y_steps = ...\n\n # Take steps according to the randomly sampled steps above\n for step in range(num_steps):\n\n # take a random step in x and y. We remove 0.5 to make it centered around 0\n x[step + 1] = x[step] + (random_x_steps[step] - 0.5)*step_size\n y[step + 1] = y[step] + (random_y_steps[step] - 0.5)*step_size\n\n # restrict to be within the 1 x 1 unit box\n x[step + 1]= min(max(x[step + 1], 0), 1)\n y[step + 1]= min(max(y[step + 1], 0), 1)\n\n return x, y\n\n# Set a random seed\nnp.random.seed(2)\n\n# Select parameters\nnum_steps = 100 # number of steps in random walk\nstep_size = 0.5 # size of each step\n\n# Generate the random walk\nx, y = generate_random_walk(num_steps, step_size)\n\n# Visualize\nplot_random_walk(x, y, \"Rat's location throughout random walk\")",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_84dec82b.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=1120.0 height=845.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial1_Solution_84dec82b_0.png>\n\n",
"_____no_output_____"
],
[
"We put a little green dot for the starting point and a red point for the ending point. ",
"_____no_output_____"
],
[
"### Interactive Demo 1.2: Varying parameters of a random walk\nIn the interactive demo below, you can examine random walks with different numbers of steps or step sizes, using the sliders. \n\n\n1. What could an increased step size mean for the actual rat's movement we are simulating?\n2. For a given number of steps, is the rat more likely to visit all general areas of the arena with a big step size or small step size?",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\n\[email protected](num_steps = widgets.IntSlider(value=100, min=0, max=500, step=1), step_size = widgets.FloatSlider(value=0.1, min=0.1, max=1, step=0.1))\ndef gen_and_plot_random_walk(num_steps, step_size):\n x, y = generate_random_walk(num_steps, step_size)\n plot_random_walk(x, y, \"Rat's location throughout random walk\")",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_6e912c89.py)\n\n",
"_____no_output_____"
],
[
"In practice a uniform random movement is too simple an assumption. Rats do not move completely randomly; even if you could assume that, you would need to approximate with a more complex probability distribution.\n\nNevertheless, this example highlights how you can use sampling to approximate behaviour. \n\n**Main course preview:** On day W3D2 we will see how random walk models can be used to also model accumulation of information in decision making.",
"_____no_output_____"
],
[
"---\n# Section 2: Discrete distributions",
"_____no_output_____"
],
[
"## Section 2.1: Binomial distributions",
"_____no_output_____"
]
],
[
[
"# @title Video 3: Binomial distribution\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV1Ev411W7mw\", width=854, height=480, fs=1)\n print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"kOXEQlmzFyw\", width=854, height=480, fs=1, rel=0)\n print('Video available at https://youtube.com/watch?v=' + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"This video covers the Bernoulli and binomial distributions.\n\n<details>\n<summary> <font color='blue'>Click here for text recap of video </font></summary>\n\nThe uniform distribution is very simple, and can only be used in some rare cases. If we only had access to this distribution, our statistical toolbox would be very empty. Thankfully we do have some more advanced distributions!\n\nThe uniform distribution that we looked at above is an example of a continuous distribution. The value of $X$ that we draw from this distribution can take **any value** between $a$ and $b$.\n\nHowever, sometimes we want to be able to look at discrete events. Imagine that the rat from before is now placed in a T-maze, with food placed at the end of both arms. Initially, we would expect the rat to be choosing randomly between the two arms, but after learning it should choose more consistently.\n\nA simple way to model such random behaviour is with a single **Bernoulli trial**, that has two outcomes, {$Left, Right$}, with probability $P(Left)=p$ and $P(Right)=1-p$ as the two mutually exclusive possibilities (whether the rat goes down the left or right arm of the maze).\n</details>\n\nThe binomial distribution simulates $n$ number of binary events, such as the $Left, Right$ choices of the random rat in the T-maze. Imagine that you have done an experiment and found that your rat turned left in 7 out of 10 trials. What is the probability of the rat indeed turning left 7 times ($k = 7$)?\n\nThis is given by the binomial probability of $k$, given $n$ trials and probability $p$:\n\n$$ P(k|n,p)= \\left( \\begin{array} \\\\n \\\\ k\\end{array} \\right) p^k (1-p)^{n-k}$$\n$$\\binom {n}{k}={\\frac {n!}{k!(n-k)!}}$$\n\nIn this formula, $p$ is the probability of turning left, $n$ is the number of binary events, or trials, and $k$ is the number of times the rat turned left. The term $\\binom {n}{k}$ is the binomial coefficient.\n\nThis is an example of a *probability mass function*, which specifies the probability that a discrete random variable is equal to each value. In other words, how large a part of the probability space (mass) is placed at each exact discrete value. We require that all probability adds up to 1, i.e. that \n\n\n$\\sum_k P(k|n,p)=1$.\n\nEssentially, if $k$ can only be one of 10 values, the probabilities of $k$ being equal to each possible value have to sum up to 1 because there is a probability of 1 it will equal one of those 10 values (no other options exist).\n\n\nIf we assume an equal chance of turning left or right, then $p=0.5$. Note that if we only have a single trial $n=1$ this is equivalent to a single Bernoulli trial (feel free to do the math!).\n\n",
"_____no_output_____"
],
[
"### Think! 2.1: Binomial distribution sampling\nWe will draw a desired number of random samples from a binomial distribution, with $n = 10$ and $p = 0.5$. Each sample returns the number of trials, $k$, a rat turns left out of $n$ trials.\n\nWe will draw 1000 samples of this (so it is as if we are observing 10 trials of the rat, 1000 different times). We can do this using numpy: `np.random.binomial(n, p, size = (n_samples,))`\n\nSee below to visualize a histogram of the different values of $k$, or the number of times the rat turned left in each of the 1000 samples. In a histogram all the data is placed into bins and the contents of each bin is counted, to give a visualisation of the distribution of data. Discuss the following questions.\n\n\n1. What are the x-axis limits of the histogram and why?\n2. What is the shape of the histogram?\n3. Looking at the histogram, how would you interpret the outcome of the simulation if you didn't know what p was? Would you have guessed p = 0.5?\n3. What do you think the histogram would look like if the probability of turning left is 0.8 ($p = 0.8$)?\n\n\n",
"_____no_output_____"
]
],
[
[
"# @markdown Execute this cell to see visualization\n\n# Select parameters for conducting binomial trials\nn = 10\np = 0.5\nn_samples = 1000\n\n# Set random seed\nnp.random.seed(1)\n\n# Now draw 1000 samples by calling the function again\nleft_turn_samples_1000 = np.random.binomial(n, p, size = (n_samples,))\n\n# Visualize\ncount, bins = plot_hist(left_turn_samples_1000, 'Number of left turns in sample')",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_06a79c9b.py)\n\n",
"_____no_output_____"
],
[
"When working with the Bernoulli and binomial distributions, there are only 2 possible outcomes (in this case, turn left or turn right). In the more general case where there are $n$ possible outcomes (our rat is an n-armed maze) each with their own associated probability $p_1, p_2, p_3, p_4, ...$ , we use a **categorical distribution**. Draws from this distribution are a simple extension of the Bernoulli trial: we now have a probability for each outcome and draw based on those probabilities. We have to make sure that the probabilities sum to one:\n\n$$\\sum_i P(x=i)=\\sum_i p_i =1$$\n\nIf we sample from this distribution multiple times, we can then describe the distribution of outcomes from each sample as the **multinomial distribution**. Essentially, the categorical distribution is the multiple outcome extension of the Bernoulli, and the multinomial distribution is the multiple outcome extension of the binomial distribution. We'll see a bit more about this in the next tutorial when we look at Markov chains.",
"_____no_output_____"
],
[
"## Section 2.2: Poisson distribution",
"_____no_output_____"
]
],
[
[
"# @title Video 4: Poisson distribution\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV1wV411x7P6\", width=854, height=480, fs=1)\n print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"E_nvNb596DY\", width=854, height=480, fs=1, rel=0)\n print('Video available at https://youtube.com/watch?v=' + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"This video covers the Poisson distribution and how it can be used to describe neural spiking.\n<details>\n<summary> <font color='blue'>Click here for text recap of video </font></summary>\n\nFor some phenomena there may not be a natural limit on the maximum number of possible events or outcomes. \n\nThe Poisson distribution is a '**point-process**', meaning that it determines the number of discrete 'point', or binary, events that happen within a fixed space or time, allowing for the occurence of a potentially infinite number of events. The Poisson distribution is specified by a single parameter $\\lambda$ that encapsulates the mean number of events that can occur in a single time or space interval (there will be more on this concept of the 'mean' later!). \n\nRelevant to us, we can model the number of times a neuron spikes within a time interval using a Poisson distribution. In fact, neuroscientists often do! As an example, if we are recording from a neuron that tends to fire at an average rate of 4 spikes per second, then the Poisson distribution specifies the distribution of recorded spikes over one second, where $\\lambda=4$.\n\n</details>\n\nThe formula for a Poisson distribution on $x$ is: \n\n$$P(x)=\\frac{\\lambda^x e^{-\\lambda}}{x!}$$\nwhere $\\lambda$ is a parameter corresponding to the average outcome of $x$.",
"_____no_output_____"
],
[
"### Coding Exercise 2.2: Poisson distribution sampling\n\nIn the exercise below we will draw some samples from the Poisson distribution and see what the histogram looks.\n\nIn the code, fill in the missing line so we draw 5 samples from a Poisson distribution with $\\lambda = 4$. Use `np.random.poisson`.",
"_____no_output_____"
]
],
[
[
"# Set random seed\nnp.random.seed(0)\n\n# Draw 5 samples from a Poisson distribution with lambda = 4\nsampled_spike_counts = ...\n\n# Print the counts\nprint(\"The samples drawn from the Poisson distribution are \" +\n str(sampled_spike_counts))",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_90422623.py)\n\n",
"_____no_output_____"
],
[
"You should see that the neuron spiked 6 times, 7 times, 1 time, 8 times, and 4 times in 5 different intervals.",
"_____no_output_____"
],
[
"### Interactive Demo 2.2: Varying parameters of Poisson distribution\n\nUse the interactive demo below to vary $\\lambda$ and the number of samples, and then visualize the resulting histogram.\n\n\n1. What effect does increasing the number of samples have? \n2. What effect does changing $\\lambda$ have?\n3. With a small lambda, why is the distribution asymmetric?\n",
"_____no_output_____"
]
],
[
[
"# @markdown Make sure you execute this cell to enable the widget!\n\[email protected](lambda_value = widgets.FloatSlider(value=4, min=0.1, max=10, step=0.1),\n n_samples = widgets.IntSlider(value=5, min=5, max=500, step=1))\n\ndef gen_and_plot_possion_samples(lambda_value, n_samples):\n sampled_spike_counts = np.random.poisson(lambda_value, n_samples)\n count, bins = plot_hist(sampled_spike_counts, 'Recorded spikes per second')",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_81eb1b08.py)\n\n",
"_____no_output_____"
],
[
"---\n# Section 3: Continuous distributions",
"_____no_output_____"
]
],
[
[
"# @title Video 5: Continuous distributions\nfrom ipywidgets import widgets\n\nout2 = widgets.Output()\nwith out2:\n from IPython.display import IFrame\n class BiliVideo(IFrame):\n def __init__(self, id, page=1, width=400, height=300, **kwargs):\n self.id=id\n src = 'https://player.bilibili.com/player.html?bvid={0}&page={1}'.format(id, page)\n super(BiliVideo, self).__init__(src, width, height, **kwargs)\n\n video = BiliVideo(id=\"BV1dq4y1L7eC\", width=854, height=480, fs=1)\n print('Video available at https://www.bilibili.com/video/{0}'.format(video.id))\n display(video)\n\nout1 = widgets.Output()\nwith out1:\n from IPython.display import YouTubeVideo\n video = YouTubeVideo(id=\"LJ4Zdokb6lc\", width=854, height=480, fs=1, rel=0)\n print('Video available at https://youtube.com/watch?v=' + video.id)\n display(video)\n\nout = widgets.Tab([out1, out2])\nout.set_title(0, 'Youtube')\nout.set_title(1, 'Bilibili')\n\ndisplay(out)",
"_____no_output_____"
]
],
[
[
"We do not have to restrict ourselves to only probabilistic models of discrete events. While some events in neuroscience are discrete (e.g. number of spikes by a neuron), many others are continuous (e.g. neuroimaging signals in EEG or fMRI, distance traveled by an animal, human pointing in a direction of a stimulus).\n\n\nWhile for discrete outcomes we can ask about the probability of an specific event (\"what is the probability this neuron will fire 4 times in the next second\"), this is not defined for a continuous distribution (\"what is the probability of the BOLD signal being exactly 4.000120141...\"). Hence we need to focus on intervals when calculating probabilities from a continuous distribution. \n\nIf we want to make predictions about possible outcomes (\"I believe the BOLD signal from the area will be in the range $x_1$ to $ x_2 $\") we can use the integral $\\int_{x_1}^{x_2} P(x)$.\n$P(x)$ is now a **probability density function**, sometimes written as $f(x)$ to distinguish it from the probability mass functions.\n\n\nWith continuous distributions we have to replace the normalising sum \n\\begin{equation}\\sum_i P(x=p_i) =1\\end{equation}\nover all possible events, with an integral\n\\begin{equation}\\int_a^b P(x) =1\\end{equation}\n\nwhere a and b are the limits of the random variable $x$ (often $-\\infty$ and $\\infty$).\n",
"_____no_output_____"
],
[
"## Section 3.1: Gaussian Distribution\n\nThe most widely used continuous distribution is probably the Gaussian (also known as Normal) distribution. It is extremely common across all kinds of statistical analyses. Because of the central limit theorem, many quantities are Gaussian distributed. Gaussians also have some nice mathematical properties that permit simple closed-form solutions to several important problems. \n\nAs a working example, imagine that a human participant is asked to point in the direction where they perceived a sound coming from. As an approximation, we can assume that the variability in the direction/orientation they point towards is Gaussian distributed.\n",
"_____no_output_____"
],
[
"### Coding Exercise 3.1A: Gaussian Distribution\n\nIn this exercise, you will implement a Gaussian by filling in the missing portions of code for the function `my_gaussian` below. Gaussians have two parameters. The **mean** $\\mu$, which sets the location of its center, and its \"scale\" or spread is controlled by its **standard deviation** $\\sigma$, or **variance** $\\sigma^2$ (i.e. the square of standard deviation). **Be careful not to use one when the other is required.**\n\nThe equation for a Gaussian probability density function is:\n$$\nf(x;\\mu,\\sigma^2)=\\mathcal{N}(\\mu,\\sigma^2) = \\frac{1}{\\sqrt{2\\pi\\sigma^2}}\\exp\\left(\\frac{-(x-\\mu)^2}{2\\sigma^2}\\right)\n$$\nIn Python $\\pi$ and $e$ can be written as `np.pi` and `np.exp` respectively.\n\nAs a probability distribution this has an integral of one when integrated from $-\\infty$ to $\\infty$, however in the following your numerical Gaussian will only be computed over a finite number of points (for the cell below we will sample from -8 to 9 in step sizes of 0.1). You therefore need to explicitly normalize it to sum to one yourself. \n\n\nTest out your implementation with a $\\mu = -1$ and $\\sigma = 1$. ",
"_____no_output_____"
]
],
[
[
"def my_gaussian(x_points, mu, sigma):\n \"\"\" Returns normalized Gaussian estimated at points `x_points`, with\n parameters: mean `mu` and standard deviation `sigma`\n\n Args:\n x_points (ndarray of floats): points at which the gaussian is evaluated\n mu (scalar): mean of the Gaussian\n sigma (scalar): standard deviation of the gaussian\n\n Returns:\n (numpy array of floats) : normalized Gaussian evaluated at `x`\n \"\"\"\n\n ###################################################################\n ## TODO for students: Implement the formula for a Gaussian\n ## Add code to calculate the gaussian px as a function of mu and sigma,\n ## for every x in x_points\n ## Function Hints: exp -> np.exp()\n ## power -> z**2\n ##\n ## Fill out the following then remove\n raise NotImplementedError(\"Student exercise: need to implement Gaussian\")\n ###################################################################\n px = ...\n\n # as we are doing numerical integration we have to remember to normalise\n # taking into account the stepsize (0.1)\n px = px/(0.1*sum(px))\n return px\n\nx = np.arange(-8, 9, 0.1)\n\n# Generate Gaussian\npx = my_gaussian(x, -1, 1)\n\n# Visualize\nmy_plot_single(x, px)",
"_____no_output_____"
]
],
[
[
"[*Click for solution*](https://github.com/NeuromatchAcademy/course-content/tree/master//tutorials/W0D5_Statistics/solutions/W0D5_Tutorial1_Solution_2730515e.py)\n\n*Example output:*\n\n<img alt='Solution hint' align='left' width=1115.0 height=828.0 src=https://raw.githubusercontent.com/NeuromatchAcademy/course-content/master/tutorials/W0D5_Statistics/static/W0D5_Tutorial1_Solution_2730515e_1.png>\n\n",
"_____no_output_____"
],
[
"### Interactive Demo 3.1: Sampling from a Gaussian distribution\n\nNow that we have gained a bit of intuition about the shape of the Gaussian, let's imagine that a human participant is asked to point in the direction of a sound source, which we then measure in horizontal degrees. To simulate that we draw samples from a Normal distribution:\n\n$$x \\sim \\mathcal{N}(\\mu,\\sigma) $$\n\n\nWe can sample from a Gaussian with mean $\\mu$ and standard deviation $\\sigma$ using `np.random.normal(mu, sigma, size = (n_samples,))`.\n\nIn the demo below, you can change the mean and standard deviation of the Gaussian, and the number of samples, we can compare the histogram of the samples to the true analytical distribution (in red).\n\n\n\n1. With what number of samples would you say that the full distribution (in red) is well approximated by the histogram? \n2. What if you just wanted to approximate the variables that defined the distribution, i.e. mean and variance?\n",
"_____no_output_____"
]
],
[
[
"#@markdown Make sure you execute this cell to enable the widget!\n\n\[email protected](mean = widgets.FloatSlider(value=0, min=-5, max=5, step=0.5),\n standard_dev = widgets.FloatSlider(value=0.5, min=0, max=10, step=0.1),\n n_samples = widgets.IntSlider(value=5, min=1, max=300, step=1))\ndef gen_and_plot_normal_samples(mean, standard_dev, n_samples):\n x = np.random.normal(mean, standard_dev, size = (n_samples,))\n xspace = np.linspace(-20, 20, 100)\n plot_gaussian_samples_true(x, xspace, mean, standard_dev,\n 'orientation (degrees)', 'probability')",
"_____no_output_____"
]
],
[
[
"**Main course preview:** Gaussian distriutions are everywhere and are critical for filtering, linear systems (W2D2), optimal control (W3D3) and almost any statistical model of continuous data (W3D1, W3D2, etc.).",
"_____no_output_____"
],
[
"---\n# Summary\n\nAcross the different exercises you should now:\n* have gotten some intuition about how stochastic randomly generated data can be\n* understand how to model data using simple distributions \n* understand the difference between discrete and continuous distributions\n* be able to plot a Gaussian distribution\n\nFor more reading on these topics see just about any statistics textbook, or take a look at the online resources at\nhttps://github.com/NeuromatchAcademy/precourse/blob/master/resources.md",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
]
]
|
ec733764cadfdfbfd7e9213a56b8074e71b26c9b | 5,274 | ipynb | Jupyter Notebook | arrays_strings/compress/compress_solution.ipynb | benkeesey/interactive-coding-challenges | 4994452a729f4bcfab5c8a4225f2b5e004b79075 | [
"Apache-2.0"
]
| 27,173 | 2015-07-06T12:36:05.000Z | 2022-03-31T23:56:41.000Z | arrays_strings/compress/compress_solution.ipynb | benkeesey/interactive-coding-challenges | 4994452a729f4bcfab5c8a4225f2b5e004b79075 | [
"Apache-2.0"
]
| 143 | 2015-07-07T05:13:11.000Z | 2021-12-07T17:05:54.000Z | arrays_strings/compress/compress_solution.ipynb | benkeesey/interactive-coding-challenges | 4994452a729f4bcfab5c8a4225f2b5e004b79075 | [
"Apache-2.0"
]
| 4,657 | 2015-07-06T13:28:02.000Z | 2022-03-31T10:11:28.000Z | 25.478261 | 229 | 0.50948 | [
[
[
"This notebook was prepared by [Donne Martin](http://donnemartin.com). Source and license info is on [GitHub](https://github.com/donnemartin/interactive-coding-challenges).",
"_____no_output_____"
],
[
"# Solution Notebook",
"_____no_output_____"
],
[
"## Problem: Compress a string such that 'AAABCCDDDD' becomes 'A3BC2D4'. Only compress the string if it saves space.\n\n* [Constraints](#Constraints)\n* [Test Cases](#Test-Cases)\n* [Algorithm](#Algorithm)\n* [Code](#Code)\n* [Unit Test](#Unit-Test)",
"_____no_output_____"
],
[
"## Constraints\n\n* Can we assume the string is ASCII?\n * Yes\n * Note: Unicode strings could require special handling depending on your language\n* Is this case sensitive?\n * Yes\n* Can we use additional data structures? \n * Yes\n* Can we assume this fits in memory?\n * Yes",
"_____no_output_____"
],
[
"## Test Cases\n\n* None -> None\n* '' -> ''\n* 'AABBCC' -> 'AABBCC'\n* 'AAABCCDDDD' -> 'A3BC2D4'",
"_____no_output_____"
],
[
"## Algorithm\n\n* For each char in string\n * If char is the same as last_char, increment count\n * Else\n * Append last_char and count to compressed_string\n * last_char = char\n * count = 1\n* Append last_char and count to compressed_string\n* If the compressed string size is < string size\n * Return compressed string\n* Else\n * Return string\n\nComplexity:\n* Time: O(n)\n* Space: O(n)\n\nComplexity Note:\n* Although strings are immutable in Python, appending to strings is optimized in CPython so that it now runs in O(n) and extends the string in-place. Refer to this [Stack Overflow post](http://stackoverflow.com/a/4435752).",
"_____no_output_____"
],
[
"## Code",
"_____no_output_____"
]
],
[
[
"class CompressString(object):\n\n def compress(self, string):\n if string is None or not string:\n return string\n result = ''\n prev_char = string[0]\n count = 0\n for char in string:\n if char == prev_char:\n count += 1\n else:\n result += self._calc_partial_result(prev_char, count)\n prev_char = char\n count = 1\n result += self._calc_partial_result(prev_char, count)\n return result if len(result) < len(string) else string\n\n def _calc_partial_result(self, prev_char, count):\n return prev_char + (str(count) if count > 1 else '')",
"_____no_output_____"
]
],
[
[
"## Unit Test",
"_____no_output_____"
]
],
[
[
"%%writefile test_compress.py\nimport unittest\n\n\nclass TestCompress(unittest.TestCase):\n\n def test_compress(self, func):\n self.assertEqual(func(None), None)\n self.assertEqual(func(''), '')\n self.assertEqual(func('AABBCC'), 'AABBCC')\n self.assertEqual(func('AAABCCDDDDE'), 'A3BC2D4E')\n self.assertEqual(func('BAAACCDDDD'), 'BA3C2D4')\n self.assertEqual(func('AAABAACCDDDD'), 'A3BA2C2D4')\n print('Success: test_compress')\n\n\ndef main():\n test = TestCompress()\n compress_string = CompressString()\n test.test_compress(compress_string.compress)\n\n\nif __name__ == '__main__':\n main()",
"Overwriting test_compress.py\n"
],
[
"%run -i test_compress.py",
"Success: test_compress\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec733cac43da78a209309db1cc4dcda012ca9129 | 299,270 | ipynb | Jupyter Notebook | EDA.ipynb | yahoyoungho/Recipy_Recommender | d74b26cc654ee79f60420daa85bdeed0c6777fa0 | [
"MIT"
]
| null | null | null | EDA.ipynb | yahoyoungho/Recipy_Recommender | d74b26cc654ee79f60420daa85bdeed0c6777fa0 | [
"MIT"
]
| null | null | null | EDA.ipynb | yahoyoungho/Recipy_Recommender | d74b26cc654ee79f60420daa85bdeed0c6777fa0 | [
"MIT"
]
| null | null | null | 140.238988 | 38,180 | 0.826237 | [
[
[
"import pandas as pd\npd.set_option(\"display.max_columns\",None)\nimport numpy as np\nimport pickle\nimport matplotlib.pyplot as plt\nimport seaborn as sns\nfrom collections import Counter",
"_____no_output_____"
],
[
"# load all the df\ningr_map_df = pd.read_pickle(\"src/data/kaggle_food_data/ingr_map.pkl\")\nraw_recipes_df = pd.read_csv(\"src/data/kaggle_food_data/RAW_recipes.csv\")\nraw_interaction_df = pd.read_csv(\"src/data/kaggle_food_data/RAW_interactions.csv\") ",
"_____no_output_____"
],
[
"raw_dfs = [raw_interaction_df, raw_recipes_df]",
"_____no_output_____"
]
],
[
[
"## observing all df",
"_____no_output_____"
]
],
[
[
"ingr_map_df.head()",
"_____no_output_____"
]
],
[
[
"id: map number to replaced variable",
"_____no_output_____"
]
],
[
[
"raw_recipes_df.head()",
"_____no_output_____"
]
],
[
[
"- name: recipe name\n- id: recipe id\n- minutes: minutes to prepare recipe\n- contributor_id: user id who submitted recipe\n- submitted: date recipe was submitted\n- tags: tags for recipe\n- nutrition: (calories (#), total fat (PDV), sugar (PDV) , sodium (PDV) , protein (PDV) , saturated fat (PDV) , and carbohydrates (PDV))\n- n_steps: num of steps in recipe\n- steps: text for recipe step, in order\n- description: user-provided description",
"_____no_output_____"
]
],
[
[
"raw_interaction_df.head()",
"_____no_output_____"
]
],
[
[
"# EDA",
"_____no_output_____"
]
],
[
[
"raw_recipes_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 231637 entries, 0 to 231636\nData columns (total 12 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 name 231636 non-null object\n 1 id 231637 non-null int64 \n 2 minutes 231637 non-null int64 \n 3 contributor_id 231637 non-null int64 \n 4 submitted 231637 non-null object\n 5 tags 231637 non-null object\n 6 nutrition 231637 non-null object\n 7 n_steps 231637 non-null int64 \n 8 steps 231637 non-null object\n 9 description 226658 non-null object\n 10 ingredients 231637 non-null object\n 11 n_ingredients 231637 non-null int64 \ndtypes: int64(5), object(7)\nmemory usage: 21.2+ MB\n"
],
[
"raw_interaction_df.info()",
"<class 'pandas.core.frame.DataFrame'>\nRangeIndex: 1132367 entries, 0 to 1132366\nData columns (total 5 columns):\n # Column Non-Null Count Dtype \n--- ------ -------------- ----- \n 0 user_id 1132367 non-null int64 \n 1 recipe_id 1132367 non-null int64 \n 2 date 1132367 non-null object\n 3 rating 1132367 non-null int64 \n 4 review 1132198 non-null object\ndtypes: int64(3), object(2)\nmemory usage: 43.2+ MB\n"
],
[
"raw_interaction_df.groupby([\"recipe_id\"])[[\"rating\"]].agg(\"mean\").plot.hist()\nplt.title(\"average rating for all recipes\".title(), fontsize = 14)\nplt.xlabel(\"ratings\".title());",
"_____no_output_____"
],
[
"plt.hist(raw_recipes_df[\"n_ingredients\"], bins=43, edgecolor = \"black\")\nplt.title(\"Histogram of number of ingredients used for each recipe\".title(), fontsize = 14);\nplt.xlabel(\"number of ingredients used\".title())\nplt.ylabel(\"frequency\".title());",
"_____no_output_____"
],
[
"plt.hist(raw_recipes_df[\"n_steps\"], bins=25, edgecolor = \"black\")\nplt.title(\"Histogram of number of steps in each recipe\".title(), fontsize = 14);\nplt.xlabel(\"number of steps\".title())\nplt.ylabel(\"frequency\".title());",
"_____no_output_____"
],
[
"len(ingr_map_df[\"id\"].unique()) == len(ingr_map_df[\"replaced\"].unique())",
"_____no_output_____"
],
[
"raw_recipes_df.head()",
"_____no_output_____"
],
[
"def prep_ingr(ingrs):\n toreturn = []\n ingrs = ingrs.strip(\"[]\").replace(\"'\",\"\").replace('\"',\"\").split(\", \")\n for ingr in ingrs:\n toreturn.append(\"_\".join(ingr.split(\" \")))\n return \" \".join(toreturn)",
"_____no_output_____"
],
[
"raw_recipes_df[\"processed_ingrs\"] = raw_recipes_df[\"ingredients\"].apply(prep_ingr)",
"_____no_output_____"
],
[
"raw_recipes_df.head()",
"_____no_output_____"
],
[
"ingr_list = []\nfor ingr in raw_recipes_df[\"processed_ingrs\"].values:\n [ingr_list.append(i) for i in ingr.split(\" \")]\ningr_counts = Counter(ingr_list)",
"_____no_output_____"
],
[
"ingr_names = list(ingr_counts.keys())",
"_____no_output_____"
],
[
"ingr_count_df = pd.DataFrame(ingr_counts, index = [\"counts\"])\ningr_count_df = ingr_count_df.T",
"_____no_output_____"
],
[
"ingr_count_df.sort_values(\"counts\", inplace = True, ascending=False)\ningr_count_df.reset_index(inplace= True)\ningr_count_df.columns = [\"ingredients\",\"counts\"]",
"_____no_output_____"
],
[
"sns.barplot(data = ingr_count_df.iloc[:20,:],x = \"counts\", y = \"ingredients\")\nplt.title(\"20 most used ingredients\".title());",
"_____no_output_____"
],
[
"sns.barplot(data = ingr_count_df.iloc[-20:,:],x = \"counts\", y = \"ingredients\")\nplt.title(\"20 least used ingredients\".title());",
"_____no_output_____"
],
[
"tags_list = []\nfor t in raw_recipes_df[\"tags\"]:\n [tags_list.append(x.replace(\"'\",\"\").replace('\"',\"\").strip(\"[]\")) for x in t.split(\", \")]\ntag_counts = Counter(tags_list)",
"_____no_output_____"
],
[
"tag_counts_df = pd.DataFrame(tag_counts, index = [\"counts\"]).T\ntag_counts_df.reset_index(inplace=True)\ntag_counts_df.columns = [\"tags\",\"counts\"]\ntag_counts_df.sort_values(\"counts\", ascending=False, inplace=True)",
"_____no_output_____"
],
[
"sns.barplot(data =tag_counts_df.iloc[:20,:], x = \"counts\", y= \"tags\")\nplt.title(\"20 most common tags\".title(), fontsize = 14);",
"_____no_output_____"
],
[
"sns.barplot(data =tag_counts_df.iloc[-20:,:], x = \"counts\", y= \"tags\")\nplt.title(\"20 least common tags\".title(), fontsize = 14);",
"_____no_output_____"
]
],
[
[
"# most popular recipes",
"_____no_output_____"
]
],
[
[
"raw_interaction_df",
"_____no_output_____"
],
[
"recipe_rating_df = raw_interaction_df.groupby(\"recipe_id\")[[\"rating\"]].count().reset_index()",
"_____no_output_____"
],
[
"recipe_rating_df.columns = [\"recipe_id\",\"counts\"]\nrecipe_rating_df",
"_____no_output_____"
],
[
"joined_recipe_df = recipe_rating_df.join(raw_recipes_df.set_index(\"id\"), how=\"inner\")",
"_____no_output_____"
],
[
"joined_recipe_df",
"_____no_output_____"
],
[
"sns.barplot(data = joined_recipe_df.sort_values(\"counts\", ascending = False).iloc[:20,:], x = \"counts\", y = \"name\");\nplt.title(\"most popular recipes by number of ratings\".title(), fontsize = 14);",
"_____no_output_____"
],
[
"sns.barplot(data = joined_recipe_df.sort_values(\"counts\").iloc[:20,:], x = \"counts\", y = \"name\");\nplt.title(\"least popular recipes by number of ratings\".title(), fontsize = 14);",
"_____no_output_____"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec7348135b200c63f26c4a8f878e15c6287ee0b7 | 82,946 | ipynb | Jupyter Notebook | C09/C09-w02-Lab2_reusable-embeddings.ipynb | pascal-p/ML_wtih_TF_GCP | 54d567fbabd0e4370dd3468d5bc2a16230422c99 | [
"Apache-2.0"
]
| null | null | null | C09/C09-w02-Lab2_reusable-embeddings.ipynb | pascal-p/ML_wtih_TF_GCP | 54d567fbabd0e4370dd3468d5bc2a16230422c99 | [
"Apache-2.0"
]
| null | null | null | C09/C09-w02-Lab2_reusable-embeddings.ipynb | pascal-p/ML_wtih_TF_GCP | 54d567fbabd0e4370dd3468d5bc2a16230422c99 | [
"Apache-2.0"
]
| null | null | null | 52.630711 | 15,640 | 0.682613 | [
[
[
"# Using pre-trained embedding with Tensorflow Hub\n\n**Learning Objectives**\n1. How to instantiate a Tensorflow Hub module\n1. How to find pretrained Tensorflow Hub module for variety of purposes\n1. How to use a pre-trained TF Hub text modules to generate sentence vectors\n1. How to incorporate a pre-trained TF-Hub module into a Keras model\n\n\n\n## Introduction\n\n\nIn this notebook, we will implement text models to recognize the probable source (Github, Tech-Crunch, or The New-York Times) of the titles we have in the title dataset.\n\nFirst, we will load and pre-process the texts and labels so that they are suitable to be fed to sequential Keras models with first layer being TF-hub pre-trained modules. Thanks to this first layer, we won't need to tokenize and integerize the text before passing it to our models. The pre-trained layer will take care of that for us, and consume directly raw text. However, we will still have to one-hot-encode each of the 3 classes into a 3 dimensional basis vector.\n\nThen we will build, train and compare simple models starting with different pre-trained TF-Hub layers.",
"_____no_output_____"
]
],
[
[
"!sudo chown -R jupyter:jupyter /home/jupyter/training-data-analyst",
"_____no_output_____"
],
[
"!pip install --user google-cloud-bigquery==1.25.0",
"Collecting google-cloud-bigquery==1.25.0\n Downloading google_cloud_bigquery-1.25.0-py2.py3-none-any.whl (169 kB)\n\u001b[2K \u001b[90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\u001b[0m \u001b[32m169.1/169.1 KB\u001b[0m \u001b[31m9.8 MB/s\u001b[0m eta \u001b[36m0:00:00\u001b[0m\n\u001b[?25hRequirement already satisfied: google-auth<2.0dev,>=1.9.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-bigquery==1.25.0) (1.35.0)\nRequirement already satisfied: protobuf>=3.6.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-bigquery==1.25.0) (3.19.4)\nCollecting google-cloud-core<2.0dev,>=1.1.0\n Downloading google_cloud_core-1.7.2-py2.py3-none-any.whl (28 kB)\nCollecting google-resumable-media<0.6dev,>=0.5.0\n Downloading google_resumable_media-0.5.1-py2.py3-none-any.whl (38 kB)\nRequirement already satisfied: six<2.0.0dev,>=1.13.0 in /opt/conda/lib/python3.7/site-packages (from google-cloud-bigquery==1.25.0) (1.15.0)\nCollecting google-api-core<2.0dev,>=1.15.0\n Downloading google_api_core-1.31.5-py2.py3-none-any.whl (93 kB)\n\u001b[2K \u001b[90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━\u001b[0m \u001b[32m93.3/93.3 KB\u001b[0m \u001b[31m5.0 MB/s\u001b[0m eta \u001b[36m0:00:00\u001b[0m\n\u001b[?25hRequirement already satisfied: setuptools>=40.3.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (59.8.0)\nRequirement already satisfied: requests<3.0.0dev,>=2.18.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (2.27.1)\nRequirement already satisfied: googleapis-common-protos<2.0dev,>=1.6.0 in /opt/conda/lib/python3.7/site-packages (from google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (1.54.0)\nRequirement already satisfied: packaging>=14.3 in /opt/conda/lib/python3.7/site-packages (from google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (21.3)\nRequirement already satisfied: pytz in /opt/conda/lib/python3.7/site-packages (from google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (2021.3)\nRequirement already satisfied: cachetools<5.0,>=2.0.0 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.9.0->google-cloud-bigquery==1.25.0) (4.2.4)\nRequirement already satisfied: pyasn1-modules>=0.2.1 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.9.0->google-cloud-bigquery==1.25.0) (0.2.7)\nRequirement already satisfied: rsa<5,>=3.1.4 in /opt/conda/lib/python3.7/site-packages (from google-auth<2.0dev,>=1.9.0->google-cloud-bigquery==1.25.0) (4.8)\nRequirement already satisfied: pyparsing!=3.0.5,>=2.0.2 in /opt/conda/lib/python3.7/site-packages (from packaging>=14.3->google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (3.0.7)\nRequirement already satisfied: pyasn1<0.5.0,>=0.4.6 in /opt/conda/lib/python3.7/site-packages (from pyasn1-modules>=0.2.1->google-auth<2.0dev,>=1.9.0->google-cloud-bigquery==1.25.0) (0.4.8)\nRequirement already satisfied: urllib3<1.27,>=1.21.1 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (1.26.8)\nRequirement already satisfied: idna<4,>=2.5 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (3.3)\nRequirement already satisfied: certifi>=2017.4.17 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (2021.10.8)\nRequirement already satisfied: charset-normalizer~=2.0.0 in /opt/conda/lib/python3.7/site-packages (from requests<3.0.0dev,>=2.18.0->google-api-core<2.0dev,>=1.15.0->google-cloud-bigquery==1.25.0) (2.0.12)\nInstalling collected packages: google-resumable-media, google-api-core, google-cloud-core, google-cloud-bigquery\n\u001b[31mERROR: pip's dependency resolver does not currently take into account all the packages that are installed. This behaviour is the source of the following dependency conflicts.\ngoogle-cloud-storage 2.2.1 requires google-resumable-media>=2.3.2, but you have google-resumable-media 0.5.1 which is incompatible.\ncloud-tpu-client 0.10 requires google-api-python-client==1.8.0, but you have google-api-python-client 2.41.0 which is incompatible.\u001b[0m\u001b[31m\n\u001b[0mSuccessfully installed google-api-core-1.31.5 google-cloud-bigquery-1.25.0 google-cloud-core-1.7.2 google-resumable-media-0.5.1\n"
]
],
[
[
"**Note**: Restart your kernel to use updated packages.",
"_____no_output_____"
],
[
"Kindly ignore the deprecation warnings and incompatibility errors related to google-cloud-storage.",
"_____no_output_____"
]
],
[
[
"import os\n\nfrom google.cloud import bigquery\nimport pandas as pd",
"_____no_output_____"
],
[
"%load_ext google.cloud.bigquery",
"The google.cloud.bigquery extension is already loaded. To reload it, use:\n %reload_ext google.cloud.bigquery\n"
]
],
[
[
"Replace the variable values in the cell below:",
"_____no_output_____"
]
],
[
[
"PROJECT = \"qwiklabs-gcp-01-7bff2816dcba\" # Replace with your PROJECT\nBUCKET = PROJECT # defaults to PROJECT\nREGION = \"australia-southeast1\" # Replace with your REGION\nSEED = 0",
"_____no_output_____"
]
],
[
[
"## Create a Dataset from BigQuery \n\nHacker news headlines are available as a BigQuery public dataset. The [dataset](https://bigquery.cloud.google.com/table/bigquery-public-data:hacker_news.stories?tab=details) contains all headlines from the sites inception in October 2006 until October 2015. \n\nHere is a sample of the dataset:",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\n\nSELECT url, title, score\nFROM `bigquery-public-data.hacker_news.stories`\nWHERE LENGTH(title) > 10\nAND score > 10\nAND LENGTH(url) > 0\nLIMIT 10",
"_____no_output_____"
]
],
[
[
"Let's do some regular expression parsing in BigQuery to get the source of the newspaper article from the URL.",
"_____no_output_____"
]
],
[
[
"%%bigquery --project $PROJECT\n\nSELECT ARRAY_REVERSE(SPLIT(REGEXP_EXTRACT(url, '.*://(.[^/]+)/'), '.'))[OFFSET(1)] AS source, COUNT(title) AS num_articles\nFROM `bigquery-public-data.hacker_news.stories`\nWHERE REGEXP_CONTAINS(REGEXP_EXTRACT(url, '.*://(.[^/]+)/'), '.com$')\nAND LENGTH(title) > 10\nGROUP BY source\nORDER BY num_articles DESC LIMIT 100",
"_____no_output_____"
]
],
[
[
"Now that we have good parsing of the URL to get the source, let's put together a dataset of source and titles. This will be our labeled dataset for machine learning.",
"_____no_output_____"
]
],
[
[
"regex = '.*://(.[^/]+)/'\n\n\nsub_query = \"\"\"\nSELECT title, ARRAY_REVERSE(SPLIT(REGEXP_EXTRACT(url, '{0}'), '.'))[OFFSET(1)] AS source\nFROM `bigquery-public-data.hacker_news.stories`\nWHERE REGEXP_CONTAINS(REGEXP_EXTRACT(url, '{0}'), '.com$')\nAND LENGTH(title) > 10\n\"\"\".format(regex)\n\n\nquery = \"\"\"\nSELECT LOWER(REGEXP_REPLACE(title, '[^a-zA-Z0-9 $.-]', ' ')) AS title, source\nFROM ({sub_query})\nWHERE (source = 'github' OR source = 'nytimes' OR source = 'techcrunch')\n\"\"\".format(sub_query=sub_query)\n\nprint(query)",
"\nSELECT LOWER(REGEXP_REPLACE(title, '[^a-zA-Z0-9 $.-]', ' ')) AS title, source\nFROM (\nSELECT title, ARRAY_REVERSE(SPLIT(REGEXP_EXTRACT(url, '.*://(.[^/]+)/'), '.'))[OFFSET(1)] AS source\nFROM `bigquery-public-data.hacker_news.stories`\nWHERE REGEXP_CONTAINS(REGEXP_EXTRACT(url, '.*://(.[^/]+)/'), '.com$')\nAND LENGTH(title) > 10\n)\nWHERE (source = 'github' OR source = 'nytimes' OR source = 'techcrunch')\n\n"
]
],
[
[
"For ML training, we usually need to split our dataset into training and evaluation datasets (and perhaps an independent test dataset if we are going to do model or feature selection based on the evaluation dataset). AutoML however figures out on its own how to create these splits, so we won't need to do that here. \n\n",
"_____no_output_____"
]
],
[
[
"bq = bigquery.Client(project=PROJECT)\ntitle_dataset = bq.query(query).to_dataframe()\ntitle_dataset.head(7)",
"_____no_output_____"
]
],
[
[
"AutoML for text classification requires that\n* the dataset be in csv form with \n* the first column being the texts to classify or a GCS path to the text \n* the last column to be the text labels\n\nThe dataset we pulled from BiqQuery satisfies these requirements.",
"_____no_output_____"
]
],
[
[
"print(\"The full dataset contains {n} titles\".format(n=len(title_dataset)))",
"The full dataset contains 96203 titles\n"
]
],
[
[
"Let's make sure we have roughly the same number of labels for each of our three labels:",
"_____no_output_____"
]
],
[
[
"title_dataset.source.value_counts()",
"_____no_output_____"
]
],
[
[
"Finally we will save our data, which is currently in-memory, to disk.\n\nWe will create a csv file containing the full dataset and another containing only 1000 articles for development.\n\n**Note:** It may take a long time to train AutoML on the full dataset, so we recommend to use the sample dataset for the purpose of learning the tool. \n",
"_____no_output_____"
]
],
[
[
"DATADIR = './data/'\n\nif not os.path.exists(DATADIR):\n os.makedirs(DATADIR)",
"_____no_output_____"
],
[
"FULL_DATASET_NAME = 'titles_full.csv'\nFULL_DATASET_PATH = os.path.join(DATADIR, FULL_DATASET_NAME)\n\n# Let's shuffle the data before writing it to disk.\ntitle_dataset = title_dataset.sample(n=len(title_dataset))\n\ntitle_dataset.to_csv(FULL_DATASET_PATH, header=False, index=False, encoding='utf-8')",
"_____no_output_____"
]
],
[
[
"Now let's sample 1000 articles from the full dataset and make sure we have enough examples for each label in our sample dataset (see [here](https://cloud.google.com/natural-language/automl/docs/beginners-guide) for further details on how to prepare data for AutoML).",
"_____no_output_____"
]
],
[
[
"sample_title_dataset = title_dataset.sample(n=1000)\nsample_title_dataset.source.value_counts()",
"_____no_output_____"
]
],
[
[
"Let's write the sample datatset to disk.",
"_____no_output_____"
]
],
[
[
"SAMPLE_DATASET_NAME = 'titles_sample.csv'\nSAMPLE_DATASET_PATH = os.path.join(DATADIR, SAMPLE_DATASET_NAME)\n\nsample_title_dataset.to_csv(SAMPLE_DATASET_PATH, header=False, index=False, encoding='utf-8')",
"_____no_output_____"
],
[
"sample_title_dataset.head()",
"_____no_output_____"
],
[
"import datetime\nimport os\nimport shutil\n\nimport pandas as pd\nimport tensorflow as tf\nfrom tensorflow.keras.callbacks import TensorBoard, EarlyStopping\nfrom tensorflow_hub import KerasLayer\nfrom tensorflow.keras.layers import Dense\nfrom tensorflow.keras.models import Sequential\nfrom tensorflow.keras.preprocessing.text import Tokenizer\nfrom tensorflow.keras.utils import to_categorical\n\n\nprint(tf.__version__)",
"2.6.3\n"
],
[
"%matplotlib inline",
"_____no_output_____"
]
],
[
[
"Let's start by specifying where the information about the trained models will be saved as well as where our dataset is located:",
"_____no_output_____"
]
],
[
[
"MODEL_DIR = \"./text_models\"\nDATA_DIR = \"./data\"",
"_____no_output_____"
]
],
[
[
"## Loading the dataset",
"_____no_output_____"
],
[
"As in the previous labs, our dataset consists of titles of articles along with the label indicating from which source these articles have been taken from (GitHub, Tech-Crunch, or the New-York Times):",
"_____no_output_____"
]
],
[
[
"ls ./data/",
"titles_full.csv titles_sample.csv\n"
],
[
"DATASET_NAME = \"titles_full.csv\"\nTITLE_SAMPLE_PATH = os.path.join(DATA_DIR, DATASET_NAME)\nCOLUMNS = ['title', 'source']\n\ntitles_df = pd.read_csv(TITLE_SAMPLE_PATH, header=None, names=COLUMNS)\ntitles_df.head()",
"_____no_output_____"
]
],
[
[
"Let's look again at the number of examples per label to make sure we have a well-balanced dataset:",
"_____no_output_____"
]
],
[
[
"titles_df.source.value_counts()",
"_____no_output_____"
]
],
[
[
"## Preparing the labels",
"_____no_output_____"
],
[
"In this lab, we will use pre-trained [TF-Hub embeddings modules for english](https://tfhub.dev/s?q=tf2%20embeddings%20text%20english) for the first layer of our models. One immediate\nadvantage of doing so is that the TF-Hub embedding module will take care for us of processing the raw text. \nThis also means that our model will be able to consume text directly instead of sequences of integers representing the words.\n\nHowever, as before, we still need to preprocess the labels into one-hot-encoded vectors:",
"_____no_output_____"
]
],
[
[
"CLASSES = {\n 'github': 0,\n 'nytimes': 1,\n 'techcrunch': 2\n}\n\nN_CLASSES = len(CLASSES)",
"_____no_output_____"
],
[
"def encode_labels(sources):\n classes = [CLASSES[source] for source in sources]\n return to_categorical(classes, num_classes=N_CLASSES)",
"_____no_output_____"
],
[
"encode_labels(titles_df.source[:4])",
"_____no_output_____"
]
],
[
[
"## Preparing the train/test splits",
"_____no_output_____"
],
[
"Let's split our data into train and test splits:",
"_____no_output_____"
]
],
[
[
"N_TRAIN = int(len(titles_df) * 0.95)\n\ntitles_train, sources_train = (titles_df.title[:N_TRAIN], titles_df.source[:N_TRAIN])\ntitles_valid, sources_valid = (titles_df.title[N_TRAIN:], titles_df.source[N_TRAIN:])",
"_____no_output_____"
]
],
[
[
"To be on the safe side, we verify that the train and test splits\nhave roughly the same number of examples per class.\n\nSince it is the case, accuracy will be a good metric to use to measure\nthe performance of our models.",
"_____no_output_____"
]
],
[
[
"sources_train.value_counts()",
"_____no_output_____"
],
[
"sources_valid.value_counts()",
"_____no_output_____"
]
],
[
[
"Now let's create the features and labels we will feed our models with:",
"_____no_output_____"
]
],
[
[
"X_train, Y_train = titles_train.values, encode_labels(sources_train)\nX_valid, Y_valid = titles_valid.values, encode_labels(sources_valid)",
"_____no_output_____"
],
[
"X_train[:3]",
"_____no_output_____"
],
[
"Y_train[:3]",
"_____no_output_____"
]
],
[
[
"## NNLM Model",
"_____no_output_____"
],
[
"We will first try a word embedding pre-trained using a [Neural Probabilistic Language Model](http://www.jmlr.org/papers/volume3/bengio03a/bengio03a.pdf). TF-Hub has a 50-dimensional one called \n[nnlm-en-dim50-with-normalization](https://tfhub.dev/google/tf2-preview/nnlm-en-dim50/1), which also\nnormalizes the vectors produced. \n\nOnce loaded from its url, the TF-hub module can be used as a normal Keras layer in a sequential or functional model. Since we have enough data to fine-tune the parameters of the pre-trained embedding itself, we will set `trainable=True` in the `KerasLayer` that loads the pre-trained embedding:",
"_____no_output_____"
]
],
[
[
"NNLM = \"https://tfhub.dev/google/nnlm-en-dim50/2\"\n\nnnlm_module = KerasLayer(NNLM, output_shape=[50], input_shape=[], dtype=tf.string, trainable=True)",
"2022-03-25 10:06:06.977844: I tensorflow/core/common_runtime/process_util.cc:146] Creating new thread pool with default inter op setting: 2. Tune using inter_op_parallelism_threads for best performance.\n2022-03-25 10:06:07.140455: I tensorflow/compiler/mlir/mlir_graph_optimization_pass.cc:185] None of the MLIR Optimization Passes are enabled (registered 2)\n"
]
],
[
[
"Note that this TF-Hub embedding produces a single 50-dimensional vector when passed a sentence:",
"_____no_output_____"
]
],
[
[
"nnlm_module(tf.constant([\"The dog is happy to see people in the street.\"]))",
"_____no_output_____"
]
],
[
[
"## Building the models",
"_____no_output_____"
],
[
"Let's write a function that \n\n* takes as input an instance of a `KerasLayer` (i.e. the `nnlm_module` we constructed above) as well as the name of the model (say `nnlm`)\n* returns a compiled Keras sequential model starting with this pre-trained TF-hub layer, adding one or more dense relu layers to it, and ending with a softmax layer giving the probability of each of the classes:",
"_____no_output_____"
]
],
[
[
"def build_model(hub_module, name):\n model = Sequential([\n hub_module, # TODO \n Dense(16, activation='relu'),\n Dense(N_CLASSES, activation='softmax')\n ], name=name)\n\n model.compile(\n optimizer='adam',\n loss='categorical_crossentropy',\n metrics=['accuracy']\n )\n return model",
"_____no_output_____"
]
],
[
[
"Let's also wrap the training code into a `train_and_evaluate` function that \n* takes as input the training and validation data, as well as the compiled model itself, and the `batch_size`\n* trains the compiled model for 100 epochs at most, and does early-stopping when the validation loss is no longer decreasing\n* returns an `history` object, which will help us to plot the learning curves",
"_____no_output_____"
]
],
[
[
"def train_and_evaluate(train_data, val_data, model, batch_size=5000):\n X_train, Y_train = train_data\n tf.random.set_seed(33)\n model_dir = os.path.join(MODEL_DIR, model.name)\n if tf.io.gfile.exists(model_dir):\n tf.io.gfile.rmtree(model_dir)\n history = model.fit(\n X_train, Y_train,\n epochs=13, # 100\n batch_size=batch_size,\n validation_data=val_data,\n callbacks=[TensorBoard(model_dir)], # EarlyStopping()\n )\n return history",
"_____no_output_____"
]
],
[
[
"## Training NNLM",
"_____no_output_____"
]
],
[
[
"data = (X_train, Y_train)\nval_data = (X_valid, Y_valid)",
"_____no_output_____"
],
[
"nnlm_model = build_model(nnlm_module, 'nnlm')\nnnlm_history = train_and_evaluate(data, val_data, nnlm_model)",
"2022-03-25 10:09:54.386050: I tensorflow/core/profiler/lib/profiler_session.cc:131] Profiler session initializing.\n2022-03-25 10:09:54.386089: I tensorflow/core/profiler/lib/profiler_session.cc:146] Profiler session started.\n2022-03-25 10:09:54.386293: I tensorflow/core/profiler/lib/profiler_session.cc:164] Profiler session tear down.\n"
],
[
"history = nnlm_history\npd.DataFrame(history.history)[['loss', 'val_loss']].plot()\npd.DataFrame(history.history)[['accuracy', 'val_accuracy']].plot()",
"_____no_output_____"
]
],
[
[
"## Bonus",
"_____no_output_____"
],
[
"Try to beat the best model by modifying the model architecture, changing the TF-Hub embedding, and tweaking the training parameters.",
"_____no_output_____"
],
[
"Copyright 2020 Google Inc. Licensed under the Apache License, Version 2.0 (the \"License\"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0 Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an \"AS IS\" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec73489c7b0a780a2e09eee1ab150a856383659e | 10,889 | ipynb | Jupyter Notebook | azmpdata_demo.ipynb | j-harbin/azmpdata | 91803e5a6924509042c85a63526054a4cc6e1c2d | [
"MIT"
]
| 10 | 2020-05-05T13:04:50.000Z | 2022-02-25T19:30:24.000Z | azmpdata_demo.ipynb | j-harbin/azmpdata | 91803e5a6924509042c85a63526054a4cc6e1c2d | [
"MIT"
]
| 43 | 2020-10-15T18:37:21.000Z | 2021-10-14T11:36:46.000Z | azmpdata_demo.ipynb | j-harbin/azmpdata | 91803e5a6924509042c85a63526054a4cc6e1c2d | [
"MIT"
]
| 12 | 2020-05-08T14:45:26.000Z | 2021-11-05T11:34:38.000Z | 26.365617 | 232 | 0.476811 | [
[
[
"<a href=\"https://colab.research.google.com/github/Echisholm21/azmpdata/blob/master/azmpdata_demo.ipynb\" target=\"_parent\"><img src=\"https://colab.research.google.com/assets/colab-badge.svg\" alt=\"Open In Colab\"/></a>",
"_____no_output_____"
],
[
"### **<font color=\"Black\">azmpdata - R Package Demo</font>**\n\nFrom **github**, you can launch this page by clicking on the \"Open in Colab\" badge.\n\n<br>\n\n### **This Tutorial uses Google Colab.**\n\n\n\n<br>\n",
"_____no_output_____"
],
[
"## **Set up your environment**\n\n- First we need to tell the notebook we will be working in R\n- Then we install azmpdata from github\n- Lastly, call the library to make sure it is loaded (you should get a message about your package being up to date)",
"_____no_output_____"
]
],
[
[
"# activate R magic\n%load_ext rpy2.ipython",
"_____no_output_____"
],
[
"%%R\n# download devtools so we can install from github\ninstall.packages('devtools')\n\n# install azmpdata\ndevtools::install_github('casaultb/azmpdata')\nlibrary(azmpdata)",
"_____no_output_____"
]
],
[
[
"## **Let's try to access some data!**\n- data can be called directly (if we know the dataframe name)\n- R auto-complete magic doesn't work in colab but in R-Studio you would get a list of dataframes if you started typing 'data(...)' slowly",
"_____no_output_____"
]
],
[
[
"%%R\n\ndata('Discrete_Annual_Broadscale')\nDiscrete_Annual_Broadscale[Discrete_Annual_Broadscale$year == 2018,]",
"_____no_output_____"
]
],
[
[
"- we can also list all the available dataframes",
"_____no_output_____"
]
],
[
[
"%%R\n\ndata(package = 'azmpdata')",
"_____no_output_____"
]
],
[
[
"## **Now you try**\n- Try calling one of the other available dataframes from the package\n- use the `summary()` function to see the data\n- Try your favourite 'data peek' function!",
"_____no_output_____"
]
],
[
[
"%%R\n\ndata('Zooplankton_Occupations_Broadscale')\n# TODO: Complete activity instructions above!\n",
"_____no_output_____"
]
],
[
[
"## **Searching azmpdata**\n\nThere is a built in search function which can help you find the data you are looking for within the azmpdata package\n\n",
"_____no_output_____"
],
[
"- This search function can:\n - Search through variable names in all dataframes\n - Search through help files ",
"_____no_output_____"
]
],
[
[
"%%R\n\nres <- variable_lookup('nitrate')\nprint(res)",
"_____no_output_____"
],
[
"%%R\n\nres <- variable_lookup('stratification', search_help = TRUE)\nprint(res)",
"_____no_output_____"
]
],
[
[
"The result you get from the search function includes the variable name that matched your search, and the dataframe in which it is contained. \n\nYou can then call the dataframe with your desired variable!",
"_____no_output_____"
]
],
[
[
"%%R\n\nhead(get(res$dataframe[1]))",
"_____no_output_____"
]
],
[
[
"## **Now you try**\n- Try searching for the term 'zooplankton'\n- Try combining your search terms (eg. 'zooplankton' and 'dry_weight')\n- Try searching through both the variable names and help files\n- Try calling the dataframe which matches your search terms and viewing the data!",
"_____no_output_____"
]
],
[
[
"%%R\n\nvariable_lookup(...)\nvariable_lookup(..., search_help = TRUE) # search through help files!",
"_____no_output_____"
]
],
[
[
"## **Analysis and Plotting**\n\n- We can use dplyr, or base R tools to manipulate, analyze and plot the data\n- We can also use some specially deisgned tools to simplify these tasks\n\n\nLet's download another package `multivaR`",
"_____no_output_____"
]
],
[
[
"%%R\n\ndevtools::install_github('echisholm21/multivaR')\nlibrary(multivaR)\nlibrary(dplyr)",
"_____no_output_____"
]
],
[
[
"- We can use multivaR to do common analysis tasks such as calculate anomalies",
"_____no_output_____"
]
],
[
[
"%%R\n\n# call in some data to play with\ndata('Derived_Occupations_Stations')\n\ndf_anom <- calculate_anomaly(data = Derived_Occupations_Stations,\n anomalyType = 'monthly', \n climatologyYears = c(1999, 2010),\n var = 'nitrate_0_50',\n normalizedAnomaly = FALSE)\nhead(df_anom)",
"_____no_output_____"
]
],
[
[
"Or even look at the spatial extent of our data",
"_____no_output_____"
]
],
[
[
"%%R\n# plot_region(name = 'HL2')\n sysreg_att<- system.file('extdata/', 'polygons_attributes.csv', package = 'multivaR', mustWork = TRUE)\n sysreg_geo<- system.file('extdata/', 'polygons_geometry.csv', package = 'multivaR', mustWork = TRUE)\n\n regtab_att <- utils::read.csv(sysreg_att)\n regtab_geo <- utils::read.csv(sysreg_geo)\n\nhead(regtab_att)\n\n\n",
"_____no_output_____"
],
[
"%%R\n# get the info just for the station of interest\nsubtab <- regtab_att[regtab_att$sname == 'P5',]\n\n# join with geographic info\nsubtab_geo <- regtab_geo[regtab_geo$record == unique(subtab$record),]\n\n\nfull_join(subtab, subtab_geo)",
"_____no_output_____"
]
],
[
[
"## **Now you try**\n- use help(package = 'multivaR') to see what else multivaR can do\n- try calculating a PCA for the 'Derived_Occupations_Stations' dataframe \n",
"_____no_output_____"
]
],
[
[
"%%R\n\nhelp(package = 'multivaR')\nhelp(..., package = 'multivaR') #HINT: Insert the function name to replace '...'\n\n# call in data\n\n# perform PCA\nPCA(...) # HINT: you should only give the PCA function data variables, not metadata!",
"_____no_output_____"
]
],
[
[
"# Thank you for participating!",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec734ce4c9c27eee15b9582347a3055b8e4e12d5 | 4,963 | ipynb | Jupyter Notebook | notebooks/13-Advanced Python Modules/04-Timing your code - timeit.ipynb | vcelis/python-3-bootcamp | 6b1fdeca6a3c8fe9e56e39ea8ef186eaeb29deba | [
"MIT"
]
| null | null | null | notebooks/13-Advanced Python Modules/04-Timing your code - timeit.ipynb | vcelis/python-3-bootcamp | 6b1fdeca6a3c8fe9e56e39ea8ef186eaeb29deba | [
"MIT"
]
| null | null | null | notebooks/13-Advanced Python Modules/04-Timing your code - timeit.ipynb | vcelis/python-3-bootcamp | 6b1fdeca6a3c8fe9e56e39ea8ef186eaeb29deba | [
"MIT"
]
| null | null | null | 25.582474 | 322 | 0.56216 | [
[
[
"# Timing your code\nSometimes it's important to know how long your code is taking to run, or at least know if a particular line of code is slowing down your entire project. Python has a built-in timing module to do this. \n\nThis module provides a simple way to time small bits of Python code. It has both a Command-Line Interface as well as a callable one. It avoids a number of common traps for measuring execution times. \n\nLet's learn about timeit!",
"_____no_output_____"
]
],
[
[
"import timeit",
"_____no_output_____"
]
],
[
[
"Let's use timeit to time various methods of creating the string '0-1-2-3-.....-99'\n\nWe'll pass two arguments: the actual line we want to test encapsulated as a string and the number of times we wish to run it. Here we'll choose 10,000 runs to get some high enough numbers to compare various methods.",
"_____no_output_____"
]
],
[
[
"# For loop\ntimeit.timeit('\"-\".join(str(n) for n in range(100))', number=10000)",
"_____no_output_____"
],
[
"# List comprehension\ntimeit.timeit('\"-\".join([str(n) for n in range(100)])', number=10000)",
"_____no_output_____"
],
[
"# Map()\ntimeit.timeit('\"-\".join(map(str, range(100)))', number=10000)",
"_____no_output_____"
]
],
[
[
"Great! We see a significant time difference by using map()! This is good to know and we should keep this in mind.\n\nNow let's introduce iPython's magic function **%timeit**<br>\n*NOTE: This method is specific to jupyter notebooks!*\n\niPython's %timeit will perform the same lines of code a certain number of times (loops) and will give you the fastest performance time (best of 3).\n\nLet's repeat the above examinations using iPython magic!",
"_____no_output_____"
]
],
[
[
"%timeit \"-\".join(str(n) for n in range(100))",
"17.6 µs ± 174 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
"%timeit \"-\".join([str(n) for n in range(100)])",
"15.1 µs ± 60.1 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
],
[
"%timeit \"-\".join(map(str, range(100)))",
"12.2 µs ± 130 ns per loop (mean ± std. dev. of 7 runs, 100000 loops each)\n"
]
],
[
[
"Great! We arrive at the same conclusion. It's also important to note that iPython will limit the amount of *real time* it will spend on its timeit procedure. For instance if running 100000 loops took 10 minutes, iPython would automatically reduce the number of loops to something more reasonable like 100 or 1000.\n\nGreat! You should now feel comfortable timing lines of your code, both in and out of iPython. Check out the documentation for more information:\nhttps://docs.python.org/3/library/timeit.html",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec735384d26333b4e57ac0b2219e608f7e9518ae | 830,866 | ipynb | Jupyter Notebook | Data Analysis/Visualizing the Gender Gap in College Degrees/Visualizing The Gender Gap In College Degrees.ipynb | linnforsman/data-science-portfolio | fd51d5b74cea7a598fe0e4e7555af48cecf4c980 | [
"MIT"
]
| null | null | null | Data Analysis/Visualizing the Gender Gap in College Degrees/Visualizing The Gender Gap In College Degrees.ipynb | linnforsman/data-science-portfolio | fd51d5b74cea7a598fe0e4e7555af48cecf4c980 | [
"MIT"
]
| null | null | null | Data Analysis/Visualizing the Gender Gap in College Degrees/Visualizing The Gender Gap In College Degrees.ipynb | linnforsman/data-science-portfolio | fd51d5b74cea7a598fe0e4e7555af48cecf4c980 | [
"MIT"
]
| null | null | null | 1,417.860068 | 176,484 | 0.942629 | [
[
[
"# Introduction",
"_____no_output_____"
]
],
[
[
"%matplotlib inline\nimport pandas as pd\nimport matplotlib.pyplot as plt\n\nwomen_degrees = pd.read_csv('percent-bachelors-degrees-women-usa.csv')\ncb_dark_blue = (0/255,107/255,164/255)\ncb_orange = (255/255, 128/255, 14/255)\nstem_cats = ['Engineering', 'Computer Science', 'Psychology', 'Biology', 'Physical Sciences', 'Math and Statistics']\n\nfig = plt.figure(figsize=(18, 3))\n\nfor sp in range(0,6):\n ax = fig.add_subplot(1,6,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[sp]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[sp]], c=cb_orange, label='Men', linewidth=3)\n ax.spines[\"right\"].set_visible(False) \n ax.spines[\"left\"].set_visible(False)\n ax.spines[\"top\"].set_visible(False) \n ax.spines[\"bottom\"].set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[sp])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\")\n \n if sp == 0:\n ax.text(2005, 87, 'Men')\n ax.text(2002, 8, 'Women')\n elif sp == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2001, 35, 'Women')\nplt.show()",
"_____no_output_____"
]
],
[
[
"# Comparing across all degrees\n",
"_____no_output_____"
]
],
[
[
"stem_cats = ['Psychology', 'Biology', 'Math and Statistics', 'Physical Sciences', 'Computer Science', 'Engineering']\nlib_arts_cats = ['Foreign Languages', 'English', 'Communications and Journalism', 'Art and Performance', 'Social Sciences and History']\nother_cats = ['Health Professions', 'Public Administration', 'Education', 'Agriculture','Business', 'Architecture']",
"_____no_output_____"
],
[
"fig = plt.figure(figsize=(16, 20))\n\n## Generate first column of line charts. STEM degrees.\nfor sp in range(0,18,3):\n cat_index = int(sp/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\")\n \n if cat_index == 0:\n ax.text(2003, 85, 'Women')\n ax.text(2005, 10, 'Men')\n elif cat_index == 5:\n ax.text(2005, 87, 'Men')\n ax.text(2003, 7, 'Women')\n\n## Generate second column of line charts. Liberal arts degrees.\nfor sp in range(1,16,3):\n cat_index = int((sp-1)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[lib_arts_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[lib_arts_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(lib_arts_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\")\n \n if cat_index == 0:\n ax.text(2003, 78, 'Women')\n ax.text(2005, 18, 'Men')\n\n## Generate third column of line charts. Other degrees.\nfor sp in range(2,20,3):\n cat_index = int((sp-2)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[other_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[other_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(other_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\")\n \n if cat_index == 0:\n ax.text(2003, 90, 'Women')\n ax.text(2005, 5, 'Men')\n elif cat_index == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2003, 30, 'Women')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"# Hiding x-axis labels\nBecause seventeen line charts in one diagram that contains non-data elements quickly clutter the field of view, we have to remove the x-axis labels for every line chart in a column except for the bottom most one. We accomplish this by modifying the call to Axes.tick_params() and setting labelbottom to off. ",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16, 16))\n\n## Generate first column of line charts. STEM degrees.\nfor sp in range(0,18,3):\n cat_index = int(sp/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n \n if cat_index == 0:\n ax.text(2003, 85, 'Women')\n ax.text(2005, 10, 'Men')\n elif cat_index == 5:\n ax.text(2005, 87, 'Men')\n ax.text(2003, 7, 'Women')\n ax.tick_params(labelbottom='on')\n\n## Generate second column of line charts. Liberal arts degrees.\nfor sp in range(1,16,3):\n cat_index = int((sp-1)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[lib_arts_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[lib_arts_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(lib_arts_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n \n if cat_index == 0:\n ax.text(2003, 78, 'Women')\n ax.text(2005, 18, 'Men')\n elif cat_index == 4:\n ax.tick_params(labelbottom='on')\n\n## Generate third column of line charts. Other degrees.\nfor sp in range(2,20,3):\n cat_index = int((sp-2)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[other_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[other_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(other_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n \n if cat_index == 0:\n ax.text(2003, 90, 'Women')\n ax.text(2005, 5, 'Men')\n elif cat_index == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2003, 30, 'Women')\n ax.tick_params(labelbottom='on')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"# Setting y-axis labels\nWe removed the x-axis labels for all but bottommost plots solved the issues we noticed with the overlapping text. ",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16, 16))\n\n## Generate first column of line charts. STEM degrees.\nfor sp in range(0,18,3):\n cat_index = int(sp/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 85, 'Women')\n ax.text(2005, 10, 'Men')\n elif cat_index == 5:\n ax.text(2005, 87, 'Men')\n ax.text(2003, 7, 'Women')\n ax.tick_params(labelbottom='on')\n\n## Generate second column of line charts. Liberal arts degrees.\nfor sp in range(1,16,3):\n cat_index = int((sp-1)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[lib_arts_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[lib_arts_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(lib_arts_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 78, 'Women')\n ax.text(2005, 18, 'Men')\n elif cat_index == 4:\n ax.tick_params(labelbottom='on')\n\n## Generate third column of line charts. Other degrees.\nfor sp in range(2,20,3):\n cat_index = int((sp-2)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[other_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[other_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(other_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 90, 'Women')\n ax.text(2005, 5, 'Men')\n elif cat_index == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2003, 30, 'Women')\n ax.tick_params(labelbottom='on')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"# Adding a horizontal line\nWe reduced clutter by removing most of the y-axis labels but it is also hard to understand which degree has close to 50-50 gender breakdown. While keeping all the y-axis labels would have made it easier, we can do one better and use a horizontal line across all of the line charts where the y-axis label 50 would have been.\n\nWe do it using the Axes.axhline() method. The only parameter required is the y-axis location for the start of the line.",
"_____no_output_____"
]
],
[
[
"fig = plt.figure(figsize=(16, 16))\n\n## Generate first column of line charts. STEM degrees.\nfor sp in range(0,18,3):\n cat_index = int(sp/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n \n if cat_index == 0:\n ax.text(2003, 85, 'Women')\n ax.text(2005, 10, 'Men')\n elif cat_index == 5:\n ax.text(2005, 87, 'Men')\n ax.text(2003, 7, 'Women')\n ax.tick_params(labelbottom='on')\n\n## Generate second column of line charts. Liberal arts degrees.\nfor sp in range(1,16,3):\n cat_index = int((sp-1)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[lib_arts_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[lib_arts_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(lib_arts_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n \n if cat_index == 0:\n ax.text(2003, 78, 'Women')\n ax.text(2005, 18, 'Men')\n elif cat_index == 4:\n ax.tick_params(labelbottom='on')\n\n## Generate third column of line charts. Other degrees.\nfor sp in range(2,20,3):\n cat_index = int((sp-2)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[other_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[other_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(other_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.set_yticks([0,100])\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n \n if cat_index == 0:\n ax.text(2003, 90, 'Women')\n ax.text(2005, 5, 'Men')\n elif cat_index == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2003, 30, 'Women')\n ax.tick_params(labelbottom='on')\n \nplt.show()",
"_____no_output_____"
]
],
[
[
"# Exporting to a file\n",
"_____no_output_____"
]
],
[
[
"# Set backend to Agg.\nfig = plt.figure(figsize=(16, 16))\n\n## Generate first column of line charts. STEM degrees.\nfor sp in range(0,18,3):\n cat_index = int(sp/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[stem_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[stem_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(stem_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 85, 'Women')\n ax.text(2005, 10, 'Men')\n elif cat_index == 5:\n ax.text(2005, 87, 'Men')\n ax.text(2003, 7, 'Women')\n ax.tick_params(labelbottom='on')\n\n## Generate second column of line charts. Liberal arts degrees.\nfor sp in range(1,16,3):\n cat_index = int((sp-1)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[lib_arts_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[lib_arts_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(lib_arts_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 75, 'Women')\n ax.text(2005, 20, 'Men')\n elif cat_index == 4:\n ax.tick_params(labelbottom='on')\n\n## Generate third column of line charts. Other degrees.\nfor sp in range(2,20,3):\n cat_index = int((sp-2)/3)\n ax = fig.add_subplot(6,3,sp+1)\n ax.plot(women_degrees['Year'], women_degrees[other_cats[cat_index]], c=cb_dark_blue, label='Women', linewidth=3)\n ax.plot(women_degrees['Year'], 100-women_degrees[other_cats[cat_index]], c=cb_orange, label='Men', linewidth=3)\n for key,spine in ax.spines.items():\n spine.set_visible(False)\n ax.set_xlim(1968, 2011)\n ax.set_ylim(0,100)\n ax.set_title(other_cats[cat_index])\n ax.tick_params(bottom=\"off\", top=\"off\", left=\"off\", right=\"off\", labelbottom='off')\n ax.axhline(50, c=(171/255, 171/255, 171/255), alpha=0.3)\n ax.set_yticks([0,100])\n \n if cat_index == 0:\n ax.text(2003, 90, 'Women')\n ax.text(2005, 5, 'Men')\n elif cat_index == 5:\n ax.text(2005, 62, 'Men')\n ax.text(2003, 30, 'Women')\n ax.tick_params(labelbottom='on')\n\n# Export file before calling pyplot.show()\nfig.savefig(\"gender_degrees.png\")\nplt.show()",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec736a6fd34e6d453c2b89553f6b286148545fe9 | 24,356 | ipynb | Jupyter Notebook | analysis/Week 6 - Use Embedding.ipynb | Chaoste/CommentBot | ba892e9a41f348be4dc299be89b7048b0056b476 | [
"MIT"
]
| null | null | null | analysis/Week 6 - Use Embedding.ipynb | Chaoste/CommentBot | ba892e9a41f348be4dc299be89b7048b0056b476 | [
"MIT"
]
| 12 | 2020-01-28T22:23:04.000Z | 2022-03-11T23:21:40.000Z | analysis/Week 6 - Use Embedding.ipynb | Chaoste/CommentBot | ba892e9a41f348be4dc299be89b7048b0056b476 | [
"MIT"
]
| null | null | null | 44.608059 | 1,471 | 0.598415 | [
[
[
"### Tutorial on Keras with Gensim\nhttps://www.depends-on-the-definition.com/guide-to-word-vectors-with-gensim-and-keras/\n\n- http://www.orbifold.net/default/2017/01/10/embedding-and-tokenizer-in-keras/\n- https://github.com/keras-team/keras/issues/853\n- http://adventuresinmachinelearning.com/gensim-word2vec-tutorial/\n- https://machinelearningmastery.com/use-word-embedding-layers-deep-learning-keras/\n- https://stats.stackexchange.com/questions/320701/how-to-use-keras-pre-trained-embedding-layer\n- https://blog.keras.io/using-pre-trained-word-embeddings-in-a-keras-model.html",
"_____no_output_____"
]
],
[
[
"import numpy as np\nimport pandas as pd\nfrom sklearn.model_selection import train_test_split\n\n# from nltk.tokenize import WordPunctTokenizer\nfrom collections import Counter\n\nfrom keras.layers import Dense, Input, LSTM, CuDNNLSTM, Embedding, Dropout,SpatialDropout1D, Bidirectional\nfrom keras.models import Sequential\nfrom keras.optimizers import Adam\nfrom keras.layers.normalization import BatchNormalization\nfrom keras.preprocessing.text import Tokenizer\n\n# Visualization\nimport matplotlib.pyplot as plt\nimport matplotlib.cm as cmap\n%matplotlib inline\n\nimport importlib\nimport utils\nimportlib.reload(utils)\nimport text_utils\nimportlib.reload(text_utils)",
"_____no_output_____"
],
[
"# Constants\n\nOUTPUT_DIR = './week-6-plots'\nORIG_COMMENTS = '../data/guardian-all/sorted_comments-standardized-pol-all.csv'\nTOKENIZED_COMMENTS = '../data/embedding/sorted_comments-standardized-tokenized.csv'",
"_____no_output_____"
],
[
"tokenized_comments = pd.read_csv(TOKENIZED_COMMENTS, ';')",
"_____no_output_____"
],
[
"tokenized_comments.shape",
"_____no_output_____"
]
],
[
[
"#### Preprocess data",
"_____no_output_____"
]
],
[
[
"data_articles, data_articles_pol, data_authors, data_comments_pol = utils.load_data()",
"_____no_output_____"
],
[
"X = data_comments_pol['comment_text']\ny = pd.get_dummies(['pos' if x > 2 else 'neg' for x in data_comments_pol['upvotes']]).values # 2 -> ? vs. ?\nX_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)",
"_____no_output_____"
]
],
[
[
"#### Load embedding",
"_____no_output_____"
]
],
[
[
"ft_model = text_utils.load_embedding()\nword_vectors = ft_model.wv\nEMBEDDING_DIM = 50\nprint(\"Number of word vectors: {}\".format(len(word_vectors.vocab)))",
"_____no_output_____"
],
[
"# Setup tokenizer\n# tokenizer = WordPunctTokenizer()\n# Use a counter for selecting the X most common words (therefore tokenize)\n# vocab = Counter()\n# comments = text_utils.process_comments(tokenizer, vocab, X, lower=True)\ntokenizer = Tokenizer()\ntokenizer.fit_on_texts(X)\nindex_dict = tokenizer.word_index",
"100%|████████████████████████████████████████████████████████| 665523/665523 [01:58<00:00, 5629.16it/s]\n"
],
[
"backtranslater = dict((i, w) for w, i in index_dict.items())",
"_____no_output_____"
],
[
"# assemble the embedding_weights in one numpy array\nn_symbols = len(index_dict) + 1 # adding 1 to account for 0th index (for masking)\nembedding_weights = np.zeros((n_symbols, EMBEDDING_DIM))\nfor word, index in index_dict.items():\n if word in word_vectors:\n embedding_weights[index, :] = word_vectors[word]\n\n# define inputs here\n# embedding_layer = Embedding(output_dim=vocab_dim, input_dim=n_symbols, weights=[embedding_weights], trainable=False)\n# embedded = embedding_layer(input_layer)\n# embedding_layer = model.wv.get_keras_embedding(train_embeddings=False)",
"_____no_output_____"
],
[
"word_vectors.most_similar_cosmul(positive=['president', 'german'])",
"_____no_output_____"
],
[
"word_vectors.most_similar_cosmul(positive=['woman', 'king'], negative=['man'])",
"_____no_output_____"
]
],
[
[
"#### Pad/Cut tokenized comments to a certain length",
"_____no_output_____"
]
],
[
[
"MAX_NB_WORDS = len(word_vectors.vocab)\nMAX_SEQUENCE_LENGTH = 200\ntrain_size = len(X_train)\nmost_common = sorted(tokenizer.word_counts.items(), key=lambda x: x[1], reverse=True)\n# word_index tells ous the\nX_train, X_test, word_index = text_utils.pad_or_cut_tokenized_comments(\n most_common, comments, train_size, MAX_NB_WORDS, MAX_SEQUENCE_LENGTH)",
"Shape of training data tensor: (532418, 200)\nShape of test_data tensor: (133105, 200)\n"
],
[
"# https://codekansas.github.io/blog/2016/gensim.html\nWV_DIM = 50\nnb_words = min(MAX_NB_WORDS, len(word_vectors.vocab))\nwv_matrix = word_vectors.syn0 # text_utils.get_weights_matrix(word_index, word_vectors, nb_words, WV_DIM)",
"_____no_output_____"
]
],
[
[
"#### Prepare Keras Model",
"_____no_output_____"
]
],
[
[
"embed_dim = 128\nlstm_out = 64\nbatch_size = 16\n\nmodel = Sequential()\nmodel.add(Embedding(nb_words,\n WV_DIM,\n mask_zero=False,\n weights=[wv_matrix],\n input_length=MAX_SEQUENCE_LENGTH,\n trainable=False))\n# model.add(Embedding(vocabulary, embed_dim, input_length = X.shape[1]))\nmodel.add(SpatialDropout1D(0.1))\n# model.add(LSTM(lstm_out, return_sequences=True, recurrent_dropout=0.3, dropout=0.3))\nmodel.add(LSTM(lstm_out, recurrent_dropout=0, dropout=0.1))\nmodel.add(Dense(2, activation='softmax'))\nmodel.compile(loss='binary_crossentropy', optimizer='adam', metrics = ['accuracy'])",
"_____no_output_____"
],
[
"model.summary()",
"_________________________________________________________________\nLayer (type) Output Shape Param # \n=================================================================\nembedding_2 (Embedding) (None, 200, 50) 64307550 \n_________________________________________________________________\nspatial_dropout1d_1 (Spatial (None, 200, 50) 0 \n_________________________________________________________________\nlstm_1 (LSTM) (None, 64) 29440 \n_________________________________________________________________\ndense_1 (Dense) (None, 2) 130 \n=================================================================\nTotal params: 64,337,120\nTrainable params: 29,570\nNon-trainable params: 64,307,550\n_________________________________________________________________\n"
],
[
"epochs = 20\nbatch_size = 32\n\n# Here we train the Network.\ntry:\n history = model.fit([X_train[:50000]], y_train[:50000], validation_split=0.1,\n batch_size = batch_size, epochs = epochs,\n verbose = 2, shuffle=True)\nexcept KeyboardInterrupt:\n print(\"Fitting stopped manually\")",
"Train on 45000 samples, validate on 5000 samples\nEpoch 1/20\n - 379s - loss: 0.6946 - acc: 0.5004 - val_loss: 0.6934 - val_acc: 0.5014\nEpoch 2/20\n - 171s - loss: 0.6933 - acc: 0.5081 - val_loss: 0.6941 - val_acc: 0.4850\nEpoch 3/20\n - 184s - loss: 0.6930 - acc: 0.5076 - val_loss: 0.6927 - val_acc: 0.5166\nEpoch 4/20\n - 188s - loss: 0.6928 - acc: 0.5095 - val_loss: 0.6943 - val_acc: 0.4840\nEpoch 5/20\n - 191s - loss: 0.6925 - acc: 0.5148 - val_loss: 0.6935 - val_acc: 0.5096\nEpoch 6/20\n - 193s - loss: 0.6921 - acc: 0.5148 - val_loss: 0.6932 - val_acc: 0.5148\nEpoch 7/20\n - 184s - loss: 0.6916 - acc: 0.5204 - val_loss: 0.6948 - val_acc: 0.5086\nEpoch 8/20\n - 168s - loss: 0.6910 - acc: 0.5231 - val_loss: 0.6960 - val_acc: 0.5072\nEpoch 9/20\n - 148s - loss: 0.6903 - acc: 0.5282 - val_loss: 0.6961 - val_acc: 0.5076\nEpoch 10/20\n - 144s - loss: 0.6887 - acc: 0.5340 - val_loss: 0.6982 - val_acc: 0.5076\nEpoch 11/20\n - 145s - loss: 0.6874 - acc: 0.5364 - val_loss: 0.6992 - val_acc: 0.5066\nEpoch 12/20\n - 140s - loss: 0.6866 - acc: 0.5412 - val_loss: 0.7003 - val_acc: 0.5050\nEpoch 13/20\n - 132s - loss: 0.6838 - acc: 0.5500 - val_loss: 0.7019 - val_acc: 0.5016\nEpoch 14/20\n - 128s - loss: 0.6825 - acc: 0.5559 - val_loss: 0.7039 - val_acc: 0.5090\nEpoch 15/20\n - 127s - loss: 0.6807 - acc: 0.5552 - val_loss: 0.7031 - val_acc: 0.4988\nEpoch 16/20\n - 127s - loss: 0.6769 - acc: 0.5652 - val_loss: 0.7121 - val_acc: 0.4936\nEpoch 17/20\n - 127s - loss: 0.6749 - acc: 0.5699 - val_loss: 0.7168 - val_acc: 0.4894\nEpoch 18/20\n - 127s - loss: 0.6716 - acc: 0.5763 - val_loss: 0.7172 - val_acc: 0.4952\nEpoch 19/20\n - 127s - loss: 0.6696 - acc: 0.5798 - val_loss: 0.7182 - val_acc: 0.4890\nEpoch 20/20\n - 127s - loss: 0.6660 - acc: 0.5869 - val_loss: 0.7195 - val_acc: 0.4890\n"
],
[
"def inspect_preprocessed_comment(data_comments_pol, X_train, backtranslater, idx):\n print('Original:')\n print(data_comments_pol.iloc[idx])\n print('\\nAfter preprocessing (& backtranslating):')\n print(' '.join([backtranslater[x] for x in X_train[idx] if x in backtranslater]))\n\ninspect_preprocessed_comment(data_comments_pol, X_train, backtranslater, 0)",
"Original:\nUnnamed: 0 159895\narticle_id 1731\nauthor_id 10860\ncomment_id 61083280\ncomment_text Actually it has - it was last night, I was the...\ntimestamp 2015-10-09T08:30:36Z\nparent_comment_id 6.10705e+07\nupvotes 2\nName: 0, dtype: object\n\nAfter preprocessing (& backtranslating):\nactually it has it was last night i was there unless he ' s going to give another speech at the university of sheffield in which he says these things\n"
],
[
"utils.plot_history(history)",
"_____no_output_____"
],
[
"# Measuring score and accuracy on test set\n\nscore, acc = model.evaluate([X:test], y_test, verbose = 2,\n batch_size = batch_size)\nprint(\"Logloss score: %.2f\" % (score))\nprint(\"Test set Accuracy: %.2f\" % (acc))",
"_____no_output_____"
],
[
"plt.hist(data_comments_pol['comment_text'].str.split().len)",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec7382485032c919e68808c1d405f12e33801c40 | 27,905 | ipynb | Jupyter Notebook | doc/ipython-notebooks/classification/HashedDocDotFeatures.ipynb | LiuYuHui/shogun | aab9e644d0be2bc4a9a3f40810c6010a987956c0 | [
"BSD-3-Clause"
]
| 1 | 2020-05-12T10:37:30.000Z | 2020-05-12T10:37:30.000Z | doc/ipython-notebooks/classification/HashedDocDotFeatures.ipynb | LiuYuHui/shogun | aab9e644d0be2bc4a9a3f40810c6010a987956c0 | [
"BSD-3-Clause"
]
| null | null | null | doc/ipython-notebooks/classification/HashedDocDotFeatures.ipynb | LiuYuHui/shogun | aab9e644d0be2bc4a9a3f40810c6010a987956c0 | [
"BSD-3-Clause"
]
| null | null | null | 60.927948 | 921 | 0.68253 | [
[
[
"# Large scale document classification with the Shogun Machine Learning Toolbox",
"_____no_output_____"
],
[
"#### By Evangelos Anagnostopoulos (GitHub ID: <a href=\"http://github.com/van51\">van51</a>)",
"_____no_output_____"
],
[
"This notebook demonstrates <a href=\"http://en.wikipedia.org/wiki/Document_classification\">Document classification</a> in Shogun. After providing a semi-formal introduction to the <a href=\"http://en.wikipedia.org/wiki/Bag-of-words_model\">Bag of Words</a> model and its limitations, we illustrate the <a href=\"http://en.wikipedia.org/wiki/Feature_hashing\">hashing trick</a>. This is consolidated by performing experiments on the large-scale webspam data set.",
"_____no_output_____"
],
[
"## Document Classification",
"_____no_output_____"
],
[
"<h3>Background</h3>\nDocument classification consists of assigning documents to specific predefined categories. This usually works by transforming the documents into a vector space that is acceptable by common classifiers, like SVMs, and then learning a model on that new representation.\n\nThe most common and the most widely used representation of document collections is the <a href=\"http://en.wikipedia.org/wiki/Bag-of-words_model\">Bag of Words</a> model.The BoW representation considers each document as a collection of tokens. A token is defined as the minimum arbitrary sequence of characters that can be considered as atomic. Tokens depending on the choice of the user can be either whole words or n-grams, etc. A simple approach is to consider every possible token in your document collection as a different dimension in a feature space and then vectorize each document by assigning 1 to every dimension that corresponds to a token contained in that document and 0 to every other. This is the simpler approach and is known as the Boolean Vector Space Model. Another approach is, instead of using {1,0}, to use a weight for every term, like their frequencies, but this is out of our scope.<br>\n\nLet's now consider a document collection consisting of the following documents:<br>\n <b>Document 1</b> = \"This is the first document\"<br>\n <b>Document 2</b> = \"Document classification: Introduction\"<br>\n <b>Document 3</b> = \"A third document about classification\"<br>\n\nand suppose that we consider as tokens, every word that appears in the collection.<br>\nThen we would arrive to the following representation:\n<table><tr><td></td><td><b>Document 1</b></td><td><b>Document 2</b></td><td><b>Document 3</b></td></tr>\n <tr><td>[0] this</td><td>1</td><td>0</td><td>0</td></tr>\n <tr><td>[1] is</td><td>1</td><td>0</td><td>0</td></tr>\n <tr><td>[2] the</td><td>1</td><td>0</td><td>0</td></tr>\n <tr><td>[3] first</td><td>1</td><td>0</td><td>0</td></tr>\n <tr><td>[4] document</td><td>1</td><td>1</td><td>1</td></tr>\n <tr><td>[5] classification</td><td>0</td><td>1</td><td>1</td></tr>\n <tr><td>[6] introduction</td><td>0</td><td>1</td><td>0</td></tr>\n <tr><td>[7] a</td><td>0</td><td>0</td><td>1</td></tr>\n <tr><td>[8] third</td><td>0</td><td>0</td><td>1</td></tr>\n <tr><td>[9] about</td><td>0</td><td>0</td><td>1</td></tr>\n</table>\nThe above matrix is called the <a href=\"http://en.wikipedia.org/wiki/Document-term_matrix\">document-term matrix</a> and is widely used in Information Retrieval applications. In our case since every document is now represented by a numerical vector, we can just pass this as a dataset to common classifiers, including kernel machines. <br>\n<h3>Limitations on large scale collections</h3>\nAlthough the aforementioned procedure is pretty intuitive and straight-forward it has some drawbacks when considered on large collections of documents.<br>\n<ul><li>First of all, the dimensionality of the feature space (or else, the number of distinct tokens in the collection) is usually <b>not known beforehand</b> and requires a pass over the entire dataset to be calculated. This can make the creation of the document-term matrix trickier, especially in online learning scenarios.</li>\n <li>Secondly, this dimensionality can grow to be <b>very large</b>; imagine considering as tokens the set of every possible word, or every possible 8-gram.</li>\n <li>Thirdly, this approach requires an <b>additional dictionary</b> in order to work that maps every token to its appropriate dimension, for instance every appearance of 'this' above should always be mapped to dimension 0, and this mapping data structure will also grow to be <b>very large.</b></li></ul><br>\n<h3>The hashing response</h3>\nA convenient approach that eliminates all the problems that the BoW representation introduced is to use a hash function to transform all the tokens into numerical entities and then restrict those values into a range [0, n-1] using the modulo function. The values in the [0, n-1] range will correspond to indices on an n-dimensional document-term matrix.<br>This way: <ul>\n<li>the dimensionality of the target feature space is now <b>known beforehand</b>, as it's simply the number of all possible outcomes of the hash functions, restricted to a specific range [0, n-1] by using the modulo function. No extra passes over the dataset are needed this way.</li>\n<li>In addition, the dimension can now be <b>restricted to a certain size</b> that is convenient depending on the limitations of the available hardware</li>\n<li>The dictionary is also <b>not needed</b> anymore since each token is directly mapped to the appropriate dimension through the outcome of the hash function.</li></ul>\n\nThe above approach will result in a hashed document-term matrix, similar to the one shown above, of a pre-defined dimension size (number of rows).<br>\nEach token will now be assigned to a random dimension, although always the same for the same token, that is determined from the hash function.<br> This will probably incur some collisions, but if the hash functions are good enough and our choice for the dimension size is sufficiently large it will not be a problem. Especially if we are considering large collections. <br>\n\nAfter the creation of the hashed document-term matrix, the intuition is to pass it as a dataset to a linear classifier that will take care of the rest!<br>\n<h3>On-the-fly Hashing</h3>\nA naive way to implement the hashing procedure is to pre-hash every token of the collection and store it to the disk for later access. However, this approach is not very convenient since it will require space analogous to the size of the original dimension. It will also make the experimentation on different parameters, like the size of the hash function or the choice of tokens, a bit trickier since one would have to re-convert the entire collection based on the choice of those parameters.<br>\nThe response to that is to read our collection as it is and compute the hash of every token only when it's required, on-the-fly.<br>\n<h3>On-the-fly Hashing with Shogun</h3>\nWe will now have a look at how the above idea is represented in the Shogun Toolbox. That is we will see how we can load our document collection in memory and consider a hashed document-term matrix with the hashing of every document (or token more specifically) happening on-the-fly, only when it's required to be computed. Although it may sound a bit tricky, it's actually pretty straightforward and here is how.<br><br>\nFirst of all we import the required components from the shogun library.",
"_____no_output_____"
]
],
[
[
"import os\nimport shogun as sg\nimport numpy as np\nimport matplotlib.pyplot as plt\n%matplotlib inline\n\nSHOGUN_DATA_DIR=os.getenv('SHOGUN_DATA_DIR', '../../../data')",
"_____no_output_____"
]
],
[
[
"The <a href=\"http://shogun-toolbox.org/doc/en/current/classshogun_1_1CStringFeatures.html\">StringCharFeatures</a> are nothing more than a collection of char vectors, where each one represents a single document.<br>\n<a href=\"https://www.shogun-toolbox.org/api/latest/namespaceshogun.html#a7fffbfce3d76cf49bde3916d23186b03\">RAWBYTE</a> is simply an enum that specifies that every possible character can be found in the collection.<br>\nThe <a href=\"https://www.shogun.ml/api/latest/classshogun_1_1CHashedDocDotFeatures.html\">HashedDocDotFeatures</a> is where all the magic happens! This class is responsible for encapsulating the document collection and providing the calculation of the hashed representation of each document whenever it is required. <br>\nThe <a href=\"https://www.shogun.ml/api/latest/classshogun_1_1CNGramTokenizer.html\">NGramTokenizer</a> is the tokenizer we will use on our collection. Its job is to parse a document and create the tokens. As its name suggests, it creates a sequence of ngrams. Another option would be the <a href=\"https://www.shogun.ml/api/latest/classshogun_1_1CDelimiterTokenizer.html\">DelimiterTokenizer</a>, which allows the user to specify the delimiter that will be used in order to create the tokens. <br><br><br>\nSuppose now that we have the following documents:",
"_____no_output_____"
]
],
[
[
"doc_1 = \"this is the first document\"\ndoc_2 = \"document classification introduction\"\ndoc_3 = \"a third document about classification\"\ndocument_collection = [doc_1, doc_2, doc_3]",
"_____no_output_____"
]
],
[
[
"We will take some time off now to assign each document to a category, to ease our work later on. Since the two last documents refer to classification we will label them as \"Relevant\" and the first document as \"Not relevant\". Since we only have two categories, this makes it a binary problem and we will represent \"Relevant\" as 1 and \"Not relevant\" as -1.",
"_____no_output_____"
]
],
[
[
"labels = sg.BinaryLabels(np.array([-1, 1, 1], dtype=np.int64))",
"_____no_output_____"
]
],
[
[
"We now create our document collection:",
"_____no_output_____"
]
],
[
[
"string_features = sg.StringCharFeatures(document_collection, sg.RAWBYTE)",
"_____no_output_____"
]
],
[
[
"Now we will create the object responsible for the hashed BoW representation. We are going to specify that we want a hash size of 8 bits, which will be translated to a dimension of size 2^8 = 256 (powers of 2 are considered to speed up computations) and a tokenizer that creates 5-grams. We will also specify that we want to\nnormalize each hashed vector with regards to the square root of the document length, which is a common approach:",
"_____no_output_____"
]
],
[
[
"hash_size = 8\ntokenizer = sg.NGramTokenizer(5)\nnormalize = True\n\nhashed_feats = sg.HashedDocDotFeatures(hash_size, string_features, tokenizer, normalize)",
"_____no_output_____"
]
],
[
[
"And that was it!<br>\nThe hashed_feats object now has all the required parameters and knows how to communicate and provide the hashed representation <b>only</b> when it is needed by the various algorithms of Shogun. <br><br>\nSo how do we proceed to actually learn a model that can classify documents?<br>\nWe said before that the idea after we have taken care of the hashing is to use a linear classifier.<br>\nShogun has many <a href=\"https://www.shogun-toolbox.org/api/latest/classshogun_1_1CLinearMachine.html\">linear</a> classifiers available, but for the moment we will consider SVMOcas and we will see how to use it:",
"_____no_output_____"
]
],
[
[
"C = 0.1\nepsilon = 0.01\nsvm = sg.create_machine(\"SVMOcas\", C1=C, C2=C, labels=labels, epsilon=epsilon)",
"_____no_output_____"
]
],
[
[
"We have now created our svm. The parameter C specifies the regularization constant. The best choice for this parameter will usually be selected after a model selection process.<br>\nNow, let's train our svm:",
"_____no_output_____"
]
],
[
[
"_=svm.train(hashed_feats)",
"_____no_output_____"
]
],
[
[
"When the execution finishes, we will have learned our so desired linear model! Mind that for large collections the above call can take hours.<br>\nNow that we have a model ready, we would like to see how well it does. Let's see what it predicts for our training data:",
"_____no_output_____"
]
],
[
[
"predicted_labels = svm.apply()\nprint(predicted_labels.get(\"labels\"))",
"_____no_output_____"
]
],
[
[
"We can see that it misclassified the first document. This has to do with the nature of our overly-simplified toy dataset which doesn't provide enough information. However, another option of the HashedDocDotFeatures class will allow us to extract some more information from the same dataset!<br>\n<br>\n<h3>Quadratic Features</h3>\nGenerally, the quadratic features refer to the production of new features from our current dataset, by multiplying currently existing features.\n<br>For example, if we have a dataset consisting of the following example vector:\n<table><tr><td></td><td>A</td><td>B</td><td>C</td></tr>\n <tr><td>x:</td><td>2</td><td>3</td><td>1</td></tr></table>\nWe can create the following extra quadratic features:\n<table><tr><td></td><td>A</td><td>B</td><td>C</td><td>AA</td><td>AB</td><td>AC</td><td>BB</td><td>BC</td><td>CC</td></tr>\n <tr><td>x:</td><td>2</td><td>3</td><td>1</td><td>2*2=4</td><td>2*3=6</td><td>2*1=2</td><td>3*3=9</td><td>3*1=3</td><td>1*1=1</td></tr></table>\nThe creation of these features can be very useful, however, it should be used with caution since it will also increase the training time and the size requirements. <br>\nThe idea behind the quadratic features is very simple and can be easily implemented for numerical features. However, in the case of text collections, where we have tokens instead of numbers, things are not so straightforward.<br><br>\nThe approach we have selected in Shogun for the text collections does not compute all the quadratic features, but only a subset of them defined using a k-skip n-grams rule. N-grams in this context refer to a collection of up to n consecutive tokens and should not be confused with our choice for what a token consists of, where it can be whole words or n-grams themselves. The k-skip in front allows us to skip up to k tokens when we group them in up-to n sized groups. Although this is a simple idea it can be a bit confusing when introduced through a definition, so let's have a look at an example:\nSuppose we have the following <b>consecutive</b> tokens:<br>\nTokens : [\"a\", \"b\", \"c\", \"d\"]<br>\nand that we want to consider a 2-skip 2-grams approach. This means that we can combine up to 2 tokens (that is a single token or two tokens) while skipping up to 2 tokens between them (that means we can skip 0,1 and 2 tokens). Therefore, we obtain the following combinations: <br>\n[\"a\" (0-skips, 1-gram), \"ab\" (0-skips, 2-gram), \"ac\" (1-skip 2-gram), \"ad\" (2-skips, 2-gram), \"b\" (0-skips, 1-gram), \"bc\" (0-skips, 2-gram),\n\"bd\" (1-skip, 2-gram), \"c\" (0-skip, 1-gram), \"cd\" (0-skip, 2-gram), \"d\" (0-skip, 1-gram)] <br>\n<br>These newly created tokens are then treated like our regular ones and hashed to find their appropriate dimension.\n<br>As discussed above this idea may enhance our information on the dataset, but it also requires more time to be computed. So if used, the choice of k and n should be kept to small numbers.<br>\n<h3>Quadratic Features with HashedDocDotFeatures</h3>\n\nIn order to use the k-skip n-gram approach explained above, one only has to specify his choice for k and n to the HashedDocDotFeatures object he is creating.<br>\nSo, by keeping our previous choice of parameters intact and introducing the k and n we obtain the following piece of code :",
"_____no_output_____"
]
],
[
[
"k = 3 # number of tokens, up to which we allow it to skip\nn = 3 # number of tokens, up to which we allow it to combine\n\nhashed_feats_quad = sg.HashedDocDotFeatures(hash_size, string_features, tokenizer, normalize, n, k)",
"_____no_output_____"
]
],
[
[
"If we do not specify these numbers, as we did not do before, then, (you maybe have guessed it!) they are set by default to the following values, n=1, k=0!\n<br>\nLet's see if there was any improvement in our predicting capabilities.<br>\nBy keeping the same svm settings, we will train on the new HashedDocDotFeatures object we have just created. Although we have the same document collection underneath, we are now considering a different representation model because of the quadratic features.",
"_____no_output_____"
]
],
[
[
"svm.train(hashed_feats_quad)\npredicted_labels = svm.apply()\nprint(predicted_labels.get(\"labels\"))",
"_____no_output_____"
]
],
[
[
"Better!<br>\n\n<b>Attention!</b> Some clarifications now will follow on the Quadratic Features to clear any misunderstandings:\n<ul><li>The term \"ngram\" can come up on two different levels. <br>The first one is in the tokenization process of the documents, where we have a raw stream of characters and our job there is to split the document on sequences of characters that are considered atomic. For instance, we may decide that sequences of three characters are the basic units which make up a document and therefore we are considering 3-grams (for tokens).<br>\nThe second level, is when we are combining the tokens generated in the previous phase to create quadratic features, by following the aforementioned k-skip n-grams rule. Now we are not considering anymore the document as a sequence of characters, but rather as a sequence of tokens. And it is on those tokens that we are now applying the rule. Therefore, if we decide to apply a 0-skip 3-grams rule then we may combine up to 3 tokens (not characters).\n In conclusion, the way the quadratic features work is completely independent of the Tokenizer used on the documents. We could have chosen a DelimiterTokenizer and still consider ngrams (of tokens) on the quadratic level.</li>\n <li>The \"quadratic\" in front of \"quadratic features\" also refers to the way the computing time increases when we select this approach! So it should only be used when it is necessary.</li></ul>",
"_____no_output_____"
],
[
"## Experiments on real datasets",
"_____no_output_____"
],
[
"The example that was demonstrated before was very small and simple and was manipulated for demonstration purposes, to show the capabilities of the HashedDocDotFeatures class. <br> <br><h3>Webspam</h3>Now let's have a look at the results the HashedDocDotFeatures class together with SVMOcas produced when faced against the <a href=\"ftp://largescale.ml.tu-berlin.de/largescale/webspam/\">webspam</a> dataset, which consists of 350000 documents labeled either as \"Spam\" or as \"Not spam\".<br>We demonstrate results when compared against SVMLight, using the Spectrum Kernel. <br>\nWe consider the following settings: <br>number of bits in hash = 16, 8-grams for tokens, SVM epsilon = 0.01.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\n\n# HashedDocDotFeatures results\nhashed_training_examples = [5000, 10000, 15000, 20000, 25000, 30000, 50000, 100000]\n\n# For C=1\nhashed_C_1_sec = [2682.750000,5202.690000,8120.460000,10846.410000,13944.200000,17016.840000,30496.720000,66302.950000]\nhashed_C_1_roc = [0.980730,0.986382,0.988894,0.990666,0.991602,0.991957,0.993680,0.995184]\n\n# For C=0.1\nhashed_C_01_sec = [1074.130000,2142.390000,3434.710000,4641.380000,5984.530000,7206.040000,12864.270000,28393.540000]\nhashed_C_01_roc = [0.976560,0.982660,0.985251,0.987380,0.988368,0.989022,0.990950,0.993197]\n\n# Spectrum kernel results\nkernel_training_examples = [5000, 10000, 15000, 20000, 25000]\n\n# For C=1\nkernel_C_1_sec = [2912.410000,6543.220000,10840.550000,16108.360000,19899.610000]\nkernel_C_1_roc = [0.971284,0.976628,0.979715,0.982084,0.984355]\n\n# For C=0.1\nkernel_C_01_sec = [1441.380000,3261.870000,5071.040000,7568.130000,10436.430000]\nkernel_C_01_roc = [0.946308,0.955245,0.961576,0.965204,0.968264]\n\nplt.figure(figsize=(12,6))\nplt.subplot(1,2,1)\nplt.plot(hashed_training_examples, hashed_C_1_sec, 'b')\nplt.plot(kernel_training_examples, kernel_C_1_sec, 'r')\nplt.title(\"Time comparison for C=1\")\nplt.xlabel(\"Number of examples\")\nplt.ylabel(\"Time in seconds\")\nplt.legend([\"HashedDocDotFeatures\", \"Spectrum Kernel\"], loc=2)\n\nplt.subplot(1,2,2)\nplt.plot(hashed_training_examples, hashed_C_1_roc, 'b')\nplt.plot(kernel_training_examples, kernel_C_1_roc, 'r')\nplt.title(\"Area under ROC comparison for C=1\")\nplt.xlabel(\"Number of examples\")\nplt.ylabel(\"auROC\")\n_=plt.legend([\"HashedDocDotFeatures\", \"Spectrum Kernel\"], loc=4)",
"_____no_output_____"
],
[
"plt.clf\nplt.figure(figsize=(12,6))\nplt.subplot(1,2,1)\nplt.plot(hashed_training_examples, hashed_C_01_sec, 'b')\nplt.plot(kernel_training_examples, kernel_C_01_sec, 'r')\nplt.title(\"Time comparison for C=0.1\")\nplt.xlabel(\"Number of examples\")\nplt.ylabel(\"Time in seconds\")\nplt.ylim((0,70000))\nplt.legend([\"HashedDocDotFeatures\", \"Spectrum Kernel\"], loc=2)\n\nplt.subplot(1,2,2)\nplt.plot(hashed_training_examples, hashed_C_01_roc, 'b')\nplt.plot(kernel_training_examples, kernel_C_01_roc, 'r')\nplt.title(\"Area under ROC comparison for C=0.1\")\nplt.xlabel(\"Number of examples\")\nplt.ylabel(\"auROC\")\n_=plt.legend([\"HashedDocDotFeatures\", \"Spectrum Kernel\"], loc=4)",
"_____no_output_____"
]
],
[
[
"<h3>Language detection</h3><br>\nAnother experiment we conducted was to create a language detection application. For that purpose we created a dataset consisting of 10,000 documents for 5 different languages; English, Greek, German, Italian and Spanish. <br>The demo uses underneath it a svm model trained on 30,000 documents with Ngrams of size 4, bits of hash size = 18, quadratic features with options: 2-skips 3-grams and svm C = 0.1. The demo can be found here and you can test it on your own how well it does!",
"_____no_output_____"
],
[
"<h3>Extra stuff</h3>\n\nThe hashing trick has also been implemented in Shogun for the DenseFeatures and SparseFeatures classes, as HashedDenseFeatures and HashedSparseFeatures classes. These classes also support quadratic features and provide the option to maintain or drop the linear terms. <br>\nFor online algorithms or for cases when the data do not fit in memory there exist similar classes with the prefix \"Streaming\" that support reading examples from the disk and providing them to some algorithms one at a time. The classes specifically are StreamingHashedDocDotFeatures, StreamingHashedDenseFeatures and StreamingHashedSparseFeatures. If one has mixed features, that are not just numerical or not just text, then he can use the CombinedDotFeatures class to combine objects of the aforementioned classes!<br>Another option is to use the Vw* algorithms and the VwExample class that requires the input to be in vw format and are a bit trickier to use.<br>\nIn addition, a HashedDocConverter class exists that allows the user to get the hashed BoW representation of a document collection as a <a href=\"http://shogun-toolbox.org/doc/en/current/classshogun_1_1SparseFeatures.html\">SparseFeatures</a> object.",
"_____no_output_____"
],
[
"## References",
"_____no_output_____"
],
[
"<ol>\n<li>Kilian Weinberger, Anirban Dasgupta, Josh Attenberg, John Langford, Alex Smola : <a href=\"http://arxiv.org/pdf/0902.2206.pdf\">Feature Hashing for Large Scale Multitask Learning</a></li>\n<li>Olivier Chapelle, Eren Manavoglu, Romer Rosales : <a href=\"http://people.csail.mit.edu/romer/papers/TISTRespPredAds.pdf\">Simple and scalable response prediction for display advertising</a></li>\n<li>S̈oren Sonnenburg, Gunnar R̈atsch, Konrad Rieck : <a href=\"http://sonnenburgs.de/soeren/publications/SonRaeRie07.pdf\">Large Scale Learning with String Kernels</a></li>\n<li><a href=\"https://github.com/JohnLangford/vowpal_wabbit/wiki\">Vowpal Wabbit</a></li>\n<li>A <a href=\"http://metaoptimize.com/qa/questions/6943/what-is-the-hashing-trick<\">post</a> in metaoptimize</li></ol><br/>\n<b>Special thanks to my mentors for this project on GSoC 2013 : Soeren Sonnenburg, Olivier Chapelle and Benoit Rostykus</b>",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec738f6ecbe4e9bc46e032b03d8f8e9f0d99d1fa | 4,160 | ipynb | Jupyter Notebook | talkmap.ipynb | samscibelli/samscibelli.github.io | 8bc5650cab9d90d3e53d523392c41438554fe305 | [
"MIT"
]
| null | null | null | talkmap.ipynb | samscibelli/samscibelli.github.io | 8bc5650cab9d90d3e53d523392c41438554fe305 | [
"MIT"
]
| null | null | null | talkmap.ipynb | samscibelli/samscibelli.github.io | 8bc5650cab9d90d3e53d523392c41438554fe305 | [
"MIT"
]
| null | null | null | 26.163522 | 274 | 0.548798 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec73b11e581065fdc58bc6f1245c398db70713a4 | 23,997 | ipynb | Jupyter Notebook | scripts/Tool_Overlap_Scripts_Phaster_Only.ipynb | Strong-Lab/Prophage_In_NTM | c7bacefbfac80dc7d36f8ea32acbe094e603059c | [
"MIT"
]
| 1 | 2020-12-13T20:43:04.000Z | 2020-12-13T20:43:04.000Z | scripts/Tool_Overlap_Scripts_Phaster_Only.ipynb | Strong-Lab/Prophage_In_NTM | c7bacefbfac80dc7d36f8ea32acbe094e603059c | [
"MIT"
]
| null | null | null | scripts/Tool_Overlap_Scripts_Phaster_Only.ipynb | Strong-Lab/Prophage_In_NTM | c7bacefbfac80dc7d36f8ea32acbe094e603059c | [
"MIT"
]
| null | null | null | 40.467116 | 164 | 0.513856 | [
[
[
"### Define Tool Overlap and Load Data",
"_____no_output_____"
]
],
[
[
"import pandas as pd \n\n### Import Data\nIVS = pd.read_csv(\"../data/Prophage_Tool_Predictions/Isolates_Virsorter.csv\")\nIP = pd.read_csv(\"../data/Prophage_Tool_Predictions/Isolates_Phaster.csv\")\nCVS = pd.read_csv(\"../data/Prophage_Tool_Predictions/Complete_Genomes_VirSorter.csv\")\nCP = pd.read_csv(\"../data/Prophage_Tool_Predictions/Complete_Genomes_Phaster.csv\")",
"_____no_output_____"
],
[
"def range_intersect(r1_s, r1_e, r2_s, r2_e):\n return len(range(max(r1_s,r2_s), min(r1_e,r2_e))) or None",
"_____no_output_____"
],
[
"### Unique Identifiers\n\n#### IVS\nIVS[\"Contig\"] = IVS[\"Contig\"].astype(str)\nIVS[\"Start\"] = IVS[\"Start\"].astype(str)\nIVS[\"End\"] = IVS['End'].astype(str)\nIVS[\"Unique\"] = IVS[\"Identifier\"] + IVS[\"Contig\"] + IVS[\"Start\"] + IVS[\"End\"]\nIVS[\"Contig\"] = IVS[\"Contig\"].astype(int)\nIVS[\"Start\"] = IVS[\"Start\"].astype(int)\nIVS[\"End\"] = IVS['End'].astype(int)\n\n### IP \nIP[\"Contig\"] = IP[\"Contig\"].astype(str)\nIP[\"Start\"] = IP[\"Start\"].astype(str)\nIP[\"End\"] = IP['End'].astype(str)\nIP[\"Unique\"] = IP[\"Identifier\"] + IP[\"Contig\"] + IP[\"Start\"] + IP[\"End\"]\nIP[\"Contig\"] = IP[\"Contig\"].astype(int)\nIP[\"Start\"] = IP[\"Start\"].astype(int)\nIP[\"End\"] = IP['End'].astype(int)\n\n### CP\nCP[\"Start\"] = CP[\"Start\"].astype(str)\nCP[\"End\"] = CP['End'].astype(str)\nCP[\"Unique\"] = CP[\"ID\"] + CP[\"Start\"] + CP[\"End\"]\nCP[\"Start\"] = CP[\"Start\"].astype(int)\nCP[\"End\"] = CP['End'].astype(int)\n\n#### IVS\nCVS[\"Start\"] = CVS[\"Start\"].astype(str)\nCVS[\"End\"] = CVS['End'].astype(str)\nCVS[\"Unique\"] = CVS[\"ID\"] + CVS[\"Start\"] + CVS[\"End\"]\nCVS[\"Start\"] = CVS[\"Start\"].astype(int)\nCVS[\"End\"] = CVS['End'].astype(int)\n\n\n## Replace Contigs Scoring to Prophage\nIVS[\"Category\"] = IVS[\"Category\"].astype(str)\nIVS['Category'] = IVS['Category'].str.replace('1','7')\nIVS['Category'] = IVS['Category'].str.replace('2','8')\nIVS['Category'] = IVS['Category'].str.replace('3','9')\n\n## Replace Phaster Naming Convention w/ groups \nCP['Category'] = CP['Category'].str.replace('intact','4')\nCP['Category'] = CP['Category'].str.replace('questionable','5')\nCP['Category'] = CP['Category'].str.replace('incomplete','6')\nCP[\"Category\"] = pd.to_numeric(CP[\"Category\"])\nCVS[\"Category\"] = pd.to_numeric(CVS[\"Category\"])\n\n## Replace Phaster Naming Convention w/ groups \nIP['Category'] = IP['Category'].str.replace('intact','4')\nIP['Category'] = IP['Category'].str.replace('questionable','5')\nIP['Category'] = IP['Category'].str.replace('incomplete','6')\nIP[\"Category\"] = pd.to_numeric(IP[\"Category\"])\nIVS[\"Category\"] = pd.to_numeric(IVS[\"Category\"])\n\nx = len(IVS)+len(IP)+len(CP)+len(CVS)\nprint(\"Total number of prophages predicted across both tools in all confidence levels: \" + str(x))",
"Total number of prophages predicted across both tools in all confidence levels: 2072\n"
]
],
[
[
"### Complete Genomes",
"_____no_output_____"
]
],
[
[
"print('Genomes with predicted prophages from Phaster any quality:' + str(len(set(CP[\"ID\"]))))\nprint('Genomes with predicted prophages from VirSorter any quality:' + str(len(set(CVS[\"GCF\"]))))\nin_both_complete = set(CP[\"ID\"])&set(CVS[\"GCF\"])\nmissing = set(CP[\"ID\"])-set(CVS[\"GCF\"])\n\nCPB = CP[CP[\"ID\"].isin(in_both_complete)]\nCVSB = CVS[CVS[\"GCF\"].isin(in_both_complete)]\n\n## Split dataframe into list of smaller dataframes by GCF IDs\ngb = CVSB.groupby('GCF')\ny = [gb.get_group(x) for x in gb.groups]\n\ngb = CPB.groupby('ID')\nz = [gb.get_group(x) for x in gb.groups]\n\nmy_set = set()\nmy_other_set = set()\n\ncount = 0 \n\nfor item in range(len(z)):\n name_z = list(z[item][\"ID\"])[0]\n name_y = list(y[item][\"GCF\"])[0]\n if name_z != name_y:\n print(\"IDs do not match: \")\n df_y=y[item]\n df_z=z[item]\n \n #def overlap remaining\n\n for item in range(len(df_y)):\n ### Get Start and End Position of Y\n unique_y = df_y.iloc[item].Unique\n start_y = df_y.iloc[item].Start\n end_y = df_y.iloc[item].End\n \n for j in range(len(df_z)):\n unique_z = df_z.iloc[j].Unique\n start_z = df_z.iloc[j].Start\n end_z = df_z.iloc[j].End\n \n out = range_intersect(start_y, end_y, start_z, end_z)\n \n if out != None:\n count+=1\n if df_y.iloc[item].Category >= df_z.iloc[j].Category:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n ## If categories are equal, take longer predicted phage\n elif df_y.iloc[item].Category == df_z.iloc[j].Category:\n if df_y.iloc[item].Length > df_z.iloc[j].Length:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n else:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n else:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\ncombined_set = set(list(my_set)+list(my_other_set))\n\n\noverlapped_retained_CVSB = CVSB[CVSB[\"Unique\"].isin(my_set)]\noverlapped_discarded_CVSB = CVSB[CVSB[\"Unique\"].isin(my_other_set)]\noverlapped_combined_CVSB = CVSB[CVSB[\"Unique\"].isin(combined_set)]\nnonoverlap_CVSB = CVSB[~CVSB[\"Unique\"].isin(combined_set)]\n## -------------------------\noverlapped_retained_CPB = CPB[CPB[\"Unique\"].isin(my_set)]\noverlapped_discarded_CPB = CPB[CPB[\"Unique\"].isin(my_other_set)]\noverlapped_combined_CPB = CPB[CPB[\"Unique\"].isin(combined_set)]\nnonoverlap_CPB = CPB[~CPB[\"Unique\"].isin(combined_set)]",
"Genomes with predicted prophages from Phaster any quality:53\nGenomes with predicted prophages from VirSorter any quality:47\n"
],
[
"df_overlapped = overlapped_retained_CVSB[[\"ID\", \"Category\", \"Start\", \"End\", \"Length\",\"Species.1\"]]\ndf_overlapped.columns = [\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\"]\ndf_overlapped['AttP'] = \"Not Applicable\"\ndf_y_o2 = overlapped_retained_CPB[[\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\", \"AttP Site\"]]\ndf_y_o2.columns = [\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\", \"AttP\"]\ndf_overlapped = df_overlapped.append(df_y_o2)\n\n\n## Subset and combine tools\ndf_total = nonoverlap_CVSB[[\"ID\", \"Category\", \"Start\", \"End\", \"Length\",\"Species.1\"]]\ndf_total.columns = [\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\"]\ndf_total['AttP'] = \"Not Applicable\"\ndf_y_o2 = nonoverlap_CPB[[\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\", \"AttP Site\"]]\ndf_y_o2.columns = [\"ID\", \"Category\", \"Start\", \"End\", \"Length\", \"Species\", \"AttP\"]\ndf_total = df_total.append(df_y_o2)\n\n## Combined overlap and non overlapping predictions\ndf_total = df_total.append(df_overlapped)\n\ntest = CVS[[\"ID\", \"GCF\"]]\ntest = test.set_index(\"ID\")\ntest = test.drop_duplicates()\ny = test.to_dict()\ny = y.get(\"GCF\")\ndf_overlapped[\"ID\"] = df_overlapped[\"ID\"].replace(y)\ndf_total[\"ID\"] = df_total[\"ID\"].replace(y)\n\n\ndf_overlapped[\"Start\"] = df_overlapped[\"Start\"].astype(str)\ndf_overlapped[\"End\"] = df_overlapped['End'].astype(str)\ndf_overlapped[\"Unique\"] = df_overlapped[\"ID\"] + \"-\"+ df_overlapped[\"Start\"] + \"-\"+ df_overlapped[\"End\"]\ndf_overlapped[\"Start\"] = df_overlapped[\"Start\"].astype(int)\ndf_overlapped[\"End\"] = df_overlapped['End'].astype(int)\n\ndf_overlapped = df_overlapped.reset_index()\ndf_overlapped = df_overlapped.drop(\"index\", axis=1)\n\nmy_set = set()\n\nfor item in range(len(df_overlapped)):\n ## Split Row from Dataframe\n row_of_interest = df_overlapped.iloc[item]\n df_without_row = df_overlapped.drop(item)\n ## Perform Comparison of Row Characterisitics \n ## Identifier is a column that I want to compare\n for j in range(len(df_without_row)):\n if df_without_row.iloc[j][\"ID\"] == row_of_interest[\"ID\"]:\n \n ## \n ds = df_without_row.iloc[j][\"Start\"]\n de = df_without_row.iloc[j][\"End\"]\n dc = df_without_row.iloc[j][\"Category\"]\n dl = df_without_row.iloc[j][\"Length\"]\n du = df_without_row.iloc[j][\"Unique\"]\n \n ## \n rs = row_of_interest[\"Start\"]\n re = row_of_interest[\"End\"]\n rc = row_of_interest[\"Category\"]\n rl = row_of_interest[\"Length\"]\n ru = row_of_interest[\"Unique\"]\n \n out = range_intersect(rs, re, ds, de)\n \n if out != None:\n if rc == dc:\n if rl <= dl:\n my_set.add(ru)\n else:\n my_set.add(du)\n elif rc < dc:\n my_set.add(du)\n \n else:\n my_set.add(ru)\ndf_overlapped = df_overlapped[~df_overlapped[\"Unique\"].isin(list(my_set))]",
"/Users/stronglab2/anaconda2/envs/py36/lib/python3.6/site-packages/ipykernel_launcher.py:12: SettingWithCopyWarning: \nA value is trying to be set on a copy of a slice from a DataFrame.\nTry using .loc[row_indexer,col_indexer] = value instead\n\nSee the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy\n if sys.path[0] == '':\n"
],
[
"print(\"Percentage of overlapping prophages retained from Phaster (with attachment site predictions): \" + str(round(((33.0/38)*100),2)) + \"%\")\nprint(\"Percentage of overlapping prophages with AttP sites: \" + str(round(((22.0/38)*100),2)) + \"%\")\nprint(\"Percentage of overlapping prophages with AttP sites only from Phaster predictions: \" + str(round(((22.0/33)*100),2)) + \"%\")",
"Percentage of overlapping prophages retained from Phaster (with attachment site predictions): 86.84%\nPercentage of overlapping prophages with AttP sites: 57.89%\nPercentage of overlapping prophages with AttP sites only from Phaster predictions: 66.67%\n"
],
[
"## Save Files \ndf_overlapped.to_csv(\"../data/Tables/Overlapped_Prophages_Complete_Phaster.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"### Isolates",
"_____no_output_____"
]
],
[
[
"print('Genomes with predicted prophages from Phaster any quality:' + str(len(set(IP[\"Identifier\"]))))\nprint('Genomes with predicted prophages from VirSorter any quality:' + str(len(set(IVS[\"Identifier\"]))))\nin_both_complete = set(IP[\"Identifier\"])&set(IVS[\"Identifier\"])\nmissing = set(IP[\"Identifier\"])-set(IVS[\"Identifier\"])\n\nIPB = IP[IP[\"Identifier\"].isin(in_both_complete)]\nIVSB = IVS[IVS[\"Identifier\"].isin(in_both_complete)]\n\n## Split dataframe into list of smaller dataframes by GCF IDs\ngb = IVSB.groupby('Identifier')\ny = [gb.get_group(x) for x in gb.groups]\n\ngb = IPB.groupby('Identifier')\nz = [gb.get_group(x) for x in gb.groups]\n\nmy_set = set()\nmy_other_set = set()\n\ncount = 0 \n\nfor item in range(len(z)):\n name_z = list(z[item][\"Identifier\"])[0]\n name_y = list(y[item][\"Identifier\"])[0]\n if name_z != name_y:\n print(\"IDs do not match: \")\n df_y=y[item]\n df_z=z[item]\n \n\n\n for item in range(len(df_y)):\n unique_y = df_y.iloc[item].Unique\n start_y = df_y.iloc[item].Start\n end_y = df_y.iloc[item].End\n contig_y = df_y.iloc[item].Contig\n \n for j in range(len(df_z)):\n unique_z = df_z.iloc[j].Unique\n start_z = df_z.iloc[j].Start\n end_z = df_z.iloc[j].End\n contig_z = df_z.iloc[j].Contig\n \n if contig_z == contig_y:\n \n out = range_intersect(start_y, end_y, start_z, end_z)\n \n if out != None:\n count+=1\n if df_y.iloc[item].Category >= df_z.iloc[j].Category:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n ## If categories are equal, take longer predicted phage\n elif df_y.iloc[item].Category == df_z.iloc[j].Category:\n if df_y.iloc[item].Length > df_z.iloc[j].Length:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n else:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\n else:\n my_set.add(unique_z)\n my_other_set.add(unique_y)\ncombined_set = set(list(my_set)+list(my_other_set))\n\n\noverlapped_retained_IVSB = IVSB[IVSB[\"Unique\"].isin(my_set)]\noverlapped_discarded_IVSB = IVSB[IVSB[\"Unique\"].isin(my_other_set)]\noverlapped_combined_IVSB = IVSB[IVSB[\"Unique\"].isin(combined_set)]\nnonoverlap_IVSB = IVSB[~IVSB[\"Unique\"].isin(combined_set)]\n## -------------------------\noverlapped_retained_IPB = IPB[IPB[\"Unique\"].isin(my_set)]\noverlapped_discarded_IPB = IPB[IPB[\"Unique\"].isin(my_other_set)]\noverlapped_combined_IPB = IPB[IPB[\"Unique\"].isin(combined_set)]\nnonoverlap_IPB = IPB[~IPB[\"Unique\"].isin(combined_set)]",
"Genomes with predicted prophages from Phaster any quality:179\nGenomes with predicted prophages from VirSorter any quality:180\n"
],
[
"## Remove Overlapping Identifiers\nnonoverlap_IP = IP[~IP[\"Unique\"].isin(combined_set)]\nnonoverlap_IVS = IVS[~IVS[\"Unique\"].isin(combined_set)]\n\n## \noverlapped_combined_IVSB\ndf_overlapped = overlapped_retained_IVSB[[\"Identifier\", \"Category\", \"Contig\", \"Start\", \"End\", \"Length\",\"Species\"]]\ndf_overlapped.columns = [\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\", \"Species\"]\ndf_overlapped['AttP'] = \"Not Applicable\"\ndf_y_o2 = overlapped_retained_IPB[[\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\", \"Species\", \"AttP Site\"]]\ndf_y_o2.columns = [\"Identifier\", \"Category\", \"Contig\", \"Start\", \"End\", \"Length\", \"Species\", \"AttP\"]\ndf_overlapped = df_overlapped.append(df_y_o2)\n\n## Subset and combine tools\ndf_total = nonoverlap_IVS[[\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\",\"Species\"]]\ndf_total.columns = [\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\", \"Species\"]\ndf_total['AttP'] = \"Not Applicable\"\ndf_y_o2 = nonoverlap_IP[[\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\", \"Species\", \"AttP Site\"]]\ndf_y_o2.columns = [\"Identifier\", \"Category\", \"Contig\",\"Start\", \"End\", \"Length\", \"Species\", \"AttP\"]\ndf_total = df_total.append(df_y_o2)\n\n## Combined overlap and non overlapping predictions\ndf_total = df_total.append(df_overlapped)",
"_____no_output_____"
],
[
"df_overlapped[\"Contig\"] = df_overlapped[\"Contig\"].astype(str)\ndf_overlapped[\"Start\"] = df_overlapped[\"Start\"].astype(str)\ndf_overlapped[\"End\"] = df_overlapped['End'].astype(str)\ndf_overlapped[\"Unique\"] = df_overlapped['Identifier'] + \"-\" + df_overlapped['Contig'] + \"-\" + df_overlapped['Start'] + \"-\" + df_overlapped[\"End\"]\ndf_overlapped[\"Contig\"] = df_overlapped[\"Contig\"].astype(int)\ndf_overlapped[\"Start\"] = df_overlapped[\"Start\"].astype(int)\ndf_overlapped[\"End\"] = df_overlapped['End'].astype(int)\n\ndf_overlapped = df_overlapped.reset_index()\ndf_overlapped = df_overlapped.drop(\"index\", axis=1)\n\nmy_set = set()\n\nfor item in range(len(df_overlapped)):\n ## Split Row from Dataframe\n row_of_interest = df_overlapped.iloc[item]\n df_without_row = df_overlapped.drop(item)\n ## Perform Comparison of Row Characterisitics \n ## Identifier is a column that I want to compare\n for j in range(len(df_without_row)):\n if df_without_row.iloc[j][\"Identifier\"] == row_of_interest[\"Identifier\"]:\n if df_without_row.iloc[j][\"Contig\"] == row_of_interest[\"Contig\"]:\n \n ## \n ds = df_without_row.iloc[j][\"Start\"]\n de = df_without_row.iloc[j][\"End\"]\n dc = df_without_row.iloc[j][\"Category\"]\n dl = df_without_row.iloc[j][\"Length\"]\n du = df_without_row.iloc[j][\"Unique\"]\n \n ## \n rs = row_of_interest[\"Start\"]\n re = row_of_interest[\"End\"]\n rc = row_of_interest[\"Category\"]\n rl = row_of_interest[\"Length\"]\n ru = row_of_interest[\"Unique\"]\n \n out = range_intersect(rs, re, ds, de)\n \n if out != None:\n if rc == dc:\n if rl <= dl:\n my_set.add(ru)\n else:\n my_set.add(du)\n elif rc < dc:\n my_set.add(du)\n \n else:\n my_set.add(ru)\ndf_overlapped = df_overlapped[~df_overlapped[\"Unique\"].isin(list(my_set))]",
"_____no_output_____"
],
[
"print(\"Percentage of overlapping prophages retained from Phaster (with attachment site predictions): \" + str(round(((92.0/150)*100),2)) + \"%\")\nprint(\"Percentage of overlapping prophages with AttP sites: \" + str(round(((47.0/150)*100),2)) + \"%\")\nprint(\"Percentage of overlapping prophages with AttP sites only from Phaster predictions: \" + str(round(((47.0/92)*100),2)) + \"%\")",
"Percentage of overlapping prophages retained from Phaster (with attachment site predictions): 61.33%\nPercentage of overlapping prophages with AttP sites: 31.33%\nPercentage of overlapping prophages with AttP sites only from Phaster predictions: 51.09%\n"
],
[
"## Save Files \ndf_overlapped.to_csv(\"../data/Tables/Overlapped_Prophages_Isolates_Phaster.csv\", index=False)",
"_____no_output_____"
]
],
[
[
"### End of Tool Overlap Script",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
ec73c2f8fce9e9c88fa408cbcf0978876a51770d | 31,652 | ipynb | Jupyter Notebook | tensorflow_graphics/projects/point_convolutions/pylib/notebooks/ModelNet40.ipynb | schellmi42/graphics | 2e705622c2a6b0007347d2db154ccdf5a0eb73d4 | [
"Apache-2.0"
]
| null | null | null | tensorflow_graphics/projects/point_convolutions/pylib/notebooks/ModelNet40.ipynb | schellmi42/graphics | 2e705622c2a6b0007347d2db154ccdf5a0eb73d4 | [
"Apache-2.0"
]
| null | null | null | tensorflow_graphics/projects/point_convolutions/pylib/notebooks/ModelNet40.ipynb | schellmi42/graphics | 2e705622c2a6b0007347d2db154ccdf5a0eb73d4 | [
"Apache-2.0"
]
| 1 | 2021-10-11T09:10:56.000Z | 2021-10-11T09:10:56.000Z | 38.459295 | 431 | 0.512448 | [
[
[
"##### Copyright 2020 Google LLC.",
"_____no_output_____"
]
],
[
[
"#@title Licensed under the Apache License, Version 2.0 (the \"License\");\n# you may not use this file except in compliance with the License.\n# You may obtain a copy of the License at\n#\n# https://www.apache.org/licenses/LICENSE-2.0\n#\n# Unless required by applicable law or agreed to in writing, software\n# distributed under the License is distributed on an \"AS IS\" BASIS,\n# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.\n# See the License for the specific language governing permissions and\n# limitations under the License.",
"_____no_output_____"
]
],
[
[
"# Point Clouds for tensorflow_graphics\n<table class=\"tfo-notebook-buttons\" align=\"left\">\n <td>\n <a target=\"_blank\" href=\"https://colab.research.google.com/github/schellmi42/graphics/blob/point_convolutions/tensorflow_graphics/projects/point_convolutions/pylib/notebooks/ModelNet40.ipynb\"><img src=\"https://www.tensorflow.org/images/colab_logo_32px.png\" />Run in Google Colab</a>\n </td>\n <td>\n <a target=\"_blank\" href=\"https://github.com/schellmi42/graphics/blob/point_convolutions/tensorflow_graphics/projects/point_convolutions/pylib/notebooks/ModelNet40.ipynb\"><img src=\"https://www.tensorflow.org/images/GitHub-Mark-32px.png\" />View source on GitHub</a>\n </td>\n</table>",
"_____no_output_____"
],
[
"## Initialization",
"_____no_output_____"
],
[
"\n### Clone repositories, install requirements and custom_op package",
"_____no_output_____"
]
],
[
[
"# Clone repositories\n!rm -r graphics\n!git clone https://github.com/schellmi42/graphics\n\n# install requirements and load tfg module \n!pip install -r graphics/requirements.txt\n\n# install custom ops\n!pip install graphics/tensorflow_graphics/projects/point_convolutions/custom_ops/pkg_builds/tf_2.2.0/*.whl\n",
"_____no_output_____"
]
],
[
[
"### Load modules",
"_____no_output_____"
]
],
[
[
"import sys\n# (this is equivalent to export PYTHONPATH='$HOME/graphics:/content/graphics:$PYTHONPATH', but adds path to running session)\nsys.path.append(\"/content/graphics\")\n\n# load point cloud module \n# (this is equivalent to export PYTHONPATH='/content/graphics/tensorflow_graphics/projects/point_convolutions:$PYTHONPATH', but adds path to running session)\nsys.path.append(\"/content/graphics/tensorflow_graphics/projects/point_convolutions\")",
"_____no_output_____"
]
],
[
[
"Check if it loads without errors",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport tensorflow_graphics as tfg\nimport pylib.pc as pc\nimport numpy as np\n\nprint('TensorFlow version: %s'%tf.__version__)\nprint('TensorFlow-Graphics version: %s'%tfg.__version__)\nprint('Point Cloud Module: ', pc)",
"_____no_output_____"
]
],
[
[
"## Classification on ModelNet40",
"_____no_output_____"
],
[
"### Data preparation",
"_____no_output_____"
],
[
"First we load the data consisting of 10k points per model.",
"_____no_output_____"
]
],
[
[
"!wget --no-check-certificate https://shapenet.cs.stanford.edu/media/modelnet40_normal_resampled.zip \n!echo '---unzipping---'\n!unzip -q modelnet40_normal_resampled.zip \n!echo '[done]'",
"_____no_output_____"
]
],
[
[
"Next we load the data, using the input function in the `io` module.\n\nTo speed up this tutorial, we only load a subset of the points per model.\n\n",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf \nimport pylib.pc as pc\nimport pylib.io as io\nimport numpy as np\nimport tensorflow_graphics\nimport os, time\n\n\nquick_test = False # only load 100 models\n\n# -- loading data ---\n\ndata_dir = 'modelnet40_normal_resampled/'\nnum_classes = 40 # modelnet 10 or 40\npoints_per_file = 1024 # number of points loaded per model\n\n# load category names\ncategory_names = []\nwith open(data_dir + f'modelnet{num_classes}_shape_names.txt') as inFile:\n for line in inFile:\n category_names.append(line.replace('\\n', ''))\n\n# load names of training files\ntrain_set = []\ntrain_labels = []\nwith open(data_dir + f'modelnet{num_classes}_train.txt') as inFile:\n for line in inFile:\n line = line.replace('\\n', '')\n category = line[:-5]\n train_set.append(data_dir + category + '/' + line + '.txt')\n if category not in category_names:\n raise ValueError('Unknown category ' + category)\n train_labels.append(category_names.index(category))\n\n# load names of test files\ntest_set = []\ntest_labels = []\nwith open(data_dir + f'modelnet{num_classes}_test.txt') as inFile:\n for line in inFile:\n line = line.replace('\\n', '')\n category = line[:-5]\n test_set.append(data_dir + category + '/' + line + '.txt')\n if category not in category_names:\n raise ValueError('Unknown category ' + category)\n test_labels.append(category_names.index(category))\n\n# load training data\ntrain_data_points = np.empty([len(train_set), points_per_file, 3])\n\nprint(f'### loading modelnet{num_classes} train ###')\nfor i, filename in enumerate(train_set):\n points, _ = \\\n io.load_points_from_file_to_numpy(filename,\n max_num_points=points_per_file)\n train_data_points[i] = points\n if i % 500 == 0:\n print(f'{i}/{len(train_set)}')\n if quick_test and i > 100:\n break\n\n# load test data\ntest_data_points = np.empty([len(test_set), points_per_file, 3])\n\nprint(f'### loading modelnet{num_classes} test ###')\nfor i, filename in enumerate(test_set):\n points, _ = \\\n io.load_points_from_file_to_numpy(filename,\n max_num_points=points_per_file)\n test_data_points[i] = points\n if i % 500 == 0:\n print(f'{i}/{len(test_set)}')\n if quick_test and i > 100:\n break",
"_____no_output_____"
]
],
[
[
"Now let's define a small data loader.\n\nTo make the network evaluation faster, we randomly samples the point clouds to reduce the input size.\n\nAs we don't want to provide any additional features other than the point location to the network, we will use a constant `1` as input feature.\n\n\n\n",
"_____no_output_____"
]
],
[
[
"class modelnet_data_generator(tf.keras.utils.Sequence):\n ''' Small generator of batched data.\n '''\n def __init__(self,\n points,\n labels,\n batch_size,\n augment):\n self.points = points\n self.labels = np.array(labels, dtype=int)\n self.batch_size = batch_size\n self.epoch_size = len(self.points)\n self.augment = augment\n\n self.ids = np.arange(0, points_per_file)\n # shuffle data before training\n self.on_epoch_end()\n\n def __len__(self):\n # number of batches per epoch\n return(int(np.floor(self.epoch_size / self.batch_size)))\n\n def __call__(self):\n ''' Loads batch and increases batch index.\n '''\n data = self.__getitem__(self.index)\n self.index += 1\n return data\n\n def __getitem__(self, index, samples_per_model=1024):\n ''' Loads data of current batch and samples random subset of the points.\n '''\n labels = \\\n self.labels[index * self.batch_size:(index + 1) * self.batch_size]\n points = \\\n self.points[index * self.batch_size:(index + 1) * self.batch_size]\n # constant input feature\n features = tf.ones([self.batch_size, samples_per_model, 1])\n\n # sample points\n sampled_points = np.empty([self.batch_size, samples_per_model, 3])\n for batch in range(self.batch_size):\n selection = np.random.choice(self.ids, samples_per_model)\n sampled_points[batch] = points[batch][selection]\n \n if self.augment:\n # Data augmentation - Anisotropic scale.\n cur_scaling = np.random.uniform(size=(1, 3)) * 0.2 + 0.9\n sampled_points[batch] = sampled_points[batch] * cur_scaling\n\n return sampled_points, features, labels\n\n def on_epoch_end(self):\n ''' Shuffles data and resets batch index.\n '''\n shuffle = np.random.permutation(np.arange(0, len(self.points)))\n self.points = self.points[shuffle]\n self.labels = self.labels[shuffle]\n self.index = 0",
"_____no_output_____"
]
],
[
[
"### Network architecture\n\nLet's build a simple classification network, which uses point convolutions for encoding the shape, and two dense layers for predicting the class.\n\nThe following model contains example calls for the three different available point convolutions in the `layer` module:\n\n\n* [Monte-Carlo convolutions](https://www.uni-ulm.de/fileadmin/website_uni_ulm/iui.inst.100/institut/Papers/viscom/2018/hermosilla2018montecarlo.pdf), which uses MLPs for representing the convolutional kernel, and aggregates the features inside the convolution radius using [Monte-Carlo integration](https://en.wikipedia.org/wiki/Monte_Carlo_integration), where each feature is weighted by a point density estimation.\n* [Kernel Point convolutions](https://arxiv.org/pdf/1904.08889.pdf), where the convolutional kernel is represented by a set of weights on kernel points, which are interpolated. \n(Note: We use rigid kernel points in the example below but deformable kernel points are also supported)\n\n* [PointConv convolutions](https://openaccess.thecvf.com/content_CVPR_2019/papers/Wu_PointConv_Deep_Convolutional_Networks_on_3D_Point_Clouds_CVPR_2019_paper.pdf)\n, which uses a single MLP for representing the convolutional kernel, and aggregates the features using an integration, where each feature is weighted by a learned inverse density estimation.\n\n\nNote that different to an image convolution layer, a point convolution layer needs additional input about the spatial location of the features, i.e. point coordinates.\nIn case of a 'strided' point convolution, where the output features are defined on different points than the input, we have to provide two point clouds.\n\nIn between the point convolutions 1x1 convolutions are used for more expressiveness.\n\nFor sampling the point clouds to lower densities, we use the `PointHierarchy` class.\n\nAt the end of the encoder we use a point convolution to a single point to aggregate a global feature.",
"_____no_output_____"
]
],
[
[
"from pylib.pc import layers\n\nclass mymodel(tf.Module):\n ''' Model architecture with `L` convolutional layers followed by\n two dense layers.\n\n Args:\n features_sizes: A `list` of `ints`, the feature dimensions. Shape `[L+3]`.\n sample_radii: A `list` of `floats, the radii used for sampling\n of the point clouds. Shape `[L]`.\n conv_radii: A `list` of `floats`, the radii used by the convolution\n layers. Shape `[L]`.\n layer_type: A `string`, the type of convolution used,\n can be 'MCConv', 'KPConv', 'PointConv'.\n sampling_method: 'poisson disk' or 'cell average'.\n dropout_rate: An `float`, percentage of neurons dropped in the \n classification head.\n '''\n\n def __init__(self,\n feature_sizes,\n sample_radii,\n conv_radii,\n layer_type='MCConv',\n sampling_method='poisson disk',\n dropout_rate=0.5):\n\n super().__init__(name=None)\n\n self.num_layers = len(sample_radii)\n self.sample_radii = sample_radii.reshape(-1, 1)\n self.conv_radii = conv_radii\n self.sampling_method = sampling_method\n self.conv_layers = []\n self.batch_layers = []\n self.dense_layers = []\n self.activations = []\n self.dropouts = []\n # encoder\n for i in range(self.num_layers):\n # convolutional downsampling layers\n # 'strided' convolutions\n if layer_type == 'MCConv':\n self.conv_layers.append(layers.MCConv(\n num_features_in=feature_sizes[i],\n num_features_out=feature_sizes[i + 1],\n num_dims=3,\n num_mlps=4,\n mlp_size=[8]))\n elif layer_type == 'PointConv':\n self.conv_layers.append(layers.PointConv(\n num_features_in=feature_sizes[i],\n num_features_out=feature_sizes[i + 1],\n num_dims=3,\n size_hidden=32))\n elif layer_type == 'KPConv':\n self.conv_layers.append(layers.KPConv(\n num_features_in=feature_sizes[i],\n num_features_out=feature_sizes[i + 1],\n num_dims=3,\n num_kernel_points=15))\n else:\n raise ValueError(\"Unknown layer type!\")\n if i < self.num_layers - 1:\n # batch normalization and activation function\n self.batch_layers.append(tf.keras.layers.BatchNormalization())\n self.activations.append(tf.keras.layers.LeakyReLU())\n # 1x1 Convolution between 'strided' convolutions\n self.conv_layers.append(layers.Conv1x1(\n num_features_in=feature_sizes[i + 1],\n num_features_out=feature_sizes[i + 1]))\n self.batch_layers.append(tf.keras.layers.BatchNormalization())\n self.activations.append(tf.keras.layers.LeakyReLU())\n # MLP\n self.batch_layers.append(tf.keras.layers.BatchNormalization())\n self.activations.append(tf.keras.layers.LeakyReLU())\n self.dropouts.append(tf.keras.layers.Dropout(dropout_rate))\n self.dense_layers.append(tf.keras.layers.Dense(feature_sizes[-2]))\n self.batch_layers.append(tf.keras.layers.BatchNormalization())\n self.activations.append(tf.keras.layers.LeakyReLU())\n self.dropouts.append(tf.keras.layers.Dropout(dropout_rate))\n self.dense_layers.append(tf.keras.layers.Dense(feature_sizes[-1]))\n\n def __call__(self,\n points,\n features,\n training):\n ''' Evaluates network.\n\n Args:\n points: The point coordinates. Shape `[B, N, 3]`.\n features: Input features. Shape `[B, N, C]`.\n training: A `bool`, passed to the batch norm layers.\n\n Returns:\n The logits per class.\n '''\n sample_radii = self.sample_radii\n conv_radii = self.conv_radii\n sampling_method = self.sampling_method\n # input point cloud\n # Note: Here all point clouds have the same number of points, so no `sizes`\n # or `batch_ids` are passed.\n point_cloud = pc.PointCloud(points)\n # spatial downsampling\n point_hierarchy = pc.PointHierarchy(point_cloud,\n sample_radii,\n sampling_method)\n # network evaluation\n for i in range(self.num_layers):\n if i < self.num_layers - 1:\n # 'strided' conv\n features = self.conv_layers[i * 2](\n features,\n point_hierarchy[i],\n point_hierarchy[i + 1],\n conv_radii[i])\n features = self.batch_layers[i * 2](features, training=training)\n features = self.activations[i * 2](features)\n # 1x1 conv\n features = self.conv_layers[i * 2 + 1](\n features,\n point_hierarchy[i + 1])\n features = self.batch_layers[i * 2 + 1](features, training=training)\n features = self.activations[i * 2 + 1](features)\n else:\n # the last layer has one point pe rpoint cloud, to ensure correctly sorted output features by batch id, we use `return_sorted = True`\n features = self.conv_layers[i * 2](\n features,\n point_hierarchy[i],\n point_hierarchy[i + 1],\n conv_radii[i],\n return_sorted=True)\n # classification head\n features = self.batch_layers[-2](features, training=training)\n features = self.activations[-2](features)\n features = self.dropouts[-2](features, training=training)\n features = self.dense_layers[-2](features)\n features = self.batch_layers[-1](features, training=training)\n features = self.activations[-1](features)\n features = self.dropouts[-1](features, training=training)\n return self.dense_layers[-1](features)",
"_____no_output_____"
]
],
[
[
"### Model parameters",
"_____no_output_____"
]
],
[
[
"batch_size = 16\n\nfeature_sizes = [1, 128, 256, 512, 1024, 1024, num_classes]\n# np.sqrt(3) * 2 ensures sampling to a single point.\nsample_radii = np.array([0.1, 0.2, 0.4, np.sqrt(3) * 2])\nconv_radii = sample_radii\n\n# initialize data generators\ngen_train = modelnet_data_generator(\n train_data_points, train_labels, batch_size, augment=True)\ngen_test = modelnet_data_generator(\n test_data_points, test_labels, batch_size, augment=False)\n\n# loss function and optimizer\nlr_decay = tf.keras.optimizers.schedules.Exponent\nialDecay(\n initial_learning_rate=0.001,\n decay_steps=20 * len(gen_train), # every 20th epoch\n decay_rate=0.7,\n staircase=True)\n\noptimizer = tf.keras.optimizers.RMSprop(learning_rate=lr_decay)\n\nloss_function = tf.keras.losses.SparseCategoricalCrossentropy()\n",
"_____no_output_____"
]
],
[
[
"### Training Loop",
"_____no_output_____"
]
],
[
[
"def training(model,\n optimizer,\n loss_function,\n num_epochs=400,\n epochs_print=1):\n train_loss_results = []\n train_accuracy_results = []\n test_loss_results = []\n test_accuracy_results = []\n epoch_loss_avg = tf.keras.metrics.Mean()\n epoch_accuracy = tf.keras.metrics.SparseCategoricalAccuracy()\n\n for epoch in range(num_epochs):\n time_epoch_start = time.time()\n\n # --- Training ---\n epoch_loss_avg.reset_states()\n epoch_accuracy.reset_states()\n for points, features, labels in gen_train:\n # evaluate model; forward pass\n with tf.GradientTape() as tape:\n logits = model(points, features, training=True)\n pred = tf.nn.softmax(logits, axis=-1)\n loss = loss_function(y_true=labels, y_pred=pred)\n # backpropagation\n grads = tape.gradient(loss, model.trainable_variables)\n\n optimizer.apply_gradients(zip(grads, model.trainable_variables))\n\n epoch_loss_avg.update_state(loss)\n epoch_accuracy.update_state(labels, pred)\n\n train_loss_results.append(epoch_loss_avg.result())\n train_accuracy_results.append(epoch_accuracy.result())\n\n # --- Validation ---\n epoch_loss_avg.reset_states()\n epoch_accuracy.reset_states()\n\n for points, features, labels in gen_test:\n # evaluate model; forward pass\n logits = model(points, features, training=False)\n pred = tf.nn.softmax(logits, axis=-1)\n loss = loss_function(y_true=labels, y_pred=pred)\n\n epoch_loss_avg.update_state(loss)\n epoch_accuracy.update_state(labels, pred)\n\n test_loss_results.append(epoch_loss_avg.result())\n test_accuracy_results.append(epoch_accuracy.result())\n\n time_epoch_end = time.time()\n\n if epoch % epochs_print == 0:\n # End epoch\n print('Epoch {:03d} Time: {:.3f}s'.format(\n epoch,\n time_epoch_end - time_epoch_start))\n print('Training: Loss: {:.3f}, Accuracy: {:.3%}'.format(\n train_loss_results[-1],\n train_accuracy_results[-1]))\n print('Validation: Loss: {:.3f}, Accuracy: {:.3%}'.format(\n test_loss_results[-1],\n test_accuracy_results[-1]))\n",
"_____no_output_____"
]
],
[
[
"#### Train with [Monte-Carlo convolutions](https://www.uni-ulm.de/fileadmin/website_uni_ulm/iui.inst.100/institut/Papers/viscom/2018/hermosilla2018montecarlo.pdf).\n\nNote: After 300 epochs this network should achieve >90% validation accuracy.",
"_____no_output_____"
]
],
[
[
"model_MC = mymodel(feature_sizes, sample_radii, conv_radii,\n layer_type='MCConv', dropout_rate=0.5)\ntraining(model_MC, \n optimizer=optimizer,\n loss_function=loss_function,\n num_epochs=10)",
"_____no_output_____"
]
],
[
[
"#### Train with [Kernel Point convolutions](https://arxiv.org/pdf/1904.08889.pdf).\n\nTo use the cell average sampling used in the paper, we can simply change the sampling method, which is passed to the point hierarchy constructor.",
"_____no_output_____"
]
],
[
[
"model_KP = mymodel(feature_sizes, sample_radii, conv_radii,\n layer_type='KPConv', sampling_method='cell average',\n dropout_rate=0.5)\ntraining(model_KP, \n optimizer=optimizer,\n loss_function=loss_function,\n num_epochs=10)",
"_____no_output_____"
]
],
[
[
"#### Train with [PointConv convolutions](https://openaccess.thecvf.com/content_CVPR_2019/papers/Wu_PointConv_Deep_Convolutional_Networks_on_3D_Point_Clouds_CVPR_2019_paper.pdf).",
"_____no_output_____"
]
],
[
[
"model_PC = mymodel(feature_sizes, sample_radii, conv_radii,\n layer_type='PointConv', dropout_rate=0.5)\ntraining(model_PC, \n optimizer=optimizer,\n loss_function=loss_function,\n num_epochs=10)",
"_____no_output_____"
],
[
"",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
]
]
|
ec73d0c36458b77a82f0b4f6b8d27153e1fe16dc | 119,860 | ipynb | Jupyter Notebook | notebooks/usecase_graph_embedding_and_clustering_in_hyperbolic_space.ipynb | doublestrong/geomstats | 67d8b63f2ef62cc9eac1a6b47ee7fb895dc19487 | [
"MIT"
]
| 1 | 2022-01-10T07:22:11.000Z | 2022-01-10T07:22:11.000Z | notebooks/usecase_graph_embedding_and_clustering_in_hyperbolic_space.ipynb | doublestrong/geomstats | 67d8b63f2ef62cc9eac1a6b47ee7fb895dc19487 | [
"MIT"
]
| null | null | null | notebooks/usecase_graph_embedding_and_clustering_in_hyperbolic_space.ipynb | doublestrong/geomstats | 67d8b63f2ef62cc9eac1a6b47ee7fb895dc19487 | [
"MIT"
]
| 1 | 2021-03-14T06:54:09.000Z | 2021-03-14T06:54:09.000Z | 102.0954 | 42,600 | 0.84003 | [
[
[
"# Tutorial: Hyperbolic Embedding of Graphs and Clustering",
"_____no_output_____"
],
[
"## Introduction",
"_____no_output_____"
],
[
"From social networks to parse trees, knowledge graphs to protein interaction networks, Graph-Structured Data is endemic to a wide variety of natural and engineered systems. Often, understanding the structure and/or dynamics of these graphs yields insight into the systems under investigation. Take, for example, the problems of finding key influencers or distinct communities within social networks. \n\nThe goal of graph embedding is to find a way of representing the graph in a space which more readily lends itself to analysis/investigation. One approach is to identify points in a vector space with nodes of the graph in such a way that important relations between nodes are preserved via relations between their corresponding points.\n\nThere are a wide variety of methods which approach this problem in different ways and for different aims, say for clustering or for link prediction. Recently, the embedding of Graph Structured Data (GSD) on manifolds has received considerable attention. In particular, much work has shown that hyperbolic spaces are beneficial for a wide variety of tasks with GSD [[ND2017]](#References). This tutorial shows how to learn such embeddings using the Poincaré Ball manifold and the well-known 'Karate Club' social network dataset with `geomstats`. This data and several others can be found in the `datasets.data` module of the project's github repository. ",
"_____no_output_____"
],
[
"\n*Learning a Poincaré disk embedding of the Karate club graph dataset*",
"_____no_output_____"
],
[
"## Setup\n\n\nWe start by importing standard tools for logging and visualization, allowing us to draw the embedding of the GSD on the manifold. Next, we import the manifold of interest, visualization tools, and other methods from `geomstats`.",
"_____no_output_____"
]
],
[
[
"import os\nimport sys\nimport warnings\n\nsys.path.append(os.path.dirname(os.getcwd()))\nwarnings.filterwarnings('ignore')",
"_____no_output_____"
],
[
"import logging\nimport matplotlib.pyplot as plt\n\nimport geomstats.backend as gs\nimport geomstats.visualization as visualization\n\nfrom geomstats.datasets.utils import load_karate_graph\nfrom geomstats.geometry.poincare_ball import PoincareBall",
"INFO: Using numpy backend\n"
]
],
[
[
"## Parameters and Initialization\n\nWe define the following parameters needed for embedding:\n\n| Parameter | Description |\n|:------|:------|\n| random.seed | An initial manually set number for generating pseudorandom numbers|\n| dim | Dimensions of the manifold used for embedding |\n|max_epochs|Number of iterations for learning the embedding |\n|lr| Learning rate|\n|n_negative| Number of negative samples|\n|context_size| Size of the considered context for each node of the graph|\n\nLet us discuss a few things about the parameters of the above table.\nThe number of dimensions should be high (i.e., 10+) for large datasets \n(i.e., where the number of nodes/edges is significantly large). \nIn this tutorial we consider a dataset that is quite small with only 34 nodes. The Poincaré disk of only two dimensions is therefore sufficient to\ncapture the complexity of the graph and provide a faithful representation.\nSome parameters are hard to know in advance, such as `max_epochs` and `lr`. These should be tuned specifically for each dataset.\nVisualization can help with tuning the parameters. Also, one can perform a grid search to find values of these parameters which maximize some performance function. In learning embeddings, one can consider performance metrics such as\na measure for cluster seperability or normalized mutual \ninformation (NMI) or others.\nSimilarly, the number of negative samples and context size can also be thought of as hyperparameters and will\nbe further discussed in the sequel. An instance of the `Graph` class is created\n and set to the Karate club dataset.",
"_____no_output_____"
]
],
[
[
"gs.random.seed(1234)\ndim = 2\nmax_epochs = 100\nlr = .05\nn_negative = 2\ncontext_size = 1\nkarate_graph = load_karate_graph()",
"_____no_output_____"
]
],
[
[
"The Zachary karate club network was collected from\nthe members of a university karate club by Wayne Zachary\nin 1977. Each node represents a member of the club,\nand each edge represents an undirected relation between\ntwo members. An often discussed problem using this dataset\nis to find the two groups of people into which the\nkarate club split after an argument between two teachers.\n<img src=\"figures/karate_graph.png\" width=\"60%\">\nSome information about the dataset is displayed to provide\ninsight into its complexity.",
"_____no_output_____"
]
],
[
[
"nb_vertices_by_edges =\\\n [len(e_2) for _, e_2 in karate_graph.edges.items()]\nlogging.info('Number of vertices: %s', len(karate_graph.edges))\nlogging.info(\n 'Mean edge-vertex ratio: %s',\n (sum(nb_vertices_by_edges, 0) / len(karate_graph.edges)))",
"INFO: Number of vertices: 34\nINFO: Mean edge-vertex ratio: 4.588235294117647\n"
]
],
[
[
"Denote $V$ as the set of nodes and $E \\subset V\\times V$ the set \nof edges. The goal of embedding GSD is to provide a faithful and exploitable representation \nof the graph structure. It is mainly achieved by preserving *first-order* proximity \nthat enforces nodes sharing edges to be close to each other. It can additionally \npreserve *second-order* proximity that enforces two nodes sharing the same context \n(i.e., nodes that share neighbors but are not necessarily directly connected) to be close. \nLet $\\mathbb{B}^m$ be the Poincaré Ball of dimension $m$ equipped with the distance function $d$.\nThe below figure shows geodesics between pairs of points on $\\mathbb{B}^2$. Geodesics are\nthe shortest path between two points. The distance function $d$ of two points is\nthe length of the geodesic that links them.\n\n<img src=\"figures/geodesics.png\" width=\"40%\">\n",
"_____no_output_____"
],
[
"Declaring an instance of the `PoincareBall` manifold of two dimensions in `geomstats` is straightforward:",
"_____no_output_____"
]
],
[
[
"hyperbolic_manifold = PoincareBall(2)",
"_____no_output_____"
]
],
[
[
"*first* and *second-order* proximities can be achieved by optimising the following loss functions:",
"_____no_output_____"
],
[
"## Loss function.\n\nTo preserve first and second-order proximities we adopt a loss function similar to (Nickel, 2017) and consider the negative sampling approach as in (Mikolov, 2013) :\n\n$$ \\mathcal{L} = - \\sum_{v_i\\in V} \\sum_{v_j \\in C_i} \\bigg[ log(\\sigma(-d^2(\\phi_i, \\phi_j'))) + \\sum_{v_k\\sim \\mathcal{P}_n} log(\\sigma(d^2(\\phi_i, \\phi_k'))) \\bigg]$$\n\nwhere $\\sigma(x)=\\frac{1}{1+e^{-x}}$ is the sigmoid function and $\\phi_i \\in \\mathbb{B}^m$ \nis the embedding of the $i$-th node of $V$, $C_i$ the nodes in the context of the \n$i$-th node, $\\phi_j'\\in \\mathbb{B}^m$ the embedding of $v_j\\in C_i$ and \n$\\mathcal{P}_n$ the negative sampling distribution over $V$: \n$\\mathcal{P}_n(v)=\\frac{deg(v)^{3/4}}{\\sum_{v_i\\in V}deg(v_i)^{3/4}}$. \nIntuitively one can see that to minimizing $L$, the distance between $v_i$ and $v_j$ should get smaller, while the one between\n$v_i$ and $v_k$ would get larger.\n<img src=\"figures/notations.png\" width=\"40%\">",
"_____no_output_____"
],
[
"## Riemannian optimization.\nFollowing the idea of (Ganea, 2018) we use the following formula to optimize $L$:\n\n$$ \\phi^{t+1} = \\text{Exp}_{\\phi^t} \\left( -lr \\frac{\\partial L}{\\partial \\phi} \\right) $$\n\nwhere $\\phi$ is a parameter of $L$, $t\\in\\{1,2,\\cdots\\}$ is the epoch iteration number \nand $lr$ is the learning rate. The formula consists of first computing the usual gradient of the loss function\ngiving the direction in which the parameter should move. \nThe Riemannian exponential map $\\text{Exp}$ is a function that takes a base point $\\phi^t$ and some \ndirection vector $T$ and returns the point $\\phi^{t+1}$ such that $\\phi^{t+1}$ belongs to the geodesic\ninitiated from $\\phi{t}$ in the direction of $T$ and the length of the geoedesic curve between $\\phi^t$ and $\\phi^{t+1}$ is of 1 unit. \nThe Riemannian exponential map is implemented as a method of the `PoincareBallMetric` class in the `geometry` module of `geomstats`.\n\nTherefore to minimize $L$ we will need to compute its gradient. Several steps are required to do so,\n1. Compute the gradient of the squared distance\n2. Compute the gradient of the log sigmoid\n3. Compute the gradient of the composision of 1. and 2.",
"_____no_output_____"
],
[
"For 1., we use the formula proposed by (Arnaudon, 2013) which uses the Riemannian logarithmic map to compute the gradient of the distance.\nThis is implemented as",
"_____no_output_____"
]
],
[
[
"def grad_squared_distance(point_a, point_b):\n \"\"\"Gradient of squared hyperbolic distance.\n\n Gradient of the squared distance based on the\n Ball representation according to point_a\n\n Parameters\n ----------\n point_a : array-like, shape=[n_samples, dim]\n First point in hyperbolic space.\n point_b : array-like, shape=[n_samples, dim]\n Second point in hyperbolic space.\n\n Returns\n -------\n dist : array-like, shape=[n_samples, 1]\n Geodesic squared distance between the two points.\n \"\"\"\n hyperbolic_metric = PoincareBall(2).metric\n log_map = hyperbolic_metric.log(point_b, point_a)\n\n return -2 * log_map",
"_____no_output_____"
]
],
[
[
"For 2. define the `log_sigmoid` corresponding as follows:",
"_____no_output_____"
]
],
[
[
"def log_sigmoid(vector):\n \"\"\"Logsigmoid function.\n\n Apply log sigmoid function\n\n Parameters\n ----------\n vector : array-like, shape=[n_samples, dim]\n\n Returns\n -------\n result : array-like, shape=[n_samples, dim]\n \"\"\"\n return gs.log((1 / (1 + gs.exp(-vector))))",
"_____no_output_____"
]
],
[
[
"The gradient of the logarithm of sigmoid function is implemented as:",
"_____no_output_____"
]
],
[
[
"def grad_log_sigmoid(vector):\n \"\"\"Gradient of log sigmoid function.\n\n Parameters\n ----------\n vector : array-like, shape=[n_samples, dim]\n\n Returns\n -------\n gradient : array-like, shape=[n_samples, dim]\n \"\"\"\n return 1 / (1 + gs.exp(vector))",
"_____no_output_____"
]
],
[
[
"For 3., apply the composition rule to obtain the gradient of $L$. The following function given $\\phi_i$, $\\phi'_j$ and $\\phi'_k$ returns the total value of $L$ and its gradient vector at $\\phi_i$. For the value of $L$ the loss function formula is simply applied. For the gradient, we apply the composition of `grad_log_sigmoid` with `grad_squared_distance` while paying attention to the signs.",
"_____no_output_____"
]
],
[
[
"def loss(example_embedding, context_embedding, negative_embedding,\n manifold):\n \"\"\"Compute loss and grad.\n\n Compute loss and grad given embedding of the current example,\n embedding of the context and negative sampling embedding.\n \"\"\"\n n_edges, dim =\\\n negative_embedding.shape[0], example_embedding.shape[-1]\n example_embedding = gs.expand_dims(example_embedding, 0)\n context_embedding = gs.expand_dims(context_embedding, 0)\n positive_distance =\\\n manifold.metric.squared_dist(\n example_embedding, context_embedding)\n positive_loss =\\\n log_sigmoid(-positive_distance)\n\n reshaped_example_embedding =\\\n gs.repeat(example_embedding, n_edges, axis=0)\n negative_distance =\\\n manifold.metric.squared_dist(\n reshaped_example_embedding, negative_embedding)\n negative_loss = log_sigmoid(negative_distance)\n\n total_loss = -(positive_loss + negative_loss.sum())\n\n positive_log_sigmoid_grad =\\\n -grad_log_sigmoid(-positive_distance)\n\n positive_distance_grad =\\\n grad_squared_distance(example_embedding, context_embedding)\n\n positive_grad =\\\n gs.repeat(positive_log_sigmoid_grad, dim, axis=-1)\\\n * positive_distance_grad\n\n negative_distance_grad =\\\n grad_squared_distance(reshaped_example_embedding, negative_embedding)\n\n negative_distance = gs.to_ndarray(negative_distance,\n to_ndim=2, axis=-1)\n negative_log_sigmoid_grad =\\\n grad_log_sigmoid(negative_distance)\n\n negative_grad = negative_log_sigmoid_grad\\\n * negative_distance_grad\n example_grad = -(positive_grad + negative_grad.sum(axis=0))\n\n return total_loss, example_grad",
"_____no_output_____"
]
],
[
[
"## Capturing the graph structure\nAt this point we have the necessary bricks to compute the resulting gradient of $L$. We are ready to prepare\nthe nodes $v_i$, $v_j$ and $v_k$ and initialise their embeddings $\\phi_i$, $\\phi^{'}_j$ and $\\phi^{'}_k$.\nFirst, initialize an array that will hold embeddings $\\phi_i$ of each node $v_i\\in V$ with random points belonging to the Poincaré disk. ",
"_____no_output_____"
]
],
[
[
"embeddings = gs.random.normal(size=(karate_graph.n_nodes, dim))\nembeddings = embeddings * 0.2",
"_____no_output_____"
]
],
[
[
"Next, to prepare the context nodes $v_j$ for each node $v_i$, we compute random walks initialised from each $v_i$ \nup to some length (5 by default). The latter is done via a special function within the `Graph` class. The nodes $v_j$ will be later picked from the random walk of $v_i$.\n\n",
"_____no_output_____"
]
],
[
[
"random_walks = karate_graph.random_walk()",
"_____no_output_____"
]
],
[
[
"Negatively sampled nodes $v_k$ are chosen according to the previously defined probability distribution function\n$\\mathcal{P}_n(v_k)$ implemented as",
"_____no_output_____"
]
],
[
[
"negative_table_parameter = 5\nnegative_sampling_table = []\n\nfor i, nb_v in enumerate(nb_vertices_by_edges):\n negative_sampling_table +=\\\n ([i] * int((nb_v**(3. / 4.))) * negative_table_parameter)\n\nnegative_sampling_table = gs.array(negative_sampling_table)",
"_____no_output_____"
]
],
[
[
"## Numerically optimizing the loss function\nOptimising the loss function is performed numerically over the number of epochs. \nAt each iteration, we will compute the gradient of $L$. Then the graph nodes are moved in the direction\npointed by the gradient. The movement of the nodes is performed by following geodesics in the gradient direction.\nThe key to obtain an embedding representing accurately the dataset, is to move the nodes smoothly rather\nthan brutal movements. This is done by tuning the learning rate, such as at each epoch\nall the nodes made small movements.",
"_____no_output_____"
],
[
"A *first level* loop iterates over the epochs, the table `total_loss` will record the value of $L$ at each iteration\nand help us track the minimization of $L$.",
"_____no_output_____"
],
[
"A *second level* nested loop iterates over each path in the previously computed random walks. Observing these walks, notice that nodes having \nmany edges appear more often. Such nodes\ncan be considered as important crossroads and will therefore be subject to a greater number of embedding updates. \nThis is one of the main reasons why random walks have proven to be effective\nin capturing the structure of graphs. The context of each $v_i$ will be the set of nodes $v_j$ belonging \nto the random walk from $v_i$. The `context_size` specified earlier will limit the length of the walk to be considered. Similarly, we use\nthe same `context_size` to limit the number of negative samples. We find $\\phi_i$ from the `embeddings` array.\n",
"_____no_output_____"
],
[
"\nA *third level* nested loop will iterate on each $v_j$ and $v_k$. From within, we find $\\phi'_j$ and $\\phi'_k$ then call the `loss` function to compute the gradient.\nThen the Riemannian exponential map is applied to find the new value of $\\phi_i$ as we mentioned before.",
"_____no_output_____"
]
],
[
[
"for epoch in range(max_epochs):\n total_loss = []\n for path in random_walks:\n\n for example_index, one_path in enumerate(path):\n context_index = path[max(0, example_index - context_size):\n min(example_index + context_size,\n len(path))]\n negative_index =\\\n gs.random.randint(negative_sampling_table.shape[0],\n size=(len(context_index),\n n_negative))\n negative_index = negative_sampling_table[negative_index]\n\n example_embedding = embeddings[one_path]\n for one_context_i, one_negative_i in zip(context_index,\n negative_index):\n context_embedding = embeddings[one_context_i]\n negative_embedding = embeddings[one_negative_i]\n l, g_ex = loss(\n example_embedding,\n context_embedding,\n negative_embedding,\n hyperbolic_manifold)\n total_loss.append(l)\n\n example_to_update = embeddings[one_path]\n embeddings[one_path] = hyperbolic_manifold.metric.exp(\n -lr * g_ex, example_to_update)\n logging.info(\n 'iteration %d loss_value %f',\n epoch, sum(total_loss, 0) / len(total_loss))",
"INFO: iteration 0 loss_value 1.826876\nINFO: iteration 1 loss_value 1.774560\nINFO: iteration 2 loss_value 1.725700\nINFO: iteration 3 loss_value 1.663358\nINFO: iteration 4 loss_value 1.655706\nINFO: iteration 5 loss_value 1.615405\nINFO: iteration 6 loss_value 1.581097\nINFO: iteration 7 loss_value 1.526418\nINFO: iteration 8 loss_value 1.507913\nINFO: iteration 9 loss_value 1.505934\nINFO: iteration 10 loss_value 1.466526\nINFO: iteration 11 loss_value 1.453769\nINFO: iteration 12 loss_value 1.443878\nINFO: iteration 13 loss_value 1.451272\nINFO: iteration 14 loss_value 1.397864\nINFO: iteration 15 loss_value 1.396170\nINFO: iteration 16 loss_value 1.373677\nINFO: iteration 17 loss_value 1.390120\nINFO: iteration 18 loss_value 1.382397\nINFO: iteration 19 loss_value 1.404103\nINFO: iteration 20 loss_value 1.395782\nINFO: iteration 21 loss_value 1.389617\nINFO: iteration 22 loss_value 1.410152\nINFO: iteration 23 loss_value 1.390600\nINFO: iteration 24 loss_value 1.374832\nINFO: iteration 25 loss_value 1.367194\nINFO: iteration 26 loss_value 1.323190\nINFO: iteration 27 loss_value 1.389616\nINFO: iteration 28 loss_value 1.361034\nINFO: iteration 29 loss_value 1.384930\nINFO: iteration 30 loss_value 1.340814\nINFO: iteration 31 loss_value 1.349682\nINFO: iteration 32 loss_value 1.317423\nINFO: iteration 33 loss_value 1.346869\nINFO: iteration 34 loss_value 1.327198\nINFO: iteration 35 loss_value 1.363809\nINFO: iteration 36 loss_value 1.352347\nINFO: iteration 37 loss_value 1.317670\nINFO: iteration 38 loss_value 1.320039\nINFO: iteration 39 loss_value 1.323888\nINFO: iteration 40 loss_value 1.341444\nINFO: iteration 41 loss_value 1.312259\nINFO: iteration 42 loss_value 1.315983\nINFO: iteration 43 loss_value 1.305483\nINFO: iteration 44 loss_value 1.325384\nINFO: iteration 45 loss_value 1.328024\nINFO: iteration 46 loss_value 1.306958\nINFO: iteration 47 loss_value 1.303357\nINFO: iteration 48 loss_value 1.303790\nINFO: iteration 49 loss_value 1.324749\nINFO: iteration 50 loss_value 1.328376\nINFO: iteration 51 loss_value 1.313816\nINFO: iteration 52 loss_value 1.325978\nINFO: iteration 53 loss_value 1.317516\nINFO: iteration 54 loss_value 1.353495\nINFO: iteration 55 loss_value 1.331988\nINFO: iteration 56 loss_value 1.346874\nINFO: iteration 57 loss_value 1.348946\nINFO: iteration 58 loss_value 1.324719\nINFO: iteration 59 loss_value 1.330355\nINFO: iteration 60 loss_value 1.331077\nINFO: iteration 61 loss_value 1.305729\nINFO: iteration 62 loss_value 1.311746\nINFO: iteration 63 loss_value 1.347637\nINFO: iteration 64 loss_value 1.326300\nINFO: iteration 65 loss_value 1.309570\nINFO: iteration 66 loss_value 1.313999\nINFO: iteration 67 loss_value 1.346287\nINFO: iteration 68 loss_value 1.300901\nINFO: iteration 69 loss_value 1.323723\nINFO: iteration 70 loss_value 1.320784\nINFO: iteration 71 loss_value 1.313709\nINFO: iteration 72 loss_value 1.312143\nINFO: iteration 73 loss_value 1.309172\nINFO: iteration 74 loss_value 1.320642\nINFO: iteration 75 loss_value 1.308333\nINFO: iteration 76 loss_value 1.325884\nINFO: iteration 77 loss_value 1.316740\nINFO: iteration 78 loss_value 1.325933\nINFO: iteration 79 loss_value 1.316672\nINFO: iteration 80 loss_value 1.312291\nINFO: iteration 81 loss_value 1.332372\nINFO: iteration 82 loss_value 1.317499\nINFO: iteration 83 loss_value 1.329194\nINFO: iteration 84 loss_value 1.305926\nINFO: iteration 85 loss_value 1.304747\nINFO: iteration 86 loss_value 1.342343\nINFO: iteration 87 loss_value 1.331992\nINFO: iteration 88 loss_value 1.295439\nINFO: iteration 89 loss_value 1.332853\nINFO: iteration 90 loss_value 1.332004\nINFO: iteration 91 loss_value 1.357248\nINFO: iteration 92 loss_value 1.342234\nINFO: iteration 93 loss_value 1.329379\nINFO: iteration 94 loss_value 1.313617\nINFO: iteration 95 loss_value 1.310320\nINFO: iteration 96 loss_value 1.320590\nINFO: iteration 97 loss_value 1.315822\nINFO: iteration 98 loss_value 1.328819\nINFO: iteration 99 loss_value 1.339718\n"
]
],
[
[
"## Plotting results\nOnce the `max_epochs` iterations of epochs is achieved, we can plot the resulting `embeddings` array and the true labels shown\nas two colors. At 100 epochs we can see that the two group of nodes with different labels are moving away from each other\non the manifold. If one increases the `max_epochs`, then further separability is achieved.",
"_____no_output_____"
]
],
[
[
"import matplotlib.patches as mpatches\n\ncolors = {1: 'b', 2: 'r'}\ngroup_1 = mpatches.Patch(color=colors[1], label='Group 1')\ngroup_2 = mpatches.Patch(color=colors[2], label='Group 2')\n\ncircle = visualization.PoincareDisk(point_type='ball')\n\nfig, ax = plt.subplots(figsize=(8, 8))\nax.axes.xaxis.set_visible(False)\nax.axes.yaxis.set_visible(False)\ncircle.set_ax(ax)\ncircle.draw(ax=ax)\nfor i_embedding, embedding in enumerate(embeddings):\n x = embedding[0]\n y = embedding[1]\n pt_id = i_embedding\n plt.scatter(\n x, y,\n c=colors[karate_graph.labels[pt_id][0]],\n s = 150\n )\n ax.annotate(pt_id, (x,y))\n\nplt.tick_params(\nwhich='both')\nplt.title('Poincare Ball Embedding of the Karate Club Network')\nplt.legend(handles=[group_1, group_2])\nplt.show()",
"_____no_output_____"
]
],
[
[
"In `geomstats`, several unsupervized clustering algorithms on manifolds are implemented such as $K$-means and Expectation-Maximization. \n\nLet us apply $K$-means to learn the node belonging of the two groups and see how well we predicted the true\nlabels.\nLets first import $K$-means",
"_____no_output_____"
]
],
[
[
"from geomstats.learning.kmeans import RiemannianKMeans",
"_____no_output_____"
]
],
[
[
"Set the number of groups to 2.",
"_____no_output_____"
]
],
[
[
"n_clusters = 2",
"_____no_output_____"
]
],
[
[
"Initialize an instance of $K$-means.",
"_____no_output_____"
]
],
[
[
"kmeans = RiemannianKMeans(metric= hyperbolic_manifold.metric,\n n_clusters=n_clusters,\n init='random',\n mean_method='frechet-poincare-ball'\n )",
"_____no_output_____"
]
],
[
[
"Fit the embedded nodes",
"_____no_output_____"
]
],
[
[
"centroids = kmeans.fit(X=embeddings, max_iter=100)\nlabels = kmeans.predict(X=embeddings)",
"_____no_output_____"
]
],
[
[
"And plot the resulting labels provided by $K$-means",
"_____no_output_____"
]
],
[
[
"colors = ['g', 'c', 'm']\ncircle = visualization.PoincareDisk(point_type='ball')\nfig2, ax2 = plt.subplots(figsize=(8, 8))\ncircle.set_ax(ax2)\ncircle.draw(ax=ax2)\nax2.axes.xaxis.set_visible(False)\nax2.axes.yaxis.set_visible(False)\ngroup_1_predicted = mpatches.Patch(color=colors[0], label='Predicted Group 1')\ngroup_2_predicted = mpatches.Patch(color=colors[1], label='Predicted Group 2')\ngroup_centroids = mpatches.Patch(color=colors[2], label='Cluster centroids')\n\nfor i in range(n_clusters):\n for i_embedding, embedding in enumerate(embeddings):\n x = embedding[0]\n y = embedding[1]\n pt_id = i_embedding\n if labels[i_embedding] == 0:\n color = colors[0]\n else:\n color = colors[1]\n plt.scatter(\n x, y,\n c=color,\n s = 150\n )\n ax2.annotate(pt_id, (x,y))\n\nfor i_centroid, centroid in enumerate(centroids): \n x = centroid[0]\n y = centroid[1]\n plt.scatter(\n x, y,\n c=colors[2],\n marker='*',\n s = 150,\n )\n\nplt.title('K-means applied to Karate club embedding')\nplt.legend(handles = [group_1_predicted, group_2_predicted, group_centroids])\nplt.show()",
"_____no_output_____"
]
],
[
[
"By comparing the $K$-means labels and the true labels, notice how $K$-means \naccurately finds the two groups of nodes (not perfectly, e.g., nodes 2 and 8). We therefore achieved good performances in\npredicting the belonging of each member of the Karate club to one of the two groups.",
"_____no_output_____"
],
[
"## References",
"_____no_output_____"
],
[
".. [ABY2013] Arnaudon, Marc, Frédéric Barbaresco, and Le Yang. \"Riemannian medians and means with applications to radar signal processing.\" IEEE Journal of Selected Topics in Signal Processing 7.4 (2013): 595-604.\n\n.. [GBH2018] Ganea, Octavian, Gary Bécigneul, and Thomas Hofmann. \"Hyperbolic neural networks.\" Advances in neural information processing systems. 2018.\n\n.. [M2013] Mikolov, Tomas, et al. \"Distributed representations of words and phrases and their compositionality.\" Advances in neural information processing systems. 2013.\n\n.. [ND2017] Nickel, Maximillian, and Douwe Kiela. \"Poincaré embeddings for learning hierarchical representations.\" Advances in neural information processing systems. 2017.\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
]
]
|
ec73d650446749befe862f60b9007759cfac0025 | 20,334 | ipynb | Jupyter Notebook | Letter Recognition Dataset Analysis.ipynb | sashah12/COGS-118A-Final | 2121eae54add34ded69f002d12b9fb1e19a443e0 | [
"MIT"
]
| null | null | null | Letter Recognition Dataset Analysis.ipynb | sashah12/COGS-118A-Final | 2121eae54add34ded69f002d12b9fb1e19a443e0 | [
"MIT"
]
| null | null | null | Letter Recognition Dataset Analysis.ipynb | sashah12/COGS-118A-Final | 2121eae54add34ded69f002d12b9fb1e19a443e0 | [
"MIT"
]
| null | null | null | 49.354369 | 334 | 0.627717 | [
[
[
"import numpy as np\nfrom sklearn import preprocessing\nfrom sklearn.model_selection import GridSearchCV\nfrom sklearn import svm\nfrom sklearn.model_selection import train_test_split\nfrom sklearn.neighbors import KNeighborsClassifier, KernelDensity\nfrom sklearn.ensemble import RandomForestClassifier\nfrom sklearn.utils import shuffle\nimport pandas as pd",
"_____no_output_____"
],
[
"#Load dataset into Pandas Dataframe\ndf3 = pd.read_csv('letter-recognition.data')",
"_____no_output_____"
]
],
[
[
"# Letter Dataset P.2",
"_____no_output_____"
]
],
[
[
"#Rename Column\nletter_df = df3\nletter_df = letter_df.rename(columns={'T': 'Letter'})\n#Initialize Column\nletter_df['Class'] = 0\nletters = ['A', 'B', 'C', 'D', 'E', 'F', 'G', 'H', 'I', 'J', 'K', 'L', 'M']\n#For each row in the dataframe, if the alphabet letter in that row is not in the list above,\n# set the class of that data point to 0. Otherwise, it's 1.\nfor index, row in letter_df.iterrows():\n if row['Letter'] in letters:\n letter_df.at[index, 'Class'] = 1\nletter_df = letter_df.drop(['Letter'], axis = 1)\n#Converting dataframe into array.\nletter_arr = letter_df.to_numpy()",
"_____no_output_____"
],
[
"#Accuracy function. Compares predicted labels vs actual labels and counts accuracy.\ndef prediction(predictions, Y_given):\n wrong = 0\n counter = 0\n for test, train in zip(predictions, Y_given):\n if test == train:\n wrong = wrong\n else:\n wrong = wrong + 1\n counter = counter + 1\n accuracy = 1 - (wrong/counter)\n return accuracy",
"_____no_output_____"
]
],
[
[
"# SVM",
"_____no_output_____"
]
],
[
[
"#Three lists to store training accuracy for each trial, test accuracy for each trial, and best parameters for each trial.\nSVM_train_accuracy = []\nSVM_test_accuracy = []\nSVM_best_params = []\n\n#For each Trial\nfor i in range(3):\n data_svm = shuffle(letter_arr)\n#Data splitting, train-test-split.\n X_svm = data_svm[:, 0:-1]\n Y_svm = data_svm[:, -1]\n X_train_svm, X_test_svm, Y_train_svm, Y_test_svm = train_test_split(X_svm, Y_svm, test_size = 14999/19999, random_state=42)\n#Scaling training data using StandardScaler\n scaler_svm = preprocessing.StandardScaler().fit(X_train_svm)\n X_train_svm = scaler_svm.transform(X_train_svm)\n#Param-grid, setting SVM class,grid search parameters, and fitting training data.\n parameters = [{'kernel': ['rbf'], 'gamma': [0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1.0, 2.0], 'C': [10**-7, 10**-6, 10**-5,\n 10**-4, 10**-3, 10**-2,\n 10**-1]},\n {'kernel': ['poly'], 'degree': [2, 3], 'C': [10**-7, 10**-6, 10**-5, 10**-4, 10**-3, 10**-2, 10**-1]},\n {'kernel': ['linear'], 'C': [10**-7, 10**-6, 10**-5, 10**-4, 10**-3, 10**-2, 10**-1]}]\n svc = svm.SVC(gamma = 'auto')\n grid_search_svm = GridSearchCV(svc, parameters, cv = 5, error_score = np.nan)\n grid_search2_svm = grid_search_svm.fit(X_train_svm, Y_train_svm)\n best_params_svm = grid_search2_svm.best_params_\n#Printing and storing best params into list.\n print(\"Best params: {}\".format(best_params_svm))\n SVM_best_params.append('Trial ' + str(i + 1) + ': ' + str(best_params_svm))\n#Using best_estimator to predict training data labels.\n#Using prediction function to calculate training accuracy.\n#Printing and storing training accuracies into list.\n train_predictions_svm = grid_search2_svm.best_estimator_.predict(X_train_svm) #.best_estimator_\n train_accuracy_svm = prediction(train_predictions_svm, Y_train_svm)\n print(\"Train accuracy: {}\".format(train_accuracy_svm))\n SVM_train_accuracy.append('Trial ' + str(i + 1) + ': ' + str(train_accuracy_svm))\n#Scaling testing data with my training data scaler. This ensures training and testing data are scaled the same.\n#Using prediction function to calculate testing accuracy.\n#Printing and storing test accuracy into list.\n X_test_svm = scaler_svm.transform(X_test_svm)\n test_predictions_svm = grid_search2_svm.best_estimator_.predict(X_test_svm) #.best_estimator_\n test_accuracy_svm = prediction(test_predictions_svm, Y_test_svm)\n print(\"Test accuracy: {}\".format(test_accuracy_svm))\n SVM_test_accuracy.append('Trial ' + str(i + 1) + ': ' + str(test_accuracy_svm))",
"Best params: {'C': 0.1, 'gamma': 0.5, 'kernel': 'rbf'}\nTrain accuracy: 0.9566\nTest accuracy: 0.8923261550770052\nBest params: {'C': 0.1, 'gamma': 0.5, 'kernel': 'rbf'}\nTrain accuracy: 0.9672\nTest accuracy: 0.9193279551970132\nBest params: {'C': 0.1, 'gamma': 0.5, 'kernel': 'rbf'}\nTrain accuracy: 0.9502\nTest accuracy: 0.8862590839389293\n"
]
],
[
[
"# kNN",
"_____no_output_____"
]
],
[
[
"#Three lists to store training accuracy for each trial, test accuracy for each trial, and best parameters for each trial.\nkNN_train_accuracy = []\nkNN_test_accuracy = []\nkNN_best_params = []\n\n#Setting K parameters\np = (np.linspace(1,500,25))\np = p.astype('int64')\n\n#For each trial\nfor i in range(3):\n data_knn = shuffle(letter_arr)\n#Data splitting, train-test-split\n X_knn = data_knn[:, 0:-1]\n Y_knn = data_knn[:, -1]\n X_train_knn, X_test_knn, Y_train_knn, Y_test_knn = train_test_split(X_knn, Y_knn, test_size = 14999/19999, random_state=42)\n# Scaling training data using StandardScaler()\n scaler_knn = preprocessing.StandardScaler().fit(X_train_knn)\n X_train_knn = scaler_knn.transform(X_train_knn)\n#Param-grid, initializing kNN class, grid search parameters and fitting training data.\n# Printing and storing best parameters in list.\n params = [{'weights' : ['uniform', 'distance'], 'metric' : ['minkowski'],'n_neighbors': p}]\n neighbor = KNeighborsClassifier()\n grid_search_knn = GridSearchCV(neighbor, params, cv=5, error_score = np.nan)\n grid_search_knn2 = grid_search_knn.fit(X_train_knn, Y_train_knn)\n best_params_knn = grid_search_knn2.best_params_\n print(\"Best params: {}\".format(best_params_knn))\n kNN_best_params.append('Trial ' + str(i + 1) + ': ' + str(best_params_knn))\n#Using best_estimator to predict training data labels.\n#Using prediction function to calculate training accuracy\n#Printing and storing training accuracy into list.\n train_predictions_knn = grid_search_knn2.best_estimator_.predict(X_train_knn)\n train_accuracy_knn = prediction(train_predictions_knn, Y_train_knn)\n print(\"Train accuracy: {}\".format(train_accuracy_knn))\n kNN_train_accuracy.append('Trial ' + str(i + 1) + ': ' + str(train_accuracy_knn))\n#Scaling testing data with my training data scaler. This ensures training and testing data are scaled the same.\n#Using prediction function to calculate testing accuracy.\n#Printing and storing test accuracy into list.\n X_test_knn = scaler_knn.transform(X_test_knn)\n test_predictions_knn = grid_search_knn2.best_estimator_.predict(X_test_knn)\n test_accuracy_knn = prediction(test_predictions_knn, Y_test_knn)\n print(\"Test accuracy: {}\".format(test_accuracy_knn))\n kNN_test_accuracy.append('Trial ' + str(i + 1) + ': ' + str(test_accuracy_knn))",
"Best params: {'metric': 'minkowski', 'n_neighbors': 1, 'weights': 'uniform'}\nTrain accuracy: 1.0\nTest accuracy: 0.950530035335689\nBest params: {'metric': 'minkowski', 'n_neighbors': 1, 'weights': 'uniform'}\nTrain accuracy: 1.0\nTest accuracy: 0.9535302353490233\nBest params: {'metric': 'minkowski', 'n_neighbors': 1, 'weights': 'uniform'}\nTrain accuracy: 1.0\nTest accuracy: 0.9537302486832455\n"
]
],
[
[
"# Random Forest",
"_____no_output_____"
]
],
[
[
"#Three lists to store training accuracy for each trial, test accuracy for each trial, and best parameters for each trial.\nrf_train_accuracy = []\nrf_test_accuracy = []\nrf_best_params = []\n\n#For each trial\nfor i in range(3):\n data_rf = shuffle(letter_arr)\n#Data splitting, train-test-split\n X_rf = data_rf[:, 0:-1]\n Y_rf = data_rf[:, -1]\n X_train_rf, X_test_rf, Y_train_rf, Y_test_rf = train_test_split(X_rf, Y_rf, test_size = 14999/19999, random_state=42)\n#Param-grid, initialize random forest class, and fitting training data with grid search.\n params = [{'n_estimators' : [1024], 'max_features' : [1, 2, 4, 6, 8, 12, 16, 20]}]\n forest = RandomForestClassifier()\n grid_search_rf = GridSearchCV(forest, params, cv=5, error_score = np.nan)\n grid_search_rf2 = grid_search_rf.fit(X_train_rf, Y_train_rf)\n#Printing and storing best params into list.\n best_params_rf = grid_search_rf2.best_params_\n print(\"Best params: {}\".format(best_params_rf))\n rf_best_params.append('Trial ' + str(i + 1) + ': ' + str(best_params_rf))\n#Using best_estimator to predict training data.\n#Storing and printing training accuracy into list.\n#Using defined prediction function to calculate total accuracy.\n train_predictions_rf = grid_search_rf2.best_estimator_.predict(X_train_rf)\n train_accuracy_rf = prediction(train_predictions_rf, Y_train_rf)\n print(\"Train accuracy: {}\".format(train_accuracy_rf))\n rf_train_accuracy.append('Trial ' + str(i + 1) + ': ' + str(train_accuracy_rf))\n#Predicting Test data with best_estimator.predict.\n#Using prediction function to calculate accuracy.\n#Printing and storing Test accuracy into list.\n test_predictions_rf = grid_search_rf2.best_estimator_.predict(X_test_rf)\n test_accuracy_rf = prediction(test_predictions_rf, Y_test_rf)\n print(\"Test accuracy: {}\".format(test_accuracy_rf))\n rf_test_accuracy.append('Trial ' + str(i + 1) + ': ' + str(test_accuracy_rf))",
"c:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:545: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details: \nValueError: max_features must be in (0, n_features]\n\n FitFailedWarning)\nc:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:545: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details: \nValueError: max_features must be in (0, n_features]\n\n FitFailedWarning)\nc:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:545: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details: \nValueError: max_features must be in (0, n_features]\n\n FitFailedWarning)\nc:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:545: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details: \nValueError: max_features must be in (0, n_features]\n\n FitFailedWarning)\nc:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_validation.py:545: FitFailedWarning: Estimator fit failed. The score on this train-test partition for these parameters will be set to nan. Details: \nValueError: max_features must be in (0, n_features]\n\n FitFailedWarning)\nc:\\users\\zingb\\appdata\\local\\programs\\python\\python37-32\\lib\\site-packages\\sklearn\\model_selection\\_search.py:813: DeprecationWarning: The default of the `iid` parameter will change from True to False in version 0.22 and will be removed in 0.24. This will change numeric results when test-set sizes are unequal.\n DeprecationWarning)\n"
]
]
]
| [
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec73d86d5989a9c4b1971d8ae18273b6e09094ee | 4,826 | ipynb | Jupyter Notebook | Quantification/.ipynb_checkpoints/DNA_quant_four_plates-checkpoint.ipynb | spencer-siddons/Moeller_Opentrons_protocol_library | 28190dd5793ff891cf467112512e02c89c2e5761 | [
"MIT"
]
| 9 | 2020-07-16T13:48:35.000Z | 2022-01-31T14:12:31.000Z | Quantification/.ipynb_checkpoints/DNA_quant_four_plates-checkpoint.ipynb | spencer-siddons/Moeller_Opentrons_protocol_library | 28190dd5793ff891cf467112512e02c89c2e5761 | [
"MIT"
]
| null | null | null | Quantification/.ipynb_checkpoints/DNA_quant_four_plates-checkpoint.ipynb | spencer-siddons/Moeller_Opentrons_protocol_library | 28190dd5793ff891cf467112512e02c89c2e5761 | [
"MIT"
]
| 9 | 2020-05-20T19:11:24.000Z | 2021-07-17T22:27:45.000Z | 35.485294 | 105 | 0.451513 | [
[
[
"from opentrons import protocol_api\nfrom opentrons.protocols.types import APIVersion\n\nmetadata = {\n 'apiLevel': '2.2',\n 'author': 'Jon Sanders'}\n\n\napi_version = APIVersion(2, 2)\n\ndef get_96_from_384_wells(method='interleaved', start=1):\n \n if method == 'interleaved':\n rows = [chr(64 + x*2 - (start % 2)) for x in range(1,9)]\n cols = [x*2 - int((start+1) / 2)%2 for x in range(1,13)]\n \n for col in cols:\n for row in rows:\n yield('%s%s' % (row, col))\n \n if method == 'packed':\n for col in range((start-1)*6+1, (start-1)*6 + 7):\n for row in [chr(x+65) for x in range(0,16,2)]:\n yield('%s%s' % (row,col))\n for col in range((start-1)*6+1, (start-1)*6 + 7):\n for row in [chr(x+65) for x in range(1,17,2)]:\n yield('%s%s' % (row,col))\n\ndef run(protocol: protocol_api.ProtocolContext(api_version=api_version)):\n \n # define deck positions and labware\n \n # tips\n tiprack_300 = protocol.load_labware('opentrons_96_tiprack_300ul', 6)\n \n tipracks_10f = [protocol.load_labware('opentrons_96_filtertiprack_10ul', x)\n for x in [1, 4, 7, 10]]\n \n # plates\n reagents = protocol.load_labware('nest_12_reservoir_15ml', 3, 'reagents')\n assay = protocol.load_labware('corning_384_wellplate_112ul_flat', 9, 'assay')\n \n samples = [protocol.load_labware('biorad_96_wellplate_200ul_pcr', x, 'samples')\n for x in [2, 5, 8, 11]]\n \n # initialize pipettes\n pipette_left = protocol.load_instrument('p300_multi', \n 'left',\n tip_racks=[tiprack_300])\n\n pipette_right = protocol.load_instrument('p10_multi', \n 'right',\n tip_racks=tipracks_10f)\n \n \n # # home instrument\n protocol.home()\n \n # distribute 38 µL of quantification reagent into each well of the assay plate. \n # Use the same tip for the entirety of these transfers, then replace it in the rack.\n \n pipette_left.distribute(38,\n [reagents.wells_by_name()['A1'],\n reagents.wells_by_name()['A2']],\n assay.wells(),\n disposal_volume=10,\n trash=False,\n blow_out=False,\n new_tip='once')\n \n # add 2 µL of each sample to each of the wells. Mix after dispensing. Dispose of these tips.\n \n for i, plate in enumerate(samples):\n start = i + 1\n assay_wells = get_96_from_384_wells(method='interleaved', start=start)\n pipette_right.transfer(2, \n plate.wells(), \n [assay[x] for x in assay_wells],\n mix_after=(1, 10),\n touch_tip=True,\n trash=True,\n new_tip='always')\n \n \n\n\n",
"/Users/jgs286/.opentrons/deck_calibration.json not found. Loading defaults\n/Users/jgs286/.opentrons/robot_settings.json not found. Loading defaults\n"
]
]
]
| [
"code"
]
| [
[
"code"
]
]
|
ec73dc56d165ef91840918e49eef739e7cc14bb6 | 21,726 | ipynb | Jupyter Notebook | physics/classical/Lab1/Model2NB.ipynb | soundofives/hpu-classes | 8c690b64e2de1ff7ffadf7aa9aabbfe634cf36a8 | [
"MIT"
]
| null | null | null | physics/classical/Lab1/Model2NB.ipynb | soundofives/hpu-classes | 8c690b64e2de1ff7ffadf7aa9aabbfe634cf36a8 | [
"MIT"
]
| null | null | null | physics/classical/Lab1/Model2NB.ipynb | soundofives/hpu-classes | 8c690b64e2de1ff7ffadf7aa9aabbfe634cf36a8 | [
"MIT"
]
| null | null | null | 142.934211 | 18,160 | 0.88493 | [
[
[
"from __future__ import division, print_function\nimport numpy as np\nimport matplotlib.pyplot as plt",
"_____no_output_____"
],
[
"M = 1 #\"big\" particle mass\nm = 0.0001 #mediating particle mass\n#c = 1 #speed of mediating particle\nro = 1 #initial separation\nvo = 0 #initial speed (of massive particles)\n\nN = 50 #number of collisions to track\n #WARNING: only finite number actually occur\n #so this will break down if N chosen too large",
"_____no_output_____"
],
[
"t = np.zeros(N) #times collision cycles begin\nv = np.zeros(N) #particle speeds at start of collision cycle\nr = np.zeros(N) #spatial separation at start of collision cycle \nc = np.zeros(N)\n\nr[0] = ro #set initial values in arrays\nv[0] = vo\nt[0] = 0\nc[0] = 1",
"_____no_output_____"
],
[
"i = 0\nwhile i<N-1:\n c[i+1] = ((-2*M)/(M+m))*v[i] + ((M-m)/(M+m))*c[i]\n v[i+1] = ((M-m)/(M+m))*v[i] + (2*m/(M+m))*c[i]\n t[i+1] = t[i] + r[i]/(2*(c[i]-v[i]))*(1+(c[i]/c[i+1]))\n r[i+1] = r[i] + (v[i]*r[i]/(c[i]-v[i]))+(c[i]/c[i+1])*(r[i]*v[i+1]/(c[i]-v[i]))\n \n \n i=i+1",
"_____no_output_____"
],
[
"#Acceleration obtained by finite-difference\na = (v[1:N]-v[0:N-1])/(t[1:N]-t[0:N-1])\n\n#Acceleration applies to \"midpoint\" so to plot vs r we need the r\n#values mid-collision cycle\nra = r[0:N-1] + 0.5*(r[1:N]-r[0:N-1])",
"_____no_output_____"
],
[
"plt.plot(np.log(ra),np.log(a))\nplt.xlabel('log(r)')\nplt.ylabel('log(a)')\nplt.title('acceleration vs position midpoints')\nplt.show()\n",
"_____no_output_____"
],
[
"#linear curve fit p[0] is slope, p[1] is y-int\np = np.polyfit(np.log(ra),np.log(a),1)\nprint(p[0])",
"-2.999323116169414\n"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code",
"code",
"code",
"code",
"code"
]
]
|
ec73e106fd35c8e2764a2f930832caf57b5bdc17 | 13,930 | ipynb | Jupyter Notebook | containers/jupyter-datascience-notebooks/test-project/test-r.ipynb | Jkumarraj/vscode-dev-containers | d768e00477585be01341e42601b4edc0017cbb35 | [
"MIT"
]
| 3,087 | 2019-05-06T22:47:37.000Z | 2022-03-31T15:10:05.000Z | containers/jupyter-datascience-notebooks/test-project/test-r.ipynb | Jkumarraj/vscode-dev-containers | d768e00477585be01341e42601b4edc0017cbb35 | [
"MIT"
]
| 1,044 | 2019-05-06T21:09:19.000Z | 2022-03-31T21:13:48.000Z | containers/jupyter-datascience-notebooks/test-project/test-r.ipynb | Jkumarraj/vscode-dev-containers | d768e00477585be01341e42601b4edc0017cbb35 | [
"MIT"
]
| 1,275 | 2019-05-07T12:31:12.000Z | 2022-03-31T22:48:46.000Z | 180.909091 | 11,489 | 0.900072 | [
[
[
"runif(10)",
"_____no_output_____"
],
[
"plot(runif(10))",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code"
]
]
|
ec73e11b74dacd71806a266242542b5de44a9c2d | 6,661 | ipynb | Jupyter Notebook | examples/jdbc-rdbms-with-spark.ipynb | nmukerje/emr-studio-notebook-examples | e1136f8be37481c9c988a5dbf312658c8584b952 | [
"MIT-0"
]
| null | null | null | examples/jdbc-rdbms-with-spark.ipynb | nmukerje/emr-studio-notebook-examples | e1136f8be37481c9c988a5dbf312658c8584b952 | [
"MIT-0"
]
| null | null | null | examples/jdbc-rdbms-with-spark.ipynb | nmukerje/emr-studio-notebook-examples | e1136f8be37481c9c988a5dbf312658c8584b952 | [
"MIT-0"
]
| null | null | null | 26.967611 | 181 | 0.539709 | [
[
[
"# Connect RDBS using jdbc connector from EMR Studio Notebook using Pyspark, Spark Scala, and SparkR\n\n#### Topics covered in this example\n\n* Configuring jdbc driver\n* Connecting to database using jdbc to read data\n* Connecting to database using jdbc to write data",
"_____no_output_____"
],
[
"## Table of Contents:\n\n1. [Prerequisites](#Prerequisites)\n2. [Introduction](#Introduction)\n3. [Upload the MySQL jdbc driver in S3 and declare the path](#Upload-the-MySQL-jdbc-driver-in-S3-and-declare-the-path)\n4. [Read and write data using Pyspark](#Read-and-write-data-using-Pyspark)\n5. [Read and write data using Scala](#Read-and-write-data-using-Scala)\n6. [Read and write data using SparkR](#Read-and-write-data-using-SparkR)\n",
"_____no_output_____"
],
[
"## Prerequisites",
"_____no_output_____"
],
[
"Download jdbc driver and upload it on S3 which is accessible from the Amazon EMR cluster attached to the Amazon EMR Studio. ",
"_____no_output_____"
],
[
"## Introduction",
"_____no_output_____"
],
[
"This notebooks shows how to connect RDBS using jdbc connector from Amazon EMR Studio Notebook. ",
"_____no_output_____"
],
[
"## Upload the MySQL jdbc driver in S3 and declare the path",
"_____no_output_____"
]
],
[
[
"%%configure -f\n{\n \"conf\": {\n \"spark.jars\": \"s3://<S3 Bucket>/jars/mysql-connector-java-8.0.19.jar\" \n }\n}",
"_____no_output_____"
]
],
[
[
"## Read and write data using Pyspark",
"_____no_output_____"
]
],
[
[
"%%pyspark\n\n#Declare the variables and replace the variables values as appropiate\n\nstr_jdbc_url='jdbc:mysql://<Database endpoint>:3306/<Database name>'\nstr_Query= '<Select query>'\nstr_dbname='<Database name>'\nstr_username='<Username>'\nstr_password='<Password>'\nstr_tgt_table='<Target Table>'\n\n# Read data from source table\n\njdbcDF = spark.read \\\n .format(\"jdbc\") \\\n .option(\"url\", str_jdbc_url) \\\n .option(\"query\", str_Query) \\\n .option(\"user\", str_username) \\\n .option(\"password\", str_password) \\\n .load()\n\njdbcDF.limit(5).show()\n\n# Write data to the target database\n\njdbcDF.write \\\n .format(\"jdbc\") \\\n .option(\"url\", str_jdbc_url) \\\n .option(\"dbtable\", str_tgt_table) \\\n .option(\"user\", str_username) \\\n .option(\"password\", str_password).mode('append').save()\n",
"_____no_output_____"
]
],
[
[
"## Read and write data using Scala",
"_____no_output_____"
]
],
[
[
"%%scalaspark\n\n#Declare the variables and replace the variables values as appropiate\n\nval str_jdbc_url=\"jdbc:mysql://<Database endpoint>:3306/<Database name>\"\nval str_Query= \"<Select Query>\"\nval str_dbname=\"<Database name>\"\nval str_username=\"<Username>\"\nval str_password=\"<Password>\"\nval str_tgt_table=\"<Target table>\"\n\n# Read data from source table\n\nval jdbcDF = (spark.read.format(\"jdbc\")\n .option(\"url\", str_jdbc_url)\n .option(\"query\", str_Query)\n .option(\"user\", str_username)\n .option(\"password\", str_password)\n .load())\n\njdbcDF.limit(5).show()\n\n# Write data to the target database\n\nval connectionProperties = new java.util.Properties\nconnectionProperties.put(\"user\", str_username)\nconnectionProperties.put(\"password\", str_password)\n\njdbcDF.write.mode(\"append\").jdbc(str_jdbc_url, str_tgt_table, connectionProperties)\n",
"_____no_output_____"
]
],
[
[
"## Read and write data using SparkR",
"_____no_output_____"
]
],
[
[
"%%rspark\n\n#Declare the variables and replace the variables values as appropiate\n\nstr_jdbc_url=\"jdbc:mysql://<Database endpoint>:3306/<Database name>\"\nstr_dbname=\"<Database name>\"\nstr_username=\"<Username>\"\nstr_password=\"<Password>\"\nstr_tgt_table=\"<Target table>\"\n\n# Read data from source database\n\ndf <- read.jdbc(str_jdbc_url, \n \"(select employee_id, first_name, last_name, email, dept_name from notebook.employee e, notebook.dept d where e.department_id = d.department_id) AS tmp\", \n user = str_username, \n password = str_password)\n\nshowDF(df)\n\njdbcDF.limit(5).show()\n\n# Write data to the target database\n\nwrite.jdbc(df, \n str_jdbc_url, \n str_tgt_table,\n user = str_username,\n password = str_password,\n mode = \"append\")\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
]
]
|
ec73e4ec6a714ae2e81fa6e7cb33b5803f060ffb | 8,852 | ipynb | Jupyter Notebook | maps/index.ipynb | vikineema/deafrica-docs | 3e82b1dee56508fe31341263cd1ddb6857532715 | [
"Apache-2.0"
]
| null | null | null | maps/index.ipynb | vikineema/deafrica-docs | 3e82b1dee56508fe31341263cd1ddb6857532715 | [
"Apache-2.0"
]
| null | null | null | maps/index.ipynb | vikineema/deafrica-docs | 3e82b1dee56508fe31341263cd1ddb6857532715 | [
"Apache-2.0"
]
| null | null | null | 47.848649 | 348 | 0.660416 | [
[
[
"# DE Africa Map",
"_____no_output_____"
],
[
"This guide provides detailed instructions on how to use the different features of the [Digital Earth Africa Map](https://maps.digitalearth.africa/).",
"_____no_output_____"
],
[
"## Get Started\n\nTo launch the DE Africa Map and display some basic data follow these steps.\n\n\n* Launch the DE Africa Map at [maps.digitalearth.africa](https://maps.digitalearth.africa/).\n* In the left panel click the **Add Data** button to open the data catalogue (see image below).\n* Find a dataset of interest such as water.\n* Select the dataset to see a preview of that data and description.\n* To view your selected dataset on the map, click the **Add to the map** button.\n\n\n<img align=\"middle\" src=\"../_static/maps/DataCatalogue.jpg\" alt=\"DE Africa maps image\" width=\"800\">\n\nVisualise and analyse your dataset:\n\n* The spatial data will be displayed in the map view, and a visual legend will appear in the Data workbench, on the left side of the page.\n\n* It may not be immediately obvious where your selected spatial data has loaded on the map if it does not cover a large part of Africa. To locate loaded data on the map, go to the workbench, and select **Zoom to extent**.\n\n* The dataset should now be visible in the map view.\n\n* To add additional datasets to the map, repeat the above steps.\n\n* Zoom manually by moving your mouse pointer over the map and using your mouse wheel to zoom in or out further.\n\n* Click and drag the map to further show the region in which you are interested.\n",
"_____no_output_____"
],
[
"## Explore the workbench\n\n<img align=\"middle\" src=\"../_static/maps/Workbench.jpg\" alt=\"DE Africa maps image\" width=\"800\">\n\n\nWhen a dataset is added to the map via the data catalogue, a legend for that dataset will appear in the workbench. From the workbench you can:\n\n* Set the order data is shown on the map. To do this, click the title of a dataset and drag it to a new position in your workbench.\n* Toggle the visibility of added datasets by selecting the checkbox opposite your prefered dataset title\n* Zoom to the geographical extent of an added data set\n* Set the opacity of individual data sets\n* Remove data sets from the map. Note: they can be re-added via the data catalogue.",
"_____no_output_____"
],
[
"## Navigate the Map\n\n<img align=\"middle\" src=\"../_static/maps/Navigate.jpg\" alt=\"DE Africa maps image\" width=\"800\">\n\nThere are multiple ways to navigate DE Africa Map's map view:\n\n### Zooming\n\nMove your mouse pointer over the map and use the scroll wheel to zoom in or out. The location at the centre of the map display is the centre of the zooming. Right-click and drag upwards or downwards over the map to zoom about the centre point. Select the zoom control to zoom in or out quickly by a set amount.\n\n### Panning\n\nClick anywhere on the map and drag it to the required location.\n\n### Rotate the map\n\nUse the compass control to rotate the map so North is no longer at the top. Select the “gyroscope” in the centre of the compass control and drag slowly to the left or right to rotate the map clockwise or anti-clockwise respectively. The further you drag, the faster it rotates. Release the mouse button when you reach the desired rotation.\n\nSelect the North Point or outer ring of the Compass Control and drag it around to set the desired rotation directly.\n\nControl + left-click and drag left or right over the map to rotate the view about the centre.\n\n### Perspective view\n\nSelect the “gyroscope” in the centre of the compass control and drag slowly upwards to tilt the view into a perspective view. Drag downwards to tilt the view back to vertical. The further you drag, the faster it tilts. Release the mouse button when you reach the desired view.\n\nControl + left-click and drag upwards or downwards over the map to enter or adjust the perspective view.\n\nWhen you are in perspective view, control + left-click and drag left or right over the map to “orbit” around the centre of the view. You can also click and drag left or right over the “gyroscope” at the centre of the Compass Control to orbit about the centre of the view.\n\nDouble-click the “gyroscope” in the centre of the Compass Control to return the view quickly to a vertical view with North to the top at the current location and scale.\n\nDragging to pan and using the mouse wheel to zoom still work while showing a perspective view.\n\n### Use the splitter functionality\n\nThe splitter functionality allows you to compare a dataset for different time periods. To use it:\n\n* Select an area/location\n* Select a dataset from the catalogue to add it to the workbench\n* Select the **Split** link in the workbench to create a copy of the data already selected. Note that you can use the splitter with two different datasets, not just with copies of the same data\n* Select different times (using the date picker) for the “left” and “right” sections of the screen\n* Use the back and forward arrows to explore imagery and select cloud-free views\n* Drag the splitter on the screen to observe the differences",
"_____no_output_____"
],
[
"This video shows an example of the splitter used over a map of Australia, but the functionality is the same.",
"_____no_output_____"
],
[
"[](http://www.youtube.com/watch?v=H3htpdYAE7w \"Split functionality\")",
"_____no_output_____"
],
[
"## Upload Data to the Map\n\nThere are two ways to load your data:\n\n1. Drag your data file onto the DE Africa Map map view. The format of the data file will be auto-detected.\n1. Select **Add Data** in the left panel. This will launch the data catalogue. Select the **My Data** tab at the top of the modal window and follow the provided instructions.\n\nAs for DE Africa Map data sets, you can select regions or points to see the data available for that location. If the file is a CSV file, the data from all columns will be shown in the feature information dialogue.\n\nYou can also use all of the features of the workbench on the data you have loaded as well.\n\nTo share a view of your data with others, you must first publish it to the web somewhere with a URL, and then load it from there.\n\nDE Africa Map can display two kinds of spreadsheets:\n\n1. Spreadsheets with a point location (latitude and longitude) for each row, expressed as two columns: lat and lon. These will be displayed as points (circles).\n2. Spreadsheets where each row refers to a region such as a local government area (council), state, postcode, or ABS statistical unit such as an SA2 or CED (Commonwealth Electoral Division). Columns must be named according to the CSV-geo-au standard. These will be displayed as regions, highlighting the actual shape of each area.\n\nSpreadsheets must be saved as CSV (comma-separated values).\n\nOther standard spatial data types such as GeoJSON and KML are also supported.\n\n[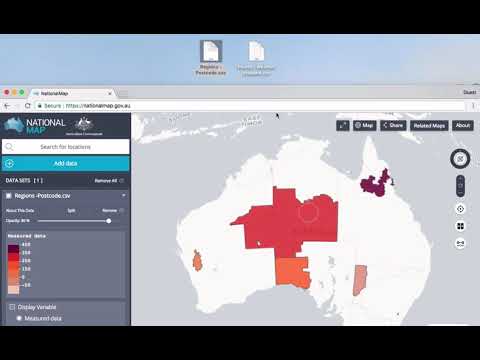](http://www.youtube.com/watch?v=XKP90TcBq6A \"Upload Location\")",
"_____no_output_____"
]
]
]
| [
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
]
]
|
ec73efb1ebd424c3a426ad6e70b36c965e5fd1b0 | 35,139 | ipynb | Jupyter Notebook | PS3-mapping-bash-new.ipynb | Scaburet/RNAseq-part1 | 4070bc147742e30b51b275138836a44cce5b1846 | [
"BSD-3-Clause"
]
| null | null | null | PS3-mapping-bash-new.ipynb | Scaburet/RNAseq-part1 | 4070bc147742e30b51b275138836a44cce5b1846 | [
"BSD-3-Clause"
]
| null | null | null | PS3-mapping-bash-new.ipynb | Scaburet/RNAseq-part1 | 4070bc147742e30b51b275138836a44cce5b1846 | [
"BSD-3-Clause"
]
| null | null | null | 32.809524 | 514 | 0.608526 | [
[
[
"empty"
]
]
]
| [
"empty"
]
| [
[
"empty"
]
]
|
ec73f550860d9e506eefd5ec4883d5764455dada | 58,376 | ipynb | Jupyter Notebook | aulas/aula05_01_metricas.ipynb | flaviobp/introducao-aprendizado-maquina | 87c2f57352a03dd39e2507a6f6ff33f5dd280aa9 | [
"MIT"
]
| null | null | null | aulas/aula05_01_metricas.ipynb | flaviobp/introducao-aprendizado-maquina | 87c2f57352a03dd39e2507a6f6ff33f5dd280aa9 | [
"MIT"
]
| null | null | null | aulas/aula05_01_metricas.ipynb | flaviobp/introducao-aprendizado-maquina | 87c2f57352a03dd39e2507a6f6ff33f5dd280aa9 | [
"MIT"
]
| null | null | null | 109.93597 | 34,416 | 0.82856 | [
[
[
"# Métricas para avaliação de modelos\n\nA escolha da métrica a ser otimizada depende da aplicação!\n\n#### Métricas para regressão\n\nMSE (mean squared error)\n\n$\n\\begin{align}\n\\frac{1}{n} \\sum_{i=1}^{n}(y_i - \\hat{y}_i)^2\n\\end{align}\n$\n\nMAE (mean absolute error)\n\n$\n\\begin{align}\n\\frac{1}{n} \\sum_{i=1}^{n}(y_i - \\hat{y}_i)\n\\end{align}\n$\n\n#### Métricas para classificação\n\nSejam:\n\n$\n\\begin{align}\nTP = true \\ positives \\\\\nTN = true \\ negatives \\\\\nFP = false \\ positives \\ (erro \\ tipo \\ I)\\\\\nFN = false \\ negatives \\ (erro \\ tipo \\ II)\n\\end{align}\n$\n\nPrecisão\n\n$\n\\begin{align}\n\\frac{TP}{TP+FP}\n\\end{align}\n$\n\nRecall / sensibilidade\n\n$\n\\begin{align}\n\\frac{TP}{TP+FN}\n\\end{align}\n$\n\nAcurácia\n\n$\n\\begin{align}\n\\frac{TP+TN}{TP+TN+FP+FN}\n\\end{align}\n$\n\n\nF1 score: média harmônica de precisão e recall\n\n$\n\\begin{align}\n2 \\cdot \\frac{precision \\cdot recall}{precision + recall}\n\\end{align}\n$",
"_____no_output_____"
],
[
"## Validação cruzada\n\nConsiste em particionar o dataset em k subconjuntos e treinar o modelo k vezes, usando um subconjunto diferente para validação por vez e os k-1 restantes para treino.\n\nVamos usar uma função auxiliar que recebe uma instância de um modelo, a quantidade de partições $k$, a matriz de *features* $X$, o vetor de *targets* $y$ e uma função que computa a métrica desejada.\n\nExperimente variar $k$ e a função de métrica nos exemplos abaixo: classificação de flores e regressão de valores de casas.",
"_____no_output_____"
]
],
[
[
"import matplotlib.pyplot as plt\nimport numpy as np\nimport pandas as pd\nfrom sklearn.datasets import load_boston, load_iris\nfrom sklearn.linear_model import LinearRegression, LogisticRegression\nfrom sklearn.metrics import accuracy_score, recall_score, f1_score, mean_absolute_error, mean_squared_error\nfrom sklearn.model_selection import KFold\n\n\ndef train_model(model, k, X, y, score_func):\n kfolds = KFold(k)\n scores = []\n for train_idx, test_idx in kfolds.split(X, y):\n X_train, y_train = X[train_idx], y[train_idx]\n X_test, y_test = X[test_idx], y[test_idx]\n model = model.fit(X_train, y_train)\n y_hat = model.predict(X_test)\n scores.append(score_func(y_test, y_hat))\n print('scores: ', np.round(scores,4), '\\nmean: ', np.round(np.mean(scores),4))",
"_____no_output_____"
]
],
[
[
"#### Classificação de flores\n\nDataset Iris.",
"_____no_output_____"
]
],
[
[
"data_iris = load_iris()\ndf_iris = pd.DataFrame(data_iris.data, columns=data_iris.feature_names)\n\nX_iris = df_iris.values\ny_iris = data_iris.target\ncols_iris = data_iris.feature_names\n\ndf_iris.head()",
"_____no_output_____"
]
],
[
[
"Podemos visualizar as *features* duas a duas, como no *snippet* abaixo. Experimente variar as *features* (i.e. `X_iris[:,0]` e `X_iris[:,1]`). Não se esqueça de também trocar as labels do gráfico.",
"_____no_output_____"
]
],
[
[
"plt.scatter(X_iris[:,0], X_iris[:,1], c=y_iris, alpha=0.7)\nplt.xlabel(cols_iris[0])\nplt.ylabel(cols_iris[1])",
"_____no_output_____"
],
[
"lr_iris = LogisticRegression(solver='lbfgs', max_iter=200, multi_class='multinomial')\ntrain_model(lr_iris, 5, X_iris, y_iris, accuracy_score)",
"scores: [1. 1. 0.8667 0.9333 0.8333] \nmean: 0.9267\n"
]
],
[
[
"#### Regressão de valores de casas\n\nDataset *Boston Housing*.",
"_____no_output_____"
]
],
[
[
"data_boston = load_boston()\ndf_boston = pd.DataFrame(data_boston.data, columns=data_boston.feature_names)\n\nX_boston = df_boston.values\ny_boston = data_boston.target\ncols_boston = data_boston.feature_names\n\ndf_boston.head()",
"_____no_output_____"
],
[
"plt.scatter(X_boston[:,0], X_boston[:,1], alpha=0.7)\nplt.xlabel(cols_boston[0])\nplt.ylabel(cols_boston[1])",
"_____no_output_____"
],
[
"lr_boston = LinearRegression()\ntrain_model(lr_boston, 10, X_boston, y_boston, mean_absolute_error)",
"scores: [2.2069 2.8968 2.7867 4.5985 4.1099 3.5647 2.6697 9.6564 5.0227 2.5373] \nmean: 4.0049\n"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code"
]
| [
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code",
"code",
"code"
]
]
|
ec73f7cd09dd453bd72e29aa83ce522edbc766f8 | 867,842 | ipynb | Jupyter Notebook | face recognition.ipynb | dasthagirikancharla/Face-Recognition | 630a94940c9d6ba0415c7835e4286b3ac5e70235 | [
"CC0-1.0"
]
| 1 | 2021-11-16T13:22:29.000Z | 2021-11-16T13:22:29.000Z | face recognition.ipynb | prasanna-chintamaneni/FACE-RECOGNITION | d09d9a0a4ef1e3888759e8501121176ff5c967aa | [
"CC0-1.0"
]
| null | null | null | face recognition.ipynb | prasanna-chintamaneni/FACE-RECOGNITION | d09d9a0a4ef1e3888759e8501121176ff5c967aa | [
"CC0-1.0"
]
| null | null | null | 867,842 | 867,842 | 0.964672 | [
[
[
"!pip install face-recognition",
"Collecting face-recognition\n Downloading face_recognition-1.3.0-py2.py3-none-any.whl (15 kB)\nRequirement already satisfied: numpy in /usr/local/lib/python3.7/dist-packages (from face-recognition) (1.19.5)\nRequirement already satisfied: Pillow in /usr/local/lib/python3.7/dist-packages (from face-recognition) (7.1.2)\nRequirement already satisfied: Click>=6.0 in /usr/local/lib/python3.7/dist-packages (from face-recognition) (7.1.2)\nRequirement already satisfied: dlib>=19.7 in /usr/local/lib/python3.7/dist-packages (from face-recognition) (19.18.0)\nCollecting face-recognition-models>=0.3.0\n Downloading face_recognition_models-0.3.0.tar.gz (100.1 MB)\n\u001b[K |████████████████████████████████| 100.1 MB 8.5 kB/s \n\u001b[?25hBuilding wheels for collected packages: face-recognition-models\n Building wheel for face-recognition-models (setup.py) ... \u001b[?25l\u001b[?25hdone\n Created wheel for face-recognition-models: filename=face_recognition_models-0.3.0-py2.py3-none-any.whl size=100566185 sha256=bc8c682d23b1a74236cde6aff9edb8ebf36de2700b41bca2988dc8451b95cd8f\n Stored in directory: /root/.cache/pip/wheels/d6/81/3c/884bcd5e1c120ff548d57c2ecc9ebf3281c9a6f7c0e7e7947a\nSuccessfully built face-recognition-models\nInstalling collected packages: face-recognition-models, face-recognition\nSuccessfully installed face-recognition-1.3.0 face-recognition-models-0.3.0\n"
],
[
"import cv2\nimport face_recognition\nfrom google.colab.patches import cv2_imshow\nimgprasanna = face_recognition.load_image_file('/content/camera.jpg')\nimgprasanna = cv2.cvtColor(imgprasanna, cv2.COLOR_BGR2RGB)\nimgTest = face_recognition.load_image_file('')\nimgTest = cv2.cvtColor(imgTest, cv2.COLOR_BGR2RGB)\nfaceLoc = face_recognition.face_locations(imgprasanna)[0]\nencodeprasanna = face_recognition.face_encodings(imgprasanna)[0]\ncv2.rectangle(imgprasanna, (faceLoc[3], faceLoc[0]), (faceLoc[1], faceLoc[2]), (255, 0, 255), 2)\nfaceLocTest = face_recognition.face_locations(imgTest)[0]\nencodeTest = face_recognition.face_encodings(imgTest)[0]\ncv2.rectangle(imgTest, (faceLocTest[3], faceLocTest[0]), (faceLocTest[1], faceLocTest[2]), (255, 0, 255), 2)\nresults = face_recognition.compare_faces([encodeprasanna], encodeTest)\nfaceDis = face_recognition.face_distance([encodeprasanna], encodeTest)\nprint(results, faceDis)\ncv2.putText(imgTest, f'{results} {round(faceDis[0],2)}', (50, 50), cv2.FONT_HERSHEY_COMPLEX, 1, (0, 0, 255), 2)\n# cv2.imshow('prasanna', imgprasanna)\ncv2_imshow(imgprasanna)\n# cv2.imshow('prasanna Test', imgTest)\ncv2_imshow(imgTest)\n# cv2.waitKey(0)",
"[False] [0.63417429]\n"
],
[
"",
"_____no_output_____"
]
]
]
| [
"code"
]
| [
[
"code",
"code",
"code"
]
]
|
ec7404f116cacd7a35ea2397d35a641ff0165648 | 153,500 | ipynb | Jupyter Notebook | Residual_Networks.ipynb | Anjoom765/CNN_1 | f49ca0e33ece46dc69803c96732ee7776fe114a7 | [
"MIT"
]
| null | null | null | Residual_Networks.ipynb | Anjoom765/CNN_1 | f49ca0e33ece46dc69803c96732ee7776fe114a7 | [
"MIT"
]
| null | null | null | Residual_Networks.ipynb | Anjoom765/CNN_1 | f49ca0e33ece46dc69803c96732ee7776fe114a7 | [
"MIT"
]
| null | null | null | 79.823193 | 21,692 | 0.673231 | [
[
[
"# Residual Networks\n\nWelcome to the first assignment of this week! You'll be building a very deep convolutional network, using Residual Networks (ResNets). In theory, very deep networks can represent very complex functions; but in practice, they are hard to train. Residual Networks, introduced by [He et al.](https://arxiv.org/pdf/1512.03385.pdf), allow you to train much deeper networks than were previously feasible.\n\n**By the end of this assignment, you'll be able to:**\n\n- Implement the basic building blocks of ResNets in a deep neural network using Keras\n- Put together these building blocks to implement and train a state-of-the-art neural network for image classification\n- Implement a skip connection in your network\n\nFor this assignment, you'll use Keras. \n\nBefore jumping into the problem, run the cell below to load the required packages.",
"_____no_output_____"
],
[
"## Table of Content\n\n- [1 - Packages](#1)\n- [2 - The Problem of Very Deep Neural Networks](#2)\n- [3 - Building a Residual Network](#3)\n - [3.1 - The Identity Block](#3-1)\n - [Exercise 1 - identity_block](#ex-1)\n - [3.2 - The Convolutional Block](#3-2)\n - [Exercise 2 - convolutional_block](#ex-2)\n- [4 - Building Your First ResNet Model (50 layers)](#4)\n - [Exercise 3 - ResNet50](#ex-3)\n- [5 - Test on Your Own Image (Optional/Ungraded)](#5)\n- [6 - Bibliography](#6)",
"_____no_output_____"
],
[
"<a name='1'></a>\n## 1 - Packages",
"_____no_output_____"
]
],
[
[
"import tensorflow as tf\nimport numpy as np\nimport scipy.misc\nfrom tensorflow.keras.applications.resnet_v2 import ResNet50V2\nfrom tensorflow.keras.preprocessing import image\nfrom tensorflow.keras.applications.resnet_v2 import preprocess_input, decode_predictions\nfrom tensorflow.keras import layers\nfrom tensorflow.keras.layers import Input, Add, Dense, Activation, ZeroPadding2D, BatchNormalization, Flatten, Conv2D, AveragePooling2D, MaxPooling2D, GlobalMaxPooling2D\nfrom tensorflow.keras.models import Model, load_model\nfrom resnets_utils import *\nfrom tensorflow.keras.initializers import random_uniform, glorot_uniform, constant, identity\nfrom tensorflow.python.framework.ops import EagerTensor\nfrom matplotlib.pyplot import imshow\n\nfrom test_utils import summary, comparator\nimport public_tests\n\n%matplotlib inline",
"_____no_output_____"
]
],
[
[
"<a name='2'></a>\n## 2 - The Problem of Very Deep Neural Networks\n\nLast week, you built your first convolutional neural networks: first manually with numpy, then using Tensorflow and Keras. \n\nIn recent years, neural networks have become much deeper, with state-of-the-art networks evolving from having just a few layers (e.g., AlexNet) to over a hundred layers.\n\n* The main benefit of a very deep network is that it can represent very complex functions. It can also learn features at many different levels of abstraction, from edges (at the shallower layers, closer to the input) to very complex features (at the deeper layers, closer to the output). \n\n* However, using a deeper network doesn't always help. A huge barrier to training them is vanishing gradients: very deep networks often have a gradient signal that goes to zero quickly, thus making gradient descent prohibitively slow.\n\n* More specifically, during gradient descent, as you backpropagate from the final layer back to the first layer, you are multiplying by the weight matrix on each step, and thus the gradient can decrease exponentially quickly to zero (or, in rare cases, grow exponentially quickly and \"explode,\" from gaining very large values). \n\n* During training, you might therefore see the magnitude (or norm) of the gradient for the shallower layers decrease to zero very rapidly as training proceeds, as shown below: ",
"_____no_output_____"
],
[
"<img src=\"images/vanishing_grad_kiank.png\" style=\"width:450px;height:220px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 1</b> </u><font color='purple'> : <b>Vanishing gradient</b> <br> The speed of learning decreases very rapidly for the shallower layers as the network trains </center></caption>\n\nNot to worry! You are now going to solve this problem by building a Residual Network!",
"_____no_output_____"
],
[
"<a name='3'></a>\n## 3 - Building a Residual Network\n\nIn ResNets, a \"shortcut\" or a \"skip connection\" allows the model to skip layers: \n\n<img src=\"images/skip_connection_kiank.png\" style=\"width:650px;height:200px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 2</b> </u><font color='purple'> : A ResNet block showing a skip-connection <br> </center></caption>\n\nThe image on the left shows the \"main path\" through the network. The image on the right adds a shortcut to the main path. By stacking these ResNet blocks on top of each other, you can form a very deep network. \n\nThe lecture mentioned that having ResNet blocks with the shortcut also makes it very easy for one of the blocks to learn an identity function. This means that you can stack on additional ResNet blocks with little risk of harming training set performance. \n \nOn that note, there is also some evidence that the ease of learning an identity function accounts for ResNets' remarkable performance even more than skip connections help with vanishing gradients.\n\nTwo main types of blocks are used in a ResNet, depending mainly on whether the input/output dimensions are the same or different. You are going to implement both of them: the \"identity block\" and the \"convolutional block.\"",
"_____no_output_____"
],
[
"<a name='3-1'></a>\n### 3.1 - The Identity Block\n\nThe identity block is the standard block used in ResNets, and corresponds to the case where the input activation (say $a^{[l]}$) has the same dimension as the output activation (say $a^{[l+2]}$). To flesh out the different steps of what happens in a ResNet's identity block, here is an alternative diagram showing the individual steps:\n\n<img src=\"images/idblock2_kiank.png\" style=\"width:650px;height:150px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 3</b> </u><font color='purple'> : <b>Identity block.</b> Skip connection \"skips over\" 2 layers. </center></caption>\n\nThe upper path is the \"shortcut path.\" The lower path is the \"main path.\" In this diagram, notice the CONV2D and ReLU steps in each layer. To speed up training, a BatchNorm step has been added. Don't worry about this being complicated to implement--you'll see that BatchNorm is just one line of code in Keras! \n\nIn this exercise, you'll actually implement a slightly more powerful version of this identity block, in which the skip connection \"skips over\" 3 hidden layers rather than 2 layers. It looks like this: \n\n<img src=\"images/idblock3_kiank.png\" style=\"width:650px;height:150px;\">\n <caption><center> <u> <font color='purple'> <b>Figure 4</b> </u><font color='purple'> : <b>Identity block.</b> Skip connection \"skips over\" 3 layers.</center></caption>",
"_____no_output_____"
],
[
"These are the individual steps:\n\nFirst component of main path: \n- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\". Use 0 as the seed for the random uniform initialization: `kernel_initializer = initializer(seed=0)`. \n- The first BatchNorm is normalizing the 'channels' axis.\n- Then apply the ReLU activation function. This has no hyperparameters. \n\nSecond component of main path:\n- The second CONV2D has $F_2$ filters of shape $(f,f)$ and a stride of (1,1). Its padding is \"same\". Use 0 as the seed for the random uniform initialization: `kernel_initializer = initializer(seed=0)`.\n- The second BatchNorm is normalizing the 'channels' axis.\n- Then apply the ReLU activation function. This has no hyperparameters.\n\nThird component of main path:\n- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\". Use 0 as the seed for the random uniform initialization: `kernel_initializer = initializer(seed=0)`. \n- The third BatchNorm is normalizing the 'channels' axis.\n- Note that there is **no** ReLU activation function in this component. \n\nFinal step: \n- The `X_shortcut` and the output from the 3rd layer `X` are added together.\n- **Hint**: The syntax will look something like `Add()([var1,var2])`\n- Then apply the ReLU activation function. This has no hyperparameters. \n\n<a name='ex-1'></a>\n### Exercise 1 - identity_block\n\nImplement the ResNet identity block. The first component of the main path has been implemented for you already! First, you should read these docs carefully to make sure you understand what's happening. Then, implement the rest. \n- To implement the Conv2D step: [Conv2D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)\n- To implement BatchNorm: [BatchNormalization](https://www.tensorflow.org/api_docs/python/tf/keras/layers/BatchNormalization) `BatchNormalization(axis = 3)(X, training = training)`. If training is set to False, its weights are not updated with the new examples. I.e when the model is used in prediction mode.\n- For the activation, use: `Activation('relu')(X)`\n- To add the value passed forward by the shortcut: [Add](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Add)\n\nWe have added the initializer argument to our functions. This parameter receives an initializer function like the ones included in the package [tensorflow.keras.initializers](https://www.tensorflow.org/api_docs/python/tf/keras/initializers) or any other custom initializer. By default it will be set to [random_uniform](https://www.tensorflow.org/api_docs/python/tf/keras/initializers/RandomUniform)\n\nRemember that these functions accept a `seed` argument that can be any value you want, but that in this notebook must set to 0 for **grading purposes**.",
"_____no_output_____"
],
[
" Here is where you're actually using the power of the Functional API to create a shortcut path: ",
"_____no_output_____"
]
],
[
[
"# UNQ_C1\n# GRADED FUNCTION: identity_block\n\ndef identity_block(X, f, filters, training=True, initializer=random_uniform):\n \"\"\"\n Implementation of the identity block as defined in Figure 4\n \n Arguments:\n X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)\n f -- integer, specifying the shape of the middle CONV's window for the main path\n filters -- python list of integers, defining the number of filters in the CONV layers of the main path\n training -- True: Behave in training mode\n False: Behave in inference mode\n initializer -- to set up the initial weights of a layer. Equals to random uniform initializer\n \n Returns:\n X -- output of the identity block, tensor of shape (m, n_H, n_W, n_C)\n \"\"\"\n \n # Retrieve Filters\n F1, F2, F3 = filters\n \n # Save the input value. You'll need this later to add back to the main path. \n X_shortcut = X\n \n # First component of main path\n X = Conv2D(filters = F1, kernel_size = 1, strides = (1,1), padding = 'valid', kernel_initializer = initializer(seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training = training) # Default axis\n X = Activation('relu')(X)\n \n ### START CODE HERE\n ## Second component of main path (≈3 lines)\n X = Conv2D(filters = F2, kernel_size = f, strides = (1,1), padding = 'same', kernel_initializer = initializer(seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training = training)\n X = Activation('relu')(X)\n\n ## Third component of main path (≈2 lines)\n X = Conv2D(filters = F3, kernel_size = 1, strides = (1,1), padding = 'valid', kernel_initializer = initializer(seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training = training)\n \n ## Final step: Add shortcut value to main path, and pass it through a RELU activation (≈2 lines)\n X = Add()([X, X_shortcut])\n X = Activation('relu')(X)\n ### END CODE HERE\n\n return X",
"_____no_output_____"
],
[
"np.random.seed(1)\nX1 = np.ones((1, 4, 4, 3)) * -1\nX2 = np.ones((1, 4, 4, 3)) * 1\nX3 = np.ones((1, 4, 4, 3)) * 3\n\nX = np.concatenate((X1, X2, X3), axis = 0).astype(np.float32)\n\nA3 = identity_block(X, f=2, filters=[4, 4, 3],\n initializer=lambda seed=0:constant(value=1),\n training=False)\nprint('\\033[1mWith training=False\\033[0m\\n')\nA3np = A3.numpy()\nprint(np.around(A3.numpy()[:,(0,-1),:,:].mean(axis = 3), 5))\nresume = A3np[:,(0,-1),:,:].mean(axis = 3)\nprint(resume[1, 1, 0])\n\nprint('\\n\\033[1mWith training=True\\033[0m\\n')\nnp.random.seed(1)\nA4 = identity_block(X, f=2, filters=[3, 3, 3],\n initializer=lambda seed=0:constant(value=1),\n training=True)\nprint(np.around(A4.numpy()[:,(0,-1),:,:].mean(axis = 3), 5))\n\npublic_tests.identity_block_test(identity_block)",
"\u001b[1mWith training=False\u001b[0m\n\n[[[ 0. 0. 0. 0. ]\n [ 0. 0. 0. 0. ]]\n\n [[192.71234 192.71234 192.71234 96.85617]\n [ 96.85617 96.85617 96.85617 48.92808]]\n\n [[578.1371 578.1371 578.1371 290.5685 ]\n [290.5685 290.5685 290.5685 146.78426]]]\n96.85617\n\n\u001b[1mWith training=True\u001b[0m\n\n[[[0. 0. 0. 0. ]\n [0. 0. 0. 0. ]]\n\n [[0.40739 0.40739 0.40739 0.40739]\n [0.40739 0.40739 0.40739 0.40739]]\n\n [[4.99991 4.99991 4.99991 3.25948]\n [3.25948 3.25948 3.25948 2.40739]]]\n\u001b[32mAll tests passed!\u001b[0m\n"
]
],
[
[
"**Expected value**\n\n```\nWith training=False\n\n[[[ 0. 0. 0. 0. ]\n [ 0. 0. 0. 0. ]]\n\n [[192.71234 192.71234 192.71234 96.85617]\n [ 96.85617 96.85617 96.85617 48.92808]]\n\n [[578.1371 578.1371 578.1371 290.5685 ]\n [290.5685 290.5685 290.5685 146.78426]]]\n96.85617\n\nWith training=True\n\n[[[0. 0. 0. 0. ]\n [0. 0. 0. 0. ]]\n\n [[0.40739 0.40739 0.40739 0.40739]\n [0.40739 0.40739 0.40739 0.40739]]\n\n [[4.99991 4.99991 4.99991 3.25948]\n [3.25948 3.25948 3.25948 2.40739]]]\n```",
"_____no_output_____"
],
[
"<a name='3-2'></a>\n### 3.2 - The Convolutional Block\n\nThe ResNet \"convolutional block\" is the second block type. You can use this type of block when the input and output dimensions don't match up. The difference with the identity block is that there is a CONV2D layer in the shortcut path: \n\n<img src=\"images/convblock_kiank.png\" style=\"width:650px;height:150px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 4</b> </u><font color='purple'> : <b>Convolutional block</b> </center></caption>\n\n* The CONV2D layer in the shortcut path is used to resize the input $x$ to a different dimension, so that the dimensions match up in the final addition needed to add the shortcut value back to the main path. (This plays a similar role as the matrix $W_s$ discussed in lecture.) \n* For example, to reduce the activation dimensions's height and width by a factor of 2, you can use a 1x1 convolution with a stride of 2. \n* The CONV2D layer on the shortcut path does not use any non-linear activation function. Its main role is to just apply a (learned) linear function that reduces the dimension of the input, so that the dimensions match up for the later addition step. \n* As for the previous exercise, the additional `initializer` argument is required for grading purposes, and it has been set by default to [glorot_uniform](https://www.tensorflow.org/api_docs/python/tf/keras/initializers/GlorotUniform)\n\nThe details of the convolutional block are as follows. \n\nFirst component of main path:\n- The first CONV2D has $F_1$ filters of shape (1,1) and a stride of (s,s). Its padding is \"valid\". Use 0 as the `glorot_uniform` seed `kernel_initializer = initializer(seed=0)`.\n- The first BatchNorm is normalizing the 'channels' axis.\n- Then apply the ReLU activation function. This has no hyperparameters. \n\nSecond component of main path:\n- The second CONV2D has $F_2$ filters of shape (f,f) and a stride of (1,1). Its padding is \"same\". Use 0 as the `glorot_uniform` seed `kernel_initializer = initializer(seed=0)`.\n- The second BatchNorm is normalizing the 'channels' axis.\n- Then apply the ReLU activation function. This has no hyperparameters. \n\nThird component of main path:\n- The third CONV2D has $F_3$ filters of shape (1,1) and a stride of (1,1). Its padding is \"valid\". Use 0 as the `glorot_uniform` seed `kernel_initializer = initializer(seed=0)`.\n- The third BatchNorm is normalizing the 'channels' axis. Note that there is no ReLU activation function in this component. \n\nShortcut path:\n- The CONV2D has $F_3$ filters of shape (1,1) and a stride of (s,s). Its padding is \"valid\". Use 0 as the `glorot_uniform` seed `kernel_initializer = initializer(seed=0)`.\n- The BatchNorm is normalizing the 'channels' axis. \n\nFinal step: \n- The shortcut and the main path values are added together.\n- Then apply the ReLU activation function. This has no hyperparameters. \n \n<a name='ex-2'></a> \n### Exercise 2 - convolutional_block\n \nImplement the convolutional block. The first component of the main path is already implemented; then it's your turn to implement the rest! As before, always use 0 as the seed for the random initialization, to ensure consistency with the grader.\n- [Conv2D](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)\n- [BatchNormalization](https://www.tensorflow.org/api_docs/python/tf/keras/layers/BatchNormalization) (axis: Integer, the axis that should be normalized (typically the features axis)) `BatchNormalization(axis = 3)(X, training = training)`. If training is set to False, its weights are not updated with the new examples. I.e when the model is used in prediction mode.\n- For the activation, use: `Activation('relu')(X)`\n- [Add](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Add)\n \nWe have added the initializer argument to our functions. This parameter receives an initializer function like the ones included in the package [tensorflow.keras.initializers](https://www.tensorflow.org/api_docs/python/tf/keras/initializers) or any other custom initializer. By default it will be set to [random_uniform](https://www.tensorflow.org/api_docs/python/tf/keras/initializers/RandomUniform)\n\nRemember that these functions accept a `seed` argument that can be any value you want, but that in this notebook must set to 0 for **grading purposes**.",
"_____no_output_____"
]
],
[
[
"# UNQ_C2\n# GRADED FUNCTION: convolutional_block\n\ndef convolutional_block(X, f, filters, s = 2, training=True, initializer=glorot_uniform):\n \"\"\"\n Implementation of the convolutional block as defined in Figure 4\n \n Arguments:\n X -- input tensor of shape (m, n_H_prev, n_W_prev, n_C_prev)\n f -- integer, specifying the shape of the middle CONV's window for the main path\n filters -- python list of integers, defining the number of filters in the CONV layers of the main path\n s -- Integer, specifying the stride to be used\n training -- True: Behave in training mode\n False: Behave in inference mode\n initializer -- to set up the initial weights of a layer. Equals to Glorot uniform initializer, \n also called Xavier uniform initializer.\n \n Returns:\n X -- output of the convolutional block, tensor of shape (n_H, n_W, n_C)\n \"\"\"\n \n # Retrieve Filters\n F1, F2, F3 = filters\n \n # Save the input value\n X_shortcut = X\n\n\n ##### MAIN PATH #####\n \n # First component of main path glorot_uniform(seed=0)\n X = Conv2D(filters = F1, kernel_size = 1, strides = (s, s), padding='valid', kernel_initializer = glorot_uniform (seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training=training)\n X = Activation('relu')(X)\n\n ### START CODE HERE\n \n ## Second component of main path (≈3 lines)\n X = Conv2D(filters = F2, kernel_size = f, strides = (1, 1), padding='same', kernel_initializer =glorot_uniform (seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training=training)\n X = Activation('relu')(X)\n\n ## Third component of main path (≈2 lines)\n X = Conv2D(filters = F3, kernel_size = 1, strides = (1, 1), padding='valid', kernel_initializer = glorot_uniform (seed=0))(X)\n X = BatchNormalization(axis = 3)(X, training=training)\n \n ##### SHORTCUT PATH ##### (≈2 lines)\n X_shortcut = Conv2D(filters = F3, kernel_size = 1, strides = (s, s), padding='valid', kernel_initializer =glorot_uniform (seed=0))(X_shortcut) \n X_shortcut = BatchNormalization(axis = 3)(X_shortcut, training=training)\n \n ### END CODE HERE\n\n # Final step: Add shortcut value to main path (Use this order [X, X_shortcut]), and pass it through a RELU activation\n X = Add()([X, X_shortcut])\n X = Activation('relu')(X)\n \n return X",
"_____no_output_____"
],
[
"from outputs import convolutional_block_output1, convolutional_block_output2\nnp.random.seed(1)\n#X = np.random.randn(3, 4, 4, 6).astype(np.float32)\nX1 = np.ones((1, 4, 4, 3)) * -1\nX2 = np.ones((1, 4, 4, 3)) * 1\nX3 = np.ones((1, 4, 4, 3)) * 3\n\nX = np.concatenate((X1, X2, X3), axis = 0).astype(np.float32)\n\nA = convolutional_block(X, f = 2, filters = [2, 4, 6], training=False)\n\nassert type(A) == EagerTensor, \"Use only tensorflow and keras functions\"\nassert tuple(tf.shape(A).numpy()) == (3, 2, 2, 6), \"Wrong shape.\"\nassert np.allclose(A.numpy(), convolutional_block_output1), \"Wrong values when training=False.\"\nprint(A[0])\n\nB = convolutional_block(X, f = 2, filters = [2, 4, 6], training=True)\nassert np.allclose(B.numpy(), convolutional_block_output2), \"Wrong values when training=True.\"\n\nprint('\\033[92mAll tests passed!')\n",
"tf.Tensor(\n[[[0. 0.66683817 0. 0. 0.88853896 0.5274254 ]\n [0. 0.65053666 0. 0. 0.89592844 0.49965227]]\n\n [[0. 0.6312079 0. 0. 0.8636247 0.47643146]\n [0. 0.5688321 0. 0. 0.85534114 0.41709304]]], shape=(2, 2, 6), dtype=float32)\n\u001b[92mAll tests passed!\n"
]
],
[
[
"**Expected value**\n\n```\ntf.Tensor(\n[[[0. 0.66683817 0. 0. 0.88853896 0.5274254 ]\n [0. 0.65053666 0. 0. 0.89592844 0.49965227]]\n\n [[0. 0.6312079 0. 0. 0.8636247 0.47643146]\n [0. 0.5688321 0. 0. 0.85534114 0.41709304]]], shape=(2, 2, 6), dtype=float32)\n```",
"_____no_output_____"
],
[
"<a name='4'></a> \n## 4 - Building Your First ResNet Model (50 layers)\n\nYou now have the necessary blocks to build a very deep ResNet. The following figure describes in detail the architecture of this neural network. \"ID BLOCK\" in the diagram stands for \"Identity block,\" and \"ID BLOCK x3\" means you should stack 3 identity blocks together.\n\n<img src=\"images/resnet_kiank.png\" style=\"width:850px;height:150px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 5</b> </u><font color='purple'> : <b>ResNet-50 model</b> </center></caption>\n\nThe details of this ResNet-50 model are:\n- Zero-padding pads the input with a pad of (3,3)\n- Stage 1:\n - The 2D Convolution has 64 filters of shape (7,7) and uses a stride of (2,2). \n - BatchNorm is applied to the 'channels' axis of the input.\n - ReLU activation is applied.\n - MaxPooling uses a (3,3) window and a (2,2) stride.\n- Stage 2:\n - The convolutional block uses three sets of filters of size [64,64,256], \"f\" is 3, and \"s\" is 1.\n - The 2 identity blocks use three sets of filters of size [64,64,256], and \"f\" is 3.\n- Stage 3:\n - The convolutional block uses three sets of filters of size [128,128,512], \"f\" is 3 and \"s\" is 2.\n - The 3 identity blocks use three sets of filters of size [128,128,512] and \"f\" is 3.\n- Stage 4:\n - The convolutional block uses three sets of filters of size [256, 256, 1024], \"f\" is 3 and \"s\" is 2.\n - The 5 identity blocks use three sets of filters of size [256, 256, 1024] and \"f\" is 3.\n- Stage 5:\n - The convolutional block uses three sets of filters of size [512, 512, 2048], \"f\" is 3 and \"s\" is 2.\n - The 2 identity blocks use three sets of filters of size [512, 512, 2048] and \"f\" is 3.\n- The 2D Average Pooling uses a window of shape (2,2).\n- The 'flatten' layer doesn't have any hyperparameters.\n- The Fully Connected (Dense) layer reduces its input to the number of classes using a softmax activation.\n\n \n<a name='ex-3'></a> \n### Exercise 3 - ResNet50 \n \nImplement the ResNet with 50 layers described in the figure above. We have implemented Stages 1 and 2. Please implement the rest. (The syntax for implementing Stages 3-5 should be quite similar to that of Stage 2) Make sure you follow the naming convention in the text above. \n\nYou'll need to use this function: \n- Average pooling [see reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/AveragePooling2D)\n\nHere are some other functions we used in the code below:\n- Conv2D: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Conv2D)\n- BatchNorm: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/BatchNormalization) (axis: Integer, the axis that should be normalized (typically the features axis))\n- Zero padding: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/ZeroPadding2D)\n- Max pooling: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/MaxPool2D)\n- Fully connected layer: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dense)\n- Addition: [See reference](https://www.tensorflow.org/api_docs/python/tf/keras/layers/Add)",
"_____no_output_____"
]
],
[
[
"# UNQ_C3\n# GRADED FUNCTION: ResNet50\n\ndef ResNet50(input_shape = (64, 64, 3), classes = 6):\n \"\"\"\n Stage-wise implementation of the architecture of the popular ResNet50:\n CONV2D -> BATCHNORM -> RELU -> MAXPOOL -> CONVBLOCK -> IDBLOCK*2 -> CONVBLOCK -> IDBLOCK*3\n -> CONVBLOCK -> IDBLOCK*5 -> CONVBLOCK -> IDBLOCK*2 -> AVGPOOL -> FLATTEN -> DENSE \n\n Arguments:\n input_shape -- shape of the images of the dataset\n classes -- integer, number of classes\n\n Returns:\n model -- a Model() instance in Keras\n \"\"\"\n \n # Define the input as a tensor with shape input_shape\n X_input = Input(input_shape)\n\n \n # Zero-Padding\n X = ZeroPadding2D((3, 3))(X_input)\n \n # Stage 1\n X = Conv2D(64, (7, 7), strides = (2, 2), kernel_initializer = glorot_uniform(seed=0))(X)\n X = BatchNormalization(axis = 3)(X)\n X = Activation('relu')(X)\n X = MaxPooling2D((3, 3), strides=(2, 2))(X)\n\n # Stage 2\n X = convolutional_block(X, f = 3, filters = [64, 64, 256], s = 1)\n X = identity_block(X, 3, [64, 64, 256])\n X = identity_block(X, 3, [64, 64, 256])\n\n ### START CODE HERE\n \n ## Stage 3 (≈4 lines)\n X = convolutional_block(X,f=3,filters=[128,128,512],s=2)\n X = identity_block(X,3,[128,128,512])\n X = identity_block(X,3,[128,128,512])\n X = identity_block(X,3,[128,128,512])\n \n ## Stage 4 (≈6 lines)\n X = convolutional_block(X,f=3,filters=[256,256,1024],s=2)\n X = identity_block(X,3,[256,256,1024])\n X = identity_block(X,3,[256,256,1024])\n X = identity_block(X,3,[256,256,1024])\n X = identity_block(X,3,[256,256,1024])\n X = identity_block(X,3,[256,256,1024])\n ## Stage 5 (≈3 lines)\n X = convolutional_block(X,f=3,filters=[512,512,2048],s=2)\n X = identity_block(X, 3, [512, 512, 2048])\n X = identity_block(X, 3, [512, 512, 2048])\n\n ## AVGPOOL (≈1 line). Use \"X = AveragePooling2D(...)(X)\"\n X = AveragePooling2D(pool_size=(2,2),padding='same')(X)\n \n ### END CODE HERE\n\n # output layer\n X = Flatten()(X)\n X = Dense(classes, activation='softmax', kernel_initializer = glorot_uniform(seed=0))(X)\n \n \n # Create model\n model = Model(inputs = X_input, outputs = X)\n\n return model",
"_____no_output_____"
]
],
[
[
"Run the following code to build the model's graph. If your implementation is incorrect, you'll know it by checking your accuracy when running `model.fit(...)` below.",
"_____no_output_____"
]
],
[
[
"model = ResNet50(input_shape = (64, 64, 3), classes = 6)\nprint(model.summary())",
"Model: \"functional_1\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 64, 64, 3)] 0 \n__________________________________________________________________________________________________\nzero_padding2d (ZeroPadding2D) (None, 70, 70, 3) 0 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 32, 32, 64) 9472 zero_padding2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 32, 32, 64) 256 conv2d_20[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 32, 32, 64) 0 batch_normalization_20[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 15, 15, 64) 0 activation_18[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 15, 15, 64) 4160 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 15, 15, 64) 256 conv2d_21[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 15, 15, 64) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 15, 15, 64) 36928 activation_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 15, 15, 64) 256 conv2d_22[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 15, 15, 64) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 15, 15, 256) 16640 activation_20[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 15, 15, 256) 16640 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 15, 15, 256) 1024 conv2d_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 15, 15, 256) 1024 conv2d_24[0][0] \n__________________________________________________________________________________________________\nadd_6 (Add) (None, 15, 15, 256) 0 batch_normalization_23[0][0] \n batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 15, 15, 256) 0 add_6[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 15, 15, 64) 16448 activation_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 15, 15, 64) 256 conv2d_25[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 15, 15, 64) 0 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 15, 15, 64) 36928 activation_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 15, 15, 64) 256 conv2d_26[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 15, 15, 64) 0 batch_normalization_26[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 15, 15, 256) 16640 activation_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 15, 15, 256) 1024 conv2d_27[0][0] \n__________________________________________________________________________________________________\nadd_7 (Add) (None, 15, 15, 256) 0 batch_normalization_27[0][0] \n activation_21[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 15, 15, 256) 0 add_7[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 15, 15, 64) 16448 activation_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 15, 15, 64) 256 conv2d_28[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 15, 15, 64) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 15, 15, 64) 36928 activation_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 15, 15, 64) 256 conv2d_29[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 15, 15, 64) 0 batch_normalization_29[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 15, 15, 256) 16640 activation_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 15, 15, 256) 1024 conv2d_30[0][0] \n__________________________________________________________________________________________________\nadd_8 (Add) (None, 15, 15, 256) 0 batch_normalization_30[0][0] \n activation_24[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 15, 15, 256) 0 add_8[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 8, 8, 128) 32896 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 8, 8, 128) 512 conv2d_31[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 8, 8, 128) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 8, 8, 128) 147584 activation_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 8, 8, 128) 512 conv2d_32[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 8, 8, 128) 0 batch_normalization_32[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 8, 8, 512) 66048 activation_29[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 8, 8, 512) 131584 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 8, 8, 512) 2048 conv2d_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 8, 8, 512) 2048 conv2d_34[0][0] \n__________________________________________________________________________________________________\nadd_9 (Add) (None, 8, 8, 512) 0 batch_normalization_33[0][0] \n batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 8, 8, 512) 0 add_9[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 8, 8, 128) 65664 activation_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 8, 8, 128) 512 conv2d_35[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 8, 8, 128) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 8, 8, 128) 147584 activation_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 8, 8, 128) 512 conv2d_36[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 8, 8, 128) 0 batch_normalization_36[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 8, 8, 512) 66048 activation_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 8, 8, 512) 2048 conv2d_37[0][0] \n__________________________________________________________________________________________________\nadd_10 (Add) (None, 8, 8, 512) 0 batch_normalization_37[0][0] \n activation_30[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 8, 8, 512) 0 add_10[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 8, 8, 128) 65664 activation_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 8, 8, 128) 512 conv2d_38[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 8, 8, 128) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 8, 8, 128) 147584 activation_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 8, 8, 128) 512 conv2d_39[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 8, 8, 128) 0 batch_normalization_39[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 8, 8, 512) 66048 activation_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 8, 8, 512) 2048 conv2d_40[0][0] \n__________________________________________________________________________________________________\nadd_11 (Add) (None, 8, 8, 512) 0 batch_normalization_40[0][0] \n activation_33[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 8, 8, 512) 0 add_11[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 8, 8, 128) 65664 activation_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 8, 8, 128) 512 conv2d_41[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 8, 8, 128) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 8, 8, 128) 147584 activation_37[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 8, 8, 128) 512 conv2d_42[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 8, 8, 128) 0 batch_normalization_42[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 8, 8, 512) 66048 activation_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 8, 8, 512) 2048 conv2d_43[0][0] \n__________________________________________________________________________________________________\nadd_12 (Add) (None, 8, 8, 512) 0 batch_normalization_43[0][0] \n activation_36[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 8, 8, 512) 0 add_12[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 4, 4, 256) 131328 activation_39[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 4, 4, 256) 1024 conv2d_44[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 4, 4, 256) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 4, 4, 256) 590080 activation_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 4, 4, 256) 1024 conv2d_45[0][0] \n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 4, 4, 256) 0 batch_normalization_45[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 4, 4, 1024) 263168 activation_41[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 4, 4, 1024) 525312 activation_39[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 4, 4, 1024) 4096 conv2d_46[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 4, 4, 1024) 4096 conv2d_47[0][0] \n__________________________________________________________________________________________________\nadd_13 (Add) (None, 4, 4, 1024) 0 batch_normalization_46[0][0] \n batch_normalization_47[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 4, 4, 1024) 0 add_13[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 4, 4, 256) 262400 activation_42[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 4, 4, 256) 1024 conv2d_48[0][0] \n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 4, 4, 256) 0 batch_normalization_48[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 4, 4, 256) 590080 activation_43[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 4, 4, 256) 1024 conv2d_49[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 4, 4, 256) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 4, 4, 1024) 263168 activation_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 4, 4, 1024) 4096 conv2d_50[0][0] \n__________________________________________________________________________________________________\nadd_14 (Add) (None, 4, 4, 1024) 0 batch_normalization_50[0][0] \n activation_42[0][0] \n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 4, 4, 1024) 0 add_14[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 4, 4, 256) 262400 activation_45[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 4, 4, 256) 1024 conv2d_51[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 4, 4, 256) 0 batch_normalization_51[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 4, 4, 256) 590080 activation_46[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 4, 4, 256) 1024 conv2d_52[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 4, 4, 256) 0 batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 4, 4, 1024) 263168 activation_47[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 4, 4, 1024) 4096 conv2d_53[0][0] \n__________________________________________________________________________________________________\nadd_15 (Add) (None, 4, 4, 1024) 0 batch_normalization_53[0][0] \n activation_45[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 4, 4, 1024) 0 add_15[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 4, 4, 256) 262400 activation_48[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 4, 4, 256) 1024 conv2d_54[0][0] \n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 4, 4, 256) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 4, 4, 256) 590080 activation_49[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 4, 4, 256) 1024 conv2d_55[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 4, 4, 256) 0 batch_normalization_55[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 4, 4, 1024) 263168 activation_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 4, 4, 1024) 4096 conv2d_56[0][0] \n__________________________________________________________________________________________________\nadd_16 (Add) (None, 4, 4, 1024) 0 batch_normalization_56[0][0] \n activation_48[0][0] \n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 4, 4, 1024) 0 add_16[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 4, 4, 256) 262400 activation_51[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 4, 4, 256) 1024 conv2d_57[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 4, 4, 256) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 4, 4, 256) 590080 activation_52[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 4, 4, 256) 1024 conv2d_58[0][0] \n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 4, 4, 256) 0 batch_normalization_58[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 4, 4, 1024) 263168 activation_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_59 (BatchNo (None, 4, 4, 1024) 4096 conv2d_59[0][0] \n__________________________________________________________________________________________________\nadd_17 (Add) (None, 4, 4, 1024) 0 batch_normalization_59[0][0] \n activation_51[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 4, 4, 1024) 0 add_17[0][0] \n__________________________________________________________________________________________________\nconv2d_60 (Conv2D) (None, 4, 4, 256) 262400 activation_54[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_60 (BatchNo (None, 4, 4, 256) 1024 conv2d_60[0][0] \n__________________________________________________________________________________________________\nactivation_55 (Activation) (None, 4, 4, 256) 0 batch_normalization_60[0][0] \n__________________________________________________________________________________________________\nconv2d_61 (Conv2D) (None, 4, 4, 256) 590080 activation_55[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_61 (BatchNo (None, 4, 4, 256) 1024 conv2d_61[0][0] \n__________________________________________________________________________________________________\nactivation_56 (Activation) (None, 4, 4, 256) 0 batch_normalization_61[0][0] \n__________________________________________________________________________________________________\nconv2d_62 (Conv2D) (None, 4, 4, 1024) 263168 activation_56[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_62 (BatchNo (None, 4, 4, 1024) 4096 conv2d_62[0][0] \n__________________________________________________________________________________________________\nadd_18 (Add) (None, 4, 4, 1024) 0 batch_normalization_62[0][0] \n activation_54[0][0] \n__________________________________________________________________________________________________\nactivation_57 (Activation) (None, 4, 4, 1024) 0 add_18[0][0] \n__________________________________________________________________________________________________\nconv2d_63 (Conv2D) (None, 2, 2, 512) 524800 activation_57[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_63 (BatchNo (None, 2, 2, 512) 2048 conv2d_63[0][0] \n__________________________________________________________________________________________________\nactivation_58 (Activation) (None, 2, 2, 512) 0 batch_normalization_63[0][0] \n__________________________________________________________________________________________________\nconv2d_64 (Conv2D) (None, 2, 2, 512) 2359808 activation_58[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_64 (BatchNo (None, 2, 2, 512) 2048 conv2d_64[0][0] \n__________________________________________________________________________________________________\nactivation_59 (Activation) (None, 2, 2, 512) 0 batch_normalization_64[0][0] \n__________________________________________________________________________________________________\nconv2d_65 (Conv2D) (None, 2, 2, 2048) 1050624 activation_59[0][0] \n__________________________________________________________________________________________________\nconv2d_66 (Conv2D) (None, 2, 2, 2048) 2099200 activation_57[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_65 (BatchNo (None, 2, 2, 2048) 8192 conv2d_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_66 (BatchNo (None, 2, 2, 2048) 8192 conv2d_66[0][0] \n__________________________________________________________________________________________________\nadd_19 (Add) (None, 2, 2, 2048) 0 batch_normalization_65[0][0] \n batch_normalization_66[0][0] \n__________________________________________________________________________________________________\nactivation_60 (Activation) (None, 2, 2, 2048) 0 add_19[0][0] \n__________________________________________________________________________________________________\nconv2d_67 (Conv2D) (None, 2, 2, 512) 1049088 activation_60[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_67 (BatchNo (None, 2, 2, 512) 2048 conv2d_67[0][0] \n__________________________________________________________________________________________________\nactivation_61 (Activation) (None, 2, 2, 512) 0 batch_normalization_67[0][0] \n__________________________________________________________________________________________________\nconv2d_68 (Conv2D) (None, 2, 2, 512) 2359808 activation_61[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_68 (BatchNo (None, 2, 2, 512) 2048 conv2d_68[0][0] \n__________________________________________________________________________________________________\nactivation_62 (Activation) (None, 2, 2, 512) 0 batch_normalization_68[0][0] \n__________________________________________________________________________________________________\nconv2d_69 (Conv2D) (None, 2, 2, 2048) 1050624 activation_62[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_69 (BatchNo (None, 2, 2, 2048) 8192 conv2d_69[0][0] \n__________________________________________________________________________________________________\nadd_20 (Add) (None, 2, 2, 2048) 0 batch_normalization_69[0][0] \n activation_60[0][0] \n__________________________________________________________________________________________________\nactivation_63 (Activation) (None, 2, 2, 2048) 0 add_20[0][0] \n__________________________________________________________________________________________________\nconv2d_70 (Conv2D) (None, 2, 2, 512) 1049088 activation_63[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_70 (BatchNo (None, 2, 2, 512) 2048 conv2d_70[0][0] \n__________________________________________________________________________________________________\nactivation_64 (Activation) (None, 2, 2, 512) 0 batch_normalization_70[0][0] \n__________________________________________________________________________________________________\nconv2d_71 (Conv2D) (None, 2, 2, 512) 2359808 activation_64[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_71 (BatchNo (None, 2, 2, 512) 2048 conv2d_71[0][0] \n__________________________________________________________________________________________________\nactivation_65 (Activation) (None, 2, 2, 512) 0 batch_normalization_71[0][0] \n__________________________________________________________________________________________________\nconv2d_72 (Conv2D) (None, 2, 2, 2048) 1050624 activation_65[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_72 (BatchNo (None, 2, 2, 2048) 8192 conv2d_72[0][0] \n__________________________________________________________________________________________________\nadd_21 (Add) (None, 2, 2, 2048) 0 batch_normalization_72[0][0] \n activation_63[0][0] \n__________________________________________________________________________________________________\nactivation_66 (Activation) (None, 2, 2, 2048) 0 add_21[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d (AveragePooli (None, 1, 1, 2048) 0 activation_66[0][0] \n__________________________________________________________________________________________________\nflatten (Flatten) (None, 2048) 0 average_pooling2d[0][0] \n__________________________________________________________________________________________________\ndense (Dense) (None, 6) 12294 flatten[0][0] \n==================================================================================================\nTotal params: 23,600,006\nTrainable params: 23,546,886\nNon-trainable params: 53,120\n__________________________________________________________________________________________________\nNone\n"
],
[
"from outputs import ResNet50_summary\n\nmodel = ResNet50(input_shape = (64, 64, 3), classes = 6)\n\ncomparator(summary(model), ResNet50_summary)\n",
"\u001b[32mAll tests passed!\u001b[0m\n"
]
],
[
[
"As shown in the Keras Tutorial Notebook, prior to training a model, you need to configure the learning process by compiling the model.",
"_____no_output_____"
]
],
[
[
"model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['accuracy'])",
"_____no_output_____"
]
],
[
[
"The model is now ready to be trained. The only thing you need now is a dataset!",
"_____no_output_____"
],
[
"Let's load your old friend, the SIGNS dataset.\n\n<img src=\"images/signs_data_kiank.png\" style=\"width:450px;height:250px;\">\n<caption><center> <u> <font color='purple'> <b>Figure 6</b> </u><font color='purple'> : <b>SIGNS dataset</b> </center></caption>\n",
"_____no_output_____"
]
],
[
[
"X_train_orig, Y_train_orig, X_test_orig, Y_test_orig, classes = load_dataset()\n\n# Normalize image vectors\nX_train = X_train_orig / 255.\nX_test = X_test_orig / 255.\n\n# Convert training and test labels to one hot matrices\nY_train = convert_to_one_hot(Y_train_orig, 6).T\nY_test = convert_to_one_hot(Y_test_orig, 6).T\n\nprint (\"number of training examples = \" + str(X_train.shape[0]))\nprint (\"number of test examples = \" + str(X_test.shape[0]))\nprint (\"X_train shape: \" + str(X_train.shape))\nprint (\"Y_train shape: \" + str(Y_train.shape))\nprint (\"X_test shape: \" + str(X_test.shape))\nprint (\"Y_test shape: \" + str(Y_test.shape))",
"number of training examples = 1080\nnumber of test examples = 120\nX_train shape: (1080, 64, 64, 3)\nY_train shape: (1080, 6)\nX_test shape: (120, 64, 64, 3)\nY_test shape: (120, 6)\n"
]
],
[
[
"Run the following cell to train your model on 10 epochs with a batch size of 32. On a GPU, it should take less than 2 minutes. ",
"_____no_output_____"
]
],
[
[
"model.fit(X_train, Y_train, epochs = 10, batch_size = 32)",
"Epoch 1/10\n34/34 [==============================] - 1s 29ms/step - loss: 1.8387 - accuracy: 0.5046\nEpoch 2/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.5694 - accuracy: 0.8019\nEpoch 3/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.3907 - accuracy: 0.8713\nEpoch 4/10\n34/34 [==============================] - 1s 24ms/step - loss: 0.2689 - accuracy: 0.9194\nEpoch 5/10\n34/34 [==============================] - 1s 24ms/step - loss: 0.3365 - accuracy: 0.8944\nEpoch 6/10\n34/34 [==============================] - 1s 26ms/step - loss: 0.2075 - accuracy: 0.9361\nEpoch 7/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.1412 - accuracy: 0.9528\nEpoch 8/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.1446 - accuracy: 0.9528\nEpoch 9/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.1769 - accuracy: 0.9491\nEpoch 10/10\n34/34 [==============================] - 1s 23ms/step - loss: 0.1370 - accuracy: 0.9565\n"
]
],
[
[
"**Expected Output**:\n\n```\nEpoch 1/10\n34/34 [==============================] - 1s 34ms/step - loss: 1.9241 - accuracy: 0.4620\nEpoch 2/10\n34/34 [==============================] - 2s 57ms/step - loss: 0.6403 - accuracy: 0.7898\nEpoch 3/10\n34/34 [==============================] - 1s 24ms/step - loss: 0.3744 - accuracy: 0.8731\nEpoch 4/10\n34/34 [==============================] - 2s 44ms/step - loss: 0.2220 - accuracy: 0.9231\nEpoch 5/10\n34/34 [==============================] - 2s 57ms/step - loss: 0.1333 - accuracy: 0.9583\nEpoch 6/10\n34/34 [==============================] - 2s 52ms/step - loss: 0.2243 - accuracy: 0.9444\nEpoch 7/10\n34/34 [==============================] - 2s 48ms/step - loss: 0.2913 - accuracy: 0.9102\nEpoch 8/10\n34/34 [==============================] - 1s 30ms/step - loss: 0.2269 - accuracy: 0.9306\nEpoch 9/10\n34/34 [==============================] - 2s 46ms/step - loss: 0.1113 - accuracy: 0.9630\nEpoch 10/10\n34/34 [==============================] - 2s 57ms/step - loss: 0.0709 - accuracy: 0.9778\n```\n\nThe exact values could not match, but don't worry about that. The important thing that you must see is that the loss value decreases, and the accuracy increases for the firsts 5 epochs.",
"_____no_output_____"
],
[
"Let's see how this model (trained on only two epochs) performs on the test set.",
"_____no_output_____"
]
],
[
[
"preds = model.evaluate(X_test, Y_test)\nprint (\"Loss = \" + str(preds[0]))\nprint (\"Test Accuracy = \" + str(preds[1]))",
"4/4 [==============================] - 0s 7ms/step - loss: 0.1846 - accuracy: 0.9333\nLoss = 0.184574156999588\nTest Accuracy = 0.9333333373069763\n"
]
],
[
[
"**Expected Output**:\n\n<table>\n <tr>\n <td>\n <b>Test Accuracy</b>\n </td>\n <td>\n >0.80\n </td>\n </tr>\n\n</table>",
"_____no_output_____"
],
[
"For the purposes of this assignment, you've been asked to train the model for ten epochs. You can see that it performs well. The online grader will only run your code for a small number of epochs as well. Please go ahead and submit your assignment. ",
"_____no_output_____"
],
[
"After you have finished this official (graded) part of this assignment, you can also optionally train the ResNet for more iterations, if you want. It tends to get much better performance when trained for ~20 epochs, but this does take more than an hour when training on a CPU. \n\nUsing a GPU, this ResNet50 model's weights were trained on the SIGNS dataset. You can load and run the trained model on the test set in the cells below. It may take ≈1min to load the model. Have fun! ",
"_____no_output_____"
]
],
[
[
"pre_trained_model = tf.keras.models.load_model('resnet50.h5')",
"_____no_output_____"
],
[
"preds = pre_trained_model.evaluate(X_test, Y_test)\nprint (\"Loss = \" + str(preds[0]))\nprint (\"Test Accuracy = \" + str(preds[1]))",
"4/4 [==============================] - 0s 7ms/step - loss: 0.1596 - accuracy: 0.9500\nLoss = 0.15958689153194427\nTest Accuracy = 0.949999988079071\n"
]
],
[
[
"**Congratulations** on finishing this assignment! You've now implemented a state-of-the-art image classification system! Woo hoo! \n\nResNet50 is a powerful model for image classification when it's trained for an adequate number of iterations. Hopefully, from this point, you can use what you've learned and apply it to your own classification problem to perform state-of-the-art accuracy.",
"_____no_output_____"
],
[
"<font color = 'blue'>\n\n**What you should remember**:\n\n- Very deep \"plain\" networks don't work in practice because vanishing gradients make them hard to train. \n- Skip connections help address the Vanishing Gradient problem. They also make it easy for a ResNet block to learn an identity function. \n- There are two main types of blocks: The **identity block** and the **convolutional block**. \n- Very deep Residual Networks are built by stacking these blocks together.",
"_____no_output_____"
],
[
"<a name='5'></a> \n## 5 - Test on Your Own Image (Optional/Ungraded)",
"_____no_output_____"
],
[
"If you wish, you can also take a picture of your own hand and see the output of the model. To do this:\n 1. Click on \"File\" in the upper bar of this notebook, then click \"Open\" to go on your Coursera Hub.\n 2. Add your image to this Jupyter Notebook's directory, in the \"images\" folder\n 3. Write your image's name in the following code\n 4. Run the code and check if the algorithm is right! ",
"_____no_output_____"
]
],
[
[
"img_path = 'images/my_image.jpg'\nimg = image.load_img(img_path, target_size=(64, 64))\nx = image.img_to_array(img)\nx = np.expand_dims(x, axis=0)\nx = x/255.0\nprint('Input image shape:', x.shape)\nimshow(img)\nprediction = pre_trained_model.predict(x)\nprint(\"Class prediction vector [p(0), p(1), p(2), p(3), p(4), p(5)] = \", prediction)\nprint(\"Class:\", np.argmax(prediction))\n",
"Input image shape: (1, 64, 64, 3)\nClass prediction vector [p(0), p(1), p(2), p(3), p(4), p(5)] = [[9.2633512e-05 4.2629417e-02 9.2548847e-01 4.1606426e-04 3.1337839e-02\n 3.5633519e-05]]\nClass: 2\n"
]
],
[
[
"Even though the model has high accuracy, it might be performing poorly on your own set of images. Notice that, the shape of the pictures, the lighting where the photos were taken, and all of the preprocessing steps can have an impact on the performance of the model. Considering everything you have learned in this specialization so far, what do you think might be the cause here?\n\n*Hint*: It might be related to some distributions. Can you come up with a potential solution ?",
"_____no_output_____"
],
[
"You can also print a summary of your model by running the following code.",
"_____no_output_____"
]
],
[
[
"pre_trained_model.summary()",
"Model: \"ResNet50\"\n__________________________________________________________________________________________________\nLayer (type) Output Shape Param # Connected to \n==================================================================================================\ninput_1 (InputLayer) [(None, 64, 64, 3)] 0 \n__________________________________________________________________________________________________\nzero_padding2d (ZeroPadding2D) (None, 70, 70, 3) 0 input_1[0][0] \n__________________________________________________________________________________________________\nconv2d_7 (Conv2D) (None, 32, 32, 64) 9472 zero_padding2d[0][0] \n__________________________________________________________________________________________________\nbn_conv1 (BatchNormalization) (None, 32, 32, 64) 256 conv2d_7[0][0] \n__________________________________________________________________________________________________\nactivation_6 (Activation) (None, 32, 32, 64) 0 bn_conv1[0][0] \n__________________________________________________________________________________________________\nmax_pooling2d (MaxPooling2D) (None, 15, 15, 64) 0 activation_6[0][0] \n__________________________________________________________________________________________________\nconv2d_8 (Conv2D) (None, 15, 15, 64) 4160 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_7 (BatchNor (None, 15, 15, 64) 256 conv2d_8[0][0] \n__________________________________________________________________________________________________\nactivation_7 (Activation) (None, 15, 15, 64) 0 batch_normalization_7[0][0] \n__________________________________________________________________________________________________\nconv2d_9 (Conv2D) (None, 15, 15, 64) 36928 activation_7[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_8 (BatchNor (None, 15, 15, 64) 256 conv2d_9[0][0] \n__________________________________________________________________________________________________\nactivation_8 (Activation) (None, 15, 15, 64) 0 batch_normalization_8[0][0] \n__________________________________________________________________________________________________\nconv2d_10 (Conv2D) (None, 15, 15, 256) 16640 activation_8[0][0] \n__________________________________________________________________________________________________\nconv2d_11 (Conv2D) (None, 15, 15, 256) 16640 max_pooling2d[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_9 (BatchNor (None, 15, 15, 256) 1024 conv2d_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_10 (BatchNo (None, 15, 15, 256) 1024 conv2d_11[0][0] \n__________________________________________________________________________________________________\nadd_2 (Add) (None, 15, 15, 256) 0 batch_normalization_9[0][0] \n batch_normalization_10[0][0] \n__________________________________________________________________________________________________\nactivation_9 (Activation) (None, 15, 15, 256) 0 add_2[0][0] \n__________________________________________________________________________________________________\nconv2d_12 (Conv2D) (None, 15, 15, 64) 16448 activation_9[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_11 (BatchNo (None, 15, 15, 64) 256 conv2d_12[0][0] \n__________________________________________________________________________________________________\nactivation_10 (Activation) (None, 15, 15, 64) 0 batch_normalization_11[0][0] \n__________________________________________________________________________________________________\nconv2d_13 (Conv2D) (None, 15, 15, 64) 36928 activation_10[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_12 (BatchNo (None, 15, 15, 64) 256 conv2d_13[0][0] \n__________________________________________________________________________________________________\nactivation_11 (Activation) (None, 15, 15, 64) 0 batch_normalization_12[0][0] \n__________________________________________________________________________________________________\nconv2d_14 (Conv2D) (None, 15, 15, 256) 16640 activation_11[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_13 (BatchNo (None, 15, 15, 256) 1024 conv2d_14[0][0] \n__________________________________________________________________________________________________\nadd_3 (Add) (None, 15, 15, 256) 0 batch_normalization_13[0][0] \n activation_9[0][0] \n__________________________________________________________________________________________________\nactivation_12 (Activation) (None, 15, 15, 256) 0 add_3[0][0] \n__________________________________________________________________________________________________\nconv2d_15 (Conv2D) (None, 15, 15, 64) 16448 activation_12[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_14 (BatchNo (None, 15, 15, 64) 256 conv2d_15[0][0] \n__________________________________________________________________________________________________\nactivation_13 (Activation) (None, 15, 15, 64) 0 batch_normalization_14[0][0] \n__________________________________________________________________________________________________\nconv2d_16 (Conv2D) (None, 15, 15, 64) 36928 activation_13[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_15 (BatchNo (None, 15, 15, 64) 256 conv2d_16[0][0] \n__________________________________________________________________________________________________\nactivation_14 (Activation) (None, 15, 15, 64) 0 batch_normalization_15[0][0] \n__________________________________________________________________________________________________\nconv2d_17 (Conv2D) (None, 15, 15, 256) 16640 activation_14[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_16 (BatchNo (None, 15, 15, 256) 1024 conv2d_17[0][0] \n__________________________________________________________________________________________________\nadd_4 (Add) (None, 15, 15, 256) 0 batch_normalization_16[0][0] \n activation_12[0][0] \n__________________________________________________________________________________________________\nactivation_15 (Activation) (None, 15, 15, 256) 0 add_4[0][0] \n__________________________________________________________________________________________________\nconv2d_18 (Conv2D) (None, 8, 8, 128) 32896 activation_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_17 (BatchNo (None, 8, 8, 128) 512 conv2d_18[0][0] \n__________________________________________________________________________________________________\nactivation_16 (Activation) (None, 8, 8, 128) 0 batch_normalization_17[0][0] \n__________________________________________________________________________________________________\nconv2d_19 (Conv2D) (None, 8, 8, 128) 147584 activation_16[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_18 (BatchNo (None, 8, 8, 128) 512 conv2d_19[0][0] \n__________________________________________________________________________________________________\nactivation_17 (Activation) (None, 8, 8, 128) 0 batch_normalization_18[0][0] \n__________________________________________________________________________________________________\nconv2d_20 (Conv2D) (None, 8, 8, 512) 66048 activation_17[0][0] \n__________________________________________________________________________________________________\nconv2d_21 (Conv2D) (None, 8, 8, 512) 131584 activation_15[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_19 (BatchNo (None, 8, 8, 512) 2048 conv2d_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_20 (BatchNo (None, 8, 8, 512) 2048 conv2d_21[0][0] \n__________________________________________________________________________________________________\nadd_5 (Add) (None, 8, 8, 512) 0 batch_normalization_19[0][0] \n batch_normalization_20[0][0] \n__________________________________________________________________________________________________\nactivation_18 (Activation) (None, 8, 8, 512) 0 add_5[0][0] \n__________________________________________________________________________________________________\nconv2d_22 (Conv2D) (None, 8, 8, 128) 65664 activation_18[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_21 (BatchNo (None, 8, 8, 128) 512 conv2d_22[0][0] \n__________________________________________________________________________________________________\nactivation_19 (Activation) (None, 8, 8, 128) 0 batch_normalization_21[0][0] \n__________________________________________________________________________________________________\nconv2d_23 (Conv2D) (None, 8, 8, 128) 147584 activation_19[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_22 (BatchNo (None, 8, 8, 128) 512 conv2d_23[0][0] \n__________________________________________________________________________________________________\nactivation_20 (Activation) (None, 8, 8, 128) 0 batch_normalization_22[0][0] \n__________________________________________________________________________________________________\nconv2d_24 (Conv2D) (None, 8, 8, 512) 66048 activation_20[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_23 (BatchNo (None, 8, 8, 512) 2048 conv2d_24[0][0] \n__________________________________________________________________________________________________\nadd_6 (Add) (None, 8, 8, 512) 0 batch_normalization_23[0][0] \n activation_18[0][0] \n__________________________________________________________________________________________________\nactivation_21 (Activation) (None, 8, 8, 512) 0 add_6[0][0] \n__________________________________________________________________________________________________\nconv2d_25 (Conv2D) (None, 8, 8, 128) 65664 activation_21[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_24 (BatchNo (None, 8, 8, 128) 512 conv2d_25[0][0] \n__________________________________________________________________________________________________\nactivation_22 (Activation) (None, 8, 8, 128) 0 batch_normalization_24[0][0] \n__________________________________________________________________________________________________\nconv2d_26 (Conv2D) (None, 8, 8, 128) 147584 activation_22[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_25 (BatchNo (None, 8, 8, 128) 512 conv2d_26[0][0] \n__________________________________________________________________________________________________\nactivation_23 (Activation) (None, 8, 8, 128) 0 batch_normalization_25[0][0] \n__________________________________________________________________________________________________\nconv2d_27 (Conv2D) (None, 8, 8, 512) 66048 activation_23[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_26 (BatchNo (None, 8, 8, 512) 2048 conv2d_27[0][0] \n__________________________________________________________________________________________________\nadd_7 (Add) (None, 8, 8, 512) 0 batch_normalization_26[0][0] \n activation_21[0][0] \n__________________________________________________________________________________________________\nactivation_24 (Activation) (None, 8, 8, 512) 0 add_7[0][0] \n__________________________________________________________________________________________________\nconv2d_28 (Conv2D) (None, 8, 8, 128) 65664 activation_24[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_27 (BatchNo (None, 8, 8, 128) 512 conv2d_28[0][0] \n__________________________________________________________________________________________________\nactivation_25 (Activation) (None, 8, 8, 128) 0 batch_normalization_27[0][0] \n__________________________________________________________________________________________________\nconv2d_29 (Conv2D) (None, 8, 8, 128) 147584 activation_25[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_28 (BatchNo (None, 8, 8, 128) 512 conv2d_29[0][0] \n__________________________________________________________________________________________________\nactivation_26 (Activation) (None, 8, 8, 128) 0 batch_normalization_28[0][0] \n__________________________________________________________________________________________________\nconv2d_30 (Conv2D) (None, 8, 8, 512) 66048 activation_26[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_29 (BatchNo (None, 8, 8, 512) 2048 conv2d_30[0][0] \n__________________________________________________________________________________________________\nadd_8 (Add) (None, 8, 8, 512) 0 batch_normalization_29[0][0] \n activation_24[0][0] \n__________________________________________________________________________________________________\nactivation_27 (Activation) (None, 8, 8, 512) 0 add_8[0][0] \n__________________________________________________________________________________________________\nconv2d_31 (Conv2D) (None, 4, 4, 256) 131328 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_30 (BatchNo (None, 4, 4, 256) 1024 conv2d_31[0][0] \n__________________________________________________________________________________________________\nactivation_28 (Activation) (None, 4, 4, 256) 0 batch_normalization_30[0][0] \n__________________________________________________________________________________________________\nconv2d_32 (Conv2D) (None, 4, 4, 256) 590080 activation_28[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_31 (BatchNo (None, 4, 4, 256) 1024 conv2d_32[0][0] \n__________________________________________________________________________________________________\nactivation_29 (Activation) (None, 4, 4, 256) 0 batch_normalization_31[0][0] \n__________________________________________________________________________________________________\nconv2d_33 (Conv2D) (None, 4, 4, 1024) 263168 activation_29[0][0] \n__________________________________________________________________________________________________\nconv2d_34 (Conv2D) (None, 4, 4, 1024) 525312 activation_27[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_32 (BatchNo (None, 4, 4, 1024) 4096 conv2d_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_33 (BatchNo (None, 4, 4, 1024) 4096 conv2d_34[0][0] \n__________________________________________________________________________________________________\nadd_9 (Add) (None, 4, 4, 1024) 0 batch_normalization_32[0][0] \n batch_normalization_33[0][0] \n__________________________________________________________________________________________________\nactivation_30 (Activation) (None, 4, 4, 1024) 0 add_9[0][0] \n__________________________________________________________________________________________________\nconv2d_35 (Conv2D) (None, 4, 4, 256) 262400 activation_30[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_34 (BatchNo (None, 4, 4, 256) 1024 conv2d_35[0][0] \n__________________________________________________________________________________________________\nactivation_31 (Activation) (None, 4, 4, 256) 0 batch_normalization_34[0][0] \n__________________________________________________________________________________________________\nconv2d_36 (Conv2D) (None, 4, 4, 256) 590080 activation_31[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_35 (BatchNo (None, 4, 4, 256) 1024 conv2d_36[0][0] \n__________________________________________________________________________________________________\nactivation_32 (Activation) (None, 4, 4, 256) 0 batch_normalization_35[0][0] \n__________________________________________________________________________________________________\nconv2d_37 (Conv2D) (None, 4, 4, 1024) 263168 activation_32[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_36 (BatchNo (None, 4, 4, 1024) 4096 conv2d_37[0][0] \n__________________________________________________________________________________________________\nadd_10 (Add) (None, 4, 4, 1024) 0 batch_normalization_36[0][0] \n activation_30[0][0] \n__________________________________________________________________________________________________\nactivation_33 (Activation) (None, 4, 4, 1024) 0 add_10[0][0] \n__________________________________________________________________________________________________\nconv2d_38 (Conv2D) (None, 4, 4, 256) 262400 activation_33[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_37 (BatchNo (None, 4, 4, 256) 1024 conv2d_38[0][0] \n__________________________________________________________________________________________________\nactivation_34 (Activation) (None, 4, 4, 256) 0 batch_normalization_37[0][0] \n__________________________________________________________________________________________________\nconv2d_39 (Conv2D) (None, 4, 4, 256) 590080 activation_34[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_38 (BatchNo (None, 4, 4, 256) 1024 conv2d_39[0][0] \n__________________________________________________________________________________________________\nactivation_35 (Activation) (None, 4, 4, 256) 0 batch_normalization_38[0][0] \n__________________________________________________________________________________________________\nconv2d_40 (Conv2D) (None, 4, 4, 1024) 263168 activation_35[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_39 (BatchNo (None, 4, 4, 1024) 4096 conv2d_40[0][0] \n__________________________________________________________________________________________________\nadd_11 (Add) (None, 4, 4, 1024) 0 batch_normalization_39[0][0] \n activation_33[0][0] \n__________________________________________________________________________________________________\nactivation_36 (Activation) (None, 4, 4, 1024) 0 add_11[0][0] \n__________________________________________________________________________________________________\nconv2d_41 (Conv2D) (None, 4, 4, 256) 262400 activation_36[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_40 (BatchNo (None, 4, 4, 256) 1024 conv2d_41[0][0] \n__________________________________________________________________________________________________\nactivation_37 (Activation) (None, 4, 4, 256) 0 batch_normalization_40[0][0] \n__________________________________________________________________________________________________\nconv2d_42 (Conv2D) (None, 4, 4, 256) 590080 activation_37[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_41 (BatchNo (None, 4, 4, 256) 1024 conv2d_42[0][0] \n__________________________________________________________________________________________________\nactivation_38 (Activation) (None, 4, 4, 256) 0 batch_normalization_41[0][0] \n__________________________________________________________________________________________________\nconv2d_43 (Conv2D) (None, 4, 4, 1024) 263168 activation_38[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_42 (BatchNo (None, 4, 4, 1024) 4096 conv2d_43[0][0] \n__________________________________________________________________________________________________\nadd_12 (Add) (None, 4, 4, 1024) 0 batch_normalization_42[0][0] \n activation_36[0][0] \n__________________________________________________________________________________________________\nactivation_39 (Activation) (None, 4, 4, 1024) 0 add_12[0][0] \n__________________________________________________________________________________________________\nconv2d_44 (Conv2D) (None, 4, 4, 256) 262400 activation_39[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_43 (BatchNo (None, 4, 4, 256) 1024 conv2d_44[0][0] \n__________________________________________________________________________________________________\nactivation_40 (Activation) (None, 4, 4, 256) 0 batch_normalization_43[0][0] \n__________________________________________________________________________________________________\nconv2d_45 (Conv2D) (None, 4, 4, 256) 590080 activation_40[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_44 (BatchNo (None, 4, 4, 256) 1024 conv2d_45[0][0] \n__________________________________________________________________________________________________\nactivation_41 (Activation) (None, 4, 4, 256) 0 batch_normalization_44[0][0] \n__________________________________________________________________________________________________\nconv2d_46 (Conv2D) (None, 4, 4, 1024) 263168 activation_41[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_45 (BatchNo (None, 4, 4, 1024) 4096 conv2d_46[0][0] \n__________________________________________________________________________________________________\nadd_13 (Add) (None, 4, 4, 1024) 0 batch_normalization_45[0][0] \n activation_39[0][0] \n__________________________________________________________________________________________________\nactivation_42 (Activation) (None, 4, 4, 1024) 0 add_13[0][0] \n__________________________________________________________________________________________________\nconv2d_47 (Conv2D) (None, 4, 4, 256) 262400 activation_42[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_46 (BatchNo (None, 4, 4, 256) 1024 conv2d_47[0][0] \n__________________________________________________________________________________________________\nactivation_43 (Activation) (None, 4, 4, 256) 0 batch_normalization_46[0][0] \n__________________________________________________________________________________________________\nconv2d_48 (Conv2D) (None, 4, 4, 256) 590080 activation_43[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_47 (BatchNo (None, 4, 4, 256) 1024 conv2d_48[0][0] \n__________________________________________________________________________________________________\nactivation_44 (Activation) (None, 4, 4, 256) 0 batch_normalization_47[0][0] \n__________________________________________________________________________________________________\nconv2d_49 (Conv2D) (None, 4, 4, 1024) 263168 activation_44[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_48 (BatchNo (None, 4, 4, 1024) 4096 conv2d_49[0][0] \n__________________________________________________________________________________________________\nadd_14 (Add) (None, 4, 4, 1024) 0 batch_normalization_48[0][0] \n activation_42[0][0] \n__________________________________________________________________________________________________\nactivation_45 (Activation) (None, 4, 4, 1024) 0 add_14[0][0] \n__________________________________________________________________________________________________\nconv2d_50 (Conv2D) (None, 2, 2, 512) 524800 activation_45[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_49 (BatchNo (None, 2, 2, 512) 2048 conv2d_50[0][0] \n__________________________________________________________________________________________________\nactivation_46 (Activation) (None, 2, 2, 512) 0 batch_normalization_49[0][0] \n__________________________________________________________________________________________________\nconv2d_51 (Conv2D) (None, 2, 2, 512) 2359808 activation_46[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_50 (BatchNo (None, 2, 2, 512) 2048 conv2d_51[0][0] \n__________________________________________________________________________________________________\nactivation_47 (Activation) (None, 2, 2, 512) 0 batch_normalization_50[0][0] \n__________________________________________________________________________________________________\nconv2d_52 (Conv2D) (None, 2, 2, 2048) 1050624 activation_47[0][0] \n__________________________________________________________________________________________________\nconv2d_53 (Conv2D) (None, 2, 2, 2048) 2099200 activation_45[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_51 (BatchNo (None, 2, 2, 2048) 8192 conv2d_52[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_52 (BatchNo (None, 2, 2, 2048) 8192 conv2d_53[0][0] \n__________________________________________________________________________________________________\nadd_15 (Add) (None, 2, 2, 2048) 0 batch_normalization_51[0][0] \n batch_normalization_52[0][0] \n__________________________________________________________________________________________________\nactivation_48 (Activation) (None, 2, 2, 2048) 0 add_15[0][0] \n__________________________________________________________________________________________________\nconv2d_54 (Conv2D) (None, 2, 2, 512) 1049088 activation_48[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_53 (BatchNo (None, 2, 2, 512) 2048 conv2d_54[0][0] \n__________________________________________________________________________________________________\nactivation_49 (Activation) (None, 2, 2, 512) 0 batch_normalization_53[0][0] \n__________________________________________________________________________________________________\nconv2d_55 (Conv2D) (None, 2, 2, 512) 2359808 activation_49[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_54 (BatchNo (None, 2, 2, 512) 2048 conv2d_55[0][0] \n__________________________________________________________________________________________________\nactivation_50 (Activation) (None, 2, 2, 512) 0 batch_normalization_54[0][0] \n__________________________________________________________________________________________________\nconv2d_56 (Conv2D) (None, 2, 2, 2048) 1050624 activation_50[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_55 (BatchNo (None, 2, 2, 2048) 8192 conv2d_56[0][0] \n__________________________________________________________________________________________________\nadd_16 (Add) (None, 2, 2, 2048) 0 batch_normalization_55[0][0] \n activation_48[0][0] \n__________________________________________________________________________________________________\nactivation_51 (Activation) (None, 2, 2, 2048) 0 add_16[0][0] \n__________________________________________________________________________________________________\nconv2d_57 (Conv2D) (None, 2, 2, 512) 1049088 activation_51[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_56 (BatchNo (None, 2, 2, 512) 2048 conv2d_57[0][0] \n__________________________________________________________________________________________________\nactivation_52 (Activation) (None, 2, 2, 512) 0 batch_normalization_56[0][0] \n__________________________________________________________________________________________________\nconv2d_58 (Conv2D) (None, 2, 2, 512) 2359808 activation_52[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_57 (BatchNo (None, 2, 2, 512) 2048 conv2d_58[0][0] \n__________________________________________________________________________________________________\nactivation_53 (Activation) (None, 2, 2, 512) 0 batch_normalization_57[0][0] \n__________________________________________________________________________________________________\nconv2d_59 (Conv2D) (None, 2, 2, 2048) 1050624 activation_53[0][0] \n__________________________________________________________________________________________________\nbatch_normalization_58 (BatchNo (None, 2, 2, 2048) 8192 conv2d_59[0][0] \n__________________________________________________________________________________________________\nadd_17 (Add) (None, 2, 2, 2048) 0 batch_normalization_58[0][0] \n activation_51[0][0] \n__________________________________________________________________________________________________\nactivation_54 (Activation) (None, 2, 2, 2048) 0 add_17[0][0] \n__________________________________________________________________________________________________\naverage_pooling2d (AveragePooli (None, 1, 1, 2048) 0 activation_54[0][0] \n__________________________________________________________________________________________________\nflatten (Flatten) (None, 2048) 0 average_pooling2d[0][0] \n__________________________________________________________________________________________________\nfc6 (Dense) (None, 6) 12294 flatten[0][0] \n==================================================================================================\nTotal params: 23,600,006\nTrainable params: 23,546,886\nNon-trainable params: 53,120\n__________________________________________________________________________________________________\n"
]
],
[
[
"<a name='6'></a> \n## 6 - Bibliography\n\nThis notebook presents the ResNet algorithm from He et al. (2015). The implementation here also took significant inspiration and follows the structure given in the GitHub repository of Francois Chollet: \n\n- Kaiming He, Xiangyu Zhang, Shaoqing Ren, Jian Sun - [Deep Residual Learning for Image Recognition (2015)](https://arxiv.org/abs/1512.03385)\n- Francois Chollet's GitHub repository: https://github.com/fchollet/deep-learning-models/blob/master/resnet50.py\n",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown",
"code",
"markdown"
]
| [
[
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code",
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown",
"markdown"
],
[
"code",
"code"
],
[
"markdown",
"markdown",
"markdown",
"markdown"
],
[
"code"
],
[
"markdown",
"markdown"
],
[
"code"
],
[
"markdown"
]
]
|
ec740550198e2c632e6085076325c1cb8e4929cb | 62,333 | ipynb | Jupyter Notebook | UGESCO_MAIN_SIMPLIFIED.ipynb | DalavanCloud/UGESCO | 03f037b8c0e48b7a2fbefdac570d12241deb64e7 | [
"MIT"
]
| 1 | 2018-09-23T14:43:40.000Z | 2018-09-23T14:43:40.000Z | UGESCO_MAIN_SIMPLIFIED.ipynb | DalavanCloud/UGESCO | 03f037b8c0e48b7a2fbefdac570d12241deb64e7 | [
"MIT"
]
| null | null | null | UGESCO_MAIN_SIMPLIFIED.ipynb | DalavanCloud/UGESCO | 03f037b8c0e48b7a2fbefdac570d12241deb64e7 | [
"MIT"
]
| null | null | null | 42.723098 | 134 | 0.452938 | [
[
[
"# Ugesco : enrich and desambiguate the Samnang-Hans' JSON",
"_____no_output_____"
]
],
[
[
"from ugesco import *\nimport warnings\nwarnings.filterwarnings('ignore') #évite les alertes lors de l'import d'Ugesco.py\n",
"_____no_output_____"
],
[
"file = \"data/jsondata_ugesco.json\"\ndata = json_to_df(file)",
"_____no_output_____"
],
[
"# exclude image classifications and temporal values for the moment\ndata = data[data.columns.drop(list(data.filter(regex='imageclassification|temporal')))]",
"_____no_output_____"
],
[
"# reshape the dataframe verticaly\ndata = data.set_index('beeldid')\ndata.columns = data.columns.str.split('_', expand=True)\ndata = data.stack().reset_index(level=1, drop=True).reset_index()\ndata.columns = ['beeldid', 'spatial_value', 'spatial_key', 'to_drop']\ndata.drop('to_drop', 1, inplace=True)",
"_____no_output_____"
],
[
"#join/merge with phototheque_pallas to get the locations. \n#The csv is gzipped to get around the 100MB file weigth limitation of Github\nphototeque = pd.read_csv(\"data/phototheque_pallas.csv.gz\", compression=\"gzip\", encoding=\"utf8\", dtype={'beeldid': str})\n\ndata = pd.merge(data, phototeque, how='left', on=['beeldid'], suffixes=['', '_x'])",
"_____no_output_____"
],
[
"# Pictures that contains more than a loc in the thesaurus descriptors\n\ndata[data['loc_qid'].str.contains(\",\", na = False)]",
"_____no_output_____"
],
[
"#test Rosette API. Max 10 000 calls a month and 1000 a day\n \n#data['rosette'] = data['LEGEND'].apply(rosette)",
"_____no_output_____"
],
[
"# Some descriptive stats on columns\n#data.describe(include=\"all\")",
"_____no_output_____"
],
[
"# More data profiling\n#import pandas_profiling\n#pandas_profiling.ProfileReport(data)",
"_____no_output_____"
],
[
"#data.to_csv(\"C:/Users/ettor/Desktop/ugesco_file_temp_simplified.csv\", encoding=\"utf-8\")",
"_____no_output_____"
],
[
"# Merge with NER_classes (spatial_keys matched with wikidata)\nner_classes = pd.read_csv(\"data/ner_classes.csv\")\n\ndata = pd.merge(data, ner_classes, how='left', left_on = ['spatial_key'], right_on = ['ner_class'])",
"_____no_output_____"
],
[
"# rename the new columns merged\ndata.rename(columns={'wiki_qid': 'ner_class_qid', 'wiki_class':'ner_class_name'}, inplace=True)",
"_____no_output_____"
],
[
"# remove articles in spatial_value\ndata['spatial_value'] = data['spatial_value'].str.replace(pat=r\"^\\s?(le|la|l\\s+'|l'|les)\\s+\", repl=\"\", n=1, case=False)",
"_____no_output_____"
],
[
"data.head()",
"_____no_output_____"
],
[
"data.shape",
"_____no_output_____"
],
[
"data.to_csv(r\"C:\\Users\\ettor\\Desktop\\data_ugesco.csv\", header=True, index=False, encoding=\"utf-8\")",
"_____no_output_____"
],
[
"#test de stanford ner (lent)\n#test = data.LEGEND[0:100].apply(get_ner)",
"_____no_output_____"
],
[
"#data.LEGEND[0:100]",
"_____no_output_____"
],
[
"#test_stanford = pd.concat([data.LEGEND[0:100], test], axis=1).reset_index()",
"_____no_output_____"
],
[
"#test_stanford.to_csv(\"C:/Users/ettor/Desktop/test_stanford.csv\")",
"_____no_output_____"
]
],
[
[
"#apply the algorithm\nfor index, rows in data.iterrows():\n dic = {}\n dic['index'] = index\n dic['beeldid'] = rows.beeldid\n if (\",\" not in str(rows.loc_wikiname) \n and str(rows.loc_wikiname) != \"nan\"\n and isQidACity(rows.loc_qid)): \n dic['city_thes_name'] = rows.loc_wikiname\n dic['city_qid_name'] = rows.loc_qid\n #dic['country_name'] = rows.country_name\n #dic['country_qid'] = rows.country_qid\n #elif (rows.spatial_key_0 == \"LOC_PLACE\" \n #and isStringACity(rows.spatial_value_0)):\n #dic['city'] = rows.spatial_value_0\n print(dic)\n\n \n ",
"_____no_output_____"
]
]
]
| [
"markdown",
"code",
"markdown"
]
| [
[
"markdown"
],
[
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code",
"code"
],
[
"markdown"
]
]
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.